prompt
stringlengths 19
1.03M
| completion
stringlengths 4
2.12k
| api
stringlengths 8
90
|
|---|---|---|
########################## Fuzzy Discernibility Matrix: Reduct ###############################
####################### Dr. <NAME> 25-01-21, version: 1.0 ###########################
import warnings
warnings.filterwarnings('ignore')
import pandas as pd
from sklearn import preprocessing
from sklearn.preprocessing import KBinsDiscretizer
import numpy as np
def gaussmf(x, mean, sigma):
"""
Gaussian fuzzy membership function.
Parameters
----------
x : 1d array or iterable
Independent variable.
mean : float
Gaussian parameter for center (mean) value.
sigma : float
Gaussian parameter for standard deviation.
Returns
-------
y : 1d array
Gaussian membership function for x
"""
eps=1e-16
return np.exp(-((x - mean) ** 2.) / (float(sigma) ** 2.+eps))
def fuzzy_distance10(x,y):
eps = 1e-16
d = 1-(sum(min(x,y))/(sum(min(x,y))+eps))
return d
def getthefuzzyvalues(F,p,t,cls):
temp=[]
for i in range(cls):
temp.append(F[p][((t)*cls)+i])
return temp
def my_fuzzy_discrn_mat(df,best):
r,c=df.shape
dff=df.copy()
if best<c:
oldf=df.columns #check if actual column names are in 0 to ...
newf=list(range(c))
df.rename(columns=dict(zip(oldf, newf)), inplace=True)
dc=list(df[newf[-1]])
#print('\nAfter changing the column names to 0 to ...\n',df)
# creating instance of labelencoder
labelencoder = preprocessing.LabelEncoder()
labelencoder.fit(df[newf[-1]])
#print(list(labelencoder.classes_))
# Assigning numerical values and storing in another column
df[newf[-1]] = labelencoder.fit_transform(df[newf[-1]])
#df
datasets = {}
by_class = df.groupby(newf[-1])
for groups, data in by_class:
datasets[groups] = data
#len(datasets)
meand=[]
stdd=[]
for i in range(len(datasets)):
#print(datasets[i])
meand.append(list(datasets[i].mean(axis = 0)))
#print('mean',meand)
stdd.append(list(datasets[i].std(axis = 0)))
#print('std',stdd)
X=df.to_numpy()
#r,c=X.shape
#oldf=df.columns
#newf=list(range(c))
#df.rename(columns=dict(zip(oldf, newf)), inplace=True) #renaming column
D=list(df[newf[-1]])
labelencoder.fit(df[newf[-1]])
#list(labelencoder.classes_)
classes=len(list(labelencoder.classes_))
FD=np.zeros((r,(c-1)*classes))
'''for i in range(len(datasets)):
oldf=datasets[i].columns
newf=list(range(c))
datasets[i].rename(columns=dict(zip(oldf, newf)), inplace=True) #renaming column'''
#print(datasets[i])
for j in range(c-1):
for i in range(r):
l=(j*classes)
for k in range(classes):
g=gaussmf(X[i][j], np.array(meand[k][j]), np.array(stdd[k][j]))
#print(g)
FD[i][l]=g #float(str(round(g, 8)))
l = l+1
D=np.array(D)
D=D.reshape(D.shape[0],1)
F=np.concatenate((FD, D), axis=1)
# Create Fuzzyfied DataFrame
fdf = | pd.DataFrame(F) | pandas.DataFrame |
#!/usr/bin/env python
import json
import pandas
import os
series_description_map = {
'TORAX AP': 'AP',
'PORTATIL': 'AP',
'CHEST': 'UNK',
'W034 TÓRAX LAT.': 'LAT',
'AP HORIZONTAL': 'AP SUPINE',
'TÓRAX PA H': 'PA',
'BUCKY PA': 'PA',
'ESCAPULA Y': 'UNK',
'LATERAL IZQ.': 'LAT',
'TORAX SUPINE AP': 'AP SUPINE',
'DIRECTO AP': 'AP',
'T034 TÓRAX LAT': 'LAT',
'PECHO AP': 'AP',
'TORAX AP DIRECTO': 'AP',
'W034 TÓRAX LAT. *': 'LAT',
'TÓRAX LAT': 'LAT',
'ERECT LAT': 'LAT',
'TORAX LAT': 'LAT',
'TÓRAX AP H': 'AP SUPINE',
'TÒRAX AP': 'AP',
'TORAX PORTATIL': 'AP',
'DEC. SUPINO AP': 'AP SUPINE',
'SUPINE AP': 'AP SUPINE',
'TÓRAX': 'UNK',
'RX TORAX CON PORTATIL': 'AP',
'TORAX PA': 'PA',
'TORAX ERECT PA': 'PA',
'DIRECTO PA': 'PA',
'RX TORAX CON PORTATIL PED': 'AP',
'LATERAL': 'LAT',
'TORAX BIPE PA': 'PA',
'SUP.AP PORTABLE': 'AP SUPINE',
'TORAX CAMILLA': 'AP',
'TORAX-ABD PA': 'PA',
'TORAX SEDE AP': 'AP',
'BUCKY LAT': 'LAT',
'ERECT PA': 'PA',
'TORAX SUPINO AP': 'AP SUPINE',
'W033 TÓRAX AP': 'AP',
'PORTÁTIL AP': 'AP',
'TORAX ERECT LAT': 'LAT',
'PA': 'PA',
'W033 TÓRAX PA *': 'PA',
'TÓRAX PA': 'PA',
'TòRAX AP': 'PA',
'RX TORAX PA Y LAT': 'UNK',
'AP': 'AP',
'T035 TÓRAX PA': 'PA',
'RX TORAX, PA O AP': 'UNK',
'W033 TÓRAX PA': 'PA',
'TORAX PA': 'PA'}
ENFORCE_LATERAL = [
"bimcv+/sub-S04079/ses-E08254/mod-rx/sub-S04079_ses-E08254_acq-2_run-1_bp-chest_vp-pa_dx.png",
"bimcv+/sub-S03582/ses-E07281/mod-rx/sub-S03582_ses-E07281_run-1_bp-chest_vp-ap_cr.png",
"bimcv+/sub-S03585/ses-E07287/mod-rx/sub-S03585_ses-E07287_acq-2_run-1_bp-chest_vp-pa_dx.png",
"bimcv+/sub-S03585/ses-E07911/mod-rx/sub-S03585_ses-E07911_acq-2_run-1_bp-chest_vp-pa_dx.png",
"bimcv+/sub-S03996/ses-E08157/mod-rx/sub-S03996_ses-E08157_run-1_bp-chest_vp-ap_cr.png",
"bimcv+/sub-S04334/ses-E08628/mod-rx/sub-S04334_ses-E08628_acq-2_run-1_bp-chest_vp-pa_cr.png",
"bimcv+/sub-S04489/ses-E08918/mod-rx/sub-S04489_ses-E08918_acq-2_run-1_bp-chest_vp-pa_dx.png",
"bimcv+/sub-S04224/ses-E08468/mod-rx/sub-S04224_ses-E08468_acq-2_run-1_bp-chest_vp-pa_dx.png",
"bimcv+/sub-S04084/ses-E08261/mod-rx/sub-S04084_ses-E08261_acq-2_run-1_bp-chest_vp-pa_cr.png",
"bimcv+/sub-S03898/ses-E07949/mod-rx/sub-S03898_ses-E07949_acq-2_run-1_bp-chest_vp-pa_dx.png",
"bimcv+/sub-S04190/ses-E08422/mod-rx/sub-S04190_ses-E08422_acq-2_run-1_bp-chest_vp-pa_dx.png",
"bimcv+/sub-S04195/ses-E08429/mod-rx/sub-S04195_ses-E08429_acq-2_run-1_bp-chest_vp-pa_dx.png",
"bimcv+/sub-S04075/ses-E08435/mod-rx/sub-S04075_ses-E08435_acq-2_run-1_bp-chest_vp-pa_dx.png",
"bimcv+/sub-S04075/ses-E08250/mod-rx/sub-S04075_ses-E08250_acq-2_run-1_bp-chest_vp-pa_dx.png",
"bimcv+/sub-S04484/ses-E08905/mod-rx/sub-S04484_ses-E08905_run-1_bp-chest_vp-ap_cr.png",
"bimcv+/sub-S03736/ses-E07772/mod-rx/sub-S03736_ses-E07772_acq-2_run-1_bp-chest_vp-pa_dx.png",
"bimcv+/sub-S04021/ses-E08189/mod-rx/sub-S04021_ses-E08189_acq-2_run-1_bp-chest_vp-pa_dx.png",
"bimcv+/sub-S04275/ses-E08532/mod-rx/sub-S04275_ses-E08532_acq-2_run-1_bp-chest_vp-pa_dx.png",
"bimcv+/sub-S03939/ses-E08092/mod-rx/sub-S03939_ses-E08092_acq-2_run-1_bp-chest_vp-pa_dx.png",
"bimcv+/sub-S04298/ses-E08566/mod-rx/sub-S04298_ses-E08566_acq-2_run-1_bp-chest_vp-pa_cr.png",
"bimcv+/sub-S04101/ses-E08526/mod-rx/sub-S04101_ses-E08526_acq-2_run-1_bp-chest_vp-pa_dx.png",
"bimcv+/sub-S04101/ses-E08453/mod-rx/sub-S04101_ses-E08453_acq-2_run-1_bp-chest_vp-pa_dx.png",
"bimcv+/sub-S03699/ses-E07505/mod-rx/sub-S03699_ses-E07505_acq-2_run-1_bp-chest_vp-pa_dx.png",
"bimcv+/sub-S04203/ses-E08442/mod-rx/sub-S04203_ses-E08442_acq-2_run-1_bp-chest_vp-pa_dx.png",
"bimcv+/sub-S04316/ses-E08597/mod-rx/sub-S04316_ses-E08597_acq-2_run-1_bp-chest_vp-pa_dx.png",
"bimcv+/sub-S03610/ses-E07319/mod-rx/sub-S03610_ses-E07319_acq-2_run-1_bp-chest_vp-pa_dx.png",
"bimcv+/sub-S04402/ses-E08747/mod-rx/sub-S04402_ses-E08747_acq-2_run-1_bp-chest_vp-pa_dx.png",
"bimcv+/sub-S03563/ses-E07251/mod-rx/sub-S03563_ses-E07251_acq-2_run-1_bp-chest_vp-pa_dx.png",
"bimcv+/sub-S03936/ses-E08089/mod-rx/sub-S03936_ses-E08089_acq-2_run-1_bp-chest_vp-pa_dx.png",
"bimcv+/sub-S03931/ses-E08648/mod-rx/sub-S03931_ses-E08648_acq-2_run-1_bp-chest_vp-pa_dx.png"
]
def main():
datapath = 'bimcv+'
patientdf = pandas.read_csv(os.path.join(datapath, 'participants.tsv'),
sep='\t')
data = {}
series_descriptions = set()
idx = -1
for _, row in patientdf.iterrows():
subject = row.participant
modalities = row.modality_dicom
modalities = eval(modalities)
if 'CR' in modalities or 'DX' in modalities:
contents = os.listdir(os.path.join(datapath, subject))
for sessionfile in contents:
if os.path.isdir(os.path.join(datapath, subject, sessionfile)):
image_candidates = os.listdir(os.path.join(datapath, subject, sessionfile, 'mod-rx'))
for i in image_candidates:
if i.lower().endswith('.png'):
idx += 1
entry = {}
path = os.path.join(datapath, subject, sessionfile, 'mod-rx', i)
entry['path'] = path
entry['participant'] = subject
jsonpath = path[:-4] + '.json'
try:
with open(jsonpath, 'r') as handle:
metadata = json.load(handle)
except OSError:
entry['projection'] = 'UNK'
data[idx] = entry
break
entry['modality'] = metadata['00080060']['Value'][0]
entry['manufacturer'] = metadata['00080070']['Value'][0]
entry['sex'] = metadata['00100040']['Value'][0]
try:
photometric_interpretation = metadata['00280004']['Value'][0]
entry['photometric_interpretation'] = photometric_interpretation
except KeyError:
print('no photometric_interpretation for: ', path)
try:
entry['rotation'] = metadata['00181140']['Value'][0]
print(entry['rotation'])
except KeyError:
pass
try:
entry['lut'] = metadata['00283010']['Value'][0]['00283006']['Value']
entry['lut_min'] = metadata['00283010']['Value'][0]['00283002']['Value'][1]
try:
entry['rescale_slope'] = metadata['00281053']['Value'][0]
entry['rescale_intercept'] = metadata['00281052']['Value'][0]
except KeyError:
pass
try:
entry['bits_stored'] = metadata['00280101']['Value'][0]
except KeyError:
try:
entry['bits_stored'] = metadata['00283010']['Value'][0]['00283002']['Value'][2]
except KeyError:
pass
except KeyError:
try:
entry['window_center'] = metadata['00281050']['Value'][0]
entry['window_width'] = metadata['00281051']['Value'][0]
except KeyError:
print("No window information for : ", path)
try:
entry['study_date'] = int(metadata['00080020']['Value'][0])
except KeyError:
pass
try:
entry['study_time'] = float(metadata['00080030']['Value'][0])
except KeyError:
pass
try:
entry['age'] = int(metadata['00101010']['Value'][0][:-1])
except KeyError:
pass
try:
series_description = metadata['0008103E']['Value'][0]
except Exception as e:
try:
series_description = metadata['00081032']['Value'][0]['00080104']['Value'][0]
except Exception as e:
raise e
series_description = series_description.upper()
series_descriptions.add(series_description)
projection = series_description_map[series_description]
entry['projection'] = projection
# these images are manually set to lateral
if path.strip() in ENFORCE_LATERAL:
print("enforcing lateral projection for {:s}".format(path))
entry['projection'] = 'LAT'
data[idx] = entry
df = | pandas.DataFrame.from_dict(data, orient='index') | pandas.DataFrame.from_dict |
import torch, sys, math, pickle, datetime
import numpy as np
import numpy.random as npr
from collections import OrderedDict
plot_path = './'
use_cuda = torch.cuda.is_available()
npr.seed(1234)
if use_cuda :
torch.set_default_tensor_type('torch.cuda.DoubleTensor')
torch.cuda.manual_seed(1234)
else :
torch.set_default_tensor_type('torch.DoubleTensor')
torch.manual_seed(1234)
### Load carat-data
fname_dpvi = '2019-03-26'
import pandas as pd
ds = [8,16,32,64,96]
dpvi_err = []
dpvi_times = []
learn = 0
if learn:
sys.path.append('../../dpvi/')
from sampler import fast_sample
for d in ds:
app_data = pd.read_csv('../../data/subsets/carat_apps_sub{}.dat'.format(d), sep=' ', header=None)\
.astype('float').values
N = len(app_data)
models = pickle.load(open('../../dpvi/models_{0}/models_{0}_{1}.p'.format(fname_dpvi, d), 'rb'))
for model in models:
syn_app_data = fast_sample(model, N)
syn_cov = np.cov(syn_app_data.T)
orig_cov = np.cov(app_data.T)
dpvi_err.append(np.linalg.norm(orig_cov-syn_cov))
log = open('logs_{0}/out_file_{0}_{1}.txt'.format(fname_dpvi, d), 'r')
wall_time, cpu_time = log.readlines()[-2:]
log.close()
wall_time = float(wall_time.strip('Wall time').strip('\n'))
cpu_time = float(cpu_time.strip('CPU time').strip('\n'))
dpvi_times.append((wall_time, cpu_time))
pd.DataFrame(dpvi_err).to_csv('../plot_data/dpvi_cov_err_(8,16,32,64,96)_{}.csv'\
.format(fname_dpvi), sep=';', header=None, index=False)
| pd.DataFrame(dpvi_times) | pandas.DataFrame |
import psycopg2
import psycopg2
import sqlalchemy as salc
import numpy as np
import warnings
import datetime
import pandas as pd
import json
from math import pi
from flask import request, send_file, Response
# import visualization libraries
from bokeh.io import export_png
from bokeh.embed import json_item
from bokeh.plotting import figure
from bokeh.models import Label, LabelSet, ColumnDataSource, Legend
from bokeh.palettes import Colorblind
from bokeh.layouts import gridplot
from bokeh.transform import cumsum
warnings.filterwarnings('ignore')
def create_routes(server):
def quarters(month, year):
if 1 <= month <= 3:
return '01' + '/' + year
elif 4 <= month <= 6:
return '04' + '/' + year
elif 5 <= month <= 9:
return '07' + '/' + year
elif 10 <= month <= 12:
return '10' + '/' + year
def new_contributor_data_collection(repo_id, required_contributions):
rank_list = []
for num in range(1, required_contributions + 1):
rank_list.append(num)
rank_tuple = tuple(rank_list)
contributor_query = salc.sql.text(f"""
SELECT * FROM (
SELECT ID AS
cntrb_id,
A.created_at AS created_at,
date_part('month', A.created_at::DATE) AS month,
date_part('year', A.created_at::DATE) AS year,
A.repo_id,
repo_name,
full_name,
login,
ACTION,
rank() OVER (
PARTITION BY id
ORDER BY A.created_at ASC
)
FROM
(
(
SELECT
canonical_id AS ID,
created_at AS created_at,
repo_id,
'issue_opened' AS ACTION,
contributors.cntrb_full_name AS full_name,
contributors.cntrb_login AS login
FROM
augur_data.issues
LEFT OUTER JOIN augur_data.contributors ON contributors.cntrb_id = issues.reporter_id
LEFT OUTER JOIN (
SELECT DISTINCT ON ( cntrb_canonical ) cntrb_full_name,
cntrb_canonical AS canonical_email,
data_collection_date,
cntrb_id AS canonical_id
FROM augur_data.contributors
WHERE cntrb_canonical = cntrb_email ORDER BY cntrb_canonical
) canonical_full_names ON canonical_full_names.canonical_email =contributors.cntrb_canonical
WHERE
repo_id = {repo_id}
AND pull_request IS NULL
GROUP BY
canonical_id,
repo_id,
issues.created_at,
contributors.cntrb_full_name,
contributors.cntrb_login
) UNION ALL
(
SELECT
canonical_id AS ID,
TO_TIMESTAMP( cmt_author_date, 'YYYY-MM-DD' ) AS created_at,
repo_id,
'commit' AS ACTION,
contributors.cntrb_full_name AS full_name,
contributors.cntrb_login AS login
FROM
augur_data.commits
LEFT OUTER JOIN augur_data.contributors ON cntrb_email = cmt_author_email
LEFT OUTER JOIN (
SELECT DISTINCT ON ( cntrb_canonical ) cntrb_full_name,
cntrb_canonical AS canonical_email,
data_collection_date, cntrb_id AS canonical_id
FROM augur_data.contributors
WHERE cntrb_canonical = cntrb_email ORDER BY cntrb_canonical
) canonical_full_names ON canonical_full_names.canonical_email =contributors.cntrb_canonical
WHERE
repo_id = {repo_id}
GROUP BY
repo_id,
canonical_email,
canonical_id,
commits.cmt_author_date,
contributors.cntrb_full_name,
contributors.cntrb_login
) UNION ALL
(
SELECT
message.cntrb_id AS ID,
created_at AS created_at,
commits.repo_id,
'commit_comment' AS ACTION,
contributors.cntrb_full_name AS full_name,
contributors.cntrb_login AS login
FROM
augur_data.commit_comment_ref,
augur_data.commits,
augur_data.message
LEFT OUTER JOIN augur_data.contributors ON contributors.cntrb_id = message.cntrb_id
LEFT OUTER JOIN (
SELECT DISTINCT ON ( cntrb_canonical ) cntrb_full_name,
cntrb_canonical AS canonical_email,
data_collection_date, cntrb_id AS canonical_id
FROM augur_data.contributors
WHERE cntrb_canonical = cntrb_email ORDER BY cntrb_canonical
) canonical_full_names ON canonical_full_names.canonical_email =contributors.cntrb_canonical
WHERE
commits.cmt_id = commit_comment_ref.cmt_id
AND commits.repo_id = {repo_id}
AND commit_comment_ref.msg_id = message.msg_id
GROUP BY
ID,
commits.repo_id,
commit_comment_ref.created_at,
contributors.cntrb_full_name,
contributors.cntrb_login
) UNION ALL
(
SELECT
issue_events.cntrb_id AS ID,
issue_events.created_at AS created_at,
issues.repo_id,
'issue_closed' AS ACTION,
contributors.cntrb_full_name AS full_name,
contributors.cntrb_login AS login
FROM
augur_data.issues,
augur_data.issue_events
LEFT OUTER JOIN augur_data.contributors ON contributors.cntrb_id = issue_events.cntrb_id
LEFT OUTER JOIN (
SELECT DISTINCT ON ( cntrb_canonical ) cntrb_full_name,
cntrb_canonical AS canonical_email,
data_collection_date,
cntrb_id AS canonical_id
FROM augur_data.contributors
WHERE cntrb_canonical = cntrb_email ORDER BY cntrb_canonical
) canonical_full_names ON canonical_full_names.canonical_email =contributors.cntrb_canonical
WHERE
issues.repo_id = {repo_id}
AND issues.issue_id = issue_events.issue_id
AND issues.pull_request IS NULL
AND issue_events.cntrb_id IS NOT NULL
AND ACTION = 'closed'
GROUP BY
issue_events.cntrb_id,
issues.repo_id,
issue_events.created_at,
contributors.cntrb_full_name,
contributors.cntrb_login
) UNION ALL
(
SELECT
pr_augur_contributor_id AS ID,
pr_created_at AS created_at,
pull_requests.repo_id,
'open_pull_request' AS ACTION,
contributors.cntrb_full_name AS full_name,
contributors.cntrb_login AS login
FROM
augur_data.pull_requests
LEFT OUTER JOIN augur_data.contributors ON pull_requests.pr_augur_contributor_id = contributors.cntrb_id
LEFT OUTER JOIN (
SELECT DISTINCT ON ( cntrb_canonical ) cntrb_full_name,
cntrb_canonical AS canonical_email,
data_collection_date,
cntrb_id AS canonical_id
FROM augur_data.contributors
WHERE cntrb_canonical = cntrb_email ORDER BY cntrb_canonical
) canonical_full_names ON canonical_full_names.canonical_email =contributors.cntrb_canonical
WHERE
pull_requests.repo_id = {repo_id}
GROUP BY
pull_requests.pr_augur_contributor_id,
pull_requests.repo_id,
pull_requests.pr_created_at,
contributors.cntrb_full_name,
contributors.cntrb_login
) UNION ALL
(
SELECT
message.cntrb_id AS ID,
msg_timestamp AS created_at,
pull_requests.repo_id as repo_id,
'pull_request_comment' AS ACTION,
contributors.cntrb_full_name AS full_name,
contributors.cntrb_login AS login
FROM
augur_data.pull_requests,
augur_data.pull_request_message_ref,
augur_data.message
LEFT OUTER JOIN augur_data.contributors ON contributors.cntrb_id = message.cntrb_id
LEFT OUTER JOIN (
SELECT DISTINCT ON ( cntrb_canonical ) cntrb_full_name,
cntrb_canonical AS canonical_email,
data_collection_date,
cntrb_id AS canonical_id
FROM augur_data.contributors
WHERE cntrb_canonical = cntrb_email ORDER BY cntrb_canonical
) canonical_full_names ON canonical_full_names.canonical_email =contributors.cntrb_canonical
WHERE
pull_requests.repo_id = {repo_id}
AND pull_request_message_ref.pull_request_id = pull_requests.pull_request_id
AND pull_request_message_ref.msg_id = message.msg_id
GROUP BY
message.cntrb_id,
pull_requests.repo_id,
message.msg_timestamp,
contributors.cntrb_full_name,
contributors.cntrb_login
) UNION ALL
(
SELECT
issues.reporter_id AS ID,
msg_timestamp AS created_at,
issues.repo_id as repo_id,
'issue_comment' AS ACTION,
contributors.cntrb_full_name AS full_name,
contributors.cntrb_login AS login
FROM
issues,
issue_message_ref,
message
LEFT OUTER JOIN augur_data.contributors ON contributors.cntrb_id = message.cntrb_id
LEFT OUTER JOIN (
SELECT DISTINCT ON ( cntrb_canonical ) cntrb_full_name,
cntrb_canonical AS canonical_email,
data_collection_date,
cntrb_id AS canonical_id
FROM augur_data.contributors
WHERE cntrb_canonical = cntrb_email ORDER BY cntrb_canonical
) canonical_full_names ON canonical_full_names.canonical_email =contributors.cntrb_canonical
WHERE
issues.repo_id = {repo_id}
AND issue_message_ref.msg_id = message.msg_id
AND issues.issue_id = issue_message_ref.issue_id
AND issues.pull_request_id = NULL
GROUP BY
issues.reporter_id,
issues.repo_id,
message.msg_timestamp,
contributors.cntrb_full_name,
contributors.cntrb_login
)
) A,
repo
WHERE
ID IS NOT NULL
AND A.repo_id = repo.repo_id
GROUP BY
A.ID,
A.repo_id,
A.ACTION,
A.created_at,
repo.repo_name,
A.full_name,
A.login
ORDER BY
cntrb_id
) b
WHERE RANK IN {rank_tuple}
""")
df = pd.read_sql(contributor_query, server.augur_app.database)
df = df.loc[~df['full_name'].str.contains('bot', na=False)]
df = df.loc[~df['login'].str.contains('bot', na=False)]
df = df.loc[~df['cntrb_id'].isin(df[df.duplicated(['cntrb_id', 'created_at', 'repo_id', 'rank'])]['cntrb_id'])]
# add yearmonths to contributor
df[['month', 'year']] = df[['month', 'year']].astype(int).astype(str)
df['yearmonth'] = df['month'] + '/' + df['year']
df['yearmonth'] = pd.to_datetime(df['yearmonth'])
# add column with every value being one, so when the contributor df is concatenated
# with the months df, the filler months won't be counted in the sums
df['new_contributors'] = 1
# add quarters to contributor dataframe
df['month'] = df['month'].astype(int)
df['quarter'] = df.apply(lambda x: quarters(x['month'], x['year']), axis=1, result_type='reduce')
df['quarter'] = pd.to_datetime(df['quarter'])
return df
def months_data_collection(start_date, end_date):
# months_query makes a df of years and months, this is used to fill
# the months with no data in the visualizations
months_query = salc.sql.text(f"""
SELECT *
FROM
(
SELECT
date_part( 'year', created_month :: DATE ) AS year,
date_part( 'month', created_month :: DATE ) AS MONTH
FROM
(SELECT *
FROM (
SELECT created_month :: DATE
FROM generate_series (TIMESTAMP '{start_date}', TIMESTAMP '{end_date}', INTERVAL '1 month' ) created_month ) d ) x
) y
""")
months_df = pd.read_sql(months_query, server.augur_app.database)
# add yearmonths to months_df
months_df[['year', 'month']] = months_df[['year', 'month']].astype(float).astype(int).astype(str)
months_df['yearmonth'] = months_df['month'] + '/' + months_df['year']
months_df['yearmonth'] = pd.to_datetime(months_df['yearmonth'])
# filter months_df with start_date and end_date, the contributor df is filtered in the visualizations
months_df = months_df.set_index(months_df['yearmonth'])
months_df = months_df.loc[start_date: end_date].reset_index(drop=True)
# add quarters to months dataframe
months_df['month'] = months_df['month'].astype(int)
months_df['quarter'] = months_df.apply(lambda x: quarters(x['month'], x['year']), axis=1)
months_df['quarter'] = pd.to_datetime(months_df['quarter'])
return months_df
def get_repo_id_start_date_and_end_date():
now = datetime.datetime.now()
repo_id = int(request.args.get('repo_id'))
start_date = str(request.args.get('start_date', "{}-01-01".format(now.year - 1)))
end_date = str(request.args.get('end_date', "{}-{}-{}".format(now.year, now.month, now.day)))
return repo_id, start_date, end_date
def filter_out_repeats_without_required_contributions_in_required_time(repeat_list, repeats_df, required_time,
first_list):
differences = []
for i in range(0, len(repeat_list)):
time_difference = repeat_list[i] - first_list[i]
total = time_difference.days * 86400 + time_difference.seconds
differences.append(total)
repeats_df['differences'] = differences
# remove contributions who made enough contributions, but not in a short enough time
repeats_df = repeats_df.loc[repeats_df['differences'] <= required_time * 86400]
return repeats_df
def compute_fly_by_and_returning_contributors_dfs(input_df, required_contributions, required_time, start_date):
# create a copy of contributor dataframe
driver_df = input_df.copy()
# remove first time contributors before begin date, along with their second contribution
mask = (driver_df['yearmonth'] < start_date)
driver_df = driver_df[~driver_df['cntrb_id'].isin(driver_df.loc[mask]['cntrb_id'])]
# determine if contributor is a drive by by finding all the cntrb_id's that do not have a second contribution
repeats_df = driver_df.copy()
repeats_df = repeats_df.loc[repeats_df['rank'].isin([1, required_contributions])]
# removes all the contributors that only have a first contirbution
repeats_df = repeats_df[
repeats_df['cntrb_id'].isin(repeats_df.loc[driver_df['rank'] == required_contributions]['cntrb_id'])]
repeat_list = repeats_df.loc[driver_df['rank'] == required_contributions]['created_at'].tolist()
first_list = repeats_df.loc[driver_df['rank'] == 1]['created_at'].tolist()
repeats_df = repeats_df.loc[driver_df['rank'] == 1]
repeats_df['type'] = 'repeat'
repeats_df = filter_out_repeats_without_required_contributions_in_required_time(
repeat_list, repeats_df, required_time, first_list)
repeats_df = repeats_df.loc[repeats_df['differences'] <= required_time * 86400]
repeat_cntrb_ids = repeats_df['cntrb_id'].to_list()
drive_by_df = driver_df.loc[~driver_df['cntrb_id'].isin(repeat_cntrb_ids)]
drive_by_df = drive_by_df.loc[driver_df['rank'] == 1]
drive_by_df['type'] = 'drive_by'
return drive_by_df, repeats_df
def add_caption_to_visualizations(caption, required_contributions, required_time, plot_width):
caption_plot = figure(width=plot_width, height=200, margin=(0, 0, 0, 0))
caption_plot.add_layout(Label(
x=0,
y=160,
x_units='screen',
y_units='screen',
text='{}'.format(caption.format(required_contributions, required_time)),
text_font='times',
text_font_size='15pt',
render_mode='css'
))
caption_plot.outline_line_color = None
return caption_plot
def format_new_cntrb_bar_charts(plot, rank, group_by_format_string):
plot.xgrid.grid_line_color = None
plot.y_range.start = 0
plot.axis.minor_tick_line_color = None
plot.outline_line_color = None
plot.title.align = "center"
plot.title.text_font_size = "18px"
plot.yaxis.axis_label = 'Second Time Contributors' if rank == 2 else 'New Contributors'
plot.xaxis.axis_label = group_by_format_string
plot.xaxis.axis_label_text_font_size = "18px"
plot.yaxis.axis_label_text_font_size = "16px"
plot.xaxis.major_label_text_font_size = "16px"
plot.xaxis.major_label_orientation = 45.0
plot.yaxis.major_label_text_font_size = "16px"
return plot
def add_charts_and_captions_to_correct_positions(chart_plot, caption_plot, rank, contributor_type,
row_1, row_2, row_3, row_4):
if rank == 1 and (contributor_type == 'All' or contributor_type == 'repeat'):
row_1.append(chart_plot)
row_2.append(caption_plot)
elif rank == 2 or contributor_type == 'drive_by':
row_3.append(chart_plot)
row_4.append(caption_plot)
def get_new_cntrb_bar_chart_query_params():
group_by = str(request.args.get('group_by', "quarter"))
required_contributions = int(request.args.get('required_contributions', 4))
required_time = int(request.args.get('required_time', 365))
return group_by, required_contributions, required_time
def remove_rows_before_start_date(df, start_date):
mask = (df['yearmonth'] < start_date)
result_df = df[~df['cntrb_id'].isin(df.loc[mask]['cntrb_id'])]
return result_df
def remove_rows_with_null_values(df, not_null_columns=[]):
"""Remove null data from pandas df
Parameters
-- df
description: the dataframe that will be modified
type: Pandas Dataframe
-- list_of_columns
description: columns that are searched for NULL values
type: list
default: [] (means all columns will be checked for NULL values)
IMPORTANT: if an empty list is passed or nothing is passed it will check all columns for NULL values
Return Value
-- Modified Pandas Dataframe
"""
if len(not_null_columns) == 0:
not_null_columns = df.columns.to_list()
total_rows_removed = 0
for col in not_null_columns:
rows_removed = len(df.loc[df[col].isnull() == True])
if rows_removed > 0:
print(f"{rows_removed} rows have been removed because of null values in column {col}")
total_rows_removed += rows_removed
df = df.loc[df[col].isnull() == False]
if total_rows_removed > 0:
print(f"\nTotal rows removed because of null data: {total_rows_removed}");
else:
print("No null data found")
return df
def get_needed_columns(df, list_of_columns):
"""Get only a specific list of columns from a Pandas Dataframe
Parameters
-- df
description: the dataframe that will be modified
type: Pandas Dataframe
-- list_of_columns
description: columns that will be kept in dataframe
type: list
Return Value
-- Modified Pandas Dataframe
"""
return df[list_of_columns]
def filter_data(df, needed_columns, not_null_columns=[]):
"""Filters out the unneeded rows in the df, and removed NULL data from df
Parameters
-- df
description: the dataframe that will be modified
type: Pandas Dataframe
-- needed_columns
description: the columns to keep in the dataframe
-- not_null_columns
description: columns that will be searched for NULL data,
if NULL values are found those rows will be removed
default: [] (means all columns in needed_columns list will be checked for NULL values)
IMPORTANT: if an empty list is passed or nothing is passed it will check
all columns in needed_columns list for NULL values
Return Value
-- Modified Pandas Dataframe
"""
if all(x in needed_columns for x in not_null_columns):
df = get_needed_columns(df, needed_columns)
df = remove_rows_with_null_values(df, not_null_columns)
return df
else:
print("Developer error, not null columns should be a subset of needed columns")
return df
@server.app.route('/{}/contributor_reports/new_contributors_bar/'.format(server.api_version), methods=["GET"])
def new_contributors_bar():
repo_id, start_date, end_date = get_repo_id_start_date_and_end_date()
group_by, required_contributions, required_time = get_new_cntrb_bar_chart_query_params()
input_df = new_contributor_data_collection(repo_id=repo_id, required_contributions=required_contributions)
months_df = months_data_collection(start_date=start_date, end_date=end_date)
# TODO remove full_name from data for all charts since it is not needed in vis generation
not_null_columns = ['cntrb_id', 'created_at', 'month', 'year', 'repo_id', 'repo_name', 'login', 'action',
'rank', 'yearmonth', 'new_contributors', 'quarter']
input_df = remove_rows_with_null_values(input_df, not_null_columns)
if len(input_df) == 0:
return Response(response="There is no data for this repo, in the database you are accessing",
mimetype='application/json',
status=200)
repo_dict = {repo_id: input_df.loc[input_df['repo_id'] == repo_id].iloc[0]['repo_name']}
contributor_types = ['All', 'repeat', 'drive_by']
ranks = [1, 2]
row_1, row_2, row_3, row_4 = [], [], [], []
all_df = remove_rows_before_start_date(input_df, start_date)
drive_by_df, repeats_df = compute_fly_by_and_returning_contributors_dfs(input_df, required_contributions,
required_time, start_date)
for rank in ranks:
for contributor_type in contributor_types:
# do not display these visualizations since drive-by's do not have second contributions, and the
# second contribution of a repeat contributor is the same thing as the all the second time contributors
if (rank == 2 and contributor_type == 'drive_by') or (rank == 2 and contributor_type == 'repeat'):
continue
if contributor_type == 'repeat':
driver_df = repeats_df
caption = """This graph shows repeat contributors in the specified time period. Repeat contributors
are contributors who have made {} or more contributions in {} days and their first contribution is
in the specified time period. New contributors are individuals who make their first contribution
in the specified time period."""
elif contributor_type == 'drive_by':
driver_df = drive_by_df
caption = """This graph shows fly by contributors in the specified time period. Fly by contributors
are contributors who make less than the required {} contributions in {} days. New contributors are
individuals who make their first contribution in the specified time period. Of course, then, “All
fly-by’s are by definition first time contributors”. However, not all first time contributors are
fly-by’s."""
elif contributor_type == 'All':
if rank == 1:
driver_df = all_df
# makes df with all first time contributors
driver_df = driver_df.loc[driver_df['rank'] == 1]
caption = """This graph shows all the first time contributors, whether they contribute once, or
contribute multiple times. New contributors are individuals who make their first contribution
in the specified time period."""
if rank == 2:
driver_df = all_df
# creates df with all second time contributors
driver_df = driver_df.loc[driver_df['rank'] == 2]
caption = """This graph shows the second contribution of all
first time contributors in the specified time period."""
# y_axis_label = 'Second Time Contributors'
# filter by end_date, this is not done with the begin date filtering because a repeat contributor
# will look like drive-by if the second contribution is removed by end_date filtering
mask = (driver_df['yearmonth'] < end_date)
driver_df = driver_df.loc[mask]
# adds all months to driver_df so the lists of dates will include all months and years
driver_df = pd.concat([driver_df, months_df])
data = pd.DataFrame()
if group_by == 'year':
data['dates'] = driver_df[group_by].unique()
# new contributor counts for y-axis
data['new_contributor_counts'] = driver_df.groupby([group_by]).sum().reset_index()[
'new_contributors']
# used to format x-axis and title
group_by_format_string = "Year"
elif group_by == 'quarter' or group_by == 'month':
# set variables to group the data by quarter or month
if group_by == 'quarter':
date_column = 'quarter'
group_by_format_string = "Quarter"
elif group_by == 'month':
date_column = 'yearmonth'
group_by_format_string = "Month"
# modifies the driver_df[date_column] to be a string with year and month,
# then finds all the unique values
data['dates'] = np.unique(np.datetime_as_string(driver_df[date_column], unit='M'))
# new contributor counts for y-axis
data['new_contributor_counts'] = driver_df.groupby([date_column]).sum().reset_index()[
'new_contributors']
# if the data set is large enough it will dynamically assign the width, if the data set is
# too small it will by default set to 870 pixel so the title fits
if len(data['new_contributor_counts']) >= 15:
plot_width = 46 * len(data['new_contributor_counts'])
else:
plot_width = 870
# create a dict convert an integer number into a word
# used to turn the rank into a word, so it is nicely displayed in the title
numbers = ['Zero', 'First', 'Second']
num_conversion_dict = {}
for i in range(1, len(numbers)):
num_conversion_dict[i] = numbers[i]
number = '{}'.format(num_conversion_dict[rank])
# define pot for bar chart
p = figure(x_range=data['dates'], plot_height=400, plot_width=plot_width,
title="{}: {} {} Time Contributors Per {}".format(repo_dict[repo_id],
contributor_type.capitalize(), number,
group_by_format_string),
y_range=(0, max(data['new_contributor_counts']) * 1.15), margin=(0, 0, 10, 0))
p.vbar(x=data['dates'], top=data['new_contributor_counts'], width=0.8)
source = ColumnDataSource(
data=dict(dates=data['dates'], new_contributor_counts=data['new_contributor_counts']))
# add contributor_count labels to chart
p.add_layout(LabelSet(x='dates', y='new_contributor_counts', text='new_contributor_counts', y_offset=4,
text_font_size="13pt", text_color="black",
source=source, text_align='center'))
plot = format_new_cntrb_bar_charts(p, rank, group_by_format_string)
caption_plot = add_caption_to_visualizations(caption, required_contributions, required_time, plot_width)
add_charts_and_captions_to_correct_positions(plot, caption_plot, rank, contributor_type, row_1,
row_2, row_3, row_4)
# puts plots together into a grid
grid = gridplot([row_1, row_2, row_3, row_4])
filename = export_png(grid)
return send_file(filename)
@server.app.route('/{}/contributor_reports/new_contributors_stacked_bar/'.format(server.api_version),
methods=["GET"])
def new_contributors_stacked_bar():
repo_id, start_date, end_date = get_repo_id_start_date_and_end_date()
group_by, required_contributions, required_time = get_new_cntrb_bar_chart_query_params()
input_df = new_contributor_data_collection(repo_id=repo_id, required_contributions=required_contributions)
months_df = months_data_collection(start_date=start_date, end_date=end_date)
needed_columns = ['cntrb_id', 'created_at', 'month', 'year', 'repo_id', 'repo_name', 'login', 'action',
'rank', 'yearmonth', 'new_contributors', 'quarter']
input_df = filter_data(input_df, needed_columns)
if len(input_df) == 0:
return Response(response="There is no data for this repo, in the database you are accessing",
mimetype='application/json',
status=200)
repo_dict = {repo_id: input_df.loc[input_df['repo_id'] == repo_id].iloc[0]['repo_name']}
contributor_types = ['All', 'repeat', 'drive_by']
ranks = [1, 2]
row_1, row_2, row_3, row_4 = [], [], [], []
all_df = remove_rows_before_start_date(input_df, start_date)
drive_by_df, repeats_df = compute_fly_by_and_returning_contributors_dfs(input_df, required_contributions,
required_time, start_date)
for rank in ranks:
for contributor_type in contributor_types:
# do not display these visualizations since drive-by's do not have second contributions,
# and the second contribution of a repeat contributor is the same thing as the all the
# second time contributors
if (rank == 2 and contributor_type == 'drive_by') or (rank == 2 and contributor_type == 'repeat'):
continue
if contributor_type == 'repeat':
driver_df = repeats_df
caption = """This graph shows repeat contributors in the specified time period. Repeat contributors
are contributors who have made {} or more contributions in {} days and their first contribution is
in the specified time period. New contributors are individuals who make their first contribution in
the specified time period."""
elif contributor_type == 'drive_by':
driver_df = drive_by_df
caption = """This graph shows fly by contributors in the specified time period. Fly by contributors
are contributors who make less than the required {} contributions in {} days. New contributors are
individuals who make their first contribution in the specified time period. Of course, then, “All
fly-by’s are by definition first time contributors”. However, not all first time contributors are
fly-by’s."""
elif contributor_type == 'All':
if rank == 1:
driver_df = all_df
# makes df with all first time contributors
driver_df = driver_df.loc[driver_df['rank'] == 1]
caption = """This graph shows all the first time contributors, whether they contribute once, or
contribute multiple times. New contributors are individuals who make their first contribution in
the specified time period."""
if rank == 2:
driver_df = all_df
# creates df with all second time contributor
driver_df = driver_df.loc[driver_df['rank'] == 2]
caption = """This graph shows the second contribution of all first time
contributors in the specified time period."""
# y_axis_label = 'Second Time Contributors'
# filter by end_date, this is not done with the begin date filtering because a repeat contributor will
# look like drive-by if the second contribution is removed by end_date filtering
mask = (driver_df['yearmonth'] < end_date)
driver_df = driver_df.loc[mask]
# adds all months to driver_df so the lists of dates will include all months and years
driver_df = pd.concat([driver_df, months_df])
actions = ['open_pull_request', 'pull_request_comment', 'commit', 'issue_closed', 'issue_opened',
'issue_comment']
data = pd.DataFrame()
if group_by == 'year':
# x-axis dates
data['dates'] = driver_df[group_by].unique()
for contribution_type in actions:
data[contribution_type] = \
pd.concat([driver_df.loc[driver_df['action'] == contribution_type], months_df]).groupby(
group_by).sum().reset_index()['new_contributors']
# new contributor counts for all actions
data['new_contributor_counts'] = driver_df.groupby([group_by]).sum().reset_index()[
'new_contributors']
# used to format x-axis and graph title
group_by_format_string = "Year"
elif group_by == 'quarter' or group_by == 'month':
# set variables to group the data by quarter or month
if group_by == 'quarter':
date_column = 'quarter'
group_by_format_string = "Quarter"
elif group_by == 'month':
date_column = 'yearmonth'
group_by_format_string = "Month"
# modifies the driver_df[date_column] to be a string with year and month,
# then finds all the unique values
data['dates'] = np.unique(np.datetime_as_string(driver_df[date_column], unit='M'))
# new_contributor counts for each type of action
for contribution_type in actions:
data[contribution_type] = \
pd.concat([driver_df.loc[driver_df['action'] == contribution_type], months_df]).groupby(
date_column).sum().reset_index()['new_contributors']
print(data.to_string())
# new contributor counts for all actions
data['new_contributor_counts'] = driver_df.groupby([date_column]).sum().reset_index()[
'new_contributors']
# if the data set is large enough it will dynamically assign the width, if the data set is too small it
# will by default set to 870 pixel so the title fits
if len(data['new_contributor_counts']) >= 15:
plot_width = 46 * len(data['new_contributor_counts']) + 200
else:
plot_width = 870
# create list of values for data source dict
actions_df_references = []
for action in actions:
actions_df_references.append(data[action])
# created dict with the actions as the keys, and the values as the values from the df
data_source = {actions[i]: actions_df_references[i] for i in range(len(actions))}
data_source.update({'dates': data['dates'], 'New Contributor Counts': data['new_contributor_counts']})
colors = Colorblind[len(actions)]
source = ColumnDataSource(data=data_source)
# create a dict convert an integer number into a word
# used to turn the rank into a word, so it is nicely displayed in the title
numbers = ['Zero', 'First', 'Second']
num_conversion_dict = {}
for i in range(1, len(numbers)):
num_conversion_dict[i] = numbers[i]
number = '{}'.format(num_conversion_dict[rank])
# y_max = 20
# creates plot to hold chart
p = figure(x_range=data['dates'], plot_height=400, plot_width=plot_width,
title='{}: {} {} Time Contributors Per {}'.format(repo_dict[repo_id],
contributor_type.capitalize(), number,
group_by_format_string),
toolbar_location=None, y_range=(0, max(data['new_contributor_counts']) * 1.15))
# max(data['new_contributor_counts'])* 1.15), margin = (0, 0, 0, 0))
vbar = p.vbar_stack(actions, x='dates', width=0.8, color=colors, source=source)
# add total count labels
p.add_layout(LabelSet(x='dates', y='New Contributor Counts', text='New Contributor Counts', y_offset=4,
text_font_size="14pt",
text_color="black", source=source, text_align='center'))
# add legend
legend = Legend(items=[(date, [action]) for (date, action) in zip(actions, vbar)], location=(0, 120),
label_text_font_size="16px")
p.add_layout(legend, 'right')
plot = format_new_cntrb_bar_charts(p, rank, group_by_format_string)
caption_plot = add_caption_to_visualizations(caption, required_contributions, required_time, plot_width)
add_charts_and_captions_to_correct_positions(plot, caption_plot, rank, contributor_type, row_1,
row_2, row_3, row_4)
# puts plots together into a grid
grid = gridplot([row_1, row_2, row_3, row_4])
filename = export_png(grid)
return send_file(filename)
@server.app.route('/{}/contributor_reports/returning_contributors_pie_chart/'.format(server.api_version),
methods=["GET"])
def returning_contributor_pie_chart():
repo_id, start_date, end_date = get_repo_id_start_date_and_end_date()
required_contributions = int(request.args.get('required_contributions', 4))
required_time = int(request.args.get('required_time', 365))
input_df = new_contributor_data_collection(repo_id=repo_id, required_contributions=required_contributions)
needed_columns = ['cntrb_id', 'created_at', 'month', 'year', 'repo_id', 'repo_name', 'login', 'action',
'rank', 'yearmonth', 'new_contributors', 'quarter']
input_df = filter_data(input_df, needed_columns)
if len(input_df) == 0:
return Response(response="There is no data for this repo, in the database you are accessing",
mimetype='application/json',
status=200)
repo_dict = {repo_id: input_df.loc[input_df['repo_id'] == repo_id].iloc[0]['repo_name']}
drive_by_df, repeats_df = compute_fly_by_and_returning_contributors_dfs(input_df, required_contributions,
required_time, start_date)
print(repeats_df.to_string())
driver_df = pd.concat([drive_by_df, repeats_df])
# filter df by end date
mask = (driver_df['yearmonth'] < end_date)
driver_df = driver_df.loc[mask]
# first and second time contributor counts
drive_by_contributors = driver_df.loc[driver_df['type'] == 'drive_by'].count()['new_contributors']
repeat_contributors = driver_df.loc[driver_df['type'] == 'repeat'].count()['new_contributors']
# create a dict with the # of drive-by and repeat contributors
x = {'Drive_By': drive_by_contributors,
'Repeat': repeat_contributors}
# turn dict 'x' into a dataframe with columns 'contributor_type', and 'counts'
data = | pd.Series(x) | pandas.Series |
from .._common import *
import pandas as pd
import numpy as np
class ToDataframe(yo_fluq.agg.PushQueryElement):
def __init__(self, **kwargs):
self.kwargs = kwargs
def on_enter(factory,instance):
instance.lst = []
def on_process(factory, instance, element):
instance.lst.append(element)
def on_report(factory, instance):
return | pd.DataFrame(instance.lst,**factory.kwargs) | pandas.DataFrame |
import pandas as pd
import numpy as np
class PreviousValuesGenerator:
transactions = None
def __init__(self, transactions_path):
print(f'leyendo fichero {transactions_path}')
self.transactions = pd.read_csv(transactions_path, sep=';')
print(f'Existen {self.transactions.shape[0]} registros de venta desde {self.transactions.date.min()} hasta {self.transactions.date.max()}')
def __transactions_with_previous_dates(self):
result = self.transactions.copy()
result['last_year'] = result.date - pd.offsets.DateOffset(years=1)
result['last_quarter'] = result.date - pd.offsets.DateOffset(months=3)
result['last_month'] = result.date - pd.offsets.DateOffset(months=1)
result['last_week'] = result.date - pd.offsets.DateOffset(weeks=1)
result['last_day'] = result.date - pd.offsets.DateOffset(days=1)
result['last_52_weeks'] = result.date - pd.offsets.DateOffset(weeks=52)
result['last_12_weeks'] = result.date - pd.offsets.DateOffset(weeks=12)
result['last_8_weeks'] = result.date - pd.offsets.DateOffset(weeks=8)
result['last_4_weeks'] = result.date - pd.offsets.DateOffset(weeks=4)
return result
def __set_previous_values(self, t):
transactions = self.transactions
y = transactions[(transactions.date == t.last_year) & (transactions.sku == t.sku)].units_sold.max()
q = transactions[(transactions.date == t.last_quarter) & (transactions.sku == t.sku)].units_sold.max()
m = transactions[(transactions.date == t.last_month) & (transactions.sku == t.sku)].units_sold.max()
w = transactions[(transactions.date == t.last_week) & (transactions.sku == t.sku)].units_sold.max()
w52 = transactions[(transactions.date == t.last_52_weeks) & (transactions.sku == t.sku)].units_sold.max()
w12 = transactions[(transactions.date == t.last_12_weeks) & (transactions.sku == t.sku)].units_sold.max()
w8 = transactions[(transactions.date == t.last_8_weeks) & (transactions.sku == t.sku)].units_sold.max()
w4 = transactions[(transactions.date == t.last_4_weeks) & (transactions.sku == t.sku)].units_sold.max()
d = transactions[(transactions.date == t.last_day) & (transactions.sku == t.sku)].units_sold.max()
if not pd.isna(y):
y = np.int64(y)
if not pd.isna(q):
q = np.int64(q)
if not pd.isna(m):
m = np.int64(m)
if not pd.isna(w):
w = np.int64(w)
if not pd.isna(w52):
w52 = np.int64(w52)
if not pd.isna(w12):
w12 = np.int64(w12)
if not | pd.isna(w8) | pandas.isna |
from pandas import DataFrame
# State abbreviation -> Full Name and visa versa. FL -> Florida, etc.
# (Handle Washington DC and territories like Puerto Rico etc.)
def add_state_names(my_df):
new_df = my_df.copy()
names_map = {"CA":"Cali", "CO":"Colo", "CT":"Conn"}
new_df["name"] = new_df["abbrev"].map(names_map)
breakpoint()
return my_df
if __name__ == "__main__":
df = | DataFrame({"abbrev":["CA","CO","CT","DC","TX"]}) | pandas.DataFrame |
"""
General utility functions that are used in a variety of contexts.
The functions in this module are used in various stages of the ETL and post-etl
processes. They are usually not dataset specific, but not always. If a function
is designed to be used as a general purpose tool, applicable in multiple
scenarios, it should probably live here. There are lost of transform type
functions in here that help with cleaning and restructing dataframes.
"""
import itertools
import logging
import pathlib
import re
import shutil
from functools import partial
import addfips
import numpy as np
import pandas as pd
import requests
import sqlalchemy as sa
import timezonefinder
from sqlalchemy.engine import reflection
import pudl
from pudl import constants as pc
logger = logging.getLogger(__name__)
# This is a little abbreviated function that allows us to propagate the NA
# values through groupby aggregations, rather than using inefficient lambda
# functions in each one.
sum_na = partial(pd.Series.sum, skipna=False)
# Initializing this TimezoneFinder opens a bunch of geography files and holds
# them open for efficiency. I want to avoid doing that for every call to find
# the timezone, so this is global.
tz_finder = timezonefinder.TimezoneFinder()
def download_zip_url(url, save_path, chunk_size=128):
"""
Download and save a Zipfile locally.
Useful for acquiring and storing non-PUDL data locally.
Args:
url (str): The URL from which to download the Zipfile
save_path (pathlib.Path): The location to save the file.
chunk_size (int): Data chunk in bytes to use while downloading.
Returns:
None
"""
# This is a temporary hack to avoid being filtered as a bot:
headers = {
'User-Agent': 'Mozilla/5.0 (Macintosh; Intel Mac OS X 10.15; rv:77.0) Gecko/20100101 Firefox/77.0',
'Accept': 'text/html,application/xhtml+xml,application/xml;q=0.9,image/webp,*/*;q=0.8',
'Accept-Language': 'en-US,en;q=0.5',
'DNT': '1',
'Connection': 'keep-alive',
'Upgrade-Insecure-Requests': '1',
}
r = requests.get(url, stream=True, headers=headers)
with save_path.open(mode='wb') as fd:
for chunk in r.iter_content(chunk_size=chunk_size):
fd.write(chunk)
def add_fips_ids(df, state_col="state", county_col="county", vintage=2015):
"""Add State and County FIPS IDs to a dataframe."""
# force the columns to be the nullable string types so we have a consistent
# null value to filter out before feeding to addfips
df = df.astype({
state_col: pd.StringDtype(),
county_col: pd.StringDtype(),
})
af = addfips.AddFIPS(vintage=vintage)
# Lookup the state and county FIPS IDs and add them to the dataframe:
df["state_id_fips"] = df.apply(
lambda x: (af.get_state_fips(state=x[state_col])
if pd.notnull(x[state_col]) else pd.NA),
axis=1)
logger.info(
f"Assigned state FIPS codes for "
f"{len(df[df.state_id_fips.notnull()])/len(df):.2%} of records."
)
df["county_id_fips"] = df.apply(
lambda x: (af.get_county_fips(state=x[state_col], county=x[county_col])
if pd.notnull(x[county_col]) else pd.NA),
axis=1)
# force the code columns to be nullable strings - the leading zeros are
# important
df = df.astype({
"county_id_fips": pd.StringDtype(),
"state_id_fips": pd.StringDtype(),
})
logger.info(
f"Assigned county FIPS codes for "
f"{len(df[df.county_id_fips.notnull()])/len(df):.2%} of records."
)
return df
def clean_eia_counties(df, fixes, state_col="state", county_col="county"):
"""Replace non-standard county names with county nmes from US Census."""
df = df.copy()
df[county_col] = (
df[county_col].str.strip()
.str.replace(r"\s+", " ", regex=True) # Condense multiple whitespace chars.
.str.replace(r"^St ", "St. ", regex=True) # Standardize abbreviation.
.str.replace(r"^Ste ", "Ste. ", regex=True) # Standardize abbreviation.
.str.replace("Kent & New Castle", "Kent, New Castle") # Two counties
# Fix ordering, remove comma
.str.replace("Borough, Kodiak Island", "Kodiak Island Borough")
# Turn comma-separated counties into lists
.str.replace(r",$", "", regex=True).str.split(',')
)
# Create new records for each county in a multi-valued record
df = df.explode(county_col)
df[county_col] = df[county_col].str.strip()
# Yellowstone county is in MT, not WY
df.loc[(df[state_col] == "WY") &
(df[county_col] == "Yellowstone"), state_col] = "MT"
# Replace individual bad county names with identified correct names in fixes:
for fix in fixes.itertuples():
state_mask = df[state_col] == fix.state
county_mask = df[county_col] == fix.eia_county
df.loc[state_mask & county_mask, county_col] = fix.fips_county
return df
def oob_to_nan(df, cols, lb=None, ub=None):
"""
Set non-numeric values and those outside of a given rage to NaN.
Args:
df (pandas.DataFrame): The dataframe containing values to be altered.
cols (iterable): Labels of the columns whose values are to be changed.
lb: (number): Lower bound, below which values are set to NaN. If None,
don't use a lower bound.
ub: (number): Upper bound, below which values are set to NaN. If None,
don't use an upper bound.
Returns:
pandas.DataFrame: The altered DataFrame.
"""
out_df = df.copy()
for col in cols:
# Force column to be numeric if possible, NaN otherwise:
out_df.loc[:, col] = pd.to_numeric(out_df[col], errors="coerce")
if lb is not None:
out_df.loc[out_df[col] < lb, col] = np.nan
if ub is not None:
out_df.loc[out_df[col] > ub, col] = np.nan
return out_df
def prep_dir(dir_path, clobber=False):
"""
Create (or delete and recreate) a directory.
Args:
dir_path (path-like): path to the directory that you are trying to
clean and prepare.
clobber (bool): If True and dir_path exists, it will be removed and
replaced with a new, empty directory.
Raises:
FileExistsError: if a file or directory already exists at dir_path.
Returns:
pathlib.Path: Path to the created directory.
"""
dir_path = pathlib.Path(dir_path)
if dir_path.exists():
if clobber:
shutil.rmtree(dir_path)
else:
raise FileExistsError(
f'{dir_path} exists and clobber is {clobber}')
dir_path.mkdir(parents=True)
return dir_path
def is_doi(doi):
"""
Determine if a string is a valid digital object identifier (DOI).
Function simply checks whether the offered string matches a regular
expresssion -- it doesn't check whether the DOI is actually registered
with the relevant authority.
Args:
doi (str): String to validate.
Returns:
bool: True if doi matches the regex for valid DOIs, False otherwise.
"""
doi_regex = re.compile(
r'(doi:\s*|(?:https?://)?(?:dx\.)?doi\.org/)?(10\.\d+(.\d+)*/.+)$',
re.IGNORECASE | re.UNICODE)
return bool(re.match(doi_regex, doi))
def is_annual(df_year, year_col='report_date'):
"""
Determine whether a DataFrame contains consistent annual time-series data.
Some processes will only work with consistent yearly reporting. This means
if you have two non-contiguous years of data or the datetime reporting is
inconsistent, the process will break. This function attempts to infer the
temporal frequency of the dataframe, or if that is impossible, to at least
see whether the data would be consistent with annual reporting -- e.g. if
there is only a single year of data, it should all have the same date, and
that date should correspond to January 1st of a given year.
This function is known to be flaky and needs to be re-written to deal with
the edge cases better.
Args:
df_year (pandas.DataFrame): A pandas DataFrame that might
contain time-series data at annual resolution.
year_col (str): The column of the DataFrame in which the year is
reported.
Returns:
bool: True if df_year is found to be consistent with continuous annual
time resolution, False otherwise.
"""
year_index = pd.DatetimeIndex(df_year[year_col].unique()).sort_values()
if len(year_index) >= 3:
date_freq = pd.infer_freq(year_index)
assert date_freq == 'AS-JAN', "infer_freq() not AS-JAN"
elif len(year_index) == 2:
min_year = year_index.min()
max_year = year_index.max()
assert year_index.min().month == 1, "min year not Jan"
assert year_index.min().day == 1, "min day not 1st"
assert year_index.max().month == 1, "max year not Jan"
assert year_index.max().day == 1, "max day not 1st"
delta_year = pd.Timedelta(max_year - min_year)
assert delta_year / pd.Timedelta(days=1) >= 365.0
assert delta_year / pd.Timedelta(days=1) <= 366.0
elif len(year_index) == 1:
assert year_index.min().month == 1, "only month not Jan"
assert year_index.min().day == 1, "only day not 1st"
else:
assert False, "Zero dates found!"
return True
def merge_on_date_year(df_date, df_year, on=(), how='inner',
date_col='report_date',
year_col='report_date'):
"""Merge two dataframes based on a shared year.
Some of our data is annual, and has an integer year column (e.g. FERC 1).
Some of our data is annual, and uses a Date column (e.g. EIA 860), and
some of our data has other temporal resolutions, and uses date columns
(e.g. EIA 923 fuel receipts are monthly, EPA CEMS data is hourly). This
function takes two data frames and merges them based on the year that the
data pertains to. It requires one of the dataframes to have annual
resolution, and allows the annual time to be described as either an integer
year or a Date. The non-annual dataframe must have a Date column.
By default, it is assumed that both the date and annual columns to be
merged on are called 'report_date' since that's the common case when
bringing together EIA860 and EIA923 data.
Args:
df_date: the dataframe with a more granular date column, the label of
which is specified by date_col (report_date by default)
df_year: the dataframe with a column containing annual dates, the label
of which is specified by year_col (report_date by default)
on: The list of columns to merge on, other than the year and date
columns.
date_col: name of the date column to use to find the year to merge on.
Must be a Date.
year_col: name of the year column to merge on. Must be a Date
column with annual resolution.
Returns:
pandas.DataFrame: a dataframe with a date column, but no year
columns, and only one copy of any shared columns that were not part of
the list of columns to be merged on. The values from df1 are the ones
which are retained for any shared, non-merging columns
Raises:
ValueError: if the date or year columns are not found, or if the year
column is found to be inconsistent with annual reporting.
Todo: Right mergers will result in null values in the resulting date
column. The final output includes the date_col from the date_df and thus
if there are any entity records (records being merged on) in the
year_df but not in the date_df, a right merge will result in nulls in
the date_col. And when we drop the 'year_temp' column, the year from
the year_df will be gone. Need to determine how to deal with this.
Should we generate a montly record in each year? Should we generate
full time serires? Should we restrict right merges in this function?
"""
if date_col not in df_date.columns.tolist():
raise ValueError(f"Date column {date_col} not found in df_date.")
if year_col not in df_year.columns.tolist():
raise ValueError(f"Year column {year_col} not found in df_year.")
if not is_annual(df_year, year_col=year_col):
raise ValueError(f"df_year is not annual, based on column {year_col}.")
first_date = pd.to_datetime(df_date[date_col].min())
all_dates = pd.DatetimeIndex(df_date[date_col]).unique().sort_values()
if not len(all_dates) > 0:
raise ValueError("Didn't find any dates in DatetimeIndex.")
if len(all_dates) > 1:
if len(all_dates) == 2:
second_date = all_dates.max()
elif len(all_dates) > 2:
date_freq = pd.infer_freq(all_dates)
rng = pd.date_range(start=first_date, periods=2, freq=date_freq)
second_date = rng[1]
if (second_date - first_date) / pd.Timedelta(days=366) > 1.0:
raise ValueError("Consecutive annual dates >1 year apart.")
# Create a temporary column in each dataframe with the year
df_year = df_year.copy()
df_date = df_date.copy()
df_year['year_temp'] = pd.to_datetime(df_year[year_col]).dt.year
# Drop the yearly report_date column: this way there won't be duplicates
# and the final df will have the more granular report_date.
df_year = df_year.drop([year_col], axis=1)
df_date['year_temp'] = pd.to_datetime(df_date[date_col]).dt.year
full_on = on + ['year_temp']
unshared_cols = [col for col in df_year.columns.tolist()
if col not in df_date.columns.tolist()]
cols_to_use = unshared_cols + full_on
# Merge and drop the temp
merged = pd.merge(df_date, df_year[cols_to_use], how=how, on=full_on)
merged = merged.drop(['year_temp'], axis=1)
return merged
def organize_cols(df, cols):
"""
Organize columns into key ID & name fields & alphabetical data columns.
For readability, it's nice to group a few key columns at the beginning
of the dataframe (e.g. report_year or report_date, plant_id...) and then
put all the rest of the data columns in alphabetical order.
Args:
df: The DataFrame to be re-organized.
cols: The columns to put first, in their desired output ordering.
Returns:
pandas.DataFrame: A dataframe with the same columns as the input
DataFrame df, but with cols first, in the same order as they
were passed in, and the remaining columns sorted alphabetically.
"""
# Generate a list of all the columns in the dataframe that are not
# included in cols
data_cols = [c for c in df.columns.tolist() if c not in cols]
data_cols.sort()
organized_cols = cols + data_cols
return df[organized_cols]
def simplify_strings(df, columns):
"""
Simplify the strings contained in a set of dataframe columns.
Performs several operations to simplify strings for comparison and parsing purposes.
These include removing Unicode control characters, stripping leading and trailing
whitespace, using lowercase characters, and compacting all internal whitespace to a
single space.
Leaves null values unaltered. Casts other values with astype(str).
Args:
df (pandas.DataFrame): DataFrame whose columns are being cleaned up.
columns (iterable): The labels of the string columns to be simplified.
Returns:
pandas.DataFrame: The whole DataFrame that was passed in, with
the string columns cleaned up.
"""
out_df = df.copy()
for col in columns:
if col in out_df.columns:
out_df.loc[out_df[col].notnull(), col] = (
out_df.loc[out_df[col].notnull(), col]
.astype(str)
.str.replace(r"[\x00-\x1f\x7f-\x9f]", "", regex=True)
.str.strip()
.str.lower()
.str.replace(r'\s+', ' ', regex=True)
)
return out_df
def cleanstrings_series(col, str_map, unmapped=None, simplify=True):
"""Clean up the strings in a single column/Series.
Args:
col (pandas.Series): A pandas Series, typically a single column of a
dataframe, containing the freeform strings that are to be cleaned.
str_map (dict): A dictionary of lists of strings, in which the keys are
the simplified canonical strings, witch which each string found in
the corresponding list will be replaced.
unmapped (str): A value with which to replace any string found in col
that is not found in one of the lists of strings in map. Typically
the null string ''. If None, these strings will not be replaced.
simplify (bool): If True, strip and compact whitespace, and lowercase
all strings in both the list of values to be replaced, and the
values found in col. This can reduce the number of strings that
need to be kept track of.
Returns:
pandas.Series: The cleaned up Series / column, suitable for
replacing the original messy column in a :class:`pandas.DataFrame`.
"""
if simplify:
col = (
col.astype(str).
str.strip().
str.lower().
str.replace(r'\s+', ' ', regex=True)
)
for k in str_map:
str_map[k] = [re.sub(r'\s+', ' ', s.lower().strip())
for s in str_map[k]]
for k in str_map:
if str_map[k]:
col = col.replace(str_map[k], k)
if unmapped is not None:
badstrings = np.setdiff1d(col.unique(), list(str_map.keys()))
# This call to replace can only work if there are actually some
# leftover strings to fix -- otherwise it runs forever because we
# are replacing nothing with nothing.
if len(badstrings) > 0:
col = col.replace(badstrings, unmapped)
return col
def cleanstrings(df, columns, stringmaps, unmapped=None, simplify=True):
"""Consolidate freeform strings in several dataframe columns.
This function will consolidate freeform strings found in `columns` into
simplified categories, as defined by `stringmaps`. This is useful when
a field contains many different strings that are really meant to represent
a finite number of categories, e.g. a type of fuel. It can also be used to
create simplified categories that apply to similar attributes that are
reported in various data sources from different agencies that use their own
taxonomies.
The function takes and returns a pandas.DataFrame, making it suitable for
use with the :func:`pandas.DataFrame.pipe` method in a chain.
Args:
df (pandas.DataFrame): the DataFrame containing the string columns to
be cleaned up.
columns (list): a list of string column labels found in the column
index of df. These are the columns that will be cleaned.
stringmaps (list): a list of dictionaries. The keys of these
dictionaries are strings, and the values are lists of strings. Each
dictionary in the list corresponds to a column in columns. The
keys of the dictionaries are the values with which every string in
the list of values will be replaced.
unmapped (str, None): the value with which strings not found in the
stringmap dictionary will be replaced. Typically the null string
''. If None, then strings found in the columns but not in the
stringmap will be left unchanged.
simplify (bool): If true, strip whitespace, remove duplicate
whitespace, and force lower-case on both the string map and the
values found in the columns to be cleaned. This can reduce the
overall number of string values that need to be tracked.
Returns:
pandas.DataFrame: The function returns a new DataFrame containing the
cleaned strings.
"""
out_df = df.copy()
for col, str_map in zip(columns, stringmaps):
out_df[col] = cleanstrings_series(
out_df[col], str_map, unmapped=unmapped, simplify=simplify)
return out_df
def fix_int_na(df, columns, float_na=np.nan, int_na=-1, str_na=''):
"""Convert NA containing integer columns from float to string.
Numpy doesn't have a real NA value for integers. When pandas stores integer
data which has NA values, it thus upcasts integers to floating point
values, using np.nan values for NA. However, in order to dump some of our
dataframes to CSV files for use in data packages, we need to write out
integer formatted numbers, with empty strings as the NA value. This
function replaces np.nan values with a sentinel value, converts the column
to integers, and then to strings, finally replacing the sentinel value with
the desired NA string.
This is an interim solution -- now that pandas extension arrays have been
implemented, we need to go back through and convert all of these integer
columns that contain NA values to Nullable Integer types like Int64.
Args:
df (pandas.DataFrame): The dataframe to be fixed. This argument allows
method chaining with the pipe() method.
columns (iterable of strings): A list of DataFrame column labels
indicating which columns need to be reformatted for output.
float_na (float): The floating point value to be interpreted as NA and
replaced in col.
int_na (int): Sentinel value to substitute for float_na prior to
conversion of the column to integers.
str_na (str): sa.String value to substitute for int_na after the column
has been converted to strings.
Returns:
df (pandas.DataFrame): a new DataFrame, with the selected columns
converted to strings that look like integers, compatible with
the postgresql COPY FROM command.
"""
return (
df.replace({c: float_na for c in columns}, int_na)
.astype({c: int for c in columns})
.astype({c: str for c in columns})
.replace({c: str(int_na) for c in columns}, str_na)
)
def month_year_to_date(df):
"""Convert all pairs of year/month fields in a dataframe into Date fields.
This function finds all column names within a dataframe that match the
regular expression '_month$' and '_year$', and looks for pairs that have
identical prefixes before the underscore. These fields are assumed to
describe a date, accurate to the month. The two fields are used to
construct a new _date column (having the same prefix) and the month/year
columns are then dropped.
Todo:
This function needs to be combined with convert_to_date, and improved:
* find and use a _day$ column as well
* allow specification of default month & day values, if none are found.
* allow specification of lists of year, month, and day columns to be
combined, rather than automataically finding all the matching ones.
* Do the Right Thing when invalid or NA values are encountered.
Args:
df (pandas.DataFrame): The DataFrame in which to convert year/months
fields to Date fields.
Returns:
pandas.DataFrame: A DataFrame in which the year/month fields have been
converted into Date fields.
"""
df = df.copy()
month_regex = "_month$"
year_regex = "_year$"
# Columns that match our month or year patterns.
month_cols = list(df.filter(regex=month_regex).columns)
year_cols = list(df.filter(regex=year_regex).columns)
# Base column names that don't include the month or year pattern
months_base = [re.sub(month_regex, '', m) for m in month_cols]
years_base = [re.sub(year_regex, '', y) for y in year_cols]
# We only want to retain columns that have BOTH month and year
# matches -- otherwise there's no point in creating a Date.
date_base = [base for base in months_base if base in years_base]
# For each base column that DOES have both a month and year,
# We need to grab the real column names corresponding to each,
# so we can access the values in the data frame, and use them
# to create a corresponding Date column named [BASE]_date
month_year_date = []
for base in date_base:
base_month_regex = f'^{base}{month_regex}'
month_col = list(df.filter(regex=base_month_regex).columns)
if not len(month_col) == 1:
raise AssertionError()
month_col = month_col[0]
base_year_regex = f'^{base}{year_regex}'
year_col = list(df.filter(regex=base_year_regex).columns)
if not len(year_col) == 1:
raise AssertionError()
year_col = year_col[0]
date_col = f'{base}_date'
month_year_date.append((month_col, year_col, date_col))
for month_col, year_col, date_col in month_year_date:
df = fix_int_na(df, columns=[year_col, month_col])
date_mask = (df[year_col] != '') & (df[month_col] != '')
years = df.loc[date_mask, year_col]
months = df.loc[date_mask, month_col]
df.loc[date_mask, date_col] = pd.to_datetime({
'year': years,
'month': months,
'day': 1}, errors='coerce')
# Now that we've replaced these fields with a date, we drop them.
df = df.drop([month_col, year_col], axis=1)
return df
def convert_to_date(df,
date_col="report_date",
year_col="report_year",
month_col="report_month",
day_col="report_day",
month_value=1,
day_value=1):
"""
Convert specified year, month or day columns into a datetime object.
If the input ``date_col`` already exists in the input dataframe, then no
conversion is applied, and the original dataframe is returned unchanged.
Otherwise the constructed date is placed in that column, and the columns
which were used to create the date are dropped.
Args:
df (pandas.DataFrame): dataframe to convert
date_col (str): the name of the column you want in the output.
year_col (str): the name of the year column in the original table.
month_col (str): the name of the month column in the original table.
day_col: the name of the day column in the original table.
month_value (int): generated month if no month exists.
day_value (int): generated day if no month exists.
Returns:
pandas.DataFrame: A DataFrame in which the year, month, day columns
values have been converted into datetime objects.
Todo:
Update docstring.
"""
df = df.copy()
if date_col in df.columns:
return df
year = df[year_col]
if month_col not in df.columns:
month = month_value
else:
month = df[month_col]
if day_col not in df.columns:
day = day_value
else:
day = df[day_col]
df[date_col] = pd.to_datetime({'year': year,
'month': month,
'day': day})
cols_to_drop = [x for x in [
day_col, year_col, month_col] if x in df.columns]
df.drop(cols_to_drop, axis="columns", inplace=True)
return df
def fix_eia_na(df):
"""
Replace common ill-posed EIA NA spreadsheet values with np.nan.
Args:
df (pandas.DataFrame): The DataFrame to clean.
Returns:
pandas.DataFrame: The cleaned DataFrame.
Todo:
Update docstring.
"""
return df.replace(to_replace=[r'^\.$', r'^\s$', r'^$'],
value=np.nan, regex=True)
def simplify_columns(df):
"""
Simplify column labels for use as snake_case database fields.
All columns will be re-labeled by:
* Replacing all non-alphanumeric characters with spaces.
* Forcing all letters to be lower case.
* Compacting internal whitespace to a single " ".
* Stripping leading and trailing whitespace.
* Replacing all remaining whitespace with underscores.
Args:
df (pandas.DataFrame): The DataFrame to clean.
Returns:
pandas.DataFrame: The cleaned DataFrame.
Todo:
Update docstring.
"""
df.columns = (
df.columns.
str.replace(r'[^0-9a-zA-Z]+', ' ', regex=True).
str.strip().
str.lower().
str.replace(r'\s+', ' ', regex=True).
str.replace(' ', '_')
)
return df
def find_timezone(*, lng=None, lat=None, state=None, strict=True):
"""Find the timezone associated with the a specified input location.
Note that this function requires named arguments. The names are lng, lat,
and state. lng and lat must be provided, but they may be NA. state isn't
required, and isn't used unless lng/lat are NA or timezonefinder can't find
a corresponding timezone.
Timezones based on states are imprecise, so it's far better to use lng/lat
if possible. If `strict` is True, state will not be used.
More on state-to-timezone conversion here:
https://en.wikipedia.org/wiki/List_of_time_offsets_by_U.S._state_and_territory
Args:
lng (int or float in [-180,180]): Longitude, in decimal degrees
lat (int or float in [-90, 90]): Latitude, in decimal degrees
state (str): Abbreviation for US state or Canadian province
strict (bool): Raise an error if no timezone is found?
Returns:
str: The timezone (as an IANA string) for that location.
Todo:
Update docstring.
"""
try:
tz = tz_finder.timezone_at(lng=lng, lat=lat)
if tz is None: # Try harder
# Could change the search radius as well
tz = tz_finder.closest_timezone_at(lng=lng, lat=lat)
# For some reason w/ Python 3.6 we get a ValueError here, but with
# Python 3.7 we get an OverflowError...
except (OverflowError, ValueError):
# If we're being strict, only use lng/lat, not state
if strict:
raise ValueError(
f"Can't find timezone for: lng={lng}, lat={lat}, state={state}"
)
# If, e.g., the coordinates are missing, try looking in the
# state_tz_approx dictionary.
try:
tz = pudl.constants.state_tz_approx[state]
except KeyError:
tz = None
return tz
def drop_tables(engine,
clobber=False):
"""Drops all tables from a SQLite database.
Creates an sa.schema.MetaData object reflecting the structure of the
database that the passed in ``engine`` refers to, and uses that schema to
drop all existing tables.
Todo:
Treat DB connection as a context manager (with/as).
Args:
engine (sa.engine.Engine): An SQL Alchemy SQLite database Engine
pointing at an exising SQLite database to be deleted.
Returns:
None
"""
md = sa.MetaData()
md.reflect(engine)
insp = reflection.Inspector.from_engine(engine)
if len(insp.get_table_names()) > 0 and not clobber:
raise AssertionError(
f'You are attempting to drop your database without setting clobber to {clobber}')
md.drop_all(engine)
conn = engine.connect()
conn.execute("VACUUM")
conn.close()
def merge_dicts(list_of_dicts):
"""
Merge multipe dictionaries together.
Given any number of dicts, shallow copy and merge into a new dict,
precedence goes to key value pairs in latter dicts.
Args:
dict_args (list): a list of dictionaries.
Returns:
dict
"""
merge_dict = {}
for dictionary in list_of_dicts:
merge_dict.update(dictionary)
return merge_dict
def convert_cols_dtypes(df, data_source, name=None):
"""
Convert the data types for a dataframe.
This function will convert a PUDL dataframe's columns to the correct data
type. It uses a dictionary in constants.py called column_dtypes to assign
the right type. Within a given data source (e.g. eia923, ferc1) each column
name is assumed to *always* have the same data type whenever it is found.
Boolean type conversions created a special problem, because null values in
boolean columns get converted to True (which is bonkers!)... we generally
want to preserve the null values and definitely don't want them to be True,
so we are keeping those columns as objects and preforming a simple mask for
the boolean columns.
The other exception in here is with the `utility_id_eia` column. It is
often an object column of strings. All of the strings are numbers, so it
should be possible to convert to :func:`pandas.Int32Dtype` directly, but it
is requiring us to convert to int first. There will probably be other
columns that have this problem... and hopefully pandas just enables this
direct conversion.
Args:
df (pandas.DataFrame): dataframe with columns that appear in the PUDL
tables.
data_source (str): the name of the datasource (eia, ferc1, etc.)
name (str): name of the table (for logging only!)
Returns:
pandas.DataFrame: a dataframe with columns as specified by the
:mod:`pudl.constants` ``column_dtypes`` dictionary.
"""
# get me all of the columns for the table in the constants dtype dict
col_dtypes = {col: col_dtype for col, col_dtype
in pc.column_dtypes[data_source].items()
if col in list(df.columns)}
# grab only the boolean columns (we only need their names)
bool_cols = {col: col_dtype for col, col_dtype
in col_dtypes.items()
if col_dtype == pd.BooleanDtype()}
# Grab only the string columns...
string_cols = {col: col_dtype for col, col_dtype
in col_dtypes.items()
if col_dtype == pd.StringDtype()}
# grab all of the non boolean columns
non_bool_cols = {col: col_dtype for col, col_dtype
in col_dtypes.items()
if col_dtype != pd.BooleanDtype()}
# If/when we have the columns exhaustively typed, we can do it like this,
# but right now we don't have the FERC columns done, so we can't:
# get me all of the columns for the table in the constants dtype dict
# col_types = {
# col: pc.column_dtypes[data_source][col] for col in df.columns
# }
# grab only the boolean columns (we only need their names)
# bool_cols = {col for col in col_types if col_types[col] is bool}
# grab all of the non boolean columns
# non_bool_cols = {
# col: col_types[col] for col in col_types if col_types[col] is not bool
# }
for col in bool_cols:
# Bc the og bool values were sometimes coming across as actual bools or
# strings, for some reason we need to map both types (I'm not sure
# why!). We use na_action to preserve the og NaN's. I've also added in
# the string version of a null value bc I am sure it will exist.
df[col] = df[col].map({'False': False,
'True': True,
False: False,
True: True,
'nan': pd.NA})
if name:
logger.debug(f'Converting the dtypes of: {name}')
# unfortunately, the pd.Int32Dtype() doesn't allow a conversion from object
# columns to this nullable int type column. `utility_id_eia` shows up as a
# column of strings (!) of numbers so it is an object column, and therefor
# needs to be converted beforehand.
if 'utility_id_eia' in df.columns:
# we want to be able to use this dtype cleaning at many stages, and
# sometimes this column has been converted to a float and therefor
# we need to skip this conversion
if df.utility_id_eia.dtypes is np.dtype('object'):
df = df.astype({'utility_id_eia': 'float'})
df = (
df.replace(to_replace="<NA>", value={
col: pd.NA for col in string_cols})
.replace(to_replace="nan", value={col: pd.NA for col in string_cols})
.astype(non_bool_cols)
.astype(bool_cols)
)
# Zip codes are highly coorelated with datatype. If they datatype gets
# converted at any point it may mess up the accuracy of the data. For
# example: 08401.0 or 8401 are both incorrect versions of 080401 that a
# simple datatype conversion cannot fix. For this reason, we use the
# zero_pad_zips function.
if any('zip_code' for col in df.columns):
zip_cols = [col for col in df.columns if 'zip_code' in col]
for col in zip_cols:
if '4' in col:
df[col] = zero_pad_zips(df[col], 4)
else:
df[col] = zero_pad_zips(df[col], 5)
return df
def convert_dfs_dict_dtypes(dfs_dict, data_source):
"""Convert the data types of a dictionary of dataframes.
This is a wrapper for :func:`pudl.helpers.convert_cols_dtypes` which loops
over an entire dictionary of dataframes, assuming they are all from the
specified data source, and appropriately assigning data types to each
column based on the data source specific type map stored in pudl.constants
"""
cleaned_dfs_dict = {}
for name, df in dfs_dict.items():
cleaned_dfs_dict[name] = convert_cols_dtypes(df, data_source, name)
return cleaned_dfs_dict
def generate_rolling_avg(df, group_cols, data_col, window, **kwargs):
"""
Generate a rolling average.
For a given dataframe with a ``report_date`` column, generate a monthly
rolling average and use this rolling average to impute missing values.
Args:
df (pandas.DataFrame): Original dataframe. Must have group_cols
column, a data_col column and a ``report_date`` column.
group_cols (iterable): a list of columns to groupby.
data_col (str): the name of the data column.
window (int): window from :func:`pandas.Series.rolling`.
kwargs : Additional arguments to pass to
:func:`pandas.Series.rolling`.
Returns:
pandas.DataFrame
"""
df = df.astype({'report_date': 'datetime64[ns]'})
# create a full date range for this df
date_range = (pd.DataFrame(pd.date_range(
start=min(df['report_date']),
end=max(df['report_date']), freq='MS',
name='report_date')).
# assiging a temp column to merge on
assign(tmp=1))
groups = (df[group_cols + ['report_date']].
drop_duplicates().
# assiging a temp column to merge on
assign(tmp=1))
# merge the date range and the groups together
# to get the backbone/complete date range/groups
bones = (date_range.merge(groups).
# drop the temp column
drop('tmp', axis=1).
# then merge the actual data onto the
merge(df, on=group_cols + ['report_date']).
set_index(group_cols + ['report_date']).
groupby(by=group_cols + ['report_date']).
mean())
# with the aggregated data, get a rolling average
roll = (bones.rolling(window=window, center=True, **kwargs).
agg({data_col: 'mean'})
)
# return the merged
return bones.merge(roll,
on=group_cols + ['report_date'],
suffixes=('', '_rolling')).reset_index()
def fillna_w_rolling_avg(df_og, group_cols, data_col, window=12, **kwargs):
"""
Filling NaNs with a rolling average.
Imputes null values from a dataframe on a rolling monthly average. To note,
this was designed to work with the PudlTabl object's tables.
Args:
df_og (pandas.DataFrame): Original dataframe. Must have group_cols
column, a data_col column and a 'report_date' column.
group_cols (iterable): a list of columns to groupby.
data_col (str): the name of the data column.
window (int): window from pandas.Series.rolling
kwargs : Additional arguments to pass to
:class:`pandas.Series.rolling`.
Returns:
pandas.DataFrame: dataframe with nulls filled in.
"""
df_og = df_og.astype({'report_date': 'datetime64[ns]'})
df_roll = generate_rolling_avg(df_og, group_cols, data_col,
window, **kwargs)
df_roll[data_col] = df_roll[data_col].fillna(
df_roll[f'{data_col}_rolling'])
df_new = df_og.merge(df_roll,
how='left',
on=group_cols + ['report_date'],
suffixes=('', '_rollfilled'))
df_new[data_col] = df_new[data_col].fillna(
df_new[f'{data_col}_rollfilled'])
return df_new.drop(columns=[f'{data_col}_rollfilled', f'{data_col}_rolling'])
def count_records(df, cols, new_count_col_name):
"""
Count the number of unique records in group in a dataframe.
Args:
df (panda.DataFrame) : dataframe you would like to groupby and count.
cols (iterable) : list of columns to group and count by.
new_count_col_name (string) : the name that will be assigned to the
column that will contain the count.
Returns:
pandas.DataFrame: dataframe with only the `cols` definted and the
`new_count_col_name`.
"""
return (df.assign(count_me=1).
groupby(cols).
agg({'count_me': 'count'}).
reset_index().
rename(columns={'count_me': new_count_col_name}))
def cleanstrings_snake(df, cols):
"""
Clean the strings in a columns in a dataframe with snake case.
Args:
df (panda.DataFrame) : original dataframe.
cols (list): list of columns in `df` to apply snake case to.
"""
for col in cols:
df.loc[:, col] = (
df[col].astype(str).
str.strip().
str.lower().
str.replace(r'\s+', '_', regex=True)
)
return df
def zero_pad_zips(zip_series, n_digits):
"""
Retain prefix zeros in zipcodes.
Args:
zip_series (pd.Series) : series containing the zipcode values.
n_digits(int) : zipcode length (likely 4 or 5 digits).
Returns:
pandas.Series: a series containing zipcodes with their prefix zeros
intact and invalid zipcodes rendered as na.
"""
# Add preceeding zeros where necessary and get rid of decimal zeros
def get_rid_of_decimal(series):
return series.str.replace(r'[\.]+\d*', '', regex=True)
zip_series = (
zip_series
.astype(pd.StringDtype())
.replace('nan', np.nan)
.fillna("0")
.pipe(get_rid_of_decimal)
.str.zfill(n_digits)
.replace({n_digits * "0": pd.NA}) # All-zero Zip codes aren't valid.
)
return zip_series
def iterate_multivalue_dict(**kwargs):
"""Make dicts from dict with main dict key and one value of main dict."""
single_valued = {k: v for k,
v in kwargs.items()
if not (isinstance(v, list) or isinstance(v, tuple))}
# Transform multi-valued {k: vlist} into {k1: [{k1: v1}, {k1: v2}, ...], k2: [...], ...}
multi_valued = {k: [{k: v} for v in vlist]
for k, vlist in kwargs.items()
if (isinstance(vlist, list) or isinstance(vlist, tuple))}
for value_assignments in itertools.product(*multi_valued.values()):
result = dict(single_valued)
for k_v in value_assignments:
result.update(k_v)
yield result
def get_working_eia_dates():
"""Get all working EIA dates as a DatetimeIndex."""
dates = pd.DatetimeIndex([])
for dataset_name, dataset in pc.working_partitions.items():
if 'eia' in dataset_name:
for name, partition in dataset.items():
if name == 'years':
dates = dates.append(
| pd.to_datetime(partition, format='%Y') | pandas.to_datetime |
import numpy as np
import pandas as pd
import os
import csv
import scipy
import torch
import torch.nn as nn
from torch_geometric.data import Data, Batch
from torch_geometric.nn import graclus, max_pool
from sklearn.preprocessing import StandardScaler
from sklearn.impute import SimpleImputer
def get_genes_graph(genes_path, save_path, method='pearson', thresh=0.95, p_value=False):
"""
determining adjaceny matrix based on correlation
:param genes_exp_path:
:return:
"""
if not os.path.exists(save_path):
os.makedirs(save_path)
genes_exp_df = pd.read_csv(os.path.join(genes_path, 'exp.csv'), index_col=0)
# calculate correlation matrix
genes_exp_corr = genes_exp_df.corr(method=method)
genes_exp_corr = genes_exp_corr.apply(lambda x: abs(x))
n = genes_exp_df.shape[0]
# binarize
if p_value == True:
dist = scipy.stats.beta(n / 2 - 1, n / 2 - 1, loc=-1, scale=2)
thresh = dist.isf(0.05)
adj = np.where(genes_exp_corr > thresh, 1, 0)
adj = adj - np.eye(genes_exp_corr.shape[0], dtype=np.int)
edge_index = np.nonzero(adj)
np.save(os.path.join(save_path, 'edge_index_{}_{}.npy').format(method, thresh), edge_index)
return n, edge_index
def ensp_to_hugo_map():
with open('./data/9606.protein.info.v11.0.txt') as csv_file:
next(csv_file) # Skip first line
csv_reader = csv.reader(csv_file, delimiter='\t')
ensp_map = {row[0]: row[1] for row in csv_reader if row[0] != ""}
return ensp_map
def hugo_to_ncbi_map():
with open('./data/enterez_NCBI_to_hugo_gene_symbol_march_2019.txt') as csv_file:
next(csv_file) # Skip first line
csv_reader = csv.reader(csv_file, delimiter='\t')
hugo_map = {row[0]: int(row[1]) for row in csv_reader if row[1] != ""}
return hugo_map
def save_cell_graph(genes_path, save_path):
if not os.path.exists(save_path):
os.makedirs(save_path)
exp = pd.read_csv(os.path.join(genes_path, 'exp.csv'), index_col=0)
cn = pd.read_csv(os.path.join(genes_path, 'cn.csv'), index_col=0)
mu = pd.read_csv(os.path.join(genes_path, 'mu.csv'), index_col=0)
# me = pd.read_csv(os.path.join(genes_path, 'me.csv'), index_col=0)
# print('Miss values:{},{},{}, {}'.format(exp.isna().sum().sum(), cn.isna().sum().sum(), mu.isna().sum().sum(),
# me.isna().sum().sum()))
index = exp.index
columns = exp.columns
scaler = StandardScaler()
exp = scaler.fit_transform(exp)
cn = scaler.fit_transform(cn)
# me = scaler.fit_transform(me)
imp_mean = SimpleImputer()
exp = imp_mean.fit_transform(exp)
exp = pd.DataFrame(exp, index=index, columns=columns)
cn = pd.DataFrame(cn, index=index, columns=columns)
mu = pd.DataFrame(mu, index=index, columns=columns)
# me = pd.DataFrame(me, index=index, columns=columns)
cell_names = exp.index
# print('Miss values:{},{},{}, {}'.format(exp.isna().sum().sum(), cn.isna().sum().sum(), mu.isna().sum().sum(),
# me.isna().sum().sum()))
cell_dict = {}
for i in cell_names:
# cell_dict[i] = Data(x=torch.tensor([exp.loc[i]], dtype=torch.float).T)
# cell_dict[i] = Data(x=torch.tensor([cn.loc[i]], dtype=torch.float).T)
# cell_dict[i] = Data(x=torch.tensor([mu.loc[i]], dtype=torch.float).T)
cell_dict[i] = Data(x=torch.tensor([exp.loc[i], cn.loc[i], mu.loc[i]], dtype=torch.float).T)
# cell_dict[i] = Data(x=torch.tensor([exp.loc[i], cn.loc[i], mu.loc[i], me.loc[i]], dtype=torch.float).T)
# cell_dict[i] = [np.array(exp.loc[i], dtype=np.float32), np.array(cn.loc[i], dtype=np.float32),
# np.array(mu.loc[i], dtype=np.float32)]
np.save(os.path.join(save_path, 'cell_feature_cn_std.npy'), cell_dict)
print("finish saving cell mut data!")
def get_STRING_graph(genes_path, thresh=0.95):
save_path = os.path.join(genes_path, 'edge_index_PPI_{}.npy'.format(thresh))
if not os.path.exists(save_path):
# gene_list
exp = pd.read_csv(os.path.join(genes_path, 'exp.csv'), index_col=0)
gene_list = exp.columns.to_list()
gene_list = [int(gene[1:-1]) for gene in gene_list]
# load STRING
ensp_map = ensp_to_hugo_map()
hugo_map = hugo_to_ncbi_map()
edges = | pd.read_csv('./data/9606.protein.links.detailed.v11.0.txt', sep=' ') | pandas.read_csv |
# Data source: College Scorecard
import ssl
import pandas as pd
from ._data_processing import DataProcessor, MisValueFiller
class Dataset:
def __init__(self, path='https://raw.githubusercontent.com/alisoltanirad/'
'CDA/main/cda/college_scorecard/'
'college_scorecard.csv'):
ssl._create_default_https_context = ssl._create_unverified_context
self._dataset = pd.read_csv(path, dtype='unicode')
self._data_processor = DataProcessor()
self._mis_value_filler = MisValueFiller()
self.college_names = self._dataset['instnm']
self.ownership = self._data_processor._ownership_types(
self._dataset['control']
)
class CollegeData(Dataset):
def __init__(self, path='https://raw.githubusercontent.com/alisoltanirad/'
'CDA/main/cda/college_scorecard/'
'college_scorecard.csv'):
Dataset.__init__(self, path)
self._set_general_info()
self._set_fiscal_info()
self._set_evaluation_metrics()
def get_info(self):
data = {
'Name': self.college_names,
'Ownership': self.ownership,
'State': self.state,
'Student_Size': self.student_size,
'Is_Online_Only': self.online_only,
'Is_Men_Only': self.men_only,
'Is_Women_Only': self.women_only,
'Is_Religious_Affiliate': self.religious_affiliate,
'Is_For_Profit': self.for_profit,
'Tuition_Revenue': self.tuition_revenue,
'Instructional_Expenditure': self.instructional_expenditure,
'Faculty_Salary': self.faculty_salary,
'Faculty_Full_Time_Rate': self.faculty_fulltime_rate,
'Highest_Degree': self.highest_degrees,
}
return pd.DataFrame(data)
def get_evaluation_metrics(self):
data = {
'Name': self.college_names,
'Admission_Rate': self.admission_rates,
'Completion_Rate_Overall': self.completion_rate_avg,
'SAT_Scores_Overall': self.sat_scores,
}
return pd.DataFrame(data)
def _set_general_info(self):
self.state = self._dataset['stabbr']
self.student_size = self._mis_value_filler.mean(
self._dataset['ugds'], int
)
self.online_only = self._dataset['distanceonly'].fillna('false')
self.men_only = self._dataset['menonly'].fillna('false')
self.women_only = self._dataset['womenonly'].fillna('false')
self.religious_affiliate = self._data_processor._is_religious_affiliate(
self._dataset['relaffil']
)
self.for_profit = self._data_processor._is_for_profit(
self._dataset['control']
)
self.highest_degrees = self._data_processor._degree_types(
self._dataset['highdeg']
)
def _set_fiscal_info(self):
self.tuition_revenue = self._mis_value_filler.mean(
self._dataset['tuitfte'], int
)
self.instructional_expenditure = self._mis_value_filler.mean(
self._dataset['inexpfte'], int
)
self.faculty_salary = self._mis_value_filler.mean(
self._dataset['avgfacsal'], int
)
self.faculty_fulltime_rate = self._mis_value_filler.mean(
self._dataset['pftfac'], float
)
def _set_evaluation_metrics(self):
self.admission_rates = self._dataset['adm_rate']
completion_2yr = self._dataset['overall_yr2_n']
completion_3yr = self._dataset['overall_yr3_n']
completion_4yr = self._dataset['overall_yr4_n']
completion_6yr = self._dataset['overall_yr6_n']
completion_8yr = self._dataset['overall_yr8_n']
self.completion_rate_avg = self._data_processor._list_average([
completion_2yr,
completion_3yr,
completion_4yr,
completion_6yr,
completion_8yr
])
self.sat_scores = self._dataset['sat_avg']
class StudentData(Dataset):
def __init__(self, path='https://raw.githubusercontent.com/alisoltanirad/'
'CDA/main/cda/college_scorecard/'
'college_scorecard.csv'):
Dataset.__init__(self, path)
self._set_general_info()
self._set_race_info()
self._set_family_info()
def get_info(self):
data = {
'Name': self.college_names,
'Ownership': self.ownership,
'Part_Time_Share': self.part_time_share,
'Race_White': self.race_white,
'Race_Black': self.race_black,
'Race_Hispanic': self.race_hispanic,
'Race_Asian': self.race_asian,
'Race_AIAN': self.race_aian,
'Race_NHPI': self.race_nhpi,
'Race_Mixed': self.race_mixed,
'Family_Income_Dependent': self.family_income_dependent,
'Family_Income_Independent': self.family_income_independent,
}
return pd.DataFrame(data)
def _set_general_info(self):
self.part_time_share = self._mis_value_filler.mean(
self._dataset['pptug_ef'], float
)
def _set_race_info(self):
self.race_white = self._mis_value_filler.mean(
self._dataset['ugds_white'], float
)
self.race_black = self._mis_value_filler.mean(
self._dataset['ugds_black'], float
)
self.race_hispanic = self._mis_value_filler.mean(
self._dataset['ugds_hisp'], float
)
self.race_asian = self._mis_value_filler.mean(
self._dataset['ugds_asian'], float
)
self.race_aian = self._mis_value_filler.mean(
self._dataset['ugds_aian'], float
)
self.race_nhpi = self._mis_value_filler.mean(
self._dataset['ugds_nhpi'], float
)
self.race_mixed = self._mis_value_filler.mean(
self._dataset['ugds_2mor'], float
)
def _set_family_info(self):
self.family_income_dependent = self._mis_value_filler.mean(
self._dataset['dep_inc_n'], int
)
self.family_income_independent = self._mis_value_filler.mean(
self._dataset['ind_inc_n'], int
)
class FinancialData(Dataset):
def __init__(self, path='https://raw.githubusercontent.com/alisoltanirad/'
'CDA/main/cda/college_scorecard/'
'college_scorecard.csv'):
Dataset.__init__(self, path)
self._set_cost_info()
self._set_aid_info()
self._set_family_info()
def get_cost_info(self):
data = {
'Name': self.college_names,
'Ownership': self.ownership,
'Net_Price': self.net_price,
'Attendance_Cost': self.attendance_cost,
'Tuition_In_State': self.tuition_in_state,
'Tuition_Out_State': self.tuition_out_state,
}
return pd.DataFrame(data)
def get_aid_info(self):
data = {
'Name': self.college_names,
'Ownership': self.ownership,
'Title_IV': self.title_iv,
'Federal_Loan_Rate': self.federal_loan_rate,
'Debt_Overall': self.debt_overall,
'Debt_Completers': self.debt_completers,
'Debt_NonCompleters': self.debt_noncompleters,
'Debt_Dependent': self.debt_dependent,
'Debt_Independent': self.debt_independent,
'Family_Income_Dependent': self.family_income_dependent,
'Family_Income_Independent': self.family_income_independent,
}
return | pd.DataFrame(data) | pandas.DataFrame |
import pandas as pd
import numpy as np
from sklearn.ensemble.forest import RandomForestRegressor
from sklearn.model_selection import train_test_split
from sklearn.metrics import mean_squared_error, mean_absolute_error, r2_score, median_absolute_error
from sklearn.preprocessing import MinMaxScaler, StandardScaler
import pickle
chicago_bike_data_Q1= pd.read_csv('datasets/Divvy_Trips_2018_Q1.csv')
chicago_bike_data_Q2= pd.read_csv('datasets/Divvy_Trips_2018_Q2.csv')
chicago_bike_data_Q3= pd.read_csv('datasets/Divvy_Trips_2018_Q3.csv')
chicago_bike_data_Q4= pd.read_csv('datasets/Divvy_Trips_2018_Q4.csv')
chicago_bike_data_Q1 = chicago_bike_data_Q1.rename(columns={"03 - Rental Start Station Name":"from_station_name",
"02 - Rental End Station Name":"to_station_name","01 - Rental Details Rental ID":"trip_id"})
data_groupby_day_out_Q1 = pd.DataFrame(chicago_bike_data_Q1.groupby(['from_station_name'])['trip_id'].count()).reset_index()
data_groupby_day_in_Q1 = pd.DataFrame(chicago_bike_data_Q1.groupby(['to_station_name'])['trip_id'].count()).reset_index()
data_groupby_day_out_Q1= data_groupby_day_out_Q1.rename(columns={"trip_id": "Number Of Outgoing Trips",
"from_station_name":"Station Name"})
data_groupby_day_in_Q1 = data_groupby_day_in_Q1.rename(columns={"trip_id": "Number Of Incoming Trips",
"to_station_name":"Station Name"})
new_df_Q1=pd.merge(data_groupby_day_out_Q1,data_groupby_day_in_Q1,on=['Station Name'],how='outer')
data_groupby_day_out_Q2 = pd.DataFrame(chicago_bike_data_Q2.groupby(['from_station_name'])['trip_id'].count()).reset_index()
data_groupby_day_in_Q2 = pd.DataFrame(chicago_bike_data_Q2.groupby(['to_station_name'])['trip_id'].count()).reset_index()
data_groupby_day_out_Q2= data_groupby_day_out_Q2.rename(columns={"trip_id": "Number Of Outgoing Trips",
"from_station_name":"Station Name"})
data_groupby_day_in_Q2 = data_groupby_day_in_Q2.rename(columns={"trip_id": "Number Of Incoming Trips",
"to_station_name":"Station Name"})
new_df_Q2=pd.merge(data_groupby_day_out_Q2,data_groupby_day_in_Q2,on=['Station Name'],how='outer')
data_groupby_day_out_Q3 = pd.DataFrame(chicago_bike_data_Q3.groupby(['from_station_name'])['trip_id'].count()).reset_index()
data_groupby_day_in_Q3 = pd.DataFrame(chicago_bike_data_Q3.groupby(['to_station_name'])['trip_id'].count()).reset_index()
data_groupby_day_out_Q3= data_groupby_day_out_Q3.rename(columns={"trip_id": "Number Of Outgoing Trips",
"from_station_name":"Station Name"})
data_groupby_day_in_Q3 = data_groupby_day_in_Q3.rename(columns={"trip_id": "Number Of Incoming Trips",
"to_station_name":"Station Name"})
new_df_Q3=pd.merge(data_groupby_day_out_Q3,data_groupby_day_in_Q3,on=['Station Name'],how='outer')
data_groupby_day_out_Q4 = pd.DataFrame(chicago_bike_data_Q4.groupby(['from_station_name'])['trip_id'].count()).reset_index()
data_groupby_day_in_Q4 = pd.DataFrame(chicago_bike_data_Q4.groupby(['to_station_name'])['trip_id'].count()).reset_index()
data_groupby_day_out_Q4= data_groupby_day_out_Q4.rename(columns={"trip_id": "Number Of Outgoing Trips",
"from_station_name":"Station Name"})
data_groupby_day_in_Q4 = data_groupby_day_in_Q4.rename(columns={"trip_id": "Number Of Incoming Trips",
"to_station_name":"Station Name"})
new_df_Q4=pd.merge(data_groupby_day_out_Q4,data_groupby_day_in_Q4,on=['Station Name'],how='outer')
new_df = pd.concat([new_df_Q1,new_df_Q2,new_df_Q3,new_df_Q4])
top_rentals = new_df.sort_values(['Number Of Outgoing Trips'], ascending=False).iloc[0:10,0:2].reset_index()
chicago_bike_data_Q1['day'] = [int(str(starttime).split(" ")[0].split("-")[2]) for starttime in chicago_bike_data_Q1['01 - Rental Details Local Start Time']]
chicago_bike_data_Q1['month'] = [int(str(starttime).split(" ")[0].split("-")[1]) for starttime in chicago_bike_data_Q1['01 - Rental Details Local Start Time']]
chicago_bike_data_Q1['hour'] = [int(str(starttime).split(" ")[1].split(":")[0]) for starttime in chicago_bike_data_Q1['01 - Rental Details Local Start Time']]
chicago_bike_data_Q2['day'] = [int(str(starttime).split(" ")[0].split("-")[2]) for starttime in chicago_bike_data_Q2['start_time']]
chicago_bike_data_Q2['month'] = [int(str(starttime).split(" ")[0].split("-")[1]) for starttime in chicago_bike_data_Q2['start_time']]
chicago_bike_data_Q2['hour'] = [int(str(starttime).split(" ")[1].split(":")[0]) for starttime in chicago_bike_data_Q2['start_time']]
chicago_bike_data_Q3['day'] = [int(str(starttime).split(" ")[0].split("-")[2]) for starttime in chicago_bike_data_Q3['start_time']]
chicago_bike_data_Q3['month'] = [int(str(starttime).split(" ")[0].split("-")[1]) for starttime in chicago_bike_data_Q3['start_time']]
chicago_bike_data_Q3['hour'] = [int(str(starttime).split(" ")[1].split(":")[0]) for starttime in chicago_bike_data_Q3['start_time']]
chicago_bike_data_Q4['day'] = [int(str(starttime).split(" ")[0].split("-")[2]) for starttime in chicago_bike_data_Q4['start_time']]
chicago_bike_data_Q4['month'] = [int(str(starttime).split(" ")[0].split("-")[1]) for starttime in chicago_bike_data_Q4['start_time']]
chicago_bike_data_Q4['hour'] = [int(str(starttime).split(" ")[1].split(":")[0]) for starttime in chicago_bike_data_Q4['start_time']]
data_groupby_day_out_Q1 = pd.DataFrame(chicago_bike_data_Q1.groupby(['from_station_name', 'month','day','hour'])['trip_id'].count()).reset_index()
data_groupby_day_in_Q1 = pd.DataFrame(chicago_bike_data_Q1.groupby(['to_station_name','month','day','hour'])['trip_id'].count()).reset_index()
data_groupby_day_out_Q1= data_groupby_day_out_Q1.rename(columns={"trip_id": "Number Of Outgoing Trips",
"from_station_name":"Station Name"})
data_groupby_day_in_Q1 = data_groupby_day_in_Q1.rename(columns={"trip_id": "Number Of Incoming Trips",
"to_station_name":"Station Name"})
new_df_Q1=pd.merge(data_groupby_day_out_Q1,data_groupby_day_in_Q1,on=['Station Name','month','day','hour'],how='outer')
new_df_Q1 = new_df_Q1.sort_values(['month','day','hour'], ascending=True).reset_index()
data_groupby_day_out_Q2 = pd.DataFrame(chicago_bike_data_Q2.groupby(['from_station_name', 'month','day','hour'])['trip_id'].count()).reset_index()
data_groupby_day_in_Q2 = pd.DataFrame(chicago_bike_data_Q2.groupby(['to_station_name','month','day','hour'])['trip_id'].count()).reset_index()
data_groupby_day_out_Q2= data_groupby_day_out_Q2.rename(columns={"trip_id": "Number Of Outgoing Trips",
"from_station_name":"Station Name"})
data_groupby_day_in_Q2 = data_groupby_day_in_Q2.rename(columns={"trip_id": "Number Of Incoming Trips",
"to_station_name":"Station Name"})
new_df_Q2=pd.merge(data_groupby_day_out_Q2,data_groupby_day_in_Q2,on=['Station Name','month','day','hour'],how='outer')
new_df_Q2 = new_df_Q2.sort_values(['month','day','hour'], ascending=True).reset_index()
data_groupby_day_out_Q3 = pd.DataFrame(chicago_bike_data_Q3.groupby(['from_station_name', 'month','day','hour'])['trip_id'].count()).reset_index()
data_groupby_day_in_Q3 = pd.DataFrame(chicago_bike_data_Q3.groupby(['to_station_name','month','day','hour'])['trip_id'].count()).reset_index()
data_groupby_day_out_Q3= data_groupby_day_out_Q3.rename(columns={"trip_id": "Number Of Outgoing Trips",
"from_station_name":"Station Name"})
data_groupby_day_in_Q3 = data_groupby_day_in_Q3.rename(columns={"trip_id": "Number Of Incoming Trips",
"to_station_name":"Station Name"})
new_df_Q3=pd.merge(data_groupby_day_out_Q3,data_groupby_day_in_Q3,on=['Station Name','month','day','hour'],how='outer')
new_df_Q3 = new_df_Q3.sort_values(['month','day','hour'], ascending=True).reset_index()
data_groupby_day_out_Q4 = pd.DataFrame(chicago_bike_data_Q4.groupby(['from_station_name', 'month','day','hour'])['trip_id'].count()).reset_index()
data_groupby_day_in_Q4 = pd.DataFrame(chicago_bike_data_Q4.groupby(['to_station_name','month','day','hour'])['trip_id'].count()).reset_index()
data_groupby_day_out_Q4= data_groupby_day_out_Q4.rename(columns={"trip_id": "Number Of Outgoing Trips",
"from_station_name":"Station Name"})
data_groupby_day_in_Q4 = data_groupby_day_in_Q4.rename(columns={"trip_id": "Number Of Incoming Trips",
"to_station_name":"Station Name"})
new_df_Q4=pd.merge(data_groupby_day_out_Q4,data_groupby_day_in_Q4,on=['Station Name','month','day','hour'],how='outer')
new_df_Q4 = new_df_Q4.sort_values(['month','day','hour'], ascending=True).reset_index()
new_df = pd.concat([new_df_Q1,new_df_Q2,new_df_Q3,new_df_Q4])
new_df['Number Of Outgoing Trips'] = new_df['Number Of Outgoing Trips'].fillna(0)
new_df['Number Of Incoming Trips'] = new_df['Number Of Incoming Trips'].fillna(0)
def get_processed_station_data(new_df, station_name):
new_single_station_df = new_df.loc[new_df['Station Name']==station_name]
new_single_station_df = new_single_station_df.sort_values(['month','day'], ascending=True)
for month in set(new_single_station_df['month']):
new_single_station_month_df = new_single_station_df.loc[new_single_station_df['month']==month]
for day in set(new_single_station_month_df['day']):
new_single_station_day_df = new_single_station_month_df.loc[new_single_station_month_df['day']==day]
list_of_hours = new_single_station_day_df['hour']
for i in range(0,24):
if(i not in set(list_of_hours)):
app_df = | pd.DataFrame([(month,day,i,0)], columns=['month','day','hour','Number Of Outgoing Trips']) | pandas.DataFrame |
#!/usr/bin/env python
# -*- coding: utf-8 -*-
"""DeepPrecip Module
<NAME> 2022
This is the alternate main executable for DeepPrecip that includes the code necessary for running the model on GraphCore IPUs.
You can adjust model hyperparams in the global variable definition section.
For more information on how to run the model, please view our GitHub page: https://github.com/frasertheking/DeepPrecip
"""
####################################################################################################################################
############ Imports
import sys,os,io
import time
import pandas as pd
import numpy as np
import seaborn as sb
import matplotlib.pyplot as plt
import sklearn
import tensorflow.keras as keras
from tensorflow.keras.callbacks import ModelCheckpoint
from sklearn import metrics
from sklearn.model_selection import train_test_split
from sklearn.preprocessing import StandardScaler
from tensorflow.python import ipu
####################################################################################################################################
############ Global Variables
DATA_PATH = "data/"
MIN_AVG = 20
USE_SHUFFLE = False
RANDOM_STATE = None
BATCH_SIZE = 128
MAX_EPOCHS = 3000
PRED_COUNT = 256
RUN_TIME = str(int(time.time()))
os.mkdir('runs/' + RUN_TIME)
os.environ["TF_POPLAR_FLAGS"] = "--executable_cache_path=./cache"
####################################################################################################################################
############ Class Definitions
# Used for holding ERA5-L data
class model_metrics(object):
name = ""
index_val = []
y_test = []
y_pred = []
def __init__(self, name, index_val, y_test, y_pred):
self.name = name
self.index_val = index_val
self.y_test = y_test
self.y_pred = y_pred
def mse(self):
return metrics.mean_squared_error(self.y_test, self.y_pred)
def corr(self):
return np.corrcoef(self.y_test, self.y_pred)[0][1]
def mae(self):
return metrics.mean_absolute_error(self.y_test, self.y_pred)
def r2(self):
return metrics.r2_score(self.y_test, self.y_pred)
def export_metrics():
return self.mse(), self.corr(), self.mae(), self.r2()
def data_length(self):
return len(self.y_pred)
def max_val(self):
max_val = max(self.y_pred)
if (max(self.y_test)) > max_val:
max_val = max(self.y_test)
return max_val
def summary(self):
print("\n####################\n")
print(self.name + " STATS (n=" + str(self.data_length()) + "):" + "\nMSE: " + str(round(self.mse(),5)) + \
"\nCorrelation: " + str(round(self.corr(),5)) +\
"\nMean Absolute Error: " + str(round(self.mae(),5)) +\
"\nR-Squared: " + str(round(self.r2(),5)))
print("\n####################\n")
def scatter(self):
stats = self.name + " STATS (n=" + str(self.data_length()) + "):" + "\nMSE: " + str(round(self.mse(),5)) + \
"\nCorrelation: " + str(round(self.corr(),5)) +\
"\nMean Absolute Error: " + str(round(self.mae(),5)) +\
"\nR-Squared: " + str(round(self.r2(),5))
fig, ax=plt.subplots(figsize=(10,10))
plt.grid(linestyle='--')
plt.title(self.name + ' Actual vs Predicted Values')
plt.xlabel('Predicted Accumulation (mm SWE)')
plt.ylabel('Observed Accumulation (mm SWE)')
plt.xlim((0, self.max_val()))
plt.ylim((0, self.max_val()))
plt.scatter(self.y_pred, self.y_test,color='red', alpha=0.25)
plt.plot([0, self.max_val()], [0, self.max_val()], linestyle='--', color='black')
plt.text(0.02, 0.9, stats, horizontalalignment='left', verticalalignment='center', transform=ax.transAxes, fontsize=16)
plt.savefig('runs/' + RUN_TIME + '/scatter_full_column.png', DPI=300)
def timeseries(self):
roll_y_test = | pd.Series(self.y_test) | pandas.Series |
import pandas as pd
pd.set_option('display.max_rows', 500)
pd.set_option('display.max_columns', 500)
from elecsim.constants import ROOT_DIR, KW_TO_MW
import numpy as np
from scipy.optimize import root
KTOE_TO_MWH = 11630
investment_mechanism = "future_price_fit"
# investment_mechanism = "projection_fit"
potential_plants_to_invest = ['CCGT H Class', 'CCGT F Class', 'CCGT CHP mode', 'Coal - CCS ASC NH3 FOAK', 'Coal - CCS ASC Partial FOAK', 'Coal - CCS ASC FOAK', 'Nuclear - PWR FOAK', 'Onshore UK>5MW', 'Offshore R2', 'Offshore R3', 'PV>5MW', 'Recip Gas 2000 hr', 'RECIP GAS 500 hrs', 'Recip Diesel 2000 hr', 'Recip Diesel 500 hrs', 'Recip Diesel 90 hrs']
multi_year_data = pd.read_csv('{}/data/processed/multi_day_data/4_medoids.csv'.format(ROOT_DIR))
multi_year_data_scaled = pd.read_csv('{}/data/processed/multi_day_data/4_medoids_scaled.csv'.format(ROOT_DIR))
# BEIS Final Electricity Consumption
electricity_ktoe = [24822.84204, 24470.32991, 24301.10466, 24288.25851, 24438.20398, 24717.02662, 24995.8011, 25361.32776, 25813.71342, 26342.63171, 26893.41806, 27471.23698, 27867.29557, 28262.51807, 28754.44771, 29191.80642, 29648.75735]
renewables_ktoe = [7609.872531, 8007.503459, 8179.451416, 8267.036507, 8314.81648, 8323.254324, 8317.588187, 8318.702017, 8325.651362, 8328.676807, 8335.161717, 8333.378709, 8327.276614, 8303.48562, 8198.764019, 7988.909411, 7737.627294]
final_consumption = [(elec+renew) * 11630 for elec, renew in zip(electricity_ktoe, renewables_ktoe)]
def get_difference(scaler, df, required_mwh):
load_dat = df[df.data_type=='load']
load_total = load_dat.capacity_factor * scaler
return load_total.sum() - required_mwh
demand_sizes = [root(fun=get_difference, x0=50000, args=(multi_year_data_scaled, consumption)).x for consumption in final_consumption]
demand_sizes_repeated = np.repeat(demand_sizes, 8).tolist()
# Demand per segment of load duration function
segment_demand_diff = [17568, 21964, 23127, 24327, 25520, 26760, 27888, 28935, 29865, 30721, 31567, 32315, 33188, 34182, 35505, 37480, 39585, 42206, 45209, 52152]
segment_demand = [52152, 45209, 42206, 39585, 37480, 35505, 34182, 33188, 32315, 31567, 30721, 29865, 28935, 27888, 26760, 25520, 24327, 23127, 21964, 17568]
# Time of load duration function
segment_time = [8752.5, 8291.83, 7831.17, 7370.5, 6909.92, 6449.25, 5988.58, 5527.92, 5067.25, 4606.58, 4146, 3685.33, 3224.67, 2764, 2303.33, 1842.67, 1382.08, 921.42, 460.75, 0.08]
# Change in load duration function by year
# yearly_demand_change = [0.949620, 0.959511, 0.979181, 0.984285, 0.987209, 0.983118]
yearly_demand_change = [1-((b[0] - a[0]) / a[0]) for a, b in zip(demand_sizes[::1], demand_sizes[1::1])]
# First year maximum demand size
initial_max_demand_size = demand_sizes[0]
# Electricity Prices
electricity_volume_weighted = [58, 56, 53, 52, 53, 54, 57, 58, 58, 60, 58, 58, 59, 61, 60, 63, 60, 58]
electricity_baseload = [58, 55, 52, 51, 53, 53, 56, 57, 57, 59, 57, 57, 58, 59, 59, 61, 58, 56]
multi_year_data = pd.read_csv('{}/data/processed/multi_day_data/4_medoids.csv'.format(ROOT_DIR))
multi_year_data_scaled = pd.read_csv('{}/data/processed/multi_day_data/4_medoids_scaled.csv'.format(ROOT_DIR))
# Fuel prices (£/MWh)
# Historical fuel prices of coal, oil and gas Source: Average prices of fuels purchased by the major UK power producers, BEIS UK Government, table_321.xlsx
historical_fuel_prices_long = pd.read_csv('{}/data/processed/fuel/fuel_costs/historical_fuel_costs/historical_fuel_costs_converted_long.csv'.format(ROOT_DIR))
historical_fuel_prices_mw = pd.read_csv('{}/data/processed/fuel/fuel_costs/historical_fuel_costs/fuel_costs_per_mwh.csv'.format(ROOT_DIR))
# Future $/GBP exchange rate
dollar_gbp_exchange_rate = [1.36, 1.38, 1.40, 1.40, 1.40, 1.40, 1.40, 1.40, 1.40, 1.40, 1.40, 1.40, 1.40, 1.40, 1.40, 1.40, 1.40]
# Future fuel prices
gas_scenario = [53.0, 48.0, 49.0, 51.0, 52.0, 54.0, 56.0, 57.0, 59.0, 60.0, 62.0, 63.0, 63.0, 63.0, 63.0, 63.0, 63.0]
gas_scenario = [price / 0.0293001 / 100 for price in gas_scenario]
# gas_price = np.repeat(gas_scenario, 8).tolist()
gas_price = gas_scenario
coal_scenario = [85.7, 85.7, 85.7, 85.7, 85.7, 85.7, 85.7, 86.7, 86.7, 86.7, 86.7, 86.7, 86.7, 86.7, 86.7, 86.7, 86.7]
coal_price = [price/8.141/exchange for price, exchange in zip(coal_scenario, dollar_gbp_exchange_rate)]
# coal_price = np.repeat(coal_scenario, 8).tolist()
oil_scenario = [70.7, 71.7, 72.7, 74.7, 75.7, 76.7, 77.7, 79.7, 80.7, 81.7, 83.7, 84.7, 84.7, 84.7, 84.7, 84.7, 84.7]
oil_scenario = [price/1.69941/exchange for price, exchange in zip(oil_scenario, dollar_gbp_exchange_rate)]
# gas_price = [KW_TO_MW * 0.01909] * 60 # Source: Average prices of fuels purchased by the major UK power producers: table_321.xls
# coal_price = [KW_TO_MW * 0.01106] * 60 # Source: Average prices of fuels purchased by the major UK power producers: table_321.xls
uranium_price = [KW_TO_MW * 0.0039] * 17 # Source: The Economics of Nuclear Power: EconomicsNP.pdf
oil_price = [KW_TO_MW * 0.02748] * 17 # Source: Average prices of fuels purchased by the major UK power producers: table_321.xls
diesel_price = [KW_TO_MW * 0.1] * 17 # Source: https://www.racfoundation.org/data/wholesale-fuel-prices-v-pump-prices-data
woodchip_price = [KW_TO_MW * 0.0252] * 17 # Source: Biomass for Power Generation: IRENA BiomassCost.pdf
poultry_litter_price = [KW_TO_MW * 0.01139] * 17 # Source: How much is poultry litter worth?: sp06ca08.pdf
straw_price = [KW_TO_MW * 0.016488] * 17 # Source: https://dairy.ahdb.org.uk/market-information/farm-expenses/hay-straw-prices/#.W6JnFJNKiYU
meat_price = [KW_TO_MW * 0.01] * 17 # Assumption: Low price due to plant_type being a waste product
waste_price_post_2000 = [KW_TO_MW * -0.0252] * 17 # Source: Gate fees report 2017 Comparing the costs of waste treatment options: Gate Fees report 2017_FINAL_clean.pdf
waste_price_pre_2000 = [KW_TO_MW * -0.01551] * 17 # Source: Gate fees report 2017 Comparing the costs of waste treatment options: Gate Fees report 2017_FINAL_clean.pdf
# Joining historical and future fuel prices for simulation purposes.
fuel_prices = pd.DataFrame(data=[coal_price, oil_price, gas_price, uranium_price, diesel_price, woodchip_price,
poultry_litter_price, straw_price, meat_price, waste_price_post_2000,
waste_price_pre_2000],
columns=[str(i) for i in range(2019, (2019+len(gas_price)))])
fuel_prices = | pd.concat([historical_fuel_prices_mw, fuel_prices], axis=1) | pandas.concat |
from collections import OrderedDict
import math
from auto_ml import utils
import pandas as pd
from sklearn.ensemble import GradientBoostingRegressor, GradientBoostingClassifier
from sklearn.metrics import mean_squared_error, make_scorer, brier_score_loss, accuracy_score, explained_variance_score, mean_absolute_error, median_absolute_error, r2_score, log_loss, roc_auc_score
import numpy as np
from tabulate import tabulate
bad_vals_as_strings = set([str(float('nan')), str(float('inf')), str(float('-inf')), 'None', 'none', 'NaN', 'NAN', 'nan', 'NULL', 'null', '', 'inf', '-inf', 'np.nan', 'numpy.nan'])
def advanced_scoring_classifiers(probas, actuals, name=None):
# pandas Series don't play nice here. Make sure our actuals list is indeed a list
actuals = list(actuals)
predictions = list(probas)
print('Here is our brier-score-loss, which is the default value we optimized for while training, and is the value returned from .score() unless you requested a custom scoring metric')
print('It is a measure of how close the PROBABILITY predictions are.')
if name != None:
print(name)
# Sometimes we will be given "flattened" probabilities (only the probability of our positive label), while other times we might be given "nested" probabilities (probabilities of both positive and negative, in a list, for each item).
try:
probas = [proba[1] for proba in probas]
except:
pass
brier_score = brier_score_loss(actuals, probas)
print(format(brier_score, '.4f'))
print('\nHere is the trained estimator\'s overall accuracy (when it predicts a label, how frequently is that the correct label?)')
predicted_labels = []
for pred in probas:
if pred >= 0.5:
predicted_labels.append(1)
else:
predicted_labels.append(0)
print(format(accuracy_score(y_true=actuals, y_pred=predicted_labels) * 100, '.1f') + '%')
print('\nHere is a confusion matrix showing predictions vs. actuals by label:')
#it would make sense to use sklearn's confusion_matrix here but it apparently has no labels
#took this idea instead from: http://stats.stackexchange.com/a/109015
conf = pd.crosstab(pd.Series(actuals), pd.Series(predicted_labels), rownames=['v Actual v'], colnames=['Predicted >'], margins=True)
print(conf)
#I like knowing the per class accuracy to see if the model is mishandling imbalanced data.
#For example, if it is predicting 100% of observations to one class just because it is the majority
#Wikipedia seems to call that Positive/negative predictive value
print('\nHere is predictive value by class:')
df = pd.concat([ | pd.Series(actuals,name='actuals') | pandas.Series |
from pandas import DataFrame, Series
def avg_medal_count():
"""
Compute the average number of bronze medals earned by countries who
earned at least one gold medal.
Save this to a variable named avg_bronze_at_least_one_gold. You do not
need to call the function in your code when running it in the browser -
the grader will do that automatically when you submit or test it.
HINT-1:
You can retrieve all of the values of a Pandas column from a
data frame, "df", as follows:
df['column_name']
HINT-2:
The numpy.mean function can accept as an argument a single
Pandas column.
For example, numpy.mean(df["col_name"]) would return the
mean of the values located in "col_name" of a dataframe df.
"""
countries = ['Russian Fed.', 'Norway', 'Canada', 'United States',
'Netherlands', 'Germany', 'Switzerland', 'Belarus',
'Austria', 'France', 'Poland', 'China', 'Korea',
'Sweden', 'Czech Republic', 'Slovenia', 'Japan',
'Finland', 'Great Britain', 'Ukraine', 'Slovakia',
'Italy', 'Latvia', 'Australia', 'Croatia', 'Kazakhstan']
gold = [13, 11, 10, 9, 8, 8, 6, 5, 4, 4, 4, 3, 3, 2, 2, 2, 1, 1, 1, 1, 1, 0, 0, 0, 0, 0]
silver = [11, 5, 10, 7, 7, 6, 3, 0, 8, 4, 1, 4, 3, 7, 4, 2, 4, 3, 1, 0, 0, 2, 2, 2, 1, 0]
bronze = [9, 10, 5, 12, 9, 5, 2, 1, 5, 7, 1, 2, 2, 6, 2, 4, 3, 1, 2, 1, 0, 6, 2, 1, 0, 1]
olympic_medal_counts = {'country_name': Series(countries),
'gold': Series(gold),
'silver': | Series(silver) | pandas.Series |
import pandas as pd
import numpy as np
import pytest
from kgextension.caching_helper import freeze_unhashable, unfreeze_unhashable
class TestFreezeUnfreezeUnhashable:
def test1_arg_series(self):
@freeze_unhashable(freeze_by="argument", freeze_argument="the_arg")
def test_fun(a, b, c=12, the_arg=[]):
the_arg = unfreeze_unhashable(the_arg, frozen_type="series")
if a == 10 and b == 11 and c == 12:
return the_arg
else:
return None
df = pd.DataFrame({"a": [1,2,3,np.nan], "b": ["x", "y", "z", np.nan]})
s = df["a"]
s_unfrozen = test_fun(10, 11, the_arg=s)
pd.testing.assert_series_equal(s, s_unfrozen)
def test2_arg_series_kwargs(self):
@freeze_unhashable(freeze_by="argument", freeze_argument="the_arg")
def test_fun(a, b, c=12, the_arg=[]):
the_arg = unfreeze_unhashable(the_arg, frozen_type="series")
if a == 10 and b == 11 and c == 12:
return the_arg
else:
return None
df = | pd.DataFrame({"a": [1,2,3,np.nan], "b": ["x", "y", "z", np.nan]}) | pandas.DataFrame |
#!/usr/bin/env python
# coding: utf-8
# ## This is a YOLOv4 training pipeline with Pytorch.
# I use coco pre-trained weights.
# Have fun and feel free to leave any comment!
# ## Reference
# https://github.com/Tianxiaomo/pytorch-YOLOv4
# https://www.kaggle.com/orkatz2/yolov5-train
# In[20]:
# !pip install torch==1.4.0 torchvision==0.5.0
# !git clone https://github.com/Tianxiaomo/pytorch-YOLOv4
# !rm ./* -r
# !cp -r pytorch-YOLOv4/* ./
# !pip install -U -r requirements.txt
# In[21]:
import os
import random
import numpy as np
import pandas as pd
#from tqdm.notebook import tqdm # tqdm_notebook as tqdm
from tqdm import tqdm
#from tqdm.contrib.concurrent import process_map
import shutil as sh
import cv2
from PIL import Image
def convert_dataset_to_coco(dataset_name: str = 'widerface', det_threshold: int = 3):
PATH = os.getcwd()
df = ''
if dataset_name == 'wheat':
# Wheat train.csv sample
df = pd.read_csv(os.path.join(PATH, 'data', dataset_name, 'train.csv')) # dataset_name
bboxs = np.stack(df['bbox'].apply(lambda x: np.fromstring(x[1:-1], sep=',')))
for i, column in enumerate(['x', 'y', 'w', 'h']):
df[column] = bboxs[:,i]
df.drop(columns=['bbox'], inplace=True)
df['x1'] = df['x'] + df['w']
df['y1'] = df['y'] + df['h']
df['classes'] = 0
df = df[['image_id','x', 'y', 'w', 'h','x1','y1','classes']]
print(df.shape)
print(df.head())
print(f"Unique classes: {df['classes'].unique()}")
else:
modes = ['train', 'test', 'val']
label_path = dict([(mode, os.path.join(PATH, 'data', dataset_name, mode, 'label.txt')) for mode in modes])
data_dict = {}
for mode in modes:
data_dict[mode] = {'label_path': label_path[mode]}
data = {'image_id': [], 'x': [], 'y': [], 'w': [], 'h': [],
'x1': [], 'y1': [], 'classes': [], 'landmarks': []}
def txt_to_list(path_to_file: str, mode: str):
file = open(path_to_file,'r')
lines = file.readlines()
isFirst = True
labels = []
words = []
imgs_path = []
anno_folder = os.path.join(PATH, 'data', dataset_name)
for line in lines:
line = line.rstrip()
if line.startswith('#'):
if isFirst is True:
isFirst = False
else:
labels_copy = labels.copy()
words.append(labels_copy)
labels.clear()
img_path = line[2:]
full_path = os.path.join(anno_folder, mode, 'images', img_path)
imgs_path.append(full_path)
else:
line = line.split(' ')
label = [float(x) for x in line]
visible = True if (label[2] >= det_threshold and label[3] >= det_threshold) else False
if visible:
labels.append(label)
words.append(labels)
if mode == 'test':
return imgs_path
else:
return words, labels, imgs_path
for mode in modes:
if mode != 'test':
words, labels, imgs_path = txt_to_list(label_path[mode], mode)
data_dict[mode]['words'] = words
data_dict[mode]['labels'] = labels
data_dict[mode]['imgs_path'] = imgs_path
else:
imgs_path = txt_to_list(label_path[mode], mode)
# print(imgs_path)
data_dict[mode]['imgs_path'] = imgs_path
for mode in modes:
if mode != 'test':
new_words_list = []
for i, word in enumerate(tqdm(data_dict[mode]['words'])):
for string in word:
new_words_list.append(tuple([data_dict[mode]['imgs_path'][i], string]))
data_dict[mode]['new_words_list'] = new_words_list
def convert_data(tuple_datum):
path, string = tuple_datum
file_name = f"{path.split('/')[-2]}/{path.split('/')[-1]}" # .split('.')[0]
x, y, w, h = string[0:4]
x1, y1 = x + w, y + h
landmarks = string[5:]
data = {'image_id': file_name, 'x': x, 'y': y,
'w': w, 'h': h, 'x1': x1, 'y1': y1,
'classes': 0,
'landmarks': landmarks}
return data
def convert_data_val(path):
file_name = f"{path.split('/')[-2]}/{path.split('/')[-1]}" # .split('.')[0]
data = {'image_id': file_name}
return data
for mode in modes:
csv_path = os.path.join(PATH, 'data', dataset_name, mode, f'{mode}.csv')
if mode != 'test':
data_dict[mode]['csv_path'] = csv_path
datum = []
for path in tqdm(data_dict[mode]['new_words_list']):
datum.append(convert_data(path))
df = pd.DataFrame(data=datum)
df.to_csv(csv_path)
else:
data_dict[mode]['csv_path'] = csv_path
#datum = {}
#datum = process_map(convert_data_val, data_dict[mode]['imgs_path'],
# max_workers=12)
datum = []
for path in tqdm(data_dict[mode]['imgs_path']):
datum.append(convert_data_val(path))
df = pd.DataFrame(data=datum)
df.to_csv(csv_path)
df = | pd.read_csv(data_dict['train']['csv_path'], index_col=0) | pandas.read_csv |
# Functions to estimate cost for each lambda, by voxel:
from __future__ import division
from numpy.linalg import inv, svd
import numpy as np
from sklearn.model_selection import KFold
from sklearn.linear_model import Ridge, RidgeCV
import time
import scipy as sp
from sklearn.kernel_ridge import KernelRidge
import pickle as pkl
import pandas as pd
def load_dict(name):
with open(name + '.pkl', 'rb') as f:
return pkl.load(f)
def save_dict(dictionary, name):
with open(name + '.pkl', 'wb') as f:
pkl.dump(dictionary, f)
def save_dict_greater_than_4gb(dictionary, name):
with open(name + '.pkl', 'wb') as f:
pkl.dump(dictionary, f, protocol=4)
def get_sub_by_roi(data_to_reshape, train_subs):
pred_performance = np.zeros((len(train_subs),268))
for subIdx in np.arange(0, len(train_subs)):
pred_performance[subIdx, :] = data_to_reshape[subIdx* 268 : (subIdx *268) + 268]
return pred_performance
def drop_subjects_without_behavior(behavior_data, predictive_performance, behavior_data_all):
if np.sum(np.isnan(np.asarray(behavior_data))) == 0:
return behavior_data, predictive_performance, train_subs
else:
behav_without_nans = behavior_data.drop(behavior_data_all.loc[pd.isna(behavior_data), :].index)
sub_to_drop = []
for sub in behavior_data_all.loc[pd.isna(behavior_data), :].index:
sub_to_drop.append(train_subs.index(sub))
dropped_sub_predictive_performance = np.delete(predictive_performance,sub_to_drop, axis = 0)
sub_to_keep = np.delete(behavior_data.index, sub_to_drop)
return behav_without_nans, dropped_sub_predictive_performance, sub_to_keep
def drop_subjects_without_behavior_3T(behavior_data, predictive_performance, behavior_data_all, train_subs_3T):
if np.sum(np.isnan(np.asarray(behavior_data))) == 0: #no nan's
return behavior_data, predictive_performance, train_subs_3T
else:
behav_without_nans = behavior_data.drop(behavior_data_all.loc[pd.isna(behavior_data), :].index)
sub_to_drop = []
for sub in behavior_data_all.loc[ | pd.isna(behavior_data) | pandas.isna |
"""プロットサンプルページの管理データと挙動を実装するクラス."""
import numpy as np
import pandas as pd
from use_cases.linear_function_interactor import LinearFunctionInteractor
from view_models.plot_sample_view_model import PlotSampleViewModel
class PlotSampleController:
"""サンプルプロットページ制御クラス."""
def __init__(self, use_case: LinearFunctionInteractor):
"""コントローラークラスのコンストラクタの一例.
Args:
use_case (ExampleInteractor): コントローラークラスで使用するユースケース
"""
self.view_model = PlotSampleViewModel(gradient=0.0, intercept=0.0)
self.__use_case = use_case
def create_plot_data(self) -> pd.DataFrame:
"""サンプルデータを作成する."""
data = self.__use_case.create_data(
self.view_model.gradient, self.view_model.intercept)
df = | pd.DataFrame(data) | pandas.DataFrame |
import unittest
import qteasy as qt
import pandas as pd
from pandas import Timestamp
import numpy as np
import math
from numpy import int64
import itertools
import datetime
from qteasy.utilfuncs import list_to_str_format, regulate_date_format, time_str_format, str_to_list
from qteasy.utilfuncs import maybe_trade_day, is_market_trade_day, prev_trade_day, next_trade_day
from qteasy.utilfuncs import next_market_trade_day, unify, mask_to_signal, list_or_slice, labels_to_dict
from qteasy.utilfuncs import weekday_name, prev_market_trade_day, is_number_like, list_truncate, input_to_list
from qteasy.space import Space, Axis, space_around_centre, ResultPool
from qteasy.core import apply_loop
from qteasy.built_in import SelectingFinanceIndicator, TimingDMA, TimingMACD, TimingCDL, TimingTRIX
from qteasy.tsfuncs import income, indicators, name_change, get_bar
from qteasy.tsfuncs import stock_basic, trade_calendar, new_share, get_index
from qteasy.tsfuncs import balance, cashflow, top_list, index_indicators, composite
from qteasy.tsfuncs import future_basic, future_daily, options_basic, options_daily
from qteasy.tsfuncs import fund_basic, fund_net_value, index_basic, stock_company
from qteasy.evaluate import eval_alpha, eval_benchmark, eval_beta, eval_fv
from qteasy.evaluate import eval_info_ratio, eval_max_drawdown, eval_sharp
from qteasy.evaluate import eval_volatility
from qteasy.tafuncs import bbands, dema, ema, ht, kama, ma, mama, mavp, mid_point
from qteasy.tafuncs import mid_price, sar, sarext, sma, t3, tema, trima, wma, adx, adxr
from qteasy.tafuncs import apo, bop, cci, cmo, dx, macd, macdext, aroon, aroonosc
from qteasy.tafuncs import macdfix, mfi, minus_di, minus_dm, mom, plus_di, plus_dm
from qteasy.tafuncs import ppo, roc, rocp, rocr, rocr100, rsi, stoch, stochf, stochrsi
from qteasy.tafuncs import trix, ultosc, willr, ad, adosc, obv, atr, natr, trange
from qteasy.tafuncs import avgprice, medprice, typprice, wclprice, ht_dcperiod
from qteasy.tafuncs import ht_dcphase, ht_phasor, ht_sine, ht_trendmode, cdl2crows
from qteasy.tafuncs import cdl3blackcrows, cdl3inside, cdl3linestrike, cdl3outside
from qteasy.tafuncs import cdl3starsinsouth, cdl3whitesoldiers, cdlabandonedbaby
from qteasy.tafuncs import cdladvanceblock, cdlbelthold, cdlbreakaway, cdlclosingmarubozu
from qteasy.tafuncs import cdlconcealbabyswall, cdlcounterattack, cdldarkcloudcover
from qteasy.tafuncs import cdldoji, cdldojistar, cdldragonflydoji, cdlengulfing
from qteasy.tafuncs import cdleveningdojistar, cdleveningstar, cdlgapsidesidewhite
from qteasy.tafuncs import cdlgravestonedoji, cdlhammer, cdlhangingman, cdlharami
from qteasy.tafuncs import cdlharamicross, cdlhighwave, cdlhikkake, cdlhikkakemod
from qteasy.tafuncs import cdlhomingpigeon, cdlidentical3crows, cdlinneck
from qteasy.tafuncs import cdlinvertedhammer, cdlkicking, cdlkickingbylength
from qteasy.tafuncs import cdlladderbottom, cdllongleggeddoji, cdllongline, cdlmarubozu
from qteasy.tafuncs import cdlmatchinglow, cdlmathold, cdlmorningdojistar, cdlmorningstar
from qteasy.tafuncs import cdlonneck, cdlpiercing, cdlrickshawman, cdlrisefall3methods
from qteasy.tafuncs import cdlseparatinglines, cdlshootingstar, cdlshortline, cdlspinningtop
from qteasy.tafuncs import cdlstalledpattern, cdlsticksandwich, cdltakuri, cdltasukigap
from qteasy.tafuncs import cdlthrusting, cdltristar, cdlunique3river, cdlupsidegap2crows
from qteasy.tafuncs import cdlxsidegap3methods, beta, correl, linearreg, linearreg_angle
from qteasy.tafuncs import linearreg_intercept, linearreg_slope, stddev, tsf, var, acos
from qteasy.tafuncs import asin, atan, ceil, cos, cosh, exp, floor, ln, log10, sin, sinh
from qteasy.tafuncs import sqrt, tan, tanh, add, div, max, maxindex, min, minindex, minmax
from qteasy.tafuncs import minmaxindex, mult, sub, sum
from qteasy.history import get_financial_report_type_raw_data, get_price_type_raw_data
from qteasy.history import stack_dataframes, dataframe_to_hp, HistoryPanel
from qteasy.database import DataSource
from qteasy.strategy import Strategy, SimpleTiming, RollingTiming, SimpleSelecting, FactoralSelecting
from qteasy._arg_validators import _parse_string_kwargs, _valid_qt_kwargs
from qteasy.blender import _exp_to_token, blender_parser, signal_blend
class TestCost(unittest.TestCase):
def setUp(self):
self.amounts = np.array([10000., 20000., 10000.])
self.op = np.array([0., 1., -0.33333333])
self.amounts_to_sell = np.array([0., 0., -3333.3333])
self.cash_to_spend = np.array([0., 20000., 0.])
self.prices = np.array([10., 20., 10.])
self.r = qt.Cost(0.0)
def test_rate_creation(self):
"""测试对象生成"""
print('testing rates objects\n')
self.assertIsInstance(self.r, qt.Cost, 'Type should be Rate')
self.assertEqual(self.r.buy_fix, 0)
self.assertEqual(self.r.sell_fix, 0)
def test_rate_operations(self):
"""测试交易费率对象"""
self.assertEqual(self.r['buy_fix'], 0.0, 'Item got is incorrect')
self.assertEqual(self.r['sell_fix'], 0.0, 'Item got is wrong')
self.assertEqual(self.r['buy_rate'], 0.003, 'Item got is incorrect')
self.assertEqual(self.r['sell_rate'], 0.001, 'Item got is incorrect')
self.assertEqual(self.r['buy_min'], 5., 'Item got is incorrect')
self.assertEqual(self.r['sell_min'], 0.0, 'Item got is incorrect')
self.assertEqual(self.r['slipage'], 0.0, 'Item got is incorrect')
self.assertEqual(np.allclose(self.r.calculate(self.amounts),
[0.003, 0.003, 0.003]),
True,
'fee calculation wrong')
def test_rate_fee(self):
"""测试买卖交易费率"""
self.r.buy_rate = 0.003
self.r.sell_rate = 0.001
self.r.buy_fix = 0.
self.r.sell_fix = 0.
self.r.buy_min = 0.
self.r.sell_min = 0.
self.r.slipage = 0.
print('\nSell result with fixed rate = 0.001 and moq = 0:')
print(self.r.get_selling_result(self.prices, self.amounts_to_sell))
test_rate_fee_result = self.r.get_selling_result(self.prices, self.amounts_to_sell)
self.assertIs(np.allclose(test_rate_fee_result[0], [0., 0., -3333.3333]), True, 'result incorrect')
self.assertAlmostEqual(test_rate_fee_result[1], 33299.999667, msg='result incorrect')
self.assertAlmostEqual(test_rate_fee_result[2], 33.333332999999996, msg='result incorrect')
print('\nSell result with fixed rate = 0.001 and moq = 1:')
print(self.r.get_selling_result(self.prices, self.amounts_to_sell, 1.))
test_rate_fee_result = self.r.get_selling_result(self.prices, self.amounts_to_sell, 1)
self.assertIs(np.allclose(test_rate_fee_result[0], [0., 0., -3333]), True, 'result incorrect')
self.assertAlmostEqual(test_rate_fee_result[1], 33296.67, msg='result incorrect')
self.assertAlmostEqual(test_rate_fee_result[2], 33.33, msg='result incorrect')
print('\nSell result with fixed rate = 0.001 and moq = 100:')
print(self.r.get_selling_result(self.prices, self.amounts_to_sell, 100))
test_rate_fee_result = self.r.get_selling_result(self.prices, self.amounts_to_sell, 100)
self.assertIs(np.allclose(test_rate_fee_result[0], [0., 0., -3300]), True, 'result incorrect')
self.assertAlmostEqual(test_rate_fee_result[1], 32967.0, msg='result incorrect')
self.assertAlmostEqual(test_rate_fee_result[2], 33, msg='result incorrect')
print('\nPurchase result with fixed rate = 0.003 and moq = 0:')
print(self.r.get_purchase_result(self.prices, self.cash_to_spend, 0))
test_rate_fee_result = self.r.get_purchase_result(self.prices, self.cash_to_spend, 0)
self.assertIs(np.allclose(test_rate_fee_result[0], [0., 997.00897308, 0.]), True, 'result incorrect')
self.assertAlmostEqual(test_rate_fee_result[1], -20000.0, msg='result incorrect')
self.assertAlmostEqual(test_rate_fee_result[2], 59.82053838484547, msg='result incorrect')
print('\nPurchase result with fixed rate = 0.003 and moq = 1:')
print(self.r.get_purchase_result(self.prices, self.cash_to_spend, 1))
test_rate_fee_result = self.r.get_purchase_result(self.prices, self.cash_to_spend, 1)
self.assertIs(np.allclose(test_rate_fee_result[0], [0., 997., 0.]), True, 'result incorrect')
self.assertAlmostEqual(test_rate_fee_result[1], -19999.82, msg='result incorrect')
self.assertAlmostEqual(test_rate_fee_result[2], 59.82, msg='result incorrect')
print('\nPurchase result with fixed rate = 0.003 and moq = 100:')
print(self.r.get_purchase_result(self.prices, self.cash_to_spend, 100))
test_rate_fee_result = self.r.get_purchase_result(self.prices, self.cash_to_spend, 100)
self.assertIs(np.allclose(test_rate_fee_result[0], [0., 900., 0.]), True, 'result incorrect')
self.assertAlmostEqual(test_rate_fee_result[1], -18054., msg='result incorrect')
self.assertAlmostEqual(test_rate_fee_result[2], 54.0, msg='result incorrect')
def test_min_fee(self):
"""测试最低交易费用"""
self.r.buy_rate = 0.
self.r.sell_rate = 0.
self.r.buy_fix = 0.
self.r.sell_fix = 0.
self.r.buy_min = 300
self.r.sell_min = 300
self.r.slipage = 0.
print('\npurchase result with fixed cost rate with min fee = 300 and moq = 0:')
print(self.r.get_purchase_result(self.prices, self.cash_to_spend, 0))
test_min_fee_result = self.r.get_purchase_result(self.prices, self.cash_to_spend, 0)
self.assertIs(np.allclose(test_min_fee_result[0], [0., 985, 0.]), True, 'result incorrect')
self.assertAlmostEqual(test_min_fee_result[1], -20000.0, msg='result incorrect')
self.assertAlmostEqual(test_min_fee_result[2], 300.0, msg='result incorrect')
print('\npurchase result with fixed cost rate with min fee = 300 and moq = 10:')
print(self.r.get_purchase_result(self.prices, self.cash_to_spend, 10))
test_min_fee_result = self.r.get_purchase_result(self.prices, self.cash_to_spend, 10)
self.assertIs(np.allclose(test_min_fee_result[0], [0., 980, 0.]), True, 'result incorrect')
self.assertAlmostEqual(test_min_fee_result[1], -19900.0, msg='result incorrect')
self.assertAlmostEqual(test_min_fee_result[2], 300.0, msg='result incorrect')
print('\npurchase result with fixed cost rate with min fee = 300 and moq = 100:')
print(self.r.get_purchase_result(self.prices, self.cash_to_spend, 100))
test_min_fee_result = self.r.get_purchase_result(self.prices, self.cash_to_spend, 100)
self.assertIs(np.allclose(test_min_fee_result[0], [0., 900, 0.]), True, 'result incorrect')
self.assertAlmostEqual(test_min_fee_result[1], -18300.0, msg='result incorrect')
self.assertAlmostEqual(test_min_fee_result[2], 300.0, msg='result incorrect')
print('\nselling result with fixed cost rate with min fee = 300 and moq = 0:')
print(self.r.get_selling_result(self.prices, self.amounts_to_sell))
test_min_fee_result = self.r.get_selling_result(self.prices, self.amounts_to_sell)
self.assertIs(np.allclose(test_min_fee_result[0], [0, 0, -3333.3333]), True, 'result incorrect')
self.assertAlmostEqual(test_min_fee_result[1], 33033.333)
self.assertAlmostEqual(test_min_fee_result[2], 300.0)
print('\nselling result with fixed cost rate with min fee = 300 and moq = 1:')
print(self.r.get_selling_result(self.prices, self.amounts_to_sell, 1))
test_min_fee_result = self.r.get_selling_result(self.prices, self.amounts_to_sell, 1)
self.assertIs(np.allclose(test_min_fee_result[0], [0, 0, -3333]), True, 'result incorrect')
self.assertAlmostEqual(test_min_fee_result[1], 33030)
self.assertAlmostEqual(test_min_fee_result[2], 300.0)
print('\nselling result with fixed cost rate with min fee = 300 and moq = 100:')
print(self.r.get_selling_result(self.prices, self.amounts_to_sell, 100))
test_min_fee_result = self.r.get_selling_result(self.prices, self.amounts_to_sell, 100)
self.assertIs(np.allclose(test_min_fee_result[0], [0, 0, -3300]), True, 'result incorrect')
self.assertAlmostEqual(test_min_fee_result[1], 32700)
self.assertAlmostEqual(test_min_fee_result[2], 300.0)
def test_rate_with_min(self):
"""测试最低交易费用对其他交易费率参数的影响"""
self.r.buy_rate = 0.0153
self.r.sell_rate = 0.01
self.r.buy_fix = 0.
self.r.sell_fix = 0.
self.r.buy_min = 300
self.r.sell_min = 333
self.r.slipage = 0.
print('\npurchase result with fixed cost rate with buy_rate = 0.0153, min fee = 300 and moq = 0:')
print(self.r.get_purchase_result(self.prices, self.cash_to_spend, 0))
test_rate_with_min_result = self.r.get_purchase_result(self.prices, self.cash_to_spend, 0)
self.assertIs(np.allclose(test_rate_with_min_result[0], [0., 984.9305624, 0.]), True, 'result incorrect')
self.assertAlmostEqual(test_rate_with_min_result[1], -20000.0, msg='result incorrect')
self.assertAlmostEqual(test_rate_with_min_result[2], 301.3887520929774, msg='result incorrect')
print('\npurchase result with fixed cost rate with buy_rate = 0.0153, min fee = 300 and moq = 10:')
print(self.r.get_purchase_result(self.prices, self.cash_to_spend, 10))
test_rate_with_min_result = self.r.get_purchase_result(self.prices, self.cash_to_spend, 10)
self.assertIs(np.allclose(test_rate_with_min_result[0], [0., 980, 0.]), True, 'result incorrect')
self.assertAlmostEqual(test_rate_with_min_result[1], -19900.0, msg='result incorrect')
self.assertAlmostEqual(test_rate_with_min_result[2], 300.0, msg='result incorrect')
print('\npurchase result with fixed cost rate with buy_rate = 0.0153, min fee = 300 and moq = 100:')
print(self.r.get_purchase_result(self.prices, self.cash_to_spend, 100))
test_rate_with_min_result = self.r.get_purchase_result(self.prices, self.cash_to_spend, 100)
self.assertIs(np.allclose(test_rate_with_min_result[0], [0., 900, 0.]), True, 'result incorrect')
self.assertAlmostEqual(test_rate_with_min_result[1], -18300.0, msg='result incorrect')
self.assertAlmostEqual(test_rate_with_min_result[2], 300.0, msg='result incorrect')
print('\nselling result with fixed cost rate with sell_rate = 0.01, min fee = 333 and moq = 0:')
print(self.r.get_selling_result(self.prices, self.amounts_to_sell))
test_rate_with_min_result = self.r.get_selling_result(self.prices, self.amounts_to_sell)
self.assertIs(np.allclose(test_rate_with_min_result[0], [0, 0, -3333.3333]), True, 'result incorrect')
self.assertAlmostEqual(test_rate_with_min_result[1], 32999.99967)
self.assertAlmostEqual(test_rate_with_min_result[2], 333.33333)
print('\nselling result with fixed cost rate with sell_rate = 0.01, min fee = 333 and moq = 1:')
print(self.r.get_selling_result(self.prices, self.amounts_to_sell, 1))
test_rate_with_min_result = self.r.get_selling_result(self.prices, self.amounts_to_sell, 1)
self.assertIs(np.allclose(test_rate_with_min_result[0], [0, 0, -3333]), True, 'result incorrect')
self.assertAlmostEqual(test_rate_with_min_result[1], 32996.7)
self.assertAlmostEqual(test_rate_with_min_result[2], 333.3)
print('\nselling result with fixed cost rate with sell_rate = 0.01, min fee = 333 and moq = 100:')
print(self.r.get_selling_result(self.prices, self.amounts_to_sell, 100))
test_rate_with_min_result = self.r.get_selling_result(self.prices, self.amounts_to_sell, 100)
self.assertIs(np.allclose(test_rate_with_min_result[0], [0, 0, -3300]), True, 'result incorrect')
self.assertAlmostEqual(test_rate_with_min_result[1], 32667.0)
self.assertAlmostEqual(test_rate_with_min_result[2], 333.0)
def test_fixed_fee(self):
"""测试固定交易费用"""
self.r.buy_rate = 0.
self.r.sell_rate = 0.
self.r.buy_fix = 200
self.r.sell_fix = 150
self.r.buy_min = 0
self.r.sell_min = 0
self.r.slipage = 0
print('\nselling result of fixed cost with fixed fee = 150 and moq=0:')
print(self.r.get_selling_result(self.prices, self.amounts_to_sell, 0))
test_fixed_fee_result = self.r.get_selling_result(self.prices, self.amounts_to_sell)
self.assertIs(np.allclose(test_fixed_fee_result[0], [0, 0, -3333.3333]), True, 'result incorrect')
self.assertAlmostEqual(test_fixed_fee_result[1], 33183.333, msg='result incorrect')
self.assertAlmostEqual(test_fixed_fee_result[2], 150.0, msg='result incorrect')
print('\nselling result of fixed cost with fixed fee = 150 and moq=100:')
print(self.r.get_selling_result(self.prices, self.amounts_to_sell, 100))
test_fixed_fee_result = self.r.get_selling_result(self.prices, self.amounts_to_sell, 100)
self.assertIs(np.allclose(test_fixed_fee_result[0], [0, 0, -3300.]), True,
f'result incorrect, {test_fixed_fee_result[0]} does not equal to [0,0,-3400]')
self.assertAlmostEqual(test_fixed_fee_result[1], 32850., msg='result incorrect')
self.assertAlmostEqual(test_fixed_fee_result[2], 150., msg='result incorrect')
print('\npurchase result of fixed cost with fixed fee = 200:')
print(self.r.get_purchase_result(self.prices, self.cash_to_spend, 0))
test_fixed_fee_result = self.r.get_purchase_result(self.prices, self.cash_to_spend, 0)
self.assertIs(np.allclose(test_fixed_fee_result[0], [0., 990., 0.]), True, 'result incorrect')
self.assertAlmostEqual(test_fixed_fee_result[1], -20000.0, msg='result incorrect')
self.assertAlmostEqual(test_fixed_fee_result[2], 200.0, msg='result incorrect')
print('\npurchase result of fixed cost with fixed fee = 200:')
print(self.r.get_purchase_result(self.prices, self.cash_to_spend, 100))
test_fixed_fee_result = self.r.get_purchase_result(self.prices, self.cash_to_spend, 100)
self.assertIs(np.allclose(test_fixed_fee_result[0], [0., 900., 0.]), True, 'result incorrect')
self.assertAlmostEqual(test_fixed_fee_result[1], -18200.0, msg='result incorrect')
self.assertAlmostEqual(test_fixed_fee_result[2], 200.0, msg='result incorrect')
def test_slipage(self):
"""测试交易滑点"""
self.r.buy_fix = 0
self.r.sell_fix = 0
self.r.buy_min = 0
self.r.sell_min = 0
self.r.buy_rate = 0.003
self.r.sell_rate = 0.001
self.r.slipage = 1E-9
print('\npurchase result of fixed rate = 0.003 and slipage = 1E-10 and moq = 0:')
print(self.r.get_purchase_result(self.prices, self.cash_to_spend, 0))
print('\npurchase result of fixed rate = 0.003 and slipage = 1E-10 and moq = 100:')
print(self.r.get_purchase_result(self.prices, self.cash_to_spend, 100))
print('\nselling result with fixed rate = 0.001 and slipage = 1E-10:')
print(self.r.get_selling_result(self.prices, self.amounts_to_sell))
test_fixed_fee_result = self.r.get_selling_result(self.prices, self.amounts_to_sell)
self.assertIs(np.allclose(test_fixed_fee_result[0], [0, 0, -3333.3333]), True,
f'{test_fixed_fee_result[0]} does not equal to [0, 0, -10000]')
self.assertAlmostEqual(test_fixed_fee_result[1], 33298.88855591,
msg=f'{test_fixed_fee_result[1]} does not equal to 99890.')
self.assertAlmostEqual(test_fixed_fee_result[2], 34.44444409,
msg=f'{test_fixed_fee_result[2]} does not equal to -36.666663.')
test_fixed_fee_result = self.r.get_purchase_result(self.prices, self.cash_to_spend, 0)
self.assertIs(np.allclose(test_fixed_fee_result[0], [0., 996.98909294, 0.]), True, 'result incorrect')
self.assertAlmostEqual(test_fixed_fee_result[1], -20000.0, msg='result incorrect')
self.assertAlmostEqual(test_fixed_fee_result[2], 60.21814121353513, msg='result incorrect')
test_fixed_fee_result = self.r.get_purchase_result(self.prices, self.cash_to_spend, 100)
self.assertIs(np.allclose(test_fixed_fee_result[0], [0., 900., 0.]), True, 'result incorrect')
self.assertAlmostEqual(test_fixed_fee_result[1], -18054.36, msg='result incorrect')
self.assertAlmostEqual(test_fixed_fee_result[2], 54.36, msg='result incorrect')
class TestSpace(unittest.TestCase):
def test_creation(self):
"""
test if creation of space object is fine
"""
# first group of inputs, output Space with two discr axis from [0,10]
print('testing space objects\n')
# pars_list = [[(0, 10), (0, 10)],
# [[0, 10], [0, 10]]]
#
# types_list = ['discr',
# ['discr', 'discr']]
#
# input_pars = itertools.product(pars_list, types_list)
# for p in input_pars:
# # print(p)
# s = qt.Space(*p)
# b = s.boes
# t = s.types
# # print(s, t)
# self.assertIsInstance(s, qt.Space)
# self.assertEqual(b, [(0, 10), (0, 10)], 'boes incorrect!')
# self.assertEqual(t, ['discr', 'discr'], 'types incorrect')
#
pars_list = [[(0, 10), (0, 10)],
[[0, 10], [0, 10]]]
types_list = ['foo, bar',
['foo', 'bar']]
input_pars = itertools.product(pars_list, types_list)
for p in input_pars:
# print(p)
s = Space(*p)
b = s.boes
t = s.types
# print(s, t)
self.assertEqual(b, [(0, 10), (0, 10)], 'boes incorrect!')
self.assertEqual(t, ['enum', 'enum'], 'types incorrect')
pars_list = [[(0, 10), (0, 10)],
[[0, 10], [0, 10]]]
types_list = [['discr', 'foobar']]
input_pars = itertools.product(pars_list, types_list)
for p in input_pars:
# print(p)
s = Space(*p)
b = s.boes
t = s.types
# print(s, t)
self.assertEqual(b, [(0, 10), (0, 10)], 'boes incorrect!')
self.assertEqual(t, ['discr', 'enum'], 'types incorrect')
pars_list = [(0., 10), (0, 10)]
s = Space(pars=pars_list, par_types=None)
self.assertEqual(s.types, ['conti', 'discr'])
self.assertEqual(s.dim, 2)
self.assertEqual(s.size, (10.0, 11))
self.assertEqual(s.shape, (np.inf, 11))
self.assertEqual(s.count, np.inf)
self.assertEqual(s.boes, [(0., 10), (0, 10)])
pars_list = [(0., 10), (0, 10)]
s = Space(pars=pars_list, par_types='conti, enum')
self.assertEqual(s.types, ['conti', 'enum'])
self.assertEqual(s.dim, 2)
self.assertEqual(s.size, (10.0, 2))
self.assertEqual(s.shape, (np.inf, 2))
self.assertEqual(s.count, np.inf)
self.assertEqual(s.boes, [(0., 10), (0, 10)])
pars_list = [(1, 2), (2, 3), (3, 4)]
s = Space(pars=pars_list)
self.assertEqual(s.types, ['discr', 'discr', 'discr'])
self.assertEqual(s.dim, 3)
self.assertEqual(s.size, (2, 2, 2))
self.assertEqual(s.shape, (2, 2, 2))
self.assertEqual(s.count, 8)
self.assertEqual(s.boes, [(1, 2), (2, 3), (3, 4)])
pars_list = [(1, 2, 3), (2, 3, 4), (3, 4, 5)]
s = Space(pars=pars_list)
self.assertEqual(s.types, ['enum', 'enum', 'enum'])
self.assertEqual(s.dim, 3)
self.assertEqual(s.size, (3, 3, 3))
self.assertEqual(s.shape, (3, 3, 3))
self.assertEqual(s.count, 27)
self.assertEqual(s.boes, [(1, 2, 3), (2, 3, 4), (3, 4, 5)])
pars_list = [((1, 2, 3), (2, 3, 4), (3, 4, 5))]
s = Space(pars=pars_list)
self.assertEqual(s.types, ['enum'])
self.assertEqual(s.dim, 1)
self.assertEqual(s.size, (3,))
self.assertEqual(s.shape, (3,))
self.assertEqual(s.count, 3)
pars_list = ((1, 2, 3), (2, 3, 4), (3, 4, 5))
s = Space(pars=pars_list)
self.assertEqual(s.types, ['enum', 'enum', 'enum'])
self.assertEqual(s.dim, 3)
self.assertEqual(s.size, (3, 3, 3))
self.assertEqual(s.shape, (3, 3, 3))
self.assertEqual(s.count, 27)
self.assertEqual(s.boes, [(1, 2, 3), (2, 3, 4), (3, 4, 5)])
def test_extract(self):
"""
:return:
"""
pars_list = [(0, 10), (0, 10)]
types_list = ['discr', 'discr']
s = Space(pars=pars_list, par_types=types_list)
extracted_int, count = s.extract(3, 'interval')
extracted_int_list = list(extracted_int)
print('extracted int\n', extracted_int_list)
self.assertEqual(count, 16, 'extraction count wrong!')
self.assertEqual(extracted_int_list, [(0, 0), (0, 3), (0, 6), (0, 9), (3, 0), (3, 3),
(3, 6), (3, 9), (6, 0), (6, 3), (6, 6), (6, 9),
(9, 0), (9, 3), (9, 6), (9, 9)],
'space extraction wrong!')
extracted_rand, count = s.extract(10, 'rand')
extracted_rand_list = list(extracted_rand)
self.assertEqual(count, 10, 'extraction count wrong!')
print('extracted rand\n', extracted_rand_list)
for point in list(extracted_rand_list):
self.assertEqual(len(point), 2)
self.assertLessEqual(point[0], 10)
self.assertGreaterEqual(point[0], 0)
self.assertLessEqual(point[1], 10)
self.assertGreaterEqual(point[1], 0)
pars_list = [(0., 10), (0, 10)]
s = Space(pars=pars_list, par_types=None)
extracted_int2, count = s.extract(3, 'interval')
self.assertEqual(count, 16, 'extraction count wrong!')
extracted_int_list2 = list(extracted_int2)
self.assertEqual(extracted_int_list2, [(0, 0), (0, 3), (0, 6), (0, 9), (3, 0), (3, 3),
(3, 6), (3, 9), (6, 0), (6, 3), (6, 6), (6, 9),
(9, 0), (9, 3), (9, 6), (9, 9)],
'space extraction wrong!')
print('extracted int list 2\n', extracted_int_list2)
self.assertIsInstance(extracted_int_list2[0][0], float)
self.assertIsInstance(extracted_int_list2[0][1], (int, int64))
extracted_rand2, count = s.extract(10, 'rand')
self.assertEqual(count, 10, 'extraction count wrong!')
extracted_rand_list2 = list(extracted_rand2)
print('extracted rand list 2:\n', extracted_rand_list2)
for point in extracted_rand_list2:
self.assertEqual(len(point), 2)
self.assertIsInstance(point[0], float)
self.assertLessEqual(point[0], 10)
self.assertGreaterEqual(point[0], 0)
self.assertIsInstance(point[1], (int, int64))
self.assertLessEqual(point[1], 10)
self.assertGreaterEqual(point[1], 0)
pars_list = [(0., 10), ('a', 'b')]
s = Space(pars=pars_list, par_types='enum, enum')
extracted_int3, count = s.extract(1, 'interval')
self.assertEqual(count, 4, 'extraction count wrong!')
extracted_int_list3 = list(extracted_int3)
self.assertEqual(extracted_int_list3, [(0., 'a'), (0., 'b'), (10, 'a'), (10, 'b')],
'space extraction wrong!')
print('extracted int list 3\n', extracted_int_list3)
self.assertIsInstance(extracted_int_list3[0][0], float)
self.assertIsInstance(extracted_int_list3[0][1], str)
extracted_rand3, count = s.extract(3, 'rand')
self.assertEqual(count, 3, 'extraction count wrong!')
extracted_rand_list3 = list(extracted_rand3)
print('extracted rand list 3:\n', extracted_rand_list3)
for point in extracted_rand_list3:
self.assertEqual(len(point), 2)
self.assertIsInstance(point[0], (float, int))
self.assertLessEqual(point[0], 10)
self.assertGreaterEqual(point[0], 0)
self.assertIsInstance(point[1], str)
self.assertIn(point[1], ['a', 'b'])
pars_list = [((0, 10), (1, 'c'), ('a', 'b'), (1, 14))]
s = Space(pars=pars_list, par_types='enum')
extracted_int4, count = s.extract(1, 'interval')
self.assertEqual(count, 4, 'extraction count wrong!')
extracted_int_list4 = list(extracted_int4)
it = zip(extracted_int_list4, [(0, 10), (1, 'c'), (0, 'b'), (1, 14)])
for item, item2 in it:
print(item, item2)
self.assertTrue(all([tuple(ext_item) == item for ext_item, item in it]))
print('extracted int list 4\n', extracted_int_list4)
self.assertIsInstance(extracted_int_list4[0], tuple)
extracted_rand4, count = s.extract(3, 'rand')
self.assertEqual(count, 3, 'extraction count wrong!')
extracted_rand_list4 = list(extracted_rand4)
print('extracted rand list 4:\n', extracted_rand_list4)
for point in extracted_rand_list4:
self.assertEqual(len(point), 2)
self.assertIsInstance(point[0], (int, str))
self.assertIn(point[0], [0, 1, 'a'])
self.assertIsInstance(point[1], (int, str))
self.assertIn(point[1], [10, 14, 'b', 'c'])
self.assertIn(point, [(0., 10), (1, 'c'), ('a', 'b'), (1, 14)])
pars_list = [((0, 10), (1, 'c'), ('a', 'b'), (1, 14)), (1, 4)]
s = Space(pars=pars_list, par_types='enum, discr')
extracted_int5, count = s.extract(1, 'interval')
self.assertEqual(count, 16, 'extraction count wrong!')
extracted_int_list5 = list(extracted_int5)
for item, item2 in extracted_int_list5:
print(item, item2)
self.assertTrue(all([tuple(ext_item) == item for ext_item, item in it]))
print('extracted int list 5\n', extracted_int_list5)
self.assertIsInstance(extracted_int_list5[0], tuple)
extracted_rand5, count = s.extract(5, 'rand')
self.assertEqual(count, 5, 'extraction count wrong!')
extracted_rand_list5 = list(extracted_rand5)
print('extracted rand list 5:\n', extracted_rand_list5)
for point in extracted_rand_list5:
self.assertEqual(len(point), 2)
self.assertIsInstance(point[0], tuple)
print(f'type of point[1] is {type(point[1])}')
self.assertIsInstance(point[1], (int, np.int64))
self.assertIn(point[0], [(0., 10), (1, 'c'), ('a', 'b'), (1, 14)])
print(f'test incremental extraction')
pars_list = [(10., 250), (10., 250), (10., 250), (10., 250), (10., 250), (10., 250)]
s = Space(pars_list)
ext, count = s.extract(64, 'interval')
self.assertEqual(count, 4096)
points = list(ext)
# 已经取出所有的点,围绕其中10个点生成十个subspaces
# 检查是否每个subspace都为Space,是否都在s范围内,使用32生成点集,检查生成数量是否正确
for point in points[1000:1010]:
subspace = s.from_point(point, 64)
self.assertIsInstance(subspace, Space)
self.assertTrue(subspace in s)
self.assertEqual(subspace.dim, 6)
self.assertEqual(subspace.types, ['conti', 'conti', 'conti', 'conti', 'conti', 'conti'])
ext, count = subspace.extract(32)
points = list(ext)
self.assertGreaterEqual(count, 512)
self.assertLessEqual(count, 4096)
print(f'\n---------------------------------'
f'\nthe space created around point <{point}> is'
f'\n{subspace.boes}'
f'\nand extracted {count} points, the first 5 are:'
f'\n{points[:5]}')
def test_axis_extract(self):
# test axis object with conti type
axis = Axis((0., 5))
self.assertIsInstance(axis, Axis)
self.assertEqual(axis.axis_type, 'conti')
self.assertEqual(axis.axis_boe, (0., 5.))
self.assertEqual(axis.count, np.inf)
self.assertEqual(axis.size, 5.0)
self.assertTrue(np.allclose(axis.extract(1, 'int'), [0., 1., 2., 3., 4.]))
self.assertTrue(np.allclose(axis.extract(0.5, 'int'), [0., 0.5, 1., 1.5, 2., 2.5, 3., 3.5, 4., 4.5]))
extracted = axis.extract(8, 'rand')
self.assertEqual(len(extracted), 8)
self.assertTrue(all([(0 <= item <= 5) for item in extracted]))
# test axis object with discrete type
axis = Axis((1, 5))
self.assertIsInstance(axis, Axis)
self.assertEqual(axis.axis_type, 'discr')
self.assertEqual(axis.axis_boe, (1, 5))
self.assertEqual(axis.count, 5)
self.assertEqual(axis.size, 5)
self.assertTrue(np.allclose(axis.extract(1, 'int'), [1, 2, 3, 4, 5]))
self.assertRaises(ValueError, axis.extract, 0.5, 'int')
extracted = axis.extract(8, 'rand')
self.assertEqual(len(extracted), 8)
self.assertTrue(all([(item in [1, 2, 3, 4, 5]) for item in extracted]))
# test axis object with enumerate type
axis = Axis((1, 5, 7, 10, 'A', 'F'))
self.assertIsInstance(axis, Axis)
self.assertEqual(axis.axis_type, 'enum')
self.assertEqual(axis.axis_boe, (1, 5, 7, 10, 'A', 'F'))
self.assertEqual(axis.count, 6)
self.assertEqual(axis.size, 6)
self.assertEqual(axis.extract(1, 'int'), [1, 5, 7, 10, 'A', 'F'])
self.assertRaises(ValueError, axis.extract, 0.5, 'int')
extracted = axis.extract(8, 'rand')
self.assertEqual(len(extracted), 8)
self.assertTrue(all([(item in [1, 5, 7, 10, 'A', 'F']) for item in extracted]))
def test_from_point(self):
"""测试从一个点生成一个space"""
# 生成一个space,指定space中的一个点以及distance,生成一个sub-space
pars_list = [(0., 10), (0, 10)]
s = Space(pars=pars_list, par_types=None)
self.assertEqual(s.types, ['conti', 'discr'])
self.assertEqual(s.dim, 2)
self.assertEqual(s.size, (10., 11))
self.assertEqual(s.shape, (np.inf, 11))
self.assertEqual(s.count, np.inf)
self.assertEqual(s.boes, [(0., 10), (0, 10)])
print('create subspace from a point in space')
p = (3, 3)
distance = 2
subspace = s.from_point(p, distance)
self.assertIsInstance(subspace, Space)
self.assertEqual(subspace.types, ['conti', 'discr'])
self.assertEqual(subspace.dim, 2)
self.assertEqual(subspace.size, (4.0, 5))
self.assertEqual(subspace.shape, (np.inf, 5))
self.assertEqual(subspace.count, np.inf)
self.assertEqual(subspace.boes, [(1, 5), (1, 5)])
print('create subspace from a 6 dimensional discrete space')
s = Space(pars=[(10, 250), (10, 250), (10, 250), (10, 250), (10, 250), (10, 250)])
p = (15, 200, 150, 150, 150, 150)
d = 10
subspace = s.from_point(p, d)
self.assertIsInstance(subspace, Space)
self.assertEqual(subspace.types, ['discr', 'discr', 'discr', 'discr', 'discr', 'discr'])
self.assertEqual(subspace.dim, 6)
self.assertEqual(subspace.volume, 65345616)
self.assertEqual(subspace.size, (16, 21, 21, 21, 21, 21))
self.assertEqual(subspace.shape, (16, 21, 21, 21, 21, 21))
self.assertEqual(subspace.count, 65345616)
self.assertEqual(subspace.boes, [(10, 25), (190, 210), (140, 160), (140, 160), (140, 160), (140, 160)])
print('create subspace from a 6 dimensional continuous space')
s = Space(pars=[(10., 250), (10., 250), (10., 250), (10., 250), (10., 250), (10., 250)])
p = (15, 200, 150, 150, 150, 150)
d = 10
subspace = s.from_point(p, d)
self.assertIsInstance(subspace, Space)
self.assertEqual(subspace.types, ['conti', 'conti', 'conti', 'conti', 'conti', 'conti'])
self.assertEqual(subspace.dim, 6)
self.assertEqual(subspace.volume, 48000000)
self.assertEqual(subspace.size, (15.0, 20.0, 20.0, 20.0, 20.0, 20.0))
self.assertEqual(subspace.shape, (np.inf, np.inf, np.inf, np.inf, np.inf, np.inf))
self.assertEqual(subspace.count, np.inf)
self.assertEqual(subspace.boes, [(10, 25), (190, 210), (140, 160), (140, 160), (140, 160), (140, 160)])
print('create subspace with different distances on each dimension')
s = Space(pars=[(10., 250), (10., 250), (10., 250), (10., 250), (10., 250), (10., 250)])
p = (15, 200, 150, 150, 150, 150)
d = [10, 5, 5, 10, 10, 5]
subspace = s.from_point(p, d)
self.assertIsInstance(subspace, Space)
self.assertEqual(subspace.types, ['conti', 'conti', 'conti', 'conti', 'conti', 'conti'])
self.assertEqual(subspace.dim, 6)
self.assertEqual(subspace.volume, 6000000)
self.assertEqual(subspace.size, (15.0, 10.0, 10.0, 20.0, 20.0, 10.0))
self.assertEqual(subspace.shape, (np.inf, np.inf, np.inf, np.inf, np.inf, np.inf))
self.assertEqual(subspace.count, np.inf)
self.assertEqual(subspace.boes, [(10, 25), (195, 205), (145, 155), (140, 160), (140, 160), (145, 155)])
class TestCashPlan(unittest.TestCase):
def setUp(self):
self.cp1 = qt.CashPlan(['2012-01-01', '2010-01-01'], [10000, 20000], 0.1)
self.cp1.info()
self.cp2 = qt.CashPlan(['20100501'], 10000)
self.cp2.info()
self.cp3 = qt.CashPlan(pd.date_range(start='2019-01-01',
freq='Y',
periods=12),
[i * 1000 + 10000 for i in range(12)],
0.035)
self.cp3.info()
def test_creation(self):
self.assertIsInstance(self.cp1, qt.CashPlan, 'CashPlan object creation wrong')
self.assertIsInstance(self.cp2, qt.CashPlan, 'CashPlan object creation wrong')
self.assertIsInstance(self.cp3, qt.CashPlan, 'CashPlan object creation wrong')
# test __repr__()
print(self.cp1)
print(self.cp2)
print(self.cp3)
# test __str__()
self.cp1.info()
self.cp2.info()
self.cp3.info()
# test assersion errors
self.assertRaises(AssertionError, qt.CashPlan, '2016-01-01', [10000, 10000])
self.assertRaises(KeyError, qt.CashPlan, '2020-20-20', 10000)
def test_properties(self):
self.assertEqual(self.cp1.amounts, [20000, 10000], 'property wrong')
self.assertEqual(self.cp1.first_day, Timestamp('2010-01-01'))
self.assertEqual(self.cp1.last_day, Timestamp('2012-01-01'))
self.assertEqual(self.cp1.investment_count, 2)
self.assertEqual(self.cp1.period, 730)
self.assertEqual(self.cp1.dates, [Timestamp('2010-01-01'), Timestamp('2012-01-01')])
self.assertEqual(self.cp1.ir, 0.1)
self.assertAlmostEqual(self.cp1.closing_value, 34200)
self.assertAlmostEqual(self.cp2.closing_value, 10000)
self.assertAlmostEqual(self.cp3.closing_value, 220385.3483685)
self.assertIsInstance(self.cp1.plan, pd.DataFrame)
self.assertIsInstance(self.cp2.plan, pd.DataFrame)
self.assertIsInstance(self.cp3.plan, pd.DataFrame)
def test_operation(self):
cp_self_add = self.cp1 + self.cp1
cp_add = self.cp1 + self.cp2
cp_add_int = self.cp1 + 10000
cp_mul_int = self.cp1 * 2
cp_mul_float = self.cp2 * 1.5
cp_mul_time = 3 * self.cp2
cp_mul_time2 = 2 * self.cp1
cp_mul_time3 = 2 * self.cp3
cp_mul_float2 = 2. * self.cp3
self.assertIsInstance(cp_self_add, qt.CashPlan)
self.assertEqual(cp_self_add.amounts, [40000, 20000])
self.assertEqual(cp_add.amounts, [20000, 10000, 10000])
self.assertEqual(cp_add_int.amounts, [30000, 20000])
self.assertEqual(cp_mul_int.amounts, [40000, 20000])
self.assertEqual(cp_mul_float.amounts, [15000])
self.assertEqual(cp_mul_float.dates, [Timestamp('2010-05-01')])
self.assertEqual(cp_mul_time.amounts, [10000, 10000, 10000])
self.assertEqual(cp_mul_time.dates, [Timestamp('2010-05-01'),
Timestamp('2011-05-01'),
Timestamp('2012-04-30')])
self.assertEqual(cp_mul_time2.amounts, [20000, 10000, 20000, 10000])
self.assertEqual(cp_mul_time2.dates, [Timestamp('2010-01-01'),
Timestamp('2012-01-01'),
Timestamp('2014-01-01'),
Timestamp('2016-01-01')])
self.assertEqual(cp_mul_time3.dates, [Timestamp('2019-12-31'),
Timestamp('2020-12-31'),
Timestamp('2021-12-31'),
Timestamp('2022-12-31'),
Timestamp('2023-12-31'),
Timestamp('2024-12-31'),
Timestamp('2025-12-31'),
Timestamp('2026-12-31'),
Timestamp('2027-12-31'),
| Timestamp('2028-12-31') | pandas.Timestamp |
""" I/O functions of the aecg package: tools for annotated ECG HL7 XML files
This module implements helper functions to parse and read annotated
electrocardiogram (ECG) stored in XML files following HL7
specification.
See authors, license and disclaimer at the top level directory of this project.
"""
# Imports =====================================================================
from typing import Dict, Tuple
from lxml import etree
from aecg import validate_xpath, new_validation_row, VALICOLS, \
TIME_CODES, SEQUENCE_CODES, \
Aecg, AecgLead, AecgAnnotationSet
import copy
import logging
import pandas as pd
import re
import zipfile
# Python logging ==============================================================
logger = logging.getLogger(__name__)
def parse_annotations(xml_filename: str,
zip_filename: str,
aecg_doc: etree._ElementTree,
aecgannset: AecgAnnotationSet,
path_prefix: str,
annsset_xmlnode_path: str,
valgroup: str = "RHYTHM",
log_validation: bool = False) -> Tuple[
AecgAnnotationSet, pd.DataFrame]:
"""Parses `aecg_doc` XML document and extracts annotations
Args:
xml_filename (str): Filename of the aECG XML file.
zip_filename (str): Filename of zip file containint the aECG XML file.
If '', then xml file is not stored in a zip file.
aecg_doc (etree._ElementTree): XML document of the aECG XML file.
aecgannset (AecgAnnotationSet): Annotation set to which append found
annotations.
path_prefix (str): Prefix of xml path from which start searching for
annotations.
annsset_xmlnode_path (str): Path to xml node of the annotation set
containing the annotations.
valgroup (str, optional): Indicates whether to search annotations in
rhythm or derived waveform. Defaults to "RHYTHM".
log_validation (bool, optional): Indicates whether to maintain the
validation results in `aecg.validatorResults`. Defaults to
False.
Returns:
Tuple[AecgAnnotationSet, pd.DataFrame]: Annotation set updated with
found annotations and dataframe with results of validation.
"""
anngrpid = 0
# Annotations stored within a beat
beatnodes = aecg_doc.xpath((
path_prefix +
"/component/annotation/code[@code=\'MDC_ECG_BEAT\']").replace(
'/', '/ns:'), namespaces={'ns': 'urn:hl7-org:v3'})
beatnum = 0
valpd = pd.DataFrame()
if len(beatnodes) > 0:
logger.info(
f'{xml_filename},{zip_filename},'
f'{valgroup} {len(beatnodes)} annotated beats found')
for beatnode in beatnodes:
for rel_path in ["../component/annotation/"
"code[contains(@code, \"MDC_ECG_\")]"]:
annsnodes = beatnode.xpath(rel_path.replace('/', '/ns:'),
namespaces={'ns': 'urn:hl7-org:v3'})
rel_path2 = "../value"
for annsnode in annsnodes:
ann = {"anngrpid": anngrpid, "beatnum": "", "code": "",
"codetype": "",
"wavecomponent": "", "wavecomponent2": "",
"timecode": "",
"value": "", "value_unit": "",
"low": "", "low_unit": "",
"high": "", "high_unit": "",
"lead": ""}
# Annotation code
valrow2 = validate_xpath(
annsnode,
".",
"urn:hl7-org:v3",
"code",
new_validation_row(xml_filename,
valgroup,
"ANNSET_BEAT_ANNS"),
failcat="WARNING")
valrow2["XPATH"] = annsset_xmlnode_path + "/" + rel_path
if valrow2["VALIOUT"] == "PASSED":
ann["code"] = valrow2["VALUE"]
# Annotation type from top level value
valrow2 = validate_xpath(annsnode,
"../value",
"urn:hl7-org:v3",
"code",
new_validation_row(
xml_filename, valgroup,
"ANNSET_BEAT_ANNS"),
failcat="WARNING")
valrow2["XPATH"] = annsset_xmlnode_path + "/value"
if log_validation:
valpd = valpd.append(pd.DataFrame(
[valrow2], columns=VALICOLS), ignore_index=True)
if valrow2["VALIOUT"] == "PASSED":
ann["codetype"] = valrow2["VALUE"]
# Annotations type
valrow2 = validate_xpath(
annsnode,
rel_path2,
"urn:hl7-org:v3",
"code",
new_validation_row(xml_filename,
valgroup,
"ANNSET_BEAT_ANNS"),
failcat="WARNING")
valrow2["XPATH"] = annsset_xmlnode_path + "/" + rel_path + \
"/" + rel_path2
if valrow2["VALIOUT"] == "PASSED":
ann["beatnum"] = beatnum
ann["codetype"] = valrow2["VALUE"]
if log_validation:
valpd = valpd.append(
pd.DataFrame([valrow2], columns=VALICOLS),
ignore_index=True)
subannsnodes = annsnode.xpath(
rel_path.replace('/', '/ns:'),
namespaces={'ns': 'urn:hl7-org:v3'})
if len(subannsnodes) == 0:
subannsnodes = [annsnode]
else:
subannsnodes += [annsnode]
# Exclude annotations reporting interval values only
subannsnodes = [
sa for sa in subannsnodes
if not sa.get("code").startswith("MDC_ECG_TIME_PD_")]
for subannsnode in subannsnodes:
# Annotations type
valrow2 = validate_xpath(subannsnode,
rel_path2,
"urn:hl7-org:v3",
"code",
new_validation_row(
xml_filename,
valgroup,
"ANNSET_BEAT_ANNS"),
failcat="WARNING")
valrow2["XPATH"] = annsset_xmlnode_path + "/" + \
rel_path + "/" + rel_path2
if valrow2["VALIOUT"] == "PASSED":
ann["wavecomponent"] = valrow2["VALUE"]
if log_validation:
valpd = valpd.append(
pd.DataFrame([valrow2], columns=VALICOLS),
ignore_index=True)
# Annotations value
valrow2 = validate_xpath(subannsnode,
rel_path2,
"urn:hl7-org:v3",
"value",
new_validation_row(
xml_filename,
valgroup,
"ANNSET_BEAT_ANNS"),
failcat="WARNING")
valrow2["XPATH"] = annsset_xmlnode_path + "/" + \
rel_path + "/" + rel_path2
if valrow2["VALIOUT"] == "PASSED":
ann["value"] = valrow2["VALUE"]
if log_validation:
valpd = valpd.append(
pd.DataFrame([valrow2], columns=VALICOLS),
ignore_index=True)
# Annotations value units
valrow2 = validate_xpath(subannsnode,
rel_path2,
"urn:hl7-org:v3",
"unit",
new_validation_row(
xml_filename,
valgroup,
"ANNSET_BEAT_ANNS"),
failcat="WARNING")
valrow2["XPATH"] = annsset_xmlnode_path + "/" + \
rel_path + "/" + rel_path2
if valrow2["VALIOUT"] == "PASSED":
ann["value_unit"] = valrow2["VALUE"]
if log_validation:
valpd = valpd.append(
pd.DataFrame([valrow2], columns=VALICOLS),
ignore_index=True)
# annotations info from supporting ROI
rel_path3 = "../support/supportingROI/component/"\
"boundary/value"
for n in ["", "low", "high"]:
if n != "":
rp = rel_path3 + "/" + n
else:
rp = rel_path3
valrow3 = validate_xpath(
subannsnode,
rp,
"urn:hl7-org:v3",
"value",
new_validation_row(xml_filename,
valgroup,
"ANNSET_BEAT_ANNS"),
failcat="WARNING")
valrow3["XPATH"] = annsset_xmlnode_path + "/" + \
rel_path + "/" + rp
if valrow3["VALIOUT"] == "PASSED":
if n != "":
ann[n] = valrow3["VALUE"]
else:
ann["value"] = valrow3["VALUE"]
if log_validation:
valpd = valpd.append(
pd.DataFrame([valrow3], columns=VALICOLS),
ignore_index=True)
valrow3 = validate_xpath(
subannsnode,
rp,
"urn:hl7-org:v3",
"unit",
new_validation_row(xml_filename,
valgroup,
"ANNSET_BEAT_ANNS"),
failcat="WARNING")
valrow3["XPATH"] = annsset_xmlnode_path + "/" + \
rel_path + "/" + rp
if valrow3["VALIOUT"] == "PASSED":
if n != "":
ann[n + "_unit"] = valrow3["VALUE"]
else:
ann["value_unit"] = valrow3["VALUE"]
if log_validation:
valpd = valpd.append(
pd.DataFrame([valrow3], columns=VALICOLS),
ignore_index=True)
# annotations time encoding, lead and other info used
# by value and supporting ROI
rel_path4 = "../support/supportingROI/component/"\
"boundary/code"
roinodes = subannsnode.xpath(
rel_path4.replace('/', '/ns:'),
namespaces={'ns': 'urn:hl7-org:v3'})
for roinode in roinodes:
valrow4 = validate_xpath(
roinode,
".",
"urn:hl7-org:v3",
"code",
new_validation_row(xml_filename,
valgroup,
"ANNSET_BEAT_ANNS"),
failcat="WARNING")
valrow4["XPATH"] = annsset_xmlnode_path + "/" + \
rel_path + "/" + rel_path4
if valrow4["VALIOUT"] == "PASSED":
if valrow4["VALUE"] in ["TIME_ABSOLUTE",
"TIME_RELATIVE"]:
ann["timecode"] = valrow4["VALUE"]
else:
ann["lead"] = valrow4["VALUE"]
if log_validation:
valpd = valpd.append(
pd.DataFrame([valrow4], columns=VALICOLS),
ignore_index=True)
aecgannset.anns.append(copy.deepcopy(ann))
else:
# Annotations type
valrow2 = validate_xpath(annsnode,
".",
"urn:hl7-org:v3",
"code",
new_validation_row(xml_filename,
valgroup,
"ANNSET_BEAT_"
"ANNS"),
failcat="WARNING")
valrow2["XPATH"] = annsset_xmlnode_path + "/" + rel_path +\
"/" + rel_path2
if valrow2["VALIOUT"] == "PASSED":
ann["beatnum"] = beatnum
ann["codetype"] = valrow2["VALUE"]
if log_validation:
valpd = valpd.append(
pd.DataFrame([valrow2], columns=VALICOLS),
ignore_index=True)
# Annotations value
valrow2 = validate_xpath(annsnode,
rel_path2,
"urn:hl7-org:v3",
"value",
new_validation_row(
xml_filename,
valgroup,
"ANNSET_BEAT_ANNS"),
failcat="WARNING")
valrow2["XPATH"] = annsset_xmlnode_path + "/" + \
rel_path + "/" + rel_path2
if valrow2["VALIOUT"] == "PASSED":
ann["value"] = valrow2["VALUE"]
if log_validation:
valpd = valpd.append(
pd.DataFrame([valrow2], columns=VALICOLS),
ignore_index=True)
# Annotations value units
valrow2 = validate_xpath(annsnode,
rel_path2,
"urn:hl7-org:v3",
"unit",
new_validation_row(
xml_filename,
valgroup,
"ANNSET_BEAT_ANNS"),
failcat="WARNING")
valrow2["XPATH"] = annsset_xmlnode_path + "/" + \
rel_path + "/" + rel_path2
if valrow2["VALIOUT"] == "PASSED":
ann["value_unit"] = valrow2["VALUE"]
if log_validation:
valpd = valpd.append(
pd.DataFrame([valrow2], columns=VALICOLS),
ignore_index=True)
# annotations time encoding, lead and other info used
# by value and supporting ROI
rel_path4 = "../support/supportingROI/component/" \
"boundary/code"
roinodes = annsnode.xpath(
rel_path4.replace('/', '/ns:'),
namespaces={'ns': 'urn:hl7-org:v3'})
for roinode in roinodes:
valrow4 = validate_xpath(roinode,
".",
"urn:hl7-org:v3",
"code",
new_validation_row(
xml_filename,
valgroup,
"ANNSET_BEAT_ANNS"),
failcat="WARNING")
valrow4["XPATH"] = annsset_xmlnode_path + "/" + \
rel_path + "/" + rel_path4
if valrow4["VALIOUT"] == "PASSED":
if valrow4["VALUE"] in ["TIME_ABSOLUTE",
"TIME_RELATIVE"]:
ann["timecode"] = valrow4["VALUE"]
else:
ann["lead"] = valrow4["VALUE"]
if log_validation:
valpd = valpd.append(
pd.DataFrame([valrow4],
columns=VALICOLS),
ignore_index=True)
aecgannset.anns.append(copy.deepcopy(ann))
else:
if log_validation:
valpd = valpd.append(
pd.DataFrame([valrow2], columns=VALICOLS),
ignore_index=True)
anngrpid = anngrpid + 1
beatnum = beatnum + 1
if len(beatnodes) > 0:
logger.info(
f'{xml_filename},{zip_filename},'
f'{valgroup} {beatnum} annotated beats and {anngrpid} '
f'annotations groups found')
anngrpid_from_beats = anngrpid
# Annotations stored without an associated beat
for codetype_path in ["/component/annotation/code["
"(contains(@code, \"MDC_ECG_\") and"
" not (@code=\'MDC_ECG_BEAT\'))]"]:
annsnodes = aecg_doc.xpath(
(path_prefix + codetype_path).replace('/', '/ns:'),
namespaces={'ns': 'urn:hl7-org:v3'})
rel_path2 = "../value"
for annsnode in annsnodes:
ann = {"anngrpid": anngrpid, "beatnum": "", "code": "",
"codetype": "",
"wavecomponent": "", "wavecomponent2": "",
"timecode": "",
"value": "", "value_unit": "",
"low": "", "low_unit": "",
"high": "", "high_unit": "",
"lead": ""}
# Annotations code
valrow2 = validate_xpath(annsnode,
".",
"urn:hl7-org:v3",
"code",
new_validation_row(xml_filename, valgroup,
"ANNSET_NOBEAT_ANNS"),
failcat="WARNING")
valrow2["XPATH"] = annsset_xmlnode_path
if log_validation:
valpd = valpd.append(pd.DataFrame([valrow2], columns=VALICOLS),
ignore_index=True)
if valrow2["VALIOUT"] == "PASSED":
ann["code"] = valrow2["VALUE"]
# Annotation type from top level value
valrow2 = validate_xpath(annsnode,
"../value",
"urn:hl7-org:v3",
"code",
new_validation_row(xml_filename, valgroup,
"ANNSET_NOBEAT_ANNS"),
failcat="WARNING")
valrow2["XPATH"] = annsset_xmlnode_path + "/value"
if log_validation:
valpd = valpd.append(pd.DataFrame([valrow2], columns=VALICOLS),
ignore_index=True)
if valrow2["VALIOUT"] == "PASSED":
ann["codetype"] = valrow2["VALUE"]
subannsnodes = annsnode.xpath(
(".." + codetype_path).replace('/', '/ns:'),
namespaces={'ns': 'urn:hl7-org:v3'})
if len(subannsnodes) == 0:
subannsnodes = [annsnode]
for subannsnode in subannsnodes:
subsubannsnodes = subannsnode.xpath(
(".." + codetype_path).replace('/', '/ns:'),
namespaces={'ns': 'urn:hl7-org:v3'})
tmpnodes = [subannsnode]
if len(subsubannsnodes) > 0:
tmpnodes = tmpnodes + subsubannsnodes
for subsubannsnode in tmpnodes:
ann["wavecomponent"] = ""
ann["wavecomponent2"] = ""
ann["timecode"] = ""
ann["value"] = ""
ann["value_unit"] = ""
ann["low"] = ""
ann["low_unit"] = ""
ann["high"] = ""
ann["high_unit"] = ""
roi_base = "../support/supportingROI/component/boundary"
rel_path3 = roi_base + "/value"
valrow2 = validate_xpath(
subsubannsnode,
".",
"urn:hl7-org:v3",
"code",
new_validation_row(xml_filename,
valgroup,
"ANNSET_NOBEAT_"
"ANNS"),
failcat="WARNING")
valrow2["XPATH"] = annsset_xmlnode_path + "/.." + \
codetype_path + "/code"
if valrow2["VALIOUT"] == "PASSED":
if not ann["codetype"].endswith("WAVE"):
ann["codetype"] = valrow2["VALUE"]
if log_validation:
valpd = valpd.append(
pd.DataFrame([valrow2], columns=VALICOLS),
ignore_index=True)
# Annotations type
valrow2 = validate_xpath(
subsubannsnode,
rel_path2,
"urn:hl7-org:v3",
"code",
new_validation_row(xml_filename,
valgroup,
"ANNSET_NOBEAT_"
"ANNS"),
failcat="WARNING")
valrow2["XPATH"] = annsset_xmlnode_path + "/.." + \
codetype_path + "/" + rel_path2
if valrow2["VALIOUT"] == "PASSED":
ann["wavecomponent"] = valrow2["VALUE"]
# if ann["wavecomponent"] == "":
# ann["wavecomponent"] = valrow2["VALUE"]
# else:
# ann["wavecomponent2"] = valrow2["VALUE"]
if log_validation:
valpd = valpd.append(
pd.DataFrame([valrow2], columns=VALICOLS),
ignore_index=True)
# Annotations value
valrow2 = validate_xpath(
subsubannsnode,
rel_path2,
"urn:hl7-org:v3",
"",
new_validation_row(xml_filename,
valgroup,
"ANNSET_NOBEAT_"
"ANNS"),
failcat="WARNING")
valrow2["XPATH"] = annsset_xmlnode_path + "/.." + \
codetype_path + "/" + rel_path2
if valrow2["VALIOUT"] == "PASSED":
ann["value"] = valrow2["VALUE"]
if log_validation:
valpd = valpd.append(
pd.DataFrame([valrow2], columns=VALICOLS),
ignore_index=True)
# Annotations value as attribute
valrow2 = validate_xpath(
subsubannsnode,
rel_path2,
"urn:hl7-org:v3",
"value",
new_validation_row(xml_filename,
valgroup,
"ANNSET_NOBEAT_"
"ANNS"),
failcat="WARNING")
valrow2["XPATH"] = annsset_xmlnode_path + "/.." + \
codetype_path + "/" + rel_path2
if valrow2["VALIOUT"] == "PASSED":
ann["value"] = valrow2["VALUE"]
if log_validation:
valpd = valpd.append(
pd.DataFrame([valrow2], columns=VALICOLS),
ignore_index=True)
# Annotations value units
valrow2 = validate_xpath(
subsubannsnode,
rel_path2,
"urn:hl7-org:v3",
"unit",
new_validation_row(xml_filename,
valgroup,
"ANNSET_NOBEAT_"
"ANNS"),
failcat="WARNING")
valrow2["XPATH"] = annsset_xmlnode_path + "/.." + \
codetype_path + "/" + rel_path2
if valrow2["VALIOUT"] == "PASSED":
ann["value_unit"] = valrow2["VALUE"]
if log_validation:
valpd = valpd.append(
pd.DataFrame([valrow2], columns=VALICOLS),
ignore_index=True)
# annotations info from supporting ROI
for n in ["", "low", "high"]:
if n != "":
rp = rel_path3 + "/" + n
else:
rp = rel_path3
valrow3 = validate_xpath(
subsubannsnode,
rp,
"urn:hl7-org:v3",
"value",
new_validation_row(xml_filename,
valgroup,
"ANNSET_NOBEAT_"
"ANNS"),
failcat="WARNING")
valrow3["XPATH"] = annsset_xmlnode_path + "/.." + \
codetype_path + "/" + rp
if valrow3["VALIOUT"] == "PASSED":
if n != "":
ann[n] = valrow3["VALUE"]
else:
ann["value"] = valrow3["VALUE"]
else:
roi_base = "../component/annotation/support/"\
"supportingROI/component/boundary"
# Annotations type
valrow2 = validate_xpath(subsubannsnode,
"../component/annotation/"
"value",
"urn:hl7-org:v3",
"code",
new_validation_row(
xml_filename,
valgroup,
"ANNSET_NOBEAT_ANNS"),
failcat="WARNING")
valrow2["XPATH"] = annsset_xmlnode_path + "/.." + \
codetype_path + "/" + \
"../component/annotation/value"
if valrow2["VALIOUT"] == "PASSED":
ann["wavecomponent2"] = valrow2["VALUE"]
if log_validation:
valpd = valpd.append(
pd.DataFrame([valrow2], columns=VALICOLS),
ignore_index=True)
# annotation values
if n != "":
rp = roi_base + "/value/" + n
else:
rp = roi_base + "/value"
valrow3 = validate_xpath(subsubannsnode,
rp,
"urn:hl7-org:v3",
"value",
new_validation_row(
xml_filename,
valgroup,
"ANNSET_NOBEAT_ANNS"),
failcat="WARNING")
valrow3["XPATH"] = annsset_xmlnode_path + "/.." + \
codetype_path + "/" + rp
if valrow3["VALIOUT"] == "PASSED":
if n != "":
ann[n] = valrow3["VALUE"]
else:
ann["value"] = valrow3["VALUE"]
if log_validation:
valpd = valpd.append(
pd.DataFrame([valrow3], columns=VALICOLS),
ignore_index=True)
valrow3 = validate_xpath(
subsubannsnode,
rp,
"urn:hl7-org:v3",
"unit",
new_validation_row(xml_filename,
valgroup,
"ANNSET_NOBEAT"
"_ANNS"),
failcat="WARNING")
valrow3["XPATH"] = annsset_xmlnode_path + "/.." + \
codetype_path + "/" + rp
if valrow3["VALIOUT"] == "PASSED":
if n != "":
ann[n + "_unit"] = valrow3["VALUE"]
else:
ann["value_unit"] = valrow3["VALUE"]
if log_validation:
valpd = valpd.append(
pd.DataFrame([valrow3], columns=VALICOLS),
ignore_index=True)
# annotations time encoding, lead and other info used by
# value and supporting ROI
for rel_path4 in ["../support/supportingROI/component/"
"boundary",
"../component/annotation/support/"
"supportingROI/component/boundary"]:
roinodes = subsubannsnode.xpath(
rel_path4.replace('/', '/ns:'),
namespaces={'ns': 'urn:hl7-org:v3'})
for roinode in roinodes:
valrow4 = validate_xpath(roinode,
"./code",
"urn:hl7-org:v3",
"code",
new_validation_row(
xml_filename,
valgroup,
"ANNSET_NOBEAT_ANNS"),
failcat="WARNING")
valrow4["XPATH"] = annsset_xmlnode_path + "/.." + \
codetype_path + "/" + rel_path4
if valrow4["VALIOUT"] == "PASSED":
if valrow4["VALUE"] in ["TIME_ABSOLUTE",
"TIME_RELATIVE"]:
ann["timecode"] = valrow4["VALUE"]
else:
ann["lead"] = valrow4["VALUE"]
if log_validation:
valpd = valpd.append(
pd.DataFrame([valrow4], columns=VALICOLS),
ignore_index=True)
aecgannset.anns.append(copy.deepcopy(ann))
anngrpid = anngrpid + 1
logger.info(
f'{xml_filename},{zip_filename},'
f'{valgroup} {anngrpid-anngrpid_from_beats} annotations groups'
f' without an associated beat found')
return aecgannset, valpd
def parse_generalinfo(aecg_doc: etree._ElementTree,
aecg: Aecg,
log_validation: bool = False) -> Aecg:
"""Parses `aecg_doc` XML document and extracts general information
This function parses the `aecg_doc` xml document searching for general
information that includes in the returned `Aecg`: unique identifier (UUID),
ECG date and time of collection (EGDTC), and device information.
Args:
aecg_doc (etree._ElementTree): aECG XML document
aecg (Aecg): The aECG object to update
log_validation (bool, optional): Indicates whether to maintain the
validation results in `aecg.validatorResults`. Defaults to
False.
Returns:
Aecg: `aecg` updated with the information found in the xml document.
"""
# =======================================
# UUID
# =======================================
valrow = validate_xpath(aecg_doc,
"./*[local-name() = \"id\"]",
"",
"root",
new_validation_row(aecg.filename,
"GENERAL",
"UUID"))
if log_validation:
aecg.validatorResults = aecg.validatorResults.append(
pd.DataFrame([valrow], columns=VALICOLS), ignore_index=True)
if valrow["VALIOUT"] == "PASSED":
logger.info(
f'{aecg.filename},{aecg.zipContainer},'
f'UUID found: {valrow["VALUE"]}')
aecg.UUID = valrow["VALUE"]
else:
logger.critical(
f'{aecg.filename},{aecg.zipContainer},'
f'UUID not found')
valrow = validate_xpath(aecg_doc,
"./*[local-name() = \"id\"]",
"",
"extension",
new_validation_row(aecg.filename,
"GENERAL",
"UUID"))
if log_validation:
aecg.validatorResults = aecg.validatorResults.append(
pd.DataFrame([valrow], columns=VALICOLS), ignore_index=True)
if valrow["VALIOUT"] == "PASSED":
logger.debug(
f'{aecg.filename},{aecg.zipContainer},'
f'UUID extension found: {valrow["VALUE"]}')
aecg.UUID += valrow["VALUE"]
logger.info(
f'{aecg.filename},{aecg.zipContainer},'
f'UUID updated to: {aecg.UUID}')
else:
logger.debug(
f'{aecg.filename},{aecg.zipContainer},'
f'UUID extension not found')
# =======================================
# EGDTC
# =======================================
valpd = pd.DataFrame()
egdtc_found = False
for n in ["low", "center", "high"]:
valrow = validate_xpath(aecg_doc,
"./*[local-name() = \"effectiveTime\"]/"
"*[local-name() = \"" + n + "\"]",
"",
"value",
new_validation_row(aecg.filename, "GENERAL",
"EGDTC_" + n),
"WARNING")
if valrow["VALIOUT"] == "PASSED":
egdtc_found = True
logger.info(
f'{aecg.filename},{aecg.zipContainer},'
f'EGDTC {n} found: {valrow["VALUE"]}')
aecg.EGDTC[n] = valrow["VALUE"]
if log_validation:
valpd = valpd.append(pd.DataFrame([valrow], columns=VALICOLS),
ignore_index=True)
if not egdtc_found:
logger.critical(
f'{aecg.filename},{aecg.zipContainer},'
f'EGDTC not found')
if log_validation:
aecg.validatorResults = aecg.validatorResults.append(valpd,
ignore_index=True)
# =======================================
# DEVICE
# =======================================
# DEVICE = {"manufacturer": "", "model": "", "software": ""}
valrow = validate_xpath(aecg_doc,
"./component/series/author/"
"seriesAuthor/manufacturerOrganization/name",
"urn:hl7-org:v3",
"",
new_validation_row(aecg.filename, "GENERAL",
"DEVICE_manufacturer"),
"WARNING")
if valrow["VALIOUT"] == "PASSED":
tmp = valrow["VALUE"].replace("\n", "|")
logger.info(
f'{aecg.filename},{aecg.zipContainer},'
f'DEVICE manufacturer found: {tmp}')
aecg.DEVICE["manufacturer"] = valrow["VALUE"]
else:
logger.warning(
f'{aecg.filename},{aecg.zipContainer},'
f'DEVICE manufacturer not found')
if log_validation:
aecg.validatorResults = aecg.validatorResults.append(
pd.DataFrame([valrow], columns=VALICOLS), ignore_index=True)
valrow = validate_xpath(aecg_doc,
"./component/series/author/"
"seriesAuthor/manufacturedSeriesDevice/"
"manufacturerModelName",
"urn:hl7-org:v3",
"",
new_validation_row(aecg.filename, "GENERAL",
"DEVICE_model"),
"WARNING")
if valrow["VALIOUT"] == "PASSED":
tmp = valrow["VALUE"].replace("\n", "|")
logger.info(
f'{aecg.filename},{aecg.zipContainer},'
f'DEVICE model found: {tmp}')
aecg.DEVICE["model"] = valrow["VALUE"]
else:
logger.warning(
f'{aecg.filename},{aecg.zipContainer},'
f'DEVICE model not found')
if log_validation:
aecg.validatorResults = aecg.validatorResults.append(
pd.DataFrame([valrow], columns=VALICOLS), ignore_index=True)
valrow = validate_xpath(aecg_doc,
"./component/series/author/"
"seriesAuthor/manufacturedSeriesDevice/"
"softwareName",
"urn:hl7-org:v3",
"",
new_validation_row(aecg.filename, "GENERAL",
"DEVICE_software"),
"WARNING")
if valrow["VALIOUT"] == "PASSED":
tmp = valrow["VALUE"].replace("\n", "|")
logger.info(
f'{aecg.filename},{aecg.zipContainer},'
f'DEVICE software found: {tmp}')
aecg.DEVICE["software"] = valrow["VALUE"]
else:
logger.warning(
f'{aecg.filename},{aecg.zipContainer},'
f'DEVICE software not found')
if log_validation:
aecg.validatorResults = aecg.validatorResults.append(
pd.DataFrame([valrow], columns=VALICOLS), ignore_index=True)
return aecg
def parse_subjectinfo(aecg_doc: etree._ElementTree,
aecg: Aecg,
log_validation: bool = False) -> Aecg:
"""Parses `aecg_doc` XML document and extracts subject information
This function parses the `aecg_doc` xml document searching for subject
information that includes in the returned `Aecg`: subject unique identifier
(USUBJID), gender, birthtime, and race.
Args:
aecg_doc (etree._ElementTree): aECG XML document
aecg (Aecg): The aECG object to update
log_validation (bool, optional): Indicates whether to maintain the
validation results in `aecg.validatorResults`. Defaults to
False.
Returns:
Aecg: `aecg` updated with the information found in the xml document.
"""
# =======================================
# USUBJID
# =======================================
valpd = pd.DataFrame()
for n in ["root", "extension"]:
valrow = validate_xpath(aecg_doc,
"./componentOf/timepointEvent/componentOf/"
"subjectAssignment/subject/trialSubject/id",
"urn:hl7-org:v3",
n,
new_validation_row(aecg.filename,
"SUBJECTINFO",
"USUBJID_" + n))
if valrow["VALIOUT"] == "PASSED":
logger.info(
f'{aecg.filename},{aecg.zipContainer},'
f'DM.USUBJID ID {n} found: {valrow["VALUE"]}')
aecg.USUBJID[n] = valrow["VALUE"]
else:
if n == "root":
logger.warning(
f'{aecg.filename},{aecg.zipContainer},'
f'DM.USUBJID ID {n} not found')
else:
logger.warning(
f'{aecg.filename},{aecg.zipContainer},'
f'DM.USUBJID ID {n} not found')
if log_validation:
valpd = valpd.append(pd.DataFrame([valrow], columns=VALICOLS),
ignore_index=True)
if (aecg.USUBJID["root"] == "") and (aecg.USUBJID["extension"] == ""):
logger.error(
f'{aecg.filename},{aecg.zipContainer},'
f'DM.USUBJID cannot be established.')
if log_validation:
aecg.validatorResults = aecg.validatorResults.append(valpd,
ignore_index=True)
# =======================================
# SEX / GENDER
# =======================================
valrow = validate_xpath(aecg_doc,
"./componentOf/timepointEvent/componentOf/"
"subjectAssignment/subject/trialSubject/"
"subjectDemographicPerson/"
"administrativeGenderCode",
"urn:hl7-org:v3",
"code",
new_validation_row(aecg.filename, "SUBJECTINFO",
"SEX"),
failcat="WARNING")
if valrow["VALIOUT"] == "PASSED":
logger.info(
f'{aecg.filename},{aecg.zipContainer},'
f'DM.SEX found: {valrow["VALUE"]}')
aecg.SEX = valrow["VALUE"]
else:
logger.debug(
f'{aecg.filename},{aecg.zipContainer},'
f'DM.SEX not found')
if log_validation:
aecg.validatorResults = aecg.validatorResults.append(
pd.DataFrame([valrow], columns=VALICOLS), ignore_index=True)
# =======================================
# BIRTHTIME
# =======================================
valrow = validate_xpath(aecg_doc,
"./componentOf/timepointEvent/componentOf/"
"subjectAssignment/subject/trialSubject/"
"subjectDemographicPerson/birthTime",
"urn:hl7-org:v3",
"value",
new_validation_row(aecg.filename, "SUBJECTINFO",
"BIRTHTIME"),
failcat="WARNING")
if valrow["VALIOUT"] == "PASSED":
logger.info(
f'{aecg.filename},{aecg.zipContainer},'
f'DM.BIRTHTIME found.')
aecg.BIRTHTIME = valrow["VALUE"]
# age_in_years = aecg.subject_age_in_years()
else:
logger.debug(
f'{aecg.filename},{aecg.zipContainer},'
f'DM.BIRTHTIME not found')
if log_validation:
aecg.validatorResults = aecg.validatorResults.append(
pd.DataFrame([valrow], columns=VALICOLS), ignore_index=True)
# =======================================
# RACE
# =======================================
valrow = validate_xpath(aecg_doc,
"./componentOf/timepointEvent/componentOf/"
"subjectAssignment/subject/trialSubject/"
"subjectDemographicPerson/raceCode",
"urn:hl7-org:v3",
"code",
new_validation_row(aecg.filename, "SUBJECTINFO",
"RACE"),
failcat="WARNING")
if valrow["VALIOUT"] == "PASSED":
logger.info(
f'{aecg.filename},{aecg.zipContainer},'
f'DM.RACE found: {valrow["VALUE"]}')
else:
logger.debug(
f'{aecg.filename},{aecg.zipContainer},'
f'DM.RACE not found')
aecg.RACE = valrow["VALUE"]
if log_validation:
aecg.validatorResults = aecg.validatorResults.append(
pd.DataFrame([valrow], columns=VALICOLS), ignore_index=True)
return aecg
def parse_trtainfo(aecg_doc: etree._ElementTree,
aecg: Aecg,
log_validation: bool = False) -> Aecg:
"""Parses `aecg_doc` XML document and extracts subject information
This function parses the `aecg_doc` xml document searching for treatment
information that includes in the returned `Aecg`.
Args:
aecg_doc (etree._ElementTree): aECG XML document
aecg (Aecg): The aECG object to update
log_validation (bool, optional): Indicates whether to maintain the
validation results in `aecg.validatorResults`. Defaults to
False.
Returns:
Aecg: `aecg` updated with the information found in the xml document.
"""
valrow = validate_xpath(aecg_doc,
"./componentOf/timepointEvent/componentOf/"
"subjectAssignment/definition/"
"treatmentGroupAssignment/code",
"urn:hl7-org:v3",
"code",
new_validation_row(aecg.filename, "STUDYINFO",
"TRTA"),
failcat="WARNING")
if valrow["VALIOUT"] == "PASSED":
logger.info(
f'{aecg.filename},{aecg.zipContainer},'
f'TRTA information found: {valrow["VALUE"]}')
aecg.TRTA = valrow["VALUE"]
else:
logger.debug(
f'{aecg.filename},{aecg.zipContainer},'
f'TRTA information not found')
if log_validation:
aecg.validatorResults = aecg.validatorResults.append(
pd.DataFrame([valrow], columns=VALICOLS), ignore_index=True)
return aecg
def parse_studyinfo(aecg_doc: etree._ElementTree,
aecg: Aecg,
log_validation: bool = False) -> Aecg:
"""Parses `aecg_doc` XML document and extracts study information
This function parses the `aecg_doc` xml document searching for study
information that includes in the returned `Aecg`: study unique identifier
(STUDYID), and study title.
Args:
aecg_doc (etree._ElementTree): aECG XML document
aecg (Aecg): The aECG object to update
log_validation (bool, optional): Indicates whether to maintain the
validation results in `aecg.validatorResults`. Defaults to
False.
Returns:
Aecg: `aecg` updated with the information found in the xml document.
"""
valpd = pd.DataFrame()
for n in ["root", "extension"]:
valrow = validate_xpath(aecg_doc,
"./componentOf/timepointEvent/componentOf/"
"subjectAssignment/componentOf/"
"clinicalTrial/id",
"urn:hl7-org:v3",
n,
new_validation_row(aecg.filename,
"STUDYINFO",
"STUDYID_" + n),
failcat="WARNING")
if valrow["VALIOUT"] == "PASSED":
logger.info(
f'{aecg.filename},{aecg.zipContainer},'
f'STUDYID {n} found: {valrow["VALUE"]}')
aecg.STUDYID[n] = valrow["VALUE"]
else:
logger.debug(
f'{aecg.filename},{aecg.zipContainer},'
f'STUDYID {n} not found')
if log_validation:
valpd = valpd.append(pd.DataFrame([valrow], columns=VALICOLS),
ignore_index=True)
if log_validation:
aecg.validatorResults = \
aecg.validatorResults.append(valpd, ignore_index=True)
valrow = validate_xpath(aecg_doc,
"./componentOf/timepointEvent/componentOf/"
"subjectAssignment/componentOf/"
"clinicalTrial/title",
"urn:hl7-org:v3",
"",
new_validation_row(aecg.filename, "STUDYINFO",
"STUDYTITLE"),
failcat="WARNING")
if valrow["VALIOUT"] == "PASSED":
tmp = valrow["VALUE"].replace("\n", "")
logger.info(
f'{aecg.filename},{aecg.zipContainer},'
f'STUDYTITLE found: {tmp}')
aecg.STUDYTITLE = valrow["VALUE"]
else:
logger.debug(
f'{aecg.filename},{aecg.zipContainer},'
f'STUDYTITLE not found')
if log_validation:
aecg.validatorResults = aecg.validatorResults.append(
pd.DataFrame([valrow], columns=VALICOLS), ignore_index=True)
return aecg
def parse_timepoints(aecg_doc: etree._ElementTree,
aecg: Aecg,
log_validation: bool = False) -> Aecg:
"""Parses `aecg_doc` XML document and extracts timepoints information
This function parses the `aecg_doc` xml document searching for timepoints
information that includes in the returned `Aecg`: absolute timepoint or
study event information (TPT), relative timepoint or study event relative
to a reference event (RTPT), and protocol timepoint information (PTPT).
Args:
aecg_doc (etree._ElementTree): aECG XML document
aecg (Aecg): The aECG object to update
log_validation (bool, optional): Indicates whether to maintain the
validation results in `aecg.validatorResults`. Defaults to
False.
Returns:
Aecg: `aecg` updated with the information found in the xml document.
"""
# =======================================
# TPT
# =======================================
valpd = pd.DataFrame()
for n in ["code", "displayName"]:
valrow = validate_xpath(aecg_doc,
"./componentOf/timepointEvent/code",
"urn:hl7-org:v3",
n,
new_validation_row(aecg.filename,
"STUDYINFO",
"TPT_" + n),
failcat="WARNING")
if valrow["VALIOUT"] == "PASSED":
logger.info(
f'{aecg.filename},{aecg.zipContainer},'
f'TPT {n} found: {valrow["VALUE"]}')
aecg.TPT[n] = valrow["VALUE"]
else:
logger.debug(
f'{aecg.filename},{aecg.zipContainer},'
f'TPT {n} not found')
if log_validation:
valpd = valpd.append(pd.DataFrame([valrow], columns=VALICOLS),
ignore_index=True)
if log_validation:
aecg.validatorResults = \
aecg.validatorResults.append(valpd, ignore_index=True)
valrow = validate_xpath(aecg_doc,
"./componentOf/timepointEvent/reasonCode",
"urn:hl7-org:v3",
"code",
new_validation_row(aecg.filename, "STUDYINFO",
"TPT_reasonCode"),
failcat="WARNING")
if valrow["VALIOUT"] == "PASSED":
logger.info(
f'{aecg.filename},{aecg.zipContainer},'
f'TPT reasonCode found: {valrow["VALUE"]}')
aecg.TPT["reasonCode"] = valrow["VALUE"]
else:
logger.debug(
f'{aecg.filename},{aecg.zipContainer},'
f'TPT reasonCode not found')
if log_validation:
aecg.validatorResults = aecg.validatorResults.append(
pd.DataFrame([valrow], columns=VALICOLS), ignore_index=True)
valpd = pd.DataFrame()
for n in ["low", "high"]:
valrow = validate_xpath(aecg_doc,
"./componentOf/timepointEvent/"
"effectiveTime/" + n,
"urn:hl7-org:v3",
"value",
new_validation_row(aecg.filename,
"STUDYINFO",
"TPT_" + n),
failcat="WARNING")
if valrow["VALIOUT"] == "PASSED":
logger.info(
f'{aecg.filename},{aecg.zipContainer},'
f'TPT {n} found: {valrow["VALUE"]}')
aecg.TPT[n] = valrow["VALUE"]
else:
logger.debug(
f'{aecg.filename},{aecg.zipContainer},'
f'TPT {n} not found')
if log_validation:
valpd = valpd.append(pd.DataFrame([valrow], columns=VALICOLS),
ignore_index=True)
if log_validation:
aecg.validatorResults = \
aecg.validatorResults.append(valpd, ignore_index=True)
# =======================================
# RTPT
# =======================================
valpd = pd.DataFrame()
for n in ["code", "displayName"]:
valrow = validate_xpath(aecg_doc,
"./definition/relativeTimepoint/code",
"urn:hl7-org:v3",
"code",
new_validation_row(aecg.filename,
"STUDYINFO",
"RTPT_" + n),
failcat="WARNING")
if valrow["VALIOUT"] == "PASSED":
logger.info(
f'{aecg.filename},{aecg.zipContainer},'
f'RTPT {n} found: {valrow["VALUE"]}')
aecg.RTPT[n] = valrow["VALUE"]
else:
logger.debug(
f'{aecg.filename},{aecg.zipContainer},'
f'RTPT {n} not found')
if log_validation:
valpd = valpd.append(pd.DataFrame([valrow], columns=VALICOLS),
ignore_index=True)
if log_validation:
aecg.validatorResults = \
aecg.validatorResults.append(valpd, ignore_index=True)
valrow = validate_xpath(aecg_doc,
"./definition/relativeTimepoint/componentOf/"
"pauseQuantity",
"urn:hl7-org:v3",
"value",
new_validation_row(aecg.filename, "STUDYINFO",
"RTPT_pauseQuantity"),
failcat="WARNING")
if valrow["VALIOUT"] == "PASSED":
logger.info(
f'{aecg.filename},{aecg.zipContainer},'
f'RTPT pauseQuantity value found: {valrow["VALUE"]}')
aecg.RTPT["pauseQuantity"] = valrow["VALUE"]
else:
logger.debug(
f'{aecg.filename},{aecg.zipContainer},'
f'RTPT pauseQuantity value not found')
if log_validation:
aecg.validatorResults = \
aecg.validatorResults.append(pd.DataFrame([valrow],
columns=VALICOLS),
ignore_index=True)
valrow = validate_xpath(aecg_doc,
"./definition/relativeTimepoint/componentOf/"
"pauseQuantity",
"urn:hl7-org:v3",
"unit",
new_validation_row(aecg.filename, "STUDYINFO",
"RTPT_pauseQuantity_unit"),
failcat="WARNING")
if valrow["VALIOUT"] == "PASSED":
logger.info(
f'{aecg.filename},{aecg.zipContainer},'
f'RTPT pauseQuantity unit found: {valrow["VALUE"]}')
aecg.RTPT["pauseQuantity_unit"] = valrow["VALUE"]
else:
logger.debug(
f'{aecg.filename},{aecg.zipContainer},'
f'RTPT pauseQuantity unit not found')
if log_validation:
aecg.validatorResults = \
aecg.validatorResults.append(pd.DataFrame([valrow],
columns=VALICOLS),
ignore_index=True)
# =======================================
# PTPT
# =======================================
valpd = pd.DataFrame()
for n in ["code", "displayName"]:
valrow = validate_xpath(aecg_doc,
"./definition/relativeTimepoint/"
"componentOf/protocolTimepointEvent/code",
"urn:hl7-org:v3",
n,
new_validation_row(aecg.filename,
"STUDYINFO",
"PTPT_" + n),
failcat="WARNING")
if valrow["VALIOUT"] == "PASSED":
logger.info(
f'{aecg.filename},{aecg.zipContainer},'
f'PTPT {n} found: {valrow["VALUE"]}')
aecg.PTPT[n] = valrow["VALUE"]
else:
logger.debug(
f'{aecg.filename},{aecg.zipContainer},'
f'PTPT {n} not found')
if log_validation:
valpd = valpd.append(pd.DataFrame([valrow], columns=VALICOLS),
ignore_index=True)
if log_validation:
aecg.validatorResults = \
aecg.validatorResults.append(valpd, ignore_index=True)
valrow = validate_xpath(aecg_doc,
"./definition/relativeTimepoint/componentOf/"
"protocolTimepointEvent/component/"
"referenceEvent/code",
"urn:hl7-org:v3",
"code",
new_validation_row(aecg.filename, "STUDYINFO",
"PTPT_referenceEvent"),
failcat="WARNING")
if valrow["VALIOUT"] == "PASSED":
logger.info(
f'{aecg.filename},{aecg.zipContainer},'
f'PTPT referenceEvent code found: {valrow["VALUE"]}')
aecg.PTPT["referenceEvent"] = valrow["VALUE"]
else:
logger.debug(
f'{aecg.filename},{aecg.zipContainer},'
f'PTPT referenceEvent code not found')
if log_validation:
aecg.validatorResults = \
aecg.validatorResults.append(pd.DataFrame([valrow],
columns=VALICOLS),
ignore_index=True)
valrow = validate_xpath(aecg_doc,
"./definition/relativeTimepoint/componentOf/"
"protocolTimepointEvent/component/"
"referenceEvent/code",
"urn:hl7-org:v3",
"displayName",
new_validation_row(aecg.filename, "STUDYINFO",
"PTPT_referenceEvent_"
"displayName"),
failcat="WARNING")
if valrow["VALIOUT"] == "PASSED":
logger.info(
f'{aecg.filename},{aecg.zipContainer},'
f'PTPT referenceEvent displayName found: '
f'{valrow["VALUE"]}')
aecg.PTPT["referenceEvent_displayName"] = valrow["VALUE"]
else:
logger.debug(
f'{aecg.filename},{aecg.zipContainer},'
f'PTPT referenceEvent displayName not found')
if log_validation:
aecg.validatorResults = \
aecg.validatorResults.append(pd.DataFrame([valrow],
columns=VALICOLS),
ignore_index=True)
return aecg
def parse_rhythm_waveform_info(aecg_doc: etree._ElementTree,
aecg: Aecg,
log_validation: bool = False) -> Aecg:
"""Parses `aecg_doc` XML document and extracts rhythm waveform information
This function parses the `aecg_doc` xml document searching for rhythm
waveform information that includes in the returned `Aecg`: waveform
identifier, code, display name, and date and time of collection.
Args:
aecg_doc (etree._ElementTree): aECG XML document
aecg (Aecg): The aECG object to update
log_validation (bool, optional): Indicates whether to maintain the
validation results in `aecg.validatorResults`. Defaults to
False.
Returns:
Aecg: `aecg` updated with the information found in the xml document.
"""
valpd = pd.DataFrame()
for n in ["root", "extension"]:
valrow = validate_xpath(aecg_doc,
"./component/series/id",
"urn:hl7-org:v3",
n,
new_validation_row(aecg.filename, "RHYTHM",
"ID_" + n),
failcat="WARNING")
if valrow["VALIOUT"] == "PASSED":
logger.info(
f'{aecg.filename},{aecg.zipContainer},'
f'RHYTHM ID {n} found: {valrow["VALUE"]}')
aecg.RHYTHMID[n] = valrow["VALUE"]
else:
if n == "root":
logger.warning(
f'{aecg.filename},{aecg.zipContainer},'
f'RHYTHM ID {n} not found')
else:
logger.warning(
f'{aecg.filename},{aecg.zipContainer},'
f'RHYTHM ID {n} not found')
if log_validation:
valpd = valpd.append(pd.DataFrame([valrow], columns=VALICOLS),
ignore_index=True)
if log_validation:
aecg.validatorResults = \
aecg.validatorResults.append(valpd, ignore_index=True)
valrow = validate_xpath(aecg_doc,
"./component/series/code",
"urn:hl7-org:v3",
"code",
new_validation_row(aecg.filename, "RHYTHM",
"CODE"),
failcat="WARNING")
if valrow["VALIOUT"] == "PASSED":
logger.debug(
f'{aecg.filename},{aecg.zipContainer},'
f'RHYTHM code found: {valrow["VALUE"]}')
aecg.RHYTHMCODE["code"] = valrow["VALUE"]
if aecg.RHYTHMCODE["code"] != "RHYTHM":
logger.warning(
f'{aecg.filename},{aecg.zipContainer},'
f'RHYTHM unexpected code found: {valrow["VALUE"]}')
valrow["VALIOUT"] = "WARNING"
valrow["VALIMSG"] = "Unexpected value found"
else:
logger.warning(
f'{aecg.filename},{aecg.zipContainer},'
f'RHYTHM code not found')
if log_validation:
aecg.validatorResults = aecg.validatorResults.append(
pd.DataFrame([valrow], columns=VALICOLS), ignore_index=True)
valrow = validate_xpath(aecg_doc,
"./component/series/code",
"urn:hl7-org:v3",
"displayName",
new_validation_row(aecg.filename, "RHYTHM",
"CODE_displayName"),
failcat="WARNING")
if valrow["VALIOUT"] == "PASSED":
logger.debug(
f'{aecg.filename},{aecg.zipContainer},'
f'RHYTHM displayName found: {valrow["VALUE"]}')
aecg.RHYTHMCODE["displayName"] = valrow["VALUE"]
else:
logger.debug(
f'{aecg.filename},{aecg.zipContainer},'
f'RHYTHM displayName not found')
if log_validation:
aecg.validatorResults = aecg.validatorResults.append(
pd.DataFrame([valrow], columns=VALICOLS), ignore_index=True)
valpd = pd.DataFrame()
for n in ["low", "high"]:
valrow = validate_xpath(aecg_doc,
"./component/series/effectiveTime/" + n,
"urn:hl7-org:v3",
"value",
new_validation_row(aecg.filename, "RHYTHM",
"EGDTC_" + n),
failcat="WARNING")
if valrow["VALIOUT"] == "PASSED":
logger.info(
f'{aecg.filename},{aecg.zipContainer},'
f'RHYTHMEGDTC {n} found: {valrow["VALUE"]}')
aecg.RHYTHMEGDTC[n] = valrow["VALUE"]
else:
logger.debug(
f'{aecg.filename},{aecg.zipContainer},'
f'RHYTHMEGDTC {n} not found')
if log_validation:
valpd = valpd.append(pd.DataFrame([valrow], columns=VALICOLS),
ignore_index=True)
if log_validation:
aecg.validatorResults = \
aecg.validatorResults.append(valpd, ignore_index=True)
return aecg
def parse_derived_waveform_info(aecg_doc: etree._ElementTree,
aecg: Aecg,
log_validation: bool = False) -> Aecg:
"""Parses `aecg_doc` XML document and extracts derived waveform information
This function parses the `aecg_doc` xml document searching for derived
waveform information that includes in the returned `Aecg`: waveform
identifier, code, display name, and date and time of collection.
Args:
aecg_doc (etree._ElementTree): aECG XML document
aecg (Aecg): The aECG object to update
log_validation (bool, optional): Indicates whether to maintain the
validation results in `aecg.validatorResults`. Defaults to
False.
Returns:
Aecg: `aecg` updated with the information found in the xml document.
"""
valpd = pd.DataFrame()
for n in ["root", "extension"]:
valrow = validate_xpath(aecg_doc,
"./component/series/derivation/"
"derivedSeries/id",
"urn:hl7-org:v3",
n,
new_validation_row(aecg.filename, "DERIVED",
"ID_" + n),
failcat="WARNING")
if valrow["VALIOUT"] == "PASSED":
logger.info(
f'{aecg.filename},{aecg.zipContainer},'
f'DERIVED ID {n} found: {valrow["VALUE"]}')
aecg.DERIVEDID[n] = valrow["VALUE"]
else:
if n == "root":
logger.warning(
f'{aecg.filename},{aecg.zipContainer},'
f'DERIVED ID {n} not found')
else:
logger.warning(
f'{aecg.filename},{aecg.zipContainer},'
f'DERIVED ID {n} not found')
if log_validation:
valpd = valpd.append(pd.DataFrame([valrow], columns=VALICOLS),
ignore_index=True)
if log_validation:
aecg.validatorResults = \
aecg.validatorResults.append(valpd, ignore_index=True)
valrow = validate_xpath(aecg_doc,
"./component/series/derivation/"
"derivedSeries/code",
"urn:hl7-org:v3",
"code",
new_validation_row(aecg.filename, "DERIVED",
"CODE"),
failcat="WARNING")
if valrow["VALIOUT"] == "PASSED":
logger.debug(
f'{aecg.filename},{aecg.zipContainer},'
f'DERIVED code found: {valrow["VALUE"]}')
aecg.DERIVEDCODE["code"] = valrow["VALUE"]
if aecg.DERIVEDCODE["code"] != "REPRESENTATIVE_BEAT":
logger.warning(
f'{aecg.filename},{aecg.zipContainer},'
f'DERIVED unexpected code found: {valrow["VALUE"]}')
valrow["VALIOUT"] = "WARNING"
valrow["VALIMSG"] = "Unexpected value found"
else:
logger.warning(
f'{aecg.filename},{aecg.zipContainer},'
f'DERIVED code not found')
if log_validation:
aecg.validatorResults = aecg.validatorResults.append(
pd.DataFrame([valrow], columns=VALICOLS), ignore_index=True)
valrow = validate_xpath(aecg_doc,
"./component/series/derivation/"
"derivedSeries/code",
"urn:hl7-org:v3",
"displayName",
new_validation_row(aecg.filename, "DERIVED",
"CODE_displayName"),
failcat="WARNING")
if valrow["VALIOUT"] == "PASSED":
logger.debug(
f'{aecg.filename},{aecg.zipContainer},'
f'DERIVED displayName found: {valrow["VALUE"]}')
aecg.DERIVEDCODE["displayName"] = valrow["VALUE"]
else:
logger.debug(
f'{aecg.filename},{aecg.zipContainer},'
f'DERIVED displayName not found')
if log_validation:
aecg.validatorResults = aecg.validatorResults.append(
pd.DataFrame([valrow], columns=VALICOLS), ignore_index=True)
valpd = pd.DataFrame()
for n in ["low", "high"]:
valrow = validate_xpath(aecg_doc,
"./component/series/derivation/"
"derivedSeries/effectiveTime/" + n,
"urn:hl7-org:v3",
"value",
new_validation_row(aecg.filename, "DERIVED",
"EGDTC_" + n),
failcat="WARNING")
if valrow["VALIOUT"] == "PASSED":
logger.info(
f'{aecg.filename},{aecg.zipContainer},'
f'DERIVEDEGDTC {n} found: {valrow["VALUE"]}')
aecg.DERIVEDEGDTC[n] = valrow["VALUE"]
else:
logger.debug(
f'{aecg.filename},{aecg.zipContainer},'
f'DERIVEDEGDTC {n} not found')
if log_validation:
valpd = valpd.append(pd.DataFrame([valrow], columns=VALICOLS),
ignore_index=True)
if log_validation:
aecg.validatorResults = \
aecg.validatorResults.append(valpd, ignore_index=True)
return aecg
def parse_rhythm_waveform_timeseries(aecg_doc: etree._ElementTree,
aecg: Aecg,
include_digits: bool = False,
log_validation: bool = False) -> Aecg:
"""Parses `aecg_doc` XML document and extracts rhythm's timeseries
This function parses the `aecg_doc` xml document searching for rhythm
waveform timeseries (sequences) information that includes in the returned
:any:`Aecg`. Each found sequence is stored as an :any:`AecgLead` in the
:any:`Aecg.RHYTHMLEADS` list of the returned :any:`Aecg`.
Args:
aecg_doc (etree._ElementTree): aECG XML document
aecg (Aecg): The aECG object to update
include_digits (bool, optional): Indicates whether to include the
digits information in the returned `Aecg`.
log_validation (bool, optional): Indicates whether to maintain the
validation results in `aecg.validatorResults`. Defaults to
False.
Returns:
Aecg: `aecg` updated with the information found in the xml document.
"""
path_prefix = './component/series/component/sequenceSet/' \
'component/sequence'
seqnodes = aecg_doc.xpath((path_prefix + '/code').replace('/', '/ns:'),
namespaces={'ns': 'urn:hl7-org:v3'})
if len(seqnodes) > 0:
logger.info(
f'{aecg.filename},{aecg.zipContainer},'
f'RHYTHM sequenceSet(s) found: '
f'{len(seqnodes)} sequenceSet nodes')
else:
logger.warning(
f'{aecg.filename},{aecg.zipContainer},'
f'RHYTHM sequenceSet not found')
for xmlnode in seqnodes:
xmlnode_path = aecg_doc.getpath(xmlnode)
valrow = validate_xpath(aecg_doc,
xmlnode_path,
"urn:hl7-org:v3",
"code",
new_validation_row(aecg.filename, "RHYTHM",
"SEQUENCE_CODE"),
failcat="WARNING")
valpd = pd.DataFrame()
if valrow["VALIOUT"] == "PASSED":
if not valrow["VALUE"] in SEQUENCE_CODES:
logger.warning(
f'{aecg.filename},{aecg.zipContainer},'
f'RHYTHM unexpected sequenceSet code '
f'found: {valrow["VALUE"]}')
valrow["VALIOUT"] = "WARNING"
valrow["VALIMSG"] = "Unexpected sequence code found"
if valrow["VALUE"] in TIME_CODES:
logger.info(
f'{aecg.filename},{aecg.zipContainer},'
f'RHYTHM sequenceSet code found: {valrow["VALUE"]}')
aecg.RHYTHMTIME["code"] = valrow["VALUE"]
# Retrieve time head info from value node
rel_path = "../value/head"
valrow2 = validate_xpath(
xmlnode,
rel_path,
"urn:hl7-org:v3",
"value",
new_validation_row(
aecg.filename, "RHYTHM", "SEQUENCE_TIME_HEAD"),
failcat="WARNING")
valrow2["XPATH"] = xmlnode_path + "/" + rel_path
if valrow2["VALIOUT"] == "PASSED":
logger.info(
f'{aecg.filename},{aecg.zipContainer},'
f'RHYTHM SEQUENCE_TIME_HEAD found: {valrow2["VALUE"]}')
aecg.RHYTHMTIME["head"] = valrow2["VALUE"]
else:
logger.debug(
f'{aecg.filename},{aecg.zipContainer},'
f'RHYTHM SEQUENCE_TIME_HEAD not found')
if log_validation:
valpd = valpd.append(
pd.DataFrame([valrow2], columns=VALICOLS),
ignore_index=True)
# Retrieve time increment info from value node
rel_path = "../value/increment"
for n in ["value", "unit"]:
valrow2 = validate_xpath(
xmlnode,
rel_path,
"urn:hl7-org:v3",
n,
new_validation_row(
aecg.filename, "RHYTHM", "SEQUENCE_TIME_" + n),
failcat="WARNING")
valrow2["XPATH"] = xmlnode_path + "/" + rel_path
if valrow2["VALIOUT"] == "PASSED":
logger.info(
f'{aecg.filename},{aecg.zipContainer},'
f'RHYTHM SEQUENCE_TIME_{n} found: '
f'{valrow2["VALUE"]}')
if n == "value":
aecg.RHYTHMTIME["increment"] = float(
valrow2["VALUE"])
else:
aecg.RHYTHMTIME[n] = valrow2["VALUE"]
if log_validation:
valpd = \
valpd.append(pd.DataFrame([valrow2],
columns=VALICOLS),
ignore_index=True)
else:
logger.info(
f'{aecg.filename},{aecg.zipContainer},'
f'RHYTHM sequenceSet code found: '
f'{valrow["VALUE"]}')
logger.info(
f'{aecg.filename},{aecg.zipContainer},'
f'LEADNAME from RHYTHM sequenceSet code: '
f'{valrow["VALUE"]}')
# Assume is a lead
aecglead = AecgLead()
aecglead.leadname = valrow["VALUE"]
# Inherit last parsed RHYTHMTIME
aecglead.LEADTIME = copy.deepcopy(aecg.RHYTHMTIME)
# Retrive lead origin info
rel_path = "../value/origin"
for n in ["value", "unit"]:
valrow2 = validate_xpath(
xmlnode,
rel_path,
"urn:hl7-org:v3",
n,
new_validation_row(
aecg.filename, "RHYTHM",
"SEQUENCE_LEAD_ORIGIN_" + n),
failcat="WARNING")
valrow2["XPATH"] = xmlnode_path + "/" + rel_path
if valrow2["VALIOUT"] == "PASSED":
logger.info(
f'{aecg.filename},{aecg.zipContainer},'
f'RHYTHM SEQUENCE_LEAD_ORIGIN_{n} '
f'found: {valrow2["VALUE"]}')
if n == "value":
try:
aecglead.origin = float(valrow2["VALUE"])
except Exception as ex:
valrow2["VALIOUT"] == "ERROR"
valrow2["VALIMSG"] = "SEQUENCE_LEAD_"\
"ORIGIN is not a "\
"number"
else:
aecglead.origin_unit = valrow2["VALUE"]
else:
logger.debug(
f'{aecg.filename},{aecg.zipContainer},'
f'RHYTHM SEQUENCE_LEAD_ORIGIN_{n} not found')
if log_validation:
valpd = valpd.append(
pd.DataFrame([valrow2], columns=VALICOLS),
ignore_index=True)
# Retrive lead scale info
rel_path = "../value/scale"
for n in ["value", "unit"]:
valrow2 = validate_xpath(
xmlnode,
rel_path,
"urn:hl7-org:v3",
n,
new_validation_row(
aecg.filename, "RHYTHM",
"SEQUENCE_LEAD_SCALE_" + n),
failcat="WARNING")
valrow2["XPATH"] = xmlnode_path + "/" + rel_path
if valrow2["VALIOUT"] == "PASSED":
logger.info(
f'{aecg.filename},{aecg.zipContainer},'
f'RHYTHM SEQUENCE_LEAD_SCALE_{n} '
f'found: {valrow2["VALUE"]}')
if n == "value":
try:
aecglead.scale = float(valrow2["VALUE"])
except Exception as ex:
logger.error(
f'{aecg.filename},{aecg.zipContainer},'
f'RHYTHM SEQUENCE_LEAD_SCALE '
f'value is not a valid number: \"{ex}\"')
valrow2["VALIOUT"] == "ERROR"
valrow2["VALIMSG"] = "SEQUENCE_LEAD_"\
"SCALE is not a "\
"number"
else:
aecglead.scale_unit = valrow2["VALUE"]
else:
logger.debug(
f'{aecg.filename},{aecg.zipContainer},'
f'RHYTHM SEQUENCE_LEAD_SCALE_{n} not found')
if log_validation:
valpd = valpd.append(
pd.DataFrame([valrow2], columns=VALICOLS),
ignore_index=True)
# Include digits if requested
if include_digits:
rel_path = "../value/digits"
valrow2 = validate_xpath(
xmlnode,
rel_path,
"urn:hl7-org:v3",
"",
new_validation_row(
aecg.filename, "RHYTHM", "SEQUENCE_LEAD_DIGITS"),
failcat="WARNING")
valrow2["XPATH"] = xmlnode_path + "/" + rel_path
if valrow2["VALIOUT"] == "PASSED":
try:
# Convert string of digits to list of integers
# remove new lines
sdigits = valrow2["VALUE"].replace("\n", " ")
# remove carriage retruns
sdigits = sdigits.replace("\r", " ")
# remove tabs
sdigits = sdigits.replace("\t", " ")
# collapse 2 or more spaces into 1 space char
# and remove leading/trailing white spaces
sdigits = re.sub("\\s+", " ", sdigits).strip()
# Convert string into list of integers
aecglead.digits = [int(s) for s in
sdigits.split(' ')]
logger.info(
f'{aecg.filename},{aecg.zipContainer},'
f'DIGITS added to lead'
f' {aecglead.leadname} (n: '
f'{len(aecglead.digits)})')
except Exception as ex:
logger.error(
f'{aecg.filename},{aecg.zipContainer},'
f'Error parsing DIGITS from '
f'string to list of integers: \"{ex}\"')
valrow2["VALIOUT"] == "ERROR"
valrow2["VALIMSG"] = "Error parsing SEQUENCE_"\
"LEAD_DIGITS from string"\
" to list of integers"
else:
logger.error(
f'{aecg.filename},{aecg.zipContainer},'
f'DIGITS not found for lead {aecglead.leadname}')
if log_validation:
valpd = valpd.append(
pd.DataFrame([valrow2], columns=VALICOLS),
ignore_index=True)
else:
logger.info(
f'{aecg.filename},{aecg.zipContainer},'
f'DIGITS were not requested by the user')
aecg.RHYTHMLEADS.append(copy.deepcopy(aecglead))
else:
logger.warning(
f'{aecg.filename},{aecg.zipContainer},'
f'RHYTHM sequenceSet code not found')
if log_validation:
aecg.validatorResults = aecg.validatorResults.append(
pd.DataFrame([valrow], columns=VALICOLS),
ignore_index=True)
if valpd.shape[0] > 0:
aecg.validatorResults = \
aecg.validatorResults.append(valpd, ignore_index=True)
return aecg
def parse_derived_waveform_timeseries(aecg_doc: etree._ElementTree,
aecg: Aecg,
include_digits: bool = False,
log_validation: bool = False):
"""Parses `aecg_doc` XML document and extracts derived's timeseries
This function parses the `aecg_doc` xml document searching for derived
waveform timeseries (sequences) information that includes in the returned
:any:`Aecg`. Each found sequence is stored as an :any:`AecgLead` in the
:any:`Aecg.DERIVEDLEADS` list of the returned :any:`Aecg`.
Args:
aecg_doc (etree._ElementTree): aECG XML document
aecg (Aecg): The aECG object to update
include_digits (bool, optional): Indicates whether to include the
digits information in the returned `Aecg`.
log_validation (bool, optional): Indicates whether to maintain the
validation results in `aecg.validatorResults`. Defaults to
False.
Returns:
Aecg: `aecg` updated with the information found in the xml document.
"""
path_prefix = './component/series/derivation/derivedSeries/component'\
'/sequenceSet/component/sequence'
seqnodes = aecg_doc.xpath((path_prefix + '/code').replace('/', '/ns:'),
namespaces={'ns': 'urn:hl7-org:v3'})
if len(seqnodes) > 0:
logger.info(
f'{aecg.filename},{aecg.zipContainer},'
f'DERIVED sequenceSet(s) found: '
f'{len(seqnodes)} sequenceSet nodes')
else:
logger.warning(
f'{aecg.filename},{aecg.zipContainer},'
f'DERIVED sequenceSet not found')
for xmlnode in seqnodes:
xmlnode_path = aecg_doc.getpath(xmlnode)
valrow = validate_xpath(aecg_doc,
xmlnode_path,
"urn:hl7-org:v3",
"code",
new_validation_row(aecg.filename, "DERIVED",
"SEQUENCE_CODE"),
failcat="WARNING")
valpd = pd.DataFrame()
if valrow["VALIOUT"] == "PASSED":
if not valrow["VALUE"] in SEQUENCE_CODES:
logger.warning(
f'{aecg.filename},{aecg.zipContainer},'
f'DERIVED unexpected sequenceSet code '
f'found: {valrow["VALUE"]}')
valrow["VALIOUT"] = "WARNING"
valrow["VALIMSG"] = "Unexpected sequence code found"
if valrow["VALUE"] in TIME_CODES:
logger.info(
f'{aecg.filename},{aecg.zipContainer},'
f'DERIVED sequenceSet code found: {valrow["VALUE"]}')
aecg.DERIVEDTIME["code"] = valrow["VALUE"]
# Retrieve time head info from value node
rel_path = "../value/head"
valrow2 = validate_xpath(
xmlnode,
rel_path,
"urn:hl7-org:v3",
"value",
new_validation_row(aecg.filename, "DERIVED",
"SEQUENCE_TIME_HEAD"),
failcat="WARNING")
valrow2["XPATH"] = xmlnode_path + "/" + rel_path
if valrow2["VALIOUT"] == "PASSED":
logger.info(
f'{aecg.filename},{aecg.zipContainer},'
f'DERIVED SEQUENCE_TIME_HEAD found: '
f'{valrow2["VALUE"]}')
aecg.DERIVEDTIME["head"] = valrow2["VALUE"]
else:
logger.debug(
f'{aecg.filename},{aecg.zipContainer},'
f'DERIVED SEQUENCE_TIME_HEAD not found')
if log_validation:
valpd = valpd.append(
pd.DataFrame([valrow2], columns=VALICOLS),
ignore_index=True)
# Retrieve time increment info from value node
rel_path = "../value/increment"
for n in ["value", "unit"]:
valrow2 = validate_xpath(
xmlnode,
rel_path,
"urn:hl7-org:v3",
n,
new_validation_row(aecg.filename, "DERIVED",
"SEQUENCE_TIME_" + n),
failcat="WARNING")
valrow2["XPATH"] = xmlnode_path + "/" + rel_path
if valrow2["VALIOUT"] == "PASSED":
logger.info(
f'{aecg.filename},{aecg.zipContainer},'
f'DERIVED SEQUENCE_TIME_{n} found: '
f'{valrow2["VALUE"]}')
if n == "value":
aecg.DERIVEDTIME["increment"] =\
float(valrow2["VALUE"])
else:
aecg.DERIVEDTIME[n] = valrow2["VALUE"]
if log_validation:
valpd = valpd.append(
pd.DataFrame([valrow2], columns=VALICOLS),
ignore_index=True)
else:
logger.debug(
f'{aecg.filename},{aecg.zipContainer},'
f'DERIVED sequenceSet code found: {valrow["VALUE"]}')
logger.info(
f'{aecg.filename},{aecg.zipContainer},'
f'LEADNAME from DERIVED sequenceSet code: '
f'{valrow["VALUE"]}')
# Assume is a lead
aecglead = AecgLead()
aecglead.leadname = valrow["VALUE"]
# Inherit last parsed DERIVEDTIME
aecglead.LEADTIME = copy.deepcopy(aecg.DERIVEDTIME)
# Retrive lead origin info
rel_path = "../value/origin"
for n in ["value", "unit"]:
valrow2 = validate_xpath(
xmlnode,
rel_path,
"urn:hl7-org:v3",
n,
new_validation_row(aecg.filename, "DERIVED",
"SEQUENCE_LEAD_ORIGIN_" + n),
failcat="WARNING")
valrow2["XPATH"] = xmlnode_path + "/" + rel_path
if valrow2["VALIOUT"] == "PASSED":
logger.info(
f'{aecg.filename},{aecg.zipContainer},'
f'DERIVED SEQUENCE_LEAD_ORIGIN_{n} '
f'found: {valrow2["VALUE"]}')
if n == "value":
try:
aecglead.origin = float(valrow2["VALUE"])
except Exception as ex:
valrow2["VALIOUT"] == "ERROR"
valrow2["VALIMSG"] = \
"SEQUENCE_LEAD_ORIGIN is not a number"
else:
aecglead.origin_unit = valrow2["VALUE"]
else:
logger.debug(
f'{aecg.filename},{aecg.zipContainer},'
f'DERIVED SEQUENCE_LEAD_ORIGIN_{n} not found')
if log_validation:
valpd = valpd.append(
pd.DataFrame([valrow2], columns=VALICOLS),
ignore_index=True)
# Retrive lead scale info
rel_path = "../value/scale"
for n in ["value", "unit"]:
valrow2 = validate_xpath(
xmlnode,
rel_path,
"urn:hl7-org:v3",
n,
new_validation_row(aecg.filename, "DERIVED",
"SEQUENCE_LEAD_SCALE_" + n),
failcat="WARNING")
valrow2["XPATH"] = xmlnode_path + "/" + rel_path
if valrow2["VALIOUT"] == "PASSED":
logger.info(
f'{aecg.filename},{aecg.zipContainer},'
f'DERIVED SEQUENCE_LEAD_SCALE_{n} '
f'found: {valrow2["VALUE"]}')
if n == "value":
try:
aecglead.scale = float(valrow2["VALUE"])
except Exception as ex:
logger.error(
f'{aecg.filename},{aecg.zipContainer},'
f'DERIVED SEQUENCE_LEAD_SCALE'
f' value is not a valid number: \"{ex}\"')
valrow2["VALIOUT"] == "ERROR"
valrow2["VALIMSG"] = "SEQUENCE_LEAD_SCALE"\
" is not a number"
else:
aecglead.scale_unit = valrow2["VALUE"]
else:
logger.debug(
f'{aecg.filename},{aecg.zipContainer},'
f'DERIVED SEQUENCE_LEAD_SCALE_{n} not found')
if log_validation:
valpd = valpd.append(
pd.DataFrame([valrow2], columns=VALICOLS),
ignore_index=True)
# Include digits if requested
if include_digits:
rel_path = "../value/digits"
valrow2 = validate_xpath(
xmlnode,
rel_path,
"urn:hl7-org:v3",
"",
new_validation_row(aecg.filename, "DERIVED",
"SEQUENCE_LEAD_DIGITS"),
failcat="WARNING")
valrow2["XPATH"] = xmlnode_path + "/" + rel_path
if valrow2["VALIOUT"] == "PASSED":
try:
# Convert string of digits to list of integers
# remove new lines
sdigits = valrow2["VALUE"].replace("\n", " ")
# remove carriage retruns
sdigits = sdigits.replace("\r", " ")
# remove tabs
sdigits = sdigits.replace("\t", " ")
# collapse 2 or more spaces into 1 space char
# and remove leading/trailing white spaces
sdigits = re.sub("\\s+", " ", sdigits).strip()
# Convert string into list of integers
aecglead.digits = [int(s) for s in
sdigits.split(' ')]
logger.info(
f'{aecg.filename},{aecg.zipContainer},'
f'DIGITS added to lead'
f' {aecglead.leadname} (n: '
f'{len(aecglead.digits)})')
except Exception as ex:
logger.error(
f'{aecg.filename},{aecg.zipContainer},'
f'Error parsing DIGITS from '
f'string to list of integers: \"{ex}\"')
valrow2["VALIOUT"] == "ERROR"
valrow2["VALIMSG"] = "Error parsing SEQUENCE_"\
"LEAD_DIGITS from string"\
" to list of integers"
else:
logger.error(
f'{aecg.filename},{aecg.zipContainer},'
f'DIGITS not found for lead {aecglead.leadname}')
if log_validation:
valpd = valpd.append(
pd.DataFrame([valrow2], columns=VALICOLS),
ignore_index=True)
else:
logger.info(
f'{aecg.filename},{aecg.zipContainer},'
f'DIGITS were not requested by the user')
aecg.DERIVEDLEADS.append(copy.deepcopy(aecglead))
else:
logger.warning(
f'{aecg.filename},{aecg.zipContainer},'
f'RHYTHM sequenceSet code not found')
if log_validation:
aecg.validatorResults = aecg.validatorResults.append(
| pd.DataFrame([valrow], columns=VALICOLS) | pandas.DataFrame |
""" Module for data preprocessing.
"""
import datetime
import warnings
from typing import Any
from typing import Callable
from typing import Dict
from typing import List
from typing import Optional
from typing import Set
from typing import Union
import numpy as np
import pandas as pd
from sklearn.base import BaseEstimator
from sklearn.base import TransformerMixin
from sklearn.utils.validation import check_is_fitted
__all__ = [
'ColumnSelector',
'ColumnDropper',
'ColumnRename',
'NaDropper',
'Clip',
'DatetimeTransformer',
'NumericTransformer',
'TimeframeExtractor',
'DateExtractor',
'ValueMapper',
'Sorter',
'Fill',
'TimeOffsetTransformer',
'ConditionedDropper',
'ZeroVarianceDropper',
'SignalSorter',
'ColumnSorter',
'DifferentialCreator'
]
class ColumnSelector(BaseEstimator, TransformerMixin):
"""Transformer to select a list of columns by their name.
Example:
>>> data = pd.DataFrame({'a': [0], 'b': [0]})
>>> ColumnSelector(keys=['a']).transform(data)
pd.DataFrame({'a': [0]})
"""
def __init__(self, keys: List[str]):
"""Creates ColumnSelector.
Transformer to select a list of columns for further processing.
Args:
keys (List[str]): List of columns to extract.
"""
self._keys = keys
def fit(self, X, y=None):
return self
def transform(self, X):
"""Extracts the columns from `X`.
Args:
X (pd.DataFrame): Dataframe.
Returns:
pd.DataFrame: Returns a DataFrame only containing the selected
features.
"""
return X.loc[:, self._keys]
class ColumnDropper(BaseEstimator, TransformerMixin):
"""Transformer to drop a list of columns by their name.
Example:
>>> data = pd.DataFrame({'a': [0], 'b': [0]})
>>> ColumnDropper(columns=['b']).transform(data)
pd.DataFrame({'a': [0]})
"""
def __init__(self,
*,
columns: Union[List[str], Set[str]],
verbose: bool = False):
"""Creates ColumnDropper.
Transformer to drop a list of columns from the data frame.
Args:
keys (list): List of columns names to drop.
"""
self.columns = set(columns)
self.verbose = verbose
def fit(self, X, y=None):
return self
def transform(self, X):
"""Drops a list of columns of `X`.
Args:
X (pd.DataFrame): Dataframe.
Returns:
pd.DataFrame: Returns the dataframe without the dropped features.
"""
cols = set(X.columns.to_list())
if len(m := self.columns - cols) > 0:
warnings.warn(f'Columns {m} not found in dataframe.')
if self.verbose:
print(f'New columns: {cols - self.columns}. '
f'Removed: {self.columns}.')
return X.drop(self.columns, axis=1, errors='ignore')
class ColumnRename(BaseEstimator, TransformerMixin):
"""Transformer to rename column with a function.
Example:
>>> data = pd.DataFrame({'a.b.c': [0], 'd.e.f': [0]})
>>> ColumnRename(lambda x: x.split('.')[-1]).transform(data)
pd.DataFrame({'c': [0], 'f': [0]})
"""
def __init__(self, mapper: Callable[[str], str]):
"""Create ColumnRename.
Transformer to rename columns by a mapper function.
Args:
mapper (lambda): Mapper rename function.
Example:
Given column with name: a.b.c
lambda x: x.split('.')[-1]
Returns c
"""
self.mapper = mapper
def fit(self, X, y=None):
return self
def transform(self, X):
"""Renames a columns in `X` with a mapper function.
Args:
X (pd.DataFrame): Dataframe.
Returns:
pd.DataFrame: Returns the dataframe with the renamed columns.
"""
# split the column name
# use the last item as new name
return X.rename(columns=self.mapper)
class NaDropper(BaseEstimator, TransformerMixin):
"""Transformer that drops rows with na values.
Example:
>>> data = pd.DataFrame({'a': [0, 1], 'b': [0, np.nan]})
>>> NaDropper().transform(data)
pd.DataFrame({'a': [0], 'b': [0]})
"""
def __init__(self):
pass
def fit(self, X, y=None):
return self
def transform(self, X):
return X.dropna()
class Clip(BaseEstimator, TransformerMixin):
"""Transformer that clips values by a lower and upper bound.
Example:
>>> data = pd.DataFrame({'a': [-0.1, 1.2], 'b': [0.5, 0.6]})
>>> Clip().transform(data)
pd.DataFrame({'a': [0, 1], 'b': [0.5, 0.6]})
"""
def __init__(self, lower: float = 0.0, upper: float = 1.0):
"""Creates Clip.
Transformer that clips a numeric column to the treshold if the
threshold is exceeded. Works with an upper and lower threshold. Wrapper
for pd.DataFrame.clip.
Args:
lower (float, optional): lower limit. Defaults to 0.
upper (float, optional): upper limit. Defaults to 1.
"""
self.upper = upper
self.lower = lower
def fit(self, X, y=None):
return self
def transform(self, X):
return X.clip(lower=self.lower, upper=self.upper, axis=0)
class ColumnTSMapper(BaseEstimator, TransformerMixin):
def __init__(self,
cols: List[str],
timedelta: pd.Timedelta = pd.Timedelta(250, 'ms'),
classes: List[str] = None,
verbose: bool = False):
"""Creates ColumnTSMapper.
Expects the timestamp column to be of type pd.Timestamp.
Args:
cols (List[str]): names of [0] timestamp column, [1] sensor names,
[2] sensor values.
timedelta (pd.Timedelta): Timedelta to resample with.
classes (List[str]): List of sensor names.
verbose (bool, optional): Whether to allow prints.
"""
super().__init__()
self._cols = cols
self._timedelta = timedelta
self._verbose = verbose
if classes is not None:
self.classes_ = classes
def fit(self, X, y=None):
"""Gets the unique values in the sensor name column that
are needed to expand the dataframe.
Args:
X (pd.DataFrame): Dataframe.
y (array-like, optional): Labels. Defaults to None.
Returns:
ColumnTSMapper: Returns this.
"""
classes = X[self._cols[1]].unique()
self.classes_ = np.hstack(['Timestamp', classes])
return self
def transform(self, X):
"""Performs the mapping to equidistant timestamps.
Args:
X (pd.DataFrame): Dataframe.
Raises:
ValueError: Raised if column is not found in `X`.
Returns:
pd.DataFrame: Returns the remapped dataframe.
"""
# check is fit had been called
check_is_fitted(self)
# check if all columns exist
if not all([item in X.columns for item in self._cols]):
raise ValueError(
f'Columns {self._cols} not found in DataFrame '
f'{X.columns.to_list()}.')
# split sensors into individual columns
# create new dataframe with all _categories
# use timestamp index, to use resample later on
# initialized with na
sensors = pd.DataFrame(
None, columns=self.classes_, index=X[self._cols[0]])
# group by sensor
groups = X.groupby([self._cols[1]])
# write sensor values to sensors which is indexed by the timestamp
for g in groups:
sensors.loc[g[1][self._cols[0]], g[0]
] = g[1][self._cols[2]].to_numpy()
sensors = sensors.apply(pd.to_numeric, errors='ignore')
# fill na, important before resampling
# otherwise mean affects more samples than necessary
# first: forward fill to next valid observation
# second: backward fill first missing rows
sensors = sensors.fillna(method='ffill').fillna(method='bfill')
# resamples to equidistant timeframe
# take avg if multiple samples in the same timeframe
sensors = sensors.resample(self._timedelta).mean()
sensors = sensors.fillna(method='ffill').fillna(method='bfill')
# FIXME: to avoid nans in model, but needs better fix
sensors = sensors.fillna(value=0.0)
# move index to column and use rangeindex
sensors['Timestamp'] = sensors.index
sensors.index = pd.RangeIndex(stop=sensors.shape[0])
if self._verbose:
start, end = sensors.iloc[0, 0], sensors.iloc[-1, 0]
print('ColumnTSMapper: ')
print(f'{sensors.shape[0]} rows. '
f'Mapped to {self._timedelta.total_seconds()}s interval '
f'from {start} to {end}.')
return sensors
class DatetimeTransformer(BaseEstimator, TransformerMixin):
"""Transforms a list of columns to datetime.
Example:
>>> data = pd.DataFrame({'dt': ['2021-07-02 16:30:00']})
>>> data = DatetimeTransformer(columns=['dt']).transform(data)
>>> data.dtypes
dt datetime64[ns]
"""
def __init__(self, *, columns: List[str], dt_format: str = None):
"""Creates DatetimeTransformer.
Parses a list of column to pd.Timestamp.
Args:
columns (list): List of columns names.
dt_format (str): Optional format string.
"""
super().__init__()
self._columns = columns
self._format = dt_format
def fit(self, X, y=None):
return self
def transform(self, X):
"""Parses `columns` to datetime.
Args:
X (pd.DataFrame): Dataframe.
Raises:
ValueError: Raised if columns are missing in `X`.
Returns:
pd.DataFrame: Returns the dataframe with datetime columns.
"""
X = X.copy()
# check if columns in dataframe
if len(diff := set(self._columns) - set(X.columns)):
raise ValueError(
f'Columns {diff} not found in DataFrame with columns'
f'{X.columns.to_list()}.')
# parse to pd.Timestamp
X[self._columns] = X[self._columns].apply(
lambda x: pd.to_datetime(x, format=self._format), axis=0)
# column wise
return X
class NumericTransformer(BaseEstimator, TransformerMixin):
"""Transforms a list of columns to numeric datatype.
Example:
>>> data = pd.DataFrame({'a': [0], 'b': ['1']})
>>> data.dtypes
a int64
b object
>>> data = NumericTransformer().transform(data)
>>> data.dtypes
a int64
b int64
"""
def __init__(self, *, columns: Optional[List[str]] = None):
"""Creates NumericTransformer.
Parses a list of column to numeric datatype. If None, all are
attempted to be parsed.
Args:
columns (list): List of columns names.
dt_format (str): Optional format string.
"""
super().__init__()
self._columns = columns
def fit(self, X, y=None):
return self
def transform(self, X):
"""Parses `columns` to numeric.
Args:
X (pd.DataFrame): Dataframe.
Raises:
ValueError: Raised if columns are missing in `X`.
Returns:
pd.DataFrame: Returns the dataframe with datetime columns.
"""
X = X.copy()
# transform all columns
if self._columns is None:
columns = X.columns.to_list()
else:
columns = self._columns
if len((diff := list(set(columns) - set(cols := X.columns)))):
raise ValueError(f'Columns found: {cols.to_list()}. '
f'Columns missing: {diff}.')
# parse to numeric
# column wise
X[columns] = X[columns].apply(pd.to_numeric, axis=0)
return X
class TimeframeExtractor(BaseEstimator, TransformerMixin):
"""Drops sampes that are not between a given start and end time.
Limits are inclusive.
Example:
>>> data = pd.DataFrame(
{'dates': [datetime.datetime(2021, 7, 2, 9, 50, 0),
datetime.datetime(2021, 7, 2, 11, 0, 0),
datetime.datetime(2021, 7, 2, 12, 10, 0)],
'values': [0, 1, 2]})
>>> TimeframeExtractor(time_column='dates',
start_time= datetime.time(10, 0, 0),
end_time=datetime.time(12, 0, 0)
).transform(data)
pd.DataFrame({'dates': datetime.datetime(2021, 7, 2, 11, 0, 0),
'values': [1]})
"""
def __init__(self,
*,
time_column: str,
start_time: datetime.time,
end_time: datetime.time,
invert: bool = False,
verbose: bool = False):
"""Creates TimeframeExtractor.
Drops samples that are not in between `start_time` and `end_time` in
`time_column`.
Args:
time_column (str): Column name of the timestamp column.
start_time (datetime.time): Start time.
end_time (datetime.time): End time.
invert(bool): Whether to invert the range.
verbose (bool, optional): Whether to allow prints.
"""
super().__init__()
self._start = start_time
self._end = end_time
self._column = time_column
self._negate = invert
self._verbose = verbose
def fit(self, X, y=None):
return self
def transform(self, X):
"""Drops rows from the dataframe if they are not in between
`start_time` and `end_time`. Limits are inclusive. Reindexes the
dataframe.
Args:
X (pd.DataFrame): Dataframe.
Returns:
pd.DataFrame: Returns the new dataframe.
"""
X = X.copy()
rows_before = X.shape[0]
dates = pd.to_datetime(X[self._column])
if self._negate:
X = X.loc[~((dates.dt.time >= self._start) &
(dates.dt.time <= self._end)), :]
else:
X = X.loc[(dates.dt.time >= self._start) &
(dates.dt.time <= self._end), :]
X.index = pd.RangeIndex(0, X.shape[0])
rows_after = X.shape[0]
if self._verbose:
print(
'TimeframeExtractor: \n'
f'{rows_after} rows. Dropped {rows_before - rows_after} '
f'rows which are {"in" if self._negate else "not in"} between '
f'{self._start} and {self._end}.'
)
return X
class DateExtractor(BaseEstimator, TransformerMixin):
""" Drops rows that are not between a start and end date.
Limits are inclusive.
Example:
>>> data = pd.DataFrame(
{'dates': [datetime.datetime(2021, 7, 1, 9, 50, 0),
datetime.datetime(2021, 7, 2, 11, 0, 0),
datetime.datetime(2021, 7, 3, 12, 10, 0)],
'values': [0, 1, 2]})
>>> DateExtractor(date_column='dates',
start_date=datetime.date(2021, 7, 2),
end_date=datetime.date(2021, 7, 2)).transform(data)
pd.DataFrame({'dates': datetime.datetime(2021, 07, 2, 11, 0, 0),
'values': [1]})
"""
def __init__(self,
*,
date_column: str,
start_date: datetime.date,
end_date: datetime.date,
invert: bool = False,
verbose: bool = False):
"""Initializes `DateExtractor`.
Args:
date_column (str): Name of timestamp column.
start_date (datetime.date): Start date.
end_date (datetime.date): End date.
invert (bool): Whether to invert the range.
verbose (bool, optional): Whether to allow prints.
"""
super().__init__()
self._start = start_date
self._end = end_date
self._column = date_column
self._negate = invert
self._verbose = verbose
def fit(self, X, y=None):
return self
def transform(self, X):
"""Drops rows which date is not between `start` and end date.
Bounds are inclusive. Dataframe is reindexed.
Args:
X (pd.Dataframe): Dataframe.
Returns:
pd.Dataframe: Returns the new dataframe.
"""
rows_before = X.shape[0]
dates = pd.to_datetime(X[self._column])
if self._negate:
X = X.loc[~((dates.dt.date >= self._start) &
(dates.dt.date <= self._end)), :]
else:
X = X.loc[(dates.dt.date >= self._start) &
(dates.dt.date <= self._end), :]
X.index = pd.RangeIndex(0, X.shape[0])
rows_after = X.shape[0]
if self._verbose:
print(
'DateExtractor: \n'
f'{rows_after} rows. Dropped {rows_before - rows_after} rows '
f'which are {"in" if self._negate else "not in"} between '
f'{self._start} and {self._end}.'
)
return X
class ValueMapper(BaseEstimator, TransformerMixin):
"""Maps values in `column` according to `classes`. Wrapper for
pd.DataFrame.replace.
Example:
>>> data = pd.DataFrame({'a': [0.0, 1.0, 2.0]})
>>> ValueMapper(columns=['a'], classes={2.0: 1.0}).transform(data)
pd.DataFrame({'a': [0.0, 1.0, 1.0]})
"""
def __init__(self,
*,
columns: List[str],
classes: Dict,
verbose: bool = False):
"""Initialize `ValueMapper`.
Args:
columns (List[str]): Names of columns to remap.
classes (Dict): Dictionary of old and new value.
verbose (bool, optional): Whether to allow prints.
"""
super().__init__()
self._columns = columns
self._classes = classes
self._verbose = verbose
def fit(self, X, y=None):
return self
def transform(self, X):
"""Remaps values in `column` according to `classes`.
Gives UserWarning if unmapped values are found.
Args:
X (pd.DataFrame): Dataframe.
Returns:
pd.DataFrame: Returns the new dataframe with remapped values.
"""
X = X.copy()
# warning if unmapped values
values = pd.unique(X[self._columns].values.ravel('K'))
if not set(self._classes.keys()).issuperset(values):
warnings.warn(
f'Classes {set(self._classes.keys()) - set(values)} ignored.')
X[self._columns] = X[self._columns].replace(self._classes)
return X
class Sorter(BaseEstimator, TransformerMixin):
"""Sorts the dataframe by a list of columns. Wrapper for
pd.DataFrame.sort_values.
Example:
>>> data = pd.DataFrame({'a': [0, 1], 'b': [1, 0]})
>>> Sorter(columns=['b'], ascending=True).transform(data)
pd.DataFrame({'a': [1, 0], 'b': [0, 1]})
"""
def __init__(self,
*,
columns: List[str],
ascending: bool = True,
axis: int = 0):
"""Initialize `Sorter`.
Args:
columns (List[str]): List of column names to sort by.
ascending (bool): Whether to sort ascending.
axis (int): Axis to sort by.
"""
super().__init__()
self._columns = columns
self._ascending = ascending
self._axis = axis
def fit(self, X, y=None):
return self
def transform(self, X):
"""Sorts `X` by `columns`.
Args:
X (pd.DataFrame): Dataframe.
Returns:
pd.DataFrame: Returns the sorted Dataframe.
"""
X = X.copy()
return X.sort_values(by=self._columns,
ascending=self._ascending,
axis=self._axis)
class Fill(BaseEstimator, TransformerMixin):
"""Fills NA values with a constant or 'bfill' / 'ffill'.
Wrapper for df.fillna.
Example:
>>> data = pd.DataFrame({'a': [0.0, np.nan]})
>>> Fill(value=1.0).transform(data)
pd.DataFrame({'a': [0.0, 1.0]})
"""
def __init__(self,
*,
value: Any,
method: str = None):
"""Initialize `Fill`.
Args:
value (Any): Constant to fill NAs.
method (str): method: 'ffill' or 'bfill'.
"""
super().__init__()
self._value = value
self._method = method
def fit(self, X, y=None):
return self
def transform(self, X):
"""Fills NAs.
Args:
X (pd.DataFrame): Dataframe.
Returns:
pd.DataFrame: Returns the filled dataframe.
"""
X = X.copy()
return X.fillna(self._value, method=self._method)
class TimeOffsetTransformer(BaseEstimator, TransformerMixin):
"""`TimeOffsetTransformer` offsets a datetime by `timedelta`.
Example:
>>> data = pd.DataFrame(
{'dates': [datetime.datetime(2021, 7, 1, 16, 0, 0)]})
>>> TimeOffsetTransformer(time_columns=['dates'],
timedelta=pd.Timedelta(1, 'h')
).transform(data)
pd.DataFrame({'dates': datetime.datetime(2021, 07, 2, 17, 0, 0)})
"""
def __init__(self, *, time_columns: List[str], timedelta: pd.Timedelta):
"""
Initialize `TimeOffsetTransformer`.
Args:
time_column (List[str]): List of names of columns with timestamps
to offset.
timedelta (pd.Timedelta): Offset.
"""
super().__init__()
self._time_columns = time_columns
self._timedelta = timedelta
def fit(self, X, y=None):
return self
def transform(self, X):
"""Offsets the timestamps in `time_columns` by `timedelta`-
Args:
X (pd.DataFrame): Dataframe.
Returns:
pd.DataFrame: Returns the dataframe.
"""
X = X.copy()
for column in self._time_columns:
X[column] = pd.to_datetime(X[column]) + self._timedelta
return X
class ConditionedDropper(BaseEstimator, TransformerMixin):
"""Module to drop rows in `column` that contain numeric values and are
above `threshold`. If `inverted` is true, values below `threshold` are
dropped.
Example:
>>> data = pd.DataFrame({'a': [0.0, 1.2, 0.5]})
>>> ConditionedDropper(column='a', threshold=0.5).transform(data)
pd.DataFrame({'a': [0.0, 0.5]})
"""
def __init__(self,
*,
column: str,
threshold: float,
invert: bool = False):
"""Initializes `ConditionedDropper`.
Args:
column (str): Column to match condition in.
threshold (float): Threshold.
inverted (bool, optional): If false, all values below `threshold`
are dropped, otherwise all values above are dropped.
"""
super().__init__()
self.column = column
self.threshold = threshold
self.inverted = invert
def fit(self, X, y=None):
return self
def transform(self, X):
"""Drops rows if below or above a threshold.
Args:
X (pd.DataFrame): Dataframe.
Returns:
pd.DataFrame: Returns the dataframe.
"""
X = X.copy()
if not self.inverted:
X = X.drop(X[X[self.column] > self.threshold].index)
else:
X = X.drop(X[X[self.column] < self.threshold].index)
X.index = pd.RangeIndex(X.shape[0])
return X
class ZeroVarianceDropper(BaseEstimator, TransformerMixin):
"""Removes all columns that are numeric and have zero variance.
Needs to be fitted first. Gives a warning if a column that was
registered as zero variance deviates.
Example:
>>> data = pd.DataFrame({'a': [0.0, 0.0], 'b': [1.0, 0.0]})
>>> ZeroVarianceDropper().fit_transform(data)
pd.DataFrame({'b': [1.0, 0.0]})
"""
def __init__(self, verbose: bool = False):
"""Initialize `ZeroVarianceDropper`.
Args:
verbose (bool, optional): Whether to print status messages.
"""
super().__init__()
self._verbose = verbose
def _get_zero_variance_columns(self, X: pd.DataFrame) -> List[str]:
"""Finds all columns with zero variance.
Args:
X (pd.DataFrame): Dataframe.
Returns:
List[str]: Returns a list of column names.
"""
var = X.var()
# get columns with zero variance
return [k for k, v in var.iteritems() if v == .0]
def fit(self, X, y=None):
"""Finds all columns with zero variance.
Args:
X (pd.DataFrame): Dataframe.
y (array-like, optional): Labels. Defaults to None.
Returns:
ZeroVarianceDropper: Returns self.
"""
self.columns_ = self._get_zero_variance_columns(X)
if self._verbose:
print(
f'Found {len(self.columns_)} columns with 0 variance '
f'({self.columns_}).')
return self
def transform(self, X):
"""Drops all columns found by fit with zero variance.
Args:
X (pd.DataFrame): Dataframe.
Returns:
pd.DataFrame: Returns the new dataframe.
"""
check_is_fitted(self, 'columns_')
X = X.copy()
# check if columns match
columns = self._get_zero_variance_columns(X)
disj = {*columns} ^ {*self.columns_}
if len(disj) > 0:
warnings.warn(f'Found column with higher variance: {disj}.')
before = X.shape[-1]
X = X.drop(self.columns_, axis=1)
if self._verbose:
after = X.shape[-1]
print(f'Dropped {before - after} columns.')
return X
class SignalSorter(BaseEstimator, TransformerMixin):
"""Sorts the signals into continuous and binary signals. First the
continuous, then the binary signals.
Example:
>>> data = pd.DataFrame({'a': [0.0, 1.0], 'b': [0.0, 0.2]})
>>> SignalSorter().fit_transform(data)
pd.DataFrame({'b': [1.0, 0.0], 'a': [0.0, 1.0]})
"""
def __init__(self, verbose: bool = False):
"""Initialize `SignalSorter`.
Args:
False: [binary, continuous]
verbose (bool, optional): Whether to print status.
"""
super().__init__()
self.verbose = verbose
def fit(self, X, y=None):
# find signals that are binary
uniques = {col: self._is_binary(X[col]) for col in X.columns}
self.order_ = sorted(uniques.items(), key=lambda v: v[1])
if self.verbose:
print(f'Binary: {self.order_}')
return self
def _is_binary(self, X: pd.Series) -> bool:
"""
Args:
X (pd.Series): Column of a data frame.
Returns:
bool: Whether `x` is a binary series.
"""
unique = X.unique()
if len(unique) > 2:
return False
if len(unique) == 1:
return True
try:
if set(unique.astype('float')) != {1., 0.}:
return False
return True
except Exception:
return False
def transform(self, X):
"""Sorts `x` into to a block of continuous and binary signals.
Args:
X (pd.DataFrame): Dataframe.
Returns:
pd.DataFrame: Returns the sorted dataframe.
"""
check_is_fitted(self, [])
X = X.copy()
return X[[c[0] for c in self.order_]]
class ColumnSorter(BaseEstimator, TransformerMixin):
"""Sorts the dataframe in the same order as the fitted dataframe.
Example:
>>> data = pd.DataFrame({'a': [0.0, 1.0], 'b': [0.0, 0.2]})
>>> (sorter := ColumnSorter()).fit(data)
>>> sorter.transform(pd.DataFrame({'b': [0.2, 1.0], 'a': [0.0, 0.1]}))
pd.DataFrame({'a': [0.0, 0.1], 'b': [0.2, 1.0]})
"""
def __init__(self, *, raise_on_error: bool = True, verbose: bool = False):
"""Initialize `ColumnSorter`.
Attributes:
raise_on_error (bool): Whether to raise an exception if additional
columns that were not fitted are found.
verbose (bool): Whether to print the status.
"""
super().__init__()
self.raise_on_error = raise_on_error
self.verbose = verbose
def fit(self, X, y=None):
self.columns_ = X.columns.to_numpy()
if self.verbose:
print(f'Sorting in order {self.columns_}.')
return self
def transform(self, X):
"""Sorts `X` by `columns`.
Args:
X (pd.DataFrame): Dataframe.
Returns:
pd.DataFrame: Returns the sorted Dataframe.
"""
check_is_fitted(self)
if len((diff := list(set(self.columns_) - set(X.columns)))):
raise ValueError(f'Columns missing: {diff}.')
if len((diff := list(set(X.columns) - set(self.columns_)))):
if self.raise_on_error:
raise ValueError(f'Found additional columns: {diff}.')
else:
warnings.warn(f'Found additional columns: {diff}.')
return X.loc[:, self.columns_]
class DifferentialCreator(BaseEstimator, TransformerMixin):
"""Calculates signal differences between subsequent time points.
Concatenates the new information with the dataframe.
Example:
>>> data = pd.DataFrame({'a': [1.0, 2.0, 1.0]})
>>> dcreator = DifferentialCreator(columns=['a'])
>>> dcreator.transform(pd.DataFrame(data)
pd.DataFrame({'a': [1.0, 2.0, 1.0], 'a_dif': [1.0, -1.0, 0.0]})
"""
def __init__(self, *, columns: List[str]):
"""Initialize `DifferentialCreator`.
Attributes:
keys: List[str]: Columns to create derivatives
"""
super().__init__()
self._columns = columns
def fit(self, X, y=None):
return self
def transform(self, X):
"""Calculate differences between subsequent points. Fill NaN with zero.
Args:
X (pd.DataFrame): Dataframe.
Returns:
pd.DataFrame: Returns the concatenated DataFrame.
"""
X_dif = (X[self._columns]
.diff(axis=0)
.fillna(0)
.add_suffix('_dif'))
return | pd.concat([X, X_dif], axis=1) | pandas.concat |
#codes to for analyse the model.
import re
import os
from astropy import units as u
from tardis import constants
import numpy as np
import pandas as pd
class LastLineInteraction(object):
@classmethod
def from_model(cls, model):
return cls(model.runner.last_line_interaction_in_id,
model.runner.last_line_interaction_out_id,
model.runner.last_line_interaction_shell_id,
model.runner.output_nu, model.plasma.atomic_data.lines)
def __init__(self, last_line_interaction_in_id,
last_line_interaction_out_id, last_line_interaction_shell_id,
output_nu, lines, packet_filter_mode='packet_nu'):
# mask out packets which did not perform a line interaction
# TODO mask out packets which do not escape to observer?
mask = last_line_interaction_out_id != -1
self.last_line_interaction_in_id = last_line_interaction_in_id[mask]
self.last_line_interaction_out_id = last_line_interaction_out_id[mask]
self.last_line_interaction_shell_id = last_line_interaction_shell_id[mask]
self.last_line_interaction_angstrom = output_nu.to(
u.Angstrom, equivalencies=u.spectral())[mask]
self.lines = lines
self._wavelength_start = 0 * u.angstrom
self._wavelength_end = np.inf * u.angstrom
self._atomic_number = None
self._ion_number = None
self.packet_filter_mode = packet_filter_mode
self.update_last_interaction_filter()
@property
def wavelength_start(self):
return self._wavelength_start.to('angstrom')
@wavelength_start.setter
def wavelength_start(self, value):
if not isinstance(value, u.Quantity):
raise ValueError('needs to be a Quantity')
self._wavelength_start = value
self.update_last_interaction_filter()
@property
def wavelength_end(self):
return self._wavelength_end.to('angstrom')
@wavelength_end.setter
def wavelength_end(self, value):
if not isinstance(value, u.Quantity):
raise ValueError('needs to be a Quantity')
self._wavelength_end = value
self.update_last_interaction_filter()
@property
def atomic_number(self):
return self._atomic_number
@atomic_number.setter
def atomic_number(self, value):
self._atomic_number = value
self.update_last_interaction_filter()
@property
def ion_number(self):
return self._ion_number
@ion_number.setter
def ion_number(self, value):
self._ion_number = value
self.update_last_interaction_filter()
def update_last_interaction_filter(self):
if self.packet_filter_mode == 'packet_nu':
packet_filter = (
(self.last_line_interaction_angstrom >
self.wavelength_start) &
(self.last_line_interaction_angstrom <
self.wavelength_end))
elif self.packet_filter_mode == 'line_in_nu':
line_in_nu = (
self.lines.wavelength.iloc[
self.last_line_interaction_in_id].values)
packet_filter = (
(line_in_nu > self.wavelength_start.to(u.angstrom).value) &
(line_in_nu < self.wavelength_end.to(u.angstrom).value))
self.last_line_in = self.lines.iloc[
self.last_line_interaction_in_id[packet_filter]]
self.last_line_out = self.lines.iloc[
self.last_line_interaction_out_id[packet_filter]]
if self.atomic_number is not None:
self.last_line_in = self.last_line_in.xs(
self.atomic_number, level='atomic_number', drop_level=False)
self.last_line_out = self.last_line_out.xs(
self.atomic_number, level='atomic_number', drop_level=False)
if self.ion_number is not None:
self.last_line_in = self.last_line_in.xs(
self.ion_number, level='ion_number', drop_level=False)
self.last_line_out = self.last_line_out.xs(
self.ion_number, level='ion_number', drop_level=False)
last_line_in_count = self.last_line_in.line_id.value_counts()
last_line_out_count = self.last_line_out.line_id.value_counts()
self.last_line_in_table = self.last_line_in.reset_index()[
[
'wavelength', 'atomic_number', 'ion_number',
'level_number_lower', 'level_number_upper']]
self.last_line_in_table['count'] = last_line_in_count
self.last_line_in_table.sort_values(by='count', ascending=False,
inplace=True)
self.last_line_out_table = self.last_line_out.reset_index()[
[
'wavelength', 'atomic_number', 'ion_number',
'level_number_lower', 'level_number_upper']]
self.last_line_out_table['count'] = last_line_out_count
self.last_line_out_table.sort_values(by='count', ascending=False,
inplace=True)
def plot_wave_in_out(self, fig, do_clf=True, plot_resonance=True):
if do_clf:
fig.clf()
ax = fig.add_subplot(111)
wave_in = self.last_line_list_in['wavelength']
wave_out = self.last_line_list_out['wavelength']
if plot_resonance:
min_wave = np.min([wave_in.min(), wave_out.min()])
max_wave = np.max([wave_in.max(), wave_out.max()])
ax.plot([min_wave, max_wave], [min_wave, max_wave], 'b-')
ax.plot(wave_in, wave_out, 'b.', picker=True)
ax.set_xlabel('Last interaction Wave in')
ax.set_ylabel('Last interaction Wave out')
def onpick(event):
print("-" * 80)
print("Line_in (%d/%d):\n%s" % (
len(event.ind), self.current_no_packets,
self.last_line_list_in.ix[event.ind]))
print("\n\n")
print("Line_out (%d/%d):\n%s" % (
len(event.ind), self.current_no_packets,
self.last_line_list_in.ix[event.ind]))
print("^" * 80)
def onpress(event):
pass
fig.canvas.mpl_connect('pick_event', onpick)
fig.canvas.mpl_connect('on_press', onpress)
class TARDISHistory(object):
"""
Records the history of the model
"""
def __init__(self, hdf5_fname, iterations=None):
self.hdf5_fname = hdf5_fname
if iterations is None:
iterations = []
hdf_store = pd.HDFStore(self.hdf5_fname, 'r')
for key in hdf_store.keys():
if key.split('/')[1] == 'atom_data':
continue
iterations.append(
int(re.match(r'model(\d+)', key.split('/')[1]).groups()[0]))
self.iterations = np.sort(np.unique(iterations))
hdf_store.close()
else:
self.iterations=iterations
self.levels = None
self.lines = None
def load_atom_data(self):
if self.levels is None or self.lines is None:
hdf_store = pd.HDFStore(self.hdf5_fname, 'r')
self.levels = hdf_store['atom_data/levels']
self.lines = hdf_store['atom_data/lines']
hdf_store.close()
def load_t_inner(self, iterations=None):
t_inners = []
hdf_store = pd.HDFStore(self.hdf5_fname, 'r')
if iterations is None:
iterations = self.iterations
elif np.isscalar(iterations):
iterations = [self.iterations[iterations]]
else:
iterations = self.iterations[iterations]
for iter in iterations:
t_inners.append(hdf_store['model%03d/configuration' %iter].ix['t_inner'])
hdf_store.close()
t_inners = np.array(t_inners)
return t_inners
def load_t_rads(self, iterations=None):
t_rads_dict = {}
hdf_store = pd.HDFStore(self.hdf5_fname, 'r')
if iterations is None:
iterations = self.iterations
elif np.isscalar(iterations):
iterations = [self.iterations[iterations]]
else:
iterations = self.iterations[iterations]
for iter in iterations:
current_iter = 'iter%03d' % iter
t_rads_dict[current_iter] = hdf_store['model%03d/t_rads' % iter]
t_rads = pd.DataFrame(t_rads_dict)
hdf_store.close()
return t_rads
def load_ws(self, iterations=None):
ws_dict = {}
hdf_store = pd.HDFStore(self.hdf5_fname, 'r')
if iterations is None:
iterations = self.iterations
elif np.isscalar(iterations):
iterations = [self.iterations[iterations]]
else:
iterations = self.iterations[iterations]
for iter in iterations:
current_iter = 'iter{:03d}'.format(iter)
ws_dict[current_iter] = hdf_store['model{:03d}/ws'.format(iter)]
hdf_store.close()
return pd.DataFrame(ws_dict)
def load_level_populations(self, iterations=None):
level_populations_dict = {}
hdf_store = pd.HDFStore(self.hdf5_fname, 'r')
is_scalar = False
if iterations is None:
iterations = self.iterations
elif np.isscalar(iterations):
is_scalar = True
iterations = [self.iterations[iterations]]
else:
iterations = self.iterations[iterations]
for iter in iterations:
current_iter = 'iter%03d' % iter
level_populations_dict[current_iter] = hdf_store[
'model{:03d}/level_populations'.format(iter)]
hdf_store.close()
if is_scalar:
return pd.DataFrame(level_populations_dict.values()[0])
else:
return pd.Panel(level_populations_dict)
def load_jblues(self, iterations=None):
jblues_dict = {}
hdf_store = pd.HDFStore(self.hdf5_fname, 'r')
is_scalar = False
if iterations is None:
iterations = self.iterations
elif np.isscalar(iterations):
is_scalar = True
iterations = [self.iterations[iterations]]
else:
iterations = self.iterations[iterations]
for iter in iterations:
current_iter = 'iter{:03d}'.format(iter)
jblues_dict[current_iter] = hdf_store[
'model{:03d}/j_blues'.format(iter)]
hdf_store.close()
if is_scalar:
return pd.DataFrame(jblues_dict.values()[0])
else:
return pd.Panel(jblues_dict)
def load_ion_populations(self, iterations=None):
ion_populations_dict = {}
hdf_store = pd.HDFStore(self.hdf5_fname, 'r')
is_scalar = False
if iterations is None:
iterations = self.iterations
elif np.isscalar(iterations):
is_scalar = True
iterations = [self.iterations[iterations]]
else:
iterations = self.iterations[iterations]
for iter in iterations:
current_iter = 'iter{:03d}'.format(iter)
ion_populations_dict[current_iter] = hdf_store[
'model{:03d}/ion_populations'.format(iter)]
hdf_store.close()
if is_scalar:
return pd.DataFrame(ion_populations_dict.values()[0])
else:
return | pd.Panel(ion_populations_dict) | pandas.Panel |
"""
This module contains the classes for testing the exodata of mpcpy.
"""
from mpcpy import exodata
from mpcpy import utility
from mpcpy import units
from mpcpy import variables
from testing import TestCaseMPCPy
import unittest
import numpy as np
import pickle
import copy
import os
import pandas as pd
import datetime
import pytz
#%% Weather Tests
class WeatherFromEPW(TestCaseMPCPy):
'''Test the collection of weather data from an EPW.
'''
def setUp(self):
self.epw_filepath = os.path.join(self.get_unittest_path(), 'resources', 'weather', \
'USA_IL_Chicago-OHare.Intl.AP.725300_TMY3.epw');
self.weather = exodata.WeatherFromEPW(self.epw_filepath);
def tearDown(self):
del self.weather
def test_instantiate(self):
self.assertEqual(self.weather.name, 'weather_from_epw');
self.assertEqual(self.weather.file_path, self.epw_filepath);
self.assertAlmostEqual(self.weather.lat.display_data(), 41.980, places=4);
self.assertAlmostEqual(self.weather.lon.display_data(), -87.92, places=4);
self.assertEqual(self.weather.tz_name, 'America/Chicago');
def test_collect_data(self):
start_time = '1/1/2015';
final_time = '1/1/2016';
self.weather.collect_data(start_time, final_time);
# Check reference
df_test = self.weather.display_data();
self.check_df(df_test, 'collect_data.csv');
def test_collect_data_partial(self):
start_time = '10/2/2015 06:00:00';
final_time = '11/13/2015 16:00:00';
self.weather.collect_data(start_time, final_time);
# Check references
df_test = self.weather.display_data();
self.check_df(df_test, 'collect_data_partial_display.csv');
df_test = self.weather.get_base_data();
self.check_df(df_test, 'collect_data_partial_base.csv');
def test_standard_time(self):
start_time = '1/1/2015';
final_time = '1/1/2016';
weather = exodata.WeatherFromEPW(self.epw_filepath, standard_time=True)
weather.collect_data(start_time, final_time);
# Check instantiation
self.assertAlmostEqual(weather.lat.display_data(), 41.980, places=4);
self.assertAlmostEqual(weather.lon.display_data(), -87.92, places=4);
self.assertEqual(weather.tz_name, 'utc');
# Check reference
df_test = weather.display_data();
self.check_df(df_test, 'collect_data_standard_time.csv');
class CalSolRad(TestCaseMPCPy):
'''Test the method of calculate_solar_radiation
'''
def setUp(self):
self.csv_filepath = os.path.join(self.get_unittest_path(), 'resources', 'weather', 'calSolRadCSV.csv');
self.geography = [37.8716, -122.2727];
self.variable_map = {'Solar Altitude' : ('weaSolAlt', units.rad), \
'Cloud Cover' : ('weaNTot', units.unit1), \
'Relative Humidity' : ('weaRelHum', units.percent), \
'Wind Speed' : ('weaWinSpe', units.m_s)}
self.start_time = '10/19/2016 12:53:00 PM'
self.final_time = '10/19/2016 11:53:00 PM'
self.time_header = 'TimePDT'
def tearDown(self):
del self.csv_filepath
del self.geography
del self.variable_map
del self.start_time
del self.final_time
del self.time_header
def test_calculate(self):
# Instantiate weather object
weather = exodata.WeatherFromCSV(self.csv_filepath, \
self.variable_map, \
self.geography, \
time_header = self.time_header, \
tz_name = 'from_geography')
# Get weather data
weather.collect_data(self.start_time, self.final_time)
# Calculate solar radiation
weather.calculate_solar_radiation(method = 'Zhang-Huang')
# Check reference
df_test = weather.display_data();
self.check_df(df_test, 'calculate_solar_radiation.csv')
def test_catch_method_error(self):
# Instantiate weather object
with self.assertRaises(NameError):
weather.calculate_solar_radiation(method = 'test')
def test_catch_value_missing_error(self):
# Instantiate weather object
self.variable_map_missingRH = {'Solar Altitude' : ('weaSolAlt', units.rad), \
'Cloud Cover' : ('weaNTot', units.unit1), \
'Wind Speed' : ('weaWinSpe', units.m_s)}
# Instantiate weather object
weather = exodata.WeatherFromCSV(self.csv_filepath, \
self.variable_map_missingRH, \
self.geography, \
time_header = self.time_header, \
tz_name = 'from_geography')
# Get weather data
weather.collect_data(self.start_time, self.final_time)
with self.assertRaises(KeyError):
weather.calculate_solar_radiation(method = 'Zhang-Huang')
class WeatherFromCSV(TestCaseMPCPy):
'''Test the collection of weather data from a CSV file.
'''
def setUp(self):
self.csv_filepath = os.path.join(self.get_unittest_path(), 'resources', 'weather', 'BerkeleyCSV.csv');
self.geography = [37.8716, -122.2727];
self.variable_map = {'TemperatureF' : ('weaTDryBul', units.degF), \
'Dew PointF' : ('weaTDewPoi', units.degF), \
'Humidity' : ('weaRelHum', units.percent), \
'Sea Level PressureIn' : ('weaPAtm', units.inHg), \
'WindDirDegrees' : ('weaWinDir', units.deg)};
def tearDown(self):
del self.csv_filepath
del self.geography
del self.variable_map
def test_instantiate(self):
weather = exodata.WeatherFromCSV(self.csv_filepath, \
self.variable_map,
self.geography);
self.assertEqual(weather.name, 'weather_from_csv');
self.assertEqual(weather.file_path, self.csv_filepath);
self.assertAlmostEqual(weather.lat.display_data(), 37.8716, places=4);
self.assertAlmostEqual(weather.lon.display_data(), -122.2727, places=4);
self.assertEqual(weather.tz_name, 'UTC');
def test_instantiate_without_geography(self):
with self.assertRaises(TypeError):
weather = exodata.WeatherFromCSV(self.csv_filepath,
self.variable_map);
def test_collect_data_default_time(self):
start_time = '2016-10-19 19:53:00';
final_time = '2016-10-20 06:53:00';
time_header = 'DateUTC';
# Instantiate weather object
weather = exodata.WeatherFromCSV(self.csv_filepath, \
self.variable_map, \
self.geography, \
time_header = time_header);
# Get weather data
weather.collect_data(start_time, final_time);
# Check reference
df_test = weather.display_data();
self.check_df(df_test, 'collect_data_default_time.csv');
def test_collect_data_local_time_from_geography(self):
start_time = '10/19/2016 12:53:00 PM';
final_time = '10/19/2016 11:53:00 PM';
time_header = 'TimePDT';
# Instantiate weather object
weather = exodata.WeatherFromCSV(self.csv_filepath, \
self.variable_map, \
self.geography, \
time_header = time_header, \
tz_name = 'from_geography');
# Get weather data
weather.collect_data(start_time, final_time);
# Check reference
df_test = weather.display_data();
self.check_df(df_test, 'collect_data_local_time_from_geography.csv');
def test_collect_data_local_time_from_tz_name(self):
start_time = '10/19/2016 12:53:00 PM';
final_time = '10/19/2016 11:53:00 PM';
time_header = 'TimePDT';
# Instantiate weather object
weather = exodata.WeatherFromCSV(self.csv_filepath, \
self.variable_map, \
self.geography, \
time_header = time_header, \
tz_name = 'America/Los_Angeles');
# Get weather data
weather.collect_data(start_time, final_time);
# Check reference
df_test = weather.display_data();
self.check_df(df_test, 'collect_data_local_time_from_tz_name.csv');
def test_collect_data_clean_data(self):
start_time = '2016-10-19 19:53:00';
final_time = '2016-10-20 06:53:00';
time_header = 'DateUTC';
variable_map = {'TemperatureF' : ('weaTDryBul', units.degF), \
'Dew PointF' : ('weaTDewPoi', units.degF), \
'Humidity' : ('weaRelHum', units.percent), \
'Sea Level PressureIn' : ('weaPAtm', units.inHg), \
'WindDirDegrees' : ('weaWinDir', units.deg), \
'Wind SpeedMPH' : ('weaWinSpe', units.mph)};
clean_data = {'Wind SpeedMPH' : {'cleaning_type' : variables.Timeseries.cleaning_replace, \
'cleaning_args' : ('Calm', 0)}};
# Instantiate weather object
weather = exodata.WeatherFromCSV(self.csv_filepath, \
variable_map, \
self.geography, \
time_header = time_header,
clean_data = clean_data);
# Get weather data
weather.collect_data(start_time, final_time);
# Check reference
df_test = weather.display_data();
self.check_df(df_test, 'collect_data_clean_data.csv');
class WeatherFromDF(TestCaseMPCPy):
'''Test the collection of weather data from a pandas DataFrame object.
'''
def setUp(self):
self.df = pd.read_csv(os.path.join(self.get_unittest_path(), 'resources', 'weather', 'BerkeleyCSV.csv'));
self.geography = [37.8716, -122.2727];
self.variable_map = {'TemperatureF' : ('weaTDryBul', units.degF), \
'Dew PointF' : ('weaTDewPoi', units.degF), \
'Humidity' : ('weaRelHum', units.percent), \
'Sea Level PressureIn' : ('weaPAtm', units.inHg), \
'WindDirDegrees' : ('weaWinDir', units.deg)};
def tearDown(self):
del self.df
del self.geography
del self.variable_map
def test_instantiate(self):
time = pd.to_datetime(self.df['DateUTC']);
self.df.set_index(time, inplace=True);
weather = exodata.WeatherFromDF(self.df, \
self.variable_map,
self.geography);
self.assertEqual(weather.name, 'weather_from_df');
self.assertAlmostEqual(weather.lat.display_data(), 37.8716, places=4);
self.assertAlmostEqual(weather.lon.display_data(), -122.2727, places=4);
self.assertEqual(weather.tz_name, 'UTC');
def test_instantiate_without_geography(self):
with self.assertRaises(TypeError):
weather = exodata.WeatherFromDF(self.df,
self.variable_map)
def test_collect_data_default_time(self):
start_time = '2016-10-19 19:53:00';
final_time = '2016-10-20 06:53:00';
time = pd.to_datetime(self.df['DateUTC']);
self.df.set_index(time, inplace=True);
# Instantiate weather object
weather = exodata.WeatherFromDF(self.df, \
self.variable_map, \
self.geography);
# Get weather data
weather.collect_data(start_time, final_time);
# Check reference
df_test = weather.display_data();
self.check_df(df_test, 'collect_data_default_time.csv');
def test_collect_data_local_time_from_geography(self):
start_time = '10/19/2016 12:53:00 PM';
final_time = '10/19/2016 11:53:00 PM';
time = pd.to_datetime(self.df['TimePDT']);
self.df.set_index(time, inplace=True);
# Instantiate weather object
weather = exodata.WeatherFromDF(self.df, \
self.variable_map, \
self.geography,
tz_name = 'from_geography');
# Get weather data
weather.collect_data(start_time, final_time);
# Check reference
df_test = weather.display_data();
self.check_df(df_test, 'collect_data_local_time_from_geography.csv');
def test_collect_data_local_time_from_tz_name(self):
start_time = '10/19/2016 12:53:00 PM';
final_time = '10/19/2016 11:53:00 PM';
time = pd.to_datetime(self.df['TimePDT']);
self.df.set_index(time, inplace=True);
# Instantiate weather object
weather = exodata.WeatherFromDF(self.df, \
self.variable_map, \
self.geography,
tz_name = 'America/Los_Angeles');
# Get weather data
weather.collect_data(start_time, final_time);
# Check reference
df_test = weather.display_data();
self.check_df(df_test, 'collect_data_local_time_from_tz_name.csv');
def test_collect_data_tz_handling(self):
start_time = '2016-10-19 19:53:00';
final_time = '2016-10-20 06:53:00';
time = pd.to_datetime(self.df['DateUTC']);
self.df.set_index(time, inplace=True);
# Localize timezone
self.df = self.df.tz_localize('UTC')
# Instantiate weather object
with self.assertRaises(TypeError):
weather = exodata.WeatherFromDF(self.df, \
self.variable_map, \
self.geography);
# Remove timezone
self.df = self.df.tz_convert(None)
# Instantiate weather object
weather = exodata.WeatherFromDF(self.df, \
self.variable_map, \
self.geography);
# Get weather data
weather.collect_data(start_time, final_time);
# Collect twice
weather.collect_data(start_time, final_time);
# Check reference
df_test = weather.display_data();
self.check_df(df_test, 'collect_data_default_time.csv');
class WeatherFromNOAA(TestCaseMPCPy):
'''Test the collection of weather data from NOAA.
'''
def setUp(self):
self.geography = [37.8716, -122.2727]
self.ins_model_name = 'GFS'
self.weather = exodata.WeatherFromNOAA(self.geography, self.ins_model_name);
self.start_time_pre = pd.Timestamp(datetime.datetime.now(pytz.timezone(self.weather.tz_name)))
self.final_time_pre = self.start_time_pre + pd.Timedelta(days=7)
self.start_time_his = self.start_time_pre - pd.Timedelta(days=7)
self.final_time_his = self.start_time_pre
def tearDown(self):
del self.geography
del self.ins_model_name
del self.start_time_his
del self.final_time_his
del self.start_time_pre
del self.final_time_pre
def valueTest(self, df_test):
'''The function to test the weather values are reasonable:
1. Contains the prediction of weaHDifHor, weaHDirNor, weaHGloHor, weaNTot, weaTDryBul, weaWinSpe
2. The values of predicted weaHDifHor, weaHDirNor, weaHGloHor are in the range of (0,2000)
3. The values of predicted weaNTot are in the range of (0,100)
4. The values of predicted weaTDryBul are in the range of (200,350)
5. The values of predicted weaWinSpe are in the range of (0,20)
'''
# test 1:
fields = df_test.columns
self.assertIn('weaHDifHor', fields)
self.assertIn('weaHDirNor', fields)
self.assertIn('weaHGloHor', fields)
self.assertIn('weaNTot', fields)
self.assertIn('weaTDryBul', fields)
self.assertIn('weaWinSpe', fields)
# test 2-5:
self.assertEqual((df_test[['weaHDifHor','weaHDirNor','weaHGloHor']]<0).sum().sum(),0)
self.assertEqual((df_test[['weaHDifHor','weaHDirNor','weaHGloHor']]>2000).sum().sum(),0)
self.assertEqual((df_test['weaNTot']<0).sum(),0)
self.assertEqual((df_test['weaNTot']>100).sum(),0)
self.assertEqual((df_test['weaTDryBul']<200).sum(),0)
self.assertEqual((df_test['weaTDryBul']>350).sum(),0)
self.assertEqual((df_test['weaWinSpe']<0).sum(),0)
self.assertEqual((df_test['weaWinSpe']>20).sum(),0)
def test_instantiate(self):
weather = exodata.WeatherFromNOAA(self.geography, self.ins_model_name);
self.assertEqual(weather.name, 'weather_from_noaa');
self.assertEqual(weather.tz_name, 'America/Los_Angeles');
self.assertAlmostEqual(weather.lat.display_data(), 37.8716, places=4);
self.assertAlmostEqual(weather.lon.display_data(), -122.2727, places=4);
def test_GFS_collect_historical_data(self):
# Instantiate weather object
weather = exodata.WeatherFromNOAA(self.geography,'GFS');
# Get weather data
weather.collect_data(self.start_time_his, self.final_time_his);
self.df_test = weather.get_base_data()
# Check the fields and value range
self.valueTest(self.df_test)
# Check the length of collected data is at least (24/3)*7-1, because of 3 hours interval
self.assertGreater(self.df_test.shape[0], 55)
def test_HRRR_collect_historical_data(self):
# Instantiate weather object
weather = exodata.WeatherFromNOAA(self.geography,'HRRR');
# Get weather data
weather.collect_data(self.start_time_his, self.final_time_his);
self.df_test = weather.get_base_data()
# Check the fields and value range
self.valueTest(self.df_test)
# Check the length of collected data is at least 3 days, HRRR method only save 3 days' data
self.assertGreater(self.df_test.shape[0], 72)
def test_RAP_collect_historical_data(self):
# Instantiate weather object
weather = exodata.WeatherFromNOAA(self.geography,'RAP');
# Get weather data
weather.collect_data(self.start_time_his, self.final_time_his);
self.df_test = weather.get_base_data()
# Check the fields and value range
self.valueTest(self.df_test)
# Check the length of collected data is at least (24/1)*7-1, because of 1 hours interval
self.assertGreater(self.df_test.shape[0], 167)
def test_NAM_collect_historical_data(self):
# Instantiate weather object
weather = exodata.WeatherFromNOAA(self.geography,'NAM');
# Get weather data
weather.collect_data(self.start_time_his, self.final_time_his);
self.df_test = weather.get_base_data()
# Check the fields and value range
self.valueTest(self.df_test)
# Check the length of collected data is at least (24/1)*7-1, because of 1 hours interval
self.assertGreater(self.df_test.shape[0], 167)
def test_GFS_collect_prediction_data(self):
# Instantiate weather object
weather = exodata.WeatherFromNOAA(self.geography,'GFS');
# Get weather data
weather.collect_data(self.start_time_pre, self.final_time_pre);
self.df_test = weather.get_base_data()
# Check the fields and value range
self.valueTest(self.df_test)
# Check the first prediction is within 3 hours
self.secToFirstPre = (self.df_test.index[0] - self.start_time_pre.tz_convert('UTC')).total_seconds()
self.assertLess(self.secToFirstPre, 3600*3)
# Check the prediction is available for at least 5 days
self.secToLastPre = (self.df_test.index[-1] - self.start_time_pre.tz_convert('UTC')).total_seconds()
self.assertGreater(self.secToLastPre, 3600*24*5)
def test_HRRR_collect_prediction_data(self):
# Instantiate weather object
weather = exodata.WeatherFromNOAA(self.geography,'HRRR');
# Get weather data
weather.collect_data(self.start_time_pre, self.final_time_pre);
self.df_test = weather.get_base_data()
# Check the fields and value range
self.valueTest(self.df_test)
# Check the first prediction is within 1 hours
self.secToFirstPre = (self.df_test.index[0] - self.start_time_pre.tz_convert('UTC')).total_seconds()
self.assertLess(self.secToFirstPre, 3600*1)
# Check the prediction is available for at least 15 hours
self.secToLastPre = (self.df_test.index[-1] - self.start_time_pre.tz_convert('UTC')).total_seconds()
self.assertGreaterEqual(self.secToLastPre, 3600*15)
def test_RAP_collect_prediction_data(self):
# Instantiate weather object
weather = exodata.WeatherFromNOAA(self.geography,'RAP');
# Get weather data
weather.collect_data(self.start_time_pre, self.final_time_pre);
self.df_test = weather.get_base_data()
# Check the fields and value range
self.valueTest(self.df_test)
# Check the first prediction is within 1 hours
self.secToFirstPre = (self.df_test.index[0] - self.start_time_pre.tz_convert('UTC')).total_seconds()
self.assertLess(self.secToFirstPre, 3600*1)
# Check the prediction is available for at least 18 hours
self.secToLastPre = (self.df_test.index[-1] - self.start_time_pre.tz_convert('UTC')).total_seconds()
self.assertGreaterEqual(self.secToLastPre, 3600*18)
def test_NAM_collect_prediction_data(self):
# Instantiate weather object
weather = exodata.WeatherFromNOAA(self.geography,'NAM');
# Get weather data
weather.collect_data(self.start_time_pre, self.final_time_pre);
self.df_test = weather.get_base_data()
# Check the fields and value range
self.valueTest(self.df_test)
# Check the first prediction is within 6 hours
self.secToFirstPre = (self.df_test.index[0] - self.start_time_pre.tz_convert('UTC')).total_seconds()
self.assertLess(self.secToFirstPre, 3600*6)
# Check the prediction is available for at least 3 days
self.secToLastPre = (self.df_test.index[-1] - self.start_time_pre.tz_convert('UTC')).total_seconds()
self.assertGreaterEqual(self.secToLastPre, 3600*24*3)
def test_catch_method_error(self):
# Instantiate weather object
with self.assertRaises(NameError):
weather = exodata.WeatherFromNOAA(self.geography,'test');
#%% Internal Tests
class InternalFromCSV(TestCaseMPCPy):
'''Test the collection of internal data from a CSV file.
'''
def setUp(self):
csv_filepath = os.path.join(self.get_unittest_path(), 'resources', 'internal', 'sampleCSV.csv');
variable_map = {'intRad_wes' : ('wes', 'intRad', units.W_m2), \
'intCon_wes' : ('wes', 'intCon', units.W_m2), \
'intLat_wes' : ('wes', 'intLat', units.W_m2), \
'intRad_hal' : ('hal', 'intRad', units.W_m2), \
'intCon_hal' : ('hal', 'intCon', units.W_m2), \
'intLat_hal' : ('hal', 'intLat', units.W_m2), \
'intRad_eas' : ('eas', 'intRad', units.W_m2), \
'intCon_eas' : ('eas', 'intCon', units.W_m2), \
'intLat_eas' : ('eas', 'intLat', units.W_m2)};
# Instantiate internal object
self.internal = exodata.InternalFromCSV(csv_filepath, \
variable_map);
def tearDown(self):
del self.internal
def test_collect_data(self):
start_time = '1/2/2015';
final_time = '1/3/2015';
# Get internal data
self.internal.collect_data(start_time, final_time);
# Check reference
df_test = self.internal.display_data();
self.check_df(df_test, 'collect_data.csv');
class InternalFromOccupancyModel(TestCaseMPCPy):
'''Test the collection of internal data from an occupancy model.
'''
def setUp(self):
# Time
start_time_occupancy = '4/1/2013';
final_time_occupancy = '4/7/2013 23:55:00';
# Load occupancy models
with open(os.path.join(utility.get_MPCPy_path(), 'unittests', 'references', \
'test_models', 'OccupancyFromQueueing', \
'occupancy_model_estimated.txt'), 'r') as f:
occupancy_model = pickle.load(f);
# Define zones and loads
zone_list = ['wes', 'hal', 'eas'];
load_list = [[0.4,0.4,0.2], [0.4,0.4,0.2], [0.4,0.4,0.2]];
# Simulate occupancy models for each zone
occupancy_model_list = [];
np.random.seed(1); # start with same seed for random number generation
for zone in zone_list:
simulate_options = occupancy_model.get_simulate_options();
simulate_options['iter_num'] = 5;
occupancy_model.simulate(start_time_occupancy, final_time_occupancy)
occupancy_model_list.append(copy.deepcopy(occupancy_model));
# Instantiate internal object
self.internal = exodata.InternalFromOccupancyModel(zone_list, load_list, units.W_m2, occupancy_model_list);
def tearDown(self):
del self.internal
def test_collect_data(self):
start_time = '4/2/2013';
final_time = '4/4/2013';
# Get internal data
self.internal.collect_data(start_time, final_time);
# Check reference
df_test = self.internal.display_data();
self.check_df(df_test, 'collect_data.csv');
#%% Control Tests
class ControlFromCSV(TestCaseMPCPy):
'''Test the collection of control data from a CSV file.
'''
def setUp(self):
csv_filepath = os.path.join(self.get_unittest_path(), 'resources', 'building', 'ControlCSV_0.csv');
variable_map = {'conHeat_wes' : ('conHeat_wes', units.unit1), \
'conHeat_hal' : ('conHeat_hal', units.unit1), \
'conHeat_eas' : ('conHeat_eas', units.unit1)};
# Instantiate control object
self.control = exodata.ControlFromCSV(csv_filepath, \
variable_map);
def tearDown(self):
del self.control
def test_collect_data(self):
start_time = '1/1/2015 13:00:00';
final_time = '1/2/2015';
# Get control data
self.control.collect_data(start_time, final_time);
# Check reference
df_test = self.control.display_data();
self.check_df(df_test, 'collect_data.csv');
class ControlFromDF(TestCaseMPCPy):
'''Test the collection of control data from a pandas DataFrame object.
'''
def setUp(self):
self.df = pd.read_csv(os.path.join(self.get_unittest_path(), 'resources', 'building', 'ControlCSV_0.csv'));
time = pd.to_datetime(self.df['Time']);
self.df.set_index(time, inplace=True);
self.variable_map = {'conHeat_wes' : ('conHeat_wes', units.unit1), \
'conHeat_hal' : ('conHeat_hal', units.unit1), \
'conHeat_eas' : ('conHeat_eas', units.unit1)};
def tearDown(self):
del self.df
del self.variable_map
def test_collect_data(self):
start_time = '1/1/2015 13:00:00';
final_time = '1/2/2015';
# Instantiate control object
control = exodata.ControlFromDF(self.df, \
self.variable_map);
# Get control data
control.collect_data(start_time, final_time);
# Check reference
df_test = control.display_data();
self.check_df(df_test, 'collect_data.csv');
def test_collect_data_tz_handling(self):
start_time = '1/1/2015 13:00:00';
final_time = '1/2/2015';
# Localize timezone
self.df = self.df.tz_localize('UTC')
# Instantiate weather object
with self.assertRaises(TypeError):
control = exodata.ControlFromDF(self.df, \
self.variable_map);
# Remove timezone
self.df = self.df.tz_convert(None)
# Instantiate weather object
control = exodata.ControlFromDF(self.df, \
self.variable_map);
# Get control data
control.collect_data(start_time, final_time);
# Collect twice
control.collect_data(start_time, final_time);
# Check reference
df_test = control.display_data();
self.check_df(df_test, 'collect_data.csv');
#%% Other Input Tests
class OtherInputFromCSV(TestCaseMPCPy):
'''Test the collection of other input data from a CSV file.
'''
def setUp(self):
csv_filepath = os.path.join(self.get_unittest_path(), 'resources', 'weather', 'Tamb.csv');
variable_map = {'T' : ('Tamb', units.degC)};
# Instantiate other input object
self.otherinput = exodata.OtherInputFromCSV(csv_filepath, \
variable_map);
def tearDown(self):
del self.otherinput
def test_collect_data(self):
start_time = '1/1/2015 00:00:00';
final_time = '1/1/2015 06:00:00';
# Get other input data
self.otherinput.collect_data(start_time, final_time);
# Check reference
df_test = self.otherinput.display_data();
self.check_df(df_test, 'collect_data.csv');
class OtherInputFromDF(TestCaseMPCPy):
'''Test the collection of other input data from a pandas DataFrame object.
'''
def setUp(self):
self.df = pd.read_csv(os.path.join(self.get_unittest_path(), 'resources', 'weather', 'Tamb.csv'));
time = | pd.to_datetime(self.df['Time']) | pandas.to_datetime |
from IMLearn.utils import split_train_test
from IMLearn.learners.regressors import LinearRegression
from typing import NoReturn
import numpy as np
import pandas as pd
import plotly.graph_objects as go
import plotly.express as px
import plotly.io as pio
pio.templates.default = "simple_white"
def load_data(filename: str):
"""
Load house prices dataset and preprocess data.
Parameters
----------
filename: str
Path to house prices dataset
Returns
-------
Design matrix and response vector (prices) - either as a single
DataFrame or a Tuple[DataFrame, Series]
"""
df = pd.read_csv(filename)
# removing irrelevant data
df.drop(['id', 'date', 'lat', 'long'], axis=1, inplace=True)
# removing samples with invalid values
for col_name in ('price', 'sqft_living', 'floors',
'sqft_above', 'yr_built', 'zipcode',
'sqft_living15', 'sqft_lot15'):
df.drop(df[df[col_name] <= 0].index, inplace=True)
for col_name in ('bedrooms', 'bathrooms', 'sqft_lot',
'sqft_basement', 'yr_renovated'):
df.drop(df[df[col_name] < 0].index, inplace=True)
# removing samples with null values
df.dropna(axis=0, inplace=True)
# making sure categorical columns are in correct range
waterfront_min, waterfront_max = 0, 1
df.drop(df[(df['waterfront'] < waterfront_min)
| (df['waterfront'] > waterfront_max)].index, inplace=True)
view_min, view_max = 0, 4
df.drop(df[(df['view'] < view_min)
| (df['view'] > view_max)].index, inplace=True)
condition_min, condition_max = 1, 5
df.drop(df[(df['condition'] < condition_min)
| (df['condition'] > condition_max)].index, inplace=True)
grade_min, grade_max = 1, 13
df.drop(df[(df['grade'] < grade_min)
| (df['grade'] > grade_max)].index, inplace=True)
# add additional columns
yr_built_range = max(df['yr_built']) - min(df['yr_built'])
# add newly_built as top 10% of yr_built
yr_built_lower_bound = max(df['yr_built']) - yr_built_range / 10
df['newly_built'] = \
np.where((df['yr_built'] > yr_built_lower_bound)
| (df['yr_renovated'] > yr_built_lower_bound), 1, 0)
# dummy values for relevant columns - using one-hot encoding
df['zipcode'] = df['zipcode'].astype(int)
df = | pd.get_dummies(df, prefix='zipcode', columns=['zipcode']) | pandas.get_dummies |
# pylint: disable-msg=E1101,W0612
from datetime import datetime, timedelta
import os
import operator
import unittest
import cStringIO as StringIO
import nose
from numpy import nan
import numpy as np
import numpy.ma as ma
from pandas import Index, Series, TimeSeries, DataFrame, isnull, notnull
from pandas.core.index import MultiIndex
import pandas.core.datetools as datetools
from pandas.util import py3compat
from pandas.util.testing import assert_series_equal, assert_almost_equal
import pandas.util.testing as tm
#-------------------------------------------------------------------------------
# Series test cases
JOIN_TYPES = ['inner', 'outer', 'left', 'right']
class CheckNameIntegration(object):
def test_scalarop_preserve_name(self):
result = self.ts * 2
self.assertEquals(result.name, self.ts.name)
def test_copy_name(self):
result = self.ts.copy()
self.assertEquals(result.name, self.ts.name)
# def test_copy_index_name_checking(self):
# # don't want to be able to modify the index stored elsewhere after
# # making a copy
# self.ts.index.name = None
# cp = self.ts.copy()
# cp.index.name = 'foo'
# self.assert_(self.ts.index.name is None)
def test_append_preserve_name(self):
result = self.ts[:5].append(self.ts[5:])
self.assertEquals(result.name, self.ts.name)
def test_binop_maybe_preserve_name(self):
# names match, preserve
result = self.ts * self.ts
self.assertEquals(result.name, self.ts.name)
result = self.ts * self.ts[:-2]
self.assertEquals(result.name, self.ts.name)
# names don't match, don't preserve
cp = self.ts.copy()
cp.name = 'something else'
result = self.ts + cp
self.assert_(result.name is None)
def test_combine_first_name(self):
result = self.ts.combine_first(self.ts[:5])
self.assertEquals(result.name, self.ts.name)
def test_getitem_preserve_name(self):
result = self.ts[self.ts > 0]
self.assertEquals(result.name, self.ts.name)
result = self.ts[[0, 2, 4]]
self.assertEquals(result.name, self.ts.name)
result = self.ts[5:10]
self.assertEquals(result.name, self.ts.name)
def test_multilevel_name_print(self):
index = MultiIndex(levels=[['foo', 'bar', 'baz', 'qux'],
['one', 'two', 'three']],
labels=[[0, 0, 0, 1, 1, 2, 2, 3, 3, 3],
[0, 1, 2, 0, 1, 1, 2, 0, 1, 2]],
names=['first', 'second'])
s = Series(range(0,len(index)), index=index, name='sth')
expected = ["first second",
"foo one 0",
" two 1",
" three 2",
"bar one 3",
" two 4",
"baz two 5",
" three 6",
"qux one 7",
" two 8",
" three 9",
"Name: sth"]
expected = "\n".join(expected)
self.assertEquals(repr(s), expected)
def test_multilevel_preserve_name(self):
index = MultiIndex(levels=[['foo', 'bar', 'baz', 'qux'],
['one', 'two', 'three']],
labels=[[0, 0, 0, 1, 1, 2, 2, 3, 3, 3],
[0, 1, 2, 0, 1, 1, 2, 0, 1, 2]],
names=['first', 'second'])
s = Series(np.random.randn(len(index)), index=index, name='sth')
result = s['foo']
result2 = s.ix['foo']
self.assertEquals(result.name, s.name)
self.assertEquals(result2.name, s.name)
def test_name_printing(self):
# test small series
s = Series([0, 1, 2])
s.name = "test"
self.assert_("Name: test" in repr(s))
s.name = None
self.assert_(not "Name:" in repr(s))
# test big series (diff code path)
s = Series(range(0,1000))
s.name = "test"
self.assert_("Name: test" in repr(s))
s.name = None
self.assert_(not "Name:" in repr(s))
def test_pickle_preserve_name(self):
unpickled = self._pickle_roundtrip(self.ts)
self.assertEquals(unpickled.name, self.ts.name)
def _pickle_roundtrip(self, obj):
obj.save('__tmp__')
unpickled = Series.load('__tmp__')
os.remove('__tmp__')
return unpickled
def test_argsort_preserve_name(self):
result = self.ts.argsort()
self.assertEquals(result.name, self.ts.name)
def test_sort_index_name(self):
result = self.ts.sort_index(ascending=False)
self.assertEquals(result.name, self.ts.name)
def test_to_sparse_pass_name(self):
result = self.ts.to_sparse()
self.assertEquals(result.name, self.ts.name)
class SafeForSparse(object):
pass
class TestSeries(unittest.TestCase, CheckNameIntegration):
def setUp(self):
self.ts = tm.makeTimeSeries()
self.ts.name = 'ts'
self.series = tm.makeStringSeries()
self.series.name = 'series'
self.objSeries = tm.makeObjectSeries()
self.objSeries.name = 'objects'
self.empty = Series([], index=[])
def test_constructor(self):
# Recognize TimeSeries
self.assert_(isinstance(self.ts, TimeSeries))
# Pass in Series
derived = Series(self.ts)
self.assert_(isinstance(derived, TimeSeries))
self.assert_(tm.equalContents(derived.index, self.ts.index))
# Ensure new index is not created
self.assertEquals(id(self.ts.index), id(derived.index))
# Pass in scalar
scalar = Series(0.5)
self.assert_(isinstance(scalar, float))
# Mixed type Series
mixed = Series(['hello', np.NaN], index=[0, 1])
self.assert_(mixed.dtype == np.object_)
self.assert_(mixed[1] is np.NaN)
self.assert_(not isinstance(self.empty, TimeSeries))
self.assert_(not isinstance(Series({}), TimeSeries))
self.assertRaises(Exception, Series, np.random.randn(3, 3),
index=np.arange(3))
def test_constructor_empty(self):
empty = Series()
empty2 = Series([])
assert_series_equal(empty, empty2)
empty = Series(index=range(10))
empty2 = Series(np.nan, index=range(10))
assert_series_equal(empty, empty2)
def test_constructor_maskedarray(self):
data = ma.masked_all((3,), dtype=float)
result = Series(data)
expected = Series([nan, nan, nan])
assert_series_equal(result, expected)
data[0] = 0.0
data[2] = 2.0
index = ['a', 'b', 'c']
result = Series(data, index=index)
expected = Series([0.0, nan, 2.0], index=index)
assert_series_equal(result, expected)
def test_constructor_default_index(self):
s = Series([0, 1, 2])
assert_almost_equal(s.index, np.arange(3))
def test_constructor_corner(self):
df = tm.makeTimeDataFrame()
objs = [df, df]
s = Series(objs, index=[0, 1])
self.assert_(isinstance(s, Series))
def test_constructor_cast(self):
self.assertRaises(ValueError, Series, ['a', 'b', 'c'], dtype=float)
def test_constructor_dict(self):
d = {'a' : 0., 'b' : 1., 'c' : 2.}
result = Series(d, index=['b', 'c', 'd', 'a'])
expected = Series([1, 2, nan, 0], index=['b', 'c', 'd', 'a'])
assert_series_equal(result, expected)
def test_constructor_list_of_tuples(self):
data = [(1, 1), (2, 2), (2, 3)]
s = Series(data)
self.assertEqual(list(s), data)
def test_constructor_tuple_of_tuples(self):
data = ((1, 1), (2, 2), (2, 3))
s = Series(data)
self.assertEqual(tuple(s), data)
def test_fromDict(self):
data = {'a' : 0, 'b' : 1, 'c' : 2, 'd' : 3}
series = Series(data)
self.assert_(tm.is_sorted(series.index))
data = {'a' : 0, 'b' : '1', 'c' : '2', 'd' : datetime.now()}
series = Series(data)
self.assert_(series.dtype == np.object_)
data = {'a' : 0, 'b' : '1', 'c' : '2', 'd' : '3'}
series = Series(data)
self.assert_(series.dtype == np.object_)
data = {'a' : '0', 'b' : '1'}
series = Series(data, dtype=float)
self.assert_(series.dtype == np.float64)
def test_setindex(self):
# wrong type
series = self.series.copy()
self.assertRaises(TypeError, setattr, series, 'index', None)
# wrong length
series = self.series.copy()
self.assertRaises(AssertionError, setattr, series, 'index',
np.arange(len(series) - 1))
# works
series = self.series.copy()
series.index = np.arange(len(series))
self.assert_(isinstance(series.index, Index))
def test_array_finalize(self):
pass
def test_fromValue(self):
nans = Series(np.NaN, index=self.ts.index)
self.assert_(nans.dtype == np.float_)
self.assertEqual(len(nans), len(self.ts))
strings = Series('foo', index=self.ts.index)
self.assert_(strings.dtype == np.object_)
self.assertEqual(len(strings), len(self.ts))
d = datetime.now()
dates = Series(d, index=self.ts.index)
self.assert_(dates.dtype == np.object_)
self.assertEqual(len(dates), len(self.ts))
def test_contains(self):
tm.assert_contains_all(self.ts.index, self.ts)
def test_pickle(self):
unp_series = self._pickle_roundtrip(self.series)
unp_ts = self._pickle_roundtrip(self.ts)
assert_series_equal(unp_series, self.series)
assert_series_equal(unp_ts, self.ts)
def _pickle_roundtrip(self, obj):
obj.save('__tmp__')
unpickled = Series.load('__tmp__')
os.remove('__tmp__')
return unpickled
def test_getitem_get(self):
idx1 = self.series.index[5]
idx2 = self.objSeries.index[5]
self.assertEqual(self.series[idx1], self.series.get(idx1))
self.assertEqual(self.objSeries[idx2], self.objSeries.get(idx2))
self.assertEqual(self.series[idx1], self.series[5])
self.assertEqual(self.objSeries[idx2], self.objSeries[5])
self.assert_(self.series.get(-1) is None)
self.assertEqual(self.series[5], self.series.get(self.series.index[5]))
# missing
d = self.ts.index[0] - datetools.bday
self.assertRaises(KeyError, self.ts.__getitem__, d)
def test_iget(self):
s = Series(np.random.randn(10), index=range(0, 20, 2))
for i in range(len(s)):
result = s.iget(i)
exp = s[s.index[i]]
assert_almost_equal(result, exp)
# pass a slice
result = s.iget(slice(1, 3))
expected = s.ix[2:4]
assert_series_equal(result, expected)
def test_getitem_regression(self):
s = Series(range(5), index=range(5))
result = s[range(5)]
assert_series_equal(result, s)
def test_getitem_slice_bug(self):
s = Series(range(10), range(10))
result = s[-12:]
assert_series_equal(result, s)
result = s[-7:]
assert_series_equal(result, s[3:])
result = s[:-12]
assert_series_equal(result, s[:0])
def test_getitem_int64(self):
idx = np.int64(5)
self.assertEqual(self.ts[idx], self.ts[5])
def test_getitem_fancy(self):
slice1 = self.series[[1,2,3]]
slice2 = self.objSeries[[1,2,3]]
self.assertEqual(self.series.index[2], slice1.index[1])
self.assertEqual(self.objSeries.index[2], slice2.index[1])
self.assertEqual(self.series[2], slice1[1])
self.assertEqual(self.objSeries[2], slice2[1])
def test_getitem_boolean(self):
s = self.series
mask = s > s.median()
# passing list is OK
result = s[list(mask)]
expected = s[mask]
assert_series_equal(result, expected)
self.assert_(np.array_equal(result.index, s.index[mask]))
def test_getitem_generator(self):
gen = (x > 0 for x in self.series)
result = self.series[gen]
result2 = self.series[iter(self.series > 0)]
expected = self.series[self.series > 0]
assert_series_equal(result, expected)
assert_series_equal(result2, expected)
def test_getitem_boolean_object(self):
# using column from DataFrame
s = self.series
mask = s > s.median()
omask = mask.astype(object)
# getitem
result = s[omask]
expected = s[mask]
assert_series_equal(result, expected)
# setitem
cop = s.copy()
cop[omask] = 5
s[mask] = 5
assert_series_equal(cop, s)
# nans raise exception
omask[5:10] = np.nan
self.assertRaises(Exception, s.__getitem__, omask)
self.assertRaises(Exception, s.__setitem__, omask, 5)
def test_getitem_setitem_boolean_corner(self):
ts = self.ts
mask_shifted = ts.shift(1, offset=datetools.bday) > ts.median()
self.assertRaises(Exception, ts.__getitem__, mask_shifted)
self.assertRaises(Exception, ts.__setitem__, mask_shifted, 1)
self.assertRaises(Exception, ts.ix.__getitem__, mask_shifted)
self.assertRaises(Exception, ts.ix.__setitem__, mask_shifted, 1)
def test_getitem_setitem_slice_integers(self):
s = Series(np.random.randn(8), index=[2, 4, 6, 8, 10, 12, 14, 16])
result = s[:4]
expected = s.reindex([2, 4, 6, 8])
assert_series_equal(result, expected)
s[:4] = 0
self.assert_((s[:4] == 0).all())
self.assert_(not (s[4:] == 0).any())
def test_getitem_out_of_bounds(self):
# don't segfault, GH #495
self.assertRaises(IndexError, self.ts.__getitem__, len(self.ts))
def test_getitem_box_float64(self):
value = self.ts[5]
self.assert_(isinstance(value, np.float64))
def test_getitem_ambiguous_keyerror(self):
s = Series(range(10), index=range(0, 20, 2))
self.assertRaises(KeyError, s.__getitem__, 1)
self.assertRaises(KeyError, s.ix.__getitem__, 1)
def test_setitem_ambiguous_keyerror(self):
s = Series(range(10), index=range(0, 20, 2))
self.assertRaises(KeyError, s.__setitem__, 1, 5)
self.assertRaises(KeyError, s.ix.__setitem__, 1, 5)
def test_slice(self):
numSlice = self.series[10:20]
numSliceEnd = self.series[-10:]
objSlice = self.objSeries[10:20]
self.assert_(self.series.index[9] not in numSlice.index)
self.assert_(self.objSeries.index[9] not in objSlice.index)
self.assertEqual(len(numSlice), len(numSlice.index))
self.assertEqual(self.series[numSlice.index[0]],
numSlice[numSlice.index[0]])
self.assertEqual(numSlice.index[1], self.series.index[11])
self.assert_(tm.equalContents(numSliceEnd,
np.array(self.series)[-10:]))
# test return view
sl = self.series[10:20]
sl[:] = 0
self.assert_((self.series[10:20] == 0).all())
def test_slice_can_reorder_not_uniquely_indexed(self):
s = Series(1, index=['a', 'a', 'b', 'b', 'c'])
result = s[::-1] # it works!
def test_setitem(self):
self.ts[self.ts.index[5]] = np.NaN
self.ts[[1,2,17]] = np.NaN
self.ts[6] = np.NaN
self.assert_(np.isnan(self.ts[6]))
self.assert_(np.isnan(self.ts[2]))
self.ts[np.isnan(self.ts)] = 5
self.assert_(not np.isnan(self.ts[2]))
# caught this bug when writing tests
series = Series(tm.makeIntIndex(20).astype(float),
index=tm.makeIntIndex(20))
series[::2] = 0
self.assert_((series[::2] == 0).all())
# set item that's not contained
self.assertRaises(Exception, self.series.__setitem__,
'foobar', 1)
def test_set_value(self):
idx = self.ts.index[10]
res = self.ts.set_value(idx, 0)
self.assert_(res is self.ts)
self.assertEqual(self.ts[idx], 0)
res = self.series.set_value('foobar', 0)
self.assert_(res is not self.series)
self.assert_(res.index[-1] == 'foobar')
self.assertEqual(res['foobar'], 0)
def test_setslice(self):
sl = self.ts[5:20]
self.assertEqual(len(sl), len(sl.index))
self.assertEqual(len(sl.index.indexMap), len(sl.index))
def test_basic_getitem_setitem_corner(self):
# invalid tuples, e.g. self.ts[:, None] vs. self.ts[:, 2]
self.assertRaises(Exception, self.ts.__getitem__,
(slice(None, None), 2))
self.assertRaises(Exception, self.ts.__setitem__,
(slice(None, None), 2), 2)
# weird lists. [slice(0, 5)] will work but not two slices
result = self.ts[[slice(None, 5)]]
expected = self.ts[:5]
assert_series_equal(result, expected)
# OK
self.assertRaises(Exception, self.ts.__getitem__,
[5, slice(None, None)])
self.assertRaises(Exception, self.ts.__setitem__,
[5, slice(None, None)], 2)
def test_basic_getitem_with_labels(self):
indices = self.ts.index[[5, 10, 15]]
result = self.ts[indices]
expected = self.ts.reindex(indices)
assert_series_equal(result, expected)
result = self.ts[indices[0]:indices[2]]
expected = self.ts.ix[indices[0]:indices[2]]
assert_series_equal(result, expected)
# integer indexes, be careful
s = Series(np.random.randn(10), index=range(0, 20, 2))
inds = [0, 2, 5, 7, 8]
arr_inds = np.array([0, 2, 5, 7, 8])
result = s[inds]
expected = s.reindex(inds)
assert_series_equal(result, expected)
result = s[arr_inds]
expected = s.reindex(arr_inds)
assert_series_equal(result, expected)
def test_basic_setitem_with_labels(self):
indices = self.ts.index[[5, 10, 15]]
cp = self.ts.copy()
exp = self.ts.copy()
cp[indices] = 0
exp.ix[indices] = 0
assert_series_equal(cp, exp)
cp = self.ts.copy()
exp = self.ts.copy()
cp[indices[0]:indices[2]] = 0
exp.ix[indices[0]:indices[2]] = 0
assert_series_equal(cp, exp)
# integer indexes, be careful
s = Series(np.random.randn(10), index=range(0, 20, 2))
inds = [0, 4, 6]
arr_inds = np.array([0, 4, 6])
cp = s.copy()
exp = s.copy()
s[inds] = 0
s.ix[inds] = 0
assert_series_equal(cp, exp)
cp = s.copy()
exp = s.copy()
s[arr_inds] = 0
s.ix[arr_inds] = 0
assert_series_equal(cp, exp)
inds_notfound = [0, 4, 5, 6]
arr_inds_notfound = np.array([0, 4, 5, 6])
self.assertRaises(Exception, s.__setitem__, inds_notfound, 0)
self.assertRaises(Exception, s.__setitem__, arr_inds_notfound, 0)
def test_ix_getitem(self):
inds = self.series.index[[3,4,7]]
assert_series_equal(self.series.ix[inds], self.series.reindex(inds))
assert_series_equal(self.series.ix[5::2], self.series[5::2])
# slice with indices
d1, d2 = self.ts.index[[5, 15]]
result = self.ts.ix[d1:d2]
expected = self.ts.truncate(d1, d2)
assert_series_equal(result, expected)
# boolean
mask = self.series > self.series.median()
assert_series_equal(self.series.ix[mask], self.series[mask])
# ask for index value
self.assertEquals(self.ts.ix[d1], self.ts[d1])
self.assertEquals(self.ts.ix[d2], self.ts[d2])
def test_ix_getitem_not_monotonic(self):
d1, d2 = self.ts.index[[5, 15]]
ts2 = self.ts[::2][::-1]
self.assertRaises(KeyError, ts2.ix.__getitem__, slice(d1, d2))
self.assertRaises(KeyError, ts2.ix.__setitem__, slice(d1, d2), 0)
def test_ix_getitem_setitem_integer_slice_keyerrors(self):
s = Series(np.random.randn(10), index=range(0, 20, 2))
# this is OK
cp = s.copy()
cp.ix[4:10] = 0
self.assert_((cp.ix[4:10] == 0).all())
# so is this
cp = s.copy()
cp.ix[3:11] = 0
self.assert_((cp.ix[3:11] == 0).values.all())
result = s.ix[4:10]
result2 = s.ix[3:11]
expected = s.reindex([4, 6, 8, 10])
assert_series_equal(result, expected)
assert_series_equal(result2, expected)
# non-monotonic, raise KeyError
s2 = s[::-1]
self.assertRaises(KeyError, s2.ix.__getitem__, slice(3, 11))
self.assertRaises(KeyError, s2.ix.__setitem__, slice(3, 11), 0)
def test_ix_getitem_iterator(self):
idx = iter(self.series.index[:10])
result = self.series.ix[idx]
assert_series_equal(result, self.series[:10])
def test_ix_setitem(self):
inds = self.series.index[[3,4,7]]
result = self.series.copy()
result.ix[inds] = 5
expected = self.series.copy()
expected[[3,4,7]] = 5
assert_series_equal(result, expected)
result.ix[5:10] = 10
expected[5:10] = 10
assert_series_equal(result, expected)
# set slice with indices
d1, d2 = self.series.index[[5, 15]]
result.ix[d1:d2] = 6
expected[5:16] = 6 # because it's inclusive
assert_series_equal(result, expected)
# set index value
self.series.ix[d1] = 4
self.series.ix[d2] = 6
self.assertEquals(self.series[d1], 4)
self.assertEquals(self.series[d2], 6)
def test_ix_setitem_boolean(self):
mask = self.series > self.series.median()
result = self.series.copy()
result.ix[mask] = 0
expected = self.series
expected[mask] = 0
assert_series_equal(result, expected)
def test_ix_setitem_corner(self):
inds = list(self.series.index[[5, 8, 12]])
self.series.ix[inds] = 5
self.assertRaises(Exception, self.series.ix.__setitem__,
inds + ['foo'], 5)
def test_get_set_boolean_different_order(self):
ordered = self.series.order()
# setting
copy = self.series.copy()
copy[ordered > 0] = 0
expected = self.series.copy()
expected[expected > 0] = 0
assert_series_equal(copy, expected)
# getting
sel = self.series[ordered > 0]
exp = self.series[self.series > 0]
assert_series_equal(sel, exp)
def test_repr(self):
str(self.ts)
str(self.series)
str(self.series.astype(int))
str(self.objSeries)
str(Series(tm.randn(1000), index=np.arange(1000)))
str(Series(tm.randn(1000), index=np.arange(1000, 0, step=-1)))
# empty
str(self.empty)
# with NaNs
self.series[5:7] = np.NaN
str(self.series)
# tuple name, e.g. from hierarchical index
self.series.name = ('foo', 'bar', 'baz')
repr(self.series)
biggie = Series(tm.randn(1000), index=np.arange(1000),
name=('foo', 'bar', 'baz'))
repr(biggie)
def test_to_string(self):
from cStringIO import StringIO
buf = StringIO()
s = self.ts.to_string()
retval = self.ts.to_string(buf=buf)
self.assert_(retval is None)
self.assertEqual(buf.getvalue().strip(), s)
# pass float_format
format = '%.4f'.__mod__
result = self.ts.to_string(float_format=format)
result = [x.split()[1] for x in result.split('\n')]
expected = [format(x) for x in self.ts]
self.assertEqual(result, expected)
# empty string
result = self.ts[:0].to_string()
self.assertEqual(result, '')
result = self.ts[:0].to_string(length=0)
self.assertEqual(result, '')
# name and length
cp = self.ts.copy()
cp.name = 'foo'
result = cp.to_string(length=True, name=True)
last_line = result.split('\n')[-1].strip()
self.assertEqual(last_line, "Name: foo, Length: %d" % len(cp))
def test_to_string_mixed(self):
s = Series(['foo', np.nan, -1.23, 4.56])
result = s.to_string()
expected = ('0 foo\n'
'1 NaN\n'
'2 -1.23\n'
'3 4.56')
self.assertEqual(result, expected)
# but don't count NAs as floats
s = Series(['foo', np.nan, 'bar', 'baz'])
result = s.to_string()
expected = ('0 foo\n'
'1 NaN\n'
'2 bar\n'
'3 baz')
self.assertEqual(result, expected)
s = Series(['foo', 5, 'bar', 'baz'])
result = s.to_string()
expected = ('0 foo\n'
'1 5\n'
'2 bar\n'
'3 baz')
self.assertEqual(result, expected)
def test_to_string_float_na_spacing(self):
s = Series([0., 1.5678, 2., -3., 4.])
s[::2] = np.nan
result = s.to_string()
expected = ('0 NaN\n'
'1 1.568\n'
'2 NaN\n'
'3 -3.000\n'
'4 NaN')
self.assertEqual(result, expected)
def test_iter(self):
for i, val in enumerate(self.series):
self.assertEqual(val, self.series[i])
for i, val in enumerate(self.ts):
self.assertEqual(val, self.ts[i])
def test_keys(self):
# HACK: By doing this in two stages, we avoid 2to3 wrapping the call
# to .keys() in a list()
getkeys = self.ts.keys
self.assert_(getkeys() is self.ts.index)
def test_values(self):
self.assert_(np.array_equal(self.ts, self.ts.values))
def test_iteritems(self):
for idx, val in self.series.iteritems():
self.assertEqual(val, self.series[idx])
for idx, val in self.ts.iteritems():
self.assertEqual(val, self.ts[idx])
def test_sum(self):
self._check_stat_op('sum', np.sum)
def test_sum_inf(self):
s = Series(np.random.randn(10))
s2 = s.copy()
s[5:8] = np.inf
s2[5:8] = np.nan
assert_almost_equal(s.sum(), s2.sum())
import pandas.core.nanops as nanops
arr = np.random.randn(100, 100).astype('f4')
arr[:, 2] = np.inf
res = nanops.nansum(arr, axis=1)
expected = nanops._nansum(arr, axis=1)
assert_almost_equal(res, expected)
def test_mean(self):
self._check_stat_op('mean', np.mean)
def test_median(self):
self._check_stat_op('median', np.median)
# test with integers, test failure
int_ts = TimeSeries(np.ones(10, dtype=int), index=range(10))
self.assertAlmostEqual(np.median(int_ts), int_ts.median())
def test_prod(self):
self._check_stat_op('prod', np.prod)
def test_min(self):
self._check_stat_op('min', np.min, check_objects=True)
def test_max(self):
self._check_stat_op('max', np.max, check_objects=True)
def test_std(self):
alt = lambda x: np.std(x, ddof=1)
self._check_stat_op('std', alt)
def test_var(self):
alt = lambda x: np.var(x, ddof=1)
self._check_stat_op('var', alt)
def test_skew(self):
from scipy.stats import skew
alt =lambda x: skew(x, bias=False)
self._check_stat_op('skew', alt)
def test_argsort(self):
self._check_accum_op('argsort')
argsorted = self.ts.argsort()
self.assert_(issubclass(argsorted.dtype.type, np.integer))
def test_cumsum(self):
self._check_accum_op('cumsum')
def test_cumprod(self):
self._check_accum_op('cumprod')
def _check_stat_op(self, name, alternate, check_objects=False):
from pandas import DateRange
import pandas.core.nanops as nanops
def testit():
f = getattr(Series, name)
# add some NaNs
self.series[5:15] = np.NaN
# skipna or no
self.assert_(notnull(f(self.series)))
self.assert_(isnull(f(self.series, skipna=False)))
# check the result is correct
nona = self.series.dropna()
assert_almost_equal(f(nona), alternate(nona))
allna = self.series * nan
self.assert_(np.isnan(f(allna)))
# dtype=object with None, it works!
s = Series([1, 2, 3, None, 5])
f(s)
# check DateRange
if check_objects:
s = Series(DateRange('1/1/2000', periods=10))
res = f(s)
exp = alternate(s)
self.assertEqual(res, exp)
testit()
try:
import bottleneck as bn
nanops._USE_BOTTLENECK = False
testit()
nanops._USE_BOTTLENECK = True
except ImportError:
pass
def _check_accum_op(self, name):
func = getattr(np, name)
self.assert_(np.array_equal(func(self.ts), func(np.array(self.ts))))
# with missing values
ts = self.ts.copy()
ts[::2] = np.NaN
result = func(ts)[1::2]
expected = func(np.array(ts.valid()))
self.assert_(np.array_equal(result, expected))
def test_round(self):
# numpy.round doesn't preserve metadata, probably a numpy bug,
# re: GH #314
result = np.round(self.ts, 2)
expected = Series(np.round(self.ts.values, 2), index=self.ts.index)
assert_series_equal(result, expected)
self.assertEqual(result.name, self.ts.name)
def test_prod_numpy16_bug(self):
s = Series([1., 1., 1.] , index=range(3))
result = s.prod()
self.assert_(not isinstance(result, Series))
def test_quantile(self):
from scipy.stats import scoreatpercentile
q = self.ts.quantile(0.1)
self.assertEqual(q, scoreatpercentile(self.ts.valid(), 10))
q = self.ts.quantile(0.9)
self.assertEqual(q, scoreatpercentile(self.ts.valid(), 90))
def test_describe(self):
_ = self.series.describe()
_ = self.ts.describe()
def test_describe_objects(self):
s = Series(['a', 'b', 'b', np.nan, np.nan, np.nan, 'c', 'd', 'a', 'a'])
result = s.describe()
expected = Series({'count' : 7, 'unique' : 4,
'top' : 'a', 'freq' : 3}, index=result.index)
assert_series_equal(result, expected)
def test_append(self):
appendedSeries = self.series.append(self.ts)
for idx, value in appendedSeries.iteritems():
if idx in self.series.index:
self.assertEqual(value, self.series[idx])
elif idx in self.ts.index:
self.assertEqual(value, self.ts[idx])
else:
self.fail("orphaned index!")
self.assertRaises(Exception, self.ts.append, self.ts)
def test_append_many(self):
pieces = [self.ts[:5], self.ts[5:10], self.ts[10:]]
result = pieces[0].append(pieces[1:])
assert_series_equal(result, self.ts)
def test_all_any(self):
np.random.seed(12345)
ts = tm.makeTimeSeries()
bool_series = ts > 0
self.assert_(not bool_series.all())
self.assert_(bool_series.any())
def test_operators(self):
series = self.ts
other = self.ts[::2]
def _check_op(other, op, pos_only=False):
left = np.abs(series) if pos_only else series
right = np.abs(other) if pos_only else other
cython_or_numpy = op(left, right)
python = left.combine(right, op)
tm.assert_almost_equal(cython_or_numpy, python)
def check(other):
simple_ops = ['add', 'sub', 'mul', 'truediv', 'floordiv',
'gt', 'ge', 'lt', 'le']
for opname in simple_ops:
_check_op(other, getattr(operator, opname))
_check_op(other, operator.pow, pos_only=True)
_check_op(other, lambda x, y: operator.add(y, x))
_check_op(other, lambda x, y: operator.sub(y, x))
_check_op(other, lambda x, y: operator.truediv(y, x))
_check_op(other, lambda x, y: operator.floordiv(y, x))
_check_op(other, lambda x, y: operator.mul(y, x))
_check_op(other, lambda x, y: operator.pow(y, x),
pos_only=True)
check(self.ts * 2)
check(self.ts * 0)
check(self.ts[::2])
check(5)
def check_comparators(other):
_check_op(other, operator.gt)
_check_op(other, operator.ge)
_check_op(other, operator.eq)
_check_op(other, operator.lt)
_check_op(other, operator.le)
check_comparators(5)
check_comparators(self.ts + 1)
def test_operators_empty_int_corner(self):
s1 = Series([], [], dtype=np.int32)
s2 = Series({'x' : 0.})
# it works!
_ = s1 * s2
# NumPy limitiation =(
# def test_logical_range_select(self):
# np.random.seed(12345)
# selector = -0.5 <= self.ts <= 0.5
# expected = (self.ts >= -0.5) & (self.ts <= 0.5)
# assert_series_equal(selector, expected)
def test_idxmin(self):
# test idxmin
# _check_stat_op approach can not be used here because of isnull check.
# add some NaNs
self.series[5:15] = np.NaN
# skipna or no
self.assertEqual(self.series[self.series.idxmin()], self.series.min())
self.assert_(isnull(self.series.idxmin(skipna=False)))
# no NaNs
nona = self.series.dropna()
self.assertEqual(nona[nona.idxmin()], nona.min())
self.assertEqual(nona.index.values.tolist().index(nona.idxmin()),
nona.values.argmin())
# all NaNs
allna = self.series * nan
self.assert_(isnull(allna.idxmin()))
def test_idxmax(self):
# test idxmax
# _check_stat_op approach can not be used here because of isnull check.
# add some NaNs
self.series[5:15] = np.NaN
# skipna or no
self.assertEqual(self.series[self.series.idxmax()], self.series.max())
self.assert_(isnull(self.series.idxmax(skipna=False)))
# no NaNs
nona = self.series.dropna()
self.assertEqual(nona[nona.idxmax()], nona.max())
self.assertEqual(nona.index.values.tolist().index(nona.idxmax()),
nona.values.argmax())
# all NaNs
allna = self.series * nan
self.assert_(isnull(allna.idxmax()))
def test_operators_date(self):
result = self.objSeries + timedelta(1)
result = self.objSeries - timedelta(1)
def test_operators_corner(self):
series = self.ts
empty = Series([], index=Index([]))
result = series + empty
self.assert_(np.isnan(result).all())
result = empty + Series([], index=Index([]))
self.assert_(len(result) == 0)
# TODO: this returned NotImplemented earlier, what to do?
# deltas = Series([timedelta(1)] * 5, index=np.arange(5))
# sub_deltas = deltas[::2]
# deltas5 = deltas * 5
# deltas = deltas + sub_deltas
# float + int
int_ts = self.ts.astype(int)[:-5]
added = self.ts + int_ts
expected = self.ts.values[:-5] + int_ts.values
self.assert_(np.array_equal(added[:-5], expected))
def test_operators_reverse_object(self):
# GH 56
arr = Series(np.random.randn(10), index=np.arange(10),
dtype=object)
def _check_op(arr, op):
result = op(1., arr)
expected = op(1., arr.astype(float))
assert_series_equal(result.astype(float), expected)
_check_op(arr, operator.add)
_check_op(arr, operator.sub)
_check_op(arr, operator.mul)
_check_op(arr, operator.truediv)
_check_op(arr, operator.floordiv)
def test_series_frame_radd_bug(self):
from pandas.util.testing import rands
import operator
# GH 353
vals = Series([rands(5) for _ in xrange(10)])
result = 'foo_' + vals
expected = vals.map(lambda x: 'foo_' + x)
assert_series_equal(result, expected)
frame = DataFrame({'vals' : vals})
result = 'foo_' + frame
expected = DataFrame({'vals' : vals.map(lambda x: 'foo_' + x)})
tm.assert_frame_equal(result, expected)
# really raise this time
self.assertRaises(TypeError, operator.add, datetime.now(), self.ts)
def test_operators_frame(self):
# rpow does not work with DataFrame
df = DataFrame({'A' : self.ts})
tm.assert_almost_equal(self.ts + self.ts, (self.ts + df)['A'])
tm.assert_almost_equal(self.ts ** self.ts, (self.ts ** df)['A'])
def test_operators_combine(self):
def _check_fill(meth, op, a, b, fill_value=0):
exp_index = a.index.union(b.index)
a = a.reindex(exp_index)
b = b.reindex(exp_index)
amask = isnull(a)
bmask = isnull(b)
exp_values = []
for i in range(len(exp_index)):
if amask[i]:
if bmask[i]:
exp_values.append(nan)
continue
exp_values.append(op(fill_value, b[i]))
elif bmask[i]:
if amask[i]:
exp_values.append(nan)
continue
exp_values.append(op(a[i], fill_value))
else:
exp_values.append(op(a[i], b[i]))
result = meth(a, b, fill_value=fill_value)
expected = Series(exp_values, exp_index)
assert_series_equal(result, expected)
a = Series([nan, 1., 2., 3., nan], index=np.arange(5))
b = Series([nan, 1, nan, 3, nan, 4.], index=np.arange(6))
ops = [Series.add, Series.sub, Series.mul, Series.div]
equivs = [operator.add, operator.sub, operator.mul]
if py3compat.PY3:
equivs.append(operator.truediv)
else:
equivs.append(operator.div)
fillvals = [0, 0, 1, 1]
for op, equiv_op, fv in zip(ops, equivs, fillvals):
result = op(a, b)
exp = equiv_op(a, b)
assert_series_equal(result, exp)
_check_fill(op, equiv_op, a, b, fill_value=fv)
def test_combine_first(self):
values = tm.makeIntIndex(20).values.astype(float)
series = Series(values, index=tm.makeIntIndex(20))
series_copy = series * 2
series_copy[::2] = np.NaN
# nothing used from the input
combined = series.combine_first(series_copy)
self.assert_(np.array_equal(combined, series))
# Holes filled from input
combined = series_copy.combine_first(series)
self.assert_(np.isfinite(combined).all())
self.assert_(np.array_equal(combined[::2], series[::2]))
self.assert_(np.array_equal(combined[1::2], series_copy[1::2]))
# mixed types
index = tm.makeStringIndex(20)
floats = Series(tm.randn(20), index=index)
strings = Series(tm.makeStringIndex(10), index=index[::2])
combined = strings.combine_first(floats)
tm.assert_dict_equal(strings, combined, compare_keys=False)
tm.assert_dict_equal(floats[1::2], combined, compare_keys=False)
# corner case
s = | Series([1., 2, 3], index=[0, 1, 2]) | pandas.Series |
import pandas as pd
import yaml
import datetime
from workalendar.europe import Belgium
meta = | pd.read_csv('jouleboulevard_metadata.csv') | pandas.read_csv |
import pandas as pd
import numpy as np
from sklearn.impute import SimpleImputer
def mean():
df = pd.read_csv('../train_cuting/train_cutting2_lstm.csv')
df['Timestamp'] = pd.to_datetime(df['Timestamp'])
hour = pd.Timedelta('1h')
dt = df['Timestamp']
in_block = (dt.diff() == hour)
in_block[0] = True
temp_mean_imputer = SimpleImputer(missing_values=np.nan, strategy='mean')
temp_mean_imputer.fit(df[['Value']])
df['Value'] = temp_mean_imputer.transform(df[['Value']])
df.to_csv('../train_cuting/train_cutting2_lstm_mean.csv', index=False)
if __name__ == '__main__':
df = pd.read_csv('../train_cuting/train_cutting2_lstm_mean.csv')
df['Timestamp'] = pd.to_datetime(df['Timestamp'])
hour = | pd.Timedelta('1h') | pandas.Timedelta |
import pandas as pd
import functools
# TODO: figure out if array return hundredths or tenths of inches; apply appropriate functions
def format_df(file, col_name, cb):
df = pd.read_csv(file,
names=['station_id', 'month', 2,3,4,5,6,7,8,9,10,11,12,13,14,15,16,17,18,19,20,21,22,23,24,25,26,27,28,29,30,31,32],
delim_whitespace=True,
header=None,
)
df[col_name] = df[[2,3,4,5,6,7,8,9,10,11,12,13,14,15,16,17,18,19,20,21,22,23,24,25,26,27,28,29,30,31,32]].values.tolist()
df[col_name] = df[col_name].apply(cb)
df.drop([2,3,4,5,6,7,8,9,10,11,12,13,14,15,16,17,18,19,20,21,22,23,24,25,26,27,28,29,30,31,32], axis=1, inplace=True)
return df
def hundredths(str):
return int(str[:-1])/100
def tenths(str):
return int(str[:-1])/10
def to_int(str):
return int(str[:-1])
def map_to_hundredths(arr):
return list(map(hundredths, arr))
def map_to_tenths(arr):
return list(map(tenths, arr))
def map_to_int(arr):
return list(map(to_int, arr))
def identity(arr):
return arr
def daily_stations_df():
max_temps = format_df('../data/dly-tmax-normal.csv', 'max_temps', map_to_tenths)
# print(max_temps[100:110])
min_temps = format_df('../data/dly-tmin-normal.csv', 'min_temps', map_to_tenths)
gdd_40 = format_df('../data/dly-grdd-base40.csv', 'daily_gdd_40', map_to_int)
gdd_50 = format_df('../data/dly-grdd-base50.csv', 'daily_gdd_50', map_to_int)
mtd_precip = format_df('../data/mtd-prcp-normal.csv', 'mtd_precip', map_to_hundredths)
mtd_snow = format_df('../data/mtd-snow-normal.csv', 'mtd_snow', map_to_tenths)
ytd_precip = format_df('../data/ytd-prcp-normal.csv', 'ytd_precip', map_to_hundredths)
ytd_snow = format_df('../data/ytd-snow-normal.csv', 'ytd_snow', map_to_tenths)
dly_precip_50 = format_df('../data/dly-prcp-50pctl.csv', 'daily_precip_50', map_to_hundredths)
dly_precip_75 = format_df('../data/dly-prcp-75pctl.csv', 'daily_precip_75', map_to_hundredths)
merge_criteria = ['station_id', 'month']
temp_merge = | pd.merge(max_temps, min_temps, on=merge_criteria) | pandas.merge |
import sys
assert sys.version_info >= (3, 5) # make sure we have Python 3.5+
import pandas as pd
import numpy as np
from pathlib import Path
# init input df - fishing gear
def init_fishing_df(path):
fishing_df = pd.read_csv('../data/' + path)
# comment out for real life data--------------
fishing_df = fishing_df[fishing_df['is_fishing'] > -0.5]
fishing_df['is_fishing'] = [0 if x < 0.3 else 1 for x in fishing_df['is_fishing']]
fishing_df = fishing_df[['is_fishing', 'lat', 'lon', 'course', 'speed', 'timestamp', 'distance_from_shore', 'distance_from_port', 'mmsi', 'source']]
fishing_df['gear_type'] = Path(path).stem
#---------------------------------------------
return fishing_df
# ------------------------This section only needed when adding sst/precip data-----------------------------
# init input df - sea surface temparature
def init_sst_df(path_sst):
sst_df = pd.read_csv('../data/' + path_sst, index_col=0)
sst_df["time_bnds"] = pd.to_datetime(sst_df["time_bnds"]).dt.to_period('M')
return sst_df
# init input df - precipitation
def init_precip_df(path_precip):
precip_df = pd.read_csv('../data/' + path_precip, index_col=0)
precip_df["time"] = pd.to_datetime(precip_df["time"]).dt.to_period('M')
return precip_df
# ------------------------This section only needed when slicing lon/lat or time-----------------------------
# custom rounding functions
def custom_season(x):
return np.round(int(x)/3)
def custom_round(x):
return 0.5 + np.floor(float(x))
# ------------------------Functions to combine/add features and feature engineering-----------------------------
def time_feature(df):
df["adjust_time_date"] = pd.to_datetime(df['timestamp'], unit='s')
df["adjust_time"] = | pd.to_datetime(df["adjust_time_date"]) | pandas.to_datetime |
import pandas as pd
import pytest
import woodwork as ww
from woodwork.logical_types import Boolean, Double, Integer
from rayml.exceptions import MethodPropertyNotFoundError
from rayml.pipelines.components import (
ComponentBase,
FeatureSelector,
RFClassifierSelectFromModel,
RFRegressorSelectFromModel,
)
def make_rf_feature_selectors():
rf_classifier = RFClassifierSelectFromModel(
number_features=5,
n_estimators=10,
max_depth=7,
percent_features=0.5,
threshold=0,
)
rf_regressor = RFRegressorSelectFromModel(
number_features=5,
n_estimators=10,
max_depth=7,
percent_features=0.5,
threshold=0,
)
return rf_classifier, rf_regressor
def test_init():
rf_classifier, rf_regressor = make_rf_feature_selectors()
assert rf_classifier.name == "RF Classifier Select From Model"
assert rf_regressor.name == "RF Regressor Select From Model"
def test_component_fit(X_y_binary, X_y_multi, X_y_regression):
X_binary, y_binary = X_y_binary
X_multi, y_multi = X_y_multi
X_reg, y_reg = X_y_regression
rf_classifier, rf_regressor = make_rf_feature_selectors()
assert isinstance(rf_classifier.fit(X_binary, y_binary), ComponentBase)
assert isinstance(rf_classifier.fit(X_multi, y_multi), ComponentBase)
assert isinstance(rf_regressor.fit(X_reg, y_reg), ComponentBase)
def test_feature_selector_missing_component_obj():
class MockFeatureSelector(FeatureSelector):
name = "Mock Feature Selector"
def fit(self, X, y):
return self
mock_feature_selector = MockFeatureSelector()
mock_feature_selector.fit(pd.DataFrame(), pd.Series())
with pytest.raises(
MethodPropertyNotFoundError,
match="Feature selector requires a transform method or a component_obj that implements transform",
):
mock_feature_selector.transform( | pd.DataFrame() | pandas.DataFrame |
import pandas as pd
from sklearn.utils import resample
from sklearn.metrics import roc_auc_score, f1_score, balanced_accuracy_score, accuracy_score
from sklearn.metrics import precision_score, recall_score, confusion_matrix
import numpy as np
from scipy import stats
def bootstrap_data(dataset):
internal_val = | pd.read_csv('../../results/validation/internal/SSI_%s_y_vals.csv' % dataset) | pandas.read_csv |
from typing import Tuple
import numpy as np
import pandas as pd
import pytest
from pandas.testing import assert_frame_equal
from etna.datasets import generate_ar_df
from etna.datasets.tsdataset import TSDataset
from etna.transforms import DateFlagsTransform
from etna.transforms import TimeSeriesImputerTransform
@pytest.fixture()
def tsdf_with_exog(random_seed) -> TSDataset:
df_1 = pd.DataFrame.from_dict({"timestamp": pd.date_range("2021-02-01", "2021-07-01", freq="1d")})
df_2 = pd.DataFrame.from_dict({"timestamp": pd.date_range("2021-02-01", "2021-07-01", freq="1d")})
df_1["segment"] = "Moscow"
df_1["target"] = [x ** 2 + np.random.uniform(-2, 2) for x in list(range(len(df_1)))]
df_2["segment"] = "Omsk"
df_2["target"] = [x ** 0.5 + np.random.uniform(-2, 2) for x in list(range(len(df_2)))]
classic_df = pd.concat([df_1, df_2], ignore_index=True)
df = classic_df.pivot(index="timestamp", columns="segment")
df = df.reorder_levels([1, 0], axis=1)
df = df.sort_index(axis=1)
df.columns.names = ["segment", "feature"]
exog = generate_ar_df(start_time="2021-01-01", periods=600, n_segments=2)
exog = exog.pivot(index="timestamp", columns="segment")
exog = exog.reorder_levels([1, 0], axis=1)
exog = exog.sort_index(axis=1)
exog.columns.names = ["segment", "feature"]
exog.columns = pd.MultiIndex.from_arrays([["Moscow", "Omsk"], ["exog", "exog"]])
ts = TSDataset(df=df, df_exog=exog, freq="1D")
return ts
@pytest.fixture()
def df_and_regressors() -> Tuple[pd.DataFrame, pd.DataFrame]:
timestamp = pd.date_range("2021-01-01", "2021-02-01")
df_1 = pd.DataFrame({"timestamp": timestamp, "target": 11, "segment": "1"})
df_2 = pd.DataFrame({"timestamp": timestamp[5:], "target": 12, "segment": "2"})
df = pd.concat([df_1, df_2], ignore_index=True)
df = TSDataset.to_dataset(df)
timestamp = pd.date_range("2020-12-01", "2021-02-11")
df_1 = pd.DataFrame({"timestamp": timestamp, "regressor_1": 1, "regressor_2": 2, "segment": "1"})
df_2 = pd.DataFrame({"timestamp": timestamp[5:], "regressor_1": 3, "regressor_2": 4, "segment": "2"})
df_exog = pd.concat([df_1, df_2], ignore_index=True)
df_exog = TSDataset.to_dataset(df_exog)
return df, df_exog
@pytest.fixture()
def ts_future(example_reg_tsds):
future = example_reg_tsds.make_future(10)
return future
def test_check_endings_error_raise():
"""Check that _check_endings method raises exception if some segments end with nan."""
timestamp = pd.date_range("2021-01-01", "2021-02-01")
df1 = pd.DataFrame({"timestamp": timestamp, "target": 11, "segment": "1"})
df2 = pd.DataFrame({"timestamp": timestamp[:-5], "target": 12, "segment": "2"})
df = pd.concat([df1, df2], ignore_index=True)
df = TSDataset.to_dataset(df)
ts = TSDataset(df=df, freq="D")
with pytest.raises(ValueError):
ts._check_endings()
def test_check_endings_error_pass():
"""Check that _check_endings method passes if there is no nans at the end of all segments."""
timestamp = pd.date_range("2021-01-01", "2021-02-01")
df1 = pd.DataFrame({"timestamp": timestamp, "target": 11, "segment": "1"})
df2 = pd.DataFrame({"timestamp": timestamp, "target": 12, "segment": "2"})
df = | pd.concat([df1, df2], ignore_index=True) | pandas.concat |
import ipyleaflet
import ipywidgets
import pandas as pd
import geopandas as gpd
from shapely.geometry import Polygon, Point
import datetime
import requests
import xml.etree.ElementTree as ET
import calendar
import numpy as np
import pathlib
import os
import bqplot as bq
from functools import reduce
class ANA_interactive_map:
def __init__(self):
self.m01 = ipyleaflet.Map(zoom=4, center=(-16, -50), scroll_wheel_zoom=True,layout=ipywidgets.Layout(width='60%', height='500px'))
self.controls_on_Map()
self.out01 = ipywidgets.Output()
self.tabs = ipywidgets.Tab([self.tab00(), self.tab01(), self.tab02(),self.tab03(), self.tab04()], layout=ipywidgets.Layout(width='40%'))
self.tabs.set_title(0, 'Inventory ')
self.tabs.set_title(1, 'Tables')
self.tabs.set_title(2, 'Stats')
self.tabs.set_title(3, 'Download')
self.tabs.set_title(4, 'Graphs')
display(ipywidgets.VBox([ipywidgets.HBox([self.m01, self.tabs]),
self.out01]))
def controls_on_Map(self):
# pass
layer_control = ipyleaflet.LayersControl(position='topright')
self.m01.add_control(layer_control)
fullscreen_control = ipyleaflet.FullScreenControl()
self.m01.add_control(fullscreen_control)
self.draw_control = ipyleaflet.DrawControl()
self.m01.add_control(self.draw_control)
self.draw_control.observe(self._draw_testeObserve, 'last_draw')
self.draw_control.observe(self._output_stats, 'last_draw')
self.draw_control.observe(self._output_stats02, 'last_draw')
scale_control = ipyleaflet.ScaleControl(position='bottomleft')
self.m01.add_control(scale_control)
# Layer too slow to used
# marks = tuple([ipyleaflet.Marker(location=(lat, lon)) for lat, lon in self.df[['Latitude', 'Longitude']].to_numpy()])
# marker_cluster = ipyleaflet.MarkerCluster(markers=marks)
# self.m01.add_layer(marker_cluster)
def tab00(self):
with self.out01:
self.html_00_01 = ipywidgets.HTML(value="<h2>Inventory</h2><hr><p>In order to utilize the program, you need to insert a <b>Inventory File</b> or get it from the <b>ANA's API</b>.</p><p>After completed the upload of the Inventory, you can select which <b>Layers</b> to visualize by checking the <b>top-right widget</b> on the map.</p>")
self.radioButton_typeInvetario = ipywidgets.RadioButtons(options=['Select Path', 'Get from API'], value=None)
self.radioButton_typeInvetario.observe(self._radioButton_inventario, names='value')
self.text_pathInvetario = ipywidgets.Text(placeholder='Insert path of the Inventario')
self.button_pathInventario = ipywidgets.Button(description='Apply')
self.button_pathInventario.on_click(self._button_pathinventario)
self.button_showInventario = ipywidgets.Button(description='Show')
self.button_showInventario.on_click(self._button_showInventario)
self.floatProgress_loadingInventario = ipywidgets.FloatProgress(min=0, max=1, value=0, layout=ipywidgets.Layout(width='90%'))
self.floatProgress_loadingInventario.bar_style = 'info'
self.intSlider_01 = ipywidgets.IntSlider(description='Radius', min=1, max=50, value=15)
self.intSlider_01.observe(self._intSlider_radius, 'value')
widget_control01 = ipyleaflet.WidgetControl(widget=self.intSlider_01, position='bottomright')
self.m01.add_control(widget_control01)
self.selectionSlider_date01 = ipywidgets.SelectionSlider(options= | pd.date_range(start='2000-01-01',end='2020-01-01', freq='M') | pandas.date_range |
import numpy as np
import pytest
from anndata import AnnData
from pandas import DataFrame
from pandas.testing import assert_frame_equal
from ehrapy.api.anndata_ext import ObsEmptyError, anndata_to_df, df_to_anndata
class TestAnndataExt:
def test_df_to_anndata_simple(self):
df, col1_val, col2_val, col3_val = TestAnndataExt._setup_df_to_anndata()
expected_x = np.array([col1_val, col2_val, col3_val], dtype="object").transpose()
adata = df_to_anndata(df)
assert adata.X.dtype == "object"
assert adata.X.shape == (100, 3)
np.testing.assert_array_equal(adata.X, expected_x)
def test_df_to_anndata_index_column(self):
df, col1_val, col2_val, col3_val = TestAnndataExt._setup_df_to_anndata()
expected_x = np.array([col2_val, col3_val], dtype="object").transpose()
adata = df_to_anndata(df, index_column="col1")
assert adata.X.dtype == "object"
assert adata.X.shape == (100, 2)
np.testing.assert_array_equal(adata.X, expected_x)
assert list(adata.obs.index) == col1_val
def test_df_to_anndata_cols_obs_only(self):
df, col1_val, col2_val, col3_val = TestAnndataExt._setup_df_to_anndata()
adata = df_to_anndata(df, columns_obs_only=["col1", "col2"])
assert adata.X.dtype == "float32"
assert adata.X.shape == (100, 1)
assert_frame_equal(
adata.obs, DataFrame({"col1": col1_val, "col2": col2_val}, index=[str(idx) for idx in range(100)])
)
def test_df_to_anndata_all_num(self):
test_array = np.random.randint(0, 100, (4, 5))
df = DataFrame(test_array, columns=["col" + str(idx) for idx in range(5)])
adata = df_to_anndata(df)
assert adata.X.dtype == "float32"
np.testing.assert_array_equal(test_array, adata.X)
def test_anndata_to_df_simple(self):
col1_val, col2_val, col3_val = TestAnndataExt._setup_anndata_to_df()
expected_df = | DataFrame({"col1": col1_val, "col2": col2_val, "col3": col3_val}, dtype="object") | pandas.DataFrame |
from press_start.pipelines.data_split.nodes import category_encoder
import pandas as pd
import numpy as np
def test_category_encoder(df_categorical):
enc, df_numeric = category_encoder(
df_categorical,
{"_run": True},
{
"columns_categorical": ["buying", "maint"],
"column_target": "class",
},
)
df_exp = pd.DataFrame.from_dict(
{
"buying_high": {0: 0.0, 1: 0.0, 2: 0.0, 3: 1.0, 4: 0.0},
"buying_low": {0: 0.0, 1: 1.0, 2: 1.0, 3: 0.0, 4: 0.0},
"buying_med": {0: 1.0, 1: 0.0, 2: 0.0, 3: 0.0, 4: 1.0},
"maint_low": {0: 0.0, 1: 0.0, 2: 1.0, 3: 0.0, 4: 1.0},
"maint_med": {0: 1.0, 1: 1.0, 2: 0.0, 3: 1.0, 4: 0.0},
"year": {0: 2007, 1: 2010, 2: 2015, 3: 1989, 4: 2008},
"class": {0: "unacc", 1: "good", 2: "acc", 3: "unacc", 4: "unacc"},
}
)
enc_categories_exp = np.array(["high", "low", "med", "low", "med"])
| pd.testing.assert_frame_equal(df_exp, df_numeric, check_like=True) | pandas.testing.assert_frame_equal |
import librosa
import numpy as np
import pandas as pd
from os import listdir
from os.path import isfile, join
from audioread import NoBackendError
def extract_features(path, label, emotionId, startid):
"""
提取path目录下的音频文件的特征,使用librosa库
:param path: 文件路径
:param label: 情绪类型
:param startid: 开始的序列号
:return: 特征矩阵 pandas.DataFrame
"""
id = startid # 序列号
feature_set = pd.DataFrame() # 特征矩阵
# 单独的特征向量
labels = pd.Series()
emotion_vector = pd.Series()
songname_vector = pd.Series()
tempo_vector = pd.Series()
total_beats = pd.Series()
average_beats = pd.Series()
chroma_stft_mean = pd.Series()
# chroma_stft_std = pd.Series()
chroma_stft_var = pd.Series()
# chroma_cq_mean = pd.Series()
# chroma_cq_std = pd.Series()
# chroma_cq_var = pd.Series()
# chroma_cens_mean = pd.Series()
# chroma_cens_std = pd.Series()
# chroma_cens_var = pd.Series()
mel_mean = pd.Series()
# mel_std = pd.Series()
mel_var = pd.Series()
mfcc_mean = pd.Series()
# mfcc_std = pd.Series()
mfcc_var = pd.Series()
mfcc_delta_mean = pd.Series()
# mfcc_delta_std = pd.Series()
mfcc_delta_var = pd.Series()
rmse_mean = pd.Series()
# rmse_std = pd.Series()
rmse_var = pd.Series()
cent_mean = pd.Series()
# cent_std = pd.Series()
cent_var = pd.Series()
spec_bw_mean = pd.Series()
# spec_bw_std = pd.Series()
spec_bw_var = pd.Series()
contrast_mean = pd.Series()
# contrast_std = pd.Series()
contrast_var = pd.Series()
rolloff_mean = pd.Series()
# rolloff_std = pd.Series()
rolloff_var = pd.Series()
poly_mean = pd.Series()
# poly_std = pd.Series()
poly_var = pd.Series()
tonnetz_mean = pd.Series()
# tonnetz_std = pd.Series()
tonnetz_var = pd.Series()
zcr_mean = pd.Series()
# zcr_std = pd.Series()
zcr_var = pd.Series()
harm_mean = | pd.Series() | pandas.Series |
import operator
import re
import warnings
import numpy as np
import pytest
from pandas._libs.sparse import IntIndex
import pandas.util._test_decorators as td
import pandas as pd
from pandas import isna
from pandas.core.sparse.api import SparseArray, SparseDtype, SparseSeries
import pandas.util.testing as tm
from pandas.util.testing import assert_almost_equal
@pytest.fixture(params=["integer", "block"])
def kind(request):
return request.param
class TestSparseArray:
def setup_method(self, method):
self.arr_data = np.array([np.nan, np.nan, 1, 2, 3,
np.nan, 4, 5, np.nan, 6])
self.arr = SparseArray(self.arr_data)
self.zarr = SparseArray([0, 0, 1, 2, 3, 0, 4, 5, 0, 6], fill_value=0)
def test_constructor_dtype(self):
arr = SparseArray([np.nan, 1, 2, np.nan])
assert arr.dtype == SparseDtype(np.float64, np.nan)
assert arr.dtype.subtype == np.float64
assert np.isnan(arr.fill_value)
arr = SparseArray([np.nan, 1, 2, np.nan], fill_value=0)
assert arr.dtype == SparseDtype(np.float64, 0)
assert arr.fill_value == 0
arr = SparseArray([0, 1, 2, 4], dtype=np.float64)
assert arr.dtype == SparseDtype(np.float64, np.nan)
assert np.isnan(arr.fill_value)
arr = SparseArray([0, 1, 2, 4], dtype=np.int64)
assert arr.dtype == SparseDtype(np.int64, 0)
assert arr.fill_value == 0
arr = SparseArray([0, 1, 2, 4], fill_value=0, dtype=np.int64)
assert arr.dtype == SparseDtype(np.int64, 0)
assert arr.fill_value == 0
arr = SparseArray([0, 1, 2, 4], dtype=None)
assert arr.dtype == SparseDtype(np.int64, 0)
assert arr.fill_value == 0
arr = SparseArray([0, 1, 2, 4], fill_value=0, dtype=None)
assert arr.dtype == SparseDtype(np.int64, 0)
assert arr.fill_value == 0
def test_constructor_dtype_str(self):
result = SparseArray([1, 2, 3], dtype='int')
expected = SparseArray([1, 2, 3], dtype=int)
tm.assert_sp_array_equal(result, expected)
def test_constructor_sparse_dtype(self):
result = SparseArray([1, 0, 0, 1], dtype=SparseDtype('int64', -1))
expected = SparseArray([1, 0, 0, 1], fill_value=-1, dtype=np.int64)
tm.assert_sp_array_equal(result, expected)
assert result.sp_values.dtype == np.dtype('int64')
def test_constructor_sparse_dtype_str(self):
result = SparseArray([1, 0, 0, 1], dtype='Sparse[int32]')
expected = SparseArray([1, 0, 0, 1], dtype=np.int32)
tm.assert_sp_array_equal(result, expected)
assert result.sp_values.dtype == np.dtype('int32')
def test_constructor_object_dtype(self):
# GH 11856
arr = SparseArray(['A', 'A', np.nan, 'B'], dtype=np.object)
assert arr.dtype == SparseDtype(np.object)
assert np.isnan(arr.fill_value)
arr = SparseArray(['A', 'A', np.nan, 'B'], dtype=np.object,
fill_value='A')
assert arr.dtype == SparseDtype(np.object, 'A')
assert arr.fill_value == 'A'
# GH 17574
data = [False, 0, 100.0, 0.0]
arr = SparseArray(data, dtype=np.object, fill_value=False)
assert arr.dtype == SparseDtype(np.object, False)
assert arr.fill_value is False
arr_expected = np.array(data, dtype=np.object)
it = (type(x) == type(y) and x == y for x, y in zip(arr, arr_expected))
assert np.fromiter(it, dtype=np.bool).all()
@pytest.mark.parametrize("dtype", [SparseDtype(int, 0), int])
def test_constructor_na_dtype(self, dtype):
with pytest.raises(ValueError, match="Cannot convert"):
SparseArray([0, 1, np.nan], dtype=dtype)
def test_constructor_spindex_dtype(self):
arr = SparseArray(data=[1, 2], sparse_index=IntIndex(4, [1, 2]))
# XXX: Behavior change: specifying SparseIndex no longer changes the
# fill_value
expected = SparseArray([0, 1, 2, 0], kind='integer')
tm.assert_sp_array_equal(arr, expected)
assert arr.dtype == SparseDtype(np.int64)
assert arr.fill_value == 0
arr = SparseArray(data=[1, 2, 3],
sparse_index=IntIndex(4, [1, 2, 3]),
dtype=np.int64, fill_value=0)
exp = SparseArray([0, 1, 2, 3], dtype=np.int64, fill_value=0)
tm.assert_sp_array_equal(arr, exp)
assert arr.dtype == SparseDtype(np.int64)
assert arr.fill_value == 0
arr = SparseArray(data=[1, 2], sparse_index=IntIndex(4, [1, 2]),
fill_value=0, dtype=np.int64)
exp = SparseArray([0, 1, 2, 0], fill_value=0, dtype=np.int64)
tm.assert_sp_array_equal(arr, exp)
assert arr.dtype == SparseDtype(np.int64)
assert arr.fill_value == 0
arr = SparseArray(data=[1, 2, 3],
sparse_index=IntIndex(4, [1, 2, 3]),
dtype=None, fill_value=0)
exp = SparseArray([0, 1, 2, 3], dtype=None)
tm.assert_sp_array_equal(arr, exp)
assert arr.dtype == SparseDtype(np.int64)
assert arr.fill_value == 0
@pytest.mark.parametrize("sparse_index", [
None, IntIndex(1, [0]),
])
def test_constructor_spindex_dtype_scalar(self, sparse_index):
# scalar input
arr = SparseArray(data=1, sparse_index=sparse_index, dtype=None)
exp = SparseArray([1], dtype=None)
tm.assert_sp_array_equal(arr, exp)
assert arr.dtype == SparseDtype(np.int64)
assert arr.fill_value == 0
arr = SparseArray(data=1, sparse_index=IntIndex(1, [0]), dtype=None)
exp = SparseArray([1], dtype=None)
tm.assert_sp_array_equal(arr, exp)
assert arr.dtype == SparseDtype(np.int64)
assert arr.fill_value == 0
def test_constructor_spindex_dtype_scalar_broadcasts(self):
arr = SparseArray(data=[1, 2], sparse_index=IntIndex(4, [1, 2]),
fill_value=0, dtype=None)
exp = SparseArray([0, 1, 2, 0], fill_value=0, dtype=None)
tm.assert_sp_array_equal(arr, exp)
assert arr.dtype == SparseDtype(np.int64)
assert arr.fill_value == 0
@pytest.mark.parametrize('data, fill_value', [
(np.array([1, 2]), 0),
(np.array([1.0, 2.0]), np.nan),
([True, False], False),
([pd.Timestamp('2017-01-01')], pd.NaT),
])
def test_constructor_inferred_fill_value(self, data, fill_value):
result = SparseArray(data).fill_value
if pd.isna(fill_value):
assert pd.isna(result)
else:
assert result == fill_value
@pytest.mark.parametrize('format', ['coo', 'csc', 'csr'])
@pytest.mark.parametrize('size', [
pytest.param(0,
marks=td.skip_if_np_lt("1.16",
reason='NumPy-11383')),
10
])
@td.skip_if_no_scipy
def test_from_spmatrix(self, size, format):
import scipy.sparse
mat = scipy.sparse.random(size, 1, density=0.5, format=format)
result = SparseArray.from_spmatrix(mat)
result = np.asarray(result)
expected = mat.toarray().ravel()
tm.assert_numpy_array_equal(result, expected)
@td.skip_if_no_scipy
def test_from_spmatrix_raises(self):
import scipy.sparse
mat = scipy.sparse.eye(5, 4, format='csc')
with pytest.raises(ValueError, match="not '4'"):
SparseArray.from_spmatrix(mat)
@pytest.mark.parametrize('scalar,dtype', [
(False, SparseDtype(bool, False)),
(0.0, SparseDtype('float64', 0)),
(1, SparseDtype('int64', 1)),
('z', SparseDtype('object', 'z'))])
def test_scalar_with_index_infer_dtype(self, scalar, dtype):
# GH 19163
arr = SparseArray(scalar, index=[1, 2, 3], fill_value=scalar)
exp = SparseArray([scalar, scalar, scalar], fill_value=scalar)
tm.assert_sp_array_equal(arr, exp)
assert arr.dtype == dtype
assert exp.dtype == dtype
@pytest.mark.parametrize("fill", [1, np.nan, 0])
@pytest.mark.filterwarnings("ignore:Sparse:FutureWarning")
def test_sparse_series_round_trip(self, kind, fill):
# see gh-13999
arr = SparseArray([np.nan, 1, np.nan, 2, 3],
kind=kind, fill_value=fill)
res = SparseArray(SparseSeries(arr))
tm.assert_sp_array_equal(arr, res)
arr = SparseArray([0, 0, 0, 1, 1, 2], dtype=np.int64,
kind=kind, fill_value=fill)
res = SparseArray(SparseSeries(arr), dtype=np.int64)
tm.assert_sp_array_equal(arr, res)
res = SparseArray(SparseSeries(arr))
tm.assert_sp_array_equal(arr, res)
@pytest.mark.parametrize("fill", [True, False, np.nan])
@pytest.mark.filterwarnings("ignore:Sparse:FutureWarning")
def test_sparse_series_round_trip2(self, kind, fill):
# see gh-13999
arr = SparseArray([True, False, True, True], dtype=np.bool,
kind=kind, fill_value=fill)
res = SparseArray(SparseSeries(arr))
tm.assert_sp_array_equal(arr, res)
res = SparseArray(SparseSeries(arr))
tm.assert_sp_array_equal(arr, res)
def test_get_item(self):
assert np.isnan(self.arr[1])
assert self.arr[2] == 1
assert self.arr[7] == 5
assert self.zarr[0] == 0
assert self.zarr[2] == 1
assert self.zarr[7] == 5
errmsg = re.compile("bounds")
with pytest.raises(IndexError, match=errmsg):
self.arr[11]
with pytest.raises(IndexError, match=errmsg):
self.arr[-11]
assert self.arr[-1] == self.arr[len(self.arr) - 1]
def test_take_scalar_raises(self):
msg = "'indices' must be an array, not a scalar '2'."
with pytest.raises(ValueError, match=msg):
self.arr.take(2)
def test_take(self):
exp = SparseArray(np.take(self.arr_data, [2, 3]))
tm.assert_sp_array_equal(self.arr.take([2, 3]), exp)
exp = SparseArray(np.take(self.arr_data, [0, 1, 2]))
tm.assert_sp_array_equal(self.arr.take([0, 1, 2]), exp)
def test_take_fill_value(self):
data = np.array([1, np.nan, 0, 3, 0])
sparse = SparseArray(data, fill_value=0)
exp = SparseArray(np.take(data, [0]), fill_value=0)
tm.assert_sp_array_equal(sparse.take([0]), exp)
exp = SparseArray(np.take(data, [1, 3, 4]), fill_value=0)
tm.assert_sp_array_equal(sparse.take([1, 3, 4]), exp)
def test_take_negative(self):
exp = SparseArray(np.take(self.arr_data, [-1]))
tm.assert_sp_array_equal(self.arr.take([-1]), exp)
exp = SparseArray(np.take(self.arr_data, [-4, -3, -2]))
tm.assert_sp_array_equal(self.arr.take([-4, -3, -2]), exp)
@pytest.mark.parametrize('fill_value', [0, None, np.nan])
def test_shift_fill_value(self, fill_value):
# GH #24128
sparse = SparseArray(np.array([1, 0, 0, 3, 0]),
fill_value=8.0)
res = sparse.shift(1, fill_value=fill_value)
if isna(fill_value):
fill_value = res.dtype.na_value
exp = SparseArray(np.array([fill_value, 1, 0, 0, 3]),
fill_value=8.0)
tm.assert_sp_array_equal(res, exp)
def test_bad_take(self):
with pytest.raises(IndexError, match="bounds"):
self.arr.take([11])
def test_take_filling(self):
# similar tests as GH 12631
sparse = SparseArray([np.nan, np.nan, 1, np.nan, 4])
result = sparse.take(np.array([1, 0, -1]))
expected = SparseArray([np.nan, np.nan, 4])
tm.assert_sp_array_equal(result, expected)
# XXX: test change: fill_value=True -> allow_fill=True
result = sparse.take(np.array([1, 0, -1]), allow_fill=True)
expected = SparseArray([np.nan, np.nan, np.nan])
tm.assert_sp_array_equal(result, expected)
# allow_fill=False
result = sparse.take(np.array([1, 0, -1]),
allow_fill=False, fill_value=True)
expected = SparseArray([np.nan, np.nan, 4])
tm.assert_sp_array_equal(result, expected)
msg = "Invalid value in 'indices'"
with pytest.raises(ValueError, match=msg):
sparse.take(np.array([1, 0, -2]), allow_fill=True)
with pytest.raises(ValueError, match=msg):
sparse.take(np.array([1, 0, -5]), allow_fill=True)
with pytest.raises(IndexError):
sparse.take(np.array([1, -6]))
with pytest.raises(IndexError):
sparse.take(np.array([1, 5]))
with pytest.raises(IndexError):
sparse.take(np.array([1, 5]), allow_fill=True)
def test_take_filling_fill_value(self):
# same tests as GH 12631
sparse = SparseArray([np.nan, 0, 1, 0, 4], fill_value=0)
result = sparse.take(np.array([1, 0, -1]))
expected = SparseArray([0, np.nan, 4], fill_value=0)
tm.assert_sp_array_equal(result, expected)
# fill_value
result = sparse.take(np.array([1, 0, -1]), allow_fill=True)
# XXX: behavior change.
# the old way of filling self.fill_value doesn't follow EA rules.
# It's supposed to be self.dtype.na_value (nan in this case)
expected = SparseArray([0, np.nan, np.nan], fill_value=0)
tm.assert_sp_array_equal(result, expected)
# allow_fill=False
result = sparse.take(np.array([1, 0, -1]),
allow_fill=False, fill_value=True)
expected = SparseArray([0, np.nan, 4], fill_value=0)
tm.assert_sp_array_equal(result, expected)
msg = ("Invalid value in 'indices'.")
with pytest.raises(ValueError, match=msg):
sparse.take(np.array([1, 0, -2]), allow_fill=True)
with pytest.raises(ValueError, match=msg):
sparse.take(np.array([1, 0, -5]), allow_fill=True)
with pytest.raises(IndexError):
sparse.take(np.array([1, -6]))
with pytest.raises(IndexError):
sparse.take(np.array([1, 5]))
with pytest.raises(IndexError):
sparse.take(np.array([1, 5]), fill_value=True)
def test_take_filling_all_nan(self):
sparse = SparseArray([np.nan, np.nan, np.nan, np.nan, np.nan])
# XXX: did the default kind from take change?
result = sparse.take(np.array([1, 0, -1]))
expected = SparseArray([np.nan, np.nan, np.nan], kind='block')
tm.assert_sp_array_equal(result, expected)
result = sparse.take(np.array([1, 0, -1]), fill_value=True)
expected = SparseArray([np.nan, np.nan, np.nan], kind='block')
tm.assert_sp_array_equal(result, expected)
with pytest.raises(IndexError):
sparse.take(np.array([1, -6]))
with pytest.raises(IndexError):
sparse.take(np.array([1, 5]))
with pytest.raises(IndexError):
sparse.take(np.array([1, 5]), fill_value=True)
def test_set_item(self):
def setitem():
self.arr[5] = 3
def setslice():
self.arr[1:5] = 2
with pytest.raises(TypeError, match="assignment via setitem"):
setitem()
with pytest.raises(TypeError, match="assignment via setitem"):
setslice()
def test_constructor_from_too_large_array(self):
with pytest.raises(TypeError, match="expected dimension <= 1 data"):
SparseArray(np.arange(10).reshape((2, 5)))
def test_constructor_from_sparse(self):
res = SparseArray(self.zarr)
assert res.fill_value == 0
assert_almost_equal(res.sp_values, self.zarr.sp_values)
def test_constructor_copy(self):
cp = SparseArray(self.arr, copy=True)
cp.sp_values[:3] = 0
assert not (self.arr.sp_values[:3] == 0).any()
not_copy = SparseArray(self.arr)
not_copy.sp_values[:3] = 0
assert (self.arr.sp_values[:3] == 0).all()
def test_constructor_bool(self):
# GH 10648
data = np.array([False, False, True, True, False, False])
arr = SparseArray(data, fill_value=False, dtype=bool)
assert arr.dtype == SparseDtype(bool)
tm.assert_numpy_array_equal(arr.sp_values, np.array([True, True]))
# Behavior change: np.asarray densifies.
# tm.assert_numpy_array_equal(arr.sp_values, np.asarray(arr))
tm.assert_numpy_array_equal(arr.sp_index.indices,
np.array([2, 3], np.int32))
dense = arr.to_dense()
assert dense.dtype == bool
tm.assert_numpy_array_equal(dense, data)
def test_constructor_bool_fill_value(self):
arr = SparseArray([True, False, True], dtype=None)
assert arr.dtype == SparseDtype(np.bool)
assert not arr.fill_value
arr = SparseArray([True, False, True], dtype=np.bool)
assert arr.dtype == SparseDtype(np.bool)
assert not arr.fill_value
arr = SparseArray([True, False, True], dtype=np.bool, fill_value=True)
assert arr.dtype == SparseDtype(np.bool, True)
assert arr.fill_value
def test_constructor_float32(self):
# GH 10648
data = np.array([1., np.nan, 3], dtype=np.float32)
arr = SparseArray(data, dtype=np.float32)
assert arr.dtype == SparseDtype(np.float32)
tm.assert_numpy_array_equal(arr.sp_values,
np.array([1, 3], dtype=np.float32))
# Behavior change: np.asarray densifies.
# tm.assert_numpy_array_equal(arr.sp_values, np.asarray(arr))
tm.assert_numpy_array_equal(arr.sp_index.indices,
np.array([0, 2], dtype=np.int32))
dense = arr.to_dense()
assert dense.dtype == np.float32
tm.assert_numpy_array_equal(dense, data)
def test_astype(self):
# float -> float
arr = SparseArray([None, None, 0, 2])
result = arr.astype("Sparse[float32]")
expected = SparseArray([None, None, 0, 2], dtype=np.dtype('float32'))
tm.assert_sp_array_equal(result, expected)
dtype = SparseDtype("float64", fill_value=0)
result = arr.astype(dtype)
expected = SparseArray._simple_new(np.array([0., 2.],
dtype=dtype.subtype),
IntIndex(4, [2, 3]),
dtype)
tm.assert_sp_array_equal(result, expected)
dtype = SparseDtype("int64", 0)
result = arr.astype(dtype)
expected = SparseArray._simple_new(np.array([0, 2], dtype=np.int64),
IntIndex(4, [2, 3]),
dtype)
tm.assert_sp_array_equal(result, expected)
arr = SparseArray([0, np.nan, 0, 1], fill_value=0)
with pytest.raises(ValueError, match='NA'):
arr.astype('Sparse[i8]')
def test_astype_bool(self):
a = pd.SparseArray([1, 0, 0, 1], dtype=SparseDtype(int, 0))
result = a.astype(bool)
expected = SparseArray([True, 0, 0, True],
dtype=SparseDtype(bool, 0))
tm.assert_sp_array_equal(result, expected)
# update fill value
result = a.astype(SparseDtype(bool, False))
expected = SparseArray([True, False, False, True],
dtype=SparseDtype(bool, False))
tm.assert_sp_array_equal(result, expected)
def test_astype_all(self, any_real_dtype):
vals = np.array([1, 2, 3])
arr = SparseArray(vals, fill_value=1)
typ = np.dtype(any_real_dtype)
res = arr.astype(typ)
assert res.dtype == SparseDtype(typ, 1)
assert res.sp_values.dtype == typ
tm.assert_numpy_array_equal(np.asarray(res.to_dense()),
vals.astype(typ))
@pytest.mark.parametrize('array, dtype, expected', [
(SparseArray([0, 1]), 'float',
SparseArray([0., 1.], dtype=SparseDtype(float, 0.0))),
(SparseArray([0, 1]), bool, SparseArray([False, True])),
(SparseArray([0, 1], fill_value=1), bool,
SparseArray([False, True], dtype=SparseDtype(bool, True))),
pytest.param(
SparseArray([0, 1]), 'datetime64[ns]',
SparseArray(np.array([0, 1], dtype='datetime64[ns]'),
dtype=SparseDtype('datetime64[ns]',
pd.Timestamp('1970'))),
marks=[pytest.mark.xfail(reason="NumPy-7619")],
),
(SparseArray([0, 1, 10]), str,
SparseArray(['0', '1', '10'], dtype=SparseDtype(str, '0'))),
(SparseArray(['10', '20']), float, SparseArray([10.0, 20.0])),
(SparseArray([0, 1, 0]), object,
SparseArray([0, 1, 0], dtype=SparseDtype(object, 0))),
])
def test_astype_more(self, array, dtype, expected):
result = array.astype(dtype)
tm.assert_sp_array_equal(result, expected)
def test_astype_nan_raises(self):
arr = SparseArray([1.0, np.nan])
with pytest.raises(ValueError, match='Cannot convert non-finite'):
arr.astype(int)
def test_set_fill_value(self):
arr = SparseArray([1., np.nan, 2.], fill_value=np.nan)
arr.fill_value = 2
assert arr.fill_value == 2
arr = SparseArray([1, 0, 2], fill_value=0, dtype=np.int64)
arr.fill_value = 2
assert arr.fill_value == 2
# XXX: this seems fine? You can construct an integer
# sparsearray with NaN fill value, why not update one?
# coerces to int
# msg = "unable to set fill_value 3\\.1 to int64 dtype"
# with pytest.raises(ValueError, match=msg):
arr.fill_value = 3.1
assert arr.fill_value == 3.1
# msg = "unable to set fill_value nan to int64 dtype"
# with pytest.raises(ValueError, match=msg):
arr.fill_value = np.nan
assert np.isnan(arr.fill_value)
arr = SparseArray([True, False, True], fill_value=False, dtype=np.bool)
arr.fill_value = True
assert arr.fill_value
# coerces to bool
# msg = "unable to set fill_value 0 to bool dtype"
# with pytest.raises(ValueError, match=msg):
arr.fill_value = 0
assert arr.fill_value == 0
# msg = "unable to set fill_value nan to bool dtype"
# with pytest.raises(ValueError, match=msg):
arr.fill_value = np.nan
assert np.isnan(arr.fill_value)
@pytest.mark.parametrize("val", [[1, 2, 3], np.array([1, 2]), (1, 2, 3)])
def test_set_fill_invalid_non_scalar(self, val):
arr = SparseArray([True, False, True], fill_value=False, dtype=np.bool)
msg = "fill_value must be a scalar"
with pytest.raises(ValueError, match=msg):
arr.fill_value = val
def test_copy(self):
arr2 = self.arr.copy()
assert arr2.sp_values is not self.arr.sp_values
assert arr2.sp_index is self.arr.sp_index
def test_values_asarray(self):
assert_almost_equal(self.arr.to_dense(), self.arr_data)
@pytest.mark.parametrize('data,shape,dtype', [
([0, 0, 0, 0, 0], (5,), None),
([], (0,), None),
([0], (1,), None),
(['A', 'A', np.nan, 'B'], (4,), np.object)
])
def test_shape(self, data, shape, dtype):
# GH 21126
out = SparseArray(data, dtype=dtype)
assert out.shape == shape
@pytest.mark.parametrize("vals", [
[np.nan, np.nan, np.nan, np.nan, np.nan],
[1, np.nan, np.nan, 3, np.nan],
[1, np.nan, 0, 3, 0],
])
@pytest.mark.parametrize("fill_value", [None, 0])
def test_dense_repr(self, vals, fill_value):
vals = np.array(vals)
arr = SparseArray(vals, fill_value=fill_value)
res = arr.to_dense()
tm.assert_numpy_array_equal(res, vals)
with tm.assert_produces_warning(FutureWarning):
res2 = arr.get_values()
tm.assert_numpy_array_equal(res2, vals)
def test_getitem(self):
def _checkit(i):
assert_almost_equal(self.arr[i], self.arr.to_dense()[i])
for i in range(len(self.arr)):
_checkit(i)
_checkit(-i)
def test_getitem_arraylike_mask(self):
arr = SparseArray([0, 1, 2])
result = arr[[True, False, True]]
expected = SparseArray([0, 2])
tm.assert_sp_array_equal(result, expected)
def test_getslice(self):
result = self.arr[:-3]
exp = SparseArray(self.arr.to_dense()[:-3])
tm.assert_sp_array_equal(result, exp)
result = self.arr[-4:]
exp = SparseArray(self.arr.to_dense()[-4:])
tm.assert_sp_array_equal(result, exp)
# two corner cases from Series
result = self.arr[-12:]
exp = SparseArray(self.arr)
tm.assert_sp_array_equal(result, exp)
result = self.arr[:-12]
exp = SparseArray(self.arr.to_dense()[:0])
tm.assert_sp_array_equal(result, exp)
def test_getslice_tuple(self):
dense = np.array([np.nan, 0, 3, 4, 0, 5, np.nan, np.nan, 0])
sparse = SparseArray(dense)
res = sparse[4:, ]
exp = SparseArray(dense[4:, ])
tm.assert_sp_array_equal(res, exp)
sparse = SparseArray(dense, fill_value=0)
res = sparse[4:, ]
exp = SparseArray(dense[4:, ], fill_value=0)
tm.assert_sp_array_equal(res, exp)
with pytest.raises(IndexError):
sparse[4:, :]
with pytest.raises(IndexError):
# check numpy compat
dense[4:, :]
def test_boolean_slice_empty(self):
arr = pd.SparseArray([0, 1, 2])
res = arr[[False, False, False]]
assert res.dtype == arr.dtype
@pytest.mark.parametrize("op", ["add", "sub", "mul",
"truediv", "floordiv", "pow"])
def test_binary_operators(self, op):
op = getattr(operator, op)
data1 = np.random.randn(20)
data2 = np.random.randn(20)
data1[::2] = np.nan
data2[::3] = np.nan
arr1 = SparseArray(data1)
arr2 = SparseArray(data2)
data1[::2] = 3
data2[::3] = 3
farr1 = SparseArray(data1, fill_value=3)
farr2 = SparseArray(data2, fill_value=3)
def _check_op(op, first, second):
res = op(first, second)
exp = SparseArray(op(first.to_dense(), second.to_dense()),
fill_value=first.fill_value)
assert isinstance(res, SparseArray)
assert_almost_equal(res.to_dense(), exp.to_dense())
res2 = op(first, second.to_dense())
assert isinstance(res2, SparseArray)
tm.assert_sp_array_equal(res, res2)
res3 = op(first.to_dense(), second)
assert isinstance(res3, SparseArray)
tm.assert_sp_array_equal(res, res3)
res4 = op(first, 4)
assert isinstance(res4, SparseArray)
# Ignore this if the actual op raises (e.g. pow).
try:
exp = op(first.to_dense(), 4)
exp_fv = op(first.fill_value, 4)
except ValueError:
pass
else:
assert_almost_equal(res4.fill_value, exp_fv)
assert_almost_equal(res4.to_dense(), exp)
with np.errstate(all="ignore"):
for first_arr, second_arr in [(arr1, arr2), (farr1, farr2)]:
_check_op(op, first_arr, second_arr)
def test_pickle(self):
def _check_roundtrip(obj):
unpickled = tm.round_trip_pickle(obj)
tm.assert_sp_array_equal(unpickled, obj)
_check_roundtrip(self.arr)
_check_roundtrip(self.zarr)
def test_generator_warnings(self):
sp_arr = SparseArray([1, 2, 3])
with warnings.catch_warnings(record=True) as w:
warnings.filterwarnings(action='always',
category=DeprecationWarning)
warnings.filterwarnings(action='always',
category=PendingDeprecationWarning)
for _ in sp_arr:
pass
assert len(w) == 0
def test_fillna(self):
s = SparseArray([1, np.nan, np.nan, 3, np.nan])
res = s.fillna(-1)
exp = SparseArray([1, -1, -1, 3, -1], fill_value=-1, dtype=np.float64)
tm.assert_sp_array_equal(res, exp)
s = SparseArray([1, np.nan, np.nan, 3, np.nan], fill_value=0)
res = s.fillna(-1)
exp = SparseArray([1, -1, -1, 3, -1], fill_value=0, dtype=np.float64)
tm.assert_sp_array_equal(res, exp)
s = SparseArray([1, np.nan, 0, 3, 0])
res = s.fillna(-1)
exp = SparseArray([1, -1, 0, 3, 0], fill_value=-1, dtype=np.float64)
tm.assert_sp_array_equal(res, exp)
s = SparseArray([1, np.nan, 0, 3, 0], fill_value=0)
res = s.fillna(-1)
exp = SparseArray([1, -1, 0, 3, 0], fill_value=0, dtype=np.float64)
tm.assert_sp_array_equal(res, exp)
s = SparseArray([np.nan, np.nan, np.nan, np.nan])
res = s.fillna(-1)
exp = SparseArray([-1, -1, -1, -1], fill_value=-1, dtype=np.float64)
tm.assert_sp_array_equal(res, exp)
s = SparseArray([np.nan, np.nan, np.nan, np.nan], fill_value=0)
res = s.fillna(-1)
exp = SparseArray([-1, -1, -1, -1], fill_value=0, dtype=np.float64)
tm.assert_sp_array_equal(res, exp)
# float dtype's fill_value is np.nan, replaced by -1
s = SparseArray([0., 0., 0., 0.])
res = s.fillna(-1)
exp = SparseArray([0., 0., 0., 0.], fill_value=-1)
tm.assert_sp_array_equal(res, exp)
# int dtype shouldn't have missing. No changes.
s = SparseArray([0, 0, 0, 0])
assert s.dtype == SparseDtype(np.int64)
assert s.fill_value == 0
res = s.fillna(-1)
tm.assert_sp_array_equal(res, s)
s = SparseArray([0, 0, 0, 0], fill_value=0)
assert s.dtype == SparseDtype(np.int64)
assert s.fill_value == 0
res = s.fillna(-1)
exp = SparseArray([0, 0, 0, 0], fill_value=0)
tm.assert_sp_array_equal(res, exp)
# fill_value can be nan if there is no missing hole.
# only fill_value will be changed
s = SparseArray([0, 0, 0, 0], fill_value=np.nan)
assert s.dtype == SparseDtype(np.int64, fill_value=np.nan)
assert np.isnan(s.fill_value)
res = s.fillna(-1)
exp = SparseArray([0, 0, 0, 0], fill_value=-1)
tm.assert_sp_array_equal(res, exp)
def test_fillna_overlap(self):
s = SparseArray([1, np.nan, np.nan, 3, np.nan])
# filling with existing value doesn't replace existing value with
# fill_value, i.e. existing 3 remains in sp_values
res = s.fillna(3)
exp = np.array([1, 3, 3, 3, 3], dtype=np.float64)
tm.assert_numpy_array_equal(res.to_dense(), exp)
s = SparseArray([1, np.nan, np.nan, 3, np.nan], fill_value=0)
res = s.fillna(3)
exp = SparseArray([1, 3, 3, 3, 3], fill_value=0, dtype=np.float64)
tm.assert_sp_array_equal(res, exp)
def test_nonzero(self):
# Tests regression #21172.
sa = pd.SparseArray([
float('nan'),
float('nan'),
1, 0, 0,
2, 0, 0, 0,
3, 0, 0
])
expected = np.array([2, 5, 9], dtype=np.int32)
result, = sa.nonzero()
tm.assert_numpy_array_equal(expected, result)
sa = pd.SparseArray([0, 0, 1, 0, 0, 2, 0, 0, 0, 3, 0, 0])
result, = sa.nonzero()
tm.assert_numpy_array_equal(expected, result)
class TestSparseArrayAnalytics:
@pytest.mark.parametrize('data,pos,neg', [
([True, True, True], True, False),
([1, 2, 1], 1, 0),
([1.0, 2.0, 1.0], 1.0, 0.0)
])
def test_all(self, data, pos, neg):
# GH 17570
out = SparseArray(data).all()
assert out
out = SparseArray(data, fill_value=pos).all()
assert out
data[1] = neg
out = SparseArray(data).all()
assert not out
out = SparseArray(data, fill_value=pos).all()
assert not out
@pytest.mark.parametrize('data,pos,neg', [
([True, True, True], True, False),
([1, 2, 1], 1, 0),
([1.0, 2.0, 1.0], 1.0, 0.0)
])
@td.skip_if_np_lt("1.15") # prior didn't dispatch
def test_numpy_all(self, data, pos, neg):
# GH 17570
out = np.all(SparseArray(data))
assert out
out = np.all(SparseArray(data, fill_value=pos))
assert out
data[1] = neg
out = np.all(SparseArray(data))
assert not out
out = np.all(SparseArray(data, fill_value=pos))
assert not out
# raises with a different message on py2.
msg = "the \'out\' parameter is not supported"
with pytest.raises(ValueError, match=msg):
np.all(SparseArray(data), out=np.array([]))
@pytest.mark.parametrize('data,pos,neg', [
([False, True, False], True, False),
([0, 2, 0], 2, 0),
([0.0, 2.0, 0.0], 2.0, 0.0)
])
def test_any(self, data, pos, neg):
# GH 17570
out = SparseArray(data).any()
assert out
out = SparseArray(data, fill_value=pos).any()
assert out
data[1] = neg
out = SparseArray(data).any()
assert not out
out = SparseArray(data, fill_value=pos).any()
assert not out
@pytest.mark.parametrize('data,pos,neg', [
([False, True, False], True, False),
([0, 2, 0], 2, 0),
([0.0, 2.0, 0.0], 2.0, 0.0)
])
@td.skip_if_np_lt("1.15") # prior didn't dispatch
def test_numpy_any(self, data, pos, neg):
# GH 17570
out = np.any(SparseArray(data))
assert out
out = np.any(SparseArray(data, fill_value=pos))
assert out
data[1] = neg
out = np.any(SparseArray(data))
assert not out
out = np.any(SparseArray(data, fill_value=pos))
assert not out
msg = "the \'out\' parameter is not supported"
with pytest.raises(ValueError, match=msg):
np.any(SparseArray(data), out=out)
def test_sum(self):
data = np.arange(10).astype(float)
out = SparseArray(data).sum()
assert out == 45.0
data[5] = np.nan
out = SparseArray(data, fill_value=2).sum()
assert out == 40.0
out = SparseArray(data, fill_value=np.nan).sum()
assert out == 40.0
def test_numpy_sum(self):
data = np.arange(10).astype(float)
out = np.sum(SparseArray(data))
assert out == 45.0
data[5] = np.nan
out = np.sum(SparseArray(data, fill_value=2))
assert out == 40.0
out = np.sum(SparseArray(data, fill_value=np.nan))
assert out == 40.0
msg = "the 'dtype' parameter is not supported"
with pytest.raises(ValueError, match=msg):
np.sum(SparseArray(data), dtype=np.int64)
msg = "the 'out' parameter is not supported"
with pytest.raises(ValueError, match=msg):
np.sum(SparseArray(data), out=out)
@pytest.mark.parametrize("data,expected", [
(np.array([1, 2, 3, 4, 5], dtype=float), # non-null data
SparseArray(np.array([1.0, 3.0, 6.0, 10.0, 15.0]))),
(np.array([1, 2, np.nan, 4, 5], dtype=float), # null data
SparseArray(np.array([1.0, 3.0, np.nan, 7.0, 12.0])))
])
@pytest.mark.parametrize("numpy", [True, False])
def test_cumsum(self, data, expected, numpy):
cumsum = np.cumsum if numpy else lambda s: s.cumsum()
out = cumsum(SparseArray(data))
tm.assert_sp_array_equal(out, expected)
out = cumsum(SparseArray(data, fill_value=np.nan))
tm.assert_sp_array_equal(out, expected)
out = cumsum(SparseArray(data, fill_value=2))
tm.assert_sp_array_equal(out, expected)
if numpy: # numpy compatibility checks.
msg = "the 'dtype' parameter is not supported"
with pytest.raises(ValueError, match=msg):
np.cumsum(SparseArray(data), dtype=np.int64)
msg = "the 'out' parameter is not supported"
with pytest.raises(ValueError, match=msg):
np.cumsum(SparseArray(data), out=out)
else:
axis = 1 # SparseArray currently 1-D, so only axis = 0 is valid.
msg = "axis\\(={axis}\\) out of bounds".format(axis=axis)
with pytest.raises(ValueError, match=msg):
SparseArray(data).cumsum(axis=axis)
def test_mean(self):
data = np.arange(10).astype(float)
out = SparseArray(data).mean()
assert out == 4.5
data[5] = np.nan
out = SparseArray(data).mean()
assert out == 40.0 / 9
def test_numpy_mean(self):
data = np.arange(10).astype(float)
out = np.mean(SparseArray(data))
assert out == 4.5
data[5] = np.nan
out = np.mean(SparseArray(data))
assert out == 40.0 / 9
msg = "the 'dtype' parameter is not supported"
with pytest.raises(ValueError, match=msg):
np.mean(SparseArray(data), dtype=np.int64)
msg = "the 'out' parameter is not supported"
with pytest.raises(ValueError, match=msg):
np.mean(SparseArray(data), out=out)
def test_ufunc(self):
# GH 13853 make sure ufunc is applied to fill_value
sparse = SparseArray([1, np.nan, 2, np.nan, -2])
result = SparseArray([1, np.nan, 2, np.nan, 2])
tm.assert_sp_array_equal(abs(sparse), result)
tm.assert_sp_array_equal(np.abs(sparse), result)
sparse = SparseArray([1, -1, 2, -2], fill_value=1)
result = SparseArray([1, 2, 2], sparse_index=sparse.sp_index,
fill_value=1)
tm.assert_sp_array_equal(abs(sparse), result)
tm.assert_sp_array_equal(np.abs(sparse), result)
sparse = SparseArray([1, -1, 2, -2], fill_value=-1)
result = SparseArray([1, 2, 2], sparse_index=sparse.sp_index,
fill_value=1)
tm.assert_sp_array_equal(abs(sparse), result)
tm.assert_sp_array_equal(np.abs(sparse), result)
sparse = SparseArray([1, np.nan, 2, np.nan, -2])
result = SparseArray(np.sin([1, np.nan, 2, np.nan, -2]))
tm.assert_sp_array_equal(np.sin(sparse), result)
sparse = SparseArray([1, -1, 2, -2], fill_value=1)
result = SparseArray(np.sin([1, -1, 2, -2]), fill_value=np.sin(1))
tm.assert_sp_array_equal(np.sin(sparse), result)
sparse = SparseArray([1, -1, 0, -2], fill_value=0)
result = SparseArray(np.sin([1, -1, 0, -2]), fill_value=np.sin(0))
tm.assert_sp_array_equal(np.sin(sparse), result)
def test_ufunc_args(self):
# GH 13853 make sure ufunc is applied to fill_value, including its arg
sparse = SparseArray([1, np.nan, 2, np.nan, -2])
result = SparseArray([2, np.nan, 3, np.nan, -1])
tm.assert_sp_array_equal(np.add(sparse, 1), result)
sparse = SparseArray([1, -1, 2, -2], fill_value=1)
result = SparseArray([2, 0, 3, -1], fill_value=2)
tm.assert_sp_array_equal(np.add(sparse, 1), result)
sparse = SparseArray([1, -1, 0, -2], fill_value=0)
result = SparseArray([2, 0, 1, -1], fill_value=1)
tm.assert_sp_array_equal(np.add(sparse, 1), result)
@pytest.mark.parametrize('fill_value', [0.0, np.nan])
def test_modf(self, fill_value):
# https://github.com/pandas-dev/pandas/issues/26946
sparse = pd.SparseArray([fill_value] * 10 + [1.1, 2.2],
fill_value=fill_value)
r1, r2 = np.modf(sparse)
e1, e2 = np.modf(np.asarray(sparse))
tm.assert_sp_array_equal(r1, pd.SparseArray(e1, fill_value=fill_value))
tm.assert_sp_array_equal(r2, pd.SparseArray(e2, fill_value=fill_value))
def test_nbytes_integer(self):
arr = SparseArray([1, 0, 0, 0, 2], kind='integer')
result = arr.nbytes
# (2 * 8) + 2 * 4
assert result == 24
def test_nbytes_block(self):
arr = SparseArray([1, 2, 0, 0, 0], kind='block')
result = arr.nbytes
# (2 * 8) + 4 + 4
# sp_values, blocs, blenghts
assert result == 24
def test_asarray_datetime64(self):
s = pd.SparseArray(
pd.to_datetime(['2012', None, None, '2013'])
)
np.asarray(s)
def test_density(self):
arr = SparseArray([0, 1])
assert arr.density == 0.5
def test_npoints(self):
arr = SparseArray([0, 1])
assert arr.npoints == 1
@pytest.mark.filterwarnings("ignore:Sparse:FutureWarning")
class TestAccessor:
@pytest.mark.parametrize('attr', [
'npoints', 'density', 'fill_value', 'sp_values',
])
def test_get_attributes(self, attr):
arr = SparseArray([0, 1])
ser = pd.Series(arr)
result = getattr(ser.sparse, attr)
expected = getattr(arr, attr)
assert result == expected
@td.skip_if_no_scipy
def test_from_coo(self):
import scipy.sparse
row = [0, 3, 1, 0]
col = [0, 3, 1, 2]
data = [4, 5, 7, 9]
sp_array = scipy.sparse.coo_matrix((data, (row, col)))
result = pd.Series.sparse.from_coo(sp_array)
index = pd.MultiIndex.from_arrays([[0, 0, 1, 3], [0, 2, 1, 3]])
expected = pd.Series([4, 9, 7, 5], index=index, dtype='Sparse[int]')
tm.assert_series_equal(result, expected)
@td.skip_if_no_scipy
def test_to_coo(self):
import scipy.sparse
ser = pd.Series([1, 2, 3],
index=pd.MultiIndex.from_product([[0], [1, 2, 3]],
names=['a', 'b']),
dtype='Sparse[int]')
A, _, _ = ser.sparse.to_coo()
assert isinstance(A, scipy.sparse.coo.coo_matrix)
def test_non_sparse_raises(self):
ser = pd.Series([1, 2, 3])
with pytest.raises(AttributeError, match='.sparse'):
ser.sparse.density
def test_setting_fill_value_fillna_still_works():
# This is why letting users update fill_value / dtype is bad
# astype has the same problem.
arr = SparseArray([1., np.nan, 1.0], fill_value=0.0)
arr.fill_value = np.nan
result = arr.isna()
# Can't do direct comparison, since the sp_index will be different
# So let's convert to ndarray and check there.
result = np.asarray(result)
expected = np.array([False, True, False])
tm.assert_numpy_array_equal(result, expected)
def test_setting_fill_value_updates():
arr = SparseArray([0.0, np.nan], fill_value=0)
arr.fill_value = np.nan
# use private constructor to get the index right
# otherwise both nans would be un-stored.
expected = SparseArray._simple_new(
sparse_array=np.array([np.nan]),
sparse_index=IntIndex(2, [1]),
dtype=SparseDtype(float, np.nan),
)
tm.assert_sp_array_equal(arr, expected)
@pytest.mark.parametrize("arr, loc", [
([None, 1, 2], 0),
([0, None, 2], 1),
([0, 1, None], 2),
([0, 1, 1, None, None], 3),
([1, 1, 1, 2], -1),
([], -1),
])
def test_first_fill_value_loc(arr, loc):
result = SparseArray(arr)._first_fill_value_loc()
assert result == loc
@pytest.mark.parametrize('arr', [
[1, 2, np.nan, np.nan],
[1, np.nan, 2, np.nan],
[1, 2, np.nan],
])
@pytest.mark.parametrize("fill_value", [
np.nan, 0, 1
])
def test_unique_na_fill(arr, fill_value):
a = pd.SparseArray(arr, fill_value=fill_value).unique()
b = pd.Series(arr).unique()
assert isinstance(a, SparseArray)
a = np.asarray(a)
tm.assert_numpy_array_equal(a, b)
def test_unique_all_sparse():
# https://github.com/pandas-dev/pandas/issues/23168
arr = SparseArray([0, 0])
result = arr.unique()
expected = SparseArray([0])
tm.assert_sp_array_equal(result, expected)
def test_map():
arr = SparseArray([0, 1, 2])
expected = SparseArray([10, 11, 12], fill_value=10)
# dict
result = arr.map({0: 10, 1: 11, 2: 12})
tm.assert_sp_array_equal(result, expected)
# series
result = arr.map(pd.Series({0: 10, 1: 11, 2: 12}))
tm.assert_sp_array_equal(result, expected)
# function
result = arr.map(pd.Series({0: 10, 1: 11, 2: 12}))
expected = SparseArray([10, 11, 12], fill_value=10)
tm.assert_sp_array_equal(result, expected)
def test_map_missing():
arr = SparseArray([0, 1, 2])
expected = SparseArray([10, 11, None], fill_value=10)
result = arr.map({0: 10, 1: 11})
| tm.assert_sp_array_equal(result, expected) | pandas.util.testing.assert_sp_array_equal |
import sklearn.neighbors._base
import sys
sys.modules['sklearn.neighbors.base'] = sklearn.neighbors._base
import pandas as pd
from sklearn.base import TransformerMixin
import numpy as np
from sklearn.impute import SimpleImputer, KNNImputer
from missingpy import MissForest
class prepross(TransformerMixin):
def __init__(self, **kwargs):
super().__init__(**kwargs)
def fit(self, X, y=None):
self.fill = pd.Series([X[c].value_counts().index[0]
if X[c].dtype == np.dtype('O') else X[c].mean() for c in X], index=X.columns)
return self
def transform(self, X, y=None):
return X.fillna(self.fill)
def rm_rows_cols(df, row_thresh=0.8, col_thresh=0.8):
if "Index" in df.columns:
df.drop("Index", axis=1, inplace=True)
df.columns = [col.strip() for col in df.columns]
df = df.drop_duplicates()
df = df.dropna(axis=0,how='all').dropna(axis=1,how='all').dropna(axis=1,how='all').dropna(axis=0,thresh= int(len(df.columns)*0.8)).dropna(axis=1,thresh=int(len(df)*0.8))
df = df.infer_objects()
return df
def replace_special_character(df,usr_char=None,do=None, ignore_col=None):
spec_chars = ["!", '"', "#", "%", "&", "'", "(", ")",
"*", "+", ",", "-", ".", "/", ":", ";", "<",
"=", ">", "?", "@", "[", "\\", "]", "^", "_",
"`", "{", "|", "}", "~", "–", "//", "%*", ":/", ".;", "Ø", "§",'$',"£"]
if do== 'remove':
for chactr in usr_char:
spec_chars.remove(chactr)
elif do=='add':
for chactr in usr_char:
spec_chars.append(chactr)
if len(ignore_col)>0:
df_to_concat = df[ignore_col]
df = df[list(set(df.columns)-set(ignore_col))]
else:
df_to_concat = | pd.DataFrame() | pandas.DataFrame |
import warnings
from datetime import datetime
from functools import partial
import numpy as np
import pandas as pd
import pandas.api.types as pdtypes
from featuretools import variable_types
from featuretools.entityset.relationship import RelationshipPath
from featuretools.exceptions import UnknownFeature
from featuretools.feature_base import (
AggregationFeature,
DirectFeature,
GroupByTransformFeature,
IdentityFeature,
TransformFeature
)
from featuretools.utils import Trie, is_python_2
from featuretools.utils.gen_utils import get_relationship_variable_id
warnings.simplefilter('ignore', np.RankWarning)
warnings.simplefilter("ignore", category=RuntimeWarning)
class FeatureSetCalculator(object):
"""
Calculates the values of a set of features for given instance ids.
"""
def __init__(self, entityset, feature_set, time_last=None,
training_window=None, precalculated_features=None, ignored=None):
"""
Args:
feature_set (FeatureSet): The features to calculate values for.
time_last (pd.Timestamp, optional): Last allowed time. Data from exactly this
time not allowed.
training_window (Timedelta, optional): Window defining how much time before the cutoff time data
can be used when calculating features. If None, all data before cutoff time is used.
precalculated_features (Trie[RelationshipPath -> pd.DataFrame]):
Maps RelationshipPaths to dataframes of precalculated_features
ignored (set[str], optional): Unique names of precalculated features.
"""
self.entityset = entityset
self.feature_set = feature_set
self.training_window = training_window
self.ignored = ignored
if time_last is None:
time_last = datetime.now()
self.time_last = time_last
if precalculated_features is None:
precalculated_features = Trie(path_constructor=RelationshipPath)
self.precalculated_features = precalculated_features
def run(self, instance_ids):
"""
Calculate values of features for the given instances of the target
entity.
Summary of algorithm:
1. Construct a trie where the edges are relationships and each node
contains a set of features for a single entity. See
FeatureSet._build_feature_trie.
2. Initialize a trie for storing dataframes.
3. Traverse the trie using depth first search. At each node calculate
the features and store the resulting dataframe in the dataframe
trie (so that its values can be used by features which depend on
these features). See _calculate_features_for_entity.
4. Get the dataframe at the root of the trie (for the target entity) and
return the columns corresponding to the requested features.
Args:
instance_ids (list): List of instance id for which to build features.
Returns:
pd.DataFrame : Pandas DataFrame of calculated feature values.
Indexed by instance_ids. Columns in same order as features
passed in.
"""
assert len(instance_ids) > 0, "0 instance ids provided"
feature_trie = self.feature_set.feature_trie
df_trie = Trie(path_constructor=RelationshipPath)
full_entity_df_trie = Trie(path_constructor=RelationshipPath)
target_entity = self.entityset[self.feature_set.target_eid]
self._calculate_features_for_entity(entity_id=self.feature_set.target_eid,
feature_trie=feature_trie,
df_trie=df_trie,
full_entity_df_trie=full_entity_df_trie,
precalculated_trie=self.precalculated_features,
filter_variable=target_entity.index,
filter_values=instance_ids)
# The dataframe for the target entity should be stored at the root of
# df_trie.
df = df_trie.value
if df.empty:
return self.generate_default_df(instance_ids=instance_ids)
# fill in empty rows with default values
missing_ids = [i for i in instance_ids if i not in
df[target_entity.index]]
if missing_ids:
default_df = self.generate_default_df(instance_ids=missing_ids,
extra_columns=df.columns)
df = df.append(default_df, sort=True)
df.index.name = self.entityset[self.feature_set.target_eid].index
column_list = []
for feat in self.feature_set.target_features:
column_list.extend(feat.get_feature_names())
return df[column_list]
def _calculate_features_for_entity(self, entity_id, feature_trie, df_trie,
full_entity_df_trie,
precalculated_trie,
filter_variable, filter_values,
parent_data=None):
"""
Generate dataframes with features calculated for this node of the trie,
and all descendant nodes. The dataframes will be stored in df_trie.
Args:
entity_id (str): The name of the entity to calculate features for.
feature_trie (Trie): the trie with sets of features to calculate.
The root contains features for the given entity.
df_trie (Trie): a parallel trie for storing dataframes. The
dataframe with features calculated will be placed in the root.
full_entity_df_trie (Trie): a trie storing dataframes will all entity
rows, for features that are uses_full_entity.
precalculated_trie (Trie): a parallel trie containing dataframes
with precalculated features. The dataframe for this entity will
be at the root.
filter_variable (str): The name of the variable to filter this
dataframe by.
filter_values (pd.Series): The values to filter the filter_variable
to.
parent_data (tuple[Relationship, list[str], pd.DataFrame]): Data
related to the parent of this trie. This will only be present if
the relationship points from this entity to the parent entity. A
3 tuple of (parent_relationship,
ancestor_relationship_variables, parent_df).
ancestor_relationship_variables is the names of variables which
link the parent entity to its ancestors.
"""
# Step 1: Get a dataframe for the given entity, filtered by the given
# conditions.
need_full_entity, full_entity_features, not_full_entity_features = feature_trie.value
if self.ignored:
full_entity_features -= self.ignored
not_full_entity_features -= self.ignored
all_features = full_entity_features | not_full_entity_features
entity = self.entityset[entity_id]
columns = self._necessary_columns(entity, all_features)
# If we need the full entity then don't filter by filter_values.
if need_full_entity:
query_variable = None
query_values = None
else:
query_variable = filter_variable
query_values = filter_values
df = entity.query_by_values(query_values,
variable_id=query_variable,
columns=columns,
time_last=self.time_last,
training_window=self.training_window)
# Step 2: Add variables to the dataframe linking it to all ancestors.
new_ancestor_relationship_variables = []
if parent_data:
parent_relationship, ancestor_relationship_variables, parent_df = \
parent_data
if ancestor_relationship_variables:
df, new_ancestor_relationship_variables = self._add_ancestor_relationship_variables(
df, parent_df, ancestor_relationship_variables, parent_relationship)
# Add the variable linking this entity to its parent, so that
# descendants get linked to the parent.
new_ancestor_relationship_variables.append(parent_relationship.child_variable.id)
# Step 3: Recurse on children.
# Pass filtered values, even if we are using a full df.
if need_full_entity:
filtered_df = df[df[filter_variable].isin(filter_values)]
else:
filtered_df = df
for edge, sub_trie in feature_trie.children():
is_forward, relationship = edge
if is_forward:
sub_entity = relationship.parent_entity.id
sub_filter_variable = relationship.parent_variable.id
sub_filter_values = filtered_df[relationship.child_variable.id]
parent_data = None
else:
sub_entity = relationship.child_entity.id
sub_filter_variable = relationship.child_variable.id
sub_filter_values = filtered_df[relationship.parent_variable.id]
parent_data = (relationship,
new_ancestor_relationship_variables,
df)
sub_df_trie = df_trie.get_node([edge])
sub_full_entity_df_trie = full_entity_df_trie.get_node([edge])
sub_precalc_trie = precalculated_trie.get_node([edge])
self._calculate_features_for_entity(
entity_id=sub_entity,
feature_trie=sub_trie,
df_trie=sub_df_trie,
full_entity_df_trie=sub_full_entity_df_trie,
precalculated_trie=sub_precalc_trie,
filter_variable=sub_filter_variable,
filter_values=sub_filter_values,
parent_data=parent_data)
# Step 4: Calculate the features for this entity.
#
# All dependencies of the features for this entity have been calculated
# by the above recursive calls, and their results stored in df_trie.
# Add any precalculated features.
precalculated_features_df = precalculated_trie.value
if precalculated_features_df is not None:
# Left outer merge to keep all rows of df.
df = df.merge(precalculated_features_df,
how='left',
left_index=True,
right_index=True,
suffixes=('', '_precalculated'))
# First, calculate any features that require the full entity. These can
# be calculated first because all of their dependents are included in
# full_entity_features.
if need_full_entity:
df = self._calculate_features(df, full_entity_df_trie, full_entity_features)
# Store full entity df.
full_entity_df_trie.value = df
# Filter df so that features that don't require the full entity are
# only calculated on the necessary instances.
df = df[df[filter_variable].isin(filter_values)]
# Calculate all features that don't require the full entity.
df = self._calculate_features(df, df_trie, not_full_entity_features)
# Step 5: Store the dataframe for this entity at the root of df_trie, so
# that it can be accessed by the caller.
df_trie.value = df
def _calculate_features(self, df, df_trie, features):
# Group the features so that each group can be calculated together.
# The groups must also be in topological order (if A is a transform of B
# then B must be in a group before A).
feature_groups = self.feature_set.group_features(features)
for group in feature_groups:
representative_feature = group[0]
handler = self._feature_type_handler(representative_feature)
df = handler(group, df, df_trie)
return df
def _add_ancestor_relationship_variables(self, child_df, parent_df,
ancestor_relationship_variables,
relationship):
"""
Merge ancestor_relationship_variables from parent_df into child_df, adding a prefix to
each column name specifying the relationship.
Return the updated df and the new relationship variable names.
Args:
child_df (pd.DataFrame): The dataframe to add relationship variables to.
parent_df (pd.DataFrame): The dataframe to copy relationship variables from.
ancestor_relationship_variables (list[str]): The names of
relationship variables in the parent_df to copy into child_df.
relationship (Relationship): the relationship through which the
child is connected to the parent.
"""
relationship_name = relationship.parent_name
new_relationship_variables = ['%s.%s' % (relationship_name, var)
for var in ancestor_relationship_variables]
# create an intermediate dataframe which shares a column
# with the child dataframe and has a column with the
# original parent's id.
col_map = {relationship.parent_variable.id: relationship.child_variable.id}
for child_var, parent_var in zip(new_relationship_variables, ancestor_relationship_variables):
col_map[parent_var] = child_var
merge_df = parent_df[list(col_map.keys())].rename(columns=col_map)
merge_df.index.name = None # change index name for merge
# Merge the dataframe, adding the relationship variables to the child.
# Left outer join so that all rows in child are kept (if it contains
# all rows of the entity then there may not be corresponding rows in the
# parent_df).
df = child_df.merge(merge_df,
how='left',
left_on=relationship.child_variable.id,
right_on=relationship.child_variable.id)
return df, new_relationship_variables
def generate_default_df(self, instance_ids, extra_columns=None):
default_row = []
default_cols = []
for f in self.feature_set.target_features:
for name in f.get_feature_names():
default_cols.append(name)
default_row.append(f.default_value)
default_matrix = [default_row] * len(instance_ids)
default_df = pd.DataFrame(default_matrix,
columns=default_cols,
index=instance_ids)
index_name = self.entityset[self.feature_set.target_eid].index
default_df.index.name = index_name
if extra_columns is not None:
for c in extra_columns:
if c not in default_df.columns:
default_df[c] = [np.nan] * len(instance_ids)
return default_df
def _feature_type_handler(self, f):
if type(f) == TransformFeature:
return self._calculate_transform_features
elif type(f) == GroupByTransformFeature:
return self._calculate_groupby_features
elif type(f) == DirectFeature:
return self._calculate_direct_features
elif type(f) == AggregationFeature:
return self._calculate_agg_features
elif type(f) == IdentityFeature:
return self._calculate_identity_features
else:
raise UnknownFeature(u"{} feature unknown".format(f.__class__))
def _calculate_identity_features(self, features, df, _df_trie):
for f in features:
assert f.get_name() in df, (
'Column "%s" missing frome dataframe' % f.get_name())
return df
def _calculate_transform_features(self, features, frame, _df_trie):
for f in features:
# handle when no data
if frame.shape[0] == 0:
set_default_column(frame, f)
continue
# collect only the variables we need for this transformation
variable_data = [frame[bf.get_name()]
for bf in f.base_features]
feature_func = f.get_function()
# apply the function to the relevant dataframe slice and add the
# feature row to the results dataframe.
if f.primitive.uses_calc_time:
values = feature_func(*variable_data, time=self.time_last)
else:
values = feature_func(*variable_data)
# if we don't get just the values, the assignment breaks when indexes don't match
if f.number_output_features > 1:
values = [strip_values_if_series(value) for value in values]
else:
values = [strip_values_if_series(values)]
update_feature_columns(f, frame, values)
return frame
def _calculate_groupby_features(self, features, frame, _df_trie):
for f in features:
set_default_column(frame, f)
# handle when no data
if frame.shape[0] == 0:
return frame
groupby = features[0].groupby.get_name()
grouped = frame.groupby(groupby)
groups = frame[groupby].unique() # get all the unique group name to iterate over later
for f in features:
feature_vals = []
for group in groups:
# skip null key if it exists
if pd.isnull(group):
continue
column_names = [bf.get_name() for bf in f.base_features]
# exclude the groupby variable from being passed to the function
variable_data = [grouped[name].get_group(group) for name in column_names[:-1]]
feature_func = f.get_function()
# apply the function to the relevant dataframe slice and add the
# feature row to the results dataframe.
if f.primitive.uses_calc_time:
values = feature_func(*variable_data, time=self.time_last)
else:
values = feature_func(*variable_data)
# make sure index is aligned
if isinstance(values, pd.Series):
values.index = variable_data[0].index
else:
values = | pd.Series(values, index=variable_data[0].index) | pandas.Series |
import numpy as np
import os
import pandas as pd
import sqlite3
from datetime import date
from dotenv import load_dotenv
from email.mime.text import MIMEText
from email.mime.multipart import MIMEMultipart
from pretty_html_table import build_table
from smtplib import SMTP
load_dotenv()
SQLITE_DB_PATH = os.getenv('SQLITE_DB_PATH')
EMAIL_ACCOUNT = os.getenv('EMAIL_ACCOUNT')
EMAIL_PASSWORD = os.getenv('EMAIL_PASSWORD')
RECIPIENT_EMAIL = os.getenv('RECIPIENT_EMAIL')
DIR_PATH = os.path.dirname(os.path.realpath(__file__))
SQLITE_DB_FULL_PATH = f'{DIR_PATH}/{SQLITE_DB_PATH}'
def get_listing_data(today_filter=True):
conn = sqlite3.connect(SQLITE_DB_FULL_PATH)
cur = conn.cursor()
cur.execute("PRAGMA table_info(listing_full_details)")
table_info = cur.fetchall()
date_filter_str = ''
if today_filter:
today = date.today().strftime('%Y/%m/%d')
date_filter_str = f"AND DATE='{today}' "
select_batch_query = f"SELECT * FROM listing_full_details WHERE STATUS='Active' {date_filter_str}AND NUMBER_ROOMS>=3 AND NUMBER_BATHROOMS>=1.5"
cur.execute(select_batch_query)
query_results = cur.fetchall()
if not query_results:
return None
column_names = [info[1] for info in table_info]
df = | pd.DataFrame(query_results, columns=column_names) | pandas.DataFrame |
from django.db import models
# Create your models here.
class Stock(models.Model):
stock_id = models.CharField(max_length=1000)
stock_value = models.CharField(max_length=100)
# checkbox
# enter_your_portfolio = models.BooleanField()
def get_stock_id():
return stock_id
def get_stock_value():
return stock_value
# def get_enter_your_portfolio():
# return enter_your_portfolio
def stock_volatility(stock_name):
from yahoofinancials import YahooFinancials
from datetime import date, timedelta
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
fig, ax = plt.subplots(1, 1, figsize=(7, 5))
# stock_id = str(Stock.stock_id)
# stock_id.replace(" ", "")
stock_id = stock_name.replace(" ", "")
list_of_stocks = stock_id.split(",")
print(list_of_stocks)
for stock_symbol in list_of_stocks:
# set date range for historical prices
end_time = date.today()
start_time = end_time - timedelta(days=365)
# format date range
end = end_time.strftime('%Y-%m-%d')
start = start_time.strftime('%Y-%m-%d')
json_prices = YahooFinancials(
stock_symbol).get_historical_price_data(start, end, 'daily')
# print(json_prices)
# json -> dataframe
prices = | pd.DataFrame(json_prices[stock_symbol]['prices']) | pandas.DataFrame |
#!/usr/bin/env python3
# -*- coding: utf-8 -*-
"""
Created on Thu Nov 21 14:08:43 2019
to produce X and y use combine_pos_neg_from_nc_file or
prepare_X_y_for_holdout_test
@author: ziskin
"""
from PW_paths import savefig_path
from PW_paths import work_yuval
from pathlib import Path
cwd = Path().cwd()
hydro_path = work_yuval / 'hydro'
axis_path = work_yuval/'axis'
gis_path = work_yuval / 'gis'
ims_path = work_yuval / 'IMS_T'
hydro_ml_path = hydro_path / 'hydro_ML'
gnss_path = work_yuval / 'GNSS_stations'
# 'tela': 17135
hydro_pw_dict = {'nizn': 25191, 'klhv': 21105, 'yrcm': 55165,
'ramo': 56140, 'drag': 48125, 'dsea': 48192,
'spir': 56150, 'nrif': 60105, 'elat': 60190
}
hydro_st_name_dict = {25191: 'Lavan - new nizana road',
21105: 'Shikma - Tel milcha',
55165: 'Mamsheet',
56140: 'Ramon',
48125: 'Draga',
48192: 'Chiemar - down the cliff',
46150: 'Nekrot - Top',
60105: 'Yaelon - Kibutz Yahel',
60190: 'Solomon - Eilat'}
best_hp_models_dict = {'SVC': {'kernel': 'rbf', 'C': 1.0, 'gamma': 0.02,
'coef0': 0.0, 'degree': 1},
'RF': {'max_depth': 5, 'max_features': 'auto',
'min_samples_leaf': 1, 'min_samples_split': 2,
'n_estimators': 400},
'MLP': {'alpha': 0.1, 'activation': 'relu',
'hidden_layer_sizes': (10,10,10), 'learning_rate': 'constant',
'solver': 'lbfgs'}}
scorer_order = ['precision', 'recall', 'f1', 'accuracy', 'tss', 'hss']
tsafit_dict = {'lat': 30.985556, 'lon': 35.263056,
'alt': -35.75, 'dt_utc': '2018-04-26T10:15:00'}
axis_southern_stations = ['Dimo', 'Ohad', 'Ddse', 'Yotv', 'Elat', 'Raha', 'Yaha']
soi_axis_dict = {'yrcm': 'Dimo',
'slom': 'Ohad',
'dsea': 'Ddse',
'nrif': 'Yotv',
'elat': 'Elat',
'klhv': 'Raha',
'spir': 'Yaha'}
def plot_mean_abs_shap_values_features(SV, fix_xticklabels=True):
import matplotlib.pyplot as plt
import numpy as np
import seaborn as sns
from natsort import natsorted
features = ['pwv', 'pressure', 'DOY']
# sns.set_palette('Dark2', 6)
sns.set_theme(style='ticks', font_scale=1.5)
# sns.set_style('whitegrid')
# sns.set_style('ticks')
sv = np.abs(SV).mean('sample').sel(clas=0).reset_coords(drop=True)
gr_spec = [20, 20, 1]
fig, axes = plt.subplots(1, 3, sharey=True, figsize=(17, 5), gridspec_kw={'width_ratios': gr_spec})
try:
axes.flatten()
except AttributeError:
axes = [axes]
for i, f in enumerate(features):
fe = [x for x in sv['feature'].values if f in x]
dsf = sv.sel(feature=fe).reset_coords(drop=True).to_dataframe()
title = '{}'.format(f.upper())
dsf.plot.bar(ax=axes[i], title=title, rot=0, legend=False, zorder=20,
width=.8, color='k', alpha=0.8)
axes[i].set_title(title)
dsf_sum = dsf.sum().tolist()
handles, labels = axes[i].get_legend_handles_labels()
labels = [
'{} ({:.1f} %)'.format(
x, y) for x, y in zip(
labels, dsf_sum)]
# axes[i].legend(handles=handles, labels=labels, prop={'size': fontsize-3}, loc='upper center')
axes[i].set_ylabel('mean(|SHAP value|)\n(average impact\non model output magnitude)')
axes[i].grid(axis='y', zorder=1)
if fix_xticklabels:
# n = sum(['pwv' in x for x in sv.feature.values])
axes[2].xaxis.set_ticklabels('')
axes[2].set_xlabel('')
hrs = np.arange(-1, -25, -1)
axes[0].set_xticklabels(hrs, rotation=30, ha="center", fontsize=12)
axes[1].set_xticklabels(hrs, rotation=30, ha="center", fontsize=12)
axes[2].tick_params()
axes[0].set_xlabel('Hours prior to flood')
axes[1].set_xlabel('Hours prior to flood')
fig.tight_layout()
filename = 'RF_shap_values_{}.png'.format('+'.join(features))
plt.savefig(savefig_path / filename, bbox_inches='tight')
return fig
def read_binary_classification_shap_values_to_pandas(shap_values, X):
import xarray as xr
SV0 = X.copy(data=shap_values[0])
SV1 = X.copy(data=shap_values[1])
SV = xr.concat([SV0, SV1], dim='clas')
SV['clas'] = [0, 1]
return SV
def get_shap_values_RF_classifier(plot=True):
import shap
X, y = combine_pos_neg_from_nc_file()
ml = ML_Classifier_Switcher()
rf = ml.pick_model('RF')
rf.set_params(**best_hp_models_dict['RF'])
X = select_doy_from_feature_list(X, features=['pwv', 'pressure', 'doy'])
rf.fit(X, y)
explainer = shap.TreeExplainer(rf)
shap_values = explainer.shap_values(X.values)
if plot:
shap.summary_plot(shap_values, X, feature_names=[
x for x in X.feature.values], max_display=49, sort=False)
return shap_values
def interpolate_pwv_to_tsafit_event(path=work_yuval, savepath=work_yuval):
import pandas as pd
import xarray as xr
from PW_stations import produce_geo_gnss_solved_stations
from interpolation_routines import interpolate_var_ds_at_multiple_dts
from aux_gps import save_ncfile
# get gnss soi-apn pwv data and geo-meta data:
geo_df = produce_geo_gnss_solved_stations(plot=False)
pw = xr.load_dataset(work_yuval/'GNSS_PW_thresh_50.nc')
pw = pw[[x for x in pw if '_error' not in x]]
pw = pw.sel(time=slice('2018-04-25', '2018-04-26'))
pw = pw.drop_vars(['elat', 'elro', 'csar', 'slom'])
# get tsafit data:
predict_df = pd.DataFrame(tsafit_dict, index=['tsafit'])
df_inter = interpolate_var_ds_at_multiple_dts(pw, geo_df, predict_df)
da=df_inter['interpolated_lr_fixed'].to_xarray()
da.name = 'pwv'
da.attrs['operation'] = 'interploated from SOI-APN PWV data'
da.attrs['WV scale height'] = 'variable from SOI-APN data'
da.attrs.update(**tsafit_dict)
if savepath is not None:
filename = 'Tsafit_PWV_event.nc'
save_ncfile(da, savepath, filename)
return da
def plot_tsafit_event(path=work_yuval):
import xarray as xr
import pandas as pd
import matplotlib.pyplot as plt
import seaborn as sns
sns.set_theme(style='ticks', font_scale=1.5)
da = xr.load_dataarray(path / 'Tsafit_PWV_event.nc')
fig, ax = plt.subplots(figsize=(11, 8))
da_sliced = da.sel(time=slice('2018-04-26T00:00:00', '2018-04-26T12:00:00'))
# da_sliced.name = 'PWV [mm]'
da_sliced = da_sliced.rename({'time': 'Time [UTC]'})
da_sliced.to_dataframe().plot(ax=ax, ylabel='PWV [mm]', linewidth=2, marker='o', legend=False)
dt = pd.to_datetime(da.attrs['dt_utc'])
ax.axvline(dt, color='r', linestyle='--', linewidth=2, label='T')
handles, labels = ax.get_legend_handles_labels()
plt.legend(handles=handles, labels=['PWV', 'Tsafit Flood Event'])
ax.grid(True)
# ax.set_xlabel('Time [UTC]')
fig.tight_layout()
fig.suptitle('PWV from SOI-APN over Tsafit area on 2018-04-26')
fig.subplots_adjust(top=0.941)
return fig
# TODO: treat all pwv from events as follows:
# For each station:
# 0) rolling mean to all pwv 1 hour
# 1) take 288 points before events, if < 144 gone then drop
# 2) interpolate them 12H using spline/other
# 3) then, check if dts coinside 1 day before, if not concat all dts+pwv for each station
# 4) prepare features, such as pressure, doy, try to get pressure near the stations and remove the longterm hour dayofyear
# pressure in BD anoms is highly correlated with SEDOM (0.9) and ELAT (0.88) so no need for local pressure features
# fixed filling with jerusalem centre since 2 drag events dropped due to lack of data 2018-11 2019-02 in pressure
# 5) feature addition: should be like pwv steps 1-3,
# 6) negative events should be sampled separtely, for
# 7) now prepare pwv and pressure to single ds with 1 hourly sample rate
# 8) produce positives and save them to file!
# 9) produce a way to get negatives considering the positives
# maybe implement permutaion importance to pwv ? see what is more important to
# the model in 24 hours ? only on SVC and MLP ?
# implemetn TSS and HSS scores and test them (make_scorer from confusion matrix)
# redo results but with inner and outer splits of 4, 4
# plot and see best_score per refit-scorrer - this is the best score of GridSearchCV on the entire
# train/validation subset per each outerfold - basically see if the test_metric increased after the gridsearchcv as it should
# use holdout set
# implement repeatedstratifiedkfold and run it...
# check for stability of the gridsearch CV...also run with 4-folds ?
# finalize the permutation_importances and permutation_test_scores
def prepare_tide_events_GNSS_dataset(hydro_path=hydro_path):
import xarray as xr
import pandas as pd
import numpy as np
from aux_gps import xr_reindex_with_date_range
feats = xr.load_dataset(
hydro_path/'hydro_tides_hourly_features_with_positives.nc')
ds = feats['Tides'].to_dataset('GNSS').rename({'tide_event': 'time'})
da_list = []
for da in ds:
time = ds[da].dropna('time')
daa = time.copy(data=np.ones(time.shape))
daa['time'] = pd.to_datetime(time.values)
daa.name = time.name + '_tide'
da_list.append(daa)
ds = xr.merge(da_list)
li = [xr_reindex_with_date_range(ds[x], freq='H') for x in ds]
ds = xr.merge(li)
return ds
def select_features_from_X(X, features='pwv'):
if isinstance(features, str):
f = [x for x in X.feature.values if features in x]
X = X.sel(feature=f)
elif isinstance(features, list):
fs = []
for f in features:
fs += [x for x in X.feature.values if f in x]
X = X.sel(feature=fs)
return X
def combine_pos_neg_from_nc_file(hydro_path=hydro_path,
negative_sample_num=1,
seed=1, std=True):
from aux_gps import path_glob
from sklearn.utils import resample
import xarray as xr
import numpy as np
# import pandas as pd
if std:
file = path_glob(
hydro_path, 'hydro_tides_hourly_features_with_positives_negatives_std*.nc')[-1]
else:
file = path_glob(
hydro_path, 'hydro_tides_hourly_features_with_positives_negatives_*.nc')[-1]
ds = xr.open_dataset(file)
# get the positive features and produce target:
X_pos = ds['X_pos'].rename({'positive_sample': 'sample'})
y_pos = xr.DataArray(np.ones(X_pos['sample'].shape), dims=['sample'])
y_pos['sample'] = X_pos['sample']
# choose at random y_pos size of negative class:
X_neg = ds['X_neg'].rename({'negative_sample': 'sample'})
pos_size = y_pos['sample'].size
np.random.seed(seed)
# negatives = []
for n_samples in [x for x in range(negative_sample_num)]:
# dts = np.random.choice(X_neg['sample'], size=y_pos['sample'].size,
# replace=False)
# print(np.unique(dts).shape)
# negatives.append(X_neg.sel(sample=dts))
negative = resample(X_neg, replace=False,
n_samples=pos_size * negative_sample_num,
random_state=seed)
negatives = np.split(negative, negative_sample_num, axis=0)
Xs = []
ys = []
for X_negative in negatives:
y_neg = xr.DataArray(np.zeros(X_negative['sample'].shape), dims=['sample'])
y_neg['sample'] = X_negative['sample']
# now concat all X's and y's:
X = xr.concat([X_pos, X_negative], 'sample')
y = xr.concat([y_pos, y_neg], 'sample')
X.name = 'X'
Xs.append(X)
ys.append(y)
if len(negatives) == 1:
return Xs[0], ys[0]
else:
return Xs, ys
def drop_hours_in_pwv_pressure_features(X, last_hours=7, verbose=True):
import numpy as np
Xcopy = X.copy()
pwvs_to_drop = ['pwv_{}'.format(x) for x in np.arange(24-last_hours + 1, 25)]
if set(pwvs_to_drop).issubset(set(X.feature.values)):
if verbose:
print('dropping {} from X.'.format(', '.join(pwvs_to_drop)))
Xcopy = Xcopy.drop_sel(feature=pwvs_to_drop)
pressures_to_drop = ['pressure_{}'.format(x) for x in np.arange(24-last_hours + 1, 25)]
if set(pressures_to_drop).issubset(set(X.feature.values)):
if verbose:
print('dropping {} from X.'.format(', '.join(pressures_to_drop)))
Xcopy = Xcopy.drop_sel(feature=pressures_to_drop)
return Xcopy
def check_if_negatives_are_within_positives(neg_da, hydro_path=hydro_path):
import xarray as xr
import pandas as pd
pos_da = xr.open_dataset(
hydro_path / 'hydro_tides_hourly_features_with_positives.nc')['X']
dt_pos = pos_da.sample.to_dataframe()
dt_neg = neg_da.sample.to_dataframe()
dt_all = dt_pos.index.union(dt_neg.index)
dff = pd.DataFrame(dt_all, index=dt_all)
dff = dff.sort_index()
samples_within = dff[(dff.diff()['sample'] <= pd.Timedelta(1, unit='D'))]
num = samples_within.size
print('samples that are within a day of each other: {}'.format(num))
print('samples are: {}'.format(samples_within))
return dff
def produce_negatives_events_from_feature_file(hydro_path=hydro_path, seed=42,
batches=1, verbose=1, std=True):
# do the same thing for pressure (as for pwv), but not for
import xarray as xr
import numpy as np
import pandas as pd
from aux_gps import save_ncfile
feats = xr.load_dataset(hydro_path / 'hydro_tides_hourly_features.nc')
feats = feats.rename({'doy': 'DOY'})
if std:
pos_filename = 'hydro_tides_hourly_features_with_positives_std.nc'
else:
pos_filename = 'hydro_tides_hourly_features_with_positives.nc'
all_tides = xr.open_dataset(
hydro_path / pos_filename)['X_pos']
# pos_tides = xr.open_dataset(hydro_path / 'hydro_tides_hourly_features_with_positives.nc')['tide_datetimes']
tides = xr.open_dataset(
hydro_path / pos_filename)['Tides']
# get the positives (tide events) for each station:
df_stns = tides.to_dataset('GNSS').to_dataframe()
# get all positives (tide events) for all stations:
df = all_tides.positive_sample.to_dataframe()['positive_sample']
df.columns = ['sample']
stns = [x for x in hydro_pw_dict.keys()]
other_feats = ['DOY', 'doy_sin', 'doy_cos']
# main stns df features (pwv)
pwv_df = feats[stns].to_dataframe()
pressure = feats['bet-dagan'].to_dataframe()['bet-dagan']
# define the initial no_choice_dt_range from the positive dt_range:
no_choice_dt_range = [pd.date_range(
start=dt, periods=48, freq='H') for dt in df]
no_choice_dt_range = pd.DatetimeIndex(
np.unique(np.hstack(no_choice_dt_range)))
dts_to_choose_from = pwv_df.index.difference(no_choice_dt_range)
# dts_to_choose_from_pressure = pwv_df.index.difference(no_choice_dt_range)
# loop over all stns and produce negative events:
np.random.seed(seed)
neg_batches = []
for i in np.arange(1, batches + 1):
if verbose >= 0:
print('preparing batch {}:'.format(i))
neg_stns = []
for stn in stns:
dts_df = df_stns[stn].dropna()
pwv = pwv_df[stn].dropna()
# loop over all events in on stn:
negatives = []
negatives_pressure = []
# neg_samples = []
if verbose >= 1:
print('finding negatives for station {}, events={}'.format(
stn, len(dts_df)))
# print('finding negatives for station {}, dt={}'.format(stn, dt.strftime('%Y-%m-%d %H:%M')))
cnt = 0
while cnt < len(dts_df):
# get random number from each stn pwv:
# r = np.random.randint(low=0, high=len(pwv.index))
# random_dt = pwv.index[r]
random_dt = np.random.choice(dts_to_choose_from)
negative_dt_range = pd.date_range(
start=random_dt, periods=24, freq='H')
if not (no_choice_dt_range.intersection(negative_dt_range)).empty:
# print('#')
if verbose >= 2:
print('Overlap!')
continue
# get the actual pwv and check it is full (24hours):
negative = pwv.loc[pwv.index.intersection(negative_dt_range)]
neg_pressure = pressure.loc[pwv.index.intersection(
negative_dt_range)]
if len(negative.dropna()) != 24 or len(neg_pressure.dropna()) != 24:
# print('!')
if verbose >= 2:
print('NaNs!')
continue
if verbose >= 2:
print('number of dts that are already chosen: {}'.format(
len(no_choice_dt_range)))
negatives.append(negative)
negatives_pressure.append(neg_pressure)
# now add to the no_choice_dt_range the negative dt_range we just aquired:
negative_dt_range_with_padding = pd.date_range(
start=random_dt-pd.Timedelta(24, unit='H'), end=random_dt+pd.Timedelta(23, unit='H'), freq='H')
no_choice_dt_range = pd.DatetimeIndex(
np.unique(np.hstack([no_choice_dt_range, negative_dt_range_with_padding])))
dts_to_choose_from = dts_to_choose_from.difference(
no_choice_dt_range)
if verbose >= 2:
print('number of dts to choose from: {}'.format(
len(dts_to_choose_from)))
cnt += 1
neg_da = xr.DataArray(negatives, dims=['sample', 'feature'])
neg_da['feature'] = ['{}_{}'.format(
'pwv', x) for x in np.arange(1, 25)]
neg_samples = [x.index[0] for x in negatives]
neg_da['sample'] = neg_samples
neg_pre_da = xr.DataArray(
negatives_pressure, dims=['sample', 'feature'])
neg_pre_da['feature'] = ['{}_{}'.format(
'pressure', x) for x in np.arange(1, 25)]
neg_pre_samples = [x.index[0] for x in negatives_pressure]
neg_pre_da['sample'] = neg_pre_samples
neg_da = xr.concat([neg_da, neg_pre_da], 'feature')
neg_da = neg_da.sortby('sample')
neg_stns.append(neg_da)
da_stns = xr.concat(neg_stns, 'sample')
da_stns = da_stns.sortby('sample')
# now loop over the remaining features (which are stns agnostic)
# and add them with the same negative datetimes of the pwv already aquired:
dts = [pd.date_range(x.item(), periods=24, freq='H')
for x in da_stns['sample']]
dts_samples = [x[0] for x in dts]
other_feat_list = []
for feat in feats[other_feats]:
# other_feat_sample_list = []
da_other = xr.DataArray(feats[feat].sel(time=dts_samples).values, dims=['sample'])
# for dt in dts_samples:
# da_other = xr.DataArray(feats[feat].sel(
# time=dt).values, dims=['feature'])
da_other['sample'] = dts_samples
other_feat_list.append(da_other)
# other_feat_da = xr.concat(other_feat_sample_list, 'feature')
da_other_feats = xr.concat(other_feat_list, 'feature')
da_other_feats['feature'] = other_feats
da_stns = xr.concat([da_stns, da_other_feats], 'feature')
neg_batches.append(da_stns)
neg_batch_da = xr.concat(neg_batches, 'sample')
# neg_batch_da['batch'] = np.arange(1, batches + 1)
neg_batch_da.name = 'X_neg'
feats['X_neg'] = neg_batch_da
feats['X_pos'] = all_tides
feats['X_pwv_stns'] = tides
# feats['tide_datetimes'] = pos_tides
feats = feats.rename({'sample': 'negative_sample'})
if std:
filename = 'hydro_tides_hourly_features_with_positives_negatives_std_{}.nc'.format(
batches)
else:
filename = 'hydro_tides_hourly_features_with_positives_negatives_{}.nc'.format(
batches)
save_ncfile(feats, hydro_path, filename)
return neg_batch_da
def produce_positives_from_feature_file(hydro_path=hydro_path, std=True):
import xarray as xr
import pandas as pd
import numpy as np
from aux_gps import save_ncfile
# load features:
if std:
file = hydro_path / 'hydro_tides_hourly_features_std.nc'
else:
file = hydro_path / 'hydro_tides_hourly_features.nc'
feats = xr.load_dataset(file)
feats = feats.rename({'doy': 'DOY'})
# load positive event for each station:
dfs = [read_station_from_tide_database(hydro_pw_dict.get(
x), rounding='1H') for x in hydro_pw_dict.keys()]
dfs = check_if_tide_events_from_stations_are_within_time_window(
dfs, days=1, rounding=None, return_hs_list=True)
da_list = []
positives_per_station = []
for i, feat in enumerate(feats):
try:
_, _, pr = produce_pwv_days_before_tide_events(feats[feat], dfs[i],
plot=False, rolling=None,
days_prior=1,
drop_thresh=0.75,
max_gap='6H',
verbose=0)
print('getting positives from station {}'.format(feat))
positives = [pd.to_datetime(
(x[-1].time + pd.Timedelta(1, unit='H')).item()) for x in pr]
da = xr.DataArray(pr, dims=['sample', 'feature'])
da['sample'] = positives
positives_per_station.append(positives)
da['feature'] = ['pwv_{}'.format(x) for x in np.arange(1, 25)]
da_list.append(da)
except IndexError:
continue
da_pwv = xr.concat(da_list, 'sample')
da_pwv = da_pwv.sortby('sample')
# now add more features:
da_list = []
for feat in ['bet-dagan']:
print('getting positives from feature {}'.format(feat))
positives = []
for dt_end in da_pwv.sample:
dt_st = pd.to_datetime(dt_end.item()) - pd.Timedelta(24, unit='H')
dt_end_end = pd.to_datetime(
dt_end.item()) - pd.Timedelta(1, unit='H')
positive = feats[feat].sel(time=slice(dt_st, dt_end_end))
positives.append(positive)
da = xr.DataArray(positives, dims=['sample', 'feature'])
da['sample'] = da_pwv.sample
if feat == 'bet-dagan':
feat_name = 'pressure'
else:
feat_name = feat
da['feature'] = ['{}_{}'.format(feat_name, x)
for x in np.arange(1, 25)]
da_list.append(da)
da_f = xr.concat(da_list, 'feature')
da_list = []
for feat in ['DOY', 'doy_sin', 'doy_cos']:
print('getting positives from feature {}'.format(feat))
positives = []
for dt in da_pwv.sample:
positive = feats[feat].sel(time=dt)
positives.append(positive)
da = xr.DataArray(positives, dims=['sample'])
da['sample'] = da_pwv.sample
# da['feature'] = feat
da_list.append(da)
da_ff = xr.concat(da_list, 'feature')
da_ff['feature'] = ['DOY', 'doy_sin', 'doy_cos']
da = xr.concat([da_pwv, da_f, da_ff], 'feature')
if std:
filename = 'hydro_tides_hourly_features_with_positives_std.nc'
else:
filename = 'hydro_tides_hourly_features_with_positives.nc'
feats['X_pos'] = da
# now add positives per stations:
pdf = pd.DataFrame(positives_per_station).T
pdf.index.name = 'tide_event'
pos_da = pdf.to_xarray().to_array('GNSS')
pos_da['GNSS'] = [x for x in hydro_pw_dict.keys()]
pos_da.attrs['info'] = 'contains the datetimes of the tide events per GNSS station.'
feats['Tides'] = pos_da
# rename sample to positive sample:
feats = feats.rename({'sample': 'positive_sample'})
save_ncfile(feats, hydro_path, filename)
return feats
def prepare_features_and_save_hourly(work_path=work_yuval, ims_path=ims_path,
savepath=hydro_path, std=True):
import xarray as xr
from aux_gps import save_ncfile
import numpy as np
# pwv = xr.load_dataset(
if std:
pwv_filename = 'GNSS_PW_thresh_0_hour_dayofyear_anoms_sd.nc'
pre_filename = 'IMS_BD_hourly_anoms_std_ps_1964-2020.nc'
else:
pwv_filename = 'GNSS_PW_thresh_0_hour_dayofyear_anoms.nc'
pre_filename = 'IMS_BD_hourly_anoms_ps_1964-2020.nc'
# work_path / 'GNSS_PW_thresh_0_hour_dayofyear_anoms.nc')
pwv = xr.load_dataset(work_path / pwv_filename)
pwv_stations = [x for x in hydro_pw_dict.keys()]
pwv = pwv[pwv_stations]
# pwv = pwv.rolling(time=12, keep_attrs=True).mean(keep_attrs=True)
pwv = pwv.resample(time='1H', keep_attrs=True).mean(keep_attrs=True)
# bd = xr.load_dataset(ims_path / 'IMS_BD_anoms_5min_ps_1964-2020.nc')
bd = xr.load_dataset(ims_path / pre_filename)
# min_time = pwv.dropna('time')['time'].min()
# bd = bd.sel(time=slice('1996', None)).resample(time='1H').mean()
bd = bd.sel(time=slice('1996', None))
pressure = bd['bet-dagan']
doy = pwv['time'].copy(data=pwv['time'].dt.dayofyear)
doy.name = 'doy'
doy_sin = np.sin(doy * np.pi / 183)
doy_sin.name = 'doy_sin'
doy_cos = np.cos(doy * np.pi / 183)
doy_cos.name = 'doy_cos'
ds = xr.merge([pwv, pressure, doy, doy_sin, doy_cos])
if std:
filename = 'hydro_tides_hourly_features_std.nc'
else:
filename = 'hydro_tides_hourly_features.nc'
save_ncfile(ds, savepath, filename)
return ds
def plot_all_decompositions(X, y, n=2):
import xarray as xr
models = [
'PCA',
'LDA',
'ISO_MAP',
'LLE',
'LLE-modified',
'LLE-hessian',
'LLE-ltsa',
'MDA',
'RTE',
'SE',
'TSNE',
'NCA']
names = [
'Principal Components',
'Linear Discriminant',
'Isomap',
'Locally Linear Embedding',
'Modified LLE',
'Hessian LLE',
'Local Tangent Space Alignment',
'MDS embedding',
'Random forest',
'Spectral embedding',
't-SNE',
'NCA embedding']
name_dict = dict(zip(models, names))
da = xr.DataArray(models, dims=['model'])
da['model'] = models
fg = xr.plot.FacetGrid(da, col='model', col_wrap=4,
sharex=False, sharey=False)
for model_str, ax in zip(da['model'].values, fg.axes.flatten()):
model = model_str.split('-')[0]
method = model_str.split('-')[-1]
if model == method:
method = None
try:
ax = scikit_decompose(X, y, model=model, n=n, method=method, ax=ax)
except ValueError:
pass
ax.set_title(name_dict[model_str])
ax.set_xlabel('')
ax.set_ylabel('')
fg.fig.suptitle('various decomposition projections (n={})'.format(n))
return
def scikit_decompose(X, y, model='PCA', n=2, method=None, ax=None):
from sklearn import (manifold, decomposition, ensemble,
discriminant_analysis, neighbors)
import matplotlib.pyplot as plt
import pandas as pd
# from mpl_toolkits.mplot3d import Axes3D
n_neighbors = 30
if model == 'PCA':
X_decomp = decomposition.TruncatedSVD(n_components=n).fit_transform(X)
elif model == 'LDA':
X2 = X.copy()
X2.values.flat[::X.shape[1] + 1] += 0.01
X_decomp = discriminant_analysis.LinearDiscriminantAnalysis(n_components=n
).fit_transform(X2, y)
elif model == 'ISO_MAP':
X_decomp = manifold.Isomap(
n_neighbors, n_components=n).fit_transform(X)
elif model == 'LLE':
# method = 'standard', 'modified', 'hessian' 'ltsa'
if method is None:
method = 'standard'
clf = manifold.LocallyLinearEmbedding(n_neighbors, n_components=2,
method=method)
X_decomp = clf.fit_transform(X)
elif model == 'MDA':
clf = manifold.MDS(n_components=n, n_init=1, max_iter=100)
X_decomp = clf.fit_transform(X)
elif model == 'RTE':
hasher = ensemble.RandomTreesEmbedding(n_estimators=200, random_state=0,
max_depth=5)
X_transformed = hasher.fit_transform(X)
pca = decomposition.TruncatedSVD(n_components=n)
X_decomp = pca.fit_transform(X_transformed)
elif model == 'SE':
embedder = manifold.SpectralEmbedding(n_components=n, random_state=0,
eigen_solver="arpack")
X_decomp = embedder.fit_transform(X)
elif model == 'TSNE':
tsne = manifold.TSNE(n_components=n, init='pca', random_state=0)
X_decomp = tsne.fit_transform(X)
elif model == 'NCA':
nca = neighbors.NeighborhoodComponentsAnalysis(init='random',
n_components=n, random_state=0)
X_decomp = nca.fit_transform(X, y)
df = pd.DataFrame(X_decomp)
df.columns = [
'{}_{}'.format(
model,
x +
1) for x in range(
X_decomp.shape[1])]
df['flood'] = y
df['flood'] = df['flood'].astype(int)
df_1 = df[df['flood'] == 1]
df_0 = df[df['flood'] == 0]
if X_decomp.shape[1] == 1:
if ax is not None:
df_1.plot.scatter(ax=ax,
x='{}_1'.format(model),
y='{}_1'.format(model),
color='b', marker='s', alpha=0.3,
label='1',
s=50)
else:
ax = df_1.plot.scatter(
x='{}_1'.format(model),
y='{}_1'.format(model),
color='b',
label='1',
s=50)
df_0.plot.scatter(
ax=ax,
x='{}_1'.format(model),
y='{}_1'.format(model),
color='r', marker='x',
label='0',
s=50)
elif X_decomp.shape[1] == 2:
if ax is not None:
df_1.plot.scatter(ax=ax,
x='{}_1'.format(model),
y='{}_2'.format(model),
color='b', marker='s', alpha=0.3,
label='1',
s=50)
else:
ax = df_1.plot.scatter(
x='{}_1'.format(model),
y='{}_2'.format(model),
color='b',
label='1',
s=50)
df_0.plot.scatter(
ax=ax,
x='{}_1'.format(model),
y='{}_2'.format(model),
color='r',
label='0',
s=50)
elif X_decomp.shape[1] == 3:
ax = plt.figure().gca(projection='3d')
# df_1.plot.scatter(x='{}_1'.format(model), y='{}_2'.format(model), z='{}_3'.format(model), color='b', label='1', s=50, ax=threedee)
ax.scatter(df_1['{}_1'.format(model)],
df_1['{}_2'.format(model)],
df_1['{}_3'.format(model)],
color='b',
label='1',
s=50)
ax.scatter(df_0['{}_1'.format(model)],
df_0['{}_2'.format(model)],
df_0['{}_3'.format(model)],
color='r',
label='0',
s=50)
ax.set_xlabel('{}_1'.format(model))
ax.set_ylabel('{}_2'.format(model))
ax.set_zlabel('{}_3'.format(model))
return ax
def permutation_scikit(X, y, cv=False, plot=True):
import matplotlib.pyplot as plt
from sklearn.svm import SVC
from sklearn.discriminant_analysis import LinearDiscriminantAnalysis
from sklearn.model_selection import KFold
from sklearn.model_selection import StratifiedKFold
from sklearn.model_selection import GridSearchCV
from sklearn.model_selection import permutation_test_score
from sklearn.model_selection import train_test_split
from sklearn.metrics import classification_report, confusion_matrix
import numpy as np
if not cv:
clf = SVC(C=0.01, break_ties=False, cache_size=200, class_weight=None, coef0=0.0,
decision_function_shape='ovr', degree=3, gamma=0.032374575428176434,
kernel='poly', max_iter=-1, probability=False, random_state=None,
shrinking=True, tol=0.001, verbose=False)
clf = SVC(kernel='linear')
# clf = LinearDiscriminantAnalysis()
cv = StratifiedKFold(4, shuffle=True)
# cv = KFold(4, shuffle=True)
n_classes = 2
score, permutation_scores, pvalue = permutation_test_score(
clf, X, y, scoring="f1", cv=cv, n_permutations=1000, n_jobs=-1, verbose=2)
print("Classification score %s (pvalue : %s)" % (score, pvalue))
plt.hist(permutation_scores, 20, label='Permutation scores',
edgecolor='black')
ylim = plt.ylim()
plt.plot(2 * [score], ylim, '--g', linewidth=3,
label='Classification Score'
' (pvalue %s)' % pvalue)
plt.plot(2 * [1. / n_classes], ylim, '--k', linewidth=3, label='Luck')
plt.ylim(ylim)
plt.legend()
plt.xlabel('Score')
plt.show()
else:
X_train, X_test, y_train, y_test = train_test_split(
X, y, test_size=0.2, shuffle=True, random_state=42)
param_grid = {
'C': np.logspace(-2, 3, 50), 'gamma': np.logspace(-2, 3, 50),
'kernel': ['rbf', 'poly', 'sigmoid']}
grid = GridSearchCV(SVC(), param_grid, refit=True, verbose=2)
grid.fit(X_train, y_train)
print(grid.best_estimator_)
grid_predictions = grid.predict(X_test)
print(confusion_matrix(y_test, grid_predictions))
print(classification_report(y_test, grid_predictions))
return
def grab_y_true_and_predict_from_sklearn_model(model, X, y, cv,
kfold_name='inner_kfold'):
from sklearn.model_selection import GridSearchCV
import xarray as xr
import numpy as np
if isinstance(model, GridSearchCV):
model = model.best_estimator_
ds_list = []
for i, (train, val) in enumerate(cv.split(X, y)):
model.fit(X[train], y[train])
y_true = y[val]
y_pred = model.predict(X[val])
try:
lr_probs = model.predict_proba(X[val])
# keep probabilities for the positive outcome only
lr_probs = lr_probs[:, 1]
except AttributeError:
lr_probs = model.decision_function(X[val])
y_true_da = xr.DataArray(y_true, dims=['sample'])
y_pred_da = xr.DataArray(y_pred, dims=['sample'])
y_prob_da = xr.DataArray(lr_probs, dims=['sample'])
ds = xr.Dataset()
ds['y_true'] = y_true_da
ds['y_pred'] = y_pred_da
ds['y_prob'] = y_prob_da
ds['sample'] = np.arange(0, len(X[val]))
ds_list.append(ds)
ds = xr.concat(ds_list, kfold_name)
ds[kfold_name] = np.arange(1, cv.n_splits + 1)
return ds
def produce_ROC_curves_from_model(model, X, y, cv, kfold_name='inner_kfold'):
import numpy as np
import xarray as xr
from sklearn.metrics import roc_curve
from sklearn.metrics import roc_auc_score
from sklearn.model_selection import GridSearchCV
from sklearn.metrics import precision_recall_curve
from sklearn.metrics import average_precision_score
# TODO: collect all predictions and y_tests from this, also predict_proba
# and save, then calculte everything elsewhere.
if isinstance(model, GridSearchCV):
model = model.best_estimator_
tprs = []
aucs = []
pr = []
pr_aucs = []
mean_fpr = np.linspace(0, 1, 100)
for i, (train, val) in enumerate(cv.split(X, y)):
model.fit(X[train], y[train])
y_pred = model.predict(X[val])
try:
lr_probs = model.predict_proba(X[val])
# keep probabilities for the positive outcome only
lr_probs = lr_probs[:, 1]
except AttributeError:
lr_probs = model.decision_function(X[val])
fpr, tpr, _ = roc_curve(y[val], y_pred)
interp_tpr = np.interp(mean_fpr, fpr, tpr)
interp_tpr[0] = 0.0
tprs.append(interp_tpr)
aucs.append(roc_auc_score(y[val], y_pred))
precision, recall, _ = precision_recall_curve(y[val], lr_probs)
pr.append(recall)
average_precision = average_precision_score(y[val], y_pred)
pr_aucs.append(average_precision)
# mean_tpr = np.mean(tprs, axis=0)
# mean_tpr[-1] = 1.0
# mean_auc = auc(mean_fpr, mean_tpr)
# std_auc = np.std(aucs)
# std_tpr = np.std(tprs, axis=0)
tpr_da = xr.DataArray(tprs, dims=[kfold_name, 'fpr'])
auc_da = xr.DataArray(aucs, dims=[kfold_name])
ds = xr.Dataset()
ds['TPR'] = tpr_da
ds['AUC'] = auc_da
ds['fpr'] = mean_fpr
ds[kfold_name] = np.arange(1, cv.n_splits + 1)
# variability for each tpr is ds['TPR'].std('kfold')
return ds
def cross_validation_with_holdout(X, y, model_name='SVC', features='pwv',
n_splits=3, test_ratio=0.25,
scorers=['f1', 'recall', 'tss', 'hss',
'precision', 'accuracy'],
seed=42, savepath=None, verbose=0,
param_grid='normal', n_jobs=-1,
n_repeats=None):
# from sklearn.model_selection import cross_validate
from sklearn.model_selection import StratifiedKFold
from sklearn.model_selection import RepeatedStratifiedKFold
from sklearn.model_selection import GridSearchCV
from sklearn.model_selection import train_test_split
from sklearn.metrics import make_scorer
# from string import digits
import numpy as np
# import xarray as xr
scores_dict = {s: s for s in scorers}
if 'tss' in scorers:
scores_dict['tss'] = make_scorer(tss_score)
if 'hss' in scorers:
scores_dict['hss'] = make_scorer(hss_score)
X = select_doy_from_feature_list(X, model_name, features)
if param_grid == 'light':
print(np.unique(X.feature.values))
# first take out the hold-out set:
X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=test_ratio,
random_state=seed,
stratify=y)
if n_repeats is None:
# configure the cross-validation procedure
cv = StratifiedKFold(n_splits=n_splits, shuffle=True,
random_state=seed)
print('CV StratifiedKfolds of {}.'.format(n_splits))
# define the model and search space:
else:
cv = RepeatedStratifiedKFold(n_splits=n_splits, n_repeats=n_repeats,
random_state=seed)
print('CV RepeatedStratifiedKFold of {} with {} repeats.'.format(n_splits, n_repeats))
ml = ML_Classifier_Switcher()
print('param grid group is set to {}.'.format(param_grid))
sk_model = ml.pick_model(model_name, pgrid=param_grid)
search_space = ml.param_grid
# define search
gr_search = GridSearchCV(estimator=sk_model, param_grid=search_space,
cv=cv, n_jobs=n_jobs,
scoring=scores_dict,
verbose=verbose,
refit=False, return_train_score=True)
gr_search.fit(X, y)
if isinstance(features, str):
features = [features]
if savepath is not None:
filename = 'GRSRCHCV_holdout_{}_{}_{}_{}_{}_{}_{}.pkl'.format(
model_name, '+'.join(features), '+'.join(scorers), n_splits,
int(test_ratio*100), param_grid, seed)
save_gridsearchcv_object(gr_search, savepath, filename)
# gr, _ = process_gridsearch_results(
# gr_search, model_name, split_dim='kfold', features=X.feature.values)
# remove_digits = str.maketrans('', '', digits)
# features = list(set([x.translate(remove_digits).split('_')[0]
# for x in X.feature.values]))
# # add more attrs, features etc:
# gr.attrs['features'] = features
return gr_search
def select_doy_from_feature_list(X, model_name='RF', features='pwv'):
# first if RF chosen, replace the cyclic coords of DOY (sin and cos) with
# the DOY itself.
if isinstance(features, list):
feats = features.copy()
else:
feats = features
if model_name == 'RF' and 'doy' in features:
if isinstance(features, list):
feats.remove('doy')
feats.append('DOY')
elif isinstance(features, str):
feats = 'DOY'
elif model_name != 'RF' and 'doy' in features:
if isinstance(features, list):
feats.remove('doy')
feats.append('doy_sin')
feats.append('doy_cos')
elif isinstance(features, str):
feats = ['doy_sin']
feats.append('doy_cos')
X = select_features_from_X(X, feats)
return X
def single_cross_validation(X_val, y_val, model_name='SVC', features='pwv',
n_splits=4, scorers=['f1', 'recall', 'tss', 'hss',
'precision', 'accuracy'],
seed=42, savepath=None, verbose=0,
param_grid='normal', n_jobs=-1,
n_repeats=None, outer_split='1-1'):
# from sklearn.model_selection import cross_validate
from sklearn.model_selection import StratifiedKFold
from sklearn.model_selection import RepeatedStratifiedKFold
from sklearn.model_selection import GridSearchCV
# from sklearn.model_selection import train_test_split
from sklearn.metrics import make_scorer
# from string import digits
import numpy as np
# import xarray as xr
scores_dict = {s: s for s in scorers}
if 'tss' in scorers:
scores_dict['tss'] = make_scorer(tss_score)
if 'hss' in scorers:
scores_dict['hss'] = make_scorer(hss_score)
X = select_doy_from_feature_list(X_val, model_name, features)
y = y_val
if param_grid == 'light':
print(np.unique(X.feature.values))
if n_repeats is None:
# configure the cross-validation procedure
cv = StratifiedKFold(n_splits=n_splits, shuffle=True,
random_state=seed)
print('CV StratifiedKfolds of {}.'.format(n_splits))
# define the model and search space:
else:
cv = RepeatedStratifiedKFold(n_splits=n_splits, n_repeats=n_repeats,
random_state=seed)
print('CV RepeatedStratifiedKFold of {} with {} repeats.'.format(
n_splits, n_repeats))
ml = ML_Classifier_Switcher()
print('param grid group is set to {}.'.format(param_grid))
if outer_split == '1-1':
cv_type = 'holdout'
print('holdout cv is selected.')
else:
cv_type = 'nested'
print('nested cv {} out of {}.'.format(
outer_split.split('-')[0], outer_split.split('-')[1]))
sk_model = ml.pick_model(model_name, pgrid=param_grid)
search_space = ml.param_grid
# define search
gr_search = GridSearchCV(estimator=sk_model, param_grid=search_space,
cv=cv, n_jobs=n_jobs,
scoring=scores_dict,
verbose=verbose,
refit=False, return_train_score=True)
gr_search.fit(X, y)
if isinstance(features, str):
features = [features]
if savepath is not None:
filename = 'GRSRCHCV_{}_{}_{}_{}_{}_{}_{}_{}.pkl'.format(cv_type,
model_name, '+'.join(features), '+'.join(
scorers), n_splits,
outer_split, param_grid, seed)
save_gridsearchcv_object(gr_search, savepath, filename)
return gr_search
def save_cv_params_to_file(cv_obj, path, name):
import pandas as pd
di = vars(cv_obj)
splitter_type = cv_obj.__repr__().split('(')[0]
di['splitter_type'] = splitter_type
(pd.DataFrame.from_dict(data=di, orient='index')
.to_csv(path / '{}.csv'.format(name), header=False))
print('{}.csv saved to {}.'.format(name, path))
return
def read_cv_params_and_instantiate(filepath):
import pandas as pd
from sklearn.model_selection import StratifiedKFold
df = pd.read_csv(filepath, header=None, index_col=0)
d = {}
for row in df.iterrows():
dd = pd.to_numeric(row[1], errors='ignore')
if dd.item() == 'True' or dd.item() == 'False':
dd = dd.astype(bool)
d[dd.to_frame().columns.item()] = dd.item()
s_type = d.pop('splitter_type')
if s_type == 'StratifiedKFold':
cv = StratifiedKFold(**d)
return cv
def nested_cross_validation_procedure(X, y, model_name='SVC', features='pwv',
outer_splits=4, inner_splits=2,
refit_scorer='roc_auc',
scorers=['f1', 'recall', 'tss', 'hss',
'roc_auc', 'precision',
'accuracy'],
seed=42, savepath=None, verbose=0,
param_grid='normal', n_jobs=-1):
from sklearn.model_selection import cross_validate
from sklearn.model_selection import StratifiedKFold
from sklearn.model_selection import GridSearchCV
from sklearn.metrics import make_scorer
from sklearn.inspection import permutation_importance
from string import digits
import numpy as np
import xarray as xr
assert refit_scorer in scorers
scores_dict = {s: s for s in scorers}
if 'tss' in scorers:
scores_dict['tss'] = make_scorer(tss_score)
if 'hss' in scorers:
scores_dict['hss'] = make_scorer(hss_score)
X = select_doy_from_feature_list(X, model_name, features)
# if model_name == 'RF':
# doy = X['sample'].dt.dayofyear
# sel_doy = [x for x in X.feature.values if 'doy_sin' in x]
# doy_X = doy.broadcast_like(X.sel(feature=sel_doy))
# doy_X['feature'] = [
# 'doy_{}'.format(x) for x in range(
# doy_X.feature.size)]
# no_doy = [x for x in X.feature.values if 'doy' not in x]
# X = X.sel(feature=no_doy)
# X = xr.concat([X, doy_X], 'feature')
# else:
# # first slice X for features:
# if isinstance(features, str):
# f = [x for x in X.feature.values if features in x]
# X = X.sel(feature=f)
# elif isinstance(features, list):
# fs = []
# for f in features:
# fs += [x for x in X.feature.values if f in x]
# X = X.sel(feature=fs)
if param_grid == 'light':
print(np.unique(X.feature.values))
# configure the cross-validation procedure
cv_inner = StratifiedKFold(n_splits=inner_splits, shuffle=True,
random_state=seed)
print('Inner CV StratifiedKfolds of {}.'.format(inner_splits))
# define the model and search space:
ml = ML_Classifier_Switcher()
if param_grid == 'light':
print('disgnostic mode light.')
sk_model = ml.pick_model(model_name, pgrid=param_grid)
search_space = ml.param_grid
# define search
gr_search = GridSearchCV(estimator=sk_model, param_grid=search_space,
cv=cv_inner, n_jobs=n_jobs,
scoring=scores_dict,
verbose=verbose,
refit=refit_scorer, return_train_score=True)
# gr.fit(X, y)
# configure the cross-validation procedure
cv_outer = StratifiedKFold(
n_splits=outer_splits, shuffle=True, random_state=seed)
# execute the nested cross-validation
scores_est_dict = cross_validate(gr_search, X, y,
scoring=scores_dict,
cv=cv_outer, n_jobs=n_jobs,
return_estimator=True, verbose=verbose)
# perm = []
# for i, (train, val) in enumerate(cv_outer.split(X, y)):
# gr_model = scores_est_dict['estimator'][i]
# gr_model.fit(X[train], y[train])
# r = permutation_importance(gr_model, X[val], y[val],scoring='f1',
# n_repeats=30, n_jobs=-1,
# random_state=0)
# perm.append(r)
# get the test scores:
test_keys = [x for x in scores_est_dict.keys() if 'test' in x]
ds = xr.Dataset()
for key in test_keys:
ds[key] = xr.DataArray(scores_est_dict[key], dims=['outer_kfold'])
preds_ds = []
gr_ds = []
for est in scores_est_dict['estimator']:
gr, _ = process_gridsearch_results(
est, model_name, split_dim='inner_kfold', features=X.feature.values)
# somehow save gr:
gr_ds.append(gr)
preds_ds.append(
grab_y_true_and_predict_from_sklearn_model(est, X, y, cv_inner))
# tpr_ds.append(produce_ROC_curves_from_model(est, X, y, cv_inner))
dss = xr.concat(preds_ds, 'outer_kfold')
gr_dss = xr.concat(gr_ds, 'outer_kfold')
dss['outer_kfold'] = np.arange(1, cv_outer.n_splits + 1)
gr_dss['outer_kfold'] = np.arange(1, cv_outer.n_splits + 1)
# aggragate results:
dss = xr.merge([ds, dss])
dss = xr.merge([dss, gr_dss])
dss.attrs = gr_dss.attrs
dss.attrs['outer_kfold_splits'] = outer_splits
remove_digits = str.maketrans('', '', digits)
features = list(set([x.translate(remove_digits).split('_')[0]
for x in X.feature.values]))
# add more attrs, features etc:
dss.attrs['features'] = features
# rename major data_vars with model name:
# ys = [x for x in dss.data_vars if 'y_' in x]
# new_ys = [y + '_{}'.format(model_name) for y in ys]
# dss = dss.rename(dict(zip(ys, new_ys)))
# new_test_keys = [y + '_{}'.format(model_name) for y in test_keys]
# dss = dss.rename(dict(zip(test_keys, new_test_keys)))
# if isinstance(X.attrs['pwv_id'], list):
# dss.attrs['pwv_id'] = '-'.join(X.attrs['pwv_id'])
# else:
# dss.attrs['pwv_id'] = X.attrs['pwv_id']
# if isinstance(y.attrs['hydro_station_id'], list):
# dss.attrs['hs_id'] = '-'.join([str(x) for x in y.attrs['hydro_station_id']])
# else:
# dss.attrs['hs_id'] = y.attrs['hydro_station_id']
# dss.attrs['hydro_max_flow'] = y.attrs['max_flow']
# dss.attrs['neg_pos_ratio'] = y.attrs['neg_pos_ratio']
# save results to file:
if savepath is not None:
save_cv_results(dss, savepath=savepath)
return dss
# def ML_main_procedure(X, y, estimator=None, model_name='SVC', features='pwv',
# val_size=0.18, n_splits=None, test_size=0.2, seed=42, best_score='f1',
# savepath=None, plot=True):
# """split the X,y for train and test, either do HP tuning using HP_tuning
# with val_size or use already tuned (or not) estimator.
# models to play with = MLP, RF and SVC.
# n_splits = 2, 3, 4.
# features = pwv, pressure.
# best_score = f1, roc_auc, accuracy.
# can do loop on them. RF takes the most time to tune."""
# X = select_features_from_X(X, features)
# X_train, X_test, y_train, y_test = train_test_split(X, y,
# test_size=test_size,
# shuffle=True,
# random_state=seed)
# # do HP_tuning:
# if estimator is None:
# cvr, model = HP_tuning(X_train, y_train, model_name=model_name, val_size=val_size, test_size=test_size,
# best_score=best_score, seed=seed, savepath=savepath, n_splits=n_splits)
# else:
# model = estimator
# if plot:
# ax = plot_many_ROC_curves(model, X_test, y_test, name=model_name,
# ax=None)
# return ax
# else:
# return model
def plot_hyper_parameters_heatmaps_from_nested_CV_model(dss, path=hydro_path, model_name='MLP',
features='pwv+pressure+doy', save=True):
import matplotlib.pyplot as plt
ds = dss.sel(features=features).reset_coords(drop=True)
non_hp_vars = ['mean_score', 'std_score',
'test_score', 'roc_auc_score', 'TPR']
if model_name == 'RF':
non_hp_vars.append('feature_importances')
ds = ds[[x for x in ds if x not in non_hp_vars]]
seq = 'Blues'
cat = 'Dark2'
cmap_hp_dict = {
'alpha': seq, 'activation': cat,
'hidden_layer_sizes': cat, 'learning_rate': cat,
'solver': cat, 'kernel': cat, 'C': seq,
'gamma': seq, 'degree': seq, 'coef0': seq,
'max_depth': seq, 'max_features': cat,
'min_samples_leaf': seq, 'min_samples_split': seq,
'n_estimators': seq
}
# fix stuff for SVC:
if model_name == 'SVC':
ds['degree'] = ds['degree'].where(ds['kernel']=='poly')
ds['coef0'] = ds['coef0'].where(ds['kernel']=='poly')
# da = ds.to_arrray('hyper_parameters')
# fg = xr.plot.FacetGrid(
# da,
# col='hyper_parameters',
# sharex=False,
# sharey=False, figsize=(16, 10))
fig, axes = plt.subplots(5, 1, sharex=True, figsize=(4, 10))
for i, da in enumerate(ds):
df = ds[da].reset_coords(drop=True).to_dataset('scorer').to_dataframe()
df.index.name = 'Outer Split'
try:
df = df.astype(float).round(2)
except ValueError:
pass
cmap = cmap_hp_dict.get(da, 'Set1')
plot_heatmap_for_hyper_parameters_df(df, ax=axes[i], title=da, cmap=cmap)
fig.tight_layout()
if save:
filename = 'Hyper-parameters_nested_{}.png'.format(
model_name)
plt.savefig(savefig_path / filename, bbox_inches='tight')
return
def plot_heatmaps_for_hyper_parameters_data_splits(df1, df2, axes=None,
cmap='colorblind',
title=None, fig=None,
cbar_params=[.92, .12, .03, .75],
fontsize=12,
val_type='float'):
import pandas as pd
import seaborn as sns
import numpy as np
# from mpl_toolkits.axes_grid1 import make_axes_locatable
from matplotlib.colors import Normalize
import matplotlib
import matplotlib.pyplot as plt
import matplotlib.cm as cm
sns.set_style('ticks')
sns.set_style('whitegrid')
sns.set(font_scale=1.2)
df1 = df1.astype(eval(val_type))
df2 = df2.astype(eval(val_type))
arr = pd.concat([df1, df2], axis=0).values.ravel()
value_to_int = {j: i for i, j in enumerate(
np.unique(arr))} # like you did
# try:
# sorted_v_to_i = dict(sorted(value_to_int.items()))
# except TypeError:
# sorted_v_to_i = value_to_int
# print(value_to_int)
n = len(value_to_int)
# discrete colormap (n samples from a given cmap)
cmap_list = sns.color_palette(cmap, n)
if val_type == 'float':
# print([value_to_int.keys()])
cbar_ticklabels = ['{:.2g}'.format(x) for x in value_to_int.keys()]
elif val_type == 'int':
cbar_ticklabels = [int(x) for x in value_to_int.keys()]
elif val_type == 'str':
cbar_ticklabels = [x for x in value_to_int.keys()]
if 'nan' in value_to_int.keys():
cmap_list[-1] = (0.5, 0.5, 0.5)
new_value_to_int = {}
for key, val in value_to_int.items():
try:
new_value_to_int[str(int(float(key)))] = val
except ValueError:
new_value_to_int['NR'] = val
cbar_ticklabels = [x for x in new_value_to_int.keys()]
# u1 = np.unique(df1.replace(value_to_int)).astype(int)
# cmap1 = [cmap_list[x] for x in u1]
# u2 = np.unique(df2.replace(value_to_int)).astype(int)
# cmap2 = [cmap_list[x] for x in u2]
# prepare normalizer
## Prepare bins for the normalizer
norm_bins = np.sort([*value_to_int.values()]) + 0.5
norm_bins = np.insert(norm_bins, 0, np.min(norm_bins) - 1.0)
# print(norm_bins)
## Make normalizer and formatter
norm = matplotlib.colors.BoundaryNorm(norm_bins, n, clip=True)
# normalizer = Normalize(np.array([x for x in value_to_int.values()])[0],np.array([x for x in value_to_int.values()])[-1])
# im=cm.ScalarMappable(norm=normalizer)
if axes is None:
fig, axes = plt.subplots(2, 1, sharex=True, sharey=False)
# divider = make_axes_locatable([axes[0], axes[1]])
# cbar_ax = divider.append_axes('right', size='5%', pad=0.05)
cbar_ax = fig.add_axes(cbar_params)
sns.heatmap(df1.replace(value_to_int), cmap=cmap_list, cbar=False,
ax=axes[0], linewidth=0.7, linecolor='k', square=True,
cbar_kws={"shrink": .9}, cbar_ax=cbar_ax, norm=norm)
sns.heatmap(df2.replace(value_to_int), cmap=cmap_list, cbar=False,
ax=axes[1], linewidth=0.7, linecolor='k', square=True,
cbar_kws={"shrink": .9}, cbar_ax=cbar_ax, norm=norm)
# else:
# ax = sns.heatmap(df.replace(sorted_v_to_i), cmap=cmap,
# ax=ax, linewidth=1, linecolor='k',
# square=False, cbar_kws={"shrink": .9})
if title is not None:
axes[0].set_title(title, fontsize=fontsize)
for ax in axes:
ax.set_xticklabels(ax.get_xticklabels(), ha='right', va='top', rotation=45)
ax.set_yticklabels(ax.get_yticklabels(), rotation=0)
ax.tick_params(labelsize=fontsize, direction='out', bottom=True,
left=True, length=2)
ax.set_ylabel(ax.get_ylabel(), fontsize=fontsize)
ax.set_xlabel(ax.get_xlabel(), fontsize=fontsize)
# colorbar = axes[0].collections[0].colorbar
# diff = norm_bins[1:] - norm_bins[:-1]
# tickz = norm_bins[:-1] + diff / 2
colorbar = fig.colorbar(cm.ScalarMappable(norm=norm, cmap=matplotlib.colors.ListedColormap(cmap_list)), ax=[axes[0], axes[1]],
shrink=1, pad=0.05, cax=cbar_ax)
# colorbar = plt.gca().images[-1].colorbar
r = colorbar.vmax - colorbar.vmin
colorbar.set_ticks([colorbar.vmin + r / n * (0.5 + i) for i in range(n)])
colorbar.ax.set_yticklabels(cbar_ticklabels, fontsize=fontsize-2)
return axes
def plot_hyper_parameters_heatmap_data_splits_per_model(dss4, dss5, fontsize=14,
save=True, model_name='SVC',
features='pwv+pressure+doy'):
import matplotlib.pyplot as plt
# import seaborn as sns
fig, axes = plt.subplots(2, 5, sharex=True, sharey=False ,figsize=(16, 5))
ds4 = dss4.sel(features=features).reset_coords(drop=True)
ds5 = dss5.sel(features=features).reset_coords(drop=True)
ds4 = ds4.reindex(scorer=scorer_order)
ds5 = ds5.reindex(scorer=scorer_order)
non_hp_vars = ['mean_score', 'std_score',
'test_score', 'roc_auc_score', 'TPR']
if model_name == 'RF':
non_hp_vars.append('feature_importances')
if model_name == 'MLP':
adj_dict=dict(
top=0.946,
bottom=0.145,
left=0.046,
right=0.937,
hspace=0.121,
wspace=0.652)
cb_st = 0.167
cb_mul = 0.193
else:
adj_dict=dict(
wspace = 0.477,
top=0.921,
bottom=0.17,
left=0.046,
right=0.937,
hspace=0.121)
cb_st = 0.18
cb_mul = 0.19
ds4 = ds4[[x for x in ds4 if x not in non_hp_vars]]
ds5 = ds5[[x for x in ds5 if x not in non_hp_vars]]
seq = 'Blues'
cat = 'Dark2'
hp_dict = {
'alpha': ['Reds', 'float'], 'activation': ['Set1_r', 'str'],
'hidden_layer_sizes': ['Paired', 'str'], 'learning_rate': ['Spectral_r', 'str'],
'solver': ['Dark2', 'str'], 'kernel': ['Dark2', 'str'], 'C': ['Blues', 'float'],
'gamma': ['Oranges', 'float'], 'degree': ['Greens', 'str'], 'coef0': ['Spectral', 'str'],
'max_depth': ['Blues', 'int'], 'max_features': ['Dark2', 'str'],
'min_samples_leaf': ['Greens', 'int'], 'min_samples_split': ['Reds', 'int'],
'n_estimators': ['Oranges', 'int']
}
# fix stuff for SVC:
if model_name == 'SVC':
ds4['degree'] = ds4['degree'].where(ds4['kernel']=='poly')
ds4['coef0'] = ds4['coef0'].where(ds4['kernel']=='poly')
ds5['degree'] = ds5['degree'].where(ds5['kernel']=='poly')
ds5['coef0'] = ds5['coef0'].where(ds5['kernel']=='poly')
for i, (da4, da5) in enumerate(zip(ds4, ds5)):
df4 = ds4[da4].reset_coords(drop=True).to_dataset('scorer').to_dataframe()
df5 = ds5[da5].reset_coords(drop=True).to_dataset('scorer').to_dataframe()
df4.index.name = 'Outer Split'
df5.index.name = 'Outer Split'
# try:
# df4 = df4.astype(float).round(2)
# df5 = df5.astype(float).round(2)
# except ValueError:
# pass
cmap = hp_dict.get(da4, 'Set1')[0]
val_type = hp_dict.get(da4, 'int')[1]
cbar_params = [cb_st + cb_mul*float(i), .175, .01, .71]
plot_heatmaps_for_hyper_parameters_data_splits(df4,
df5,
axes=[axes[0, i], axes[1, i]],
fig=fig,
title=da4,
cmap=cmap,
cbar_params=cbar_params,
fontsize=fontsize,
val_type=val_type)
if i > 0 :
axes[0, i].set_ylabel('')
axes[0, i].yaxis.set_tick_params(labelleft=False)
axes[1, i].set_ylabel('')
axes[1, i].yaxis.set_tick_params(labelleft=False)
fig.tight_layout()
fig.subplots_adjust(**adj_dict)
if save:
filename = 'Hyper-parameters_nested_{}.png'.format(
model_name)
plt.savefig(savefig_path / filename, bbox_inches='tight')
return fig
def plot_heatmap_for_hyper_parameters_df(df, ax=None, cmap='colorblind',
title=None, fontsize=12):
import pandas as pd
import seaborn as sns
import numpy as np
sns.set_style('ticks')
sns.set_style('whitegrid')
sns.set(font_scale=1.2)
value_to_int = {j: i for i, j in enumerate(
sorted(pd.unique(df.values.ravel())))} # like you did
# for key in value_to_int.copy().keys():
# try:
# if np.isnan(key):
# value_to_int['NA'] = value_to_int.pop(key)
# df = df.fillna('NA')
# except TypeError:
# pass
try:
sorted_v_to_i = dict(sorted(value_to_int.items()))
except TypeError:
sorted_v_to_i = value_to_int
n = len(value_to_int)
# discrete colormap (n samples from a given cmap)
cmap = sns.color_palette(cmap, n)
if ax is None:
ax = sns.heatmap(df.replace(sorted_v_to_i), cmap=cmap,
linewidth=1, linecolor='k', square=False,
cbar_kws={"shrink": .9})
else:
ax = sns.heatmap(df.replace(sorted_v_to_i), cmap=cmap,
ax=ax, linewidth=1, linecolor='k',
square=False, cbar_kws={"shrink": .9})
if title is not None:
ax.set_title(title, fontsize=fontsize)
ax.set_xticklabels(ax.get_xticklabels(), rotation=30)
ax.set_yticklabels(ax.get_yticklabels(), rotation=0)
ax.tick_params(labelsize=fontsize)
ax.set_ylabel(ax.get_ylabel(), fontsize=fontsize)
ax.set_xlabel(ax.get_xlabel(), fontsize=fontsize)
colorbar = ax.collections[0].colorbar
r = colorbar.vmax - colorbar.vmin
colorbar.set_ticks([colorbar.vmin + r / n * (0.5 + i) for i in range(n)])
colorbar.set_ticklabels(list(value_to_int.keys()))
return ax
# def plot_ROC_curves_for_all_models_and_scorers(dss, save=False,
# fontsize=24, fig_split=1,
# feat=['pwv', 'pwv+pressure', 'pwv+pressure+doy']):
# import xarray as xr
# import seaborn as sns
# import matplotlib.pyplot as plt
# import pandas as pd
# cmap = sns.color_palette('tab10', len(feat))
# sns.set_style('whitegrid')
# sns.set_style('ticks')
# if fig_split == 1:
# dss = dss.sel(scorer=['precision', 'recall', 'f1'])
# elif fig_split == 2:
# dss = dss.sel(scorer=['accuracy', 'tss', 'hss'])
# fg = xr.plot.FacetGrid(
# dss,
# col='model',
# row='scorer',
# sharex=True,
# sharey=True, figsize=(20, 20))
# for i in range(fg.axes.shape[0]): # i is rows
# for j in range(fg.axes.shape[1]): # j is cols
# ax = fg.axes[i, j]
# modelname = dss['model'].isel(model=j).item()
# scorer = dss['scorer'].isel(scorer=i).item()
# chance_plot = [False for x in feat]
# chance_plot[-1] = True
# for k, f in enumerate(feat):
# # name = '{}-{}-{}'.format(modelname, scoring, feat)
# # model = dss.isel({'model': j, 'scoring': i}).sel(
# # {'features': feat})
# model = dss.isel({'model': j, 'scorer': i}
# ).sel({'features': f})
# # return model
# title = 'ROC of {} model ({})'.format(modelname.replace('SVC', 'SVM'), scorer)
# try:
# ax = plot_ROC_curve_from_dss_nested_CV(model, outer_dim='outer_split',
# plot_chance=[k],
# main_label=f,
# ax=ax,
# color=cmap[k], title=title,
# fontsize=fontsize)
# except ValueError:
# ax.grid('on')
# continue
# handles, labels = ax.get_legend_handles_labels()
# lh_ser = pd.Series(labels, index=handles).drop_duplicates()
# lh_ser = lh_ser.sort_values(ascending=False)
# hand = lh_ser.index.values
# labe = lh_ser.values
# ax.legend(handles=hand.tolist(), labels=labe.tolist(), loc="lower right",
# fontsize=fontsize-7)
# ax.grid('on')
# if j >= 1:
# ax.set_ylabel('')
# if fig_split == 1:
# ax.set_xlabel('')
# ax.tick_params(labelbottom=False)
# else:
# if i <= 1:
# ax.set_xlabel('')
# # title = '{} station: {} total events'.format(
# # station.upper(), events)
# # if max_flow > 0:
# # title = '{} station: {} total events (max flow = {} m^3/sec)'.format(
# # station.upper(), events, max_flow)
# # fg.fig.suptitle(title, fontsize=fontsize)
# fg.fig.tight_layout()
# fg.fig.subplots_adjust(top=0.937,
# bottom=0.054,
# left=0.039,
# right=0.993,
# hspace=0.173,
# wspace=0.051)
# if save:
# filename = 'ROC_curves_nested_{}_figsplit_{}.png'.format(
# dss['outer_split'].size, fig_split)
# plt.savefig(savefig_path / filename, bbox_inches='tight')
# return fg
def plot_hydro_ML_models_results_from_dss(dss, std_on='outer',
save=False, fontsize=16,
plot_type='ROC', split=1,
feat=['pwv', 'pressure+pwv', 'doy+pressure+pwv']):
import xarray as xr
import seaborn as sns
import matplotlib.pyplot as plt
import pandas as pd
cmap = sns.color_palette("colorblind", len(feat))
if split == 1:
dss = dss.sel(scoring=['f1', 'precision', 'recall'])
elif split == 2:
dss = dss.sel(scoring=['tss', 'hss', 'roc-auc', 'accuracy'])
fg = xr.plot.FacetGrid(
dss,
col='model',
row='scoring',
sharex=True,
sharey=True, figsize=(20, 20))
for i in range(fg.axes.shape[0]): # i is rows
for j in range(fg.axes.shape[1]): # j is cols
ax = fg.axes[i, j]
modelname = dss['model'].isel(model=j).item()
scoring = dss['scoring'].isel(scoring=i).item()
chance_plot = [False for x in feat]
chance_plot[-1] = True
for k, f in enumerate(feat):
# name = '{}-{}-{}'.format(modelname, scoring, feat)
# model = dss.isel({'model': j, 'scoring': i}).sel(
# {'features': feat})
model = dss.isel({'model': j, 'scoring': i}
).sel({'features': f})
title = '{} of {} model ({})'.format(
plot_type, modelname, scoring)
try:
plot_ROC_PR_curve_from_dss(model, outer_dim='outer_kfold',
inner_dim='inner_kfold',
plot_chance=[k],
main_label=f, plot_type=plot_type,
plot_std_legend=False, ax=ax,
color=cmap[k], title=title,
std_on=std_on, fontsize=fontsize)
except ValueError:
ax.grid('on')
continue
handles, labels = ax.get_legend_handles_labels()
hand = pd.Series(
labels, index=handles).drop_duplicates().index.values
labe = pd.Series(labels, index=handles).drop_duplicates().values
ax.legend(handles=hand.tolist(), labels=labe.tolist(), loc="lower right",
fontsize=14)
ax.grid('on')
# title = '{} station: {} total events'.format(
# station.upper(), events)
# if max_flow > 0:
# title = '{} station: {} total events (max flow = {} m^3/sec)'.format(
# station.upper(), events, max_flow)
# fg.fig.suptitle(title, fontsize=fontsize)
fg.fig.tight_layout()
fg.fig.subplots_adjust(top=0.937,
bottom=0.054,
left=0.039,
right=0.993,
hspace=0.173,
wspace=0.051)
if save:
filename = 'hydro_models_on_{}_{}_std_on_{}_{}.png'.format(
dss['inner_kfold'].size, dss['outer_kfold'].size,
std_on, plot_type)
plt.savefig(savefig_path / filename, bbox_inches='tight')
return fg
# def plot_hydro_ML_models_result(model_da, nsplits=2, station='drag',
# test_size=20, n_splits_plot=None, save=False):
# import xarray as xr
# import seaborn as sns
# import matplotlib.pyplot as plt
# from sklearn.model_selection import train_test_split
# # TODO: add plot_roc_curve(model, X_other_station, y_other_station)
# # TODO: add pw_station, hs_id
# cmap = sns.color_palette("colorblind", 3)
# X, y = produce_X_y(station, hydro_pw_dict[station], neg_pos_ratio=1)
# events = int(y[y == 1].sum().item())
# model_da = model_da.sel(
# splits=nsplits,
# test_size=test_size).reset_coords(
# drop=True)
## just_pw = [x for x in X.feature.values if 'pressure' not in x]
## X_pw = X.sel(feature=just_pw)
# fg = xr.plot.FacetGrid(
# model_da,
# col='model',
# row='scoring',
# sharex=True,
# sharey=True, figsize=(20, 20))
# for i in range(fg.axes.shape[0]): # i is rows
# for j in range(fg.axes.shape[1]): # j is cols
# ax = fg.axes[i, j]
# modelname = model_da['model'].isel(model=j).item()
# scoring = model_da['scoring'].isel(scoring=i).item()
# chance_plot = [False, False, True]
# for k, feat in enumerate(model_da['feature'].values):
# name = '{}-{}-{}'.format(modelname, scoring, feat)
# model = model_da.isel({'model': j, 'scoring': i}).sel({'feature': feat}).item()
# title = 'ROC of {} model ({})'.format(modelname, scoring)
# if not '+' in feat:
# f = [x for x in X.feature.values if feat in x]
# X_f = X.sel(feature=f)
# else:
# X_f = X
# X_train, X_test, y_train, y_test = train_test_split(
# X_f, y, test_size=test_size/100, shuffle=True, random_state=42)
#
# plot_many_ROC_curves(model, X_f, y, name=name,
# color=cmap[k], ax=ax,
# plot_chance=chance_plot[k],
# title=title, n_splits=n_splits_plot)
# fg.fig.suptitle('{} station: {} total_events, test_events = {}, n_splits = {}'.format(station.upper(), events, int(events* test_size/100), nsplits))
# fg.fig.tight_layout()
# fg.fig.subplots_adjust(top=0.937,
# bottom=0.054,
# left=0.039,
# right=0.993,
# hspace=0.173,
# wspace=0.051)
# if save:
# plt.savefig(savefig_path / 'try.png', bbox_inches='tight')
# return fg
def order_features_list(flist):
""" order the feature list in load_ML_run_results
so i don't get duplicates"""
import pandas as pd
import numpy as np
# first get all features:
li = [x.split('+') for x in flist]
flat_list = [item for sublist in li for item in sublist]
f = list(set(flat_list))
nums = np.arange(1, len(f)+1)
# now assagin a number for each entry:
inds = []
for x in flist:
for fe, num in zip(f, nums):
x = x.replace(fe, str(10**num))
inds.append(eval(x))
ser = pd.Series(inds)
ser.index = flist
ser1 = ser.drop_duplicates()
di = dict(zip(ser1.values, ser1.index))
new_flist = []
for ind, feat in zip(inds, flist):
new_flist.append(di.get(ind))
return new_flist
def smart_add_dataarray_to_ds_list(dsl, da_name='feature_importances'):
"""add data array to ds_list even if it does not exist, use shape of
data array that exists in other part of ds list"""
import numpy as np
import xarray as xr
# print(da_name)
fi = [x for x in dsl if da_name in x][0]
print(da_name, fi[da_name].shape)
fi = fi[da_name].copy(data=np.zeros(shape=fi[da_name].shape))
new_dsl = []
for ds in dsl:
if da_name not in ds:
ds = xr.merge([ds, fi], combine_attrs='no_conflicts')
new_dsl.append(ds)
return new_dsl
def load_ML_run_results(path=hydro_ml_path, prefix='CVR',
change_DOY_to_doy=True):
from aux_gps import path_glob
import xarray as xr
# from aux_gps import save_ncfile
import pandas as pd
import numpy as np
print('loading hydro ML results for all models and features')
# print('loading hydro ML results for station {}'.format(pw_station))
model_files = path_glob(path, '{}_*.nc'.format(prefix))
model_files = sorted(model_files)
# model_files = [x for x in model_files if pw_station in x.as_posix()]
ds_list = [xr.load_dataset(x) for x in model_files]
if change_DOY_to_doy:
for ds in ds_list:
if 'DOY' in ds.features:
new_feats = [x.replace('DOY', 'doy') for x in ds['feature'].values]
ds['feature'] = new_feats
ds.attrs['features'] = [x.replace('DOY', 'doy') for x in ds.attrs['features']]
model_as_str = [x.as_posix().split('/')[-1].split('.')[0]
for x in model_files]
model_names = [x.split('_')[1] for x in model_as_str]
model_scores = [x.split('_')[3] for x in model_as_str]
model_features = [x.split('_')[2] for x in model_as_str]
if change_DOY_to_doy:
model_features = [x.replace('DOY', 'doy') for x in model_features]
new_model_features = order_features_list(model_features)
ind = pd.MultiIndex.from_arrays(
[model_names,
new_model_features,
model_scores],
names=(
'model',
'features',
'scoring'))
# ind1 = pd.MultiIndex.from_product([model_names, model_scores, model_features], names=[
# 'model', 'scoring', 'feature'])
# ds_list = [x[data_vars] for x in ds_list]
# complete non-existant fields like best and fi for all ds:
data_vars = [x for x in ds_list[0] if x.startswith('test')]
# data_vars += ['AUC', 'TPR']
data_vars += [x for x in ds_list[0] if x.startswith('y_')]
bests = [[x for x in y if x.startswith('best')] for y in ds_list]
data_vars += list(set([y for x in bests for y in x]))
if 'RF' in model_names:
data_vars += ['feature_importances']
new_ds_list = []
for dvar in data_vars:
ds_list = smart_add_dataarray_to_ds_list(ds_list, dvar)
# # check if all data vars are in each ds and merge them:
new_ds_list = [xr.merge([y[x] for x in data_vars if x in y],
combine_attrs='no_conflicts') for y in ds_list]
# concat all
dss = xr.concat(new_ds_list, dim='dim_0')
dss['dim_0'] = ind
dss = dss.unstack('dim_0')
# dss.attrs['pwv_id'] = pw_station
# fix roc_auc to roc-auc in dss datavars
dss = dss.rename_vars({'test_roc_auc': 'test_roc-auc'})
# dss['test_roc_auc'].name = 'test_roc-auc'
print('calculating ROC, PR metrics.')
dss = calculate_metrics_from_ML_dss(dss)
print('Done!')
return dss
def plot_nested_CV_test_scores(dss, feats=None, fontsize=16,
save=True, wv_label='pwv'):
import seaborn as sns
import matplotlib.pyplot as plt
from aux_gps import convert_da_to_long_form_df
import numpy as np
import xarray as xr
def change_width(ax, new_value) :
for patch in ax.patches :
current_width = patch.get_width()
diff = current_width - new_value
# we change the bar width
patch.set_width(new_value)
# we recenter the bar
patch.set_x(patch.get_x() + diff * .5)
def show_values_on_bars(axs, fs=12, fw='bold', exclude_bar_num=None):
import numpy as np
def _show_on_single_plot(ax, exclude_bar_num=3):
for i, p in enumerate(ax.patches):
if i != exclude_bar_num and exclude_bar_num is not None:
_x = p.get_x() + p.get_width() / 2
_y = p.get_y() + p.get_height()
value = '{:.2f}'.format(p.get_height())
ax.text(_x, _y, value, ha="right",
fontsize=fs, fontweight=fw, zorder=20)
if isinstance(axs, np.ndarray):
for idx, ax in np.ndenumerate(axs):
_show_on_single_plot(ax, exclude_bar_num)
else:
_show_on_single_plot(axs, exclude_bar_num)
splits = dss['outer_split'].size
try:
assert 'best' in dss.attrs['comment']
best = True
except AssertionError:
best = False
except KeyError:
best = False
if 'neg_sample' in dss.dims:
neg = dss['neg_sample'].size
else:
neg = 1
if 'model' not in dss.dims:
dss = dss.expand_dims('model')
dss['model'] = [dss.attrs['model']]
dss = dss.sortby('model', ascending=False)
dss = dss.reindex(scorer=scorer_order)
if feats is None:
feats = ['pwv', 'pwv+pressure', 'pwv+pressure+doy']
dst = dss.sel(features=feats) # .reset_coords(drop=True)
# df = dst['test_score'].to_dataframe()
# df['scorer'] = df.index.get_level_values(3)
# df['model'] = df.index.get_level_values(0)
# df['features'] = df.index.get_level_values(1)
# df['outer_splits'] = df.index.get_level_values(2)
# df['model'] = df['model'].str.replace('SVC', 'SVM')
# df = df.melt(value_vars='test_score', id_vars=[
# 'features', 'model', 'scorer', 'outer_splits'], var_name='test_score',
# value_name='score')
da = dst['test_score']
if len(feats) == 5:
da_empty = da.isel(features=0).copy(
data=np.zeros(da.isel(features=0).shape))
da_empty['features'] = 'empty'
da = xr.concat([da, da_empty], 'features')
da = da.reindex(features=['doy', 'pressure', 'pwv',
'empty', 'pwv+pressure', 'pwv+pressure+doy'])
da.name = 'feature groups'
df = convert_da_to_long_form_df(da, value_name='score',
var_name='feature groups')
sns.set(font_scale=1.5)
sns.set_style('whitegrid')
sns.set_style('ticks')
cmap = sns.color_palette('tab10', n_colors=len(feats))
if len(feats) == 5:
cmap = ['tab:purple', 'tab:brown', 'tab:blue', 'tab:blue',
'tab:orange', 'tab:green']
fg = sns.FacetGrid(data=df, row='model', col='scorer', height=4, aspect=0.9)
# fg.map_dataframe(sns.stripplot, x="test_score", y="score", hue="features",
# data=df, dodge=True, alpha=1, zorder=1, palette=cmap)
# fg.map_dataframe(sns.pointplot, x="test_score", y="score", hue="features",
# data=df, dodge=True, join=False, palette=cmap,
# markers="o", scale=.75, ci=None)
fg.map_dataframe(sns.barplot, x='feature groups', y="score", hue='features',
ci='sd', capsize=None, errwidth=2, errcolor='k',
palette=cmap, dodge=True)
# g = sns.catplot(x='test_score', y="score", hue='features',
# col="scorer", row='model', ci='sd',
# data=df, kind="bar", capsize=0.25,
# height=4, aspect=1.5, errwidth=1.5)
#fg.set_xticklabels(rotation=45)
# fg.set_yticklabels([0, 0.2, 0.4, 0.6, 0.8, 1.0], fontsize=fontsize)
fg.set_ylabels('score')
[x.grid(True) for x in fg.axes.flatten()]
handles, labels = fg.axes[0, 0].get_legend_handles_labels()
if len(feats) == 5:
del handles[3]
del labels[3]
show_values_on_bars(fg.axes, fs=fontsize-4, exclude_bar_num=3)
for i in range(fg.axes.shape[0]): # i is rows
model = dss['model'].isel(model=i).item()
if model == 'SVC':
model = 'SVM'
for j in range(fg.axes.shape[1]): # j is cols
ax = fg.axes[i, j]
scorer = dss['scorer'].isel(scorer=j).item()
title = '{} | scorer={}'.format(model, scorer)
ax.set_title(title, fontsize=fontsize)
ax.set_xlabel('')
ax.set_ylim(0, 1)
change_width(ax, 0.110)
fg.set_xlabels(' ')
if wv_label is not None:
labels = [x.replace('pwv', wv_label) for x in labels]
fg.fig.legend(handles=handles, labels=labels, prop={'size': fontsize}, edgecolor='k',
framealpha=0.5, fancybox=True, facecolor='white',
ncol=len(feats), fontsize=fontsize, loc='upper center', bbox_to_anchor=(0.5, 1.005),
bbox_transform=plt.gcf().transFigure)
# true_scores = dst.sel(scorer=scorer, model=model)['true_score']
# dss['permutation_score'].plot.hist(ax=ax, bins=25, color=color)
# ymax = ax.get_ylim()[-1] - 0.2
# ax.vlines(x=true_scores.values, ymin=0, ymax=ymax, linestyle='--', color=cmap)
fg.fig.tight_layout()
fg.fig.subplots_adjust(top=0.92)
if save:
if best:
filename = 'ML_scores_models_nested_CV_best_hp_{}_{}_neg_{}.png'.format('_'.join(feats), splits, neg)
else:
filename = 'ML_scores_models_nested_CV_{}_{}_neg_{}.png'.format('_'.join(feats), splits, neg)
plt.savefig(savefig_path / filename, bbox_inches='tight')
return fg
def plot_holdout_test_scores(dss, feats='pwv+pressure+doy'):
import seaborn as sns
import matplotlib.pyplot as plt
def show_values_on_bars(axs, fs=12, fw='bold'):
import numpy as np
def _show_on_single_plot(ax):
for p in ax.patches:
_x = p.get_x() + p.get_width() / 2
_y = p.get_y() + p.get_height()
value = '{:.2f}'.format(p.get_height())
ax.text(_x, _y, value, ha="center", fontsize=fs, fontweight=fw)
if isinstance(axs, np.ndarray):
for idx, ax in np.ndenumerate(axs):
_show_on_single_plot(ax)
else:
_show_on_single_plot(axs)
if feats is None:
feats = ['pwv', 'pwv+pressure', 'pwv+pressure+doy']
dst = dss.sel(features=feats) # .reset_coords(drop=True)
df = dst['holdout_test_scores'].to_dataframe()
df['scorer'] = df.index.droplevel(1).droplevel(0)
df['model'] = df.index.droplevel(2).droplevel(1)
df['features'] = df.index.droplevel(2).droplevel(0)
df['model'] = df['model'].str.replace('SVC', 'SVM')
df = df.melt(value_vars='holdout_test_scores', id_vars=[
'features', 'model', 'scorer'], var_name='test_score')
sns.set(font_scale=1.5)
sns.set_style('whitegrid')
sns.set_style('ticks')
g = sns.catplot(x="model", y="value", hue='features',
col="scorer", ci='sd', row=None,
col_wrap=3,
data=df, kind="bar", capsize=0.15,
height=4, aspect=1.5, errwidth=0.8)
g.set_xticklabels(rotation=45)
[x.grid(True) for x in g.axes.flatten()]
show_values_on_bars(g.axes)
filename = 'ML_scores_models_holdout_{}.png'.format('_'.join(feats))
plt.savefig(savefig_path / filename, bbox_inches='tight')
return df
def prepare_test_df_to_barplot_from_dss(dss, feats='doy+pwv+pressure',
plot=True, splitfigs=True):
import seaborn as sns
import matplotlib.pyplot as plt
dvars = [x for x in dss if 'test_' in x]
scores = [x.split('_')[-1] for x in dvars]
dst = dss[dvars]
# dst['scoring'] = [x+'_inner' for x in dst['scoring'].values]
# for i, ds in enumerate(dst):
# dst[ds] = dst[ds].sel(scoring=scores[i]).reset_coords(drop=True)
if feats is None:
feats = ['pwv', 'pressure+pwv', 'doy+pressure+pwv']
dst = dst.sel(features=feats) # .reset_coords(drop=True)
dst = dst.rename_vars(dict(zip(dvars, scores)))
# dst = dst.drop('scoring')
df = dst.to_dataframe()
# dfu = df
df['inner score'] = df.index.droplevel(2).droplevel(1).droplevel(0)
df['features'] = df.index.droplevel(2).droplevel(2).droplevel(1)
df['model'] = df.index.droplevel(2).droplevel(0).droplevel(1)
df = df.melt(value_vars=scores, id_vars=[
'features', 'model', 'inner score'], var_name='outer score')
# return dfu
# dfu.columns = dfu.columns.droplevel(1)
# dfu = dfu.T
# dfu['score'] = dfu.index
# dfu = dfu.reset_index()
# df = dfu.melt(value_vars=['MLP', 'RF', 'SVC'], id_vars=['score'])
df1 = df[(df['inner score']=='f1') | (df['inner score']=='precision') | (df['inner score']=='recall')]
df2 = df[(df['inner score']=='hss') | (df['inner score']=='tss') | (df['inner score']=='roc-auc') | (df['inner score']=='accuracy')]
if plot:
sns.set(font_scale = 1.5)
sns.set_style('whitegrid')
sns.set_style('ticks')
if splitfigs:
g = sns.catplot(x="outer score", y="value", hue='features',
col="inner score", ci='sd',row='model',
data=df1, kind="bar", capsize=0.15,
height=4, aspect=1.5,errwidth=0.8)
g.set_xticklabels(rotation=45)
filename = 'ML_scores_models_{}_1.png'.format('_'.join(feats))
plt.savefig(savefig_path / filename, bbox_inches='tight')
g = sns.catplot(x="outer score", y="value", hue='features',
col="inner score", ci='sd',row='model',
data=df2, kind="bar", capsize=0.15,
height=4, aspect=1.5,errwidth=0.8)
g.set_xticklabels(rotation=45)
filename = 'ML_scores_models_{}_2.png'.format('_'.join(feats))
plt.savefig(savefig_path / filename, bbox_inches='tight')
else:
g = sns.catplot(x="outer score", y="value", hue='features',
col="inner score", ci='sd',row='model',
data=df, kind="bar", capsize=0.15,
height=4, aspect=1.5,errwidth=0.8)
g.set_xticklabels(rotation=45)
filename = 'ML_scores_models_{}.png'.format('_'.join(feats))
plt.savefig(savefig_path / filename, bbox_inches='tight')
return df
def calculate_metrics_from_ML_dss(dss):
from sklearn.metrics import roc_curve
from sklearn.metrics import roc_auc_score
from sklearn.metrics import auc
from sklearn.metrics import precision_recall_curve
import xarray as xr
import numpy as np
import pandas as pd
mean_fpr = np.linspace(0, 1, 100)
# fpr = dss['y_true'].copy(deep=False).values
# tpr = dss['y_true'].copy(deep=False).values
# y_true = dss['y_true'].values
# y_prob = dss['y_prob'].values
ok = [x for x in dss['outer_kfold'].values]
ik = [x for x in dss['inner_kfold'].values]
m = [x for x in dss['model'].values]
sc = [x for x in dss['scoring'].values]
f = [x for x in dss['features'].values]
# r = [x for x in dss['neg_pos_ratio'].values]
ind = pd.MultiIndex.from_product(
[ok, ik, m, sc, f],
names=[
'outer_kfold',
'inner_kfold',
'model',
'scoring',
'features']) # , 'station'])
okn = [x for x in range(dss['outer_kfold'].size)]
ikn = [x for x in range(dss['inner_kfold'].size)]
mn = [x for x in range(dss['model'].size)]
scn = [x for x in range(dss['scoring'].size)]
fn = [x for x in range(dss['features'].size)]
ds_list = []
for i in okn:
for j in ikn:
for k in mn:
for n in scn:
for m in fn:
ds = xr.Dataset()
y_true = dss['y_true'].isel(
outer_kfold=i, inner_kfold=j, model=k, scoring=n, features=m).reset_coords(drop=True).squeeze()
y_prob = dss['y_prob'].isel(
outer_kfold=i, inner_kfold=j, model=k, scoring=n, features=m).reset_coords(drop=True).squeeze()
y_true = y_true.dropna('sample')
y_prob = y_prob.dropna('sample')
if y_prob.size == 0:
# in case of NaNs in the results:
fpr_da = xr.DataArray(
np.nan*np.ones((1)), dims=['sample'])
fpr_da['sample'] = [
x for x in range(fpr_da.size)]
tpr_da = xr.DataArray(
np.nan*np.ones((1)), dims=['sample'])
tpr_da['sample'] = [
x for x in range(tpr_da.size)]
prn_da = xr.DataArray(
np.nan*np.ones((1)), dims=['sample'])
prn_da['sample'] = [
x for x in range(prn_da.size)]
rcll_da = xr.DataArray(
np.nan*np.ones((1)), dims=['sample'])
rcll_da['sample'] = [
x for x in range(rcll_da.size)]
tpr_fpr = xr.DataArray(
np.nan*np.ones((100)), dims=['FPR'])
tpr_fpr['FPR'] = mean_fpr
prn_rcll = xr.DataArray(
np.nan*np.ones((100)), dims=['RCLL'])
prn_rcll['RCLL'] = mean_fpr
pr_auc_da = xr.DataArray(np.nan)
roc_auc_da = xr.DataArray(np.nan)
no_skill_da = xr.DataArray(np.nan)
else:
no_skill = len(
y_true[y_true == 1]) / len(y_true)
no_skill_da = xr.DataArray(no_skill)
fpr, tpr, _ = roc_curve(y_true, y_prob)
interp_tpr = np.interp(mean_fpr, fpr, tpr)
interp_tpr[0] = 0.0
roc_auc = roc_auc_score(y_true, y_prob)
prn, rcll, _ = precision_recall_curve(
y_true, y_prob)
interp_prn = np.interp(
mean_fpr, rcll[::-1], prn[::-1])
interp_prn[0] = 1.0
pr_auc_score = auc(rcll, prn)
roc_auc_da = xr.DataArray(roc_auc)
pr_auc_da = xr.DataArray(pr_auc_score)
prn_da = xr.DataArray(prn, dims=['sample'])
prn_da['sample'] = [x for x in range(len(prn))]
rcll_da = xr.DataArray(rcll, dims=['sample'])
rcll_da['sample'] = [
x for x in range(len(rcll))]
fpr_da = xr.DataArray(fpr, dims=['sample'])
fpr_da['sample'] = [x for x in range(len(fpr))]
tpr_da = xr.DataArray(tpr, dims=['sample'])
tpr_da['sample'] = [x for x in range(len(tpr))]
tpr_fpr = xr.DataArray(
interp_tpr, dims=['FPR'])
tpr_fpr['FPR'] = mean_fpr
prn_rcll = xr.DataArray(
interp_prn, dims=['RCLL'])
prn_rcll['RCLL'] = mean_fpr
ds['fpr'] = fpr_da
ds['tpr'] = tpr_da
ds['roc-auc'] = roc_auc_da
ds['pr-auc'] = pr_auc_da
ds['prn'] = prn_da
ds['rcll'] = rcll_da
ds['TPR'] = tpr_fpr
ds['PRN'] = prn_rcll
ds['no_skill'] = no_skill_da
ds_list.append(ds)
ds = xr.concat(ds_list, 'dim_0')
ds['dim_0'] = ind
ds = ds.unstack()
ds.attrs = dss.attrs
ds['fpr'].attrs['long_name'] = 'False positive rate'
ds['tpr'].attrs['long_name'] = 'True positive rate'
ds['prn'].attrs['long_name'] = 'Precision'
ds['rcll'].attrs['long_name'] = 'Recall'
ds['roc-auc'].attrs['long_name'] = 'ROC or FPR-TPR Area under curve'
ds['pr-auc'].attrs['long_name'] = 'Precition-Recall Area under curve'
ds['PRN'].attrs['long_name'] = 'Precision-Recall'
ds['TPR'].attrs['long_name'] = 'TPR-FPR (ROC)'
dss = xr.merge([dss, ds], combine_attrs='no_conflicts')
return dss
#
# def load_ML_models(path=hydro_ml_path, station='drag', prefix='CVM', suffix='.pkl'):
# from aux_gps import path_glob
# import joblib
# import matplotlib.pyplot as plt
# import seaborn as sns
# import xarray as xr
# import pandas as pd
# model_files = path_glob(path, '{}_*{}'.format(prefix, suffix))
# model_files = sorted(model_files)
# model_files = [x for x in model_files if station in x.as_posix()]
# m_list = [joblib.load(x) for x in model_files]
# model_files = [x.as_posix().split('/')[-1].split('.')[0] for x in model_files]
# # fix roc-auc:
# model_files = [x.replace('roc_auc', 'roc-auc') for x in model_files]
# print('loading {} station only.'.format(station))
# model_names = [x.split('_')[3] for x in model_files]
## model_pw_stations = [x.split('_')[1] for x in model_files]
## model_hydro_stations = [x.split('_')[2] for x in model_files]
# model_nsplits = [x.split('_')[6] for x in model_files]
# model_scores = [x.split('_')[5] for x in model_files]
# model_features = [x.split('_')[4] for x in model_files]
# model_test_sizes = []
# for file in model_files:
# try:
# model_test_sizes.append(int(file.split('_')[7]))
# except IndexError:
# model_test_sizes.append(20)
## model_pwv_hs_id = list(zip(model_pw_stations, model_hydro_stations))
## model_pwv_hs_id = ['_'.join(x) for filename = 'CVR_{}_{}_{}_{}_{}.nc'.format(
# name, features, refitted_scorer, ikfolds, okfolds)
# x in model_pwv_hs_id]
# # transform model_dict to dataarray:
# tups = [tuple(x) for x in zip(model_names, model_scores, model_nsplits, model_features, model_test_sizes)] #, model_pwv_hs_id)]
# ind = pd.MultiIndex.from_tuples((tups), names=['model', 'scoring', 'splits', 'feature', 'test_size']) #, 'station'])
# da = xr.DataArray(m_list, dims='dim_0')
# da['dim_0'] = ind
# da = da.unstack('dim_0')
# da['splits'] = da['splits'].astype(int)
# da['test_size'].attrs['units'] = '%'
# return da
def plot_heatmaps_for_all_models_and_scorings(dss, var='roc-auc'): # , save=True):
import xarray as xr
import seaborn as sns
import matplotlib.pyplot as plt
# assert station == dss.attrs['pwv_id']
cmaps = {'roc-auc': sns.color_palette("Blues", as_cmap=True),
'pr-auc': sns.color_palette("Greens", as_cmap=True)}
fg = xr.plot.FacetGrid(
dss,
col='model',
row='scoring',
sharex=True,
sharey=True, figsize=(10, 20))
dss = dss.mean('inner_kfold', keep_attrs=True)
vmin, vmax = dss[var].min(), 1
norm = plt.Normalize(vmin=vmin, vmax=vmax)
for i in range(fg.axes.shape[0]): # i is rows
for j in range(fg.axes.shape[1]): # j is cols
ax = fg.axes[i, j]
modelname = dss['model'].isel(model=j).item()
scoring = dss['scoring'].isel(scoring=i).item()
model = dss[var].isel(
{'model': j, 'scoring': i}).reset_coords(drop=True)
df = model.to_dataframe()
title = '{} model ({})'.format(modelname, scoring)
df = df.unstack()
mean = df.mean()
mean.name = 'mean'
df = df.append(mean).T.droplevel(0)
ax = sns.heatmap(df, annot=True, cmap=cmaps[var], cbar=False,
ax=ax, norm=norm)
ax.set_title(title)
ax.vlines([4], 0, 10, color='r', linewidth=2)
if j > 0:
ax.set_ylabel('')
if i < 2:
ax.set_xlabel('')
cax = fg.fig.add_axes([0.1, 0.025, .8, .015])
fg.fig.colorbar(ax.get_children()[0], cax=cax, orientation="horizontal")
fg.fig.suptitle('{}'.format(
dss.attrs[var].upper()), fontweight='bold')
fg.fig.tight_layout()
fg.fig.subplots_adjust(top=0.937,
bottom=0.099,
left=0.169,
right=0.993,
hspace=0.173,
wspace=0.051)
# if save:
# filename = 'hydro_models_heatmaps_on_{}_{}_{}.png'.format(
# station, dss['outer_kfold'].size, var)
# plt.savefig(savefig_path / filename, bbox_inches='tight')
return fg
def plot_ROC_from_dss(dss, feats=None, fontsize=16, save=True, wv_label='pwv',
best=False):
import seaborn as sns
import matplotlib.pyplot as plt
import pandas as pd
from aux_gps import convert_da_to_long_form_df
sns.set_style('whitegrid')
sns.set_style('ticks')
sns.set(font_scale=1.0)
cmap = sns.color_palette('tab10', n_colors=3)
splits = dss['outer_split'].size
if 'neg_sample' in dss.dims:
neg = dss['neg_sample'].size
else:
neg = 1
dss = dss.reindex(scorer=scorer_order)
if feats is None:
feats = ['pwv', 'pwv+pressure', 'pwv+pressure+doy']
if 'model' not in dss.dims:
dss = dss.expand_dims('model')
dss['model'] = [dss.attrs['model']]
dss = dss.sortby('model', ascending=False)
dst = dss.sel(features=feats) # .reset_coords(drop=True)
# df = dst['TPR'].to_dataframe()
# if 'neg_sample' in dss.dims:
# fpr_lnum = 5
# model_lnum = 0
# scorer_lnum = 4
# features_lnum = 1
# else:
# fpr_lnum = 4
# model_lnum = 0
# scorer_lnum = 3
# features_lnum = 1
# df['FPR'] = df.index.get_level_values(fpr_lnum)
# df['model'] = df.index.get_level_values(model_lnum)
# df['scorer'] = df.index.get_level_values(scorer_lnum)
# df['features'] = df.index.get_level_values(features_lnum)
df = convert_da_to_long_form_df(dst['TPR'], var_name='score')
# df = df.melt(value_vars='TPR', id_vars=[
# 'features', 'model', 'scorer', 'FPR'], var_name='score')
if best is not None:
if best == 'compare_negs':
df1 = df.copy()[df['neg_sample'] == 1]
df2 = df.copy()
df2.drop('neg_sample', axis=1, inplace=True)
df1.drop('neg_sample', axis=1, inplace=True)
df1['neg_group'] = 1
df2['neg_group'] = 25
df = pd.concat([df1, df2])
col = 'neg_group'
titles = ['Neg=1', 'Neg=25']
else:
col=None
else:
col = 'scorer'
df['model'] = df['model'].str.replace('SVC', 'SVM')
fg = sns.FacetGrid(df, col=col, row='model', aspect=1)
fg.map_dataframe(sns.lineplot, x='FPR', y='value',
hue='features', ci='sd', palette=cmap, n_boot=None,
estimator='mean')
for i in range(fg.axes.shape[0]): # i is rows
model = dss['model'].isel(model=i).item()
auc_model = dst.sel(model=model)
if model == 'SVC':
model = 'SVM'
for j in range(fg.axes.shape[1]): # j is cols
scorer = dss['scorer'].isel(scorer=j).item()
auc_scorer_df = auc_model['roc_auc_score'].sel(scorer=scorer).reset_coords(drop=True).to_dataframe()
auc_scorer_mean = [auc_scorer_df.loc[x].mean() for x in feats]
auc_scorer_std = [auc_scorer_df.loc[x].std() for x in feats]
auc_mean = [x.item() for x in auc_scorer_mean]
auc_std = [x.item() for x in auc_scorer_std]
if j == 0 and best is not None:
scorer = dss['scorer'].isel(scorer=j).item()
auc_scorer_df = auc_model['roc_auc_score'].sel(scorer=scorer).isel(neg_sample=0).reset_coords(drop=True).to_dataframe()
auc_scorer_mean = [auc_scorer_df.loc[x].mean() for x in feats]
auc_scorer_std = [auc_scorer_df.loc[x].std() for x in feats]
auc_mean = [x.item() for x in auc_scorer_mean]
auc_std = [x.item() for x in auc_scorer_std]
ax = fg.axes[i, j]
ax.plot([0, 1], [0, 1], color='tab:red', linestyle='--', lw=2,
label='chance')
if best is not None:
if best == 'compare_negs':
title = '{} | {}'.format(model, titles[j])
else:
title = '{}'.format(model)
else:
title = '{} | scorer={}'.format(model, scorer)
ax.set_title(title, fontsize=fontsize)
handles, labels = ax.get_legend_handles_labels()
hands = handles[0:3]
# labes = labels[0:3]
new_labes = []
for auc, auc_sd in zip(auc_mean, auc_std):
l = r'{:.2}$\pm${:.1}'.format(auc, auc_sd)
new_labes.append(l)
ax.legend(handles=hands, labels=new_labes, loc='lower right',
title='AUCs', prop={'size': fontsize-4})
ax.set_xticks([0, 0.2, 0.4, 0.6, 0.8, 1])
ax.grid(True)
# return handles, labels
fg.set_ylabels('True Positive Rate', fontsize=fontsize)
fg.set_xlabels('False Positive Rate', fontsize=fontsize)
if wv_label is not None:
labels = [x.replace('pwv', wv_label) for x in labels]
if best is not None:
if best == 'compare_negs':
fg.fig.legend(handles=handles, labels=labels, prop={'size': fontsize},
edgecolor='k',
framealpha=0.5, fancybox=True, facecolor='white',
ncol=2, fontsize=fontsize, loc='upper center', bbox_to_anchor=(0.5, 1.005),
bbox_transform=plt.gcf().transFigure)
fg.fig.tight_layout()
fg.fig.subplots_adjust(top=0.865,
bottom=0.079,
left=0.144,
right=0.933,
hspace=0.176,
wspace=0.2)
else:
fg.fig.legend(handles=handles, labels=labels, prop={'size': fontsize},
edgecolor='k',
framealpha=0.5, fancybox=True, facecolor='white',
ncol=1, fontsize=fontsize, loc='upper center', bbox_to_anchor=(0.5, 1.005),
bbox_transform=plt.gcf().transFigure)
fg.fig.tight_layout()
fg.fig.subplots_adjust(top=0.825,
bottom=0.079,
left=0.184,
right=0.933,
hspace=0.176,
wspace=0.2)
else:
fg.fig.legend(handles=handles, labels=labels, prop={'size': fontsize}, edgecolor='k',
framealpha=0.5, fancybox=True, facecolor='white',
ncol=5, fontsize=fontsize, loc='upper center', bbox_to_anchor=(0.5, 1.005),
bbox_transform=plt.gcf().transFigure)
# true_scores = dst.sel(scorer=scorer, model=model)['true_score']
# dss['permutation_score'].plot.hist(ax=ax, bins=25, color=color)
# ymax = ax.get_ylim()[-1] - 0.2
# ax.vlines(x=true_scores.values, ymin=0, ymax=ymax, linestyle='--', color=cmap)
fg.fig.tight_layout()
fg.fig.subplots_adjust(top=0.915)
if save:
if best is not None:
filename = 'ROC_plots_models_nested_CV_best_hp_{}_{}_neg_{}.png'.format('_'.join(feats), splits, neg)
else:
filename = 'ROC_plots_models_nested_CV_{}_{}_neg_{}.png'.format('_'.join(feats), splits, neg)
plt.savefig(savefig_path / filename, bbox_inches='tight')
return fg
def plot_permutation_importances_from_dss(dss, feat_dim='features',
outer_dim='outer_split',
features='pwv+pressure+doy',
fix_xticklabels=True,split=1,
axes=None, save=True):
import matplotlib.pyplot as plt
import numpy as np
import seaborn as sns
from natsort import natsorted
sns.set_palette('Dark2', 6)
sns.set_style('whitegrid')
sns.set_style('ticks')
model = dss.attrs['model']
# use dss.sel(model='RF') first as input
dss['feature'] = dss['feature'].str.replace('DOY', 'doy')
dss = dss.sel({feat_dim: features})
# tests_ds = dss['test_score']
# tests_ds = tests_ds.sel(scorer=scorer)
# max_score_split = int(tests_ds.idxmax(outer_dim).item())
# use mean outer split:
# dss = dss.mean(outer_dim)
dss = dss.sel({outer_dim: split})
feats = features.split('+')
fn = len(feats)
if fn == 1:
gr_spec = None
fix_xticklabels = False
elif fn == 2:
gr_spec = [1, 1]
elif fn == 3:
gr_spec = [2, 5, 5]
if axes is None:
fig, axes = plt.subplots(1, fn, sharey=True, figsize=(17, 5), gridspec_kw={'width_ratios': gr_spec})
try:
axes.flatten()
except AttributeError:
axes = [axes]
for i, f in enumerate(sorted(feats)):
fe = [x for x in dss['feature'].values if f in x]
dsf = dss['PI_mean'].sel(
feature=fe).reset_coords(
drop=True)
sorted_feat = natsorted([x for x in dsf.feature.values])
dsf = dsf.reindex(feature=sorted_feat)
print([x for x in dsf.feature.values])
# dsf = dss['PI_mean'].sel(
# feature=fe).reset_coords(
# drop=True)
dsf = dsf.to_dataset('scorer').to_dataframe(
).reset_index(drop=True)
title = '{}'.format(f.upper())
dsf.plot.bar(ax=axes[i], title=title, rot=0, legend=False, zorder=20,
width=.8)
dsf_sum = dsf.sum().tolist()
handles, labels = axes[i].get_legend_handles_labels()
labels = [
'{} ({:.1f})'.format(
x, y) for x, y in zip(
labels, dsf_sum)]
axes[i].legend(handles=handles, labels=labels, prop={'size': 10}, loc='upper left')
axes[i].set_ylabel('Scores')
axes[i].grid(axis='y', zorder=1)
if fix_xticklabels:
n = sum(['pwv' in x for x in dss.feature.values])
axes[0].xaxis.set_ticklabels('')
hrs = np.arange(-24, -24+n)
axes[1].set_xticklabels(hrs, rotation=30, ha="center", fontsize=12)
axes[2].set_xticklabels(hrs, rotation=30, ha="center", fontsize=12)
axes[1].set_xlabel('Hours prior to flood')
axes[2].set_xlabel('Hours prior to flood')
fig.tight_layout()
fig.suptitle('permutation importance scores for {} model split #{}'.format(model, split))
fig.subplots_adjust(top=0.904)
if save:
filename = 'permutation_importances_{}_split_{}_all_scorers_{}.png'.format(model, split, features)
plt.savefig(savefig_path / filename, bbox_inches='tight')
return
def plot_feature_importances_from_dss(
dss,
feat_dim='features', outer_dim='outer_split',
features='pwv+pressure+doy', fix_xticklabels=True,
axes=None, save=True, ylim=[0, 12], fontsize=16):
import matplotlib.pyplot as plt
import numpy as np
import seaborn as sns
from natsort import natsorted
sns.set_palette('Dark2', 6)
# sns.set_style('whitegrid')
# sns.set_style('ticks')
sns.set_theme(style='ticks', font_scale=1.5)
# use dss.sel(model='RF') first as input
dss['feature'] = dss['feature'].str.replace('DOY', 'doy')
dss = dss.sel({feat_dim: features})
# tests_ds = dss['test_score']
# tests_ds = tests_ds.sel(scorer=scorer)
# max_score_split = int(tests_ds.idxmax(outer_dim).item())
# use mean outer split:
dss = dss.mean(outer_dim)
feats = features.split('+')
fn = len(feats)
if fn == 1:
gr_spec = None
fix_xticklabels = False
elif fn == 2:
gr_spec = [1, 1]
elif fn == 3:
gr_spec = [5, 5, 2]
if axes is None:
fig, axes = plt.subplots(1, fn, sharey=True, figsize=(17, 5), gridspec_kw={'width_ratios': gr_spec})
try:
axes.flatten()
except AttributeError:
axes = [axes]
for i, f in enumerate(feats):
fe = [x for x in dss['feature'].values if f in x]
dsf = dss['feature_importances'].sel(
feature=fe).reset_coords(
drop=True)
# dsf = dss['PI_mean'].sel(
# feature=fe).reset_coords(
# drop=True)
sorted_feat = natsorted([x for x in dsf.feature.values])
# sorted_feat = [x for x in dsf.feature.values]
print(sorted_feat)
dsf = dsf.reindex(feature=sorted_feat)
dsf = dsf.to_dataset('scorer').to_dataframe(
).reset_index(drop=True) * 100
title = '{}'.format(f.upper())
dsf.plot.bar(ax=axes[i], title=title, rot=0, legend=False, zorder=20,
width=.8)
axes[i].set_title(title, fontsize=fontsize)
dsf_sum = dsf.sum().tolist()
handles, labels = axes[i].get_legend_handles_labels()
labels = [
'{} ({:.1f} %)'.format(
x, y) for x, y in zip(
labels, dsf_sum)]
axes[i].legend(handles=handles, labels=labels, prop={'size': 12}, loc='upper center')
axes[i].set_ylabel('Feature importances [%]')
axes[i].grid(axis='y', zorder=1)
if ylim is not None:
[ax.set_ylim(*ylim) for ax in axes]
if fix_xticklabels:
n = sum(['pwv' in x for x in dss.feature.values])
axes[2].xaxis.set_ticklabels('')
hrs = np.arange(-1, -25, -1)
axes[0].set_xticklabels(hrs, rotation=30, ha="center", fontsize=14)
axes[1].set_xticklabels(hrs, rotation=30, ha="center", fontsize=14)
axes[2].tick_params(labelsize=fontsize)
axes[0].set_xlabel('Hours prior to flood')
axes[1].set_xlabel('Hours prior to flood')
fig.tight_layout()
if save:
filename = 'RF_feature_importances_all_scorers_{}.png'.format(features)
plt.savefig(savefig_path / filename, bbox_inches='tight')
return
def plot_feature_importances(
dss,
feat_dim='features',
features='pwv+pressure+doy',
scoring='f1', fix_xticklabels=True,
axes=None, save=True):
# use dss.sel(model='RF') first as input
import matplotlib.pyplot as plt
import numpy as np
dss = dss.sel({feat_dim: features})
tests_ds = dss[[x for x in dss if 'test' in x]]
tests_ds = tests_ds.sel(scoring=scoring)
score_ds = tests_ds['test_{}'.format(scoring)]
max_score = score_ds.idxmax('outer_kfold').values
feats = features.split('+')
fn = len(feats)
if axes is None:
fig, axes = plt.subplots(1, fn, sharey=True, figsize=(17, 5), gridspec_kw={'width_ratios': [1, 4, 4]})
try:
axes.flatten()
except AttributeError:
axes = [axes]
for i, f in enumerate(feats):
fe = [x for x in dss['feature'].values if f in x]
dsf = dss['feature_importances'].sel(
feature=fe,
outer_kfold=max_score).reset_coords(
drop=True)
dsf = dsf.to_dataset('scoring').to_dataframe(
).reset_index(drop=True) * 100
title = '{} ({})'.format(f.upper(), scoring)
dsf.plot.bar(ax=axes[i], title=title, rot=0, legend=False, zorder=20,
width=.8)
dsf_sum = dsf.sum().tolist()
handles, labels = axes[i].get_legend_handles_labels()
labels = [
'{} ({:.1f} %)'.format(
x, y) for x, y in zip(
labels, dsf_sum)]
axes[i].legend(handles=handles, labels=labels, prop={'size': 8})
axes[i].set_ylabel('Feature importance [%]')
axes[i].grid(axis='y', zorder=1)
if fix_xticklabels:
axes[0].xaxis.set_ticklabels('')
hrs = np.arange(-24,0)
axes[1].set_xticklabels(hrs, rotation = 30, ha="center", fontsize=12)
axes[2].set_xticklabels(hrs, rotation = 30, ha="center", fontsize=12)
axes[1].set_xlabel('Hours prior to flood')
axes[2].set_xlabel('Hours prior to flood')
if save:
fig.tight_layout()
filename = 'RF_feature_importances_{}.png'.format(scoring)
plt.savefig(savefig_path / filename, bbox_inches='tight')
return
def plot_feature_importances_for_all_scorings(dss,
features='doy+pwv+pressure',
model='RF', splitfigs=True):
import matplotlib.pyplot as plt
# station = dss.attrs['pwv_id'].upper()
dss = dss.sel(model=model).reset_coords(drop=True)
fns = len(features.split('+'))
scores = dss['scoring'].values
scores1 = ['f1', 'precision', 'recall']
scores2 = ['hss', 'tss', 'accuracy','roc-auc']
if splitfigs:
fig, axes = plt.subplots(len(scores1), fns, sharey=True, figsize=(15, 20))
for i, score in enumerate(scores1):
plot_feature_importances(
dss, features=features, scoring=score, axes=axes[i, :])
fig.suptitle(
'feature importances of {} model'.format(model))
fig.tight_layout()
fig.subplots_adjust(top=0.935,
bottom=0.034,
left=0.039,
right=0.989,
hspace=0.19,
wspace=0.027)
filename = 'RF_feature_importances_1.png'
plt.savefig(savefig_path / filename, bbox_inches='tight')
fig, axes = plt.subplots(len(scores2), fns, sharey=True, figsize=(15, 20))
for i, score in enumerate(scores2):
plot_feature_importances(
dss, features=features, scoring=score, axes=axes[i, :])
fig.suptitle(
'feature importances of {} model'.format(model))
fig.tight_layout()
fig.subplots_adjust(top=0.935,
bottom=0.034,
left=0.039,
right=0.989,
hspace=0.19,
wspace=0.027)
filename = 'RF_feature_importances_2.png'
plt.savefig(savefig_path / filename, bbox_inches='tight')
else:
fig, axes = plt.subplots(len(scores), fns, sharey=True, figsize=(15, 20))
for i, score in enumerate(scores):
plot_feature_importances(
dss, features=features, scoring=score, axes=axes[i, :])
fig.suptitle(
'feature importances of {} model'.format(model))
fig.tight_layout()
fig.subplots_adjust(top=0.935,
bottom=0.034,
left=0.039,
right=0.989,
hspace=0.19,
wspace=0.027)
filename = 'RF_feature_importances.png'
plt.savefig(savefig_path / filename, bbox_inches='tight')
return dss
def plot_ROC_curve_from_dss_nested_CV(dss, outer_dim='outer_split',
plot_chance=True, color='tab:blue',
fontsize=14, plot_legend=True,
title=None,
ax=None, main_label=None):
import matplotlib.pyplot as plt
import numpy as np
if ax is None:
fig, ax = plt.subplots()
if title is None:
title = "Receiver operating characteristic"
mean_fpr = dss['FPR'].values
mean_tpr = dss['TPR'].mean(outer_dim).values
mean_auc = dss['roc_auc_score'].mean().item()
if np.isnan(mean_auc):
return ValueError
std_auc = dss['roc_auc_score'].std().item()
field = 'TPR'
xlabel = 'False Positive Rate'
ylabel = 'True Positive Rate'
if main_label is None:
main_label = r'Mean ROC (AUC={:.2f}$\pm${:.2f})'.format(mean_auc, std_auc)
textstr = '\n'.join(['{}'.format(
main_label), r'(AUC={:.2f}$\pm${:.2f})'.format(mean_auc, std_auc)])
main_label = textstr
ax.plot(mean_fpr, mean_tpr, color=color,
lw=3, alpha=.8, label=main_label)
std_tpr = dss[field].std(outer_dim).values
n = dss[outer_dim].size
tprs_upper = np.minimum(mean_tpr + std_tpr, 1)
tprs_lower = np.maximum(mean_tpr - std_tpr, 0)
# plot Chance line:
if plot_chance:
ax.plot([0, 1], [0, 1], linestyle='--', lw=2, color='r',
label='Chance', alpha=.8, zorder=206)
stdlabel = r'$\pm$ 1 Std. dev.'
stdstr = '\n'.join(['{}'.format(stdlabel), r'({} outer splits)'.format(n)])
ax.fill_between(
mean_fpr,
tprs_lower,
tprs_upper,
color='grey',
alpha=.2, label=stdstr)
ax.grid()
ax.set(xlim=[-0.05, 1.05], ylim=[-0.05, 1.05])
# ax.set_title(title, fontsize=fontsize)
ax.tick_params(axis='y', labelsize=fontsize)
ax.tick_params(axis='x', labelsize=fontsize)
ax.set_xlabel(xlabel, fontsize=fontsize)
ax.set_ylabel(ylabel, fontsize=fontsize)
ax.set_title(title, fontsize=fontsize)
return ax
def plot_ROC_PR_curve_from_dss(
dss,
outer_dim='outer_kfold',
inner_dim='inner_kfold',
plot_chance=True,
ax=None,
color='b',
title=None,
std_on='inner',
main_label=None,
fontsize=14,
plot_type='ROC',
plot_std_legend=True):
"""plot classifier metrics, plot_type=ROC or PR"""
import matplotlib.pyplot as plt
import numpy as np
if ax is None:
fig, ax = plt.subplots()
if title is None:
title = "Receiver operating characteristic"
if plot_type == 'ROC':
mean_fpr = dss['FPR'].values
mean_tpr = dss['TPR'].mean(outer_dim).mean(inner_dim).values
mean_auc = dss['roc-auc'].mean().item()
if np.isnan(mean_auc):
return ValueError
std_auc = dss['roc-auc'].std().item()
field = 'TPR'
xlabel = 'False Positive Rate'
ylabel = 'True Positive Rate'
elif plot_type == 'PR':
mean_fpr = dss['RCLL'].values
mean_tpr = dss['PRN'].mean(outer_dim).mean(inner_dim).values
mean_auc = dss['pr-auc'].mean().item()
if np.isnan(mean_auc):
return ValueError
std_auc = dss['pr-auc'].std().item()
no_skill = dss['no_skill'].mean(outer_dim).mean(inner_dim).item()
field = 'PRN'
xlabel = 'Recall'
ylabel = 'Precision'
# plot mean ROC:
if main_label is None:
main_label = r'Mean {} (AUC={:.2f}$\pm${:.2f})'.format(
plot_type, mean_auc, std_auc)
else:
textstr = '\n'.join(['Mean ROC {}'.format(
main_label), r'(AUC={:.2f}$\pm${:.2f})'.format(mean_auc, std_auc)])
main_label = textstr
ax.plot(mean_fpr, mean_tpr, color=color,
lw=2, alpha=.8, label=main_label)
if std_on == 'inner':
std_tpr = dss[field].mean(outer_dim).std(inner_dim).values
n = dss[inner_dim].size
elif std_on == 'outer':
std_tpr = dss[field].mean(inner_dim).std(outer_dim).values
n = dss[outer_dim].size
elif std_on == 'all':
std_tpr = dss[field].stack(
dumm=[inner_dim, outer_dim]).std('dumm').values
n = dss[outer_dim].size * dss[inner_dim].size
tprs_upper = np.minimum(mean_tpr + std_tpr, 1)
tprs_lower = np.maximum(mean_tpr - std_tpr, 0)
# plot Chance line:
if plot_chance:
if plot_type == 'ROC':
ax.plot([0, 1], [0, 1], linestyle='--', lw=2, color='r',
label='Chance', alpha=.8)
elif plot_type == 'PR':
ax.plot([0, 1], [no_skill, no_skill], linestyle='--', color='r',
lw=2, label='No Skill', alpha=.8)
# plot ROC STD range:
ax.fill_between(
mean_fpr,
tprs_lower,
tprs_upper,
color='grey',
alpha=.2, label=r'$\pm$ 1 std. dev. ({} {} splits)'.format(n, std_on))
ax.grid()
ax.set(xlim=[-0.05, 1.05], ylim=[-0.05, 1.05])
ax.set_title(title, fontsize=fontsize)
ax.tick_params(axis='y', labelsize=fontsize)
ax.tick_params(axis='x', labelsize=fontsize)
ax.set_xlabel(xlabel, fontsize=fontsize)
ax.set_ylabel(ylabel, fontsize=fontsize)
# handles, labels = ax.get_legend_handles_labels()
# if not plot_std_legend:
# if len(handles) == 7:
# handles = handles[:-2]
# labels = labels[:-2]
# else:
# handles = handles[:-1]
# labels = labels[:-1]
# ax.legend(handles=handles, labels=labels, loc="lower right",
# fontsize=fontsize)
return ax
def load_cv_splits_from_pkl(savepath):
import joblib
from aux_gps import path_glob
file = path_glob(savepath, 'CV_inds_*.pkl')[0]
n_splits = int(file.as_posix().split('/')[-1].split('_')[2])
shuffle = file.as_posix().split('/')[-1].split('.')[0].split('=')[-1]
cv_dict = joblib.load(file)
spl = len([x for x in cv_dict.keys()])
assert spl == n_splits
print('loaded {} with {} splits.'.format(file, n_splits))
return cv_dict
def save_cv_splits_to_dict(X, y, cv, train_key='train', test_key='test',
savepath=None):
import joblib
cv_dict = {}
for i, (train, test) in enumerate(cv.split(X, y)):
cv_dict[i+1] = {train_key: train, test_key: test}
# check for completness:
all_train = [x['train'] for x in cv_dict.values()]
flat_train = set([item for sublist in all_train for item in sublist])
all_test = [x['test'] for x in cv_dict.values()]
flat_test = set([item for sublist in all_test for item in sublist])
assert flat_test == flat_train
if savepath is not None:
filename = 'CV_inds_{}_splits_shuffle={}.pkl'.format(cv.n_splits, cv.shuffle)
joblib.dump(cv_dict, savepath / filename)
print('saved {} to {}.'.format(filename, savepath))
return cv_dict
def plot_many_ROC_curves(model, X, y, name='', color='b', ax=None,
plot_chance=True, title=None, n_splits=None):
from sklearn.metrics import plot_roc_curve
import matplotlib.pyplot as plt
from sklearn.metrics import roc_auc_score
from sklearn.metrics import f1_score
from sklearn.metrics import roc_curve
from sklearn.metrics import auc
import numpy as np
from sklearn.model_selection import StratifiedKFold
if ax is None:
fig, ax = plt.subplots()
if title is None:
title = "Receiver operating characteristic"
# just plot the ROC curve for X, y, no nsplits and stats:
if n_splits is None:
viz = plot_roc_curve(model, X, y, color=color, ax=ax, name=name)
else:
cv = StratifiedKFold(n_splits=n_splits, shuffle=True, random_state=42)
tprs = []
aucs = []
mean_fpr = np.linspace(0, 1, 100)
for i, (train, val) in enumerate(cv.split(X, y)):
model.fit(X[train], y[train])
# y_score = model.fit(X[train], y[train]).predict_proba(X[val])[:, 1]
y_pred = model.predict(X[val])
fpr, tpr, _ = roc_curve(y[val], y_pred)
# viz = plot_roc_curve(model, X[val], y[val],
# name='ROC fold {}'.format(i),
# alpha=0.3, lw=1, ax=ax)
# fpr = viz.fpr
# tpr = viz.tpr
interp_tpr = np.interp(mean_fpr, fpr, tpr)
interp_tpr[0] = 0.0
tprs.append(interp_tpr)
aucs.append(roc_auc_score(y[val], y_pred))
# scores.append(f1_score(y[val], y_pred))
# scores = np.array(scores)
mean_tpr = np.mean(tprs, axis=0)
mean_tpr[-1] = 1.0
mean_auc = auc(mean_fpr, mean_tpr)
std_auc = np.std(aucs)
ax.plot(mean_fpr, mean_tpr, color=color,
label=r'Mean ROC (AUC = %0.2f $\pm$ %0.2f)' % (
mean_auc, std_auc),
lw=2, alpha=.8)
std_tpr = np.std(tprs, axis=0)
tprs_upper = np.minimum(mean_tpr + std_tpr, 1)
tprs_lower = np.maximum(mean_tpr - std_tpr, 0)
ax.fill_between(mean_fpr, tprs_lower, tprs_upper, color='grey', alpha=.2,
label=r'$\pm$ 1 std. dev.')
if plot_chance:
ax.plot([0, 1], [0, 1], linestyle='--', lw=2, color='r',
label='Chance', alpha=.8)
ax.set(xlim=[-0.05, 1.05], ylim=[-0.05, 1.05],
title=title)
ax.legend(loc="lower right")
return ax
def HP_tuning(X, y, model_name='SVC', val_size=0.18, n_splits=None,
test_size=None,
best_score='f1', seed=42, savepath=None):
from sklearn.model_selection import GridSearchCV
from sklearn.model_selection import StratifiedKFold
""" do HP tuning with ML_Classfier_Switcher object and return a DataSet of
results. note that the X, y are already after split to val/test"""
# first get the features from X:
features = list(set(['_'.join(x.split('_')[0:2])
for x in X['feature'].values]))
ml = ML_Classifier_Switcher()
sk_model = ml.pick_model(model_name)
param_grid = ml.param_grid
if n_splits is None and val_size is not None:
n_splits = int((1 // val_size) - 1)
elif val_size is not None and n_splits is not None:
raise('Both val_size and n_splits are defined, choose either...')
print('StratifiedKfolds of {}.'.format(n_splits))
cv = StratifiedKFold(n_splits=n_splits, shuffle=True)
gr = GridSearchCV(estimator=sk_model, param_grid=param_grid, cv=cv,
n_jobs=-1, scoring=['f1', 'roc_auc', 'accuracy'], verbose=1,
refit=best_score, return_train_score=True)
gr.fit(X, y)
if best_score is not None:
ds, best_model = process_gridsearch_results(gr, model_name,
features=features, pwv_id=X.attrs['pwv_id'], hs_id=y.attrs['hydro_station_id'], test_size=test_size)
else:
ds = process_gridsearch_results(gr, model_name, features=features,
pwv_id=X.attrs['pwv_id'], hs_id=y.attrs['hydro_station_id'], test_size=test_size)
best_model = None
if savepath is not None:
save_cv_results(ds, best_model=best_model, savepath=savepath)
return ds, best_model
def save_gridsearchcv_object(GridSearchCV, savepath, filename):
import joblib
print('{} was saved to {}'.format(filename, savepath))
joblib.dump(GridSearchCV, savepath / filename)
return
def run_RF_feature_importance_on_all_features(path=hydro_path, gr_path=hydro_ml_path/'holdout'):
import xarray as xr
from aux_gps import get_all_possible_combinations_from_list
feats = get_all_possible_combinations_from_list(
['pwv', 'pressure', 'doy'], reduce_single_list=True, combine_by_sep='+')
feat_list = []
for feat in feats:
da = holdout_test(model_name='RF', return_RF_FI=True, features=feat)
feat_list.append(da)
daa = xr.concat(feat_list, 'features')
daa['features'] = feats
return daa
def load_nested_CV_test_results_from_all_models(path=hydro_ml_path, best=False,
neg=1, splits=4,
permutation=False):
from aux_gps import path_glob
import xarray as xr
if best:
if splits is not None:
file_str = 'nested_CV_test_results_*_all_features_with_hyper_params_best_hp_neg_{}_{}a.nc'.format(neg, splits)
if permutation:
file_str = 'nested_CV_test_results_*_all_features_permutation_tests_best_hp_neg_{}_{}a.nc'.format(neg, splits)
else:
if splits is not None:
file_str = 'nested_CV_test_results_*_all_features_with_hyper_params_neg_{}_{}a.nc'.format(neg, splits)
if permutation:
file_str = 'nested_CV_test_results_*_all_features_permutation_tests_neg_{}_{}a.nc'.format(neg, splits)
files = path_glob(path, file_str)
print(files)
models = [x.as_posix().split('/')[-1].split('_')[4] for x in files]
print('loading CV test results only for {} models'.format(', '.join(models)))
dsl = [xr.load_dataset(x) for x in files]
if not permutation:
dsl = [x[['mean_score', 'std_score', 'test_score', 'roc_auc_score', 'TPR']] for x in dsl]
dss = xr.concat(dsl, 'model')
dss['model'] = models
return dss
# def plot_all_permutation_test_results(dss, feats=None):
# import xarray as xr
# fg = xr.plot.FacetGrid(
# dss,
# col='scorer',
# row='model',
# sharex=True,
# sharey=True, figsize=(20, 20))
# for i in range(fg.axes.shape[0]): # i is rows
# model = dss['model'].isel(model=i).item()
# for j in range(fg.axes.shape[1]): # j is cols
# ax = fg.axes[i, j]
# scorer = dss['scorer'].isel(scorer=j).item()
# ax = plot_single_permutation_test_result(dss, feats=feats,
# scorer=scorer,
# model=model,
# ax=ax)
# fg.fig.tight_layout()
# return fg
def plot_permutation_test_results_from_dss(dss, feats=None, fontsize=14,
save=True, wv_label='pwv'):
# ax=None, scorer='f1', model='MLP'):
import matplotlib.pyplot as plt
import seaborn as sns
from PW_from_gps_figures import get_legend_labels_handles_title_seaborn_histplot
from aux_gps import convert_da_to_long_form_df
sns.set_style('whitegrid')
sns.set_style('ticks')
try:
splits = dss['outer_split'].size
except KeyError:
splits = 5
try:
assert 'best' in dss.attrs['comment']
best = True
except AssertionError:
best = False
if 'neg_sample' in dss.dims:
neg = dss['neg_sample'].size
else:
neg = 1
if 'model' not in dss.dims:
dss = dss.expand_dims('model')
dss['model'] = [dss.attrs['model']]
dss = dss.reindex(scorer=scorer_order)
# dss = dss.mean('outer_split')
cmap = sns.color_palette('tab10', n_colors=3)
if feats is None:
feats = ['pwv', 'pwv+pressure', 'pwv+pressure+doy']
dss = dss.sortby('model', ascending=False)
dst = dss.sel(features=feats) # .reset_coords(drop=True)
# df = dst[['permutation_score', 'true_score', 'pvalue']].to_dataframe()
# df['permutations'] = df.index.get_level_values(2)
# df['scorer'] = df.index.get_level_values(3)
# df['features'] = df.index.get_level_values(0)
# df['model'] = df.index.get_level_values(1)
# df['model'] = df['model'].str.replace('SVC', 'SVM')
# df = df.melt(value_vars=['permutation_score', 'true_score', 'pvalue'], id_vars=[
# 'features', 'model', 'scorer'], var_name='scores')
df = convert_da_to_long_form_df(dst[['permutation_score', 'true_score', 'pvalue']], var_name='scores')
df_p = df[df['scores'] == 'permutation_score']
df_pval = df[df['scores'] == 'pvalue']
# if ax is None:
# fig, ax = plt.subplots(figsize=(6, 8))
fg = sns.FacetGrid(df_p, col='scorer', row='model', legend_out=True,
sharex=False)
fg.map_dataframe(sns.histplot, x="value", hue="features",
legend=True, palette=cmap,
stat='density', kde=True,
element='bars', bins=10)
# pvals = dst.sel(scorer=scorer, model=model)[
# 'pvalue'].reset_coords(drop=True)
# pvals = pvals.values
# handles, labels, title = get_legend_labels_handles_title_seaborn_histplot(ax)
# new_labels = []
# for pval, label in zip(pvals, labels):
# label += ' (p={:.1})'.format(pval)
# new_labels.append(label)
# ax.legend(handles, new_labels, title=title)
df_t = df[df['scores'] == 'true_score']
for i in range(fg.axes.shape[0]): # i is rows
model = dss['model'].isel(model=i).item()
df_model = df_t[df_t['model'] == model]
df_pval_model = df_pval[df_pval['model'] == model]
for j in range(fg.axes.shape[1]): # j is cols
scorer = dss['scorer'].isel(scorer=j).item()
df1 = df_model[df_model['scorer'] == scorer]
df2 = df_pval_model[df_pval_model['scorer'] == scorer]
ax = fg.axes[i, j]
ymax = ax.get_ylim()[-1] - 0.2
plabels = []
for k, feat in enumerate(feats):
val = df1[df1['features']==feat]['value'].unique().item()
pval = df2[df2['features']==feat]['value'].unique().item()
plabels.append('pvalue: {:.2g}'.format(pval))
# print(i, val, feat, scorer, model)
ax.axvline(x=val, ymin=0, ymax=ymax, linestyle='--', color=cmap[k],
label=feat)
handles, labels = ax.get_legend_handles_labels()
ax.legend(handles=handles, labels=plabels,
prop={'size': fontsize-4}, loc='upper left')
if 'hss' in scorer or 'tss' in scorer:
ax.set_xlim(-0.35, 1)
else:
ax.set_xlim(0.15, 1)
# ax.set_xticks([0.3, 0.4, 0.5, 0.6, 0.7, 0.8, 0.9, 0.1])
# handles, labels, title = get_legend_labels_handles_title_seaborn_histplot(ax)
if model == 'SVC':
model = 'SVM'
title = '{} | scorer={}'.format(model, scorer)
ax.set_title(title, fontsize=fontsize)
# ax.set_xlim(-0.3, 1)
fg.set_ylabels('Density', fontsize=fontsize)
fg.set_xlabels('Score', fontsize=fontsize)
if wv_label is not None:
labels = [x.replace('pwv', wv_label) for x in labels]
fg.fig.legend(handles=handles, labels=labels, prop={'size': fontsize}, edgecolor='k',
framealpha=0.5, fancybox=True, facecolor='white',
ncol=5, fontsize=fontsize, loc='upper center', bbox_to_anchor=(0.5, 1.005),
bbox_transform=plt.gcf().transFigure)
# true_scores = dst.sel(scorer=scorer, model=model)['true_score']
# dss['permutation_score'].plot.hist(ax=ax, bins=25, color=color)
# ymax = ax.get_ylim()[-1] - 0.2
# ax.vlines(x=true_scores.values, ymin=0, ymax=ymax, linestyle='--', color=cmap)
fg.fig.tight_layout()
fg.fig.subplots_adjust(top=0.92)
if save:
if best:
filename = 'permutation_test_models_nested_CV_best_hp_{}_{}_neg_{}.png'.format('_'.join(feats), splits, neg)
else:
filename = 'permutation_test_models_nested_CV_{}_{}_neg_{}.png'.format('_'.join(feats), splits, neg)
plt.savefig(savefig_path / filename, bbox_inches='tight')
return fg
def run_CV_nested_tests_on_all_features(path=hydro_path, gr_path=hydro_ml_path/'nested4',
verbose=False, model_name='SVC', params=None,
savepath=None, drop_hours=None, PI=30, Ptest=None,
suffix=None, sample_from_negatives=1):
"""returns the nested CV test results for all scorers, features and models,
if model is chosen, i.e., model='MLP', returns just this model results
and its hyper-parameters per each outer split"""
import xarray as xr
from aux_gps import get_all_possible_combinations_from_list
from aux_gps import save_ncfile
feats = get_all_possible_combinations_from_list(
['pwv', 'pressure', 'doy'], reduce_single_list=True, combine_by_sep='+')
feat_list = []
for feat in feats:
print('Running CV on feature {}'.format(feat))
ds = CV_test_after_GridSearchCV(path=path, gr_path=gr_path,
model_name=model_name, params=params,
features=feat, PI=PI, Ptest=Ptest,
verbose=verbose, drop_hours=drop_hours,
sample_from_negatives=sample_from_negatives)
feat_list.append(ds)
dsf = xr.concat(feat_list, 'features')
dsf['features'] = feats
dss = dsf
dss.attrs['model'] = model_name
if Ptest is not None:
filename = 'nested_CV_test_results_{}_all_features_permutation_tests'.format(model_name)
else:
filename = 'nested_CV_test_results_{}_all_features_with_hyper_params'.format(model_name)
if params is not None:
dss.attrs['comment'] = 'using best hyper parameters for all features and outer splits'
filename += '_best_hp'
filename += '_neg_{}'.format(sample_from_negatives)
if suffix is not None:
filename += '_{}'.format(suffix)
filename += '.nc'
if savepath is not None:
save_ncfile(dss, savepath, filename)
return dss
def run_holdout_test_on_all_models_and_features(path=hydro_path, gr_path=hydro_ml_path/'holdout'):
import xarray as xr
from aux_gps import get_all_possible_combinations_from_list
feats = get_all_possible_combinations_from_list(
['pwv', 'pressure', 'doy'], reduce_single_list=True, combine_by_sep='+')
models = ['MLP', 'SVC', 'RF']
model_list = []
model_list2 = []
for model in models:
feat_list = []
feat_list2 = []
for feat in feats:
best, roc = holdout_test(path=path, gr_path=gr_path,
model_name=model, features=feat)
best.index.name = 'scorer'
ds = best[['mean_score', 'std_score', 'holdout_test_scores']].to_xarray()
roc.index.name = 'FPR'
roc_da = roc.to_xarray().to_array('scorer')
feat_list.append(ds)
feat_list2.append(roc_da)
dsf = xr.concat(feat_list, 'features')
dsf2 = xr.concat(feat_list2, 'features')
dsf['features'] = feats
dsf2['features'] = feats
model_list.append(dsf)
model_list2.append(dsf2)
dss = xr.concat(model_list, 'model')
rocs = xr.concat(model_list2, 'model')
dss['model'] = models
rocs['model'] = models
dss['roc'] = rocs
return dss
def prepare_X_y_for_holdout_test(features='pwv+doy', model_name='SVC',
path=hydro_path, drop_hours=None,
negative_samples=1):
# combine X,y and split them according to test ratio and seed:
X, y = combine_pos_neg_from_nc_file(path, negative_sample_num=negative_samples)
# re arange X features according to model:
feats = features.split('+')
if model_name == 'RF' and 'doy' in feats:
if isinstance(feats, list):
feats.remove('doy')
feats.append('DOY')
elif isinstance(feats, str):
feats = 'DOY'
elif model_name != 'RF' and 'doy' in feats:
if isinstance(feats, list):
feats.remove('doy')
feats.append('doy_sin')
feats.append('doy_cos')
elif isinstance(feats, str):
feats = ['doy_sin']
feats.append('doy_cos')
if isinstance(X, list):
Xs = []
for X1 in X:
Xs.append(select_features_from_X(X1, feats))
X = Xs
else:
X = select_features_from_X(X, feats)
if drop_hours is not None:
if isinstance(X, list):
Xs = []
for X1 in X:
Xs.append(drop_hours_in_pwv_pressure_features(X1, drop_hours,
verbose=True))
X = Xs
else:
X = drop_hours_in_pwv_pressure_features(X, drop_hours, verbose=True)
return X, y
def CV_test_after_GridSearchCV(path=hydro_path, gr_path=hydro_ml_path/'nested4',
model_name='SVC', features='pwv', params=None,
verbose=False, drop_hours=None, PI=None,
Ptest=None, sample_from_negatives=1):
"""do cross_validate with all scorers on all gridsearchcv folds,
reads the nested outer splits CV file in gr_path"""
import xarray as xr
import numpy as np
# cv = read_cv_params_and_instantiate(gr_path/'CV_outer.csv')
cv_dict = load_cv_splits_from_pkl(gr_path)
if verbose:
print(cv_dict)
param_df_dict = load_one_gridsearchcv_object(path=gr_path,
cv_type='nested',
features=features,
model_name=model_name,
verbose=verbose)
Xs, ys = prepare_X_y_for_holdout_test(features, model_name, path,
drop_hours=drop_hours,
negative_samples=sample_from_negatives)
bests = []
for i, negative_sample in enumerate(np.arange(1, sample_from_negatives + 1)):
print('running with negative sample #{} out of {}'.format(
negative_sample, sample_from_negatives))
if isinstance(Xs, list):
X = Xs[i]
y = ys[i]
else:
X = Xs
y = ys
if Ptest is not None:
print('Permutation Test is in progress!')
ds = run_permutation_classifier_test(X, y, 5, param_df_dict, Ptest=Ptest,
params=params,
model_name=model_name, verbose=verbose)
return ds
if params is not None:
if verbose:
print('running with custom hyper parameters: ', params)
outer_bests = []
outer_rocs = []
fis = []
pi_means = []
pi_stds = []
n_splits = len([x for x in cv_dict.keys()])
for split, tt in cv_dict.items():
X_train = X[tt['train']]
y_train = y[tt['train']]
X_test = X[tt['test']]
y_test = y[tt['test']]
outer_split = '{}-{}'.format(split, n_splits)
# for i, (train_index, test_index) in enumerate(cv.split(X, y)):
# X_train = X[train_index]
# y_train = y[train_index]
# X_test = X[test_index]
# y_test = y[test_index]
# outer_split = '{}-{}'.format(i+1, cv.n_splits)
best_params_df = param_df_dict.get(outer_split)
if params is not None:
for key, value in params.items():
if isinstance(value, tuple):
for ind in best_params_df.index:
best_params_df.at[ind, key] = value
else:
best_params_df[key] = value
if model_name == 'RF':
if PI is not None:
bdf, roc, fi, pi_mean, pi_std = run_test_on_CV_split(X_train, y_train, X_test, y_test,
best_params_df, PI=PI, Ptest=Ptest,
model_name=model_name, verbose=verbose)
else:
bdf, roc, fi = run_test_on_CV_split(X_train, y_train, X_test, y_test,
best_params_df, PI=PI, Ptest=Ptest,
model_name=model_name, verbose=verbose)
fis.append(fi)
else:
if PI is not None:
bdf, roc, pi_mean, pi_std = run_test_on_CV_split(X_train, y_train, X_test, y_test,
best_params_df, PI=PI,
model_name=model_name, verbose=verbose)
else:
bdf, roc = run_test_on_CV_split(X_train, y_train, X_test, y_test,
best_params_df, PI=PI,
model_name=model_name, verbose=verbose)
if PI is not None:
pi_means.append(pi_mean)
pi_stds.append(pi_std)
bdf.index.name = 'scorer'
roc.index.name = 'FPR'
if 'hidden_layer_sizes' in bdf.columns:
bdf['hidden_layer_sizes'] = bdf['hidden_layer_sizes'].astype(str)
bdf_da = bdf.to_xarray()
roc_da = roc.to_xarray().to_array('scorer')
roc_da.name = 'TPR'
outer_bests.append(bdf_da)
outer_rocs.append(roc_da)
best_da = xr.concat(outer_bests, 'outer_split')
roc_da = xr.concat(outer_rocs, 'outer_split')
best = xr.merge([best_da, roc_da])
best['outer_split'] = np.arange(1, n_splits + 1)
if model_name == 'RF':
fi_da = xr.concat(fis, 'outer_split')
best['feature_importances'] = fi_da
if PI is not None:
pi_mean_da = xr.concat(pi_means, 'outer_split')
pi_std_da = xr.concat(pi_stds, 'outer_split')
best['PI_mean'] = pi_mean_da
best['PI_std'] = pi_std_da
bests.append(best)
if len(bests) == 1:
return bests[0]
else:
best_ds = xr.concat(bests, 'neg_sample')
best_ds['neg_sample'] = np.arange(1, sample_from_negatives + 1)
return best_ds
def run_permutation_classifier_test(X, y, cv, best_params_df, Ptest=100,
model_name='SVC', verbose=False, params=None):
from sklearn.model_selection import permutation_test_score
import xarray as xr
import numpy as np
def run_one_permutation_test(X=X, y=y, cv=cv, bp_df=best_params_df,
model_name=model_name, n_perm=Ptest,
verbose=verbose):
true_scores = []
pvals = []
perm_scores = []
for scorer in bp_df.index:
sk_model = ml.pick_model(model_name)
# get best params (drop two last cols since they are not params):
b_params = bp_df.T[scorer][:-2].to_dict()
if verbose:
print('{} scorer, params:{}'.format(scorer, b_params))
true, perm_scrs, pval = permutation_test_score(sk_model, X, y,
cv=cv,
n_permutations=Ptest,
scoring=scorers(scorer),
random_state=0,
n_jobs=-1)
true_scores.append(true)
pvals.append(pval)
perm_scores.append(perm_scrs)
true_da = xr.DataArray(true_scores, dims=['scorer'])
true_da['scorer'] = [x for x in bp_df.index.values]
true_da.name = 'true_score'
pval_da = xr.DataArray(pvals, dims=['scorer'])
pval_da['scorer'] = [x for x in bp_df.index.values]
pval_da.name = 'pvalue'
perm_da = xr.DataArray(perm_scores, dims=['scorer', 'permutations'])
perm_da['scorer'] = [x for x in bp_df.index.values]
perm_da['permutations'] = np.arange(1, Ptest+1)
perm_da.name = 'permutation_score'
ds = xr.merge([true_da, pval_da, perm_da])
return ds
ml = ML_Classifier_Switcher()
if params is not None:
best_p_df = best_params_df['1-{}'.format(len(best_params_df))]
for key, value in params.items():
if isinstance(value, tuple):
for ind in best_p_df.index:
best_p_df.at[ind, key] = value
else:
best_p_df[key] = value
dss = run_one_permutation_test(bp_df=best_p_df)
else:
if verbose:
print('Picking {} model with best params'.format(model_name))
splits = []
for i, df in enumerate(best_params_df.values()):
if verbose:
print('running on split #{}'.format(i+1))
ds = run_one_permutation_test()
splits.append(ds)
dss = xr.concat(splits, dim='outer_split')
dss['outer_split'] = np.arange(1, len(best_params_df)+ 1)
return dss
def run_test_on_CV_split(X_train, y_train, X_test, y_test, param_df,
model_name='SVC', verbose=False, PI=None,
Ptest=None):
import numpy as np
import xarray as xr
import pandas as pd
from sklearn.metrics import roc_curve
from sklearn.metrics import roc_auc_score
from sklearn.inspection import permutation_importance
best_df = param_df.copy()
ml = ML_Classifier_Switcher()
if verbose:
print('Picking {} model with best params'.format(model_name))
# print('Features are: {}'.format(features))
test_scores = []
fi_list = []
mean_fpr = np.linspace(0, 1, 100)
tprs = []
roc_aucs = []
pi_mean_list = []
pi_std_list = []
for scorer in best_df.index:
sk_model = ml.pick_model(model_name)
# get best params (drop two last cols since they are not params):
params = best_df.T[scorer][:-2].to_dict()
if verbose:
print('{} scorer, params:{}'.format(scorer, params))
sk_model.set_params(**params)
sk_model.fit(X_train, y_train)
if hasattr(sk_model, 'feature_importances_'):
# print(X_train['feature'])
# input('press any key')
FI = xr.DataArray(sk_model.feature_importances_, dims=['feature'])
FI['feature'] = X_train['feature']
fi_list.append(FI)
y_pred = sk_model.predict(X_test)
fpr, tpr, _ = roc_curve(y_test, y_pred)
interp_tpr = np.interp(mean_fpr, fpr, tpr)
interp_tpr[0] = 0.0
roc_auc = roc_auc_score(y_test, y_pred)
roc_aucs.append(roc_auc)
tprs.append(interp_tpr)
score = scorer_function(scorer, y_test, y_pred)
test_scores.append(score)
if PI is not None:
pi = permutation_importance(sk_model, X_test, y_test,
n_repeats=PI,
scoring=scorers(scorer),
random_state=0, n_jobs=-1)
pi_mean = xr.DataArray(pi['importances_mean'], dims='feature')
pi_std = xr.DataArray(pi['importances_std'], dims='feature')
pi_mean.name = 'PI_mean'
pi_std.name = 'PI_std'
pi_mean['feature'] = X_train['feature']
pi_std['feature'] = X_train['feature']
pi_mean_list.append(pi_mean)
pi_std_list.append(pi_std)
if PI is not None:
pi_mean_da = xr.concat(pi_mean_list, 'scorer')
pi_std_da = xr.concat(pi_std_list, 'scorer')
pi_mean_da['scorer'] = [x for x in best_df.index.values]
pi_std_da['scorer'] = [x for x in best_df.index.values]
roc_df = pd.DataFrame(tprs).T
roc_df.columns = [x for x in best_df.index]
roc_df.index = mean_fpr
best_df['test_score'] = test_scores
best_df['roc_auc_score'] = roc_aucs
if hasattr(sk_model, 'feature_importances_'):
fi = xr.concat(fi_list, 'scorer')
fi['scorer'] = [x for x in best_df.index.values]
if PI is not None:
return best_df, roc_df, fi, pi_mean_da, pi_std_da
else:
return best_df, roc_df, fi
elif PI is not None:
return best_df, roc_df, pi_mean_da, pi_std_da
else:
return best_df, roc_df
def holdout_test(path=hydro_path, gr_path=hydro_ml_path/'holdout',
model_name='SVC', features='pwv', return_RF_FI=False,
verbose=False):
"""do a holdout test with best model from gridsearchcv
with all scorers"""
from sklearn.model_selection import train_test_split
from sklearn.metrics import roc_curve
from sklearn.metrics import roc_auc_score
import xarray as xr
import pandas as pd
import numpy as np
# process gridsearchcv results:
best_df, test_ratio, seed = load_one_gridsearchcv_object(path=gr_path,
cv_type='holdout',
features=features,
model_name=model_name,
verbose=False)
print('Using random seed of {} and {}% test ratio'.format(seed, test_ratio))
ts = int(test_ratio) / 100
X, y = prepare_X_y_for_holdout_test(features, model_name, path)
# split using test_size and seed:
X_train, X_test, y_train, y_test = train_test_split(X, y,
test_size=ts,
random_state=int(seed),
stratify=y)
if verbose:
print('y train pos/neg:{}, {}'.format((y_train==1).sum().item(),(y_train==0).sum().item()))
print('y test pos/neg:{}, {}'.format((y_test==1).sum().item(),(y_test==0).sum().item()))
# pick model and set the params to best from gridsearchcv:
ml = ML_Classifier_Switcher()
print('Picking {} model with best params'.format(model_name))
print('Features are: {}'.format(features))
test_scores = []
fi_list = []
mean_fpr = np.linspace(0, 1, 100)
tprs = []
roc_aucs = []
for scorer in best_df.index:
sk_model = ml.pick_model(model_name)
# get best params (drop two last cols since they are not params):
params = best_df.T[scorer][:-2].to_dict()
if verbose:
print('{} scorer, params:{}'.format(scorer, params))
sk_model.set_params(**params)
sk_model.fit(X_train, y_train)
if hasattr(sk_model, 'feature_importances_'):
FI = xr.DataArray(sk_model.feature_importances_, dims=['feature'])
FI['feature'] = X_train['feature']
fi_list.append(FI)
y_pred = sk_model.predict(X_test)
fpr, tpr, _ = roc_curve(y_test, y_pred)
interp_tpr = np.interp(mean_fpr, fpr, tpr)
interp_tpr[0] = 0.0
roc_auc = roc_auc_score(y_test, y_pred)
roc_aucs.append(roc_auc)
tprs.append(interp_tpr)
score = scorer_function(scorer, y_test, y_pred)
test_scores.append(score)
roc_df = pd.DataFrame(tprs).T
roc_df.columns = [x for x in best_df.index]
roc_df.index = mean_fpr
best_df['holdout_test_scores'] = test_scores
best_df['roc_auc_score'] = roc_aucs
if fi_list and return_RF_FI:
da = xr.concat(fi_list, 'scorer')
da['scorer'] = best_df.index.values
da.name = 'RF_feature_importances'
return da
return best_df, roc_df
def load_one_gridsearchcv_object(path=hydro_ml_path, cv_type='holdout', features='pwv',
model_name='SVC', verbose=True):
"""load one gridsearchcv obj with model_name and features and run read_one_gridsearchcv_object"""
from aux_gps import path_glob
import joblib
# first filter for model name:
if verbose:
print('loading GridsearchCVs results for {} model with {} cv type'.format(model_name, cv_type))
model_files = path_glob(path, 'GRSRCHCV_{}_*.pkl'.format(cv_type))
model_files = [x for x in model_files if model_name in x.as_posix()]
# now select features:
if verbose:
print('loading GridsearchCVs results with {} features'.format(features))
model_features = [x.as_posix().split('/')[-1].split('_')[3] for x in model_files]
feat_ind = get_feature_set_from_list(model_features, features)
# also get the test ratio and seed number:
if len(feat_ind) > 1:
if verbose:
print('found {} GR objects.'.format(len(feat_ind)))
files = sorted([model_files[x] for x in feat_ind])
outer_splits = [x.as_posix().split('/')[-1].split('.')[0].split('_')[-3] for x in files]
grs = [joblib.load(x) for x in files]
best_dfs = [read_one_gridsearchcv_object(x) for x in grs]
di = dict(zip(outer_splits, best_dfs))
return di
else:
file = model_files[feat_ind]
seed = file.as_posix().split('/')[-1].split('.')[0].split('_')[-1]
outer_splits = file.as_posix().split('/')[-1].split('.')[0].split('_')[-3]
# load and produce best_df:
gr = joblib.load(file)
best_df = read_one_gridsearchcv_object(gr)
return best_df, outer_splits, seed
def get_feature_set_from_list(model_features_list, features, sep='+'):
"""select features from model_features_list,
return the index in the model_features_list and the entry itself"""
# first find if features is a single or multiple features:
if isinstance(features, str) and sep not in features:
try:
ind = [i for i, e in enumerate(model_features_list) if e == features]
# ind = model_features_list.index(features)
except ValueError:
raise ValueError('{} is not in {}'.format(features, ', '.join(model_features_list)))
elif isinstance(features, str) and sep in features:
features_split = features.split(sep)
mf = [x.split(sep) for x in model_features_list]
bool_list = [set(features_split) == (set(x)) for x in mf]
ind = [i for i, x in enumerate(bool_list) if x]
# print(len(ind))
# ind = ind[0]
# feat = model_features_list[ind]
# feat = model_features_list[ind]
return ind
def read_one_gridsearchcv_object(gr):
"""read one gridsearchcv multimetric object and
get the best params, best mean/std scores"""
import pandas as pd
# first get all the scorers used:
scorers = [x for x in gr.scorer_.keys()]
# now loop over the scorers:
best_params = []
best_mean_scores = []
best_std_scores = []
for scorer in scorers:
df_mean = pd.concat([pd.DataFrame(gr.cv_results_["params"]), pd.DataFrame(
gr.cv_results_["mean_test_{}".format(scorer)], columns=[scorer])], axis=1)
df_std = pd.concat([pd.DataFrame(gr.cv_results_["params"]), pd.DataFrame(
gr.cv_results_["std_test_{}".format(scorer)], columns=[scorer])], axis=1)
# best index = highest score:
best_ind = df_mean[scorer].idxmax()
best_mean_scores.append(df_mean.iloc[best_ind][scorer])
best_std_scores.append(df_std.iloc[best_ind][scorer])
best_params.append(df_mean.iloc[best_ind].to_frame().T.iloc[:, :-1])
best_df = pd.concat(best_params)
best_df['mean_score'] = best_mean_scores
best_df['std_score'] = best_std_scores
best_df.index = scorers
return best_df
# # param grid dict:
# params = gr.param_grid
# # scorer names:
# scoring = [x for x in gr.scoring.keys()]
# # df:
# df = pd.DataFrame().from_dict(gr.cv_results_)
# # produce multiindex from param_grid dict:
# param_names = [x for x in params.keys()]
# # unpack param_grid vals to list of lists:
# pro = [[y for y in x] for x in params.values()]
# ind = pd.MultiIndex.from_product((pro), names=param_names)
# df.index = ind
# best_params = []
# best_mean_scores = []
# best_std_scores = []
# for scorer in scoring:
# best_params.append(df[df['rank_test_{}'.format(scorer)]==1]['mean_test_{}'.format(scorer)].index[0])
# best_mean_scores.append(df[df['rank_test_{}'.format(scorer)]==1]['mean_test_{}'.format(scorer)].iloc[0])
# best_std_scores.append(df[df['rank_test_{}'.format(scorer)]==1]['std_test_{}'.format(scorer)].iloc[0])
# best_df = pd.DataFrame(best_params, index=scoring, columns=param_names)
# best_df['mean_score'] = best_mean_scores
# best_df['std_score'] = best_std_scores
# return best_df, best_df_1
def process_gridsearch_results(GridSearchCV, model_name,
split_dim='inner_kfold', features=None,
pwv_id=None, hs_id=None, test_size=None):
import xarray as xr
import pandas as pd
import numpy as np
# finish getting best results from all scorers togather
"""takes GridSreachCV object with cv_results and xarray it into dataarray"""
params = GridSearchCV.param_grid
scoring = GridSearchCV.scoring
results = GridSearchCV.cv_results_
# for scorer in scoring:
# for sample in ['train', 'test']:
# sample_score_mean = results['mean_{}_{}'.format(sample, scorer)]
# sample_score_std = results['std_{}_{}'.format(sample, scorer)]
# best_index = np.nonzero(results['rank_test_{}'.format(scorer)] == 1)[0][0]
# best_score = results['mean_test_{}'.format(scorer)][best_index]
names = [x for x in params.keys()]
# unpack param_grid vals to list of lists:
pro = [[y for y in x] for x in params.values()]
ind = pd.MultiIndex.from_product((pro), names=names)
# result_names = [x for x in GridSearchCV.cv_results_.keys() if 'split'
# not in x and 'time' not in x and 'param' not in x and
# 'rank' not in x]
result_names = [
x for x in results.keys() if 'param' not in x]
ds = xr.Dataset()
for da_name in result_names:
da = xr.DataArray(results[da_name])
ds[da_name] = da
ds = ds.assign(dim_0=ind).unstack('dim_0')
for dim in ds.dims:
if ds[dim].dtype == 'O':
try:
ds[dim] = ds[dim].astype(str)
except ValueError:
ds = ds.assign_coords({dim: [str(x) for x in ds[dim].values]})
if ('True' in ds[dim]) and ('False' in ds[dim]):
ds[dim] = ds[dim] == 'True'
# get all splits data and concat them along number of splits:
all_splits = [x for x in ds.data_vars if 'split' in x]
train_splits = [x for x in all_splits if 'train' in x]
test_splits = [x for x in all_splits if 'test' in x]
# loop over scorers:
trains = []
tests = []
for scorer in scoring:
train_splits_scorer = [x for x in train_splits if scorer in x]
trains.append(xr.concat([ds[x]
for x in train_splits_scorer], split_dim))
test_splits_scorer = [x for x in test_splits if scorer in x]
tests.append(xr.concat([ds[x] for x in test_splits_scorer], split_dim))
splits_scorer = np.arange(1, len(train_splits_scorer) + 1)
train_splits = xr.concat(trains, 'scoring')
test_splits = xr.concat(tests, 'scoring')
# splits = [x for x in range(len(train_splits))]
# train_splits = xr.concat([ds[x] for x in train_splits], 'split')
# test_splits = xr.concat([ds[x] for x in test_splits], 'split')
# replace splits data vars with newly dataarrays:
ds = ds[[x for x in ds.data_vars if x not in all_splits]]
ds['split_train_score'] = train_splits
ds['split_test_score'] = test_splits
ds[split_dim] = splits_scorer
if isinstance(scoring, list):
ds['scoring'] = scoring
elif isinstance(scoring, dict):
ds['scoring'] = [x for x in scoring.keys()]
ds.attrs['name'] = 'CV_results'
ds.attrs['param_names'] = names
ds.attrs['model_name'] = model_name
ds.attrs['{}_splits'.format(split_dim)] = ds[split_dim].size
if GridSearchCV.refit:
if hasattr(GridSearchCV.best_estimator_, 'feature_importances_'):
f_import = xr.DataArray(
GridSearchCV.best_estimator_.feature_importances_,
dims=['feature'])
f_import['feature'] = features
ds['feature_importances'] = f_import
ds['best_score'] = GridSearchCV.best_score_
# ds['best_model'] = GridSearchCV.best_estimator_
ds.attrs['refitted_scorer'] = GridSearchCV.refit
for name in names:
if isinstance(GridSearchCV.best_params_[name], tuple):
GridSearchCV.best_params_[name] = ','.join(
map(str, GridSearchCV.best_params_[name]))
ds['best_{}'.format(name)] = GridSearchCV.best_params_[name]
return ds, GridSearchCV.best_estimator_
else:
return ds, None
def save_cv_results(cvr, savepath=hydro_path):
from aux_gps import save_ncfile
features = '+'.join(cvr.attrs['features'])
# pwv_id = cvr.attrs['pwv_id']
# hs_id = cvr.attrs['hs_id']
# neg_pos_ratio = cvr.attrs['neg_pos_ratio']
ikfolds = cvr.attrs['inner_kfold_splits']
okfolds = cvr.attrs['outer_kfold_splits']
name = cvr.attrs['model_name']
refitted_scorer = cvr.attrs['refitted_scorer'].replace('_', '-')
# filename = 'CVR_{}_{}_{}_{}_{}_{}_{}_{}.nc'.format(pwv_id, hs_id,
# name, features, refitted_scorer, ikfolds, okfolds, neg_pos_ratio)
filename = 'CVR_{}_{}_{}_{}_{}.nc'.format(
name, features, refitted_scorer, ikfolds, okfolds)
save_ncfile(cvr, savepath, filename)
return
def scikit_fit_predict(X, y, seed=42, with_pressure=True, n_splits=7,
plot=True):
# step1: CV for train/val (80% from 80-20 test). display results with
# model and scores(AUC, f1), use StratifiedKFold
# step 2: use validated model with test (20%) and build ROC curve
# step 3: add features (pressure) but check for correlation
# check permutations with scikit learn
from sklearn.model_selection import train_test_split
from sklearn.discriminant_analysis import LinearDiscriminantAnalysis, QuadraticDiscriminantAnalysis
from sklearn.metrics import f1_score
from sklearn.metrics import plot_roc_curve
from sklearn.svm import SVC
from numpy import interp
from sklearn.metrics import auc
import numpy as np
import matplotlib.pyplot as plt
from sklearn.model_selection import KFold
from sklearn.model_selection import LeaveOneOut
from sklearn.metrics import confusion_matrix
from sklearn.metrics import roc_auc_score
from sklearn.model_selection import StratifiedKFold
if not with_pressure:
just_pw = [x for x in X.feature.values if 'pressure' not in x]
X = X.sel(feature=just_pw)
X_tt, X_test, y_tt, y_test = train_test_split(
X, y, test_size=0.2, shuffle=True, random_state=seed)
cv = StratifiedKFold(n_splits=n_splits, shuffle=True, random_state=seed)
# cv = LeaveOneOut()
classifier = SVC(kernel='rbf', probability=False,
random_state=seed)
# classifier = LinearDiscriminantAnalysis()
# clf = QuadraticDiscriminantAnalysis()
scores = []
fig, ax = plt.subplots()
tprs = []
aucs = []
mean_fpr = np.linspace(0, 1, 100)
for i, (train, val) in enumerate(cv.split(X_tt, y_tt)):
# for i in range(100):
# X_train, X_val, y_train, y_val = train_test_split(
# X_tt, y_tt, shuffle=True, test_size=0.5, random_state=i)
# clf.fit(X_train, y_train)
classifier.fit(X_tt[train], y_tt[train])
# viz = plot_roc_curve(clf, X_val, y_val,
# name='ROC run {}'.format(i),
# alpha=0.3, lw=1, ax=ax)
viz = plot_roc_curve(classifier, X_tt[val], y_tt[val],
name='ROC fold {}'.format(i),
alpha=0.3, lw=1, ax=ax)
interp_tpr = interp(mean_fpr, viz.fpr, viz.tpr)
interp_tpr[0] = 0.0
tprs.append(interp_tpr)
# aucs.append(viz.roc_auc)
# y_pred = clf.predict(X_val)
y_pred = classifier.predict(X_tt[val])
aucs.append(roc_auc_score(y_tt[val], y_pred))
# scores.append(clf.score(X_val, y_val))
scores.append(f1_score(y_tt[val], y_pred))
scores = np.array(scores)
ax.plot([0, 1], [0, 1], linestyle='--', lw=2, color='r',
label='Chance', alpha=.8)
mean_tpr = np.mean(tprs, axis=0)
mean_tpr[-1] = 1.0
mean_auc = auc(mean_fpr, mean_tpr)
std_auc = np.std(aucs)
ax.plot(mean_fpr, mean_tpr, color='b',
label=r'Mean ROC (AUC = %0.2f $\pm$ %0.2f)' % (mean_auc, std_auc),
lw=2, alpha=.8)
std_tpr = np.std(tprs, axis=0)
tprs_upper = np.minimum(mean_tpr + std_tpr, 1)
tprs_lower = np.maximum(mean_tpr - std_tpr, 0)
ax.fill_between(mean_fpr, tprs_lower, tprs_upper, color='grey', alpha=.2,
label=r'$\pm$ 1 std. dev.')
ax.set(xlim=[-0.05, 1.05], ylim=[-0.05, 1.05],
title="Receiver operating characteristic example")
ax.legend(loc="lower right")
ax.set_title(
'ROC curve for KFold={}, with pressure anomalies.'.format(n_splits))
if not with_pressure:
ax.set_title(
'ROC curve for KFold={}, without pressure anomalies.'.format(n_splits))
y_test_predict = classifier.predict(X_test)
print('final test predict score:')
print(f1_score(y_test, y_test_predict))
if plot:
plt.figure()
plt.hist(scores, bins=15, edgecolor='k')
return scores
# clf.fit(X,y)
def produce_X_y_from_list(pw_stations=['drag', 'dsea', 'elat'],
hs_ids=[48125, 48199, 60170],
pressure_station='bet-dagan', max_flow=0,
window=25, neg_pos_ratio=1, path=work_yuval,
ims_path=ims_path, hydro_path=hydro_path,
concat_Xy=False):
if isinstance(hs_ids, int):
hs_ids = [hs_ids for x in range(len(pw_stations))]
kwargs = locals()
[kwargs.pop(x) for x in ['pw_stations', 'hs_ids', 'concat_Xy']]
Xs = []
ys = []
for pw_station, hs_id in list(zip(pw_stations, hs_ids)):
X, y = produce_X_y(pw_station, hs_id, **kwargs)
Xs.append(X)
ys.append(y)
if concat_Xy:
print('concatenating pwv stations {}, with hydro_ids {}.'.format(
pw_stations, hs_ids))
X, y = concat_X_y(Xs, ys)
return X, y
else:
return Xs, ys
def concat_X_y(Xs, ys):
import xarray as xr
import pandas as pd
X_attrs = [x.attrs for x in Xs]
X_com_attrs = dict(zip(pd.DataFrame(X_attrs).T.index.values,
pd.DataFrame(X_attrs).T.values.tolist()))
y_attrs = [x.attrs for x in ys]
y_com_attrs = dict(zip(pd.DataFrame(y_attrs).T.index.values,
pd.DataFrame(y_attrs).T.values.tolist()))
for X in Xs:
feat = [x.replace('_' + X.attrs['pwv_id'], '')
for x in X.feature.values]
X['feature'] = feat
X = xr.concat(Xs, 'sample')
X.attrs = X_com_attrs
y = xr.concat(ys, 'sample')
y.attrs = y_com_attrs
return X, y
def produce_X_y(pw_station='drag', hs_id=48125, pressure_station='bet-dagan',
window=25, seed=42,
max_flow=0, neg_pos_ratio=1, path=work_yuval,
ims_path=ims_path, hydro_path=hydro_path):
import xarray as xr
from aux_gps import anomalize_xr
from PW_stations import produce_geo_gnss_solved_stations
import numpy as np
# call preprocess_hydro_station
hdf, y_meta = preprocess_hydro_station(
hs_id, hydro_path, max_flow=max_flow)
# load PWV and other features and combine them to fdf:
pw = xr.open_dataset(path / 'GNSS_PW_anom_hourly_50_hour_dayofyear.nc')
fdf = pw[pw_station].to_dataframe(name='pwv_{}'.format(pw_station))
# add Day of year to fdf:
doy = fdf.index.dayofyear
# scale doy to cyclic with amp ~1:
fdf['doy_sin'] = np.sin(doy * np.pi / 183)
fdf['doy_cos'] = np.cos(doy * np.pi / 183)
if pressure_station is not None:
p = xr.load_dataset(
ims_path /
'IMS_BD_hourly_ps_1964-2020.nc')[pressure_station]
p_attrs = p.attrs
p_attrs = {'pressure_{}'.format(
key): val for key, val in p_attrs.items()}
p = p.sel(time=slice('1996', None))
p = anomalize_xr(p, freq='MS')
fdf['pressure_{}'.format(pressure_station)] = p.to_dataframe()
# check the the last date of hdf is bigger than the first date of fdf,
# i.e., there is at least one overlapping event in the data:
if hdf.index[-1] < fdf.index[0]:
raise KeyError('Data not overlapping, hdf for {} stops at {} and fdf starts at {}'.format(
hs_id, hdf.index[-1], fdf.index[0]))
# finally, call add_features_and_produce_X_y
X, y = add_features_and_produce_X_y(hdf, fdf, window_size=window,
seed=seed,
neg_pos_ratio=neg_pos_ratio)
# add meta data:
gps = produce_geo_gnss_solved_stations(plot=False)
pwv_attrs = gps.loc[pw_station, :][['lat', 'lon', 'alt', 'name']].to_dict()
pwv_attrs = {'pwv_{}'.format(key): val for key, val in pwv_attrs.items()}
X.attrs = pwv_attrs
if pressure_station is not None:
X.attrs.update(p_attrs)
y.attrs = y_meta
y.attrs['hydro_station_id'] = hs_id
y.attrs['neg_pos_ratio'] = neg_pos_ratio
# calculate distance to hydro station:
lat1 = X.attrs['pwv_lat']
lon1 = X.attrs['pwv_lon']
lat2 = y.attrs['lat']
lon2 = y.attrs['lon']
y.attrs['max_flow'] = max_flow
distance = calculate_distance_between_two_latlons_israel(
lat1, lon1, lat2, lon2)
X.attrs['distance_to_hydro_station_in_km'] = distance / 1000.0
y.attrs['distance_to_pwv_station_in_km'] = distance / 1000.0
X.attrs['pwv_id'] = pw_station
return X, y
# def produce_X_y(station='drag', hs_id=48125, lag=25, anoms=True,
# neg_pos_ratio=2, add_pressure=False,
# path=work_yuval, hydro_path=hydro_path, with_ends=False,
# seed=42,
# verbose=True, return_xarray=False, pressure_anoms=None):
# import pandas as pd
# import numpy as np
# import xarray as xr
#
# def produce_da_from_list(event_list, feature='pwv'):
# X_da = xr.DataArray(event_list, dims=['sample', 'feature'])
# X_da['feature'] = ['{}_{}'.format(feature, x) for x in np.arange(0, 24, 1)]
# X_df = pd.concat(event_list)
# X_da['sample'] = [x for x in X_df.index[::24]]
# return X_da
#
# df = preprocess_hydro_pw(
# pw_station=station,
# hs_id=hs_id,
# path=path,
# hydro_path=hydro_path,
# with_tide_ends=with_ends, anoms=anoms,
# pressure_anoms=pressure_anoms,
# add_pressure=add_pressure)
# if pressure_anoms is not None:
# station = pressure_anoms.name
# # first produce all the positives:
# # get the tides datetimes:
# y_pos = df[df['tides'] == 1]['tides']
# # get the datetimes of 24 hours before tide event (not inclusive):
# y_lag_pos = y_pos.index - pd.Timedelta(lag, unit='H')
# masks = [(df.index > start) & (df.index < end)
# for start, end in zip(y_lag_pos, y_pos.index)]
# # also drop event if less than 24 hour before available:
# pw_pos_list = []
# pressure_pos_list = []
# ind = []
# bad_ind = []
# for i, tide in enumerate(masks):
# if len(df['tides'][tide]) == (lag - 1):
# pw_pos_list.append(df[station][tide])
# pressure_pos_list.append(df['pressure'][tide])
# ind.append(i)
# else:
# bad_ind.append(i)
# # get the indices of the dropped events:
# # ind = [x[0] for x in pw_pos_list]
# if bad_ind:
# if verbose:
# print('{} are without full 24 hours before record.'.format(
# ','.join([x for x in df.iloc[bad_ind].index.strftime('%Y-%m-%d:%H:00:00')])))
# # drop the events in y so len(y) == in each x from tides_list:
# y_pos_arr = y_pos.iloc[ind].values
# # now get the negative y's with neg_pos_ratio (set to 1 if the same pos=neg):
# y_neg_arr = np.zeros(y_pos_arr.shape[0] * neg_pos_ratio)
# cnt = 0
# pw_neg_list = []
# pressure_neg_list = []
# np.random.seed(seed)
# while cnt < len(y_neg_arr):
# # get a random date from df:
# r = np.random.randint(low=0, high=len(df))
# # slice -24 to 24 range with t=0 being the random date:
# # update: extend the range to -72 hours to 72 hours:
# lag_factor = 72 / lag
# slice_range = int(lag * lag_factor)
# sliced = df.iloc[r - slice_range:r + slice_range]
# # if tides inside this date range, continue:
# if y_pos.iloc[ind].index in sliced.index:
# if verbose:
# print('found positive tide in randomly sliced 48 window')
# continue
# # now if no 24 items exist, also continue:
# negative = df.iloc[r - lag:r - 1][station]
# if len(negative) != (lag-1):
# if verbose:
# print('didnt find full {} hours sliced negative'.format(lag-1))
# continue
# # else, append to pw_neg_list and increase cnt
# pw_neg_list.append(negative)
# pressure_neg_list.append(df.iloc[r - lag:r - 1]['pressure'])
# cnt += 1
# # lastly, assemble for X, y using np.columnstack:
# y = np.concatenate([y_pos_arr, y_neg_arr])
# X = np.stack([[x.values for x in pw_pos_list] +
# [x.values for x in pw_neg_list]])
# X = X.squeeze()
# pw_pos_da = produce_da_from_list(pw_pos_list, feature='pwv')
# pw_neg_da = produce_da_from_list(pw_neg_list, feature='pwv')
# pr_pos_da = produce_da_from_list(pressure_pos_list, feature='pressure')
# pr_neg_da = produce_da_from_list(pressure_neg_list, feature='pressure')
# if return_xarray:
# y = xr.DataArray(y, dims='sample')
# X_pwv = xr.concat([pw_pos_da, pw_neg_da], 'sample')
# X_pressure = xr.concat([pr_pos_da, pr_neg_da], 'sample')
# X = xr.concat([X_pwv, X_pressure], 'feature')
# X.name = 'X'
# y['sample'] = X['sample']
# y.name = 'y'
# X.attrs['PWV_station'] = station
# X.attrs['hydro_station_id'] = hs_id
# y.attrs = X.attrs
# return X, y
# else:
# return X, y
def plot_Xpos_Xneg_mean_std(X_pos_da, X_neg_da):
import matplotlib.pyplot as plt
from PW_from_gps_figures import plot_field_with_fill_between
fig, ax = plt.subplots(figsize=(8, 6))
posln = plot_field_with_fill_between(X_pos_da, ax=ax, mean_dim='event',
dim='time', color='b', marker='s')
negln = plot_field_with_fill_between(X_neg_da, ax=ax, mean_dim='event',
dim='time', color='r', marker='o')
ax.legend(posln+negln, ['Positive tide events', 'Negative tide events'])
ax.grid()
return fig
def preprocess_hydro_station(hs_id=48125, hydro_path=hydro_path, max_flow=0,
with_tide_ends=False):
"""load hydro station tide events with max_flow and round it up to
hourly sample rate, with_tide_ends, puts the value 2 at the datetime of
tide end. regardless 1 is the datetime for tide event."""
import xarray as xr
import pandas as pd
import numpy as np
# first load tides data:
all_tides = xr.open_dataset(hydro_path / 'hydro_tides.nc')
# get all tides for specific station without nans:
sta_slice = [x for x in all_tides.data_vars if str(hs_id) in x]
sta_slice = [
x for x in sta_slice if 'max_flow' in x or 'tide_end' in x or 'tide_max' in x]
if not sta_slice:
raise KeyError('hydro station {} not found in database'.format(hs_id))
tides = all_tides[sta_slice].dropna('tide_start')
max_flow_tide = tides['TS_{}_max_flow'.format(hs_id)]
max_flow_attrs = max_flow_tide.attrs
tide_starts = tides['tide_start'].where(
~tides.isnull()).where(max_flow_tide > max_flow).dropna('tide_start')['tide_start']
tide_ends = tides['TS_{}_tide_end'.format(hs_id)].where(
~tides.isnull()).where(max_flow_tide > max_flow).dropna('tide_start')['TS_{}_tide_end'.format(hs_id)]
max_flows = max_flow_tide.where(
max_flow_tide > max_flow).dropna('tide_start')
# round all tide_starts to hourly:
ts = tide_starts.dt.round('1H')
max_flows = max_flows.sel(tide_start=ts, method='nearest')
max_flows['tide_start'] = ts
ts_end = tide_ends.dt.round('1H')
time_dt = pd.date_range(
start=ts.min().values,
end=ts_end.max().values,
freq='1H')
df = pd.DataFrame(data=np.zeros(time_dt.shape), index=time_dt)
df.loc[ts.values, 0] = 1
df.loc[ts.values, 1] = max_flows.loc[ts.values]
df.columns = ['tides', 'max_flow']
df = df.fillna(0)
if with_tide_ends:
df.loc[ts_end.values, :] = 2
return df, max_flow_attrs
def add_features_and_produce_X_y(hdf, fdf, window_size=25, seed=42,
neg_pos_ratio=1, plot=False):
"""hdf is the hydro events df and fdf is the features df in 'H' freq.
This function checks the fdf for window-sized data and hour before
each positive event.
returns the combined df (hdf+fdf) the positive events labels and features.
"""
import pandas as pd
import numpy as np
import xarray as xr
# first add check_window_size of 0's to hdf:
st = hdf.index[0] - pd.Timedelta(window_size, unit='H')
en = hdf.index[0]
dts = pd.date_range(st, en - pd.Timedelta(1, unit='H'), freq='H')
mdf = pd.DataFrame(
np.zeros(window_size),
index=dts,
columns=['tides'])
hdf = pd.concat([hdf, mdf], axis=0)
# check for hourly sample rate and concat:
if not pd.infer_freq(fdf.index) == 'H':
raise('pls resample fdf to hourly...')
feature = [x for x in fdf.columns]
df = pd.concat([hdf, fdf], axis=1)
# get the tides(positive events) datetimes:
y_pos = df[df['tides'] == 1]['tides']
# get the datetimes of 24 hours before tide event (not inclusive):
y_lag_pos = y_pos.index - pd.Timedelta(window_size, unit='H')
masks = [(df.index > start) & (df.index < end)
for start, end in zip(y_lag_pos, y_pos.index)]
# first check how many full periods of data the feature has:
avail = [window_size - 1 - df[feature][masks[x]].isnull().sum()
for x in range(len(masks))]
adf = pd.DataFrame(avail, index=y_pos.index, columns=feature)
if plot:
adf.plot(kind='bar')
# produce the positive events datetimes for which all the features have
# window sized data and hour before the event:
good_dts = adf[adf.loc[:, feature] == window_size - 1].dropna().index
# y array of positives (1's):
y_pos_arr = y_pos.loc[good_dts].values
# now produce the feature list itself:
good_inds_for_masks = [adf.index.get_loc(x) for x in good_dts]
good_masks = [masks[x] for x in good_inds_for_masks]
feature_pos_list = [df[feature][x].values for x in good_masks]
dts_pos_list = [df[feature][x].index[-1] +
pd.Timedelta(1, unit='H') for x in good_masks]
# TODO: add diagnostic mode for how and where are missing features
# now get the negative y's with neg_pos_ratio
# (set to 1 if the same pos=neg):
y_neg_arr = np.zeros(y_pos_arr.shape[0] * neg_pos_ratio)
cnt = 0
feature_neg_list = []
dts_neg_list = []
np.random.seed(seed)
while cnt < len(y_neg_arr):
# get a random date from df:
r = np.random.randint(low=0, high=len(df))
# slice -24 to 24 range with t=0 being the random date:
# update: extend the range to -72 hours to 72 hours:
window_factor = 72 / window_size
slice_range = int(window_size * window_factor)
sliced = df.iloc[r - slice_range:r + slice_range]
# if tides inside this date range, continue:
# try:
if not (y_pos.loc[good_dts].index.intersection(sliced.index)).empty:
# print('#')
continue
# except TypeError:
# return y_pos, good_dts, sliced
# now if no 24 items exist, also continue:
negative = df.iloc[r - window_size:r - 1][feature].dropna().values
if len(negative) != (window_size - 1):
# print('!')
continue
# get the negative datetimes (last record)
neg_dts = df.iloc[r - window_size:r -
1][feature].dropna().index[-1] + pd.Timedelta(1, unit='H')
# else, append to pw_neg_list and increase cnt
feature_neg_list.append(negative)
dts_neg_list.append(neg_dts)
cnt += 1
# print(cnt)
# lastly, assemble for X, y using np.columnstack:
y = np.concatenate([y_pos_arr, y_neg_arr])
# TODO: add exception where no features exist, i.e., there is no
# pw near flood events at all...
Xpos_da = xr.DataArray(feature_pos_list, dims=['sample', 'window', 'feat'])
Xpos_da['window'] = np.arange(0, window_size - 1)
Xpos_da['feat'] = adf.columns
Xpos_da['sample'] = dts_pos_list
Xneg_da = xr.DataArray(feature_neg_list, dims=['sample', 'window', 'feat'])
Xneg_da['window'] = np.arange(0, window_size - 1)
Xneg_da['feat'] = adf.columns
Xneg_da['sample'] = dts_neg_list
X = xr.concat([Xpos_da, Xneg_da], 'sample')
# if feature_pos_list[0].shape[1] > 0 and feature_neg_list[0].shape[1] > 0:
# xpos = [x.ravel() for x in feature_pos_list]
# xneg = [x.ravel() for x in feature_neg_list]
# X = np.column_stack([[x for x in xpos] +
# [x for x in xneg]])
y_dts = np.stack([[x for x in dts_pos_list]+[x for x in dts_neg_list]])
y_dts = y_dts.squeeze()
X_da = X.stack(feature=['feat', 'window'])
feature = ['_'.join([str(x), str(y)]) for x, y in X_da.feature.values]
X_da['feature'] = feature
y_da = xr.DataArray(y, dims=['sample'])
y_da['sample'] = y_dts
# feats = []
# for f in feature:
# feats.append(['{}_{}'.format(f, x) for x in np.arange(0, window_size
# - 1, 1)])
# X_da['feature'] = [item for sublist in feats for item in sublist]
return X_da, y_da
# def preprocess_hydro_pw(pw_station='drag', hs_id=48125, path=work_yuval,
# ims_path=ims_path,
# anoms=True, hydro_path=hydro_path, max_flow=0,
# with_tide_ends=False, pressure_anoms=None,
# add_pressure=False):
# import xarray as xr
# import pandas as pd
# import numpy as np
# from aux_gps import anomalize_xr
# # df.columns = ['tides']
# # now load pw:
# if anoms:
# pw = xr.load_dataset(path / 'GNSS_PW_anom_hourly_50_hour_dayofyear.nc')[pw_station]
# else:
# pw = xr.load_dataset(path / 'GNSS_PW_hourly_thresh_50.nc')[pw_station]
# if pressure_anoms is not None:
# pw = pressure_anoms
# pw_df = pw.dropna('time').to_dataframe()
# # now align the both dataframes:
# pw_df['tides'] = df['tides']
# pw_df['max_flow'] = df['max_flow']
# if add_pressure:
# pressure = xr.load_dataset(ims_path / 'IMS_BP_israeli_hourly.nc')['JERUSALEM-CENTRE']
# pressure = anomalize_xr(pressure, freq='MS')
# pr_df = pressure.dropna('time').to_dataframe()
# pw_df['pressure'] = pr_df
# pw_df = pw_df.fillna(0)
# return pw_df
def loop_over_gnss_hydro_and_aggregate(sel_hydro, pw_anom=False,
pressure_anoms=None,
max_flow_thresh=None,
hydro_path=hydro_path,
work_yuval=work_yuval, ndays=5,
ndays_forward=1,
plot=True, plot_all=False):
import xarray as xr
import matplotlib.pyplot as plt
from aux_gps import path_glob
filename = 'PW_tide_sites_{}_{}.nc'.format(ndays, ndays_forward)
if pw_anom:
filename = 'PW_tide_sites_anom_{}_{}.nc'.format(ndays, ndays_forward)
gnss_stations = []
if (hydro_path / filename).is_file():
print('loading {}...'.format(filename))
ds = xr.load_dataset(hydro_path / filename)
else:
if pw_anom:
file = path_glob(work_yuval, 'GNSS_PW_anom_*.nc')[-1]
gnss_pw = xr.open_dataset(file)
else:
gnss_pw = xr.open_dataset(
work_yuval / 'GNSS_PW_thresh_50_homogenized.nc')
just_pw = [x for x in gnss_pw.data_vars if '_error' not in x]
gnss_pw = gnss_pw[just_pw]
da_list = []
for i, gnss_sta in enumerate(just_pw):
print('proccessing station {}'.format(gnss_sta))
sliced = sel_hydro[~sel_hydro[gnss_sta].isnull()]
hydro_ids = [x for x in sliced.id.values]
if not hydro_ids:
print(
'skipping {} station since no close hydro stations...'.format(gnss_sta))
continue
else:
try:
if pressure_anoms is not None:
pname = pressure_anoms.name
dass = aggregate_get_ndays_pw_hydro(
pressure_anoms,
hydro_ids,
max_flow_thresh=max_flow_thresh,
ndays=ndays, ndays_forward=ndays_forward,
plot=plot_all)
gnss_stations.append(gnss_sta)
dass.name = '{}_{}'.format(pname, i)
else:
dass = aggregate_get_ndays_pw_hydro(
gnss_pw[gnss_sta],
hydro_ids,
max_flow_thresh=max_flow_thresh,
ndays=ndays, ndays_forward=ndays_forward,
plot=plot_all)
da_list.append(dass)
except ValueError as e:
print('skipping {} because {}'.format(gnss_sta, e))
continue
ds = xr.merge(da_list)
ds.to_netcdf(hydro_path / filename, 'w')
if plot:
names = [x for x in ds.data_vars]
fig, ax = plt.subplots()
for name in names:
ds.mean('station').mean('tide_start')[name].plot.line(
marker='.', linewidth=0., ax=ax)
if pressure_anoms is not None:
names = [x.split('_')[0] for x in ds.data_vars]
names = [x + ' ({})'.format(y)
for x, y in zip(names, gnss_stations)]
ax.set_xlabel('Days before tide event')
ax.grid()
hstations = [ds[x].attrs['hydro_stations'] for x in ds.data_vars]
events = [ds[x].attrs['total_events'] for x in ds.data_vars]
fmt = list(zip(names, hstations, events))
ax.legend(['{} with {} stations ({} total events)'.format(x, y, z)
for x, y, z in fmt])
if pw_anom:
title = 'Mean PWV anomalies for tide stations near all GNSS stations'
ylabel = 'PWV anomalies [mm]'
else:
title = 'Mean PWV for tide stations near all GNSS stations'
ylabel = 'PWV [mm]'
if max_flow_thresh is not None:
title += ' (max_flow > {} m^3/sec)'.format(max_flow_thresh)
if pressure_anoms is not None:
ylabel = 'Surface pressure anomalies [hPa]'
title = 'Mean surface pressure anomaly in {} for all tide stations near GNSS stations'.format(
pname)
ax.set_title(title)
ax.set_ylabel(ylabel)
return ds
def aggregate_get_ndays_pw_hydro(pw_da, hs_ids, max_flow_thresh=None,
hydro_path=hydro_path, ndays=5,
ndays_forward=1, plot=True):
import xarray as xr
import matplotlib.pyplot as plt
das = []
max_flows_list = []
pw_ndays_list = []
if not isinstance(hs_ids, list):
hs_ids = [int(hs_ids)]
else:
hs_ids = [int(x) for x in hs_ids]
used_ids = []
events = []
for sid in hs_ids:
print('proccessing hydro station {}'.format(sid))
try:
max_flows, pw_ndays, da = get_n_days_pw_hydro_all(pw_da, sid,
max_flow_thresh=max_flow_thresh,
hydro_path=hydro_path,
ndays=ndays, ndays_forward=ndays_forward,
return_max_flows=True,
plot=False)
das.append(da)
pw_ndays_list.append(pw_ndays)
max_flows_list.append(max_flows)
used_ids.append(sid)
events.append(max_flows.size)
except KeyError as e:
print('{}, skipping...'.format(e))
continue
except ValueError as e:
print('{}, skipping...'.format(e))
continue
pw_ndays = xr.concat(pw_ndays_list, 'time')
dass = xr.concat(das, 'station')
dass['station'] = used_ids
dass.name = pw_da.name
dass.attrs['hydro_stations'] = len(used_ids)
dass.attrs['total_events'] = sum(events)
if plot:
fig, ax = plt.subplots(figsize=(20, 4))
color = 'tab:blue'
pw_ndays.plot.line(marker='.', linewidth=0., color=color, ax=ax)
ax.tick_params(axis='y', labelcolor=color)
ax.set_ylabel('PW [mm]', color=color)
ax2 = ax.twinx()
color = 'tab:red'
for mf in max_flows_list:
mf.plot.line(marker='X', linewidth=0., color=color, ax=ax2)
ax2.tick_params(axis='y', labelcolor=color)
ax.grid()
ax2.set_title(
'PW in station {} {} days before tide events ({} total)'.format(
pw_da.name, ndays, sum(events)))
ax2.set_ylabel('max_flow [m^3/sec]', color=color)
fig.tight_layout()
fig, ax = plt.subplots()
for sid in used_ids:
dass.sel(
station=sid).mean('tide_start').plot.line(
marker='.', linewidth=0., ax=ax)
ax.set_xlabel('Days before tide event')
ax.set_ylabel('PW [mm]')
ax.grid()
fmt = list(zip(used_ids, events))
ax.legend(['station #{} ({} events)'.format(x, y) for x, y in fmt])
ax.set_title(
'Mean PW for tide stations near {} station'.format(pw_da.name))
if max_flow_thresh is not None:
ax.set_title(
'Mean PW for tide stations (above {} m^3/sec) near {} station'.format(
max_flow_thresh, pw_da.name))
return dass
def produce_pwv_days_before_tide_events(pw_da, hs_df, days_prior=1, drop_thresh=0.5,
days_after=1, plot=False, verbose=0,
max_gap='12H', rolling=12):
"""
takes pwv and hydro tide dates from one station and
rounds the hydro tides dates to 5 min
selects the tides dates that are at least the first date of pwv available
then if no pwv data prior to 1 day of tides date - drops
if more than half day missing - drops
then interpolates the missing pwv data points using spline
returns the dataframes contains pwv 1 day before and after tides
and pwv's 1 day prior to event and 1 day after.
Parameters
----------
pw_da : TYPE
pwv of station.
hs_df : TYPE
hydro tide dataframe for one station.
days_prior : TYPE, optional
DESCRIPTION. The default is 1.
drop_thresh : TYPE, optional
DESCRIPTION. The default is 0.5.
days_after : TYPE, optional
DESCRIPTION. The default is 1.
plot : TYPE, optional
DESCRIPTION. The default is False.
verbose : TYPE, optional
DESCRIPTION. The default is 0.
max_gap : TYPE, optional
DESCRIPTION. The default is '12H'.
rolling : TYPE, optional
DESCRIPTION. The default is 12.
Returns
-------
df : TYPE
DESCRIPTION.
pwv_after_list : TYPE
DESCRIPTION.
pwv_prior_list : TYPE
DESCRIPTION.
"""
import pandas as pd
import xarray as xr
import numpy as np
import matplotlib.pyplot as plt
if rolling is not None:
pw_da = pw_da.rolling(time=rolling, center=True).mean(keep_attrs=True)
if drop_thresh is None:
drop_thresh = 0
# first infer time freq of pw_da:
freq = xr.infer_freq(pw_da['time'])
if freq == '5T':
pts_per_day = 288
timedelta = pd.Timedelta(5, unit='min')
if freq == '1H' or freq == 'H':
pts_per_day = 24
timedelta = pd.Timedelta(1, unit='H')
# get the minimum dt of the pwv station:
min_dt = pw_da.dropna('time').time.min().values
# round the hs_df to 5 mins, and find the closest min_dt:
hs_df.index = hs_df.index.round(freq)
hs_df = hs_df[~hs_df.index.duplicated(keep='first')]
hs_df = hs_df.sort_index()
min_ind = hs_df.index.get_loc(min_dt, method='nearest')
# slice the tides data accordinaly:
hs_df = hs_df.iloc[min_ind:].dropna()
# loop over each tide start and grab the datetimes
pwv_prior_list = []
pwv_after_list = []
# se_list = []
tot_events = hs_df.index.size
event_cnt = 0
dropped_thresh = 0
dropped_no_data = 0
for ts in hs_df.index:
dt_prior = ts - pd.Timedelta(days_prior, unit='d')
dt_after = ts + pd.Timedelta(days_after, unit='d')
after_da = pw_da.sel(time=slice(ts, dt_after))
prior_da = pw_da.sel(time=slice(dt_prior, ts - timedelta))
if prior_da.dropna('time').size == 0:
if verbose == 1:
print('{} found no prior data for PWV {} days prior'.format(
ts.strftime('%Y-%m-%d %H:%M'), days_prior))
dropped_no_data += 1
continue
elif prior_da.dropna('time').size < pts_per_day*drop_thresh:
if verbose == 1:
print('{} found less than {} a day prior data for PWV {} days prior'.format(
ts.strftime('%Y-%m-%d %H:%M'), drop_thresh, days_prior))
dropped_thresh += 1
continue
if max_gap is not None:
prior_da = prior_da.interpolate_na(
'time', method='spline', max_gap=max_gap, keep_attrs=True)
event_cnt += 1
# if rolling is not None:
# after_da = after_da.rolling(time=rolling, center=True, keep_attrs=True).mean(keep_attrs=True)
# prior_da = prior_da.rolling(time=rolling, center=True, keep_attrs=True).mean(keep_attrs=True)
# after_da.name = pw_da.name + '_{}'.format(i)
pwv_after_list.append(after_da)
pwv_prior_list.append(prior_da)
# se = da.reset_index('time', drop=True).to_dataframe()[da.name]
# se_list.append(se)
se_list = []
for i, (prior, after) in enumerate(zip(pwv_prior_list, pwv_after_list)):
# return prior, after
# df_p = prior.to_dataframe()
# df_a = after.to_dataframe()
# return df_p, df_a
da = xr.concat([prior, after], 'time')
# print(da)
se = da.reset_index('time', drop=True).to_dataframe()
se.columns = [da.name + '_{}'.format(i)]
# print(se)
# [da.name + '_{}'.format(i)]
se_list.append(se)
df = pd.concat(se_list, axis=1)
df = df.iloc[:-1]
df.index = np.arange(-days_prior, days_after, 1/pts_per_day)
if verbose >= 0:
print('total events with pwv:{} , dropped due to no data: {}, dropped due to thresh:{}, left events: {}'.format(
tot_events, dropped_no_data, dropped_thresh, event_cnt))
if plot:
ax = df.T.mean().plot()
ax.grid()
ax.axvline(color='k', linestyle='--')
ax.set_xlabel('Days before tide event')
ax.set_ylabel('PWV anomalies [mm]')
ax.set_title('GNSS station: {} with {} events'.format(
pw_da.name.upper(), event_cnt))
better = df.copy()
better.index = pd.to_timedelta(better.index, unit='d')
better = better.resample('15S').interpolate(
method='cubic').T.mean().resample('5T').mean()
better = better.reset_index(drop=True)
better.index = np.linspace(-days_prior, days_after, better.index.size)
better.plot(ax=ax)
# fig, ax = plt.subplots(figsize=(20, 7))
# [pwv.plot.line(ax=ax) for pwv in pwv_list]
return df, pwv_after_list, pwv_prior_list
def get_n_days_pw_hydro_all(pw_da, hs_id, max_flow_thresh=None,
hydro_path=hydro_path, ndays=5, ndays_forward=1,
return_max_flows=False, plot=True):
"""calculate the mean of the PW ndays before all tide events in specific
hydro station. can use max_flow_thresh to get only event with al least
this max_flow i.e., big tide events"""
# important, DO NOT dropna pw_da!
import xarray as xr
import matplotlib.pyplot as plt
import pandas as pd
def get_n_days_pw_hydro_one_event(pw_da, tide_start, ndays=ndays, ndays_forward=0):
freq = pd.infer_freq(pw_da.time.values)
# for now, work with 5 mins data:
if freq == '5T':
points = int(ndays) * 24 * 12
points_forward = int(ndays_forward) * 24 * 12
elif freq == '10T':
points = int(ndays) * 24 * 6
points_forward = int(ndays_forward) * 24 * 6
elif freq == 'H':
points = int(ndays) * 24
points_forward = int(ndays_forward) * 24
lag = pd.timedelta_range(end=0, periods=points, freq=freq)
forward_lag = pd.timedelta_range(
start=0, periods=points_forward, freq=freq)
lag = lag.union(forward_lag)
time_arr = pd.to_datetime(pw_da.time.values)
tide_start = pd.to_datetime(tide_start).round(freq)
ts_loc = time_arr.get_loc(tide_start)
# days = pd.Timedelta(ndays, unit='D')
# time_slice = [tide_start - days, tide_start]
# pw = pw_da.sel(time=slice(*time_slice))
pw = pw_da.isel(time=slice(ts_loc - points,
ts_loc + points_forward - 1))
return pw, lag
# first load tides data:
all_tides = xr.open_dataset(hydro_path / 'hydro_tides.nc')
# get all tides for specific station without nans:
sta_slice = [x for x in all_tides.data_vars if str(hs_id) in x]
if not sta_slice:
raise KeyError('hydro station {} not found in database'.format(hs_id))
tides = all_tides[sta_slice].dropna('tide_start')
tide_starts = tides['tide_start'].where(
~tides.isnull()).dropna('tide_start')['tide_start']
# get max flow tides data:
mf = [x for x in tides.data_vars if 'max_flow' in x]
max_flows = tides[mf].dropna('tide_start').to_array('max_flow').squeeze()
# also get tide end and tide max data:
# te = [x for x in tides.data_vars if 'tide_end' in x]
# tide_ends = tides[te].dropna('tide_start').to_array('tide_end').squeeze()
# tm = [x for x in tides.data_vars if 'tide_max' in x]
# tide_maxs = tides[tm].dropna('tide_start').to_array('tide_max').squeeze()
# slice minmum time for convenience:
min_pw_time = pw_da.dropna('time').time.min().values
tide_starts = tide_starts.sel(tide_start=slice(min_pw_time, None))
max_flows = max_flows.sel(tide_start=slice(min_pw_time, None))
# filter if hydro station data ends before gnss pw:
if tide_starts.size == 0:
raise ValueError('tides data end before gnss data begin')
if max_flow_thresh is not None:
# pick only big events:
max_flows = max_flows.where(
max_flows > max_flow_thresh).dropna('tide_start')
tide_starts = tide_starts.where(
max_flows > max_flow_thresh).dropna('tide_start')
pw_list = []
for ts in tide_starts.values:
# te = tide_ends.sel(tide_start=ts).values
# tm = tide_maxs.sel(tide_start=ts).values
pw, lag = get_n_days_pw_hydro_one_event(
pw_da, ts, ndays=ndays, ndays_forward=ndays_forward)
pw.attrs['ts'] = ts
pw_list.append(pw)
# filter events that no PW exists:
pw_list = [x for x in pw_list if x.dropna('time').size > 0]
da = xr.DataArray([x.values for x in pw_list], dims=['tide_start', 'lag'])
da['tide_start'] = [x.attrs['ts'] for x in pw_list] # tide_starts
da['lag'] = lag
# da.name = pw_da.name + '_tide_events'
da.attrs = pw_da.attrs
if max_flow_thresh is not None:
da.attrs['max_flow_minimum'] = max_flow_thresh
pw_ndays = xr.concat(pw_list, 'time')
if plot:
fig, ax = plt.subplots(figsize=(20, 4))
color = 'tab:blue'
pw_ndays.plot.line(marker='.', linewidth=0., color=color, ax=ax)
ax.tick_params(axis='y', labelcolor=color)
ax.set_ylabel('PW [mm]', color=color)
ax2 = ax.twinx()
color = 'tab:red'
max_flows.plot.line(marker='X', linewidth=0., color=color, ax=ax2)
ax2.tick_params(axis='y', labelcolor=color)
ax.grid()
ax2.set_title(
'PW in station {} {} days before tide events'.format(
pw_da.name, ndays))
ax2.set_ylabel('max_flow [m^3/sec]', color=color)
fig.tight_layout()
fig, ax = plt.subplots()
da.mean('tide_start').plot.line(marker='.', linewidth=0., ax=ax)
ax.set_xlabel('Days before tide event')
ax.set_ylabel('PW [mm]')
ax.grid()
ax.set_title(
'Mean PW for {} tide events near {} station'.format(
da.tide_start.size, pw_da.name))
if max_flow_thresh is not None:
ax.set_title(
'Mean PW for {} tide events (above {} m^3/sec) near {} station'.format(
da.tide_start.size, max_flow_thresh, pw_da.name))
if return_max_flows:
return max_flows, pw_ndays, da
else:
return da
def calculate_distance_between_two_latlons_israel(lat1, lon1, lat2, lon2):
import geopandas as gpd
import numpy as np
import pandas as pd
points = np.array(([lat1, lon1], [lat2, lon2]))
df = pd.DataFrame(points, columns=['lat', 'lon'])
pdf = gpd.GeoDataFrame(df, geometry=gpd.points_from_xy(df.lon, df.lat),
crs={'init': 'epsg:4326'})
pdf_meters = pdf.to_crs({'init': 'epsg:6991'})
# distance in meters:
distance = pdf_meters.geometry[0].distance(pdf_meters.geometry[1])
return distance
def get_hydro_near_GNSS(radius=5, n=5, hydro_path=hydro_path,
gis_path=gis_path, plot=True):
import pandas as pd
import geopandas as gpd
from pathlib import Path
import xarray as xr
import matplotlib.pyplot as plt
df = pd.read_csv(Path().cwd() / 'israeli_gnss_coords.txt',
delim_whitespace=True)
df = df[['lon', 'lat']]
gnss = gpd.GeoDataFrame(df, geometry=gpd.points_from_xy(df.lon, df.lat),
crs={'init': 'epsg:4326'})
gnss = gnss.to_crs({'init': 'epsg:2039'})
hydro_meta = read_hydro_metadata(hydro_path, gis_path, plot=False)
hydro_meta = hydro_meta.to_crs({'init': 'epsg:2039'})
for index, row in gnss.iterrows():
# hdict[index] = hydro_meta.geometry.distance(row['geometry'])
hydro_meta[index] = hydro_meta.geometry.distance(row['geometry'])
hydro_meta[index] = hydro_meta[index].where(
hydro_meta[index] <= radius * 1000)
gnss_list = [x for x in gnss.index]
# get only stations within desired radius
mask = ~hydro_meta.loc[:, gnss_list].isnull().all(axis=1)
sel_hydro = hydro_meta.copy()[mask] # pd.concat(hydro_list)
# filter unexisting stations:
tides = xr.load_dataset(hydro_path / 'hydro_tides.nc')
to_remove = []
for index, row in sel_hydro.iterrows():
sid = row['id']
try:
tides['TS_{}_max_flow'.format(sid)]
except KeyError:
print('{} hydro station non-existant in database'.format(sid))
to_remove.append(index)
sel_hydro.drop(to_remove, axis=0, inplace=True)
if plot:
isr = gpd.read_file(gis_path / 'Israel_and_Yosh.shp')
isr.crs = {'init': 'epsg:4326'}
gnss = gnss.to_crs({'init': 'epsg:4326'})
sel_hydro = sel_hydro.to_crs({'init': 'epsg:4326'})
ax = isr.plot(figsize=(10, 16))
sel_hydro.plot(ax=ax, color='yellow', edgecolor='black')
gnss.plot(ax=ax, color='green', edgecolor='black', alpha=0.7)
for x, y, label in zip(gnss.lon, gnss.lat, gnss.index):
ax.annotate(label, xy=(x, y), xytext=(3, 3),
textcoords="offset points")
plt.legend(['hydro-tide stations', 'GNSS stations'], loc='upper left')
plt.suptitle(
'hydro-tide stations within {} km of a GNSS station'.format(radius), fontsize=14)
plt.tight_layout()
plt.subplots_adjust(top=0.95)
# for x, y, label in zip(sel_hydro.lon, sel_hydro.lat,
# sel_hydro.id):
# ax.annotate(label, xy=(x, y), xytext=(3, 3),
# textcoords="offset points")
return sel_hydro
def read_hydro_metadata(path=hydro_path, gis_path=gis_path, plot=True):
import pandas as pd
import geopandas as gpd
import xarray as xr
df = pd.read_excel(hydro_path / 'hydro_stations_metadata.xlsx',
header=4)
# drop last row:
df.drop(df.tail(1).index, inplace=True) # drop last n rows
df.columns = [
'id',
'name',
'active',
'agency',
'type',
'X',
'Y',
'area']
df.loc[:, 'active'][df['active'] == 'פעילה'] = 1
df.loc[:, 'active'][df['active'] == 'לא פעילה'] = 0
df.loc[:, 'active'][df['active'] == 'לא פעילה זמנית'] = 0
df['active'] = df['active'].astype(float)
df = df[~df.X.isnull()]
df = df[~df.Y.isnull()]
# now, geopandas part:
geo_df = gpd.GeoDataFrame(df, geometry=gpd.points_from_xy(df.X, df.Y),
crs={'init': 'epsg:2039'})
# geo_df.crs = {'init': 'epsg:2039'}
geo_df = geo_df.to_crs({'init': 'epsg:4326'})
isr_dem = xr.open_rasterio(gis_path / 'israel_dem.tif')
alt_list = []
for index, row in geo_df.iterrows():
lat = row.geometry.y
lon = row.geometry.x
alt = isr_dem.sel(band=1, x=lon, y=lat, method='nearest').values.item()
alt_list.append(float(alt))
geo_df['alt'] = alt_list
geo_df['lat'] = geo_df.geometry.y
geo_df['lon'] = geo_df.geometry.x
isr = gpd.read_file(gis_path / 'Israel_and_Yosh.shp')
isr.crs = {'init': 'epsg:4326'}
geo_df = gpd.sjoin(geo_df, isr, op='within')
if plot:
ax = isr.plot()
geo_df.plot(ax=ax, edgecolor='black', legend=True)
return geo_df
def read_tides(path=hydro_path):
from aux_gps import path_glob
import pandas as pd
import xarray as xr
from aux_gps import get_unique_index
files = path_glob(path, 'tide_report*.xlsx')
df_list = []
for file in files:
df = pd.read_excel(file, header=4)
df.drop(df.columns[len(df.columns) - 1], axis=1, inplace=True)
df.columns = [
'id',
'name',
'hydro_year',
'tide_start_hour',
'tide_start_date',
'tide_end_hour',
'tide_end_date',
'tide_duration',
'tide_max_hour',
'tide_max_date',
'max_height',
'max_flow[m^3/sec]',
'tide_vol[MCM]']
df = df[~df.hydro_year.isnull()]
df['id'] = df['id'].astype(int)
df['tide_start'] = pd.to_datetime(
df['tide_start_date'], dayfirst=True) + pd.to_timedelta(
df['tide_start_hour'].add(':00'), unit='m', errors='coerce')
# tides are in local Israeli winter clock (no DST):
# dst = np.zeros(df['tide_start'].shape)
# df['tide_start'] = df['tide_start'].dt.tz_localize('Asia/Jerusalem', ambiguous=dst).dt.tz_convert('UTC')
df['tide_start'] = df['tide_start'] - pd.Timedelta(2, unit='H')
df['tide_end'] = pd.to_datetime(
df['tide_end_date'], dayfirst=True) + pd.to_timedelta(
df['tide_end_hour'].add(':00'),
unit='m',
errors='coerce')
# also to tide ends:
df['tide_end'] = df['tide_end'] - pd.Timedelta(2, unit='H')
# df['tide_end'] = df['tide_end'].dt.tz_localize('Asia/Jerusalem', ambiguous=dst).dt.tz_convert('UTC')
df['tide_max'] = pd.to_datetime(
df['tide_max_date'], dayfirst=True) + pd.to_timedelta(
df['tide_max_hour'].add(':00'),
unit='m',
errors='coerce')
# also to tide max:
# df['tide_max'] = df['tide_max'].dt.tz_localize('Asia/Jerusalem', ambiguous=dst).dt.tz_convert('UTC')
df['tide_max'] = df['tide_max'] - pd.Timedelta(2, unit='H')
df['tide_duration'] = pd.to_timedelta(
df['tide_duration'] + ':00', unit='m', errors='coerce')
df.loc[:,
'max_flow[m^3/sec]'][df['max_flow[m^3/sec]'].str.contains('<',
na=False)] = 0
df.loc[:, 'tide_vol[MCM]'][df['tide_vol[MCM]'].str.contains(
'<', na=False)] = 0
df['max_flow[m^3/sec]'] = df['max_flow[m^3/sec]'].astype(float)
df['tide_vol[MCM]'] = df['tide_vol[MCM]'].astype(float)
to_drop = ['tide_start_hour', 'tide_start_date', 'tide_end_hour',
'tide_end_date', 'tide_max_hour', 'tide_max_date']
df = df.drop(to_drop, axis=1)
df_list.append(df)
df = pd.concat(df_list)
dfs = [x for _, x in df.groupby('id')]
ds_list = []
meta_df = read_hydro_metadata(path, gis_path, False)
for df in dfs:
st_id = df['id'].iloc[0]
st_name = df['name'].iloc[0]
print('proccessing station number: {}, {}'.format(st_id, st_name))
meta = meta_df[meta_df['id'] == st_id]
ds = xr.Dataset()
df.set_index('tide_start', inplace=True)
attrs = {}
attrs['station_name'] = st_name
if not meta.empty:
attrs['lon'] = meta.lon.values.item()
attrs['lat'] = meta.lat.values.item()
attrs['alt'] = meta.alt.values.item()
attrs['drainage_basin_area'] = meta.area.values.item()
attrs['active'] = meta.active.values.item()
attrs['units'] = 'm'
max_height = df['max_height'].to_xarray()
max_height.name = 'TS_{}_max_height'.format(st_id)
max_height.attrs = attrs
max_flow = df['max_flow[m^3/sec]'].to_xarray()
max_flow.name = 'TS_{}_max_flow'.format(st_id)
attrs['units'] = 'm^3/sec'
max_flow.attrs = attrs
attrs['units'] = 'MCM'
tide_vol = df['tide_vol[MCM]'].to_xarray()
tide_vol.name = 'TS_{}_tide_vol'.format(st_id)
tide_vol.attrs = attrs
attrs.pop('units')
# tide_start = df['tide_start'].to_xarray()
# tide_start.name = 'TS_{}_tide_start'.format(st_id)
# tide_start.attrs = attrs
tide_end = df['tide_end'].to_xarray()
tide_end.name = 'TS_{}_tide_end'.format(st_id)
tide_end.attrs = attrs
tide_max = df['tide_max'].to_xarray()
tide_max.name = 'TS_{}_tide_max'.format(st_id)
tide_max.attrs = attrs
ds['{}'.format(max_height.name)] = max_height
ds['{}'.format(max_flow.name)] = max_flow
ds['{}'.format(tide_vol.name)] = tide_vol
# ds['{}'.format(tide_start.name)] = tide_start
ds['{}'.format(tide_end.name)] = tide_end
ds['{}'.format(tide_max.name)] = tide_max
ds_list.append(ds)
dsu = [get_unique_index(x, dim='tide_start') for x in ds_list]
print('merging...')
ds = xr.merge(dsu)
ds.attrs['time'] = 'UTC'
filename = 'hydro_tides.nc'
print('saving {} to {}'.format(filename, path))
comp = dict(zlib=True, complevel=9) # best compression
encoding = {var: comp for var in ds.data_vars}
ds.to_netcdf(path / filename, 'w', encoding=encoding)
print('Done!')
return ds
def plot_hydro_events(hs_id, path=hydro_path, field='max_flow', min_flow=10):
import xarray as xr
import matplotlib.pyplot as plt
tides = xr.open_dataset(path/'hydro_tides.nc')
sta_slice = [x for x in tides.data_vars if str(hs_id) in x]
tide = tides[sta_slice]['TS_{}_{}'.format(hs_id, field)]
tide = tide.dropna('tide_start')
fig, ax = plt.subplots()
tide.plot.line(linewidth=0., marker='x', color='r', ax=ax)
if min_flow is not None:
tide[tide > min_flow].plot.line(
linewidth=0., marker='x', color='b', ax=ax)
print('min flow of {} m^3/sec: {}'.format(min_flow,
tide[tide > min_flow].dropna('tide_start').size))
return tide
def text_process_hydrographs(path=hydro_path, gis_path=gis_path):
from aux_gps import path_glob
files = path_glob(path, 'hydro_flow*.txt')
for i, file in enumerate(files):
print(file)
with open(file, 'r') as f:
big_list = f.read().splitlines()
# for small_list in big_list:
# flat_list = [item for sublist in l7 for item in sublist]
big = [x.replace(',', ' ') for x in big_list]
big = big[6:]
big = [x.replace('\t', ',') for x in big]
filename = 'hydro_graph_{}.txt'.format(i)
with open(path / filename, 'w') as fs:
for item in big:
fs.write('{}\n'.format(item))
print('{} saved to {}'.format(filename, path))
return
def read_hydrographs(path=hydro_path):
from aux_gps import path_glob
import pandas as pd
import xarray as xr
from aux_gps import get_unique_index
files = path_glob(path, 'hydro_graph*.txt')
df_list = []
for file in files:
print(file)
df = pd.read_csv(file, header=0, sep=',')
df.columns = [
'id',
'name',
'time',
'tide_height[m]',
'flow[m^3/sec]',
'data_type',
'flow_type',
'record_type',
'record_code']
# make sure the time is in UTC since database is in ISR winter clock (no DST)
df['time'] = pd.to_datetime(df['time'], dayfirst=True) - pd.Timedelta(2, unit='H')
df['tide_height[m]'] = df['tide_height[m]'].astype(float)
df['flow[m^3/sec]'] = df['flow[m^3/sec]'].astype(float)
df.loc[:, 'data_type'][df['data_type'].str.contains(
'מדודים', na=False)] = 'measured'
df.loc[:, 'data_type'][df['data_type'].str.contains(
'משוחזרים', na=False)] = 'reconstructed'
df.loc[:, 'flow_type'][df['flow_type'].str.contains(
'תקין', na=False)] = 'normal'
df.loc[:, 'flow_type'][df['flow_type'].str.contains(
'גאות', na=False)] = 'tide'
df.loc[:, 'record_type'][df['record_type'].str.contains(
'נקודה פנימית', na=False)] = 'inner_point'
df.loc[:, 'record_type'][df['record_type'].str.contains(
'נקודה פנימית', na=False)] = 'inner_point'
df.loc[:, 'record_type'][df['record_type'].str.contains(
'התחלת קטע', na=False)] = 'section_begining'
df.loc[:, 'record_type'][df['record_type'].str.contains(
'סיום קטע', na=False)] = 'section_ending'
df_list.append(df)
df = pd.concat(df_list)
dfs = [x for _, x in df.groupby('id')]
ds_list = []
meta_df = read_hydro_metadata(path, gis_path, False)
for df in dfs:
st_id = df['id'].iloc[0]
st_name = df['name'].iloc[0]
print('proccessing station number: {}, {}'.format(st_id, st_name))
meta = meta_df[meta_df['id'] == st_id]
ds = xr.Dataset()
df.set_index('time', inplace=True)
attrs = {}
attrs['station_name'] = st_name
if not meta.empty:
attrs['lon'] = meta.lon.values.item()
attrs['lat'] = meta.lat.values.item()
attrs['alt'] = meta.alt.values.item()
attrs['drainage_basin_area'] = meta.area.values.item()
attrs['active'] = meta.active.values.item()
attrs['units'] = 'm'
tide_height = df['tide_height[m]'].to_xarray()
tide_height.name = 'HS_{}_tide_height'.format(st_id)
tide_height.attrs = attrs
flow = df['flow[m^3/sec]'].to_xarray()
flow.name = 'HS_{}_flow'.format(st_id)
attrs['units'] = 'm^3/sec'
flow.attrs = attrs
ds['{}'.format(tide_height.name)] = tide_height
ds['{}'.format(flow.name)] = flow
ds_list.append(ds)
dsu = [get_unique_index(x) for x in ds_list]
print('merging...')
ds = xr.merge(dsu)
ds.attrs['time'] = 'UTC'
filename = 'hydro_graphs.nc'
print('saving {} to {}'.format(filename, path))
comp = dict(zlib=True, complevel=9) # best compression
encoding = {var: comp for var in ds.data_vars}
ds.to_netcdf(path / filename, 'w', encoding=encoding)
print('Done!')
return ds
def read_station_from_tide_database(hs_id=48125, rounding='1H',
hydro_path=hydro_path):
import xarray as xr
all_tides = xr.open_dataset(hydro_path / 'hydro_tides.nc')
# get all tides for specific station without nans:
sta_slice = [x for x in all_tides.data_vars if str(hs_id) in x]
if not sta_slice:
raise KeyError('hydro station {} not found in database'.format(hs_id))
# tides = all_tides[sta_slice].dropna('tide_start')
df = all_tides[sta_slice].to_dataframe()
df.columns = ['max_height', 'max_flow', 'tide_vol', 'tide_end', 'tide_max']
df = df[df['max_flow'] != 0]
df['hydro_station_id'] = hs_id
if rounding is not None:
print('rounding to {}'.format(rounding))
df.index = df.index.round(rounding)
return df
# tide_starts = tides['tide_start'].where(
# ~tides.isnull()).dropna('tide_start')['tide_start']
def check_if_tide_events_from_stations_are_within_time_window(df_list, rounding='H',
days=1, return_hs_list=False):
import pandas as pd
dfs = []
for i, df in enumerate(df_list):
df.dropna(inplace=True)
if rounding is not None:
df.index = df.index.round(rounding)
dfs.append(df['hydro_station_id'])
df = pd.concat(dfs, axis=0).to_frame()
df['time'] = df.index
df = df.sort_index()
# filter co-tide events:
df = df.loc[~df.index.duplicated()]
print('found {} co-tide events'.format(df.index.duplicated().sum()))
# secondly check for events that are within days period of each other and filter:
dif = df['time'].diff()
mask = abs(dif) <= pd.Timedelta(days, unit='D')
dupes = dif[mask].index
print('found {} tide events that are within {} of each other.'.format(
dupes.size, days))
print(df.loc[dupes, 'hydro_station_id'])
df = df.loc[~mask]
if return_hs_list:
hs_ids = [x['hydro_station_id'].iloc[0] for x in df_list]
df_list = [df[df['hydro_station_id'] == x] for x in hs_ids]
return df_list
else:
return df
def standertize_pwv_using_long_term_stat(axis_ds, hydro_path=hydro_path, filename='axis_southern_stations_stats.nc'):
from aux_gps import transform_time_series_groups_agg_to_time_series
import xarray as xr
import pandas as pd
stats = xr.load_dataset(hydro_path/filename)
da_list = []
for da in axis_ds:
df_mean = transform_time_series_groups_agg_to_time_series(axis_ds[da], stats, stat='mean')
df_std = transform_time_series_groups_agg_to_time_series(axis_ds[da], stats, stat='std')
if df_mean is None or df_std is None:
print('No stats for {}, skipping...'.format(da))
continue
df = pd.concat([df_mean, df_std], axis=1)
df = df.loc[:, ~df.columns.duplicated()]
df['anomalies'] = (df[da] - df['mean'])/df['std']
da_anom = df['anomalies'].to_xarray()
da_anom.name = da
da_anom.attrs = axis_ds[da].attrs
da_anom.attrs['action'] = 'standertized by hour and day of year'
da_list.append(da_anom)
ds = xr.merge(da_list)
return ds
def get_closest_southern_axis_station_to_SOI_and_produce_long_term_stats(axis_path=axis_path, soi_path=cwd,
gnss_path=gnss_path, savepath=hydro_path):
from axis_process import read_axis_stations
from PW_stations import load_gipsyx_PWV_time_series
import geopandas as gpd
import pandas as pd
import xarray as xr
from aux_gps import save_ncfile
def min_dist(point, gpd2):
gpd2['Dist'] = gpd2.apply(
lambda row: point.distance(
row.geometry), axis=1)
geoseries = gpd2.iloc[gpd2['Dist'].values.argmin()]
geoseries.loc['distance'] = gpd2['Dist'].values.min()
return geoseries
# read axis stations:
df = read_axis_stations(axis_path)
axis = gpd.GeoDataFrame(df, geometry=gpd.points_from_xy(df['lon'], df['lat']))
axis.crs = {'init': 'epsg:4326'}
# select only southern stations:
axis = axis.loc[axis_southern_stations]
# convert to Israeli grid (meters):
axis = axis.to_crs(epsg='2039')
# read soi stations:
soi = pd.read_csv(soi_path/'israeli_gnss_coords.txt', delim_whitespace=True)
soi.drop(soi.tail(4).index,axis=0, inplace=True)
soi.drop(['lhav', 'gilb'],axis=0, inplace=True)
soi = gpd.GeoDataFrame(soi, geometry=gpd.points_from_xy(soi['lon'], soi['lat']))
soi.crs = {'init': 'epsg:4326'}
# convert to Israeli grid (meters):
soi = soi.to_crs(epsg='2039')
# now, iterate over axis (8 southern stations) and find the min distance to each soi station:
min_list = []
for gps_rows in axis.iterrows():
ims_min_series = min_dist(gps_rows[1]['geometry'], soi)
min_list.append(ims_min_series)
soi_df = pd.concat(min_list, axis=1).T
axis_sub = axis.reset_index()[['station','alt']].set_index(soi_df.index)
axis_sub.columns = ['axis_station', 'axis_alt']
soi_df = pd.concat([soi_df, axis_sub], axis=1)
# now find the bias between two sets of stations, using lapse_rate:
height = soi_df[['axis_station', 'axis_alt']]
Hdf = find_pwv_at_surface_and_scale_height_soi_and_fix_with_height(height_df=height)
soi_df['bias'] = Hdf['bias'].dropna()
# now, iterate over soi_df stations and produce stats:
das = []
for i, soi_sta in enumerate(soi_df.index):
# load soi station:
sta = load_gipsyx_PWV_time_series(station=soi_sta, gnss_path=gnss_path)
# take only PWV and add the bias:
bias = soi_df.iloc[i]['bias']
sta = sta[soi_sta] + bias
print('fixed {} station by {} mm.'.format(soi_sta, bias))
df = sta.to_dataframe()
# df['month'] = df.index.month
df['dayofyear'] = df.index.dayofyear
df['hour'] = df.index.hour
da_mean = df.groupby(['dayofyear', 'hour']).mean().to_xarray()
da_mean[soi_sta].attrs = sta.attrs
da_std = df.groupby(['dayofyear', 'hour']).std().to_xarray()
da_std[soi_sta].attrs = sta.attrs
da = xr.concat([da_mean, da_std], 'agg')
da['agg'] = ['mean', 'std']
das.append(da)
soi_stats = xr.merge(das)
soi_stats = soi_stats.rename(soi_df['axis_station'].to_dict())
if savepath is not None:
filename = 'axis_southern_stations_stats.nc'
save_ncfile(soi_stats, savepath, filename)
return soi_df, soi_stats
def find_pwv_at_surface_and_scale_height_soi_and_fix_with_height(path=cwd,
gnss_path=gnss_path,
height_df=None):
import pandas as pd
from PW_stations import load_gipsyx_PWV_time_series
from interpolation_routines import get_var_lapse_rate
from interpolation_routines import apply_lapse_rate_change
import numpy as np
soi = pd.read_csv(path/'israeli_gnss_coords.txt', delim_whitespace=True)
soi.drop(soi.tail(4).index,axis=0, inplace=True)
soi.drop(['lhav', 'gilb', 'hrmn'],axis=0, inplace=True)
ds = load_gipsyx_PWV_time_series(station=None, gnss_path=gnss_path)
hdf=ds.mean('time').expand_dims('time').to_dataframe().T
hdf = hdf.join(soi[['lat', 'lon', 'alt']])
hdf = hdf.reset_index()
hdf = hdf.set_index('alt')
hdf = hdf.sort_index().dropna()
hdf.columns = ['name', 'pwv', 'lat', 'lon']
hdf = hdf[['pwv', 'lat', 'lon', 'name']]
H = get_var_lapse_rate(hdf, plot=True)
hdf_at_surface = apply_lapse_rate_change(hdf, H)
hdf['pwv_sur'] = hdf_at_surface['pwv']
df = hdf.reset_index()
df = df.set_index('name')
if height_df is not None:
df = | pd.concat([df, height_df], axis=1) | pandas.concat |
#
# Licensed to the Apache Software Foundation (ASF) under one or more
# contributor license agreements. See the NOTICE file distributed with
# this work for additional information regarding copyright ownership.
# The ASF licenses this file to You under the Apache License, Version 2.0
# (the "License"); you may not use this file except in compliance with
# the License. You may obtain a copy of the License at
#
# http://www.apache.org/licenses/LICENSE-2.0
#
# Unless required by applicable law or agreed to in writing, software
# distributed under the License is distributed on an "AS IS" BASIS,
# WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.
# See the License for the specific language governing permissions and
# limitations under the License.
#
import decimal
from datetime import datetime
from distutils.version import LooseVersion
import inspect
import sys
import unittest
from io import StringIO
from typing import List
import numpy as np
import pandas as pd
from pandas.tseries.offsets import DateOffset
from pyspark import StorageLevel
from pyspark.ml.linalg import SparseVector
from pyspark.sql.types import StructType
from pyspark import pandas as ps
from pyspark.pandas.config import option_context
from pyspark.pandas.exceptions import PandasNotImplementedError
from pyspark.pandas.frame import CachedDataFrame
from pyspark.pandas.missing.frame import _MissingPandasLikeDataFrame
from pyspark.pandas.typedef.typehints import (
extension_dtypes,
extension_dtypes_available,
extension_float_dtypes_available,
extension_object_dtypes_available,
)
from pyspark.testing.pandasutils import (
have_tabulate,
PandasOnSparkTestCase,
SPARK_CONF_ARROW_ENABLED,
tabulate_requirement_message,
)
from pyspark.testing.sqlutils import SQLTestUtils
from pyspark.pandas.utils import name_like_string
class DataFrameTest(PandasOnSparkTestCase, SQLTestUtils):
@property
def pdf(self):
return pd.DataFrame(
{"a": [1, 2, 3, 4, 5, 6, 7, 8, 9], "b": [4, 5, 6, 3, 2, 1, 0, 0, 0]},
index=np.random.rand(9),
)
@property
def psdf(self):
return ps.from_pandas(self.pdf)
@property
def df_pair(self):
pdf = self.pdf
psdf = ps.from_pandas(pdf)
return pdf, psdf
def test_dataframe(self):
pdf, psdf = self.df_pair
self.assert_eq(psdf["a"] + 1, pdf["a"] + 1)
self.assert_eq(psdf.columns, pd.Index(["a", "b"]))
self.assert_eq(psdf[psdf["b"] > 2], pdf[pdf["b"] > 2])
self.assert_eq(-psdf[psdf["b"] > 2], -pdf[pdf["b"] > 2])
self.assert_eq(psdf[["a", "b"]], pdf[["a", "b"]])
self.assert_eq(psdf.a, pdf.a)
self.assert_eq(psdf.b.mean(), pdf.b.mean())
self.assert_eq(psdf.b.var(), pdf.b.var())
self.assert_eq(psdf.b.std(), pdf.b.std())
pdf, psdf = self.df_pair
self.assert_eq(psdf[["a", "b"]], pdf[["a", "b"]])
self.assertEqual(psdf.a.notnull().rename("x").name, "x")
# check ps.DataFrame(ps.Series)
pser = pd.Series([1, 2, 3], name="x", index=np.random.rand(3))
psser = ps.from_pandas(pser)
self.assert_eq(pd.DataFrame(pser), ps.DataFrame(psser))
# check psdf[pd.Index]
pdf, psdf = self.df_pair
column_mask = pdf.columns.isin(["a", "b"])
index_cols = pdf.columns[column_mask]
self.assert_eq(psdf[index_cols], pdf[index_cols])
def _check_extension(self, psdf, pdf):
if LooseVersion("1.1") <= LooseVersion(pd.__version__) < LooseVersion("1.2.2"):
self.assert_eq(psdf, pdf, check_exact=False)
for dtype in psdf.dtypes:
self.assertTrue(isinstance(dtype, extension_dtypes))
else:
self.assert_eq(psdf, pdf)
@unittest.skipIf(not extension_dtypes_available, "pandas extension dtypes are not available")
def test_extension_dtypes(self):
pdf = pd.DataFrame(
{
"a": pd.Series([1, 2, None, 4], dtype="Int8"),
"b": pd.Series([1, None, None, 4], dtype="Int16"),
"c": pd.Series([1, 2, None, None], dtype="Int32"),
"d": pd.Series([None, 2, None, 4], dtype="Int64"),
}
)
psdf = ps.from_pandas(pdf)
self._check_extension(psdf, pdf)
self._check_extension(psdf + psdf, pdf + pdf)
@unittest.skipIf(not extension_dtypes_available, "pandas extension dtypes are not available")
def test_astype_extension_dtypes(self):
pdf = pd.DataFrame(
{
"a": [1, 2, None, 4],
"b": [1, None, None, 4],
"c": [1, 2, None, None],
"d": [None, 2, None, 4],
}
)
psdf = ps.from_pandas(pdf)
astype = {"a": "Int8", "b": "Int16", "c": "Int32", "d": "Int64"}
self._check_extension(psdf.astype(astype), pdf.astype(astype))
@unittest.skipIf(
not extension_object_dtypes_available, "pandas extension object dtypes are not available"
)
def test_extension_object_dtypes(self):
pdf = pd.DataFrame(
{
"a": pd.Series(["a", "b", None, "c"], dtype="string"),
"b": pd.Series([True, None, False, True], dtype="boolean"),
}
)
psdf = ps.from_pandas(pdf)
self._check_extension(psdf, pdf)
@unittest.skipIf(
not extension_object_dtypes_available, "pandas extension object dtypes are not available"
)
def test_astype_extension_object_dtypes(self):
pdf = pd.DataFrame({"a": ["a", "b", None, "c"], "b": [True, None, False, True]})
psdf = ps.from_pandas(pdf)
astype = {"a": "string", "b": "boolean"}
self._check_extension(psdf.astype(astype), pdf.astype(astype))
@unittest.skipIf(
not extension_float_dtypes_available, "pandas extension float dtypes are not available"
)
def test_extension_float_dtypes(self):
pdf = pd.DataFrame(
{
"a": pd.Series([1.0, 2.0, None, 4.0], dtype="Float32"),
"b": pd.Series([1.0, None, 3.0, 4.0], dtype="Float64"),
}
)
psdf = ps.from_pandas(pdf)
self._check_extension(psdf, pdf)
self._check_extension(psdf + 1, pdf + 1)
self._check_extension(psdf + psdf, pdf + pdf)
@unittest.skipIf(
not extension_float_dtypes_available, "pandas extension float dtypes are not available"
)
def test_astype_extension_float_dtypes(self):
pdf = pd.DataFrame({"a": [1.0, 2.0, None, 4.0], "b": [1.0, None, 3.0, 4.0]})
psdf = ps.from_pandas(pdf)
astype = {"a": "Float32", "b": "Float64"}
self._check_extension(psdf.astype(astype), pdf.astype(astype))
def test_insert(self):
#
# Basic DataFrame
#
pdf = pd.DataFrame([1, 2, 3])
psdf = ps.from_pandas(pdf)
psdf.insert(1, "b", 10)
pdf.insert(1, "b", 10)
self.assert_eq(psdf.sort_index(), pdf.sort_index(), almost=True)
psdf.insert(2, "c", 0.1)
pdf.insert(2, "c", 0.1)
self.assert_eq(psdf.sort_index(), pdf.sort_index(), almost=True)
psdf.insert(3, "d", psdf.b + 1)
pdf.insert(3, "d", pdf.b + 1)
self.assert_eq(psdf.sort_index(), pdf.sort_index(), almost=True)
psser = ps.Series([4, 5, 6])
self.assertRaises(ValueError, lambda: psdf.insert(0, "y", psser))
self.assertRaisesRegex(
ValueError, "cannot insert b, already exists", lambda: psdf.insert(1, "b", 10)
)
self.assertRaisesRegex(
TypeError,
'"column" should be a scalar value or tuple that contains scalar values',
lambda: psdf.insert(0, list("abc"), psser),
)
self.assertRaisesRegex(
TypeError,
"loc must be int",
lambda: psdf.insert((1,), "b", 10),
)
self.assertRaisesRegex(
NotImplementedError,
"Assigning column name as tuple is only supported for MultiIndex columns for now.",
lambda: psdf.insert(0, ("e",), 10),
)
self.assertRaises(ValueError, lambda: psdf.insert(0, "e", [7, 8, 9, 10]))
self.assertRaises(ValueError, lambda: psdf.insert(0, "f", ps.Series([7, 8])))
self.assertRaises(AssertionError, lambda: psdf.insert(100, "y", psser))
self.assertRaises(AssertionError, lambda: psdf.insert(1, "y", psser, allow_duplicates=True))
#
# DataFrame with MultiIndex as columns
#
pdf = pd.DataFrame({("x", "a", "b"): [1, 2, 3]})
psdf = ps.from_pandas(pdf)
psdf.insert(1, "b", 10)
pdf.insert(1, "b", 10)
self.assert_eq(psdf.sort_index(), pdf.sort_index(), almost=True)
psdf.insert(2, "c", 0.1)
pdf.insert(2, "c", 0.1)
self.assert_eq(psdf.sort_index(), pdf.sort_index(), almost=True)
psdf.insert(3, "d", psdf.b + 1)
pdf.insert(3, "d", pdf.b + 1)
self.assert_eq(psdf.sort_index(), pdf.sort_index(), almost=True)
self.assertRaisesRegex(
ValueError, "cannot insert d, already exists", lambda: psdf.insert(4, "d", 11)
)
self.assertRaisesRegex(
ValueError,
r"cannot insert \('x', 'a', 'b'\), already exists",
lambda: psdf.insert(4, ("x", "a", "b"), 11),
)
self.assertRaisesRegex(
ValueError,
'"column" must have length equal to number of column levels.',
lambda: psdf.insert(4, ("e",), 11),
)
def test_inplace(self):
pdf, psdf = self.df_pair
pser = pdf.a
psser = psdf.a
pdf["a"] = pdf["a"] + 10
psdf["a"] = psdf["a"] + 10
self.assert_eq(psdf, pdf)
self.assert_eq(psser, pser)
def test_assign_list(self):
pdf, psdf = self.df_pair
pser = pdf.a
psser = psdf.a
pdf["x"] = [10, 20, 30, 40, 50, 60, 70, 80, 90]
psdf["x"] = [10, 20, 30, 40, 50, 60, 70, 80, 90]
self.assert_eq(psdf.sort_index(), pdf.sort_index())
self.assert_eq(psser, pser)
with self.assertRaisesRegex(ValueError, "Length of values does not match length of index"):
psdf["z"] = [10, 20, 30, 40, 50, 60, 70, 80]
def test_dataframe_multiindex_columns(self):
pdf = pd.DataFrame(
{
("x", "a", "1"): [1, 2, 3],
("x", "b", "2"): [4, 5, 6],
("y.z", "c.d", "3"): [7, 8, 9],
("x", "b", "4"): [10, 11, 12],
},
index=np.random.rand(3),
)
psdf = ps.from_pandas(pdf)
self.assert_eq(psdf, pdf)
self.assert_eq(psdf["x"], pdf["x"])
self.assert_eq(psdf["y.z"], pdf["y.z"])
self.assert_eq(psdf["x"]["b"], pdf["x"]["b"])
self.assert_eq(psdf["x"]["b"]["2"], pdf["x"]["b"]["2"])
self.assert_eq(psdf.x, pdf.x)
self.assert_eq(psdf.x.b, pdf.x.b)
self.assert_eq(psdf.x.b["2"], pdf.x.b["2"])
self.assertRaises(KeyError, lambda: psdf["z"])
self.assertRaises(AttributeError, lambda: psdf.z)
self.assert_eq(psdf[("x",)], pdf[("x",)])
self.assert_eq(psdf[("x", "a")], pdf[("x", "a")])
self.assert_eq(psdf[("x", "a", "1")], pdf[("x", "a", "1")])
def test_dataframe_column_level_name(self):
column = pd.Index(["A", "B", "C"], name="X")
pdf = pd.DataFrame([[1, 2, 3], [4, 5, 6]], columns=column, index=np.random.rand(2))
psdf = ps.from_pandas(pdf)
self.assert_eq(psdf, pdf)
self.assert_eq(psdf.columns.names, pdf.columns.names)
self.assert_eq(psdf.to_pandas().columns.names, pdf.columns.names)
def test_dataframe_multiindex_names_level(self):
columns = pd.MultiIndex.from_tuples(
[("X", "A", "Z"), ("X", "B", "Z"), ("Y", "C", "Z"), ("Y", "D", "Z")],
names=["lvl_1", "lvl_2", "lv_3"],
)
pdf = pd.DataFrame(
[[1, 2, 3, 4], [5, 6, 7, 8], [9, 10, 11, 12], [13, 14, 15, 16], [17, 18, 19, 20]],
columns=columns,
index=np.random.rand(5),
)
psdf = ps.from_pandas(pdf)
self.assert_eq(psdf.columns.names, pdf.columns.names)
self.assert_eq(psdf.to_pandas().columns.names, pdf.columns.names)
psdf1 = ps.from_pandas(pdf)
self.assert_eq(psdf1.columns.names, pdf.columns.names)
self.assertRaises(
AssertionError,
lambda: ps.DataFrame(psdf1._internal.copy(column_label_names=("level",))),
)
self.assert_eq(psdf["X"], pdf["X"])
self.assert_eq(psdf["X"].columns.names, pdf["X"].columns.names)
self.assert_eq(psdf["X"].to_pandas().columns.names, pdf["X"].columns.names)
self.assert_eq(psdf["X"]["A"], pdf["X"]["A"])
self.assert_eq(psdf["X"]["A"].columns.names, pdf["X"]["A"].columns.names)
self.assert_eq(psdf["X"]["A"].to_pandas().columns.names, pdf["X"]["A"].columns.names)
self.assert_eq(psdf[("X", "A")], pdf[("X", "A")])
self.assert_eq(psdf[("X", "A")].columns.names, pdf[("X", "A")].columns.names)
self.assert_eq(psdf[("X", "A")].to_pandas().columns.names, pdf[("X", "A")].columns.names)
self.assert_eq(psdf[("X", "A", "Z")], pdf[("X", "A", "Z")])
def test_itertuples(self):
pdf = pd.DataFrame({"num_legs": [4, 2], "num_wings": [0, 2]}, index=["dog", "hawk"])
psdf = ps.from_pandas(pdf)
for ptuple, ktuple in zip(
pdf.itertuples(index=False, name="Animal"), psdf.itertuples(index=False, name="Animal")
):
self.assert_eq(ptuple, ktuple)
for ptuple, ktuple in zip(pdf.itertuples(name=None), psdf.itertuples(name=None)):
self.assert_eq(ptuple, ktuple)
pdf.index = pd.MultiIndex.from_arrays(
[[1, 2], ["black", "brown"]], names=("count", "color")
)
psdf = ps.from_pandas(pdf)
for ptuple, ktuple in zip(pdf.itertuples(name="Animal"), psdf.itertuples(name="Animal")):
self.assert_eq(ptuple, ktuple)
pdf.columns = pd.MultiIndex.from_arrays(
[["CA", "WA"], ["age", "children"]], names=("origin", "info")
)
psdf = ps.from_pandas(pdf)
for ptuple, ktuple in zip(pdf.itertuples(name="Animal"), psdf.itertuples(name="Animal")):
self.assert_eq(ptuple, ktuple)
pdf = pd.DataFrame([1, 2, 3])
psdf = ps.from_pandas(pdf)
for ptuple, ktuple in zip(
(pdf + 1).itertuples(name="num"), (psdf + 1).itertuples(name="num")
):
self.assert_eq(ptuple, ktuple)
# DataFrames with a large number of columns (>254)
pdf = pd.DataFrame(np.random.random((1, 255)))
psdf = ps.from_pandas(pdf)
for ptuple, ktuple in zip(pdf.itertuples(name="num"), psdf.itertuples(name="num")):
self.assert_eq(ptuple, ktuple)
def test_iterrows(self):
pdf = pd.DataFrame(
{
("x", "a", "1"): [1, 2, 3],
("x", "b", "2"): [4, 5, 6],
("y.z", "c.d", "3"): [7, 8, 9],
("x", "b", "4"): [10, 11, 12],
},
index=np.random.rand(3),
)
psdf = ps.from_pandas(pdf)
for (pdf_k, pdf_v), (psdf_k, psdf_v) in zip(pdf.iterrows(), psdf.iterrows()):
self.assert_eq(pdf_k, psdf_k)
self.assert_eq(pdf_v, psdf_v)
def test_reset_index(self):
pdf = pd.DataFrame({"a": [1, 2, 3], "b": [4, 5, 6]}, index=np.random.rand(3))
psdf = ps.from_pandas(pdf)
self.assert_eq(psdf.reset_index(), pdf.reset_index())
self.assert_eq(psdf.reset_index().index, pdf.reset_index().index)
self.assert_eq(psdf.reset_index(drop=True), pdf.reset_index(drop=True))
pdf.index.name = "a"
psdf.index.name = "a"
with self.assertRaisesRegex(ValueError, "cannot insert a, already exists"):
psdf.reset_index()
self.assert_eq(psdf.reset_index(drop=True), pdf.reset_index(drop=True))
# inplace
pser = pdf.a
psser = psdf.a
pdf.reset_index(drop=True, inplace=True)
psdf.reset_index(drop=True, inplace=True)
self.assert_eq(psdf, pdf)
self.assert_eq(psser, pser)
pdf.columns = ["index", "b"]
psdf.columns = ["index", "b"]
self.assert_eq(psdf.reset_index(), pdf.reset_index())
def test_reset_index_with_default_index_types(self):
pdf = pd.DataFrame({"a": [1, 2, 3], "b": [4, 5, 6]}, index=np.random.rand(3))
psdf = ps.from_pandas(pdf)
with ps.option_context("compute.default_index_type", "sequence"):
self.assert_eq(psdf.reset_index(), pdf.reset_index())
with ps.option_context("compute.default_index_type", "distributed-sequence"):
self.assert_eq(psdf.reset_index(), pdf.reset_index())
with ps.option_context("compute.default_index_type", "distributed"):
# the index is different.
self.assert_eq(psdf.reset_index().to_pandas().reset_index(drop=True), pdf.reset_index())
def test_reset_index_with_multiindex_columns(self):
index = pd.MultiIndex.from_tuples(
[("bird", "falcon"), ("bird", "parrot"), ("mammal", "lion"), ("mammal", "monkey")],
names=["class", "name"],
)
columns = pd.MultiIndex.from_tuples([("speed", "max"), ("species", "type")])
pdf = pd.DataFrame(
[(389.0, "fly"), (24.0, "fly"), (80.5, "run"), (np.nan, "jump")],
index=index,
columns=columns,
)
psdf = ps.from_pandas(pdf)
self.assert_eq(psdf, pdf)
self.assert_eq(psdf.reset_index(), pdf.reset_index())
self.assert_eq(psdf.reset_index(level="class"), pdf.reset_index(level="class"))
self.assert_eq(
psdf.reset_index(level="class", col_level=1),
pdf.reset_index(level="class", col_level=1),
)
self.assert_eq(
psdf.reset_index(level="class", col_level=1, col_fill="species"),
pdf.reset_index(level="class", col_level=1, col_fill="species"),
)
self.assert_eq(
psdf.reset_index(level="class", col_level=1, col_fill="genus"),
pdf.reset_index(level="class", col_level=1, col_fill="genus"),
)
with self.assertRaisesRegex(IndexError, "Index has only 2 levels, not 3"):
psdf.reset_index(col_level=2)
pdf.index.names = [("x", "class"), ("y", "name")]
psdf.index.names = [("x", "class"), ("y", "name")]
self.assert_eq(psdf.reset_index(), pdf.reset_index())
with self.assertRaisesRegex(ValueError, "Item must have length equal to number of levels."):
psdf.reset_index(col_level=1)
def test_index_to_frame_reset_index(self):
def check(psdf, pdf):
self.assert_eq(psdf.reset_index(), pdf.reset_index())
self.assert_eq(psdf.reset_index(drop=True), pdf.reset_index(drop=True))
pdf.reset_index(drop=True, inplace=True)
psdf.reset_index(drop=True, inplace=True)
self.assert_eq(psdf, pdf)
pdf, psdf = self.df_pair
check(psdf.index.to_frame(), pdf.index.to_frame())
check(psdf.index.to_frame(index=False), pdf.index.to_frame(index=False))
check(psdf.index.to_frame(name="a"), pdf.index.to_frame(name="a"))
check(psdf.index.to_frame(index=False, name="a"), pdf.index.to_frame(index=False, name="a"))
check(psdf.index.to_frame(name=("x", "a")), pdf.index.to_frame(name=("x", "a")))
check(
psdf.index.to_frame(index=False, name=("x", "a")),
pdf.index.to_frame(index=False, name=("x", "a")),
)
def test_multiindex_column_access(self):
columns = pd.MultiIndex.from_tuples(
[
("a", "", "", "b"),
("c", "", "d", ""),
("e", "", "f", ""),
("e", "g", "", ""),
("", "", "", "h"),
("i", "", "", ""),
]
)
pdf = pd.DataFrame(
[
(1, "a", "x", 10, 100, 1000),
(2, "b", "y", 20, 200, 2000),
(3, "c", "z", 30, 300, 3000),
],
columns=columns,
index=np.random.rand(3),
)
psdf = ps.from_pandas(pdf)
self.assert_eq(psdf, pdf)
self.assert_eq(psdf["a"], pdf["a"])
self.assert_eq(psdf["a"]["b"], pdf["a"]["b"])
self.assert_eq(psdf["c"], pdf["c"])
self.assert_eq(psdf["c"]["d"], pdf["c"]["d"])
self.assert_eq(psdf["e"], pdf["e"])
self.assert_eq(psdf["e"][""]["f"], pdf["e"][""]["f"])
self.assert_eq(psdf["e"]["g"], pdf["e"]["g"])
self.assert_eq(psdf[""], pdf[""])
self.assert_eq(psdf[""]["h"], pdf[""]["h"])
self.assert_eq(psdf["i"], pdf["i"])
self.assert_eq(psdf[["a", "e"]], pdf[["a", "e"]])
self.assert_eq(psdf[["e", "a"]], pdf[["e", "a"]])
self.assert_eq(psdf[("a",)], pdf[("a",)])
self.assert_eq(psdf[("e", "g")], pdf[("e", "g")])
# self.assert_eq(psdf[("i",)], pdf[("i",)])
self.assert_eq(psdf[("i", "")], pdf[("i", "")])
self.assertRaises(KeyError, lambda: psdf[("a", "b")])
def test_repr_cache_invalidation(self):
# If there is any cache, inplace operations should invalidate it.
df = ps.range(10)
df.__repr__()
df["a"] = df["id"]
self.assertEqual(df.__repr__(), df.to_pandas().__repr__())
def test_repr_html_cache_invalidation(self):
# If there is any cache, inplace operations should invalidate it.
df = ps.range(10)
df._repr_html_()
df["a"] = df["id"]
self.assertEqual(df._repr_html_(), df.to_pandas()._repr_html_())
def test_empty_dataframe(self):
pdf = pd.DataFrame({"a": pd.Series([], dtype="i1"), "b": pd.Series([], dtype="str")})
psdf = ps.from_pandas(pdf)
self.assert_eq(psdf, pdf)
with self.sql_conf({SPARK_CONF_ARROW_ENABLED: False}):
psdf = ps.from_pandas(pdf)
self.assert_eq(psdf, pdf)
def test_all_null_dataframe(self):
pdf = pd.DataFrame(
{
"a": [None, None, None, "a"],
"b": [None, None, None, 1],
"c": [None, None, None] + list(np.arange(1, 2).astype("i1")),
"d": [None, None, None, 1.0],
"e": [None, None, None, True],
"f": [None, None, None] + list(pd.date_range("20130101", periods=1)),
},
)
psdf = ps.from_pandas(pdf)
self.assert_eq(psdf.iloc[:-1], pdf.iloc[:-1])
with self.sql_conf({SPARK_CONF_ARROW_ENABLED: False}):
self.assert_eq(psdf.iloc[:-1], pdf.iloc[:-1])
pdf = pd.DataFrame(
{
"a": pd.Series([None, None, None], dtype="float64"),
"b": pd.Series([None, None, None], dtype="str"),
},
)
psdf = ps.from_pandas(pdf)
self.assert_eq(psdf, pdf)
with self.sql_conf({SPARK_CONF_ARROW_ENABLED: False}):
psdf = ps.from_pandas(pdf)
self.assert_eq(psdf, pdf)
def test_nullable_object(self):
pdf = pd.DataFrame(
{
"a": list("abc") + [np.nan, None],
"b": list(range(1, 4)) + [np.nan, None],
"c": list(np.arange(3, 6).astype("i1")) + [np.nan, None],
"d": list(np.arange(4.0, 7.0, dtype="float64")) + [np.nan, None],
"e": [True, False, True, np.nan, None],
"f": list(pd.date_range("20130101", periods=3)) + [np.nan, None],
},
index=np.random.rand(5),
)
psdf = ps.from_pandas(pdf)
self.assert_eq(psdf, pdf)
with self.sql_conf({SPARK_CONF_ARROW_ENABLED: False}):
psdf = ps.from_pandas(pdf)
self.assert_eq(psdf, pdf)
def test_assign(self):
pdf, psdf = self.df_pair
psdf["w"] = 1.0
pdf["w"] = 1.0
self.assert_eq(psdf, pdf)
psdf.w = 10.0
pdf.w = 10.0
self.assert_eq(psdf, pdf)
psdf[1] = 1.0
pdf[1] = 1.0
self.assert_eq(psdf, pdf)
psdf = psdf.assign(a=psdf["a"] * 2)
pdf = pdf.assign(a=pdf["a"] * 2)
self.assert_eq(psdf, pdf)
# multi-index columns
columns = pd.MultiIndex.from_tuples([("x", "a"), ("x", "b"), ("y", "w"), ("y", "v")])
pdf.columns = columns
psdf.columns = columns
psdf[("a", "c")] = "def"
pdf[("a", "c")] = "def"
self.assert_eq(psdf, pdf)
psdf = psdf.assign(Z="ZZ")
pdf = pdf.assign(Z="ZZ")
self.assert_eq(psdf, pdf)
psdf["x"] = "ghi"
pdf["x"] = "ghi"
self.assert_eq(psdf, pdf)
def test_head(self):
pdf, psdf = self.df_pair
self.assert_eq(psdf.head(2), pdf.head(2))
self.assert_eq(psdf.head(3), pdf.head(3))
self.assert_eq(psdf.head(0), pdf.head(0))
self.assert_eq(psdf.head(-3), pdf.head(-3))
self.assert_eq(psdf.head(-10), pdf.head(-10))
with option_context("compute.ordered_head", True):
self.assert_eq(psdf.head(), pdf.head())
def test_attributes(self):
psdf = self.psdf
self.assertIn("a", dir(psdf))
self.assertNotIn("foo", dir(psdf))
self.assertRaises(AttributeError, lambda: psdf.foo)
psdf = ps.DataFrame({"a b c": [1, 2, 3]})
self.assertNotIn("a b c", dir(psdf))
psdf = ps.DataFrame({"a": [1, 2], 5: [1, 2]})
self.assertIn("a", dir(psdf))
self.assertNotIn(5, dir(psdf))
def test_column_names(self):
pdf, psdf = self.df_pair
self.assert_eq(psdf.columns, pdf.columns)
self.assert_eq(psdf[["b", "a"]].columns, pdf[["b", "a"]].columns)
self.assert_eq(psdf["a"].name, pdf["a"].name)
self.assert_eq((psdf["a"] + 1).name, (pdf["a"] + 1).name)
self.assert_eq((psdf.a + psdf.b).name, (pdf.a + pdf.b).name)
self.assert_eq((psdf.a + psdf.b.rename("a")).name, (pdf.a + pdf.b.rename("a")).name)
self.assert_eq((psdf.a + psdf.b.rename()).name, (pdf.a + pdf.b.rename()).name)
self.assert_eq((psdf.a.rename() + psdf.b).name, (pdf.a.rename() + pdf.b).name)
self.assert_eq(
(psdf.a.rename() + psdf.b.rename()).name, (pdf.a.rename() + pdf.b.rename()).name
)
def test_rename_columns(self):
pdf = pd.DataFrame(
{"a": [1, 2, 3, 4, 5, 6, 7], "b": [7, 6, 5, 4, 3, 2, 1]}, index=np.random.rand(7)
)
psdf = ps.from_pandas(pdf)
psdf.columns = ["x", "y"]
pdf.columns = ["x", "y"]
self.assert_eq(psdf.columns, pd.Index(["x", "y"]))
self.assert_eq(psdf, pdf)
self.assert_eq(psdf._internal.data_spark_column_names, ["x", "y"])
self.assert_eq(psdf.to_spark().columns, ["x", "y"])
self.assert_eq(psdf.to_spark(index_col="index").columns, ["index", "x", "y"])
columns = pdf.columns
columns.name = "lvl_1"
psdf.columns = columns
self.assert_eq(psdf.columns.names, ["lvl_1"])
self.assert_eq(psdf, pdf)
msg = "Length mismatch: Expected axis has 2 elements, new values have 4 elements"
with self.assertRaisesRegex(ValueError, msg):
psdf.columns = [1, 2, 3, 4]
# Multi-index columns
pdf = pd.DataFrame(
{("A", "0"): [1, 2, 2, 3], ("B", "1"): [1, 2, 3, 4]}, index=np.random.rand(4)
)
psdf = ps.from_pandas(pdf)
columns = pdf.columns
self.assert_eq(psdf.columns, columns)
self.assert_eq(psdf, pdf)
pdf.columns = ["x", "y"]
psdf.columns = ["x", "y"]
self.assert_eq(psdf.columns, pd.Index(["x", "y"]))
self.assert_eq(psdf, pdf)
self.assert_eq(psdf._internal.data_spark_column_names, ["x", "y"])
self.assert_eq(psdf.to_spark().columns, ["x", "y"])
self.assert_eq(psdf.to_spark(index_col="index").columns, ["index", "x", "y"])
pdf.columns = columns
psdf.columns = columns
self.assert_eq(psdf.columns, columns)
self.assert_eq(psdf, pdf)
self.assert_eq(psdf._internal.data_spark_column_names, ["(A, 0)", "(B, 1)"])
self.assert_eq(psdf.to_spark().columns, ["(A, 0)", "(B, 1)"])
self.assert_eq(psdf.to_spark(index_col="index").columns, ["index", "(A, 0)", "(B, 1)"])
columns.names = ["lvl_1", "lvl_2"]
psdf.columns = columns
self.assert_eq(psdf.columns.names, ["lvl_1", "lvl_2"])
self.assert_eq(psdf, pdf)
self.assert_eq(psdf._internal.data_spark_column_names, ["(A, 0)", "(B, 1)"])
self.assert_eq(psdf.to_spark().columns, ["(A, 0)", "(B, 1)"])
self.assert_eq(psdf.to_spark(index_col="index").columns, ["index", "(A, 0)", "(B, 1)"])
def test_rename_dataframe(self):
pdf1 = pd.DataFrame({"A": [1, 2, 3], "B": [4, 5, 6]})
psdf1 = ps.from_pandas(pdf1)
self.assert_eq(
psdf1.rename(columns={"A": "a", "B": "b"}), pdf1.rename(columns={"A": "a", "B": "b"})
)
result_psdf = psdf1.rename(index={1: 10, 2: 20})
result_pdf = pdf1.rename(index={1: 10, 2: 20})
self.assert_eq(result_psdf, result_pdf)
# inplace
pser = result_pdf.A
psser = result_psdf.A
result_psdf.rename(index={10: 100, 20: 200}, inplace=True)
result_pdf.rename(index={10: 100, 20: 200}, inplace=True)
self.assert_eq(result_psdf, result_pdf)
self.assert_eq(psser, pser)
def str_lower(s) -> str:
return str.lower(s)
self.assert_eq(
psdf1.rename(str_lower, axis="columns"), pdf1.rename(str_lower, axis="columns")
)
def mul10(x) -> int:
return x * 10
self.assert_eq(psdf1.rename(mul10, axis="index"), pdf1.rename(mul10, axis="index"))
self.assert_eq(
psdf1.rename(columns=str_lower, index={1: 10, 2: 20}),
pdf1.rename(columns=str_lower, index={1: 10, 2: 20}),
)
idx = pd.MultiIndex.from_tuples([("X", "A"), ("X", "B"), ("Y", "C"), ("Y", "D")])
pdf2 = pd.DataFrame([[1, 2, 3, 4], [5, 6, 7, 8]], columns=idx)
psdf2 = ps.from_pandas(pdf2)
self.assert_eq(psdf2.rename(columns=str_lower), pdf2.rename(columns=str_lower))
self.assert_eq(
psdf2.rename(columns=str_lower, level=0), pdf2.rename(columns=str_lower, level=0)
)
self.assert_eq(
psdf2.rename(columns=str_lower, level=1), pdf2.rename(columns=str_lower, level=1)
)
pdf3 = pd.DataFrame([[1, 2], [3, 4], [5, 6], [7, 8]], index=idx, columns=list("ab"))
psdf3 = ps.from_pandas(pdf3)
self.assert_eq(psdf3.rename(index=str_lower), pdf3.rename(index=str_lower))
self.assert_eq(
psdf3.rename(index=str_lower, level=0), pdf3.rename(index=str_lower, level=0)
)
self.assert_eq(
psdf3.rename(index=str_lower, level=1), pdf3.rename(index=str_lower, level=1)
)
pdf4 = pdf2 + 1
psdf4 = psdf2 + 1
self.assert_eq(psdf4.rename(columns=str_lower), pdf4.rename(columns=str_lower))
pdf5 = pdf3 + 1
psdf5 = psdf3 + 1
self.assert_eq(psdf5.rename(index=str_lower), pdf5.rename(index=str_lower))
msg = "Either `index` or `columns` should be provided."
with self.assertRaisesRegex(ValueError, msg):
psdf1.rename()
msg = "`mapper` or `index` or `columns` should be either dict-like or function type."
with self.assertRaisesRegex(ValueError, msg):
psdf1.rename(mapper=[str_lower], axis=1)
msg = "Mapper dict should have the same value type."
with self.assertRaisesRegex(ValueError, msg):
psdf1.rename({"A": "a", "B": 2}, axis=1)
msg = r"level should be an integer between \[0, column_labels_level\)"
with self.assertRaisesRegex(ValueError, msg):
psdf2.rename(columns=str_lower, level=2)
def test_rename_axis(self):
index = pd.Index(["A", "B", "C"], name="index")
columns = pd.Index(["numbers", "values"], name="cols")
pdf = pd.DataFrame([[1.0, 2.0], [3.0, 4.0], [5.0, 6.0]], index=index, columns=columns)
psdf = ps.from_pandas(pdf)
for axis in [0, "index"]:
self.assert_eq(
pdf.rename_axis("index2", axis=axis).sort_index(),
psdf.rename_axis("index2", axis=axis).sort_index(),
)
self.assert_eq(
pdf.rename_axis(["index2"], axis=axis).sort_index(),
psdf.rename_axis(["index2"], axis=axis).sort_index(),
)
for axis in [1, "columns"]:
self.assert_eq(
pdf.rename_axis("cols2", axis=axis).sort_index(),
psdf.rename_axis("cols2", axis=axis).sort_index(),
)
self.assert_eq(
pdf.rename_axis(["cols2"], axis=axis).sort_index(),
psdf.rename_axis(["cols2"], axis=axis).sort_index(),
)
pdf2 = pdf.copy()
psdf2 = psdf.copy()
pdf2.rename_axis("index2", axis="index", inplace=True)
psdf2.rename_axis("index2", axis="index", inplace=True)
self.assert_eq(pdf2.sort_index(), psdf2.sort_index())
self.assertRaises(ValueError, lambda: psdf.rename_axis(["index2", "index3"], axis=0))
self.assertRaises(ValueError, lambda: psdf.rename_axis(["cols2", "cols3"], axis=1))
self.assertRaises(TypeError, lambda: psdf.rename_axis(mapper=["index2"], index=["index3"]))
self.assert_eq(
pdf.rename_axis(index={"index": "index2"}, columns={"cols": "cols2"}).sort_index(),
psdf.rename_axis(index={"index": "index2"}, columns={"cols": "cols2"}).sort_index(),
)
self.assert_eq(
pdf.rename_axis(index={"missing": "index2"}, columns={"missing": "cols2"}).sort_index(),
psdf.rename_axis(
index={"missing": "index2"}, columns={"missing": "cols2"}
).sort_index(),
)
self.assert_eq(
pdf.rename_axis(index=str.upper, columns=str.upper).sort_index(),
psdf.rename_axis(index=str.upper, columns=str.upper).sort_index(),
)
index = pd.MultiIndex.from_tuples(
[("A", "B"), ("C", "D"), ("E", "F")], names=["index1", "index2"]
)
columns = pd.MultiIndex.from_tuples(
[("numbers", "first"), ("values", "second")], names=["cols1", "cols2"]
)
pdf = pd.DataFrame([[1.0, 2.0], [3.0, 4.0], [5.0, 6.0]], index=index, columns=columns)
psdf = ps.from_pandas(pdf)
for axis in [0, "index"]:
self.assert_eq(
pdf.rename_axis(["index3", "index4"], axis=axis).sort_index(),
psdf.rename_axis(["index3", "index4"], axis=axis).sort_index(),
)
for axis in [1, "columns"]:
self.assert_eq(
pdf.rename_axis(["cols3", "cols4"], axis=axis).sort_index(),
psdf.rename_axis(["cols3", "cols4"], axis=axis).sort_index(),
)
self.assertRaises(
ValueError, lambda: psdf.rename_axis(["index3", "index4", "index5"], axis=0)
)
self.assertRaises(ValueError, lambda: psdf.rename_axis(["cols3", "cols4", "cols5"], axis=1))
self.assert_eq(
pdf.rename_axis(index={"index1": "index3"}, columns={"cols1": "cols3"}).sort_index(),
psdf.rename_axis(index={"index1": "index3"}, columns={"cols1": "cols3"}).sort_index(),
)
self.assert_eq(
pdf.rename_axis(index={"missing": "index3"}, columns={"missing": "cols3"}).sort_index(),
psdf.rename_axis(
index={"missing": "index3"}, columns={"missing": "cols3"}
).sort_index(),
)
self.assert_eq(
pdf.rename_axis(
index={"index1": "index3", "index2": "index4"},
columns={"cols1": "cols3", "cols2": "cols4"},
).sort_index(),
psdf.rename_axis(
index={"index1": "index3", "index2": "index4"},
columns={"cols1": "cols3", "cols2": "cols4"},
).sort_index(),
)
self.assert_eq(
pdf.rename_axis(index=str.upper, columns=str.upper).sort_index(),
psdf.rename_axis(index=str.upper, columns=str.upper).sort_index(),
)
def test_dot(self):
psdf = self.psdf
with self.assertRaisesRegex(TypeError, "Unsupported type DataFrame"):
psdf.dot(psdf)
def test_dot_in_column_name(self):
self.assert_eq(
ps.DataFrame(ps.range(1)._internal.spark_frame.selectExpr("1L as `a.b`"))["a.b"],
ps.Series([1], name="a.b"),
)
def test_aggregate(self):
pdf = pd.DataFrame(
[[1, 2, 3], [4, 5, 6], [7, 8, 9], [np.nan, np.nan, np.nan]], columns=["A", "B", "C"]
)
psdf = ps.from_pandas(pdf)
self.assert_eq(
psdf.agg(["sum", "min"])[["A", "B", "C"]].sort_index(), # TODO?: fix column order
pdf.agg(["sum", "min"])[["A", "B", "C"]].sort_index(),
)
self.assert_eq(
psdf.agg({"A": ["sum", "min"], "B": ["min", "max"]})[["A", "B"]].sort_index(),
pdf.agg({"A": ["sum", "min"], "B": ["min", "max"]})[["A", "B"]].sort_index(),
)
self.assertRaises(KeyError, lambda: psdf.agg({"A": ["sum", "min"], "X": ["min", "max"]}))
# multi-index columns
columns = pd.MultiIndex.from_tuples([("X", "A"), ("X", "B"), ("Y", "C")])
pdf.columns = columns
psdf.columns = columns
self.assert_eq(
psdf.agg(["sum", "min"])[[("X", "A"), ("X", "B"), ("Y", "C")]].sort_index(),
pdf.agg(["sum", "min"])[[("X", "A"), ("X", "B"), ("Y", "C")]].sort_index(),
)
self.assert_eq(
psdf.agg({("X", "A"): ["sum", "min"], ("X", "B"): ["min", "max"]})[
[("X", "A"), ("X", "B")]
].sort_index(),
pdf.agg({("X", "A"): ["sum", "min"], ("X", "B"): ["min", "max"]})[
[("X", "A"), ("X", "B")]
].sort_index(),
)
self.assertRaises(TypeError, lambda: psdf.agg({"X": ["sum", "min"], "Y": ["min", "max"]}))
# non-string names
pdf = pd.DataFrame(
[[1, 2, 3], [4, 5, 6], [7, 8, 9], [np.nan, np.nan, np.nan]], columns=[10, 20, 30]
)
psdf = ps.from_pandas(pdf)
self.assert_eq(
psdf.agg(["sum", "min"])[[10, 20, 30]].sort_index(),
pdf.agg(["sum", "min"])[[10, 20, 30]].sort_index(),
)
self.assert_eq(
psdf.agg({10: ["sum", "min"], 20: ["min", "max"]})[[10, 20]].sort_index(),
pdf.agg({10: ["sum", "min"], 20: ["min", "max"]})[[10, 20]].sort_index(),
)
columns = pd.MultiIndex.from_tuples([("X", 10), ("X", 20), ("Y", 30)])
pdf.columns = columns
psdf.columns = columns
self.assert_eq(
psdf.agg(["sum", "min"])[[("X", 10), ("X", 20), ("Y", 30)]].sort_index(),
pdf.agg(["sum", "min"])[[("X", 10), ("X", 20), ("Y", 30)]].sort_index(),
)
self.assert_eq(
psdf.agg({("X", 10): ["sum", "min"], ("X", 20): ["min", "max"]})[
[("X", 10), ("X", 20)]
].sort_index(),
pdf.agg({("X", 10): ["sum", "min"], ("X", 20): ["min", "max"]})[
[("X", 10), ("X", 20)]
].sort_index(),
)
pdf = pd.DataFrame(
[datetime(2019, 2, 2, 0, 0, 0, 0), datetime(2019, 2, 3, 0, 0, 0, 0)],
columns=["timestamp"],
)
psdf = ps.from_pandas(pdf)
self.assert_eq(psdf.timestamp.min(), pdf.timestamp.min())
self.assert_eq(psdf.timestamp.max(), pdf.timestamp.max())
self.assertRaises(ValueError, lambda: psdf.agg(("sum", "min")))
def test_droplevel(self):
pdf = (
pd.DataFrame([[1, 2, 3, 4], [5, 6, 7, 8], [9, 10, 11, 12]])
.set_index([0, 1])
.rename_axis(["a", "b"])
)
pdf.columns = pd.MultiIndex.from_tuples(
[("c", "e"), ("d", "f")], names=["level_1", "level_2"]
)
psdf = ps.from_pandas(pdf)
self.assertRaises(ValueError, lambda: psdf.droplevel(["a", "b"]))
self.assertRaises(ValueError, lambda: psdf.droplevel([1, 1, 1, 1, 1]))
self.assertRaises(IndexError, lambda: psdf.droplevel(2))
self.assertRaises(IndexError, lambda: psdf.droplevel(-3))
self.assertRaises(KeyError, lambda: psdf.droplevel({"a"}))
self.assertRaises(KeyError, lambda: psdf.droplevel({"a": 1}))
self.assertRaises(ValueError, lambda: psdf.droplevel(["level_1", "level_2"], axis=1))
self.assertRaises(IndexError, lambda: psdf.droplevel(2, axis=1))
self.assertRaises(IndexError, lambda: psdf.droplevel(-3, axis=1))
self.assertRaises(KeyError, lambda: psdf.droplevel({"level_1"}, axis=1))
self.assertRaises(KeyError, lambda: psdf.droplevel({"level_1": 1}, axis=1))
self.assert_eq(pdf.droplevel("a"), psdf.droplevel("a"))
self.assert_eq(pdf.droplevel(["a"]), psdf.droplevel(["a"]))
self.assert_eq(pdf.droplevel(("a",)), psdf.droplevel(("a",)))
self.assert_eq(pdf.droplevel(0), psdf.droplevel(0))
self.assert_eq(pdf.droplevel(-1), psdf.droplevel(-1))
self.assert_eq(pdf.droplevel("level_1", axis=1), psdf.droplevel("level_1", axis=1))
self.assert_eq(pdf.droplevel(["level_1"], axis=1), psdf.droplevel(["level_1"], axis=1))
self.assert_eq(pdf.droplevel(("level_1",), axis=1), psdf.droplevel(("level_1",), axis=1))
self.assert_eq(pdf.droplevel(0, axis=1), psdf.droplevel(0, axis=1))
self.assert_eq(pdf.droplevel(-1, axis=1), psdf.droplevel(-1, axis=1))
# Tupled names
pdf.columns.names = [("level", 1), ("level", 2)]
pdf.index.names = [("a", 10), ("x", 20)]
psdf = ps.from_pandas(pdf)
self.assertRaises(KeyError, lambda: psdf.droplevel("a"))
self.assertRaises(KeyError, lambda: psdf.droplevel(("a", 10)))
self.assert_eq(pdf.droplevel([("a", 10)]), psdf.droplevel([("a", 10)]))
self.assert_eq(
pdf.droplevel([("level", 1)], axis=1), psdf.droplevel([("level", 1)], axis=1)
)
# non-string names
pdf = (
pd.DataFrame([[1, 2, 3, 4], [5, 6, 7, 8], [9, 10, 11, 12]])
.set_index([0, 1])
.rename_axis([10.0, 20.0])
)
pdf.columns = pd.MultiIndex.from_tuples([("c", "e"), ("d", "f")], names=[100.0, 200.0])
psdf = ps.from_pandas(pdf)
self.assert_eq(pdf.droplevel(10.0), psdf.droplevel(10.0))
self.assert_eq(pdf.droplevel([10.0]), psdf.droplevel([10.0]))
self.assert_eq(pdf.droplevel((10.0,)), psdf.droplevel((10.0,)))
self.assert_eq(pdf.droplevel(0), psdf.droplevel(0))
self.assert_eq(pdf.droplevel(-1), psdf.droplevel(-1))
self.assert_eq(pdf.droplevel(100.0, axis=1), psdf.droplevel(100.0, axis=1))
self.assert_eq(pdf.droplevel(0, axis=1), psdf.droplevel(0, axis=1))
def test_drop(self):
pdf = pd.DataFrame({"x": [1, 2], "y": [3, 4], "z": [5, 6]}, index=np.random.rand(2))
psdf = ps.from_pandas(pdf)
# Assert 'labels' or 'columns' parameter is set
expected_error_message = "Need to specify at least one of 'labels' or 'columns'"
with self.assertRaisesRegex(ValueError, expected_error_message):
psdf.drop()
#
# Drop columns
#
# Assert using a str for 'labels' works
self.assert_eq(psdf.drop("x", axis=1), pdf.drop("x", axis=1))
self.assert_eq((psdf + 1).drop("x", axis=1), (pdf + 1).drop("x", axis=1))
# Assert using a list for 'labels' works
self.assert_eq(psdf.drop(["y", "z"], axis=1), pdf.drop(["y", "z"], axis=1))
self.assert_eq(psdf.drop(["x", "y", "z"], axis=1), pdf.drop(["x", "y", "z"], axis=1))
# Assert using 'columns' instead of 'labels' produces the same results
self.assert_eq(psdf.drop(columns="x"), pdf.drop(columns="x"))
self.assert_eq(psdf.drop(columns=["y", "z"]), pdf.drop(columns=["y", "z"]))
self.assert_eq(psdf.drop(columns=["x", "y", "z"]), pdf.drop(columns=["x", "y", "z"]))
self.assert_eq(psdf.drop(columns=[]), pdf.drop(columns=[]))
columns = pd.MultiIndex.from_tuples([(1, "x"), (1, "y"), (2, "z")])
pdf.columns = columns
psdf = ps.from_pandas(pdf)
self.assert_eq(psdf.drop(columns=1), pdf.drop(columns=1))
self.assert_eq(psdf.drop(columns=(1, "x")), pdf.drop(columns=(1, "x")))
self.assert_eq(psdf.drop(columns=[(1, "x"), 2]), pdf.drop(columns=[(1, "x"), 2]))
self.assert_eq(
psdf.drop(columns=[(1, "x"), (1, "y"), (2, "z")]),
pdf.drop(columns=[(1, "x"), (1, "y"), (2, "z")]),
)
self.assertRaises(KeyError, lambda: psdf.drop(columns=3))
self.assertRaises(KeyError, lambda: psdf.drop(columns=(1, "z")))
pdf.index = pd.MultiIndex.from_tuples([("i", 0), ("j", 1)])
psdf = ps.from_pandas(pdf)
self.assert_eq(
psdf.drop(columns=[(1, "x"), (1, "y"), (2, "z")]),
pdf.drop(columns=[(1, "x"), (1, "y"), (2, "z")]),
)
# non-string names
pdf = pd.DataFrame({10: [1, 2], 20: [3, 4], 30: [5, 6]}, index=np.random.rand(2))
psdf = ps.from_pandas(pdf)
self.assert_eq(psdf.drop(10, axis=1), pdf.drop(10, axis=1))
self.assert_eq(psdf.drop([20, 30], axis=1), pdf.drop([20, 30], axis=1))
#
# Drop rows
#
pdf = pd.DataFrame({"X": [1, 2, 3], "Y": [4, 5, 6], "Z": [7, 8, 9]}, index=["A", "B", "C"])
psdf = ps.from_pandas(pdf)
# Given labels (and axis = 0)
self.assert_eq(psdf.drop(labels="A", axis=0), pdf.drop(labels="A", axis=0))
self.assert_eq(psdf.drop(labels="A"), pdf.drop(labels="A"))
self.assert_eq((psdf + 1).drop(labels="A"), (pdf + 1).drop(labels="A"))
self.assert_eq(psdf.drop(labels=["A", "C"], axis=0), pdf.drop(labels=["A", "C"], axis=0))
self.assert_eq(
psdf.drop(labels=["A", "B", "C"], axis=0), pdf.drop(labels=["A", "B", "C"], axis=0)
)
with ps.option_context("compute.isin_limit", 2):
self.assert_eq(
psdf.drop(labels=["A", "B", "C"], axis=0), pdf.drop(labels=["A", "B", "C"], axis=0)
)
# Given index
self.assert_eq(psdf.drop(index="A"), pdf.drop(index="A"))
self.assert_eq(psdf.drop(index=["A", "C"]), pdf.drop(index=["A", "C"]))
self.assert_eq(psdf.drop(index=["A", "B", "C"]), pdf.drop(index=["A", "B", "C"]))
self.assert_eq(psdf.drop(index=[]), pdf.drop(index=[]))
with ps.option_context("compute.isin_limit", 2):
self.assert_eq(psdf.drop(index=["A", "B", "C"]), pdf.drop(index=["A", "B", "C"]))
# Non-string names
pdf.index = [10, 20, 30]
psdf = ps.from_pandas(pdf)
self.assert_eq(psdf.drop(labels=10, axis=0), pdf.drop(labels=10, axis=0))
self.assert_eq(psdf.drop(labels=[10, 30], axis=0), pdf.drop(labels=[10, 30], axis=0))
self.assert_eq(
psdf.drop(labels=[10, 20, 30], axis=0), pdf.drop(labels=[10, 20, 30], axis=0)
)
with ps.option_context("compute.isin_limit", 2):
self.assert_eq(
psdf.drop(labels=[10, 20, 30], axis=0), pdf.drop(labels=[10, 20, 30], axis=0)
)
# MultiIndex
pdf.index = pd.MultiIndex.from_tuples([("a", "x"), ("b", "y"), ("c", "z")])
psdf = ps.from_pandas(pdf)
self.assertRaises(NotImplementedError, lambda: psdf.drop(labels=[("a", "x")]))
#
# Drop rows and columns
#
pdf = pd.DataFrame({"X": [1, 2, 3], "Y": [4, 5, 6], "Z": [7, 8, 9]}, index=["A", "B", "C"])
psdf = ps.from_pandas(pdf)
self.assert_eq(psdf.drop(index="A", columns="X"), pdf.drop(index="A", columns="X"))
self.assert_eq(
psdf.drop(index=["A", "C"], columns=["X", "Z"]),
pdf.drop(index=["A", "C"], columns=["X", "Z"]),
)
self.assert_eq(
psdf.drop(index=["A", "B", "C"], columns=["X", "Z"]),
pdf.drop(index=["A", "B", "C"], columns=["X", "Z"]),
)
with ps.option_context("compute.isin_limit", 2):
self.assert_eq(
psdf.drop(index=["A", "B", "C"], columns=["X", "Z"]),
pdf.drop(index=["A", "B", "C"], columns=["X", "Z"]),
)
self.assert_eq(
psdf.drop(index=[], columns=["X", "Z"]),
pdf.drop(index=[], columns=["X", "Z"]),
)
self.assert_eq(
psdf.drop(index=["A", "B", "C"], columns=[]),
pdf.drop(index=["A", "B", "C"], columns=[]),
)
self.assert_eq(
psdf.drop(index=[], columns=[]),
pdf.drop(index=[], columns=[]),
)
self.assertRaises(
ValueError,
lambda: psdf.drop(labels="A", axis=0, columns="X"),
)
def _test_dropna(self, pdf, axis):
psdf = ps.from_pandas(pdf)
self.assert_eq(psdf.dropna(axis=axis), pdf.dropna(axis=axis))
self.assert_eq(psdf.dropna(axis=axis, how="all"), pdf.dropna(axis=axis, how="all"))
self.assert_eq(psdf.dropna(axis=axis, subset=["x"]), pdf.dropna(axis=axis, subset=["x"]))
self.assert_eq(psdf.dropna(axis=axis, subset="x"), pdf.dropna(axis=axis, subset=["x"]))
self.assert_eq(
psdf.dropna(axis=axis, subset=["y", "z"]), pdf.dropna(axis=axis, subset=["y", "z"])
)
self.assert_eq(
psdf.dropna(axis=axis, subset=["y", "z"], how="all"),
pdf.dropna(axis=axis, subset=["y", "z"], how="all"),
)
self.assert_eq(psdf.dropna(axis=axis, thresh=2), pdf.dropna(axis=axis, thresh=2))
self.assert_eq(
psdf.dropna(axis=axis, thresh=1, subset=["y", "z"]),
pdf.dropna(axis=axis, thresh=1, subset=["y", "z"]),
)
pdf2 = pdf.copy()
psdf2 = psdf.copy()
pser = pdf2[pdf2.columns[0]]
psser = psdf2[psdf2.columns[0]]
pdf2.dropna(inplace=True, axis=axis)
psdf2.dropna(inplace=True, axis=axis)
self.assert_eq(psdf2, pdf2)
self.assert_eq(psser, pser)
# multi-index
columns = pd.MultiIndex.from_tuples([("a", "x"), ("a", "y"), ("b", "z")])
if axis == 0:
pdf.columns = columns
else:
pdf.index = columns
psdf = ps.from_pandas(pdf)
self.assert_eq(psdf.dropna(axis=axis), pdf.dropna(axis=axis))
self.assert_eq(psdf.dropna(axis=axis, how="all"), pdf.dropna(axis=axis, how="all"))
self.assert_eq(
psdf.dropna(axis=axis, subset=[("a", "x")]), pdf.dropna(axis=axis, subset=[("a", "x")])
)
self.assert_eq(
psdf.dropna(axis=axis, subset=("a", "x")), pdf.dropna(axis=axis, subset=[("a", "x")])
)
self.assert_eq(
psdf.dropna(axis=axis, subset=[("a", "y"), ("b", "z")]),
pdf.dropna(axis=axis, subset=[("a", "y"), ("b", "z")]),
)
self.assert_eq(
psdf.dropna(axis=axis, subset=[("a", "y"), ("b", "z")], how="all"),
pdf.dropna(axis=axis, subset=[("a", "y"), ("b", "z")], how="all"),
)
self.assert_eq(psdf.dropna(axis=axis, thresh=2), pdf.dropna(axis=axis, thresh=2))
self.assert_eq(
psdf.dropna(axis=axis, thresh=1, subset=[("a", "y"), ("b", "z")]),
pdf.dropna(axis=axis, thresh=1, subset=[("a", "y"), ("b", "z")]),
)
def test_dropna_axis_index(self):
pdf = pd.DataFrame(
{
"x": [np.nan, 2, 3, 4, np.nan, 6],
"y": [1, 2, np.nan, 4, np.nan, np.nan],
"z": [1, 2, 3, 4, np.nan, np.nan],
},
index=np.random.rand(6),
)
psdf = ps.from_pandas(pdf)
self._test_dropna(pdf, axis=0)
# empty
pdf = pd.DataFrame(index=np.random.rand(6))
psdf = ps.from_pandas(pdf)
self.assert_eq(psdf.dropna(), pdf.dropna())
self.assert_eq(psdf.dropna(how="all"), pdf.dropna(how="all"))
self.assert_eq(psdf.dropna(thresh=0), pdf.dropna(thresh=0))
self.assert_eq(psdf.dropna(thresh=1), pdf.dropna(thresh=1))
with self.assertRaisesRegex(ValueError, "No axis named foo"):
psdf.dropna(axis="foo")
self.assertRaises(KeyError, lambda: psdf.dropna(subset="1"))
with self.assertRaisesRegex(ValueError, "invalid how option: 1"):
psdf.dropna(how=1)
with self.assertRaisesRegex(TypeError, "must specify how or thresh"):
psdf.dropna(how=None)
def test_dropna_axis_column(self):
pdf = pd.DataFrame(
{
"x": [np.nan, 2, 3, 4, np.nan, 6],
"y": [1, 2, np.nan, 4, np.nan, np.nan],
"z": [1, 2, 3, 4, np.nan, np.nan],
},
index=[str(r) for r in np.random.rand(6)],
).T
self._test_dropna(pdf, axis=1)
psdf = ps.from_pandas(pdf)
with self.assertRaisesRegex(
ValueError, "The length of each subset must be the same as the index size."
):
psdf.dropna(subset=(["x", "y"]), axis=1)
# empty
pdf = pd.DataFrame({"x": [], "y": [], "z": []})
psdf = ps.from_pandas(pdf)
self.assert_eq(psdf.dropna(axis=1), pdf.dropna(axis=1))
self.assert_eq(psdf.dropna(axis=1, how="all"), pdf.dropna(axis=1, how="all"))
self.assert_eq(psdf.dropna(axis=1, thresh=0), pdf.dropna(axis=1, thresh=0))
self.assert_eq(psdf.dropna(axis=1, thresh=1), pdf.dropna(axis=1, thresh=1))
def test_dtype(self):
pdf = pd.DataFrame(
{
"a": list("abc"),
"b": list(range(1, 4)),
"c": np.arange(3, 6).astype("i1"),
"d": np.arange(4.0, 7.0, dtype="float64"),
"e": [True, False, True],
"f": pd.date_range("20130101", periods=3),
},
index=np.random.rand(3),
)
psdf = ps.from_pandas(pdf)
self.assert_eq(psdf, pdf)
self.assertTrue((psdf.dtypes == pdf.dtypes).all())
# multi-index columns
columns = pd.MultiIndex.from_tuples(zip(list("xxxyyz"), list("abcdef")))
pdf.columns = columns
psdf.columns = columns
self.assertTrue((psdf.dtypes == pdf.dtypes).all())
def test_fillna(self):
pdf = pd.DataFrame(
{
"x": [np.nan, 2, 3, 4, np.nan, 6],
"y": [1, 2, np.nan, 4, np.nan, np.nan],
"z": [1, 2, 3, 4, np.nan, np.nan],
},
index=np.random.rand(6),
)
psdf = ps.from_pandas(pdf)
self.assert_eq(psdf, pdf)
self.assert_eq(psdf.fillna(-1), pdf.fillna(-1))
self.assert_eq(
psdf.fillna({"x": -1, "y": -2, "z": -5}), pdf.fillna({"x": -1, "y": -2, "z": -5})
)
self.assert_eq(pdf.fillna(method="ffill"), psdf.fillna(method="ffill"))
self.assert_eq(pdf.fillna(method="ffill", limit=2), psdf.fillna(method="ffill", limit=2))
self.assert_eq(pdf.fillna(method="bfill"), psdf.fillna(method="bfill"))
self.assert_eq(pdf.fillna(method="bfill", limit=2), psdf.fillna(method="bfill", limit=2))
pdf = pdf.set_index(["x", "y"])
psdf = ps.from_pandas(pdf)
# check multi index
self.assert_eq(psdf.fillna(-1), pdf.fillna(-1))
self.assert_eq(pdf.fillna(method="bfill"), psdf.fillna(method="bfill"))
self.assert_eq(pdf.fillna(method="ffill"), psdf.fillna(method="ffill"))
pser = pdf.z
psser = psdf.z
pdf.fillna({"x": -1, "y": -2, "z": -5}, inplace=True)
psdf.fillna({"x": -1, "y": -2, "z": -5}, inplace=True)
self.assert_eq(psdf, pdf)
self.assert_eq(psser, pser)
s_nan = pd.Series([-1, -2, -5], index=["x", "y", "z"], dtype=int)
self.assert_eq(psdf.fillna(s_nan), pdf.fillna(s_nan))
with self.assertRaisesRegex(NotImplementedError, "fillna currently only"):
psdf.fillna(-1, axis=1)
with self.assertRaisesRegex(NotImplementedError, "fillna currently only"):
psdf.fillna(-1, axis="columns")
with self.assertRaisesRegex(ValueError, "limit parameter for value is not support now"):
psdf.fillna(-1, limit=1)
with self.assertRaisesRegex(TypeError, "Unsupported.*DataFrame"):
psdf.fillna(pd.DataFrame({"x": [-1], "y": [-1], "z": [-1]}))
with self.assertRaisesRegex(TypeError, "Unsupported.*int64"):
psdf.fillna({"x": np.int64(-6), "y": np.int64(-4), "z": -5})
with self.assertRaisesRegex(ValueError, "Expecting 'pad', 'ffill', 'backfill' or 'bfill'."):
psdf.fillna(method="xxx")
with self.assertRaisesRegex(
ValueError, "Must specify a fillna 'value' or 'method' parameter."
):
psdf.fillna()
# multi-index columns
pdf = pd.DataFrame(
{
("x", "a"): [np.nan, 2, 3, 4, np.nan, 6],
("x", "b"): [1, 2, np.nan, 4, np.nan, np.nan],
("y", "c"): [1, 2, 3, 4, np.nan, np.nan],
},
index=np.random.rand(6),
)
psdf = ps.from_pandas(pdf)
self.assert_eq(psdf.fillna(-1), pdf.fillna(-1))
self.assert_eq(
psdf.fillna({("x", "a"): -1, ("x", "b"): -2, ("y", "c"): -5}),
pdf.fillna({("x", "a"): -1, ("x", "b"): -2, ("y", "c"): -5}),
)
self.assert_eq(pdf.fillna(method="ffill"), psdf.fillna(method="ffill"))
self.assert_eq(pdf.fillna(method="ffill", limit=2), psdf.fillna(method="ffill", limit=2))
self.assert_eq(pdf.fillna(method="bfill"), psdf.fillna(method="bfill"))
self.assert_eq(pdf.fillna(method="bfill", limit=2), psdf.fillna(method="bfill", limit=2))
self.assert_eq(psdf.fillna({"x": -1}), pdf.fillna({"x": -1}))
self.assert_eq(
psdf.fillna({"x": -1, ("x", "b"): -2}), pdf.fillna({"x": -1, ("x", "b"): -2})
)
self.assert_eq(
psdf.fillna({("x", "b"): -2, "x": -1}), pdf.fillna({("x", "b"): -2, "x": -1})
)
# check multi index
pdf = pdf.set_index([("x", "a"), ("x", "b")])
psdf = ps.from_pandas(pdf)
self.assert_eq(psdf.fillna(-1), pdf.fillna(-1))
self.assert_eq(
psdf.fillna({("x", "a"): -1, ("x", "b"): -2, ("y", "c"): -5}),
pdf.fillna({("x", "a"): -1, ("x", "b"): -2, ("y", "c"): -5}),
)
def test_isnull(self):
pdf = pd.DataFrame(
{"x": [1, 2, 3, 4, None, 6], "y": list("abdabd")}, index=np.random.rand(6)
)
psdf = ps.from_pandas(pdf)
self.assert_eq(psdf.notnull(), pdf.notnull())
self.assert_eq(psdf.isnull(), pdf.isnull())
def test_to_datetime(self):
pdf = pd.DataFrame(
{"year": [2015, 2016], "month": [2, 3], "day": [4, 5]}, index=np.random.rand(2)
)
psdf = ps.from_pandas(pdf)
self.assert_eq(pd.to_datetime(pdf), ps.to_datetime(psdf))
def test_nunique(self):
pdf = pd.DataFrame({"A": [1, 2, 3], "B": [np.nan, 3, np.nan]}, index=np.random.rand(3))
psdf = ps.from_pandas(pdf)
# Assert NaNs are dropped by default
self.assert_eq(psdf.nunique(), pdf.nunique())
# Assert including NaN values
self.assert_eq(psdf.nunique(dropna=False), pdf.nunique(dropna=False))
# Assert approximate counts
self.assert_eq(
ps.DataFrame({"A": range(100)}).nunique(approx=True),
pd.Series([103], index=["A"]),
)
self.assert_eq(
ps.DataFrame({"A": range(100)}).nunique(approx=True, rsd=0.01),
pd.Series([100], index=["A"]),
)
# Assert unsupported axis value yet
msg = 'axis should be either 0 or "index" currently.'
with self.assertRaisesRegex(NotImplementedError, msg):
psdf.nunique(axis=1)
# multi-index columns
columns = pd.MultiIndex.from_tuples([("X", "A"), ("Y", "B")], names=["1", "2"])
pdf.columns = columns
psdf.columns = columns
self.assert_eq(psdf.nunique(), pdf.nunique())
self.assert_eq(psdf.nunique(dropna=False), pdf.nunique(dropna=False))
def test_sort_values(self):
pdf = pd.DataFrame(
{"a": [1, 2, 3, 4, 5, None, 7], "b": [7, 6, 5, 4, 3, 2, 1]}, index=np.random.rand(7)
)
psdf = ps.from_pandas(pdf)
self.assert_eq(psdf.sort_values("b"), pdf.sort_values("b"))
for ascending in [True, False]:
for na_position in ["first", "last"]:
self.assert_eq(
psdf.sort_values("a", ascending=ascending, na_position=na_position),
pdf.sort_values("a", ascending=ascending, na_position=na_position),
)
self.assert_eq(psdf.sort_values(["a", "b"]), pdf.sort_values(["a", "b"]))
self.assert_eq(
psdf.sort_values(["a", "b"], ascending=[False, True]),
pdf.sort_values(["a", "b"], ascending=[False, True]),
)
self.assertRaises(ValueError, lambda: psdf.sort_values(["b", "a"], ascending=[False]))
self.assert_eq(
psdf.sort_values(["a", "b"], na_position="first"),
pdf.sort_values(["a", "b"], na_position="first"),
)
self.assertRaises(ValueError, lambda: psdf.sort_values(["b", "a"], na_position="invalid"))
pserA = pdf.a
psserA = psdf.a
self.assert_eq(psdf.sort_values("b", inplace=True), pdf.sort_values("b", inplace=True))
self.assert_eq(psdf, pdf)
self.assert_eq(psserA, pserA)
# multi-index columns
pdf = pd.DataFrame(
{("X", 10): [1, 2, 3, 4, 5, None, 7], ("X", 20): [7, 6, 5, 4, 3, 2, 1]},
index=np.random.rand(7),
)
psdf = ps.from_pandas(pdf)
self.assert_eq(psdf.sort_values(("X", 20)), pdf.sort_values(("X", 20)))
self.assert_eq(
psdf.sort_values([("X", 20), ("X", 10)]), pdf.sort_values([("X", 20), ("X", 10)])
)
self.assertRaisesRegex(
ValueError,
"For a multi-index, the label must be a tuple with elements",
lambda: psdf.sort_values(["X"]),
)
# non-string names
pdf = pd.DataFrame(
{10: [1, 2, 3, 4, 5, None, 7], 20: [7, 6, 5, 4, 3, 2, 1]}, index=np.random.rand(7)
)
psdf = ps.from_pandas(pdf)
self.assert_eq(psdf.sort_values(20), pdf.sort_values(20))
self.assert_eq(psdf.sort_values([20, 10]), pdf.sort_values([20, 10]))
def test_sort_index(self):
pdf = pd.DataFrame(
{"A": [2, 1, np.nan], "B": [np.nan, 0, np.nan]}, index=["b", "a", np.nan]
)
psdf = ps.from_pandas(pdf)
# Assert invalid parameters
self.assertRaises(NotImplementedError, lambda: psdf.sort_index(axis=1))
self.assertRaises(NotImplementedError, lambda: psdf.sort_index(kind="mergesort"))
self.assertRaises(ValueError, lambda: psdf.sort_index(na_position="invalid"))
# Assert default behavior without parameters
self.assert_eq(psdf.sort_index(), pdf.sort_index())
# Assert sorting descending
self.assert_eq(psdf.sort_index(ascending=False), pdf.sort_index(ascending=False))
# Assert sorting NA indices first
self.assert_eq(psdf.sort_index(na_position="first"), pdf.sort_index(na_position="first"))
# Assert sorting descending and NA indices first
self.assert_eq(
psdf.sort_index(ascending=False, na_position="first"),
pdf.sort_index(ascending=False, na_position="first"),
)
# Assert sorting inplace
pserA = pdf.A
psserA = psdf.A
self.assertEqual(psdf.sort_index(inplace=True), pdf.sort_index(inplace=True))
self.assert_eq(psdf, pdf)
self.assert_eq(psserA, pserA)
# Assert multi-indices
pdf = pd.DataFrame(
{"A": range(4), "B": range(4)[::-1]}, index=[["b", "b", "a", "a"], [1, 0, 1, 0]]
)
psdf = ps.from_pandas(pdf)
self.assert_eq(psdf.sort_index(), pdf.sort_index())
self.assert_eq(psdf.sort_index(level=[1, 0]), pdf.sort_index(level=[1, 0]))
self.assert_eq(psdf.reset_index().sort_index(), pdf.reset_index().sort_index())
# Assert with multi-index columns
columns = pd.MultiIndex.from_tuples([("X", "A"), ("X", "B")])
pdf.columns = columns
psdf.columns = columns
self.assert_eq(psdf.sort_index(), pdf.sort_index())
def test_swaplevel(self):
# MultiIndex with two levels
arrays = [[1, 1, 2, 2], ["red", "blue", "red", "blue"]]
pidx = pd.MultiIndex.from_arrays(arrays, names=("number", "color"))
pdf = pd.DataFrame({"x1": ["a", "b", "c", "d"], "x2": ["a", "b", "c", "d"]}, index=pidx)
psdf = ps.from_pandas(pdf)
self.assert_eq(pdf.swaplevel(), psdf.swaplevel())
self.assert_eq(pdf.swaplevel(0, 1), psdf.swaplevel(0, 1))
self.assert_eq(pdf.swaplevel(1, 1), psdf.swaplevel(1, 1))
self.assert_eq(pdf.swaplevel("number", "color"), psdf.swaplevel("number", "color"))
# MultiIndex with more than two levels
arrays = [[1, 1, 2, 2], ["red", "blue", "red", "blue"], ["l", "m", "s", "xs"]]
pidx = pd.MultiIndex.from_arrays(arrays, names=("number", "color", "size"))
pdf = pd.DataFrame({"x1": ["a", "b", "c", "d"], "x2": ["a", "b", "c", "d"]}, index=pidx)
psdf = ps.from_pandas(pdf)
self.assert_eq(pdf.swaplevel(), psdf.swaplevel())
self.assert_eq(pdf.swaplevel(0, 1), psdf.swaplevel(0, 1))
self.assert_eq(pdf.swaplevel(0, 2), psdf.swaplevel(0, 2))
self.assert_eq(pdf.swaplevel(1, 2), psdf.swaplevel(1, 2))
self.assert_eq(pdf.swaplevel(1, 1), psdf.swaplevel(1, 1))
self.assert_eq(pdf.swaplevel(-1, -2), psdf.swaplevel(-1, -2))
self.assert_eq(pdf.swaplevel("number", "color"), psdf.swaplevel("number", "color"))
self.assert_eq(pdf.swaplevel("number", "size"), psdf.swaplevel("number", "size"))
self.assert_eq(pdf.swaplevel("color", "size"), psdf.swaplevel("color", "size"))
self.assert_eq(
pdf.swaplevel("color", "size", axis="index"),
psdf.swaplevel("color", "size", axis="index"),
)
self.assert_eq(
pdf.swaplevel("color", "size", axis=0), psdf.swaplevel("color", "size", axis=0)
)
pdf = pd.DataFrame(
{
"x1": ["a", "b", "c", "d"],
"x2": ["a", "b", "c", "d"],
"x3": ["a", "b", "c", "d"],
"x4": ["a", "b", "c", "d"],
}
)
pidx = pd.MultiIndex.from_arrays(arrays, names=("number", "color", "size"))
pdf.columns = pidx
psdf = ps.from_pandas(pdf)
self.assert_eq(pdf.swaplevel(axis=1), psdf.swaplevel(axis=1))
self.assert_eq(pdf.swaplevel(0, 1, axis=1), psdf.swaplevel(0, 1, axis=1))
self.assert_eq(pdf.swaplevel(0, 2, axis=1), psdf.swaplevel(0, 2, axis=1))
self.assert_eq(pdf.swaplevel(1, 2, axis=1), psdf.swaplevel(1, 2, axis=1))
self.assert_eq(pdf.swaplevel(1, 1, axis=1), psdf.swaplevel(1, 1, axis=1))
self.assert_eq(pdf.swaplevel(-1, -2, axis=1), psdf.swaplevel(-1, -2, axis=1))
self.assert_eq(
pdf.swaplevel("number", "color", axis=1), psdf.swaplevel("number", "color", axis=1)
)
self.assert_eq(
pdf.swaplevel("number", "size", axis=1), psdf.swaplevel("number", "size", axis=1)
)
self.assert_eq(
pdf.swaplevel("color", "size", axis=1), psdf.swaplevel("color", "size", axis=1)
)
self.assert_eq(
pdf.swaplevel("color", "size", axis="columns"),
psdf.swaplevel("color", "size", axis="columns"),
)
# Error conditions
self.assertRaises(AssertionError, lambda: ps.DataFrame([1, 2]).swaplevel())
self.assertRaises(IndexError, lambda: psdf.swaplevel(0, 9, axis=1))
self.assertRaises(KeyError, lambda: psdf.swaplevel("not_number", "color", axis=1))
self.assertRaises(ValueError, lambda: psdf.swaplevel(axis=2))
def test_swapaxes(self):
pdf = pd.DataFrame(
[[1, 2, 3], [4, 5, 6], [7, 8, 9]], index=["x", "y", "z"], columns=["a", "b", "c"]
)
psdf = ps.from_pandas(pdf)
self.assert_eq(psdf.swapaxes(0, 1), pdf.swapaxes(0, 1))
self.assert_eq(psdf.swapaxes(1, 0), pdf.swapaxes(1, 0))
self.assert_eq(psdf.swapaxes("index", "columns"), pdf.swapaxes("index", "columns"))
self.assert_eq(psdf.swapaxes("columns", "index"), pdf.swapaxes("columns", "index"))
self.assert_eq((psdf + 1).swapaxes(0, 1), (pdf + 1).swapaxes(0, 1))
self.assertRaises(AssertionError, lambda: psdf.swapaxes(0, 1, copy=False))
self.assertRaises(ValueError, lambda: psdf.swapaxes(0, -1))
def test_nlargest(self):
pdf = pd.DataFrame(
{"a": [1, 2, 3, 4, 5, None, 7], "b": [7, 6, 5, 4, 3, 2, 1]}, index=np.random.rand(7)
)
psdf = ps.from_pandas(pdf)
self.assert_eq(psdf.nlargest(n=5, columns="a"), pdf.nlargest(5, columns="a"))
self.assert_eq(psdf.nlargest(n=5, columns=["a", "b"]), pdf.nlargest(5, columns=["a", "b"]))
def test_nsmallest(self):
pdf = pd.DataFrame(
{"a": [1, 2, 3, 4, 5, None, 7], "b": [7, 6, 5, 4, 3, 2, 1]}, index=np.random.rand(7)
)
psdf = ps.from_pandas(pdf)
self.assert_eq(psdf.nsmallest(n=5, columns="a"), pdf.nsmallest(5, columns="a"))
self.assert_eq(
psdf.nsmallest(n=5, columns=["a", "b"]), pdf.nsmallest(5, columns=["a", "b"])
)
def test_xs(self):
d = {
"num_legs": [4, 4, 2, 2],
"num_wings": [0, 0, 2, 2],
"class": ["mammal", "mammal", "mammal", "bird"],
"animal": ["cat", "dog", "bat", "penguin"],
"locomotion": ["walks", "walks", "flies", "walks"],
}
pdf = pd.DataFrame(data=d)
pdf = pdf.set_index(["class", "animal", "locomotion"])
psdf = ps.from_pandas(pdf)
self.assert_eq(psdf.xs("mammal"), pdf.xs("mammal"))
self.assert_eq(psdf.xs(("mammal",)), pdf.xs(("mammal",)))
self.assert_eq(psdf.xs(("mammal", "dog", "walks")), pdf.xs(("mammal", "dog", "walks")))
self.assert_eq(
ps.concat([psdf, psdf]).xs(("mammal", "dog", "walks")),
pd.concat([pdf, pdf]).xs(("mammal", "dog", "walks")),
)
self.assert_eq(psdf.xs("cat", level=1), pdf.xs("cat", level=1))
self.assert_eq(psdf.xs("flies", level=2), pdf.xs("flies", level=2))
self.assert_eq(psdf.xs("mammal", level=-3), pdf.xs("mammal", level=-3))
msg = 'axis should be either 0 or "index" currently.'
with self.assertRaisesRegex(NotImplementedError, msg):
psdf.xs("num_wings", axis=1)
with self.assertRaises(KeyError):
psdf.xs(("mammal", "dog", "walk"))
msg = r"'Key length \(4\) exceeds index depth \(3\)'"
with self.assertRaisesRegex(KeyError, msg):
psdf.xs(("mammal", "dog", "walks", "foo"))
msg = "'key' should be a scalar value or tuple that contains scalar values"
with self.assertRaisesRegex(TypeError, msg):
psdf.xs(["mammal", "dog", "walks", "foo"])
self.assertRaises(IndexError, lambda: psdf.xs("foo", level=-4))
self.assertRaises(IndexError, lambda: psdf.xs("foo", level=3))
self.assertRaises(KeyError, lambda: psdf.xs(("dog", "walks"), level=1))
# non-string names
pdf = pd.DataFrame(data=d)
pdf = pdf.set_index(["class", "animal", "num_legs", "num_wings"])
psdf = ps.from_pandas(pdf)
self.assert_eq(psdf.xs(("mammal", "dog", 4)), pdf.xs(("mammal", "dog", 4)))
self.assert_eq(psdf.xs(2, level=2), pdf.xs(2, level=2))
self.assert_eq((psdf + "a").xs(("mammal", "dog", 4)), (pdf + "a").xs(("mammal", "dog", 4)))
self.assert_eq((psdf + "a").xs(2, level=2), (pdf + "a").xs(2, level=2))
def test_missing(self):
psdf = self.psdf
missing_functions = inspect.getmembers(_MissingPandasLikeDataFrame, inspect.isfunction)
unsupported_functions = [
name for (name, type_) in missing_functions if type_.__name__ == "unsupported_function"
]
for name in unsupported_functions:
with self.assertRaisesRegex(
PandasNotImplementedError,
"method.*DataFrame.*{}.*not implemented( yet\\.|\\. .+)".format(name),
):
getattr(psdf, name)()
deprecated_functions = [
name for (name, type_) in missing_functions if type_.__name__ == "deprecated_function"
]
for name in deprecated_functions:
with self.assertRaisesRegex(
PandasNotImplementedError, "method.*DataFrame.*{}.*is deprecated".format(name)
):
getattr(psdf, name)()
missing_properties = inspect.getmembers(
_MissingPandasLikeDataFrame, lambda o: isinstance(o, property)
)
unsupported_properties = [
name
for (name, type_) in missing_properties
if type_.fget.__name__ == "unsupported_property"
]
for name in unsupported_properties:
with self.assertRaisesRegex(
PandasNotImplementedError,
"property.*DataFrame.*{}.*not implemented( yet\\.|\\. .+)".format(name),
):
getattr(psdf, name)
deprecated_properties = [
name
for (name, type_) in missing_properties
if type_.fget.__name__ == "deprecated_property"
]
for name in deprecated_properties:
with self.assertRaisesRegex(
PandasNotImplementedError, "property.*DataFrame.*{}.*is deprecated".format(name)
):
getattr(psdf, name)
def test_to_numpy(self):
pdf = pd.DataFrame(
{
"a": [4, 2, 3, 4, 8, 6],
"b": [1, 2, 9, 4, 2, 4],
"c": ["one", "three", "six", "seven", "one", "5"],
},
index=np.random.rand(6),
)
psdf = ps.from_pandas(pdf)
self.assert_eq(psdf.to_numpy(), pdf.values)
def test_to_pandas(self):
pdf, psdf = self.df_pair
self.assert_eq(psdf.to_pandas(), pdf)
def test_isin(self):
pdf = pd.DataFrame(
{
"a": [4, 2, 3, 4, 8, 6],
"b": [1, 2, 9, 4, 2, 4],
"c": ["one", "three", "six", "seven", "one", "5"],
},
index=np.random.rand(6),
)
psdf = ps.from_pandas(pdf)
self.assert_eq(psdf.isin([4, "six"]), pdf.isin([4, "six"]))
# Seems like pandas has a bug when passing `np.array` as parameter
self.assert_eq(psdf.isin(np.array([4, "six"])), pdf.isin([4, "six"]))
self.assert_eq(
psdf.isin({"a": [2, 8], "c": ["three", "one"]}),
pdf.isin({"a": [2, 8], "c": ["three", "one"]}),
)
self.assert_eq(
psdf.isin({"a": np.array([2, 8]), "c": ["three", "one"]}),
pdf.isin({"a": np.array([2, 8]), "c": ["three", "one"]}),
)
msg = "'DataFrame' object has no attribute {'e'}"
with self.assertRaisesRegex(AttributeError, msg):
psdf.isin({"e": [5, 7], "a": [1, 6]})
msg = "DataFrame and Series are not supported"
with self.assertRaisesRegex(NotImplementedError, msg):
psdf.isin(pdf)
msg = "Values should be iterable, Series, DataFrame or dict."
with self.assertRaisesRegex(TypeError, msg):
psdf.isin(1)
pdf = pd.DataFrame(
{
"a": [4, 2, 3, 4, 8, 6],
"b": [1, None, 9, 4, None, 4],
"c": [None, 5, None, 3, 2, 1],
},
)
psdf = ps.from_pandas(pdf)
if LooseVersion(pd.__version__) >= LooseVersion("1.2"):
self.assert_eq(psdf.isin([4, 3, 1, 1, None]), pdf.isin([4, 3, 1, 1, None]))
else:
expected = pd.DataFrame(
{
"a": [True, False, True, True, False, False],
"b": [True, False, False, True, False, True],
"c": [False, False, False, True, False, True],
}
)
self.assert_eq(psdf.isin([4, 3, 1, 1, None]), expected)
if LooseVersion(pd.__version__) >= LooseVersion("1.2"):
self.assert_eq(
psdf.isin({"b": [4, 3, 1, 1, None]}), pdf.isin({"b": [4, 3, 1, 1, None]})
)
else:
expected = pd.DataFrame(
{
"a": [False, False, False, False, False, False],
"b": [True, False, False, True, False, True],
"c": [False, False, False, False, False, False],
}
)
self.assert_eq(psdf.isin({"b": [4, 3, 1, 1, None]}), expected)
def test_merge(self):
left_pdf = pd.DataFrame(
{
"lkey": ["foo", "bar", "baz", "foo", "bar", "l"],
"value": [1, 2, 3, 5, 6, 7],
"x": list("abcdef"),
},
columns=["lkey", "value", "x"],
)
right_pdf = pd.DataFrame(
{
"rkey": ["baz", "foo", "bar", "baz", "foo", "r"],
"value": [4, 5, 6, 7, 8, 9],
"y": list("efghij"),
},
columns=["rkey", "value", "y"],
)
right_ps = pd.Series(list("defghi"), name="x", index=[5, 6, 7, 8, 9, 10])
left_psdf = ps.from_pandas(left_pdf)
right_psdf = ps.from_pandas(right_pdf)
right_psser = ps.from_pandas(right_ps)
def check(op, right_psdf=right_psdf, right_pdf=right_pdf):
k_res = op(left_psdf, right_psdf)
k_res = k_res.to_pandas()
k_res = k_res.sort_values(by=list(k_res.columns))
k_res = k_res.reset_index(drop=True)
p_res = op(left_pdf, right_pdf)
p_res = p_res.sort_values(by=list(p_res.columns))
p_res = p_res.reset_index(drop=True)
self.assert_eq(k_res, p_res)
check(lambda left, right: left.merge(right))
check(lambda left, right: left.merge(right, on="value"))
check(lambda left, right: left.merge(right, on=("value",)))
check(lambda left, right: left.merge(right, left_on="lkey", right_on="rkey"))
check(lambda left, right: left.set_index("lkey").merge(right.set_index("rkey")))
check(
lambda left, right: left.set_index("lkey").merge(
right, left_index=True, right_on="rkey"
)
)
check(
lambda left, right: left.merge(
right.set_index("rkey"), left_on="lkey", right_index=True
)
)
check(
lambda left, right: left.set_index("lkey").merge(
right.set_index("rkey"), left_index=True, right_index=True
)
)
# MultiIndex
check(
lambda left, right: left.merge(
right, left_on=["lkey", "value"], right_on=["rkey", "value"]
)
)
check(
lambda left, right: left.set_index(["lkey", "value"]).merge(
right, left_index=True, right_on=["rkey", "value"]
)
)
check(
lambda left, right: left.merge(
right.set_index(["rkey", "value"]), left_on=["lkey", "value"], right_index=True
)
)
# TODO: when both left_index=True and right_index=True with multi-index
# check(lambda left, right: left.set_index(['lkey', 'value']).merge(
# right.set_index(['rkey', 'value']), left_index=True, right_index=True))
# join types
for how in ["inner", "left", "right", "outer"]:
check(lambda left, right: left.merge(right, on="value", how=how))
check(lambda left, right: left.merge(right, left_on="lkey", right_on="rkey", how=how))
# suffix
check(
lambda left, right: left.merge(
right, left_on="lkey", right_on="rkey", suffixes=["_left", "_right"]
)
)
# Test Series on the right
check(lambda left, right: left.merge(right), right_psser, right_ps)
check(
lambda left, right: left.merge(right, left_on="x", right_on="x"), right_psser, right_ps
)
check(
lambda left, right: left.set_index("x").merge(right, left_index=True, right_on="x"),
right_psser,
right_ps,
)
# Test join types with Series
for how in ["inner", "left", "right", "outer"]:
check(lambda left, right: left.merge(right, how=how), right_psser, right_ps)
check(
lambda left, right: left.merge(right, left_on="x", right_on="x", how=how),
right_psser,
right_ps,
)
# suffix with Series
check(
lambda left, right: left.merge(
right,
suffixes=["_left", "_right"],
how="outer",
left_index=True,
right_index=True,
),
right_psser,
right_ps,
)
# multi-index columns
left_columns = pd.MultiIndex.from_tuples([(10, "lkey"), (10, "value"), (20, "x")])
left_pdf.columns = left_columns
left_psdf.columns = left_columns
right_columns = pd.MultiIndex.from_tuples([(10, "rkey"), (10, "value"), (30, "y")])
right_pdf.columns = right_columns
right_psdf.columns = right_columns
check(lambda left, right: left.merge(right))
check(lambda left, right: left.merge(right, on=[(10, "value")]))
check(
lambda left, right: (left.set_index((10, "lkey")).merge(right.set_index((10, "rkey"))))
)
check(
lambda left, right: (
left.set_index((10, "lkey")).merge(
right.set_index((10, "rkey")), left_index=True, right_index=True
)
)
)
# TODO: when both left_index=True and right_index=True with multi-index columns
# check(lambda left, right: left.merge(right,
# left_on=[('a', 'lkey')], right_on=[('a', 'rkey')]))
# check(lambda left, right: (left.set_index(('a', 'lkey'))
# .merge(right, left_index=True, right_on=[('a', 'rkey')])))
# non-string names
left_pdf.columns = [10, 100, 1000]
left_psdf.columns = [10, 100, 1000]
right_pdf.columns = [20, 100, 2000]
right_psdf.columns = [20, 100, 2000]
check(lambda left, right: left.merge(right))
check(lambda left, right: left.merge(right, on=[100]))
check(lambda left, right: (left.set_index(10).merge(right.set_index(20))))
check(
lambda left, right: (
left.set_index(10).merge(right.set_index(20), left_index=True, right_index=True)
)
)
def test_merge_same_anchor(self):
pdf = pd.DataFrame(
{
"lkey": ["foo", "bar", "baz", "foo", "bar", "l"],
"rkey": ["baz", "foo", "bar", "baz", "foo", "r"],
"value": [1, 1, 3, 5, 6, 7],
"x": list("abcdef"),
"y": list("efghij"),
},
columns=["lkey", "rkey", "value", "x", "y"],
)
psdf = ps.from_pandas(pdf)
left_pdf = pdf[["lkey", "value", "x"]]
right_pdf = pdf[["rkey", "value", "y"]]
left_psdf = psdf[["lkey", "value", "x"]]
right_psdf = psdf[["rkey", "value", "y"]]
def check(op, right_psdf=right_psdf, right_pdf=right_pdf):
k_res = op(left_psdf, right_psdf)
k_res = k_res.to_pandas()
k_res = k_res.sort_values(by=list(k_res.columns))
k_res = k_res.reset_index(drop=True)
p_res = op(left_pdf, right_pdf)
p_res = p_res.sort_values(by=list(p_res.columns))
p_res = p_res.reset_index(drop=True)
self.assert_eq(k_res, p_res)
check(lambda left, right: left.merge(right))
check(lambda left, right: left.merge(right, on="value"))
check(lambda left, right: left.merge(right, left_on="lkey", right_on="rkey"))
check(lambda left, right: left.set_index("lkey").merge(right.set_index("rkey")))
check(
lambda left, right: left.set_index("lkey").merge(
right, left_index=True, right_on="rkey"
)
)
check(
lambda left, right: left.merge(
right.set_index("rkey"), left_on="lkey", right_index=True
)
)
check(
lambda left, right: left.set_index("lkey").merge(
right.set_index("rkey"), left_index=True, right_index=True
)
)
def test_merge_retains_indices(self):
left_pdf = pd.DataFrame({"A": [0, 1]})
right_pdf = pd.DataFrame({"B": [1, 2]}, index=[1, 2])
left_psdf = ps.from_pandas(left_pdf)
right_psdf = ps.from_pandas(right_pdf)
self.assert_eq(
left_psdf.merge(right_psdf, left_index=True, right_index=True),
left_pdf.merge(right_pdf, left_index=True, right_index=True),
)
self.assert_eq(
left_psdf.merge(right_psdf, left_on="A", right_index=True),
left_pdf.merge(right_pdf, left_on="A", right_index=True),
)
self.assert_eq(
left_psdf.merge(right_psdf, left_index=True, right_on="B"),
left_pdf.merge(right_pdf, left_index=True, right_on="B"),
)
self.assert_eq(
left_psdf.merge(right_psdf, left_on="A", right_on="B"),
left_pdf.merge(right_pdf, left_on="A", right_on="B"),
)
def test_merge_how_parameter(self):
left_pdf = pd.DataFrame({"A": [1, 2]})
right_pdf = pd.DataFrame({"B": ["x", "y"]}, index=[1, 2])
left_psdf = ps.from_pandas(left_pdf)
right_psdf = ps.from_pandas(right_pdf)
psdf = left_psdf.merge(right_psdf, left_index=True, right_index=True)
pdf = left_pdf.merge(right_pdf, left_index=True, right_index=True)
self.assert_eq(
psdf.sort_values(by=list(psdf.columns)).reset_index(drop=True),
pdf.sort_values(by=list(pdf.columns)).reset_index(drop=True),
)
psdf = left_psdf.merge(right_psdf, left_index=True, right_index=True, how="left")
pdf = left_pdf.merge(right_pdf, left_index=True, right_index=True, how="left")
self.assert_eq(
psdf.sort_values(by=list(psdf.columns)).reset_index(drop=True),
pdf.sort_values(by=list(pdf.columns)).reset_index(drop=True),
)
psdf = left_psdf.merge(right_psdf, left_index=True, right_index=True, how="right")
pdf = left_pdf.merge(right_pdf, left_index=True, right_index=True, how="right")
self.assert_eq(
psdf.sort_values(by=list(psdf.columns)).reset_index(drop=True),
pdf.sort_values(by=list(pdf.columns)).reset_index(drop=True),
)
psdf = left_psdf.merge(right_psdf, left_index=True, right_index=True, how="outer")
pdf = left_pdf.merge(right_pdf, left_index=True, right_index=True, how="outer")
self.assert_eq(
psdf.sort_values(by=list(psdf.columns)).reset_index(drop=True),
pdf.sort_values(by=list(pdf.columns)).reset_index(drop=True),
)
def test_merge_raises(self):
left = ps.DataFrame(
{"value": [1, 2, 3, 5, 6], "x": list("abcde")},
columns=["value", "x"],
index=["foo", "bar", "baz", "foo", "bar"],
)
right = ps.DataFrame(
{"value": [4, 5, 6, 7, 8], "y": list("fghij")},
columns=["value", "y"],
index=["baz", "foo", "bar", "baz", "foo"],
)
with self.assertRaisesRegex(ValueError, "No common columns to perform merge on"):
left[["x"]].merge(right[["y"]])
with self.assertRaisesRegex(ValueError, "not a combination of both"):
left.merge(right, on="value", left_on="x")
with self.assertRaisesRegex(ValueError, "Must pass right_on or right_index=True"):
left.merge(right, left_on="x")
with self.assertRaisesRegex(ValueError, "Must pass right_on or right_index=True"):
left.merge(right, left_index=True)
with self.assertRaisesRegex(ValueError, "Must pass left_on or left_index=True"):
left.merge(right, right_on="y")
with self.assertRaisesRegex(ValueError, "Must pass left_on or left_index=True"):
left.merge(right, right_index=True)
with self.assertRaisesRegex(
ValueError, "len\\(left_keys\\) must equal len\\(right_keys\\)"
):
left.merge(right, left_on="value", right_on=["value", "y"])
with self.assertRaisesRegex(
ValueError, "len\\(left_keys\\) must equal len\\(right_keys\\)"
):
left.merge(right, left_on=["value", "x"], right_on="value")
with self.assertRaisesRegex(ValueError, "['inner', 'left', 'right', 'full', 'outer']"):
left.merge(right, left_index=True, right_index=True, how="foo")
with self.assertRaisesRegex(KeyError, "id"):
left.merge(right, on="id")
def test_append(self):
pdf = pd.DataFrame([[1, 2], [3, 4]], columns=list("AB"))
psdf = ps.from_pandas(pdf)
other_pdf = pd.DataFrame([[3, 4], [5, 6]], columns=list("BC"), index=[2, 3])
other_psdf = ps.from_pandas(other_pdf)
self.assert_eq(psdf.append(psdf), pdf.append(pdf))
self.assert_eq(psdf.append(psdf, ignore_index=True), pdf.append(pdf, ignore_index=True))
# Assert DataFrames with non-matching columns
self.assert_eq(psdf.append(other_psdf), pdf.append(other_pdf))
# Assert appending a Series fails
msg = "DataFrames.append() does not support appending Series to DataFrames"
with self.assertRaises(TypeError, msg=msg):
psdf.append(psdf["A"])
# Assert using the sort parameter raises an exception
msg = "The 'sort' parameter is currently not supported"
with self.assertRaises(NotImplementedError, msg=msg):
psdf.append(psdf, sort=True)
# Assert using 'verify_integrity' only raises an exception for overlapping indices
self.assert_eq(
psdf.append(other_psdf, verify_integrity=True),
pdf.append(other_pdf, verify_integrity=True),
)
msg = "Indices have overlapping values"
with self.assertRaises(ValueError, msg=msg):
psdf.append(psdf, verify_integrity=True)
# Skip integrity verification when ignore_index=True
self.assert_eq(
psdf.append(psdf, ignore_index=True, verify_integrity=True),
pdf.append(pdf, ignore_index=True, verify_integrity=True),
)
# Assert appending multi-index DataFrames
multi_index_pdf = pd.DataFrame([[1, 2], [3, 4]], columns=list("AB"), index=[[2, 3], [4, 5]])
multi_index_psdf = ps.from_pandas(multi_index_pdf)
other_multi_index_pdf = pd.DataFrame(
[[5, 6], [7, 8]], columns=list("AB"), index=[[2, 3], [6, 7]]
)
other_multi_index_psdf = ps.from_pandas(other_multi_index_pdf)
self.assert_eq(
multi_index_psdf.append(multi_index_psdf), multi_index_pdf.append(multi_index_pdf)
)
# Assert DataFrames with non-matching columns
self.assert_eq(
multi_index_psdf.append(other_multi_index_psdf),
multi_index_pdf.append(other_multi_index_pdf),
)
# Assert using 'verify_integrity' only raises an exception for overlapping indices
self.assert_eq(
multi_index_psdf.append(other_multi_index_psdf, verify_integrity=True),
multi_index_pdf.append(other_multi_index_pdf, verify_integrity=True),
)
with self.assertRaises(ValueError, msg=msg):
multi_index_psdf.append(multi_index_psdf, verify_integrity=True)
# Skip integrity verification when ignore_index=True
self.assert_eq(
multi_index_psdf.append(multi_index_psdf, ignore_index=True, verify_integrity=True),
multi_index_pdf.append(multi_index_pdf, ignore_index=True, verify_integrity=True),
)
# Assert trying to append DataFrames with different index levels
msg = "Both DataFrames have to have the same number of index levels"
with self.assertRaises(ValueError, msg=msg):
psdf.append(multi_index_psdf)
# Skip index level check when ignore_index=True
self.assert_eq(
psdf.append(multi_index_psdf, ignore_index=True),
pdf.append(multi_index_pdf, ignore_index=True),
)
columns = pd.MultiIndex.from_tuples([("A", "X"), ("A", "Y")])
pdf.columns = columns
psdf.columns = columns
self.assert_eq(psdf.append(psdf), pdf.append(pdf))
def test_clip(self):
pdf = pd.DataFrame(
{"A": [0, 2, 4], "B": [4, 2, 0], "X": [-1, 10, 0]}, index=np.random.rand(3)
)
psdf = ps.from_pandas(pdf)
# Assert list-like values are not accepted for 'lower' and 'upper'
msg = "List-like value are not supported for 'lower' and 'upper' at the moment"
with self.assertRaises(TypeError, msg=msg):
psdf.clip(lower=[1])
with self.assertRaises(TypeError, msg=msg):
psdf.clip(upper=[1])
# Assert no lower or upper
self.assert_eq(psdf.clip(), pdf.clip())
# Assert lower only
self.assert_eq(psdf.clip(1), pdf.clip(1))
# Assert upper only
self.assert_eq(psdf.clip(upper=3), pdf.clip(upper=3))
# Assert lower and upper
self.assert_eq(psdf.clip(1, 3), pdf.clip(1, 3))
pdf["clip"] = pdf.A.clip(lower=1, upper=3)
psdf["clip"] = psdf.A.clip(lower=1, upper=3)
self.assert_eq(psdf, pdf)
# Assert behavior on string values
str_psdf = ps.DataFrame({"A": ["a", "b", "c"]}, index=np.random.rand(3))
self.assert_eq(str_psdf.clip(1, 3), str_psdf)
def test_binary_operators(self):
pdf = pd.DataFrame(
{"A": [0, 2, 4], "B": [4, 2, 0], "X": [-1, 10, 0]}, index=np.random.rand(3)
)
psdf = ps.from_pandas(pdf)
self.assert_eq(psdf + psdf.copy(), pdf + pdf.copy())
self.assert_eq(psdf + psdf.loc[:, ["A", "B"]], pdf + pdf.loc[:, ["A", "B"]])
self.assert_eq(psdf.loc[:, ["A", "B"]] + psdf, pdf.loc[:, ["A", "B"]] + pdf)
self.assertRaisesRegex(
ValueError,
"it comes from a different dataframe",
lambda: ps.range(10).add(ps.range(10)),
)
self.assertRaisesRegex(
TypeError,
"add with a sequence is currently not supported",
lambda: ps.range(10).add(ps.range(10).id),
)
psdf_other = psdf.copy()
psdf_other.columns = pd.MultiIndex.from_tuples([("A", "Z"), ("B", "X"), ("C", "C")])
self.assertRaisesRegex(
ValueError,
"cannot join with no overlapping index names",
lambda: psdf.add(psdf_other),
)
def test_binary_operator_add(self):
# Positive
pdf = pd.DataFrame({"a": ["x"], "b": ["y"], "c": [1], "d": [2]})
psdf = ps.from_pandas(pdf)
self.assert_eq(psdf["a"] + psdf["b"], pdf["a"] + pdf["b"])
self.assert_eq(psdf["c"] + psdf["d"], pdf["c"] + pdf["d"])
# Negative
ks_err_msg = "Addition can not be applied to given types"
self.assertRaisesRegex(TypeError, ks_err_msg, lambda: psdf["a"] + psdf["c"])
self.assertRaisesRegex(TypeError, ks_err_msg, lambda: psdf["c"] + psdf["a"])
self.assertRaisesRegex(TypeError, ks_err_msg, lambda: psdf["c"] + "literal")
self.assertRaisesRegex(TypeError, ks_err_msg, lambda: "literal" + psdf["c"])
self.assertRaisesRegex(TypeError, ks_err_msg, lambda: 1 + psdf["a"])
self.assertRaisesRegex(TypeError, ks_err_msg, lambda: psdf["a"] + 1)
def test_binary_operator_sub(self):
# Positive
pdf = pd.DataFrame({"a": [2], "b": [1]})
psdf = ps.from_pandas(pdf)
self.assert_eq(psdf["a"] - psdf["b"], pdf["a"] - pdf["b"])
# Negative
psdf = ps.DataFrame({"a": ["x"], "b": [1]})
ks_err_msg = "Subtraction can not be applied to given types"
self.assertRaisesRegex(TypeError, ks_err_msg, lambda: psdf["b"] - psdf["a"])
self.assertRaisesRegex(TypeError, ks_err_msg, lambda: psdf["b"] - "literal")
self.assertRaisesRegex(TypeError, ks_err_msg, lambda: "literal" - psdf["b"])
ks_err_msg = "Subtraction can not be applied to strings"
self.assertRaisesRegex(TypeError, ks_err_msg, lambda: psdf["a"] - psdf["b"])
self.assertRaisesRegex(TypeError, ks_err_msg, lambda: 1 - psdf["a"])
self.assertRaisesRegex(TypeError, ks_err_msg, lambda: psdf["a"] - 1)
psdf = ps.DataFrame({"a": ["x"], "b": ["y"]})
self.assertRaisesRegex(TypeError, ks_err_msg, lambda: psdf["a"] - psdf["b"])
def test_binary_operator_truediv(self):
# Positive
pdf = pd.DataFrame({"a": [3], "b": [2]})
psdf = ps.from_pandas(pdf)
self.assert_eq(psdf["a"] / psdf["b"], pdf["a"] / pdf["b"])
# Negative
psdf = ps.DataFrame({"a": ["x"], "b": [1]})
ks_err_msg = "True division can not be applied to given types"
self.assertRaisesRegex(TypeError, ks_err_msg, lambda: psdf["b"] / psdf["a"])
self.assertRaisesRegex(TypeError, ks_err_msg, lambda: psdf["b"] / "literal")
self.assertRaisesRegex(TypeError, ks_err_msg, lambda: "literal" / psdf["b"])
ks_err_msg = "True division can not be applied to strings"
self.assertRaisesRegex(TypeError, ks_err_msg, lambda: psdf["a"] / psdf["b"])
self.assertRaisesRegex(TypeError, ks_err_msg, lambda: 1 / psdf["a"])
def test_binary_operator_floordiv(self):
psdf = ps.DataFrame({"a": ["x"], "b": [1]})
ks_err_msg = "Floor division can not be applied to strings"
self.assertRaisesRegex(TypeError, ks_err_msg, lambda: psdf["a"] // psdf["b"])
self.assertRaisesRegex(TypeError, ks_err_msg, lambda: 1 // psdf["a"])
ks_err_msg = "Floor division can not be applied to given types"
self.assertRaisesRegex(TypeError, ks_err_msg, lambda: psdf["b"] // psdf["a"])
self.assertRaisesRegex(TypeError, ks_err_msg, lambda: psdf["b"] // "literal")
self.assertRaisesRegex(TypeError, ks_err_msg, lambda: "literal" // psdf["b"])
def test_binary_operator_mod(self):
# Positive
pdf = pd.DataFrame({"a": [3], "b": [2]})
psdf = ps.from_pandas(pdf)
self.assert_eq(psdf["a"] % psdf["b"], pdf["a"] % pdf["b"])
# Negative
psdf = ps.DataFrame({"a": ["x"], "b": [1]})
ks_err_msg = "Modulo can not be applied to given types"
self.assertRaisesRegex(TypeError, ks_err_msg, lambda: psdf["b"] % psdf["a"])
self.assertRaisesRegex(TypeError, ks_err_msg, lambda: psdf["b"] % "literal")
ks_err_msg = "Modulo can not be applied to strings"
self.assertRaisesRegex(TypeError, ks_err_msg, lambda: psdf["a"] % psdf["b"])
self.assertRaisesRegex(TypeError, ks_err_msg, lambda: 1 % psdf["a"])
def test_binary_operator_multiply(self):
# Positive
pdf = pd.DataFrame({"a": ["x", "y"], "b": [1, 2], "c": [3, 4]})
psdf = ps.from_pandas(pdf)
self.assert_eq(psdf["b"] * psdf["c"], pdf["b"] * pdf["c"])
self.assert_eq(psdf["c"] * psdf["b"], pdf["c"] * pdf["b"])
self.assert_eq(psdf["a"] * psdf["b"], pdf["a"] * pdf["b"])
self.assert_eq(psdf["b"] * psdf["a"], pdf["b"] * pdf["a"])
self.assert_eq(psdf["a"] * 2, pdf["a"] * 2)
self.assert_eq(psdf["b"] * 2, pdf["b"] * 2)
self.assert_eq(2 * psdf["a"], 2 * pdf["a"])
self.assert_eq(2 * psdf["b"], 2 * pdf["b"])
# Negative
psdf = ps.DataFrame({"a": ["x"], "b": [2]})
ks_err_msg = "Multiplication can not be applied to given types"
self.assertRaisesRegex(TypeError, ks_err_msg, lambda: psdf["b"] * "literal")
self.assertRaisesRegex(TypeError, ks_err_msg, lambda: "literal" * psdf["b"])
self.assertRaisesRegex(TypeError, ks_err_msg, lambda: psdf["a"] * "literal")
self.assertRaisesRegex(TypeError, ks_err_msg, lambda: psdf["a"] * psdf["a"])
self.assertRaisesRegex(TypeError, ks_err_msg, lambda: psdf["a"] * 0.1)
self.assertRaisesRegex(TypeError, ks_err_msg, lambda: 0.1 * psdf["a"])
self.assertRaisesRegex(TypeError, ks_err_msg, lambda: "literal" * psdf["a"])
def test_sample(self):
pdf = pd.DataFrame({"A": [0, 2, 4]})
psdf = ps.from_pandas(pdf)
# Make sure the tests run, but we can't check the result because they are non-deterministic.
psdf.sample(frac=0.1)
psdf.sample(frac=0.2, replace=True)
psdf.sample(frac=0.2, random_state=5)
psdf["A"].sample(frac=0.2)
psdf["A"].sample(frac=0.2, replace=True)
psdf["A"].sample(frac=0.2, random_state=5)
with self.assertRaises(ValueError):
psdf.sample()
with self.assertRaises(NotImplementedError):
psdf.sample(n=1)
def test_add_prefix(self):
pdf = pd.DataFrame({"A": [1, 2, 3, 4], "B": [3, 4, 5, 6]}, index=np.random.rand(4))
psdf = ps.from_pandas(pdf)
self.assert_eq(pdf.add_prefix("col_"), psdf.add_prefix("col_"))
columns = pd.MultiIndex.from_tuples([("X", "A"), ("X", "B")])
pdf.columns = columns
psdf.columns = columns
self.assert_eq(pdf.add_prefix("col_"), psdf.add_prefix("col_"))
def test_add_suffix(self):
pdf = pd.DataFrame({"A": [1, 2, 3, 4], "B": [3, 4, 5, 6]}, index=np.random.rand(4))
psdf = ps.from_pandas(pdf)
self.assert_eq(pdf.add_suffix("first_series"), psdf.add_suffix("first_series"))
columns = pd.MultiIndex.from_tuples([("X", "A"), ("X", "B")])
pdf.columns = columns
psdf.columns = columns
self.assert_eq(pdf.add_suffix("first_series"), psdf.add_suffix("first_series"))
def test_join(self):
# check basic function
pdf1 = pd.DataFrame(
{"key": ["K0", "K1", "K2", "K3"], "A": ["A0", "A1", "A2", "A3"]}, columns=["key", "A"]
)
pdf2 = pd.DataFrame(
{"key": ["K0", "K1", "K2"], "B": ["B0", "B1", "B2"]}, columns=["key", "B"]
)
psdf1 = ps.from_pandas(pdf1)
psdf2 = ps.from_pandas(pdf2)
join_pdf = pdf1.join(pdf2, lsuffix="_left", rsuffix="_right")
join_pdf.sort_values(by=list(join_pdf.columns), inplace=True)
join_psdf = psdf1.join(psdf2, lsuffix="_left", rsuffix="_right")
join_psdf.sort_values(by=list(join_psdf.columns), inplace=True)
self.assert_eq(join_pdf, join_psdf)
# join with duplicated columns in Series
with self.assertRaisesRegex(ValueError, "columns overlap but no suffix specified"):
ks1 = ps.Series(["A1", "A5"], index=[1, 2], name="A")
psdf1.join(ks1, how="outer")
# join with duplicated columns in DataFrame
with self.assertRaisesRegex(ValueError, "columns overlap but no suffix specified"):
psdf1.join(psdf2, how="outer")
# check `on` parameter
join_pdf = pdf1.join(pdf2.set_index("key"), on="key", lsuffix="_left", rsuffix="_right")
join_pdf.sort_values(by=list(join_pdf.columns), inplace=True)
join_psdf = psdf1.join(psdf2.set_index("key"), on="key", lsuffix="_left", rsuffix="_right")
join_psdf.sort_values(by=list(join_psdf.columns), inplace=True)
self.assert_eq(join_pdf.reset_index(drop=True), join_psdf.reset_index(drop=True))
join_pdf = pdf1.set_index("key").join(
pdf2.set_index("key"), on="key", lsuffix="_left", rsuffix="_right"
)
join_pdf.sort_values(by=list(join_pdf.columns), inplace=True)
join_psdf = psdf1.set_index("key").join(
psdf2.set_index("key"), on="key", lsuffix="_left", rsuffix="_right"
)
join_psdf.sort_values(by=list(join_psdf.columns), inplace=True)
self.assert_eq(join_pdf.reset_index(drop=True), join_psdf.reset_index(drop=True))
# multi-index columns
columns1 = pd.MultiIndex.from_tuples([("x", "key"), ("Y", "A")])
columns2 = pd.MultiIndex.from_tuples([("x", "key"), ("Y", "B")])
pdf1.columns = columns1
pdf2.columns = columns2
psdf1.columns = columns1
psdf2.columns = columns2
join_pdf = pdf1.join(pdf2, lsuffix="_left", rsuffix="_right")
join_pdf.sort_values(by=list(join_pdf.columns), inplace=True)
join_psdf = psdf1.join(psdf2, lsuffix="_left", rsuffix="_right")
join_psdf.sort_values(by=list(join_psdf.columns), inplace=True)
self.assert_eq(join_pdf, join_psdf)
# check `on` parameter
join_pdf = pdf1.join(
pdf2.set_index(("x", "key")), on=[("x", "key")], lsuffix="_left", rsuffix="_right"
)
join_pdf.sort_values(by=list(join_pdf.columns), inplace=True)
join_psdf = psdf1.join(
psdf2.set_index(("x", "key")), on=[("x", "key")], lsuffix="_left", rsuffix="_right"
)
join_psdf.sort_values(by=list(join_psdf.columns), inplace=True)
self.assert_eq(join_pdf.reset_index(drop=True), join_psdf.reset_index(drop=True))
join_pdf = pdf1.set_index(("x", "key")).join(
pdf2.set_index(("x", "key")), on=[("x", "key")], lsuffix="_left", rsuffix="_right"
)
join_pdf.sort_values(by=list(join_pdf.columns), inplace=True)
join_psdf = psdf1.set_index(("x", "key")).join(
psdf2.set_index(("x", "key")), on=[("x", "key")], lsuffix="_left", rsuffix="_right"
)
join_psdf.sort_values(by=list(join_psdf.columns), inplace=True)
self.assert_eq(join_pdf.reset_index(drop=True), join_psdf.reset_index(drop=True))
# multi-index
midx1 = pd.MultiIndex.from_tuples(
[("w", "a"), ("x", "b"), ("y", "c"), ("z", "d")], names=["index1", "index2"]
)
midx2 = pd.MultiIndex.from_tuples(
[("w", "a"), ("x", "b"), ("y", "c")], names=["index1", "index2"]
)
pdf1.index = midx1
pdf2.index = midx2
psdf1 = ps.from_pandas(pdf1)
psdf2 = ps.from_pandas(pdf2)
join_pdf = pdf1.join(pdf2, on=["index1", "index2"], rsuffix="_right")
join_pdf.sort_values(by=list(join_pdf.columns), inplace=True)
join_psdf = psdf1.join(psdf2, on=["index1", "index2"], rsuffix="_right")
join_psdf.sort_values(by=list(join_psdf.columns), inplace=True)
self.assert_eq(join_pdf, join_psdf)
with self.assertRaisesRegex(
ValueError, r'len\(left_on\) must equal the number of levels in the index of "right"'
):
psdf1.join(psdf2, on=["index1"], rsuffix="_right")
def test_replace(self):
pdf = pd.DataFrame(
{
"name": ["Ironman", "Captain America", "Thor", "Hulk"],
"weapon": ["Mark-45", "Shield", "Mjolnir", "Smash"],
},
index=np.random.rand(4),
)
psdf = ps.from_pandas(pdf)
with self.assertRaisesRegex(
NotImplementedError, "replace currently works only for method='pad"
):
psdf.replace(method="bfill")
with self.assertRaisesRegex(
NotImplementedError, "replace currently works only when limit=None"
):
psdf.replace(limit=10)
with self.assertRaisesRegex(
NotImplementedError, "replace currently doesn't supports regex"
):
psdf.replace(regex="")
with self.assertRaisesRegex(ValueError, "Length of to_replace and value must be same"):
psdf.replace(to_replace=["Ironman"], value=["Spiderman", "Doctor Strange"])
with self.assertRaisesRegex(TypeError, "Unsupported type function"):
psdf.replace("Ironman", lambda x: "Spiderman")
with self.assertRaisesRegex(TypeError, "Unsupported type function"):
psdf.replace(lambda x: "Ironman", "Spiderman")
self.assert_eq(psdf.replace("Ironman", "Spiderman"), pdf.replace("Ironman", "Spiderman"))
self.assert_eq(
psdf.replace(["Ironman", "Captain America"], ["Rescue", "Hawkeye"]),
pdf.replace(["Ironman", "Captain America"], ["Rescue", "Hawkeye"]),
)
self.assert_eq(
psdf.replace(("Ironman", "Captain America"), ("Rescue", "Hawkeye")),
pdf.replace(("Ironman", "Captain America"), ("Rescue", "Hawkeye")),
)
# inplace
pser = pdf.name
psser = psdf.name
pdf.replace("Ironman", "Spiderman", inplace=True)
psdf.replace("Ironman", "Spiderman", inplace=True)
self.assert_eq(psdf, pdf)
self.assert_eq(psser, pser)
pdf = pd.DataFrame(
{"A": [0, 1, 2, 3, np.nan], "B": [5, 6, 7, 8, np.nan], "C": ["a", "b", "c", "d", None]},
index=np.random.rand(5),
)
psdf = ps.from_pandas(pdf)
self.assert_eq(psdf.replace([0, 1, 2, 3, 5, 6], 4), pdf.replace([0, 1, 2, 3, 5, 6], 4))
self.assert_eq(
psdf.replace([0, 1, 2, 3, 5, 6], [6, 5, 4, 3, 2, 1]),
pdf.replace([0, 1, 2, 3, 5, 6], [6, 5, 4, 3, 2, 1]),
)
self.assert_eq(psdf.replace({0: 10, 1: 100, 7: 200}), pdf.replace({0: 10, 1: 100, 7: 200}))
self.assert_eq(
psdf.replace({"A": [0, np.nan], "B": [5, np.nan]}, 100),
pdf.replace({"A": [0, np.nan], "B": [5, np.nan]}, 100),
)
self.assert_eq(
psdf.replace({"A": {0: 100, 4: 400, np.nan: 700}}),
pdf.replace({"A": {0: 100, 4: 400, np.nan: 700}}),
)
self.assert_eq(
psdf.replace({"X": {0: 100, 4: 400, np.nan: 700}}),
pdf.replace({"X": {0: 100, 4: 400, np.nan: 700}}),
)
self.assert_eq(psdf.replace({"C": ["a", None]}, "e"), pdf.replace({"C": ["a", None]}, "e"))
# multi-index columns
columns = pd.MultiIndex.from_tuples([("X", "A"), ("X", "B"), ("Y", "C")])
pdf.columns = columns
psdf.columns = columns
self.assert_eq(psdf.replace([0, 1, 2, 3, 5, 6], 4), pdf.replace([0, 1, 2, 3, 5, 6], 4))
self.assert_eq(
psdf.replace([0, 1, 2, 3, 5, 6], [6, 5, 4, 3, 2, 1]),
pdf.replace([0, 1, 2, 3, 5, 6], [6, 5, 4, 3, 2, 1]),
)
self.assert_eq(psdf.replace({0: 10, 1: 100, 7: 200}), pdf.replace({0: 10, 1: 100, 7: 200}))
self.assert_eq(
psdf.replace({("X", "A"): [0, np.nan], ("X", "B"): 5}, 100),
pdf.replace({("X", "A"): [0, np.nan], ("X", "B"): 5}, 100),
)
self.assert_eq(
psdf.replace({("X", "A"): {0: 100, 4: 400, np.nan: 700}}),
pdf.replace({("X", "A"): {0: 100, 4: 400, np.nan: 700}}),
)
self.assert_eq(
psdf.replace({("X", "B"): {0: 100, 4: 400, np.nan: 700}}),
pdf.replace({("X", "B"): {0: 100, 4: 400, np.nan: 700}}),
)
self.assert_eq(
psdf.replace({("Y", "C"): ["a", None]}, "e"),
pdf.replace({("Y", "C"): ["a", None]}, "e"),
)
def test_update(self):
# check base function
def get_data(left_columns=None, right_columns=None):
left_pdf = pd.DataFrame(
{"A": ["1", "2", "3", "4"], "B": ["100", "200", np.nan, np.nan]}, columns=["A", "B"]
)
right_pdf = pd.DataFrame(
{"B": ["x", np.nan, "y", np.nan], "C": ["100", "200", "300", "400"]},
columns=["B", "C"],
)
left_psdf = ps.DataFrame(
{"A": ["1", "2", "3", "4"], "B": ["100", "200", None, None]}, columns=["A", "B"]
)
right_psdf = ps.DataFrame(
{"B": ["x", None, "y", None], "C": ["100", "200", "300", "400"]}, columns=["B", "C"]
)
if left_columns is not None:
left_pdf.columns = left_columns
left_psdf.columns = left_columns
if right_columns is not None:
right_pdf.columns = right_columns
right_psdf.columns = right_columns
return left_psdf, left_pdf, right_psdf, right_pdf
left_psdf, left_pdf, right_psdf, right_pdf = get_data()
pser = left_pdf.B
psser = left_psdf.B
left_pdf.update(right_pdf)
left_psdf.update(right_psdf)
self.assert_eq(left_pdf.sort_values(by=["A", "B"]), left_psdf.sort_values(by=["A", "B"]))
self.assert_eq(psser.sort_index(), pser.sort_index())
left_psdf, left_pdf, right_psdf, right_pdf = get_data()
left_pdf.update(right_pdf, overwrite=False)
left_psdf.update(right_psdf, overwrite=False)
self.assert_eq(left_pdf.sort_values(by=["A", "B"]), left_psdf.sort_values(by=["A", "B"]))
with self.assertRaises(NotImplementedError):
left_psdf.update(right_psdf, join="right")
# multi-index columns
left_columns = pd.MultiIndex.from_tuples([("X", "A"), ("X", "B")])
right_columns = pd.MultiIndex.from_tuples([("X", "B"), ("Y", "C")])
left_psdf, left_pdf, right_psdf, right_pdf = get_data(
left_columns=left_columns, right_columns=right_columns
)
left_pdf.update(right_pdf)
left_psdf.update(right_psdf)
self.assert_eq(
left_pdf.sort_values(by=[("X", "A"), ("X", "B")]),
left_psdf.sort_values(by=[("X", "A"), ("X", "B")]),
)
left_psdf, left_pdf, right_psdf, right_pdf = get_data(
left_columns=left_columns, right_columns=right_columns
)
left_pdf.update(right_pdf, overwrite=False)
left_psdf.update(right_psdf, overwrite=False)
self.assert_eq(
left_pdf.sort_values(by=[("X", "A"), ("X", "B")]),
left_psdf.sort_values(by=[("X", "A"), ("X", "B")]),
)
right_columns = pd.MultiIndex.from_tuples([("Y", "B"), ("Y", "C")])
left_psdf, left_pdf, right_psdf, right_pdf = get_data(
left_columns=left_columns, right_columns=right_columns
)
left_pdf.update(right_pdf)
left_psdf.update(right_psdf)
self.assert_eq(
left_pdf.sort_values(by=[("X", "A"), ("X", "B")]),
left_psdf.sort_values(by=[("X", "A"), ("X", "B")]),
)
def test_pivot_table_dtypes(self):
pdf = pd.DataFrame(
{
"a": [4, 2, 3, 4, 8, 6],
"b": [1, 2, 2, 4, 2, 4],
"e": [1, 2, 2, 4, 2, 4],
"c": [1, 2, 9, 4, 7, 4],
},
index=np.random.rand(6),
)
psdf = ps.from_pandas(pdf)
# Skip columns comparison by reset_index
res_df = psdf.pivot_table(
index=["c"], columns="a", values=["b"], aggfunc={"b": "mean"}
).dtypes.reset_index(drop=True)
exp_df = pdf.pivot_table(
index=["c"], columns="a", values=["b"], aggfunc={"b": "mean"}
).dtypes.reset_index(drop=True)
self.assert_eq(res_df, exp_df)
# Results don't have the same column's name
# Todo: self.assert_eq(psdf.pivot_table(columns="a", values="b").dtypes,
# pdf.pivot_table(columns="a", values="b").dtypes)
# Todo: self.assert_eq(psdf.pivot_table(index=['c'], columns="a", values="b").dtypes,
# pdf.pivot_table(index=['c'], columns="a", values="b").dtypes)
# Todo: self.assert_eq(psdf.pivot_table(index=['e', 'c'], columns="a", values="b").dtypes,
# pdf.pivot_table(index=['e', 'c'], columns="a", values="b").dtypes)
# Todo: self.assert_eq(psdf.pivot_table(index=['e', 'c'],
# columns="a", values="b", fill_value=999).dtypes, pdf.pivot_table(index=['e', 'c'],
# columns="a", values="b", fill_value=999).dtypes)
def test_pivot_table(self):
pdf = pd.DataFrame(
{
"a": [4, 2, 3, 4, 8, 6],
"b": [1, 2, 2, 4, 2, 4],
"e": [10, 20, 20, 40, 20, 40],
"c": [1, 2, 9, 4, 7, 4],
"d": [-1, -2, -3, -4, -5, -6],
},
index=np.random.rand(6),
)
psdf = ps.from_pandas(pdf)
# Checking if both DataFrames have the same results
self.assert_eq(
psdf.pivot_table(columns="a", values="b").sort_index(),
pdf.pivot_table(columns="a", values="b").sort_index(),
almost=True,
)
self.assert_eq(
psdf.pivot_table(index=["c"], columns="a", values="b").sort_index(),
pdf.pivot_table(index=["c"], columns="a", values="b").sort_index(),
almost=True,
)
self.assert_eq(
psdf.pivot_table(index=["c"], columns="a", values="b", aggfunc="sum").sort_index(),
pdf.pivot_table(index=["c"], columns="a", values="b", aggfunc="sum").sort_index(),
almost=True,
)
self.assert_eq(
psdf.pivot_table(index=["c"], columns="a", values=["b"], aggfunc="sum").sort_index(),
pdf.pivot_table(index=["c"], columns="a", values=["b"], aggfunc="sum").sort_index(),
almost=True,
)
self.assert_eq(
psdf.pivot_table(
index=["c"], columns="a", values=["b", "e"], aggfunc="sum"
).sort_index(),
pdf.pivot_table(
index=["c"], columns="a", values=["b", "e"], aggfunc="sum"
).sort_index(),
almost=True,
)
self.assert_eq(
psdf.pivot_table(
index=["c"], columns="a", values=["b", "e", "d"], aggfunc="sum"
).sort_index(),
pdf.pivot_table(
index=["c"], columns="a", values=["b", "e", "d"], aggfunc="sum"
).sort_index(),
almost=True,
)
self.assert_eq(
psdf.pivot_table(
index=["c"], columns="a", values=["b", "e"], aggfunc={"b": "mean", "e": "sum"}
).sort_index(),
pdf.pivot_table(
index=["c"], columns="a", values=["b", "e"], aggfunc={"b": "mean", "e": "sum"}
).sort_index(),
almost=True,
)
self.assert_eq(
psdf.pivot_table(index=["e", "c"], columns="a", values="b").sort_index(),
pdf.pivot_table(index=["e", "c"], columns="a", values="b").sort_index(),
almost=True,
)
self.assert_eq(
psdf.pivot_table(
index=["e", "c"], columns="a", values="b", fill_value=999
).sort_index(),
pdf.pivot_table(index=["e", "c"], columns="a", values="b", fill_value=999).sort_index(),
almost=True,
)
# multi-index columns
columns = pd.MultiIndex.from_tuples(
[("x", "a"), ("x", "b"), ("y", "e"), ("z", "c"), ("w", "d")]
)
pdf.columns = columns
psdf.columns = columns
self.assert_eq(
psdf.pivot_table(columns=("x", "a"), values=("x", "b")).sort_index(),
pdf.pivot_table(columns=[("x", "a")], values=[("x", "b")]).sort_index(),
almost=True,
)
self.assert_eq(
psdf.pivot_table(
index=[("z", "c")], columns=("x", "a"), values=[("x", "b")]
).sort_index(),
pdf.pivot_table(
index=[("z", "c")], columns=[("x", "a")], values=[("x", "b")]
).sort_index(),
almost=True,
)
self.assert_eq(
psdf.pivot_table(
index=[("z", "c")], columns=("x", "a"), values=[("x", "b"), ("y", "e")]
).sort_index(),
pdf.pivot_table(
index=[("z", "c")], columns=[("x", "a")], values=[("x", "b"), ("y", "e")]
).sort_index(),
almost=True,
)
self.assert_eq(
psdf.pivot_table(
index=[("z", "c")], columns=("x", "a"), values=[("x", "b"), ("y", "e"), ("w", "d")]
).sort_index(),
pdf.pivot_table(
index=[("z", "c")],
columns=[("x", "a")],
values=[("x", "b"), ("y", "e"), ("w", "d")],
).sort_index(),
almost=True,
)
self.assert_eq(
psdf.pivot_table(
index=[("z", "c")],
columns=("x", "a"),
values=[("x", "b"), ("y", "e")],
aggfunc={("x", "b"): "mean", ("y", "e"): "sum"},
).sort_index(),
pdf.pivot_table(
index=[("z", "c")],
columns=[("x", "a")],
values=[("x", "b"), ("y", "e")],
aggfunc={("x", "b"): "mean", ("y", "e"): "sum"},
).sort_index(),
almost=True,
)
def test_pivot_table_and_index(self):
# https://github.com/databricks/koalas/issues/805
pdf = pd.DataFrame(
{
"A": ["foo", "foo", "foo", "foo", "foo", "bar", "bar", "bar", "bar"],
"B": ["one", "one", "one", "two", "two", "one", "one", "two", "two"],
"C": [
"small",
"large",
"large",
"small",
"small",
"large",
"small",
"small",
"large",
],
"D": [1, 2, 2, 3, 3, 4, 5, 6, 7],
"E": [2, 4, 5, 5, 6, 6, 8, 9, 9],
},
columns=["A", "B", "C", "D", "E"],
index=np.random.rand(9),
)
psdf = ps.from_pandas(pdf)
ptable = pdf.pivot_table(
values="D", index=["A", "B"], columns="C", aggfunc="sum", fill_value=0
).sort_index()
ktable = psdf.pivot_table(
values="D", index=["A", "B"], columns="C", aggfunc="sum", fill_value=0
).sort_index()
self.assert_eq(ktable, ptable)
self.assert_eq(ktable.index, ptable.index)
self.assert_eq(repr(ktable.index), repr(ptable.index))
def test_stack(self):
pdf_single_level_cols = pd.DataFrame(
[[0, 1], [2, 3]], index=["cat", "dog"], columns=["weight", "height"]
)
psdf_single_level_cols = ps.from_pandas(pdf_single_level_cols)
self.assert_eq(
psdf_single_level_cols.stack().sort_index(), pdf_single_level_cols.stack().sort_index()
)
multicol1 = pd.MultiIndex.from_tuples(
[("weight", "kg"), ("weight", "pounds")], names=["x", "y"]
)
pdf_multi_level_cols1 = pd.DataFrame(
[[1, 2], [2, 4]], index=["cat", "dog"], columns=multicol1
)
psdf_multi_level_cols1 = ps.from_pandas(pdf_multi_level_cols1)
self.assert_eq(
psdf_multi_level_cols1.stack().sort_index(), pdf_multi_level_cols1.stack().sort_index()
)
multicol2 = pd.MultiIndex.from_tuples([("weight", "kg"), ("height", "m")])
pdf_multi_level_cols2 = pd.DataFrame(
[[1.0, 2.0], [3.0, 4.0]], index=["cat", "dog"], columns=multicol2
)
psdf_multi_level_cols2 = ps.from_pandas(pdf_multi_level_cols2)
self.assert_eq(
psdf_multi_level_cols2.stack().sort_index(), pdf_multi_level_cols2.stack().sort_index()
)
pdf = pd.DataFrame(
{
("y", "c"): [True, True],
("x", "b"): [False, False],
("x", "c"): [True, False],
("y", "a"): [False, True],
}
)
psdf = ps.from_pandas(pdf)
self.assert_eq(psdf.stack().sort_index(), pdf.stack().sort_index())
self.assert_eq(psdf[[]].stack().sort_index(), pdf[[]].stack().sort_index(), almost=True)
def test_unstack(self):
pdf = pd.DataFrame(
np.random.randn(3, 3),
index=pd.MultiIndex.from_tuples([("rg1", "x"), ("rg1", "y"), ("rg2", "z")]),
)
psdf = ps.from_pandas(pdf)
self.assert_eq(psdf.unstack().sort_index(), pdf.unstack().sort_index(), almost=True)
self.assert_eq(
psdf.unstack().unstack().sort_index(), pdf.unstack().unstack().sort_index(), almost=True
)
def test_pivot_errors(self):
psdf = ps.range(10)
with self.assertRaisesRegex(ValueError, "columns should be set"):
psdf.pivot(index="id")
with self.assertRaisesRegex(ValueError, "values should be set"):
psdf.pivot(index="id", columns="id")
def test_pivot_table_errors(self):
pdf = pd.DataFrame(
{
"a": [4, 2, 3, 4, 8, 6],
"b": [1, 2, 2, 4, 2, 4],
"e": [1, 2, 2, 4, 2, 4],
"c": [1, 2, 9, 4, 7, 4],
},
index=np.random.rand(6),
)
psdf = ps.from_pandas(pdf)
self.assertRaises(KeyError, lambda: psdf.pivot_table(index=["c"], columns="a", values=5))
msg = "index should be a None or a list of columns."
with self.assertRaisesRegex(TypeError, msg):
psdf.pivot_table(index="c", columns="a", values="b")
msg = "pivot_table doesn't support aggfunc as dict and without index."
with self.assertRaisesRegex(NotImplementedError, msg):
psdf.pivot_table(columns="a", values=["b", "e"], aggfunc={"b": "mean", "e": "sum"})
msg = "columns should be one column name."
with self.assertRaisesRegex(TypeError, msg):
psdf.pivot_table(columns=["a"], values=["b"], aggfunc={"b": "mean", "e": "sum"})
msg = "Columns in aggfunc must be the same as values."
with self.assertRaisesRegex(ValueError, msg):
psdf.pivot_table(
index=["e", "c"], columns="a", values="b", aggfunc={"b": "mean", "e": "sum"}
)
msg = "values can't be a list without index."
with self.assertRaisesRegex(NotImplementedError, msg):
psdf.pivot_table(columns="a", values=["b", "e"])
msg = "Wrong columns A."
with self.assertRaisesRegex(ValueError, msg):
psdf.pivot_table(
index=["c"], columns="A", values=["b", "e"], aggfunc={"b": "mean", "e": "sum"}
)
msg = "values should be one column or list of columns."
with self.assertRaisesRegex(TypeError, msg):
psdf.pivot_table(columns="a", values=(["b"], ["c"]))
msg = "aggfunc must be a dict mapping from column name to aggregate functions"
with self.assertRaisesRegex(TypeError, msg):
psdf.pivot_table(columns="a", values="b", aggfunc={"a": lambda x: sum(x)})
psdf = ps.DataFrame(
{
"A": ["foo", "foo", "foo", "foo", "foo", "bar", "bar", "bar", "bar"],
"B": ["one", "one", "one", "two", "two", "one", "one", "two", "two"],
"C": [
"small",
"large",
"large",
"small",
"small",
"large",
"small",
"small",
"large",
],
"D": [1, 2, 2, 3, 3, 4, 5, 6, 7],
"E": [2, 4, 5, 5, 6, 6, 8, 9, 9],
},
columns=["A", "B", "C", "D", "E"],
index=np.random.rand(9),
)
msg = "values should be a numeric type."
with self.assertRaisesRegex(TypeError, msg):
psdf.pivot_table(
index=["C"], columns="A", values=["B", "E"], aggfunc={"B": "mean", "E": "sum"}
)
msg = "values should be a numeric type."
with self.assertRaisesRegex(TypeError, msg):
psdf.pivot_table(index=["C"], columns="A", values="B", aggfunc={"B": "mean"})
def test_transpose(self):
# TODO: what if with random index?
pdf1 = pd.DataFrame(data={"col1": [1, 2], "col2": [3, 4]}, columns=["col1", "col2"])
psdf1 = ps.from_pandas(pdf1)
pdf2 = pd.DataFrame(
data={"score": [9, 8], "kids": [0, 0], "age": [12, 22]},
columns=["score", "kids", "age"],
)
psdf2 = ps.from_pandas(pdf2)
self.assert_eq(pdf1.transpose().sort_index(), psdf1.transpose().sort_index())
self.assert_eq(pdf2.transpose().sort_index(), psdf2.transpose().sort_index())
with option_context("compute.max_rows", None):
self.assert_eq(pdf1.transpose().sort_index(), psdf1.transpose().sort_index())
self.assert_eq(pdf2.transpose().sort_index(), psdf2.transpose().sort_index())
pdf3 = pd.DataFrame(
{
("cg1", "a"): [1, 2, 3],
("cg1", "b"): [4, 5, 6],
("cg2", "c"): [7, 8, 9],
("cg3", "d"): [9, 9, 9],
},
index=pd.MultiIndex.from_tuples([("rg1", "x"), ("rg1", "y"), ("rg2", "z")]),
)
psdf3 = ps.from_pandas(pdf3)
self.assert_eq(pdf3.transpose().sort_index(), psdf3.transpose().sort_index())
with option_context("compute.max_rows", None):
self.assert_eq(pdf3.transpose().sort_index(), psdf3.transpose().sort_index())
def _test_cummin(self, pdf, psdf):
self.assert_eq(pdf.cummin(), psdf.cummin())
self.assert_eq(pdf.cummin(skipna=False), psdf.cummin(skipna=False))
self.assert_eq(pdf.cummin().sum(), psdf.cummin().sum())
def test_cummin(self):
pdf = pd.DataFrame(
[[2.0, 1.0], [5, None], [1.0, 0.0], [2.0, 4.0], [4.0, 9.0]],
columns=list("AB"),
index=np.random.rand(5),
)
psdf = ps.from_pandas(pdf)
self._test_cummin(pdf, psdf)
def test_cummin_multiindex_columns(self):
arrays = [np.array(["A", "A", "B", "B"]), np.array(["one", "two", "one", "two"])]
pdf = pd.DataFrame(np.random.randn(3, 4), index=["A", "C", "B"], columns=arrays)
pdf.at["C", ("A", "two")] = None
psdf = ps.from_pandas(pdf)
self._test_cummin(pdf, psdf)
def _test_cummax(self, pdf, psdf):
self.assert_eq(pdf.cummax(), psdf.cummax())
self.assert_eq(pdf.cummax(skipna=False), psdf.cummax(skipna=False))
self.assert_eq(pdf.cummax().sum(), psdf.cummax().sum())
def test_cummax(self):
pdf = pd.DataFrame(
[[2.0, 1.0], [5, None], [1.0, 0.0], [2.0, 4.0], [4.0, 9.0]],
columns=list("AB"),
index=np.random.rand(5),
)
psdf = ps.from_pandas(pdf)
self._test_cummax(pdf, psdf)
def test_cummax_multiindex_columns(self):
arrays = [np.array(["A", "A", "B", "B"]), np.array(["one", "two", "one", "two"])]
pdf = pd.DataFrame(np.random.randn(3, 4), index=["A", "C", "B"], columns=arrays)
pdf.at["C", ("A", "two")] = None
psdf = ps.from_pandas(pdf)
self._test_cummax(pdf, psdf)
def _test_cumsum(self, pdf, psdf):
self.assert_eq(pdf.cumsum(), psdf.cumsum())
self.assert_eq(pdf.cumsum(skipna=False), psdf.cumsum(skipna=False))
self.assert_eq(pdf.cumsum().sum(), psdf.cumsum().sum())
def test_cumsum(self):
pdf = pd.DataFrame(
[[2.0, 1.0], [5, None], [1.0, 0.0], [2.0, 4.0], [4.0, 9.0]],
columns=list("AB"),
index=np.random.rand(5),
)
psdf = ps.from_pandas(pdf)
self._test_cumsum(pdf, psdf)
def test_cumsum_multiindex_columns(self):
arrays = [np.array(["A", "A", "B", "B"]), np.array(["one", "two", "one", "two"])]
pdf = pd.DataFrame(np.random.randn(3, 4), index=["A", "C", "B"], columns=arrays)
pdf.at["C", ("A", "two")] = None
psdf = ps.from_pandas(pdf)
self._test_cumsum(pdf, psdf)
def _test_cumprod(self, pdf, psdf):
self.assert_eq(pdf.cumprod(), psdf.cumprod(), almost=True)
self.assert_eq(pdf.cumprod(skipna=False), psdf.cumprod(skipna=False), almost=True)
self.assert_eq(pdf.cumprod().sum(), psdf.cumprod().sum(), almost=True)
def test_cumprod(self):
pdf = pd.DataFrame(
[[2.0, 1.0, 1], [5, None, 2], [1.0, -1.0, -3], [2.0, 0, 4], [4.0, 9.0, 5]],
columns=list("ABC"),
index=np.random.rand(5),
)
psdf = ps.from_pandas(pdf)
self._test_cumprod(pdf, psdf)
def test_cumprod_multiindex_columns(self):
arrays = [np.array(["A", "A", "B", "B"]), np.array(["one", "two", "one", "two"])]
pdf = pd.DataFrame(np.random.rand(3, 4), index=["A", "C", "B"], columns=arrays)
pdf.at["C", ("A", "two")] = None
psdf = ps.from_pandas(pdf)
self._test_cumprod(pdf, psdf)
def test_drop_duplicates(self):
pdf = pd.DataFrame(
{"a": [1, 2, 2, 2, 3], "b": ["a", "a", "a", "c", "d"]}, index=np.random.rand(5)
)
psdf = ps.from_pandas(pdf)
# inplace is False
for keep in ["first", "last", False]:
with self.subTest(keep=keep):
self.assert_eq(
pdf.drop_duplicates(keep=keep).sort_index(),
psdf.drop_duplicates(keep=keep).sort_index(),
)
self.assert_eq(
pdf.drop_duplicates("a", keep=keep).sort_index(),
psdf.drop_duplicates("a", keep=keep).sort_index(),
)
self.assert_eq(
pdf.drop_duplicates(["a", "b"], keep=keep).sort_index(),
psdf.drop_duplicates(["a", "b"], keep=keep).sort_index(),
)
self.assert_eq(
pdf.set_index("a", append=True).drop_duplicates(keep=keep).sort_index(),
psdf.set_index("a", append=True).drop_duplicates(keep=keep).sort_index(),
)
self.assert_eq(
pdf.set_index("a", append=True).drop_duplicates("b", keep=keep).sort_index(),
psdf.set_index("a", append=True).drop_duplicates("b", keep=keep).sort_index(),
)
columns = pd.MultiIndex.from_tuples([("x", "a"), ("y", "b")])
pdf.columns = columns
psdf.columns = columns
# inplace is False
for keep in ["first", "last", False]:
with self.subTest("multi-index columns", keep=keep):
self.assert_eq(
pdf.drop_duplicates(keep=keep).sort_index(),
psdf.drop_duplicates(keep=keep).sort_index(),
)
self.assert_eq(
pdf.drop_duplicates(("x", "a"), keep=keep).sort_index(),
psdf.drop_duplicates(("x", "a"), keep=keep).sort_index(),
)
self.assert_eq(
pdf.drop_duplicates([("x", "a"), ("y", "b")], keep=keep).sort_index(),
psdf.drop_duplicates([("x", "a"), ("y", "b")], keep=keep).sort_index(),
)
# inplace is True
subset_list = [None, "a", ["a", "b"]]
for subset in subset_list:
pdf = pd.DataFrame(
{"a": [1, 2, 2, 2, 3], "b": ["a", "a", "a", "c", "d"]}, index=np.random.rand(5)
)
psdf = ps.from_pandas(pdf)
pser = pdf.a
psser = psdf.a
pdf.drop_duplicates(subset=subset, inplace=True)
psdf.drop_duplicates(subset=subset, inplace=True)
self.assert_eq(psdf.sort_index(), pdf.sort_index())
self.assert_eq(psser.sort_index(), pser.sort_index())
# multi-index columns, inplace is True
subset_list = [None, ("x", "a"), [("x", "a"), ("y", "b")]]
for subset in subset_list:
pdf = pd.DataFrame(
{"a": [1, 2, 2, 2, 3], "b": ["a", "a", "a", "c", "d"]}, index=np.random.rand(5)
)
psdf = ps.from_pandas(pdf)
columns = pd.MultiIndex.from_tuples([("x", "a"), ("y", "b")])
pdf.columns = columns
psdf.columns = columns
pser = pdf[("x", "a")]
psser = psdf[("x", "a")]
pdf.drop_duplicates(subset=subset, inplace=True)
psdf.drop_duplicates(subset=subset, inplace=True)
self.assert_eq(psdf.sort_index(), pdf.sort_index())
self.assert_eq(psser.sort_index(), pser.sort_index())
# non-string names
pdf = pd.DataFrame(
{10: [1, 2, 2, 2, 3], 20: ["a", "a", "a", "c", "d"]}, index=np.random.rand(5)
)
psdf = ps.from_pandas(pdf)
self.assert_eq(
pdf.drop_duplicates(10, keep=keep).sort_index(),
psdf.drop_duplicates(10, keep=keep).sort_index(),
)
self.assert_eq(
pdf.drop_duplicates([10, 20], keep=keep).sort_index(),
psdf.drop_duplicates([10, 20], keep=keep).sort_index(),
)
def test_reindex(self):
index = pd.Index(["A", "B", "C", "D", "E"])
columns = pd.Index(["numbers"])
pdf = pd.DataFrame([1.0, 2.0, 3.0, 4.0, None], index=index, columns=columns)
psdf = ps.from_pandas(pdf)
columns2 = pd.Index(["numbers", "2", "3"], name="cols2")
self.assert_eq(
pdf.reindex(columns=columns2).sort_index(),
psdf.reindex(columns=columns2).sort_index(),
)
columns = pd.Index(["numbers"], name="cols")
pdf.columns = columns
psdf.columns = columns
self.assert_eq(
pdf.reindex(["A", "B", "C"], columns=["numbers", "2", "3"]).sort_index(),
psdf.reindex(["A", "B", "C"], columns=["numbers", "2", "3"]).sort_index(),
)
self.assert_eq(
pdf.reindex(["A", "B", "C"], index=["numbers", "2", "3"]).sort_index(),
psdf.reindex(["A", "B", "C"], index=["numbers", "2", "3"]).sort_index(),
)
self.assert_eq(
pdf.reindex(index=["A", "B"]).sort_index(), psdf.reindex(index=["A", "B"]).sort_index()
)
self.assert_eq(
pdf.reindex(index=["A", "B", "2", "3"]).sort_index(),
psdf.reindex(index=["A", "B", "2", "3"]).sort_index(),
)
self.assert_eq(
pdf.reindex(index=["A", "E", "2", "3"], fill_value=0).sort_index(),
psdf.reindex(index=["A", "E", "2", "3"], fill_value=0).sort_index(),
)
self.assert_eq(
pdf.reindex(columns=["numbers"]).sort_index(),
psdf.reindex(columns=["numbers"]).sort_index(),
)
self.assert_eq(
pdf.reindex(columns=["numbers"], copy=True).sort_index(),
psdf.reindex(columns=["numbers"], copy=True).sort_index(),
)
# Using float as fill_value to avoid int64/32 clash
self.assert_eq(
pdf.reindex(columns=["numbers", "2", "3"], fill_value=0.0).sort_index(),
psdf.reindex(columns=["numbers", "2", "3"], fill_value=0.0).sort_index(),
)
columns2 = pd.Index(["numbers", "2", "3"])
self.assert_eq(
pdf.reindex(columns=columns2).sort_index(),
psdf.reindex(columns=columns2).sort_index(),
)
columns2 = pd.Index(["numbers", "2", "3"], name="cols2")
self.assert_eq(
pdf.reindex(columns=columns2).sort_index(),
psdf.reindex(columns=columns2).sort_index(),
)
# Reindexing single Index on single Index
pindex2 = pd.Index(["A", "C", "D", "E", "0"], name="index2")
kindex2 = ps.from_pandas(pindex2)
for fill_value in [None, 0]:
self.assert_eq(
pdf.reindex(index=pindex2, fill_value=fill_value).sort_index(),
psdf.reindex(index=kindex2, fill_value=fill_value).sort_index(),
)
pindex2 = pd.DataFrame({"index2": ["A", "C", "D", "E", "0"]}).set_index("index2").index
kindex2 = ps.from_pandas(pindex2)
for fill_value in [None, 0]:
self.assert_eq(
pdf.reindex(index=pindex2, fill_value=fill_value).sort_index(),
psdf.reindex(index=kindex2, fill_value=fill_value).sort_index(),
)
# Reindexing MultiIndex on single Index
pindex = pd.MultiIndex.from_tuples(
[("A", "B"), ("C", "D"), ("F", "G")], names=["name1", "name2"]
)
kindex = ps.from_pandas(pindex)
self.assert_eq(
pdf.reindex(index=pindex, fill_value=0.0).sort_index(),
psdf.reindex(index=kindex, fill_value=0.0).sort_index(),
)
self.assertRaises(TypeError, lambda: psdf.reindex(columns=["numbers", "2", "3"], axis=1))
self.assertRaises(TypeError, lambda: psdf.reindex(columns=["numbers", "2", "3"], axis=2))
self.assertRaises(TypeError, lambda: psdf.reindex(columns="numbers"))
self.assertRaises(TypeError, lambda: psdf.reindex(index=["A", "B", "C"], axis=1))
self.assertRaises(TypeError, lambda: psdf.reindex(index=123))
# Reindexing MultiIndex on MultiIndex
pdf = pd.DataFrame({"numbers": [1.0, 2.0, None]}, index=pindex)
psdf = ps.from_pandas(pdf)
pindex2 = pd.MultiIndex.from_tuples(
[("A", "G"), ("C", "D"), ("I", "J")], names=["name1", "name2"]
)
kindex2 = ps.from_pandas(pindex2)
for fill_value in [None, 0.0]:
self.assert_eq(
pdf.reindex(index=pindex2, fill_value=fill_value).sort_index(),
psdf.reindex(index=kindex2, fill_value=fill_value).sort_index(),
)
pindex2 = (
pd.DataFrame({"index_level_1": ["A", "C", "I"], "index_level_2": ["G", "D", "J"]})
.set_index(["index_level_1", "index_level_2"])
.index
)
kindex2 = ps.from_pandas(pindex2)
for fill_value in [None, 0.0]:
self.assert_eq(
pdf.reindex(index=pindex2, fill_value=fill_value).sort_index(),
psdf.reindex(index=kindex2, fill_value=fill_value).sort_index(),
)
columns = pd.MultiIndex.from_tuples([("X", "numbers")], names=["cols1", "cols2"])
pdf.columns = columns
psdf.columns = columns
# Reindexing MultiIndex index on MultiIndex columns and MultiIndex index
for fill_value in [None, 0.0]:
self.assert_eq(
pdf.reindex(index=pindex2, fill_value=fill_value).sort_index(),
psdf.reindex(index=kindex2, fill_value=fill_value).sort_index(),
)
index = pd.Index(["A", "B", "C", "D", "E"])
pdf = pd.DataFrame(data=[1.0, 2.0, 3.0, 4.0, None], index=index, columns=columns)
psdf = ps.from_pandas(pdf)
pindex2 = pd.Index(["A", "C", "D", "E", "0"], name="index2")
kindex2 = ps.from_pandas(pindex2)
# Reindexing single Index on MultiIndex columns and single Index
for fill_value in [None, 0.0]:
self.assert_eq(
pdf.reindex(index=pindex2, fill_value=fill_value).sort_index(),
psdf.reindex(index=kindex2, fill_value=fill_value).sort_index(),
)
for fill_value in [None, 0.0]:
self.assert_eq(
pdf.reindex(
columns=[("X", "numbers"), ("Y", "2"), ("Y", "3")], fill_value=fill_value
).sort_index(),
psdf.reindex(
columns=[("X", "numbers"), ("Y", "2"), ("Y", "3")], fill_value=fill_value
).sort_index(),
)
columns2 = pd.MultiIndex.from_tuples(
[("X", "numbers"), ("Y", "2"), ("Y", "3")], names=["cols3", "cols4"]
)
self.assert_eq(
pdf.reindex(columns=columns2).sort_index(),
psdf.reindex(columns=columns2).sort_index(),
)
self.assertRaises(TypeError, lambda: psdf.reindex(columns=["X"]))
self.assertRaises(ValueError, lambda: psdf.reindex(columns=[("X",)]))
def test_reindex_like(self):
data = [[1.0, 2.0], [3.0, None], [None, 4.0]]
index = pd.Index(["A", "B", "C"], name="index")
columns = pd.Index(["numbers", "values"], name="cols")
pdf = pd.DataFrame(data=data, index=index, columns=columns)
psdf = ps.from_pandas(pdf)
# Reindexing single Index on single Index
data2 = [[5.0, None], [6.0, 7.0], [8.0, None]]
index2 = pd.Index(["A", "C", "D"], name="index2")
columns2 = pd.Index(["numbers", "F"], name="cols2")
pdf2 = pd.DataFrame(data=data2, index=index2, columns=columns2)
psdf2 = ps.from_pandas(pdf2)
self.assert_eq(
pdf.reindex_like(pdf2).sort_index(),
psdf.reindex_like(psdf2).sort_index(),
)
pdf2 = pd.DataFrame({"index_level_1": ["A", "C", "I"]})
psdf2 = ps.from_pandas(pdf2)
self.assert_eq(
pdf.reindex_like(pdf2.set_index(["index_level_1"])).sort_index(),
psdf.reindex_like(psdf2.set_index(["index_level_1"])).sort_index(),
)
# Reindexing MultiIndex on single Index
index2 = pd.MultiIndex.from_tuples(
[("A", "G"), ("C", "D"), ("I", "J")], names=["name3", "name4"]
)
pdf2 = pd.DataFrame(data=data2, index=index2)
psdf2 = ps.from_pandas(pdf2)
self.assert_eq(
pdf.reindex_like(pdf2).sort_index(),
psdf.reindex_like(psdf2).sort_index(),
)
self.assertRaises(TypeError, lambda: psdf.reindex_like(index2))
self.assertRaises(AssertionError, lambda: psdf2.reindex_like(psdf))
# Reindexing MultiIndex on MultiIndex
columns2 = pd.MultiIndex.from_tuples(
[("numbers", "third"), ("values", "second")], names=["cols3", "cols4"]
)
pdf2.columns = columns2
psdf2.columns = columns2
columns = pd.MultiIndex.from_tuples(
[("numbers", "first"), ("values", "second")], names=["cols1", "cols2"]
)
index = pd.MultiIndex.from_tuples(
[("A", "B"), ("C", "D"), ("E", "F")], names=["name1", "name2"]
)
pdf = pd.DataFrame(data=data, index=index, columns=columns)
psdf = ps.from_pandas(pdf)
self.assert_eq(
pdf.reindex_like(pdf2).sort_index(),
psdf.reindex_like(psdf2).sort_index(),
)
def test_melt(self):
pdf = pd.DataFrame(
{"A": [1, 3, 5], "B": [2, 4, 6], "C": [7, 8, 9]}, index=np.random.rand(3)
)
psdf = ps.from_pandas(pdf)
self.assert_eq(
psdf.melt().sort_values(["variable", "value"]).reset_index(drop=True),
pdf.melt().sort_values(["variable", "value"]),
)
self.assert_eq(
psdf.melt(id_vars="A").sort_values(["variable", "value"]).reset_index(drop=True),
pdf.melt(id_vars="A").sort_values(["variable", "value"]),
)
self.assert_eq(
psdf.melt(id_vars=["A", "B"]).sort_values(["variable", "value"]).reset_index(drop=True),
pdf.melt(id_vars=["A", "B"]).sort_values(["variable", "value"]),
)
self.assert_eq(
psdf.melt(id_vars=("A", "B")).sort_values(["variable", "value"]).reset_index(drop=True),
pdf.melt(id_vars=("A", "B")).sort_values(["variable", "value"]),
)
self.assert_eq(
psdf.melt(id_vars=["A"], value_vars=["C"])
.sort_values(["variable", "value"])
.reset_index(drop=True),
pdf.melt(id_vars=["A"], value_vars=["C"]).sort_values(["variable", "value"]),
)
self.assert_eq(
psdf.melt(id_vars=["A"], value_vars=["B"], var_name="myVarname", value_name="myValname")
.sort_values(["myVarname", "myValname"])
.reset_index(drop=True),
pdf.melt(
id_vars=["A"], value_vars=["B"], var_name="myVarname", value_name="myValname"
).sort_values(["myVarname", "myValname"]),
)
self.assert_eq(
psdf.melt(value_vars=("A", "B"))
.sort_values(["variable", "value"])
.reset_index(drop=True),
pdf.melt(value_vars=("A", "B")).sort_values(["variable", "value"]),
)
self.assertRaises(KeyError, lambda: psdf.melt(id_vars="Z"))
self.assertRaises(KeyError, lambda: psdf.melt(value_vars="Z"))
# multi-index columns
TEN = 10.0
TWELVE = 20.0
columns = pd.MultiIndex.from_tuples([(TEN, "A"), (TEN, "B"), (TWELVE, "C")])
pdf.columns = columns
psdf.columns = columns
self.assert_eq(
psdf.melt().sort_values(["variable_0", "variable_1", "value"]).reset_index(drop=True),
pdf.melt().sort_values(["variable_0", "variable_1", "value"]),
)
self.assert_eq(
psdf.melt(id_vars=[(TEN, "A")])
.sort_values(["variable_0", "variable_1", "value"])
.reset_index(drop=True),
pdf.melt(id_vars=[(TEN, "A")])
.sort_values(["variable_0", "variable_1", "value"])
.rename(columns=name_like_string),
)
self.assert_eq(
psdf.melt(id_vars=[(TEN, "A")], value_vars=[(TWELVE, "C")])
.sort_values(["variable_0", "variable_1", "value"])
.reset_index(drop=True),
pdf.melt(id_vars=[(TEN, "A")], value_vars=[(TWELVE, "C")])
.sort_values(["variable_0", "variable_1", "value"])
.rename(columns=name_like_string),
)
self.assert_eq(
psdf.melt(
id_vars=[(TEN, "A")],
value_vars=[(TEN, "B")],
var_name=["myV1", "myV2"],
value_name="myValname",
)
.sort_values(["myV1", "myV2", "myValname"])
.reset_index(drop=True),
pdf.melt(
id_vars=[(TEN, "A")],
value_vars=[(TEN, "B")],
var_name=["myV1", "myV2"],
value_name="myValname",
)
.sort_values(["myV1", "myV2", "myValname"])
.rename(columns=name_like_string),
)
columns.names = ["v0", "v1"]
pdf.columns = columns
psdf.columns = columns
self.assert_eq(
psdf.melt().sort_values(["v0", "v1", "value"]).reset_index(drop=True),
pdf.melt().sort_values(["v0", "v1", "value"]),
)
self.assertRaises(ValueError, lambda: psdf.melt(id_vars=(TEN, "A")))
self.assertRaises(ValueError, lambda: psdf.melt(value_vars=(TEN, "A")))
self.assertRaises(KeyError, lambda: psdf.melt(id_vars=[TEN]))
self.assertRaises(KeyError, lambda: psdf.melt(id_vars=[(TWELVE, "A")]))
self.assertRaises(KeyError, lambda: psdf.melt(value_vars=[TWELVE]))
self.assertRaises(KeyError, lambda: psdf.melt(value_vars=[(TWELVE, "A")]))
# non-string names
pdf.columns = [10.0, 20.0, 30.0]
psdf.columns = [10.0, 20.0, 30.0]
self.assert_eq(
psdf.melt().sort_values(["variable", "value"]).reset_index(drop=True),
pdf.melt().sort_values(["variable", "value"]),
)
self.assert_eq(
psdf.melt(id_vars=10.0).sort_values(["variable", "value"]).reset_index(drop=True),
pdf.melt(id_vars=10.0).sort_values(["variable", "value"]),
)
self.assert_eq(
psdf.melt(id_vars=[10.0, 20.0])
.sort_values(["variable", "value"])
.reset_index(drop=True),
pdf.melt(id_vars=[10.0, 20.0]).sort_values(["variable", "value"]),
)
self.assert_eq(
psdf.melt(id_vars=(10.0, 20.0))
.sort_values(["variable", "value"])
.reset_index(drop=True),
pdf.melt(id_vars=(10.0, 20.0)).sort_values(["variable", "value"]),
)
self.assert_eq(
psdf.melt(id_vars=[10.0], value_vars=[30.0])
.sort_values(["variable", "value"])
.reset_index(drop=True),
pdf.melt(id_vars=[10.0], value_vars=[30.0]).sort_values(["variable", "value"]),
)
self.assert_eq(
psdf.melt(value_vars=(10.0, 20.0))
.sort_values(["variable", "value"])
.reset_index(drop=True),
pdf.melt(value_vars=(10.0, 20.0)).sort_values(["variable", "value"]),
)
def test_all(self):
pdf = pd.DataFrame(
{
"col1": [False, False, False],
"col2": [True, False, False],
"col3": [0, 0, 1],
"col4": [0, 1, 2],
"col5": [False, False, None],
"col6": [True, False, None],
},
index=np.random.rand(3),
)
psdf = ps.from_pandas(pdf)
self.assert_eq(psdf.all(), pdf.all())
columns = pd.MultiIndex.from_tuples(
[
("a", "col1"),
("a", "col2"),
("a", "col3"),
("b", "col4"),
("b", "col5"),
("c", "col6"),
]
)
pdf.columns = columns
psdf.columns = columns
self.assert_eq(psdf.all(), pdf.all())
columns.names = ["X", "Y"]
pdf.columns = columns
psdf.columns = columns
self.assert_eq(psdf.all(), pdf.all())
with self.assertRaisesRegex(
NotImplementedError, 'axis should be either 0 or "index" currently.'
):
psdf.all(axis=1)
def test_any(self):
pdf = pd.DataFrame(
{
"col1": [False, False, False],
"col2": [True, False, False],
"col3": [0, 0, 1],
"col4": [0, 1, 2],
"col5": [False, False, None],
"col6": [True, False, None],
},
index=np.random.rand(3),
)
psdf = ps.from_pandas(pdf)
self.assert_eq(psdf.any(), pdf.any())
columns = pd.MultiIndex.from_tuples(
[
("a", "col1"),
("a", "col2"),
("a", "col3"),
("b", "col4"),
("b", "col5"),
("c", "col6"),
]
)
pdf.columns = columns
psdf.columns = columns
self.assert_eq(psdf.any(), pdf.any())
columns.names = ["X", "Y"]
pdf.columns = columns
psdf.columns = columns
self.assert_eq(psdf.any(), pdf.any())
with self.assertRaisesRegex(
NotImplementedError, 'axis should be either 0 or "index" currently.'
):
psdf.any(axis=1)
def test_rank(self):
pdf = pd.DataFrame(
data={"col1": [1, 2, 3, 1], "col2": [3, 4, 3, 1]},
columns=["col1", "col2"],
index=np.random.rand(4),
)
psdf = ps.from_pandas(pdf)
self.assert_eq(pdf.rank().sort_index(), psdf.rank().sort_index())
self.assert_eq(pdf.rank().sum(), psdf.rank().sum())
self.assert_eq(
pdf.rank(ascending=False).sort_index(), psdf.rank(ascending=False).sort_index()
)
self.assert_eq(pdf.rank(method="min").sort_index(), psdf.rank(method="min").sort_index())
self.assert_eq(pdf.rank(method="max").sort_index(), psdf.rank(method="max").sort_index())
self.assert_eq(
pdf.rank(method="first").sort_index(), psdf.rank(method="first").sort_index()
)
self.assert_eq(
pdf.rank(method="dense").sort_index(), psdf.rank(method="dense").sort_index()
)
msg = "method must be one of 'average', 'min', 'max', 'first', 'dense'"
with self.assertRaisesRegex(ValueError, msg):
psdf.rank(method="nothing")
# multi-index columns
columns = pd.MultiIndex.from_tuples([("x", "col1"), ("y", "col2")])
pdf.columns = columns
psdf.columns = columns
self.assert_eq(pdf.rank().sort_index(), psdf.rank().sort_index())
def test_round(self):
pdf = pd.DataFrame(
{
"A": [0.028208, 0.038683, 0.877076],
"B": [0.992815, 0.645646, 0.149370],
"C": [0.173891, 0.577595, 0.491027],
},
columns=["A", "B", "C"],
index=np.random.rand(3),
)
psdf = ps.from_pandas(pdf)
pser = pd.Series([1, 0, 2], index=["A", "B", "C"])
psser = ps.Series([1, 0, 2], index=["A", "B", "C"])
self.assert_eq(pdf.round(2), psdf.round(2))
self.assert_eq(pdf.round({"A": 1, "C": 2}), psdf.round({"A": 1, "C": 2}))
self.assert_eq(pdf.round({"A": 1, "D": 2}), psdf.round({"A": 1, "D": 2}))
self.assert_eq(pdf.round(pser), psdf.round(psser))
msg = "decimals must be an integer, a dict-like or a Series"
with self.assertRaisesRegex(TypeError, msg):
psdf.round(1.5)
# multi-index columns
columns = pd.MultiIndex.from_tuples([("X", "A"), ("X", "B"), ("Y", "C")])
pdf.columns = columns
psdf.columns = columns
pser = pd.Series([1, 0, 2], index=columns)
psser = ps.Series([1, 0, 2], index=columns)
self.assert_eq(pdf.round(2), psdf.round(2))
self.assert_eq(
pdf.round({("X", "A"): 1, ("Y", "C"): 2}), psdf.round({("X", "A"): 1, ("Y", "C"): 2})
)
self.assert_eq(pdf.round({("X", "A"): 1, "Y": 2}), psdf.round({("X", "A"): 1, "Y": 2}))
self.assert_eq(pdf.round(pser), psdf.round(psser))
# non-string names
pdf = pd.DataFrame(
{
10: [0.028208, 0.038683, 0.877076],
20: [0.992815, 0.645646, 0.149370],
30: [0.173891, 0.577595, 0.491027],
},
index=np.random.rand(3),
)
psdf = ps.from_pandas(pdf)
self.assert_eq(pdf.round({10: 1, 30: 2}), psdf.round({10: 1, 30: 2}))
def test_shift(self):
pdf = pd.DataFrame(
{
"Col1": [10, 20, 15, 30, 45],
"Col2": [13, 23, 18, 33, 48],
"Col3": [17, 27, 22, 37, 52],
},
index=np.random.rand(5),
)
psdf = ps.from_pandas(pdf)
self.assert_eq(pdf.shift(3), psdf.shift(3))
self.assert_eq(pdf.shift().shift(-1), psdf.shift().shift(-1))
self.assert_eq(pdf.shift().sum().astype(int), psdf.shift().sum())
# Need the expected result since pandas 0.23 does not support `fill_value` argument.
pdf1 = pd.DataFrame(
{"Col1": [0, 0, 0, 10, 20], "Col2": [0, 0, 0, 13, 23], "Col3": [0, 0, 0, 17, 27]},
index=pdf.index,
)
self.assert_eq(pdf1, psdf.shift(periods=3, fill_value=0))
msg = "should be an int"
with self.assertRaisesRegex(TypeError, msg):
psdf.shift(1.5)
# multi-index columns
columns = pd.MultiIndex.from_tuples([("x", "Col1"), ("x", "Col2"), ("y", "Col3")])
pdf.columns = columns
psdf.columns = columns
self.assert_eq(pdf.shift(3), psdf.shift(3))
self.assert_eq(pdf.shift().shift(-1), psdf.shift().shift(-1))
def test_diff(self):
pdf = pd.DataFrame(
{"a": [1, 2, 3, 4, 5, 6], "b": [1, 1, 2, 3, 5, 8], "c": [1, 4, 9, 16, 25, 36]},
index=np.random.rand(6),
)
psdf = ps.from_pandas(pdf)
self.assert_eq(pdf.diff(), psdf.diff())
self.assert_eq(pdf.diff().diff(-1), psdf.diff().diff(-1))
self.assert_eq(pdf.diff().sum().astype(int), psdf.diff().sum())
msg = "should be an int"
with self.assertRaisesRegex(TypeError, msg):
psdf.diff(1.5)
msg = 'axis should be either 0 or "index" currently.'
with self.assertRaisesRegex(NotImplementedError, msg):
psdf.diff(axis=1)
# multi-index columns
columns = pd.MultiIndex.from_tuples([("x", "Col1"), ("x", "Col2"), ("y", "Col3")])
pdf.columns = columns
psdf.columns = columns
self.assert_eq(pdf.diff(), psdf.diff())
def test_duplicated(self):
pdf = pd.DataFrame(
{"a": [1, 1, 2, 3], "b": [1, 1, 1, 4], "c": [1, 1, 1, 5]}, index=np.random.rand(4)
)
psdf = ps.from_pandas(pdf)
self.assert_eq(pdf.duplicated().sort_index(), psdf.duplicated().sort_index())
self.assert_eq(
pdf.duplicated(keep="last").sort_index(),
psdf.duplicated(keep="last").sort_index(),
)
self.assert_eq(
pdf.duplicated(keep=False).sort_index(),
psdf.duplicated(keep=False).sort_index(),
)
self.assert_eq(
pdf.duplicated(subset="b").sort_index(),
psdf.duplicated(subset="b").sort_index(),
)
self.assert_eq(
pdf.duplicated(subset=["b"]).sort_index(),
psdf.duplicated(subset=["b"]).sort_index(),
)
with self.assertRaisesRegex(ValueError, "'keep' only supports 'first', 'last' and False"):
psdf.duplicated(keep="false")
with self.assertRaisesRegex(KeyError, "'d'"):
psdf.duplicated(subset=["d"])
pdf.index.name = "x"
psdf.index.name = "x"
self.assert_eq(pdf.duplicated().sort_index(), psdf.duplicated().sort_index())
# multi-index
self.assert_eq(
pdf.set_index("a", append=True).duplicated().sort_index(),
psdf.set_index("a", append=True).duplicated().sort_index(),
)
self.assert_eq(
pdf.set_index("a", append=True).duplicated(keep=False).sort_index(),
psdf.set_index("a", append=True).duplicated(keep=False).sort_index(),
)
self.assert_eq(
pdf.set_index("a", append=True).duplicated(subset=["b"]).sort_index(),
psdf.set_index("a", append=True).duplicated(subset=["b"]).sort_index(),
)
# mutli-index columns
columns = pd.MultiIndex.from_tuples([("x", "a"), ("x", "b"), ("y", "c")])
pdf.columns = columns
psdf.columns = columns
self.assert_eq(pdf.duplicated().sort_index(), psdf.duplicated().sort_index())
self.assert_eq(
pdf.duplicated(subset=("x", "b")).sort_index(),
psdf.duplicated(subset=("x", "b")).sort_index(),
)
self.assert_eq(
pdf.duplicated(subset=[("x", "b")]).sort_index(),
psdf.duplicated(subset=[("x", "b")]).sort_index(),
)
# non-string names
pdf = pd.DataFrame(
{10: [1, 1, 2, 3], 20: [1, 1, 1, 4], 30: [1, 1, 1, 5]}, index=np.random.rand(4)
)
psdf = ps.from_pandas(pdf)
self.assert_eq(pdf.duplicated().sort_index(), psdf.duplicated().sort_index())
self.assert_eq(
pdf.duplicated(subset=10).sort_index(),
psdf.duplicated(subset=10).sort_index(),
)
def test_ffill(self):
idx = np.random.rand(6)
pdf = pd.DataFrame(
{
"x": [np.nan, 2, 3, 4, np.nan, 6],
"y": [1, 2, np.nan, 4, np.nan, np.nan],
"z": [1, 2, 3, 4, np.nan, np.nan],
},
index=idx,
)
psdf = ps.from_pandas(pdf)
self.assert_eq(psdf.ffill(), pdf.ffill())
self.assert_eq(psdf.ffill(limit=1), pdf.ffill(limit=1))
pser = pdf.y
psser = psdf.y
psdf.ffill(inplace=True)
pdf.ffill(inplace=True)
self.assert_eq(psdf, pdf)
self.assert_eq(psser, pser)
self.assert_eq(psser[idx[2]], pser[idx[2]])
def test_bfill(self):
idx = np.random.rand(6)
pdf = pd.DataFrame(
{
"x": [np.nan, 2, 3, 4, np.nan, 6],
"y": [1, 2, np.nan, 4, np.nan, np.nan],
"z": [1, 2, 3, 4, np.nan, np.nan],
},
index=idx,
)
psdf = ps.from_pandas(pdf)
self.assert_eq(psdf.bfill(), pdf.bfill())
self.assert_eq(psdf.bfill(limit=1), pdf.bfill(limit=1))
pser = pdf.x
psser = psdf.x
psdf.bfill(inplace=True)
pdf.bfill(inplace=True)
self.assert_eq(psdf, pdf)
self.assert_eq(psser, pser)
self.assert_eq(psser[idx[0]], pser[idx[0]])
def test_filter(self):
pdf = pd.DataFrame(
{
"aa": ["aa", "bd", "bc", "ab", "ce"],
"ba": [1, 2, 3, 4, 5],
"cb": [1.0, 2.0, 3.0, 4.0, 5.0],
"db": [1.0, np.nan, 3.0, np.nan, 5.0],
}
)
pdf = pdf.set_index("aa")
psdf = ps.from_pandas(pdf)
self.assert_eq(
psdf.filter(items=["ab", "aa"], axis=0).sort_index(),
pdf.filter(items=["ab", "aa"], axis=0).sort_index(),
)
with option_context("compute.isin_limit", 0):
self.assert_eq(
psdf.filter(items=["ab", "aa"], axis=0).sort_index(),
pdf.filter(items=["ab", "aa"], axis=0).sort_index(),
)
self.assert_eq(
psdf.filter(items=["ba", "db"], axis=1).sort_index(),
pdf.filter(items=["ba", "db"], axis=1).sort_index(),
)
self.assert_eq(psdf.filter(like="b", axis="index"), pdf.filter(like="b", axis="index"))
self.assert_eq(psdf.filter(like="c", axis="columns"), pdf.filter(like="c", axis="columns"))
self.assert_eq(
psdf.filter(regex="b.*", axis="index"), pdf.filter(regex="b.*", axis="index")
)
self.assert_eq(
psdf.filter(regex="b.*", axis="columns"), pdf.filter(regex="b.*", axis="columns")
)
pdf = pdf.set_index("ba", append=True)
psdf = ps.from_pandas(pdf)
self.assert_eq(
psdf.filter(items=[("aa", 1), ("bd", 2)], axis=0).sort_index(),
pdf.filter(items=[("aa", 1), ("bd", 2)], axis=0).sort_index(),
)
with self.assertRaisesRegex(TypeError, "Unsupported type list"):
psdf.filter(items=[["aa", 1], ("bd", 2)], axis=0)
with self.assertRaisesRegex(ValueError, "The item should not be empty."):
psdf.filter(items=[(), ("bd", 2)], axis=0)
self.assert_eq(psdf.filter(like="b", axis=0), pdf.filter(like="b", axis=0))
self.assert_eq(psdf.filter(regex="b.*", axis=0), pdf.filter(regex="b.*", axis=0))
with self.assertRaisesRegex(ValueError, "items should be a list-like object"):
psdf.filter(items="b")
with self.assertRaisesRegex(ValueError, "No axis named"):
psdf.filter(regex="b.*", axis=123)
with self.assertRaisesRegex(TypeError, "Must pass either `items`, `like`"):
psdf.filter()
with self.assertRaisesRegex(TypeError, "mutually exclusive"):
psdf.filter(regex="b.*", like="aaa")
# multi-index columns
pdf = pd.DataFrame(
{
("x", "aa"): ["aa", "ab", "bc", "bd", "ce"],
("x", "ba"): [1, 2, 3, 4, 5],
("y", "cb"): [1.0, 2.0, 3.0, 4.0, 5.0],
("z", "db"): [1.0, np.nan, 3.0, np.nan, 5.0],
}
)
pdf = pdf.set_index(("x", "aa"))
psdf = ps.from_pandas(pdf)
self.assert_eq(
psdf.filter(items=["ab", "aa"], axis=0).sort_index(),
pdf.filter(items=["ab", "aa"], axis=0).sort_index(),
)
self.assert_eq(
psdf.filter(items=[("x", "ba"), ("z", "db")], axis=1).sort_index(),
pdf.filter(items=[("x", "ba"), ("z", "db")], axis=1).sort_index(),
)
self.assert_eq(psdf.filter(like="b", axis="index"), pdf.filter(like="b", axis="index"))
self.assert_eq(psdf.filter(like="c", axis="columns"), pdf.filter(like="c", axis="columns"))
self.assert_eq(
psdf.filter(regex="b.*", axis="index"), pdf.filter(regex="b.*", axis="index")
)
self.assert_eq(
psdf.filter(regex="b.*", axis="columns"), pdf.filter(regex="b.*", axis="columns")
)
def test_pipe(self):
psdf = ps.DataFrame(
{"category": ["A", "A", "B"], "col1": [1, 2, 3], "col2": [4, 5, 6]},
columns=["category", "col1", "col2"],
)
self.assertRaisesRegex(
ValueError,
"arg is both the pipe target and a keyword argument",
lambda: psdf.pipe((lambda x: x, "arg"), arg="1"),
)
def test_transform(self):
pdf = pd.DataFrame(
{
"a": [1, 2, 3, 4, 5, 6] * 100,
"b": [1.0, 1.0, 2.0, 3.0, 5.0, 8.0] * 100,
"c": [1, 4, 9, 16, 25, 36] * 100,
},
columns=["a", "b", "c"],
index=np.random.rand(600),
)
psdf = ps.DataFrame(pdf)
self.assert_eq(
psdf.transform(lambda x: x + 1).sort_index(),
pdf.transform(lambda x: x + 1).sort_index(),
)
self.assert_eq(
psdf.transform(lambda x, y: x + y, y=2).sort_index(),
pdf.transform(lambda x, y: x + y, y=2).sort_index(),
)
with option_context("compute.shortcut_limit", 500):
self.assert_eq(
psdf.transform(lambda x: x + 1).sort_index(),
pdf.transform(lambda x: x + 1).sort_index(),
)
self.assert_eq(
psdf.transform(lambda x, y: x + y, y=1).sort_index(),
pdf.transform(lambda x, y: x + y, y=1).sort_index(),
)
with self.assertRaisesRegex(AssertionError, "the first argument should be a callable"):
psdf.transform(1)
# multi-index columns
columns = pd.MultiIndex.from_tuples([("x", "a"), ("x", "b"), ("y", "c")])
pdf.columns = columns
psdf.columns = columns
self.assert_eq(
psdf.transform(lambda x: x + 1).sort_index(),
pdf.transform(lambda x: x + 1).sort_index(),
)
with option_context("compute.shortcut_limit", 500):
self.assert_eq(
psdf.transform(lambda x: x + 1).sort_index(),
pdf.transform(lambda x: x + 1).sort_index(),
)
def test_apply(self):
pdf = pd.DataFrame(
{
"a": [1, 2, 3, 4, 5, 6] * 100,
"b": [1.0, 1.0, 2.0, 3.0, 5.0, 8.0] * 100,
"c": [1, 4, 9, 16, 25, 36] * 100,
},
columns=["a", "b", "c"],
index=np.random.rand(600),
)
psdf = ps.DataFrame(pdf)
self.assert_eq(
psdf.apply(lambda x: x + 1).sort_index(), pdf.apply(lambda x: x + 1).sort_index()
)
self.assert_eq(
psdf.apply(lambda x, b: x + b, args=(1,)).sort_index(),
pdf.apply(lambda x, b: x + b, args=(1,)).sort_index(),
)
self.assert_eq(
psdf.apply(lambda x, b: x + b, b=1).sort_index(),
pdf.apply(lambda x, b: x + b, b=1).sort_index(),
)
with option_context("compute.shortcut_limit", 500):
self.assert_eq(
psdf.apply(lambda x: x + 1).sort_index(), pdf.apply(lambda x: x + 1).sort_index()
)
self.assert_eq(
psdf.apply(lambda x, b: x + b, args=(1,)).sort_index(),
pdf.apply(lambda x, b: x + b, args=(1,)).sort_index(),
)
self.assert_eq(
psdf.apply(lambda x, b: x + b, b=1).sort_index(),
pdf.apply(lambda x, b: x + b, b=1).sort_index(),
)
# returning a Series
self.assert_eq(
psdf.apply(lambda x: len(x), axis=1).sort_index(),
pdf.apply(lambda x: len(x), axis=1).sort_index(),
)
self.assert_eq(
psdf.apply(lambda x, c: len(x) + c, axis=1, c=100).sort_index(),
pdf.apply(lambda x, c: len(x) + c, axis=1, c=100).sort_index(),
)
with option_context("compute.shortcut_limit", 500):
self.assert_eq(
psdf.apply(lambda x: len(x), axis=1).sort_index(),
pdf.apply(lambda x: len(x), axis=1).sort_index(),
)
self.assert_eq(
psdf.apply(lambda x, c: len(x) + c, axis=1, c=100).sort_index(),
pdf.apply(lambda x, c: len(x) + c, axis=1, c=100).sort_index(),
)
with self.assertRaisesRegex(AssertionError, "the first argument should be a callable"):
psdf.apply(1)
with self.assertRaisesRegex(TypeError, "The given function.*1 or 'column'; however"):
def f1(_) -> ps.DataFrame[int]:
pass
psdf.apply(f1, axis=0)
with self.assertRaisesRegex(TypeError, "The given function.*0 or 'index'; however"):
def f2(_) -> ps.Series[int]:
pass
psdf.apply(f2, axis=1)
# multi-index columns
columns = pd.MultiIndex.from_tuples([("x", "a"), ("x", "b"), ("y", "c")])
pdf.columns = columns
psdf.columns = columns
self.assert_eq(
psdf.apply(lambda x: x + 1).sort_index(), pdf.apply(lambda x: x + 1).sort_index()
)
with option_context("compute.shortcut_limit", 500):
self.assert_eq(
psdf.apply(lambda x: x + 1).sort_index(), pdf.apply(lambda x: x + 1).sort_index()
)
# returning a Series
self.assert_eq(
psdf.apply(lambda x: len(x), axis=1).sort_index(),
pdf.apply(lambda x: len(x), axis=1).sort_index(),
)
with option_context("compute.shortcut_limit", 500):
self.assert_eq(
psdf.apply(lambda x: len(x), axis=1).sort_index(),
pdf.apply(lambda x: len(x), axis=1).sort_index(),
)
def test_apply_with_type(self):
pdf = self.pdf
psdf = ps.from_pandas(pdf)
def identify1(x) -> ps.DataFrame[int, int]:
return x
# Type hints set the default column names, and we use default index for
# pandas API on Spark. Here we ignore both diff.
actual = psdf.apply(identify1, axis=1)
expected = pdf.apply(identify1, axis=1)
self.assert_eq(sorted(actual["c0"].to_numpy()), sorted(expected["a"].to_numpy()))
self.assert_eq(sorted(actual["c1"].to_numpy()), sorted(expected["b"].to_numpy()))
def identify2(x) -> ps.DataFrame[slice("a", int), slice("b", int)]: # noqa: F405
return x
actual = psdf.apply(identify2, axis=1)
expected = pdf.apply(identify2, axis=1)
self.assert_eq(sorted(actual["a"].to_numpy()), sorted(expected["a"].to_numpy()))
self.assert_eq(sorted(actual["b"].to_numpy()), sorted(expected["b"].to_numpy()))
def test_apply_batch(self):
pdf = pd.DataFrame(
{
"a": [1, 2, 3, 4, 5, 6] * 100,
"b": [1.0, 1.0, 2.0, 3.0, 5.0, 8.0] * 100,
"c": [1, 4, 9, 16, 25, 36] * 100,
},
columns=["a", "b", "c"],
index=np.random.rand(600),
)
psdf = ps.DataFrame(pdf)
self.assert_eq(
psdf.pandas_on_spark.apply_batch(lambda pdf, a: pdf + a, args=(1,)).sort_index(),
(pdf + 1).sort_index(),
)
with option_context("compute.shortcut_limit", 500):
self.assert_eq(
psdf.pandas_on_spark.apply_batch(lambda pdf: pdf + 1).sort_index(),
(pdf + 1).sort_index(),
)
self.assert_eq(
psdf.pandas_on_spark.apply_batch(lambda pdf, b: pdf + b, b=1).sort_index(),
(pdf + 1).sort_index(),
)
with self.assertRaisesRegex(AssertionError, "the first argument should be a callable"):
psdf.pandas_on_spark.apply_batch(1)
with self.assertRaisesRegex(TypeError, "The given function.*frame as its type hints"):
def f2(_) -> ps.Series[int]:
pass
psdf.pandas_on_spark.apply_batch(f2)
with self.assertRaisesRegex(ValueError, "The given function should return a frame"):
psdf.pandas_on_spark.apply_batch(lambda pdf: 1)
# multi-index columns
columns = pd.MultiIndex.from_tuples([("x", "a"), ("x", "b"), ("y", "c")])
pdf.columns = columns
psdf.columns = columns
self.assert_eq(
psdf.pandas_on_spark.apply_batch(lambda x: x + 1).sort_index(), (pdf + 1).sort_index()
)
with option_context("compute.shortcut_limit", 500):
self.assert_eq(
psdf.pandas_on_spark.apply_batch(lambda x: x + 1).sort_index(),
(pdf + 1).sort_index(),
)
def test_apply_batch_with_type(self):
pdf = self.pdf
psdf = ps.from_pandas(pdf)
def identify1(x) -> ps.DataFrame[int, int]:
return x
# Type hints set the default column names, and we use default index for
# pandas API on Spark. Here we ignore both diff.
actual = psdf.pandas_on_spark.apply_batch(identify1)
expected = pdf
self.assert_eq(sorted(actual["c0"].to_numpy()), sorted(expected["a"].to_numpy()))
self.assert_eq(sorted(actual["c1"].to_numpy()), sorted(expected["b"].to_numpy()))
def identify2(x) -> ps.DataFrame[slice("a", int), slice("b", int)]: # noqa: F405
return x
actual = psdf.pandas_on_spark.apply_batch(identify2)
expected = pdf
self.assert_eq(sorted(actual["a"].to_numpy()), sorted(expected["a"].to_numpy()))
self.assert_eq(sorted(actual["b"].to_numpy()), sorted(expected["b"].to_numpy()))
pdf = pd.DataFrame(
{"a": [1, 2, 3, 4, 5, 6, 7, 8, 9], "b": [[e] for e in [4, 5, 6, 3, 2, 1, 0, 0, 0]]},
index=np.random.rand(9),
)
psdf = ps.from_pandas(pdf)
def identify3(x) -> ps.DataFrame[float, [int, List[int]]]:
return x
actual = psdf.pandas_on_spark.apply_batch(identify3)
actual.columns = ["a", "b"]
self.assert_eq(actual, pdf)
# For NumPy typing, NumPy version should be 1.21+ and Python version should be 3.8+
if sys.version_info >= (3, 8) and LooseVersion(np.__version__) >= LooseVersion("1.21"):
import numpy.typing as ntp
psdf = ps.from_pandas(pdf)
def identify4(
x,
) -> ps.DataFrame[float, [int, ntp.NDArray[int]]]: # type: ignore[name-defined]
return x
actual = psdf.pandas_on_spark.apply_batch(identify4)
actual.columns = ["a", "b"]
self.assert_eq(actual, pdf)
arrays = [[1, 2, 3, 4, 5, 6, 7, 8, 9], ["a", "b", "c", "d", "e", "f", "g", "h", "i"]]
idx = pd.MultiIndex.from_arrays(arrays, names=("number", "color"))
pdf = pd.DataFrame(
{"a": [1, 2, 3, 4, 5, 6, 7, 8, 9], "b": [[e] for e in [4, 5, 6, 3, 2, 1, 0, 0, 0]]},
index=idx,
)
psdf = ps.from_pandas(pdf)
def identify4(x) -> ps.DataFrame[[int, str], [int, List[int]]]:
return x
actual = psdf.pandas_on_spark.apply_batch(identify4)
actual.index.names = ["number", "color"]
actual.columns = ["a", "b"]
self.assert_eq(actual, pdf)
def identify5(
x,
) -> ps.DataFrame[
[("number", int), ("color", str)], [("a", int), ("b", List[int])] # noqa: F405
]:
return x
actual = psdf.pandas_on_spark.apply_batch(identify5)
self.assert_eq(actual, pdf)
def test_transform_batch(self):
pdf = pd.DataFrame(
{
"a": [1, 2, 3, 4, 5, 6] * 100,
"b": [1.0, 1.0, 2.0, 3.0, 5.0, 8.0] * 100,
"c": [1, 4, 9, 16, 25, 36] * 100,
},
columns=["a", "b", "c"],
index=np.random.rand(600),
)
psdf = ps.DataFrame(pdf)
self.assert_eq(
psdf.pandas_on_spark.transform_batch(lambda pdf: pdf.c + 1).sort_index(),
(pdf.c + 1).sort_index(),
)
self.assert_eq(
psdf.pandas_on_spark.transform_batch(lambda pdf, a: pdf + a, 1).sort_index(),
(pdf + 1).sort_index(),
)
self.assert_eq(
psdf.pandas_on_spark.transform_batch(lambda pdf, a: pdf.c + a, a=1).sort_index(),
(pdf.c + 1).sort_index(),
)
with option_context("compute.shortcut_limit", 500):
self.assert_eq(
psdf.pandas_on_spark.transform_batch(lambda pdf: pdf + 1).sort_index(),
(pdf + 1).sort_index(),
)
self.assert_eq(
psdf.pandas_on_spark.transform_batch(lambda pdf: pdf.b + 1).sort_index(),
(pdf.b + 1).sort_index(),
)
self.assert_eq(
psdf.pandas_on_spark.transform_batch(lambda pdf, a: pdf + a, 1).sort_index(),
(pdf + 1).sort_index(),
)
self.assert_eq(
psdf.pandas_on_spark.transform_batch(lambda pdf, a: pdf.c + a, a=1).sort_index(),
(pdf.c + 1).sort_index(),
)
with self.assertRaisesRegex(AssertionError, "the first argument should be a callable"):
psdf.pandas_on_spark.transform_batch(1)
with self.assertRaisesRegex(ValueError, "The given function should return a frame"):
psdf.pandas_on_spark.transform_batch(lambda pdf: 1)
with self.assertRaisesRegex(
ValueError, "transform_batch cannot produce aggregated results"
):
psdf.pandas_on_spark.transform_batch(lambda pdf: pd.Series(1))
# multi-index columns
columns = pd.MultiIndex.from_tuples([("x", "a"), ("x", "b"), ("y", "c")])
pdf.columns = columns
psdf.columns = columns
self.assert_eq(
psdf.pandas_on_spark.transform_batch(lambda x: x + 1).sort_index(),
(pdf + 1).sort_index(),
)
with option_context("compute.shortcut_limit", 500):
self.assert_eq(
psdf.pandas_on_spark.transform_batch(lambda x: x + 1).sort_index(),
(pdf + 1).sort_index(),
)
def test_transform_batch_with_type(self):
pdf = self.pdf
psdf = ps.from_pandas(pdf)
def identify1(x) -> ps.DataFrame[int, int]:
return x
# Type hints set the default column names, and we use default index for
# pandas API on Spark. Here we ignore both diff.
actual = psdf.pandas_on_spark.transform_batch(identify1)
expected = pdf
self.assert_eq(sorted(actual["c0"].to_numpy()), sorted(expected["a"].to_numpy()))
self.assert_eq(sorted(actual["c1"].to_numpy()), sorted(expected["b"].to_numpy()))
def identify2(x) -> ps.DataFrame[slice("a", int), slice("b", int)]: # noqa: F405
return x
actual = psdf.pandas_on_spark.transform_batch(identify2)
expected = pdf
self.assert_eq(sorted(actual["a"].to_numpy()), sorted(expected["a"].to_numpy()))
self.assert_eq(sorted(actual["b"].to_numpy()), sorted(expected["b"].to_numpy()))
def test_transform_batch_same_anchor(self):
psdf = ps.range(10)
psdf["d"] = psdf.pandas_on_spark.transform_batch(lambda pdf: pdf.id + 1)
self.assert_eq(
psdf,
pd.DataFrame({"id": list(range(10)), "d": list(range(1, 11))}, columns=["id", "d"]),
)
psdf = ps.range(10)
def plus_one(pdf) -> ps.Series[np.int64]:
return pdf.id + 1
psdf["d"] = psdf.pandas_on_spark.transform_batch(plus_one)
self.assert_eq(
psdf,
pd.DataFrame({"id": list(range(10)), "d": list(range(1, 11))}, columns=["id", "d"]),
)
psdf = ps.range(10)
def plus_one(ser) -> ps.Series[np.int64]:
return ser + 1
psdf["d"] = psdf.id.pandas_on_spark.transform_batch(plus_one)
self.assert_eq(
psdf,
pd.DataFrame({"id": list(range(10)), "d": list(range(1, 11))}, columns=["id", "d"]),
)
def test_empty_timestamp(self):
pdf = pd.DataFrame(
{
"t": [
datetime(2019, 1, 1, 0, 0, 0),
datetime(2019, 1, 2, 0, 0, 0),
datetime(2019, 1, 3, 0, 0, 0),
]
},
index=np.random.rand(3),
)
psdf = ps.from_pandas(pdf)
self.assert_eq(psdf[psdf["t"] != psdf["t"]], pdf[pdf["t"] != pdf["t"]])
self.assert_eq(psdf[psdf["t"] != psdf["t"]].dtypes, pdf[pdf["t"] != pdf["t"]].dtypes)
def test_to_spark(self):
psdf = ps.from_pandas(self.pdf)
with self.assertRaisesRegex(ValueError, "'index_col' cannot be overlapped"):
psdf.to_spark(index_col="a")
with self.assertRaisesRegex(ValueError, "length of index columns.*1.*3"):
psdf.to_spark(index_col=["x", "y", "z"])
def test_keys(self):
pdf = pd.DataFrame(
[[1, 2], [4, 5], [7, 8]],
index=["cobra", "viper", "sidewinder"],
columns=["max_speed", "shield"],
)
psdf = ps.from_pandas(pdf)
self.assert_eq(psdf.keys(), pdf.keys())
def test_quantile(self):
pdf, psdf = self.df_pair
self.assert_eq(psdf.quantile(0.5), pdf.quantile(0.5))
self.assert_eq(psdf.quantile([0.25, 0.5, 0.75]), pdf.quantile([0.25, 0.5, 0.75]))
self.assert_eq(psdf.loc[[]].quantile(0.5), pdf.loc[[]].quantile(0.5))
self.assert_eq(
psdf.loc[[]].quantile([0.25, 0.5, 0.75]), pdf.loc[[]].quantile([0.25, 0.5, 0.75])
)
with self.assertRaisesRegex(
NotImplementedError, 'axis should be either 0 or "index" currently.'
):
psdf.quantile(0.5, axis=1)
with self.assertRaisesRegex(TypeError, "accuracy must be an integer; however"):
psdf.quantile(accuracy="a")
with self.assertRaisesRegex(TypeError, "q must be a float or an array of floats;"):
psdf.quantile(q="a")
with self.assertRaisesRegex(TypeError, "q must be a float or an array of floats;"):
psdf.quantile(q=["a"])
with self.assertRaisesRegex(
ValueError, r"percentiles should all be in the interval \[0, 1\]"
):
psdf.quantile(q=[1.1])
self.assert_eq(
psdf.quantile(0.5, numeric_only=False), pdf.quantile(0.5, numeric_only=False)
)
self.assert_eq(
psdf.quantile([0.25, 0.5, 0.75], numeric_only=False),
pdf.quantile([0.25, 0.5, 0.75], numeric_only=False),
)
# multi-index column
columns = pd.MultiIndex.from_tuples([("x", "a"), ("y", "b")])
pdf.columns = columns
psdf.columns = columns
self.assert_eq(psdf.quantile(0.5), pdf.quantile(0.5))
self.assert_eq(psdf.quantile([0.25, 0.5, 0.75]), pdf.quantile([0.25, 0.5, 0.75]))
pdf = pd.DataFrame({"x": ["a", "b", "c"]})
psdf = ps.from_pandas(pdf)
self.assert_eq(psdf.quantile(0.5), pdf.quantile(0.5))
self.assert_eq(psdf.quantile([0.25, 0.5, 0.75]), pdf.quantile([0.25, 0.5, 0.75]))
with self.assertRaisesRegex(TypeError, "Could not convert object \\(string\\) to numeric"):
psdf.quantile(0.5, numeric_only=False)
with self.assertRaisesRegex(TypeError, "Could not convert object \\(string\\) to numeric"):
psdf.quantile([0.25, 0.5, 0.75], numeric_only=False)
def test_pct_change(self):
pdf = pd.DataFrame(
{"a": [1, 2, 3, 2], "b": [4.0, 2.0, 3.0, 1.0], "c": [300, 200, 400, 200]},
index=np.random.rand(4),
)
pdf.columns = pd.MultiIndex.from_tuples([("a", "x"), ("b", "y"), ("c", "z")])
psdf = ps.from_pandas(pdf)
self.assert_eq(psdf.pct_change(2), pdf.pct_change(2), check_exact=False)
self.assert_eq(psdf.pct_change().sum(), pdf.pct_change().sum(), check_exact=False)
def test_where(self):
pdf, psdf = self.df_pair
# pandas requires `axis` argument when the `other` is Series.
# `axis` is not fully supported yet in pandas-on-Spark.
self.assert_eq(
psdf.where(psdf > 2, psdf.a + 10, axis=0), pdf.where(pdf > 2, pdf.a + 10, axis=0)
)
with self.assertRaisesRegex(TypeError, "type of cond must be a DataFrame or Series"):
psdf.where(1)
def test_mask(self):
psdf = ps.from_pandas(self.pdf)
with self.assertRaisesRegex(TypeError, "type of cond must be a DataFrame or Series"):
psdf.mask(1)
def test_query(self):
pdf = pd.DataFrame({"A": range(1, 6), "B": range(10, 0, -2), "C": range(10, 5, -1)})
psdf = ps.from_pandas(pdf)
exprs = ("A > B", "A < C", "C == B")
for expr in exprs:
self.assert_eq(psdf.query(expr), pdf.query(expr))
# test `inplace=True`
for expr in exprs:
dummy_psdf = psdf.copy()
dummy_pdf = pdf.copy()
pser = dummy_pdf.A
psser = dummy_psdf.A
dummy_pdf.query(expr, inplace=True)
dummy_psdf.query(expr, inplace=True)
self.assert_eq(dummy_psdf, dummy_pdf)
self.assert_eq(psser, pser)
# invalid values for `expr`
invalid_exprs = (1, 1.0, (exprs[0],), [exprs[0]])
for expr in invalid_exprs:
with self.assertRaisesRegex(
TypeError,
"expr must be a string to be evaluated, {} given".format(type(expr).__name__),
):
psdf.query(expr)
# invalid values for `inplace`
invalid_inplaces = (1, 0, "True", "False")
for inplace in invalid_inplaces:
with self.assertRaisesRegex(
TypeError,
'For argument "inplace" expected type bool, received type {}.'.format(
type(inplace).__name__
),
):
psdf.query("a < b", inplace=inplace)
# doesn't support for MultiIndex columns
columns = pd.MultiIndex.from_tuples([("A", "Z"), ("B", "X"), ("C", "C")])
psdf.columns = columns
with self.assertRaisesRegex(TypeError, "Doesn't support for MultiIndex columns"):
psdf.query("('A', 'Z') > ('B', 'X')")
def test_take(self):
pdf = pd.DataFrame(
{"A": range(0, 50000), "B": range(100000, 0, -2), "C": range(100000, 50000, -1)}
)
psdf = ps.from_pandas(pdf)
# axis=0 (default)
self.assert_eq(psdf.take([1, 2]).sort_index(), pdf.take([1, 2]).sort_index())
self.assert_eq(psdf.take([-1, -2]).sort_index(), pdf.take([-1, -2]).sort_index())
self.assert_eq(
psdf.take(range(100, 110)).sort_index(), pdf.take(range(100, 110)).sort_index()
)
self.assert_eq(
psdf.take(range(-110, -100)).sort_index(), pdf.take(range(-110, -100)).sort_index()
)
self.assert_eq(
psdf.take([10, 100, 1000, 10000]).sort_index(),
pdf.take([10, 100, 1000, 10000]).sort_index(),
)
self.assert_eq(
psdf.take([-10, -100, -1000, -10000]).sort_index(),
pdf.take([-10, -100, -1000, -10000]).sort_index(),
)
# axis=1
self.assert_eq(
psdf.take([1, 2], axis=1).sort_index(), pdf.take([1, 2], axis=1).sort_index()
)
self.assert_eq(
psdf.take([-1, -2], axis=1).sort_index(), pdf.take([-1, -2], axis=1).sort_index()
)
self.assert_eq(
psdf.take(range(1, 3), axis=1).sort_index(),
pdf.take(range(1, 3), axis=1).sort_index(),
)
self.assert_eq(
psdf.take(range(-1, -3), axis=1).sort_index(),
pdf.take(range(-1, -3), axis=1).sort_index(),
)
self.assert_eq(
psdf.take([2, 1], axis=1).sort_index(),
pdf.take([2, 1], axis=1).sort_index(),
)
self.assert_eq(
psdf.take([-1, -2], axis=1).sort_index(),
pdf.take([-1, -2], axis=1).sort_index(),
)
# MultiIndex columns
columns = pd.MultiIndex.from_tuples([("A", "Z"), ("B", "X"), ("C", "C")])
psdf.columns = columns
pdf.columns = columns
# MultiIndex columns with axis=0 (default)
self.assert_eq(psdf.take([1, 2]).sort_index(), pdf.take([1, 2]).sort_index())
self.assert_eq(psdf.take([-1, -2]).sort_index(), pdf.take([-1, -2]).sort_index())
self.assert_eq(
psdf.take(range(100, 110)).sort_index(), pdf.take(range(100, 110)).sort_index()
)
self.assert_eq(
psdf.take(range(-110, -100)).sort_index(), pdf.take(range(-110, -100)).sort_index()
)
self.assert_eq(
psdf.take([10, 100, 1000, 10000]).sort_index(),
pdf.take([10, 100, 1000, 10000]).sort_index(),
)
self.assert_eq(
psdf.take([-10, -100, -1000, -10000]).sort_index(),
pdf.take([-10, -100, -1000, -10000]).sort_index(),
)
# axis=1
self.assert_eq(
psdf.take([1, 2], axis=1).sort_index(), pdf.take([1, 2], axis=1).sort_index()
)
self.assert_eq(
psdf.take([-1, -2], axis=1).sort_index(), pdf.take([-1, -2], axis=1).sort_index()
)
self.assert_eq(
psdf.take(range(1, 3), axis=1).sort_index(),
pdf.take(range(1, 3), axis=1).sort_index(),
)
self.assert_eq(
psdf.take(range(-1, -3), axis=1).sort_index(),
pdf.take(range(-1, -3), axis=1).sort_index(),
)
self.assert_eq(
psdf.take([2, 1], axis=1).sort_index(),
pdf.take([2, 1], axis=1).sort_index(),
)
self.assert_eq(
psdf.take([-1, -2], axis=1).sort_index(),
pdf.take([-1, -2], axis=1).sort_index(),
)
# Checking the type of indices.
self.assertRaises(TypeError, lambda: psdf.take(1))
self.assertRaises(TypeError, lambda: psdf.take("1"))
self.assertRaises(TypeError, lambda: psdf.take({1, 2}))
self.assertRaises(TypeError, lambda: psdf.take({1: None, 2: None}))
def test_axes(self):
pdf = self.pdf
psdf = ps.from_pandas(pdf)
self.assert_eq(pdf.axes, psdf.axes)
# multi-index columns
columns = pd.MultiIndex.from_tuples([("x", "a"), ("y", "b")])
pdf.columns = columns
psdf.columns = columns
self.assert_eq(pdf.axes, psdf.axes)
def test_udt(self):
sparse_values = {0: 0.1, 1: 1.1}
sparse_vector = SparseVector(len(sparse_values), sparse_values)
pdf = pd.DataFrame({"a": [sparse_vector], "b": [10]})
psdf = ps.from_pandas(pdf)
self.assert_eq(psdf, pdf)
def test_eval(self):
pdf = pd.DataFrame({"A": range(1, 6), "B": range(10, 0, -2)})
psdf = ps.from_pandas(pdf)
# operation between columns (returns Series)
self.assert_eq(pdf.eval("A + B"), psdf.eval("A + B"))
self.assert_eq(pdf.eval("A + A"), psdf.eval("A + A"))
# assignment (returns DataFrame)
self.assert_eq(pdf.eval("C = A + B"), psdf.eval("C = A + B"))
self.assert_eq(pdf.eval("A = A + A"), psdf.eval("A = A + A"))
# operation between scalars (returns scalar)
self.assert_eq(pdf.eval("1 + 1"), psdf.eval("1 + 1"))
# complicated operations with assignment
self.assert_eq(
pdf.eval("B = A + B // (100 + 200) * (500 - B) - 10.5"),
psdf.eval("B = A + B // (100 + 200) * (500 - B) - 10.5"),
)
# inplace=True (only support for assignment)
pdf.eval("C = A + B", inplace=True)
psdf.eval("C = A + B", inplace=True)
self.assert_eq(pdf, psdf)
pser = pdf.A
psser = psdf.A
pdf.eval("A = B + C", inplace=True)
psdf.eval("A = B + C", inplace=True)
self.assert_eq(pdf, psdf)
self.assert_eq(pser, psser)
# doesn't support for multi-index columns
columns = pd.MultiIndex.from_tuples([("x", "a"), ("y", "b"), ("z", "c")])
psdf.columns = columns
self.assertRaises(TypeError, lambda: psdf.eval("x.a + y.b"))
@unittest.skipIf(not have_tabulate, tabulate_requirement_message)
def test_to_markdown(self):
pdf = pd.DataFrame(data={"animal_1": ["elk", "pig"], "animal_2": ["dog", "quetzal"]})
psdf = ps.from_pandas(pdf)
self.assert_eq(pdf.to_markdown(), psdf.to_markdown())
def test_cache(self):
pdf = pd.DataFrame(
[(0.2, 0.3), (0.0, 0.6), (0.6, 0.0), (0.2, 0.1)], columns=["dogs", "cats"]
)
psdf = ps.from_pandas(pdf)
with psdf.spark.cache() as cached_df:
self.assert_eq(isinstance(cached_df, CachedDataFrame), True)
self.assert_eq(
repr(cached_df.spark.storage_level), repr(StorageLevel(True, True, False, True))
)
def test_persist(self):
pdf = pd.DataFrame(
[(0.2, 0.3), (0.0, 0.6), (0.6, 0.0), (0.2, 0.1)], columns=["dogs", "cats"]
)
psdf = ps.from_pandas(pdf)
storage_levels = [
StorageLevel.DISK_ONLY,
StorageLevel.MEMORY_AND_DISK,
StorageLevel.MEMORY_ONLY,
StorageLevel.OFF_HEAP,
]
for storage_level in storage_levels:
with psdf.spark.persist(storage_level) as cached_df:
self.assert_eq(isinstance(cached_df, CachedDataFrame), True)
self.assert_eq(repr(cached_df.spark.storage_level), repr(storage_level))
self.assertRaises(TypeError, lambda: psdf.spark.persist("DISK_ONLY"))
def test_squeeze(self):
axises = [None, 0, 1, "rows", "index", "columns"]
# Multiple columns
pdf = pd.DataFrame([[1, 2], [3, 4]], columns=["a", "b"], index=["x", "y"])
psdf = ps.from_pandas(pdf)
for axis in axises:
self.assert_eq(pdf.squeeze(axis), psdf.squeeze(axis))
# Multiple columns with MultiIndex columns
columns = pd.MultiIndex.from_tuples([("A", "Z"), ("B", "X")])
pdf.columns = columns
psdf.columns = columns
for axis in axises:
self.assert_eq(pdf.squeeze(axis), psdf.squeeze(axis))
# Single column with single value
pdf = pd.DataFrame([[1]], columns=["a"], index=["x"])
psdf = ps.from_pandas(pdf)
for axis in axises:
self.assert_eq(pdf.squeeze(axis), psdf.squeeze(axis))
# Single column with single value with MultiIndex column
columns = pd.MultiIndex.from_tuples([("A", "Z")])
pdf.columns = columns
psdf.columns = columns
for axis in axises:
self.assert_eq(pdf.squeeze(axis), psdf.squeeze(axis))
# Single column with multiple values
pdf = pd.DataFrame([1, 2, 3, 4], columns=["a"])
psdf = ps.from_pandas(pdf)
for axis in axises:
self.assert_eq(pdf.squeeze(axis), psdf.squeeze(axis))
# Single column with multiple values with MultiIndex column
pdf.columns = columns
psdf.columns = columns
for axis in axises:
self.assert_eq(pdf.squeeze(axis), psdf.squeeze(axis))
def test_rfloordiv(self):
pdf = pd.DataFrame(
{"angles": [0, 3, 4], "degrees": [360, 180, 360]},
index=["circle", "triangle", "rectangle"],
columns=["angles", "degrees"],
)
psdf = ps.from_pandas(pdf)
expected_result = pdf.rfloordiv(10)
self.assert_eq(psdf.rfloordiv(10), expected_result)
def test_truncate(self):
pdf1 = pd.DataFrame(
{
"A": ["a", "b", "c", "d", "e", "f", "g"],
"B": ["h", "i", "j", "k", "l", "m", "n"],
"C": ["o", "p", "q", "r", "s", "t", "u"],
},
index=[-500, -20, -1, 0, 400, 550, 1000],
)
psdf1 = ps.from_pandas(pdf1)
pdf2 = pd.DataFrame(
{
"A": ["a", "b", "c", "d", "e", "f", "g"],
"B": ["h", "i", "j", "k", "l", "m", "n"],
"C": ["o", "p", "q", "r", "s", "t", "u"],
},
index=[1000, 550, 400, 0, -1, -20, -500],
)
psdf2 = ps.from_pandas(pdf2)
self.assert_eq(psdf1.truncate(), pdf1.truncate())
self.assert_eq(psdf1.truncate(before=-20), pdf1.truncate(before=-20))
self.assert_eq(psdf1.truncate(after=400), pdf1.truncate(after=400))
self.assert_eq(psdf1.truncate(copy=False), pdf1.truncate(copy=False))
self.assert_eq(psdf1.truncate(-20, 400, copy=False), pdf1.truncate(-20, 400, copy=False))
# The bug for these tests has been fixed in pandas 1.1.0.
if LooseVersion(pd.__version__) >= LooseVersion("1.1.0"):
self.assert_eq(psdf2.truncate(0, 550), pdf2.truncate(0, 550))
self.assert_eq(psdf2.truncate(0, 550, copy=False), pdf2.truncate(0, 550, copy=False))
else:
expected_psdf = ps.DataFrame(
{"A": ["b", "c", "d"], "B": ["i", "j", "k"], "C": ["p", "q", "r"]},
index=[550, 400, 0],
)
self.assert_eq(psdf2.truncate(0, 550), expected_psdf)
self.assert_eq(psdf2.truncate(0, 550, copy=False), expected_psdf)
# axis = 1
self.assert_eq(psdf1.truncate(axis=1), pdf1.truncate(axis=1))
self.assert_eq(psdf1.truncate(before="B", axis=1), pdf1.truncate(before="B", axis=1))
self.assert_eq(psdf1.truncate(after="A", axis=1), pdf1.truncate(after="A", axis=1))
self.assert_eq(psdf1.truncate(copy=False, axis=1), pdf1.truncate(copy=False, axis=1))
self.assert_eq(psdf2.truncate("B", "C", axis=1), pdf2.truncate("B", "C", axis=1))
self.assert_eq(
psdf1.truncate("B", "C", copy=False, axis=1),
pdf1.truncate("B", "C", copy=False, axis=1),
)
# MultiIndex columns
columns = pd.MultiIndex.from_tuples([("A", "Z"), ("B", "X"), ("C", "Z")])
pdf1.columns = columns
psdf1.columns = columns
pdf2.columns = columns
psdf2.columns = columns
self.assert_eq(psdf1.truncate(), pdf1.truncate())
self.assert_eq(psdf1.truncate(before=-20), pdf1.truncate(before=-20))
self.assert_eq(psdf1.truncate(after=400), pdf1.truncate(after=400))
self.assert_eq(psdf1.truncate(copy=False), pdf1.truncate(copy=False))
self.assert_eq(psdf1.truncate(-20, 400, copy=False), pdf1.truncate(-20, 400, copy=False))
# The bug for these tests has been fixed in pandas 1.1.0.
if LooseVersion(pd.__version__) >= LooseVersion("1.1.0"):
self.assert_eq(psdf2.truncate(0, 550), pdf2.truncate(0, 550))
self.assert_eq(psdf2.truncate(0, 550, copy=False), pdf2.truncate(0, 550, copy=False))
else:
expected_psdf.columns = columns
self.assert_eq(psdf2.truncate(0, 550), expected_psdf)
self.assert_eq(psdf2.truncate(0, 550, copy=False), expected_psdf)
# axis = 1
self.assert_eq(psdf1.truncate(axis=1), pdf1.truncate(axis=1))
self.assert_eq(psdf1.truncate(before="B", axis=1), pdf1.truncate(before="B", axis=1))
self.assert_eq(psdf1.truncate(after="A", axis=1), pdf1.truncate(after="A", axis=1))
self.assert_eq(psdf1.truncate(copy=False, axis=1), pdf1.truncate(copy=False, axis=1))
self.assert_eq(psdf2.truncate("B", "C", axis=1), pdf2.truncate("B", "C", axis=1))
self.assert_eq(
psdf1.truncate("B", "C", copy=False, axis=1),
pdf1.truncate("B", "C", copy=False, axis=1),
)
# Exceptions
psdf = ps.DataFrame(
{
"A": ["a", "b", "c", "d", "e", "f", "g"],
"B": ["h", "i", "j", "k", "l", "m", "n"],
"C": ["o", "p", "q", "r", "s", "t", "u"],
},
index=[-500, 100, 400, 0, -1, 550, -20],
)
msg = "truncate requires a sorted index"
with self.assertRaisesRegex(ValueError, msg):
psdf.truncate()
psdf = ps.DataFrame(
{
"A": ["a", "b", "c", "d", "e", "f", "g"],
"B": ["h", "i", "j", "k", "l", "m", "n"],
"C": ["o", "p", "q", "r", "s", "t", "u"],
},
index=[-500, -20, -1, 0, 400, 550, 1000],
)
msg = "Truncate: -20 must be after 400"
with self.assertRaisesRegex(ValueError, msg):
psdf.truncate(400, -20)
msg = "Truncate: B must be after C"
with self.assertRaisesRegex(ValueError, msg):
psdf.truncate("C", "B", axis=1)
def test_explode(self):
pdf = pd.DataFrame({"A": [[-1.0, np.nan], [0.0, np.inf], [1.0, -np.inf]], "B": 1})
pdf.index.name = "index"
pdf.columns.name = "columns"
psdf = ps.from_pandas(pdf)
expected_result1 = pdf.explode("A")
expected_result2 = pdf.explode("B")
self.assert_eq(psdf.explode("A"), expected_result1, almost=True)
self.assert_eq(psdf.explode("B"), expected_result2)
self.assert_eq(psdf.explode("A").index.name, expected_result1.index.name)
self.assert_eq(psdf.explode("A").columns.name, expected_result1.columns.name)
self.assertRaises(TypeError, lambda: psdf.explode(["A", "B"]))
# MultiIndex
midx = pd.MultiIndex.from_tuples(
[("x", "a"), ("x", "b"), ("y", "c")], names=["index1", "index2"]
)
pdf.index = midx
psdf = ps.from_pandas(pdf)
expected_result1 = pdf.explode("A")
expected_result2 = pdf.explode("B")
self.assert_eq(psdf.explode("A"), expected_result1, almost=True)
self.assert_eq(psdf.explode("B"), expected_result2)
self.assert_eq(psdf.explode("A").index.names, expected_result1.index.names)
self.assert_eq(psdf.explode("A").columns.name, expected_result1.columns.name)
self.assertRaises(TypeError, lambda: psdf.explode(["A", "B"]))
# MultiIndex columns
columns = pd.MultiIndex.from_tuples([("A", "Z"), ("B", "X")], names=["column1", "column2"])
pdf.columns = columns
psdf.columns = columns
expected_result1 = pdf.explode(("A", "Z"))
expected_result2 = pdf.explode(("B", "X"))
expected_result3 = pdf.A.explode("Z")
self.assert_eq(psdf.explode(("A", "Z")), expected_result1, almost=True)
self.assert_eq(psdf.explode(("B", "X")), expected_result2)
self.assert_eq(psdf.explode(("A", "Z")).index.names, expected_result1.index.names)
self.assert_eq(psdf.explode(("A", "Z")).columns.names, expected_result1.columns.names)
self.assert_eq(psdf.A.explode("Z"), expected_result3, almost=True)
self.assertRaises(TypeError, lambda: psdf.explode(["A", "B"]))
self.assertRaises(ValueError, lambda: psdf.explode("A"))
def test_spark_schema(self):
psdf = ps.DataFrame(
{
"a": list("abc"),
"b": list(range(1, 4)),
"c": np.arange(3, 6).astype("i1"),
"d": np.arange(4.0, 7.0, dtype="float64"),
"e": [True, False, True],
"f": pd.date_range("20130101", periods=3),
},
columns=["a", "b", "c", "d", "e", "f"],
)
actual = psdf.spark.schema()
expected = (
StructType()
.add("a", "string", False)
.add("b", "long", False)
.add("c", "byte", False)
.add("d", "double", False)
.add("e", "boolean", False)
.add("f", "timestamp", False)
)
self.assertEqual(actual, expected)
actual = psdf.spark.schema("index")
expected = (
StructType()
.add("index", "long", False)
.add("a", "string", False)
.add("b", "long", False)
.add("c", "byte", False)
.add("d", "double", False)
.add("e", "boolean", False)
.add("f", "timestamp", False)
)
self.assertEqual(actual, expected)
def test_print_schema(self):
psdf = ps.DataFrame(
{"a": list("abc"), "b": list(range(1, 4)), "c": np.arange(3, 6).astype("i1")},
columns=["a", "b", "c"],
)
prev = sys.stdout
try:
out = StringIO()
sys.stdout = out
psdf.spark.print_schema()
actual = out.getvalue().strip()
self.assertTrue("a: string" in actual, actual)
self.assertTrue("b: long" in actual, actual)
self.assertTrue("c: byte" in actual, actual)
out = StringIO()
sys.stdout = out
psdf.spark.print_schema(index_col="index")
actual = out.getvalue().strip()
self.assertTrue("index: long" in actual, actual)
self.assertTrue("a: string" in actual, actual)
self.assertTrue("b: long" in actual, actual)
self.assertTrue("c: byte" in actual, actual)
finally:
sys.stdout = prev
def test_explain_hint(self):
psdf1 = ps.DataFrame(
{"lkey": ["foo", "bar", "baz", "foo"], "value": [1, 2, 3, 5]},
columns=["lkey", "value"],
)
psdf2 = ps.DataFrame(
{"rkey": ["foo", "bar", "baz", "foo"], "value": [5, 6, 7, 8]},
columns=["rkey", "value"],
)
merged = psdf1.merge(psdf2.spark.hint("broadcast"), left_on="lkey", right_on="rkey")
prev = sys.stdout
try:
out = StringIO()
sys.stdout = out
merged.spark.explain()
actual = out.getvalue().strip()
self.assertTrue("Broadcast" in actual, actual)
finally:
sys.stdout = prev
def test_mad(self):
pdf = pd.DataFrame(
{
"A": [1, 2, None, 4, np.nan],
"B": [-0.1, 0.2, -0.3, np.nan, 0.5],
"C": ["a", "b", "c", "d", "e"],
}
)
psdf = ps.from_pandas(pdf)
self.assert_eq(psdf.mad(), pdf.mad())
self.assert_eq(psdf.mad(axis=1), pdf.mad(axis=1))
with self.assertRaises(ValueError):
psdf.mad(axis=2)
# MultiIndex columns
columns = pd.MultiIndex.from_tuples([("A", "X"), ("A", "Y"), ("A", "Z")])
pdf.columns = columns
psdf.columns = columns
self.assert_eq(psdf.mad(), pdf.mad())
self.assert_eq(psdf.mad(axis=1), pdf.mad(axis=1))
pdf = pd.DataFrame({"A": [True, True, False, False], "B": [True, False, False, True]})
psdf = ps.from_pandas(pdf)
self.assert_eq(psdf.mad(), pdf.mad())
self.assert_eq(psdf.mad(axis=1), pdf.mad(axis=1))
def test_abs(self):
pdf = pd.DataFrame({"a": [-2, -1, 0, 1]})
psdf = ps.from_pandas(pdf)
self.assert_eq(abs(psdf), abs(pdf))
self.assert_eq(np.abs(psdf), np.abs(pdf))
def test_iteritems(self):
pdf = pd.DataFrame(
{"species": ["bear", "bear", "marsupial"], "population": [1864, 22000, 80000]},
index=["panda", "polar", "koala"],
columns=["species", "population"],
)
psdf = ps.from_pandas(pdf)
for (p_name, p_items), (k_name, k_items) in zip(pdf.iteritems(), psdf.iteritems()):
self.assert_eq(p_name, k_name)
self.assert_eq(p_items, k_items)
def test_tail(self):
pdf = pd.DataFrame({"x": range(1000)})
psdf = ps.from_pandas(pdf)
self.assert_eq(pdf.tail(), psdf.tail())
self.assert_eq(pdf.tail(10), psdf.tail(10))
self.assert_eq(pdf.tail(-990), psdf.tail(-990))
self.assert_eq(pdf.tail(0), psdf.tail(0))
self.assert_eq(pdf.tail(-1001), psdf.tail(-1001))
self.assert_eq(pdf.tail(1001), psdf.tail(1001))
self.assert_eq((pdf + 1).tail(), (psdf + 1).tail())
self.assert_eq((pdf + 1).tail(10), (psdf + 1).tail(10))
self.assert_eq((pdf + 1).tail(-990), (psdf + 1).tail(-990))
self.assert_eq((pdf + 1).tail(0), (psdf + 1).tail(0))
self.assert_eq((pdf + 1).tail(-1001), (psdf + 1).tail(-1001))
self.assert_eq((pdf + 1).tail(1001), (psdf + 1).tail(1001))
with self.assertRaisesRegex(TypeError, "bad operand type for unary -: 'str'"):
psdf.tail("10")
def test_last_valid_index(self):
pdf = pd.DataFrame(
{"a": [1, 2, 3, None], "b": [1.0, 2.0, 3.0, None], "c": [100, 200, 400, None]},
index=["Q", "W", "E", "R"],
)
psdf = ps.from_pandas(pdf)
self.assert_eq(pdf.last_valid_index(), psdf.last_valid_index())
self.assert_eq(pdf[[]].last_valid_index(), psdf[[]].last_valid_index())
# MultiIndex columns
pdf.columns = pd.MultiIndex.from_tuples([("a", "x"), ("b", "y"), ("c", "z")])
psdf = ps.from_pandas(pdf)
self.assert_eq(pdf.last_valid_index(), psdf.last_valid_index())
# Empty DataFrame
pdf = pd.Series([]).to_frame()
psdf = ps.Series([]).to_frame()
self.assert_eq(pdf.last_valid_index(), psdf.last_valid_index())
def test_last(self):
index = pd.date_range("2018-04-09", periods=4, freq="2D")
pdf = pd.DataFrame([1, 2, 3, 4], index=index)
psdf = ps.from_pandas(pdf)
self.assert_eq(pdf.last("1D"), psdf.last("1D"))
self.assert_eq(pdf.last(DateOffset(days=1)), psdf.last(DateOffset(days=1)))
with self.assertRaisesRegex(TypeError, "'last' only supports a DatetimeIndex"):
ps.DataFrame([1, 2, 3, 4]).last("1D")
def test_first(self):
index = pd.date_range("2018-04-09", periods=4, freq="2D")
pdf = pd.DataFrame([1, 2, 3, 4], index=index)
psdf = ps.from_pandas(pdf)
self.assert_eq(pdf.first("1D"), psdf.first("1D"))
self.assert_eq(pdf.first(DateOffset(days=1)), psdf.first(DateOffset(days=1)))
with self.assertRaisesRegex(TypeError, "'first' only supports a DatetimeIndex"):
ps.DataFrame([1, 2, 3, 4]).first("1D")
def test_first_valid_index(self):
pdf = pd.DataFrame(
{"a": [None, 2, 3, 2], "b": [None, 2.0, 3.0, 1.0], "c": [None, 200, 400, 200]},
index=["Q", "W", "E", "R"],
)
psdf = ps.from_pandas(pdf)
self.assert_eq(pdf.first_valid_index(), psdf.first_valid_index())
self.assert_eq(pdf[[]].first_valid_index(), psdf[[]].first_valid_index())
# MultiIndex columns
pdf.columns = pd.MultiIndex.from_tuples([("a", "x"), ("b", "y"), ("c", "z")])
psdf = ps.from_pandas(pdf)
self.assert_eq(pdf.first_valid_index(), psdf.first_valid_index())
# Empty DataFrame
pdf = pd.Series([]).to_frame()
psdf = ps.Series([]).to_frame()
self.assert_eq(pdf.first_valid_index(), psdf.first_valid_index())
pdf = pd.DataFrame(
{"a": [None, 2, 3, 2], "b": [None, 2.0, 3.0, 1.0], "c": [None, 200, 400, 200]},
index=[
datetime(2021, 1, 1),
datetime(2021, 2, 1),
datetime(2021, 3, 1),
datetime(2021, 4, 1),
],
)
psdf = ps.from_pandas(pdf)
self.assert_eq(pdf.first_valid_index(), psdf.first_valid_index())
def test_product(self):
pdf = pd.DataFrame(
{"A": [1, 2, 3, 4, 5], "B": [10, 20, 30, 40, 50], "C": ["a", "b", "c", "d", "e"]}
)
psdf = ps.from_pandas(pdf)
self.assert_eq(pdf.prod(), psdf.prod().sort_index())
# Named columns
pdf.columns.name = "Koalas"
psdf = ps.from_pandas(pdf)
self.assert_eq(pdf.prod(), psdf.prod().sort_index())
# MultiIndex columns
pdf.columns = pd.MultiIndex.from_tuples([("a", "x"), ("b", "y"), ("c", "z")])
psdf = ps.from_pandas(pdf)
self.assert_eq(pdf.prod(), psdf.prod().sort_index())
# Named MultiIndex columns
pdf.columns.names = ["Hello", "Koalas"]
psdf = ps.from_pandas(pdf)
self.assert_eq(pdf.prod(), psdf.prod().sort_index())
# No numeric columns
pdf = pd.DataFrame({"key": ["a", "b", "c"], "val": ["x", "y", "z"]})
psdf = ps.from_pandas(pdf)
self.assert_eq(pdf.prod(), psdf.prod().sort_index())
# No numeric named columns
pdf.columns.name = "Koalas"
psdf = ps.from_pandas(pdf)
self.assert_eq(pdf.prod(), psdf.prod().sort_index(), almost=True)
# No numeric MultiIndex columns
pdf.columns = pd.MultiIndex.from_tuples([("a", "x"), ("b", "y")])
psdf = ps.from_pandas(pdf)
self.assert_eq(pdf.prod(), psdf.prod().sort_index(), almost=True)
# No numeric named MultiIndex columns
pdf.columns.names = ["Hello", "Koalas"]
psdf = ps.from_pandas(pdf)
self.assert_eq(pdf.prod(), psdf.prod().sort_index(), almost=True)
# All NaN columns
pdf = pd.DataFrame(
{
"A": [np.nan, np.nan, np.nan, np.nan, np.nan],
"B": [10, 20, 30, 40, 50],
"C": ["a", "b", "c", "d", "e"],
}
)
psdf = ps.from_pandas(pdf)
self.assert_eq(pdf.prod(), psdf.prod().sort_index(), check_exact=False)
# All NaN named columns
pdf.columns.name = "Koalas"
psdf = ps.from_pandas(pdf)
self.assert_eq(pdf.prod(), psdf.prod().sort_index(), check_exact=False)
# All NaN MultiIndex columns
pdf.columns = pd.MultiIndex.from_tuples([("a", "x"), ("b", "y"), ("c", "z")])
psdf = ps.from_pandas(pdf)
self.assert_eq(pdf.prod(), psdf.prod().sort_index(), check_exact=False)
# All NaN named MultiIndex columns
pdf.columns.names = ["Hello", "Koalas"]
psdf = ps.from_pandas(pdf)
self.assert_eq(pdf.prod(), psdf.prod().sort_index(), check_exact=False)
def test_from_dict(self):
data = {"row_1": [3, 2, 1, 0], "row_2": [10, 20, 30, 40]}
pdf = pd.DataFrame.from_dict(data)
psdf = ps.DataFrame.from_dict(data)
self.assert_eq(pdf, psdf)
pdf = pd.DataFrame.from_dict(data, dtype="int8")
psdf = ps.DataFrame.from_dict(data, dtype="int8")
self.assert_eq(pdf, psdf)
pdf = pd.DataFrame.from_dict(data, orient="index", columns=["A", "B", "C", "D"])
psdf = ps.DataFrame.from_dict(data, orient="index", columns=["A", "B", "C", "D"])
self.assert_eq(pdf, psdf)
def test_pad(self):
pdf = pd.DataFrame(
{
"A": [None, 3, None, None],
"B": [2, 4, None, 3],
"C": [None, None, None, 1],
"D": [0, 1, 5, 4],
},
columns=["A", "B", "C", "D"],
)
psdf = ps.from_pandas(pdf)
if LooseVersion(pd.__version__) >= LooseVersion("1.1"):
self.assert_eq(pdf.pad(), psdf.pad())
# Test `inplace=True`
pdf.pad(inplace=True)
psdf.pad(inplace=True)
self.assert_eq(pdf, psdf)
else:
expected = ps.DataFrame(
{
"A": [None, 3, 3, 3],
"B": [2.0, 4.0, 4.0, 3.0],
"C": [None, None, None, 1],
"D": [0, 1, 5, 4],
},
columns=["A", "B", "C", "D"],
)
self.assert_eq(expected, psdf.pad())
# Test `inplace=True`
psdf.pad(inplace=True)
self.assert_eq(expected, psdf)
def test_backfill(self):
pdf = pd.DataFrame(
{
"A": [None, 3, None, None],
"B": [2, 4, None, 3],
"C": [None, None, None, 1],
"D": [0, 1, 5, 4],
},
columns=["A", "B", "C", "D"],
)
psdf = ps.from_pandas(pdf)
if LooseVersion(pd.__version__) >= LooseVersion("1.1"):
self.assert_eq(pdf.backfill(), psdf.backfill())
# Test `inplace=True`
pdf.backfill(inplace=True)
psdf.backfill(inplace=True)
self.assert_eq(pdf, psdf)
else:
expected = ps.DataFrame(
{
"A": [3.0, 3.0, None, None],
"B": [2.0, 4.0, 3.0, 3.0],
"C": [1.0, 1.0, 1.0, 1.0],
"D": [0, 1, 5, 4],
},
columns=["A", "B", "C", "D"],
)
self.assert_eq(expected, psdf.backfill())
# Test `inplace=True`
psdf.backfill(inplace=True)
self.assert_eq(expected, psdf)
def test_align(self):
pdf1 = pd.DataFrame({"a": [1, 2, 3], "b": ["a", "b", "c"]}, index=[10, 20, 30])
psdf1 = ps.from_pandas(pdf1)
for join in ["outer", "inner", "left", "right"]:
for axis in [None, 0, 1]:
psdf_l, psdf_r = psdf1.align(psdf1[["b"]], join=join, axis=axis)
pdf_l, pdf_r = pdf1.align(pdf1[["b"]], join=join, axis=axis)
self.assert_eq(psdf_l, pdf_l)
self.assert_eq(psdf_r, pdf_r)
psdf_l, psdf_r = psdf1[["a"]].align(psdf1[["b", "a"]], join=join, axis=axis)
pdf_l, pdf_r = pdf1[["a"]].align(pdf1[["b", "a"]], join=join, axis=axis)
self.assert_eq(psdf_l, pdf_l)
self.assert_eq(psdf_r, pdf_r)
psdf_l, psdf_r = psdf1[["b", "a"]].align(psdf1[["a"]], join=join, axis=axis)
pdf_l, pdf_r = pdf1[["b", "a"]].align(pdf1[["a"]], join=join, axis=axis)
self.assert_eq(psdf_l, pdf_l)
self.assert_eq(psdf_r, pdf_r)
psdf_l, psdf_r = psdf1.align(psdf1["b"], axis=0)
pdf_l, pdf_r = pdf1.align(pdf1["b"], axis=0)
self.assert_eq(psdf_l, pdf_l)
self.assert_eq(psdf_r, pdf_r)
psdf_l, psser_b = psdf1[["a"]].align(psdf1["b"], axis=0)
pdf_l, pser_b = pdf1[["a"]].align(pdf1["b"], axis=0)
self.assert_eq(psdf_l, pdf_l)
self.assert_eq(psser_b, pser_b)
self.assertRaises(ValueError, lambda: psdf1.align(psdf1, join="unknown"))
self.assertRaises(ValueError, lambda: psdf1.align(psdf1["b"]))
self.assertRaises(TypeError, lambda: psdf1.align(["b"]))
self.assertRaises(NotImplementedError, lambda: psdf1.align(psdf1["b"], axis=1))
pdf2 = pd.DataFrame({"a": [4, 5, 6], "d": ["d", "e", "f"]}, index=[10, 11, 12])
psdf2 = ps.from_pandas(pdf2)
for join in ["outer", "inner", "left", "right"]:
psdf_l, psdf_r = psdf1.align(psdf2, join=join, axis=1)
pdf_l, pdf_r = pdf1.align(pdf2, join=join, axis=1)
self.assert_eq(psdf_l.sort_index(), pdf_l.sort_index())
self.assert_eq(psdf_r.sort_index(), pdf_r.sort_index())
def test_between_time(self):
idx = pd.date_range("2018-04-09", periods=4, freq="1D20min")
pdf = pd.DataFrame({"A": [1, 2, 3, 4]}, index=idx)
psdf = ps.from_pandas(pdf)
self.assert_eq(
pdf.between_time("0:15", "0:45").sort_index(),
psdf.between_time("0:15", "0:45").sort_index(),
)
pdf.index.name = "ts"
psdf = ps.from_pandas(pdf)
self.assert_eq(
pdf.between_time("0:15", "0:45").sort_index(),
psdf.between_time("0:15", "0:45").sort_index(),
)
# Column label is 'index'
pdf.columns = pd.Index(["index"])
psdf = ps.from_pandas(pdf)
self.assert_eq(
pdf.between_time("0:15", "0:45").sort_index(),
psdf.between_time("0:15", "0:45").sort_index(),
)
# Both index name and column label are 'index'
pdf.index.name = "index"
psdf = ps.from_pandas(pdf)
self.assert_eq(
pdf.between_time("0:15", "0:45").sort_index(),
psdf.between_time("0:15", "0:45").sort_index(),
)
# Index name is 'index', column label is ('X', 'A')
pdf.columns = pd.MultiIndex.from_arrays([["X"], ["A"]])
psdf = ps.from_pandas(pdf)
self.assert_eq(
pdf.between_time("0:15", "0:45").sort_index(),
psdf.between_time("0:15", "0:45").sort_index(),
)
with self.assertRaisesRegex(
NotImplementedError, "between_time currently only works for axis=0"
):
psdf.between_time("0:15", "0:45", axis=1)
psdf = ps.DataFrame({"A": [1, 2, 3, 4]})
with self.assertRaisesRegex(TypeError, "Index must be DatetimeIndex"):
psdf.between_time("0:15", "0:45")
def test_at_time(self):
idx = pd.date_range("2018-04-09", periods=4, freq="1D20min")
pdf = pd.DataFrame({"A": [1, 2, 3, 4]}, index=idx)
psdf = ps.from_pandas(pdf)
psdf.at_time("0:20")
self.assert_eq(
pdf.at_time("0:20").sort_index(),
psdf.at_time("0:20").sort_index(),
)
# Index name is 'ts'
pdf.index.name = "ts"
psdf = ps.from_pandas(pdf)
self.assert_eq(
pdf.at_time("0:20").sort_index(),
psdf.at_time("0:20").sort_index(),
)
# Index name is 'ts', column label is 'index'
pdf.columns = pd.Index(["index"])
psdf = ps.from_pandas(pdf)
self.assert_eq(
pdf.at_time("0:40").sort_index(),
psdf.at_time("0:40").sort_index(),
)
# Both index name and column label are 'index'
pdf.index.name = "index"
psdf = ps.from_pandas(pdf)
self.assert_eq(
pdf.at_time("0:40").sort_index(),
psdf.at_time("0:40").sort_index(),
)
# Index name is 'index', column label is ('X', 'A')
pdf.columns = pd.MultiIndex.from_arrays([["X"], ["A"]])
psdf = ps.from_pandas(pdf)
self.assert_eq(
pdf.at_time("0:40").sort_index(),
psdf.at_time("0:40").sort_index(),
)
with self.assertRaisesRegex(NotImplementedError, "'asof' argument is not supported"):
psdf.at_time("0:15", asof=True)
with self.assertRaisesRegex(NotImplementedError, "at_time currently only works for axis=0"):
psdf.at_time("0:15", axis=1)
psdf = ps.DataFrame({"A": [1, 2, 3, 4]})
with self.assertRaisesRegex(TypeError, "Index must be DatetimeIndex"):
psdf.at_time("0:15")
def test_astype(self):
psdf = self.psdf
msg = "Only a column name can be used for the key in a dtype mappings argument."
with self.assertRaisesRegex(KeyError, msg):
psdf.astype({"c": float})
def test_describe(self):
pdf, psdf = self.df_pair
# numeric columns
self.assert_eq(psdf.describe(), pdf.describe())
psdf.a += psdf.a
pdf.a += pdf.a
self.assert_eq(psdf.describe(), pdf.describe())
# string columns
psdf = ps.DataFrame({"A": ["a", "b", "b", "c"], "B": ["d", "e", "f", "f"]})
pdf = psdf.to_pandas()
self.assert_eq(psdf.describe(), pdf.describe().astype(str))
psdf.A += psdf.A
pdf.A += pdf.A
self.assert_eq(psdf.describe(), pdf.describe().astype(str))
# timestamp columns
psdf = ps.DataFrame(
{
"A": [
pd.Timestamp("2020-10-20"),
pd.Timestamp("2021-06-02"),
pd.Timestamp("2021-06-02"),
pd.Timestamp("2022-07-11"),
],
"B": [
pd.Timestamp("2021-11-20"),
pd.Timestamp("2023-06-02"),
pd.Timestamp("2026-07-11"),
pd.Timestamp("2026-07-11"),
],
}
)
pdf = psdf.to_pandas()
# NOTE: Set `datetime_is_numeric=True` for pandas:
# FutureWarning: Treating datetime data as categorical rather than numeric in `.describe` is deprecated
# and will be removed in a future version of pandas. Specify `datetime_is_numeric=True` to silence this
# warning and adopt the future behavior now.
# NOTE: Compare the result except percentiles, since we use approximate percentile
# so the result is different from pandas.
if LooseVersion(pd.__version__) >= LooseVersion("1.1.0"):
self.assert_eq(
psdf.describe().loc[["count", "mean", "min", "max"]],
pdf.describe(datetime_is_numeric=True)
.astype(str)
.loc[["count", "mean", "min", "max"]],
)
else:
self.assert_eq(
psdf.describe(),
ps.DataFrame(
{
"A": [
"4",
"2021-07-16 18:00:00",
"2020-10-20 00:00:00",
"2020-10-20 00:00:00",
"2021-06-02 00:00:00",
"2021-06-02 00:00:00",
"2022-07-11 00:00:00",
],
"B": [
"4",
"2024-08-02 18:00:00",
"2021-11-20 00:00:00",
"2021-11-20 00:00:00",
"2023-06-02 00:00:00",
"2026-07-11 00:00:00",
"2026-07-11 00:00:00",
],
},
index=["count", "mean", "min", "25%", "50%", "75%", "max"],
),
)
# String & timestamp columns
psdf = ps.DataFrame(
{
"A": ["a", "b", "b", "c"],
"B": [
| pd.Timestamp("2021-11-20") | pandas.Timestamp |
""" County info extractor
TODO describe
"""
import glob
from multiprocessing import Pool
import pandas as pd
import matplotlib.pyplot as plt
import lasio
from tqdm import tqdm
import geopandas as gpd
# Unused
##import numpy as np
##from textwrap import wrap # for making pretty well names
##from functools import partial
##from multiprocessing import Queue
def get_well(well_log):
""" get well log
TODO describe
"""
try:
return well_log.well["WELL"]
except:
return well_log.well["WELL:1"]
def get_county(well_log):
""" get well log
TODO describe
"""
if "CNTY" in well_log.well:
return well_log.well["CNTY"]
if "CNTY." in well_log.well:
return well_log.well["CNTY."]
if "CNTY ." in well_log.well:
return well_log.well["CNTY ."]
if "API" in well_log.well:
print(well_log.well["API"])
return f"NA"
def add_log(file):
""" Add well Log
TODO describe
"""
try:
return lasio.read(file)
except:
return None
if __name__ == "__main__":
wells = []
year = 2016
counties = []
well_logs = []
for name in glob.glob(f"logs/{year}/*.las"):
wells.append(name)
pool = Pool()
print(f"Queue 'em up")
well_logs = list(pool.imap(add_log, wells))
pool.close()
pool.join()
well_logs = filter(lambda x: x is not None, well_logs) # Remove nulls
for log in well_logs:
counties.append(get_county(log))
print(f"Contains {len(counties)}")
for i, county in enumerate(counties):
if type(county) != type(""):
counties[i] = county.value
counties[i] = counties[i].upper()
# Extract all the counties into a dataframe
kwargs = dict(County=counties)
final_df = pd.DataFrame(kwargs)
# There were a bunch of errors and typos in this county data
# Time to fix the typos
corrections = {
"ANDERSON SEC. 22 TWP. 20S RGE. 20E": "ANDERSON",
"STATON": "STANTON",
"KEARNEY": "KEARNY",
"<NAME>": "NA",
"LGAN": "LOGAN",
"SALINA": "SALINE",
"<NAME>": "HARPER",
"HARPER CO.": "HARPER",
"SUMMER": "SUMNER",
"SEDGWICH": "SEDGWICK",
"SEDOWICK": "SEDGWICK",
"SEDGEWICK": "SEDGWICK",
"LORRAINE": "ELLSWORTH", # Lorrained is a city in Ellsworth CO.
"HASKEL": "HASKELL",
"DECTAUR": "DECATUR",
"TRGO": "TREGO",
"ELLS": "ELLIS",
"NESS CO.": "NESS",
"OSBOURNE": "OSBORNE",
"": "NA",
"HODGMAN": "HODGEMAN",
"USA": "NA",
"KANSAS": "NA",
"RUSSEL": "RUSSELL",
"PRATT COUNTY": "PRATT",
"WITCHITA": "WICHITA",
"RUCH": "RUSH",
"RAWLINGS": "RAWLINS",
"RENO CO": "RENO",
"RENO CO.": "RENO",
}
# Apply colrrections
for key, value in tqdm(corrections.items(), desc="Corrections"):
final_df.loc[final_df["County"] == key] = value
freq_count = final_df["County"].value_counts()
freq_df = pd.DataFrame(
{"County": freq_count.keys(), "Frequency": freq_count.values}
)
freq_df = freq_df.sort_values(by="Frequency", ascending=False)
freq_df = freq_df.reset_index(drop=True)
print(freq_df.head()) # Lets see the Frequencies in order
freq_df["Percent"] = freq_df["Frequency"] / freq_df["Frequency"].sum() * 100
print(f"Number of NA's {freq_df[freq_df['County'] == 'NA']}")
fig = plt.figure(figsize=(6, 17))
plt.yticks(range(len(freq_df)), freq_df["County"])
# Lets plot the frequency
plt.barh(range(len(freq_df)), freq_df["Frequency"])
kansas_map = gpd.read_file("kansas.zip")
# To merge the county data with the kansas map we must use fips numbers
fpis = | pd.read_csv("fips.csv") | pandas.read_csv |
import re
import numpy as np
import pandas as pd
import pytest
from woodwork import DataTable
from woodwork.logical_types import (
URL,
Boolean,
Categorical,
CountryCode,
Datetime,
Double,
Filepath,
FullName,
Integer,
IPAddress,
LatLong,
NaturalLanguage,
Ordinal,
PhoneNumber,
SubRegionCode,
ZIPCode
)
def test_datatable_physical_types(sample_df):
dt = DataTable(sample_df)
assert isinstance(dt.physical_types, dict)
assert set(dt.physical_types.keys()) == set(sample_df.columns)
for k, v in dt.physical_types.items():
assert isinstance(k, str)
assert v == sample_df[k].dtype
def test_sets_category_dtype_on_init():
column_name = 'test_series'
series_list = [
pd.Series(['a', 'b', 'c'], name=column_name),
pd.Series(['a', None, 'c'], name=column_name),
pd.Series(['a', np.nan, 'c'], name=column_name),
pd.Series(['a', pd.NA, 'c'], name=column_name),
pd.Series(['a', pd.NaT, 'c'], name=column_name),
]
logical_types = [
Categorical,
CountryCode,
Ordinal(order=['a', 'b', 'c']),
SubRegionCode,
ZIPCode,
]
for series in series_list:
series = series.astype('object')
for logical_type in logical_types:
ltypes = {
column_name: logical_type,
}
dt = DataTable(pd.DataFrame(series), logical_types=ltypes)
assert dt.columns[column_name].logical_type == logical_type
assert dt.columns[column_name].dtype == logical_type.pandas_dtype
assert dt.to_dataframe()[column_name].dtype == logical_type.pandas_dtype
def test_sets_category_dtype_on_update():
column_name = 'test_series'
series = pd.Series(['a', 'b', 'c'], name=column_name)
series = series.astype('object')
logical_types = [
Categorical,
CountryCode,
Ordinal(order=['a', 'b', 'c']),
SubRegionCode,
ZIPCode,
]
for logical_type in logical_types:
ltypes = {
column_name: NaturalLanguage,
}
dt = DataTable(pd.DataFrame(series), logical_types=ltypes)
dt = dt.set_types(logical_types={column_name: logical_type})
assert dt.columns[column_name].logical_type == logical_type
assert dt.columns[column_name].dtype == logical_type.pandas_dtype
assert dt.to_dataframe()[column_name].dtype == logical_type.pandas_dtype
def test_sets_object_dtype_on_init(latlong_df):
for column_name in latlong_df.columns:
ltypes = {
column_name: LatLong,
}
dt = DataTable(latlong_df.loc[:, [column_name]], logical_types=ltypes)
assert dt.columns[column_name].logical_type == LatLong
assert dt.columns[column_name].dtype == LatLong.pandas_dtype
assert dt.to_dataframe()[column_name].dtype == LatLong.pandas_dtype
def test_sets_object_dtype_on_update(latlong_df):
for column_name in latlong_df.columns:
ltypes = {
column_name: NaturalLanguage
}
dt = DataTable(latlong_df.loc[:, [column_name]], logical_types=ltypes)
dt = dt.set_types(logical_types={column_name: LatLong})
assert dt.columns[column_name].logical_type == LatLong
assert dt.columns[column_name].dtype == LatLong.pandas_dtype
assert dt.to_dataframe()[column_name].dtype == LatLong.pandas_dtype
def test_sets_string_dtype_on_init():
column_name = 'test_series'
series_list = [
pd.Series(['a', 'b', 'c'], name=column_name),
pd.Series(['a', None, 'c'], name=column_name),
pd.Series(['a', np.nan, 'c'], name=column_name),
pd.Series(['a', pd.NA, 'c'], name=column_name),
]
logical_types = [
Filepath,
FullName,
IPAddress,
NaturalLanguage,
PhoneNumber,
URL,
]
for series in series_list:
series = series.astype('object')
for logical_type in logical_types:
ltypes = {
column_name: logical_type,
}
dt = DataTable( | pd.DataFrame(series) | pandas.DataFrame |
# http://www.vdh.virginia.gov/coronavirus/
from bs4 import BeautifulSoup
import csv
from datetime import datetime
from io import StringIO
import os
import requests
import pandas as pd
# Remove empty rows
def filtered(rows):
return [x for x in rows if "".join([(x[y] or "").strip() for y in x]) != ""]
def run_VA(args):
# Parameters
raw_name = '../VA/raw'
data_name = '../VA/data/data_%s.csv'
now = datetime.now()
links = [("locality", "https://data.virginia.gov/resource/bre9-aqqr.csv"),
("conf", "https://data.virginia.gov/resource/uqs3-x7zh.csv"),
("dist", "https://data.virginia.gov/resource/v5a8-4ahw.csv"),
("age", "https://data.virginia.gov/resource/uktn-mwig.csv"),
("sex", "https://data.virginia.gov/resource/tdt3-q47w.csv"),
("race_ethnicity", "https://data.virginia.gov/resource/9sba-m86n.csv")]
for link in links:
most_recent = ""
exists = os.path.exists(data_name % link[0])
out = []
# If current data file does not exist
if not exists:
version = 0
v_exists = True
while v_exists:
version += 1
v_exists = os.path.exists((data_name % (link[0] + "_V" + str(version))))
version = version - 1
v_df = pd.read_csv((data_name % (link[0] + "_V" + str(version))))
date_col = ""
for col in v_df.columns:
if "date" in col.lower() and "report" in col.lower():
date_col = col
break
# Getting most recent date
dates = (pd.to_datetime(v_df[date_col])).to_list()
most_recent = max(dt for dt in dates if dt < now)
# Getting new dates
new_df = pd.read_csv(link[1])
new_df.to_csv(raw_name + "/" + link[0] + "_" + str(now) + ".csv")
new_date_col = ""
for col in new_df.columns:
if "date" in col.lower() and "report" in col.lower():
new_date_col = col
break
new_df[new_date_col] = pd.to_datetime(new_df[new_date_col])
rows = new_df.to_dict(orient="records")
for row in rows:
if row[new_date_col] <= most_recent:
continue
else:
out.append(row)
else:
curr_df = | pd.read_csv(data_name % link[0]) | pandas.read_csv |
import sys
sys.path.append("./log_helper")
import tensorflow as tf
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
from matplotlib.ticker import PercentFormatter
import math
import random
import argparse
import time
import logging
import glob
from os.path import isfile, join, splitext
from datetime import datetime, timedelta
from pytz import timezone, utc
plt.rcParams["font.size"] = 12
plt.rcParams["patch.force_edgecolor"] = True
sys.path.append("./model")
from process_raw_prices import *
import trading_env
import rl_load_data
import rl_constants
from log_helper import LogHelper
tf.enable_eager_execution()
########################## functions ##############################
def sample_action(logits, batch_size, random=False):
if random:
dist = tf.distributions.Categorical(logits=tf.zeros([batch_size, a_num]))
else:
dist = tf.distributions.Categorical(logits=logits)
# 1-D Tensor where the i-th element correspond to a sample from
# the i-th categorical distribution
return dist.sample()
def discount_rewards(r):
"""
r is a numpy array in the shape of (n, batch_size).
return the discounted and cumulative rewards"""
result = np.zeros_like(r, dtype=float)
n = r.shape[0]
sum_ = np.zeros_like(r[0], dtype=float)
for i in range(n-1,-1,-1):
sum_ *= gamma
sum_ += r[i]
result[i] = sum_
return result
def loss(all_logits, all_actions, all_advantages):
neg_log_select_prob = tf.nn.sparse_softmax_cross_entropy_with_logits(logits=all_logits, labels=all_actions)
# 0 axis is the time axis. 1 axis is the batch axis
return tf.reduce_mean(neg_log_select_prob * all_advantages, 0)
def extract_pair_name(s):
return '_'.join(s.split('-')[:2])
def extract_pair_index(s):
return int(s.split('-')[-1])
def save_model():
hkg_time = get_hkg_time()
checkpoint_name = hkg_time.strftime("%Y%m%d_%H%M%S")
# change the dir name to separate different models...
checkpoint_prefix = checkpoint_dir+checkpoint_name+"/"
path = root.save(checkpoint_prefix)
_logger.info('checkpoint path: {}'.format(path))
def restore_model(checkpoint_dir):
root.restore(tf.train.latest_checkpoint(checkpoint_dir))
def get_hkg_time():
utc_dt = utc.localize(datetime.utcnow())
my_tz = timezone("Asia/Hong_Kong")
hkg_time = utc_dt.astimezone(my_tz)
return hkg_time
def run_rl_backtest(stock1, stock2, period_index):
pair_name = "-".join([stock1, stock2])
job_name = {1: '0412_train_0_test_1', 2: '0412_train_01_test_2', 3: '0412_train_012_test_3'}
config = generate_parser().parse_args(['--job_name', job_name[period_index], '--run_mode', 'plot_distribution'])
copy_config(config)
main_global_setup(config, filter_pairs=[pair_name])
model_paths = sorted(glob.glob(checkpoint_dir+"*"))
_logger.info("restore model from {}".format(model_paths[-1]))
_logger.info("evaluate return distribution")
restore_model(model_paths[-1])
return evaluate_a_pair([period_index], pair_name)
def evaluate_a_pair(data_indices, pair_name):
done = False
s = env.reset(data_indices, pair_name)
saved_a = [0]
saved_portfolio_val = [env.port_val[0]]
y_quantity = [env.quantity['y'][0]]
x_quantity = [env.quantity['x'][0]]
softmax = [np.ones(3)/3] # dummy
# for accumalting episode statistics
act_batch_size = tf.shape(s).numpy()[0]
_logger.info("this batch size should be 1: {}".format(act_batch_size))
total_r = np.zeros(act_batch_size)
# internally the episode length is fixed by trading_period
while not done:
logits = pi(s)
# softmax
softmax.append(tf.nn.softmax(logits).numpy()[0])
a = sample_action(logits, act_batch_size)
saved_a.append(a[0].numpy())
y_quantity.append(env.quantity['y'][0])
x_quantity.append(env.quantity['x'][0])
# get immediate reward, update state, and get done
r, s, done = env.step(a.numpy())
saved_portfolio_val.append(env.port_val[0])
date = env.history[:,-1,0]
yclose = env.history[:, rl_load_data.col_name_to_ind["y_close"], 0]
xclose = env.history[:, rl_load_data.col_name_to_ind["x_close"], 0]
normalized_data0 = env.history[:, 2, 0]
normalized_data1 = env.history[:, 3, 0]
y_quantity = np.array(y_quantity)
x_quantity = np.array(x_quantity)
y_val = yclose*y_quantity
x_val = xclose*x_quantity
softmax = np.array(softmax)
# plt.figure()
# plt.plot(env.history[:,-1,0], env.history[:,rl_load_data.col_name_to_ind["spread"],0])
# plt.plot(env.history[:,-1,0], saved_a)
# plt.savefig(join(plot_folder_path, 'spread_action_{}.png'.format(pair_name)))
# plt.figure()
# plt.plot(env.history[:,-1,0], saved_portfolio_val)
# plt.savefig(join(plot_folder_path, 'portfolio_val_{}.png'.format(pair_name)))
result_df = pd.DataFrame({'spread': env.history[:,rl_load_data.col_name_to_ind["spread"],0],
'date': env.history[:,-1,0],
'latest_trade_action': saved_a,
'portfolio_value': saved_portfolio_val,
'softmax_0': softmax[:, 0],
'softmax_1': softmax[:, 1],
'softmax_2': softmax[:, 2],
'data0': yclose,
'data1': xclose,
'normalized_data0': normalized_data0,
'normalized_data1': normalized_data1,
'quantity0': y_quantity,
'quantity1': x_quantity
})
result_df['quantity0'] = result_df['quantity0'].diff().values
result_df['quantity1'] = result_df['quantity1'].diff().values
columns_required = ['spread', 'date', 'latest_trade_action']
action_df = result_df.loc[result_df['latest_trade_action'].diff() != 0][columns_required]
dic = {0: "exit_spread", 1: "long_spread", 2: "short_spread"}
action_df = action_df.replace({'latest_trade_action': dic})
action_df = action_df.iloc[1:]
action_df["sell_stk"] = None
action_df["buy_stk"] = None
action_df["buy_amt"] = np.nan
action_df["sell_amt"] = np.nan
action_df = action_df.reset_index()
action_df['spread'] = pd.to_numeric(action_df['spread'])
action_df['date'] = | pd.to_datetime(action_df['date']) | pandas.to_datetime |
import ast
import collections
import glob
import inspect
import math
import os
import random
import shutil
import subprocess
import time
import warnings
from concurrent.futures import ThreadPoolExecutor
from contextlib import suppress
from datetime import datetime
from typing import Any, Dict, Tuple, Sequence, List, Optional, Callable, Union
import adaptive
import toolz
from adaptive.notebook_integration import in_ipynb
from ipyparallel import Client
from tqdm import tqdm, tqdm_notebook
MAX_LINE_LENGTH = 100
def shuffle_list(*lists, seed=0):
"""Shuffle multiple lists in the same order."""
combined = list(zip(*lists))
random.Random(seed).shuffle(combined)
return zip(*combined)
def _split(seq, n_parts):
lst = list(seq)
n = math.ceil(len(lst) / n_parts)
return toolz.partition_all(n, lst)
def split_in_balancing_learners(
learners: List[adaptive.BaseLearner],
fnames: List[str],
n_parts: int,
strategy: str = "npoints",
) -> Tuple[List[adaptive.BaseLearner], List[str]]:
r"""Split a list of learners and fnames into `adaptive.BalancingLearner`\s.
Parameters
----------
learners : list
List of learners.
fnames : list
List of filenames.
n_parts : int
Total number of `~adaptive.BalancingLearner`\s.
strategy : str
Learning strategy of the `~adaptive.BalancingLearner`.
Returns
-------
new_learners, new_fnames
"""
new_learners = []
new_fnames = []
for x in _split(zip(learners, fnames), n_parts):
learners_part, fnames_part = zip(*x)
learner = adaptive.BalancingLearner(learners_part, strategy=strategy)
new_learners.append(learner)
new_fnames.append(fnames_part)
return new_learners, new_fnames
def _progress(seq: Sequence, with_progress_bar: bool = True, desc: str = ""):
if not with_progress_bar:
return seq
else:
if in_ipynb():
return tqdm_notebook(list(seq), desc=desc)
else:
return tqdm(list(seq), desc=desc)
def _cancel_function(cancel_cmd: str, queue_function: Callable) -> Callable:
def cancel(
job_names: List[str], with_progress_bar: bool = True, max_tries: int = 5
) -> Callable:
"""Cancel all jobs in `job_names`.
Parameters
----------
job_names : list
List of job names.
with_progress_bar : bool, default: True
Display a progress bar using `tqdm`.
max_tries : int, default: 5
Maximum number of attempts to cancel a job.
"""
def to_cancel(job_names):
return [
job_id
for job_id, info in queue_function().items()
if info["name"] in job_names
]
def cancel_jobs(job_ids):
for job_id in _progress(job_ids, with_progress_bar, "Canceling jobs"):
cmd = f"{cancel_cmd} {job_id}".split()
returncode = subprocess.run(cmd, stderr=subprocess.PIPE).returncode
if returncode != 0:
warnings.warn(f"Couldn't cancel '{job_id}'.", UserWarning)
job_names = set(job_names)
for _ in range(max_tries):
job_ids = to_cancel(job_names)
if not job_ids:
# no more running jobs
break
cancel_jobs(job_ids)
return cancel
def combo_to_fname(combo: Dict[str, Any], folder: Optional[str] = None) -> str:
"""Converts a dict into a human readable filename."""
fname = "__".join(f"{k}_{v}" for k, v in combo.items()) + ".pickle"
if folder is None:
return fname
return os.path.join(folder, fname)
def cleanup_files(
job_names: List[str],
extensions: List[str] = ("sbatch", "out", "batch", "e*", "o*"),
with_progress_bar: bool = True,
move_to: Optional[str] = None,
log_file_folder: str = "",
) -> None:
"""Cleanup the scheduler log-files files.
Parameters
----------
job_names : list
List of job names.
extensions : list
List of file extensions to be removed.
with_progress_bar : bool, default: True
Display a progress bar using `tqdm`.
move_to : str, default: None
Move the file to a different directory.
If None the file is removed.
log_file_folder : str, default: ''
The folder in which to delete the log-files.
"""
# Finding the files
fnames = []
for job in job_names:
for ext in extensions:
pattern = f"{job}*.{ext}"
fnames += glob.glob(pattern)
if log_file_folder:
# The log-files might be in a different folder, but we're
# going to loop over every extension anyway.
fnames += glob.glob(os.path.join(log_file_folder, pattern))
_remove_or_move_files(fnames, with_progress_bar, move_to)
def _remove_or_move_files(
fnames: List[str], with_progress_bar: bool = True, move_to: Optional[str] = None
) -> None:
"""Remove files by filename.
Parameters
----------
fnames : list
List of filenames.
with_progress_bar : bool, default: True
Display a progress bar using `tqdm`.
move_to : str, default None
Move the file to a different directory.
If None the file is removed.
"""
n_failed = 0
for fname in _progress(fnames, with_progress_bar, "Removing files"):
try:
if move_to is None:
os.remove(fname)
else:
os.makedirs(move_to, exist_ok=True)
shutil.move(fname, move_to)
except Exception:
n_failed += 1
if n_failed:
warnings.warn(f"Failed to remove {n_failed} files.")
def load_parallel(
learners: List[adaptive.BaseLearner],
fnames: List[str],
*,
with_progress_bar: bool = True,
) -> None:
r"""Load a sequence of learners in parallel.
Parameters
----------
learners : sequence of `adaptive.BaseLearner`\s
The learners to be loaded.
fnames : sequence of str
A list of filenames corresponding to `learners`.
with_progress_bar : bool, default True
Display a progress bar using `tqdm`.
"""
def load(learner, fname):
learner.load(fname)
with ThreadPoolExecutor() as ex:
futs = []
iterator = zip(learners, fnames)
pbar = _progress(iterator, with_progress_bar, "Submitting loading tasks")
futs = [ex.submit(load, *args) for args in pbar]
for fut in _progress(futs, with_progress_bar, "Finishing loading"):
fut.result()
def save_parallel(
learners: List[adaptive.BaseLearner],
fnames: List[str],
*,
with_progress_bar: bool = True,
) -> None:
r"""Save a sequence of learners in parallel.
Parameters
----------
learners : sequence of `adaptive.BaseLearner`\s
The learners to be saved.
fnames : sequence of str
A list of filenames corresponding to `learners`.
with_progress_bar : bool, default True
Display a progress bar using `tqdm`.
"""
def save(learner, fname):
learner.save(fname)
with ThreadPoolExecutor() as ex:
futs = []
iterator = zip(learners, fnames)
pbar = _progress(iterator, with_progress_bar, "Submitting saving tasks")
futs = [ex.submit(save, *args) for args in pbar]
for fut in _progress(futs, with_progress_bar, "Finishing saving"):
fut.result()
def _get_status_prints(fname: str, only_last: bool = True):
status_lines = []
with open(fname) as f:
lines = f.readlines()
if not lines:
return status_lines
for line in reversed(lines):
if "current status" in line:
status_lines.append(line)
if only_last:
return status_lines
return status_lines
def parse_log_files(
job_names: List[str],
only_last: bool = True,
db_fname: Optional[str] = None,
log_file_folder: str = "",
):
"""Parse the log-files and convert it to a `~pandas.core.frame.DataFrame`.
This only works if you use `adaptive_scheduler.client_support.log_info`
inside your ``run_script``.
Parameters
----------
job_names : list
List of job names.
only_last : bool, default: True
Only look use the last printed status message.
db_fname : str, optional
The database filename. If passed, ``fname`` will be populated.
log_file_folder : str, default: ""
The folder in which the log-files are.
Returns
-------
`~pandas.core.frame.DataFrame`
"""
# XXX: it could be that the job_id and the logfile don't match up ATM! This
# probably happens when a job got canceled and is pending now.
try:
import pandas as pd
with_pandas = True
except ImportError:
with_pandas = False
warnings.warn("`pandas` is not installed, a list of dicts will be returned.")
# import here to avoid circular imports
from adaptive_scheduler.server_support import queue, get_database
def convert_type(k, v):
if k == "elapsed_time":
return pd.to_timedelta(v)
elif k == "overhead":
return float(v[:-1])
else:
return ast.literal_eval(v)
def join_str(info):
"""Turns an incorrectly split string
["elapsed_time=1", "day,", "0:20:57.330515", "nlearners=31"]
back the correct thing
['elapsed_time=1 day, 0:20:57.330515', 'nlearners=31']
"""
_info = []
for x in info:
if "=" in x:
_info.append(x)
else:
_info[-1] += f" {x}"
return _info
infos = []
for job in job_names:
fnames = glob.glob(os.path.join(log_file_folder, f"{job}-*.out"))
if not fnames:
continue
fname = fnames[-1] # take the last file
statuses = _get_status_prints(fname, only_last)
if statuses is None:
continue
for status in statuses:
time, info = status.split("current status")
info = join_str(info.strip().split(" "))
info = dict([x.split("=") for x in info])
info = {k: convert_type(k, v) for k, v in info.items()}
info["job"] = job
info["time"] = datetime.strptime(time.strip(), "%Y-%m-%d %H:%M.%S")
info["log_file"] = fname
infos.append(info)
# Polulate state and job_id from the queue
mapping = {
info["name"]: (job_id, info["state"]) for job_id, info in queue().items()
}
for info in infos:
info["job_id"], info["state"] = mapping.get(info["job"], (None, None))
if db_fname is not None:
# populate job_id
db = get_database(db_fname)
fnames = {info["job_id"]: info["fname"] for info in db}
id_done = {info["job_id"]: info["is_done"] for info in db}
for info in infos:
info["fname"] = fnames.get(info["job_id"], "UNKNOWN")
info["is_done"] = id_done.get(info["job_id"], "UNKNOWN")
return | pd.DataFrame(infos) | pandas.DataFrame |
#!/usr/bin/python
import argparse
import pandas as pd
import logging
from pandas.io.json import json_normalize
import os
f = '%(asctime)s %(name)-12s %(levelname)-8s %(message)s'
logging.basicConfig(filename = "conversion.log", filemode='a', level=logging.DEBUG, format=f)
console = logging.StreamHandler()
formatter = logging.Formatter(f)
console.setFormatter(formatter)
console.setLevel(logging.DEBUG)
logging.getLogger().addHandler(console)
logger = logging.getLogger(__name__)
def parse_args():
parser = argparse.ArgumentParser(description='Digitraffic portnet location data parser')
parser.add_argument('-i', '--input_file', help='Input JSON file', required=True)
parser.add_argument('-o', '--output_file', help='Output CSV file', required=True)
args = vars(parser.parse_args())
return args
def append_columns(df, column_name):
df_add = | json_normalize(df[column_name][0]) | pandas.io.json.json_normalize |
from typing import Union, Optional, List, Dict, Tuple, Any
import pandas as pd
import numpy as np
from .common.validators import validate_integer
from .macro import Inflation
from .common.helpers import Float, Frame, Date, Index
from .settings import default_ticker, PeriodLength, _MONTHS_PER_YEAR
from .api.data_queries import QueryData
from .api.namespaces import get_assets_namespaces
class Asset:
"""
A financial asset, that could be used in a list of assets or in portfolio.
Parameters
----------
symbol: str, default "SPY.US"
Symbol is an asset ticker with namespace after dot. The default value is "SPY.US" (SPDR S&P 500 ETF Trust).
Examples
--------
>>> asset = ok.Asset()
>>> asset
symbol SPY.US
name SPDR S&P 500 ETF Trust
country USA
exchange NYSE ARCA
currency USD
type ETF
first date 1993-02
last date 2021-03
period length 28.1
dtype: object
An Asset object could be easy created whithout specifying a symbol Asset() using the default symbol.
"""
def __init__(self, symbol: str = default_ticker):
if symbol is None or len(str(symbol).strip()) == 0:
raise ValueError("Symbol can not be empty")
self._symbol = str(symbol).strip()
self._check_namespace()
self._get_symbol_data(symbol)
self.ror: pd.Series = QueryData.get_ror(symbol)
self.first_date: pd.Timestamp = self.ror.index[0].to_timestamp()
self.last_date: pd.Timestamp = self.ror.index[-1].to_timestamp()
self.period_length: float = round(
(self.last_date - self.first_date) / np.timedelta64(365, "D"), ndigits=1
)
def __repr__(self):
dic = {
"symbol": self.symbol,
"name": self.name,
"country": self.country,
"exchange": self.exchange,
"currency": self.currency,
"type": self.type,
"first date": self.first_date.strftime("%Y-%m"),
"last date": self.last_date.strftime("%Y-%m"),
"period length": "{:.2f}".format(self.period_length),
}
return repr(pd.Series(dic))
def _check_namespace(self):
namespace = self._symbol.split(".", 1)[-1]
allowed_namespaces = get_assets_namespaces()
if namespace not in allowed_namespaces:
raise ValueError(
f"{namespace} is not in allowed assets namespaces: {allowed_namespaces}"
)
@property
def symbol(self) -> str:
"""
Return a symbol of the asset.
Returns
-------
str
"""
return self._symbol
def _get_symbol_data(self, symbol) -> None:
x = QueryData.get_symbol_info(symbol)
self.ticker: str = x["code"]
self.name: str = x["name"]
self.country: str = x["country"]
self.exchange: str = x["exchange"]
self.currency: str = x["currency"]
self.type: str = x["type"]
self.inflation: str = f"{self.currency}.INFL"
@property
def price(self) -> Optional[float]:
"""
Return live price of an asset.
Live price is delayed (15-20 minutes).
For certain namespaces (FX, INDX, PIF etc.) live price is not supported.
Returns
-------
float, None
Live price of the asset. Returns None if not defined.
"""
return QueryData.get_live_price(self.symbol)
@property
def dividends(self) -> pd.Series:
"""
Return dividends time series historical daily data.
Returns
-------
Series
Time series of dividends historical data (daily).
Examples
--------
>>> x = ok.Asset('VNQ.US')
>>> x.dividends
Date
2004-12-22 1.2700
2005-03-24 0.6140
2005-06-27 0.6440
2005-09-26 0.6760
...
2020-06-25 0.7590
2020-09-25 0.5900
2020-12-24 1.3380
2021-03-25 0.5264
Freq: D, Name: VNQ.US, Length: 66, dtype: float64
"""
div = QueryData.get_dividends(self.symbol)
if div.empty:
# Zero time series for assets where dividend yield is not defined.
index = pd.date_range(
start=self.first_date, end=self.last_date, freq="MS", closed=None
)
period = index.to_period("D")
div = pd.Series(data=0, index=period)
div.rename(self.symbol, inplace=True)
return div
@property
def nav_ts(self) -> Optional[pd.Series]:
"""
Return NAV time series (monthly) for mutual funds.
"""
if self.exchange == "PIF":
return QueryData.get_nav(self.symbol)
return np.nan
class AssetList:
"""
The list of financial assets implementation.
"""
def __init__(
self,
symbols: Optional[List[str]] = None,
*,
first_date: Optional[str] = None,
last_date: Optional[str] = None,
ccy: str = "USD",
inflation: bool = True,
):
self.__symbols = symbols
self.__tickers: List[str] = [x.split(".", 1)[0] for x in self.symbols]
self.__currency: Asset = Asset(symbol=f"{ccy}.FX")
self.__make_asset_list(self.symbols)
if inflation:
self.inflation: str = f"{ccy}.INFL"
self._inflation_instance: Inflation = Inflation(
self.inflation, self.first_date, self.last_date
)
self.inflation_ts: pd.Series = self._inflation_instance.values_ts
self.inflation_first_date: pd.Timestamp = self._inflation_instance.first_date
self.inflation_last_date: pd.Timestamp = self._inflation_instance.last_date
self.first_date = max(self.first_date, self.inflation_first_date)
self.last_date: pd.Timestamp = min(self.last_date, self.inflation_last_date)
# Add inflation to the date range dict
self.assets_first_dates.update({self.inflation: self.inflation_first_date})
self.assets_last_dates.update({self.inflation: self.inflation_last_date})
if first_date:
self.first_date = max(self.first_date, pd.to_datetime(first_date))
self.ror = self.ror[self.first_date :]
if last_date:
self.last_date = min(self.last_date, pd.to_datetime(last_date))
self.ror: pd.DataFrame = self.ror[self.first_date: self.last_date]
self.period_length: float = round(
(self.last_date - self.first_date) / np.timedelta64(365, "D"), ndigits=1
)
self.pl = PeriodLength(
self.ror.shape[0] // _MONTHS_PER_YEAR, self.ror.shape[0] % _MONTHS_PER_YEAR
)
self._pl_txt = f"{self.pl.years} years, {self.pl.months} months"
self._dividend_yield: pd.DataFrame = pd.DataFrame(dtype=float)
self._dividends_ts: pd.DataFrame = pd.DataFrame(dtype=float)
def __repr__(self):
dic = {
"symbols": self.symbols,
"currency": self.currency.ticker,
"first date": self.first_date.strftime("%Y-%m"),
"last_date": self.last_date.strftime("%Y-%m"),
"period length": self._pl_txt,
"inflation": self.inflation if hasattr(self, "inflation") else "None",
}
return repr(pd.Series(dic))
def __len__(self):
return len(self.symbols)
def __make_asset_list(self, ls: list) -> None:
"""
Make an asset list from a list of symbols.
"""
first_dates: Dict[str, pd.Timestamp] = {}
last_dates: Dict[str, pd.Timestamp] = {}
names: Dict[str, str] = {}
currencies: Dict[str, str] = {}
df = pd.DataFrame()
for i, x in enumerate(ls):
asset = Asset(x)
if i == 0: # required to use pd.concat below (df should not be empty).
if asset.currency == self.currency.name:
df = asset.ror
else:
df = self._set_currency(
returns=asset.ror, asset_currency=asset.currency
)
else:
if asset.currency == self.currency.name:
new = asset.ror
else:
new = self._set_currency(
returns=asset.ror, asset_currency=asset.currency
)
df = | pd.concat([df, new], axis=1, join="inner", copy="false") | pandas.concat |
"""
library for simulating semi-analytic mock maps of CMB secondary anisotropies
"""
__author__ = ["<NAME>", "<NAME>"]
__email__ = ["<EMAIL>", "<EMAIL>"]
import os
import warnings
from sys import getsizeof
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
from matplotlib import cm
from warnings import warn
import inspect
from itertools import product
import operator
import re
from functools import partial
from tqdm.auto import tqdm
from astropaint.lib.log import CMBAlreadyAdded, NoiseAlreadyAdded
try:
import healpy as hp
except ModuleNotFoundError:
warn("Healpy is not installed. You cannot use the full sky canvas without it.")
from astropy.coordinates import cartesian_to_spherical
from .lib import transform, utils
# find the package path; same as __path__
path_dir = os.path.dirname(os.path.abspath(__file__))
#########################################################
# Halo Catalog Object
#########################################################
class Catalog:
"""halo catalog containing halo masses, locations, velocities, and redshifts
Units
-----
x, y, z: [Mpc]
v_x, v_y, v_z: [km/s]
M_200c: [M_sun]
"""
def __init__(self,
data=None,
calculate_redshifts=False,
default_redshift=0,
):
"""
Parameters
----------
data: dataframe or str
Input data can be either a pandas dataframe or any table with the
following columns:
["x", "y", "z", "v_x", "v_y", "M_200c"]
Alternatively data can be set to a string indicating the name of
a halo catalog to be loaded. There are various options for the input
string:
"random box" and "random shell" (case insensitive) respectively call
.generate_random_box() and .generate_random_shell() methods with the
default arguments.
"test" generates 6 test halos in the positive and negative x, y, z
directions. This is useful for testing and building prototypes.
Any other string will be looked up as the name of a csv file under
astropaint/data/
e.g. "websky", "MICE", or "Sehgal"
calculate_redshifts: bool
if True, redshifts of objects will be calculated from the comoving
distance according to the latest Planck cosmology (astropy.cosmo.Planck18_arXiv_v2)
This can be numerically expensive for large catalogs so if your
catalog already comes with redshifts, set this to False to save time.
default_redshift: float
If calculate_redshift is set to False, this value will be used as the
default redshift for all the halos.
"""
#TODO: define attribute dictionary with __slots__
self._build_counter = 0
self.calculate_redshifts = calculate_redshifts
# if calculate_redshifts==False, assume this redshift for everything
self.default_redshift = default_redshift
# if no input is provided generate a random catalog
if data is None:
self.data = self._initialize_catalog(1)
#self.generate_random_box()
elif isinstance(data, str):
if re.match(".*random.*box", data, re.IGNORECASE):
self.generate_random_box()
elif re.match(".*random.*shell", data, re.IGNORECASE):
self.generate_random_shell()
elif re.match(".*test.*", data, re.IGNORECASE):
self.generate_test_box(configuration=["all"])
else:
self.load_from_csv(data)
else:
#FIXME: check data type and columns
self.data = data
# .................
# octant signatures
# .................
# (x,y,z) signatures for each octant e.g. (+,+,+) , (+,+,-) etc.
self.octant_signature = self._get_octant_signatures(mode="user")
# same thing but for use in calculations
self._octant_shift_signature = self._get_octant_signatures(mode="shift")
self._octant_mirror_signature = self._get_octant_signatures(mode="mirror")
self._octant_rotate_signature = self._get_octant_signatures(mode="rotate")
# TODO: check input type/columns/etc
# ------------------------
# properties
# ------------------------
@property
def data(self):
return self._data
@data.setter
def data(self, val):
self._data = val
self._data = pd.DataFrame(self.data).reset_index(drop=True)
self.size = len(self._data)
self.box_size = self._get_box_size()
if self._build_counter>0:
print("Catalog data has been modified...\n")
# build the complete data frame
# e.g. angular distances, radii, etc.
self.build_dataframe(calculate_redshifts=self.calculate_redshifts,
default_redshift=self.default_redshift)
# ------------------------
# sample data
# ------------------------
#TODO: support inputs other than csv
def load_from_csv(self, sample_name="MICE"):
"""load sample data using the name of dataset"""
if not sample_name.endswith(".csv"):
sample_name += ".csv"
fname = os.path.join(path_dir, "data", f"{sample_name}")
print(f"Catalog loaded from:\n{fname}")
self.data = pd.read_csv(fname, index_col=0)
def save_to_csv(self, sample_name):
"""load sample data using the name of dataset"""
if not sample_name.endswith(".csv"):
sample_name += ".csv"
fname = os.path.join(path_dir, "data", f"{sample_name}")
self.data.to_csv(fname)
print(f"Catalog saved to:\n{fname}")
def generate_random_box(self,
box_size=50,
v_max=100,
mass_min=1E14,
mass_max=1E15,
n_tot=50000,
put_on_shell=False,
inplace=True,
):
catalog = self._initialize_catalog(n_tot)
print("generating random catalog...\n")
# generate random positions
x, y, z = np.random.uniform(low=-box_size/2,
high=box_size/2,
size=(3, n_tot))
if put_on_shell:
(x, y, z) = box_size * np.true_divide((x, y, z), np.linalg.norm((x, y, z), axis=0))
catalog["x"], catalog["y"], catalog["z"] = x, y, z
# generate random velocities
v_x, v_y, v_z = np.random.uniform(low=-v_max,
high=v_max,
size=(3, n_tot))
catalog["v_x"], catalog["v_y"], catalog["v_z"] = v_x, v_y, v_z
# generate random log uniform masses
catalog["M_200c"] = np.exp(np.random.uniform(low=np.log(mass_min),
high=np.log(mass_max),
size=n_tot))
if inplace:
self.data = pd.DataFrame(catalog)
else:
return pd.DataFrame(catalog) # convert catalog to pandas data frame
def generate_random_shell(self,
shell_radius=50,
v_max=100,
mass_min=1E14,
mass_max=1E15,
n_tot=50000,
inplace=True,
):
catalog = self._initialize_catalog(n_tot)
print("generating random catalog...\n")
# generate random points according to http://mathworld.wolfram.com/SpherePointPicking.html
u,v = np.random.uniform(low=0,
high=1,
size=(2, n_tot))
phi = 2 * np.pi * u
theta = np.arccos(2 * v -1)
# (x, y, z) = box_size * np.true_divide((x, y, z), np.linalg.norm((x, y, z), axis=0))
catalog["x"], catalog["y"], catalog["z"] = np.sin(theta) * np.cos(phi),\
np.sin(theta) * np.sin(phi),\
np.cos(theta)
catalog[["x", "y", "z"]] *= shell_radius
# generate random velocities
v_x, v_y, v_z = np.random.uniform(low=-v_max,
high=v_max,
size=(3, n_tot))
catalog["v_x"], catalog["v_y"], catalog["v_z"] = v_x, v_y, v_z
# generate random log uniform masses
catalog["M_200c"] = np.exp(np.random.uniform(low=np.log(mass_min),
high=np.log(mass_max),
size=n_tot))
if inplace:
self.data = pd.DataFrame(catalog)
else:
return pd.DataFrame(catalog) # convert catalog to pandas data frame
def generate_test_box(self,
configuration=["all"],
distance=100,
mass=1E15,
inplace=True,
):
catalog = pd.DataFrame(self._initialize_catalog(0))
config_dict = {"front": (1, 0, 0),
"back": (-1, 0, 0),
"left": (0, 1, 0),
"right": (0, -1, 0),
"top": (0, 0, 1),
"bottom": (0, 0, -1),
}
# set configuration for "all" keyword
if "all" in configuration:
configuration = config_dict.keys()
for key in configuration:
# get the coordinates from config_dic and load it in a dataframe
x, y, z = config_dict[key]
df = pd.DataFrame(Catalog._initialize_catalog(1))
df["x"], df["y"], df["z"] = x, y, z
df[["x", "y", "z"]] *= distance
# set the mass
df["M_200c"] = mass
# append the test case to the catalog
catalog = catalog.append(df, ignore_index=True)
if inplace:
self.data = pd.DataFrame(catalog)
else:
return | pd.DataFrame(catalog) | pandas.DataFrame |
from __future__ import print_function
from datetime import datetime, timedelta
import numpy as np
import pandas as pd
from pandas import (Series, Index, Int64Index, Timestamp, Period,
DatetimeIndex, PeriodIndex, TimedeltaIndex,
Timedelta, timedelta_range, date_range, Float64Index,
_np_version_under1p10)
import pandas.tslib as tslib
import pandas.tseries.period as period
import pandas.util.testing as tm
from pandas.tests.test_base import Ops
class TestDatetimeIndexOps(Ops):
tz = [None, 'UTC', 'Asia/Tokyo', 'US/Eastern', 'dateutil/Asia/Singapore',
'dateutil/US/Pacific']
def setUp(self):
super(TestDatetimeIndexOps, self).setUp()
mask = lambda x: (isinstance(x, DatetimeIndex) or
isinstance(x, PeriodIndex))
self.is_valid_objs = [o for o in self.objs if mask(o)]
self.not_valid_objs = [o for o in self.objs if not mask(o)]
def test_ops_properties(self):
self.check_ops_properties(
['year', 'month', 'day', 'hour', 'minute', 'second', 'weekofyear',
'week', 'dayofweek', 'dayofyear', 'quarter'])
self.check_ops_properties(['date', 'time', 'microsecond', 'nanosecond',
'is_month_start', 'is_month_end',
'is_quarter_start',
'is_quarter_end', 'is_year_start',
'is_year_end', 'weekday_name'],
lambda x: isinstance(x, DatetimeIndex))
def test_ops_properties_basic(self):
# sanity check that the behavior didn't change
# GH7206
for op in ['year', 'day', 'second', 'weekday']:
self.assertRaises(TypeError, lambda x: getattr(self.dt_series, op))
# attribute access should still work!
s = Series(dict(year=2000, month=1, day=10))
self.assertEqual(s.year, 2000)
self.assertEqual(s.month, 1)
self.assertEqual(s.day, 10)
self.assertRaises(AttributeError, lambda: s.weekday)
def test_asobject_tolist(self):
idx = pd.date_range(start='2013-01-01', periods=4, freq='M',
name='idx')
expected_list = [Timestamp('2013-01-31'),
Timestamp('2013-02-28'),
Timestamp('2013-03-31'),
Timestamp('2013-04-30')]
expected = pd.Index(expected_list, dtype=object, name='idx')
result = idx.asobject
self.assertTrue(isinstance(result, Index))
self.assertEqual(result.dtype, object)
self.assert_index_equal(result, expected)
self.assertEqual(result.name, expected.name)
self.assertEqual(idx.tolist(), expected_list)
idx = pd.date_range(start='2013-01-01', periods=4, freq='M',
name='idx', tz='Asia/Tokyo')
expected_list = [Timestamp('2013-01-31', tz='Asia/Tokyo'),
Timestamp('2013-02-28', tz='Asia/Tokyo'),
Timestamp('2013-03-31', tz='Asia/Tokyo'),
Timestamp('2013-04-30', tz='Asia/Tokyo')]
expected = pd.Index(expected_list, dtype=object, name='idx')
result = idx.asobject
self.assertTrue(isinstance(result, Index))
self.assertEqual(result.dtype, object)
self.assert_index_equal(result, expected)
self.assertEqual(result.name, expected.name)
self.assertEqual(idx.tolist(), expected_list)
idx = DatetimeIndex([datetime(2013, 1, 1), datetime(2013, 1, 2),
pd.NaT, datetime(2013, 1, 4)], name='idx')
expected_list = [Timestamp('2013-01-01'),
Timestamp('2013-01-02'), pd.NaT,
Timestamp('2013-01-04')]
expected = pd.Index(expected_list, dtype=object, name='idx')
result = idx.asobject
self.assertTrue(isinstance(result, Index))
self.assertEqual(result.dtype, object)
self.assert_index_equal(result, expected)
self.assertEqual(result.name, expected.name)
self.assertEqual(idx.tolist(), expected_list)
def test_minmax(self):
for tz in self.tz:
# monotonic
idx1 = pd.DatetimeIndex(['2011-01-01', '2011-01-02',
'2011-01-03'], tz=tz)
self.assertTrue(idx1.is_monotonic)
# non-monotonic
idx2 = pd.DatetimeIndex(['2011-01-01', pd.NaT, '2011-01-03',
'2011-01-02', pd.NaT], tz=tz)
self.assertFalse(idx2.is_monotonic)
for idx in [idx1, idx2]:
self.assertEqual(idx.min(), Timestamp('2011-01-01', tz=tz))
self.assertEqual(idx.max(), Timestamp('2011-01-03', tz=tz))
self.assertEqual(idx.argmin(), 0)
self.assertEqual(idx.argmax(), 2)
for op in ['min', 'max']:
# Return NaT
obj = DatetimeIndex([])
self.assertTrue(pd.isnull(getattr(obj, op)()))
obj = DatetimeIndex([pd.NaT])
self.assertTrue(pd.isnull(getattr(obj, op)()))
obj = DatetimeIndex([pd.NaT, pd.NaT, pd.NaT])
self.assertTrue(pd.isnull(getattr(obj, op)()))
def test_numpy_minmax(self):
dr = pd.date_range(start='2016-01-15', end='2016-01-20')
self.assertEqual(np.min(dr),
Timestamp('2016-01-15 00:00:00', freq='D'))
self.assertEqual(np.max(dr),
Timestamp('2016-01-20 00:00:00', freq='D'))
errmsg = "the 'out' parameter is not supported"
tm.assertRaisesRegexp(ValueError, errmsg, np.min, dr, out=0)
tm.assertRaisesRegexp(ValueError, errmsg, np.max, dr, out=0)
self.assertEqual(np.argmin(dr), 0)
self.assertEqual(np.argmax(dr), 5)
if not _np_version_under1p10:
errmsg = "the 'out' parameter is not supported"
tm.assertRaisesRegexp(ValueError, errmsg, np.argmin, dr, out=0)
tm.assertRaisesRegexp(ValueError, errmsg, np.argmax, dr, out=0)
def test_round(self):
for tz in self.tz:
rng = pd.date_range(start='2016-01-01', periods=5,
freq='30Min', tz=tz)
elt = rng[1]
expected_rng = DatetimeIndex([
Timestamp('2016-01-01 00:00:00', tz=tz, freq='30T'),
Timestamp('2016-01-01 00:00:00', tz=tz, freq='30T'),
Timestamp('2016-01-01 01:00:00', tz=tz, freq='30T'),
Timestamp('2016-01-01 02:00:00', tz=tz, freq='30T'),
Timestamp('2016-01-01 02:00:00', tz=tz, freq='30T'),
])
expected_elt = expected_rng[1]
tm.assert_index_equal(rng.round(freq='H'), expected_rng)
self.assertEqual(elt.round(freq='H'), expected_elt)
msg = pd.tseries.frequencies._INVALID_FREQ_ERROR
with tm.assertRaisesRegexp(ValueError, msg):
rng.round(freq='foo')
with tm.assertRaisesRegexp(ValueError, msg):
elt.round(freq='foo')
msg = "<MonthEnd> is a non-fixed frequency"
tm.assertRaisesRegexp(ValueError, msg, rng.round, freq='M')
tm.assertRaisesRegexp(ValueError, msg, elt.round, freq='M')
def test_repeat_range(self):
rng = date_range('1/1/2000', '1/1/2001')
result = rng.repeat(5)
self.assertIsNone(result.freq)
self.assertEqual(len(result), 5 * len(rng))
for tz in self.tz:
index = pd.date_range('2001-01-01', periods=2, freq='D', tz=tz)
exp = pd.DatetimeIndex(['2001-01-01', '2001-01-01',
'2001-01-02', '2001-01-02'], tz=tz)
for res in [index.repeat(2), np.repeat(index, 2)]:
tm.assert_index_equal(res, exp)
self.assertIsNone(res.freq)
index = pd.date_range('2001-01-01', periods=2, freq='2D', tz=tz)
exp = pd.DatetimeIndex(['2001-01-01', '2001-01-01',
'2001-01-03', '2001-01-03'], tz=tz)
for res in [index.repeat(2), np.repeat(index, 2)]:
tm.assert_index_equal(res, exp)
self.assertIsNone(res.freq)
index = pd.DatetimeIndex(['2001-01-01', 'NaT', '2003-01-01'],
tz=tz)
exp = pd.DatetimeIndex(['2001-01-01', '2001-01-01', '2001-01-01',
'NaT', 'NaT', 'NaT',
'2003-01-01', '2003-01-01', '2003-01-01'],
tz=tz)
for res in [index.repeat(3), np.repeat(index, 3)]:
tm.assert_index_equal(res, exp)
self.assertIsNone(res.freq)
def test_repeat(self):
reps = 2
msg = "the 'axis' parameter is not supported"
for tz in self.tz:
rng = pd.date_range(start='2016-01-01', periods=2,
freq='30Min', tz=tz)
expected_rng = DatetimeIndex([
Timestamp('2016-01-01 00:00:00', tz=tz, freq='30T'),
Timestamp('2016-01-01 00:00:00', tz=tz, freq='30T'),
Timestamp('2016-01-01 00:30:00', tz=tz, freq='30T'),
Timestamp('2016-01-01 00:30:00', tz=tz, freq='30T'),
])
res = rng.repeat(reps)
tm.assert_index_equal(res, expected_rng)
self.assertIsNone(res.freq)
tm.assert_index_equal(np.repeat(rng, reps), expected_rng)
tm.assertRaisesRegexp(ValueError, msg, np.repeat,
rng, reps, axis=1)
def test_representation(self):
idx = []
idx.append(DatetimeIndex([], freq='D'))
idx.append(DatetimeIndex(['2011-01-01'], freq='D'))
idx.append(DatetimeIndex(['2011-01-01', '2011-01-02'], freq='D'))
idx.append(DatetimeIndex(
['2011-01-01', '2011-01-02', '2011-01-03'], freq='D'))
idx.append(DatetimeIndex(
['2011-01-01 09:00', '2011-01-01 10:00', '2011-01-01 11:00'
], freq='H', tz='Asia/Tokyo'))
idx.append(DatetimeIndex(
['2011-01-01 09:00', '2011-01-01 10:00', pd.NaT], tz='US/Eastern'))
idx.append(DatetimeIndex(
['2011-01-01 09:00', '2011-01-01 10:00', pd.NaT], tz='UTC'))
exp = []
exp.append("""DatetimeIndex([], dtype='datetime64[ns]', freq='D')""")
exp.append("DatetimeIndex(['2011-01-01'], dtype='datetime64[ns]', "
"freq='D')")
exp.append("DatetimeIndex(['2011-01-01', '2011-01-02'], "
"dtype='datetime64[ns]', freq='D')")
exp.append("DatetimeIndex(['2011-01-01', '2011-01-02', '2011-01-03'], "
"dtype='datetime64[ns]', freq='D')")
exp.append("DatetimeIndex(['2011-01-01 09:00:00+09:00', "
"'2011-01-01 10:00:00+09:00', '2011-01-01 11:00:00+09:00']"
", dtype='datetime64[ns, Asia/Tokyo]', freq='H')")
exp.append("DatetimeIndex(['2011-01-01 09:00:00-05:00', "
"'2011-01-01 10:00:00-05:00', 'NaT'], "
"dtype='datetime64[ns, US/Eastern]', freq=None)")
exp.append("DatetimeIndex(['2011-01-01 09:00:00+00:00', "
"'2011-01-01 10:00:00+00:00', 'NaT'], "
"dtype='datetime64[ns, UTC]', freq=None)""")
with pd.option_context('display.width', 300):
for indx, expected in zip(idx, exp):
for func in ['__repr__', '__unicode__', '__str__']:
result = getattr(indx, func)()
self.assertEqual(result, expected)
def test_representation_to_series(self):
idx1 = DatetimeIndex([], freq='D')
idx2 = DatetimeIndex(['2011-01-01'], freq='D')
idx3 = DatetimeIndex(['2011-01-01', '2011-01-02'], freq='D')
idx4 = DatetimeIndex(
['2011-01-01', '2011-01-02', '2011-01-03'], freq='D')
idx5 = DatetimeIndex(['2011-01-01 09:00', '2011-01-01 10:00',
'2011-01-01 11:00'], freq='H', tz='Asia/Tokyo')
idx6 = DatetimeIndex(['2011-01-01 09:00', '2011-01-01 10:00', pd.NaT],
tz='US/Eastern')
idx7 = DatetimeIndex(['2011-01-01 09:00', '2011-01-02 10:15'])
exp1 = """Series([], dtype: datetime64[ns])"""
exp2 = """0 2011-01-01
dtype: datetime64[ns]"""
exp3 = """0 2011-01-01
1 2011-01-02
dtype: datetime64[ns]"""
exp4 = """0 2011-01-01
1 2011-01-02
2 2011-01-03
dtype: datetime64[ns]"""
exp5 = """0 2011-01-01 09:00:00+09:00
1 2011-01-01 10:00:00+09:00
2 2011-01-01 11:00:00+09:00
dtype: datetime64[ns, Asia/Tokyo]"""
exp6 = """0 2011-01-01 09:00:00-05:00
1 2011-01-01 10:00:00-05:00
2 NaT
dtype: datetime64[ns, US/Eastern]"""
exp7 = """0 2011-01-01 09:00:00
1 2011-01-02 10:15:00
dtype: datetime64[ns]"""
with pd.option_context('display.width', 300):
for idx, expected in zip([idx1, idx2, idx3, idx4,
idx5, idx6, idx7],
[exp1, exp2, exp3, exp4,
exp5, exp6, exp7]):
result = repr(Series(idx))
self.assertEqual(result, expected)
def test_summary(self):
# GH9116
idx1 = DatetimeIndex([], freq='D')
idx2 = DatetimeIndex(['2011-01-01'], freq='D')
idx3 = DatetimeIndex(['2011-01-01', '2011-01-02'], freq='D')
idx4 = DatetimeIndex(
['2011-01-01', '2011-01-02', '2011-01-03'], freq='D')
idx5 = DatetimeIndex(['2011-01-01 09:00', '2011-01-01 10:00',
'2011-01-01 11:00'],
freq='H', tz='Asia/Tokyo')
idx6 = DatetimeIndex(['2011-01-01 09:00', '2011-01-01 10:00', pd.NaT],
tz='US/Eastern')
exp1 = """DatetimeIndex: 0 entries
Freq: D"""
exp2 = """DatetimeIndex: 1 entries, 2011-01-01 to 2011-01-01
Freq: D"""
exp3 = """DatetimeIndex: 2 entries, 2011-01-01 to 2011-01-02
Freq: D"""
exp4 = """DatetimeIndex: 3 entries, 2011-01-01 to 2011-01-03
Freq: D"""
exp5 = ("DatetimeIndex: 3 entries, 2011-01-01 09:00:00+09:00 "
"to 2011-01-01 11:00:00+09:00\n"
"Freq: H")
exp6 = """DatetimeIndex: 3 entries, 2011-01-01 09:00:00-05:00 to NaT"""
for idx, expected in zip([idx1, idx2, idx3, idx4, idx5, idx6],
[exp1, exp2, exp3, exp4, exp5, exp6]):
result = idx.summary()
self.assertEqual(result, expected)
def test_resolution(self):
for freq, expected in zip(['A', 'Q', 'M', 'D', 'H', 'T',
'S', 'L', 'U'],
['day', 'day', 'day', 'day', 'hour',
'minute', 'second', 'millisecond',
'microsecond']):
for tz in self.tz:
idx = pd.date_range(start='2013-04-01', periods=30, freq=freq,
tz=tz)
self.assertEqual(idx.resolution, expected)
def test_union(self):
for tz in self.tz:
# union
rng1 = pd.date_range('1/1/2000', freq='D', periods=5, tz=tz)
other1 = pd.date_range('1/6/2000', freq='D', periods=5, tz=tz)
expected1 = pd.date_range('1/1/2000', freq='D', periods=10, tz=tz)
rng2 = pd.date_range('1/1/2000', freq='D', periods=5, tz=tz)
other2 = pd.date_range('1/4/2000', freq='D', periods=5, tz=tz)
expected2 = pd.date_range('1/1/2000', freq='D', periods=8, tz=tz)
rng3 = pd.date_range('1/1/2000', freq='D', periods=5, tz=tz)
other3 = pd.DatetimeIndex([], tz=tz)
expected3 = pd.date_range('1/1/2000', freq='D', periods=5, tz=tz)
for rng, other, expected in [(rng1, other1, expected1),
(rng2, other2, expected2),
(rng3, other3, expected3)]:
result_union = rng.union(other)
tm.assert_index_equal(result_union, expected)
def test_add_iadd(self):
for tz in self.tz:
# offset
offsets = [pd.offsets.Hour(2), timedelta(hours=2),
np.timedelta64(2, 'h'), Timedelta(hours=2)]
for delta in offsets:
rng = pd.date_range('2000-01-01', '2000-02-01', tz=tz)
result = rng + delta
expected = pd.date_range('2000-01-01 02:00',
'2000-02-01 02:00', tz=tz)
tm.assert_index_equal(result, expected)
rng += delta
tm.assert_index_equal(rng, expected)
# int
rng = pd.date_range('2000-01-01 09:00', freq='H', periods=10,
tz=tz)
result = rng + 1
expected = pd.date_range('2000-01-01 10:00', freq='H', periods=10,
tz=tz)
tm.assert_index_equal(result, expected)
rng += 1
tm.assert_index_equal(rng, expected)
idx = DatetimeIndex(['2011-01-01', '2011-01-02'])
msg = "cannot add a datelike to a DatetimeIndex"
with tm.assertRaisesRegexp(TypeError, msg):
idx + Timestamp('2011-01-01')
with tm.assertRaisesRegexp(TypeError, msg):
Timestamp('2011-01-01') + idx
def test_add_dti_dti(self):
# previously performed setop (deprecated in 0.16.0), now raises
# TypeError (GH14164)
dti = date_range('20130101', periods=3)
dti_tz = date_range('20130101', periods=3).tz_localize('US/Eastern')
with tm.assertRaises(TypeError):
dti + dti
with tm.assertRaises(TypeError):
dti_tz + dti_tz
with tm.assertRaises(TypeError):
dti_tz + dti
with tm.assertRaises(TypeError):
dti + dti_tz
def test_difference(self):
for tz in self.tz:
# diff
rng1 = pd.date_range('1/1/2000', freq='D', periods=5, tz=tz)
other1 = pd.date_range('1/6/2000', freq='D', periods=5, tz=tz)
expected1 = pd.date_range('1/1/2000', freq='D', periods=5, tz=tz)
rng2 = pd.date_range('1/1/2000', freq='D', periods=5, tz=tz)
other2 = pd.date_range('1/4/2000', freq='D', periods=5, tz=tz)
expected2 = pd.date_range('1/1/2000', freq='D', periods=3, tz=tz)
rng3 = pd.date_range('1/1/2000', freq='D', periods=5, tz=tz)
other3 = pd.DatetimeIndex([], tz=tz)
expected3 = pd.date_range('1/1/2000', freq='D', periods=5, tz=tz)
for rng, other, expected in [(rng1, other1, expected1),
(rng2, other2, expected2),
(rng3, other3, expected3)]:
result_diff = rng.difference(other)
tm.assert_index_equal(result_diff, expected)
def test_sub_isub(self):
for tz in self.tz:
# offset
offsets = [pd.offsets.Hour(2), timedelta(hours=2),
np.timedelta64(2, 'h'), Timedelta(hours=2)]
for delta in offsets:
rng = pd.date_range('2000-01-01', '2000-02-01', tz=tz)
expected = pd.date_range('1999-12-31 22:00',
'2000-01-31 22:00', tz=tz)
result = rng - delta
tm.assert_index_equal(result, expected)
rng -= delta
tm.assert_index_equal(rng, expected)
# int
rng = pd.date_range('2000-01-01 09:00', freq='H', periods=10,
tz=tz)
result = rng - 1
expected = pd.date_range('2000-01-01 08:00', freq='H', periods=10,
tz=tz)
tm.assert_index_equal(result, expected)
rng -= 1
tm.assert_index_equal(rng, expected)
def test_sub_dti_dti(self):
# previously performed setop (deprecated in 0.16.0), now changed to
# return subtraction -> TimeDeltaIndex (GH ...)
dti = date_range('20130101', periods=3)
dti_tz = date_range('20130101', periods=3).tz_localize('US/Eastern')
dti_tz2 = date_range('20130101', periods=3).tz_localize('UTC')
expected = TimedeltaIndex([0, 0, 0])
result = dti - dti
tm.assert_index_equal(result, expected)
result = dti_tz - dti_tz
tm.assert_index_equal(result, expected)
with tm.assertRaises(TypeError):
dti_tz - dti
with tm.assertRaises(TypeError):
dti - dti_tz
with tm.assertRaises(TypeError):
dti_tz - dti_tz2
# isub
dti -= dti
tm.assert_index_equal(dti, expected)
# different length raises ValueError
dti1 = date_range('20130101', periods=3)
dti2 = date_range('20130101', periods=4)
with tm.assertRaises(ValueError):
dti1 - dti2
# NaN propagation
dti1 = DatetimeIndex(['2012-01-01', np.nan, '2012-01-03'])
dti2 = DatetimeIndex(['2012-01-02', '2012-01-03', np.nan])
expected = TimedeltaIndex(['1 days', np.nan, np.nan])
result = dti2 - dti1
tm.assert_index_equal(result, expected)
def test_sub_period(self):
# GH 13078
# not supported, check TypeError
p = pd.Period('2011-01-01', freq='D')
for freq in [None, 'D']:
idx = pd.DatetimeIndex(['2011-01-01', '2011-01-02'], freq=freq)
with tm.assertRaises(TypeError):
idx - p
with tm.assertRaises(TypeError):
p - idx
def test_comp_nat(self):
left = pd.DatetimeIndex([pd.Timestamp('2011-01-01'), pd.NaT,
pd.Timestamp('2011-01-03')])
right = pd.DatetimeIndex([pd.NaT, pd.NaT, pd.Timestamp('2011-01-03')])
for l, r in [(left, right), (left.asobject, right.asobject)]:
result = l == r
expected = np.array([False, False, True])
tm.assert_numpy_array_equal(result, expected)
result = l != r
expected = np.array([True, True, False])
tm.assert_numpy_array_equal(result, expected)
expected = np.array([False, False, False])
tm.assert_numpy_array_equal(l == pd.NaT, expected)
tm.assert_numpy_array_equal(pd.NaT == r, expected)
expected = np.array([True, True, True])
tm.assert_numpy_array_equal(l != pd.NaT, expected)
tm.assert_numpy_array_equal(pd.NaT != l, expected)
expected = np.array([False, False, False])
tm.assert_numpy_array_equal(l < pd.NaT, expected)
tm.assert_numpy_array_equal(pd.NaT > l, expected)
def test_value_counts_unique(self):
# GH 7735
for tz in self.tz:
idx = pd.date_range('2011-01-01 09:00', freq='H', periods=10)
# create repeated values, 'n'th element is repeated by n+1 times
idx = DatetimeIndex(np.repeat(idx.values, range(1, len(idx) + 1)),
tz=tz)
exp_idx = pd.date_range('2011-01-01 18:00', freq='-1H', periods=10,
tz=tz)
expected = Series(range(10, 0, -1), index=exp_idx, dtype='int64')
for obj in [idx, Series(idx)]:
tm.assert_series_equal(obj.value_counts(), expected)
expected = pd.date_range('2011-01-01 09:00', freq='H', periods=10,
tz=tz)
tm.assert_index_equal(idx.unique(), expected)
idx = DatetimeIndex(['2013-01-01 09:00', '2013-01-01 09:00',
'2013-01-01 09:00', '2013-01-01 08:00',
'2013-01-01 08:00', pd.NaT], tz=tz)
exp_idx = DatetimeIndex(['2013-01-01 09:00', '2013-01-01 08:00'],
tz=tz)
expected = Series([3, 2], index=exp_idx)
for obj in [idx, Series(idx)]:
tm.assert_series_equal(obj.value_counts(), expected)
exp_idx = DatetimeIndex(['2013-01-01 09:00', '2013-01-01 08:00',
pd.NaT], tz=tz)
expected = Series([3, 2, 1], index=exp_idx)
for obj in [idx, Series(idx)]:
tm.assert_series_equal(obj.value_counts(dropna=False),
expected)
tm.assert_index_equal(idx.unique(), exp_idx)
def test_nonunique_contains(self):
# GH 9512
for idx in map(DatetimeIndex,
([0, 1, 0], [0, 0, -1], [0, -1, -1],
['2015', '2015', '2016'], ['2015', '2015', '2014'])):
tm.assertIn(idx[0], idx)
def test_order(self):
# with freq
idx1 = DatetimeIndex(['2011-01-01', '2011-01-02',
'2011-01-03'], freq='D', name='idx')
idx2 = DatetimeIndex(['2011-01-01 09:00', '2011-01-01 10:00',
'2011-01-01 11:00'], freq='H',
tz='Asia/Tokyo', name='tzidx')
for idx in [idx1, idx2]:
ordered = idx.sort_values()
self.assert_index_equal(ordered, idx)
self.assertEqual(ordered.freq, idx.freq)
ordered = idx.sort_values(ascending=False)
expected = idx[::-1]
self.assert_index_equal(ordered, expected)
self.assertEqual(ordered.freq, expected.freq)
self.assertEqual(ordered.freq.n, -1)
ordered, indexer = idx.sort_values(return_indexer=True)
self.assert_index_equal(ordered, idx)
self.assert_numpy_array_equal(indexer,
np.array([0, 1, 2]),
check_dtype=False)
self.assertEqual(ordered.freq, idx.freq)
ordered, indexer = idx.sort_values(return_indexer=True,
ascending=False)
expected = idx[::-1]
self.assert_index_equal(ordered, expected)
self.assert_numpy_array_equal(indexer,
np.array([2, 1, 0]),
check_dtype=False)
self.assertEqual(ordered.freq, expected.freq)
self.assertEqual(ordered.freq.n, -1)
# without freq
for tz in self.tz:
idx1 = DatetimeIndex(['2011-01-01', '2011-01-03', '2011-01-05',
'2011-01-02', '2011-01-01'],
tz=tz, name='idx1')
exp1 = DatetimeIndex(['2011-01-01', '2011-01-01', '2011-01-02',
'2011-01-03', '2011-01-05'],
tz=tz, name='idx1')
idx2 = DatetimeIndex(['2011-01-01', '2011-01-03', '2011-01-05',
'2011-01-02', '2011-01-01'],
tz=tz, name='idx2')
exp2 = DatetimeIndex(['2011-01-01', '2011-01-01', '2011-01-02',
'2011-01-03', '2011-01-05'],
tz=tz, name='idx2')
idx3 = DatetimeIndex([pd.NaT, '2011-01-03', '2011-01-05',
'2011-01-02', pd.NaT], tz=tz, name='idx3')
exp3 = DatetimeIndex([pd.NaT, pd.NaT, '2011-01-02', '2011-01-03',
'2011-01-05'], tz=tz, name='idx3')
for idx, expected in [(idx1, exp1), (idx2, exp2), (idx3, exp3)]:
ordered = idx.sort_values()
self.assert_index_equal(ordered, expected)
self.assertIsNone(ordered.freq)
ordered = idx.sort_values(ascending=False)
self.assert_index_equal(ordered, expected[::-1])
self.assertIsNone(ordered.freq)
ordered, indexer = idx.sort_values(return_indexer=True)
self.assert_index_equal(ordered, expected)
exp = np.array([0, 4, 3, 1, 2])
self.assert_numpy_array_equal(indexer, exp, check_dtype=False)
self.assertIsNone(ordered.freq)
ordered, indexer = idx.sort_values(return_indexer=True,
ascending=False)
self.assert_index_equal(ordered, expected[::-1])
exp = np.array([2, 1, 3, 4, 0])
self.assert_numpy_array_equal(indexer, exp, check_dtype=False)
self.assertIsNone(ordered.freq)
def test_getitem(self):
idx1 = pd.date_range('2011-01-01', '2011-01-31', freq='D', name='idx')
idx2 = pd.date_range('2011-01-01', '2011-01-31', freq='D',
tz='Asia/Tokyo', name='idx')
for idx in [idx1, idx2]:
result = idx[0]
self.assertEqual(result, Timestamp('2011-01-01', tz=idx.tz))
result = idx[0:5]
expected = pd.date_range('2011-01-01', '2011-01-05', freq='D',
tz=idx.tz, name='idx')
self.assert_index_equal(result, expected)
self.assertEqual(result.freq, expected.freq)
result = idx[0:10:2]
expected = pd.date_range('2011-01-01', '2011-01-09', freq='2D',
tz=idx.tz, name='idx')
self.assert_index_equal(result, expected)
self.assertEqual(result.freq, expected.freq)
result = idx[-20:-5:3]
expected = pd.date_range('2011-01-12', '2011-01-24', freq='3D',
tz=idx.tz, name='idx')
self.assert_index_equal(result, expected)
self.assertEqual(result.freq, expected.freq)
result = idx[4::-1]
expected = DatetimeIndex(['2011-01-05', '2011-01-04', '2011-01-03',
'2011-01-02', '2011-01-01'],
freq='-1D', tz=idx.tz, name='idx')
self.assert_index_equal(result, expected)
self.assertEqual(result.freq, expected.freq)
def test_drop_duplicates_metadata(self):
# GH 10115
idx = pd.date_range('2011-01-01', '2011-01-31', freq='D', name='idx')
result = idx.drop_duplicates()
self.assert_index_equal(idx, result)
self.assertEqual(idx.freq, result.freq)
idx_dup = idx.append(idx)
self.assertIsNone(idx_dup.freq) # freq is reset
result = idx_dup.drop_duplicates()
self.assert_index_equal(idx, result)
self.assertIsNone(result.freq)
def test_drop_duplicates(self):
# to check Index/Series compat
base = pd.date_range('2011-01-01', '2011-01-31', freq='D', name='idx')
idx = base.append(base[:5])
res = idx.drop_duplicates()
tm.assert_index_equal(res, base)
res = Series(idx).drop_duplicates()
tm.assert_series_equal(res, Series(base))
res = idx.drop_duplicates(keep='last')
exp = base[5:].append(base[:5])
tm.assert_index_equal(res, exp)
res = Series(idx).drop_duplicates(keep='last')
tm.assert_series_equal(res, Series(exp, index=np.arange(5, 36)))
res = idx.drop_duplicates(keep=False)
tm.assert_index_equal(res, base[5:])
res = Series(idx).drop_duplicates(keep=False)
tm.assert_series_equal(res, Series(base[5:], index=np.arange(5, 31)))
def test_take(self):
# GH 10295
idx1 = pd.date_range('2011-01-01', '2011-01-31', freq='D', name='idx')
idx2 = pd.date_range('2011-01-01', '2011-01-31', freq='D',
tz='Asia/Tokyo', name='idx')
for idx in [idx1, idx2]:
result = idx.take([0])
self.assertEqual(result, Timestamp('2011-01-01', tz=idx.tz))
result = idx.take([0, 1, 2])
expected = pd.date_range('2011-01-01', '2011-01-03', freq='D',
tz=idx.tz, name='idx')
self.assert_index_equal(result, expected)
self.assertEqual(result.freq, expected.freq)
result = idx.take([0, 2, 4])
expected = pd.date_range('2011-01-01', '2011-01-05', freq='2D',
tz=idx.tz, name='idx')
self.assert_index_equal(result, expected)
self.assertEqual(result.freq, expected.freq)
result = idx.take([7, 4, 1])
expected = pd.date_range('2011-01-08', '2011-01-02', freq='-3D',
tz=idx.tz, name='idx')
self.assert_index_equal(result, expected)
self.assertEqual(result.freq, expected.freq)
result = idx.take([3, 2, 5])
expected = DatetimeIndex(['2011-01-04', '2011-01-03',
'2011-01-06'],
freq=None, tz=idx.tz, name='idx')
self.assert_index_equal(result, expected)
self.assertIsNone(result.freq)
result = idx.take([-3, 2, 5])
expected = DatetimeIndex(['2011-01-29', '2011-01-03',
'2011-01-06'],
freq=None, tz=idx.tz, name='idx')
self.assert_index_equal(result, expected)
self.assertIsNone(result.freq)
def test_take_invalid_kwargs(self):
idx = pd.date_range('2011-01-01', '2011-01-31', freq='D', name='idx')
indices = [1, 6, 5, 9, 10, 13, 15, 3]
msg = r"take\(\) got an unexpected keyword argument 'foo'"
tm.assertRaisesRegexp(TypeError, msg, idx.take,
indices, foo=2)
msg = "the 'out' parameter is not supported"
tm.assertRaisesRegexp(ValueError, msg, idx.take,
indices, out=indices)
msg = "the 'mode' parameter is not supported"
tm.assertRaisesRegexp(ValueError, msg, idx.take,
indices, mode='clip')
def test_infer_freq(self):
# GH 11018
for freq in ['A', '2A', '-2A', 'Q', '-1Q', 'M', '-1M', 'D', '3D',
'-3D', 'W', '-1W', 'H', '2H', '-2H', 'T', '2T', 'S',
'-3S']:
idx = pd.date_range('2011-01-01 09:00:00', freq=freq, periods=10)
result = pd.DatetimeIndex(idx.asi8, freq='infer')
tm.assert_index_equal(idx, result)
self.assertEqual(result.freq, freq)
def test_nat_new(self):
idx = pd.date_range('2011-01-01', freq='D', periods=5, name='x')
result = idx._nat_new()
exp = pd.DatetimeIndex([pd.NaT] * 5, name='x')
tm.assert_index_equal(result, exp)
result = idx._nat_new(box=False)
exp = np.array([tslib.iNaT] * 5, dtype=np.int64)
tm.assert_numpy_array_equal(result, exp)
def test_shift(self):
# GH 9903
for tz in self.tz:
idx = pd.DatetimeIndex([], name='xxx', tz=tz)
tm.assert_index_equal(idx.shift(0, freq='H'), idx)
tm.assert_index_equal(idx.shift(3, freq='H'), idx)
idx = pd.DatetimeIndex(['2011-01-01 10:00', '2011-01-01 11:00'
'2011-01-01 12:00'], name='xxx', tz=tz)
tm.assert_index_equal(idx.shift(0, freq='H'), idx)
exp = pd.DatetimeIndex(['2011-01-01 13:00', '2011-01-01 14:00'
'2011-01-01 15:00'], name='xxx', tz=tz)
tm.assert_index_equal(idx.shift(3, freq='H'), exp)
exp = pd.DatetimeIndex(['2011-01-01 07:00', '2011-01-01 08:00'
'2011-01-01 09:00'], name='xxx', tz=tz)
tm.assert_index_equal(idx.shift(-3, freq='H'), exp)
def test_nat(self):
self.assertIs(pd.DatetimeIndex._na_value, pd.NaT)
self.assertIs(pd.DatetimeIndex([])._na_value, pd.NaT)
for tz in [None, 'US/Eastern', 'UTC']:
idx = pd.DatetimeIndex(['2011-01-01', '2011-01-02'], tz=tz)
self.assertTrue(idx._can_hold_na)
tm.assert_numpy_array_equal(idx._isnan, np.array([False, False]))
self.assertFalse(idx.hasnans)
tm.assert_numpy_array_equal(idx._nan_idxs,
np.array([], dtype=np.intp))
idx = pd.DatetimeIndex(['2011-01-01', 'NaT'], tz=tz)
self.assertTrue(idx._can_hold_na)
tm.assert_numpy_array_equal(idx._isnan, np.array([False, True]))
self.assertTrue(idx.hasnans)
tm.assert_numpy_array_equal(idx._nan_idxs,
np.array([1], dtype=np.intp))
def test_equals(self):
# GH 13107
for tz in [None, 'UTC', 'US/Eastern', 'Asia/Tokyo']:
idx = pd.DatetimeIndex(['2011-01-01', '2011-01-02', 'NaT'])
self.assertTrue(idx.equals(idx))
self.assertTrue(idx.equals(idx.copy()))
self.assertTrue(idx.equals(idx.asobject))
self.assertTrue(idx.asobject.equals(idx))
self.assertTrue(idx.asobject.equals(idx.asobject))
self.assertFalse(idx.equals(list(idx)))
self.assertFalse(idx.equals(pd.Series(idx)))
idx2 = pd.DatetimeIndex(['2011-01-01', '2011-01-02', 'NaT'],
tz='US/Pacific')
self.assertFalse(idx.equals(idx2))
self.assertFalse(idx.equals(idx2.copy()))
self.assertFalse(idx.equals(idx2.asobject))
self.assertFalse(idx.asobject.equals(idx2))
self.assertFalse(idx.equals(list(idx2)))
self.assertFalse(idx.equals(pd.Series(idx2)))
# same internal, different tz
idx3 = pd.DatetimeIndex._simple_new(idx.asi8, tz='US/Pacific')
tm.assert_numpy_array_equal(idx.asi8, idx3.asi8)
self.assertFalse(idx.equals(idx3))
self.assertFalse(idx.equals(idx3.copy()))
self.assertFalse(idx.equals(idx3.asobject))
self.assertFalse(idx.asobject.equals(idx3))
self.assertFalse(idx.equals(list(idx3)))
self.assertFalse(idx.equals(pd.Series(idx3)))
class TestTimedeltaIndexOps(Ops):
def setUp(self):
super(TestTimedeltaIndexOps, self).setUp()
mask = lambda x: isinstance(x, TimedeltaIndex)
self.is_valid_objs = [o for o in self.objs if mask(o)]
self.not_valid_objs = []
def test_ops_properties(self):
self.check_ops_properties(['days', 'hours', 'minutes', 'seconds',
'milliseconds'])
self.check_ops_properties(['microseconds', 'nanoseconds'])
def test_asobject_tolist(self):
idx = timedelta_range(start='1 days', periods=4, freq='D', name='idx')
expected_list = [Timedelta('1 days'), Timedelta('2 days'),
Timedelta('3 days'), Timedelta('4 days')]
expected = pd.Index(expected_list, dtype=object, name='idx')
result = idx.asobject
self.assertTrue(isinstance(result, Index))
self.assertEqual(result.dtype, object)
self.assert_index_equal(result, expected)
self.assertEqual(result.name, expected.name)
self.assertEqual(idx.tolist(), expected_list)
idx = TimedeltaIndex([timedelta(days=1), timedelta(days=2), pd.NaT,
timedelta(days=4)], name='idx')
expected_list = [Timedelta('1 days'), Timedelta('2 days'), pd.NaT,
Timedelta('4 days')]
expected = pd.Index(expected_list, dtype=object, name='idx')
result = idx.asobject
self.assertTrue(isinstance(result, Index))
self.assertEqual(result.dtype, object)
self.assert_index_equal(result, expected)
self.assertEqual(result.name, expected.name)
self.assertEqual(idx.tolist(), expected_list)
def test_minmax(self):
# monotonic
idx1 = TimedeltaIndex(['1 days', '2 days', '3 days'])
self.assertTrue(idx1.is_monotonic)
# non-monotonic
idx2 = TimedeltaIndex(['1 days', np.nan, '3 days', 'NaT'])
self.assertFalse(idx2.is_monotonic)
for idx in [idx1, idx2]:
self.assertEqual(idx.min(), Timedelta('1 days')),
self.assertEqual(idx.max(), Timedelta('3 days')),
self.assertEqual(idx.argmin(), 0)
self.assertEqual(idx.argmax(), 2)
for op in ['min', 'max']:
# Return NaT
obj = TimedeltaIndex([])
self.assertTrue(pd.isnull(getattr(obj, op)()))
obj = TimedeltaIndex([pd.NaT])
self.assertTrue(pd.isnull(getattr(obj, op)()))
obj = TimedeltaIndex([pd.NaT, pd.NaT, pd.NaT])
self.assertTrue(pd.isnull(getattr(obj, op)()))
def test_numpy_minmax(self):
dr = pd.date_range(start='2016-01-15', end='2016-01-20')
td = TimedeltaIndex(np.asarray(dr))
self.assertEqual(np.min(td), Timedelta('16815 days'))
self.assertEqual(np.max(td), Timedelta('16820 days'))
errmsg = "the 'out' parameter is not supported"
tm.assertRaisesRegexp(ValueError, errmsg, np.min, td, out=0)
tm.assertRaisesRegexp(ValueError, errmsg, np.max, td, out=0)
self.assertEqual(np.argmin(td), 0)
self.assertEqual(np.argmax(td), 5)
if not _np_version_under1p10:
errmsg = "the 'out' parameter is not supported"
tm.assertRaisesRegexp(ValueError, errmsg, np.argmin, td, out=0)
tm.assertRaisesRegexp(ValueError, errmsg, np.argmax, td, out=0)
def test_round(self):
td = pd.timedelta_range(start='16801 days', periods=5, freq='30Min')
elt = td[1]
expected_rng = TimedeltaIndex([
Timedelta('16801 days 00:00:00'),
Timedelta('16801 days 00:00:00'),
Timedelta('16801 days 01:00:00'),
Timedelta('16801 days 02:00:00'),
Timedelta('16801 days 02:00:00'),
])
expected_elt = expected_rng[1]
tm.assert_index_equal(td.round(freq='H'), expected_rng)
self.assertEqual(elt.round(freq='H'), expected_elt)
msg = pd.tseries.frequencies._INVALID_FREQ_ERROR
with self.assertRaisesRegexp(ValueError, msg):
td.round(freq='foo')
with tm.assertRaisesRegexp(ValueError, msg):
elt.round(freq='foo')
msg = "<MonthEnd> is a non-fixed frequency"
tm.assertRaisesRegexp(ValueError, msg, td.round, freq='M')
tm.assertRaisesRegexp(ValueError, msg, elt.round, freq='M')
def test_representation(self):
idx1 = TimedeltaIndex([], freq='D')
idx2 = TimedeltaIndex(['1 days'], freq='D')
idx3 = TimedeltaIndex(['1 days', '2 days'], freq='D')
idx4 = TimedeltaIndex(['1 days', '2 days', '3 days'], freq='D')
idx5 = TimedeltaIndex(['1 days 00:00:01', '2 days', '3 days'])
exp1 = """TimedeltaIndex([], dtype='timedelta64[ns]', freq='D')"""
exp2 = ("TimedeltaIndex(['1 days'], dtype='timedelta64[ns]', "
"freq='D')")
exp3 = ("TimedeltaIndex(['1 days', '2 days'], "
"dtype='timedelta64[ns]', freq='D')")
exp4 = ("TimedeltaIndex(['1 days', '2 days', '3 days'], "
"dtype='timedelta64[ns]', freq='D')")
exp5 = ("TimedeltaIndex(['1 days 00:00:01', '2 days 00:00:00', "
"'3 days 00:00:00'], dtype='timedelta64[ns]', freq=None)")
with pd.option_context('display.width', 300):
for idx, expected in zip([idx1, idx2, idx3, idx4, idx5],
[exp1, exp2, exp3, exp4, exp5]):
for func in ['__repr__', '__unicode__', '__str__']:
result = getattr(idx, func)()
self.assertEqual(result, expected)
def test_representation_to_series(self):
idx1 = TimedeltaIndex([], freq='D')
idx2 = TimedeltaIndex(['1 days'], freq='D')
idx3 = TimedeltaIndex(['1 days', '2 days'], freq='D')
idx4 = TimedeltaIndex(['1 days', '2 days', '3 days'], freq='D')
idx5 = TimedeltaIndex(['1 days 00:00:01', '2 days', '3 days'])
exp1 = """Series([], dtype: timedelta64[ns])"""
exp2 = """0 1 days
dtype: timedelta64[ns]"""
exp3 = """0 1 days
1 2 days
dtype: timedelta64[ns]"""
exp4 = """0 1 days
1 2 days
2 3 days
dtype: timedelta64[ns]"""
exp5 = """0 1 days 00:00:01
1 2 days 00:00:00
2 3 days 00:00:00
dtype: timedelta64[ns]"""
with pd.option_context('display.width', 300):
for idx, expected in zip([idx1, idx2, idx3, idx4, idx5],
[exp1, exp2, exp3, exp4, exp5]):
result = repr(pd.Series(idx))
self.assertEqual(result, expected)
def test_summary(self):
# GH9116
idx1 = TimedeltaIndex([], freq='D')
idx2 = TimedeltaIndex(['1 days'], freq='D')
idx3 = TimedeltaIndex(['1 days', '2 days'], freq='D')
idx4 = TimedeltaIndex(['1 days', '2 days', '3 days'], freq='D')
idx5 = TimedeltaIndex(['1 days 00:00:01', '2 days', '3 days'])
exp1 = """TimedeltaIndex: 0 entries
Freq: D"""
exp2 = """TimedeltaIndex: 1 entries, 1 days to 1 days
Freq: D"""
exp3 = """TimedeltaIndex: 2 entries, 1 days to 2 days
Freq: D"""
exp4 = """TimedeltaIndex: 3 entries, 1 days to 3 days
Freq: D"""
exp5 = ("TimedeltaIndex: 3 entries, 1 days 00:00:01 to 3 days "
"00:00:00")
for idx, expected in zip([idx1, idx2, idx3, idx4, idx5],
[exp1, exp2, exp3, exp4, exp5]):
result = idx.summary()
self.assertEqual(result, expected)
def test_add_iadd(self):
# only test adding/sub offsets as + is now numeric
# offset
offsets = [pd.offsets.Hour(2), timedelta(hours=2),
np.timedelta64(2, 'h'), Timedelta(hours=2)]
for delta in offsets:
rng = timedelta_range('1 days', '10 days')
result = rng + delta
expected = timedelta_range('1 days 02:00:00', '10 days 02:00:00',
freq='D')
tm.assert_index_equal(result, expected)
rng += delta
tm.assert_index_equal(rng, expected)
# int
rng = timedelta_range('1 days 09:00:00', freq='H', periods=10)
result = rng + 1
expected = timedelta_range('1 days 10:00:00', freq='H', periods=10)
tm.assert_index_equal(result, expected)
rng += 1
tm.assert_index_equal(rng, expected)
def test_sub_isub(self):
# only test adding/sub offsets as - is now numeric
# offset
offsets = [pd.offsets.Hour(2), timedelta(hours=2),
np.timedelta64(2, 'h'), Timedelta(hours=2)]
for delta in offsets:
rng = timedelta_range('1 days', '10 days')
result = rng - delta
expected = timedelta_range('0 days 22:00:00', '9 days 22:00:00')
tm.assert_index_equal(result, expected)
rng -= delta
tm.assert_index_equal(rng, expected)
# int
rng = timedelta_range('1 days 09:00:00', freq='H', periods=10)
result = rng - 1
expected = timedelta_range('1 days 08:00:00', freq='H', periods=10)
tm.assert_index_equal(result, expected)
rng -= 1
tm.assert_index_equal(rng, expected)
idx = TimedeltaIndex(['1 day', '2 day'])
msg = "cannot subtract a datelike from a TimedeltaIndex"
with tm.assertRaisesRegexp(TypeError, msg):
idx - Timestamp('2011-01-01')
result = Timestamp('2011-01-01') + idx
expected = DatetimeIndex(['2011-01-02', '2011-01-03'])
tm.assert_index_equal(result, expected)
def test_ops_compat(self):
offsets = [pd.offsets.Hour(2), timedelta(hours=2),
np.timedelta64(2, 'h'), Timedelta(hours=2)]
rng = timedelta_range('1 days', '10 days', name='foo')
# multiply
for offset in offsets:
self.assertRaises(TypeError, lambda: rng * offset)
# divide
expected = Int64Index((np.arange(10) + 1) * 12, name='foo')
for offset in offsets:
result = rng / offset
tm.assert_index_equal(result, expected, exact=False)
# divide with nats
rng = TimedeltaIndex(['1 days', pd.NaT, '2 days'], name='foo')
expected = Float64Index([12, np.nan, 24], name='foo')
for offset in offsets:
result = rng / offset
tm.assert_index_equal(result, expected)
# don't allow division by NaT (make could in the future)
self.assertRaises(TypeError, lambda: rng / pd.NaT)
def test_subtraction_ops(self):
# with datetimes/timedelta and tdi/dti
tdi = TimedeltaIndex(['1 days', pd.NaT, '2 days'], name='foo')
dti = date_range('20130101', periods=3, name='bar')
td = Timedelta('1 days')
dt = Timestamp('20130101')
self.assertRaises(TypeError, lambda: tdi - dt)
self.assertRaises(TypeError, lambda: tdi - dti)
self.assertRaises(TypeError, lambda: td - dt)
self.assertRaises(TypeError, lambda: td - dti)
result = dt - dti
expected = TimedeltaIndex(['0 days', '-1 days', '-2 days'], name='bar')
tm.assert_index_equal(result, expected)
result = dti - dt
expected = TimedeltaIndex(['0 days', '1 days', '2 days'], name='bar')
tm.assert_index_equal(result, expected)
result = tdi - td
expected = TimedeltaIndex(['0 days', pd.NaT, '1 days'], name='foo')
tm.assert_index_equal(result, expected, check_names=False)
result = td - tdi
expected = TimedeltaIndex(['0 days', pd.NaT, '-1 days'], name='foo')
tm.assert_index_equal(result, expected, check_names=False)
result = dti - td
expected = DatetimeIndex(
['20121231', '20130101', '20130102'], name='bar')
tm.assert_index_equal(result, expected, check_names=False)
result = dt - tdi
expected = DatetimeIndex(['20121231', pd.NaT, '20121230'], name='foo')
tm.assert_index_equal(result, expected)
def test_subtraction_ops_with_tz(self):
# check that dt/dti subtraction ops with tz are validated
dti = date_range('20130101', periods=3)
ts = Timestamp('20130101')
dt = ts.to_pydatetime()
dti_tz = date_range('20130101', periods=3).tz_localize('US/Eastern')
ts_tz = Timestamp('20130101').tz_localize('US/Eastern')
ts_tz2 = Timestamp('20130101').tz_localize('CET')
dt_tz = ts_tz.to_pydatetime()
td = Timedelta('1 days')
def _check(result, expected):
self.assertEqual(result, expected)
self.assertIsInstance(result, Timedelta)
# scalars
result = ts - ts
expected = Timedelta('0 days')
_check(result, expected)
result = dt_tz - ts_tz
expected = Timedelta('0 days')
_check(result, expected)
result = ts_tz - dt_tz
expected = Timedelta('0 days')
_check(result, expected)
# tz mismatches
self.assertRaises(TypeError, lambda: dt_tz - ts)
self.assertRaises(TypeError, lambda: dt_tz - dt)
self.assertRaises(TypeError, lambda: dt_tz - ts_tz2)
self.assertRaises(TypeError, lambda: dt - dt_tz)
self.assertRaises(TypeError, lambda: ts - dt_tz)
self.assertRaises(TypeError, lambda: ts_tz2 - ts)
self.assertRaises(TypeError, lambda: ts_tz2 - dt)
self.assertRaises(TypeError, lambda: ts_tz - ts_tz2)
# with dti
self.assertRaises(TypeError, lambda: dti - ts_tz)
self.assertRaises(TypeError, lambda: dti_tz - ts)
self.assertRaises(TypeError, lambda: dti_tz - ts_tz2)
result = dti_tz - dt_tz
expected = TimedeltaIndex(['0 days', '1 days', '2 days'])
tm.assert_index_equal(result, expected)
result = dt_tz - dti_tz
expected = TimedeltaIndex(['0 days', '-1 days', '-2 days'])
tm.assert_index_equal(result, expected)
result = dti_tz - ts_tz
expected = TimedeltaIndex(['0 days', '1 days', '2 days'])
tm.assert_index_equal(result, expected)
result = ts_tz - dti_tz
expected = TimedeltaIndex(['0 days', '-1 days', '-2 days'])
tm.assert_index_equal(result, expected)
result = td - td
expected = Timedelta('0 days')
_check(result, expected)
result = dti_tz - td
expected = DatetimeIndex(
['20121231', '20130101', '20130102'], tz='US/Eastern')
tm.assert_index_equal(result, expected)
def test_dti_tdi_numeric_ops(self):
# These are normally union/diff set-like ops
tdi = TimedeltaIndex(['1 days', pd.NaT, '2 days'], name='foo')
dti = date_range('20130101', periods=3, name='bar')
# TODO(wesm): unused?
# td = Timedelta('1 days')
# dt = Timestamp('20130101')
result = tdi - tdi
expected = TimedeltaIndex(['0 days', pd.NaT, '0 days'], name='foo')
tm.assert_index_equal(result, expected)
result = tdi + tdi
expected = TimedeltaIndex(['2 days', pd.NaT, '4 days'], name='foo')
tm.assert_index_equal(result, expected)
result = dti - tdi # name will be reset
expected = DatetimeIndex(['20121231', pd.NaT, '20130101'])
tm.assert_index_equal(result, expected)
def test_sub_period(self):
# GH 13078
# not supported, check TypeError
p = pd.Period('2011-01-01', freq='D')
for freq in [None, 'H']:
idx = pd.TimedeltaIndex(['1 hours', '2 hours'], freq=freq)
with tm.assertRaises(TypeError):
idx - p
with tm.assertRaises(TypeError):
p - idx
def test_addition_ops(self):
# with datetimes/timedelta and tdi/dti
tdi = TimedeltaIndex(['1 days', pd.NaT, '2 days'], name='foo')
dti = date_range('20130101', periods=3, name='bar')
td = Timedelta('1 days')
dt = Timestamp('20130101')
result = tdi + dt
expected = DatetimeIndex(['20130102', pd.NaT, '20130103'], name='foo')
tm.assert_index_equal(result, expected)
result = dt + tdi
expected = DatetimeIndex(['20130102', pd.NaT, '20130103'], name='foo')
tm.assert_index_equal(result, expected)
result = td + tdi
expected = TimedeltaIndex(['2 days', pd.NaT, '3 days'], name='foo')
tm.assert_index_equal(result, expected)
result = tdi + td
expected = TimedeltaIndex(['2 days', pd.NaT, '3 days'], name='foo')
tm.assert_index_equal(result, expected)
# unequal length
self.assertRaises(ValueError, lambda: tdi + dti[0:1])
self.assertRaises(ValueError, lambda: tdi[0:1] + dti)
# random indexes
self.assertRaises(TypeError, lambda: tdi + Int64Index([1, 2, 3]))
# this is a union!
# self.assertRaises(TypeError, lambda : Int64Index([1,2,3]) + tdi)
result = tdi + dti # name will be reset
expected = DatetimeIndex(['20130102', pd.NaT, '20130105'])
tm.assert_index_equal(result, expected)
result = dti + tdi # name will be reset
expected = DatetimeIndex(['20130102', pd.NaT, '20130105'])
tm.assert_index_equal(result, expected)
result = dt + td
expected = Timestamp('20130102')
self.assertEqual(result, expected)
result = td + dt
expected = Timestamp('20130102')
self.assertEqual(result, expected)
def test_comp_nat(self):
left = pd.TimedeltaIndex([pd.Timedelta('1 days'), pd.NaT,
pd.Timedelta('3 days')])
right = pd.TimedeltaIndex([pd.NaT, pd.NaT, pd.Timedelta('3 days')])
for l, r in [(left, right), (left.asobject, right.asobject)]:
result = l == r
expected = np.array([False, False, True])
tm.assert_numpy_array_equal(result, expected)
result = l != r
expected = np.array([True, True, False])
tm.assert_numpy_array_equal(result, expected)
expected = np.array([False, False, False])
tm.assert_numpy_array_equal(l == pd.NaT, expected)
tm.assert_numpy_array_equal(pd.NaT == r, expected)
expected = np.array([True, True, True])
tm.assert_numpy_array_equal(l != pd.NaT, expected)
tm.assert_numpy_array_equal(pd.NaT != l, expected)
expected = np.array([False, False, False])
tm.assert_numpy_array_equal(l < pd.NaT, expected)
tm.assert_numpy_array_equal(pd.NaT > l, expected)
def test_value_counts_unique(self):
# GH 7735
idx = timedelta_range('1 days 09:00:00', freq='H', periods=10)
# create repeated values, 'n'th element is repeated by n+1 times
idx = TimedeltaIndex(np.repeat(idx.values, range(1, len(idx) + 1)))
exp_idx = timedelta_range('1 days 18:00:00', freq='-1H', periods=10)
expected = Series(range(10, 0, -1), index=exp_idx, dtype='int64')
for obj in [idx, Series(idx)]:
tm.assert_series_equal(obj.value_counts(), expected)
expected = timedelta_range('1 days 09:00:00', freq='H', periods=10)
tm.assert_index_equal(idx.unique(), expected)
idx = TimedeltaIndex(['1 days 09:00:00', '1 days 09:00:00',
'1 days 09:00:00', '1 days 08:00:00',
'1 days 08:00:00', pd.NaT])
exp_idx = TimedeltaIndex(['1 days 09:00:00', '1 days 08:00:00'])
expected = Series([3, 2], index=exp_idx)
for obj in [idx, Series(idx)]:
tm.assert_series_equal(obj.value_counts(), expected)
exp_idx = TimedeltaIndex(['1 days 09:00:00', '1 days 08:00:00',
pd.NaT])
expected = Series([3, 2, 1], index=exp_idx)
for obj in [idx, Series(idx)]:
tm.assert_series_equal(obj.value_counts(dropna=False), expected)
tm.assert_index_equal(idx.unique(), exp_idx)
def test_nonunique_contains(self):
# GH 9512
for idx in map(TimedeltaIndex, ([0, 1, 0], [0, 0, -1], [0, -1, -1],
['00:01:00', '00:01:00', '00:02:00'],
['00:01:00', '00:01:00', '00:00:01'])):
tm.assertIn(idx[0], idx)
def test_unknown_attribute(self):
# GH 9680
tdi = pd.timedelta_range(start=0, periods=10, freq='1s')
ts = pd.Series(np.random.normal(size=10), index=tdi)
self.assertNotIn('foo', ts.__dict__.keys())
self.assertRaises(AttributeError, lambda: ts.foo)
def test_order(self):
# GH 10295
idx1 = TimedeltaIndex(['1 day', '2 day', '3 day'], freq='D',
name='idx')
idx2 = TimedeltaIndex(
['1 hour', '2 hour', '3 hour'], freq='H', name='idx')
for idx in [idx1, idx2]:
ordered = idx.sort_values()
self.assert_index_equal(ordered, idx)
self.assertEqual(ordered.freq, idx.freq)
ordered = idx.sort_values(ascending=False)
expected = idx[::-1]
self.assert_index_equal(ordered, expected)
self.assertEqual(ordered.freq, expected.freq)
self.assertEqual(ordered.freq.n, -1)
ordered, indexer = idx.sort_values(return_indexer=True)
self.assert_index_equal(ordered, idx)
self.assert_numpy_array_equal(indexer,
np.array([0, 1, 2]),
check_dtype=False)
self.assertEqual(ordered.freq, idx.freq)
ordered, indexer = idx.sort_values(return_indexer=True,
ascending=False)
self.assert_index_equal(ordered, idx[::-1])
self.assertEqual(ordered.freq, expected.freq)
self.assertEqual(ordered.freq.n, -1)
idx1 = TimedeltaIndex(['1 hour', '3 hour', '5 hour',
'2 hour ', '1 hour'], name='idx1')
exp1 = TimedeltaIndex(['1 hour', '1 hour', '2 hour',
'3 hour', '5 hour'], name='idx1')
idx2 = TimedeltaIndex(['1 day', '3 day', '5 day',
'2 day', '1 day'], name='idx2')
# TODO(wesm): unused?
# exp2 = TimedeltaIndex(['1 day', '1 day', '2 day',
# '3 day', '5 day'], name='idx2')
# idx3 = TimedeltaIndex([pd.NaT, '3 minute', '5 minute',
# '2 minute', pd.NaT], name='idx3')
# exp3 = TimedeltaIndex([pd.NaT, pd.NaT, '2 minute', '3 minute',
# '5 minute'], name='idx3')
for idx, expected in [(idx1, exp1), (idx1, exp1), (idx1, exp1)]:
ordered = idx.sort_values()
self.assert_index_equal(ordered, expected)
self.assertIsNone(ordered.freq)
ordered = idx.sort_values(ascending=False)
self.assert_index_equal(ordered, expected[::-1])
self.assertIsNone(ordered.freq)
ordered, indexer = idx.sort_values(return_indexer=True)
self.assert_index_equal(ordered, expected)
exp = np.array([0, 4, 3, 1, 2])
self.assert_numpy_array_equal(indexer, exp, check_dtype=False)
self.assertIsNone(ordered.freq)
ordered, indexer = idx.sort_values(return_indexer=True,
ascending=False)
self.assert_index_equal(ordered, expected[::-1])
exp = np.array([2, 1, 3, 4, 0])
self.assert_numpy_array_equal(indexer, exp, check_dtype=False)
self.assertIsNone(ordered.freq)
def test_getitem(self):
idx1 = pd.timedelta_range('1 day', '31 day', freq='D', name='idx')
for idx in [idx1]:
result = idx[0]
self.assertEqual(result, pd.Timedelta('1 day'))
result = idx[0:5]
expected = pd.timedelta_range('1 day', '5 day', freq='D',
name='idx')
self.assert_index_equal(result, expected)
self.assertEqual(result.freq, expected.freq)
result = idx[0:10:2]
expected = pd.timedelta_range('1 day', '9 day', freq='2D',
name='idx')
self.assert_index_equal(result, expected)
self.assertEqual(result.freq, expected.freq)
result = idx[-20:-5:3]
expected = pd.timedelta_range('12 day', '24 day', freq='3D',
name='idx')
self.assert_index_equal(result, expected)
self.assertEqual(result.freq, expected.freq)
result = idx[4::-1]
expected = TimedeltaIndex(['5 day', '4 day', '3 day',
'2 day', '1 day'],
freq='-1D', name='idx')
self.assert_index_equal(result, expected)
self.assertEqual(result.freq, expected.freq)
def test_drop_duplicates_metadata(self):
# GH 10115
idx = pd.timedelta_range('1 day', '31 day', freq='D', name='idx')
result = idx.drop_duplicates()
self.assert_index_equal(idx, result)
self.assertEqual(idx.freq, result.freq)
idx_dup = idx.append(idx)
self.assertIsNone(idx_dup.freq) # freq is reset
result = idx_dup.drop_duplicates()
self.assert_index_equal(idx, result)
self.assertIsNone(result.freq)
def test_drop_duplicates(self):
# to check Index/Series compat
base = pd.timedelta_range('1 day', '31 day', freq='D', name='idx')
idx = base.append(base[:5])
res = idx.drop_duplicates()
tm.assert_index_equal(res, base)
res = Series(idx).drop_duplicates()
tm.assert_series_equal(res, Series(base))
res = idx.drop_duplicates(keep='last')
exp = base[5:].append(base[:5])
tm.assert_index_equal(res, exp)
res = Series(idx).drop_duplicates(keep='last')
tm.assert_series_equal(res, Series(exp, index=np.arange(5, 36)))
res = idx.drop_duplicates(keep=False)
tm.assert_index_equal(res, base[5:])
res = Series(idx).drop_duplicates(keep=False)
tm.assert_series_equal(res, Series(base[5:], index=np.arange(5, 31)))
def test_take(self):
# GH 10295
idx1 = pd.timedelta_range('1 day', '31 day', freq='D', name='idx')
for idx in [idx1]:
result = idx.take([0])
self.assertEqual(result, pd.Timedelta('1 day'))
result = idx.take([-1])
self.assertEqual(result, pd.Timedelta('31 day'))
result = idx.take([0, 1, 2])
expected = pd.timedelta_range('1 day', '3 day', freq='D',
name='idx')
self.assert_index_equal(result, expected)
self.assertEqual(result.freq, expected.freq)
result = idx.take([0, 2, 4])
expected = pd.timedelta_range('1 day', '5 day', freq='2D',
name='idx')
self.assert_index_equal(result, expected)
self.assertEqual(result.freq, expected.freq)
result = idx.take([7, 4, 1])
expected = pd.timedelta_range('8 day', '2 day', freq='-3D',
name='idx')
self.assert_index_equal(result, expected)
self.assertEqual(result.freq, expected.freq)
result = idx.take([3, 2, 5])
expected = TimedeltaIndex(['4 day', '3 day', '6 day'], name='idx')
self.assert_index_equal(result, expected)
self.assertIsNone(result.freq)
result = idx.take([-3, 2, 5])
expected = TimedeltaIndex(['29 day', '3 day', '6 day'], name='idx')
self.assert_index_equal(result, expected)
self.assertIsNone(result.freq)
def test_take_invalid_kwargs(self):
idx = pd.timedelta_range('1 day', '31 day', freq='D', name='idx')
indices = [1, 6, 5, 9, 10, 13, 15, 3]
msg = r"take\(\) got an unexpected keyword argument 'foo'"
tm.assertRaisesRegexp(TypeError, msg, idx.take,
indices, foo=2)
msg = "the 'out' parameter is not supported"
tm.assertRaisesRegexp(ValueError, msg, idx.take,
indices, out=indices)
msg = "the 'mode' parameter is not supported"
tm.assertRaisesRegexp(ValueError, msg, idx.take,
indices, mode='clip')
def test_infer_freq(self):
# GH 11018
for freq in ['D', '3D', '-3D', 'H', '2H', '-2H', 'T', '2T', 'S', '-3S'
]:
idx = pd.timedelta_range('1', freq=freq, periods=10)
result = pd.TimedeltaIndex(idx.asi8, freq='infer')
tm.assert_index_equal(idx, result)
self.assertEqual(result.freq, freq)
def test_nat_new(self):
idx = pd.timedelta_range('1', freq='D', periods=5, name='x')
result = idx._nat_new()
exp = pd.TimedeltaIndex([pd.NaT] * 5, name='x')
tm.assert_index_equal(result, exp)
result = idx._nat_new(box=False)
exp = np.array([tslib.iNaT] * 5, dtype=np.int64)
tm.assert_numpy_array_equal(result, exp)
def test_shift(self):
# GH 9903
idx = pd.TimedeltaIndex([], name='xxx')
tm.assert_index_equal(idx.shift(0, freq='H'), idx)
tm.assert_index_equal(idx.shift(3, freq='H'), idx)
idx = pd.TimedeltaIndex(['5 hours', '6 hours', '9 hours'], name='xxx')
tm.assert_index_equal(idx.shift(0, freq='H'), idx)
exp = pd.TimedeltaIndex(['8 hours', '9 hours', '12 hours'], name='xxx')
tm.assert_index_equal(idx.shift(3, freq='H'), exp)
exp = | pd.TimedeltaIndex(['2 hours', '3 hours', '6 hours'], name='xxx') | pandas.TimedeltaIndex |
"""
Test output formatting for Series/DataFrame, including to_string & reprs
"""
from datetime import datetime
from io import StringIO
import itertools
from operator import methodcaller
import os
from pathlib import Path
import re
from shutil import get_terminal_size
import sys
import textwrap
import dateutil
import numpy as np
import pytest
import pytz
from pandas.compat import (
IS64,
is_platform_windows,
)
import pandas.util._test_decorators as td
import pandas as pd
from pandas import (
DataFrame,
Index,
MultiIndex,
NaT,
Series,
Timestamp,
date_range,
get_option,
option_context,
read_csv,
reset_option,
set_option,
)
import pandas._testing as tm
import pandas.io.formats.format as fmt
import pandas.io.formats.printing as printing
use_32bit_repr = is_platform_windows() or not IS64
@pytest.fixture(params=["string", "pathlike", "buffer"])
def filepath_or_buffer_id(request):
"""
A fixture yielding test ids for filepath_or_buffer testing.
"""
return request.param
@pytest.fixture
def filepath_or_buffer(filepath_or_buffer_id, tmp_path):
"""
A fixture yielding a string representing a filepath, a path-like object
and a StringIO buffer. Also checks that buffer is not closed.
"""
if filepath_or_buffer_id == "buffer":
buf = StringIO()
yield buf
assert not buf.closed
else:
assert isinstance(tmp_path, Path)
if filepath_or_buffer_id == "pathlike":
yield tmp_path / "foo"
else:
yield str(tmp_path / "foo")
@pytest.fixture
def assert_filepath_or_buffer_equals(
filepath_or_buffer, filepath_or_buffer_id, encoding
):
"""
Assertion helper for checking filepath_or_buffer.
"""
def _assert_filepath_or_buffer_equals(expected):
if filepath_or_buffer_id == "string":
with open(filepath_or_buffer, encoding=encoding) as f:
result = f.read()
elif filepath_or_buffer_id == "pathlike":
result = filepath_or_buffer.read_text(encoding=encoding)
elif filepath_or_buffer_id == "buffer":
result = filepath_or_buffer.getvalue()
assert result == expected
return _assert_filepath_or_buffer_equals
def curpath():
pth, _ = os.path.split(os.path.abspath(__file__))
return pth
def has_info_repr(df):
r = repr(df)
c1 = r.split("\n")[0].startswith("<class")
c2 = r.split("\n")[0].startswith(r"<class") # _repr_html_
return c1 or c2
def has_non_verbose_info_repr(df):
has_info = has_info_repr(df)
r = repr(df)
# 1. <class>
# 2. Index
# 3. Columns
# 4. dtype
# 5. memory usage
# 6. trailing newline
nv = len(r.split("\n")) == 6
return has_info and nv
def has_horizontally_truncated_repr(df):
try: # Check header row
fst_line = np.array(repr(df).splitlines()[0].split())
cand_col = np.where(fst_line == "...")[0][0]
except IndexError:
return False
# Make sure each row has this ... in the same place
r = repr(df)
for ix, l in enumerate(r.splitlines()):
if not r.split()[cand_col] == "...":
return False
return True
def has_vertically_truncated_repr(df):
r = repr(df)
only_dot_row = False
for row in r.splitlines():
if re.match(r"^[\.\ ]+$", row):
only_dot_row = True
return only_dot_row
def has_truncated_repr(df):
return has_horizontally_truncated_repr(df) or has_vertically_truncated_repr(df)
def has_doubly_truncated_repr(df):
return has_horizontally_truncated_repr(df) and has_vertically_truncated_repr(df)
def has_expanded_repr(df):
r = repr(df)
for line in r.split("\n"):
if line.endswith("\\"):
return True
return False
@pytest.mark.filterwarnings("ignore::FutureWarning:.*format")
class TestDataFrameFormatting:
def test_eng_float_formatter(self, float_frame):
df = float_frame
df.loc[5] = 0
fmt.set_eng_float_format()
repr(df)
fmt.set_eng_float_format(use_eng_prefix=True)
repr(df)
fmt.set_eng_float_format(accuracy=0)
repr(df)
tm.reset_display_options()
def test_show_null_counts(self):
df = DataFrame(1, columns=range(10), index=range(10))
df.iloc[1, 1] = np.nan
def check(show_counts, result):
buf = StringIO()
df.info(buf=buf, show_counts=show_counts)
assert ("non-null" in buf.getvalue()) is result
with option_context(
"display.max_info_rows", 20, "display.max_info_columns", 20
):
check(None, True)
check(True, True)
check(False, False)
with option_context("display.max_info_rows", 5, "display.max_info_columns", 5):
check(None, False)
check(True, False)
check(False, False)
# GH37999
with tm.assert_produces_warning(
FutureWarning, match="null_counts is deprecated.+"
):
buf = StringIO()
df.info(buf=buf, null_counts=True)
assert "non-null" in buf.getvalue()
# GH37999
with pytest.raises(ValueError, match=r"null_counts used with show_counts.+"):
df.info(null_counts=True, show_counts=True)
def test_repr_truncation(self):
max_len = 20
with option_context("display.max_colwidth", max_len):
df = DataFrame(
{
"A": np.random.randn(10),
"B": [
tm.rands(np.random.randint(max_len - 1, max_len + 1))
for i in range(10)
],
}
)
r = repr(df)
r = r[r.find("\n") + 1 :]
adj = fmt.get_adjustment()
for line, value in zip(r.split("\n"), df["B"]):
if adj.len(value) + 1 > max_len:
assert "..." in line
else:
assert "..." not in line
with option_context("display.max_colwidth", 999999):
assert "..." not in repr(df)
with option_context("display.max_colwidth", max_len + 2):
assert "..." not in repr(df)
def test_repr_deprecation_negative_int(self):
# TODO(2.0): remove in future version after deprecation cycle
# Non-regression test for:
# https://github.com/pandas-dev/pandas/issues/31532
width = get_option("display.max_colwidth")
with | tm.assert_produces_warning(FutureWarning) | pandas._testing.assert_produces_warning |
"""
calculate the option dividend yield per atm strikes per day
- winsorize each strike
- average the strike yield
- boxplot each day yield range
"""
import matplotlib.pyplot as plt
import pandas as pd
import numpy as np
import helpers.step_functions as sf
from scipy.stats import mstats
# import call and put mids
def getLogFile(logFile, startTime="09:30:00.000"):
sdf = pd.read_csv(logFile, index_col=0)
sdf.index = pd.to_datetime(sdf.index).strftime("%H:%M:%S.%f")
sdf = sdf[sdf.index > startTime]
return sdf
def getDividendYield(C, P, K, S, r, tau):
return -1/tau * np.log( (C-P + K*np.exp(-r*tau)) / S)
def getDailyStrikeDividendYield(date, logPath="~/logs/", winsorize=False):
"""
date: 0620 (month-day)
"""
# get strike log
sdf = getLogFile("%s/strikes_%s_1809.csv" %(logPath, date))
# get dividend yield log
div_df = getLogFile("%s/div_yield_%s_1809.csv" %(logPath, date))
rate = div_df.rate.iloc[0]
tau = div_df.tau.iloc[0]
# get spot and add to strike df
spot_sf = sf.makeStepFunction(div_df.index.values, div_df.spot.values)
# print("spot sf ", spot_sf)
sdf.loc[:, "spot"] = spot_sf.getValue(sdf.index.values)
strikes = sdf.strike.unique()
strikes.sort()
# print("strikes ", strikes)
# calc dy per strike
dy_dict = {}
for K in strikes:
df = sdf[sdf.strike == K]
dy = getDividendYield(df.call_mid, df.put_mid, K, df.spot, rate, tau)
dy_dict[K] = dy
dy_df = | pd.DataFrame(dy_dict, index=df.index.values) | pandas.DataFrame |
import datetime
from typing import Any, Dict
import pandas as pd
import pytest
from ruamel.yaml import YAML
from great_expectations.execution_engine.execution_engine import MetricDomainTypes
from great_expectations.rule_based_profiler import RuleBasedProfiler
from great_expectations.rule_based_profiler.config.base import RuleBasedProfilerConfig
from great_expectations.rule_based_profiler.domain_builder import ColumnDomainBuilder
from great_expectations.rule_based_profiler.expectation_configuration_builder import (
DefaultExpectationConfigurationBuilder,
)
from great_expectations.rule_based_profiler.rule import Rule
from great_expectations.rule_based_profiler.types import (
Domain,
ParameterContainer,
ParameterNode,
)
from tests.conftest import skip_if_python_below_minimum_version
yaml = YAML()
# Measure of "closeness" between "actual" and "desired" is computed as: atol + rtol * abs(desired)
# (see "https://numpy.org/doc/stable/reference/generated/numpy.testing.assert_allclose.html" for details).
RTOL: float = 1.0e-7
ATOL: float = 5.0e-2
@pytest.fixture
def pandas_test_df():
skip_if_python_below_minimum_version()
df: pd.DataFrame = pd.DataFrame(
{
"Age": pd.Series(
[
7,
15,
21,
39,
None,
],
dtype="float64",
),
"Date": pd.Series(
[
datetime.date(2020, 12, 31),
datetime.date(2021, 1, 1),
datetime.date(2021, 2, 21),
datetime.date(2021, 3, 20),
None,
],
dtype="object",
),
"Description": pd.Series(
[
"child",
"teenager",
"young adult",
"adult",
None,
],
dtype="object",
),
}
)
df["Date"] = | pd.to_datetime(df["Date"]) | pandas.to_datetime |
# -*- coding: utf-8 -*-
"""
Created on 2018-09-13
@author: <NAME>
"""
import numpy as np
import pandas as pd
CURRENT_ROUND = 38
# Load data from all 2018 rounds
# Data from https://github.com/henriquepgomide/caRtola
rounds = []
rounds.append(pd.read_csv('data/rodada-1.csv'))
rounds.append(pd.read_csv('2018/data/rodada-2.csv'))
rounds.append(pd.read_csv('2018/data/rodada-3.csv'))
rounds.append(pd.read_csv('2018/data/rodada-4.csv'))
rounds.append(pd.read_csv('2018/data/rodada-5.csv'))
rounds.append(pd.read_csv('2018/data/rodada-6.csv'))
rounds.append(pd.read_csv('2018/data/rodada-7.csv'))
rounds.append(pd.read_csv('2018/data/rodada-8.csv'))
rounds.append(pd.read_csv('2018/data/rodada-9.csv'))
rounds.append(pd.read_csv('2018/data/rodada-10.csv'))
rounds.append(pd.read_csv('2018/data/rodada-11.csv'))
rounds.append(pd.read_csv('2018/data/rodada-12.csv'))
rounds.append(pd.read_csv('2018/data/rodada-13.csv'))
rounds.append(pd.read_csv('2018/data/rodada-14.csv'))
rounds.append(pd.read_csv('2018/data/rodada-15.csv'))
rounds.append(pd.read_csv('2018/data/rodada-16.csv'))
rounds.append(pd.read_csv('2018/data/rodada-17.csv'))
rounds.append(pd.read_csv('2018/data/rodada-18.csv'))
rounds.append(pd.read_csv('2018/data/rodada-19.csv'))
rounds.append(pd.read_csv('2018/data/rodada-20.csv'))
rounds.append(pd.read_csv('2018/data/rodada-21.csv'))
rounds.append(pd.read_csv('2018/data/rodada-22.csv'))
rounds.append(pd.read_csv('2018/data/rodada-23.csv'))
rounds.append(pd.read_csv('2018/data/rodada-24.csv'))
rounds.append(pd.read_csv('2018/data/rodada-25.csv'))
rounds.append( | pd.read_csv('2018/data/rodada-26.csv') | pandas.read_csv |
import numpy as np
import pandas as pd
import pickle
def create_distance_matrix():
distance_path = 'distance.csv'
# ids_path = os.path.join(data_path, dataset_name, 'graph_sensor_ids.txt')
# nodes and indexs
# with open(ids_path) as f:
# ids = f.read().strip().split(',')
# # print(ids)
num_ids = 307
id_to_index = {}
index_to_id = {}
ids = []
for i in range(num_ids):
ID = str(i)
ids.append(ID)
id_to_index[ID] = i
index_to_id[i] = ID
# create matrix
dist_matrix = np.zeros((num_ids, num_ids), dtype=np.float32)
# dist_matrix[:] = np.inf
distance = pd.read_csv(distance_path, dtype={'from': 'str', 'to': 'str', 'cost': float})
for row in distance.values:
if row[0] not in ids or row[1] not in ids:
continue
dist_matrix[id_to_index[row[0]], id_to_index[row[1]]] = row[2] / 70000.0 * 60.0
adj = dist_matrix
print(adj.shape)
# # save
# path1 = os.path.join(data_path, dataset_name, 'id_to_index.json')
# path2 = os.path.join(data_path, dataset_name, 'index_to_id.json')
path3 = 'connection.xlsx'
# print(path3)
# with open(path1, 'w') as f:
# json.dump(id_to_index, f)
# with open(path2, 'w') as f:
# json.dump(index_to_id, f)
df = | pd.DataFrame(adj, index=ids, columns=ids) | pandas.DataFrame |
"""
Sleep features.
This file calculates a set of features from the PSG sleep data.
These include:
- Spectral power (with and without adjustement for 1/f)
- Spindles and slow-waves detection
- Slow-waves / spindles phase-amplitude coupling
- Entropy and fractal dimension
Author: Dr <NAME> <<EMAIL>>, UC Berkeley.
Date: March 2021
DANGER: This function has not been extensively debugged and validated.
Use at your own risk.
"""
import mne
import yasa
import logging
import numpy as np
import pandas as pd
import antropy as ant
import scipy.signal as sp_sig
import scipy.stats as sp_stats
logger = logging.getLogger('yasa')
__all__ = ['compute_features_stage']
def compute_features_stage(raw, hypno, max_freq=35, spindles_params=dict(),
sw_params=dict(), do_1f=True):
"""Calculate a set of features for each sleep stage from PSG data.
Features are calculated for N2, N3, NREM (= N2 + N3) and REM sleep.
Parameters
----------
raw : :py:class:`mne.io.BaseRaw`
An MNE Raw instance.
hypno : array_like
Sleep stage (hypnogram). The hypnogram must have the exact same
number of samples as ``data``. To upsample your hypnogram,
please refer to :py:func:`yasa.hypno_upsample_to_data`.
.. note::
The default hypnogram format in YASA is a 1D integer
vector where:
- -2 = Unscored
- -1 = Artefact / Movement
- 0 = Wake
- 1 = N1 sleep
- 2 = N2 sleep
- 3 = N3 sleep
- 4 = REM sleep
max_freq : int
Maximum frequency. This will be used to bandpass-filter the data and
to calculate 1 Hz bins bandpower.
kwargs_sp : dict
Optional keywords arguments that are passed to the
:py:func:`yasa.spindles_detect` function. We strongly recommend
adapting the thresholds to your population (e.g. more liberal for
older adults).
kwargs_sw : dict
Optional keywords arguments that are passed to the
:py:func:`yasa.sw_detect` function. We strongly recommend
adapting the thresholds to your population (e.g. more liberal for
older adults).
Returns
-------
feature : pd.DataFrame
A long-format dataframe with stage and channel as index and
all the calculated metrics as columns.
"""
# #########################################################################
# 1) PREPROCESSING
# #########################################################################
# Safety checks
assert isinstance(max_freq, int), "`max_freq` must be int."
assert isinstance(raw, mne.io.BaseRaw), "`raw` must be a MNE Raw object."
assert isinstance(spindles_params, dict)
assert isinstance(sw_params, dict)
# Define 1 Hz bins frequency bands for bandpower
# Similar to [(0.5, 1, "0.5-1"), (1, 2, "1-2"), ..., (34, 35, "34-35")]
bands = []
freqs = [0.5] + list(range(1, max_freq + 1))
for i, b in enumerate(freqs[:-1]):
bands.append(tuple((b, freqs[i + 1], "%s-%s" % (b, freqs[i + 1]))))
# Append traditional bands
bands_classic = [
(0.5, 1, 'slowdelta'), (1, 4, 'fastdelta'), (0.5, 4, 'delta'),
(4, 8, 'theta'), (8, 12, 'alpha'), (12, 16, 'sigma'), (16, 30, 'beta'),
(30, max_freq, 'gamma')]
bands = bands_classic + bands
# Find min and maximum frequencies. These will be used for bandpass-filter
# and 1/f adjustement of bandpower. l_freq = 0.5 / h_freq = 35 Hz.
all_freqs_sorted = np.sort(np.unique(
[b[0] for b in bands] + [b[1] for b in bands]))
l_freq = all_freqs_sorted[0]
h_freq = all_freqs_sorted[-1]
# Mapping dictionnary integer to string for sleep stages (2 --> N2)
stage_mapping = {
-2: 'Unscored',
-1: 'Artefact',
0: 'Wake',
1: 'N1',
2: 'N2',
3: 'N3',
4: 'REM',
6: 'NREM',
7: 'WN' # Whole night = N2 + N3 + REM
}
# Hypnogram check + calculate NREM hypnogram
hypno = np.asarray(hypno, dtype=int)
assert hypno.ndim == 1, 'Hypno must be one dimensional.'
unique_hypno = np.unique(hypno)
logger.info('Number of unique values in hypno = %i', unique_hypno.size)
# IMPORTANT: NREM is defined as N2 + N3, excluding N1 sleep.
hypno_NREM = pd.Series(hypno).replace({2: 6, 3: 6}).to_numpy()
minutes_of_NREM = (hypno_NREM == 6).sum() / (60 * raw.info['sfreq'])
# WN = Whole night = N2 + N3 + REM (excluding N1)
hypno_WN = | pd.Series(hypno) | pandas.Series |
import pandas as pd
from openpyxl import Workbook
import cx_Oracle
import sys
from sqlalchemy import create_engine
from PyQt6 import QtCore, QtGui, QtWidgets
import ctypes
import time
import threading
import qdarktheme
import cgitb
cgitb.enable(format = 'text')
dsn_tns = cx_Oracle.makedsn('ip-banco-oracle', 'porta', service_name='nomedoservico')
conn = cx_Oracle.connect(user=r'usuario', password='<PASSWORD>', dsn=dsn_tns)
c = conn.cursor()
engine = create_engine('sqlite://', echo=False)
class Ui_ConferenciadeNotas(object):
def setupUi(self, ConferenciadeNotas):
ConferenciadeNotas.setObjectName("ConferenciadeNotas")
ConferenciadeNotas.resize(868, 650)
ConferenciadeNotas.setWindowIcon(QtGui.QIcon("icone.ico"))
self.localArquivo = QtWidgets.QTextEdit(ConferenciadeNotas)
self.localArquivo.setGeometry(QtCore.QRect(100, 60, 590, 30))
self.localArquivo.setObjectName("localArquivo")
self.label = QtWidgets.QLabel(ConferenciadeNotas)
self.label.setGeometry(QtCore.QRect(0, 0, 870, 40))
font = QtGui.QFont()
font.setFamily("Century Gothic")
font.setPointSize(18)
self.label.setFont(font)
self.label.setAlignment(QtCore.Qt.AlignmentFlag.AlignCenter)
self.label.setObjectName("label")
self.label_2 = QtWidgets.QLabel(ConferenciadeNotas)
self.label_2.setGeometry(QtCore.QRect(10, 60, 90, 30))
font = QtGui.QFont()
font.setFamily("Century Gothic")
font.setPointSize(16)
font.setBold(False)
font.setWeight(50)
self.label_2.setFont(font)
self.label_2.setAlignment(QtCore.Qt.AlignmentFlag.AlignLeading|QtCore.Qt.AlignmentFlag.AlignLeft|QtCore.Qt.AlignmentFlag.AlignVCenter)
self.label_2.setObjectName("label_2")
self.localizarArquivoBT = QtWidgets.QPushButton(ConferenciadeNotas)
self.localizarArquivoBT.setGeometry(QtCore.QRect(700, 60, 160, 30))
font = QtGui.QFont()
font.setFamily("Century Gothic")
font.setPointSize(12)
self.localizarArquivoBT.setFont(font)
self.localizarArquivoBT.setObjectName("localizarArquivoBT")
self.localizarArquivoBT.clicked.connect(self.locArquivo)
self.conferidoFiliais = QtWidgets.QTableWidget(ConferenciadeNotas)
self.conferidoFiliais.setGeometry(QtCore.QRect(20, 130, 180, 440))
font = QtGui.QFont()
font.setFamily("Century Gothic")
self.conferidoFiliais.setFont(font)
self.conferidoFiliais.setRowCount(16)
self.conferidoFiliais.setObjectName("conferidoFiliais")
self.conferidoFiliais.setColumnCount(3)
item = QtWidgets.QTableWidgetItem()
item.setTextAlignment(QtCore.Qt.AlignmentFlag.AlignCenter)
self.conferidoFiliais.setVerticalHeaderItem(0, item)
item = QtWidgets.QTableWidgetItem()
self.conferidoFiliais.setVerticalHeaderItem(1, item)
item = QtWidgets.QTableWidgetItem()
self.conferidoFiliais.setVerticalHeaderItem(2, item)
item = QtWidgets.QTableWidgetItem()
self.conferidoFiliais.setVerticalHeaderItem(3, item)
item = QtWidgets.QTableWidgetItem()
self.conferidoFiliais.setVerticalHeaderItem(4, item)
item = QtWidgets.QTableWidgetItem()
self.conferidoFiliais.setVerticalHeaderItem(5, item)
item = QtWidgets.QTableWidgetItem()
self.conferidoFiliais.setVerticalHeaderItem(6, item)
item = QtWidgets.QTableWidgetItem()
self.conferidoFiliais.setVerticalHeaderItem(7, item)
item = QtWidgets.QTableWidgetItem()
self.conferidoFiliais.setVerticalHeaderItem(8, item)
item = QtWidgets.QTableWidgetItem()
self.conferidoFiliais.setVerticalHeaderItem(9, item)
item = QtWidgets.QTableWidgetItem()
self.conferidoFiliais.setVerticalHeaderItem(10, item)
item = QtWidgets.QTableWidgetItem()
self.conferidoFiliais.setVerticalHeaderItem(11, item)
item = QtWidgets.QTableWidgetItem()
self.conferidoFiliais.setVerticalHeaderItem(12, item)
item = QtWidgets.QTableWidgetItem()
self.conferidoFiliais.setVerticalHeaderItem(13, item)
item = QtWidgets.QTableWidgetItem()
self.conferidoFiliais.setVerticalHeaderItem(14, item)
item = QtWidgets.QTableWidgetItem()
self.conferidoFiliais.setVerticalHeaderItem(15, item)
item = QtWidgets.QTableWidgetItem()
item.setTextAlignment(QtCore.Qt.AlignmentFlag.AlignCenter)
self.conferidoFiliais.setHorizontalHeaderItem(0, item)
item = QtWidgets.QTableWidgetItem()
item.setTextAlignment(QtCore.Qt.AlignmentFlag.AlignCenter)
self.conferidoFiliais.setHorizontalHeaderItem(1, item)
item = QtWidgets.QTableWidgetItem()
item.setTextAlignment(QtCore.Qt.AlignmentFlag.AlignCenter)
self.conferidoFiliais.setHorizontalHeaderItem(2, item)
item = QtWidgets.QTableWidgetItem()
self.conferidoFiliais.setItem(0, 0, item)
item = QtWidgets.QTableWidgetItem()
font = QtGui.QFont()
font.setKerning(True)
item.setFont(font)
item.setCheckState(QtCore.Qt.CheckState.Unchecked)
self.conferidoFiliais.setItem(0, 1, item)
item = QtWidgets.QTableWidgetItem()
item.setCheckState(QtCore.Qt.CheckState.Unchecked)
self.conferidoFiliais.setItem(0, 2, item)
item = QtWidgets.QTableWidgetItem()
self.conferidoFiliais.setItem(1, 0, item)
item = QtWidgets.QTableWidgetItem()
item.setTextAlignment(QtCore.Qt.AlignmentFlag.AlignCenter)
item.setCheckState(QtCore.Qt.CheckState.Unchecked)
self.conferidoFiliais.setItem(1, 1, item)
item = QtWidgets.QTableWidgetItem()
item.setCheckState(QtCore.Qt.CheckState.Unchecked)
self.conferidoFiliais.setItem(1, 2, item)
item = QtWidgets.QTableWidgetItem()
self.conferidoFiliais.setItem(2, 0, item)
item = QtWidgets.QTableWidgetItem()
item.setCheckState(QtCore.Qt.CheckState.Unchecked)
self.conferidoFiliais.setItem(2, 1, item)
item = QtWidgets.QTableWidgetItem()
item.setCheckState(QtCore.Qt.CheckState.Unchecked)
self.conferidoFiliais.setItem(2, 2, item)
item = QtWidgets.QTableWidgetItem()
self.conferidoFiliais.setItem(3, 0, item)
item = QtWidgets.QTableWidgetItem()
item.setCheckState(QtCore.Qt.CheckState.Unchecked)
self.conferidoFiliais.setItem(3, 1, item)
item = QtWidgets.QTableWidgetItem()
item.setCheckState(QtCore.Qt.CheckState.Unchecked)
self.conferidoFiliais.setItem(3, 2, item)
item = QtWidgets.QTableWidgetItem()
self.conferidoFiliais.setItem(4, 0, item)
item = QtWidgets.QTableWidgetItem()
item.setCheckState(QtCore.Qt.CheckState.Unchecked)
self.conferidoFiliais.setItem(4, 1, item)
item = QtWidgets.QTableWidgetItem()
item.setCheckState(QtCore.Qt.CheckState.Unchecked)
self.conferidoFiliais.setItem(4, 2, item)
item = QtWidgets.QTableWidgetItem()
self.conferidoFiliais.setItem(5, 0, item)
item = QtWidgets.QTableWidgetItem()
item.setCheckState(QtCore.Qt.CheckState.Unchecked)
self.conferidoFiliais.setItem(5, 1, item)
item = QtWidgets.QTableWidgetItem()
item.setCheckState(QtCore.Qt.CheckState.Unchecked)
self.conferidoFiliais.setItem(5, 2, item)
item = QtWidgets.QTableWidgetItem()
self.conferidoFiliais.setItem(6, 0, item)
item = QtWidgets.QTableWidgetItem()
item.setCheckState(QtCore.Qt.CheckState.Unchecked)
self.conferidoFiliais.setItem(6, 1, item)
item = QtWidgets.QTableWidgetItem()
item.setCheckState(QtCore.Qt.CheckState.Unchecked)
self.conferidoFiliais.setItem(6, 2, item)
item = QtWidgets.QTableWidgetItem()
self.conferidoFiliais.setItem(7, 0, item)
item = QtWidgets.QTableWidgetItem()
item.setCheckState(QtCore.Qt.CheckState.Unchecked)
self.conferidoFiliais.setItem(7, 1, item)
item = QtWidgets.QTableWidgetItem()
item.setCheckState(QtCore.Qt.CheckState.Unchecked)
self.conferidoFiliais.setItem(7, 2, item)
item = QtWidgets.QTableWidgetItem()
self.conferidoFiliais.setItem(8, 0, item)
item = QtWidgets.QTableWidgetItem()
item.setCheckState(QtCore.Qt.CheckState.Unchecked)
self.conferidoFiliais.setItem(8, 1, item)
item = QtWidgets.QTableWidgetItem()
item.setCheckState(QtCore.Qt.CheckState.Unchecked)
self.conferidoFiliais.setItem(8, 2, item)
item = QtWidgets.QTableWidgetItem()
self.conferidoFiliais.setItem(9, 0, item)
item = QtWidgets.QTableWidgetItem()
item.setCheckState(QtCore.Qt.CheckState.Unchecked)
self.conferidoFiliais.setItem(9, 1, item)
item = QtWidgets.QTableWidgetItem()
item.setCheckState(QtCore.Qt.CheckState.Unchecked)
self.conferidoFiliais.setItem(9, 2, item)
item = QtWidgets.QTableWidgetItem()
self.conferidoFiliais.setItem(10, 0, item)
item = QtWidgets.QTableWidgetItem()
item.setCheckState(QtCore.Qt.CheckState.Unchecked)
self.conferidoFiliais.setItem(10, 1, item)
item = QtWidgets.QTableWidgetItem()
item.setCheckState(QtCore.Qt.CheckState.Unchecked)
self.conferidoFiliais.setItem(10, 2, item)
item = QtWidgets.QTableWidgetItem()
self.conferidoFiliais.setItem(11, 0, item)
item = QtWidgets.QTableWidgetItem()
item.setCheckState(QtCore.Qt.CheckState.Unchecked)
self.conferidoFiliais.setItem(11, 1, item)
item = QtWidgets.QTableWidgetItem()
item.setCheckState(QtCore.Qt.CheckState.Unchecked)
self.conferidoFiliais.setItem(11, 2, item)
item = QtWidgets.QTableWidgetItem()
self.conferidoFiliais.setItem(12, 0, item)
item = QtWidgets.QTableWidgetItem()
item.setCheckState(QtCore.Qt.CheckState.Unchecked)
self.conferidoFiliais.setItem(12, 1, item)
item = QtWidgets.QTableWidgetItem()
item.setCheckState(QtCore.Qt.CheckState.Unchecked)
self.conferidoFiliais.setItem(12, 2, item)
item = QtWidgets.QTableWidgetItem()
self.conferidoFiliais.setItem(13, 0, item)
item = QtWidgets.QTableWidgetItem()
item.setCheckState(QtCore.Qt.CheckState.Unchecked)
self.conferidoFiliais.setItem(13, 1, item)
item = QtWidgets.QTableWidgetItem()
item.setCheckState(QtCore.Qt.CheckState.Unchecked)
self.conferidoFiliais.setItem(13, 2, item)
item = QtWidgets.QTableWidgetItem()
self.conferidoFiliais.setItem(14, 0, item)
item = QtWidgets.QTableWidgetItem()
item.setCheckState(QtCore.Qt.CheckState.Unchecked)
self.conferidoFiliais.setItem(14, 1, item)
item = QtWidgets.QTableWidgetItem()
item.setCheckState(QtCore.Qt.CheckState.Unchecked)
self.conferidoFiliais.setItem(14, 2, item)
item = QtWidgets.QTableWidgetItem()
self.conferidoFiliais.setItem(15, 0, item)
item = QtWidgets.QTableWidgetItem()
item.setCheckState(QtCore.Qt.CheckState.Unchecked)
self.conferidoFiliais.setItem(15, 1, item)
item = QtWidgets.QTableWidgetItem()
item.setCheckState(QtCore.Qt.CheckState.Unchecked)
self.conferidoFiliais.setItem(15, 2, item)
self.conferidoFiliais.horizontalHeader().setDefaultSectionSize(50)
self.conferidoFiliais.horizontalHeader().setMinimumSectionSize(50)
self.conferidoFiliais.verticalHeader().setDefaultSectionSize(23)
self.conferidoFiliais.verticalHeader().setMinimumSectionSize(23)
self.nfsComErro = QtWidgets.QTableWidget(ConferenciadeNotas)
self.nfsComErro.setGeometry(QtCore.QRect(200, 130, 651, 440))
font = QtGui.QFont()
font.setFamily("Century Gothic")
self.nfsComErro.setFont(font)
#self.nfsComErro.setRowCount(100)
self.nfsComErro.setObjectName("nfsComErro")
self.nfsComErro.setColumnCount(6)
item = QtWidgets.QTableWidgetItem()
self.nfsComErro.setHorizontalHeaderItem(0, item)
item = QtWidgets.QTableWidgetItem()
self.nfsComErro.setHorizontalHeaderItem(1, item)
item = QtWidgets.QTableWidgetItem()
self.nfsComErro.setHorizontalHeaderItem(2, item)
item = QtWidgets.QTableWidgetItem()
self.nfsComErro.setHorizontalHeaderItem(3, item)
item = QtWidgets.QTableWidgetItem()
self.nfsComErro.setHorizontalHeaderItem(4, item)
item = QtWidgets.QTableWidgetItem()
self.nfsComErro.setHorizontalHeaderItem(5, item)
self.nfsComErro.setSelectionMode(QtWidgets.QAbstractItemView.SelectionMode.ExtendedSelection)
self.nfsComErro.setSelectionBehavior(QtWidgets.QAbstractItemView.SelectionBehavior.SelectItems)
self.label_3 = QtWidgets.QLabel(ConferenciadeNotas)
self.label_3.setGeometry(QtCore.QRect(0, 100, 870, 20))
font = QtGui.QFont()
font.setFamily("Century Gothic")
font.setPointSize(16)
self.label_3.setFont(font)
self.label_3.setAlignment(QtCore.Qt.AlignmentFlag.AlignCenter)
self.label_3.setObjectName("label_3")
self.exportResult = QtWidgets.QPushButton(ConferenciadeNotas)
self.exportResult.setGeometry(QtCore.QRect(703, 600, 150, 30))
font = QtGui.QFont()
font.setFamily("Century Gothic")
font.setPointSize(12)
self.exportResult.setFont(font)
self.exportResult.setObjectName("exportResult")
self.exportResult.setText('Exportar')
self.exportResult.clicked.connect(self.exportExcel)
self.retranslateUi(ConferenciadeNotas)
QtCore.QMetaObject.connectSlotsByName(ConferenciadeNotas)
self.rows = 0
self.conferidoFiliais.horizontalHeader().setStretchLastSection(True)
self.nfsComErro.horizontalHeader().setStretchLastSection(True)
self.conferidoFiliais.horizontalHeader().setStyleSheet(""" QHeaderView::section {padding-left: 2;
padding-right: -10;
}""")
self.nfsComErro.horizontalHeader().setStyleSheet(""" QHeaderView::section {padding-left: 2;
padding-right: -10;
}""")
def retranslateUi(self, ConferenciadeNotas):
_translate = QtCore.QCoreApplication.translate
ConferenciadeNotas.setWindowTitle(_translate("ConferenciadeNotas", "Conferência de Notas CIGAMxSEFAZ"))
self.label.setText(_translate("ConferenciadeNotas", "Conferência de Notas CIGAM x SEFAZ"))
self.label_2.setText(_translate("ConferenciadeNotas", "Arquivo:"))
self.localizarArquivoBT.setText(_translate("ConferenciadeNotas", "Localizar Arquivo"))
item = self.conferidoFiliais.verticalHeaderItem(0)
item.setText(_translate("ConferenciadeNotas", " "))
item = self.conferidoFiliais.verticalHeaderItem(1)
item.setText(_translate("ConferenciadeNotas", " "))
item = self.conferidoFiliais.verticalHeaderItem(2)
item.setText(_translate("ConferenciadeNotas", " "))
item = self.conferidoFiliais.verticalHeaderItem(3)
item.setText(_translate("ConferenciadeNotas", " "))
item = self.conferidoFiliais.verticalHeaderItem(4)
item.setText(_translate("ConferenciadeNotas", " "))
item = self.conferidoFiliais.verticalHeaderItem(5)
item.setText(_translate("ConferenciadeNotas", " "))
item = self.conferidoFiliais.verticalHeaderItem(6)
item.setText(_translate("ConferenciadeNotas", " "))
item = self.conferidoFiliais.verticalHeaderItem(7)
item.setText(_translate("ConferenciadeNotas", " "))
item = self.conferidoFiliais.verticalHeaderItem(8)
item.setText(_translate("ConferenciadeNotas", " "))
item = self.conferidoFiliais.verticalHeaderItem(9)
item.setText(_translate("ConferenciadeNotas", " "))
item = self.conferidoFiliais.verticalHeaderItem(10)
item.setText(_translate("ConferenciadeNotas", " "))
item = self.conferidoFiliais.verticalHeaderItem(11)
item.setText(_translate("ConferenciadeNotas", " "))
item = self.conferidoFiliais.verticalHeaderItem(12)
item.setText(_translate("ConferenciadeNotas", " "))
item = self.conferidoFiliais.verticalHeaderItem(13)
item.setText(_translate("ConferenciadeNotas", " "))
item = self.conferidoFiliais.verticalHeaderItem(14)
item.setText(_translate("ConferenciadeNotas", " "))
item = self.conferidoFiliais.verticalHeaderItem(15)
item.setText(_translate("ConferenciadeNotas", " "))
item = self.conferidoFiliais.horizontalHeaderItem(0)
item.setText(_translate("ConferenciadeNotas", "UN"))
item = self.conferidoFiliais.horizontalHeaderItem(1)
item.setText(_translate("ConferenciadeNotas", "NFE"))
item = self.conferidoFiliais.horizontalHeaderItem(2)
item.setText(_translate("ConferenciadeNotas", "NFCE"))
__sortingEnabled = self.conferidoFiliais.isSortingEnabled()
self.conferidoFiliais.setSortingEnabled(False)
item = self.conferidoFiliais.item(0, 0)
item.setText(_translate("ConferenciadeNotas", "001"))
item = self.conferidoFiliais.item(1, 0)
item.setText(_translate("ConferenciadeNotas", "002"))
item = self.conferidoFiliais.item(2, 0)
item.setText(_translate("ConferenciadeNotas", "003"))
item = self.conferidoFiliais.item(3, 0)
item.setText(_translate("ConferenciadeNotas", "004"))
item = self.conferidoFiliais.item(4, 0)
item.setText(_translate("ConferenciadeNotas", "005"))
item = self.conferidoFiliais.item(5, 0)
item.setText(_translate("ConferenciadeNotas", "006"))
item = self.conferidoFiliais.item(6, 0)
item.setText(_translate("ConferenciadeNotas", "007"))
item = self.conferidoFiliais.item(7, 0)
item.setText(_translate("ConferenciadeNotas", "008"))
item = self.conferidoFiliais.item(8, 0)
item.setText(_translate("ConferenciadeNotas", "009"))
item = self.conferidoFiliais.item(9, 0)
item.setText(_translate("ConferenciadeNotas", "010"))
item = self.conferidoFiliais.item(10, 0)
item.setText(_translate("ConferenciadeNotas", "011"))
item = self.conferidoFiliais.item(11, 0)
item.setText(_translate("ConferenciadeNotas", "013"))
item = self.conferidoFiliais.item(12, 0)
item.setText(_translate("ConferenciadeNotas", "014"))
item = self.conferidoFiliais.item(13, 0)
item.setText(_translate("ConferenciadeNotas", "016"))
item = self.conferidoFiliais.item(14, 0)
item.setText(_translate("ConferenciadeNotas", "100"))
item = self.conferidoFiliais.item(15, 0)
item.setText(_translate("ConferenciadeNotas", "200"))
self.conferidoFiliais.setSortingEnabled(__sortingEnabled)
item = self.nfsComErro.horizontalHeaderItem(0)
item.setText(_translate("ConferenciadeNotas", "UN"))
item = self.nfsComErro.horizontalHeaderItem(1)
item.setText(_translate("ConferenciadeNotas", "SERIE"))
item = self.nfsComErro.horizontalHeaderItem(2)
item.setText(_translate("ConferenciadeNotas", "NOTA"))
item = self.nfsComErro.horizontalHeaderItem(3)
item.setText(_translate("ConferenciadeNotas", "DATA"))
item = self.nfsComErro.horizontalHeaderItem(4)
item.setText(_translate("ConferenciadeNotas", "SITUACAO"))
item = self.nfsComErro.horizontalHeaderItem(5)
item.setText(_translate("ConferenciadeNotas", "TEM"))
self.label_3.setText(_translate("ConferenciadeNotas", "Unidade: Série: Data: até "))
def locArquivo(self):
arquivoLocal = QtWidgets.QFileDialog.getOpenFileNames(filter='*.xls')[0]
if (arquivoLocal == []):
def Mbox(title, text, style):
return ctypes.windll.user32.MessageBoxW(0, text, title, style)
Mbox('Erro arquivo', 'Arquivo não localizado ou invalido!', 0)
for files in arquivoLocal:
self.localArquivo.setText(' ')
self.localArquivo.setText(files)
self.file = files
df = pd.read_excel(self.file, skiprows=lambda x: x not in list(range(6, 9999)))
sqlSerie = " SELECT DISTINCT(A.SERIE) FROM (select CASE WHEN [SÉRIE] = '3' THEN 'NFE' WHEN [SÉRIE] = '7' THEN 'NFCE' WHEN [SÉRIE] = '8' THEN '2NFCE' ELSE 'NFCE' END AS SERIE \
FROM NFSEFAZ) A "
try:
df.to_sql('NFSEFAZ', engine, if_exists='replace', index=False)
except:
pass
def Mbox(title, text, style):
return ctypes.windll.user32.MessageBoxW(0, text, title, style)
Mbox('Erro arquivo', 'Arquivo '+ self.file + ' invalido, favor verificar!', 0)
try:
serieDf = engine.execute(sqlSerie)
except:
pass
def Mbox(title, text, style):
return ctypes.windll.user32.MessageBoxW(0, text, title, style)
Mbox('Erro arquivo', 'Arquivo '+ self.file + ' invalido, favor verificar!', 0)
serieFim = pd.DataFrame(serieDf, columns=['SERIE'])
self.serieTxt = serieFim.iloc[0]['SERIE']
try:
self.serieTxt2 = serieFim.iloc[1]['SERIE']
except:
pass
self.serieTxt2 = serieFim.iloc[0]['SERIE']
if(self.serieTxt in ['NFCE','2NFCE']):
file = self.file
dff = pd.read_excel(file, skiprows=lambda x: x not in list(range(0, 6)))
dff.to_sql('NFCESEFAZ', engine, if_exists='replace', index=False)
ie_un = engine.execute('SELECT REPLACE(SUBSTR("SECRETARIA DE ESTADO DE FAZENDA",21,10),"-","") FROM NFCESEFAZ WHERE "SECRETARIA DE ESTADO DE FAZENDA" LIKE "%INSCRIÇÃO ESTADUAL%"')
ie_un = ie_un.first()[0]
df = pd.read_excel(file, skiprows=lambda x: x not in list(range(6, 9999)))
sqlsefaz = (" select CASE WHEN {} = 130241750 THEN '001' \
WHEN {} = 131817086 THEN '002'\
WHEN {} = 131838245 THEN '003'\
WHEN {} = 131875523 THEN '004'\
WHEN {} = 131980203 THEN '005'\
WHEN {} = 132009412 THEN '006'\
WHEN {} = 132894939 THEN '007'\
WHEN {} = 132702371 THEN '008'\
WHEN {} = 133644065 THEN '009'\
WHEN {} = 131537326 THEN '010'\
WHEN {} = 133446565 THEN '011'\
WHEN {} = 132124726 THEN '013'\
WHEN {} = 133779416 THEN '014'\
WHEN {} = 133830900 THEN '016'\
WHEN {} = 133762033 THEN '100'\
WHEN {} = 131847031 THEN '200' ELSE {} END AS UN,\
CASE WHEN [SÉRIE] = '3' THEN 'NFE' WHEN [SÉRIE] = '7' THEN 'NFCE' WHEN [SÉRIE] = '8' THEN '2NFCE' ELSE 'NFCE' END AS SERIE,\
[NUMERO NOTA FISCAL] as NF, SUBSTR([DATA EMISSÃO],0,11) as DT_NF, \
CASE WHEN upper([SITUAÇÃO]) = 'CANCELADA FORA DO PRAZO' THEN 'CANCELADA' \
WHEN upper([SITUAÇÃO]) = 'AUTORIZADA FORA PRAZO' THEN 'AUTORIZADA' ELSE upper([SITUAÇÃO]) END AS SITUACAO\
FROM NFSEFAZ ").format(ie_un, ie_un, ie_un, ie_un, ie_un, ie_un, ie_un, ie_un, ie_un, ie_un,
ie_un, ie_un, ie_un, ie_un, ie_un, ie_un, ie_un)
df.to_sql('NFSEFAZ', engine, if_exists='replace', index=False)
results = engine.execute(sqlsefaz)
final = pd.DataFrame(results, columns=['UN', 'SERIE', 'NF', 'DT_NF', 'SITUACAO'])
final.to_sql('NOTASSEFAZ', engine, if_exists='replace', index=False)
dt_inicio = engine.execute('SELECT MIN(SUBSTR([DATA EMISSÃO],0,11)) FROM NFSEFAZ')
dt_fim = engine.execute('SELECT MAX(SUBSTR([DATA EMISSÃO],0,11)) FROM NFSEFAZ')
un_neg = engine.execute('SELECT distinct(UN) FROM NOTASSEFAZ')
serie_nf = engine.execute('SELECT distinct(SERIE) FROM NOTASSEFAZ')
dt_inicio = dt_inicio.first()[0]
dt_fim = dt_fim.first()[0]
un_neg = un_neg.first()[0]
#serie_nf = [dict(row) for row in serie_nf]
list_serie = []
for row in serie_nf:
list_serie.append(row[0])
list_serie = str(list_serie)[1:-1]
self.label_3.setText("Unidade: " + un_neg + " Série: " + list_serie.replace("'",'').replace(",",' e') + " Data: " + dt_inicio + " até " + dt_fim)
#self.dtLabel["text"] = " Unidade: "+ un_neg + " Série: " + self.serieTxt + " Data: "+ dt_inicio+ " até " + dt_fim
sql = ("""SELECT F.CD_UNIDADE_DE_N,\
F.SERIE,F.NF,TO_CHAR(F.DT_EMISSAO, 'DD/MM/YYYY') AS DT,\
CASE WHEN F.ESPECIE_NOTA = 'S' THEN 'AUTORIZADA' \
WHEN F.ESPECIE_NOTA = 'N' THEN 'CANCELADA' \
WHEN F.ESPECIE_NOTA = 'E' THEN 'AUTORIZADA' \
END AS STATUS \
FROM FANFISCA F \
WHERE F.SERIE in ({}) \
AND F.CD_UNIDADE_DE_N = '{}' \
AND F.DT_EMISSAO BETWEEN '{}' AND '{}' \
""").format(list_serie, un_neg, dt_inicio, dt_fim)
nfbanco = pd.read_sql(sql, conn)
nfbanco.to_sql('NFCIGAM', engine, if_exists='replace', index=False)
comparaNfSefaz = engine.execute(" SELECT S.*,'SEFAZ' AS TEM FROM NOTASSEFAZ S LEFT JOIN NFCIGAM C ON (S.UN = C.CD_UNIDADE_DE_N AND S.SERIE = C.SERIE AND S.NF = C.NF) WHERE C.NF IS NULL")
resultComparaNfSefaz = pd.DataFrame(comparaNfSefaz, columns=['UN', 'SERIE', 'NOTA', 'DATA', 'SITUACAO', 'TEM'])
comparaNfCigam = engine.execute(" SELECT C.*,'CIGAM' AS TEM FROM NFCIGAM C LEFT JOIN NOTASSEFAZ S ON ( C.CD_UNIDADE_DE_N = S.UN AND C.SERIE = S.SERIE AND C.NF = S.NF) WHERE S.NF IS NULL")
resultComparaNfCigam = pd.DataFrame(comparaNfCigam, columns=['UN', 'SERIE', 'NOTA', 'DATA', 'SITUACAO', 'TEM'])
comparaNfCigamXSefaz = engine.execute( " SELECT C.CD_UNIDADE_DE_N,C.SERIE,C.NF,C.DT,C.STATUS || ' x ' || S.SITUACAO,'CIGAM e SEFAZ' as TEM FROM NFCIGAM C INNER JOIN NOTASSEFAZ S ON ( C.CD_UNIDADE_DE_N = S.UN AND C.SERIE = S.SERIE AND C.NF = S.NF) WHERE S.SITUACAO <> C.STATUS")
resultComparaNfCigamXSefaz = pd.DataFrame(comparaNfCigamXSefaz, columns=['UN', 'SERIE', 'NOTA', 'DATA', 'SITUACAO','TEM'])
for index, row in resultComparaNfSefaz.iterrows():
#print(row[0])
self.nfsComErro.setRowCount(self.rows+1)
self.nfsComErro.setItem(self.rows, 0, QtWidgets.QTableWidgetItem(str(row["UN"])))
self.nfsComErro.setItem(self.rows, 1, QtWidgets.QTableWidgetItem(str(row["SERIE"])))
self.nfsComErro.setItem(self.rows, 2, QtWidgets.QTableWidgetItem(str(row["NOTA"])))
self.nfsComErro.setItem(self.rows, 3, QtWidgets.QTableWidgetItem(str(row["DATA"])))
self.nfsComErro.setItem(self.rows, 4, QtWidgets.QTableWidgetItem(str(row["SITUACAO"])))
self.nfsComErro.setItem(self.rows, 5, QtWidgets.QTableWidgetItem(str(row["TEM"])))
self.rows=self.rows+1
for index, row in resultComparaNfCigam.iterrows():
#print(row[0])
self.nfsComErro.setRowCount(self.rows+1)
self.nfsComErro.setItem(self.rows, 0, QtWidgets.QTableWidgetItem(str(row["UN"])))
self.nfsComErro.setItem(self.rows, 1, QtWidgets.QTableWidgetItem(str(row["SERIE"])))
self.nfsComErro.setItem(self.rows, 2, QtWidgets.QTableWidgetItem(str(row["NOTA"])))
self.nfsComErro.setItem(self.rows, 3, QtWidgets.QTableWidgetItem(str(row["DATA"])))
self.nfsComErro.setItem(self.rows, 4, QtWidgets.QTableWidgetItem(str(row["SITUACAO"])))
self.nfsComErro.setItem(self.rows, 5, QtWidgets.QTableWidgetItem(str(row["TEM"])))
self.rows=self.rows+1
for index, row in resultComparaNfCigamXSefaz.iterrows():
#print(row[0])
self.nfsComErro.setRowCount(self.rows+1)
self.nfsComErro.setItem(self.rows, 0, QtWidgets.QTableWidgetItem(str(row["UN"])))
self.nfsComErro.setItem(self.rows, 1, QtWidgets.QTableWidgetItem(str(row["SERIE"])))
self.nfsComErro.setItem(self.rows, 2, QtWidgets.QTableWidgetItem(str(row["NOTA"])))
self.nfsComErro.setItem(self.rows, 3, QtWidgets.QTableWidgetItem(str(row["DATA"])))
self.nfsComErro.setItem(self.rows, 4, QtWidgets.QTableWidgetItem(str(row["SITUACAO"])))
self.nfsComErro.setItem(self.rows, 5, QtWidgets.QTableWidgetItem(str(row["TEM"])))
self.rows=self.rows+1
item = QtWidgets.QTableWidgetItem()
item.setCheckState(QtCore.Qt.CheckState.Checked)
if(un_neg == '001'):
self.conferidoFiliais.setItem(0, 2, item)
if(un_neg == '002'):
self.conferidoFiliais.setItem(1, 2, item)
if(un_neg == '003'):
self.conferidoFiliais.setItem(2, 2, item)
if(un_neg == '004'):
self.conferidoFiliais.setItem(3, 2, item)
if(un_neg == '005'):
self.conferidoFiliais.setItem(4, 2, item)
if(un_neg == '006'):
self.conferidoFiliais.setItem(5, 2, item)
if(un_neg == '007'):
self.conferidoFiliais.setItem(6, 2, item)
if(un_neg == '008'):
self.conferidoFiliais.setItem(7, 2, item)
if(un_neg == '009'):
self.conferidoFiliais.setItem(8, 2, item)
if(un_neg == '010'):
self.conferidoFiliais.setItem(9, 2, item)
if(un_neg == '011'):
self.conferidoFiliais.setItem(10, 2, item)
if(un_neg == '013'):
self.conferidoFiliais.setItem(11, 2, item)
if(un_neg == '014'):
self.conferidoFiliais.setItem(12, 2, item)
if(un_neg == '016'):
self.conferidoFiliais.setItem(13, 2, item)
if(un_neg == '100'):
self.conferidoFiliais.setItem(14, 2, item)
if(un_neg == '200'):
self.conferidoFiliais.setItem(15, 2, item)
def worker(title, close_until_seconds):
time.sleep(close_until_seconds)
wd = ctypes.windll.user32.FindWindowW(0, title)
ctypes.windll.user32.SendMessageW(wd, 0x0010, 0, 0)
return
def AutoCloseMessageBoxW(text, title, close_until_seconds):
t = threading.Thread(target=worker, args=(title, close_until_seconds))
t.start()
ctypes.windll.user32.MessageBoxW(0, text, title, 0)
AutoCloseMessageBoxW('Conferido NFCe UN:'+un_neg, 'NFCe Conferida', 0.5)
#print(resultComparaNfSefaz, "\n", "\n", resultComparaNfCigam, "\n", "\n", resultComparaNfCigamXSefaz)
if (self.serieTxt == 'NFE'):
file = self.file
df = pd.read_excel(file, skiprows=lambda x: x not in list(range(6, 9999)))
sqlsefaz = " select CASE WHEN [INSCRIÇÃO ESTADUAL] = '130241750' THEN '001' \
WHEN [INSCRIÇÃO ESTADUAL] = '131817086' THEN '002'\
WHEN [INSCRIÇÃO ESTADUAL] = '131838245' THEN '003'\
WHEN [INSCRIÇÃO ESTADUAL] = '131875523' THEN '004'\
WHEN [INSCRIÇÃO ESTADUAL] = '131980203' THEN '005'\
WHEN [INSCRIÇÃO ESTADUAL] = '132009412' THEN '006'\
WHEN [INSCRIÇÃO ESTADUAL] = '132894939' THEN '007'\
WHEN [INSCRIÇÃO ESTADUAL] = '132702371' THEN '008'\
WHEN [INSCRIÇÃO ESTADUAL] = '133644065' THEN '009'\
WHEN [INSCRIÇÃO ESTADUAL] = '131537326' THEN '010'\
WHEN [INSCRIÇÃO ESTADUAL] = '133446565' THEN '011'\
WHEN [INSCRIÇÃO ESTADUAL] = '132124726' THEN '013'\
WHEN [INSCRIÇÃO ESTADUAL] = '133779416' THEN '014'\
WHEN [INSCRIÇÃO ESTADUAL] = '133830900' THEN '016'\
WHEN [INSCRIÇÃO ESTADUAL] = '133762033' THEN '100'\
WHEN [INSCRIÇÃO ESTADUAL] = '131847031' THEN '200' ELSE [INSCRIÇÃO ESTADUAL] END AS UN,\
CASE WHEN [SÉRIE] = '3' THEN 'NFE' WHEN [SÉRIE] = '7' THEN 'NFCE' WHEN [SÉRIE] = '8' THEN '2NFCE' ELSE 'NFCE' END AS SERIE,\
[NUMERO NOTA FISCAL] as NF, SUBSTR([DATA EMISSÃO],0,11) as DT_NF, \
CASE WHEN [SITUAÇÃO] = 'CANCELADA FORA DO PRAZO' THEN 'CANCELADA' \
WHEN [SITUAÇÃO] = 'AUTORIZADA FORA DO PRAZO' THEN 'AUTORIZADA' \
ELSE [SITUAÇÃO]\
END AS SITUACAO\
FROM NFSEFAZ "
df.to_sql('NFSEFAZ', engine, if_exists='replace', index=False)
results = engine.execute(sqlsefaz)
final = pd.DataFrame(results, columns=['UN', 'SERIE', 'NF', 'DT_NF', 'SITUACAO'])
final.to_sql('NOTASSEFAZ', engine, if_exists='replace', index=False)
dt_inicio = engine.execute('SELECT MIN(SUBSTR([DATA EMISSÃO],0,11)) FROM NFSEFAZ')
dt_fim = engine.execute('SELECT MAX(SUBSTR([DATA EMISSÃO],0,11)) FROM NFSEFAZ')
un_neg = engine.execute('SELECT distinct(UN) FROM NOTASSEFAZ')
serie_nf = engine.execute('SELECT distinct(SERIE) FROM NOTASSEFAZ')
dt_inicio = dt_inicio.first()[0]
dt_fim = dt_fim.first()[0]
un_neg = un_neg.first()[0]
serie_nf = serie_nf.first()[0]
self.label_3.setText("Unidade: " + un_neg + " Série: " + serie_nf + " Data: " + dt_inicio + " até " + dt_fim)
#self.dtLabel["text"] = " Unidade: " + un_neg + " Série: " + self.serieTxt + " Data: " + dt_inicio + " até " + dt_fim
sql = ("""SELECT F.CD_UNIDADE_DE_N,\
F.SERIE,F.NF,TO_CHAR(F.DT_EMISSAO, 'DD/MM/YYYY') AS DT,\
CASE WHEN F.ESPECIE_NOTA = 'S' THEN 'AUTORIZADA' \
WHEN F.ESPECIE_NOTA = 'N' THEN 'CANCELADA' \
WHEN F.ESPECIE_NOTA = 'E' THEN 'AUTORIZADA' \
END AS STATUS \
FROM FANFISCA F \
WHERE F.SERIE = '{}' \
AND F.CD_UNIDADE_DE_N = '{}' \
AND F.DT_EMISSAO BETWEEN '{}' AND '{}' \
""").format(serie_nf, un_neg, dt_inicio, dt_fim)
nfbanco = pd.read_sql(sql, conn)
nfbanco.to_sql('NFCIGAM', engine, if_exists='replace', index=False)
comparaNfSefaz = engine.execute(" SELECT S.*,'SEFAZ' AS TEM FROM NOTASSEFAZ S LEFT JOIN NFCIGAM C ON (S.UN = C.CD_UNIDADE_DE_N AND S.SERIE = C.SERIE AND S.NF = C.NF) WHERE C.NF IS NULL")
resultComparaNfSefaz = pd.DataFrame(comparaNfSefaz, columns=['UN', 'SERIE', 'NOTA', 'DATA', 'SITUACAO', 'TEM'])
comparaNfCigam = engine.execute(" SELECT C.*,'CIGAM' AS TEM FROM NFCIGAM C LEFT JOIN NOTASSEFAZ S ON ( C.CD_UNIDADE_DE_N = S.UN AND C.SERIE = S.SERIE AND C.NF = S.NF) WHERE S.NF IS NULL")
resultComparaNfCigam = | pd.DataFrame(comparaNfCigam, columns=['UN', 'SERIE', 'NOTA', 'DATA', 'SITUACAO', 'TEM']) | pandas.DataFrame |
import biom
import skbio
import numpy as np
import pandas as pd
from deicode.matrix_completion import MatrixCompletion
from deicode.preprocessing import rclr
from deicode._rpca_defaults import (DEFAULT_RANK, DEFAULT_MSC, DEFAULT_MFC,
DEFAULT_ITERATIONS)
from scipy.linalg import svd
def rpca(table: biom.Table,
n_components: int = DEFAULT_RANK,
min_sample_count: int = DEFAULT_MSC,
min_feature_count: int = DEFAULT_MFC,
max_iterations: int = DEFAULT_ITERATIONS) -> (
skbio.OrdinationResults,
skbio.DistanceMatrix):
"""Runs RPCA with an rclr preprocessing step.
This code will be run by both the standalone and QIIME 2 versions of
DEICODE.
"""
# filter sample to min depth
def sample_filter(val, id_, md): return sum(val) > min_sample_count
def observation_filter(val, id_, md): return sum(val) > min_feature_count
# filter and import table
table = table.filter(observation_filter, axis='observation')
table = table.filter(sample_filter, axis='sample')
table = table.to_dataframe().T
if len(table.index) != len(set(table.index)):
raise ValueError('Data-table contains duplicate indices')
if len(table.columns) != len(set(table.columns)):
raise ValueError('Data-table contains duplicate columns')
# rclr preprocessing and OptSpace (RPCA)
opt = MatrixCompletion(n_components=n_components,
max_iterations=max_iterations).fit(rclr(table))
rename_cols = ['PC' + str(i+1) for i in range(n_components)]
X = opt.sample_weights @ opt.s @ opt.feature_weights.T
X = X - X.mean(axis=0)
X = X - X.mean(axis=1).reshape(-1, 1)
u, s, v = svd(X)
u = u[:, :n_components]
v = v.T[:, :n_components]
p = s**2 / np.sum(s**2)
p = p[:n_components]
s = s[:n_components]
feature_loading = pd.DataFrame(v, index=table.columns, columns=rename_cols)
sample_loading = | pd.DataFrame(u, index=table.index, columns=rename_cols) | pandas.DataFrame |
from flask import Flask, render_template, jsonify, request
import pandas as pd
import pickle
import os
import sklearn
from sklearn import preprocessing
app = Flask(__name__)
@app.route("/")
def home():
return render_template("index.html")
@app.route('/predict', methods=['POST'])
def predict():
json = request.json
weather_df = | pd.DataFrame.from_dict(json, orient='index') | pandas.DataFrame.from_dict |
import pandas
from enum import Enum
# urls of CSV, from which the tickers will be extracted
_NYSE_URL = 'https://old.nasdaq.com/screening/companies-by-name.aspx?letter=0&exchange=nyse&render=download'
_NASDAQ_URL = 'https://old.nasdaq.com/screening/companies-by-name.aspx?letter=0&exchange=nasdaq&render=download'
_AMEX_URL = 'https://old.nasdaq.com/screening/companies-by-name.aspx?letter=0&exchange=amex&render=download'
_URL_LIST = [_NYSE_URL, _NASDAQ_URL, _AMEX_URL]
class Region(Enum):
AFRICA = 'AFRICA'
EUROPE = 'EUROPE'
ASIA = 'ASIA'
AUSTRALIA_SOUTH_PACIFIC = 'AUSTRALIA+AND+SOUTH+PACIFIC'
CARIBBEAN = 'CARIBBEAN'
SOUTH_AMERICA = 'SOUTH+AMERICA'
MIDDLE_EAST = 'MIDDLE+EAST'
NORTH_AMERICA = 'NORTH+AMERICA'
# get tickers from chosen exchanges (default all) as a list
def get_tickers(NYSE=True, NASDAQ=True, AMEX=True):
tickers_list = []
if NYSE:
tickers_list.extend(__url2list(_NYSE_URL))
if NASDAQ:
tickers_list.extend(__url2list(_NASDAQ_URL))
if AMEX:
tickers_list.extend(__url2list(_AMEX_URL))
return tickers_list
def get_tickers_filtered(mktcap_min=None, mktcap_max=None):
tickers_list = []
for url in _URL_LIST:
tickers_list.extend(__url2list_filtered(url, mktcap_min=mktcap_min, mktcap_max=mktcap_max))
return tickers_list
def get_tickers_by_region(region):
if region in Region:
return __url2list(f'https://old.nasdaq.com/screening/'
f'companies-by-region.aspx?region={region.value}&render=download')
else:
raise ValueError('Please enter a valid region (use a Region.REGION as the argument, e.g. Region.AFRICA)')
def __url2list(url):
df = | pandas.read_csv(url) | pandas.read_csv |
import json
import multiprocessing
import warnings
from pathlib import PurePosixPath, Path
from typing import Optional, List, Tuple, Dict, Union
import numpy as np
import pandas as pd
from joblib._multiprocessing_helpers import mp
from rdkit import Chem
from rdkit.Chem import AllChem, Mol, MACCSkeys
from sklearn.feature_extraction import DictVectorizer
from sklearn.metrics import roc_auc_score
from tqdm import tqdm
from datasets.utils import process_map
def _init_molecule(molecule: Union[str, Mol, bytes]) -> Mol:
if isinstance(molecule, bytes):
mol = Mol(molecule)
elif isinstance(molecule, Mol):
mol = molecule
else:
mol = Chem.MolFromSmiles(molecule)
return mol
class ECFC_featurizer():
"""based on the implemenation provided by <NAME>"""
def __init__(self, radius=6, min_fragm_occur=50, useChirality=True, useFeatures=False):
self.v = DictVectorizer(sparse=True, dtype=np.uint16)
self.min_fragm_occur = min_fragm_occur
self.idx_col = None
self.radius = radius
self.useChirality = useChirality
self.useFeatures = useFeatures
def compute_fp_list(self, smiles_list):
fp_list = []
for smiles in smiles_list:
try:
if isinstance(smiles, list):
smiles = smiles[0]
mol = Chem.MolFromSmiles(smiles) # TODO small hack only applicable here!!!
fp_list.append(AllChem.GetMorganFingerprint(mol, self.radius, useChirality=self.useChirality,
useFeatures=self.useFeatures).GetNonzeroElements()) # returns a dict
except:
fp_list.append({})
return fp_list
def fit(self, x_train):
fp_list = self.compute_fp_list(x_train)
Xraw = self.v.fit_transform(fp_list)
idx_col = np.array((Xraw > 0).sum(axis=0) >= self.min_fragm_occur).flatten()
self.idx_col = idx_col
return Xraw[:, self.idx_col].toarray()
def transform(self, x_test):
fp_list = self.compute_fp_list(x_test)
X_raw = self.v.transform(fp_list)
return X_raw[:, self.idx_col].toarray()
class ECFPFeaturizer():
def __init__(self,
radius: int = 2,
fold: Optional[int] = None,
use_chirality: bool = True,
use_features: bool = True,
return_count: bool = True,
map_dict: Optional[dict] = None,
n_jobs: int = multiprocessing.cpu_count(),
mp_context: str = "spawn",
chunksize: int = None,
):
self.radius = radius
self.fold = fold
self.use_chirality = use_chirality
self.use_features = use_features
self.map_dict = map_dict
self.return_count = return_count
self.n_jobs = n_jobs
self.mp_context = mp_context
self.chunksize = chunksize
@property
def n_features(self) -> int:
if self.fold is None:
return len(self.map_dict) if self.map_dict else -1
else:
return self.fold
def _ecfp(self, smile: str) -> Union[Tuple[Dict, Dict, Mol], Tuple[None, None, None]]:
mol = Chem.MolFromSmiles(smile)
if mol is None:
warnings.warn(f"could not parse smile: {smile}")
return None, None, None
else:
bit_info = {}
fingerprint = AllChem.GetMorganFingerprint(mol, radius=self.radius, useChirality=self.use_chirality,
useFeatures=self.use_features, bitInfo=bit_info).GetNonzeroElements()
return fingerprint, bit_info, mol
def ecfp(self, smiles: List[str]) -> Tuple[List[dict], List[dict], List[Mol]]:
if self.n_jobs > 1:
fingerprints, bit_infos, mols = zip(
*process_map(
self._ecfp, smiles,
chunksize=(len(smiles) // self.n_jobs) + 1 if self.chunksize is None else self.chunksize,
# chunksize=1,
max_workers=self.n_jobs, desc="_ecfp",
mp_context=mp.get_context(self.mp_context)
)
)
else:
fingerprints, bit_infos, mols = zip(*list(map(self._ecfp, tqdm(smiles, total=len(smiles), desc="_ecfp"))))
return fingerprints, bit_infos, mols
def _fit(self, fingerprints: List[dict]):
if self.map_dict is None:
features = sorted(list(set.union(*[set(s.keys()) for s in fingerprints])))
if self.fold is None:
self.map_dict = dict(zip(features, range(len(features))))
else:
self.map_dict = {f: f % self.fold for f in features}
def fit_transform(self, smiles: List[str]) -> np.ndarray:
fingerprints, *_ = self.ecfp(smiles)
self._fit(fingerprints)
desc_mat = np.zeros((len(fingerprints), self.n_features), dtype=np.uint8)
for i, fp in enumerate(fingerprints):
for f, cnt in fp.items():
if f in self.map_dict:
desc_mat[i, self.map_dict[f]] = cnt
else:
warnings.warn(f"feature {f} not in map")
return desc_mat
def __call__(self, smiles: List[str]) -> np.ndarray:
features = self.fit_transform(smiles)
return features if self.return_count else np.where(features > 0, 1, 0).astype(features.dtype)
def _atomic_mapping(self,
molecule: Union[str, Mol, bytes],
num_atoms: Optional[int] = None,
bit_info: Optional[dict] = None
) -> List[List[Tuple[int, int]]]:
"""
gets the individual atomic mapping for one molecule - mapping indicates the feature idx + factor which contributes
"""
mol = _init_molecule(molecule)
num_atoms = mol.GetNumAtoms() if not num_atoms else num_atoms
if bit_info is None:
bit_info = {}
AllChem.GetMorganFingerprint(mol, radius=self.radius, useChirality=self.use_chirality, useFeatures=self.use_features,
bitInfo=bit_info)
atomic_mapping = [[] for _ in range(num_atoms)]
for feature, value in bit_info.items():
# feature mapping to account for e.g. folding
feature_idx = self.map_dict[feature]
for center_atom, radius in value:
mapping_submol = [[] for _ in range(num_atoms)]
count_atoms = 0
if radius > 0:
env_mol = Chem.FindAtomEnvironmentOfRadiusN(mol, radius, center_atom)
atom_map = {}
Chem.PathToSubmol(mol, env_mol, atomMap=atom_map)
for atom_k in atom_map.keys():
mapping_submol[atom_k].append(feature_idx)
count_atoms += 1
else:
mapping_submol[center_atom].append(feature_idx)
count_atoms = 1
for i in range(num_atoms):
if len(mapping_submol[i]) > 0:
for _feature_idx in mapping_submol[i]:
atomic_mapping[i].append((_feature_idx, count_atoms))
return atomic_mapping
def _atomic_attribution(self,
mol: Mol,
feature_attribution: np.ndarray,
num_atoms: Optional[int] = None,
bit_info: Optional[dict] = None) -> np.ndarray:
"""
gets the individual atomic contribution for one molecule based on the feature attribution
based and adapted from the implementation provided by <NAME>
"""
num_atoms = mol.GetNumAtoms() if not num_atoms else num_atoms
if bit_info is None:
bit_info = {}
AllChem.GetMorganFingerprint(mol, radius=self.radius, useChirality=self.use_chirality, useFeatures=self.use_features,
bitInfo=bit_info)
atomic_attribution = np.zeros(num_atoms)
for f, value in bit_info.items():
# feature mapping to account for e.g. folding
f = self.map_dict[f]
attribution_value = feature_attribution[f]
for center_atom, radius in value:
attribution_submol = np.zeros(num_atoms)
count_atoms = 0
if radius > 0:
env_mol = Chem.FindAtomEnvironmentOfRadiusN(mol, radius, center_atom)
atom_map = {}
Chem.PathToSubmol(mol, env_mol, atomMap=atom_map)
for atom_k in atom_map.keys():
attribution_submol[atom_k] += attribution_value
count_atoms += 1
else:
attribution_submol[center_atom] += attribution_value
count_atoms = 1
attribution_submol /= count_atoms
atomic_attribution += attribution_submol
return atomic_attribution
def atomic_attributions(self, smiles: List[str], feature_attributions: np.ndarray) -> List[np.ndarray]:
assert len(smiles) == len(
feature_attributions), f"provided number of smiles {len(smiles)} does not match number of features {len(feature_attributions)}"
fingerprints, bit_infos, mols = self.ecfp(smiles)
if self.map_dict is None:
self._fit(fingerprints)
atomic_attributions = []
for i, (smile, fingerprint, bit_info, mol) in tqdm(enumerate(zip(smiles, fingerprints, bit_infos, mols)), total=len(smiles),
desc="_ecfp_atomic_attributions"):
if mol is None:
raise ValueError(f"could not process smile/molecule {i}: {smile}")
atomic_attribution = self._atomic_attribution(mol, feature_attributions[i], bit_info=bit_info)
atomic_attributions.append(atomic_attribution)
return atomic_attributions
def atomic_mappings(self, smiles: List[str]) -> List[List[List[Tuple[int, int]]]]:
fingerprints, bit_infos, mols = self.ecfp(smiles)
if self.map_dict is None:
self._fit(fingerprints)
atomic_mappings = []
for i, (smile, fingerprint, bit_info, mol) in tqdm(enumerate(zip(smiles, fingerprints, bit_infos, mols)), total=len(smiles),
desc="_ecfp_atomic_mappings"):
if mol is None:
raise ValueError(f"could not process smile/molecule {i}: {smile}")
atomic_mapping = self._atomic_mapping(mol, bit_info=bit_info)
atomic_mappings.append(atomic_mapping)
return atomic_mappings
def _smarts_substr() -> Dict[int, Mol]:
with open(Path(PurePosixPath(__file__)).parent / "resources/maccs_smarts_substr.json") as file:
data = json.load(file)
return {int(k): Chem.MolFromSmarts(smile) for k, smile in data.items()}
class MACCSFeaturizer():
SMARTS_ATOMIC_NUMBER = {
2: [104], # atomic num >103 Not complete, RDKit only accepts up to #104
3: [32, 33, 34, 50, 51, 52, 82, 83, 84], # Group IVa,Va,VIa Rows 4-6
4: [89, 90, 91, 92, 93, 94, 95, 96, 97, 98, 99, 100, 101, 102, 103], # actinide
5: [21, 22, 39, 40, 72], # Group IIIB,IVB
6: [57, 58, 59, 60, 61, 62, 63, 64, 65, 66, 67, 68, 69, 70, 71], # Lanthanide
7: [23, 24, 25, 41, 42, 43, 73, 74, 75], # Group VB,VIB,VIIB
9: [26, 27, 28, 44, 45, 46, 76, 77, 78], # Group VIII
10: [4, 12, 20, 38, 56, 88], # Group II
12: [29, 30, 47, 48, 79, 80], # Group IB,IIB
18: [5, 13, 31, 49, 81], # Group III
27: [53], # I
29: [15], # P
35: [3, 11, 19, 37, 55, 87], # Group I
42: [9], # Fluor
46: [35], # Br
88: [16], # S
103: [17], # Cl
134: [9, 17, 35, 53] # Halogen: F,Cl,Br,I
}
SMARTS_SUBSTR = _smarts_substr()
def __init__(self, n_jobs: int = multiprocessing.cpu_count(), mp_context: str = "spawn", chunksize: int = None, ):
super(MACCSFeaturizer).__init__()
self.n_jobs = n_jobs
self.mp_context = mp_context
self.chunksize = chunksize
@property
def n_features(self) -> int:
return 167
def _macc(self, molecule: Union[str, Mol, bytes]) -> np.ndarray:
mol = _init_molecule(molecule)
_maccs = MACCSkeys.GenMACCSKeys(mol)
return np.array(_maccs)
def _maccs(self, smiles: List[str]) -> Tuple[np.ndarray, List[Mol]]:
maccs, mols = [], []
for i, smile in enumerate(tqdm(smiles, desc="_mol_maccs")):
mol = Chem.MolFromSmiles(smile)
mols.append(mol)
if mol is None:
warnings.warn(f"could not parse smile {i}: {smile}")
_mols = [m.ToBinary() for m in mols if m]
if self.n_jobs > 1:
maccs = process_map(self._macc, _mols,
chunksize=(len(smiles) // self.n_jobs) + 1 if self.chunksize is None else self.chunksize,
# chunksize=1,
max_workers=self.n_jobs,
desc="_maccs",
mp_context=mp.get_context(self.mp_context))
else:
maccs = list(map(self._macc, _mols))
return np.stack(maccs), mols
def __call__(self, smiles: List[str]) -> np.ndarray:
return self._maccs(smiles)[0]
def _atomic_mapping(self, molecule: Union[str, Mol, bytes],
num_atoms: Optional[int] = None) -> List[List[Tuple[int, int]]]:
mol = _init_molecule(molecule)
num_atoms = mol.GetNumAtoms() if not num_atoms else num_atoms
idx_maccs = list(MACCSFeaturizer.SMARTS_SUBSTR.keys())
idx_maccs_atomnumbs = list(MACCSFeaturizer.SMARTS_ATOMIC_NUMBER.keys())
atomic_attribution = np.zeros(num_atoms)
atomic_mapping = [[] for _ in range(num_atoms)]
for maccs_idx in idx_maccs:
# Substructure features
pattern = MACCSFeaturizer.SMARTS_SUBSTR[maccs_idx]
feature_idx = maccs_idx
substructures = mol.GetSubstructMatches(pattern)
mapping_submol = [[] for _ in range(num_atoms)]
count_atoms = 0
for atom_idx in range(num_atoms):
for structure in substructures:
if atom_idx in structure:
mapping_submol[atom_idx].append(feature_idx)
count_atoms += 1
count_atoms = 1 if count_atoms == 0 else count_atoms
for i in range(num_atoms):
if len(mapping_submol[i]) > 0:
for _feature_idx in mapping_submol[i]:
atomic_mapping[i].append((_feature_idx, count_atoms))
# Count features
# MACCS feature: 130
atomic_mapping = MACCSFeaturizer.maccs_count_features_mapping(maccs_idx, substructures, num_atoms,
atomic_mapping, feature_idx1=124,
feature_idx2=130)
# MACCS feature: 127
atomic_mapping = MACCSFeaturizer.maccs_count_features_mapping(maccs_idx, substructures, num_atoms,
atomic_mapping, feature_idx1=143,
feature_idx2=127)
# MACCS feature: 138:
atomic_mapping = MACCSFeaturizer.maccs_count_features_mapping(maccs_idx, substructures, num_atoms,
atomic_mapping, feature_idx1=153,
feature_idx2=138)
# MACCS features: 140, 146, 159
## 159
if maccs_idx == 164 and len(substructures) > 1:
mapping_submol = [[] for _ in range(num_atoms)]
count_atoms = 0
for atom_idx in range(num_atoms):
for structure in substructures:
if atom_idx in structure:
mapping_submol[atom_idx].append(159)
count_atoms += 1
count_atoms = 1 if count_atoms == 0 else count_atoms
for i in range(num_atoms):
if len(mapping_submol[i]) > 0:
for _feature_idx in mapping_submol[i]:
atomic_mapping[i].append((_feature_idx, count_atoms))
## 146
if len(substructures) > 2:
mapping_submol = [[] for _ in range(num_atoms)]
count_atoms = 0
for atom_idx in range(num_atoms):
for structure in substructures:
if atom_idx in structure:
mapping_submol[atom_idx].append(146)
count_atoms += 1
count_atoms = 1 if count_atoms == 0 else count_atoms
for i in range(num_atoms):
if len(mapping_submol[i]) > 0:
for _feature_idx in mapping_submol[i]:
atomic_mapping[i].append((_feature_idx, count_atoms))
## 140
if len(substructures) > 3:
mapping_submol = [[] for _ in range(num_atoms)]
count_atoms = 0
for atom_idx in range(num_atoms):
for structure in substructures:
if atom_idx in structure:
mapping_submol[atom_idx].append(140)
count_atoms += 1
count_atoms = 1 if count_atoms == 0 else count_atoms
for i in range(num_atoms):
if len(mapping_submol[i]) > 0:
for _feature_idx in mapping_submol[i]:
atomic_mapping[i].append((_feature_idx, count_atoms))
# MACCS feature 142
atomic_mapping = MACCSFeaturizer.maccs_count_features_mapping(maccs_idx, substructures, num_atoms,
atomic_mapping, feature_idx1=161,
feature_idx2=142)
# MACCS feature 145
atomic_mapping = MACCSFeaturizer.maccs_count_features_mapping(maccs_idx, substructures, num_atoms,
atomic_mapping, feature_idx1=163,
feature_idx2=145)
# MACCS feature 149
atomic_mapping = MACCSFeaturizer.maccs_count_features_mapping(maccs_idx, substructures, num_atoms,
atomic_mapping, feature_idx1=160,
feature_idx2=149)
# Atomic number features
for idx_maccs_atomnumb in idx_maccs_atomnumbs:
maccs_feature = MACCSFeaturizer.SMARTS_ATOMIC_NUMBER[idx_maccs_atomnumb]
feature_idx = idx_maccs_atomnumb
mapping_submol = [[] for _ in range(num_atoms)]
count_atoms = 0
for atom_idx in range(num_atoms):
if atom_idx in maccs_feature:
mapping_submol[atom_idx].append(feature_idx)
count_atoms += 1
count_atoms = 1 if count_atoms == 0 else count_atoms
for i in range(num_atoms):
if len(mapping_submol[i]) > 0:
for _feature_idx in mapping_submol[i]:
atomic_mapping[i].append((_feature_idx, count_atoms))
# MACCS 125: Aromatic rings
atomic_mapping = MACCSFeaturizer.maccs_125_aromaticrings_mapping(mol, num_atoms, atomic_mapping)
# MACCS 166: Fragments
atomic_mapping = MACCSFeaturizer.maccs_166_fragments_mapping(mol, num_atoms, atomic_mapping)
return atomic_mapping
def _atomic_attribution(self, molecule: Union[str, Mol, bytes], feature_attribution: np.ndarray,
num_atoms: Optional[int] = None) -> np.ndarray:
"""adapted/based on the implementation by <NAME>"""
mol = _init_molecule(molecule)
num_atoms = mol.GetNumAtoms() if not num_atoms else num_atoms
idx_maccs = list(MACCSFeaturizer.SMARTS_SUBSTR.keys())
idx_maccs_atomnumbs = list(MACCSFeaturizer.SMARTS_ATOMIC_NUMBER.keys())
atomic_attribution = np.zeros(num_atoms)
for maccs_idx in idx_maccs:
# Substructure features
pattern = MACCSFeaturizer.SMARTS_SUBSTR[maccs_idx]
attribution_value = feature_attribution[maccs_idx]
substructures = mol.GetSubstructMatches(pattern)
attribution_submol = np.zeros(num_atoms)
count_atoms = 0
for atom_idx in range(num_atoms):
for structure in substructures:
if atom_idx in structure:
attribution_submol[atom_idx] += attribution_value
count_atoms += 1
if count_atoms != 0:
attribution_submol = attribution_submol / count_atoms
atomic_attribution += attribution_submol
# Count features
# MACCS feature: 130
atomic_attribution = MACCSFeaturizer.maccs_count_features(maccs_idx, substructures, feature_attribution, num_atoms,
atomic_attribution,
feature_idx1=124, feature_idx2=130)
# MACCS feature: 127
atomic_attribution = MACCSFeaturizer.maccs_count_features(maccs_idx, substructures, feature_attribution, num_atoms,
atomic_attribution,
feature_idx1=143, feature_idx2=127)
# MACCS feature: 138:
atomic_attribution = MACCSFeaturizer.maccs_count_features(maccs_idx, substructures, feature_attribution, num_atoms,
atomic_attribution,
feature_idx1=153, feature_idx2=138)
# MACCS features: 140, 146, 159
## 159
if maccs_idx == 164 and len(substructures) > 1:
attribution_value = feature_attribution[159]
attribution_submol = np.zeros(num_atoms)
count_atoms = 0
for atom_idx in range(num_atoms):
for structure in substructures:
if atom_idx in structure:
attribution_submol[atom_idx] += attribution_value
count_atoms += 1
if count_atoms != 0:
attribution_submol = attribution_submol / count_atoms
atomic_attribution += attribution_submol
## 146
if len(substructures) > 2:
attribution_value = feature_attribution[146]
attribution_submol = np.zeros(num_atoms)
count_atoms = 0
for atom_idx in range(num_atoms):
for structure in substructures:
if atom_idx in structure:
attribution_submol[atom_idx] += attribution_value
count_atoms += 1
if count_atoms != 0:
attribution_submol = attribution_submol / count_atoms
atomic_attribution += attribution_submol
## 140
if len(substructures) > 3:
attribution_value = feature_attribution[140]
attribution_submol = np.zeros(num_atoms)
count_atoms = 0
for atom_idx in range(num_atoms):
for structure in substructures:
if atom_idx in structure:
attribution_submol[atom_idx] += attribution_value
count_atoms += 1
if count_atoms != 0:
attribution_submol = attribution_submol / count_atoms
atomic_attribution += attribution_submol
# MACCS feature 142
atomic_attribution = MACCSFeaturizer.maccs_count_features(maccs_idx, substructures, feature_attribution, num_atoms,
atomic_attribution,
feature_idx1=161, feature_idx2=142)
# MACCS feature 145
atomic_attribution = MACCSFeaturizer.maccs_count_features(maccs_idx, substructures, feature_attribution, num_atoms,
atomic_attribution,
feature_idx1=163, feature_idx2=145)
# MACCS feature 149
atomic_attribution = MACCSFeaturizer.maccs_count_features(maccs_idx, substructures, feature_attribution, num_atoms,
atomic_attribution,
feature_idx1=160, feature_idx2=149)
# Atomic number features
for idx_maccs_atomnumb in idx_maccs_atomnumbs:
maccs_feature = MACCSFeaturizer.SMARTS_ATOMIC_NUMBER[idx_maccs_atomnumb]
attribution_value = feature_attribution[idx_maccs_atomnumb]
attribution_submol = np.zeros(num_atoms)
count_atoms = 0
for atom_idx in range(num_atoms):
if atom_idx in maccs_feature:
attribution_submol[atom_idx] += attribution_value
count_atoms += 1
if count_atoms != 0:
attribution_submol = attribution_submol / count_atoms
atomic_attribution += attribution_submol
# MACCS 125: Aromatic rings
atomic_attribution = MACCSFeaturizer.maccs_125_aromaticrings(mol, feature_attribution, num_atoms, atomic_attribution)
# MACCS 166: Fragments
atomic_attribution = MACCSFeaturizer.maccs_166_fragments(mol, feature_attribution, num_atoms, atomic_attribution)
return atomic_attribution
def atomic_mappings(self, smiles: List[str]) -> List[List[List[Tuple[int, int]]]]:
_, mols = self._maccs(smiles)
_mols = [m.ToBinary() for m in mols if m]
if self.n_jobs > 1:
atomic_mappings = process_map(self._atomic_mapping, _mols,
chunksize=(len(smiles) // self.n_jobs) + 1 if self.chunksize is None else self.chunksize,
# chunksize=1,
max_workers=self.n_jobs,
desc="_maccs_atomic_mappings",
mp_context=mp.get_context(self.mp_context))
else:
atomic_mappings = list(
map(self._atomic_mapping, tqdm(_mols, total=len(smiles), desc="_maccs_atomic_mappings")))
return atomic_mappings
def atomic_attributions(self, smiles: List[str], feature_attributions: np.ndarray) -> List[np.ndarray]:
assert len(smiles) == len(
feature_attributions), f"provided number of smiles {len(smiles)} does not match number of features {len(feature_attributions)}"
_, mols = self._maccs(smiles)
_mols = [m.ToBinary() for m in mols if m]
if self.n_jobs > 1:
atomic_attributions = process_map(self._atomic_attribution, _mols, feature_attributions,
chunksize=(len(smiles) // self.n_jobs) + 1 if self.chunksize is None else self.chunksize,
# chunksize=1,
max_workers=self.n_jobs,
desc="_maccs_atomic_attributions",
mp_context=mp.get_context(self.mp_context))
else:
atomic_attributions = list(
map(self._atomic_attribution, tqdm(_mols, total=len(smiles), desc="_maccs_atomic_attributions"), feature_attributions))
return atomic_attributions
@staticmethod
def maccs_count_features_mapping(maccs_idx: int, substructures, num_atoms: int,
atomic_mapping: List[List[Tuple[int, int]]], feature_idx1: int, feature_idx2: int
) -> List[List[Tuple[int, int]]]:
if maccs_idx == feature_idx1 and len(substructures) > 1:
mapping_submol = [[] for _ in range(num_atoms)]
count_atoms = 0
for atom_idx in range(num_atoms):
for structure in substructures:
if atom_idx in structure:
mapping_submol[atom_idx].append(feature_idx2)
count_atoms += 1
count_atoms = 1 if count_atoms == 0 else count_atoms
for i in range(num_atoms):
if len(mapping_submol[i]) > 0:
for _feature_idx in mapping_submol[i]:
atomic_mapping[i].append((_feature_idx, count_atoms))
return atomic_mapping
@staticmethod
def maccs_count_features(maccs_idx: int, substructures, feature_attribution: np.ndarray, num_atoms: int, atomic_attribution: np.ndarray,
feature_idx1: int, feature_idx2: int) -> np.ndarray:
"""based on the implementation by <NAME>"""
if maccs_idx == feature_idx1 and len(substructures) > 1:
attribution_value = feature_attribution[feature_idx2]
weights_submol = np.zeros(num_atoms)
count_atoms = 0
for atom_idx in range(num_atoms):
for structure in substructures:
if atom_idx in structure:
weights_submol[atom_idx] += attribution_value
count_atoms += 1
if count_atoms != 0:
weights_submol = weights_submol / count_atoms
atomic_attribution += weights_submol
return atomic_attribution
@staticmethod
def isRingAromatic(mol: Mol, ringbond: Tuple[int, ...]) -> bool:
"""based on the implementation by <NAME>"""
for id in ringbond:
if not mol.GetBondWithIdx(id).GetIsAromatic():
return False
return True
@staticmethod
def maccs_125_aromaticrings_mapping(mol: Mol,
num_atoms: int, atomic_mapping: List[List[Tuple[int, int]]]):
substructure = list()
ri = mol.GetRingInfo()
ringcount = ri.NumRings()
rings = ri.AtomRings()
ringbonds = ri.BondRings()
if ringcount > 1:
for ring_idx in range(ringcount):
ring = rings[ring_idx]
ringbond = ringbonds[ring_idx]
is_aromatic = MACCSFeaturizer.isRingAromatic(mol, ringbond)
if is_aromatic == True:
substructure.append(ring)
mapping_submol = [[] for _ in range(num_atoms)]
count_atoms = 0
for atom_idx in range(num_atoms):
for structure in substructure:
if atom_idx in structure:
mapping_submol[atom_idx].append(125)
count_atoms += 1
count_atoms = 1 if count_atoms == 0 else count_atoms
for i in range(num_atoms):
if len(mapping_submol[i]) > 0:
for _feature_idx in mapping_submol[i]:
atomic_mapping[i].append((_feature_idx, count_atoms))
return atomic_mapping
@staticmethod
def maccs_125_aromaticrings(mol: Mol, feature_attribution: np.ndarray, num_atoms: int, atomic_attribution: np.ndarray) -> np.ndarray:
"""based on the implementation by <NAME>"""
attribution_value = feature_attribution[125]
substructure = list()
ri = mol.GetRingInfo()
ringcount = ri.NumRings()
rings = ri.AtomRings()
ringbonds = ri.BondRings()
if ringcount > 1:
for ring_idx in range(ringcount):
ring = rings[ring_idx]
ringbond = ringbonds[ring_idx]
is_aromatic = MACCSFeaturizer.isRingAromatic(mol, ringbond)
if is_aromatic == True:
substructure.append(ring)
weights_submol = np.zeros(num_atoms)
count_atoms = 0
for atom_idx in range(num_atoms):
for structure in substructure:
if atom_idx in structure:
weights_submol[atom_idx] += attribution_value
count_atoms += 1
if count_atoms != 0:
weights_submol = weights_submol / count_atoms
atomic_attribution += weights_submol
return atomic_attribution
@staticmethod
def maccs_166_fragments_mapping(mol: Mol, num_atoms: int, atomic_mapping: List[List[Tuple[int, int]]]) -> List[
List[Tuple[int, int]]]:
frags = Chem.GetMolFrags(mol)
if len(frags) > 1:
mapping_submol = [[] for _ in range(num_atoms)]
count_atoms = 0
for atom_idx in range(num_atoms):
for structure in frags:
if atom_idx in structure:
mapping_submol[atom_idx].append(166)
count_atoms += 1
count_atoms = 1 if count_atoms == 0 else count_atoms
for i in range(num_atoms):
if len(mapping_submol[i]) > 0:
for _feature_idx in mapping_submol[i]:
atomic_mapping[i].append((_feature_idx, count_atoms))
return atomic_mapping
@staticmethod
def maccs_166_fragments(mol: Mol, feature_attribution: np.ndarray, num_atoms: int, atomic_attribution: np.ndarray) -> np.ndarray:
"""based on the implementation by <NAME>"""
attribution_value = feature_attribution[166]
frags = Chem.GetMolFrags(mol)
if len(frags) > 1:
weights_submol = np.zeros(num_atoms)
count_atoms = 0
for atom_idx in range(num_atoms):
for structure in frags:
if atom_idx in structure:
weights_submol[atom_idx] += attribution_value
count_atoms += 1
if count_atoms != 0:
weights_submol = weights_submol / count_atoms
atomic_attribution += weights_submol
return atomic_attribution
def _all_patterns():
"""based/adapted on the implementation by <NAME>"""
with open(Path(PurePosixPath(__file__)).parent / "resources/tox_smarts.json") as file:
smarts_list = [s[1] for s in json.load(file)]
# Code does not work for this case
assert len([s for s in smarts_list if ("AND" in s) and ("OR" in s)]) == 0
# Chem.MolFromSmarts takes a long time so it pays of to parse all the smarts first
# and then use them for all molecules. This gives a huge speedup over existing code.
# a list of patterns, whether to negate the match result and how to join them to obtain one boolean value
all_patterns = []
for smarts in smarts_list:
patterns = [] # list of smarts-patterns
# value for each of the patterns above. Negates the values of the above later.
negations = []
if " AND " in smarts:
smarts = smarts.split(" AND ")
merge_any = False # If an " AND " is found all "subsmarts" have to match
else:
# If there is an " OR " present it"s enough is any of the "subsmarts" match.
# This also accumulates smarts where neither " OR " nor " AND " occur
smarts = smarts.split(" OR ")
merge_any = True
# for all subsmarts check if they are preceded by "NOT "
for s in smarts:
neg = s.startswith("NOT ")
if neg:
s = s[4:]
patterns.append(Chem.MolFromSmarts(s))
negations.append(neg)
all_patterns.append((patterns, negations, merge_any))
return all_patterns
class ToxFeaturizer():
ALL_PATTERNS = _all_patterns()
def __init__(self, n_jobs: int = multiprocessing.cpu_count(), mp_context: str = "spawn", chunksize: int = None, ):
super(ToxFeaturizer, self).__init__()
self.n_jobs = n_jobs
self.mp_context = mp_context
self.chunksize = chunksize
@property
def n_features(self) -> int:
return 826
def _tox(self, molecule: Union[str, Mol, bytes]) -> np.ndarray:
"""
based/adapted on the implementation by <NAME>
Matches the tox patterns against a molecule. Returns a boolean array
"""
mol = _init_molecule(molecule)
mol_features = []
for patts, negations, merge_any in ToxFeaturizer.ALL_PATTERNS:
matches = [mol.HasSubstructMatch(p) for p in patts]
matches = [m != n for m, n in zip(matches, negations)]
if merge_any:
pres = any(matches)
else:
pres = all(matches)
mol_features.append(pres)
return np.array(mol_features)
def _toxs(self, smiles: List[str]) -> Tuple[np.ndarray, List[Mol]]:
toxs, mols = [], []
for i, smile in enumerate(tqdm(smiles, desc="_mols_toxs")):
mol = Chem.MolFromSmiles(smile)
mols.append(mol)
if mol is None:
warnings.warn(f"could not parse smile {i}: {smile}")
_mols = [m.ToBinary() for m in mols if m]
if self.n_jobs > 1:
toxs = process_map(self._tox, _mols,
chunksize=(len(smiles) // self.n_jobs) + 1 if self.chunksize is None else self.chunksize,
# chunksize=1,
max_workers=self.n_jobs,
desc="_toxs",
mp_context=mp.get_context(self.mp_context))
else:
toxs = list(map(self._tox, _mols))
return np.stack([t for t in toxs if not None]), mols
def __call__(self, smiles: List[str]) -> np.ndarray:
"""returns a binary numpy array"""
return self._toxs(smiles)[0]
def _atomic_mapping(self,
molecule: Union[str, Mol, bytes],
num_atoms: Optional[int] = None,
) -> List[List[Tuple[int, int]]]:
"""
gets the individual atomic mapping for one molecule - mapping indicates the feature idx + factor which contributes
"""
mol = _init_molecule(molecule)
num_atoms = mol.GetNumAtoms() if not num_atoms else num_atoms
atomic_mapping = [[] for _ in range(num_atoms)]
for feature_idx, (patts, negations, merge_any) in enumerate(ToxFeaturizer.ALL_PATTERNS):
mapping_submol = [[] for _ in range(num_atoms)]
count_atoms = 0
for atom_idx in range(num_atoms):
for i in range(len(negations)):
neg = negations[i]
pattern = patts[i]
substructures = mol.GetSubstructMatches(pattern)
for structure in substructures:
atom_in_sub = list()
if str(neg) == "False":
if atom_idx in structure:
atom_in_sub.append("y")
elif str(neg) == "True":
if atom_idx not in structure:
atom_in_sub.append("y")
if "y" in str(atom_in_sub):
mapping_submol[atom_idx].append(feature_idx)
count_atoms += 1
if count_atoms != 0:
for i in range(num_atoms):
if len(mapping_submol[i]) > 0:
for _feature_idx in mapping_submol[i]:
atomic_mapping[i].append((_feature_idx, count_atoms))
return atomic_mapping
def _atomic_attribution(self, molecule: Union[str, Mol, bytes], feature_attribution: np.ndarray,
num_atoms: Optional[int] = None) -> np.ndarray:
"""adapted/based on the implementation by <NAME>"""
mol = _init_molecule(molecule)
num_atoms = mol.GetNumAtoms() if not num_atoms else num_atoms
atomic_attribution = np.zeros(num_atoms)
tox_idx = 0
for patts, negations, merge_any in ToxFeaturizer.ALL_PATTERNS:
attribution_value = feature_attribution[tox_idx]
attribution_submol = np.zeros(num_atoms)
count_atoms = 0
for atom_idx in range(num_atoms):
for i in range(len(negations)):
neg = negations[i]
pattern = patts[i]
substructures = mol.GetSubstructMatches(pattern)
for structure in substructures:
atom_in_sub = list()
if str(neg) == "False":
if atom_idx in structure:
atom_in_sub.append("y")
elif str(neg) == "True":
if atom_idx not in structure:
atom_in_sub.append("y")
if "y" in str(atom_in_sub):
attribution_submol[atom_idx] += attribution_value
count_atoms += 1
if count_atoms != 0:
attribution_submol = attribution_submol / count_atoms
atomic_attribution += attribution_submol
tox_idx += 1
return atomic_attribution
def atomic_attributions(self, smiles: List[str], feature_attributions: np.ndarray) -> List[np.ndarray]:
assert len(smiles) == len(
feature_attributions), f"provided number of smiles {len(smiles)} does not match number of features {len(feature_attributions)}"
_, mols = self._toxs(smiles)
_mols = [m.ToBinary() for m in mols if m]
if self.n_jobs > 1:
atomic_attributions = process_map(self._atomic_attribution, _mols, feature_attributions,
chunksize=(len(smiles) // self.n_jobs) + 1 if self.chunksize is None else self.chunksize,
# chunksize=1,
max_workers=self.n_jobs,
desc="_tox_atomic_attributions",
mp_context=mp.get_context(self.mp_context))
else:
atomic_attributions = list(
map(self._atomic_attribution, tqdm(_mols, total=len(smiles), desc="_tox_atomic_attributions"), feature_attributions))
return atomic_attributions
def atomic_mappings(self, smiles: List[str]) -> List[List[List[Tuple[int, int]]]]:
_, mols = self._toxs(smiles)
_mols = [m.ToBinary() for m in mols if m]
if self.n_jobs > 1:
atomic_mappings = process_map(self._atomic_mapping, _mols,
chunksize=(len(smiles) // self.n_jobs) + 1 if self.chunksize is None else self.chunksize,
# chunksize=1,
max_workers=self.n_jobs,
desc="_tox_atomic_mappings",
mp_context=mp.get_context(self.mp_context))
else:
atomic_mappings = list(
map(self._atomic_mapping, tqdm(_mols, total=len(smiles), desc="_tox_atomic_mappings")))
return atomic_mappings
def _atomic_attribution_from_mapping(atomic_mapping: List[List[Tuple[int, int]]], feature_attribution: np.ndarray) -> np.ndarray:
"""calculate atomic attribution for single molecule based on provided mapping and features attributions"""
num_atoms = len(atomic_mapping)
atomic_attribution = np.zeros(num_atoms)
for atom_idx, atom_map in enumerate(atomic_mapping):
for feature_idx, count_atoms in atom_map:
atomic_attribution[atom_idx] += feature_attribution[feature_idx] * 1 / count_atoms
return atomic_attribution
def atomic_attributions_from_mappings(atomic_mappings: List[List[List[Tuple[int, int]]]],
feature_attributions: np.ndarray,
n_jobs: int = multiprocessing.cpu_count(),
mp_context: str = "fork",
chunksize: int = None) -> List[np.ndarray]:
"""calculate atomic attributions for multiple molecules based on provided mappings and features attributions"""
if n_jobs > 1:
atomic_attributions = process_map(_atomic_attribution_from_mapping, atomic_mappings, feature_attributions,
chunksize=(len(atomic_mappings) // n_jobs) + 1 if chunksize is None else chunksize,
max_workers=n_jobs,
desc="_attributions_from_atomic_mappings",
mp_context=mp.get_context(mp_context))
else:
atomic_attributions = list(
map(_atomic_attribution_from_mapping,
tqdm(atomic_mappings, total=len(atomic_mappings), desc="_attributions_from_atomic_mappings"), feature_attributions))
return atomic_attributions
def calculate_ranking_scores(smiles: List[str],
references: Union[List[str], List[Tuple[str, int]]],
atomic_attributions: List[np.ndarray],
labels: Optional[np.ndarray] = None,
preds: Optional[np.ndarray] = None,
) -> Tuple[Dict, List[Dict], pd.DataFrame]:
"""
Function calculates the score to rank atoms of reference smiles/atomic substructures according to the provided atomic attribution/weights
Args:
smiles (): List of smile strings
references (): List of tuples of provided reference smiles and if they are supposed to be active or not
Scores are calculated per reference smile
atomic_attributions (): List of atomic attributions/weights
labels (): Optional provide binary true labels
preds (): Optional provide binary predictions
Returns:
Tuple containing
- Dictionary of mean calculated scores for all provided reference smiles
- List of dictionaries per reference score with mean scores per reference smile
- Dataframe containing table with full details per smile and per reference smile with all matches, scores, etc.
"""
assert len(smiles) == len(
atomic_attributions), f"length of provided smiles {len(smiles)} must match length of provided attributions {len(atomic_attributions)}"
if labels is not None:
assert labels.ndim == 1, f"nr of dimensions of provided labels must be 1 but is {labels.ndim}"
assert len(labels) == len(smiles), f"nr of labels {len(labels)} must match number of smiles {len(smiles)}"
if preds is not None:
assert preds.ndim == 1, f"nr of dimensions of provided predictions must be 1 but is {preds.ndim}"
assert len(preds) == len(smiles), f"nr of predictions {len(preds)} must match number of smiles {len(smiles)}"
df = | pd.DataFrame() | pandas.DataFrame |
import unittest
import qteasy as qt
import pandas as pd
from pandas import Timestamp
import numpy as np
from numpy import int64
import itertools
import datetime
from qteasy.utilfuncs import list_to_str_format, regulate_date_format, time_str_format, str_to_list
from qteasy.utilfuncs import maybe_trade_day, is_market_trade_day, prev_trade_day, next_trade_day, prev_market_trade_day
from qteasy.utilfuncs import next_market_trade_day
from qteasy.space import Space, Axis, space_around_centre, ResultPool
from qteasy.core import apply_loop
from qteasy.built_in import SelectingFinanceIndicator
from qteasy.history import stack_dataframes
from qteasy.tsfuncs import income, indicators, name_change, get_bar
from qteasy.tsfuncs import stock_basic, trade_calendar, new_share, get_index
from qteasy.tsfuncs import balance, cashflow, top_list, index_indicators, composite
from qteasy.tsfuncs import future_basic, future_daily, options_basic, options_daily
from qteasy.tsfuncs import fund_basic, fund_net_value, index_basic
from qteasy.evaluate import eval_alpha, eval_benchmark, eval_beta, eval_fv
from qteasy.evaluate import eval_info_ratio, eval_max_drawdown, eval_sharp
from qteasy.evaluate import eval_volatility
from qteasy.tafuncs import bbands, dema, ema, ht, kama, ma, mama, mavp, mid_point
from qteasy.tafuncs import mid_price, sar, sarext, sma, t3, tema, trima, wma, adx, adxr
from qteasy.tafuncs import apo, bop, cci, cmo, dx, macd, macdext, aroon, aroonosc
from qteasy.tafuncs import macdfix, mfi, minus_di, minus_dm, mom, plus_di, plus_dm
from qteasy.tafuncs import ppo, roc, rocp, rocr, rocr100, rsi, stoch, stochf, stochrsi
from qteasy.tafuncs import trix, ultosc, willr, ad, adosc, obv, atr, natr, trange
from qteasy.tafuncs import avgprice, medprice, typprice, wclprice, ht_dcperiod
from qteasy.tafuncs import ht_dcphase, ht_phasor, ht_sine, ht_trendmode, cdl2crows
from qteasy.tafuncs import cdl3blackcrows, cdl3inside, cdl3linestrike, cdl3outside
from qteasy.tafuncs import cdl3starsinsouth, cdl3whitesoldiers, cdlabandonedbaby
from qteasy.tafuncs import cdladvanceblock, cdlbelthold, cdlbreakaway, cdlclosingmarubozu
from qteasy.tafuncs import cdlconcealbabyswall, cdlcounterattack, cdldarkcloudcover
from qteasy.tafuncs import cdldoji, cdldojistar, cdldragonflydoji, cdlengulfing
from qteasy.tafuncs import cdleveningdojistar, cdleveningstar, cdlgapsidesidewhite
from qteasy.tafuncs import cdlgravestonedoji, cdlhammer, cdlhangingman, cdlharami
from qteasy.tafuncs import cdlharamicross, cdlhighwave, cdlhikkake, cdlhikkakemod
from qteasy.tafuncs import cdlhomingpigeon, cdlidentical3crows, cdlinneck
from qteasy.tafuncs import cdlinvertedhammer, cdlkicking, cdlkickingbylength
from qteasy.tafuncs import cdlladderbottom, cdllongleggeddoji, cdllongline, cdlmarubozu
from qteasy.tafuncs import cdlmatchinglow, cdlmathold, cdlmorningdojistar, cdlmorningstar
from qteasy.tafuncs import cdlonneck, cdlpiercing, cdlrickshawman, cdlrisefall3methods
from qteasy.tafuncs import cdlseparatinglines, cdlshootingstar, cdlshortline, cdlspinningtop
from qteasy.tafuncs import cdlstalledpattern, cdlsticksandwich, cdltakuri, cdltasukigap
from qteasy.tafuncs import cdlthrusting, cdltristar, cdlunique3river, cdlupsidegap2crows
from qteasy.tafuncs import cdlxsidegap3methods, beta, correl, linearreg, linearreg_angle
from qteasy.tafuncs import linearreg_intercept, linearreg_slope, stddev, tsf, var, acos
from qteasy.tafuncs import asin, atan, ceil, cos, cosh, exp, floor, ln, log10, sin, sinh
from qteasy.tafuncs import sqrt, tan, tanh, add, div, max, maxindex, min, minindex, minmax
from qteasy.tafuncs import minmaxindex, mult, sub, sum
from qteasy.history import get_financial_report_type_raw_data, get_price_type_raw_data
from qteasy.database import DataSource
from qteasy._arg_validators import _parse_string_kwargs, _valid_qt_kwargs
class TestCost(unittest.TestCase):
def setUp(self):
self.amounts = np.array([10000, 20000, 10000])
self.op = np.array([0, 1, -0.33333333])
self.prices = np.array([10, 20, 10])
self.r = qt.Cost()
def test_rate_creation(self):
print('testing rates objects\n')
self.assertIsInstance(self.r, qt.Cost, 'Type should be Rate')
def test_rate_operations(self):
self.assertEqual(self.r['buy_fix'], 0.0, 'Item got is incorrect')
self.assertEqual(self.r['sell_fix'], 0.0, 'Item got is wrong')
self.assertEqual(self.r['buy_rate'], 0.003, 'Item got is incorrect')
self.assertEqual(self.r['sell_rate'], 0.001, 'Item got is incorrect')
self.assertEqual(self.r['buy_min'], 5., 'Item got is incorrect')
self.assertEqual(self.r['sell_min'], 0.0, 'Item got is incorrect')
self.assertEqual(self.r['slipage'], 0.0, 'Item got is incorrect')
self.assertEqual(np.allclose(self.r(self.amounts), [0.003, 0.003, 0.003]), True, 'fee calculation wrong')
def test_rate_fee(self):
self.r.buy_rate = 0.003
self.r.sell_rate = 0.001
self.r.buy_fix = 0
self.r.sell_fix = 0
self.r.buy_min = 0
self.r.sell_min = 0
self.r.slipage = 0
print('\nSell result with fixed rate = 0.001 and moq = 0:')
print(self.r.get_selling_result(self.prices, self.op, self.amounts))
test_rate_fee_result = self.r.get_selling_result(self.prices, self.op, self.amounts)
self.assertIs(np.allclose(test_rate_fee_result[0], [0., 0., -3333.3333]), True, 'result incorrect')
self.assertAlmostEqual(test_rate_fee_result[1], 33299.999667, msg='result incorrect')
self.assertAlmostEqual(test_rate_fee_result[2], 33.333332999999996, msg='result incorrect')
print('\nSell result with fixed rate = 0.001 and moq = 1:')
print(self.r.get_selling_result(self.prices, self.op, self.amounts, 1))
test_rate_fee_result = self.r.get_selling_result(self.prices, self.op, self.amounts, 1)
self.assertIs(np.allclose(test_rate_fee_result[0], [0., 0., -3333]), True, 'result incorrect')
self.assertAlmostEqual(test_rate_fee_result[1], 33296.67, msg='result incorrect')
self.assertAlmostEqual(test_rate_fee_result[2], 33.33, msg='result incorrect')
print('\nSell result with fixed rate = 0.001 and moq = 100:')
print(self.r.get_selling_result(self.prices, self.op, self.amounts, 100))
test_rate_fee_result = self.r.get_selling_result(self.prices, self.op, self.amounts, 100)
self.assertIs(np.allclose(test_rate_fee_result[0], [0., 0., -3300]), True, 'result incorrect')
self.assertAlmostEqual(test_rate_fee_result[1], 32967.0, msg='result incorrect')
self.assertAlmostEqual(test_rate_fee_result[2], 33, msg='result incorrect')
print('\nPurchase result with fixed rate = 0.003 and moq = 0:')
print(self.r.get_purchase_result(self.prices, self.op, self.amounts, 0))
test_rate_fee_result = self.r.get_purchase_result(self.prices, self.op, self.amounts, 0)
self.assertIs(np.allclose(test_rate_fee_result[0], [0., 997.00897308, 0.]), True, 'result incorrect')
self.assertAlmostEqual(test_rate_fee_result[1], -20000.0, msg='result incorrect')
self.assertAlmostEqual(test_rate_fee_result[2], 59.82053838484547, msg='result incorrect')
print('\nPurchase result with fixed rate = 0.003 and moq = 1:')
print(self.r.get_purchase_result(self.prices, self.op, self.amounts, 1))
test_rate_fee_result = self.r.get_purchase_result(self.prices, self.op, self.amounts, 1)
self.assertIs(np.allclose(test_rate_fee_result[0], [0., 997., 0.]), True, 'result incorrect')
self.assertAlmostEqual(test_rate_fee_result[1], -19999.82, msg='result incorrect')
self.assertAlmostEqual(test_rate_fee_result[2], 59.82, msg='result incorrect')
print('\nPurchase result with fixed rate = 0.003 and moq = 100:')
print(self.r.get_purchase_result(self.prices, self.op, self.amounts, 100))
test_rate_fee_result = self.r.get_purchase_result(self.prices, self.op, self.amounts, 100)
self.assertIs(np.allclose(test_rate_fee_result[0], [0., 900., 0.]), True, 'result incorrect')
self.assertAlmostEqual(test_rate_fee_result[1], -18054., msg='result incorrect')
self.assertAlmostEqual(test_rate_fee_result[2], 54.0, msg='result incorrect')
def test_min_fee(self):
self.r.buy_rate = 0.
self.r.sell_rate = 0.
self.r.buy_fix = 0.
self.r.sell_fix = 0.
self.r.buy_min = 300
self.r.sell_min = 300
self.r.slipage = 0.
print('\npurchase result with fixed cost rate with min fee = 300 and moq = 0:')
print(self.r.get_purchase_result(self.prices, self.op, self.amounts, 0))
test_min_fee_result = self.r.get_purchase_result(self.prices, self.op, self.amounts, 0)
self.assertIs(np.allclose(test_min_fee_result[0], [0., 985, 0.]), True, 'result incorrect')
self.assertAlmostEqual(test_min_fee_result[1], -20000.0, msg='result incorrect')
self.assertAlmostEqual(test_min_fee_result[2], 300.0, msg='result incorrect')
print('\npurchase result with fixed cost rate with min fee = 300 and moq = 10:')
print(self.r.get_purchase_result(self.prices, self.op, self.amounts, 10))
test_min_fee_result = self.r.get_purchase_result(self.prices, self.op, self.amounts, 10)
self.assertIs(np.allclose(test_min_fee_result[0], [0., 980, 0.]), True, 'result incorrect')
self.assertAlmostEqual(test_min_fee_result[1], -19900.0, msg='result incorrect')
self.assertAlmostEqual(test_min_fee_result[2], 300.0, msg='result incorrect')
print('\npurchase result with fixed cost rate with min fee = 300 and moq = 100:')
print(self.r.get_purchase_result(self.prices, self.op, self.amounts, 100))
test_min_fee_result = self.r.get_purchase_result(self.prices, self.op, self.amounts, 100)
self.assertIs(np.allclose(test_min_fee_result[0], [0., 900, 0.]), True, 'result incorrect')
self.assertAlmostEqual(test_min_fee_result[1], -18300.0, msg='result incorrect')
self.assertAlmostEqual(test_min_fee_result[2], 300.0, msg='result incorrect')
print('\nselling result with fixed cost rate with min fee = 300 and moq = 0:')
print(self.r.get_selling_result(self.prices, self.op, self.amounts))
test_min_fee_result = self.r.get_selling_result(self.prices, self.op, self.amounts)
self.assertIs(np.allclose(test_min_fee_result[0], [0, 0, -3333.3333]), True, 'result incorrect')
self.assertAlmostEqual(test_min_fee_result[1], 33033.333)
self.assertAlmostEqual(test_min_fee_result[2], 300.0)
print('\nselling result with fixed cost rate with min fee = 300 and moq = 1:')
print(self.r.get_selling_result(self.prices, self.op, self.amounts, 1))
test_min_fee_result = self.r.get_selling_result(self.prices, self.op, self.amounts, 1)
self.assertIs(np.allclose(test_min_fee_result[0], [0, 0, -3333]), True, 'result incorrect')
self.assertAlmostEqual(test_min_fee_result[1], 33030)
self.assertAlmostEqual(test_min_fee_result[2], 300.0)
print('\nselling result with fixed cost rate with min fee = 300 and moq = 100:')
print(self.r.get_selling_result(self.prices, self.op, self.amounts, 100))
test_min_fee_result = self.r.get_selling_result(self.prices, self.op, self.amounts, 100)
self.assertIs(np.allclose(test_min_fee_result[0], [0, 0, -3300]), True, 'result incorrect')
self.assertAlmostEqual(test_min_fee_result[1], 32700)
self.assertAlmostEqual(test_min_fee_result[2], 300.0)
def test_rate_with_min(self):
"""Test transaction cost calculated by rate with min_fee"""
self.r.buy_rate = 0.0153
self.r.sell_rate = 0.01
self.r.buy_fix = 0.
self.r.sell_fix = 0.
self.r.buy_min = 300
self.r.sell_min = 333
self.r.slipage = 0.
print('\npurchase result with fixed cost rate with buy_rate = 0.0153, min fee = 300 and moq = 0:')
print(self.r.get_purchase_result(self.prices, self.op, self.amounts, 0))
test_rate_with_min_result = self.r.get_purchase_result(self.prices, self.op, self.amounts, 0)
self.assertIs(np.allclose(test_rate_with_min_result[0], [0., 984.9305624, 0.]), True, 'result incorrect')
self.assertAlmostEqual(test_rate_with_min_result[1], -20000.0, msg='result incorrect')
self.assertAlmostEqual(test_rate_with_min_result[2], 301.3887520929774, msg='result incorrect')
print('\npurchase result with fixed cost rate with buy_rate = 0.0153, min fee = 300 and moq = 10:')
print(self.r.get_purchase_result(self.prices, self.op, self.amounts, 10))
test_rate_with_min_result = self.r.get_purchase_result(self.prices, self.op, self.amounts, 10)
self.assertIs(np.allclose(test_rate_with_min_result[0], [0., 980, 0.]), True, 'result incorrect')
self.assertAlmostEqual(test_rate_with_min_result[1], -19900.0, msg='result incorrect')
self.assertAlmostEqual(test_rate_with_min_result[2], 300.0, msg='result incorrect')
print('\npurchase result with fixed cost rate with buy_rate = 0.0153, min fee = 300 and moq = 100:')
print(self.r.get_purchase_result(self.prices, self.op, self.amounts, 100))
test_rate_with_min_result = self.r.get_purchase_result(self.prices, self.op, self.amounts, 100)
self.assertIs(np.allclose(test_rate_with_min_result[0], [0., 900, 0.]), True, 'result incorrect')
self.assertAlmostEqual(test_rate_with_min_result[1], -18300.0, msg='result incorrect')
self.assertAlmostEqual(test_rate_with_min_result[2], 300.0, msg='result incorrect')
print('\nselling result with fixed cost rate with sell_rate = 0.01, min fee = 333 and moq = 0:')
print(self.r.get_selling_result(self.prices, self.op, self.amounts))
test_rate_with_min_result = self.r.get_selling_result(self.prices, self.op, self.amounts)
self.assertIs(np.allclose(test_rate_with_min_result[0], [0, 0, -3333.3333]), True, 'result incorrect')
self.assertAlmostEqual(test_rate_with_min_result[1], 32999.99967)
self.assertAlmostEqual(test_rate_with_min_result[2], 333.33333)
print('\nselling result with fixed cost rate with sell_rate = 0.01, min fee = 333 and moq = 1:')
print(self.r.get_selling_result(self.prices, self.op, self.amounts, 1))
test_rate_with_min_result = self.r.get_selling_result(self.prices, self.op, self.amounts, 1)
self.assertIs(np.allclose(test_rate_with_min_result[0], [0, 0, -3333]), True, 'result incorrect')
self.assertAlmostEqual(test_rate_with_min_result[1], 32996.7)
self.assertAlmostEqual(test_rate_with_min_result[2], 333.3)
print('\nselling result with fixed cost rate with sell_rate = 0.01, min fee = 333 and moq = 100:')
print(self.r.get_selling_result(self.prices, self.op, self.amounts, 100))
test_rate_with_min_result = self.r.get_selling_result(self.prices, self.op, self.amounts, 100)
self.assertIs(np.allclose(test_rate_with_min_result[0], [0, 0, -3300]), True, 'result incorrect')
self.assertAlmostEqual(test_rate_with_min_result[1], 32667.0)
self.assertAlmostEqual(test_rate_with_min_result[2], 333.0)
def test_fixed_fee(self):
self.r.buy_rate = 0.
self.r.sell_rate = 0.
self.r.buy_fix = 200
self.r.sell_fix = 150
self.r.buy_min = 0
self.r.sell_min = 0
self.r.slipage = 0
print('\nselling result of fixed cost with fixed fee = 150 and moq=0:')
print(self.r.get_selling_result(self.prices, self.op, self.amounts, 0))
test_fixed_fee_result = self.r.get_selling_result(self.prices, self.op, self.amounts)
self.assertIs(np.allclose(test_fixed_fee_result[0], [0, 0, -3333.3333]), True, 'result incorrect')
self.assertAlmostEqual(test_fixed_fee_result[1], 33183.333, msg='result incorrect')
self.assertAlmostEqual(test_fixed_fee_result[2], 150.0, msg='result incorrect')
print('\nselling result of fixed cost with fixed fee = 150 and moq=100:')
print(self.r.get_selling_result(self.prices, self.op, self.amounts, 100))
test_fixed_fee_result = self.r.get_selling_result(self.prices, self.op, self.amounts, 100)
self.assertIs(np.allclose(test_fixed_fee_result[0], [0, 0, -3300.]), True,
f'result incorrect, {test_fixed_fee_result[0]} does not equal to [0,0,-3400]')
self.assertAlmostEqual(test_fixed_fee_result[1], 32850., msg='result incorrect')
self.assertAlmostEqual(test_fixed_fee_result[2], 150., msg='result incorrect')
print('\npurchase result of fixed cost with fixed fee = 200:')
print(self.r.get_purchase_result(self.prices, self.op, self.amounts, 0))
test_fixed_fee_result = self.r.get_purchase_result(self.prices, self.op, self.amounts, 0)
self.assertIs(np.allclose(test_fixed_fee_result[0], [0., 990., 0.]), True, 'result incorrect')
self.assertAlmostEqual(test_fixed_fee_result[1], -20000.0, msg='result incorrect')
self.assertAlmostEqual(test_fixed_fee_result[2], 200.0, msg='result incorrect')
print('\npurchase result of fixed cost with fixed fee = 200:')
print(self.r.get_purchase_result(self.prices, self.op, self.amounts, 100))
test_fixed_fee_result = self.r.get_purchase_result(self.prices, self.op, self.amounts, 100)
self.assertIs(np.allclose(test_fixed_fee_result[0], [0., 900., 0.]), True, 'result incorrect')
self.assertAlmostEqual(test_fixed_fee_result[1], -18200.0, msg='result incorrect')
self.assertAlmostEqual(test_fixed_fee_result[2], 200.0, msg='result incorrect')
def test_slipage(self):
self.r.buy_fix = 0
self.r.sell_fix = 0
self.r.buy_min = 0
self.r.sell_min = 0
self.r.buy_rate = 0.003
self.r.sell_rate = 0.001
self.r.slipage = 1E-9
print('\npurchase result of fixed rate = 0.003 and slipage = 1E-10 and moq = 0:')
print(self.r.get_purchase_result(self.prices, self.op, self.amounts, 0))
print('\npurchase result of fixed rate = 0.003 and slipage = 1E-10 and moq = 100:')
print(self.r.get_purchase_result(self.prices, self.op, self.amounts, 100))
print('\nselling result with fixed rate = 0.001 and slipage = 1E-10:')
print(self.r.get_selling_result(self.prices, self.op, self.amounts))
test_fixed_fee_result = self.r.get_selling_result(self.prices, self.op, self.amounts)
self.assertIs(np.allclose(test_fixed_fee_result[0], [0, 0, -3333.3333]), True,
f'{test_fixed_fee_result[0]} does not equal to [0, 0, -10000]')
self.assertAlmostEqual(test_fixed_fee_result[1], 33298.88855591,
msg=f'{test_fixed_fee_result[1]} does not equal to 99890.')
self.assertAlmostEqual(test_fixed_fee_result[2], 34.44444409,
msg=f'{test_fixed_fee_result[2]} does not equal to -36.666663.')
test_fixed_fee_result = self.r.get_purchase_result(self.prices, self.op, self.amounts, 0)
self.assertIs(np.allclose(test_fixed_fee_result[0], [0., 996.98909294, 0.]), True, 'result incorrect')
self.assertAlmostEqual(test_fixed_fee_result[1], -20000.0, msg='result incorrect')
self.assertAlmostEqual(test_fixed_fee_result[2], 60.21814121353513, msg='result incorrect')
test_fixed_fee_result = self.r.get_purchase_result(self.prices, self.op, self.amounts, 100)
self.assertIs(np.allclose(test_fixed_fee_result[0], [0., 900., 0.]), True, 'result incorrect')
self.assertAlmostEqual(test_fixed_fee_result[1], -18054.36, msg='result incorrect')
self.assertAlmostEqual(test_fixed_fee_result[2], 54.36, msg='result incorrect')
class TestSpace(unittest.TestCase):
def test_creation(self):
"""
test if creation of space object is fine
"""
# first group of inputs, output Space with two discr axis from [0,10]
print('testing space objects\n')
# pars_list = [[(0, 10), (0, 10)],
# [[0, 10], [0, 10]]]
#
# types_list = ['discr',
# ['discr', 'discr']]
#
# input_pars = itertools.product(pars_list, types_list)
# for p in input_pars:
# # print(p)
# s = qt.Space(*p)
# b = s.boes
# t = s.types
# # print(s, t)
# self.assertIsInstance(s, qt.Space)
# self.assertEqual(b, [(0, 10), (0, 10)], 'boes incorrect!')
# self.assertEqual(t, ['discr', 'discr'], 'types incorrect')
#
pars_list = [[(0, 10), (0, 10)],
[[0, 10], [0, 10]]]
types_list = ['foo, bar',
['foo', 'bar']]
input_pars = itertools.product(pars_list, types_list)
for p in input_pars:
# print(p)
s = Space(*p)
b = s.boes
t = s.types
# print(s, t)
self.assertEqual(b, [(0, 10), (0, 10)], 'boes incorrect!')
self.assertEqual(t, ['enum', 'enum'], 'types incorrect')
pars_list = [[(0, 10), (0, 10)],
[[0, 10], [0, 10]]]
types_list = [['discr', 'foobar']]
input_pars = itertools.product(pars_list, types_list)
for p in input_pars:
# print(p)
s = Space(*p)
b = s.boes
t = s.types
# print(s, t)
self.assertEqual(b, [(0, 10), (0, 10)], 'boes incorrect!')
self.assertEqual(t, ['discr', 'enum'], 'types incorrect')
pars_list = [(0., 10), (0, 10)]
s = Space(pars=pars_list, par_types=None)
self.assertEqual(s.types, ['conti', 'discr'])
self.assertEqual(s.dim, 2)
self.assertEqual(s.size, (10.0, 11))
self.assertEqual(s.shape, (np.inf, 11))
self.assertEqual(s.count, np.inf)
self.assertEqual(s.boes, [(0., 10), (0, 10)])
pars_list = [(0., 10), (0, 10)]
s = Space(pars=pars_list, par_types='conti, enum')
self.assertEqual(s.types, ['conti', 'enum'])
self.assertEqual(s.dim, 2)
self.assertEqual(s.size, (10.0, 2))
self.assertEqual(s.shape, (np.inf, 2))
self.assertEqual(s.count, np.inf)
self.assertEqual(s.boes, [(0., 10), (0, 10)])
pars_list = [(1, 2), (2, 3), (3, 4)]
s = Space(pars=pars_list)
self.assertEqual(s.types, ['discr', 'discr', 'discr'])
self.assertEqual(s.dim, 3)
self.assertEqual(s.size, (2, 2, 2))
self.assertEqual(s.shape, (2, 2, 2))
self.assertEqual(s.count, 8)
self.assertEqual(s.boes, [(1, 2), (2, 3), (3, 4)])
pars_list = [(1, 2, 3), (2, 3, 4), (3, 4, 5)]
s = Space(pars=pars_list)
self.assertEqual(s.types, ['enum', 'enum', 'enum'])
self.assertEqual(s.dim, 3)
self.assertEqual(s.size, (3, 3, 3))
self.assertEqual(s.shape, (3, 3, 3))
self.assertEqual(s.count, 27)
self.assertEqual(s.boes, [(1, 2, 3), (2, 3, 4), (3, 4, 5)])
pars_list = [((1, 2, 3), (2, 3, 4), (3, 4, 5))]
s = Space(pars=pars_list)
self.assertEqual(s.types, ['enum'])
self.assertEqual(s.dim, 1)
self.assertEqual(s.size, (3,))
self.assertEqual(s.shape, (3,))
self.assertEqual(s.count, 3)
pars_list = ((1, 2, 3), (2, 3, 4), (3, 4, 5))
s = Space(pars=pars_list)
self.assertEqual(s.types, ['enum', 'enum', 'enum'])
self.assertEqual(s.dim, 3)
self.assertEqual(s.size, (3, 3, 3))
self.assertEqual(s.shape, (3, 3, 3))
self.assertEqual(s.count, 27)
self.assertEqual(s.boes, [(1, 2, 3), (2, 3, 4), (3, 4, 5)])
def test_extract(self):
"""
:return:
"""
pars_list = [(0, 10), (0, 10)]
types_list = ['discr', 'discr']
s = Space(pars=pars_list, par_types=types_list)
extracted_int, count = s.extract(3, 'interval')
extracted_int_list = list(extracted_int)
print('extracted int\n', extracted_int_list)
self.assertEqual(count, 16, 'extraction count wrong!')
self.assertEqual(extracted_int_list, [(0, 0), (0, 3), (0, 6), (0, 9), (3, 0), (3, 3),
(3, 6), (3, 9), (6, 0), (6, 3), (6, 6), (6, 9),
(9, 0), (9, 3), (9, 6), (9, 9)],
'space extraction wrong!')
extracted_rand, count = s.extract(10, 'rand')
extracted_rand_list = list(extracted_rand)
self.assertEqual(count, 10, 'extraction count wrong!')
print('extracted rand\n', extracted_rand_list)
for point in list(extracted_rand_list):
self.assertEqual(len(point), 2)
self.assertLessEqual(point[0], 10)
self.assertGreaterEqual(point[0], 0)
self.assertLessEqual(point[1], 10)
self.assertGreaterEqual(point[1], 0)
pars_list = [(0., 10), (0, 10)]
s = Space(pars=pars_list, par_types=None)
extracted_int2, count = s.extract(3, 'interval')
self.assertEqual(count, 16, 'extraction count wrong!')
extracted_int_list2 = list(extracted_int2)
self.assertEqual(extracted_int_list2, [(0, 0), (0, 3), (0, 6), (0, 9), (3, 0), (3, 3),
(3, 6), (3, 9), (6, 0), (6, 3), (6, 6), (6, 9),
(9, 0), (9, 3), (9, 6), (9, 9)],
'space extraction wrong!')
print('extracted int list 2\n', extracted_int_list2)
self.assertIsInstance(extracted_int_list2[0][0], float)
self.assertIsInstance(extracted_int_list2[0][1], (int, int64))
extracted_rand2, count = s.extract(10, 'rand')
self.assertEqual(count, 10, 'extraction count wrong!')
extracted_rand_list2 = list(extracted_rand2)
print('extracted rand list 2:\n', extracted_rand_list2)
for point in extracted_rand_list2:
self.assertEqual(len(point), 2)
self.assertIsInstance(point[0], float)
self.assertLessEqual(point[0], 10)
self.assertGreaterEqual(point[0], 0)
self.assertIsInstance(point[1], (int, int64))
self.assertLessEqual(point[1], 10)
self.assertGreaterEqual(point[1], 0)
pars_list = [(0., 10), ('a', 'b')]
s = Space(pars=pars_list, par_types='enum, enum')
extracted_int3, count = s.extract(1, 'interval')
self.assertEqual(count, 4, 'extraction count wrong!')
extracted_int_list3 = list(extracted_int3)
self.assertEqual(extracted_int_list3, [(0., 'a'), (0., 'b'), (10, 'a'), (10, 'b')],
'space extraction wrong!')
print('extracted int list 3\n', extracted_int_list3)
self.assertIsInstance(extracted_int_list3[0][0], float)
self.assertIsInstance(extracted_int_list3[0][1], str)
extracted_rand3, count = s.extract(3, 'rand')
self.assertEqual(count, 3, 'extraction count wrong!')
extracted_rand_list3 = list(extracted_rand3)
print('extracted rand list 3:\n', extracted_rand_list3)
for point in extracted_rand_list3:
self.assertEqual(len(point), 2)
self.assertIsInstance(point[0], (float, int))
self.assertLessEqual(point[0], 10)
self.assertGreaterEqual(point[0], 0)
self.assertIsInstance(point[1], str)
self.assertIn(point[1], ['a', 'b'])
pars_list = [((0, 10), (1, 'c'), ('a', 'b'), (1, 14))]
s = Space(pars=pars_list, par_types='enum')
extracted_int4, count = s.extract(1, 'interval')
self.assertEqual(count, 4, 'extraction count wrong!')
extracted_int_list4 = list(extracted_int4)
it = zip(extracted_int_list4, [(0, 10), (1, 'c'), (0, 'b'), (1, 14)])
for item, item2 in it:
print(item, item2)
self.assertTrue(all([tuple(ext_item) == item for ext_item, item in it]))
print('extracted int list 4\n', extracted_int_list4)
self.assertIsInstance(extracted_int_list4[0], tuple)
extracted_rand4, count = s.extract(3, 'rand')
self.assertEqual(count, 3, 'extraction count wrong!')
extracted_rand_list4 = list(extracted_rand4)
print('extracted rand list 4:\n', extracted_rand_list4)
for point in extracted_rand_list4:
self.assertEqual(len(point), 2)
self.assertIsInstance(point[0], (int, str))
self.assertIn(point[0], [0, 1, 'a'])
self.assertIsInstance(point[1], (int, str))
self.assertIn(point[1], [10, 14, 'b', 'c'])
self.assertIn(point, [(0., 10), (1, 'c'), ('a', 'b'), (1, 14)])
pars_list = [((0, 10), (1, 'c'), ('a', 'b'), (1, 14)), (1, 4)]
s = Space(pars=pars_list, par_types='enum, discr')
extracted_int5, count = s.extract(1, 'interval')
self.assertEqual(count, 16, 'extraction count wrong!')
extracted_int_list5 = list(extracted_int5)
for item, item2 in extracted_int_list5:
print(item, item2)
self.assertTrue(all([tuple(ext_item) == item for ext_item, item in it]))
print('extracted int list 5\n', extracted_int_list5)
self.assertIsInstance(extracted_int_list5[0], tuple)
extracted_rand5, count = s.extract(5, 'rand')
self.assertEqual(count, 5, 'extraction count wrong!')
extracted_rand_list5 = list(extracted_rand5)
print('extracted rand list 5:\n', extracted_rand_list5)
for point in extracted_rand_list5:
self.assertEqual(len(point), 2)
self.assertIsInstance(point[0], tuple)
print(f'type of point[1] is {type(point[1])}')
self.assertIsInstance(point[1], (int, np.int64))
self.assertIn(point[0], [(0., 10), (1, 'c'), ('a', 'b'), (1, 14)])
print(f'test incremental extraction')
pars_list = [(10., 250), (10., 250), (10., 250), (10., 250), (10., 250), (10., 250)]
s = Space(pars_list)
ext, count = s.extract(64, 'interval')
self.assertEqual(count, 4096)
points = list(ext)
# 已经取出所有的点,围绕其中10个点生成十个subspaces
# 检查是否每个subspace都为Space,是否都在s范围内,使用32生成点集,检查生成数量是否正确
for point in points[1000:1010]:
subspace = s.from_point(point, 64)
self.assertIsInstance(subspace, Space)
self.assertTrue(subspace in s)
self.assertEqual(subspace.dim, 6)
self.assertEqual(subspace.types, ['conti', 'conti', 'conti', 'conti', 'conti', 'conti'])
ext, count = subspace.extract(32)
points = list(ext)
self.assertGreaterEqual(count, 512)
self.assertLessEqual(count, 4096)
print(f'\n---------------------------------'
f'\nthe space created around point <{point}> is'
f'\n{subspace.boes}'
f'\nand extracted {count} points, the first 5 are:'
f'\n{points[:5]}')
def test_axis_extract(self):
# test axis object with conti type
axis = Axis((0., 5))
self.assertIsInstance(axis, Axis)
self.assertEqual(axis.axis_type, 'conti')
self.assertEqual(axis.axis_boe, (0., 5.))
self.assertEqual(axis.count, np.inf)
self.assertEqual(axis.size, 5.0)
self.assertTrue(np.allclose(axis.extract(1, 'int'), [0., 1., 2., 3., 4.]))
self.assertTrue(np.allclose(axis.extract(0.5, 'int'), [0., 0.5, 1., 1.5, 2., 2.5, 3., 3.5, 4., 4.5]))
extracted = axis.extract(8, 'rand')
self.assertEqual(len(extracted), 8)
self.assertTrue(all([(0 <= item <= 5) for item in extracted]))
# test axis object with discrete type
axis = Axis((1, 5))
self.assertIsInstance(axis, Axis)
self.assertEqual(axis.axis_type, 'discr')
self.assertEqual(axis.axis_boe, (1, 5))
self.assertEqual(axis.count, 5)
self.assertEqual(axis.size, 5)
self.assertTrue(np.allclose(axis.extract(1, 'int'), [1, 2, 3, 4, 5]))
self.assertRaises(ValueError, axis.extract, 0.5, 'int')
extracted = axis.extract(8, 'rand')
self.assertEqual(len(extracted), 8)
self.assertTrue(all([(item in [1, 2, 3, 4, 5]) for item in extracted]))
# test axis object with enumerate type
axis = Axis((1, 5, 7, 10, 'A', 'F'))
self.assertIsInstance(axis, Axis)
self.assertEqual(axis.axis_type, 'enum')
self.assertEqual(axis.axis_boe, (1, 5, 7, 10, 'A', 'F'))
self.assertEqual(axis.count, 6)
self.assertEqual(axis.size, 6)
self.assertEqual(axis.extract(1, 'int'), [1, 5, 7, 10, 'A', 'F'])
self.assertRaises(ValueError, axis.extract, 0.5, 'int')
extracted = axis.extract(8, 'rand')
self.assertEqual(len(extracted), 8)
self.assertTrue(all([(item in [1, 5, 7, 10, 'A', 'F']) for item in extracted]))
def test_from_point(self):
"""测试从一个点生成一个space"""
# 生成一个space,指定space中的一个点以及distance,生成一个sub-space
pars_list = [(0., 10), (0, 10)]
s = Space(pars=pars_list, par_types=None)
self.assertEqual(s.types, ['conti', 'discr'])
self.assertEqual(s.dim, 2)
self.assertEqual(s.size, (10., 11))
self.assertEqual(s.shape, (np.inf, 11))
self.assertEqual(s.count, np.inf)
self.assertEqual(s.boes, [(0., 10), (0, 10)])
print('create subspace from a point in space')
p = (3, 3)
distance = 2
subspace = s.from_point(p, distance)
self.assertIsInstance(subspace, Space)
self.assertEqual(subspace.types, ['conti', 'discr'])
self.assertEqual(subspace.dim, 2)
self.assertEqual(subspace.size, (4.0, 5))
self.assertEqual(subspace.shape, (np.inf, 5))
self.assertEqual(subspace.count, np.inf)
self.assertEqual(subspace.boes, [(1, 5), (1, 5)])
print('create subspace from a 6 dimensional discrete space')
s = Space(pars=[(10, 250), (10, 250), (10, 250), (10, 250), (10, 250), (10, 250)])
p = (15, 200, 150, 150, 150, 150)
d = 10
subspace = s.from_point(p, d)
self.assertIsInstance(subspace, Space)
self.assertEqual(subspace.types, ['discr', 'discr', 'discr', 'discr', 'discr', 'discr'])
self.assertEqual(subspace.dim, 6)
self.assertEqual(subspace.volume, 65345616)
self.assertEqual(subspace.size, (16, 21, 21, 21, 21, 21))
self.assertEqual(subspace.shape, (16, 21, 21, 21, 21, 21))
self.assertEqual(subspace.count, 65345616)
self.assertEqual(subspace.boes, [(10, 25), (190, 210), (140, 160), (140, 160), (140, 160), (140, 160)])
print('create subspace from a 6 dimensional continuous space')
s = Space(pars=[(10., 250), (10., 250), (10., 250), (10., 250), (10., 250), (10., 250)])
p = (15, 200, 150, 150, 150, 150)
d = 10
subspace = s.from_point(p, d)
self.assertIsInstance(subspace, Space)
self.assertEqual(subspace.types, ['conti', 'conti', 'conti', 'conti', 'conti', 'conti'])
self.assertEqual(subspace.dim, 6)
self.assertEqual(subspace.volume, 48000000)
self.assertEqual(subspace.size, (15.0, 20.0, 20.0, 20.0, 20.0, 20.0))
self.assertEqual(subspace.shape, (np.inf, np.inf, np.inf, np.inf, np.inf, np.inf))
self.assertEqual(subspace.count, np.inf)
self.assertEqual(subspace.boes, [(10, 25), (190, 210), (140, 160), (140, 160), (140, 160), (140, 160)])
print('create subspace with different distances on each dimension')
s = Space(pars=[(10., 250), (10., 250), (10., 250), (10., 250), (10., 250), (10., 250)])
p = (15, 200, 150, 150, 150, 150)
d = [10, 5, 5, 10, 10, 5]
subspace = s.from_point(p, d)
self.assertIsInstance(subspace, Space)
self.assertEqual(subspace.types, ['conti', 'conti', 'conti', 'conti', 'conti', 'conti'])
self.assertEqual(subspace.dim, 6)
self.assertEqual(subspace.volume, 6000000)
self.assertEqual(subspace.size, (15.0, 10.0, 10.0, 20.0, 20.0, 10.0))
self.assertEqual(subspace.shape, (np.inf, np.inf, np.inf, np.inf, np.inf, np.inf))
self.assertEqual(subspace.count, np.inf)
self.assertEqual(subspace.boes, [(10, 25), (195, 205), (145, 155), (140, 160), (140, 160), (145, 155)])
class TestCashPlan(unittest.TestCase):
def setUp(self):
self.cp1 = qt.CashPlan(['2012-01-01', '2010-01-01'], [10000, 20000], 0.1)
self.cp1.info()
self.cp2 = qt.CashPlan(['20100501'], 10000)
self.cp2.info()
self.cp3 = qt.CashPlan(pd.date_range(start='2019-01-01',
freq='Y',
periods=12),
[i * 1000 + 10000 for i in range(12)],
0.035)
self.cp3.info()
def test_creation(self):
self.assertIsInstance(self.cp1, qt.CashPlan, 'CashPlan object creation wrong')
self.assertIsInstance(self.cp2, qt.CashPlan, 'CashPlan object creation wrong')
self.assertIsInstance(self.cp3, qt.CashPlan, 'CashPlan object creation wrong')
# test __repr__()
print(self.cp1)
print(self.cp2)
print(self.cp3)
# test __str__()
self.cp1.info()
self.cp2.info()
self.cp3.info()
# test assersion errors
self.assertRaises(AssertionError, qt.CashPlan, '2016-01-01', [10000, 10000])
self.assertRaises(KeyError, qt.CashPlan, '2020-20-20', 10000)
def test_properties(self):
self.assertEqual(self.cp1.amounts, [20000, 10000], 'property wrong')
self.assertEqual(self.cp1.first_day, Timestamp('2010-01-01'))
self.assertEqual(self.cp1.last_day, Timestamp('2012-01-01'))
self.assertEqual(self.cp1.investment_count, 2)
self.assertEqual(self.cp1.period, 730)
self.assertEqual(self.cp1.dates, [Timestamp('2010-01-01'), Timestamp('2012-01-01')])
self.assertEqual(self.cp1.ir, 0.1)
self.assertAlmostEqual(self.cp1.closing_value, 34200)
self.assertAlmostEqual(self.cp2.closing_value, 10000)
self.assertAlmostEqual(self.cp3.closing_value, 220385.3483685)
self.assertIsInstance(self.cp1.plan, pd.DataFrame)
self.assertIsInstance(self.cp2.plan, pd.DataFrame)
self.assertIsInstance(self.cp3.plan, pd.DataFrame)
def test_operation(self):
cp_self_add = self.cp1 + self.cp1
cp_add = self.cp1 + self.cp2
cp_add_int = self.cp1 + 10000
cp_mul_int = self.cp1 * 2
cp_mul_float = self.cp2 * 1.5
cp_mul_time = 3 * self.cp2
cp_mul_time2 = 2 * self.cp1
cp_mul_time3 = 2 * self.cp3
cp_mul_float2 = 2. * self.cp3
self.assertIsInstance(cp_self_add, qt.CashPlan)
self.assertEqual(cp_self_add.amounts, [40000, 20000])
self.assertEqual(cp_add.amounts, [20000, 10000, 10000])
self.assertEqual(cp_add_int.amounts, [30000, 20000])
self.assertEqual(cp_mul_int.amounts, [40000, 20000])
self.assertEqual(cp_mul_float.amounts, [15000])
self.assertEqual(cp_mul_float.dates, [Timestamp('2010-05-01')])
self.assertEqual(cp_mul_time.amounts, [10000, 10000, 10000])
self.assertEqual(cp_mul_time.dates, [Timestamp('2010-05-01'),
Timestamp('2011-05-01'),
Timestamp('2012-04-30')])
self.assertEqual(cp_mul_time2.amounts, [20000, 10000, 20000, 10000])
self.assertEqual(cp_mul_time2.dates, [Timestamp('2010-01-01'),
Timestamp('2012-01-01'),
Timestamp('2014-01-01'),
Timestamp('2016-01-01')])
self.assertEqual(cp_mul_time3.dates, [Timestamp('2019-12-31'),
Timestamp('2020-12-31'),
Timestamp('2021-12-31'),
Timestamp('2022-12-31'),
Timestamp('2023-12-31'),
Timestamp('2024-12-31'),
Timestamp('2025-12-31'),
Timestamp('2026-12-31'),
Timestamp('2027-12-31'),
Timestamp('2028-12-31'),
Timestamp('2029-12-31'),
Timestamp('2030-12-31'),
Timestamp('2031-12-29'),
Timestamp('2032-12-29'),
Timestamp('2033-12-29'),
Timestamp('2034-12-29'),
Timestamp('2035-12-29'),
Timestamp('2036-12-29'),
Timestamp('2037-12-29'),
Timestamp('2038-12-29'),
Timestamp('2039-12-29'),
Timestamp('2040-12-29'),
Timestamp('2041-12-29'),
Timestamp('2042-12-29')])
self.assertEqual(cp_mul_float2.dates, [Timestamp('2019-12-31'),
Timestamp('2020-12-31'),
Timestamp('2021-12-31'),
Timestamp('2022-12-31'),
Timestamp('2023-12-31'),
Timestamp('2024-12-31'),
Timestamp('2025-12-31'),
Timestamp('2026-12-31'),
Timestamp('2027-12-31'),
Timestamp('2028-12-31'),
Timestamp('2029-12-31'),
Timestamp('2030-12-31')])
self.assertEqual(cp_mul_float2.amounts, [20000.0,
22000.0,
24000.0,
26000.0,
28000.0,
30000.0,
32000.0,
34000.0,
36000.0,
38000.0,
40000.0,
42000.0])
class TestPool(unittest.TestCase):
def setUp(self):
self.p = ResultPool(5)
self.items = ['first', 'second', (1, 2, 3), 'this', 24]
self.perfs = [1, 2, 3, 4, 5]
self.additional_result1 = ('abc', 12)
self.additional_result2 = ([1, 2], -1)
self.additional_result3 = (12, 5)
def test_create(self):
self.assertIsInstance(self.p, ResultPool)
def test_operation(self):
self.p.in_pool(self.additional_result1[0], self.additional_result1[1])
self.p.cut()
self.assertEqual(self.p.item_count, 1)
self.assertEqual(self.p.items, ['abc'])
for item, perf in zip(self.items, self.perfs):
self.p.in_pool(item, perf)
self.assertEqual(self.p.item_count, 6)
self.assertEqual(self.p.items, ['abc', 'first', 'second', (1, 2, 3), 'this', 24])
self.p.cut()
self.assertEqual(self.p.items, ['second', (1, 2, 3), 'this', 24, 'abc'])
self.assertEqual(self.p.perfs, [2, 3, 4, 5, 12])
self.p.in_pool(self.additional_result2[0], self.additional_result2[1])
self.p.in_pool(self.additional_result3[0], self.additional_result3[1])
self.assertEqual(self.p.item_count, 7)
self.p.cut(keep_largest=False)
self.assertEqual(self.p.items, [[1, 2], 'second', (1, 2, 3), 'this', 24])
self.assertEqual(self.p.perfs, [-1, 2, 3, 4, 5])
class TestCoreSubFuncs(unittest.TestCase):
"""Test all functions in core.py"""
def setUp(self):
pass
def test_input_to_list(self):
print('Testing input_to_list() function')
input_str = 'first'
self.assertEqual(qt.utilfuncs.input_to_list(input_str, 3), ['first', 'first', 'first'])
self.assertEqual(qt.utilfuncs.input_to_list(input_str, 4), ['first', 'first', 'first', 'first'])
self.assertEqual(qt.utilfuncs.input_to_list(input_str, 2, None), ['first', 'first'])
input_list = ['first', 'second']
self.assertEqual(qt.utilfuncs.input_to_list(input_list, 3), ['first', 'second', None])
self.assertEqual(qt.utilfuncs.input_to_list(input_list, 4, 'padder'), ['first', 'second', 'padder', 'padder'])
self.assertEqual(qt.utilfuncs.input_to_list(input_list, 1), ['first', 'second'])
self.assertEqual(qt.utilfuncs.input_to_list(input_list, -5), ['first', 'second'])
def test_point_in_space(self):
sp = Space([(0., 10.), (0., 10.), (0., 10.)])
p1 = (5.5, 3.2, 7)
p2 = (-1, 3, 10)
self.assertTrue(p1 in sp)
print(f'point {p1} is in space {sp}')
self.assertFalse(p2 in sp)
print(f'point {p2} is not in space {sp}')
sp = Space([(0., 10.), (0., 10.), range(40, 3, -2)], 'conti, conti, enum')
p1 = (5.5, 3.2, 8)
self.assertTrue(p1 in sp)
print(f'point {p1} is in space {sp}')
def test_space_in_space(self):
print('test if a space is in another space')
sp = Space([(0., 10.), (0., 10.), (0., 10.)])
sp2 = Space([(0., 10.), (0., 10.), (0., 10.)])
self.assertTrue(sp2 in sp)
self.assertTrue(sp in sp2)
print(f'space {sp2} is in space {sp}\n'
f'and space {sp} is in space {sp2}\n'
f'they are equal to each other\n')
sp2 = Space([(0, 5.), (2, 7.), (3., 9.)])
self.assertTrue(sp2 in sp)
self.assertFalse(sp in sp2)
print(f'space {sp2} is in space {sp}\n'
f'and space {sp} is not in space {sp2}\n'
f'{sp2} is a sub space of {sp}\n')
sp2 = Space([(0, 5), (2, 7), (3., 9)])
self.assertFalse(sp2 in sp)
self.assertFalse(sp in sp2)
print(f'space {sp2} is not in space {sp}\n'
f'and space {sp} is not in space {sp2}\n'
f'they have different types of axes\n')
sp = Space([(0., 10.), (0., 10.), range(40, 3, -2)])
self.assertFalse(sp in sp2)
self.assertFalse(sp2 in sp)
print(f'space {sp2} is not in space {sp}\n'
f'and space {sp} is not in space {sp2}\n'
f'they have different types of axes\n')
def test_space_around_centre(self):
sp = Space([(0., 10.), (0., 10.), (0., 10.)])
p1 = (5.5, 3.2, 7)
ssp = space_around_centre(space=sp, centre=p1, radius=1.2)
print(ssp.boes)
print('\ntest multiple diameters:')
self.assertEqual(ssp.boes, [(4.3, 6.7), (2.0, 4.4), (5.8, 8.2)])
ssp = space_around_centre(space=sp, centre=p1, radius=[1, 2, 1])
print(ssp.boes)
self.assertEqual(ssp.boes, [(4.5, 6.5), (1.2000000000000002, 5.2), (6.0, 8.0)])
print('\ntest points on edge:')
p2 = (5.5, 3.2, 10)
ssp = space_around_centre(space=sp, centre=p1, radius=3.9)
print(ssp.boes)
self.assertEqual(ssp.boes, [(1.6, 9.4), (0.0, 7.1), (3.1, 10.0)])
print('\ntest enum spaces')
sp = Space([(0, 100), range(40, 3, -2)], 'discr, enum')
p1 = [34, 12]
ssp = space_around_centre(space=sp, centre=p1, radius=5, ignore_enums=False)
self.assertEqual(ssp.boes, [(29, 39), (22, 20, 18, 16, 14, 12, 10, 8, 6, 4)])
print(ssp.boes)
print('\ntest enum space and ignore enum axis')
ssp = space_around_centre(space=sp, centre=p1, radius=5)
self.assertEqual(ssp.boes, [(29, 39),
(40, 38, 36, 34, 32, 30, 28, 26, 24, 22, 20, 18, 16, 14, 12, 10, 8, 6, 4)])
print(sp.boes)
def test_time_string_format(self):
print('Testing qt.time_string_format() function:')
t = 3.14
self.assertEqual(time_str_format(t), '3s 140.0ms')
self.assertEqual(time_str_format(t, estimation=True), '3s ')
self.assertEqual(time_str_format(t, short_form=True), '3"140')
self.assertEqual(time_str_format(t, estimation=True, short_form=True), '3"')
t = 300.14
self.assertEqual(time_str_format(t), '5min 140.0ms')
self.assertEqual(time_str_format(t, estimation=True), '5min ')
self.assertEqual(time_str_format(t, short_form=True), "5'140")
self.assertEqual(time_str_format(t, estimation=True, short_form=True), "5'")
t = 7435.0014
self.assertEqual(time_str_format(t), '2hrs 3min 55s 1.4ms')
self.assertEqual(time_str_format(t, estimation=True), '2hrs ')
self.assertEqual(time_str_format(t, short_form=True), "2H3'55\"001")
self.assertEqual(time_str_format(t, estimation=True, short_form=True), "2H")
t = 88425.0509
self.assertEqual(time_str_format(t), '1days 33min 45s 50.9ms')
self.assertEqual(time_str_format(t, estimation=True), '1days ')
self.assertEqual(time_str_format(t, short_form=True), "1D33'45\"051")
self.assertEqual(time_str_format(t, estimation=True, short_form=True), "1D")
def test_get_stock_pool(self):
print(f'start test building stock pool function\n')
share_basics = stock_basic(fields='ts_code,symbol,name,area,industry,market,list_date,exchange')
print(f'\nselect all stocks by area')
stock_pool = qt.get_stock_pool(area='上海')
print(f'{len(stock_pool)} shares selected, first 5 are: {stock_pool[0:5]}\n'
f'check if all stock areas are "上海"\n'
f'{share_basics[np.isin(share_basics.ts_code, stock_pool)].head()}')
self.assertTrue(share_basics[np.isin(share_basics.ts_code, stock_pool)]['area'].eq('上海').all())
print(f'\nselect all stocks by multiple areas')
stock_pool = qt.get_stock_pool(area='贵州,北京,天津')
print(f'\n{len(stock_pool)} shares selected, first 5 are: {stock_pool[0:5]}\n'
f'check if all stock areas are in list of ["贵州", "北京", "天津"]\n'
f'{share_basics[np.isin(share_basics.ts_code, stock_pool)].head()}')
self.assertTrue(share_basics[np.isin(share_basics.ts_code, stock_pool)]['area'].isin(['贵州',
'北京',
'天津']).all())
print(f'\nselect all stocks by area and industry')
stock_pool = qt.get_stock_pool(area='四川', industry='银行, 金融')
print(f'\n{len(stock_pool)} shares selected, first 5 are: {stock_pool[0:5]}\n'
f'check if all stock areas are "四川", and industry in ["银行", "金融"]\n'
f'{share_basics[np.isin(share_basics.ts_code, stock_pool)].head()}')
self.assertTrue(share_basics[np.isin(share_basics.ts_code, stock_pool)]['industry'].isin(['银行', '金融']).all())
self.assertTrue(share_basics[np.isin(share_basics.ts_code, stock_pool)]['area'].isin(['四川']).all())
print(f'\nselect all stocks by industry')
stock_pool = qt.get_stock_pool(industry='银行, 金融')
print(f'\n{len(stock_pool)} shares selected, first 5 are: {stock_pool[0:5]}\n'
f'check if all stocks industry in ["银行", "金融"]\n'
f'{share_basics[np.isin(share_basics.ts_code, stock_pool)].head()}')
self.assertTrue(share_basics[np.isin(share_basics.ts_code, stock_pool)]['industry'].isin(['银行', '金融']).all())
print(f'\nselect all stocks by market')
stock_pool = qt.get_stock_pool(market='主板')
print(f'\n{len(stock_pool)} shares selected, first 5 are: {stock_pool[0:5]}\n'
f'check if all stock market is "主板"\n'
f'{share_basics[np.isin(share_basics.ts_code, stock_pool)].head()}')
self.assertTrue(share_basics[np.isin(share_basics.ts_code, stock_pool)]['market'].isin(['主板']).all())
print(f'\nselect all stocks by market and list date')
stock_pool = qt.get_stock_pool(date='2000-01-01', market='主板')
print(f'\n{len(stock_pool)} shares selected, first 5 are: {stock_pool[0:5]}\n'
f'check if all stock market is "主板", and list date after "2000-01-01"\n'
f'{share_basics[np.isin(share_basics.ts_code, stock_pool)].head()}')
self.assertTrue(share_basics[np.isin(share_basics.ts_code, stock_pool)]['market'].isin(['主板']).all())
self.assertTrue(share_basics[np.isin(share_basics.ts_code, stock_pool)]['list_date'].le('2000-01-01').all())
print(f'\nselect all stocks by list date')
stock_pool = qt.get_stock_pool(date='1997-01-01')
print(f'\n{len(stock_pool)} shares selected, first 5 are: {stock_pool[0:5]}\n'
f'check if all list date after "1997-01-01"\n'
f'{share_basics[np.isin(share_basics.ts_code, stock_pool)].head()}')
self.assertTrue(share_basics[np.isin(share_basics.ts_code, stock_pool)]['list_date'].le('1997-01-01').all())
print(f'\nselect all stocks by exchange')
stock_pool = qt.get_stock_pool(exchange='SSE')
print(f'\n{len(stock_pool)} shares selected, first 5 are: {stock_pool[0:5]}\n'
f'check if all exchanges are "SSE"\n'
f'{share_basics[np.isin(share_basics.ts_code, stock_pool)].head()}')
self.assertTrue(share_basics[np.isin(share_basics.ts_code, stock_pool)]['exchange'].eq('SSE').all())
print(f'\nselect all stocks by industry, area and list date')
industry_list = ['银行', '全国地产', '互联网', '环境保护', '区域地产',
'酒店餐饮', '运输设备', '综合类', '建筑工程', '玻璃',
'家用电器', '文教休闲', '其他商业', '元器件', 'IT设备',
'其他建材', '汽车服务', '火力发电', '医药商业', '汽车配件',
'广告包装', '轻工机械', '新型电力', '多元金融', '饲料']
area_list = ['深圳', '北京', '吉林', '江苏', '辽宁', '广东',
'安徽', '四川', '浙江', '湖南', '河北', '新疆',
'山东', '河南', '山西', '江西', '青海', '湖北',
'内蒙', '海南', '重庆', '陕西', '福建', '广西',
'上海']
stock_pool = qt.get_stock_pool(date='19980101',
industry=industry_list,
area=area_list)
print(f'\n{len(stock_pool)} shares selected, first 5 are: {stock_pool[0:5]}\n'
f'check if all exchanges are "SSE"\n'
f'{share_basics[np.isin(share_basics.ts_code, stock_pool)].head()}')
self.assertTrue(share_basics[np.isin(share_basics.ts_code, stock_pool)]['list_date'].le('1998-01-01').all())
self.assertTrue(share_basics[np.isin(share_basics.ts_code, stock_pool)]['industry'].isin(industry_list).all())
self.assertTrue(share_basics[np.isin(share_basics.ts_code, stock_pool)]['area'].isin(area_list).all())
self.assertRaises(KeyError, qt.get_stock_pool, industry=25)
self.assertRaises(KeyError, qt.get_stock_pool, share_name='000300.SH')
self.assertRaises(KeyError, qt.get_stock_pool, markets='SSE')
class TestEvaluations(unittest.TestCase):
"""Test all evaluation functions in core.py"""
# 以下手动计算结果在Excel文件中
def setUp(self):
"""用np.random生成测试用数据,使用cumsum()模拟股票走势"""
self.test_data1 = pd.DataFrame([5.34892759, 5.65768696, 5.79227076, 5.56266871, 5.88189632,
6.24795001, 5.92755558, 6.38748165, 6.31331899, 5.86001665,
5.61048472, 5.30696736, 5.40406792, 5.03180571, 5.37886353,
5.78608307, 6.26540339, 6.59348026, 6.90943801, 6.70911677,
6.33015954, 6.06697417, 5.9752499, 6.45786408, 6.95273763,
6.7691991, 6.70355481, 6.28048969, 6.61344541, 6.24620003,
6.47409983, 6.4522311, 6.8773094, 6.99727832, 6.59262674,
6.59014938, 6.63758237, 6.38331869, 6.09902105, 6.35390109,
6.51993567, 6.87244592, 6.83963485, 7.08797815, 6.88003144,
6.83657323, 6.97819483, 7.01600276, 7.12554256, 7.58941523,
7.61014457, 7.21224091, 7.48174399, 7.66490854, 7.51371968,
7.11586198, 6.97147399, 6.67453301, 6.2042138, 6.33967015,
6.22187938, 5.98426993, 6.37096079, 6.55897161, 6.26422645,
6.69363762, 7.12668015, 6.83232926, 7.30524081, 7.4262041,
7.54031383, 7.17545919, 7.20659257, 7.44886016, 7.37094393,
6.88011022, 7.08142491, 6.74992833, 6.5967097, 6.21336693,
6.35565105, 6.82347596, 6.44773408, 6.84538053, 6.47966466,
6.09699528, 5.63927014, 6.01081024, 6.20585303, 6.60528206,
7.01594726, 7.03684251, 6.76574977, 7.08740846, 6.65336462,
7.07126686, 6.80058956, 6.79241977, 6.47843472, 6.39245474],
columns=['value'])
self.test_data2 = pd.DataFrame([5.09276527, 4.83828592, 4.6000911, 4.63170487, 4.63566451,
4.50546921, 4.96390044, 4.64557907, 4.25787855, 3.76585551,
3.38826334, 3.76243422, 4.06365426, 3.87084726, 3.91400935,
4.13438822, 4.27064542, 4.56776104, 5.03800296, 5.31070529,
5.39902276, 5.21186286, 5.05683114, 4.68842046, 5.11895168,
5.27151571, 5.72294993, 6.09961056, 6.26569635, 6.48806151,
6.16058885, 6.2582459, 6.38934791, 6.57831057, 6.19508831,
5.70155153, 5.20435735, 5.36538825, 5.40450056, 5.2227697,
5.37828693, 5.53058991, 6.02996797, 5.76802181, 5.66166713,
6.07988994, 5.61794367, 5.63218151, 6.10728013, 6.0324168,
6.27164431, 6.27551239, 6.52329665, 7.00470007, 7.34163113,
7.33699083, 7.67661334, 8.09395749, 7.68086668, 7.58341161,
7.46219819, 7.58671899, 7.19348298, 7.40088323, 7.47562005,
7.93342043, 8.2286081, 8.3521632, 8.43590025, 8.34977395,
8.57563095, 8.81586328, 9.08738649, 9.01542031, 8.8653815,
9.21763111, 9.04233017, 8.59533999, 8.47590075, 8.70857222,
8.78890756, 8.92697606, 9.35743773, 9.68280866, 10.15622021,
10.55908549, 10.6337894, 10.55197128, 10.65435176, 10.54611045,
10.19432562, 10.48320884, 10.36176768, 10.03186854, 10.23656092,
10.0062843, 10.13669686, 10.30758958, 9.87904176, 10.05126375],
columns=['value'])
self.test_data3 = pd.DataFrame([5.02851874, 5.20700348, 5.02410709, 5.49836387, 5.06834371,
5.10956737, 5.15314979, 5.02256472, 5.09746382, 5.23909247,
4.93410336, 4.96316186, 5.40026682, 5.7353255, 5.53438319,
5.79092139, 5.67528173, 5.89840855, 5.75379463, 6.10855386,
5.77322365, 5.84538021, 5.6103973, 5.7518655, 5.49729695,
5.13610628, 5.30524121, 5.68093462, 5.73251319, 6.04420783,
6.26929843, 6.59610234, 6.09872345, 6.25475121, 6.72927396,
6.91395783, 7.00693283, 7.36217783, 7.71516676, 7.67580263,
7.62477511, 7.73600568, 7.53457914, 7.46170277, 7.83658014,
8.11481319, 8.03705544, 7.64948845, 7.52043731, 7.67247943,
7.46511982, 7.43541798, 7.58856517, 7.9392717, 8.25406287,
7.77031632, 8.03223447, 7.86799055, 7.57630999, 7.33230519,
7.22378732, 6.85972264, 7.17548456, 7.5387846, 7.2392632,
6.8455644, 6.59557185, 6.6496796, 6.73685623, 7.18598015,
7.13619128, 6.88060157, 7.1399681, 7.30308077, 6.94942434,
7.0247815, 7.37567798, 7.50080197, 7.59719284, 7.14520561,
7.29913484, 7.79551341, 8.15497781, 8.40456095, 8.86516528,
8.53042688, 8.94268762, 8.52048006, 8.80036284, 8.91602364,
9.19953385, 8.70828953, 8.24613093, 8.18770453, 7.79548389,
7.68627967, 7.23205036, 6.98302636, 7.06515819, 6.95068113],
columns=['value'])
self.test_data4 = pd.DataFrame([4.97926539, 5.44016005, 5.45122915, 5.74485615, 5.45600553,
5.44858945, 5.2435413, 5.47315161, 5.58464303, 5.36179749,
5.38236326, 5.29614981, 5.76523508, 5.75102892, 6.15316618,
6.03852528, 6.01442228, 5.70510182, 5.22748133, 5.46762379,
5.78926267, 5.8221362, 5.61236849, 5.30615725, 5.24200611,
5.41042642, 5.59940342, 5.28306781, 4.99451932, 5.08799266,
5.38865647, 5.58229139, 5.33492845, 5.48206276, 5.09721379,
5.39190493, 5.29965087, 5.0374415, 5.50798022, 5.43107577,
5.22759507, 4.991809, 5.43153084, 5.39966868, 5.59916352,
5.66412137, 6.00611838, 5.63564902, 5.66723484, 5.29863863,
4.91115153, 5.3749929, 5.75082334, 6.08308148, 6.58091182,
6.77848803, 7.19588758, 7.64862286, 7.99818347, 7.91824794,
8.30341071, 8.45984973, 7.98700002, 8.18924931, 8.60755649,
8.66233396, 8.91018407, 9.0782739, 9.33515448, 8.95870245,
8.98426422, 8.50340317, 8.64916085, 8.93592407, 8.63145745,
8.65322862, 8.39543204, 8.37969997, 8.23394504, 8.04062872,
7.91259763, 7.57252171, 7.72670114, 7.74486117, 8.06908188,
7.99166889, 7.92155906, 8.39956136, 8.80181323, 8.47464091,
8.06557064, 7.87145573, 8.0237959, 8.39481998, 8.68525692,
8.81185461, 8.98632237, 9.0989835, 8.89787405, 8.86508591],
columns=['value'])
self.test_data5 = pd.DataFrame([4.50258923, 4.35142568, 4.07459514, 3.87791297, 3.73715985,
3.98455684, 4.07587908, 4.00042472, 4.28276612, 4.01362051,
4.13713565, 4.49312372, 4.48633159, 4.4641207, 4.13444605,
3.79107217, 4.22941629, 4.56548511, 4.92472163, 5.27723158,
5.67409193, 6.00176917, 5.88889928, 5.55256103, 5.39308314,
5.2610492, 5.30738908, 5.22222408, 4.90332238, 4.57499908,
4.96097146, 4.81531011, 4.39115442, 4.63200662, 5.04588813,
4.67866025, 5.01705123, 4.83562258, 4.60381702, 4.66187576,
4.41292828, 4.86604507, 4.42280124, 4.07517294, 4.16317319,
4.10316596, 4.42913598, 4.06609666, 3.96725913, 4.15965746,
4.12379564, 4.04054068, 3.84342851, 3.45902867, 3.17649855,
3.09773586, 3.5502119, 3.66396995, 3.66306483, 3.29131401,
2.79558533, 2.88319542, 3.03671098, 3.44645857, 3.88167161,
3.57961874, 3.60180276, 3.96702102, 4.05429995, 4.40056979,
4.05653231, 3.59600456, 3.60792477, 4.09989922, 3.73503663,
4.01892626, 3.94597242, 3.81466605, 3.71417992, 3.93767156,
4.42806557, 4.06988106, 4.03713636, 4.34408673, 4.79810156,
5.18115011, 4.89798406, 5.3960077, 5.72504875, 5.61894017,
5.1958197, 4.85275896, 5.17550207, 4.71548987, 4.62408567,
4.55488535, 4.36532649, 4.26031979, 4.25225607, 4.58627048],
columns=['value'])
self.test_data6 = pd.DataFrame([5.08639513, 5.05761083, 4.76160923, 4.62166504, 4.62923183,
4.25070173, 4.13447513, 3.90890013, 3.76687608, 3.43342482,
3.67648224, 3.6274775, 3.9385404, 4.39771627, 4.03199346,
3.93265288, 3.50059789, 3.3851961, 3.29743973, 3.2544872,
2.93692949, 2.70893003, 2.55461976, 2.20922332, 2.29054475,
2.2144714, 2.03726827, 2.39007617, 2.29866155, 2.40607111,
2.40440444, 2.79374649, 2.66541922, 2.27018079, 2.08505127,
2.55478864, 2.22415625, 2.58517923, 2.58802256, 2.94870959,
2.69301739, 2.19991535, 2.69473146, 2.64704637, 2.62753542,
2.14240825, 2.38565154, 1.94592117, 2.32243877, 2.69337246,
2.51283854, 2.62484451, 2.15559054, 2.35410875, 2.31219177,
1.96018265, 2.34711266, 2.58083322, 2.40290041, 2.20439791,
2.31472425, 2.16228248, 2.16439749, 2.20080737, 1.73293206,
1.9264407, 2.25089861, 2.69269101, 2.59296687, 2.1420998,
1.67819153, 1.98419023, 2.14479494, 1.89055376, 1.96720648,
1.9916694, 2.37227761, 2.14446036, 2.34573903, 1.86162546,
2.1410721, 2.39204939, 2.52529064, 2.47079939, 2.9299031,
3.09452923, 2.93276708, 3.21731309, 3.06248964, 2.90413406,
2.67844632, 2.45621213, 2.41463398, 2.7373913, 3.14917045,
3.4033949, 3.82283446, 4.02285451, 3.7619638, 4.10346795],
columns=['value'])
self.test_data7 = pd.DataFrame([4.75233583, 4.47668283, 4.55894263, 4.61765848, 4.622892,
4.58941116, 4.32535872, 3.88112797, 3.47237806, 3.50898953,
3.82530406, 3.6718017, 3.78918195, 4.1800752, 4.01818557,
4.40822582, 4.65474654, 4.89287256, 4.40879274, 4.65505126,
4.36876403, 4.58418934, 4.75687172, 4.3689799, 4.16126498,
4.0203982, 3.77148242, 3.38198096, 3.07261764, 2.9014741,
2.5049543, 2.756105, 2.28779058, 2.16986991, 1.8415962,
1.83319008, 2.20898291, 2.00128981, 1.75747025, 1.26676663,
1.40316876, 1.11126484, 1.60376367, 1.22523829, 1.58816681,
1.49705679, 1.80244138, 1.55128293, 1.35339409, 1.50985759,
1.0808451, 1.05892796, 1.43414812, 1.43039101, 1.73631655,
1.43940867, 1.82864425, 1.71088265, 2.12015154, 2.45417128,
2.84777618, 2.7925612, 2.90975121, 3.25920745, 3.13801182,
3.52733677, 3.65468491, 3.69395211, 3.49862035, 3.24786017,
3.64463138, 4.00331929, 3.62509565, 3.78013949, 3.4174012,
3.76312271, 3.62054004, 3.67206716, 3.60596058, 3.38636199,
3.42580676, 3.32921095, 3.02976759, 3.28258676, 3.45760838,
3.24917528, 2.94618304, 2.86980011, 2.63191259, 2.39566759,
2.53159917, 2.96273967, 3.25626185, 2.97425402, 3.16412191,
3.58280763, 3.23257727, 3.62353556, 3.12806399, 2.92532313],
columns=['value'])
# 建立一个长度为 500 个数据点的测试数据, 用于测试数据点多于250个的情况下的评价过程
self.long_data = pd.DataFrame([ 9.879, 9.916, 10.109, 10.214, 10.361, 10.768, 10.594, 10.288,
10.082, 9.994, 10.125, 10.126, 10.384, 10.734, 10.4 , 10.87 ,
11.338, 11.061, 11.415, 11.724, 12.077, 12.196, 12.064, 12.423,
12.19 , 11.729, 11.677, 11.448, 11.485, 10.989, 11.242, 11.239,
11.113, 11.075, 11.471, 11.745, 11.754, 11.782, 12.079, 11.97 ,
12.178, 11.95 , 12.438, 12.612, 12.804, 12.952, 12.612, 12.867,
12.832, 12.832, 13.015, 13.315, 13.249, 12.904, 12.776, 12.64 ,
12.543, 12.287, 12.225, 11.844, 11.985, 11.945, 11.542, 11.871,
12.245, 12.228, 12.362, 11.899, 11.962, 12.374, 12.816, 12.649,
12.252, 12.579, 12.3 , 11.988, 12.177, 12.312, 12.744, 12.599,
12.524, 12.82 , 12.67 , 12.876, 12.986, 13.271, 13.606, 13.82 ,
14.161, 13.833, 13.831, 14.137, 13.705, 13.414, 13.037, 12.759,
12.642, 12.948, 13.297, 13.483, 13.836, 14.179, 13.709, 13.655,
13.198, 13.508, 13.953, 14.387, 14.043, 13.987, 13.561, 13.391,
12.923, 12.555, 12.503, 12.292, 11.877, 12.34 , 12.141, 11.687,
11.992, 12.458, 12.131, 11.75 , 11.739, 11.263, 11.762, 11.976,
11.578, 11.854, 12.136, 12.422, 12.311, 12.56 , 12.879, 12.861,
12.973, 13.235, 13.53 , 13.531, 13.137, 13.166, 13.31 , 13.103,
13.007, 12.643, 12.69 , 12.216, 12.385, 12.046, 12.321, 11.9 ,
11.772, 11.816, 11.871, 11.59 , 11.518, 11.94 , 11.803, 11.924,
12.183, 12.136, 12.361, 12.406, 11.932, 11.684, 11.292, 11.388,
11.874, 12.184, 12.002, 12.16 , 11.741, 11.26 , 11.123, 11.534,
11.777, 11.407, 11.275, 11.679, 11.62 , 11.218, 11.235, 11.352,
11.366, 11.061, 10.661, 10.582, 10.899, 11.352, 11.792, 11.475,
11.263, 11.538, 11.183, 10.936, 11.399, 11.171, 11.214, 10.89 ,
10.728, 11.191, 11.646, 11.62 , 11.195, 11.178, 11.18 , 10.956,
11.205, 10.87 , 11.098, 10.639, 10.487, 10.507, 10.92 , 10.558,
10.119, 9.882, 9.573, 9.515, 9.845, 9.852, 9.495, 9.726,
10.116, 10.452, 10.77 , 11.225, 10.92 , 10.824, 11.096, 11.542,
11.06 , 10.568, 10.585, 10.884, 10.401, 10.068, 9.964, 10.285,
10.239, 10.036, 10.417, 10.132, 9.839, 9.556, 9.084, 9.239,
9.304, 9.067, 8.587, 8.471, 8.007, 8.321, 8.55 , 9.008,
9.138, 9.088, 9.434, 9.156, 9.65 , 9.431, 9.654, 10.079,
10.411, 10.865, 10.51 , 10.205, 10.519, 10.367, 10.855, 10.642,
10.298, 10.622, 10.173, 9.792, 9.995, 9.904, 9.771, 9.597,
9.506, 9.212, 9.688, 10.032, 9.723, 9.839, 9.918, 10.332,
10.236, 9.989, 10.192, 10.685, 10.908, 11.275, 11.72 , 12.158,
12.045, 12.244, 12.333, 12.246, 12.552, 12.958, 13.11 , 13.53 ,
13.123, 13.138, 13.57 , 13.389, 13.511, 13.759, 13.698, 13.744,
13.467, 13.795, 13.665, 13.377, 13.423, 13.772, 13.295, 13.073,
12.718, 12.388, 12.399, 12.185, 11.941, 11.818, 11.465, 11.811,
12.163, 11.86 , 11.935, 11.809, 12.145, 12.624, 12.768, 12.321,
12.277, 11.889, 12.11 , 12.606, 12.943, 12.945, 13.112, 13.199,
13.664, 14.051, 14.189, 14.339, 14.611, 14.656, 15.112, 15.086,
15.263, 15.021, 15.346, 15.572, 15.607, 15.983, 16.151, 16.215,
16.096, 16.089, 16.32 , 16.59 , 16.657, 16.752, 16.583, 16.743,
16.373, 16.662, 16.243, 16.163, 16.491, 16.958, 16.977, 17.225,
17.637, 17.344, 17.684, 17.892, 18.036, 18.182, 17.803, 17.588,
17.101, 17.538, 17.124, 16.787, 17.167, 17.138, 16.955, 17.148,
17.135, 17.635, 17.718, 17.675, 17.622, 17.358, 17.754, 17.729,
17.576, 17.772, 18.239, 18.441, 18.729, 18.319, 18.608, 18.493,
18.069, 18.122, 18.314, 18.423, 18.709, 18.548, 18.384, 18.391,
17.988, 17.986, 17.653, 17.249, 17.298, 17.06 , 17.36 , 17.108,
17.348, 17.596, 17.46 , 17.635, 17.275, 17.291, 16.933, 17.337,
17.231, 17.146, 17.148, 16.751, 16.891, 17.038, 16.735, 16.64 ,
16.231, 15.957, 15.977, 16.077, 16.054, 15.797, 15.67 , 15.911,
16.077, 16.17 , 15.722, 15.258, 14.877, 15.138, 15. , 14.811,
14.698, 14.407, 14.583, 14.704, 15.153, 15.436, 15.634, 15.453,
15.877, 15.696, 15.563, 15.927, 16.255, 16.696, 16.266, 16.698,
16.365, 16.493, 16.973, 16.71 , 16.327, 16.605, 16.486, 16.846,
16.935, 17.21 , 17.389, 17.546, 17.773, 17.641, 17.485, 17.794,
17.354, 16.904, 16.675, 16.43 , 16.898, 16.819, 16.921, 17.201,
17.617, 17.368, 17.864, 17.484],
columns=['value'])
self.long_bench = pd.DataFrame([ 9.7 , 10.179, 10.321, 9.855, 9.936, 10.096, 10.331, 10.662,
10.59 , 11.031, 11.154, 10.945, 10.625, 10.233, 10.284, 10.252,
10.221, 10.352, 10.444, 10.773, 10.904, 11.104, 10.797, 10.55 ,
10.943, 11.352, 11.641, 11.983, 11.696, 12.138, 12.365, 12.379,
11.969, 12.454, 12.947, 13.119, 13.013, 12.763, 12.632, 13.034,
12.681, 12.561, 12.938, 12.867, 13.202, 13.132, 13.539, 13.91 ,
13.456, 13.692, 13.771, 13.904, 14.069, 13.728, 13.97 , 14.228,
13.84 , 14.041, 13.963, 13.689, 13.543, 13.858, 14.118, 13.987,
13.611, 14.028, 14.229, 14.41 , 14.74 , 15.03 , 14.915, 15.207,
15.354, 15.665, 15.877, 15.682, 15.625, 15.175, 15.105, 14.893,
14.86 , 15.097, 15.178, 15.293, 15.238, 15. , 15.283, 14.994,
14.907, 14.664, 14.888, 15.297, 15.313, 15.368, 14.956, 14.802,
14.506, 14.257, 14.619, 15.019, 15.049, 14.625, 14.894, 14.978,
15.434, 15.578, 16.038, 16.107, 16.277, 16.365, 16.204, 16.465,
16.401, 16.895, 17.057, 16.621, 16.225, 16.075, 15.863, 16.292,
16.551, 16.724, 16.817, 16.81 , 17.192, 16.86 , 16.745, 16.707,
16.552, 16.133, 16.301, 16.08 , 15.81 , 15.75 , 15.909, 16.127,
16.457, 16.204, 16.329, 16.748, 16.624, 17.011, 16.548, 16.831,
16.653, 16.791, 16.57 , 16.778, 16.928, 16.932, 17.22 , 16.876,
17.301, 17.422, 17.689, 17.316, 17.547, 17.534, 17.409, 17.669,
17.416, 17.859, 17.477, 17.307, 17.245, 17.352, 17.851, 17.412,
17.144, 17.138, 17.085, 16.926, 16.674, 16.854, 17.064, 16.95 ,
16.609, 16.957, 16.498, 16.552, 16.175, 15.858, 15.697, 15.781,
15.583, 15.36 , 15.558, 16.046, 15.968, 15.905, 16.358, 16.783,
17.048, 16.762, 17.224, 17.363, 17.246, 16.79 , 16.608, 16.423,
15.991, 15.527, 15.147, 14.759, 14.792, 15.206, 15.148, 15.046,
15.429, 14.999, 15.407, 15.124, 14.72 , 14.713, 15.022, 15.092,
14.982, 15.001, 14.734, 14.713, 14.841, 14.562, 15.005, 15.483,
15.472, 15.277, 15.503, 15.116, 15.12 , 15.442, 15.476, 15.789,
15.36 , 15.764, 16.218, 16.493, 16.642, 17.088, 16.816, 16.645,
16.336, 16.511, 16.2 , 15.994, 15.86 , 15.929, 16.316, 16.416,
16.746, 17.173, 17.531, 17.627, 17.407, 17.49 , 17.768, 17.509,
17.795, 18.147, 18.63 , 18.945, 19.021, 19.518, 19.6 , 19.744,
19.63 , 19.32 , 18.933, 19.297, 19.598, 19.446, 19.236, 19.198,
19.144, 19.159, 19.065, 19.032, 18.586, 18.272, 18.119, 18.3 ,
17.894, 17.744, 17.5 , 17.083, 17.092, 16.864, 16.453, 16.31 ,
16.681, 16.342, 16.447, 16.715, 17.068, 17.067, 16.822, 16.673,
16.675, 16.592, 16.686, 16.397, 15.902, 15.597, 15.357, 15.162,
15.348, 15.603, 15.283, 15.257, 15.082, 14.621, 14.366, 14.039,
13.957, 14.141, 13.854, 14.243, 14.414, 14.033, 13.93 , 14.104,
14.461, 14.249, 14.053, 14.165, 14.035, 14.408, 14.501, 14.019,
14.265, 14.67 , 14.797, 14.42 , 14.681, 15.16 , 14.715, 14.292,
14.411, 14.656, 15.094, 15.366, 15.055, 15.198, 14.762, 14.294,
13.854, 13.811, 13.549, 13.927, 13.897, 13.421, 13.037, 13.32 ,
13.721, 13.511, 13.999, 13.529, 13.418, 13.881, 14.326, 14.362,
13.987, 14.015, 13.599, 13.343, 13.307, 13.689, 13.851, 13.404,
13.577, 13.395, 13.619, 13.195, 12.904, 12.553, 12.294, 12.649,
12.425, 11.967, 12.062, 11.71 , 11.645, 12.058, 12.136, 11.749,
11.953, 12.401, 12.044, 11.901, 11.631, 11.396, 11.036, 11.244,
10.864, 11.207, 11.135, 11.39 , 11.723, 12.084, 11.8 , 11.471,
11.33 , 11.504, 11.295, 11.3 , 10.901, 10.494, 10.825, 11.054,
10.866, 10.713, 10.875, 10.846, 10.947, 11.422, 11.158, 10.94 ,
10.521, 10.36 , 10.411, 10.792, 10.472, 10.305, 10.525, 10.853,
10.556, 10.72 , 10.54 , 10.583, 10.299, 10.061, 10.004, 9.903,
9.796, 9.472, 9.246, 9.54 , 9.456, 9.177, 9.484, 9.557,
9.493, 9.968, 9.536, 9.39 , 8.922, 8.423, 8.518, 8.686,
8.771, 9.098, 9.281, 8.858, 9.027, 8.553, 8.784, 8.996,
9.379, 9.846, 9.855, 9.502, 9.608, 9.761, 9.409, 9.4 ,
9.332, 9.34 , 9.284, 8.844, 8.722, 8.376, 8.775, 8.293,
8.144, 8.63 , 8.831, 8.957, 9.18 , 9.601, 9.695, 10.018,
9.841, 9.743, 9.292, 8.85 , 9.316, 9.288, 9.519, 9.738,
9.289, 9.785, 9.804, 10.06 , 10.188, 10.095, 9.739, 9.881,
9.7 , 9.991, 10.391, 10.002],
columns=['value'])
def test_performance_stats(self):
"""test the function performance_statistics()
"""
pass
def test_fv(self):
print(f'test with test data and empty DataFrame')
self.assertAlmostEqual(eval_fv(self.test_data1), 6.39245474)
self.assertAlmostEqual(eval_fv(self.test_data2), 10.05126375)
self.assertAlmostEqual(eval_fv(self.test_data3), 6.95068113)
self.assertAlmostEqual(eval_fv(self.test_data4), 8.86508591)
self.assertAlmostEqual(eval_fv(self.test_data5), 4.58627048)
self.assertAlmostEqual(eval_fv(self.test_data6), 4.10346795)
self.assertAlmostEqual(eval_fv(self.test_data7), 2.92532313)
self.assertAlmostEqual(eval_fv(pd.DataFrame()), -np.inf)
print(f'Error testing')
self.assertRaises(AssertionError, eval_fv, 15)
self.assertRaises(KeyError,
eval_fv,
pd.DataFrame([1, 2, 3], columns=['non_value']))
def test_max_drawdown(self):
print(f'test with test data and empty DataFrame')
self.assertAlmostEqual(eval_max_drawdown(self.test_data1)[0], 0.264274308)
self.assertEqual(eval_max_drawdown(self.test_data1)[1], 53)
self.assertEqual(eval_max_drawdown(self.test_data1)[2], 86)
self.assertTrue(np.isnan(eval_max_drawdown(self.test_data1)[3]))
self.assertAlmostEqual(eval_max_drawdown(self.test_data2)[0], 0.334690849)
self.assertEqual(eval_max_drawdown(self.test_data2)[1], 0)
self.assertEqual(eval_max_drawdown(self.test_data2)[2], 10)
self.assertEqual(eval_max_drawdown(self.test_data2)[3], 19)
self.assertAlmostEqual(eval_max_drawdown(self.test_data3)[0], 0.244452899)
self.assertEqual(eval_max_drawdown(self.test_data3)[1], 90)
self.assertEqual(eval_max_drawdown(self.test_data3)[2], 99)
self.assertTrue(np.isnan(eval_max_drawdown(self.test_data3)[3]))
self.assertAlmostEqual(eval_max_drawdown(self.test_data4)[0], 0.201849684)
self.assertEqual(eval_max_drawdown(self.test_data4)[1], 14)
self.assertEqual(eval_max_drawdown(self.test_data4)[2], 50)
self.assertEqual(eval_max_drawdown(self.test_data4)[3], 54)
self.assertAlmostEqual(eval_max_drawdown(self.test_data5)[0], 0.534206456)
self.assertEqual(eval_max_drawdown(self.test_data5)[1], 21)
self.assertEqual(eval_max_drawdown(self.test_data5)[2], 60)
self.assertTrue(np.isnan(eval_max_drawdown(self.test_data5)[3]))
self.assertAlmostEqual(eval_max_drawdown(self.test_data6)[0], 0.670062689)
self.assertEqual(eval_max_drawdown(self.test_data6)[1], 0)
self.assertEqual(eval_max_drawdown(self.test_data6)[2], 70)
self.assertTrue(np.isnan(eval_max_drawdown(self.test_data6)[3]))
self.assertAlmostEqual(eval_max_drawdown(self.test_data7)[0], 0.783577449)
self.assertEqual(eval_max_drawdown(self.test_data7)[1], 17)
self.assertEqual(eval_max_drawdown(self.test_data7)[2], 51)
self.assertTrue(np.isnan(eval_max_drawdown(self.test_data7)[3]))
self.assertEqual(eval_max_drawdown(pd.DataFrame()), -np.inf)
print(f'Error testing')
self.assertRaises(AssertionError, eval_fv, 15)
self.assertRaises(KeyError,
eval_fv,
pd.DataFrame([1, 2, 3], columns=['non_value']))
# test max drawdown == 0:
# TODO: investigate: how does divide by zero change?
self.assertAlmostEqual(eval_max_drawdown(self.test_data4 - 5)[0], 1.0770474121951792)
self.assertEqual(eval_max_drawdown(self.test_data4 - 5)[1], 14)
self.assertEqual(eval_max_drawdown(self.test_data4 - 5)[2], 50)
def test_info_ratio(self):
reference = self.test_data1
self.assertAlmostEqual(eval_info_ratio(self.test_data2, reference, 'value'), 0.075553316)
self.assertAlmostEqual(eval_info_ratio(self.test_data3, reference, 'value'), 0.018949457)
self.assertAlmostEqual(eval_info_ratio(self.test_data4, reference, 'value'), 0.056328143)
self.assertAlmostEqual(eval_info_ratio(self.test_data5, reference, 'value'), -0.004270068)
self.assertAlmostEqual(eval_info_ratio(self.test_data6, reference, 'value'), 0.009198027)
self.assertAlmostEqual(eval_info_ratio(self.test_data7, reference, 'value'), -0.000890283)
def test_volatility(self):
self.assertAlmostEqual(eval_volatility(self.test_data1), 0.748646166)
self.assertAlmostEqual(eval_volatility(self.test_data2), 0.75527442)
self.assertAlmostEqual(eval_volatility(self.test_data3), 0.654188853)
self.assertAlmostEqual(eval_volatility(self.test_data4), 0.688375814)
self.assertAlmostEqual(eval_volatility(self.test_data5), 1.089989522)
self.assertAlmostEqual(eval_volatility(self.test_data6), 1.775419308)
self.assertAlmostEqual(eval_volatility(self.test_data7), 1.962758406)
self.assertAlmostEqual(eval_volatility(self.test_data1, logarithm=False), 0.750993311)
self.assertAlmostEqual(eval_volatility(self.test_data2, logarithm=False), 0.75571473)
self.assertAlmostEqual(eval_volatility(self.test_data3, logarithm=False), 0.655331424)
self.assertAlmostEqual(eval_volatility(self.test_data4, logarithm=False), 0.692683021)
self.assertAlmostEqual(eval_volatility(self.test_data5, logarithm=False), 1.09602969)
self.assertAlmostEqual(eval_volatility(self.test_data6, logarithm=False), 1.774789504)
self.assertAlmostEqual(eval_volatility(self.test_data7, logarithm=False), 2.003329156)
self.assertEqual(eval_volatility(pd.DataFrame()), -np.inf)
self.assertRaises(AssertionError, eval_volatility, [1, 2, 3])
# 测试长数据的Volatility计算
expected_volatility = np.array([ np.nan, np.nan, np.nan, np.nan, np.nan,
np.nan, np.nan, np.nan, np.nan, np.nan,
np.nan, np.nan, np.nan, np.nan, np.nan,
np.nan, np.nan, np.nan, np.nan, np.nan,
np.nan, np.nan, np.nan, np.nan, np.nan,
np.nan, np.nan, np.nan, np.nan, np.nan,
np.nan, np.nan, np.nan, np.nan, np.nan,
np.nan, np.nan, np.nan, np.nan, np.nan,
np.nan, np.nan, np.nan, np.nan, np.nan,
np.nan, np.nan, np.nan, np.nan, np.nan,
np.nan, np.nan, np.nan, np.nan, np.nan,
np.nan, np.nan, np.nan, np.nan, np.nan,
np.nan, np.nan, np.nan, np.nan, np.nan,
np.nan, np.nan, np.nan, np.nan, np.nan,
np.nan, np.nan, np.nan, np.nan, np.nan,
np.nan, np.nan, np.nan, np.nan, np.nan,
np.nan, np.nan, np.nan, np.nan, np.nan,
np.nan, np.nan, np.nan, np.nan, np.nan,
np.nan, np.nan, np.nan, np.nan, np.nan,
np.nan, np.nan, np.nan, np.nan, np.nan,
np.nan, np.nan, np.nan, np.nan, np.nan,
np.nan, np.nan, np.nan, np.nan, np.nan,
np.nan, np.nan, np.nan, np.nan, np.nan,
np.nan, np.nan, np.nan, np.nan, np.nan,
np.nan, np.nan, np.nan, np.nan, np.nan,
np.nan, np.nan, np.nan, np.nan, np.nan,
np.nan, np.nan, np.nan, np.nan, np.nan,
np.nan, np.nan, np.nan, np.nan, np.nan,
np.nan, np.nan, np.nan, np.nan, np.nan,
np.nan, np.nan, np.nan, np.nan, np.nan,
np.nan, np.nan, np.nan, np.nan, np.nan,
np.nan, np.nan, np.nan, np.nan, np.nan,
np.nan, np.nan, np.nan, np.nan, np.nan,
np.nan, np.nan, np.nan, np.nan, np.nan,
np.nan, np.nan, np.nan, np.nan, np.nan,
np.nan, np.nan, np.nan, np.nan, np.nan,
np.nan, np.nan, np.nan, np.nan, np.nan,
np.nan, np.nan, np.nan, np.nan, np.nan,
np.nan, np.nan, np.nan, np.nan, np.nan,
np.nan, np.nan, np.nan, np.nan, np.nan,
np.nan, np.nan, np.nan, np.nan, np.nan,
np.nan, np.nan, np.nan, np.nan, np.nan,
np.nan, np.nan, np.nan, np.nan, np.nan,
np.nan, np.nan, np.nan, np.nan, np.nan,
np.nan, np.nan, np.nan, np.nan, np.nan,
np.nan, np.nan, np.nan, np.nan, np.nan,
np.nan, np.nan, np.nan, np.nan, np.nan,
np.nan, np.nan, np.nan, np.nan, np.nan,
np.nan, np.nan, np.nan, np.nan, np.nan,
np.nan, np.nan, np.nan, np.nan, np.nan,
0.39955371, 0.39974258, 0.40309866, 0.40486593, 0.4055514 ,
0.40710639, 0.40708157, 0.40609006, 0.4073625 , 0.40835305,
0.41155304, 0.41218193, 0.41207489, 0.41300276, 0.41308415,
0.41292392, 0.41207645, 0.41238397, 0.41229291, 0.41164056,
0.41316317, 0.41348842, 0.41462249, 0.41474574, 0.41652625,
0.41649176, 0.41701556, 0.4166593 , 0.41684221, 0.41491689,
0.41435209, 0.41549087, 0.41849338, 0.41998049, 0.41959106,
0.41907311, 0.41916103, 0.42120773, 0.42052391, 0.42111225,
0.42124589, 0.42356445, 0.42214672, 0.42324022, 0.42476639,
0.42621689, 0.42549439, 0.42533678, 0.42539414, 0.42545038,
0.42593637, 0.42652095, 0.42665489, 0.42699563, 0.42798159,
0.42784512, 0.42898006, 0.42868781, 0.42874188, 0.42789631,
0.4277768 , 0.42776827, 0.42685216, 0.42660989, 0.42563155,
0.42618281, 0.42606281, 0.42505222, 0.42653242, 0.42555378,
0.42500842, 0.42561939, 0.42442059, 0.42395414, 0.42384356,
0.42319135, 0.42397497, 0.42488579, 0.42449729, 0.42508766,
0.42509878, 0.42456616, 0.42535577, 0.42681884, 0.42688552,
0.42779918, 0.42706058, 0.42792887, 0.42762114, 0.42894045,
0.42977398, 0.42919859, 0.42829041, 0.42780946, 0.42825318,
0.42858952, 0.42858315, 0.42805601, 0.42764751, 0.42744107,
0.42775518, 0.42707283, 0.4258592 , 0.42615335, 0.42526286,
0.4248906 , 0.42368986, 0.4232565 , 0.42265079, 0.42263954,
0.42153046, 0.42132051, 0.41995353, 0.41916605, 0.41914271,
0.41876945, 0.41740175, 0.41583884, 0.41614026, 0.41457908,
0.41472411, 0.41310876, 0.41261041, 0.41212369, 0.41211677,
0.4100645 , 0.40852504, 0.40860297, 0.40745338, 0.40698661,
0.40644546, 0.40591375, 0.40640744, 0.40620663, 0.40656649,
0.40727154, 0.40797605, 0.40807137, 0.40808913, 0.40809676,
0.40711767, 0.40724628, 0.40713077, 0.40772698, 0.40765157,
0.40658297, 0.4065991 , 0.405011 , 0.40537645, 0.40432626,
0.40390177, 0.40237701, 0.40291623, 0.40301797, 0.40324145,
0.40312864, 0.40328316, 0.40190955, 0.40246506, 0.40237663,
0.40198407, 0.401969 , 0.40185623, 0.40198313, 0.40005643,
0.39940743, 0.39850438, 0.39845398, 0.39695093, 0.39697295,
0.39663201, 0.39675444, 0.39538699, 0.39331959, 0.39326074,
0.39193287, 0.39157266, 0.39021327, 0.39062591, 0.38917591,
0.38976991, 0.38864187, 0.38872158, 0.38868096, 0.38868377,
0.38842057, 0.38654784, 0.38649517, 0.38600464, 0.38408115,
0.38323049, 0.38260215, 0.38207663, 0.38142669, 0.38003262,
0.37969367, 0.37768092, 0.37732108, 0.37741991, 0.37617779,
0.37698504, 0.37606784, 0.37499276, 0.37533731, 0.37350437,
0.37375172, 0.37385382, 0.37384003, 0.37338938, 0.37212288,
0.37273075, 0.370559 , 0.37038506, 0.37062153, 0.36964661,
0.36818564, 0.3656634 , 0.36539259, 0.36428672, 0.36502487,
0.3647148 , 0.36551435, 0.36409919, 0.36348181, 0.36254383,
0.36166601, 0.36142665, 0.35954942, 0.35846915, 0.35886759,
0.35813867, 0.35642888, 0.35375231, 0.35061783, 0.35078463,
0.34995508, 0.34688918, 0.34548257, 0.34633158, 0.34622833,
0.34652111, 0.34622774, 0.34540951, 0.34418809, 0.34276593,
0.34160916, 0.33811193, 0.33822709, 0.3391685 , 0.33883381])
test_volatility = eval_volatility(self.long_data)
test_volatility_roll = self.long_data['volatility'].values
self.assertAlmostEqual(test_volatility, np.nanmean(expected_volatility))
self.assertTrue(np.allclose(expected_volatility, test_volatility_roll, equal_nan=True))
def test_sharp(self):
self.assertAlmostEqual(eval_sharp(self.test_data1, 5, 0), 0.06135557)
self.assertAlmostEqual(eval_sharp(self.test_data2, 5, 0), 0.167858667)
self.assertAlmostEqual(eval_sharp(self.test_data3, 5, 0), 0.09950547)
self.assertAlmostEqual(eval_sharp(self.test_data4, 5, 0), 0.154928241)
self.assertAlmostEqual(eval_sharp(self.test_data5, 5, 0.002), 0.007868673)
self.assertAlmostEqual(eval_sharp(self.test_data6, 5, 0.002), 0.018306537)
self.assertAlmostEqual(eval_sharp(self.test_data7, 5, 0.002), 0.006259971)
# 测试长数据的sharp率计算
expected_sharp = np.array([ np.nan, np.nan, np.nan, np.nan, np.nan,
np.nan, np.nan, np.nan, np.nan, np.nan,
np.nan, np.nan, np.nan, np.nan, np.nan,
np.nan, np.nan, np.nan, np.nan, np.nan,
np.nan, np.nan, np.nan, np.nan, np.nan,
np.nan, np.nan, np.nan, np.nan, np.nan,
np.nan, np.nan, np.nan, np.nan, np.nan,
np.nan, np.nan, np.nan, np.nan, np.nan,
np.nan, np.nan, np.nan, np.nan, np.nan,
np.nan, np.nan, np.nan, np.nan, np.nan,
np.nan, np.nan, np.nan, np.nan, np.nan,
np.nan, np.nan, np.nan, np.nan, np.nan,
np.nan, np.nan, np.nan, np.nan, np.nan,
np.nan, np.nan, np.nan, np.nan, np.nan,
np.nan, np.nan, np.nan, np.nan, np.nan,
np.nan, np.nan, np.nan, np.nan, np.nan,
np.nan, np.nan, np.nan, np.nan, np.nan,
np.nan, np.nan, np.nan, np.nan, np.nan,
np.nan, np.nan, np.nan, np.nan, np.nan,
np.nan, np.nan, np.nan, np.nan, np.nan,
np.nan, np.nan, np.nan, np.nan, np.nan,
np.nan, np.nan, np.nan, np.nan, np.nan,
np.nan, np.nan, np.nan, np.nan, np.nan,
np.nan, np.nan, np.nan, np.nan, np.nan,
np.nan, np.nan, np.nan, np.nan, np.nan,
np.nan, np.nan, np.nan, np.nan, np.nan,
np.nan, np.nan, np.nan, np.nan, np.nan,
np.nan, np.nan, np.nan, np.nan, np.nan,
np.nan, np.nan, np.nan, np.nan, np.nan,
np.nan, np.nan, np.nan, np.nan, np.nan,
np.nan, np.nan, np.nan, np.nan, np.nan,
np.nan, np.nan, np.nan, np.nan, np.nan,
np.nan, np.nan, np.nan, np.nan, np.nan,
np.nan, np.nan, np.nan, np.nan, np.nan,
np.nan, np.nan, np.nan, np.nan, np.nan,
np.nan, np.nan, np.nan, np.nan, np.nan,
np.nan, np.nan, np.nan, np.nan, np.nan,
np.nan, np.nan, np.nan, np.nan, np.nan,
np.nan, np.nan, np.nan, np.nan, np.nan,
np.nan, np.nan, np.nan, np.nan, np.nan,
np.nan, np.nan, np.nan, np.nan, np.nan,
np.nan, np.nan, np.nan, np.nan, np.nan,
np.nan, np.nan, np.nan, np.nan, np.nan,
np.nan, np.nan, np.nan, np.nan, np.nan,
np.nan, np.nan, np.nan, np.nan, np.nan,
np.nan, np.nan, np.nan, np.nan, np.nan,
np.nan, np.nan, np.nan, np.nan, np.nan,
np.nan, np.nan, np.nan, np.nan, np.nan,
np.nan, np.nan, np.nan, np.nan, np.nan,
np.nan, np.nan, np.nan, np.nan, np.nan,
-0.02346815, -0.02618783, -0.03763912, -0.03296276, -0.03085698,
-0.02851101, -0.02375842, -0.02016746, -0.01107885, -0.01426613,
-0.00787204, -0.01135784, -0.01164232, -0.01003481, -0.00022512,
-0.00046792, -0.01209378, -0.01278892, -0.01298135, -0.01938214,
-0.01671044, -0.02120509, -0.0244281 , -0.02416067, -0.02763238,
-0.027579 , -0.02372774, -0.02215294, -0.02467094, -0.02091266,
-0.02590194, -0.03049876, -0.02077131, -0.01483653, -0.02488144,
-0.02671638, -0.02561547, -0.01957986, -0.02479803, -0.02703162,
-0.02658087, -0.01641755, -0.01946472, -0.01647757, -0.01280889,
-0.00893643, -0.00643275, -0.00698457, -0.00549962, -0.00654677,
-0.00494757, -0.0035633 , -0.00109037, 0.00750654, 0.00451208,
0.00625502, 0.01221367, 0.01326454, 0.01535037, 0.02269538,
0.02028715, 0.02127712, 0.02333264, 0.02273159, 0.01670643,
0.01376513, 0.01265342, 0.02211647, 0.01612449, 0.00856706,
-0.00077147, -0.00268848, 0.00210993, -0.00443934, -0.00411912,
-0.0018756 , -0.00867461, -0.00581601, -0.00660835, -0.00861137,
-0.00678614, -0.01188408, -0.00589617, -0.00244323, -0.00201891,
-0.01042846, -0.01471016, -0.02167034, -0.02258554, -0.01306809,
-0.00909086, -0.01233746, -0.00595166, -0.00184208, 0.00750497,
0.01481886, 0.01761972, 0.01562886, 0.01446414, 0.01285826,
0.01357719, 0.00967613, 0.01636272, 0.01458437, 0.02280183,
0.02151903, 0.01700276, 0.01597368, 0.02114336, 0.02233297,
0.02585631, 0.02768459, 0.03519235, 0.04204535, 0.04328161,
0.04672855, 0.05046191, 0.04619848, 0.04525853, 0.05381529,
0.04598861, 0.03947394, 0.04665006, 0.05586077, 0.05617728,
0.06495018, 0.06205172, 0.05665466, 0.06500615, 0.0632062 ,
0.06084328, 0.05851466, 0.05659229, 0.05159347, 0.0432977 ,
0.0474047 , 0.04231723, 0.03613176, 0.03618391, 0.03591012,
0.03885674, 0.0402686 , 0.03846423, 0.04534014, 0.04721458,
0.05130912, 0.05026281, 0.05394312, 0.05529349, 0.05949243,
0.05463304, 0.06195165, 0.06767606, 0.06880985, 0.07048996,
0.07078815, 0.07420767, 0.06773439, 0.0658441 , 0.06470875,
0.06302349, 0.06456876, 0.06411282, 0.06216669, 0.067094 ,
0.07055075, 0.07254976, 0.07119253, 0.06173308, 0.05393352,
0.05681246, 0.05250643, 0.06099845, 0.0655544 , 0.06977334,
0.06636514, 0.06177949, 0.06869908, 0.06719767, 0.06178738,
0.05915714, 0.06882277, 0.06756821, 0.06507994, 0.06489791,
0.06553941, 0.073123 , 0.07576757, 0.06805446, 0.06063571,
0.05033801, 0.05206971, 0.05540306, 0.05249118, 0.05755587,
0.0586174 , 0.05051288, 0.0564852 , 0.05757284, 0.06358355,
0.06130082, 0.04925482, 0.03834472, 0.04163981, 0.04648316,
0.04457858, 0.04324626, 0.04328791, 0.04156207, 0.04818652,
0.04972634, 0.06024123, 0.06489556, 0.06255485, 0.06069815,
0.06466389, 0.07081163, 0.07895358, 0.0881782 , 0.09374151,
0.08336506, 0.08764795, 0.09080174, 0.08808926, 0.08641158,
0.07811943, 0.06885318, 0.06479503, 0.06851185, 0.07382819,
0.07047903, 0.06658251, 0.07638379, 0.08667974, 0.08867918,
0.08245323, 0.08961866, 0.09905298, 0.0961908 , 0.08562706,
0.0839014 , 0.0849072 , 0.08338395, 0.08783487, 0.09463609,
0.10332336, 0.11806497, 0.11220297, 0.11589097, 0.11678405])
test_sharp = eval_sharp(self.long_data, 5, 0.00035)
self.assertAlmostEqual(np.nanmean(expected_sharp), test_sharp)
self.assertTrue(np.allclose(self.long_data['sharp'].values, expected_sharp, equal_nan=True))
def test_beta(self):
reference = self.test_data1
self.assertAlmostEqual(eval_beta(self.test_data2, reference, 'value'), -0.017148939)
self.assertAlmostEqual(eval_beta(self.test_data3, reference, 'value'), -0.042204233)
self.assertAlmostEqual(eval_beta(self.test_data4, reference, 'value'), -0.15652986)
self.assertAlmostEqual(eval_beta(self.test_data5, reference, 'value'), -0.049195532)
self.assertAlmostEqual(eval_beta(self.test_data6, reference, 'value'), -0.026995082)
self.assertAlmostEqual(eval_beta(self.test_data7, reference, 'value'), -0.01147809)
self.assertRaises(TypeError, eval_beta, [1, 2, 3], reference, 'value')
self.assertRaises(TypeError, eval_beta, self.test_data3, [1, 2, 3], 'value')
self.assertRaises(KeyError, eval_beta, self.test_data3, reference, 'not_found_value')
# 测试长数据的beta计算
expected_beta = np.array([ np.nan, np.nan, np.nan, np.nan, np.nan,
np.nan, np.nan, np.nan, np.nan, np.nan,
np.nan, np.nan, np.nan, np.nan, np.nan,
np.nan, np.nan, np.nan, np.nan, np.nan,
np.nan, np.nan, np.nan, np.nan, np.nan,
np.nan, np.nan, np.nan, np.nan, np.nan,
np.nan, np.nan, np.nan, np.nan, np.nan,
np.nan, np.nan, np.nan, np.nan, np.nan,
np.nan, np.nan, np.nan, np.nan, np.nan,
np.nan, np.nan, np.nan, np.nan, np.nan,
np.nan, np.nan, np.nan, np.nan, np.nan,
np.nan, np.nan, np.nan, np.nan, np.nan,
np.nan, np.nan, np.nan, np.nan, np.nan,
np.nan, np.nan, np.nan, np.nan, np.nan,
np.nan, np.nan, np.nan, np.nan, np.nan,
np.nan, np.nan, np.nan, np.nan, np.nan,
np.nan, np.nan, np.nan, np.nan, np.nan,
np.nan, np.nan, np.nan, np.nan, np.nan,
np.nan, np.nan, np.nan, np.nan, np.nan,
np.nan, np.nan, np.nan, np.nan, np.nan,
np.nan, np.nan, np.nan, np.nan, np.nan,
np.nan, np.nan, np.nan, np.nan, np.nan,
np.nan, np.nan, np.nan, np.nan, np.nan,
np.nan, np.nan, np.nan, np.nan, np.nan,
np.nan, np.nan, np.nan, np.nan, np.nan,
np.nan, np.nan, np.nan, np.nan, np.nan,
np.nan, np.nan, np.nan, np.nan, np.nan,
np.nan, np.nan, np.nan, np.nan, np.nan,
np.nan, np.nan, np.nan, np.nan, np.nan,
np.nan, np.nan, np.nan, np.nan, np.nan,
np.nan, np.nan, np.nan, np.nan, np.nan,
np.nan, np.nan, np.nan, np.nan, np.nan,
np.nan, np.nan, np.nan, np.nan, np.nan,
np.nan, np.nan, np.nan, np.nan, np.nan,
np.nan, np.nan, np.nan, np.nan, np.nan,
np.nan, np.nan, np.nan, np.nan, np.nan,
np.nan, np.nan, np.nan, np.nan, np.nan,
np.nan, np.nan, np.nan, np.nan, np.nan,
np.nan, np.nan, np.nan, np.nan, np.nan,
np.nan, np.nan, np.nan, np.nan, np.nan,
np.nan, np.nan, np.nan, np.nan, np.nan,
np.nan, np.nan, np.nan, np.nan, np.nan,
np.nan, np.nan, np.nan, np.nan, np.nan,
np.nan, np.nan, np.nan, np.nan, np.nan,
np.nan, np.nan, np.nan, np.nan, np.nan,
np.nan, np.nan, np.nan, np.nan, np.nan,
np.nan, np.nan, np.nan, np.nan, np.nan,
np.nan, np.nan, np.nan, np.nan, np.nan,
np.nan, np.nan, np.nan, np.nan, np.nan,
np.nan, np.nan, np.nan, np.nan, np.nan,
-0.04988841, -0.05127618, -0.04692104, -0.04272652, -0.04080598,
-0.0493347 , -0.0460858 , -0.0416761 , -0.03691527, -0.03724924,
-0.03678865, -0.03987324, -0.03488321, -0.02567672, -0.02690303,
-0.03010128, -0.02437967, -0.02571932, -0.02455681, -0.02839811,
-0.03358653, -0.03396697, -0.03466321, -0.03050966, -0.0247583 ,
-0.01629325, -0.01880895, -0.01480403, -0.01348783, -0.00544294,
-0.00648176, -0.00467036, -0.01135331, -0.0156841 , -0.02340763,
-0.02615705, -0.02730771, -0.02906174, -0.02860664, -0.02412914,
-0.02066416, -0.01744816, -0.02185133, -0.02145285, -0.02681765,
-0.02827694, -0.02394581, -0.02744096, -0.02778825, -0.02703065,
-0.03160023, -0.03615371, -0.03681072, -0.04265126, -0.04344738,
-0.04232421, -0.04705272, -0.04533344, -0.04605934, -0.05272737,
-0.05156463, -0.05134196, -0.04730733, -0.04425352, -0.03869831,
-0.04159571, -0.04223998, -0.04346747, -0.04229844, -0.04740093,
-0.04992507, -0.04621232, -0.04477644, -0.0486915 , -0.04598224,
-0.04943463, -0.05006391, -0.05362256, -0.04994067, -0.05464769,
-0.05443275, -0.05513493, -0.05173594, -0.04500994, -0.04662891,
-0.03903505, -0.0419592 , -0.04307773, -0.03925718, -0.03711574,
-0.03992631, -0.0433058 , -0.04533641, -0.0461183 , -0.05600344,
-0.05758377, -0.05959874, -0.05605942, -0.06002859, -0.06253002,
-0.06747014, -0.06427915, -0.05931947, -0.05769974, -0.04791515,
-0.05175088, -0.05748039, -0.05385232, -0.05072975, -0.05052637,
-0.05125567, -0.05005785, -0.05325104, -0.04977727, -0.04947867,
-0.05148544, -0.05739156, -0.05742069, -0.06047279, -0.0558414 ,
-0.06086126, -0.06265151, -0.06411129, -0.06828052, -0.06781762,
-0.07083409, -0.07211207, -0.06799162, -0.06913295, -0.06775162,
-0.0696265 , -0.06678248, -0.06867502, -0.06581961, -0.07055823,
-0.06448184, -0.06097973, -0.05795587, -0.0618383 , -0.06130145,
-0.06050652, -0.05936661, -0.05749424, -0.0499 , -0.05050495,
-0.04962687, -0.05033439, -0.05070116, -0.05422009, -0.05369759,
-0.05548943, -0.05907353, -0.05933035, -0.05927918, -0.06227663,
-0.06011455, -0.05650432, -0.05828134, -0.05620949, -0.05715323,
-0.05482478, -0.05387113, -0.05095559, -0.05377999, -0.05334267,
-0.05220438, -0.04001521, -0.03892434, -0.03660782, -0.04282708,
-0.04324623, -0.04127048, -0.04227559, -0.04275226, -0.04347049,
-0.04125853, -0.03806295, -0.0330632 , -0.03155531, -0.03277152,
-0.03304518, -0.03878731, -0.03830672, -0.03727434, -0.0370571 ,
-0.04509224, -0.04207632, -0.04116198, -0.04545179, -0.04584584,
-0.05287341, -0.05417433, -0.05175836, -0.05005509, -0.04268674,
-0.03442321, -0.03457309, -0.03613426, -0.03524391, -0.03629479,
-0.04361312, -0.02626705, -0.02406115, -0.03046384, -0.03181044,
-0.03375164, -0.03661673, -0.04520779, -0.04926951, -0.05726738,
-0.0584486 , -0.06220608, -0.06800563, -0.06797431, -0.07562211,
-0.07481996, -0.07731229, -0.08413381, -0.09031826, -0.09691925,
-0.11018071, -0.11952675, -0.10826026, -0.11173895, -0.10756359,
-0.10775916, -0.11664559, -0.10505051, -0.10606547, -0.09855355,
-0.10004159, -0.10857084, -0.12209301, -0.11605758, -0.11105113,
-0.1155195 , -0.11569505, -0.10513348, -0.09611072, -0.10719791,
-0.10843965, -0.11025856, -0.10247839, -0.10554044, -0.10927647,
-0.10645088, -0.09982498, -0.10542734, -0.09631372, -0.08229695])
test_beta_mean = eval_beta(self.long_data, self.long_bench, 'value')
test_beta_roll = self.long_data['beta'].values
self.assertAlmostEqual(test_beta_mean, np.nanmean(expected_beta))
self.assertTrue(np.allclose(test_beta_roll, expected_beta, equal_nan=True))
def test_alpha(self):
reference = self.test_data1
self.assertAlmostEqual(eval_alpha(self.test_data2, 5, reference, 'value', 0.5), 11.63072977)
self.assertAlmostEqual(eval_alpha(self.test_data3, 5, reference, 'value', 0.5), 1.886590071)
self.assertAlmostEqual(eval_alpha(self.test_data4, 5, reference, 'value', 0.5), 6.827021872)
self.assertAlmostEqual(eval_alpha(self.test_data5, 5, reference, 'value', 0.92), -1.192265168)
self.assertAlmostEqual(eval_alpha(self.test_data6, 5, reference, 'value', 0.92), -1.437142359)
self.assertAlmostEqual(eval_alpha(self.test_data7, 5, reference, 'value', 0.92), -1.781311545)
# 测试长数据的alpha计算
expected_alpha = np.array([ np.nan, np.nan, np.nan, np.nan, np.nan,
np.nan, np.nan, np.nan, np.nan, np.nan,
np.nan, np.nan, np.nan, np.nan, np.nan,
np.nan, np.nan, np.nan, np.nan, np.nan,
np.nan, np.nan, np.nan, np.nan, np.nan,
np.nan, np.nan, np.nan, np.nan, np.nan,
np.nan, np.nan, np.nan, np.nan, np.nan,
np.nan, np.nan, np.nan, np.nan, np.nan,
np.nan, np.nan, np.nan, np.nan, np.nan,
np.nan, np.nan, np.nan, np.nan, np.nan,
np.nan, np.nan, np.nan, np.nan, np.nan,
np.nan, np.nan, np.nan, np.nan, np.nan,
np.nan, np.nan, np.nan, np.nan, np.nan,
np.nan, np.nan, np.nan, np.nan, np.nan,
np.nan, np.nan, np.nan, np.nan, np.nan,
np.nan, np.nan, np.nan, np.nan, np.nan,
np.nan, np.nan, np.nan, np.nan, np.nan,
np.nan, np.nan, np.nan, np.nan, np.nan,
np.nan, np.nan, np.nan, np.nan, np.nan,
np.nan, np.nan, np.nan, np.nan, np.nan,
np.nan, np.nan, np.nan, np.nan, np.nan,
np.nan, np.nan, np.nan, np.nan, np.nan,
np.nan, np.nan, np.nan, np.nan, np.nan,
np.nan, np.nan, np.nan, np.nan, np.nan,
np.nan, np.nan, np.nan, np.nan, np.nan,
np.nan, np.nan, np.nan, np.nan, np.nan,
np.nan, np.nan, np.nan, np.nan, np.nan,
np.nan, np.nan, np.nan, np.nan, np.nan,
np.nan, np.nan, np.nan, np.nan, np.nan,
np.nan, np.nan, np.nan, np.nan, np.nan,
np.nan, np.nan, np.nan, np.nan, np.nan,
np.nan, np.nan, np.nan, np.nan, np.nan,
np.nan, np.nan, np.nan, np.nan, np.nan,
np.nan, np.nan, np.nan, np.nan, np.nan,
np.nan, np.nan, np.nan, np.nan, np.nan,
np.nan, np.nan, np.nan, np.nan, np.nan,
np.nan, np.nan, np.nan, np.nan, np.nan,
np.nan, np.nan, np.nan, np.nan, np.nan,
np.nan, np.nan, np.nan, np.nan, np.nan,
np.nan, np.nan, np.nan, np.nan, np.nan,
np.nan, np.nan, np.nan, np.nan, np.nan,
np.nan, np.nan, np.nan, np.nan, np.nan,
np.nan, np.nan, np.nan, np.nan, np.nan,
np.nan, np.nan, np.nan, np.nan, np.nan,
np.nan, np.nan, np.nan, np.nan, np.nan,
np.nan, np.nan, np.nan, np.nan, np.nan,
np.nan, np.nan, np.nan, np.nan, np.nan,
np.nan, np.nan, np.nan, np.nan, np.nan,
np.nan, np.nan, np.nan, np.nan, np.nan,
np.nan, np.nan, np.nan, np.nan, np.nan,
-0.09418119, -0.11188463, -0.17938358, -0.15588172, -0.1462678 ,
-0.13089586, -0.10780125, -0.09102891, -0.03987585, -0.06075686,
-0.02459503, -0.04104284, -0.0444565 , -0.04074585, 0.02191275,
0.02255955, -0.05583375, -0.05875539, -0.06055551, -0.09648245,
-0.07913737, -0.10627829, -0.12320965, -0.12368335, -0.1506743 ,
-0.15768033, -0.13638829, -0.13065298, -0.14537834, -0.127428 ,
-0.15504529, -0.18184636, -0.12652146, -0.09190138, -0.14847221,
-0.15840648, -0.1525789 , -0.11859418, -0.14700954, -0.16295761,
-0.16051645, -0.10364859, -0.11961134, -0.10258267, -0.08090148,
-0.05727746, -0.0429945 , -0.04672356, -0.03581408, -0.0439215 ,
-0.03429495, -0.0260362 , -0.01075022, 0.04931808, 0.02779388,
0.03984083, 0.08311951, 0.08995566, 0.10522428, 0.16159058,
0.14238174, 0.14759783, 0.16257712, 0.158908 , 0.11302115,
0.0909566 , 0.08272888, 0.15261884, 0.10546376, 0.04990313,
-0.01284111, -0.02720704, 0.00454725, -0.03965491, -0.03818265,
-0.02186992, -0.06574751, -0.04846454, -0.05204211, -0.06316498,
-0.05095099, -0.08502656, -0.04681162, -0.02362027, -0.02205091,
-0.07706374, -0.10371841, -0.14434688, -0.14797935, -0.09055402,
-0.06739549, -0.08824959, -0.04855888, -0.02291244, 0.04027138,
0.09370505, 0.11472939, 0.10243593, 0.0921445 , 0.07662648,
0.07946651, 0.05450718, 0.10497677, 0.09068334, 0.15462924,
0.14231034, 0.10544952, 0.09980256, 0.14035223, 0.14942974,
0.17624102, 0.19035477, 0.2500807 , 0.30724652, 0.31768915,
0.35007521, 0.38412975, 0.34356521, 0.33614463, 0.41206165,
0.33999177, 0.28045963, 0.34076789, 0.42220356, 0.42314636,
0.50790423, 0.47713348, 0.42520169, 0.50488411, 0.48705211,
0.46252601, 0.44325578, 0.42640573, 0.37986783, 0.30652822,
0.34503393, 0.2999069 , 0.24928617, 0.24730218, 0.24326897,
0.26657905, 0.27861168, 0.26392824, 0.32552649, 0.34177792,
0.37837011, 0.37025267, 0.4030612 , 0.41339361, 0.45076809,
0.40383354, 0.47093422, 0.52505036, 0.53614256, 0.5500943 ,
0.55319293, 0.59021451, 0.52358459, 0.50605947, 0.49359168,
0.47895956, 0.49320243, 0.4908336 , 0.47310767, 0.51821564,
0.55105932, 0.57291504, 0.5599809 , 0.46868842, 0.39620087,
0.42086934, 0.38317217, 0.45934108, 0.50048866, 0.53941991,
0.50676751, 0.46500915, 0.52993663, 0.51668366, 0.46405428,
0.44100603, 0.52726147, 0.51565458, 0.49186248, 0.49001081,
0.49367648, 0.56422294, 0.58882785, 0.51334664, 0.44386256,
0.35056709, 0.36490029, 0.39205071, 0.3677061 , 0.41134736,
0.42315067, 0.35356394, 0.40324562, 0.41340007, 0.46503322,
0.44355762, 0.34854314, 0.26412842, 0.28633753, 0.32335224,
0.30761141, 0.29709569, 0.29570487, 0.28000063, 0.32802547,
0.33967726, 0.42511212, 0.46252357, 0.44244974, 0.42152907,
0.45436727, 0.50482359, 0.57339198, 0.6573356 , 0.70912003,
0.60328917, 0.6395092 , 0.67015805, 0.64241557, 0.62779142,
0.55028063, 0.46448736, 0.43709245, 0.46777983, 0.51789439,
0.48594916, 0.4456216 , 0.52008189, 0.60548684, 0.62792473,
0.56645031, 0.62766439, 0.71829315, 0.69481356, 0.59550329,
0.58133754, 0.59014148, 0.58026655, 0.61719273, 0.67373203,
0.75573056, 0.89501633, 0.8347253 , 0.87964685, 0.89015835])
test_alpha_mean = eval_alpha(self.long_data, 100, self.long_bench, 'value')
test_alpha_roll = self.long_data['alpha'].values
self.assertAlmostEqual(test_alpha_mean, np.nanmean(expected_alpha))
self.assertTrue(np.allclose(test_alpha_roll, expected_alpha, equal_nan=True))
def test_calmar(self):
"""test evaluate function eval_calmar()"""
pass
def test_benchmark(self):
reference = self.test_data1
tr, yr = eval_benchmark(self.test_data2, reference, 'value')
self.assertAlmostEqual(tr, 0.19509091)
self.assertAlmostEqual(yr, 0.929154957)
tr, yr = eval_benchmark(self.test_data3, reference, 'value')
self.assertAlmostEqual(tr, 0.19509091)
self.assertAlmostEqual(yr, 0.929154957)
tr, yr = eval_benchmark(self.test_data4, reference, 'value')
self.assertAlmostEqual(tr, 0.19509091)
self.assertAlmostEqual(yr, 0.929154957)
tr, yr = eval_benchmark(self.test_data5, reference, 'value')
self.assertAlmostEqual(tr, 0.19509091)
self.assertAlmostEqual(yr, 0.929154957)
tr, yr = eval_benchmark(self.test_data6, reference, 'value')
self.assertAlmostEqual(tr, 0.19509091)
self.assertAlmostEqual(yr, 0.929154957)
tr, yr = eval_benchmark(self.test_data7, reference, 'value')
self.assertAlmostEqual(tr, 0.19509091)
self.assertAlmostEqual(yr, 0.929154957)
def test_evaluate(self):
pass
class TestLoop(unittest.TestCase):
"""通过一个假设但精心设计的例子来测试loop_step以及loop方法的正确性"""
def setUp(self):
self.shares = ['share1', 'share2', 'share3', 'share4', 'share5', 'share6', 'share7']
self.dates = ['2016/07/01', '2016/07/04', '2016/07/05', '2016/07/06', '2016/07/07',
'2016/07/08', '2016/07/11', '2016/07/12', '2016/07/13', '2016/07/14',
'2016/07/15', '2016/07/18', '2016/07/19', '2016/07/20', '2016/07/21',
'2016/07/22', '2016/07/25', '2016/07/26', '2016/07/27', '2016/07/28',
'2016/07/29', '2016/08/01', '2016/08/02', '2016/08/03', '2016/08/04',
'2016/08/05', '2016/08/08', '2016/08/09', '2016/08/10', '2016/08/11',
'2016/08/12', '2016/08/15', '2016/08/16', '2016/08/17', '2016/08/18',
'2016/08/19', '2016/08/22', '2016/08/23', '2016/08/24', '2016/08/25',
'2016/08/26', '2016/08/29', '2016/08/30', '2016/08/31', '2016/09/01',
'2016/09/02', '2016/09/05', '2016/09/06', '2016/09/07', '2016/09/08',
'2016/09/09', '2016/09/12', '2016/09/13', '2016/09/14', '2016/09/15',
'2016/09/16', '2016/09/19', '2016/09/20', '2016/09/21', '2016/09/22',
'2016/09/23', '2016/09/26', '2016/09/27', '2016/09/28', '2016/09/29',
'2016/09/30', '2016/10/10', '2016/10/11', '2016/10/12', '2016/10/13',
'2016/10/14', '2016/10/17', '2016/10/18', '2016/10/19', '2016/10/20',
'2016/10/21', '2016/10/23', '2016/10/24', '2016/10/25', '2016/10/26',
'2016/10/27', '2016/10/29', '2016/10/30', '2016/10/31', '2016/11/01',
'2016/11/02', '2016/11/05', '2016/11/06', '2016/11/07', '2016/11/08',
'2016/11/09', '2016/11/12', '2016/11/13', '2016/11/14', '2016/11/15',
'2016/11/16', '2016/11/19', '2016/11/20', '2016/11/21', '2016/11/22']
self.dates = [pd.Timestamp(date_text) for date_text in self.dates]
self.prices = np.array([[5.35, 5.09, 5.03, 4.98, 4.50, 5.09, 4.75],
[5.66, 4.84, 5.21, 5.44, 4.35, 5.06, 4.48],
[5.79, 4.60, 5.02, 5.45, 4.07, 4.76, 4.56],
[5.56, 4.63, 5.50, 5.74, 3.88, 4.62, 4.62],
[5.88, 4.64, 5.07, 5.46, 3.74, 4.63, 4.62],
[6.25, 4.51, 5.11, 5.45, 3.98, 4.25, 4.59],
[5.93, 4.96, 5.15, 5.24, 4.08, 4.13, 4.33],
[6.39, 4.65, 5.02, 5.47, 4.00, 3.91, 3.88],
[6.31, 4.26, 5.10, 5.58, 4.28, 3.77, 3.47],
[5.86, 3.77, 5.24, 5.36, 4.01, 3.43, 3.51],
[5.61, 3.39, 4.93, 5.38, 4.14, 3.68, 3.83],
[5.31, 3.76, 4.96, 5.30, 4.49, 3.63, 3.67],
[5.40, 4.06, 5.40, 5.77, 4.49, 3.94, 3.79],
[5.03, 3.87, 5.74, 5.75, 4.46, 4.40, 4.18],
[5.38, 3.91, 5.53, 6.15, 4.13, 4.03, 4.02],
[5.79, 4.13, 5.79, 6.04, 3.79, 3.93, 4.41],
[6.27, 4.27, 5.68, 6.01, 4.23, 3.50, 4.65],
[6.59, 4.57, 5.90, 5.71, 4.57, 3.39, 4.89],
[6.91, 5.04, 5.75, 5.23, 4.92, 3.30, 4.41],
[6.71, 5.31, 6.11, 5.47, 5.28, 3.25, 4.66],
[6.33, 5.40, 5.77, 5.79, 5.67, 2.94, 4.37],
[6.07, 5.21, 5.85, 5.82, 6.00, 2.71, 4.58],
[5.98, 5.06, 5.61, 5.61, 5.89, 2.55, 4.76],
[6.46, 4.69, 5.75, 5.31, 5.55, 2.21, 4.37],
[6.95, 5.12, 5.50, 5.24, 5.39, 2.29, 4.16],
[6.77, 5.27, 5.14, 5.41, 5.26, 2.21, 4.02],
[6.70, 5.72, 5.31, 5.60, 5.31, 2.04, 3.77],
[6.28, 6.10, 5.68, 5.28, 5.22, 2.39, 3.38],
[6.61, 6.27, 5.73, 4.99, 4.90, 2.30, 3.07],
[6.25, 6.49, 6.04, 5.09, 4.57, 2.41, 2.90],
[6.47, 6.16, 6.27, 5.39, 4.96, 2.40, 2.50],
[6.45, 6.26, 6.60, 5.58, 4.82, 2.79, 2.76],
[6.88, 6.39, 6.10, 5.33, 4.39, 2.67, 2.29],
[7.00, 6.58, 6.25, 5.48, 4.63, 2.27, 2.17],
[6.59, 6.20, 6.73, 5.10, 5.05, 2.09, 1.84],
[6.59, 5.70, 6.91, 5.39, 4.68, 2.55, 1.83],
[6.64, 5.20, 7.01, 5.30, 5.02, 2.22, 2.21],
[6.38, 5.37, 7.36, 5.04, 4.84, 2.59, 2.00],
[6.10, 5.40, 7.72, 5.51, 4.60, 2.59, 1.76],
[6.35, 5.22, 7.68, 5.43, 4.66, 2.95, 1.27],
[6.52, 5.38, 7.62, 5.23, 4.41, 2.69, 1.40],
[6.87, 5.53, 7.74, 4.99, 4.87, 2.20, 1.11],
[6.84, 6.03, 7.53, 5.43, 4.42, 2.69, 1.60],
[7.09, 5.77, 7.46, 5.40, 4.08, 2.65, 1.23],
[6.88, 5.66, 7.84, 5.60, 4.16, 2.63, 1.59],
[6.84, 6.08, 8.11, 5.66, 4.10, 2.14, 1.50],
[6.98, 5.62, 8.04, 6.01, 4.43, 2.39, 1.80],
[7.02, 5.63, 7.65, 5.64, 4.07, 1.95, 1.55],
[7.13, 6.11, 7.52, 5.67, 3.97, 2.32, 1.35],
[7.59, 6.03, 7.67, 5.30, 4.16, 2.69, 1.51],
[7.61, 6.27, 7.47, 4.91, 4.12, 2.51, 1.08],
[7.21, 6.28, 7.44, 5.37, 4.04, 2.62, 1.06],
[7.48, 6.52, 7.59, 5.75, 3.84, 2.16, 1.43],
[7.66, 7.00, 7.94, 6.08, 3.46, 2.35, 1.43],
[7.51, 7.34, 8.25, 6.58, 3.18, 2.31, 1.74],
[7.12, 7.34, 7.77, 6.78, 3.10, 1.96, 1.44],
[6.97, 7.68, 8.03, 7.20, 3.55, 2.35, 1.83],
[6.67, 8.09, 7.87, 7.65, 3.66, 2.58, 1.71],
[6.20, 7.68, 7.58, 8.00, 3.66, 2.40, 2.12],
[6.34, 7.58, 7.33, 7.92, 3.29, 2.20, 2.45],
[6.22, 7.46, 7.22, 8.30, 2.80, 2.31, 2.85],
[5.98, 7.59, 6.86, 8.46, 2.88, 2.16, 2.79],
[6.37, 7.19, 7.18, 7.99, 3.04, 2.16, 2.91],
[6.56, 7.40, 7.54, 8.19, 3.45, 2.20, 3.26],
[6.26, 7.48, 7.24, 8.61, 3.88, 1.73, 3.14],
[6.69, 7.93, 6.85, 8.66, 3.58, 1.93, 3.53],
[7.13, 8.23, 6.60, 8.91, 3.60, 2.25, 3.65],
[6.83, 8.35, 6.65, 9.08, 3.97, 2.69, 3.69],
[7.31, 8.44, 6.74, 9.34, 4.05, 2.59, 3.50],
[7.43, 8.35, 7.19, 8.96, 4.40, 2.14, 3.25],
[7.54, 8.58, 7.14, 8.98, 4.06, 1.68, 3.64],
[7.18, 8.82, 6.88, 8.50, 3.60, 1.98, 4.00],
[7.21, 9.09, 7.14, 8.65, 3.61, 2.14, 3.63],
[7.45, 9.02, 7.30, 8.94, 4.10, 1.89, 3.78],
[7.37, 8.87, 6.95, 8.63, 3.74, 1.97, 3.42],
[6.88, 9.22, 7.02, 8.65, 4.02, 1.99, 3.76],
[7.08, 9.04, 7.38, 8.40, 3.95, 2.37, 3.62],
[6.75, 8.60, 7.50, 8.38, 3.81, 2.14, 3.67],
[6.60, 8.48, 7.60, 8.23, 3.71, 2.35, 3.61],
[6.21, 8.71, 7.15, 8.04, 3.94, 1.86, 3.39],
[6.36, 8.79, 7.30, 7.91, 4.43, 2.14, 3.43],
[6.82, 8.93, 7.80, 7.57, 4.07, 2.39, 3.33],
[6.45, 9.36, 8.15, 7.73, 4.04, 2.53, 3.03],
[6.85, 9.68, 8.40, 7.74, 4.34, 2.47, 3.28],
[6.48, 10.16, 8.87, 8.07, 4.80, 2.93, 3.46],
[6.10, 10.56, 8.53, 7.99, 5.18, 3.09, 3.25],
[5.64, 10.63, 8.94, 7.92, 4.90, 2.93, 2.95],
[6.01, 10.55, 8.52, 8.40, 5.40, 3.22, 2.87],
[6.21, 10.65, 8.80, 8.80, 5.73, 3.06, 2.63],
[6.61, 10.55, 8.92, 8.47, 5.62, 2.90, 2.40],
[7.02, 10.19, 9.20, 8.07, 5.20, 2.68, 2.53],
[7.04, 10.48, 8.71, 7.87, 4.85, 2.46, 2.96],
[6.77, 10.36, 8.25, 8.02, 5.18, 2.41, 3.26],
[7.09, 10.03, 8.19, 8.39, 4.72, 2.74, 2.97],
[6.65, 10.24, 7.80, 8.69, 4.62, 3.15, 3.16],
[7.07, 10.01, 7.69, 8.81, 4.55, 3.40, 3.58],
[6.80, 10.14, 7.23, 8.99, 4.37, 3.82, 3.23],
[6.79, 10.31, 6.98, 9.10, 4.26, 4.02, 3.62],
[6.48, 9.88, 7.07, 8.90, 4.25, 3.76, 3.13],
[6.39, 10.05, 6.95, 8.87, 4.59, 4.10, 2.93]])
self.op_signals = np.array([[0, 0, 0, 0, 0.25, 0, 0],
[0, 0, 0, 0, 0, 0, 0],
[0, 0, 0, 0, 0, 0.1, 0.15],
[0.2, 0.2, 0, 0, 0, 0, 0],
[0, 0, 0.1, 0, 0, 0, 0],
[0, 0, 0, 0, -0.75, 0, 0],
[0, 0, 0, 0, 0, 0, 0],
[0, 0, 0, 0, 0, 0, 0],
[0, 0, 0, 0, 0, 0, 0],
[0, 0, 0, 0, 0, 0, 0],
[0, 0, 0, 0, 0, 0, 0],
[0, 0, 0, 0, 0, 0, 0],
[0, 0, 0, 0, 0, 0, 0],
[-0.333, 0, 0, 0, 0, 0, 0],
[0, 0, 0, 0, 0, 0, 0],
[0, 0, 0, 0, 0, 0, 0],
[0, 0, 0, 0, 0, 0, 0],
[0, 0, -0.5, 0, 0, 0, 0],
[0, 0, 0, 0, 0, 0, 0],
[0, 0, 0, 0, 0, 0, 0],
[0, 0, 0, 0, 0, 0, 0],
[0, 0, 0, 0, 0, 0, -1],
[0, 0, 0, 0, 0.2, 0, 0],
[0, 0, 0, 0, 0, 0, 0],
[0, 0, 0, 0, 0, 0, 0],
[-0.5, 0, 0, 0.15, 0, 0, 0],
[0, 0, 0, 0, 0, 0, 0],
[0, 0, 0, 0, 0, 0, 0],
[0, 0, 0, 0, 0, 0, 0],
[0, 0, 0, 0, 0, 0, 0],
[0, 0, 0.2, 0, -1, 0.2, 0],
[0.5, 0, 0, 0, 0, 0, 0],
[0, 0, 0, 0, 0, 0, 0],
[0, 0, 0, 0, 0, 0, 0],
[0, 0, 0, 0, 0, 0, 0],
[0, 0, 0, 0, 0, 0, 0],
[0, 0, 0, 0, 0, 0, 0],
[0, 0, 0, 0, 0, 0, 0],
[0, 0, 0, 0, 0, 0, 0],
[0, 0, 0, 0, 0, 0, 0],
[0, 0, 0, 0, 0, 0, 0],
[0, 0, 0, 0, 0, 0, 0],
[0.2, 0, 0, 0, 0, 0, 0],
[0, 0, 0, 0, 0, 0, 0],
[0, 0, 0, 0, 0, 0, 0],
[0, 0, 0, 0, 0, -0.5, 0.2],
[0, 0, 0, 0, 0, 0, 0],
[0, 0, 0, 0, 0, 0, 0],
[0, 0, 0, 0, 0, 0, 0],
[0, 0, 0, 0, 0, 0, 0],
[0, 0.2, 0, 0, 0, 0, 0],
[0, 0, 0, 0, 0, 0, 0],
[0, 0, 0, 0, 0, 0, 0],
[0, 0, 0, 0, 0, 0, 0],
[0, 0, 0, 0, 0, 0, 0],
[0, 0, 0, 0, 0, 0, 0],
[0, 0, 0, 0, 0, 0, 0],
[0, 0, 0, 0, 0, 0, 0],
[0, 0, 0, 0, 0, 0, 0],
[0, 0, 0, 0, 0, 0, 0],
[0, 0, 0, 0, 0.15, 0, 0],
[-1, 0, 0.25, 0.25, 0, 0.25, 0],
[0, 0, 0, 0, 0, 0, 0],
[0.25, 0, 0, 0, 0, 0, 0],
[0, 0, 0, 0, 0, 0, 0],
[0, 0, 0, 0, 0, 0, 0],
[0, 0, 0, 0, 0, 0, 0],
[0, 0, 0, 0, 0, 0, 0],
[0, 0, 0, 0, 0, 0, 0],
[0, 0, 0, 0, 0, 0, 0],
[0, 0, 0, 0, 0, 0, 0],
[0, 0, 0, 0, 0, 0, 0],
[0, 0, 0, 0, 0, 0, 0],
[0, 0, 0, 0, 0, 0, 0],
[0, 0, 0, 0, 0, 0, 0],
[0, 0, 0, 0, 0, 0, 0],
[0, 0, 0, 0, 0, 0, 0],
[0, 0, 0, 0, 0, 0, 0],
[0, 0, 0, 0, 0, 0, 0],
[0, 0, 0, 0, 0, 0, 0],
[0, 0, 0, 0, 0, 0, 0],
[0, 0.2, 0, 0, 0, 0, 0],
[0, 0, 0, 0, 0, 0, 0],
[0, 0, 0, 0, 0, 0, 0],
[0, 0, 0, 0, 0, 0, 0],
[0, 0, 0, 0, 0, 0, 0],
[0, 0, 0, 0, 0, 0, 0],
[0, 0, 0, 0, -1, 0, 0.2],
[0, 0, 0, 0, 0, 0, 0],
[0, 0, 0, 0, 0, 0, 0],
[0, 0, 0, 0, 0, 0, 0],
[0, 0, 0, 0, 0, 0, 0],
[0, 0, 0, 0, 0, 0, 0],
[0, 0, 0, 0, 0, 0, 0],
[0, 0, 0, 0, 0, 0, 0],
[0, 0, 0, 0, 0, 0, 0],
[0, -1, 0, 0, 0, 0, 0],
[-1, 0, 0.15, 0, 0, 0, 0],
[0, 0, 0, 0, 0, 0, 0],
[0, 0, 0, 0, 0, 0, 0]])
self.cash = qt.CashPlan(['2016/07/01', '2016/08/12', '2016/09/23'], [10000, 10000, 10000])
self.rate = qt.Cost(buy_fix=0,
sell_fix=0,
buy_rate=0,
sell_rate=0,
buy_min=0,
sell_min=0,
slipage=0)
self.rate2 = qt.Cost(buy_fix=0,
sell_fix=0,
buy_rate=0,
sell_rate=0,
buy_min=10,
sell_min=5,
slipage=0)
self.op_signal_df = pd.DataFrame(self.op_signals, index=self.dates, columns=self.shares)
self.history_list = pd.DataFrame(self.prices, index=self.dates, columns=self.shares)
self.res = np.array([[0.000, 0.000, 0.000, 0.000, 555.556, 0.000, 0.000, 7500.000, 0.000, 10000.000],
[0.000, 0.000, 0.000, 0.000, 555.556, 0.000, 0.000, 7500.000, 0.000, 9916.667],
[0.000, 0.000, 0.000, 0.000, 555.556, 205.065, 321.089, 5059.722, 0.000, 9761.111],
[346.982, 416.679, 0.000, 0.000, 555.556, 205.065, 321.089, 1201.278, 0.000, 9646.112],
[346.982, 416.679, 191.037, 0.000, 555.556, 205.065, 321.089, 232.719, 0.000, 9685.586],
[346.982, 416.679, 191.037, 0.000, 138.889, 205.065, 321.089, 1891.052, 0.000, 9813.218],
[346.982, 416.679, 191.037, 0.000, 138.889, 205.065, 321.089, 1891.052, 0.000, 9803.129],
[346.982, 416.679, 191.037, 0.000, 138.889, 205.065, 321.089, 1891.052, 0.000, 9608.020],
[346.982, 416.679, 191.037, 0.000, 138.889, 205.065, 321.089, 1891.052, 0.000, 9311.573],
[346.982, 416.679, 191.037, 0.000, 138.889, 205.065, 321.089, 1891.052, 0.000, 8883.625],
[346.982, 416.679, 191.037, 0.000, 138.889, 205.065, 321.089, 1891.052, 0.000, 8751.390],
[346.982, 416.679, 191.037, 0.000, 138.889, 205.065, 321.089, 1891.052, 0.000, 8794.181],
[346.982, 416.679, 191.037, 0.000, 138.889, 205.065, 321.089, 1891.052, 0.000, 9136.570],
[231.437, 416.679, 191.037, 0.000, 138.889, 205.065, 321.089, 2472.244, 0.000, 9209.359],
[231.437, 416.679, 191.037, 0.000, 138.889, 205.065, 321.089, 2472.244, 0.000, 9093.829],
[231.437, 416.679, 191.037, 0.000, 138.889, 205.065, 321.089, 2472.244, 0.000, 9387.554],
[231.437, 416.679, 191.037, 0.000, 138.889, 205.065, 321.089, 2472.244, 0.000, 9585.959],
[231.437, 416.679, 95.519, 0.000, 138.889, 205.065, 321.089, 3035.804, 0.000, 9928.777],
[231.437, 416.679, 95.519, 0.000, 138.889, 205.065, 321.089, 3035.804, 0.000, 10060.381],
[231.437, 416.679, 95.519, 0.000, 138.889, 205.065, 321.089, 3035.804, 0.000, 10281.002],
[231.437, 416.679, 95.519, 0.000, 138.889, 205.065, 321.089, 3035.804, 0.000, 10095.561],
[231.437, 416.679, 95.519, 0.000, 138.889, 205.065, 0.000, 4506.393, 0.000, 10029.957],
[231.437, 416.679, 95.519, 0.000, 474.224, 205.065, 0.000, 2531.270, 0.000, 9875.613],
[231.437, 416.679, 95.519, 0.000, 474.224, 205.065, 0.000, 2531.270, 0.000, 9614.946],
[231.437, 416.679, 95.519, 0.000, 474.224, 205.065, 0.000, 2531.270, 0.000, 9824.172],
[115.719, 416.679, 95.519, 269.850, 474.224, 205.065, 0.000, 1854.799, 0.000, 9732.574],
[115.719, 416.679, 95.519, 269.850, 474.224, 205.065, 0.000, 1854.799, 0.000, 9968.339],
[115.719, 416.679, 95.519, 269.850, 474.224, 205.065, 0.000, 1854.799, 0.000, 10056.158],
[115.719, 416.679, 95.519, 269.850, 474.224, 205.065, 0.000, 1854.799, 0.000, 9921.492],
[115.719, 416.679, 95.519, 269.850, 474.224, 205.065, 0.000, 1854.799, 0.000, 9894.162],
[115.719, 416.679, 735.644, 269.850, 0.000, 1877.393, 0.000, 6179.774, 0.000, 20067.937],
[1073.823, 416.679, 735.644, 269.850, 0.000, 1877.393, 0.000, 0.000, 0.000, 21133.508],
[1073.823, 416.679, 735.644, 269.850, 0.000, 1877.393, 0.000, 0.000, 0.000, 20988.848],
[1073.823, 416.679, 735.644, 269.850, 0.000, 1877.393, 0.000, 0.000, 0.000, 20596.743],
[1073.823, 416.679, 735.644, 269.850, 0.000, 1877.393, 0.000, 0.000, 0.000, 19910.773],
[1073.823, 416.679, 735.644, 269.850, 0.000, 1877.393, 0.000, 0.000, 0.000, 20776.707],
[1073.823, 416.679, 735.644, 269.850, 0.000, 1877.393, 0.000, 0.000, 0.000, 20051.797],
[1073.823, 416.679, 735.644, 269.850, 0.000, 1877.393, 0.000, 0.000, 0.000, 20725.388],
[1073.823, 416.679, 735.644, 269.850, 0.000, 1877.393, 0.000, 0.000, 0.000, 20828.880],
[1073.823, 416.679, 735.644, 269.850, 0.000, 1877.393, 0.000, 0.000, 0.000, 21647.181],
[1073.823, 416.679, 735.644, 269.850, 0.000, 1877.393, 0.000, 0.000, 0.000, 21310.169],
[1073.823, 416.679, 735.644, 269.850, 0.000, 1877.393, 0.000, 0.000, 0.000, 20852.099],
[1073.823, 416.679, 735.644, 269.850, 0.000, 1877.393, 0.000, 0.000, 0.000, 21912.395],
[1073.823, 416.679, 735.644, 269.850, 0.000, 1877.393, 0.000, 0.000, 0.000, 21937.828],
[1073.823, 416.679, 735.644, 269.850, 0.000, 1877.393, 0.000, 0.000, 0.000, 21962.458],
[1073.823, 416.679, 735.644, 269.850, 0.000, 938.697, 1339.207, 0.000, 0.000, 21389.402],
[1073.823, 416.679, 735.644, 269.850, 0.000, 938.697, 1339.207, 0.000, 0.000, 22027.453],
[1073.823, 416.679, 735.644, 269.850, 0.000, 938.697, 1339.207, 0.000, 0.000, 20939.999],
[1073.823, 416.679, 735.644, 269.850, 0.000, 938.697, 1339.207, 0.000, 0.000, 21250.064],
[1073.823, 416.679, 735.644, 269.850, 0.000, 938.697, 1339.207, 0.000, 0.000, 22282.781],
[1073.823, 416.679, 735.644, 269.850, 0.000, 938.697, 1339.207, 0.000, 0.000, 21407.066],
[1073.823, 416.679, 735.644, 269.850, 0.000, 938.697, 1339.207, 0.000, 0.000, 21160.237],
[1073.823, 416.679, 735.644, 269.850, 0.000, 938.697, 1339.207, 0.000, 0.000, 21826.768],
[1073.823, 416.679, 735.644, 269.850, 0.000, 938.697, 1339.207, 0.000, 0.000, 22744.940],
[1073.823, 416.679, 735.644, 269.850, 0.000, 938.697, 1339.207, 0.000, 0.000, 23466.118],
[1073.823, 416.679, 735.644, 269.850, 0.000, 938.697, 1339.207, 0.000, 0.000, 22017.882],
[1073.823, 416.679, 735.644, 269.850, 0.000, 938.697, 1339.207, 0.000, 0.000, 23191.466],
[1073.823, 416.679, 735.644, 269.850, 0.000, 938.697, 1339.207, 0.000, 0.000, 23099.082],
[1073.823, 416.679, 735.644, 269.850, 0.000, 938.697, 1339.207, 0.000, 0.000, 22684.767],
[1073.823, 416.679, 735.644, 269.850, 0.000, 938.697, 1339.207, 0.000, 0.000, 22842.135],
[1073.823, 416.679, 735.644, 269.850, 1785.205, 938.697, 1339.207, 5001.425, 0, 33323.836],
[0.000, 416.679, 1290.692, 719.924, 1785.205, 2701.488, 1339.207, 0.000, 0.000, 32820.290],
[0.000, 416.679, 1290.692, 719.924, 1785.205, 2701.488, 1339.207, 0.000, 0.000, 33174.614],
[0.000, 416.679, 1290.692, 719.924, 1785.205, 2701.488, 1339.207, 0.000, 0.000, 35179.466],
[0.000, 416.679, 1290.692, 719.924, 1785.205, 2701.488, 1339.207, 0.000, 0.000, 34465.195],
[0.000, 416.679, 1290.692, 719.924, 1785.205, 2701.488, 1339.207, 0.000, 0.000, 34712.354],
[0.000, 416.679, 1290.692, 719.924, 1785.205, 2701.488, 1339.207, 0.000, 0.000, 35755.550],
[0.000, 416.679, 1290.692, 719.924, 1785.205, 2701.488, 1339.207, 0.000, 0.000, 37895.223],
[0.000, 416.679, 1290.692, 719.924, 1785.205, 2701.488, 1339.207, 0.000, 0.000, 37854.284],
[0.000, 416.679, 1290.692, 719.924, 1785.205, 2701.488, 1339.207, 0.000, 0.000, 37198.374],
[0.000, 416.679, 1290.692, 719.924, 1785.205, 2701.488, 1339.207, 0.000, 0.000, 35916.711],
[0.000, 416.679, 1290.692, 719.924, 1785.205, 2701.488, 1339.207, 0.000, 0.000, 35806.937],
[0.000, 416.679, 1290.692, 719.924, 1785.205, 2701.488, 1339.207, 0.000, 0.000, 36317.592],
[0.000, 416.679, 1290.692, 719.924, 1785.205, 2701.488, 1339.207, 0.000, 0.000, 37103.973],
[0.000, 416.679, 1290.692, 719.924, 1785.205, 2701.488, 1339.207, 0.000, 0.000, 35457.883],
[0.000, 416.679, 1290.692, 719.924, 1785.205, 2701.488, 1339.207, 0.000, 0.000, 36717.685],
[0.000, 416.679, 1290.692, 719.924, 1785.205, 2701.488, 1339.207, 0.000, 0.000, 37641.463],
[0.000, 416.679, 1290.692, 719.924, 1785.205, 2701.488, 1339.207, 0.000, 0.000, 36794.298],
[0.000, 416.679, 1290.692, 719.924, 1785.205, 2701.488, 1339.207, 0.000, 0.000, 37073.817],
[0.000, 416.679, 1290.692, 719.924, 1785.205, 2701.488, 1339.207, 0.000, 0.000, 35244.299],
[0.000, 416.679, 1290.692, 719.924, 1785.205, 2701.488, 1339.207, 0.000, 0.000, 37062.382],
[0.000, 416.679, 1290.692, 719.924, 1785.205, 2701.488, 1339.207, 0.000, 0.000, 37420.067],
[0.000, 416.679, 1290.692, 719.924, 1785.205, 2701.488, 1339.207, 0.000, 0.000, 38089.058],
[0.000, 416.679, 1290.692, 719.924, 1785.205, 2701.488, 1339.207, 0.000, 0.000, 39260.542],
[0.000, 416.679, 1290.692, 719.924, 1785.205, 2701.488, 1339.207, 0.000, 0.000, 42609.684],
[0.000, 416.679, 1290.692, 719.924, 1785.205, 2701.488, 1339.207, 0.000, 0.000, 43109.309],
[0.000, 416.679, 1290.692, 719.924, 1785.205, 2701.488, 1339.207, 0.000, 0.000, 42283.408],
[0.000, 416.679, 1290.692, 719.924, 0.000, 2701.488, 4379.099, 915.621, 0.000, 43622.444],
[0.000, 416.679, 1290.692, 719.924, 0.000, 2701.488, 4379.099, 915.621, 0.000, 42830.254],
[0.000, 416.679, 1290.692, 719.924, 0.000, 2701.488, 4379.099, 915.621, 0.000, 41266.463],
[0.000, 416.679, 1290.692, 719.924, 0.000, 2701.488, 4379.099, 915.621, 0.000, 41164.839],
[0.000, 416.679, 1290.692, 719.924, 0.000, 2701.488, 4379.099, 915.621, 0.000, 41797.937],
[0.000, 416.679, 1290.692, 719.924, 0.000, 2701.488, 4379.099, 915.621, 0.000, 42440.861],
[0.000, 416.679, 1290.692, 719.924, 0.000, 2701.488, 4379.099, 915.621, 0.000, 42113.839],
[0.000, 416.679, 1290.692, 719.924, 0.000, 2701.488, 4379.099, 915.621, 0.000, 43853.588],
[0.000, 416.679, 1290.692, 719.924, 0.000, 2701.488, 4379.099, 915.621, 0.000, 46216.760],
[0.000, 0.000, 1290.692, 719.924, 0.000, 2701.488, 4379.099, 5140.743, 0.000, 45408.737],
[0.000, 0.000, 2027.188, 719.924, 0.000, 2701.488, 4379.099, 0.000, 0.000, 47413.401],
[0.000, 0.000, 2027.188, 719.924, 0.000, 2701.488, 4379.099, 0.000, 0.000, 44603.718],
[0.000, 0.000, 2027.188, 719.924, 0.000, 2701.488, 4379.099, 0.000, 0.000, 44381.544]])
def test_loop_step(self):
cash, amounts, fee, value = qt.core._loop_step(pre_cash=10000,
pre_amounts=np.zeros(7, dtype='float'),
op=self.op_signals[0],
prices=self.prices[0],
rate=self.rate,
moq_buy=0,
moq_sell=0,
print_log=True)
print(f'day 1 result in complete looping: \n'
f'cash: {cash}\n'
f'amounts: {np.round(amounts, 2)}\n'
f'value: {value}')
self.assertAlmostEqual(cash, 7500)
self.assertTrue(np.allclose(amounts, np.array([0, 0, 0, 0, 555.5555556, 0, 0])))
self.assertAlmostEqual(value, 10000.00)
cash, amounts, fee, value = qt.core._loop_step(pre_cash=5059.722222,
pre_amounts=np.array([0, 0, 0, 0, 555.5555556,
205.0653595, 321.0891813]),
op=self.op_signals[3],
prices=self.prices[3],
rate=self.rate,
moq_buy=0,
moq_sell=0,
print_log=True)
print(f'day 4 result in complete looping: \n'
f'cash: {cash}\n'
f'amounts: {np.round(amounts, 2)}\n'
f'value: {value}')
self.assertAlmostEqual(cash, 1201.2775195, 5)
self.assertTrue(np.allclose(amounts, np.array([346.9824373, 416.6786936, 0, 0,
555.5555556, 205.0653595, 321.0891813])))
self.assertAlmostEqual(value, 9646.111756, 5)
cash, amounts, fee, value = qt.core._loop_step(pre_cash=6179.77423,
pre_amounts=np.array([115.7186428, 416.6786936, 735.6441811,
269.8495646, 0, 1877.393446, 0]),
op=self.op_signals[31],
prices=self.prices[31],
rate=self.rate,
moq_buy=0,
moq_sell=0,
print_log=True)
print(f'day 32 result in complete looping: \n'
f'cash: {cash}\n'
f'amounts: {np.round(amounts, 2)}\n'
f'value: {value}')
self.assertAlmostEqual(cash, 0, 5)
self.assertTrue(np.allclose(amounts, np.array([1073.823175, 416.6786936, 735.6441811,
269.8495646, 0, 1877.393446, 0])))
self.assertAlmostEqual(value, 21133.50798, 5)
cash, amounts, fee, value = qt.core._loop_step(pre_cash=10000,
pre_amounts=np.array([1073.823175, 416.6786936, 735.6441811,
269.8495646, 0, 938.6967231, 1339.207325]),
op=self.op_signals[60],
prices=self.prices[60],
rate=self.rate,
moq_buy=0,
moq_sell=0,
print_log=True)
print(f'day 61 result in complete looping: \n'
f'cash: {cash}\n'
f'amounts: {np.round(amounts, 2)}\n'
f'value: {value}')
self.assertAlmostEqual(cash, 5001.424618, 5)
self.assertTrue(np.allclose(amounts, np.array([1073.823175, 416.6786936, 735.6441811, 269.8495646,
1785.205494, 938.6967231, 1339.207325])))
self.assertAlmostEqual(value, 33323.83588, 5)
cash, amounts, fee, value = qt.core._loop_step(pre_cash=cash,
pre_amounts=amounts,
op=self.op_signals[61],
prices=self.prices[61],
rate=self.rate,
moq_buy=0,
moq_sell=0,
print_log=True)
print(f'day 62 result in complete looping: \n'
f'cash: {cash}\n'
f'amounts: {np.round(amounts, 2)}\n'
f'value: {value}')
self.assertAlmostEqual(cash, 0, 5)
self.assertTrue(np.allclose(amounts, np.array([0, 416.6786936, 1290.69215, 719.9239224,
1785.205494, 2701.487958, 1339.207325])))
self.assertAlmostEqual(value, 32820.29007, 5)
cash, amounts, fee, value = qt.core._loop_step(pre_cash=915.6208259,
pre_amounts=np.array([0, 416.6786936, 1290.69215, 719.9239224,
0, 2701.487958, 4379.098907]),
op=self.op_signals[96],
prices=self.prices[96],
rate=self.rate,
moq_buy=0,
moq_sell=0,
print_log=True)
print(f'day 97 result in complete looping: \n'
f'cash: {cash}\n'
f'amounts: {np.round(amounts, 2)}\n'
f'value: {value}')
self.assertAlmostEqual(cash, 5140.742779, 5)
self.assertTrue(np.allclose(amounts, np.array([0, 0, 1290.69215, 719.9239224, 0, 2701.487958, 4379.098907])))
self.assertAlmostEqual(value, 45408.73655, 4)
cash, amounts, fee, value = qt.core._loop_step(pre_cash=cash,
pre_amounts=amounts,
op=self.op_signals[97],
prices=self.prices[97],
rate=self.rate,
moq_buy=0,
moq_sell=0,
print_log=True)
print(f'day 98 result in complete looping: \n'
f'cash: {cash}\n'
f'amounts: {np.round(amounts, 2)}\n'
f'value: {value}')
self.assertAlmostEqual(cash, 0, 5)
self.assertTrue(np.allclose(amounts, np.array([0, 0, 2027.18825, 719.9239224, 0, 2701.487958, 4379.098907])))
self.assertAlmostEqual(value, 47413.40131, 4)
def test_loop(self):
res = apply_loop(op_list=self.op_signal_df,
history_list=self.history_list,
cash_plan=self.cash,
cost_rate=self.rate,
moq_buy=0,
moq_sell=0,
inflation_rate=0,
print_log=True)
self.assertIsInstance(res, pd.DataFrame)
print(f'in test_loop:\nresult of loop test is \n{res}')
self.assertTrue(np.allclose(res.values, self.res, 5))
print(f'test assertion errors in apply_loop: detect moqs that are not compatible')
self.assertRaises(AssertionError,
apply_loop,
self.op_signal_df,
self.history_list,
self.cash,
self.rate,
0, 1,
0,
False)
self.assertRaises(AssertionError,
apply_loop,
self.op_signal_df,
self.history_list,
self.cash,
self.rate,
1, 5,
0,
False)
print(f'test loop results with moq equal to 100')
res = apply_loop(op_list=self.op_signal_df,
history_list=self.history_list,
cash_plan=self.cash,
cost_rate=self.rate2,
moq_buy=100,
moq_sell=1,
inflation_rate=0,
print_log=True)
self.assertIsInstance(res, pd.DataFrame)
print(f'in test_loop:\nresult of loop test is \n{res}')
class TestOperatorSubFuncs(unittest.TestCase):
def setUp(self):
mask_list = [[0.0, 0.0, 0.0, 0.0],
[0.5, 0.0, 0.0, 1.0],
[0.5, 0.0, 0.3, 1.0],
[0.5, 0.0, 0.3, 0.5],
[0.5, 0.5, 0.3, 0.5],
[0.5, 0.5, 0.3, 1.0],
[0.3, 0.5, 0.0, 1.0],
[0.3, 1.0, 0.0, 1.0]]
signal_list = [[0.0, 0.0, 0.0, 0.0],
[0.5, 0.0, 0.0, 1.0],
[0.0, 0.0, 0.3, 0.0],
[0.0, 0.0, 0.0, -0.5],
[0.0, 0.5, 0.0, 0.0],
[0.0, 0.0, 0.0, 0.5],
[-0.4, 0.0, -1.0, 0.0],
[0.0, 0.5, 0.0, 0.0]]
mask_multi = [[[0, 0, 1, 1, 0],
[0, 1, 1, 1, 0],
[1, 1, 1, 1, 0],
[1, 1, 1, 1, 1],
[1, 1, 1, 1, 1],
[1, 1, 0, 1, 1],
[0, 1, 0, 0, 1],
[1, 1, 0, 0, 1],
[1, 1, 0, 0, 1],
[1, 0, 0, 0, 1]],
[[0, 0, 1, 0, 1],
[0, 1, 1, 1, 1],
[1, 1, 0, 1, 1],
[1, 1, 1, 0, 0],
[1, 1, 0, 0, 0],
[1, 0, 0, 0, 0],
[1, 0, 0, 0, 0],
[1, 1, 0, 0, 0],
[1, 1, 0, 1, 0],
[0, 1, 0, 1, 0]],
[[0, 0, 0., 0, 1],
[0, 0, 1., 0, 1],
[0, 0, 1., 0, 1],
[1, 0, 1., 0, 1],
[1, 1, .5, 1, 1],
[1, 0, .5, 1, 0],
[1, 1, .5, 1, 0],
[0, 1, 0., 0, 0],
[1, 0, 0., 0, 0],
[0, 1, 0., 0, 0]]]
signal_multi = [[[0., 0., 1., 1., 0.],
[0., 1., 0., 0., 0.],
[1., 0., 0., 0., 0.],
[0., 0., 0., 0., 1.],
[0., 0., 0., 0., 0.],
[0., 0., -1., 0., 0.],
[-1., 0., 0., -1., 0.],
[1., 0., 0., 0., 0.],
[0., 0., 0., 0., 0.],
[0., -1., 0., 0., 0.]],
[[0., 0., 1., 0., 1.],
[0., 1., 0., 1., 0.],
[1., 0., -1., 0., 0.],
[0., 0., 1., -1., -1.],
[0., 0., -1., 0., 0.],
[0., -1., 0., 0., 0.],
[0., 0., 0., 0., 0.],
[0., 1., 0., 0., 0.],
[0., 0., 0., 1., 0.],
[-1., 0., 0., 0., 0.]],
[[0., 0., 0., 0., 1.],
[0., 0., 1., 0., 0.],
[0., 0., 0., 0., 0.],
[1., 0., 0., 0., 0.],
[0., 1., -0.5, 1., 0.],
[0., -1., 0., 0., -1.],
[0., 1., 0., 0., 0.],
[-1., 0., -1., -1., 0.],
[1., -1., 0., 0., 0.],
[-1., 1., 0., 0., 0.]]]
self.mask = np.array(mask_list)
self.multi_mask = np.array(mask_multi)
self.correct_signal = np.array(signal_list)
self.correct_multi_signal = np.array(signal_multi)
self.op = qt.Operator()
def test_ls_blend(self):
"""测试多空蒙板的混合器,三种混合方式均需要测试"""
ls_mask1 = [[0.0, 0.0, 0.0, -0.0],
[1.0, 0.0, 0.0, -1.0],
[1.0, 0.0, 1.0, -1.0],
[1.0, 0.0, 1.0, -1.0],
[1.0, 1.0, 1.0, -1.0],
[1.0, 1.0, 1.0, -1.0],
[0.0, 1.0, 0.0, -1.0],
[0.0, 1.0, 0.0, -1.0]]
ls_mask2 = [[0.0, 0.0, 0.5, -0.5],
[0.0, 0.0, 0.5, -0.3],
[0.0, 0.5, 0.5, -0.0],
[0.5, 0.5, 0.3, -0.0],
[0.5, 0.5, 0.3, -0.3],
[0.5, 0.5, 0.0, -0.5],
[0.3, 0.5, 0.0, -1.0],
[0.3, 1.0, 0.0, -1.0]]
ls_mask3 = [[0.5, 0.0, 1.0, -0.4],
[0.4, 0.0, 1.0, -0.3],
[0.3, 0.0, 0.8, -0.2],
[0.2, 0.0, 0.6, -0.1],
[0.1, 0.2, 0.4, -0.2],
[0.1, 0.3, 0.2, -0.5],
[0.1, 0.4, 0.0, -0.5],
[0.1, 0.5, 0.0, -1.0]]
# result with blender 'avg'
ls_blnd_avg = [[0.16666667, 0.00000000, 0.50000000, -0.3],
[0.46666667, 0.00000000, 0.50000000, -0.53333333],
[0.43333333, 0.16666667, 0.76666667, -0.4],
[0.56666667, 0.16666667, 0.63333333, -0.36666667],
[0.53333333, 0.56666667, 0.56666667, -0.5],
[0.53333333, 0.60000000, 0.40000000, -0.66666667],
[0.13333333, 0.63333333, 0.00000000, -0.83333333],
[0.13333333, 0.83333333, 0.00000000, -1.]]
# result with blender 'str-1.5'
ls_blnd_str_15 = [[0, 0, 1, 0],
[0, 0, 1, -1],
[0, 0, 1, 0],
[1, 0, 1, 0],
[1, 1, 1, -1],
[1, 1, 0, -1],
[0, 1, 0, -1],
[0, 1, 0, -1]]
# result with blender 'pos-2' == 'pos-2-0'
ls_blnd_pos_2 = [[0, 0, 1, -1],
[1, 0, 1, -1],
[1, 0, 1, -1],
[1, 0, 1, -1],
[1, 1, 1, -1],
[1, 1, 1, -1],
[1, 1, 0, -1],
[1, 1, 0, -1]]
# result with blender 'pos-2-0.25'
ls_blnd_pos_2_25 = [[0, 0, 1, -1],
[1, 0, 1, -1],
[1, 0, 1, 0],
[1, 0, 1, 0],
[1, 1, 1, -1],
[1, 1, 0, -1],
[0, 1, 0, -1],
[0, 1, 0, -1]]
# result with blender 'avg_pos-2' == 'pos-2-0'
ls_blnd_avg_pos_2 = [[0.00000000, 0.00000000, 0.50000000, -0.3],
[0.46666667, 0.00000000, 0.50000000, -0.53333333],
[0.43333333, 0.00000000, 0.76666667, -0.4],
[0.56666667, 0.00000000, 0.63333333, -0.36666667],
[0.53333333, 0.56666667, 0.56666667, -0.5],
[0.53333333, 0.60000000, 0.40000000, -0.66666667],
[0.13333333, 0.63333333, 0.00000000, -0.83333333],
[0.13333333, 0.83333333, 0.00000000, -1.]]
# result with blender 'avg_pos-2-0.25'
ls_blnd_avg_pos_2_25 = [[0.00000000, 0.00000000, 0.50000000, -0.3],
[0.46666667, 0.00000000, 0.50000000, -0.53333333],
[0.43333333, 0.00000000, 0.76666667, 0.00000000],
[0.56666667, 0.00000000, 0.63333333, 0.00000000],
[0.53333333, 0.56666667, 0.56666667, -0.5],
[0.53333333, 0.60000000, 0.00000000, -0.66666667],
[0.00000000, 0.63333333, 0.00000000, -0.83333333],
[0.00000000, 0.83333333, 0.00000000, -1.]]
# result with blender 'combo'
ls_blnd_combo = [[0.5, 0., 1.5, -0.9],
[1.4, 0., 1.5, -1.6],
[1.3, 0.5, 2.3, -1.2],
[1.7, 0.5, 1.9, -1.1],
[1.6, 1.7, 1.7, -1.5],
[1.6, 1.8, 1.2, -2.],
[0.4, 1.9, 0., -2.5],
[0.4, 2.5, 0., -3.]]
ls_masks = np.array([np.array(ls_mask1), np.array(ls_mask2), np.array(ls_mask3)])
# test A: the ls_blender 'str-T'
self.op.set_blender('ls', 'str-1.5')
res = qt.Operator._ls_blend(self.op, ls_masks)
print(f'test A: result of ls_blender: str-1.5: \n{res}')
self.assertTrue(np.allclose(res, ls_blnd_str_15))
# test B: the ls_blender 'pos-N-T'
self.op.set_blender('ls', 'pos-2')
res = qt.Operator._ls_blend(self.op, ls_masks)
print(f'Test B-1: result of ls_blender: pos-2: \n{res}')
self.assertTrue(np.allclose(res, ls_blnd_pos_2))
self.op.set_blender('ls', 'pos-2-0.25')
res = qt.Operator._ls_blend(self.op, ls_masks)
print(f'Test B-2: result of ls_blender: pos-2-0.25: \n{res}')
self.assertTrue(np.allclose(res, ls_blnd_pos_2_25))
# test C: the ls_blender 'avg_pos-N-T'
self.op.set_blender('ls', 'avg_pos-2')
res = qt.Operator._ls_blend(self.op, ls_masks)
print(f'Test C-1: result of ls_blender: avg_pos-2: \n{res}')
self.assertTrue(np.allclose(res, ls_blnd_pos_2, 5))
self.op.set_blender('ls', 'avg_pos-2-0.25')
res = qt.Operator._ls_blend(self.op, ls_masks)
print(f'Test C-2: result of ls_blender: avg_pos-2-0.25: \n{res}')
self.assertTrue(np.allclose(res, ls_blnd_pos_2_25, 5))
# test D: the ls_blender 'avg'
self.op.set_blender('ls', 'avg')
res = qt.Operator._ls_blend(self.op, ls_masks)
print(f'Test D: result of ls_blender: avg: \n{res}')
self.assertTrue(np.allclose(res, ls_blnd_avg))
# test E: the ls_blender 'combo'
self.op.set_blender('ls', 'combo')
res = qt.Operator._ls_blend(self.op, ls_masks)
print(f'Test E: result of ls_blender: combo: \n{res}')
self.assertTrue(np.allclose(res, ls_blnd_combo))
def test_sel_blend(self):
"""测试选股蒙板的混合器,包括所有的混合模式"""
# step2, test blending of sel masks
pass
def test_bs_blend(self):
"""测试买卖信号混合模式"""
# step3, test blending of op signals
pass
def test_unify(self):
print('Testing Unify functions\n')
l1 = np.array([[3, 2, 5], [5, 3, 2]])
res = qt.unify(l1)
target = np.array([[0.3, 0.2, 0.5], [0.5, 0.3, 0.2]])
self.assertIs(np.allclose(res, target), True, 'sum of all elements is 1')
l1 = np.array([[1, 1, 1, 1, 1], [2, 2, 2, 2, 2]])
res = qt.unify(l1)
target = np.array([[0.2, 0.2, 0.2, 0.2, 0.2], [0.2, 0.2, 0.2, 0.2, 0.2]])
self.assertIs(np.allclose(res, target), True, 'sum of all elements is 1')
def test_mask_to_signal(self):
signal = qt.mask_to_signal(self.mask)
print(f'Test A: single mask to signal, result: \n{signal}')
self.assertTrue(np.allclose(signal, self.correct_signal))
signal = qt.mask_to_signal(self.multi_mask)
print(f'Test A: single mask to signal, result: \n{signal}')
self.assertTrue(np.allclose(signal, self.correct_multi_signal))
class TestLSStrategy(qt.RollingTiming):
"""用于test测试的简单多空蒙板生成策略。基于RollingTiming滚动择时方法生成
该策略有两个参数,N与Price
N用于计算OHLC价格平均值的N日简单移动平均,判断,当移动平均值大于等于Price时,状态为看多,否则为看空
"""
def __init__(self):
super().__init__(stg_name='test_LS',
stg_text='test long/short strategy',
par_count=2,
par_types='discr, conti',
par_bounds_or_enums=([1, 5], [2, 10]),
data_types='close, open, high, low',
data_freq='d',
window_length=5)
pass
def _realize(self, hist_data: np.ndarray, params: tuple):
n, price = params
h = hist_data.T
avg = (h[0] + h[1] + h[2] + h[3]) / 4
ma = sma(avg, n)
if ma[-1] < price:
return 0
else:
return 1
class TestSelStrategy(qt.SimpleSelecting):
"""用于Test测试的简单选股策略,基于Selecting策略生成
策略没有参数,选股周期为5D
在每个选股周期内,从股票池的三只股票中选出今日变化率 = (今收-昨收)/平均股价(OHLC平均股价)最高的两支,放入中选池,否则落选。
选股比例为平均分配
"""
def __init__(self):
super().__init__(stg_name='test_SEL',
stg_text='test portfolio selection strategy',
par_count=0,
par_types='',
par_bounds_or_enums=(),
data_types='high, low, close',
data_freq='d',
sample_freq='10d',
window_length=5)
pass
def _realize(self, hist_data: np.ndarray, params: tuple):
avg = np.nanmean(hist_data, axis=(1, 2))
dif = (hist_data[:, :, 2] - np.roll(hist_data[:, :, 2], 1, 1))
dif_no_nan = np.array([arr[~np.isnan(arr)][-1] for arr in dif])
difper = dif_no_nan / avg
large2 = difper.argsort()[1:]
chosen = np.zeros_like(avg)
chosen[large2] = 0.5
return chosen
class TestSelStrategyDiffTime(qt.SimpleSelecting):
"""用于Test测试的简单选股策略,基于Selecting策略生成
策略没有参数,选股周期为5D
在每个选股周期内,从股票池的三只股票中选出今日变化率 = (今收-昨收)/平均股价(OHLC平均股价)最高的两支,放入中选池,否则落选。
选股比例为平均分配
"""
def __init__(self):
super().__init__(stg_name='test_SEL',
stg_text='test portfolio selection strategy',
par_count=0,
par_types='',
par_bounds_or_enums=(),
data_types='close, low, open',
data_freq='d',
sample_freq='w',
window_length=2)
pass
def _realize(self, hist_data: np.ndarray, params: tuple):
avg = hist_data.mean(axis=1).squeeze()
difper = (hist_data[:, :, 0] - np.roll(hist_data[:, :, 0], 1))[:, -1] / avg
large2 = difper.argsort()[0:2]
chosen = np.zeros_like(avg)
chosen[large2] = 0.5
return chosen
class TestSigStrategy(qt.SimpleTiming):
"""用于Test测试的简单信号生成策略,基于SimpleTiming策略生成
策略有三个参数,第一个参数为ratio,另外两个参数为price1以及price2
ratio是k线形状比例的阈值,定义为abs((C-O)/(H-L))。当这个比值小于ratio阈值时,判断该K线为十字交叉(其实还有丁字等多种情形,但这里做了
简化处理。
信号生成的规则如下:
1,当某个K线出现十字交叉,且昨收与今收之差大于price1时,买入信号
2,当某个K线出现十字交叉,且昨收与今收之差小于price2时,卖出信号
"""
def __init__(self):
super().__init__(stg_name='test_SIG',
stg_text='test signal creation strategy',
par_count=3,
par_types='conti, conti, conti',
par_bounds_or_enums=([2, 10], [0, 3], [0, 3]),
data_types='close, open, high, low',
window_length=2)
pass
def _realize(self, hist_data: np.ndarray, params: tuple):
r, price1, price2 = params
h = hist_data.T
ratio = np.abs((h[0] - h[1]) / (h[3] - h[2]))
diff = h[0] - np.roll(h[0], 1)
sig = np.where((ratio < r) & (diff > price1),
1,
np.where((ratio < r) & (diff < price2), -1, 0))
return sig
class TestOperator(unittest.TestCase):
"""全面测试Operator对象的所有功能。包括:
1, Strategy 参数的设置
2, 历史数据的获取与分配提取
3, 策略优化参数的批量设置和优化空间的获取
4, 策略输出值的正确性验证
5, 策略结果的混合结果确认
"""
def setUp(self):
"""prepare data for Operator test"""
print('start testing HistoryPanel object\n')
# build up test data: a 4-type, 3-share, 50-day matrix of prices that contains nan values in some days
# for some share_pool
# for share1:
data_rows = 50
share1_close = [10.04, 10, 10, 9.99, 9.97, 9.99, 10.03, 10.03, 10.06, 10.06, 10.11,
10.09, 10.07, 10.06, 10.09, 10.03, 10.03, 10.06, 10.08, 10, 9.99,
10.03, 10.03, 10.06, 10.03, 9.97, 9.94, 9.83, 9.77, 9.84, 9.91, 9.93,
9.96, 9.91, 9.91, 9.88, 9.91, 9.64, 9.56, 9.57, 9.55, 9.57, 9.61, 9.61,
9.55, 9.57, 9.63, 9.64, 9.65, 9.62]
share1_open = [10.02, 10, 9.98, 9.97, 9.99, 10.01, 10.04, 10.06, 10.06, 10.11,
10.11, 10.07, 10.06, 10.09, 10.03, 10.02, 10.06, 10.08, 9.99, 10,
10.03, 10.02, 10.06, 10.03, 9.97, 9.94, 9.83, 9.78, 9.77, 9.91, 9.92,
9.97, 9.91, 9.9, 9.88, 9.91, 9.63, 9.64, 9.57, 9.55, 9.58, 9.61, 9.62,
9.55, 9.57, 9.61, 9.63, 9.64, 9.61, 9.56]
share1_high = [10.07, 10, 10, 10, 10.03, 10.03, 10.04, 10.09, 10.1, 10.14, 10.11, 10.1,
10.09, 10.09, 10.1, 10.05, 10.07, 10.09, 10.1, 10, 10.04, 10.04, 10.06,
10.09, 10.05, 9.97, 9.96, 9.86, 9.77, 9.92, 9.94, 9.97, 9.97, 9.92, 9.92,
9.92, 9.93, 9.64, 9.58, 9.6, 9.58, 9.62, 9.62, 9.64, 9.59, 9.62, 9.63,
9.7, 9.66, 9.64]
share1_low = [9.99, 10, 9.97, 9.97, 9.97, 9.98, 9.99, 10.03, 10.03, 10.04, 10.11, 10.07,
10.05, 10.03, 10.03, 10.01, 9.99, 10.03, 9.95, 10, 9.95, 10, 10.01, 9.99,
9.96, 9.89, 9.83, 9.77, 9.77, 9.8, 9.9, 9.91, 9.89, 9.89, 9.87, 9.85, 9.6,
9.64, 9.53, 9.55, 9.54, 9.55, 9.58, 9.54, 9.53, 9.53, 9.63, 9.64, 9.59, 9.56]
# for share2:
share2_close = [9.68, 9.87, 9.86, 9.87, 9.79, 9.82, 9.8, 9.66, 9.62, 9.58, 9.69, 9.78, 9.75,
9.96, 9.9, 10.04, 10.06, 10.08, 10.24, 10.24, 10.24, 9.86, 10.13, 10.12,
10.1, 10.25, 10.24, 10.22, 10.75, 10.64, 10.56, 10.6, 10.42, 10.25, 10.24,
10.49, 10.57, 10.63, 10.48, 10.37, 10.96, 11.02, np.nan, np.nan, 10.88, 10.87, 11.01,
11.01, 11.58, 11.8]
share2_open = [9.88, 9.88, 9.89, 9.75, 9.74, 9.8, 9.62, 9.65, 9.58, 9.67, 9.81, 9.8, 10,
9.95, 10.1, 10.06, 10.14, 9.9, 10.2, 10.29, 9.86, 9.48, 10.01, 10.24, 10.26,
10.24, 10.12, 10.65, 10.64, 10.56, 10.42, 10.43, 10.29, 10.3, 10.44, 10.6,
10.67, 10.46, 10.39, 10.9, 11.01, 11.01, np.nan, np.nan, 10.82, 11.02, 10.96,
11.55, 11.74, 11.8]
share2_high = [9.91, 10.04, 9.93, 10.04, 9.84, 9.88, 9.99, 9.7, 9.67, 9.71, 9.85, 9.9, 10,
10.2, 10.11, 10.18, 10.21, 10.26, 10.38, 10.47, 10.42, 10.07, 10.24, 10.27,
10.38, 10.43, 10.39, 10.65, 10.84, 10.65, 10.73, 10.63, 10.51, 10.35, 10.46,
10.63, 10.74, 10.76, 10.54, 11.02, 11.12, 11.17, np.nan, np.nan, 10.92, 11.15,
11.11, 11.55, 11.95, 11.93]
share2_low = [9.63, 9.84, 9.81, 9.74, 9.67, 9.72, 9.57, 9.54, 9.51, 9.47, 9.68, 9.63, 9.75,
9.65, 9.9, 9.93, 10.03, 9.8, 10.14, 10.09, 9.78, 9.21, 9.11, 9.68, 10.05,
10.12, 9.89, 9.89, 10.59, 10.43, 10.34, 10.32, 10.21, 10.2, 10.18, 10.36,
10.51, 10.41, 10.32, 10.37, 10.87, 10.95, np.nan, np.nan, 10.65, 10.71, 10.75,
10.91, 11.31, 11.58]
# for share3:
share3_close = [6.64, 7.26, 7.03, 6.87, np.nan, 6.64, 6.85, 6.7, 6.39, 6.22, 5.92, 5.91, 6.11,
5.91, 6.23, 6.28, 6.28, 6.27, np.nan, 5.56, 5.67, 5.16, 5.69, 6.32, 6.14, 6.25,
5.79, 5.26, 5.05, 5.45, 6.06, 6.21, 5.69, 5.46, 6.02, 6.69, 7.43, 7.72, 8.16,
7.83, 8.7, 8.71, 8.88, 8.54, 8.87, 8.87, 8.18, 7.8, 7.97, 8.25]
share3_open = [7.26, 7, 6.88, 6.91, np.nan, 6.81, 6.63, 6.45, 6.16, 6.24, 5.96, 5.97, 5.96,
6.2, 6.35, 6.11, 6.37, 5.58, np.nan, 5.65, 5.19, 5.42, 6.3, 6.15, 6.05, 5.89,
5.22, 5.2, 5.07, 6.04, 6.12, 5.85, 5.67, 6.02, 6.04, 7.07, 7.64, 7.99, 7.59,
8.73, 8.72, 8.97, 8.58, 8.71, 8.77, 8.4, 7.95, 7.76, 8.25, 7.51]
share3_high = [7.41, 7.31, 7.14, 7, np.nan, 6.82, 6.96, 6.85, 6.5, 6.34, 6.04, 6.02, 6.12, 6.38,
6.43, 6.46, 6.43, 6.27, np.nan, 6.01, 5.67, 5.67, 6.35, 6.32, 6.43, 6.36, 5.79,
5.47, 5.65, 6.04, 6.14, 6.23, 5.83, 6.25, 6.27, 7.12, 7.82, 8.14, 8.27, 8.92,
8.76, 9.15, 8.9, 9.01, 9.16, 9, 8.27, 7.99, 8.33, 8.25]
share3_low = [6.53, 6.87, 6.83, 6.7, np.nan, 6.63, 6.57, 6.41, 6.15, 6.07, 5.89, 5.82, 5.73, 5.81,
6.1, 6.06, 6.16, 5.57, np.nan, 5.51, 5.19, 5.12, 5.69, 6.01, 5.97, 5.86, 5.18, 5.19,
4.96, 5.45, 5.84, 5.85, 5.28, 5.42, 6.02, 6.69, 7.28, 7.64, 7.25, 7.83, 8.41, 8.66,
8.53, 8.54, 8.73, 8.27, 7.95, 7.67, 7.8, 7.51]
# for sel_finance test
shares_eps = np.array([[np.nan, np.nan, np.nan],
[0.1, np.nan, np.nan],
[np.nan, 0.2, np.nan],
[np.nan, np.nan, 0.3],
[np.nan, np.nan, np.nan],
[np.nan, np.nan, np.nan],
[np.nan, np.nan, np.nan],
[np.nan, np.nan, 0.2],
[0.1, np.nan, np.nan],
[np.nan, 0.3, np.nan],
[np.nan, np.nan, np.nan],
[np.nan, np.nan, np.nan],
[0.3, np.nan, np.nan],
[np.nan, np.nan, np.nan],
[np.nan, 0.3, np.nan],
[np.nan, np.nan, np.nan],
[np.nan, np.nan, 0.3],
[np.nan, np.nan, np.nan],
[np.nan, np.nan, np.nan],
[np.nan, np.nan, np.nan],
[np.nan, np.nan, np.nan],
[np.nan, 0, 0.2],
[np.nan, np.nan, np.nan],
[np.nan, np.nan, np.nan],
[0.1, np.nan, np.nan],
[np.nan, np.nan, np.nan],
[np.nan, np.nan, np.nan],
[np.nan, np.nan, np.nan],
[np.nan, np.nan, np.nan],
[np.nan, np.nan, 0.2],
[np.nan, np.nan, np.nan],
[np.nan, np.nan, np.nan],
[np.nan, np.nan, np.nan],
[0.15, np.nan, np.nan],
[np.nan, 0.1, np.nan],
[np.nan, np.nan, np.nan],
[0.1, np.nan, np.nan],
[np.nan, np.nan, np.nan],
[np.nan, np.nan, 0.3],
[np.nan, np.nan, np.nan],
[np.nan, np.nan, np.nan],
[np.nan, np.nan, np.nan],
[np.nan, np.nan, np.nan],
[0.2, np.nan, np.nan],
[np.nan, 0.5, np.nan],
[0.4, np.nan, 0.3],
[np.nan, np.nan, np.nan],
[np.nan, 0.3, np.nan],
[0.9, np.nan, np.nan],
[np.nan, np.nan, 0.1]])
self.date_indices = ['2016-07-01', '2016-07-04', '2016-07-05', '2016-07-06',
'2016-07-07', '2016-07-08', '2016-07-11', '2016-07-12',
'2016-07-13', '2016-07-14', '2016-07-15', '2016-07-18',
'2016-07-19', '2016-07-20', '2016-07-21', '2016-07-22',
'2016-07-25', '2016-07-26', '2016-07-27', '2016-07-28',
'2016-07-29', '2016-08-01', '2016-08-02', '2016-08-03',
'2016-08-04', '2016-08-05', '2016-08-08', '2016-08-09',
'2016-08-10', '2016-08-11', '2016-08-12', '2016-08-15',
'2016-08-16', '2016-08-17', '2016-08-18', '2016-08-19',
'2016-08-22', '2016-08-23', '2016-08-24', '2016-08-25',
'2016-08-26', '2016-08-29', '2016-08-30', '2016-08-31',
'2016-09-01', '2016-09-02', '2016-09-05', '2016-09-06',
'2016-09-07', '2016-09-08']
self.shares = ['000010', '000030', '000039']
self.types = ['close', 'open', 'high', 'low']
self.sel_finance_tyeps = ['eps']
self.test_data_3D = np.zeros((3, data_rows, 4))
self.test_data_2D = np.zeros((data_rows, 3))
self.test_data_2D2 = np.zeros((data_rows, 4))
self.test_data_sel_finance = np.empty((3, data_rows, 1))
# Build up 3D data
self.test_data_3D[0, :, 0] = share1_close
self.test_data_3D[0, :, 1] = share1_open
self.test_data_3D[0, :, 2] = share1_high
self.test_data_3D[0, :, 3] = share1_low
self.test_data_3D[1, :, 0] = share2_close
self.test_data_3D[1, :, 1] = share2_open
self.test_data_3D[1, :, 2] = share2_high
self.test_data_3D[1, :, 3] = share2_low
self.test_data_3D[2, :, 0] = share3_close
self.test_data_3D[2, :, 1] = share3_open
self.test_data_3D[2, :, 2] = share3_high
self.test_data_3D[2, :, 3] = share3_low
self.test_data_sel_finance[:, :, 0] = shares_eps.T
self.hp1 = qt.HistoryPanel(values=self.test_data_3D,
levels=self.shares,
columns=self.types,
rows=self.date_indices)
print(f'in test Operator, history panel is created for timing test')
self.hp1.info()
self.hp2 = qt.HistoryPanel(values=self.test_data_sel_finance,
levels=self.shares,
columns=self.sel_finance_tyeps,
rows=self.date_indices)
print(f'in test_Operator, history panel is created for selection finance test:')
self.hp2.info()
self.op = qt.Operator(selecting_types=['all'], timing_types='dma', ricon_types='urgent')
def test_info(self):
"""Test information output of Operator"""
print(f'test printing information of operator object')
# self.op.info()
def test_operator_ready(self):
"""test the method ready of Operator"""
pass
# print(f'operator is ready? "{self.op.ready}"')
def test_operator_add_strategy(self):
"""test adding strategies to Operator"""
pass
# self.assertIsInstance(self.op, qt.Operator)
# self.assertIsInstance(self.op.timing[0], qt.TimingDMA)
# self.assertIsInstance(self.op.selecting[0], qt.SelectingAll)
# self.assertIsInstance(self.op.ricon[0], qt.RiconUrgent)
# self.assertEqual(self.op.selecting_count, 1)
# self.assertEqual(self.op.strategy_count, 3)
# self.assertEqual(self.op.ricon_count, 1)
# self.assertEqual(self.op.timing_count, 1)
# self.assertEqual(self.op.ls_blender, 'pos-1')
# print(f'test adding strategies into existing op')
# print('test adding strategy by string')
# self.op.add_strategy('macd', 'timing')
# self.assertIsInstance(self.op.timing[0], qt.TimingDMA)
# self.assertIsInstance(self.op.timing[1], qt.TimingMACD)
# self.assertEqual(self.op.selecting_count, 1)
# self.assertEqual(self.op.strategy_count, 4)
# self.assertEqual(self.op.ricon_count, 1)
# self.assertEqual(self.op.timing_count, 2)
# self.assertEqual(self.op.ls_blender, 'pos-1')
# self.op.add_strategy('random', 'selecting')
# self.assertIsInstance(self.op.selecting[0], qt.TimingDMA)
# self.assertIsInstance(self.op.selecting[1], qt.TimingMACD)
# self.assertEqual(self.op.selecting_count, 2)
# self.assertEqual(self.op.strategy_count, 5)
# self.assertEqual(self.op.ricon_count, 1)
# self.assertEqual(self.op.timing_count, 2)
# self.assertEqual(self.op.selecting_blender, '0 or 1')
# self.op.add_strategy('none', 'ricon')
# self.assertIsInstance(self.op.ricon[0], qt.TimingDMA)
# self.assertIsInstance(self.op.ricon[1], qt.TimingMACD)
# self.assertEqual(self.op.selecting_count, 2)
# self.assertEqual(self.op.strategy_count, 6)
# self.assertEqual(self.op.ricon_count, 2)
# self.assertEqual(self.op.timing_count, 2)
# print('test adding strategy by list')
# self.op.add_strategy(['dma', 'macd'], 'timing')
# print('test adding strategy by object')
# test_ls = TestLSStrategy()
# self.op.add_strategy(test_ls, 'timing')
def test_operator_remove_strategy(self):
"""test removing strategies from Operator"""
pass
# self.op.remove_strategy(stg='macd')
def test_property_get(self):
self.assertIsInstance(self.op, qt.Operator)
self.assertIsInstance(self.op.timing[0], qt.TimingDMA)
self.assertIsInstance(self.op.selecting[0], qt.SelectingAll)
self.assertIsInstance(self.op.ricon[0], qt.RiconUrgent)
self.assertEqual(self.op.selecting_count, 1)
self.assertEqual(self.op.strategy_count, 3)
self.assertEqual(self.op.ricon_count, 1)
self.assertEqual(self.op.timing_count, 1)
print(self.op.strategies, '\n', [qt.TimingDMA, qt.SelectingAll, qt.RiconUrgent])
print(f'info of Timing strategy: \n{self.op.strategies[0].info()}')
self.assertEqual(len(self.op.strategies), 3)
self.assertIsInstance(self.op.strategies[0], qt.TimingDMA)
self.assertIsInstance(self.op.strategies[1], qt.SelectingAll)
self.assertIsInstance(self.op.strategies[2], qt.RiconUrgent)
self.assertEqual(self.op.strategy_count, 3)
self.assertEqual(self.op.op_data_freq, 'd')
self.assertEqual(self.op.op_data_types, ['close'])
self.assertEqual(self.op.opt_space_par, ([], []))
self.assertEqual(self.op.max_window_length, 270)
self.assertEqual(self.op.ls_blender, 'pos-1')
self.assertEqual(self.op.selecting_blender, '0')
self.assertEqual(self.op.ricon_blender, 'add')
self.assertEqual(self.op.opt_types, [0, 0, 0])
def test_prepare_data(self):
test_ls = TestLSStrategy()
test_sel = TestSelStrategy()
test_sig = TestSigStrategy()
self.op = qt.Operator(timing_types=[test_ls],
selecting_types=[test_sel],
ricon_types=[test_sig])
too_early_cash = qt.CashPlan(dates='2016-01-01', amounts=10000)
early_cash = qt.CashPlan(dates='2016-07-01', amounts=10000)
on_spot_cash = qt.CashPlan(dates='2016-07-08', amounts=10000)
no_trade_cash = qt.CashPlan(dates='2016-07-08, 2016-07-30, 2016-08-11, 2016-09-03',
amounts=[10000, 10000, 10000, 10000])
late_cash = qt.CashPlan(dates='2016-12-31', amounts=10000)
multi_cash = qt.CashPlan(dates='2016-07-08, 2016-08-08', amounts=[10000, 10000])
self.op.set_parameter(stg_id='t-0',
pars={'000300': (5, 10.),
'000400': (5, 10.),
'000500': (5, 6.)})
self.op.set_parameter(stg_id='s-0',
pars=())
self.op.set_parameter(stg_id='r-0',
pars=(0.2, 0.02, -0.02))
self.op.prepare_data(hist_data=self.hp1,
cash_plan=on_spot_cash)
self.assertIsInstance(self.op._selecting_history_data, list)
self.assertIsInstance(self.op._timing_history_data, list)
self.assertIsInstance(self.op._ricon_history_data, list)
self.assertEqual(len(self.op._selecting_history_data), 1)
self.assertEqual(len(self.op._timing_history_data), 1)
self.assertEqual(len(self.op._ricon_history_data), 1)
sel_hist_data = self.op._selecting_history_data[0]
tim_hist_data = self.op._timing_history_data[0]
ric_hist_data = self.op._ricon_history_data[0]
print(f'in test_prepare_data in TestOperator:')
print('selecting history data:\n', sel_hist_data)
print('originally passed data in correct sequence:\n', self.test_data_3D[:, 3:, [2, 3, 0]])
print('difference is \n', sel_hist_data - self.test_data_3D[:, :, [2, 3, 0]])
self.assertTrue(np.allclose(sel_hist_data, self.test_data_3D[:, :, [2, 3, 0]], equal_nan=True))
self.assertTrue(np.allclose(tim_hist_data, self.test_data_3D, equal_nan=True))
self.assertTrue(np.allclose(ric_hist_data, self.test_data_3D[:, 3:, :], equal_nan=True))
# raises Value Error if empty history panel is given
empty_hp = qt.HistoryPanel()
correct_hp = qt.HistoryPanel(values=np.random.randint(10, size=(3, 50, 4)),
columns=self.types,
levels=self.shares,
rows=self.date_indices)
too_many_shares = qt.HistoryPanel(values=np.random.randint(10, size=(5, 50, 4)))
too_many_types = qt.HistoryPanel(values=np.random.randint(10, size=(3, 50, 5)))
# raises Error when history panel is empty
self.assertRaises(ValueError,
self.op.prepare_data,
empty_hp,
on_spot_cash)
# raises Error when first investment date is too early
self.assertRaises(AssertionError,
self.op.prepare_data,
correct_hp,
early_cash)
# raises Error when last investment date is too late
self.assertRaises(AssertionError,
self.op.prepare_data,
correct_hp,
late_cash)
# raises Error when some of the investment dates are on no-trade-days
self.assertRaises(ValueError,
self.op.prepare_data,
correct_hp,
no_trade_cash)
# raises Error when number of shares in history data does not fit
self.assertRaises(AssertionError,
self.op.prepare_data,
too_many_shares,
on_spot_cash)
# raises Error when too early cash investment date
self.assertRaises(AssertionError,
self.op.prepare_data,
correct_hp,
too_early_cash)
# raises Error when number of d_types in history data does not fit
self.assertRaises(AssertionError,
self.op.prepare_data,
too_many_types,
on_spot_cash)
# test the effect of data type sequence in strategy definition
def test_operator_generate(self):
"""
:return:
"""
test_ls = TestLSStrategy()
test_sel = TestSelStrategy()
test_sig = TestSigStrategy()
self.op = qt.Operator(timing_types=[test_ls],
selecting_types=[test_sel],
ricon_types=[test_sig])
self.assertIsInstance(self.op, qt.Operator, 'Operator Creation Error')
self.op.set_parameter(stg_id='t-0',
pars={'000300': (5, 10.),
'000400': (5, 10.),
'000500': (5, 6.)})
self.op.set_parameter(stg_id='s-0',
pars=())
# 在所有策略的参数都设置好之前调用prepare_data会发生assertion Error
self.assertRaises(AssertionError,
self.op.prepare_data,
hist_data=self.hp1,
cash_plan=qt.CashPlan(dates='2016-07-08', amounts=10000))
self.op.set_parameter(stg_id='r-0',
pars=(0.2, 0.02, -0.02))
self.op.prepare_data(hist_data=self.hp1,
cash_plan=qt.CashPlan(dates='2016-07-08', amounts=10000))
self.op.info()
op_list = self.op.create_signal(hist_data=self.hp1)
print(f'operation list is created: as following:\n {op_list}')
self.assertTrue(isinstance(op_list, pd.DataFrame))
self.assertEqual(op_list.shape, (26, 3))
# 删除去掉重复信号的code后,信号从原来的23条变为26条,包含三条重复信号,但是删除重复信号可能导致将不应该删除的信号删除,详见
# operator.py的create_signal()函数注释836行
target_op_dates = ['2016/07/08', '2016/07/12', '2016/07/13', '2016/07/14',
'2016/07/18', '2016/07/20', '2016/07/22', '2016/07/26',
'2016/07/27', '2016/07/28', '2016/08/02', '2016/08/03',
'2016/08/04', '2016/08/05', '2016/08/08', '2016/08/10',
'2016/08/16', '2016/08/18', '2016/08/24', '2016/08/26',
'2016/08/29', '2016/08/30', '2016/08/31', '2016/09/05',
'2016/09/06', '2016/09/08']
target_op_values = np.array([[0.0, 1.0, 0.0],
[0.5, -1.0, 0.0],
[1.0, 0.0, 0.0],
[0.0, 0.0, -1.0],
[0.0, 1.0, 0.0],
[0.0, 1.0, 0.0],
[0.0, 1.0, 0.0],
[0.0, 0.5, 0.0],
[0.0, 1.0, 0.0],
[0.0, 0.0, -1.0],
[0.0, 1.0, 0.0],
[0.0, -1.0, 0.0],
[0.0, 0.0, -1.0],
[0.0, 1.0, 0.0],
[-1.0, 0.0, 0.0],
[0.0, 0.0, -1.0],
[0.0, 0.0, -1.0],
[0.0, 0.0, 1.0],
[-1.0, 0.0, 0.0],
[0.0, 1.0, 1.0],
[0.0, 1.0, 0.5],
[0.0, 1.0, 0.0],
[0.0, 1.0, 0.0],
[0.0, 1.0, 0.0],
[0.0, 0.0, -1.0],
[0.0, 1.0, 0.0]])
target_op = pd.DataFrame(data=target_op_values, index=target_op_dates, columns=['000010', '000030', '000039'])
target_op = target_op.rename(index=pd.Timestamp)
print(f'target operation list is as following:\n {target_op}')
dates_pairs = [[date1, date2, date1 == date2]
for date1, date2
in zip(target_op.index.strftime('%m-%d'), op_list.index.strftime('%m-%d'))]
signal_pairs = [[list(sig1), list(sig2), all(sig1 == sig2)]
for sig1, sig2
in zip(list(target_op.values), list(op_list.values))]
print(f'dates side by side:\n '
f'{dates_pairs}')
print(f'signals side by side:\n'
f'{signal_pairs}')
print([item[2] for item in dates_pairs])
print([item[2] for item in signal_pairs])
self.assertTrue(np.allclose(target_op.values, op_list.values, equal_nan=True))
self.assertTrue(all([date1 == date2
for date1, date2
in zip(target_op.index.strftime('%m-%d'), op_list.index.strftime('%m-%d'))]))
def test_operator_parameter_setting(self):
"""
:return:
"""
new_op = qt.Operator(selecting_types=['all'], timing_types='dma', ricon_types='urgent')
print(new_op.strategies, '\n', [qt.TimingDMA, qt.SelectingAll, qt.RiconUrgent])
print(f'info of Timing strategy in new op: \n{new_op.strategies[0].info()}')
self.op.set_parameter('t-0',
pars=(5, 10, 5),
opt_tag=1,
par_boes=((5, 10), (5, 15), (10, 15)),
window_length=10,
data_types=['close', 'open', 'high'])
self.op.set_parameter(stg_id='s-0',
pars=None,
opt_tag=1,
sample_freq='10d',
window_length=10,
data_types='close, open')
self.op.set_parameter(stg_id='r-0',
pars=None,
opt_tag=0,
sample_freq='d',
window_length=20,
data_types='close, open')
self.assertEqual(self.op.timing[0].pars, (5, 10, 5))
self.assertEqual(self.op.timing[0].par_boes, ((5, 10), (5, 15), (10, 15)))
self.assertEqual(self.op.op_data_freq, 'd')
self.assertEqual(self.op.op_data_types, ['close', 'high', 'open'])
self.assertEqual(self.op.opt_space_par,
([(5, 10), (5, 15), (10, 15), (0, 1)], ['discr', 'discr', 'discr', 'conti']))
self.assertEqual(self.op.max_window_length, 20)
self.assertRaises(AssertionError, self.op.set_parameter, stg_id='t-1', pars=(1, 2))
self.assertRaises(AssertionError, self.op.set_parameter, stg_id='t1', pars=(1, 2))
self.assertRaises(AssertionError, self.op.set_parameter, stg_id=32, pars=(1, 2))
self.op.set_blender('selecting', '0 and 1 or 2')
self.op.set_blender('ls', 'str-1.2')
self.assertEqual(self.op.ls_blender, 'str-1.2')
self.assertEqual(self.op.selecting_blender, '0 and 1 or 2')
self.assertEqual(self.op.selecting_blender_expr, ['or', 'and', '0', '1', '2'])
self.assertEqual(self.op.ricon_blender, 'add')
self.assertRaises(ValueError, self.op.set_blender, 'select', '0and1')
self.assertRaises(TypeError, self.op.set_blender, 35, '0 and 1')
self.assertEqual(self.op.opt_space_par,
([(5, 10), (5, 15), (10, 15), (0, 1)], ['discr', 'discr', 'discr', 'conti']))
self.assertEqual(self.op.opt_types, [1, 1, 0])
def test_exp_to_blender(self):
self.op.set_blender('selecting', '0 and 1 or 2')
self.assertEqual(self.op.selecting_blender_expr, ['or', 'and', '0', '1', '2'])
self.op.set_blender('selecting', '0 and ( 1 or 2 )')
self.assertEqual(self.op.selecting_blender_expr, ['and', '0', 'or', '1', '2'])
self.assertRaises(ValueError, self.op.set_blender, 'selecting', '0 and (1 or 2)')
def test_set_opt_par(self):
self.op.set_parameter('t-0',
pars=(5, 10, 5),
opt_tag=1,
par_boes=((5, 10), (5, 15), (10, 15)),
window_length=10,
data_types=['close', 'open', 'high'])
self.op.set_parameter(stg_id='s-0',
pars=(0.5,),
opt_tag=0,
sample_freq='10d',
window_length=10,
data_types='close, open')
self.op.set_parameter(stg_id='r-0',
pars=(9, -0.23),
opt_tag=1,
sample_freq='d',
window_length=20,
data_types='close, open')
self.assertEqual(self.op.timing[0].pars, (5, 10, 5))
self.assertEqual(self.op.selecting[0].pars, (0.5,))
self.assertEqual(self.op.ricon[0].pars, (9, -0.23))
self.assertEqual(self.op.opt_types, [1, 0, 1])
self.op.set_opt_par((5, 12, 9, 8, -0.1))
self.assertEqual(self.op.timing[0].pars, (5, 12, 9))
self.assertEqual(self.op.selecting[0].pars, (0.5,))
self.assertEqual(self.op.ricon[0].pars, (8, -0.1))
# test set_opt_par when opt_tag is set to be 2 (enumerate type of parameters)
self.assertRaises(ValueError, self.op.set_opt_par, (5, 12, 9, 8))
def test_stg_attribute_get_and_set(self):
self.stg = qt.TimingCrossline()
self.stg_type = 'TIMING'
self.stg_name = "CROSSLINE STRATEGY"
self.stg_text = 'Moving average crossline strategy, determine long/short position according to the cross ' \
'point' \
' of long and short term moving average prices '
self.pars = (35, 120, 10, 'buy')
self.par_boes = [(10, 250), (10, 250), (1, 100), ('buy', 'sell', 'none')]
self.par_count = 4
self.par_types = ['discr', 'discr', 'conti', 'enum']
self.opt_tag = 0
self.data_types = ['close']
self.data_freq = 'd'
self.sample_freq = 'd'
self.window_length = 270
self.assertEqual(self.stg.stg_type, self.stg_type)
self.assertEqual(self.stg.stg_name, self.stg_name)
self.assertEqual(self.stg.stg_text, self.stg_text)
self.assertEqual(self.stg.pars, self.pars)
self.assertEqual(self.stg.par_types, self.par_types)
self.assertEqual(self.stg.par_boes, self.par_boes)
self.assertEqual(self.stg.par_count, self.par_count)
self.assertEqual(self.stg.opt_tag, self.opt_tag)
self.assertEqual(self.stg.data_freq, self.data_freq)
self.assertEqual(self.stg.sample_freq, self.sample_freq)
self.assertEqual(self.stg.data_types, self.data_types)
self.assertEqual(self.stg.window_length, self.window_length)
self.stg.stg_name = 'NEW NAME'
self.stg.stg_text = 'NEW TEXT'
self.assertEqual(self.stg.stg_name, 'NEW NAME')
self.assertEqual(self.stg.stg_text, 'NEW TEXT')
self.stg.pars = (1, 2, 3, 4)
self.assertEqual(self.stg.pars, (1, 2, 3, 4))
self.stg.par_count = 3
self.assertEqual(self.stg.par_count, 3)
self.stg.par_boes = [(1, 10), (1, 10), (1, 10), (1, 10)]
self.assertEqual(self.stg.par_boes, [(1, 10), (1, 10), (1, 10), (1, 10)])
self.stg.par_types = ['conti', 'conti', 'discr', 'enum']
self.assertEqual(self.stg.par_types, ['conti', 'conti', 'discr', 'enum'])
self.stg.par_types = 'conti, conti, discr, conti'
self.assertEqual(self.stg.par_types, ['conti', 'conti', 'discr', 'conti'])
self.stg.data_types = 'close, open'
self.assertEqual(self.stg.data_types, ['close', 'open'])
self.stg.data_types = ['close', 'high', 'low']
self.assertEqual(self.stg.data_types, ['close', 'high', 'low'])
self.stg.data_freq = 'w'
self.assertEqual(self.stg.data_freq, 'w')
self.stg.window_length = 300
self.assertEqual(self.stg.window_length, 300)
def test_rolling_timing(self):
stg = TestLSStrategy()
stg_pars = {'000100': (5, 10),
'000200': (5, 10),
'000300': (5, 6)}
stg.set_pars(stg_pars)
history_data = self.hp1.values
output = stg.generate(hist_data=history_data)
self.assertIsInstance(output, np.ndarray)
self.assertEqual(output.shape, (45, 3))
lsmask = np.array([[0., 0., 1.],
[0., 0., 1.],
[1., 0., 1.],
[1., 0., 1.],
[1., 0., 1.],
[1., 0., 1.],
[1., 0., 1.],
[1., 0., 1.],
[1., 0., 1.],
[1., 0., 1.],
[1., 0., 1.],
[1., 0., 1.],
[1., 1., 1.],
[1., 1., 1.],
[1., 1., 1.],
[1., 1., 0.],
[1., 1., 0.],
[1., 1., 0.],
[1., 0., 0.],
[1., 0., 0.],
[1., 1., 0.],
[0., 1., 0.],
[0., 1., 0.],
[0., 1., 0.],
[0., 1., 0.],
[0., 1., 0.],
[0., 1., 0.],
[0., 1., 0.],
[0., 1., 0.],
[0., 1., 0.],
[0., 1., 1.],
[0., 1., 1.],
[0., 1., 1.],
[0., 1., 1.],
[0., 1., 1.],
[0., 1., 1.],
[0., 1., 1.],
[0., 1., 1.],
[0., 1., 1.],
[0., 1., 1.],
[0., 1., 1.],
[0., 1., 1.],
[0., 1., 1.],
[0., 1., 1.],
[0., 1., 1.]])
# TODO: Issue to be solved: the np.nan value are converted to 0 in the lsmask,这样做可能会有意想不到的后果
# TODO: 需要解决nan值的问题
self.assertEqual(output.shape, lsmask.shape)
self.assertTrue(np.allclose(output, lsmask, equal_nan=True))
def test_sel_timing(self):
stg = TestSelStrategy()
stg_pars = ()
stg.set_pars(stg_pars)
history_data = self.hp1['high, low, close', :, :]
seg_pos, seg_length, seg_count = stg._seg_periods(dates=self.hp1.hdates, freq=stg.sample_freq)
self.assertEqual(list(seg_pos), [0, 5, 11, 19, 26, 33, 41, 47, 49])
self.assertEqual(list(seg_length), [5, 6, 8, 7, 7, 8, 6, 2])
self.assertEqual(seg_count, 8)
output = stg.generate(hist_data=history_data, shares=self.hp1.shares, dates=self.hp1.hdates)
self.assertIsInstance(output, np.ndarray)
self.assertEqual(output.shape, (45, 3))
selmask = np.array([[0.5, 0.5, 0.0],
[0.5, 0.5, 0.0],
[0.5, 0.5, 0.0],
[0.5, 0.5, 0.0],
[0.5, 0.5, 0.0],
[0.5, 0.5, 0.0],
[0.5, 0.5, 0.0],
[0.5, 0.5, 0.0],
[0.5, 0.5, 0.0],
[0.5, 0.5, 0.0],
[0.5, 0.5, 0.0],
[0.5, 0.5, 0.0],
[0.5, 0.5, 0.0],
[0.5, 0.5, 0.0],
[0.5, 0.5, 0.0],
[0.5, 0.5, 0.0],
[0.5, 0.5, 0.0],
[0.5, 0.5, 0.0],
[0.5, 0.5, 0.0],
[0.5, 0.5, 0.0],
[0.5, 0.5, 0.0],
[0.0, 0.5, 0.5],
[0.0, 0.5, 0.5],
[0.0, 0.5, 0.5],
[0.0, 0.5, 0.5],
[0.0, 0.5, 0.5],
[0.0, 0.5, 0.5],
[0.0, 0.5, 0.5],
[0.5, 0.5, 0.0],
[0.5, 0.5, 0.0],
[0.5, 0.5, 0.0],
[0.5, 0.5, 0.0],
[0.5, 0.5, 0.0],
[0.5, 0.5, 0.0],
[0.5, 0.5, 0.0],
[0.5, 0.5, 0.0],
[0.0, 0.5, 0.5],
[0.0, 0.5, 0.5],
[0.0, 0.5, 0.5],
[0.0, 0.5, 0.5],
[0.0, 0.5, 0.5],
[0.0, 0.5, 0.5],
[0.5, 0.5, 0.0],
[0.5, 0.5, 0.0],
[0.5, 0.5, 0.0]])
self.assertEqual(output.shape, selmask.shape)
self.assertTrue(np.allclose(output, selmask))
def test_simple_timing(self):
stg = TestSigStrategy()
stg_pars = (0.2, 0.02, -0.02)
stg.set_pars(stg_pars)
history_data = self.hp1['close, open, high, low', :, 3:50]
output = stg.generate(hist_data=history_data, shares=self.shares, dates=self.date_indices)
self.assertIsInstance(output, np.ndarray)
self.assertEqual(output.shape, (45, 3))
sigmatrix = np.array([[0.0, 1.0, 0.0],
[0.0, 0.0, 0.0],
[0.0, -1.0, 0.0],
[1.0, 0.0, 0.0],
[0.0, 0.0, -1.0],
[0.0, 0.0, 0.0],
[0.0, 1.0, 0.0],
[0.0, 0.0, 0.0],
[0.0, 1.0, 0.0],
[0.0, 0.0, 0.0],
[0.0, 1.0, 0.0],
[0.0, 0.0, 0.0],
[0.0, 0.0, 0.0],
[0.0, 1.0, 0.0],
[0.0, 0.0, -1.0],
[0.0, 0.0, 0.0],
[0.0, 0.0, 0.0],
[0.0, 1.0, 0.0],
[0.0, 0.0, 0.0],
[0.0, 0.0, -1.0],
[0.0, 1.0, 0.0],
[0.0, 0.0, 0.0],
[0.0, 0.0, 0.0],
[0.0, 0.0, -1.0],
[0.0, 0.0, 0.0],
[0.0, 0.0, 0.0],
[0.0, 0.0, 0.0],
[0.0, 0.0, -1.0],
[0.0, 0.0, 0.0],
[0.0, 0.0, 1.0],
[0.0, 0.0, 0.0],
[0.0, 0.0, 0.0],
[0.0, 0.0, 0.0],
[-1.0, 0.0, 0.0],
[0.0, 0.0, 0.0],
[0.0, 1.0, 1.0],
[0.0, 1.0, 0.0],
[0.0, 1.0, 0.0],
[0.0, 1.0, 0.0],
[0.0, 0.0, 0.0],
[0.0, 0.0, 0.0],
[0.0, 1.0, 0.0],
[0.0, 0.0, -1.0],
[0.0, 0.0, 0.0],
[0.0, 1.0, 0.0]])
side_by_side_array = np.array([[i, out_line, sig_line]
for
i, out_line, sig_line
in zip(range(len(output)), output, sigmatrix)])
print(f'output and signal matrix lined up side by side is \n'
f'{side_by_side_array}')
self.assertEqual(sigmatrix.shape, output.shape)
self.assertTrue(np.allclose(output, sigmatrix))
def test_sel_finance(self):
"""Test selecting_finance strategy, test all built-in strategy parameters"""
stg = SelectingFinanceIndicator()
stg_pars = (False, 'even', 'greater', 0, 0, 0.67)
stg.set_pars(stg_pars)
stg.window_length = 5
stg.data_freq = 'd'
stg.sample_freq = '10d'
stg.sort_ascending = False
stg.condition = 'greater'
stg.lbound = 0
stg.ubound = 0
stg._poq = 0.67
history_data = self.hp2.values
print(f'Start to test financial selection parameter {stg_pars}')
seg_pos, seg_length, seg_count = stg._seg_periods(dates=self.hp1.hdates, freq=stg.sample_freq)
self.assertEqual(list(seg_pos), [0, 5, 11, 19, 26, 33, 41, 47, 49])
self.assertEqual(list(seg_length), [5, 6, 8, 7, 7, 8, 6, 2])
self.assertEqual(seg_count, 8)
output = stg.generate(hist_data=history_data, shares=self.hp1.shares, dates=self.hp1.hdates)
self.assertIsInstance(output, np.ndarray)
self.assertEqual(output.shape, (45, 3))
selmask = np.array([[0.0, 0.5, 0.5],
[0.0, 0.5, 0.5],
[0.0, 0.5, 0.5],
[0.0, 0.5, 0.5],
[0.0, 0.5, 0.5],
[0.0, 0.5, 0.5],
[0.0, 0.5, 0.5],
[0.0, 0.5, 0.5],
[0.0, 0.5, 0.5],
[0.0, 0.5, 0.5],
[0.0, 0.5, 0.5],
[0.0, 0.5, 0.5],
[0.0, 0.5, 0.5],
[0.0, 0.5, 0.5],
[0.0, 0.5, 0.5],
[0.0, 0.5, 0.5],
[0.0, 0.5, 0.5],
[0.0, 0.5, 0.5],
[0.0, 0.5, 0.5],
[0.0, 0.5, 0.5],
[0.0, 0.5, 0.5],
[0.5, 0.0, 0.5],
[0.5, 0.0, 0.5],
[0.5, 0.0, 0.5],
[0.5, 0.0, 0.5],
[0.5, 0.0, 0.5],
[0.5, 0.0, 0.5],
[0.5, 0.0, 0.5],
[0.0, 0.0, 1.0],
[0.0, 0.0, 1.0],
[0.0, 0.0, 1.0],
[0.0, 0.0, 1.0],
[0.0, 0.0, 1.0],
[0.0, 0.0, 1.0],
[0.0, 0.0, 1.0],
[0.0, 0.0, 1.0],
[0.5, 0.0, 0.5],
[0.5, 0.0, 0.5],
[0.5, 0.0, 0.5],
[0.5, 0.0, 0.5],
[0.5, 0.0, 0.5],
[0.5, 0.0, 0.5],
[0.5, 0.5, 0.0],
[0.5, 0.5, 0.0],
[0.5, 0.5, 0.0]])
self.assertEqual(output.shape, selmask.shape)
self.assertTrue(np.allclose(output, selmask))
# test single factor, get mininum factor
stg_pars = (True, 'even', 'less', 1, 1, 0.67)
stg.sort_ascending = True
stg.condition = 'less'
stg.lbound = 1
stg.ubound = 1
stg.set_pars(stg_pars)
print(f'Start to test financial selection parameter {stg_pars}')
output = stg.generate(hist_data=history_data, shares=self.hp1.shares, dates=self.hp1.hdates)
selmask = np.array([[0.5, 0.5, 0.0],
[0.5, 0.5, 0.0],
[0.5, 0.5, 0.0],
[0.5, 0.5, 0.0],
[0.5, 0.5, 0.0],
[0.5, 0.5, 0.0],
[0.5, 0.0, 0.5],
[0.5, 0.0, 0.5],
[0.5, 0.0, 0.5],
[0.5, 0.0, 0.5],
[0.5, 0.0, 0.5],
[0.5, 0.0, 0.5],
[0.5, 0.0, 0.5],
[0.5, 0.0, 0.5],
[0.5, 0.5, 0.0],
[0.5, 0.5, 0.0],
[0.5, 0.5, 0.0],
[0.5, 0.5, 0.0],
[0.5, 0.5, 0.0],
[0.5, 0.5, 0.0],
[0.5, 0.5, 0.0],
[0.5, 0.5, 0.0],
[0.5, 0.5, 0.0],
[0.5, 0.5, 0.0],
[0.5, 0.5, 0.0],
[0.5, 0.5, 0.0],
[0.5, 0.5, 0.0],
[0.5, 0.5, 0.0],
[0.0, 0.0, 1.0],
[0.0, 0.0, 1.0],
[0.0, 0.0, 1.0],
[0.0, 0.0, 1.0],
[0.0, 0.0, 1.0],
[0.0, 0.0, 1.0],
[0.0, 0.0, 1.0],
[0.0, 0.0, 1.0],
[0.5, 0.5, 0.0],
[0.5, 0.5, 0.0],
[0.5, 0.5, 0.0],
[0.5, 0.5, 0.0],
[0.5, 0.5, 0.0],
[0.5, 0.5, 0.0],
[0.5, 0.0, 0.5],
[0.5, 0.0, 0.5],
[0.5, 0.0, 0.5]])
self.assertEqual(output.shape, selmask.shape)
self.assertTrue(np.allclose(output, selmask))
# test single factor, get max factor in linear weight
stg_pars = (False, 'linear', 'greater', 0, 0, 0.67)
stg.sort_ascending = False
stg.weighting = 'linear'
stg.condition = 'greater'
stg.lbound = 0
stg.ubound = 0
stg.set_pars(stg_pars)
print(f'Start to test financial selection parameter {stg_pars}')
output = stg.generate(hist_data=history_data, shares=self.hp1.shares, dates=self.hp1.hdates)
selmask = np.array([[0.00000, 0.33333, 0.66667],
[0.00000, 0.33333, 0.66667],
[0.00000, 0.33333, 0.66667],
[0.00000, 0.33333, 0.66667],
[0.00000, 0.33333, 0.66667],
[0.00000, 0.33333, 0.66667],
[0.00000, 0.66667, 0.33333],
[0.00000, 0.66667, 0.33333],
[0.00000, 0.66667, 0.33333],
[0.00000, 0.66667, 0.33333],
[0.00000, 0.66667, 0.33333],
[0.00000, 0.66667, 0.33333],
[0.00000, 0.66667, 0.33333],
[0.00000, 0.66667, 0.33333],
[0.00000, 0.33333, 0.66667],
[0.00000, 0.33333, 0.66667],
[0.00000, 0.33333, 0.66667],
[0.00000, 0.33333, 0.66667],
[0.00000, 0.33333, 0.66667],
[0.00000, 0.33333, 0.66667],
[0.00000, 0.33333, 0.66667],
[0.33333, 0.00000, 0.66667],
[0.33333, 0.00000, 0.66667],
[0.33333, 0.00000, 0.66667],
[0.33333, 0.00000, 0.66667],
[0.33333, 0.00000, 0.66667],
[0.33333, 0.00000, 0.66667],
[0.33333, 0.00000, 0.66667],
[0.00000, 0.00000, 1.00000],
[0.00000, 0.00000, 1.00000],
[0.00000, 0.00000, 1.00000],
[0.00000, 0.00000, 1.00000],
[0.00000, 0.00000, 1.00000],
[0.00000, 0.00000, 1.00000],
[0.00000, 0.00000, 1.00000],
[0.00000, 0.00000, 1.00000],
[0.33333, 0.00000, 0.66667],
[0.33333, 0.00000, 0.66667],
[0.33333, 0.00000, 0.66667],
[0.33333, 0.00000, 0.66667],
[0.33333, 0.00000, 0.66667],
[0.33333, 0.00000, 0.66667],
[0.33333, 0.66667, 0.00000],
[0.33333, 0.66667, 0.00000],
[0.33333, 0.66667, 0.00000]])
self.assertEqual(output.shape, selmask.shape)
self.assertTrue(np.allclose(output, selmask))
# test single factor, get max factor in linear weight
stg_pars = (False, 'proportion', 'greater', 0, 0, 0.67)
stg.sort_ascending = False
stg.weighting = 'proportion'
stg.condition = 'greater'
stg.lbound = 0
stg.ubound = 0
stg.set_pars(stg_pars)
print(f'Start to test financial selection parameter {stg_pars}')
output = stg.generate(hist_data=history_data, shares=self.hp1.shares, dates=self.hp1.hdates)
selmask = np.array([[0.00000, 0.08333, 0.91667],
[0.00000, 0.08333, 0.91667],
[0.00000, 0.08333, 0.91667],
[0.00000, 0.08333, 0.91667],
[0.00000, 0.08333, 0.91667],
[0.00000, 0.08333, 0.91667],
[0.00000, 0.91667, 0.08333],
[0.00000, 0.91667, 0.08333],
[0.00000, 0.91667, 0.08333],
[0.00000, 0.91667, 0.08333],
[0.00000, 0.91667, 0.08333],
[0.00000, 0.91667, 0.08333],
[0.00000, 0.91667, 0.08333],
[0.00000, 0.91667, 0.08333],
[0.00000, 0.50000, 0.50000],
[0.00000, 0.50000, 0.50000],
[0.00000, 0.50000, 0.50000],
[0.00000, 0.50000, 0.50000],
[0.00000, 0.50000, 0.50000],
[0.00000, 0.50000, 0.50000],
[0.00000, 0.50000, 0.50000],
[0.08333, 0.00000, 0.91667],
[0.08333, 0.00000, 0.91667],
[0.08333, 0.00000, 0.91667],
[0.08333, 0.00000, 0.91667],
[0.08333, 0.00000, 0.91667],
[0.08333, 0.00000, 0.91667],
[0.08333, 0.00000, 0.91667],
[0.00000, 0.00000, 1.00000],
[0.00000, 0.00000, 1.00000],
[0.00000, 0.00000, 1.00000],
[0.00000, 0.00000, 1.00000],
[0.00000, 0.00000, 1.00000],
[0.00000, 0.00000, 1.00000],
[0.00000, 0.00000, 1.00000],
[0.00000, 0.00000, 1.00000],
[0.08333, 0.00000, 0.91667],
[0.08333, 0.00000, 0.91667],
[0.08333, 0.00000, 0.91667],
[0.08333, 0.00000, 0.91667],
[0.08333, 0.00000, 0.91667],
[0.08333, 0.00000, 0.91667],
[0.08333, 0.91667, 0.00000],
[0.08333, 0.91667, 0.00000],
[0.08333, 0.91667, 0.00000]])
self.assertEqual(output.shape, selmask.shape)
self.assertTrue(np.allclose(output, selmask, 0.001))
# test single factor, get max factor in linear weight, threshold 0.2
stg_pars = (False, 'even', 'greater', 0.2, 0.2, 0.67)
stg.sort_ascending = False
stg.weighting = 'even'
stg.condition = 'greater'
stg.lbound = 0.2
stg.ubound = 0.2
stg.set_pars(stg_pars)
print(f'Start to test financial selection parameter {stg_pars}')
output = stg.generate(hist_data=history_data, shares=self.hp1.shares, dates=self.hp1.hdates)
selmask = np.array([[0.0, 0.5, 0.5],
[0.0, 0.5, 0.5],
[0.0, 0.5, 0.5],
[0.0, 0.5, 0.5],
[0.0, 0.5, 0.5],
[0.0, 0.5, 0.5],
[0.0, 0.5, 0.5],
[0.0, 0.5, 0.5],
[0.0, 0.5, 0.5],
[0.0, 0.5, 0.5],
[0.0, 0.5, 0.5],
[0.0, 0.5, 0.5],
[0.0, 0.5, 0.5],
[0.0, 0.5, 0.5],
[0.0, 0.5, 0.5],
[0.0, 0.5, 0.5],
[0.0, 0.5, 0.5],
[0.0, 0.5, 0.5],
[0.0, 0.5, 0.5],
[0.0, 0.5, 0.5],
[0.0, 0.5, 0.5],
[0.0, 0.0, 1.0],
[0.0, 0.0, 1.0],
[0.0, 0.0, 1.0],
[0.0, 0.0, 1.0],
[0.0, 0.0, 1.0],
[0.0, 0.0, 1.0],
[0.0, 0.0, 1.0],
[0.0, 0.0, 1.0],
[0.0, 0.0, 1.0],
[0.0, 0.0, 1.0],
[0.0, 0.0, 1.0],
[0.0, 0.0, 1.0],
[0.0, 0.0, 1.0],
[0.0, 0.0, 1.0],
[0.0, 0.0, 1.0],
[0.0, 0.0, 1.0],
[0.0, 0.0, 1.0],
[0.0, 0.0, 1.0],
[0.0, 0.0, 1.0],
[0.0, 0.0, 1.0],
[0.0, 0.0, 1.0],
[0.5, 0.5, 0.0],
[0.5, 0.5, 0.0],
[0.5, 0.5, 0.0]])
self.assertEqual(output.shape, selmask.shape)
self.assertTrue(np.allclose(output, selmask, 0.001))
class TestLog(unittest.TestCase):
def test_init(self):
pass
class TestConfig(unittest.TestCase):
"""测试Config对象以及QT_CONFIG变量的设置和获取值"""
def test_init(self):
pass
def test_invest(self):
pass
def test_pars_string_to_type(self):
_parse_string_kwargs('000300', 'asset_pool', _valid_qt_kwargs())
class TestHistoryPanel(unittest.TestCase):
def setUp(self):
print('start testing HistoryPanel object\n')
self.data = np.random.randint(10, size=(5, 10, 4))
self.index = pd.date_range(start='20200101', freq='d', periods=10)
self.index2 = ['2016-07-01', '2016-07-04', '2016-07-05', '2016-07-06',
'2016-07-07', '2016-07-08', '2016-07-11', '2016-07-12',
'2016-07-13', '2016-07-14']
self.index3 = '2016-07-01, 2016-07-04, 2016-07-05, 2016-07-06, 2016-07-07, ' \
'2016-07-08, 2016-07-11, 2016-07-12, 2016-07-13, 2016-07-14'
self.shares = '000100,000101,000102,000103,000104'
self.htypes = 'close,open,high,low'
self.data2 = np.random.randint(10, size=(10, 5))
self.data3 = np.random.randint(10, size=(10, 4))
self.data4 = np.random.randint(10, size=(10))
self.hp = qt.HistoryPanel(values=self.data, levels=self.shares, columns=self.htypes, rows=self.index)
self.hp2 = qt.HistoryPanel(values=self.data2, levels=self.shares, columns='close', rows=self.index)
self.hp3 = qt.HistoryPanel(values=self.data3, levels='000100', columns=self.htypes, rows=self.index2)
self.hp4 = qt.HistoryPanel(values=self.data4, levels='000100', columns='close', rows=self.index3)
self.hp5 = qt.HistoryPanel(values=self.data)
self.hp6 = qt.HistoryPanel(values=self.data, levels=self.shares, rows=self.index3)
def test_properties(self):
""" test all properties of HistoryPanel
"""
self.assertFalse(self.hp.is_empty)
self.assertEqual(self.hp.row_count, 10)
self.assertEqual(self.hp.column_count, 4)
self.assertEqual(self.hp.level_count, 5)
self.assertEqual(self.hp.shape, (5, 10, 4))
self.assertSequenceEqual(self.hp.htypes, ['close', 'open', 'high', 'low'])
self.assertSequenceEqual(self.hp.shares, ['000100', '000101', '000102', '000103', '000104'])
self.assertSequenceEqual(list(self.hp.hdates), list(self.index))
self.assertDictEqual(self.hp.columns, {'close': 0, 'open': 1, 'high': 2, 'low': 3})
self.assertDictEqual(self.hp.levels, {'000100': 0, '000101': 1, '000102': 2, '000103': 3, '000104': 4})
row_dict = {Timestamp('2020-01-01 00:00:00', freq='D'): 0,
Timestamp('2020-01-02 00:00:00', freq='D'): 1,
Timestamp('2020-01-03 00:00:00', freq='D'): 2,
Timestamp('2020-01-04 00:00:00', freq='D'): 3,
Timestamp('2020-01-05 00:00:00', freq='D'): 4,
Timestamp('2020-01-06 00:00:00', freq='D'): 5,
Timestamp('2020-01-07 00:00:00', freq='D'): 6,
Timestamp('2020-01-08 00:00:00', freq='D'): 7,
Timestamp('2020-01-09 00:00:00', freq='D'): 8,
Timestamp('2020-01-10 00:00:00', freq='D'): 9}
self.assertDictEqual(self.hp.rows, row_dict)
def test_len(self):
""" test the function len(HistoryPanel)
:return:
"""
self.assertEqual(len(self.hp), 10)
def test_empty_history_panel(self):
"""测试空HP或者特殊HP如维度标签为纯数字的HP"""
test_hp = qt.HistoryPanel(self.data)
self.assertFalse(test_hp.is_empty)
self.assertIsInstance(test_hp, qt.HistoryPanel)
self.assertEqual(test_hp.shape[0], 5)
self.assertEqual(test_hp.shape[1], 10)
self.assertEqual(test_hp.shape[2], 4)
self.assertEqual(test_hp.level_count, 5)
self.assertEqual(test_hp.row_count, 10)
self.assertEqual(test_hp.column_count, 4)
self.assertEqual(test_hp.shares, list(range(5)))
self.assertEqual(test_hp.hdates, list(pd.date_range(start='20200730', periods=10, freq='d')))
self.assertEqual(test_hp.htypes, list(range(4)))
self.assertTrue(np.allclose(test_hp.values, self.data))
print(f'shares: {test_hp.shares}\nhtypes: {test_hp.htypes}')
print(test_hp)
empty_hp = qt.HistoryPanel()
self.assertTrue(empty_hp.is_empty)
self.assertIsInstance(empty_hp, qt.HistoryPanel)
self.assertEqual(empty_hp.shape[0], 0)
self.assertEqual(empty_hp.shape[1], 0)
self.assertEqual(empty_hp.shape[2], 0)
self.assertEqual(empty_hp.level_count, 0)
self.assertEqual(empty_hp.row_count, 0)
self.assertEqual(empty_hp.column_count, 0)
def test_create_history_panel(self):
""" test the creation of a HistoryPanel object by passing all data explicitly
"""
self.assertIsInstance(self.hp, qt.HistoryPanel)
self.assertEqual(self.hp.shape[0], 5)
self.assertEqual(self.hp.shape[1], 10)
self.assertEqual(self.hp.shape[2], 4)
self.assertEqual(self.hp.level_count, 5)
self.assertEqual(self.hp.row_count, 10)
self.assertEqual(self.hp.column_count, 4)
self.assertEqual(list(self.hp.levels.keys()), self.shares.split(','))
self.assertEqual(list(self.hp.columns.keys()), self.htypes.split(','))
self.assertEqual(list(self.hp.rows.keys())[0], pd.Timestamp('20200101'))
self.assertIsInstance(self.hp2, qt.HistoryPanel)
self.assertEqual(self.hp2.shape[0], 5)
self.assertEqual(self.hp2.shape[1], 10)
self.assertEqual(self.hp2.shape[2], 1)
self.assertEqual(self.hp2.level_count, 5)
self.assertEqual(self.hp2.row_count, 10)
self.assertEqual(self.hp2.column_count, 1)
self.assertEqual(list(self.hp2.levels.keys()), self.shares.split(','))
self.assertEqual(list(self.hp2.columns.keys()), ['close'])
self.assertEqual(list(self.hp2.rows.keys())[0], pd.Timestamp('20200101'))
self.assertIsInstance(self.hp3, qt.HistoryPanel)
self.assertEqual(self.hp3.shape[0], 1)
self.assertEqual(self.hp3.shape[1], 10)
self.assertEqual(self.hp3.shape[2], 4)
self.assertEqual(self.hp3.level_count, 1)
self.assertEqual(self.hp3.row_count, 10)
self.assertEqual(self.hp3.column_count, 4)
self.assertEqual(list(self.hp3.levels.keys()), ['000100'])
self.assertEqual(list(self.hp3.columns.keys()), self.htypes.split(','))
self.assertEqual(list(self.hp3.rows.keys())[0], pd.Timestamp('2016-07-01'))
self.assertIsInstance(self.hp4, qt.HistoryPanel)
self.assertEqual(self.hp4.shape[0], 1)
self.assertEqual(self.hp4.shape[1], 10)
self.assertEqual(self.hp4.shape[2], 1)
self.assertEqual(self.hp4.level_count, 1)
self.assertEqual(self.hp4.row_count, 10)
self.assertEqual(self.hp4.column_count, 1)
self.assertEqual(list(self.hp4.levels.keys()), ['000100'])
self.assertEqual(list(self.hp4.columns.keys()), ['close'])
self.assertEqual(list(self.hp4.rows.keys())[0], pd.Timestamp('2016-07-01'))
self.hp5.info()
self.assertIsInstance(self.hp5, qt.HistoryPanel)
self.assertTrue(np.allclose(self.hp5.values, self.data))
self.assertEqual(self.hp5.shape[0], 5)
self.assertEqual(self.hp5.shape[1], 10)
self.assertEqual(self.hp5.shape[2], 4)
self.assertEqual(self.hp5.level_count, 5)
self.assertEqual(self.hp5.row_count, 10)
self.assertEqual(self.hp5.column_count, 4)
self.assertEqual(list(self.hp5.levels.keys()), [0, 1, 2, 3, 4])
self.assertEqual(list(self.hp5.columns.keys()), [0, 1, 2, 3])
self.assertEqual(list(self.hp5.rows.keys())[0], pd.Timestamp('2020-07-30'))
self.hp6.info()
self.assertIsInstance(self.hp6, qt.HistoryPanel)
self.assertTrue(np.allclose(self.hp6.values, self.data))
self.assertEqual(self.hp6.shape[0], 5)
self.assertEqual(self.hp6.shape[1], 10)
self.assertEqual(self.hp6.shape[2], 4)
self.assertEqual(self.hp6.level_count, 5)
self.assertEqual(self.hp6.row_count, 10)
self.assertEqual(self.hp6.column_count, 4)
self.assertEqual(list(self.hp6.levels.keys()), ['000100', '000101', '000102', '000103', '000104'])
self.assertEqual(list(self.hp6.columns.keys()), [0, 1, 2, 3])
self.assertEqual(list(self.hp6.rows.keys())[0], pd.Timestamp('2016-07-01'))
# Error testing during HistoryPanel creating
# shape does not match
self.assertRaises(AssertionError,
qt.HistoryPanel,
self.data,
levels=self.shares, columns='close', rows=self.index)
# valus is not np.ndarray
self.assertRaises(AssertionError,
qt.HistoryPanel,
list(self.data))
# dimension/shape does not match
self.assertRaises(AssertionError,
qt.HistoryPanel,
self.data2,
levels='000100', columns=self.htypes, rows=self.index)
# value dimension over 3
self.assertRaises(AssertionError,
qt.HistoryPanel,
np.random.randint(10, size=(5, 10, 4, 2)))
# lebel value not valid
self.assertRaises(ValueError,
qt.HistoryPanel,
self.data2,
levels=self.shares, columns='close',
rows='a,b,c,d,e,f,g,h,i,j')
def test_history_panel_slicing(self):
"""测试HistoryPanel的各种切片方法
包括通过标签名称切片,通过数字切片,通过逗号分隔的标签名称切片,通过冒号分隔的标签名称切片等切片方式"""
self.assertTrue(np.allclose(self.hp['close'], self.data[:, :, 0:1]))
self.assertTrue(np.allclose(self.hp['close,open'], self.data[:, :, 0:2]))
self.assertTrue(np.allclose(self.hp[['close', 'open']], self.data[:, :, 0:2]))
self.assertTrue(np.allclose(self.hp['close:high'], self.data[:, :, 0:3]))
self.assertTrue(np.allclose(self.hp['close,high'], self.data[:, :, [0, 2]]))
self.assertTrue(np.allclose(self.hp[:, '000100'], self.data[0:1, :, ]))
self.assertTrue(np.allclose(self.hp[:, '000100,000101'], self.data[0:2, :]))
self.assertTrue(np.allclose(self.hp[:, ['000100', '000101']], self.data[0:2, :]))
self.assertTrue(np.allclose(self.hp[:, '000100:000102'], self.data[0:3, :]))
self.assertTrue(np.allclose(self.hp[:, '000100,000102'], self.data[[0, 2], :]))
self.assertTrue(np.allclose(self.hp['close,open', '000100,000102'], self.data[[0, 2], :, 0:2]))
print('start testing HistoryPanel')
data = np.random.randint(10, size=(10, 5))
# index = pd.date_range(start='20200101', freq='d', periods=10)
shares = '000100,000101,000102,000103,000104'
dtypes = 'close'
df = pd.DataFrame(data)
print('=========================\nTesting HistoryPanel creation from DataFrame')
hp = qt.dataframe_to_hp(df=df, shares=shares, htypes=dtypes)
hp.info()
hp = qt.dataframe_to_hp(df=df, shares='000100', htypes='close, open, high, low, middle', column_type='htypes')
hp.info()
print('=========================\nTesting HistoryPanel creation from initialization')
data = np.random.randint(10, size=(5, 10, 4)).astype('float')
index = pd.date_range(start='20200101', freq='d', periods=10)
dtypes = 'close, open, high,low'
data[0, [5, 6, 9], [0, 1, 3]] = np.nan
data[1:4, [4, 7, 6, 2], [1, 1, 3, 0]] = np.nan
data[4:5, [2, 9, 1, 2], [0, 3, 2, 1]] = np.nan
hp = qt.HistoryPanel(data, levels=shares, columns=dtypes, rows=index)
hp.info()
print('==========================\n输出close类型的所有历史数据\n')
self.assertTrue(np.allclose(hp['close', :, :], data[:, :, 0:1], equal_nan=True))
print(f'==========================\n输出close和open类型的所有历史数据\n')
self.assertTrue(np.allclose(hp[[0, 1], :, :], data[:, :, 0:2], equal_nan=True))
print(f'==========================\n输出第一只股票的所有类型历史数据\n')
self.assertTrue(np.allclose(hp[:, [0], :], data[0:1, :, :], equal_nan=True))
print('==========================\n输出第0、1、2个htype对应的所有股票全部历史数据\n')
self.assertTrue(np.allclose(hp[[0, 1, 2]], data[:, :, 0:3], equal_nan=True))
print('==========================\n输出close、high两个类型的所有历史数据\n')
self.assertTrue(np.allclose(hp[['close', 'high']], data[:, :, [0, 2]], equal_nan=True))
print('==========================\n输出0、1两个htype的所有历史数据\n')
self.assertTrue(np.allclose(hp[[0, 1]], data[:, :, 0:2], equal_nan=True))
print('==========================\n输出close、high两个类型的所有历史数据\n')
self.assertTrue(np.allclose(hp['close,high'], data[:, :, [0, 2]], equal_nan=True))
print('==========================\n输出close起到high止的三个类型的所有历史数据\n')
self.assertTrue(np.allclose(hp['close:high'], data[:, :, 0:3], equal_nan=True))
print('==========================\n输出0、1、3三个股票的全部历史数据\n')
self.assertTrue(np.allclose(hp[:, [0, 1, 3]], data[[0, 1, 3], :, :], equal_nan=True))
print('==========================\n输出000100、000102两只股票的所有历史数据\n')
self.assertTrue(np.allclose(hp[:, ['000100', '000102']], data[[0, 2], :, :], equal_nan=True))
print('==========================\n输出0、1、2三个股票的历史数据\n', hp[:, 0: 3])
self.assertTrue(np.allclose(hp[:, 0: 3], data[0:3, :, :], equal_nan=True))
print('==========================\n输出000100、000102两只股票的所有历史数据\n')
self.assertTrue(np.allclose(hp[:, '000100, 000102'], data[[0, 2], :, :], equal_nan=True))
print('==========================\n输出所有股票的0-7日历史数据\n')
self.assertTrue(np.allclose(hp[:, :, 0:8], data[:, 0:8, :], equal_nan=True))
print('==========================\n输出000100股票的0-7日历史数据\n')
self.assertTrue(np.allclose(hp[:, '000100', 0:8], data[0, 0:8, :], equal_nan=True))
print('==========================\nstart testing multy axis slicing of HistoryPanel object')
print('==========================\n输出000100、000120两只股票的close、open两组历史数据\n',
hp['close,open', ['000100', '000102']])
print('==========================\n输出000100、000120两只股票的close到open三组历史数据\n',
hp['close,open', '000100, 000102'])
print(f'historyPanel: hp:\n{hp}')
print(f'data is:\n{data}')
hp.htypes = 'open,high,low,close'
hp.info()
hp.shares = ['000300', '600227', '600222', '000123', '000129']
hp.info()
def test_relabel(self):
new_shares_list = ['000001', '000002', '000003', '000004', '000005']
new_shares_str = '000001, 000002, 000003, 000004, 000005'
new_htypes_list = ['close', 'volume', 'value', 'exchange']
new_htypes_str = 'close, volume, value, exchange'
temp_hp = self.hp.copy()
temp_hp.re_label(shares=new_shares_list)
print(temp_hp.info())
print(temp_hp.htypes)
self.assertTrue(np.allclose(self.hp.values, temp_hp.values))
self.assertEqual(self.hp.htypes, temp_hp.htypes)
self.assertEqual(self.hp.hdates, temp_hp.hdates)
self.assertEqual(temp_hp.shares, new_shares_list)
temp_hp = self.hp.copy()
temp_hp.re_label(shares=new_shares_str)
self.assertTrue(np.allclose(self.hp.values, temp_hp.values))
self.assertEqual(self.hp.htypes, temp_hp.htypes)
self.assertEqual(self.hp.hdates, temp_hp.hdates)
self.assertEqual(temp_hp.shares, new_shares_list)
temp_hp = self.hp.copy()
temp_hp.re_label(htypes=new_htypes_list)
self.assertTrue(np.allclose(self.hp.values, temp_hp.values))
self.assertEqual(self.hp.shares, temp_hp.shares)
self.assertEqual(self.hp.hdates, temp_hp.hdates)
self.assertEqual(temp_hp.htypes, new_htypes_list)
temp_hp = self.hp.copy()
temp_hp.re_label(htypes=new_htypes_str)
self.assertTrue(np.allclose(self.hp.values, temp_hp.values))
self.assertEqual(self.hp.shares, temp_hp.shares)
self.assertEqual(self.hp.hdates, temp_hp.hdates)
self.assertEqual(temp_hp.htypes, new_htypes_list)
print(f'test errors raising')
temp_hp = self.hp.copy()
self.assertRaises(AssertionError, temp_hp.re_label, htypes=new_shares_str)
self.assertRaises(TypeError, temp_hp.re_label, htypes=123)
self.assertRaises(AssertionError, temp_hp.re_label, htypes='wrong input!')
def test_csv_to_hp(self):
pass
def test_hdf_to_hp(self):
pass
def test_hp_join(self):
# TODO: 这里需要加强,需要用具体的例子确认hp_join的结果正确
# TODO: 尤其是不同的shares、htypes、hdates,以及它们在顺
# TODO: 序不同的情况下是否能正确地组合
print(f'join two simple HistoryPanels with same shares')
temp_hp = self.hp.join(self.hp2, same_shares=True)
self.assertIsInstance(temp_hp, qt.HistoryPanel)
def test_df_to_hp(self):
print(f'test converting DataFrame to HistoryPanel')
data = np.random.randint(10, size=(10, 5))
df1 = pd.DataFrame(data)
df2 = pd.DataFrame(data, columns=qt.str_to_list(self.shares))
df3 = pd.DataFrame(data[:, 0:4])
df4 = pd.DataFrame(data[:, 0:4], columns=qt.str_to_list(self.htypes))
hp = qt.dataframe_to_hp(df1, htypes='close')
self.assertIsInstance(hp, qt.HistoryPanel)
self.assertEqual(hp.shares, [0, 1, 2, 3, 4])
self.assertEqual(hp.htypes, ['close'])
self.assertEqual(hp.hdates, [pd.Timestamp('1970-01-01 00:00:00'),
pd.Timestamp('1970-01-01 00:00:00.000000001'),
pd.Timestamp('1970-01-01 00:00:00.000000002'),
pd.Timestamp('1970-01-01 00:00:00.000000003'),
pd.Timestamp('1970-01-01 00:00:00.000000004'),
pd.Timestamp('1970-01-01 00:00:00.000000005'),
pd.Timestamp('1970-01-01 00:00:00.000000006'),
pd.Timestamp('1970-01-01 00:00:00.000000007'),
pd.Timestamp('1970-01-01 00:00:00.000000008'),
pd.Timestamp('1970-01-01 00:00:00.000000009')])
hp = qt.dataframe_to_hp(df2, shares=self.shares, htypes='close')
self.assertIsInstance(hp, qt.HistoryPanel)
self.assertEqual(hp.shares, qt.str_to_list(self.shares))
self.assertEqual(hp.htypes, ['close'])
hp = qt.dataframe_to_hp(df3, shares='000100', column_type='htypes')
self.assertIsInstance(hp, qt.HistoryPanel)
self.assertEqual(hp.shares, ['000100'])
self.assertEqual(hp.htypes, [0, 1, 2, 3])
hp = qt.dataframe_to_hp(df4, shares='000100', htypes=self.htypes, column_type='htypes')
self.assertIsInstance(hp, qt.HistoryPanel)
self.assertEqual(hp.shares, ['000100'])
self.assertEqual(hp.htypes, qt.str_to_list(self.htypes))
hp.info()
self.assertRaises(KeyError, qt.dataframe_to_hp, df1)
def test_to_dataframe(self):
""" 测试HistoryPanel对象的to_dataframe方法
"""
print(f'START TEST == test_to_dataframe')
print(f'test converting test hp to dataframe with share == "000102":')
df_test = self.hp.to_dataframe(share='000102')
self.assertIsInstance(df_test, pd.DataFrame)
self.assertEqual(list(self.hp.hdates), list(df_test.index))
self.assertEqual(list(self.hp.htypes), list(df_test.columns))
values = df_test.values
self.assertTrue(np.allclose(self.hp[:, '000102'], values))
print(f'test DataFrame conversion with share == "000100"')
df_test = self.hp.to_dataframe(share='000100')
self.assertIsInstance(df_test, pd.DataFrame)
self.assertEqual(list(self.hp.hdates), list(df_test.index))
self.assertEqual(list(self.hp.htypes), list(df_test.columns))
values = df_test.values
self.assertTrue(np.allclose(self.hp[:, '000100'], values))
print(f'test DataFrame conversion error: type incorrect')
self.assertRaises(AssertionError, self.hp.to_dataframe, share=3.0)
print(f'test DataFrame error raising with share not found error')
self.assertRaises(KeyError, self.hp.to_dataframe, share='000300')
print(f'test DataFrame conversion with htype == "close"')
df_test = self.hp.to_dataframe(htype='close')
self.assertIsInstance(df_test, pd.DataFrame)
self.assertEqual(list(self.hp.hdates), list(df_test.index))
self.assertEqual(list(self.hp.shares), list(df_test.columns))
values = df_test.values
self.assertTrue(np.allclose(self.hp['close'].T, values))
print(f'test DataFrame conversion with htype == "high"')
df_test = self.hp.to_dataframe(htype='high')
self.assertIsInstance(df_test, pd.DataFrame)
self.assertEqual(list(self.hp.hdates), list(df_test.index))
self.assertEqual(list(self.hp.shares), list(df_test.columns))
values = df_test.values
self.assertTrue(np.allclose(self.hp['high'].T, values))
print(f'test DataFrame conversion with htype == "high" and dropna')
v = self.hp.values.astype('float')
v[:, 3, :] = np.nan
v[:, 4, :] = np.inf
test_hp = qt.HistoryPanel(v, levels=self.shares, columns=self.htypes, rows=self.index)
df_test = test_hp.to_dataframe(htype='high', dropna=True)
self.assertIsInstance(df_test, pd.DataFrame)
self.assertEqual(list(self.hp.hdates[:3]) + list(self.hp.hdates[4:]), list(df_test.index))
self.assertEqual(list(self.hp.shares), list(df_test.columns))
values = df_test.values
target_values = test_hp['high'].T
target_values = target_values[np.where(~np.isnan(target_values))].reshape(9, 5)
self.assertTrue(np.allclose(target_values, values))
print(f'test DataFrame conversion with htype == "high", dropna and treat infs as na')
v = self.hp.values.astype('float')
v[:, 3, :] = np.nan
v[:, 4, :] = np.inf
test_hp = qt.HistoryPanel(v, levels=self.shares, columns=self.htypes, rows=self.index)
df_test = test_hp.to_dataframe(htype='high', dropna=True, inf_as_na=True)
self.assertIsInstance(df_test, pd.DataFrame)
self.assertEqual(list(self.hp.hdates[:3]) + list(self.hp.hdates[5:]), list(df_test.index))
self.assertEqual(list(self.hp.shares), list(df_test.columns))
values = df_test.values
target_values = test_hp['high'].T
target_values = target_values[np.where(~np.isnan(target_values) & ~np.isinf(target_values))].reshape(8, 5)
self.assertTrue(np.allclose(target_values, values))
print(f'test DataFrame conversion error: type incorrect')
self.assertRaises(AssertionError, self.hp.to_dataframe, htype=pd.DataFrame())
print(f'test DataFrame error raising with share not found error')
self.assertRaises(KeyError, self.hp.to_dataframe, htype='non_type')
print(f'Raises ValueError when both or none parameter is given')
self.assertRaises(KeyError, self.hp.to_dataframe)
self.assertRaises(KeyError, self.hp.to_dataframe, share='000100', htype='close')
def test_to_df_dict(self):
"""测试HistoryPanel公有方法to_df_dict"""
print('test convert history panel slice by share')
df_dict = self.hp.to_df_dict('share')
self.assertEqual(self.hp.shares, list(df_dict.keys()))
df_dict = self.hp.to_df_dict()
self.assertEqual(self.hp.shares, list(df_dict.keys()))
print('test convert historypanel slice by htype ')
df_dict = self.hp.to_df_dict('htype')
self.assertEqual(self.hp.htypes, list(df_dict.keys()))
print('test raise assertion error')
self.assertRaises(AssertionError, self.hp.to_df_dict, by='random text')
self.assertRaises(AssertionError, self.hp.to_df_dict, by=3)
print('test empty hp')
df_dict = qt.HistoryPanel().to_df_dict('share')
self.assertEqual(df_dict, {})
def test_stack_dataframes(self):
print('test stack dataframes in a list')
df1 = pd.DataFrame({'a': [1, 2, 3, 4], 'b': [2, 3, 4, 5], 'c': [3, 4, 5, 6]})
df1.index = ['20200101', '20200102', '20200103', '20200104']
df2 = pd.DataFrame({'b': [4, 3, 2, 1], 'd': [1, 1, 1, 1], 'c': [6, 5, 4, 3]})
df2.index = ['20200101', '20200102', '20200104', '20200105']
df3 = pd.DataFrame({'a': [6, 6, 6, 6], 'd': [4, 4, 4, 4], 'b': [2, 4, 6, 8]})
df3.index = ['20200101', '20200102', '20200103', '20200106']
values1 = np.array([[[1., 2., 3., np.nan],
[2., 3., 4., np.nan],
[3., 4., 5., np.nan],
[4., 5., 6., np.nan],
[np.nan, np.nan, np.nan, np.nan],
[np.nan, np.nan, np.nan, np.nan]],
[[np.nan, 4., 6., 1.],
[np.nan, 3., 5., 1.],
[np.nan, np.nan, np.nan, np.nan],
[np.nan, 2., 4., 1.],
[np.nan, 1., 3., 1.],
[np.nan, np.nan, np.nan, np.nan]],
[[6., 2., np.nan, 4.],
[6., 4., np.nan, 4.],
[6., 6., np.nan, 4.],
[np.nan, np.nan, np.nan, np.nan],
[np.nan, np.nan, np.nan, np.nan],
[6., 8., np.nan, 4.]]])
values2 = np.array([[[1., np.nan, 6.],
[2., np.nan, 6.],
[3., np.nan, 6.],
[4., np.nan, np.nan],
[np.nan, np.nan, np.nan],
[np.nan, np.nan, 6.]],
[[2., 4., 2.],
[3., 3., 4.],
[4., np.nan, 6.],
[5., 2., np.nan],
[np.nan, 1., np.nan],
[np.nan, np.nan, 8.]],
[[3., 6., np.nan],
[4., 5., np.nan],
[5., np.nan, np.nan],
[6., 4., np.nan],
[np.nan, 3., np.nan],
[np.nan, np.nan, np.nan]],
[[np.nan, 1., 4.],
[np.nan, 1., 4.],
[np.nan, np.nan, 4.],
[np.nan, 1., np.nan],
[np.nan, 1., np.nan],
[np.nan, np.nan, 4.]]])
print(df1.rename(index=pd.to_datetime))
print(df2.rename(index=pd.to_datetime))
print(df3.rename(index=pd.to_datetime))
hp1 = stack_dataframes([df1, df2, df3], stack_along='shares',
shares=['000100', '000200', '000300'])
hp2 = stack_dataframes([df1, df2, df3], stack_along='shares',
shares='000100, 000300, 000200')
print('hp1 is:\n', hp1)
print('hp2 is:\n', hp2)
self.assertEqual(hp1.htypes, ['a', 'b', 'c', 'd'])
self.assertEqual(hp1.shares, ['000100', '000200', '000300'])
self.assertTrue(np.allclose(hp1.values, values1, equal_nan=True))
self.assertEqual(hp2.htypes, ['a', 'b', 'c', 'd'])
self.assertEqual(hp2.shares, ['000100', '000300', '000200'])
self.assertTrue(np.allclose(hp2.values, values1, equal_nan=True))
hp3 = stack_dataframes([df1, df2, df3], stack_along='htypes',
htypes=['close', 'high', 'low'])
hp4 = stack_dataframes([df1, df2, df3], stack_along='htypes',
htypes='open, close, high')
print('hp3 is:\n', hp3.values)
print('hp4 is:\n', hp4.values)
self.assertEqual(hp3.htypes, ['close', 'high', 'low'])
self.assertEqual(hp3.shares, ['a', 'b', 'c', 'd'])
self.assertTrue(np.allclose(hp3.values, values2, equal_nan=True))
self.assertEqual(hp4.htypes, ['open', 'close', 'high'])
self.assertEqual(hp4.shares, ['a', 'b', 'c', 'd'])
self.assertTrue(np.allclose(hp4.values, values2, equal_nan=True))
print('test stack dataframes in a dict')
df1 = pd.DataFrame({'a': [1, 2, 3, 4], 'b': [2, 3, 4, 5], 'c': [3, 4, 5, 6]})
df1.index = ['20200101', '20200102', '20200103', '20200104']
df2 = pd.DataFrame({'b': [4, 3, 2, 1], 'd': [1, 1, 1, 1], 'c': [6, 5, 4, 3]})
df2.index = ['20200101', '20200102', '20200104', '20200105']
df3 = pd.DataFrame({'a': [6, 6, 6, 6], 'd': [4, 4, 4, 4], 'b': [2, 4, 6, 8]})
df3.index = ['20200101', '20200102', '20200103', '20200106']
values1 = np.array([[[1., 2., 3., np.nan],
[2., 3., 4., np.nan],
[3., 4., 5., np.nan],
[4., 5., 6., np.nan],
[np.nan, np.nan, np.nan, np.nan],
[np.nan, np.nan, np.nan, np.nan]],
[[np.nan, 4., 6., 1.],
[np.nan, 3., 5., 1.],
[np.nan, np.nan, np.nan, np.nan],
[np.nan, 2., 4., 1.],
[np.nan, 1., 3., 1.],
[np.nan, np.nan, np.nan, np.nan]],
[[6., 2., np.nan, 4.],
[6., 4., np.nan, 4.],
[6., 6., np.nan, 4.],
[np.nan, np.nan, np.nan, np.nan],
[np.nan, np.nan, np.nan, np.nan],
[6., 8., np.nan, 4.]]])
values2 = np.array([[[1., np.nan, 6.],
[2., np.nan, 6.],
[3., np.nan, 6.],
[4., np.nan, np.nan],
[np.nan, np.nan, np.nan],
[np.nan, np.nan, 6.]],
[[2., 4., 2.],
[3., 3., 4.],
[4., np.nan, 6.],
[5., 2., np.nan],
[np.nan, 1., np.nan],
[np.nan, np.nan, 8.]],
[[3., 6., np.nan],
[4., 5., np.nan],
[5., np.nan, np.nan],
[6., 4., np.nan],
[np.nan, 3., np.nan],
[np.nan, np.nan, np.nan]],
[[np.nan, 1., 4.],
[np.nan, 1., 4.],
[np.nan, np.nan, 4.],
[np.nan, 1., np.nan],
[np.nan, 1., np.nan],
[np.nan, np.nan, 4.]]])
print(df1.rename(index=pd.to_datetime))
print(df2.rename(index=pd.to_datetime))
print(df3.rename(index=pd.to_datetime))
hp1 = stack_dataframes(dfs={'000001.SZ': df1, '000002.SZ': df2, '000003.SZ': df3},
stack_along='shares')
hp2 = stack_dataframes(dfs={'000001.SZ': df1, '000002.SZ': df2, '000003.SZ': df3},
stack_along='shares',
shares='000100, 000300, 000200')
print('hp1 is:\n', hp1)
print('hp2 is:\n', hp2)
self.assertEqual(hp1.htypes, ['a', 'b', 'c', 'd'])
self.assertEqual(hp1.shares, ['000001.SZ', '000002.SZ', '000003.SZ'])
self.assertTrue(np.allclose(hp1.values, values1, equal_nan=True))
self.assertEqual(hp2.htypes, ['a', 'b', 'c', 'd'])
self.assertEqual(hp2.shares, ['000100', '000300', '000200'])
self.assertTrue(np.allclose(hp2.values, values1, equal_nan=True))
hp3 = stack_dataframes(dfs={'close': df1, 'high': df2, 'low': df3},
stack_along='htypes')
hp4 = stack_dataframes(dfs={'close': df1, 'low': df2, 'high': df3},
stack_along='htypes',
htypes='open, close, high')
print('hp3 is:\n', hp3.values)
print('hp4 is:\n', hp4.values)
self.assertEqual(hp3.htypes, ['close', 'high', 'low'])
self.assertEqual(hp3.shares, ['a', 'b', 'c', 'd'])
self.assertTrue(np.allclose(hp3.values, values2, equal_nan=True))
self.assertEqual(hp4.htypes, ['open', 'close', 'high'])
self.assertEqual(hp4.shares, ['a', 'b', 'c', 'd'])
self.assertTrue(np.allclose(hp4.values, values2, equal_nan=True))
def test_to_csv(self):
pass
def test_to_hdf(self):
pass
def test_fill_na(self):
print(self.hp)
new_values = self.hp.values.astype(float)
new_values[[0, 1, 3, 2], [1, 3, 0, 2], [1, 3, 2, 2]] = np.nan
print(new_values)
temp_hp = qt.HistoryPanel(values=new_values, levels=self.hp.levels, rows=self.hp.rows, columns=self.hp.columns)
self.assertTrue(np.allclose(temp_hp.values[[0, 1, 3, 2], [1, 3, 0, 2], [1, 3, 2, 2]], np.nan, equal_nan=True))
temp_hp.fillna(2.3)
new_values[[0, 1, 3, 2], [1, 3, 0, 2], [1, 3, 2, 2]] = 2.3
self.assertTrue(np.allclose(temp_hp.values,
new_values, equal_nan=True))
def test_get_history_panel(self):
# TODO: implement this test case
# test get only one line of data
pass
def test_get_price_type_raw_data(self):
shares = '000039.SZ, 600748.SH, 000040.SZ'
start = '20200101'
end = '20200131'
htypes = 'open, high, low, close'
target_price_000039 = [[9.45, 9.49, 9.12, 9.17],
[9.46, 9.56, 9.4, 9.5],
[9.7, 9.76, 9.5, 9.51],
[9.7, 9.75, 9.7, 9.72],
[9.73, 9.77, 9.7, 9.73],
[9.83, 9.85, 9.71, 9.72],
[9.85, 9.85, 9.75, 9.79],
[9.96, 9.96, 9.83, 9.86],
[9.87, 9.94, 9.77, 9.93],
[9.82, 9.9, 9.76, 9.87],
[9.8, 9.85, 9.77, 9.82],
[9.84, 9.86, 9.71, 9.72],
[9.83, 9.93, 9.81, 9.86],
[9.7, 9.87, 9.7, 9.82],
[9.83, 9.86, 9.69, 9.79],
[9.8, 9.94, 9.8, 9.86]]
target_price_600748 = [[5.68, 5.68, 5.32, 5.37],
[5.62, 5.68, 5.46, 5.65],
[5.72, 5.72, 5.61, 5.62],
[5.76, 5.77, 5.6, 5.73],
[5.78, 5.84, 5.73, 5.75],
[5.89, 5.91, 5.76, 5.77],
[6.03, 6.04, 5.87, 5.89],
[5.94, 6.07, 5.94, 6.02],
[5.96, 5.98, 5.88, 5.97],
[6.04, 6.06, 5.95, 5.96],
[5.98, 6.04, 5.96, 6.03],
[6.1, 6.11, 5.89, 5.94],
[6.02, 6.12, 6., 6.1],
[5.96, 6.05, 5.88, 6.01],
[6.03, 6.03, 5.95, 5.99],
[6.02, 6.12, 5.99, 5.99]]
target_price_000040 = [[3.63, 3.83, 3.63, 3.65],
[3.99, 4.07, 3.97, 4.03],
[4.1, 4.11, 3.93, 3.95],
[4.12, 4.13, 4.06, 4.11],
[4.13, 4.19, 4.07, 4.13],
[4.27, 4.28, 4.11, 4.12],
[4.37, 4.38, 4.25, 4.29],
[4.34, 4.5, 4.32, 4.41],
[4.28, 4.35, 4.2, 4.34],
[4.41, 4.43, 4.29, 4.31],
[4.42, 4.45, 4.36, 4.41],
[4.51, 4.56, 4.33, 4.35],
[4.35, 4.55, 4.31, 4.55],
[4.3, 4.41, 4.22, 4.36],
[4.27, 4.44, 4.23, 4.34],
[4.23, 4.27, 4.18, 4.25]]
print(f'test get price type raw data with single thread')
df_list = get_price_type_raw_data(start=start, end=end, shares=shares, htypes=htypes, freq='d')
self.assertIsInstance(df_list, dict)
self.assertEqual(len(df_list), 3)
self.assertTrue(np.allclose(df_list['000039.SZ'].values, np.array(target_price_000039)))
self.assertTrue(np.allclose(df_list['600748.SH'].values, np.array(target_price_600748)))
self.assertTrue(np.allclose(df_list['000040.SZ'].values, np.array(target_price_000040)))
print(f'in get financial report type raw data, got DataFrames: \n"000039.SZ":\n'
f'{df_list["000039.SZ"]}\n"600748.SH":\n'
f'{df_list["600748.SH"]}\n"000040.SZ":\n{df_list["000040.SZ"]}')
print(f'test get price type raw data with with multi threads')
df_list = get_price_type_raw_data(start=start, end=end, shares=shares, htypes=htypes, freq='d', parallel=10)
self.assertIsInstance(df_list, dict)
self.assertEqual(len(df_list), 3)
self.assertTrue(np.allclose(df_list['000039.SZ'].values, np.array(target_price_000039)))
self.assertTrue(np.allclose(df_list['600748.SH'].values, np.array(target_price_600748)))
self.assertTrue(np.allclose(df_list['000040.SZ'].values, np.array(target_price_000040)))
print(f'in get financial report type raw data, got DataFrames: \n"000039.SZ":\n'
f'{df_list["000039.SZ"]}\n"600748.SH":\n'
f'{df_list["600748.SH"]}\n"000040.SZ":\n{df_list["000040.SZ"]}')
def test_get_financial_report_type_raw_data(self):
shares = '000039.SZ, 600748.SH, 000040.SZ'
start = '20160101'
end = '20201231'
htypes = 'eps,basic_eps,diluted_eps,total_revenue,revenue,total_share,' \
'cap_rese,undistr_porfit,surplus_rese,net_profit'
target_eps_000039 = [[1.41],
[0.1398],
[-0.0841],
[-0.1929],
[0.37],
[0.1357],
[0.1618],
[0.1191],
[1.11],
[0.759],
[0.3061],
[0.1409],
[0.81],
[0.4187],
[0.2554],
[0.1624],
[0.14],
[-0.0898],
[-0.1444],
[0.1291]]
target_eps_600748 = [[0.41],
[0.22],
[0.22],
[0.09],
[0.42],
[0.23],
[0.22],
[0.09],
[0.36],
[0.16],
[0.15],
[0.07],
[0.47],
[0.19],
[0.12],
[0.07],
[0.32],
[0.22],
[0.14],
[0.07]]
target_eps_000040 = [[-0.6866],
[-0.134],
[-0.189],
[-0.036],
[-0.6435],
[0.05],
[0.062],
[0.0125],
[0.8282],
[1.05],
[0.985],
[0.811],
[0.41],
[0.242],
[0.113],
[0.027],
[0.19],
[0.17],
[0.17],
[0.064]]
target_basic_eps_000039 = [[1.3980000e-01, 1.3980000e-01, 6.3591954e+10, 6.3591954e+10],
[-8.4100000e-02, -8.4100000e-02, 3.9431807e+10, 3.9431807e+10],
[-1.9290000e-01, -1.9290000e-01, 1.5852177e+10, 1.5852177e+10],
[3.7000000e-01, 3.7000000e-01, 8.5815341e+10, 8.5815341e+10],
[1.3570000e-01, 1.3430000e-01, 6.1660271e+10, 6.1660271e+10],
[1.6180000e-01, 1.6040000e-01, 4.2717729e+10, 4.2717729e+10],
[1.1910000e-01, 1.1900000e-01, 1.9099547e+10, 1.9099547e+10],
[1.1100000e+00, 1.1000000e+00, 9.3497622e+10, 9.3497622e+10],
[7.5900000e-01, 7.5610000e-01, 6.6906147e+10, 6.6906147e+10],
[3.0610000e-01, 3.0380000e-01, 4.3560398e+10, 4.3560398e+10],
[1.4090000e-01, 1.4050000e-01, 1.9253639e+10, 1.9253639e+10],
[8.1000000e-01, 8.1000000e-01, 7.6299930e+10, 7.6299930e+10],
[4.1870000e-01, 4.1710000e-01, 5.3962706e+10, 5.3962706e+10],
[2.5540000e-01, 2.5440000e-01, 3.3387152e+10, 3.3387152e+10],
[1.6240000e-01, 1.6200000e-01, 1.4675987e+10, 1.4675987e+10],
[1.4000000e-01, 1.4000000e-01, 5.1111652e+10, 5.1111652e+10],
[-8.9800000e-02, -8.9800000e-02, 3.4982614e+10, 3.4982614e+10],
[-1.4440000e-01, -1.4440000e-01, 2.3542843e+10, 2.3542843e+10],
[1.2910000e-01, 1.2860000e-01, 1.0412416e+10, 1.0412416e+10],
[7.2000000e-01, 7.1000000e-01, 5.8685804e+10, 5.8685804e+10]]
target_basic_eps_600748 = [[2.20000000e-01, 2.20000000e-01, 5.29423397e+09, 5.29423397e+09],
[2.20000000e-01, 2.20000000e-01, 4.49275653e+09, 4.49275653e+09],
[9.00000000e-02, 9.00000000e-02, 1.59067065e+09, 1.59067065e+09],
[4.20000000e-01, 4.20000000e-01, 8.86555586e+09, 8.86555586e+09],
[2.30000000e-01, 2.30000000e-01, 5.44850143e+09, 5.44850143e+09],
[2.20000000e-01, 2.20000000e-01, 4.34978927e+09, 4.34978927e+09],
[9.00000000e-02, 9.00000000e-02, 1.73793793e+09, 1.73793793e+09],
[3.60000000e-01, 3.60000000e-01, 8.66375241e+09, 8.66375241e+09],
[1.60000000e-01, 1.60000000e-01, 4.72875116e+09, 4.72875116e+09],
[1.50000000e-01, 1.50000000e-01, 3.76879016e+09, 3.76879016e+09],
[7.00000000e-02, 7.00000000e-02, 1.31785454e+09, 1.31785454e+09],
[4.70000000e-01, 4.70000000e-01, 7.23391685e+09, 7.23391685e+09],
[1.90000000e-01, 1.90000000e-01, 3.76072215e+09, 3.76072215e+09],
[1.20000000e-01, 1.20000000e-01, 2.35845364e+09, 2.35845364e+09],
[7.00000000e-02, 7.00000000e-02, 1.03831865e+09, 1.03831865e+09],
[3.20000000e-01, 3.20000000e-01, 6.48880919e+09, 6.48880919e+09],
[2.20000000e-01, 2.20000000e-01, 3.72209142e+09, 3.72209142e+09],
[1.40000000e-01, 1.40000000e-01, 2.22563924e+09, 2.22563924e+09],
[7.00000000e-02, 7.00000000e-02, 8.96647052e+08, 8.96647052e+08],
[4.80000000e-01, 4.80000000e-01, 6.61917508e+09, 6.61917508e+09]]
target_basic_eps_000040 = [[-1.34000000e-01, -1.34000000e-01, 2.50438755e+09, 2.50438755e+09],
[-1.89000000e-01, -1.89000000e-01, 1.32692347e+09, 1.32692347e+09],
[-3.60000000e-02, -3.60000000e-02, 5.59073338e+08, 5.59073338e+08],
[-6.43700000e-01, -6.43700000e-01, 6.80576162e+09, 6.80576162e+09],
[5.00000000e-02, 5.00000000e-02, 6.38891620e+09, 6.38891620e+09],
[6.20000000e-02, 6.20000000e-02, 5.23267082e+09, 5.23267082e+09],
[1.25000000e-02, 1.25000000e-02, 2.22420874e+09, 2.22420874e+09],
[8.30000000e-01, 8.30000000e-01, 8.67628947e+09, 8.67628947e+09],
[1.05000000e+00, 1.05000000e+00, 5.29431716e+09, 5.29431716e+09],
[9.85000000e-01, 9.85000000e-01, 3.56822382e+09, 3.56822382e+09],
[8.11000000e-01, 8.11000000e-01, 1.06613439e+09, 1.06613439e+09],
[4.10000000e-01, 4.10000000e-01, 8.13102532e+09, 8.13102532e+09],
[2.42000000e-01, 2.42000000e-01, 5.17971521e+09, 5.17971521e+09],
[1.13000000e-01, 1.13000000e-01, 3.21704120e+09, 3.21704120e+09],
[2.70000000e-02, 2.70000000e-02, 8.41966738e+08, 8.24272235e+08],
[1.90000000e-01, 1.90000000e-01, 3.77350171e+09, 3.77350171e+09],
[1.70000000e-01, 1.70000000e-01, 2.38643892e+09, 2.38643892e+09],
[1.70000000e-01, 1.70000000e-01, 1.29127117e+09, 1.29127117e+09],
[6.40000000e-02, 6.40000000e-02, 6.03256858e+08, 6.03256858e+08],
[1.30000000e-01, 1.30000000e-01, 1.66572918e+09, 1.66572918e+09]]
target_total_share_000039 = [[3.5950140e+09, 4.8005360e+09, 2.1573660e+10, 3.5823430e+09],
[3.5860750e+09, 4.8402300e+09, 2.0750827e+10, 3.5823430e+09],
[3.5860750e+09, 4.9053550e+09, 2.0791307e+10, 3.5823430e+09],
[3.5845040e+09, 4.8813110e+09, 2.1482857e+10, 3.5823430e+09],
[3.5831490e+09, 4.9764250e+09, 2.0926816e+10, 3.2825850e+09],
[3.5825310e+09, 4.8501270e+09, 2.1020418e+10, 3.2825850e+09],
[2.9851110e+09, 5.4241420e+09, 2.2438350e+10, 3.2825850e+09],
[2.9849890e+09, 4.1284000e+09, 2.2082769e+10, 3.2825850e+09],
[2.9849610e+09, 4.0838010e+09, 2.1045994e+10, 3.2815350e+09],
[2.9849560e+09, 4.2491510e+09, 1.9694345e+10, 3.2815350e+09],
[2.9846970e+09, 4.2351600e+09, 2.0016361e+10, 3.2815350e+09],
[2.9828890e+09, 4.2096630e+09, 1.9734494e+10, 3.2815350e+09],
[2.9813960e+09, 3.4564240e+09, 1.8562738e+10, 3.2793790e+09],
[2.9803530e+09, 3.0759650e+09, 1.8076208e+10, 3.2793790e+09],
[2.9792680e+09, 3.1376690e+09, 1.7994776e+10, 3.2793790e+09],
[2.9785770e+09, 3.1265850e+09, 1.7495053e+10, 3.2793790e+09],
[2.9783640e+09, 3.1343850e+09, 1.6740840e+10, 3.2035780e+09],
[2.9783590e+09, 3.1273880e+09, 1.6578389e+10, 3.2035780e+09],
[2.9782780e+09, 3.1169280e+09, 1.8047639e+10, 3.2035780e+09],
[2.9778200e+09, 3.1818630e+09, 1.7663145e+10, 3.2035780e+09]]
target_total_share_600748 = [[1.84456289e+09, 2.60058426e+09, 5.72443733e+09, 4.58026529e+08],
[1.84456289e+09, 2.60058426e+09, 5.72096899e+09, 4.58026529e+08],
[1.84456289e+09, 2.60058426e+09, 5.65738237e+09, 4.58026529e+08],
[1.84456289e+09, 2.60058426e+09, 5.50257806e+09, 4.58026529e+08],
[1.84456289e+09, 2.59868164e+09, 5.16741523e+09, 4.44998882e+08],
[1.84456289e+09, 2.59684471e+09, 5.14677280e+09, 4.44998882e+08],
[1.84456289e+09, 2.59684471e+09, 4.94955591e+09, 4.44998882e+08],
[1.84456289e+09, 2.59684471e+09, 4.79001451e+09, 4.44998882e+08],
[1.84456289e+09, 3.11401684e+09, 4.46326988e+09, 4.01064256e+08],
[1.84456289e+09, 3.11596723e+09, 4.45419136e+09, 4.01064256e+08],
[1.84456289e+09, 3.11596723e+09, 4.39652948e+09, 4.01064256e+08],
[1.84456289e+09, 3.18007783e+09, 4.26608403e+09, 4.01064256e+08],
[1.84456289e+09, 3.10935622e+09, 3.78417688e+09, 3.65651701e+08],
[1.84456289e+09, 3.10935622e+09, 3.65806574e+09, 3.65651701e+08],
[1.84456289e+09, 3.10935622e+09, 3.62063090e+09, 3.65651701e+08],
[1.84456289e+09, 3.10935622e+09, 3.50063915e+09, 3.65651701e+08],
[1.41889453e+09, 3.55940850e+09, 3.22272993e+09, 3.62124939e+08],
[1.41889453e+09, 3.56129650e+09, 3.11477476e+09, 3.62124939e+08],
[1.41889453e+09, 3.59632888e+09, 3.06836903e+09, 3.62124939e+08],
[1.08337087e+09, 3.37400726e+07, 3.00918704e+09, 3.62124939e+08]]
target_total_share_000040 = [[1.48687387e+09, 1.06757900e+10, 8.31900755e+08, 2.16091994e+08],
[1.48687387e+09, 1.06757900e+10, 7.50177302e+08, 2.16091994e+08],
[1.48687387e+09, 1.06757899e+10, 9.90255974e+08, 2.16123282e+08],
[1.48687387e+09, 1.06757899e+10, 1.03109866e+09, 2.16091994e+08],
[1.48687387e+09, 1.06757910e+10, 2.07704745e+09, 2.16123282e+08],
[1.48687387e+09, 1.06757910e+10, 2.09608665e+09, 2.16123282e+08],
[1.48687387e+09, 1.06803833e+10, 2.13354083e+09, 2.16123282e+08],
[1.48687387e+09, 1.06804090e+10, 2.11489364e+09, 2.16123282e+08],
[1.33717327e+09, 8.87361727e+09, 2.42939924e+09, 1.88489589e+08],
[1.33717327e+09, 8.87361727e+09, 2.34220254e+09, 1.88489589e+08],
[1.33717327e+09, 8.87361727e+09, 2.16390368e+09, 1.88489589e+08],
[1.33717327e+09, 8.87361727e+09, 1.07961915e+09, 1.88489589e+08],
[1.33717327e+09, 8.87361727e+09, 8.58866066e+08, 1.88489589e+08],
[1.33717327e+09, 8.87361727e+09, 6.87024393e+08, 1.88489589e+08],
[1.33717327e+09, 8.87361727e+09, 5.71554565e+08, 1.88489589e+08],
[1.33717327e+09, 8.87361727e+09, 5.54241222e+08, 1.88489589e+08],
[1.33717327e+09, 8.87361726e+09, 5.10059576e+08, 1.88489589e+08],
[1.33717327e+09, 8.87361726e+09, 4.59351639e+08, 1.88489589e+08],
[4.69593364e+08, 2.78355875e+08, 4.13430814e+08, 1.88489589e+08],
[4.69593364e+08, 2.74235459e+08, 3.83557678e+08, 1.88489589e+08]]
target_net_profit_000039 = [[np.nan],
[2.422180e+08],
[np.nan],
[2.510113e+09],
[np.nan],
[1.102220e+09],
[np.nan],
[4.068455e+09],
[np.nan],
[1.315957e+09],
[np.nan],
[3.158415e+09],
[np.nan],
[1.066509e+09],
[np.nan],
[7.349830e+08],
[np.nan],
[-5.411600e+08],
[np.nan],
[2.271961e+09]]
target_net_profit_600748 = [[np.nan],
[4.54341757e+08],
[np.nan],
[9.14476670e+08],
[np.nan],
[5.25360283e+08],
[np.nan],
[9.24502415e+08],
[np.nan],
[4.66560302e+08],
[np.nan],
[9.15265285e+08],
[np.nan],
[2.14639674e+08],
[np.nan],
[7.45093049e+08],
[np.nan],
[2.10967312e+08],
[np.nan],
[6.04572711e+08]]
target_net_profit_000040 = [[np.nan],
[-2.82458846e+08],
[np.nan],
[-9.57130872e+08],
[np.nan],
[9.22114527e+07],
[np.nan],
[1.12643819e+09],
[np.nan],
[1.31715269e+09],
[np.nan],
[5.39940093e+08],
[np.nan],
[1.51440838e+08],
[np.nan],
[1.75339071e+08],
[np.nan],
[8.04740415e+07],
[np.nan],
[6.20445815e+07]]
print('test get financial data, in multi thread mode')
df_list = get_financial_report_type_raw_data(start=start, end=end, shares=shares, htypes=htypes, parallel=4)
self.assertIsInstance(df_list, tuple)
self.assertEqual(len(df_list), 4)
self.assertEqual(len(df_list[0]), 3)
self.assertEqual(len(df_list[1]), 3)
self.assertEqual(len(df_list[2]), 3)
self.assertEqual(len(df_list[3]), 3)
# 检查确认所有数据类型正确
self.assertTrue(all(isinstance(item, pd.DataFrame) for subdict in df_list for item in subdict.values()))
# 检查是否有空数据
print(all(item.empty for subdict in df_list for item in subdict.values()))
# 检查获取的每组数据正确,且所有数据的顺序一致, 如果取到空数据,则忽略
if df_list[0]['000039.SZ'].empty:
print(f'income data for "000039.SZ" is empty')
else:
self.assertTrue(np.allclose(df_list[0]['000039.SZ'].values, target_basic_eps_000039))
if df_list[0]['600748.SH'].empty:
print(f'income data for "600748.SH" is empty')
else:
self.assertTrue(np.allclose(df_list[0]['600748.SH'].values, target_basic_eps_600748))
if df_list[0]['000040.SZ'].empty:
print(f'income data for "000040.SZ" is empty')
else:
self.assertTrue(np.allclose(df_list[0]['000040.SZ'].values, target_basic_eps_000040))
if df_list[1]['000039.SZ'].empty:
print(f'indicator data for "000039.SZ" is empty')
else:
self.assertTrue(np.allclose(df_list[1]['000039.SZ'].values, target_eps_000039))
if df_list[1]['600748.SH'].empty:
print(f'indicator data for "600748.SH" is empty')
else:
self.assertTrue(np.allclose(df_list[1]['600748.SH'].values, target_eps_600748))
if df_list[1]['000040.SZ'].empty:
print(f'indicator data for "000040.SZ" is empty')
else:
self.assertTrue(np.allclose(df_list[1]['000040.SZ'].values, target_eps_000040))
if df_list[2]['000039.SZ'].empty:
print(f'balance data for "000039.SZ" is empty')
else:
self.assertTrue(np.allclose(df_list[2]['000039.SZ'].values, target_total_share_000039))
if df_list[2]['600748.SH'].empty:
print(f'balance data for "600748.SH" is empty')
else:
self.assertTrue(np.allclose(df_list[2]['600748.SH'].values, target_total_share_600748))
if df_list[2]['000040.SZ'].empty:
print(f'balance data for "000040.SZ" is empty')
else:
self.assertTrue(np.allclose(df_list[2]['000040.SZ'].values, target_total_share_000040))
if df_list[3]['000039.SZ'].empty:
print(f'cash flow data for "000039.SZ" is empty')
else:
self.assertTrue(np.allclose(df_list[3]['000039.SZ'].values, target_net_profit_000039, equal_nan=True))
if df_list[3]['600748.SH'].empty:
print(f'cash flow data for "600748.SH" is empty')
else:
self.assertTrue(np.allclose(df_list[3]['600748.SH'].values, target_net_profit_600748, equal_nan=True))
if df_list[3]['000040.SZ'].empty:
print(f'cash flow data for "000040.SZ" is empty')
else:
self.assertTrue(np.allclose(df_list[3]['000040.SZ'].values, target_net_profit_000040, equal_nan=True))
print('test get financial data, in single thread mode')
df_list = get_financial_report_type_raw_data(start=start, end=end, shares=shares, htypes=htypes, parallel=0)
self.assertIsInstance(df_list, tuple)
self.assertEqual(len(df_list), 4)
self.assertEqual(len(df_list[0]), 3)
self.assertEqual(len(df_list[1]), 3)
self.assertEqual(len(df_list[2]), 3)
self.assertEqual(len(df_list[3]), 3)
# 检查确认所有数据类型正确
self.assertTrue(all(isinstance(item, pd.DataFrame) for subdict in df_list for item in subdict.values()))
# 检查是否有空数据,因为网络问题,有可能会取到空数据
self.assertFalse(all(item.empty for subdict in df_list for item in subdict.values()))
# 检查获取的每组数据正确,且所有数据的顺序一致, 如果取到空数据,则忽略
if df_list[0]['000039.SZ'].empty:
print(f'income data for "000039.SZ" is empty')
else:
self.assertTrue(np.allclose(df_list[0]['000039.SZ'].values, target_basic_eps_000039))
if df_list[0]['600748.SH'].empty:
print(f'income data for "600748.SH" is empty')
else:
self.assertTrue(np.allclose(df_list[0]['600748.SH'].values, target_basic_eps_600748))
if df_list[0]['000040.SZ'].empty:
print(f'income data for "000040.SZ" is empty')
else:
self.assertTrue(np.allclose(df_list[0]['000040.SZ'].values, target_basic_eps_000040))
if df_list[1]['000039.SZ'].empty:
print(f'indicator data for "000039.SZ" is empty')
else:
self.assertTrue(np.allclose(df_list[1]['000039.SZ'].values, target_eps_000039))
if df_list[1]['600748.SH'].empty:
print(f'indicator data for "600748.SH" is empty')
else:
self.assertTrue(np.allclose(df_list[1]['600748.SH'].values, target_eps_600748))
if df_list[1]['000040.SZ'].empty:
print(f'indicator data for "000040.SZ" is empty')
else:
self.assertTrue(np.allclose(df_list[1]['000040.SZ'].values, target_eps_000040))
if df_list[2]['000039.SZ'].empty:
print(f'balance data for "000039.SZ" is empty')
else:
self.assertTrue(np.allclose(df_list[2]['000039.SZ'].values, target_total_share_000039))
if df_list[2]['600748.SH'].empty:
print(f'balance data for "600748.SH" is empty')
else:
self.assertTrue(np.allclose(df_list[2]['600748.SH'].values, target_total_share_600748))
if df_list[2]['000040.SZ'].empty:
print(f'balance data for "000040.SZ" is empty')
else:
self.assertTrue(np.allclose(df_list[2]['000040.SZ'].values, target_total_share_000040))
if df_list[3]['000039.SZ'].empty:
print(f'cash flow data for "000039.SZ" is empty')
else:
self.assertTrue(np.allclose(df_list[3]['000039.SZ'].values, target_net_profit_000039, equal_nan=True))
if df_list[3]['600748.SH'].empty:
print(f'cash flow data for "600748.SH" is empty')
else:
self.assertTrue(np.allclose(df_list[3]['600748.SH'].values, target_net_profit_600748, equal_nan=True))
if df_list[3]['000040.SZ'].empty:
print(f'cash flow data for "000040.SZ" is empty')
else:
self.assertTrue(np.allclose(df_list[3]['000040.SZ'].values, target_net_profit_000040, equal_nan=True))
def test_get_composite_type_raw_data(self):
pass
class TestUtilityFuncs(unittest.TestCase):
def setUp(self):
pass
def test_str_to_list(self):
self.assertEqual(str_to_list('a,b,c,d,e'), ['a', 'b', 'c', 'd', 'e'])
self.assertEqual(str_to_list('a, b, c '), ['a', 'b', 'c'])
self.assertEqual(str_to_list('a, b: c', sep_char=':'), ['a,b', 'c'])
def test_list_or_slice(self):
str_dict = {'close': 0, 'open': 1, 'high': 2, 'low': 3}
self.assertEqual(qt.list_or_slice(slice(1, 2, 1), str_dict), slice(1, 2, 1))
self.assertEqual(qt.list_or_slice('open', str_dict), [1])
self.assertEqual(list(qt.list_or_slice('close, high, low', str_dict)), [0, 2, 3])
self.assertEqual(list(qt.list_or_slice('close:high', str_dict)), [0, 1, 2])
self.assertEqual(list(qt.list_or_slice(['open'], str_dict)), [1])
self.assertEqual(list(qt.list_or_slice(['open', 'high'], str_dict)), [1, 2])
self.assertEqual(list(qt.list_or_slice(0, str_dict)), [0])
self.assertEqual(list(qt.list_or_slice([0, 2], str_dict)), [0, 2])
self.assertEqual(list(qt.list_or_slice([True, False, True, False], str_dict)), [0, 2])
def test_label_to_dict(self):
target_list = [0, 1, 10, 100]
target_dict = {'close': 0, 'open': 1, 'high': 2, 'low': 3}
target_dict2 = {'close': 0, 'open': 2, 'high': 1, 'low': 3}
self.assertEqual(qt.labels_to_dict('close, open, high, low', target_list), target_dict)
self.assertEqual(qt.labels_to_dict(['close', 'open', 'high', 'low'], target_list), target_dict)
self.assertEqual(qt.labels_to_dict('close, high, open, low', target_list), target_dict2)
self.assertEqual(qt.labels_to_dict(['close', 'high', 'open', 'low'], target_list), target_dict2)
def test_regulate_date_format(self):
self.assertEqual(regulate_date_format('2019/11/06'), '20191106')
self.assertEqual(regulate_date_format('2019-11-06'), '20191106')
self.assertEqual(regulate_date_format('20191106'), '20191106')
self.assertEqual(regulate_date_format('191106'), '20061119')
self.assertEqual(regulate_date_format('830522'), '19830522')
self.assertEqual(regulate_date_format(datetime.datetime(2010, 3, 15)), '20100315')
self.assertEqual(regulate_date_format(pd.Timestamp('2010.03.15')), '20100315')
self.assertRaises(ValueError, regulate_date_format, 'abc')
self.assertRaises(ValueError, regulate_date_format, '2019/13/43')
def test_list_to_str_format(self):
self.assertEqual(list_to_str_format(['close', 'open', 'high', 'low']),
'close,open,high,low')
self.assertEqual(list_to_str_format(['letters', ' ', '123 4', 123, ' kk l']),
'letters,,1234,kkl')
self.assertEqual(list_to_str_format('a string input'),
'a,string,input')
self.assertEqual(list_to_str_format('already,a,good,string'),
'already,a,good,string')
self.assertRaises(AssertionError, list_to_str_format, 123)
def test_is_trade_day(self):
"""test if the funcion maybe_trade_day() and is_market_trade_day() works properly
"""
date_trade = '20210401'
date_holiday = '20210102'
date_weekend = '20210424'
date_seems_trade_day = '20210217'
date_too_early = '19890601'
date_too_late = '20230105'
date_christmas = '20201225'
self.assertTrue(maybe_trade_day(date_trade))
self.assertFalse(maybe_trade_day(date_holiday))
self.assertFalse(maybe_trade_day(date_weekend))
self.assertTrue(maybe_trade_day(date_seems_trade_day))
self.assertTrue(maybe_trade_day(date_too_early))
self.assertTrue(maybe_trade_day(date_too_late))
self.assertTrue(maybe_trade_day(date_christmas))
self.assertTrue(is_market_trade_day(date_trade))
self.assertFalse(is_market_trade_day(date_holiday))
self.assertFalse(is_market_trade_day(date_weekend))
self.assertFalse(is_market_trade_day(date_seems_trade_day))
self.assertFalse(is_market_trade_day(date_too_early))
self.assertFalse(is_market_trade_day(date_too_late))
self.assertTrue(is_market_trade_day(date_christmas))
self.assertFalse(is_market_trade_day(date_christmas, exchange='XHKG'))
date_trade = pd.to_datetime('20210401')
date_holiday = pd.to_datetime('20210102')
date_weekend = pd.to_datetime('20210424')
self.assertTrue(maybe_trade_day(date_trade))
self.assertFalse(maybe_trade_day(date_holiday))
self.assertFalse(maybe_trade_day(date_weekend))
def test_prev_trade_day(self):
"""test the function prev_trade_day()
"""
date_trade = '20210401'
date_holiday = '20210102'
prev_holiday = pd.to_datetime(date_holiday) - pd.Timedelta(2, 'd')
date_weekend = '20210424'
prev_weekend = pd.to_datetime(date_weekend) - pd.Timedelta(1, 'd')
date_seems_trade_day = '20210217'
prev_seems_trade_day = '20210217'
date_too_early = '19890601'
date_too_late = '20230105'
date_christmas = '20201225'
self.assertEqual(pd.to_datetime(prev_trade_day(date_trade)),
pd.to_datetime(date_trade))
self.assertEqual(pd.to_datetime(prev_trade_day(date_holiday)),
pd.to_datetime(prev_holiday))
self.assertEqual(pd.to_datetime(prev_trade_day(date_weekend)),
pd.to_datetime(prev_weekend))
self.assertEqual(pd.to_datetime(prev_trade_day(date_seems_trade_day)),
pd.to_datetime(prev_seems_trade_day))
self.assertEqual(pd.to_datetime(prev_trade_day(date_too_early)),
pd.to_datetime(date_too_early))
self.assertEqual(pd.to_datetime(prev_trade_day(date_too_late)),
pd.to_datetime(date_too_late))
self.assertEqual(pd.to_datetime(prev_trade_day(date_christmas)),
pd.to_datetime(date_christmas))
def test_next_trade_day(self):
""" test the function next_trade_day()
"""
date_trade = '20210401'
date_holiday = '20210102'
next_holiday = pd.to_datetime(date_holiday) + pd.Timedelta(2, 'd')
date_weekend = '20210424'
next_weekend = pd.to_datetime(date_weekend) + pd.Timedelta(2, 'd')
date_seems_trade_day = '20210217'
next_seems_trade_day = '20210217'
date_too_early = '19890601'
date_too_late = '20230105'
date_christmas = '20201225'
self.assertEqual(pd.to_datetime(next_trade_day(date_trade)),
pd.to_datetime(date_trade))
self.assertEqual(pd.to_datetime(next_trade_day(date_holiday)),
pd.to_datetime(next_holiday))
self.assertEqual(pd.to_datetime(next_trade_day(date_weekend)),
pd.to_datetime(next_weekend))
self.assertEqual(pd.to_datetime(next_trade_day(date_seems_trade_day)),
pd.to_datetime(next_seems_trade_day))
self.assertEqual(pd.to_datetime(next_trade_day(date_too_early)),
pd.to_datetime(date_too_early))
self.assertEqual(pd.to_datetime(next_trade_day(date_too_late)),
pd.to_datetime(date_too_late))
self.assertEqual(pd.to_datetime(next_trade_day(date_christmas)),
pd.to_datetime(date_christmas))
def test_prev_market_trade_day(self):
""" test the function prev_market_trade_day()
"""
date_trade = '20210401'
date_holiday = '20210102'
prev_holiday = pd.to_datetime(date_holiday) - pd.Timedelta(2, 'd')
date_weekend = '20210424'
prev_weekend = pd.to_datetime(date_weekend) - pd.Timedelta(1, 'd')
date_seems_trade_day = '20210217'
prev_seems_trade_day = pd.to_datetime(date_seems_trade_day) - | pd.Timedelta(7, 'd') | pandas.Timedelta |
# Copyright 2016 Quantopian, Inc.
#
# Licensed under the Apache License, Version 2.0 (the "License");
# you may not use this file except in compliance with the License.
# You may obtain a copy of the License at
#
# http://www.apache.org/licenses/LICENSE-2.0
#
# Unless required by applicable law or agreed to in writing, software
# distributed under the License is distributed on an "AS IS" BASIS,
# WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.
# See the License for the specific language governing permissions and
# limitations under the License.
from collections import OrderedDict
from abc import ABCMeta, abstractmethod
import numpy as np
import pandas as pd
from six import with_metaclass
from zipline.data._resample import (
_minute_to_session_open,
_minute_to_session_high,
_minute_to_session_low,
_minute_to_session_close,
_minute_to_session_volume,
)
from zipline.data.minute_bars import MinuteBarReader
from zipline.data.session_bars import SessionBarReader
from zipline.utils.memoize import lazyval
_MINUTE_TO_SESSION_OHCLV_HOW = OrderedDict((
('open', 'first'),
('high', 'max'),
('low', 'min'),
('close', 'last'),
('volume', 'sum'),
))
def minute_frame_to_session_frame(minute_frame, calendar):
"""
Resample a DataFrame with minute data into the frame expected by a
BcolzDailyBarWriter.
Parameters
----------
minute_frame : pd.DataFrame
A DataFrame with the columns `open`, `high`, `low`, `close`, `volume`,
and `dt` (minute dts)
calendar : zipline.utils.calendars.trading_calendar.TradingCalendar
A TradingCalendar on which session labels to resample from minute
to session.
Return
------
session_frame : pd.DataFrame
A DataFrame with the columns `open`, `high`, `low`, `close`, `volume`,
and `day` (datetime-like).
"""
how = OrderedDict((c, _MINUTE_TO_SESSION_OHCLV_HOW[c])
for c in minute_frame.columns)
return minute_frame.groupby(calendar.minute_to_session_label).agg(how)
def minute_to_session(column, close_locs, data, out):
"""
Resample an array with minute data into an array with session data.
This function assumes that the minute data is the exact length of all
minutes in the sessions in the output.
Parameters
----------
column : str
The `open`, `high`, `low`, `close`, or `volume` column.
close_locs : array[intp]
The locations in `data` which are the market close minutes.
data : array[float64|uint32]
The minute data to be sampled into session data.
The first value should align with the market open of the first session,
containing values for all minutes for all sessions. With the last value
being the market close of the last session.
out : array[float64|uint32]
The output array into which to write the sampled sessions.
"""
if column == 'open':
_minute_to_session_open(close_locs, data, out)
elif column == 'high':
_minute_to_session_high(close_locs, data, out)
elif column == 'low':
_minute_to_session_low(close_locs, data, out)
elif column == 'close':
_minute_to_session_close(close_locs, data, out)
elif column == 'volume':
_minute_to_session_volume(close_locs, data, out)
return out
class DailyHistoryAggregator(object):
"""
Converts minute pricing data into a daily summary, to be used for the
last slot in a call to history with a frequency of `1d`.
This summary is the same as a daily bar rollup of minute data, with the
distinction that the summary is truncated to the `dt` requested.
i.e. the aggregation slides forward during a the course of simulation day.
Provides aggregation for `open`, `high`, `low`, `close`, and `volume`.
The aggregation rules for each price type is documented in their respective
"""
def __init__(self, market_opens, minute_reader, trading_calendar):
self._market_opens = market_opens
self._minute_reader = minute_reader
self._trading_calendar = trading_calendar
# The caches are structured as (date, market_open, entries), where
# entries is a dict of asset -> (last_visited_dt, value)
#
# Whenever an aggregation method determines the current value,
# the entry for the respective asset should be overwritten with a new
# entry for the current dt.value (int) and aggregation value.
#
# When the requested dt's date is different from date the cache is
# flushed, so that the cache entries do not grow unbounded.
#
# Example cache:
# cache = (date(2016, 3, 17),
# pd.Timestamp('2016-03-17 13:31', tz='UTC'),
# {
# 1: (1458221460000000000, np.nan),
# 2: (1458221460000000000, 42.0),
# })
self._caches = {
'open': None,
'high': None,
'low': None,
'close': None,
'volume': None
}
# The int value is used for deltas to avoid extra computation from
# creating new Timestamps.
self._one_min = pd.Timedelta('1 min').value
def _prelude(self, dt, field):
session = self._trading_calendar.minute_to_session_label(dt)
dt_value = dt.value
cache = self._caches[field]
if cache is None or cache[0] != session:
market_open = self._market_opens.loc[session]
cache = self._caches[field] = (session, market_open, {})
_, market_open, entries = cache
market_open = market_open.tz_localize('UTC')
if dt != market_open:
prev_dt = dt_value - self._one_min
else:
prev_dt = None
return market_open, prev_dt, dt_value, entries
def opens(self, assets, dt):
"""
The open field's aggregation returns the first value that occurs
for the day, if there has been no data on or before the `dt` the open
is `nan`.
Once the first non-nan open is seen, that value remains constant per
asset for the remainder of the day.
Returns
-------
np.array with dtype=float64, in order of assets parameter.
"""
market_open, prev_dt, dt_value, entries = self._prelude(dt, 'open')
opens = []
session_label = self._trading_calendar.minute_to_session_label(dt)
for asset in assets:
if not asset.is_alive_for_session(session_label):
opens.append(np.NaN)
continue
if prev_dt is None:
val = self._minute_reader.get_value(asset, dt, 'open')
entries[asset] = (dt_value, val)
opens.append(val)
continue
else:
try:
last_visited_dt, first_open = entries[asset]
if last_visited_dt == dt_value:
opens.append(first_open)
continue
elif not pd.isnull(first_open):
opens.append(first_open)
entries[asset] = (dt_value, first_open)
continue
else:
after_last = pd.Timestamp(
last_visited_dt + self._one_min, tz='UTC')
window = self._minute_reader.load_raw_arrays(
['open'],
after_last,
dt,
[asset],
)[0]
nonnan = window[~pd.isnull(window)]
if len(nonnan):
val = nonnan[0]
else:
val = np.nan
entries[asset] = (dt_value, val)
opens.append(val)
continue
except KeyError:
window = self._minute_reader.load_raw_arrays(
['open'],
market_open,
dt,
[asset],
)[0]
nonnan = window[~pd.isnull(window)]
if len(nonnan):
val = nonnan[0]
else:
val = np.nan
entries[asset] = (dt_value, val)
opens.append(val)
continue
return np.array(opens)
def highs(self, assets, dt):
"""
The high field's aggregation returns the largest high seen between
the market open and the current dt.
If there has been no data on or before the `dt` the high is `nan`.
Returns
-------
np.array with dtype=float64, in order of assets parameter.
"""
market_open, prev_dt, dt_value, entries = self._prelude(dt, 'high')
highs = []
session_label = self._trading_calendar.minute_to_session_label(dt)
for asset in assets:
if not asset.is_alive_for_session(session_label):
highs.append(np.NaN)
continue
if prev_dt is None:
val = self._minute_reader.get_value(asset, dt, 'high')
entries[asset] = (dt_value, val)
highs.append(val)
continue
else:
try:
last_visited_dt, last_max = entries[asset]
if last_visited_dt == dt_value:
highs.append(last_max)
continue
elif last_visited_dt == prev_dt:
curr_val = self._minute_reader.get_value(
asset, dt, 'high')
if pd.isnull(curr_val):
val = last_max
elif | pd.isnull(last_max) | pandas.isnull |
import collections
import gc
import os
from typing import List, Optional, Union, Tuple, Dict, Any
import albumentations
import numpy as np
import pandas as pd
import torch
from hydra.utils import instantiate
from pytorch_toolbelt.inference import (
ApplySigmoidTo,
ApplySoftmaxTo,
Ensembler,
GeneralizedTTA,
MultiscaleTTA,
d2_image_augment,
d4_image_augment,
d4_image_deaugment,
d2_image_deaugment,
flips_image_deaugment,
flips_image_augment,
fliplr_image_augment,
fliplr_image_deaugment,
)
from pytorch_toolbelt.utils import to_numpy, fs
from torch import nn
from torch.utils.data import DistributedSampler
from tqdm import tqdm
from pytorch_toolbelt.utils.distributed import is_main_process, get_rank, get_world_size, all_gather
from xview3.centernet.bboxer import (
MultilabelCircleNetDecodeResult,
MultilabelCircleNetCoder,
)
from xview3.centernet.constants import (
CENTERNET_OUTPUT_SIZE,
CENTERNET_OUTPUT_OFFSET,
CENTERNET_OUTPUT_OBJECTNESS_MAP,
CENTERNET_OUTPUT_VESSEL_MAP,
CENTERNET_OUTPUT_FISHING_MAP,
)
from xview3.centernet.models.inference import (
multilabel_centernet_tiled_inference,
get_box_coder_from_model,
)
from xview3.dataset import (
XView3DataModule,
read_multichannel_image,
stack_multichannel_image,
)
__all__ = [
"average_checkpoints",
"ensemble_from_checkpoints",
"model_from_checkpoint",
"ensemble_from_config",
"wrap_multilabel_model_with_tta",
"maybe_run_inference",
]
def average_checkpoints(inputs):
"""Loads checkpoints from inputs and returns a model with averaged weights. Original implementation taken from:
https://github.com/pytorch/fairseq/blob/a48f235636557b8d3bc4922a6fa90f3a0fa57955/scripts/average_checkpoints.py#L16
Args:
inputs (List[str]): An iterable of string paths of checkpoints to load from.
Returns:
A dict of string keys mapping to various values. The 'model' key
from the returned dict should correspond to an OrderedDict mapping
string parameter names to torch Tensors.
"""
params_dict = collections.OrderedDict()
params_keys = None
new_state = None
num_models = len(inputs)
for fpath in inputs:
with open(fpath, "rb") as f:
state = torch.load(
f,
map_location="cpu",
)
# Copies over the settings from the first checkpoint
if new_state is None:
new_state = state
model_params = state["model_state_dict"]
model_params_keys = list(model_params.keys())
if params_keys is None:
params_keys = model_params_keys
elif params_keys != model_params_keys:
raise KeyError("For checkpoint {}, expected list of params: {}, " "but found: {}".format(f, params_keys, model_params_keys))
for k in params_keys:
p = model_params[k]
if isinstance(p, torch.HalfTensor):
p = p.float()
if k not in params_dict:
params_dict[k] = p.clone()
# NOTE: clone() is needed in case of p is a shared parameter
else:
params_dict[k] += p
averaged_params = collections.OrderedDict()
for k, v in params_dict.items():
averaged_params[k] = v
if averaged_params[k].is_floating_point():
averaged_params[k].div_(num_models)
else:
averaged_params[k] //= num_models
new_state["model_state_dict"] = averaged_params
return new_state
def model_from_checkpoint(checkpoint_config: Union[str, Dict], **kwargs) -> Tuple[nn.Module, Dict]:
if isinstance(checkpoint_config, collections.Mapping):
if "average_checkpoints" in checkpoint_config:
checkpoint = average_checkpoints(checkpoint_config["average_checkpoints"])
else:
checkpoint_name = checkpoint_config["checkpoint"]
if os.path.isfile(checkpoint_name):
checkpoint = torch.load(checkpoint_name, map_location="cpu")
else:
checkpoint = torch.hub.load_state_dict_from_url(checkpoint_name)
model_config = checkpoint["checkpoint_data"]["config"]["model"]
else:
checkpoint_name = checkpoint_config
if os.path.isfile(checkpoint_name):
checkpoint = torch.load(checkpoint_name, map_location="cpu")
else:
checkpoint = torch.hub.load_state_dict_from_url(checkpoint_name)
model_config = checkpoint["checkpoint_data"]["config"]["model"]
model_state_dict = checkpoint["model_state_dict"]
model = instantiate(model_config, _recursive_=False)
model.load_state_dict(model_state_dict, strict=False)
return model.eval(), checkpoint
def wrap_multilabel_model_with_tta(model, tta_mode, with_offset=True, size_offsets=(0, -32, -64, +32, +64)):
from xview3.centernet import (
CENTERNET_OUTPUT_VESSEL_MAP,
CENTERNET_OUTPUT_FISHING_MAP,
CENTERNET_OUTPUT_OBJECTNESS_MAP,
CENTERNET_OUTPUT_OFFSET,
CENTERNET_OUTPUT_SIZE,
)
keys_to_deaug = [
CENTERNET_OUTPUT_VESSEL_MAP,
CENTERNET_OUTPUT_FISHING_MAP,
CENTERNET_OUTPUT_OBJECTNESS_MAP,
CENTERNET_OUTPUT_SIZE,
]
if with_offset:
keys_to_deaug.append(CENTERNET_OUTPUT_OFFSET)
def _make_deaug_dict(keys, fn):
return dict((key, fn) for key in keys)
if tta_mode == "d4":
return GeneralizedTTA(model, augment_fn=d4_image_augment, deaugment_fn=_make_deaug_dict(keys_to_deaug, d4_image_deaugment))
elif tta_mode == "ms":
return MultiscaleTTA(model, size_offsets)
elif tta_mode == "d2-ms":
return MultiscaleTTA(GeneralizedTTA(model, d2_image_augment, d2_image_deaugment), size_offsets)
elif tta_mode == "d2":
model = GeneralizedTTA(model, d2_image_augment, deaugment_fn=_make_deaug_dict(keys_to_deaug, d2_image_deaugment))
elif tta_mode == "flips":
model = GeneralizedTTA(model, flips_image_augment, deaugment_fn=_make_deaug_dict(keys_to_deaug, flips_image_deaugment))
elif tta_mode == "fliplr":
model = GeneralizedTTA(model, fliplr_image_augment, deaugment_fn=_make_deaug_dict(keys_to_deaug, fliplr_image_deaugment))
elif tta_mode is None:
return model
else:
raise KeyError("Unusupported TTA mode '" + tta_mode + "'")
return model
def ensemble_from_checkpoints(
checkpoint_fnames: List[str],
strict=True,
sigmoid_outputs=None,
softmax_outputs=None,
activation: str = "after_model",
tta: Optional[str] = None,
with_offset=True,
):
if activation not in {None, "None", "after_model", "after_tta", "after_ensemble"}:
raise KeyError(activation)
models = []
checkpoints = []
for ck in checkpoint_fnames:
model, checkpoint = model_from_checkpoint(ck, strict=strict)
models.append(model)
checkpoints.append(checkpoint)
if activation == "after_model":
if sigmoid_outputs is not None:
models = [ApplySigmoidTo(m, output_key=sigmoid_outputs) for m in models]
print("Applying sigmoid activation to", sigmoid_outputs, "after each model", len(models))
if softmax_outputs is not None:
models = [ApplySoftmaxTo(m, output_key=softmax_outputs) for m in models]
print("Applying softmax activation to", softmax_outputs, "after each model", len(models))
if len(models) > 1:
model = Ensembler(models)
if activation == "after_ensemble":
if sigmoid_outputs is not None:
model = ApplySigmoidTo(model, output_key=sigmoid_outputs)
print("Applying sigmoid activation to", sigmoid_outputs, "after ensemble")
if softmax_outputs is not None:
model = ApplySoftmaxTo(model, output_key=softmax_outputs)
print("Applying softmax activation to", softmax_outputs, "after ensemble")
else:
assert len(models) == 1
model = models[0]
if tta not in {None, "None"}:
model = wrap_multilabel_model_with_tta(model, tta, with_offset=with_offset)
print("Wrapping models with TTA", tta)
if activation == "after_tta":
if sigmoid_outputs is not None:
model = ApplySigmoidTo(model, output_key=sigmoid_outputs)
print("Applying sigmoid activation to ", sigmoid_outputs, " after TTA")
if softmax_outputs is not None:
model = ApplySoftmaxTo(model, output_key=softmax_outputs)
print("Applying softmax activation to", softmax_outputs, "after TTA")
return model.eval(), checkpoints
def ensemble_from_config(config: Dict[str, Any]):
model, checkpoints = ensemble_from_checkpoints(
checkpoint_fnames=config["ensemble"]["models"],
strict=True,
activation=config["ensemble"]["activation_after"],
tta=config["ensemble"]["tta"],
sigmoid_outputs=config["ensemble"]["sigmoid_outputs"],
softmax_outputs=config["ensemble"]["softmax_outputs"],
with_offset=config["ensemble"]["with_offset"],
)
box_coder = get_box_coder_from_model(model)
model = model.eval().cuda()
return model, checkpoints, box_coder
@torch.jit.optimized_execution(False)
def predict_multilabel_scenes(
model,
box_coder: MultilabelCircleNetCoder,
scenes: List[str],
channels: List[str],
tile_step: int,
tile_size: int,
objectness_thresholds_lower_bound: float,
normalization: Dict[str, albumentations.ImageOnlyTransform],
accumulate_on_gpu: bool,
fp16: bool,
batch_size: int,
apply_activation: bool,
save_raw_predictions: bool,
max_objects: int,
channels_last: bool,
output_predictions_dir=None,
) -> pd.DataFrame:
if output_predictions_dir is not None:
os.makedirs(output_predictions_dir, exist_ok=True)
all_predictions = []
scenes = np.array(scenes)
world_size, local_rank = get_world_size(), get_rank()
if world_size > 1:
sampler = DistributedSampler(scenes, world_size, local_rank, shuffle=False)
rank_local_indexes = np.array(list(iter(sampler)))
scenes = scenes[rank_local_indexes]
print("Node", local_rank, "got", len(scenes), "to process")
torch.distributed.barrier()
for scene in tqdm(scenes, desc=f"Inference at Node {local_rank}/{world_size}", position=local_rank):
gc.collect()
scene_id = fs.id_from_fname(scene)
predictions = maybe_run_inference(
model=model,
box_coder=box_coder,
scene=scene,
output_predictions_dir=output_predictions_dir,
accumulate_on_gpu=accumulate_on_gpu,
tile_size=tile_size,
tile_step=tile_step,
fp16=fp16,
batch_size=batch_size,
save_raw_predictions=save_raw_predictions,
apply_activation=apply_activation,
max_objects=max_objects,
channels_last=channels_last,
normalization=normalization,
channels=channels,
objectness_thresholds_lower_bound=objectness_thresholds_lower_bound,
)
all_predictions.append(predictions)
if output_predictions_dir is not None:
predictions.to_csv(os.path.join(output_predictions_dir, scene_id + ".csv"), index=False)
all_predictions = pd.concat(all_predictions).reset_index(drop=True)
if world_size > 1:
torch.distributed.barrier()
all_predictions = pd.concat(all_gather(all_predictions)).reset_index(drop=True)
return all_predictions
def maybe_run_inference(
model,
box_coder,
scene,
output_predictions_dir,
channels,
normalization,
objectness_thresholds_lower_bound: float,
tile_size,
tile_step,
accumulate_on_gpu,
fp16,
batch_size,
save_raw_predictions,
apply_activation,
max_objects,
channels_last,
):
scene_id = fs.id_from_fname(scene)
predictions_computed_offline = False
if output_predictions_dir is not None:
raw_predictions_file = os.path.join(output_predictions_dir, scene_id + ".npz")
decoded_predictions_file = os.path.join(output_predictions_dir, scene_id + ".csv")
if os.path.isfile(decoded_predictions_file):
try:
predictions = pd.read_csv(decoded_predictions_file)
return predictions
except Exception as e:
print(e)
predictions_computed_offline = False
elif os.path.isfile(raw_predictions_file):
try:
saved_predictions = np.load(raw_predictions_file, allow_pickle=True)
outputs = dict(
CENTERNET_OUTPUT_OBJECTNESS_MAP=torch.from_numpy(saved_predictions[CENTERNET_OUTPUT_OBJECTNESS_MAP]),
CENTERNET_OUTPUT_VESSEL_MAP=torch.from_numpy(saved_predictions[CENTERNET_OUTPUT_VESSEL_MAP]),
CENTERNET_OUTPUT_FISHING_MAP=torch.from_numpy(saved_predictions[CENTERNET_OUTPUT_FISHING_MAP]),
CENTERNET_OUTPUT_SIZE=torch.from_numpy(saved_predictions[CENTERNET_OUTPUT_SIZE]),
CENTERNET_OUTPUT_OFFSET=torch.from_numpy(saved_predictions[CENTERNET_OUTPUT_OFFSET])
if CENTERNET_OUTPUT_OFFSET in saved_predictions
else None,
)
predictions_computed_offline = True
except Exception as e:
print(e)
predictions_computed_offline = False
if not predictions_computed_offline:
image = read_multichannel_image(scene, channels)
for channel_name in set(channels):
image[channel_name] = normalization[channel_name](image=image[channel_name])["image"]
image = stack_multichannel_image(image, channels)
outputs = multilabel_centernet_tiled_inference(
model,
image,
box_coder=box_coder,
tile_size=tile_size,
tile_step=tile_step,
accumulate_on_gpu=accumulate_on_gpu,
fp16=fp16,
batch_size=batch_size,
channels_last=channels_last,
)
if save_raw_predictions and output_predictions_dir is not None:
raw_predictions_file = os.path.join(output_predictions_dir, scene_id + ".npz")
predictions_dict = dict(
CENTERNET_OUTPUT_OBJECTNESS_MAP=to_numpy(outputs[CENTERNET_OUTPUT_OBJECTNESS_MAP]),
CENTERNET_OUTPUT_VESSEL_MAP=to_numpy(outputs[CENTERNET_OUTPUT_VESSEL_MAP]),
CENTERNET_OUTPUT_FISHING_MAP=to_numpy(outputs[CENTERNET_OUTPUT_FISHING_MAP]),
CENTERNET_OUTPUT_SIZE=to_numpy(outputs[CENTERNET_OUTPUT_SIZE]),
)
if CENTERNET_OUTPUT_OFFSET in outputs:
predictions_dict[CENTERNET_OUTPUT_OFFSET] = to_numpy(outputs[CENTERNET_OUTPUT_OFFSET])
np.savez(raw_predictions_file, **predictions_dict)
preds: MultilabelCircleNetDecodeResult = box_coder.decode(
objectness_map=outputs[CENTERNET_OUTPUT_OBJECTNESS_MAP],
is_vessel_map=outputs[CENTERNET_OUTPUT_VESSEL_MAP],
is_fishing_map=outputs[CENTERNET_OUTPUT_FISHING_MAP],
length_map=outputs[CENTERNET_OUTPUT_SIZE],
offset_map=outputs.get(CENTERNET_OUTPUT_OFFSET, None),
apply_activation=apply_activation,
max_objects=max_objects,
)
pos_mask = preds.scores[0] >= objectness_thresholds_lower_bound
centers = to_numpy(preds.centers[0][pos_mask]).astype(int)
scores = to_numpy(preds.scores[0, pos_mask]).astype(np.float32)
lengths = XView3DataModule.decode_lengths(preds.lengths[0, pos_mask])
is_vessel_prob = to_numpy(preds.is_vessel[0, pos_mask]).astype(np.float32)
is_fishing_prob = to_numpy(preds.is_fishing[0, pos_mask]).astype(np.float32)
predictions = collections.defaultdict(list)
for (
(detect_scene_column, detect_scene_row),
objectness_score,
is_vessel_p,
is_fishing_p,
vessel_length_m,
) in zip(centers, scores, is_vessel_prob, is_fishing_prob, lengths):
predictions["vessel_length_m"].append(vessel_length_m)
predictions["detect_scene_row"].append(detect_scene_row)
predictions["detect_scene_column"].append(detect_scene_column)
predictions["scene_id"].append(scene_id)
# Scores
predictions["objectness_p"].append(objectness_score)
predictions["is_vessel_p"].append(is_vessel_p)
predictions["is_fishing_p"].append(is_fishing_p)
# Thresholds
predictions["objectness_threshold"].append(objectness_thresholds_lower_bound)
predictions = | pd.DataFrame.from_dict(predictions) | pandas.DataFrame.from_dict |
import numpy as np
import pandas as pd
import theano.tensor as T
from random import shuffle
from theano import shared, function
from patsy import dmatrix
from collections import defaultdict
class MainClauseModel(object):
def __init__(self, nlatfeats=8, alpha=1., discount=None, beta=0.5, gamma=0.9,
delta=2., orthogonality_penalty=0., nonparametric=False):
'''
Parameters
----------
nlatfeats : int
Number of latent features for each verb; the default of 8 is
the number of unique subcat frames in the data
alpha : float (positive)
Beta process hyperparameter as specified in Teh et al. 2007
"Stick-breaking Construction for the Indian Buffet Process";
changes meaning based on Pitman-Yor discount hyperparameter
(see Teh et al. 2007, p.3)
discount : float (unit) or None
If discount is a float, it must satisfy alpha > -discount
beta : float (positive)
If parametric=True, concetration parameter for verb-specific
beta draws based on beta process sample; if nonparametric=False,
hyperparameter of a Beta(beta, beta); in the latter case, beta
should be on (0,1), otherwise the verb representations are
unidentifiable, since their is a flat prior on the selection
probability
gamma : float (positive)
Hyperparameter of a beta distribution on the projection matrix
delta : float (positive)
Hyperparameter of a beta distribution on the verb feature
probability matrix
orthogonality_penalty : float (positive)
How much to penalize for singularity
nonparametric : bool
Whether to use a nonparametric prior
divergence_weight : float (0 to negative infinity) (ADDED)
How much to weight the either-or bias. If 0, no either-or bias.
'''
self.nlatfeats = nlatfeats
self.alpha = alpha
self.discount = discount
self.beta = beta
self.gamma = gamma
self.delta = delta
self.orthogonality_penalty = orthogonality_penalty
self.nonparametric = nonparametric
self.divergence_weight = -1
self._validate_params()
self._ident = ''.join(np.random.choice(9, size=10).astype(str))
def _validate_params(self):
if self.discount is not None:
self._pitman_yor = True
try:
assert self.alpha > -self.discount
except AssertionError:
raise ValueError('alpha must be greater than -discount')
else:
self._pitman_yor = False
def _initialize_model(self, data, stochastic):
self.data = data
self._initialize_counter()
self._initialize_reps()
self._initialize_loss()
self._initialize_updaters(stochastic)
def _initialize_counter(self):
self._verbcount = T.zeros(self.data.n('verb'))
self._verbeye = T.eye(self.data.n('verb'))
def _initialize_reps(self):
self._reps = {}
if self.nonparametric:
# nu_aux = np.array([2.]+[-1.]*(self.nlatfeats-1))
nu_aux = np.array([0.]*self.nlatfeats)
self._reps['nu'] = shared(nu_aux, name='nu')
self._nu = T.nnet.sigmoid(self._reps['nu'])
self._mu = T.cumprod(self._nu)
verbreps_aux = np.random.normal(0., 1e-2, size=[self.data.n('verb')-self.data.n('clausetype'),
self.nlatfeats])
projection_aux = np.random.normal(0., 1e-2, size=[self.nlatfeats, self.data.n('feature')])
verbfeatprob_aux = np.zeros([self.data.n('verb')-self.data.n('clausetype'), self.nlatfeats])-4.
if self.data.n('clausetype'):
try:
assert self.data.n('clausetype') <= self.nlatfeats
except AssertionError:
raise ValueError('nlatfeats must be greater than or equal to the number of clausetypes')
ctype_ident = (1.-1e-10)*np.eye(self.data.n('clausetype'))
ct_aux_vr = np.log(ctype_ident)-np.log(1.-ctype_ident)
ct_aux_vr = np.concatenate([ct_aux_vr, -np.inf*np.ones([self.data.n('clausetype'),
self.nlatfeats-self.data.n('clausetype')])],
axis=1)
ct_aux_vfp = np.inf*np.ones([self.data.n('clausetype'), self.nlatfeats])
verbreps_aux = np.concatenate([ct_aux_vr, verbreps_aux])
verbfeatprob_aux = np.concatenate([ct_aux_vfp, verbfeatprob_aux])
self._reps['verbreps'] = shared(verbreps_aux, name='verbreps')
self._reps['projection'] = shared(projection_aux, name='projection')
self._reps['verbfeatprob'] = shared(verbfeatprob_aux, name='verbfeatprob')
self._verbreps = T.nnet.sigmoid(self._reps['verbreps'])
self._projection = T.nnet.sigmoid(self._reps['projection'])
self._verbfeatprob = T.nnet.sigmoid(self._reps['verbfeatprob'])
softand = self._verbfeatprob[:,:,None]*self._verbreps[:,:,None]*self._projection[None,:,:]
self._featureprob = 1.-T.prod(1.-softand, axis=1)
# Added to White et al. model: divergence function. Calculates JS-divergence (cf. SciPy version which yields the square root value)
def _get_js_divergence(self):
vr = self._verbreps
assertProb = vr[:, 0] #s_v,belief
requestProb = vr[:, 1] #s_v,desire
m0 = (assertProb + 1-requestProb)/2
m1 = (1-assertProb + requestProb)/2
kl_assert = (assertProb * T.log(assertProb / m0)
+ (1-assertProb) * T.log((1-assertProb) / m1))
kl_request = ((1-requestProb) * T.log((1-requestProb) / m0)
+ requestProb * T.log(requestProb / m1))
js = ((kl_assert + kl_request) / 2 )**1
# Above code leads to NaN error for verbs 0 and 1 (DECLARATIVE & IMPERATIVE), probably because of how Theano deals with floating point representations
# These should be 0. Stipulate them as such.
# cf. https://stackoverflow.com/questions/31919818/theano-sqrt-returning-nan-values.
js = T.set_subtensor(js[0], 0.) # try ... js[tuple([0,])], 0...
js = T.set_subtensor(js[1], 0.)
return js
# Added to White et al. model: divergence function. Calculates KL-divergence
def _get_kl_divergence(self):
vr = self._verbreps
assertProb = vr[:, 0]
requestProb = vr[:, 1]
kl_assert = (assertProb * T.log(assertProb / (1-requestProb))
+ (1-assertProb) * T.log((1-assertProb) / requestProb))
kl_request = ((1-requestProb) * T.log((1-requestProb) / assertProb)
+ requestProb * T.log(requestProb / (1-assertProb)))
kl = ((kl_assert + kl_request) / 2 )**1
# Above code leads to NaN error for verbs 0 and 1 (DECLARATIVE & IMPERATIVE), probably because of how Theano deals with floating point representations
# These should be 0. Stipulate them as such.
# cf. https://stackoverflow.com/questions/31919818/theano-sqrt-returning-nan-values.
kl = T.set_subtensor(kl[0], 0.) # try ... js[tuple([0,])], 0...
kl = T.set_subtensor(kl[1], 0.)
return kl
def _initialize_loss(self):
self._log_projection_prior = (self.gamma-1.)*T.log(self._projection) +\
(self.gamma-1.)*T.log(1.-self._projection)
self._log_verbfeatureprob_prior = (self.delta-1.)*T.log(self._verbfeatprob) +\
(self.delta-1.)*T.log(1.-self._verbfeatprob)
# self._log_verbfeatureprob_prior = -T.log(self._verbfeatprob)/T.log(self._verbreps)
if self.nonparametric:
def betaln(alpha, beta):
return T.gammaln(alpha) + T.gammaln(beta) - T.gammaln(alpha+beta)
if self._pitman_yor:
upper_a = self.alpha + self.nlatfeats*self.discount
upper_b = 1.-self.discount
else:
upper_a = 1.
upper_b = self.alpha
self._log_upper_prior = (upper_a-1.)*T.log(self._nu) +\
(upper_b-1.)*T.log(1.-self._nu) -\
betaln(upper_a, upper_b)
lower_a = self.beta*self._mu
lower_b = self.beta*(1.-self._mu)
self._log_lower_prior = (lower_a-1.)[None,:]*T.log(self._verbreps) +\
(lower_b-1.)[None,:]*T.log(1.-self._verbreps) -\
betaln(lower_a, lower_b)[None,:]
self._prior = T.sum(self._log_upper_prior)/self.nlatfeats +\
T.sum(self._log_lower_prior)/(self.data.n('verb')*self.nlatfeats) +\
T.sum(self._log_projection_prior)/(self.data.n('feature')*self.nlatfeats)+\
T.sum(self._log_verbfeatureprob_prior)/(self.data.n('verb')*self.nlatfeats)
else:
self._log_verbreps_prior = (self.beta-1.)*T.log(self._verbreps) +\
(self.beta-1.)*T.log(1.-self._verbreps)
self._prior = T.sum(self._log_verbreps_prior)/(self.data.n('verb')*self.nlatfeats) +\
T.sum(self._log_projection_prior)/(self.data.n('feature')*self.nlatfeats)+\
T.sum(self._log_verbfeatureprob_prior)/(self.data.n('verb')*self.nlatfeats)
if self.orthogonality_penalty:
verbrep2 = T.dot(self._verbreps.T, self._verbreps)
verbrep2_rawsum = T.sum(T.square(verbrep2 - verbrep2*T.identity_like(verbrep2)))
self._orthogonality_penalty = -self.orthogonality_penalty*\
verbrep2_rawsum/(self.nlatfeats*self.data.n('verb'))
else:
self._orthogonality_penalty = 0.
p = self._featureprob[self.data.verb]
k = self.data.features
# r = 1./self._verbreps.sum(axis=1)[self.data.verb,None]
#self._ll_per_feature = k*T.log(p)+r*T.log(1.-p)+T.gammaln(k+r)-T.gammaln(k+1)-T.gammaln(r)
self._ll_per_feature = k*T.log(p)+(1.-k)*T.log(1.-p) # log likelihood, by defn. negative (log 1 = 0)
self._total_ll = T.sum(self._ll_per_feature)/(self.data.verb.shape[0]*\
self.data.n('feature'))
self._total_loss = self._prior+self._orthogonality_penalty+self._total_ll
self._itr = T.ivector('itr')
## Added to White et al. model
# Option A: mean of JS divergence for observed verbs
self._divergence = T.mean(self._get_js_divergence()[self.data.verb][self._itr])*self.divergence_weight
# Option B: mean of KL divergence for observed verbs
# self._divergence = T.mean(self._get_kl_divergence()[self.data.verb][self._itr])*self.divergence_weight
# Other options:
# T.mean(self._get_js_divergence()) # Option A1: mean of JS divergence for ALL verbs, regardless of verbs observed for the particular utterance
# T.mean(self._get_kl_divergence()) # Option B1: mean of KL divergence for ALL verbs, regardless of verbs observed for the particular utterance
self._itr_ll = T.sum(self._ll_per_feature[self._itr])/self.data.n('feature')
self._itr_loss = self._prior+self._orthogonality_penalty+self._itr_ll + self._divergence
# Subtract divergence. Effectively, we are taking the raw log-likelihood (_ll_per_feature), a negative value, and adjusting it by this divergence score. Both JSD and KLD yield a positive value. Since the model tries to maximize log-likelihood, we want the adjusted log-likelihood to be lower when the divergence score is high. One way to do so is adjust divergence with a negative weight, effectively subtracting divergence from log-likelihood.
def _initialize_updaters(self, stochastic):
update_dict_ada = []
self.rep_grad_hist_t = {}
for name, rep in self._reps.items():
if stochastic:
rep_grad = T.grad(-self._itr_loss, rep)
else:
rep_grad = T.grad(-self._total_loss, rep)
if name in ['verbreps', 'projection', 'verbfeatprob']:
rep_grad = T.switch((rep>10)*(rep_grad<0),
T.zeros_like(rep_grad),
rep_grad)
rep_grad = T.switch((rep<-10)*(rep_grad>0),
T.zeros_like(rep_grad),
rep_grad)
# Incorporating divergence causes verbreps gradients for DECLARATIVE and IMPERATIVE to equal NaN; so replace NaN with 0s (declaratives and imperative gradients don't change)
rep_grad = T.switch(T.isnan(rep_grad), 0., rep_grad)
self.rep_grad_hist_t[name] = shared(np.ones(rep.shape.eval()),
name=name+'_hist'+self._ident)
rep_grad_adj = rep_grad / (T.sqrt(self.rep_grad_hist_t[name]))
learning_rate = 2.# if name != 'nu' else 1e-20
update_dict_ada += [(self.rep_grad_hist_t[name], self.rep_grad_hist_t[name] +\
T.power(rep_grad, 2)),
(rep, rep - learning_rate*rep_grad_adj)]
self.updater_ada = function(inputs=[self._itr],
outputs=[self._total_ll, self._itr_ll,
self._verbreps, self._projection, self._divergence],
updates=update_dict_ada,
name='updater_ada'+self._ident)
def _fit(self, sentid, nupdates, verbose):
for j, sid in enumerate(sentid):
idx = self.data.sentence(sid)
for i in range(nupdates):
total_loss, itr_loss, verbreps, projection, divergence = self.updater_ada(idx)
if not j % 10:
self._verbreps_hist.append(verbreps)
self._projection_hist.append(projection)
if verbose:
verb_list = list(self.data.categories('verb')[np.array(self.data.verb)[idx]])
print('\n', j, '\tloss', np.round(total_loss, 3), '\titr_loss',\
np.round(itr_loss,3), '\tdiverge', np.round(divergence, 7), '\t', verb_list,'\n',
'\t', verb_list,'\t verb ID', np.array(self.data.verb)[idx]
)
def fit(self, data, nepochs=0, niters=20000, nupdates=1,
stochastic=True, verbose=True):
self._initialize_model(data, stochastic)
sentid = list(self.data.categories('sentenceid'))
self._verbreps_hist = []
self._projection_hist = []
if nepochs:
for e in range(nepochs):
shuffle(sentid)
if verbose:
print(e)
self._fit(sentid, nupdates, verbose)
else:
order = np.random.choice(sentid, size=niters)
self._fit(order, nupdates, verbose)
return self
@property
def verbreps(self):
return pd.DataFrame(T.nnet.sigmoid(self._reps['verbreps']).eval(),
index=self.data.categories('verb'))
@property
def verbfeatprob(self):
return pd.DataFrame(T.nnet.sigmoid(self._reps['verbfeatprob']).eval(),
index=self.data.categories('verb'))
@property
def projection(self):
return pd.DataFrame(T.nnet.sigmoid(self._reps['projection']).eval(),
columns=self.data.feature_names)
@property
def verbreps_history(self):
reps = []
for t, r in enumerate(self._verbreps_hist):
r = pd.DataFrame(r)
r['verb'] = self.data.categories('verb')
r['sentence'] = t
reps.append(r)
return | pd.concat(reps) | pandas.concat |
"""<NAME>0.
MLearner Machine Learning Library Extensions
Author:<NAME><www.linkedin.com/in/jaisenbe>
License: MIT
"""
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
import os
import datetime
import time
import joblib
from sklearn.base import BaseEstimator, ClassifierMixin
from sklearn.model_selection import train_test_split
from sklearn.metrics import accuracy_score
import xgboost as xgb
from xgboost import XGBClassifier
import seaborn as sns
from mlearner.training import Training
from mlearner.utils import ParamsManager
import warnings
warnings.filterwarnings("ignore")
param_file = "mlearner/classifier/config/models.json"
class modelXGBoost(Training, BaseEstimator, ClassifierMixin):
"""
XGBoost is an optimized distributed gradient boosting library designed to be highly efficient,
flexible and portable. It implements machine learning algorithms under the Gradient Boosting framework.
XGBoost provides a parallel tree boosting (also known as GBDT, GBM) that solve many data science
problems in a fast and accurate way. The same code runs on major distributed environment
(Hadoop, SGE, MPI) and can solve problems beyond billions of examples.
Parameters
----------
"min_child_weight": [ Minimum sum of instance weight (hessian) needed in a child.
"objective": learning task.
"eval_metric": Evaluation metrics for validation data.
"max_depth": Maximum depth of a tree. Increasing this value will make the model more complex and more likely to overfit
"max_delta_step": /Maximum delta step we allow each leaf output to be. If the value is set to 0, it means there is no constraint.
"sampling_method": The method to use to sample the training instances.
"subsample": Subsample ratio of the training instances. Setting it to 0.5 means that XGBoost would randomly sample half of the training data prior to growing trees. and this will prevent overfitting.
"eta": tep size shrinkage used in update to prevents overfitting.
"gamma": Minimum loss reduction required to make a further partition on a leaf node of the tree.
"lambda": L2 regularization term on weights. Increasing this value will make model more conservative.
"alpha": L1 regularization term on weights. Increasing this value will make model more conservative.
"tree_method": he tree construction algorithm used in XGBoost.
"predictor": The type of predictor algorithm to use.
"num_parallel_tree": umber of parallel trees constructed during each iteration.
...
Documentation
-------------
https://xgboost.readthedocs.io/en/latest/
https://www.analyticsvidhya.com/blog/2016/03/complete-guide-parameter-tuning-xgboost-with-codes-python/
"""
def __init__(self, name="XGB", random_state=99, train_dir="", params=None, *args, **kwargs):
self.name = name
self.train_dir = train_dir + "/" + "model_" + str(self.name) + "/"
self.random_state = random_state
if params is None:
self.get_params_json()
self.params.update({
'model_dir': self.train_dir,
"seed": self.random_state})
else:
# if isinstance(params)
self.params = params
self.model = XGBClassifier(**self.params)
super().__init__(self.model, random_state=self.random_state)
def get_params_json(self):
self.manager_models = ParamsManager(param_file, key_read="Models")
self.params = self.manager_models.get_params()["XGBoost"]
self.manager_finetune = ParamsManager(param_file, key_read="FineTune")
self.params_finetune = self.manager_finetune.get_params()["XGBoost"]
def dataset(self, X, y, categorical_columns_indices=None, test_size=0.2, *args, **kwarg):
self.categorical_columns_indices = categorical_columns_indices
self.X = X
self.columns = list(X)
self.y, self.cat_replace = self.replace_multiclass(y)
self.X_train, self.X_test, self.y_train, self.y_test = train_test_split(self.X, self.y, test_size=test_size, random_state=self.random_state)
self.dtrain = xgb.DMatrix(self.X_train, label=self.y_train)
self.dvalid = xgb.DMatrix(self.X_test, label=self.y_test)
self.all_train_data = xgb.DMatrix(self.X, label=self.y)
def set_dataset_nosplit(self, X_train, X_test, y_train, y_test, categorical_columns_indices=None, *args, **kwarg):
self.categorical_columns_indices = categorical_columns_indices
self.columns = list(X_train)
_ytrain, _ = self.replace_multiclass(y_train)
_ytest, _ = self.replace_multiclass(y_test)
self.X_train = X_train
self.X_test = X_test
self.y_train = y_train
self.y_test = y_test
self.X = | pd.concat([X_train, X_test], axis=0) | pandas.concat |
import os
import csv
import pandas as pd
# Receives the STS-B and creates gender-occupation datasets
# Inspired by counterfactual data augmentation method as introduced here: https://arxiv.org/pdf/1807.11714.pdf
class CreateGenderStsb():
def __init__(self, lang=None, data_dir=None, occupation=None, multilingual=None):
self.lang = lang
self.data_dir = data_dir
self.occupation = occupation
self.multilinugal = multilingual
def create_gendered_dataframes(self):
"""
Creates one dataframe for "he" and one for "she".
Each dataset consists of 173 pairs of sentences:
each pair contains one gendered sentence and one that contains the occupation.
"""
df = self.create_dataframe()
# create men and women dataframes
women = df[df['gender'] == 'woman']
men = df[df['gender'] == 'man']
# create copies of men and women dataframes
men2 = women.copy()
women2 = men.copy()
# transform the copies to opposite gender ones
if self.lang == 'sv':
men2['sentence1'] = men2['sentence1'].apply(lambda x: self.replace_with(x, 'man'))
women2['sentence1'] = women2['sentence1'].apply(lambda x: self.replace_with(x, 'kvinna'))
if self.lang == 'en':
men2['sentence1'] = men2['sentence1'].apply(lambda x: self.replace_with(x, 'man'))
women2['sentence1'] = women2['sentence1'].apply(lambda x: self.replace_with(x, 'woman'))
# concatenate dataframes of same gender
women_df = | pd.concat([women, women2]) | pandas.concat |
from os.path import join, exists
from os import mkdir, remove
from io import StringIO
from subprocess import call, run as r, DEVNULL, STDOUT
from Bio import SeqIO
from Bio.SeqRecord import SeqRecord
import pandas as pd
from .plotting import plot_alignment, plot_genbank
class Insertion():
def __init__(self, args):
# Parsed files
self.out_dir = args.out_dir
self.reference = args.ancestral
self.mutant_gbk = args.mutant
# Step and window size for chunking
self.step = 100
self.window = 500
# Mutant contigs and features
self.mutant_contigs = [contig for contig in SeqIO.parse(
self.mutant_gbk, 'genbank')]
self.mutant_features = self.parse_genbank()
self.mutant_fasta = join(self.out_dir, 'mutant.fasta')
with open(join(self.out_dir, 'mutant.fasta'), 'w') as handle:
SeqIO.write(self.mutant_contigs, handle, 'fasta')
# Reference contigs
self.reference_contigs = [contig for contig in SeqIO.parse(
self.reference, 'fasta')]
# Chunked mutant
self.chunks = join(self.out_dir,
"chunked_sequences.fasta")
# Mutant reference alignments
self.bam = join(self.out_dir, "aligned.sorted.bam")
# Out dataframes
self. insertions = | pd.DataFrame(columns=['chromosome', 'position', 'length']) | pandas.DataFrame |
import math
import shapely
import param
import panel as pn
from holoviews import streams
import geopandas as gpd
import geoviews as gv
import pandas as pd
from pydsm.hydroh5 import HydroH5
import datetime
import holoviews as hv
from holoviews import opts
import hvplot.pandas
hv.extension('bokeh')
gv.extension('bokeh')
pn.extension()
# code
def calc_angle(coords):
c2 = coords[1]
c1 = coords[0]
return math.degrees(math.atan2(c2[1] - c1[1], c2[0] - c1[0]))
assert calc_angle([(0, 0), (1, 1)]) == 45
def line(e, n, length, angle):
# for a size of length 10 and angle 0 as a basic shape
pa = shapely.geometry.LineString([(0, 0), (10, 0)])
pa = shapely.affinity.rotate(pa, angle, origin=(0, 0))
pa = shapely.affinity.scale(pa, length, length, origin=(0, 0))
pa = shapely.affinity.translate(pa, e, n)
return pa
def arrow(e, n, angle):
# for side 10 and equilateral triangle ie. 60/2 = 30 degs
s = 10
a = math.radians(30)
pa = shapely.geometry.Polygon(
[(2 / 3. * s * math.sin(a), 0),
(-s / 3 * math.cos(a), -s * math.sin(a)),
(-s / 3 * math.cos(a), s * math.sin(a))])
scale = 100
pa = shapely.affinity.rotate(pa, angle, origin=(0, 0))
pa = shapely.affinity.scale(pa, scale, scale, origin=(0, 0))
pa = shapely.affinity.translate(pa, e, n)
return pa
class InputStage(param.Parameterized):
base_dir = param.String('.')
@param.output(('filename_base', param.String), ('filename_alt', param.String))
def output(self):
self.filename_base = self.file_selector.value[0]
self.filename_alt = self.file_selector.value[1]
return self.filename_base, self.filename_alt
def panel(self):
self.file_selector = pn.widgets.FileSelector(directory=self.base_dir)
return pn.Column(self.file_selector)
class StudyComparisionDashboard(param.Parameterized):
filename_base = param.String()
filename_alt = param.String()
twstr = param.String()
base_dir = param.String('.')
# all these are hardwired
tw_list_file = param.String(f'timeperiods.txt')
channels_flow_file = param.String(f'channels_flow.txt')
stage_channels_file = param.String(f'channels_stage.txt')
dsm2_channels_gis_file = param.String(
f'v8.2-opendata/gisgridmapv8.2channelstraightlines/dsm2_channels_straightlines_8_2.shp')
ltrb_bounds = (-121.63, 37.93, -121.27, 37.76) # south delta bounds
time_periods = param.List()
tw_selector = param.Selector()
def __init__(self, **kwargs):
super().__init__(**kwargs)
self.view_pane = pn.Column("Building view")
@param.depends('base_dir', watch=True, on_init=True)
def _update_base_dir(self):
self.channels_flow_file = f'{self.base_dir}/channels_flow.txt'
self.stage_channels_file = f'{self.base_dir}/channels_stage.txt'
self.dsm2_channels_gis_file = f'{self.base_dir}/v8.2-opendata/gisgridmapv8.2channelstraightlines/dsm2_channels_straightlines_8_2.shp'
self.tw_list_file = f'{self.base_dir}/timeperiods.txt'
self.time_periods = self.load_time_periods(self.tw_list_file)
def load_time_periods(self, time_periods_file):
time_periods = pd.read_csv(time_periods_file)
# time_periods
time_periods['start_time'] = time_periods.iloc[:, 0:3].apply(
lambda x: datetime.datetime(*x.values), axis="columns")
time_periods['end_time'] = time_periods.iloc[:, 3:6].apply(
lambda x: datetime.datetime(*x.values), axis="columns")
time_periods = time_periods.assign(twstr=time_periods['start_time'].dt.strftime(
'%d%b%Y').str.upper() + '-' + time_periods['end_time'].dt.strftime('%d%b%Y').str.upper())
return time_periods['twstr'].to_list()
def set_map_bounds(self, map):
# set bounds
e_lt, n_lt = hv.util.transform.lon_lat_to_easting_northing(
longitude=self.ltrb_bounds[0], latitude=self.ltrb_bounds[1])
e_rb, n_rb = hv.util.transform.lon_lat_to_easting_northing(
longitude=self.ltrb_bounds[2], latitude=self.ltrb_bounds[3])
return map.opts(xlim=(e_lt, e_rb), ylim=(n_rb, n_lt)).opts(width=900, height=500)
def create_dsm2_channel_map(self):
"""
ltrb: left, top, right, bottom longitudes and latitudes
"""
self.dsm2_channels = gpd.read_file(self.dsm2_channels_gis_file)
# epsg 3857 for web mercator which is what tiles are based on
self.dsm2_channels = self.dsm2_channels.to_crs(epsg=3857)
map = self.dsm2_channels.hvplot(tiles='CartoLight', hover_cols='all')
# set bounds
map = self.set_map_bounds(map)
map = map.opts(opts.Path(line_width=5, line_color="black"))
return map
def get_data(hydro, channel_id, location, twstr, vartype):
if vartype == 'flow':
data = hydro.get_channel_flow(str(channel_id), location, twstr)
else:
data = hydro.get_channel_stage(str(channel_id), location, twstr)
return data
def get_mean_flow_data(hydro, twstr, channel_ids):
return [StudyComparisionDashboard.get_data(hydro, channel_id, 'upstream', twstr, 'flow').mean().values[0] for channel_id in channel_ids]
def load_stage_channels(self):
self.stage_channels = pd.read_csv(
self.stage_channels_file, header=None, names=['channel_id'])
# stage_channels
self.stage_channels = self.dsm2_channels[self.dsm2_channels['id'].isin(
self.stage_channels.channel_id)]
self.stage_channels = self.stage_channels.reset_index(
drop=True).rename(columns={'id': 'channel_id'})
def get_all_stage_data(self, filename_h5):
hydro = HydroH5(filename_h5)
location = 'upstream'
vartype = 'stage' # 'flow' or 'stage'
data = [StudyComparisionDashboard.get_data(hydro, channel_id, location, self.twstr, vartype)
for channel_id in self.stage_channels['channel_id']]
data = pd.concat(data, axis=1)
return data
def load_all_stage_data(self):
self.load_stage_channels()
self.hydrow = HydroH5(self.filename_base)
self.hydrowo = HydroH5(self.filename_alt)
#location = 'upstream'
# vartype = 'stage' # 'flow' or 'stage'
self.dataw = self.get_all_stage_data(self.filename_base)
self.datawo = self.get_all_stage_data(self.filename_alt)
def create_mean_flow_barplot(self):
flow_channels = pd.read_csv(
self.channels_flow_file, header=None, names=['channel_id'])
# flow_channels
#
sd_channels = self.dsm2_channels[self.dsm2_channels['id'].isin(
flow_channels.channel_id)].rename(columns={'id': 'channel_id'})
sd_channel_map = sd_channels.hvplot(
tiles='CartoLight', hover_cols='all').opts(opts.Path(line_width=3))
# arrows
downstream_end = sd_channels.geometry.apply(lambda x: x.coords[1])
# downstream_end
channel_angles = sd_channels.geometry.apply(
lambda x: calc_angle(x.coords))
# channel_angles
arrow_geoms = [arrow(d[0][0], d[0][1], d[1])
for d in zip(downstream_end.values, channel_angles.values)]
arrow_map = gpd.GeoDataFrame(
geometry=arrow_geoms).hvplot().opts(line_alpha=0)
# labels
labelsgdf = gpd.GeoDataFrame(geometry=sd_channels.geometry.centroid).assign(
text=sd_channels.channel_id.values)
labelsgdf = labelsgdf.assign(
x=labelsgdf.geometry.x, y=labelsgdf.geometry.y)
labelsmap = labelsgdf.hvplot.labels(x='x', y='y', text='text', hover=None).opts(
opts.Labels(text_font_size='8pt', text_align='left'))
#
# sd_channel_map*labelsmap*arrow_map
flow_mean_values = []
# for channel_id in flow_channels.iloc[1:2, :].channel_id:
# fw = get_data(hydrow, channel_id, 'upstream', twstr, 'flow')
# fwo = get_data(hydrowo, channel_id, 'upstream', twstr, 'flow')
flow_mean_w = StudyComparisionDashboard.get_mean_flow_data(
self.hydrow, self.twstr, flow_channels.channel_id.to_list())
flow_mean_wo = StudyComparisionDashboard.get_mean_flow_data(
self.hydrowo, self.twstr, flow_channels.channel_id.to_list())
flow_table = flow_channels.assign(
**{"without": flow_mean_wo, "with": flow_mean_w})
flows_bar_plot = flow_table.set_index('channel_id').hvplot.bar().opts(
xrotation=45, title=f'Mean flows with and without barriers: {self.twstr}')
plot = flows_bar_plot + (sd_channel_map * labelsmap *
arrow_map).opts(xaxis=None, yaxis=None)
plot.cols(1).opts(width=1000)
return plot
def show_plot(self, index):
if index == None or len(index) == 0:
index = [0]
tsplotw = self.dataw.iloc[:, index].hvplot(label='with barriers')
vplotw = self.dataw.iloc[:, index].hvplot.violin()
tsplotwo = self.datawo.iloc[:, index].hvplot(label='without barriers')
vplotwo = self.datawo.iloc[:, index].hvplot.violin()
plot = (tsplotwo * tsplotw).opts(legend_position='top_right') + \
(vplotwo * vplotw).opts(width=200)
return plot.opts(title=f'Stage Impact for with and without barriers scenario: {self.twstr}', toolbar=None)
def create_stage_interactive(self):
self.stage_channel_map = self.stage_channels.hvplot(
tiles='CartoLight', hover_cols='all').opts(opts.Path(line_width=5))
self.stage_channel_map = self.stage_channel_map.opts(
title='Stage Channels for analysis')
selection = streams.Selection1D(
source=self.stage_channel_map.Path.I, index=[0])
dmap_show_data = hv.DynamicMap(self.show_plot, streams=[selection])
self.stage_channel_map = self.stage_channel_map.opts(opts.Path(tools=['tap', 'hover'], color='green',
nonselection_color='blue', nonselection_alpha=0.3,
frame_width=650))
self.stage_channel_map = self.set_map_bounds(
self.stage_channel_map).opts(xaxis=None, yaxis=None, toolbar=None)
return pn.Column(self.stage_channel_map, dmap_show_data)
def create_violin_plots(self):
vopts = opts.Violin(violin_selection_alpha=0, outline_alpha=0,
stats_alpha=1, violin_alpha=0.0, box_alpha=0.0, )
vplotw = self.dataw.hvplot.violin(
color='red', group_label='with').opts(vopts).opts(xrotation=45)
vplotw.redim(value='stage').opts(ylabel='stage')
vplotwo = self.datawo.hvplot.violin(
color='blue', group_label='without').opts(vopts)
vplotwo = vplotwo.opts(opts.Violin(violin_fill_alpha=0.1, outline_color='blue',
outline_alpha=0.7)).opts(xrotation=45)
vplotwo.redim(value='stage').opts(ylabel='stage')
vplots = vplotw * vplotwo
vplots = vplots.opts(
height=400, title=f'Stage Distribution with (red) & without (subdued blue): {self.twstr}')
return vplots
def create_box_plots(self):
from holoviews import dim
dfwb = self.dataw.rename(columns=dict(zip(self.dataw.columns,
| pd.Series(self.dataw.columns) | pandas.Series |
import pandas as pd
from skimage import io
import json
import numpy as np
def createCountMatrix(assigned_genes:str):
original_df = pd.read_csv(assigned_genes)
original_df = original_df[original_df.Cell_Label != 0]
df1 = pd.crosstab(original_df.Gene,original_df.Cell_Label,original_df.Cell_Label,aggfunc='count').fillna(0)
df2 = original_df.groupby('Gene')['Cell_Label'].value_counts().unstack('Cell_Label', fill_value=0).reset_index()
return df1
# we're gonna write it like we're assigning globally, not on tiles, so the input is the transformed decoded df, not the one with Tile X and Y coordinates
def createPatchCountMatrix(decoded_df_csv, patch_labeled_image_path, neighbour_dict_path):
decoded_df = | pd.read_csv(decoded_df_csv) | pandas.read_csv |
#Lib for Streamlit
# Copyright(c) 2021 - AilluminateX LLC
# This is main Sofware... Screening and Tirage
# Customized to general Major Activities
# Make all the School Activities- st.write(DataFrame) ==> (outputs) Commented...
# The reason, since still we need the major calculations.
# Also the Computing is not that expensive.. So, no need to optimize at this point
import streamlit as st
import pandas as pd
#Change website title (set_page_config)
#==============
from PIL import Image
image_favicon=Image.open('Logo_AiX.jpg')
st.set_page_config(page_title='AilluminateX - Covid Platform', page_icon = 'Logo_AiX.jpg') #, layout = 'wide', initial_sidebar_state = 'auto'), # layout = 'wide',)
# favicon being an object of the same kind as the one you should provide st.image() with
#(ie. a PIL array for example) or a string (url or local file path)
#==============
#Hide footer and customize the text
#=========================
hide_streamlit_style = """
<style>
#MainMenu {visibility: hidden;}
footer {visibility: hidden;}
footer:after {
content:'Copyright(c) 2021 - AilluminateX LLC and Ailysium - Covid19 Bio-Forecasting Platform | https://www.aillumiante.com';
visibility: visible;
display: block;
position: relative;
#background-color: gray;
padding: 5px;
top: 2px;
}
</style>
"""
st.markdown(hide_streamlit_style, unsafe_allow_html=True)
#==============================
from sklearn.ensemble import RandomForestClassifier
from sklearn.metrics import confusion_matrix
from sklearn.metrics import classification_report
from yellowbrick.classifier import ClassificationReport
from sklearn.metrics import accuracy_score
#import numpy as np
from sklearn.metrics import accuracy_score
import matplotlib.pyplot as plt
import plotly.express as px
import numpy as np
import plotly
import plotly.graph_objects as go
from plotly.subplots import make_subplots
import altair as alt
import plotly.figure_factory as ff
import matplotlib
from matplotlib import cm
import seaborn as sns; sns.set()
from PIL import Image
import statsmodels.api as sm
import statsmodels.formula.api as smf
#from sklearn import model_selection, preprocessing, metrics, svm,linear_model
from sklearn.model_selection import train_test_split, GridSearchCV, cross_val_score, cross_validate, StratifiedKFold
from sklearn.feature_selection import SelectKBest, chi2
#from sklearn.decomposition import PCA
from sklearn.ensemble import RandomForestClassifier
from sklearn.metrics import auc, roc_auc_score, roc_curve, explained_variance_score, precision_recall_curve,average_precision_score,accuracy_score, classification_report
#from sklearn.preprocessing import StandardScaler
from sklearn.neural_network import MLPRegressor
from sklearn.datasets import make_regression
from sklearn.model_selection import train_test_split
from sklearn.preprocessing import StandardScaler
from sklearn.neighbors import KNeighborsClassifier
from sklearn.metrics import accuracy_score
from sklearn.ensemble import RandomForestClassifier
from sklearn.tree import DecisionTreeClassifier
from scipy.stats import boxcox
from matplotlib import pyplot
import pickle
#from sklearn.externals import joblib
import joblib
# Load Image & Logo
#====================
st.image("Logo_AiX.jpg") # Change to MSpace Logo
#st.write("https://www.ailluminate.com")
#st.image("LogoAiX1.jpg") # Change to MSpace Logo
st.markdown("<h1 style='text-align: left; color: turquoise;'>Ailysium: BioForecast Platform</h1>", unsafe_allow_html=True)
#st.markdown("<h1 style='text-align: left; color: turquoise;'>Train AI BioForecast Model (Realtime)</h1>", unsafe_allow_html=True)
#st.markdown("<h1 style='text-align: left; color: turquoise;'>Opening-Economy & Society</h1>", unsafe_allow_html=True)
#df_forecast= pd.read_csv("2021-03-27-all-forecasted-cases-model-data.csv", engine='python', dtype={'fips': str})
df_forecast=pd.read_csv("recent-all-forecasted-cases-model-data.csv", engine='python', dtype={'fips': str})
#Load Data - The last/most recent Forecast and latest Data
#=====================
# The last two, most recent forecast
#df_forecast= pd.read_csv("2021-03-15-all-forecasted-cases-model-data.csv", engine='python', dtype={'fips': str})
#Forcast_date="2021-03-15"
#Forecasted_dates=["3/20/2021", "3/27/2021", "4/03/2021", "4/10/2021" ]
#df_forecast= pd.read_csv("2021-03-22-all-forecasted-cases-model-data.csv", engine='python', dtype={'fips': str})
#Forcast_date="2021-03-22"
#Forecasted_dates=["3/27/2021", "4/03/2021", "4/10/2021", "4/17/2021" ]
#==========================================
df_forecast_previous= pd.read_csv("previous-all-forecasted-cases-model-data.csv", engine='python', dtype={'fips': str})
Forcast_date="2021-03-22"
Forecasted_dates=["3/27/2021", "4/03/2021", "4/10/2021", "4/17/2021" ]
df_forecast_recent=pd.read_csv("recent-all-forecasted-cases-model-data.csv", engine='python', dtype={'fips': str})
Forcast_date="2021-03-29"
Forecasted_dates=["4/03/2021", "4/10/2021", "4/17/2021", "4/24/2021" ]
#================
#initialize the data
#=======================
#Models
#====================
#st.success("What Forecast Model Data to Load?")
forecast_model_Options= ['Reference Model',
'Ensemble',
'UGA-CEID',
'Columbia',
'ISU',
'UVA',
'LNQ',
'Facebook',
'JHU-APL',
'UpstateSU',
'JHU-IDD',
'LANL',
'Ensemble']
#st.success("What Date Forecast Data to Load?")
data_dates_options=['2021-01-04', '2021-01-11', '2021-01-18',
'2021-01-25', '2021-02-01', '2021-02-08',
'2021-02-15', '2021-02-22', '2021-03-01',
'2021-03-08', '2021-03-15', '2021-03-22',
'2021-03-29']
data_dates_options=['2021-03-29',
'2021-03-22', '2021-03-15', '2021-03-08',
'2021-03-01', '2021-02-22', '2021-02-15',
'2021-02-08', '2021-02-01', '2021-01-25',
'2021-01-18', '2021-01-11', '2021-01-04']
data_dates_options=['2021-04-14']
load_ai_model_options=['Reference Model',
'AI Model 1',
'AI Model 2 (L)',
'AI Model 3 (Fast)',
'AI Model 4 (Fast) (L)',
'AI Model 5',
'AI Model 6',
'AI Model 7 (VERY Slow- Do Not Use, if You have too!)',
'AI Model 8',
'AI Model 9 (Slow)',
'AI Model 10',
'AI Model 11 (L)',
'AI Model 12',
'AI Model 13',
'AI Model 14 (L)',
'AI Model 15',
'AI Model 16 (L)',
'AI Model (aggregator)']
train_ai_model_options=load_ai_model_options
#===========================
#Selectt Option Section
#============================
select_options=["AiX-ai-Forecast-Platform",
"Load Forecast Data", #Simply Check the Forecast Data
"Load AI Model",
"Train AI Model",
"AiX-Platform"]
select_options=["AiX-ai-Forecast-Platform"]
your_option=select_options
st.sidebar.success("Please Select your Option" )
option_selectbox = st.sidebar.selectbox( "Select your Option:", your_option)
select_Name=option_selectbox
#if option_selectbox=='Load Forecast Data' or option_selectbox!='Load Forecast Data':
#if select_Name=='Load Forecast Data' or select_Name!='Load Forecast Data':
if select_Name=='AiX-ai-Forecast-Platform' or select_Name!='AiX-ai-Forecast-Platform':
#Models
#====================
#st.success("What Forecast Model Data to Load?")
your_option=forecast_model_Options
st.sidebar.info("Please Select Forecast Model" )
option_selectbox = st.sidebar.selectbox( "Select Forecast Model:", your_option)
if option_selectbox =='Reference Model':
option_selectbox='Reference Model'
option_selectbox='Ensemble'
forecast_model_Name=option_selectbox
#if option_selectbox=='Load Forecast Data' or option_selectbox!='Load Forecast Data':
if select_Name=='Load Forecast Data' or select_Name!='Load Forecast Data':
#st.success("What Date Forecast Data to Load?")
your_option=data_dates_options
st.sidebar.warning("Please Select Forecast Date" )
option_selectbox = st.sidebar.selectbox( "Select Forecast Date:", your_option)
#if option_selectbox=='2021-03-22':
# option_selectbox= '2021-03-15'
data_dates_Name=option_selectbox
if option_selectbox==data_dates_Name:
your_option=["One(1) Week Ahead", "Two(2) Weeks Ahead", "Three(3) Weeks Ahead", "Four(4) Weeks Ahead"]
st.sidebar.warning("Please Select Forecast Week" )
option_selectbox = st.sidebar.selectbox( "Select Forecast Weeks Ahead:", your_option)
data_week_Name=option_selectbox
if data_week_Name !="One(1) Week Ahead":
st.write("Two(2), Three(3), and Four(4) Weeks Ahead are being calculated offline currently and are not presented as realtime")
#if option_selectbox=='Load AI Model':
if select_Name=='Load AI Model':
your_option=load_ai_model_options
st.sidebar.error("Please Select AI Model to load" )
option_selectbox = st.sidebar.selectbox( "Select AI-Model to Load:", your_option)
ai_load_Name=option_selectbox
#if option_selectbox=='Train AI Model':
if select_Name=='Train AI Model':
your_option=train_ai_model_options
st.sidebar.success("Please Select AI Model to Train" )
option_selectbox = st.sidebar.selectbox( "Select AI-Model to Train:", your_option)
ai_train_Name=option_selectbox
#load_data_csv=data_dates_Name+"-all-forecasted-cases-model-data.csv"
#st.write("Data to load: ", load_data_csv)
#Load Models and Sidebar Selection
#===================================================================================# Load AI Models
#if option_selectbox=='AiX Platform':
if select_Name=='AiX Platform':
model2load=pd.read_csv('model2load.csv', engine='python', dtype=str) # dtype={"Index": int})
model_index=model2load
model_names_option=model_index.AI_Models.values
st.sidebar.success("Please Select your AI Model!" )
model_selectbox = st.sidebar.selectbox( "Select AI Model", model_names_option)
Model_Name=model_selectbox
Index_model=model2load.Index[model2load.AI_Models==Model_Name].values[0]
Index_model=int(Index_model)
pkl_model_load=model2load.Pkl_Model[model2load.AI_Models==Model_Name].values[0]
#Load Data and Model
Pkl_Filename = pkl_model_load #"Pickle_RForest.pkl"
#st.write(Pkl_Filename)
# Load the Model back from file
#****with open(Pkl_Filename, 'rb') as file: # This line to load the file
#*** Pickle_LoadModel = pickle.load(file) # This line to load the file
# Pickle_RForest = pickle.load(file)
#RForest=Pickle_RForest
load_data_csv=data_dates_Name+"-all-forecasted-cases-model-data.csv"
#st.write('Load CDC Model Data- Data to load:', ' ', load_data_csv)
load_data_csv="recent-all-forecasted-cases-model-data.csv"
#st.write('Load CDC Model Data- Data to load:', ' ', load_data_csv)
#Forecast Data is being loaded and alll sort of sidebars also created.
#===================================================
#import pandas as pd
# Load Reference Model Forecast Ensemble - Only For Visualization Purpose
#=============================================================================
#df_forecast= pd.read_csv("2021-03-15-all-forecasted-cases-model-data.csv", engine='python', dtype={'fips': str})
#df_forecast= pd.read_csv(load_data_csv, engine='python', dtype={'fips': str})
df_forecast_ref=pd.DataFrame()
df_forecast_ref=pd.read_csv("previous-all-forecasted-cases-model-data.csv", engine='python', dtype={'fips': str})
Forcast_date="2021-03-22"
Forecasted_dates=["3/27/2021", "4/03/2021", "4/10/2021", "4/17/2021" ]
df_forecast=pd.DataFrame()
df_forecast= df_forecast_ref.copy()
df=pd.DataFrame()
df=df_forecast.copy()
# Drop all the States. We are only interested in Counties
df_drop=df[df.location_name!=df.State]
#df_drop1 = df.query("location_name != State")
#df_drop.fips= df_drop.fips.astype(str)
df_forecast=df_drop.copy()
#df_drop.fips= df_drop.fips.astype(str)
#df_forecast_Ensemble=df_forecast[df_forecast.model=="Ensemble"]
#forecast_model_Name="Ensemble"
df_forecast_Ensemble=pd.DataFrame()
df_forecast_Ensemble=df_forecast[df_forecast.model=="Ensemble"]
df_forecast_Ensemble=df_forecast_Ensemble[df_forecast_Ensemble.target=="1 wk ahead inc case"]
df_forecast_Ensemble_ref=pd.DataFrame()
df_forecast_Ensemble_ref=df_forecast_Ensemble.copy()
# Load Previous Forecast
#=========================
#df_forecast= pd.read_csv("2021-03-15-all-forecasted-cases-model-data.csv", engine='python', dtype={'fips': str})
#df_forecast= pd.read_csv(load_data_csv, engine='python', dtype={'fips': str})
df_forecast_previous=pd.DataFrame()
df_forecast_previous=pd.read_csv("previous-all-forecasted-cases-model-data.csv", engine='python', dtype={'fips': str})
Forcast_date="2021-03-22"
Forecasted_dates=["3/27/2021", "4/03/2021", "4/10/2021", "4/17/2021" ]
df_forecast=pd.DataFrame()
df_forecast= df_forecast_previous.copy()
df=pd.DataFrame()
df=df_forecast.copy()
# Drop all the States. We are only interested in Counties
df_drop=df[df.location_name!=df.State]
#df_drop1 = df.query("location_name != State")
#df_drop.fips= df_drop.fips.astype(str)
df_forecast=df_drop.copy()
#df_drop.fips= df_drop.fips.astype(str)
#df_forecast_Ensemble=df_forecast[df_forecast.model=="Ensemble"]
df_forecast_Ensemble=pd.DataFrame()
df_forecast_Ensemble=df_forecast[df_forecast.model==forecast_model_Name]
df_forecast_Ensemble=df_forecast_Ensemble[df_forecast_Ensemble.target=="1 wk ahead inc case"]
df_forecast_Ensemble_previous=pd.DataFrame()
df_forecast_Ensemble_previous=df_forecast_Ensemble.copy()
#Load Most Recent Forecast
#====================
#df_forecast= pd.read_csv(load_data_csv, engine='python', dtype={'fips': str})
df_forecast_recent=pd.DataFrame()
df_forecast_recent=pd.read_csv("recent-all-forecasted-cases-model-data.csv", engine='python', dtype={'fips': str})
Forcast_date="2021-03-29"
Forecasted_dates=["4/03/2021", "4/10/2021", "4/17/2021", "4/24/2021" ]
df_forecast=pd.DataFrame()
df_forecast= df_forecast_recent.copy()
df=pd.DataFrame()
df=df_forecast.copy()
# Drop all the States. We are only interested in Counties
df_drop=df[df.location_name!=df.State]
#df_drop1 = df.query("location_name != State")
#df_drop.fips= df_drop.fips.astype(str)
#df_drop.fips= df_drop.fips.astype(str)
#df_forecast_Ensemble=df_forecast[df_forecast.model=="Ensemble"]
df_forecast_Ensemble=pd.DataFrame()
df_forecast_Ensemble=df_forecast[df_forecast.model==forecast_model_Name]
df_forecast_Ensemble=df_forecast_Ensemble[df_forecast_Ensemble.target=="1 wk ahead inc case"]
df_forecast_Ensemble_recent=pd.DataFrame()
df_forecast_Ensemble_recent=df_forecast_Ensemble.copy()
#Load Actual Cases
#==========================
df_actual_cases=pd.DataFrame()
df_actual_cases=pd.read_csv("covid_confirmed_usafacts_forecast.csv", engine='python', dtype={'fips': str})
#======================Visulaization of data =======================
# ======================Compare the Forecast with actula data ================"
df_ref_temp=pd.DataFrame(np.array(df_forecast_Ensemble_ref.iloc[:,[6,7]].values), columns=["fips", "Forecast_Reference"]) # 6,7: fips and point
df_model_temp=pd.DataFrame(np.array(df_forecast_Ensemble_previous.iloc[:,[6,7]].values), columns=["fips", "Forecast_Model"]) # 6,7: fips and point
df_actual_temp=pd.DataFrame(np.array(df_actual_cases.iloc[:,[0,-2]].values), columns=["fips", "Actual_Target"]) # 0, -2: fips and most recent actual-target
df_actual_temp=pd.DataFrame(np.array(df_actual_cases.iloc[:,[0,-7,-6,-5,-4,-3, -2]].values),
columns=["fips", "TimeN5", "TimeN4", "TimeN3", "TimeN2", "TimeN1", "Actual_Target"]) # 0, -2: fips and most recent actual-target
#st.write("Last 6 Total Weekly Cases, ", df_actual_temp.head(20))
data_merge= pd.DataFrame() #df_ref_temp.copy()
data_merge= pd.merge(df_ref_temp, df_model_temp, on="fips")
data_merge_left=data_merge.copy()
data_merge= pd.merge(data_merge_left, df_actual_temp, on="fips")
#st.write("df_actual_temp:, ", data_merge.head())
#st.error("Stop for checking how many is loaded")
data_merge.iloc[:,1:] = data_merge.iloc[:,1:].astype(float)
#st.write("Data Merged: ", data_merge.head())
#data_merge = data_merge.iloc[:,[1,2,3]].astype(float)
df_forecast_target=data_merge.copy()
#df_forecast_target_Scaled = df_forecast_target_Scaled.astype(float)
len_data=len(df_forecast_target)
df_population= pd.read_csv("covid_county_population_usafacts.csv", engine='python', dtype={'fips': str, 'fips_1': str})
df_forecast_target_Scaled = df_forecast_target.copy()
i=0
while i <len_data:
fips=df_forecast_target['fips'].iloc[0]
population=df_population.population[df_population.fips==fips].values[0]
df_forecast_target_Scaled.iloc[i,1:]=df_forecast_target.iloc[i,1:]/population*1000
i=i+1
df_forecast_target_Scaled.iloc[:,1:] = df_forecast_target_Scaled.iloc[:,1:].astype(float)
#st.write("df_forecast_target_Scaled", df_forecast_target_Scaled.head())
data_viz=df_forecast_target_Scaled.copy()
#Delete All The Data Frames that we do not need!
#=======================Delete all the DataFrame we do not need ==================
df_forecast_target_Scaled=pd.DataFrame()
data_merge=pd.DataFrame()
df_forecast_target=pd.DataFrame()
df_forecast_Ensemble_previous=pd.DataFrame()
df_forecast_Ensemble_recent=pd.DataFrame()
df_forecast_Ensemble_ref=pd.DataFrame()
df_forecast=pd.DataFrame()
df_ref_temp=pd.DataFrame()
df_model_temp=pd.DataFrame()
df_actual_temp=pd.DataFrame()
df_drop=pd.DataFrame()
#===================End of Delete ==========================
#data_viz.to_csv("data_viz.csv", index=False)
data_viz= data_viz.drop(data_viz.columns[[0]], axis=1)
data_viz= data_viz*100
data_viz= data_viz.astype(float)
#st.write("Data viz: head ", data_viz.head())
#st.write("Data viz: Stat ", data_viz.describe())
data_viz.drop( data_viz[ data_viz.Forecast_Reference >4500 ].index , inplace=True)
data_viz.drop( data_viz[ data_viz.Forecast_Model >4500 ].index , inplace=True)
data_viz.drop( data_viz[ data_viz.Actual_Target >5000 ].index , inplace=True)
data_viz.drop( data_viz[ data_viz.TimeN1>5000 ].index , inplace=True)
data_viz.drop( data_viz[ data_viz.TimeN2>5000 ].index , inplace=True)
data_viz.drop( data_viz[ data_viz.TimeN3>5000 ].index , inplace=True)
data_viz.drop( data_viz[ data_viz.TimeN4>5000 ].index , inplace=True)
data_viz.drop( data_viz[ data_viz.TimeN5>5000 ].index , inplace=True)
#st.write("Data viz: Stat 2- after cut off of 4500-5000 ", data_viz.describe())
#st.success("Stop")
#data_viz= data_viz*100
#data_viz["Forecast_Reference"]=data_viz["Forecast_Reference"].apply(np.ceil)
data_viz.drop( data_viz[ data_viz.Forecast_Reference <1 ].index , inplace=True)
#data_viz["Forecast_Model"]=data_viz["Forecast_Model"].apply(np.ceil)
data_viz.drop( data_viz[ data_viz.Forecast_Model <1 ].index , inplace=True)
#data_viz["Actual_Target"]=data_viz["Actual_Target"].apply(np.ceil)
data_viz.drop( data_viz[ data_viz.Actual_Target <1 ].index , inplace=True)
data_viz.drop( data_viz[ data_viz.TimeN1<1 ].index , inplace=True)
data_viz.drop( data_viz[ data_viz.TimeN2<1 ].index , inplace=True)
data_viz.drop( data_viz[ data_viz.TimeN3<1 ].index , inplace=True)
data_viz.drop( data_viz[ data_viz.TimeN4<1 ].index , inplace=True)
data_viz.drop( data_viz[ data_viz.TimeN5<1 ].index , inplace=True)
#data_viz= np.around(data_viz)
#data_viz=data_viz[data_viz.Actual_Target>=0]
#data_viz= data_viz*100 #np.around(data_viz+)
#data_viz=data_viz[data_viz.Actual_Target<5000]
#data_viz=data_viz[data_viz.Forecast_Reference<4200]
#data_viz=data_viz[data_viz.Forecast_Model<4200]
#data_viz_temp=data_viz[data_viz<5000]
if data_viz.empty:
st.error("No Data matches our criteria both for AI Model and Visualization!")
st.warning("Please select another option!")
st.stop("The Program stopped here!")
#data_viz.drop( data_viz[ data_viz >5000 ].index , inplace=True)
#st.write("describe data -2")
#st.write(data_viz.describe())
#================= Visualization
#sns.jointplot(data=data_viz, x="target", y="Ensemble")
#sns.pairplot(data=data_viz, hue='color')
#data_viz=pd.read_csv("data_viz.csv", engine='python')
i=0.2
data_viz=(data_viz**i-1)/i
#data_viz=np.log(data_viz)
#st.write("Data viz: Stat3333333333333 ", data_viz.describe())
huecolor=data_viz.Actual_Target.values
huecolor=huecolor.astype(int)
data_viz["huecolor"]=huecolor.astype(int)
#data_viz=data_viz[data_viz>0]
#st.write("describe data -2")
#st.write(data_viz.describe())
huecolor=data_viz.Actual_Target.values.astype(int)
data_viz["huecolor"]=huecolor.astype(int)
#st.title("Hello")
#fig = sns.pairplot(penguins, hue="species")
#st.pyplot(fig)
data_vis=data_viz.copy()
st.write(" ")
st.markdown("<h1 style='text-align: left; color: turquoise;'>Forecast: Reference vs Selected Model </h1>", unsafe_allow_html=True)
# 2-D plot of images
#fig=sns.pairplot(data_viz, hue="huecolor", diag_kind="hist")
#st.pyplot(fig)
data_vis= data_vis.drop(data_vis.columns[[2,3,4,5,6]], axis=1)
#fig=sns.pairplot(data_vis, hue="huecolor", diag_kind="hist")
#st.pyplot(fig)
#data_vis=pd.DataFrame()
#import numpy as np
#import pandas as pd
#import statsmodels.api as sm
#import statsmodels.formula.api as smf
#import matplotlib.pyplot as plt
mod = smf.quantreg('Forecast_Model ~ Actual_Target', data_viz)
res = mod.fit(q=.5)
#st.write(res.summary())
#LRresult = (res.summary2().tables[0])
#st.write(LRresult)
#LRresult = (res.summary2().tables[1])
#st.write(LRresult)
#import statsmodels.api as sm
#model = sm.OLS(y,x)
#res = model.fit()
results_summary = res.summary()
results_as_html = results_summary.tables[0].as_html()
df_res=pd.read_html(results_as_html, header=0, index_col=0)[0]
st.write(df_res)
# Note that tables is a list. The table at index 1 is the "core" table. Additionally, read_html puts dfs in a list, so we want index 0
results_as_html = results_summary.tables[1].as_html()
df_res= | pd.read_html(results_as_html, header=0, index_col=0) | pandas.read_html |
from numpy import *
import nlopt
import numpy as np
import matplotlib.pyplot as plt
import numbers
import math
import pandas as pd
import random
import autograd.numpy as ag
from autograd import grad
from mpl_toolkits.mplot3d import Axes3D
from numpy.lib.function_base import vectorize
from autograd import value_and_grad
##### Define the test problems discussed in Arnoud, Guvenen and Kleineberg as classes
##### These allow us to evaluate the function for a vector x, and a value a -> Guvenen et al. specified a=200 in their Analysis -> we do this too
class griewank:
def __init__(self,x,a):
self.arg=x
self.dim=x.size # allows to check the dimension of the function
def function_value(x,a): # function that gives us the function value of the griewank function as specified in Guvenen et al.
input = np.array(x)
sum = (1 / a) * np.dot(input,input)
prod = np.prod( np.cos( input / np.sqrt( np.arange(1,input.size + 1,1) ) ) )
out = sum - prod + 2
return out
self.function_val=function_value(x,a) # returns the function value for the griewank function evaluated at x
#domain=([-100]*self.dim,[100]*self.dim)
#self.domain=domain
#self.lower_bound=([-100]*self.dim)
#self.upper_bound=([100]*self.dim)
#name= 'Griewank Function'
#self.name=name
#problem_solver=np.array([0]*self.dim) # best known global minimum
#self.solver=problem_solver
#self.solver_function_value=function_value(problem_solver,a)
class griewank_info: ##### This class stores the general information for a griewank function
def __init__(self,dim,a): ### arguments are the number of dimensions of the problem and the parameter a
domain=([-100]*dim,[100]*dim)
self.domain=domain ### returns the domain of the function
self.lower_bound=([-100]*dim) ### returns thw lower bound of the function
self.upper_bound=([100]*dim) ### returns the upper bound of the function
name= 'Griewank Function'
self.name=name
problem_solver=np.array([0]*dim)
self.solver=problem_solver ### returns the known solution to the problem
def function_value(x,a):
input = np.array(x)
sum = (1 / a) * np.dot(input,input)
prod = np.prod( np.cos( input / np.sqrt( np.arange(1,input.size + 1,1) ) ) )
out = sum - prod + 2
return out
self.solver_function_value=function_value(problem_solver,a) ### returns the function value of the known solution to the problem
##### Now we define the griewank function such that it fits the interface from nlopt
def problem_griewank(x,grad):
return (1 / 200) * np.dot(x,x)- np.prod( np.cos( x / np.sqrt( np.arange(1,x.size + 1,1) ) ) )+2 #### returns the griewank problem
### x is the input vector
### grad is the gradient vector -> for derivative free algorithms we do not need this argument
### nlopt algorithms also work if grad=NONE
### Most of our algorithms are derivative free except for StoGO
def get_starting_points(n,problem_info_object,p):
### n: number of desired dimensions of the problem
### problem_info_object: object that contains the known information of the problem e.g.: g_1=griewank_info(n,a=200)
### p: desired number of starting points you want to draw
## as Guvenen et al. do not specify how they generate the random starting points I will choose a method
### Method:
# as the starting point has to be a vector fo dimension = dimension of the function, I draw every coordinate
# for the vector from a uniform distribution
# repeat this until you get 100 vectors of dimension = dim of function which are randomly generated
data=[]
lower_b=problem_info_object.lower_bound
upper_b=problem_info_object.upper_bound
for i in range(n):
v=np.random.uniform(lower_b[i],upper_b[i],p)
data.append(v)
df= | pd.DataFrame(data) | pandas.DataFrame |
# pylint: disable=too-many-lines
"""Statistical functions in ArviZ."""
import warnings
import logging
from collections import OrderedDict
import numpy as np
import pandas as pd
import scipy.stats as st
from scipy.optimize import minimize
import xarray as xr
from ..data import convert_to_inference_data, convert_to_dataset
from .diagnostics import _multichain_statistics, _mc_error, ess
from .stats_utils import make_ufunc as _make_ufunc, logsumexp as _logsumexp
from ..utils import _var_names
_log = logging.getLogger(__name__)
__all__ = ["compare", "hpd", "loo", "psislw", "r2_score", "summary", "waic"]
def compare(
dataset_dict,
ic="waic",
method="BB-pseudo-BMA",
b_samples=1000,
alpha=1,
seed=None,
scale="deviance",
):
r"""Compare models based on WAIC or LOO cross-validation.
WAIC is the widely applicable information criterion, and LOO is leave-one-out
(LOO) cross-validation. Read more theory here - in a paper by some of the
leading authorities on model selection - dx.doi.org/10.1111/1467-9868.00353
Parameters
----------
dataset_dict : dict[str] -> InferenceData
A dictionary of model names and InferenceData objects
ic : str
Information Criterion (WAIC or LOO) used to compare models. Default WAIC.
method : str
Method used to estimate the weights for each model. Available options are:
- 'stacking' : stacking of predictive distributions.
- 'BB-pseudo-BMA' : (default) pseudo-Bayesian Model averaging using Akaike-type
weighting. The weights are stabilized using the Bayesian bootstrap.
- 'pseudo-BMA': pseudo-Bayesian Model averaging using Akaike-type
weighting, without Bootstrap stabilization (not recommended).
For more information read https://arxiv.org/abs/1704.02030
b_samples: int
Number of samples taken by the Bayesian bootstrap estimation.
Only useful when method = 'BB-pseudo-BMA'.
alpha : float
The shape parameter in the Dirichlet distribution used for the Bayesian bootstrap. Only
useful when method = 'BB-pseudo-BMA'. When alpha=1 (default), the distribution is uniform
on the simplex. A smaller alpha will keeps the final weights more away from 0 and 1.
seed : int or np.random.RandomState instance
If int or RandomState, use it for seeding Bayesian bootstrap. Only
useful when method = 'BB-pseudo-BMA'. Default None the global
np.random state is used.
scale : str
Output scale for IC. Available options are:
- `deviance` : (default) -2 * (log-score)
- `log` : 1 * log-score (after Vehtari et al. (2017))
- `negative_log` : -1 * (log-score)
Returns
-------
A DataFrame, ordered from lowest to highest IC. The index reflects the key with which the
models are passed to this function. The columns are:
IC : Information Criteria (WAIC or LOO).
Smaller IC indicates higher out-of-sample predictive fit ("better" model). Default WAIC.
If `scale == log` higher IC indicates higher out-of-sample predictive fit ("better" model).
pIC : Estimated effective number of parameters.
dIC : Relative difference between each IC (WAIC or LOO) and the lowest IC (WAIC or LOO).
It's always 0 for the top-ranked model.
weight: Relative weight for each model.
This can be loosely interpreted as the probability of each model (among the compared model)
given the data. By default the uncertainty in the weights estimation is considered using
Bayesian bootstrap.
SE : Standard error of the IC estimate.
If method = BB-pseudo-BMA these values are estimated using Bayesian bootstrap.
dSE : Standard error of the difference in IC between each model and the top-ranked model.
It's always 0 for the top-ranked model.
warning : A value of 1 indicates that the computation of the IC may not be reliable.
This could be indication of WAIC/LOO starting to fail see
http://arxiv.org/abs/1507.04544 for details.
scale : Scale used for the IC.
Examples
--------
Compare the centered and non centered models of the eight school problem:
.. ipython::
In [1]: import arviz as az
...: data1 = az.load_arviz_data("non_centered_eight")
...: data2 = az.load_arviz_data("centered_eight")
...: compare_dict = {"non centered": data1, "centered": data2}
...: az.compare(compare_dict)
Compare the models using LOO-CV, returning the IC in log scale and calculating the
weights using the stacking method.
.. ipython::
In [1]: az.compare(compare_dict, ic="loo", method="stacking", scale="log")
"""
names = list(dataset_dict.keys())
scale = scale.lower()
if scale == "log":
scale_value = 1
ascending = False
else:
if scale == "negative_log":
scale_value = -1
else:
scale_value = -2
ascending = True
ic = ic.lower()
if ic == "waic":
ic_func = waic
df_comp = pd.DataFrame(
index=names,
columns=["waic", "p_waic", "d_waic", "weight", "se", "dse", "warning", "waic_scale"],
)
scale_col = "waic_scale"
elif ic == "loo":
ic_func = loo
df_comp = pd.DataFrame(
index=names,
columns=["loo", "p_loo", "d_loo", "weight", "se", "dse", "warning", "loo_scale"],
)
scale_col = "loo_scale"
else:
raise NotImplementedError("The information criterion {} is not supported.".format(ic))
if method.lower() not in ["stacking", "bb-pseudo-bma", "pseudo-bma"]:
raise ValueError("The method {}, to compute weights, is not supported.".format(method))
ic_se = "{}_se".format(ic)
p_ic = "p_{}".format(ic)
ic_i = "{}_i".format(ic)
ics = pd.DataFrame()
names = []
for name, dataset in dataset_dict.items():
names.append(name)
ics = ics.append([ic_func(dataset, pointwise=True, scale=scale)])
ics.index = names
ics.sort_values(by=ic, inplace=True, ascending=ascending)
if method.lower() == "stacking":
rows, cols, ic_i_val = _ic_matrix(ics, ic_i)
exp_ic_i = np.exp(ic_i_val / scale_value)
last_col = cols - 1
def w_fuller(weights):
return np.concatenate((weights, [max(1.0 - np.sum(weights), 0.0)]))
def log_score(weights):
w_full = w_fuller(weights)
score = 0.0
for i in range(rows):
score += np.log(np.dot(exp_ic_i[i], w_full))
return -score
def gradient(weights):
w_full = w_fuller(weights)
grad = np.zeros(last_col)
for k in range(last_col - 1):
for i in range(rows):
grad[k] += (exp_ic_i[i, k] - exp_ic_i[i, last_col]) / np.dot(
exp_ic_i[i], w_full
)
return -grad
theta = np.full(last_col, 1.0 / cols)
bounds = [(0.0, 1.0) for _ in range(last_col)]
constraints = [
{"type": "ineq", "fun": lambda x: 1.0 - np.sum(x)},
{"type": "ineq", "fun": np.sum},
]
weights = minimize(
fun=log_score, x0=theta, jac=gradient, bounds=bounds, constraints=constraints
)
weights = w_fuller(weights["x"])
ses = ics[ic_se]
elif method.lower() == "bb-pseudo-bma":
rows, cols, ic_i_val = _ic_matrix(ics, ic_i)
ic_i_val = ic_i_val * rows
b_weighting = st.dirichlet.rvs(alpha=[alpha] * rows, size=b_samples, random_state=seed)
weights = np.zeros((b_samples, cols))
z_bs = np.zeros_like(weights)
for i in range(b_samples):
z_b = np.dot(b_weighting[i], ic_i_val)
u_weights = np.exp((z_b - np.min(z_b)) / scale_value)
z_bs[i] = z_b # pylint: disable=unsupported-assignment-operation
weights[i] = u_weights / np.sum(u_weights)
weights = weights.mean(axis=0)
ses = pd.Series(z_bs.std(axis=0), index=names) # pylint: disable=no-member
elif method.lower() == "pseudo-bma":
min_ic = ics.iloc[0][ic]
z_rv = np.exp((ics[ic] - min_ic) / scale_value)
weights = z_rv / np.sum(z_rv)
ses = ics[ic_se]
if np.any(weights):
min_ic_i_val = ics[ic_i].iloc[0]
for idx, val in enumerate(ics.index):
res = ics.loc[val]
if scale_value < 0:
diff = res[ic_i] - min_ic_i_val
else:
diff = min_ic_i_val - res[ic_i]
d_ic = np.sum(diff)
d_std_err = np.sqrt(len(diff) * np.var(diff))
std_err = ses.loc[val]
weight = weights[idx]
df_comp.at[val] = (
res[ic],
res[p_ic],
d_ic,
weight,
std_err,
d_std_err,
res["warning"],
res[scale_col],
)
return df_comp.sort_values(by=ic, ascending=ascending)
def _ic_matrix(ics, ic_i):
"""Store the previously computed pointwise predictive accuracy values (ics) in a 2D matrix."""
cols, _ = ics.shape
rows = len(ics[ic_i].iloc[0])
ic_i_val = np.zeros((rows, cols))
for idx, val in enumerate(ics.index):
ic = ics.loc[val][ic_i]
if len(ic) != rows:
raise ValueError("The number of observations should be the same across all models")
ic_i_val[:, idx] = ic
return rows, cols, ic_i_val
def hpd(ary, credible_interval=0.94, circular=False):
"""
Calculate highest posterior density (HPD) of array for given credible_interval.
The HPD is the minimum width Bayesian credible interval (BCI). This implementation works only
for unimodal distributions.
Parameters
----------
x : Numpy array
An array containing posterior samples
credible_interval : float, optional
Credible interval to compute. Defaults to 0.94.
circular : bool, optional
Whether to compute the hpd taking into account `x` is a circular variable
(in the range [-np.pi, np.pi]) or not. Defaults to False (i.e non-circular variables).
Returns
-------
np.ndarray
lower and upper value of the interval.
Examples
--------
Calculate the hpd of a Normal random variable:
.. ipython::
In [1]: import arviz as az
...: import numpy as np
...: data = np.random.normal(size=2000)
...: az.hpd(data, credible_interval=.68)
"""
if ary.ndim > 1:
hpd_array = np.array(
[hpd(row, credible_interval=credible_interval, circular=circular) for row in ary.T]
)
return hpd_array
# Make a copy of trace
ary = ary.copy()
n = len(ary)
if circular:
mean = st.circmean(ary, high=np.pi, low=-np.pi)
ary = ary - mean
ary = np.arctan2(np.sin(ary), np.cos(ary))
ary = np.sort(ary)
interval_idx_inc = int(np.floor(credible_interval * n))
n_intervals = n - interval_idx_inc
interval_width = ary[interval_idx_inc:] - ary[:n_intervals]
if len(interval_width) == 0:
raise ValueError(
"Too few elements for interval calculation. "
"Check that credible_interval meets condition 0 =< credible_interval < 1"
)
min_idx = np.argmin(interval_width)
hdi_min = ary[min_idx]
hdi_max = ary[min_idx + interval_idx_inc]
if circular:
hdi_min = hdi_min + mean
hdi_max = hdi_max + mean
hdi_min = np.arctan2(np.sin(hdi_min), np.cos(hdi_min))
hdi_max = np.arctan2(np.sin(hdi_max), np.cos(hdi_max))
return np.array([hdi_min, hdi_max])
def loo(data, pointwise=False, reff=None, scale="deviance"):
"""Pareto-smoothed importance sampling leave-one-out cross-validation.
Calculates leave-one-out (LOO) cross-validation for out of sample predictive model fit,
following Vehtari et al. (2017). Cross-validation is computed using Pareto-smoothed
importance sampling (PSIS).
Parameters
----------
data : obj
Any object that can be converted to an az.InferenceData object. Refer to documentation
of az.convert_to_inference_data for details
pointwise : bool, optional
if True the pointwise predictive accuracy will be returned. Defaults to False
reff : float, optional
Relative MCMC efficiency, `ess / n` i.e. number of effective samples divided by
the number of actual samples. Computed from trace by default.
scale : str
Output scale for loo. Available options are:
- `deviance` : (default) -2 * (log-score)
- `log` : 1 * log-score (after Vehtari et al. (2017))
- `negative_log` : -1 * (log-score)
Returns
-------
pandas.Series with the following columns:
loo : approximated Leave-one-out cross-validation
loo_se : standard error of loo
p_loo : effective number of parameters
shape_warn : bool
True if the estimated shape parameter of
Pareto distribution is greater than 0.7 for one or more samples
loo_i : array of pointwise predictive accuracy, only if pointwise True
pareto_k : array of Pareto shape values, only if pointwise True
loo_scale : scale of the loo results
Examples
--------
Calculate the LOO-CV of a model:
.. ipython::
In [1]: import arviz as az
...: data = az.load_arviz_data("centered_eight")
...: az.loo(data, pointwise=True)
"""
inference_data = convert_to_inference_data(data)
for group in ("posterior", "sample_stats"):
if not hasattr(inference_data, group):
raise TypeError(
"Must be able to extract a {group} group from data!".format(group=group)
)
if "log_likelihood" not in inference_data.sample_stats:
raise TypeError("Data must include log_likelihood in sample_stats")
posterior = inference_data.posterior
log_likelihood = inference_data.sample_stats.log_likelihood
n_samples = log_likelihood.chain.size * log_likelihood.draw.size
new_shape = (n_samples, np.product(log_likelihood.shape[2:]))
log_likelihood = log_likelihood.values.reshape(*new_shape)
if scale.lower() == "deviance":
scale_value = -2
elif scale.lower() == "log":
scale_value = 1
elif scale.lower() == "negative_log":
scale_value = -1
else:
raise TypeError('Valid scale values are "deviance", "log", "negative_log"')
if reff is None:
n_chains = len(posterior.chain)
if n_chains == 1:
reff = 1.0
else:
ess_p = ess(posterior, method="mean")
# this mean is over all data variables
reff = (
np.hstack([ess_p[v].values.flatten() for v in ess_p.data_vars]).mean() / n_samples
)
log_weights, pareto_shape = psislw(-log_likelihood, reff)
log_weights += log_likelihood
warn_mg = False
if np.any(pareto_shape > 0.7):
warnings.warn(
"Estimated shape parameter of Pareto distribution is greater than 0.7 for "
"one or more samples. You should consider using a more robust model, this is because "
"importance sampling is less likely to work well if the marginal posterior and "
"LOO posterior are very different. This is more likely to happen with a non-robust "
"model and highly influential observations."
)
warn_mg = True
loo_lppd_i = scale_value * _logsumexp(log_weights, axis=0)
loo_lppd = loo_lppd_i.sum()
loo_lppd_se = (len(loo_lppd_i) * np.var(loo_lppd_i)) ** 0.5
lppd = np.sum(_logsumexp(log_likelihood, axis=0, b_inv=log_likelihood.shape[0]))
p_loo = lppd - loo_lppd / scale_value
if pointwise:
if np.equal(loo_lppd, loo_lppd_i).all(): # pylint: disable=no-member
warnings.warn(
"""The point-wise LOO is the same with the sum LOO, please double check
the Observed RV in your model to make sure it returns element-wise logp.
"""
)
return pd.Series(
data=[loo_lppd, loo_lppd_se, p_loo, warn_mg, loo_lppd_i, pareto_shape, scale],
index=["loo", "loo_se", "p_loo", "warning", "loo_i", "pareto_k", "loo_scale"],
)
else:
return pd.Series(
data=[loo_lppd, loo_lppd_se, p_loo, warn_mg, scale],
index=["loo", "loo_se", "p_loo", "warning", "loo_scale"],
)
def psislw(log_weights, reff=1.0):
"""
Pareto smoothed importance sampling (PSIS).
Parameters
----------
log_weights : array
Array of size (n_samples, n_observations)
reff : float
relative MCMC efficiency, `ess / n`
Returns
-------
lw_out : array
Smoothed log weights
kss : array
Pareto tail indices
"""
rows, cols = log_weights.shape
log_weights_out = np.copy(log_weights, order="F")
kss = np.empty(cols)
# precalculate constants
cutoff_ind = -int(np.ceil(min(rows / 5.0, 3 * (rows / reff) ** 0.5))) - 1
cutoffmin = np.log(np.finfo(float).tiny) # pylint: disable=no-member, assignment-from-no-return
k_min = 1.0 / 3
# loop over sets of log weights
for i, x in enumerate(log_weights_out.T):
# improve numerical accuracy
x -= np.max(x)
# sort the array
x_sort_ind = np.argsort(x)
# divide log weights into body and right tail
xcutoff = max(x[x_sort_ind[cutoff_ind]], cutoffmin)
expxcutoff = np.exp(xcutoff)
tailinds, = np.where(x > xcutoff) # pylint: disable=unbalanced-tuple-unpacking
x_tail = x[tailinds]
tail_len = len(x_tail)
if tail_len <= 4:
# not enough tail samples for gpdfit
k = np.inf
else:
# order of tail samples
x_tail_si = np.argsort(x_tail)
# fit generalized Pareto distribution to the right tail samples
x_tail = np.exp(x_tail) - expxcutoff
k, sigma = _gpdfit(x_tail[x_tail_si])
if k >= k_min:
# no smoothing if short tail or GPD fit failed
# compute ordered statistic for the fit
sti = np.arange(0.5, tail_len) / tail_len
smoothed_tail = _gpinv(sti, k, sigma)
smoothed_tail = np.log( # pylint: disable=assignment-from-no-return
smoothed_tail + expxcutoff
)
# place the smoothed tail into the output array
x[tailinds[x_tail_si]] = smoothed_tail
# truncate smoothed values to the largest raw weight 0
x[x > 0] = 0
# renormalize weights
x -= _logsumexp(x)
# store tail index k
kss[i] = k
return log_weights_out, kss
def _gpdfit(ary):
"""Estimate the parameters for the Generalized Pareto Distribution (GPD).
Empirical Bayes estimate for the parameters of the generalized Pareto
distribution given the data.
Parameters
----------
ary : array
sorted 1D data array
Returns
-------
k : float
estimated shape parameter
sigma : float
estimated scale parameter
"""
prior_bs = 3
prior_k = 10
n = len(ary)
m_est = 30 + int(n ** 0.5)
b_ary = 1 - np.sqrt(m_est / (np.arange(1, m_est + 1, dtype=float) - 0.5))
b_ary /= prior_bs * ary[int(n / 4 + 0.5) - 1]
b_ary += 1 / ary[-1]
k_ary = np.log1p(-b_ary[:, None] * ary).mean(axis=1) # pylint: disable=no-member
len_scale = n * (np.log(-(b_ary / k_ary)) - k_ary - 1)
weights = 1 / np.exp(len_scale - len_scale[:, None]).sum(axis=1)
# remove negligible weights
real_idxs = weights >= 10 * np.finfo(float).eps
if not np.all(real_idxs):
weights = weights[real_idxs]
b_ary = b_ary[real_idxs]
# normalise weights
weights /= weights.sum()
# posterior mean for b
b_post = np.sum(b_ary * weights)
# estimate for k
k_post = np.log1p(-b_post * ary).mean() # pylint: disable=invalid-unary-operand-type,no-member
# add prior for k_post
k_post = (n * k_post + prior_k * 0.5) / (n + prior_k)
sigma = -k_post / b_post
return k_post, sigma
def _gpinv(probs, kappa, sigma):
"""Inverse Generalized Pareto distribution function."""
# pylint: disable=unsupported-assignment-operation, invalid-unary-operand-type
x = np.full_like(probs, np.nan)
if sigma <= 0:
return x
ok = (probs > 0) & (probs < 1)
if np.all(ok):
if np.abs(kappa) < np.finfo(float).eps:
x = -np.log1p(-probs)
else:
x = np.expm1(-kappa * np.log1p(-probs)) / kappa
x *= sigma
else:
if np.abs(kappa) < np.finfo(float).eps:
x[ok] = -np.log1p(-probs[ok])
else:
x[ok] = np.expm1(-kappa * np.log1p(-probs[ok])) / kappa
x *= sigma
x[probs == 0] = 0
if kappa >= 0:
x[probs == 1] = np.inf
else:
x[probs == 1] = -sigma / kappa
return x
def r2_score(y_true, y_pred):
"""R² for Bayesian regression models. Only valid for linear models.
Parameters
----------
y_true : array-like of shape = (n_samples) or (n_samples, n_outputs)
Ground truth (correct) target values.
y_pred : array-like of shape = (n_samples) or (n_samples, n_outputs)
Estimated target values.
Returns
-------
Pandas Series with the following indices:
r2: Bayesian R²
r2_std: standard deviation of the Bayesian R².
"""
if y_pred.ndim == 1:
var_y_est = np.var(y_pred)
var_e = np.var(y_true - y_pred)
else:
var_y_est = np.var(y_pred.mean(0))
var_e = np.var(y_true - y_pred, 0)
r_squared = var_y_est / (var_y_est + var_e)
return pd.Series([np.mean(r_squared), np.std(r_squared)], index=["r2", "r2_std"])
def summary(
data,
var_names=None,
fmt="wide",
round_to=None,
include_circ=None,
stat_funcs=None,
extend=True,
credible_interval=0.94,
order="C",
index_origin=0,
):
"""Create a data frame with summary statistics.
Parameters
----------
data : obj
Any object that can be converted to an az.InferenceData object
Refer to documentation of az.convert_to_dataset for details
var_names : list
Names of variables to include in summary
include_circ : bool
Whether to include circular statistics
fmt : {'wide', 'long', 'xarray'}
Return format is either pandas.DataFrame {'wide', 'long'} or xarray.Dataset {'xarray'}.
round_to : int
Number of decimals used to round results. Defaults to 2. Use "none" to return raw numbers.
stat_funcs : dict
A list of functions or a dict of functions with function names as keys used to calculate
statistics. By default, the mean, standard deviation, simulation standard error, and
highest posterior density intervals are included.
The functions will be given one argument, the samples for a variable as an nD array,
The functions should be in the style of a ufunc and return a single number. For example,
`np.mean`, or `scipy.stats.var` would both work.
extend : boolean
If True, use the statistics returned by `stat_funcs` in addition to, rather than in place
of, the default statistics. This is only meaningful when `stat_funcs` is not None.
credible_interval : float, optional
Credible interval to plot. Defaults to 0.94. This is only meaningful when `stat_funcs` is
None.
order : {"C", "F"}
If fmt is "wide", use either C or F unpacking order. Defaults to C.
index_origin : int
If fmt is "wide, select n-based indexing for multivariate parameters. Defaults to 0.
Returns
-------
pandas.DataFrame
With summary statistics for each variable. Defaults statistics are: `mean`, `sd`,
`hpd_3%`, `hpd_97%`, `mcse_mean`, `mcse_sd`, `ess_bulk`, `ess_tail` and `r_hat`.
`r_hat` is only computed for traces with 2 or more chains.
Examples
--------
.. ipython::
In [1]: import arviz as az
...: data = az.load_arviz_data("centered_eight")
...: az.summary(data, var_names=["mu", "tau"])
Other statistics can be calculated by passing a list of functions
or a dictionary with key, function pairs.
.. ipython::
In [1]: import numpy as np
...: def median_sd(x):
...: median = np.percentile(x, 50)
...: sd = np.sqrt(np.mean((x-median)**2))
...: return sd
...:
...: func_dict = {
...: "std": np.std,
...: "median_std": median_sd,
...: "5%": lambda x: np.percentile(x, 5),
...: "median": lambda x: np.percentile(x, 50),
...: "95%": lambda x: np.percentile(x, 95),
...: }
...: az.summary(
...: data,
...: var_names=["mu", "tau"],
...: stat_funcs=func_dict,
...: extend=False
...: )
"""
posterior = convert_to_dataset(data, group="posterior")
var_names = _var_names(var_names, posterior)
posterior = posterior if var_names is None else posterior[var_names]
fmt_group = ("wide", "long", "xarray")
if not isinstance(fmt, str) or (fmt.lower() not in fmt_group):
raise TypeError("Invalid format: '{}'! Formatting options are: {}".format(fmt, fmt_group))
unpack_order_group = ("C", "F")
if not isinstance(order, str) or (order.upper() not in unpack_order_group):
raise TypeError(
"Invalid order: '{}'! Unpacking options are: {}".format(order, unpack_order_group)
)
alpha = 1 - credible_interval
extra_metrics = []
extra_metric_names = []
if stat_funcs is not None:
if isinstance(stat_funcs, dict):
for stat_func_name, stat_func in stat_funcs.items():
extra_metrics.append(
xr.apply_ufunc(
_make_ufunc(stat_func), posterior, input_core_dims=(("chain", "draw"),)
)
)
extra_metric_names.append(stat_func_name)
else:
for stat_func in stat_funcs:
extra_metrics.append(
xr.apply_ufunc(
_make_ufunc(stat_func), posterior, input_core_dims=(("chain", "draw"),)
)
)
extra_metric_names.append(stat_func.__name__)
if extend:
mean = posterior.mean(dim=("chain", "draw"))
sd = posterior.std(dim=("chain", "draw"))
hpd_lower, hpd_higher = xr.apply_ufunc(
_make_ufunc(hpd, n_output=2),
posterior,
kwargs=dict(credible_interval=credible_interval),
input_core_dims=(("chain", "draw"),),
output_core_dims=tuple([] for _ in range(2)),
)
if include_circ:
circ_mean = xr.apply_ufunc(
_make_ufunc(st.circmean),
posterior,
kwargs=dict(high=np.pi, low=-np.pi),
input_core_dims=(("chain", "draw"),),
)
circ_sd = xr.apply_ufunc(
_make_ufunc(st.circstd),
posterior,
kwargs=dict(high=np.pi, low=-np.pi),
input_core_dims=(("chain", "draw"),),
)
circ_mcse = xr.apply_ufunc(
_make_ufunc(_mc_error),
posterior,
kwargs=dict(circular=True),
input_core_dims=(("chain", "draw"),),
)
circ_hpd_lower, circ_hpd_higher = xr.apply_ufunc(
_make_ufunc(hpd, n_output=2),
posterior,
kwargs=dict(credible_interval=credible_interval, circular=True),
input_core_dims=(("chain", "draw"),),
output_core_dims=tuple([] for _ in range(2)),
)
mcse_mean, mcse_sd, ess_mean, ess_sd, ess_bulk, ess_tail, r_hat = xr.apply_ufunc(
_make_ufunc(_multichain_statistics, n_output=7, ravel=False),
posterior,
input_core_dims=(("chain", "draw"),),
output_core_dims=tuple([] for _ in range(7)),
)
# Combine metrics
metrics = []
metric_names = []
if extend:
metrics.extend(
(
mean,
sd,
mcse_mean,
mcse_sd,
hpd_lower,
hpd_higher,
ess_mean,
ess_sd,
ess_bulk,
ess_tail,
r_hat,
)
)
metric_names.extend(
(
"mean",
"sd",
"mcse_mean",
"mcse_sd",
"hpd_{:g}%".format(100 * alpha / 2),
"hpd_{:g}%".format(100 * (1 - alpha / 2)),
"ess_mean",
"ess_sd",
"ess_bulk",
"ess_tail",
"r_hat",
)
)
if include_circ:
metrics.extend((circ_mean, circ_sd, circ_mcse, circ_hpd_lower, circ_hpd_higher))
metric_names.extend(
(
"circular_mean",
"circular_sd",
"circular_mcse",
"circular_hpd_{:g}%".format(100 * alpha / 2),
"circular_hpd_{:g}%".format(100 * (1 - alpha / 2)),
)
)
metrics.extend(extra_metrics)
metric_names.extend(extra_metric_names)
joined = xr.concat(metrics, dim="metric").assign_coords(metric=metric_names)
if fmt.lower() == "wide":
dfs = []
for var_name, values in joined.data_vars.items():
if len(values.shape[1:]):
metric = list(values.metric.values)
data_dict = OrderedDict()
for idx in np.ndindex(values.shape[1:] if order == "C" else values.shape[1:][::-1]):
if order == "F":
idx = tuple(idx[::-1])
ser = pd.Series(values[(Ellipsis, *idx)].values, index=metric)
key_index = ",".join(map(str, (i + index_origin for i in idx)))
key = "{}[{}]".format(var_name, key_index)
data_dict[key] = ser
df = | pd.DataFrame.from_dict(data_dict, orient="index") | pandas.DataFrame.from_dict |
"""
Computes the fingerprint similarity of molecules in the validation and test set to
molecules in the training set.
"""
import numpy as np
import pandas as pd
from syn_net.utils.data_utils import *
from rdkit import Chem
from rdkit.Chem import AllChem
import multiprocessing as mp
from scripts._mp_search_similar import func
if __name__ == '__main__':
ncpu = 64
data_path = '/pool001/whgao/data/synth_net/st_hb/st_train.json.gz'
st_set = SyntheticTreeSet()
st_set.load(data_path)
data = st_set.sts
data_train = [t.root.smiles for t in data]
data_path = '/pool001/whgao/data/synth_net/st_hb/st_test.json.gz'
st_set = SyntheticTreeSet()
st_set.load(data_path)
data = st_set.sts
data_test = [t.root.smiles for t in data]
data_path = '/pool001/whgao/data/synth_net/st_hb/st_valid.json.gz'
st_set = SyntheticTreeSet()
st_set.load(data_path)
data = st_set.sts
data_valid = [t.root.smiles for t in data]
fps_valid = [AllChem.GetMorganFingerprintAsBitVect(Chem.MolFromSmiles(smi), 2, nBits=1024) for smi in data_valid]
fps_test = [AllChem.GetMorganFingerprintAsBitVect(Chem.MolFromSmiles(smi), 2, nBits=1024) for smi in data_test]
with mp.Pool(processes=ncpu) as pool:
results = pool.map(func, fps_valid)
similaritys = [r[0] for r in results]
indices = [data_train[r[1]] for r in results]
df1 = | pd.DataFrame({'smiles': data_valid, 'split': 'valid', 'most similar': indices, 'similarity': similaritys}) | pandas.DataFrame |
######################################################################
# This file contains utility functions to load test data from file, #
# and invoke DeepAR predictor and plot the observed and target data. #
######################################################################
import io
import os
import json
import pandas as pd
import sagemaker
def series_to_json_obj(ts, target_column=None, dyn_feat=None, start=None):
"""Returns a dictionary of values in DeepAR, JSON format.
:param dyn_feat: array of dynamic features
:param ts: A time series dataframe containing stock prices data features.
:param target_column: A single feature time series to be predicted.
:param start: A datetime start value to be used as beginning of time series used as prediction context
:return: A dictionary of values with "start", "target" and "dynamic_feat" keys if any
"""
# get start time and target from the time series, ts
if start is not None:
start_index = start
ts = ts.loc[start_index:]
if not dyn_feat:
if isinstance(ts, pd.DataFrame):
json_obj = {"start": str(pd.to_datetime(start_index)),
"target": list(ts.loc[:, target_column])}
elif isinstance(ts, pd.Series):
json_obj = {"start": str(pd.to_datetime(start_index)),
"target": list(ts.values)}
else:
# populating dynamic features array
dyn_feat_list = generate_dyn_feat_list(dyn_feat, ts)
# creating json object
json_obj = {"start": str(pd.to_datetime(start_index)),
"target": list(ts.loc[:, target_column]),
"dynamic_feat": list(dyn_feat_list)}
else:
if not dyn_feat:
if isinstance(ts, pd.DataFrame):
json_obj = {"start": str(ts.index[0]),
"target": list(ts.loc[:, target_column])}
elif isinstance(ts, pd.Series):
json_obj = {"start": str(ts.index[0]),
"target": list(ts.values)}
else:
# populating dynamic features array
dyn_feat_list = generate_dyn_feat_list(dyn_feat, ts)
# creating json object
json_obj = {"start": str(ts.index[0]), "target": list(ts.loc[:, target_column]),
"dynamic_feat": list(dyn_feat_list)}
return json_obj
def generate_dyn_feat_list(dyn_feat, ts):
df_dyn_feat = ts.loc[:, dyn_feat]
dyn_feat_list = []
for feat in df_dyn_feat:
dyn_feat_list.append(list(df_dyn_feat[feat]))
return dyn_feat_list
# TODO check for start value usage
def future_date_to_json_obj(start_date):
"""Returns a dictionary of values in DeepAR, JSON format.
:param start_date: start date of the json to be produced
:return: A json dictionary of values with "start" date and an empty "target" value list.
"""
json_obj = {
"start": pd.to_datetime(start_date).strftime(format="%Y-%m-%d"),
"target": []
}
return json_obj
def ts2dar_json(ts, saving_path, file_name, dyn_feat=[], start=None):
"""
Serializes a dataframe containing time series data into a json ready
to be processed by DeepAR
"""
if isinstance(ts, pd.DataFrame):
json_obj = series_to_json_obj(ts=ts, target_column='Adj Close',
dyn_feat=dyn_feat, start=start)
elif isinstance(ts, pd.Series):
json_obj = series_to_json_obj(ts=ts, start=start)
with open(os.path.join(saving_path, file_name), 'w') as fp:
json.dump(json_obj, fp)
# Class that allows making requests using pandas Series objects rather than raw JSON strings
class DeepARPredictor(sagemaker.predictor.Predictor):
def __init__(self, endpoint_name, sagemaker_session):
super().__init__(endpoint_name=endpoint_name, sagemaker_session=sagemaker_session)
self.__freq = 'D'
self.__prediction_length = 20
def set_prediction_parameters(self, freq, prediction_length):
"""
Set the time frequency and prediction length parameters. This method **must** be called
before being able to use `predict`, otherwise, default values of 'D' and `20` wil be used.
Parameters:
freq -- string indicating the time frequency
prediction_length -- integer, number of predicted time points
Return value: none.
"""
self.__freq = freq
self.__prediction_length = prediction_length
def predict(self, ts, cat=None, encoding="utf-8", num_samples=100, quantiles=["0.1", "0.5", "0.9"],
content_type="application/json"):
"""Requests the prediction of for the time series listed in `ts`, each with the (optional)
corresponding category listed in `cat`.
Parameters:
ts -- Time series to predict from. Can be either a list of dataframes,
a single dataframe or a json S3 file path.
cat -- list of integers (default: None)
encoding -- string, encoding to use for the request (default: "utf-8")
num_samples -- integer, number of samples to compute at prediction time (default: 100)
quantiles -- list of strings specifying the quantiles to compute (default: ["0.1", "0.5", "0.9"])
Return value: list of `pandas.DataFrame` objects, each containing the predictions
"""
if isinstance(ts, list):
prediction_times = [x.index[-1] + pd.Timedelta(1, unit=self.__freq) for x in ts]
req = self.__encode_request(ts, cat, encoding, num_samples, quantiles)
elif isinstance(ts, pd.DataFrame):
prediction_times = ts.index[-1] + pd.Timedelta(1, unit=self.__freq)
req = self.__encode_request(ts, cat, encoding, num_samples, quantiles)
elif isinstance(ts, str):
# TODO add code to process ts as an S3 path to a json file coded time series
if ts.upper() == 'IBM':
# TODO add code to feed predictor with IBM data starting from last value of test set
pass
elif ts.upper() == 'AAPL':
# TODO add code to feed predictor with AAPL data starting from last value of test set
pass
elif ts.upper() == 'AMZN':
# TODO add code to feed predictor with AMZN data starting from last value of test set
pass
elif ts.upper() == 'GOOGL':
# TODO add code to feed predictor with GOOGL data starting from last value of test set
pass
else:
pass
req = None
else:
# TODO add code to handle error in input format
req = None
res = super(DeepARPredictor, self).predict(req, initial_args={"ContentType": content_type})
return self.__decode_response(res, prediction_times, encoding)
@staticmethod
def __encode_request(ts, cat, encoding, num_samples, quantiles) -> object:
"""
This function encodes a json request for the endpoint, that accepts
:param ts: time series to be predicted
:param cat: categorical features
:param encoding: encoding to be used
:param num_samples: number of samples to be used by DeepAR
:param quantiles: list of quantiles to be used by
:return:
"""
instances = [series_to_json_obj(ts[k], target_column='Adj Close',
dyn_feat=[], start=None) for k in range(len(ts))]
configuration = {
"num_samples": num_samples,
"output_types": ["quantiles"],
"quantiles": quantiles,
}
http_request_data = {"instances": instances, "configuration": configuration}
return json.dumps(http_request_data).encode(encoding)
@staticmethod
def __encode_future_request(start_times, cat, encoding, num_samples, quantiles):
instances = [{"start": st.strftime(format="%Y-%m-%d"), "target": []} for k, st in enumerate(start_times)]
configuration = {
"num_samples": num_samples,
"output_types": ["quantiles"],
"quantiles": quantiles,
}
http_request_data = {"instances": instances, "configuration": configuration}
return json.dumps(http_request_data).encode(encoding)
def __decode_response(self, response, prediction_times, encoding):
response_data = json.loads(response.decode(encoding))
list_of_df = []
for k in range(len(prediction_times)):
prediction_index = pd.date_range(
start=prediction_times[k], freq=self.__freq, periods=self.__prediction_length
)
list_of_df.append(
pd.DataFrame(data=response_data["predictions"][k]["quantiles"], index=prediction_index)
)
return list_of_df
def predict_future(self, start_times, cat=None, encoding="utf-8",
num_samples=100, quantiles=["0.1", "0.5", "0.9"], content_type="application/json") -> list:
"""Requests the prediction of future time series values for the time series from `start_date`, each with the (optional)
corresponding category listed in `cat`.
Parameters:
start_times -- start date of the future prediction
cat -- list of integers (default: None)
encoding -- string, encoding to use for the request (default: "utf-8")
num_samples -- integer, number of samples to compute at prediction time (default: 100)
quantiles -- list of strings specifying the quantiles to compute (default: ["0.1", "0.5", "0.9"])
Return value: list of `pandas.DataFrame` objects, each containing the predictions
"""
prediction_times = [st + | pd.Timedelta(1, unit=self.__freq) | pandas.Timedelta |
#!/usr/bin/env python3
# coding: utf-8
import csv
import numpy as np
import pandas as pd
## I/O configuration
# column delimiters for input and output files
input_sep = '\t'
output_sep = ','
output_type = '_peptides.csv'
# print row names/indices?
write_row_names=False
# print the column titles?
write_header=True
# quoting?
#quoting=csv.QUOTE_NONNUMERIC
quoting=csv.QUOTE_NONE
# leave empty to not print
additional_header = []
# separate by this value
sep_by = 'Raw file'
def __pep_001(df):
# get PEP, ceil to 1
pep = df['PEP'].values
pep[pep > 1] = 1
return (pep > 0.01)
def __fdr_001(df):
# get PEP, ceil to 1
#pep = df['PEP'].values
pep = df['PEP'].values
pep[pep > 1] = 1
# magic!!!
# basically, we need to cumulatively sum the PEP to get the FDR
# and then map back the cumulatively summed FDR to its original PEP
qval = (np.cumsum(pep[np.argsort(pep)]) / np.arange(1, df.shape[0]+1))[np.argsort(np.argsort(pep))]
return (qval > 0.01)
# filter out observations w/o any mass error value
def __missing_mass_error(df):
mass_error = df['Mass error [ppm]'].values
simple_mass_error = df['Simple mass error [ppm]'].values
return (pd.isnull(mass_error) & pd.isnull(simple_mass_error))
# filter out mass error above 20 PPM
def __large_mass_error(df):
mass_error = df['Mass error [ppm]'].values
simple_mass_error = df['Simple mass error [ppm]'].values
nan_inds = pd.isnull(mass_error)
mass_error[nan_inds] = simple_mass_error[nan_inds]
return np.abs(mass_error) > 20
filters = {
'remove_decoy': (lambda df: df['Proteins'].str.contains('REV__').values),
'remove_contaminant': (lambda df: df['Proteins'].str.contains('CON__').values),
'remove_no_protein': (lambda df: | pd.isnull(df['Proteins']) | pandas.isnull |
# -*- coding: utf-8 -*-
"""
Autor: <NAME>
Revisó: <NAME>
Aprobó: <NAME>
versión 0.0
"""
import powerfactory as pf
import pandas as pd
import numpy as np
from xlsxwriter.utility import xl_rowcol_to_cell
##### Inicia la aplicación #####
app=pf.GetApplication()
app.ClearOutputWindow()
app.EchoOff()
##### Obtener los datos dentro del ComPython ######
script=app.GetCurrentScript()
Tipo_corto=script.Tipo_corto
Directorio=script.Directorio
sPhase=script.sPhase #### Fase fallada (Falla monofásica)
bPhase=script.bPhase #### Fases falladas
filename=script.file_name
###### Fases falladas por defecto ############
if bPhase not in (0,1,2):
app.PrintInfo('Seleccione una fase fallada válida, se ejecutará la falla sobre la fase A')
bPhase=0
if sPhase not in (0,1,2):
sPhase=0
#### Casos de estudio ###
shc_cases=script.GetContents('Cases')[0].GetAll('IntScenario')
if shc_cases==[]:
app.PrintError('No hay casos de estudio seleccionados para ejecutar el script, se ejecutará el caso activo')
### Crear el archivo de excel
try:
writer=pd.ExcelWriter(Directorio+'\\'+filename+'.xlsx',engine='xlsxwriter')
except:
app.PrintError('Seleccione un directorio válido para guardar los resultados!!')
##### Objeto encargado de realizar los cortos circuitos
class Shc_cases:
def __init__(self):
###### Parámetros por defecto del corto circuito a ejecutar #####
self.script=app.GetCurrentScript()
self.Tipo_corto=self.script.Tipo_corto
self.Directorio=self.script.Directorio
self.shc_cases=self.script.GetContents('Cases')[0].GetAll('IntScenario')
self.Barras=self.script.GetContents('Elementos')[0].GetAll('ElmTerm')
self.shc=self.script.GetContents('Sch')[0]
self.script.GetContents('Elementos')[0].Clear()
self.script.GetContents('Elementos')[0].AddRef(self.Barras)
self.fase_rara=0
# self.Tipo_corto=3 #### Método completo
self.Tipo_falla='spgf' #### Falla monofásica a tierra
self.iopt_cur=0 #### Máxima corriente de corto circuito
##### Variables a devolver ####
@property
def I0x3(self):
return self._I0x3
@property
def Ikss(self):
return self._Ikss
@property
def IkssA(self):
return self._IkssA
@property
def IkssB(self):
return self._IkssB
@property
def IkssC(self):
return self._IkssC
@property
def nombre(self):
return self._nombre
@property
def tension(self):
return self._tension
###### Activar corto circuito ----- Extraer resultados ###
def short_circuit(self,shc_case):
shc_case.Activate() ### Activar el caso de estudio
ldf=app.GetFromStudyCase('ComLdf')
ldf.iopt_net=0
self.shc.iopt_mde=self.Tipo_corto #### Método de cortocircuito
self.shc.iopt_cur=self.iopt_cur
self.shc.iopt_allbus=0
# if self.Tipo_corto==3:
# self.shc.c_ldf.iopt_net=0 ### Forzar flujo de carga balanceado
self.shc.iopt_shc=self.Tipo_falla
self.shc.shcobj=self.script.GetContents('Elementos')[0]
if self.Tipo_falla=='2psc':
self.shc.i_p2psc=self.fase_rara
elif self.Tipo_falla=='2pgf':
self.shc.i_p2pgf=self.fase_rara
elif self.Tipo_falla=='spgf':
self.shc.i_pspgf=self.fase_rara
self.shc.Execute() ### Correr el corto circuito
#### Extrae el nombre de las barras falladas ####
self._nombre=[]
for self.Barra in self.Barras:
# self._nombre.append(self.Barra.loc_name)
try:
self._nombre.append(self.Barra.cpSubstat.loc_name+" "+self.Barra.loc_name)
except:
self._nombre.append(self.Barra.loc_name)
#### Extrae la tensión
self._tension=[self.Barra.uknom for self.Barra in self.Barras]
#### Extrae corriente 3I0 y Ikss para fallas monofásicas
if self.Tipo_falla=='spgf':
self._I0x3=[]
self._Ikss=[]
for self.Barra in self.Barras:
try:
self._I0x3.append(self.Barra.GetAttribute('m:I0x3'))
except:
self._I0x3.append(0)
try:
self._Ikss.append(self.Barra.GetAttribute('m:Ikss'))
except:
self._Ikss.append(0)
##### Ikss para fallas trifásicas
if self.Tipo_falla=='3psc':
self._Ikss=[]
for self.Barra in self.Barras:
try:
self._Ikss.append(self.Barra.GetAttribute('m:Ikss'))
# self._Ikss=[self.Barra.GetAttribute('m:Ikss') for self.Barra in self.Barras ]
except:
self._Ikss.append(0)
##### Extraer datos fallas bifásicas aisladas
if self.Tipo_falla=='2psc':
self._IkssA=[]
self._IkssB=[]
self._IkssC=[]
for self.Barra in self.Barras:
try:
self._IkssA.append(self.Barra.GetAttribute('m:Ikss:A'))
except:
self._IkssA.append(0)
try:
self._IkssB.append(self.Barra.GetAttribute('m:Ikss:B'))
except:
self._IkssB.append(0)
try:
self._IkssC.append(self.Barra.GetAttribute('m:Ikss:C'))
except:
self._IkssC.append(0)
#### Extraer datos de fallas bifásicas a tierra ####
if self.Tipo_falla=='2pgf':
self._IkssA=[]
self._IkssB=[]
self._IkssC=[]
self._I0x3=[]
for self.Barra in self.Barras:
try:
self._IkssA.append(self.Barra.GetAttribute('m:Ikss:A'))
except:
self._IkssA.append(0)
try:
self._IkssB.append(self.Barra.GetAttribute('m:Ikss:B'))
except:
self._IkssB.append(0)
try:
self._IkssC.append(self.Barra.GetAttribute('m:Ikss:C'))
except:
self._IkssC.append(0)
try:
self._I0x3.append(self.Barra.GetAttribute('m:I0x3'))
except:
self._I0x3.append(0)
### Guardar datos corto monofásico en un dataframe
def corto_mono_keep(Nombre,I0x3='',Ikss=''):
datos_mono=pd.DataFrame()
datos_mono['Nombre']=list(Nombre)
# try:
# datos_mono['Ikss']=list(Ikss)
# except:
# pass
try:
datos_mono['3I0']=list(I0x3)
except:
pass
datos_mono=datos_mono.sort_values(by='Nombre')
datos_mono=datos_mono.drop_duplicates(subset='Nombre')
datos_mono=datos_mono.drop(['Nombre'],axis=1)
return(datos_mono)
### Guardar datos corto bifásico a tierra en un dataframe
def corto_bg_keep(Nombre,I0x3,IkssA,IkssB,IkssC):
datos_corto_bg=pd.DataFrame()
datos_corto_bg['Nombre']=list(Nombre)
# try:
# datos_corto_bg['Ikss']=list(IkssA)
# # app.PrintPlain('El promedio de la fila A es:'+str(datos_corto_bg['Ikss A'].mean()))
# if datos_corto_bg['Ikss'].mean()==0:
# datos_corto_bg.drop(['Ikss A'],axis=1)
# datos_corto_bg=datos_corto_bg.drop(['Ikss A'],axis=1)
# except:
# pass
# try:
# datos_corto_bg['Ikss B']=list(IkssB)
# # app.PrintPlain('El promedio de la fila B es:'+str(datos_corto_bg['Ikss B'].mean()))
# if datos_corto_bg['Ikss B'].mean()==0:
# datos_corto_bg.drop(['Ikss B'],axis=1)
# datos_corto_bg=datos_corto_bg.drop(['Ikss B'],axis=1)
# except:
# pass
# try:
# datos_corto_bg['Ikss C']=list(IkssC)
# # app.PrintPlain('El promedio de la fila C es:'+str(datos_corto_bg['Ikss C'].mean()))
# if datos_corto_bg['Ikss C'].mean()==0:
# datos_corto_bg.drop(['Ikss C'],axis=1)
# datos_corto_bg=datos_corto_bg.drop(['Ikss C'],axis=1)
# except:
# pass
datos_corto_bg['3I0']=list(I0x3)
datos_corto_bg=datos_corto_bg.sort_values(by='Nombre')
datos_corto_bg=datos_corto_bg.drop_duplicates(subset='Nombre')
datos_corto_bg=datos_corto_bg.drop(['Nombre'],axis=1)
return(datos_corto_bg)
### Guardar datos corto bifásico en un dataframe
def corto_bi_keep(Nombre,IkssA='',IkssB='',IkssC=''):
datos_corto_bi=pd.DataFrame()
datos_corto_bi['Nombre']=list(Nombre)
try:
datos_corto_bi['Ikss A']=list(IkssA)
if datos_corto_bi['Ikss A'].mean()==0:
datos_corto_bi.drop(['Ikss A'],axis=1)
datos_corto_bi=datos_corto_bi.drop(['Ikss A'],axis=1)
except:
pass
try:
datos_corto_bi['Ikss B']=list(IkssB)
if datos_corto_bi['Ikss B'].mean()==0:
datos_corto_bi.drop(['Ikss B'],axis=1)
datos_corto_bi=datos_corto_bi.drop(['Ikss B'],axis=1)
except:
pass
try:
datos_corto_bi['Ikss C']=list(IkssC)
if datos_corto_bi['Ikss C'].mean()==0:
datos_corto_bi.drop(['Ikss C'],axis=1)
datos_corto_bi=datos_corto_bi.drop(['Ikss C'],axis=1)
except:
pass
datos_corto_bi=datos_corto_bi.sort_values(by='Nombre')
datos_corto_bi=datos_corto_bi.drop_duplicates(subset='Nombre')
datos_corto_bi=datos_corto_bi.drop(['Nombre'],axis=1)
return(datos_corto_bi[datos_corto_bi.columns[0]])
### Guardar datos corto trifásico en un dataframe
def corto_tri_keep(Nombre,Ikss,tension):
datos_corto_tri=pd.DataFrame()
datos_corto_tri['Nombre']=list(Nombre)
datos_corto_tri['Tensión [kV]']=list(tension)
try:
datos_corto_tri['Ikss [kA]']=list(Ikss)
except:
datos_corto_tri['Ikss [kA]']=''
datos_corto_tri=datos_corto_tri.sort_values(by='Nombre')
datos_corto_tri=datos_corto_tri.drop_duplicates(subset='Nombre')
return(datos_corto_tri)
### unir dataframes
def join_dataframe(data1,dato2,dato3,dato4):
joined_dataframe= | pd.concat([data1,dato2,dato3,dato4], axis=1) | pandas.concat |
import codecs
import datetime
import functools
import json
import os
import re
import shutil
import pandas as pd
from dateutil.relativedelta import relativedelta
from requests.exceptions import ConnectionError
from utils_pandas import add_data
from utils_pandas import cut_ages
from utils_pandas import export
from utils_pandas import fuzzy_join
from utils_pandas import import_csv
from utils_scraping import logger
from utils_scraping import s
from utils_scraping import web_files
from utils_thai import DISTRICT_RANGE
from utils_thai import join_provinces
from utils_thai import to_thaiyear
from utils_thai import today
#################################
# Cases Apis
#################################
def get_cases_old():
logger.info("========Covid19 Timeline==========")
# https://covid19.th-stat.com/json/covid19v2/getTimeline.json
# https://covid19.ddc.moph.go.th/api/Cases/round-1to2-all
# https://covid19.ddc.moph.go.th/api/Cases/timeline-cases-all
# {"Date":"01\/01\/2020","NewConfirmed":0,"NewRecovered":0,"NewHospitalized":0,"NewDeaths":0,"Confirmed":0,"Recovered":0,"Hospitalized":0,"Deaths":0}
# {"txn_date":"2021-03-31","new_case":42,"total_case":28863,"new_case_excludeabroad":24,"total_case_excludeabroad":25779,"new_death":0,"total_death":94,"new_recovered":47,"total_recovered":27645}
# "txn_date":"2021-04-01","new_case":26,"total_case":28889,"new_case_excludeabroad":21,"total_case_excludeabroad":25800,"new_death":0,"total_death":94,"new_recovered":122,"total_recovered":27767,"update_date":"2021-09-01 07:40:49"}
try:
file, text, url = next(
web_files("https://covid19.th-stat.com/json/covid19v2/getTimeline.json", dir="inputs/json", check=True))
except ConnectionError:
# I think we have all this data covered by other sources. It's a little unreliable.
return pd.DataFrame()
data = pd.DataFrame(json.loads(text)['Data'])
data['Date'] = pd.to_datetime(data['Date'])
data = data.set_index("Date")
cases = data[["NewConfirmed", "NewDeaths", "NewRecovered", "Hospitalized"]]
cases = cases.rename(columns=dict(NewConfirmed="Cases", NewDeaths="Deaths", NewRecovered="Recovered"))
cases["Source Cases"] = url
return cases
def get_cases():
logger.info("========Covid19 Timeline==========")
# https://covid19.th-stat.com/json/covid19v2/getTimeline.json
# https://covid19.ddc.moph.go.th/api/Cases/round-1to2-all
# https://covid19.ddc.moph.go.th/api/Cases/timeline-cases-all
# {"Date":"01\/01\/2020","NewConfirmed":0,"NewRecovered":0,"NewHospitalized":0,"NewDeaths":0,"Confirmed":0,"Recovered":0,"Hospitalized":0,"Deaths":0}
# {"txn_date":"2021-03-31","new_case":42,"total_case":28863,"new_case_excludeabroad":24,"total_case_excludeabroad":25779,"new_death":0,"total_death":94,"new_recovered":47,"total_recovered":27645}
# "txn_date":"2021-04-01","new_case":26,"total_case":28889,"new_case_excludeabroad":21,"total_case_excludeabroad":25800,"new_death":0,"total_death":94,"new_recovered":122,"total_recovered":27767,"update_date":"2021-09-01 07:40:49"}
url1 = "https://covid19.ddc.moph.go.th/api/Cases/round-1to2-all"
url2 = "https://covid19.ddc.moph.go.th/api/Cases/timeline-cases-all"
try:
_, json1, url = next(web_files(url1, dir="inputs/json", check=False))
_, json2, url = next(web_files(url2, dir="inputs/json", check=True))
except ConnectionError:
# I think we have all this data covered by other sources. It's a little unreliable.
return pd.DataFrame()
data = pd.read_json(json1).append(pd.read_json(json2))
data['Date'] = pd.to_datetime(data['txn_date'])
data = data.set_index("Date")
data = data.rename(columns=dict(new_case="Cases", new_death="Deaths", new_recovered="Recovered"))
cases = data[["Cases", "Deaths", "Recovered"]]
cases["Source Cases"] = url
# 2021-12-28 had duplicate because cases went up 4610 from 2305. Why? Google says 4610
cases = cases[~cases.index.duplicated(keep='first')]
return cases
@functools.lru_cache(maxsize=100, typed=False)
def get_case_details_csv():
if False:
return get_case_details_api()
cols = "No.,announce_date,Notified date,sex,age,Unit,nationality,province_of_isolation,risk,province_of_onset,district_of_onset".split(
",")
url = "https://data.go.th/dataset/covid-19-daily"
file, text, _ = next(web_files(url, dir="inputs/json", check=True))
data = re.search(r"packageApp\.value\('meta',([^;]+)\);", text.decode("utf8")).group(1)
apis = json.loads(data)
links = [api['url'] for api in apis if "รายงานจำนวนผู้ติดเชื้อ COVID-19 ประจำวัน" in api['name']]
# get earlier one first
links = sorted([link for link in links if '.php' not in link and '.xlsx' not in link], reverse=True)
# 'https://data.go.th/dataset/8a956917-436d-4afd-a2d4-59e4dd8e906e/resource/be19a8ad-ab48-4081-b04a-8035b5b2b8d6/download/confirmed-cases.csv'
cases = pd.DataFrame()
for link, check in zip(links, ([False] * len(links))[:-1] + [True]):
for file, _, _ in web_files(link, dir="inputs/json", check=check, strip_version=True, appending=True):
if file.endswith(".xlsx"):
continue
#cases = pd.read_excel(file)
elif file.endswith(".csv"):
confirmedcases = pd.read_csv(file)
if "risk" not in confirmedcases.columns:
confirmedcases.columns = cols
if '�' in confirmedcases.loc[0]['risk']:
# bad encoding
with codecs.open(file, encoding="tis-620") as fp:
confirmedcases = pd.read_csv(fp)
first, last, ldate = confirmedcases["No."].iloc[0], confirmedcases["No."].iloc[-1], confirmedcases["announce_date"].iloc[-1]
logger.info("Covid19daily: rows={} {}={} {} {}", len(confirmedcases), last - first, last - first, ldate, file)
cases = cases.combine_first(confirmedcases.set_index("No."))
else:
raise Exception(f"Unknown filetype for covid19daily {file}")
cases = cases.reset_index("No.")
cases['announce_date'] = pd.to_datetime(cases['announce_date'], dayfirst=True)
cases['Notified date'] = pd.to_datetime(cases['Notified date'], dayfirst=True, errors="coerce")
cases = cases.rename(columns=dict(announce_date="Date"))
cases['age'] = pd.to_numeric(cases['age'], downcast="integer", errors="coerce")
#assert cases.index.max() <
# Fix typos in Nationality columns
# This won't include every possible misspellings and need some further improvement
mapping = pd.DataFrame([['Thai', 'Thailand'],
['Thai', 'Thai'],
['Thai', 'India-Thailand'],
['Thai', 'ไทยใหญ่'],
['Lao', 'laotian / Lao'],
['Lao', 'Lao'],
['Lao', 'Laotian/Lao'],
['Lao', 'Laotian / Lao'],
['Lao', 'laos'],
['Lao', 'Laotian'],
['Lao', 'Laos'],
['Lao', 'ลาว'],
['Indian', 'Indian'],
['Indian', 'India'],
['Indian', 'indian'],
['Cambodian', 'Cambodian'],
['Cambodian', 'cambodian'],
['Cambodian', 'Cambodia'],
['South Korean', 'South Korean'],
['South Korean', 'Korea, South'],
['South Korean', 'Korean'],
['Burmese', 'Burmese'],
['Burmese', 'พม่า'],
['Burmese', 'burmese'],
['Burmese', 'Burma'],
['Chinese', 'Chinese'],
['Chinese', 'จีน'],
['Chinese', 'China'],
],
columns=['Nat Main', 'Nat Alt']).set_index('Nat Alt')
cases = fuzzy_join(cases, mapping, 'nationality')
cases['nationality'] = cases['Nat Main'].fillna(cases['nationality'])
return cases
def get_case_details_api():
rid = "67d43695-8626-45ad-9094-dabc374925ab"
chunk = 10000
url = f"https://data.go.th/api/3/action/datastore_search?resource_id={rid}&limit={chunk}&q=&offset="
records = []
cases = import_csv("covid-19", ["_id"], dir="inputs/json")
lastid = cases.last_valid_index() if cases.last_valid_index() else 0
data = None
while data is None or len(data) == chunk:
r = s.get(f"{url}{lastid}")
data = json.loads(r.content)['result']['records']
df = pd.DataFrame(data)
df['announce_date'] = pd.to_datetime(df['announce_date'], dayfirst=True)
df['Notified date'] = pd.to_datetime(df['Notified date'], dayfirst=True, errors="coerce")
df = df.rename(columns=dict(announce_date="Date"))
# df['age'] = pd.to_numeric(df['age'], downcast="integer", errors="coerce")
cases = cases.combine_first(df.set_index("_id"))
lastid += chunk - 1
export(cases, "covid-19", csv_only=True, dir="inputs/json")
cases = cases.set_index("Date")
logger.info("Covid19daily: covid-19 {}", cases.last_valid_index())
# # they screwed up the date conversion. d and m switched sometimes
# # TODO: bit slow. is there way to do this in pandas?
# for record in records:
# record['announce_date'] = to_switching_date(record['announce_date'])
# record['Notified date'] = to_switching_date(record['Notified date'])
# cases = pd.DataFrame(records)
return cases
@functools.lru_cache(maxsize=100, typed=False)
def get_cases_by_demographics_api():
logger.info("========Covid19Daily Demographics==========")
cases = get_case_details_csv()
age_groups = cut_ages(cases, ages=[10, 20, 30, 40, 50, 60, 70], age_col="age", group_col="Age Group")
case_ages = pd.crosstab(age_groups['Date'], age_groups['Age Group'])
case_ages.columns = [f"Cases Age {a}" for a in case_ages.columns.tolist()]
#labels2 = ["Age 0-14", "Age 15-39", "Age 40-59", "Age 60-"]
#age_groups2 = pd.cut(cases['age'], bins=[0, 14, 39, 59, np.inf], right=True, labels=labels2)
age_groups2 = cut_ages(cases, ages=[15, 40, 60], age_col="age", group_col="Age Group")
case_ages2 = pd.crosstab(age_groups2['Date'], age_groups2['Age Group'])
case_ages2.columns = [f"Cases Age {a}" for a in case_ages2.columns.tolist()]
cases['risk'].value_counts()
risks = {}
risks['สถานบันเทิง'] = "Entertainment"
risks['อยู่ระหว่างการสอบสวน'] = "Investigating" # Under investigation
risks['การค้นหาผู้ป่วยเชิงรุกและค้นหาผู้ติดเชื้อในชุมชน'] = "Proactive Search"
risks['State Quarantine'] = 'Imported'
risks['ไปสถานที่ชุมชน เช่น ตลาดนัด สถานที่ท่องเที่ยว'] = "Community"
risks['Cluster ผับ Thonglor'] = "Entertainment"
risks['ผู้ที่เดินทางมาจากต่างประเทศ และเข้า ASQ/ALQ'] = 'Imported'
risks['Cluster บางแค'] = "Community" # bangkhee
risks['Cluster ตลาดพรพัฒน์'] = "Community" # market
risks['Cluster ระยอง'] = "Entertainment" # Rayong
# work with foreigners
risks['อาชีพเสี่ยง เช่น ทำงานในสถานที่แออัด หรือทำงานใกล้ชิดสัมผัสชาวต่างชาติ เป็นต้น'] = "Work"
risks['ศูนย์กักกัน ผู้ต้องกัก'] = "Prison" # detention
risks['คนไทยเดินทางกลับจากต่างประเทศ'] = "Imported"
risks['สนามมวย'] = "Entertainment" # Boxing
risks['ไปสถานที่แออัด เช่น งานแฟร์ คอนเสิร์ต'] = "Community" # fair/market
risks['คนต่างชาติเดินทางมาจากต่างประเทศ'] = "Imported"
risks['บุคลากรด้านการแพทย์และสาธารณสุข'] = "Work"
risks['ระบุไม่ได้'] = "Unknown"
risks['อื่นๆ'] = "Unknown"
risks['พิธีกรรมทางศาสนา'] = "Community" # Religious
risks['Cluster บ่อนพัทยา/ชลบุรี'] = "Entertainment" # gambling rayong
risks['ผู้ที่เดินทางมาจากต่างประเทศ และเข้า HQ/AHQ'] = "Imported"
risks['Cluster บ่อนไก่อ่างทอง'] = "Entertainment" # cockfighting
risks['Cluster จันทบุรี'] = "Entertainment" # Chanthaburi - gambling?
risks['Cluster โรงงาน Big Star'] = "Work" # Factory
r = {
27: 'Cluster ชลบุรี:Entertainment', # Chonburi - gambling
28: 'Cluster เครือคัสเซ่อร์พีคโฮลดิ้ง (CPG,CPH):Work',
29: 'ตรวจก่อนทำหัตถการ:Unknown', # 'Check before the procedure'
30: 'สัมผัสผู้เดินทางจากต่างประเทศ:Contact', # 'touch foreign travelers'
31: "Cluster Memory 90's กรุงเทพมหานคร:Entertainment",
32: 'สัมผัสผู้ป่วยยืนยัน:Contact',
33: 'ปอดอักเสบ (Pneumonia):Pneumonia',
34: 'Cluster New Jazz กรุงเทพมหานคร:Entertainment',
35: 'Cluster มหาสารคาม:Entertainment', # Cluster Mahasarakham
36: 'ผู้ที่เดินทางมาจากต่างประเทศ และเข้า OQ:Imported',
37: 'Cluster สมุทรปราการ (โรงงาน บริษัทเมทัล โปรดักส์):Work',
38: 'สัมผัสใกล้ชิดผู้ป่วยยันยันก่อนหน้า:Contact',
39: 'Cluster ตลาดบางพลี:Work',
40: 'Cluster บ่อนเทพารักษ์:Community', # Bangplee Market'
41: 'Cluster Icon siam:Community',
42: 'Cluster The Lounge Salaya:Entertainment',
43: 'Cluster ชลบุรี โรงเบียร์ 90:Entertainment',
44: 'Cluster โรงงาน standard can:Work',
45: 'Cluster ตราด :Community', # Trat?
46: 'Cluster สถานบันเทิงย่านทองหล่อ:Entertainment',
47: 'ไปยังพื้นที่ที่มีการระบาด:Community',
48: 'Cluster สมุทรสาคร:Work', # Samut Sakhon
49: 'สัมผัสใกล้ชิดกับผู้ป่วยยืนยันรายก่อนหน้านี้:Contact',
51: 'อยู่ระหว่างสอบสวน:Unknown',
20210510.1: 'Cluster คลองเตย:Community', # Cluster Klongtoey, 77
# Go to a community / crowded place, 17
20210510.2: 'ไปแหล่งชุมชน/สถานที่คนหนาแน่น:Community',
20210510.3: 'สัมผัสใกล้ชิดผู้ป่วยยืนยันก่อนหน้า:Contact',
# Cluster Chonburi Daikin Company, 3
20210510.4: 'Cluster ชลบุรี บริษัทไดกิ้น:Work',
20210510.5: 'ร้านอาหาร:Entertainment', # restaurant
# touch the infected person confirm Under investigation, 5
20210510.6: 'สัมผัสผู้ติดเชื้อยืนยัน อยู่ระหว่างสอบสวน:Contact',
# touch the infected person confirm Under investigation, 5
20210510.7: 'สัมผัสผู้ป่วยยืนยัน อยู่ระหว่างสอบสวน:Contact',
# Travelers from high-risk areas Bangkok, 2
20210510.8: 'ผู้เดินทางมาจากพื้นที่เสี่ยง กรุงเทพมหานคร:Community',
# to / from Epidemic area, Bangkok Metropolis, 1
20210510.9: 'ไปยัง/มาจาก พื้นที่ระบาดกรุงเทพมหานครมหานคร:Community',
20210510.11: 'ระหว่างสอบสวน:Investigating',
# party pakchong https://www.bangkokpost.com/thailand/general/2103827/5-covid-clusters-in-nakhon-ratchasima
20210510.12: 'Cluster ปากช่อง:Entertainment',
20210512.1: 'Cluster คลองเตย:Community', # klongtoey cluster
20210512.2: 'อยู่ระหว่างสอบสวนโรค:Investigating',
20210512.3: 'อื่น ๆ:Unknown', # Other
# African gem merchants dining after Ramadan
20210512.4: 'Cluster จันทบุรี (ชาวกินี ):Entertainment',
20210516.0: 'Cluster เรือนจำกลางคลองเปรม:Prison', # 894
20210516.1: 'Cluster ตลาดสี่มุมเมือง:Community', # 344 Four Corners Market
20210516.2: 'Cluster สมุทรปราการ GRP Hightech:Work', # 130
20210516.3: 'Cluster ตลาดนนทบุรี:Community', # Cluster Talat Nonthaburi, , 85
20210516.4: 'Cluster โรงงาน QPP ประจวบฯ:Work', # 69
# 41 Cluster Special Prison Thonburi,
20210516.5: 'Cluster เรือนจำพิเศษธนบุรี:Prison',
# 26 Cluster Chanthaburi (Guinea),
20210516.6: 'Cluster จันทบุรี (ชาวกินี):Entertainment',
# 20210516.7: 'Cluster บริษัทศรีสวัสดิ์,Work', #16
20210516.8: 'อื่น:Unknown', # 10
20210516.9: 'Cluster เรือนจำพิเศษมีนบุรี:Prison', # 5
20210516.11: 'Cluster จนท. สนามบินสุวรรณภูมิ:Work', # 4
20210516.12: 'สัมผัสผู้ป่วยที่ติดโควิด:Contact', # 4
20210531.0: 'Cluster เรือนจำพิเศษกรุงเทพ:Prison',
20210531.1: 'Cluster บริษัทศรีสวัสดิ์:Work',
20210531.2: "สัมผัสผู้ป่วยยืนยัน อยู่ระหว่างสอบสวน:Contact",
20210531.3: 'Cluster ตราด:Community',
20210531.4: 'ผู้ที่เดินทางมาจากต่างประเทศ และเข้า AOQ:Imported',
20210531.5: 'ผู้เดินทางมาจากพื้นที่เสี่ยง กรุงเทพมหานคร:Community',
20210531.6: 'Cluster กรุงเทพมหานคร. คลองเตย:Community',
20210622.0: 'อยู่ระหว่างการสอบสวน\n:Investigating',
20210622.1: 'Cluster ตราด:Community',
20210622.2: "สัมผัสผู้ป่วยยืนยัน \n อยู่ระหว่างสอบสวน:Contact",
20210622.3: "ผู้เดินทางมาจากพื้นที่เสี่ยง กรุงเทพมหานคร.:Community",
20210622.4: "อาศัย/เดินทางไปในพื้นที่ที่มีการระบาด:Community",
20210622.5: "อยุ่ระหว่างสอบสวน:Unknown",
20210622.6: "สัมผัสผู้ป่วยยืนยัน อยุ๋ระหว่างสอบสวน:Contact",
20210622.7: "สัมผัสผู้ติดเชื้อยืนยัน\nอยู่ระหว่างสอบสวน:Contact",
20210622.8: "ระหว่างการสอบสวนโรค:Investigating",
20210622.9: "ปอดอักเสบ Pneumonia:Pneumonia",
20210622.01: "Cluster ตลาดบางแค:Community",
20210622.11: "คนไทยเดินทางมาจากต่างประเทศ:Imported",
20210622.12: "คนไทยมาจากพื้นที่เสี่ยง:Community",
20210622.13: "cluster ชลบุรี\n(อยู่ระหว่างการสอบสวน):Investigating",
20210622.14: "Cluster โรงงาน Big Star:Work",
20210622.15: "Cluster สมุทรปราการ ตลาดเคหะบางพลี:Work",
20210622.16: "Cluster ระยอง วิริยะประกันภัย:Work",
20210622.17: "Cluster ตลาดบางแค/คลองขวาง:Work",
20210622.18: "เดินทางมาจากพื้นที่มีการระบาดของโรค:Community",
20210622.19: "Cluster งานมอเตอร์ โชว์:Community",
20210622.02: "ทัณฑสถาน/เรือนจำ:Prison",
20210622.21: "สถานที่ทำงาน:Work",
20210622.22: "รอประสาน:Unknown",
20210622.23: "ผู้ติดเชื้อในประเทศ:Contact",
20210622.24: "ค้นหาเชิงรุก:Proactive Search",
20210622.25: "Cluster ทัณฑสถานโรงพยาบาลราชทัณฑ์:Prison",
20210622.26: "2.สัมผัสผู้ติดเชื้อ:Contact",
20210622.27: "Cluster ระยอง:Community",
20210622.28: "ตรวจสุขภาพแรงงานต่างด้าว:Work",
20210622.29: "สัมผัสในสถานพยาบาล:Work", # contact in hospital
20210622.03: "ไปเที่ยวสถานบันเทิงในอุบลที่พบการระบาดของโรค Ubar:Entertainment",
20210622.31: "ไปสถานที่เสี่ยง เช่น ตลาด สถานที่ชุมชน:Community",
20210622.32: "Cluster ทัณฑสถานหญิงกลาง:Prison",
20210622.33: "ACF สนามกีฬาไทย-ญี่ปุ่น:Entertainment",
20210622.34: "ACF สีลม:Entertainment",
20210622.35: "ACF รองเมือง:Entertainment",
20210622.36: "ACF สนามกีฬาธูปะเตมีย์:Entertainment",
20210622.37: "Cluster ห้างแสงทอง (สายล่าง):Community",
20210622.38: "Cluster ทันฑสถานบำบัดพิเศษกลาง:Community",
20210714.01: "Sandbox:Imported",
20210731.01: "Samui plus:Imported",
20210731.02: "ACF เคหะหลักสี่:Work",
20210731.03: "เดินทางมาจากพื้นที่เสี่ยงที่มีการระบาดของโรค:Community",
20210806.01: "ท้ายบ้าน:Unknown",
20210806.02: "อื่นๆ:Unknown", # Other
20211113.01: "Phuket Sandbox:Imported",
20211113.02: "Chonburi Sandbox:Imported",
20211113.03: "Test and Go:Imported",
20211113.04: "ผู้ที่เดินทางมาจากต่างประเทศ และเข้า AQ:Imported",
20211113.05: "สถานศึกษา:Work", # educational institutions
20211113.06: "สัมผัสผู้ป่วยยืนยัน ภายในครอบครัว/ชุมชน/เพื่อน:Contact",
20211113.07: "10.อื่นๆ:Unknown",
}
for v in r.values():
key, cat = v.split(":")
risks[key] = cat
risks = pd.DataFrame(risks.items(), columns=[
"risk", "risk_group"]).set_index("risk")
cases_risks, unmatched = fuzzy_join(cases, risks, on="risk", return_unmatched=True)
# dump mappings to file so can be inspected
matched = cases_risks[["risk", "risk_group"]]
export(matched.value_counts().to_frame("count"), "risk_groups", csv_only=True)
export(unmatched, "risk_groups_unmatched", csv_only=True)
case_risks_daily = | pd.crosstab(cases_risks['Date'], cases_risks["risk_group"]) | pandas.crosstab |
# -*- coding: utf-8 -*-
"""
Created on Sun Sep 20 00:24:43 2020
@author: Ray
@email: <EMAIL>
@wechat: RayTing0305
"""
import pandas as pd
import numpy as np
import re
'''
Quiz
'''
index1 = ['James', 'Mike', 'Sally']
col1 = ['Business', 'Law', 'Engineering']
student_df = pd.DataFrame(col1, index1)
student_df.index.name='Name'
student_df.columns = ['School']
index2 = ['Kelly', 'Sally', 'James']
col2 = ['Director of HR', 'Course liasion', 'Grader']
staff_df = pd.DataFrame(col2, index2)
staff_df.index.name = 'Name'
staff_df.columns = ['Role']
df = pd.DataFrame({'P2010':[100.1, 200.1],
'P2011':[100.1, 200.1],
'P2012':[100.1, 200.1],
'P2013':[100.1, 200.1],
'P2014':[100.1, 200.1],
'P2015':[100.1, 200.1]})
frames = ['P2010', 'P2011', 'P2012', 'P2013','P2014', 'P2015']
df['AVG'] = df[frames].apply(lambda z: np.mean(z), axis=1)
result_df = df.drop(frames,axis=1)
df = pd.DataFrame(['A+', 'A', 'A-', 'B+', 'B', 'B-', 'C+', 'C', 'C-', 'D+', 'D'], index=['excellent', 'excellent', 'excellent', 'good', 'good', 'good', 'ok', 'ok', 'ok', 'poor', 'poor'], columns = ['Grades'])
my_categories= pd.CategoricalDtype(categories=['A+', 'A', 'A-', 'B+', 'B', 'B-', 'C+', 'C', 'C-', 'D+', 'D'], ordered=True)
grades = df['Grades'].astype(my_categories)
result = grades[(grades>'B') & (grades<'A')]
(pd.Timestamp('11/29/2019') + pd.offsets.MonthEnd()).weekday()
pd.Period('01/12/2019', 'M') + 5
'''
Assignment
'''
def answer_one():
# YOUR CODE HERE
x = pd.ExcelFile('assets/Energy Indicators.xls')
energy = x.parse(skiprows=17,skip_footer=(38))
energy = energy[['Unnamed: 1','Petajoules','Gigajoules','%']]
energy.columns = ['Country', 'Energy Supply', 'Energy Supply per Capita', '% Renewable']
energy[['Energy Supply', 'Energy Supply per Capita', '% Renewable']] = energy[['Energy Supply', 'Energy Supply per Capita', '% Renewable']].replace('...',np.NaN).apply(pd.to_numeric)
energy['Energy Supply'] = energy['Energy Supply']*1000000
energy['Country'] = energy['Country'].replace({
"Republic of Korea": "South Korea",
"United States of America": "United States",
"United Kingdom of Great Britain and Northern Ireland": "United Kingdom",
"China, Hong Kong Special Administrative Region": "Hong Kong",
'Iran (Islamic Republic of)':'Iran',
'Bolivia (Plurinational State of)':'Bolivia'
})
energy['Country'] = energy['Country'].str.replace(r" \(.*\)","")
GDP = pd.read_csv('assets/world_bank.csv',skiprows=4)
GDP['Country Name'] = GDP['Country Name'].replace({
"Korea, Rep.": "South Korea",
"Iran, Islamic Rep.": "Iran",
"Hong Kong SAR, China": "Hong Kong"
})
GDP = GDP[['Country Name','2006','2007','2008','2009','2010','2011','2012','2013','2014','2015']]
GDP.columns = ['Country','2006','2007','2008','2009','2010','2011','2012','2013','2014','2015']
ScimEn = pd.read_excel(io='assets/scimagojr-3.xlsx')
ScimEn_m = ScimEn[:15]
df1 = pd.merge(ScimEn_m, energy, how='inner', left_on='Country', right_on='Country')
df2 = pd.merge(df1, GDP, how='inner', left_on='Country', right_on='Country')
res = df2.set_index('Country')
return res
raise NotImplementedError()
def answer_two():
# YOUR CODE HERE
x = pd.ExcelFile('assets/Energy Indicators.xls')
energy = x.parse(skiprows=17,skip_footer=(38))
energy = energy[['Unnamed: 1','Petajoules','Gigajoules','%']]
energy.columns = ['Country', 'Energy Supply', 'Energy Supply per Capita', '% Renewable']
energy[['Energy Supply', 'Energy Supply per Capita', '% Renewable']] = energy[['Energy Supply', 'Energy Supply per Capita', '% Renewable']].replace('...',np.NaN).apply(pd.to_numeric)
energy['Energy Supply'] = energy['Energy Supply']*1000000
energy['Country'] = energy['Country'].replace({
"Republic of Korea": "South Korea",
"United States of America": "United States",
"United Kingdom of Great Britain and Northern Ireland": "United Kingdom",
"China, Hong Kong Special Administrative Region": "Hong Kong",
'Iran (Islamic Republic of)':'Iran',
'Bolivia (Plurinational State of)':'Bolivia'
})
energy['Country'] = energy['Country'].str.replace(r" \(.*\)","")
GDP = | pd.read_csv('assets/world_bank.csv',skiprows=4) | pandas.read_csv |
import pytest
import pandas as pd
import pickle
from hashlib import sha256
from tempfile import NamedTemporaryFile
from ketl.loader.Loader import (
BaseLoader, DatabaseLoader, HashLoader, DelimitedFileLoader, ParquetLoader,
LocalFileLoader, PickleLoader
)
from ketl.db.settings import get_engine
@pytest.fixture
def data_frame():
return pd.DataFrame.from_records([(1, 2, 3), (4, 5, 6)], columns=['x', 'y', 'z'])
def test_base_loader(data_frame):
loader = BaseLoader('foo')
assert loader.destination == 'foo'
with pytest.raises(NotImplementedError):
loader.load(data_frame)
with pytest.raises(NotImplementedError):
loader.finalize()
def test_hash_loader(data_frame, tmp_path):
tf = NamedTemporaryFile(dir=tmp_path, delete=False)
tf.close()
loader = HashLoader(tf.name)
expected_hash = sha256(pd.util.hash_pandas_object(data_frame).values).hexdigest()
loader.load(data_frame)
with open(tf.name, 'r') as f:
data = f.read().strip()
assert expected_hash == data
loader.finalize()
def test_local_file_loader_csv(data_frame, tmp_path):
csv_file = tmp_path / 'df.csv'
with open(csv_file, 'w') as f:
f.write('hello world')
loader = DelimitedFileLoader(csv_file, index=False)
assert loader.destination == csv_file
assert loader.kwargs == {'index': False}
assert not csv_file.exists()
loader.load(data_frame)
assert csv_file.exists()
df_out = pd.read_csv(csv_file, index_col=None)
assert df_out.equals(data_frame)
def test_local_file_loader_parquet(data_frame, tmp_path):
parquet_file = tmp_path / 'df.parquet'
loader = ParquetLoader(parquet_file, index=False)
assert loader.destination == parquet_file
assert loader.kwargs == {'index': False}
assert not parquet_file.exists()
loader.load(data_frame)
loader.finalize()
assert parquet_file.exists()
df_out = pd.read_parquet(parquet_file)
assert df_out.equals(data_frame)
df1 = data_frame.copy(deep=True)
df1.attrs['name'] = 'df1'
df2 = data_frame.copy(deep=True)
df2.attrs['name'] = 'df2'
def naming_function(df):
return df.attrs['name'] + '.parquet'
pq_file1 = tmp_path / 'df1.parquet'
pq_file2 = tmp_path / 'df2.parquet'
loader = ParquetLoader(tmp_path, naming_func=naming_function, index=False)
loader.load(df1)
loader.load(df2)
loader.finalize()
df1_out = pd.read_parquet(pq_file1)
df2_out = | pd.read_parquet(pq_file2) | pandas.read_parquet |
import matplotlib.pyplot as plt
import matplotlib.dates as mdates
from datetime import datetime
from pandas.plotting import register_matplotlib_converters
register_matplotlib_converters()
import numpy as np
import operator
import pandas as pd
from Abstract import Conference
# ================================== read conferences csv ==================================
data = pd.DataFrame()
df = pd.read_csv('conferences.csv')
df['date'] = | pd.to_datetime(df['date']) | pandas.to_datetime |
from tea.ast import ( Node, Variable, Literal,
Equal, NotEqual, LessThan,
LessThanEqual, GreaterThan, GreaterThanEqual,
Relate, PositiveRelationship
)
from tea.runtimeDataStructures.dataset import Dataset
from tea.runtimeDataStructures.varData import VarData
from tea.runtimeDataStructures.bivariateData import BivariateData
from tea.runtimeDataStructures.multivariateData import MultivariateData
from tea.runtimeDataStructures.resultData import ResultData
from tea.helpers.evaluateHelperMethods import determine_study_type, assign_roles, add_paired_property, execute_test
from tea.z3_solver.solver import synthesize_tests
import attr
from typing import Any
from types import SimpleNamespace # allows for dot notation access for dictionaries
from typing import Dict
from scipy import stats # Stats library used
import statsmodels.api as sm
import statsmodels.formula.api as smf
import numpy as np # Use some stats from numpy instead
import pandas as pd
# TODO: Pass participant_id as part of experimental design, not load_data
def evaluate(dataset: Dataset, expr: Node, assumptions: Dict[str, str], design: Dict[str, str]=None):
if isinstance(expr, Variable):
# dataframe = dataset[expr.name] # I don't know if we want this. We may want to just store query (in metadata?) and
# then use query to get raw data later....(for user, not interpreter?)
metadata = dataset.get_variable_data(expr.name) # (dtype, categories)
# if expr.name == 'strategy':
# import pdb; pdb.set_trace()
metadata['var_name'] = expr.name
metadata['query'] = ''
return VarData(metadata)
elif isinstance(expr, Literal):
data = pd.Series([expr.value] * len(dataset.data), index=dataset.data.index) # Series filled with literal value
# metadata = None # metadata=None means literal
metadata = dict() # metadata=None means literal
metadata['var_name'] = '' # because not a var in the dataset
metadata['query'] = ''
metadata['value'] = expr.value
return VarData(data, metadata)
elif isinstance(expr, Equal):
lhs = evaluate(dataset, expr.lhs)
rhs = evaluate(dataset, expr.rhs)
assert isinstance(lhs, VarData)
assert isinstance(rhs, VarData)
dataframe = lhs.dataframe[lhs.dataframe == rhs.dataframe]
metadata = lhs.metadata
if (isinstance(expr.rhs, Literal)):
metadata['query'] = f" == \'{rhs.metadata['value']}\'" # override lhs metadata for query
elif (isinstance(expr.rhs, Variable)):
metadata['query'] = f" == {rhs.metadata['var_name']}"
else:
raise ValueError(f"Not implemented for {rhs}")
return VarData(metadata)
elif isinstance(expr, NotEqual):
rhs = evaluate(dataset, expr.rhs)
lhs = evaluate(dataset, expr.lhs)
assert isinstance(rhs, VarData)
assert isinstance(lhs, VarData)
dataframe = lhs.dataframe[lhs.dataframe != rhs.dataframe]
metadata = lhs.metadata
if (isinstance(expr.rhs, Literal)):
metadata['query'] = " != \'\'" # override lhs metadata for query
elif (isinstance(expr.rhs, Variable)):
metadata['query'] = f" != {rhs.metadata['var_name']}"
else:
raise ValueError(f"Not implemented for {rhs}")
return VarData(metadata)
elif isinstance(expr, LessThan):
lhs = evaluate(dataset, expr.lhs)
rhs = evaluate(dataset, expr.rhs)
assert isinstance(lhs, VarData)
assert isinstance(rhs, VarData)
dataframe = None
metadata = rhs.metadata
if (not lhs.metadata):
raise ValueError('Malformed Relation. Filter on Variables must have variable as rhs')
elif (lhs.metadata['dtype'] is DataType.NOMINAL):
raise ValueError('Cannot compare nominal values with Less Than')
elif (lhs.metadata['dtype'] is DataType.ORDINAL):
# TODO May want to add a case should RHS and LHS both be variables
# assert (rhs.metadata is None)
comparison = rhs.dataframe.iloc[0]
if (isinstance(comparison, str)):
categories = lhs.metadata['categories'] # OrderedDict
# Get raw Pandas Series indices for desired data
ids = [i for i,x in enumerate(lhs.dataframe) if categories[x] < categories[comparison]]
# Get Pandas Series set indices for desired data
p_ids = [lhs.dataframe.index.values[i] for i in ids]
# Create new Pandas Series with only the desired data, using set indices
dataframe = pd.Series(lhs.dataframe, p_ids)
dataframe.index.name = dataset.pid_col_name
elif (np.issubdtype(comparison, np.integer)):
categories = lhs.metadata['categories'] # OrderedDict
# Get raw Pandas Series indices for desired data
ids = [i for i,x in enumerate(lhs.dataframe) if categories[x] < comparison]
# Get Pandas Series set indices for desired data
p_ids = [lhs.dataframe.index.values[i] for i in ids]
# Create new Pandas Series with only the desired data, using set indices
dataframe = | pd.Series(lhs.dataframe, p_ids) | pandas.Series |
from __future__ import division
from datetime import datetime
import sys
if sys.version_info < (3, 3):
import mock
else:
from unittest import mock
import pandas as pd
import numpy as np
from nose.tools import assert_almost_equal as aae
import bt
import bt.algos as algos
def test_algo_name():
class TestAlgo(algos.Algo):
pass
actual = TestAlgo()
assert actual.name == 'TestAlgo'
class DummyAlgo(algos.Algo):
def __init__(self, return_value=True):
self.return_value = return_value
self.called = False
def __call__(self, target):
self.called = True
return self.return_value
def test_algo_stack():
algo1 = DummyAlgo(return_value=True)
algo2 = DummyAlgo(return_value=False)
algo3 = DummyAlgo(return_value=True)
target = mock.MagicMock()
stack = bt.AlgoStack(algo1, algo2, algo3)
actual = stack(target)
assert not actual
assert algo1.called
assert algo2.called
assert not algo3.called
def test_run_once():
algo = algos.RunOnce()
assert algo(None)
assert not algo(None)
assert not algo(None)
def test_run_period():
target = mock.MagicMock()
dts = pd.date_range('2010-01-01', periods=35)
data = pd.DataFrame(index=dts, columns=['c1', 'c2'], data=100)
algo = algos.RunPeriod()
# adds the initial day
backtest = bt.Backtest(
bt.Strategy('', [algo]),
data
)
target.data = backtest.data
dts = target.data.index
target.now = None
assert not algo(target)
# run on first date
target.now = dts[0]
assert not algo(target)
# run on first supplied date
target.now = dts[1]
assert algo(target)
# run on last date
target.now = dts[len(dts) - 1]
assert not algo(target)
algo = algos.RunPeriod(
run_on_first_date=False,
run_on_end_of_period=True,
run_on_last_date=True
)
# adds the initial day
backtest = bt.Backtest(
bt.Strategy('', [algo]),
data
)
target.data = backtest.data
dts = target.data.index
# run on first date
target.now = dts[0]
assert not algo(target)
# first supplied date
target.now = dts[1]
assert not algo(target)
# run on last date
target.now = dts[len(dts) - 1]
assert algo(target)
# date not in index
target.now = datetime(2009, 2, 15)
assert not algo(target)
def test_run_daily():
target = mock.MagicMock()
dts = pd.date_range('2010-01-01', periods=35)
data = pd.DataFrame(index=dts, columns=['c1', 'c2'], data=100)
algo = algos.RunDaily()
# adds the initial day
backtest = bt.Backtest(
bt.Strategy('',[algo]),
data
)
target.data = backtest.data
target.now = dts[1]
assert algo(target)
def test_run_weekly():
dts = pd.date_range('2010-01-01', periods=367)
data = pd.DataFrame(index=dts, columns=['c1', 'c2'], data=100)
target = mock.MagicMock()
target.data = data
algo = algos.RunWeekly()
# adds the initial day
backtest = bt.Backtest(
bt.Strategy('', [algo]),
data
)
target.data = backtest.data
# end of week
target.now = dts[2]
assert not algo(target)
# new week
target.now = dts[3]
assert algo(target)
algo = algos.RunWeekly(
run_on_first_date=False,
run_on_end_of_period=True,
run_on_last_date=True
)
# adds the initial day
backtest = bt.Backtest(
bt.Strategy('', [algo]),
data
)
target.data = backtest.data
# end of week
target.now = dts[2]
assert algo(target)
# new week
target.now = dts[3]
assert not algo(target)
dts = pd.DatetimeIndex([datetime(2016, 1, 3), datetime(2017, 1, 8),datetime(2018, 1, 7)])
data = pd.DataFrame(index=dts, columns=['c1', 'c2'], data=100)
# adds the initial day
backtest = bt.Backtest(
bt.Strategy('', [algo]),
data
)
target.data = backtest.data
# check next year
target.now = dts[1]
assert algo(target)
def test_run_monthly():
dts = pd.date_range('2010-01-01', periods=367)
data = pd.DataFrame(index=dts, columns=['c1', 'c2'], data=100)
target = mock.MagicMock()
target.data = data
algo = algos.RunMonthly()
# adds the initial day
backtest = bt.Backtest(
bt.Strategy('', [algo]),
data
)
target.data = backtest.data
# end of month
target.now = dts[30]
assert not algo(target)
# new month
target.now = dts[31]
assert algo(target)
algo = algos.RunMonthly(
run_on_first_date=False,
run_on_end_of_period=True,
run_on_last_date=True
)
# adds the initial day
backtest = bt.Backtest(
bt.Strategy('', [algo]),
data
)
target.data = backtest.data
# end of month
target.now = dts[30]
assert algo(target)
# new month
target.now = dts[31]
assert not algo(target)
dts = pd.DatetimeIndex([datetime(2016, 1, 3), datetime(2017, 1, 8), datetime(2018, 1, 7)])
data = pd.DataFrame(index=dts, columns=['c1', 'c2'], data=100)
# adds the initial day
backtest = bt.Backtest(
bt.Strategy('', [algo]),
data
)
target.data = backtest.data
# check next year
target.now = dts[1]
assert algo(target)
def test_run_quarterly():
dts = pd.date_range('2010-01-01', periods=367)
data = pd.DataFrame(index=dts, columns=['c1', 'c2'], data=100)
target = mock.MagicMock()
target.data = data
algo = algos.RunQuarterly()
# adds the initial day
backtest = bt.Backtest(
bt.Strategy('', [algo]),
data
)
target.data = backtest.data
# end of quarter
target.now = dts[89]
assert not algo(target)
# new quarter
target.now = dts[90]
assert algo(target)
algo = algos.RunQuarterly(
run_on_first_date=False,
run_on_end_of_period=True,
run_on_last_date=True
)
# adds the initial day
backtest = bt.Backtest(
bt.Strategy('', [algo]),
data
)
target.data = backtest.data
# end of quarter
target.now = dts[89]
assert algo(target)
# new quarter
target.now = dts[90]
assert not algo(target)
dts = pd.DatetimeIndex([datetime(2016, 1, 3), datetime(2017, 1, 8), datetime(2018, 1, 7)])
data = pd.DataFrame(index=dts, columns=['c1', 'c2'], data=100)
# adds the initial day
backtest = bt.Backtest(
bt.Strategy('', [algo]),
data
)
target.data = backtest.data
# check next year
target.now = dts[1]
assert algo(target)
def test_run_yearly():
dts = pd.date_range('2010-01-01', periods=367)
data = pd.DataFrame(index=dts, columns=['c1', 'c2'], data=100)
target = mock.MagicMock()
target.data = data
algo = algos.RunYearly()
# adds the initial day
backtest = bt.Backtest(
bt.Strategy('', [algo]),
data
)
target.data = backtest.data
# end of year
target.now = dts[364]
assert not algo(target)
# new year
target.now = dts[365]
assert algo(target)
algo = algos.RunYearly(
run_on_first_date=False,
run_on_end_of_period=True,
run_on_last_date=True
)
# adds the initial day
backtest = bt.Backtest(
bt.Strategy('', [algo]),
data
)
target.data = backtest.data
# end of year
target.now = dts[364]
assert algo(target)
# new year
target.now = dts[365]
assert not algo(target)
def test_run_on_date():
target = mock.MagicMock()
target.now = pd.to_datetime('2010-01-01')
algo = algos.RunOnDate('2010-01-01', '2010-01-02')
assert algo(target)
target.now = | pd.to_datetime('2010-01-02') | pandas.to_datetime |
# Databricks notebook source
# MAGIC %md
# MAGIC
# MAGIC # Databricks - Credit Scoring
# MAGIC
# MAGIC ## Introduction
# MAGIC
# MAGIC Banks play a crucial role in market economies. They decide who can get finance and on what terms and can make or break investment decisions. For markets and society to function, individuals and companies need access to credit.
# MAGIC
# MAGIC Credit scoring algorithms, which make a guess at the probability of default, are the method banks use to determine whether or not a loan should be granted.
# MAGIC
# MAGIC ## The problem
# MAGIC
# MAGIC Down below you will find a possible solution to the challenge described in [c/GiveMeSomeCredit](https://www.kaggle.com/c/GiveMeSomeCredit) where participants where required to improve on the state of the art in credit scoring, by predicting the probability that somebody will experience financial distress in the next two years.
# MAGIC
# MAGIC ## The data
# MAGIC
# MAGIC The training data contains the following variables:
# MAGIC
# MAGIC
# MAGIC | **Variable Name** | **Description** | **Type** |
# MAGIC |--------------------------------------|--------------------------------------------------------------------------------------------------------------------------------------------------------------|------------|
# MAGIC | SeriousDlqin2yrs | Person experienced 90 days past due delinquency or worse | *Y/N* |
# MAGIC | RevolvingUtilizationOfUnsecuredLines | Total balance on credit cards and personal lines of credit except real estate and no installment debt like car loans divided by the sum of credit limits | percentage |
# MAGIC | age | Age of borrower in years | integer |
# MAGIC | NumberOfTime30-59DaysPastDueNotWorse | Number of times borrower has been 30-59 days past due but no worse in the last 2 years. | integer |
# MAGIC | DebtRatio | Monthly debt payments, alimony,living costs divided by monthy gross income | percentage |
# MAGIC | MonthlyIncome | Monthly income | real |
# MAGIC | NumberOfOpenCreditLinesAndLoans | Number of Open loans (installment like car loan or mortgage) and Lines of credit (e.g. credit cards) | integer |
# MAGIC | NumberOfTimes90DaysLate | Number of times borrower has been 90 days or more past due. | integer |
# MAGIC | NumberRealEstateLoansOrLines | Number of mortgage and real estate loans including home equity lines of credit | integer |
# MAGIC | NumberOfTime60-89DaysPastDueNotWorse | Number of times borrower has been 60-89 days past due but no worse in the last 2 years. | integer |
# MAGIC | NumberOfDependents | Number of dependents in family excluding themselves (spouse, children etc.) | integer |
# MAGIC
# MAGIC The **SeriousDlqin2yrs** is the dependent variable of the dataset, or better named the **label**. This is a boolean value which details if a certain individual has experienced a deliquency of 90 days past due or worse in the last 2 years.
# MAGIC
# MAGIC You can get the training data from [here](https://github.com/dlawrences/GlobalAINightBucharest/blob/master/data/cs-training.csv).
# MAGIC
# MAGIC This dataset should be used for:
# MAGIC - creating two smaller sets, one for the actual training (e.g. 80%) and one for testing (e.g. 20%)
# MAGIC - during cross validation, if you want to do the validation on multiple different folds of data to manage better the bias and the variance
# MAGIC
# MAGIC The benchmark/real unseen data you could use to test your model predictions may be downloaded from [here](https://github.com/dlawrences/GlobalAINightBucharest/blob/master/data/cs-test.csv).
# MAGIC
# MAGIC ## The Data Science Process
# MAGIC
# MAGIC This is the outline of the process we'll be following in this workshop.
# MAGIC
# MAGIC 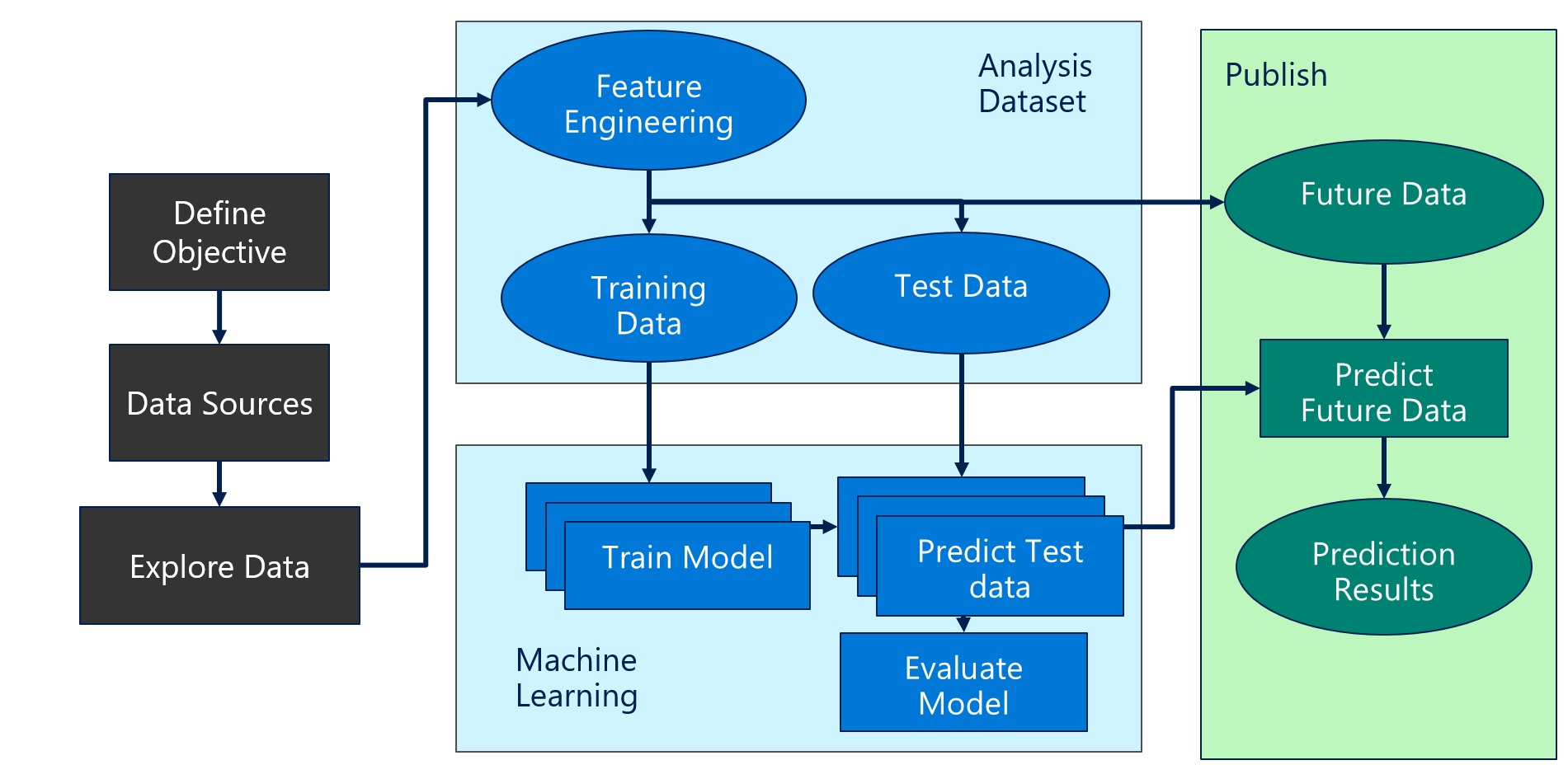
# COMMAND ----------
# MAGIC %md
# MAGIC ## Data import
# MAGIC
# MAGIC Before starting to do anything else, we need to import the data. First, let's download both datasets and store them in DBFS.
# COMMAND ----------
import urllib.request
training_data_url = "https://raw.githubusercontent.com/dlawrences/GlobalAINightBucharest/master/data/cs-training.csv"
training_data_filename = "cs_training.csv"
test_data_url = "https://raw.githubusercontent.com/dlawrences/GlobalAINightBucharest/master/data/cs-test.csv"
test_data_filename = "cs_test.csv"
dbfs_data_folder = "dbfs/FileStore/data/"
project_name = 'credit-scoring'
dbfs_project_folder = dbfs_data_folder + project_name + "/"
# Download files and move them to the final directory in DBFS
urllib.request.urlretrieve(training_data_url, "/tmp/" + training_data_filename)
urllib.request.urlretrieve(test_data_url, "/tmp/" + test_data_filename)
# Create the project directory if it does not exist and move files to it
dbutils.fs.mkdirs(dbfs_project_folder)
dbutils.fs.mv("file:/tmp/" + training_data_filename, dbfs_project_folder)
dbutils.fs.mv("file:/tmp/" + test_data_filename, dbfs_project_folder)
# List the contents of the directory
dbutils.fs.ls(dbfs_project_folder)
# COMMAND ----------
import numpy as np # library for linear algebra and stuff
import pandas as pd # library for data processing, I/O on csvs etc
import matplotlib.pyplot as plt # library for plotting
import seaborn as sns # a library which is better for plotting
# File location and type
file_location = dbfs_project_folder + training_data_filename
file_type = "csv"
# CSV options
infer_schema = "true"
first_row_is_header = "true"
delimiter = ","
# The applied options are for CSV files. For other file types, these will be ignored.
creditSDF = spark.read.format(file_type) \
.option("inferSchema", infer_schema) \
.option("header", first_row_is_header) \
.option("sep", delimiter) \
.load(file_location)
display(creditSDF)
# COMMAND ----------
# MAGIC %md
# MAGIC
# MAGIC Now that we've loaded the dataset, the first thing we're going to do is set aside a part of it - 25 to 30 percent is a usual percentage - and not touch it until it's time to test our models.
# COMMAND ----------
temp_table_name = "trainingSDF"
# Split the data into training and test sets (25% held out for testing)
(trainingSDF, testingSDF) = creditSDF.randomSplit([0.75, 0.25], seed=1)
# Make the dataframe available in the SQL context
trainingSDF.createOrReplaceTempView(temp_table_name)
# COMMAND ----------
# Sample out 10 rows of the dataset
display(trainingSDF.sample(False, 0.1, seed=0).limit(10))
# COMMAND ----------
# Inspect the schema
trainingSDF.printSchema()
# COMMAND ----------
# Check of the summary statistics of the features
display(trainingSDF.describe())
# COMMAND ----------
# highlight how many missing values we have for every feature
from pyspark.sql.functions import lit, col
rows = trainingSDF.count()
summary = trainingSDF.describe().filter(col("summary") == "count")
display(summary.select(*((lit(rows)-col(c)).alias(c) for c in trainingSDF.columns)))
# COMMAND ----------
# MAGIC %md
# MAGIC Quick conclusions:
# MAGIC - there are a lot of null values for **MonthlyIncome** and **NumberOfDependents**; we will analyse how to impute these next
# MAGIC - the minimum value for the **age** variable is 0 and it presents an outlier/bad data; this will be imputed with the median
# MAGIC - the maximum value of **329664** for the **DebtRatio** variable is rather weird given this variable is a mere percentage; from a modelling perspective, thus we will need to understand why there are such big values and decide what to do with them
# MAGIC - the maximum value of **50708** for the **RevolvingUtilizationOfUnsecuredLines** variable is rather weird given this variable is a mere percentage; rom a modelling perspective, thus we will need to understand why there are such big values and decide what to do with them
# COMMAND ----------
# MAGIC %md
# MAGIC ## Exploratory Data Analysis & Data Cleaning
# MAGIC
# MAGIC We are going to take step by step most of the interesting columns that need visualizing and cleansing to be done.
# MAGIC
# MAGIC ### Target class - SeriousDlqin2yrs
# MAGIC
# MAGIC Let's understand the distribution of our target class (**SeriousDlqin2yrs**). This could very well influence the algorithm we will want to use to model the problem.
# COMMAND ----------
# MAGIC %sql
# MAGIC
# MAGIC select SeriousDlqin2yrs, count(*) as TotalCount from trainingSDF group by SeriousDlqin2yrs
# COMMAND ----------
# MAGIC %md
# MAGIC
# MAGIC There seems to be a lot of **class imbalance** going on. Let's understand the positive event rate in our target class.
# COMMAND ----------
class_0 = trainingSDF.filter(trainingSDF.SeriousDlqin2yrs == 0).count()
class_1 = trainingSDF.filter(trainingSDF.SeriousDlqin2yrs == 1).count()
print("Total number of observations with a class of 0: {}".format(class_0))
print("Total number of observations with a class of 1: {}".format(class_1))
print("Positive event rate: {} %".format(class_1/(class_0+class_1) * 100))
# COMMAND ----------
# MAGIC %md
# MAGIC
# MAGIC A positive event rate of 6.6% is by no means ideal. Going through with this distribution for the target class may mean that the minorit class will be ignored by the algorithm we are going to use to model the problem, thus the model will be biased to customers which are not likely to default.
# MAGIC
# MAGIC A couple of ideas which we are going to take into consideration going further to go around this problem:
# MAGIC - given we have a lot of training data (100k+ observations), we may actually considering resampling the dataset.
# MAGIC - we are going to use an evaluation metric which compensates the imbalance between classes, e.g. **ROC AUC**
# COMMAND ----------
# MAGIC %md
# MAGIC ### Age variable
# MAGIC
# MAGIC We are interested in knowing the distribution of the **age** variable.
# MAGIC
# MAGIC We are not looking for customers under the legal age of 18 years. If any, we will impute the age of these with the median of the column.
# COMMAND ----------
import matplotlib.ticker as ticker
# spark.sql does not have any histogram method, however the RDD api does
age_histogram = trainingSDF.select('age').rdd.flatMap(lambda x: x).histogram(10)
fig, ax = plt.subplots()
# the computed histogram needs to be loaded in a pandas dataframe so we will be able to plot it using sns
age_histogram_df = pd.DataFrame(
list(zip(*age_histogram)),
columns=['bin', 'frequency']
)
ax = sns.barplot(x = "bin", y = "frequency", data = age_histogram_df)
ax.get_xaxis().set_major_formatter(ticker.FuncFormatter(lambda x, p: format(age_histogram_df.iloc[x]['bin'], '.1f')))
display(fig)
# COMMAND ----------
# MAGIC %md
# MAGIC It seems there may be customers under the legal age. Let's see how many.
# COMMAND ----------
# We can use the filter method to understand what are the observations for which the customers falls under the legal age.
display(trainingSDF.filter(trainingSDF.age < 18))
# COMMAND ----------
# MAGIC %md
# MAGIC Fortunately there is only one. Let's impute this value with the median.
# COMMAND ----------
# Import functions which will help us code an if statement
from pyspark.sql import functions as F
def imputeAgeWithMedian(df, medianAge):
# Update with the median for the rows where the age columnis equal to 0
df = df.withColumn('age',
F.when(
F.col('age') == 0,
medianAge
).otherwise(
F.col('age')
)
)
return df
# Compute the median of the age variable
trainingMedianAge = np.median(trainingSDF.select('age').dropna().collect())
trainingSDF = imputeAgeWithMedian(trainingSDF, trainingMedianAge)
# Check to see that the only row shown above has a new age value
display(trainingSDF.filter(trainingSDF.Idx == 65696))
# COMMAND ----------
# MAGIC %md
# MAGIC Finally, let's check the distribution of the age for each group, based on the values for the **SeriousDlqin2yrs** target variable.
# MAGIC
# MAGIC We're going to use a [box and whiskers plot](https://towardsdatascience.com/understanding-boxplots-5e2df7bcbd51?gi=9e6b6042f263) to better visualize the distribution.
# COMMAND ----------
fig, ax = plt.subplots()
ax = sns.boxplot(x="SeriousDlqin2yrs", y="age", data = trainingSDF.toPandas())
display(fig)
# COMMAND ----------
# MAGIC %sql
# MAGIC
# MAGIC SELECT SeriousDlqin2yrs, age FROM trainingSDF
# COMMAND ----------
# MAGIC %md
# MAGIC Based on the cleaned age column, let's create an age banding column (bins) which might be better predictors to credit risk.
# MAGIC
# MAGIC For this example, we are going to use the bins included in this paper: [figure in paper](https://www.researchgate.net/figure/Percentage-of-default-risk-among-different-age-groups_fig2_268345909).
# MAGIC
# MAGIC > NOTE: For simplicity we are using a [Spark UDF](https://databricks.com/blog/2017/10/30/introducing-vectorized-udfs-for-pyspark.html) (User-defined function), although that may pose performance problems if the dataset is large. Consider using Scala for the production data preparation pipeline once the data scientist has defined and tested one that should be used in production.
# COMMAND ----------
from pyspark.sql.functions import udf
from pyspark.sql.types import StringType
def bandingFunction(age):
if (age < 25):
return '18-25'
elif (age >= 25 and age < 30):
return '25-29'
elif (age >= 30 and age < 35):
return '30-34'
elif (age >= 35 and age < 40):
return '35-39'
elif (age >= 40 and age < 45):
return '40-44'
elif (age >= 45 and age < 50):
return '45-49'
elif (age >= 50 and age < 55):
return '50-54'
elif (age >= 55 and age < 60):
return '55-59'
elif (age >= 60 and age < 65):
return '60-64'
elif (age >= 65 and age < 70):
return '65-69'
elif (age >= 70 and age < 75):
return '70-74'
elif (age >= 75):
return '75+'
else:
return ''
age_banding_udf = udf(bandingFunction, StringType() )
def addAgeBanding(df):
df = df.withColumn('age_banding', age_banding_udf(df.age))
return df.drop('age')
trainingSDF = addAgeBanding(trainingSDF)
trainingSDF.createOrReplaceTempView(temp_table_name)
# COMMAND ----------
# MAGIC %md
# MAGIC Let's now visualize the distribution.
# MAGIC
# MAGIC NOTE: as an alternative to Python-based plotting libraries like *seaborn* or *pyplot* we can also use Databricks' built-in visualizations. Click on the Visualization button below the results of this cell to select a **Bar** chart.
# COMMAND ----------
# MAGIC %sql
# MAGIC select age_banding, count(*) as Counts from trainingSDF group by age_banding order by age_banding
# COMMAND ----------
# MAGIC %md
# MAGIC ### MonthlyIncome variable
# MAGIC
# MAGIC In credit scoring, the income of the individual - besides the other debt that he is into - is of greater importance than other things when it comes to the final decision.
# MAGIC
# MAGIC Let's see how the distribution of this variable looks.
# COMMAND ----------
fig, ax = plt.subplots()
ax = sns.boxplot(x="SeriousDlqin2yrs", y="MonthlyIncome", data = trainingSDF.toPandas())
display(fig)
# COMMAND ----------
# MAGIC %md
# MAGIC
# MAGIC Hmm, the chart isn't that useful, probably because we have some very large outliers (large *MonthlyIncome* values) skewing the plot. Let's try using a log scale for the y axis:
# COMMAND ----------
fig, ax = plt.subplots()
sns.set(style="whitegrid")
ax = sns.boxplot(x="SeriousDlqin2yrs", y="MonthlyIncome", data = trainingSDF.toPandas())
ax.set_yscale("log")
display(fig)
# COMMAND ----------
# MAGIC %md
# MAGIC We can also display the quartile values of MonthlyIncome for each class, using Spark SQL.
# COMMAND ----------
# MAGIC %sql
# MAGIC
# MAGIC SELECT SeriousDlqin2yrs,
# MAGIC percentile(MonthlyIncome,0.25) AS Q1,
# MAGIC percentile(MonthlyIncome,0.5) AS Q2_Median,
# MAGIC percentile(MonthlyIncome,0.75) AS Q3
# MAGIC FROM trainingSDF
# MAGIC GROUP BY SeriousDlqin2yrs
# COMMAND ----------
# MAGIC %md
# MAGIC
# MAGIC That's better. One thing is certain - people which have gone through issues usually have a lower income. However, from the original summary statistics it also looked like the dataset contained really low values - like 5$ or less a month which is really odd.
# MAGIC
# MAGIC For our reference, let's view the [Characteristics of Minimum Wage Workers in the US: 2010](https://www.bls.gov/cps/minwage2010.htm). In this article, it is stated that the prevailing Federal minimum wage was $7.25 per hour.
# MAGIC
# MAGIC In this case, considering an individual would work on a full-time basis for 52 weeks straight in a year, that individual would earn **$7.25 X 40 hrs X 52 weeks** = **_$15,080_**.
# MAGIC
# MAGIC This translates to approximately **_$1,256_** a month. For a part-time worker, this would mean a wage of **_$628_**. For an individual working only a quarter of the total time, that would mean a wage of only **_$314_**.
# MAGIC
# MAGIC According to the [US Census Bureau, Current Population Survey 2016](https://en.wikipedia.org/wiki/Personal_income_in_the_United_States#cite_note-CPS_2015-2), 6.48% of people earned **_$2,500_** or less in a full year. This translates to only **_$208_** a month. Median personal income comes to about **_$31,099_** a year, which is about **_$2,592_** dollars a month.
# MAGIC
# MAGIC Given all this information, let's do some more exploratory data analysis to see where this odd **MonthlyIncome** needs patching a bit.
# COMMAND ----------
# MAGIC %md
# MAGIC Off the bat, there is weirdness in having NULL **MonthlyIncome** data, but being able to calculate **DebtRatio**.
# COMMAND ----------
# MAGIC %sql
# MAGIC select avg(DebtRatio), count(1) as Instances from trainingSDF where MonthlyIncome is null
# COMMAND ----------
# MAGIC %md
# MAGIC It may be the case than whoever gathered this data have replaced **NULL** in this column to 1 to be able to calculate the **DebtRatio** using the data about the **TotalDebt** of the individual they had. This will be need to be treated this way:
# MAGIC - impute the **NULL** values with the median of the dataset
# MAGIC - recalculate the **DebtRatio** given we know that the **TotalDebt** is currently equal for those individuals to the value of the **DebtRatio**
# COMMAND ----------
# MAGIC %md
# MAGIC A very low **MonthlyIncome** between $1 and $7 is again a bit suspicious (having worked under 1hr per month). Let's see a list of people with very small monthly incomes:
# COMMAND ----------
# MAGIC %sql
# MAGIC select MonthlyIncome, count(1) as Instances, avg(DebtRatio) from trainingSDF where MonthlyIncome between 1 and 100 group by MonthlyIncome order by 1
# COMMAND ----------
# MAGIC %md
# MAGIC Given the number of records where **MonthlyIncome** is equal to 1 is suspiciously high, we are going to impute it like we do for the **NULL** values. However, for the other values, there isn't just too much wrong data to draw any conclusions. If we extend the window up to 208:
# COMMAND ----------
# MAGIC %sql
# MAGIC select count(1) as Instances from trainingSDF where MonthlyIncome between 2 and 208
# COMMAND ----------
# MAGIC %md
# MAGIC 100-odd rows is a low percentage of samples from the whole dataset, so for now we will be keeping these as they are.
# COMMAND ----------
# MAGIC %md
# MAGIC That's quite a lot of information, so let's wrap up what we are going to do:
# MAGIC
# MAGIC For the specifics of this lab, we are going to consider that:
# MAGIC - observations with a MonthlyIncome of 1 will be processed to get the median MonthlyIncome
# MAGIC - observations with a MonthlyIncome of null will be processed to get the median MonthlyIncome
# MAGIC
# MAGIC Given the **DebtRatio** has been computed as the overall **Debt** divided by the **MonthlyIncome**, we are going to regenerate the initial debt first so we can use it later to recompute the **DebtRatio** based on the then cleaned **MonthlyIncome**.
# COMMAND ----------
# MAGIC %md
# MAGIC First, we save the initial **Debt** so we are able to recompute the updated DebtRatio afterwards.
# COMMAND ----------
from pyspark.sql import functions as F
def addInitialDebtColumn(df):
df = df.withColumn(
'initialDebt',
F.when(
(((F.col('MonthlyIncome') >= 0) & (F.col('MonthlyIncome') <= 1)) | (F.col('MonthlyIncome').isNull())),
F.col('DebtRatio')
).otherwise(
F.col('MonthlyIncome') * F.col('DebtRatio')
)
)
return df
trainingSDF = addInitialDebtColumn(trainingSDF)
# COMMAND ----------
display(trainingSDF)
# COMMAND ----------
# MAGIC %md
# MAGIC After the initial **Debt** has been saved, we are good to start imputing the **MonthlyIncome** column.
# MAGIC If the actual value is <= $7 or missing, we manually impute using the **numpy**-calculated median.
# COMMAND ----------
def imputeMonthlyIncome(df, incomeMedian):
# Apply income median if the MonthlyIncome is <=7, or null
df = df.withColumn('MonthlyIncome',
F.when(
(((F.col('MonthlyIncome') >= 0) & (F.col('MonthlyIncome') <= 7)) | (F.col('MonthlyIncome').isNull())),
incomeMedian
).otherwise(
F.col('MonthlyIncome')
)
)
return df
trainingIncomeMedian = np.median(trainingSDF.select('MonthlyIncome').dropna().collect())
trainingSDF = imputeMonthlyIncome(trainingSDF, trainingIncomeMedian)
# COMMAND ----------
# MAGIC %md
# MAGIC Now that the **MonthlyIncome** variable has been imputed, let's recalculate a more correct **DebtRatio** based on the initial **Debt** we have saved previously.
# COMMAND ----------
def recalculateDebtRatio(df):
df = df.withColumn(
'DebtRatio',
df.initialDebt/df.MonthlyIncome
)
return df
trainingSDF = recalculateDebtRatio(trainingSDF)
trainingSDF.createOrReplaceTempView(temp_table_name)
# COMMAND ----------
# MAGIC %md
# MAGIC Let's see how many values in this column are actually exceeding the threshold of **1** now.
# COMMAND ----------
# MAGIC %sql
# MAGIC select count(1) from trainingSDF where DebtRatio > 1
# COMMAND ----------
# MAGIC %md
# MAGIC Let's see how it looks from a distribution point of view.
# COMMAND ----------
fig, ax = plt.subplots()
ax = sns.boxplot(x="DebtRatio", data = trainingSDF.toPandas())
ax.set_xscale("log")
display(fig)
# COMMAND ----------
# MAGIC %md
# MAGIC It seems this values are going up into the hundreds. Individuals may exceed a **DebtRatio** of 1 whenever they are lending more than they are earning (and some people in difficult scenarios tend to do that).
# MAGIC
# MAGIC Let's default the higher values to a threshold of **1.5**.
# COMMAND ----------
def defaultDebtRatioToThreshold(df):
df = df.withColumn('DebtRatio',
F.when(
(F.col('DebtRatio') > 1.5),
1.5
).otherwise(
F.col('DebtRatio')
)
)
return df
trainingSDF = defaultDebtRatioToThreshold(trainingSDF)
trainingSDF.createOrReplaceTempView(temp_table_name)
# COMMAND ----------
# MAGIC %md
# MAGIC ### RevolvingUtilizationOfUnsecuredLines variable
# MAGIC Let's understand how many values exceed 1 for this column and default them to this max value.
# COMMAND ----------
# MAGIC %sql
# MAGIC select count(1) from trainingSDF where RevolvingUtilizationOfUnsecuredLines > 1
# COMMAND ----------
# MAGIC %md
# MAGIC Some records have a **RevolvingUtilizationOfUnsecuredLines** value higher than 1. Given the total balance on credit cards and personal lines of credit is divided to the sum of credit limits, this should not exceed 1.
# MAGIC
# MAGIC Let's view the distribution of it and then default the weird records to this threshold.
# COMMAND ----------
fig, ax = plt.subplots()
ax = sns.boxplot(x="RevolvingUtilizationOfUnsecuredLines", data = trainingSDF.toPandas())
ax.set_xscale("log")
display(fig)
# COMMAND ----------
def defaultRevolvingUtilizationToThreshold(df):
df = df.withColumn('RevolvingUtilizationOfUnsecuredLines',
F.when(
(F.col('RevolvingUtilizationOfUnsecuredLines') > 1),
1
).otherwise(
F.col('RevolvingUtilizationOfUnsecuredLines')
)
)
return df
trainingSDF = defaultRevolvingUtilizationToThreshold(trainingSDF)
trainingSDF.createOrReplaceTempView(temp_table_name)
# COMMAND ----------
# MAGIC %md
# MAGIC ### NumberOfDependents variable
# MAGIC
# MAGIC Let's understand how many missing values this column has.
# COMMAND ----------
# MAGIC %sql
# MAGIC select count(1) from trainingSDF where NumberOfDependents is null
# COMMAND ----------
# MAGIC %md
# MAGIC About 3000 missing values out of the total number of rows is not bad at all.
# MAGIC
# MAGIC Let's see how the distribution of this variable looks. We will understand the mode from it and will be able to impute using it.
# COMMAND ----------
# spark.sql does not have any histogram method, however the RDD api does
dependents_histogram = trainingSDF.select('NumberOfDependents').rdd.flatMap(lambda x: x).histogram(10)
fig, ax = plt.subplots()
# the computed histogram needs to be loaded in a pandas dataframe so we will be able to plot it using sns
dependents_histogram_df = pd.DataFrame(
list(zip(*dependents_histogram)),
columns=['bin', 'count']
)
ax = sns.barplot(x = "bin", y = "count", data = dependents_histogram_df)
display(fig)
# COMMAND ----------
# MAGIC %md
# MAGIC We can tell from the barplot above that the mode (most frequent value) of this column is 0. Let's impute the missing values with it.
# COMMAND ----------
def imputeNumberOfDependents(df):
df = df.withColumn('NumberOfDependents',
F.when(
(F.col('NumberOfDependents').isNull()),
0
).otherwise(
F.col('NumberOfDependents')
)
)
return df
trainingSDF = imputeNumberOfDependents(trainingSDF)
trainingSDF.createOrReplaceTempView(temp_table_name)
# COMMAND ----------
# Check of the summary statistics of the features now
display(trainingSDF.describe())
# COMMAND ----------
# MAGIC %md
# MAGIC ## Building our first model
# MAGIC
# MAGIC For our first attempt at building a model we will use a relatively simple algorithm, Decision Trees.
# MAGIC
# MAGIC 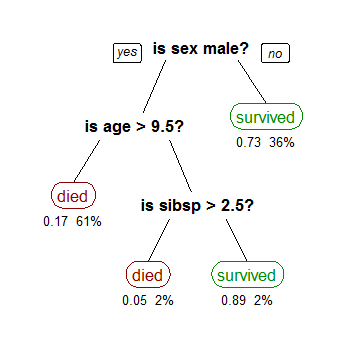
# MAGIC
# MAGIC [Click here](https://www.youtube.com/watch?v=7VeUPuFGJHk) for a straightforward video explanation of how Decision Trees work, and how we can build one using Gini Impurity.
# COMMAND ----------
from pyspark.ml import Pipeline
from pyspark.ml.classification import DecisionTreeClassifier
from pyspark.ml.feature import VectorAssembler, StringIndexer, MinMaxScaler
# Index categorical features
categorical_indexer = StringIndexer(inputCol="age_banding", outputCol="age_banding_indexed")
# assemble all features into a features vector
feature_assembler = VectorAssembler(
inputCols=[
'RevolvingUtilizationOfUnsecuredLines',
'NumberOfTime30-59DaysPastDueNotWorse',
'NumberOfOpenCreditLinesAndLoans',
'NumberOfTimes90DaysLate',
'NumberRealEstateLoansOrLines',
'NumberOfTime60-89DaysPastDueNotWorse',
'NumberOfDependents',
'age_banding_indexed',
'initialDebt',
'DebtRatio',
'MonthlyIncome'],
outputCol="features")
# Train a DecisionTree model.
decision_tree_classifier = DecisionTreeClassifier(labelCol="SeriousDlqin2yrs", featuresCol="features",
impurity="gini", maxDepth=5, seed=1)
# Chain assembler and model in a Pipeline
dtc_pipeline = Pipeline(stages=[categorical_indexer, feature_assembler, decision_tree_classifier])
# Train model.
dtc_model = dtc_pipeline.fit(trainingSDF)
print(dtc_model.stages[2])
# COMMAND ----------
#let's get a text-based representation of the tree
print(dtc_model.stages[2].toDebugString)
# COMMAND ----------
# MAGIC %md
# MAGIC # Testing the model
# MAGIC
# MAGIC We will now test the model by predicting on the test data. Once we obtain predictions, we use the predictions and the ground truth values from the test dataset to compute binary classification evaluation metrics.
# MAGIC
# MAGIC Before we can use the model, we need to apply to the test data the same transformations we did in the preprocessing stage.
# MAGIC
# MAGIC Notice that se are using statistical values like `trainingMedianAge` which were computed on the training data set. It's good practice to treat the test dataset as completely new information, and not use it in any way except actually testing our ML pipeline.
# COMMAND ----------
testingSDF = imputeAgeWithMedian(testingSDF, trainingMedianAge)
testingSDF = addAgeBanding(testingSDF)
testingSDF = addInitialDebtColumn(testingSDF)
testingSDF = imputeMonthlyIncome(testingSDF, trainingIncomeMedian)
testingSDF = recalculateDebtRatio(testingSDF)
testingSDF = defaultDebtRatioToThreshold(testingSDF)
testingSDF = defaultRevolvingUtilizationToThreshold(testingSDF)
testingSDF = imputeNumberOfDependents(testingSDF)
# Make the dataframe available in the SQL context
test_temp_table_name = "testingSDF"
# Make the dataframe available in the SQL context
testingSDF.createOrReplaceTempView(test_temp_table_name)
display(testingSDF)
# COMMAND ----------
# Make predictions.
dtc_predictions = dtc_model.transform(testingSDF)
# Select example rows to display.
display(dtc_predictions.select("probability", "prediction", "SeriousDlqin2yrs"))
# COMMAND ----------
# MAGIC %md
# MAGIC
# MAGIC First, we're going to calculate and display the [Confusion Matrix](https://en.wikipedia.org/wiki/Confusion_matrix) for our binary classifier.
# COMMAND ----------
# display the confusion matrix
from sklearn.metrics import confusion_matrix
def plotConfusionMatrix(confusion_matrix):
fig, ax = plt.subplots()
plt.imshow(confusion_matrix, interpolation='nearest', cmap=plt.cm.Wistia)
classNames = ['Negative','Positive']
ax.set_title(f'Confusion Matrix')
ax.set_ylabel('True label')
ax.set_xlabel('Predicted label')
tick_marks = np.arange(len(classNames))
ax.set_xticks(tick_marks)
ax.set_yticks(tick_marks)
s = [['TN','FP'], ['FN', 'TP']]
for i in range(2):
for j in range(2):
ax.text(j,i, str(s[i][j])+" = "+str(confusion_matrix[i][j]))
display(fig)
dtc_confusion_matrix = confusion_matrix(dtc_predictions.select("SeriousDlqin2yrs").collect(), dtc_predictions.select("prediction").collect())
plotConfusionMatrix(dtc_confusion_matrix)
# COMMAND ----------
# MAGIC %md
# MAGIC
# MAGIC ###Precision and Recall
# MAGIC
# MAGIC 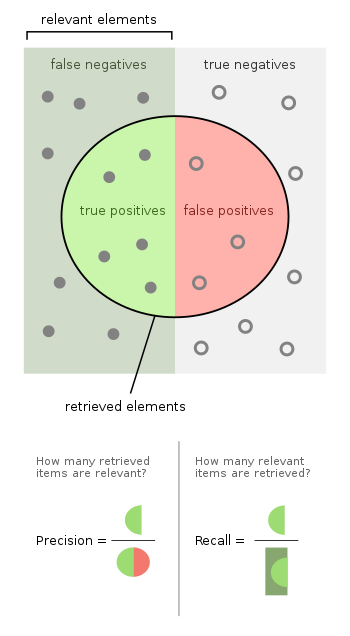
# COMMAND ----------
tn, fp, fn, tp = dtc_confusion_matrix.ravel()
print(f"Accuracy = (TP + TN) / (TP + FP + TN + FN) = {(tp+tn)/(tp+fp+tn+fn)}")
print(f"Precision = TP / (TP + FP) = {tp/(tp+fp)}")
print(f"Recall = TP / (TP + FN) = {tp/(tp+fn)}")
# COMMAND ----------
# MAGIC %md
# MAGIC
# MAGIC ### Sensitivity and Specificity, and the ROC Curve
# MAGIC
# MAGIC 
# COMMAND ----------
# plot the ROC curve
from sklearn.metrics import roc_curve, auc
def plotROCCurve(predictions, show_thresholds=False):
results = predictions.select(['probability', 'SeriousDlqin2yrs']).collect()
y_score = [float(i[0][1]) for i in results]
y_true = [float(i[1]) for i in results]
fpr, tpr, thresholds = roc_curve(y_true, y_score, pos_label = 1)
roc_auc = auc(fpr, tpr)
fig, ax = plt.subplots()
ax.plot(fpr, tpr, label='ROC curve (area = %0.4f)' % roc_auc)
ax.plot([0, 1], [0, 1], 'k--')
if show_thresholds:
tr_idx = np.arange(385, len(thresholds), 700)
for i in tr_idx:
ax.plot(fpr[i], tpr[i], "xr")
ax.annotate(xy=(fpr[i], tpr[i]), s="%0.3f" % thresholds[i])
ax.set_xlim([0.0, 1.0])
ax.set_ylim([0.0, 1.0])
ax.set_xlabel('False Positive Rate (1 - Specificity)')
ax.set_ylabel('True Positive Rate (Sensitivity)')
ax.set_title('Receiver operating characteristic')
ax.legend(loc="lower right")
display(fig)
plotROCCurve(dtc_predictions)
# COMMAND ----------
# MAGIC %md
# MAGIC
# MAGIC ## Gradient Boosted Trees
# MAGIC
# MAGIC Now we're going to try an ensemble model: [Gradient Boosted Trees](https://en.wikipedia.org/wiki/Gradient_boosting).
# MAGIC
# MAGIC 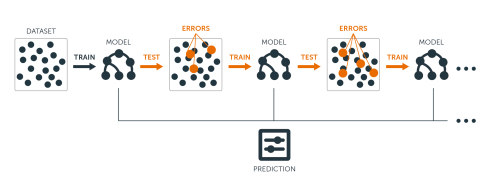
# MAGIC
# MAGIC [Click here](https://www.youtube.com/watch?v=3CC4N4z3GJc) for a nice visual explanation of how Gradient Boosting works.
# COMMAND ----------
from pyspark.ml.classification import GBTClassifier
# scale features
scaler = MinMaxScaler(inputCol="features", outputCol="scaledFeatures")
# Train a Gradient-boosted tree classifier model.
gbt_classifier = GBTClassifier(labelCol="SeriousDlqin2yrs", featuresCol="features",
maxIter=35, seed=1)
# Chain assembler and model in a Pipeline
gbt_pipeline = Pipeline(stages=[categorical_indexer, feature_assembler, scaler, gbt_classifier])
# Train model.
gbt_model = gbt_pipeline.fit(trainingSDF)
print(gbt_model.stages[3])
# COMMAND ----------
print(gbt_model.stages[3].toDebugString)
# COMMAND ----------
# Make predictions.
gbt_predictions = gbt_model.transform(testingSDF)
# Select example rows to display.
display(gbt_predictions.select("probability", "prediction", "SeriousDlqin2yrs"))
# COMMAND ----------
gbt_confusion_matrix = confusion_matrix(gbt_predictions.select("SeriousDlqin2yrs").collect(), gbt_predictions.select("prediction").collect())
plotConfusionMatrix(gbt_confusion_matrix)
# COMMAND ----------
tn, fp, fn, tp = gbt_confusion_matrix.ravel()
print(f"Precision = TP / (TP + FP) = {tp/(tp+fp)}")
print(f"Recall = TP / (TP + FN) = {tp/(tp+fn)}")
print(f"Sensitivity = TP / (TP + FN) = {tp/(tp+fn)}")
print(f"Specificity = TN / (TN + FP) = {tn/(tn+fp)}")
# COMMAND ----------
plotROCCurve(gbt_predictions)
# COMMAND ----------
# MAGIC %md
# MAGIC ### Selecting a better threshold for class separation
# COMMAND ----------
plotROCCurve(gbt_predictions, show_thresholds = True)
# COMMAND ----------
# select a different threshold for class separation, make predictions based on that threshold, and recalculate Precision, Recall, Sensitivity and Specificity.
from pyspark.sql.types import FloatType
get_positive_probability=udf(lambda v:float(v[1]),FloatType())
selected_threshold = 0.11
pred_colname = f'prediction-threshold'
gbt_predictions_threshold = gbt_predictions.withColumn(pred_colname,
F.when(get_positive_probability('probability') <= selected_threshold,0)
.otherwise(1))
display(gbt_predictions_threshold.select("probability", "prediction", pred_colname, "SeriousDlqin2yrs"))
# COMMAND ----------
gbt_threshold_confusion_matrix = confusion_matrix(gbt_predictions_threshold.select("SeriousDlqin2yrs").collect(), gbt_predictions_threshold.select("prediction-threshold").collect())
plotConfusionMatrix(gbt_threshold_confusion_matrix)
# COMMAND ----------
tn, fp, fn, tp = gbt_threshold_confusion_matrix.ravel()
print(f"Precision = TP / (TP + FP) = {tp/(tp+fp)}")
print(f"Recall = TP / (TP + FN) = {tp/(tp+fn)}")
print(f"Sensitivity = TP / (TP + FN) = {tp/(tp+fn)}")
print(f"Specificity = TN / (TN + FP) = {tn/(tn+fp)}")
# COMMAND ----------
# MAGIC %md
# MAGIC
# MAGIC ## Hyperparameter Tuning
# MAGIC
# MAGIC Until now we've built a Gradient Boosted classfier with just the default parameters (except `maxIter` which we set to 35).
# MAGIC It would be useful to optimize the parameters for the algorithm (also called hyperparameters).
# MAGIC
# MAGIC [Hyperparameter optimization](https://en.wikipedia.org/wiki/Hyperparameter_optimization) or tuning is the problem of choosing a set of optimal hyperparameters for a learning algorithm. A hyperparameter is a parameter whose value is used to control the learning process. By contrast, the values of other parameters (typically node weights) are learned.
# MAGIC
# MAGIC We will combine Hyperparameter Tuning with [Cross-Validation](https://en.wikipedia.org/wiki/Cross-validation_(statistics)) on the training dataset, so we are able to even out the noise in the training data. It is mainly used in settings where the goal is prediction, and one wants to estimate how accurately a predictive model will perform in practice.
# MAGIC
# MAGIC 
# COMMAND ----------
print(gbt_classifier.explainParams())
# COMMAND ----------
from pyspark.ml.tuning import ParamGridBuilder, CrossValidator
from pyspark.ml.evaluation import BinaryClassificationEvaluator
paramGrid = (ParamGridBuilder()
.addGrid(gbt_classifier.maxDepth, [5, 8])
.addGrid(gbt_classifier.maxIter, [25, 40])
.addGrid(gbt_classifier.stepSize, [0.1, 0.2])
.build())
evaluator = BinaryClassificationEvaluator(
rawPredictionCol="prediction", labelCol="SeriousDlqin2yrs", metricName="areaUnderROC")
cv = CrossValidator(estimator=gbt_pipeline, estimatorParamMaps=paramGrid, evaluator=evaluator, numFolds=3)
# Train model.
gbt_models_cv = cv.fit(trainingSDF)
# COMMAND ----------
best_model = gbt_models_cv.bestModel.stages[3]
print(best_model.explainParams())
# COMMAND ----------
# Make predictions.
gbt_cv_predictions = gbt_models_cv.transform(testingSDF)
# Select example rows to display.
display(gbt_cv_predictions.select("probability", "prediction", "SeriousDlqin2yrs"))
# COMMAND ----------
plotROCCurve(gbt_cv_predictions, show_thresholds = True)
# COMMAND ----------
selected_threshold = 0.11
pred_colname = f'prediction-threshold'
gbt_cv_predictions_threshold = gbt_cv_predictions.withColumn(pred_colname,
F.when(get_positive_probability('probability') < selected_threshold,0)
.otherwise(1))
display(gbt_cv_predictions_threshold.select("probability", "prediction", pred_colname, "SeriousDlqin2yrs"))
# COMMAND ----------
gbt_cv_threshold_confusion_matrix = confusion_matrix(gbt_cv_predictions_threshold.select("SeriousDlqin2yrs").collect(), gbt_cv_predictions_threshold.select("prediction-threshold").collect())
plotConfusionMatrix(gbt_cv_threshold_confusion_matrix)
# COMMAND ----------
tn, fp, fn, tp = gbt_cv_threshold_confusion_matrix.ravel()
print(f"Precision = TP / (TP + FP) = {tp/(tp+fp)}")
print(f"Recall = TP / (TP + FN) = {tp/(tp+fn)}")
print(f"Sensitivity = TP / (TP + FN) = {tp/(tp+fn)}")
print(f"Specificity = TN / (TN + FP) = {tn/(tn+fp)}")
# COMMAND ----------
# MAGIC %md
# MAGIC ## Using Automated ML from Azure ML Service
# MAGIC
# MAGIC We are now going to use the [AutoML feature](https://docs.microsoft.com/en-us/azure/machine-learning/service/concept-automated-ml) from the Azure Machine Learning Service SDK.
# MAGIC
# MAGIC Automated machine learning, also referred to as automated ML, is the process of automating the time consuming, iterative tasks of machine learning model development. It allows data scientists, analysts, and developers to build ML models with high scale, efficiency, and productivity all while sustaining model quality. Automated ML is based on a breakthrough from our Microsoft Research division.
# MAGIC
# MAGIC Traditional machine learning model development is resource-intensive, requiring significant domain knowledge and time to produce and compare dozens of models. Apply automated ML when you want Azure Machine Learning to train and tune a model for you using the target metric you specify. The service then iterates through ML algorithms paired with feature selections, where each iteration produces a model with a training score. The higher the score, the better the model is considered to "fit" your data.
# MAGIC
# MAGIC
# MAGIC We will provide the cleansed training data to Azure ML which will test multiple types of algorithms in order to maximize a certain evaluation criteria we define. As per the [initial challenge from kaggle](https://www.kaggle.com/c/GiveMeSomeCredit), the criteria of choice is AUC (Area Under Curve).
# MAGIC
# MAGIC The validation during training is done by using cross validation in 5 folds.
# MAGIC
# MAGIC After we are done, the best trained model will be evaluated against a separated dataset (the test dataset) in order to understand real _performance_.
# MAGIC
# MAGIC ### Training using AutoML
# MAGIC
# MAGIC In order to get things going, we first initialize our Workspace...
# COMMAND ----------
subscription_id = "6787a35f-386b-4845-91d1-695f24e0924b" # the Azure subscription ID you are using
azureml_resource_group = "spark-ml-workshop-25" #you should be owner or contributor
azureml_workspace_name = "azureml-lab-25" #your Azure Machine Learning workspace name
import azureml.core
# Check core SDK version number - based on build number of preview/master.
print("Azure ML SDK version:", azureml.core.VERSION)
from azureml.core import Workspace
ws = Workspace(workspace_name = azureml_workspace_name,
subscription_id = subscription_id,
resource_group = azureml_resource_group)
# Persist the subscription id, resource group name, and workspace name in aml_config/config.json.
ws.write_config()
# COMMAND ----------
ws = Workspace.from_config()
print('Workspace name: ' + ws.name,
'Azure region: ' + ws.location,
'Subscription id: ' + ws.subscription_id,
'Resource group: ' + ws.resource_group, sep = '\n')
# COMMAND ----------
# MAGIC %md
# MAGIC And then we make sure we have all the important libraries in place.
# COMMAND ----------
import logging
import os
import random
import time
from matplotlib import pyplot as plt
from matplotlib.pyplot import imshow
import numpy as np
import pandas as pd
import azureml.core
from azureml.core.experiment import Experiment
from azureml.core.workspace import Workspace
from azureml.train.automl import AutoMLConfig
from azureml.train.automl.run import AutoMLRun
# COMMAND ----------
# MAGIC %md
# MAGIC We prepare the experiment properties which will be provided once we issue a training request.
# COMMAND ----------
# Get the last seven letters of the username which will be used to build up exp name
import re
regexStr = r'^([^@]+)@[^@]+$'
emailStr = dbutils.notebook.entry_point.getDbutils().notebook().getContext().tags().apply("user")
matchobj = re.search(regexStr, emailStr)
if not matchobj is None:
if len(matchobj.group(1)) > 10:
notebook_username = matchobj.group(1)[-10:]
else:
notebook_username = matchobj.group(1)
print(notebook_username)
else:
print("Did not match")
# COMMAND ----------
# Choose a name for the experiment and specify the project folder.
experiment_base_name = 'automl-scoring-'
experiment_suffix_name = notebook_username.replace(".", "") + "-" + str(random.randint(1000, 9999))
experiment_name = experiment_base_name + experiment_suffix_name
project_folder = './globalainight_projects/automl-credit-scring'
print(experiment_name)
experiment = Experiment(ws, experiment_name)
output = {}
output['SDK version'] = azureml.core.VERSION
output['Subscription ID'] = ws.subscription_id
output['Workspace Name'] = ws.name
output['Resource Group'] = ws.resource_group
output['Location'] = ws.location
output['Project Directory'] = project_folder
output['Experiment Name'] = experiment.name
| pd.set_option('display.max_colwidth', -1) | pandas.set_option |
# -*- coding: utf-8 -*-
from datetime import timedelta
import operator
import numpy as np
import pytest
import pandas as pd
from pandas import Series, compat
from pandas.core.indexes.period import IncompatibleFrequency
import pandas.util.testing as tm
def _permute(obj):
return obj.take(np.random.permutation(len(obj)))
class TestSeriesFlexArithmetic(object):
@pytest.mark.parametrize(
'ts',
[
(lambda x: x, lambda x: x * 2, False),
(lambda x: x, lambda x: x[::2], False),
(lambda x: x, lambda x: 5, True),
(lambda x: tm.makeFloatSeries(),
lambda x: tm.makeFloatSeries(),
True)
])
@pytest.mark.parametrize('opname', ['add', 'sub', 'mul', 'floordiv',
'truediv', 'div', 'pow'])
def test_flex_method_equivalence(self, opname, ts):
# check that Series.{opname} behaves like Series.__{opname}__,
tser = tm.makeTimeSeries().rename('ts')
series = ts[0](tser)
other = ts[1](tser)
check_reverse = ts[2]
if opname == 'div' and compat.PY3:
pytest.skip('div test only for Py3')
op = getattr(Series, opname)
if op == 'div':
alt = operator.truediv
else:
alt = getattr(operator, opname)
result = op(series, other)
expected = alt(series, other)
tm.assert_almost_equal(result, expected)
if check_reverse:
rop = getattr(Series, "r" + opname)
result = rop(series, other)
expected = alt(other, series)
tm.assert_almost_equal(result, expected)
class TestSeriesArithmetic(object):
# Some of these may end up in tests/arithmetic, but are not yet sorted
def test_empty_series_add_sub(self):
# GH#13844
a = Series(dtype='M8[ns]')
b = Series(dtype='m8[ns]')
tm.assert_series_equal(a, a + b)
tm.assert_series_equal(a, a - b)
tm.assert_series_equal(a, b + a)
with pytest.raises(TypeError):
b - a
def test_add_series_with_period_index(self):
rng = pd.period_range('1/1/2000', '1/1/2010', freq='A')
ts = Series(np.random.randn(len(rng)), index=rng)
result = ts + ts[::2]
expected = ts + ts
expected[1::2] = np.nan
tm.assert_series_equal(result, expected)
result = ts + _permute(ts[::2])
tm.assert_series_equal(result, expected)
msg = "Input has different freq=D from PeriodIndex\\(freq=A-DEC\\)"
with tm.assert_raises_regex(IncompatibleFrequency, msg):
ts + ts.asfreq('D', how="end")
def test_operators_datetimelike(self):
# ## timedelta64 ###
td1 = Series([timedelta(minutes=5, seconds=3)] * 3)
td1.iloc[2] = np.nan
# ## datetime64 ###
dt1 = Series([pd.Timestamp('20111230'), pd.Timestamp('20120101'),
pd.Timestamp('20120103')])
dt1.iloc[2] = np.nan
dt2 = Series([pd.Timestamp('20111231'), pd.Timestamp('20120102'),
| pd.Timestamp('20120104') | pandas.Timestamp |
import array
import os
import pandas as pd
import pymongo
import json
import pandas_ta as ta
from bson import json_util, ObjectId
from bson.json_util import loads
from Sma2019 import data
myclient = pymongo.MongoClient("mongodb://localhost:27017/")
df=pd.Series(data)
pd.ewma(df, span=5)
| pd.ewma(df, span=5, min_periods=5) | pandas.ewma |
from collections import OrderedDict
import datetime
from datetime import timedelta
from io import StringIO
import json
import os
import numpy as np
import pytest
from pandas.compat import is_platform_32bit, is_platform_windows
import pandas.util._test_decorators as td
import pandas as pd
from pandas import DataFrame, DatetimeIndex, Series, Timestamp, read_json
import pandas._testing as tm
_seriesd = tm.getSeriesData()
_tsd = tm.getTimeSeriesData()
_frame = DataFrame(_seriesd)
_intframe = DataFrame({k: v.astype(np.int64) for k, v in _seriesd.items()})
_tsframe = DataFrame(_tsd)
_cat_frame = _frame.copy()
cat = ["bah"] * 5 + ["bar"] * 5 + ["baz"] * 5 + ["foo"] * (len(_cat_frame) - 15)
_cat_frame.index = pd.CategoricalIndex(cat, name="E")
_cat_frame["E"] = list(reversed(cat))
_cat_frame["sort"] = np.arange(len(_cat_frame), dtype="int64")
_mixed_frame = _frame.copy()
def assert_json_roundtrip_equal(result, expected, orient):
if orient == "records" or orient == "values":
expected = expected.reset_index(drop=True)
if orient == "values":
expected.columns = range(len(expected.columns))
tm.assert_frame_equal(result, expected)
@pytest.mark.filterwarnings("ignore:the 'numpy' keyword is deprecated:FutureWarning")
class TestPandasContainer:
@pytest.fixture(autouse=True)
def setup(self):
self.intframe = _intframe.copy()
self.tsframe = _tsframe.copy()
self.mixed_frame = _mixed_frame.copy()
self.categorical = _cat_frame.copy()
yield
del self.intframe
del self.tsframe
del self.mixed_frame
def test_frame_double_encoded_labels(self, orient):
df = DataFrame(
[["a", "b"], ["c", "d"]],
index=['index " 1', "index / 2"],
columns=["a \\ b", "y / z"],
)
result = read_json(df.to_json(orient=orient), orient=orient)
expected = df.copy()
assert_json_roundtrip_equal(result, expected, orient)
@pytest.mark.parametrize("orient", ["split", "records", "values"])
def test_frame_non_unique_index(self, orient):
df = DataFrame([["a", "b"], ["c", "d"]], index=[1, 1], columns=["x", "y"])
result = read_json(df.to_json(orient=orient), orient=orient)
expected = df.copy()
assert_json_roundtrip_equal(result, expected, orient)
@pytest.mark.parametrize("orient", ["index", "columns"])
def test_frame_non_unique_index_raises(self, orient):
df = DataFrame([["a", "b"], ["c", "d"]], index=[1, 1], columns=["x", "y"])
msg = f"DataFrame index must be unique for orient='{orient}'"
with pytest.raises(ValueError, match=msg):
df.to_json(orient=orient)
@pytest.mark.parametrize("orient", ["split", "values"])
@pytest.mark.parametrize(
"data",
[
[["a", "b"], ["c", "d"]],
[[1.5, 2.5], [3.5, 4.5]],
[[1, 2.5], [3, 4.5]],
[[Timestamp("20130101"), 3.5], [Timestamp("20130102"), 4.5]],
],
)
def test_frame_non_unique_columns(self, orient, data):
df = DataFrame(data, index=[1, 2], columns=["x", "x"])
result = read_json(
df.to_json(orient=orient), orient=orient, convert_dates=["x"]
)
if orient == "values":
expected = pd.DataFrame(data)
if expected.iloc[:, 0].dtype == "datetime64[ns]":
# orient == "values" by default will write Timestamp objects out
# in milliseconds; these are internally stored in nanosecond,
# so divide to get where we need
# TODO: a to_epoch method would also solve; see GH 14772
expected.iloc[:, 0] = expected.iloc[:, 0].astype(np.int64) // 1000000
elif orient == "split":
expected = df
tm.assert_frame_equal(result, expected)
@pytest.mark.parametrize("orient", ["index", "columns", "records"])
def test_frame_non_unique_columns_raises(self, orient):
df = DataFrame([["a", "b"], ["c", "d"]], index=[1, 2], columns=["x", "x"])
msg = f"DataFrame columns must be unique for orient='{orient}'"
with pytest.raises(ValueError, match=msg):
df.to_json(orient=orient)
def test_frame_default_orient(self, float_frame):
assert float_frame.to_json() == float_frame.to_json(orient="columns")
@pytest.mark.parametrize("dtype", [False, float])
@pytest.mark.parametrize("convert_axes", [True, False])
@pytest.mark.parametrize("numpy", [True, False])
def test_roundtrip_simple(self, orient, convert_axes, numpy, dtype, float_frame):
data = float_frame.to_json(orient=orient)
result = pd.read_json(
data, orient=orient, convert_axes=convert_axes, numpy=numpy, dtype=dtype
)
expected = float_frame
assert_json_roundtrip_equal(result, expected, orient)
@pytest.mark.parametrize("dtype", [False, np.int64])
@pytest.mark.parametrize("convert_axes", [True, False])
@pytest.mark.parametrize("numpy", [True, False])
def test_roundtrip_intframe(self, orient, convert_axes, numpy, dtype):
data = self.intframe.to_json(orient=orient)
result = pd.read_json(
data, orient=orient, convert_axes=convert_axes, numpy=numpy, dtype=dtype
)
expected = self.intframe.copy()
if (
numpy
and (is_platform_32bit() or is_platform_windows())
and not dtype
and orient != "split"
):
# TODO: see what is causing roundtrip dtype loss
expected = expected.astype(np.int32)
assert_json_roundtrip_equal(result, expected, orient)
@pytest.mark.parametrize("dtype", [None, np.float64, np.int, "U3"])
@pytest.mark.parametrize("convert_axes", [True, False])
@pytest.mark.parametrize("numpy", [True, False])
def test_roundtrip_str_axes(self, orient, convert_axes, numpy, dtype):
df = DataFrame(
np.zeros((200, 4)),
columns=[str(i) for i in range(4)],
index=[str(i) for i in range(200)],
dtype=dtype,
)
# TODO: do we even need to support U3 dtypes?
if numpy and dtype == "U3" and orient != "split":
pytest.xfail("Can't decode directly to array")
data = df.to_json(orient=orient)
result = pd.read_json(
data, orient=orient, convert_axes=convert_axes, numpy=numpy, dtype=dtype
)
expected = df.copy()
if not dtype:
expected = expected.astype(np.int64)
# index columns, and records orients cannot fully preserve the string
# dtype for axes as the index and column labels are used as keys in
# JSON objects. JSON keys are by definition strings, so there's no way
# to disambiguate whether those keys actually were strings or numeric
# beforehand and numeric wins out.
# TODO: Split should be able to support this
if convert_axes and (orient in ("split", "index", "columns")):
expected.columns = expected.columns.astype(np.int64)
expected.index = expected.index.astype(np.int64)
elif orient == "records" and convert_axes:
expected.columns = expected.columns.astype(np.int64)
assert_json_roundtrip_equal(result, expected, orient)
@pytest.mark.parametrize("convert_axes", [True, False])
@pytest.mark.parametrize("numpy", [True, False])
def test_roundtrip_categorical(self, orient, convert_axes, numpy):
# TODO: create a better frame to test with and improve coverage
if orient in ("index", "columns"):
pytest.xfail(f"Can't have duplicate index values for orient '{orient}')")
data = self.categorical.to_json(orient=orient)
if numpy and orient in ("records", "values"):
pytest.xfail(f"Orient {orient} is broken with numpy=True")
result = pd.read_json(
data, orient=orient, convert_axes=convert_axes, numpy=numpy
)
expected = self.categorical.copy()
expected.index = expected.index.astype(str) # Categorical not preserved
expected.index.name = None # index names aren't preserved in JSON
if not numpy and orient == "index":
expected = expected.sort_index()
assert_json_roundtrip_equal(result, expected, orient)
@pytest.mark.parametrize("convert_axes", [True, False])
@pytest.mark.parametrize("numpy", [True, False])
def test_roundtrip_empty(self, orient, convert_axes, numpy, empty_frame):
data = empty_frame.to_json(orient=orient)
result = pd.read_json(
data, orient=orient, convert_axes=convert_axes, numpy=numpy
)
expected = empty_frame.copy()
# TODO: both conditions below are probably bugs
if convert_axes:
expected.index = expected.index.astype(float)
expected.columns = expected.columns.astype(float)
if numpy and orient == "values":
expected = expected.reindex([0], axis=1).reset_index(drop=True)
tm.assert_frame_equal(result, expected)
@pytest.mark.parametrize("convert_axes", [True, False])
@pytest.mark.parametrize("numpy", [True, False])
def test_roundtrip_timestamp(self, orient, convert_axes, numpy):
# TODO: improve coverage with date_format parameter
data = self.tsframe.to_json(orient=orient)
result = pd.read_json(
data, orient=orient, convert_axes=convert_axes, numpy=numpy
)
expected = self.tsframe.copy()
if not convert_axes: # one off for ts handling
# DTI gets converted to epoch values
idx = expected.index.astype(np.int64) // 1000000
if orient != "split": # TODO: handle consistently across orients
idx = idx.astype(str)
expected.index = idx
assert_json_roundtrip_equal(result, expected, orient)
@pytest.mark.parametrize("convert_axes", [True, False])
@pytest.mark.parametrize("numpy", [True, False])
def test_roundtrip_mixed(self, orient, convert_axes, numpy):
if numpy and orient != "split":
pytest.xfail("Can't decode directly to array")
index = pd.Index(["a", "b", "c", "d", "e"])
values = {
"A": [0.0, 1.0, 2.0, 3.0, 4.0],
"B": [0.0, 1.0, 0.0, 1.0, 0.0],
"C": ["foo1", "foo2", "foo3", "foo4", "foo5"],
"D": [True, False, True, False, True],
}
df = DataFrame(data=values, index=index)
data = df.to_json(orient=orient)
result = pd.read_json(
data, orient=orient, convert_axes=convert_axes, numpy=numpy
)
expected = df.copy()
expected = expected.assign(**expected.select_dtypes("number").astype(np.int64))
if not numpy and orient == "index":
expected = expected.sort_index()
assert_json_roundtrip_equal(result, expected, orient)
@pytest.mark.parametrize(
"data,msg,orient",
[
('{"key":b:a:d}', "Expected object or value", "columns"),
# too few indices
(
'{"columns":["A","B"],'
'"index":["2","3"],'
'"data":[[1.0,"1"],[2.0,"2"],[null,"3"]]}',
r"Shape of passed values is \(3, 2\), indices imply \(2, 2\)",
"split",
),
# too many columns
(
'{"columns":["A","B","C"],'
'"index":["1","2","3"],'
'"data":[[1.0,"1"],[2.0,"2"],[null,"3"]]}',
"3 columns passed, passed data had 2 columns",
"split",
),
# bad key
(
'{"badkey":["A","B"],'
'"index":["2","3"],'
'"data":[[1.0,"1"],[2.0,"2"],[null,"3"]]}',
r"unexpected key\(s\): badkey",
"split",
),
],
)
def test_frame_from_json_bad_data_raises(self, data, msg, orient):
with pytest.raises(ValueError, match=msg):
read_json(StringIO(data), orient=orient)
@pytest.mark.parametrize("dtype", [True, False])
@pytest.mark.parametrize("convert_axes", [True, False])
@pytest.mark.parametrize("numpy", [True, False])
def test_frame_from_json_missing_data(self, orient, convert_axes, numpy, dtype):
num_df = DataFrame([[1, 2], [4, 5, 6]])
result = read_json(
num_df.to_json(orient=orient),
orient=orient,
convert_axes=convert_axes,
dtype=dtype,
)
assert np.isnan(result.iloc[0, 2])
obj_df = DataFrame([["1", "2"], ["4", "5", "6"]])
result = read_json(
obj_df.to_json(orient=orient),
orient=orient,
convert_axes=convert_axes,
dtype=dtype,
)
if not dtype: # TODO: Special case for object data; maybe a bug?
assert result.iloc[0, 2] is None
else:
assert np.isnan(result.iloc[0, 2])
@pytest.mark.parametrize("inf", [np.inf, np.NINF])
@pytest.mark.parametrize("dtype", [True, False])
def test_frame_infinity(self, orient, inf, dtype):
# infinities get mapped to nulls which get mapped to NaNs during
# deserialisation
df = DataFrame([[1, 2], [4, 5, 6]])
df.loc[0, 2] = inf
result = read_json(df.to_json(), dtype=dtype)
assert np.isnan(result.iloc[0, 2])
@pytest.mark.skipif(
is_platform_32bit(), reason="not compliant on 32-bit, xref #15865"
)
@pytest.mark.parametrize(
"value,precision,expected_val",
[
(0.95, 1, 1.0),
(1.95, 1, 2.0),
(-1.95, 1, -2.0),
(0.995, 2, 1.0),
(0.9995, 3, 1.0),
(0.99999999999999944, 15, 1.0),
],
)
def test_frame_to_json_float_precision(self, value, precision, expected_val):
df = pd.DataFrame([dict(a_float=value)])
encoded = df.to_json(double_precision=precision)
assert encoded == f'{{"a_float":{{"0":{expected_val}}}}}'
def test_frame_to_json_except(self):
df = DataFrame([1, 2, 3])
msg = "Invalid value 'garbage' for option 'orient'"
with pytest.raises(ValueError, match=msg):
df.to_json(orient="garbage")
def test_frame_empty(self):
df = DataFrame(columns=["jim", "joe"])
assert not df._is_mixed_type
tm.assert_frame_equal(
read_json(df.to_json(), dtype=dict(df.dtypes)), df, check_index_type=False
)
# GH 7445
result = pd.DataFrame({"test": []}, index=[]).to_json(orient="columns")
expected = '{"test":{}}'
assert result == expected
def test_frame_empty_mixedtype(self):
# mixed type
df = DataFrame(columns=["jim", "joe"])
df["joe"] = df["joe"].astype("i8")
assert df._is_mixed_type
tm.assert_frame_equal(
read_json(df.to_json(), dtype=dict(df.dtypes)), df, check_index_type=False
)
def test_frame_mixedtype_orient(self): # GH10289
vals = [
[10, 1, "foo", 0.1, 0.01],
[20, 2, "bar", 0.2, 0.02],
[30, 3, "baz", 0.3, 0.03],
[40, 4, "qux", 0.4, 0.04],
]
df = DataFrame(
vals, index=list("abcd"), columns=["1st", "2nd", "3rd", "4th", "5th"]
)
assert df._is_mixed_type
right = df.copy()
for orient in ["split", "index", "columns"]:
inp = df.to_json(orient=orient)
left = read_json(inp, orient=orient, convert_axes=False)
tm.assert_frame_equal(left, right)
right.index = np.arange(len(df))
inp = df.to_json(orient="records")
left = read_json(inp, orient="records", convert_axes=False)
tm.assert_frame_equal(left, right)
right.columns = np.arange(df.shape[1])
inp = df.to_json(orient="values")
left = read_json(inp, orient="values", convert_axes=False)
tm.assert_frame_equal(left, right)
def test_v12_compat(self, datapath):
df = DataFrame(
[
[1.56808523, 0.65727391, 1.81021139, -0.17251653],
[-0.2550111, -0.08072427, -0.03202878, -0.17581665],
[1.51493992, 0.11805825, 1.629455, -1.31506612],
[-0.02765498, 0.44679743, 0.33192641, -0.27885413],
[0.05951614, -2.69652057, 1.28163262, 0.34703478],
],
columns=["A", "B", "C", "D"],
index=pd.date_range("2000-01-03", "2000-01-07"),
)
df["date"] = pd.Timestamp("19920106 18:21:32.12")
df.iloc[3, df.columns.get_loc("date")] = pd.Timestamp("20130101")
df["modified"] = df["date"]
df.iloc[1, df.columns.get_loc("modified")] = pd.NaT
dirpath = datapath("io", "json", "data")
v12_json = os.path.join(dirpath, "tsframe_v012.json")
df_unser = pd.read_json(v12_json)
tm.assert_frame_equal(df, df_unser)
df_iso = df.drop(["modified"], axis=1)
v12_iso_json = os.path.join(dirpath, "tsframe_iso_v012.json")
df_unser_iso = pd.read_json(v12_iso_json)
tm.assert_frame_equal(df_iso, df_unser_iso)
def test_blocks_compat_GH9037(self):
index = pd.date_range("20000101", periods=10, freq="H")
df_mixed = DataFrame(
OrderedDict(
float_1=[
-0.92077639,
0.77434435,
1.25234727,
0.61485564,
-0.60316077,
0.24653374,
0.28668979,
-2.51969012,
0.95748401,
-1.02970536,
],
int_1=[
19680418,
75337055,
99973684,
65103179,
79373900,
40314334,
21290235,
4991321,
41903419,
16008365,
],
str_1=[
"78c608f1",
"64a99743",
"13d2ff52",
"ca7f4af2",
"97236474",
"bde7e214",
"1a6bde47",
"b1190be5",
"7a669144",
"8d64d068",
],
float_2=[
-0.0428278,
-1.80872357,
3.36042349,
-0.7573685,
-0.48217572,
0.86229683,
1.08935819,
0.93898739,
-0.03030452,
1.43366348,
],
str_2=[
"14f04af9",
"d085da90",
"4bcfac83",
"81504caf",
"2ffef4a9",
"08e2f5c4",
"07e1af03",
"addbd4a7",
"1f6a09ba",
"4bfc4d87",
],
int_2=[
86967717,
98098830,
51927505,
20372254,
12601730,
20884027,
34193846,
10561746,
24867120,
76131025,
],
),
index=index,
)
# JSON deserialisation always creates unicode strings
df_mixed.columns = df_mixed.columns.astype("unicode")
df_roundtrip = pd.read_json(df_mixed.to_json(orient="split"), orient="split")
tm.assert_frame_equal(
df_mixed,
df_roundtrip,
check_index_type=True,
check_column_type=True,
by_blocks=True,
check_exact=True,
)
def test_frame_nonprintable_bytes(self):
# GH14256: failing column caused segfaults, if it is not the last one
class BinaryThing:
def __init__(self, hexed):
self.hexed = hexed
self.binary = bytes.fromhex(hexed)
def __str__(self) -> str:
return self.hexed
hexed = "574b4454ba8c5eb4f98a8f45"
binthing = BinaryThing(hexed)
# verify the proper conversion of printable content
df_printable = DataFrame({"A": [binthing.hexed]})
assert df_printable.to_json() == f'{{"A":{{"0":"{hexed}"}}}}'
# check if non-printable content throws appropriate Exception
df_nonprintable = DataFrame({"A": [binthing]})
msg = "Unsupported UTF-8 sequence length when encoding string"
with pytest.raises(OverflowError, match=msg):
df_nonprintable.to_json()
# the same with multiple columns threw segfaults
df_mixed = DataFrame({"A": [binthing], "B": [1]}, columns=["A", "B"])
with pytest.raises(OverflowError):
df_mixed.to_json()
# default_handler should resolve exceptions for non-string types
result = df_nonprintable.to_json(default_handler=str)
expected = f'{{"A":{{"0":"{hexed}"}}}}'
assert result == expected
assert (
df_mixed.to_json(default_handler=str)
== f'{{"A":{{"0":"{hexed}"}},"B":{{"0":1}}}}'
)
def test_label_overflow(self):
# GH14256: buffer length not checked when writing label
result = pd.DataFrame({"bar" * 100000: [1], "foo": [1337]}).to_json()
expected = f'{{"{"bar" * 100000}":{{"0":1}},"foo":{{"0":1337}}}}'
assert result == expected
def test_series_non_unique_index(self):
s = Series(["a", "b"], index=[1, 1])
msg = "Series index must be unique for orient='index'"
with pytest.raises(ValueError, match=msg):
s.to_json(orient="index")
tm.assert_series_equal(
s, read_json(s.to_json(orient="split"), orient="split", typ="series")
)
unser = read_json(s.to_json(orient="records"), orient="records", typ="series")
tm.assert_numpy_array_equal(s.values, unser.values)
def test_series_default_orient(self, string_series):
assert string_series.to_json() == string_series.to_json(orient="index")
@pytest.mark.parametrize("numpy", [True, False])
def test_series_roundtrip_simple(self, orient, numpy, string_series):
data = string_series.to_json(orient=orient)
result = pd.read_json(data, typ="series", orient=orient, numpy=numpy)
expected = string_series
if orient in ("values", "records"):
expected = expected.reset_index(drop=True)
if orient != "split":
expected.name = None
tm.assert_series_equal(result, expected)
@pytest.mark.parametrize("dtype", [False, None])
@pytest.mark.parametrize("numpy", [True, False])
def test_series_roundtrip_object(self, orient, numpy, dtype, object_series):
data = object_series.to_json(orient=orient)
result = pd.read_json(
data, typ="series", orient=orient, numpy=numpy, dtype=dtype
)
expected = object_series
if orient in ("values", "records"):
expected = expected.reset_index(drop=True)
if orient != "split":
expected.name = None
tm.assert_series_equal(result, expected)
@pytest.mark.parametrize("numpy", [True, False])
def test_series_roundtrip_empty(self, orient, numpy, empty_series):
data = empty_series.to_json(orient=orient)
result = pd.read_json(data, typ="series", orient=orient, numpy=numpy)
expected = empty_series
if orient in ("values", "records"):
expected = expected.reset_index(drop=True)
else:
expected.index = expected.index.astype(float)
tm.assert_series_equal(result, expected)
@pytest.mark.parametrize("numpy", [True, False])
def test_series_roundtrip_timeseries(self, orient, numpy, datetime_series):
data = datetime_series.to_json(orient=orient)
result = pd.read_json(data, typ="series", orient=orient, numpy=numpy)
expected = datetime_series
if orient in ("values", "records"):
expected = expected.reset_index(drop=True)
if orient != "split":
expected.name = None
tm.assert_series_equal(result, expected)
@pytest.mark.parametrize("dtype", [np.float64, np.int])
@pytest.mark.parametrize("numpy", [True, False])
def test_series_roundtrip_numeric(self, orient, numpy, dtype):
s = Series(range(6), index=["a", "b", "c", "d", "e", "f"])
data = s.to_json(orient=orient)
result = pd.read_json(data, typ="series", orient=orient, numpy=numpy)
expected = s.copy()
if orient in ("values", "records"):
expected = expected.reset_index(drop=True)
tm.assert_series_equal(result, expected)
def test_series_to_json_except(self):
s = Series([1, 2, 3])
msg = "Invalid value 'garbage' for option 'orient'"
with pytest.raises(ValueError, match=msg):
s.to_json(orient="garbage")
def test_series_from_json_precise_float(self):
s = Series([4.56, 4.56, 4.56])
result = read_json(s.to_json(), typ="series", precise_float=True)
tm.assert_series_equal(result, s, check_index_type=False)
def test_series_with_dtype(self):
# GH 21986
s = Series([4.56, 4.56, 4.56])
result = read_json(s.to_json(), typ="series", dtype=np.int64)
expected = Series([4] * 3)
tm.assert_series_equal(result, expected)
@pytest.mark.parametrize(
"dtype,expected",
[
(True, Series(["2000-01-01"], dtype="datetime64[ns]")),
(False, Series([946684800000])),
],
)
def test_series_with_dtype_datetime(self, dtype, expected):
s = Series(["2000-01-01"], dtype="datetime64[ns]")
data = s.to_json()
result = pd.read_json(data, typ="series", dtype=dtype)
tm.assert_series_equal(result, expected)
def test_frame_from_json_precise_float(self):
df = DataFrame([[4.56, 4.56, 4.56], [4.56, 4.56, 4.56]])
result = read_json(df.to_json(), precise_float=True)
tm.assert_frame_equal(
result, df, check_index_type=False, check_column_type=False
)
def test_typ(self):
s = Series(range(6), index=["a", "b", "c", "d", "e", "f"], dtype="int64")
result = read_json(s.to_json(), typ=None)
tm.assert_series_equal(result, s)
def test_reconstruction_index(self):
df = DataFrame([[1, 2, 3], [4, 5, 6]])
result = read_json(df.to_json())
tm.assert_frame_equal(result, df)
df = DataFrame({"a": [1, 2, 3], "b": [4, 5, 6]}, index=["A", "B", "C"])
result = read_json(df.to_json())
tm.assert_frame_equal(result, df)
def test_path(self, float_frame):
with tm.ensure_clean("test.json") as path:
for df in [
float_frame,
self.intframe,
self.tsframe,
self.mixed_frame,
]:
df.to_json(path)
read_json(path)
def test_axis_dates(self, datetime_series):
# frame
json = self.tsframe.to_json()
result = read_json(json)
tm.assert_frame_equal(result, self.tsframe)
# series
json = datetime_series.to_json()
result = read_json(json, typ="series")
tm.assert_series_equal(result, datetime_series, check_names=False)
assert result.name is None
def test_convert_dates(self, datetime_series):
# frame
df = self.tsframe.copy()
df["date"] = Timestamp("20130101")
json = df.to_json()
result = read_json(json)
tm.assert_frame_equal(result, df)
df["foo"] = 1.0
json = df.to_json(date_unit="ns")
result = read_json(json, convert_dates=False)
expected = df.copy()
expected["date"] = expected["date"].values.view("i8")
expected["foo"] = expected["foo"].astype("int64")
tm.assert_frame_equal(result, expected)
# series
ts = Series(Timestamp("20130101"), index=datetime_series.index)
json = ts.to_json()
result = read_json(json, typ="series")
tm.assert_series_equal(result, ts)
@pytest.mark.parametrize("date_format", ["epoch", "iso"])
@pytest.mark.parametrize("as_object", [True, False])
@pytest.mark.parametrize(
"date_typ", [datetime.date, datetime.datetime, pd.Timestamp]
)
def test_date_index_and_values(self, date_format, as_object, date_typ):
data = [date_typ(year=2020, month=1, day=1), pd.NaT]
if as_object:
data.append("a")
ser = pd.Series(data, index=data)
result = ser.to_json(date_format=date_format)
if date_format == "epoch":
expected = '{"1577836800000":1577836800000,"null":null}'
else:
expected = (
'{"2020-01-01T00:00:00.000Z":"2020-01-01T00:00:00.000Z","null":null}'
)
if as_object:
expected = expected.replace("}", ',"a":"a"}')
assert result == expected
@pytest.mark.parametrize(
"infer_word",
[
"trade_time",
"date",
"datetime",
"sold_at",
"modified",
"timestamp",
"timestamps",
],
)
def test_convert_dates_infer(self, infer_word):
# GH10747
from pandas.io.json import dumps
data = [{"id": 1, infer_word: 1036713600000}, {"id": 2}]
expected = DataFrame(
[[1, Timestamp("2002-11-08")], [2, pd.NaT]], columns=["id", infer_word]
)
result = read_json(dumps(data))[["id", infer_word]]
tm.assert_frame_equal(result, expected)
@pytest.mark.parametrize(
"date,date_unit",
[
("20130101 20:43:42.123", None),
("20130101 20:43:42", "s"),
("20130101 20:43:42.123", "ms"),
("20130101 20:43:42.123456", "us"),
("20130101 20:43:42.123456789", "ns"),
],
)
def test_date_format_frame(self, date, date_unit):
df = self.tsframe.copy()
df["date"] = Timestamp(date)
df.iloc[1, df.columns.get_loc("date")] = pd.NaT
df.iloc[5, df.columns.get_loc("date")] = pd.NaT
if date_unit:
json = df.to_json(date_format="iso", date_unit=date_unit)
else:
json = df.to_json(date_format="iso")
result = read_json(json)
expected = df.copy()
expected.index = expected.index.tz_localize("UTC")
expected["date"] = expected["date"].dt.tz_localize("UTC")
tm.assert_frame_equal(result, expected)
def test_date_format_frame_raises(self):
df = self.tsframe.copy()
msg = "Invalid value 'foo' for option 'date_unit'"
with pytest.raises(ValueError, match=msg):
df.to_json(date_format="iso", date_unit="foo")
@pytest.mark.parametrize(
"date,date_unit",
[
("20130101 20:43:42.123", None),
("20130101 20:43:42", "s"),
("20130101 20:43:42.123", "ms"),
("20130101 20:43:42.123456", "us"),
("20130101 20:43:42.123456789", "ns"),
],
)
def test_date_format_series(self, date, date_unit, datetime_series):
ts = Series(Timestamp(date), index=datetime_series.index)
ts.iloc[1] = pd.NaT
ts.iloc[5] = pd.NaT
if date_unit:
json = ts.to_json(date_format="iso", date_unit=date_unit)
else:
json = ts.to_json(date_format="iso")
result = read_json(json, typ="series")
expected = ts.copy()
expected.index = expected.index.tz_localize("UTC")
expected = expected.dt.tz_localize("UTC")
tm.assert_series_equal(result, expected)
def test_date_format_series_raises(self, datetime_series):
ts = Series(Timestamp("20130101 20:43:42.123"), index=datetime_series.index)
msg = "Invalid value 'foo' for option 'date_unit'"
with pytest.raises(ValueError, match=msg):
ts.to_json(date_format="iso", date_unit="foo")
@pytest.mark.parametrize("unit", ["s", "ms", "us", "ns"])
def test_date_unit(self, unit):
df = self.tsframe.copy()
df["date"] = Timestamp("20130101 20:43:42")
dl = df.columns.get_loc("date")
df.iloc[1, dl] = Timestamp("19710101 20:43:42")
df.iloc[2, dl] = Timestamp("21460101 20:43:42")
df.iloc[4, dl] = pd.NaT
json = df.to_json(date_format="epoch", date_unit=unit)
# force date unit
result = read_json(json, date_unit=unit)
tm.assert_frame_equal(result, df)
# detect date unit
result = read_json(json, date_unit=None)
tm.assert_frame_equal(result, df)
def test_weird_nested_json(self):
# this used to core dump the parser
s = r"""{
"status": "success",
"data": {
"posts": [
{
"id": 1,
"title": "A blog post",
"body": "Some useful content"
},
{
"id": 2,
"title": "Another blog post",
"body": "More content"
}
]
}
}"""
read_json(s)
def test_doc_example(self):
dfj2 = DataFrame(np.random.randn(5, 2), columns=list("AB"))
dfj2["date"] = Timestamp("20130101")
dfj2["ints"] = range(5)
dfj2["bools"] = True
dfj2.index = pd.date_range("20130101", periods=5)
json = dfj2.to_json()
result = read_json(json, dtype={"ints": np.int64, "bools": np.bool_})
tm.assert_frame_equal(result, result)
def test_misc_example(self):
# parsing unordered input fails
result = read_json('[{"a": 1, "b": 2}, {"b":2, "a" :1}]', numpy=True)
expected = DataFrame([[1, 2], [1, 2]], columns=["a", "b"])
error_msg = """DataFrame\\.index are different
DataFrame\\.index values are different \\(100\\.0 %\\)
\\[left\\]: Index\\(\\['a', 'b'\\], dtype='object'\\)
\\[right\\]: RangeIndex\\(start=0, stop=2, step=1\\)"""
with pytest.raises(AssertionError, match=error_msg):
tm.assert_frame_equal(result, expected, check_index_type=False)
result = read_json('[{"a": 1, "b": 2}, {"b":2, "a" :1}]')
expected = DataFrame([[1, 2], [1, 2]], columns=["a", "b"])
tm.assert_frame_equal(result, expected)
@tm.network
@pytest.mark.single
def test_round_trip_exception_(self):
# GH 3867
csv = "https://raw.github.com/hayd/lahman2012/master/csvs/Teams.csv"
df = pd.read_csv(csv)
s = df.to_json()
result = pd.read_json(s)
tm.assert_frame_equal(result.reindex(index=df.index, columns=df.columns), df)
@tm.network
@pytest.mark.single
@pytest.mark.parametrize(
"field,dtype",
[
["created_at", pd.DatetimeTZDtype(tz="UTC")],
["closed_at", "datetime64[ns]"],
["updated_at", pd.DatetimeTZDtype(tz="UTC")],
],
)
def test_url(self, field, dtype):
url = "https://api.github.com/repos/pandas-dev/pandas/issues?per_page=5" # noqa
result = read_json(url, convert_dates=True)
assert result[field].dtype == dtype
def test_timedelta(self):
converter = lambda x: pd.to_timedelta(x, unit="ms")
s = Series([timedelta(23), timedelta(seconds=5)])
assert s.dtype == "timedelta64[ns]"
result = pd.read_json(s.to_json(), typ="series").apply(converter)
tm.assert_series_equal(result, s)
s = Series([timedelta(23), timedelta(seconds=5)], index=pd.Index([0, 1]))
assert s.dtype == "timedelta64[ns]"
result = pd.read_json(s.to_json(), typ="series").apply(converter)
tm.assert_series_equal(result, s)
frame = DataFrame([timedelta(23), timedelta(seconds=5)])
assert frame[0].dtype == "timedelta64[ns]"
tm.assert_frame_equal(frame, pd.read_json(frame.to_json()).apply(converter))
frame = DataFrame(
{
"a": [timedelta(days=23), timedelta(seconds=5)],
"b": [1, 2],
"c": pd.date_range(start="20130101", periods=2),
}
)
result = pd.read_json(frame.to_json(date_unit="ns"))
result["a"] = pd.to_timedelta(result.a, unit="ns")
result["c"] = pd.to_datetime(result.c)
tm.assert_frame_equal(frame, result)
def test_mixed_timedelta_datetime(self):
frame = DataFrame(
{"a": [timedelta(23), pd.Timestamp("20130101")]}, dtype=object
)
expected = DataFrame(
{"a": [pd.Timedelta(frame.a[0]).value, pd.Timestamp(frame.a[1]).value]}
)
result = pd.read_json(frame.to_json(date_unit="ns"), dtype={"a": "int64"})
tm.assert_frame_equal(result, expected, check_index_type=False)
@pytest.mark.parametrize("as_object", [True, False])
@pytest.mark.parametrize("date_format", ["iso", "epoch"])
@pytest.mark.parametrize("timedelta_typ", [pd.Timedelta, timedelta])
def test_timedelta_to_json(self, as_object, date_format, timedelta_typ):
# GH28156: to_json not correctly formatting Timedelta
data = [timedelta_typ(days=1), timedelta_typ(days=2), pd.NaT]
if as_object:
data.append("a")
ser = pd.Series(data, index=data)
if date_format == "iso":
expected = (
'{"P1DT0H0M0S":"P1DT0H0M0S","P2DT0H0M0S":"P2DT0H0M0S","null":null}'
)
else:
expected = '{"86400000":86400000,"172800000":172800000,"null":null}'
if as_object:
expected = expected.replace("}", ',"a":"a"}')
result = ser.to_json(date_format=date_format)
assert result == expected
def test_default_handler(self):
value = object()
frame = DataFrame({"a": [7, value]})
expected = DataFrame({"a": [7, str(value)]})
result = pd.read_json(frame.to_json(default_handler=str))
tm.assert_frame_equal(expected, result, check_index_type=False)
def test_default_handler_indirect(self):
from pandas.io.json import dumps
def default(obj):
if isinstance(obj, complex):
return [("mathjs", "Complex"), ("re", obj.real), ("im", obj.imag)]
return str(obj)
df_list = [
9,
DataFrame(
{"a": [1, "STR", complex(4, -5)], "b": [float("nan"), None, "N/A"]},
columns=["a", "b"],
),
]
expected = (
'[9,[[1,null],["STR",null],[[["mathjs","Complex"],'
'["re",4.0],["im",-5.0]],"N\\/A"]]]'
)
assert dumps(df_list, default_handler=default, orient="values") == expected
def test_default_handler_numpy_unsupported_dtype(self):
# GH12554 to_json raises 'Unhandled numpy dtype 15'
df = DataFrame(
{"a": [1, 2.3, complex(4, -5)], "b": [float("nan"), None, complex(1.2, 0)]},
columns=["a", "b"],
)
expected = (
'[["(1+0j)","(nan+0j)"],'
'["(2.3+0j)","(nan+0j)"],'
'["(4-5j)","(1.2+0j)"]]'
)
assert df.to_json(default_handler=str, orient="values") == expected
def test_default_handler_raises(self):
msg = "raisin"
def my_handler_raises(obj):
raise TypeError(msg)
with pytest.raises(TypeError, match=msg):
DataFrame({"a": [1, 2, object()]}).to_json(
default_handler=my_handler_raises
)
with pytest.raises(TypeError, match=msg):
DataFrame({"a": [1, 2, complex(4, -5)]}).to_json(
default_handler=my_handler_raises
)
def test_categorical(self):
# GH4377 df.to_json segfaults with non-ndarray blocks
df = DataFrame({"A": ["a", "b", "c", "a", "b", "b", "a"]})
df["B"] = df["A"]
expected = df.to_json()
df["B"] = df["A"].astype("category")
assert expected == df.to_json()
s = df["A"]
sc = df["B"]
assert s.to_json() == sc.to_json()
def test_datetime_tz(self):
# GH4377 df.to_json segfaults with non-ndarray blocks
tz_range = pd.date_range("20130101", periods=3, tz="US/Eastern")
tz_naive = tz_range.tz_convert("utc").tz_localize(None)
df = DataFrame({"A": tz_range, "B": pd.date_range("20130101", periods=3)})
df_naive = df.copy()
df_naive["A"] = tz_naive
expected = df_naive.to_json()
assert expected == df.to_json()
stz = Series(tz_range)
s_naive = Series(tz_naive)
assert stz.to_json() == s_naive.to_json()
def test_sparse(self):
# GH4377 df.to_json segfaults with non-ndarray blocks
df = pd.DataFrame(np.random.randn(10, 4))
df.loc[:8] = np.nan
sdf = df.astype("Sparse")
expected = df.to_json()
assert expected == sdf.to_json()
s = pd.Series(np.random.randn(10))
s.loc[:8] = np.nan
ss = s.astype("Sparse")
expected = s.to_json()
assert expected == ss.to_json()
@pytest.mark.parametrize(
"ts",
[
Timestamp("2013-01-10 05:00:00Z"),
Timestamp("2013-01-10 00:00:00", tz="US/Eastern"),
Timestamp("2013-01-10 00:00:00-0500"),
],
)
def test_tz_is_utc(self, ts):
from pandas.io.json import dumps
exp = '"2013-01-10T05:00:00.000Z"'
assert dumps(ts, iso_dates=True) == exp
dt = ts.to_pydatetime()
assert dumps(dt, iso_dates=True) == exp
@pytest.mark.parametrize(
"tz_range",
[
pd.date_range("2013-01-01 05:00:00Z", periods=2),
pd.date_range("2013-01-01 00:00:00", periods=2, tz="US/Eastern"),
pd.date_range("2013-01-01 00:00:00-0500", periods=2),
],
)
def test_tz_range_is_utc(self, tz_range):
from pandas.io.json import dumps
exp = '["2013-01-01T05:00:00.000Z","2013-01-02T05:00:00.000Z"]'
dfexp = (
'{"DT":{'
'"0":"2013-01-01T05:00:00.000Z",'
'"1":"2013-01-02T05:00:00.000Z"}}'
)
assert dumps(tz_range, iso_dates=True) == exp
dti = pd.DatetimeIndex(tz_range)
assert dumps(dti, iso_dates=True) == exp
df = DataFrame({"DT": dti})
result = dumps(df, iso_dates=True)
assert result == dfexp
def test_read_inline_jsonl(self):
# GH9180
result = read_json('{"a": 1, "b": 2}\n{"b":2, "a" :1}\n', lines=True)
expected = DataFrame([[1, 2], [1, 2]], columns=["a", "b"])
tm.assert_frame_equal(result, expected)
@td.skip_if_not_us_locale
def test_read_s3_jsonl(self, s3_resource):
# GH17200
result = read_json("s3n://pandas-test/items.jsonl", lines=True)
expected = DataFrame([[1, 2], [1, 2]], columns=["a", "b"])
tm.assert_frame_equal(result, expected)
def test_read_local_jsonl(self):
# GH17200
with tm.ensure_clean("tmp_items.json") as path:
with open(path, "w") as infile:
infile.write('{"a": 1, "b": 2}\n{"b":2, "a" :1}\n')
result = read_json(path, lines=True)
expected = DataFrame([[1, 2], [1, 2]], columns=["a", "b"])
tm.assert_frame_equal(result, expected)
def test_read_jsonl_unicode_chars(self):
# GH15132: non-ascii unicode characters
# \u201d == RIGHT DOUBLE QUOTATION MARK
# simulate file handle
json = '{"a": "foo”", "b": "bar"}\n{"a": "foo", "b": "bar"}\n'
json = StringIO(json)
result = read_json(json, lines=True)
expected = DataFrame([["foo\u201d", "bar"], ["foo", "bar"]], columns=["a", "b"])
tm.assert_frame_equal(result, expected)
# simulate string
json = '{"a": "foo”", "b": "bar"}\n{"a": "foo", "b": "bar"}\n'
result = read_json(json, lines=True)
expected = DataFrame([["foo\u201d", "bar"], ["foo", "bar"]], columns=["a", "b"])
tm.assert_frame_equal(result, expected)
def test_read_json_large_numbers(self):
# GH18842
json = '{"articleId": "1404366058080022500245"}'
json = StringIO(json)
result = read_json(json, typ="series")
expected = Series(1.404366e21, index=["articleId"])
tm.assert_series_equal(result, expected)
json = '{"0": {"articleId": "1404366058080022500245"}}'
json = StringIO(json)
result = read_json(json)
expected = DataFrame(1.404366e21, index=["articleId"], columns=[0])
tm.assert_frame_equal(result, expected)
def test_to_jsonl(self):
# GH9180
df = DataFrame([[1, 2], [1, 2]], columns=["a", "b"])
result = df.to_json(orient="records", lines=True)
expected = '{"a":1,"b":2}\n{"a":1,"b":2}'
assert result == expected
df = DataFrame([["foo}", "bar"], ['foo"', "bar"]], columns=["a", "b"])
result = df.to_json(orient="records", lines=True)
expected = '{"a":"foo}","b":"bar"}\n{"a":"foo\\"","b":"bar"}'
assert result == expected
tm.assert_frame_equal(pd.read_json(result, lines=True), df)
# GH15096: escaped characters in columns and data
df = DataFrame([["foo\\", "bar"], ['foo"', "bar"]], columns=["a\\", "b"])
result = df.to_json(orient="records", lines=True)
expected = '{"a\\\\":"foo\\\\","b":"bar"}\n{"a\\\\":"foo\\"","b":"bar"}'
assert result == expected
tm.assert_frame_equal(pd.read_json(result, lines=True), df)
# TODO: there is a near-identical test for pytables; can we share?
def test_latin_encoding(self):
# GH 13774
pytest.skip("encoding not implemented in .to_json(), xref #13774")
values = [
[b"E\xc9, 17", b"", b"a", b"b", b"c"],
[b"E\xc9, 17", b"a", b"b", b"c"],
[b"EE, 17", b"", b"a", b"b", b"c"],
[b"E\xc9, 17", b"\xf8\xfc", b"a", b"b", b"c"],
[b"", b"a", b"b", b"c"],
[b"\xf8\xfc", b"a", b"b", b"c"],
[b"A\xf8\xfc", b"", b"a", b"b", b"c"],
[np.nan, b"", b"b", b"c"],
[b"A\xf8\xfc", np.nan, b"", b"b", b"c"],
]
values = [
[x.decode("latin-1") if isinstance(x, bytes) else x for x in y]
for y in values
]
examples = []
for dtype in ["category", object]:
for val in values:
examples.append(Series(val, dtype=dtype))
def roundtrip(s, encoding="latin-1"):
with tm.ensure_clean("test.json") as path:
s.to_json(path, encoding=encoding)
retr = read_json(path, encoding=encoding)
tm.assert_series_equal(s, retr, check_categorical=False)
for s in examples:
roundtrip(s)
def test_data_frame_size_after_to_json(self):
# GH15344
df = DataFrame({"a": [str(1)]})
size_before = df.memory_usage(index=True, deep=True).sum()
df.to_json()
size_after = df.memory_usage(index=True, deep=True).sum()
assert size_before == size_after
@pytest.mark.parametrize(
"index", [None, [1, 2], [1.0, 2.0], ["a", "b"], ["1", "2"], ["1.", "2."]]
)
@pytest.mark.parametrize("columns", [["a", "b"], ["1", "2"], ["1.", "2."]])
def test_from_json_to_json_table_index_and_columns(self, index, columns):
# GH25433 GH25435
expected = DataFrame([[1, 2], [3, 4]], index=index, columns=columns)
dfjson = expected.to_json(orient="table")
result = pd.read_json(dfjson, orient="table")
tm.assert_frame_equal(result, expected)
def test_from_json_to_json_table_dtypes(self):
# GH21345
expected = pd.DataFrame({"a": [1, 2], "b": [3.0, 4.0], "c": ["5", "6"]})
dfjson = expected.to_json(orient="table")
result = pd.read_json(dfjson, orient="table")
tm.assert_frame_equal(result, expected)
@pytest.mark.parametrize("dtype", [True, {"b": int, "c": int}])
def test_read_json_table_dtype_raises(self, dtype):
# GH21345
df = pd.DataFrame({"a": [1, 2], "b": [3.0, 4.0], "c": ["5", "6"]})
dfjson = df.to_json(orient="table")
msg = "cannot pass both dtype and orient='table'"
with pytest.raises(ValueError, match=msg):
pd.read_json(dfjson, orient="table", dtype=dtype)
def test_read_json_table_convert_axes_raises(self):
# GH25433 GH25435
df = DataFrame([[1, 2], [3, 4]], index=[1.0, 2.0], columns=["1.", "2."])
dfjson = df.to_json(orient="table")
msg = "cannot pass both convert_axes and orient='table'"
with pytest.raises(ValueError, match=msg):
pd.read_json(dfjson, orient="table", convert_axes=True)
@pytest.mark.parametrize(
"data, expected",
[
(
DataFrame([[1, 2], [4, 5]], columns=["a", "b"]),
{"columns": ["a", "b"], "data": [[1, 2], [4, 5]]},
),
(
DataFrame([[1, 2], [4, 5]], columns=["a", "b"]).rename_axis("foo"),
{"columns": ["a", "b"], "data": [[1, 2], [4, 5]]},
),
(
DataFrame(
[[1, 2], [4, 5]], columns=["a", "b"], index=[["a", "b"], ["c", "d"]]
),
{"columns": ["a", "b"], "data": [[1, 2], [4, 5]]},
),
(Series([1, 2, 3], name="A"), {"name": "A", "data": [1, 2, 3]}),
(
Series([1, 2, 3], name="A").rename_axis("foo"),
{"name": "A", "data": [1, 2, 3]},
),
(
Series([1, 2], name="A", index=[["a", "b"], ["c", "d"]]),
{"name": "A", "data": [1, 2]},
),
],
)
def test_index_false_to_json_split(self, data, expected):
# GH 17394
# Testing index=False in to_json with orient='split'
result = data.to_json(orient="split", index=False)
result = json.loads(result)
assert result == expected
@pytest.mark.parametrize(
"data",
[
(DataFrame([[1, 2], [4, 5]], columns=["a", "b"])),
(DataFrame([[1, 2], [4, 5]], columns=["a", "b"]).rename_axis("foo")),
(
DataFrame(
[[1, 2], [4, 5]], columns=["a", "b"], index=[["a", "b"], ["c", "d"]]
)
),
(Series([1, 2, 3], name="A")),
(Series([1, 2, 3], name="A").rename_axis("foo")),
(Series([1, 2], name="A", index=[["a", "b"], ["c", "d"]])),
],
)
def test_index_false_to_json_table(self, data):
# GH 17394
# Testing index=False in to_json with orient='table'
result = data.to_json(orient="table", index=False)
result = json.loads(result)
expected = {
"schema": pd.io.json.build_table_schema(data, index=False),
"data": DataFrame(data).to_dict(orient="records"),
}
assert result == expected
@pytest.mark.parametrize("orient", ["records", "index", "columns", "values"])
def test_index_false_error_to_json(self, orient):
# GH 17394
# Testing error message from to_json with index=False
df = pd.DataFrame([[1, 2], [4, 5]], columns=["a", "b"])
msg = "'index=False' is only valid when 'orient' is 'split' or 'table'"
with pytest.raises(ValueError, match=msg):
df.to_json(orient=orient, index=False)
@pytest.mark.parametrize("orient", ["split", "table"])
@pytest.mark.parametrize("index", [True, False])
def test_index_false_from_json_to_json(self, orient, index):
# GH25170
# Test index=False in from_json to_json
expected = DataFrame({"a": [1, 2], "b": [3, 4]})
dfjson = expected.to_json(orient=orient, index=index)
result = read_json(dfjson, orient=orient)
tm.assert_frame_equal(result, expected)
def test_read_timezone_information(self):
# GH 25546
result = read_json(
'{"2019-01-01T11:00:00.000Z":88}', typ="series", orient="index"
)
expected = Series([88], index=DatetimeIndex(["2019-01-01 11:00:00"], tz="UTC"))
tm.assert_series_equal(result, expected)
@pytest.mark.parametrize(
"date_format,key", [("epoch", 86400000), ("iso", "P1DT0H0M0S")]
)
def test_timedelta_as_label(self, date_format, key):
df = pd.DataFrame([[1]], columns=[pd.Timedelta("1D")])
expected = f'{{"{key}":{{"0":1}}}}'
result = df.to_json(date_format=date_format)
assert result == expected
@pytest.mark.parametrize(
"orient,expected",
[
("index", "{\"('a', 'b')\":{\"('c', 'd')\":1}}"),
("columns", "{\"('c', 'd')\":{\"('a', 'b')\":1}}"),
# TODO: the below have separate encoding procedures
# They produce JSON but not in a consistent manner
pytest.param("split", "", marks=pytest.mark.skip),
pytest.param("table", "", marks=pytest.mark.skip),
],
)
def test_tuple_labels(self, orient, expected):
# GH 20500
df = pd.DataFrame([[1]], index=[("a", "b")], columns=[("c", "d")])
result = df.to_json(orient=orient)
assert result == expected
@pytest.mark.parametrize("indent", [1, 2, 4])
def test_to_json_indent(self, indent):
# GH 12004
df = | pd.DataFrame([["foo", "bar"], ["baz", "qux"]], columns=["a", "b"]) | pandas.DataFrame |
# -*- coding: utf-8 -*-
"""
@authors: <NAME> and <NAME>
Functions for Generative Language Model Project
"""
#####################################
# imports
#####################################
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
import seaborn as sns
from keras.models import Sequential
from keras.layers import Dense, Dropout, Activation, Conv2D, Flatten, MaxPool2D, BatchNormalization
from keras.callbacks import ModelCheckpoint
from keras import optimizers, backend
from keras.models import load_model
import time
import pickle
import os
import sqlite3
import re
from unicodedata import normalize
#####################################
# functions
#####################################
#####
# for data import & preprocessing
#####
def create_connection(db):
""" connect to a sqlite database
:param db: database file
:return: a sqlite db connection object,
none if error
"""
try:
conn = sqlite3.connect(db)
return conn
except Error as e:
print(e)
return None
def _include_parents(df, db):
""" for a given sample of posts,
make sure their parent post is
included in the sample, and remove
any duplicates.
:param df: dataframe with samples
:param db: a sqlite db connection object
:return comp_df: a dataframe complete with
original posts and their parents
"""
unique_parent_ids_in_df = set(df.parent_id.unique())
sample_parents_sql = "SELECT subreddit, ups, downs, score, body, id, name, link_id, parent_id \
FROM May2015 \
WHERE subreddit != '' AND body != '' AND id != '' AND \
id IN ('{}') \
;".format("', '".join(unique_parent_ids_in_df))
sample_parents_df = | pd.read_sql(sample_parents_sql, db) | pandas.read_sql |
import numpy as np
import sklearn
import pandas as pd
import scipy.spatial.distance as ssd
from scipy.cluster import hierarchy
from scipy.stats import chi2_contingency
from sklearn.base import BaseEstimator
from sklearn.ensemble import RandomForestClassifier
from sklearn.feature_extraction.text import CountVectorizer
from sklearn.feature_selection import SelectKBest, SelectorMixin
from sklearn.pipeline import Pipeline
class SelectHierarchicalClustering(SelectorMixin, BaseEstimator):
"""
A transformer that clusters the features in X according to dist_matrix, and selects a feature from each cluster with
the highest chi2 score of X[feature] versus y
"""
def __init__(self, dist_matrix=None, threshold=1):
self.dist_matrix = dist_matrix
self.threshold = threshold
def _phi_coef(self, x, y):
"""
Calculates phi coefficient between features
Parameters
----------
x - feature x column
y - feature y column
Returns
----------
phi coefficient value
"""
confusion_matrix = | pd.crosstab(x, y) | pandas.crosstab |
## ~~~~~ Imports ~~~~~
## Data Manipulation
import pandas as pd
import numpy as np
## Plotting
import seaborn as sns
import matplotlib.pyplot as plt
## Scraping
import requests
import xmltodict
## OS Related
import os
from os import listdir
from os.path import isfile, join
## Datetime Handling
from datetime import timedelta, datetime, date
import time
## Miscellaneous
import time
import warnings
import collections
from ipypb import track
## Stream Data
from ElexonDataPortal import stream_info
## ~~~~~ Helper Functions/Classes ~~~~~
class RequestError(Exception):
def __init__(self, http_code, error_type, description):
self.message = f'{http_code} - {error_type}\n{description}'
def __str__(self):
return self.message
## ~~~~~ Core Wrapper Class ~~~~~
class Wrapper:
def dt_rng_2_SPs(self, start_date:datetime, end_date:datetime, freq='30T', tz='Europe/London'):
dt_rng = pd.date_range(start_date, end_date, freq=freq, tz=tz)
dt_strs = dt_rng.strftime('%Y-%m-%d')
dt_SP_counts = pd.Series(dt_strs).groupby(dt_strs).count()
SPs = []
for num_SPs in dt_SP_counts.values:
SPs += [SP+1 for SP in list(range(num_SPs))]
df_dates_SPs = pd.DataFrame({'date':dt_strs, 'SP':SPs}, index=dt_rng)
return df_dates_SPs
def add_local_datetime(self, df:pd.DataFrame, start_date:str, end_date:str, stream:str):
"""
Accepts a dataframe, start and end date, and date and SP columns in the dataframe
creates a mapping from date and SP columns to the local datetime,
then maps the data and adds the new column to the dataframe.
"""
stream_metadata = stream_info.streams[stream]
assert all(col in stream_metadata.keys() for col in ['date_col', 'SP_col']), f'{stream}\'s metadata does not contain the required date_col and SP_col parameters'
## Adding End-Date Margin
end_date = (pd.to_datetime(end_date) + pd.Timedelta(days=1)).strftime('%Y-%m-%d')
## Creating Date & SP to Timestamp Map
ts_2_dt_SPs = self.dt_rng_2_SPs(start_date, end_date)
date_SP_tuples = list(zip(ts_2_dt_SPs['date'], ts_2_dt_SPs['SP']))
dt_SP_2_ts = dict(zip(date_SP_tuples, ts_2_dt_SPs.index))
## Mapping & Setting Datetimes
s_dt_SPs = pd.Series(zip(df[stream_info.streams[stream]['date_col']], df[stream_info.streams[stream]['SP_col']].astype(int)), index=df.index)
df['local_datetime'] = s_dt_SPs.map(dt_SP_2_ts)
return df
def expand_cols(self, df, cols_2_expand=[]):
for col in cols_2_expand:
new_df_cols = df[col].apply(pd.Series)
df[new_df_cols.columns] = new_df_cols
df = df.drop(columns=col)
s_cols_2_expand = df.iloc[0].apply(type).isin([collections.OrderedDict, dict, list, tuple])
if s_cols_2_expand.sum() > 0:
cols_2_expand = s_cols_2_expand[s_cols_2_expand].index
df = self.expand_cols(df, cols_2_expand)
return df
def check_response(self, r_metadata):
if r_metadata['httpCode'] != '200':
raise RequestError(r_metadata['httpCode'], r_metadata['errorType'], r_metadata['description'])
if 'cappingApplied' in r_metadata.keys():
if r_metadata['cappingApplied'] == 'Yes':
self.capping_applied = True
else:
self.capping_applied = False
else:
self.capping_applied = 'Could not be determined'
def check_and_parse_query_args(self, query_args, stream):
## ~~~~~ Parsing args ~~~~~
## Creating new params dictionary
stream_params = dict()
stream_params.update({'APIKey':'APIKey', 'ServiceType':'ServiceType'})
## If dictionary of query parameter mappings exist then add it to the stream_params
for param_type in ['required_params', 'optional_params']:
if stream_info.streams[stream][param_type]:
stream_params.update(stream_info.streams[stream][param_type])
## ~~~~~ Checking args ~~~~~
extra_args = list(set(query_args.keys()) - set(stream_params.keys()))
missing_args = list(set(stream_params.keys()) - set(query_args.keys()) - set(['APIKey', 'ServiceType']))
assert(len(missing_args)) == 0, f'The following arguments were needed but not provided: {", ".join(missing_args)}'
if len(extra_args) != 0:
warnings.warn(f'The following arguments were provided but not needed: {", ".join(extra_args)}')
for key in extra_args:
query_args.pop(key, None)
## ~~~~ Mapping args ~~~~~
## Mapping the generalised wrapper parameters to the parameter names expected by the API
parsed_query_args = dict((stream_params[key], val) for key, val in query_args.items())
return parsed_query_args
def parse_response(self, response, stream):
r_dict = xmltodict.parse(response.text)
r_metadata = r_dict['response']['responseMetadata']
self.last_request_metadata = r_metadata
if r_metadata['httpCode'] == '204':
warnings.warn(f'Data request was succesful but no content was returned')
return pd.DataFrame()
self.check_response(r_metadata)
content_dict = r_dict['response']['responseBody']['responseList']['item']
data_parse_type = stream_info.streams[stream]['data_parse_type']
data = self.data_parse_types[data_parse_type](content_dict)
if data_parse_type == 'dataframe':
df = self.expand_cols(data)
if data_parse_type == 'series':
df = pd.DataFrame(data).T
return df
def make_request(self, stream, query_args, service_type='xml'):
## Checking inputs
assert stream in stream_info.streams.keys(), f'Data stream should be one of: {", ".join(stream_info.streams.keys())}'
query_args = self.check_and_parse_query_args(query_args, stream)
## Forming url and request parameters
url_endpoint = f'https://api.bmreports.com/BMRS/{stream}/v{stream_info.streams[stream]["API_version"]}'
query_args.update({
'APIKey' : self.API_key,
'ServiceType' : self.service_type,
})
## Making request
response = requests.get(url_endpoint, params=query_args)
return response
def query(self, stream, query_args, service_type='xml'):
response = self.make_request(stream, query_args, service_type)
df = self.parse_response(response, stream)
return df
def query_orchestrator(self, stream, query_args, service_type='xml', track_label=None, wait_time=0):
check_date_rng_args = lambda query_args: all(x in query_args.keys() for x in ['start_date', 'end_date'])
df = pd.DataFrame()
if stream_info.streams[stream]['request_type'] == 'date_range':
## Dealing with date range requests - main concern is whether capping has been applied
assert check_date_rng_args(query_args), 'All date range queries should be provided with a "start_date" and "end_date".'
self.capping_applied = True
date_col = stream_info.streams[stream]['date_col']
start_date, end_date = query_args['start_date'], query_args['end_date']
absolute_start_date = start_date
while self.capping_applied == True:
response = self.make_request(stream, query_args, service_type)
df_new = self.parse_response(response, stream)
df = df.append(df_new)
assert self.capping_applied != None, 'Whether or not capping limits had been breached could not be found in the response metadata'
if self.capping_applied == True:
start_date = pd.to_datetime(df[date_col]).max().tz_localize(None)
warnings.warn(f'Response was capped, request is rerunning for missing data from {start_date}')
if pd.to_datetime(start_date) >= pd.to_datetime(end_date):
warnings.warn(f'End data ({end_date}) was earlier than start date ({start_date})\nThe start date will be set one day earlier.')
start_date = ( | pd.to_datetime(end_date) | pandas.to_datetime |
from pymongo import *
from url import URL
import statistics as stat
from time import strptime, mktime
import pandas as pd
import sys
import re
client = MongoClient(URL)
db = client.crypto_wallet
def checkLen(a, b):
if len(a) == len(b):
return True
else:
return f'DB Objs:{len(a)} < Clean Arr Items:{len(b)}' \
if len(a) < len(b) else f'Clean Arr Items:{len(b)} < DB Objs:{len(a)}'
def p(o):
return print(o)
def filterData(coll, st, narr):
for obj in coll:
try:
tmp = obj.get(st)
narr.append(tmp)
except Exception as e:
print(e, obj['_id'])
return narr
def datetime_converter(dtstr):
tmstmp = strptime(dtstr, '%Y-%m-%d %H:%M:%S')
epoch = mktime(tmstmp)
return int(epoch)
def updateCSV():
BTC_Tickers_Collection = db.BTC_Tickers
BTC_Tickers_Objs = list(BTC_Tickers_Collection.find())
BTC_epochs = []
BTC_prices = []
BTC_volumes = []
BTC_highs = []
BTC_lows = []
for obj in BTC_Tickers_Objs:
dt = re.sub(r'\..*', '', obj.get('time')).replace('T', ' ').rstrip('Z')
BTC_epochs.append(datetime_converter(dt))
BTC_prices.append(float(obj.get('price')))
BTC_volumes.append(obj.get('volume'))
BTC_highs.append(obj.get('ask'))
BTC_lows.append(obj.get('bid'))
for i, e in enumerate(BTC_epochs):
if i == 0:
pass
else:
BTC_epochs[i] = BTC_epochs[i - 1] + 60
p(checkLen(BTC_Tickers_Objs, BTC_prices))
p(checkLen(BTC_epochs, BTC_prices))
p(checkLen(BTC_Tickers_Objs, BTC_volumes))
p(checkLen(BTC_Tickers_Objs, BTC_highs))
p(checkLen(BTC_Tickers_Objs, BTC_lows))
BTC_RSI_Collection = db.BTC_RSI14_Data
BTC_RSI_Objs = list(BTC_RSI_Collection.find())
BTC_RSIs = []
Errors = []
for rsio in BTC_RSI_Objs:
RSI = rsio.get('RSI')
try:
if type(RSI) == float:
BTC_RSIs.append(int(RSI))
elif type(RSI) == list:
if RSI[0] == None:
pass
else:
BTC_RSIs.append(int(stat.mean(RSI)))
else:
BTC_RSIs.append(RSI)
except Exception as e:
Errors.append(rsio['_id'])
print(e, rsio['_id'])
sys.exit(1)
if len(Errors) > 0:
print(Errors)
p(checkLen(BTC_RSI_Objs, BTC_RSIs))
BTC_ADL_Collection = db.BTC_ADL_Data
BTC_ADL_Objs = list(BTC_ADL_Collection.find())
BTC_ADLs = []
BTC_ADL_slope = []
for o in BTC_ADL_Objs:
ADL = o.get('ADL')
slope = o.get('slope')
try:
if type(ADL) == float:
BTC_ADLs.append(int(ADL))
elif type(ADL) == list:
BTC_ADLs.append(int(stat.mean(ADL)))
else:
BTC_ADLs.append(ADL)
if type(slope) == int:
BTC_ADL_slope.append(float(slope))
elif type(slope) == list:
BTC_ADL_slope.append(int(stat.mean(slope)))
else:
BTC_ADL_slope.append(slope)
except Exception as e:
print(e, o['_id'])
sys.exit(1)
p(checkLen(BTC_ADL_Objs, BTC_ADLs))
p(checkLen(BTC_ADL_slope, BTC_ADLs))
BTC_OBV_Collection = db.BTC_OBV_Data
BTC_OBV_Objs = list(BTC_OBV_Collection.find())
BTC_OBVs = []
BTC_OBV_slope = []
for o in BTC_OBV_Collection.find():
OBV = o.get('OBV')
slope = o.get('slope')
try:
if type(OBV) == float:
BTC_OBVs.append(int(OBV))
elif type(OBV) == list:
BTC_OBVs.append(int(stat.mean(OBV)))
else:
BTC_OBVs.append(OBV)
if type(slope) == int:
BTC_OBV_slope.append(float(slope))
elif type(slope) == list:
BTC_ADL_slope.append(int(stat.mean(slope)))
else:
BTC_OBV_slope.append(slope)
except Exception as e:
print(e, o['_id'])
sys.exit(1)
p(checkLen(BTC_OBV_Objs, BTC_OBVs))
p(checkLen(BTC_OBV_slope, BTC_OBVs))
print(f'datetime: {len(BTC_epochs)}\nprices: {len(BTC_prices)}')
collection_lengths = [len(BTC_volumes), len(BTC_highs),
len(BTC_lows), len(BTC_ADLs),
len(BTC_ADL_slope), len(BTC_OBVs), len(BTC_OBV_slope)]
print(f'Volumes: {len(BTC_RSIs)}\nHighs: {len(BTC_RSIs)}\n'
f'Lows: {len(BTC_RSIs)}\nRSI: {len(BTC_RSIs)}'
f'\nADL_slp: {len(BTC_ADL_slope)}\nOBV_slp: {len(BTC_OBV_slope)}')
min = collection_lengths[0]
for i in range(1, len(collection_lengths)):
if collection_lengths[i] < min:
min = collection_lengths[i]
BTC_Data = {'Datetime': BTC_epochs[0:min],
'Prices': BTC_prices[0:min],
'Volumes':BTC_volumes[0:min],
'High':BTC_highs[0:min],
'Low':BTC_lows[0:min],
'RSI': BTC_RSIs[0:min],
'ADL' : BTC_ADLs[0:min],
'ADL_slope': BTC_ADL_slope[0:min], 'OBV' : BTC_OBVs[0:min], 'OBV_slope': BTC_OBV_slope[0:min]}
BTC_df = | pd.DataFrame(BTC_Data) | pandas.DataFrame |
import geopandas
import pandas as pd
import requests
class WrakkenBankData:
"""
"""
def __init__(self):
url = 'https://wrakkendatabank.api.afdelingkust.be/v1/wrecks'
response = requests.get(url)
if response.status_code == 200:
wrecks_json = response.json()['wrecks']
df = | pd.DataFrame(wrecks_json) | pandas.DataFrame |
import argparse
import json
import logging
import os
import pickle
import subprocess
import sys
import tarfile
logger = logging.getLogger(__name__)
logger.setLevel(logging.DEBUG)
def install(package):
subprocess.check_call([sys.executable, "-m", "pip", "install", package])
os.system("conda install -c sebp scikit-survival -y")
install("xgboost")
install("smdebug==1.0.5")
install("shap==0.39.0")
install("scikit-learn==0.24.1")
install("matplotlib")
import matplotlib.pyplot as plt
import numpy as np
import pandas as pd
import shap
import xgboost
from sklearn.metrics import accuracy_score, classification_report
from sksurv.datasets import get_x_y
from sksurv.metrics import brier_score, concordance_index_ipcw
from smdebug.trials import create_trial
def main(args):
"""
Runs evaluation for the data set
1. Loads model from tar.gz
2. Reads in test features
3. Runs an accuracy report
4. Generates feature importance with SHAP
Args:
model-name (str): Name of the trained model, default xgboost
test-features (str): preprocessed test features for
evaluation, default test_features.csv
train-features (str): preproceed train features for SHAP,
default train_features.csv
test-features (str): preproceed test features for SHAP,
default test_features.csv
report-name (str): Name of the evaluation output
, default evaluation.json
shap-name (str): Name of the SHAP feature importance
output file, default shap.csv
threshold (float): Threshold to cut probablities at
, default 0.5
tau (int): time range for the c-index will be from 0 to tau
, default 100
"""
model_path = os.path.join("/opt/ml/processing/model", "model.tar.gz")
logger.info(f"Extracting model from path: {model_path}")
with tarfile.open(model_path) as tar:
tar.extractall(path=".")
logger.info("Loading model")
with open(args.model_name, "rb") as f:
model = pickle.load(f)
logger.info("Loading train and test data")
test_features_data = os.path.join("/opt/ml/processing/test", args.test_features)
train_features_data = os.path.join("/opt/ml/processing/train", args.train_features)
X_test = | pd.read_csv(test_features_data, header=0) | pandas.read_csv |
"""
Tests for Timestamp timezone-related methods
"""
from datetime import (
date,
datetime,
timedelta,
)
import dateutil
from dateutil.tz import (
gettz,
tzoffset,
)
import pytest
import pytz
from pytz.exceptions import (
AmbiguousTimeError,
NonExistentTimeError,
)
from pandas._libs.tslibs import timezones
from pandas.errors import OutOfBoundsDatetime
import pandas.util._test_decorators as td
from pandas import (
NaT,
Timestamp,
)
class TestTimestampTZOperations:
# --------------------------------------------------------------
# Timestamp.tz_localize
def test_tz_localize_pushes_out_of_bounds(self):
# GH#12677
# tz_localize that pushes away from the boundary is OK
msg = (
f"Converting {Timestamp.min.strftime('%Y-%m-%d %H:%M:%S')} "
f"underflows past {Timestamp.min}"
)
pac = Timestamp.min.tz_localize("US/Pacific")
assert pac.value > Timestamp.min.value
pac.tz_convert("Asia/Tokyo") # tz_convert doesn't change value
with pytest.raises(OutOfBoundsDatetime, match=msg):
Timestamp.min.tz_localize("Asia/Tokyo")
# tz_localize that pushes away from the boundary is OK
msg = (
f"Converting {Timestamp.max.strftime('%Y-%m-%d %H:%M:%S')} "
f"overflows past {Timestamp.max}"
)
tokyo = Timestamp.max.tz_localize("Asia/Tokyo")
assert tokyo.value < Timestamp.max.value
tokyo.tz_convert("US/Pacific") # tz_convert doesn't change value
with pytest.raises(OutOfBoundsDatetime, match=msg):
Timestamp.max.tz_localize("US/Pacific")
def test_tz_localize_ambiguous_bool(self):
# make sure that we are correctly accepting bool values as ambiguous
# GH#14402
ts = Timestamp("2015-11-01 01:00:03")
expected0 = Timestamp("2015-11-01 01:00:03-0500", tz="US/Central")
expected1 = Timestamp("2015-11-01 01:00:03-0600", tz="US/Central")
msg = "Cannot infer dst time from 2015-11-01 01:00:03"
with pytest.raises(pytz.AmbiguousTimeError, match=msg):
ts.tz_localize("US/Central")
result = ts.tz_localize("US/Central", ambiguous=True)
assert result == expected0
result = ts.tz_localize("US/Central", ambiguous=False)
assert result == expected1
def test_tz_localize_ambiguous(self):
ts = Timestamp("2014-11-02 01:00")
ts_dst = ts.tz_localize("US/Eastern", ambiguous=True)
ts_no_dst = ts.tz_localize("US/Eastern", ambiguous=False)
assert (ts_no_dst.value - ts_dst.value) / 1e9 == 3600
msg = "Cannot infer offset with only one time"
with pytest.raises(ValueError, match=msg):
ts.tz_localize("US/Eastern", ambiguous="infer")
# GH#8025
msg = "Cannot localize tz-aware Timestamp, use tz_convert for conversions"
with pytest.raises(TypeError, match=msg):
Timestamp("2011-01-01", tz="US/Eastern").tz_localize("Asia/Tokyo")
msg = "Cannot convert tz-naive Timestamp, use tz_localize to localize"
with pytest.raises(TypeError, match=msg):
Timestamp("2011-01-01").tz_convert("Asia/Tokyo")
@pytest.mark.parametrize(
"stamp, tz",
[
("2015-03-08 02:00", "US/Eastern"),
("2015-03-08 02:30", "US/Pacific"),
("2015-03-29 02:00", "Europe/Paris"),
("2015-03-29 02:30", "Europe/Belgrade"),
],
)
def test_tz_localize_nonexistent(self, stamp, tz):
# GH#13057
ts = Timestamp(stamp)
with pytest.raises(NonExistentTimeError, match=stamp):
ts.tz_localize(tz)
# GH 22644
with pytest.raises(NonExistentTimeError, match=stamp):
ts.tz_localize(tz, nonexistent="raise")
assert ts.tz_localize(tz, nonexistent="NaT") is NaT
def test_tz_localize_ambiguous_raise(self):
# GH#13057
ts = Timestamp("2015-11-1 01:00")
msg = "Cannot infer dst time from 2015-11-01 01:00:00,"
with pytest.raises(AmbiguousTimeError, match=msg):
ts.tz_localize("US/Pacific", ambiguous="raise")
def test_tz_localize_nonexistent_invalid_arg(self):
# GH 22644
tz = "Europe/Warsaw"
ts = Timestamp("2015-03-29 02:00:00")
msg = (
"The nonexistent argument must be one of 'raise', 'NaT', "
"'shift_forward', 'shift_backward' or a timedelta object"
)
with pytest.raises(ValueError, match=msg):
ts.tz_localize(tz, nonexistent="foo")
@pytest.mark.parametrize(
"stamp",
[
"2014-02-01 09:00",
"2014-07-08 09:00",
"2014-11-01 17:00",
"2014-11-05 00:00",
],
)
def test_tz_localize_roundtrip(self, stamp, tz_aware_fixture):
tz = tz_aware_fixture
ts = Timestamp(stamp)
localized = ts.tz_localize(tz)
assert localized == Timestamp(stamp, tz=tz)
msg = "Cannot localize tz-aware Timestamp"
with pytest.raises(TypeError, match=msg):
localized.tz_localize(tz)
reset = localized.tz_localize(None)
assert reset == ts
assert reset.tzinfo is None
def test_tz_localize_ambiguous_compat(self):
# validate that pytz and dateutil are compat for dst
# when the transition happens
naive = Timestamp("2013-10-27 01:00:00")
pytz_zone = "Europe/London"
dateutil_zone = "dateutil/Europe/London"
result_pytz = naive.tz_localize(pytz_zone, ambiguous=0)
result_dateutil = naive.tz_localize(dateutil_zone, ambiguous=0)
assert result_pytz.value == result_dateutil.value
assert result_pytz.value == 1382835600000000000
# fixed ambiguous behavior
# see gh-14621
assert result_pytz.to_pydatetime().tzname() == "GMT"
assert result_dateutil.to_pydatetime().tzname() == "BST"
assert str(result_pytz) == str(result_dateutil)
# 1 hour difference
result_pytz = naive.tz_localize(pytz_zone, ambiguous=1)
result_dateutil = naive.tz_localize(dateutil_zone, ambiguous=1)
assert result_pytz.value == result_dateutil.value
assert result_pytz.value == 1382832000000000000
# see gh-14621
assert str(result_pytz) == str(result_dateutil)
assert (
result_pytz.to_pydatetime().tzname()
== result_dateutil.to_pydatetime().tzname()
)
@pytest.mark.parametrize(
"tz",
[
pytz.timezone("US/Eastern"),
gettz("US/Eastern"),
"US/Eastern",
"dateutil/US/Eastern",
],
)
def test_timestamp_tz_localize(self, tz):
stamp = Timestamp("3/11/2012 04:00")
result = stamp.tz_localize(tz)
expected = Timestamp("3/11/2012 04:00", tz=tz)
assert result.hour == expected.hour
assert result == expected
@pytest.mark.parametrize(
"start_ts, tz, end_ts, shift",
[
["2015-03-29 02:20:00", "Europe/Warsaw", "2015-03-29 03:00:00", "forward"],
[
"2015-03-29 02:20:00",
"Europe/Warsaw",
"2015-03-29 01:59:59.999999999",
"backward",
],
[
"2015-03-29 02:20:00",
"Europe/Warsaw",
"2015-03-29 03:20:00",
timedelta(hours=1),
],
[
"2015-03-29 02:20:00",
"Europe/Warsaw",
"2015-03-29 01:20:00",
timedelta(hours=-1),
],
["2018-03-11 02:33:00", "US/Pacific", "2018-03-11 03:00:00", "forward"],
[
"2018-03-11 02:33:00",
"US/Pacific",
"2018-03-11 01:59:59.999999999",
"backward",
],
[
"2018-03-11 02:33:00",
"US/Pacific",
"2018-03-11 03:33:00",
timedelta(hours=1),
],
[
"2018-03-11 02:33:00",
"US/Pacific",
"2018-03-11 01:33:00",
timedelta(hours=-1),
],
],
)
@pytest.mark.parametrize("tz_type", ["", "dateutil/"])
def test_timestamp_tz_localize_nonexistent_shift(
self, start_ts, tz, end_ts, shift, tz_type
):
# GH 8917, 24466
tz = tz_type + tz
if isinstance(shift, str):
shift = "shift_" + shift
ts = Timestamp(start_ts)
result = ts.tz_localize(tz, nonexistent=shift)
expected = Timestamp(end_ts).tz_localize(tz)
assert result == expected
@pytest.mark.parametrize("offset", [-1, 1])
@pytest.mark.parametrize("tz_type", ["", "dateutil/"])
def test_timestamp_tz_localize_nonexistent_shift_invalid(self, offset, tz_type):
# GH 8917, 24466
tz = tz_type + "Europe/Warsaw"
ts = Timestamp("2015-03-29 02:20:00")
msg = "The provided timedelta will relocalize on a nonexistent time"
with pytest.raises(ValueError, match=msg):
ts.tz_localize(tz, nonexistent=timedelta(seconds=offset))
@pytest.mark.parametrize("tz", ["Europe/Warsaw", "dateutil/Europe/Warsaw"])
def test_timestamp_tz_localize_nonexistent_NaT(self, tz):
# GH 8917
ts = Timestamp("2015-03-29 02:20:00")
result = ts.tz_localize(tz, nonexistent="NaT")
assert result is NaT
@pytest.mark.parametrize("tz", ["Europe/Warsaw", "dateutil/Europe/Warsaw"])
def test_timestamp_tz_localize_nonexistent_raise(self, tz):
# GH 8917
ts = Timestamp("2015-03-29 02:20:00")
msg = "2015-03-29 02:20:00"
with pytest.raises(pytz.NonExistentTimeError, match=msg):
ts.tz_localize(tz, nonexistent="raise")
msg = (
"The nonexistent argument must be one of 'raise', 'NaT', "
"'shift_forward', 'shift_backward' or a timedelta object"
)
with pytest.raises(ValueError, match=msg):
ts.tz_localize(tz, nonexistent="foo")
# ------------------------------------------------------------------
# Timestamp.tz_convert
@pytest.mark.parametrize(
"stamp",
[
"2014-02-01 09:00",
"2014-07-08 09:00",
"2014-11-01 17:00",
"2014-11-05 00:00",
],
)
def test_tz_convert_roundtrip(self, stamp, tz_aware_fixture):
tz = tz_aware_fixture
ts = Timestamp(stamp, tz="UTC")
converted = ts.tz_convert(tz)
reset = converted.tz_convert(None)
assert reset == Timestamp(stamp)
assert reset.tzinfo is None
assert reset == converted.tz_convert("UTC").tz_localize(None)
@pytest.mark.parametrize("tzstr", ["US/Eastern", "dateutil/US/Eastern"])
def test_astimezone(self, tzstr):
# astimezone is an alias for tz_convert, so keep it with
# the tz_convert tests
utcdate = Timestamp("3/11/2012 22:00", tz="UTC")
expected = utcdate.tz_convert(tzstr)
result = utcdate.astimezone(tzstr)
assert expected == result
assert isinstance(result, Timestamp)
@td.skip_if_windows
def test_tz_convert_utc_with_system_utc(self):
# from system utc to real utc
ts = Timestamp("2001-01-05 11:56", tz=timezones.maybe_get_tz("dateutil/UTC"))
# check that the time hasn't changed.
assert ts == ts.tz_convert(dateutil.tz.tzutc())
# from system utc to real utc
ts = Timestamp("2001-01-05 11:56", tz=timezones.maybe_get_tz("dateutil/UTC"))
# check that the time hasn't changed.
assert ts == ts.tz_convert(dateutil.tz.tzutc())
# ------------------------------------------------------------------
# Timestamp.__init__ with tz str or tzinfo
def test_timestamp_constructor_tz_utc(self):
utc_stamp = Timestamp("3/11/2012 05:00", tz="utc")
assert utc_stamp.tzinfo is pytz.utc
assert utc_stamp.hour == 5
utc_stamp = Timestamp("3/11/2012 05:00").tz_localize("utc")
assert utc_stamp.hour == 5
def test_timestamp_to_datetime_tzoffset(self):
tzinfo = tzoffset(None, 7200)
expected = Timestamp("3/11/2012 04:00", tz=tzinfo)
result = Timestamp(expected.to_pydatetime())
assert expected == result
def test_timestamp_constructor_near_dst_boundary(self):
# GH#11481 & GH#15777
# Naive string timestamps were being localized incorrectly
# with tz_convert_from_utc_single instead of tz_localize_to_utc
for tz in ["Europe/Brussels", "Europe/Prague"]:
result = Timestamp("2015-10-25 01:00", tz=tz)
expected = Timestamp("2015-10-25 01:00").tz_localize(tz)
assert result == expected
msg = "Cannot infer dst time from 2015-10-25 02:00:00"
with pytest.raises(pytz.AmbiguousTimeError, match=msg):
Timestamp("2015-10-25 02:00", tz=tz)
result = Timestamp("2017-03-26 01:00", tz="Europe/Paris")
expected = | Timestamp("2017-03-26 01:00") | pandas.Timestamp |
from pathlib import Path
import pandas as pd
# Directory of this file
this_dir = Path(__file__).resolve().parent
# Read in all Excel files from all subfolders of sales_data
parts = []
for path in (this_dir / "sales_data").rglob("*.xls*"):
print(f'Reading {path.name}')
part = pd.read_excel(path, index_col="transaction_id")
parts.append(part)
# Combine the DataFrames from each file into a single DataFrame
# pandas takes care of properly aligning the columns
df = | pd.concat(parts) | pandas.concat |
# Import your libraries
import pandas as pd
import numpy as np
# Start writing code
max_users = len(list(set(list(facebook_friends.user2.unique() ) + list(facebook_friends.user1.unique()))))
revert = facebook_friends.rename(columns = {'user1' : 'user2', 'user2':'user1'})
Grouped = | pd.concat([revert, facebook_friends]) | pandas.concat |
# -*- coding: utf-8 -*-
"""
Created on Wed Dec 15 20:41:19 2021
@author: DELL
"""
# This Python 3 environment comes with many helpful analytics libraries installed
# It is defined by the kaggle/python docker image: https://github.com/kaggle/docker-python
# For example, here's several helpful packages to load in
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
import matplotlib.pyplot as plt
import seaborn as sns # visualization tool
data = pd.read_csv('pokemon.csv')
data.head()
data.info()
data.corr()
#%%
#corellation map
f,ax =plt.subplots(figsize=(18,18))
sns.heatmap(data.corr(),annot = True, linewidths=.5 , fmt= '.1f', ax=ax)
plt.show()
data.head(10)
data.columns
#%%
data.Speed.plot(kind="line", color="g", label="speed",linewidth=1,alpha=0.5,grid=True, linestyle=":")
data.Defense.plot(kind="line", color="r", label="Defense",linewidth=1,alpha=0.5,grid=True, linestyle="-.")
plt.legend(loc="upper right")
plt.title("line plot")
plt.xlabel("x axis")
plt.ylabel("y axis")
plt.show()
#scatter plot
#x =Attack y= Defense
data.plot(kind="scatter",x="Attack", y="Defense", alpha=0.5,color= "red")
plt.xlabel("Attack")
plt.ylabel("Defense")
plt.title("Attack Defense Scatter Plot")
plt.show()
#histogram
data.Speed.plot(kind="hist", bins= 50, figsize=(15,15))
#clf()
data.Speed.plot(kind="hist",bins=50)
plt.clf()
#%%
#create dictionary and look its keys and values
dictionary ={"Spain":"Madrid","Usa":"Washington"}
print(dictionary.keys())
print(dictionary.values())
# Keys have to be immutable objects like string, boolean, float, integer or tubles
# List is not immutable
# Keys are unique
dictionary["Spain"]= " Barcelona" # update exiting entry= çıkan girişi güncelle
print(dictionary)
dictionary["France"]= " Paris" # add new dictionary = yeni giriş ekle
print(dictionary)
print("France" in dictionary) # check inculede or not = içerip içermediğini kontrol et
del dictionary["Spain"]
print(dictionary) #remove entry with key "spain" = ispanya anahtarını sil
dictionary.clear()
print(dictionary)
# In order to run all code you need to take comment this line = Tüm kodu çalıştırmak için bu satırı yorumlamanız gerekir.
#del dictionary # delete entire dictionary = Tüm sözlüğü sil
print(dictionary) # it gives error because dictionary is deleted = Sözlük silindiği için hata veriyor
#%%PANDAS
data = pd.read_csv("C:/Users/DELL/myfirstreactproject/pokemon.csv")
series = data["Defense"] #data["Defense"] =series = Bu bir seridir
data_frame= data[["Defense"]] # data["defense] = data frame
print(type(data_frame))
# Comparison operator
print(3 > 2)
print(3!=2)
# Boolean operators
print(True and False)
print(True or False)
# 1 - Filtering Pandas data frame
x = data["Defense"]> 200 # There are only 3 pokemons who have higher defense value than 200 = 200'den daha yüksek savunma değerine sahip sadece 3 pokemon var
data[x]
data[np.logical_and(data["Defense"]> 200 , data["Attack"]>100)] #There are only 2 pokemon with defense value higher than 2oo and attack value higher than 100.#2oo'dan daha yüksek savunma değerine ve 100'den daha yüksek saldırı değerine sahip sadece 2 pokemon var.
data[(data['Defense']>200) & (data['Attack']>100)] # This is the same as the previous line of code. So we can also use '&' for filtering. # Bu, önceki kod satırıyla da aynıdır. Bu nedenle filtreleme için '&' da kullanabiliriz.
#%% USER DEFINED FUNCTION
# What do we need to know about functions:
# docstrings: documentation for functions. Example:
# for f():
# """This is docstring for documentation of function f"""
# tuple: sequence of immutable python objects.
# cant modify values
# tuple uses paranthesis like tuble = (1,2,3)
# unpack tuple into several variables like a,b,c = tuple
#Fonksiyonlar hakkında bilmemiz gerekenler:
# docstrings: işlevler için belgeler. Örnek:
# f() için:
# """Bu, f fonksiyonunun dokümantasyonu için doküman dizisidir"""
# Tuple: değişmez python nesnelerinin dizisi.
# değerleri değiştiremez
# Tuple, tuble = (1,2,3) gibi parantez kullanır
# Tuple'ı a,b,c = tuple gibi çeşitli değişkenlere ayırın
def tuple_ex():
"""return defined tuple"""
t=(1,2,3)
return t
a,b,c= tuple_ex()
print(a,b,c)
#%%
#SCOPE
# What we need to know about scope:
# global: defined main body in script
# local: defined in a function
# built in scope: names in predefined built in scope module such as print, len
# Lets make some basic examples
# KAPSAM
# Kapsam hakkında bilmemiz gerekenler:
# global: komut dosyasında tanımlanmış ana gövde
# yerel: bir fonksiyonda tanımlanmış
# yerleşik kapsam: print, len gibi önceden tanımlanmış yerleşik kapsam modülündeki adlar
# Bazı temel örnekler yapalım
# guess prints what # tahmin et ne yazdırır
x = 2
def f():
x =3
return x
print(x) # x = 2 global scope = global kapsam
print(f()) # x = 3 local scope = yerel kapsam
# What if there is no local scope? = Yerel kapsam yoksa ne olur?
x = 5
def f():
y = 2*x # there is no local scope x = yerel kapsam yok x
return y
print(f()) # it uses global scope x = global kapsam x kullanır
# First local scope searched, then global scope searched, if two of them cannot be found lastly built in scope searched.
#Önce yerel kapsam aranır, ardından global kapsam aranır, bunlardan ikisi bulunamazsa son olarak kapsam içinde yerleşik olarak aranır.
# How can we learn what is built in scope = yerleşik kapsamları nasıl bulabiliriz?# bunlar daha önce python tarafından tanımlanmış belirli kapsamlardır
import builtins
dir (builtins)
#%%NESTED FUNCTION İÇ İÇE FONKSİYONLAR
# function inside function. = Fonksiyon içinde fonksiyon
# There is a LEGB rule that is search local scope, enclosing function, global and built in scopes, respectively. = # Sırasıyla yerel kapsam arama, kapsama işlevi, global ve yerleşik kapsam arama şeklinde bir LEGB kuralı vardır.
def square():
""" return square of value """
def add():
""" add two local variable """
x = 2
y = 3
z = x + y
return z
return add()**2
print(square())
#%%Default Argumans = varsayılan argümanlar
# Default argument example:
# def f(a, b=1):
# """ b = 1 is default argument"""
# Flexible argument example:
# def f(*args):
# """ *args can be one or more"""
# def f(** kwargs)
# """ **kwargs is a dictionary"""
def f(a, b=1, c=2):
y =a+b+c
return y
print(f(5)) # a değeri yerine 5 yazılıyor
# what if we want to change default arguments # ya varsayılan argümanları değiştirmek istersek
print(f(5,4,3))
# flexible arguments *args
def f(*args):
for i in args:
print(i)
f(1)
print("")
f(1,2,3,4)
# flexible arguments **kwargs that is dictionary
def f(**kwargs):
""" print key and value of dictionary"""
for key, value in kwargs.items():
print(key, " ", value)
f(country = 'spain', capital = 'madrid', population = 123456)
#%%
#User Defined Function(long way)
def square(x):
return x**2
print(square(5))
#User dDefined Function(short way)
square = lambda x: x**2 #where x is name of argument
print(square(4))
tot = lambda x,y,z : x+y+z #where x,y,z is name of argument
print(tot(1,2,3))
#%%ANONYMOUS FUNCTİON
#Like lambda function but it can take more than one arguments.
#map(func,seq) : applies a function to all the items in a list
number_list = [1,2,3]
y = map(lambda x : x**2, number_list) # map fonksiyonu bize listedeki elemanların tek tek karelerini hesaplamamızda yardımcı oluyor yani lambda fonksiyonunu her bir parametre için kullanmamızı sağlıyor
print(list(y))
#%%# iteration example
name = "ronaldo"
it = iter(name)
print(next(it)) # print next iteration
print(*it) # print remaining iteration
#%% zip example
list1 = [1,2,3,4]
list2 = [5,6,7,8]
z = zip(list1, list2)
print(z)
z_list = list(z)
print(z_list)
un_zip = zip(*z_list)
un_list1,un_list2 =list(un_zip) #un_list return tuple
print(un_list1)
print(un_list2)
print(type(un_list2))
#%%
# LIST COMPREHENSİON
# One of the most important topic of this kernel
# We use list comprehension for data analysis often.
# list comprehension: collapse for loops for building lists into a single line
# Ex: num1 = [1,2,3] and we want to make it num2 = [2,3,4]. This can be done with for loop. However it is unnecessarily long. We can make it one line code that is list comprehension.
num1 = [1,2,3]
num2 = [i+1 for i in num1]
print(num2)
# [i + 1 for i in num1 ]: list of comprehension
# i +1: list comprehension syntax
# for i in num1: for loop syntax
# i: iterator
# num1: iterable object
# [num1'deki i için i + 1): kavrama listesi
# i +1: liste anlama sözdizimi
# num1'deki i için: döngü sözdizimi için
# i: yineleyici
# num1: yinelenebilir nesne
num1 = [5,10,15]
num2 =[i**2 if i==10 else i-5 if i < 7 else i +5 for i in num1]
print(num2)
#%%
import pandas as pd
data = pd.read_csv('pokemon.csv')
data.head()
data.tail()
data.columns # columns gives column names of features # başlık isimleri
data.shape # shape gives number of rows and columns in a tuble # satır ve sütun sayısı
# info gives data type like dataframe, number of sample or row, number of feature or column, feature types and memory usage
data.info()
# For example lets look frequency of pokemom types # Örneğin pokemon türlerinin sıklığına bakalım
print(data["Type 1"].value_counts(dropna = False))
#%%
# EXPLORATORY DATA ANALYSIS
# value_counts(): Frequency counts
# outliers: the value that is considerably higher or lower from rest of the data
# Lets say value at 75% is Q3 and value at 25% is Q1.
# Outlier are smaller than Q1 - 1.5(Q3-Q1) and bigger than Q3 + 1.5(Q3-Q1). (Q3-Q1) = IQR
# We will use describe() method. Describe method includes:
# count: number of entries
# mean: average of entries
# std: standart deviation
# min: minimum entry
# 25%: first quantile
# 50%: median or second quantile
# 75%: third quantile
# max: maximum entry
# KEŞİF VERİ ANALİZİ
# value_counts(): Frekans sayıları
# aykırı değerler: verilerin geri kalanından önemli ölçüde yüksek veya düşük olan değer
# Diyelim ki %75'teki değer Q3 ve %25'teki değer Q1.
# Outlier, Q1 - 1.5(Q3-Q1)'den küçük ve Q3 + 1.5(Q3-Q1)'den daha büyük. (Q3-Q1) = IQR
# tarif() yöntemini kullanacağız. Açıklama yöntemi şunları içerir:
# sayı: giriş sayısı
# ortalama: girişlerin ortalaması
# standart: standart sapma
# dak: minimum giriş
# %25: ilk nicelik
# %50: medyan veya ikinci nicelik
# %75: üçüncü nicelik
# max: maksimum giriş
# What is quantile?
# 1,4,5,6,8,9,11,12,13,14,15,16,17
# The median is the number that is in middle of the sequence. In this case it would be 11.
# The lower quartile is the median in between the smallest number and the median i.e. in between 1 and 11, which is 6.
# The upper quartile, you find the median between the median and the largest number i.e. between 11 and 17, which will be 14 according to the question above.
# quantile nedir?
# 1,4,5,6,8,9,11,12,13,14,15,16,17
# Medyan, dizinin ortasındaki sayıdır. Bu durumda 11 olur.
# Alt çeyrek, en küçük sayı ile medyan arasındaki medyandır, yani 1 ile 11 arasında, yani 6'dır.
# Üst çeyrek, medyanı medyan ile en büyük sayı arasında yani yukarıdaki soruya göre 14 olacak olan 11 ile 17 arasında buluyorsunuz.
#%%VISUAL EXPLORATORY DATA ANALYSIS
#Box plots: visualize basic statistics like outliers, min/max or quantiles
#Kutu grafikleri: aykırı değerler, min/maks veya nicelikler gibi temel istatistikleri görselleştirin
# For example: compare attack of pokemons that are legendary or not
# Black line at top is max
# Blue line at top is 75%
# Green line is median (50%)
# Blue line at bottom is 25%
# Black line at bottom is min
# There are no outliers
data.describe()
data.boxplot(column= "Attack", by="Legendary")
#%%TIDY DATA
#We tidy data with melt(). Describing melt is confusing. Therefore lets make example to understand it.
#DÜZENLİ VERİLER
#Verileri melt() ile düzenliyoruz. Erimeyi tanımlamak kafa karıştırıcıdır. Bu yüzden anlamak için örnek yapalım.
# Firstly I create new data from pokemons data to explain melt nore easily. = # İlk olarak melt nore'u kolayca açıklamak için pokemon verilerinden yeni veriler oluşturuyorum.
data_new = data.head()
data_new
# lets melt
# id_vars = what we do not wish to melt
# value_vars = what we want to melt
# eritelim
# id_vars = eritmek istemediğimiz şey
# value_vars = eritmek istediğimiz şey
melted = pd.melt(frame= data_new, id_vars="Name", value_vars=["Attack","Defense"])
melted
#%%PIVOTING DATA
#Reverse of melting.
# Index is name
# I want to make that columns are variable
# Finally values in columns are value
# İndeks isimdir
# Bu sütunları değişken yapmak istiyorum
# Son olarak sütunlardaki değerler değerdir
melted.pivot(index = 'Name', columns = 'variable',values='value')
#%%CONCATENATING DATA
#We can concatenate two dataframe =#İki veri çerçevesini birleştirebiliriz
# Firstly lets create 2 data frame # Öncelikle 2 data frame oluşturalım
data1 = data.head()
data2= data.tail()
conc_data_row =pd.concat([data1,data2],axis=0,ignore_index= True) #axis=0 verileri yatay eksende sıralar
conc_data_row
data1 =data["Attack"].head()
data2 =data["Defense"].head()
conc_data_col = pd.concat([data1,data2],axis=1) #verileri dikey eksende sıralar
conc_data_col
#%%
# DATA TYPES There are 5 basic data types: object(string),boolean, integer, float and categorical. We can make conversion data types like from str to categorical or from int to float Why is category important:
# make dataframe smaller in memory can be utilized for anlaysis especially for sklearn.
# VERİ TÜRLERİ 5 temel veri türü vardır: nesne(string), boolean, tamsayı, kayan nokta ve kategorik. str'den kategoriye veya int'den float'a gibi dönüşüm veri türlerini yapabiliriz. Kategori neden önemlidir:
# veri çerçevesini bellekte küçültme, analiz için özellikle sklearn için kullanılabilir.
# lets convert object(str) to categorical and int to float.
data['Type 1'] = data['Type 1'].astype('category')
data['Speed'] = data['Speed'].astype('float')
# As you can see Type 1 is converted from object to categorical = # Gördüğünüz gibi Type 1 nesneden kategoriye dönüştürülür
# And Speed ,s converted from int to float =# Ve Hız, int'den float'a dönüştürülür
data.dtypes
#%%MISSING DATA and TESTING WITH ASSERT
# If we encounter with missing data, what we can do:
# leave as is
# drop them with dropna()
# fill missing value with fillna()
# fill missing values with test statistics like mean
# Assert statement: check that you can turn on or turn off when you are done with your testing of the program
# EKSİK VERİLER VE ASSERT İLE TEST EDİLMESİ
# Eksik verilerle karşılaşırsak ne yapabiliriz:
# olduğu gibi bırak
# onları dropna() ile bırak
# eksik değeri fillna() ile doldurun
# ortalama gibi test istatistikleri ile eksik değerleri doldurun
# Assert deyimi: programı test etmeniz bittiğinde açıp kapatabileceğinizi kontrol edin
data.info()
# Lets chech Type 2
data["Type 2"].value_counts(dropna =False)
# As you can see, there are 386 NAN value (Gördüğünüz gibi 386 NAN değeri var)
# Lets drop nan values (nan değerlerini bırakalım)
data1=data # also we will use data to fill missing value so I assign it to data1 variable #ayrıca eksik değeri doldurmak için verileri kullanacağız, bu yüzden onu data1 değişkenine atadım
data1["Type 2"].dropna(inplace = True) # inplace = True means we do not assign it to new variable. Changes automatically assigned to data # inplace = True, onu yeni değişkene atamadığımız anlamına gelir. Verilere otomatik olarak atanan değişiklikler
# So does it work ?
# Lets check with assert statement
# Assert statement:
assert 1==1 # return nothing because it is true
# In order to run all code, we need to make this line comment # Kodun tamamını çalıştırabilmemiz için bu satırın yorumunu yapmamız gerekiyor.
# assert 1==2 # return error because it is false # assert 1==2 # false olduğu için hata döndür
assert data['Type 2'].notnull().all() # returns nothing because we drop nan values =# nan değerlerini düşürdüğümüz için hiçbir şey döndürmez
data["Type 2"].fillna('empty',inplace = True)
assert data['Type 2'].notnull().all() # returns nothing because we do not have nan values= #nan değerlerimiz olmadığı için hiçbir şey döndürmez
# # With assert statement we can check a lot of thing. For example
# assert data.columns[1] == 'Name'
# assert data.Speed.dtypes == np.int
## Assert deyimi ile bir çok şeyi kontrol edebiliriz. Örneğin
# assert data.columns[1] == 'Name'
# assert data.Speed.dtypes == np.int
#%%PANDAS FOUNDATION
# REVİEW of PANDAS
# As you notice, I do not give all idea in a same time. Although, we learn some basics of pandas, we will go deeper in pandas.
# single column = series
# NaN = not a number
# dataframe.values = numpy
# <NAME>
# Fark ettiğiniz gibi tüm fikirleri aynı anda vermiyorum. Pandasın bazı temellerini öğrenmiş olsak da , pandasda daha derine ineceğiz.
# tek sütun = seri
# NaN = değer yok
# dataframe.values = numpy
# data frames from dictionary
import pandas as pd
country = ["spain","France"]
population =["11","12"]
list_label = ["country","population"]
list_col = [country,population]
zipped= list(zip(list_label,list_col))
data_dict= dict(zipped)
df= pd.DataFrame(data_dict)
df
# listeden başlayarak önce dictionary oluşturduk daha sonra dataframe oluşturduk
# add new columns # yeni sütun ekleme
df["capital"]= ["madrid","paris"]
df
#Broadcasting entire column
df["income"] = 0
df
#%%# Plotting all data
data1 = data.loc[:,["Attack","Defense","Speed"]]
data1.plot()
# it is confusing
#%%
# subplots
data1.plot(subplots = True)
plt.show()
#%%
# scatter plot
data1.plot(kind = "scatter",x="Attack",y = "Defense")
plt.show()
#%%
# hist plot
data1.plot(kind = "hist",y = "Defense",bins = 50,range= (0,250),normed = True)
#%%
# histogram subplot with non cumulative and cumulative
fig, axes = plt.subplots(nrows=2,ncols=1)
data1.plot(kind = "hist",y = "Defense",bins = 50,range= (0,250),normed = True,ax = axes[0])
data1.plot(kind = "hist",y = "Defense",bins = 50,range= (0,250),normed = True,ax = axes[1],cumulative = True)
plt.savefig('graph.png')
plt
#%%
# INDEXING PANDAS TIME SERIES
# datetime = object
# parse_dates(boolean): Transform date to ISO 8601 (yyyy-mm-dd hh:mm:ss ) format
# PANDAS ZAMAN SERİSİ ENDEKSİ
# tarihsaat = nesne
# parse_dates(boolean): Tarihi ISO 8601 (yyyy-aa-gg ss:dd:ss ) biçimine dönüştürün
time_list = ["1992-03-08","1992-04-12"]
print(type(time_list[1])) # As you can see date is string # Gördüğünüz gibi tarih string
# however we want it to be datetime object ## ancak bunun datetime nesnesi olmasını istiyoruz
datetime_object = pd.to_datetime(time_list)
print(type(datetime_object))
# close warning
import warnings
warnings.filterwarnings("ignore")
# In order to practice lets take head of pokemon data and add it a time list ## Pratik yapmak için pokemon verilerinin başını alıp bir zaman listesi ekleyelim
data2 = data.head()
#%%
date_list = ["1992-01-10","1992-02-10","1992-03-10","1993-03-15","1993-03-16"]
datetime_object = pd.to_datetime(date_list)
data2["date"] = datetime_object
# lets make date as index = indeks olarak tarih yapalım
data2= data2.set_index("date")
data2
# Now we can select according to our date index # Artık tarih dizinimize göre seçim yapabiliriz
print(data2.loc["1993-03-16"])
print(data2.loc["1992-03-10":"1993-03-16"])
#RESAMPLING PANDAS TIME SERIES
# Resampling: statistical method over different time intervals
# Needs string to specify frequency like "M" = month or "A" = year
# Downsampling: reduce date time rows to slower frequency like from daily to weekly
# Upsampling: increase date time rows to faster frequency like from daily to hourly
# Interpolate: Interpolate values according to different methods like ‘linear’, ‘time’ or index’
# https://pandas.pydata.org/pandas-docs/stable/generated/pandas.Series.interpolate.html
#PANDAS ZAMAN SERİSİNİN YENİDEN ÖRNEKLENMESİ
# Yeniden örnekleme: farklı zaman aralıklarında istatistiksel yöntem
# "M" = ay veya "A" = yıl gibi bir sıklık belirtmek için dize gerekiyor
# Altörnekleme: tarih saat satırlarını günlükten haftalık gibi daha yavaş sıklığa düşürün
# Örnekleme: tarih saat satırlarını günlükten saatlik gibi daha hızlı sıklığa yükseltin
# Interpolate: Değerleri 'doğrusal', 'zaman' veya dizin' gibi farklı yöntemlere göre enterpolasyon yapın
# https://pandas.pydata.org/pandas-docs/stable/generated/pandas.Series.interpolate.html
# We will use data2 that we create at previous part ## Bir önceki bölümde oluşturduğumuz data2'yi kullanacağız
data2.resample("A").mean()
# We will use data2 that we create at previous part # bir önceki bölümde oluşturduğumuz data2 yi kullanacağız
data2.resample("M").mean()
# In real life (data is real. Not created from us like data2) we can solve this problem with interpolate = # Gerçek hayatta (veriler gerçektir. data2 gibi bizden yaratılmamışlardır) bu sorunu enterpolasyon ile çözebiliriz.
# We can interpolete from first value = # İlk değerden enterpolasyon yapabiliriz.
data2.resample("M").first().interpolate("linear")
# Or we can interpolate with mean()
data2.resample("M").mean().interpolate("linear")
#%%MANIPULATING DATA FRAMES WITH PANDAS
data = pd.read_csv('pokemon.csv')
data= data.set_index("#")
data.head()
data["HP"][1] #OR
data.HP[1]
# using loc accessor
data.loc[1,["HP"]]
# Selecting only some columns
data[["HP","Attack"]]
# Difference between selecting columns: series and dataframes = # Sütun seçme arasındaki fark: seriler ve veri çerçeveleri
print(type(data["HP"])) # series
print(type(data[["HP"]])) # data frames
# Slicing and indexing series
data.loc[1:10,"HP":"Defense"] # 10 and "Defense" are inclusive
# Reverse slicing
data.loc[10:1:-1,"HP":"Defense"]
# From something to end
data.loc[1:10,"Speed":]
#%%
# FILTERING DATA FRAMES = VERİ ÇERÇEVELERİNİN FİLTRELENMESİ
# Creating boolean series Combining filters Filtering column based others = # Boolean serisi oluşturma Filtreleri birleştirme Sütun bazında diğerlerini filtreleme
# Creating boolean series
boolean = data.HP > 200
data[boolean]
# Combining filters
first_filter = data.HP > 150
second_filter = data.Speed > 35
data[first_filter & second_filter]
# Filtering column based others
data.HP[data.Speed<15]
#%%DÖNÜŞÜM VERİLERİ
# Düz PYTHON işlevleri
# Lambda işlevi: her öğeye isteğe bağlı python işlevi uygulamak için
# Diğer sütunları kullanarak sütun tanımlama
# TRANSFORMING DATA
# Plain python functions
# Lambda function: to apply arbitrary python function to every element
# Defining column using other columns
data.HP.apply(lambda n : n/2)
# Defining column using other columns # farklı bir sütun oluşturmak istediğimizde
data["total_power"] = data.Attack + data.Defense
data.head()
#%%
# INDEX OBJECTS AND LABELED DATA İNDEKS NESNELERİ VE ETİKETLİ VERİLER
# index: sequence of label = # ındex: etiket dizisi
# our index name is this:
print(data.index.name)
# lets change it = değiştirelim
data.index.name = "index_name"
data.head()
# Overwrite index # ındex in üzerine yaz
# if we want to modify index we need to change all of them. # indeksi değiştirmek istiyorsak hepsini değiştirmemiz gerekiyor.
data.head()
# first copy of our data to data3 then change index # önce verilerimizin data3'e kopyalanması, ardından ındex i değiştirme
data3 = data.copy()
# lets make index start from 100. It is not remarkable change but it is just example # indeksi 100'den başlatalım. Dikkate değer bir değişiklik değil, sadece örnek
data3.index = range(100,900,1)
data3.head()
#%%HIERARCHICAL INDEXING Hiyerarşik indeksleme
# Setting indexing # İndekslemeyi ayarlama
# lets read data frame one more time to start from beginning =# baştan başlamak için veri çerçevesini bir kez daha okuyalım
data = pd.read_csv('pokemon.csv')
data.head()
# As you can see there is index. However we want to set one or more column to be index # Gördüğünüz gibi indeks var. Ancak bir veya daha fazla sütunu dizin olarak ayarlamak istiyoruz.
# Setting index : type 1 is outer type 2 is inner index # Ayar indeksi : tip 1 dış tip 2 iç indekstir
data1 = data.set_index(["Type 1","Type 2"])
data1.head(100)
# data1.loc["Fire","Flying"] # howw to use indexes # data1.loc["Fire","Flying"] # dizinlerin nasıl kullanılacağı
#%%
# PIVOTING DATA FRAMES
# pivoting: reshape tool
dic = {"treatment":["A","A","B","B"],"gender":["F","M","F","M"],"response":[10,45,5,9],"age":[15,4,72,65]}
df = | pd.DataFrame(dic) | pandas.DataFrame |
Subsets and Splits
No community queries yet
The top public SQL queries from the community will appear here once available.