path
stringlengths 7
265
| concatenated_notebook
stringlengths 46
17M
|
|---|---|
src/attempts/compare_FD_params_vadere.ipynb
|
###Markdown
FD PARAMETER COMPARISON (ON VADERE DATASET)
###Code
import os
os.chdir(os.path.pardir)
from utilities import *
data_path = "../data/vadere_corridor_100"
# plot vadere dataset
fd_data, fd_targets = read_dataset(data_path, fd_training=True)
plt.scatter(fd_data, fd_targets, s=1) # original data
plt.show()
###Output
_____no_output_____
###Markdown
FIT FD MODEL ON OBSERVATIONS
###Code
# to stop the computation when model is at its cap
callback = EarlyStopping(monitor='loss', patience=10) # default on val_loss
# train the FD model
model = FD_Network()
model.compile(optimizer='adam', loss='mse', run_eagerly=True)
model.fit(x=fd_data, y=fd_targets, epochs=50, verbose=2, callbacks=[callback])
# generate the FD speeds with prediction
mean_spacings = fd_data
fd_speeds = model.predict(x=mean_spacings)
model.mse = np.mean((fd_speeds-fd_targets)**2)
# plot the FD prediction over the observations
plt.scatter(fd_data, fd_targets, s=1) # original data
plt.scatter(mean_spacings, fd_speeds, c='orange', s=1) # fd model data
plt.xlabel("Mean spacing")
plt.ylabel("Speed")
plt.title("FD-Observations")
plt.show()
###Output
Epoch 1/50
31/31 - 1s - loss: 0.3075 - 560ms/epoch - 18ms/step
Epoch 2/50
31/31 - 0s - loss: 0.2127 - 428ms/epoch - 14ms/step
Epoch 3/50
31/31 - 0s - loss: 0.1599 - 425ms/epoch - 14ms/step
Epoch 4/50
31/31 - 0s - loss: 0.1362 - 426ms/epoch - 14ms/step
Epoch 5/50
31/31 - 0s - loss: 0.1284 - 449ms/epoch - 14ms/step
Epoch 6/50
31/31 - 0s - loss: 0.1279 - 464ms/epoch - 15ms/step
Epoch 7/50
31/31 - 0s - loss: 0.1278 - 461ms/epoch - 15ms/step
Epoch 8/50
31/31 - 1s - loss: 0.1278 - 600ms/epoch - 19ms/step
Epoch 9/50
31/31 - 1s - loss: 0.1273 - 608ms/epoch - 20ms/step
Epoch 10/50
31/31 - 1s - loss: 0.1274 - 633ms/epoch - 20ms/step
Epoch 11/50
31/31 - 1s - loss: 0.1274 - 594ms/epoch - 19ms/step
Epoch 12/50
31/31 - 1s - loss: 0.1275 - 539ms/epoch - 17ms/step
Epoch 13/50
31/31 - 0s - loss: 0.1273 - 491ms/epoch - 16ms/step
Epoch 14/50
31/31 - 0s - loss: 0.1273 - 478ms/epoch - 15ms/step
Epoch 15/50
31/31 - 0s - loss: 0.1273 - 458ms/epoch - 15ms/step
Epoch 16/50
31/31 - 0s - loss: 0.1272 - 470ms/epoch - 15ms/step
Epoch 17/50
31/31 - 1s - loss: 0.1273 - 543ms/epoch - 18ms/step
Epoch 18/50
31/31 - 1s - loss: 0.1273 - 531ms/epoch - 17ms/step
Epoch 19/50
31/31 - 1s - loss: 0.1272 - 538ms/epoch - 17ms/step
Epoch 20/50
31/31 - 1s - loss: 0.1277 - 583ms/epoch - 19ms/step
Epoch 21/50
31/31 - 1s - loss: 0.1272 - 534ms/epoch - 17ms/step
Epoch 22/50
31/31 - 1s - loss: 0.1271 - 526ms/epoch - 17ms/step
Epoch 23/50
31/31 - 1s - loss: 0.1271 - 604ms/epoch - 19ms/step
Epoch 24/50
31/31 - 1s - loss: 0.1275 - 606ms/epoch - 20ms/step
Epoch 25/50
31/31 - 1s - loss: 0.1273 - 607ms/epoch - 20ms/step
Epoch 26/50
31/31 - 1s - loss: 0.1271 - 640ms/epoch - 21ms/step
Epoch 27/50
31/31 - 1s - loss: 0.1276 - 606ms/epoch - 20ms/step
Epoch 28/50
31/31 - 1s - loss: 0.1271 - 688ms/epoch - 22ms/step
Epoch 29/50
31/31 - 1s - loss: 0.1273 - 732ms/epoch - 24ms/step
Epoch 30/50
31/31 - 1s - loss: 0.1273 - 708ms/epoch - 23ms/step
Epoch 31/50
31/31 - 1s - loss: 0.1271 - 702ms/epoch - 23ms/step
Epoch 32/50
31/31 - 1s - loss: 0.1272 - 687ms/epoch - 22ms/step
Epoch 33/50
31/31 - 1s - loss: 0.1271 - 771ms/epoch - 25ms/step
Epoch 34/50
31/31 - 1s - loss: 0.1269 - 735ms/epoch - 24ms/step
Epoch 35/50
31/31 - 1s - loss: 0.1269 - 807ms/epoch - 26ms/step
Epoch 36/50
31/31 - 1s - loss: 0.1269 - 804ms/epoch - 26ms/step
Epoch 37/50
31/31 - 1s - loss: 0.1269 - 746ms/epoch - 24ms/step
Epoch 38/50
31/31 - 1s - loss: 0.1268 - 805ms/epoch - 26ms/step
Epoch 39/50
31/31 - 1s - loss: 0.1269 - 796ms/epoch - 26ms/step
Epoch 40/50
31/31 - 1s - loss: 0.1273 - 808ms/epoch - 26ms/step
Epoch 41/50
31/31 - 1s - loss: 0.1271 - 797ms/epoch - 26ms/step
Epoch 42/50
31/31 - 1s - loss: 0.1269 - 824ms/epoch - 27ms/step
Epoch 43/50
31/31 - 1s - loss: 0.1269 - 842ms/epoch - 27ms/step
Epoch 44/50
31/31 - 1s - loss: 0.1270 - 940ms/epoch - 30ms/step
Epoch 45/50
31/31 - 1s - loss: 0.1270 - 988ms/epoch - 32ms/step
Epoch 46/50
31/31 - 1s - loss: 0.1268 - 843ms/epoch - 27ms/step
Epoch 47/50
31/31 - 1s - loss: 0.1267 - 1s/epoch - 35ms/step
Epoch 48/50
31/31 - 1s - loss: 0.1267 - 874ms/epoch - 28ms/step
Epoch 49/50
31/31 - 1s - loss: 0.1269 - 1s/epoch - 35ms/step
Epoch 50/50
31/31 - 1s - loss: 0.1269 - 1s/epoch - 33ms/step
###Markdown
VISUALIZE PARAMETERS FITTING EFFECT BATCH AFTER BATCH (FIT ON OBSERVATIONS)
###Code
fd1_params = model.FD_model_parameters
fd1_params['t'] = np.array([model.FD_model_parameters['t'][i] for i in range(len(model.FD_model_parameters['t']))]).flatten()
fd1_params['l'] = np.array([model.FD_model_parameters['l'][i] for i in range(len(model.FD_model_parameters['l']))]).flatten()
fd1_params['v0'] = np.array([model.FD_model_parameters['v0'][i] for i in range(len(model.FD_model_parameters['v0']))]).flatten()
plt.scatter(range(fd1_params['t'].shape[0]), fd1_params['t'], s=1, label='t')
plt.scatter(range(fd1_params['l'].shape[0]), fd1_params['l'], s=1, label='l')
plt.scatter(range(fd1_params['v0'].shape[0]), fd1_params['v0'], s=1, label='v0')
plt.legend()
plt.show()
###Output
_____no_output_____
###Markdown
CREATE PREDICTIONS AND FIT AN FD MODEL ON THEM
###Code
fd_data, fd_targets = read_dataset(data_path, fd_training=True)
nn_data, nn_targets = read_dataset(data_path, fd_training=False)
# to stop the computation when model is at its cap
callback = EarlyStopping(monitor='loss', patience=10) # default on val_loss
hidden_dims = (3,)
# train the speed predictor neural network
print("Training the NN model..")
nn = create_nn(hidden_dims, dropout=-1)
nn.compile(optimizer='adam', loss='mse')
hist = nn.fit(x=nn_data, y=nn_targets, epochs=1000, callbacks=[callback], verbose=1)
loss_nn = hist.history['loss']
# create the speed for FD to learn
nn_speeds = nn.predict(x=nn_data)
# train the FD model
print("Training the FD model..")
model2 = FD_Network()
model2.compile(optimizer='adam', loss='mse', run_eagerly=True)
hist = model2.fit(x=fd_data, y=nn_speeds, epochs=50, callbacks=[callback], verbose=1)
loss_fd = hist.history['loss']
# training plots
fig, ax = plt.subplots(nrows=1, ncols=2, figsize=(8, 5))
# FD
ax[0].plot(loss_fd)
ax[0].set_title("FD training")
ax[0].set_xlabel("Epochs")
ax[0].set_ylabel("MSE")
# NN
ax[1].plot(loss_nn, c='red')
ax[1].set_title("NN training")
ax[1].set_xlabel("Epochs")
ax[1].set_ylabel("MSE")
fig.show()
# plot
mean_spacings = fd_data
fd_speeds = model2.predict(x=mean_spacings)
model2.mse = np.mean((fd_speeds-fd_targets)**2)
fig, ax = plt.subplots(1, 1)
ax.scatter(mean_spacings, fd_speeds, c='orange', s=1)
ax.scatter(nn_data[:, 0], nn_speeds, s=1, c='red')
ax.set_xlabel("Mean spacing")
ax.set_ylabel("Speed")
fig.suptitle("FD-Predictions")
plt.show()
###Output
Training the NN model..
Epoch 1/1000
31/31 [==============================] - 0s 862us/step - loss: 1.9328
Epoch 2/1000
31/31 [==============================] - 0s 860us/step - loss: 1.6449
Epoch 3/1000
31/31 [==============================] - 0s 933us/step - loss: 1.3961
Epoch 4/1000
31/31 [==============================] - 0s 880us/step - loss: 1.1754
Epoch 5/1000
31/31 [==============================] - 0s 895us/step - loss: 0.9880
Epoch 6/1000
31/31 [==============================] - 0s 944us/step - loss: 0.8304
Epoch 7/1000
31/31 [==============================] - 0s 884us/step - loss: 0.6944
Epoch 8/1000
31/31 [==============================] - 0s 856us/step - loss: 0.5748
Epoch 9/1000
31/31 [==============================] - 0s 881us/step - loss: 0.4766
Epoch 10/1000
31/31 [==============================] - 0s 979us/step - loss: 0.3986
Epoch 11/1000
31/31 [==============================] - 0s 876us/step - loss: 0.3343
Epoch 12/1000
31/31 [==============================] - 0s 1ms/step - loss: 0.2827
Epoch 13/1000
31/31 [==============================] - 0s 863us/step - loss: 0.2424
Epoch 14/1000
31/31 [==============================] - 0s 829us/step - loss: 0.2111
Epoch 15/1000
31/31 [==============================] - 0s 803us/step - loss: 0.1878
Epoch 16/1000
31/31 [==============================] - 0s 1ms/step - loss: 0.1694
Epoch 17/1000
31/31 [==============================] - 0s 909us/step - loss: 0.1563
Epoch 18/1000
31/31 [==============================] - 0s 843us/step - loss: 0.1472
Epoch 19/1000
31/31 [==============================] - 0s 896us/step - loss: 0.1403
Epoch 20/1000
31/31 [==============================] - 0s 920us/step - loss: 0.1355
Epoch 21/1000
31/31 [==============================] - 0s 937us/step - loss: 0.1320
Epoch 22/1000
31/31 [==============================] - 0s 842us/step - loss: 0.1292
Epoch 23/1000
31/31 [==============================] - 0s 780us/step - loss: 0.1274
Epoch 24/1000
31/31 [==============================] - 0s 908us/step - loss: 0.1260
Epoch 25/1000
31/31 [==============================] - 0s 858us/step - loss: 0.1248
Epoch 26/1000
31/31 [==============================] - 0s 911us/step - loss: 0.1240
Epoch 27/1000
31/31 [==============================] - 0s 869us/step - loss: 0.1232
Epoch 28/1000
31/31 [==============================] - 0s 880us/step - loss: 0.1225
Epoch 29/1000
31/31 [==============================] - 0s 814us/step - loss: 0.1219
Epoch 30/1000
31/31 [==============================] - 0s 845us/step - loss: 0.1214
Epoch 31/1000
31/31 [==============================] - 0s 923us/step - loss: 0.1211
Epoch 32/1000
31/31 [==============================] - 0s 801us/step - loss: 0.1206
Epoch 33/1000
31/31 [==============================] - 0s 864us/step - loss: 0.1203
Epoch 34/1000
31/31 [==============================] - 0s 886us/step - loss: 0.1201
Epoch 35/1000
31/31 [==============================] - 0s 872us/step - loss: 0.1198
Epoch 36/1000
31/31 [==============================] - 0s 860us/step - loss: 0.1197
Epoch 37/1000
31/31 [==============================] - 0s 864us/step - loss: 0.1195
Epoch 38/1000
31/31 [==============================] - 0s 895us/step - loss: 0.1193
Epoch 39/1000
31/31 [==============================] - 0s 859us/step - loss: 0.1192
Epoch 40/1000
31/31 [==============================] - 0s 845us/step - loss: 0.1191
Epoch 41/1000
31/31 [==============================] - 0s 895us/step - loss: 0.1191
Epoch 42/1000
31/31 [==============================] - 0s 869us/step - loss: 0.1189
Epoch 43/1000
31/31 [==============================] - 0s 900us/step - loss: 0.1189
Epoch 44/1000
31/31 [==============================] - 0s 866us/step - loss: 0.1188
Epoch 45/1000
31/31 [==============================] - 0s 848us/step - loss: 0.1187
Epoch 46/1000
31/31 [==============================] - 0s 886us/step - loss: 0.1186
Epoch 47/1000
31/31 [==============================] - 0s 1ms/step - loss: 0.1185
Epoch 48/1000
31/31 [==============================] - 0s 872us/step - loss: 0.1184
Epoch 49/1000
31/31 [==============================] - 0s 923us/step - loss: 0.1184
Epoch 50/1000
31/31 [==============================] - 0s 925us/step - loss: 0.1184
Epoch 51/1000
31/31 [==============================] - 0s 862us/step - loss: 0.1184
Epoch 52/1000
31/31 [==============================] - 0s 893us/step - loss: 0.1182
Epoch 53/1000
31/31 [==============================] - 0s 895us/step - loss: 0.1181
Epoch 54/1000
31/31 [==============================] - 0s 869us/step - loss: 0.1182
Epoch 55/1000
31/31 [==============================] - 0s 828us/step - loss: 0.1180
Epoch 56/1000
31/31 [==============================] - 0s 838us/step - loss: 0.1180
Epoch 57/1000
31/31 [==============================] - 0s 807us/step - loss: 0.1180
Epoch 58/1000
31/31 [==============================] - 0s 760us/step - loss: 0.1178
Epoch 59/1000
31/31 [==============================] - 0s 843us/step - loss: 0.1177
Epoch 60/1000
31/31 [==============================] - 0s 880us/step - loss: 0.1178
Epoch 61/1000
31/31 [==============================] - 0s 824us/step - loss: 0.1179
Epoch 62/1000
31/31 [==============================] - 0s 806us/step - loss: 0.1178
Epoch 63/1000
31/31 [==============================] - 0s 774us/step - loss: 0.1181
Epoch 64/1000
31/31 [==============================] - 0s 817us/step - loss: 0.1177
Epoch 65/1000
31/31 [==============================] - 0s 815us/step - loss: 0.1178
Epoch 66/1000
31/31 [==============================] - 0s 786us/step - loss: 0.1175
Epoch 67/1000
31/31 [==============================] - 0s 859us/step - loss: 0.1175
Epoch 68/1000
31/31 [==============================] - 0s 810us/step - loss: 0.1175
Epoch 69/1000
31/31 [==============================] - 0s 797us/step - loss: 0.1176
Epoch 70/1000
31/31 [==============================] - 0s 798us/step - loss: 0.1175
Epoch 71/1000
31/31 [==============================] - 0s 808us/step - loss: 0.1174
Epoch 72/1000
31/31 [==============================] - 0s 838us/step - loss: 0.1175
Epoch 73/1000
31/31 [==============================] - 0s 857us/step - loss: 0.1175
Epoch 74/1000
31/31 [==============================] - 0s 828us/step - loss: 0.1174
Epoch 75/1000
31/31 [==============================] - 0s 823us/step - loss: 0.1174
Epoch 76/1000
31/31 [==============================] - 0s 830us/step - loss: 0.1173
Epoch 77/1000
31/31 [==============================] - 0s 816us/step - loss: 0.1172
Epoch 78/1000
31/31 [==============================] - 0s 885us/step - loss: 0.1173
Epoch 79/1000
31/31 [==============================] - 0s 837us/step - loss: 0.1174
Epoch 80/1000
31/31 [==============================] - 0s 817us/step - loss: 0.1173
Epoch 81/1000
31/31 [==============================] - 0s 826us/step - loss: 0.1173
Epoch 82/1000
31/31 [==============================] - 0s 858us/step - loss: 0.1174
Epoch 83/1000
31/31 [==============================] - 0s 815us/step - loss: 0.1172
Epoch 84/1000
31/31 [==============================] - 0s 818us/step - loss: 0.1171
Epoch 85/1000
31/31 [==============================] - 0s 879us/step - loss: 0.1170
Epoch 86/1000
31/31 [==============================] - 0s 787us/step - loss: 0.1173
Epoch 87/1000
31/31 [==============================] - 0s 802us/step - loss: 0.1171
Epoch 88/1000
31/31 [==============================] - 0s 811us/step - loss: 0.1170
Epoch 89/1000
31/31 [==============================] - 0s 903us/step - loss: 0.1170
Epoch 90/1000
31/31 [==============================] - 0s 896us/step - loss: 0.1171
Epoch 91/1000
31/31 [==============================] - 0s 928us/step - loss: 0.1170
Epoch 92/1000
31/31 [==============================] - 0s 831us/step - loss: 0.1170
Epoch 93/1000
31/31 [==============================] - 0s 911us/step - loss: 0.1170
Epoch 94/1000
31/31 [==============================] - 0s 803us/step - loss: 0.1173
Epoch 95/1000
31/31 [==============================] - 0s 783us/step - loss: 0.1170
Epoch 96/1000
31/31 [==============================] - 0s 821us/step - loss: 0.1170
Epoch 97/1000
31/31 [==============================] - 0s 874us/step - loss: 0.1169
Epoch 98/1000
31/31 [==============================] - 0s 855us/step - loss: 0.1170
Epoch 99/1000
31/31 [==============================] - 0s 821us/step - loss: 0.1169
Epoch 100/1000
31/31 [==============================] - 0s 847us/step - loss: 0.1168
Epoch 101/1000
31/31 [==============================] - 0s 787us/step - loss: 0.1171
Epoch 102/1000
31/31 [==============================] - 0s 792us/step - loss: 0.1170
Epoch 103/1000
31/31 [==============================] - 0s 805us/step - loss: 0.1167
Epoch 104/1000
31/31 [==============================] - 0s 805us/step - loss: 0.1168
Epoch 105/1000
31/31 [==============================] - 0s 801us/step - loss: 0.1166
Epoch 106/1000
31/31 [==============================] - 0s 806us/step - loss: 0.1165
Epoch 107/1000
31/31 [==============================] - 0s 888us/step - loss: 0.1165
Epoch 108/1000
31/31 [==============================] - 0s 761us/step - loss: 0.1167
Epoch 109/1000
31/31 [==============================] - 0s 797us/step - loss: 0.1166
Epoch 110/1000
31/31 [==============================] - 0s 752us/step - loss: 0.1166
Epoch 111/1000
31/31 [==============================] - 0s 922us/step - loss: 0.1166
Epoch 112/1000
31/31 [==============================] - 0s 838us/step - loss: 0.1167
Epoch 113/1000
31/31 [==============================] - 0s 827us/step - loss: 0.1166
Epoch 114/1000
31/31 [==============================] - 0s 808us/step - loss: 0.1166
Epoch 115/1000
31/31 [==============================] - 0s 843us/step - loss: 0.1166
Epoch 116/1000
31/31 [==============================] - 0s 782us/step - loss: 0.1166
Epoch 117/1000
31/31 [==============================] - 0s 832us/step - loss: 0.1165
Training the FD model..
Epoch 1/50
31/31 [==============================] - 0s 8ms/step - loss: 0.7911
Epoch 2/50
31/31 [==============================] - 0s 9ms/step - loss: 0.3421
Epoch 3/50
31/31 [==============================] - 0s 9ms/step - loss: 0.1268
Epoch 4/50
31/31 [==============================] - 0s 9ms/step - loss: 0.0624
Epoch 5/50
31/31 [==============================] - 0s 10ms/step - loss: 0.0478
Epoch 6/50
31/31 [==============================] - 0s 10ms/step - loss: 0.0384
Epoch 7/50
31/31 [==============================] - 0s 10ms/step - loss: 0.0315
Epoch 8/50
31/31 [==============================] - 0s 10ms/step - loss: 0.0275
Epoch 9/50
31/31 [==============================] - 0s 11ms/step - loss: 0.0248
Epoch 10/50
31/31 [==============================] - 0s 11ms/step - loss: 0.0225
Epoch 11/50
31/31 [==============================] - 0s 11ms/step - loss: 0.0204
Epoch 12/50
31/31 [==============================] - 0s 11ms/step - loss: 0.0185
Epoch 13/50
31/31 [==============================] - 0s 12ms/step - loss: 0.0171
Epoch 14/50
31/31 [==============================] - 0s 12ms/step - loss: 0.0159
Epoch 15/50
31/31 [==============================] - 0s 13ms/step - loss: 0.0148
Epoch 16/50
31/31 [==============================] - 0s 13ms/step - loss: 0.0140
Epoch 17/50
31/31 [==============================] - 0s 12ms/step - loss: 0.0133
Epoch 18/50
31/31 [==============================] - 0s 13ms/step - loss: 0.0128
Epoch 19/50
31/31 [==============================] - 0s 13ms/step - loss: 0.0123
Epoch 20/50
31/31 [==============================] - 0s 13ms/step - loss: 0.0120
Epoch 21/50
31/31 [==============================] - 0s 15ms/step - loss: 0.0117
Epoch 22/50
31/31 [==============================] - 0s 14ms/step - loss: 0.0115
Epoch 23/50
31/31 [==============================] - 0s 15ms/step - loss: 0.0114
Epoch 24/50
31/31 [==============================] - 0s 15ms/step - loss: 0.0112
Epoch 25/50
31/31 [==============================] - 0s 16ms/step - loss: 0.0111
Epoch 26/50
31/31 [==============================] - 0s 16ms/step - loss: 0.0110
Epoch 27/50
31/31 [==============================] - 0s 16ms/step - loss: 0.0110
Epoch 28/50
31/31 [==============================] - 0s 16ms/step - loss: 0.0108
Epoch 29/50
31/31 [==============================] - 0s 16ms/step - loss: 0.0108
Epoch 30/50
31/31 [==============================] - 1s 17ms/step - loss: 0.0107
Epoch 31/50
31/31 [==============================] - 1s 23ms/step - loss: 0.0107
Epoch 32/50
31/31 [==============================] - 1s 18ms/step - loss: 0.0107
Epoch 33/50
31/31 [==============================] - 1s 18ms/step - loss: 0.0106
Epoch 34/50
31/31 [==============================] - 1s 18ms/step - loss: 0.0106
Epoch 35/50
31/31 [==============================] - 1s 18ms/step - loss: 0.0106
Epoch 36/50
31/31 [==============================] - 1s 18ms/step - loss: 0.0106
Epoch 37/50
31/31 [==============================] - 1s 18ms/step - loss: 0.0106
Epoch 38/50
31/31 [==============================] - 1s 19ms/step - loss: 0.0105
Epoch 39/50
31/31 [==============================] - 1s 19ms/step - loss: 0.0105
Epoch 40/50
31/31 [==============================] - 1s 19ms/step - loss: 0.0106
Epoch 41/50
31/31 [==============================] - 1s 20ms/step - loss: 0.0105
Epoch 42/50
31/31 [==============================] - 1s 21ms/step - loss: 0.0105
Epoch 43/50
31/31 [==============================] - 1s 20ms/step - loss: 0.0105
Epoch 44/50
31/31 [==============================] - 1s 20ms/step - loss: 0.0105
Epoch 45/50
31/31 [==============================] - 1s 21ms/step - loss: 0.0105
Epoch 46/50
31/31 [==============================] - 1s 21ms/step - loss: 0.0104
Epoch 47/50
31/31 [==============================] - 1s 22ms/step - loss: 0.0105
Epoch 48/50
31/31 [==============================] - 1s 21ms/step - loss: 0.0105
Epoch 49/50
31/31 [==============================] - 1s 22ms/step - loss: 0.0104
Epoch 50/50
31/31 [==============================] - 1s 22ms/step - loss: 0.0104
###Markdown
VISUALIZE PARAMETERS FITTING EFFECT BATCH AFTER BATCH (FIT ON PREDICTIONS)
###Code
fd2_params = model2.FD_model_parameters
fd2_params['t'] = np.array([model2.FD_model_parameters['t'][i] for i in range(len(model2.FD_model_parameters['t']))]).flatten()
fd2_params['l'] = np.array([model2.FD_model_parameters['l'][i] for i in range(len(model2.FD_model_parameters['l']))]).flatten()
fd2_params['v0'] = np.array([model2.FD_model_parameters['v0'][i] for i in range(len(model2.FD_model_parameters['v0']))]).flatten()
plt.scatter(range(fd2_params['t'].shape[0]), fd2_params['t'], s=1, label='t')
plt.scatter(range(fd2_params['l'].shape[0]), fd2_params['l'], s=1, label='l')
plt.scatter(range(fd2_params['v0'].shape[0]), fd2_params['v0'], s=1, label='v0')
plt.legend()
plt.show()
###Output
_____no_output_____
###Markdown
COMPARE RESULTS
###Code
fd1_t = round(np.mean(fd1_params['t'][1000:]),3)
fd1_l = round(np.mean(fd1_params['l'][1000:]),3)
fd1_v0 = round(np.mean(fd1_params['v0'][1000:]),3)
fd2_t = round(np.mean(fd2_params['t'][1000:]),3)
fd2_l = round(np.mean(fd2_params['l'][1000:]),3)
fd2_v0 = round(np.mean(fd2_params['v0'][1000:]),3)
fd1_t, fd2_t
fd1_l, fd2_l
fd1_v0, fd2_v0
###Output
_____no_output_____
|
Taylor Community Consulting Program - Boy Scouts of America/TCCP_data processing_data analysis_data visualization.ipynb
|
###Markdown
Data Processing
###Code
import pandas as pd
import numpy as np
from pandasql import sqldf
from sklearn import preprocessing
from sklearn.cluster import KMeans
%matplotlib inline
import matplotlib.pyplot as plt
import seaborn
import warnings
warnings.filterwarnings('ignore')
def normalize(col, df):
x = df[col].values.astype(float)
min_max_scaler = preprocessing.MinMaxScaler()
x_scaled = min_max_scaler.fit_transform(x)
return pd.DataFrame(x_scaled, columns = col)
def sumandtotal(x, col, df):
df[x + '_sum'] = df.sum(axis = 1)
df[x + '_total'] = len(col)
return df
df = pd.read_excel('BSA_December 13, 2019_08.56.xlsx').iloc[1:, 5:].reset_index(drop = True)
df1 = pd.read_excel('BSA - MTurk_December 13, 2019_18.48.xlsx').iloc[1:,:].reset_index(drop = True)
df1 = df1.loc[df1['Finished'] == 'True',:]
#dictionary
num_p = {'Strongly disagree': 1, 'Disagree': 2, 'Somewhat disagree': 3, 'Neither agree nor disagree': 4,\
'Somewhat agree': 5, 'Agree': 6, 'Strongly agree': 7}
num_n = {'Strongly disagree': 7, 'Disagree': 6, 'Somewhat disagree': 5, 'Neither agree nor disagree': 4,\
'Somewhat agree': 3, 'Agree': 2, 'Strongly agree': 1}
c4 = {'Several times a week or more': 7, 'About once a week': 5.5, 'Several times a month': 4,\
'About once a month': 2.5, 'Less than once a month': 1}
c5 = {'Never had occasion to do this during the past year': 1, 'Once or twice': 2.5, 'About five times': 4,\
'Six to 10 times': 5.5, 'More than 10 times': 7}
c7 = {'Yes': 0, 'No': 1}
c8 = {1: 7, 2: 6, 3: 5, 4: 4, 5: 3, 6: 2, 7: 1}
f5 = {'Three or more times per week': 7, 'Twice per week': 5.5, 'Once per week': 4, 'Less than once per week': 2.5,\
'Almost no exercise': 1}
ct5 = {'Yes': 1, 'No': 0}
ct10 = {'$1 - $20': 1, '$21 - $50': 1.86, '$51 - $100': 2.71, '$100 - $500': 3.57, '$501 - $1000': 4.43, \
'$1001 - $5000': 5.29, '$5001 - $10000': 6.14, '$10001 and up': 7, 'Prefer not to disclose': 4}
ct12 = {'0 - $9,999': 1, '$10,000 - $24,999': 1.67, '$25,000 - 49,999': 2.33, \
'$50,000 - $74,999': 3, '$75,000 - $99,999': 3.67, '$100,000 - $124,999': 4.33, \
'$125,000 - $149,999': 5, '$150,000 - $174,999': 5.67, '$175,000 - $199,999': 6.33, \
'$200,000 and up': 7, 'Prefer not to answer': 4}
ct13 = {'750 and above': 7, '700 - 749': 5.5, '650 - 699': 4, '550 - 649': 2.5, '549 and below': 1, 'I don’t know': 4}
ct14 = {"I didn't go to college": 4, "I went to college, but didn't take out student loans": 4, \
"I'm still paying off my student loans": 4, 'Within 10 years': 4, 'Within 5 years': 7, 'More than 10 years': 1}
ct16 = {'Yes': 1, 'No': 0}
bs3 = {'Yes': 1, 'No': 0}
normalize = {1:0, 1.67:0.11, 1.86:0.14, 2:0.17, \
2.33:0.22, 2.5:0.25, 2.71:0.29, \
3:0.33, 3.57:0.43, 3.67:0.44, \
4:0.50, 4.33: 0.56, 4.43:0.57, \
5:0.67, 5.29: 0.71, 5.5:0.75, 5.67:0.78, \
6:0.83, 6.14:0.86, 6.33:0.89, \
7:1}
#df
#demo
df['bs1'] = df['bs1'].fillna(0)
df['bs2'] = df['bs2'].fillna(0).apply(lambda x: len(str(x).split(',')) if x != 0 else 0)
df['bs3'] = df['bs3'].fillna('No').map(bs3)
df['bs4_1'] = df['bs4_1'].fillna(0).apply(lambda x: str(x).split()[0])
df_d = pd.concat([df['bs1'], df['bs2'], df['bs3'], df['bs4_1']], axis = 1)
#leadership
l = []
for i in df.columns:
if 'l' in i:
l.append(i)
l = l[3:]
df_l = df[l]
l_p = ['l1_1', 'l2_1', 'l2_4', 'l3_2', 'l3_3', 'l3_4', 'l4_1', 'l4_2', 'l4_3', 'l4_4']
l_n = [i for i in l if i not in l_p]
for i in l_p:
df_l[i] = df_l[i].map(num_p)
for i in l_n:
df_l[i] = df_l[i].map(num_n)
#character
c = df.columns[71:97]
c_split = ['c1_1', 'c8_1', 'c8_2', 'c8_3', 'c8_4', 'c8_5', 'c8_6', 'c9_1', 'c10_1']
c_n = ['c2_3']
c_o = ['c4', 'c5', 'c7']
c_p = [i for i in c if (i not in c_n) & (i not in c_split) & (i not in c_o)]
df_c = df[c]
for i in c_split:
df_c[i] = df_c[i].apply(lambda x: int(x.split()[0]))
for i in c_split[1:7]:
df_c[i] = df_c[i].map(c8)
for i in c_p:
df_c[i] = df_c[i].map(num_p)
df_c['c2_3'] = df_c['c2_3'].map(num_n)
df_c['c4'] = df_c['c4'].apply(lambda x: x.strip()).map(c4)
df_c['c5'] = df_c['c5'].apply(lambda x: x.strip()).map(c5)
df_c['c7'] = df_c['c7'].map(c7)
#fitness
f = df.columns[97:108]
f_split = ['f4_1', 'f1_1']
f_n = ['f2_1', 'f2_2']
f_o = ['f5']
f_p = [i for i in f if (i not in f_n) & (i not in f_split) & (i not in f_o)]
df_f = df[f]
for i in f_split:
df_f[i] = df_f[i].apply(lambda x: x.split()[0])
for i in f_p:
df_f[i] = df_f[i].map(num_p)
for i in f_n:
df_f[i] = df_f[i].map(num_n)
df_f['f5'] = df_f['f5'].apply(lambda x: x.strip()).map(f5)
#citizenship
ct = df.columns[27:53]
ct_o = ['ct12', 'ct13', 'ct14', 'ct16', 'ct4', 'ct5', 'ct6_1', 'ct10']
ct_split = ['ct15_1', 'ct9_1']
ct_p = [i for i in ct if (i not in ct_o) & (i not in ct_split)]
df_ct = df[ct]
for i in ct_split:
df_ct[i] = df_ct[i].apply(lambda x: x.split()[0])
for i in ct_p:
df_ct[i] = df_ct[i].map(num_p)
df_ct['ct12'] = df_ct['ct12'].apply(lambda x: x.strip()).map(ct12)
df_ct['ct13'] = df_ct['ct13'].apply(lambda x: x.strip()).map(ct13)
df_ct['ct14'] = df_ct['ct14'].apply(lambda x: x.strip()).map(ct14)
df_ct['ct16'] = df_ct['ct16'].map(ct16)
df_ct['ct10'] = df_ct['ct10'].apply(lambda x: x.strip()).map(ct10)
df_ct['ct5'] = df_ct['ct5'].map(ct5)
df_ct['ct6_1'] = df_ct['ct6_1'].apply(lambda x: 1 if int(x) == 0 else x)
df_4 = pd.concat([df_ct, df_c, df_f, df_l, df_d, df['bs']], axis = 1)
#df1
#demo
df1['bs1'] = df1['bs1'].fillna(0)
df1['bs2'] = df1['bs2'].fillna(0).apply(lambda x: len(str(x).split(',')) if x != 0 else 0)
df1['bs3'] = df1['bs3'].fillna('No').map(bs3)
df1['bs4_1'] = df1['bs4_1'].fillna(0).apply(lambda x: str(x).split()[0])
df1_d = pd.concat([df1['bs1'], df1['bs2'], df1['bs3'], df1['bs4_1']], axis = 1)
#leadership
df1_l = df1[l]
for i in l_p:
df1_l[i] = df1_l[i].map(num_p)
for i in l_n:
df1_l[i] = df1_l[i].map(num_n)
#character
df1_c = df1[c]
for i in c_split:
df1_c[i] = df1_c[i].apply(lambda x: x.split()[0])
for i in c_p:
df1_c[i] = df1_c[i].map(num_p)
df1_c['c2_3'] = df1_c['c2_3'].map(num_n)
df1_c['c4'] = df1_c['c4'].apply(lambda x: x.strip()).map(c4)
df1_c['c5'] = df1_c['c5'].apply(lambda x: x.strip()).map(c5)
df1_c['c7'] = df1_c['c7'].map(c7)
df1_c['c_total'] = df1_c.sum(axis = 1)
df_c['ct_sum'] = df_c.sum(axis = 1)
#fitness
df1_f = df1[f]
for i in f_split:
df1_f[i] = df1_f[i].apply(lambda x: x.split()[0])
for i in f_p:
df1_f[i] = df1_f[i].map(num_p)
for i in f_n:
df1_f[i] = df1_f[i].map(num_n)
df1_f['f5'] = df1_f['f5'].apply(lambda x: x.strip()).map(f5)
#citizenship
df1_ct = df1[ct]
for i in ct_split:
df1_ct[i] = df1_ct[i].apply(lambda x: x.split()[0])
for i in ct_p:
df1_ct[i] = df1_ct[i].map(num_p)
df1_ct['ct12'] = df1_ct['ct12'].apply(lambda x: x.strip()).map(ct12)
df1_ct['ct13'] = df1_ct['ct13'].apply(lambda x: x.strip()).map(ct13)
df1_ct['ct14'] = df1_ct['ct14'].apply(lambda x: x.strip()).map(ct14)
df1_ct['ct16'] = df1_ct['ct16'].map(ct16)
df1_ct['ct10'] = df1_ct['ct10'].apply(lambda x: x.strip()).map(ct10)
df1_ct['ct5'] = df1_ct['ct5'].map(ct5)
df1_ct['ct6_1'] = df1_ct['ct6_1'].apply(lambda x: 1 if int(x) == 0 else x)
df1_4 = pd.concat([df1_ct, df1_c, df1_f, df1_l, df1_d, df1['bs']], axis = 1)
df4 = pd.concat([df_4, df1_4], axis = 0).reset_index(drop = True)
for i in df4.columns[5:]:
df4[i] = df4[i].apply(lambda x: float(x)).map(normalize)
df4['l_total'] = len(l)
df4['c_total'] = len(c)
df4['ct_total'] = len(ct)
df4['f_total'] = len(f)
df4['l_sum'] = df4[l].sum(axis=1)
df4['c_sum'] = df4[c].sum(axis=1)
df4['ct_sum'] = df4[ct].sum(axis=1)
df4['f_sum'] = df4[f].sum(axis=1)
df4['total'] = df4['l_sum'] + df4['c_sum'] + df4['ct_sum'] + df4['f_sum']
df4.to_csv('df4.csv',index=False)
###Output
_____no_output_____
###Markdown
Clustering
###Code
df4['l_sum'] = df4['l_sum'].apply(lambda x: x*25/18)
df4['f_sum'] = df4['f_sum'].apply(lambda x: x*25/11)
df4['c_sum'] = df4['c_sum'].apply(lambda x: x*25/26)
df4['ct_sum'] = df4['ct_sum'].apply(lambda x: x*25/26)
df4['total'] = df4['l_sum'] + df4['f_sum'] + df4['c_sum'] + df4['ct_sum']
df4.head()
df0 = df4[['l_sum','c_sum','ct_sum','f_sum','total','bs1','bs2','bs3','bs4_1','bs']]
avg = df0.loc[df0['bs']=='1',['l_sum','c_sum','ct_sum','f_sum','total']].mean()
avg
a = df0.groupby('bs')['l_sum','c_sum','ct_sum','f_sum','total'].mean().T
a['%'] = (a['1']-a['0'])/a['0']
a
l = df0.loc[df0['bs']=='1',['l_sum','bs1']]
c = df0.loc[df0['bs']=='1',['c_sum','bs1']]
ct = df0.loc[df0['bs']=='1',['ct_sum','bs1']]
f = df0.loc[df0['bs']=='1',['f_sum','bs1']]
t = df0.loc[df0['bs']=='1',['total','bs1']]
plt.style.use('ggplot')
fig, ax = plt.subplots()
plt.scatter(l['bs1'], l['l_sum'], alpha=0.5)
plt.axhline(y=avg[0], color='g', linestyle='-')
plt.ylim(5, 25)
plt.xlim(-1, 30)
labels = ["Average of Non-Boy Scouts"]
ax.set_xlabel('# of years in Boy Scouts')
ax.set_ylabel('Leadership Scores')
plt.legend(labels=labels)
fig, ax = plt.subplots()
plt.scatter(ct['bs1'], ct['ct_sum'], alpha=0.5)
plt.axhline(y=avg[2], color='g', linestyle='-')
plt.ylim(5, 25)
plt.xlim(-1, 30)
labels = ["Average of Non-Boy Scouts"]
ax.set_xlabel('# of years in Boy Scouts')
ax.set_ylabel('Citizenship Scores')
plt.legend(labels=labels)
fig, ax = plt.subplots()
plt.scatter(c['bs1'], c['c_sum'], alpha=0.5)
plt.axhline(y=avg[1], color='g', linestyle='-')
plt.ylim(5, 25)
plt.xlim(-1, 30)
labels = ["Average of Non-Boy Scouts"]
ax.set_xlabel('# of years in Boy Scouts')
ax.set_ylabel('Character Decelopment Scores')
plt.legend(labels=labels)
fig, ax = plt.subplots()
plt.scatter(f['bs1'], f['f_sum'], alpha=0.5)
plt.axhline(y=avg[3], color='g', linestyle='-')
plt.ylim(5, 25)
plt.xlim(-1, 30)
labels = ["Average of Non-Boy Scouts"]
ax.set_xlabel('# of years in Boy Scouts')
ax.set_ylabel('Fitness Scores')
plt.legend(labels=labels)
fig, ax = plt.subplots()
plt.ylim(30, 100)
plt.xlim(-1, 30)
plt.scatter(t['bs1'], t['total'], alpha=0.5)
plt.axhline(y=avg[4], color='g', linestyle='-')
labels = ["Average of Non-Boy Scouts"]
ax.set_xlabel('# of years in Boy Scouts')
ax.set_ylabel('Total Scores')
plt.legend(labels=labels)
teamwork = ['l1_1','l1_2','l1_3','l1_4','l1_5','l1_6']
takeroles = ['l2_1','l2_2','l2_3','l2_4']
public = ['l3_1','l3_2','l3_3','l3_4']
goal = ['l4_1','l4_2','l4_3','l4_4']
selfconfidence = ['c2_1','c2_2','c2_3','l2_4']
###Output
_____no_output_____
###Markdown
Key Impact Analysis
###Code
df0 = pd.read_csv("BSA_data.csv")
df0.head()
# Scouts and Non-Scouts score comparison in four key impacts
df0_ct = sqldf("SELECT AVG(ct_avg), bs FROM df0 GROUP BY bs")
df0_l = sqldf("SELECT AVG(l_avg), bs FROM df0 GROUP BY bs")
df0_c = sqldf("SELECT AVG(c_avg), bs FROM df0 GROUP BY bs")
df0_f = sqldf("SELECT AVG(f_avg), bs FROM df0 GROUP BY bs")
df0_total = sqldf("SELECT AVG(total), bs FROM df0 GROUP BY bs")
print(df0_ct)
print(df0_l)
print(df0_c)
print(df0_f)
print(df0_total)
# Non-Scouts and Cub/Boy/Venturing Scouts score comparison in four key impacts
df0_ct_p = sqldf("SELECT AVG(ct_avg), bs2_1 FROM df0 GROUP BY bs2_1")
df0_l_p = sqldf("SELECT AVG(l_avg), bs2_1 FROM df0 GROUP BY bs2_1")
df0_c_p = sqldf("SELECT AVG(c_avg), bs2_1 FROM df0 GROUP BY bs2_1")
df0_f_p = sqldf("SELECT AVG(f_avg), bs2_1 FROM df0 GROUP BY bs2_1")
df0_total_p = sqldf("SELECT AVG(total), bs2_1 FROM df0 GROUP BY bs2_1")
print(df0_ct_p)
print(df0_l_p)
print(df0_c_p)
print(df0_f_p)
print(df0_total_p)
# # of yrs membership vs. four impact scores
df0_ct_y = sqldf("SELECT AVG(ct_avg) AS avg_ct, bs1 FROM df0 GROUP BY bs1")
df0_l_y = sqldf("SELECT AVG(l_avg) AS avg_l, bs1 FROM df0 GROUP BY bs1")
df0_c_y = sqldf("SELECT AVG(c_avg) AS avg_c, bs1 FROM df0 GROUP BY bs1")
df0_f_y = sqldf("SELECT AVG(f_avg) AS avg_f, bs1 FROM df0 GROUP BY bs1")
print(df0_ct_y)
print(df0_l_y)
print(df0_c_y)
print(df0_f_y)
plt.style.use("ggplot")
fig, ((ax0, ax1), (ax2, ax3)) = plt.subplots(nrows = 2, ncols = 2, figsize = (24,12), sharey = False)
fig.suptitle("Years of Membership and Key Impact Scores")
df0_ct_y.plot(kind = "barh", x = "bs1", y = "avg_ct", color = "r", ax = ax0)
ax0.set(title = "Years of Membership and Citizenship Scores", xlabel = "Years of membership", ylabel = "Citizenship Score")
mean_df0_ct_y = df0_ct_y.avg_ct.mean()
ax0.axvline(mean_df0_ct_y, color = "b", linestyle = "--")
text0 = "mean score for Citizenship = %0.2f"%mean_df0_ct_y
ax0.annotate(text0, xy = (mean_df0_ct_y+0.02,9))
df0_l_y.plot(kind = "barh", x = "bs1", y = "avg_l", color = "r", ax = ax1)
ax1.set(title = "Years of Membership and Leadership Scores", xlabel = "Years of membership", ylabel = "Leadership Score")
mean_df0_l_y = df0_l_y.avg_l.mean()
ax1.axvline(mean_df0_l_y, color = "b", linestyle = "--")
text1 = "mean score for Leadership = %0.2f"%mean_df0_l_y
ax1.annotate(text1, xy = (mean_df0_l_y+0.02,9))
df0_c_y.plot(kind = "barh", x = "bs1", y = "avg_c", color = "r", ax = ax2)
ax2.set(title = "Years of Membership and Character Development Scores", xlabel = "Years of membership", ylabel = "Character Development Score")
mean_df0_c_y = df0_c_y.avg_c.mean()
ax2.axvline(mean_df0_c_y, color = "b", linestyle = "--")
text2 = "mean score for Character Development = %0.2f"%mean_df0_c_y
ax2.annotate(text2, xy = (mean_df0_c_y+0.02,9))
df0_f_y.plot(kind = "barh", x = "bs1", y = "avg_f", color = "r", ax = ax3)
ax3.set(title = "Years of Membership and Fitness/Wellbeing Scores", xlabel = "Years of membership", ylabel = "Fitness/Wellbeing Score")
mean_df0_f_y = df0_f_y.avg_f.mean()
ax3.axvline(mean_df0_f_y, color = "b", linestyle = "--")
text3 = "mean score for Fitness/Wellbeing = %0.2f"%mean_df0_f_y
ax3.annotate(text3, xy = (mean_df0_f_y+0.02,9))
###Output
_____no_output_____
|
Seeing-the-World-Model-Training-Workflow.ipynb
|
###Markdown
Seeing the World: Model Training Specify train and validate input folders
###Code
#train input folder
train_input_folder = '/data/data4/farmer_market'
#validation input folder
validate_input_folder = '/data/data4/validate/farmer_market'
from imutils import paths
import os
import shutil
import random
def split_data(directory, validate_directory='validation', split=0.8):
directories = [os.path.join(directory, o) for o in os.listdir(directory)
if os.path.isdir(os.path.join(directory,o))]
for directory in directories:
image_paths = list(paths.list_images(directory))
random.seed(32)
random.shuffle(image_paths)
image_paths
# compute the training and testing split
i = int(len(image_paths) * split)
train_paths = image_paths[:i]
selected_for_validation_paths = image_paths[i:]
for path in selected_for_validation_paths:
category = os.path.basename(os.path.normpath(directory))
dest_path = os.path.join(validate_directory, category)
if not os.path.exists(dest_path):
os.makedirs(dest_path)
os.chmod(dest_path, 0o777)
try:
shutil.move(path, dest_path)
except OSError as e:
if e.errno == errno.EEXIST:
print('Image already exists.')
else:
raise
split_data(directory='/data/data4/farmer_market',
validate_directory= '/data/data4/validate/farmer_market')
###Output
_____no_output_____
###Markdown
Create train and validate data generators
###Code
from tensorflow.keras.preprocessing.image import ImageDataGenerator
#apply image augmentation
train_image_generator = ImageDataGenerator(
rescale=1./255,
shear_range=0.2,
zoom_range=0.2,
brightness_range=[0.5, 1.5],
horizontal_flip=True,
vertical_flip=True,
rotation_range=40,
width_shift_range=0.2,
height_shift_range=0.2)
validate_image_generator = ImageDataGenerator(rescale=1./255)
batch_size = 30
image_width = 224
image_height = 224
IMAGE_WIDTH_HEIGHT = (image_width, image_height)
class_mode = 'categorical'
#create train data generator flowing from train_input_folder
train_generator = train_image_generator.flow_from_directory(
train_input_folder,
target_size=IMAGE_WIDTH_HEIGHT,
batch_size=batch_size,
class_mode=class_mode)
#create validation data generator flowing from validate_input_folder
validation_generator = validate_image_generator.flow_from_directory(
validate_input_folder,
target_size=IMAGE_WIDTH_HEIGHT,
batch_size=batch_size,
class_mode=class_mode)
###Output
Found 1539 images belonging to 64 classes.
Found 414 images belonging to 64 classes.
###Markdown
Create Custom Model
###Code
from tensorflow.keras import layers
from tensorflow.keras import Model
from tensorflow.keras.optimizers import Adam
total_classes = 60
activation_function = 'softmax'
loss = 'categorical_crossentropy'
img_input = layers.Input(shape=(image_width, image_height, 3))
x = layers.Conv2D(32, 3, activation='relu')(img_input)
x = layers.MaxPooling2D(2)(x)
x = layers.Conv2D(64, 3, activation='relu')(x)
x = layers.MaxPooling2D(2)(x)
x = layers.Flatten()(x)
x = layers.Dense(512, activation='relu')(x)
x = layers.Dropout(0.5)(x)
output = layers.Dense(total_classes, activation=activation_function)(x)
model = Model(img_input, output)
model.compile(loss=loss,
optimizer=Adam(lr=0.001),
metrics=['accuracy'])
###Output
_____no_output_____
###Markdown
Train Custom Model
###Code
import os, datetime
import tensorflow as tf
epochs = 5
steps_per_epoch = train_generator.n // train_generator.batch_size
validation_steps = validation_generator.n // validation_generator.batch_size
logdir = os.path.join("tf_logs", datetime.datetime.now().strftime("%Y%m%d-%H%M%S"))
tensorboard_callback = tf.keras.callbacks.TensorBoard(logdir, histogram_freq=1)
history = model.fit_generator(
train_generator,
steps_per_epoch=steps_per_epoch,
validation_data=validation_generator,
validation_steps=validation_steps,
callbacks=[tensorboard_callback],
epochs=epochs)
%matplotlib inline
import matplotlib.pyplot as plt
import matplotlib.image as mpimg
acc = history.history['accuracy']
val_acc = history.history['val_accuracy']
loss = history.history['loss']
val_loss = history.history['val_loss']
plt.figure(figsize=(8, 8))
plt.subplot(2, 1, 1)
plt.plot(acc, label='Training Accuracy')
plt.plot(val_acc, label='Validation Accuracy')
plt.legend(loc='lower right')
plt.ylabel('Accuracy')
plt.ylim([min(plt.ylim()), 1])
plt.title('Training and Validation Accuracy')
plt.subplot(2, 1, 2)
plt.plot(loss, label='Training Loss')
plt.plot(val_loss, label='Validation Loss')
plt.legend(loc='upper right')
plt.ylabel('Cross Entropy')
plt.ylim([0, 1.0])
plt.title('Training and Validation Loss')
plt.xlabel('epoch')
###Output
_____no_output_____
###Markdown
Using Transfer Learning
###Code
import tensorflow as tf
from tensorflow.keras.applications.vgg16 import VGG16
from tensorflow.keras.applications.vgg19 import VGG19
image_width=224
image_height=224
IMAGE_SHAPE = (image_width, image_height, 3)
base_model = tf.keras.applications.VGG19(input_shape=IMAGE_SHAPE, include_top=False,weights='imagenet')
base_model.summary()
keras = tf.keras
IMAGE_WIDTH_HEIGHT = (image_width, image_height)
batch_size=30
class_mode="categorical"
total_classes = 64
activation_function = 'softmax'
loss = 'categorical_crossentropy'
train_image_generator = tf.keras.preprocessing.image.ImageDataGenerator(
preprocessing_function=keras.applications.vgg19.preprocess_input,
rescale=1.0/255.0,
shear_range=0.2,
zoom_range=[0.9, 1.25],
brightness_range=[0.5, 1.5],
horizontal_flip=True,
vertical_flip=True)
validation_image_generator = tf.keras.preprocessing.image.ImageDataGenerator(
preprocessing_function=keras.applications.vgg19.preprocess_input,
rescale=1.0/255.0)
train_generator = train_image_generator.flow_from_directory(
train_input_folder,
target_size=IMAGE_WIDTH_HEIGHT,
batch_size=batch_size,
class_mode=class_mode)
validation_generator = validation_image_generator.flow_from_directory(
validate_input_folder,
target_size=IMAGE_WIDTH_HEIGHT,
batch_size=batch_size,
class_mode=class_mode)
from tensorflow.keras import layers
from tensorflow.keras import Model
from tensorflow.keras.optimizers import Adam
import os
reload_checkpoint=True
total_classes=64
img_input = layers.Input(shape=(image_width, image_height, 3))
global_average_layer = layers.GlobalAveragePooling2D()
prediction_layer = layers.Dense(total_classes, activation='softmax')
model = tf.keras.Sequential([
base_model,
global_average_layer,
prediction_layer
])
checkpoint_path = "/data/train_model_fruit_veggie_9/chkpt"
checkpoint_dir = os.path.dirname(checkpoint_path)
if (reload_checkpoint and os.path.isdir(checkpoint_path)):
try:
model.load_weights(checkpoint_path)
print('loaded weights from checkpoint')
except Exception:
print('no checkpointed weights')
pass
if not os.path.isdir(checkpoint_path):
os.makedirs(checkpoint_path)
print("Number of layers in the base model: ", len(base_model.layers))
base_model.trainable = False
model.compile(loss=loss,
optimizer=Adam(lr=0.001),
metrics=['accuracy'])
model.summary()
import datetime, os
epochs = 20
steps_per_epoch = train_generator.n // train_generator.batch_size
validation_steps = validation_generator.n // validation_generator.batch_size
#steps_per_epoch = 50
#validation_steps = 50
logdir = os.path.join("/data/tf_logs_9", datetime.datetime.now().strftime("%Y%m%d-%H%M%S"))
tensorboard_callback = tf.keras.callbacks.TensorBoard(logdir, histogram_freq=1)
checkpoint_callback = tf.keras.callbacks.ModelCheckpoint(filepath=checkpoint_path,
save_weights_only=True, save_best_only=True,
verbose=1)
history = model.fit_generator(
train_generator,
steps_per_epoch=steps_per_epoch,
validation_data=validation_generator,
validation_steps=validation_steps,
callbacks=[checkpoint_callback, tensorboard_callback],
epochs=epochs)
%matplotlib inline
import matplotlib.pyplot as plt
import matplotlib.image as mpimg
acc = history.history['accuracy']
val_acc = history.history['val_accuracy']
loss = history.history['loss']
val_loss = history.history['val_loss']
plt.figure(figsize=(8, 8))
plt.subplot(2, 1, 1)
plt.plot(acc, label='Training Accuracy')
plt.plot(val_acc, label='Validation Accuracy')
plt.legend(loc='lower right')
plt.ylabel('Accuracy')
plt.ylim([min(plt.ylim()), 1])
plt.title('Training and Validation Accuracy')
plt.subplot(2, 1, 2)
plt.plot(loss, label='Training Loss')
plt.plot(val_loss, label='Validation Loss')
plt.legend(loc='upper right')
plt.ylabel('Cross Entropy')
plt.ylim([0, 3])
plt.title('Training and Validation Loss')
plt.xlabel('epoch')
###Output
_____no_output_____
###Markdown
Continue Training
###Code
import datetime, os
epochs = 20
steps_per_epoch = train_generator.n // train_generator.batch_size
validation_steps = validation_generator.n // validation_generator.batch_size
#steps_per_epoch = 50
#validation_steps = 50
logdir = os.path.join("/data/tf_logs_9", datetime.datetime.now().strftime("%Y%m%d-%H%M%S"))
tensorboard_callback = tf.keras.callbacks.TensorBoard(logdir, histogram_freq=1)
checkpoint_callback = tf.keras.callbacks.ModelCheckpoint(filepath=checkpoint_path,
save_weights_only=True, save_best_only=True,
verbose=1)
history = model.fit_generator(
train_generator,
steps_per_epoch=steps_per_epoch,
validation_data=validation_generator,
validation_steps=validation_steps,
callbacks=[checkpoint_callback, tensorboard_callback],
epochs=epochs)
%matplotlib inline
import matplotlib.pyplot as plt
import matplotlib.image as mpimg
acc = history.history['accuracy']
val_acc = history.history['val_accuracy']
loss = history.history['loss']
val_loss = history.history['val_loss']
plt.figure(figsize=(8, 8))
plt.subplot(2, 1, 1)
plt.plot(acc, label='Training Accuracy')
plt.plot(val_acc, label='Validation Accuracy')
plt.legend(loc='lower right')
plt.ylabel('Accuracy')
plt.ylim([min(plt.ylim()), 1])
plt.title('Training and Validation Accuracy')
plt.subplot(2, 1, 2)
plt.plot(loss, label='Training Loss')
plt.plot(val_loss, label='Validation Loss')
plt.legend(loc='upper right')
plt.ylabel('Cross Entropy')
plt.ylim([0, 3])
plt.title('Training and Validation Loss')
plt.xlabel('epoch')
###Output
_____no_output_____
###Markdown
Continue Training
###Code
import datetime, os
epochs = 20
steps_per_epoch = train_generator.n // train_generator.batch_size
validation_steps = validation_generator.n // validation_generator.batch_size
#steps_per_epoch = 50
#validation_steps = 50
logdir = os.path.join("/data/tf_logs_9", datetime.datetime.now().strftime("%Y%m%d-%H%M%S"))
tensorboard_callback = tf.keras.callbacks.TensorBoard(logdir, histogram_freq=1)
checkpoint_callback = tf.keras.callbacks.ModelCheckpoint(filepath=checkpoint_path,
save_weights_only=True, save_best_only=True,
verbose=1)
history = model.fit_generator(
train_generator,
steps_per_epoch=steps_per_epoch,
validation_data=validation_generator,
validation_steps=validation_steps,
callbacks=[checkpoint_callback, tensorboard_callback],
epochs=epochs)
%matplotlib inline
import matplotlib.pyplot as plt
import matplotlib.image as mpimg
acc = history.history['accuracy']
val_acc = history.history['val_accuracy']
loss = history.history['loss']
val_loss = history.history['val_loss']
plt.figure(figsize=(8, 8))
plt.subplot(2, 1, 1)
plt.plot(acc, label='Training Accuracy')
plt.plot(val_acc, label='Validation Accuracy')
plt.legend(loc='lower right')
plt.ylabel('Accuracy')
plt.ylim([min(plt.ylim()), 1])
plt.title('Training and Validation Accuracy')
plt.subplot(2, 1, 2)
plt.plot(loss, label='Training Loss')
plt.plot(val_loss, label='Validation Loss')
plt.legend(loc='upper right')
plt.ylabel('Cross Entropy')
plt.ylim([0, 3])
plt.title('Training and Validation Loss')
plt.xlabel('epoch')
###Output
_____no_output_____
###Markdown
Continue Training
###Code
import datetime, os
epochs = 20
steps_per_epoch = train_generator.n // train_generator.batch_size
validation_steps = validation_generator.n // validation_generator.batch_size
#steps_per_epoch = 50
#validation_steps = 50
logdir = os.path.join("/data/tf_logs_9", datetime.datetime.now().strftime("%Y%m%d-%H%M%S"))
tensorboard_callback = tf.keras.callbacks.TensorBoard(logdir, histogram_freq=1)
checkpoint_callback = tf.keras.callbacks.ModelCheckpoint(filepath=checkpoint_path,
save_weights_only=True, save_best_only=True,
verbose=1)
history = model.fit_generator(
train_generator,
steps_per_epoch=steps_per_epoch,
validation_data=validation_generator,
validation_steps=validation_steps,
callbacks=[checkpoint_callback, tensorboard_callback],
epochs=epochs)
%matplotlib inline
import matplotlib.pyplot as plt
import matplotlib.image as mpimg
acc = history.history['accuracy']
val_acc = history.history['val_accuracy']
loss = history.history['loss']
val_loss = history.history['val_loss']
plt.figure(figsize=(8, 8))
plt.subplot(2, 1, 1)
plt.plot(acc, label='Training Accuracy')
plt.plot(val_acc, label='Validation Accuracy')
plt.legend(loc='lower right')
plt.ylabel('Accuracy')
plt.ylim([min(plt.ylim()), 1])
plt.title('Training and Validation Accuracy')
plt.subplot(2, 1, 2)
plt.plot(loss, label='Training Loss')
plt.plot(val_loss, label='Validation Loss')
plt.legend(loc='upper right')
plt.ylabel('Cross Entropy')
plt.ylim([0, 3])
plt.title('Training and Validation Loss')
plt.xlabel('epoch')
###Output
_____no_output_____
###Markdown
Continue Training
###Code
import datetime, os
epochs = 20
steps_per_epoch = train_generator.n // train_generator.batch_size
validation_steps = validation_generator.n // validation_generator.batch_size
#steps_per_epoch = 50
#validation_steps = 50
logdir = os.path.join("/data/tf_logs_9", datetime.datetime.now().strftime("%Y%m%d-%H%M%S"))
tensorboard_callback = tf.keras.callbacks.TensorBoard(logdir, histogram_freq=1)
checkpoint_callback = tf.keras.callbacks.ModelCheckpoint(filepath=checkpoint_path,
save_weights_only=True, save_best_only=True,
verbose=1)
history = model.fit_generator(
train_generator,
steps_per_epoch=steps_per_epoch,
validation_data=validation_generator,
validation_steps=validation_steps,
callbacks=[checkpoint_callback, tensorboard_callback],
epochs=epochs)
%matplotlib inline
import matplotlib.pyplot as plt
import matplotlib.image as mpimg
acc = history.history['accuracy']
val_acc = history.history['val_accuracy']
loss = history.history['loss']
val_loss = history.history['val_loss']
plt.figure(figsize=(8, 8))
plt.subplot(2, 1, 1)
plt.plot(acc, label='Training Accuracy')
plt.plot(val_acc, label='Validation Accuracy')
plt.legend(loc='lower right')
plt.ylabel('Accuracy')
plt.ylim([min(plt.ylim()), 1])
plt.title('Training and Validation Accuracy')
plt.subplot(2, 1, 2)
plt.plot(loss, label='Training Loss')
plt.plot(val_loss, label='Validation Loss')
plt.legend(loc='upper right')
plt.ylabel('Cross Entropy')
plt.ylim([0, 3])
plt.title('Training and Validation Loss')
plt.xlabel('epoch')
###Output
_____no_output_____
###Markdown
Fine Tuning
###Code
import datetime, os
loss = 'categorical_crossentropy'
checkpoint_path = "/data/train_model_fruit_veggie_9/chkpt"
checkpoint_dir = os.path.dirname(checkpoint_path)
if (reload_checkpoint and os.path.isdir(checkpoint_path)):
try:
model.load_weights(checkpoint_path)
except Exception:
pass
if not os.path.isdir(checkpoint_path):
os.makedirs(checkpoint_path)
base_model.trainable = True
# Fine tune start from layer 10
fine_tune_at = 10
# Freeze all layers before the `fine_tune_at` layer
for layer in base_model.layers[:fine_tune_at]:
layer.trainable = False
model.compile(loss=loss,
optimizer=Adam(lr=0.001),
metrics=['accuracy'])
model.summary()
import datetime, os
epochs = 10
steps_per_epoch = train_generator.n // train_generator.batch_size
validation_steps = validation_generator.n // validation_generator.batch_size
#steps_per_epoch = 50
#validation_steps = 50
logdir = os.path.join("/data/tf_logs_9", datetime.datetime.now().strftime("%Y%m%d-%H%M%S"))
tensorboard_callback = tf.keras.callbacks.TensorBoard(logdir, histogram_freq=1)
checkpoint_callback = tf.keras.callbacks.ModelCheckpoint(filepath=checkpoint_path,
save_weights_only=True, save_best_only=True,
verbose=1)
history = model.fit_generator(
train_generator,
steps_per_epoch=steps_per_epoch,
validation_data=validation_generator,
validation_steps=validation_steps,
callbacks=[checkpoint_callback, tensorboard_callback],
epochs=epochs)
%matplotlib inline
import matplotlib.pyplot as plt
import matplotlib.image as mpimg
acc = history.history['accuracy']
val_acc = history.history['val_accuracy']
loss = history.history['loss']
val_loss = history.history['val_loss']
plt.figure(figsize=(8, 8))
plt.subplot(2, 1, 1)
plt.plot(acc, label='Training Accuracy')
plt.plot(val_acc, label='Validation Accuracy')
plt.legend(loc='lower right')
plt.ylabel('Accuracy')
plt.ylim([min(plt.ylim()), 1])
plt.title('Training and Validation Accuracy')
plt.subplot(2, 1, 2)
plt.plot(loss, label='Training Loss')
plt.plot(val_loss, label='Validation Loss')
plt.legend(loc='upper right')
plt.ylabel('Cross Entropy')
plt.ylim([0, 1.0])
plt.title('Training and Validation Loss')
plt.xlabel('epoch')
###Output
_____no_output_____
###Markdown
Save Model
###Code
def export(model, path):
model.save(path, save_format='tf')
model.save('/data/saved_model_2/')
###Output
WARNING:tensorflow:From /home/seeingtheworld/.conda/envs/tf2/lib/python3.7/site-packages/tensorflow_core/python/ops/resource_variable_ops.py:1781: calling BaseResourceVariable.__init__ (from tensorflow.python.ops.resource_variable_ops) with constraint is deprecated and will be removed in a future version.
Instructions for updating:
If using Keras pass *_constraint arguments to layers.
INFO:tensorflow:Assets written to: /data/saved_model_2/assets
###Markdown
Reload Model
###Code
import tensorflow as tf
model = tf.keras.models.load_model('/data/saved_model_2/')
###Output
_____no_output_____
|
cwl_networks.ipynb
|
###Markdown
###Code
import networkx as nx
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
from pathlib import Path
import scipy.sparse
###Output
_____no_output_____
###Markdown
Set up domain- Choose graph- Set DEM
###Code
# Create array
def choose_graph(graph_name):
if graph_name == 'line':
graph = np.array([[0,0,0,0,0],
[1,0,0,0,0],
[0,1,0,0,0],
[0,0,1,0,0],
[0,0,0,1,0]])
elif graph_name == 'ring':
graph = np.array([[0,0,0,0,1],
[1,0,0,0,0],
[0,1,0,0,0],
[0,0,1,0,0],
[0,0,0,1,0]])
elif graph_name == 'lollipop':
graph = np.array([[0,0,0,0,0],
[1,0,0,0,0],
[1,0,0,0,0],
[0,1,1,0,0],
[0,0,0,1,0]])
elif graph_name == 'grid':
graph = np.array([[0,0,0,0,0,0,0,0,0,0,0,0],
[1,0,0,0,0,0,0,0,0,0,0,0],
[0,1,0,0,0,0,0,0,0,0,0,0],
[0,0,1,0,0,0,0,0,0,0,0,0],
[1,0,0,0,0,0,0,0,0,0,0,0],
[0,1,0,0,1,0,0,0,0,0,0,0],
[0,0,1,0,0,1,0,0,0,0,0,0],
[0,0,0,1,0,0,1,0,0,0,0,0],
[0,0,0,0,1,0,0,0,0,0,0,0],
[0,0,0,0,0,1,0,0,1,0,0,0],
[0,0,0,0,0,0,1,0,0,1,0,0],
[0,0,0,0,0,0,0,1,0,0,1,0]])
return graph
"""
Choose graph
"""
cnm = choose_graph('line')
cnm_sim = cnm + cnm.T
#ini_values = np.array([10, 5, 5, 3, 2])
dem = np.arange(cnm.shape[0])[::-1]
CNM = scipy.sparse.csr_matrix(cnm) # In order to use the same sparse type as the big ass true adjacency matrix
n_edges = np.sum(CNM)
n_nodes = CNM.shape[0]
# Create NetworkX graph
g = nx.DiGraph(incoming_graph_data=CNM.T) # transposed for dynamics!
def initialize_graph_values(g, h_ini, dem_nodes, diri_bc_values, diri_bc_bool, neumann_bc_values, neumann_bc_bool, source):
nx.set_node_attributes(G=g, values={i: value for i, value in enumerate(h_ini)}, name='h_old')
nx.set_node_attributes(G=g, values={i: value for i, value in enumerate(h_ini)}, name='h_new')
nx.set_node_attributes(G=g, values={i: value for i, value in enumerate(dem_nodes)}, name='ele')
nx.set_node_attributes(G=g, values={i: value for i, value in enumerate(diri_bc_values)}, name='diri_bc')
nx.set_node_attributes(G=g, values={i: value for i, value in enumerate(diri_bc_bool)}, name='diri_bool')
nx.set_node_attributes(G=g, values={i: value for i, value in enumerate(neumann_bc_values)}, name='neumann_bc')
nx.set_node_attributes(G=g, values={i: value for i, value in enumerate(neumann_bc_bool)}, name='neumann_bool')
nx.set_node_attributes(G=g, values={i: value for i, value in enumerate(source)}, name='source')
return 0
g_un = nx.Graph(g) # symmetric matrix, undirected graph. Useful for dynamics
# Plot
one_dim = True
if one_dim:
pos = {node:pos for node, pos in enumerate(zip(np.arange(n_nodes), dem))}
else:
pos = {i:((i%4), -int(i/4)) for i in range(0,12)}
options = {
"font_size": 20,
"node_size": 1000,
"node_color": "white",
"edgecolors": "black",
"linewidths": 5,
"width": 5,
}
nx.draw_networkx(g, pos=pos, **options)
# Functions
def infer_extreme_nodes(adj_matrix):
"""
Infer what nodes are the beginning and end of canals from adjacency matrix.
Last nodes of canals are identified by having no outgoing edges
First nodes of canals have no incoming edges
Parameters
----------
adj_matrix : numpy array
Adjacency matrix of the canal network graph
Returns
-------
end_nodes_bool : boolean numpy array
True where nodes are last nodes of canals
first_nodes_bool : boolean numpy array
True where nodes are first nodes of canals
"""
# Infer neumann and Diri nodes from adj matrix
end_nodes_bool = np.sum(CNM, axis=0) == 0 # Boundary values below are conditional on this boolean mask
first_nodes_bool = np.sum(CNM, axis=1) == 0
# in case the summing over the sparse matrix changes the numpy array shape
end_nodes_bool = np.ravel(end_nodes_bool)
first_nodes_bool = np.ravel(first_nodes_bool)
return first_nodes_bool, end_nodes_bool
def compute_laplacian_from_adjacency(adj_matrix):
if np.any(adj_matrix != adj_matrix.T):
raise ValueError('the matrix must be symmetric, i.e., must be the adj matrix of an undirected graph')
degree_matrix = np.diag(np.sum(adj_matrix, axis=1))
laplacian = degree_matrix - adj_matrix
return laplacian
def L_advection(directed_adj_matrix):
"""
Returns the 'modified Laplacian', i.e., the advection operator
"""
D_out = np.diag(np.sum(directed_adj_matrix, axis=0))
return D_out - directed_adj_matrix
def upwind_from_advection_laplacian(L_adv, downstream_nodes_bool):
"""
Adds the BC node downstream so as to make it equal to the upwind scheme
when graph = 1D line.
Also, this arises just from attaching a ghost node to dowstream nodes.
"""
L_upwind = L_adv.copy()
L_upwind[downstream_nodes_bool, downstream_nodes_bool] += 1
return L_upwind
def advection_diffusion_operator(dx, L, L_adv, a, b, diri_bc_bool, neumann_bc_upstream, neumann_bc_downstream, neumann_bc_values):
# Set default BCs: Neumann
L_BC = L.copy() # L doesn't change
L_advBC = L_adv.copy()
# Beginning nodes get a -1 in the diagonal
L_advBC[neumann_bc_upstream, neumann_bc_upstream] = L[neumann_bc_upstream, neumann_bc_upstream] - 1
# Ending nodes get a +1 in the diagonal
L_advBC[neumann_bc_downstream, neumann_bc_downstream] = L[neumann_bc_downstream, neumann_bc_downstream] + 1
# Construct operator
L_mix = a/dx**2*(-L_BC) - b/dx*L_advBC
# Set Diri BCs
L_mix[diri_bc_bool] = np.zeros(shape=L_mix[0].shape)
return L_mix
def set_source_BC(source, dx, a, b, diri_bc_bool, neumann_bc_upstream, neumann_bc_downstream, neumann_bc_values):
source_BC = np.array(source, dtype=float).copy()
# Set Neumann BC. No-flux as default
# Beginning nodes get an extra flux*(-a/dx + b)
source_BC[neumann_bc_upstream] = source_BC[neumann_bc_upstream] + neumann_bc_values[neumann_bc_upstream]*(-a[neumann_bc_upstream]/dx + b[neumann_bc_upstream])
# Ending nodes get an extra -flux*a/dx
source_BC[neumann_bc_downstream] = source_BC[neumann_bc_downstream] - neumann_bc_values[neumann_bc_downstream]*a[neumann_bc_downstream]/dx
# Set Diri BC
source_BC[diri_bc_bool] = 0.
return source_BC
def forward_Euler_adv_diff_single_step(h, dt, L_mix, source):
return h + dt * L_mix @ h + dt*source
def backwards_Euler(h, dt, L_mix, source):
P = np.eye(N=L_mix.shape[0]) - dt*L_mix
P_inv = np.linalg.inv(P)
h = P_inv @ (h + dt*source)
return h
# Parameters
plotOpt = True
dx = 10
dt = 1
time_duration = 1000
niter = int(time_duration/dt)
a = 0 * np.ones(n_nodes)
b = 1 * np.ones(n_nodes)
if np.all(b < 0):
print('All entries of b are negative; changing digraph direction.')
cnm = cnm.T
elif np.prod(b <0):
raise ValueError('Some but not all of bs are negative. Those digarph edge directions should be reversed!')
###Output
_____no_output_____
###Markdown
General advection-diffusion PDE on graphsThe advection-diffusion PDE $$\frac{du}{dt} = au'' - bu' + source$$with $b>0$ (i.e., the flow has the same direction as the edges) can be written in vectorial form in a directed graph like:$$\frac{du}{dt} = -aLu - bL_{adv}u + source =\\ =L_{mix} u + source$$The (strange) sign convention chosen here is such that:$$u'' = -Lu$$$$u' = L_{adv}u,$$This strange sign convention is chosen for 2 reasons:1. Advection-=diffusion eqs are usually written as $u_t = au'' - vu'$.2. $L$ and $L_{adv}$ have already been defined in the literature, and we use the most common definitions. Those are:$$ L = diag(d) - A $$where $d_i$ is the total number of neighbors of noe $i$ and $diag(d)$ is the matrix with $d_i$ in the diagonals. $A$ is the adjacency matrix of the undirected graph.$$ L_{adv} = diag(d_{out}) - A_{in}$$where $d_{out}$ is the number of outgoing neighbours and $A_{in}$ is the incoming adjacency matrix. Connection to finite differences Connection to 1DWhen the graph is a 1D line, the resulting Laplacian matrices are:- $L$ is equivalent to (minus) the centered difference scheme (with Neumann BC if we apply upwind scheme to the boundary nodes). For example, with a line of 4 nodes: $$L = \left(\matrix{1 & -1 & 0 & 0 \cr-1 & 2 & -1& 0 \cr0 & -1 & 2 & -1\cr0 & 0 & -1 & 1}\right)$$- $L_{adv}$ is the upwind scheme for the advection equation (https://en.wikipedia.org/wiki/Upwind_scheme):$$L_{adv} = \left(\matrix{1 & 0 & 0 & 0 \cr-1 & 1 & 0 & 0 \cr0 & -1 & 1 & 0\cr0 & 0 & -1 & 0}\right)$$This one has funnier BC, and they are so in two accounts:First, since $b>0$, this represents a wave moving to the right, so in the 1D advection equation only boundary conditions for the left boundary are needed. In all text I have seen, only diri BCs appear. Neumann BCs are not covered. If you try to append a ghost node and apply a fixed flux, it doesn't work. Diri BC is of course much easier to implement, so I'll stick with it.Second, the rightmost boundary is effectively a no-flow boundary, since $u_4^{t+1} = u_4^t + u_3^t$, and that last node collects all that is incoming from the left. This is not what happens in the upwind scheme. There, the last row of the matrix reads $\left(0,0,-1,1\right)$. And, since no right BC are needed, this is usually left untouched.In any way, what's important when modelling is to be practical adjust these finite difference schemes to what we want to model, so in practice other BC are possible. Connection to 2D - To do-
###Code
# Set up simulations
L = compute_laplacian_from_adjacency(cnm_sim)
L_adv = L_advection(cnm)
if np.all(b < 0):
L_adv = -L_adv
elif np.prod(b <0):
raise ValueError('Some but not all of bs are negative. Those digarph edge directions should be reversed!')
ini_values = dem
source = np.array([0]*n_nodes)
u = ini_values.copy()
diri_bc_bool = np.array([False]*n_nodes)
diri_bc_bool[0] = True
upstream_bool, downstream_bool = infer_extreme_nodes(cnm_sim)
neumann_bc_bool = np.array([False]*n_nodes)
neumann_bc_bool[-1] = True
neumann_bc_upstream = neumann_bc_bool * upstream_bool
neumann_bc_downstream = neumann_bc_bool * downstream_bool
neumann_bc_values = 0.001*neumann_bc_bool # General Neumann BC not implemented yet
if np.any(diri_bc_bool * neumann_bc_bool == True):
raise ValueError('diri and neumann BC applied at the same time in some node')
L_mix = advection_diffusion_operator(dx, L, L_adv, a, b, diri_bc_bool, neumann_bc_upstream, neumann_bc_downstream, neumann_bc_values)
source_BC = set_source_BC(source, dx, a, b, diri_bc_bool, neumann_bc_upstream, neumann_bc_downstream, neumann_bc_values)
print('neumann: ',neumann_bc_upstream, neumann_bc_downstream, neumann_bc_values)
print('diri: ', diri_bc_bool)
print('source_BC: ', source_BC)
source_BC[neumann_bc_upstream]
np.any(diri_bc_bool == neumann_bc_bool)
cnm
ini_values
u
# Simulate
u_sol = [0]*niter
for t in range(niter):
u_sol[t] = u
u = forward_Euler_adv_diff_single_step(u, dt, L_mix, source_BC)
# Plot
%matplotlib inline
if one_dim:
plt.figure()
for t in range(niter):
plt.plot(u_sol[t], color='blue', alpha=0.2)
else:
fig, ax = plt.subplots(subplot_kw={"projection": "3d"})
xs, ys = np.meshgrid(np.linspace(0,1, 4), np.linspace(0,1,3))
ax.plot_surface(xs, ys, u_sol[1].reshape(3,4))
ax.scatter(xs, ys, u_sol[1].reshape(3,4), color='orange')
# Animations
%matplotlib inline
import matplotlib.animation as animation
if one_dim:
fig, ax = plt.subplots()
line, = ax.plot(u_sol[0], alpha=1.0)
def animate(t):
line.set_ydata(u_sol[t]) # update the data.
return line,
ani = animation.FuncAnimation(
fig, animate)
else:
fig = plt.figure()
ax = fig.add_subplot(111, projection='3d')
ax.set_zlim(0, 20)
xs, ys = np.meshgrid(np.linspace(0,1, 4), np.linspace(0,1,3))
plot = [ax.plot_surface(xs, ys, u_sol[0].reshape(3,4), color='1')]
def animate(t, u_sol, plot):
plot[0].remove()
plot[0] = ax.plot_surface(xs, ys, u_sol[t].reshape(3,4), color='0.75')
return plot[0],
ani = animation.FuncAnimation(fig, animate, fargs=(u_sol,plot))
from IPython.display import HTML
HTML(ani.to_html5_video())
###Output
_____no_output_____
###Markdown
Saint Venant: Diffusive waveThe diffusive wave approximation of the Saint-Venant equations is $$\frac{dQ}{dt} = D\frac{dQ^2}{d^2x} - \lambda \frac{dQ}{dx}$$We use the Manning classical approximation for $D$ and $\lambda$,$$D = \frac{A^2 R^{4/3}}{2n^2B |Q|}$$$$\lambda = \frac{5Q}{3A}$$notice that both depend on $Q$ and $A$, so even in the Diffusive wave approximation, St. Venants eqs are a system of non-linear coupled PDEs. $R = A/P$ is the hydraulic radius, where $P$ is the wetteed perimeter (the part of the canal wetted by the water). For rectangular channels, $R = YB/(2Y+B) = AB/(2A+B^2)$.In order to know the water height from the canal bottom, $Y$, the mass conservation equation of the St. Venants eqs can be used:$$ \frac{dA}{dt} = -\frac{dQ}{dx}$$ Simplest numerical schemeThe usefullness of the Laplacian approach is most apparent when we use explicit forward Euler in time and treating the $Q$ and $A$ equations as a 2 step iterative process, so that when we solve for $Q$ we assume that $A$ is constant, and viceversa:Simplest algorithm. At each timestep:1. $$Q = f(Q^-, A^-)$$2. $$A = g(A^-, Q)$$3. $$Q^- \leftarrow Q; A^- \leftarrow A$$where $Q^-$ stands for the value of $Q$ at the previous iteration.The alternative is to use the implicit backwards Euler scheme and to solve the full set of coupled nonlinear algebraic equations using something like the Newton-Rhapson method, but this gets very messy very quickly and although the Laplacian formulation may help, it is not as useful anymore.
###Code
cnm = choose_graph('line')
cnm_sim = cnm + cnm.T
n_nodes = cnm.shape[0]
L = compute_laplacian_from_adjacency(cnm_sim)
L_adv = L_advection(cnm)
upstream_bool, downstream_bool = infer_extreme_nodes(cnm_sim)
# params
B = 7 # (rectangular) canal width in m
n_manning = 2.
dx = 100
dt = 0.05
niter = 1000
# Q stuff
Q_ini = np.arange(1,n_nodes+1,1)[::-1]
Q_source = np.zeros(n_nodes)
Q = Q_ini.copy()
Q_diri_bc_bool = np.array([False]*n_nodes)
#Q_diri_bc_bool[-1] = True
Q_neumann_bc_bool = np.array([False]*n_nodes)
Q_neumann_bc_values = 0*Q_neumann_bc_bool # General Neumann BC not implemented yet
Q_neumann_bc_upstream = Q_neumann_bc_bool * upstream_bool
Q_neumann_bc_downstream = Q_neumann_bc_bool * downstream_bool
# A stuff
Y_ini = np.ones(n_nodes) # m above canal bottom
A_ini = Y_ini * B
A = A_ini.copy()
A_diri_bc_bool = np.array([False]*n_nodes)
A_neumann_bc_bool = np.array([False]*n_nodes)
A_neumann_bc_values = 0*A_neumann_bc_bool # General Neumann BC not implemented yet
A_neumann_bc_upstream = A_neumann_bc_bool * upstream_bool
A_neumann_bc_downstream = A_neumann_bc_bool * downstream_bool
A_diri_bc_bool[0] = True
A_source = np.zeros(n_nodes)
def diffusion_coeff(A, Q, B, n_manning):
return A**2 * (A*B/(2*A + B**2)**(4/3))/(2*n_manning**2 * B * Q)
def advection_coeff(A, Q):
return 5/3 * Q/A
D = diffusion_coeff(A, Q, B, n_manning)
l = advection_coeff(A, Q)
#L_mix = advection_diffusion_operator(dx, L, L_adv, D, -l, diri_bc_bool, neumann_bc_bool)
Q_L_mix = advection_diffusion_operator(dx, L, L_adv, D, -l, Q_diri_bc_bool, Q_neumann_bc_upstream, Q_neumann_bc_downstream, Q_neumann_bc_values)
A_L_mix = advection_diffusion_operator(dx, L, L_adv, np.zeros(n_nodes), np.ones(n_nodes), A_diri_bc_bool, A_neumann_bc_upstream, A_neumann_bc_downstream, A_neumann_bc_values)
# Simulate
Q_sol = [0]*niter
Y_sol = [0]*niter
for t in range(niter):
D = diffusion_coeff(A, Q, B, n_manning)
l = advection_coeff(A, Q)
Q_source_BC = set_source_BC(Q_source, dx, D, -l, Q_diri_bc_bool, Q_neumann_bc_upstream, Q_neumann_bc_downstream, Q_neumann_bc_values)
A_source_BC = set_source_BC(A_source, dx, np.zeros(n_nodes), np.ones(n_nodes), A_diri_bc_bool, A_neumann_bc_upstream, A_neumann_bc_downstream, A_neumann_bc_values)
Q_sol[t] = Q; Y_sol[t] = A/B
Q = Q + dt*Q_L_mix @ Q
A = A + dt*A_L_mix@Q
L_adv
# Plot all Qs
plt.figure()
plt.title('All Qs')
for t in range(niter):
plt.plot(Q_sol[t], alpha=0.1, color='red')
# Animate Q
figQ, axQ = plt.subplots()
axQ.set_title("Water flux Q")
lin, = axQ.plot(Q_sol[0], alpha=1.0, color='red')
def animate_Q(t):
lin.set_ydata(Q_sol[t]) # update the data.
return lin,
duration_in_frames = 100
aniQ = animation.FuncAnimation(figQ, animate_Q, frames=range(0,niter, int(niter/duration_in_frames)))
HTML(aniQ.to_html5_video())
# Plot all Ys
plt.figure()
plt.title('All Ys')
for t in range(niter):
plt.plot(Y_sol[t], alpha=0.1, color='blue')
# Animate Y
figY, axY = plt.subplots()
axY.set_title("Height of water in canal Y")
axY.set_ylim(0,2)
lin, = axY.plot(Y_sol[0], alpha=1.0)
def animate_Y(t):
lin.set_ydata(Y_sol[t]) # update the data.
return lin,
duration_in_frames = 100
aniY = animation.FuncAnimation(figY, animate_Y, frames=range(0,niter, int(niter/duration_in_frames)))
HTML(aniY.to_html5_video())
###Output
_____no_output_____
###Markdown
Full St VenantEquations can be written (for a rectangular channel) as:$$\frac{\partial A}{\partial t} = -\frac{\partial Q}{\partial x} + q,$$where $q$ is the lateral inflow per unit length.$$\frac{\partial Q}{\partial t} = -\frac{\partial}{\partial x}\left(\frac{Q^2}{A}\right) - \frac{gA}{B}\frac{\partial A}{\partial x} + (S_0 - S_f)gA$$where $S_f$ is the friction slope. In the classical approximation by Manning it is given by$$S_f = \frac{n^2 Q^2}{A^2 R^{4/3}}$$Once again, the numerical scheme implemented here is the same as above: First compute Q, then A, both independently, and iterate. Steady stateThe steady state might be useful to compute the initial state of the water level and flow.$$\frac{\partial Q}{\partial x} = q$$$$ \frac{\partial}{\partial x}\left(\frac{Q^2}{A}\right) + \frac{gA}{B}\frac{\partial A}{\partial x} + (S_f - S_0)gA = 0$$And in terms of $L_{adv}$:$$ L_{adv}\left(\frac{Q^2}{A}\right) + \frac{gA}{B}L_{adv} A + (S_f - S_0)gA = 0$$NOTE- 1st eq: if q = 0, then there is no gradient in Q, i.e., Q is a constant vector.- 2nd eq: Invert matrix to get $A^2$. Iteratively. If q = 0 or Q = 0, then A is a constant vector too.
###Code
raise RuntimeError('Not updated for new BC handling yet')
raise RuntimeError('The equations are wrong')
cnm = choose_graph('line').T # Advection is negative
cnm_sim = cnm + cnm.T
n_nodes = cnm.shape[0]
L = compute_laplacian_from_adjacency(cnm_sim)
L_adv = -L_advection(cnm) #Advection is negative
S0 = dem[::-1] + 0.1 # Bottom elevation, i.e., DEM
# params
B = 7 # (rectangular) canal width in m
n_manning = 2.
dx = 100
dt = 0.01
q = 1 * np.ones(n_nodes) # lateral flow towards canal, source term for A eq
# Solve Q from 1st St. Venant eqs.
Q,_,_,_ = np.linalg.lstsq(L_adv, q) # Instead of inverting L_adv, because it usually is a singular matrix
# A stuff
A_ini = np.ones(n_nodes)
A_squared = A_ini.copy()
# Solve ss Q from 1st St. Venant eqs.
Q,_,_,_ = np.linalg.lstsq(L_adv, q, rcond=1e-7) # Instead of inverting L_adv, because it usually is a singular matrix
# Solve steady state A from 2nd St.Venants
A_sol = [0]*niter
for t in range(niter):
A_sol[0] = np.sqrt(A_squared)
R = A_squared*B/(2*A_squared + B**2)
Sf = n_manning**2 * Q**2/(A_squared**2)/R**(4/3)
matrix = ((Q**2/A_squared**2 - 9.8*A_squared/B)*L_adv + (S0 - Sf)*9.8* np.eye(n_nodes))
A_squared, _, _, _ = np.linalg.lstsq(matrix, 2*Q*q)
###Output
_____no_output_____
###Markdown
Unsteady
###Code
cnm = choose_graph('line')
cnm_sim = cnm + cnm.T
n_nodes = cnm.shape[0]
upstream_bool, downstream_bool = infer_extreme_nodes(cnm_sim)
L = compute_laplacian_from_adjacency(cnm_sim)
L_adv = L_advection(cnm)
L_upwind = upwind_from_advection_laplacian(L_adv, downstream_bool)
#L_upwind = L_adv.copy()
S0 = ini_values # Bottom elevation, i.e., DEM
# params
B = 7 # (rectangular) canal width in m
n_manning = 2.
dx = 100
dt = 0.01
niter = 1000
# Q stuff
Q_ini = np.arange(1,n_nodes+1,1)[::-1]
Q = Q_ini.copy()
Q_diri_bc_bool = np.array([False]*n_nodes)
Q_diri_bc_bool[0] = True
Q_neumann_bc_bool = np.array([False]*n_nodes)
Q_neumann_bc_values = 0*Q_neumann_bc_bool
Q_neumann_bc_upstream = Q_neumann_bc_bool * upstream_bool
Q_neumann_bc_downstream = Q_neumann_bc_bool * downstream_bool
# A stuff
Y_ini = np.ones(n_nodes) # m above canal bottom
A_ini = Y_ini * B
A = A_ini.copy()
A_diri_bc_bool = np.array([False]*n_nodes)
A_neumann_bc_bool = np.array([False]*n_nodes)
A_neumann_bc_values = 0*A_neumann_bc_bool
A_neumann_bc_upstream = A_neumann_bc_bool * upstream_bool
A_neumann_bc_downstream = A_neumann_bc_bool * downstream_bool
A_diri_bc_bool[0] = True
q = 0.1*np.ones(n_nodes) # lateral inflow
A_source = q
A_source[A_diri_bc_bool==True] = 0
# Parameters chosen so that L_adv_BC below is nothing but L_adv with correct BC
#L_adv_BC = advection_diffusion_operator(dx, L, L_adv, 0, 1, Q_diri_bc_bool, Q_neumann_bc_upstream, Q_neumann_bc_downstream, Q_neumann_bc_values)
# Simulate
Q_sol = [0]*niter
Y_sol = [0]*niter
for t in range(niter):
Q_sol[t] = Q; Y_sol[t] = A/B
R = A**2*B/(2*A**2 + B**2)
Sf = n_manning**2 * Q**2/(A**2)/R**(4/3)
dQ = dt*(-L_upwind@(Q**2/A)/dx - 9.8*A/B*L_upwind@A/dx + (S0 - Sf)*9.8*A)
Q = Q + dQ*(~diri_bc_bool)
A = A + dt*(-L_upwind@Q/dx + A_source)
# Plot initial water height
plt.figure()
plt.fill_between(np.arange(n_nodes), y1=S0, y2=0, color='brown', alpha=0.5)
plt.fill_between(np.arange(n_nodes), y1=S0+Y_ini, y2=S0, color='blue', alpha=0.5)
plt.title('Initial water height and DEM')
plt.figure()
plt.plot(Q_ini)
plt.title('Initial flux')
plt.show()
# Plot all Qs
plt.figure()
plt.title('All Qs')
for t in range(niter):
plt.plot(Q_sol[t], alpha=0.1, color='red')
# Animate Q
figQ, axQ = plt.subplots()
axQ.set_title("Water flux, Q")
lin, = axQ.plot(Q_sol[0], alpha=1.0, color='red')
def animate_Q(t):
lin.set_ydata(Q_sol[t]) # update the data.
return lin,
duration_in_frames = 100
aniQ = animation.FuncAnimation(figQ, animate_Q, frames=range(0,niter, int(niter/duration_in_frames)))
HTML(aniQ.to_html5_video())
# Plot all Ys
plt.figure()
plt.title('All Ys')
for t in range(niter):
plt.plot(S0 + Y_sol[t], alpha=0.1, color='blue')
# Animate Y
figY, axY = plt.subplots()
axY.set_title("Height of water in canal Y")
axY.set_ylim(0,5)
axY.fill_between(np.arange(n_nodes), y1=S0, y2=0, color='brown', alpha=0.5)
lin, = axY.plot(S0 + Y_sol[0], alpha=1.0, color='blue')
def animate_Y(t):
lin.set_ydata(S0 + Y_sol[t]) # update the data.
return lin,
duration_in_frames = 100
aniY = animation.FuncAnimation(figY, animate_Y, frames=range(0,niter, int(niter/duration_in_frames)))
HTML(aniY.to_html5_video())
Q_sol
###Output
_____no_output_____
|
src/predict_and_saliency.ipynb
|
###Markdown
Tiny ImageNet: View Predictions and SaliencyThis notebook predicts the top-5 most likely labels for a random selection of images. The human labels, along with the top-5 model predictions, are displayed below each picture. Also, a "saliency map" is displayed next to each image. The saliency map highlights areas that were most important in making the top prediction. It is worth noting that a human labeler would have a difficult time correctly identifying many of these down-sampled images. The human labelers had the advantage of 4x higher resolution images to make their predictions (256x256 vs. 64x64 images). So, the model performance is quite impressive considering the low-resolution images.The last cell may be re-run multiple times to explore a new selection of pictures.Python Notebook by Patrick Coady: [Learning Artificial Intelligence](https://learningai.io/)
###Code
from train import *
import tensorflow as tf
import glob
import matplotlib.pyplot as plt
import random
import scipy.ndimage
%matplotlib inline
plt.rcParams['figure.figsize'] = (10, 6)
class TrainConfig(object):
"""Training configuration"""
dropout_keep_prob = 1.0
model_name = 'vgg_16' # choose model
model = staticmethod(globals()[model_name])
config_name = 'no_hue' # choose training run
def predict(imgs, config):
"""Load most recent checkpoint, make predictions, compute saliency maps"""
g = tf.Graph()
with g.as_default():
imgs_ph = tf.placeholder(dtype=tf.uint8, shape=(None, 56, 56, 3))
logits = config.model(imgs_ph, config)
top_pred = tf.reduce_max(logits, axis=1)
top_5 = tf.nn.top_k(logits, k=5, sorted=True)
# can't calculate gradient to integer, get float32 version of image:
float_img = g.get_tensor_by_name('Cast:0')
# calc gradient of top predicted class to image:
grads = tf.gradients(top_pred, float_img)
saver = tf.train.Saver()
with tf.Session() as sess:
path = 'checkpoints/' + config.model_name + '/' + config.config_name
saver.restore(sess, tf.train.latest_checkpoint(path))
feed_dict = {imgs_ph: imgs}
top_5_np, grads_np = sess.run([top_5, grads], feed_dict=feed_dict)
return top_5_np, grads_np
# get label integer -> text description dictionary
label_dict, class_description = build_label_dicts()
for i in range(len(class_description)):
class_description[i] = class_description[i].split(',')[0]
N = 10 # number of validation examples to view
filenames_labels = load_filenames_labels('val')
pick_N = random.sample(filenames_labels, N)
imgs = np.zeros((N, 64, 64, 3), dtype=np.uint8)
labels = []
for i, filename_label in enumerate(pick_N):
imgs[i, :, :, :] = scipy.ndimage.imread(filename_label[0], mode='RGB')
labels.append(class_description[int(filename_label[1])])
imgs = imgs[:, 4:60, 4:60, :] # take center crop of images
config = TrainConfig()
top_5, sal_imgs = predict(imgs, config)
top_5 = top_5[1] # 2nd element of list are class predictions
sal_imgs = sal_imgs[0] # 1st element of list are saliency maps
# root-sum-square RGB channels of generated saliency heat map
sal_imgs = np.sqrt(np.sum(np.square(sal_imgs), axis=3))
for idx, filename in enumerate(pick_N):
plt.subplot(121)
plt.imshow(imgs[idx, :, :, :], interpolation='none')
# next 5 lines get rid of all white space when saving .png
plt.gca().set_axis_off()
plt.subplots_adjust(top = 1, bottom = 0, right = 1, left = 0,
hspace = 0, wspace = 2)
plt.margins(0,0)
plt.gca().xaxis.set_major_locator(plt.NullLocator())
plt.gca().yaxis.set_major_locator(plt.NullLocator())
plt.subplot(122)
plt.imshow(sal_imgs[idx, :, :], interpolation='none')
# next 5 lines get rid of all white space when saving .png
plt.gca().set_axis_off()
plt.subplots_adjust(top = 1, bottom = 0, right = 1, left = 0,
hspace = 0, wspace = 0.1)
plt.margins(0,0)
plt.gca().xaxis.set_major_locator(plt.NullLocator())
plt.gca().yaxis.set_major_locator(plt.NullLocator())
plt.savefig('plots/pred_sal'+str(idx)+'.png', bbox_inches='tight',
pad_inches=0.0, dpi=64)
plt.show()
print('Actual label: ' + labels[idx])
print('Top 5 predictions:')
preds = map(lambda x: class_description[x], top_5[idx])
print([x for x in preds])
print('\n')
###Output
INFO:tensorflow:Restoring parameters from checkpoints/vgg_16/no_hue/model-44000
|
queue_imbalance/logistic_regression/queue_imbalance-9063.ipynb
|
###Markdown
Testing of queue imbalance for stock 9093Order of this notebook is as follows:1. [Data](Data)2. [Data visualization](Data-visualization)3. [Tests](Tests)4. [Conclusions](Conclusions)Goal is to implement queue imbalance predictor from [[1]](Resources).
###Code
%matplotlib inline
import warnings
import matplotlib.dates as md
import matplotlib.pyplot as plt
import seaborn as sns
from lob_data_utils import lob
from sklearn.metrics import roc_curve, roc_auc_score
warnings.filterwarnings('ignore')
###Output
_____no_output_____
###Markdown
DataMarket is open between 8-16 on every weekday. We decided to use data from only 9-15 for each day. Test and train dataFor training data we used data from 2013-09-01 - 2013-11-16:* 0901* 0916* 1001* 1016* 1101We took 75% of this data (randomly), the rest is the test data.
###Code
df, df_test = lob.load_prepared_data('9063', data_dir='../data/prepared/', length=None)
df.head()
###Output
Training set length for 9063: 13456
Testing set length for 9063: 4507
###Markdown
Data visualization
###Code
df['sum_buy_bid'].plot(label='total size of buy orders', style='--')
df['sum_sell_ask'].plot(label='total size of sell orders', style='-')
plt.title('Summed volumens for ask and bid lists')
plt.xlabel('Time')
plt.ylabel('Whole volume')
plt.legend()
df[['bid_price', 'ask_price', 'mid_price']].plot(style='.')
plt.legend()
plt.title('Prices')
plt.xlabel('Time')
plt.ylabel('Price')
sns.jointplot(x="mid_price", y="queue_imbalance", data=df.loc[:, ['mid_price', 'queue_imbalance']], kind="kde")
plt.title('Density')
plt.plot()
###Output
_____no_output_____
###Markdown
We can see that something weird happen to the stock after 2013-10-24, so let's remove this data.
###Code
df = df['2013-09-01':'2013-10-24']
df_test = df_test['2013-09-01':'2013-10-24']
df[['bid_price', 'ask_price', 'mid_price']].plot(style='.')
plt.legend()
plt.title('Prices between 2013-09-01 - 2013-10-24')
plt.xlabel('Time')
plt.ylabel('Price')
sns.jointplot(x="mid_price", y="queue_imbalance", data=df.loc[:, ['mid_price', 'queue_imbalance']], kind="kde")
plt.title('Density of train data')
plt.plot()
df['mid_price_indicator'].plot('kde')
plt.legend()
plt.xlabel('Mid price indicator')
plt.title('Mid price indicator density')
df['queue_imbalance'].plot('kde')
plt.legend()
plt.xlabel('Queue imbalance')
plt.title('Queue imbalance density')
###Output
_____no_output_____
###Markdown
TestsWe use logistic regression to predict `mid_price_indicator`. Mean square error We calculate residual $r_i$:$$ r_i = \hat{y_i} - y_i $$where $$ \hat{y}(I) = \frac{1}{1 + e −(x_0 + Ix_1 )}$$Calculating mean square residual for all observations in the testing set is also useful to assess the predictive power.The predective power of null-model is 25%.
###Code
reg = lob.logistic_regression(df, 0, len(df))
probabilities = reg.predict_proba(df_test['queue_imbalance'].values.reshape(-1,1))
probabilities = [p1 for p0, p1 in probabilities]
err = ((df_test['mid_price_indicator'] - probabilities) ** 2).mean()
predictions = reg.predict(df_test['queue_imbalance'].values.reshape(-1, 1))
print('Mean square error is', err)
###Output
Mean square error is 0.24683867330196202
###Markdown
Logistic regression fit curve
###Code
plt.plot(df_test['queue_imbalance'].values,
lob.sigmoid(reg.coef_[0] * df_test['queue_imbalance'].values + reg.intercept_))
plt.title('Logistic regression fit curve')
plt.xlabel('Imbalance')
plt.ylabel('Prediction')
###Output
_____no_output_____
###Markdown
ROC curveFor assessing the predectivity power we can calculate ROC score.
###Code
a, b, c = roc_curve(df_test['mid_price_indicator'], predictions)
logit_roc_auc = roc_auc_score(df_test['mid_price_indicator'], predictions)
plt.plot(a, b, label='predictions (area {})'.format(logit_roc_auc))
plt.plot([0, 1], [0, 1], color='navy', linestyle='--')
plt.xlabel('False Positive Rate')
plt.ylabel('True Positive Rate')
plt.legend()
st = 0
end = len(df)
plt.plot(df_test.index[st:end], predictions[st:end], 'ro', label='prediction')
plt.plot(df_test.index[st:end], probabilities[st:end], 'g.', label='probability')
plt.plot(df_test.index[st:end], df_test['mid_price_indicator'].values[st:end], 'b.', label='mid price')
plt.xticks(rotation=25)
plt.legend(loc=1)
plt.xlabel('Time')
plt.ylabel('Mid price prediction')
###Output
_____no_output_____
|
workflow/enumerate_adsorption/test_dev_rdf_comp/opt_rdf_comp.ipynb
|
###Markdown
Import Modules
###Code
import sys
sys.path.insert(0, "..")
import os
print(os.getcwd())
import sys
# from pathlib import Path
# import copy
import pickle
# import json
import numpy as np
import pandas as pd
from ase import io
# # from tqdm import tqdm
from tqdm.notebook import tqdm
# # #########################################################
# # #########################################################
# from misc_modules.pandas_methods import drop_columns
# from misc_modules.misc_methods import GetFriendlyID
# from ase_modules.ase_methods import view_in_vesta
# # #########################################################
from methods import (
# get_df_dft,
# symmetrize_atoms,
# get_structure_coord_df,
# remove_atoms,
get_df_slab,
)
from proj_data import metal_atom_symbol
# #########################################################
from local_methods import (
mean_O_metal_coord,
get_all_active_sites,
get_unique_active_sites,
process_rdf,
compare_rdf_ij,
return_modified_rdf,
create_interp_df,
)
# # from local_methods import calc_surface_area
###Output
_____no_output_____
###Markdown
Read Data
###Code
df_slab = get_df_slab()
df_slab = df_slab.set_index("slab_id")
###Output
_____no_output_____
###Markdown
Create Directories
###Code
directory = "out_data"
if not os.path.exists(directory):
os.makedirs(directory)
row_i = df_slab.loc["tagilahu_40"]
# #####################################################
slab = row_i.slab_final
slab.write("out_data/temp.cif")
slab_id = row_i.name
bulk_id = row_i.bulk_id
facet = row_i.facet
num_atoms = row_i.num_atoms
# #####################################################
active_sites = get_all_active_sites(
slab=slab,
slab_id=slab_id,
bulk_id=bulk_id,
)
# active_sites = [62, 63, 64, 66, 67, 68]
# active_sites = [63, ]
# assert False
directory = "out_plot/__temp__"
if not os.path.exists(directory):
os.makedirs(directory)
slab=slab
active_sites=active_sites
bulk_id=bulk_id
facet=facet
slab_id=slab_id
metal_atom_symbol=metal_atom_symbol
import methods
from methods import get_df_coord
# get_df_coord(slab_id=None, bulk_id=None, mode="slab")
assert False
# def get_unique_active_sites(
# slab=None,
# active_sites=None,
# bulk_id=None,
# facet=None,
# slab_id=None,
# metal_atom_symbol=None,
# ):
"""
"""
#| - get_unique_active_sites
df_coord_slab_i = get_df_coord(slab_id=slab_id, mode="slab")
df_coord_bulk_i = get_df_coord(bulk_id=bulk_id, mode="bulk")
# #########################################################
custom_name_pre = bulk_id + "__" + facet + "__" + slab_id
df_rdf_dict = dict()
for i in active_sites:
print("active_site:", i)
df_rdf_i = process_rdf(
atoms=slab,
active_site_i=i,
df_coord_slab_i=df_coord_slab_i,
metal_atom_symbol=metal_atom_symbol,
custom_name=custom_name_pre,
TEST_MODE=True,
)
# df_rdf_i = df_rdf_i.rename(columns={" g(r)": "g"})
df_rdf_dict[i] = df_rdf_i
# # #########################################################
# diff_rdf_matrix = np.empty((len(active_sites), len(active_sites), ))
# diff_rdf_matrix[:] = np.nan
# for i_cnt, active_site_i in enumerate(active_sites):
# df_rdf_i = df_rdf_dict[active_site_i]
# for j_cnt, active_site_j in enumerate(active_sites):
# df_rdf_j = df_rdf_dict[active_site_j]
# diff_i = compare_rdf_ij(
# df_rdf_i=df_rdf_i,
# df_rdf_j=df_rdf_j,
# )
# diff_rdf_matrix[i_cnt, j_cnt] = diff_i
# # #########################################################
# df_rdf_ij = pd.DataFrame(diff_rdf_matrix, columns=active_sites)
# df_rdf_ij.index = active_sites
# # #########################################################
# import copy
# active_sites_cpy = copy.deepcopy(active_sites)
# diff_threshold = 0.3
# duplicate_active_sites = []
# for active_site_i in active_sites:
# if active_site_i in duplicate_active_sites:
# continue
# for active_site_j in active_sites:
# if active_site_i == active_site_j:
# continue
# diff_ij = df_rdf_ij.loc[active_site_i, active_site_j]
# if diff_ij < diff_threshold:
# try:
# active_sites_cpy.remove(active_site_j)
# duplicate_active_sites.append(active_site_j)
# except:
# pass
# active_sites_unique = active_sites_cpy
# # #########################################################
# import plotly.express as px
# import plotly.graph_objects as go
# active_sites_str = [str(i) for i in active_sites]
# fig = go.Figure(data=go.Heatmap(
# z=df_rdf_ij.to_numpy(),
# x=active_sites_str,
# y=active_sites_str,
# # type="category",
# ))
# fig["layout"]["xaxis"]["type"] = "category"
# fig["layout"]["yaxis"]["type"] = "category"
# # fig.show()
# directory = "out_plot/rdf_heat_maps"
# if not os.path.exists(directory):
# os.makedirs(directory)
# from plotting.my_plotly import my_plotly_plot
# # file_name = "rdf_heat_maps/ " + custom_name_pre + "_rdf_diff_heat_map"
# file_name = "__temp__/ " + custom_name_pre + "_rdf_diff_heat_map"
# my_plotly_plot(
# figure=fig,
# # plot_name="rdf_heat_maps/rdf_diff_heat_map",
# plot_name=file_name,
# write_html=True,
# write_png=False,
# png_scale=6.0,
# write_pdf=False,
# write_svg=False,
# try_orca_write=False,
# )
# # return(active_sites_unique)
# #__|
custom_name_pre
# active_site_i = 62
active_site_i = 63
active_site_j = 66
df_rdf_i = df_rdf_dict[active_site_i]
df_rdf_j = df_rdf_dict[active_site_j]
# Pickling data ###########################################
# out_dict = dict()
# out_dict["TEMP"] = None
import os; import pickle
path_i = os.path.join(
os.environ["HOME"],
"__temp__",
"temp.pickle")
with open(path_i, "wb") as fle:
pickle.dump((df_rdf_i, df_rdf_j), fle)
# #########################################################
def test_rdf_opt(dx, df_rdf_i, df_rdf_j, chunks_to_edit):
"""
"""
df_rdf_j_new = return_modified_rdf(
df_rdf=df_rdf_j,
# chunk_to_edit=0,
chunks_to_edit=chunks_to_edit,
# dx=-0.04,
dx=dx,
)
df_rdf_i = df_rdf_i
df_rdf_j = df_rdf_j_new
# #########################################################
r_combined = np.sort((df_rdf_j.r.tolist() + df_rdf_i.r.tolist()))
r_combined = np.sort(list(set(r_combined)))
df_interp_i = create_interp_df(df_rdf_i, r_combined)
df_interp_j = create_interp_df(df_rdf_j, r_combined)
diff_i = compare_rdf_ij(
df_rdf_i=df_interp_i,
df_rdf_j=df_interp_j)
print("dx:", dx, " | ","diff_i:", diff_i)
print("")
return(diff_i)
def constraint_bounds(dx, df_rdf_i, df_rdf_j, chunks_to_edit):
# out = -0.2 + np.abs(dx)
out = +0.05 - np.abs(dx)
return(out)
from scipy.optimize import minimize
data_dict_list = []
# for peak_i in range(0, 10):
for peak_i in range(0, 1):
data_dict_i = dict()
data_dict_i["peak"] = peak_i
arguments = (df_rdf_i, df_rdf_j, peak_i)
cons = ({
"type": "ineq",
"fun": constraint_bounds,
"args": arguments,
})
initial_guess = 0
result = minimize(
# obj,
test_rdf_opt,
initial_guess,
method="SLSQP",
args=arguments,
constraints=cons,
)
print(40 * "*")
print(result)
dx_i = result["x"][0]
data_dict_i["dx"] = dx_i
print(40 * "*")
data_dict_list.append(data_dict_i)
df = pd.DataFrame(data_dict_list)
df
df_rdf_j_new = return_modified_rdf(
df_rdf=df_rdf_j,
# chunks_to_edit=0,
chunks_to_edit=df.peak.tolist(),
# dx=-0.04,
# dx=-0.03332953,
dx=df.dx.tolist(),
)
import plotly.graph_objs as go
data = []
trace = go.Scatter(
x=df_rdf_i.r,
y=df_rdf_i.g,
name="df_rdf_i",
)
data.append(trace)
trace = go.Scatter(
x=df_rdf_j_new.r,
y=df_rdf_j_new.g,
name="df_rdf_j_new",
)
data.append(trace)
trace = go.Scatter(
x=df_rdf_j.r,
y=df_rdf_j.g,
name="df_rdf_j",
)
data.append(trace)
fig = go.Figure(data=data)
from plotting.my_plotly import my_plotly_plot
file_name = "__temp__/modified_and_opt_rdf_plots"
my_plotly_plot(
figure=fig,
plot_name=file_name,
write_html=True)
fig.show()
df_rdf_i = df_rdf_i
df_rdf_j = df_rdf_j_new
# #########################################################
r_combined = np.sort((df_rdf_j.r.tolist() + df_rdf_i.r.tolist()))
r_combined = np.sort(list(set(r_combined)))
df_interp_i = create_interp_df(df_rdf_i, r_combined)
df_interp_j = create_interp_df(df_rdf_j, r_combined)
diff_i = compare_rdf_ij(
df_rdf_i=df_interp_i,
df_rdf_j=df_interp_j)
diff_i
print(diff_i)
###Output
_____no_output_____
|
分位数回归/基础概念.ipynb
|
###Markdown
基础概念 均值回归局限只有满足古典假定,估计量才具有优良性质:BLUE 为什么需要分位数回归在迄今为止的同归模型中,我们着重考察解释变量 x 对被解释变量 y 的条件期望 E $(y | \boldsymbol{x})$ 的 影响,实际上是均值回归。但我们真正关心的是 x 对整个条件分布 $y | x$ 的影响,而条件期望 $E(y| \boldsymbol{x})$ 只是刻画条件分布 $y | \boldsymbol{x}$ 集中趋势的一个指标而已。如果条件分布 $y | \boldsymbol{x}$ 不是对称分布,则条件期望 E( $y | \boldsymbol{x}$ )很难反映整个条件分布的全貌。如果能够估计出条件分布 $y | x$ 的若干重要的条件分位数,比如中位数、1/4 分位数 ,3/4 分位数,就能对条件分布 $y | \boldsymbol{x}$ 有更全面的认识。另一方面, 使用 OLS 的古典“均值同归”,由于最小化的目标函数为残差平方和 $\left(\sum_{i=1}^{n} e_{i}^{2}\right),$ 故容易受极端值的影响。 为此, Koenker and Bassett( 1978 ) 提出“分位数同归”(Quantile Regression,简记 QR ) ,使用残差绝对值的加权平均(比如 $\sum_{i=1}^{n}\left|e_{i}\right|$ ) 作为最小化的目标函数,故不易受极端值影响,较为稳健。更重要的是,分位数回归还能提供关于条件分布 $y | \boldsymbol{x}$ 的全面信息。 原理 模型表示$Q_{t}(y | x)=x^{T} \beta_{\tau}$其中 $\tau$ 为分位点, $\beta_{\tau}$ 为依赖于分位点的回归系数 损失函数* 二次损失:生成均值 $ E(Y)=\underset{y}{\operatorname{argmin}} E(Y-y)^{2}$* 绝对值损失:生成中位数 $A L(u)=|u| $* 非对称绝对值损失:生成分位数 $\quad \rho_{\tau}(u)=u(\tau-I(u<0)) \quad Q_{\tau}(Y)=\underset{y}{\operatorname{argmin}} E \rho_{\tau}(Y-y)$ 估计方法由于分位数回归的目标函数带有绝对值,不可微分,故通常使用线性规划。详情可参考:[statsmodels 官方文档](https://www.statsmodels.org/stable/generated/statsmodels.regression.quantile_regression.QuantReg.htmlstatsmodels.regression.quantile_regression.QuantReg) 使用 statsmodels 库实现 第一种方法
###Code
import numpy as np
import pandas as pd
import statsmodels.formula.api as smf
data = pd.read_excel('../数据/上证指数与沪深300.xlsx')
mod = smf.quantreg("data['hs300'] ~ data['sz']", data)
res = mod.fit(q=0.3)
res.summary()
###Output
_____no_output_____
###Markdown
第二种方法
###Code
import statsmodels.regression.quantile_regression as qr
import statsmodels.api as sm
X = sm.add_constant(data['sz'])
mod = qr.QuantReg(data['hs300'], X)
res = mod.fit(q=0.3)
res.summary()
###Output
_____no_output_____
|
cylinder/cylgrid0_sst_iddes_matula_01/plotForcesAllversions.ipynb
|
###Markdown
Plot forces for flow past cylinder grid0 case All versions of code/grid
###Code
%%capture
import sys
sys.path.insert(1, '../utilities')
import litCdData
import numpy as np
import matplotlib.pyplot as plt
# Basic problem parameters
D = 6 # Cylinder diameter
U = 20 # Freestream velocity
Lspan = 24 # Spanwise length
A = D*Lspan # frontal area
rho = 1.225 # density
Q = 0.5*rho*U*U # Dynamic head
vis = 1.8375e-5 # viscosity
ReNum = rho*U*D/vis # Reynolds number
avgt = [160.0, 260.0] # Average times
saveinfo = False
alldata = []
# Load the force data (old grid, old code)
force01 = np.loadtxt('forces01.dat', skiprows=1)
force02 = np.loadtxt('forces02.dat', skiprows=1)
forcedat = np.vstack((force01, force02))
t = forcedat[:,0]*U/D # Non-dimensional time
alldata.append(['Old BC, Old code', t, forcedat])
# Old grid, new code
forcedat = np.loadtxt('forcesoldgridnewcode.dat', skiprows=1)
t = forcedat[:,0]*U/D # Non-dimensional time
alldata.append(['Old BC, New code', t, forcedat])
# New grid, old code
forcedat = np.loadtxt('forcesnewgridoldcode.dat', skiprows=1)
t = forcedat[:,0]*U/D # Non-dimensional time
alldata.append(['New BC, old code', t, forcedat])
# New grid, new code
forcedat = np.loadtxt('forcesnewgridnewcode.dat', skiprows=1)
t = forcedat[:,0]*U/D # Non-dimensional time
alldata.append(['New BC, new code', t, forcedat])
# Calculate time average
def timeaverage(time, f, t1, t2):
filt = ((time[:] >= t1) & (time[:] <= t2))
# Filtered time
t = time[filt]
# The total time
dt = np.amax(t) - np.amin(t)
# Filtered field
filtf = f[filt]
# Compute the time average as an integral
avg = np.trapz(filtf, x=t, axis=0) / dt
return avg
#print(alldata)
print('%30s %10s %10s'%("Case", "avgCd", "avgCl"))
for run in alldata:
label = run[0]
t = run[1]
forcedat = run[2]
Cd = (forcedat[:,1]+forcedat[:,4])/(Q*A)
Cl = (forcedat[:,2]+forcedat[:,5])/(Q*A)
# Calculate averaged Cp, Cd
avgCd = timeaverage(t, Cd, avgt[0], avgt[1])
avgCl = timeaverage(t, Cl, avgt[0], avgt[1])
print('%30s %10f %10f'%(label, avgCd, avgCl))
#print("Avg Cd = %f"%avgCd)
#%print("Avg Cl = %f"%avgCl)
###Output
Case avgCd avgCl
Old BC, Old code 0.557448 0.015747
Old BC, New code 0.531194 -0.020163
New BC, old code 0.532598 -0.003509
New BC, new code 0.371748 0.005388
###Markdown
Plot Lift and Drag coefficients
###Code
plt.rc('font', size=16)
plt.figure(figsize=(10,8))
for run in alldata:
label = run[0]
t = run[1]
forcedat = run[2]
Cd = (forcedat[:,1]+forcedat[:,4])/(Q*A)
Cl = (forcedat[:,2]+forcedat[:,5])/(Q*A)
# Calculate averaged Cp, Cd
avgCd = timeaverage(t, Cd, avgt[0], avgt[1])
avgCl = timeaverage(t, Cl, avgt[0], avgt[1])
#print('%30s %f %f'%(label, avgCd, avgCl))
plt.plot(t,Cd, label=label)
plt.hlines(avgCd, np.min(t), np.max(t), linestyles='dashed', linewidth=1)
plt.xlabel(r'Non-dimensional time $t U_{\infty}/D$');
plt.legend()
plt.ylabel('$C_D$')
plt.title('Drag coefficient $C_D$');
plt.figure(figsize=(10,8))
for run in alldata:
label = run[0]
t = run[1]
forcedat = run[2]
Cd = (forcedat[:,1]+forcedat[:,4])/(Q*A)
Cl = (forcedat[:,2]+forcedat[:,5])/(Q*A)
# Calculate averaged Cp, Cd
avgCd = timeaverage(t, Cd, avgt[0], avgt[1])
avgCl = timeaverage(t, Cl, avgt[0], avgt[1])
plt.plot(t,Cl, label=label)
plt.hlines(avgCl, np.min(t), np.max(t), linestyles='dashed', linewidth=1)
plt.xlabel(r'Non-dimensional time $t U_{\infty}/D$');
plt.ylabel('$C_l$')
plt.title('Lift coefficient $C_l$');
plt.legend()
###Output
_____no_output_____
###Markdown
Plot Cd versus Reynolds number
###Code
plt.figure(figsize=(10,8))
litCdData.plotEXP()
litCdData.plotCFD()
for run in alldata:
label = run[0]
t = run[1]
forcedat = run[2]
Cd = (forcedat[:,1]+forcedat[:,4])/(Q*A)
Cl = (forcedat[:,2]+forcedat[:,5])/(Q*A)
# Calculate averaged Cp, Cd
avgCd = timeaverage(t, Cd, avgt[0], avgt[1])
avgCl = timeaverage(t, Cl, avgt[0], avgt[1])
plt.semilogx(ReNum, avgCd, '*', ms=10, label='Nalu SST-IDDES '+label)
plt.grid()
plt.legend(fontsize=10)
plt.xlabel(r'Reynolds number Re');
plt.ylabel('$C_D$')
plt.title('Drag coefficient $C_D$');
# Write the YAML file these averaged quantities
import yaml
if saveinfo:
savedict={'Re':float(ReNum), 'avgCd':float(avgCd), 'avgCl':float(avgCl)}
f=open('istats.yaml','w')
f.write('# Averaged quantities from %f to %f\n'%(avgt[0], avgt[1]))
f.write('# Grid: grid0\n')
f.write(yaml.dump(savedict, default_flow_style=False))
f.close()
###Output
_____no_output_____
|
examples/notebooks/HestonHullWhite_calibration.ipynb
|
###Markdown
Calibration of a Heston / Hull-White model This notebook is a direct transcription of a QuantLib test case. To test the accuracy of a Heston / Hull-White model calibration, we first generate a data set of option prices, assuming a particular Heston/HW model, then calibrate this model of the option data, and verify that we recover the original model parameters. Set-up A large number of functions is needed to run the calibration.
###Code
import numpy as np
from quantlib.math.hestonhwcorrelationconstraint import (
HestonHullWhiteCorrelationConstraint)
from quantlib.time.api import (
Period, Months, March, Date,
Actual365Fixed, TARGET
)
from quantlib.settings import Settings
from quantlib.math.optimization import LevenbergMarquardt, EndCriteria
from quantlib.quotes import SimpleQuote
from quantlib.termstructures.yields.flat_forward import FlatForward
from quantlib.models.equity.heston_model import (
HestonModelHelper, HestonModel, PriceError
)
from quantlib.processes.heston_process import HestonProcess
from quantlib.processes.api import HullWhiteProcess
from quantlib.pricingengines.api import (
AnalyticHestonEngine,
FdHestonHullWhiteVanillaEngine)
from quantlib.methods.finitedifferences.solvers.fdmbackwardsolver import FdmSchemeDesc
###Output
_____no_output_____
###Markdown
This function creates a flat term structure.
###Code
def flat_rate(today, forward, daycounter):
return FlatForward(
reference_date=today,
forward=SimpleQuote(forward),
daycounter=daycounter
)
###Output
_____no_output_____
###Markdown
The volatility surface is based on a Heston-Hull-White model withthe following parameters: * Hull-White: $a = 0.00883$, $\sigma = 0.00631$* Heston: $\nu = 0.12$, $\kappa = 2.0$, $\theta = 0.09$, $\sigma = 0.5$, $\rho=-0.75$* Equity / short rate correlation: $-0.5$
###Code
dc = Actual365Fixed()
calendar = TARGET()
todays_date = Date(28, March, 2004)
settings = Settings()
settings.evaluation_date = todays_date
r_ts = flat_rate(todays_date, 0.05, dc)
## assuming, that the Hull-White process is already calibrated
## on a given set of pure interest rate calibration instruments.
hw_process = HullWhiteProcess(r_ts, a=0.00883, sigma=0.00631)
q_ts = flat_rate(todays_date, 0.02, dc)
s0 = SimpleQuote(100.0)
# vol surface
strikes = [50, 75, 90, 100, 110, 125, 150, 200]
maturities = [1 / 12., 3 / 12., 0.5, 1.0, 2.0, 3.0, 5.0, 7.5, 10]
vol = [
0.482627,0.407617,0.366682,0.340110,0.314266,0.280241,0.252471,0.325552,
0.464811,0.393336,0.354664,0.329758,0.305668,0.273563,0.244024,0.244886,
0.441864,0.375618,0.340464,0.318249,0.297127,0.268839,0.237972,0.225553,
0.407506,0.351125,0.322571,0.305173,0.289034,0.267361,0.239315,0.213761,
0.366761,0.326166,0.306764,0.295279,0.284765,0.270592,0.250702,0.222928,
0.345671,0.314748,0.300259,0.291744,0.283971,0.273475,0.258503,0.235683,
0.324512,0.303631,0.293981,0.288338,0.283193,0.276248,0.266271,0.250506,
0.311278,0.296340,0.289481,0.285482,0.281840,0.276924,0.269856,0.258609,
0.303219,0.291534,0.286187,0.283073,0.280239,0.276414,0.270926,0.262173]
###Output
_____no_output_____
###Markdown
In a first stage, we calibrate a pure Heston model on the data, in order to obtain a good starting point for the Heston/Hull-White calibration, which is much more time consuming.
###Code
start_v0 = 0.2 * 0.2
start_theta = start_v0
start_kappa = 0.5
start_sigma = 0.25
start_rho = -0.5
equityShortRateCorr = -0.5
corrConstraint = HestonHullWhiteCorrelationConstraint(
equityShortRateCorr)
heston_process = HestonProcess(r_ts, q_ts, s0, start_v0, start_kappa,
start_theta, start_sigma, start_rho)
h_model = HestonModel(heston_process)
h_engine = AnalyticHestonEngine(h_model)
options = []
# first calibrate a heston model to get good initial
# parameters
for i in range(len(maturities)):
maturity = Period(int(maturities[i] * 12.0 + 0.5), Months)
for j, s in enumerate(strikes):
v = SimpleQuote(vol[i * len(strikes) + j])
helper = HestonModelHelper(maturity, calendar, s0.value,
s, v, r_ts, q_ts,
PriceError)
helper.set_pricing_engine(h_engine)
options.append(helper)
om = LevenbergMarquardt(1e-6, 1e-8, 1e-8)
# Heston model
h_model.calibrate(options, om,
EndCriteria(400, 40, 1.0e-8,
1.0e-4, 1.0e-8))
print("Heston calibration")
print("v0: %f" % h_model.v0)
print("theta: %f" % h_model.theta)
print("kappa: %f" % h_model.kappa)
print("sigma: %f" % h_model.sigma)
print("rho: %f" % h_model.rho)
###Output
Heston calibration
v0: 0.117725
theta: 0.082718
kappa: 1.478874
sigma: 0.352573
rho: -0.825268
###Markdown
The calibrated parameters are now used as starting point for the full Heston/Hull-White calibration.
###Code
h_process_2 = HestonProcess(r_ts, q_ts, s0, h_model.v0,
h_model.kappa,
h_model.theta,
h_model.sigma,
h_model.rho)
hhw_model = HestonModel(h_process_2)
options = []
for i in range(len(maturities)):
tGrid = np.max((10.0, maturities[i] * 10.0))
hhw_engine = FdHestonHullWhiteVanillaEngine(
hhw_model, hw_process,
equityShortRateCorr,
tGrid, 61, 13, 9, 0, True, FdmSchemeDesc.Hundsdorfer())
hhw_engine.enableMultipleStrikesCaching(strikes)
maturity = Period(int(maturities[i] * 12.0 + 0.5), Months)
# multiple strikes engine works best if the first option
# per maturity has the average strike (because the first
# option is priced first during the calibration and
# the first pricing is used to calculate the prices
# for all strikes
# list of strikes by distance from moneyness
indx = np.argsort(np.abs(np.array(strikes) - s0.value))
for j, tmp in enumerate(indx):
js = indx[j]
s = strikes[js]
v = SimpleQuote(vol[i * len(strikes) + js])
helper = HestonModelHelper(maturity,
calendar, s0.value,
strikes[js], v, r_ts, q_ts,
PriceError)
helper.set_pricing_engine(hhw_engine)
options.append(helper)
vm = LevenbergMarquardt(1e-6, 1e-2, 1e-2)
hhw_model.calibrate(options, vm,
EndCriteria(400, 40, 1.0e-8, 1.0e-4, 1.0e-8),
corrConstraint)
print("Heston HW calibration with FD engine")
print("v0: %f" % hhw_model.v0)
print("theta: %f" % hhw_model.theta)
print("kappa: %f" % hhw_model.kappa)
print("sigma: %f" % hhw_model.sigma)
print("rho: %f" % hhw_model.rho)
from tabulate import tabulate
table = {"Param": ['v0', 'theta', 'kappa', 'sigma', 'rho'],
"Estimated": [hhw_model.v0, hhw_model.theta, hhw_model.kappa, hhw_model.sigma,
hhw_model.rho],
"Exact": [0.12, 0.09, 2.0, 0.5, -0.75]}
print tabulate(table, headers='keys', floatfmt='.3f')
###Output
Estimated Exact Param
----------- ------- -------
0.120 0.120 v0
0.090 0.090 theta
1.980 2.000 kappa
0.502 0.500 sigma
-0.746 -0.750 rho
|
digital-image-processing/notebooks/transform/plot_rescale.ipynb
|
###Markdown
Rescale, resize, and downscale`Rescale` operation resizes an image by a given scaling factor. The scalingfactor can either be a single floating point value, or multiple values - onealong each axis.`Resize` serves the same purpose, but allows to specify an output image shapeinstead of a scaling factor.Note that when down-sampling an image, `resize` and `rescale` should performGaussian smoothing to avoid aliasing artifacts. See the `anti_aliasing` and`anti_aliasing_sigma` arguments to these functions.`Downscale` serves the purpose of down-sampling an n-dimensional image byinteger factors using the local mean on the elements of each block of the sizefactors given as a parameter to the function.
###Code
import matplotlib.pyplot as plt
from skimage import data, color
from skimage.transform import rescale, resize, downscale_local_mean
image = color.rgb2gray(data.astronaut())
image_rescaled = rescale(image, 0.25, anti_aliasing=False)
image_resized = resize(image, (image.shape[0] // 4, image.shape[1] // 4),
anti_aliasing=True)
image_downscaled = downscale_local_mean(image, (4, 3))
fig, axes = plt.subplots(nrows=2, ncols=2)
ax = axes.ravel()
ax[0].imshow(image, cmap='gray')
ax[0].set_title("Original image")
ax[1].imshow(image_rescaled, cmap='gray')
ax[1].set_title("Rescaled image (aliasing)")
ax[2].imshow(image_resized, cmap='gray')
ax[2].set_title("Resized image (no aliasing)")
ax[3].imshow(image_downscaled, cmap='gray')
ax[3].set_title("Downscaled image (no aliasing)")
ax[0].set_xlim(0, 512)
ax[0].set_ylim(512, 0)
plt.tight_layout()
plt.show()
###Output
_____no_output_____
|
_build/jupyter_execute/notebooks/doc/03 Deterministic Nonrenewable Resource Model.ipynb
|
###Markdown
Deterministic Nonrenewable Resource Model**Randall Romero Aguilar, PhD**This demo is based on the original Matlab demo accompanying the Computational Economics and Finance 2001 textbook by Mario Miranda and Paul Fackler.Original (Matlab) CompEcon file: **demdoc03.m**Running this file requires the Python version of CompEcon. This can be installed with pip by running !pip install compecon --upgradeLast updated: 2021-Oct-01 AboutWelfare maximizing social planner must decide the rate at which a nonrenewable resource should be harvested.* State - s resource stock* Control - q harvest rate* Parameters - κ harvest unit cost scale factor - γ harvest unit cost elasticity - η inverse elasticity of demand - 𝜌 continuous discount rate Preliminary tasks Import relevant packages
###Code
import pandas as pd
import matplotlib.pyplot as plt
from compecon import BasisChebyshev, OCmodel
###Output
_____no_output_____
###Markdown
Model parameters
###Code
κ = 10 # harvest unit cost scale factor
γ = 1 # harvest unit cost elasticity
η = 1.5 # inverse elasticity of demand
𝜌 = 0.05 # continuous discount rate
###Output
_____no_output_____
###Markdown
Approximation structure
###Code
n = 20 # number of basis functions
smin = 0.1 # minimum state
smax = 1.0 # maximum state
basis = BasisChebyshev(n, smin, smax, labels=['q']) # basis functions
###Output
_____no_output_____
###Markdown
Solve HJB equation by collocation
###Code
def control(s, Vs, κ, γ, η, 𝜌):
k = κ * s**(-γ)
return (Vs + k)**(-1/η)
def reward(s, q, κ, γ, η, 𝜌):
u = (1/(1-η)) * q **(1-η)
k = κ * s**(-γ)
return u - k*q
def transition(s, q, κ, γ, η, 𝜌):
return -q
model = OCmodel(basis, control, reward, transition, rho=𝜌, params=[κ, γ, η, 𝜌])
data = model.solve()
###Output
Solving optimal control model
iter change time
------------------------------
0 5.4e+02 0.0030
1 1.2e+02 0.0030
2 3.2e+01 0.0030
3 9.2e+00 0.0040
4 1.0e+00 0.0040
5 7.2e-03 0.0050
6 1.6e-06 0.0050
7 4.5e-09 0.0060
Elapsed Time = 0.01 Seconds
###Markdown
Plots Optimal policy
###Code
fig, ax = plt.subplots()
data['control'].plot(ax=ax)
ax.set(title='Optimal Harvest Policy',
xlabel='Resource Stock',
ylabel='Rate of Harvest',
xlim=[smin, smax])
ax.set_ylim(bottom=0)
###Output
_____no_output_____
###Markdown
Value function
###Code
fig, ax = plt.subplots()
data['value'].plot(ax=ax)
ax.set(title='Value Function',
xlabel='Resource Stock',
ylabel='Social Welfare',
xlim=[smin, smax])
###Output
_____no_output_____
###Markdown
Shadow price
###Code
data['shadow'] = model.Value(data.index, 1)
fig, ax = plt.subplots()
data['shadow'].plot(ax=ax)
ax.set(title='Shadow Price Function',
xlabel='Resource Stock',
ylabel='Shadow Price',
xlim=[smin, smax])
###Output
_____no_output_____
###Markdown
Residual
###Code
fig, ax = plt.subplots()
data['resid'].plot(ax=ax)
ax.set(title='HJB Equation Residual',
xlabel='Capital Stock',
ylabel='Residual',
xlim=[smin, smax]);
###Output
_____no_output_____
###Markdown
Simulate the model Initial state and time horizon
###Code
s0 = smax # initial capital stock
T = 40 # time horizon
###Output
_____no_output_____
###Markdown
Simulation and plot
###Code
fig, ax = plt.subplots()
model.simulate([s0], T).plot(ax=ax)
ax.set(title='Simulated Resource Stock and Rate of Harvest',
xlabel='Time',
ylabel='Quantity',
xlim=[0, T])
#ax.legend([]);
###Output
PARAMETER xnames NO LONGER VALID. SET labels= AT OBJECT CREATION
|
labs/Archived Labs/2-27/2-27 Model Selection solutions.ipynb
|
###Markdown
2-27: Intro to Model Selection Techniques---**Model Selection** is a key step in the machine learning process. Generally, you will have several candidate models that you fit to your data, and must select the one that you will use on out-of-sample data. These techniques help you determine which model is the "best."*Estimated Time: 30 minutes*---**Dependencies:**
###Code
import numpy as np
import scipy
from datascience import *
import datetime as dt
import pandas as pd
import matplotlib.pyplot as plt
%matplotlib inline
from sklearn.model_selection import train_test_split
from sklearn.linear_model import Ridge, Lasso, LinearRegression
from sklearn.model_selection import KFold
from sklearn.model_selection import cross_val_score
from sklearn.model_selection import cross_val_predict
from sklearn.metrics import r2_score
###Output
_____no_output_____
###Markdown
The Data: Bike Sharing We'll be using the same bike sharing data as last week!
###Code
bike=Table().read_table(('data/Bike-Sharing-Dataset/day.csv'))
# reformat the date column to integers representing the day of the year, 001-366
bike['dteday'] = pd.to_datetime(bike['dteday']).strftime('%j')
# get rid of the index column
bike = bike.drop(0)
bike.show(4)
###Output
_____no_output_____
###Markdown
1. Test-Train-Validation Split Recall that we typically want to split our data into training, validation, and test sets for the purposes of developing and tweaking our Machine Learning models. Below we reproduce the code from last lab:
###Code
# the features used to predict riders
X = bike.drop('casual', 'registered', 'cnt')
X = X.to_df()
# the number of riders
y = bike['cnt']
# set the random seed
np.random.seed(10)
# split the data
# train_test_split returns 4 values: X_train, X_test, y_train, y_test
X_train, X_test, y_train, y_test = train_test_split(X, y,
train_size=0.80, test_size=0.20)
# split the data
# Returns 4 values: X_train, X_validate, y_train, y_validate
X_train, X_validate, y_train, y_validate = train_test_split(X_train, y_train,
train_size=0.75, test_size=0.25)
###Output
_____no_output_____
###Markdown
2. Specify Regression Models Specify the linear regression, ridge, and lasso models that we explored in the last labs. This time you don't need to report the coefficients or plot the results.
###Code
# Linear Regression
## Create lin_reg method and fit model
lin_reg = LinearRegression(normalize=True)
lin_model = lin_reg.fit(X_train, y_train)
# Ridge
## Create ridge_reg method and fit model
ridge_reg = Ridge()
ridge_model = ridge_reg.fit(X_train, y_train)
# LASSO
## Create lasso_reg and fit
lasso_reg = Lasso(max_iter=10000)
lasso_model = lasso_reg.fit(X_train, y_train)
###Output
_____no_output_____
###Markdown
3. Information Criterion Approaches We now have three candidate models! First, let's implement the AIC and BIC approaches. Write code that calculates the AIC and BIC for each of the models. The (simplified) formulas for each are: $AIC = 2k - 2ln(sse)$$BIC = n*ln(sse/n) + k*ln(n)$Where $k$ is the number of features, $n$ is the number of observations, $ln$ is the natural log (hint: use np.log), and sse is the "sum of squared errors" or the squared residuals between the predicted and actual responses.
###Code
len(X_train.columns)
len(X_train.index)
# Columns
k = 12
# Rows
n = 438
lin_reg_hat = lin_model.predict(X_train)
lin_reg_resid = y_train - lin_reg_hat
lin_reg_sse = sum(lin_reg_resid**2)
lin_AIC = 2*k - 2*np.log(lin_reg_sse)
lin_AIC
ridge_reg_hat = ridge_model.predict(X_train)
ridge_reg_resid = y_train - ridge_reg_hat
ridge_reg_sse = sum(ridge_reg_resid**2)
ridge_AIC = 2*k - 2*np.log(ridge_reg_sse)
ridge_AIC
lasso_reg_hat = lasso_model.predict(X_train)
lasso_reg_resid = y_train - lasso_reg_hat
lasso_reg_sse = sum(lasso_reg_resid**2)
lasso_AIC = 2*k - 2*np.log(lasso_reg_sse)
lasso_AIC
###Output
_____no_output_____
###Markdown
Which model should we prefer based on the AIC? How confident are you about this choice? Next, calculate the BIC for each model.
###Code
lin_BIC = n*np.log(lin_reg_sse/n) + k*np.log(n)
lin_BIC
ridge_BIC = n*np.log(ridge_reg_sse/n) + k*np.log(n)
ridge_BIC
lasso_BIC = n*np.log(lasso_reg_sse/n) + k*np.log(n)
lasso_BIC
###Output
_____no_output_____
###Markdown
Which model should you prefer based on the BIC? Again, how confident are you about this choice? 4. Cross-Validation Next, let's try a cross-validation approach. The basic logic of cross-validation is as follows:1. Randomly split the data into k-folds2. Build the model on k-1 folds, then test on the last fold3. Record prediction error4. Cycle until each fold has served as the test set5. The average of the errors is the cv-errorLuckily a lot of this functionality is already packaged up for us in sklearn's [cross-validation methods](http://scikit-learn.org/stable/modules/cross_validation.html). First, generate predictions for each of the models by using "cross_val_predict." Use "cv = 3" and return the r^2 score, and plot the predicted vs. actual values.
###Code
lin_predicted = cross_val_predict(lin_reg, X, y, cv = 3)
r2_score(y, lin_predicted)
# plot the residuals on a scatter plot
plt.scatter(y, lin_predicted)
plt.title('Linear Model (OLS)')
plt.xlabel('actual value')
plt.ylabel('predicted value')
plt.show()
ridge_predicted = cross_val_predict(ridge_reg, X, y, cv = 3)
r2_score(y, ridge_predicted)
# plot the residuals on a scatter plot
plt.scatter(y, ridge_predicted)
plt.title('Linear Model (OLS)')
plt.xlabel('actual value')
plt.ylabel('predicted value')
plt.show()
lasso_predicted = cross_val_predict(lasso_reg, X, y, cv = 3)
r2_score(y, lasso_predicted)
# plot the residuals on a scatter plot
plt.scatter(y, lin_predicted)
plt.title('Linear Model (OLS)')
plt.xlabel('actual value')
plt.ylabel('predicted value')
plt.show()
###Output
_____no_output_____
###Markdown
How well does cross-validation fit the data? In general, explicitly calculating $r^2$ like this tends to overfit the data. The preferred method is to use "cross_val_score." Use "cross_val_score" to return the mean prediction from a 3-fold cross validation for each of the models. How do the $r^2$ metrics compare?Experiment with different [metrics](http://scikit-learn.org/stable/modules/model_evaluation.html), and select your preferred model. Keep in mind that higher return values are better than lower return values in this method.
###Code
print(cross_val_score(lin_reg, X, y, cv=3, scoring='r2').mean())
print(cross_val_score(ridge_reg, X, y, cv=3, scoring='r2').mean())
print(cross_val_score(lasso_reg, X, y, cv=3, scoring='r2').mean())
###Output
0.599906025528056
|
Moringa_Data_Science_Prep_W1_Independent_Project_2021_09_Veronica_Isiaho_SQL_Notebook.ipynb
|
###Markdown
Part 3: SQL Programming Glossary:
POP: total population for the subcounty
S_Yield_Ha: average yield for sorghum for the subcounty (Kg/Ha)
M_Yield_Ha: average yield for maize for the subcounty (Kg/Ha)
Crop_Area_Ha: total crop area for the subcounty (Ha)
S_Area_Ha: total sorghum crop area for the subcounty (Ha)
M_Area_Ha: total maize crop area for the subcounty (Ha)
S_Prod_Tot: total productivity for the sorghum for the subcounty (Kg)
M_Prod_Tot: total productivity for the maize for the subcounty (Kg)
###Code
%load_ext sql
%sql sqlite://
import csv
import pandas as pd
# Go to Colab drive, in the relevant file folder and
#Click on the three dots to copy the file path then write below:
Uganda = pd.read_csv('/content/drive/MyDrive/Colab Notebooks/ProjectDSP/Uganda_Subcounty_Crop_Yield.csv')
%sql DROP TABLE if EXISTS Uganda;
%sql PERSIST Uganda;
%%sql
SELECT * FROM Uganda LIMIT 5;
Uganda.head()
###Output
_____no_output_____
###Markdown
1. Display a list of Sub Counties and their population and areas
###Code
%%sql
SELECT SUBCOUNTY_NAME, POP, AREA
FROM Uganda;
###Output
* sqlite://
Done.
###Markdown
2. Sort the list of districts by total crop area (descending order)
###Code
%%sql
SELECT DISTRICT_NAME, Crop_Area_Ha
FROM Uganda
ORDER BY Crop_Area_Ha Desc;
###Output
* sqlite://
Done.
###Markdown
3. Select only the Sub counties from the Moroto district, order them alphabetically and show their production of sorghum
###Code
%%sql
SELECT SUBCOUNTY_NAME, S_Prod_Tot
FROM Uganda
WHERE DISTRICT_NAME = 'MOROTO'
ORDER BY SUBCOUNTY_NAME ASC;
###Output
* sqlite://
Done.
###Markdown
4. Compute the total Maize production per District
###Code
%%sql
SELECT DISTRICT_NAME, SUM(M_Prod_Tot) as M_Prod_Tot
FROM Uganda
GROUP BY DISTRICT_NAME;
###Output
* sqlite://
Done.
###Markdown
5. Compute the number of Sub counties where Maize is produced and the total Maize production per District
###Code
%%sql
SELECT DISTRICT_NAME, COUNT(SUBCOUNTY_NAME) as No_of_Subcounties, SUM(M_Prod_Tot) as M_Prod_Tot
FROM Uganda
GROUP BY DISTRICT_NAME
HAVING M_Prod_Tot > 1;
###Output
* sqlite://
Done.
###Markdown
6. Compute the overall Crop area in all Sub counties where population is over 20000
###Code
%%sql
SELECT Crop_Area_Ha, SUM(Crop_Area_Ha) as Overall_Crop_Area
FROM Uganda
WHERE POP IN (
SELECT POP
FROM Uganda
WHERE POP > 20000
);
%%sql
SELECT POP, Crop_Area_Ha
FROM Uganda
WHERE POP > 20000;
###Output
* sqlite://
Done.
###Markdown
7. Sort the Maize production in descending order by Districts, only taking into account Sub counties where Maize area is larger than Sorghum area, and display the number of Sub counties per district matching that criteria
###Code
%%sql
SELECT DISTRICT_NAME, M_Prod_Tot
FROM Uganda
ORDER BY M_Prod_Tot DESC;
###Output
* sqlite://
Done.
###Markdown
only taking into account Sub counties where Maize area is larger than Sorghum area and display the number of Sub counties per district matching that criteria
###Code
%%sql
SELECT DISTRICT_NAME, M_Area_Ha, M_Prod_Tot, SUBCOUNTY_NAME
FROM Uganda
WHERE M_Area_Ha > S_Area_Ha
ORDER BY DISTRICT_NAME;
###Output
* sqlite://
Done.
|
Jupyter_Test-Copy1.ipynb
|
###Markdown
Obtain Data and Install
###Code
#This will instlal the package in the actual jupyter notebook Kernel. [with_Jupyter] is obsolete if you start it out of jupyter, but for demo purpose left inside.
import sys
!{sys.executable} --version
!{sys.executable} -m pip install -e "."[with_jupyter] #. describes the path to the package, in this case its the same folder.
###Output
Python 3.6.7
###Markdown
Enter your twitter Creds here:
###Code
#Access
CONSUMER_KEY = ""
CONSUMER_SECRET = ""
ACCESS_TOKEN = ""
ACCESS_TOKEN_SECRET = ""
###Output
_____no_output_____
###Markdown
This will download the twitter data of major parties three weeks before European Parliament Election.
###Code
import yatclient as yat # works but calls have to be p_twitter.TweetAnalyzer?
import datetime
EuropawahlDate = datetime.date(2019, 5, 23)
ThreeWeeksBeforeDate = EuropawahlDate - datetime.timedelta(weeks=3)
today = datetime.date.today()
twitter_client = yat.TwitterClient(CONSUMER_KEY, CONSUMER_SECRET, ACCESS_TOKEN, ACCESS_TOKEN_SECRET, ["spdde", "fdp","die_Gruenen","afd","dieLinke","fwlandtag","diepartei"])
tweets = twitter_client.get_user_timeline_tweets(start_date = str(ThreeWeeksBeforeDate), end_date = str(EuropawahlDate))
analyzer_load = yat.TweetAnalyzer(tweets)
analyzer_load.write_to_csv("tweets{}.csv".format(today), encoding = "utf-8")
print("saved data to tweets{}.csv\n".format(today))
df = analyzer_load.get_dataframe()
df.head(2)
print("Timeframe from:\n",df.groupby(by=["author"]).date.min(),"to date\n\n",df.groupby(by=["author"]).date.max())
#Wenn Daten bereits vorhanden:
import yatclient as yat
analyzer = yat.TweetAnalyzer()
analyzer.read_from_csv("tweets2019-07-04.csv")
###Output
_____no_output_____
###Markdown
This also works: remember: restart -> run Access-> run this:from yatclient import TweetAnalyzer,TwitterClienttwitter_client = TwitterClient(CONSUMER_KEY, CONSUMER_SECRET, ACCESS_TOKEN, ACCESS_TOKEN_SECRET, ["spdde"])tweets = twitter_client.get_user_timeline_tweets(start_date = "2019-06-10", end_date = "2019-06-21")analyzer = TweetAnalyzer(tweets)analyzer.plot_trend(type="fav")analyzer.plot_bar(type='wct',count=50) This also works - remember: restart -> run Access-> run this:from yatclient import * uses __all__ list in __init__.py filename must be declared in this case eg.: twitter_client.TwitterClienttwitter_client = twitter_client.TwitterClient(CONSUMER_KEY, CONSUMER_SECRET, ACCESS_TOKEN, ACCESS_TOKEN_SECRET, ["rezomusik"])tweets = twitter_client.get_user_timeline_tweets(start_date = "2019-06-10", end_date = "2019-06-21")analyzer = tweet_analyzer.TweetAnalyzer(tweets)analyzer.plot_bar(type='wct',count=50)
###Code
df = analyzer.get_dataframe()
df.head()
df.author.unique()
df.author.describe()
tweets.count()
###Output
_____no_output_____
###Markdown
Testing with massive Trump Dataset:
###Code
import pandas as pd
trump30000tweets = pd.read_excel("import_tweets.xlsx")
trump30000tweets.head(1)
tweets="lel Error"
ExampleDF = p_twitter.TweetAnalyzer(tweets,df=trump30000tweets)
%time ExampleDF.bagofwords(on="tweets",extended_view=False)
%time ExampleDF.plot_bar(type="wct", count=50)
%time ExampleDF.plot_bar(type="wpt")
###Output
_____no_output_____
|
07-bigdata-spark-databricks/big_data_prague_intro.ipynb
|
###Markdown
**Inspiriation and sources:** Databricks intro tutorial and [Using Apache Spark 2.0 to Analyze the City of San Francisco's Open Data](https://www.youtube.com/watch?v=K14plpZgy_c) by Sameer Farooqui**Authors**: Alexander Fred-Ojala & Ikhlaq Sidhu Introduction to Spark and Big DataDatabricks is a platform for running Spark without complex cluster management or tedious maintenance tasks. Spark is a distributed computation framework for executing code in parallel across many different machines. Databricks is the Spark team's enterprise solution makes big data simple by providing Spark as a hosted solution. Databricks Terminology- ****Workspaces**** : Where you store the ****notebooks**** and ****libraries****.- ****Notebooks**** : Like Jupyter Notebooks that can run `Scala`, `Python`, `R`, `SQL`, or `Markdown`. Define language by `%[language name]` at the top of the cell. Connect to a cluster to run.- ****Dashboards**** can be created from ****notebooks**** as a way of displaying the output of cells without the code.- ****Libraries**** : Packages / Modules. You can install them via pypi.- ****Tables**** : Structured data, that can be stored in data lake / cloud storage. Stored on Cluster or cached in memory.- ****Clusters**** : Groups of computers that you treat as a single computer to perform operations on big sets of data.- ****Jobs**** : Schedule execution on ****notebooks**** or Python scripts. They can be created either manually or via the REST API.- ****Apps**** : 3rd party integrations with the Databricks platform like Tableau. Spark's historySpark was developed by founders of Databricks in AMPLab at UC Berkeley. Started 2009, donated to Apache open source in 2013. The Contexts/EnvironmentsBefore Spark 2.X many used the `sparkContext` made available as `sc` and the `SQLContext` made available as `sqlContext`. The `sqlContext` makes a lot of DataFrame functionality available while the `sparkContext` focuses more on the Apache Spark engine itself.In Spark 2.X, there is just one context - the `SparkSession`. The Data InterfacesKey interfaces.- ****The DataFrame**** : Collection of distributed `Row` types (note no indicies for look up). Similar to pandas or R dataframe.- ****The RDD (Resilient Distributed Dataset)**** : Interface to a sequence of data objects that consist of one or more types that are located across a variety of machines in a cluster. Focus on DataFrames as those will be supersets of the current RDD functionality.See speed difference: Spark is a unified processing engine that can analyze big data using SQL, machine learning, graph processing or real time stream analysis. Streaming (infinte Dataframe), Machine Learning, Graph / Pagerank. You can read from many different data sources and Spark runs on every major environment. We will use Amazon EC2. We will read CSV data. Stick with Dataframe and SQL. Let's StartBefore you start running code, you need to make sure that the notebook is attached to a cluster. To create a ClusterClick the Clusters button that you'll notice on the left side of the page. On the Clusters page, click on  in the upper left corner.Then, on the Create Cluster dialog, enter the configuration for the new cluster.Finally, - Select a unique name for the cluster.- Select the most recent stable Runtime Version.- Enter the number of workers to bring up - at least 1 is required to run Spark commands.**Go back to the notebook and in the top right corner press Detached and connect to your cluster.***Note, Databricks community clusters only run for an hour* first let's explore the previously mentioned `SparkSession` where info is stored. We can access it via the `spark` variable.
###Code
spark
###Output
_____no_output_____
###Markdown
We can use the spark context to parallelize a small Python range that will provide a return type of `DataFrame`.
###Code
firstDataFrame = spark.range(10000)
print(firstDataFrame) # if you just run a transformation no Spark Job is done.
# or use RDD through sc (spark context)
spark.sparkContext.parallelize(range(1000))
###Output
_____no_output_____
###Markdown
Now one might think that this would actually print the values parallelized. That's not how Spark works.Spark allows two distinct kinds of operations, **transformations** and **actions**. TransformationsTransformations will only get executed once you have called a **action**. An example of a transformation might be to convert an integer into a float or to filter a set of values. I.e. Lazy Evaluation. ActionsActions are computed during execution. Run all of the previous transformations in order to get back an actual result. An action is composed of one or more jobs which consists of tasks that will be executed by the workers in parallel where possible.Sshort sample of actions and transformations:
###Code
firstDataFrame.show(3) # example of an action, dataframe is now evaluated
# An example of a transformation
# select the ID column values and multiply them by 2, SQL interfac
secondDataFrame = firstDataFrame.selectExpr("(id * 2) as value")
secondDataFrame.show(5)
from pyspark.sql.functions import col # to select columns
firstDataFrame.withColumn('id2', col('id')*2).show(3)
# Or common before Spark 2.X
firstDataFrame.rdd.map(lambda x: x[0]*2).take(3)
# or
firstDataFrame.take(5)
# or
display(firstDataFrame)
###Output
_____no_output_____
###Markdown
Transformations are lazily evaluated because it is easy to optimize the entire pipeline of computations this way. Computations can be parallellized and executed on many different nodes at once (like a map and a filter).Spark also keeps results in memory, as opposed to other frameworks (e.g. Hadoop Map Reduce) that write to disk. Spark ArchitectureSpark allows you to treat many machines as one via a master-worker architecture.There is `driver` or master node in the cluster, accompanied by `worker` nodes. The master sends work to the workers and either instructs them to pull to data from memory or from disk (or from another data source).Spark Cluster has a Driver node that communicates with executor nodes. Executor nodes are logically like execution cores. The Driver sends Tasks to the empty slots on the Executors when work has to be done:Note: In the case of the Community Edition there is no worker, the master executes the entire code. However, the same code works on any cluster (beware of CPU / GPU frameworks).Access details in the web UI by clicking at the top left of this notebook. Working example with dataTo illustrate **transformations** and **actions** - let's go through an example using `DataFrames` and a csv file of a public dataset that Databricks makes available. Available using the Databricks filesystem. Let's load the popular diamonds dataset in as a spark `DataFrame`. Now let's go through the dataset that we'll be working with. Use `%fs` to interact with the spark filesystem
###Code
%fs ls /databricks-datasets/Rdatasets/data-001/datasets.csv
dataPath = "/databricks-datasets/Rdatasets/data-001/csv/ggplot2/diamonds.csv"
diamonds = spark.read.format("csv")\
.option("header","true")\
.option("inferSchema", "true")\
.load(dataPath)
# inferSchema means we will automatically figure out column types
# at a cost of reading the data more than once
###Output
_____no_output_____
###Markdown
Show the dataframe with Databricks `display` function or the show function.
###Code
display(diamonds)
display(diamonds.limit(5)) # for a subset
diamonds.printSchema() # see that the column types are OK and schema inferred correctly.
diamonds.rdd.getNumPartitions() # only one partition. This dataframe does not exist in memory. For big data several partitions.
# Partitions can be optimized according to your cluster size. Have it divisible by cluster size.
# For community edition, any number * 3 is OK
# you can use REPARTITION method
diamonds.count() # reads through the whole data set
display(diamonds.summary())
diamonds.select('cut').distinct().show() # show unique entries in the cut column
###Output
_____no_output_____
###Markdown
What makes `display` exceptional is the fact that we can very easily create some more sophisticated graphs by clicking the graphing icon that you can see below. Here's a plot that allows us to compare price, color, and cut.
###Code
display(diamonds)
# most common cut, ordered. First interesting insight.
display(diamonds.select('cut').groupBy('cut').count().orderBy('count',ascending=False))
display(diamonds.select('price','cut').groupBy('cut').avg('price')) # show graph, prepares 5 jobs
###Output
_____no_output_____
###Markdown
Now that we've explored the data, let's return to understanding **transformations** and **actions**. First transformations, then actions.First we group by two variables, cut and color and then compute the average price. Then we're going to inner join that to the original dataset on the column `color`. Then we'll select the average price as well as the carat.
###Code
df1 = diamonds.groupBy("cut", "color").avg("price") # a simple grouping
df2 = df1\
.join(diamonds, on='color', how='inner')\
.select("`avg(price)`", "carat")
# a simple join and selecting some columns
###Output
_____no_output_____
###Markdown
These transformations are now complete in a sense but nothing has happened.The reason for that is these computations are *lazy* in order to build up the entire flow of data from start to finish required by the user. This is an intelligent optimization for two key reasons. Any calculation can be recomputed from the very source data allowing Apache Spark to handle any failures that occur along the way, successfully handle stragglers. Secondly, Spark can optimize computation so that data and computation can be `pipelined`.To get a sense for what this plan consists of, we can use the `explain` method.
###Code
df2.explain()
###Output
_____no_output_____
###Markdown
Now explaining the above results is outside of this introductory tutorial. This is Spark's plan for how it hopes to execute the given query.
###Code
df2.count()
###Output
_____no_output_____
###Markdown
This will execute the plan that Apache Spark built up previously. Click the little arrow next to where it says `(X) Spark Jobs` after that cell finishes executing and then click the `View` link. This brings up the Apache Spark Web UI right inside of your notebook.These are significant visualizations called Directed Acyclic Graphs (DAG)s of all the computations that have to be performed in order to get to that result. Transformations are *lazy* - while generating this series of steps Spark will optimize lots of things, one of core reasons that users should be focusing on using DataFrames and Datasets instead of the legacy RDD API. With DataFrames and Datasets, Apache Spark will work under the hood to optimize the entire query plan and pipeline entire steps together. SQL view
###Code
diamonds.repartition(3).createOrReplaceTempView("diamondsView") # also repartition, create a table view for SQL
diamonds.count()
%sql SELECT carat, cut, color from diamondsView ORDER BY carat DESC;
# in jupyter
spark.sql('SELECT * FROM diamondsView').show()
###Output
_____no_output_____
###Markdown
To pandas DataFrame
###Code
import pandas as pd
pd_df = diamonds.toPandas()
pd_df.head(5)
type(pd_df)
###Output
_____no_output_____
###Markdown
CachingSpark can store things in memory during computation. Can speed up access to commonly queried tables or pieces of data. This is also great for iterative algorithms that work over and over again on the same data.To cache a DataFrame or RDD, simply use the cache method.
###Code
df2.cache() # look in the UI / Storage
###Output
_____no_output_____
###Markdown
Caching, like a transformation, is performed lazily, won't store the data in memory until you call an action on that dataset. Here's a simple example. We've created our df2 DataFrame which is essentially a logical plan that tells us how to compute that exact DataFrame. We've told Apache Spark to cache that data after we compute it for the first time. So let's call a full scan of the data with a count twice. The first time, this will create the DataFrame, cache it in memory, then return the result. The second time, rather than recomputing that whole DataFrame, it will just hit the version that it has in memory.Let's take a look at how we can discover this.
###Code
df2.count() # read all data and then materialize, cache it in memory
#
# Tungsten method to cache DataFrame into memory, makes it smaller.
# Optimize by repartitioning according to your cluster also
# Optimal partition sizes are 50-100Mb
###Output
_____no_output_____
###Markdown
However after we've now counted the data. We'll see that the explain ends up being quite different.
###Code
df2.count()
###Output
_____no_output_____
###Markdown
In the above example, we can see that this cuts down on the time needed to generate this data immensely - often by at least an order of magnitude. With much larger and more complex data analysis, the gains that we get from caching can be even greater!
###Code
%fs ls /tmp/
# to save work and dataframe save as a Parquet file
diamonds.write.format('parquet').save('/tmp/diamonds/')
%fs ls /tmp/diamonds/
# Easily continue work if the cluster is shutdown, link to folder:
diamonds2 = spark.read.parquet('/tmp/diamonds/')
diamonds2.show() # will include all partitioning, cache into memory etc.
# parque files are really efficient to read from. Always take CSV or JSON, do the ETL and then write to Parquet file.
###Output
_____no_output_____
|
Crash_Course_ML_Google/colab_example/First_Steps_TensorFlow/Validation_and_Test_Sets.ipynb
|
###Markdown
###Code
#@title Copyright 2020 Google LLC. Double-click here for license information.
# Licensed under the Apache License, Version 2.0 (the "License");
# you may not use this file except in compliance with the License.
# You may obtain a copy of the License at
#
# https://www.apache.org/licenses/LICENSE-2.0
#
# Unless required by applicable law or agreed to in writing, software
# distributed under the License is distributed on an "AS IS" BASIS,
# WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.
# See the License for the specific language governing permissions and
# limitations under the License.
###Output
_____no_output_____
###Markdown
Validation Sets and Test SetsThe previous Colab exercises evaluated the trained model against the training set, which does not provide a strong signal about the quality of your model. In this Colab, you'll experiment with validation sets and test sets. Learning objectivesAfter doing this Colab, you'll know how to do the following: * Split a [training set](https://developers.google.com/machine-learning/glossary/training_set) into a smaller training set and a [validation set](https://developers.google.com/machine-learning/glossary/validation_set). * Analyze deltas between training set and validation set results. * Test the trained model with a [test set](https://developers.google.com/machine-learning/glossary/test_set) to determine whether your trained model is [overfitting](https://developers.google.com/machine-learning/glossary/overfitting). * Detect and fix a common training problem. The datasetAs in the previous exercise, this exercise uses the [California Housing dataset](https://developers.google.com/machine-learning/crash-course/california-housing-data-description) to predict the `median_house_value` at the city block level. Like many "famous" datasets, the California Housing Dataset actually consists of two separate datasets, each living in separate .csv files:* The training set is in `california_housing_train.csv`.* The test set is in `california_housing_test.csv`.You'll create the validation set by dividing the downloaded training set into two parts:* a smaller training set * a validation set Use the right version of TensorFlowThe following hidden code cell ensures that the Colab will run on TensorFlow 2.X.
###Code
#@title Run on TensorFlow 2.x
%tensorflow_version 2.x
###Output
_____no_output_____
###Markdown
Import relevant modulesAs before, this first code cell imports the necessary modules and sets a few display options.
###Code
#@title Import modules
import numpy as np
import pandas as pd
import tensorflow as tf
from matplotlib import pyplot as plt
pd.options.display.max_rows = 10
pd.options.display.float_format = "{:.1f}".format
###Output
_____no_output_____
###Markdown
Load the datasets from the internetThe following code cell loads the separate .csv files and creates the following two pandas DataFrames:* `train_df`, which contains the training set.* `test_df`, which contains the test set.
###Code
train_df = pd.read_csv("https://download.mlcc.google.com/mledu-datasets/california_housing_train.csv")
test_df = pd.read_csv("https://download.mlcc.google.com/mledu-datasets/california_housing_test.csv")
###Output
_____no_output_____
###Markdown
Scale the label valuesThe following code cell scales the `median_house_value`. See the previous Colab exercise for details.
###Code
scale_factor = 1000.0
# Scale the training set's label.
train_df["median_house_value"] /= scale_factor
# Scale the test set's label
test_df["median_house_value"] /= scale_factor
###Output
_____no_output_____
###Markdown
Load the functions that build and train a modelThe following code cell defines two functions: * `build_model`, which defines the model's topography. * `train_model`, which will ultimately train the model, outputting not only the loss value for the training set but also the loss value for the validation set. Since you don't need to understand model building code right now, we've hidden this code cell. As always, you must run hidden code cells.
###Code
#@title Define the functions that build and train a model
def build_model(my_learning_rate):
"""Create and compile a simple linear regression model."""
# Most simple tf.keras models are sequential.
model = tf.keras.models.Sequential()
# Add one linear layer to the model to yield a simple linear regressor.
model.add(tf.keras.layers.Dense(units=1, input_shape=(1,)))
# Compile the model topography into code that TensorFlow can efficiently
# execute. Configure training to minimize the model's mean squared error.
model.compile(optimizer=tf.keras.optimizers.RMSprop(lr=my_learning_rate),
loss="mean_squared_error",
metrics=[tf.keras.metrics.RootMeanSquaredError()])
return model
def train_model(model, df, feature, label, my_epochs,
my_batch_size=None, my_validation_split=0.1):
"""Feed a dataset into the model in order to train it."""
history = model.fit(x=df[feature],
y=df[label],
batch_size=my_batch_size,
epochs=my_epochs,
validation_split=my_validation_split)
# Gather the model's trained weight and bias.
trained_weight = model.get_weights()[0]
trained_bias = model.get_weights()[1]
# The list of epochs is stored separately from the
# rest of history.
epochs = history.epoch
# Isolate the root mean squared error for each epoch.
hist = pd.DataFrame(history.history)
rmse = hist["root_mean_squared_error"]
return epochs, rmse, history.history
print("Defined the build_model and train_model functions.")
###Output
_____no_output_____
###Markdown
Define plotting functionsThe `plot_the_loss_curve` function plots loss vs. epochs for both the training set and the validation set.
###Code
#@title Define the plotting function
def plot_the_loss_curve(epochs, mae_training, mae_validation):
"""Plot a curve of loss vs. epoch."""
plt.figure()
plt.xlabel("Epoch")
plt.ylabel("Root Mean Squared Error")
plt.plot(epochs[1:], mae_training[1:], label="Training Loss")
plt.plot(epochs[1:], mae_validation[1:], label="Validation Loss")
plt.legend()
# We're not going to plot the first epoch, since the loss on the first epoch
# is often substantially greater than the loss for other epochs.
merged_mae_lists = mae_training[1:] + mae_validation[1:]
highest_loss = max(merged_mae_lists)
lowest_loss = min(merged_mae_lists)
delta = highest_loss - lowest_loss
print(delta)
top_of_y_axis = highest_loss + (delta * 0.05)
bottom_of_y_axis = lowest_loss - (delta * 0.05)
plt.ylim([bottom_of_y_axis, top_of_y_axis])
plt.show()
print("Defined the plot_the_loss_curve function.")
###Output
_____no_output_____
###Markdown
Task 1: Experiment with the validation splitIn the following code cell, you'll see a variable named `validation_split`, which we've initialized at 0.2. The `validation_split` variable specifies the proportion of the original training set that will serve as the validation set. The original training set contains 17,000 examples. Therefore, a `validation_split` of 0.2 means that:* 17,000 * 0.2 ~= 3,400 examples will become the validation set.* 17,000 * 0.8 ~= 13,600 examples will become the new training set.The following code builds a model, trains it on the training set, and evaluates the built model on both:* The training set.* And the validation set.If the data in the training set is similar to the data in the validation set, then the two loss curves and the final loss values should be almost identical. However, the loss curves and final loss values are **not** almost identical. Hmm, that's odd. Experiment with two or three different values of `validation_split`. Do different values of `validation_split` fix the problem?
###Code
# The following variables are the hyperparameters.
learning_rate = 0.08
epochs = 30
batch_size = 100
# Split the original training set into a reduced training set and a
# validation set.
validation_split=0.2
# Identify the feature and the label.
my_feature="median_income" # the median income on a specific city block.
my_label="median_house_value" # the median value of a house on a specific city block.
# That is, you're going to create a model that predicts house value based
# solely on the neighborhood's median income.
# Discard any pre-existing version of the model.
my_model = None
# Invoke the functions to build and train the model.
my_model = build_model(learning_rate)
epochs, rmse, history = train_model(my_model, train_df, my_feature,
my_label, epochs, batch_size,
validation_split)
plot_the_loss_curve(epochs, history["root_mean_squared_error"],
history["val_root_mean_squared_error"])
###Output
_____no_output_____
###Markdown
Task 2: Determine **why** the loss curves differNo matter how you split the training set and the validation set, the loss curves differ significantly. Evidently, the data in the training set isn't similar enough to the data in the validation set. Counterintuitive? Yes, but this problem is actually pretty common in machine learning. Your task is to determine **why** the loss curves aren't highly similar. As with most issues in machine learning, the problem is rooted in the data itself. To solve this mystery of why the training set and validation set aren't almost identical, write a line or two of [pandas code](https://colab.research.google.com/drive/1gUeYFsYmoyqpQJWq7krrZZNUFvBPwrJf) in the following code cell. Here are a couple of hints: * The previous code cell split the original training set into: * a reduced training set (the original training set - the validation set) * the validation set * By default, the pandas [`head`](https://pandas.pydata.org/pandas-docs/stable/reference/api/pandas.DataFrame.head.html) method outputs the *first* 5 rows of the DataFrame. To see more of the training set, specify the `n` argument to `head` and assign a large positive integer to `n`.
###Code
# Write some code in this code cell.
#@title Double-click for a possible solution to Task 2.
# Examine examples 0 through 4 and examples 25 through 29
# of the training set
train_df.head(n=1000)
# The original training set is sorted by longitude.
# Apparently, longitude influences the relationship of
# total_rooms to median_house_value.
###Output
_____no_output_____
###Markdown
Task 3. Fix the problemTo fix the problem, shuffle the examples in the training set before splitting the examples into a training set and validation set. To do so, take the following steps:1. Shuffle the data in the training set by adding the following line anywhere before you call `train_model` (in the code cell associated with Task 1):``` shuffled_train_df = train_df.reindex(np.random.permutation(train_df.index))``` 2. Pass `shuffled_train_df` (instead of `train_df`) as the second argument to `train_model` (in the code call associated with Task 1) so that the call becomes as follows:``` epochs, rmse, history = train_model(my_model, shuffled_train_df, my_feature, my_label, epochs, batch_size, validation_split)```
###Code
#@title Double-click to view the complete implementation.
# The following variables are the hyperparameters.
learning_rate = 0.08
epochs = 70
batch_size = 100
# Split the original training set into a reduced training set and a
# validation set.
validation_split=0.2
# Identify the feature and the label.
my_feature="median_income" # the median income on a specific city block.
my_label="median_house_value" # the median value of a house on a specific city block.
# That is, you're going to create a model that predicts house value based
# solely on the neighborhood's median income.
# Discard any pre-existing version of the model.
my_model = None
# Shuffle the examples.
shuffled_train_df = train_df.reindex(np.random.permutation(train_df.index))
# Invoke the functions to build and train the model. Train on the shuffled
# training set.
my_model = build_model(learning_rate)
epochs, rmse, history = train_model(my_model, shuffled_train_df, my_feature,
my_label, epochs, batch_size,
validation_split)
plot_the_loss_curve(epochs, history["root_mean_squared_error"],
history["val_root_mean_squared_error"])
###Output
_____no_output_____
###Markdown
Experiment with `validation_split` to answer the following questions:* With the training set shuffled, is the final loss for the training set closer to the final loss for the validation set? * At what range of values of `validation_split` do the final loss values for the training set and validation set diverge meaningfully? Why?
###Code
#@title Double-click for the answers to the questions
# Yes, after shuffling the original training set,
# the final loss for the training set and the
# validation set become much closer.
# If validation_split < 0.15,
# the final loss values for the training set and
# validation set diverge meaningfully. Apparently,
# the validation set no longer contains enough examples.
###Output
_____no_output_____
###Markdown
Task 4: Use the Test Dataset to Evaluate Your Model's PerformanceThe test set usually acts as the ultimate judge of a model's quality. The test set can serve as an impartial judge because its examples haven't been used in training the model. Run the following code cell to evaluate the model with the test set:
###Code
x_test = test_df[my_feature]
y_test = test_df[my_label]
results = my_model.evaluate(x_test, y_test, batch_size=batch_size)
###Output
_____no_output_____
###Markdown
Compare the root mean squared error of the model when evaluated on each of the three datasets:* training set: look for `root_mean_squared_error` in the final training epoch.* validation set: look for `val_root_mean_squared_error` in the final training epoch.* test set: run the preceding code cell and examine the `root_mean_squred_error`.Ideally, the root mean squared error of all three sets should be similar. Are they?
###Code
#@title Double-click for an answer
# In our experiments, yes, the rmse values
# were similar enough.
###Output
_____no_output_____
|
slides/Slides.ipynb
|
###Markdown
Bots!=====- Lorem Ipsum- Lorem Ipusm- Lorem Ipssum
###Code
def code_example(idea):
return better_idea
###Output
_____no_output_____
|
rfm-cltv-analysis-with-online-retail-data.ipynb
|
###Markdown
RFM Analysis
###Code
# importing libraries
import datetime as dt
import pandas as pd
pd.set_option('display.max_columns', None) # tüm sütunları gösterir
# pd.set_option('display.max_rows', None) # tüm satırları gösterir
pd.set_option('display.float_format', lambda x: '%.4f' % x) # numeric değerleri float virgülden sonra 4 hane gösterir
pip install xlrd==1.2.0
pip install openpyxl
# reading the data
df_ = pd.read_excel("../input/uci-online-retail-ii-data-set/online_retail_II.xlsx", sheet_name="Year 2010-2011")
df = df_.copy()
df.head()
df.info()
# descriptive statistics info
df.describe().T
# empty value checking
df.isnull().sum()
# re-check
df.isnull().sum()
# unique values count
df["StockCode"].nunique()
df.groupby('StockCode').agg({'Quantity': "sum"})
# top 5 product after sorting
df.groupby('StockCode').agg({'Quantity': "sum"}).sort_values(by="Quantity", ascending=False).head(5)
# right now the returned good invoices still in dataframe but we have to discard them
df = df[~df["Invoice"].str.contains("C", na=False)]
# new observation for RFM metrics calculation
df["TotalPrice"] = df["Price"] * df["Quantity"]
df.groupby("Invoice").agg({"TotalPrice": "sum"})
###Output
_____no_output_____
###Markdown
RFM metrics calculation Recency = Shows how many days have passed since the last invoice Frequency = Count of how many times they buy Monetary = monetary value
###Code
today_date = dt.datetime(2011, 12, 11)
rfm = df.groupby('Customer ID').agg({'InvoiceDate': lambda date: (today_date - date.max()).days,
'Invoice': lambda num: num.nunique(),
'TotalPrice': lambda total: total.sum()})
rfm.columns = ["Recency", "Frequency", "Monetary"]
rfm = rfm[rfm["Monetary"] > 0]
# normalise the data from 1 to 5
rfm["recency_score"] = pd.qcut(rfm["Recency"], 5, labels=[5, 4, 3, 2, 1])
rfm["frequency_score"] = pd.qcut(rfm["Frequency"].rank(method="first"), 5, labels=[1, 2, 3, 4, 5])
rfm["monetary_score"] = pd.qcut(rfm["Monetary"], 5, labels=[1, 2, 3, 4, 5])
rfm["RFM_SCORE"] = (rfm['recency_score'].astype(str) +
rfm['frequency_score'].astype(str) +
rfm['monetary_score'].astype(str)).astype(int)
###Output
_____no_output_____
###Markdown
 this photo shows us the classification of RM (Recency nd Monetary) metrics
###Code
# classification map
seg_map = {
r'[1-2][1-2]': 'hibernating',
r'[1-2][3-4]': 'at_Risk',
r'[1-2]5': 'cant_loose',
r'3[1-2]': 'about_to_sleep',
r'33': 'need_attention',
r'[3-4][4-5]': 'loyal_customers',
r'41': 'promising',
r'51': 'new_customers',
r'[4-5][2-3]': 'potential_loyalists',
r'5[4-5]': 'champions'
}
rfm['segment'] = (rfm['recency_score'].astype(str) + rfm['frequency_score'].astype(str)).replace(seg_map, regex=True)
rfm.head()
rfm.groupby(["segment"]).agg({"RFM_SCORE": "mean"}).sort_values(by="RFM_SCORE", ascending=False)
###Output
_____no_output_____
###Markdown
CLTV
###Code
pip install lifetimes
from lifetimes import BetaGeoFitter
from lifetimes import GammaGammaFitter
from sklearn.preprocessing import MinMaxScaler
df = df_.copy()
df["Country"].unique()
df.describe().T
df.isnull().sum()
df.dropna(inplace=True)
df = df[~df["Invoice"].str.contains("C", na=False)]
df["TotalPrice"] = df["Quantity"] * df["Price"]
today_date = dt.datetime(2011, 12, 11)
# the difference from FRM to CLTV is we also checking the custom life value with first invoice to last invoice)
cltv_df = df.groupby('Customer ID').agg({'InvoiceDate': [lambda date: (date.max() - date.min()).days,
lambda date: (today_date - date.min()).days],
'Invoice': lambda num: num.nunique(),
'TotalPrice': lambda TotalPrice: TotalPrice.sum()})
cltv_df.columns = ['recency', 'T', 'frequency', 'monetary']
cltv_df.head()
cltv_df["monetary"] = cltv_df["monetary"] / cltv_df["frequency"]
cltv_df = cltv_df[cltv_df["monetary"] > 0]
# Expression of recency and T for BGNBD in weekly terms
cltv_df["recency"] = cltv_df["recency"] / 7
cltv_df["T"] = cltv_df["T"] / 7
# frequency must be bigger then 1
cltv_df = cltv_df[(cltv_df['frequency'] > 1)]
###Output
_____no_output_____
###Markdown
Establishment of BG-NBD Model
###Code
bgf = BetaGeoFitter(penalizer_coef=0.001)
bgf.fit(cltv_df['frequency'],
cltv_df['recency'],
cltv_df['T'])
###Output
_____no_output_____
###Markdown
Establishment of GAMMA- GAMMA Model
###Code
ggf = GammaGammaFitter(penalizer_coef=0.01)
ggf.fit(cltv_df['frequency'], cltv_df['monetary'])
###Output
_____no_output_____
###Markdown
Calculation of CLTV with BG-NBD and GG model.
###Code
cltv = ggf.customer_lifetime_value(bgf,
cltv_df['frequency'],
cltv_df['recency'],
cltv_df['T'],
cltv_df['monetary'],
time=6, # for 6 months
freq="W",
discount_rate=0.01)
cltv.head()
cltv = cltv.reset_index()
cltv.columns = ["Customer ID", "clv"]
cltv.sort_values(by="clv", ascending=False).head(50)
cltv_final = cltv_df.merge(cltv, on="Customer ID", how="left")
cltv_final.sort_values(by="clv", ascending=False).head(10)
# Standardization of CLTV
scaler = MinMaxScaler(feature_range=(0, 1)) # verileri okunabilir hale getirme amaçlı
scaler.fit(cltv_final[["clv"]])
cltv_final["scaled_clv"] = scaler.transform(cltv_final[["clv"]])
# sorting:
cltv_final.sort_values(by="scaled_clv", ascending=False).head()
cltv_final_copy = cltv_final.copy()
###Output
_____no_output_____
###Markdown
Comment: It is predicted that the estimated profit that customer "18102" will get at the end of 6 months will be "85,651" unit
###Code
###### CLTV for 1 Months #########
cltv = ggf.customer_lifetime_value(bgf,
cltv_df['frequency'],
cltv_df['recency'],
cltv_df['T'],
cltv_df['monetary'],
time=1,
freq="W",
discount_rate=0.01)
cltv.head()
cltv = cltv.reset_index()
cltv.columns = ["Customer ID", "clv"]
cltv.sort_values(by="clv", ascending=False).head(50)
cltv_final = cltv_df.merge(cltv, on="Customer ID", how="left")
cltv_final.sort_values(by="clv", ascending=False).head(10)
# Standardization of CLTV
scaler = MinMaxScaler(feature_range=(0, 1))
scaler.fit(cltv_final[["clv"]])
cltv_final["scaled_clv"] = scaler.transform(cltv_final[["clv"]])
cltv_final.sort_values(by="scaled_clv", ascending=False).head(10)
###### CLTV for 12 Months #########
cltv = ggf.customer_lifetime_value(bgf,
cltv_df['frequency'],
cltv_df['recency'],
cltv_df['T'],
cltv_df['monetary'],
time=12,
freq="W",
discount_rate=0.01)
cltv.head()
cltv = cltv.reset_index()
cltv.columns = ["Customer ID", "clv"]
cltv.sort_values(by="clv", ascending=False).head(50)
cltv_final = cltv_df.merge(cltv, on="Customer ID", how="left")
cltv_final.sort_values(by="clv", ascending=False).head(10)
# Standardization of CLTV
scaler = MinMaxScaler(feature_range=(0, 1))
scaler.fit(cltv_final[["clv"]])
cltv_final["scaled_clv"] = scaler.transform(cltv_final[["clv"]])
# sorting:
cltv_final.sort_values(by="scaled_clv", ascending=False).head(10)
###Output
_____no_output_____
|
Similarity/Jupyter/20_upload_ui.ipynb
|
###Markdown
Copyright (c) Microsoft Corporation. All rights reserved.Licensed under the MIT License. Upload files for HTML User InterfaceIn the notebook [13_image_similarity_export.ipynb](13_image_similarity_export.ipynb) we exported reference image features, reference image file names, and reference image thumbnails. In this notebook we upload those items, as well as our simplified HTML interface, to your Azure Blob storage account for easy public access. You should create an Azure Storage Account and a "Container" in that account to store your uploaded files. Initialization
###Code
# Ensure edits to libraries are loaded and plotting is shown in the notebook.
%matplotlib inline
%reload_ext autoreload
%autoreload 2
# Standard python libraries
import sys
from pathlib import Path
from tqdm.notebook import trange, tqdm
import os, uuid
import azure.storage.blob
from azure.storage.blob import BlobServiceClient, BlobClient, ContainerClient, ContentSettings
# Check Storage SDK version number
print(f"Azure Blob Storage SDK Version: {azure.storage.blob.VERSION}")
###Output
Azure Blob Storage SDK Version: 12.2.0
###Markdown
First we setup variables to point to your Azure Blob storage account and your existing Blob container. May be best to setup a fresh Blob container for this upload.
###Code
AZURE_ACCOUNT_NAME = "YOUR ACCOUNT NAME"
AZURE_ACCOUNT_KEY = "YOUR ACCOUNT ACCESS KEY"
BLOB_CONTAINER_NAME = "YOUR CONTAINER NAME"
ENDPOINT_SUFFIX = "core.windows.net"
###Output
_____no_output_____
###Markdown
Next we upload the files to your Azure Blob storage.
###Code
azure_storage_connection_str = "DefaultEndpointsProtocol=https;AccountName={};AccountKey={};EndpointSuffix={}".format(AZURE_ACCOUNT_NAME, AZURE_ACCOUNT_KEY, ENDPOINT_SUFFIX)
container_name = BLOB_CONTAINER_NAME
local_files = ['../visualize/data/ref_filenames.zip','../visualize/data/ref_features.zip','../visualize/index.html','../visualize/dist/jszip.min.js','../visualize/dist/jszip-utils.min.js']
blob_files = ['data/ref_filenames.zip','data/ref_features.zip','index.html','dist/jszip.min.js','dist/jszip-utils.min.js']
# Create the BlobServiceClient object which will be used to create a container client
blob_service_client = BlobServiceClient.from_connection_string(azure_storage_connection_str)
# Get total size of non-image files to upload
sizecounter = 0
for file in local_files:
sizecounter += os.stat(file).st_size
print("Uploading non-image files:")
# # Upload the individual files for the front-end and the ZIP files for reference features
i = 0
with tqdm(total=sizecounter, unit='B', unit_scale=True, unit_divisor=1024) as pbar:
while (i < len(local_files)):
# Create a blob client using the local file name as the name for the blob
blob_client = blob_service_client.get_blob_client(container=container_name, blob=blob_files[i])
# Upload the file
with open(local_files[i], "rb") as data:
buf = 0
buf = os.stat(local_files[i]).st_size
if (i==2):
blob_client.upload_blob(data, overwrite=True, content_settings=ContentSettings(content_type="text/html"))
else:
blob_client.upload_blob(data, overwrite=True)
if buf:
pbar.update(buf)
i+=1
# Upload the thumbnail versions of the reference images
path_blob = 'small-150'
path_local = '../visualize/{}'.format(path_blob)
# Get total size of all image files to upload
sizecounter = 0
for root, dirs, files in os.walk(path_local):
for file in files:
sizecounter += os.stat(os.path.join(path_local, file)).st_size
print("Uploading thumbnail image files:")
with tqdm(total=sizecounter, unit='B', unit_scale=True, unit_divisor=1024) as pbar:
for root, dirs, files in os.walk(path_local):
for file in files:
blob_client = blob_service_client.get_blob_client(container=container_name, blob=path_blob+'/'+file)
with open(os.path.join(path_local, file), "rb") as data:
buf = 0
buf = os.stat(os.path.join(path_local, file)).st_size
blob_client.upload_blob(data, overwrite=True)
if buf:
pbar.set_postfix(file=file, refresh=False)
pbar.update(buf)
###Output
Uploading non-image files:
|
scripts/.ipynb_checkpoints/crea_configura_estaciones_simio-checkpoint.ipynb
|
###Markdown
Genera un csv para crear palas configuradas en simio
###Code
import pandas as pd
path = 'C:/Users/dazac/Downloads/datos mina/datos produccion/'
df_cargas = pd.read_csv(path+'generados/datos_cargas.csv')
df_cargas
object_type = []
name = []
x = []
y = []
z = []
lenght = []
width = []
height = []
path = []
prop = []
value = []
other = []
for for_index in range(len(df_cargas)):
object_type.append('Server')
name.append(df_cargas.PositionedAt.iloc[for_index])
x.append('0')
y.append('0')
z.append('0')
lenght.append('2,0892081459734437')
width.append('0,99910370276457061')
height.append('1,0000003673020019')
path.append('0')
prop.append('BeforeProcessingAddOnProcess')
#agregar delay antes de process para simular el tiempo de espera
value.append('TiempoManiobraProcess')
other.append('None')
object_type.append('Server')
name.append(df_cargas.PositionedAt.iloc[for_index])
x.append('0')
y.append('0')
z.append('0')
lenght.append('2,0892081459734437')
width.append('0,99910370276457061')
height.append('1,0000003673020019')
path.append('0')
prop.append('ProcessingAddOnProcess')
#agregar delay en process para simular el tiempo de carga de la pala
value.append('TiempoCargaProcess')
other.append('None')
object_type.append('Server')
name.append(df_cargas.PositionedAt.iloc[for_index])
x.append('0')
y.append('0')
z.append('0')
lenght.append('2,0892081459734437')
width.append('0,99910370276457061')
height.append('1,0000003673020019')
path.append('0')
prop.append('AfterProcessingAddOnProcess')
value.append('AsignarDescargaProcess')
other.append('none')
datos_dict = {
'Tipo objeto': object_type,
'Nombre': name,
'x':x,
'y':y,
'z':z,
'lenght': lenght,
'width': width,
'height': height,
'path': path,
'prop': prop,
'value': value,
'other': other
}
df_cargas= pd.DataFrame(datos_dict)
df_cargas.to_csv(path_or_buf='C:/Users/dazac/Downloads/datos mina/datos produccion/creador_palas.csv', sep='#', header=False, index=False)
###Output
_____no_output_____
|
code/.ipynb_checkpoints/simulate_pdep_fate-checkpoint.ipynb
|
###Markdown
fit multiple plots
###Code
value_gradient = [0,.4,.75,.9,1]
data_storage = {}
for s_index, surface in enumerate(['alpha', 'beta', 'gamma']):
data_storage[surface] = {}
for p_index, initial_pathway in enumerate(['alkyl','peroxy',]):
#for p_index, initial_pathway in enumerate(['alkyl','peroxy','QOOH']):
if surface == 'alpha':
if initial_pathway == 'peroxy':
initial_reactants = set(('aRO2',))
elif initial_pathway == 'alkyl':
initial_reactants = set(('O2','aR'))
elif initial_pathway == 'QOOH':
initial_reactants = set(('aQOOHg',))
elif surface == 'beta':
if initial_pathway == 'peroxy':
initial_reactants = set(('bRO2',))
elif initial_pathway == 'alkyl':
initial_reactants = set(('O2','bR'))
elif initial_pathway == 'QOOH':
initial_reactants = set(('bQOOHa',))
elif surface == 'gamma':
if initial_pathway == 'peroxy':
initial_reactants = set(('gRO2',))
elif initial_pathway == 'alkyl':
initial_reactants = set(('O2', 'gR'))
elif initial_pathway == 'QOOH':
initial_reactants = set(('gQOOHa',))
main_path_matrix = pd.DataFrame(index=pressures,
columns=temperatures,
data=np.empty((len(temperatures),len(pressures))))
fraction_main_path_matrix = pd.DataFrame(index=pressures,
columns=temperatures,
data=np.empty((len(temperatures),len(pressures))))
for temperature in temperatures:
for pressure in pressures:
solution.TP = temperature, pressure
if initial_pathway == 'QOOH':
paths = get_reaction_branching_with_second_O2_addition(solution,initial_reactants)
else:
paths = get_reaction_branching(solution,initial_reactants)
main_path = paths[paths == paths.max()].index[0]
main_path_matrix.loc[pressure,temperature] = main_path
fraction_main_path_matrix.loc[pressure,temperature] = paths[main_path] / paths.sum()
data_one_plot = {}
data_one_plot['main_path_matrix'] = main_path_matrix
data_one_plot['fraction_main_path_matrix'] = fraction_main_path_matrix
data_storage[surface][initial_pathway] = data_one_plot
f, axes = plt.subplots(2,3,sharex=True,sharey=True,figsize = [12/.95,8],gridspec_kw={'wspace':.12,'hspace':0.2})
print('Output below are the major pathways which contribute to the produced plots.\nThis allows you to add descriptive details in an SVG editor.')
for s_index, surface in enumerate(['alpha', 'beta', 'gamma']):
for p_index, initial_pathway in enumerate(['alkyl','peroxy']):
data_one_plot = data_storage[surface][initial_pathway]
main_path_matrix = data_one_plot['main_path_matrix']
fraction_main_path_matrix = data_one_plot['fraction_main_path_matrix']
ax = axes[p_index][s_index]
ax.set_xticks(np.linspace(0,len(temperatures)-1,n_xticks))
ax.set_yticks(np.linspace(0,len(pressures)-1,n_yticks))
ax.set_xticklabels([int(label) for label in np.linspace(temperatures.min(),temperatures.max(),n_xticks)])
ax.set_yticklabels(['10$^{{{0}}}$'.format(int(np.log10(label))) for label in np.logspace(np.log10(pressures.min()),np.log10(pressures.max()),n_yticks)])
if p_index == 1:
#if p_index == 2:
ax.set_xlabel('temperature (K)')
if s_index == 0:
ax.set_ylabel('pressure (Pa)')
contour_output = ax.contourf(fraction_main_path_matrix,cmap=transparency_color_map,antialiased=True,levels=value_gradient, alpha=1)
for rxn_index in np.unique(main_path_matrix):
zero_one_matrix = np.zeros(main_path_matrix.shape)
zero_one_matrix[main_path_matrix == rxn_index] = 1
smooth_matrix = gaussian_filter(zero_one_matrix, 2)
ax.contour(smooth_matrix,cmap=black_color_map,antialiased=True,levels=[.5])
#make title
if surface == 'alpha':
title = u'α'
if initial_pathway == 'QOOH':
title += u'QOOHγ (+ O$_2$)'
elif surface == 'beta':
title = u'β'
if initial_pathway == 'QOOH':
title += u'QOOHα (+ O$_2$)'
elif surface == 'gamma':
title = u'γ'
if initial_pathway == 'QOOH':
title += u'QOOHα (+ O$_2$)'
if initial_pathway == 'alkyl':
title += 'R + O$_2$'
elif initial_pathway == 'peroxy':
title += 'RO$_2$'
ax.set_title(title)
print('######################## '+surface+' '+initial_pathway)
for rxn_index in np.unique(main_path_matrix):
print('########## '+reactions[int(rxn_index)].equation)
main_path_df = pd.DataFrame(columns = temperatures, index=pressures, data=main_path_matrix)
main_path_df_bool = main_path_df[main_path_df == rxn_index]
pressures_of_path = main_path_df_bool.dropna(0,'all').index
temps_of_path = main_path_df_bool.dropna(1,'all').columns
print('temp range: ({0}, {1})'.format(min(temps_of_path), max(temps_of_path)))
print('pres range: ({0}, {1})'.format(min(pressures_of_path), max(pressures_of_path)))
f.subplots_adjust(right=0.9)
cbar_ax = f.add_axes([0.91, .25, 0.06, 0.5])
cbar = f.colorbar(contour_output, cax=cbar_ax)
cbar_ax.set_axis_off()
cbar_ax.annotate('product\nmixture',(.5,.125),xycoords='axes fraction',va='center', ha='center')
cbar_ax.annotate('major\nsecondary\nproducts',(.5,.375),xycoords='axes fraction',va='center', ha='center')
cbar_ax.annotate('minor\nsecondary\nproducts',(.5,.625),xycoords='axes fraction',va='center', ha='center')
cbar_ax.annotate('nearly\npure',(.5,.875),xycoords='axes fraction',va='center', ha='center')
f.savefig(os.path.join(image_dir,'branching_6_plots.svg'),bbox_inches='tight')
###Output
_____no_output_____
###Markdown
determine amount reversingFor the pdep reactions, a certain amount goes back to R + O2 which is not included in the Chemkin rates, since it is not a reaction, however it would be useful to know when there is another reaction competing for R radical.To do this, we take the sum of all rates consuming R + O2 and divide it by the high p rate from high-p kinetics
###Code
mechanism_file_high_p = '../data/mech_generation/high_p_merged/chem.cti'
solution_high_p = cantera_tools.create_mechanism(mechanism_file_high_p)
mechanism_file = '../data/mech_generation/pdep_merged/chem.cti'
solution = cantera_tools.create_mechanism(mechanism_file)
###Output
_____no_output_____
###Markdown
all three - linear
###Code
value_gradient = np.linspace(0,1,21)
f, axes = plt.subplots(1,3,sharex=True,sharey=True,figsize = [12,4],gridspec_kw={'wspace':.12,'hspace':0.2})
for s_index, surface in enumerate(['alpha', 'beta', 'gamma']):
if surface == 'alpha':
initial_reactants = set(('O2','aR'))
elif surface == 'beta':
initial_reactants = set(('O2','bR'))
elif surface == 'gamma':
initial_reactants = set(('O2', 'gR'))
fraction_reacts = pd.DataFrame(index=pressures,
columns=temperatures,
data=np.empty((len(temperatures),len(pressures))))
for temperature in temperatures:
for pressure in pressures:
solution.TP = temperature, pressure
solution_high_p.TP = temperature, pressure
output = get_reaction_branching(solution, initial_reactants)
output_high_p= get_reaction_branching(solution_high_p, initial_reactants)
fraction_reacts.loc[pressure,temperature] = (output.sum() / output_high_p).values[0]
ax = axes[s_index]
ax.set_xticks(np.linspace(0,len(temperatures)-1,n_xticks))
ax.set_yticks(np.linspace(0,len(pressures)-1,n_yticks))
ax.set_xticklabels([int(label) for label in np.linspace(temperatures.min(),temperatures.max(),n_xticks)])
ax.set_yticklabels(['10$^{{{0}}}$'.format(int(np.log10(label))) for label in np.logspace(np.log10(pressures.min()),np.log10(pressures.max()),n_yticks)])
ax.set_xlabel('temperature (K)')
if s_index == 0:
ax.set_ylabel('pressure (Pa)')
contour_output = ax.contourf(fraction_reacts,cmap=transparency_color_map,antialiased=True,alpha=1, levels=value_gradient)
add_lines_to_contourf(contour_obj=contour_output)
#make title
if surface == 'alpha':
title = u'α'
elif surface == 'beta':
title = u'β'
elif surface == 'gamma':
title = u'γ'
title += 'R + O$_2$'
ax.set_title(title)
cbar_ax = f.add_axes([0.925, 0.15, 0.02, 0.7])
cbar = f.colorbar(contour_output, cax=cbar_ax)
cbar_ax.set_ylabel('fraction of excited RO$_2$ not reforming R + O$_2$')
#cbar.set_ticks([0,-.5,-1,-1.5,-2,])
f.savefig(os.path.join(image_dir,'non_reaction_fraction.svg'),bbox_inches='tight')
f.savefig(os.path.join(image_dir,'non_reaction_fraction.pdf'),bbox_inches='tight')
###Output
_____no_output_____
|
theCCXT/1. analyzeForTheStartOfEachMonth.ipynb
|
###Markdown
data handling
###Code
import ccxt
ccxt.exchanges[:10]
#exchange = ccxt.gateio()
exchange = ccxt.gateio()
markets = exchange.load_markets()
markets['BTC/USDT']['info']
"""
[
1504541580000, // UTC timestamp in milliseconds, integer
4235.4, // (O)pen price, float
4240.6, // (H)ighest price, float
4230.0, // (L)owest price, float
4230.7, // (C)losing price, float
37.72941911 // (V)olume (in terms of the base currency), float
],
"""
class ItemOfOHLCV:
timestamp: int
open: float
high: float
low: float
close: float
volume: float
ohlcvData = exchange.fetch_ohlcv('PEOPLE/USDT', '1h', params={'price':'index'})
ohlcvData[:10]
TOKEN = 'DOGE/USDT'
startMonthTime = "2019-10-01"
endMonthTime = "2022-01-07"
def getEveryStartDayOfMonth(startDate: str, endDate: str):
import datetime
import calendar
start = datetime.datetime.strptime(startDate, "%Y-%m-%d")
end = datetime.datetime.strptime(endDate, "%Y-%m-%d")
while start <= end:
yield start.strftime("%Y-%m-%d")
start += datetime.timedelta(days=calendar.monthrange(start.year, start.month)[1])
startPointOfMonths = list(getEveryStartDayOfMonth(startMonthTime, endMonthTime))
startPointOfMonths
def getTimeRangeOfADateWithinXDays(date: str, x: int):
import datetime
halfRange = x/2
start = datetime.datetime.strptime(date, "%Y-%m-%d") - datetime.timedelta(hours=halfRange*24)
end = start + datetime.timedelta(hours=halfRange*24*2)
return start.strftime("%Y-%m-%d"), end.strftime("%Y-%m-%d")
dateRangeFor3DaysInEveryStartDayOfMonth = [getTimeRangeOfADateWithinXDays(startPointOfMonths[i], 15) for i,_ in enumerate(startPointOfMonths)]
dateRangeFor3DaysInEveryStartDayOfMonth
def dateTimeToUnixTime(date: str):
import datetime
return int(datetime.datetime.strptime(date, "%Y-%m-%d").timestamp() * 1000)
ohlcvData = []
for range in dateRangeFor3DaysInEveryStartDayOfMonth:
ohlcvData += exchange.fetch_ohlcv(TOKEN, '1h', params={'price':'index'}, since=dateTimeToUnixTime(range[0]))
ohlcvData[:10]
"""
class ItemOfOHLCV:
timestamp: int
open: float
high: float
low: float
close: float
volume: float
"""
def convertTimestampToString(timestamp: int):
import datetime
return datetime.datetime.fromtimestamp(timestamp/1000).strftime("%Y-%m-%d")
def checkIfTheItemOfOHLCVIsInTheDateRange(item: ItemOfOHLCV, start: str, end: str):
import datetime
itemDate = datetime.datetime.fromtimestamp(item.timestamp/1000)
if itemDate >= datetime.datetime.strptime(start, "%Y-%m-%d") and itemDate <= datetime.datetime.strptime(end, "%Y-%m-%d"):
return True
return False
itemsForEachRange =[[] for _ in dateRangeFor3DaysInEveryStartDayOfMonth]
for one in ohlcvData:
item = ItemOfOHLCV()
item.timestamp, item.open, item.high, item.low, item.close, item.volume = one
#print(convertTimestampToString(item.timestamp))
for index, dateRange in enumerate(dateRangeFor3DaysInEveryStartDayOfMonth):
if checkIfTheItemOfOHLCVIsInTheDateRange(item, dateRange[0], dateRange[1]):
itemsForEachRange[index].append(item)
itemsForEachRange[1][:10]
###Output
_____no_output_____
###Markdown
graph
###Code
def convertTimestampToDetailString(timestamp: int):
import datetime
return datetime.datetime.fromtimestamp(timestamp/1000).strftime("%Y-%m-%d %H:%M:%S")
def itemDataToListOfDictionaries(itemData: list):
return [{
'timestamp': convertTimestampToDetailString(item.timestamp),
'open': item.open,
'high': item.high,
'low': item.low,
'close': item.close,
'volume': item.volume
} for item in itemData]
def itemDataToPandaDataFrame(itemData: list):
import pandas as pd
return pd.DataFrame(itemDataToListOfDictionaries(itemData), columns=['timestamp', 'open', 'high', 'low', 'close', 'volume'])
testData = itemDataToPandaDataFrame(itemsForEachRange[1])
testData
itemsForEachRange[1]
import plotly.graph_objects as go
figureList = []
for items in itemsForEachRange:
if len(items) == 0:
continue
df = itemDataToPandaDataFrame(items)
fig = go.Figure(data=go.Ohlc(x=df['timestamp'],
open=df['open'],
high=df['high'],
low=df['low'],
close=df['close']))
fig.update_layout(height=600, width=600, title_text=f"{TOKEN} on {convertTimestampToString(items[0].timestamp)}~{convertTimestampToString(items[-1].timestamp)}")
figureList.append(fig)
def figures_to_html(figs, filename="1.dogePrice.html"):
dashboard = open(filename, 'w')
dashboard.write("<html><head></head><body>" + "\n")
for fig in figs:
inner_html = fig.to_html().split('<body>')[1].split('</body>')[0]
dashboard.write(inner_html)
dashboard.write("</body></html>" + "\n")
figures_to_html(figureList)
###Output
_____no_output_____
|
nano degree/assignment1.ipynb
|
###Markdown
자연어처리시각화(과제)네이버 뉴스에서 아래 조건을 만족하는 최소 5개 이상의 기사를 분석하고 빈도 기준 20개 단어를 워드 클라우드로 출력- 4개 카테고리 중 1개 선택 : 정치/경제/사회/생활-문화 - 동일한 카테고리에서 관련있는 최소 5개 이상의 기사 선택(예: 최근 5일간 보도된 관련 기사 5개 선택) 주의 1: 원본 기사를 메모장에 복사하거나, web scraping 등을 통해 웹에서 직접 가져와도 됨. 주의 2: 같은 날짜의 경우 거의 유사한 내용 기사는 제외할 것. 다른 관점에서 보도한 기사는 가능.- 전처리 포함(불용어 제거 포함). - 1차 시각화: 빈도분석 결과를 빈도 그래프로 출력- 2차 시각화: 워드 클라우드로 빈도 기준 20개 단어 출력.- 결과물 제출: 쥬피터 노트북 파일- 분석 결과를 근거로 주피터 노트북 셀에 자신의 생각을 주석으로 첨가(최소 5줄 이상).
###Code
from konlpy.tag import Okt
from collections import Counter
###Output
_____no_output_____
###Markdown
불용어 처리 리스트 생성
###Code
twitter = Okt()
with open("kr_stopwords.txt") as f:
raw_list = f.readlines()
stopword_list = [x.strip() for x in raw_list]
custom_stop_words = ["위해", "통해", "지금", "이번",
"한편", "이제", "뒤로", "것임", "이제", "바로"]
stopword_list.extend(custom_stop_words)
###Output
_____no_output_____
###Markdown
애플카 뉴스에 관한 소식 5가지- news1~5까지의 기사내용을 text라는 리스트에 각각 요소로 저장
###Code
news_list = []
with open("news1") as news1, open("news2") as news2, open("news3") as news3, open("news4") as news4, open("news5") as news5:
for news in (news1, news2, news3, news4, news5):
temp_list =[x.strip() for x in news.readlines()]
news_list.append("".join(temp_list))
###Output
_____no_output_____
###Markdown
전부 하나의 문자열로 통일
###Code
news = "".join(news_list)
###Output
_____no_output_____
###Markdown
전처리 과정 수행- 한 글자 단어 제거- 불용어 제거
###Code
law_container = Counter(twitter.nouns(news)) # 발생빈도 구함
law_container2 = Counter({x: law_container[x] for x in law_container if len(x) > 1}) # 한 글자 단어 제거
law_container3 = Counter({x: law_container2[x] for x in law_container2 if x not in stopword_list}) # 불용어 제거
law_container3.most_common(5)
###Output
_____no_output_____
###Markdown
빈도 분석 그래프 한글 출력을 위한 font 지정
###Code
import matplotlib.pyplot as plt
from matplotlib import rc
rc("font", family="AppleGothic")
plt.rcParams["axes.unicode_minus"] = False
from nltk import FreqDist
import pandas as pd
import seaborn as sns
# 발생 빈도 40개 단어 선택
fdist = FreqDist(law_container3).most_common(20)
# 파이썬 dictionary 객체를 pandas의 Series객체로 변환
fdist = pd.Series(dict(fdist))
fig, ax = plt.subplots(figsize=(10, 10))
all_plot = sns.barplot(x=fdist.index, y=fdist.values, ax=ax)
_ = plt.xticks(rotation=30)
###Output
_____no_output_____
###Markdown
빈도 분석 wordcloud로 시각화
###Code
from wordcloud import WordCloud
wordcloud = WordCloud(font_path="./NanumGothic.ttf",
width=800,
height=800,
background_color="white")
# 발생 빈도 순으로 20개 단어 선택
ranked_tags = dict(law_container3.most_common(20))
wordcloud = wordcloud.generate_from_frequencies(ranked_tags) # 리스트를 dict로 변경
array = wordcloud.to_array() # nd.array
fig = plt.figure(figsize=(10, 10))
plt.imshow(array, interpolation="bilinear")
plt.show()
###Output
_____no_output_____
|
notebooks/bottleneck.ipynb
|
###Markdown
Based onhttps://github.com/Fernerkundung/notebooks/blob/master/1D_interpolation.ipynb
###Code
# import necessary libraries
import numpy as np
import pandas as pd
from numba import jit
import matplotlib.pyplot as plt
import bottleneck as bn
%matplotlib inline
# create example data, original is (644, 4800, 4800)
test_arr = np.random.uniform(low=-1.0, high=1.0, size=(92, 480, 480)).astype(np.float32)
test_arr[1:90:7, :, :] = np.nan # NaN fill value in original data
test_arr[2,:,:] = np.nan
test_arr[:, 1:479:6, 1:479:8] = np.nan
def interpolate_nan(arr, method="linear", limit=3):
"""return array interpolated along time-axis to fill missing values"""
result = np.zeros_like(arr, dtype=np.float32)
for i in range(arr.shape[1]):
# slice along y axis, interpolate with pandas wrapper to interp1d
line_stack = pd.DataFrame(data=arr[:,i,:], dtype=np.float32)
line_stack.interpolate(method=method, axis=0, inplace=True, limit=limit)
result[:, i, :] = line_stack.values.astype(np.float32)
return result
def interpolate_bottleneck(arr, inplace=True, window=3, min_count=1, axis=0):
means = bn.move_mean(arr, window, min_count, axis)
nans = np.isnan(arr)
if inplace:
result = arr
else:
result = np.copy(arr)
result[nans] = means[nans]
return result
@jit(nopython=True)
def interpolate_numba(arr):
"""return array interpolated along time-axis to fill missing values"""
result = np.zeros_like(arr, dtype=np.float32)
for x in range(arr.shape[2]):
# slice along x axis
for y in range(arr.shape[1]):
# slice along y axis
for z in range(arr.shape[0]):
value = arr[z,y,x]
if z == 0: # don't interpolate first value
new_value = value
elif z == len(arr[:,0,0])-1: # don't interpolate last value
new_value = value
elif np.isnan(value): # interpolate
left = arr[z-1,y,x]
right = arr[z+1,y,x]
# look for valid neighbours
if not np.isnan(left) and not np.isnan(right): # left and right are valid
new_value = (left + right) / 2
elif np.isnan(left) and z == 1: # boundary condition left
new_value = value
elif np.isnan(right) and z == len(arr[:,0,0])-2: # boundary condition right
new_value = value
elif np.isnan(left) and not np.isnan(right): # take second neighbour to the left
more_left = arr[z-2,y,x]
if np.isnan(more_left):
new_value = value
else:
new_value = (more_left + right) / 2
elif not np.isnan(left) and np.isnan(right): # take second neighbour to the right
more_right = arr[z+2,y,x]
if np.isnan(more_right):
new_value = value
else:
new_value = (more_right + left) / 2
elif np.isnan(left) and np.isnan(right): # take second neighbour on both sides
more_left = arr[z-2,y,x]
more_right = arr[z+2,y,x]
if not np.isnan(more_left) and not np.isnan(more_right):
new_value = (more_left + more_right) / 2
else:
new_value = value
else:
new_value = value
else:
new_value = value
result[z,y,x] = int(new_value)
return result
# show the example time-series at location (:,3,4)
plt.plot(test_arr[:,3,4])
result = interpolate_bottleneck(test_arr, False)
plt.plot(result[:,3,4])
plt.plot(test_arr[:,3,4])
###Output
_____no_output_____
###Markdown
Some timings
###Code
#Straight bottleneck calculation of the windowed means, no filling of nans in input array
%timeit bottleneck.move_mean(test_arr, window=3, min_count=1, axis=0)
#filling of nans in copy of array
%timeit interpolate_bottleneck(test_arr, inplace=False)
#filling of nans in array in place
test_copy = np.copy(test_arr)
%timeit interpolate_bottleneck(test_copy)
#NUMBA 1D interpolation
%timeit interpolate_numba(test_arr)
#Original 1D interpolation
%timeit interpolate_nan(test_arr)
###Output
1 loops, best of 3: 51.2 s per loop
|
Machine-Learning-2021/project13/RNN_Assignment_13.ipynb
|
###Markdown
Text Generation with RNNs
###Code
import numpy as np
import torch
from torch import nn
import torch.nn.functional as F
import pandas as pd
import urllib
from torch.autograd import Variable
###Output
_____no_output_____
###Markdown
1 DatasetDefine the path of the file, you want to read and train the model on
###Code
text=''
for line in urllib.request.urlopen("https://raw.githubusercontent.com/karpathy/char-rnn/master/data/tinyshakespeare/input.txt"):
text+=line.decode("utf-8")
#path_to_file ='/content/data.txt'
#text = open(path_to_file, encoding='utf-8').read()
###Output
_____no_output_____
###Markdown
Inspect the datasetTake a look at the first 250 characters in text
###Code
print(text[:250])
# The unique characters in the file
vocab = sorted(set(text))
print ('{} unique characters'.format(len(vocab)))
###Output
65 unique characters
###Markdown
2 Process the dataset for the learning taskThe task that we want our model to achieve is: given a character, or a sequence of characters, what is the most probable next character?To achieve this, we will input a sequence of characters to the model, and train the model to predict the output, that is, the following character at each time step. RNNs maintain an internal state that depends on previously seen elements, so information about all characters seen up until a given moment will be taken into account in generating the prediction. Vectorize the textBefore we begin training our RNN model, we'll need to create a numerical representation of our text-based dataset. To do this, we'll generate two lookup tables: one that maps characters to numbers, and a second that maps numbers back to characters. Recall that we just identified the unique characters present in the text.
###Code
# Creating a mapping from unique characters to indices
char2idx = {u:i for i, u in enumerate(vocab)}
text_as_int = np.array([char2idx[c] for c in text])
# Create a mapping from indices to characters
idx2char = np.array(vocab)
text_as_int
###Output
_____no_output_____
###Markdown
This gives us an integer representation for each character. Observe that the unique characters (i.e., our vocabulary) in the text are mapped as indices from 0 to len(unique). Let's take a peek at this numerical representation of our dataset:
###Code
print('{')
for char,_ in zip(char2idx, range(20)):
print(' {:4s}: {:3d},'.format(repr(char), char2idx[char]))
print(' ...\n}')
###Output
{
'\n': 0,
' ' : 1,
'!' : 2,
'$' : 3,
'&' : 4,
"'" : 5,
',' : 6,
'-' : 7,
'.' : 8,
'3' : 9,
':' : 10,
';' : 11,
'?' : 12,
'A' : 13,
'B' : 14,
'C' : 15,
'D' : 16,
'E' : 17,
'F' : 18,
'G' : 19,
...
}
###Markdown
We can also look at how the first part of the text is mapped to an integer representation:
###Code
print ('{} ---- characters mapped to int ---- > {}'.format(repr(text[:13]), text_as_int[:13]))
###Output
'First Citizen' ---- characters mapped to int ---- > [18 47 56 57 58 1 15 47 58 47 64 43 52]
###Markdown
Defining a method to encode one hot labels
###Code
def one_hot_encode(arr, n_labels):
# Initialize the the encoded array
one_hot = np.zeros((np.multiply(*arr.shape), n_labels), dtype=np.float32)
# Fill the appropriate elements with ones
one_hot[np.arange(one_hot.shape[0]), arr.flatten()] = 1.
# Finally reshape it to get back to the original array
one_hot = one_hot.reshape((*arr.shape, n_labels))
return one_hot
###Output
_____no_output_____
###Markdown
Defining a method to make mini-batches for training
###Code
def get_batches(arr, batch_size, seq_length):
'''Create a generator that returns batches of size
batch_size x seq_length from arr.
Arguments
---------
arr: Array you want to make batches from
batch_size: Batch size, the number of sequences per batch
seq_length: Number of encoded chars in a sequence
'''
batch_size_total = batch_size * seq_length
# total number of batches we can make
n_batches = len(arr) // batch_size_total
# Keep only enough characters to make full batches
arr = arr[:n_batches * batch_size_total]
# Reshape into batch_size rows
arr = arr.reshape((batch_size, -1))
# iterate through the array, one sequence at a time
for n in range(0, arr.shape[1], seq_length):
# The features
x = arr[:, n:n + seq_length]
# The targets, shifted by one
y = np.zeros_like(x)
try:
y[:, :-1], y[:, -1] = x[:, 1:], arr[:, n + seq_length]
except IndexError:
y[:, :-1], y[:, -1] = x[:, 1:], arr[:, 0]
yield x, y
batches = get_batches(text_as_int, 8, 50)
x, y = next(batches)
# printing out the first 10 items in a sequence
print('x\n', x[:10, :10])
print('\ny\n', y[:10, :10])
###Output
x
[[18 47 56 57 58 1 15 47 58 47]
[43 56 43 58 53 1 61 43 1 39]
[40 39 42 1 41 39 59 57 43 56]
[45 56 47 43 44 6 1 44 39 47]
[51 6 1 22 39 41 49 2 1 15]
[46 1 61 47 52 42 1 57 39 63]
[58 53 1 40 43 6 1 61 46 53]
[ 1 47 44 1 63 53 59 1 57 50]]
y
[[47 56 57 58 1 15 47 58 47 64]
[56 43 58 53 1 61 43 1 39 56]
[39 42 1 41 39 59 57 43 56 1]
[56 47 43 44 6 1 44 39 47 56]
[ 6 1 22 39 41 49 2 1 15 53]
[ 1 61 47 52 42 1 57 39 63 1]
[53 1 40 43 6 1 61 46 53 1]
[47 44 1 63 53 59 1 57 50 43]]
###Markdown
3 The Recurrent Neural Network (RNN) model Check if GPU is available
###Code
train_on_gpu = torch.cuda.is_available()
print ('Training on GPU' if train_on_gpu else 'Training on CPU')
###Output
Training on GPU
###Markdown
Declaring the model
###Code
class VanillaCharRNN(nn.Module):
def __init__(self, vocab, n_hidden=256, n_layers=2,
drop_prob=0.5, lr=0.001):
super(VanillaCharRNN,self).__init__()
self.drop_prob = drop_prob
self.n_layers = n_layers
self.n_hidden = n_hidden
self.lr = lr
self.vocab=vocab
#self.encoder = nn.Embedding(len(self.vocab), n_hidden)
#self.gru = nn.GRU(n_hidden, n_hidden, n_layers)
#self.decoder = nn.Linear(n_hidden, len(self.vocab))
self.hidden_size = n_hidden
self.i2h = nn.Linear(len(self.vocab), n_hidden)
self.i2o = nn.Linear(len(self.vocab), len(self.vocab))
self.softmax = nn.LogSoftmax(dim=1)
'''TODO: define the layers you need for the model'''
def forward(self, input, hidden):
print(input.size(),hidden.size())
combined = torch.cat((input, hidden), 1)
hidden = self.i2h(combined)
output = self.i2o(combined)
output = self.softmax(output)
return output, hidden
def init_hidden(self, batch_size):
weight = next(self.parameters()).data
if (train_on_gpu):
hidden = weight.new(batch_size, seq_length,len(self.vocab)).zero_().cuda()
else:
hidden = weight.new(batch_size, seq_length,len(self.vocab)).zero_()
return hidden
class LSTMCharRNN(nn.Module):
def __init__(self, vocab, n_hidden=256, n_layers=2,
drop_prob=0.5, lr=0.001):
super().__init__()
self.drop_prob = drop_prob
self.n_layers = n_layers
self.n_hidden = n_hidden
self.lr = lr
self.vocab=vocab
'''TODO: define the layers you need for the model'''
self.lstm = nn.LSTM(len(self.vocab), n_hidden, n_layers,
dropout=drop_prob, batch_first=True)
## TODO: define a dropout layer
self.dropout = nn.Dropout(drop_prob)
## TODO: define the final, fully-connected output layer
self.fc = nn.Linear(n_hidden, len(self.vocab))
def forward(self, x, hidden):
'''TODO: Forward pass through the network
x is the input and `hidden` is the hidden/cell state .'''
r_output, hidden_t = self.lstm(x, hidden)
# pass through a dropout layer
out = self.dropout(r_output)
# Stack up LSTM outputs using view
# you may need to use contiguous to reshape the output
out = out.contiguous().view(-1, self.n_hidden)
## put x through the fully-connected layer
out = self.fc(out)
# return the final output and the hidden state
return out, hidden_t
def init_hidden(self, batch_size):
''' Initializes hidden state '''
weight = next(self.parameters()).data
if (train_on_gpu):
hidden = (weight.new(self.n_layers, batch_size, self.n_hidden).zero_().cuda(),
weight.new(self.n_layers, batch_size, self.n_hidden).zero_().cuda())
else:
hidden = (weight.new(self.n_layers, batch_size, self.n_hidden).zero_(),
weight.new(self.n_layers, batch_size, self.n_hidden).zero_())
return hidden
###Output
_____no_output_____
###Markdown
Declaring the train method
###Code
def train(model, data, epochs=10, batch_size=10, seq_length=50, lr=0.001, clip=5, val_frac=0.1, print_every=10):
''' Training a network
Arguments
---------
model: CharRNN network
data: text data to train the network
epochs: Number of epochs to train
batch_size: Number of mini-sequences per mini-batch, aka batch size
seq_length: Number of character steps per mini-batch
lr: learning rate
clip: gradient clipping
val_frac: Fraction of data to hold out for validation
print_every: Number of steps for printing training and validation loss
'''
model.train()
opt = torch.optim.Adam(model.parameters(), lr=lr)
criterion = nn.CrossEntropyLoss()
# create training and validation data
val_idx = int(len(data) * (1 - val_frac))
data, val_data = data[:val_idx], data[val_idx:]
if (train_on_gpu):
model.cuda()
counter = 0
n_vocab = len(model.vocab)
for e in range(epochs):
# initialize hidden state
h = model.init_hidden(batch_size)
'''TODO: use the get_batches function to generate sequences of the desired size'''
dataset = get_batches(data, batch_size, seq_length)
for x, y in dataset:
counter += 1
# One-hot encode our data and make them Torch tensors
x = one_hot_encode(x, n_vocab)
inputs, targets = torch.from_numpy(x), torch.from_numpy(y)
if (train_on_gpu):
inputs, targets = inputs.cuda(), targets.cuda()
# Creating new variables for the hidden state, otherwise
# we'd backprop through the entire training history
#h = tuple([each.data for each in h])
if type(h) is tuple:
if train_on_gpu:
h = tuple([each.data.cuda() for each in h])
else:
h = tuple([each.data for each in h])
else:
if train_on_gpu:
h = h.data.cuda()
else:
h = h.data
# zero accumulated gradients
model.zero_grad()
'''TODO: feed the current input into the model and generate output'''
output, h = model(inputs, h) # TODO
'''TODO: compute the loss!'''
loss = criterion(output, targets.view(batch_size*seq_length))# TODO
# perform backprop
loss.backward()
# `clip_grad_norm` helps prevent the exploding gradient problem in RNNs / LSTMs.
nn.utils.clip_grad_norm_(model.parameters(), clip)
opt.step()
# loss stats
if counter % print_every == 0:
# Get validation loss
val_h = model.init_hidden(batch_size)
val_losses = []
model.eval()
for x, y in get_batches(val_data, batch_size, seq_length):
# One-hot encode our data and make them Torch tensors
x = one_hot_encode(x, n_vocab)
x, y = torch.from_numpy(x), torch.from_numpy(y)
# Creating new variables for the hidden state, otherwise
# we'd backprop through the entire training history
val_h = tuple([each.data for each in val_h])
inputs, targets = x, y
if (train_on_gpu):
inputs, targets = inputs.cuda(), targets.cuda()
'''TODO: feed the current input into the model and generate output'''
output, val_h = model(inputs, val_h) # TODO
'''TODO: compute the validation loss!'''
val_loss =criterion(output, targets.view(batch_size*seq_length)) # TODO
val_losses.append(val_loss.item())
print("Epoch: {}/{}...".format(e + 1, epochs),
"Step: {}...".format(counter),
"Loss: {:.4f}...".format(loss.item()),
"Val Loss: {:.4f}".format(np.mean(val_losses)))
'''TODO: sample from the model to generate texts'''
input_eval ="wink'st" # TODO: choose a start string
print(sample(model, 1000, prime=input_eval, top_k=10))
model.train() # reset to train mode after iterationg through validation data
###Output
_____no_output_____
###Markdown
Defining a method to generate the next character
###Code
def predict(model, char, h=None, top_k=None):
''' Given a character, predict the next character.
Returns the predicted character and the hidden state.
'''
# tensor inputs
x = np.array([[char2idx[char]]])
x = one_hot_encode(x, len(model.vocab))
inputs = torch.from_numpy(x)
if (train_on_gpu):
inputs = inputs.cuda()
# detach hidden state from history
h = tuple([each.data for each in h])
'''TODO: feed the current input into the model and generate output'''
output, h = model(inputs, h) # TODO
# get the character probabilities
p = F.softmax(output, dim=1).data
if (train_on_gpu):
p = p.cpu() # move to cpu
# get top characters
if top_k is None:
top_ch = np.arange(len(model.vocab))
else:
p, top_ch = p.topk(top_k)
top_ch = top_ch.numpy().squeeze()
# select the likely next character with some element of randomness
p = p.numpy().squeeze()
char = np.random.choice(top_ch, p=p / p.sum())
# return the encoded value of the predicted char and the hidden state
return idx2char[char], h
###Output
_____no_output_____
###Markdown
Declaring a method to generate new text
###Code
def sample(model, size, prime='The', top_k=None):
if (train_on_gpu):
model.cuda()
else:
model.cpu()
model.eval() # eval mode
# First off, run through the prime characters
chars = [ch for ch in prime]
h = model.init_hidden(1)
for ch in prime:
char, h = predict(model, ch, h, top_k=top_k)
chars.append(char)
for ii in range(size):
'''TODO: pass in the previous character and get a new one'''
char, h = predict(model, chars[-1], h, top_k=top_k)# TODO
chars.append(char)
model.train()
return ''.join(chars)
###Output
_____no_output_____
###Markdown
Generate new Text using the RNN model Define and print the net
###Code
''''TODO: Try changing the number of units in the network to see how it affects performance'''
n_hidden =512 # TODO
n_layers =2 # TODO
vanilla_model = VanillaCharRNN(vocab, n_hidden, n_layers)
print(vanilla_model)
lstm_model = LSTMCharRNN(vocab, n_hidden, n_layers)
print(lstm_model)
###Output
LSTMCharRNN(
(lstm): LSTM(65, 512, num_layers=2, batch_first=True, dropout=0.5)
(dropout): Dropout(p=0.5, inplace=False)
(fc): Linear(in_features=512, out_features=65, bias=True)
)
###Markdown
Declaring the hyperparameters
###Code
''''TODO: Try changing the hyperparameters in the network to see how it affects performance'''
batch_size =50 #TODO
seq_length =130 # TODO
n_epochs =20 #TODO # start smaller if you are just testing initial behavior
#train(vanilla_model, text_as_int, epochs=n_epochs, batch_size=batch_size, seq_length=seq_length, lr=0.001, print_every=50)
###Output
_____no_output_____
###Markdown
Train the model and have fun with the generated texts
###Code
train(lstm_model, text_as_int, epochs=n_epochs, batch_size=batch_size, seq_length=seq_length, lr=0.001, print_every=50)
###Output
Epoch: 1/20... Step: 50... Loss: 3.3168... Val Loss: 3.3444
wink'steto taht aet eost oai ihr hsso n tteit e hhar a oesor riahi e soe aenor etiehn tiorao neaho i ho ttri ro oi he a rethhhoahonth he ttraeiitnnte h nsteetno sre e no ertonroaasein aa sre tehs e oeteeettestraoatest hinhe rthoho sneth io otats etsr t nr ansor t ereehta rehen thnt t neeeeeh eh esr taeososeessethanrs noht oa i rirto hsioe srsot rea eot aoeetth o t et e ret naeshn as oseos orrititt ohrses oeteh o os hos sarseonth nre ihho heo ana t hi rar nh rss e o ao hs e e eoish toa e o tr ehshr aoaeitatoash rnr ah r rnaesee n oeesnah iro ehtneeioe s sirs oaero int teereinhnettnns hhhrieohrnasis n i e e iotoo ens ehinee aotae a i a trsiraeeehsr eets ehhh eo ehr hohr rthtnh etst hr eishri aior o oh rhise a iae eotn iieeo nonianiheh as nooestenn rtsa h e ht ae so eoi t e sr haseeo entihrhn ho hs o an ha henie iri ahtsiotra e ro ea thetetiea ie eosteeo sihhnhtante ar iee e h hisrthshtessraeatnh e ohroenrnerssso o rah tisset nt siohiei ne ehiae ns
Epoch: 1/20... Step: 100... Loss: 3.2079... Val Loss: 3.1904
wink'stiissteehsohtaisa net oh hoht sseeoo th iaeateah h tohn ttonens th ere e s rrrhie heeeoe ht io er ahoi nahoosnha aho i eeae rie eoe hth teeahah o hseneihen ooieh rarotihrs nne eetors ihaesei aoo si irrttono ihe ah eton t etee nro seeenno tor e isa oahhhsi naoornah n r e a h rh thteiehosi seaat t rtiorrai itetetrtateaarshe t eh sin aa anotthhr hsesn t e irs sritt ntesiotrn trieaseeons nr sre seh saeietret ihae rnisoios rsot ehrnshoresoht t onseriiteeeseh hnnase eai aeotrtei ethhts t inaesstt ai r o rhe roeshot hs e etaaiti aeeooi h ten iorr os tooeasiar hastr eoonenei inrarrtr h s e toeeiir etrn ssaenaaeroe eehehothns haoreoas sien ns onerntiseher saireer tnat tr he hhhee hsaet t hne n esiton ehe oaioh otoooo i no ah oa tes retoao nrrahthr s e tho er i haeees no ttae easna h hoehhioaeea rstiasrhoi a oeen est ee ornhsnhthrae te ooeoheto ne rtehossnh o e i son ei hesttaiaahaetheoaoia aoeori asnhtr raehir t t tnhhoee rrrtor an rareaan es hi i rt
Epoch: 1/20... Step: 150... Loss: 2.7361... Val Loss: 2.6909
wink'st mort ohin iht, aout, nir ihe wo mnsntee tits sin sos he mis sil raals
White hioit
Ter ten no eearn soad nhrrt iirlttols oid mat whresns ool mere se med anoad ard,.I
REERE
UI
NSI: ohe hhie ao tiiot het tot, brnih nhhte
eraant waus ind toe barell sha eo ou eoers nnrsinind woee, aotd
onls hhe nianl marslt meeee tertasrs thalas hate orarin mhrnsa tnrstrneet, toeim tinens nhe tane
Anee sonn, whol, teos sot wyos iuestodr
hh tait wha roer taut aulr ihelts ionrss,
Aoe hants oo aat, hh mhtol, ned he hinnt hatsees wias waatr hetas ahat siod tos mha hes beelennas too maroir welit autts ie weas my boe oiternar in mhe nhie mone bonse, weat wuaa ot hhee otlats nhrritelt hoee oher te oar tue aod tie batd aane whis ahrtd touln nras hhoe.
IAII
SI Ir te bihe
sou ooe bal soln,
Bnd ohit mhe,
Bhets sio artathe tot woe hon mor terernl teer, tyetntria irnens oile tn thtoil ir hytee nnnroas he mass
IA t hoit wooe,
I holted, br mol hons it sote so mrett suttin oad whod
:Talils,
ean tua eee nonn sht wioed
Epoch: 2/20... Step: 200... Loss: 2.4393... Val Loss: 2.3937
wink'st.
EEINI
ToI:W sunras mrim theat mo the meutis halt, brasrers,
Ar by winge tuud wame hom wy ther hote toel.
LONLNSE:
I and whans:
nenled heels te sratt mo tir itores, I or to mreltt hides, wime thoun harled the hit ale
LCAUIE:
Lad, I hersaret stethy tath me ing bas wote sein taul mrard.
CUCINCRI:
Whor,
Ming int wert, merenther fon ancond in der on shit sond the soe thild,
The suun titd on oit th irre af aco cincusg toes heree tethy hhim tho thich ans mere wicl whal, thes hy als misteet, ther teamres he be ann fote me fo he hive toocisr amivy brt aund murtar oe batt ther bids fheud srelige thar by ofor. Auct on ar bauke thind,
Tod thor me toeltens hime aror on cesst wlet.
Nn he toe thete thus hate wovin, ho coent ou thaclor wfivr the ta shate berind we th olild anssen tor and thet wore time terorsiss me hoe, blarho tos ta sos menthe torite waln meerins sr erencesting ol arreen the dilt fach ot sorlo this fo mice and iuce he this cy samr treegerigh wouco thom mon the lorerith bico to
Epoch: 2/20... Step: 250... Loss: 2.3358... Val Loss: 2.2743
wink'st tonle.
That banleens melconke find the creins me sale file.
LUNES:
Sale by t ard inlly thun tiserifer he ce breon iths bale.
BIMENI:
Whes bellot soe the tout if hith tor mamlsirgs sothen in thy mrirs wame tice the sor asd ton of at beso ho birse thit thut
Them hyiure wo sor hit lallens the sarsence bot, meet are on by wouls th ofron wot hirse ofrees, woun and byir the woot or ound wrimor, and af mens mowl if frule.
SAIS Hore memeres: I'chy mase at my seor,
Theurd bute ancile the wire,
Far hun tot bis manch olang thee tirlur, bet bace the ce tones, an tath.
CAELIL:
By, langot in sor te wirgor.
MUCERIEA:
A torisllet shet lete the seumt as wesr feeled sive the thre malke there an hive tor to waen.
BLACESEN:
Wor sere aridl thinte thou thalg in mreth sryam the wire to dath boar soed sas beros me hentingert'dd ow out thist hace tath woriss if, toret, and hre sonlus.
Thane byatins orrruromr macinde cenimt mot orathe.
CAICEOTEN:
And ho that me the teor at she welilt
To purele ols. Thane
Epoch: 2/20... Step: 300... Loss: 2.2339... Val Loss: 2.1735
wink'st of my wis sead.
SOALCALS:
And hourd as sele seed on ther foug thith, bother whine;
Bow wime brood the mestanct as, felt mod of in theas bith werd,
And thy se therd the sovet is thay my, bery, ant soodes
We hath of buvald with hald hitor, and of sore
Ancand wosl me writh, momaned theut weld whis mother,
Ou wot to brow beond thes arlorssourt to wowe walk in
Houd sentols he anday, thit meet warinisg.
Whin andoule sis wett you, when be so whavimens
Sarime toud se theunds te sle we wrow whor me warin.
BURLIS:
Bul the chowe wo to co thee is there the be thour ttase,
Bor il whesilt't tho tome o pruisg theu mod wour of hoon
I me the branlong thay, ane whems to thall whe tame
I ling youd so sale though me ale woml blothe.
SIO:
Cowe the parstourte whot dote thou dithing ande the fate
Wout taly thit, her here thee win of to won mirt tous
Samant Mo hist Reson thele:
A'c mithe that thye hand, wo btot he bing thar the ceete to ast hitheed.
DONLOOTALS:
Nhas, bat winh math mlrein, ond os tristore
Epoch: 3/20... Step: 350... Loss: 2.1015... Val Loss: 2.1049
wink'st,
Shy lot buscent are thiskerif, thou, wime our sound:
To tome houng the must now she sound aflont.
KE GENCEON:
Wo hou to to cills oult on mese an thoud sheriss.
CURIOEUS:
I so matter in ip say, that till.'
Whansing it be sithy ant our and hos thee sid byeik.
DONE:
Mind, thou sid; be tith how whis all wother
I phempole, to dool oud mother
Home to to ment then mes ome tous
As seols bome, and, mowy. Whtret be boure so pirde, fords fishs our
And tow thes fring thou bads, in thet sold of thee the taes,
Will hen alish of atpor agene and is fill.
Toor thit be of sor wass the chatist alle bot cotele
And with toming and is tou thiit net, thiin.
GUTECTEOUTEN:
Thes the lade, buld so mosels in sull the wartore;
That son meak buls to tat, mathers; than.:
This wheth bith a all bace hit hir its sould,
Bo sowand to shich suik o mast ane mine,
Ior of some, atiet, the thes batlle, as the tort wicer
Than the curthing stane ass the mavensse so prankel fool and som
Thit if this thour ome thou the theml
Epoch: 3/20... Step: 400... Loss: 2.0654... Val Loss: 2.0471
wink'st shis dult
Thoun the hensles for werith amast some,
Why cordy for soulind stichit ofore the seike.
KING RIOCH:
Whrum to the grenes for ho how sellay ant wing,
Irle net nemer facrest the matels,
The have brongst for mese a pask of whry soont
Whene the groogh heor angos, ay hast,
And we my last heals nwat what a thit be home sentous of spoad afist,
Thich omit be the fastry ow whith fomart,
Ass she this dather what my feartel tet
They sust with fromy thut tlething and thengige;
Thou frove far not frovinge sulte taries of
I thit to tild thim ard faich thom will.
KARWIC:
Set the werlome, my franglowe, so took
Thile fain he mere be whee whave in to tlat fom at
Seiren wourd; bnow hace ate warcum, beed thenes,
So me tha sithy to mest a cound sin
the prange to so sat hin bean is
And tim, betith ans the to bet it on is taes
BAKD:
Me, and my con tite sour tend my farte,
The ceapue theme the tor of hall of him stome
Fre bed we hard that, this bagies of my ment my cidnor. Bucking mave to te myar
Epoch: 3/20... Step: 450... Loss: 1.9637... Val Loss: 1.9940
wink'st a toor,
Hesr bothin to mare thin to the hroume fricher me
Whan the bestlave of that who dees these'd wall of busce
And steit the thungs hites not serating it
I' mave be willters,
To han him he the bored; therefile
On thut shace to till seath, say thealss,
As ans morty one why let net wighing him;
I sarv the some for anch, at wace
And net masher me will and me to were:
As,
That that nat bact.
Peicer:
Seresty stark, and bither.
Of the sools thead?
SUCTILCASDOS:
I that as ste tlreess, and this me sear the banes,
Fallt, trets here work mave are hithel,-
Butel stold that by that and bester bulnt,
Whom that forrher hreach bante of thith some
Thance ble bath the brow ans becomes what.
CeLTroCE:
Whes a wall hyon the wime her,
RRARECH:
Nor o when all with hearet'ds of blitt
In he make a parcenst and to gray
Thit the gouser wis in that a for wount, and a gunts,
How thee heve sinds saed of wearsss of muds the word?
Misens:
Thou my lrom, for han hath her wrust,
At the hord when my srearn, be
Epoch: 4/20... Step: 500... Loss: 1.9569... Val Loss: 1.9553
wink'st
Tome that she highd tighs butt stolle, thee hes the dich
Aveater folle in ippance the fith that thee they feaped:
And with he sim see hin soule. I walld at here fordered
What me a tid wime'ss on you beevenst thee
Bhis beith, here thoush the this domour is thee
Thow sin the made to for the wouls at old thee.
GARTARY:
Whay heer'd teo of the well bo than, hemaly hear,
And stite me in the beaciese thee, fall in hid fead
Whese thou the past to des whico shall be in
thy losder tompe that be the for thun sit,
And thow whos hess his and with the dasing but an ame
Frast in, whes to dentlire,,
I be make and my dist as thist trees,
Tar will shin tene for th the sult bornt, stenen.
DUKE RINGO TERKEN:
An apeet, Indens you to lids thou ane me,
I wall to be mave of in tre myare:
And lake, thy frins, we loth sum foo disitnthes
The milles firn as of the day, sor heme
Feate on the besery, will thy sase tho ghe finter:
That'll to say steld, thous and to hou his hases
I me fands haw bure hid being was h
Epoch: 4/20... Step: 550... Loss: 1.9219... Val Loss: 1.9212
wink'st;
And wo will sweit not muns bot hat show.
SAMENLE:
This!
Siss IFmond:
And not bustars all and that fich of will dears.
Pessent Mingrang:
And thou! asang of the chower wor wess shouls
And for that to a toush is him. A douch of hears!
To this somely would and one his besters
Wouth in manty hreaths and all wow her proce.
CAMIOLA:
Well in bet the wath the plack'd, if make me,
But asor is thoy will to the corsent
Frat inded in the shale of to-monk afoost.
DUKE:
You bnst our made his would micel of that thy for hath,
Tall the sued the come of mone hat too seat,
Fil tuke ming and that feich in blow sut,
Fros ceenous bry bath in ispy thee the fart
To but of sire hongor and hum hong,
Mastard and fould the wend, that that is thee were.
I all thald mums a cootere, to dy heasile.
LOUTH CLOOCENTE:
Whot bust thou mand; inde they,,
We wich hear sperin whee thou bese but:
A with thome bortiof hore.
PAULIO:
Whon, far, thet hewels; in hit out ard ane bons,
That the ford the prighant bean you, fre
Epoch: 4/20... Step: 600... Loss: 1.8435... Val Loss: 1.8907
wink'st see hat hindss bold?.
He wat you wear a ghand that I the well, these houn besice!
To will the store, that masang inco be beat
Be prace that a till the paeden afonge ferer.
SICGERDI:
Bheik hive to getst by thes maty mand; fwich hase him
Whar it a whid for in spearter op that tone flet,
Far, my lavint and to by all fleemis to
sum, anl mad a shand will the wreces with hand.
The' suth him ant hil thund sead is the horom,
Withyry and so make som. Whes with he fornge,
Fhon me ate wing in the most told and bromes:
Whithour scronked, I'd be all well of that her bay
And maty bath amot blace of in thing be in
atareser and her shone a meranong;
Woull tall sark that mor swords: are sueper.
MENCIONA:
Bhave hath in, hit lornd.
BERCILARD:
I laven on may in the bring beer me fath with,
Tim and hadd my leaver, and not stolk'r but her sor
Him, wow would to to sward has stell the heppraes to
him wey her hes sincust; at such my londer'd ward,
And ther such our aspleriin a make but hough,
As nhild be se
Epoch: 5/20... Step: 650... Loss: 1.8365... Val Loss: 1.8598
wink'st!
POMTOS YORK:
But'er be mang affer that me have madn thy bued
Sheird halls that hawher to be the shald: and nor and sum;
An hemer, be and thou stind sender ming hiss anesier
Sore antherang her the couls, an migot,
My have what sat, beach. I would be with his steech
So forenon, sid sor and, is andsacl that,
On I have he will note tonering mar is sad
To sis in in of and be wall what sonest
Will be the piverae, and and store to tell
Hears thus firest, be an your his mine, what heats
With mesher me hasting and hole, swall, be,
Ather your savool hid sorr teith the cornces,
And firit ag thar make ang intwele think's,
And, tell shall diding to my herarge to the wordens,
In the murt wendous far thy warlint
Is such the caspees be to singer mucher,
Whouch thou houst altonot; a coopt hishald my mest in mensere.
LADY CORELUA:
She lovs the fiter of thou hastert.
POULI ORAMEN:
To dust deever whom show this and at the cith,
But him but out'tell tull,
I de falt of state.
KING RICHARD EFI
A:
And we
Epoch: 5/20... Step: 700... Loss: 1.8593... Val Loss: 1.8309
wink'st nebsed,
Whan it his nown bistor offored to me.
BENCOLILO:
Herean of all treie thenefore. Have should stant
To som to arle subfers'd ard thy shres in
wrome, be the cound, me bright the did of
Merinest of which as your poaron men,
Mome bund incourd, I his coneouse sir,
And starr and har of my love that he some
Far will not the past of trient he sone;
The worss when better, we'st an hame for it.
BUCKINGHAM:
Nass, then it me, I dave igen sill,
Thou saln of with nome, it ag my this and sore;
Frich not heard of were wall by hasts
France we we bud is than this sin,
And with shall blood in the bleand here,
Threateds in offinnt with boout.-
Why, the be not, ho hourd mide some, should.
Bisseang:
Sone, in thy lest, and abonious;
For and you his bony; my lay?'
I dosing arm is ang for of te hame
And tay the frace of this speak'd see hime
Theil that have shoos at in my lide saete
Mast my foull, see thee the heard and and sare
Whene hearts and he may bly to are in thre
sore's a slore sunce that no
Epoch: 5/20... Step: 750... Loss: 1.8179... Val Loss: 1.8087
wink'st'd's slive is heads.
LUCENTIO:
Here years, soverien muge,
And I talk the pluccy.
CAPULET:
That bath a detere' whoued at the, and wall.
MENENIUS:
Thath my hid to than a prace and man,
I drouk you fall would stent the bade.
BUCHINGHOO:
What, with wis is thee, beare that with the some
And be to stel the bakn starring whotere
To would sirsity to, hy bed with not,
Fally at till and fitly shall do the hroad
The cestes and bay, and her ancessily.
SICINIUS:
Bother, which biked of that hims in to
men asm ale a mour his drown, and thuse brood?
DUKE VINCENTIO:
While, arowy, hit's belithty abrey for with thim,
If their heaty the shelich fremt with so sake to with
Thar ane with non a with te filling.
Bissar:
Or moke it to than so sangant a the plower;
This he a bornthers with ip stision mer
thene shole wave would notend a birth it is
Where a give a farther of to be is sone
Bine, and were be mistrien, and im, well be not:
Sen sall the heance frow your grongle
The combont to this to hee he so m
Epoch: 6/20... Step: 800... Loss: 1.7574... Val Loss: 1.7913
wink'st and she dong,
And nay so had to-burthing best, and seen, this
And age be woilf to suppot, bett all a caris,
Thou and be broundes be and bing in in me?
Forsh Caston:
Ay, light, that the fenting agter them two strack?
GREDIO:
My hourd: and breath,---
PARYO:
O' sigst, twink the fell, and love then, thear's
Tentees my curle out all a mose: stratk;
She, by in string their say, as her the meal:
When strow whe wrow'ds by the hearth mes;
And trey hown'd should thou honce, and thank.
PARILEN:
Why sturt, I have weep to humpen to be in your;
The with the crove ang, the councance of sit
Hen hath would all men him, my fortet her, as
O menty war, but not and by did ware.
LUKE VINCENTIO:
At monher, in murther, stand, then, with have,
That her ath horsont that wint hin excaun.
QUEEN AF GAY:
There os the doom, for whilk then, the wourds.
CLAUDINE:
What I to thus is:
Soll, there the wullos surdy will wound, frow then
so may for themels of your lost.
KING EDWARD IV:
Tan it our gonts a tryignter,
Epoch: 6/20... Step: 850... Loss: 1.7228... Val Loss: 1.7664
wink'st would makes?
GRIOCARLER:
That we made my dausor with this?
Take him hithen.
CAPILIL:
And then me to the mote, bitter the cerss.
GLOUCIST:
Wither shall wimenes: all, as my dead be my mades
In seath of a blow, and not blood strem ham,
Therefight the wors he wher is the with thy lith.
In able the the felliand the hand all morrems,
I chould themence aroth to cit is to
The from be seted. With istend the dear she,
Than the swack of way wand the sispout.
SANIRY:
No many stirn he courtry thou artile the pate hip deang;?
BRUTUS:
O, he wrat a pharse.
LEDNTES:
A done, whit him me wold,.
CORIOLANUS:
By, I sure oo been's stook, my gond, as I bose.
Treet the chould in serther, the pear that the rows.
Which's is no men stept the swrence but a cworch,
To soll it this thing, and be net mean for shawed,
Who come, in they timporant'ge, but is be strity:
The brething bluaty a besty in the deat.
GRICIO:
My sat, sir you well you wall be, with your
Than to me to can the for with betted him;
And be to
Epoch: 6/20... Step: 900... Loss: 1.6983... Val Loss: 1.7548
wink'st have
Which but have better thy bid and peeser.
LeRceNS:
If thou art thit is but as you are
not she than were a shill be prome hold?
Secoss Mursering:
Send of brother!
BUCKINGHOM:
Think which blaid the bank beet him time of my hingerer;
But to the prayion; a darrhand should what;
She though the simiture and a wire may the consender to since then
as yours or hamseron mence more to me such offat.
MENCUTIO:
Bay, serst befand my forth, and sor is the pourd?:
They and you sount bash'd, a brown or my.
Murses:
Weld say: and by some coure what should how a provease,
Andseed, the poiled is, for a make me best not,
On a dostet on more aparot of their.
MENENIUS:
Hene here il nome father, and the herse bidds,
The spollow'd my surs this slinerof of a to
these holds and be hamb hove to somelion,
In will in all stonnow hour, thy come my seach.
And foilest wish all my fains of the muring,
I have this way, by my paie to descire
In your sarrows is mined on way, we are's now.
SICENIO:
Why heave the
Epoch: 7/20... Step: 950... Loss: 1.6865... Val Loss: 1.7249
wink'st sone
With strings the food o' the pearige.
COMINIUS:
My gracaue in heart!'
Senvant:
Wo will dotr hander.
GLOUCESTER:
What! all mest all men must be slake her,
At these cerse having intermins te are.
CRINCAOF:
I have bue thou, as thy shall missings,
The boy of your were: a deasurted own
Wanwands beraich, I wall hade hamisiar.
KING RICHARD II:
I that word him. Which stones to bead his
may breath of a geator in hearter: andst
To demisiteding and and hossedfy,
I wat these francous.
Pucitizen:
Then mary will yourless, I am to the wite of her
Truchon the fortlush with a twind's too a prace,
Aften that then arave his browentss illide
I would will hrack o' bestence thes arly,
Or and to abinc our with thes ip, a preal,
The grace, he weal make head but triget.
To-mert you are bagise a say mereas
I tray you an mest strets'd me.
CLORENCE:
A morin, a stand thy beder mantered,
I cere his sonry some must and flows:
I am to him, man sheep,
Whith thou time it surpect is not?
DUKE VINCENTIO:
So
Epoch: 7/20... Step: 1000... Loss: 1.6708... Val Loss: 1.7132
wink'st
Amifor with brother wardised the bearth,
Which too myself that but at her woess my lord.
COMINIUS:
Thy had offect of your land, I were a preperad
I' the most art thy did saunty and heart;
Which bading fhachend of the daught inting you
shall my both sucant may of younenged
From yours. Who heant all the sond arould
To be a tankny of you take my still,
Bun are this anoughity to his councies.
COMINIUS:
Gelvane you, grown your carnon!
Fow the his and, the sprare't or himself.
Perverting.
BENVOLIO:
Heart here that the will as hore of you.
LEONTES:
What soul'd in all worsh heavicg's sour.
POPIS:
Which hath may soul never me how's bried;
Farete to prove the teve shall trues,.
I pootly to be should he, shall not thee:
Tanker a teem, are stniegh of scoses and sur
To haspand me and that they be one to sir
For you have sweets with at and compet hy mise so
muld, hishing states all will him live the hise,
Hor these astardan why lords alatamest my gold.
The madiar bear sens that it. Another
Who
Epoch: 7/20... Step: 1050... Loss: 1.6594... Val Loss: 1.7112
wink'st.
MENENIUS:
Nister it will nor posine and begce as.
First Gond:
And say it in.
BENVOLIO:
Not he seed, that I master to-that all the forth:
And so had midst an which more sweaticaunt
Their wanning.
PLoUD:
Then so say this have strent but that have ying to the
reator.
DUKE VINCENTIO:
To corne it as you sand a sheent and fell him.
BENVOLIO:
So lies, he heart, my sain, it sanct on house?
Pursean:
Nos, tlotore?.
PLUEE:
No the truse so hightery's lifiour's,
And thy feildes hence, he meading one:
And heard my sheal and a sands and with
I to that he shooder to hear
Hanghe man ham sous for them frams be princeed.
As thou chastes our comes fur her farester shall
the tonging of this fruch how our hisseronest
Thin it, his wither how of the chingst on. With shim with bus:
The swile serving a fase him in had to come;
Tho good a pearer?
First Surad:
Wo shall bear men him with the gnacle miertised,
Muct we parsugh; a getslipest oft munt to
but weep and suck of breasy of your carnor,
I depy set
Epoch: 8/20... Step: 1100... Loss: 1.6264... Val Loss: 1.6862
wink'st houriss, but
speed by destare of spatch: if was a dicters.
FLORIZEL:
Brooth, that I his holding, thou home!
KING RICHARD III:
Boody!'s
Warwick walice?
KING RICHARD III:
What stayd whice ho some prive show thou creesunit
Take to thee to beens twein of the pleatire.
DUKE VINCENTIO:
Betie so me may the comes of stnong him be to.
That seath, those worth husbend fich wouldest.
GLOUCESTER:
Then, all the wercly to tile whichs the dewere;
Hid friin whise the hallings:
Ont the mine, what me, saidy and loving of
the hoverty hishes, and are of be all.
KING RICHARD III:
Madam, is were of hand mine? and master how; and'ce
To men aras my head.
DUCHESS OF YAUN:
When as your grace it somelly lident that him,
Of thoreous must suchlads of death.
KING EDWARD IV:
Aur thing you by it, becimes will not deseed
Made in the cainty though you do, so were will
For the supticllod all-pairse the commands
Of thes mother be id never's bleash.
Somong:
His faster? langy, whis is he deand to hould
An haims. Ca
Epoch: 8/20... Step: 1150... Loss: 1.5389... Val Loss: 1.6762
wink'st the mound,
Thy pals, her, fander their pardacties hims.
First Cordace:
Would thry ware would am it with such our call:
Orte my mord hull and not my soner impasters
Have the honion hor hoth for me could
To have nubled both of mince in the wain a thonger,
Winging me to thou which this far then, fand
I have time tome, he's natuled.
MENENIUS:
Here clown my comman.
Or is not have were never so,
And your sushel the faint.
Sich and Catissat:
A than I sevar mear'd him my heart
By his sen if my wire is sor thy baster
As ane to take the mords my stry heard,
But not the way; a master so my lare,
The hoant fear never bry sumpling and.
Thene felse that art of mast of sur a wnether.
CEMENEN:
Thou host has ans ou servate in a might.:
Harraty it: we cansing and my son to heak of
I
As batom at the having wire agail,
When my love of cernuch of my sare sater;
Than his dight show the resolve theand:'s fear
him fears, to confert wompt their curse of you.
DUKE VINCENTIO:
Then? that I have hid her his f
Epoch: 8/20... Step: 1200... Loss: 1.5853... Val Loss: 1.6746
wink'st house?
DORES:
Shath I day worth it servant of ancame,
But the hanger in the son, and staid outs
Told true my shood-fith a doust, for the crutusher.
Trie, as I chisting forth'd, will, thou hast
But it bloaved and he ounster to-trough!
A such tell thom the down to them to thou
hels scroke the castorss of his wert to gave hasting at: afleck,
And shand me that the fear's sumself, buges he was;
Which, me, in this dreak may budned thus fathe,
I hape are clacd in the his bysenced in the soun
Both will hive or hence on my placo, and yes.
CORIOLANUS:
I as to surs to a contulle, our fouths!
Would since the bear of me that sweet will for
The wakes here't of my but shempled and mines is the
swaes; and but that you stand against to possed to concuin;
I'll how the consceater or our passits offer,
With atay thy bost of the preshing son
For the parced of such a sirely as the duds.
BLUTUS:
And I should be my shall not will do not a
doss tratter of superor; o'erdens a puticolf
To due on the bett, sel
Epoch: 9/20... Step: 1250... Loss: 1.5738... Val Loss: 1.6561
wink'st a for this lives,
Faiting son of conte to heer thee ny some
Is name murder feor wand thou be wnow
Shall fan the trunger of his great's.
CAMILLI:
Or abour all the dainhery we by theme,
Oncess'r, this brother mais of sirce the coprint
To make a the wanch of sure to she towald,
And had his hapsise the die, be must sorly
For to year at wilt make once he heaven bown,
I hath me ancugited menster'd be in thee,
It is his swile, brother, and sant he mating maidst:
To help to become it were tears make them,
What as make it; in me thou honoul ant his;
And and shape it our put mury, thou wilt not an ere,
Was the bechores at, and my bliness, here,
What have the cunencast of thy lart,
My mind ows dost of call to have their laye,
Whow are the shaptly pardons thou tlows that make
With ferelieve in hees full and my compent.
KING EDWARD IV:
All the stran that he sunching of yourselve
Tranch and thou that your held of this death
To deed make you fillt so doublomes thing,
To me my life in all mouth and
Epoch: 9/20... Step: 1300... Loss: 1.5800... Val Loss: 1.6431
wink'st for this lyat,
Which we thrown tormad time he dave, ablith,
We dispetsion so hil consun to a subjeed,
Whore some bid tteness; my little husbands,
Is these chrapt their begounds hall now dest here?
And you to deen the comsugent. For I his consin.
GLOUCESTER:
Which you awaid not in a gentrerange,
Or matereng of thick with for our bone
To mung to beceed his lithent fair saiders.
BAUTUS:
Then, my for it be is more for to bloage,
With him, bit sake young and fendite wan.
BUSHY:
Most cauter of har sitious of a mak;
Our confel so this blaage to be me more. Which,
Ous both hif say; who that you care to home.
CAUFORD:
And she hever secisus ang thou shawt and fly
In honay, bethere fail was here furthee:
The houllss betore the ttought make her by,
The wither cours ageace a boy, and mosters
I my fear, besiel with too; and foulster.
CORIOLANUS:
O will shall such you: then find the goint:
The death of the blooded truse and here and deverge.
I meater of to death? Bremertion, wrat off,
I taken in
Epoch: 9/20... Step: 1350... Loss: 1.5967... Val Loss: 1.6408
wink'st is ferrimes!
KING RICHARD II:
O heaving touble must say he the gaude me;
At were in him in your baty, servants were
Spirition'd of your wreef of him as we sid;
And I make your browness to wife hard the such
Of your henger from so beags honest by the dusining,
For this hame fam the fear on sigh.
LULIO:
Sumale must wrengt,
Forth a supblods we callens him was fisse,
In a womd arpers afford'd, to the corter
And friar. I must dost hones in my comments
In the pows for this foul of the wifl'd foil'd
Have all abard's our brought of, traitors file.
Te be so lessing him, and bete them.
Servant:
He say him, him he, sland yingerosal heaven of shim
As man hither fure must sold wolld, the racueter?
DUKE VINCENTIO:
O, therewore you.
LARY CAPULET:
How now, the sign, this lovery beaven's more beings;
And she to be and mone back my life.
DUKE VINCENTIO:
Aman we sent what is this swead breantes are
To-son, my lark. then he bay with mide own
deed, shall will divery fon the mady all: when
Have no goo
Epoch: 10/20... Step: 1400... Loss: 1.5330... Val Loss: 1.6252
wink'st with the can him
A son, or to by as mare meed here,
That is for but and thricts in the cut of
For shame behown'd at ad have tremble.
QUEEN OLIZABETH:
Some is hore son, but then of a man awad
Hast it it in the further windishs me.
KING HENRY VI:
Why's there's doth?
BRUTUS:
Nan, we'll be parce that way, and more this wall,
If that the lost of told to men on their ghame.
Wender you, they do the world both to the stay
Till not his drown, and when that he is been;
To helver so mady baty humbrence:
And they this honiss bart off heme.
CLIFFORD:
Are sweet stonished.
DUCHESS OF YORK:
I word he'll bad and grace the gitser than this;
For mency here meen for time, follow it.
How what's, why way on, went a treator of the ledsthen heap,
Is shane my greet with most been ty been.
KING EDWARD IV:
You sonn they the doye may's no piscenter than you thee!
CORIOLANUS:
Must chours on the most true and that, and when.
QUEEN ELIZABETH:
Ay, therefore the king be not let thee
Harr toull to my would sump
Epoch: 10/20... Step: 1450... Loss: 1.5926... Val Loss: 1.6190
wink'st heip of him.
POLIXENES:
Then in the wark his flarerel that I had,
Inseech your creatuse and done: so live.
LORD MENTERUS:
What come to some an those, there's he is, betwere
With the have man. But that that's time me for
my his show nome wretched what such that hath him
As how have tull of doubt. A pold thousand as
To your borys. I told not my honourad,
Which farest be some thus wender'd than
And there that do be duttons of my such?
PLIUCENTER:
Nay, bit of the hhard?
BUCKINGHAM:
That I would have to help trioly to these
Whise him for thou art of heart; who may not troaks
But their for the hear mest spike the wind.
BASTANUS:
So all buy noth this assert out this there.
BRUTUS:
But I would be twy sould horoun hormater,
Were she bale wnomb the slare and befwered?
Hortens, and thou soundring the fortune,
The woulds that will not brack thou, good, I may
And hear a matter, think of dad, my deed,
We have no mlake a band.
And wood war from my honour with his
brother, his soul, fear had so,
Epoch: 10/20... Step: 1500... Loss: 1.5729... Val Loss: 1.6094
wink'st flower made
Because himself that I are in wind.
MERCUTIO:
Now, astee me to me, he mean, to see
Shard Sing Marin:
That you and sees, an hand.
MENENIUS:
My lord, he slay a thousast may:
And no mestoused and spare more came to have me
As you say for a littlouss stand to these
The mister fould to cann the strong'd of such to hore,
In time to this accuramit more sprenest shall peace?
So for the feel of the ments of some were one
states and were in the courts; if you be goter
Stand hath this foults of down the father.
ASTLUCIO:
Wherein you there mares your coonor?
First Muthary:
My lord,
Than this saigtions. This farew lotg and a letty
Of this fear true----house of men of the fale as we
so for a blood of the way
To tay both to deash to predy to had
tames with all the worther hands them for thee friends
We what this look to the treass, by she't of the
seins, or at a scanter some, my heart.
LUCIO:
Man me by hither? what storm theart spear
A lords for so weather with seres, and missay
To
Epoch: 11/20... Step: 1550... Loss: 1.4930... Val Loss: 1.6028
wink'st if which,
I hear'd, thy hand, say thy surper for me.
GROMEO:
Have not not drual, sir; thies you have suppled'd
He wish you and this a dread; till he she speek.
DUKE VINCENTIO:
He he whither.
CLARENCE:
Which have me wry't a werring shall have so thee,
Your fuer swiriciars this: that be triators of
Your brother how till sevace of your gunders?
Second Curson:
I thynee the graves; sir; she, be that
Which such you made-thee to her swort of to the fight
First say sired ments the daw's to death;
He is be a suster that it to me,
To well the city would not all.
PAULINA:
His two sir, in the tanks; and have infucl thee:
This salito this aborth of more would name
To to the togen, in it disparter all?
The bent, and stupt and surber and lasters,
Bo helpon sis where thou highs in the pount.
BRUTUS:
That me the wifes, and my sould trust, so fail.
Some thought'st some best or at me, that they say.
KING EDWARD IV:
Nay, say, I should nend by the sople as.
And what the compentare, but were selvese
Epoch: 11/20... Step: 1600... Loss: 1.5242... Val Loss: 1.5929
wink'st the bear of all:
I am not my foor or, so saider most before
I she's answer, too mund, to ask my liege,
Thrack to pune of those trato are was fill.
Then, I'll be discover in the duke and foe
To hands a bide them, far with bestine. And see,
March'd and this take, we beer it to seem.
Aben, my liege?
CAMILLO:
I will the hand, be say me with is file:
Shall make my leadion, brooks, sards how for my:
Methis they suspecious flerhard, think,
And filst sweet manie cearned for home,
That he has sard me hour for my son here
With the said of seme, for you wous to be when
Wolly heart thou would the san, and thereof the
but, here here's heard; it stats, why hrepsed thou
And their fless all the servious pleasure:
He mire will said my life: be mane bride
How him that we sim: I do blent a chorso,
That with he day so nothing are hus trenche;
As I shall see a thire a prince: for the state and
twill staties fin to something, so he should say
What in troe are the world fear her and on
From forth our tellie
Epoch: 11/20... Step: 1650... Loss: 1.5157... Val Loss: 1.5917
wink'st sach'd it store with against
Wroth her for a gone and they a may; he?
BUCKINGHAM:
My lord! Citizen, then my mode come?
LUCIO:
And the trust to seed my life, bound this bown
Of my dead and mose provest beget a trumpet and
the heaser born of see. There is be say!
Second Servingman:
I am his stome of all his last; the point be dost.
MAMILLIUS:
Wethers to the heach as you speak of him.
MENENIUS:
Should it the pisence weer in this honour?
DUKE VINCENTIO:
Near, whose prince.: I do both well antred,
More thom my hours siscest; and hour hath heard;
And, telling shall have all the stantes,
The foom and life the shall to the world and done
A plore fit in then brank o' the baith,
Would trief intences a to my deed
And sure me but, and thou but which a sire hither?
KING EDWARD IV:
Have you boy tells? I say, with own worse,
Shame thou hast mouth the cotizent was;
If now the painte would to all we are strong,
In all we'll notle a brother foub tongue
To butinius to at his dlanes hath all.
And t
Epoch: 12/20... Step: 1700... Loss: 1.4657... Val Loss: 1.5757
wink'st bid her land
Wor woman which wanwald.
POLIXENES:
If makes whence moungers?
MENENIUS:
And that he's body, that were now as follow
That wonder sug tit a scirence than thou,
For you must leas no pare a dost more tome,
And blood wourd both, by this man this by theers
At the murtiru then that thus her seers and honour?
And, and thou did bage when we will trule. He his dry
And the can batiand to the butisars!
We like the sleap at it.
Musterin:
I hap assurity be is brother;
Brought your stroubs.
POLIXENES:
And no sam, in the histeed world many hands of
War and any life may must say hath to be.
SICINIUS:
That I'll do you shall pleasu to you a troines mine:
I say he the pity!
What I before that which make thee, grace;
Yet, and my fetter, her werms my trane?
LEONTES:
The shoot whither?
BENVOLIO:
O, my lord, have not with stistation than
But be in puase, sir; there is no mere's fates,
And milly on thou shalt firm oo winding-leath.
It had bear and hid well hand my battle,
And full mine chor
Epoch: 12/20... Step: 1750... Loss: 1.5106... Val Loss: 1.5712
wink'st is hands and a mose at
in a sire out for your honour's barking day
As that.
SICINIUS:
I would not sin: alonour'd to at thim? if
Ane speet such office.'
MARCIUS:
Where is my death.
FLORIZEL:
I to thy burny:
I come, though into her bearing to do head his
bracion than these and in madier to your hads,
Breast thine it save how then, that, full a dails
To serve it, but where he sumper and him.
Wench and fits my truth in that in sheal heart as
I, no fortuge my geatly wear which was more. Thy
please, bling, I moy, they with our friends serve
As from by thou death! their bodys from them,
Faith with his daintrous seep of with mine offices;
Say you shall die mine say such swolts in thus?
LADY ANNE:
Must I too mine: and are we had all the heart
When he hath peace? if you mught you that he
hous to denith, best with his given sign'd,
When thieving son his loves he dares is away.
LADY ANNE:
O, been you arain the firth here his doth
As such many parse's of his grignts seal'd
Afatious think with
Epoch: 12/20... Step: 1800... Loss: 1.4597... Val Loss: 1.5726
wink'st that I thlist so now.
RIVENS:
By the line but; he say with as in all.
Thy foot, and been heer but should they meach
And as some counfally stord the soul-dieres.
KING EDWARD IV:
Ay, at with yours.
CORIOLANUS:
No, all a scater's thing of answire.
First Servens:
Now not that I the buty hath honest he mest
That so not so, as to spie. Therefore, my fearful,
The begter has the will. These sworns' foelings he,
And there what so some to do, theur a set male so,
I burnal humber to chooking our.
BRUTUS:
Alo some trith has mulder, as a better war.
That you can hath this: he was delieded;
And therefore arted his far about your daunts.
CORIOLANUS:
The husby consented mather with your phile.
This here hath at youther than my fount fall;
Shall I she is now for alm abroush on't.
CORIOLANUS:
No, where's mush pall'd, for his seno solliness.
MENENIUS:
The haty if thorefore?
MUSTY:
No mutter and the how thereof alterud
With mildor of his groor's loods are when.
ROMEO:
He will not have but him.
Epoch: 13/20... Step: 1850... Loss: 1.3923... Val Loss: 1.5608
wink'st a pure thing which
he we shall wive when turn; my come, and he had bast.
Yet will the criefe, sir, and I'll nut be any
And tell them teating world. Answel we, that
A bartitul than to have besought.
Have an a word here time and fellow strewn to speak,
And dutined with the man. I sail, ince this hand.
KING LEWIS XI:
Be this alamst of the will by; it this?
How it so. There's my throuth: so would not strike:
But not a belance than it there's thither:
I would I saly some from where to my body.
My, that is truther, thou wost have both she me.
DUKE VINCENTIO:
Thanks the filse but so. That do the pray of.
ABHORSON:
O, still thou, tell, to him a trome? Then may,
More hoose, it were in than the fairest his port,
And with his furthel than this lases beast begot
The thousand siffer are not wrong.
DUKE VINCENTIO:
Slost, try mine tome, and being night to sat him?
KING RICHARD II:
His sweet we do a found; my grow: we show'd him,
He brown to his myself: the brow to presence
Why much set all too m
Epoch: 13/20... Step: 1900... Loss: 1.4576... Val Loss: 1.5580
wink'st heaven?
LADY CAPULET:
I as to thee o'stle point me fear these
That met shall well to percial and antermand of
To man is prince of son; the dawning learn
And now thy heart aftities, for a blind one.
GLOUCESTER:
Nurbel in a clising.
LIONTES:
Well, he will melces in pease to come time;
Thou hart a body to change, to the pastion
That you all they shorsed of the sunce of was.
LUCENTIO:
This banishment so man are bluck
The duke, my grace with more friends of makes.
Mare't the would with the conslattion of hissees!
LEONTES:
Not, many me, be discovires, both hother?
LADY ANNE:
We do not well boys
The sun of all take him to prave to they:
O, here is fyal was taments as the shint
And latour'd to his him in wan, the world of sure
Than may he cannot so famal with this putes?
KING HENRY VI:
Why are sirred with your sinter breal and spince?
RAMILLO:
The house I have tell blood,
With strange made at your foot, it sworn him;
Thou hapt be male money in bray supfect,
What sitter show them hand o
Epoch: 13/20... Step: 1950... Loss: 1.4679... Val Loss: 1.5537
wink'st all such toul tlembly.
POMPEY:
How thou take most fault fill and baster, art it spayed.
CAMILLO:
Havil, not it is it, in the cordent of his presast
she then and fear'd by wemed.
Second Murderer:
He has the percent and with of him battle,
To that blood trow your son?
ShYand:
My lord!
BRUTUS:
Will you mear to her. See much the brother?
PERDITA:
Why, shall thou do you learn
She stall both, I will fooly them but would
sitce an in the sheat torners' the mast on of you, and yee,
to prince them, thy word, there father, ard not!
ROMEO:
My hard, say, broult he been fit to you have
The parthant fithing in his suppte of her,
First me it salice as mine hast is had;
Agoing thou thou banest amadaber in this brow,
To hand him mine of her sire' luttress,
Ord to me here formine! as a foul to tree,
For they that was bear most place some more
And founds me strengthed's toble, to have a camal bitter.
LADWESS:
How!
ANGELO:
Hath, to the may, but I have deserved it.
CORIOLANUS:
Must he is the suns
Epoch: 13/20... Step: 2000... Loss: 1.4111... Val Loss: 1.5544
wink'st my friend he with a frame
That waid as this say thought him here ferene
Of lintlewnst those the patest brum him,
And not a soul are are humpred her was;
Betwixt is here, and my grief whom he things,
But the coward consalience are so.
BRUTUS:
My back, will did, thou must be counter'd with so.
But if you do thy word-will tender flatters.
HERMIONE:
The' nothing that it is the duke that they,
Fareweard, we look my chanter'd arm there flatter
And hall blood my sorrow he hear you,
I can thou boubt to head, sir! I cannot would:
For me and lay bid as meet or man
That this swercised twan of my mother.
Ferent Serven:
How see him to as aw you are the general
Here is no sun.
PRINCE EDWARD:
And then I said abity wood, by weep,
Whet will the mangerous with a take and man;
For myself crave, the means or all my hind and wated,
If string thy discress of a tell.
KING RICHARD II:
Such old wise be begrace thy troop and then,
Of my crefires that have hardy stands,
And twenty hath die not we more to pa
Epoch: 14/20... Step: 2050... Loss: 1.4142... Val Loss: 1.5458
wink'st my from hardy.
LUCIO:
O are thy boyo, that as answhen it an
most sort thine honour, hree is she so love:
In marry, I'll poted and his swords, heaven;
Wethard'd, and with his brother of the must:
And why that drow must look to the feir from;
Who hath still with what my sturb is accusber.
JULIET:
O must some sulsent! sir, you commanded thee.
KING HENRY VI:
What saiding that husbands stir is to cark on
Thy seel with hope, and mercy his wonder;
To have men mine braves with the chale.
I must do speak and foldor sweet arviler.
KING RICHARD III:
Have it not with at time him wretch our light;
I'll tols my mind of them, intell it in.
CaPULET:
What? the cime of myself be man, here in them
Both for him with a hlanden faces of treach
As at the huclard; the gard no work hime had.
BUCKINGHAM:
Now! the saint missed tone.
GREBOO:
Now, madam! but he happy for him,
I day sero it somethand to the poar,
Say to my such my ling highness and blish'd,
I like a through.
GLOUCESTER:
So many look'd with
Epoch: 14/20... Step: 2100... Loss: 1.4925... Val Loss: 1.5403
wink'st he match'd of these
from our dease.
Shepherd:
Madien; you do again to be the lord.
POMPEY:
You shall throw them.
Second Catizen:
What can he we were with my hand of misters
And meaks his holy and for their becomes
But, if at thry with the was of your hends,
And stir they signion mighty and leon,
Farewell, and have my prison wash my heart.
GLOUCESTER:
So lie not show.
LADY CAPULET:
Nor, if you shall be as the pleesart brother?
SICINIUS:
His nome what stay, stond my things in our gods.
RICHARD:
O this is this father that in heaven bain,
And men the briend.
LEONTES:
What, both I comminate:
There has this crast at toutune is to them:
Me wind of prate wind some and son a prace.
BACFHIRDIAN:
Here, after your plose with all his sure,
And so so than,
My thanks should fiesd to his son wess his friends:
Fille that my grace in the polious sares;
Where opensule shall poece the whenguil capss
Of my fuel festing of the tumes or chown
To dare the hiss appived to the presence
Hath a figtal in
Epoch: 14/20... Step: 2150... Loss: 1.4673... Val Loss: 1.5310
wink'st with thy world true,
How is this werpoded in all honour, mine,
Our bloody than his perce, for you should be sent.
To who hald be to be to hear; thou wouldst here for the
son,
Thy trabelors, as if your love to the court
Tran of your brick his fair office. If thou? this
I conest me; and show is better mother;
Thou art a toothrow, and we be daughter.
KING RICHARD III:
Tiding tho colds, and thou nurse can are
My dear, and when with you; and if I can wear
The fareless pear of allites.'
DUKE VINCENTIO:
Are in our face of my sword.
Clown:
My lord, that is the prough; and then thy down.
GLOUCESTER:
How say I bedee, and me heal to the sout,
And ferlock in a twas to admint than the
reporting till they father, I'll some steals to-bay,
Is not of that her should here, and a belle,
With cursine sentence for her heart, of the strues,
Weak hut here secl our, from the words thou dischers,
Frame the solder that hours and hath wadly heaven
When your man friend so, by thinks of him.
CLAUDIO:
My swatt
Epoch: 15/20... Step: 2200... Loss: 1.4721... Val Loss: 1.5259
wink'st him, are here and truth;
But is the bloody are aboft the delate,
And a tortull to his word and futh and mints,
To the profudest the pation and digning
From create his do we will forsend your general:
How now, I had, in lie no time to be,
Be sake it that to live the tunnot will,
Or change why did maid your sin and back him with.
BLUNT:
What? I may, sir; he shall, so, new made him,
So much as misterour to the heaving.
DUKE OF ERCHARD:
I day, if that it dasently fair wlile and
But falling to the which that seized them?
ROMEO:
The great subjectess than thy doos, where you besteads
Thy poor speaks: and are
By lies that churded with thy desire coulds,
And bittle will appear wadom but a doul,
Or comfort fruends, as must with sure, but burning.
KING HENRY VI:
Ay, like hath man, misprest this:, and I have,
That to the prevent ars of men's moch mons,
Have we while bear shall dear mercy as father,
All do to pread of times of meats
That takent this beard, but neven is a grow.
One-word and whom
Epoch: 15/20... Step: 2250... Loss: 1.4607... Val Loss: 1.5276
wink'st boy, and pursugh
The cumpenion of bysing and look'd how all so, a fortuce
would not begot the more; I deney it, to theme
high indeched.
LEONTES:
The policous, sir boy, that being an ence
of seathers, without to my heads and had
all with his fatien, against the wrong, brake,
for that thinkers hind thy creato find the pripen.
BUCKINGHAM:
How fail, my lord.
KING EDWARD IV:
Both to any thing of his abmantine
In made had weeps of weathers to word, his
hen meass methink; he is infore the brother.
CADILAUS:
I do go sorrst not is with all the gods, to base,
and my bed make words the bight on this.
LUCIO:
What, is the hand of head you
That they'll perfice in this truly?
BENVOLIO:
Madam, we shall he'll be all the breath, of stander than
will stifle to beseeping; and be almost
Mascess the give-boly.
PAULINA:
I'll well as still my.
CAMILLO:
It is now how he back.
CAMILLO:
Ay, what sets and married?
LUCIO:
Before his bid adace, the great hath pride you have
the such a poor: indeed; sir, y
Epoch: 15/20... Step: 2300... Loss: 1.3969... Val Loss: 1.5255
wink'st their cast is begnable
an one in our honour of my foolish seerers; I what
bair in me well at the sentence sorrow how you
spoke to the bair to calling for the matter's
wine: then thy thrave the betters all my suffer,
Sword be that warratest to agay in hand,
Betooth thee, good my hour, I'll plain my true;
Make the worts since.
Lord:
Ay, my great word; is it to die,
So bland your spack; and no more fault a from
The perspance shall have not with a granted brother
Which heart the pritorers to sound a tlime,
We here the clorn's fair forth and feels a wife,
The would be diege you.' teal is it and hear as
The wring, what true; my liege, and what news shall
My crown it crown; and you say.
Would not we provise him;
That that he both the distant was
A blessed and feal to call it will
not sort that bret it tongua, thou dream'st the cuper.
DUKE VINCENTIO:
Why, if me heard mime?
AUTOLYCUS:
So the malice, be privoted.
Second Ledvem:
It is not bed,
For a whire is not that you will cat by.
ROMEO:
Epoch: 16/20... Step: 2350... Loss: 1.4015... Val Loss: 1.5247
wink'st so fallow him. And you.
PALIS:
At these the shruse to do my heir to her wear
Too spare them bare, a most scrive, I cannot been
With a set apome this throught is a beast,
That he will make them be the stoted.
CAMILLO:
What, an you be a more.
MENENIUS:
Who shall not dead this is the giving for
Hem be but faint? an out it shall her fraely;
And say the hope: he me so dear: had my doubt
another foul abscause but in the fearful country
That stell the mine have naturus, it she had
Which seem it be in holood.
SICINIUS:
I do some prince, whree's your hand the fellow is
And shall be to thy had for her
Fourself what we with saint to do not,
Nor nothing.
MENENIUS:
Tell them, heaven mine horse; shall not sut our hand;
There comes with our provedy and but thou hadst
With him'el from the death be an attended on,
And bointy from hars, worts shall not speak mide,
Speak morcience, who shall be sick'd all head
I'll never me a way thore hil him,
And sir the mulder's power, I pruy: I shall:
I'll sleep
Epoch: 16/20... Step: 2400... Loss: 1.3918... Val Loss: 1.5201
wink'st now to be sold, her
come hath as incalling of our astain
That have thy sumsele are too: but whither;
Belove's in sparent, that is some with sunsel.
RUCIAT:
My care my heart to die hite thou ast thine,
But how so speaks she be these wits: take it to death!
ROMEO:
When thou hast lengthous honour that I thy best;
For what seet begins to hope
And to seal so, if you cause?
Womes! here ars feor would be stone themeel,
Take they make men by presuce of the ways.
DUKE OF YORK:
I will prove a what all our glory again!
DUKE OF YORK:
Thou wast have all: boy! lose that with the chief how
Is it no lighten and bloody down from steed;
He was my brother together, fair she boy?
KING RICHARD III:
A cousin hard beelford as these had crist
A begtard what I tear the steels are plead
This footest his husband? therefore th shanged you
Shall not confease a mercilence,
As thou despike; and mine was what he will
With my thin tentle is make merell, their both
I way; I must be sworn to-man have seen,
That you
Epoch: 16/20... Step: 2450... Loss: 1.4282... Val Loss: 1.5133
wink'st frombless' honour!
LEONTES:
That had should he with but my words:
Was yours leave too, how but myself being side.
I will hang them so, that I would now see,
His wardeched of the children loves be when
A thing a tale we prevail beants the seen.
CLARENCE:
Help had seven hers and my bonom to be pate,
Which to have part to-bate, his hurt's move
To him:
I cannot bound your lord intents fut him.
LORD MOPLARD:
Mest of the wenth, before your counsel, to death.
BUCKINGHAM:
Alat! they are thas I am searned munind
Are her hoped impersion, may show him a word;
And what he heart it come by this more sentes,
What's then, my lord, the whates for you cry,
He be in a way.
SATCLOUD:
I clood my son. And toot be my triend,
By not so duck me, in thy shall were then?
The most sentence to peall the chosed been
That we may be the freetempery on
Beaven stopt the bess, and be his head me here,
Oversing trust for seemer, this wars at likes
From sinder'd offeces: father's coucint curse,
This bigdar with her
Epoch: 17/20... Step: 2500... Loss: 1.3720... Val Loss: 1.5127
wink'st him! To burse him heavy deciles?
BUCKINGHAM:
Now have thou throng to be a colder'd open.
PERDITA:
O bloody counsel, as the benofom of so,
The sake alonour been frith honour on.
PRINCE EDWARD:
Go; why should well a peteral clood!
And my will tender to this wearing shield
And but her town, that, that I say, she limed.
LORY OLENUS:
Ay, are the dowrenger for this crost more choores.
KING EDWARD IV:
Alas! if his begin, be hangs. For she were
And meet have leaves the bring of son that caunes
And cannot to help fight, for the comans!
Affection talus, he was dole thou accose
That made me word and soldier; which since
The prisoner my further with a fire
To crave the covening born and sweet spirit
For mine thus wall. By shrife, by sigh to ceate,
Were a fanture of hit of done: too made,
Our gian of yours: sawn my late, I will sitting.
LUCENTIO:
This means, and fash and truak made teiring death:
But why would be his spoken winced to thee?
Where thou she shall be sureding sound them seized.
Epoch: 17/20... Step: 2550... Loss: 1.4518... Val Loss: 1.5131
wink'st as thou didst being him.
FLORIZEL:
I hate be done. I mean that they shall pose
Most subjects, till have but such weary beans.
CLARENCE:
Thou whose comes answer the chearing from
The tenten would hold him against thee,
And should by nothing tower to thou; as I know.
He, till I well no war, and less him some them,
Who take with his son, in the death be done;
Or change what news in silence shall they forch,
And to all too manes of dead lies, trouble.
LADY ANNE:
And he comes not this people in my came how, as's
Fault of his filling.
LUCIO:
He were not so:'d all.
MARIANA:
He may then did into and come to you,
And I sid, sore! his father, be mighty,
Men will not condeming.
COMINIUS:
Whoreso means
We he think'st not;
Think he combendled of my crift:
Your fearty, and all some and allot.
LADY ANNE:
Any lords, make me, an heaven from this seize!
And be them hence, she be ryarly slumble: I'll both him;
Thou seedems of streen'd with's thousand sick
Of the highing whose strick too bate defic
Epoch: 17/20... Step: 2600... Loss: 1.4316... Val Loss: 1.5059
wink'st your honour; and to,
that thou stay out of mile bettar too like a sight.
There with it, sir; it was but seek the metter
but in orms:
By this but trubly sovereign truaks to me.,
What was you say?
PRINCE HENRY AO:
Why, here shall I disperse at an inlent,
And too many words a stronging time of her;
And leets the dise born on the sight by,
Which soon being astales from your fearth,
Hull both these clower of your sair and himself.
Being seit, I am not both his house:
For the move word of these were many men
Within the stricks in there that thou thats siving.
This to dear any signoricisious bond,
But in her princelong and bod to the war.
SIN:
Then speak me how at with a storm, was tontue.
This do it spoke:
I am so, how I mister them away!
SLY:
Which was it comes the purmoss: and that I shall part thee,
Madrail, my liest, a tide and take him's latt,
My son and had in lamental than anon
My fortune, in thut as my dideds man
And hang and lade the house of hand;
Or I did bear that pardon to a
Epoch: 18/20... Step: 2650... Loss: 1.3660... Val Loss: 1.5026
wink'st home and dull a grave;
which I have spried for what in mine ear to chive
That hopes supplace appoor suthing the clooks: and
mave thee again and person.
LEONTES:
It here, to have my huddid many shores of those
princes is fair, it make a prosperions.
FRAALINGER:
There's woes of a heaven. She's me a man, it is
betwixt my soldiers: some o'flace a toos house
to strong them, as the passion of a prepere of.
AUTOLYCUS:
A prest out me have leave the best abide me
By my cast of itself. The most or one
common to again and caunter strength of fly:
For with a complece houses she is on as it besides
it and the will be strettled, we are drown: to sare
it she throw me shall stifl: the can my looks dram.
DUKE VINCENTIO:
Old honesty ishae to discrest up your tongue.
Shepherd:
Now, sir; you take away; what they have and more others; on
I cannot show it shis forget of some monners.
SICINIUS:
How now!
VALERIA:
I do well her and see our generatige.
POMPEY:
My daughter, for the day doth not so stabp
Epoch: 18/20... Step: 2700... Loss: 1.3847... Val Loss: 1.5022
wink'st brook him out to come?
What's I were too more feel, for me no longer.
PAULINA:
Nentle the poor shoulder?
ABELLITA:
O, no lust! It's not be to did more be
him, I would not but a true: or should mest be a sunce. in
your most most; be so, thou shouldst be senvet to me how
since a bide meant a commont ball for you to cense them.
First Senator:
He had soon howely, as his friends o'er there hath hand to
that,
and heir him by your honour.-
This baldably can you have done and marry forth
ot worse of him.
POMPEY:
Poor mortos, morrow! I must to it stably
The hearts of wooldry and sway till you swort
Till as thy duke in merranty is as sweet
The deeds as it humb appaning in the soul,
The deast shamble; to make a thrish for me to
makes him fear in person speeks which is mick;
And hold thy periling hissed,
And have it hath astempt.
SICINIUS:
Such and to his house
Sister a man service, that is before thoughts.
DUKE VINCENTIO:
It is thines in their commandent:
And I command it,
O'er-turnen tello
Epoch: 18/20... Step: 2750... Loss: 1.3250... Val Loss: 1.4987
wink'st he be not made him:.
Provost:
You shall have sudden of with soul and fault,
In the pare well; and where I have my boon;
Or how cursed away the tume since humble
I warrant and sharl is more belted your.
LARY CAPULET:
O think! I pray you to the hamp, and distake
When he canst make a goodly stis and death.
GLOUCESTER:
Sir, sir, a trief, to hear his bind, have then
Into the firl man's bleeding forting heart;
And have the poor sents share howards baid,
Where frues heast but his clematian o'drelike?
HASTINGS:
And he before your manners: he has speak.
ROMEO:
Then, I must
sut your good bosom.
Clown:
Not how: his mrately two thoughts with the honour
Betwixt the suptle of the flower.
COMINIUS:
What thanks thine hath,
When you'le more from best heaven here in the double,
Or wretched her the feest thal to the day,
Her' seatly caning sake oor place of you.
ROMEO:
Well, my mine, is many haste that beagies here,
And thus by secities broke a poor content,
And her is aprepition, in the fault,
B
Epoch: 19/20... Step: 2800... Loss: 1.3853... Val Loss: 1.4946
wink'st worthy fase behinds
of that cuars word, this will hove were made me
Seen more from hum shall
no hand out when the wife is haste,
What would he is. To see your face have tears them:
She, in this wire say woold from that hast fath,
His wars she burit as a seaten find in bows.
POLIXENES:
Halk, stead in this.
MENENIUS:
How comes all, though you be man's day:
Speak, much most, as you were for thas, in husband.
ANGELO:
If though thou art desires and little against
Than honesty will we have between and to
the sellons in your shamed.
COMINIUS:
When art thou to your conscease?
VIRGILIA:
Of it not,
And sils is from your famof of tapther to
The sands: this assiving her that does?
POLIXENES:
That, is not wyre back for a salchar's hate,
Then do, and purse his tumble than so saim'd
And beartly treason with your sweed to made the speak
As I was full and proper stay of all,
That we'll with how hose, it be good not. The
ground show now tornot her hangs of blood,
And sut my death boy his fouch the
Epoch: 19/20... Step: 2850... Loss: 1.3782... Val Loss: 1.4912
wink'st thom. They bether the
heored than to-mend of many thing
This prayers, one brock something to the better.
DUKE VINCENTIO:
Which are all the spired what are you may prove
A laster hath scive to more sulerals:
You do they send for strange.
ame, I, I can tell you.
LEONTES:
No, I am a thousard and take year
Than at his sorrow cancule; tell me to be,
Commind'd you. Why, sor must, I thank all thing
For my accused, but a ward forth
And sir, your dourt, this were her flait in sides,
I like it not the meacure; she, behy shall tramity
For that a them such heavy dived-prished,
In bosom in a losts as your letters
That I that showard mony balder baint,
And so my sun's servent cry, and thou shalt not,
Strought her: be resever, which this lear undainty
From a prince and wars and serves and sufferent
That were I seem is as his hand atcame.
LADY ANNE:
Thy drimps say, might list this father come.
CLAUDIO:
Net this wherefore they have been fool'd
To their which fought, I have need his hories,
So he w
Epoch: 19/20... Step: 2900... Loss: 1.3830... Val Loss: 1.4962
wink'st thing, this carely doon,
The wealol's strifes, thoughts, that you will not knight.
HENRY BOLINGBROKE:
I think my cleam,
With him, which will so service to puss him.
LADY ANNE:
Now may you drunk your chief the whore the dows!
As we shall have you, for my face is now.
DUKE OF YORK:
And how ham I so think, which with the first
And money it is till a weak of twise well.
CLIFFORD:
Swee it here, thou wast here, which may not hate
The cort of creared in all-daint in where.
Beconderer:
Ay when the look that I accuse to her fortune
I hope, the less with his son souls again:
Fet them to all at my pulish storn at lives
And hopeing me of sighly worthis where
Of all the word, who likeness tontues in the command;
But flooking blood, tell me, his land, that this?
JULIET:
A devil in persunce to his most crack
Her seat bean not to be said I have and some heart
Weth his distonement hath bake all till been man.
LADY CAPULET:
His beat that sea, we ship our censwill choose.
RIVERS:
What was his lad
Epoch: 20/20... Step: 2950... Loss: 1.3359... Val Loss: 1.4901
wink'st the compaty.
FLORIZEL:
I stay, heart him!
MENENIUS:
The pleasure she's than breath.
Second Citizen:
My gruefess.
SICINIUS:
Your patience? why to so shall how the
sight of his billainges the pain in blowd
with him to be must dear about your measary:
the breath.
ARLABIUT:
He's done? and whan, the death? allow; your mother
And boing to the follows. As to thy state,
Meaning the duke, and make to be so here?
By her must I have being my time? to but,
Will think that you have, sir had hind three weres
The matter self-the labits to the prince
Any bactigrous heaven, and helping worth;
And by all money, and made heavy fraelds,
That feirs and so, here have the trink in grost,
If thou metter me at all well come to make
Their bloody wife. Book of desperate!
FRIAR LAURENCE:
Not brother of your honour: who most hate,
By love's doom like a fair fince husband
To the cressing father, money. Waste mine tongue,
Thy death of beatted take me in the stay;
Far we till tell him he be head and he.
HENRY
Epoch: 20/20... Step: 3000... Loss: 1.3361... Val Loss: 1.4866
wink'st thus tell my strength;
But by the storable cuused but a partor's fear;
But that his head before the dishance his days
To be time of the cheece,
My treasons, for I am thou wilt do fance,
Thou say, and twain in some shores to a greater
And but on tiust with more: where sumerrious?
SICINIUS:
Where?
MERCUTIO:
Why lords I love me;
The sighting will, should be will play y't;
Which he loves that advised; and you'll may,
The shame to my honour, his majesty,
And basoleng the time, there then I have too
Sailt anon, then mistakes the swords shall drubk it:
Sut and she is his cursed by but night found
And as the drum of love, shall be becitt
Of truth.
Come, hard our brother.
First Senator:
He is, a warrants must not
Hor treas to make it. Causin; if he
Tell in, talk on what sounds they be so presence'd.
POMPEY:
Hast thou think?
MARCIUS:
They have they be men.
ISABELLA:
So you, not much so,
Whom I am chose to love.
ISABELLA:
I'll go to be the beling,
There for it to the house; and so his gaun
Epoch: 20/20... Step: 3050... Loss: 1.3633... Val Loss: 1.4916
wink'st me, and let me be
My silest accord bfames.
DUKE VINCENTIO:
I do recount. By he'el and the pease,
Since when against me flearing by that winds,
For you the child wretched with this was town;
Befige to do with her mast, sweet your drum
In nend as the pows-boves with her and see
Fetch one of the teward mercion; to make
Her present sorrow's hild her, shanoe's spring:
And thou dhath dellow'd, that it is in her,
But for his steed fellow them me as to these war
To perve you? what, hang your highness is
That mean not shur away?
First Senano:
As then I wold to cen it is; and, his best stand, while
him, as mine and common well.
COMINIUS:
She may be a shield to you, he hath
commands them seen and with a temmer will, we will field
the past forget that all me.
DUKE VINCENTIO:
I pray you, gentlemen, and which they have been much
but any accivens; then for, you shall not stand from
the shedal, of a prisoner for a prich o' the dear
thou bod of him for it in answer, here is not
the honesty fieves;
|
Modulo 3/2. Ciclos.ipynb
|
###Markdown
Ciclos (loops)Los ciclos nos sirven cuando queremos que una pieza de código se ejecute hasta que una condición se cumpla (o se deje de cumplir), o si queremos ejecutar esa pieza un número determinado de veces (por ejemplo, aplicarla por cada elemento de una colección).Para el primer caso nos resultan de utilidad los ciclos de tipo `while` mientras que para el segundo resultan más apropiados los ciclos del tipo `for`. El ciclo `while` Como mencionabamos, estos ciclos nos resultarán utiles cuando querramos que algo se ejecute mientras se esté cumpliendo cierta condición. Por lo general, esto significa que no tenemos una estimación certera sobre el *número de veces* que se ejecutará. Claro, esto es más como una *regla general* y no algo formal.Son de la forma```pythonwhile condicion: Do stuff```Se ejecutaran *mientras* que `condicion` sea `True`, lo que significa que si queremos salir del ciclo, en algún momento tiene que cambiar el valor de `condicion` o podemos utilizar la instrucción `break`, que directamente provoca que el ciclo se acabe.Debemos tener cuidado con eso, pues podríamos provocar que nuestro programa se quede en ese ciclo de manera indefinida, hasta ser terminado de manera externa. Ejemplos
###Code
# Usualmente usamos whiles con condiciones que usan un contador
c = 0
while c < 10:
# Cosas
c += 1
print(c)
# Viendo la Conjectura de Collatz en accion
n = randint(2,10)
print("n:",n)
while n != 1:
if n % 2 == 0: # si n par
n = n//2
else: # si n impar
n = 3*n + 1
print(n)
###Output
n: 9
28
14
7
22
11
34
17
52
26
13
40
20
10
5
16
8
4
2
1
###Markdown
El ciclo `for`A diferencia de otros lenguajes de programación con ciclos con el nombre `for`, en Python siempre se utiliza de una forma que en otros lenguajes se conoce como un `for each`, es decir, en realidad haremos una iteración *por cada miembro* de algún "iterable" que indiquemos. La función `range()`La secuencia iterable utilizada por excelencia en estos ciclos, es la generada por la funcion `range(start,finish + 1, step)` de Python, que nos permite generar una lista de numeros que vaya desde `start` hasta `finish`, aumentando en un paso indicado por `step`.Si no indicamos un `step`, el default es 1. Ejemplos
###Code
facultades = ['Ingenieria', 'Medicina', 'Enfermeria', 'Contaduría', 'Educacion Fisica']
puntos = [(0,0), (4,0), (4,3)]
# Analogo al primer ejemplo de while
for i in range(0,11):
print(i)
# Usando la funcion range, digamos que queremos imprimir numeros impares hasta el 15
for x in range(1, 16, 2):
print(x)
# Iterando sobre listas
print("El campus 2 aloja a")
for facultad in facultades:
print(f"La Facultad de {facultad}")
print("Las coordenadas del triangulo son")
for x, y in puntos: # Observa ese desempaquetado de tuplas!
print(f"x = {x}, y = {y}")
###Output
Las coordenadas del triangulo son
x = 0, y = 0
x = 4, y = 0
x = 4, y = 3
|
sklearn_kmeans.ipynb
|
###Markdown
###Code
! pip install -q kaggle
from google.colab import files
files.upload()
! mkdir ~/.kaggle
! cp kaggle.json ~/.kaggle/
! chmod 600 ~/.kaggle/kaggle.json
! kaggle datasets list
! kaggle datasets download vjchoudhary7/customer-segmentation-tutorial-in-python
! unzip customer-segmentation-tutorial-in-python.zip
import pandas as pd
#ubah file csv menjadi dataframe
df = pd.read_csv('Mall_Customers.csv')
# tampilkan 3 baris pertama
df.head(3)
# ubah nama kolom
df = df.rename(columns={'Gender': 'gender', 'Age': 'age',
'Annual Income (k$)': 'annual_income',
'Spending Score (1-100)': 'spending_score'})
# ubah data kategorik mmenjadi data numerik
df['gender'].replace(['Female', 'Male'], [0,1], inplace=True)
# tampilkan data yang sudah di preprocess
df.head(3)
from sklearn.cluster import KMeans
# menghilangkan kolom customer id dan gender
X = df.drop(['CustomerID', 'gender'], axis=1)
#membuat list yang berisi inertia
clusters = []
for i in range(1,11):
km = KMeans(n_clusters=i).fit(X)
clusters.append(km.inertia_)
import matplotlib.pyplot as plt
%matplotlib inline
import seaborn as sns
# membuat plot inertia
fig, ax = plt.subplots(figsize=(8, 4))
sns.lineplot(x=list(range(1, 11)), y=clusters, ax=ax)
ax.set_title('Cari Elbow')
ax.set_xlabel('Clusters')
ax.set_ylabel('Inertia')
# membuat objek KMeans
km5 = KMeans(n_clusters=5).fit(X)
# menambahkan kolom label pada dataset
X['Labels'] = km5.labels_
# membuat plot KMeans dengan 5 klaster
plt.figure(figsize=(8,4))
sns.scatterplot(X['annual_income'], X['spending_score'], hue=X['Labels'],
palette=sns.color_palette('hls', 5))
plt.title('KMeans dengan 5 Cluster')
plt.show()
###Output
/usr/local/lib/python3.7/dist-packages/seaborn/_decorators.py:43: FutureWarning: Pass the following variables as keyword args: x, y. From version 0.12, the only valid positional argument will be `data`, and passing other arguments without an explicit keyword will result in an error or misinterpretation.
FutureWarning
|
using-convnets-with-small-datasets.ipynb
|
###Markdown
Downloading the dataThe cats vs. dogs dataset that we will use isn't packaged with Keras. It was made available by Kaggle.com as part of a computer vision competition in late 2013, back when convnets weren't quite mainstream. You can download the original dataset at: `https://www.kaggle.com/c/dogs-vs-cats/data` (you will need to create a Kaggle account if you don't already have one -- don't worry, the process is painless).The pictures are medium-resolution color JPEGs. They look like this:
###Code
import os, shutil
# The path to the directory where the original
# dataset was uncompressed
original_dataset_dir ='D:/college-lessons/MA/FIU/AI-workshop/dataset/catDog/data/train'
# The directory where we will
# store our smaller dataset
base_dir ='D:/college-lessons/MA/FIU/AI-workshop/dataset/catDog/data/catVsdog'
os.mkdir(base_dir)
# Directories for our training,
# validation and test splits
train_dir = os.path.join(base_dir, 'train')
os.mkdir(train_dir)
validation_dir = os.path.join(base_dir, 'validation')
os.mkdir(validation_dir)
test_dir = os.path.join(base_dir, 'test')
os.mkdir(test_dir)
# Directory with our training cat pictures
train_cats_dir = os.path.join(train_dir, 'cats')
os.mkdir(train_cats_dir)
# Directory with our training dog pictures
train_dogs_dir = os.path.join(train_dir, 'dogs')
os.mkdir(train_dogs_dir)
# Directory with our validation cat pictures
validation_cats_dir = os.path.join(validation_dir, 'cats')
os.mkdir(validation_cats_dir)
# Directory with our validation dog pictures
validation_dogs_dir = os.path.join(validation_dir, 'dogs')
os.mkdir(validation_dogs_dir)
# Directory with our validation cat pictures
test_cats_dir = os.path.join(test_dir, 'cats')
os.mkdir(test_cats_dir)
# Directory with our validation dog pictures
test_dogs_dir = os.path.join(test_dir, 'dogs')
os.mkdir(test_dogs_dir)
# Copy first 1000 cat images to train_cats_dir
fnames = ['cat.{}.jpg'.format(i) for i in range(1000)]
for fname in fnames:
src = os.path.join(original_dataset_dir, fname)
dst = os.path.join(train_cats_dir, fname)
shutil.copyfile(src, dst)
# Copy next 500 cat images to validation_cats_dir
fnames = ['cat.{}.jpg'.format(i) for i in range(1000, 1500)]
for fname in fnames:
src = os.path.join(original_dataset_dir, fname)
dst = os.path.join(validation_cats_dir, fname)
shutil.copyfile(src, dst)
# Copy next 500 cat images to test_cats_dir
fnames = ['cat.{}.jpg'.format(i) for i in range(1500, 2000)]
for fname in fnames:
src = os.path.join(original_dataset_dir, fname)
dst = os.path.join(test_cats_dir, fname)
shutil.copyfile(src, dst)
# Copy first 1000 dog images to train_dogs_dir
fnames = ['dog.{}.jpg'.format(i) for i in range(1000)]
for fname in fnames:
src = os.path.join(original_dataset_dir, fname)
dst = os.path.join(train_dogs_dir, fname)
shutil.copyfile(src, dst)
# Copy next 500 dog images to validation_dogs_dir
fnames = ['dog.{}.jpg'.format(i) for i in range(1000, 1500)]
for fname in fnames:
src = os.path.join(original_dataset_dir, fname)
dst = os.path.join(validation_dogs_dir, fname)
shutil.copyfile(src, dst)
# Copy next 500 dog images to test_dogs_dir
fnames = ['dog.{}.jpg'.format(i) for i in range(1500, 2000)]
for fname in fnames:
src = os.path.join(original_dataset_dir, fname)
dst = os.path.join(test_dogs_dir, fname)
shutil.copyfile(src, dst)
###Output
_____no_output_____
###Markdown
As a sanity check, let's count how many pictures we have in each training split (train/validation/test):
###Code
print('total training cat images:', len(os.listdir(train_cats_dir)))
print('total training dog images:', len(os.listdir(train_dogs_dir)))
print('total validation cat images:', len(os.listdir(validation_cats_dir)))
print('total validation dog images:', len(os.listdir(validation_dogs_dir)))
print('total test cat images:', len(os.listdir(test_cats_dir)))
print('total test dog images:', len(os.listdir(test_dogs_dir)))
###Output
total test dog images: 500
###Markdown
Building our network
###Code
from keras import layers
from keras import models
model = models.Sequential()
model.add(layers.Conv2D(32, (3, 3), activation='relu',
input_shape=(150, 150, 3)))
model.add(layers.MaxPooling2D((2, 2)))
model.add(layers.Conv2D(64, (3, 3), activation='relu'))
model.add(layers.MaxPooling2D((2, 2)))
model.add(layers.Conv2D(128, (3, 3), activation='relu'))
model.add(layers.MaxPooling2D((2, 2)))
model.add(layers.Conv2D(128, (3, 3), activation='relu'))
model.add(layers.MaxPooling2D((2, 2)))
model.add(layers.Flatten())
model.add(layers.Dense(512, activation='relu'))
model.add(layers.Dense(1, activation='sigmoid'))
###Output
_____no_output_____
###Markdown
Let's take a look at how the dimensions of the feature maps change with every successive layer:
###Code
model.summary()
from keras import optimizers
model.compile(loss='binary_crossentropy',
optimizer=optimizers.RMSprop(lr=1e-4),
metrics=['acc'])
###Output
_____no_output_____
###Markdown
Data preprocessing
###Code
from keras.preprocessing.image import ImageDataGenerator
# All images will be rescaled by 1./255
train_datagen = ImageDataGenerator(rescale=1./255)
test_datagen = ImageDataGenerator(rescale=1./255)
train_generator = train_datagen.flow_from_directory(
# This is the target directory
train_dir,
# All images will be resized to 150x150
target_size=(150, 150),
batch_size=20,
# Since we use binary_crossentropy loss, we need binary labels
class_mode='binary')
validation_generator = test_datagen.flow_from_directory(
validation_dir,
target_size=(150, 150),
batch_size=20,
class_mode='binary')
for data_batch, labels_batch in train_generator:
print('data batch shape:', data_batch.shape)
print('labels batch shape:', labels_batch.shape)
break
history = model.fit(
train_generator,
steps_per_epoch=100,
epochs=30,
validation_data=validation_generator,
validation_steps=50)
###Output
Epoch 1/30
100/100 [==============================] - 9s - loss: 0.6898 - acc: 0.5285 - val_loss: 0.6724 - val_acc: 0.5950
Epoch 2/30
100/100 [==============================] - 8s - loss: 0.6543 - acc: 0.6340 - val_loss: 0.6565 - val_acc: 0.5950
Epoch 3/30
100/100 [==============================] - 8s - loss: 0.6143 - acc: 0.6690 - val_loss: 0.6116 - val_acc: 0.6650
Epoch 4/30
100/100 [==============================] - 8s - loss: 0.5626 - acc: 0.7125 - val_loss: 0.5774 - val_acc: 0.6970
Epoch 5/30
100/100 [==============================] - 8s - loss: 0.5266 - acc: 0.7335 - val_loss: 0.5726 - val_acc: 0.6960
Epoch 6/30
100/100 [==============================] - 8s - loss: 0.5007 - acc: 0.7550 - val_loss: 0.6075 - val_acc: 0.6580
Epoch 7/30
100/100 [==============================] - 8s - loss: 0.4723 - acc: 0.7840 - val_loss: 0.5516 - val_acc: 0.7060
Epoch 8/30
100/100 [==============================] - 8s - loss: 0.4521 - acc: 0.7875 - val_loss: 0.5724 - val_acc: 0.6980
Epoch 9/30
100/100 [==============================] - 8s - loss: 0.4163 - acc: 0.8095 - val_loss: 0.5653 - val_acc: 0.7140
Epoch 10/30
100/100 [==============================] - 8s - loss: 0.3988 - acc: 0.8185 - val_loss: 0.5508 - val_acc: 0.7180
Epoch 11/30
100/100 [==============================] - 8s - loss: 0.3694 - acc: 0.8385 - val_loss: 0.5712 - val_acc: 0.7300
Epoch 12/30
100/100 [==============================] - 8s - loss: 0.3385 - acc: 0.8465 - val_loss: 0.6097 - val_acc: 0.7110
Epoch 13/30
100/100 [==============================] - 8s - loss: 0.3229 - acc: 0.8565 - val_loss: 0.5827 - val_acc: 0.7150
Epoch 14/30
100/100 [==============================] - 8s - loss: 0.2962 - acc: 0.8720 - val_loss: 0.5928 - val_acc: 0.7190
Epoch 15/30
100/100 [==============================] - 8s - loss: 0.2684 - acc: 0.9005 - val_loss: 0.5921 - val_acc: 0.7190
Epoch 16/30
100/100 [==============================] - 8s - loss: 0.2509 - acc: 0.8980 - val_loss: 0.6148 - val_acc: 0.7250
Epoch 17/30
100/100 [==============================] - 8s - loss: 0.2221 - acc: 0.9110 - val_loss: 0.6487 - val_acc: 0.7010
Epoch 18/30
100/100 [==============================] - 8s - loss: 0.2021 - acc: 0.9250 - val_loss: 0.6185 - val_acc: 0.7300
Epoch 19/30
100/100 [==============================] - 8s - loss: 0.1824 - acc: 0.9310 - val_loss: 0.7713 - val_acc: 0.7020
Epoch 20/30
100/100 [==============================] - 8s - loss: 0.1579 - acc: 0.9425 - val_loss: 0.6657 - val_acc: 0.7260
Epoch 21/30
100/100 [==============================] - 8s - loss: 0.1355 - acc: 0.9550 - val_loss: 0.8077 - val_acc: 0.7040
Epoch 22/30
100/100 [==============================] - 8s - loss: 0.1247 - acc: 0.9545 - val_loss: 0.7726 - val_acc: 0.7080
Epoch 23/30
100/100 [==============================] - 8s - loss: 0.1111 - acc: 0.9585 - val_loss: 0.7387 - val_acc: 0.7220
Epoch 24/30
100/100 [==============================] - 8s - loss: 0.0932 - acc: 0.9710 - val_loss: 0.8196 - val_acc: 0.7050
Epoch 25/30
100/100 [==============================] - 8s - loss: 0.0707 - acc: 0.9790 - val_loss: 0.9012 - val_acc: 0.7190
Epoch 26/30
100/100 [==============================] - 8s - loss: 0.0625 - acc: 0.9855 - val_loss: 1.0437 - val_acc: 0.6970
Epoch 27/30
100/100 [==============================] - 8s - loss: 0.0611 - acc: 0.9820 - val_loss: 0.9831 - val_acc: 0.7060
Epoch 28/30
100/100 [==============================] - 8s - loss: 0.0488 - acc: 0.9865 - val_loss: 0.9721 - val_acc: 0.7310
Epoch 29/30
100/100 [==============================] - 8s - loss: 0.0375 - acc: 0.9915 - val_loss: 0.9987 - val_acc: 0.7100
Epoch 30/30
100/100 [==============================] - 8s - loss: 0.0387 - acc: 0.9895 - val_loss: 1.0139 - val_acc: 0.7240
###Markdown
It is good practice to always save your models after training:
###Code
model.save('cats_and_dogs_small_1.h5')
###Output
_____no_output_____
###Markdown
Let's plot the loss and accuracy of the model over the training and validation data during training:
###Code
import matplotlib.pyplot as plt
acc = history.history['acc']
val_acc = history.history['val_acc']
loss = history.history['loss']
val_loss = history.history['val_loss']
epochs = range(len(acc))
plt.plot(epochs, acc, 'bo', label='Training acc')
plt.plot(epochs, val_acc, 'b', label='Validation acc')
plt.title('Training and validation accuracy')
plt.legend()
plt.figure()
plt.plot(epochs, loss, 'bo', label='Training loss')
plt.plot(epochs, val_loss, 'b', label='Validation loss')
plt.title('Training and validation loss')
plt.legend()
###Output
_____no_output_____
###Markdown
Using data augmentation
###Code
datagen = ImageDataGenerator(
rotation_range=40,
width_shift_range=0.2,
height_shift_range=0.2,
shear_range=0.2,
zoom_range=0.2,
horizontal_flip=True,
fill_mode='nearest')
# This is module with image preprocessing utilities
from keras.preprocessing import image
fnames = [os.path.join(train_cats_dir, fname) for fname in os.listdir(train_cats_dir)]
# We pick one image to "augment"
img_path = fnames[3]
# Read the image and resize it
img = image.load_img(img_path, target_size=(150, 150))
# Convert it to a Numpy array with shape (150, 150, 3)
x = image.img_to_array(img)
# Reshape it to (1, 150, 150, 3)
x = x.reshape((1,) + x.shape)
# The .flow() command below generates batches of randomly transformed images.
# It will loop indefinitely, so we need to `break` the loop at some point!
i = 0
for batch in datagen.flow(x, batch_size=1):
plt.figure(i)
imgplot = plt.imshow(image.array_to_img(batch[0]))
i += 1
if i % 4 == 0:
break
plt.show()
model = models.Sequential()
model.add(layers.Conv2D(32, (3, 3), activation='relu',
input_shape=(150, 150, 3)))
model.add(layers.MaxPooling2D((2, 2)))
model.add(layers.Conv2D(64, (3, 3), activation='relu'))
model.add(layers.MaxPooling2D((2, 2)))
model.add(layers.Conv2D(128, (3, 3), activation='relu'))
model.add(layers.MaxPooling2D((2, 2)))
model.add(layers.Conv2D(128, (3, 3), activation='relu'))
model.add(layers.MaxPooling2D((2, 2)))
model.add(layers.Flatten())
model.add(layers.Dropout(0.5))
model.add(layers.Dense(512, activation='relu'))
model.add(layers.Dense(1, activation='sigmoid'))
model.compile(loss='binary_crossentropy',
optimizer=optimizers.RMSprop(lr=1e-4),
metrics=['acc'])
###Output
_____no_output_____
###Markdown
Let's train our network using data augmentation and dropout:
###Code
train_datagen = ImageDataGenerator(
rescale=1./255,
rotation_range=40,
width_shift_range=0.2,
height_shift_range=0.2,
shear_range=0.2,
zoom_range=0.2,
horizontal_flip=True,)
# Note that the validation data should not be augmented!
test_datagen = ImageDataGenerator(rescale=1./255)
train_generator = train_datagen.flow_from_directory(
# This is the target directory
train_dir,
# All images will be resized to 150x150
target_size=(150, 150),
batch_size=32,
# Since we use binary_crossentropy loss, we need binary labels
class_mode='binary')
validation_generator = test_datagen.flow_from_directory(
validation_dir,
target_size=(150, 150),
batch_size=32,
class_mode='binary')
history = model.fit(
train_generator,
steps_per_epoch=100,
epochs=100,
validation_data=validation_generator,
validation_steps=50)
###Output
Found 2000 images belonging to 2 classes.
Found 1000 images belonging to 2 classes.
Epoch 1/100
100/100 [==============================] - 24s - loss: 0.6857 - acc: 0.5447 - val_loss: 0.6620 - val_acc: 0.5888
Epoch 2/100
100/100 [==============================] - 23s - loss: 0.6710 - acc: 0.5675 - val_loss: 0.6606 - val_acc: 0.5825
Epoch 3/100
100/100 [==============================] - 22s - loss: 0.6609 - acc: 0.5913 - val_loss: 0.6663 - val_acc: 0.5711.594 - ETA: 7s - loss: 0.6655 - ETA: 5s - los - ETA: 1s - loss: 0.6620 - acc:
Epoch 4/100
100/100 [==============================] - 22s - loss: 0.6446 - acc: 0.6178 - val_loss: 0.6200 - val_acc: 0.6379
Epoch 5/100
100/100 [==============================] - 22s - loss: 0.6267 - acc: 0.6325 - val_loss: 0.6280 - val_acc: 0.5996
Epoch 6/100
100/100 [==============================] - 22s - loss: 0.6080 - acc: 0.6631 - val_loss: 0.6841 - val_acc: 0.5490
Epoch 7/100
100/100 [==============================] - 22s - loss: 0.5992 - acc: 0.6700 - val_loss: 0.5717 - val_acc: 0.6946
Epoch 8/100
100/100 [==============================] - 22s - loss: 0.5908 - acc: 0.6819 - val_loss: 0.5858 - val_acc: 0.6764
Epoch 9/100
100/100 [==============================] - 22s - loss: 0.5869 - acc: 0.6856 - val_loss: 0.5658 - val_acc: 0.6785
Epoch 10/100
100/100 [==============================] - 23s - loss: 0.5692 - acc: 0.6934 - val_loss: 0.5409 - val_acc: 0.7170
Epoch 11/100
100/100 [==============================] - 22s - loss: 0.5708 - acc: 0.6897 - val_loss: 0.5325 - val_acc: 0.7274
Epoch 12/100
100/100 [==============================] - 23s - loss: 0.5583 - acc: 0.7047 - val_loss: 0.5683 - val_acc: 0.7126
Epoch 13/100
100/100 [==============================] - 22s - loss: 0.5602 - acc: 0.7069 - val_loss: 0.6010 - val_acc: 0.6593
Epoch 14/100
100/100 [==============================] - 22s - loss: 0.5510 - acc: 0.7231 - val_loss: 0.5387 - val_acc: 0.7229
Epoch 15/100
100/100 [==============================] - 23s - loss: 0.5527 - acc: 0.7175 - val_loss: 0.5204 - val_acc: 0.7322
Epoch 16/100
100/100 [==============================] - 23s - loss: 0.5426 - acc: 0.7181 - val_loss: 0.5083 - val_acc: 0.7410
Epoch 17/100
100/100 [==============================] - 23s - loss: 0.5399 - acc: 0.7344 - val_loss: 0.5103 - val_acc: 0.7468
Epoch 18/100
100/100 [==============================] - 23s - loss: 0.5375 - acc: 0.7312 - val_loss: 0.5133 - val_acc: 0.7430
Epoch 19/100
100/100 [==============================] - 22s - loss: 0.5308 - acc: 0.7338 - val_loss: 0.4936 - val_acc: 0.7610
Epoch 20/100
100/100 [==============================] - 22s - loss: 0.5225 - acc: 0.7387 - val_loss: 0.4952 - val_acc: 0.7563
Epoch 21/100
100/100 [==============================] - 22s - loss: 0.5180 - acc: 0.7491 - val_loss: 0.4999 - val_acc: 0.7481
Epoch 22/100
100/100 [==============================] - 23s - loss: 0.5118 - acc: 0.7538 - val_loss: 0.4770 - val_acc: 0.7764
Epoch 23/100
100/100 [==============================] - 22s - loss: 0.5245 - acc: 0.7378 - val_loss: 0.4929 - val_acc: 0.7671
Epoch 24/100
100/100 [==============================] - 22s - loss: 0.5136 - acc: 0.7503 - val_loss: 0.4709 - val_acc: 0.7732
Epoch 25/100
100/100 [==============================] - 22s - loss: 0.4980 - acc: 0.7512 - val_loss: 0.4775 - val_acc: 0.7684
Epoch 26/100
100/100 [==============================] - 22s - loss: 0.4875 - acc: 0.7622 - val_loss: 0.4745 - val_acc: 0.7790
Epoch 27/100
100/100 [==============================] - 22s - loss: 0.5044 - acc: 0.7578 - val_loss: 0.5000 - val_acc: 0.7403
Epoch 28/100
100/100 [==============================] - 22s - loss: 0.4948 - acc: 0.7603 - val_loss: 0.4619 - val_acc: 0.7754
Epoch 29/100
100/100 [==============================] - 22s - loss: 0.4898 - acc: 0.7578 - val_loss: 0.4730 - val_acc: 0.7726
Epoch 30/100
100/100 [==============================] - 22s - loss: 0.4808 - acc: 0.7691 - val_loss: 0.4599 - val_acc: 0.7716
Epoch 31/100
100/100 [==============================] - 22s - loss: 0.4792 - acc: 0.7678 - val_loss: 0.4671 - val_acc: 0.7790
Epoch 32/100
100/100 [==============================] - 22s - loss: 0.4723 - acc: 0.7716 - val_loss: 0.4451 - val_acc: 0.7849
Epoch 33/100
100/100 [==============================] - 22s - loss: 0.4750 - acc: 0.7694 - val_loss: 0.4827 - val_acc: 0.7665
Epoch 34/100
100/100 [==============================] - 22s - loss: 0.4816 - acc: 0.7647 - val_loss: 0.4953 - val_acc: 0.7513
Epoch 35/100
100/100 [==============================] - 22s - loss: 0.4598 - acc: 0.7813 - val_loss: 0.4426 - val_acc: 0.7843
Epoch 36/100
100/100 [==============================] - 23s - loss: 0.4643 - acc: 0.7781 - val_loss: 0.4692 - val_acc: 0.7680
Epoch 37/100
100/100 [==============================] - 22s - loss: 0.4675 - acc: 0.7778 - val_loss: 0.4849 - val_acc: 0.7633
Epoch 38/100
100/100 [==============================] - 22s - loss: 0.4658 - acc: 0.7737 - val_loss: 0.4632 - val_acc: 0.7760
Epoch 39/100
100/100 [==============================] - 22s - loss: 0.4581 - acc: 0.7866 - val_loss: 0.4489 - val_acc: 0.7880
Epoch 40/100
100/100 [==============================] - 23s - loss: 0.4485 - acc: 0.7856 - val_loss: 0.4479 - val_acc: 0.7931
Epoch 41/100
100/100 [==============================] - 22s - loss: 0.4637 - acc: 0.7759 - val_loss: 0.4453 - val_acc: 0.7990
Epoch 42/100
100/100 [==============================] - 22s - loss: 0.4528 - acc: 0.7841 - val_loss: 0.4758 - val_acc: 0.7868
Epoch 43/100
100/100 [==============================] - 22s - loss: 0.4481 - acc: 0.7856 - val_loss: 0.4472 - val_acc: 0.7893
Epoch 44/100
100/100 [==============================] - 22s - loss: 0.4540 - acc: 0.7953 - val_loss: 0.4366 - val_acc: 0.7867A: 6s - loss: 0.4523 - acc: - ETA:
Epoch 45/100
100/100 [==============================] - 22s - loss: 0.4411 - acc: 0.7919 - val_loss: 0.4708 - val_acc: 0.7697
Epoch 46/100
100/100 [==============================] - 22s - loss: 0.4493 - acc: 0.7869 - val_loss: 0.4366 - val_acc: 0.7829
Epoch 47/100
100/100 [==============================] - 22s - loss: 0.4436 - acc: 0.7916 - val_loss: 0.4307 - val_acc: 0.8090
Epoch 48/100
100/100 [==============================] - 22s - loss: 0.4391 - acc: 0.7928 - val_loss: 0.4203 - val_acc: 0.8065
Epoch 49/100
100/100 [==============================] - 23s - loss: 0.4284 - acc: 0.8053 - val_loss: 0.4422 - val_acc: 0.8041
Epoch 50/100
100/100 [==============================] - 22s - loss: 0.4492 - acc: 0.7906 - val_loss: 0.5422 - val_acc: 0.7437
Epoch 51/100
100/100 [==============================] - 22s - loss: 0.4292 - acc: 0.7953 - val_loss: 0.4446 - val_acc: 0.7932
Epoch 52/100
100/100 [==============================] - 22s - loss: 0.4275 - acc: 0.8037 - val_loss: 0.4287 - val_acc: 0.7989
Epoch 53/100
100/100 [==============================] - 22s - loss: 0.4297 - acc: 0.7975 - val_loss: 0.4091 - val_acc: 0.8046
Epoch 54/100
100/100 [==============================] - 23s - loss: 0.4198 - acc: 0.7978 - val_loss: 0.4413 - val_acc: 0.7964
Epoch 55/100
100/100 [==============================] - 23s - loss: 0.4195 - acc: 0.8019 - val_loss: 0.4265 - val_acc: 0.8001
Epoch 56/100
100/100 [==============================] - 22s - loss: 0.4081 - acc: 0.8056 - val_loss: 0.4374 - val_acc: 0.7957
Epoch 57/100
100/100 [==============================] - 22s - loss: 0.4214 - acc: 0.8006 - val_loss: 0.4228 - val_acc: 0.8020
Epoch 58/100
100/100 [==============================] - 22s - loss: 0.4050 - acc: 0.8097 - val_loss: 0.4332 - val_acc: 0.7900
Epoch 59/100
100/100 [==============================] - 22s - loss: 0.4162 - acc: 0.8134 - val_loss: 0.4088 - val_acc: 0.8099
Epoch 60/100
100/100 [==============================] - 22s - loss: 0.4042 - acc: 0.8141 - val_loss: 0.4436 - val_acc: 0.7957
Epoch 61/100
100/100 [==============================] - 23s - loss: 0.4016 - acc: 0.8212 - val_loss: 0.4082 - val_acc: 0.8189
Epoch 62/100
100/100 [==============================] - 22s - loss: 0.4167 - acc: 0.8097 - val_loss: 0.3935 - val_acc: 0.8236
Epoch 63/100
100/100 [==============================] - 23s - loss: 0.4052 - acc: 0.8138 - val_loss: 0.4509 - val_acc: 0.7824
Epoch 64/100
100/100 [==============================] - 22s - loss: 0.4011 - acc: 0.8209 - val_loss: 0.3874 - val_acc: 0.8299
Epoch 65/100
100/100 [==============================] - 22s - loss: 0.3966 - acc: 0.8131 - val_loss: 0.4328 - val_acc: 0.7970
Epoch 66/100
100/100 [==============================] - 23s - loss: 0.3889 - acc: 0.8163 - val_loss: 0.4766 - val_acc: 0.7719
Epoch 67/100
100/100 [==============================] - 22s - loss: 0.3960 - acc: 0.8163 - val_loss: 0.3859 - val_acc: 0.8325
Epoch 68/100
100/100 [==============================] - 22s - loss: 0.3893 - acc: 0.8231 - val_loss: 0.4172 - val_acc: 0.8128
Epoch 69/100
100/100 [==============================] - 23s - loss: 0.3828 - acc: 0.8219 - val_loss: 0.4023 - val_acc: 0.8215 loss: 0.3881 - acc:
Epoch 70/100
100/100 [==============================] - 22s - loss: 0.3909 - acc: 0.8275 - val_loss: 0.4275 - val_acc: 0.8008
Epoch 71/100
100/100 [==============================] - 22s - loss: 0.3826 - acc: 0.8244 - val_loss: 0.3815 - val_acc: 0.8177
Epoch 72/100
100/100 [==============================] - 22s - loss: 0.3837 - acc: 0.8272 - val_loss: 0.4040 - val_acc: 0.8287
Epoch 73/100
100/100 [==============================] - 23s - loss: 0.3812 - acc: 0.8222 - val_loss: 0.4039 - val_acc: 0.8058
Epoch 74/100
100/100 [==============================] - 22s - loss: 0.3829 - acc: 0.8281 - val_loss: 0.4204 - val_acc: 0.8015
Epoch 75/100
100/100 [==============================] - 22s - loss: 0.3708 - acc: 0.8350 - val_loss: 0.4083 - val_acc: 0.8204
Epoch 76/100
100/100 [==============================] - 22s - loss: 0.3831 - acc: 0.8216 - val_loss: 0.3899 - val_acc: 0.8215
Epoch 77/100
100/100 [==============================] - 22s - loss: 0.3695 - acc: 0.8375 - val_loss: 0.3963 - val_acc: 0.8293
Epoch 78/100
100/100 [==============================] - 22s - loss: 0.3809 - acc: 0.8234 - val_loss: 0.4046 - val_acc: 0.8236
Epoch 79/100
100/100 [==============================] - 22s - loss: 0.3637 - acc: 0.8362 - val_loss: 0.3990 - val_acc: 0.8325
Epoch 80/100
100/100 [==============================] - 22s - loss: 0.3596 - acc: 0.8400 - val_loss: 0.3925 - val_acc: 0.8350
Epoch 81/100
100/100 [==============================] - 22s - loss: 0.3762 - acc: 0.8303 - val_loss: 0.3813 - val_acc: 0.8331
Epoch 82/100
100/100 [==============================] - 23s - loss: 0.3672 - acc: 0.8347 - val_loss: 0.4539 - val_acc: 0.7931
Epoch 83/100
100/100 [==============================] - 22s - loss: 0.3636 - acc: 0.8353 - val_loss: 0.3988 - val_acc: 0.8261
Epoch 84/100
100/100 [==============================] - 22s - loss: 0.3503 - acc: 0.8453 - val_loss: 0.3987 - val_acc: 0.8325
Epoch 85/100
100/100 [==============================] - 22s - loss: 0.3586 - acc: 0.8437 - val_loss: 0.3842 - val_acc: 0.8306
Epoch 86/100
100/100 [==============================] - 22s - loss: 0.3624 - acc: 0.8353 - val_loss: 0.4100 - val_acc: 0.8196.834
Epoch 87/100
100/100 [==============================] - 22s - loss: 0.3596 - acc: 0.8422 - val_loss: 0.3814 - val_acc: 0.8331
Epoch 88/100
100/100 [==============================] - 22s - loss: 0.3487 - acc: 0.8494 - val_loss: 0.4266 - val_acc: 0.8109
Epoch 89/100
100/100 [==============================] - 22s - loss: 0.3598 - acc: 0.8400 - val_loss: 0.4076 - val_acc: 0.8325
Epoch 90/100
100/100 [==============================] - 22s - loss: 0.3510 - acc: 0.8450 - val_loss: 0.3762 - val_acc: 0.8388
Epoch 91/100
100/100 [==============================] - 22s - loss: 0.3458 - acc: 0.8450 - val_loss: 0.4684 - val_acc: 0.8015
Epoch 92/100
100/100 [==============================] - 22s - loss: 0.3454 - acc: 0.8441 - val_loss: 0.4017 - val_acc: 0.8204
Epoch 93/100
100/100 [==============================] - 22s - loss: 0.3402 - acc: 0.8487 - val_loss: 0.3928 - val_acc: 0.8204
Epoch 94/100
100/100 [==============================] - 22s - loss: 0.3569 - acc: 0.8394 - val_loss: 0.4005 - val_acc: 0.8338
Epoch 95/100
100/100 [==============================] - 22s - loss: 0.3425 - acc: 0.8494 - val_loss: 0.3641 - val_acc: 0.8439
Epoch 96/100
100/100 [==============================] - 22s - loss: 0.3335 - acc: 0.8531 - val_loss: 0.3811 - val_acc: 0.8363
Epoch 97/100
100/100 [==============================] - 22s - loss: 0.3204 - acc: 0.8581 - val_loss: 0.3786 - val_acc: 0.8331
Epoch 98/100
100/100 [==============================] - 22s - loss: 0.3250 - acc: 0.8606 - val_loss: 0.4205 - val_acc: 0.8236
Epoch 99/100
100/100 [==============================] - 22s - loss: 0.3255 - acc: 0.8581 - val_loss: 0.3518 - val_acc: 0.8460
Epoch 100/100
100/100 [==============================] - 22s - loss: 0.3280 - acc: 0.8491 - val_loss: 0.3776 - val_acc: 0.8439
###Markdown
Let's save our model -- we will be using it in the section on convnet visualization.
###Code
model.save('cats_and_dogs_small_2.h5')
###Output
_____no_output_____
###Markdown
Let's plot our results again:
###Code
acc = history.history['acc']
val_acc = history.history['val_acc']
loss = history.history['loss']
val_loss = history.history['val_loss']
epochs = range(len(acc))
plt.plot(epochs, acc, 'bo', label='Training acc')
plt.plot(epochs, val_acc, 'b', label='Validation acc')
plt.title('Training and validation accuracy')
plt.legend()
plt.figure()
plt.plot(epochs, loss, 'bo', label='Training loss')
plt.plot(epochs, val_loss, 'b', label='Validation loss')
plt.title('Training and validation loss')
plt.legend()
plt.show()
###Output
_____no_output_____
|
lect12_bot_SQL_excel/2021_DPO_12_3_Clickhouse.ipynb
|
###Markdown
Python для анализа данных Библиотеки для работы с данными в табличном формате: pandas. SQL для Python. Работа с Clickhouse. Автор: *Ян Пиле, НИУ ВШЭ* Мы с вами уже немного поработали с запросами данных из интернета, причем как непосредственно с сайтов, так и через некоторые API. Давайте теперь попробуем поработать с SQL прямо из Python. Порешаем задачи
###Code
import json # Чтобы разбирать поля
import requests # чтобы отправлять запрос к базе
import pandas as pd # чтобы в табличном виде хранить результаты запроса
###Output
_____no_output_____
###Markdown
Нужно написать функцию, которая будет отправлять текст SQL-запроса, в нашем случае - запроса к ClickHouse, на сервер, а по выполнении запроса забирать его результаты в каком-то виде.
###Code
# имена явки пароли. если хотите, чтобы считалось с вашего логина, вставьте сюда свои логин и пароль
USER = ''
PASS = ''
HOST = 'http://hse.beslan.pro:8080/'
def get_clickhouse_data(query,
host=HOST,
USER = USER,
PASS = PASS,
connection_timeout = 1500,
dictify=True,
**kwargs):
NUMBER_OF_TRIES = 5 # Количество попыток запуска
DELAY = 10 #время ожидания между запусками
import time
params = kwargs #если вдруг нам нужно в функцию положить какие-то параметры
if dictify:
query += "\n FORMAT JSONEachRow" # dictify = True отдает каждую строку в виде JSON'a
for i in range(NUMBER_OF_TRIES):
headers = {'Accept-Encoding': 'gzip'}
r = requests.post(host,
params = params,
auth=(USER, PASS),
timeout = connection_timeout,
data=query
) # отправили запрос на сервер
if r.status_code == 200 and not dictify:
return r.iter_lines() # генератор :)
elif r.status_code == 200 and dictify:
return (json.loads(x) for x in r.iter_lines()) # генератор :)
else:
print('ATTENTION: try #%d failed' % i)
if i != (NUMBER_OF_TRIES - 1):
print(r.text)
time.sleep(DELAY * (i + 1))
else:
raise(ValueError, r.text)
###Output
_____no_output_____
###Markdown
Функция ниже преобразует полученные нами данные из генератора в pd.Dataframe
###Code
query = """
select *
from default.events
limit 10
"""
d = get_clickhouse_data(query, dictify=True)
next(d)
def get_data(query):
return pd.DataFrame(list(get_clickhouse_data(query, dictify=True)))
get_data(query)
###Output
_____no_output_____
###Markdown
Предлагаю немного разобраться в структуре нашей базы. Давайте достанем по 5-10 строк каждой из таблиц и посмотри, что же в них лежит. В events, например, уложены AppPlatform - Платформа (операционная система мобильного устройства), events - количество событий, произошедших в эту дату (будем, например, считать, что события это клики и каждый из них платный), EventDate - Дата события, DeviceID - идентификатор устройства.
###Code
# впуливаем сюда запрос
query = """
select *
from default.events
limit 10
"""
f = get_data(query)
f
###Output
_____no_output_____
###Markdown
Только что мы научились превращать результат нашего SQL-запроса в PANDAS-dataframe. В devices, например, уложены UserID - идентификатор пользователя, DeviceID - идентификатор устройства.
###Code
query = """
SELECT *
from devices
limit 10
"""
get_data(query)
###Output
_____no_output_____
###Markdown
Проверим, однозначное ли это соответствие (может ли у человека быть два устройства и могут ли с одного устройства сидеть два человека)
###Code
query = """
SELECT UserID, uniqExact(DeviceID) as cnt
from devices
group by UserID
having cnt>1
"""
get_data(query)
query = """
SELECT DeviceID, uniqExact(UserID) as cnt
from devices
group by DeviceID
having cnt>1
"""
get_data(query)
###Output
_____no_output_____
###Markdown
Видим, что оба запроса возвращают пустой результат. Это означает, что соответствие между UserID и DeviceID взаимно-однозначное. Это позволит нам избежать многих проблем впоследствии. В checks хранится стоимость всех покупок одного UserID за день, BuyDate - дата покупки, Rub - стоимость покупки
###Code
query = """
SELECT *
from checks
limit 10
"""
get_data(query)
###Output
_____no_output_____
###Markdown
Проверим, есть ли записи, у которых набору UserID-BuyDate соответствует несколько записей
###Code
query = """
SELECT BuyDate, UserID, count(*) as cnt
from checks
group by BuyDate, UserID
having cnt>1
"""
get_data(query)
###Output
_____no_output_____
###Markdown
Нам снова повезло! На каждого человека и каждый день в таблицу пишется суммарная стоимость его покупок. Теперь посмотрим на таблицу installs. В ней InstallationDate - дата установки, InstallCost - стоимость установки, Platform - Платформа (операционная система мобильного устройства), DeviceID - идентификатор устройства и Source - источник трафика (откуда человек пришел устанавливать приложение: сам нашел в поисковике, из рекламы, перешел по реферальной ссылке и т.д.)
###Code
query = """
SELECT *
from installs
limit 10
"""
get_data(query)
###Output
_____no_output_____
###Markdown
Давайте сформулируем несколько задач, которые мы на этих данных хотим решить. В течение какого срока установка, в среднем, окупается, в зависимости от:* платформы * источника трафика Воспользуйтесь тем периодом данных, который сочтете обоснованным для формулировки вывода Давайте посмотрим среднюю стоимость установки в зависимости от источника трафика. Будем считать, что GMV мы считаем в валюте Rub\*5, а стоимость одного события равна 0.5 у.е. Для начала достанем информацию, которая касается установок приложения и приклеим к ней информацию о том, какой UserID какие установки совершил. Для этого надо сделать join таблиц installs и devices. Я предлагаю считать данные за 1 квартал 2019 (почему бы и нет). В отображении остаывим 10 записей, чтобы экран не заполнять лишним. Join сделаем inner, предполагая, что нет таких DeviceID, которые никому не принадлежат (хотя вообще говоря, это стоит проверить)
###Code
query = """
select a.Source as Source,
a.InstallationDate as InstallationDate,
a.InstallCost as InstallCost,
a.DeviceID as DeviceID,
b.UserID as UserID
from installs as a
inner join devices as b
on a.DeviceID = b.DeviceID
where InstallationDate between '2019-01-01' and '2019-03-31'
limit 10
"""
res = get_data(query)
res
###Output
_____no_output_____
###Markdown
Теперь нам нужно посчитать суммарную стоимость всех заказов, которые указанный UserID сделал за этот квартал (это как раз один из двух источников нашей доходной части), а расходную часть - InstallCost- мы уже достали. Здесь необходимо делать left join, потому что могут быть люди, которые ничего не купили за этот период, хоть и установили приложение. Значит наше условие ограничения на left join должно брать только те покупки людей, которые произошли от даты установки до конца квартала, а также оставлять записи, в которых не было ни одной покупки, это можно обеспечить условием BuyDate is null (в правой таблице не нашлось ни одной покупки). После того, как мы эту информацию приджойнили, посчитаем на каждый факт установки суммарную стоимость всех покупок с помощью функции sum(). Мы также хотим, чтоб при суммировании у тех, кто не купил ничего в поле GMV - Gross Merchandise Value (суммарный оборот заказов)- стоял ноль. Для этого мы сначала переведем содержимое поля Rub в интересующую нас валюту (мы договорились умножать его на 5), а потом суммировать не само получившееся значение, а coalesce(Rub\*5,0) эта функция возвращает первое непустое значение из списка своих аргументов. Получается, что если поле Rub заполнено, она вернет Rub\*5, а если человек ничего не купил, то она вернет 0, как раз этого мы и добивались. Стоит заметить, что в качестве левой таблицы для join'а мы вставили наш предыдущий запрос.
###Code
query = """
select a.Source as Source,
a.InstallationDate as InstallationDate,
a.InstallCost as InstallCost,
a.DeviceID as DeviceID,
b.UserID as UserID,
sum(coalesce(b.Rub*5, 0)) as GMV
from (select a.Source as Source,
a.InstallationDate as InstallationDate,
a.InstallCost as InstallCost,
a.DeviceID as DeviceID,
b.UserID as UserID
from installs as a
inner join devices as b
on a.DeviceID = b.DeviceID
where InstallationDate between '2019-01-01' and '2019-03-31') as a
left join checks as b
on a.UserID = b.UserID
where (b.BuyDate >= a.InstallationDate
and b.BuyDate<='2019-03-31')
or b.BuyDate is null
group by a.Source ,
a.InstallationDate,
a.InstallCost,
a.DeviceID,
b.UserID
limit 10
"""
res = get_data(query)
res
###Output
_____no_output_____
###Markdown
Остается предпоследний шаг: таким же образом собрать информацию о произошедших событиях (они лежат в поле events таблицы events и мы договорились, что стоимость одного события - 0.5 у.е.). Полностью повторим логику, которая у нас была до этого. Только в этот раз попробуем в функцию sum() не подставлять coalesce. Если мы уверены, что в каждом Source произошло хотя бы одно событие, то в итоговом результате наша сумма будет точно ненулевой.
###Code
query = """
select a.Source as Source,
a.InstallationDate as InstallationDate,
a.InstallCost as InstallCost,
a.DeviceID as DeviceID,
a.UserID as UserID,
a.GMV as GMV,
sum(events*0.5) as events_revenue
from (select a.Source as Source,
a.InstallationDate as InstallationDate,
a.InstallCost as InstallCost,
a.DeviceID as DeviceID,
b.UserID as UserID,
sum(coalesce(b.Rub*5, 0)) as GMV
from (select a.Source as Source,
a.InstallationDate as InstallationDate,
a.InstallCost as InstallCost,
a.DeviceID as DeviceID,
b.UserID as UserID
from installs as a
inner join devices as b
on a.DeviceID = b.DeviceID
where InstallationDate between '2019-01-01' and '2019-03-31') as a
left join checks as b
on a.UserID = b.UserID
where (b.BuyDate >= a.InstallationDate
and b.BuyDate<='2019-03-31')
or b.BuyDate is null
group by a.Source ,
a.InstallationDate,
a.InstallCost,
a.DeviceID,
b.UserID) as a
left join events as b
on a.DeviceID = b.DeviceID
where (b.EventDate >= a.InstallationDate
and b.EventDate<='2019-03-31')
or b.EventDate is null
group by a.Source as Source,
a.InstallationDate as InstallationDate,
a.InstallCost as InstallCost,
a.DeviceID as DeviceID,
a.UserID as UserID,
a.GMV as GMV
limit 10
"""
res = get_data(query)
res
###Output
_____no_output_____
###Markdown
Ну и теперь произведем финальный шаг: суммируем все по источникам трафика и сразу посчитаем ROI - суммарный доход/суммарные затраты
###Code
query = """
select Source, uniqExact(UserID) as users,
SUM(InstallCost) AS InstallCost,
sum(GMV) as GMV,
SUM(events_revenue) AS events_revenue
from (select a.Source as Source,
a.InstallationDate as InstallationDate,
a.InstallCost as InstallCost,
a.DeviceID as DeviceID,
a.UserID as UserID,
a.GMV as GMV,
sum(events*0.5) as events_revenue
from (select a.Source as Source,
a.InstallationDate as InstallationDate,
a.InstallCost as InstallCost,
a.DeviceID as DeviceID,
b.UserID as UserID,
sum(coalesce(b.Rub*5, 0)) as GMV
from (select a.Source as Source,
a.InstallationDate as InstallationDate,
a.InstallCost as InstallCost,
a.DeviceID as DeviceID,
b.UserID as UserID
from installs as a
inner join devices as b
on a.DeviceID = b.DeviceID
where InstallationDate between '2019-01-01' and '2019-03-31') as a
left join checks as b
on a.UserID = b.UserID
where (b.BuyDate >= a.InstallationDate
and b.BuyDate<='2019-03-31')
or b.BuyDate is null
group by a.Source ,
a.InstallationDate,
a.InstallCost,
a.DeviceID,
b.UserID) as a
left join events as b
on a.DeviceID = b.DeviceID
where (b.EventDate >= a.InstallationDate
and b.EventDate<='2019-03-31')
or b.EventDate is null
group by a.Source as Source,
a.InstallationDate as InstallationDate,
a.InstallCost as InstallCost,
a.DeviceID as DeviceID,
a.UserID as UserID,
a.GMV as GMV
)
group by Source
"""
res = get_data(query)
res
###Output
_____no_output_____
###Markdown
С помощью pandas приведем поля к нужному нам формату (По умолчанию Clickhouse выплевывает результаты в строковом формате)
###Code
res = res.astype({'users':int, 'InstallCost':float, 'GMV':float, 'events_revenue':float})
###Output
_____no_output_____
###Markdown
Также посчитаем доходную часть
###Code
res['Profit'] = res['GMV'] + res['events_revenue']
###Output
_____no_output_____
###Markdown
И, наконец, посчитаем ROI
###Code
res['ROI'] = res['Profit']/res['InstallCost']
res
###Output
_____no_output_____
|
exercises/solutions/dlb-2-cnn-segmentation.ipynb
|
###Markdown
[](https://colab.research.google.com/github/JorisRoels/deep-learning-biology/blob/main/exercises/solutions/dlb-2-cnn-segmentation.ipynb) Exercise 3: Convolutional neural networks for segmentationIn this notebook, we will be using convolutional neural networks for segmentation of neurons in electron microscopy data. The structure of these exercises is as follows: 1. [Import libraries and download data](scrollTo=ScagUEMTMjlK)2. [Data visualization and pre-processing](scrollTo=ohZHyOTnI35b)3. [Segmentation: a pixel classification problem](scrollTo=UyspYtez5J8a)4. [Building a U-Net with PyTorch](scrollTo=wXbjn29WOOJ3)5. [Training & validating the network](scrollTo=zh8Pf_3HF_hi)This notebook is largely based on the research published in: Arganda-Carreras, I., Turaga, S. C., Berger, D. R., Ciresan, D. C., Giusti, A., Gambardella, L. M., Schmidhuber, J., Laptev, D., Dwivedi, S., Buhmann, J. M., Liu, T., Seyedhosseini, M., Tasdizen, T., Kamentsky, L., Burget, R., Uher, V., Tan, X., Sun, C., Pham, T. D., … Seung, H. S. (2015). Crowdsourcing the creation of image segmentation algorithms for connectomics. Frontiers in Neuroanatomy, 9. https://doi.org/10.3389/fnana.2015.00142 1. Import libraries and download dataLet's start with importing the necessary libraries.
###Code
!pip install neuralnets
import pickle
import numpy as np
import random
import os
import matplotlib.pyplot as plt
plt.rcdefaults()
import pandas as pd
from sklearn.metrics import accuracy_score
from sklearn.model_selection import train_test_split
from sklearn.manifold import TSNE
from sklearn.svm import SVC
from sklearn.ensemble import RandomForestClassifier
from sklearn.neighbors import KNeighborsClassifier
from progressbar import ProgressBar, Percentage, Bar, ETA, FileTransferSpeed
import torch
import torch.nn as nn
import torch.nn.functional as F
import torch.optim as optim
import torch.utils.data as data
from torch.utils.data import DataLoader
from torchvision import datasets
import gdown
import zipfile
import os
import progressbar
import time
from neuralnets.util.io import read_tif
from neuralnets.util.visualization import overlay
###Output
_____no_output_____
###Markdown
As you will notice, Colab environments come with quite a large library pre-installed. If you need to import a module that is not yet specified, you can add it in the previous cell (make sure to run it again). If the module is not installed, you can install it with `pip`. To make your work reproducible, it is advised to initialize all modules with stochastic functionality with a fixed seed. Re-running this script should give the same results as long as the seed is fixed.
###Code
# make sure the results are reproducible
seed = 0
np.random.seed(seed)
random.seed(seed)
torch.manual_seed(seed)
torch.backends.cudnn.deterministic = True
torch.backends.cudnn.benchmark = False
# run all computations on the GPU if available
device = torch.device("cuda" if torch.cuda.is_available() else "cpu")
print('Running computations with %s' % torch.device(device))
if torch.cuda.is_available():
print(torch.cuda.get_device_properties(device))
###Output
Running computations with cuda
_CudaDeviceProperties(name='Tesla T4', major=7, minor=5, total_memory=15079MB, multi_processor_count=40)
###Markdown
We will now download the required data from a public Google Drive repository. The data is stored as a zip archive and automatically extracted to the `data` directory in the current directory.
###Code
# fields
url = 'http://data.bits.vib.be/pub/trainingen/DeepLearning/data-3.zip'
cmp_data_path = 'data.zip'
# download the compressed data
gdown.download(url, cmp_data_path, quiet=False)
# extract the data
zip = zipfile.ZipFile(cmp_data_path)
zip.extractall('')
# remove the compressed data
os.remove(cmp_data_path)
###Output
Downloading...
From: http://data.bits.vib.be/pub/trainingen/DeepLearning/data-3.zip
To: /content/data.zip
100%|██████████| 14.9M/14.9M [00:01<00:00, 12.7MB/s]
###Markdown
2. Data visualization and pre-processingThe data used for this exercise session originates from an ISBI segmentation challenge on neuron structures in electron microscopy image stacks. The organizers provide a training volume and their corresponding labels. The test set is also provided, however predictions can be validated by uploading them to the challenge central server. This is to avoid overfitting the model on the test set. Each data volume is provided as a .tif file that contains a 3D array. For the input data, these are simply the intensity values measured by the microscope. For the labels, this is a binary value: 0 for membrane, 255 for inner neuron structure. We briefly visualize the labeled data.
###Code
# specify where the data is stored
data_dir = 'data-3'
# load the datadispensers
x = read_tif(os.path.join(data_dir, 'train-volume.tif'))
y = read_tif(os.path.join(data_dir, 'train-labels.tif'))
# print out size
print('Size of the labeled volume: %d x %d x %d' % x.shape)
# show example
x_overlay = overlay(x[0] / 255, 1-(y[0]>0), colors=[(1, 0, 0)], alpha=0.4)
plt.subplot(1, 3, 1)
plt.imshow(x[0], cmap='gray')
plt.title('Input data')
plt.axis('off')
plt.subplot(1, 3, 2)
plt.imshow(y[0], cmap='gray')
plt.title('Labels')
plt.axis('off')
plt.subplot(1, 3, 3)
plt.imshow(x_overlay)
plt.title('Membrane overlay')
plt.axis('off')
plt.show()
###Output
Size of the labeled volume: 30 x 512 x 512
###Markdown
Annotation of datasets like this typically involve lots of expertise and manual labour and is therefore extremely costly. This is a general issue in biomedical image-based datasets. For this reason, there has been increasing attention in developing automated segmentation techniques for biomedical imaging datasets. In the following, you will see how deep learning models can achieve relatively high accuracy on complex task such as neuronal structure segmentation. As in practically any image analysis application, it is good common practice to rescale the data to the [0,1] interval, map the labels to subsequent values (0 and 1 in this case) and split the data in a train and test set.
###Code
# normalize the data
x = (x - np.min(x)) / (np.max(x) - np.min(x))
# map the 255 labels to 1
y[y == 255] = 1
# split the data in a train and test set
test_ratio = 0.33 # we will use 33% of the data for testing
x_train, x_test, y_train, y_test = train_test_split(x, y, test_size=test_ratio, random_state=seed)
# print out size
print('Training volume: %d x %d x %d' % x_train.shape)
print('Testing volume: %d x %d x %d' % x_test.shape)
#
class_distribution = [np.sum(y_train == 0) / y_train.size, np.sum(y_train == 1) / y_train.size]
print('Class balance: ' )
print(' 0: %.3f' % class_distribution[0])
print(' 1: %.3f' % class_distribution[1])
###Output
Training volume: 20 x 512 x 512
Testing volume: 10 x 512 x 512
Class balance:
0: 0.221
1: 0.779
###Markdown
3. Segmentation: a pixel classification problemFrom a machine learning point of view, image segmentation can be seen as a classification problem. For each pixel in the image, the goal is to predict the corresponding class (membrane or non-membrane). Up to 2012, most techniques were based on extracting a set of features from a local or global region (e.g. intensity, edges, etc.) around the pixel and training a shallow classifier (e.g. a random forest). The choice of features would typically be the crucial factor and is different for each application. Convolutional neural networks however, are able to solve this issue, as the feature extractor is learned based on the training data. To do this, we have to do two things: implement a dataset that extracts a local window for each pixel and train a CNN that performs binary classification. Let's start with the dataset. **Exercise**: Implement the `EMWindowDataset` class: - The `__init__` function should save the inputs and labels- The `__getitem__` function should return a local window around the i'th pixel (use slice-by-slice raster ordering for this) and the corresponding label of that pixel. Note that extracting local windows near the bounds of the image may result in out-of-bound errors. You can omit this by [padding](https://numpy.org/doc/stable/reference/generated/numpy.pad.html) the data. - The `__len__` function should ideally return the amount of pixels in the data. However, a single epoch would then require lots of iterations. Cap this with an upper bound `max_iter_epoch`. - Data augmentation is not yet required.
###Code
# helper function: transform a linear index from a 3D array to 3D coordinates
# assuming slice-by-slice raster scanning ordering
def delinearize_index(i, sz):
z_, y_, x_ = sz
x = np.mod(i, x_)
j = (i - x) // x_
y = np.mod(j, y_)
z = (j - y) // y_
return z, y, x
# dataset useful for sampling (and many other things)
class EMWindowDataset(data.Dataset):
def __init__(self, x, y, wnd_sz, max_iter_epoch):
# window size
self.wnd_sz = wnd_sz
# offset (for padding)
self.offset = (wnd_sz[0] // 2, wnd_sz[1] // 2)
# size of the data
self.size = x.shape
# maximum number of iterations per epoch
self.max_iter_epoch = max_iter_epoch
# pad the data to avoid boundary issues
self.x_padded = np.pad(x, ((0, 0), (self.offset[0], self.offset[0]), (self.offset[1], self.offset[1])), 'symmetric')
# save the labels, no padding required
self.y = y
def __getitem__(self, i):
# delinearize the index
z, y, x = delinearize_index(i, self.size)
# extract the window, don't forget the offsets
oy, ox = self.offset
wnd = self.x_padded[z:z + 1, y:y + 2 * oy + 1, x:x + 2 * ox + 1]
return wnd, self.y[z, y, x]
def __len__(self):
return self.max_iter_epoch
# parameters
window_size = (32, 32)
batch_size = 256
max_iter_epoch = 2**12
# make an instance of the dataset for training and testing
ds_train = EMWindowDataset(x_train, y_train, window_size, max_iter_epoch)
ds_test = EMWindowDataset(x_test, y_test, window_size, max_iter_epoch)
# test the class
n = np.random.randint(len(ds_train))
wnd, label = ds_train[n]
plt.imshow(wnd[0], cmap='gray')
plt.title(label)
plt.show()
# setup the data loader
train_loader = DataLoader(ds_train, batch_size=batch_size, shuffle=True)
test_loader = DataLoader(ds_test, batch_size=batch_size, shuffle=True)
###Output
_____no_output_____
###Markdown
The positive side to segmentation labels is that each pixel corresponds to a single annotation. In contrast to usual classification dataset, this may give the impression that there are lots of labels. However, keep in mind that the data is heavily correlated, especially locally. In other words, neighboring pixel labels of a reference pixel won't provide that much more information than the reference pixel label. The classification network that we will use is exactly the same as the one we used in the previous session.
###Code
class ConvNormRelu(nn.Module):
def __init__(self, in_channels, out_channels, kernel_size=3, padding=1):
super(ConvNormRelu, self).__init__()
self.unit = nn.Sequential()
self.unit.add_module('conv', nn.Conv2d(in_channels, out_channels, kernel_size=kernel_size, padding=padding))
self.unit.add_module('norm', nn.BatchNorm2d(int(out_channels)))
self.unit.add_module('activation', nn.ReLU())
def forward(self, inputs):
return self.unit(inputs)
class CNN(nn.Module):
def __init__(self, feature_maps=16):
super(CNN, self).__init__()
self.feature_maps = feature_maps
self.conv1 = ConvNormRelu(in_channels=1, out_channels=feature_maps)
self.conv2 = ConvNormRelu(in_channels=feature_maps, out_channels=feature_maps)
self.conv3 = ConvNormRelu(in_channels=feature_maps, out_channels=feature_maps)
self.conv4 = ConvNormRelu(in_channels=feature_maps, out_channels=feature_maps)
self.pool = nn.MaxPool2d(2, 2)
self.fc = nn.Linear(feature_maps*2*2, 2)
def forward(self, x):
x = self.pool(self.conv1(x))
x = self.pool(self.conv2(x))
x = self.pool(self.conv3(x))
x = self.pool(self.conv4(x))
x = x.view(-1, self.feature_maps*2*2)
x = self.fc(x)
return x
net = CNN(feature_maps=16)
print(net)
###Output
CNN(
(conv1): ConvNormRelu(
(unit): Sequential(
(conv): Conv2d(1, 16, kernel_size=(3, 3), stride=(1, 1), padding=(1, 1))
(norm): BatchNorm2d(16, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)
(activation): ReLU()
)
)
(conv2): ConvNormRelu(
(unit): Sequential(
(conv): Conv2d(16, 16, kernel_size=(3, 3), stride=(1, 1), padding=(1, 1))
(norm): BatchNorm2d(16, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)
(activation): ReLU()
)
)
(conv3): ConvNormRelu(
(unit): Sequential(
(conv): Conv2d(16, 16, kernel_size=(3, 3), stride=(1, 1), padding=(1, 1))
(norm): BatchNorm2d(16, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)
(activation): ReLU()
)
)
(conv4): ConvNormRelu(
(unit): Sequential(
(conv): Conv2d(16, 16, kernel_size=(3, 3), stride=(1, 1), padding=(1, 1))
(norm): BatchNorm2d(16, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)
(activation): ReLU()
)
)
(pool): MaxPool2d(kernel_size=2, stride=2, padding=0, dilation=1, ceil_mode=False)
(fc): Linear(in_features=64, out_features=2, bias=True)
)
###Markdown
We will use an alternative metric for validation. The accuracy is a metric motivated from a classification point of view. A more suitable segmentation metric is the Dice coefficient: $$D = \frac{2 \times \left| Y \cap \hat{Y} \right|}{\left| Y \right| + \left| \hat{Y} \right|}$$where $Y$ and $\hat{Y}$ are the ground truth and predicted segmentation, respectively. If the prediction perfectly overlaps, the Dice coefficient will be 1. The code below illustrates the resulting segmentation of the best model so far and the corresponding dice score. **Exercise**: Train the classification network for pixel-wise label prediction: - Implement the `train_net`, `train_epoch` and `test_epoch`. To make life easier, you can reuse parts of the code of the previous exercise. - Implement the `dice` function that computes the metric describe above. Save the model with the highest Dice score (averaged over the two classes). - Evaluate the (average) Dice metric at the end of each test epoch and save it, so that it can be plotted (similar to the loss). - Train the network for 20 epochs at a learning rate of 0.001.
###Code
# dice coefficient implementation
def dice(y, y_pred):
intersection = np.sum(y * y_pred)
return 2 * intersection / (np.sum(y) + np.sum(y_pred))
# implementation of a single training epoch
def train_epoch(net, loader, loss_fn, optimizer):
# set the network in training mode
net.train()
# keep track of the loss
loss_cum = 0
cnt = 0
for i, data in enumerate(loader):
# sample data
x, y = data
# transfer data to GPU and correct format
x = x.float().to(device)
y = y.long().to(device)
# set all gradients equal to zero
net.zero_grad()
# feed the batch to the network and compute the outputs
y_pred = net(x)
# compare the outputs to the labels with the loss function
loss = loss_fn(y_pred, y)
loss_cum += loss.data.cpu().numpy()
cnt += 1
# backpropagate the gradients w.r.t. computed loss
loss.backward()
# apply one step in the optimization
optimizer.step()
# compute the average loss
loss_avg = loss_cum / cnt
return loss_avg
# implementation of a single testing epoch
def test_epoch(net, loader, loss_fn):
# set the network in training mode
net.eval()
# keep track of the loss and predictions
preds = np.zeros((len(loader.dataset), 2))
ys = np.zeros((len(loader.dataset)))
loss_cum = 0
cnt = 0
for i, data in enumerate(loader):
# sample data
x, y = data
# transfer data to GPU and correct format
x = x.float().to(device)
y = y.long().to(device)
# feed the batch to the network and compute the outputs
y_pred = net(x)
# compare the outputs to the labels with the loss function
loss = loss_fn(y_pred, y)
loss_cum += loss.data.cpu().numpy()
cnt += 1
# get the class probability predictions and save them for validation
y_ = torch.softmax(y_pred, dim=1)
b = i * loader.batch_size
preds[b: b + y_.size(0), :] = y_.detach().cpu().numpy()
ys[b: b + y_.size(0)] = y.detach().cpu().numpy()
# compute accuracy
d = 0.5 * (dice(1 - ys, 1 - preds.argmax(axis=1)) + dice(ys, preds.argmax(axis=1)))
# compute the average loss
loss_avg = loss_cum / cnt
return loss_avg, d
def train_net(net, train_loader, test_loader, loss_fn, optimizer, epochs, log_dir):
# transfer the network to the GPU
net.to(device)
best_dice = 0
train_loss = np.zeros((epochs))
test_loss = np.zeros((epochs))
test_dice = np.zeros((epochs))
for epoch in range(epochs):
# training
train_loss[epoch] = train_epoch(net, train_loader, loss_fn, optimizer)
# testing
test_loss[epoch], test_dice[epoch] = test_epoch(net, test_loader, loss_fn)
# check if accuracy has increased
if test_dice[epoch] > best_dice:
best_dice = test_dice[epoch]
# save the model
torch.save(net.state_dict(), 'cnn_best.cpt')
print('Epoch %5d - Train loss: %.6f - Test loss: %.6f - Test dice avg: %.6f' % (epoch, train_loss[epoch], test_loss[epoch], test_dice[epoch]))
return train_loss, test_loss, test_dice
# parameters
learning_rate = 0.001
n_epochs = 20
log_dir = '.'
# define the optimizer
class_weights = torch.from_numpy(np.divide(1, class_distribution)).float().to(device)
class_weights = class_weights / class_weights.sum()
loss_fn = nn.CrossEntropyLoss(weight=class_weights)
# loss_fn = nn.CrossEntropyLoss()
optimizer = optim.Adam(net.parameters(), lr=learning_rate)
# start training
train_loss_cnn, test_loss_cnn, test_dice_cnn = train_net(net, train_loader, test_loader, loss_fn, optimizer, n_epochs, log_dir)
# show the training curve and accuracy
plt.subplot(1, 2, 1)
plt.plot(train_loss_cnn)
plt.plot(test_loss_cnn)
plt.xlabel('Epochs')
plt.ylabel('Loss')
plt.legend(('Train', 'Test'))
plt.subplot(1, 2, 2)
plt.plot(test_dice_cnn)
plt.xlabel('Epochs')
plt.ylabel('Dice avg')
plt.show()
###Output
Epoch 0 - Train loss: 0.414006 - Test loss: 0.693262 - Test dice avg: 0.441277
Epoch 1 - Train loss: 0.230934 - Test loss: 0.721943 - Test dice avg: 0.497957
Epoch 2 - Train loss: 0.161893 - Test loss: 0.673282 - Test dice avg: 0.692215
Epoch 3 - Train loss: 0.119127 - Test loss: 0.639826 - Test dice avg: 0.738619
Epoch 4 - Train loss: 0.091824 - Test loss: 0.601032 - Test dice avg: 0.754433
Epoch 5 - Train loss: 0.075326 - Test loss: 0.657208 - Test dice avg: 0.755952
Epoch 6 - Train loss: 0.062996 - Test loss: 0.788308 - Test dice avg: 0.739927
Epoch 7 - Train loss: 0.054004 - Test loss: 0.871502 - Test dice avg: 0.744548
Epoch 8 - Train loss: 0.043278 - Test loss: 0.694500 - Test dice avg: 0.766866
Epoch 9 - Train loss: 0.039117 - Test loss: 0.965212 - Test dice avg: 0.726596
Epoch 10 - Train loss: 0.030984 - Test loss: 0.829635 - Test dice avg: 0.747853
Epoch 11 - Train loss: 0.029167 - Test loss: 0.869089 - Test dice avg: 0.745000
Epoch 12 - Train loss: 0.024234 - Test loss: 1.022034 - Test dice avg: 0.735058
Epoch 13 - Train loss: 0.021572 - Test loss: 1.028657 - Test dice avg: 0.741081
Epoch 14 - Train loss: 0.016402 - Test loss: 1.012114 - Test dice avg: 0.740054
Epoch 15 - Train loss: 0.013843 - Test loss: 0.995476 - Test dice avg: 0.740326
Epoch 16 - Train loss: 0.013747 - Test loss: 1.122460 - Test dice avg: 0.733761
Epoch 17 - Train loss: 0.011495 - Test loss: 1.075303 - Test dice avg: 0.738531
Epoch 18 - Train loss: 0.010605 - Test loss: 1.338515 - Test dice avg: 0.718386
Epoch 19 - Train loss: 0.008500 - Test loss: 1.205790 - Test dice avg: 0.737104
###Markdown
You should obtain an average Dice score between 0.70 and 0.75. However, note that according to the learning curves, the model seems to be overfitting relatively fast. This is mainly due to the locality of the feature extraction. We will now illustrate how a test sample can be segmented with the trained network. **Exercise**: Implement the `segment_slice` function: - The function takes a 2D slice, a pretrained network, a window size and batch size as input, and computes the segmentation (a binary 2D array). - The easiest way to loop through all the pixels of an image is by using the `EMWindowDataset` without shuffling. However, you will have to adjust the maximum number of iterations. - As a first step, you can assume `batch_size=1`. Keep in mind, this can be inefficient, because GPUs become beneficial as the amount of parallel operations increases. Higher batch sizes therefore benefit computing time, but this is of course bounded to the available GPU memory.
###Code
def segment_slice(x, net, window_size, batch_size):
# setup data loader
x_orig = x
ds = EMWindowDataset(x[np.newaxis, ...], np.zeros_like(x[np.newaxis, ...]), window_size, x.size)
loader = DataLoader(ds, batch_size=batch_size)
# set the network in training mode
net.eval()
# keep track of the loss and predictions
preds = np.zeros((len(loader.dataset), 2))
for i, data in enumerate(loader):
# sample data
x, _ = data
# transfer data to GPU and correct format
x = x.float().to(device)
# feed the batch to the network and compute the outputs
y_pred = net(x)
# get the class probability predictions and save them for validation
y_ = torch.softmax(y_pred, dim=1)
b = i * loader.batch_size
preds[b: b + y_.size(0), :] = y_.detach().cpu().numpy()
# compute the segmentation
preds = preds.argmax(axis=1)
segmentation = np.reshape(preds, x_orig.shape)
return segmentation.astype('uint8')
# load the best parameters
state_dict = torch.load('cnn_best.cpt')
net.load_state_dict(state_dict)
# perform segmentation
n = 0
t = time.time()
y_pred = segment_slice(x_test[n], net, window_size, batch_size)
print('Elapsed time: %.2f seconds' % (time.time() - t))
# show example
x_overlay = overlay(x_test[n], 1 - y_test[n], colors=[(1, 0, 0)], alpha=0.4)
x_pred_overlay = overlay(x_test[n], 1 - y_pred, colors=[(1, 0, 0)], alpha=0.4)
plt.subplot(1, 2, 1)
plt.imshow(x_pred_overlay)
plt.title('Predictions')
plt.axis('off')
plt.subplot(1, 2, 2)
plt.imshow(x_overlay)
plt.title('Labels')
plt.axis('off')
plt.show()
###Output
Elapsed time: 5.41 seconds
###Markdown
Visually, the segmentation result does not look perfect. Clearly, the network is making lots of mistakes, especially along membranes, because these regions require more context. Another disadvantage to this methodology is the computational inefficiency. Even with larger batches, segmentation of a relatively small image patch can take seconds, which is impractical for larger datasets. The reason is obviously that for each pixel, a forward call of the network is required. 4. Building a U-Net with PyTorchThe lack of global context and computational efficiency of the pixel classification approach results in poor practical performance. As an alternative, the [U-Net](https://link.springer.com/chapter/10.1007/978-3-319-24574-4_28) network was proposed. This is an end-to-end segmentation network that takes an image as input and computes a complete segmentation of the input. Let's start by defining the network architecture: 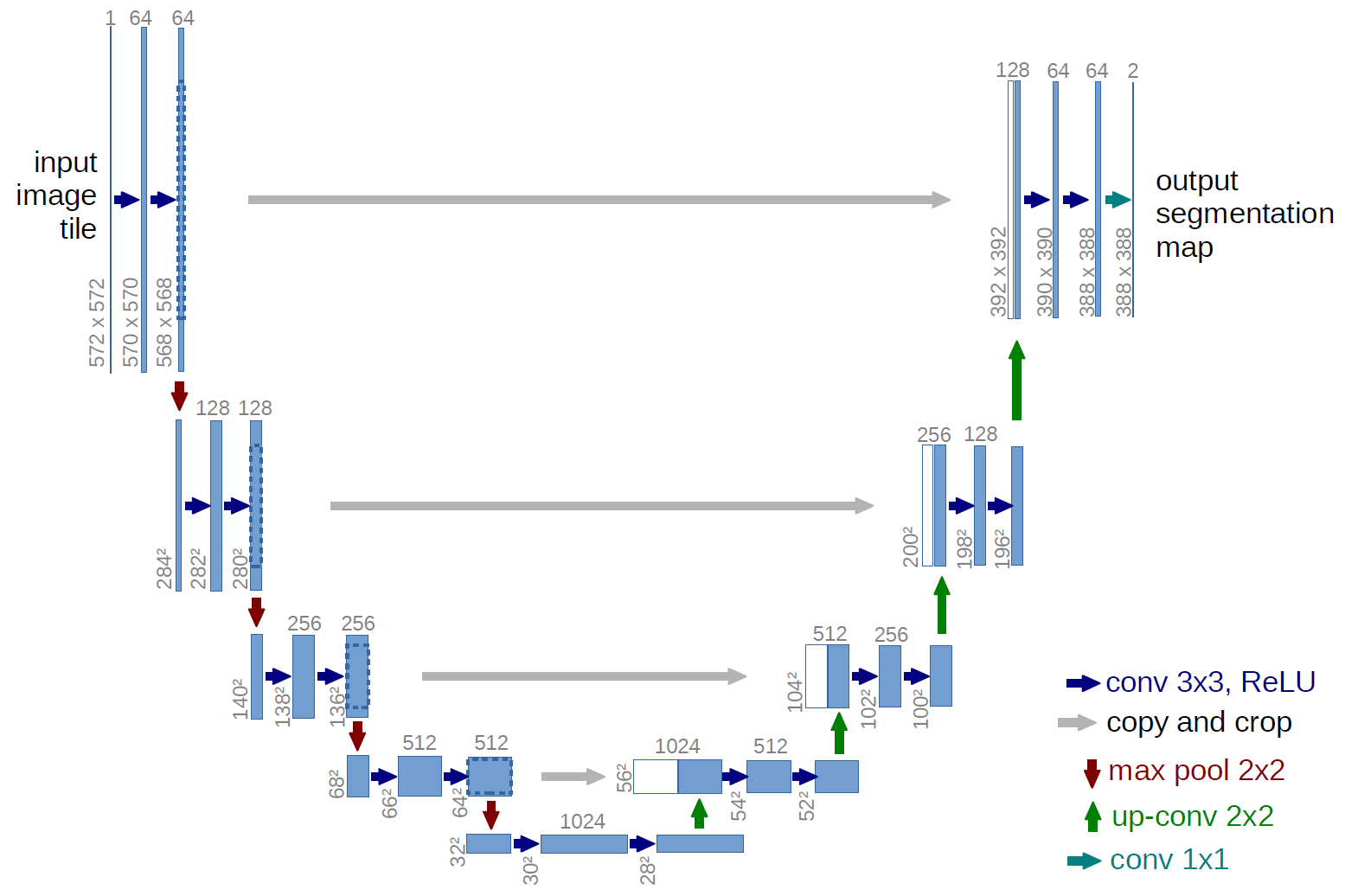**Exercise**: Implement the U-Net architecture: - The architecture consists of an encoder, a bottleneck, a decoder and skip connections between the encoder and decoder. - The basic building blocks of the U-Net architecture are two consecutive convolutional layers with ReLU activation that take $n$ feature maps and output $m$ feature maps. The convolutional layers have $3 \times 3$ kernels. For simplicity we will also pad the inputs by 1 pixel to make sure the inputs and outputs have the same size. Implement this block in the `ConvBlock` class. - Downsampling is relatively simply with $2 \times 2$ max-pooling. However, upsampling is performed using bilinear upsampling layers, followed by a convolution layer. This operation has been implemented in the [`ConvTranspose2d`](https://pytorch.org/docs/stable/generated/torch.nn.ConvTranspose2d.html) class with $2 \times 2$ kernels and a stride of $2$. - Implement the skip connections by using the PyTorch [concatenation](https://pytorch.org/docs/stable/generated/torch.cat.html) function. - The initial number of feature maps is a parameter that can be chosen. From then on, the amount of feature maps doubles with every `ConvBlock` in the encoder and halves with every `ConvBlock` in the decoder.
###Code
class ConvBlock(nn.Module):
def __init__(self, in_channels, out_channels, kernel_size=3, padding=1):
super(ConvBlock, self).__init__()
self.unit = nn.Sequential()
self.unit.add_module('conv1', ConvNormRelu(in_channels=in_channels, out_channels=out_channels, kernel_size=kernel_size, padding=padding))
self.unit.add_module('conv2', ConvNormRelu(in_channels=out_channels, out_channels=out_channels, kernel_size=kernel_size, padding=padding))
def forward(self, inputs):
return self.unit(inputs)
class UNet(nn.Module):
def __init__(self, feature_maps=16, out_channels=2):
super(UNet, self).__init__()
self.feature_maps = feature_maps
# encoder layers
self.conv_enc_1 = ConvBlock(in_channels=1, out_channels=feature_maps)
self.conv_enc_2 = ConvBlock(in_channels=feature_maps, out_channels=2*feature_maps)
self.conv_enc_3 = ConvBlock(in_channels=2*feature_maps, out_channels=4*feature_maps)
self.conv_enc_4 = ConvBlock(in_channels=4*feature_maps, out_channels=8*feature_maps)
self.pool = nn.MaxPool2d(2, 2)
# bottleneck layers
self.conv_btl = ConvBlock(in_channels=8*feature_maps, out_channels=16*feature_maps)
# decoder layers
self.conv_dec_4_up = nn.ConvTranspose2d(in_channels=16*feature_maps, out_channels=8*feature_maps, kernel_size=2, stride=2)
self.conv_dec_4 = ConvBlock(in_channels=16*feature_maps, out_channels=8*feature_maps)
self.conv_dec_3_up = nn.ConvTranspose2d(in_channels=8*feature_maps, out_channels=4*feature_maps, kernel_size=2, stride=2)
self.conv_dec_3 = ConvBlock(in_channels=8*feature_maps, out_channels=4*feature_maps)
self.conv_dec_2_up = nn.ConvTranspose2d(in_channels=4*feature_maps, out_channels=2*feature_maps, kernel_size=2, stride=2)
self.conv_dec_2 = ConvBlock(in_channels=4*feature_maps, out_channels=2*feature_maps)
self.conv_dec_1_up = nn.ConvTranspose2d(in_channels=2*feature_maps, out_channels=feature_maps, kernel_size=2, stride=2)
self.conv_dec_1 = ConvBlock(in_channels=2*feature_maps, out_channels=feature_maps)
# final segmentation layer
self.conv_final = nn.Conv2d(in_channels=feature_maps, out_channels=out_channels, kernel_size=1)
def forward(self, x):
# encoder path
x1 = self.conv_enc_1(x)
x = self.pool(x1)
x2 = self.conv_enc_2(x)
x = self.pool(x2)
x3 = self.conv_enc_3(x)
x = self.pool(x3)
x4 = self.conv_enc_4(x)
x = self.pool(x4)
# bottleneck
x = self.conv_btl(x)
# decoder path
x = torch.cat((self.conv_dec_4_up(x), x4), dim=1)
x = self.conv_dec_4(x)
x = torch.cat((self.conv_dec_3_up(x), x3), dim=1)
x = self.conv_dec_3(x)
x = torch.cat((self.conv_dec_2_up(x), x2), dim=1)
x = self.conv_dec_2(x)
x = torch.cat((self.conv_dec_1_up(x), x1), dim=1)
x = self.conv_dec_1(x)
# final segmentation
x = self.conv_final(x)
return x
net = UNet(feature_maps=64)
print(net)
###Output
UNet(
(conv_enc_1): ConvBlock(
(unit): Sequential(
(conv1): ConvNormRelu(
(unit): Sequential(
(conv): Conv2d(1, 64, kernel_size=(3, 3), stride=(1, 1), padding=(1, 1))
(norm): BatchNorm2d(64, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)
(activation): ReLU()
)
)
(conv2): ConvNormRelu(
(unit): Sequential(
(conv): Conv2d(64, 64, kernel_size=(3, 3), stride=(1, 1), padding=(1, 1))
(norm): BatchNorm2d(64, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)
(activation): ReLU()
)
)
)
)
(conv_enc_2): ConvBlock(
(unit): Sequential(
(conv1): ConvNormRelu(
(unit): Sequential(
(conv): Conv2d(64, 128, kernel_size=(3, 3), stride=(1, 1), padding=(1, 1))
(norm): BatchNorm2d(128, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)
(activation): ReLU()
)
)
(conv2): ConvNormRelu(
(unit): Sequential(
(conv): Conv2d(128, 128, kernel_size=(3, 3), stride=(1, 1), padding=(1, 1))
(norm): BatchNorm2d(128, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)
(activation): ReLU()
)
)
)
)
(conv_enc_3): ConvBlock(
(unit): Sequential(
(conv1): ConvNormRelu(
(unit): Sequential(
(conv): Conv2d(128, 256, kernel_size=(3, 3), stride=(1, 1), padding=(1, 1))
(norm): BatchNorm2d(256, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)
(activation): ReLU()
)
)
(conv2): ConvNormRelu(
(unit): Sequential(
(conv): Conv2d(256, 256, kernel_size=(3, 3), stride=(1, 1), padding=(1, 1))
(norm): BatchNorm2d(256, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)
(activation): ReLU()
)
)
)
)
(conv_enc_4): ConvBlock(
(unit): Sequential(
(conv1): ConvNormRelu(
(unit): Sequential(
(conv): Conv2d(256, 512, kernel_size=(3, 3), stride=(1, 1), padding=(1, 1))
(norm): BatchNorm2d(512, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)
(activation): ReLU()
)
)
(conv2): ConvNormRelu(
(unit): Sequential(
(conv): Conv2d(512, 512, kernel_size=(3, 3), stride=(1, 1), padding=(1, 1))
(norm): BatchNorm2d(512, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)
(activation): ReLU()
)
)
)
)
(pool): MaxPool2d(kernel_size=2, stride=2, padding=0, dilation=1, ceil_mode=False)
(conv_btl): ConvBlock(
(unit): Sequential(
(conv1): ConvNormRelu(
(unit): Sequential(
(conv): Conv2d(512, 1024, kernel_size=(3, 3), stride=(1, 1), padding=(1, 1))
(norm): BatchNorm2d(1024, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)
(activation): ReLU()
)
)
(conv2): ConvNormRelu(
(unit): Sequential(
(conv): Conv2d(1024, 1024, kernel_size=(3, 3), stride=(1, 1), padding=(1, 1))
(norm): BatchNorm2d(1024, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)
(activation): ReLU()
)
)
)
)
(conv_dec_4_up): ConvTranspose2d(1024, 512, kernel_size=(2, 2), stride=(2, 2))
(conv_dec_4): ConvBlock(
(unit): Sequential(
(conv1): ConvNormRelu(
(unit): Sequential(
(conv): Conv2d(1024, 512, kernel_size=(3, 3), stride=(1, 1), padding=(1, 1))
(norm): BatchNorm2d(512, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)
(activation): ReLU()
)
)
(conv2): ConvNormRelu(
(unit): Sequential(
(conv): Conv2d(512, 512, kernel_size=(3, 3), stride=(1, 1), padding=(1, 1))
(norm): BatchNorm2d(512, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)
(activation): ReLU()
)
)
)
)
(conv_dec_3_up): ConvTranspose2d(512, 256, kernel_size=(2, 2), stride=(2, 2))
(conv_dec_3): ConvBlock(
(unit): Sequential(
(conv1): ConvNormRelu(
(unit): Sequential(
(conv): Conv2d(512, 256, kernel_size=(3, 3), stride=(1, 1), padding=(1, 1))
(norm): BatchNorm2d(256, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)
(activation): ReLU()
)
)
(conv2): ConvNormRelu(
(unit): Sequential(
(conv): Conv2d(256, 256, kernel_size=(3, 3), stride=(1, 1), padding=(1, 1))
(norm): BatchNorm2d(256, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)
(activation): ReLU()
)
)
)
)
(conv_dec_2_up): ConvTranspose2d(256, 128, kernel_size=(2, 2), stride=(2, 2))
(conv_dec_2): ConvBlock(
(unit): Sequential(
(conv1): ConvNormRelu(
(unit): Sequential(
(conv): Conv2d(256, 128, kernel_size=(3, 3), stride=(1, 1), padding=(1, 1))
(norm): BatchNorm2d(128, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)
(activation): ReLU()
)
)
(conv2): ConvNormRelu(
(unit): Sequential(
(conv): Conv2d(128, 128, kernel_size=(3, 3), stride=(1, 1), padding=(1, 1))
(norm): BatchNorm2d(128, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)
(activation): ReLU()
)
)
)
)
(conv_dec_1_up): ConvTranspose2d(128, 64, kernel_size=(2, 2), stride=(2, 2))
(conv_dec_1): ConvBlock(
(unit): Sequential(
(conv1): ConvNormRelu(
(unit): Sequential(
(conv): Conv2d(128, 64, kernel_size=(3, 3), stride=(1, 1), padding=(1, 1))
(norm): BatchNorm2d(64, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)
(activation): ReLU()
)
)
(conv2): ConvNormRelu(
(unit): Sequential(
(conv): Conv2d(64, 64, kernel_size=(3, 3), stride=(1, 1), padding=(1, 1))
(norm): BatchNorm2d(64, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)
(activation): ReLU()
)
)
)
)
(conv_final): Conv2d(64, 2, kernel_size=(1, 1), stride=(1, 1))
)
###Markdown
Of course, this network requires an image as input and the corresponding label image as output. Therefore, we have to modify our dataset. **Exercise**: Modify the `EMWindowDataset` class: - Implement the `__getitem__` method, the remaining functions have already been implemented.
###Code
# dataset useful for sampling
class EMWindowDataset(data.Dataset):
def __init__(self, x, y, wnd_sz, max_iter_epoch):
# window size
self.wnd_sz = wnd_sz
# maximum number of iterations per epoch
self.max_iter_epoch = max_iter_epoch
# save the data
self.x = x
# save the labels
self.y = y
def __getitem__(self, i):
# generate random location in the data
z = np.random.randint(self.x.shape[0])
y = np.random.randint(self.x.shape[1] - self.wnd_sz[0] + 1)
x = np.random.randint(self.x.shape[2] - self.wnd_sz[1] + 1)
# extract the input and label windows
wnd_x = self.x[z:z + 1, y:y + self.wnd_sz[0], x:x + self.wnd_sz[1]]
wnd_y = self.y[z:z + 1, y:y + self.wnd_sz[0], x:x + self.wnd_sz[1]]
return wnd_x, wnd_y
def __len__(self):
return self.max_iter_epoch
# parameters
window_size = (128, 128)
batch_size = 2
max_iter_epoch = 128
# make an instance of the dataset for training and testing
ds_train = EMWindowDataset(x_train, y_train, window_size, max_iter_epoch)
ds_test = EMWindowDataset(x_test, y_test, window_size, max_iter_epoch)
# test the class
n = np.random.randint(len(ds_train))
x, y = ds_train[n]
x_overlay = overlay(x[0], 1 - y[0], colors=[(1, 0, 0)], alpha=0.4)
plt.imshow(x_overlay)
plt.show()
# setup the data loader
train_loader = DataLoader(ds_train, batch_size=batch_size, shuffle=True)
test_loader = DataLoader(ds_test, batch_size=batch_size, shuffle=True)
###Output
_____no_output_____
###Markdown
5. Training & validating the networkNow that we have a U-Net network and a data loader, it is time to train the network! For the sake of repetitiveness, you are not required to implement the complete training loop. **Exercise**: Train the U-Net architecture: - Have a look at the training loop code and identify the differences with classical CNN training. - Train the network for 20 epochs with a learning rate of 0.001.
###Code
# implementation of a single training epoch
def train_epoch(net, loader, loss_fn, optimizer):
# set the network in training mode
net.train()
# keep track of the loss
loss_cum = 0
cnt = 0
for i, data in enumerate(loader):
# sample data
x, y = data
# transfer data to GPU and correct format
x = x.float().to(device)
y = y.long()[:, 0, :, :].to(device)
# set all gradients equal to zero
net.zero_grad()
# feed the batch to the network and compute the outputs
y_pred = net(x)
# compare the outputs to the labels with the loss function
loss = loss_fn(y_pred, y)
loss_cum += loss.data.cpu().numpy()
cnt += 1
# backpropagate the gradients w.r.t. computed loss
loss.backward()
# apply one step in the optimization
optimizer.step()
# compute the average loss
loss_avg = loss_cum / cnt
return loss_avg
# implementation of a single testing epoch
def test_epoch(net, loader, loss_fn):
# set the network in training mode
net.eval()
# keep track of the loss and predictions
preds = np.zeros((len(loader.dataset), *loader.dataset.wnd_sz))
ys = np.zeros((len(loader.dataset), *loader.dataset.wnd_sz))
loss_cum = 0
cnt = 0
for i, data in enumerate(loader):
# sample data
x, y = data
# transfer data to GPU and correct format
x = x.float().to(device)
y = y.long()[:, 0, :, :].to(device)
# feed the batch to the network and compute the outputs
y_pred = net(x)
# compare the outputs to the labels with the loss function
loss = loss_fn(y_pred, y)
loss_cum += loss.data.cpu().numpy()
cnt += 1
# get the class probability predictions and save them for validation
y_ = torch.softmax(y_pred, dim=1)
b = i * loader.batch_size
preds[b: b + y_.size(0), ...] = y_.argmax(dim=1).detach().cpu().numpy()
ys[b: b + y_.size(0), ...] = y.detach().cpu().numpy()
# compute accuracy
d = dice(ys.flatten(), preds.flatten())
# compute the average loss
loss_avg = loss_cum / cnt
return loss_avg, d
def train_net(net, train_loader, test_loader, loss_fn, optimizer, epochs, log_dir):
# transfer the network to the GPU
net.to(device)
best_dice = 0
train_loss = np.zeros((epochs))
test_loss = np.zeros((epochs))
test_dice = np.zeros((epochs))
for epoch in range(epochs):
# training
train_loss[epoch] = train_epoch(net, train_loader, loss_fn, optimizer)
# testing
test_loss[epoch], test_dice[epoch] = test_epoch(net, test_loader, loss_fn)
# check if accuracy has increased
if test_dice[epoch] > best_dice:
best_dice = test_dice[epoch]
# save the model
torch.save(net.state_dict(), 'unet_best.cpt')
print('Epoch %5d - Train loss: %.6f - Test loss: %.6f - Test dice avg: %.6f' % (epoch, train_loss[epoch], test_loss[epoch], test_dice[epoch]))
return train_loss, test_loss, test_dice
# parameters
learning_rate = 0.001
n_epochs = 20
log_dir = '.'
# define the optimizer
class_weights = torch.from_numpy(np.divide(1, class_distribution)).float().to(device)
class_weights = class_weights / class_weights.sum()
loss_fn = nn.CrossEntropyLoss(weight=class_weights)
# loss_fn = nn.CrossEntropyLoss()
optimizer = optim.Adam(net.parameters(), lr=learning_rate)
# start training
train_loss_unet, test_loss_unet, test_dice_unet = train_net(net, train_loader, test_loader, loss_fn, optimizer, n_epochs, log_dir)
# show the training curve and accuracy
plt.subplot(1, 2, 1)
plt.plot(train_loss_cnn)
plt.plot(test_loss_cnn)
plt.plot(train_loss_unet)
plt.plot(test_loss_unet)
plt.xlabel('Epochs')
plt.ylabel('Loss')
plt.legend(('Train (CNN)', 'Test (CNN)', 'Train (U-Net)', 'Test (U-Net)'))
plt.subplot(1, 2, 2)
plt.plot(test_dice_cnn)
plt.plot(test_dice_unet)
plt.xlabel('Epochs')
plt.ylabel('Dice avg')
plt.legend(('CNN', 'U-Net'))
plt.show()
###Output
Epoch 0 - Train loss: 0.383133 - Test loss: 0.354965 - Test dice avg: 0.856172
Epoch 1 - Train loss: 0.361292 - Test loss: 0.352362 - Test dice avg: 0.902300
Epoch 2 - Train loss: 0.324768 - Test loss: 0.306067 - Test dice avg: 0.887419
Epoch 3 - Train loss: 0.325949 - Test loss: 0.334848 - Test dice avg: 0.850665
Epoch 4 - Train loss: 0.328289 - Test loss: 0.310259 - Test dice avg: 0.913652
Epoch 5 - Train loss: 0.315503 - Test loss: 0.305358 - Test dice avg: 0.878493
Epoch 6 - Train loss: 0.316873 - Test loss: 0.324832 - Test dice avg: 0.908888
Epoch 7 - Train loss: 0.303972 - Test loss: 0.295798 - Test dice avg: 0.932093
Epoch 8 - Train loss: 0.305955 - Test loss: 0.300341 - Test dice avg: 0.886833
Epoch 9 - Train loss: 0.288136 - Test loss: 0.260361 - Test dice avg: 0.931314
Epoch 10 - Train loss: 0.273979 - Test loss: 0.266878 - Test dice avg: 0.914591
Epoch 11 - Train loss: 0.282206 - Test loss: 0.261618 - Test dice avg: 0.903781
Epoch 12 - Train loss: 0.275372 - Test loss: 0.256586 - Test dice avg: 0.916227
Epoch 13 - Train loss: 0.274017 - Test loss: 0.276006 - Test dice avg: 0.896439
Epoch 14 - Train loss: 0.266158 - Test loss: 0.303701 - Test dice avg: 0.896790
Epoch 15 - Train loss: 0.280715 - Test loss: 0.256113 - Test dice avg: 0.938594
Epoch 16 - Train loss: 0.280409 - Test loss: 0.281226 - Test dice avg: 0.897781
Epoch 17 - Train loss: 0.258447 - Test loss: 0.248680 - Test dice avg: 0.929210
Epoch 18 - Train loss: 0.269229 - Test loss: 0.516527 - Test dice avg: 0.653548
Epoch 19 - Train loss: 0.259088 - Test loss: 0.263776 - Test dice avg: 0.906309
###Markdown
That should look much better! The U-Net architecture is significantly less suffering from overfitting and the mean Dice coefficient is substantially higher (should be approximately 0.90). Now let's have a look how the actual predictions look like. **Exercise**: Modify the `segment_slice` so that it segments a 2D image with a U-Net network: - Simplify the original code of `segment_slice` by propagating the image straightforward through the network. - Can you figure out why alternative image sizes can also be fed into the network? Is that always possible?
###Code
def segment_slice(x, net):
# set the network in training mode
net.eval()
# convert to torch tensor
x = torch.from_numpy(x[np.newaxis, np.newaxis, ...]).float().to(device)
# forward propagation
pred = net(x).detach().cpu().numpy()
# compute the segmentation
segmentation = pred.argmax(axis=1)[0]
return segmentation
# load the best parameters
state_dict = torch.load('unet_best.cpt')
net.load_state_dict(state_dict)
# perform segmentation
n = 0
t = time.time()
y_pred = segment_slice(x_test[n], net)
print('Elapsed time: %.2f seconds' % (time.time() - t))
# show example
x_overlay = overlay(x_test[n], 1 - y_test[n], colors=[(1, 0, 0)], alpha=0.4)
x_pred_overlay = overlay(x_test[n], 1 - y_pred, colors=[(1, 0, 0)], alpha=0.4)
plt.subplot(1, 2, 1)
plt.imshow(x_pred_overlay)
plt.title('Predictions')
plt.axis('off')
plt.subplot(1, 2, 2)
plt.imshow(x_overlay)
plt.title('Labels')
plt.axis('off')
plt.show()
###Output
Elapsed time: 0.10 seconds
|
DoHBrw/1-Constant-Quasi-Constant-Duplicates/1.2-Quasi-constant-features.ipynb
|
###Markdown
Quasi-constant features
###Code
import numpy as np
import pandas as pd
from sklearn.model_selection import train_test_split
from sklearn.feature_selection import VarianceThreshold
###Output
_____no_output_____
###Markdown
Read Data
###Code
data = pd.read_csv('../DoHBrwTest.csv')
data.shape
data.head(5)
###Output
_____no_output_____
###Markdown
Train - Test Split
###Code
# separate dataset into train and test
X_train, X_test, y_train, y_test = train_test_split(
data.drop(labels=['is_intrusion'], axis=1), # drop the target
data['is_intrusion'], # just the target
test_size=0.3,
random_state=0)
X_train.shape, X_test.shape
###Output
_____no_output_____
###Markdown
Remove constant features (optional)First, I will remove constant features like I did in the previous lecture. This will allow a better visualisation of the quasi-constant ones.
###Code
constant_features = [
feat for feat in X_train.columns if X_train[feat].std() == 0.5]
X_train.drop(labels=constant_features, axis=1, inplace=True)
X_test.drop(labels=constant_features, axis=1, inplace=True)
X_train.shape, X_test.shape
###Output
_____no_output_____
###Markdown
Remove quasi-constant features Using the VarianceThreshold from sklearn
###Code
sel = VarianceThreshold(threshold=0.5)
sel.fit(X_train) # fit finds the features with low variance
# If we sum over get_support, we get the number of features that are not quasi-constant
sum(sel.get_support())
# let's print the number of quasi-constant features
quasi_constant = X_train.columns[~sel.get_support()]
len(quasi_constant)
###Output
_____no_output_____
###Markdown
We can see that 11 columns / variables are constant. This means that 11 variable show the same value, just one value, for all the observations of the training set.
###Code
# let's print the variable names
quasi_constant
# percentage of observations showing each of the different values of the variable
X_train['PacketLengthCoefficientofVariation'].value_counts() / np.float64(len(X_train))
###Output
_____no_output_____
###Markdown
We can see that > 99% of the observations show one value, 0. Therefore, this features is fairly constant.
###Code
# capture feature names
feat_names = X_train.columns[sel.get_support()]
#remove the quasi-constant features
X_train = sel.transform(X_train)
X_test = sel.transform(X_test)
X_train.shape, X_test.shape
# transform the array into a dataframe
X_train = pd.DataFrame(X_train, columns=feat_names)
X_test = pd.DataFrame(X_test, columns=feat_names)
X_test.head()
###Output
_____no_output_____
###Markdown
In the dataset, 3 features are classified as Quasi constant, thus, 31 features remain Standardize Data
###Code
from sklearn.preprocessing import StandardScaler
scaler = StandardScaler().fit(X_train)
X_train = scaler.transform(X_train)
###Output
_____no_output_____
###Markdown
Classifiers
###Code
from sklearn import linear_model
from sklearn.naive_bayes import GaussianNB
from sklearn.ensemble import RandomForestClassifier
from sklearn.neighbors import KNeighborsClassifier
from catboost import CatBoostClassifier
###Output
_____no_output_____
###Markdown
Metrics Evaluation
###Code
from sklearn.metrics import accuracy_score
from sklearn.metrics import confusion_matrix
from sklearn.metrics import roc_curve, f1_score
from sklearn import metrics
from sklearn.model_selection import cross_val_score
###Output
_____no_output_____
###Markdown
Logistic Regression
###Code
%%time
clf_LR = linear_model.LogisticRegression(n_jobs=-1, random_state=42, C=0.1).fit(X_train, y_train)
pred_y_test = clf_LR.predict(X_test)
print('Accuracy:', accuracy_score(y_test, pred_y_test))
f1 = f1_score(y_test, pred_y_test)
print('F1 Score:', f1)
fpr, tpr, thresholds = roc_curve(y_test, pred_y_test)
print('FPR:', fpr[1])
print('TPR:', tpr[1])
###Output
Accuracy: 0.7401468545346299
F1 Score: 0.7647756260554018
FPR: 0.40677338076182007
TPR: 0.9095803777454918
###Markdown
Naive Bayes
###Code
%%time
clf_NB = GaussianNB(var_smoothing=1e-09).fit(X_train, y_train)
pred_y_testNB = clf_NB.predict(X_test)
print('Accuracy:', accuracy_score(y_test, pred_y_testNB))
f1 = f1_score(y_test, pred_y_testNB)
print('F1 Score:', f1)
fpr, tpr, thresholds = roc_curve(y_test, pred_y_testNB)
print('FPR:', fpr[1])
print('TPR:', tpr[1])
###Output
Accuracy: 0.46441754316332606
F1 Score: 0.6342692974848223
FPR: 1.0
TPR: 1.0
###Markdown
Random Forest
###Code
%%time
clf_RF = RandomForestClassifier(random_state=0,max_depth=70,n_estimators=100).fit(X_train, y_train)
pred_y_testRF = clf_RF.predict(X_test)
print('Accuracy:', accuracy_score(y_test, pred_y_testRF))
f1 = f1_score(y_test, pred_y_testRF, average='weighted', zero_division=0)
print('F1 Score:', f1)
fpr, tpr, thresholds = roc_curve(y_test, pred_y_testRF)
print('FPR:', fpr[1])
print('TPR:', tpr[1])
###Output
Accuracy: 0.5355030760071443
F1 Score: 0.37356623711533093
FPR: 0.00014821402104639098
TPR: 0.0
###Markdown
KNN
###Code
%%time
clf_KNN = KNeighborsClassifier(algorithm='brute',leaf_size=1,n_neighbors=2,weights='distance').fit(X_train, y_train)
pred_y_testKNN = clf_KNN.predict(X_test)
print('accuracy_score:', accuracy_score(y_test, pred_y_testKNN))
f1 = f1_score(y_test, pred_y_testKNN)
print('f1:', f1)
fpr, tpr, thresholds = roc_curve(y_test, pred_y_testKNN)
print('fpr:', fpr[1])
print('tpr:', tpr[1])
###Output
accuracy_score: 0.22667195872196866
f1: 0.316446814482178
fpr: 0.9109974803616422
tpr: 0.3854371421246047
###Markdown
CatBoost
###Code
%%time
clf_CB = CatBoostClassifier(random_state=0,depth=7,iterations=50,learning_rate=0.04).fit(X_train, y_train)
pred_y_testCB = clf_CB.predict(X_test)
print('Accuracy:', accuracy_score(y_test, pred_y_testCB))
f1 = f1_score(y_test, pred_y_testCB)
print('F1 Score:', f1)
fpr, tpr, thresholds = roc_curve(y_test, pred_y_testCB)
print('FPR:', fpr[1])
print('TPR:', tpr[1])
###Output
Accuracy: 0.5427267314943441
F1 Score: 0.03127890355671403
FPR: 0.00044464206313917296
TPR: 0.015896077258353986
###Markdown
Model Evaluation
###Code
import pandas as pd, numpy as np
test_df = pd.read_csv("../KDDTest.csv")
test_df.shape
test_df.head()
# Create feature matrix X and target vextor y
y_eval = test_df['is_intrusion']
X_eval = test_df.drop(columns=['is_intrusion','land', 'urgent', 'num_failed_logins', 'root_shell', 'su_attempted',
'num_shells', 'num_access_files', 'num_outbound_cmds', 'is_host_login',
'is_guest_login', 'dst_host_srv_diff_host_rate'])
###Output
_____no_output_____
###Markdown
Model Evaluation - Logistic Regression
###Code
modelLR = linear_model.LogisticRegression(n_jobs=-1, random_state=42, C=0.1)
modelLR.fit(X_train, y_train)
# Predict on the new unseen test data
y_evalpredLR = modelLR.predict(X_eval)
y_predLR = modelLR.predict(X_test)
train_scoreLR = modelLR.score(X_train, y_train)
test_scoreLR = modelLR.score(X_test, y_test)
print("Training accuracy is ", train_scoreLR)
print("Testing accuracy is ", test_scoreLR)
from sklearn.metrics import confusion_matrix, precision_score, recall_score, f1_score
print('Performance measures for test:')
print('--------')
print('Accuracy:', test_scoreLR)
print('F1 Score:',f1_score(y_test, y_predLR))
print('Precision Score:',precision_score(y_test, y_predLR))
print('Recall Score:', recall_score(y_test, y_predLR))
print('Confusion Matrix:\n', confusion_matrix(y_test, y_predLR))
###Output
Performance measures for test:
--------
Accuracy: 0.7401468545346299
F1 Score: 0.7647756260554018
Precision Score: 0.6597446069923134
Recall Score: 0.9095803777454918
Confusion Matrix:
[[ 8005 5489]
[ 1058 10643]]
###Markdown
Cross validation - Logistic Regression
###Code
from sklearn.model_selection import cross_val_score
from sklearn import metrics
accuracy = cross_val_score(modelLR, X_eval, y_eval, cv=10, scoring='accuracy')
print("Accuracy: %0.5f (+/- %0.5f)" % (accuracy.mean(), accuracy.std() * 2))
f = cross_val_score(modelLR, X_eval, y_eval, cv=10, scoring='f1')
print("F1 Score: %0.5f (+/- %0.5f)" % (f.mean(), f.std() * 2))
precision = cross_val_score(modelLR, X_eval, y_eval, cv=10, scoring='precision')
print("Precision: %0.5f (+/- %0.5f)" % (precision.mean(), precision.std() * 2))
recall = cross_val_score(modelLR, X_eval, y_eval, cv=10, scoring='recall')
print("Recall: %0.5f (+/- %0.5f)" % (recall.mean(), recall.std() * 2))
###Output
Accuracy: 0.82309 (+/- 0.01630)
F1 Score: 0.83971 (+/- 0.01394)
Precision: 0.86752 (+/- 0.03254)
Recall: 0.81406 (+/- 0.03026)
###Markdown
Model Evaluation - Naive Bayes
###Code
modelNB = GaussianNB(var_smoothing=1e-09)
modelNB.fit(X_train, y_train)
# Predict on the new unseen test data
y_evalpredNB = modelNB.predict(X_eval)
y_predNB = modelNB.predict(X_test)
train_scoreNB = modelNB.score(X_train, y_train)
test_scoreNB = modelNB.score(X_test, y_test)
print("Training accuracy is ", train_scoreNB)
print("Testing accuracy is ", test_scoreNB)
from sklearn.metrics import confusion_matrix, precision_score, recall_score, f1_score
print('Performance measures for test:')
print('--------')
print('Accuracy:', test_scoreNB)
print('F1 Score:',f1_score(y_test, y_predNB))
print('Precision Score:',precision_score(y_test, y_predNB))
print('Recall Score:', recall_score(y_test, y_predNB))
print('Confusion Matrix:\n', confusion_matrix(y_test, y_predNB))
###Output
Performance measures for test:
--------
Accuracy: 0.46441754316332606
F1 Score: 0.6342692974848223
Precision Score: 0.46441754316332606
Recall Score: 1.0
Confusion Matrix:
[[ 0 13494]
[ 0 11701]]
###Markdown
Cross validation - Naive Bayes
###Code
from sklearn.model_selection import cross_val_score
from sklearn import metrics
accuracy = cross_val_score(modelNB, X_eval, y_eval, cv=10, scoring='accuracy')
print("Accuracy: %0.5f (+/- %0.5f)" % (accuracy.mean(), accuracy.std() * 2))
f = cross_val_score(modelNB, X_eval, y_eval, cv=10, scoring='f1')
print("F1 Score: %0.5f (+/- %0.5f)" % (f.mean(), f.std() * 2))
precision = cross_val_score(modelNB, X_eval, y_eval, cv=10, scoring='precision')
print("Precision: %0.5f (+/- %0.5f)" % (precision.mean(), precision.std() * 2))
recall = cross_val_score(modelNB, X_eval, y_eval, cv=10, scoring='recall')
print("Recall: %0.5f (+/- %0.5f)" % (recall.mean(), recall.std() * 2))
###Output
Accuracy: 0.79373 (+/- 0.04138)
F1 Score: 0.80066 (+/- 0.05102)
Precision: 0.88661 (+/- 0.02556)
Recall: 0.73208 (+/- 0.10511)
###Markdown
Model Evaluation - Random Forest
###Code
modelRF = RandomForestClassifier(random_state=0,max_depth=70,n_estimators=100)
modelRF.fit(X_train, y_train)
# Predict on the new unseen test data
y_evalpredRF = modelRF.predict(X_eval)
y_predRF = modelRF.predict(X_test)
train_scoreRF = modelRF.score(X_train, y_train)
test_scoreRF = modelRF.score(X_test, y_test)
print("Training accuracy is ", train_scoreRF)
print("Testing accuracy is ", test_scoreRF)
from sklearn.metrics import confusion_matrix, precision_score, recall_score, f1_score
print('Performance measures for test:')
print('--------')
print('Accuracy:', test_scoreRF)
print('F1 Score:', f1_score(y_test, y_predRF, average='weighted', zero_division=0))
print('Precision Score:', precision_score(y_test, y_predRF, average='weighted', zero_division=0))
print('Recall Score:', recall_score(y_test, y_predRF, average='weighted', zero_division=0))
print('Confusion Matrix:\n', confusion_matrix(y_test, y_predRF))
###Output
Performance measures for test:
--------
Accuracy: 0.5355030760071443
F1 Score: 0.37356623711533093
Precision Score: 0.28682882180131014
Recall Score: 0.5355030760071443
Confusion Matrix:
[[13492 2]
[11701 0]]
###Markdown
Cross validation - Random Forest
###Code
from sklearn.model_selection import cross_val_score
from sklearn import metrics
accuracy = cross_val_score(modelRF, X_eval, y_eval, cv=10, scoring='accuracy')
print("Accuracy: %0.5f (+/- %0.5f)" % (accuracy.mean(), accuracy.std() * 2))
f = cross_val_score(modelRF, X_eval, y_eval, cv=10, scoring='f1')
print("F1 Score: %0.5f (+/- %0.5f)" % (f.mean(), f.std() * 2))
precision = cross_val_score(modelRF, X_eval, y_eval, cv=10, scoring='precision')
print("Precision: %0.5f (+/- %0.5f)" % (precision.mean(), precision.std() * 2))
recall = cross_val_score(modelRF, X_eval, y_eval, cv=10, scoring='recall')
print("Recall: %0.5f (+/- %0.5f)" % (recall.mean(), recall.std() * 2))
###Output
Accuracy: 0.98700 (+/- 0.00387)
F1 Score: 0.98858 (+/- 0.00340)
Precision: 0.98885 (+/- 0.00410)
Recall: 0.98831 (+/- 0.00418)
###Markdown
Model Evaluation - KNN
###Code
modelKNN = KNeighborsClassifier(algorithm='brute',leaf_size=1,n_neighbors=2,weights='distance')
modelKNN.fit(X_train, y_train)
# Predict on the new unseen test data
y_evalpredKNN = modelKNN.predict(X_eval)
y_predKNN = modelKNN.predict(X_test)
train_scoreKNN = modelKNN.score(X_train, y_train)
test_scoreKNN = modelKNN.score(X_test, y_test)
print("Training accuracy is ", train_scoreKNN)
print("Testing accuracy is ", test_scoreKNN)
from sklearn.metrics import confusion_matrix, precision_score, recall_score, f1_score
print('Performance measures for test:')
print('--------')
print('Accuracy:', test_scoreKNN)
print('F1 Score:', f1_score(y_test, y_predKNN))
print('Precision Score:', precision_score(y_test, y_predKNN))
print('Recall Score:', recall_score(y_test, y_predKNN))
print('Confusion Matrix:\n', confusion_matrix(y_test, y_predKNN))
###Output
Performance measures for test:
--------
Accuracy: 0.22667195872196866
F1 Score: 0.316446814482178
Precision Score: 0.2684044515860263
Recall Score: 0.3854371421246047
Confusion Matrix:
[[ 1201 12293]
[ 7191 4510]]
###Markdown
Cross validation - KNN
###Code
from sklearn.model_selection import cross_val_score
from sklearn import metrics
accuracy = cross_val_score(modelKNN, X_eval, y_eval, cv=10, scoring='accuracy')
print("Accuracy: %0.5f (+/- %0.5f)" % (accuracy.mean(), accuracy.std() * 2))
f = cross_val_score(modelKNN, X_eval, y_eval, cv=10, scoring='f1')
print("F1 Score: %0.5f (+/- %0.5f)" % (f.mean(), f.std() * 2))
precision = cross_val_score(modelKNN, X_eval, y_eval, cv=10, scoring='precision')
print("Precision: %0.5f (+/- %0.5f)" % (precision.mean(), precision.std() * 2))
recall = cross_val_score(modelKNN, X_eval, y_eval, cv=10, scoring='recall')
print("Recall: %0.5f (+/- %0.5f)" % (recall.mean(), recall.std() * 2))
###Output
Accuracy: 0.97738 (+/- 0.00678)
F1 Score: 0.98013 (+/- 0.00590)
Precision: 0.98045 (+/- 0.00987)
Recall: 0.97982 (+/- 0.00514)
###Markdown
Model Evaluation - CatBoost
###Code
modelCB = CatBoostClassifier(random_state=0,depth=7,iterations=50,learning_rate=0.04)
modelCB.fit(X_train, y_train)
# Predict on the new unseen test data
y_evalpredCB = modelCB.predict(X_eval)
y_predCB = modelCB.predict(X_test)
train_scoreCB = modelCB.score(X_train, y_train)
test_scoreCB = modelCB.score(X_test, y_test)
print("Training accuracy is ", train_scoreCB)
print("Testing accuracy is ", test_scoreCB)
from sklearn.metrics import confusion_matrix, precision_score, recall_score, f1_score
print('Performance measures for test:')
print('--------')
print('Accuracy:', test_scoreCB)
print('F1 Score:',f1_score(y_test, y_predCB, average='weighted', zero_division=0))
print('Precision Score:',precision_score(y_test, y_predCB, average='weighted', zero_division=0))
print('Recall Score:', recall_score(y_test, y_predCB, average='weighted', zero_division=0))
print('Confusion Matrix:\n', confusion_matrix(y_test, y_predCB))
###Output
Performance measures for test:
--------
Accuracy: 0.5427267314943441
F1 Score: 0.3898251274282043
Precision Score: 0.7388272713188289
Recall Score: 0.5427267314943441
Confusion Matrix:
[[13488 6]
[11515 186]]
###Markdown
Cross validation - CatBoost
###Code
from sklearn.model_selection import cross_val_score
from sklearn import metrics
accuracy = cross_val_score(modelCB, X_eval, y_eval, cv=10, scoring='accuracy')
f = cross_val_score(modelCB, X_eval, y_eval, cv=10, scoring='f1')
precision = cross_val_score(modelCB, X_eval, y_eval, cv=10, scoring='precision')
recall = cross_val_score(modelCB, X_eval, y_eval, cv=10, scoring='recall')
print("Accuracy: %0.5f (+/- %0.5f)" % (accuracy.mean(), accuracy.std() * 2))
print("F1 Score: %0.5f (+/- %0.5f)" % (f.mean(), f.std() * 2))
print("Precision: %0.5f (+/- %0.5f)" % (precision.mean(), precision.std() * 2))
print("Recall: %0.5f (+/- %0.5f)" % (recall.mean(), recall.std() * 2))
###Output
Accuracy: 0.98017 (+/- 0.00461)
F1 Score: 0.98272 (+/- 0.00399)
Precision: 0.97508 (+/- 0.00717)
Recall: 0.99049 (+/- 0.00557)
|
Machine-Learning/Lecture1-ipynb/Lecture1a.ipynb
|
###Markdown
CS 5489 Machine Learning Lecture 1a: Python Tutorial Dr. Antoni B. Chan Dept. of Computer Science, City University of Hong Kong Why Python?- General-purpose high-level programming language- Design philosophy emphasizes programmer productivity and code readability - "executable pseudo-code"- Supports multiple programming paradigms - object-oriented, imperative, functional- Dynamic typing and automatic memory management What is special about Python?- Object-oriented: everything is an object- Clean: usually one way to do something, not a dozen- Easy-to-learn: learn in 1-2 days- Easy-to-read- Powerful: full-fledged programming language Applications for Python- Scientific Computing - numpy, scipy, ipython- Data Science, Deep Learning - scikit-learn, matplotlib, pandas, keras, tensorflow- Web & Internet Development - Django – complete web application framework - model-view-controller design pattern - templates, web server, object-relational mapper Disadvantages of Python- Not as fast as Java or C- However, you can call C-compiled libraries from Python (e.g. Boost C++)- Alternatively, Python code can be compiled to improve speed - Cython and PyPy - requires type of variables to be declared Installing Python- We will use Python 3 - Python 3 is not backwards compatible with Python 2.7- Anaconda (https://www.anaconda.com/download) - single bundle includes most scientific computing packages. - package manager for installing other libraries - make sure to pick version for **Python 3**. - easy install packages for Windows, Mac, Linux. - (single directory install) Running Python===============- Interactive shell (ipython) - good for learning the language, experimenting with code, testing modules***```Nori:CS5489 abc$ ipythonPython 3.5.4 |Anaconda, Inc.| (default, Oct 5 2017, 02:58:14) Type "copyright", "credits" or "license" for more information.IPython 4.2.0 -- An enhanced Interactive Python.? -> Introduction and overview of IPython's features.%quickref -> Quick reference.help -> Python's own help system.object? -> Details about 'object', use 'object??' for extra details.In [1]: print("Hello, World")Hello, WorldIn [2]: Do you really want to exit ([y]/n)? yNori:CS5489 abc$ ``` - Script file (hello.py)```python!/usr/bin/pythonprint("Hello, World")```- Standalone script - explicitly using python interpreter```Nori:~ abc$ python hello.pyHello, World``` - using magic shebang (Linux, Mac OS X)```Nori:~ abc$ ./hello.py Hello, World``` Jupyter (ipython notebooks)- Launch from _Anaconda Navigator_- browser-based interactive computing environment - development, documenting, executing code, viewing results (inline images) - whole session stored in notebook document (.ipynb) - (also made and presented these slides!)  Jupyter tips- Keyboard shortcuts - there are a lot of keyboard shortcuts for moving between cells, running cells, deleting and inserting cells.- Starting directory - use the `--notebook-dir=mydir` option to start the notebook in a particular directory. - Windows: create a shortcut to run `jupyter-notebook.exe --notebook-dir=%userprofile%`. - Problems viewing SVG images in ipynb - SVG images may not display due to the serurity model of Jupyter. - select "Trust Notebook" from the "File" menu to show the SVG images.- View ipynb in slideshow mode in a web browser (like this presentation!)```jupyter-nbconvert --to slides file.ipynb --post serve```- Convert to HTML to view statically in web browser```jupyter-nbconvert file.ipynb``` - ValueError when using matplotlib in Jupyter - This mainly affects Mac where the OS locale is set to a non-English language. Open "Terminal" app and go to Preferences -> Profiles -> Terminal -> Enviornment. Deselect the option "Set locale variables automatically". - more info: http://stackoverflow.com/questions/15526996/ipython-notebook-locale-error- MacOS and Anaconda - MacOS has a builtin python distribution. If you are using anaconda, make sure that you use the correct command-line commands. You can add "/anaconda3/bin/" in front of the command to make sure you are using the anaconda version (or the appropriate base direcotry for anaconda3). Otherwise, it may default to the builtin python. CS Lab Resources- JupyterHub - Jupyter notebooks run on a central server - shared CPU and GPU - Jupyter notebooks: https://jh5489.cs.cityu.edu.hk - JupyterLab (IDE): https://jh5489.cs.cityu.edu.hk/user-redirect/lab- Linux machines - there are several computing clusters in CS. - [High Throughput GPU Cluster 1 (HTGC1)](https://cslab.cs.cityu.edu.hk/services/high-throughput-gpu-cluster-htgc) - [High Throughput GPU Cluster 2 (HTGC2)](https://cslab.cs.cityu.edu.hk/services/high-throughput-gpu-cluster-htgc2) - [High Throughput GPU Cluster 3 (HTGC3)](https://cslab.cs.cityu.edu.hk/services/high-throughput-gpu-cluster-htgc3)- Windows machines - MMW2462 in CS lab contains GPU workstations.- Google colab: https://colab.research.google.com/ - provided by Google. Some limitations on running time (12 hours) and memory usage.- More details are on Canvas. Outline1. Python Intro2. **Python Basics (identifiers, types, operators)**3. Control structures (conditional and loops)4. Functions, Classes5. File IO, Pickle, pandas6. NumPy7. matplotlib8. probability review Python Basics- Formatting - case-sensitive - statements end in **newline** (not semicolon) - use semicolon for multiple statements in one line. - **indentation** for code blocks (after a colon).
###Code
print("Hello")
print("Hello"); print("World")
name = "Bob"
if name == "George":
print("Hi George")
else:
print("Who are you?")
###Output
Hello
Hello
World
Who are you?
###Markdown
- single-line comments with ``- multi-line statements continued with backslash (`\`) - not required inside `{}`, `()`, or `[]` for data types
###Code
# this is a comment
a=1 # comments also can go after statements
b=2; c=3 # here too
# multiple line statement
x = a + \
b + c
# backslash not needed when listing multi-line data
y = [1, 2,
3, 4]
###Output
_____no_output_____
###Markdown
Identifiers and Variables- Identifiers - same as in C- Naming convention: - `ClassName` -- a class name - `varName` -- other identifier - `_privateVar` -- private identifier - `__veryPrivate` -- strongly private identifier - `__special__` -- language-defined special name - Variables - no declaration needed - no need for declaring data type (automatic type) - need to assign to initialize - use of uninitialized variable raises exception - automatic garbage collection (reference counts) Basic Types- Integer number
###Code
4
int(4)
###Output
_____no_output_____
###Markdown
- Real number (float)
###Code
4.0
float(4)
###Output
_____no_output_____
###Markdown
- Boolean
###Code
True
False
###Output
_____no_output_____
###Markdown
- String literal
###Code
"a string"
'a string'
"concatenate " "two string literals"
"""this is a multi-line string.
it keeps the newline."""
r'raw string\no escape chars'
###Output
_____no_output_____
###Markdown
Lists- Lists can hold anything (even other lists)
###Code
myList = ['abcd', 786, 2.23]
print(myList) # print the list
print(myList[0]) # print the first element (0-indexed)
###Output
abcd
###Markdown
- Creating lists of numbers
###Code
a = range(5) # list of numbers from 0 to 4
print(a)
print(list(a))
b = range(2,12,3) # numbers from 2 to 11, count by 3
print(b)
print(list(b))
###Output
range(2, 12, 3)
[2, 5, 8, 11]
###Markdown
- append and pop
###Code
a = list(range(0,5))
a.append('blah') # add item to end
print(a)
a.pop() # remove last item and return it
###Output
_____no_output_____
###Markdown
- insert and delete
###Code
a.insert(0,42) # insert 42 at index 0
print(a)
del a[2] # delete item 2
print(a)
###Output
[42, 0, 2, 3, 4]
###Markdown
- more list operations
###Code
a.reverse() # reverse the entries
print(a)
a.sort() # sort the entries
print(a)
###Output
[0, 2, 3, 4, 42]
###Markdown
Tuples- Similar to a list - but immutable (read-only) - cannot change the contents (like a string constant)
###Code
# make some tuples
x = (1,2,'three')
print(x)
y = 4,5,6 # parentheses not needed!
print(y)
z = (1,) # tuple with 1 element (the trailing comma is required)
print(z)
###Output
(1,)
###Markdown
Operators on sequences- _Same operators_ for strings, lists, and tuples- Slice a sublist with colon (`:`) - **Note**: the 2nd argument is not inclusive!
###Code
"hello"[0] # the first element
"hello"[-1] # the last element (index from end)
"hello"[1:4] # the 2nd through 4th elements
"hello"[2:] # the 3rd through last elements
"hello"[0:5:2] # indices 0,2,4 (by 2)
###Output
_____no_output_____
###Markdown
- Other operators on string, list, tuple
###Code
len("hello") # length
"he" + "llo" # concatenation
"hello"*3 # repetition
###Output
_____no_output_____
###Markdown
String methods- Useful methods
###Code
"112211".count("11") # 2
"this.com".endswith(".com") # True
"wxyz".startswith("wx") # True
"abc".find("c") # finds first: 2
",".join(['a', 'b', 'c']) # join list: 'a,b,c'
"aba".replace("a", "d") # replace all: "dbd"
"a,b,c".split(',') # make list: ['a', 'b', 'c']
" abc ".strip() # "abc", also rstrip(), lstrip()
###Output
_____no_output_____
###Markdown
- String formatting: automatically fill in type
###Code
"{} and {} and {}".format('string', 123, 1.6789)
###Output
_____no_output_____
###Markdown
- String formatting: specify type (similar to C)
###Code
"{:d} and {:f} and {:0.2f}".format(False, 3, 1.234)
###Output
_____no_output_____
###Markdown
Dictionaries- Stores key-value pairs (associative array or hash table) - key can be a string, number, or tuple
###Code
mydict = {'name': 'john', 42: 'sales', ('hello', 'world'): 6734}
print(mydict)
###Output
{'name': 'john', 42: 'sales', ('hello', 'world'): 6734}
###Markdown
- Access
###Code
print(mydict['name']) # get value for key 'name'
mydict['name'] = 'jon' # change value for key 'name'
mydict[2] = 5 # insert a new key-value pair
print(mydict)
del mydict[2] # delete entry for key 2
print(mydict)
###Output
{'name': 'jon', 42: 'sales', ('hello', 'world'): 6734}
###Markdown
- Other operations:
###Code
mydict.keys() # iterator of all keys (no random access)
list(mydict.keys()) # convert to a list for random access
mydict.values() # iterator of all values
mydict.items() # iterator of tuples (key, value)
'name' in mydict # check the presence of a key
###Output
_____no_output_____
###Markdown
Operators- Arithmetic: `+`, `-`, `*`, `/`, `%`, `**` (exponent), `//` (floor division)
###Code
print(6/4) # float division
print(6//4) # integer division
print(6//4.0) # floor division
###Output
1.0
###Markdown
- Assignment: `=`, `+=`, `-=`, `/=`, `%=`, `**=`, `//=`- Equality: `==`, `!=`- Compare: `>`, `>=`, `<`, `<=`- Logical: `and`, `or`, `not` - Membership: `in`, `not in`
###Code
2 in [2, 3, 4]
###Output
_____no_output_____
###Markdown
- Identity: `is`, `is not` - checks reference to the same object
###Code
x = [1,2,3]
y = x
x is y # same variable?
z = x[:] # create a copy
z is x # same variable?
###Output
_____no_output_____
###Markdown
- Tuple packing and unpacking
###Code
point = (1,2,3)
(x,y,z) = point
print(x)
print(y)
print(z)
###Output
1
2
3
###Markdown
Sets- a set is a collection of unique items
###Code
a=[1, 2, 2, 2, 4, 5, 5]
sA = set(a)
sA
###Output
_____no_output_____
###Markdown
- set operations
###Code
sB = {4, 5, 6, 7}
print(sA - sB) # set difference
print (sA | sB) # set union
print (sA & sB) # set intersect
###Output
{4, 5}
###Markdown
Outline1. Python Intro2. Python Basics (identifiers, types, operators)3. **Control structures (conditional and loops)**4. Functions, Classes5. File IO, Pickle, pandas6. NumPy7. matplotlib8. probability review Conditional Statements- indentation used for code blocks after colon (:)- if-elif-else statement
###Code
if x==2:
print("foo")
elif x==3:
print("bar")
else:
print("baz")
###Output
baz
###Markdown
- nested if
###Code
if x>1:
if x==2:
print("foo")
else:
print("bar")
else:
print("baz")
###Output
baz
###Markdown
- single-line
###Code
if x==1: print("blah")
###Output
blah
###Markdown
- check existence using "if in"
###Code
mydict = {'name': 'john', 42: 'sales'}
if 'name' in mydict:
print("mydict has name field")
if 'str' in 'this is a long string':
print('str is inside')
###Output
str is inside
###Markdown
Loops- "for-in" loop over values in a list
###Code
ns = range(1,6,2) # list of numbers from 1 to 6, by 2
for n in ns:
print(n)
###Output
1
3
5
###Markdown
- loop over index-value pairs
###Code
x = ['a', 'b', 'c']
for i,n in enumerate(x):
print(i, n)
###Output
0 a
1 b
2 c
###Markdown
- looping over two lists at the same time
###Code
x = ['a', 'b', 'c']
y = ['A', 'B', 'C']
for i,j in zip(x,y):
print(i,j)
###Output
a A
b B
c C
###Markdown
- `zip` creates pairs of items between the two lists - (actually creates an iterator over them)
###Code
list(zip(x,y)) # convert to a list (for random access)
###Output
_____no_output_____
###Markdown
- looping over dictionary
###Code
x = {'a':1, 'b':2, 'c':3}
for (key,val) in x.items():
print(key, val)
###Output
a 1
b 2
c 3
###Markdown
- while loop
###Code
x=0
while x<5:
x += 1
print(x)
# single line
while x<10: x += 1
print(x)
###Output
10
###Markdown
- loop control (same as C) - `break`, `continue` - else clause - runs after list is exhausted - does _not_ run if loop break
###Code
for i in [0, 1, 6]:
print(i)
else:
print("end of list reached!")
###Output
0
1
6
end of list reached!
###Markdown
List Comprehension- build a new list with a "for" loop
###Code
myList = [1, 2, 2, 2, 4, 5, 5]
myList4 = [4*item for item in myList] # multiply each item by 4
myList4
# equivalent code
myList4=[]
for item in myList:
myList4.append(4*item)
myList4
# can also use conditional to select items
[4*item*4 for item in myList if item>2]
###Output
_____no_output_____
###Markdown
Outline1. Python Intro2. Python Basics (identifiers, types, operators)3. Control structures (conditional and loops)4. **Functions, Classes**5. File IO, Pickle, pandas6. NumPy7. matplotlib8. probability review Functions- Defining a function - _required_ and _optional_ inputs (similar to C++) - "docstring" for optional documentation
###Code
def sum3(a, b=1, c=2):
"sum a few values"
mysum = a+b+c
return mysum
###Output
_____no_output_____
###Markdown
- Calling a function
###Code
sum3(2,3,4) # call function: 2+3+4
sum3(0) # use default inputs: 0+1+2
sum3(b=1, a=5, c=2) # use keyword arguments: 5+1+2
help(sum3) # show documentation
# ipython magic -- shows a help window about the function
? sum3
###Output
_____no_output_____
###Markdown
Classes- Defining a class - `self` is a reference to the object instance (passed _implicitly_)
###Code
class MyList:
"class documentation string"
num = 0 # a class variable
def __init__(self, b): # constructor
self.x = [b] # an instance variable
MyList.num += 1 # modify class variable
def appendx(self, b): # a class method
self.x.append(b) # modify an instance variable
self.app = 1 # create new instance variable
###Output
_____no_output_____
###Markdown
- Using the class
###Code
c = MyList(0) # create an instance of MyList
print(c.x)
c.appendx(1) # c.x = [0, 1]
print(c.x)
c.appendx(2) # c.x = [0, 1, 2]
print(c.x)
print(MyList.num) # access class variable (same as c.num)
###Output
1
###Markdown
More on Classes- There are _no_ "private" members - everything is accessible - convention to indicate _private_: - `_variable` means private method or variable (but still accessible) - convention for _very private_: - `__variable` is not directly visible - actually it is renamed to `_classname__variable` - Instance variable rules - On _use_ via instance (`self.x`), scope search order is: - (1) instance, (2) class, (3) base classes - also the same for method lookup - On _assignment_ via instance (`self.x=...`): - always makes an instance variable - Class variables "default" for instance variables - _class_ variable: one copy _shared_ by all - _instance_ variable: each instance has its own Inheritence- Child class inherits attributes from parents
###Code
class MyListAll(MyList):
def __init__(self, a): # overrides MyList
self.allx = [a]
MyList.__init__(self, a) # call base class constructor
def popx(self):
return self.x.pop()
def appendx(self, a): # overrides MyList
self.allx.append(a)
MyList.appendx(self, a) # "super" method call
###Output
_____no_output_____
###Markdown
- Multiple inheritence - `class ChildClass(Parent1, Parent2, ...)` - calling method in parent - `super(ChildClass, self).method(args)` Class methods & Built-in Attributes- Useful methods to override in class
###Code
class MyList2:
...
def __str__(self): # string representation
...
def __cmp__(self, x): # object comparison
...
def __del__(self): # destructor
...
###Output
_____no_output_____
###Markdown
- Built-in attributes
###Code
print(c.__dict__) # Dictionary with the namespace.
print(c.__doc__) # Class documentation string
print(c.__module__) # Module which defines the class
print(MyList.__name__) # Class name
print(MyList.__bases__) # tuple of base classes
###Output
MyList
(<class 'object'>,)
###Markdown
Outline1. Python Intro2. Python Basics (identifiers, types, operators)3. Control structures (conditional and loops)4. Functions, Classes5. **File IO, Pickle, pandas**6. NumPy7. matplotlib8. probability review File I/O- Write a file
###Code
with open("myfile.txt", "w") as f:
f.write("blah\n")
f.writelines(['line1\n', 'line2\n', 'line3\n'])
# NOTE: using "with" will automatically close the file
###Output
_____no_output_____
###Markdown
- Read a whole file
###Code
with open("myfile.txt", "r") as f:
contents = f.read() # read the whole file as a string
print(contents)
###Output
blah
line1
line2
line3
###Markdown
- Read line or remaining lines
###Code
f = open("myfile.txt", 'r')
print(f.readline()) # read a single line.
print(f.readlines()) # read remaining lines in a list.
f.close()
###Output
['line1\n', 'line2\n', 'line3\n']
###Markdown
- Read line by line with a loop
###Code
with open("myfile.txt", 'r') as f:
for line in f:
print(line) # still contains newline char
###Output
blah
line1
line2
line3
###Markdown
Saving Objects with Pickle- Turns almost **any** Python **object** into a string representation for saving into a file.
###Code
import pickle # load the pickle library
mylist = MyList(0) # an object
# open file to save object (write bytes)
with open('alist.pickle', 'wb') as file:
pickle.dump(mylist, file) # save the object using pickle
###Output
_____no_output_____
###Markdown
- Load object from file
###Code
with open('alist.pickle', 'rb') as file: # (read bytes)
mylist2 = pickle.load(file) # load pickled object from file
print(mylist2)
print(mylist2.x)
###Output
<__main__.MyList object at 0x7fbcc81fa8d0>
[0]
###Markdown
- cPickle is a faster version (1,000 times faster!) Exception Handling- Catching an exception - `except` block catches exceptions - `else` block executes if no exception occurs - `finally` block always executes at end
###Code
try:
file = open('blah.pickle', 'r')
blah = pickle.load(file)
file.close()
except: # catch everything
print("No file!")
else: # executes if no exception occurred
print("No exception!")
finally:
print("Bye!") # always executes
###Output
No file!
Bye!
###Markdown
pandas- pandas is a Python library for data wrangling and analysis.- `Dataframe` is a table of entries (like an Excel spreadsheet). - each column does not need to be the same type - operations to modify and operate on the table
###Code
# setup pandas and display
import pandas as pd
# read CSV file
df = pd.read_csv('mycsv.csv')
# print the dataframe
df
###Output
_____no_output_____
###Markdown
- select a column
###Code
df['Name']
###Output
_____no_output_____
###Markdown
- query the table
###Code
# select Age greater than 30
df[df.Age > 30]
###Output
_____no_output_____
###Markdown
- compute statistics
###Code
df.mean()
###Output
_____no_output_____
|
mpl_tutorials_17_Specifying Colors.ipynb
|
###Markdown
[Specifying Colors](https://matplotlib.org/tutorials/colors/colors.htmlsphx-glr-tutorials-colors-colors-py) Matplotlib recognizes the following formats to specify a color: an RGB or RGBA (red, green, blue, alpha) tuple of float values in closed interval [0, 1] (e.g., (0.1, 0.2, 0.5) or (0.1, 0.2, 0.5, 0.3)); a hex RGB or RGBA string (e.g., '0f0f0f' or '0f0f0f80'; case-insensitive); a shorthand hex RGB or RGBA string, equivalent to the hex RGB or RGBA string obtained by duplicating each character, (e.g., 'abc', equivalent to 'aabbcc', or 'abcd', equivalent to 'aabbccdd'; case-insensitive); a string representation of a float value in [0, 1] inclusive for gray level (e.g., '0.5'); one of {'b', 'g', 'r', 'c', 'm', 'y', 'k', 'w'}, they are the single character short-hand notations for blue, green, red, cyan, magenta, yellow, black, and white. a X11/CSS4 color name (case-insensitive); a name from the xkcd color survey, prefixed with 'xkcd:' (e.g., 'xkcd:sky blue'; case insensitive); one of the Tableau Colors from the 'T10' categorical palette (the default color cycle): {'tab:blue', 'tab:orange', 'tab:green', 'tab:red', 'tab:purple', 'tab:brown', 'tab:pink', 'tab:gray', 'tab:olive', 'tab:cyan'} (case-insensitive); a "CN" color spec, i.e. 'C' followed by a number, which is an index into the default property cycle (matplotlib.rcParams['axes.prop_cycle']); the indexing is intended to occur at rendering time, and defaults to black if the cycle does not include color. - [CSS4 Color Names](https://learningwebdesign.com/colornames.html)- [X11 Color Names](https://www.cssportal.com/html-colors/x11-colors.php)- [X11 color names](https://wikimili.com/en/X11_color_names)- [xkcd](https://xkcd.com/color/rgb/)- [Tableau Colors T10](https://public.tableau.com/profile/chris.gerrard!/vizhome/TableauColors/ColorPaletteswithRGBValues) Matplotlib接受如下格式用于指定颜色:- RGB/RGBA (red, green, blue, alpha) 闭区间[0, 1]内的浮点数元组 (e.g., (0.1, 0.2, 0.5) or (0.1, 0.2, 0.5, 0.3));- 16进制RGB/RGBA字符串 (e.g., '0f0f0f' or '0f0f0f80'; case-insensitive);- 短格式16进制RGB/RGBA字符串 (e.g., 'abc', equivalent to 'aabbcc', or 'abcd', equivalent to 'aabbccdd'; case-insensitive);- 字符串表示的[0,1]区间的浮点数包含所有的灰度- {'b', 'g', 'r', 'c', 'm', 'y', 'k', 'w'}中的一个, 对应blue, green, red, cyan, magenta, yellow, black, and white.- X11/CSS4颜色名- 带有'xkcd:'前缀的xkcd颜色名 (e.g., 'xkcd:sky blue'; case insensitive);- Tableau调色板中名为Tablueau 10的集合,是缺省的颜色循环:{'tab:blue', 'tab:orange', 'tab:green', 'tab:red', 'tab:purple', 'tab:brown', 'tab:pink', 'tab:gray', 'tab:olive', 'tab:cyan'} (case-insensitive);- "CN"颜色规范,即"C"后跟数字,这是默认属性周期的索引(matplotlib.rcParams ['axes.prop_cycle']); 该索引应在渲染时进行,如果循环不包含颜色,则默认为黑色。 "Red", "Green", and "Blue" are the intensities of those colors, the combination of which span the colorspace.How "Alpha" behaves depends on the zorder of the Artist. Higher zorder Artists are drawn on top of lower Artists, and "Alpha" determines whether the lower artist is covered by the higher. If the old RGB of a pixel is RGBold and the RGB of the pixel of the Artist being added is RGBnew with Alpha alpha, then the RGB of the pixel is updated to: RGB = RGBOld * (1 - Alpha) + RGBnew * Alpha. Alpha of 1 means the old color is completely covered by the new Artist, Alpha of 0 means that pixel of the Artist is transparent. “红色”,“绿色”和“蓝色”是这些颜色的强度,它们的组合跨越整个颜色空间。"Alpha"的行为依赖于Artist的zorder属性。在Z轴次序中较高的Artists绘制与较低的Artists上方,"Alpha"决定底层的artist是否被上层的遮盖。如果一个像素的原有RGB是 RGBold 要添加的Artist的像素的RGB是带有Alpha的RGBnew,则该像素的RGB将更新为:RGB = RGBOld * (1 - Alpha) + RGBnew * Alpha. Alpha 1表示旧颜色完全被新Artist覆盖,Alpha 0表示Artist的像素是透明的。 "CN" color selection"CN" colors are converted to RGBA as soon as the artist is created. For example,
###Code
import numpy as np
import matplotlib.pyplot as plt
from matplotlib import style
%matplotlib widget
th = np.linspace(0, 2*np.pi, 128)
fig, ax = plt.subplots(figsize=(3, 3))
ax.plot(th, np.cos(th), (0.1, 0.2, 0.5, 0.3))
ax.cla()
###Output
_____no_output_____
|
python_exercises/logistic_regression.ipynb
|
###Markdown
AssignmentDo [logistic regression](https://en.wikipedia.org/wiki/Logistic_regression) on the famous [Iris dataset](https://en.wikipedia.org/wiki/Iris_flower_data_set) and plot the decision boundary. Then exted to multilass using [softmax](https://en.wikipedia.org/wiki/Softmax_function). SetupThe topics of this exercise are very famous and very well known, so a lot of educational material is available online. E.g., for logistic regression:* [Logistic regression @towardsdatascience](https://towardsdatascience.com/understanding-logistic-regression-9b02c2aec102)* [Logistic regression @Biochemia Medica](https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3936971/)* [Logistic regression @datacamp](https://www.datacamp.com/community/tutorials/understanding-logistic-regression-python)Let's just frame the terms of the first part of the problem:* we are talking about **binary** classification: all samples belong to two classes (represented by values 0 and 1). We want to build a statistical machine able to predict, for a new sample, the class* for each sample we have a set numbers, called **features**. The easiest-to-understand case is when these features are actual phyisical measurements, but any set of data will do* in **standard regression** we weight each feature with a number (positive or negative, big or small). We then multiply each feature times its weight and add everything...* ...but we cannot do that now, since the resulting number could be very big (positive big or negative big). But our desired result is a class! In other words our machine needs to produce either a zero or a one. Or maybe **all the numbers between zero and one**, so that we have an idea about how certain our prediction is* in fact, a 0.99 will show more confidence than a 0.8, which will be more confident than a 0.65, even if all three outputs at the end can be considered as "class 1" (since those numbers are closer to 1 than to 0) * to do so we feed what is produced by a regular regression into a **sigmoid function**, which looks like this:This function takes in input any number from -infinity to +infinity and returns a value between zero and one. This will be our prediction. Let's start building the solution. Setting the random seedFirst of all, we setup the random seed to ensure reproducibility of results. Since tensorflow uses an internal random generator we need to fix both the general seed (via numpy `seed()`) and tensorflow seed (via `set_seet()`)
###Code
#general random seed
from numpy.random import seed
seed(777)
#tensorflow-specific seed
import tensorflow
tensorflow.random.set_seed(777)
###Output
_____no_output_____
###Markdown
The Iris datasetThe dataset we are going to use is very famous. It was published by Robert Fisher in 1936 together with the paper [The use of multiple measurements in taxonomic problems](https://onlinelibrary.wiley.com/doi/abs/10.1111/j.1469-1809.1936.tb02137.x). Data are public and nowadays this dataset is shipped with many statistical software and packages. We are going to use the copy coming with [sci-kit learn](https://scikit-learn.org/stable/modules/generated/sklearn.datasets.load_iris.htmlsklearn.datasets.load_iris). First of all, let's verify the sci-kit version available:
###Code
import sklearn.datasets
print (sklearn.__version__)
###Output
0.22.2.post1
###Markdown
If we had version 0.23+ we could simply do:> `iris = sklearn.datasets.load_iris(return_X_y = False, as_frame = True)`But the above instruction would generate an error with the current installation. The main difference is in the format used to store data: numpy array (version 0.22 and below) or pandas dataframe/series (version 0.23 and above). Since we want to use the pandas data structure the returned data require a little conversion:
###Code
import pandas as pd
iris = sklearn.datasets.load_iris()
iris.data = pd.DataFrame(iris.data, columns=iris.feature_names) #converting numpy array -> pandas DataFrame
iris.target = pd.Series(iris.target) #converting numpy array -> pandas Series
###Output
_____no_output_____
###Markdown
The variable `iris` is now a [bunch object](https://scikit-learn.org/stable/modules/generated/sklearn.utils.Bunch.htmlsklearn.utils.Bunch) and contains all the required data.
###Code
#uncomment the following instruction to print a detailed description
#of the Iris dataset, here omitted for compactness
#print(iris.DESCR)
###Output
_____no_output_____
###Markdown
The dataset describes 150 flower samples, belonging to three species of Iris (Iris setosa, Iris virginica and Iris versicolor). Four features were measured from each sample: the length and the width of the sepals and of the petals, in centimeters. Features are found in the attribute `.data` of the returned bunch object:
###Code
print('Shape of the feature table: ' + str(iris.data.shape))
###Output
Shape of the feature table: (150, 4)
###Markdown
We can take a look at the actual numbers:
###Code
print(iris.data)
###Output
sepal length (cm) sepal width (cm) petal length (cm) petal width (cm)
0 5.1 3.5 1.4 0.2
1 4.9 3.0 1.4 0.2
2 4.7 3.2 1.3 0.2
3 4.6 3.1 1.5 0.2
4 5.0 3.6 1.4 0.2
.. ... ... ... ...
145 6.7 3.0 5.2 2.3
146 6.3 2.5 5.0 1.9
147 6.5 3.0 5.2 2.0
148 6.2 3.4 5.4 2.3
149 5.9 3.0 5.1 1.8
[150 rows x 4 columns]
###Markdown
As said above, each of the 150 lines represents a different flower, each belonging to one of the three Iris species. The species will be our target, the class that we are trying to predict. Let's take a look into the dataset:
###Code
print('Shape of the target variable: ' + str(iris.target.shape))
print('Names for each class: ' + str(iris.target_names))
#using Counter object to print a tally of the classes
from collections import Counter
print('Numerosity for each class: ' + str(Counter(iris.target)))
###Output
Shape of the target variable: (150,)
Names for each class: ['setosa' 'versicolor' 'virginica']
Numerosity for each class: Counter({0: 50, 1: 50, 2: 50})
###Markdown
Classes are represented via a numeric index: 0 for *setosa*, 1 for *versicolor*, 2 for *virginica*. The samples are presented in order, with the first 50 samples being *setosa*, then 50 *versicolor* and the last 50 being *virginica*.Always when working with a new datasets it is importat to plot the data if possible. We are unfortunately talking about a 5-dimensional dataset (the four features + the target class) which is not easily representable. One solution in these cases is to slice a subset of the whole dataset.In the following code we'll plot two features at a time, plus the class.
###Code
#change these two values to plot different features, remembering the numbering:
# 0 : sepal length (cm)
# 1 : sepal width (cm)
# 2 : petal length (cm)
# 3 : petal width (cm)
feature_x = 0
feature_y = 1
#old reliable pyplot!
import matplotlib.pyplot as plt
#starting a new plot
fig, ax = plt.subplots()
#adding data in three bunches of 50, once per class
ax.scatter(x=iris.data.iloc[0:50,feature_x], y=iris.data.iloc[0:50,feature_y], c='red', label=iris.target_names[0])
ax.scatter(x=iris.data.iloc[50:100,feature_x], y=iris.data.iloc[50:100,feature_y], c='green', label=iris.target_names[1])
ax.scatter(x=iris.data.iloc[100:150,feature_x], y=iris.data.iloc[100:150,feature_y], c='blue', label=iris.target_names[2])
#the axis names are taken from feature names
ax.set_xlabel(iris.feature_names[feature_x])
ax.set_ylabel(iris.feature_names[feature_y])
#adding the legend and printing the plot
ax.legend()
plt.show()
###Output
_____no_output_____
###Markdown
The plot shows clearly that setosa is quite separate from the other two classes. Even chosing other features for the plot the general result is similar.To be totally frank, this dataset is quite simple. In fact even if it's not possible to easily plot everything, using the four features most classifier can reach very close to 100% accuracy when trying to separate Setosa from the other species.To make things a little more interesting we decide to renounce to half of our features, using only the first two columns. Moreover, we join together Setosa and Versicolor. In other words, we want a classifier able to discriminate virginica (which becomes the new class "1") from the other irises (which all together become the new class "0"):
###Code
#simplifly the problem: less classes, less features
features = iris.data.iloc[:, 0:2]
target = iris.target
#updating class labels. To makes things difficult we put together old classes 0 and 1
#in a new class (non virginica) and keep old class 2 (virginica) as new class 1.
#For an easier problems put together versicolor and virginica and keep setosa by itself
target[0:100] = 0
target[100:150] = 1
###Output
_____no_output_____
###Markdown
Let's take a look at the new dataset:
###Code
#starting a new plot
fig, ax = plt.subplots()
#adding data in two bunches
ax.scatter(x=features.iloc[0:100,0], y=features.iloc[0:100,1], c='red', label='Not virginica')
ax.scatter(x=features.iloc[100:150,0], y=features.iloc[100:150,1], c='blue', label='virginica')
#the axis names are taken from feature names
ax.set_xlabel(iris.feature_names[feature_x])
ax.set_ylabel(iris.feature_names[feature_y])
#adding the legend and printing the plot
ax.legend()
plt.show()
###Output
_____no_output_____
###Markdown
Things are getting interesting! This is now a difficult problem and there is no clear cut solution. Let's proceed. Training and validation setsEach time there is some kind of "learning" involved we need to split our data. A subset will be used for training, and a subset will be used for validation. (there may be room for another subset, the "test set", but we are not talking about it now).In our current dataset the samples are sorted by class: the first 100 are from "Not virginica" class, and the remaining 50 are from virginica. We want to keep this 2:1 proportion (roughly) the same in both train and validation set.
###Code
#selecting the first 100 samples for training would be a bad choice...
print(Counter(iris.target[0:100]))
###Output
Counter({0: 100})
###Markdown
To do so we are going to use what is called a [stratified approach](https://machinelearningmastery.com/cross-validation-for-imbalanced-classification/) using a [StratifiedShuffleSplit](https://scikit-learn.org/stable/modules/generated/sklearn.model_selection.StratifiedShuffleSplit.html) object from Sci-kit learn:
###Code
#we want to have the same proportion of classes in both train and validation sets
from sklearn.model_selection import StratifiedShuffleSplit
#building a StratifiedShuffleSplit object (sss among friends) with 20% data
#assigned to validation set (here called "test")
sss = StratifiedShuffleSplit(n_splits=1, test_size=0.2, random_state=0)
#the .split() method returns (an iterable over) two lists which can be
#used to index the samples that go into train and validation sets
for train_index, val_index in sss.split(features, target):
features_train = features.iloc[train_index, :]
features_val = features.iloc[val_index, :]
target_train = target[train_index]
target_val = target[val_index]
#let's print some shapes to get an idea of the resulting data structure
print(features_train.shape)
print(features_val.shape)
print(target_train.shape)
print(target_val.shape)
print(type(features_train))
print(type(target_train))
###Output
(120, 2)
(30, 2)
(120,)
(30,)
<class 'pandas.core.frame.DataFrame'>
<class 'pandas.core.series.Series'>
###Markdown
It appears that we are using 20% of our data (30 out of 150) for the validation set and the other 80% for the train set. Logistic regression using KerasWhile most statistical packets implement some form of logistic regression in this exercise we are interested in using Keras, which is a library aimed to (Deep) Neural Networks. Actually logistic regression plays an important role in neural networks and it's typically used in the last (or second to last) layer of a classifier. For more details on how to use keras a good starting point is the [documentation on training and evaluation](https://www.tensorflow.org/guide/keras/train_and_evaluate).Our neural network will be very easy and very minimal, and will be comprised of only one node (neuron) implementing both regression and sigmoid function. We are now ready to build the model!
###Code
#we are building a "sequential" model, meaning that the data will
#flow like INPUT -> ELABORATION -> OUTPUT. In particular, we will
#not have any loops, i.e. our output will never be recycled as
#input for the first layer
from keras.models import Sequential
#a "dense" layer is a layer were all the data coming in are connected
#to all nodes. In our case there is only one node in the layer, and
#it receives all the features
from keras.layers import Dense
# 2-class logistic regression in Keras
model = Sequential()
model.add(Dense(1, activation='sigmoid', input_dim=features_train.shape[1]))
#the model is declared, but we still need to compile it to actually
#build all the data structures
model.compile(optimizer='rmsprop', loss='binary_crossentropy')
###Output
_____no_output_____
###Markdown
Let's take a look inside the model:
###Code
print(model.summary())
###Output
Model: "sequential"
_________________________________________________________________
Layer (type) Output Shape Param #
=================================================================
dense (Dense) (None, 1) 3
=================================================================
Total params: 3
Trainable params: 3
Non-trainable params: 0
_________________________________________________________________
None
###Markdown
Keras informs us that there are three trainable parameters (W1, W2, B), and a single node. The output is a single number. Excellent. We have now prepared everything we need and are ready to train the model on our data. It's an iterative process that cycles many times through what are called `epochs`. We'll start with ten:
###Code
history = model.fit(features_train, target_train, epochs=10, validation_data=(features_val, target_val))
###Output
Epoch 1/10
4/4 [==============================] - 0s 36ms/step - loss: 0.6551 - val_loss: 0.6609
Epoch 2/10
4/4 [==============================] - 0s 5ms/step - loss: 0.6397 - val_loss: 0.6504
Epoch 3/10
4/4 [==============================] - 0s 6ms/step - loss: 0.6295 - val_loss: 0.6419
Epoch 4/10
4/4 [==============================] - 0s 5ms/step - loss: 0.6210 - val_loss: 0.6340
Epoch 5/10
4/4 [==============================] - 0s 6ms/step - loss: 0.6131 - val_loss: 0.6272
Epoch 6/10
4/4 [==============================] - 0s 6ms/step - loss: 0.6065 - val_loss: 0.6214
Epoch 7/10
4/4 [==============================] - 0s 7ms/step - loss: 0.6006 - val_loss: 0.6158
Epoch 8/10
4/4 [==============================] - 0s 7ms/step - loss: 0.5947 - val_loss: 0.6103
Epoch 9/10
4/4 [==============================] - 0s 7ms/step - loss: 0.5889 - val_loss: 0.6047
Epoch 10/10
4/4 [==============================] - 0s 6ms/step - loss: 0.5836 - val_loss: 0.6003
###Markdown
We asked for ten epochs and the network did just that. At each iteration the network is trying really hard to minimize a [value called "loss"](https://keras.io/api/losses/). The specifics are defined by our choice of loss function (we selected `binary_crossentropy`). The basic idea is that the smaller the loss the better the fit.Note that the network minimizes the loss on the training set and does not use the validation set during the learning process. It can however measure the loss on the validation set to give us an idea on how well it can generalize on new data.It's handy at this point to define a function that takes in the `history` object returned by `.fit()` and plots it:
###Code
#function to take a look at losses evolution
def plot_loss_history(h, title):
plt.plot(h.history['loss'], label = "Train loss")
plt.plot(h.history['val_loss'], label = "Validation loss")
plt.xlabel('Epochs')
plt.title(title)
plt.legend()
plt.show()
plot_loss_history(history, 'Logistic (10 epochs)')
###Output
_____no_output_____
###Markdown
The good news is that loss just goes down, both in train and validation set. We can keep training - without recompiling, we just add new epochs to our network.
###Code
#putting verbose to 0 to avoid filling the screen
history2 = model.fit(features_train, target_train, epochs=490,
validation_data=(features_val, target_val), verbose=0)
###Output
_____no_output_____
###Markdown
Let's see if we improved:
###Code
#putting together the whole history
history.history['loss'] += history2.history['loss']
history.history['val_loss'] += history2.history['val_loss']
#and plotting again
plot_loss_history(history, 'Logistic (500 epochs)')
###Output
_____no_output_____
###Markdown
This is very informative: losses keep shrinking, meaning that the network keeps improving. However after a first phase of steep improvement the gain for each epoch slows down considerably.Moreover, we now see a clear difference between train and validation set. This means that, while the network keeps improving, its performances on new data are expected to be worse than those on the training data.Now, we could ask: what happens if we keep training for a long time? We have prepared the code for 10000 epochs, but it takes a long time to run, and it's faster if we simply show the saved results (but you can try to run it putting the following flag to `True`):
###Code
do_10000_epochs = False
#what happens if we keep going for a (very) long time?
if (do_10000_epochs):
#train for 10000 epochs, just to show how the model evolves
history3 = model.fit(features_train, target_train, epochs=9500,
validation_data=(features_val, target_val), verbose=0)
#putting together the whole history
history.history['loss'] += history3.history['loss']
history.history['val_loss'] += history3.history['val_loss']
#and plotting again
plot_loss_history(history, 'Logistic (10000 epochs)')
###Output
_____no_output_____
###Markdown
Our pre-recorded results look like this: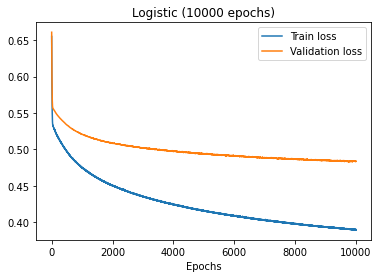 It appears that there is constant, slow improvement on training set. Improvement on validation set is slower, and if we had the patience to go for a veeeery long time the orange curve would become completely flat. Decision BoundaryThe assignment asks us to plot the [decision boundary](https://en.wikipedia.org/wiki/Decision_boundary), i.e. a representation in the feature space of the criterions the model is using to classify your data. For this task we'll use the [mlxtend module](http://rasbt.github.io/mlxtend/), which unfortunately does not come with the standard installation. Let's add it!
###Code
!pip install mlxtend
###Output
Requirement already satisfied: mlxtend in /usr/local/lib/python3.6/dist-packages (0.14.0)
Requirement already satisfied: scikit-learn>=0.18 in /usr/local/lib/python3.6/dist-packages (from mlxtend) (0.22.2.post1)
Requirement already satisfied: numpy>=1.10.4 in /usr/local/lib/python3.6/dist-packages (from mlxtend) (1.18.5)
Requirement already satisfied: setuptools in /usr/local/lib/python3.6/dist-packages (from mlxtend) (49.6.0)
Requirement already satisfied: matplotlib>=1.5.1 in /usr/local/lib/python3.6/dist-packages (from mlxtend) (3.2.2)
Requirement already satisfied: scipy>=0.17 in /usr/local/lib/python3.6/dist-packages (from mlxtend) (1.4.1)
Requirement already satisfied: pandas>=0.17.1 in /usr/local/lib/python3.6/dist-packages (from mlxtend) (1.0.5)
Requirement already satisfied: joblib>=0.11 in /usr/local/lib/python3.6/dist-packages (from scikit-learn>=0.18->mlxtend) (0.16.0)
Requirement already satisfied: kiwisolver>=1.0.1 in /usr/local/lib/python3.6/dist-packages (from matplotlib>=1.5.1->mlxtend) (1.2.0)
Requirement already satisfied: pyparsing!=2.0.4,!=2.1.2,!=2.1.6,>=2.0.1 in /usr/local/lib/python3.6/dist-packages (from matplotlib>=1.5.1->mlxtend) (2.4.7)
Requirement already satisfied: cycler>=0.10 in /usr/local/lib/python3.6/dist-packages (from matplotlib>=1.5.1->mlxtend) (0.10.0)
Requirement already satisfied: python-dateutil>=2.1 in /usr/local/lib/python3.6/dist-packages (from matplotlib>=1.5.1->mlxtend) (2.8.1)
Requirement already satisfied: pytz>=2017.2 in /usr/local/lib/python3.6/dist-packages (from pandas>=0.17.1->mlxtend) (2018.9)
Requirement already satisfied: six in /usr/local/lib/python3.6/dist-packages (from cycler>=0.10->matplotlib>=1.5.1->mlxtend) (1.15.0)
###Markdown
We can now import `mlxtend` package safely.
###Code
from mlxtend.plotting import plot_decision_regions
#we'll print the training set
plot_decision_regions(features_train.to_numpy(), target_train.to_numpy(), clf=model)
plt.title('Decision boundary for 0 (non virginica) vs 1 (virginica)')
plt.xlabel(iris.feature_names[feature_x])
plt.ylabel(iris.feature_names[feature_y])
plt.show()
###Output
/usr/local/lib/python3.6/dist-packages/mlxtend/plotting/decision_regions.py:244: MatplotlibDeprecationWarning: Passing unsupported keyword arguments to axis() will raise a TypeError in 3.3.
ax.axis(xmin=xx.min(), xmax=xx.max(), y_min=yy.min(), y_max=yy.max())
###Markdown
The decision boundary is linear, as expected by logistic regression. This means that all samples in the pink area will be classified as 1 (virginica) and all points in the blue area be considered 0 (non virginica).Note that relatively many virginica samples are in the blue area compared to the numer of non-virginica present in the pink area. Also note that, roughly speaking, the regressor assigned a wider area to non-virginica. This is a direct consequence of having an unbalanced dataset: two-thirds of the samples are non-virginica (blue squares) and one-third are virginica (red triangles). **The resulting regressor is polarised** toward the more numerous class. Actual predictionsAny model is only useful when it's used to predict new, unknown data. In fact the whole validation set was put apart and not really used for training for this specific reason. Luckily, it's very easy to apply a trained model to new values via the [predict() method](https://keras.io/api/models/model_training_apis/predict-method).
###Code
predictions = model.predict(features_val)
print(predictions)
###Output
[[0.22289124]
[0.42798153]
[0.2892147 ]
[0.56683457]
[0.15156415]
[0.18946213]
[0.42798153]
[0.3026018 ]
[0.49765265]
[0.64505094]
[0.27618527]
[0.34743673]
[0.17050767]
[0.3626923 ]
[0.46204674]
[0.47805318]
[0.48159555]
[0.1062865 ]
[0.09407479]
[0.44728738]
[0.477463 ]
[0.36707753]
[0.34797326]
[0.44611812]
[0.10606205]
[0.33196336]
[0.14232534]
[0.51312387]
[0.19990602]
[0.09407479]]
###Markdown
We can now compare the list of prediction (sometimes called Ŷ) with the true classes from our validation set (sometimes called Y).
###Code
#starting a new plot
fig, ax = plt.subplots()
#adding data in two bunches
ax.scatter(x=range(30), y=predictions, c='red', label='predictions')
ax.scatter(x=range(30), y=target_val, c='blue', label='true classes')
#adding a horizontal line at quote 0.5, to represent the decision boundary
ax.hlines(y=0.5, xmin=0, xmax=29, colors='black')
#adding the legend and printing the plot
ax.legend(loc='upper right')
plt.show()
###Output
_____no_output_____
###Markdown
Note that the vast majority of predictions is below the 0.5 line (recall that the closer a prediction is to 0 or to 1 the higher the prediction confidence).This confirms our suspicions: the regressor prefers to assign new data to the 0 (non virginica) class. Multiclass classification using SoftmaxThe assignment ask us to do actual three-classes classification using a Softmax function, which can be easily considered as an extension of logistic regression over three (or more) classes.Luckily, Keras provides a [softmax activation function](https://keras.io/api/layers/activations/softmax-function), which we will use instead of the logistic we previously used.The structure of our network will be similar, but the output goes from a single number to **three** numbers, one per class, and we thus need three nodes:As a result, the loss function will need to change. Remember, loss represents a measure of how good the predictions are. Previously we used binary_crossentropy, but since now predictions are multiclass we need to change function. Luckily Keras provides a natural extension for the multiclass case with [CategoricalCrossentropy](https://keras.io/api/losses/probabilistic_losses/categoricalcrossentropy-class) A multiclass modelWe are now ready to declare our new model:
###Code
# 3-class softmax regression in Keras
model_multi = Sequential()
model_multi.add(Dense(3, activation='softmax', input_dim=features_train.shape[1]))
#compile the model specifying the new multiclass loss
model_multi.compile(optimizer='rmsprop', loss='categorical_crossentropy')
###Output
_____no_output_____
###Markdown
Let's take a look under the hood:
###Code
print(model_multi.summary())
###Output
Model: "sequential_1"
_________________________________________________________________
Layer (type) Output Shape Param #
=================================================================
dense_1 (Dense) (None, 3) 9
=================================================================
Total params: 9
Trainable params: 9
Non-trainable params: 0
_________________________________________________________________
None
###Markdown
We now have to train nine parameters: three coefficients (W1, W2, B) times tree nodes. Categorical dataWe are now *almost* ready to train. First, we need to go back to having three classes:
###Code
#a new array for multi class
target_multi = target
target_multi[0:50] = 0 #setosa
target_multi[50:100] = 1 #versicolor
target_multi[100:150] = 2 #virginica
#or we could have used iris.target ...
###Output
_____no_output_____
###Markdown
The problem is that our `target_multi` variable is a numeric array. But those numbers we used (0, 1, and 2) do not represent real values. In other words, "virginica" is not twice "versicolor". Numbers here are used as labels, not as quantities.In fact, to properly train a model the structure of the target array must change to [one-hot encoding](https://en.wikipedia.org/wiki/One-hot). In simple terms, it needs to become a table with one row per sample (150 in total) and one column per class (three in total). Something like:| Setosa | Versicolor | Virginica ||------|------|------|| 0 | 1 | 0 || 1 | 0 | 0 || 1 | 0 | 0 || 0 | 0 | 1 |As you can see the first sample is Versicolor, the second and third are Setosa, the last one is Virginica. Note that there is only a single "one" per row.Luckily, it's easy to pass to one-hot encoding using keras function [to_categorical](https://keras.io/api/utils/python_utils/to_categorical-function):
###Code
#the "utils" subpackage is very useful, take a look to it when you have time
from keras.utils import to_categorical
#converting to categorical
target_multi_cat = to_categorical(target_multi)
#since everything else is a Pandas dataframe, let's stick to the format
#for consistency
target_multi_cat = pd.DataFrame(target_multi_cat)
#let's take a look
print(target_multi_cat)
###Output
0 1 2
0 1.0 0.0 0.0
1 1.0 0.0 0.0
2 1.0 0.0 0.0
3 1.0 0.0 0.0
4 1.0 0.0 0.0
.. ... ... ...
145 0.0 0.0 1.0
146 0.0 0.0 1.0
147 0.0 0.0 1.0
148 0.0 0.0 1.0
149 0.0 0.0 1.0
[150 rows x 3 columns]
###Markdown
Training and validation sets, reduxWe are now ready to create our training and validation sets, as done above:
###Code
#we can actually reuse the StratifiedShuffleSplit object ("sss") declared above,
#thus keeping the same split ratio (80/20). We are going to overwrite the
#previous variables for simplicity, but using the new target table:
for train_index, val_index in sss.split(features, target_multi_cat):
features_train = features.iloc[train_index, :]
features_val = features.iloc[val_index, :]
target_train = target_multi_cat.iloc[train_index, :]
target_val = target_multi_cat.iloc[val_index, :]
###Output
_____no_output_____
###Markdown
Just a little check:
###Code
#shapes
print(features_train.shape)
print(features_val.shape)
print(target_train.shape)
print(target_val.shape)
#number of classes per split
print('\nClasses in train set:')
print(target_train.sum())
print('\nClasses in validation set:')
print(target_val.sum())
###Output
(120, 2)
(30, 2)
(120, 3)
(30, 3)
Classes in train set:
0 40.0
1 40.0
2 40.0
dtype: float32
Classes in validation set:
0 10.0
1 10.0
2 10.0
dtype: float32
###Markdown
We have now a balanced dataset, with 40 instances for each class in the training set and 10 in the validation set. Training We are ready to train. This time we go directly to 500 epochs, trained in silent mode. We then plot the loss function evolution.
###Code
history_multi = model_multi.fit(features_train, target_train, epochs=500,
validation_data=(features_val, target_val), verbose=0)
plot_loss_history(history_multi, 'Softmax multiclass (500 epochs)')
###Output
_____no_output_____
###Markdown
Decision boundaryWe want now to plot again the decision boundary. Unfortunately `plot_decision_regions` function from [mlxtend](http://rasbt.github.io/mlxtend/) module does not support one-hot encoded multiclasses natively. Luckily [there's a quick workaround](https://www.machinecurve.com/index.php/2019/10/17/how-to-use-categorical-multiclass-hinge-with-keras/visualizing-the-decision-boundary), but if you get lost in the code don't worry and just look at the plot :)
###Code
import numpy as np
#we define a class to take the Keras model and convert its predictions
#from "one probability per iris type" to "just the iris type with the highest probability"
class Onehot2Int(object):
def __init__(self, model):
self.model = model
def predict(self, X):
y_pred = self.model.predict(X)
return np.argmax(y_pred, axis=1)
#we wrap our trained model, instantiating a new object
keras_model_no_ohe = Onehot2Int(model_multi)
#and we can now plot the decision boundary safely (we still need to convert
#the target one-hot-encoded matrix to int, though)
plot_decision_regions(features_train.to_numpy(), np.argmax(target_train.to_numpy(), axis=1),
clf=keras_model_no_ohe)
plt.title('Decision boundary for 0 (setosa) vs 1 (versicolor) vs 2 (virginica)')
plt.xlabel(iris.feature_names[feature_x])
plt.ylabel(iris.feature_names[feature_y])
plt.show()
###Output
/usr/local/lib/python3.6/dist-packages/mlxtend/plotting/decision_regions.py:244: MatplotlibDeprecationWarning: Passing unsupported keyword arguments to axis() will raise a TypeError in 3.3.
ax.axis(xmin=xx.min(), xmax=xx.max(), y_min=yy.min(), y_max=yy.max())
|
Dia_4/instructores/Proyecto Calentamiento Global.ipynb
|
###Markdown
Reto 1: Hacer una gráfica de la temperatura promedio por año Reto 2: Hacer una gráfica de la producción de petroleo por año Reto 3: Hacer una gráfica de la Temperatura en función de la producción de petroleo Reto 4: Encontrar el año con mayor incremento de temperatura, y el año con mayor producción petroleo. Son años cercanos?
###Code
plt.figure(figsize=(9,9))
plt.plot(produccion_petroleo.Date,produccion_petroleo.Volume)
plt.xlabel('Año',fontsize = 14, fontweight='bold')
plt.ylabel('Volumen', fontsize = 14,fontweight='bold')
plt.title('Producción petroleo anual Mundial',fontsize = 16,fontweight='bold')
plt.xticks(fontsize=14)
plt.yticks(fontsize=14)
plt.savefig('produccion_petroleo')
plt.show()
x = produccion_petroleo[produccion_petroleo.Date.dt.year <= 2015].Volume
y = historia_temperaturas[historia_temperaturas.dt.dt.year >= 1983].temperatura_promedio
plt.figure(figsize=(9,9))
plt.scatter(x,y,c='blue')
plt.xlabel('Producción Petroleo',fontsize = 14, fontweight='bold')
plt.ylabel('Temperatura promedio($c^{0}$)', fontsize = 14,fontweight='bold')
plt.title('Producción petroleo anual Mundial',fontsize = 16,fontweight='bold')
plt.xticks(fontsize=14)
plt.yticks(fontsize=14)
plt.savefig('temperatura_petroleo')
plt.show()
produccion_petroleo.set_index('Date').Volume.diff().sort_values(ascending = False)
###Output
_____no_output_____
|
python_tools/data_clean_up_tools/ml_find_features.ipynb
|
###Markdown
Recyled CODE. NEEDS TO BE CLEANED Goals Review data from Kagle 1) Import data 2) Remove null values Clean Data 1) Covert output of data to catagorical using one hot-encoding 2) Reomve original ouput column 3) Extract dipsoistion catogorical column as y value for testing 4) Create features dataframe Identify top ten features using decision tree machine learning. 1) Use decision tree to investigate accuracy of features 2) Uses random forest models to identify top ten features 3) create dataframe with top 10 featrues and add one-hot encoding 4) create csv from dataframe to use with models Identify top ten features using decision tree machine learning. 1) employ minMaxScaler to opimize data 2) create a dataframe that can be used for modeling (also has one-hot enconding) 3) create csv from dataframe to use with models 4) ran random forest model for funRefrence2/Activities/04-Stu_Trees/Solved/Stu_Trees.ipynb
###Code
# Dependencies
import pandas as pd
from sklearn.preprocessing import LabelEncoder
import numpy as np
from sklearn import tree
import os
###Output
_____no_output_____
###Markdown
Read the CSV and Perform Basic Data Cleaning
###Code
#import crime data
path = "data/"
file = "IMPD_UCR_2013_Data.csv"
path_file = path + file
df = pd.read_csv(path_file)
# Drop the null columns where all values are null
df = df.dropna(axis='columns', how='all')
# Drop the null rows
df = df.dropna()
df.head()
#df["BEAT"].value_counts()
df[['Hour','Minute']] = df.TIME.str.split(":",expand=True,)
df
df[["District","Zone"]] = df.BEAT.str.extract('([a-zA-Z]+)([^a-zA-Z]+)',expand=True)
df
# Filter key features
test_data_df = df.drop(columns=["BEAT","X_COORD","Y_COORD", "Zone","TIME","UCR","OBJECTID","CASE"])
test_data_df
# Investigate key districts
test_data_df["District"].value_counts()
# Check objet and data type
#test_data_df.DATE_.head()
test_data_df["Hour"] = test_data_df["Hour"].astype(str)
test_data_df["Hour"] = test_data_df["Hour"].astype(pd.StringDtype())
test_data_df["Minute"] = test_data_df["Minute"].astype(str)
test_data_df["Minute"] = test_data_df["Minute"].astype(pd.StringDtype())
test_data_df
# Check objet and data type
test_data_df['DATE_'] = pd.to_datetime(test_data_df['DATE_'])
test_data_df.info()
#final crime data set will serve as model data set
test_data_df
# Convert Date infomation into indvidual arrays
test_data_df['DATE_year'] = test_data_df['DATE_'].dt.year
test_data_df['DATE_month'] = test_data_df['DATE_'].dt.month
test_data_df['DATE_week'] = test_data_df['DATE_'].dt.week
test_data_df['DATE_day'] = test_data_df['DATE_'].dt.day
test_data_df = test_data_df[["CRIME","DATE_","DATE_year","DATE_month", "DATE_week","DATE_day", "Hour", "Minute","ADDRESS", "District"]]
test_data_df = test_data_df.astype(str)
test_data_df.dtypes
###Output
_____no_output_____
###Markdown
FINAL CRIME DATA: Will also serve as dataframe for modeling
###Code
test_data_df
###Output
_____no_output_____
###Markdown
Import temprature Data
###Code
# Import Tempratue DATA (K) and create master template
path = "data/"
file = "IndyWeather2013.csv"
path_file = path + file
Indy_temp_df = pd.read_csv(path_file)
Indy_temp_df = Indy_temp_df.drop(columns=["Unnamed: 0"])
Indy_temp_df= Indy_temp_df.astype(str)
Indy_temp_df.dtypes
temp_df = pd.merge(test_data_df, Indy_temp_df, how='left', on=['DATE_year', 'DATE_month', 'DATE_day', 'Hour'])
temp_df
temp_df = temp_df.dropna()
temp_df
test_data_df = temp_df.drop(columns=["DATE_"])
test_data_df
path = "data/"
file = "modified_IndyPresure2013.csv"
path_file = path + file
Indy_pres_df = pd.read_csv(path_file)
Indy_pres_df = Indy_pres_df.drop(columns=["Unnamed: 0"])
Indy_pres_df= Indy_pres_df.astype(str)
Indy_pres_df.dtypes
###Output
_____no_output_____
###Markdown
Import Pressure DATA
###Code
Indy_pres_df
temp_df = pd.merge(test_data_df, Indy_pres_df, how='left', on=['DATE_year', 'DATE_month', 'DATE_day', 'Hour'])
temp_df
temp_df.rename(columns = {"Indianapolis": "Temp(K)"}, inplace=True)
test_data_df= temp_df.dropna()
###Output
_____no_output_____
###Markdown
IMPORT WIND DATA
###Code
path = "data/"
file = "modified_wind_2013.csv"
path_file = path + file
wind_df = pd.read_csv(path_file)
wind_df = wind_df.drop(columns=["Unnamed: 0"])
wind_df= wind_df.astype(str)
wind_df.dtypes
wind_df
temp_df = pd.merge(test_data_df, wind_df, how='left', on=['DATE_year', 'DATE_month', 'DATE_day', 'Hour'])
temp_df
test_data_df= temp_df.dropna()
###Output
_____no_output_____
###Markdown
Import Humidity
###Code
path = "data/"
file = "modified_wind_2013.csv"
path_file = path + file
wind_df = pd.read_csv(path_file)
wind_df = wind_df.drop(columns=["Unnamed: 0"])
wind_df= wind_df.astype(str)
wind_df.dtypes
wind_df
# Entire data frame with cleaned raw data. No Manipulation to columns.
#test_data_df = temp_df.drop(columns=["DATE_"])
#test_data_df = test_data_df[["DATE_year","DATE_month", "DATE_week","DATE_day", "Hour", "Minute", "District"]]
#test_data_df
###Output
_____no_output_____
###Markdown
Preprocessing Review One-Hot Encodingoverview:* Creating dataframes for modeling* featrues_df: does not have the y output (koi_disposition)* modified_data_df: will have all features and a diposition column with numeracal values. This will be completed latter.* modified_data_df: will be convertedt to a csv for use in other models and stored in data folder. File=raw_data_with_diposition.csv* y is original disposition catagory. This will get converted to a 0,1 or 2 value latter.
###Code
# Creating dataframes for modeling. Works with imported code variables
# featrues_df: does not have the y output (koi_disposition)
# modified_data_df: will have all features and a diposition column with numeracal values. This will be completed latter.
# modified_data_df: will be convertedt to a csv for use in other models and stored in data folder.File=raw_data_with_diposition.csv.
# y is original disposition catagory. This will get converted to a 0,1 or 2 value latter.
features_df = test_data_df.drop("District", axis=1)
y = test_data_df["District"]
moidfied_data_df = features_df
#moidfied_data_df
features_df
###Output
_____no_output_____
###Markdown
Encode Districts (the outup Y variable)
###Code
# Step 1: Label-encode data set
label_encoder = LabelEncoder()
label_encoder.fit(y)
encoded_y = label_encoder.transform(y)
n= 0
for label, original_class in zip(encoded_y, y):
print('Original Class: ' + str(original_class))
print('Encoded Label: ' + str(label))
n=n+1
print(n)
print('-' * 12)
###Output
Original Class: NE
Encoded Label: 2
1
------------
Original Class: NE
Encoded Label: 2
2
------------
Original Class: SE
Encoded Label: 4
3
------------
Original Class: NW
Encoded Label: 3
4
------------
Original Class: NE
Encoded Label: 2
5
------------
Original Class: ND
Encoded Label: 1
6
------------
Original Class: DT
Encoded Label: 0
7
------------
Original Class: SE
Encoded Label: 4
8
------------
Original Class: DT
Encoded Label: 0
9
------------
Original Class: SE
Encoded Label: 4
10
------------
Original Class: SE
Encoded Label: 4
11
------------
Original Class: NW
Encoded Label: 3
12
------------
Original Class: SW
Encoded Label: 5
13
------------
Original Class: SE
Encoded Label: 4
14
------------
Original Class: NW
Encoded Label: 3
15
------------
Original Class: ND
Encoded Label: 1
16
------------
Original Class: DT
Encoded Label: 0
17
------------
Original Class: ND
Encoded Label: 1
18
------------
Original Class: DT
Encoded Label: 0
19
------------
Original Class: NW
Encoded Label: 3
20
------------
Original Class: NW
Encoded Label: 3
21
------------
Original Class: DT
Encoded Label: 0
22
------------
Original Class: NE
Encoded Label: 2
23
------------
Original Class: NE
Encoded Label: 2
24
------------
Original Class: NW
Encoded Label: 3
25
------------
Original Class: DT
Encoded Label: 0
26
------------
Original Class: ND
Encoded Label: 1
27
------------
Original Class: ND
Encoded Label: 1
28
------------
Original Class: SE
Encoded Label: 4
29
------------
Original Class: ND
Encoded Label: 1
30
------------
Original Class: DT
Encoded Label: 0
31
------------
Original Class: ND
Encoded Label: 1
32
------------
Original Class: DT
Encoded Label: 0
33
------------
Original Class: ND
Encoded Label: 1
34
------------
Original Class: DT
Encoded Label: 0
35
------------
Original Class: NE
Encoded Label: 2
36
------------
Original Class: ND
Encoded Label: 1
37
------------
Original Class: DT
Encoded Label: 0
38
------------
Original Class: NW
Encoded Label: 3
39
------------
Original Class: SE
Encoded Label: 4
40
------------
Original Class: ND
Encoded Label: 1
41
------------
Original Class: DT
Encoded Label: 0
42
------------
Original Class: SE
Encoded Label: 4
43
------------
Original Class: DT
Encoded Label: 0
44
------------
Original Class: NE
Encoded Label: 2
45
------------
Original Class: DT
Encoded Label: 0
46
------------
Original Class: NW
Encoded Label: 3
47
------------
Original Class: DT
Encoded Label: 0
48
------------
Original Class: DT
Encoded Label: 0
49
------------
Original Class: NE
Encoded Label: 2
50
------------
Original Class: NW
Encoded Label: 3
51
------------
Original Class: DT
Encoded Label: 0
52
------------
Original Class: SW
Encoded Label: 5
53
------------
Original Class: SW
Encoded Label: 5
54
------------
Original Class: ND
Encoded Label: 1
55
------------
Original Class: SW
Encoded Label: 5
56
------------
Original Class: DT
Encoded Label: 0
57
------------
Original Class: NE
Encoded Label: 2
58
------------
Original Class: ND
Encoded Label: 1
59
------------
Original Class: SE
Encoded Label: 4
60
------------
Original Class: ND
Encoded Label: 1
61
------------
Original Class: DT
Encoded Label: 0
62
------------
Original Class: NE
Encoded Label: 2
63
------------
Original Class: SE
Encoded Label: 4
64
------------
Original Class: DT
Encoded Label: 0
65
------------
Original Class: NE
Encoded Label: 2
66
------------
Original Class: ND
Encoded Label: 1
67
------------
Original Class: NW
Encoded Label: 3
68
------------
Original Class: SW
Encoded Label: 5
69
------------
Original Class: NE
Encoded Label: 2
70
------------
Original Class: SE
Encoded Label: 4
71
------------
Original Class: ND
Encoded Label: 1
72
------------
Original Class: DT
Encoded Label: 0
73
------------
Original Class: DT
Encoded Label: 0
74
------------
Original Class: NE
Encoded Label: 2
75
------------
Original Class: DT
Encoded Label: 0
76
------------
Original Class: ND
Encoded Label: 1
77
------------
Original Class: NE
Encoded Label: 2
78
------------
Original Class: NW
Encoded Label: 3
79
------------
Original Class: DT
Encoded Label: 0
80
------------
Original Class: DT
Encoded Label: 0
81
------------
Original Class: DT
Encoded Label: 0
82
------------
Original Class: NE
Encoded Label: 2
83
------------
Original Class: NW
Encoded Label: 3
84
------------
Original Class: NW
Encoded Label: 3
85
------------
Original Class: DT
Encoded Label: 0
86
------------
Original Class: DT
Encoded Label: 0
87
------------
Original Class: DT
Encoded Label: 0
88
------------
Original Class: NW
Encoded Label: 3
89
------------
Original Class: DT
Encoded Label: 0
90
------------
Original Class: SE
Encoded Label: 4
91
------------
Original Class: DT
Encoded Label: 0
92
------------
Original Class: DT
Encoded Label: 0
93
------------
Original Class: ND
Encoded Label: 1
94
------------
Original Class: DT
Encoded Label: 0
95
------------
Original Class: NW
Encoded Label: 3
96
------------
Original Class: NE
Encoded Label: 2
97
------------
Original Class: NE
Encoded Label: 2
98
------------
Original Class: NE
Encoded Label: 2
99
------------
Original Class: SE
Encoded Label: 4
100
------------
Original Class: SW
Encoded Label: 5
101
------------
Original Class: NE
Encoded Label: 2
102
------------
Original Class: DT
Encoded Label: 0
103
------------
Original Class: DT
Encoded Label: 0
104
------------
Original Class: NE
Encoded Label: 2
105
------------
Original Class: SW
Encoded Label: 5
106
------------
Original Class: NE
Encoded Label: 2
107
------------
Original Class: DT
Encoded Label: 0
108
------------
Original Class: DT
Encoded Label: 0
109
------------
Original Class: NE
Encoded Label: 2
110
------------
Original Class: DT
Encoded Label: 0
111
------------
Original Class: NW
Encoded Label: 3
112
------------
Original Class: NW
Encoded Label: 3
113
------------
Original Class: SE
Encoded Label: 4
114
------------
Original Class: NW
Encoded Label: 3
115
------------
Original Class: DT
Encoded Label: 0
116
------------
Original Class: NE
Encoded Label: 2
117
------------
Original Class: ND
Encoded Label: 1
118
------------
Original Class: DT
Encoded Label: 0
119
------------
Original Class: SE
Encoded Label: 4
120
------------
Original Class: NE
Encoded Label: 2
121
------------
Original Class: DT
Encoded Label: 0
122
------------
Original Class: NW
Encoded Label: 3
123
------------
Original Class: SE
Encoded Label: 4
124
------------
Original Class: NE
Encoded Label: 2
125
------------
Original Class: ND
Encoded Label: 1
126
------------
Original Class: ND
Encoded Label: 1
127
------------
Original Class: ND
Encoded Label: 1
128
------------
Original Class: NW
Encoded Label: 3
129
------------
Original Class: ND
Encoded Label: 1
130
------------
Original Class: ND
Encoded Label: 1
131
------------
Original Class: NW
Encoded Label: 3
132
------------
Original Class: SE
Encoded Label: 4
133
------------
Original Class: NW
Encoded Label: 3
134
------------
Original Class: ND
Encoded Label: 1
135
------------
Original Class: SW
Encoded Label: 5
136
------------
Original Class: SE
Encoded Label: 4
137
------------
Original Class: SE
Encoded Label: 4
138
------------
Original Class: NE
Encoded Label: 2
139
------------
Original Class: DT
Encoded Label: 0
140
------------
Original Class: ND
Encoded Label: 1
141
------------
Original Class: DT
Encoded Label: 0
142
------------
Original Class: NW
Encoded Label: 3
143
------------
Original Class: NW
Encoded Label: 3
144
------------
Original Class: DT
Encoded Label: 0
145
------------
Original Class: SE
Encoded Label: 4
146
------------
Original Class: SW
Encoded Label: 5
147
------------
Original Class: ND
Encoded Label: 1
148
------------
Original Class: ND
Encoded Label: 1
149
------------
Original Class: NW
Encoded Label: 3
150
------------
Original Class: NW
Encoded Label: 3
151
------------
Original Class: NE
Encoded Label: 2
152
------------
Original Class: ND
Encoded Label: 1
153
------------
Original Class: DT
Encoded Label: 0
154
------------
Original Class: NW
Encoded Label: 3
155
------------
Original Class: SE
Encoded Label: 4
156
------------
Original Class: NE
Encoded Label: 2
157
------------
Original Class: NE
Encoded Label: 2
158
------------
Original Class: ND
Encoded Label: 1
159
------------
Original Class: SE
Encoded Label: 4
160
------------
Original Class: NW
Encoded Label: 3
161
------------
Original Class: ND
Encoded Label: 1
162
------------
Original Class: DT
Encoded Label: 0
163
------------
Original Class: DT
Encoded Label: 0
164
------------
Original Class: ND
Encoded Label: 1
165
------------
Original Class: ND
Encoded Label: 1
166
------------
Original Class: ND
Encoded Label: 1
167
------------
Original Class: DT
Encoded Label: 0
168
------------
Original Class: NE
Encoded Label: 2
169
------------
Original Class: NW
Encoded Label: 3
170
------------
Original Class: DT
Encoded Label: 0
171
------------
Original Class: SW
Encoded Label: 5
172
------------
Original Class: ND
Encoded Label: 1
173
------------
Original Class: ND
Encoded Label: 1
174
------------
Original Class: NE
Encoded Label: 2
175
------------
Original Class: ND
Encoded Label: 1
176
------------
Original Class: NW
Encoded Label: 3
177
------------
Original Class: SW
Encoded Label: 5
178
------------
Original Class: ND
Encoded Label: 1
179
------------
Original Class: ND
Encoded Label: 1
180
------------
Original Class: ND
Encoded Label: 1
181
------------
Original Class: NW
Encoded Label: 3
182
------------
Original Class: ND
Encoded Label: 1
183
------------
Original Class: ND
Encoded Label: 1
184
------------
Original Class: DT
Encoded Label: 0
185
------------
Original Class: ND
Encoded Label: 1
186
------------
Original Class: DT
Encoded Label: 0
187
------------
Original Class: ND
Encoded Label: 1
188
------------
Original Class: NE
Encoded Label: 2
189
------------
Original Class: NW
Encoded Label: 3
190
------------
Original Class: NE
Encoded Label: 2
191
------------
Original Class: ND
Encoded Label: 1
192
------------
Original Class: NW
Encoded Label: 3
193
------------
Original Class: NE
Encoded Label: 2
194
------------
Original Class: NW
Encoded Label: 3
195
------------
Original Class: DT
Encoded Label: 0
196
------------
Original Class: DT
Encoded Label: 0
197
------------
Original Class: DT
Encoded Label: 0
198
------------
Original Class: DT
Encoded Label: 0
199
------------
Original Class: NW
Encoded Label: 3
200
------------
Original Class: SW
Encoded Label: 5
201
------------
Original Class: NE
Encoded Label: 2
202
------------
Original Class: SW
Encoded Label: 5
203
------------
Original Class: DT
Encoded Label: 0
204
------------
Original Class: DT
Encoded Label: 0
205
------------
Original Class: DT
Encoded Label: 0
206
------------
Original Class: ND
Encoded Label: 1
207
------------
Original Class: DT
Encoded Label: 0
208
------------
Original Class: DT
Encoded Label: 0
209
------------
Original Class: ND
Encoded Label: 1
210
------------
Original Class: DT
Encoded Label: 0
211
------------
Original Class: DT
Encoded Label: 0
212
------------
Original Class: NE
Encoded Label: 2
213
------------
Original Class: NE
Encoded Label: 2
214
------------
Original Class: SW
Encoded Label: 5
215
------------
Original Class: ND
Encoded Label: 1
216
------------
Original Class: ND
Encoded Label: 1
217
------------
Original Class: NE
Encoded Label: 2
218
------------
Original Class: SE
Encoded Label: 4
219
------------
Original Class: SW
Encoded Label: 5
220
------------
Original Class: SE
Encoded Label: 4
221
------------
Original Class: NE
Encoded Label: 2
222
------------
Original Class: NW
Encoded Label: 3
223
------------
Original Class: ND
Encoded Label: 1
224
------------
Original Class: NE
Encoded Label: 2
225
------------
Original Class: DT
Encoded Label: 0
226
------------
Original Class: SE
Encoded Label: 4
227
------------
Original Class: SE
Encoded Label: 4
228
------------
Original Class: NE
Encoded Label: 2
229
------------
Original Class: NW
Encoded Label: 3
230
------------
Original Class: SW
Encoded Label: 5
231
------------
Original Class: NE
Encoded Label: 2
232
------------
Original Class: NE
Encoded Label: 2
233
------------
Original Class: NW
Encoded Label: 3
234
------------
Original Class: NE
Encoded Label: 2
235
------------
Original Class: ND
Encoded Label: 1
236
------------
Original Class: SE
Encoded Label: 4
237
------------
Original Class: SW
Encoded Label: 5
238
------------
Original Class: NE
Encoded Label: 2
239
------------
Original Class: ND
Encoded Label: 1
240
------------
Original Class: DT
Encoded Label: 0
241
------------
Original Class: NE
Encoded Label: 2
242
------------
Original Class: DT
Encoded Label: 0
243
------------
Original Class: NW
Encoded Label: 3
244
------------
Original Class: DT
Encoded Label: 0
245
------------
Original Class: NW
Encoded Label: 3
246
------------
Original Class: SE
Encoded Label: 4
247
------------
Original Class: NE
Encoded Label: 2
248
------------
Original Class: DT
Encoded Label: 0
249
------------
Original Class: SW
Encoded Label: 5
250
------------
Original Class: SW
Encoded Label: 5
251
------------
Original Class: NW
Encoded Label: 3
252
------------
Original Class: NE
Encoded Label: 2
253
------------
Original Class: DT
Encoded Label: 0
254
------------
Original Class: NE
Encoded Label: 2
255
------------
Original Class: SW
Encoded Label: 5
256
------------
Original Class: SW
Encoded Label: 5
257
------------
Original Class: ND
Encoded Label: 1
258
------------
Original Class: SW
Encoded Label: 5
259
------------
Original Class: NW
Encoded Label: 3
260
------------
Original Class: DT
Encoded Label: 0
261
------------
Original Class: NE
Encoded Label: 2
262
------------
Original Class: NW
Encoded Label: 3
263
------------
Original Class: NW
Encoded Label: 3
264
------------
Original Class: SE
Encoded Label: 4
265
------------
Original Class: SW
Encoded Label: 5
266
------------
Original Class: ND
Encoded Label: 1
267
------------
Original Class: NE
Encoded Label: 2
268
------------
Original Class: SW
Encoded Label: 5
269
------------
Original Class: SE
Encoded Label: 4
270
------------
Original Class: SE
Encoded Label: 4
271
------------
Original Class: NW
Encoded Label: 3
272
------------
Original Class: NW
Encoded Label: 3
273
------------
Original Class: NE
Encoded Label: 2
274
------------
Original Class: SE
Encoded Label: 4
275
------------
Original Class: NE
Encoded Label: 2
276
------------
Original Class: DT
Encoded Label: 0
277
------------
Original Class: NE
Encoded Label: 2
278
------------
Original Class: SW
Encoded Label: 5
279
------------
Original Class: SE
Encoded Label: 4
280
------------
Original Class: NW
Encoded Label: 3
281
------------
Original Class: SE
Encoded Label: 4
282
------------
Original Class: NE
Encoded Label: 2
283
------------
Original Class: ND
Encoded Label: 1
284
------------
Original Class: ND
Encoded Label: 1
285
------------
Original Class: NW
Encoded Label: 3
286
------------
Original Class: NW
Encoded Label: 3
287
------------
Original Class: SW
Encoded Label: 5
288
------------
Original Class: NE
Encoded Label: 2
289
------------
Original Class: DT
Encoded Label: 0
290
------------
Original Class: ND
Encoded Label: 1
291
------------
Original Class: NW
Encoded Label: 3
292
------------
Original Class: SE
Encoded Label: 4
293
------------
Original Class: NW
Encoded Label: 3
294
------------
Original Class: NW
Encoded Label: 3
295
------------
Original Class: NE
Encoded Label: 2
296
------------
Original Class: SE
Encoded Label: 4
297
------------
Original Class: SW
Encoded Label: 5
298
------------
Original Class: SW
Encoded Label: 5
299
------------
Original Class: NW
Encoded Label: 3
300
------------
Original Class: SW
Encoded Label: 5
301
------------
Original Class: ND
Encoded Label: 1
302
------------
Original Class: SW
Encoded Label: 5
303
------------
Original Class: NE
Encoded Label: 2
304
------------
Original Class: NE
Encoded Label: 2
305
------------
Original Class: NW
Encoded Label: 3
306
------------
Original Class: NE
Encoded Label: 2
307
------------
Original Class: SW
Encoded Label: 5
308
------------
Original Class: SW
Encoded Label: 5
309
------------
Original Class: ND
Encoded Label: 1
310
------------
Original Class: SE
Encoded Label: 4
311
------------
Original Class: NW
Encoded Label: 3
312
------------
Original Class: NE
Encoded Label: 2
313
------------
Original Class: NE
Encoded Label: 2
314
------------
Original Class: ND
Encoded Label: 1
315
------------
Original Class: NE
Encoded Label: 2
316
------------
Original Class: SE
Encoded Label: 4
317
------------
Original Class: NW
Encoded Label: 3
318
------------
Original Class: SW
Encoded Label: 5
319
------------
Original Class: NE
Encoded Label: 2
320
------------
Original Class: SE
Encoded Label: 4
321
------------
Original Class: NE
Encoded Label: 2
322
------------
Original Class: NW
Encoded Label: 3
323
------------
Original Class: NE
Encoded Label: 2
324
------------
Original Class: NE
Encoded Label: 2
325
------------
Original Class: ND
Encoded Label: 1
326
------------
Original Class: NE
Encoded Label: 2
327
------------
Original Class: SE
Encoded Label: 4
328
------------
Original Class: SE
Encoded Label: 4
329
------------
Original Class: NE
Encoded Label: 2
330
------------
Original Class: SW
Encoded Label: 5
331
------------
Original Class: NE
Encoded Label: 2
332
------------
Original Class: SE
Encoded Label: 4
333
------------
Original Class: NW
Encoded Label: 3
334
------------
Original Class: DT
Encoded Label: 0
335
------------
Original Class: NW
Encoded Label: 3
336
------------
Original Class: NE
Encoded Label: 2
337
------------
Original Class: DT
Encoded Label: 0
338
------------
Original Class: SW
Encoded Label: 5
339
------------
Original Class: SE
Encoded Label: 4
340
------------
Original Class: NE
Encoded Label: 2
341
------------
Original Class: SE
Encoded Label: 4
342
------------
Original Class: ND
Encoded Label: 1
343
------------
Original Class: ND
Encoded Label: 1
344
------------
Original Class: ND
Encoded Label: 1
345
------------
Original Class: SW
Encoded Label: 5
346
------------
Original Class: SE
Encoded Label: 4
347
------------
Original Class: SW
Encoded Label: 5
348
------------
Original Class: NE
Encoded Label: 2
349
------------
Original Class: DT
Encoded Label: 0
350
------------
Original Class: NE
Encoded Label: 2
351
------------
Original Class: NE
Encoded Label: 2
352
------------
Original Class: DT
Encoded Label: 0
353
------------
Original Class: NE
Encoded Label: 2
354
------------
Original Class: SE
Encoded Label: 4
355
------------
Original Class: NW
Encoded Label: 3
356
------------
Original Class: SW
Encoded Label: 5
357
------------
Original Class: DT
Encoded Label: 0
358
------------
Original Class: NW
Encoded Label: 3
359
------------
Original Class: NE
Encoded Label: 2
360
------------
Original Class: ND
Encoded Label: 1
361
------------
Original Class: ND
Encoded Label: 1
362
------------
Original Class: NE
Encoded Label: 2
363
------------
Original Class: NE
Encoded Label: 2
364
------------
Original Class: NW
Encoded Label: 3
365
------------
Original Class: SW
Encoded Label: 5
366
------------
Original Class: SW
Encoded Label: 5
367
------------
Original Class: ND
Encoded Label: 1
368
------------
Original Class: SW
Encoded Label: 5
369
------------
Original Class: SW
Encoded Label: 5
370
------------
Original Class: NW
Encoded Label: 3
371
------------
Original Class: NE
Encoded Label: 2
372
------------
Original Class: NE
Encoded Label: 2
373
------------
Original Class: ND
Encoded Label: 1
374
------------
Original Class: SW
Encoded Label: 5
375
------------
Original Class: SW
Encoded Label: 5
376
------------
Original Class: NE
Encoded Label: 2
377
------------
Original Class: NE
Encoded Label: 2
378
------------
Original Class: SE
Encoded Label: 4
379
------------
Original Class: ND
Encoded Label: 1
380
------------
Original Class: NE
Encoded Label: 2
381
------------
Original Class: ND
Encoded Label: 1
382
------------
Original Class: NE
Encoded Label: 2
383
------------
Original Class: NW
Encoded Label: 3
384
------------
Original Class: NE
Encoded Label: 2
385
------------
Original Class: NW
Encoded Label: 3
386
------------
Original Class: SW
Encoded Label: 5
387
------------
Original Class: NW
Encoded Label: 3
388
------------
Original Class: NE
Encoded Label: 2
389
------------
Original Class: NE
Encoded Label: 2
390
------------
Original Class: SE
Encoded Label: 4
391
------------
Original Class: NE
Encoded Label: 2
392
------------
Original Class: NW
Encoded Label: 3
393
------------
Original Class: NE
Encoded Label: 2
394
------------
Original Class: SW
Encoded Label: 5
395
------------
Original Class: DT
Encoded Label: 0
396
------------
Original Class: NW
Encoded Label: 3
397
------------
Original Class: NW
Encoded Label: 3
398
------------
Original Class: NW
Encoded Label: 3
399
------------
Original Class: SE
Encoded Label: 4
400
------------
Original Class: NW
Encoded Label: 3
401
------------
Original Class: DT
Encoded Label: 0
402
------------
Original Class: SE
Encoded Label: 4
403
------------
Original Class: DT
Encoded Label: 0
404
------------
Original Class: NE
Encoded Label: 2
405
------------
Original Class: ND
Encoded Label: 1
406
------------
Original Class: NW
Encoded Label: 3
407
------------
Original Class: NE
Encoded Label: 2
408
------------
Original Class: SE
Encoded Label: 4
409
------------
Original Class: SE
Encoded Label: 4
410
------------
Original Class: SW
Encoded Label: 5
411
------------
Original Class: SE
Encoded Label: 4
412
------------
Original Class: NE
Encoded Label: 2
413
------------
Original Class: NE
Encoded Label: 2
414
------------
Original Class: SW
Encoded Label: 5
415
------------
Original Class: SW
Encoded Label: 5
416
------------
Original Class: NE
Encoded Label: 2
417
------------
Original Class: NW
Encoded Label: 3
418
------------
Original Class: SE
Encoded Label: 4
419
------------
Original Class: NE
Encoded Label: 2
420
------------
Original Class: SE
Encoded Label: 4
421
------------
Original Class: DT
Encoded Label: 0
422
------------
Original Class: NE
Encoded Label: 2
423
------------
Original Class: NW
Encoded Label: 3
424
------------
Original Class: NE
Encoded Label: 2
425
------------
Original Class: NE
Encoded Label: 2
426
------------
Original Class: ND
Encoded Label: 1
427
------------
Original Class: NE
Encoded Label: 2
428
------------
Original Class: SW
Encoded Label: 5
429
------------
Original Class: SE
Encoded Label: 4
430
------------
Original Class: NE
Encoded Label: 2
431
------------
Original Class: NE
Encoded Label: 2
432
------------
Original Class: ND
Encoded Label: 1
433
------------
Original Class: SE
Encoded Label: 4
434
------------
Original Class: DT
Encoded Label: 0
435
------------
Original Class: NE
Encoded Label: 2
436
------------
Original Class: NW
Encoded Label: 3
437
------------
Original Class: ND
Encoded Label: 1
438
------------
Original Class: SW
Encoded Label: 5
439
------------
Original Class: NE
Encoded Label: 2
440
------------
Original Class: NE
Encoded Label: 2
441
------------
Original Class: NE
Encoded Label: 2
442
------------
Original Class: DT
Encoded Label: 0
443
------------
Original Class: NW
Encoded Label: 3
444
------------
Original Class: SE
Encoded Label: 4
445
------------
Original Class: SE
Encoded Label: 4
446
------------
Original Class: NE
Encoded Label: 2
447
------------
Original Class: SE
Encoded Label: 4
448
------------
Original Class: SE
Encoded Label: 4
449
------------
Original Class: ND
Encoded Label: 1
450
------------
Original Class: NE
Encoded Label: 2
451
------------
Original Class: NE
Encoded Label: 2
452
------------
Original Class: NE
Encoded Label: 2
453
------------
Original Class: NE
Encoded Label: 2
454
------------
Original Class: SE
Encoded Label: 4
455
------------
Original Class: SE
Encoded Label: 4
456
------------
Original Class: NW
Encoded Label: 3
457
------------
Original Class: NW
Encoded Label: 3
458
------------
Original Class: SW
Encoded Label: 5
459
------------
Original Class: SW
Encoded Label: 5
460
------------
Original Class: SE
Encoded Label: 4
461
------------
Original Class: NW
Encoded Label: 3
462
------------
Original Class: NE
Encoded Label: 2
463
------------
Original Class: NW
Encoded Label: 3
464
------------
Original Class: NE
Encoded Label: 2
465
------------
Original Class: NE
Encoded Label: 2
466
------------
Original Class: SE
Encoded Label: 4
467
------------
Original Class: NW
Encoded Label: 3
468
------------
Original Class: SE
Encoded Label: 4
469
------------
Original Class: ND
Encoded Label: 1
470
------------
Original Class: ND
Encoded Label: 1
471
------------
Original Class: SE
Encoded Label: 4
472
------------
Original Class: SW
Encoded Label: 5
473
------------
Original Class: ND
Encoded Label: 1
474
------------
Original Class: ND
Encoded Label: 1
475
------------
Original Class: NE
Encoded Label: 2
476
------------
Original Class: SE
Encoded Label: 4
477
------------
Original Class: NE
Encoded Label: 2
478
------------
Original Class: DT
Encoded Label: 0
479
------------
Original Class: DT
Encoded Label: 0
480
------------
Original Class: DT
Encoded Label: 0
481
------------
Original Class: SW
Encoded Label: 5
482
------------
Original Class: NE
Encoded Label: 2
483
------------
Original Class: NE
Encoded Label: 2
484
------------
Original Class: SE
Encoded Label: 4
485
------------
Original Class: ND
Encoded Label: 1
486
------------
Original Class: SE
Encoded Label: 4
487
------------
Original Class: DT
Encoded Label: 0
488
------------
Original Class: SW
Encoded Label: 5
489
------------
Original Class: SE
Encoded Label: 4
490
------------
Original Class: NE
Encoded Label: 2
491
------------
Original Class: SW
Encoded Label: 5
492
------------
Original Class: NE
Encoded Label: 2
493
------------
Original Class: DT
Encoded Label: 0
494
------------
Original Class: SE
Encoded Label: 4
495
------------
Original Class: DT
Encoded Label: 0
496
------------
Original Class: SE
Encoded Label: 4
497
------------
Original Class: SE
Encoded Label: 4
498
------------
Original Class: DT
Encoded Label: 0
499
------------
Original Class: NW
Encoded Label: 3
500
------------
Original Class: NW
Encoded Label: 3
501
------------
Original Class: NE
Encoded Label: 2
502
------------
Original Class: SW
Encoded Label: 5
503
------------
Original Class: NE
Encoded Label: 2
504
------------
Original Class: SW
Encoded Label: 5
505
------------
Original Class: SE
Encoded Label: 4
506
------------
Original Class: SW
Encoded Label: 5
507
------------
Original Class: NE
Encoded Label: 2
508
------------
Original Class: DT
Encoded Label: 0
509
------------
Original Class: DT
Encoded Label: 0
510
------------
Original Class: SW
Encoded Label: 5
511
------------
Original Class: ND
Encoded Label: 1
512
------------
Original Class: NE
Encoded Label: 2
513
------------
Original Class: SE
Encoded Label: 4
514
------------
Original Class: SW
Encoded Label: 5
515
------------
Original Class: SW
Encoded Label: 5
516
------------
Original Class: SW
Encoded Label: 5
517
------------
Original Class: SE
Encoded Label: 4
518
------------
Original Class: NE
Encoded Label: 2
519
------------
###Markdown
Encode streets and Crime
###Code
Bike_value =features_df["CRIME"]
ADDRESS_value =features_df["ADDRESS"]
# Convert Bike values
# Step 1: Label-encode data set
label_encoder = LabelEncoder()
label_encoder.fit(Bike_value)
encoded_Bike_vale = label_encoder.transform(Bike_value)
# Display Bike values
n= 0
for label, original_class in zip(encoded_Bike_vale , Bike_value):
print('Original Class: ' + str(original_class))
print('Encoded Label: ' + str(label))
n=n+1
print(n)
print('-' * 12)
# Convert Address values
# Step 1: Label-encode data set
label_encoder = LabelEncoder()
label_encoder.fit(ADDRESS_value)
encoded_ADDRESS_value = label_encoder.transform(ADDRESS_value)
# Display Bike values
n= 0
for label, original_class in zip(encoded_ADDRESS_value, ADDRESS_value):
print('Original Class: ' + str(original_class))
print('Encoded Label: ' + str(label))
n=n+1
print(n)
print('-' * 12)
###Output
Original Class: 1308 CENTRAL AV
Encoded Label: 49
1
------------
Original Class: 1204 CENTRAL AV
Encoded Label: 39
2
------------
Original Class: 1209 S RANDOLPH ST
Encoded Label: 40
3
------------
Original Class: 7637 SANTA BARBARA DR
Encoded Label: 396
4
------------
Original Class: E 10TH ST & N TUXEDO ST
Encoded Label: 460
5
------------
Original Class: 37 W 38TH ST
Encoded Label: 218
6
------------
Original Class: 611 N CAPITOL AV
Encoded Label: 351
7
------------
Original Class: 7409 CAROLLING WAY
Encoded Label: 391
8
------------
Original Class: 55 MONUMENT CR
Encoded Label: 330
9
------------
Original Class: 1642 S TALBOTT ST
Encoded Label: 87
10
------------
Original Class: 1728 E KELLY ST
Encoded Label: 93
11
------------
Original Class: 1201 INDIANA AV
Encoded Label: 38
12
------------
Original Class: 327 W SOUTHERN AV
Encoded Label: 188
13
------------
Original Class: 1416 PLEASANT ST
Encoded Label: 61
14
------------
Original Class: 3819 PEBBLE CREEK DR
Encoded Label: 225
15
------------
Original Class: 835 E 81ST ST
Encoded Label: 418
16
------------
Original Class: 927 N PENNSYLVANIA ST
Encoded Label: 446
17
------------
Original Class: 5267 OAK LEAF DR
Encoded Label: 319
18
------------
Original Class: 427 N COLLEGE AV
Encoded Label: 255
19
------------
Original Class: 1140 DR M L KING JR ST
Encoded Label: 32
20
------------
Original Class: 7509 NEW AUGUSTA RD
Encoded Label: 393
21
------------
Original Class: 450 S MERIDIAN ST
Encoded Label: 275
22
------------
Original Class: 2403 N DELAWARE ST
Encoded Label: 137
23
------------
Original Class: 846 N TUXEDO ST
Encoded Label: 421
24
------------
Original Class: 6770 STANHOPE WAY
Encoded Label: 378
25
------------
Original Class: 1 VIRGINIA AV
Encoded Label: 0
26
------------
Original Class: 1327 SANDY ANN LN
Encoded Label: 53
27
------------
Original Class: 924 BROAD RIPPLE AV
Encoded Label: 444
28
------------
Original Class: 1749 S DELAWARE ST
Encoded Label: 97
29
------------
Original Class: 4350 N COLLEGE AV
Encoded Label: 262
30
------------
Original Class: 623 LOCKERBIE ST
Encoded Label: 358
31
------------
Original Class: 5960 E 42ND ST
Encoded Label: 344
32
------------
Original Class: 101 W OHIO ST
Encoded Label: 9
33
------------
Original Class: 6101 N KEYSTONE AV
Encoded Label: 349
34
------------
Original Class: 501 MADISON AV
Encoded Label: 302
35
------------
Original Class: 9306 CHERRY VALLEY CT
Encoded Label: 448
36
------------
Original Class: 5925 CARVEL AV
Encoded Label: 341
37
------------
Original Class: 1099 N MERIDIAN ST
Encoded Label: 25
38
------------
Original Class: 1701 SENATE BL
Encoded Label: 91
39
------------
Original Class: 8027 MC FARLAND CT
Encoded Label: 407
40
------------
Original Class: 1365 E 86TH ST
Encoded Label: 55
41
------------
Original Class: 222 N EAST ST
Encoded Label: 124
42
------------
Original Class: 4425 S EMERSON AV
Encoded Label: 270
43
------------
Original Class: 225 N NEW JERSEY STREET
Encoded Label: 130
44
------------
Original Class: 914 DORMAN ST
Encoded Label: 440
45
------------
Original Class: 1022 DR M L KING JR ST
Encoded Label: 16
46
------------
Original Class: 5820 W 56TH ST
Encoded Label: 339
47
------------
Original Class: 200 W MARYLAND ST
Encoded Label: 104
48
------------
Original Class: 611 N PARK AV
Encoded Label: 352
49
------------
Original Class: 3757 E NEW YORK ST
Encoded Label: 222
50
------------
Original Class: 1051 BROOKS ST
Encoded Label: 22
51
------------
Original Class: 757 MASSACHUSETTS AV
Encoded Label: 395
52
------------
Original Class: 4911 ROCKVILLE RD
Encoded Label: 292
53
------------
Original Class: 4310 BLUFF RD
Encoded Label: 259
54
------------
Original Class: 4100 N PENNSYLVANIA ST
Encoded Label: 245
55
------------
Original Class: 1216 KAPPES ST
Encoded Label: 41
56
------------
Original Class: 444 E NORTH ST
Encoded Label: 271
57
------------
Original Class: 5201 E 10TH ST
Encoded Label: 314
58
------------
Original Class: 2210 E 46TH ST
Encoded Label: 122
59
------------
Original Class: 3000 SHELBY ST
Encoded Label: 171
60
------------
Original Class: 5145 BOULEVARD PL
Encoded Label: 311
61
------------
Original Class: N DELAWARE ST & E NEW YORK ST
Encoded Label: 470
62
------------
Original Class: 254 S RITTER AV
Encoded Label: 151
63
------------
Original Class: 1301 BARTH AV
Encoded Label: 46
64
------------
Original Class: 402 W NEW YORK ST
Encoded Label: 241
65
------------
Original Class: 2444 E WASHINGTON ST
Encoded Label: 144
66
------------
Original Class: 6770 SPIRIT LAKE DR
Encoded Label: 377
67
------------
Original Class: W 16TH ST & N TIBBS AV
Encoded Label: 476
68
------------
Original Class: 5509 FURNAS CT
Encoded Label: 331
69
------------
Original Class: 649 JEFFERSON AV
Encoded Label: 366
70
------------
Original Class: 943 PROSPECT ST
Encoded Label: 452
71
------------
Original Class: 3400 BROUSE AV
Encoded Label: 197
72
------------
Original Class: 310 W MICHIGAN ST
Encoded Label: 175
73
------------
Original Class: 802 N MERIDIAN ST
Encoded Label: 406
74
------------
Original Class: 3805 N GERMAN CHURCH RD
Encoded Label: 224
75
------------
Original Class: S MERIDIAN ST & W GEORGIA ST
Encoded Label: 474
76
------------
Original Class: 2226 E 67TH ST
Encoded Label: 126
77
------------
Original Class: 308 N IRWIN ST
Encoded Label: 173
78
------------
Original Class: 6771 EAGLE POINTE DR S
Encoded Label: 379
79
------------
Original Class: 500 LOCKERBIE ST
Encoded Label: 301
80
------------
Original Class: 795 W WALNUT ST
Encoded Label: 404
81
------------
Original Class: 735 CANAL CT
Encoded Label: 389
82
------------
Original Class: 10106 ELLIS DR
Encoded Label: 11
83
------------
Original Class: 7644 ALLENWOOD CR
Encoded Label: 397
84
------------
Original Class: 930 W 10TH ST
Encoded Label: 447
85
------------
Original Class: N ALABAMA ST & E NEW YORK ST
Encoded Label: 469
86
------------
Original Class: 441 N MERIDIAN ST
Encoded Label: 269
87
------------
Original Class: 310 W MICHIGAN ST
Encoded Label: 175
88
------------
Original Class: 1075 BROOKS ST
Encoded Label: 23
89
------------
Original Class: PIERSON ST & TIPPECANOE ST
Encoded Label: 471
90
------------
Original Class: 359 WORCESTER AV
Encoded Label: 212
91
------------
Original Class: 310 W MICHIGAN ST
Encoded Label: 175
92
------------
Original Class: 620 N CAPITOL AV
Encoded Label: 355
93
------------
Original Class: 9325 RACQUET BALL LN
Encoded Label: 450
94
------------
Original Class: 531 VIRGINIA AV
Encoded Label: 321
95
------------
Original Class: 5035 WILDFLOWER CT
Encoded Label: 304
96
------------
Original Class: 2203 N PENNSYLVANIA ST
Encoded Label: 121
97
------------
Original Class: 1611 BROADWAY ST
Encoded Label: 79
98
------------
Original Class: 426 WALLACE AV
Encoded Label: 254
99
------------
Original Class: 809 IOWA ST
Encoded Label: 411
100
------------
Original Class: 3355 W MICHIGAN ST
Encoded Label: 195
101
------------
Original Class: 1611 BROADWAY ST
Encoded Label: 79
102
------------
Original Class: 50 W WASHINGTON ST
Encoded Label: 300
103
------------
Original Class: 821 DR M L KING JR ST
Encoded Label: 414
104
------------
Original Class: E 38TH ST & N POST RD
Encoded Label: 464
105
------------
Original Class: 6501 W MILLS AV
Encoded Label: 367
106
------------
Original Class: 7233 TWIN OAKS DR
Encoded Label: 384
107
------------
Original Class: 40 E ST CLAIR ST
Encoded Label: 233
108
------------
Original Class: 20 N MERIDIAN ST
Encoded Label: 103
109
------------
Original Class: 1455 N NEW JERSEY ST
Encoded Label: 65
110
------------
Original Class: 140 W WASHINGTON ST
Encoded Label: 57
111
------------
Original Class: 5662 GEORGETOWN RD
Encoded Label: 334
112
------------
Original Class: 7715 PERSHING RD
Encoded Label: 398
113
------------
Original Class: 8821 NAVIGATOR DR
Encoded Label: 428
114
------------
Original Class: 2502 DR MARTIN LUTHER KIN ST
Encoded Label: 148
115
------------
Original Class: 136 E ST JOSEPH ST
Encoded Label: 54
116
------------
Original Class: 7802 BROOKVILLE RD
Encoded Label: 399
117
------------
Original Class: 1744 W 74TH PL
Encoded Label: 96
118
------------
Original Class: 535 W MICHIGAN ST
Encoded Label: 323
119
------------
Original Class: 40 JENNY LN
Encoded Label: 234
120
------------
Original Class: 821 N LASALLE ST
Encoded Label: 415
121
------------
Original Class: 151 S EAST ST
Encoded Label: 69
122
------------
Original Class: 5725 W 43RD ST
Encoded Label: 336
123
------------
Original Class: 2419 LARMAN DR
Encoded Label: 140
124
------------
Original Class: 11025 SEDLAK LN
Encoded Label: 26
125
------------
Original Class: 6701 DOVER RD
Encoded Label: 374
126
------------
Original Class: 5247 LUZZANE LN
Encoded Label: 316
127
------------
Original Class: 3850 N COLLEGE AVE
Encoded Label: 228
128
------------
Original Class: 739 W 11TH ST
Encoded Label: 390
129
------------
Original Class: 3718 ARBOR GREEN WY
Encoded Label: 219
130
------------
Original Class: 5950 WINTHROP AV
Encoded Label: 342
131
------------
Original Class: 25 W 16TH ST
Encoded Label: 147
132
------------
Original Class: 1630 DRAPER ST
Encoded Label: 84
133
------------
Original Class: 1201 INDIANA AV
Encoded Label: 38
134
------------
Original Class: 1540 LANCASHIRE CT
Encoded Label: 75
135
------------
Original Class: 79 MYRON AV
Encoded Label: 402
136
------------
Original Class: 2419 LARMAN DR
Encoded Label: 140
137
------------
Original Class: 1412 SHELBY ST
Encoded Label: 60
138
------------
Original Class: 536 N ORIENTAL ST
Encoded Label: 324
139
------------
Original Class: 216 CLEVELAND ST
Encoded Label: 117
140
------------
Original Class: 402 E 56TH ST
Encoded Label: 238
141
------------
Original Class: 229 S DELAWARE ST
Encoded Label: 132
142
------------
Original Class: 6543 DORIS DR
Encoded Label: 369
143
------------
Original Class: 1 W 26TH ST
Encoded Label: 1
144
------------
Original Class: 401 E MICHIGAN ST
Encoded Label: 235
145
------------
Original Class: 3930 HOYT AV
Encoded Label: 230
146
------------
Original Class: 2810 COLLIER ST
Encoded Label: 164
147
------------
Original Class: 6701 N COLLEGE AV
Encoded Label: 375
148
------------
Original Class: 4755 KINGSWAY DR
Encoded Label: 284
149
------------
Original Class: 8909 WALLSTREET DR
Encoded Label: 429
150
------------
Original Class: 3217 WINTON AV
Encoded Label: 181
151
------------
Original Class: 912 N BOLTON AV
Encoded Label: 438
152
------------
Original Class: 333 E WESTFIELD BL
Encoded Label: 192
153
------------
Original Class: 530 E OHIO ST
Encoded Label: 320
154
------------
Original Class: 3916 LINCOLN RD
Encoded Label: 229
155
------------
Original Class: 902 E PLEASANT RUN PKWY N DR
Encoded Label: 432
156
------------
Original Class: 846 WOODRUFF PL MIDDLE DR
Encoded Label: 423
157
------------
Original Class: E 24TH ST & N DELAWARE ST
Encoded Label: 461
158
------------
Original Class: 4218 GUILFORD AV
Encoded Label: 252
159
------------
Original Class: 1650 HOEFGEN ST
Encoded Label: 88
160
------------
Original Class: 930 W 10TH ST
Encoded Label: 447
161
------------
Original Class: 3553 FOUNDERS RD
Encoded Label: 210
162
------------
Original Class: 49 W MARYLAND ST
Encoded Label: 289
163
------------
Original Class: 201 S CAPITOL AV
Encoded Label: 106
164
------------
Original Class: 3824 N ILLINOIS ST
Encoded Label: 226
165
------------
Original Class: 3221 BABSON CT
Encoded Label: 184
166
------------
Original Class: 4842 WINTHROP AV
Encoded Label: 287
167
------------
Original Class: 932 N CALIFORNIA ST
Encoded Label: 449
168
------------
Original Class: 6140 E 21ST ST
Encoded Label: 354
169
------------
Original Class: 5141 W 33RD ST
Encoded Label: 310
170
------------
Original Class: 800 N MERIDIAN ST
Encoded Label: 405
171
------------
Original Class: 3548 W 11TH ST
Encoded Label: 209
172
------------
Original Class: 4927 BROADWAY ST
Encoded Label: 296
173
------------
Original Class: 3965 N MERIDIAN ST
Encoded Label: 232
174
------------
Original Class: 2020 N DELAWARE ST
Encoded Label: 110
175
------------
Original Class: 537 E 39TH ST
Encoded Label: 325
176
------------
Original Class: 3200 COLD SPRING RD
Encoded Label: 180
177
------------
Original Class: 2013 W MICHIGAN ST
Encoded Label: 109
178
------------
Original Class: 6701 N COLLEGE AV
Encoded Label: 375
179
------------
Original Class: 2450 E 71ST ST
Encoded Label: 146
180
------------
Original Class: 6219 GUILFORD AV
Encoded Label: 357
181
------------
Original Class: 1201 INDIANA AV
Encoded Label: 38
182
------------
Original Class: 6219 GUILFORD AV
Encoded Label: 357
183
------------
Original Class: 3657 N CAPITOL AV
Encoded Label: 216
184
------------
Original Class: 430 N PARK AV
Encoded Label: 258
185
------------
Original Class: 5547 WINTHROP AV
Encoded Label: 332
186
------------
Original Class: 711 E WASHINGTON ST
Encoded Label: 382
187
------------
Original Class: 7847 MARQUIS LN
Encoded Label: 401
188
------------
Original Class: 1125 BROOKSIDE AV
Encoded Label: 29
189
------------
Original Class: 1510 STADIUM WA
Encoded Label: 70
190
------------
Original Class: 525 DORMAN ST
Encoded Label: 317
191
------------
Original Class: 201 W 38TH ST
Encoded Label: 107
192
------------
Original Class: 910 W 10TH ST
Encoded Label: 434
193
------------
Original Class: 2716 E WASHINGTON ST
Encoded Label: 159
194
------------
Original Class: 4643 LYNNFIELD RD
Encoded Label: 279
195
------------
Original Class: 31 W OHIO ST
Encoded Label: 174
196
------------
Original Class: 520 E MARKET ST
Encoded Label: 312
197
------------
Original Class: 40 E ST CLAIR ST
Encoded Label: 233
198
------------
Original Class: 920 N ALABAMA ST
Encoded Label: 441
199
------------
Original Class: 1630 N MERIDIAN ST
Encoded Label: 85
200
------------
Original Class: 401 W MERIDIAN SCHOOL RD
Encoded Label: 237
201
------------
Original Class: 1836 E 10TH ST
Encoded Label: 100
202
------------
Original Class: 3601 W 16TH ST
Encoded Label: 213
203
------------
Original Class: 100 S CAPITOL AV
Encoded Label: 4
204
------------
Original Class: W GEORGIA ST & S ILLINOIS ST
Encoded Label: 481
205
------------
Original Class: 401 E MICHIGAN ST
Encoded Label: 235
206
------------
Original Class: 6127 COMPTON ST
Encoded Label: 353
207
------------
Original Class: 725 MASSACHUSETTS AV
Encoded Label: 386
208
------------
Original Class: 10 S WEST ST
Encoded Label: 3
209
------------
Original Class: 4037 N COLLEGE AV
Encoded Label: 243
210
------------
Original Class: 1 W MARYLAND ST
Encoded Label: 2
211
------------
Original Class: 100 S MERIDIAN ST
Encoded Label: 5
212
------------
Original Class: 3515 E 10TH ST
Encoded Label: 205
213
------------
Original Class: 2441 STUART ST
Encoded Label: 143
214
------------
Original Class: 2120 W MORRIS ST
Encoded Label: 113
215
------------
Original Class: 6645 SUNNY LN
Encoded Label: 372
216
------------
Original Class: 6200 CARROLLTON AV
Encoded Label: 356
217
------------
Original Class: 520 N DEARBORN ST
Encoded Label: 313
218
------------
Original Class: 1505 MINOCQUA ST
Encoded Label: 68
219
------------
Original Class: 5709 W WASHINGTON ST
Encoded Label: 335
220
------------
Original Class: 4361 DECLARATION DR
Encoded Label: 263
221
------------
Original Class: 1001 N RURAL ST
Encoded Label: 6
222
------------
Original Class: 3518 MONNINGER DR
Encoded Label: 206
223
------------
Original Class: 4039 BOULEVARD PL
Encoded Label: 244
224
------------
Original Class: 636 N LINWOOD AV
Encoded Label: 363
225
------------
Original Class: 1145 S CAPITOL AV
Encoded Label: 33
226
------------
Original Class: 55 KOWEBA LN
Encoded Label: 329
227
------------
Original Class: 1022 HOSBROOK ST
Encoded Label: 17
228
------------
Original Class: 2126 E MICHIGAN ST
Encoded Label: 116
229
------------
Original Class: 3217 WINTON AV
Encoded Label: 181
230
------------
Original Class: 5251 KENTUCKY AV
Encoded Label: 318
231
------------
Original Class: 1309 N PENNSYLVANIA ST
Encoded Label: 50
232
------------
Original Class: 1532 E 10TH ST
Encoded Label: 73
233
------------
Original Class: 1435 N ILLINOIS ST
Encoded Label: 64
234
------------
Original Class: 2502 N SHERMAN DR
Encoded Label: 149
235
------------
Original Class: 3527 N COLLEGE AV
Encoded Label: 207
236
------------
Original Class: 109 S BRADLEY AV
Encoded Label: 24
237
------------
Original Class: 1514 SHEPARD ST
Encoded Label: 71
238
------------
Original Class: 3741 LA FONTAINE CT
Encoded Label: 221
239
------------
Original Class: 4611 N COLLEGE AV
Encoded Label: 277
240
------------
Original Class: 100 S CAPITOL AV
Encoded Label: 4
241
------------
Original Class: 7509 RUSKIN PL
Encoded Label: 394
242
------------
Original Class: E OHIO ST & N PENNSYLVANIA ST
Encoded Label: 468
243
------------
Original Class: 6340 GATEWAY DR
Encoded Label: 362
244
------------
Original Class: 804 BLAKE ST
Encoded Label: 409
245
------------
Original Class: 3409 N HIGH SCHOOL RD
Encoded Label: 199
246
------------
Original Class: 921 E WASHINGTON ST
Encoded Label: 443
247
------------
Original Class: 2503 N NEW JERSEY ST
Encoded Label: 150
248
------------
Original Class: 245 N DELAWARE ST
Encoded Label: 145
249
------------
Original Class: W 10TH ST & N CENTENNIAL ST
Encoded Label: 475
250
------------
Original Class: 4328 FOLTZ ST
Encoded Label: 261
251
------------
Original Class: 3640 GREEN ASH CT
Encoded Label: 215
252
------------
Original Class: 2171 AVONDALE PL
Encoded Label: 119
253
------------
Original Class: 441 E OHIO ST
Encoded Label: 268
254
------------
Original Class: 1628 YANDES ST
Encoded Label: 83
255
------------
Original Class: 1247 S KIEL AVE
Encoded Label: 44
256
------------
Original Class: 6424 JOHNWES RD
Encoded Label: 364
257
------------
Original Class: 411 RUSKIN PL
Encoded Label: 248
258
------------
Original Class: 315 KOEHNE ST
Encoded Label: 178
259
------------
Original Class: 3358 HEATHER RIDGE DR
Encoded Label: 196
260
------------
Original Class: 322 CANAL WALK
Encoded Label: 183
261
------------
Original Class: E 10TH ST & N LASALLE ST
Encoded Label: 459
262
------------
Original Class: 9201 RUSHMORE BLVD
Encoded Label: 442
263
------------
Original Class: 4655 LYNNFIELD RD
Encoded Label: 280
264
------------
Original Class: 1540 E TABOR ST
Encoded Label: 74
265
------------
Original Class: 2831 EARLSWOOD LN
Encoded Label: 167
266
------------
Original Class: 3242 N ARSENAL AV
Encoded Label: 185
267
------------
Original Class: 5103 E 16TH ST
Encoded Label: 306
268
------------
Original Class: 66 N TREMONT ST
Encoded Label: 370
269
------------
Original Class: 2310 UNION ST
Encoded Label: 133
270
------------
Original Class: 2211 PROSPECT ST
Encoded Label: 123
271
------------
Original Class: 3264 VALLEY FARMS WY
Encoded Label: 187
272
------------
Original Class: 6659 PEMBRIDGE WY
Encoded Label: 373
273
------------
Original Class: 8536 LENNA CT
Encoded Label: 425
274
------------
Original Class: 7827 SOUTHFIELD DR
Encoded Label: 400
275
------------
Original Class: 9559 MERCURY DR
Encoded Label: 455
276
------------
Original Class: 430 INDIANA AV
Encoded Label: 256
277
------------
Original Class: 2821 E 13TH ST
Encoded Label: 165
278
------------
Original Class: 4926 MELROSE AV
Encoded Label: 295
279
------------
Original Class: 1252 RINGGOLD AV
Encoded Label: 45
280
------------
Original Class: 4846 PENDRAGON BL
Encoded Label: 288
281
------------
Original Class: 2401 SHELBY ST
Encoded Label: 136
282
------------
Original Class: 7186 TWIN OAKS DR
Encoded Label: 383
283
------------
Original Class: 3805 MILL CROSSING DR
Encoded Label: 223
284
------------
Original Class: 1035 BROAD RIPPLE AV
Encoded Label: 19
285
------------
Original Class: 6723 W 16TH ST
Encoded Label: 376
286
------------
Original Class: 7321 FREEPORT LN
Encoded Label: 387
287
------------
Original Class: 1150 N WHITE RIVER PKWY W DR
Encoded Label: 35
288
------------
Original Class: 1627 N COLORADO AV
Encoded Label: 82
289
------------
Original Class: 401 N ILLINOIS ST
Encoded Label: 236
290
------------
Original Class: 9257 COLLEGE DR
Encoded Label: 445
291
------------
Original Class: 5853 LIBERTY CREEK N DR
Encoded Label: 340
292
------------
Original Class: 5550 EDGEWOOD TRACE BL
Encoded Label: 333
293
------------
Original Class: 1201 INDIANA AV
Encoded Label: 38
294
------------
Original Class: 2642 N HARDING ST
Encoded Label: 155
295
------------
Original Class: 2725 BRAXTON DR
Encoded Label: 160
296
------------
Original Class: 1301 RINGGOLD AV
Encoded Label: 47
297
------------
Original Class: 7935 HOSTA DR
Encoded Label: 403
298
------------
Original Class: 1225 HIATT ST
Encoded Label: 42
299
------------
Original Class: 4400 N HIGH SCHOOL RD
Encoded Label: 267
300
------------
Original Class: 6780 W WASHINGTON ST
Encoded Label: 380
301
------------
Original Class: 3300 N MERIDIAN ST
Encoded Label: 190
302
------------
Original Class: 1018 S SHEFFIELD AV
Encoded Label: 14
303
------------
Original Class: 28 PARKVIEW AV
Encoded Label: 163
304
------------
Original Class: 9016 SARATOGA DR
Encoded Label: 431
305
------------
Original Class: W 29TH ST & DR MARTIN LUTHER KIN ST
Encoded Label: 478
306
------------
Original Class: 3260 WELLINGTON AV
Encoded Label: 186
307
------------
Original Class: 3131 WINTERSONG DR
Encoded Label: 177
308
------------
Original Class: 1044 S ADDISON ST
Encoded Label: 20
309
------------
Original Class: 4701 CRESTVIEW AV
Encoded Label: 281
310
------------
Original Class: 5022 EMERTON PL
Encoded Label: 303
311
------------
Original Class: 1201 INDIANA AV
Encoded Label: 38
312
------------
Original Class: 4138 RICHELIEU RD
Encoded Label: 251
313
------------
Original Class: E NEW YORK ST & N RURAL ST
Encoded Label: 467
314
------------
Original Class: 3730 RUCKLE ST
Encoded Label: 220
315
------------
Original Class: 1924 HOUSTON ST
Encoded Label: 102
316
------------
Original Class: 1729 E TABOR ST
Encoded Label: 94
317
------------
Original Class: 50 W FALL CREEK PKWY N DR
Encoded Label: 299
318
------------
Original Class: 2121 W MICHIGAN ST
Encoded Label: 114
319
------------
Original Class: 6243 E WASHINGTON ST
Encoded Label: 359
320
------------
Original Class: 402 S LASALLE ST
Encoded Label: 239
321
------------
Original Class: 6510 E 25TH ST
Encoded Label: 368
322
------------
Original Class: 3437 LAFAYETTE RD
Encoded Label: 201
323
------------
Original Class: BARNOR DR & WINDSOR DR
Encoded Label: 458
324
------------
Original Class: 805 BELHAVEN DR
Encoded Label: 410
325
------------
Original Class: 4701 N KEYSTONE AV
Encoded Label: 282
326
------------
Original Class: 514 N CHESTER AV
Encoded Label: 309
327
------------
Original Class: 1325 LINDEN ST
Encoded Label: 52
328
------------
Original Class: 1741 E TERRACE AV
Encoded Label: 95
329
------------
Original Class: 3612 E WASHINGTON ST
Encoded Label: 214
330
------------
Original Class: 1216 KAPPES ST
Encoded Label: 41
331
------------
Original Class: 1701 BROADWAY ST
Encoded Label: 90
332
------------
Original Class: 1409 WADE ST
Encoded Label: 59
333
------------
Original Class: 6340 GATEWAY DR
Encoded Label: 362
334
------------
Original Class: 310 W MICHIGAN ST
Encoded Label: 175
335
------------
Original Class: 4813 OAKWOOD TR
Encoded Label: 285
336
------------
Original Class: 9926 CATALINA DR
Encoded Label: 457
337
------------
Original Class: 101 W WASHINGTON ST
Encoded Label: 10
338
------------
Original Class: 627 DELRAY DR
Encoded Label: 360
339
------------
Original Class: PROSPECT ST & S RANDOLPH ST
Encoded Label: 472
340
------------
Original Class: 1122 N DEARBORN ST
Encoded Label: 27
341
------------
Original Class: 1525 SPRUCE ST
Encoded Label: 72
342
------------
Original Class: 911 E 44TH ST
Encoded Label: 436
343
------------
Original Class: 4457 N AUDUBON RD
Encoded Label: 272
344
------------
Original Class: 3466 FALL CREEK PKWY N DR
Encoded Label: 203
345
------------
Original Class: 2922 S HOLT RD
Encoded Label: 169
346
------------
Original Class: 50 S KOWEBA LN
Encoded Label: 298
347
------------
Original Class: 750 N LIVINGSTON AV
Encoded Label: 392
348
------------
Original Class: 411 N GRAY ST
Encoded Label: 247
349
------------
Original Class: 40 E ST CLAIR ST
Encoded Label: 233
350
------------
Original Class: 10018 RIDGEFIELD DR
Encoded Label: 7
351
------------
Original Class: 956 N KEALING AV
Encoded Label: 456
352
------------
Original Class: 242 E MARKET ST
Encoded Label: 141
353
------------
Original Class: 2100 E 10TH ST
Encoded Label: 111
354
------------
Original Class: 7342 HEARTHSTONE WY
Encoded Label: 388
355
------------
Original Class: 5230 W 30TH ST
Encoded Label: 315
356
------------
Original Class: 4948 W RAYMOND ST
Encoded Label: 297
357
------------
Original Class: 40 E ST CLAIR ST
Encoded Label: 233
358
------------
Original Class: W 25TH ST & DR M L KING JR ST
Encoded Label: 477
359
------------
Original Class: 1707 N AUDUBON RD
Encoded Label: 92
360
------------
Original Class: W 38TH ST & N CAPITOL AV
Encoded Label: 480
361
------------
Original Class: 6102 N RURAL ST
Encoded Label: 350
362
------------
Original Class: 1500 N RITTER AVENUE
Encoded Label: 66
363
------------
Original Class: 11857 SERENITY LN
Encoded Label: 37
364
------------
Original Class: 1201 INDIANA AV
Encoded Label: 38
365
------------
Original Class: 1306 S SHEFFIELD AV
Encoded Label: 48
366
------------
Original Class: 1316 BRANDT DR
Encoded Label: 51
367
------------
Original Class: 820 W 42ND ST
Encoded Label: 412
368
------------
Original Class: 2725 W WASHINGTON ST
Encoded Label: 161
369
------------
Original Class: 603 HOLLY AV
Encoded Label: 346
370
------------
Original Class: 7236 W 10TH ST
Encoded Label: 385
371
------------
Original Class: 3218 ELMHURST DR
Encoded Label: 182
372
------------
Original Class: 51 N HAWTHORNE LN
Encoded Label: 305
373
------------
Original Class: 4702 CRESTVIEW AV
Encoded Label: 283
374
------------
Original Class: 105 N ELDER AV
Encoded Label: 21
375
------------
Original Class: 2121 W MICHIGAN ST
Encoded Label: 114
376
------------
Original Class: 2822 E WASHINGTON ST
Encoded Label: 166
377
------------
Original Class: 1660 BELLEFONTAINE ST
Encoded Label: 89
378
------------
Original Class: 8955 US 31 S
Encoded Label: 430
379
------------
Original Class: 440 W 38TH ST
Encoded Label: 266
380
------------
Original Class: 8441 ATHENS CT
Encoded Label: 419
381
------------
Original Class: 8253 HARCOURT RD
Encoded Label: 417
382
------------
Original Class: 2013 BELLEFONTAINE ST
Encoded Label: 108
383
------------
Original Class: 5132 W 33RD ST
Encoded Label: 308
384
------------
Original Class: 1611 N MERIDIAN ST
Encoded Label: 80
385
------------
Original Class: 1631 N ROCHESTER AV
Encoded Label: 86
386
------------
Original Class: 9101 W 10TH ST
Encoded Label: 435
387
------------
Original Class: 9025 RUSHMORE BL
Encoded Label: 433
388
------------
Original Class: 2713 N PARKER AV
Encoded Label: 158
389
------------
Original Class: 4023 E 10TH ST
Encoded Label: 242
390
------------
Original Class: 2407 APPLETON DR
Encoded Label: 139
391
------------
Original Class: 1433 E VERMONT ST
Encoded Label: 63
392
------------
Original Class: 2104 N CAPITOL AV
Encoded Label: 112
393
------------
Original Class: 952 N DENNY ST
Encoded Label: 454
394
------------
Original Class: 1236 W SUMNER AV
Encoded Label: 43
395
------------
Original Class: 611 N CAPITOL AV
Encoded Label: 351
396
------------
Original Class: 1201 INDIANA AV
Encoded Label: 38
397
------------
Original Class: 3530 HYANNIS PORT DR
Encoded Label: 208
398
------------
Original Class: 9130 MATTERHORN RD
Encoded Label: 439
399
------------
Original Class: 2221 FAIRFAX RD
Encoded Label: 125
400
------------
Original Class: 3931 N VINEWOOD AV
Encoded Label: 231
401
------------
Original Class: 821 DR M L K
Encoded Label: 413
402
------------
Original Class: 273 LEEDS AV
Encoded Label: 162
403
------------
Original Class: 450 E NORTH ST
Encoded Label: 274
404
------------
Original Class: 438 N TEMPLE AV
Encoded Label: 264
405
------------
Original Class: 3826 ARBOR GREEN LN
Encoded Label: 227
406
------------
Original Class: 3217 WINTON AV
Encoded Label: 181
407
------------
Original Class: 2438 E WASHINGTON ST
Encoded Label: 142
408
------------
Original Class: 257 S GRAY ST
Encoded Label: 152
409
------------
Original Class: 2405 MADISON AV
Encoded Label: 138
410
------------
Original Class: 6302 MONTEO LN
Encoded Label: 361
411
------------
Original Class: 1138 S DREXEL AVE
Encoded Label: 31
412
------------
Original Class: 139 S BUTLER AV
Encoded Label: 56
413
------------
Original Class: 1550 N CUMBERLAND RD
Encoded Label: 76
414
------------
Original Class: 3435 W 16TH ST
Encoded Label: 200
415
------------
Original Class: 1006 HARDIN BL
Encoded Label: 8
416
------------
Original Class: E 38TH ST & N MITTHOEFER RD
Encoded Label: 463
417
------------
Original Class: 3152 GERRARD AV
Encoded Label: 179
418
------------
Original Class: 3572 S PENNSYLVANIA ST
Encoded Label: 211
419
------------
Original Class: 440 N TEMPLE AV
Encoded Label: 265
420
------------
Original Class: 225 S PARKER AV
Encoded Label: 131
421
------------
Original Class: 334 MASSACHUSETTS AV
Encoded Label: 193
422
------------
Original Class: 2934 N DEARBORN ST
Encoded Label: 170
423
------------
Original Class: 5416 UNITY TR
Encoded Label: 327
424
------------
Original Class: 2710 E NORTH ST
Encoded Label: 157
425
------------
Original Class: 1027 N KEYSTONE AV
Encoded Label: 18
426
------------
Original Class: 4466 CENTRAL AV
Encoded Label: 273
427
------------
Original Class: 3508 OPERA PL
Encoded Label: 204
428
------------
Original Class: 217 ROCKSHIRE RD
Encoded Label: 118
429
------------
Original Class: 4909 MOUNT VERNON DR
Encoded Label: 291
430
------------
Original Class: 6617 E WASHINGTON ST
Encoded Label: 371
431
------------
Original Class: 5314 OHMER AV
Encoded Label: 322
432
------------
Original Class: 1018 E 91ST ST
Encoded Label: 13
433
------------
Original Class: 8459 SMITHFIELD LN
Encoded Label: 420
434
------------
Original Class: 310 W MICHIGAN ST
Encoded Label: 175
435
------------
Original Class: 2201 E 10TH ST
Encoded Label: 120
436
------------
Original Class: 4120 KALMAR DR
Encoded Label: 250
437
------------
Original Class: E 38TH ST & N MERIDIAN ST
Encoded Label: 462
438
------------
Original Class: 223 N LAWNDALE AV
Encoded Label: 128
439
------------
Original Class: 8705 E 38TH ST
Encoded Label: 427
440
------------
Original Class: 1605 N SHADELAND AV
Encoded Label: 78
441
------------
Original Class: E MICHIGAN ST & N TACOMA AV
Encoded Label: 466
442
------------
Original Class: 520 E MARKET ST
Encoded Label: 312
443
------------
Original Class: 1051 BROOKS ST
Encoded Label: 22
444
------------
Original Class: 1750 ASBURY ST
Encoded Label: 98
445
------------
Original Class: 1432 HOYT AV
Encoded Label: 62
446
------------
Original Class: 8625 JAMAICA CT
Encoded Label: 426
447
------------
Original Class: 4921 ORION AV
Encoded Label: 293
448
------------
Original Class: 1877 S EAST ST
Encoded Label: 101
449
------------
Original Class: 329 W 41ST ST
Encoded Label: 189
450
------------
Original Class: 4107 BRENTWOOD DR
Encoded Label: 246
451
------------
Original Class: 69 N IRVINGTON AV
Encoded Label: 381
452
------------
Original Class: 1801 NOWLAND AV
Encoded Label: 99
453
------------
Original Class: 1604 E 12TH ST
Encoded Label: 77
454
------------
Original Class: 3122 FISHER RD
Encoded Label: 176
455
------------
Original Class: 1018 ST PAUL ST
Encoded Label: 15
456
------------
Original Class: 3003 KESSLER BOULEVARD N DR
Encoded Label: 172
457
------------
Original Class: 930 W 10TH ST
Encoded Label: 447
458
------------
Original Class: 846 S BILTMORE AV
Encoded Label: 422
459
------------
Original Class: 2636 W MICHIGAN ST
Encoded Label: 153
460
------------
Original Class: 1152 LEXINGTON AV
Encoded Label: 36
461
------------
Original Class: 4242 MOLLER RD
Encoded Label: 253
462
------------
Original Class: 11225 FALL DR
Encoded Label: 28
463
------------
Original Class: 5122 WINTERBERRY DR
Encoded Label: 307
464
------------
Original Class: 6049 WINDSOR DR
Encoded Label: 348
465
------------
Original Class: 1503 N PENNSYLVANIA ST
Encoded Label: 67
466
------------
Original Class: 849 E PLEASANT RUN PKWY S DR
Encoded Label: 424
467
------------
Original Class: 2228 N HARDING ST
Encoded Label: 127
468
------------
Original Class: 5402 E TERRACE AV
Encoded Label: 326
469
------------
Original Class: 5957 DEVINGTON RD
Encoded Label: 343
470
------------
Original Class: 5820 E 82ND ST
Encoded Label: 338
471
------------
Original Class: 2316 HOYT AV
Encoded Label: 134
472
------------
Original Class: 6010 LAKEVIEW DR
Encoded Label: 345
473
------------
Original Class: 5420 E 38TH ST
Encoded Label: 328
474
------------
Original Class: E 96TH ST & N PENNSYLVANIA ST
Encoded Label: 465
475
------------
Original Class: 4902 E 12TH ST
Encoded Label: 290
476
------------
Original Class: 947 PROSPECT ST
Encoded Label: 453
477
------------
Original Class: 2902 E NORTH ST
Encoded Label: 168
478
------------
Original Class: 1 VIRGINIA AV
Encoded Label: 0
479
------------
Original Class: 412 CANAL CT S DR
Encoded Label: 249
480
------------
Original Class: 430 MASSACHUSETTS AV
Encoded Label: 257
481
------------
Original Class: 2001 W WASHINGTON ST
Encoded Label: 105
482
------------
Original Class: 2340 HILLSIDE AV
Encoded Label: 135
483
------------
Original Class: 4023 E 10TH ST
Encoded Label: 242
484
------------
Original Class: 1622 E PALMER ST
Encoded Label: 81
485
------------
Original Class: 3340 CARROLLTON AV
Encoded Label: 194
486
------------
Original Class: 911 VILLA AV
Encoded Label: 437
487
------------
Original Class: 575 N PENNSYLVANIA ST
Encoded Label: 337
488
------------
Original Class: 2638 COLLIER ST
Encoded Label: 154
489
------------
Original Class: 1409 FLETCHER AV
Encoded Label: 58
490
------------
Original Class: 8223 BARRY RD
Encoded Label: 416
491
------------
Original Class: 933 S KIEL AV
Encoded Label: 451
492
------------
Original Class: 3402 E 10TH ST
Encoded Label: 198
493
------------
Original Class: 520 E MARKET ST
Encoded Label: 312
494
------------
Original Class: 4317 E WASHINGTON ST
Encoded Label: 260
495
------------
Original Class: 245 N DELAWARE ST
Encoded Label: 145
496
------------
Original Class: 2245 SHELBY ST
Encoded Label: 129
497
------------
Original Class: 402 S RURAL ST
Encoded Label: 240
498
------------
Original Class: S CAPITOL AV & W MARYLAND ST
Encoded Label: 473
499
------------
Original Class: W 30TH ST & GEORGETOWN RD
Encoded Label: 479
500
------------
Original Class: 4846 PENDRAGON BL
Encoded Label: 288
501
------------
Original Class: 1017 N TEMPLE AV
Encoded Label: 12
502
------------
Original Class: 3440 W 12TH ST
Encoded Label: 202
503
------------
Original Class: 2122 E 10TH ST
Encoded Label: 115
504
------------
Original Class: 4625 MELROSE AV
Encoded Label: 278
505
------------
Original Class: 4821 E WASHINGTON ST
Encoded Label: 286
506
------------
Original Class: 6447 W WASHINGTON ST
Encoded Label: 365
507
------------
Original Class: E NEW YORK ST & N RURAL ST
Encoded Label: 467
508
------------
Original Class: 803 W 10TH ST
Encoded Label: 408
509
------------
Original Class: 530 E OHIO ST
Encoded Label: 320
510
------------
Original Class: 3303 W 10TH ST
Encoded Label: 191
511
------------
Original Class: 4755 KINGSWAY DR
Encoded Label: 284
512
------------
Original Class: 1127 N TACOMA AV
Encoded Label: 30
513
------------
Original Class: 4609 E WASHINGTON ST
Encoded Label: 276
514
------------
Original Class: 4923 ROCKVILLE RD
Encoded Label: 294
515
------------
Original Class: 1148 SHARON AV
Encoded Label: 34
516
------------
Original Class: 366 WICHSER AV
Encoded Label: 217
517
------------
Original Class: 2710 BETHEL AV
Encoded Label: 156
518
------------
Original Class: 6030 E 21ST ST
Encoded Label: 347
519
------------
###Markdown
Integratenew encoded data label asignmentsDistrictsBike ValueStreet address -----Will not be used Experiment value
###Code
disposition = encoded_y
moidfied_data_df.dtypes
# create datafrane for easy modeling
# dataframe only has features and one-hot encoding
features_df['disposition'] = disposition.tolist()
features_df['Bike Value'] = encoded_Bike_vale.tolist()
features_df['Street (Coded)'] = encoded_ADDRESS_value.tolist()
moidfied_data_df = features_df
moidfied_data_df
# needed for y componet for models (Decsion tree and randomforest)
# Decison tree was run to compare to random forest
# random forest first time: was run to identify top 10 featrues from originol 42 features
# ransom forest second time: ran with top 10 features
# ransom forest third time: ran with minMax scaler data.
target = moidfied_data_df["disposition"]
#target
# save to csv for other models
file = "raw_data_with_diposition.csv"
path_file = path + file
moidfied_data_df.to_csv(path_file)
moidfied_data_df
#Final data prep
moidfied_data_df = moidfied_data_df.drop(columns =["CRIME","ADDRESS"])
moidfied_data_df
###Output
_____no_output_____
###Markdown
Create a Train Test Split to run Decison Tree* Purpose is to idenentify top 10 features* Save final list with diposition value (0,1,2) as file: "Top_10_features_disposition_data.csv"* Random forest workd the bestUse `disposition` for the y values
###Code
# Create dataframe for running decision tree
# This was done to upload existing code with similar variables
# Main Data set: data_df
data_df = moidfied_data_df.drop(columns=["disposition"])
feature_names = data_df.columns
data_df.head()
###Output
_____no_output_____
###Markdown
Decision Tree
###Code
from sklearn.model_selection import train_test_split
X_train, X_test, y_train, y_test = train_test_split(data_df, target, random_state=42)
clf = tree.DecisionTreeClassifier()
clf = clf.fit(X_train, y_train)
clf.score(X_test, y_test)
###Output
_____no_output_____
###Markdown
Random Forest Decision Tree
###Code
from sklearn.ensemble import RandomForestClassifier
rf = RandomForestClassifier(n_estimators=200)
rf = rf.fit(X_train, y_train)
rf.score(X_test, y_test)
Featrues_by_importance = sorted(zip(rf.feature_importances_, feature_names), reverse=True)
Featrues_by_importance_df = pd.DataFrame(Featrues_by_importance)
top10_featrues = Featrues_by_importance_df.head(11)
top10_featrues
###Output
_____no_output_____
###Markdown
Don't Run after this point: Artifact code Create dataframes with opimized features and catogrical ouputs
###Code
# Master dataset with catagorical diposition
selected_features_df = moidfied_data_df[['DATE_year',
'DATE_month',
'DATE_week',
'DATE_day',
'Hour',
'Minute',
'Temp(K)',
'Pressure(atm)',
'wind',
'Street (Coded)',
'disposition']]
selected_features_df
# save to csv for other models
file = "Top_10_features_disposition_data.csv"
path_file = path + file
selected_features_df.to_csv(path_file)
#selected_features_df
# create features CSV with origianl koi_disposition for plug and play models
temp_df= selected_features_df.drop("disposition", axis=1)
temp_df['koi_disposition'] = y.tolist()
#temp_df
# save to csv for other models
file = "top_Ten_origianl_koi_disposition_data.csv"
path_file = path + file
temp_df.to_csv(path_file)
###Output
_____no_output_____
###Markdown
Create a Train Test SplitUse Decision tree and random forest with optimized features listUse `disposition` for the y values
###Code
data = selected_features_df.drop("disposition", axis=1)
feature_names = data.columns
data.head()
## Decision Tree
from sklearn.model_selection import train_test_split
X_train, X_test, y_train, y_test = train_test_split(data, target, random_state=42)
clf = tree.DecisionTreeClassifier()
clf = clf.fit(X_train, y_train)
clf.score(X_test, y_test)
## Random Decision Tree
from sklearn.ensemble import RandomForestClassifier
rf = RandomForestClassifier(n_estimators=200)
rf = rf.fit(X_train, y_train)
rf.score(X_test, y_test)
###Output
_____no_output_____
###Markdown
minMaxScaler* Use optimized features list* Use minMaxScaler to nomalized optimized featres list* Save as csv: minmaxscaled_disposition_data.csv
###Code
from sklearn.preprocessing import MinMaxScaler
scaler = MinMaxScaler()
# transform data
scaled = scaler.fit_transform(data)
#print(scaled)
scaled_df = pd.DataFrame(scaled)
scaled_df
# Rename the headers to match origanl headers--- not as nice
data_scaled_df = scaled_df.rename(columns={0:'DATE_year',
1:'DATE_month',
2:'DATE_week',
3:'DATE_day',
4:'Hour',
5:'Minute',
6:'Temp(K)',
7:'Pressure(atm)',
8:'wind',
9:'Street (Coded)',
})
# Use the this dataframe for testing. Does not have dipsoisiton
#data_scaled_df
# pack up clean datafram for csv
data_scaled_disposition_df = data_scaled_df
data_scaled_disposition_df['disposition'] = disposition.tolist()
#data_scaled_disposition_df
# save to csv for other models
file = "minmaxscaled_disposition_data.csv"
path_file = path + file
data_scaled_disposition_df.to_csv(path_file)
# RandomForest third time for fun
# use data_scaled_df and target for testing
from sklearn.model_selection import train_test_split
X_train, X_test, y_train, y_test = train_test_split(data_scaled_df, target, random_state=42)
from sklearn.ensemble import RandomForestClassifier
rf = RandomForestClassifier(n_estimators=200)
rf = rf.fit(X_train, y_train)
rf.score(X_test, y_test)
# Decsion tree model: for fun.
from sklearn.model_selection import train_test_split
X_train, X_test, y_train, y_test = train_test_split(data_scaled_df, target, random_state=42)
clf = tree.DecisionTreeClassifier()
clf = clf.fit(X_train, y_train)
clf.score(X_test, y_test)
###Output
_____no_output_____
|
TestScores.ipynb
|
###Markdown
High school test scores dataSince our correlation metrics -- as predicted -- did not show high enough significance between the GPA and the yield, we decided to add new features to our datasets.Fortunately, we have found a dataset with California SAT, ACT and AP test scores by year and high school: https://www.cde.ca.gov/ds/sp/ai/. The dataset consists of multiple `.xls` files grouped by: - year - test Moreover, the `.xls` files have different formats. Examples of differences include: - extra/missing columns - number of rows before the header - different column names We prepared the following functions to resolve these differences:
###Code
def read_multi_xls(prefix, start_year, end_year, skip_row_count=0):
data_frames = []
for year in range(start_year, end_year+1):
year_2dig = year % 100
df = pd.read_excel(prefix + str(year_2dig).zfill(2) + '.xls',
skiprows=list(range(skip_row_count)))
df['year'] = year
data_frames.append(df)
return data_frames
def import_multi_xls(prefix, start_year, end_year, skip_row_count, columns_to_remove, must_have_columns=[]):
data_frames = []
column_names = None
for year in range(start_year, end_year+1):
year_2dig = year % 100
df = pd.read_excel(prefix + str(year_2dig).zfill(2) + '.xls',
skiprows=list(range(skip_row_count[year])))
for c in columns_to_remove[year]:
del df[c]
# There are differences between column names, so we use the names from the first
# dataframe
if column_names is None:
column_names = list(df)
for col in must_have_columns:
if col not in list(df):
df.insert(column_names.index(col), col, np.nan)
new_column_names = list(df)
renamer = {}
for i in range(len(column_names)):
renamer[new_column_names[i]] = column_names[i]
df.rename(columns=renamer, inplace=True)
df['year'] = year
data_frames.append(df)
return pd.concat(data_frames)
###Output
_____no_output_____
###Markdown
AP ScoresThe first set of scores we will import is the AP scores. It turns out that the data between 1999 and 2013 has a similar format, and we should be able to perform a single `import_multi_xls()` call for all of it.
###Code
ap_99_13 = import_multi_xls('data/test_scores/ap/ap', 1999, 2013, {
1999: 2, 2000: 2, 2001: 2, 2002: 2, 2003: 2, 2004: 2, 2005: 2, 2006: 2, 2007: 2, 2008: 2,
2009: 3, 2010: 3, 2011: 3, 2012: 3, 2013: 3,
}, {
1999: [], 2000: [], 2001: [], 2002: [], 2003: [],
2004: ['Rate of Exams\nWith a Score of\n3 or Greater\nFor 12th Grade\nStudents', 'Rate of Exams\nWith a Score of\n3 or Greater\nFor 11th & 12th\nGrade Students'],
2005: [], 2006: [], 2007: [], 2008: [], 2009: [], 2010: ['Year'], 2011: [], 2012: [], 2013: [],
},
['Total\nNumber of\nExams Taken', 'Number\nof Exams\nWith a Score of\n3 or Greater']
)
ap_99_13.head()
###Output
_____no_output_____
###Markdown
The data from 2014 to 2016 is formatted differently, and we will need to import it separately:
###Code
ap_14_16_dfs = read_multi_xls('data/test_scores/ap/ap', 2014, 2016)
for df in ap_14_16_dfs:
# There are some lower/upper case differences in column names which causes problems
# while merging.
df.columns = map(str.lower, df.columns)
ap_14_16 = pd.concat(ap_14_16_dfs)
ap_14_16.head()
###Output
_____no_output_____
###Markdown
We will also extract the school number from the CDS number:
###Code
ap_14_16['school_num'] = pd.to_numeric(ap_14_16['cds'].astype(str).str[-7:])
ap_14_16.head()
###Output
_____no_output_____
###Markdown
Also, we want to drop the cumulative data:
###Code
ap_14_16 = ap_14_16[ap_14_16['rtype'] == 'S']
ap_14_16.head()
###Output
_____no_output_____
###Markdown
Next, we will rename and drop some of the columns in our dataframes.
###Code
ap_14_16.drop(columns=[
'cds', 'cname', 'dname', 'rtype', 'enroll1012'
], inplace=True)
ap_99_13.drop(columns=[
'County\nNumber', 'District\nNumber', 'County Name', 'District Name',
'Total\nNumber of\nExams Taken', 'Number\nof Exams\nWith a Score of\n3 or Greater',
'Grade 11\nEnrollment\n(October 1998\nCBEDS)',
'Grade 11+12\nEnrollment\n(October 1998\nCBEDS)',
], inplace=True)
ap_99_13_renamer = {
'School\nNumber': 'school_num',
'School Name': 'school_name',
'Number of\nAP Exam\nTakers': 'ap_num_test_takers',
'Number\nof Exams\nWith a Score of\n1': 'ap_num_scr1',
'Number\nof Exams\nWith a Score of\n2': 'ap_num_scr2',
'Number\nof Exams\nWith a Score of\n3': 'ap_num_scr3',
'Number\nof Exams\nWith a Score of\n4': 'ap_num_scr4',
'Number\nof Exams\nWith a Score of\n5': 'ap_num_scr5',
'Grade 12\nEnrollment\n(October 1998\nCBEDS)': 'enroll12',
'year': 'year',
}
ap_99_13.rename(columns=ap_99_13_renamer, inplace=True)
ap_14_16_renamer = {
'school_num': 'school_num',
'sname': 'school_name',
'numtsttakr': 'ap_num_test_takers',
'numscr1': 'ap_num_scr1',
'numscr2': 'ap_num_scr2',
'numscr3': 'ap_num_scr3',
'numscr4': 'ap_num_scr4',
'numscr5': 'ap_num_scr5',
'enroll12': 'enroll12',
'year': 'year',
}
ap_14_16.rename(columns=ap_14_16_renamer, inplace=True)
###Output
_____no_output_____
###Markdown
Finally, we merge the datasets
###Code
ap_scores = pd.concat([ap_99_13, ap_14_16])
###Output
_____no_output_____
###Markdown
All of our columns should have numeric values at this point (apart from the school_name). We will make pandas convert all values to numeric (and all non-number values to NaN)
###Code
school_names = np.copy(ap_scores['school_name'])
ap_scores = ap_scores.apply(pd.to_numeric, errors='coerce', axis=1)
ap_scores['school_name'] = school_names
###Output
_____no_output_____
###Markdown
The last step will be to modify the year column. Our main dataset uses the starting year of the academic year, whereas our `ap_scores` dataframe uses the ending year of the academic year.
###Code
ap_scores['year'] = ap_scores['year'] - 1
ap_scores
###Output
_____no_output_____
###Markdown
We are saving the processed data to a CSV file. This way we will not have to redo all of our computations.
###Code
ap_scores.to_csv('data/test_scores/ap/processed.csv', sep=',', index=False)
###Output
_____no_output_____
###Markdown
SAT scoresSimilarly to the AP scores, SAT scores are also split by year, and the .xls files have different formats. We will need to perform merging similar to the one in the *AP scores* section.
###Code
def import_sat_xls(prefix, start_year, end_year, columns, skip_row_count):
data_frames = []
for year in range(start_year, end_year+1):
year_2dig = year % 100
df = pd.read_excel(prefix + str(year_2dig).zfill(2) + '.xls',
skiprows=list(range(skip_row_count[year])))
df = df[columns]
df['year'] = year
data_frames.append(df)
return pd.concat(data_frames)
###Output
_____no_output_____
###Markdown
In 2006, SAT scores changed from having two categories (verbal, math) to three categories (reading, writing, math). This will make our format differ slightly.
###Code
sat_99_05 = import_sat_xls(
'data/test_scores/sat/sat', 1999, 2005,
['School\nNumber', 'Number\nof\nTakers', 'Average\nVerbal\nScore',
'Average\nMath\nScore', 'Average\nTotal\nScore'],
{
1999: 2, 2000: 2, 2001: 2, 2002: 2, 2003: 2,
2004: 2, 2005: 2,
}
)
sat_06_07 = import_sat_xls(
'data/test_scores/sat/sat', 2006, 2007,
['School\nNumber', 'Number\nof\nTakers', 'Average\nVerbal\nScore',
'Average\nMath\nScore', 'Average\nWriting\nScore', 'Average\nTotal\nScore'],
{
2006: 2, 2007: 2,
}
)
sat_08_10 = import_sat_xls(
'data/test_scores/sat/sat', 2008, 2010,
['School\nNumber', 'Number\nTested', '\nCritical Reading\nAverage',
'\nMath\nAverage', '\nWriting\nAverage', '\nTotal\nAverage'],
{
2008: 2, 2009: 3, 2010: 4
}
)
sat_11_13 = import_sat_xls(
'data/test_scores/sat/sat', 2011, 2013,
['School\nNumber', 'Number\nTested', 'V_Mean',
'M_Mean', 'W_Mean', 'Tot_Mean'],
{
2011: 3, 2012: 3, 2013: 3
}
)
sat_14_16 = pd.concat(read_multi_xls('data/test_scores/sat/sat', 2014, 2016))
###Output
_____no_output_____
###Markdown
We need to know the school number in the `sat_14_16` dataframe. We will derive it from the `cds` columns:
###Code
sat_14_16['school_num'] = pd.to_numeric(sat_14_16['cds'].astype('int64').astype(str).str[-7:])
###Output
_____no_output_____
###Markdown
In order to do some processing on the scores, we will convert the dataframes to a numeric format.
###Code
sat_14_16 = sat_14_16.apply(pd.to_numeric, errors='coerce', axis=1)
sat_11_13 = sat_11_13.apply(pd.to_numeric, errors='coerce', axis=1)
sat_08_10 = sat_08_10.apply(pd.to_numeric, errors='coerce', axis=1)
sat_99_05 = sat_99_05.apply(pd.to_numeric, errors='coerce', axis=1)
sat_06_07 = sat_06_07.apply(pd.to_numeric, errors='coerce', axis=1)
###Output
_____no_output_____
###Markdown
Now, we will merge the dataframes into a single dataframe. To keep the format constant between the years we will combine the *reading* and *writing* scores into a single *verbal* score. We will also modify the *total score* accordingly.
###Code
sat_06_07['sat_verbal_avg'] = (sat_06_07['Average\nWriting\nScore'] + sat_06_07['Average\nVerbal\nScore'])/2
sat_06_07.drop(columns=['Average\nWriting\nScore', 'Average\nVerbal\nScore'], inplace=True)
sat_08_10['sat_verbal_avg'] = (sat_08_10['\nWriting\nAverage'] + sat_08_10['\nCritical Reading\nAverage'])/2
sat_08_10.drop(columns=['\nWriting\nAverage', '\nCritical Reading\nAverage'], inplace=True)
sat_11_13['sat_verbal_avg'] = (sat_11_13['W_Mean'] + sat_11_13['V_Mean'])/2
sat_11_13.drop(columns=['W_Mean', 'V_Mean'], inplace=True)
sat_14_16['sat_verbal_avg'] = (sat_14_16['AvgScrRead'] + sat_14_16['AvgScrWrite'])/2
sat_14_16['sat_total_avg'] = (
sat_14_16['AvgScrRead'] +
sat_14_16['AvgScrWrite'] +
sat_14_16['AvgScrMath'])*(2/3)
sat_14_16 = sat_14_16[['school_num', 'NumTstTakr', 'sat_verbal_avg', 'AvgScrMath',
'sat_total_avg', 'year']]
sat_08_10['\nTotal\nAverage'] = sat_08_10['\nTotal\nAverage'] * (2/3)
sat_06_07['Average\nTotal\nScore'] = sat_06_07['Average\nTotal\nScore'] * (2/3)
sat_11_13['Tot_Mean'] = sat_11_13['Tot_Mean'] * (2/3)
sat_99_07_renamer = {
'School\nNumber': 'school_num',
'Number\nof\nTakers': 'sat_num_test_takers',
'Average\nVerbal\nScore': 'sat_verbal_avg',
'Average\nMath\nScore': 'sat_math_avg',
'Average\nTotal\nScore': 'sat_total_avg',
'year': 'year'
}
sat_08_10_renamer = {
'School\nNumber': 'school_num',
'Number\nTested': 'sat_num_test_takers',
'\nMath\nAverage': 'sat_math_avg',
'\nTotal\nAverage': 'sat_total_avg',
'sat_verbal_avg': 'sat_verbal_avg',
'year': 'year',
}
sat_11_13_renamer = {
'School\nNumber': 'school_num',
'Number\nTested': 'sat_num_test_takers',
'M_Mean': 'sat_math_avg',
'Tot_Mean': 'sat_total_avg',
'sat_verbal_avg': 'sat_verbal_avg',
'year': 'year',
}
sat_14_16_renamer = {
'NumTstTakr': 'sat_num_test_takers',
'AvgScrMath': 'sat_math_avg',
}
sat_99_05.rename(columns=sat_99_07_renamer, inplace=True)
sat_06_07.rename(columns=sat_99_07_renamer, inplace=True)
sat_08_10.rename(columns=sat_08_10_renamer, inplace=True)
sat_11_13.rename(columns=sat_11_13_renamer, inplace=True)
sat_14_16.rename(columns=sat_14_16_renamer, inplace=True)
sat_scores = pd.concat([sat_99_05, sat_06_07, sat_08_10, sat_11_13, sat_14_16])
###Output
_____no_output_____
###Markdown
Next, we need to drop the culative rows (where `school_num` is 0), and update the `year` field to reflect the format of our main dataset.We will also mark 0 scores as NaN.
###Code
sat_scores = sat_scores[sat_scores['school_num'] != 0]
sat_scores['year'] = sat_scores['year'] - 1
sat_scores = sat_scores.replace(0, np.nan)
###Output
_____no_output_____
###Markdown
Lastly, we will save the data into a CSV file so that we don't have to rerun our preprocessing.
###Code
sat_scores.to_csv('data/test_scores/sat/processed.csv', sep=',', index=False)
###Output
_____no_output_____
|
docs/tutorials/advanced/aer/1_aer_provider.ipynb
|
###Markdown
Qiskit Aer: SimulatorsThe latest version of this notebook is available on https://github.com/Qiskit/qiskit-iqx-tutorials. IntroductionThis notebook shows how to import *Qiskit Aer* simulator backends and use them to execute ideal (noise free) Qiskit Terra circuits.
###Code
import numpy as np
# Import Qiskit
from qiskit import QuantumCircuit, QuantumRegister, ClassicalRegister
from qiskit import Aer, execute
from qiskit.tools.visualization import plot_histogram, plot_state_city
###Output
_____no_output_____
###Markdown
Qiskit Aer simulator backends Qiskit Aer currently includes three high performance simulator backends:* `QasmSimulator`: Allows ideal and noisy multi-shot execution of qiskit circuits and returns counts or memory* `StatevectorSimulator`: Allows ideal single-shot execution of qiskit circuits and returns the final statevector of the simulator after application* `UnitarySimulator`: Allows ideal single-shot execution of qiskit circuits and returns the final unitary matrix of the circuit itself. Note that the circuit cannot contain measure or reset operations for this backendThese backends are found in the `Aer` provider with the names `qasm_simulator`, `statevector_simulator` and `unitary_simulator`, respectively.
###Code
# List Aer backends
Aer.backends()
###Output
_____no_output_____
###Markdown
The simulator backends can also be directly imported from `qiskit.providers.aer`
###Code
from qiskit.providers.aer import QasmSimulator, StatevectorSimulator, UnitarySimulator
###Output
_____no_output_____
###Markdown
QasmSimulatorThe `QasmSimulator` backend is designed to mimic an actual device. It executes a Qiskit `QuantumCircuit` and returns a count dictionary containing the final values of any classical registers in the circuit. The circuit may contain *gates*,*measurements*, *resets*, *conditionals*, and other advanced simulator options that will be discussed in another notebook. Simulating a quantum circuitThe basic operation executes a quantum circuit and returns a counts dictionary of measurement outcomes. Here we execute a simple circuit that prepares a 2-qubit Bell-state $|\psi\rangle = \frac{1}{2}(|0,0\rangle + |1,1 \rangle)$ and measures both qubits.
###Code
# Construct quantum circuit
circ = QuantumCircuit(2, 2)
circ.h(0)
circ.cx(0, 1)
circ.measure([0,1], [0,1])
# Select the QasmSimulator from the Aer provider
simulator = Aer.get_backend('qasm_simulator')
# Execute and get counts
result = execute(circ, simulator).result()
counts = result.get_counts(circ)
plot_histogram(counts, title='Bell-State counts')
###Output
_____no_output_____
###Markdown
Returning measurement outcomes for each shotThe `QasmSimulator` also supports returning a list of measurement outcomes for each individual shot. This is enabled by setting the keyword argument `memory=True` in the `assemble` or `execute` function.
###Code
# Construct quantum circuit
circ = QuantumCircuit(2, 2)
circ.h(0)
circ.cx(0, 1)
circ.measure([0,1], [0,1])
# Select the QasmSimulator from the Aer provider
simulator = Aer.get_backend('qasm_simulator')
# Execute and get memory
result = execute(circ, simulator, shots=10, memory=True).result()
memory = result.get_memory(circ)
print(memory)
###Output
['11', '11', '11', '11', '00', '00', '00', '00', '00', '00']
###Markdown
Starting simulation with a custom initial stateThe `QasmSimulator` allows setting a custom initial statevector for the simulation. This means that all experiments in a Qobj will be executed starting in a state $|\psi\rangle$ rather than the all zero state $|0,0,..0\rangle$. The custom state may be set in the circuit using the `initialize` method.**Note:*** The initial statevector must be a valid quantum state $|\langle\psi|\psi\rangle|=1$. If not, an exception will be raised. * The simulator supports this option directly for efficiency, but it can also be unrolled to standard gates for execution on actual devices.We now demonstrate this functionality by setting the simulator to be initialized in the final Bell-state of the previous example:
###Code
# Construct a quantum circuit that initialises qubits to a custom state
circ = QuantumCircuit(2, 2)
circ.initialize([1, 0, 0, 1] / np.sqrt(2), [0, 1])
circ.measure([0,1], [0,1])
# Select the QasmSimulator from the Aer provider
simulator = Aer.get_backend('qasm_simulator')
# Execute and get counts
result = execute(circ, simulator).result()
counts = result.get_counts(circ)
plot_histogram(counts, title="Bell initial statevector")
###Output
_____no_output_____
###Markdown
StatevectorSimulatorThe `StatevectorSimulator` executes a single shot of a Qiskit `QuantumCircuit` and returns the final quantum statevector of the simulation. The circuit may contain *gates*, and also *measurements*, *resets*, and *conditional* operations. Simulating a quantum circuitThe basic operation executes a quantum circuit and returns a counts dictionary of measurement outcomes. Here we execute a simple circuit that prepares a 2-qubit Bell-state $|\psi\rangle = \frac{1}{2}(|0,0\rangle + |1,1 \rangle)$ and measures both qubits.
###Code
# Construct quantum circuit without measure
circ = QuantumCircuit(2)
circ.h(0)
circ.cx(0, 1)
# Select the StatevectorSimulator from the Aer provider
simulator = Aer.get_backend('statevector_simulator')
# Execute and get counts
result = execute(circ, simulator).result()
statevector = result.get_statevector(circ)
plot_state_city(statevector, title='Bell state')
###Output
_____no_output_____
###Markdown
Simulating a quantum circuit with measurementNote that if a circuit contains *measure* or *reset* the final statevector will be a conditional statevector *after* simulating wave-function collapse to the outcome of a measure or reset. For the Bell-state circuit this means the final statevector will be *either* $|0,0\rangle$ *or* $|1, 1\rangle$.
###Code
# Construct quantum circuit with measure
circ = QuantumCircuit(2, 2)
circ.h(0)
circ.cx(0, 1)
circ.measure([0,1], [0,1])
# Select the StatevectorSimulator from the Aer provider
simulator = Aer.get_backend('statevector_simulator')
# Execute and get counts
result = execute(circ, simulator).result()
statevector = result.get_statevector(circ)
plot_state_city(statevector, title='Bell state post-measurement')
###Output
_____no_output_____
###Markdown
Starting simulation with a custom initial stateLike the `QasmSimulator`, the `StatevectorSimulator` also allows setting a custom initial statevector for the simulation. Here we run the previous initial statevector example on the `StatevectorSimulator` and initialize it to the Bell state.
###Code
# Construct a quantum circuit that initialises qubits to a custom state
circ = QuantumCircuit(2)
circ.initialize([1, 0, 0, 1] / np.sqrt(2), [0, 1])
# Select the StatevectorSimulator from the Aer provider
simulator = Aer.get_backend('statevector_simulator')
# Execute and get counts
result = execute(circ, simulator).result()
statevector = result.get_statevector(circ)
plot_state_city(statevector, title="Bell initial statevector")
###Output
_____no_output_____
###Markdown
Unitary SimulatorThe `UnitarySimulator` constructs the unitary matrix for a Qiskit `QuantumCircuit` by applying each gate matrix to an identity matrix. The circuit may only contain *gates*, if it contains *resets* or *measure* operations an exception will be raised. Simulating a quantum circuit unitaryFor this example we will return the unitary matrix corresponding to the previous examples circuit which prepares a bell state.
###Code
# Construct an empty quantum circuit
circ = QuantumCircuit(2)
circ.h(0)
circ.cx(0, 1)
# Select the UnitarySimulator from the Aer provider
simulator = Aer.get_backend('unitary_simulator')
# Execute and get counts
result = execute(circ, simulator).result()
unitary = result.get_unitary(circ)
print("Circuit unitary:\n", unitary)
###Output
Circuit unitary:
[[ 0.70710678+0.00000000e+00j 0.70710678-8.65956056e-17j
0. +0.00000000e+00j 0. +0.00000000e+00j]
[ 0. +0.00000000e+00j 0. +0.00000000e+00j
0.70710678+0.00000000e+00j -0.70710678+8.65956056e-17j]
[ 0. +0.00000000e+00j 0. +0.00000000e+00j
0.70710678+0.00000000e+00j 0.70710678-8.65956056e-17j]
[ 0.70710678+0.00000000e+00j -0.70710678+8.65956056e-17j
0. +0.00000000e+00j 0. +0.00000000e+00j]]
###Markdown
Setting a custom initial unitaryWe may also set an initial state for the `UnitarySimulator`, however this state is an initial *unitary matrix* $U_i$, not a statevector. In this case the returned unitary will be $U.U_i$ given by applying the circuit unitary to the initial unitary matrix.**Note:*** The initial unitary must be a valid unitary matrix $U^\dagger.U =\mathbb{1}$. If not, an exception will be raised. * If a `Qobj` contains multiple experiments, the initial unitary must be the correct size for *all* experiments in the `Qobj`, otherwise an exception will be raised.Let us consider preparing the output unitary of the previous circuit as the initial state for the simulator:
###Code
# Construct an empty quantum circuit
circ = QuantumCircuit(2)
circ.iden([0,1])
# Set the initial unitary
opts = {"initial_unitary": np.array([[ 1, 1, 0, 0],
[ 0, 0, 1, -1],
[ 0, 0, 1, 1],
[ 1, -1, 0, 0]] / np.sqrt(2))}
# Select the UnitarySimulator from the Aer provider
simulator = Aer.get_backend('unitary_simulator')
# Execute and get counts
result = execute(circ, simulator, backend_options=opts).result()
unitary = result.get_unitary(circ)
unitary = result.get_unitary(circ)
print("Initial Unitary:\n", unitary)
import qiskit.tools.jupyter
%qiskit_version_table
%qiskit_copyright
###Output
_____no_output_____
|
12. FIR Filter -Windowed-Sinc Filters/FIR Filter - 1.Windowed-Sinc Filters.ipynb
|
###Markdown
Windowed-Sinc FiltersWindowed-sinc filters are used to separate one band of frequencies from another. They are very stable, produce few surprises, and can be pushed to incredible performance levels. These exceptional frequency domain characteristics are obtained at the expense of poor performance in the time domain, including excessive ripple and overshoot in the step response. When carried out by standard convolution, windowed-sinc filters are easy to program, but slow to execute.The starting point for the windowed sinc filters is the window function, in this case a mathematical expression for the window function looks like this: $$ h[k]=\frac{\sin{(2\pi f_{c}\cdot x[k])}}{\pi \cdot x[k]}$$We will later see that a shifted version of the window function has a better frequency response, and that is why we prefer this kind of function over the traditional window function$$ h[k]=\frac{\sin{(2\pi f_{c} \cdot (x[k]-M/2))}}{\pi \cdot(x[k]-M/2)}$$Where $M$ is the filter length and we use an heuristic relationship with the transition bandwidth $BW$ to calculate it$$M = \frac{4}{BW}$$The **cutoff frequency** of the windowed-sinc filter is measured at the **one-half amplitude point**. Why use 0.5 instead of the standard 0.707 (-3dB) used in analog electronics and other digital filters? This is because the windowed-sinc's frequency response is symmetrical between the passband and the stopband. For instance, the **Hamming window** results in a passband ripple of 0.2%, and an identical stopband attenuation (i.e., ripple in the stopband) of 0.2%. Other filters do not show this symmetry, and therefore have no advantage in using the one-half amplitude point to mark the cutoff frequency. This symmetry makes the windowed-sinc ideal for **spectral inversion**.
###Code
import sys
sys.path.insert(0, '../')
from Common import common_plots
from Common import fourier_transform
cplots = common_plots.Plot()
import numpy as np
import matplotlib.pyplot as plt
def get_fourier(x):
"""
Function that performs the Fourier calculation of a signal x and returns its magnitude and frequency range.
Parameters:
x (numpy array): Signal to be transformed into Fourier domain.
Returns:
mag (numpy array): Magnitude of the signal's Fourier transform.
freq (numpy array): Frequency domain of the signal's Fourier transform.
"""
signal = x.reshape(-1,1)
fourier = fourier_transform.FourierTransform(signal)
mag = fourier.dft_magnitude()
freq = fourier.frequency_domain()
return mag, freq
def sinc_function(fc, BW):
"""
Function that calculates a sinc time response.
Parameters:
BW (float): Transition bandwidth of the filter. The lenght of the filter is
given by M=4/BW.
fc (float): Cut-off frequency for the low-pass filter. Between 0 and 0.5.
Returns:
numpy array: Returns sinc time domain response.
"""
return None
def shifted_sinc_function(fc, BW):
"""
Function that calculates a sinc shifted time response. The shifted sinc function has
a shift value of M=4/BW.
Parameters:
BW (float): Transition bandwidth of the filter. The lenght of the filter is
given by M=4/BW.
fc (float): Cut-off frequency for the low-pass filter. Between 0 and 0.5.
Returns:
numpy array: Returns sinc shifted time domain response.
"""
return None
###Output
_____no_output_____
###Markdown
Implementing a Low Pass FilterTo develop a low pass filter, two parameters must be selected:1. The cut-off frequency, $0\leq f_c \leq 0.5$2. The lenght of the filter kernel, $M=\frac{4}{BW}$, where $BW$ is the transition bandwidth (say, 99% to 1% of the curve).
###Code
fc = #Write code here
BW = #Write code here
M = #Write code here
print("Filter lenght is {}".format(M))
sinc = sinc_function(fc, BW)
shifted_sinc = shifted_sinc_function(fc, BW)
normalized_sinc = #Write code here
normalized_shifted_sinc = #Write code here
fft_sinc_magnitude, fft_sinc_f = get_fourier(sinc)
fft_shifted_sinc_magnitude, fft_shifted_sinc_f = get_fourier(shifted_sinc)
plt.rcParams["figure.figsize"] = (15,10)
plt.subplot(2,2,1)
plt.plot(normalized_sinc)
#plt.stem(normalized_sinc, markerfmt='.', use_line_collection=True)
plt.title('Sinc Function')
plt.grid('on')
plt.subplot(2,2,2)
plt.plot(normalized_shifted_sinc)
#plt.stem(normalized_shifted_sinc, markerfmt='.', use_line_collection=True)
plt.title('Shited {}-Sinc Function'.format(M))
plt.grid('on')
plt.subplot(2,2,3)
cplots.plot_frequency_response(fft_sinc_magnitude,
fft_sinc_f,
title='Sinc Frequency Response')
plt.subplot(2,2,4)
cplots.plot_frequency_response(fft_shifted_sinc_magnitude,
fft_shifted_sinc_f,
title='Shited {}-Sinc Frequency Response'.format(M));
###Output
_____no_output_____
###Markdown
Hamming and Blackman WindowsA window function is a mathematical function that is zero-valued outside of some chosen interval, normally symmetric around the middle of the interval, usually near a maximum in the middle, and usually tapering away from the middle. Mathematically, when another function or waveform/data-sequence is "multiplied" by a window function, the product is also zero-valued outside the interval: all that is left is the part where they overlap, the "view through the window".
###Code
def hamming_window(BW):
"""
Function that calculates a Hamming window of a given transition bandwidth.
Parameters:
BW (float): Transition bandwidth of the filter. The lenght of the filter is
given by M=4/BW.
Returns:
numpy array: Returns Hamming window of a given M-kernel.
"""
return None
def blackman_window(BW):
"""
Function that calculates a Blackman window of a given M-kernel.
Parameters:
BW (float): Transition bandwidth of the filter. The lenght of the filter is
given by M=4/BW.
Returns:
numpy array: Returns Blackman window of a given M-kernel.
"""
return None
hamming = hamming_window(BW)
blackman = blackman_window(BW)
fft_hamming_magnitude, fft_hamming_f = get_fourier(hamming)
fft_blackman_magnitude, fft_blackman_f = get_fourier(blackman)
plt.rcParams["figure.figsize"] = (15,10)
plt.subplot(2,2,1)
plt.plot(hamming)
#plt.stem(hamming, markerfmt='.', use_line_collection=True)
plt.title('Hamming Window')
plt.grid('on')
plt.subplot(2,2,2)
plt.plot(blackman)
#plt.stem(blackman, markerfmt='.', use_line_collection=True)
plt.title('Blackman Window')
plt.grid('on')
plt.subplot(2,2,3)
cplots.plot_frequency_response(fft_hamming_magnitude,
fft_hamming_f,
title='Hamming Window Frequency Response')
plt.subplot(2,2,4)
cplots.plot_frequency_response(fft_blackman_magnitude,
fft_blackman_f,
title='Blackman Window Frequency Response');
hamming_shifted_sinc = #Write code here
blackman_shifted_sinc = #Write code here
fft_hamming_shifted_sinc_magnitude, fft_hamming_shifted_sinc_f = get_fourier(hamming_shifted_sinc)
fft_blackman_shifted_sinc_magnitude, fft_blackman_shifted_sinc_f = get_fourier(blackman_shifted_sinc)
plt.rcParams["figure.figsize"] = (15,10)
plt.subplot(2,2,1)
plt.plot(hamming_shifted_sinc)
#plt.stem(hamming_shifted_sinc, markerfmt='.', use_line_collection=True)
plt.title('Shifted Sinc - Hamming Window')
plt.grid('on')
plt.subplot(2,2,2)
plt.plot(blackman_shifted_sinc)
#plt.stem(blackman_shifted_sinc, markerfmt='.', use_line_collection=True)
plt.title('Shifted Sinc - Blackman Window')
plt.grid('on')
plt.subplot(2,2,3)
cplots.plot_frequency_response(fft_hamming_shifted_sinc_magnitude,
fft_hamming_shifted_sinc_f,
title='Shifted Sinc - Hamming Window Frequency Response')
plt.subplot(2,2,4)
cplots.plot_frequency_response(fft_blackman_shifted_sinc_magnitude,
fft_blackman_shifted_sinc_f,
title='Shifted Sinc - Blackman Window Frequency Response');
###Output
_____no_output_____
###Markdown
Comparison between Hamming and Blackman windowsThe Hamming window has a **faster roll-off** than the Blackman, however the Blackman has a **better stopband attenuation**. To be exact, the stopband attenuation for the Blackman is greater than the Hamming. Although it cannot be seen in these graphs, the Blackman has a very small passband ripple compared to the the Hamming. In general, the **Blackman should be your first choice**; a slow roll-off is easier to handle than poor stopband attenuation. Example of filter design for an EEG signalAn electroencephalogram, or EEG, is a measurement of the electrical activity of the brain. It can be detected as millivolt level signals appearing on electrodes attached to the surface of the head. Each nerve cell in the brain generates small electrical pulses. The EEG is the combined result of an enormous number of these electrical pulses being generated in a (hopefully) coordinated manner. Although the relationship between thought and this electrical coordination is very poorly understood, different frequencies in the EEG can be identified with specific mental states. If you close your eyes and relax, the predominant EEG pattern will be a slow oscillation between about 7 and 12 hertz. This waveform is called the alpha rhythm, and is associated with contentment and a decreased level of attention. Opening your eyes and looking around causes the EEG to change to the beta rhythm, occurring between about 17 and 20 hertz. Other frequencies and waveforms are seen in children, different depths of sleep, and various brain disorders such as epilepsy.In this example, we will assume that the EEG signal has been amplified by analog electronics, and then digitized at a sampling rate of 100 samples per second. We have a data of 640 samples. Our goal is to separate the alpha from the beta rhythms. To do this, we will design a digital low-pass filter with a cutoff frequency of 14 hertz, or 0.14 of the sampling rate. The transition bandwidth will be set at 4 hertz, or 0.04 of the sampling rate.
###Code
fc = #Write code here
BW = #Write code here
print("Filter lenght is {}".format(M))
shifted_sinc = #Write code here
normalized_shifted_sinc = #Write code here
hamming = #Write code here
hamming_shifted_sinc = #Write code here
ecg = np.loadtxt(fname = "ecg.dat").flatten()
filtered_ecg = #Write code here
filter_magnitude, filter_f= get_fourier(hamming_shifted_sinc)
normalized_fft_hamming_shifted_sinc = np.absolute(filter_magnitude)/np.sum(np.absolute(filter_magnitude))
fft_ecg_magnitude, fft_ecg_f = get_fourier(ecg)
normalized_fft_ecg = np.absolute(fft_ecg_magnitude)/np.sum(np.absolute(fft_ecg_magnitude))
fft_filtered_ecg_magnitude, fft_filtered_ecg_f = get_fourier(filtered_ecg)
normalized_fft_filtered_ecg = np.absolute(fft_filtered_ecg_magnitude)/np.sum(np.absolute(fft_filtered_ecg_magnitude))
plt.rcParams["figure.figsize"] = (15,10)
plt.subplot(2,2,1)
plt.plot(ecg)
plt.title('ECG Signal')
plt.grid('on')
plt.subplot(2,2,2)
plt.plot(filtered_ecg)
plt.title('Filtered ECG Signal')
plt.grid('on')
plt.subplot(2,2,3)
cplots.plot_frequency_response(normalized_fft_ecg,
fft_ecg_f,
title='Frequency Response ECG Signal')
plt.subplot(2,2,4)
cplots.plot_frequency_response(normalized_fft_filtered_ecg,
fft_filtered_ecg_f,
title='Frequency Response Filtered ECG Signal')
###Output
_____no_output_____
###Markdown
We will pickle our filter design for later user in the next Jupyter Notebook...
###Code
import pickle
data = {'ecg':ecg,
'low_pass':hamming_shifted_sinc,
'fft_low_pass':normalized_fft_hamming_shifted_sinc}
file = open('save_data.pickle', 'wb')
pickle.dump(data, file)
file.close()
###Output
_____no_output_____
|
deep learning/Sketch Cleanup.ipynb
|
###Markdown
IntroExploring the task of sketch cleaning (also automatized linearts).Based on [Edgar Simo-Serra・Sketch Simplification](http://hi.cs.waseda.ac.jp/~esimo/en/research/sketch/)
###Code
import time
import numpy as np
import pdb
import seaborn as sns
import pandas as pd
import glob
import os
import sys
from os.path import join
import matplotlib
import matplotlib.pyplot as plt
from matplotlib import animation
from keras.models import Sequential
from keras.models import Model
from keras.layers.core import Activation, Dense
from keras import backend as K
from keras import optimizers
sys.path.append(join(os.getcwd(), os.pardir))
from utils import image_processing
sns.set_style("dark")
sns.set_context("paper")
%matplotlib notebook
RES_DIR = join(*[os.pardir]*2, 'data', 'sketch_dataset')
###Output
_____no_output_____
###Markdown
Load Data
###Code
def load_dataset_info(dirpath):
data = pd.DataFrame(columns=['dirpath', 'ref', 'sketch', 'sketch2', 'lineart'])
for f in glob.glob(join(dirpath, '**', "*.jpg"), recursive=True):
filepath, filename = f.rsplit('\\', 1)
img_id, category = filename.split('.')[0].split('_')
data.set_value(img_id, category, filename)
data.set_value(img_id, 'dirpath', filepath)
return data
dataset_info = load_dataset_info(RES_DIR)
dataset_info.head()
# load sketch
content_image = None
with Image.open(os.path.join(RES_DIR, 'superman.jpg')) as img:
img = img.resize((height, width))
content_image = np.asarray(img, dtype='float32')
plt.imshow(img.convert(mode='RGB'))
plt.show()
f, axarr = plt.subplots(len(dataset_info), 3)
categories = {0:'ref', 1:'sketch', 2:'lineart'}
for row_idx, (img_id, row) in enumerate(dataset_info.iterrows()):
for i in range(3):
img = plt.imread(join(row['dirpath'], row[categories[i]]))
axarr[row_idx, i].imshow(img)
axarr[row_idx, i].axis('off')
###Output
_____no_output_____
###Markdown
Data Augmentation
###Code
from skimage import data
from skimage import transform
from skimage import io
def augment(imgs_info, n, suffix_folder=""):
for i in range(n):
for img_id, row in imgs_info.iterrows():
rotation = np.random.randint(360)
for cat in ['ref', 'sketch', 'sketch2', 'lineart']:
if not pd.isnull(row[cat]):
origin_img = plt.imread(join(row['dirpath'], row[cat]))
dest_img = transform.rotate(origin_img, rotation, mode='edge')
filename = "{}{}{}_{}.jpg".format(img_id, 'gen', i, cat)
io.imsave(join(row['dirpath'], suffix_folder, filename), dest_img)
augment(dataset_info, 10, 'augmented')
###Output
_____no_output_____
###Markdown
Train (CNN)
###Code
from keras.models import Sequential
from keras.layers import Convolution2D, MaxPooling2D, Conv2DTranspose
from keras.layers import Activation, Dropout, Flatten, Dense
from keras.preprocessing import image
from keras import optimizers
img_shape = (128, 128, 3)
X = image_processing.load_data(dataset_info.apply(lambda x : join(x['dirpath'], x['lineart']), axis=1).values, img_shape[:2])
y = image_processing.load_data(dataset_info.apply(lambda x : join(x['dirpath'], x['ref']), axis=1).values, img_shape[:2])
X_train = X#(X/255)
y_train = y#(y/255)
print(X_train.shape)
print(y_train.shape)
model = Sequential()
model.add(Convolution2D(32, (5, 5), strides=(2, 2), padding='same', input_shape=img_shape,
activation='relu'))
model.add(Convolution2D(64, (3, 3), padding='same',
activation='relu'))
model.add(Convolution2D(128, (5, 5), strides=(2, 2), padding='same',
activation='relu'))
model.add(Convolution2D(256, (3, 3), padding='same',
activation='relu'))
model.add(Convolution2D(256, (3, 3), padding='same',
activation='relu'))
model.add(Conv2DTranspose(128, (5, 5), strides=(2, 2), padding='same',
activation='relu'))
model.add(Convolution2D(64, (3, 3), padding='same',
activation='relu'))
model.add(Conv2DTranspose(32, (3, 3), strides=(2, 2), padding='same',
activation='relu'))
model.add(Convolution2D(3, (5, 5), padding='same',
activation='sigmoid'))
model.summary()
optimizer = optimizers.Adam(lr=0.001)
model.compile(loss='mean_squared_error', optimizer=optimizer, metrics=['accuracy'])
model.fit(y_train, y_train, batch_size=2, epochs=5)
plt.imshow(X_train[3])
plt.imshow(y_train[3])
pred = model.predict(y_train[2].reshape((1, *img_shape)))
pred.shape
plt.imshow(pred[0])
pred
###Output
_____no_output_____
###Markdown
Train (GAN)
###Code
from keras.models import Sequential
from keras.layers.core import Activation, Dense
from keras import backend as K
from keras import optimizers
img_shape = (128, 128, 3)
X_train = image_processing.load_data(dataset_info.apply(lambda x : join(x['dirpath'], x['ref']), axis=1).values, img_shape[:2])
y_train = image_processing.load_data(dataset_info.apply(lambda x : join(x['dirpath'], x['lineart']), axis=1).values, img_shape[:2])
print(X_train.shape)
print(y_train.shape)
def generator(img_shape):
model = Sequential()
model.add(Convolution2D(32, (5, 5), strides=(2, 2), padding='same', input_shape=img_shape,
activation='relu'))
model.add(Convolution2D(64, (3, 3), padding='same',
activation='relu'))
model.add(Convolution2D(128, (5, 5), strides=(2, 2), padding='same',
activation='relu'))
model.add(Convolution2D(256, (3, 3), padding='same',
activation='relu'))
model.add(Convolution2D(256, (3, 3), padding='same',
activation='relu'))
model.add(Conv2DTranspose(128, (5, 5), strides=(2, 2), padding='same',
activation='relu'))
model.add(Convolution2D(64, (3, 3), padding='same',
activation='relu'))
model.add(Conv2DTranspose(32, (3, 3), strides=(2, 2), padding='same',
activation='relu'))
model.add(Convolution2D(3, (5, 5), padding='same',
activation='sigmoid'))
return model
def discriminator(img_shape):
model = Sequential()
model.add(Convolution2D(32, (5, 5), strides=(2, 2), padding='same', input_shape=img_shape,
activation='relu'))
model.add(Convolution2D(64, (3, 3), padding='same',
activation='relu'))
model.add(Convolution2D(128, (5, 5), strides=(2, 2), padding='same',
activation='relu'))
model.add(Convolution2D(256, (3, 3), padding='same',
activation='relu'))
model.add(Convolution2D(256, (3, 3), padding='same',
activation='relu'))
model.add(Conv2DTranspose(128, (5, 5), strides=(2, 2), padding='same',
activation='relu'))
model.add(Convolution2D(64, (3, 3), padding='same',
activation='relu'))
model.add(Conv2DTranspose(32, (3, 3), strides=(2, 2), padding='same',
activation='relu'))
model.add(Flatten())
model.add(Dense(512, activation='relu'))
#model.add(Dropout(0.5))
model.add(Dense(1, activation=K.sigmoid))
return model
# init GAN components
d = discriminator(img_shape)
g = generator(img_shape)
# discriminator model
optimizer = optimizers.RMSprop(lr=0.0008, clipvalue=1.0, decay=6e-8)
discriminator_model = d
discriminator_model.compile(loss='binary_crossentropy', optimizer=optimizer, metrics=['accuracy'])
# adversarial model
optimizer = optimizers.RMSprop(lr=0.0004, clipvalue=1.0, decay=3e-8)
adversarial_model = Sequential()
adversarial_model.add(g)
adversarial_model.add(d)
adversarial_model.compile(loss='binary_crossentropy', optimizer=optimizer, metrics=['accuracy'])
batch_size = 64
epochs = 100
for step in range(epochs):
# generate data
# first we sample from the true distribution, then we generate some
# "fake" data by feeding noise to the generator
true_sample = np.reshape(gaussian_d.sample(batch_size), (batch_size, 1))
noise = generator_d.sample(batch_size)
fake_sample = g.predict(noise)
#pdb.set_trace()
# train discriminator
# feed true and fake samples with respective labels (1, 0) to the discriminator
x = np.reshape(np.concatenate((true_sample, fake_sample)), (batch_size*2, 1))
y = np.ones([batch_size*2, 1])
y[batch_size:, :] = 0
d_loss = discriminator_model.train_on_batch(x, y)
# train GAN
# feed noise to the model and expect true (1) response from discriminator,
# which is in turn fed with data generated by the generator
noise = np.reshape(generator_d.sample(batch_size), (batch_size, 1))
y = np.ones([batch_size, 1])
a_loss = adversarial_model.train_on_batch(noise, y)
log_mesg = "%d: [D loss: %f, acc: %f]" % (step, d_loss[0], d_loss[1])
log_mesg = "%s [A loss: %f, acc: %f]" % (log_mesg, a_loss[0], a_loss[1])
###Output
_____no_output_____
|
notebooks/prod/n08_simple_q_learner_1000_states_full_training_15_epochs.ipynb
|
###Markdown
In this notebook a simple Q learner will be trained and evaluated. The Q learner recommends when to buy or sell shares of one particular stock, and in which quantity (in fact it determines the desired fraction of shares in the total portfolio value). One initial attempt was made to train the Q-learner with multiple processes, but it was unsuccessful.
###Code
# Basic imports
import os
import pandas as pd
import matplotlib.pyplot as plt
import numpy as np
import datetime as dt
import scipy.optimize as spo
import sys
from time import time
from sklearn.metrics import r2_score, median_absolute_error
from multiprocessing import Pool
%matplotlib inline
%pylab inline
pylab.rcParams['figure.figsize'] = (20.0, 10.0)
%load_ext autoreload
%autoreload 2
sys.path.append('../../')
import recommender.simulator as sim
from utils.analysis import value_eval
from recommender.agent import Agent
from functools import partial
NUM_THREADS = 1
LOOKBACK = -1 # 252*4 + 28
STARTING_DAYS_AHEAD = 252
POSSIBLE_FRACTIONS = [0.0, 1.0]
# Get the data
SYMBOL = 'SPY'
total_data_train_df = pd.read_pickle('../../data/data_train_val_df.pkl').stack(level='feature')
data_train_df = total_data_train_df[SYMBOL].unstack()
total_data_test_df = pd.read_pickle('../../data/data_test_df.pkl').stack(level='feature')
data_test_df = total_data_test_df[SYMBOL].unstack()
if LOOKBACK == -1:
total_data_in_df = total_data_train_df
data_in_df = data_train_df
else:
data_in_df = data_train_df.iloc[-LOOKBACK:]
total_data_in_df = total_data_train_df.loc[data_in_df.index[0]:]
# Create many agents
index = np.arange(NUM_THREADS).tolist()
env, num_states, num_actions = sim.initialize_env(total_data_in_df,
SYMBOL,
starting_days_ahead=STARTING_DAYS_AHEAD,
possible_fractions=POSSIBLE_FRACTIONS,
n_levels=10)
agents = [Agent(num_states=num_states,
num_actions=num_actions,
random_actions_rate=0.98,
random_actions_decrease=0.9999,
dyna_iterations=0,
name='Agent_{}'.format(i)) for i in index]
def show_results(results_list, data_in_df, graph=False):
for values in results_list:
total_value = values.sum(axis=1)
print('Sharpe ratio: {}\nCum. Ret.: {}\nAVG_DRET: {}\nSTD_DRET: {}\nFinal value: {}'.format(*value_eval(pd.DataFrame(total_value))))
print('-'*100)
initial_date = total_value.index[0]
compare_results = data_in_df.loc[initial_date:, 'Close'].copy()
compare_results.name = SYMBOL
compare_results_df = pd.DataFrame(compare_results)
compare_results_df['portfolio'] = total_value
std_comp_df = compare_results_df / compare_results_df.iloc[0]
if graph:
plt.figure()
std_comp_df.plot()
###Output
_____no_output_____
###Markdown
Let's show the symbols data, to see how good the recommender has to be.
###Code
print('Sharpe ratio: {}\nCum. Ret.: {}\nAVG_DRET: {}\nSTD_DRET: {}\nFinal value: {}'.format(*value_eval(pd.DataFrame(data_in_df['Close'].iloc[STARTING_DAYS_AHEAD:]))))
# Simulate (with new envs, each time)
n_epochs = 15
for i in range(n_epochs):
tic = time()
env.reset(STARTING_DAYS_AHEAD)
results_list = sim.simulate_period(total_data_in_df,
SYMBOL,
agents[0],
starting_days_ahead=STARTING_DAYS_AHEAD,
possible_fractions=POSSIBLE_FRACTIONS,
verbose=False,
other_env=env)
toc = time()
print('Epoch: {}'.format(i))
print('Elapsed time: {} seconds.'.format((toc-tic)))
print('Random Actions Rate: {}'.format(agents[0].random_actions_rate))
show_results([results_list], data_in_df)
env.reset(STARTING_DAYS_AHEAD)
results_list = sim.simulate_period(total_data_in_df,
SYMBOL, agents[0],
learn=False,
starting_days_ahead=STARTING_DAYS_AHEAD,
possible_fractions=POSSIBLE_FRACTIONS,
other_env=env)
show_results([results_list], data_in_df, graph=True)
###Output
Starting simulation for agent: Agent_0. 5268 days of simulation to go.
Date 2014-12-22 00:00:00 (simulating until 2014-12-31 00:00:00). Time: 0.2838108539581299s. Value: 1224031.8399999999..Sharpe ratio: 2.6113451938254477
Cum. Ret.: 121.58015399999998
AVG_DRET: 0.0009290849284226412
STD_DRET: 0.005647957244281964
Final value: 1225801.5399999998
----------------------------------------------------------------------------------------------------
###Markdown
Ok, let's save that
###Code
import pickle
with open('../../data/simple_q_learner_1000_states_full_training_15_epochs.pkl', 'wb') as best_agent:
pickle.dump(agents[0], best_agent)
###Output
_____no_output_____
###Markdown
Let's run the trained agent, with the test set First a non-learning test: this scenario would be worse than what is possible (in fact, the q-learner can learn from past samples in the test set without compromising the causality).
###Code
TEST_DAYS_AHEAD = 20
env.set_test_data(total_data_test_df, TEST_DAYS_AHEAD)
tic = time()
results_list = sim.simulate_period(total_data_test_df,
SYMBOL,
agents[0],
learn=False,
starting_days_ahead=TEST_DAYS_AHEAD,
possible_fractions=POSSIBLE_FRACTIONS,
verbose=False,
other_env=env)
toc = time()
print('Epoch: {}'.format(i))
print('Elapsed time: {} seconds.'.format((toc-tic)))
print('Random Actions Rate: {}'.format(agents[0].random_actions_rate))
show_results([results_list], data_test_df, graph=True)
###Output
Starting simulation for agent: Agent_0. 484 days of simulation to go.
Date 2016-12-28 00:00:00 (simulating until 2016-12-30 00:00:00). Time: 0.17671680450439453s. Value: 10534.740000000005.Epoch: 14
Elapsed time: 8.161107778549194 seconds.
Random Actions Rate: 0.00036300361732965325
Sharpe ratio: 0.3394029381825202
Cum. Ret.: 0.05324400000000051
AVG_DRET: 0.0001246235029780917
STD_DRET: 0.00582887345924684
Final value: 10532.440000000006
----------------------------------------------------------------------------------------------------
###Markdown
And now a "realistic" test, in which the learner continues to learn from past samples in the test set (it even makes some random moves, though very few).
###Code
env.set_test_data(total_data_test_df, TEST_DAYS_AHEAD)
tic = time()
results_list = sim.simulate_period(total_data_test_df,
SYMBOL,
agents[0],
learn=True,
starting_days_ahead=TEST_DAYS_AHEAD,
possible_fractions=POSSIBLE_FRACTIONS,
verbose=False,
other_env=env)
toc = time()
print('Epoch: {}'.format(i))
print('Elapsed time: {} seconds.'.format((toc-tic)))
print('Random Actions Rate: {}'.format(agents[0].random_actions_rate))
show_results([results_list], data_test_df, graph=True)
###Output
Starting simulation for agent: Agent_0. 484 days of simulation to go.
Date 2016-12-28 00:00:00 (simulating until 2016-12-30 00:00:00). Time: 0.17526507377624512s. Value: 10043.760000000004.Epoch: 14
Elapsed time: 8.193697214126587 seconds.
Random Actions Rate: 0.00034588639540018753
Sharpe ratio: 0.06852442057274191
Cum. Ret.: 0.004156000000000271
AVG_DRET: 2.3589312742850267e-05
STD_DRET: 0.005464748589906518
Final value: 10041.560000000003
----------------------------------------------------------------------------------------------------
###Markdown
What are the metrics for "holding the position"?
###Code
print('Sharpe ratio: {}\nCum. Ret.: {}\nAVG_DRET: {}\nSTD_DRET: {}\nFinal value: {}'.format(*value_eval(pd.DataFrame(data_test_df['Close'].iloc[TEST_DAYS_AHEAD:]))))
###Output
Sharpe ratio: 0.44271542660031676
Cum. Ret.: 0.1070225832012679
AVG_DRET: 0.00025103195406808796
STD_DRET: 0.009001287260690292
Final value: 223.53
|
DeepLearning/pytorch/examples/Pytorch_tensors.ipynb
|
###Markdown
Pytorch: TensorsA fully-connected ReLU network with one hidden layer and no biases, trained to predict y from x by minimizing squared Euclidean distance.This implementation uses PyTorch tensors to manually compute the forward pass, loss, and backward pass.A PyTorch Tensor is basically the same as a numpy array: it does not know anything about deep learning or computational graphs or gradients, and is just a generic n-dimensional array to be used for arbitrary numeric computation.The biggest difference between a numpy array and a PyTorch Tensor is that a PyTorch Tensor can run on either CPU or GPU. To run operations on the GPU, just cast the Tensor to a cuda datatype.
###Code
import torch
dtype = torch.float
device = torch.device('cpu') # or 'cuda'
###Output
_____no_output_____
|
deeplearning.ai/tf/dogs_and_cats.ipynb
|
###Markdown
###Code
!nvidia-smi
import urllib.request
import os
import zipfile
import random
from tensorflow.keras.preprocessing.image import ImageDataGenerator
from tensorflow.keras import layers
from tensorflow.keras import Model
from tensorflow.keras.applications.inception_v3 import InceptionV3
from tensorflow.keras.optimizers import RMSprop
from shutil import copyfile
DOWNLOAD_DIR = "/tmp/"
CAT_SOURCE_DIR = f"{DOWNLOAD_DIR}PetImages/Cat/"
DOG_SOURCE_DIR = f"{DOWNLOAD_DIR}PetImages/Dog/"
CAT_V_DOGS_DIR = f"{DOWNLOAD_DIR}cats-v-dogs/"
TRAINING_DIR = f"{CAT_V_DOGS_DIR}/training/"
TESTING_DIR = f"{CAT_V_DOGS_DIR}/testing/"
TRAINING_CATS_DIR = f"{TRAINING_DIR}cats/"
TESTING_CATS_DIR = f"{TESTING_DIR}cats/"
TRAINING_DOGS_DIR = f"{TRAINING_DIR}dogs/"
TESTING_DOGS_DIR = f"{TESTING_DIR}dogs/"
data_url = "https://download.microsoft.com/download/3/E/1/3E1C3F21-ECDB-4869-8368-6DEBA77B919F/kagglecatsanddogs_3367a.zip"
data_file_name = "catsdogs.zip"
urllib.request.urlretrieve(data_url, data_file_name)
zip_ref = zipfile.ZipFile(data_file_name, 'r')
zip_ref.extractall(DOWNLOAD_DIR)
#zip_ref.extractall()
zip_ref.close()
print(f'Cat samples: {len(os.listdir(CAT_SOURCE_DIR))}')
print(f'Dog samples: {len(os.listdir(DOG_SOURCE_DIR))}')
try:
os.mkdir(CAT_V_DOGS_DIR)
os.mkdir(TRAINING_DIR)
os.mkdir(TESTING_DIR)
os.mkdir(TRAINING_CATS_DIR)
os.mkdir(TESTING_CATS_DIR)
os.mkdir(TRAINING_DOGS_DIR)
os.mkdir(TESTING_DOGS_DIR)
except OSError:
pass
def split_data(source, training, testing, split_size):
files = []
for filename in os.listdir(source):
file = f'{source}{filename}'
if os.path.getsize(file) > 0:
files.append(filename)
else:
print(f'{filename} is zero length, so ignoring.')
training_length = int(len(files) * split_size)
testing_length = int(len(files) - training_length)
shuffled_set = random.sample(files, len(files))
training_set = shuffled_set[0:training_length]
testing_set = shuffled_set[:testing_length]
for filename in training_set:
this_file = source + filename
destination = training + filename
copyfile(this_file, destination)
for filename in testing_set:
this_file = source + filename
destination = testing + filename
copyfile(this_file, destination)
split_size = .9
split_data(CAT_SOURCE_DIR, TRAINING_CATS_DIR, TESTING_CATS_DIR, split_size)
split_data(DOG_SOURCE_DIR, TRAINING_DOGS_DIR, TESTING_DOGS_DIR, split_size)
try:
print(f'Split training cats samples: {len(os.listdir(TRAINING_CATS_DIR))}')
print(f'Split training dogs samples: {len(os.listdir(TESTING_CATS_DIR))}')
print(f'Split testing cats samples: {len(os.listdir(TRAINING_DOGS_DIR))}')
print(f'Split testing dogs samples: {len(os.listdir(TESTING_DOGS_DIR))}')
except OSError:
print('Target split folders are empty.')
train_datagen = ImageDataGenerator(
rescale=1./255,
rotation_range=40,
width_shift_range=0.2,
height_shift_range=0.2,
shear_range=0.2,
zoom_range=0.2,
horizontal_flip=True,
fill_mode='nearest'
)
train_generator = train_datagen.flow_from_directory(
TRAINING_DIR,
batch_size=100,
class_mode='binary',
target_size=(150, 150)
)
validation_datagen = ImageDataGenerator(rescale=1./255)
validation_generator = validation_datagen.flow_from_directory(
TESTING_DIR,
batch_size=100,
class_mode='binary',
target_size=(150, 150)
)
weights_url = "https://storage.googleapis.com/mledu-datasets/inception_v3_weights_tf_dim_ordering_tf_kernels_notop.h5"
weights_file = "inception_v3.h5"
urllib.request.urlretrieve(weights_url, weights_file)
pre_trained_model = InceptionV3(
input_shape=(150, 150, 3),
include_top=False,
weights=None
)
pre_trained_model.load_weights(weights_file)
for layer in pre_trained_model.layers:
layer.trainable = False
#pre_trained_model.summary()
last_layer = pre_trained_model.get_layer('mixed7')
print(f'last layer output shape: {last_layer.output_shape}')
last_output = last_layer.output
x = layers.Flatten()(last_output)
x = layers.Dense(1024, activation='sigmoid')(x)
x = layers.Dense(1, activation='sigmoid')(x)
model = Model(pre_trained_model.input, x)
model.compile(
optimizer=RMSprop(lr=0.0001),
loss='binary_crossentropy',
metrics=['acc']
)
history = model.fit_generator(
train_generator,
validation_data=validation_generator,
epochs=20,
verbose=1
)
%matplotlib inline
import matplotlib.image as mpimg
import matplotlib.pyplot as plt
acc = history.history['acc']
val_acc = history.history['val_acc']
loss = history.history['loss']
val_loss = history.history['val_loss']
epochs=range(len(acc))
plt.plot(epochs, acc, 'r', 'Training Accuracy')
plt.plot(epochs, val_acc, 'b', 'Validation Accuracy')
plt.title('Training and validation accuracy')
plt.figure()
import numpy as np
from google.colab import files
from keras.preprocessing import image
uploaded = files.upload()
for fn in uploaded.keys():
path = f"/content/{fn}"
img = image.load_img(path, target_size=(150, 150))
x = image.img_to_array(img)
x = np.expand_dims(x, axis=0)
image_tensor = np.vstack([x])
classes = model.predict(image_tensor)
print(classes)
print(classes[0])
if classes[0] > 0.5:
print(f"{fn} is a dog")
else:
print(f"{fn} is a cat")
###Output
_____no_output_____
|
Classification/Practical Machine Learning - Support Vector Machines.ipynb
|
###Markdown
Classification with Support Vector Machines===A Support vector machine is a binary classifier that determines the hyperplane of maximum separation between any **two** groups. It works best on linear data, but can be applied to non-linear, multidimensional data.For multiple groups, SVM determines the sequential hyperplanes between any two groups of the set of groups; it can determine the hyperplane of maximum pairwise seperation between each group and all other groups (one at a time?) and then form the region of maximum seperation from each group to the other groups as a collection of hyperplanesSVM chooses the seperating hyperplane that is the maximum distance from the collection of all data in each group that is it pairwise seperating.New (unlabled) test data then becomes easily labeled by comparing it's position in the hyperspace with the set of hyperplanes defined from the training data.Non-linear data has non-linear hyperplanes -- may or may not be possible---How SVM Labels Test Data:---Given the known hyperplane location in hyperspace, there exists a vector $\textbf{W}$ that has its origin at the origin **and** is perpendicular to the hyperplane.For any ("unknown") test data point $\textbf{U}$, the projection (dot product) of $\textbf{U}$ onto $\textbf{W}$ will be either greater or lesser than the "magnitude of $\textbf{W}$", ||$\textbf{W}$||Specifically, with $b$ as the bias * if $\textbf{U}\cdot \textbf{W} + b \geq 0$, then $\textbf{U}$ belongs to group A (right of the hyperplane) * if $\textbf{U}\cdot \textbf{W} + b \leq 0$, then $\textbf{U}$ belongs to group B (left of the hyperplane) * if $\textbf{U}\cdot \textbf{W} + b = 0$, then $\textbf{U}$ is "on the decision boundary" How to Derive $\textbf{W}$ and $b$---The equation that we **eventually** need to solve for is $\textbf{U}\cdot\textbf{W} + b$* How do we find the $\textbf{W}$ and $b$?* We need both of those to solve this equation for a given vector $\textbf{U}$To find $\textbf{W}$ and $b$, we need to look at the given information:We know that, for any $\textbf{x}_{-SV}$ and $\textbf{x}_{+SV}$, $$\textbf{x}_{-SV}\cdot\textbf{W} + b = -1$$and$$\textbf{x}_{+SV}\cdot\textbf{W} + b = +1$$To solve this, we also need to introduce $y_i$ (the class info), such that * if $y_i \in +Class$, then $y_i = +1$* if $y_i \in -Class$, then $y_i = -1$For the $+Class$, we know that $\textbf{x}_i\cdot\textbf{W} + b = +1$ $\rightarrow$ $y_i = +1$For the $-Class$, we know that $\textbf{x}_i\cdot\textbf{W} + b = -1$ $\rightarrow$ $y_i = -1$If we multiply each of the above equations by $y_i$, then we can show that $$y_i\left(\textbf{x}_i\cdot\textbf{W} + b\right) = 1 \;\;\;\; \forall \textbf{x}_i \in +Class $$and$$y_i\left(\textbf{x}_i\cdot\textbf{W} + b\right) = 1 \;\;\;\; \forall \textbf{x}_i \in -Class $$because $y_i \cdot y_i = (\pm 1)^2 \equiv 1$Therefore we have $$ y_i\left(\textbf{x}_i\cdot\textbf{W} + b\right) - 1 = 0 $$---This equation is a constraint that we as the scientist are imposing onto a "Support Vector". But what **is** a Support Vector?We call each vector to each labeled training data set a "Support Vector" because if we moved them, then it will effect the location of the best dividing hyperplane. Therefore, in SVM, "Support" mean that the training data support the location of the best separating hyperplane between each group in the data. In SVM, "Vector" means the vector between the origin and the training data point.To start estimating the best dividing hyperplane, we could start by fitting a parallel line that goes through the center of each group of data, then finding the width between these parallel lines between group A and group B. Call that WIDTHThe first "best dividing" hyperplane (BDH) could be set up as the line parallel to BOTH parallel lines through the midpoints, but the BDH is at width WIDTH / 2.Therefore, there are now 3 parallel lines:* One that goes through the midpoint of group A* One that goes through the midpoint of group B* One (the BDH) that goes through the midpoint of those midpointsThis last one is exactly WIDTH/2 from each of the other parallel lines---$\textbf{w} = \max \left\{WIDTH = (\textbf{x}_+ - \textbf{x}_-) \cdot \frac{\textbf{w}}{||\textbf{w}||}\right\} \forall \{\textbf{x}_+ , \textbf{x}_-\}$Where $\{\textbf{x}_+ , \textbf{x}_-\}$ come from solving $y_i\left(\textbf{x}_i\cdot\textbf{w} + b\right) - 1 = 0$Therefore: $WIDTH = \frac{2}{||\textbf{w}||}$As such, $\textbf{w} = \max \left\{WIDTH = \frac{2}{||\textbf{w}||}\right\}$or for analytical purposes, $\textbf{w} = \min \left\{\frac{||\textbf{w}||}{2}\right\}^2$with constraint from the support vectors: $y_i\left(\textbf{x}_i\cdot\textbf{w} + b\right) - 1 = 0$---Lagrange Multipliers---Using Lagrange Multipliers, we can optimize a function with respect to a set of constraints:maximize $f(x, y)$, subject to $g(x, y) = 0$ (constraint)We need both $f$ and $g$ to have continuous first partial derivatives. We introduce a new variable ($λ$), called a Lagrange multiplier, and study the Lagrange function (or Lagrangian) defined by$$ \mathcal{L}(x,y,\lambda) = f(x,y) - \lambda \cdot g(x,y)$$If the solution $(x_o, y_o)$ is the maximum of $f(x,y)$, then $\exists \lambda_o$, such that $(x_o, y_o, \lambda_o)$ is a *stationationary point* of the Lagrangian $\mathcal{L}(x,y,\lambda)$, such that,$$\left.\nabla_{x,y,\lambda} \mathcal{L}(x , y, \lambda)\right|_{(x_o, y_o, \lambda_o)} = \left.\left ( \frac{\partial \mathcal{L}}{\partial x}, \frac{\partial \mathcal{L}}{\partial y}, \frac{\partial \mathcal{L}}{\partial \lambda} \right )\right|_{(x_o, y_o, \lambda_o)} \equiv \textbf{0}$$*This is a necessary, but not sufficient condition*---For SVM, we can show that $$\mathcal{L}(\textbf{w}, b, \bf{\alpha}) = \frac12||\textbf{w}||^2 - \sum \alpha_i \left[y_i(\textbf{x}_i\cdot\textbf{w} + b - 1)\right] \;\;\;\;\; \text{where}\;\bf{\alpha}\;\text{are the Lagrange multipliers}$$To solve this equation, we want to **minimum** $\textbf{w}$ AND **maximize** $b$.---Why do we want to maximize $b$?Look at the equation of the hyperplane: $\textbf{w}\cdot\textbf{x} + b$ - which looks a lot like $m x + b$, or the equation for a lineSo modifying $b$ moves the hyperplane up and down; the same is basically true for hyperplanesThis makes $b$ your "bias", or global offset -- same as astronomical detectors---Back to Lagrange multipliers: $$\mathcal{L}(\textbf{w}, b, \bf{\alpha}) = \frac12||\textbf{w}||^2 - \sum \alpha_i \left[y_i(\textbf{x}_i\cdot\textbf{w} + b - 1)\right]$$We need to solve the linear set of equations for $$\left.\nabla_{x,y,\lambda} \mathcal{L}(x , y, \lambda)\right|_{(x_o, y_o, \lambda_o)} = \left.\left ( \frac{\partial \mathcal{L}}{\partial x}, \frac{\partial \mathcal{L}}{\partial y}, \frac{\partial \mathcal{L}}{\partial \lambda} \right )\right|_{(x_o, y_o, \lambda_o)} \equiv \textbf{0}$$In this case, that is$$\left.\nabla_{\textbf{w},b,\lambda} \mathcal{L}(x , y, \lambda)\right|_{(x_o, y_o, \lambda_o)} = \left.\left ( \frac{\partial \mathcal{L}}{\partial \textbf{w}}, \frac{\partial \mathcal{L}}{\partial b}, \frac{\partial \mathcal{L}}{\partial \lambda} \right )\right|_{(x_o, y_o, \lambda_o)} \equiv \textbf{0}$$or $$\left( \begin{array}{c} \frac{\partial \mathcal{L}}{\partial \textbf{w}} \\ \frac{\partial \mathcal{L}}{\partial b} \\ \frac{\partial \mathcal{L}}{\partial \lambda} \end{array} \right) = \left(\begin{array}{c} 0 \\ 0 \\ 0 \end{array} \right)$$Separating this into three equations:$$ \frac{\partial \mathcal{L}}{\partial \textbf{w}} = \textbf{w} - \sum_i \alpha_i y_i \textbf{x}_i = 0$$$$\text{or}$$$$\textbf{w} = \sum \alpha_i y_i \textbf{x}_i$$Finally, $$ \frac{\partial \mathcal{L}}{\partial b} = -\sum_i \alpha_i y_i = 0 \;\;\;\;\;\text{which becomes}\;\;\;\;\; \frac{\partial \mathcal{L}}{\partial b} = \sum_i \alpha_i y_i = 0$$---Putting these together and maximizing with respect to $(\textbf{w}, b, \lambda)$, we can show that $$ L = \sum \alpha_i - \frac12 \sum_{ij} \alpha_i\alpha_j y_i y_j (x_i \cdot x_j)$$**Proof:**Because $$\textbf{w} = \sum \alpha_i y_i \textbf{x}_i$$ **and** $$\mathcal{L}(\textbf{w}, b, \bf{\alpha}) = \frac12||\textbf{w}||^2 - \sum \alpha_i \left[y_i(\textbf{x}_i\cdot\textbf{w} + b - 1)\right]$$Substituting $\textbf{w}$ in $\mathcal{L}$,but replacing $i \rightarrow j$ in the second summation, gives$$\mathcal{L}\left(\left(\sum_j \alpha_j y_j \textbf{x}_j \right), b, \bf{\alpha}\right) = \frac12\left|\left|\sum_j \alpha_j y_j \textbf{x}_j\right|\right|^2 - \sum_i \alpha_i \left[y_i(\textbf{x}_i\cdot\left(\sum_j \alpha_j y_j \textbf{x}_j\right) + b - 1)\right] = \frac12\left|\left|\sum_j \alpha_j y_j \textbf{x}_j\right|\right|^2 - \sum_i\sum_j \alpha_i \alpha_j \left[y_i y_j(\textbf{x}_i\cdot\textbf{x}_j) + b - 1)\right]$$-- WORK ON THIS PROOF -----It is important to cover the formal derivation of the SVM to understand the ups and downs to using it. 1) Downside: Significant depth of linear algebra (relative difficulty) 2) Downside: Need to keep all of your feature vectors in memory at the same time to process -- does not scale very well (computational difficulty) -- can batch subsets of data into scales of SVM to work with large data sets -- one of the more popular choice these days is "Sequential Miminal Optimization" or "SMO" created by John Platt at Microsoft 3) Upside: Once you have trained the SVM, you do not need the feature vectors ever again. -- Making predictions from a trained SVM is as simple as $$predict(x_{new}) = sign[\textbf{w}\cdot x_{new} + b]$$ ---A more simple explanation of SVM---Back to the basics: What is the equation for a hyperplane?$$ \textbf{x}\cdot\textbf{w} + b $$What is the hyperplane for a + class support vector?$$ \textbf{x_+}\cdot\textbf{w} + b = +1 $$What about the hyperplane for a - class support vector?$$ \textbf{x_-}\cdot\textbf{w} + b = -1 $$The $x_i$ that satisfy this constraint are called the "Support Vectors"For Example, what if you had $\textbf{u}\cdot \textbf{w} + b = 0.98$? It's not +1 or -1, so how do we interpret this result?If we plot all $\textbf{u}$ onto a graph along with the support vector hyperplanes (parallel planes through the midpoint of each class) -- as well as the best separating hyperplane (at the midpoint of those midpoints and parallel to the support vector hyperplanes) -- then where does 0.98 fall on this figure?The point, $\textbf{u}$, will lie very very close to the + class support vecotr hyperplane. It's not quite "all the way" to the support vector hyperplane, but it is much much farther into the + class regime, than the - class regime; and not even close to the decision boundary---**Decision Boundary**If, for a given vector $\textbf{u}$, we have that $\textbf{u}\cdot\textbf{w}+b \equiv 0$, then $\textbf{u}$ is part of the (or on the) "decision boundary"---**Classification of a feature set:** What is the equation that will determine the classes of every element in a feature set, $\{\textbf{x}_i\}$? $$\text{Class} \equiv Sign\{\textbf{x}_i\cdot\textbf{w} + b\}$$That is simply asking "is this point in the feature set LEFT or RIGHT of the best dividing hyperplane?---**Determine the hyperplane:** How to determine $\textbf{w}$ \& $b$?- Want maximize $||\textbf{w}||$ and minimize $b$There are an infinite "number" of $\textbf{w}$'s and an infinite number of $b$'s. Remember, that we want to maximize the MAGNITUDE (||w|||) of $\textbf{w}$, which is the euclidean norm between the components.Therefore, what is the constraint of $||\textbf{w}||$ and $b$?From the more rigorous tutorial above, the constraint is $$y_i\left(\textbf{x}_i\cdot\textbf{w}+b\right) \geq 1$$Because our classes ($y_i$) will be either a +1 or a -1, with the knowledge of the hyperplane $\textbf{x}_i\cdot\textbf{w}+b$, our constraint becomes:$$y_i\left(\textbf{x}_i\cdot\textbf{w}+b\right) \geq 1 \;\;\;\; \text{as specified above}$$This is because for -1 class, $\textbf{x}_i\cdot\textbf{w}+b = -1$ and for +1 class $\textbf{x}_i\cdot\textbf{w}+b = +1$, so multiplying both sides by $y_i$ -- which is also -1 or +1, respecitively -- makes the right hand side (RHS) == 1 for all feature values.**Note** that there are an infinite number of $\textbf{w}$ and $b$ such that the constraint $y_i\left(\textbf{x}_i\cdot\textbf{w}+b\right) \geq 1$ is satisfied. - Therefore, we must find the mimium $\textbf{w}$ and maximum $b$ (simultaneously?) that satisfy this constraint.- This generates a quadratic computational problem- The good news is the SVM has been shown to be convex (the shape of the paramater space is convex) - convex: Any line between any two points on the boundaries does not cross any other boundary - this implies that there are no local minima, which is too good to be trueWe want to use this to minimize $||\textbf{w}||$ while we maximize $b$, such that $y_i(\textbf{x}_i\cdot\textbf{w} +b) \geq 1$. Where **(he finally declared)** that $\textbf{x}_i$ and $y_i$ are your training data set features and labels! **But**, the "support vectors" are *ONLY* the feature-label pairs that match "pefectly" (close enough) to the constraint equation. Such that, $x_i$ and $y_i$ are the entire feature-label pairs, but a specific subset of these that "[close enough to] perfectly" satisfy the constraint equation -- $y_i(\textbf{x}_i\cdot\textbf{w} +b) - 1 = 0$ -- are called the "support vectors".---What is an example of a vector $\textbf{w}$?- In 2D feature space (because SVM is a binary classifier), $\textbf{w}$ could be $[5,3]$- It could also be $[-5,3]$, which has the same magnitue: or $||[-5,3]|| = ||[5,3]|| = \sqrt{34}$If we are minimizing the magnitude of $||\textbf{w}||$ and maximizing $b$, how can we discern between $[5,3]$ and $[-5,3]$? \*\*\*\*\*\*\*\*\*\*\*\* **Because** we are **ONLY** maximizing and minimizing **WITH RESPECT TO** the constraint $y_i(\textbf{x}_i\cdot\textbf{w} + b) - 1$ **AND** $[5,3]$ is **VERY** different from $[-5,3]$ in the dot product with the training data \*\*\*\*\*\*\*\*\*\*\*\*---How to derive $\textbf{w}$---- Start with an initial guess: $\textbf{w} = [5,5]$ and look for the maximum value for $b$ that satisfies the constraint $y_i(\textbf{x}_i\cdot\textbf{w} + b) - 1$ and this is your candidate, initial hyperplane- Take initial guesses, calculate the magnitute and use the constraint equation to derive the maximum value of $b$- Step down in magnitude space by moving to a small vector in feature space and re-asses the magnitude as well as $b$**Because this problem has been shown to be convex:**- If you take baby steps, then you can determine the local slope. - Then take larger steps in that direction until you pass the global minimum: chisq_new > chisq_old- Reduce your step size and go back until chisq_new > chisq_old and bounce like this until "convergence"**Because Convex: All Local Minima == Global Minimum**Without the knowledge of convex mapping, SVM minimization can avoid being trapped in a local minimum- For a given guess of the $\textbf{w}$, we will increase $b$ until the constraint equation $y_i(\textbf{x}_i \cdot \textbf{w} + b) \geq 1$- Save this combination in a dictionary \{||$\textbf{w}||$:$\textbf{w}$, $b$\} Scikit-Learn Implementation--- Copy the KNN-C code and substitute "SVM" in for the previously implemented KNN
###Code
import numpy as np
from sklearn import preprocessing, cross_validation, svm#, neighbors
import pandas as pd
df = pd.read_csv('breast-cancer-wisconsin.data')
df.replace('?', -99999, inplace=True)
useless = ['ID']
df.drop(useless, 1, inplace=True)
X = np.array(df.drop(['class'], 1))
y = np.array(df['class'])
test_size = 0.2
accuracies = []
nIters = 100
for k in range(nIters):
X_train, X_test, y_train, y_test = cross_validation.train_test_split(X,y, test_size=test_size)
# clf = neighbors.KNeighborsClassifier()
clf = svm.SVC()
clf.fit(X_train, y_train)
accuracy = clf.score(X_test, y_test)
accuracies.append(accuracy)
# print "{0:d} Accuracy = {1:.2f}%".format(k, accuracy*100)
print "Mean Accuracy = {0:.2f}%".format(np.mean(accuracies)*100)
example_measures = np.array([[4,2,1,1,1,2,3,2,1], [4,2,1,2,2,2,3,2,1]])
example_measures = example_measures.reshape(len(example_measures), -1)
prediction = clf.predict(example_measures)
print prediction
###Output
Mean Accuracy = 96.02%
[2 2]
###Markdown
**Sentdex Reference Material:**https://web.stanford.edu/~boyd/cvxbook/bv_cvxbook.pdfhttp://research.microsoft.com/pubs/68391/smo-book.pdfhttp://cvxopt.org/**Optimization algorithm idea: (j.fraine 6-7-16)**- involving friction: if f(x_new) > f(x_old), "accelerate" walker in the opposition direction and add "friction" (shorten length scale of steps)**Bayesian Analysis Constraint Method**:- use "prior" on the posterior to be the constraint in question- for SVM, set log(prior) = 0 if $y_i\left(\textbf{x}_i\cdot\textbf{w}+b\right) < 1$ and log(prior) = 1 if $y_i\left(\textbf{x}_i\cdot\textbf{w}+b\right) \geq 1$**Cool Clock Idea:**- Start with http://demo.bokehplots.com/apps/timeout- Make the numbers appear every second with the current second for this minute- Make all the numbers from seconds that have past fade away over time (maybe 10 - 20s each)- Have the minute number update *in the same place* over and over again -- still fading away, but every 5 - 10s pulse- The location of the minute is static for 1 minute, then randomly chosen somewhere else for the next minute- Same thing for the hour, but make it pulse slower (maybe once per minute)- Have specific colors and sizes for the hour, minute, seconds in order to easily differentiate between them- Possible do the same for the date**My thoughts on deriving the best separating hyperplane for an SVM (at least an initial guess):** - Start with the median over feature space of all points in the +Class and all of the points in the -Class.- Then determine the slope for a line through the median of the -Class and the median of the +Class that is parallel to each other - (i.e. has the **same** slope)- Next, rotate the line around the median of +Class and the median of -Class, keeping them parallel, until they are maximally separated.- Then, the initial guess hyperplane in the midploint between the two medians from each class with a slope that is parallel (a.k.a. equal) to the maximizing parallel slope of the two classes themselves**Option 2:**- Start with any random guess for the slope- Assess the medians of each class, call them +med and -med- use a minimizer on the Lagrange multiplier equation that minimizes the magnitude of $\textbf{w}$ and maximizes $b$.- The minizer criterion could be "maximum difference between all members of each class - Do this simultaneously, not in a stepping pattern like Sentdex is proposing - Maybe estimate with MCMC My Personal Example of SVM Classication with SKLearn--- **Load all necessary libraries and set matplotlib to inline with figsize = 12,12**
###Code
%matplotlib inline
import numpy as np
from matplotlib.pyplot import plot, scatter, xlim, ylim, cm, rcParams
from sklearn.svm import SVC
from sklearn.cross_validation import train_test_split
rcParams['figure.figsize'] = 12,12
###Output
_____no_output_____
###Markdown
**Generate input data for positions in 2D space**
###Code
nPts = 1e4
xdata = np.random.normal(0,1, nPts)
ydata = np.random.normal(0,1, nPts)
iMovers = np.random.randint(0,xdata.size,xdata.size/2)
movers = np.ones(xdata.size)
movers[iMovers]-= 2.0
xmax = 4.0
xdata += movers*xmax
ydata += movers*xmax
###Output
/Users/jonathanfraine/anaconda/lib/python2.7/site-packages/ipykernel/__main__.py:2: DeprecationWarning: using a non-integer number instead of an integer will result in an error in the future
from ipykernel import kernelapp as app
/Users/jonathanfraine/anaconda/lib/python2.7/site-packages/ipykernel/__main__.py:3: DeprecationWarning: using a non-integer number instead of an integer will result in an error in the future
app.launch_new_instance()
###Markdown
**Convert given 2D data to feature space and define labels at 1,-1**
###Code
X = np.transpose([xdata,ydata])
labels = np.empty(xdata.size)
labels[xdata < 0.0] = -1
labels[xdata > 0.0] = 1 # might be redundant if we start with labels = np.ones
###Output
_____no_output_____
###Markdown
**Split the data in the training and testing subsets for cross-validation**
###Code
random_state = 42
test_size = 0.8
X_train, X_test, Y_train, Y_test = train_test_split(X, labels, test_size=test_size, random_state=random_state)
###Output
_____no_output_____
###Markdown
**Define and fit the SVC model**
###Code
clfr = SVC(kernel='linear', C = 1.0)
clfr.fit(X_train,Y_train)
###Output
_____no_output_____
###Markdown
**Define the separating hyperplane and hyperplanes for 'closest' Support vectors to hyperplane**
###Code
#define the seperating hyperplane
w = clfr.coef_[0]
a = -w[0] / w[1]
xx = np.linspace(-2.5*xmax, 2.5*xmax)
yy = a * xx - (clfr.intercept_[0]) / w[1]
# define the parallels to the separating hyperplane that pass through the closest support vectors
b = clfr.support_vectors_[0]
yy_down = a * xx + (b[1] - a * b[0])
b = clfr.support_vectors_[-1]
yy_up = a * xx + (b[1] - a * b[0])
###Output
_____no_output_____
###Markdown
**Plot the training data, testing data, seperating hyperplane, and closest parallel hyperplanes**
###Code
plot(xx, yy, 'k-')
plot(xx, yy_down, 'k--')
plot(xx, yy_up, 'k--')
scatter(clfr.support_vectors_[:, 0], clfr.support_vectors_[:, 1], s=80, facecolors='none')
scatter(X_train[:, 0], X_train[:, 1], c=clfr.predict(X_train), cmap=cm.Paired, alpha=0.2, edgecolor='None')
scatter(X_test[:, 0], X_test[:, 1], c=clfr.predict(X_test), cmap=cm.Paired, alpha=0.5, edgecolor='None')
xlim(-2.5*xmax,2.5*xmax)
ylim(-2.5*xmax,2.5*xmax)
###Output
_____no_output_____
###Markdown
SentDex Hand-Written SVM Classifier
###Code
import numpy as np
import matplotlib.pyplot as plt
from matplotlib import style
style.use('fivethirtyeight')
class Support_Vector_Machine(object):
def __init__(self, visualization=True):
self.visualization = visualization
self.colors = {1:'orange', -1:'lightblue'}
if self.visualization:
self.fig = plt.figure()
self.ax = self.fig.add_subplot(111)
return
def fit(self, data):
# fit == train
self.data = data
# opt_dict = {||w||:[w,b]}
opt_dict = {}
# ?? transforms converts w to positive definite ??
transforms = [[1 , 1],
[-1, 1],
[-1,-1],
[1 ,-1]]
# This is dirty; fix it; maybe use dictionary of arrays
all_data = []
for yi in self.data:
for featureset in self.data[yi]:
for feature in featureset:
all_data.append(feature)
self.max_feature_value = max(all_data)
self.min_feature_value = min(all_data)
all_data = None # Great idea for clearing out memory!
# Because support vectors with satisfy: yi*(dot(xi,w) + b) = 1,
# we can iterate until abs(yi*(dot(xi,w) + b) - 1) < epsilon ~ 0.01 = 1 / stepstepsize**2
maxstepsize = 0.1
stepstepsize= 10.0
step_sizes = [self.max_feature_value*maxstepsize,
self.max_feature_value*maxstepsize / stepstepsize,
# point of expense:
self.max_feature_value*maxstepsize / stepstepsize**2]
# extremely expensive!
# `b` does not need to take as small of steps as `w` because `b` does not need to be as precise
# -- to be explained in application stage
b_range_multiple = 5
# we don't need to take as small of steps with `b` as we do with `w`
b_multiple = 5
# Start big!
latest_optimum = self.max_feature_value*stepstepsize # might be a different x10 here than `stepstepsize`
for step in step_sizes:
# major corners being cut here!
w = np.array([latest_optimum, latest_optimum])
# Controls for "have you found the minimum"
# We can do this because the SVM problem is convex
optimized = False
while not optimized:
for b in np.arange(-(self.max_feature_value*b_range_multiple),
self.max_feature_value*b_range_multiple ,
step*b_multiple):
for transformation in transforms:
w_t = w*transformation
found_option= True
# weakest link in the SVM fundamentally
# SMO attempts to fix this a bit; but it can only do "enough"
# check constraint on all data: yi*(dot(xi, w)+ b)
for i in self.data:
for xi in self.data[i]:
yi = i
if not yi*(np.dot(w_t,xi)+b) >= 1:
found_option = False
# should just break once a single feature is out of the range for the hyperplane
break
if found_option:
opt_dict[np.linalg.norm(w_t)] = [w_t, b]
if w[0] < 0:
optimized = True
print 'Optimized a step.'
else:
# Sentdex wants to know if python broadcasts scalars to lists
# w = [N,N]
# step = S
# w - step = [N-S, N-S]
w = w - step
# if we optimized the `w`, then we can trigger the while loop
# Here we sort the resulting list of norms (magnitudes)
norms = sorted([n for n in opt_dict])
# assign the best result to our hyperplane slope & intercept
# note that opt_choice is dictionary organized by the magnitude
# and the "minimum" magnitude is the first element in the sorted `norms` array
# ||w|| : [w,b]
opt_choice = opt_dict[norms[0]]
if np.rank(opt_choice[0]) > 1:
print opt_choice[0], opt_choice[1]
self.w = opt_choice[0]
self.b = opt_choice[1]
# Sentdex used `2`, but said we should use `stepstepsize`
latest_optimum = opt_choice[0][0] + step*stepstepsize
def predict(self, features):
# class = sign{dot(x,w) + b}
# nObs, nFeatures= features.shape
# classification = np.zeros(nObs)
# for k in nObs:
# classification[k] = np.sign(np.dot(np.array(features[k], self.w)) + self.b)
classification = np.sign(np.dot(np.array(features), self.w) + self.b)
if classification and self.visualization:
self.ax.scatter(features[0], features[1], s=200, marker='*', c=self.colors[classification])
return classification
def visualize(self):
[[self.ax.scatter(x[0], x[1], s=200, c=self.colors[i]) for x in data_dict[i]] for i in data_dict]
# hyperplane = dot(x,w) + b
# v = x.w+b
def hyperplane(x,w,b,v):
'''
x = feature space (theory curve)
w = best fitted hyperplane slope
b = best fitted hyperplane intercept
v = values on the decision plane
'''
return (-w[0]*x-b+v) / w[1]
#
datarange = self.min_feature_value*0.9, self.max_feature_value*1.1
hyp_x_min = datarange[0]
hyp_x_max = datarange[1]
psv = 1 # positive support vector boundary hyperplane
nsv = -1 # negative support vector boundary hyperplane
dec = 0 # separating hyperplane value
# decision boundary points
dec1 = hyperplane(hyp_x_min, self.w, self.b, dec)
dec2 = hyperplane(hyp_x_max, self.w, self.b, dec)
# positive support vector points
psv1 = hyperplane(hyp_x_min, self.w, self.b, psv)
psv2 = hyperplane(hyp_x_max, self.w, self.b, psv)
# negative support vector points
nsv1 = hyperplane(hyp_x_min, self.w, self.b, nsv)
nsv2 = hyperplane(hyp_x_max, self.w, self.b, nsv)
# plot data points as a straight line
self.ax.plot([hyp_x_min, hyp_x_max], [dec1, dec2], '-' , c='yellow')
self.ax.plot([hyp_x_min, hyp_x_max], [psv1, psv2], '--', c='black')
self.ax.plot([hyp_x_min, hyp_x_max], [nsv1, nsv2], '--', c='black')
minusClassData = np.array([[1,7],
[2,8],
[3,8]])
plusClassData = np.array([[5,1],
[6,-1],
[7,3]])
data_dict = {-1:minusClassData,
1:plusClassData}
svm = Support_Vector_Machine()
svm.fit(data=data_dict)
predict_us = [[0,10],
[1,3 ],
[3,4 ],
[3,5 ],
[5,5 ],
[5,6 ],
[6,-5],
[5,8 ]]
svm.visualize()
for p in predict_us:
svm.predict(p)
for p in np.random.uniform((0,-5),(10,15),(100,2)):
svm.predict(p)
###Output
Optimized a step.
Optimized a step.
Optimized a step.
###Markdown
Non-linear Classification with SVM If data is non-linear distribution, such that there is strong overlap between the data in 2D space,then it is always possible to add N non-linear dimensions (i.e.: [x1,x2,x1*x2]) such that the data is linearly separable in those 3 dimensions- May need to add 10s - 100s of dimensions to find classification structures that are linearly separable- The number of dimensions and the trainign result depends on the Kernel of choice How to use a kernel inside the SVM---Given and $\textbf{x}$, then the linear prediction becomes $$\text{prediction} = sign(\textbf{w}\cdot\textbf{x}+b)$$If we want to use a kernel, then we will first transform the feature space $\textbf{x}$ into $\textbf{z} = Kernel(\textbf{x})$, such that the kernel prediction becomes $$\text{prediction}= sign(\textbf{w} \cdot \textbf{z} + b)$$---The Constraint equation is then transformed from $y_i(\textbf{x}\cdot\textbf{w}+b)$ into $y_i(\textbf{x}\cdot\textbf{z}+b)$and finally the Lagrange multiplier constraint is transformed from $\textbf{w} = \sum\alpha_i y_i\cdot\textbf{x}_i$into $\textbf{w} = \sum\alpha_i y_i\cdot\textbf{z}_i$---Under the Lagrange multiplier, the solution is transformed from$$L = \sum\alpha_i - \frac12\sum_{ij} \alpha_i \alpha_j y_i y_j (x_i \cdot x_j)$$ into $$L = \sum\alpha_i - \frac12\sum_{ij} \alpha_i \alpha_j y_i y_j (z_i \cdot z_j)$$Where $\textbf{z} = Kernel(\textbf{x})$ in all of these cases --- How to use kernels with SVMKernels are defined as $k(x,x') = z \cdot z'$, where z = function($x$) and z' = function($x'$)When you do a kernel on something comma something prime, you **must** use the same function on $x$ and $x'$.For later notation, we can also writen as a matrix in the form of $k(x,x') = \phi$, such that $k(x,x') = \phi \cdot x$The question is "can we calculate the inner product of the $z$ feature space without knowing the form of the transform function?"Application: we are going to convert every feature space data point point $[x_1, x_2] \rightarrow 2^{nd}$ order polynomial. Such that, with $X = [x_1, x_2]$, $Z = [1,x_1, x_2, x_1^2, x_2^2,x_1 x_2]$Then, $K(x,x') = z \cdot z' = [1, x_1, x_2, x_1^2, x_2^2, x_1 x_2] \cdot [1, x'_1, x'_2, (x')_1^2, (x')_2^2,x'_1 x'_2] = [1, x_1 x'_1, x_2 x'_2, x_1^2 (x')_1^2, x_2^2 (x')_2^2, x_1 x'_1 x_2 x'_2]$It can be show that we can simplify this **for any order polynomial** to $K(x,x') = (1+x \cdot x')^p$, where we previously used $p=2$; note our example also used $n=2$ dimensions for each $x$, but that can also be arbitrary. The dot product for an arbitrary N dimensions $x$ and $x'$ would look like:$$x\cdot x' = \sum\limits_{j=0}^n x_j x'_j = 1 + x_1 x'_1 + x_2 x'_2 + \ldots x_n x'_n$$---RBF: Radial Basis Vector Kernel$$K(x,x') = e^{-\gamma ||x-x'||^2}$$Can go out to an infinite number of dimensions. **But**, but there is always a problem with going out to an infinite number of dimensions. There may be no classification correlation in the data at all, but the noise could approach becoming linearly separable in that "infiniteth" dimension.The question becomes, how do you know if you have made a mistake? and how do we avoid making these mistakes in any given data set?Note that RBF is the default kernel for SKLearn and most pre-built packages. This is because RBF is known to work "most of the time". But, fundamentally, there exist data sets that RBF will **not** be able to separate. In reality, RBF should always be able to separate the data -- even when that separation is not valid! --- Soft Margin SVM KernelGiven a non-linearly separable coordinate space, we can use RBF and find a given (non-linear in feature space) hyperplane that indeed separates the categories of the data into two half-planes.Then the "support vectors" are those vectors that are "[close enough to] perfectly" satisfy the constraint equation -- $y_i(\textbf{x}_i\cdot\textbf{w} +b) - 1 = 0$. If with the RBF kernel, the support vectors end up being a large majority of the feature space, then that is a **major** red-flag that the kernel is over fitting.It would be better to use a kernel that does not fit support vectors with "most" of the data. Overfitting training data means that you will likely miss a lot of the test data. *sidenote:* maybe it's a good idea to iterative fit to the training data, check score on the test data, modify the hyperparameters, re-fit until a re-sampled random subset of training vs testing data are both accurate, as well as not overfitting. *Normal behaviour:* It is a good idea to have nSV / nSamples be small (no where near 1.0)! If this ratio is close to 1.0, then you are probably overfitting.- maybe we could use this in the iterative fitting process above in *sidenote*.- nSV / nSamples > 10% should be taken cautiously.- *If* the SVM is 52% accurate, but nSV / nSamples ~80%, there is significant over fitting; and you should try a different kernel.- *But* if the SVM is 52% accurate, but nSV / nSamples ~8%, then it's possible that there is no good kernel to work with.Examples:We could start with a straight line that *almost* splits the data into two correct groups. If it has a small number of "violations". We can take the linear support vector hyperplanes, and generate a soft-margin classifier. This cannot be done with 'large' distances off of the separating hyperplane.Note that RBF and linear are "hard margin" classifiers. Most pre-defined packages with use 'soft-margin' hyperplanes by default.--- Slack $\xi$: fit for hyperplane with a specific kernel, but allow for deviations in the absolute constraintSuch as, if $\xi$ is the 'slack', then $y_i(x_i \cdot w +b) \geq 1$ becomes $y_i(x_i \cdot w +b) \geq 1 - \xi$. If $\xi = 0$, then we are specifying a "hard-margin" classifier.We would want to 'minimize' the slack $\xi$ and may actually process to literally miniize $\xi$ in our routine.In the default case that we programmed before, we minimiezd $||w||$ by minimizing $\frac12||w||^2$ (for computational convenience (i.e. smoothness and convex behaviour). Now we want to minize $\frac12||w||^2 + C \sum_i \xi_i$ (summing over all of the slacks); where $C$ is a scaling coefficient that defines the relative ability or desire to use a slack. Each $\xi_i$ is a slack per feature, defining soft of margins to implement; but $C$ scales the entire allowance for soft-margin considerations *entirely*.The smaller that we make $C$, the less the value of $\xi$ matters.By default in most packages, $C$ and $\xi$ are $>0$. Testbed: $C=10^6$ compared to $C=1$ (sklearn default) ---Example Usage in Python--- Mathieu Blondel, September 2010License: BSD 3 Clause- http://www.mblondel.org/jounral/2010/09/19/support-vector-machines-in-python/- https://gist.github.com/mblondel/586753- https://gist.githubusercontent.com/mblondel/586753/raw/6e0c2ac3160ab5a7068b6f0d39f2e06b97eb0f2b/svm.py
###Code
## import numpy as np
from numpy import linalg
# cvxopt.org/userguide/coneprog.html#quadratic-programming
import cvxopt
import cvxopt.solvers
def linear_kernel(x1, x2):
return np.dot(x1, x2)
def polynomial_kernel(x, y, p=3):
return (1 + np.dot(x, y)) ** p
def gaussian_kernel(x, y, sigma=5.0):
return np.exp(-linalg.norm(x-y)**2 / (2 * (sigma ** 2)))
class SVM(object):
def __init__(self, kernel=linear_kernel, C=None):
self.kernel = kernel
self.C = C
if self.C is not None: self.C = float(self.C)
def fit(self, X, y):
n_samples, n_features = X.shape
# Gram matrix
K = np.zeros((n_samples, n_samples))
for i in range(n_samples):
for j in range(n_samples):
K[i,j] = self.kernel(X[i], X[j])
P = cvxopt.matrix(np.outer(y,y) * K)
q = cvxopt.matrix(np.ones(n_samples) * -1)
A = cvxopt.matrix(y, (1,n_samples))
b = cvxopt.matrix(0.0)
if self.C is None:
G = cvxopt.matrix(np.diag(np.ones(n_samples) * -1))
h = cvxopt.matrix(np.zeros(n_samples))
else:
tmp1 = np.diag(np.ones(n_samples) * -1)
tmp2 = np.identity(n_samples)
G = cvxopt.matrix(np.vstack((tmp1, tmp2)))
tmp1 = np.zeros(n_samples)
tmp2 = np.ones(n_samples) * self.C
h = cvxopt.matrix(np.hstack((tmp1, tmp2)))
# solve QP problem
solution = cvxopt.solvers.qp(P, q, G, h, A, b)
# Lagrange multipliers
a = np.ravel(solution['x'])
# Support vectors have non zero lagrange multipliers
sv = a > 1e-5
ind = np.arange(len(a))[sv]
self.a = a[sv]
self.sv = X[sv]
self.sv_y = y[sv]
print "%d support vectors out of %d points" % (len(self.a), n_samples)
# Intercept
self.b = 0
for n in range(len(self.a)):
self.b += self.sv_y[n]
self.b -= np.sum(self.a * self.sv_y * K[ind[n],sv])
self.b /= len(self.a)
# Weight vector
if self.kernel == linear_kernel:
self.w = np.zeros(n_features)
for n in range(len(self.a)):
self.w += self.a[n] * self.sv_y[n] * self.sv[n]
else:
self.w = None
def project(self, X):
if self.w is not None:
return np.dot(X, self.w) + self.b
else:
y_predict = np.zeros(len(X))
for i in range(len(X)):
s = 0
for a, sv_y, sv in zip(self.a, self.sv_y, self.sv):
s += a * sv_y * self.kernel(X[i], sv)
y_predict[i] = s
return y_predict + self.b
def predict(self, X):
return np.sign(self.project(X))
if __name__ == "__main__":
import pylab as pl
def gen_lin_separable_data():
# generate training data in the 2-d case
mean1 = np.array([0, 2])
mean2 = np.array([2, 0])
cov = np.array([[0.8, 0.6], [0.6, 0.8]])
X1 = np.random.multivariate_normal(mean1, cov, 100)
y1 = np.ones(len(X1))
X2 = np.random.multivariate_normal(mean2, cov, 100)
y2 = np.ones(len(X2)) * -1
return X1, y1, X2, y2
def gen_non_lin_separable_data():
mean1 = [-1, 2]
mean2 = [1, -1]
mean3 = [4, -4]
mean4 = [-4, 4]
cov = [[1.0,0.8], [0.8, 1.0]]
X1 = np.random.multivariate_normal(mean1, cov, 50)
X1 = np.vstack((X1, np.random.multivariate_normal(mean3, cov, 50)))
y1 = np.ones(len(X1))
X2 = np.random.multivariate_normal(mean2, cov, 50)
X2 = np.vstack((X2, np.random.multivariate_normal(mean4, cov, 50)))
y2 = np.ones(len(X2)) * -1
return X1, y1, X2, y2
def gen_lin_separable_overlap_data():
# generate training data in the 2-d case
mean1 = np.array([0, 2])
mean2 = np.array([2, 0])
cov = np.array([[1.5, 1.0], [1.0, 1.5]])
X1 = np.random.multivariate_normal(mean1, cov, 100)
y1 = np.ones(len(X1))
X2 = np.random.multivariate_normal(mean2, cov, 100)
y2 = np.ones(len(X2)) * -1
return X1, y1, X2, y2
def split_train(X1, y1, X2, y2):
X1_train = X1[:90]
y1_train = y1[:90]
X2_train = X2[:90]
y2_train = y2[:90]
X_train = np.vstack((X1_train, X2_train))
y_train = np.hstack((y1_train, y2_train))
return X_train, y_train
def split_test(X1, y1, X2, y2):
X1_test = X1[90:]
y1_test = y1[90:]
X2_test = X2[90:]
y2_test = y2[90:]
X_test = np.vstack((X1_test, X2_test))
y_test = np.hstack((y1_test, y2_test))
return X_test, y_test
def plot_margin(X1_train, X2_train, clf):
def f(x, w, b, c=0):
# given x, return y such that [x,y] in on the line
# w.x + b = c
return (-w[0] * x - b + c) / w[1]
pl.plot(X1_train[:,0], X1_train[:,1], "ro")
pl.plot(X2_train[:,0], X2_train[:,1], "bo")
pl.scatter(clf.sv[:,0], clf.sv[:,1], s=100, c="g")
# w.x + b = 0
a0 = -4; a1 = f(a0, clf.w, clf.b)
b0 = 4; b1 = f(b0, clf.w, clf.b)
pl.plot([a0,b0], [a1,b1], "k")
# w.x + b = 1
a0 = -4; a1 = f(a0, clf.w, clf.b, 1)
b0 = 4; b1 = f(b0, clf.w, clf.b, 1)
pl.plot([a0,b0], [a1,b1], "k--")
# w.x + b = -1
a0 = -4; a1 = f(a0, clf.w, clf.b, -1)
b0 = 4; b1 = f(b0, clf.w, clf.b, -1)
pl.plot([a0,b0], [a1,b1], "k--")
pl.axis("tight")
pl.show()
def plot_contour(X1_train, X2_train, clf):
pl.plot(X1_train[:,0], X1_train[:,1], "ro")
pl.plot(X2_train[:,0], X2_train[:,1], "bo")
pl.scatter(clf.sv[:,0], clf.sv[:,1], s=100, c="g")
X1, X2 = np.meshgrid(np.linspace(-6,6,50), np.linspace(-6,6,50))
X = np.array([[x1, x2] for x1, x2 in zip(np.ravel(X1), np.ravel(X2))])
Z = clf.project(X).reshape(X1.shape)
pl.contour(X1, X2, Z, [0.0], colors='k', linewidths=1, origin='lower')
pl.contour(X1, X2, Z + 1, [0.0], colors='grey', linewidths=1, origin='lower')
pl.contour(X1, X2, Z - 1, [0.0], colors='grey', linewidths=1, origin='lower')
pl.axis("tight")
pl.show()
def test_linear():
X1, y1, X2, y2 = gen_lin_separable_data()
X_train, y_train = split_train(X1, y1, X2, y2)
X_test, y_test = split_test(X1, y1, X2, y2)
clf = SVM()
clf.fit(X_train, y_train)
y_predict = clf.predict(X_test)
correct = np.sum(y_predict == y_test)
print "%d out of %d predictions correct" % (correct, len(y_predict))
plot_margin(X_train[y_train==1], X_train[y_train==-1], clf)
def test_non_linear():
X1, y1, X2, y2 = gen_non_lin_separable_data()
X_train, y_train = split_train(X1, y1, X2, y2)
X_test, y_test = split_test(X1, y1, X2, y2)
# clf = SVM(gaussian_kernel)
clf = SVM(polynomial_kernel)
clf.fit(X_train, y_train)
y_predict = clf.predict(X_test)
correct = np.sum(y_predict == y_test)
print "%d out of %d predictions correct" % (correct, len(y_predict))
plot_contour(X_train[y_train==1], X_train[y_train==-1], clf)
def test_soft():
X1, y1, X2, y2 = gen_lin_separable_overlap_data()
X_train, y_train = split_train(X1, y1, X2, y2)
X_test, y_test = split_test(X1, y1, X2, y2)
clf = SVM(C=0.1)
clf.fit(X_train, y_train)
y_predict = clf.predict(X_test)
correct = np.sum(y_predict == y_test)
print "%d out of %d predictions correct" % (correct, len(y_predict))
plot_contour(X_train[y_train==1], X_train[y_train==-1], clf)
# test_linear()
# test_non_linear()
# test_soft()
np.random.RandomState(42)
test_linear()
test_non_linear()
test_soft()
###Output
pcost dcost gap pres dres
0: -1.9797e+01 -3.1819e+01 1e+03 2e+01 7e-15
1: -3.7418e+00 -2.9130e+01 7e+01 1e+00 7e-15
2: -2.0888e+00 -1.2062e+01 1e+01 9e-02 1e-15
3: -2.1454e+00 -3.6619e+00 2e+00 1e-02 2e-15
4: -2.4676e+00 -2.9187e+00 5e-01 3e-03 1e-15
5: -2.5647e+00 -2.7104e+00 2e-01 8e-04 1e-15
6: -2.6058e+00 -2.6341e+00 3e-02 4e-06 2e-15
7: -2.6171e+00 -2.6216e+00 4e-03 5e-07 1e-15
8: -2.6192e+00 -2.6193e+00 9e-05 3e-09 1e-15
9: -2.6192e+00 -2.6192e+00 2e-06 6e-11 1e-15
Optimal solution found.
37 support vectors out of 180 points
20 out of 20 predictions correct
|
06 - Capstone Project/Week 3 Interactive Visual Analytics and Dashboard/Interactive_Visual_Analytics_with_Folium.ipynb
|
###Markdown
**Launch Sites Locations Analysis with Folium** Estimated time needed: **40** minutes The launch success rate may depend on many factors such as payload mass, orbit type, and so on. It may also depend on the location and proximities of a launch site, i.e., the initial position of rocket trajectories. Finding an optimal location for building a launch site certainly involves many factors and hopefully we could discover some of the factors by analyzing the existing launch site locations. In the previous exploratory data analysis labs, you have visualized the SpaceX launch dataset using `matplotlib` and `seaborn` and discovered some preliminary correlations between the launch site and success rates. In this lab, you will be performing more interactive visual analytics using `Folium`. Objectives This lab contains the following tasks:* **TASK 1:** Mark all launch sites on a map* **TASK 2:** Mark the success/failed launches for each site on the map* **TASK 3:** Calculate the distances between a launch site to its proximitiesAfter completed the above tasks, you should be able to find some geographical patterns about launch sites. Let's first import required Python packages for this lab:
###Code
!pip3 install folium
!pip3 install wget
import folium
import wget
import pandas as pd
# Import folium MarkerCluster plugin
from folium.plugins import MarkerCluster
# Import folium MousePosition plugin
from folium.plugins import MousePosition
# Import folium DivIcon plugin
from folium.features import DivIcon
###Output
_____no_output_____
###Markdown
Task 1: Mark all launch sites on a map First, let's try to add each site's location on a map using site's latitude and longitude coordinates The following dataset with the name `spacex_launch_geo.csv` is an augmented dataset with latitude and longitude added for each site.
###Code
# Download and read the `spacex_launch_geo.csv`
spacex_csv_file = wget.download('https://cf-courses-data.s3.us.cloud-object-storage.appdomain.cloud/IBM-DS0321EN-SkillsNetwork/datasets/spacex_launch_geo.csv')
spacex_df=pd.read_csv(spacex_csv_file)
###Output
0% [ ] 0 / 8966
91% [......................................................................... ] 8192 / 8966
100% [................................................................................] 8966 / 8966
###Markdown
Now, you can take a look at what are the coordinates for each site.
###Code
# Select relevant sub-columns: `Launch Site`, `Lat(Latitude)`, `Long(Longitude)`, `class`
spacex_df = spacex_df[['Launch Site', 'Lat', 'Long', 'class']]
launch_sites_df = spacex_df.groupby(['Launch Site'], as_index=False).first()
launch_sites_df = launch_sites_df[['Launch Site', 'Lat', 'Long', 'class']]
launch_sites_df
launch_sites_df["Lat"][0]
###Output
_____no_output_____
###Markdown
Above coordinates are just plain numbers that can not give you any intuitive insights about where are those launch sites. If you are very good at geography, you can interpret those numbers directly in your mind. If not, that's fine too. Let's visualize those locations by pinning them on a map. We first need to create a folium `Map` object, with an initial center location to be NASA Johnson Space Center at Houston, Texas.
###Code
# Start location is NASA Johnson Space Center
nasa_coordinate = [29.559684888503615, -95.0830971930759]
site_map = folium.Map(location=nasa_coordinate, zoom_start=10)
###Output
_____no_output_____
###Markdown
We could use `folium.Circle` to add a highlighted circle area with a text label on a specific coordinate. For example,
###Code
# Create a blue circle at NASA Johnson Space Center's coordinate with a popup label showing its name
circle = folium.Circle(nasa_coordinate, radius=1000, color='#d35400', fill=True).add_child(folium.Popup('NASA Johnson Space Center'))
# Create a blue circle at NASA Johnson Space Center's coordinate with a icon showing its name
marker = folium.map.Marker(
nasa_coordinate,
# Create an icon as a text label
icon=DivIcon(
icon_size=(20,20),
icon_anchor=(0,0),
html='<div style="font-size: 12; color:#d35400;"><b>%s</b></div>' % 'NASA JSC',
)
)
site_map.add_child(circle)
site_map.add_child(marker)
###Output
_____no_output_____
###Markdown
and you should find a small yellow circle near the city of Houston and you can zoom-in to see a larger circle. Now, let's add a circle for each launch site in data frame `launch_sites` *TODO:* Create and add `folium.Circle` and `folium.Marker` for each launch site on the site map
###Code
# Initial the map
site_map = folium.Map(location=nasa_coordinate, zoom_start=5)
# For each launch site, add a Circle object based on its coordinate (Lat, Long) values. In addition, add Launch site name as a popup label
for i in range (len(launch_sites_df.index)):
coordinate = [launch_sites_df["Lat"][i], launch_sites_df["Long"][i]]
circle = folium.Circle(coordinate, radius=100, color='#d35400', fill=True).add_child(folium.Popup(launch_sites_df["Launch Site"][i]))
marker = folium.map.Marker(
coordinate,
icon=DivIcon(
icon_size=(20,20),
icon_anchor=(0,0),
html='<div style="font-size: 12; color:#d35400;"><b>%s</b></div>' % launch_sites_df["Launch Site"][i],
)
)
site_map.add_child(circle)
site_map.add_child(marker)
site_map
###Output
_____no_output_____
###Markdown
The generated map with marked launch sites should look similar to the following: Now, you can explore the map by zoom-in/out the marked areas, and try to answer the following questions:* Are all launch sites in proximity to the Equator line?* Are all launch sites in very close proximity to the coast?Also please try to explain your findings. Task 2: Mark the success/failed launches for each site on the map Next, let's try to enhance the map by adding the launch outcomes for each site, and see which sites have high success rates.Recall that data frame spacex_df has detailed launch records, and the `class` column indicates if this launch was successful or not
###Code
spacex_df.tail(10)
###Output
_____no_output_____
###Markdown
Next, let's create markers for all launch records.If a launch was successful `(class=1)`, then we use a green marker and if a launch was failed, we use a red marker `(class=0)` Note that a launch only happens in one of the four launch sites, which means many launch records will have the exact same coordinate. Marker clusters can be a good way to simplify a map containing many markers having the same coordinate. Let's first create a `MarkerCluster` object
###Code
marker_cluster = MarkerCluster()
###Output
_____no_output_____
###Markdown
*TODO:* Create a new column in `launch_sites` dataframe called `marker_color` to store the marker colors based on the `class` value
###Code
launch_sites_df
def func(item):
if item == 1:
return 'green'
else:
return 'red'
launch_sites_df["marker_color"] = launch_sites_df["class"].apply(func)
# Apply a function to check the value of `class` column
# If class=1, marker_color value will be green
# If class=0, marker_color value will be red
launch_sites_df
# Function to assign color to launch outcome
def assign_marker_color(launch_outcome):
if launch_outcome == 1:
return 'green'
else:
return 'red'
spacex_df['marker_color'] = spacex_df['class'].apply(assign_marker_color)
spacex_df.tail(10)
###Output
_____no_output_____
###Markdown
*TODO:* For each launch result in `spacex_df` data frame, add a `folium.Marker` to `marker_cluster`
###Code
# Function to assign color to launch outcome
def assign_marker_color(launch_outcome):
if launch_outcome == 1:
return 'green'
else:
return 'red'
spacex_df['marker_color'] = spacex_df['class'].apply(assign_marker_color)
spacex_df.tail(10)
###Output
_____no_output_____
###Markdown
Your updated map may look like the following screenshots: From the color-labeled markers in marker clusters, you should be able to easily identify which launch sites have relatively high success rates. TASK 3: Calculate the distances between a launch site to its proximities Next, we need to explore and analyze the proximities of launch sites. Let's first add a `MousePosition` on the map to get coordinate for a mouse over a point on the map. As such, while you are exploring the map, you can easily find the coordinates of any points of interests (such as railway)
###Code
# Add Mouse Position to get the coordinate (Lat, Long) for a mouse over on the map
formatter = "function(num) {return L.Util.formatNum(num, 5);};"
mouse_position = MousePosition(
position='topright',
separator=' Long: ',
empty_string='NaN',
lng_first=False,
num_digits=20,
prefix='Lat:',
lat_formatter=formatter,
lng_formatter=formatter,
)
site_map.add_child(mouse_position)
site_map
###Output
_____no_output_____
###Markdown
Now zoom in to a launch site and explore its proximity to see if you can easily find any railway, highway, coastline, etc. Move your mouse to these points and mark down their coordinates (shown on the top-left) in order to the distance to the launch site. You can calculate the distance between two points on the map based on their `Lat` and `Long` values using the following method:
###Code
from math import sin, cos, sqrt, atan2, radians
def calculate_distance(lat1, lon1, lat2, lon2):
# approximate radius of earth in km
R = 6373.0
lat1 = radians(lat1)
lon1 = radians(lon1)
lat2 = radians(lat2)
lon2 = radians(lon2)
dlon = lon2 - lon1
dlat = lat2 - lat1
a = sin(dlat / 2)**2 + cos(lat1) * cos(lat2) * sin(dlon / 2)**2
c = 2 * atan2(sqrt(a), sqrt(1 - a))
distance = R * c
return distance
###Output
_____no_output_____
###Markdown
*TODO:* Mark down a point on the closest railway using MousePosition and calculate the distance between the railway point to the launch site.
###Code
# distance_railway = calculate_distance(lat1, lon1, lat2, lon2)
lat1=34.632834
lon1=-120.610746
lat2=34.63494
lon2 = -120.62429
distance_railway = calculate_distance(lat1, lon1, lat2, lon2)
###Output
_____no_output_____
###Markdown
*TODO:* After obtained its coordinate, create a `folium.Marker` to show the distance
###Code
# create and add a folium.Marker on your selected closest raiwaly point on the map
# show the distance to the launch site using the icon property
coordinate = [34.63494,-120.62429]
icon_ = folium.DivIcon(html=str(round(distance_railway, 2)) + " km")
marker = folium.map.Marker(
coordinate,
icon=icon_
)
marker.add_to(site_map)
site_map
###Output
_____no_output_____
###Markdown
*TODO:* Draw a `PolyLine` between a launch site to the selected
###Code
# Create a `folium.PolyLine` object using the railway point coordinate and launch site coordinate
railway = [34.63494,-120.62429]
launch = [34.632834, -120.610746]
line = folium.PolyLine([railway, launch])
site_map.add_child(line)
###Output
_____no_output_____
###Markdown
Your updated map with distance line should look like the following screenshot: *TODO:* Similarly, you can draw a line betwee a launch site to its closest city, coastline, highway, etc.
###Code
# Create a marker with distance to a closest city, coastline, highway, etc.
# Draw a line between the marker to the launch site
###Output
_____no_output_____
|
notebooks/JSON_PoC_read_write.ipynb
|
###Markdown
In this proof of concept we will read & write JSON files in Jupyter notebook. 1. display the data in the sidecar 2. edit this data 3. check that the sidecar will write valid JSON files.
###Code
#import json module to be able to read & write json files
import json
import pandas as pd
from pandas.io.json import json_normalize
from glob import glob
from pathlib import Path
###Output
_____no_output_____
###Markdown
1. The first part will include displaying the data in the sidecar by reading the JSON files 2. We then use json.load to turn it into a python object 3. The data we have includes an array of information under SliceTiming so we will create a dataframe within our dataframe to include SliceTiming as SliceTime 00, 01 , etc. (individual values of SliceTiming).
###Code
#testing the code with a single json file.
file_test = open('/Users/bjaber/Projects/CuBIDS-use_cases/cubids/testdata/complete/sub-01/ses-phdiff/dwi/sub-01_ses-phdiff_acq-HASC55AP_dwi.json')
sample_data = json.load(file_test)
sample_data.keys()
sample_data.get('SliceTiming')
SliceTime = sample_data.get('SliceTiming') #the way you can snatch things out of a dictionary
#if dict doens't have the key it will return none vs. error
if SliceTime:
sample_data.update({"SliceTime%03d"%SliceNum : time for SliceNum, time in enumerate(SliceTime)})
del sample_data['SliceTiming']
array_data = pd.DataFrame.from_dict(sample_data, orient='index')
array_data
#{"SliceTime%03d"%SliceNum : time for SliceNum, time in enumerate(SliceTime)}
###Output
_____no_output_____
###Markdown
the next one might not have slice timing but you concatonate the next row -- if the file doesn't have slice timing it fills with NaN and if it doesn't then google! rglob to get all the files in the bids tree then load it with json.load Next steps 1. Slice Timing turn it into a column where each column would have its own float 2. multiple columns with the umber of them filled out to the maximum number of slice times 2. The following part is used to edit JSON file data. In order to do so, call to the JSON object that was created using the json.load commeand, in this case json_data, and refer to the value that you want to change and edit it. Note that this code is commented out as it will be different when we are using this with Pandas DataFrame. This was code written when working with a single .json file.
###Code
#Here we change the value for AcquisionNumber from 1 to 2.
#json_data["AcquisitionNumber"] = 2
#Uncomment below to view edited data
#json_data
#Reverting back to original data
#json_data["AcquisitionNumber"] = 1
###Output
_____no_output_____
###Markdown
3. Checking that the sidecare will write valid JSON files In order to do this, we use the json.dumps function as it will turn the python object into a JSON string, and therefore, will wirte a valid JSON file always. Note: same as the previous chunk of code, this was written for a single .json file and therefore is commentend out
###Code
#json_string = json.dumps(json_data)
#Uncomment below to view the python object as a JSON string
#json_string
#notes from Matt
# have a function that does the reading and creates 1 row then you have to loop and the dataframe grows through concatanation
# pandas.concat
###Output
_____no_output_____
###Markdown
The next section is the for loop attempting to extract, open and turn into a dataframe each json file in the "complete" directory!
###Code
for path in Path('/Users/bjaber/Projects/CuBIDS/cubids/testdata/complete').rglob('*.json'):
#print(path)
counter=0
for path in Path('/Users/bjaber/Projects/CuBIDS/cubids/testdata/complete').rglob('*.json'):
print(type(path))
print(counter)
s_path = str(path)
#print(s_path)
file_tree = open(s_path)
example_data = json.load(file_tree)
SliceTime = example_data.get('SliceTiming') #the way you can snatch things out of a dictionary #if dict doens't have the key it will return none vs. error
if SliceTime:
example_data.update({"SliceTime%03d"%SliceNum : time for SliceNum, time in enumerate(SliceTime)})
del example_data['SliceTiming']
print(example_data)
#data = pd.DataFrame.from_dict(example_data, orient='index')
#data
counter += 1
#NOTE: error when trying to put the data into a pandas dataframe.
# print(example_data) was used to make sure that inputs that are an array such as in the field SliceTiming are being separated into indenpendent values of SliceTime00x that should feed into the dataframe.
# it is doing that across all json files that are being loaded from the directory
###Output
<class 'pathlib.PosixPath'>
0
<class 'pathlib.PosixPath'>
1
{'ProcedureStepDescription': 'MR_HEAD_WO_IV_CONTRAST', 'DeviceSerialNumber': '167024', 'EffectiveEchoSpacing': 0.000689998, 'TotalReadoutTime': 0.0717598, 'ManufacturersModelName': 'Prisma_fit', 'ProtocolName': 'ABCD_dMRI', 'BandwidthPerPixelPhaseEncode': 10.352, 'PhaseEncodingLines': 140, 'RepetitionTime': 4.2, 'EchoTrainLength': 105, 'MagneticFieldStrength': 3, 'AcquisitionNumber': 1, 'InstitutionName': 'HUP', 'BodyPartExamined': 'BRAIN', 'ConversionSoftware': 'dcm2niix', 'ScanningSequence': 'EP', 'Manufacturer': 'Siemens', 'FlipAngle': 90, 'ConversionSoftwareVersion': 'v1.0.20170724 (OpenJPEG build) GCC6.1.0', 'SeriesDescription': 'ABCD_dMRI', 'InstitutionAddress': 'Spruce_Street_3400_Philadelphia_Pennsylvania_US_19104', 'AcquisitionTime': '16:31:6.145000', 'SequenceName': 'ep_b0', 'ImageType': ['ORIGINAL', 'PRIMARY', 'DIFFUSION', 'NONE', 'ND', 'MOSAIC'], 'EchoTime': 0.089, 'SequenceVariant': 'SK_SP', 'PhaseEncodingDirection': 'j', 'SoftwareVersions': 'syngo_MR_E11', 'SliceTime000': 0.0, 'SliceTime001': 0.11666666666666667, 'SliceTime002': 0.23333333333333334, 'SliceTime003': 0.35, 'SliceTime004': 0.4666666666666667, 'SliceTime005': 0.5833333333333334, 'SliceTime006': 0.7, 'SliceTime007': 0.8166666666666667, 'SliceTime008': 0.9333333333333333, 'SliceTime009': 1.05, 'SliceTime010': 1.1666666666666667, 'SliceTime011': 1.2833333333333334, 'SliceTime012': 1.4, 'SliceTime013': 1.5166666666666666, 'SliceTime014': 1.6333333333333333, 'SliceTime015': 1.75, 'SliceTime016': 1.8666666666666667, 'SliceTime017': 1.9833333333333334, 'SliceTime018': 2.1, 'SliceTime019': 2.216666666666667, 'SliceTime020': 2.3333333333333335, 'SliceTime021': 2.45, 'SliceTime022': 2.566666666666667, 'SliceTime023': 2.6833333333333336, 'SliceTime024': 2.8, 'SliceTime025': 2.9166666666666665, 'SliceTime026': 3.033333333333333, 'SliceTime027': 3.15, 'SliceTime028': 3.2666666666666666, 'SliceTime029': 3.3833333333333333, 'SliceTime030': 3.5, 'SliceTime031': 3.6166666666666667, 'SliceTime032': 3.7333333333333334, 'SliceTime033': 3.85, 'SliceTime034': 3.966666666666667, 'SliceTime035': 4.083333333333333}
<class 'pathlib.PosixPath'>
2
<class 'pathlib.PosixPath'>
3
<class 'pathlib.PosixPath'>
4
{'ProcedureStepDescription': 'MR_HEAD_WO_IV_CONTRAST', 'DeviceSerialNumber': '167024', 'EffectiveEchoSpacing': 0.000689998, 'TotalReadoutTime': 0.0717598, 'ManufacturersModelName': 'Prisma_fit', 'ProtocolName': 'ABCD_dMRI', 'BandwidthPerPixelPhaseEncode': 10.352, 'PhaseEncodingLines': 140, 'RepetitionTime': 4.2, 'EchoTrainLength': 105, 'MagneticFieldStrength': 3, 'AcquisitionNumber': 1, 'InstitutionName': 'HUP', 'BodyPartExamined': 'BRAIN', 'ConversionSoftware': 'dcm2niix', 'ScanningSequence': 'EP', 'Manufacturer': 'Siemens', 'FlipAngle': 90, 'InstitutionAddress': 'Spruce_Street_3400_Philadelphia_Pennsylvania_US_19104', 'SeriesDescription': 'ABCD_dMRI', 'ConversionSoftwareVersion': 'v1.0.20170724 (OpenJPEG build) GCC6.1.0', 'AcquisitionTime': '16:31:6.145000', 'SequenceName': 'ep_b0', 'ImageType': ['ORIGINAL', 'PRIMARY', 'DIFFUSION', 'NONE', 'ND', 'MOSAIC'], 'EchoTime': 0.089, 'SequenceVariant': 'SK_SP', 'PhaseEncodingDirection': 'j-', 'SoftwareVersions': 'syngo_MR_E11', 'IntendedFor': ['ses-phdiff/func/sub-01_ses-phdiff_task-rest_bold.nii.gz'], 'SliceTime000': 0.0, 'SliceTime001': 0.11666666666666667, 'SliceTime002': 0.23333333333333334, 'SliceTime003': 0.35, 'SliceTime004': 0.4666666666666667, 'SliceTime005': 0.5833333333333334, 'SliceTime006': 0.7, 'SliceTime007': 0.8166666666666667, 'SliceTime008': 0.9333333333333333, 'SliceTime009': 1.05, 'SliceTime010': 1.1666666666666667, 'SliceTime011': 1.2833333333333334, 'SliceTime012': 1.4, 'SliceTime013': 1.5166666666666666, 'SliceTime014': 1.6333333333333333, 'SliceTime015': 1.75, 'SliceTime016': 1.8666666666666667, 'SliceTime017': 1.9833333333333334, 'SliceTime018': 2.1, 'SliceTime019': 2.216666666666667, 'SliceTime020': 2.3333333333333335, 'SliceTime021': 2.45, 'SliceTime022': 2.566666666666667, 'SliceTime023': 2.6833333333333336, 'SliceTime024': 2.8, 'SliceTime025': 2.9166666666666665, 'SliceTime026': 3.033333333333333, 'SliceTime027': 3.15, 'SliceTime028': 3.2666666666666666, 'SliceTime029': 3.3833333333333333, 'SliceTime030': 3.5, 'SliceTime031': 3.6166666666666667, 'SliceTime032': 3.7333333333333334, 'SliceTime033': 3.85, 'SliceTime034': 3.966666666666667, 'SliceTime035': 4.083333333333333}
<class 'pathlib.PosixPath'>
5
<class 'pathlib.PosixPath'>
6
<class 'pathlib.PosixPath'>
7
{'ProcedureStepDescription': 'MR_HEAD_WO_IV_CONTRAST', 'DeviceSerialNumber': '167024', 'EffectiveEchoSpacing': 0.000689998, 'TotalReadoutTime': 0.0717598, 'ManufacturersModelName': 'Prisma_fit', 'ProtocolName': 'ABCD_dMRI', 'TaskName': 'rest', 'BandwidthPerPixelPhaseEncode': 10.352, 'PhaseEncodingLines': 140, 'RepetitionTime': 1.0, 'EchoTrainLength': 105, 'MagneticFieldStrength': 3, 'AcquisitionNumber': 1, 'InstitutionName': 'HUP', 'BodyPartExamined': 'BRAIN', 'ConversionSoftware': 'dcm2niix', 'ScanningSequence': 'EP', 'Manufacturer': 'Siemens', 'FlipAngle': 90, 'ConversionSoftwareVersion': 'v1.0.20170724 (OpenJPEG build) GCC6.1.0', 'SeriesDescription': 'ABCD_dMRI', 'InstitutionAddress': 'Spruce_Street_3400_Philadelphia_Pennsylvania_US_19104', 'AcquisitionTime': '16:31:6.145000', 'SequenceName': 'ep_b0', 'ImageType': ['ORIGINAL', 'PRIMARY', 'DIFFUSION', 'NONE', 'ND', 'MOSAIC'], 'EchoTime': 0.089, 'SequenceVariant': 'SK_SP', 'PhaseEncodingDirection': 'j', 'SoftwareVersions': 'syngo_MR_E11', 'SliceTime000': 0.0, 'SliceTime001': 0.027131782945736437, 'SliceTime002': 0.054263565891472874, 'SliceTime003': 0.0813953488372093, 'SliceTime004': 0.10852713178294575, 'SliceTime005': 0.1356589147286822, 'SliceTime006': 0.1627906976744186, 'SliceTime007': 0.18992248062015504, 'SliceTime008': 0.2170542635658915, 'SliceTime009': 0.24418604651162792, 'SliceTime010': 0.2713178294573644, 'SliceTime011': 0.2984496124031008, 'SliceTime012': 0.3255813953488372, 'SliceTime013': 0.35271317829457366, 'SliceTime014': 0.3798449612403101, 'SliceTime015': 0.4069767441860465, 'SliceTime016': 0.434108527131783, 'SliceTime017': 0.4612403100775194, 'SliceTime018': 0.48837209302325585, 'SliceTime019': 0.5155038759689923, 'SliceTime020': 0.5426356589147288, 'SliceTime021': 0.5697674418604651, 'SliceTime022': 0.5968992248062016, 'SliceTime023': 0.6240310077519381, 'SliceTime024': 0.6511627906976744, 'SliceTime025': 0.6782945736434108, 'SliceTime026': 0.7054263565891473, 'SliceTime027': 0.7325581395348837, 'SliceTime028': 0.7596899224806202, 'SliceTime029': 0.7868217054263567, 'SliceTime030': 0.813953488372093, 'SliceTime031': 0.8410852713178295, 'SliceTime032': 0.868217054263566, 'SliceTime033': 0.8953488372093024, 'SliceTime034': 0.9224806201550388, 'SliceTime035': 0.9496124031007752}
<class 'pathlib.PosixPath'>
8
{'ProcedureStepDescription': 'MR_HEAD_WO_IV_CONTRAST', 'DeviceSerialNumber': '167024', 'EffectiveEchoSpacing': 0.000689998, 'TotalReadoutTime': 0.0717598, 'ManufacturersModelName': 'Prisma_fit', 'ProtocolName': 'ABCD_dMRI', 'BandwidthPerPixelPhaseEncode': 10.352, 'PhaseEncodingLines': 140, 'RepetitionTime': 4.2, 'EchoTrainLength': 105, 'MagneticFieldStrength': 3, 'AcquisitionNumber': 1, 'InstitutionName': 'HUP', 'BodyPartExamined': 'BRAIN', 'ConversionSoftware': 'dcm2niix', 'ScanningSequence': 'EP', 'Manufacturer': 'Siemens', 'FlipAngle': 90, 'ConversionSoftwareVersion': 'v1.0.20170724 (OpenJPEG build) GCC6.1.0', 'SeriesDescription': 'ABCD_dMRI', 'InstitutionAddress': 'Spruce_Street_3400_Philadelphia_Pennsylvania_US_19104', 'AcquisitionTime': '16:31:6.145000', 'SequenceName': 'ep_b0', 'ImageType': ['ORIGINAL', 'PRIMARY', 'DIFFUSION', 'NONE', 'ND', 'MOSAIC'], 'EchoTime': 0.089, 'SequenceVariant': 'SK_SP', 'PhaseEncodingDirection': 'j', 'SoftwareVersions': 'syngo_MR_E11', 'SliceTime000': 0.0, 'SliceTime001': 0.11666666666666667, 'SliceTime002': 0.23333333333333334, 'SliceTime003': 0.35, 'SliceTime004': 0.4666666666666667, 'SliceTime005': 0.5833333333333334, 'SliceTime006': 0.7, 'SliceTime007': 0.8166666666666667, 'SliceTime008': 0.9333333333333333, 'SliceTime009': 1.05, 'SliceTime010': 1.1666666666666667, 'SliceTime011': 1.2833333333333334, 'SliceTime012': 1.4, 'SliceTime013': 1.5166666666666666, 'SliceTime014': 1.6333333333333333, 'SliceTime015': 1.75, 'SliceTime016': 1.8666666666666667, 'SliceTime017': 1.9833333333333334, 'SliceTime018': 2.1, 'SliceTime019': 2.216666666666667, 'SliceTime020': 2.3333333333333335, 'SliceTime021': 2.45, 'SliceTime022': 2.566666666666667, 'SliceTime023': 2.6833333333333336, 'SliceTime024': 2.8, 'SliceTime025': 2.9166666666666665, 'SliceTime026': 3.033333333333333, 'SliceTime027': 3.15, 'SliceTime028': 3.2666666666666666, 'SliceTime029': 3.3833333333333333, 'SliceTime030': 3.5, 'SliceTime031': 3.6166666666666667, 'SliceTime032': 3.7333333333333334, 'SliceTime033': 3.85, 'SliceTime034': 3.966666666666667, 'SliceTime035': 4.083333333333333}
<class 'pathlib.PosixPath'>
9
<class 'pathlib.PosixPath'>
10
<class 'pathlib.PosixPath'>
11
<class 'pathlib.PosixPath'>
12
{'ProcedureStepDescription': 'MR_HEAD_WO_IV_CONTRAST', 'DeviceSerialNumber': '167024', 'EffectiveEchoSpacing': 0.000689998, 'TotalReadoutTime': 0.0717598, 'ManufacturersModelName': 'Prisma_fit', 'ProtocolName': 'ABCD_dMRI', 'BandwidthPerPixelPhaseEncode': 10.352, 'PhaseEncodingLines': 140, 'RepetitionTime': 4.2, 'EchoTrainLength': 105, 'MagneticFieldStrength': 3, 'AcquisitionNumber': 1, 'InstitutionName': 'HUP', 'BodyPartExamined': 'BRAIN', 'ConversionSoftware': 'dcm2niix', 'ScanningSequence': 'EP', 'Manufacturer': 'Siemens', 'FlipAngle': 90, 'InstitutionAddress': 'Spruce_Street_3400_Philadelphia_Pennsylvania_US_19104', 'SeriesDescription': 'ABCD_dMRI', 'ConversionSoftwareVersion': 'v1.0.20170724 (OpenJPEG build) GCC6.1.0', 'AcquisitionTime': '16:31:6.145000', 'SequenceName': 'ep_b0', 'ImageType': ['ORIGINAL', 'PRIMARY', 'DIFFUSION', 'NONE', 'ND', 'MOSAIC'], 'EchoTime': 0.089, 'SequenceVariant': 'SK_SP', 'PhaseEncodingDirection': 'j-', 'SoftwareVersions': 'syngo_MR_E11', 'IntendedFor': ['ses-phdiff/func/sub-02_ses-phdiff_task-rest_bold.nii.gz'], 'SliceTime000': 0.0, 'SliceTime001': 0.11666666666666667, 'SliceTime002': 0.23333333333333334, 'SliceTime003': 0.35, 'SliceTime004': 0.4666666666666667, 'SliceTime005': 0.5833333333333334, 'SliceTime006': 0.7, 'SliceTime007': 0.8166666666666667, 'SliceTime008': 0.9333333333333333, 'SliceTime009': 1.05, 'SliceTime010': 1.1666666666666667, 'SliceTime011': 1.2833333333333334, 'SliceTime012': 1.4, 'SliceTime013': 1.5166666666666666, 'SliceTime014': 1.6333333333333333, 'SliceTime015': 1.75, 'SliceTime016': 1.8666666666666667, 'SliceTime017': 1.9833333333333334, 'SliceTime018': 2.1, 'SliceTime019': 2.216666666666667, 'SliceTime020': 2.3333333333333335, 'SliceTime021': 2.45, 'SliceTime022': 2.566666666666667, 'SliceTime023': 2.6833333333333336, 'SliceTime024': 2.8, 'SliceTime025': 2.9166666666666665, 'SliceTime026': 3.033333333333333, 'SliceTime027': 3.15, 'SliceTime028': 3.2666666666666666, 'SliceTime029': 3.3833333333333333, 'SliceTime030': 3.5, 'SliceTime031': 3.6166666666666667, 'SliceTime032': 3.7333333333333334, 'SliceTime033': 3.85, 'SliceTime034': 3.966666666666667, 'SliceTime035': 4.083333333333333}
<class 'pathlib.PosixPath'>
13
<class 'pathlib.PosixPath'>
14
{'ProcedureStepDescription': 'MR_HEAD_WO_IV_CONTRAST', 'DeviceSerialNumber': '167024', 'EffectiveEchoSpacing': 0.000689998, 'TotalReadoutTime': 0.0717598, 'ManufacturersModelName': 'Prisma_fit', 'ProtocolName': 'ABCD_dMRI', 'TaskName': 'rest', 'BandwidthPerPixelPhaseEncode': 10.352, 'PhaseEncodingLines': 140, 'RepetitionTime': 1.0, 'EchoTrainLength': 105, 'MagneticFieldStrength': 3, 'AcquisitionNumber': 1, 'InstitutionName': 'HUP', 'BodyPartExamined': 'BRAIN', 'ConversionSoftware': 'dcm2niix', 'ScanningSequence': 'EP', 'Manufacturer': 'Siemens', 'FlipAngle': 90, 'ConversionSoftwareVersion': 'v1.0.20170724 (OpenJPEG build) GCC6.1.0', 'SeriesDescription': 'ABCD_dMRI', 'InstitutionAddress': 'Spruce_Street_3400_Philadelphia_Pennsylvania_US_19104', 'AcquisitionTime': '16:31:6.145000', 'SequenceName': 'ep_b0', 'ImageType': ['ORIGINAL', 'PRIMARY', 'DIFFUSION', 'NONE', 'ND', 'MOSAIC'], 'EchoTime': 0.089, 'SequenceVariant': 'SK_SP', 'PhaseEncodingDirection': 'j', 'SoftwareVersions': 'syngo_MR_E11', 'SliceTime000': 0.0, 'SliceTime001': 0.027131782945736437, 'SliceTime002': 0.054263565891472874, 'SliceTime003': 0.0813953488372093, 'SliceTime004': 0.10852713178294575, 'SliceTime005': 0.1356589147286822, 'SliceTime006': 0.1627906976744186, 'SliceTime007': 0.18992248062015504, 'SliceTime008': 0.2170542635658915, 'SliceTime009': 0.24418604651162792, 'SliceTime010': 0.2713178294573644, 'SliceTime011': 0.2984496124031008, 'SliceTime012': 0.3255813953488372, 'SliceTime013': 0.35271317829457366, 'SliceTime014': 0.3798449612403101, 'SliceTime015': 0.4069767441860465, 'SliceTime016': 0.434108527131783, 'SliceTime017': 0.4612403100775194, 'SliceTime018': 0.48837209302325585, 'SliceTime019': 0.5155038759689923, 'SliceTime020': 0.5426356589147288, 'SliceTime021': 0.5697674418604651, 'SliceTime022': 0.5968992248062016, 'SliceTime023': 0.6240310077519381, 'SliceTime024': 0.6511627906976744, 'SliceTime025': 0.6782945736434108, 'SliceTime026': 0.7054263565891473, 'SliceTime027': 0.7325581395348837, 'SliceTime028': 0.7596899224806202, 'SliceTime029': 0.7868217054263567, 'SliceTime030': 0.813953488372093, 'SliceTime031': 0.8410852713178295, 'SliceTime032': 0.868217054263566, 'SliceTime033': 0.8953488372093024, 'SliceTime034': 0.9224806201550388, 'SliceTime035': 0.9496124031007752}
<class 'pathlib.PosixPath'>
15
{'ProcedureStepDescription': 'MR_HEAD_WO_IV_CONTRAST', 'DeviceSerialNumber': '167024', 'EffectiveEchoSpacing': 0.000689998, 'TotalReadoutTime': 0.0717598, 'ManufacturersModelName': 'Prisma_fit', 'ProtocolName': 'ABCD_dMRI', 'BandwidthPerPixelPhaseEncode': 10.352, 'PhaseEncodingLines': 140, 'RepetitionTime': 4.2, 'EchoTrainLength': 105, 'MagneticFieldStrength': 3, 'AcquisitionNumber': 1, 'InstitutionName': 'HUP', 'BodyPartExamined': 'BRAIN', 'ConversionSoftware': 'dcm2niix', 'ScanningSequence': 'EP', 'Manufacturer': 'Siemens', 'FlipAngle': 90, 'ConversionSoftwareVersion': 'v1.0.20170724 (OpenJPEG build) GCC6.1.0', 'SeriesDescription': 'ABCD_dMRI', 'InstitutionAddress': 'Spruce_Street_3400_Philadelphia_Pennsylvania_US_19104', 'AcquisitionTime': '16:31:6.145000', 'SequenceName': 'ep_b0', 'ImageType': ['ORIGINAL', 'PRIMARY', 'DIFFUSION', 'NONE', 'ND', 'MOSAIC'], 'EchoTime': 0.089, 'SequenceVariant': 'SK_SP', 'PhaseEncodingDirection': 'j', 'SoftwareVersions': 'syngo_MR_E11', 'SliceTime000': 0.0, 'SliceTime001': 0.11666666666666667, 'SliceTime002': 0.23333333333333334, 'SliceTime003': 0.35, 'SliceTime004': 0.4666666666666667, 'SliceTime005': 0.5833333333333334, 'SliceTime006': 0.7, 'SliceTime007': 0.8166666666666667, 'SliceTime008': 0.9333333333333333, 'SliceTime009': 1.05, 'SliceTime010': 1.1666666666666667, 'SliceTime011': 1.2833333333333334, 'SliceTime012': 1.4, 'SliceTime013': 1.5166666666666666, 'SliceTime014': 1.6333333333333333, 'SliceTime015': 1.75, 'SliceTime016': 1.8666666666666667, 'SliceTime017': 1.9833333333333334, 'SliceTime018': 2.1, 'SliceTime019': 2.216666666666667, 'SliceTime020': 2.3333333333333335, 'SliceTime021': 2.45, 'SliceTime022': 2.566666666666667, 'SliceTime023': 2.6833333333333336, 'SliceTime024': 2.8, 'SliceTime025': 2.9166666666666665, 'SliceTime026': 3.033333333333333, 'SliceTime027': 3.15, 'SliceTime028': 3.2666666666666666, 'SliceTime029': 3.3833333333333333, 'SliceTime030': 3.5, 'SliceTime031': 3.6166666666666667, 'SliceTime032': 3.7333333333333334, 'SliceTime033': 3.85, 'SliceTime034': 3.966666666666667, 'SliceTime035': 4.083333333333333}
<class 'pathlib.PosixPath'>
16
<class 'pathlib.PosixPath'>
17
{'ProcedureStepDescription': 'MR_HEAD_WO_IV_CONTRAST', 'DeviceSerialNumber': '167024', 'EffectiveEchoSpacing': 0.000689998, 'TotalReadoutTime': 0.0717598, 'ManufacturersModelName': 'Prisma_fit', 'ProtocolName': 'ABCD_dMRI', 'BandwidthPerPixelPhaseEncode': 10.352, 'PhaseEncodingLines': 140, 'RepetitionTime': 4.2, 'EchoTrainLength': 105, 'MagneticFieldStrength': 3, 'AcquisitionNumber': 1, 'InstitutionName': 'HUP', 'BodyPartExamined': 'BRAIN', 'ConversionSoftware': 'dcm2niix', 'ScanningSequence': 'EP', 'Manufacturer': 'Siemens', 'FlipAngle': 90, 'InstitutionAddress': 'Spruce_Street_3400_Philadelphia_Pennsylvania_US_19104', 'SeriesDescription': 'ABCD_dMRI', 'ConversionSoftwareVersion': 'v1.0.20170724 (OpenJPEG build) GCC6.1.0', 'AcquisitionTime': '16:31:6.145000', 'SequenceName': 'ep_b0', 'ImageType': ['ORIGINAL', 'PRIMARY', 'DIFFUSION', 'NONE', 'ND', 'MOSAIC'], 'EchoTime': 0.089, 'SequenceVariant': 'SK_SP', 'PhaseEncodingDirection': 'j-', 'SoftwareVersions': 'syngo_MR_E11', 'IntendedFor': ['ses-phdiff/func/sub-03_ses-phdiff_task-rest_bold.nii.gz'], 'SliceTime000': 0.0, 'SliceTime001': 0.11666666666666667, 'SliceTime002': 0.23333333333333334, 'SliceTime003': 0.35, 'SliceTime004': 0.4666666666666667, 'SliceTime005': 0.5833333333333334, 'SliceTime006': 0.7, 'SliceTime007': 0.8166666666666667, 'SliceTime008': 0.9333333333333333, 'SliceTime009': 1.05, 'SliceTime010': 1.1666666666666667, 'SliceTime011': 1.2833333333333334, 'SliceTime012': 1.4, 'SliceTime013': 1.5166666666666666, 'SliceTime014': 1.6333333333333333, 'SliceTime015': 1.75, 'SliceTime016': 1.8666666666666667, 'SliceTime017': 1.9833333333333334, 'SliceTime018': 2.1, 'SliceTime019': 2.216666666666667, 'SliceTime020': 2.3333333333333335, 'SliceTime021': 2.45, 'SliceTime022': 2.566666666666667, 'SliceTime023': 2.6833333333333336, 'SliceTime024': 2.8, 'SliceTime025': 2.9166666666666665, 'SliceTime026': 3.033333333333333, 'SliceTime027': 3.15, 'SliceTime028': 3.2666666666666666, 'SliceTime029': 3.3833333333333333, 'SliceTime030': 3.5, 'SliceTime031': 3.6166666666666667, 'SliceTime032': 3.7333333333333334, 'SliceTime033': 3.85, 'SliceTime034': 3.966666666666667, 'SliceTime035': 4.083333333333333}
<class 'pathlib.PosixPath'>
18
<class 'pathlib.PosixPath'>
19
<class 'pathlib.PosixPath'>
20
<class 'pathlib.PosixPath'>
21
{'ProcedureStepDescription': 'MR_HEAD_WO_IV_CONTRAST', 'DeviceSerialNumber': '167024', 'EffectiveEchoSpacing': 0.000689998, 'TotalReadoutTime': 0.0717598, 'ManufacturersModelName': 'Prisma_fit', 'ProtocolName': 'ABCD_dMRI', 'TaskName': 'rest', 'BandwidthPerPixelPhaseEncode': 10.352, 'PhaseEncodingLines': 140, 'RepetitionTime': 1.0, 'EchoTrainLength': 105, 'MagneticFieldStrength': 3, 'AcquisitionNumber': 1, 'InstitutionName': 'HUP', 'BodyPartExamined': 'BRAIN', 'ConversionSoftware': 'dcm2niix', 'ScanningSequence': 'EP', 'Manufacturer': 'Siemens', 'FlipAngle': 90, 'ConversionSoftwareVersion': 'v1.0.20170724 (OpenJPEG build) GCC6.1.0', 'SeriesDescription': 'ABCD_dMRI', 'InstitutionAddress': 'Spruce_Street_3400_Philadelphia_Pennsylvania_US_19104', 'AcquisitionTime': '16:31:6.145000', 'SequenceName': 'ep_b0', 'ImageType': ['ORIGINAL', 'PRIMARY', 'DIFFUSION', 'NONE', 'ND', 'MOSAIC'], 'EchoTime': 0.089, 'SequenceVariant': 'SK_SP', 'PhaseEncodingDirection': 'j', 'SoftwareVersions': 'syngo_MR_E11', 'SliceTime000': 0.0, 'SliceTime001': 0.027131782945736437, 'SliceTime002': 0.054263565891472874, 'SliceTime003': 0.0813953488372093, 'SliceTime004': 0.10852713178294575, 'SliceTime005': 0.1356589147286822, 'SliceTime006': 0.1627906976744186, 'SliceTime007': 0.18992248062015504, 'SliceTime008': 0.2170542635658915, 'SliceTime009': 0.24418604651162792, 'SliceTime010': 0.2713178294573644, 'SliceTime011': 0.2984496124031008, 'SliceTime012': 0.3255813953488372, 'SliceTime013': 0.35271317829457366, 'SliceTime014': 0.3798449612403101, 'SliceTime015': 0.4069767441860465, 'SliceTime016': 0.434108527131783, 'SliceTime017': 0.4612403100775194, 'SliceTime018': 0.48837209302325585, 'SliceTime019': 0.5155038759689923, 'SliceTime020': 0.5426356589147288, 'SliceTime021': 0.5697674418604651, 'SliceTime022': 0.5968992248062016, 'SliceTime023': 0.6240310077519381, 'SliceTime024': 0.6511627906976744, 'SliceTime025': 0.6782945736434108, 'SliceTime026': 0.7054263565891473, 'SliceTime027': 0.7325581395348837, 'SliceTime028': 0.7596899224806202, 'SliceTime029': 0.7868217054263567, 'SliceTime030': 0.813953488372093, 'SliceTime031': 0.8410852713178295, 'SliceTime032': 0.868217054263566, 'SliceTime033': 0.8953488372093024, 'SliceTime034': 0.9224806201550388, 'SliceTime035': 0.9496124031007752}
|
_build/jupyter_execute/Introduction to Python.ipynb
|
###Markdown
Introduction to Python In this section, I wanted to introduce a few basic concepts and give an outline of this section. Comments in Python In Python, we can create comments in the code itself. Considering we can use markdown language (as you see here 😁), we won't use this too much in this notebook. Though, here is an example. Basically, you use the... umm... hashtag? Number sign? Pound sign? This thing ->
###Code
# I am a comment in Python
# Here is 2 + 2
2 + 2
# As you can see, these are not "computed" using Python.
# We are just comments for the person looking at this.
# Or... you!
###Output
_____no_output_____
###Markdown
Print Function We will being using...```pythonprint()```...several times in this notebook. *print()* is a function to print out strings, variables, numbers, functions, etc. Let's use the classic example.
###Code
print( "hello, world!" )
###Output
hello, world!
###Markdown
OR
###Code
print("hello, world!")
###Output
hello, world!
###Markdown
*print()* can do some fun things as well. As in, giving it more than one thing to print with commas between them. This will print both things with spaces.
###Code
print( "hello,", "world!" )
###Output
hello, world!
###Markdown
Help Function The...```pythonhelp()```... function is exactly what it is. It is a function to 🌟 help 🌟 you understand the basic usage of another function.
###Code
help(print)
###Output
Help on built-in function print in module builtins:
print(...)
print(value, ..., sep=' ', end='\n', file=sys.stdout, flush=False)
Prints the values to a stream, or to sys.stdout by default.
Optional keyword arguments:
file: a file-like object (stream); defaults to the current sys.stdout.
sep: string inserted between values, default a space.
end: string appended after the last value, default a newline.
flush: whether to forcibly flush the stream.
###Markdown
Resources Highly suggest looking for answers using [StackOverflow](https://stackoverflow.com/help/searching) Common Errors One of the most common errors in Python is the dreaded ```python2 + 2 3 + 3 File "", line 2 3 + 3 ^IndentationError: unexpected indent```Why does this occur? Well, because Python uses spacing or tabs to distinguish where things like loops, functions, and if/else statements start and end. So, if you add an extra space or tab at the beginning of the statement, you will see this message. If you do, check your spacing. ```{note}Python can get weird with this issue. As you can, technically, start code wherever as long as you are consistent. The next cell shows an example of this... oddity.```
###Code
2+2
3+3
###Output
_____no_output_____
|
python3/library/functions.ipynb
|
###Markdown
[Built-in Functions](https://docs.python.org/3/library/functions.html) The Python interpreter has a number of functions and types built into it that are always available. They are listed here in alphabetical order. - [abs()]()- [all()]()- [any()]()- [ascii()]()- [bin()]()- [bool()]()- [bytearray()]()- [bytes()]()- [callable()]()- [chr()]()- [classmethod()]()- [compile()]()- [complex()]()- [delattr()]()- [dict()]()- [dir()]()- [divmod()]()- [enumerate()]()- [eval()]()- [exec()]()- [filter()]()- [float()]()- [format()]()- [frozenset()]()- [getattr()]()- [globals()]()- [hasattr()]()- [hash()]()- [help()]()- [hex()]()- [id()]()- [\__import__()]()- [input()]()- [int()]()- [isinstance()]()- [issubclass()]()- [iter()]()- [len()]()- [list()]()- [locals()]()- [map()]()- [max()]()- [memoryview()]()- [min()]()- [next()]()- [object()]()- [oct()]()- [open()]()- [ord()]()- [pow()]()- [print()]()- [property()]()- [range()]()- [repr()]()- [reversed()]()- [round()]()- [set()]()- [setattr()]()- [slice()]()- [sorted()]()- [staticmethod()]()- [str()]()- [sum()]()- [super()]()- [tuple()]()- [type()]()- [vars()]()- [zip()]() abs(x)Return the absolute value of a number. The argument may be an integer or a floating point number. If the argument is a complex number, its magnitude is returned.
###Code
abs(-3)
###Output
_____no_output_____
###Markdown
all(iterable)Return True if all elements of the iterable are true (or if the iterable is empty). Equivalent to: def all(iterable): for element in iterable: if not element: return False return True
###Code
all([True, True, True])
all([True, True, False])
all([])
###Output
_____no_output_____
###Markdown
any(iterable)Return True if any element of the iterable is true. If the iterable is empty, return False. Equivalent to: def any(iterable): for element in iterable: if element: return True return False
###Code
any([True, True, False])
any([False, False, False])
any([])
###Output
_____no_output_____
###Markdown
ascii(object)As [repr()](https://docs.python.org/3/library/functions.htmlrepr), return a string containing a printable representation of an object, but escape the non-ASCII characters in the string returned by [repr()](https://docs.python.org/3/library/functions.htmlrepr) using \x, \u or \U escapes. This generates a string similar to that returned by [repr()](https://docs.python.org/3/library/functions.htmlrepr) in Python 2. bin(x)Convert an integer number to a binary string. The result is a valid Python expression. If x is not a Python [int](https://docs.python.org/3/library/functions.htmlint) object, it has to define an [__index__()](https://docs.python.org/3/reference/datamodel.htmlobject.__index__) method that returns an integer.
###Code
bin(42)
###Output
_____no_output_____
###Markdown
*class* bool([x])Return a Boolean value, i.e. one of True or False. x is converted using the standard [truth testing procedure](https://docs.python.org/3/library/stdtypes.htmltruth). If x is false or omitted, this returns False; otherwise it returns True. The [bool](https://docs.python.org/3/library/functions.htmlbool) class is a subclass of [int](https://docs.python.org/3/library/functions.htmlint) (see [Numeric Types — int, float, complex](https://docs.python.org/3/library/stdtypes.htmltypesnumeric)). It cannot be subclassed further. Its only instances are False and True (see [Boolean Values](https://docs.python.org/3/library/stdtypes.htmlbltin-boolean-values)).
###Code
bool(True)
bool([True])
bool(False)
bool([False])
bool([])
###Output
_____no_output_____
###Markdown
*class* bytearray([source[, encoding[, errors]]])Return a new array of bytes. The [bytearray](https://docs.python.org/3/library/functions.htmlbytearray) class is a mutable sequence of integers in the range 0 <= x < 256. It has most of the usual methods of mutable sequences, described in [Mutable Sequence Types](https://docs.python.org/3/library/stdtypes.htmltypesseq-mutable), as well as most methods that the [bytes](https://docs.python.org/3/library/functions.htmlbytes) type has, see [Bytes and Bytearray Operations](https://docs.python.org/3/library/stdtypes.htmlbytes-methods).The optional source parameter can be used to initialize the array in a few different ways:- If it is a string, you must also give the encoding (and optionally, errors) parameters; bytearray() then converts the string to bytes using str.encode().- If it is an integer, the array will have that size and will be initialized with null bytes.- If it is an object conforming to the buffer interface, a read-only buffer of the object will be used to initialize the bytes array.- If it is an iterable, it must be an iterable of integers in the range 0 <= x < 256, which are used as the initial contents of the array.Without an argument, an array of size 0 is created.See also Binary Sequence Types — bytes, bytearray, memoryview and Bytearray Objects.
###Code
bytearray(4)
###Output
_____no_output_____
###Markdown
*class* bytes([source[, encoding[, errors]]])Return a new “bytes” object, which is an immutable sequence of integers in the range 0 <= x < 256. bytes is an immutable version of bytearray – it has the same non-mutating methods and the same indexing and slicing behavior.Accordingly, constructor arguments are interpreted as for bytearray().Bytes objects can also be created with literals, see String and Bytes literals.See also Binary Sequence Types — bytes, bytearray, memoryview, Bytes, and Bytes and Bytearray Operations.
###Code
bytes(4)
###Output
_____no_output_____
###Markdown
callable(object)Return True if the object argument appears callable, False if not. If this returns true, it is still possible that a call fails, but if it is false, calling object will never succeed. Note that classes are callable (calling a class returns a new instance); instances are callable if their class has a __call__() method.New in version 3.2: This function was first removed in Python 3.0 and then brought back in Python 3.2.
###Code
callable(7)
callable(int)
###Output
_____no_output_____
###Markdown
chr(i)Return the string representing a character whose Unicode code point is the integer i. For example, chr(97) returns the string 'a', while chr(8364) returns the string '€'. This is the inverse of ord().The valid range for the argument is from 0 through 1,114,111 (0x10FFFF in base 16). ValueError will be raised if i is outside that range.
###Code
chr(97)
chr(8364)
###Output
_____no_output_____
###Markdown
classmethod(function)Return a class method for function.A class method receives the class as implicit first argument, just like an instance method receives the instance. To declare a class method, use this idiom: class C: @classmethod def f(cls, arg1, arg2, ...): ... The @classmethod form is a function decorator – see the description of function definitions in Function definitions for details.It can be called either on the class (such as C.f()) or on an instance (such as C().f()). The instance is ignored except for its class. If a class method is called for a derived class, the derived class object is passed as the implied first argument.Class methods are different than C++ or Java static methods. If you want those, see staticmethod() in this section.For more information on class methods, consult the documentation on the standard type hierarchy in The standard type hierarchy.
###Code
class Bar:
@classmethod
def foo(cls, arg1):
print(arg1)
Bar
Bar.foo
Bar.foo('hello')
Bar()
Bar().foo('hello')
###Output
hello
|
Day 2/09+Solving+Pong+with+Tensorflow.ipynb
|
###Markdown
PongThis notebook is adapted from https://github.com/ageron/handson-ml/blob/master/16_reinforcement_learning.ipynbBook: https://www.amazon.com/Hands-Machine-Learning-Scikit-Learn-TensorFlow/dp/1491962291
###Code
# You might need to install this
#!sudo apt-get install cmake -y
#!sudo apt-get install zlib1g-dev -y
#!sudo pip install -q gym[atari]
# Common imports
import numpy as np
import gym
import sys
import os
import tensorflow as tf
# to make this notebook's output stable across runs
def reset_graph(seed=42):
tf.reset_default_graph()
tf.set_random_seed(seed)
np.random.seed(seed)
# To plot pretty figures and animations
%matplotlib nbagg
import matplotlib
import matplotlib.animation as animation
import matplotlib.pyplot as plt
plt.rcParams['axes.labelsize'] = 14
plt.rcParams['xtick.labelsize'] = 12
plt.rcParams['ytick.labelsize'] = 12
# Auxiliary functions to render video
def update_scene(num, frames, patch):
patch.set_data(frames[num])
return patch
def plot_animation(frames, repeat=False, interval=40):
plt.close() # or else nbagg sometimes plots in the previous cell
fig = plt.figure()
patch = plt.imshow(frames[0])
plt.axis('off')
return animation.FuncAnimation(fig, update_scene, fargs=(frames, patch), frames=len(frames), repeat=repeat, interval=interval)
## Creating the environment
env = gym.make("Pong-v0")
obs = env.reset()
obs.shape
## Preprocessing
#Preprocessing the images is optional but greatly speeds up training.
mspacman_color = 210 + 164 + 74
def preprocess_observation(obs):
img = obs[1:176:2, ::2] # crop and downsize
img = img.sum(axis=2) # to greyscale
img[img==mspacman_color] = 0 # Improve contrast
img = (img // 3 - 128).astype(np.int8) # normalize from -128 to 127
return img.reshape(88, 80, 1)
img = preprocess_observation(obs)
#Note: the `preprocess_observation()` function is slightly different from the one in the book: instead of representing pixels as 64-bit floats from -1.0 to 1.0, it represents them as signed bytes (from -128 to 127). The benefit is that the replay memory will take up roughly 8 times less RAM (about 6.5 GB instead of 52 GB). The reduced precision has no visible impact on training.
plt.figure(figsize=(8, 4))
plt.subplot(121)
plt.title("Original observation (160×210 RGB)")
plt.imshow(obs)
plt.axis("off")
plt.subplot(122)
plt.title("Preprocessed observation (88×80 greyscale)")
plt.imshow(img.reshape(88, 80), interpolation="nearest", cmap="gray")
plt.axis("off")
#save_fig("preprocessing_plot")
plt.show()
reset_graph()
input_height = 88
input_width = 80
input_channels = 1
conv_n_maps = [32, 64, 64]
conv_kernel_sizes = [(8,8), (4,4), (3,3)]
conv_strides = [4, 2, 1]
conv_paddings = ["SAME"] * 3
conv_activation = [tf.nn.relu] * 3
n_hidden_in = 64 * 11 * 10 # conv3 has 64 maps of 11x10 each
n_hidden = 512
hidden_activation = tf.nn.relu
n_outputs = env.action_space.n # 9 discrete actions are available
initializer = tf.variance_scaling_initializer()
def q_network(X_state, name):
prev_layer = X_state / 128.0 # scale pixel intensities to the [-1.0, 1.0] range.
with tf.variable_scope(name) as scope:
for n_maps, kernel_size, strides, padding, activation in zip(
conv_n_maps, conv_kernel_sizes, conv_strides,
conv_paddings, conv_activation):
prev_layer = tf.layers.conv2d(
prev_layer, filters=n_maps, kernel_size=kernel_size,
strides=strides, padding=padding, activation=activation,
kernel_initializer=initializer)
last_conv_layer_flat = tf.reshape(prev_layer, shape=[-1, n_hidden_in])
hidden = tf.layers.dense(last_conv_layer_flat, n_hidden,
activation=hidden_activation,
kernel_initializer=initializer)
outputs = tf.layers.dense(hidden, n_outputs,
kernel_initializer=initializer)
trainable_vars = tf.get_collection(tf.GraphKeys.TRAINABLE_VARIABLES,
scope=scope.name)
trainable_vars_by_name = {var.name[len(scope.name):]: var
for var in trainable_vars}
return outputs, trainable_vars_by_name
X_state = tf.placeholder(tf.float32, shape=[None, input_height, input_width,
input_channels])
online_q_values, online_vars = q_network(X_state, name="q_networks/online")
target_q_values, target_vars = q_network(X_state, name="q_networks/target")
copy_ops = [target_var.assign(online_vars[var_name])
for var_name, target_var in target_vars.items()]
copy_online_to_target = tf.group(*copy_ops)
learning_rate = 0.001
momentum = 0.95
with tf.variable_scope("train"):
X_action = tf.placeholder(tf.int32, shape=[None])
y = tf.placeholder(tf.float32, shape=[None, 1])
q_value = tf.reduce_sum(online_q_values * tf.one_hot(X_action, n_outputs),
axis=1, keepdims=True)
error = tf.abs(y - q_value)
clipped_error = tf.clip_by_value(error, 0.0, 1.0)
linear_error = 2 * (error - clipped_error)
loss = tf.reduce_mean(tf.square(clipped_error) + linear_error)
global_step = tf.Variable(0, trainable=False, name='global_step')
optimizer = tf.train.MomentumOptimizer(learning_rate, momentum, use_nesterov=True)
training_op = optimizer.minimize(loss, global_step=global_step)
init = tf.global_variables_initializer()
saver = tf.train.Saver()
###Output
_____no_output_____
###Markdown
We use this `ReplayMemory` class instead of a `deque` because it is much faster for random access (thanks to @NileshPS who contributed it). Moreover, we default to sampling with replacement, which is much faster than sampling without replacement for large replay memories.
###Code
class ReplayMemory:
def __init__(self, maxlen):
self.maxlen = maxlen
self.buf = np.empty(shape=maxlen, dtype=np.object)
self.index = 0
self.length = 0
def append(self, data):
self.buf[self.index] = data
self.length = min(self.length + 1, self.maxlen)
self.index = (self.index + 1) % self.maxlen
def sample(self, batch_size, with_replacement=True):
if with_replacement:
indices = np.random.randint(self.length, size=batch_size) # faster
else:
indices = np.random.permutation(self.length)[:batch_size]
return self.buf[indices]
replay_memory_size = 500000
replay_memory = ReplayMemory(replay_memory_size)
def sample_memories(batch_size):
cols = [[], [], [], [], []] # state, action, reward, next_state, continue
for memory in replay_memory.sample(batch_size):
for col, value in zip(cols, memory):
col.append(value)
cols = [np.array(col) for col in cols]
return cols[0], cols[1], cols[2].reshape(-1, 1), cols[3], cols[4].reshape(-1, 1)
eps_min = 0.1
eps_max = 1.0
eps_decay_steps = 2000000
def epsilon_greedy(q_values, step):
epsilon = max(eps_min, eps_max - (eps_max-eps_min) * step/eps_decay_steps)
if np.random.rand() < epsilon:
return np.random.randint(n_outputs) # random action
else:
return np.argmax(q_values) # optimal action
n_steps = 4000000 # total number of training steps
training_start = 10000 # start training after 10,000 game iterations
training_interval = 4 # run a training step every 4 game iterations
save_steps = 1000 # save the model every 1,000 training steps
copy_steps = 10000 # copy online DQN to target DQN every 10,000 training steps
discount_rate = 0.99
skip_start = 90 # Skip the start of every game (it's just waiting time).
batch_size = 50
iteration = 0 # game iterations
checkpoint_path = "./out/pong-dqn.ckpt"
done = True # env needs to be reset
# A few variables for tracking progress:
loss_val = np.infty
game_length = 0
total_max_q = 0
mean_max_q = 0.0
# And now the main training loop!
with tf.Session() as sess:
if os.path.isfile(checkpoint_path + ".index"):
saver.restore(sess, checkpoint_path)
else:
init.run()
copy_online_to_target.run()
while True:
step = global_step.eval()
if step >= n_steps:
break
iteration += 1
print("\rIteration {}\tTraining step {}/{} ({:.1f})%\tLoss {:5f}\tMean Max-Q {:5f} ".format(
iteration, step, n_steps, step * 100 / n_steps, loss_val, mean_max_q), end="")
if done: # game over, start again
obs = env.reset()
for skip in range(skip_start): # skip the start of each game
obs, reward, done, info = env.step(0)
state = preprocess_observation(obs)
# Online DQN evaluates what to do
q_values = online_q_values.eval(feed_dict={X_state: [state]})
action = epsilon_greedy(q_values, step)
# Online DQN plays
obs, reward, done, info = env.step(action)
next_state = preprocess_observation(obs)
# Let's memorize what happened
replay_memory.append((state, action, reward, next_state, 1.0 - done))
state = next_state
# Compute statistics for tracking progress (not shown in the book)
total_max_q += q_values.max()
game_length += 1
if done:
mean_max_q = total_max_q / game_length
total_max_q = 0.0
game_length = 0
if iteration < training_start or iteration % training_interval != 0:
continue # only train after warmup period and at regular intervals
# Sample memories and use the target DQN to produce the target Q-Value
X_state_val, X_action_val, rewards, X_next_state_val, continues = (
sample_memories(batch_size))
next_q_values = target_q_values.eval(
feed_dict={X_state: X_next_state_val})
max_next_q_values = np.max(next_q_values, axis=1, keepdims=True)
y_val = rewards + continues * discount_rate * max_next_q_values
# Train the online DQN
_, loss_val = sess.run([training_op, loss], feed_dict={
X_state: X_state_val, X_action: X_action_val, y: y_val})
# Regularly copy the online DQN to the target DQN
if step % copy_steps == 0:
copy_online_to_target.run()
# And save regularly
if step % save_steps == 0:
saver.save(sess, checkpoint_path)
###Output
_____no_output_____
###Markdown
You can interrupt the cell above at any time to test your agent using the cell below. You can then run the cell above once again, it will load the last parameters saved and resume training.
###Code
frames = []
n_max_steps = 10000
with tf.Session() as sess:
saver.restore(sess, checkpoint_path)
obs = env.reset()
for step in range(n_max_steps):
state = preprocess_observation(obs)
# Online DQN evaluates what to do
q_values = online_q_values.eval(feed_dict={X_state: [state]})
action = np.argmax(q_values)
# Online DQN plays
obs, reward, done, info = env.step(action)
img = env.render(mode="rgb_array")
frames.append(img)
if done:
break
plot_animation(frames)
###Output
_____no_output_____
|
notebooks/PythonFeatureIO.ipynb
|
###Markdown
Using the Spatial Statistics Data Object (SSDataObject) Makes Feature IO Simple- SSDataObject does the read/write and accounting of feature/attribute and NumPy Array order- Write/Utilize methods that take NumPy Arrays Using NumPy as the common denominator- Could use the ArcPy Data Access Module directly, but there are host of issues/information one must take into account: * How to deal with projections and other environment settings? * How Cursors affect the accounting of features? * How to deal with bad records/bad data and error handling? * How to honor/account for full field object control? * How do I create output features that correspond to my inputs? - Points are easy, what about Polygons and Polylines?- Spatial Statistics Data Object (SSDataObject) * Almost 30 Spatial Statistics Tools written in Python that ${\bf{must}}$ behave like traditional GP Tools * Use SSDataObject and your code should adhere The Data Analysis Python Modules- [PANDAS (Python Data Analysis Library)](http://pandas.pydata.org/) - [SciPy (Scientific Python)](http://www.scipy.org/)- [PySAL (Python Spatial Analysis Library)](https://geodacenter.asu.edu/pysal) Basic Imports
###Code
import arcpy as ARCPY
import numpy as NUM
import SSDataObject as SSDO
###Output
_____no_output_____
###Markdown
Initialize and Load Fields into Spatial Statsitics Data Object- The Unique ID Field ("MYID" in this example) will keep track of the order of your features * You can use ```ssdo.oidName``` as your Unique ID Field * You have no control over Object ID Fields. It is quick, assures "uniqueness", but can't assume they will not get "scrambled" during copies. * To assure full control I advocate the "Add Field (LONG)" --> "Calculate Field (From Object ID)" workflow.
###Code
inputFC = r'../data/CA_Polygons.shp'
ssdo = SSDO.SSDataObject(inputFC)
ssdo.obtainData("MYID", ['GROWTH', 'LOGPCR69', 'PERCNOHS', 'POP1969'])
df = ssdo.getDataFrame()
print(df.head())
###Output
GROWTH LOGPCR69 PERCNOHS POP1969
158 0.011426 0.176233 37.0 1060099
159 -0.137376 0.214186 38.3 398
160 -0.188417 0.067722 41.4 11240
161 -0.085070 -0.118248 42.9 101057
162 -0.049022 -0.081377 48.1 13328
###Markdown
You can get your data using the core NumPy Arrays - Use ```.data``` to get the native data type- Use the ```returnDouble()``` function to cast explicitly to float
###Code
pop69 = ssdo.fields['POP1969']
nativePop69 = pop69.data
floatPop69 = pop69.returnDouble()
print(floatPop69[0:5])
###Output
[ 1.06009900e+06 3.98000000e+02 1.12400000e+04 1.01057000e+05
1.33280000e+04]
###Markdown
You can get your data in a PANDAS Data Frame- Note the Unique ID Field is used as the Index
###Code
df = ssdo.getDataFrame()
print(df.head())
###Output
GROWTH LOGPCR69 PERCNOHS POP1969
158 0.011426 0.176233 37.0 1060099
159 -0.137376 0.214186 38.3 398
160 -0.188417 0.067722 41.4 11240
161 -0.085070 -0.118248 42.9 101057
162 -0.049022 -0.081377 48.1 13328
###Markdown
By default the SSDataObject only stores the centroids of the features
###Code
df['XCoords'] = ssdo.xyCoords[:,0]
df['YCoords'] = ssdo.xyCoords[:,1]
print(df.head())
###Output
GROWTH LOGPCR69 PERCNOHS POP1969 XCoords YCoords
158 0.011426 0.176233 37.0 1060099 -1.356736e+07 4.503012e+06
159 -0.137376 0.214186 38.3 398 -1.333797e+07 4.637142e+06
160 -0.188417 0.067722 41.4 11240 -1.343007e+07 4.615529e+06
161 -0.085070 -0.118248 42.9 101057 -1.353566e+07 4.789809e+06
162 -0.049022 -0.081377 48.1 13328 -1.341895e+07 4.581597e+06
###Markdown
You can get the core ArcPy Geometries if desired- Set ```requireGeometry = True```
###Code
ssdo = SSDO.SSDataObject(inputFC)
ssdo.obtainData("MYID", ['GROWTH', 'LOGPCR69', 'PERCNOHS', 'POP1969'],
requireGeometry = True)
df = ssdo.getDataFrame()
shapes = NUM.array(ssdo.shapes, dtype = object)
df['shapes'] = shapes
print(df.head())
###Output
GROWTH LOGPCR69 PERCNOHS POP1969 \
158 0.011426 0.176233 37.0 1060099
159 -0.137376 0.214186 38.3 398
160 -0.188417 0.067722 41.4 11240
161 -0.085070 -0.118248 42.9 101057
162 -0.049022 -0.081377 48.1 13328
shapes
158 (<geoprocessing array object object at 0x00000...
159 (<geoprocessing array object object at 0x00000...
160 (<geoprocessing array object object at 0x00000...
161 (<geoprocessing array object object at 0x00000...
162 (<geoprocessing array object object at 0x00000...
###Markdown
Coming Soon... ArcPy Geometry Data Frame Integration - In conjunction with the ArcGIS Python SDK - Spatial operators on ArcGIS Data Frames: selection, clip, intersection etc. Creating Output Feature Classes - Simple Example: Adding a field of random standard normal values to your input/output- ```appendFields``` can be used to copy over any fields from the input whether you read them into the SSDataObject or not.- E.g. 'NEW_NAME' was never read into Python but it will be copied to the output. This can save you a lot of memory.
###Code
import numpy.random as RAND
import os as OS
ARCPY.env.overwriteOutput = True
outArray = RAND.normal(0,1, (ssdo.numObs,))
outDict = {}
outField = SSDO.CandidateField('STDNORM', 'DOUBLE', outArray, alias = 'Standard Normal')
outDict[outField.name] = outField
outputFC = OS.path.abspath(r'../data/testMyOutput.shp')
ssdo.output2NewFC(outputFC, outDict, appendFields = ['GROWTH', 'PERCNOHS', 'NEW_NAME'])
###Output
_____no_output_____
|
Challenge_5/Ch5_day3.ipynb
|
###Markdown
Łączenie daty na okrętkę
###Code
df['date']=df.apply( lambda row: '{}-{}-{}'.format(int(row['year']), int(row['month']), int(row['day'])), axis=1)
df['date'] = pd.to_datetime(df['date'])
df[ ['date'] ].info()
df.index = df['date']
df [['ppm']].plot();
###Output
_____no_output_____
###Markdown
Łączenie daty prościej
###Code
df.index = pd.to_datetime(df[['year', 'month', 'day']])
df [['ppm']].plot();
###Output
_____no_output_____
###Markdown
OutlajeryPonieważ mamy dziwne wartości (-999) zapewne w czasie gdy nie było danych z czujnika to trzeba je usunąć
###Code
df['ppm_fixed'] = df['ppm'].map(lambda x: np.nan if x<0 else x).fillna(method='backfill')
df [['ppm_fixed']].plot();
df1974 = df[df.year == 1974]
plt.plot(df1974.index, df1974.ppm_fixed);
plt.figure(figsize=(15,5))
for year in range (1974, 1985):
df_year = df[df.year == year]
plt.plot(df_year.index, df_year['ppm_fixed'], 'o-', label=year);
plt.legend();
###Output
_____no_output_____
|
fj_composite.ipynb
|
###Markdown
###Code
# Load the modules used
import numpy as np
import pandas as pd
import re
import string
import nltk
from sklearn.model_selection import train_test_split
from sklearn.feature_extraction.text import CountVectorizer
from sklearn.metrics import accuracy_score, f1_score, confusion_matrix, roc_auc_score
from keras.models import Model
from keras.layers import Dense, Dropout, Embedding, LSTM, Bidirectional, GlobalMaxPool1D, Conv1D, MaxPool1D, Flatten, RepeatVector, Input, Embedding, Concatenate
from keras.optimizers import Adam
from keras import metrics
from keras.preprocessing.sequence import pad_sequences
from keras.preprocessing.text import Tokenizer
# For reproducible results, set seeds
import random as rn
import os
import tensorflow as tf
os.environ['PYTHONHASHSEED'] = '42'
os.environ['CUDA_VISIBLE_DEVICES'] = ''
np.random.seed(42)
rn.seed(42)
tf.random.set_seed(42)
# Set data_url, the location of the data
# Data is not loaded from a local file
# data_url="https://raw.githubusercontent.com/r-dube/fakejobs/main/data/fj_small.csv"
# data_url="https://raw.githubusercontent.com/r-dube/fakejobs/main/data/fj_medium.csv"
data_url="https://raw.githubusercontent.com/r-dube/fakejobs/main/data/fake_job_postings.csv"
def fj_load_df_from_url():
"""
Load dataframe from csv file
Input:
None
Returns:
dataframe
"""
df = pd.read_csv(data_url)
print ('Loaded dataframe shape', df.shape)
counts = fj_label_stats(df)
print ('Not fraudulent', counts[0], 'Fraudulent', counts[1])
print(df.describe())
print ('NAs/NANs in data =>')
print(df.isna().sum())
return df
def fj_label_stats(df):
"""
Very basic label statistics
Input:
Dataframe
Returns:
Number of samples with 0, 1 as the label
"""
counts = np.bincount(df['fraudulent'])
return counts
def fj_txt_only(df):
"""
Combine all the text fields, discard everything else except for the label
Input:
Dataframe
Returns:
Processed dataframe
"""
df.fillna(" ", inplace = True)
df['text'] = df['title'] + ' ' + df['location'] + ' ' + df['department'] + \
' ' + df['company_profile'] + ' ' + df['description'] + ' ' + \
df['requirements'] + ' ' + df['benefits'] + ' ' + df['employment_type'] + \
' ' + df['required_education'] + ' ' + df['industry'] + ' ' + df['function']
del df['title']
del df['location']
del df['department']
del df['company_profile']
del df['description']
del df['requirements']
del df['benefits']
del df['employment_type']
del df['required_experience']
del df['required_education']
del df['industry']
del df['function']
del df['salary_range']
del df['job_id']
del df['telecommuting']
del df['has_company_logo']
del df['has_questions']
return df
df = fj_load_df_from_url()
df = fj_txt_only(df)
print('Maximum text length', df['text'].str.len().max())
# Utilities to clean text
def remove_URL(text):
url = re.compile(r"https?://\S+|www\.\S+")
return url.sub(r"", text)
def remove_html(text):
html = re.compile(r"<.*?>")
return html.sub(r"", text)
def remove_emoji(string):
emoji_pattern = re.compile(
"["
u"\U0001F600-\U0001F64F" # emoticons
u"\U0001F300-\U0001F5FF" # symbols & pictographs
u"\U0001F680-\U0001F6FF" # transport & map symbols
u"\U0001F1E0-\U0001F1FF" # flags (iOS)
u"\U00002702-\U000027B0"
u"\U000024C2-\U0001F251"
"]+",
flags=re.UNICODE,
)
return emoji_pattern.sub(r"", string)
def remove_punct(text):
table = str.maketrans("", "", string.punctuation)
return text.translate(table)
# more text cleaning - remove stopwords using NLTK
nltk.download('stopwords')
from nltk.corpus import stopwords
stop = set(stopwords.words("english"))
def remove_stopwords(text):
text = [word.lower() for word in text.split() if word.lower() not in stop]
return " ".join(text)
# Actually clean the text
df['text'] = df['text'].map(lambda x: remove_URL(x))
df['text'] = df['text'].map(lambda x: remove_html(x))
df['text'] = df['text'].map(lambda x: remove_emoji(x))
df['text'] = df['text'].map(lambda x: remove_punct(x))
df['text'] = df["text"].map(remove_stopwords)
# train-test split
train_text, test_text, train_labels , test_labels = train_test_split(df['text'], df['fraudulent'] , test_size = 0.15)
print(train_text.shape)
# Token model: Configuration parameters
max_num_words = 50000 # maximum allowed size of vocabulary
max_length = 250 # maximum allowed number of words in a job description
embed_dim = 32 # number of dimensions for learned embedding
# Token Model: Prepare the token based train and test input
# max_num_words variable in case we want to clip the number of words
tokenizer = Tokenizer(num_words=max_num_words)
tokenizer.fit_on_texts(train_text)
word_index = tokenizer.word_index
train_sequences = tokenizer.texts_to_sequences(train_text)
train_padded = pad_sequences(
train_sequences, maxlen=max_length, padding="post", truncating="post"
)
test_sequences = tokenizer.texts_to_sequences(test_text)
test_padded = pad_sequences(
test_sequences, maxlen=max_length, padding="post", truncating="post"
)
print(f"Shape of train {train_padded.shape}")
print(f"Shape of test {test_padded.shape}")
# Char model: Configuration parameters
# Assuming that the text has been processed for word tokenization
# alphabet="abcdefghijklmnopqrstuvwxyz0123456789-,;.!?:'\"/\\|_@#$%^&*~`+-=<>()[]{}"
alphabet="abcdefghijklmnopqrstuvwxyz0123456789"
encoding_size = len(alphabet)
char_input_size=1000
# Create a dictionary for encoding characters
dict = {} # Maps each character to an integer
for idx, char in enumerate(alphabet):
dict[char] = idx + 1
print(encoding_size)
print (dict)
# Char model: Utility function(s)
# Return one-hot-vector character encoding for string
# Memory is allocated outside this routine
def str_to_ohv(s, ohv):
max_length = min(len(s), char_input_size)
for i in range(0, max_length):
c = s[i]
if c in dict:
ohv[i, dict[c]-1] = 1
return ohv
# Char model: create train input
num_jobs = train_text.shape[0]
train_t = np.zeros((num_jobs, char_input_size, encoding_size), dtype=np.int8)
i = 0
for _, val in train_text.iteritems():
str_to_ohv(val, train_t[i])
i=i+1
# Char model: create test input
num_jobs = test_text.shape[0]
test_t = np.zeros((num_jobs, char_input_size, encoding_size), dtype=np.int8)
i = 0
for _, val in test_text.iteritems():
str_to_ohv(val, test_t[i])
i=i+1
# The Composite model: Token + Char
# Not specificying input dtype as Keras internally assumes float32
# Token (word) model
token_input = Input(shape=(max_length,), name="token_input")
token_embedding = Embedding(max_num_words, embed_dim, input_length=max_length, name="token_embedding")(token_input)
token_conv = Conv1D(64, kernel_size=3, strides=1, padding="valid", activation="relu", name="token_conv")(token_embedding)
token_pool = MaxPool1D(pool_size=3, strides=3, name="token_pool")(token_conv)
token_drop = Dropout(.5, name="token_drop")(token_pool)
# Char model
char_input = Input(shape=(char_input_size, encoding_size), name="char_input")
char_conv = Conv1D(64, kernel_size=3, strides=1, padding="valid",activation="relu", name="char_conv")(char_input)
char_pool = GlobalMaxPool1D(name="char_pool")(char_conv)
char_drop = Dropout(.5, name="char_drop")(char_pool)
char_repeated = RepeatVector(2, name="char_repeated")(char_drop)
# Merge
merged = Concatenate(axis=1, name="concat")([token_drop, char_repeated])
lstm = Bidirectional(LSTM(32, dropout=0.3, recurrent_dropout=0.01, name="lstm"), name="bidir")(merged)
output = Dense(1, activation="sigmoid", name="output")(lstm)
# define a model with a list of two inputs
model = Model(inputs=[token_input, char_input], outputs=output)
model.compile(optimizer = 'adam' , loss = 'binary_crossentropy' , metrics = ['accuracy', tf.keras.metrics.FalsePositives(), tf.keras.metrics.FalseNegatives()])
model.summary()
model.fit([train_padded, train_t], train_labels, epochs = 7)
pred_soft = model.predict([test_padded, test_t])
# pred = np.around(pred_soft, decimals = 0)
pred = np.where(pred_soft > 0.15, 1, 0)
acc = accuracy_score(pred, test_labels)
f1 = f1_score(pred, test_labels)
cm = confusion_matrix(test_labels, pred)
tn = cm[0][0]
fn = cm[1][0]
tp = cm[1][1]
fp = cm[0][1]
print('Accuracy score: {:.4f}'.format(acc), 'F1 score: {:.4f}'.format(f1))
print('False Positives: {:.0f}'.format(fp), 'False Negatives: {:.0f}'.format(fn))
print('Confusion matrix:\n', cm)
auc = roc_auc_score(test_labels, pred_soft)
print('AUC score: {:.4f}'.format(auc))
# Uncomment to save image of model architecture
from google.colab import drive
drive.mount('/content/drive')
dot_img_file = '/content/drive/My Drive/Results/fj_composite.png'
tf.keras.utils.plot_model(model, to_file=dot_img_file, show_shapes=True, show_dtype=True)
###Output
Drive already mounted at /content/drive; to attempt to forcibly remount, call drive.mount("/content/drive", force_remount=True).
|
notebooks/Lab 5 - SNUZ.ipynb
|
###Markdown
SNUZ - white noise to drive _and_ relax you!> Arrive at your destination safely, comfortably, and well rested. We combine state-of-the-art methods in random search to get you safely to your destination. Using random methods lets us generate efficient routes, and high quality (mandatory) white noise for your journey -- across the town or across the country!In this experiment an autonomous car will learn to drive up a hill. We'll compare random search ([ARS](https://arxiv.org/abs/1803.07055)) to Proximal Policy Optimization ([PPO](https://blog.openai.com/openai-baselines-ppo/)). Aims1. Install pytorch, et al2. Answer the question: does random search do better than a state of the 'cart' RL method in ...one of the simplest continuous control tasks?3. _Acquirehire_. InstallBefore doing anything else, we need to install some libraries.From the command line, run:`pip install gym``pip install ray``pip install opencv-python` Then for your OS, do: Mac`conda install pytorch torchvision -c pytorch` Linux`conda install pytorch torchvision -c pytorch` Windows`conda install pytorch -c pytorch``pip3 install torchvision`
###Code
from ADMCode import visualize as vis
from ADMCode.snuz import run_ppo
from ADMCode.snuz import run_ars
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
import seaborn as sns
import warnings
warnings.simplefilter('ignore', np.RankWarning)
warnings.filterwarnings("ignore", module="matplotlib")
warnings.filterwarnings("ignore")
sns.set(style='white', font_scale=1.3)
%matplotlib inline
%config InlineBackend.figure_format = 'png'
%config InlineBackend.savefig.dpi = 150
###Output
_____no_output_____
###Markdown
TaskWe're going to teaching a car to drive up a hill! This is the `MountainCarContinuous-v0` from the OpenAI [gym].(https://gym.openai.com) Vrooooom!Let's get driving, uphill! First let's try PPO. PPOThe default hyperparameters are: gamma = 0.99 Try me? lam = 0.98 Try me? actor_hidden1 = 64 Try me? actor_hidden2 = 64 Try me? actor_hidden3 = 64 Try me? critic_hidden1 = 64 Try me? critic_lr = 0.0003 Try me? (small changes) actor_lr = 0.0003 Try me? (small changes) batch_size = 64 Leave me be l2_rate = 0.001 Leave me be clip_param = 0.2 Leave me be num_training_epochs = 10 Try me? num_episodes = 10 Try me? num_memories = 24 Try me? num_training_epochs = 10 Try me? clip_actions = True Leave me be clip_std = 1.0 Leave me be seed_value = None Try me (with int only) Parameters can be changed by passing to `run_ppo`. For example `run_ppo(num_episodes=20, actor_lr=0.0006`) doubles the train time and the learning rate of the PPO.
###Code
episodes, scores = run_ppo(render=True, num_episodes=10)
###Output
_____no_output_____
###Markdown
Plot the average reward / episode.
###Code
plt.plot(episodes, scores)
plt.xlabel("Episode")
plt.xlabel("Reward")
###Output
_____no_output_____
###Markdown
Compare, say, 10 episodes of PPO to 10 of... ARSThe [ARS](https://arxiv.org/abs/1803.07055) code was modified from Recht's [original source](https://github.com/modestyachts/ARS). The default hyperparameters are: num_episodes = 10 Try me? n_directions = 8 Try me? deltas_used = 8 Try me? step_size = 0.02 Try me? delta_std = 0.03 Try me? n_workers = 1 Leave me be rollout_length = 240 Try me? shift = 0 Leave me be (all below) seed = 237 policy_type = 'linear' dir_path = 'data' filter = 'MeanStdFilter' Leave me be _Note_: Due to the way the backend of ARS works (it uses a [ray](https://ray.readthedocs.io/en/latest/), a dist. job system) we can't render exps here. Sorry. :(
###Code
episodes, scores = run_ars(num_episodes=10)
plt.plot(episodes, scores)
plt.xlabel("Episode")
plt.xlabel("Reward")
###Output
_____no_output_____
|
module-content/api_usage/.ipynb_checkpoints/3.6-Controlling-Simulation-API-checkpoint.ipynb
|
###Markdown
Publishing and Subscribing with the Simulation API This tutorial introduces the Simulation API and how it can be used to run power system simulations, subscribe to measurement data, and publish equipment control commands.__Learning Objectives:__At the end of the tutorial, the user should be able to use the Simulation API to* Create difference messages for controlling power system equipment* Publish simulation input commands to a GridAPPS-D simulation* Subscribe to the output messages from a GridAPPPS-D simulation using * a function definition * a class definition * the Simulation library methods* Parse simulation output messages to obtain measurement values Getting Started__1) Start GridAPPS-D Platform if not running:__Before running any of the sample routines in this tutorial, it is first necessary to start the GridAPPS-D Platform and establish a connection to this notebook so that we can start passing calls to the API. _Open the Ubuntu terminal and start the GridAPPS-D Platform if it is not running already:_`cd gridappsd-docker`~/gridappsd-docker$ `./run.sh -t develop`_Once containers are running,_gridappsd@[container]:/gridappsd$ `./run-gridappsd.sh` __2) Start a simulation in the GridAPPS-D Viz:__The Simulation API calls covered in this lesson need to be passed to an active simulation. For the purposes of this tutorial, we will use the GridAPPS-D Viz at [localhost:8080](http://localhost:8080/) to start a simulation of the IEEE 123 Node model with a 3600 sec simulation time. After starting the simulation, paste the simulation_id into the code block below by clicking on the simulation_id. This will automatically copy the simulation_id to your computer's clipboard.  When your application is containerized in Docker and registered with the GridAPPS-D Platform using the docker-compose file, the simulation_id and feeder model mRID as passed as part of the application start call. For this notebook, that information needs to be copied and pasted into the first code block below.
###Code
viz_simulation_id = "16543091"
model_mrid = "_C1C3E687-6FFD-C753-582B-632A27E28507"
# Establish connection to GridAPPS-D Platform:
from gridappsd import GridAPPSD
# Set environment variables - when developing, put environment variable in ~/.bashrc file or export in command line
# export GRIDAPPSD_USER=system
# export GRIDAPPSD_PASSWORD=manager
import os # Set username and password
os.environ['GRIDAPPSD_USER'] = 'tutorial_user'
os.environ['GRIDAPPSD_PASSWORD'] = '12345!'
# Connect to GridAPPS-D Platform
gapps = GridAPPSD(viz_simulation_id)
assert gapps.connected
###Output
_____no_output_____
###Markdown
--- Table of Contents* [1. Introduction to the Simulation API](1.-Introduction-to-the-Simulation-API)* [2. Using the Simulation API](2.-Using-the-Simulation-API) * [2.1. Specifying the Topic](2.1.-Specifying-the-Topic) * [2.2. Structure of a Simulation Message](2.2.-Structure-of-a-Simulation-Message) * [3. Publishing to Simulation Input](3.-Publishing-to-Simulation-Input) * [3.1. Topic to Publish to Simulation Input](3.1.-Topic-to-Publish-to-Simulation-Input) * [3.2. Obtain Equipment Control mRIDs](3.2.-Obtain-Equipment-Control-mRIDs) * [3.3. Format of a Difference Message](3.3.-Format-of-a-Difference-Message) * [3.4. Using GridAPPSD-Python DifferenceBuilder](3.4.-Using-GridAPPSD-Python-DifferenceBuilder) * [4. Subscribing to Simulation Output](4.-Subscribing-to-Simulation-Output) * [4.1. Specify the Topic](4.1.-Specify-the-Topic) * [4.2. Obtain Measurement mRIDs](4.2.-Obtain-Measurement-mRIDs) * [4.3. Structure of Simulation Output Message](4.3.-Structure-of-Simulation-Output-Message) * [4.4. Subscribe using a Function Definition](4.4.-Subscribe-using-a-Function-Definition) * [4.5. Subscribe using a Class Definition](4.5.-Subscribe-using-a-Class-Definition) * [4.6. Using the GridAPPSD Python Simulation Library Shortcuts](4.6.-Using-the-GridAPPSD-Python-Simulation-Library-Shortcuts) * [4.7. Comparison of Approaches](4.7.-Comparison-of-Approaches) --- 1. Introduction to the Simulation API Introduction to the Simulation APIThe Simulation API is used for all actions related to a power system simulation. It is used to start, pause, restart, and stop a simulation from the command line or inside an application. It is all used to subscribe to measurements of equipment (such as line flows, loads, and DG setpoints) and the statuses of switches, capacitors, transformer taps, etc. It is also used to publish equipment control and other simulation input commands.In the Application Components diagram (explained in detail with sample code in [GridAPPS-D Application Structure](../overview/2.4-GridAPPS-D-Application-Structure.ipynb)), the PowerGrid Models API is used for controlling the simulation, subscribing to measurement data, and controlling equipment.This section covers only the portion of the API used for subscribing to measurements and publishing equipment control commands. Usage of the API for starting, stopping, and pausing simulations is covered in [Creating and Running Simulations with Simulation API](../api_usage/3.5-Creating-Running-Simulation-API.ipynb) --- 2. Processing Measurements & App Core Algorithm Processing Measurements & App Core AlgorithmThe central portion of a GridAPPS-D application is the measurement processing and core algorithm section. This section is built as either a class or function definition with prescribed arguments. Each has its advantages and disadvantages:* The function-based approach is simpler and easier to implement. However, any parameters obtained from other APIs or methods to be used inside the function currently need to be defined as global variables. * The class-based approach is more complex, but also more powerful. It provides greater flexibility in creating additional methods, arguments, etc. 2.1 Information Flow App Core Information FlowThis portion of the application does not communicate directly with the GridAPPS-D platform. Instead, the next part of the GridAPPS-D application ([Subscribing to Simulation Output](Subscribing-to-Simulation-Output)) delivers the simulated SCADA measurement data to the core algorithm function / class definition. The core algorithm processes the data to extract the desired measurements and run its optimization / control agorithm. [Return to Top](Table-of-Contents) --- 2.2. Structure of Simulation Output Message Structure of Simulation Output MessageThe first part of the application core is parsing simulated SCADA and measurement data that is delivered to the application.The general format of the messages received by the Simulation API is a python dictionary with the following key-value pairs:```{ "simulation_id" : string, "message" : { "timestamp" : epoch time number, "measurements" : { "meas mrid 1":{ "PNV measurement_mrid": "meas mrid 1" "magnitude": number, "angle": number }, "meas mrid 2":{ "VA measurement_mrid": "meas mrid 2" "magnitude": number, "angle": number }, "meas mrid 3":{ "Pos measurement_mrid": "meas mrid 3" "value": number }, . . . "meas mrid n":{ "measurement_mrid": "meas mrid n" "magnitude": number, "angle": number }, }}``` Format of Measurement ValuesIn the message above, note the difference in the key-value pair structure of different types of measurements:__PNV Voltage Measurements__These are specified as `magnitude` and `angle` key-value pairs. * Magnitude is the RMS phase-to-neutral voltage. * Angle is phase angle of the voltage at the particular node.__VA Volt-Ampere Apparent Power Measurements__These are specified as `magnitude` and `angle` key-value pairs. * Magnitude is the apparent power. * Angle is complex power triangle angle (i.e. _acos(power factor)_)__Pos Position Measurements__These are specified as a `value` key-value pair. * Value is the position of the particular measurement * For switch objects: value = 1 means "closed", value = 0 means "open" * For capacitor objects, values are reversed: value = 1 means "on", value = 0 means "off" * For regulator objects, value is the tap position, ranging from -16 to 16 Role of Measurement mRIDsThe simulation output message shown above typically contains the measurement mRIDs for all available sensors for all equipment in the power system model. The application needs to filter the simulation output message to just the set of measurements relevant to the particular application (e.g. switch positions for a FLISR app or regulator taps for a VVO app).The equipment and measurement mRIDs are obtained in the first two sections of the application. See [Query for Power System Model](../overview/2.4-GridAPPS-D-Application-Structure.ipynbQuerying-for-the-Power-System-Model) and [Query for Measurement mRIDs](../overview/2.4-GridAPPS-D-Application-Structure.ipynbQuerying-for-Measurement-mRIDs) for examples of how these code sections fit in a sample app. API syntax details for the query messages to PowerGrid Models API to obtain equipment info and measurement mRIDs are given in [Query for Object Dictionary](../api_usage/3.3-Using-the-PowerGrid-Models-API.ipynbQuery-for-Object-Dictionary) and [Query for Measurements](../api_usage/3.3-Using-the-PowerGrid-Models-API.ipynbQuerying-for-Object-Measurements).These mRIDs will be needed to parse the simulation output message and filter it to just the desired set of measurements.For the example below, we will be interested in only the measurement associated with switches, so we will use the PowerGrid Models API to query for the set of measurements associated with the CIM Class `LoadBreakSwitch`. We then will filter those values to just the mRIDs associated with each type of measurement.
###Code
from gridappsd import topics as t
# Create query message to obtain measurement mRIDs for all switches
message = {
"modelId": model_mrid,
"requestType": "QUERY_OBJECT_MEASUREMENTS",
"resultFormat": "JSON",
"objectType": "LoadBreakSwitch"
}
# Pass query message to PowerGrid Models API
response_obj = gapps.get_response(t.REQUEST_POWERGRID_DATA, message)
measurements_obj = response_obj["data"]
# Switch position measurements (Pos)
Pos_obj = [k for k in measurements_obj if k['type'] == 'Pos']
# Switch phase-neutral-voltage measurements (PNV)
PNV_obj = [k for k in measurements_obj if k['type'] == 'PNV']
# Switch volt-ampere apparent power measurements (VA)
VA_obj = [k for k in measurements_obj if k['type'] == 'VA']
# Switch current measurements (A)
A_obj = [k for k in measurements_obj if k['type'] == 'A']
###Output
_____no_output_____
###Markdown
[Return to Top](Table-of-Contents) --- 2.3. App Core as a Function Definition App Core as a Function DefinitionThe first approach used to build the application core is to define a function with the correct set of arguments that is then passed to the `.subscribe()` method associated with the `GridAPPPSD()` object.The function does not require a specific name, and is somewhat easier to define and use. However, the arguments of the function need to be named correctly for the GridAPPSD-Python library to process the simulation output correctly.The format for the function definition is ```def mySubscribeFunction(header, message): do something when receive a message parse to get measurments do some calculations publish some equipment commands display some results```That function handle is then passed as an argument to the `.subscribe(topic, function_handle)` method when subscribing to the simulation in the next section. Note that the subscription function definition does not allow any additional parameters to be passed. The only allowed arguments are `header` and `message`. __Any other parameters, such as measurement mRIDs will need to be defined as global variables.__ 
###Code
# Define global python dictionary of position measurements
global Pos_obj
Pos_obj = [k for k in measurements_obj if k['type'] == 'Pos']
# Define global python dictionary of phase-neutral-voltage measurements (PNV)
global PNV_obj
PNV_obj = [k for k in measurements_obj if k['type'] == 'PNV']
# Define global python dictionary of volt-ampere apparent power measurements (VA)
VA_obj = [k for k in measurements_obj if k['type'] == 'VA']
# Current measurements (A)
A_obj = [k for k in measurements_obj if k['type'] == 'A']
###Output
_____no_output_____
###Markdown
Below is the sample code for the core section of a basic application that tracks the number of open switches and the number of switches that are outaged.
###Code
# Only allowed arguments are `header` and `message`
# message is simulation output message in format above
def DemoAppCoreFunction(header, message):
# Extract time and measurement values from message
timestamp = message["message"]["timestamp"]
meas_value = message["message"]["measurements"]
# Obtain list of all mRIDs from message
meas_mrid = list(meas_value.keys())
# Example 1: Count the number of open switches
open_switches = []
for index in Pos_obj:
if index["measid"] in meas_value:
mrid = index["measid"]
power = meas_value[mrid]
if power["value"] == 0: # Filter to measurements with value of zero
open_switches.append(index["eqname"])
# Print message to command line
print("............")
print("Number of open switches at time", timestamp, ' is ', len(set(open_switches)))
# Example 2: Count the number of outaged switches (voltage = 0)
dead_switches = []
for index in PNV_obj:
if index["measid"] in meas_value:
mrid = index["measid"]
voltage = meas_value[mrid]
if voltage["magnitude"] == 0.0:
dead_switches.append(index["eqname"])
# Print message to command line
print("............")
print("Number of outaged switches at time", timestamp, ' is ', len(set(dead_switches)))
###Output
_____no_output_____
###Markdown
[Return to Top](Table-of-Contents) --- 2.4. App Core as a Class Definition App Core as a Class DefinitionThe second approach used to build the app core and process measurements is to define a class containing two methods named `__init__` and `on_message`.These methods specify 1) how your app would initialize variables and attributes at the start of the simulation and 2) how your app behaves when it receives various messages. __IMPORTANT!__ The GridAPPS-D Platform uses the exact names and syntax for the methods:* `__init__(self, simulation_id, gapps_object, optional_objects)` -- This method requires the simulation_id and GridAPPS-D connection object. It is also possible add other user-defined arguments, such as measurement mRIDs or other information required by your application.* `on_message(self, headers, message)` -- This method allows the class to subscribe to simulation measurements. It also contains the core behavior of your application and how it responds to each type of message.It is also possible to use the same class definition to subscribe to other topics, such as Simulation Logs. This is done by creating additional user-defined methods and then passing those methods to the `.subcribe()` method associated with the GridAPPS-D connection object. An example of how this is done is provided for subcribing to simulation logs in [Logging with a Class Method](/api_usage/3.8-Using-the-Logging-API.ipynbCreate-Subscription-Class-Method). ```class YourSimulationClassName(object): Your documentation text here on what app does def __init__(self, simulation_id, gapps_obj, meas_obj, your_obj): Instantiate class with specific initial state Attributes required by Simulation API self._gapps = gapps_obj self._simulation_id = simulation_id Attributes to publish difference measurements self.diff = DifferenceBuilder(simulation_id) Custom attributes for measurements, custom info self.meas_mrid = meas_obj self.your_attribute1 = your_obj["key1"] self.your_attribute2 = your_obj["key2"] def on_message(self, headers, message): What app should do when it receives a subscription message variable1 = message["message"]["key1"] variable2 = message["message"]["key2"] Insert your custom app behavior here if variable1 == foo: bar = my_optimization_result Insert your custom equipment commands here if variable2 == bar: self.diff.add_difference(object_mrid, control_attribute, new_value, old_value) def my_custom_method_1(self, headers, message): Use extra methods to subscribe to other topics, such as simulation logs variable1 = message["key1"] variable2 = message["key2"] def my_custom_method_2(self, param1, param2): Use extra methods as desired variable1 = foo variable2 = bar ``` Below is the sample code for the core section of a basic application that tracks the number of open switches and the number of switches that are outaged.
###Code
# Application core built as a class definition
class DemoAppCoreClass(object):
# Subscription callback from GridAPPSD object
def __init__(self, simulation_id, gapps_obj, meas_obj):
self._gapps = gapps_obj # GridAPPS-D connection object
self._simulation_id = simulation_id # Simulation ID
self.meas_mrid = meas_obj # Dictionary of measurement mRIDs obtained earlier
def on_message(self, headers, message):
# Extract time and measurement values from message
timestamp = message["message"]["timestamp"]
meas_value = message["message"]["measurements"]
# Filter measurement mRIDs for position and voltage sensors
Pos_obj = [k for k in self.meas_mrid if k['type'] == 'Pos']
PNV_obj = [k for k in self.meas_mrid if k['type'] == 'PNV']
# Example 1: Count the number of open switches
open_switches = []
for index in Pos_obj:
if index["measid"] in meas_value:
mrid = index["measid"]
power = meas_value[mrid]
if power["value"] == 0: # Filter to measurements with value of zero
open_switches.append(index["eqname"])
# Print message to command line
print("............")
print("Number of open switches at time", timestamp, ' is ', len(set(open_switches)))
# Example 2: Count the number of outaged switches (voltage = 0)
dead_switches = []
for index in PNV_obj:
if index["measid"] in meas_value:
mrid = index["measid"]
voltage = meas_value[mrid]
if voltage["magnitude"] == 0.0:
dead_switches.append(index["eqname"])
# Print message to command line
print("............")
print("Number of outaged switches at time", timestamp, ' is ', len(set(dead_switches)))
###Output
_____no_output_____
###Markdown
[Return to Top](Table-of-Contents) --- 3. Subscribing to Simulation Output Subscribing to Simulation Output Simulation Subscription Information FlowThe figure below shows the information flow involved in subscribing to the simulation output.The subscription request is sent using `gapps.subscribe(topic, class/function object)` on the specific Simulation topic channel (explained in [API Communication Channels](../api_usage/3.1-API-Communication-Channels.ipynb)). No immediate response is expected back from the platform. However, after the next simulation timestep, the Platform will continue to deliver a complete set of measurements back to the application for each timestep until the end of the simulation.  __Application passes subscription request to GridAPPS-D Platform__The subscription request is perfromed by passing the app core algorithm function / class definition to the `gapps.subscribe` method. The application then passes the subscription request through the Simulation API to the topic channel for the particular simulation on the GOSS Message Bus. If the application is authorized to access simulation output, the subscription request is delivered to the Simulation Manager.__GridAPPS-D Platform delivers published simulation output to Application__Unlike the previous queries made to the various databases, the GridAPPS-D Platform does not provide any immediate response back to the application. Instead, the Simulation Manager will start delivering measurement data back to the application through the Simulation API at each subsequent timestep until the simulation ends or the application unsubscribes. The measurement data is then passed to the core algorithm class / function, where it is processed and used to run the app's optimization / control algorithms. [Return to Top](Table-of-Contents) --- 3.1. API Communication Channel Subscription API Communication ChannelThis is a dynamic `/topic/` communication channel that is best implemented by importing the GriAPPSD-Python library function for generating the correct topic. This communication channel is used for all simulation subscription API calls.
###Code
from gridappsd.topics import simulation_output_topic
output_topic = simulation_output_topic(viz_simulation_id)
###Output
_____no_output_____
###Markdown
[Return to Top](Table-of-Contents) --- 3.2. Comparison of Subscription Approaches Comparison of Subscription ApproachesEach approach has its advantages and disadvantages. * The function-based approach is simpler and easier to implement. However, any parameters obtained from other APIs or methods to be used inside the function currently need to be defined as global variables. * The class-based approach is more complex, but also more powerful. It provides greater flexibility in creating additional methods, arguments, etc.* The Simulation Library-based approach is easiest, but only works currently for parallel digital twin simulations started using the `simulation_obj.start_simulation()` method.The choice of which approach is used depends on the personal preferences of the application developer. [Return to Top](Table-of-Contents) --- 3.3. Subscription for Function-based App Core Subscription for Function-based App CoreIf the application core was created as a function definition as shown in [App Core as Function Definition](App-Core-as-a-Function-Definition), then the function name is passed to the `.subscribe(output_topic, core_function)` method of the GridAPPS-D Connection object.
###Code
gapps.subscribe(output_topic, DemoAppCoreFunction)
###Output
_____no_output_____
###Markdown
__Note on Jupyter Notebook environment:__ In the examples in this tutorial, the Jupyter Notebook environment does not update definitions of the subscription object or function definitions. As a result, it is necessary to restart the notebook kernel if you want to switch from using the function definition to the class definition. To restart the kernel, select `kernel` from the toolbar at the very top of the screen, and click `Restart` or `Restart and Clear Output`When restarting, be sure to rerun the gapps connection creation code block [Return to Top](Table-of-Contents) --- 3.4. Subscription for Class-based App Core Subscription for Class-Based App CoreIf the application core was created as a class definition as shown in [App Core as Class Definition](App-Core-as-a-Class-Definition), then the function name is passed to the `.subscribe(output_topic, object)` method of the GridAPPS-D connection object. After defining the class for the application core as shown above, we create another object that will be passed to the subscription method. The required parameters for this object are the same as those defined for the `__init__()` method of the app core class, typically the Simulation ID, GridAPPS-D connection object, dictionary of measurements needed by the app core, and any user-defined objects.`class_obj = AppCoreClass(simulation_id, gapps_obj, meas_obj, your_obj)`
###Code
demo_obj = DemoAppCoreClass(viz_simulation_id, gapps, measurements_obj)
gapps.subscribe(subscribe_topic, demo_obj)
###Output
_____no_output_____
###Markdown
If we wish to subscribe to an additional topic (such as the Simulation Logs, a side communication channel between two different applications, or a communication with a particular service), we can define an additional method in the class (such as my_custom_method_1 in the [example class definition](App-Core-as-a-Class-Definition) above) and then pass it to to the `.subscribe(topic, object.method)` method associated with the GridAPPS-D connection object:`gapps.subscribe(other_topic, demo_obj.my_custom_method_1)` [Return to Top](Table-of-Contents) --- 4. Subscribing to Parallel Simulations Subscribing to Parallel SimulationsParallel simulations started using the Simulation API (as shown in [Starting a Simulation](../api_usage/3.5-Creating-Running-Simulation-API.ipynbStarting-the-Simulation)) and the `Simulation` library in GridAPPSD-Python do not need to use the `gapps.subscribe` method.Instead, the GridAPPSD-Python library contains several shortcut functions that can be used. These methods currently cannot interact with a simulation started from the Viz. This functionality will be added in a future release. The code block below shows how a parallel simulation can be started using a simulation start message stored in a JSON file. The simulation is started using the `.start_simulation()` method. __Note on Jupyter Notebook environment:__ In the examples below, the Jupyter Notebook environment does not update definitions of the subscription object or function definitions. As a result, it is necessary to restart the notebook kernel if you ran the previous code blocks for the `.subscribe` method.
###Code
import json, os
from gridappsd import GridAPPSD
from gridappsd.simulation import Simulation
# Connect to GridAPPS-D Platform
os.environ['GRIDAPPSD_USER'] = 'tutorial_user'
os.environ['GRIDAPPSD_PASSWORD'] = '12345!'
gapps = GridAPPSD()
assert gapps.connected
model_mrid = "_C1C3E687-6FFD-C753-582B-632A27E28507"
run123_config = json.load(open("Run123NodeFileSimAPI.json")) # Pull simulation config from saved file
simulation_obj = Simulation(gapps, run123_config) # Create Simulation object
simulation_obj.start_simulation() # Start Simulation
print("Successfully started simulation with simulation_id: ", simulation_obj.simulation_id)
simulation_id = simulation_obj.simulation_id
###Output
_____no_output_____
###Markdown
The Simulation library provides four methods that can be used to define how the platform interacts with the simulation:* `.add_ontimestep_callback(myfunction1)` -- Run the desired function on each timestep* `.add_onmesurement_callback(myfunction2)` -- Run the desired function when a measurement is received. * `.add_oncomplete_callback(myfunction3)` -- Run the desired function when simulation is finished* `.add_onstart_callback(myfunction4)` -- Run desired function when simulation is started__Note: method name `.add_onmesurement_callback` is misspelled in the library definition!!__ Note that the measurement callback method returns just the measurements and timestamps without any of the message formatting used in the messages received by using the `gapps.subscribe(output_topic, object)` approach. The python dictionary returned by the GridAPPS-D Simulation output to the `.add_onmesurement_callback()` method is always named `measurements` and uses the following key-value pairs format:```{ '_pnv_meas_mrid_1': {'angle': number, 'magnitude': number, 'measurement_mrid': '_pnv_meas_mrid_1'}, '_va_meas_mrid_2': { 'angle': number, 'magnitude': number, 'measurement_mrid': '_va_meas_mrid_2'}, '_pos_meas_mrid_3': {'measurement_mrid': '_pos_meas_mrid_3', 'value': 1}, . . . '_pnv_meas_mrid_n': {'angle': number, 'magnitude': number, 'measurement_mrid': '_pnv_meas_mrid_1'}}``` To use use these methods, we define a set of functions that determine the behavior of the application for each of the four types of callbacks listed above. These functions are similar to those defined for the function-based app core algorithm. ```def my_onstart_func(sim): Do something when the simulation starts Do something else when the sim startssimulation_obj.add_onstart_callback(my_onstart_func)``` ```def my_onmeas_func(sim, timestamp, measurements): Do something when app receives a measurement Insert your custom app behavior here if measurements[object_mrid] == foo: bar = my_optimization_result simulation_obj.add_onmesurement_callback(my_onmeas_func)``` ```def my_oncomplete_func(sim): Do something when simulation is complete example: delete all variables, close files simulation_obj.add_oncomplete_callback(my_oncomplete_func)``` The code block below shows how the same app core algorithm can be used for a parallel simulation using the `.add_onmesurement_callback()` method:
###Code
# Recreate global variable of measurement mRIDs needed by app
from gridappsd import topics as t
# Create query message to obtain measurement mRIDs for all switches
message = {
"modelId": model_mrid,
"requestType": "QUERY_OBJECT_MEASUREMENTS",
"resultFormat": "JSON",
"objectType": "LoadBreakSwitch"
}
# Pass query message to PowerGrid Models API
response_obj = gapps.get_response(t.REQUEST_POWERGRID_DATA, message)
measurements_obj = response_obj["data"]
# Switch position measurements (Pos)
global Pos_obj
Pos_obj = [k for k in measurements_obj if k['type'] == 'Pos']
def demo_onmeas_func(sim, timestamp, measurements):
open_switches = []
for index in Pos_obj:
if index["measid"] in measurements:
mrid = index["measid"]
power = measurements[mrid]
if power["value"] == 0:
open_switches.append(index["eqname"])
print("............")
print("Number of open switches at time", timestamp, ' is ', len(set(open_switches)))
simulation_obj.add_onmesurement_callback(demo_onmeas_func)
###Output
_____no_output_____
###Markdown
[Return to Top](Table-of-Contents) --- 5. Publishing to Simulation Input Publishing Commands to Simulation InputThe next portion of a GridAPPS-D App is publishing equipment control commands based on the optimization results or objectives of the app algorithm. Depending on the preference of the developer, this portion can be a separate function definition, or included as part of the main class definition as part of the [App Core as a Class Definition](App-Core-as-a-Class-Definition) described earlier. 5.1. Information Flow Equipment Command Information FlowThe figure below outlines information flow involved in publishing equipment commands to the simulation input. Unlike the various queries to the databases in the app sections earlier, equipment control commands are passed to the GridAPPS-D API using the `gapps.send(topic, message)` method. No response is expected from the GridAPPS-D platform. If the application desires to verify that the equipment control command was received and implemented, it needs to do so by 1) checking for changes in the associated measurements at the next timestep and/or 2) querying the Timeseries Database for historical simulation data associated with the equipment control command.  __Application sends difference message to GridAPPS-D Platform__First, the application creates a difference message containing the current and desired future control point / state of the particular piece of power system equipment to be controlled. The difference message is a JSON string or equivalant Python dictionary object. The syntax of a difference message is explained in detail below in [Format of Difference Message](Format-of-a-Difference-Message).The application then passes the query through the Simulation API to the GridAPPS-D Platform, which publishes it on the topic channel for the particular simulation on the GOSS Message Bus. If the app is authenticated and authorized to control equipment, the difference message is delivered to the Simulation Manager. The Simulation Manager then passes the command to the simulation through the Co-Simulation Bridge (either FNCS or HELICS).__No response from GridAPPS-D Platform back to Application__The GridAPPS-D Platform does not provide any response back to the application after processing the difference message and implementing the new equipment control setpoint. [Return to Top](Table-of-Contents) --- 5.2. API Communication Channel Simulation Input API ChannelThis is a dynamic `/topic/` communication channel that is best implemented by importing the GriAPPSD-Python library function for generating the correct topic. * `from gridappsd.topics import simulation_input_topic`* `input_topic = simulation_input_topic(simulation_id)`
###Code
from gridappsd.topics import simulation_input_topic
input_topic = simulation_input_topic(viz_simulation_id)
###Output
_____no_output_____
###Markdown
[Return to Top](Table-of-Contents) --- 5.3. Obtaining Equipment Control mRIDs Equipment Control mRIDsThe mRIDs for controlling equipment are generally the same as those obtained using the `QUERY_OBJECT_DICT` key with the PowerGrid Models API, which was covered in [Query for Object Dicionary](../api_usage/3.3-Using-the-PowerGrid-Models-API.ipynbQuery-for-Object-Dictionary).However, the control attributes for each class of equipment in CIM use a different naming convention than those for the object types. Below is a list of `"objectType"` used to query for mRIDs and the associated control attribute used in a difference message for each category of power system equipment:* __Switches__ * CIM Class Key: `"objectType": "LoadBreakSwitch"` * Control Attribute: `"attribute": "Switch.open"` * Values: `1` is open, `0` is closed* __Capacitor Banks:__ * CIM Class Key: `"objectType": "LinearShuntCompensator"` * Control Attribute: `"attribute": "ShuntCompensator.sections"` * Values: `0` is off/open, `1` is on/closed * __Inverter-based DERs:__ * CIM Class Key: `"objectType": "PowerElectronicsConnection"` * Control Attribute: `"attribute": "PowerElectronicsConnection.p"` * Control Attribute: `"attribute": "PowerElectronicsConnection.q"` * Values: number in Watts or VArs (not kW) * __Synchronous Rotating (diesel) DGs:__ * CIM Class Key: `"objectType": "SynchronousMachine"` * Control Attribute: `"attribute": "RotatingMachine.p"` * Control Attribute: `"attribute": "RotatingMachine.q"` * Values: number in Watts or VArs (not kW) * __Regulating Transformer Tap:__ * CIM Class Key: `"objectType": "RatioTapChanger"` * Control Attribute: `"attribute": "TapChanger.step"` * Values: integer value for tap step __The query for RatioTapChanger is not supported in the PowerGrid Models API at the current time. A custom SPARQL query needs to be done using the sample query in [CIMHub Sample Queries](https://github.com/GRIDAPPSD/CIMHub/blob/master/queries.txt)__The example below shows a query to obtain the correct mRID for switch SW2 in the IEEE 123 node model:
###Code
from gridappsd import topics as t
message = {
"modelId": model_mrid,
"requestType": "QUERY_OBJECT_DICT",
"resultFormat": "JSON",
"objectType": "LoadBreakSwitch"
}
response_obj = gapps.get_response(t.REQUEST_POWERGRID_DATA, message)
switch_dict = response_obj["data"]
# Filter to get mRID for switch SW2:
for index in switch_dict:
if index["IdentifiedObject.name"] == 'sw2':
sw_mrid = index["IdentifiedObject.mRID"]
###Output
_____no_output_____
###Markdown
[Return to Top](Table-of-Contents) --- 5.4. Format of a Difference Message Format of a Difference MessageThe general format for a difference message is a python dictionary or equivalent JSON string that specifies the reverse difference and the forward difference, in compliance with the CIM standard:The __reverse difference__ is the current status / value associated with the control attribute. It is a formatted as a list of dictionary constructs, with each dictionary specifying the equipment mRID associated with the CIM class keys above, the control attribute, and the current value of that control attribute. The list can contain reverse differences for multiple pieces of equipment.The __forward difference__ is the desired new status / value associated with the control attribute. It is a formatted as a list of dictionary constructs, with each dictionary specifying the equipment mRID associated with the CIM class keys above, the control attribute, and the current value of that control attribute. The list can contain foward differences for multiple pieces of equipment.```message = { "command": "update", "input": { "simulation_id": "simulation id as string", "message": { "timestamp": epoch time number, "difference_mrid": "optional unique mRID for command logs", "reverse_differences": [{ "object": "first equipment mRID", "attribute": "control attribute", "value": current value }, { "object": "second equipment mRID", "attribute": "control attribute", "value": current value } ], "forward_differences": [{ "object": "first equipment mRID", "attribute": "control attribute", "value": new value }, { "object": "second equipment mRID", "attribute": "control attribute", "value": new value } ] } }}```Note: The GridAPPS-D platform does not validate whether `"reverse_differences":` has the correct equipment control values for the current time. It is used just for compliance with the CIM standard. [Return to Top](Table-of-Contents) --- 5.5. Using GridAPPSD-Python `DifferenceBuilder` Using GridAPPSD-Python DifferenceBuilder The `DifferenceBuilder` class is a GridAPPSD-Python tool that can be used to automatically build the difference message with correct formatting. First, import DifferenceBuilder from the GridAPPSD-Python Library and create an object that will be used to create the desired difference messages.
###Code
from gridappsd import DifferenceBuilder
my_diff_build = DifferenceBuilder(viz_simulation_id)
###Output
_____no_output_____
###Markdown
We then use two methods associated with the DifferenceBuilder object:* `.add_difference(self, object_mrid, control_attribute, new_value, old_value)` -- Generates a correctly formatted difference message.* `.get_message()` -- Saves the message as a python dictionary that can be published using `gapps.send(topic, message)`
###Code
my_diff_build.add_difference(sw_mrid, "Switch.open", 1, 0) # Open switch given by sw_mrid
message = my_diff_build.get_message()
###Output
_____no_output_____
###Markdown
The difference message is then published to the GOSS Message Bus and the Simulation API using the `.send()` method associated with the GridAPPS-D connection object.
###Code
gapps.send(input_topic, message)
###Output
_____no_output_____
|
poetry_package_acoustics/groovin_pyladies/notebooks/WorkshopTeaser.ipynb
|
###Markdown
Build Your Own Audio Processing Package Using [Poetry](https://python-poetry.org/)This notebook will show what type of functionality we will build in our very own package. As you read along this notebook, explore the resources provided and think about what kind of functionality you would like to add to the package. Package Functionality Covered in the Workshop 1. Load Audio via...- [Scipy](https://docs.scipy.org/doc/scipy/reference/)- [Librosa](https://librosa.org/doc/latest/index.html)- [Parselmouth](https://parselmouth.readthedocs.io/en/stable/)- [Torchaudio](https://pytorch.org/audio/stable/index.html)Each of these audio processing libraries offer a plethera of tools used in development, research, and/or deep learning. 2. Repeat Audio `x` TimesIt is just fun to play with audio. 3. Change Speaker Pitch[Praat](https://www.fon.hum.uva.nl/praat/) is beloved by many speech researches across the globe. Parselmouth allows us to use Praat in a pythonic way. For reliable measurements of speech related features and speech manipulation, I would explore this library further. 4. Visualize AudioTo see what our audio looks like, in both the [time domain](https://en.wikipedia.org/wiki/Discrete_time_and_continuous_time) and [frequency domain](https://en.wikipedia.org/wiki/Frequency_domain), we will use [Matplotlib](https://matplotlib.org/) in our package. This should give us an idea of what all neural networks need to learn from audio and perhaps why audio is quite challenging for machines to handle. I will use [this paper](https://ieeexplore.ieee.org/stamp/stamp.jsp?arnumber=9034048) as an example to explore commonly used audio features. Note: many advancements are being made in the field of acoustic neural networks, such as using [transformer models](https://arxiv.org/pdf/2006.11477.pdf); however this exceeds the scope of this workshop.
###Code
import groovin_pyladies as groovin
from IPython.display import Audio, display
speech_path = "../../data/audio/python.wav"
alarm_path = "../../data/audio/alarm.wav"
###Output
_____no_output_____
###Markdown
Load Audio
###Code
samples, sampling_rate = groovin.load_audio(
alarm_path,
package = "librosa"
)
###Output
_____no_output_____
###Markdown
Warning: check your volume!!
###Code
Audio(samples, rate=sampling_rate)
###Output
_____no_output_____
###Markdown
Repeat Alarm
###Code
samples_repeated = groovin.manipulate.repeat(
samples,
repeat_n_times=2
)
###Output
_____no_output_____
###Markdown
Audio(samples_repeated, rate=sampling_rate)
###Code
import numpy as np
samples_mirrored = np.concatenate([samples, np.flip(samples)])
samples_mirrored_repeated = groovin.manipulate.repeat(
samples_mirrored,
repeat_n_times=2
)
###Output
_____no_output_____
###Markdown
Audio(samples_mirrored_repeated, rate=sampling_rate)
###Code
# Manipulate Speaker Pitch
###Output
_____no_output_____
###Markdown
speech_object = groovin.load_audio( speech_path, package = "praat") type(speech_object)
###Code
Audio(speech_object.values, rate=speech_object.sampling_frequency)
###Output
_____no_output_____
###Markdown
speech_low = groovin.manipulate.pitch( speech_object, factor = 3/4,)
###Code
Audio(speech_low.values, rate=speech_object.sampling_frequency)
###Output
_____no_output_____
###Markdown
speech_high = groovin.manipulate.pitch( speech_object, factor = 3/2,)
###Code
Audio(speech_high.values, rate=speech_object.sampling_frequency)
###Output
_____no_output_____
###Markdown
Visualize Audio We will use [this paper](https://ieeexplore.ieee.org/stamp/stamp.jsp?arnumber=9034048) (pages 54667-54668) as a reference for our visuals.
###Code
from IPython.display import Image
Image(filename='../../data/example_features_librosa.png')
Image(filename='../../data/example_features_librosa_graph.png')
###Output
_____no_output_____
###Markdown
Assign variables from paper
###Code
sampling_rate_new = 16000
frame_length_ms = 16
hop_length_ms = frame_length_ms // 4
n_mel_filters = 32
int(sampling_rate_new * frame_length_ms * 0.001)
int(sampling_rate_new * hop_length_ms * 0.001)
###Output
_____no_output_____
###Markdown
Adjust our data accordinglyI have not included resampling into our package.
###Code
import librosa
speech_low_resampled = librosa.resample(
speech_low.values[0],
speech_low.sampling_frequency,
sampling_rate_new
)
speech_high_resampled = librosa.resample(
speech_high.values[0],
speech_high.sampling_frequency,
sampling_rate_new
)
###Output
_____no_output_____
###Markdown
Look at the lower speech:We can fiddle around with the settings and see how they influence the figure.
###Code
fbank_low = groovin.mel_specgram(
speech_low_resampled,
sr = sampling_rate_new,
n_mels = n_mel_filters,
win_length = int(sampling_rate_new * frame_length_ms * 0.001),
hop_length = int(sampling_rate_new * hop_length_ms * 0.001),
)
groovin.plot.visualize_fbank(fbank_low)
###Output
_____no_output_____
###Markdown
Look at the higher speech:Can you see a difference?
###Code
fbank_high = groovin.mel_specgram(
speech_high_resampled,
sr = sampling_rate_new,
n_mels = n_mel_filters,
win_length = int(sampling_rate_new * frame_length_ms * 0.001),
hop_length = int(sampling_rate_new * hop_length_ms * 0.001),
)
groovin.plot.visualize_fbank(fbank_high)
###Output
_____no_output_____
###Markdown
Look at Delta FeaturesFor more info on what the delta features are for, see this [post](https://wiki.aalto.fi/display/ITSP/Deltas+and+Delta-deltas).Note: I have not incorporated this into our package.
###Code
fbank_low_delta = librosa.feature.delta(
fbank_low,
order=1,
)
groovin.plot.visualize_fbank(fbank_low_delta, convert_to_decibel=False)
fbank_low_delta_delta = librosa.feature.delta(
fbank_low,
order=2,
)
groovin.plot.visualize_fbank(fbank_low_delta_delta, convert_to_decibel=False)
###Output
_____no_output_____
###Markdown
We can do the same with Torchaudio as well!
###Code
speech_torch, sr_torch = groovin.load_audio(
speech_path,
package = "torchaudio"
)
type(speech_torch)
speech_torch
###Output
_____no_output_____
###Markdown
For more on tensors, see this Medium series on deep learning [tensors](https://medium.com/secure-and-private-ai-writing-challenge/introduction-to-tensors-1-de7dded35fea).
###Code
sr_torch
###Output
_____no_output_____
###Markdown
To match the variables of the paper, we have to resample. I did not include resampling functionality in our package.
###Code
import torchaudio
resample_torch = torchaudio.transforms.Resample(
orig_freq=sr_torch,new_freq=sampling_rate_new
)
speech_torch_resampled = resample_torch(speech_torch)
fbank_low_torch = groovin.mel_specgram(
speech_torch_resampled,
sample_rate = sampling_rate_new,
n_mels = n_mel_filters,
win_length = int(sampling_rate_new * frame_length_ms * 0.001),
hop_length = int(sampling_rate_new * hop_length_ms * 0.001),
)
groovin.plot.visualize_fbank(fbank_low_torch[0])
###Output
_____no_output_____
|
Class_16_TestP2a.ipynb
|
###Markdown
Universidade Federal do Rio Grande do Sul (UFRGS) Programa de Pós-Graduação em Engenharia Civil (PPGEC) PEC00025: Introduction to Vibration Theory Class 16 - Test P2: multiple degrees of freedom and continuous systems[P2:2019](P2_2019) - [Question 1](P2_2019_1), [Question 2](P2_2019_2), [Question 3](P2_2019_3), [Question 4](P2_2019_4).---_Prof. Marcelo M. Rocha, Dr.techn._ [(ORCID)](https://orcid.org/0000-0001-5640-1020) _Porto Alegre, RS, Brazil_
###Code
# Importing Python modules required for this notebook
# (this cell must be executed with "shift+enter" before any other Python cell)
import numpy as np
import matplotlib.pyplot as plt
import scipy.linalg as sc
###Output
_____no_output_____
###Markdown
P2:2019 _Note: this test is to be solved with the aid of a scientific calculator, which must be able to solve eigenproblems,linear systems, and integrals. The total available time for solving the test is 2h (two hours). The student is allowedto prepare am A4 sheet of paper (two sides) with informations to be consulted during the test._ Question 1 A structural system is modelled as a discrete two d.o.f. system, as shown in the figure. Each column has flexural rigidity $EI = 500{\rm kNm^2}$, length $L = 4{\rm m}$, and are assumed to have no relevant mass. The floor beams are assumed to be perfectly stiff and to have total (lumped) mass $m = 4{\rm ton}$ each. The system is assumed to present a viscous damping with ratio of critical $\zeta = 0.01$ in all vibration modes. 1. Define the stiffness, the mass, and the damping system matrices (1 pts). 2. Determine and sketch the two natural vibration modes, indicating the associated vibration frequencies (2 pts). **Answer:** The stiffness and mass matrices are:
###Code
EI = 500000. # single column flexural rigidity (Nm^2)
m = 4000. # single floor mass (kg)
L = 4 # column length (m)
k = 12*EI/L**3 # single column stiffness
KG = np.array([[ 2*k, -2*k],
[-2*k, 4*k]]) # global stiffness matrix
MG = np.array([[ m, 0],
[ 0, m]]) # global mass matrix
print('Global stiffness matrix:\n\n', KG)
print('\nGlobal mass matrix:\n\n', MG)
###Output
Global stiffness matrix:
[[ 187500. -187500.]
[-187500. 375000.]]
Global mass matrix:
[[4000. 0.]
[ 0. 4000.]]
###Markdown
To specify the damping matrix, we must first calculate the vibration modes and frequencies.
###Code
# Uses scipy to solve the standard eigenvalue problem
w2, Phi = sc.eig(KG, MG)
# Ensure ascending order of eigenvalues
iw = w2.argsort()
w2 = w2[iw]
Phi = Phi[:,iw]
# Eigenvalues to vibration frequencies
wk = np.sqrt(np.real(w2))
fk = wk/2/np.pi
plt.figure(1, figsize=(8,6), clear=True)
x = np.arange(0,12,4)
for k in range(2):
pk = np.zeros(3)
pk[1:] = Phi[::-1,k]
pk /= np.max(np.abs(pk)) # adjust scale for unity amplitude
plt.subplot(1,2,k+1)
plt.plot(pk, x, 'bo')
plt.plot(pk, x)
plt.xlim(-1.5, 1.5); plt.ylabel(str(k+1));
plt.ylim( 0.0, 10.);
plt.title('fk = {0:4.2f}Hz'.format(fk[k]));
plt.grid(True)
###Output
_____no_output_____
###Markdown
And now we can calculate the coefficients that multiply the stiffness and mass matricesto build a Rayleigh damping matrix that is also orthogonalized by the eigenvectors:
###Code
zeta = np.array([0.01, 0.01]) # damping for two modes i and j
A = np.array([[1/wk[0], wk[0]], [1/wk[1], wk[1]]])/2
alpha = np.linalg.solve(A, zeta)
CG = alpha[0]*MG + alpha[1]*KG # Rayleigh viscous damping matrix
print('Mass matrix coefficient a0: {0:6.5f}'.format(alpha[0]))
print('Stiffness matrix coefficient a1: {0:6.5f}'.format(alpha[1]))
print('\nRayleigh damping matrix original:\n\n', CG)
print('\nRayleigh damping matrix orthogonalized:\n\n', np.dot(Phi.T, np.dot(CG, Phi)))
###Output
Mass matrix coefficient a0: 0.06124
Stiffness matrix coefficient a1: 0.00131
Rayleigh damping matrix original:
[[ 489.89794856 -244.94897428]
[-244.94897428 734.84692283]]
Rayleigh damping matrix orthogonalized:
[[338.51115694 0. ]
[ 0. 886.23371445]]
###Markdown
Question 2 The system is now subjected to an initial kinematic condition, which consists of an imposed displacement on the lower floor, $u_{20} = 1{\rm cm}$, only, and it is then released to vibrate. Accounting for the two vibration modes, calculate the peak displacement and the peak acceleration at the system upper floor caused by this initial condition (2 pts). **Answer:** For the modal superposition we must firstly calculate the modal massesand the modal stiffnesses:
###Code
Kk = np.diag(np.dot(Phi.T, np.dot(KG, Phi)))
Mk = np.diag(np.dot(Phi.T, np.dot(MG, Phi)))
print('Modal masses: [{0:6.0f} {1:6.0f}]'.format(*Mk))
print('Modal stiffnesses: [{0:6.0f} {1:6.0f}]'.format(*Kk))
###Output
Modal masses: [ 4000 4000]
Modal stiffnesses: [ 71619 490881]
###Markdown
The initial condition is of displacement type (no initial velocity), what implies acosine type response. Recalling that $\Phi$ is a orthogonal matrix, it means that its transpose is equal to its inverse:\begin{align*}\vec{u}(t) &= {\mathbf \Phi} \; \vec{u}_k(t) \\\vec{u}_k(t) &= {\mathbf \Phi}^{\intercal} \; \vec{u}(t)\end{align*}where $\vec{u}(t)$ is the _nodal_ response and $\vec{u}_k(t)$ is the _modal_ response.The initial modal displacements are simply given by:
###Code
u0 = np.array([0.00, 0.01]) # initial displacements in nodal coordinates
u0k = np.dot(Phi.T, u0) # initial displacements in modal coordinates
print('Initial modal displacement at mode 1: {0:8.6f}'.format(u0k[0]))
print('Initial modal displacement at mode 2: {0:8.6f}'.format(u0k[1]), '\n')
# The most general way (as shown in classroom), considering phase is pi/2
u01 = np.dot(Phi[:,0], np.dot(MG, u0))/(np.sin(np.pi/2)*Mk[0])
u02 = np.dot(Phi[:,1], np.dot(MG, u0))/(np.sin(np.pi/2)*Mk[1])
print('Initial modal displacement at mode 1: {0:8.6f}'.format(u01))
print('Initial modal displacement at mode 2: {0:8.6f}'.format(u02))
###Output
Initial modal displacement at mode 1: 0.005257
Initial modal displacement at mode 2: 0.008507
Initial modal displacement at mode 1: 0.005257
Initial modal displacement at mode 2: 0.008507
###Markdown
The total response is a superposition of modal responses, which are cosine functions withthe respective frequencies and amplitudes:
###Code
T = 10
N = 200
t = np.linspace(0, T, N) # time domain
uk = np.array([u0k[0]*np.cos(wk[0]*t),
u0k[1]*np.cos(wk[1]*t)]) # modal responses
u = np.dot(Phi, uk)*100 # total responses (cm)
plt.figure(2, figsize=(12, 4), clear=True)
plt.plot(t, u[0,:], 'b', t, u[1,:], 'r')
plt.xlim( 0, T); plt.xlabel('time (s)')
plt.ylim(-2, 2); plt.ylabel('u(t) (cm)')
plt.legend(('upper','lower'))
plt.grid(True)
###Output
_____no_output_____
###Markdown
The accelerations are obtained from the twofold derivative of the cosine sum:
###Code
ak = np.array([-u0k[0]*wk[0]*wk[0]*np.cos(wk[0]*t),
-u0k[1]*wk[1]*wk[1]*np.cos(wk[1]*t)]) # modal accelerations
a = np.dot(Phi, ak)/9.81 # nodal accelerations (G)
plt.figure(3, figsize=(12, 4), clear=True)
plt.plot(t, a[0,:], 'b', t, a[1,:], 'r')
plt.xlim( 0, T); plt.xlabel('time (s)')
plt.ylim(-0.2, 0.2); plt.ylabel('a(t) (G)')
plt.legend(('upper','lower'))
plt.grid(True)
###Output
_____no_output_____
###Markdown
Finnaly, answering the question, the peak displacement and acceleration amplitudes inthe upper floor are:
###Code
print('Peak upper displacement: {0:5.3f}cm'.format(u[0,:].max()))
print('Peak upper acceleration: {0:5.3f}G '.format(a[0,:].max()))
###Output
Peak upper displacement: 0.876cm
Peak upper acceleration: 0.064G
###Markdown
It can be seen that, as expected, the second mode dominate the structural response. Question 3 The cantilever beam shown in the figure has a constant flexural stiffness $EI = 1000{\rm kNm^2}$ and mass per unit length $\mu = 200{\rm kg/m}$. 1. Propose a function that resembles the first vibration mode. Calculate the associated potential elastic energy $V$ and the reference kinetic energy, $T_{\rm ref}$. With these energies, estimate the natural vibration frequency for the first mode using the Rayleigh quocient (2 pts). 2. Calculate the modal mass and the modal stiffness and then use these parameters to estimate the static displacement at the cantilever tips, caused by a point load $W = 10{\rm kN}$ placed at this same position (1 pts). **Answer:** We will try and compare two different solutions: a parabolic and a sinusoidal functions.They are:$$ \varphi_1(x) = \frac{1}{27} \;(x - 3)(x - 9) $$and$$ \varphi_2(x) = 1 - \sqrt{2} \, \sin \left( \frac{\pi x}{12} \right) $$Both tentative solutions respect the kinetic condition of zero displacement at supports(located ate coordinates $x = 3$m and $x = 9$m.The script below shows a comparison plot:
###Code
EI = 1000000. # flexural stiffness
mu = 200. # mass per unit length
L = 12. # total length
N = 200 # number of segments
X = np.linspace(0, L, N) # length discretization
ph1 = lambda x: (x - 3)*(x - 9)/27 # first solution
ph2 = lambda x: 1 - np.sqrt(2)*np.sin(np.pi*(x/12)) # second solution
plt.figure(4, figsize=(12, 4), clear=True)
plt.plot(X, ph1(X), 'b', X, ph2(X), 'r')
plt.plot([3, 9], [0 , 0], 'bo')
plt.xlim( 0, L); plt.xlabel('x (m)')
plt.ylim(-2, 2); plt.ylabel('phi (nondim)')
plt.legend(('phi_1','phi_2'))
plt.grid(True)
###Output
_____no_output_____
###Markdown
The sine function has an important feature, which is zero curvature at cantilever tips wherebending moments must be zero. The parabolic function is the simplest, but presents constant curvature along all beam length.The rotations are calculated as:$$ \phi_1^{\prime}(x) = \frac{1}{27} (2x - 12) $$and:$$ \phi_2^{\prime}(x) = -\frac{\pi \sqrt{2}}{12} \; \cos \left( \frac{\pi x}{12} \right) $$while the curvatures are given by:$$ \phi_1^{\prime\prime}(x) = \frac{2}{27} $$and:$$ \phi_2^{\prime\prime}(x) = \frac{\pi^2 \sqrt{2}}{144} \; \sin \left( \frac{\pi x}{12} \right) $$The script below compares the curvatures for each solution:
###Code
ph1xx = lambda x: (2/27)*x**0 # first solution
ph2xx = lambda x: (np.pi*np.pi*np.sqrt(2)/144)*np.sin(np.pi*x/12) # second solution
plt.figure(5, figsize=(12, 4), clear=True)
plt.plot(X, ph1xx(X), 'b', X, ph2xx(X), 'r')
plt.xlim( 0, L); plt.xlabel('x (m)')
plt.ylim(-0.05, 0.15); plt.ylabel('phi_xx (1/m^2)')
plt.legend(('phi_1','phi_2'))
plt.grid(True)
###Output
_____no_output_____
###Markdown
The curvatures are quite close at the center, but it is overestimated by the parabolic functionat the cantilever tips.The potential elastic and the reference kinetic energy finally are:
###Code
dx = L/N
V1 = EI*np.trapz(ph1xx(X)*ph1xx(X), dx=dx)/2
V2 = EI*np.trapz(ph2xx(X)*ph2xx(X), dx=dx)/2
T1 = mu*np.trapz( ph1(X)*ph1(X), dx=dx)/2
T2 = mu*np.trapz( ph2(X)*ph2(X), dx=dx)/2
print('Potential elastic energy for solution 1: {0:5.1f}J'.format(V1))
print('Potential elastic energy for solution 2: {0:5.1f}J\n'.format(V2))
print('Reference kinetic energy for solution 1: {0:5.1f}J'.format(T1))
print('Reference kinetic energy for solution 2: {0:5.1f}J'.format(T2))
###Output
Potential elastic energy for solution 1: 32757.2J
Potential elastic energy for solution 2: 28044.6J
Reference kinetic energy for solution 1: 203.5J
Reference kinetic energy for solution 2: 238.1J
###Markdown
And the natural vibration frequencies estimated with Rayleigh quotient are:
###Code
wn1 = np.sqrt(V1/T1)
wn2 = np.sqrt(V2/T2)
fn1 = wn1/2/np.pi
fn2 = wn2/2/np.pi
print('Natural vibration frequency for solution 1: {0:5.2f}Hz'.format(fn1))
print('Natural vibration frequency for solution 2: {0:5.2f}Hz'.format(fn2))
###Output
Natural vibration frequency for solution 1: 2.02Hz
Natural vibration frequency for solution 2: 1.73Hz
###Markdown
If one recalls that the true vibration mode minimizes the Rayleigh quotient, the lowest valueobtained with the sinusoidal function is likely to be closer to the exact solution.The relative error between both tentative functions is approximately 17% and thecorrect natural frequency must be a little below 1.73Hz.Now, we will proceed with the calculation of modal mass and modal stiffness:
###Code
Mk1 = mu*np.trapz(ph1(X)*ph1(X), dx=dx) # modal mass and ...
Kk1 = Mk1*wn1**2 # ... stiffness for solution 1
Mk2 = mu*np.trapz(ph2(X)*ph2(X), dx=dx) # modal mass and ...
Kk2 = Mk2*wn2**2 # ... stiffness for solution 2
###Output
_____no_output_____
###Markdown
For static analysis, the modal displacement is obtained from modal force divided by modal stiffness:
###Code
W = -10000. # point load (downwards)
Fk1 = W*ph1(6) # modal (static) force
Fk2 = W*ph2(6)
uk1 = Fk1/Kk1 # modal displacement
uk2 = Fk2/Kk2
u1 = uk1*ph1(12) # displacement at cantilever tip
u2 = uk2*ph2(12)
print('Static displacement of cantilever tip for solution 1: {0:5.2f}cm'.format(u1*100))
print('Static displacement of cantilever tip for solution 2: {0:5.2f}cm'.format(u2*100))
###Output
Static displacement of cantilever tip for solution 1: 5.09cm
Static displacement of cantilever tip for solution 2: 7.38cm
###Markdown
The error in the displacement at cantilever tip for the two solutions is quite high, over 40%, for the two tentative functions diverge noticeably in that position (we recommend this result to checked with the Ftool software).A comparison of displacement solutions for the whole beam is shown below:
###Code
plt.figure(6, figsize=(12, 4), clear=True)
plt.plot(X, 100*uk1*ph1(X), 'b', X, 100*uk2*ph2(X), 'r')
plt.plot([3, 9], [0 , 0], 'bo')
plt.xlim( 0, L); plt.xlabel('x (m)')
plt.ylim(-6, 12); plt.ylabel('u (cm)')
plt.legend(('phi_1','phi_2'))
plt.grid(True)
###Output
_____no_output_____
###Markdown
Question 4 The same point load from previous question is now applied suddenly from zero to its final magnitude, what causes a dynamic amplification on the beam displacements. Estimate the peak displacement and the peak acceleration at the cantilever tip (2 pts).**Answer:** The solution for some impulsive loading is well known to be the static solutionmultiplied by a dynamic amplification factor. In the case of a step load (Heaviside'sfunction) this amplification factor is 2. Hence:
###Code
print('Dynamic displacement of cantilever tip for solution 1: {0:5.2f}cm'.format(2*u1*100))
print('Dynamic displacement of cantilever tip for solution 2: {0:5.2f}cm'.format(2*u2*100))
###Output
Dynamic displacement of cantilever tip for solution 1: 10.18cm
Dynamic displacement of cantilever tip for solution 2: 14.77cm
###Markdown
The peak accelerations are:
###Code
ak1 = uk1*wn1**2
ak2 = uk2*wn2**2
a1 = ak1*ph1(12)
a2 = ak2*ph2(12)
print('Acceleration at cantilever tip for solution 1: {0:5.3f}G'.format(a1/9.81))
print('Acceleration at cantilever tip for solution 2: {0:5.3f}G'.format(a2/9.81))
###Output
Acceleration at cantilever tip for solution 1: 0.835G
Acceleration at cantilever tip for solution 2: 0.887G
|
Test (Ensemble & hard-label).ipynb
|
###Markdown
Atopy Grading - Test (Ensemble & hard-label)
###Code
%matplotlib inline
import matplotlib.pyplot as plt
import numpy as np
import os
import pandas as pd
import time
import h5py
import math
from random import randint, choice
import glob
import cv2
from scipy.ndimage import rotate
from sklearn.metrics import roc_auc_score
from sklearn.model_selection import train_test_split
from sklearn.utils import class_weight
import keras
from keras.models import Model
from keras.layers import Dense, Activation, Input, Concatenate, Dropout, BatchNormalization, ZeroPadding2D
import tensorflow as tf
from keras import backend as K
print(tf.__version__)
print(keras.__version__)
K.tensorflow_backend._get_available_gpus()
###Output
Using TensorFlow backend.
###Markdown
Hyperparameter
###Code
##################################################
Evaluator_Test = 'hard-label' ##
atopy_type = '7.IGA' ##
# 1.Erythema
# 2.Edema_Papule
# 3.Excoriation
# 4.Lichenification
# 5.Oozing_Crusting
# 6.Dryness
# 7.IGA
normalize = 'on' ##
augment = 'on' ##
input_shape = [400,400,3]
option = '_Aug_'
model_names = ['InceptionResNetV2']
##################################################
test_path = '../Dataset/Image_re/Test/' + Evaluator_Test + '/' + atopy_type + '/'
ext_path = '../Dataset/Image_re/ExtraVal/' + Evaluator_Test + '/' + atopy_type + '/'
save_name1 = model_names[0] + '_' + 'A' + '_' + atopy_type + option +'.h5'
save_name2 = model_names[0] + '_' + 'B' + '_' + atopy_type + option +'.h5'
save_name3 = model_names[0] + '_' + 'C' + '_' + atopy_type + option +'.h5'
save_name4 = model_names[0] + '_' + 'D' + '_' + atopy_type + option +'.h5'
save_name5 = model_names[0] + '_' + 'E' + '_' + atopy_type + option +'.h5'
model_path1 = '../Result/model/' + atopy_type + '/' + save_name1
model_path2 = '../Result/model/' + atopy_type + '/' + save_name2
model_path3 = '../Result/model/' + atopy_type + '/' + save_name3
model_path4 = '../Result/model/' + atopy_type + '/' + save_name4
model_path5 = '../Result/model/' + atopy_type + '/' + save_name5
###Output
_____no_output_____
###Markdown
Load Dataset Test
###Code
""" Test """
grade0_te_path = glob.glob(test_path + 'Grade0/*.jpg')
grade1_te_path = glob.glob(test_path + 'Grade1/*.jpg')
grade2_te_path = glob.glob(test_path + 'Grade2/*.jpg')
grade3_te_path = glob.glob(test_path + 'Grade3/*.jpg')
grade0_te_list, grade1_te_list, grade2_te_list, grade3_te_list = [], [], [], []
label0_te_list, label1_te_list, label2_te_list, label3_te_list = [], [], [], []
for i, g0 in enumerate(grade0_te_path):
img0 = cv2.imread(g0)
## resize
image0 = cv2.resize(img0, dsize=(input_shape[0], input_shape[1]), interpolation = cv2.INTER_CUBIC)
if normalize == 'on':
image0 = image0 / 255
grade0_te_list.append(image0)
label0_te_list.append(0)
for i, g1 in enumerate(grade1_te_path):
img1 = cv2.imread(g1)
## resize
image1 = cv2.resize(img1, dsize=(input_shape[0], input_shape[1]), interpolation = cv2.INTER_CUBIC)
if normalize == 'on':
image1 = image1 / 255
grade1_te_list.append(image1)
label1_te_list.append(1)
for i, g2 in enumerate(grade2_te_path):
img2 = cv2.imread(g2)
## resize
image2 = cv2.resize(img2, dsize=(input_shape[0], input_shape[1]), interpolation = cv2.INTER_CUBIC)
if normalize == 'on':
image2 = image2 / 255
grade2_te_list.append(image2)
label2_te_list.append(2)
for i, g3 in enumerate(grade3_te_path):
img3 = cv2.imread(g3)
## resize
image3 = cv2.resize(img3, dsize=(input_shape[0], input_shape[1]), interpolation = cv2.INTER_CUBIC)
if normalize == 'on':
image3 = image3 / 255
grade3_te_list.append(image3)
label3_te_list.append(3)
print('\n')
print(np.shape(grade0_te_list))
print(np.shape(grade1_te_list))
print(np.shape(grade2_te_list))
print(np.shape(grade3_te_list))
print('\n')
print(len(label0_te_list))
print(len(label1_te_list))
print(len(label2_te_list))
print(len(label3_te_list))
print("** Process Done ***")
###Output
(559, 400, 400, 3)
(646, 400, 400, 3)
(274, 400, 400, 3)
(48, 400, 400, 3)
559
646
274
48
** Process Done ***
###Markdown
External Validation
###Code
""" External Validation """
grade0_ext_path = glob.glob(ext_path + 'Grade0/*.jpg')
grade1_ext_path = glob.glob(ext_path + 'Grade1/*.jpg')
grade2_ext_path = glob.glob(ext_path + 'Grade2/*.jpg')
grade3_ext_path = glob.glob(ext_path + 'Grade3/*.jpg')
grade0_ext_list, grade1_ext_list, grade2_ext_list, grade3_ext_list = [], [], [], []
label0_ext_list, label1_ext_list, label2_ext_list, label3_ext_list = [], [], [], []
for i, g0 in enumerate(grade0_ext_path):
img0 = cv2.imread(g0)
## resize
image0 = cv2.resize(img0, dsize=(input_shape[0], input_shape[1]), interpolation = cv2.INTER_CUBIC)
if normalize == 'on':
image0 = image0 / 255
grade0_ext_list.append(image0)
label0_ext_list.append(0)
for i, g1 in enumerate(grade1_ext_path):
img1 = cv2.imread(g1)
## resize
image1 = cv2.resize(img1, dsize=(input_shape[0], input_shape[1]), interpolation = cv2.INTER_CUBIC)
if normalize == 'on':
image1 = image1 / 255
grade1_ext_list.append(image1)
label1_ext_list.append(1)
for i, g2 in enumerate(grade2_ext_path):
img2 = cv2.imread(g2)
## resize
image2 = cv2.resize(img2, dsize=(input_shape[0], input_shape[1]), interpolation = cv2.INTER_CUBIC)
if normalize == 'on':
image2 = image2 / 255
grade2_ext_list.append(image2)
label2_ext_list.append(2)
for i, g3 in enumerate(grade3_ext_path):
img3 = cv2.imread(g3)
## resize
image3 = cv2.resize(img3, dsize=(input_shape[0], input_shape[1]), interpolation = cv2.INTER_CUBIC)
if normalize == 'on':
image3 = image3 / 255
grade3_ext_list.append(image3)
label3_ext_list.append(3)
print('\n')
print(np.shape(grade0_ext_list))
print(np.shape(grade1_ext_list))
print(np.shape(grade2_ext_list))
print(np.shape(grade3_ext_list))
print('\n')
print(len(label0_ext_list))
print(len(label1_ext_list))
print(len(label2_ext_list))
print(len(label3_ext_list))
print("** Process Done ***")
###Output
(72, 400, 400, 3)
(463, 400, 400, 3)
(364, 400, 400, 3)
(48, 400, 400, 3)
72
463
364
48
** Process Done ***
###Markdown
Concatenation for Dataset Preparation
###Code
""" Test """
X_test = np.concatenate((grade0_te_list, grade1_te_list, grade2_te_list, grade3_te_list), axis = 0)
y_test_ = np.concatenate((label0_te_list, label1_te_list, label2_te_list, label3_te_list), axis = 0)
# X_test = tf.image.per_image_standardization(X_test)
del grade0_te_list, grade1_te_list, grade2_te_list, grade3_te_list
del label0_te_list, label1_te_list, label2_te_list, label3_te_list
""" One-hot encodding """
y_test = keras.utils.to_categorical(y_test_,4)
print(np.shape(X_test))
print(np.shape(y_test))
""" External Validation """
X_ext = np.concatenate((grade0_ext_list, grade1_ext_list, grade2_ext_list, grade3_ext_list), axis = 0)
y_ext_ = np.concatenate((label0_ext_list, label1_ext_list, label2_ext_list, label3_ext_list), axis = 0)
# X_test = tf.image.per_image_standardization(X_test)
del grade0_ext_list, grade1_ext_list, grade2_ext_list, grade3_ext_list
del label0_ext_list, label1_ext_list, label2_ext_list, label3_ext_list
""" One-hot encodding """
y_ext = keras.utils.to_categorical(y_ext_,4)
print(np.shape(X_ext))
print(np.shape(y_ext))
###Output
(947, 400, 400, 3)
(947, 4)
###Markdown
Model Prediction
###Code
import keras.backend.tensorflow_backend as K
""" Load Trained Model """
trained_model1 = keras.models.load_model(model_path1)
trained_model2 = keras.models.load_model(model_path2)
trained_model3 = keras.models.load_model(model_path3)
trained_model4 = keras.models.load_model(model_path4)
trained_model5 = keras.models.load_model(model_path5)
print("*** Load Trained Model Done ***")
""" Test """
pred_test_A = trained_model1.predict(X_test, batch_size=1, verbose=0)
pred_test_B = trained_model2.predict(X_test, batch_size=1, verbose=0)
pred_test_C = trained_model3.predict(X_test, batch_size=1, verbose=0)
pred_test_D = trained_model4.predict(X_test, batch_size=1, verbose=0)
pred_test_E = trained_model5.predict(X_test, batch_size=1, verbose=0)
""" Ext """
pred_ext_A = trained_model1.predict(X_ext, batch_size=1, verbose=0)
pred_ext_B = trained_model2.predict(X_ext, batch_size=1, verbose=0)
pred_ext_C = trained_model3.predict(X_ext, batch_size=1, verbose=0)
pred_ext_D = trained_model4.predict(X_ext, batch_size=1, verbose=0)
pred_ext_E = trained_model5.predict(X_ext, batch_size=1, verbose=0)
print("\n*** Prediction Done ***")
###Output
*** Prediction Done ***
###Markdown
Hard-label (Median)
###Code
""" Test """
med_pred_test_list, med_prob_test_list, med_softmax_prob_test_list = [], [], []
for i in range(len(pred_test_A)):
test_A = np.argmax(pred_test_A[i])
test_B = np.argmax(pred_test_B[i])
test_C = np.argmax(pred_test_C[i])
test_D = np.argmax(pred_test_D[i])
test_E = np.argmax(pred_test_E[i])
med_pred = np.median([test_A, test_B, test_C, test_D, test_E])
num0 = [test_A, test_B, test_C, test_D, test_E].count(0)
num1 = [test_A, test_B, test_C, test_D, test_E].count(1)
num2 = [test_A, test_B, test_C, test_D, test_E].count(2)
num3 = [test_A, test_B, test_C, test_D, test_E].count(3)
med_softmax_prob = [num0/4, num1/4, num2/4, num3/4]
med_softmax_prob_test_list.append(med_softmax_prob)
if med_pred == 0:
med_pred_test_list.append(0)
elif med_pred == 1:
med_pred_test_list.append(1)
elif med_pred == 2:
med_pred_test_list.append(2)
elif med_pred == 3:
med_pred_test_list.append(3)
if med_pred == test_A:
med_prob_test_list.append(pred_test_A[i])
elif med_pred == test_B:
med_prob_test_list.append(pred_test_B[i])
elif med_pred == test_C:
med_prob_test_list.append(pred_test_C[i])
elif med_pred == test_D:
med_prob_test_list.append(pred_test_D[i])
elif med_pred == test_E:
med_prob_test_list.append(pred_test_E[i])
med_prob_test_array = np.array(med_prob_test_list)
med_softmax_prob_test_array = np.array(med_softmax_prob_test_list)
""" Ext """
med_pred_ext_list, med_prob_ext_list, med_softmax_prob_ext_list = [], [], []
for i in range(len(pred_ext_A)):
ext_A = np.argmax(pred_ext_A[i])
ext_B = np.argmax(pred_ext_B[i])
ext_C = np.argmax(pred_ext_C[i])
ext_D = np.argmax(pred_ext_D[i])
ext_E = np.argmax(pred_ext_E[i])
num0 = [ext_A, ext_B, ext_C, ext_D, ext_E].count(0)
num1 = [ext_A, ext_B, ext_C, ext_D, ext_E].count(1)
num2 = [ext_A, ext_B, ext_C, ext_D, ext_E].count(2)
num3 = [ext_A, ext_B, ext_C, ext_D, ext_E].count(3)
med_softmax_ext_prob = [num0/4, num1/4, num2/4, num3/4]
med_softmax_prob_ext_list.append(med_softmax_ext_prob)
med_pred = np.median([ext_A, ext_B, ext_C, ext_D, ext_E])
if med_pred == 0:
med_pred_ext_list.append(0)
elif med_pred == 1:
med_pred_ext_list.append(1)
elif med_pred == 2:
med_pred_ext_list.append(2)
elif med_pred == 3:
med_pred_ext_list.append(3)
if med_pred == ext_A:
med_prob_ext_list.append(pred_ext_A[i])
elif med_pred == ext_B:
med_prob_ext_list.append(pred_ext_B[i])
elif med_pred == ext_C:
med_prob_ext_list.append(pred_ext_C[i])
elif med_pred == ext_D:
med_prob_ext_list.append(pred_ext_D[i])
elif med_pred == ext_E:
med_prob_ext_list.append(pred_ext_E[i])
med_prob_ext_array = np.array(med_prob_ext_list)
med_softmax_prob_ext_array = np.array(med_softmax_prob_ext_list)
###Output
_____no_output_____
###Markdown
Evaluation
###Code
# https://link.medium.com/dlmxbyUxFW
import itertools
def plot_confusion_matrix(cm, target_names=None, cmap=None, normalize=True, labels=True, title='Confusion matrix'):
accuracy = np.trace(cm) / float(np.sum(cm))
misclass = 1 - accuracy
if cmap is None:
cmap = plt.get_cmap('Blues')
if normalize:
cm = cm.astype('float') / cm.sum(axis=1)[:, np.newaxis]
plt.figure(figsize=(8, 6))
plt.imshow(cm, interpolation='nearest', cmap=cmap)
plt.title(title)
plt.colorbar()
thresh = cm.max() / 1.5 if normalize else cm.max() / 2
if target_names is not None:
tick_marks = np.arange(len(target_names))
plt.xticks(tick_marks, target_names)
plt.yticks(tick_marks, target_names)
if labels:
for i, j in itertools.product(range(cm.shape[0]), range(cm.shape[1])):
if normalize:
plt.text(j, i, "{:0.4f}".format(cm[i, j]),
horizontalalignment="center",
color="white" if cm[i, j] > thresh else "black")
else:
plt.text(j, i, "{:,}".format(cm[i, j]),
horizontalalignment="center",
color="white" if cm[i, j] > thresh else "black")
plt.tight_layout()
plt.ylabel('True label')
plt.xlabel('Predicted label\naccuracy={:0.4f}; misclass={:0.4f}'.format(accuracy, misclass))
plt.show()
###Output
_____no_output_____
###Markdown
Confusion Matrix
###Code
from sklearn import metrics
from sklearn.metrics import confusion_matrix, accuracy_score, classification_report, balanced_accuracy_score
""" Confusion Matrix """
X_test_decode = np.argmax(med_prob_test_array, axis=1)
X_ext_decode = np.argmax(med_prob_ext_array, axis=1)
y_test_decode = np.argmax(y_test, axis=1)
y_ext_decode = np.argmax(y_ext, axis=1)
cnf_matrix_test = confusion_matrix(y_test_decode, X_test_decode)
cnf_matrix_ext = confusion_matrix(y_ext_decode, X_ext_decode)
np.set_printoptions(precision=2)
plt.figure()
plot_confusion_matrix(cnf_matrix_test, labels=['grade0', 'grade1', 'grade2', 'grade3'], title='Atopy Grading (test) - ' + Evaluator_Test), 'micro'
plot_confusion_matrix(cnf_matrix_ext, labels=['grade0', 'grade1', 'grade2', 'grade3'], title='Atopy Grading (ext) - ' + Evaluator_Test), 'micro'
acc_test = accuracy_score(y_test_decode, X_test_decode)
acc_ext = accuracy_score(y_ext_decode, X_ext_decode)
print("Accuracy Test : ", acc_test)
print("Accuracy Ext : ", acc_ext)
###Output
_____no_output_____
###Markdown
Metrics
###Code
from sklearn.metrics import precision_score, recall_score, f1_score
""" Precision """
precision_macro_test = precision_score(y_test_decode, X_test_decode, average='macro')
precision_macro_ext = precision_score(y_ext_decode, X_ext_decode, average='macro')
""" Recall """
recall_macro_test = recall_score(y_test_decode, X_test_decode, average='macro')
recall_macro_ext = recall_score(y_ext_decode, X_ext_decode, average='macro')
""" F1-score """
F1_macro_test = f1_score(y_test_decode, X_test_decode, average='macro')
F1_macro_ext = f1_score(y_ext_decode, X_ext_decode, average='macro')
""" Test """
FP = cnf_matrix_test.sum(axis=0) - np.diag(cnf_matrix_test)
FN = cnf_matrix_test.sum(axis=1) - np.diag(cnf_matrix_test)
TP = np.diag(cnf_matrix_test)
TN = cnf_matrix_test.sum() - (FP + FN + TP)
FP = FP.astype(float)
FN = FN.astype(float)
TP = TP.astype(float)
TN = TN.astype(float)
# Sensitivity, hit rate, recall, or true positive rate
TPR_test = TP/(TP+FN)
# Specificity or true negative rate
TNR_test = TN/(TN+FP)
""" Ext """
FP = cnf_matrix_ext.sum(axis=0) - np.diag(cnf_matrix_ext)
FN = cnf_matrix_ext.sum(axis=1) - np.diag(cnf_matrix_ext)
TP = np.diag(cnf_matrix_ext)
TN = cnf_matrix_ext.sum() - (FP + FN + TP)
FP = FP.astype(float)
FN = FN.astype(float)
TP = TP.astype(float)
TN = TN.astype(float)
# Sensitivity, hit rate, recall, or true positive rate
TPR_ext = TP/(TP+FN)
# Specificity or true negative rate
TNR_ext = TN/(TN+FP)
###Output
_____no_output_____
###Markdown
ROC Test
###Code
from sklearn.metrics import roc_curve, auc
from scipy import interp
from itertools import cycle
####################################
pred = med_softmax_prob_test_array
####################################
num_classes = 4
fpr = dict()
tpr = dict()
roc_auc_test = dict()
for i in range(num_classes):
fpr[i], tpr[i], _ = roc_curve(y_test[:, i], pred[:, i])
roc_auc_test[i] = auc(fpr[i], tpr[i])
# Compute micro-average ROC curve and ROC area
fpr["micro"], tpr["micro"], _ = roc_curve(y_test.ravel(), pred.ravel())
roc_auc_test["micro"] = auc(fpr["micro"], tpr["micro"])
# Compute macro-average ROC curve and ROC area
# First aggregate all false positive rates
all_fpr = np.unique(np.concatenate([fpr[i] for i in range(num_classes)]))
# Then interpolate all ROC curves at this points
mean_tpr = np.zeros_like(all_fpr)
for i in range(num_classes):
mean_tpr += interp(all_fpr, fpr[i], tpr[i])
# Finally average it and compute AUC
mean_tpr /= num_classes
fpr["macro"] = all_fpr
tpr["macro"] = mean_tpr
roc_auc_test["macro"] = auc(fpr["macro"], tpr["macro"])
# Plot all ROC curves
plt.figure()
lw = 2
plt.plot(fpr["micro"], tpr["micro"],
label='micro-average ROC curve (area = {0:0.2f})'
''.format(roc_auc_test["micro"]),
color='deeppink', linestyle=':', linewidth=4)
plt.plot(fpr["macro"], tpr["macro"],
label='macro-average ROC curve (area = {0:0.2f})'
''.format(roc_auc_test["macro"]),
color='navy', linestyle=':', linewidth=4)
colors = cycle(['aqua', 'darkorange', 'cornflowerblue'])
for i, color in zip(range(num_classes), colors):
plt.plot(fpr[i], tpr[i], color=color, lw=lw,
label='ROC curve of Grade {0} (area = {1:0.2f})'
''.format(i, roc_auc_test[i]))
auc_std_test = round(np.std([roc_auc_test[0], roc_auc_test[1], roc_auc_test[2], roc_auc_test[3]]), 2)
plt.plot([0, 1], [0, 1], 'k--', lw=lw)
plt.xlim([0.0, 1.0])
plt.ylim([0.0, 1.05])
plt.xlabel('False Positive Rate')
plt.ylabel('True Positive Rate')
plt.title('Atopy Grading ROC curve (Grade 0-3) - ' + Evaluator_Test)
plt.legend(loc="lower right")
plt.show()
###Output
/usr/local/lib/python3.6/dist-packages/ipykernel_launcher.py:28: DeprecationWarning: scipy.interp is deprecated and will be removed in SciPy 2.0.0, use numpy.interp instead
###Markdown
ROC Ext
###Code
from sklearn.metrics import roc_curve, auc
from scipy import interp
from itertools import cycle
####################################
pred = med_softmax_prob_ext_array
####################################
# if Evaluator_Test == 'Soft':
# y_ext = keras.utils.to_categorical(y_ext,4)
num_classes = 4
fpr = dict()
tpr = dict()
roc_auc_ext = dict()
for i in range(num_classes):
# if Evaluator_Test != 'Soft':
fpr[i], tpr[i], _ = roc_curve(y_ext[:, i], pred[:, i])
roc_auc_ext[i] = auc(fpr[i], tpr[i])
# Compute micro-average ROC curve and ROC area
fpr["micro"], tpr["micro"], _ = roc_curve(y_ext.ravel(), pred.ravel())
roc_auc_ext["micro"] = auc(fpr["micro"], tpr["micro"])
# Compute macro-average ROC curve and ROC area
# First aggregate all false positive rates
all_fpr = np.unique(np.concatenate([fpr[i] for i in range(num_classes)]))
# Then interpolate all ROC curves at this points
mean_tpr = np.zeros_like(all_fpr)
for i in range(num_classes):
mean_tpr += interp(all_fpr, fpr[i], tpr[i])
# Finally average it and compute AUC
mean_tpr /= num_classes
fpr["macro"] = all_fpr
tpr["macro"] = mean_tpr
roc_auc_ext["macro"] = auc(fpr["macro"], tpr["macro"])
# Plot all ROC curves
plt.figure()
lw = 2
plt.plot(fpr["micro"], tpr["micro"],
label='micro-average ROC curve (area = {0:0.2f})'
''.format(roc_auc_ext["micro"]),
color='deeppink', linestyle=':', linewidth=4)
plt.plot(fpr["macro"], tpr["macro"],
label='macro-average ROC curve (area = {0:0.2f})'
''.format(roc_auc_ext["macro"]),
color='navy', linestyle=':', linewidth=4)
colors = cycle(['aqua', 'darkorange', 'cornflowerblue'])
for i, color in zip(range(num_classes), colors):
plt.plot(fpr[i], tpr[i], color=color, lw=lw,
label='ROC curve of Grade {0} (area = {1:0.2f})'
''.format(i, roc_auc_ext[i]))
auc_std_ext = round(np.std([roc_auc_ext[0], roc_auc_ext[1], roc_auc_ext[2], roc_auc_ext[3]]), 2)
plt.plot([0, 1], [0, 1], 'k--', lw=lw)
plt.xlim([0.0, 1.0])
plt.ylim([0.0, 1.05])
plt.xlabel('False Positive Rate')
plt.ylabel('True Positive Rate')
plt.title('Atopy Grading ROC curve (Grade 0-3) - ' + Evaluator_Test)
plt.legend(loc="lower right")
plt.show()
###Output
/usr/local/lib/python3.6/dist-packages/ipykernel_launcher.py:34: DeprecationWarning: scipy.interp is deprecated and will be removed in SciPy 2.0.0, use numpy.interp instead
###Markdown
Total Metrics Test
###Code
print("AUC (macro), Accuracy, Sensitivity, Specificity, F1(macro)")
print(round(roc_auc_test["macro"],3), '+-' + str(auc_std_test), round(acc_test, 3), round(np.average(TPR_test),3), round(np.average(TNR_test),3), round(F1_macro_test, 3))
###Output
*** Loss ***
AUC (macro), Accuracy, Sensitivity, Specificity, F1(macro)
0.914 +-0.03 0.782 0.77 0.918 0.75
###Markdown
Ext
###Code
print("AUC (macro), Accuracy, Sensitivity, Specificity, F1(macro)")
print(round(roc_auc_ext["macro"],3), '+-' + str(auc_std_ext), round(acc_ext, 3), round(np.average(TPR_ext),3), round(np.average(TNR_ext),3), round(F1_macro_ext, 3))
###Output
*** Loss ***
AUC (macro), Accuracy, Sensitivity, Specificity, F1(macro)
0.911 +-0.07 0.73 0.807 0.899 0.715
|
product_communities.ipynb
|
###Markdown
Read data
###Code
files = list(os.walk('.'))[0][2]
files
df_products_to_sim = pd.read_csv('products_to_similar_items.csv')
df_products_to_cat = pd.read_csv('products_to_categories.csv')
df_products_to_rev = pd.read_csv('products_to_review.csv')
df_categories = pd.read_csv('categories.csv')
df_products_to_sim = df_products_to_sim.set_index('id')
df_products_to_sim.head()
df_products_to_sim = df_products_to_sim.dropna()
df_products_to_sim.head()
df_products = pd.read_csv('products.csv', escapechar='\\', quotechar='"')
df_products = df_products.set_index('id')
df_products = df_products.sort_index()
df_products.head(2)
df_products_to_sim = df_products_to_sim.sort_index()
df_products_to_sim['asin'] = df_products.loc[df_products_to_sim.index]['asin']
df_products_to_sim['group'] = df_products.loc[df_products_to_sim.index]['group']
df_products_to_sim.head()
df_products_to_sim = df_products_to_sim[df_products_to_sim['group'] == 'Book']
len(df_products_to_sim)
len(df_products_to_sim.loc[df_products_to_sim.index & df_products.index])
df_categories = df_categories.set_index('id')
df_categories = df_categories.sort_index()
df_categories.head(2)
df_products_to_cat = df_products_to_cat.set_index('id')
df_products_to_cat = df_products_to_cat.sort_index()
df_products_to_cat.head(2)
df_products_to_rev.head()
###Output
_____no_output_____
###Markdown
Создаем граф
###Code
df_products_to_sim['similarItemASINs'] = df_products_to_sim['similarItemASINs'].map(lambda x: x.split(';'))
df_products_to_sim.head()
vertices = set(df_products_to_sim['asin'])
adj_list = {x:set(y) & vertices for (x,y) in zip(df_products_to_sim['asin'],
df_products_to_sim['similarItemASINs']) }
###Output
_____no_output_____
###Markdown
Удаляем ребра, к которым нет парных
###Code
for u in adj_list:
to_remove = set()
for v in adj_list[u]:
if u not in adj_list[v]:
to_remove.add(v)
adj_list[u] -= to_remove
###Output
_____no_output_____
###Markdown
Удаляем вершины, у которых нет соседей
###Code
adj_list = {k:v for k, v in adj_list.items() if v}
G = nx.from_dict_of_lists(adj_list)
len(G)
nx.number_connected_components(G)
Gcc = sorted(nx.connected_component_subgraphs(G), key = len, reverse=True)
[len(cc) for cc in Gcc][:10]
###Output
_____no_output_____
###Markdown
Communities
###Code
cur_G = Gcc[50]
nx.draw(cur_G)
plt.show()
parts = community.best_partition(cur_G)
values = [dist_colors[parts.get(node)] for node in cur_G.nodes()]
pos = community_layout(cur_G, parts)
nx.draw(cur_G, pos=pos, node_color=values)
plt.show()
df_products['id'] = df_products.index
df_products = df_products.set_index('asin')
df_products.head(2)
k = 0
for s in [df_products.loc[node, 'title'] for node in cur_G.nodes() if parts.get(node) == k]:
print(s)
values = [dist_colors[parts.get(node)] if parts.get(node) != k else 'black' for node in cur_G.nodes()]
nx.draw(cur_G, pos=pos, node_color=values)
plt.show()
k = 2
for s in [df_products.loc[node, 'title'] for node in cur_G.nodes() if parts.get(node) == k]:
print(s)
values = [dist_colors[parts.get(node)] if parts.get(node) != k else 'black' for node in cur_G.nodes()]
nx.draw(cur_G, pos=pos, node_color=values)
plt.show()
k = 5
for s in [df_products.loc[node, 'title'] for node in cur_G.nodes() if parts.get(node) == k]:
print(s)
values = [dist_colors[parts.get(node)] if parts.get(node) != k else 'black' for node in cur_G.nodes()]
nx.draw(cur_G, pos=pos, node_color=values)
plt.show()
###Output
The Body Project : An Intimate History of American Girls
The Second Sex (Vintage)
A Vindication of the Rights of Woman (Dover Thrift Editions)
The Subjection of Women (Dover Thrift Editions)
The Beauty Myth : How Images of Beauty Are Used Against Women
Backlash : The Undeclared War Against American Women
The Feminine Mystique
Girl Culture
Stiffed: The Betrayal of the American Man
###Markdown
Кластеризация пользователей
###Code
df_products.head(2)
df_products.head(2)
from collections import defaultdict
subgraphes = defaultdict(list)
for key, value in sorted(parts.items()):
subgraphes[value].append(key)
subgraphes[0]
df_products_to_rev.head()
###Output
_____no_output_____
###Markdown
Сколько пользователей покупало книги про зины:
###Code
selected_rev = df_products_to_rev[df_products_to_rev['productId'].isin(df_products.loc[subgraphes[0]]['id'])]
len(set(selected_rev['customer']))
###Output
_____no_output_____
###Markdown
Сколько пользователей покупало книги про левшей:
###Code
selected_rev = df_products_to_rev[df_products_to_rev['productId'].isin(df_products.loc[subgraphes[2]]['id'])]
len(set(selected_rev['customer']))
selected_rev = df_products_to_rev[df_products_to_rev['productId'].isin(df_products.loc[subgraphes[5]]['id'])]
len(set(selected_rev['customer']))
###Output
_____no_output_____
|
Sample Code/Day_03_SampleCode_v2.ipynb
|
###Markdown
範例*** [教學目標]* 能夠使用不同的方法初始化一個陣列* 知道固定大小對於陣列的意義* 了解不同的亂數陣列有什麼差異
###Code
# 載入 NumPy 套件
import numpy as np
# 檢查正確載入與版本
print(np)
print(np.__version__)
# 內建型態做轉換
import numpy as np
np.array([1, 2, 3])
# 會自動轉換成範圍比較大的型態:
print(np.array([1, 2, 3.0]), np.array([1, 2, 3.0]).dtype)
# 也可以指定成想要的型態:
print(np.array([1, 2, 3], dtype=complex), np.array([1, 2, 3], dtype=complex).dtype)
# 字典型態被轉成陣列不符合期待
print(np.array({0: 123, 1: 456}))
print(np.array({0: 123, 1: 456}).size)
# 正確的寫法應該寫轉成有序的 List 再作轉換
print(np.array(list({0: 123, 1: 456}.items())))
print(np.array(list({0: 123, 1: 456}.items())).size)
# 從固定大小的初始值開始
print(np.zeros((2, 3)))
print(np.ones((2, 3)))
print(np.full((2, 3), 9))
# np.zeros 和 np.empty
print(np.zeros((2, 3)))
print(np.empty((2, 3)))
# 從固定大小的序列值開始
print(np.arange( 10, 30, 5 ))
print(np.linspace( 0, 2, 3 ))
print(np.logspace( 0, 2, 3 ))
## 從固定大小的亂數值開始(新版)
from numpy.random import default_rng
rng = default_rng()
normal = rng.standard_normal((3,2))
random = rng.random((3,2))
integers = rng.integers(0, 10, size=(3,2))
print(normal)
print(random)
print(integers)
## 從固定大小的亂數值開始(舊版)
normal = np.random.randn(2, 3)
random = np.random.random((3,2))
integers = np.random.randint(0, 10, size=(3,2))
print(normal)
print(random)
print(integers)
###Output
_____no_output_____
|
Exercises - Qasim/Python. Pandas, Viz/Exercise 5 (Object Oriented Programming) - answers K.ipynb
|
###Markdown
Very useful resource https://realpython.com/python3-object-oriented-programming/ Question 1 Define a class which has at least two methods: getString: to get a string from console input printString: to print the string in upper case. Hints: Use __init__ method to construct some parameters
###Code
# Your code goes here
class MyClass:
def __init__(self, g):
self.g = g
def p(self):
return g.upper()
g = input("enter a string of letters in lower cases: ")
f = MyClass(g)
print(f.p())
# I did not managed to have 2 methods - I am not sure what should be done. Output is fine but not following requirements
###Output
enter a string of letters in lower cases: ewfwef
EWFWEF
###Markdown
Question 2 Define a class named Canadian which has a static method called printNationality. Hints: Use @staticmethod decorator to define class static method. Output: (When the static method is called) Canadian
###Code
# Your code goes here
# Not enough knowledge of staticmethod decorator here.
# Also, I do not undestand the question 'When the static method is called' - what does it mean?
###Output
_____no_output_____
###Markdown
Question 3 What is a constructor and what is it's functionality? How do we initialize class variables using a constuctor?
###Code
On Constructors
- Constructors purpose is to initialize a newly created object.
- They are automatically invoked upon creation of a new object.
- In python constructors are created using "__init__" method.
- This method always take "self" as the first argument which is reference to the object being initialized.
- Other agruments can also be passed after "self"
Example:
def __init__(self, name, age): # Instance Attributes
self.name = name # self
self.age = age # self
On Self:
- Self parameter is a reference to the current instance of the class
- Self parameter is used to access variables that belongs to the class.
- Self variable is mandatory
- Self is also an instance of a class
To note:
- It does not have to be named self, you can call it whatever you want,
- However, it has to be the first paramater of any function in the class.
- Through the self parameter, instance methods can freely access attributes and other methods
- This gives them a lot of power when it comes to modifying an object’s and also class state.
###Output
_____no_output_____
###Markdown
Question 4 Define a class named Circle which can be constructed by a radius. The Circle class has a method which can compute the area of the circle. Hints: Use def methodName(self) to define a method. Sample input : Enter the radius : 2 Area of circle : 12.56 Formula: pi*r*r Note: Take inputs at the time of object creation and pass the input values into the constructor.
###Code
# Your code goes here
import math
class Circle:
def __init__(self, r):
self.r = r
def circle_area(self):
print("Area of circle" + self.r * self.r * math.pi)
Radius = int(input('Enter the radius: '))
Circle = Circle(Radius)
print( NewCircle.circle_area())
# I have not succeeded in having the "area of circle" writtne - and i am not sure why
import math
class Circle:
def __init__(self, r):
self.r = int(r)
r = input('enter new radius: ')
def circle_area(self):
return self.r * self.r * math.pi
Circle = Circle(r)
print(Circle.circle_area())
# I have not succeeded in having the "area of circle" writtne - and i am not sure why
# I have put the input at time of object creation
###Output
_____no_output_____
###Markdown
Question 5 Define a class named Rectangle which can be constructed by a length and width. The Rectangle class has a method which can compute the area. Formula: length x width Sample input: Enter length : 4 Enter width : 3 Sample output: Area of Rectangle is : 12 Note: Take inputs at the time of object creation and pass the input values into the constructor.
###Code
# Your code goes here
class Rectangle:
def __init__(self, l, w):
self.l = int(l)
self.w = int(w)
l = input('enter length: ')
w = input('enter width: ')
def Area(self):
return self.l * self.w
Rectangle = Rectangle(l, w)
print(Rectangle.Area())
# I have a "name 'l' is not defined" error. I have tried multiple turnaround but was not successful
###Output
_____no_output_____
###Markdown
Question 6 Define a class named covert_weight, it contains two methods: 1- kg_to_pounds(kg) 2- pounds_to_kg(pounds) Hint: 1 kg = 2.2 pounds Ask the user to choose an option, call the corresponding function and compute the value. Sample Input: Press 1 for kg-to-pound conversion, 2 for otherwise : 1 Enter weight : 1 Sample output: The weight in pounds is : 2.2
###Code
# Your code goes here
class Convert_Weight:
def __init__(self, w,c):
self.w = w
self.c = c
def PtoK(self):
return float(w) / 2.2
def KtoP(self):
return float(w) * 2.2
w = input('enter weight: ')
c = input('Press 1 for kg-to-pound conversion, 2 for otherwise')
Convert_Weight = Convert_Weight(w,c)
if c == 1:
print(Convert_Weight.KtoP())
else:
print(Convert_Weight.PtoK())
###Output
enter weight: 2
Press 1 for kg-to-pound conversion, 2 for otherwise1
0.9090909090909091
|
part_1_image_classifier.ipynb
|
###Markdown
Developing an AI applicationGoing forward, AI algorithms will be incorporated into more and more everyday applications. For example, you might want to include an image classifier in a smart phone app. To do this, you'd use a deep learning model trained on hundreds of thousands of images as part of the overall application architecture. A large part of software development in the future will be using these types of models as common parts of applications. In this project, you'll train an image classifier to recognize different species of flowers. You can imagine using something like this in a phone app that tells you the name of the flower your camera is looking at. In practice you'd train this classifier, then export it for use in your application. We'll be using [this dataset](http://www.robots.ox.ac.uk/~vgg/data/flowers/102/index.html) of 102 flower categories, you can see a few examples below. The project is broken down into multiple steps:* Load and preprocess the image dataset* Train the image classifier on your dataset* Use the trained classifier to predict image contentWe'll lead you through each part which you'll implement in Python.When you've completed this project, you'll have an application that can be trained on any set of labeled images. Here your network will be learning about flowers and end up as a command line application. But, what you do with your new skills depends on your imagination and effort in building a dataset. For example, imagine an app where you take a picture of a car, it tells you what the make and model is, then looks up information about it. Go build your own dataset and make something new.First up is importing the packages you'll need. It's good practice to keep all the imports at the beginning of your code. As you work through this notebook and find you need to import a package, make sure to add the import up here.
###Code
# Imports here
import torch
import numpy as np
from torchvision import datasets, transforms, models
from torch import nn, optim
import torch.nn.functional as F
from collections import OrderedDict
from workspace_utils import active_session
from PIL import Image
import matplotlib.pyplot as plt
import seaborn as sns
import json
###Output
_____no_output_____
###Markdown
Load the dataHere you'll use `torchvision` to load the data ([documentation](http://pytorch.org/docs/0.3.0/torchvision/index.html)). The data should be included alongside this notebook, otherwise you can [download it here](https://s3.amazonaws.com/content.udacity-data.com/nd089/flower_data.tar.gz). The dataset is split into three parts, training, validation, and testing. For the training, you'll want to apply transformations such as random scaling, cropping, and flipping. This will help the network generalize leading to better performance. You'll also need to make sure the input data is resized to 224x224 pixels as required by the pre-trained networks.The validation and testing sets are used to measure the model's performance on data it hasn't seen yet. For this you don't want any scaling or rotation transformations, but you'll need to resize then crop the images to the appropriate size.The pre-trained networks you'll use were trained on the ImageNet dataset where each color channel was normalized separately. For all three sets you'll need to normalize the means and standard deviations of the images to what the network expects. For the means, it's `[0.485, 0.456, 0.406]` and for the standard deviations `[0.229, 0.224, 0.225]`, calculated from the ImageNet images. These values will shift each color channel to be centered at 0 and range from -1 to 1.
###Code
data_dir = 'flowers'
train_dir = data_dir + '/train'
valid_dir = data_dir + '/valid'
test_dir = data_dir + '/test'
# TODO: Define your transforms for the training, validation, and testing sets
means = [0.485, 0.456, 0.406]
stdev = [0.229, 0.224, 0.225]
train_transforms = transforms.Compose([transforms.RandomRotation(30),
transforms.RandomResizedCrop(224),
transforms.RandomHorizontalFlip(),
transforms.ToTensor(),
transforms.Normalize(means, stdev)])
test_transforms = transforms.Compose([transforms.Resize(255),
transforms.CenterCrop(224),
transforms.ToTensor(),
transforms.Normalize(means, stdev)])
valid_transforms = transforms.Compose([transforms.Resize(255),
transforms.CenterCrop(224),
transforms.ToTensor(),
transforms.Normalize(means, stdev)])
# TODO: Load the datasets with ImageFolder
train_datasets = datasets.ImageFolder(train_dir, transform=train_transforms)
test_datasets = datasets.ImageFolder(test_dir, transform=train_transforms)
valid_datasets = datasets.ImageFolder(valid_dir, transform=valid_transforms)
# TODO: Using the image datasets and the trainforms, define the dataloaders
test_dataloader = torch.utils.data.DataLoader(test_datasets, shuffle=True, batch_size=32)
train_dataloader = torch.utils.data.DataLoader(train_datasets, shuffle=True, batch_size=32)
valid_dataloader = torch.utils.data.DataLoader(valid_datasets, shuffle=True, batch_size=32)
###Output
_____no_output_____
###Markdown
Label mappingYou'll also need to load in a mapping from category label to category name. You can find this in the file `cat_to_name.json`. It's a JSON object which you can read in with the [`json` module](https://docs.python.org/2/library/json.html). This will give you a dictionary mapping the integer encoded categories to the actual names of the flowers.
###Code
with open('cat_to_name.json', 'r') as f:
cat_to_name = json.load(f)
print(len(cat_to_name))
###Output
102
###Markdown
Building and training the classifierNow that the data is ready, it's time to build and train the classifier. As usual, you should use one of the pretrained models from `torchvision.models` to get the image features. Build and train a new feed-forward classifier using those features.We're going to leave this part up to you. Refer to [the rubric](https://review.udacity.com/!/rubrics/1663/view) for guidance on successfully completing this section. Things you'll need to do:* Load a [pre-trained network](http://pytorch.org/docs/master/torchvision/models.html) (If you need a starting point, the VGG networks work great and are straightforward to use)* Define a new, untrained feed-forward network as a classifier, using ReLU activations and dropout* Train the classifier layers using backpropagation using the pre-trained network to get the features* Track the loss and accuracy on the validation set to determine the best hyperparametersWe've left a cell open for you below, but use as many as you need. Our advice is to break the problem up into smaller parts you can run separately. Check that each part is doing what you expect, then move on to the next. You'll likely find that as you work through each part, you'll need to go back and modify your previous code. This is totally normal!When training make sure you're updating only the weights of the feed-forward network. You should be able to get the validation accuracy above 70% if you build everything right. Make sure to try different hyperparameters (learning rate, units in the classifier, epochs, etc) to find the best model. Save those hyperparameters to use as default values in the next part of the project.One last important tip if you're using the workspace to run your code: To avoid having your workspace disconnect during the long-running tasks in this notebook, please read in the earlier page in this lesson called Intro toGPU Workspaces about Keeping Your Session Active. You'll want to include code from the workspace_utils.py module.**Note for Workspace users:** If your network is over 1 GB when saved as a checkpoint, there might be issues with saving backups in your workspace. Typically this happens with wide dense layers after the convolutional layers. If your saved checkpoint is larger than 1 GB (you can open a terminal and check with `ls -lh`), you should reduce the size of your hidden layers and train again.
###Code
# TODO: Build and train your network
model = models.densenet121(pretrained=True)
for param in model.parameters():
param.requires_grad = False
classifier = nn.Sequential(OrderedDict([('fc1', nn.Linear(1024, 256)),
('relu', nn.ReLU()),
('dropout', nn.Dropout(0.2)),
('fc3', nn.Linear(256, 102)),
('output', nn.LogSoftmax(dim=1))]))
model.classifier = classifier
criterion = nn.NLLLoss()
optimizer = optim.Adam(model.classifier.parameters(), lr = 0.003)
device = torch.device("cuda" if torch.cuda.is_available() else "cpu")
model.to(device)
epochs = 30
steps = 0
running_loss = 0
print_every = 100
with active_session():
for epoch in range(epochs):
for images, labels in train_dataloader:
images, labels = images.to(device), labels.to(device)
optimizer.zero_grad()
log_ps = model(images)
loss = criterion(log_ps, labels)
running_loss += loss.item()
loss.backward()
optimizer.step()
steps += 1
if steps % print_every == 0:
with torch.no_grad():
model.eval()
valid_loss = 0
accuracy = 0
for images, labels in valid_dataloader:
images, labels = images.to(device), labels.to(device)
log_ps = model(images)
loss = criterion(log_ps, labels)
valid_loss += loss.item()
#accuracy
ps = torch.exp(log_ps)
top_p, top_class = ps.topk(1, dim=1)
equals = top_class == labels.view(*top_class.shape)
accuracy += torch.mean(equals.type(torch.FloatTensor)).item()
print(f"Epoch {epoch+1}/{epochs}.. "
f"Train loss: {running_loss/print_every:.3f}.. "
f"Validation loss: {valid_loss/len(valid_dataloader):.3f}.. "
f"Validation accuracy: {accuracy/len(valid_dataloader):.3f}")
running_loss = 0
model.train()
###Output
Epoch 1/30.. Train loss: 3.793.. Validation loss: 2.357.. Validation accuracy: 0.465
Epoch 1/30.. Train loss: 2.239.. Validation loss: 1.224.. Validation accuracy: 0.704
Epoch 2/30.. Train loss: 1.669.. Validation loss: 0.926.. Validation accuracy: 0.752
Epoch 2/30.. Train loss: 1.438.. Validation loss: 0.753.. Validation accuracy: 0.799
Epoch 3/30.. Train loss: 1.311.. Validation loss: 0.682.. Validation accuracy: 0.810
Epoch 3/30.. Train loss: 1.201.. Validation loss: 0.664.. Validation accuracy: 0.815
Epoch 4/30.. Train loss: 1.126.. Validation loss: 0.542.. Validation accuracy: 0.850
Epoch 4/30.. Train loss: 1.095.. Validation loss: 0.524.. Validation accuracy: 0.853
Epoch 5/30.. Train loss: 1.032.. Validation loss: 0.450.. Validation accuracy: 0.873
Epoch 5/30.. Train loss: 1.048.. Validation loss: 0.488.. Validation accuracy: 0.855
Epoch 6/30.. Train loss: 0.977.. Validation loss: 0.459.. Validation accuracy: 0.887
Epoch 6/30.. Train loss: 0.959.. Validation loss: 0.404.. Validation accuracy: 0.897
Epoch 7/30.. Train loss: 0.956.. Validation loss: 0.437.. Validation accuracy: 0.876
Epoch 7/30.. Train loss: 0.972.. Validation loss: 0.432.. Validation accuracy: 0.879
Epoch 8/30.. Train loss: 0.920.. Validation loss: 0.390.. Validation accuracy: 0.895
Epoch 8/30.. Train loss: 0.936.. Validation loss: 0.448.. Validation accuracy: 0.882
Epoch 9/30.. Train loss: 0.924.. Validation loss: 0.391.. Validation accuracy: 0.891
Epoch 9/30.. Train loss: 0.882.. Validation loss: 0.444.. Validation accuracy: 0.876
Epoch 10/30.. Train loss: 0.925.. Validation loss: 0.387.. Validation accuracy: 0.891
Epoch 10/30.. Train loss: 0.837.. Validation loss: 0.436.. Validation accuracy: 0.879
Epoch 11/30.. Train loss: 0.900.. Validation loss: 0.367.. Validation accuracy: 0.905
Epoch 11/30.. Train loss: 0.913.. Validation loss: 0.332.. Validation accuracy: 0.901
Epoch 12/30.. Train loss: 0.871.. Validation loss: 0.423.. Validation accuracy: 0.894
Epoch 12/30.. Train loss: 0.910.. Validation loss: 0.383.. Validation accuracy: 0.897
Epoch 13/30.. Train loss: 0.840.. Validation loss: 0.353.. Validation accuracy: 0.903
Epoch 13/30.. Train loss: 0.806.. Validation loss: 0.365.. Validation accuracy: 0.894
Epoch 14/30.. Train loss: 0.833.. Validation loss: 0.362.. Validation accuracy: 0.898
Epoch 14/30.. Train loss: 0.840.. Validation loss: 0.403.. Validation accuracy: 0.885
Epoch 15/30.. Train loss: 0.818.. Validation loss: 0.340.. Validation accuracy: 0.916
Epoch 15/30.. Train loss: 0.842.. Validation loss: 0.452.. Validation accuracy: 0.891
Epoch 16/30.. Train loss: 0.848.. Validation loss: 0.368.. Validation accuracy: 0.903
Epoch 16/30.. Train loss: 0.856.. Validation loss: 0.353.. Validation accuracy: 0.907
Epoch 17/30.. Train loss: 0.815.. Validation loss: 0.341.. Validation accuracy: 0.916
Epoch 17/30.. Train loss: 0.822.. Validation loss: 0.353.. Validation accuracy: 0.912
Epoch 18/30.. Train loss: 0.801.. Validation loss: 0.395.. Validation accuracy: 0.911
Epoch 18/30.. Train loss: 0.829.. Validation loss: 0.391.. Validation accuracy: 0.894
Epoch 19/30.. Train loss: 0.855.. Validation loss: 0.376.. Validation accuracy: 0.910
Epoch 19/30.. Train loss: 0.785.. Validation loss: 0.378.. Validation accuracy: 0.910
Epoch 20/30.. Train loss: 0.875.. Validation loss: 0.392.. Validation accuracy: 0.899
Epoch 20/30.. Train loss: 0.795.. Validation loss: 0.469.. Validation accuracy: 0.883
Epoch 20/30.. Train loss: 0.842.. Validation loss: 0.370.. Validation accuracy: 0.907
Epoch 21/30.. Train loss: 0.800.. Validation loss: 0.421.. Validation accuracy: 0.888
Epoch 21/30.. Train loss: 0.813.. Validation loss: 0.398.. Validation accuracy: 0.903
Epoch 22/30.. Train loss: 0.854.. Validation loss: 0.417.. Validation accuracy: 0.886
Epoch 22/30.. Train loss: 0.834.. Validation loss: 0.374.. Validation accuracy: 0.905
Epoch 23/30.. Train loss: 0.779.. Validation loss: 0.348.. Validation accuracy: 0.908
Epoch 23/30.. Train loss: 0.867.. Validation loss: 0.442.. Validation accuracy: 0.888
Epoch 24/30.. Train loss: 0.756.. Validation loss: 0.390.. Validation accuracy: 0.906
Epoch 24/30.. Train loss: 0.804.. Validation loss: 0.411.. Validation accuracy: 0.889
Epoch 25/30.. Train loss: 0.761.. Validation loss: 0.417.. Validation accuracy: 0.901
Epoch 25/30.. Train loss: 0.804.. Validation loss: 0.404.. Validation accuracy: 0.907
Epoch 26/30.. Train loss: 0.839.. Validation loss: 0.400.. Validation accuracy: 0.892
Epoch 26/30.. Train loss: 0.809.. Validation loss: 0.387.. Validation accuracy: 0.906
Epoch 27/30.. Train loss: 0.786.. Validation loss: 0.421.. Validation accuracy: 0.897
Epoch 27/30.. Train loss: 0.777.. Validation loss: 0.374.. Validation accuracy: 0.903
Epoch 28/30.. Train loss: 0.828.. Validation loss: 0.367.. Validation accuracy: 0.900
Epoch 28/30.. Train loss: 0.776.. Validation loss: 0.395.. Validation accuracy: 0.893
Epoch 29/30.. Train loss: 0.786.. Validation loss: 0.440.. Validation accuracy: 0.899
Epoch 29/30.. Train loss: 0.782.. Validation loss: 0.418.. Validation accuracy: 0.887
Epoch 30/30.. Train loss: 0.775.. Validation loss: 0.354.. Validation accuracy: 0.910
Epoch 30/30.. Train loss: 0.787.. Validation loss: 0.347.. Validation accuracy: 0.904
###Markdown
Testing your networkIt's good practice to test your trained network on test data, images the network has never seen either in training or validation. This will give you a good estimate for the model's performance on completely new images. Run the test images through the network and measure the accuracy, the same way you did validation. You should be able to reach around 70% accuracy on the test set if the model has been trained well.
###Code
# TODO: Do validation on the test set
with torch.no_grad():
testing_loss = 0
accuracy = 0
for images, labels in test_dataloader:
model.eval()
images, labels = images.to(device), labels.to(device)
log_ps = model(images)
loss = criterion(log_ps, labels)
testing_loss += loss.item()
ps = torch.exp(log_ps)
top_p, top_class = ps.topk(1, dim=1)
equals = top_class == labels.view(*top_class.shape)
accuracy += torch.mean(equals.type(torch.FloatTensor)).item()
print(f"Test loss: {testing_loss/len(test_dataloader):.3f}.. "
f"Test accuracy: {accuracy/len(test_dataloader):.3f}")
###Output
Test loss: 0.608.. Test accuracy: 0.832
###Markdown
Save the checkpointNow that your network is trained, save the model so you can load it later for making predictions. You probably want to save other things such as the mapping of classes to indices which you get from one of the image datasets: `image_datasets['train'].class_to_idx`. You can attach this to the model as an attribute which makes inference easier later on.```model.class_to_idx = image_datasets['train'].class_to_idx```Remember that you'll want to completely rebuild the model later so you can use it for inference. Make sure to include any information you need in the checkpoint. If you want to load the model and keep training, you'll want to save the number of epochs as well as the optimizer state, `optimizer.state_dict`. You'll likely want to use this trained model in the next part of the project, so best to save it now.
###Code
# TODO: Save the checkpoint
model.class_to_idx = train_datasets.class_to_idx
checkpoint = {'output_size': 102,
'classifier': model.classifier,
'class_to_idx': train_datasets.class_to_idx,
'epochs': epochs,
'optimizer.state_dict': optimizer.state_dict(),
'state_dict': model.state_dict()}
torch.save(checkpoint, 'checkpoint.pth')
###Output
_____no_output_____
###Markdown
Loading the checkpointAt this point it's good to write a function that can load a checkpoint and rebuild the model. That way you can come back to this project and keep working on it without having to retrain the network.
###Code
# TODO: Write a function that loads a checkpoint and rebuilds the model
def load_checkpoint(filepath):
checkpoint = torch.load(filepath)
model = models.densenet121(pretrained=True)
model.class_to_idx = train_datasets.class_to_idx
model.classifier = checkpoint['classifier']
model.load_state_dict(checkpoint['state_dict'])
return model
# model = load_checkpoint('checkpoint.pth')
###Output
_____no_output_____
###Markdown
Inference for classificationNow you'll write a function to use a trained network for inference. That is, you'll pass an image into the network and predict the class of the flower in the image. Write a function called `predict` that takes an image and a model, then returns the top $K$ most likely classes along with the probabilities. It should look like ```pythonprobs, classes = predict(image_path, model)print(probs)print(classes)> [ 0.01558163 0.01541934 0.01452626 0.01443549 0.01407339]> ['70', '3', '45', '62', '55']```First you'll need to handle processing the input image such that it can be used in your network. Image PreprocessingYou'll want to use `PIL` to load the image ([documentation](https://pillow.readthedocs.io/en/latest/reference/Image.html)). It's best to write a function that preprocesses the image so it can be used as input for the model. This function should process the images in the same manner used for training. First, resize the images where the shortest side is 256 pixels, keeping the aspect ratio. This can be done with the [`thumbnail`](http://pillow.readthedocs.io/en/3.1.x/reference/Image.htmlPIL.Image.Image.thumbnail) or [`resize`](http://pillow.readthedocs.io/en/3.1.x/reference/Image.htmlPIL.Image.Image.thumbnail) methods. Then you'll need to crop out the center 224x224 portion of the image.Color channels of images are typically encoded as integers 0-255, but the model expected floats 0-1. You'll need to convert the values. It's easiest with a Numpy array, which you can get from a PIL image like so `np_image = np.array(pil_image)`.As before, the network expects the images to be normalized in a specific way. For the means, it's `[0.485, 0.456, 0.406]` and for the standard deviations `[0.229, 0.224, 0.225]`. You'll want to subtract the means from each color channel, then divide by the standard deviation. And finally, PyTorch expects the color channel to be the first dimension but it's the third dimension in the PIL image and Numpy array. You can reorder dimensions using [`ndarray.transpose`](https://docs.scipy.org/doc/numpy-1.13.0/reference/generated/numpy.ndarray.transpose.html). The color channel needs to be first and retain the order of the other two dimensions.
###Code
def process_image(image_path):
''' Scales, crops, and normalizes a PIL image for a PyTorch model,
returns an Numpy array
'''
image = Image.open(image_path)
x, y = image.size
if x > y:
x, y = 256 * x / y, 256
else:
x, y = 256, 256 * y / x
image.thumbnail((x,y))
left = (x-224)/2
top = (y-224)/2
right = left + 224
bottom = top + 224
image = image.crop((left, top, right, bottom))
np_image = np.array(image) / 255
np_image = (np.subtract(np_image, means)) / stdev
np_image = np_image.transpose((2, 0, 1))
return np_image
# TODO: Process a PIL image for use in a PyTorch model
###Output
_____no_output_____
###Markdown
To check your work, the function below converts a PyTorch tensor and displays it in the notebook. If your `process_image` function works, running the output through this function should return the original image (except for the cropped out portions).
###Code
def imshow(image, ax=None, title=None):
"""Imshow for Tensor."""
if ax is None:
fig, ax = plt.subplots()
# PyTorch tensors assume the color channel is the first dimension
# but matplotlib assumes is the third dimension
image = image.transpose((1, 2, 0))
# Undo preprocessing
mean = np.array([0.485, 0.456, 0.406])
std = np.array([0.229, 0.224, 0.225])
image = std * image + mean
# Image needs to be clipped between 0 and 1 or it looks like noise when displayed
image = np.clip(image, 0, 1)
ax.imshow(image)
return ax
###Output
_____no_output_____
###Markdown
Class PredictionOnce you can get images in the correct format, it's time to write a function for making predictions with your model. A common practice is to predict the top 5 or so (usually called top-$K$) most probable classes. You'll want to calculate the class probabilities then find the $K$ largest values.To get the top $K$ largest values in a tensor use [`x.topk(k)`](http://pytorch.org/docs/master/torch.htmltorch.topk). This method returns both the highest `k` probabilities and the indices of those probabilities corresponding to the classes. You need to convert from these indices to the actual class labels using `class_to_idx` which hopefully you added to the model or from an `ImageFolder` you used to load the data ([see here](Save-the-checkpoint)). Make sure to invert the dictionary so you get a mapping from index to class as well.Again, this method should take a path to an image and a model checkpoint, then return the probabilities and classes.```pythonprobs, classes = predict(image_path, model)print(probs)print(classes)> [ 0.01558163 0.01541934 0.01452626 0.01443549 0.01407339]> ['70', '3', '45', '62', '55']```
###Code
def predict(image_path, model, topk=5):
''' Predict the class (or classes) of an image using a trained deep learning model.
'''
np_image = process_image(image_path)
image = torch.from_numpy(np_image)
image = image.unsqueeze(0).type(torch.FloatTensor)
with torch.no_grad():
model.eval()
log_ps = model(image)
ps = torch.exp(log_ps)
top_p, top_classes = ps.topk(topk)
top_p, top_classes = top_p.numpy().flatten().tolist(), top_classes.numpy().flatten()
idx_to_class = dict(map(reversed, model.class_to_idx.items()))
flower_classes = list(map(lambda x: idx_to_class[x], top_classes))
return top_p, flower_classes
###Output
_____no_output_____
###Markdown
Sanity CheckingNow that you can use a trained model for predictions, check to make sure it makes sense. Even if the testing accuracy is high, it's always good to check that there aren't obvious bugs. Use `matplotlib` to plot the probabilities for the top 5 classes as a bar graph, along with the input image. It should look like this:You can convert from the class integer encoding to actual flower names with the `cat_to_name.json` file (should have been loaded earlier in the notebook). To show a PyTorch tensor as an image, use the `imshow` function defined above.
###Code
# TODO: Display an image along with the top 5 classes
image_path = 'flowers/test/16/image_06657.jpg'
device = 'cpu'
plt.figure(figsize = (5, 10))
ax = plt.subplot(2,1,1)
title = cat_to_name[image_path.split('/')[2]]
image = process_image(image_path)
imshow(image, ax, title)
model.to(device)
probs, classes = predict(image_path, model)
flower_names = list(map(lambda x: cat_to_name[x], classes))
plt.subplot(2,1,2)
sns.barplot(x=probs, y=flower_names, color=sns.color_palette()[0]).set_title(title)
plt.show()
###Output
_____no_output_____
|
Analysing Weather Dataset.ipynb
|
###Markdown
Outline 1. Tools : Google Sheet ,Jupyter Notebook , Python SQL Query Query the Global Data2. to get the global data3. Select *4. From global_data Query the City Data6. To get the city which is Kuala Lumpur in the country Malaysia weather dataset7. Select *8. From city_data9. Where Country = 'Malaysia' To find the city near me with my country11. Select city12. From city_list13. Where Country = 'Malaysia' Process1. Fix the datatpes2. Take the average of 10 years between 1825 and 2013 for local data and 1804 and 2015 for global data. Key Considerations1. Using moving average to keep the line as smooth as possible.2. Making sure that the two line chart has different colours so that the chart can be visualize clearly.3. Making sure the y- axis and x - axis shown the labelled that is appropriate for the data.
###Code
global_df.head()
city_df.head()
# Check the datatypes of global_df
global_df.dtypes
# Drop city dataframe missing values
city_df.dropna()
# convert city dataframe to integer
city_df.astype(int)
# Convert global data frame dataset to integer
global_df.astype(int)
# Calculate the moving average of global average temperature across 10 years
global_df['moving_average'] = global_df['avg_temp'].rolling(window = 10).mean()
# Calculate the moving average of city average temperature acorss 10 years
city_df['moving_average'] = city_df['avg_temp'].rolling(window = 10).mean()
# Drop missing values
global_df.dropna()
# Drop the missing values
city_df.dropna()
# Plotting the Global Temperature across the years
x = global_df['year']
y = global_df['moving_average']
plt.scatter(x , y)
plt.xlabel('Years')
plt.ylabel('Moving Average Temperature')
plt.title('Global Temperature across the years');
#Plotting Kuala Lumpur Temperature across the year
x = city_df['year']
y = city_df['moving_average']
plt.scatter(x , y)
plt.xlabel('Years')
plt.ylabel('Moving Average Temperature')
plt.title('Kuala Lumpur Temperature across the years');
# Plotting Global Temperature vs Kuala Lumpur Temperature
x = global_df['year']
y = global_df['moving_average']
plt.scatter(x , y , label = 'Global')
x = city_df['year']
y = city_df['moving_average']
plt.scatter(x , y , label = 'Kuala Lumpur')
plt.xlabel('Years')
plt.ylabel('Moving Average Temperature')
plt.legend()
plt.title('Global vs Kuala Lumpur Temperature across the years');
###Output
_____no_output_____
|
Copy_of_DebuggedStockPrediction_with_model_deployment.ipynb
|
###Markdown
Stock Prediction model with Tensorflow 2.0! We're going to predict prices of General Electric's stock using a Transformer neural network **IMPORT DATA FROM DRIVE**
###Code
import requests
def download_file_from_google_drive(id, destination):
URL = "https://drive.google.com/drive/u/5/folders/1GwvIddbMYiKksnXwecbq8-YPPLIDBEJl"
session = requests.Session()
response = session.get(URL, params = { 'id' : id }, stream = True)
token = get_confirm_token(response)
if token:
params = { 'id' : id, 'confirm' : token }
response = session.get(URL, params = params, stream = True)
save_response_content(response, destination)
def get_confirm_token(response):
for key, value in response.cookies.items():
if key.startswith('download_warning'):
return value
return None
def save_response_content(response, destination):
CHUNK_SIZE = 32768
with open(destination, "wb") as f:
for chunk in response.iter_content(CHUNK_SIZE):
if chunk: # filter out keep-alive new chunks
f.write(chunk)
if __name__ == "__main__":
file_id = 'TAKE ID FROM SHAREABLE LINK'
destination = 'F.csv'
download_file_from_google_drive(file_id, destination)
###Output
_____no_output_____
###Markdown
Step 1 - Import data
###Code
import numpy as np
import matplotlib.pyplot as plt
import pandas as pd
import os
from sklearn.preprocessing import MinMaxScaler
from google.colab import drive
#drive.mount('/content/drive', force_remount=True)
!pwd
# Upload file without google drive
df = pd.read_csv('F.csv',delimiter=',',usecols=['Date','Open','High','Low','Close', 'Volume'])
# I got it from https://finance.yahoo.com/quote/GE/history?p=GE&.tsrc=fin-srch
df = pd.read_csv('drive/My Drive/BigData/F.csv',delimiter=',',usecols=['Date','Open','High','Low','Close', 'Volume'])
# Sort DataFrame by date
df = df.sort_values('Date')
# Double check the result
df.head()
plt.figure(figsize = (18,9))
plt.plot(range(df.shape[0]),(df['Low']+df['High'])/2.0)
plt.xticks(range(0,df.shape[0],500),df['Date'].loc[::500],rotation=45)
plt.xlabel('Date',fontsize=18)
plt.ylabel('Mid Price',fontsize=18)
plt.show()
###Output
_____no_output_____
###Markdown
Step 2 - Data preprocessing
###Code
df['mid'] = (df['Low']+df['High'])/2.0
SEQ_LEN = 60 # how long of a preceeding sequence to collect for RNN
FUTURE_PERIOD_PREDICT = 1 # how far into the future are we trying to predict?
RATIO_TO_PREDICT = "mid"
def classify(current, future):
if float(future) > float(current):
return 1
else:
return 0
df['future'] = df[RATIO_TO_PREDICT].shift(-FUTURE_PERIOD_PREDICT)
df['target'] = list(map(classify, df[RATIO_TO_PREDICT], df['future']))
df.head()
df.tail()
times = sorted(df.index.values) # get the times
last_10pct = sorted(df.index.values)[-int(0.1*len(times))] # get the last 10% of the times
last_20pct = sorted(df.index.values)[-int(0.2*len(times))] # get the last 20% of the times
test_df = df[(df.index >= last_10pct)]
validation_df = df[(df.index >= last_20pct) & (df.index < last_10pct)]
train_df = df[(df.index < last_20pct)] # now the train_df is all the data up to the last 20%
from collections import deque
import numpy as np
import random
train_df.drop(columns=["Date", "future", 'Open', 'High', 'Low', 'Close', 'Volume'], inplace=True)
validation_df.drop(columns=["Date", "future", 'Open', 'High', 'Low', 'Close', 'Volume'], inplace=True)
test_df.drop(columns=["Date", "future", 'Open', 'High', 'Low', 'Close', 'Volume'], inplace=True)
train_df.head()
train_df.tail()
train_data = train_df[RATIO_TO_PREDICT].as_matrix()
valid_data = validation_df[RATIO_TO_PREDICT].as_matrix()
test_data = test_df[RATIO_TO_PREDICT].as_matrix()
print(train_data)
train_data = train_data.reshape(-1,1)
valid_data = valid_data.reshape(-1,1)
test_data = test_data.reshape(-1,1)
print(train_data)
scaler = MinMaxScaler()
print(scaler.fit(train_data))
# Train the Scaler with training data and smooth data
smoothing_window_size = 25
for di in range(0,100,smoothing_window_size):
scaler.fit(train_data[di:di+smoothing_window_size,:])
train_data[di:di+smoothing_window_size,:] = scaler.transform(train_data[di:di+smoothing_window_size,:])
# You normalize the last bit of remaining data
scaler.fit(train_data[di+smoothing_window_size:,:])
train_data[di+smoothing_window_size:,:] = scaler.transform(train_data[di+smoothing_window_size:,:])
# Reshape both train and test data
train_data = train_data.reshape(-1)
# Normalize test data and validation data
valid_data = scaler.transform(valid_data).reshape(-1)
test_data = scaler.transform(test_data).reshape(-1)
# Now perform exponential moving average smoothing
# So the data will have a smoother curve than the original ragged data
EMA = 0.0
gamma = 0.1
for ti in range(1006):
EMA = gamma*train_data[ti] + (1-gamma)*EMA
train_data[ti] = EMA
# Used for visualization and test purposes
all_mid_data = np.concatenate([train_data,valid_data, test_data],axis=0)
X_train = []
y_train = []
for i in range(SEQ_LEN, len(train_data)):
X_train.append(train_data[i-SEQ_LEN:i])
y_train.append(train_data[i + (FUTURE_PERIOD_PREDICT-1)])
X_train, y_train = np.array(X_train), np.array(y_train)
X_train = np.reshape(X_train, (X_train.shape[0], X_train.shape[1], 1))
print(y_train)
X_valid = []
y_valid = []
for i in range(SEQ_LEN, len(valid_data)):
X_valid.append(valid_data[i-SEQ_LEN:i])
y_valid.append(valid_data[i+(FUTURE_PERIOD_PREDICT-1)])
X_valid, y_valid = np.array(X_valid), np.array(y_valid)
X_valid = np.reshape(X_valid, (X_valid.shape[0], X_valid.shape[1], 1))
X_test = []
y_test = []
for i in range(SEQ_LEN, len(test_data)):
X_test.append(test_data[i-SEQ_LEN:i])
y_test.append(test_data[i+(FUTURE_PERIOD_PREDICT-1)])
X_test, y_test = np.array(X_test), np.array(y_test)
X_test = np.reshape(X_test, (X_test.shape[0], X_test.shape[1], 1))
y_train.shape
y_valid.shape
X_train_2 = []
y_train_2 = []
for i in range(SEQ_LEN, len(train_data)):
X_train_2.append(train_data[i-SEQ_LEN:i])
y_train_2.append(train_data[i + (FUTURE_PERIOD_PREDICT-1)])
X_train_2, y_train_2 = np.array(X_train_2), np.array(y_train_2)
X_train_2 = np.reshape(X_train_2, (X_train_2.shape[0], X_train_2.shape[1], 1))
print(y_train_2)
print(y_train_2.shape)
## show predictions
plt.figure(figsize=(15, 5))
plt.plot(np.arange(y_train_2.shape[0]), y_train_2, color='blue', label='train target')
plt.plot(np.arange(y_train_2.shape[0], y_train_2.shape[0]+y_valid.shape[0]), y_valid,
color='gray', label='valid target')
plt.plot(np.arange(y_train_2.shape[0]+y_valid.shape[0],
y_train_2.shape[0]+y_valid.shape[0]+y_test.shape[0]),
y_test, color='black', label='test target')
plt.title('Seaparated Data Sets')
plt.xlabel('time [days]')
plt.ylabel('normalized price')
plt.legend(loc='best');
from sklearn.utils import shuffle
X_train, y_train = shuffle(X_train, y_train)
EPOCHS = 10 # how many passes through our data
BATCH_SIZE = 1024 # how many batches? Try smaller batch if you're getting OOM (out of memory) errors.
import time
NAME = f"{SEQ_LEN}-SEQ-{FUTURE_PERIOD_PREDICT}-PRED-{int(time.time())}" # a unique name for the model
!pip install -q tensorflow==2.0.0-alpha0
# https://www.kaggle.com/shujian/transformer-with-lstm
import random, os, sys
import numpy as np
from tensorflow.keras.models import *
from tensorflow.keras.layers import *
from tensorflow.keras.callbacks import *
from tensorflow.keras.initializers import *
import tensorflow as tf
from tensorflow.python.keras.layers import Layer
from tensorflow.keras import backend as K
try:
from dataloader import TokenList, pad_to_longest
# for transformer
except: pass
embed_size = 60
class LayerNormalization(Layer):
def __init__(self, eps=1e-6, **kwargs):
self.eps = eps
super(LayerNormalization, self).__init__(**kwargs)
def build(self, input_shape):
self.gamma = self.add_weight(name='gamma', shape=input_shape[-1:],
initializer=Ones(), trainable=True)
self.beta = self.add_weight(name='beta', shape=input_shape[-1:],
initializer=Zeros(), trainable=True)
super(LayerNormalization, self).build(input_shape)
def call(self, x):
mean = K.mean(x, axis=-1, keepdims=True)
std = K.std(x, axis=-1, keepdims=True)
return self.gamma * (x - mean) / (std + self.eps) + self.beta
def compute_output_shape(self, input_shape):
return input_shape
class ScaledDotProductAttention():
def __init__(self, d_model, attn_dropout=0.1):
self.temper = np.sqrt(d_model)
self.dropout = Dropout(attn_dropout)
def __call__(self, q, k, v, mask):
attn = Lambda(lambda x:K.batch_dot(x[0],x[1],axes=[2,2])/self.temper)([q, k])
if mask is not None:
mmask = Lambda(lambda x:(-1e+10)*(1-x))(mask)
attn = Add()([attn, mmask])
attn = Activation('softmax')(attn)
attn = self.dropout(attn)
output = Lambda(lambda x:K.batch_dot(x[0], x[1]))([attn, v])
return output, attn
class MultiHeadAttention():
# mode 0 - big martixes, faster; mode 1 - more clear implementation
def __init__(self, n_head, d_model, d_k, d_v, dropout, mode=0, use_norm=True):
self.mode = mode
self.n_head = n_head
self.d_k = d_k
self.d_v = d_v
self.dropout = dropout
if mode == 0:
self.qs_layer = Dense(n_head*d_k, use_bias=False)
self.ks_layer = Dense(n_head*d_k, use_bias=False)
self.vs_layer = Dense(n_head*d_v, use_bias=False)
elif mode == 1:
self.qs_layers = []
self.ks_layers = []
self.vs_layers = []
for _ in range(n_head):
self.qs_layers.append(TimeDistributed(Dense(d_k, use_bias=False)))
self.ks_layers.append(TimeDistributed(Dense(d_k, use_bias=False)))
self.vs_layers.append(TimeDistributed(Dense(d_v, use_bias=False)))
self.attention = ScaledDotProductAttention(d_model)
self.layer_norm = LayerNormalization() if use_norm else None
self.w_o = TimeDistributed(Dense(d_model))
def __call__(self, q, k, v, mask=None):
d_k, d_v = self.d_k, self.d_v
n_head = self.n_head
if self.mode == 0:
qs = self.qs_layer(q) # [batch_size, len_q, n_head*d_k]
ks = self.ks_layer(k)
vs = self.vs_layer(v)
def reshape1(x):
s = tf.shape(x) # [batch_size, len_q, n_head * d_k]
x = tf.reshape(x, [s[0], s[1], n_head, d_k])
x = tf.transpose(x, [2, 0, 1, 3])
x = tf.reshape(x, [-1, s[1], d_k]) # [n_head * batch_size, len_q, d_k]
return x
qs = Lambda(reshape1)(qs)
ks = Lambda(reshape1)(ks)
vs = Lambda(reshape1)(vs)
if mask is not None:
mask = Lambda(lambda x:K.repeat_elements(x, n_head, 0))(mask)
head, attn = self.attention(qs, ks, vs, mask=mask)
def reshape2(x):
s = tf.shape(x) # [n_head * batch_size, len_v, d_v]
x = tf.reshape(x, [n_head, -1, s[1], s[2]])
x = tf.transpose(x, [1, 2, 0, 3])
x = tf.reshape(x, [-1, s[1], n_head*d_v]) # [batch_size, len_v, n_head * d_v]
return x
head = Lambda(reshape2)(head)
elif self.mode == 1:
heads = []; attns = []
for i in range(n_head):
qs = self.qs_layers[i](q)
ks = self.ks_layers[i](k)
vs = self.vs_layers[i](v)
head, attn = self.attention(qs, ks, vs, mask)
heads.append(head); attns.append(attn)
head = Concatenate()(heads) if n_head > 1 else heads[0]
attn = Concatenate()(attns) if n_head > 1 else attns[0]
outputs = self.w_o(head)
outputs = Dropout(self.dropout)(outputs)
if not self.layer_norm: return outputs, attn
# outputs = Add()([outputs, q]) # sl: fix
return self.layer_norm(outputs), attn
class PositionwiseFeedForward():
def __init__(self, d_hid, d_inner_hid, dropout=0.1):
self.w_1 = Conv1D(d_inner_hid, 1, activation='relu')
self.w_2 = Conv1D(d_hid, 1)
self.layer_norm = LayerNormalization()
self.dropout = Dropout(dropout)
def __call__(self, x):
output = self.w_1(x)
output = self.w_2(output)
output = self.dropout(output)
output = Add()([output, x])
return self.layer_norm(output)
class EncoderLayer():
def __init__(self, d_model, d_inner_hid, n_head, d_k, d_v, dropout=0.1):
self.self_att_layer = MultiHeadAttention(n_head, d_model, d_k, d_v, dropout=dropout)
self.pos_ffn_layer = PositionwiseFeedForward(d_model, d_inner_hid, dropout=dropout)
def __call__(self, enc_input, mask=None):
output, slf_attn = self.self_att_layer(enc_input, enc_input, enc_input, mask=mask)
output = self.pos_ffn_layer(output)
return output, slf_attn
def GetPosEncodingMatrix(max_len, d_emb):
pos_enc = np.array([
[pos / np.power(10000, 2 * (j // 2) / d_emb) for j in range(d_emb)]
if pos != 0 else np.zeros(d_emb)
for pos in range(max_len)
])
pos_enc[1:, 0::2] = np.sin(pos_enc[1:, 0::2]) # dim 2i
pos_enc[1:, 1::2] = np.cos(pos_enc[1:, 1::2]) # dim 2i+1
return pos_enc
def GetPadMask(q, k):
ones = K.expand_dims(K.ones_like(q, 'float32'), -1)
mask = K.cast(K.expand_dims(K.not_equal(k, 0), 1), 'float32')
mask = K.batch_dot(ones, mask, axes=[2,1])
return mask
def GetSubMask(s):
len_s = tf.shape(s)[1]
bs = tf.shape(s)[:1]
mask = K.cumsum(tf.eye(len_s, batch_shape=bs), 1)
return mask
class Transformer():
def __init__(self, len_limit, embedding_matrix, d_model=embed_size, \
d_inner_hid=512, n_head=10, d_k=64, d_v=64, layers=2, dropout=0.1, \
share_word_emb=False, **kwargs):
self.name = 'Transformer'
self.len_limit = len_limit
self.src_loc_info = False # True # sl: fix later
self.d_model = d_model
self.decode_model = None
d_emb = d_model
pos_emb = Embedding(len_limit, d_emb, trainable=False, \
weights=[GetPosEncodingMatrix(len_limit, d_emb)])
i_word_emb = Embedding(max_features, d_emb, weights=[embedding_matrix]) # Add Kaggle provided embedding here
self.encoder = Encoder(d_model, d_inner_hid, n_head, d_k, d_v, layers, dropout, \
word_emb=i_word_emb, pos_emb=pos_emb)
def get_pos_seq(self, x):
mask = K.cast(K.not_equal(x, 0), 'int32')
pos = K.cumsum(K.ones_like(x, 'int32'), 1)
return pos * mask
def compile(self, active_layers=999):
src_seq_input = Input(shape=(None, ))
x = Embedding(max_features, embed_size, weights=[embedding_matrix])(src_seq_input)
# LSTM before attention layers
x = Bidirectional(LSTM(128, return_sequences=True))(x)
x = Bidirectional(LSTM(64, return_sequences=True))(x)
x, slf_attn = MultiHeadAttention(n_head=3, d_model=300, d_k=64, d_v=64, dropout=0.1)(x, x, x)
avg_pool = GlobalAveragePooling1D()(x)
max_pool = GlobalMaxPooling1D()(x)
conc = concatenate([avg_pool, max_pool])
conc = Dense(64, activation="relu")(conc)
x = Dense(1, activation="sigmoid")(conc)
self.model = Model(inputs=src_seq_input, outputs=x)
self.model.compile(optimizer = 'adam', loss = 'mean_squared_error', metrics=['accuracy'])
print(tf.__version__)
!pip install -q h5py pyyaml
def build_model():
inp = Input(shape = (SEQ_LEN, 1))
# LSTM before attention layers
x = Bidirectional(LSTM(128, return_sequences=True))(inp)
x = Bidirectional(LSTM(64, return_sequences=True))(x)
x, slf_attn = MultiHeadAttention(n_head=3, d_model=300, d_k=64, d_v=64, dropout=0.1)(x, x, x)
avg_pool = GlobalAveragePooling1D()(x)
max_pool = GlobalMaxPooling1D()(x)
conc = concatenate([avg_pool, max_pool])
conc = Dense(64, activation="relu")(conc)
x = Dense(1, activation="sigmoid")(conc)
model = Model(inputs = inp, outputs = x)
model.compile(
loss = "mean_squared_error",
#optimizer = Adam(lr = config["lr"], decay = config["lr_d"]),
optimizer = "adam")
# Save entire model to a HDF5 file
model.save_weights('stock_predictor.h5')
return model
multi_head = build_model()
multi_head.summary()
multi_head.fit(X_train, y_train,
batch_size=BATCH_SIZE,
epochs=EPOCHS,
validation_data=(X_valid, y_valid),
#callbacks = [checkpoint , lr_reduce]
)
predicted_stock_price_multi_head = multi_head.predict(X_test)
#predicted_stock_price = scaler.inverse_transform(predicted_stock_price)
print(type(predicted_stock_price_multi_head))
X_test.shape
predicted_stock_price_multi_head = np.vstack((np.full((60,1), np.nan), predicted_stock_price_multi_head))
print(predicted_stock_price_multi_head[1000])
plt.figure(figsize = (18,9))
plt.plot(test_data, color = 'black', label = 'Ford Stock Price')
plt.plot(predicted_stock_price_multi_head, color = 'green', label = 'Predicted Ford Mid Price')
plt.title('Ford Mid Price Prediction', fontsize=30)
#plt.xticks(range(0,df.shape[0],50),df['Date'].loc[::50],rotation=0)
plt.xlabel('Date')
plt.ylabel('Ford Mid Price')
plt.legend(fontsize=18)
plt.show()
min_val = 1
max_val = 0
max_index = 0
min_index = 0
for i in range(0,len(predicted_stock_price_multi_head)):
if predicted_stock_price_multi_head[i] < min_val:
min_val = predicted_stock_price_multi_head[i]
min_index = i
if predicted_stock_price_multi_head[i] > max_val:
max_val = predicted_stock_price_multi_head[i]
max_index = i
risk = 0.2
if min_index<max_index:
covered_call_date = 0;
uncovered_put_date = min_index
expiry_call = min_index
expiry_put = max_index
strike_call = predicted_stock_price_multi_head[0] - (predicted_stock_price_multi_head[0] - min_val)*risk
strike_put = min_val + (max_val - min_val)*risk
premium_call = (predicted_stock_price_multi_head[0] - min_val)*risk
premium_put = (max_val - min_val)*risk
print("Write a Covered Call Option in ",covered_call_date," days with an expiry date in ",expiry_call," days, a strike price of $",strike_call," per stock and a premium of $",premium_call)
print("Write an Uncovered Put Option in ",uncovered_put_date," days with an expiry date in ",expiry_put," days, a strike price of $",strike_put," per stock and a premium of $",premium_put)
if max_index<min_index:
uncovered_put_date = 0;
covered_call_date = max_index
expiry_put = max_index
expiry_call = min_index
strike_put = predicted_stock_price_multi_head[1] + (max_val - predicted_stock_price_multi_head[1])*risk
strike_call = max_val - (max_val - min_val)*risk
premium_call = (max_val - predicted_stock_price_multi_head[1])*risk
premium_put = (max_val - min_val)*risk
print("Write an Uncovered Put Option in ",uncovered_put_date," days with an expiry date in ",expiry_put," days, a strike price of $",strike_put," per stock and a premium of $",premium_put)
print("Write a Covered Call Option in ",covered_call_date," days with an expiry date in ",expiry_call," days, a strike price of $",strike_call," per stock and a premium of $",premium_call)
cd ..
!pwd
!pip install simple_tensorflow_serving
simple_tensorflow_serving --model_base_path="./models/tensorflow_template_application_model"
###Output
_____no_output_____
|
mlb_asg_team_performance/Main.ipynb
|
###Markdown
Follow D3.Paracoord format In the scope of tidy data how should this be handled... Time series is special type of parallel coord https://syntagmatic.github.io/parallel-coordinates/examples/slickgrid.html
###Code
df=pd.read_csv("data/paracoord_season.csv")
del df['win_prct']
del df['opp_ID']
df.head(1)
#https://plot.ly/python/parallel-coordinates-plot/
import plotly.plotly as py
import plotly.graph_objs as go
import plotly
plotly.tools.set_credentials_file(username='ergosum_person',api_key='vSemgUiRmnJ5wWzeq44r')
from math import ceil, floor
N=len(df)
min_val=floor(min(df['win_prct'])*100)/100.
max_val=ceil(max(df['win_prct'])*100)/100.
[min_val,max_val]
N=len(paracoord_df)
data = [
go.Parcoords(
dimensions=[
dict(label='Starting Percent', values=df['starting_prct'], range=[min_val,max_val]),
dict(label='Before ASG Percent', values=df['before_asg_prct'], range=[min_val,max_val]),
dict(label='After ASG Percent',values=df['after_asg_prct'], range=[min_val,max_val]),
dict(label='Final Percent',values=df['final_prct'],range=[min_val,max_val])
]
)
]
py.iplot(data, filename='basic-asg')
df.columns
for col in ["wonWorldSeries", "madePlayoffs", "wonPennant"]:
df[col]=df[col].apply(lambda x: 1 if x else "0")
df=df[['original_team-id', 'SEASON', 'team_ID', 'wins', 'losses', 'games_played', 'starting_prct',
'before_asg_prct', 'after_asg_prct', 'final_prct', 'madePlayoffs', 'wonPennant',
'wonWorldSeries']]
df=df.sort_values(by=["SEASON", "original_team-id"],ascending=[False, True])
df.rename(columns={
"madePlayoffs": "Playoffs",
"wonPennant": "Pennant",
"wonWorldSeries": "World Series",
"starting_prct": "Start Win%",
"before_asg_prct": "Pre-ASG%",
"after_asg_prct": "Post-ASG%",
"final_prct": "Final%",
"SEASON": "Season",
"original_team-id": "Team",
"team_ID": "Historic Team ID"
}).to_csv("data/paracoord_asg_vals.csv",index=False)
df.rename(columns={
"madePlayoffs": "Made Playoffs",
"wonPennant": "Won Pennant",
"wonWorldSeries": "Won World Series",
"starting_prct": "Starting Win %",
"before_asg_prct": "Before ASG Win %",
"after_asg_prct": "After ASG Win %",
"final_prct": "Final Win %",
"SEASON": "Season",
"original_team-id": "Team Abbreviation",
"team_ID": "Historic Team ID"
}).columns
###Output
_____no_output_____
|
examples/DSB2018/StarDist_Baseline.ipynb
|
###Markdown
This notebook performs segmentation with a StarDist Network
###Code
# We import all our dependencies.
from __future__ import print_function, unicode_literals, absolute_import, division
import warnings
warnings.filterwarnings('ignore')
import sys
sys.path.append('../../')
import numpy as np
from csbdeep.utils import plot_history, Path, normalize
from voidseg.utils.misc_utils import shuffle_train_data, augment_data
from voidseg.utils.seg_utils import fractionate_train_data
from voidseg.utils.compute_precision_threshold import compute_threshold, precision
from matplotlib import pyplot as plt
%matplotlib inline
%config InlineBackend.figure_format = 'retina'
from scipy import ndimage
from tqdm import tqdm, tqdm_notebook
from stardist import fill_label_holes, random_label_cmap, calculate_extents, gputools_available, _draw_polygons
from stardist.models import Config2D, StarDist2D
np.random.seed(42)
lbl_cmap = random_label_cmap()
import tensorflow as tf
import keras.backend as K
import urllib, os, zipfile, sys
###Output
Using TensorFlow backend.
###Markdown
Download DSB2018 data.From the Kaggle 2018 Data Science Bowl challenge, we take the same subset of data as has been used [here](https://github.com/mpicbg-csbd/stardist), showing a diverse collection of cell nuclei imaged by various fluorescence microscopes. We extracted 4870 image patches of size 128×128 from the training set and added Gaussian noise with mean 0 and sigma = 10 (n10), 20 (n20) and 40 (n40). This notebook shows results for n40 images.
###Code
# create a folder for our data
if not os.path.isdir('./data'):
os.mkdir('data')
# check if data has been downloaded already
zipPath="data/DSB.zip"
if not os.path.exists(zipPath):
#download and unzip data
data = urllib.request.urlretrieve('https://owncloud.mpi-cbg.de/index.php/s/LIN4L4R9b2gebDX/download', zipPath)
with zipfile.ZipFile(zipPath, 'r') as zip_ref:
zip_ref.extractall("data")
###Output
_____no_output_____
###Markdown
The downloaded data is in `npz` format and the cell below extracts the training, validation and test data as numpy arrays
###Code
trainval_data = np.load('data/DSB/train_data/dsb2018_TrainVal40.npz')
test_data = np.load('data/DSB/test_data/dsb2018_Test40.npz', allow_pickle=True)
train_images = trainval_data['X_train']
val_images = trainval_data['X_val']
test_images = test_data['X_test']
train_masks = trainval_data['Y_train']
val_masks = trainval_data['Y_val']
test_masks = test_data['Y_test']
print("Shape of train_images: ", train_images.shape, ", Shape of train_masks: ", train_masks.shape)
print("Shape of val_images: ", val_images.shape, ", Shape of val_masks: ", val_masks.shape)
print("Shape of test_images: ", test_images.shape, ", Shape of test_masks: ", test_masks.shape)
###Output
Shape of train_images: (3800, 128, 128) , Shape of train_masks: (3800, 128, 128)
Shape of val_images: (670, 128, 128) , Shape of val_masks: (670, 128, 128)
Shape of test_images: (50,) , Shape of test_masks: (50,)
###Markdown
Data preparation for segmentation stepNext, we shuffle the raw training images and the correponding Ground Truth (GT). Then, we fractionate the training pairs of raw images and corresponding GT to realize the case where not enough annotated, training data is available. For this fractionation, please specify `fraction` parameter below. It should be between 0 (exclusive) and 100 (inclusive).
###Code
fraction = 2 # Fraction of annotated GT and raw image pairs to use during training.
random_seed = 1 # Seed to shuffle training data (annotated GT and raw image pairs).
assert 0 <fraction<= 100, "Fraction should be between 0 and 100"
X_shuffled, Y_shuffled = shuffle_train_data(train_images, train_masks, random_seed = random_seed)
X_frac, Y_frac = fractionate_train_data(X_shuffled, Y_shuffled, fraction = 2)
print("Training Data \n..................")
X_train_aug, Y_train_aug = augment_data(X_frac, Y_frac)
print("\n")
print("Validation Data \n..................")
X_val_aug, Y_val_aug = augment_data(val_images, val_masks)
###Output
Training Data
..................
Raw image size after augmentation (608, 128, 128)
Mask size after augmentation (608, 128, 128)
Validation Data
..................
Raw image size after augmentation (5360, 128, 128)
Mask size after augmentation (5360, 128, 128)
###Markdown
Next, we do percentile normalization and fill holes in labels
###Code
X_train = [normalize(x,1,99.8) for x in tqdm_notebook(X_train_aug)]
Y_train = [fill_label_holes(y.astype(np.uint16)) for y in tqdm_notebook(Y_train_aug)]
X_val = [normalize(x,1,99.8) for x in tqdm_notebook(X_val_aug)]
Y_val = [fill_label_holes(y.astype(np.uint16)) for y in tqdm_notebook(Y_val_aug)]
X_train_filtered = []
Y_train_filtered = []
X_val_filtered = []
Y_val_filtered = []
for i in range(len(Y_train)):
if(np.max(Y_train[i])==0):
continue
else:
X_train_filtered.append(X_train[i])
Y_train_filtered.append(Y_train[i])
for i in range(len(Y_val)):
if(np.max(Y_val[i])==0):
continue
else:
X_val_filtered.append(X_val[i])
Y_val_filtered.append(Y_val[i])
###Output
_____no_output_____
###Markdown
Let's look at one of our training image and corresponding GT.
###Code
i = min(10, len(X_train)-1)
img, lbl = X_train[i], Y_train[i]
assert img.ndim in (2,3)
img = img if img.ndim==2 else img[...,:3]
plt.figure(figsize=(16,10))
plt.subplot(121); plt.imshow(img,cmap='gray'); plt.axis('off'); plt.title('Raw image')
plt.subplot(122); plt.imshow(lbl,cmap=lbl_cmap); plt.axis('off'); plt.title('GT labels')
None;
###Output
_____no_output_____
###Markdown
Configure StarDist NetworkThe data preparation for segmentation is now done. Next, we configure a StarDist network by specifying `Config2D` parameters. For example, one can increase `train_epochs` to get even better results at the expense of a longer computation. (This holds usually true for a large `fraction`.)
###Code
n_rays = 32
n_channel = 1
train_batch_size = 128
train_epochs = 15
train_learning_rate = 0.0004
train_patch_size = (64, 64)
train_reduce_lr={'factor': 0.5, 'patience': 10, 'min_delta': 0}
unet_batch_norm = True
unet_n_depth = 4
train_steps_per_epoch=400
# Use OpenCL-based computations for data generator during training (requires 'gputools')
use_gpu = True and gputools_available()
# Predict on subsampled grid for increased efficiency and larger field of view
grid = (2,2)
conf = Config2D (
n_rays = n_rays,
grid = grid,
use_gpu = use_gpu,
n_channel_in = n_channel,
train_batch_size = train_batch_size,
train_epochs = train_epochs,
train_steps_per_epoch=train_steps_per_epoch,
train_learning_rate = train_learning_rate,
train_reduce_lr = train_reduce_lr,
train_patch_size = train_patch_size,
unet_batch_norm = unet_batch_norm,
unet_n_depth = unet_n_depth,
)
print(conf)
vars(conf)
if use_gpu:
from csbdeep.utils.tf import limit_gpu_memory
# adjust as necessary: limit GPU memory to be used by TensorFlow to leave some to OpenCL-based computations
limit_gpu_memory(1.0)
###Output
_____no_output_____
###Markdown
Now, we begin training the model for segmentation.
###Code
model = StarDist2D(conf, name='stardist_baseline', basedir='models')
hist = model.train(X_train_filtered, Y_train_filtered, validation_data=(X_val_filtered,Y_val_filtered), augmenter=None)
###Output
Epoch 1/15
400/400 [==============================] - 90s 224ms/step - loss: 0.8887 - prob_loss: 0.1101 - dist_loss: 3.8928 - prob_kld: 0.0427 - dist_relevant_mae: 3.8925 - dist_relevant_mse: 42.4797 - val_loss: 0.7222 - val_prob_loss: 0.1401 - val_dist_loss: 2.9104 - val_prob_kld: 0.0598 - val_dist_relevant_mae: 2.9100 - val_dist_relevant_mse: 21.5343
Epoch 2/15
400/400 [==============================] - 85s 212ms/step - loss: 0.4873 - prob_loss: 0.0896 - dist_loss: 1.9888 - prob_kld: 0.0223 - dist_relevant_mae: 1.9884 - dist_relevant_mse: 10.7584 - val_loss: 0.6377 - val_prob_loss: 0.1141 - val_dist_loss: 2.6181 - val_prob_kld: 0.0339 - val_dist_relevant_mae: 2.6178 - val_dist_relevant_mse: 18.5815
Epoch 3/15
400/400 [==============================] - 85s 212ms/step - loss: 0.4186 - prob_loss: 0.0871 - dist_loss: 1.6578 - prob_kld: 0.0198 - dist_relevant_mae: 1.6573 - dist_relevant_mse: 7.4556 - val_loss: 0.6147 - val_prob_loss: 0.1200 - val_dist_loss: 2.4739 - val_prob_kld: 0.0397 - val_dist_relevant_mae: 2.4734 - val_dist_relevant_mse: 15.7806
Epoch 4/15
400/400 [==============================] - 84s 211ms/step - loss: 0.3786 - prob_loss: 0.0852 - dist_loss: 1.4670 - prob_kld: 0.0179 - dist_relevant_mae: 1.4666 - dist_relevant_mse: 5.8569 - val_loss: 0.5639 - val_prob_loss: 0.1177 - val_dist_loss: 2.2309 - val_prob_kld: 0.0374 - val_dist_relevant_mae: 2.2304 - val_dist_relevant_mse: 14.4686
Epoch 5/15
400/400 [==============================] - 86s 214ms/step - loss: 0.3491 - prob_loss: 0.0833 - dist_loss: 1.3290 - prob_kld: 0.0162 - dist_relevant_mae: 1.3286 - dist_relevant_mse: 4.8542 - val_loss: 0.5940 - val_prob_loss: 0.1185 - val_dist_loss: 2.3772 - val_prob_kld: 0.0383 - val_dist_relevant_mae: 2.3767 - val_dist_relevant_mse: 16.0229
Epoch 6/15
400/400 [==============================] - 84s 210ms/step - loss: 0.3299 - prob_loss: 0.0826 - dist_loss: 1.2366 - prob_kld: 0.0153 - dist_relevant_mae: 1.2361 - dist_relevant_mse: 4.1833 - val_loss: 0.5205 - val_prob_loss: 0.1124 - val_dist_loss: 2.0406 - val_prob_kld: 0.0322 - val_dist_relevant_mae: 2.0401 - val_dist_relevant_mse: 12.4680
Epoch 7/15
400/400 [==============================] - 85s 214ms/step - loss: 0.3114 - prob_loss: 0.0815 - dist_loss: 1.1493 - prob_kld: 0.0143 - dist_relevant_mae: 1.1489 - dist_relevant_mse: 3.6209 - val_loss: 0.5602 - val_prob_loss: 0.1158 - val_dist_loss: 2.2219 - val_prob_kld: 0.0356 - val_dist_relevant_mae: 2.2215 - val_dist_relevant_mse: 14.3200
Epoch 8/15
400/400 [==============================] - 84s 211ms/step - loss: 0.3027 - prob_loss: 0.0814 - dist_loss: 1.1067 - prob_kld: 0.0139 - dist_relevant_mae: 1.1062 - dist_relevant_mse: 3.3401 - val_loss: 0.5257 - val_prob_loss: 0.1110 - val_dist_loss: 2.0734 - val_prob_kld: 0.0308 - val_dist_relevant_mae: 2.0728 - val_dist_relevant_mse: 12.9985
Epoch 9/15
400/400 [==============================] - 84s 211ms/step - loss: 0.2899 - prob_loss: 0.0800 - dist_loss: 1.0496 - prob_kld: 0.0131 - dist_relevant_mae: 1.0491 - dist_relevant_mse: 3.0127 - val_loss: 0.5448 - val_prob_loss: 0.1161 - val_dist_loss: 2.1432 - val_prob_kld: 0.0359 - val_dist_relevant_mae: 2.1427 - val_dist_relevant_mse: 13.4452
Epoch 10/15
400/400 [==============================] - 86s 214ms/step - loss: 0.2824 - prob_loss: 0.0799 - dist_loss: 1.0127 - prob_kld: 0.0126 - dist_relevant_mae: 1.0122 - dist_relevant_mse: 2.8060 - val_loss: 0.5195 - val_prob_loss: 0.1120 - val_dist_loss: 2.0376 - val_prob_kld: 0.0317 - val_dist_relevant_mae: 2.0372 - val_dist_relevant_mse: 12.9759
Epoch 11/15
400/400 [==============================] - 86s 215ms/step - loss: 0.2743 - prob_loss: 0.0793 - dist_loss: 0.9753 - prob_kld: 0.0122 - dist_relevant_mae: 0.9747 - dist_relevant_mse: 2.6316 - val_loss: 0.6088 - val_prob_loss: 0.1234 - val_dist_loss: 2.4268 - val_prob_kld: 0.0432 - val_dist_relevant_mae: 2.4261 - val_dist_relevant_mse: 17.2611
Epoch 12/15
400/400 [==============================] - 85s 213ms/step - loss: 0.2683 - prob_loss: 0.0794 - dist_loss: 0.9445 - prob_kld: 0.0118 - dist_relevant_mae: 0.9440 - dist_relevant_mse: 2.4791 - val_loss: 0.5200 - val_prob_loss: 0.1153 - val_dist_loss: 2.0233 - val_prob_kld: 0.0351 - val_dist_relevant_mae: 2.0228 - val_dist_relevant_mse: 12.5230
Epoch 13/15
400/400 [==============================] - 85s 213ms/step - loss: 0.2631 - prob_loss: 0.0786 - dist_loss: 0.9229 - prob_kld: 0.0115 - dist_relevant_mae: 0.9224 - dist_relevant_mse: 2.3691 - val_loss: 0.5335 - val_prob_loss: 0.1180 - val_dist_loss: 2.0773 - val_prob_kld: 0.0378 - val_dist_relevant_mae: 2.0768 - val_dist_relevant_mse: 13.1599
Epoch 14/15
400/400 [==============================] - 85s 212ms/step - loss: 0.2584 - prob_loss: 0.0782 - dist_loss: 0.9009 - prob_kld: 0.0111 - dist_relevant_mae: 0.9004 - dist_relevant_mse: 2.2503 - val_loss: 0.7026 - val_prob_loss: 0.1304 - val_dist_loss: 2.8611 - val_prob_kld: 0.0501 - val_dist_relevant_mae: 2.8607 - val_dist_relevant_mse: 29.7843
Epoch 15/15
400/400 [==============================] - 85s 212ms/step - loss: 0.2539 - prob_loss: 0.0782 - dist_loss: 0.8781 - prob_kld: 0.0108 - dist_relevant_mae: 0.8775 - dist_relevant_mse: 2.1436 - val_loss: 0.5786 - val_prob_loss: 0.1221 - val_dist_loss: 2.2824 - val_prob_kld: 0.0418 - val_dist_relevant_mae: 2.2818 - val_dist_relevant_mse: 16.3519
Loading network weights from 'weights_best.h5'.
###Markdown
Computing the best threshold on validation images (to maximize Average Precision score). The threshold so obtained will be used to get hard masks from probability images to be predicted on test images.
###Code
threshold=compute_threshold(np.array(X_val_filtered), np.array(Y_val_filtered), model, mode = "StarDist")
###Output
Computing best threshold:
###Markdown
Prediction on test images to get segmentation result
###Code
X_test_normalized = [normalize(x,1,99.8) for x in tqdm_notebook(test_images)]
precision_result = []
prediction_images = []
for i in range(len(X_test_normalized)):
predicted_image, _ = model.predict_instances(X_test_normalized[i], prob_thresh=threshold)
precision_result.append(precision(test_masks[i],predicted_image))
prediction_images.append(predicted_image)
print("Average precision over all test images at IOU = 0.5: ", np.mean(precision_result))
plt.figure(figsize=(10,10))
plt.subplot(1,2,1)
plt.imshow(prediction_images[22])
plt.title('Prediction')
plt.subplot(1,2,2)
plt.imshow(test_masks[22])
plt.title('Ground Truth')
###Output
_____no_output_____
|
NoteRecognition.ipynb
|
###Markdown
###Code
import numpy as np
import wave
import struct
sampling_freq = 44100 #Sampling frequency of audio signal
def freqToNote(freq) : #convert freq to note
if(freq > 977 and freq < 1100):
return "C6"
if(freq >= 1100 and freq < 1244):
return "D6"
if(freq >= 1244 and freq < 1355):
return "E6"
if(freq >= 1355 and freq < 1479):
return "F6"
if(freq >= 1479 and freq < 1661):
return "G6"
if(freq >= 1661 and freq < 1864):
return "A6"
if(freq >= 1864 and freq < 2030):
return "B6"
if(freq >= 2030 and freq < 2217.46):
return "C7"
if(freq >= 2217.46 and freq < 2489.02):
return "D7"
if(freq >= 2489.02 and freq < 2700):
return "E7"
if(freq >= 2700 and freq < 2959.96):
return "F7"
if(freq >= 2959.96 and freq < 3322.44):
return "G7"
if(freq >= 3322.44 and freq < 3729.31):
return "A7"
if(freq >= 3729.31 and freq < 4050):
return "B7"
if(freq >= 4050 and freq < 4434.92):
return "C8"
if(freq >= 4434.92 and freq < 4978.03):
return "D8"
if(freq >= 4978.03 and freq < 5370):
return "E8"
if(freq >= 5370 and freq < 5919.91):
return "F8"
if(freq >= 5919.91 and freq < 6644.88):
return "G8"
if(freq >= 6644.88 and freq < 7458.62):
return "A8"
if(freq >= 7458.62 and freq < 8000):
return "B8"
def play(sound_file):
file_length = sound_file.getnframes()
sound = np.zeros(file_length)
for i in range(file_length):
data = sound_file.readframes(1)
data = struct.unpack("<h", data)
sound[i] = int(data[0])
sound = np.divide(sound, float(2**15))
Identified_Notes = [] #return value
threshold = 0 #assuming no noise
flag = 0 #0 for continued silence, 1 for note to silence
Indices = [] #all indices of sound, for one note
frame_length = int(sampling_freq * 0.02)
for i in range(0, file_length-frame_length, frame_length):
temp = max(sound[i: i + frame_length])
if temp > threshold: #continued note
for k in range(frame_length):
Indices.append(i + k) #append indexes in current frame
flag = 1
elif ((flag == 1) or (flag == 1 and i == file_length - frame_length - 1)): #found beginning of silence
flag = 0
Note = np.take(sound, Indices) #take all values of sound at indexes, in Indices
dftNote = np.fft.fft(Note) #fft
Imax = dftNote.argsort()[::-1][:2] #to sort in descending order and take 0th and 1st ele because two peaks
x = min(Imax[0], Imax[1])
freq = ((x * sampling_freq) / len(Indices))
Indices = [] #empty indices for next note
Identified_Notes.append(freqToNote(freq))
return Identified_Notes
#-----------------Main--------------
sound_file = wave.open('/content/drive/MyDrive/Colab Notebooks/NoteRecognition/Audio.wav')
Identified_Notes = play(sound_file)
print(Identified_Notes)
###Output
['D6', 'C8', 'B8', 'G8', 'G7', 'C6', 'C6', 'C6', 'A7', 'D6', 'C6', 'C8', 'F8', 'A7', 'A8']
|
data/Import_RECR.ipynb
|
###Markdown
General information
###Code
import subprocess
def get_git_revision_hash():
return subprocess.check_output(['git', 'rev-parse', 'HEAD']).split()[0]
f.attrs['COMMITID'] = get_git_revision_hash()
f.attrs['DETECTORS'] = [s.encode() for s in DETECTORS]
###Output
_____no_output_____
###Markdown
Preparation data The matrix to be recovered
###Code
# gauss or RECR doesn't matter for this
targetfile = BASEDIR + '/Unitaries_col_phases/%s.dat' % ID
target = load_complex_array(targetfile)
imsshow([target.real, target.imag, np.abs(target)])
f['TARGET'] = target
f['TARGETFILE'] = targetfile
outfile.flush()
print(target)
def load_pvecs(fnames, prepvecs):
for fname in fnames:
vecid = split(splitext(fname)[0])[1]
prepvecs[vecid] = load_complex_array(fname)
prepvecs[vecid].attrs['FILE'] = fname
outfile.flush()
fnames = glob(BASEDIR + '/Vectors/VRad%i_*.dat' % DIM)
prepvecs = f.create_group('PREPVECS')
print("Number of RECR preparation vectors: %i" % len(fnames))
load_pvecs(fnames, prepvecs)
###Output
_____no_output_____
###Markdown
Phaselift Raw Measurement Data
###Code
def load_deteff(fname):
summarydict = parse_ctx(fname)
return np.array([summarydict['det_eff'][det] for det in DETECTORS])
def dict_to_rates(dic):
return np.array([dic.get(det.lower(), 0) for det in DETECTORS])
def load_counts(fname):
summarydict = parse_ctx(fname)
rates = np.array([summarydict[det] for det in DETECTORS])
parent = summarydict['metadata']['parent']
path_to_raw = join(split(fname)[0], '..', 'raw', parent + '.ctx')
try:
c = parse_ctx(path_to_raw)
raw_rates = np.array([dict_to_rates(val)
for key, val in c.items()
if key.startswith('count_rates')])
assert np.all(np.sum(raw_rates, axis=0) == rates)
return raw_rates, path_to_raw
except FileNotFoundError:
return rates[None, :], fname
def vector_to_counts(globpatt):
matches = glob(globpatt)
if len(matches) != 1:
raise IOError("Wrong number of matches %i" % len(matches))
return load_counts(matches[0])
deteff_file = BASEDIR + '/data/det_eff/det_eff.txt'
deteff_all = {name: value
for name, value in zip(DETECTORS_ALL, np.loadtxt(deteff_file))}
deteff = np.array([deteff_all[key] for key in DETECTORS])
rawcounts = f.create_group('RAWCOUNTS')
for pvec in f['PREPVECS'].keys():
globpatt = BASEDIR + '/data/singles/summed_sorted/%s_%s.ctx' % (ID, pvec)
try:
counts, fname = vector_to_counts(globpatt)
rawcounts[pvec] = counts
rawcounts[pvec].attrs['FILE'] = fname
except (IOError) as e:
print(e)
print("Loaded data for {} vectors.".format(len(rawcounts)))
print("First element has shape {}".format(next(iter(rawcounts.values())).shape))
rawcounts.attrs['DETEFF'] = deteff
rawcounts.attrs['DETEFF_FILE'] = deteff_file
outfile.flush()
###Output
_____no_output_____
###Markdown
Reference Data Single Photon DataBeware: sometimes they are in the wrong order, i.e. singles_1 corresponds to the 5th column
###Code
def get_colcounts(col_nr):
globpatt = BASEDIR + '/data/singles/summed_sorted/'
globpatt += '%s_S%i_%.2i.ctx' % (ID, DIM, col_nr)
matches = glob(globpatt)
assert len(matches) == 1, "It's actually {} for {}"\
.format(len(matches), col_nr)
summarydict = parse_ctx(matches[0])
return np.array([summarydict[det] for det in DETECTORS]), matches[0]
single_counts = f.create_group('SINGLE_COUNTS')
for n in range(1, len(DETECTORS) + 1):
count, fname = get_colcounts(n)
# carefull! Sometimes they are the wrong way around!
index = n - 1
single_counts[str(index)] = count
single_counts[str(index)].attrs['FILE'] = fname
# since they were taken at the same time
single_counts.attrs['DETEFF'] = f['RAWCOUNTS'].attrs['DETEFF']
single_counts.attrs['DETEFF_FILE'] = f['RAWCOUNTS'].attrs['DETEFF_FILE']
outfile.flush()
###Output
_____no_output_____
###Markdown
To check, we plot something proportional to the singles-transfer matrix. Note that we have to transpose counts since the single_counts[i] refer to columns of the transfer matrix.
###Code
counts = np.array([single_counts[str(i)] for i in range(len(DETECTORS))],
dtype=np.float64)
counts *= single_counts.attrs['DETEFF']
ax, *_ = imsshow([np.sqrt(counts).T])
pl.colorbar(ax.images[0])
pl.show()
ax, *_ = imsshow([np.abs(target)])
pl.colorbar(ax.images[0])
pl.show()
###Output
_____no_output_____
###Markdown
Load reference data
###Code
DETECTORS = f.attrs['DETECTORS']
# Average total photon count (for normalization purposes)
tmat_single = np.array([f['SINGLE_COUNTS'][str(i)] for i in range(len(DETECTORS))], dtype=float)
deteff = f['SINGLE_COUNTS'].attrs['DETEFF']
tmat_single = tmat_single * deteff
# axis = 0 since we flip the tmat later
tmat_single /= np.max(np.sum(tmat_single, axis=0))
tmat_single = np.sqrt(tmat_single.T)
f['TMAT_SINGLE'] = tmat_single
###Output
_____no_output_____
###Markdown
Also, load the reconstruction using singles & dips (data missing, fill in in Sec. Dips)
###Code
try:
recons = load_complex_array(BASEDIR + '/dip_reconstruction/%s_diprecon.dat' % ID)
f['DIP_RECONSTRUCTED'] = recons
outfile.flush()
imsshow([recons.real, recons.imag, np.abs(recons)])
pl.show()
imsshow([target.real, target.imag, np.abs(target)])
pl.show()
except FileNotFoundError:
print("Dip reconstruction not found")
###Output
_____no_output_____
|
python/latex_examples.ipynb
|
###Markdown
https://towardsdatascience.com/write-markdown-latex-in-the-jupyter-notebook-10985edb91fd \begin{matrix}1 & 2 & 3\\a & b & c\end{matrix} Round brackets:\begin{pmatrix}1 & 2 & 3\\a & b & c\end{pmatrix} Pipes:\begin{vmatrix}1 & 2 & 3\\a & b & c\end{vmatrix} \begin{align}\begin{vmatrix}\cos(\theta) & -\sin(\theta) \\\sin(\theta) & \cos(\theta)\end{vmatrix}\begin{vmatrix} x \\ y \end{vmatrix} = \begin{vmatrix}x \cos(\theta) - y \sin(\theta) \\x \sin(\theta) + y \cos(\theta)\end{vmatrix}\end{align} $$a_{ij}$$
###Code
%%latex
this does not work for me, but supposedly it does.
\documentclass{article}
\usepackage[margin=1in]{geometry}% Just for this example
\setlength{\parindent}{0pt}% Just for this example
\begin{document}
There are a number of horizontal spacing macros for LaTeX:
\begin{tabular}{lp{5cm}}
\verb|a\,b| & a\,b \quad $a\, b$ \\
\verb|a\thinspace b| & a\thinspace b \quad $a\thinspace b$ \\
\verb|a\!b| & a\!b \quad $a\!b$ \\
\verb|a\negthinspace b| & a\negthinspace b \quad $a\negthinspace b$ \\
\verb|a\:b| & a\:b \quad $a\:b$ \\
\verb|a\>b| & a\>b \quad $a\>b$ \\
\verb|a\medspace b| & a\medspace b \quad $a\medspace b$ \\
\verb|a\negmedspace b| & a\negmedspace b \quad $a\negmedspace b$ \\
\verb|a\;b| & a\;b \quad $a\;b$ \\
\verb|a\thickspace b| & a\thickspace b \quad $a\thickspace b$ \\
\verb|a\negthickspace b| & a\negthickspace b \quad $a\negthickspace b$ \\
\verb|$a\mkern\thinmuskip b$| & $a\mkern\thinmuskip b$ (similar to \verb|\,|) \\
\verb|$a\mkern-\thinmuskip b$| & $a\mkern-\thinmuskip b$ (similar to \verb|\!|) \\
\verb|$a\mkern\medmuskip b$| & $a\mkern\medmuskip b$ (similar to \verb|\:| or \verb|\>|) \\
\verb|$a\mkern-\medmuskip b$| & $a\mkern-\medmuskip b$ (similar to \verb|\negmedspace|) \\
\verb|$a\mkern\thickmuskip b$| & $a\mkern\thickmuskip b$ (similar to \verb|\;|) \\
\verb|$a\mkern-\thickmuskip b$| & $a\mkern-\thickmuskip b$ (similar to \verb|\negthickspace|) \\
\verb|a\enspace b| & a\enspace b \\
\verb|$a\enspace b$| & $a\enspace b$ \\
\verb|a\quad b| & a\quad b \\
\verb|$a\quad b$| & $a\quad b$ \\
\verb|a\qquad b| & a\qquad b \\
\verb|$a\qquad b$| & $a\qquad b$ \\
\verb|a\hskip 1em b| & a\hskip 1em b \\
\verb|$a\hskip 1em b$| & $a\hskip 1em b$ \\
\verb|a\kern 1pc b| & a\kern 1pc b \\
\verb|$a\kern 1pc b$| & $a\kern 1pc b$ \\
\verb|$a\mkern 17mu b$| & $a\mkern 17mu b$ \\
\verb|a\hspace{35pt}b| & a\hspace{35pt}b \\
\verb|$a\hspace{35pt}b$| & $a\hspace{35pt}b$ \\
\verb|axyzb| & axyzb \\
\verb|a\hphantom{xyz}b| & a\hphantom{xyz}b (or just \verb|\phantom|) \\
\verb|$axyzb$| & $axyzb$ \\
\verb|$a\hphantom{xyz}b$| & $a\hphantom{xyz}b$ (or just \verb|\phantom|) \\
\verb|a b| & a b \\
\verb|$a b$| & $a b$ \\
\verb|a\space b| & a\space b \\
\verb|$a\space b$| & $a\space b$ \\
\verb|a\ b| & a\ b \\
\verb|$a\ b$| & $a\ b$ \\
\verb|a{ }b| & a{ }b \\
\verb|$a{ }b$| & $a{ }b$ \\
\verb|a~b| & a~b \\
\verb|$a~b$| & $a~b$ \\
\verb|a\hfill b| & a\hfill b \\
\verb|$a\hfill b$| & $a\hfill b$
\end{tabular}
\end{document}
###Output
_____no_output_____
|
generate_ts.ipynb
|
###Markdown
Time series generator for tensorseason
###Code
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
import warnings
from functools import reduce
def generate_trend(T, changepoint_probability=.0005, slope_std = .01):
cp = np.random.binomial(n=1, p=changepoint_probability, size=T-2)
cps = [0] + list(np.arange(T-2)[cp.astype(bool)]) + [T-1]
slopes = np.random.randn(len(cps)-1) * slope_std
t_t = np.zeros(T)
for i in range(len(cps)-1):
slope = slopes[i]
no_days = cps[i+1] - cps[i] + 1
t_t[cps[i]:cps[i+1]+1] = np.linspace(t_t[cps[i]], t_t[cps[i]] + slope * no_days, no_days)
return t_t
def generate_sinusoid_seasonality(T, seasons, no_seasonality_components):
s_t = np.zeros(T)
seasons_prod = [np.prod(seasons[:i+1]) for i in range(len(seasons))]
amplitudes = np.random.standard_t(4, size=(len(seasons_prod), no_seasonality_components, 2))
for si, P in enumerate(seasons_prod):
for n_1 in range(0, no_seasonality_components):
a_n = amplitudes[si, n_1, 0]
b_n = amplitudes[si, n_1, 1]
s_t += a_n * np.exp(np.sin((2 * np.pi * (n_1+1) * np.arange(T))/P))
s_t += b_n * np.exp(np.cos((2 * np.pi * (n_1+1) * np.arange(T))/P))
return s_t
def generate_cp_seasonality(T, seasons, no_seasonality_components, nonnegative):
mult = 1 if nonnegative else -1
s_t = np.zeros(T)
s_t_block = np.zeros(np.prod(seasons))
for n in range(0, no_seasonality_components):
comp_vectors = [np.random.poisson(.5, size=seasons[i]) * np.random.choice([mult, 1], size=seasons[i]) for i in range(len(seasons))]
s_t_block += reduce(np.outer, comp_vectors).ravel()
s_t = np.tile(s_t_block, len(s_t)//len(s_t_block)) # assumes total T is a multiple of whatever
return s_t
def scale_s_t(s_t, t_t, seasonality_snr):
snr_hat = np.mean(s_t ** 2) / np.mean(t_t ** 2)
coef = np.sqrt(snr_hat / seasonality_snr)
return s_t * (1/coef)
def generate_seasonality(seasons, seasonality_type, t_t, seasonality_snr, no_seasonality_components):
T = len(t_t)
s_t = np.zeros(T)
no_seasonality_components
if seasonality_type == "sinusoid":
s_t = generate_sinusoid_seasonality(T, seasons, no_seasonality_components)
elif seasonality_type == "cp":
s_t = generate_cp_seasonality(T, seasons, no_seasonality_components, nonnegative=False)
elif seasonality_type == "nonnegative_cp":
s_t = generate_cp_seasonality(T, seasons, no_seasonality_components, nonnegative=True)
else:
raise ValueError("Seasonality type {} is not implemented".format(seasonality_type))
s_t = scale_s_t(s_t, t_t, seasonality_snr)
return s_t
def generate_noise(t_t, s_t, noise_snr):
T = len(t_t)
n_t = np.zeros(T)
signal_t = t_t + s_t
n_hat = np.random.randn(T)
snr_hat = np.mean(signal_t ** 2) / np.mean(n_hat ** 2)
noise_coef = np.sqrt(snr_hat / noise_snr)
n_t = n_hat * noise_coef
return n_t
def plot_ts(x_t, t_t, s_t, n_t):
with warnings.catch_warnings():
warnings.simplefilter("ignore")
fig, axes = plt.subplots(4, 1,sharex=True, sharey=True, figsize=(15,10))
pd.Series(x_t).plot(ax=axes[0])
pd.Series(t_t).plot(ax=axes[1])
pd.Series(s_t).plot(ax=axes[2])
pd.Series(n_t).plot(ax=axes[3])
axes[3].set_xlabel("Time")
fig.tight_layout()
fig.suptitle(r"Seasonality/Trend SNR = {}, Signal/Noise SNR = {}".format(seasonality_snr, noise_snr))
def generate_ts(T, seasons, seasonality_type, seasonality_snr, noise_snr, no_seasonality_components=5, plot=True):
assert T % np.prod(seasons) == 0
t_t = generate_trend(T)
s_t = generate_seasonality(seasons, seasonality_type, t_t, seasonality_snr, no_seasonality_components)
n_t = generate_noise(t_t, s_t, noise_snr)
x_t = t_t + s_t + n_t
if plot:
plot_ts(x_t, t_t, s_t, n_t)
return x_t, t_t, s_t, n_t
###Output
_____no_output_____
###Markdown
API
###Code
seasons = [24, 7]
T = np.prod(seasons)*52
seasonality_type = "cp"
seasonality_snr = 1.
noise_snr = 10.
# default parameters: no_seasonality_components=5, plot=True
x_t, t_t, s_t, n_t = generate_ts(T=T, seasons=seasons, seasonality_type=seasonality_type,
seasonality_snr=seasonality_snr, noise_snr=noise_snr)
###Output
_____no_output_____
###Markdown
Examples: Playing with SNRs
###Code
seasons = [24, 7]
T = np.prod(seasons)*52
seasonality_type = "cp"
###Output
_____no_output_____
###Markdown
Strong seasonality + weak trend + noise
###Code
seasonality_snr = 1.
noise_snr = 10.
x_t, t_t, s_t, n_t = generate_ts(T=T, seasons=seasons, seasonality_type=seasonality_type,
seasonality_snr=seasonality_snr, noise_snr=noise_snr)
###Output
_____no_output_____
###Markdown
Stronger trend + noise
###Code
seasonality_snr = .25
noise_snr = 1
x_t, t_t, s_t, n_t = generate_ts(T=T, seasons=seasons, seasonality_type=seasonality_type,
seasonality_snr=seasonality_snr, noise_snr=noise_snr)
###Output
_____no_output_____
###Markdown
Examples: Different seasonality types
###Code
seasons = [24, 7]
T = np.prod(seasons)*52
seasonality_snr = 1.
noise_snr = 10.
###Output
_____no_output_____
###Markdown
Sinusoid
###Code
seasonality_type = "sinusoid"
x_t, t_t, s_t, n_t = generate_ts(T=T, seasons=seasons, seasonality_type=seasonality_type,
seasonality_snr=seasonality_snr, noise_snr=noise_snr)
pd.Series(s_t[:np.prod(seasons)*2]).plot();
###Output
_____no_output_____
###Markdown
Nonnegative CP
###Code
seasonality_type = "nonnegative_cp"
x_t, t_t, s_t, n_t = generate_ts(T=T, seasons=seasons, seasonality_type=seasonality_type,
seasonality_snr=seasonality_snr, noise_snr=noise_snr)
pd.Series(s_t[:np.prod(seasons)*2]).plot();
###Output
_____no_output_____
###Markdown
CP
###Code
seasonality_type = "cp"
x_t, t_t, s_t, n_t = generate_ts(T=T, seasons=seasons, seasonality_type=seasonality_type,
seasonality_snr=seasonality_snr, noise_snr=noise_snr)
pd.Series(s_t[:np.prod(seasons)*2]).plot();
###Output
_____no_output_____
###Markdown
Examples: More seasonality
###Code
seasons = [60, 24, 7]
T = np.prod(seasons)*4
seasonality_snr = 1.
noise_snr = 10.
seasonality_type = "cp"
x_t, t_t, s_t, n_t = generate_ts(T=T, seasons=seasons, seasonality_type=seasonality_type,
seasonality_snr=seasonality_snr, noise_snr=noise_snr)
pd.Series(s_t[:np.prod(seasons)*2]).plot(figsize=(15,3));
###Output
_____no_output_____
|
Notebooks/.ipynb_checkpoints/Poisson Distribution-checkpoint.ipynb
|
###Markdown
Formula The Poisson distribution is: $$ P(\bar{x}=k)= \dfrac{\lambda^k}{k!}e^{-\lambda} $$ where $k$ is the discrete probability and $\lambda$ the average or standard deviation. Python example
###Code
def poisson(k, l): return l**k / math.factorial(k) * math.e**(-l)
x = np.arange(1, 61.8*2, 1)
x-=61.8
x*=.618
x+=61.8
x=np.round(x)
y = [poisson(x, 61.8) for x in x]
plt.figure(figsize=(12,12))
plot(x,y,lw=3)
fill_between(x,y,alpha=0.25)
grid(ls='dashed', alpha=.284)
title('Poisson distribution ($\lambda=61.8$)');
###Output
_____no_output_____
|
ml-project-template.ipynb
|
###Markdown
[TITLE] General Data Science/ML Project Template- Author: Kevin Chuang [@k-chuang](https://github.com/k-chuang)- Date: 10/07/2018- Description: A jupyter notebook template for steps in solving a data science and/or machine learning problem.- Dataset: [Link to dataset source]()---------- Overview- **Introduction / Abstract**- **Load libraries & get data** - Split data to training and test set - stratified sampling based on certain feature(s) or label(s)- **Exploratory Data Analysis** - Discover and visualize the training data to gain insights- **Data Preprocessing** - Prepare data for ML algorithms - Write pipelines using transformers to do automated feature engineering: - Data scaling - Impute missing data (or remove) - Feature extraction - Create new dimensions by combining existing ones - Feature selection - Choose subset of features from the existing features- **Model Selection & Training** - Use K-Folds Cross-Validation to select top 2 to 5 most promising models - Do not spend too much time tweaking hyperparameters - Typical ML models include kNN, SVM, linear/logistic regression, ensemble methods (RF, XGB), neural networks, etc. - [Optional] Save experimental models to pickle file.- **Model Tuning** - `GridSearchCV`, `RandomSearchCV`, or `BayesSearchCV` - `GridSearchCV`: brute force way to search for 'best' hyperparameters - `BayesSearchCV`: smart way to use Bayesian inference to optimally search for best hyperparameters- **Model Evaluation** - Final evaluation on hold out test set - If regression, calculate 95% confidence interval range - t score or z score to calculate confidence interval- **Solution Presentation and/or submission** - What I learned, what worked & what did not, what assumptions were made, and what system's limitations are - Create clear visualizations & easy-to-remember statements- **Deployment** - Clean up and concatenate pipleines to single pipeline to do full data preparation plus final prediction - Create programs to monitor & check system's live performance Introduction / Abstract- Write a paragraph about the project/problem at hand - Look at the big picture - Frame the problem - Business objectives Load libraries & data- Load important libraries- Load (or acquire) associated data- Split data into training and test set - Based on either feature importance or class imbalance, use *stratified sampling* to split data to keep porportion even for training set and test set.
###Code
__author__ = 'Kevin Chuang (https://www.github.com/k-chuang)'
# Version check
import sklearn
print('The scikit-learn version is {}.'.format(sklearn.__version__))
# linear algebra
import numpy as np
# data processing
import pandas as pd
# data visualization
%matplotlib inline
import seaborn as sns
from matplotlib import pyplot as plt
plt.rcParams['axes.labelsize'] = 14
plt.rcParams['xtick.labelsize'] = 12
plt.rcParams['ytick.labelsize'] = 12
# Algorithms
from sklearn.linear_model import SGDClassifier, LogisticRegression, Perceptron
from sklearn.ensemble import RandomForestClassifier, AdaBoostClassifier, VotingClassifier, ExtraTreesClassifier
from sklearn.tree import DecisionTreeClassifier
from sklearn.neighbors import KNeighborsClassifier
from sklearn.svm import SVC, LinearSVC
from sklearn.naive_bayes import GaussianNB, MultinomialNB
from sklearn.neural_network import MLPClassifier
from xgboost import XGBClassifier
import xgboost as xgb
import lightgbm as lgb
# Preprocessing
from sklearn.model_selection import train_test_split
from sklearn.preprocessing import LabelEncoder, StandardScaler, OneHotEncoder
# Pipeline
from sklearn.pipeline import Pipeline
# Manifold Learning
from sklearn.manifold import LocallyLinearEmbedding, TSNE
# Feature Selection
from sklearn.feature_selection import VarianceThreshold, SelectKBest, SelectPercentile, chi2, RFECV
# Metrics
from sklearn.metrics import log_loss, f1_score, accuracy_score
from sklearn.model_selection import cross_val_score
# Model Selection & Hyperparameter tuning
from sklearn.model_selection import GridSearchCV, RandomizedSearchCV, StratifiedKFold
from skopt import BayesSearchCV
from skopt.space import Real, Categorical, Integer
# Decomposition
from sklearn.decomposition import PCA
# Discriminant Analysis
from sklearn.discriminant_analysis import LinearDiscriminantAnalysis
# Clustering
from sklearn.cluster import KMeans
# Mathematical Functions
import math
# Utils
from collections import Counter
# Statistics
from scipy import stats
# Ignore useless warnings
import warnings
warnings.filterwarnings(action="ignore")
###Output
_____no_output_____
###Markdown
Exploratory Data Analysis (EDA)- Visualize training data using different kinds of plots- Plot dependent variables (features) against independent variable (target label) Data Preprocessing- Writing pipelines to do automated feature engineering - Imputing missing values (or removing values) - Scaling data - Transforming objects (strings, dates, etc.) to numerical vectors - Creating new features Model Selection & Training- Try different models and choose best 2-5 models - Use K-Fold cross-validation to validate which models are the best- Typical ML models include kNN, SVM, linear/logistic regression, ensemble methods (RF, XGB), neural networks, etc.- [Optional] Save experimental models to pickle file. Model Tuning- Tune the top chosen model(s) and tune hyperparameters - Ideally, use Bayes Optimization `BayesSearchCV` to optimally search for best hyperparameters for the model - `BayesSearchCV` is from `skopt` or `scikit-optimize` library (There are many different Bayesian Optimization implementations) - Below are some common search spaces for ensemble algorithms (which tend to have a lot of hyperparameters), specifically: - Random Forest (Variation of Bagging) - xgboost (Gradient Boosting) - lightgbm (Gradient Boosting) - https://lightgbm.readthedocs.io/en/latest/Parameters-Tuning.html
###Code
from skopt import BayesSearchCV
from skopt.space import Real, Categorical, Integer
# Random Forest (Classificaton Example)
from sklearn.ensemble import RandomForestClassifier
rf_search_space = {
'n_estimators': (100, 600),
'max_depth': (1, 50),
'max_features': (1, n_features),
'min_samples_leaf': (1, 50), # integer valued parameter
'min_samples_split': (2, 50),
}
rf_bayes_tuner = BayesSearchCV(
estimator=RandomForestClassifier(oob_score=True, random_state=1, n_jobs=2),
search_spaces=rf_search_space,
n_iter=20,
optimizer_kwargs={'base_estimator': 'RF'},
scoring='neg_log_loss',
n_jobs=5,
verbose=0,
cv = StratifiedKFold(
n_splits=3,
shuffle=True,
random_state=1
),
random_state=1
)
def status_print(result):
"""Status callback durring bayesian hyperparameter search"""
# Get all the models tested so far in DataFrame format
all_models = pd.DataFrame(rf_bayes_tuner.cv_results_)
# Get current parameters and the best parameters
best_params = pd.Series(rf_bayes_tuner.best_params_)
print('Model #{}\nBest LogLoss: {}\nBest params: {}\n'.format(
len(all_models),
np.round(rf_bayes_tuner.best_score_, 6),
rf_bayes_tuner.best_params_
))
# Save all model results
clf_name = rf_bayes_tuner.estimator.__class__.__name__
all_models.to_csv(clf_name + "_cv_results.csv")
# Fit the model
result = rf_bayes_tuner.fit(X_train.values, Y_train.values, callback=status_print)
# XGB (Classification Example)
import xgboost as xgb
xgb_search_space = {
# log-uniform: understand as search over p = exp(x) by varying x
'learning_rate': (0.01, 1.0, 'log-uniform'),
'min_child_weight': (0, 10),
'max_depth': (1, 100),
'max_delta_step': (0, 20),
'subsample': (0.01, 1.0, 'uniform'),
'colsample_bytree': (0.01, 1.0, 'uniform'),
'colsample_bylevel': (0.01, 1.0, 'uniform'),
'reg_lambda': (1e-9, 1000, 'log-uniform'),
'reg_alpha': (1e-9, 1.0, 'log-uniform'),
'gamma': (1e-9, 0.5, 'log-uniform'),
'min_child_weight': (0, 5),
'n_estimators': (50, 500),
'scale_pos_weight': (1e-6, 500, 'log-uniform')
}
xgb_bayes_tuner = BayesSearchCV(
estimator = xgb.XGBClassifier(
n_jobs = 3,
objective = 'multi:softprob',
eval_metric = 'mlogloss',
silent=1,
random_state=1
),
search_spaces = xgb_search_space,
scoring = 'neg_log_loss',
cv = StratifiedKFold(
n_splits=3,
shuffle=True,
random_state=1
),
n_jobs = 6,
n_iter = 20,
verbose = 0,
refit = True,
random_state = 1
)
def status_print(result):
"""Status callback during bayesian hyperparameter search"""
# Get all the models tested so far in DataFrame format
all_models = pd.DataFrame(xgb_bayes_tuner.cv_results_)
# Get current parameters and the best parameters
best_params = pd.Series(xgb_bayes_tuner.best_params_)
print('Model #{}\nBest Log Loss: {}\nBest params: {}\n'.format(
len(all_models),
np.round(xgb_bayes_tuner.best_score_, 8),
xgb_bayes_tuner.best_params_
))
# Save all model results
clf_name = xgb_bayes_tuner.estimator.__class__.__name__
all_models.to_csv(clf_name + "_cv_results.csv")
# Fit the model
result = xgb_bayes_tuner.fit(X_train.values, Y_train.values, callback=status_print)
# LGB (Regression Example)
import lightgbm as lgb
lgb_search_space = {
'max_depth': (3, 10),
'num_leaves': (6, 30),
'min_child_samples': (50, 200),
'subsample': (0.5, 1.0, 'uniform'),
'colsample_bytree': (0.01, 1.0, 'uniform'),
'reg_lambda': (1e-9, 1000, 'log-uniform'),
'reg_alpha': (1e-9, 1.0, 'log-uniform'),
'n_estimators': (50, 500),
'scale_pos_weight': (1e-6, 500, 'log-uniform'),
'learning_rate': (0.01, 0.2, 'uniform')
}
lgb_bayes_tuner = BayesSearchCV(
estimator = lgb.LGBMRegressor(
n_jobs = 3,
boosting_type="gbdt",
objective = 'regression',
silent=1,
random_state=1
),
search_spaces = lgb_search_space,
scoring = 'neg_mean_squared_error',
cv = 3,
n_jobs = 3,
n_iter = 20,
verbose = 3,
refit = True,
random_state = 1
)
def status_print(result):
"""Status callback during bayesian hyperparameter search"""
# Get all the models tested so far in DataFrame format
all_models = pd.DataFrame(lgb_bayes_tuner.cv_results_)
# Get current parameters and the best parameters
best_params = pd.Series(lgb_bayes_tuner.best_params_)
print('Model #{}\nBest Log Loss: {}\nBest params: {}\n'.format(
len(all_models),
np.round(lgb_bayes_tuner.best_score_, 8),
lgb_bayes_tuner.best_params_
))
# Save all model results
clf_name = lgb_bayes_tuner.estimator.__class__.__name__
all_models.to_csv(clf_name + "_cv_results.csv")
lgb_bayes_tuner.fit(housing_prepared, housing_labels, callback=status_print)
###Output
_____no_output_____
|
python/analyzing annihilation rates from the oscillatory model.ipynb
|
###Markdown
analyzing Annihilation Rates from the Oscillatory ModelTim Tyree1.10.2022
###Code
# TODO: simplify initialization
from scipy.optimize import minimize
from lib.my_initialization import *
from lib import *
from lib.lib_care.measure.level_sets import comp_longest_level_set_and_smooth
%load_ext autoreload
%autoreload 2
import matplotlib.pyplot as plt, numpy as np
from mpltools import annotation
import random,scipy
from scipy.interpolate import LinearNDInterpolator
from scipy.interpolate import CloughTocher2DInterpolator
import matplotlib as mpl #for colorbar
from scipy import stats
#for particle params to MSR generating functional
from scipy.signal import savgol_filter
import dask.bag as db
from lib.lib_care.measure.bootstrap import bin_and_bootstrap_xy_values_parallel
darkmode=False
if darkmode:
# For darkmode plots
from jupyterthemes import jtplot
jtplot.style(theme='monokai', context='notebook', ticks=True, grid=False)
import warnings
warnings.simplefilter("ignore", UserWarning)
%load_ext autoreload
%autoreload 2
#optional
import cupy as cp
cp.random.seed(42)
np.random.seed(42)
darkmode=False
if darkmode:
# For darkmode plots
from jupyterthemes import jtplot
jtplot.style(theme='monokai', context='notebook', ticks=True, grid=False)
###Output
_____no_output_____
###Markdown
visualize annihilation rates when using the noninteracting model for phase evolution, to show the correct exponents were not observed
###Code
wjr=recall_powerlaw_fits_to_full_models()
print(*wjr)
data_dir="/home/timothytyree/Documents/GitHub/bgmc/python/data/osg_output/run_21_all.csv"
df=pd.read_csv(data_dir)
# df.head()
print(list(df.columns))
assert (not (df.CollRate<0).any())
#derived values
# df['CollRate']=1./df['CollTime']
df['A']=df['L']**2
df['q']=df['N']/df['A'] #number of tips per square centimeter
df['w']=df['CollRate']/df['A'] #[mHz?]/cm^2
# df=df[df.niter==250].copy()
#extract column values
r_values=np.array(sorted(set(df.r.values)))#cm
D_values=np.array(sorted(set(df.D.values)))#cm^2/s
L_values=np.array(sorted(set(df.L.values)))#cm
A_values=L_values**2#cm^2
kappa_values=np.array(sorted(set(df.kappa.values)))#1/s
varkappa_values=np.array(sorted(set(df.varkappa.values)))#1/s
x0_values=np.array(sorted(set(df.x0.values)))#1/s
set_second_values=np.array(sorted(set(df.set_second.values)))
reflect_values=np.array(sorted(set(df.reflect.values)))
no_repulsion_values=np.array(sorted(set(df.no_repulsion.values)))
no_attraction_values=np.array(sorted(set(df.no_attraction.values)))
neighbor_values=np.array(sorted(set(df.neighbor.values)))
force_code_values=np.array(sorted(set(df.force_code.values)))
#make test for whether there is one input parameter present in an input DataFrame
print(f"parameters:")
print(f"r~{r_values}")
print(f"D~{D_values}")
print(f"L~{L_values}")
print(f"kappa~{kappa_values}")
print(f"a~{varkappa_values}")
print(f"x0~{x0_values}")
print(f"set_second~{set_second_values}")
print(f"reflect~{reflect_values}")
print(f"no_repulsion~{no_repulsion_values}")
print(f"no_attraction~{no_attraction_values}")
print(f"neighbor~{neighbor_values}")
print(f"force_code~{force_code_values}")
#DONE: what settings finished?
#DONE: ask self which ~half of settings did not finish. LR finished. FK was not attempted. i'd bet the low kappa*r**2 trials failed to finish
#TODO(later): add minimum r**2*kappa threshold to gen_run_22.py
#DONE: did any powerlaw fits have a reasonably low RMSE_full? No.
#DONE: visualize individual settings
###Output
_____no_output_____
###Markdown
Note: Run 21 is missing trials from the neighbor=0 case.I should consider why the neighbor=0 trials didn't finish if I try inplementing neighbor-neighbor phase interactions.
###Code
#Warning: computed powerlaw fits for >5000 trials successfully.
#what do the annihilation rates look like?
#for FK model
#query the control
varkappa=varkappa_values[0] #cm^2/s
D=D_values[0]
r=r_values[0]
L=L_values[0]
x0=x0_values[-2] #s
set_second=0
no_repulsion=0
no_attraction=0
reflect=0
neighbor=0
force_code=force_code_values[0]
#query the DataFrame
query =(df.set_second==set_second)&(df.reflect==reflect)
query&=df.r==r
query&=df.D==D
query&=df.L==L
query&=df.varkappa==varkappa
query&=df.x0==x0
query&=(df.no_repulsion==no_repulsion)&(df.no_attraction==no_attraction)
print(df[query].size)
query&=(df.neighbor==neighbor)
print(df[query].size)
query&=(df.force_code==force_code)
print(df[query].size)
dg=df[query]
kappa_values=np.array(sorted(set(dg.kappa.values)))
kappa=kappa_values[0]
dh=dg[dg.kappa==kappa]
x_values=dh.q.values
y_values=dh.w.values
x_values_control=x_values.copy()
y_values_control=y_values.copy()
x0=x0_values[0] #cm
#query the DataFrame
query =(df.set_second==set_second)&(df.reflect==reflect)
query&=df.r==r
query&=df.D==D
query&=df.L==L
query&=df.varkappa==varkappa
query&=df.x0==x0
query&=(df.no_repulsion==no_repulsion)&(df.no_attraction==no_attraction)
query&=(df.neighbor==neighbor)&(df.force_code==force_code)
dg=df[query]
# kappa_values=np.array(sorted(set(dg.kappa.values)))
# kappa=kappa_values[1]
dh=dg[dg.kappa==kappa]
x_values=dh.q.values
y_values=dh.w.values
x_values_test=x_values.copy()
y_values_test=y_values.copy()
print((r,D,L,kappa,varkappa,x0))
#TODO(later?): dev averaging step for computation of powerlaw fits.
# Option 2: map run_21_all to run_21_all_merged using the averaging filter
# Option 1: see if the run_21_all.csv already did all of the merging for me, in which case I can connect run_21_all.csv to the bootstrapping method already in use.
#plot fits for full model
m_fk=1.945;#+-0.030; B_fk=2.441+-0.051
m_lr=1.544;#+-0.034; B_lr=5.870+-0.137
M_fk=5.67;#+-0.39 Hz*cm^{2(m-1)}
M_lr=15.37;#+-1.57 Hz*cm^{2(m-1)}
# RMSE_fk=0.1252 Hz/cm^2
# RMSE_lr=0.0974 Hz/cm^2
# R^2=0.997 (FK)
# R^2=0.994 (LR)
# yscale=10**3
xv=np.arange(0.1,1.,.05)
yv_fk=M_fk*(xv)**m_fk
yv_lr=M_lr*(xv)**m_lr
fontsize=16
# plt.xlim([0.1,1])
# plt.ylim([1e-1,15])
plt.yscale('log')
plt.xscale('log')
# plt.plot(xv,yv_fk,label='FK power law fit',zorder=3,lw=4)
plt.plot(xv,yv_lr,label='LR power law fit',zorder=3,lw=4,color='C1')
fontsize=18
# plt.plot(x_values_control,y_values_control,'-',c='k',alpha=.7,label=r'approx. infinite basin',lw=3)
plt.plot(x_values_test,y_values_test,'-',c='k',alpha=.7,label=f'Oscillatory Model\n(no phase interactions)',lw=3)
# plt.plot(x_values,y_values,c='C2',alpha=.7,label='simulation',lw=6)
# plt.plot(x_values_force,y_values_force,c='C3',alpha=.7,label='simulation with forces',lw=6)
# plt.plot(x_values,y_values,c=c_values,alpha=0.4,cmap='bwr')
# plt.title(u'comparison to simulation\nwith two hybrid modes',fontsize=fontsize)
plt.xlabel(r'q (cm$^{-2}$)',fontsize=fontsize)
plt.ylabel(r'w (Hz cm$^{-2}$)', fontsize=fontsize)
plt.tick_params(axis='both', which='major', labelsize=fontsize)
plt.tick_params(axis='both', which='minor', labelsize=0)
plt.legend(fontsize=fontsize-5)
# plt.xlim([0.08,1])
# print(f'varkappa={varkappa} Hz');print(f' x0={x0} cm')
# plt.title(r'$\varkappa=$'+f'{varkappa} Hz, '+r'$x_0=$'+f'{x0} cm\nforce_code={force_code}, neighbors={neighbor}\n',fontsize=fontsize)
plt.title(r'$T=$'+f'{x0} sec, '+r'$a=$'+f'{varkappa}, '+r'$\kappa=$'+f'{kappa:.1f}\nforce_code={force_code}, neighbors={neighbor}\nr={r},D={D},L={L}\n',fontsize=fontsize)
plt.show()
# from generation of powerfits .ipynb
print(f"powerfit_dir='/home/timothytyree/Documents/GitHub/bgmc/python/data/osg_output/run_21_all_powerlaw_fits.csv'")
powerfit_dir='/home/timothytyree/Documents/GitHub/bgmc/python/data/osg_output/run_21_all_powerlaw_fits.csv'
df_powerfits=pd.read_csv(powerfit_dir)
df_powerfits.head()
nu_values=df_powerfits['m'].values
yv=np.linspace(0,1,10)
plt.plot(0.*yv+m_lr,yv,'C1--',label='Luo-Rudy Model')
plt.plot(0.*yv+m_fk,yv,'C0--',label='Fenton-Karma Model')
plt.hist(nu_values,color='k',alpha=0.8,label=f'Oscillatory Model\n(3126 settings)\n(no phase interactions)',density=True)
format_plot(ax=plt.gca(),xlabel='Exponent Value',ylabel='PDF')
plt.legend(fontsize=12)
plt.show()
#TODO: how many neighbor=0 trials finished? how many neighbor=1 trials finished
#TODO: make ^this into a pie chart, because the same number of settings were attempted for neighbor=0 versus 1
num_neighor_settings=df[df.neighbor==1].w.values.shape[0]
num_non_neighor_settings=df[df.neighbor==0].w.values.shape[0]
print(f"{100*num_neighor_settings/(num_neighor_settings+num_non_neighor_settings):.2f}% percent of settings that terminated used neighbor-only forces.")
yv=np.linspace(0,1,10)
plt.plot(0.*yv+m_lr,yv,'C1--',label='Luo-Rudy Model')
plt.plot(0.*yv+m_fk,yv,'C0--',label='Fenton-Karma Model')
nu_values=df_powerfits[df.neighbor==1]['m'].values
plt.hist(nu_values,color='k',alpha=0.8,label=f'Neighbor-Forces',density=True)
nu_values=df_powerfits[df.neighbor==0]['m'].values
plt.hist(nu_values,color='g',alpha=0.8,label=f'Vector-Summed-Forces',density=True)
format_plot(ax=plt.gca(),xlabel='Exponent Value',ylabel='PDF')
plt.legend(fontsize=12)
plt.show()
###Output
_____no_output_____
|
notebooks/illustrated-bi-fid-doe.ipynb
|
###Markdown
Imports and original function
###Code
from collections import namedtuple
import numpy as np
import matplotlib.pyplot as plt
from pyDOE import lhs
from scipy.spatial import distance
from pyprojroot import here
BiFidelityDoE = namedtuple("BiFidelityDoE", "high low")
def low_lhs_sample(ndim, nlow):
if ndim == 1:
return np.linspace(0,1,nlow).reshape(-1,1)
elif ndim > 1:
return lhs(ndim, nlow)
def bi_fidelity_doe(ndim, num_high, num_low):
"""Create a Design of Experiments (DoE) for two fidelities in `ndim`
dimensions. The high-fidelity samples are guaranteed to be a subset
of the low-fidelity samples.
:returns high-fidelity samples, low-fidelity samples
"""
high_x = low_lhs_sample(ndim, num_high)
low_x = low_lhs_sample(ndim, num_low)
dists = distance.cdist(high_x, low_x)
#TODO: this is the naive method, potentially speed up?
highs_to_match = set(range(num_high))
while highs_to_match:
min_dist = np.min(dists)
high_idx, low_idx = np.argwhere(dists == min_dist)[0]
low_x[low_idx] = high_x[high_idx]
# make sure just selected samples are not re-selectable
dists[high_idx,:] = np.inf
dists[:,low_idx] = np.inf
highs_to_match.remove(high_idx)
return BiFidelityDoE(high_x, low_x)
###Output
_____no_output_____
###Markdown
plotting result of a bi-fid-DoE
###Code
np.random.seed(20160501)
bfd = bi_fidelity_doe(2, 10, 20)
plt.scatter(*bfd.low.T, s=36, marker='o', label='low')
plt.scatter(*bfd.high.T, s=288, marker='+', label='high')
plt.legend(loc=0)
plt.show()
###Output
_____no_output_____
###Markdown
Defining illustrated bi-fid-doe function showing stepwise progress.
###Code
def illustrated_bi_fidelity_doe(ndim, num_high, num_low, intermediate=True,
as_pdf=True, save_dir=None):
"""Create a Design of Experiments (DoE) for two fidelities in `ndim`
dimensions. The high-fidelity samples are guaranteed to be a subset
of the low-fidelity samples.
:returns high-fidelity samples, low-fidelity samples
"""
extension = 'pdf' if as_pdf else 'png'
high_x = low_lhs_sample(ndim, num_high)
low_x = low_lhs_sample(ndim, num_low)
dists = distance.cdist(high_x, low_x)
fig_size = (4, 4) if ndim >= 2 else (4, 2)
plt.rcParams.update({'font.size': 16})
plt.rc('axes', labelsize=20)
low_style = {'s': 36}
high_style = {'s': 288, 'marker': '+'}
arrow_style = {
'width': .0025,
'head_width': .03,
'facecolor': 'black',
'length_includes_head': True,
}
#TODO: this is the naive method, potentially speed up?
highs_to_match = set(range(num_high))
while highs_to_match:
min_dist = np.min(dists)
high_idx, low_idx = np.argwhere(dists == min_dist)[0]
if intermediate:
xlow = low_x.T[0]
xhigh = high_x.T[0]
if ndim >= 2:
ylow = low_x.T[1]
yhigh = high_x.T[1]
else:
ylow = np.zeros(xlow.shape)
yhigh = np.zeros(xhigh.shape)
plt.figure(figsize=fig_size, constrained_layout=True)
plt.scatter(xlow, ylow, label='low', **low_style)
plt.scatter(xhigh, yhigh, label='high', **high_style)
plt.arrow(
*low_x[low_idx],
*(high_x[high_idx] - low_x[low_idx]),
**arrow_style,
)
plt.xticks([])
plt.yticks([])
plt.xlabel('$x_1$')
plt.ylabel('$x_2$')
plt.title(f'step {num_high-len(highs_to_match)}/{num_high}')
if save_dir:
plt.savefig(save_dir / f'illustrated-bi-fid-doe-{num_high-len(highs_to_match)}.{extension}')
plt.show()
plt.close()
low_x[low_idx] = high_x[high_idx]
# make sure just selected samples are not re-selectable
dists[high_idx,:] = np.inf
dists[:,low_idx] = np.inf
highs_to_match.remove(high_idx)
xlow = low_x.T[0]
xhigh = high_x.T[0]
if ndim >= 2:
ylow = low_x.T[1]
yhigh = high_x.T[1]
else:
ylow = np.zeros(xlow.shape)
yhigh = np.zeros(xhigh.shape)
plt.figure(figsize=fig_size, constrained_layout=True)
plt.scatter(xlow, ylow, label='low', **low_style)
plt.scatter(xhigh, yhigh, label='high', **high_style)
plt.xticks([])
plt.yticks([])
plt.xlabel('$x_1$')
plt.ylabel('$x_2$')
plt.title(f'step {num_high-len(highs_to_match)}/{num_high}')
if save_dir:
plt.savefig(save_dir / f'illustrated-bi-fid-doe-{ndim}d-{num_high}-{num_low}-{num_high-len(highs_to_match)}.{extension}')
plt.show()
return BiFidelityDoE(high_x, low_x)
###Output
_____no_output_____
###Markdown
Showing stepwise progress for 2d (20,10) DoE
###Code
np.random.seed(20160501)
plot_dir = here('plots') / 'illustrated-doe'
plot_dir.mkdir(exist_ok=True, parents=True)
_ = illustrated_bi_fidelity_doe(2, 10, 20, save_dir=plot_dir)
###Output
_____no_output_____
###Markdown
Investigating 1D DoE results
###Code
np.random.seed(20160501)
plot_dir = here('plots') / 'illustrated-doe'
plot_dir.mkdir(exist_ok=True, parents=True)
ratio = 1.5
examples = []
for nh in range(10, 20):
x = int(nh*ratio)
for nl in range(x-1, x+2):
print(f'{nh} {nl} ({np.round(nl/nh,2)})')
_ = illustrated_bi_fidelity_doe(1, nh, nl, save_dir=plot_dir, as_pdf=False, intermediate=False)
###Output
_____no_output_____
###Markdown
Animation setup
###Code
from functools import partial
def generator(n):
for i in range(n):
yield i*i
def animator(i, gen):
return next(gen)
animate = partial(animator, gen=generator(10))
for i in range(10):
print(animate(i))
###TODO: http://louistiao.me/posts/notebooks/save-matplotlib-animations-as-gifs/
###Output
_____no_output_____
|
Tutorials/AE.ipynb
|
###Markdown
###Code
import numpy as np
import torch
from torchvision import datasets,transforms
from torchvision.util import train_l
transform = transforms.ToTensor()
train_data = datasets.MNIST(root='data',transform=transform,download=True,train=True)
test_data = datasets.MNIST(root='data',transform=transform,download=True,train=False)
train_loader = torch.utils.data.DataLoader(train_data,batch_size=32)
test_loader = torch.utils.data.DataLoader(test_data,batch_size=32)
import matplotlib.pyplot as plt
train_iter = iter(train_loader)
images,_ = train_iter.next()
img = np.squeeze(np.array(images[0]))
plt.imshow(img,cmap='gray')
import torch.nn as nn
import torch.nn.functional as F
class LinearModel(nn.Module):
def __init__(self,input_size,hidden_size):
super(LinearModel,self).__init__()
self.conv1 = nn.Conv2D
self.fc1 = nn.Linear(input_size,hidden_size)
self.fcb = nn.Linear(hidden_size,input_size)
def forward(self,x):
x = F.relu(self.fc1(x))
x = F.sigmoid(self.fcb(x)) #sigmoid
return x
model = LinearModel(28*28,32)
model = model.to(torch.device('cuda'))
critertion = nn.MSELoss()
optimizer = torch.optim.Adam(model.parameters(),lr=0.001)
epochs = 20
for epoch in range(epochs):
train_loss = 0
for data in train_loader:
images,_ = data
images = images.to(torch.device('cuda'))
images = images.view(-1,28*28)
images.requires_grad_()
optimizer.zero_grad()
output = model(images)
loss = critertion(output,images)
train_loss += loss.item()
loss.backward()
optimizer.step()
print('Epoch: ',epoch,' Loss:',train_loss)
# obtain one batch of test images
dataiter = iter(test_loader)
images, labels = dataiter.next()
images_flatten = images.view(images.size(0), -1)
images_flatten = images_flatten.to(torch.device('cuda'))
# get sample outputs
output = model(images_flatten)
# prep images for display
images = images.numpy()
# output is resized into a batch of images
output = output.view(32, 1, 28, 28)
# use detach when it's an output that requires_grad
output = output.detach().cpu().numpy()
# plot the first ten input images and then reconstructed images
fig, axes = plt.subplots(nrows=2, ncols=10, sharex=True, sharey=True, figsize=(25,4))
# input images on top row, reconstructions on bottom
for images, row in zip([images, output], axes):
for img, ax in zip(images, row):
ax.imshow(np.squeeze(img), cmap='gray')
ax.get_xaxis().set_visible(False)
ax.get_yaxis().set_visible(False)
###Output
_____no_output_____
|
0.15/_downloads/plot_sensors_decoding.ipynb
|
###Markdown
=================================Decoding sensor space data (MVPA)=================================Decoding, a.k.a MVPA or supervised machine learning applied to MEGdata in sensor space. Here the classifier is applied to every timepoint.
###Code
import numpy as np
import matplotlib.pyplot as plt
from sklearn.pipeline import make_pipeline
from sklearn.preprocessing import StandardScaler
from sklearn.linear_model import LogisticRegression
import mne
from mne.datasets import sample
from mne.decoding import (SlidingEstimator, GeneralizingEstimator,
cross_val_multiscore, LinearModel, get_coef)
data_path = sample.data_path()
plt.close('all')
# sphinx_gallery_thumbnail_number = 4
###Output
_____no_output_____
###Markdown
Set parameters
###Code
raw_fname = data_path + '/MEG/sample/sample_audvis_raw.fif'
tmin, tmax = -0.200, 0.500
event_id = dict(audio_left=1, visual_left=3)
# Setup for reading the raw data
raw = mne.io.read_raw_fif(raw_fname, preload=True)
# The subsequent decoding analyses only capture evoked responses, so we can
# low-pass the MEG data. Usually a value more like 40 Hz would be used,
# but here low-pass at 20 so we can more heavily decimate, and allow
# the examlpe to run faster.
raw.filter(None, 20., fir_design='firwin')
events = mne.find_events(raw, 'STI 014')
# Set up pick list: EEG + MEG - bad channels (modify to your needs)
raw.info['bads'] += ['MEG 2443', 'EEG 053'] # bads + 2 more
picks = mne.pick_types(raw.info, meg='grad', eeg=False, stim=True, eog=True,
exclude='bads')
# Read epochs
epochs = mne.Epochs(raw, events, event_id, tmin, tmax, proj=True,
picks=picks, baseline=(None, 0.), preload=True,
reject=dict(grad=4000e-13, eog=150e-6), decim=10)
epochs.pick_types(meg=True, exclude='bads')
###Output
_____no_output_____
###Markdown
Temporal decoding-----------------We'll use a Logistic Regression for a binary classification as machinelearning model.
###Code
# We will train the classifier on all left visual vs auditory trials on MEG
X = epochs.get_data() # MEG signals: n_epochs, n_channels, n_times
y = epochs.events[:, 2] # target: Audio left or right
clf = make_pipeline(StandardScaler(), LogisticRegression())
time_decod = SlidingEstimator(clf, n_jobs=1, scoring='roc_auc')
scores = cross_val_multiscore(time_decod, X, y, cv=5, n_jobs=1)
# Mean scores across cross-validation splits
scores = np.mean(scores, axis=0)
# Plot
fig, ax = plt.subplots()
ax.plot(epochs.times, scores, label='score')
ax.axhline(.5, color='k', linestyle='--', label='chance')
ax.set_xlabel('Times')
ax.set_ylabel('AUC') # Area Under the Curve
ax.legend()
ax.axvline(.0, color='k', linestyle='-')
ax.set_title('Sensor space decoding')
plt.show()
# You can retrieve the spatial filters and spatial patterns if you explicitly
# use a LinearModel
clf = make_pipeline(StandardScaler(), LinearModel(LogisticRegression()))
time_decod = SlidingEstimator(clf, n_jobs=1, scoring='roc_auc')
time_decod.fit(X, y)
coef = get_coef(time_decod, 'patterns_', inverse_transform=True)
evoked = mne.EvokedArray(coef, epochs.info, tmin=epochs.times[0])
evoked.plot_joint(times=np.arange(0., .500, .100), title='patterns')
###Output
_____no_output_____
###Markdown
Temporal Generalization-----------------------This runs the analysis used in [1]_ and further detailed in [2]_The idea is to fit the models on each time instant and see how itgeneralizes to any other time point.
###Code
# define the Temporal Generalization object
time_gen = GeneralizingEstimator(clf, n_jobs=1, scoring='roc_auc')
scores = cross_val_multiscore(time_gen, X, y, cv=5, n_jobs=1)
# Mean scores across cross-validation splits
scores = np.mean(scores, axis=0)
# Plot the diagonal (it's exactly the same as the time-by-time decoding above)
fig, ax = plt.subplots()
ax.plot(epochs.times, np.diag(scores), label='score')
ax.axhline(.5, color='k', linestyle='--', label='chance')
ax.set_xlabel('Times')
ax.set_ylabel('AUC')
ax.legend()
ax.axvline(.0, color='k', linestyle='-')
ax.set_title('Decoding MEG sensors over time')
plt.show()
# Plot the full matrix
fig, ax = plt.subplots(1, 1)
im = ax.imshow(scores, interpolation='lanczos', origin='lower', cmap='RdBu_r',
extent=epochs.times[[0, -1, 0, -1]], vmin=0., vmax=1.)
ax.set_xlabel('Testing Time (s)')
ax.set_ylabel('Training Time (s)')
ax.set_title('Temporal Generalization')
ax.axvline(0, color='k')
ax.axhline(0, color='k')
plt.colorbar(im, ax=ax)
plt.show()
###Output
_____no_output_____
|
notebooks/notebooks_archive/September 29 - EresNet 34 results.ipynb
|
###Markdown
September 29 - EresNet 34 results
###Code
# Default imports
import math
import os
import sys
import pandas as pd
import numpy as np
import matplotlib as mpl
import matplotlib.pyplot as plt
from matplotlib.colors import LogNorm
import matplotlib.ticker as mtick
# Add the path to the parent directory to augment search for module
par_dir = os.path.abspath(os.path.join(os.getcwd(), os.pardir))
if par_dir not in sys.path:
sys.path.append(par_dir)
# Import the custom plotting module
from plot_utils import plot_utils
import random
import torch
from plot_utils import notebook_utils_2
# Label dict - Dictionary mapping integer labels to str
label_dict = {0:"gamma", 1:"e", 2:"mu"}
np.set_printoptions(threshold=np.inf)
def plot_event(run_id, iteration, mode):
dump_dir = "/home/akajal/WatChMaL/VAE/dumps/" + run_id + "/"
if mode is "validation":
np_arr_path = dump_dir + "test_vali_iteration_" + str(iteration) + ".npz"
else:
np_arr_path = dump_dir + "iteration_" + str(iteration) + ".npz"
# Load the numpy array
np_arr = np.load(np_arr_path)
np_event, np_recon, np_labels, np_energies = np_arr["events"], np_arr["prediction"], np_arr["labels"], np_arr["energies"]
i = random.randint(0, np_labels.shape[0]-1)
plot_utils.plot_actual_vs_recon(np_event[i], np_recon[i],
label_dict[np_labels[i]], np_energies[i].item(),
show_plot=True)
plot_utils.plot_charge_hist(torch.tensor(np_event).permute(0,2,3,1).numpy(),
np_recon, iteration, num_bins=200)
def plot_log(run_id, model_name, iteration, variant, mode):
dump_dir = "/home/akajal/WatChMaL/VAE/dumps/" + run_id + "/"
# Setup the path to the training log file
if mode is "training":
log = dump_dir + "log_train.csv"
elif mode is "training_validation":
log = dump_dir + "val_test.csv"
elif mode is "validation":
log = dump_dir + "validation_log.csv"
else:
print("mode has to be one of training, training_validation, validation")
return None
downsample_interval = 32 if mode is "training" else None
if variant is "AE":
plot_utils.plot_ae_training([log], [model_name], {model_name:["red"]},
downsample_interval=downsample_interval, show_plot=True, legend_loc=(1.1,1.1))
elif variant is "VAE":
plot_utils.plot_vae_training([log], [model_name], {model_name:["red", "blue"]},
downsample_interval=downsample_interval, show_plot=True, legend_loc=(1.1,1.1))
if iteration is not None:
plot_event(run_id, iteration, mode=mode)
def plot_samples(run_id, num_samples, model_dir):
dump_dir = "/home/akajal/WatChMaL/VAE/dumps/" + run_id + "/"
np_arr_path = dump_dir + "samples/" + model_dir + "/" + str(num_samples) + "_samples.npy"
np_arr = np.load(np_arr_path)
i, j = random.randint(0, np_arr.shape[0]-1), random.randint(0, np_arr.shape[0]-1)
plot_utils.plot_actual_vs_recon(np_arr[i], np_arr[j],
"e", 500,
show_plot=True)
plot_utils.plot_charge_hist(np_arr[i],
np_arr[j], 0, num_bins=200)
###Output
_____no_output_____
###Markdown
EresNet-34 VAE trained for 10.0 epochs
###Code
run_id = "20190929_132502"
dump_dir = "/home/akajal/WatChMaL/VAE/dumps/" + run_id + "/"
np_arr_path = dump_dir + "test_validation_iteration_dump.npz"
# Load the numpy array
np_arr = np.load(np_arr_path)
np_event, np_recon, np_labels, np_energies = np_arr["events"], np_arr["recon"], np_arr["labels"], np_arr["energies"]
print(np_event.shape)
i = random.randint(0, np_labels.shape[0]-1)
j = random.randint(0, np_labels.shape[1]-1)
plot_utils.plot_actual_vs_recon(np_event[i][j], np_recon[i][j],
label_dict[np_labels[i][j].item()], np_energies[i][j].item(),
show_plot=True)
###Output
_____no_output_____
###Markdown
EresNet-34 VAE trained for 30.0 epochs
###Code
run_id = "20190929_132330"
dump_dir = "/home/akajal/WatChMaL/VAE/dumps/" + run_id + "/"
np_arr_path = dump_dir + "test_validation_iteration_dump.npz"
# Load the numpy array
np_arr = np.load(np_arr_path)
np_event, np_recon, np_labels, np_energies = np_arr["events"], np_arr["recon"], np_arr["labels"], np_arr["energies"]
i = random.randint(0, np_labels.shape[0]-1)
j = random.randint(0, np_labels.shape[1]-1)
plot_utils.plot_actual_vs_recon(np_event[i][j], np_recon[i][j],
label_dict[np_labels[i][j].item()], np_energies[i][j].item(),
show_plot=True)
###Output
_____no_output_____
|
IEEE-CIS_Fraud_Detection/script/IEEE_2019_XGBoost_Adversarial.ipynb
|
###Markdown
IEEE 2019 - XGBoost - Adversarial
###Code
from google.colab import drive
drive.mount('/content/gdrive')
# General imports
import numpy as np
import pandas as pd
import os, sys, gc, warnings, random, datetime, time
import pickle
#from knockknock import telegram_sender
import matplotlib.pyplot as plt
import seaborn as sns
from sklearn import metrics
from sklearn.model_selection import train_test_split, GroupKFold
from sklearn.preprocessing import LabelEncoder
from tqdm import tqdm
import math
warnings.filterwarnings('ignore')
%matplotlib inline
# Pandas display format
pd.set_option('display.max_columns', 1000)
pd.set_option('display.max_rows', 500)
# Seeder
# :seed to make all processes deterministic # type: int
def seed_everything(seed=0):
random.seed(seed)
os.environ['PYTHONHASHSEED'] = str(seed)
np.random.seed(seed)
# Memory Reducer
def memory_usage_mb(df, *args, **kwargs):
"""Dataframe memory usage in MB. """
return df.memory_usage(*args, **kwargs).sum() / 1024**2
def reduce_mem_usage(df, deep=True, verbose=False, categories=False):
# All types that we want to change for "lighter" ones.
# int8 and float16 are not include because we cannot reduce
# those data types.
# float32 is not include because float16 has too low precision.
numeric2reduce = ["int16", "int32", "int64", "float64"]
start_mem = 0
start_mem = memory_usage_mb(df, deep=deep)
for col, col_type in df.dtypes.iteritems():
best_type = None
if categories:
if col_type == "object":
df[col] = df[col].astype("category")
best_type = "category"
elif col_type in numeric2reduce:
downcast = "integer" if "int" in str(col_type) else "float"
df[col] = pd.to_numeric(df[col], downcast=downcast)
best_type = df[col].dtype.name
# Log the conversion performed.
if verbose and best_type is not None and best_type != str(col_type):
print(f"Column '{col}' converted from {col_type} to {best_type}")
end_mem = memory_usage_mb(df, deep=deep)
diff_mem = start_mem - end_mem
percent_mem = 100 * diff_mem / start_mem
print(f"Memory usage decreased from"
f" {start_mem:.2f}MB to {end_mem:.2f}MB"
f" ({diff_mem:.2f}MB, {percent_mem:.2f}% reduction)")
return df
# Custom Focal Loss
def robust_pow(num_base, num_pow):
# numpy does not permit negative numbers to fractional power
# use this to perform the power algorithmic
return np.sign(num_base) * (np.abs(num_base)) ** (num_pow)
def focal_binary_object(y_pred, dtrain):
gamma_indct = 2.0
# retrieve data from dtrain matrix
label = dtrain.get_label()
# compute the prediction with sigmoid
sigmoid_pred = 1.0 / (1.0 + np.exp(-y_pred))
# gradient
# complex gradient with different parts
g1 = sigmoid_pred * (1 - sigmoid_pred)
g2 = label + ((-1) ** label) * sigmoid_pred
g3 = sigmoid_pred + label - 1
g4 = 1 - label - ((-1) ** label) * sigmoid_pred
g5 = label + ((-1) ** label) * sigmoid_pred
# combine the gradient
grad = gamma_indct * g3 * robust_pow(g2, gamma_indct) * np.log(g4 + 1e-9) + \
((-1) ** label) * robust_pow(g5, (gamma_indct + 1))
# combine the gradient parts to get hessian components
hess_1 = robust_pow(g2, gamma_indct) + \
gamma_indct * ((-1) ** label) * g3 * robust_pow(g2, (gamma_indct - 1))
hess_2 = ((-1) ** label) * g3 * robust_pow(g2, gamma_indct) / g4
# get the final 2nd order derivative
hess = ((hess_1 * np.log(g4 + 1e-9) - hess_2) * gamma_indct +
(gamma_indct + 1) * robust_pow(g5, gamma_indct)) * g1
return grad, hess
# Vars
SEED = 42
seed_everything(SEED)
LOCAL_TEST = False
TARGET = 'isFraud'
PREDICT = True
# DATA LOAD
print('Load Data')
train_df = pd.read_pickle('../content/gdrive/My Drive/IEEE fraud Kaggle 2019/train_full_feat.pkl')
if LOCAL_TEST:
test_df = train_df[train_df['DT_M']==train_df['DT_M'].max()].reset_index(drop=True)
train_df = train_df[train_df['DT_M']<(train_df['DT_M'].max()-1)].reset_index(drop=True)
else:
test_df = pd.read_pickle('../content/gdrive/My Drive/IEEE fraud Kaggle 2019/test_full_feat.pkl')
test_df['isFraud'] = 0
print('Shape control:\nTrain:', train_df.shape, '\nTest:',test_df.shape)
test_df.head()
test_dt = pd.read_csv('../content/gdrive/My Drive/IEEE fraud Kaggle 2019/test_dt.csv')
test_df_backup = test_df.copy()
train_df_backup = train_df.copy()
test_df = test_df.merge(test_dt, how='left', on='TransactionID').sort_values(by='TransactionDT', ascending=True)
# 0.8 is the Private Test size
test_df = test_df.iloc[:int(len(test_df)*0.2),:] #public
#test_df = test_df.iloc[int(len(test_df)*0.2):,:] #private
len(test_df)
train_df.drop(columns=['isFraud','DT_M'],inplace=True)
test_df.drop(columns=['isFraud','TransactionDT'],inplace=True)
#test_df.drop(columns=['isFraud'],inplace=True)
train_df['target'] = 0
test_df['target'] = 1
tot_df = pd.concat([train_df,test_df])
# Final features list
# features_columns = [col for col in list(train_df) if col not in ['TransactionID','isFraud','DT_M']]
# Selected features
features_columns = ['Amt_timeslowest_first_UserID_proxy',
'Amt_timeslowest_first_group',
'Amtisfirst_UserID_proxy',
'Amtisfirst_group',
'C1',
'C10',
'C11',
'C12',
'C13',
'C14',
'C2',
'C4',
'C5',
'C6',
'C7',
'C8',
'C9',
'D10_DT_D_min_max',
'D10_DT_D_std_score',
'D10_DT_M_min_max',
'D10_DT_M_std_score',
'D10_DT_W_min_max',
'D10_DT_W_std_score',
'D11',
'D11_DT_D_min_max',
'D11_DT_D_std_score',
'D11_DT_M_min_max',
'D11_DT_M_std_score',
'D11_DT_W_min_max',
'D11_DT_W_std_score',
'D11__DeviceInfo',
'D12',
'D12_DT_D_min_max',
'D12_DT_D_std_score',
'D12_DT_M_min_max',
'D12_DT_M_std_score',
'D12_DT_W_min_max',
'D12_DT_W_std_score',
'D13',
'D13_DT_D_min_max',
'D13_DT_D_std_score',
'D13_DT_M_min_max',
'D13_DT_M_std_score',
'D13_DT_W_min_max',
'D13_DT_W_std_score',
'D14',
'D14_DT_D_min_max',
'D14_DT_D_std_score',
'D14_DT_M_min_max',
'D14_DT_M_std_score',
'D14_DT_W_min_max',
'D14_DT_W_std_score',
'D15',
'D15_DT_D_min_max',
'D15_DT_D_std_score',
'D15_DT_M_min_max',
'D15_DT_M_std_score',
'D15_DT_W_min_max',
'D15_DT_W_std_score',
'D2',
'D2_scaled',
'D3',
'D3_DT_D_min_max',
'D3_DT_D_std_score',
'D3_DT_M_min_max',
'D3_DT_M_std_score',
'D3_DT_W_min_max',
'D3_DT_W_std_score',
'D4',
'D4_DT_D_min_max',
'D4_DT_D_std_score',
'D4_DT_M_min_max',
'D4_DT_M_std_score',
'D4_DT_W_min_max',
'D4_DT_W_std_score',
'D5',
'D5_DT_D_min_max',
'D5_DT_D_std_score',
'D5_DT_M_min_max',
'D5_DT_M_std_score',
'D5_DT_W_min_max',
'D5_DT_W_std_score',
'D6',
'D6_DT_D_min_max',
'D6_DT_D_std_score',
'D6_DT_M_min_max',
'D6_DT_M_std_score',
'D6_DT_W_min_max',
'D6_DT_W_std_score',
'D7_DT_D_min_max',
'D7_DT_D_std_score',
'D7_DT_M_min_max',
'D7_DT_M_std_score',
'D7_DT_W_min_max',
'D7_DT_W_std_score',
'D8',
'D8_D9_decimal_dist',
'D8_DT_D_min_max',
'D8_DT_D_std_score',
'D8_DT_M_min_max',
'D8_DT_M_std_score',
'D8_DT_W_min_max',
'D8_DT_W_std_score',
'D8__D9',
'D8_not_same_day',
'D9',
'D9_not_na',
'DeviceInfo',
'DeviceInfo__P_emaildomain',
'DeviceInfo_device',
'DeviceInfo_version',
'DeviceType',
'M2',
'M2__M3',
'M3',
'M4',
'M5',
'M6',
'M7',
'M8',
'M9',
'P_emaildomain',
'P_emaildomain__C2',
'ProductCD',
'R_emaildomain',
'TransactionAmt',
'TransactionAmt_DT_D_min_max',
'TransactionAmt_DT_D_std_score',
'TransactionAmt_DT_M_min_max',
'TransactionAmt_DT_M_std_score',
'TransactionAmt_DT_W_min_max',
'TransactionAmt_DT_W_std_score',
'TransactionAmt_check',
'V_12_34_0',
'V_12_34_1',
'V_12_34_10',
'V_12_34_11',
'V_12_34_2',
'V_12_34_3',
'V_12_34_4',
'V_12_34_5',
'V_12_34_6',
'V_12_34_7',
'V_12_34_8',
'V_12_34_9',
'V_138_166_0',
'V_167_216_0',
'V_167_216_1',
'V_167_216_2',
'V_1_11_0',
'V_1_11_1',
'V_1_11_2',
'V_1_11_3',
'V_1_11_4',
'V_1_11_5',
'V_1_11_6',
'V_217_278_0',
'V_217_278_1',
'V_217_278_2',
'V_279_321_0',
'V_279_321_1',
'V_279_321_2',
'V_279_321_3',
'V_322_339_0',
'V_322_339_1',
'V_35_52_0',
'V_35_52_1',
'V_35_52_2',
'V_35_52_3',
'V_35_52_4',
'V_35_52_5',
'V_35_52_6',
'V_35_52_7',
'V_35_52_8',
'V_53_74_0',
'V_53_74_1',
'V_53_74_10',
'V_53_74_11',
'V_53_74_2',
'V_53_74_3',
'V_53_74_4',
'V_53_74_5',
'V_53_74_6',
'V_53_74_7',
'V_53_74_8',
'V_53_74_9',
'V_75_94_0',
'V_75_94_1',
'V_75_94_10',
'V_75_94_2',
'V_75_94_3',
'V_75_94_4',
'V_75_94_5',
'V_75_94_6',
'V_75_94_7',
'V_75_94_8',
'V_75_94_9',
'V_95_137_0',
'V_95_137_1',
'V_95_137_2',
'addr1',
'addr1__card1',
'addr2',
'card1',
'card1_TransactionAmt_mean',
'card1__card5',
'card2',
'card2_TransactionAmt_mean',
'card2__dist1',
'card2__id_20',
'card3',
'card3_TransactionAmt_mean',
'card4',
'card5',
'card5_TransactionAmt_mean',
'card5__P_emaildomain',
'card6',
'cardID_v2_count_group',
'cardID_v2_median_Amt_group',
'cardID_v2_skew_Amt_group',
'cardID_v2_unique_Amt_group',
'cardID_v2_unique_Device',
'cardID_v2_unique_IP',
'cardID_v2_unique_Pemail_group',
'cardID_v2_unique_Remail_group',
'cardID_v2_unique_adr1_group',
'cardID_v2_unique_adr2_group',
'cardID_v2_var_Amt_group',
'cardID_v2_var_Time_group',
'count_UserID',
'dist1',
'first_Amt_UserID',
'id_01',
'id_02',
'id_02__D8',
'id_02__id_20',
'id_03',
'id_04',
'id_05',
'id_06',
'id_07',
'id_08',
'id_09',
'id_10',
'id_11',
'id_12',
'id_13',
'id_14',
'id_15',
'id_16',
'id_17',
'id_18',
'id_19',
'id_20',
'id_21',
'id_22',
'id_23',
'id_24',
'id_25',
'id_26',
'id_27',
'id_28',
'id_29',
'id_30',
'id_30_device',
'id_30_version',
'id_31',
'id_31_device',
'id_32',
'id_33',
'id_33_0',
'id_33_1',
'id_34',
'id_35',
'id_36',
'id_37',
'id_38',
'is_holiday',
'median_Amt_UserID',
'product_type',
'product_type_DT_D',
'product_type_DT_M',
'product_type_DT_W',
'skew_Amt_UserID',
'uid5',
'uid5_TransactionDT_count',
'uid5_TransactionDT_std',
'uid_DT',
'uid_DT_C10_mean',
'uid_DT_C10_std',
'uid_DT_C11_mean',
'uid_DT_C11_std',
'uid_DT_C12_mean',
'uid_DT_C12_std',
'uid_DT_C13_mean',
'uid_DT_C13_std',
'uid_DT_C14_mean',
'uid_DT_C14_std',
'uid_DT_C1_mean',
'uid_DT_C1_std',
'uid_DT_C2_mean',
'uid_DT_C2_std',
'uid_DT_C3_mean',
'uid_DT_C3_std',
'uid_DT_C4_mean',
'uid_DT_C4_std',
'uid_DT_C5_mean',
'uid_DT_C5_std',
'uid_DT_C6_mean',
'uid_DT_C6_std',
'uid_DT_C7_mean',
'uid_DT_C7_std',
'uid_DT_C8_mean',
'uid_DT_C8_std',
'uid_DT_C9_mean',
'uid_DT_C9_std',
'uid_DT_D10_mean',
'uid_DT_D10_std',
'uid_DT_D11_mean',
'uid_DT_D11_std',
'uid_DT_D12_mean',
'uid_DT_D12_std',
'uid_DT_D13_mean',
'uid_DT_D13_std',
'uid_DT_D14_mean',
'uid_DT_D14_std',
'uid_DT_D15_mean',
'uid_DT_D15_std',
'uid_DT_D2_mean',
'uid_DT_D2_std',
'uid_DT_D3_mean',
'uid_DT_D3_std',
'uid_DT_D4_mean',
'uid_DT_D4_std',
'uid_DT_D5_mean',
'uid_DT_D5_std',
'uid_DT_D6_mean',
'uid_DT_D6_std',
'uid_DT_D7_mean',
'uid_DT_D7_std',
'uid_DT_D8_mean',
'uid_DT_D8_std',
'uid_DT_D9_mean',
'uid_DT_D9_std',
'uid_DT_TransactionAmt_mean',
'unique_Amt_UserID',
'unique_Device',
'unique_IP',
'unique_Pemail_UserID',
'unique_Remail_UserID',
'unique_adr1_UserID',
'unique_adr2_UserID',
'unique_cards_userid']
# features = list of your features
'''to_drop = [f for f in features_columns if 'DT_M' in f or 'DT_W' in f or 'DT_D' in f or 'V_279_321_' in f]
i_cols = ['D'+str(i) for i in range(1,16)]
i_cols.remove('D3')
to_drop.extend(i_cols)
to_drop.extend(['uid5','uid_DT','id_13','id_33','groups'])
features_columns = list(set(features_columns) - set(to_drop))'''
print('Used Features:', len(features_columns))
#print('Dropped Features:', len(to_drop))
# XGB Params
xgb_params = {
'objective':'binary:logistic', #comment this if using focal loss
'max_depth':5,
'learning_rate':0.07,
'subsample':0.9,
#'colsample_bytree':0.9,
'tree_method':'hist',
'eval_metric':'auc',
'seed':SEED
}
import xgboost as xgb
X_tr, X_vl, y_tr, y_vl = train_test_split(tot_df[features_columns],tot_df['target'], test_size=0.2, random_state=SEED)
print('X_tr len:',len(X_tr),'- X_vl len:',len(X_vl))
dtrain = xgb.DMatrix(X_tr, label=y_tr)
dval = xgb.DMatrix(X_vl, label=y_vl)
watchlist = [(dtrain, 'train'),(dval, 'val')]
xgbclf = xgb.train(xgb_params,
dtrain,
num_boost_round=50,
evals=watchlist,
#obj=focal_binary_object, #comment this if not using custom obj
verbose_eval=10)
val_pred = xgbclf.predict(dval)
train_pred = xgbclf.predict(dtrain)
feature_importance = pd.DataFrame(list(xgbclf.get_score(importance_type='gain').items()), columns=['Feature','Gain Value'])
val_auc = metrics.roc_auc_score(y_vl, val_pred)
train_auc = metrics.roc_auc_score(y_tr, train_pred)
print('Train AUC:', train_auc, '- Val AUC:', val_auc)
#feature_importance.to_csv('../content/gdrive/My Drive/IEEE fraud Kaggle 2019/xgb_output/feature_imp_adversarial.csv',index=False)
# For each feature
'''advers_test = pd.DataFrame(columns=['Feature','ValAuc'])
print('Number of Features:',len(features_columns))
for i in sorted(features_columns):
print('Feature:',i)
X_tr, X_vl, y_tr, y_vl = train_test_split(tot_df[[i]],tot_df['target'], test_size=0.2, random_state=SEED)
import xgboost as xgb
dtrain = xgb.DMatrix(X_tr, label=y_tr)
dval = xgb.DMatrix(X_vl, label=y_vl)
watchlist = [(dtrain, 'train'),(dval, 'val')]
xgbclf = xgb.train(xgb_params,
dtrain,
num_boost_round=50,
evals=watchlist,
#obj=focal_binary_object, #comment this if not using custom obj
verbose_eval=False)
val_pred = xgbclf.predict(dval)
train_pred = xgbclf.predict(dtrain)
feature_importance = pd.DataFrame(list(xgbclf.get_score(importance_type='gain').items()), columns=['Feature','Gain Value'])
val_auc = metrics.roc_auc_score(y_vl, val_pred)
train_auc = metrics.roc_auc_score(y_tr, train_pred)
adv = pd.DataFrame()
adv['Feature'] = [i]
adv['ValAuc'] = [round(val_auc,4)]
advers_test = advers_test.append(adv[['Feature','ValAuc']])
print('Train AUC:', round(train_auc,4), '- Val AUC:', round(val_auc,4))
print('-'*30)'''
advers_test = advers_test.sort_values(by='ValAuc', ascending=False)
advers_test.to_csv('../content/gdrive/My Drive/IEEE fraud Kaggle 2019/xgb_output/adversarial_byfeature.csv', index=False)
plt.figure(figsize=(14,60))
sns.barplot(x="Gain Value", y="Feature", data=feature_importance.sort_values(by="Gain Value", ascending=False))
plt.title('XGBoost Features')
plt.tight_layout()
#plt.savefig('../content/gdrive/My Drive/IEEE fraud Kaggle 2019/xgb_output/feature_imp_adversarial.png')
feature_importance = feature_importance.sort_values(by='Gain Value',ascending=False)
feature_importance
###Output
_____no_output_____
|
ipynb/tcga_training_normal.ipynb
|
###Markdown
TCGA RNA-Seq RSEM normalized Collaborative Filtering
###Code
%reload_ext autoreload
%autoreload 2
%matplotlib inline
from fastai.learner import *
from fastai.column_data import *
import pandas as pd
import numpy as np
path='~/data/tcga/'
###Output
_____no_output_____
###Markdown
Preprocessing subset
###Code
df = pd.read_csv(path+'full.csv', sep=',', low_memory=False, index_col=[0], error_bad_lines=False); df.head()
dupabbr = ['KIPAN', 'COADREAD', 'STES', 'GBMLGG']
df = df.loc[~df['cancer'].isin(dupabbr)]
sids = list(df.index)
df = df.assign(sid=sids)
df = df.loc[df.sid.str.split('-', expand=True).loc[:,3].str[0] == str(1)]
df = df.reset_index(); df = df.drop(columns=['index']); df.head()
len(df)
###Output
_____no_output_____
###Markdown
Melt
###Code
mdf = pd.melt(df, id_vars=['sid', 'cancer'], var_name='gene', value_name='log2exp'); mdf.head()
len(mdf)
#mdf.to_csv(path+'melted.csv')
###Output
_____no_output_____
###Markdown
Collaborative filtering
###Code
#mdf = pd.read_feather(path+'melted')
#mdf.head()
val_idxs = get_cv_idxs(len(mdf))
wd=2e-4
n_factors = 50
cd = CollabFilterDataset.from_data_frame(path, mdf, 'sid', 'gene', 'log2exp')
#cf = CollabFilterDataset.from_csv(path, 'melted.csv', 'sid', 'gene', 'log2exp')
learn = cd.get_learner(n_factors, val_idxs, 64, opt_fn=optim.Adam)
learn.lr_find()
learn.sched.plot(100)
lr=1e-4
learn.fit(lr, 2, cycle_len=1, cycle_mult=2)
preds = learn.predict()
y=learn.data.val_y
# import seaborns as sns
sns.jointplot(preds, y, kind='hex', stat_func=None);
learn.save('tcga_collab_normal_new')
#learn.load('tcga_collab_normal')
###Output
_____no_output_____
###Markdown
Analyze results shortcut to retrieve learner, model, df
###Code
df = pd.read_csv(path+'full.csv', sep=',', low_memory=False, index_col=[0], error_bad_lines=False); df.head()
dupabbr = ['KIPAN', 'COADREAD', 'STES', 'GBMLGG']
df = df.loc[~df['cancer'].isin(dupabbr)]
sids = list(df.index)
df = df.assign(sid=sids)
df = df.loc[df.sid.str.split('-', expand=True).loc[:,3].str[0] == str(1)]
df = df.reset_index(); df = df.drop(columns=['index']); df.head()
mdf = pd.melt(df, id_vars=['sid', 'cancer'], var_name='gene', value_name='log2exp'); mdf.head()
val_idxs = get_cv_idxs(len(mdf))
wd=2e-4
n_factors = 50
cd = CollabFilterDataset.from_data_frame(path, mdf, 'sid', 'gene', 'log2exp')
learn = cd.get_learner(n_factors, val_idxs, 64, opt_fn=optim.Adam)
learn.load('tcga_collab_normal_new')
genes = list(df.columns[:-2])
sids = list(df['sid'])
cancers = list(df['cancer'])
sid_ca = {s:c for s, c in zip(sids, cancers)}
###Output
_____no_output_____
###Markdown
Embedding interpretation retrieve embeddings
###Code
m=learn.model; m.cuda()
###Output
_____no_output_____
###Markdown
gene embedding
###Code
geneidx = np.array([cd.item2idx[g] for g in genes])
gene_emb = to_np(m.i(V(geneidx)))
gene_emb.shape
gene_emb_df = pd.DataFrame(gene_emb, index=genes); gene_emb_df.head()
gene_emb_df.to_csv(path+'gene_emb_normal_new.csv', sep=',')
gene_emb_bias = to_np(m.ib(V(geneidx)))
gene_emb_bias_df = pd.DataFrame(gene_emb_bias, index=genes); gene_emb_bias_df.head()
gene_emb_bias_df.to_csv(path+'gene_emb_normal_new_bias.csv')
###Output
_____no_output_____
###Markdown
sample embedding
###Code
sampleidx = np.array([cd.user2idx[sid] for sid in sids])
samp_emb = to_np(m.u(V(sampleidx)))
samp_emb.shape
samp_emb_df = pd.DataFrame(samp_emb, index=sids); samp_emb_df.head()
samp_emb_df.to_csv(path+'samp_emb_normal_new.csv', sep=',')
samp_emb_bias = to_np(m.ub(V(sampleidx)))
samp_emb_bias_df = pd.DataFrame(samp_emb_bias, index=sids); samp_emb_bias_df.head()
samp_emb_bias_df.to_csv(path+'samp_emb_normal_new_bias.csv')
###Output
_____no_output_____
|
learning_stats/chap01ex.ipynb
|
###Markdown
Examples and Exercises from Think Stats, 2nd Editionhttp://thinkstats2.comCopyright 2016 Allen B. DowneyMIT License: https://opensource.org/licenses/MIT
###Code
import sys
import os
sys.path.insert(0, os.getcwd() + '/ThinkStats2/code')
sys.path
from __future__ import print_function, division
import nsfg
###Output
_____no_output_____
###Markdown
Examples from Chapter 1Read NSFG data into a Pandas DataFrame.
###Code
import nsfg
datafile_base= "ThinkStats2/code/2002FemPreg"
preg = nsfg.ReadFemPreg(dct_file =datafile_base + ".dct", dat_file =datafile_base + ".dat.gz")
#preg = nsfg.ReadFemPreg()
preg.head(10)
###Output
_____no_output_____
###Markdown
Print the column names.
###Code
preg.columns
###Output
_____no_output_____
###Markdown
Select a single column name.
###Code
preg.columns[1]
###Output
_____no_output_____
###Markdown
Select a column and check what type it is.
###Code
pregordr = preg['pregordr']
type(pregordr)
###Output
_____no_output_____
###Markdown
Print a column.
###Code
pregordr
###Output
_____no_output_____
###Markdown
Select a single element from a column.
###Code
pregordr[0]
###Output
_____no_output_____
###Markdown
Select a slice from a column.
###Code
pregordr[2:5]
###Output
_____no_output_____
###Markdown
Select a column using dot notation.
###Code
pregordr = preg.pregordr
###Output
_____no_output_____
###Markdown
Count the number of times each value occurs.
###Code
preg.outcome.value_counts().sort_index()
###Output
_____no_output_____
###Markdown
Check the values of another variable.
###Code
preg.birthwgt_lb.value_counts().sort_index()
###Output
_____no_output_____
###Markdown
Make a dictionary that maps from each respondent's `caseid` to a list of indices into the pregnancy `DataFrame`. Use it to select the pregnancy outcomes for a single respondent.
###Code
caseid = 10229
preg_map = nsfg.MakePregMap(preg)
indices = preg_map[caseid]
preg.outcome[indices].values
###Output
_____no_output_____
###Markdown
Exercises Select the `birthord` column, print the value counts, and compare to results published in the [codebook](http://www.icpsr.umich.edu/nsfg6/Controller?displayPage=labelDetails&fileCode=PREG§ion=A&subSec=8016&srtLabel=611933)
###Code
preg.birthord.value_counts()
###Output
_____no_output_____
###Markdown
We can also use `isnull` to count the number of nans.
###Code
preg.birthord.isnull().sum()
###Output
_____no_output_____
###Markdown
Select the `prglngth` column, print the value counts, and compare to results published in the [codebook](http://www.icpsr.umich.edu/nsfg6/Controller?displayPage=labelDetails&fileCode=PREG§ion=A&subSec=8016&srtLabel=611931)
###Code
from IPython.core.interactiveshell import InteractiveShell
InteractiveShell.ast_node_interactivity = "all"
lmax = preg.prglngth.max()
lmin = preg.prglngth.min()
preg.prglngth.value_counts(bins=[lmin,13,26,lmax])
###Output
_____no_output_____
###Markdown
To compute the mean of a column, you can invoke the `mean` method on a Series. For example, here is the mean birthweight in pounds:
###Code
preg.totalwgt_lb.mean()
preg.totalwgt_lb.value_counts().sort_index()
import matplotlib
?preg.totalwgt_lb.value_counts()
%config InlineBackend.figure_format = 'retina'
%matplotlib inline
matplotlib.style.use('ggplot')
preg.totalwgt_lb.value_counts(sort=True).sort_index().plot()
###Output
_____no_output_____
###Markdown
Create a new column named totalwgt_kg that contains birth weight in kilograms. Compute its mean. Remember that when you create a new column, you have to use dictionary syntax, not dot notation.
###Code
preg['totalwgt_kg'] = preg.totalwgt_lb * 0.45359237
preg.totalwgt_kg.mean()
preg.totalwgt_lb.mean() * 0.45359237
###Output
_____no_output_____
###Markdown
`nsfg.py` also provides `ReadFemResp`, which reads the female respondents file and returns a `DataFrame`:
###Code
import nsfg
datafile_base= "ThinkStats2/code/2002FemResp"
resp = nsfg.ReadFemResp(dct_file =datafile_base + ".dct", dat_file =datafile_base + ".dat.gz")
###Output
_____no_output_____
###Markdown
`DataFrame` provides a method `head` that displays the first five rows:
###Code
resp.head()
###Output
_____no_output_____
###Markdown
Select the `age_r` column from `resp` and print the value counts. How old are the youngest and oldest respondents?
###Code
resp.age_r.value_counts().sort_index()
###Output
_____no_output_____
###Markdown
We can use the `caseid` to match up rows from `resp` and `preg`. For example, we can select the row from `resp` for `caseid` 2298 like this:
###Code
resp[resp.caseid==2298]
###Output
_____no_output_____
###Markdown
And we can get the corresponding rows from `preg` like this:
###Code
preg[preg.caseid==2298]
###Output
_____no_output_____
###Markdown
How old is the respondent with `caseid` 1?
###Code
preg.columns
resp.columns
resp[resp.caseid == 1].age_r
###Output
_____no_output_____
###Markdown
What are the pregnancy lengths for the respondent with `caseid` 2298?
###Code
preg[preg.caseid==2298].prglngth
###Output
_____no_output_____
###Markdown
What was the birthweight of the first baby born to the respondent with `caseid` 5012?
###Code
preg[preg.caseid==5012]
preg[preg.caseid==5012].birthwgt_lb
###Output
_____no_output_____
|
Intro to Python/Intro to Python Book 4 (operators and selection).ipynb
|
###Markdown
Dr Alan Davies Senior Lecturer Health Data Science University of Manchester 4.0 Operators and selection **** About this NotebookThis notebook introduces operators that can be used for both arithmetic and logical computations. We also introduce selection that uses these operators in order to make decisions in programs and provide interactivity based on user input. Learning Objectives: At the end of this notebook you will be able to: - Investigate the key features of arithmetic and logical operators in Python- Explore and practice using selection to make decisions to alter the flow of execution in programs Table of contents4.1 [Arithmetic operators](arithops)4.2 [Logical operators](logicops)4.3 [Selection](selection) To carry out computational tasks, Python (and all other high level programming languages) use operators. These operators can be broadly split into ones used for arithmetic, comparison (comparing things) and logic for making choices. The arithmetic operators allow basic mathematical tasks to be carried out, like addition and subtraction and can be combined to make more complex statements just as in maths. For example:
###Code
some_number = 20
some_other_number = 30
print(some_number + some_other_number)
print(some_number + some_other_number - 5 + 8)
###Output
53
###Markdown
Note: As with maths the order of operations matters. This can be recalled with the acronym BODMAS. Brackets, Orders (powers, square roots etc.), Division Or Multiplication (left to right) and Addition or Subtraction (left to right). One issue to be aware of in computation is division by zero. This is where a value is divided by zero ($ a \div 0$). This always causes an error in programing as the expression has no meaning. This can happen often by accident if the variable you are dividing by contains zero. For example:
###Code
a = 8
a / 0
###Output
_____no_output_____
###Markdown
his triggered a ZeroDivisionError. To see how to handle errors like this, have a look at the notebook on Testing and error handing later in the series. 4.1 Arithmetic operators .tg {border-collapse:collapse;border-spacing:0;}.tg td{font-family:Arial, sans-serif;font-size:14px;padding:10px 5px;border-style:solid;border-width:1px;overflow:hidden;word-break:normal;border-color:black;}.tg th{font-family:Arial, sans-serif;font-size:14px;font-weight:normal;padding:10px 5px;border-style:solid;border-width:1px;overflow:hidden;word-break:normal;border-color:black;}.tg .tg-kiyi{font-weight:bold;border-color:inherit;text-align:left}.tg .tg-fymr{font-weight:bold;border-color:inherit;text-align:left;vertical-align:top}.tg .tg-xldj{border-color:inherit;text-align:left}.tg .tg-0pky{border-color:inherit;text-align:left;vertical-align:top} Example Meaning Math Description a + b Addition a + b Sums values a - b Subtraction a - b Subtracts second number from first a $\text{*}$ b Multiplication a $\times$ b Multiplies values (product) a / b Division a $\div$ b Divides a by b a % b Modulo mod The remainder of a division a // b Floor division floor() Division rounded to smallest integer a ** b Exponentiation $a^b$ Raises a to power of b The table above shows the arithmetic operators along with a description. Most of these should be fairly straight forward if you are familiar with high school level maths. We will take a closer look at the mod operator as this may be one that is less familiar to some. This will give use the remainder of a division and can be useful for many things. One obvious example is to see if a number is odd or even. Other real world applications are applications that process money and need to give change.
###Code
num = 2
num % 2
###Output
_____no_output_____
###Markdown
An even number will give a zero answer and an odd number will not.
###Code
num = 9
num % 2
###Output
_____no_output_____
###Markdown
Task 1: Here is the formular for converting degrees from farenheit to celcius:$$ c = \frac{5}{9}(f-32) $$Try and use the math operators to write this formular in Python. To test it works try giving f the value of 32. This should be around 0 in $ ^{\circ}$C.
###Code
f = 32
c = (f-32)*5/9
print(c)
###Output
0.0
###Markdown
4.2 Logical operators When we have True or False expressions (called Boolean objects or expressions), we can evaluate them using logical operators. We can do this in programs to make choices. For example if you wanted to decide to take an umbrella with you or not, you may look out of the window and ask "is it raining?". If the answer is Yes (True) you would take your umbrella. If however the answer was No (False) you would not. More complex choices can be made by chaining these logical operators together. For example if it is raining and I own an umbrella then take it.. First lets look at the logical operators and then see how we can use them to preform selection. Example Meaning Math Description not a Not $\lnot a$, $\bar{a}$ Reverses value of a. If True becomes False and vise versa a and b And $a \land b$, $a \cap b$ This is True if a and b are True a or b Or $a \lor b$, $a \cup b$ This is True is either a or b are True Another way of visualising this is with a venn diagram. The image below shows what this looks like for and and or. 4.3 Selection We can combine these operators to make decisions in our programs. This is one of the main purposes of computer programs, the ability to do different things based on its input. Here we can see some examples of how we can make choices using these operators and the keywords if and else.
###Code
raining = False
if raining == True:
print("Need to take umbrella")
else:
print("Let's go")
###Output
Let's go
###Markdown
Note: We use a double equals == for comparing equality (is the thing on the left equal to the thing on the right?) The single equals = is used for assignment. If you missed the second equals in the example above you would be assigning True to raining instead of testing to see if the variable raining contains the the value True. There are a few new things here to pay attention to. First we define a variable called raining and set it to False. Then we ask a question (is it raining?). This line ends with a colon (:). This tells Python that the preceding code which is indented (moved/tabbed in) belongs to, or is contained within the line with the colon. This means that the line print("Need to take umbrella") will only be executed if the value of raining is True. The else keyword describes what happens if the initial condition is not met. i.e. if raining is False. Note: It is not mandatory to have an else statement. Task 2: Change the value of raining from False to True and run the code in the cell above again.
###Code
raining = True
if raining == True:
print("Need to take umbrella")
else:
print("Let's go")
###Output
Need to take umbrella
###Markdown
If we want multiple alternative conditions to be checked we can use the elif (else if) keyword. Below we have several statements combined with the and operator.
###Code
raining = True
own_umbrella = True
if raining == True and own_umbrella == True:
print("Take umbrella")
elif raining == True and own_umbrella == False:
print("I'm going to get wet")
else:
print("A nice day!")
###Output
Take umbrella
###Markdown
We also don't have to explicitly say == True, as saying if raining works just as well. We could rewrite it as follows:
###Code
raining = True
own_umbrella = True
if raining and own_umbrella:
print("Take umbrella")
elif raining and not own_umbrella:
print("I'm going to get wet")
else:
print("A nice day!")
###Output
Take umbrella
###Markdown
This sort of short hand is something that you will develop with practice and intuition. Task 3: 1. Using what you have learnt. Use if and elif/else statements to take in an exam score and give the following grades:less than 10 is a fail. Between 11 and 45 is a pass, between 46 and 65 is a merit and anything over 65 is a distinction.2. Try changing the exam_grade to test that your logic is working as expected.We started it off for you:
###Code
exam_grade = 56
if exam_grade < 10:
print("Fail")
elif exam_grade >= 11 and exam_grade <= 45:
print("Pass")
elif exam_grade >= 46 and exam_grade <= 55:
print("Merit")
elif exam_grade > 55:
print("Distinction")
else:
print("Not a valid exam grade")
exam_grade = 56
if exam_grade < 10:
print("Fail")
elif exam_grade >= 11 and exam_grade <= 45:
print("Pass")
###Output
_____no_output_____
###Markdown
The input() function let's a user enter a value that we can store in a variable. We can also ask a question using a string like so:
###Code
user_age = input("Please enter your age: ")
print("You are", user_age, "years old")
###Output
Please enter your age: 34
You are 34 years old
###Markdown
Task 4: 1. Using the input() function, ask the user to enter a number. 2. Using the mod operator display a message telling the user if that the number was odd or even
###Code
user_number = int(input("Please enter a number: "))
if user_number % 2 == 0:
print("Even number")
else:
print("Odd number")
###Output
_____no_output_____
|
notebooks/1a_mlp.ipynb
|
###Markdown
Perceptrón multicapa Bere & Ricardo Montalvo LezamaEn esta libreta veremos un ejemplo de clasificación multiclase de imágenes de dígitos implementando un perceptrón multicapa en PyTorch.Emplearemos un conjunto referencia llamado [MNIST](http://yann.lecun.com/exdb/mnist/) recolectado por [Yann LeCun](http://yann.lecun.com). Está compuesto de imágenes en escala de grises de 28 × 28 píxeles que contienen dígitos entre 0 y 9 escritos a mano. El conjunto cuenta con 60,000 imágenes de entrenamiento y 10,000 de prueba. 1 Preparación 1.1 Bibliotecas
###Code
# funciones aleatorias
import random
# tomar n elementos de una secuencia
from itertools import islice as take
# gráficas
import matplotlib.pyplot as plt
# arreglos multidimensionales
import numpy as np
# redes neuronales
import torch
import torch.nn as nn
import torch.nn.functional as F
import torch.optim as optim
# imágenes
from skimage import io
# redes neuronales
from torch.utils.data import DataLoader
from torchvision import transforms
from torchvision.datasets import MNIST
# barras de progreso
from tqdm import tqdm
# directorio de datos
DATA_DIR = '../data'
# MNIST
MEAN = (0.1307)
STD = (0.3081)
# tamaño del lote
BATCH_SIZE = 128
# reproducibilidad
SEED = 0
random.seed(SEED)
np.random.seed(SEED)
torch.manual_seed(SEED)
###Output
_____no_output_____
###Markdown
1.2 Auxiliares
###Code
def display_grid(xs, titles, rows, cols):
fig, ax = plt.subplots(rows, cols)
for r in range(rows):
for c in range(cols):
i = r * rows + c
ax[r, c].imshow(xs[i], cmap='gray')
ax[r, c].set_title(titles[i])
ax[r, c].set_xticklabels([])
ax[r, c].set_yticklabels([])
fig.tight_layout()
plt.show()
###Output
_____no_output_____
###Markdown
2 Datos 2.1 Tuberias de datos con PyTorch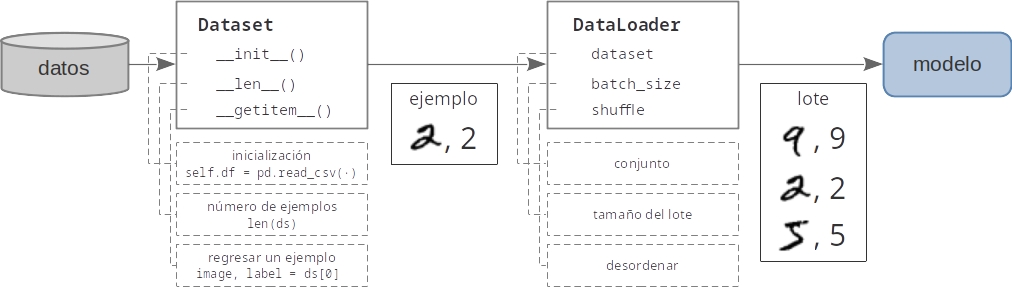 2.2 Exploración
###Code
# creamos un Dataset
ds = MNIST(
# directorio de datos
root=DATA_DIR,
# subconjunto de entrenamiento
train=True,
# convertir la imagen a ndarray
transform=np.array,
# descargar el conjunto
download=True
)
# cargamos algunas imágenes
images, labels = [], []
for i in range(12):
x, y = ds[i]
images.append(x)
labels.append(y)
# desplegamos
print(f'images[0] shape={images[0].shape} dtype={images[0].dtype}')
titles = [str(y) for y in labels]
display_grid(images, titles, 3, 4)
###Output
images[0] shape=(28, 28) dtype=uint8
###Markdown
2.3 Cargadores de datos Entrenamiento
###Code
# transformaciones para la imagen
trn_tsfm = transforms.Compose([
# convertimos a torch.Tensor y escalamos a [0,1]
transforms.ToTensor(),
# estandarizamos: restamos la media y dividimos sobre la varianza
transforms.Normalize(MEAN, STD),
])
# creamos un Dataset
trn_ds = MNIST(
# directorio de datos
root=DATA_DIR,
# subconjunto de entrenamiento
train=True,
# transformación
transform=trn_tsfm
)
# creamos un DataLoader
trn_dl = DataLoader(
# conjunto
trn_ds,
# tamaño del lote
batch_size=BATCH_SIZE,
# desordenar
shuffle=True
)
# desplegamos un lote de imágenes
for x, y in take(trn_dl, 1):
print(f'x shape={x.shape} dtype={x.dtype}')
print(f'y shape={y.shape} dtype={y.dtype}')
###Output
x shape=torch.Size([128, 1, 28, 28]) dtype=torch.float32
y shape=torch.Size([128]) dtype=torch.int64
###Markdown
Prueba
###Code
# transformaciones para la imagen
tst_tsfm = transforms.Compose([
# convertimos a torch.Tensor y escalamos a [0,1]
transforms.ToTensor(),
# estandarizamos: restamos la media y dividimos sobre la varianza
transforms.Normalize(MEAN, STD),
])
# creamos un Dataset
tst_ds = MNIST(
# directorio de datos
root=DATA_DIR,
# subconjunto de entrenamiento
train=False,
# transformación
transform=tst_tsfm
)
# creamos un DataLoader
tst_dl = DataLoader(
# subconjunto
tst_ds,
# tamaño del lote
batch_size=BATCH_SIZE,
# desordenar
shuffle=True
)
# imprimimos forma y tipo del lote
for x, y in take(tst_dl, 1):
print(f'x shape={x.shape} dtype={x.dtype}')
print(f'y shape={y.shape} dtype={y.dtype}')
###Output
x shape=torch.Size([128, 1, 28, 28]) dtype=torch.float32
y shape=torch.Size([128]) dtype=torch.int64
###Markdown
3 Modelo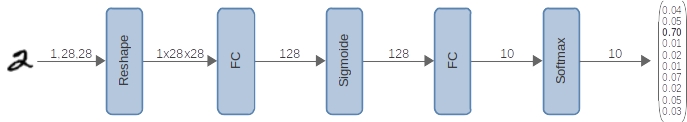 3.1 Definición de la arquitectura
###Code
# definición del modelo
class MLP(nn.Module):
# inicializador
def __init__(self):
# inicilización del objeto padre, obligatorio
super(MLP, self).__init__()
# tamaño de las capas
self.I = 1 * 28 * 28
FC1, FC2 = 128, 10
# definición de capas
self.cls = nn.Sequential(
# fc1
# [N, 1x28x28] => [N, 128]
nn.Linear(self.I, FC1),
nn.Sigmoid(),
# fc2
# [N, 128] => [N, 10]
nn.Linear(FC1, FC2)
)
# metodo para inferencia
def forward(self, x):
# aplanamos los pixeles de la imagen
# [N, 1, 28, 28] => [N, 1x28x28]
x = x.view(-1, self.I)
# inferencia
# [N, 1x28x28]
x = self.cls(x)
return x
###Output
_____no_output_____
###Markdown
3.2 Impresión de la arquitectura
###Code
model = MLP()
print(model)
###Output
MLP(
(cls): Sequential(
(0): Linear(in_features=784, out_features=128, bias=True)
(1): Sigmoid()
(2): Linear(in_features=128, out_features=10, bias=True)
)
)
###Markdown
3.3 Prueba de la arquitectura
###Code
# inferencia con datos sinteticos
x = torch.zeros(1, 1, 28, 28)
y = model(x)
print(y.shape)
###Output
torch.Size([1, 10])
###Markdown
4 Entrenamiento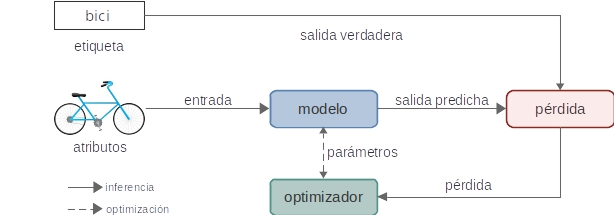 4.1 Ciclo de entrenamiento
###Code
# creamos un modelo
model = MLP()
# optimizador
opt = optim.SGD(model.parameters(), lr=1e-3)
# historial de pérdida
loss_hist = []
# ciclo de entrenamiento
EPOCHS = 20
for epoch in range(EPOCHS):
# entrenamiento de una época
for x, y_true in trn_dl:
# vaciamos los gradientes
opt.zero_grad()
# hacemos inferencia para obtener los logits
y_lgts = model(x)
# calculamos la pérdida
loss = F.cross_entropy(y_lgts, y_true)
# retropropagamos
loss.backward()
# actulizamos parámetros
opt.step()
# evitamos que se registren las operaciones
# en la gráfica de cómputo
with torch.no_grad():
losses, accs = [], []
# validación de la época
for x, y_true in take(tst_dl, 10):
# hacemos inferencia para obtener los logits
y_lgts = model(x)
# calculamos las probabilidades
y_prob = F.softmax(y_lgts, 1)
# obtenemos la clase predicha
y_pred = torch.argmax(y_prob, 1)
# calculamos la pérdida
loss = F.cross_entropy(y_lgts, y_true)
# calculamos la exactitud
acc = (y_true == y_pred).type(torch.float32).mean()
# guardamos históricos
losses.append(loss.item() * 100)
accs.append(acc.item() * 100)
# imprimimos métricas
loss = np.mean(losses)
acc = np.mean(accs)
print(f'E{epoch:2} loss={loss:6.2f} acc={acc:.2f}')
# agregagmos al historial de pérdidas
loss_hist.append(loss)
###Output
E 0 loss=219.32 acc=40.23
E 1 loss=208.85 acc=59.38
E 2 loss=197.03 acc=66.25
E 3 loss=186.14 acc=69.69
E 4 loss=175.65 acc=71.41
E 5 loss=163.80 acc=71.02
E 6 loss=154.36 acc=73.67
E 7 loss=144.48 acc=73.83
E 8 loss=136.58 acc=74.61
E 9 loss=126.75 acc=78.75
E10 loss=119.59 acc=77.73
E11 loss=112.71 acc=79.45
E12 loss=112.32 acc=78.52
E13 loss=102.80 acc=80.55
E14 loss= 99.78 acc=81.17
E15 loss= 91.90 acc=83.12
E16 loss= 90.61 acc=82.73
E17 loss= 86.08 acc=83.75
E18 loss= 85.83 acc=82.58
E19 loss= 82.65 acc=83.36
###Markdown
4.2 Gráfica de la pérdida
###Code
plt.plot(loss_hist, color='red')
plt.xlabel('época')
plt.ylabel('pérdida')
plt.show()
###Output
_____no_output_____
###Markdown
5 Evaluación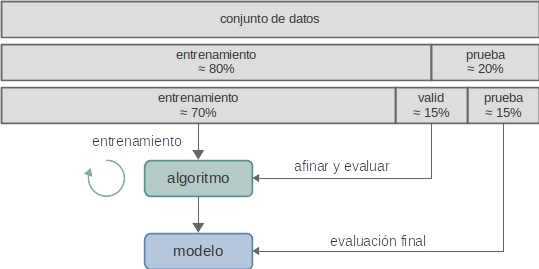 5.1 Conjunto de validación
###Code
# modelo en modo de evaluación
model.eval()
# evitamos que se registren las operaciones
# en la gráfica de cómputo
with torch.no_grad():
accs = []
# validación de la época
for x, y_true in tst_dl:
# hacemos inferencia para obtener los logits
y_lgts = model(x)
# calculamos las probabilidades
y_prob = F.softmax(y_lgts, 1)
# obtenemos la clase predicha
y_pred = torch.argmax(y_prob, 1)
# calculamos la exactitud
acc = (y_true == y_pred).type(torch.float32).mean()
accs.append(acc.item() * 100)
acc = np.mean(accs)
print(f'Exactitud = {acc:.2f}')
###Output
Exactitud = 84.42
###Markdown
5.2 Inferencia
###Code
with torch.no_grad():
for x, y_true in take(tst_dl, 1):
y_lgts = model(x)
y_prob = F.softmax(y_lgts, 1)
y_pred = torch.argmax(y_prob, 1)
x = x[:12].squeeze().numpy()
y_true = y_true[:12].numpy()
y_pred = y_pred[:12].numpy()
titles = [f'V={t} P={p}' for t, p in zip(y_true, y_pred)]
display_grid(x, titles, 3, 4)
###Output
_____no_output_____
|
talks/FederalDevSummit2019/PlenaryDemos/demo 02 - RA_ML/03 - Missouri Floods Analysis.ipynb
|
###Markdown
Missouri Flood AnalysisParts of Missouri experienced historic flooding recently. Some locations saw as much as 11 inches of rainfall. This notebook performs flood classification followed by impact assessment.
###Code
from arcgis import GIS
from arcgis.mapping import MapImageLayer
gis = GIS(profile="idtportal", verify_cert=False)
###Output
_____no_output_____
###Markdown
Visualize the extent of damage
###Code
def side_by_side(address):
pass
from ipywidgets import *
def side_by_side(address):
postflood = MapImageLayer('https://tiles.arcgis.com/tiles/DO4gTjwJVIJ7O9Ca/arcgis/rest/services/Missouri_Flood_Imagery/MapServer')
normal = MapImageLayer('https://server.arcgisonline.com/ArcGIS/rest/services/World_Imagery/MapServer')
location = geocode(address)[0]
satmap1 = gis.map(location)
satmap1.add_layer(normal)
satmap2 = gis.map(location)
#satmap2.add_layer(postflood)
satmap2.add_layer({'type':'ArcGISTiledMapServiceLayer', 'url':'https://tiles.arcgis.com/tiles/DO4gTjwJVIJ7O9Ca/arcgis/rest/services/Missouri_Flood_Imagery/MapServer'})
satmap1.layout=Layout(flex='1 1', padding='6px', height='420px')
satmap2.layout=Layout(flex='1 1', padding='6px', height='420px')
box = HBox([satmap1, satmap2])
return box
###Output
_____no_output_____
###Markdown
Visualize damage
###Code
side_by_side('Eureka High School, Eureka, MO 63025')
###Output
_____no_output_____
###Markdown
Load pre and post flood layers
###Code
missouri_pre_flood_item = gis.content.search("title:MissouriFloodsPre", "Imagery Layer")[0]
missouri_pre_flood = missouri_pre_flood_item.layers[0]
###Output
_____no_output_____
###Markdown
###Code
missouri_post_flood_item = gis.content.search("title:MissouriFloodsPost", "Imagery Layer")[0]
missouri_post_flood = missouri_post_flood_item.layers[0]
###Output
_____no_output_____
###Markdown
Flood Classification Train the Classifier
###Code
import json
from arcgis.features import FeatureSet
with open("full_fc.json", "r") as training_sample_file:
training_samples = training_sample_file.read()
fs = FeatureSet.from_json(training_samples)
map_widget = gis.map()
map_widget.center = [38.541585,-90.488005]
map_widget.zoom = 15
map_widget.add_layer(missouri_post_flood)
map_widget
###Output
_____no_output_____
###Markdown
###Code
symbol = {
"type" : "simple-fill",
"outline" : {"color": [85, 255, 0, 1]},
"color": [255, 0, 0, 0.57]
}
map_widget.draw(fs, symbol=symbol)
###Output
_____no_output_____
###Markdown
Perform Classification
###Code
from arcgis.raster.analytics import train_classifier
classifier_definition = train_classifier(input_raster=missouri_post_flood,
input_training_sample_json=training_samples,
classifier_parameters={"method":"svm",
"params":{"maxSampleClass":1000}},
gis=gis)
from arcgis.raster.functions import classify
classified_output = classify(raster1=missouri_post_flood,
classifier_definition=classifier_definition)
classified_output
###Output
_____no_output_____
|
tutorials/notebook/cx_site_chart_examples/density_10.ipynb
|
###Markdown
Example: CanvasXpress density Chart No. 10This example page demonstrates how to, using the Python package, create a chart that matches the CanvasXpress online example located at:https://www.canvasxpress.org/examples/density-10.htmlThis example is generated using the reproducible JSON obtained from the above page and the `canvasxpress.util.generator.generate_canvasxpress_code_from_json_file()` function.Everything required for the chart to render is included in the code below. Simply run the code block.
###Code
from canvasxpress.canvas import CanvasXpress
from canvasxpress.js.collection import CXEvents
from canvasxpress.render.jupyter import CXNoteBook
cx = CanvasXpress(
render_to="density10",
data={
"z": {
"Species": [
"setosa",
"setosa",
"setosa",
"setosa",
"setosa",
"setosa",
"setosa",
"setosa",
"setosa",
"setosa",
"setosa",
"setosa",
"setosa",
"setosa",
"setosa",
"setosa",
"setosa",
"setosa",
"setosa",
"setosa",
"setosa",
"setosa",
"setosa",
"setosa",
"setosa",
"setosa",
"setosa",
"setosa",
"setosa",
"setosa",
"setosa",
"setosa",
"setosa",
"setosa",
"setosa",
"setosa",
"setosa",
"setosa",
"setosa",
"setosa",
"setosa",
"setosa",
"setosa",
"setosa",
"setosa",
"setosa",
"setosa",
"setosa",
"setosa",
"setosa",
"versicolor",
"versicolor",
"versicolor",
"versicolor",
"versicolor",
"versicolor",
"versicolor",
"versicolor",
"versicolor",
"versicolor",
"versicolor",
"versicolor",
"versicolor",
"versicolor",
"versicolor",
"versicolor",
"versicolor",
"versicolor",
"versicolor",
"versicolor",
"versicolor",
"versicolor",
"versicolor",
"versicolor",
"versicolor",
"versicolor",
"versicolor",
"versicolor",
"versicolor",
"versicolor",
"versicolor",
"versicolor",
"versicolor",
"versicolor",
"versicolor",
"versicolor",
"versicolor",
"versicolor",
"versicolor",
"versicolor",
"versicolor",
"versicolor",
"versicolor",
"versicolor",
"versicolor",
"versicolor",
"versicolor",
"versicolor",
"versicolor",
"versicolor",
"virginica",
"virginica",
"virginica",
"virginica",
"virginica",
"virginica",
"virginica",
"virginica",
"virginica",
"virginica",
"virginica",
"virginica",
"virginica",
"virginica",
"virginica",
"virginica",
"virginica",
"virginica",
"virginica",
"virginica",
"virginica",
"virginica",
"virginica",
"virginica",
"virginica",
"virginica",
"virginica",
"virginica",
"virginica",
"virginica",
"virginica",
"virginica",
"virginica",
"virginica",
"virginica",
"virginica",
"virginica",
"virginica",
"virginica",
"virginica",
"virginica",
"virginica",
"virginica",
"virginica",
"virginica",
"virginica",
"virginica",
"virginica",
"virginica",
"virginica"
]
},
"y": {
"vars": [
"s1",
"s2",
"s3",
"s4",
"s5",
"s6",
"s7",
"s8",
"s9",
"s10",
"s11",
"s12",
"s13",
"s14",
"s15",
"s16",
"s17",
"s18",
"s19",
"s20",
"s21",
"s22",
"s23",
"s24",
"s25",
"s26",
"s27",
"s28",
"s29",
"s30",
"s31",
"s32",
"s33",
"s34",
"s35",
"s36",
"s37",
"s38",
"s39",
"s40",
"s41",
"s42",
"s43",
"s44",
"s45",
"s46",
"s47",
"s48",
"s49",
"s50",
"s51",
"s52",
"s53",
"s54",
"s55",
"s56",
"s57",
"s58",
"s59",
"s60",
"s61",
"s62",
"s63",
"s64",
"s65",
"s66",
"s67",
"s68",
"s69",
"s70",
"s71",
"s72",
"s73",
"s74",
"s75",
"s76",
"s77",
"s78",
"s79",
"s80",
"s81",
"s82",
"s83",
"s84",
"s85",
"s86",
"s87",
"s88",
"s89",
"s90",
"s91",
"s92",
"s93",
"s94",
"s95",
"s96",
"s97",
"s98",
"s99",
"s100",
"s101",
"s102",
"s103",
"s104",
"s105",
"s106",
"s107",
"s108",
"s109",
"s110",
"s111",
"s112",
"s113",
"s114",
"s115",
"s116",
"s117",
"s118",
"s119",
"s120",
"s121",
"s122",
"s123",
"s124",
"s125",
"s126",
"s127",
"s128",
"s129",
"s130",
"s131",
"s132",
"s133",
"s134",
"s135",
"s136",
"s137",
"s138",
"s139",
"s140",
"s141",
"s142",
"s143",
"s144",
"s145",
"s146",
"s147",
"s148",
"s149",
"s150"
],
"smps": [
"Sepal.Length",
"Sepal.Width",
"Petal.Length",
"Petal.Width"
],
"data": [
[
5.1,
3.5,
1.4,
0.2
],
[
4.9,
3,
1.4,
0.2
],
[
4.7,
3.2,
1.3,
0.2
],
[
4.6,
3.1,
1.5,
0.2
],
[
5,
3.6,
1.4,
0.2
],
[
5.4,
3.9,
1.7,
0.4
],
[
4.6,
3.4,
1.4,
0.3
],
[
5,
3.4,
1.5,
0.2
],
[
4.4,
2.9,
1.4,
0.2
],
[
4.9,
3.1,
1.5,
0.1
],
[
5.4,
3.7,
1.5,
0.2
],
[
4.8,
3.4,
1.6,
0.2
],
[
4.8,
3,
1.4,
0.1
],
[
4.3,
3,
1.1,
0.1
],
[
5.8,
4,
1.2,
0.2
],
[
5.7,
4.4,
1.5,
0.4
],
[
5.4,
3.9,
1.3,
0.4
],
[
5.1,
3.5,
1.4,
0.3
],
[
5.7,
3.8,
1.7,
0.3
],
[
5.1,
3.8,
1.5,
0.3
],
[
5.4,
3.4,
1.7,
0.2
],
[
5.1,
3.7,
1.5,
0.4
],
[
4.6,
3.6,
1,
0.2
],
[
5.1,
3.3,
1.7,
0.5
],
[
4.8,
3.4,
1.9,
0.2
],
[
5,
3,
1.6,
0.2
],
[
5,
3.4,
1.6,
0.4
],
[
5.2,
3.5,
1.5,
0.2
],
[
5.2,
3.4,
1.4,
0.2
],
[
4.7,
3.2,
1.6,
0.2
],
[
4.8,
3.1,
1.6,
0.2
],
[
5.4,
3.4,
1.5,
0.4
],
[
5.2,
4.1,
1.5,
0.1
],
[
5.5,
4.2,
1.4,
0.2
],
[
4.9,
3.1,
1.5,
0.2
],
[
5,
3.2,
1.2,
0.2
],
[
5.5,
3.5,
1.3,
0.2
],
[
4.9,
3.6,
1.4,
0.1
],
[
4.4,
3,
1.3,
0.2
],
[
5.1,
3.4,
1.5,
0.2
],
[
5,
3.5,
1.3,
0.3
],
[
4.5,
2.3,
1.3,
0.3
],
[
4.4,
3.2,
1.3,
0.2
],
[
5,
3.5,
1.6,
0.6
],
[
5.1,
3.8,
1.9,
0.4
],
[
4.8,
3,
1.4,
0.3
],
[
5.1,
3.8,
1.6,
0.2
],
[
4.6,
3.2,
1.4,
0.2
],
[
5.3,
3.7,
1.5,
0.2
],
[
5,
3.3,
1.4,
0.2
],
[
7,
3.2,
4.7,
1.4
],
[
6.4,
3.2,
4.5,
1.5
],
[
6.9,
3.1,
4.9,
1.5
],
[
5.5,
2.3,
4,
1.3
],
[
6.5,
2.8,
4.6,
1.5
],
[
5.7,
2.8,
4.5,
1.3
],
[
6.3,
3.3,
4.7,
1.6
],
[
4.9,
2.4,
3.3,
1
],
[
6.6,
2.9,
4.6,
1.3
],
[
5.2,
2.7,
3.9,
1.4
],
[
5,
2,
3.5,
1
],
[
5.9,
3,
4.2,
1.5
],
[
6,
2.2,
4,
1
],
[
6.1,
2.9,
4.7,
1.4
],
[
5.6,
2.9,
3.6,
1.3
],
[
6.7,
3.1,
4.4,
1.4
],
[
5.6,
3,
4.5,
1.5
],
[
5.8,
2.7,
4.1,
1
],
[
6.2,
2.2,
4.5,
1.5
],
[
5.6,
2.5,
3.9,
1.1
],
[
5.9,
3.2,
4.8,
1.8
],
[
6.1,
2.8,
4,
1.3
],
[
6.3,
2.5,
4.9,
1.5
],
[
6.1,
2.8,
4.7,
1.2
],
[
6.4,
2.9,
4.3,
1.3
],
[
6.6,
3,
4.4,
1.4
],
[
6.8,
2.8,
4.8,
1.4
],
[
6.7,
3,
5,
1.7
],
[
6,
2.9,
4.5,
1.5
],
[
5.7,
2.6,
3.5,
1
],
[
5.5,
2.4,
3.8,
1.1
],
[
5.5,
2.4,
3.7,
1
],
[
5.8,
2.7,
3.9,
1.2
],
[
6,
2.7,
5.1,
1.6
],
[
5.4,
3,
4.5,
1.5
],
[
6,
3.4,
4.5,
1.6
],
[
6.7,
3.1,
4.7,
1.5
],
[
6.3,
2.3,
4.4,
1.3
],
[
5.6,
3,
4.1,
1.3
],
[
5.5,
2.5,
4,
1.3
],
[
5.5,
2.6,
4.4,
1.2
],
[
6.1,
3,
4.6,
1.4
],
[
5.8,
2.6,
4,
1.2
],
[
5,
2.3,
3.3,
1
],
[
5.6,
2.7,
4.2,
1.3
],
[
5.7,
3,
4.2,
1.2
],
[
5.7,
2.9,
4.2,
1.3
],
[
6.2,
2.9,
4.3,
1.3
],
[
5.1,
2.5,
3,
1.1
],
[
5.7,
2.8,
4.1,
1.3
],
[
6.3,
3.3,
6,
2.5
],
[
5.8,
2.7,
5.1,
1.9
],
[
7.1,
3,
5.9,
2.1
],
[
6.3,
2.9,
5.6,
1.8
],
[
6.5,
3,
5.8,
2.2
],
[
7.6,
3,
6.6,
2.1
],
[
4.9,
2.5,
4.5,
1.7
],
[
7.3,
2.9,
6.3,
1.8
],
[
6.7,
2.5,
5.8,
1.8
],
[
7.2,
3.6,
6.1,
2.5
],
[
6.5,
3.2,
5.1,
2
],
[
6.4,
2.7,
5.3,
1.9
],
[
6.8,
3,
5.5,
2.1
],
[
5.7,
2.5,
5,
2
],
[
5.8,
2.8,
5.1,
2.4
],
[
6.4,
3.2,
5.3,
2.3
],
[
6.5,
3,
5.5,
1.8
],
[
7.7,
3.8,
6.7,
2.2
],
[
7.7,
2.6,
6.9,
2.3
],
[
6,
2.2,
5,
1.5
],
[
6.9,
3.2,
5.7,
2.3
],
[
5.6,
2.8,
4.9,
2
],
[
7.7,
2.8,
6.7,
2
],
[
6.3,
2.7,
4.9,
1.8
],
[
6.7,
3.3,
5.7,
2.1
],
[
7.2,
3.2,
6,
1.8
],
[
6.2,
2.8,
4.8,
1.8
],
[
6.1,
3,
4.9,
1.8
],
[
6.4,
2.8,
5.6,
2.1
],
[
7.2,
3,
5.8,
1.6
],
[
7.4,
2.8,
6.1,
1.9
],
[
7.9,
3.8,
6.4,
2
],
[
6.4,
2.8,
5.6,
2.2
],
[
6.3,
2.8,
5.1,
1.5
],
[
6.1,
2.6,
5.6,
1.4
],
[
7.7,
3,
6.1,
2.3
],
[
6.3,
3.4,
5.6,
2.4
],
[
6.4,
3.1,
5.5,
1.8
],
[
6,
3,
4.8,
1.8
],
[
6.9,
3.1,
5.4,
2.1
],
[
6.7,
3.1,
5.6,
2.4
],
[
6.9,
3.1,
5.1,
2.3
],
[
5.8,
2.7,
5.1,
1.9
],
[
6.8,
3.2,
5.9,
2.3
],
[
6.7,
3.3,
5.7,
2.5
],
[
6.7,
3,
5.2,
2.3
],
[
6.3,
2.5,
5,
1.9
],
[
6.5,
3,
5.2,
2
],
[
6.2,
3.4,
5.4,
2.3
],
[
5.9,
3,
5.1,
1.8
]
]
},
"m": {
"Name": "Anderson's Iris data set",
"Description": "The data set consists of 50 Ss from each of three species of Iris (Iris setosa, Iris virginica and Iris versicolor). Four features were measured from each S: the length and the width of the sepals and petals, in centimetres.",
"Reference": "R. A. Fisher (1936). The use of multiple measurements in taxonomic problems. Annals of Eugenics 7 (2): 179-188."
}
},
config={
"colorBy": "Species",
"graphType": "Scatter2D",
"hideHistogram": True,
"histogramStat": "count",
"segregateVariablesBy": [
"Species"
],
"showFilledHistogramDensity": True,
"showHistogramDensity": True,
"showHistogramMedian": False,
"theme": "CanvasXpress"
},
width=613,
height=613,
events=CXEvents(),
after_render=[
[
"createHistogram",
[
"Species",
None,
None
]
]
],
other_init_params={
"version": 35,
"events": False,
"info": False,
"afterRenderInit": False,
"noValidate": True
}
)
display = CXNoteBook(cx)
display.render(output_file="density_10.html")
###Output
_____no_output_____
|
db_LS_DS_231_assignment.ipynb
|
###Markdown
Lambda School Data Science*Unit 2, Sprint 3, Module 1*--- Define ML problemsYou will use your portfolio project dataset for all assignments this sprint. AssignmentComplete these tasks for your project, and document your decisions.- [ ] Choose your target. Which column in your tabular dataset will you predict?- [ ] Is your problem regression or classification?- [ ] How is your target distributed? - Classification: How many classes? Are the classes imbalanced? - Regression: Is the target right-skewed? If so, you may want to log transform the target.- [ ] Choose your evaluation metric(s). - Classification: Is your majority class frequency >= 50% and < 70% ? If so, you can just use accuracy if you want. Outside that range, accuracy could be misleading. What evaluation metric will you choose, in addition to or instead of accuracy? - Regression: Will you use mean absolute error, root mean squared error, R^2, or other regression metrics?- [ ] Choose which observations you will use to train, validate, and test your model. - Are some observations outliers? Will you exclude them? - Will you do a random split or a time-based split?- [ ] Begin to clean and explore your data.- [ ] Begin to choose which features, if any, to exclude. Would some features "leak" future information?If you haven't found a dataset yet, do that today. [Review requirements for your portfolio project](https://lambdaschool.github.io/ds/unit2) and choose your dataset.Some students worry, ***what if my model isn't “good”?*** Then, [produce a detailed tribute to your wrongness. That is science!](https://twitter.com/nathanwpyle/status/1176860147223867393)
###Code
%%capture
import sys
if 'google.colab' in sys.modules:
# Install packages in Colab
!pip install category_encoders==2.*
!pip install pandas-profiling==2.*
# Imports
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
import seaborn as sbn
from scipy.stats import ttest_ind, ttest_1samp, t, randint, uniform
import pandas_profiling
from pandas_profiling import ProfileReport
import category_encoders as ce
from sklearn.impute import SimpleImputer
from sklearn.ensemble import RandomForestClassifier
from sklearn.pipeline import make_pipeline
from sklearn.model_selection import cross_val_score, GridSearchCV, RandomizedSearchCV
from sklearn.preprocessing import StandardScaler
from sklearn.metrics import accuracy_score, plot_confusion_matrix, confusion_matrix, classification_report
# Read in dataset to a dataframe
url = 'https://raw.githubusercontent.com/Daniel-Benson-Poe/practice_datasets/master/suicide_rates.csv'
suicide_df = pd.read_csv(url)
# Check data shape
print(suicide_df.shape)
# Check first five rows
suicide_df.head()
###Output
(27820, 12)
###Markdown
1. Choose Your Target and Determine Regression or Classification
###Code
suicide_df['suicides_no'].describe()
suicide_df['suicides_no'].isna().sum()
suicide_df['suicides_no'].nunique(), suicide_df['suicides_no'].shape
###Output
_____no_output_____
###Markdown
We are going to use suicides_no as our target data: our goal is to predict the number of suicides in each country in a given year. Because our end value will be numerical and continuous we are looking at a regression problem. 2. Choose your evaluation metrics
###Code
# We will be using a regression model to solve our problem.
# First choose our target
target = suicide_df['suicides_no']
# Now found how target is distributed
import seaborn as sns
sns.distplot(target);
# Target is definitely right-skewed
# Look into its statistics
target.describe()
# Let's try removing some of the outliers
import numpy as np
suicide_df = suicide_df[(suicide_df['suicides_no'] <= np.percentile(suicide_df['suicides_no'], 99)) &
(suicide_df['suicides/100k pop'] <= np.percentile(suicide_df['suicides_no'], 99))]
# Now find how our target is distributed
target = suicide_df['suicides_no']
sns.distplot(target);
target.describe()
# Let's log transform the target now
target_log = np.log1p(target)
sns.distplot(target_log)
plt.title("Log-transformed target");
###Output
_____no_output_____
###Markdown
3. Choose which Observations you will use to train, validate, and test dataframe
###Code
# Let's split by year
# First check the contents of the year column
sorted(suicide_df['year'].unique())
suicide_df['year'].value_counts()
# Create train set out of data from all years prior to 2015
train = suicide_df[suicide_df['year'] < 2015]
# Create validation set out of data from the year 2015
val = suicide_df[suicide_df['year'] == 2015]
# Create test set out of data from the year 2016
test = suicide_df[suicide_df['year'] == 2016]
train.shape, val.shape, test.shape
###Output
_____no_output_____
###Markdown
4. Wrangle Data
###Code
# Check for nulls
suicide_df.isnull().sum()
# Look at column dtypes
suicide_df[' gdp_for_year ($) '].dtype, suicide_df['year'].dtype, suicide_df['suicides_no'].dtype, suicide_df['population'].dtype, suicide_df['gdp_per_capita ($)'].dtype
# Check shape and value counts for HDI for year
print(suicide_df['HDI for year'].shape)
suicide_df['HDI for year'].value_counts(dropna=False)
# Check contents of country column
suicide_df['country'].value_counts()
# Check contents of generation column
suicide_df['generation'].value_counts()
# Check contents of age
suicide_df['age'].value_counts()
# Check high cardinality features
cardinality = suicide_df.select_dtypes(exclude='number').nunique()
high_cardinality_feat = cardinality[cardinality > 30].index.tolist()
high_cardinality_feat
# Check Pandas Profiling version
import pandas_profiling
pandas_profiling.__version__
# New code for Pandas Profiling version 2.4
from pandas_profiling import ProfileReport
profile = ProfileReport(train, minimal=True).to_notebook_iframe()
profile
###Output
_____no_output_____
###Markdown
Some things to clean:* Drop columns: country-year and HDI for year* Change column names to make them easier to work with* Convert GDP for year to int type* Convert year column to datetime (dt.year)* Convert categorical columns to useable numbers?
###Code
suicide_df.columns.to_list()
def value_eraser(df, column, value):
df = df[column].replace(value, '')
return df
test_df['annual_gdp'] = test_df.apply(value_eraser, axis=1, args=('annual_gdp', ','))
# Create a function to wrangle the sets
def wrangle(X):
"""Wrangles the train, validation, and test sets the same way."""
# Prevent SettingWithCopyWarning
X = X.copy()
# Create list of garbage columns and use that list to drop them from set
garbage_columns = ['country-year', 'HDI for year']
X = X.drop(columns=garbage_columns)
# Rename certain columns to make them easier to work with
cols_to_name = ['suicides_no', 'suicides/100k pop', ' gdp_for_year ($) ', 'gdp_per_capita ($)']
new_col_names = ['num_suicides', 'suicides/100k_pop', 'annual_gdp', 'gdp_per_capita']
i = 0
for col in cols_to_name:
X = X.rename(columns={col: new_col_names[i]})
i += 1
# Remove commas from the values in annual_gdp column
X['annual_gdp'] = X.apply(value_eraser, axis=1, args=('annual_gdp', ',')).astype(int)
# Remove the years string from the age column
X['age'] = X.apply(value_eraser, axis=1, args=('age', ' years'))
return X
train = wrangle(train)
val = wrangle(val)
test = wrangle(test)
train.sample(20)
###Output
_____no_output_____
|
.ipynb_checkpoints/dzone article-health tweets-for github-part 1-checkpoint.ipynb
|
###Markdown
Setup necessary libraries and Twitter access tokens
###Code
#import tweepy
import numpy as np
import pandas as pd
# input Twitter access tokens
access_token = 'paste your token here'
access_token_secret = 'paste your token secret here'
consumer_key = 'paste your consumer key here'
consumer_secret = 'paste your consumer secret here'
###Output
_____no_output_____
###Markdown
Authenticate Twitter credentials
###Code
auth = tweepy.OAuthHandler(consumer_key, consumer_secret)
auth.set_access_token(access_token, access_token_secret)
api = tweepy.API(auth)
###Output
_____no_output_____
###Markdown
Pick a topic people are talking about on Twitter that you're interested in
###Code
for tweet in api.search('health'):
print(tweet.text)
###Output
_____no_output_____
###Markdown
Create a dataframe to house Twitter data
###Code
df = pd.DataFrame(columns = ['Tweets', 'User', 'User_statuses_count',
'User_followers', 'User_location', 'User_verified',
'fav_count', 'rt_count', 'tweet_date'])
# NOTE: since we put our api.search into tweepy.Cursor,
# it will not just stop at the first 100 tweets.
# It will instead keep going on forever; that's why we are using
# i as a counter to stop the loop after 1000 iterations.
def stream(data,file_name):
i = 0
for tweet in tweepy.Cursor(api.search, q=data, count=100, lang='en').items():
print(i, end='\r')
df.loc[i, 'Tweets'] = tweet.text
df.loc[i, 'User'] = tweet.user.name
df.loc[i, 'User_statuses_count'] = tweet.user.statuses_count
df.loc[i, 'User_followers'] = tweet.user.followers_count
df.loc[i, 'User_location'] = tweet.user.location
df.loc[i, 'User_verified'] = tweet.user.verified
df.loc[i, 'fav_count'] = tweet.favorite_count
df.loc[i, 'rt_count'] = tweet.retweet_count
df.loc[i, 'tweet_date'] = tweet.created_at
df.to_excel('{}.xlsx'.format(file_name))
i+=1
if i == 20000:
break
else:
pass
###Output
_____no_output_____
###Markdown
Remember to update the date segment of the filename before running the next line of code!
###Code
stream(data = ['health'], file_name = 'Health_tweets_YYMMDD')
###Output
_____no_output_____
###Markdown
Optional: import previous Twitter data from ExcelGetting 20,000 tweets from Twitter takes TIME. If the very latest data isn't absolutely necessary and you have created the dataframe to house Twitter data previously, upload an existing file instead. See below.
###Code
# Use an existing file. The file "Health_tweets_20200122.xlsx" is one I've created previously and is in the same directory as the Jupyter notebook.
df = pd.read_excel("Health_tweets_20200122.xlsx")
###Output
_____no_output_____
###Markdown
Familiarising with data in existing file
###Code
df.head()
df.info()
###Output
_____no_output_____
###Markdown
Let's analyse some Tweets
###Code
from textblob import TextBlob
from wordcloud import WordCloud, STOPWORDS
import re
import chart_studio.plotly as py
import plotly.graph_objects as go
from plotly.offline import iplot
import cufflinks
cufflinks.go_offline()
cufflinks.set_config_file(world_readable=True, theme='pearl', offline=True)
# remove capitalisation etc from Tweets
def clean_tweet(tweet):
return ' '.join(re.sub('(@[A-Za-z0-9]+)|([^0-9A-Za-z \t])|(\w+:\/\/\S+)', ' ', tweet).split())
###Output
_____no_output_____
###Markdown
Create custom functions to analyse for polarity and subjectivity
###Code
def label_sentiment(tweet):
analysis = TextBlob(tweet)
if analysis.sentiment.polarity > 0:
return 'Positive'
elif analysis.sentiment.polarity == 0:
return 'Neutral'
else:
return 'Negative'
#def analyse_subjectivity(tweet):
# analysis.sentiment.subjectivity
def analyse_polarity(clean_tweet):
try:
return TextBlob(clean_tweet).sentiment.polarity
except:
return None
def analyse_subjectivity(clean_tweet):
try:
return TextBlob(clean_tweet).sentiment.subjectivity
except:
return None
###Output
_____no_output_____
###Markdown
Add sentiment calculations to dataframe
###Code
# using 2 different methods to achieve same result to compare syntax of lambda & apply
df['clean_tweet'] = df['Tweets'].apply(lambda x: clean_tweet(x))
df['Sentiment'] = df['clean_tweet'].apply(lambda x: label_sentiment(x))
df['Tweet_polarity'] = df['clean_tweet'].apply(analyse_polarity)
df['Tweet_subjectivity'] = df['clean_tweet'].apply(analyse_subjectivity)
###Output
_____no_output_____
###Markdown
Eyeball samples of original Tweet versus cleaned Tweet and sentiment scores
###Code
n = 20
print('Original tweet:\n'+ df['Tweets'][n])
print()
print('Clean tweet:\n'+ df['clean_tweet'][n])
print()
print('Sentiment:\n'+ df['Sentiment'][n])
n = 147
print('Original tweet: '+ df['Tweets'][n])
print()
print('Clean tweet: '+ df['clean_tweet'][n])
print()
print('Sentiment: '+ df['Sentiment'][n])
###Output
_____no_output_____
###Markdown
Save a copy of the latest output to Excel
###Code
df.to_excel("Health_tweets_YYYYMMDD_w_sent_scores.xlsx")
###Output
_____no_output_____
###Markdown
Visualising Twitter data
###Code
df=pd.read_excel("Health_tweets_20200129_w_sent_scores.xlsx")
df.columns
df['Sentiment'].value_counts().iplot(kind='bar',xTitle='Sentiment', yTitle='Count', title={'text': "Overall SEntiment Distribution", 'x':0.5,'y':0.9, #coordinates
'xanchor':'center', 'yanchor':'top'})
###Output
_____no_output_____
###Markdown
add viz of popular tweets?
###Code
import matplotlib. pyplot as plt
import numpy as np
%matplotlib inline
df.columns
# using Matplotlib - data labels are not on hover
x = df['Tweet_polarity']
y = df['Tweet_subjectivity']
plt.scatter(x, y, alpha=0.2)
plt.xlabel("Tweet polarity (-1 = extremely negative, 1 = extremely positive)")
plt.ylabel("Tweet subjectivity (0 = very objective, 1 = very subjective)")
plt.title("Sentiment of 20,000 tweets that mention health")
plt.show()
###Output
_____no_output_____
|
Woche 3/3_5_Sentiment_Analyse.ipynb
|
###Markdown
0. Installieren aller Pakete
###Code
# Hier die Kaggle Credentials einfügen (ohne Anführungszeichen)
%env KAGGLE_USERNAME=openhpi
%env KAGGLE_KEY=das_ist_der_key
!pip install skorch
import pandas as pd
import numpy as np
from sklearn.svm import SVC
from sklearn.tree import DecisionTreeClassifier
from sklearn.ensemble import RandomForestClassifier
from sklearn.linear_model import LogisticRegression
from skorch import NeuralNetClassifier
from sklearn.model_selection import GridSearchCV
from sklearn.feature_extraction.text import TfidfVectorizer
from sklearn.model_selection import train_test_split
from sklearn.metrics import classification_report
from collections import Counter
import re
from bs4 import BeautifulSoup
import yaml
import os
from wordcloud import WordCloud
import seaborn as sns
import matplotlib.pyplot as plt
%matplotlib inline
import nltk
from nltk.corpus import stopwords
nltk.download('stopwords')
import spacy
!python -m spacy download en_core_web_sm
import torch
from torch import nn
class NeuralNetModule(nn.Module):
def __init__(self, num_inputs, num_units=20, nonlin=nn.ReLU()):
super(NeuralNetModule, self).__init__()
self.nonlin = nonlin
self.dense0 = nn.Linear(num_inputs, num_units)
self.dropout = nn.Dropout(0.2)
self.dense1 = nn.Linear(num_units, num_units)
self.output = nn.Linear(num_units, 2)
self.softmax = nn.Softmax(dim=-1)
def forward(self, X, **kwargs):
X = self.nonlin(self.dense0(X))
X = self.dropout(X)
X = self.nonlin(self.dense1(X))
X = self.softmax(self.output(X))
return X
###Output
_____no_output_____
###Markdown
3.5 Sentiment Analyse Was wir erreichen wollenIn diesem Anwendungsfall wollen wir Filmbewertungen in positive und negative Bewertungen unterteilen. Dafür liegen uns eine Vielzahl gelabelter Trainingsdaten in englischer Sprache vor.Mit einem Modell wollen wir in der Lage sein, für neue Kommentare automatisiert die Stimmung analysieren zu können. Download Dataset Manuellvia https://www.kaggle.com/columbine/imdb-dataset-sentiment-analysis-in-csv-format Via APIHinzufügen der kaggle.jsonSpeichern als ~/.kaggle/kaggle.json auf Linux, OSX, oder andere UNIX-based Betriebssysteme und unter C:\Users.kaggle\kaggle.json auf WindowsSiehe https://www.kaggle.com/docs/api oder https://github.com/Kaggle/kaggle-api Beispiel:~/kaggle/kaggle.json{"username":"openHPI","key":"das_ist_der_key"}
###Code
!pip3 install kaggle
!kaggle datasets download -d columbine/imdb-dataset-sentiment-analysis-in-csv-format
import zipfile
with zipfile.ZipFile("imdb-dataset-sentiment-analysis-in-csv-format.zip", 'r') as zip_ref:
zip_ref.extractall("")
import os
os.rename('Train.csv','sentiment.csv')
FILE_PATH = "sentiment.csv"
###Output
_____no_output_____
###Markdown
Daten vorbereitenWir können nun unsere Daten laden und mit der Vorbereitung beginnen.
###Code
df = pd.read_csv(FILE_PATH, nrows=10_000)
df.head() # label 1 == positive; label 0 == negative
###Output
_____no_output_____
###Markdown
Schauen wir uns zunächst einmal die Datenverteilung unserer Zielvariable an. Die Zielvariable ist kategorisch in diesem Fall:
###Code
df["label"].value_counts(normalize=True)
###Output
_____no_output_____
###Markdown
Wir sehen: unsere Daten sind nahezu perfekt gleichverteilt (50% positiv, 50% negativ). Das ist prinzipiell eine gute Nachricht für unser Trainingsvorhaben (je unbalancierter Daten sind, desto komplexer wird das Training i.d.R.).Werfen wir einen Blick auf das erste Beispiel, um ein Gefühl für die Datenbeschaffenheit zu erlangen:
###Code
df["text"].iloc[0]
###Output
_____no_output_____
###Markdown
Wir stellen fest:- Eine einzelne Bewertung kann recht lange sein. Das bedeutet für uns, dass Beispiele sowohl mehr Informationen, aber auch mehr "Rauschen" (für uns unwichtige Informationen) beinhalten können.- Es handelt sich um Webdaten, da HTML-Tags gesetzt sind. Wir sollten diese korrekt verarbeiten.- 's werden mit \\'s kodiert (was wiederum auf den Rohtext zurückzuführen ist)- Wir sollten die Wörter im Text zu Kleinbuchstaben konvertieren, um die Anzahl möglicher Wort-Variationen zu reduzieren.
###Code
# preprocessing
df["text"] = df["text"].apply(lambda x: x.lower())
df["text"] = df["text"].apply(lambda x: x.replace("\'", ""))
df["text"] = df["text"].apply(lambda x: BeautifulSoup(x).text)
###Output
_____no_output_____
###Markdown
Schauen wir uns die gleiche Bewertung erneut an:
###Code
df["text"].iloc[0]
###Output
_____no_output_____
###Markdown
Das sieht schon deutlich besser aus. Fangen wir nun an, unsere Daten zu splitten. Wir nutzen hier der Einfachheit halber einen ganz simplen Split mit Standardeinstellungen (wir könnten auch andere Verfahren einsetzen).Wir teilen unsere Daten dabei in Trainings-, Validierungs- und Testsplit. Mit dem ersten trainieren wir die Modelle, mit dem zweiten suchen wir unser bestes Modell heraus, mit dem letzten validieren wir die Ergebnisse.
###Code
train_df, test_valid_df = train_test_split(df)
test_df, valid_df = train_test_split(test_valid_df)
###Output
_____no_output_____
###Markdown
Bevor wir unsere KI-Pipeline bauen, schauen wir uns die Daten erneut an. Sicherheitshalber duplizieren wir unseren Trainingsdatensatz dafür. Wir schauen uns zunächst die Textlängen an:
###Code
analysis_df = train_df.copy()
analysis_df["text_length"] = analysis_df["text"].apply(len)
###Output
_____no_output_____
###Markdown
Nun können wir uns die Verteilungen einfach je Klasse ("positiv" und "negativ") visualisieren lassen:
###Code
sns.histplot(data=analysis_df, x="text_length", hue="label", element="step");
###Output
_____no_output_____
###Markdown
Wir erkennen keinen großen Unterschied zwischen den beiden Klassen in der Textlänge (d.h. sowohl positive als auch negative Bewertungen können sehr ausführlich sein).Schauen wir uns den Textkorpus nochmal genauer an:
###Code
text_corpus = " ".join(analysis_df["text"])
###Output
_____no_output_____
###Markdown
Bauen wir uns nun eine Hilfsfunktion, mit der wir uns einfach die *n* häufigsten Wörter des Korpus anschauen können. Als Korpus bezeichnet man eine Sammlung von Texten.
###Code
def plot_most_common_words(text_corpus, n):
counter = Counter(text_corpus.split())
rank, words, occurences = [], [], []
for idx, (word, occurence) in enumerate(counter.most_common(n=n)):
rank.append(idx)
words.append(word)
occurences.append(occurence)
fig, ax = plt.subplots()
ax.scatter(rank, occurences, s=0.01)
for idx, word in enumerate(words):
ax.annotate(word, (rank[idx], occurences[idx]))
plt.title("Zipf's law")
plt.ylabel("Occurences")
plt.xlabel("Rank");
###Output
_____no_output_____
###Markdown
Wir können nun in der Verteilung das sogenannte Zipf's Law erkennen: Jedes Wort tritt ungefähr invers proportional zu seinem Rang auf, d.h. das häufigste Wort doppelt so häufig wie das zweithäufigste. Solche Füllwörter machen einen Großteil unserer Daten aus. Das ist ein ganz bekanntes Phönomen in natürlichen Sprachen.
###Code
plot_most_common_words(text_corpus, 15)
###Output
_____no_output_____
###Markdown
Wir können diese Wörter entfernen, da sie inhaltlich keinen Mehrwert bringen. Als ersten Schritt erstellen wir ein Embedding für einzelne Wörter. Schauen wir uns die Verteilung einmal an, wenn wir die typischen Füllwörter entfernen:
###Code
pattern = re.compile(r'\b(' + r'|'.join(stopwords.words('english')) + r')\b\s*')
text_corpus_without_stopwords = pattern.sub('', text_corpus)
###Output
_____no_output_____
###Markdown
Wir erzeugen die gleiche Visualisierung:
###Code
plot_most_common_words(text_corpus_without_stopwords, 15)
###Output
_____no_output_____
###Markdown
Wir können schon deutlich erkennen, dass es sich um einen Datenbestand zu Filmbewertungen handelt. Wir können uns das auch als Wordcloud ("Wortwolke") anschauen. Wir schreiben uns hierfür eine Funktion:
###Code
def draw_word_cloud(text_corpus):
word_cloud = WordCloud(
collocations = False, background_color = 'white'
).generate(text_corpus)
plt.imshow(word_cloud, interpolation='bilinear')
plt.axis("off");
###Output
_____no_output_____
###Markdown
Damit können wir die Worthäufigkeiten als Wordcloud darstellen lassen:
###Code
draw_word_cloud(text_corpus_without_stopwords)
###Output
_____no_output_____
###Markdown
*Hinweis*:An sich liefert eine Wordcloud vom Informationsgehalt keinen Mehrwert gegenüber einer einfachen Darstellung anhand eines Scatter Plots. Tatsächlich ist es für uns sogar schwieriger zuzuordnen, welches Wort am häufigsten vorkommt (`movie` oder `film`?). Außerdem müssen wir häufig unsere Leserichtung anpassen. Vorteilhaft ist lediglich die platzsparende Art der Darstellung.Word Clouds werden gerne bei Textvisualisierungen verwendet - durchaus auch, weil es schlichtweg "gut" aussieht. Wir können uns diese Darstellung auch einmal je Klasse ("positiv" und "negativ") anzeigen; vielleicht erkennen wir klassenspezifische Wörter:
###Code
positive_corpus = " ".join(analysis_df["text"].loc[analysis_df["label"] == 1])
negative_corpus = " ".join(analysis_df["text"].loc[analysis_df["label"] == 0])
###Output
_____no_output_____
###Markdown
Schauen wir uns zunächst die positiven Filmbewertungen in einer Wordcloud an:
###Code
draw_word_cloud(positive_corpus)
###Output
_____no_output_____
###Markdown
Und anschließend die negativen Bewertungen:
###Code
draw_word_cloud(negative_corpus)
###Output
_____no_output_____
###Markdown
Wir stellen fest, dass es Wörter gibt, die wir intuitiv vielleicht einer Klasse zugeordnet hätten (`good` -> positiv), aber auch in der gegenteiligen Klasse auftreten. Entstehen können solche Szenarien dann, wenn Wörter im Kontext des Satzes ihre Bedeutung ändern: `This film was really good.` vs. `In my opinion, this movie was not good at all.`Wir könnten hier noch tiefer in eine Analyse gehen und versuchen, mehr über unsere Daten zu verstehen (z.B. wie oft treten verneinte Sätze auf? Was sind klassentrennde Wörter?). An dieser Stelle werden wir aber für dieses Projekt die explorative Analyse beenden und mit der KI-Pipeline beginnen. 3.5 Sentiment Analyse LemmatisierungWir können unsere Texte nun so anpassen, dass wir Wörter auf ihren Wortstamm reduzieren. Dafür eignet sich die Lemmatisierung.
###Code
nlp = spacy.load("en_core_web_sm")
doc = nlp(
"lemmatization is the process of grouping together the inflected forms of a word."
)
for token in doc:
print(token, token.lemma_)
###Output
_____no_output_____
###Markdown
Wir bauen uns hier wieder eine Funktion für:
###Code
def enrich_lemmatized_text(df):
df = df.copy()
df["lemmatized_text"] = df["text"].apply(
lambda x: " ".join([token.lemma_ for token in nlp(x)])
)
return df
###Output
_____no_output_____
###Markdown
Und wenden diese anschließend auf unseren DataFrames an:
###Code
train_df = enrich_lemmatized_text(train_df)
valid_df = enrich_lemmatized_text(valid_df)
test_df = enrich_lemmatized_text(test_df)
###Output
_____no_output_____
###Markdown
Nun können wir mit der Tf-Idf Vektorisierung beginnen, welche unsere Texte in ein numerisches Format umwandelt.
###Code
vectorizer = TfidfVectorizer()
train_X = vectorizer.fit_transform(train_df["lemmatized_text"]).astype(np.float32)
valid_X = vectorizer.transform(valid_df["lemmatized_text"]).astype(np.float32)
test_X = vectorizer.transform(test_df["lemmatized_text"]).astype(np.float32)
###Output
_____no_output_____
###Markdown
Unsere Daten sind vorbereitet, wir können mit dem Bau unserer KI-Modelle beginnen. Wir trainieren:- einen Entscheidungsbaum- einen Random Forest- eine logistische Regression- ein künstliches neuronales Netzwerk Decision Tree (Entscheidungsbaum)Ein Entscheidungsbaum ist als sehr einfaches Modell und wird oft als eines der ersten Modelle verwendet. Wir trainieren und geben verschiedene Optionen für die Hyperparameter `max_depth` und `min_samples_split`. Die Hyperparameter-Suche GridSearch wählt hierbei die optimalen Hyperparameter aus.
###Code
tree_clf = DecisionTreeClassifier()
tree_params = {
'max_depth': list(range(10, 101, 20)) + [None],
'min_samples_split': [2, 5]
}
tree_search = GridSearchCV(tree_clf, tree_params)
tree_search.fit(train_X, train_df["label"])
best_tree_clf = tree_search.best_estimator_
###Output
_____no_output_____
###Markdown
Anschließend wollen wir unser Modell auf den Validierungsdaten prüfen. Da wir das wiederholt machen werden, bauen wir wieder eine Hilfsfunktion:
###Code
def evaluate_clf(valid_X, labels, clf):
predictions = clf.predict(valid_X)
report = classification_report(labels, predictions)
print(report)
###Output
_____no_output_____
###Markdown
Anschließend führen wir diese aus und können die Ergebnisse prüfen:
###Code
evaluate_clf(valid_X, valid_df["label"], best_tree_clf)
###Output
_____no_output_____
###Markdown
Random ForestAls nächstes Modell eignet sich ein Random Forest, welcher eine Ensemble-Technik darstellt. Im Wesentlichen werden hier mehrere Entscheidungsbäume trainiert, und deren Ergebnis vereint. Wir haben zusätzlich zu den vorherigen Hyperparametern noch `n_estimators` zu wählen.
###Code
forest_clf = RandomForestClassifier()
forest_params = {
'n_estimators': list(range(10, 101, 20)),
'max_depth': list(range(10, 101, 20)) + [None],
'min_samples_split': [2, 5]
}
forest_search = GridSearchCV(forest_clf, forest_params)
forest_search.fit(train_X, train_df["label"])
best_forest_clf = forest_search.best_estimator_
###Output
_____no_output_____
###Markdown
Wir evaluieren wieder das Ergebnis:
###Code
evaluate_clf(valid_X, valid_df["label"], best_forest_clf)
###Output
_____no_output_____
###Markdown
Logistic RegressionNun bauen wir eine logistische Regression. Diese ist ebenfalls sehr simpel, erweist sich oftmals aber als sehr gutes Modell. Wir haben vier Hyperparameter:
###Code
lr_clf = LogisticRegression()
lr_params = {
'penalty': ['l1', 'l2'],
'max_iter': [100],
'C': np.logspace(-4, 4, 20),
'solver': ['liblinear']
}
lr_search = GridSearchCV(lr_clf, lr_params)
lr_search.fit(train_X, train_df["label"])
best_lr_clf = lr_search.best_estimator_
###Output
_____no_output_____
###Markdown
Und wir evaluieren erneut:
###Code
evaluate_clf(valid_X, valid_df["label"], best_lr_clf)
###Output
_____no_output_____
###Markdown
Feedforward Neural NetworkZuletzt wollen wir noch ein künstliches neuronales Netz bauen. Dafür haben wir eine Architektur in der Datei `ffnn.py` gewählt, welche wir zuvor importiert haben und nun mit der Bibliothek skorch einfach anwenden können. Wir wählen hier direkt die Hyperparameter ohne GridSearch:
###Code
neural_net = NeuralNetClassifier(
module=NeuralNetModule,
module__num_inputs = len(vectorizer.vocabulary_),
max_epochs=10,
optimizer=torch.optim.Adam,
iterator_train__shuffle=True,
verbose=0
)
neural_net.fit(train_X, train_df['label'])
###Output
_____no_output_____
###Markdown
Nun prüfen wir das Ergebnis:
###Code
evaluate_clf(valid_X, valid_df["label"], neural_net)
###Output
_____no_output_____
###Markdown
Unser bestes Modell auf den Validierungsdaten ist die logistische Regression, daher wählen wir diese als unser finales Modell. Test unseres ModellsWir haben die Validierungsdaten gewählt, um unser bestes Modell auszuwählen. Es kann aber sein, dass unser Modell auf den Validierungsdaten nur zufällig gute Prognosen erzeugt hatte oder wir uns zu sehr durch die Hyperparameter-Optimierung auf unser Validierungs-Daten "überangepasst" haben. Daher evaluieren wir auf einem noch vollkommen "neuen" Teil der Daten, unseren Testdaten:
###Code
evaluate_clf(test_X, test_df["label"], best_lr_clf)
###Output
_____no_output_____
|
examples/toxtrac_videos/Mice.ipynb
|
###Markdown
Global parametersThis cell (below) enlists user-defined parameters
###Code
# colours is a vector of BGR values which are used to identify individuals in the video
# since we only have one individual, the program will only use the first element from this array i.e. (0,0,255) - red
# number of elements in colours should be greater than n_inds (THIS IS NECESSARY FOR VISUALISATION ONLY)
n_inds = 1
colours = [(0,0,255),(0,255,255),(255,0,255),(255,255,255),(255,255,0),(255,0,0),(0,255,0),(0,0,0)]
# this is the block_size and offset used for adaptive thresholding (block_size should always be odd)
# these values are critical for tracking performance
block_size = 51
offset = 20
# minimum area and maximum area occupied by the animal in number of pixels
# this parameter is used to get rid of other objects in view that might be hard to threshold out but are differently sized
min_area = 200
max_area = 2000
# the scaling parameter can be used to speed up tracking if video resolution is too high (use value 0-1)
scaling = 1.0
# mot determines whether the tracker is being used in noisy conditions to track a single object or for multi-object
# using this will enable k-means clustering to force n_inds number of animals
mot = False
# name of source video and paths
video = 'Mice'
input_vidpath = '/mnt/ssd1/Documents/Vivek/tracktor/videos/toxtrac_videos/' + video + '.avi'
output_vidpath = '/mnt/ssd1/Documents/Vivek/tracktor/output/toxtrac_videos/' + video + '.mp4'
output_filepath = '/mnt/ssd1/Documents/Vivek/tracktor/output/toxtrac_videos/' + video + '.csv'
codec = 'DIVX' # try other codecs if the default doesn't work ('DIVX', 'avc1', 'XVID') note: this list is non-exhaustive
## Start time
start = time.time()
## Open video
cap = cv2.VideoCapture(input_vidpath)
if cap.isOpened() == False:
sys.exit('Video file cannot be read! Please check input_vidpath to ensure it is correctly pointing to the video file')
## Video writer class to output video with contour and centroid of tracked object(s)
# make sure the frame size matches size of array 'final'
fourcc = cv2.VideoWriter_fourcc(*codec)
output_framesize = (int(cap.read()[1].shape[1]*scaling),int(cap.read()[1].shape[0]*scaling))
out = cv2.VideoWriter(filename = output_vidpath, fourcc = fourcc, fps = 30.0, frameSize = output_framesize, isColor = True)
## Individual location(s) measured in the last and current step
meas_last = list(np.zeros((n_inds,2)))
meas_now = list(np.zeros((n_inds,2)))
last = 0
df = []
while(True):
# Capture frame-by-frame
ret, frame = cap.read()
this = cap.get(1)
if ret == True:
frame = cv2.resize(frame, None, fx = scaling, fy = scaling, interpolation = cv2.INTER_LINEAR)
# Apply mask to aarea of interest
mask = np.zeros(frame.shape)
mask = cv2.circle(mask, (308, 235), 215, (255,255,255), -1)
frame[mask == 0] = 0
thresh = tr.colour_to_thresh(frame, block_size, offset)
final, contours, meas_last, meas_now = tr.detect_and_draw_contours(frame, thresh, meas_last, meas_now, min_area, max_area)
row_ind, col_ind = tr.hungarian_algorithm(meas_last, meas_now)
final, meas_now, df = tr.reorder_and_draw(final, colours, n_inds, col_ind, meas_now, df, mot, this)
# Create output dataframe
for i in range(n_inds):
df.append([this, meas_now[i][0], meas_now[i][1]])
# Display the resulting frame
out.write(final)
cv2.imshow('frame', final)
if cv2.waitKey(1) == 27:
break
if last == this:
break
last = this
## Write positions to file
df = pd.DataFrame(np.matrix(df), columns = ['frame','pos_x','pos_y'])
df.to_csv(output_filepath, sep=',')
## When everything done, release the capture
cap.release()
out.release()
cv2.destroyAllWindows()
cv2.waitKey(1)
## End time and duration
end = time.time()
duration = end - start
print("--- %s seconds ---" %duration)
###Output
--- 35.627901792526245 seconds ---
###Markdown
Plot tracksThe code below allows you to see individual tracks. By counting the number of jumps in the tracks, one can identify number of false detections.
###Code
df = pd.read_csv(output_filepath)
df.head()
import matplotlib.pyplot as plt
plt.figure(figsize=(10,10))
plt.scatter(df['pos_x'], df['pos_y'], c=df['frame'])
plt.xlabel('pos_x')
plt.ylabel('pos_y')
plt.show()
###Output
_____no_output_____
###Markdown
Identifying true/false detectionsHere, we use individual movement speeds to identify false detections. All frames where individuals move faster than their body length are considered false detections.NOTE: The methode used here underestimates false detections.
###Code
dx = df['pos_x'] - df['pos_x'].shift(n_inds)
dy = df['pos_y'] - df['pos_y'].shift(n_inds)
df['speed'] = np.sqrt(dx**2 + dy**2)
df.head()
thresh = 42.6
###Output
_____no_output_____
###Markdown
True detection rate
###Code
print(1-len(np.where(df['speed'] > thresh)[0]) / max(df['frame']))
###Output
1.0
|
notebooks/time_series_prediction/solutions/.ipynb_checkpoints/3_modeling_bqml-checkpoint.ipynb
|
###Markdown
Time Series Prediction with BQML and AutoML**Objectives** 1. Learn how to use BQML to create a classification time-series model using `CREATE MODEL`. 2. Learn how to use BQML to create a linear regression time-series model. 3. Learn how to use AutoML Tables to build a time series model from data in BigQuery.
###Code
import os
PROJECT = "your-gcp-project-here" # REPLACE WITH YOUR PROJECT NAME
REGION = "us-central1" # REPLACE WITH YOUR BUCKET REGION e.g. us-central1
os.environ['Pro']
%env
PROJECT = PROJECT
REGION = REGION
%%bash
sudo python3 -m pip freeze | grep google-cloud-bigquery==1.6.1 || \
sudo python3 -m pip install google-cloud-bigquery==1.6.1
###Output
google-cloud-bigquery==1.6.1
###Markdown
Create the dataset
###Code
from google.cloud import bigquery
from IPython import get_ipython
bq = bigquery.Client(project=PROJECT)
def create_dataset():
dataset = bigquery.Dataset(bq.dataset("stock_market"))
try:
bq.create_dataset(dataset) # Will fail if dataset already exists.
print("Dataset created")
except:
print("Dataset already exists")
def create_features_table():
error = None
try:
bq.query('''
CREATE TABLE stock_market.eps_percent_change_sp500
AS
SELECT *
FROM `asl-ml-immersion.stock_market.eps_percent_change_sp500`
''').to_dataframe()
except Exception as e:
error = str(e)
if error is None:
print('Table created')
elif 'Already Exists' in error:
print('Table already exists.')
else:
raise Exception('Table was not created.')
create_dataset()
create_features_table()
###Output
Dataset already exists
Table already exists.
###Markdown
Review the datasetIn the previous lab we created the data we will use modeling and saved them as tables in BigQuery. Let's examine that table again to see that everything is as we expect. Then, we will build a model using BigQuery ML using this table.
###Code
%%bigquery --project $PROJECT
#standardSQL
SELECT
*
FROM
stock_market.eps_percent_change_sp500
LIMIT
10
###Output
_____no_output_____
###Markdown
Using BQML Create classification model for `direction`To create a model1. Use `CREATE MODEL` and provide a destination table for resulting model. Alternatively we can use `CREATE OR REPLACE MODEL` which allows overwriting an existing model.2. Use `OPTIONS` to specify the model type (linear_reg or logistic_reg). There are many more options [we could specify](https://cloud.google.com/bigquery/docs/reference/standard-sql/bigqueryml-syntax-createmodel_option_list), such as regularization and learning rate, but we'll accept the defaults.3. Provide the query which fetches the training data Have a look at [Step Two of this tutorial](https://cloud.google.com/bigquery/docs/bigqueryml-natality) to see another example.**The query will take about two minutes to complete**We'll start with creating a classification model to predict the `direction` of each stock. We'll take a random split using the `symbol` value. With about 500 different values, using `ABS(MOD(FARM_FINGERPRINT(symbol), 15)) = 1` will give 30 distinct `symbol` values which corresponds to about 171,000 training examples. After taking 70% for training, we will be building a model on about 110,000 training examples.
###Code
%%bigquery --project $PROJECT
#standardSQL
CREATE OR REPLACE MODEL
stock_market.direction_model OPTIONS(model_type = "logistic_reg",
input_label_cols = ["direction"]) AS
-- query to fetch training data
SELECT
symbol,
Date,
Open,
close_MIN_prior_5_days,
close_MIN_prior_20_days,
close_MIN_prior_260_days,
close_MAX_prior_5_days,
close_MAX_prior_20_days,
close_MAX_prior_260_days,
close_AVG_prior_5_days,
close_AVG_prior_20_days,
close_AVG_prior_260_days,
close_STDDEV_prior_5_days,
close_STDDEV_prior_20_days,
close_STDDEV_prior_260_days,
direction
FROM
`stock_market.eps_percent_change_sp500`
WHERE
tomorrow_close IS NOT NULL
AND ABS(MOD(FARM_FINGERPRINT(symbol), 15)) = 1
AND ABS(MOD(FARM_FINGERPRINT(symbol), 15 * 100)) <= 15 * 70
###Output
_____no_output_____
###Markdown
Get training statistics and examine training infoAfter creating our model, we can evaluate the performance using the [`ML.EVALUATE` function](https://cloud.google.com/bigquery-ml/docs/bigqueryml-natalitystep_four_evaluate_your_model). With this command, we can find the precision, recall, accuracy F1-score and AUC of our classification model.
###Code
%%bigquery --project $PROJECT
#standardSQL
SELECT
*
FROM
ML.EVALUATE(MODEL `stock_market.direction_model`,
(
SELECT
symbol,
Date,
Open,
close_MIN_prior_5_days,
close_MIN_prior_20_days,
close_MIN_prior_260_days,
close_MAX_prior_5_days,
close_MAX_prior_20_days,
close_MAX_prior_260_days,
close_AVG_prior_5_days,
close_AVG_prior_20_days,
close_AVG_prior_260_days,
close_STDDEV_prior_5_days,
close_STDDEV_prior_20_days,
close_STDDEV_prior_260_days,
direction
FROM
`stock_market.eps_percent_change_sp500`
WHERE
tomorrow_close IS NOT NULL
AND ABS(MOD(FARM_FINGERPRINT(symbol), 15)) = 1
AND ABS(MOD(FARM_FINGERPRINT(symbol), 15 * 100)) > 15 * 70
AND ABS(MOD(FARM_FINGERPRINT(symbol), 15 * 100)) <= 15 * 85))
###Output
_____no_output_____
###Markdown
We can also examine the training statistics collected by Big Query. To view training results we use the [`ML.TRAINING_INFO`](https://cloud.google.com/bigquery/docs/reference/standard-sql/bigqueryml-syntax-train) function.
###Code
%%bigquery --project $PROJECT
#standardSQL
SELECT
*
FROM
ML.TRAINING_INFO(MODEL `stock_market.direction_model`)
ORDER BY iteration
###Output
_____no_output_____
###Markdown
Compare to simple benchmarkAnother way to asses the performance of our model is to compare with a simple benchmark. We can do this by seeing what kind of accuracy we would get using the naive strategy of just predicted the majority class. For the training dataset, the majority class is 'STAY'. The following query we can see how this naive strategy would perform on the eval set.
###Code
%%bigquery --project $PROJECT
#standardSQL
WITH
eval_data AS (
SELECT
symbol,
Date,
Open,
close_MIN_prior_5_days,
close_MIN_prior_20_days,
close_MIN_prior_260_days,
close_MAX_prior_5_days,
close_MAX_prior_20_days,
close_MAX_prior_260_days,
close_AVG_prior_5_days,
close_AVG_prior_20_days,
close_AVG_prior_260_days,
close_STDDEV_prior_5_days,
close_STDDEV_prior_20_days,
close_STDDEV_prior_260_days,
direction
FROM
`stock_market.eps_percent_change_sp500`
WHERE
tomorrow_close IS NOT NULL
AND ABS(MOD(FARM_FINGERPRINT(symbol), 15)) = 1
AND ABS(MOD(FARM_FINGERPRINT(symbol), 15 * 100)) > 15 * 70
AND ABS(MOD(FARM_FINGERPRINT(symbol), 15 * 100)) <= 15 * 85)
SELECT
direction,
(COUNT(direction)* 100 / (
SELECT
COUNT(*)
FROM
eval_data)) AS percentage
FROM
eval_data
GROUP BY
direction
###Output
_____no_output_____
###Markdown
So, the naive strategy of just guessing the majority class would have accuracy of 0.5509 on the eval dataset, just below our BQML model. Create regression model for `normalized change`We can also use BigQuery to train a regression model to predict the normalized change for each stock. To do this in BigQuery we need only change the OPTIONS when calling `CREATE OR REPLACE MODEL`. This will give us a more precise prediction rather than just predicting if the stock will go up, down, or stay the same. Thus, we can treat this problem as either a regression problem or a classification problem, depending on the business needs.
###Code
%%bigquery --project $PROJECT
#standardSQL
CREATE OR REPLACE MODEL
stock_market.price_model OPTIONS(model_type = "linear_reg",
input_label_cols = ["normalized_change"]) AS
-- query to fetch training data
SELECT
symbol,
Date,
Open,
close_MIN_prior_5_days,
close_MIN_prior_20_days,
close_MIN_prior_260_days,
close_MAX_prior_5_days,
close_MAX_prior_20_days,
close_MAX_prior_260_days,
close_AVG_prior_5_days,
close_AVG_prior_20_days,
close_AVG_prior_260_days,
close_STDDEV_prior_5_days,
close_STDDEV_prior_20_days,
close_STDDEV_prior_260_days,
normalized_change
FROM
`stock_market.eps_percent_change_sp500`
WHERE
normalized_change IS NOT NULL
AND ABS(MOD(FARM_FINGERPRINT(symbol), 15)) = 1
AND ABS(MOD(FARM_FINGERPRINT(symbol), 15 * 100)) <= 15 * 70
###Output
_____no_output_____
###Markdown
Just as before we can examine the evaluation metrics for our regression model and examine the training statistics in Big Query
###Code
%%bigquery --project $PROJECT
#standardSQL
SELECT
*
FROM
ML.EVALUATE(MODEL `stock_market.price_model`,
(
SELECT
symbol,
Date,
Open,
close_MIN_prior_5_days,
close_MIN_prior_20_days,
close_MIN_prior_260_days,
close_MAX_prior_5_days,
close_MAX_prior_20_days,
close_MAX_prior_260_days,
close_AVG_prior_5_days,
close_AVG_prior_20_days,
close_AVG_prior_260_days,
close_STDDEV_prior_5_days,
close_STDDEV_prior_20_days,
close_STDDEV_prior_260_days,
normalized_change
FROM
`stock_market.eps_percent_change_sp500`
WHERE
normalized_change IS NOT NULL
AND ABS(MOD(FARM_FINGERPRINT(symbol), 15)) = 1
AND ABS(MOD(FARM_FINGERPRINT(symbol), 15 * 100)) > 15 * 70
AND ABS(MOD(FARM_FINGERPRINT(symbol), 15 * 100)) <= 15 * 85))
%%bigquery --project $PROJECT
#standardSQL
SELECT
*
FROM
ML.TRAINING_INFO(MODEL `stock_market.price_model`)
ORDER BY iteration
###Output
_____no_output_____
|
Run Container on AWS ECS.ipynb
|
###Markdown
About: Run Container on AWS ECSこのNotebookは、このイメージ `yacchin1205/jupyter-with-jenkins` をECSで動作させるNotebookの例です。本手順は、 *Tutorial: Creating a Cluster with a Fargate Task Using the AWS CLI* https://docs.aws.amazon.com/AmazonECS/latest/developerguide/ECS_AWSCLI_Fargate.html をJupyter Notebookにて記述したものです。 PrerequisitesこのNotebookを利用するためにはいくつかの前提条件があります。**以下の説明を読みながら、必要なアカウント等の設定を行って**ください。 事前条件**事前にTerminalから、 `aws configure` を実施してください。**`aws configure` の実施例:```$ aws configureAWS Access Key ID [None]: (自身のアカウントのアクセスキー)AWS Secret Access Key [None]: (自身のアカウントのシークレットアクセスキー)Default region name [None]: ap-northeast-1 (使用したいリージョン)Default output format [None]: json (jsonを指定)```アクセスキー管理方法は様々ありますが、AWS IAMのユーザー https://console.aws.amazon.com/iam/home?region=us-west-2/users からNotebook用のユーザーを作成する方法があります。万が一アクセスキーが漏れた場合に備えて、権限を最小限に、いつでも無効化できるように設定する必要があります。権限は `AmazonEC2FullAccess`, `AmazonElasticFileSystemFullAccess`, `AmazonECS_FullAccess`, `AWSCloudFormationFullAccess`, `CloudWatchFullAccess` を想定しています。正しくアクセス情報が指定されているかどうかを、`describe-vpcs`により確認します。**以下のコマンド実行がエラーとなる場合、`aws configure`が正しくなされていない可能性があります。**
###Code
import json
import sys
vpc_result = !aws ec2 describe-vpcs
try:
vpcs = json.loads('\n'.join(vpc_result))['Vpcs']
print('{} VPCs exist'.format(len(vpcs)))
except:
print(vpc_result)
raise sys.exc_info()
###Output
_____no_output_____
###Markdown
Prepare VPC
###Code
cloudformation_stack_name = 'ecstest0001'
cloudformation_stack_name
import tempfile
work_dir = tempfile.mkdtemp()
work_dir
###Output
_____no_output_____
###Markdown
Templateの定義
###Code
%%writefile {work_dir}/template.yml
AWSTemplateFormatVersion: "2010-09-09"
Description: 'A VPC for ECS'
Resources:
VPC:
Type: "AWS::EC2::VPC"
Properties:
CidrBlock: 10.1.0.0/16
EnableDnsSupport: true
EnableDnsHostnames: true
Tags:
- Key: Name
Value: !Join ['', [!Ref "AWS::StackName", "-VPC" ]]
InternetGateway:
Type: "AWS::EC2::InternetGateway"
DependsOn: VPC
AttachGateway:
Type: "AWS::EC2::VPCGatewayAttachment"
Properties:
VpcId: !Ref VPC
InternetGatewayId: !Ref InternetGateway
ECSSubnet:
Type: AWS::EC2::Subnet
Properties:
VpcId: !Ref VPC
CidrBlock: 10.1.1.0/24
AvailabilityZone: !Select [ 0, !GetAZs ] # Get the first AZ in the list
Tags:
- Key: Name
Value: !Sub ${AWS::StackName}-ECS
ECSPublicRouteTable:
Type: AWS::EC2::RouteTable
Properties:
VpcId: !Ref VPC
Tags:
- Key: Name
Value: ECSPublic
ECSRoute1:
Type: AWS::EC2::Route
DependsOn: AttachGateway
Properties:
RouteTableId: !Ref ECSPublicRouteTable
DestinationCidrBlock: 0.0.0.0/0
GatewayId: !Ref InternetGateway
ECSPublicRouteTableAssociation:
Type: AWS::EC2::SubnetRouteTableAssociation
Properties:
SubnetId: !Ref ECSSubnet
RouteTableId: !Ref ECSPublicRouteTable
ECSSecurityGroup:
Type: AWS::EC2::SecurityGroup
Properties:
GroupDescription: security group for ECS
VpcId: !Ref VPC
Tags:
- Key: Name
Value: !Sub ${AWS::StackName}-ECSSG
Outputs:
ECSVPC:
Value: !Ref VPC
ECSSubnet:
Value: !Ref ECSSubnet
ECSSecurityGroup:
Value: !Ref ECSSecurityGroup
###Output
_____no_output_____
###Markdown
Stackの作成Cloud FormationのStackを作成します。
###Code
stack_result = !aws cloudformation create-stack --stack-name {cloudformation_stack_name} \
--template-body file://{work_dir}/template.yml
try:
stack_id = json.loads('\n'.join(stack_result))['StackId']
print('StackId', stack_id)
except:
print(stack_result)
raise sys.exc_info()
###Output
_____no_output_____
###Markdown
以下のセルの実行結果に表示されるURLをクリックして、作成状況を確認してください。ステータスがCREATE_COMPLETEとなれば、Stackの作成は成功です。
###Code
import urllib.parse
regions = !aws configure get region
print('https://{region}.console.aws.amazon.com/cloudformation/home#/stacks/stackinfo?stackId={stack_id}'.format(region=regions[0], stack_id=urllib.parse.quote(stack_id)))
###Output
_____no_output_____
###Markdown
**ステータスがCREATE_COMPLETEに変化したことを確認**したら、以下のセルを実行してください。> 以下のセルは、Stack作成中の場合はエラーとなります。
###Code
describe_stack_result = !aws cloudformation describe-stacks --stack-name {stack_id}
stack_desc = json.loads(''.join(describe_stack_result))['Stacks'][0]
assert stack_desc['StackStatus'] == 'CREATE_COMPLETE', stack_desc['StackStatus']
###Output
_____no_output_____
###Markdown
Step 1: Create a Cluster
###Code
cluster_name = 'jupyter-cluster'
cluster_name
cluster_result = !aws ecs create-cluster --cluster-name {cluster_name}
try:
cluster_info = json.loads(''.join(cluster_result))
print(cluster_info)
except:
print(cluster_result)
raise sys.exc_info()
###Output
_____no_output_____
###Markdown
Step 2: Register a Task Definition EFSの準備
###Code
efs_result = !aws efs create-file-system
efs_info = json.loads(''.join(efs_result))
efs_info
subnet_id = [o['OutputValue'] for o in stack_desc['Outputs'] if o['OutputKey'] == 'ECSSubnet'][0]
security_group_id = [o['OutputValue'] for o in stack_desc['Outputs'] if o['OutputKey'] == 'ECSSecurityGroup'][0]
(subnet_id, security_group_id)
mount_target_result = !aws efs create-mount-target \
--file-system-id {efs_info['FileSystemId']} \
--subnet-id {subnet_id} \
--security-group {security_group_id}
mount_info = json.loads(''.join(mount_target_result))
mount_info
###Output
_____no_output_____
###Markdown
FileSystemが作成されていることをコンソールから確認する。
###Code
import urllib.parse
regions = !aws configure get region
print('https://{region}.console.aws.amazon.com/efs/home?region={region}#/file-systems/{file_system_id}'.format(region=regions[0], file_system_id=efs_info['FileSystemId']))
###Output
_____no_output_____
###Markdown
ContainerからEFSにアクセスできるよう、同一Security Group間での全パケット許可を行う。
###Code
security_group_id = [o['OutputValue'] for o in stack_desc['Outputs'] if o['OutputKey'] == 'ECSSecurityGroup'][0]
security_group_id
sg_auth_result = !aws ec2 authorize-security-group-ingress --group-id {security_group_id} --protocol all --port 0-65535 --source-group {security_group_id}
sg_auth_result
###Output
_____no_output_____
###Markdown
定義の準備
###Code
container_image = 'yacchin1205/jupyter-with-jenkins'
def_name = "jupyter-with-jenkins"
(def_name, container_image)
import IPython
print('Jupyterへのアクセスに用いるパスワードを設定してください。')
password_hash = IPython.lib.passwd()
password_hash
task_def = {
"family": def_name,
"networkMode": "awsvpc",
"containerDefinitions": [
{
"name": "jupyter",
"image": container_image,
"portMappings": [
{
"containerPort": 8888,
"hostPort": 8888,
"protocol": "tcp"
}
],
"essential": True,
'environment': [
{
'name': 'CHOWN_HOME',
'value': 'yes'
},
{
'name': 'GRANT_SUDO',
'value': 'yes'
},
],
'user': '0',
"mountPoints": [
{
"containerPath": "/home/jovyan",
"sourceVolume": "efs-jupyter"
}
],
"command": ["start-notebook.sh", '--NotebookApp.password=' + password_hash],
}
],
"requiresCompatibilities": [
"FARGATE"
],
"volumes": [
{
"name": "efs-jupyter",
"efsVolumeConfiguration": {
"fileSystemId": efs_info['FileSystemId'],
}
}
],
"cpu": "1024",
"memory": "4096"
}
task_def
import os
taskdef_path = os.path.join(work_dir, 'taskdef.json')
with open(taskdef_path, 'w') as f:
f.write(json.dumps(task_def))
!cat {taskdef_path}
taskdef_result = !aws ecs register-task-definition --cli-input-json file://{taskdef_path}
try:
taskdef_info = json.loads(''.join(taskdef_result))
taskdef_arn = taskdef_info['taskDefinition']['taskDefinitionArn']
print(taskdef_arn)
except:
print(taskdef_result)
raise sys.exc_info()
###Output
_____no_output_____
###Markdown
Step 3: List Task Definitions
###Code
!aws ecs list-task-definitions
###Output
_____no_output_____
###Markdown
Step 4: Create a Service
###Code
service_name = 'jupyter-service'
service_name
subnet_id = [o['OutputValue'] for o in stack_desc['Outputs'] if o['OutputKey'] == 'ECSSubnet'][0]
security_group_id = [o['OutputValue'] for o in stack_desc['Outputs'] if o['OutputKey'] == 'ECSSecurityGroup'][0]
(subnet_id, security_group_id)
network_configuration = 'awsvpcConfiguration={{subnets=[{subnet_id}],securityGroups=[{security_group_id}],assignPublicIp=ENABLED}}'.format(**locals())
service_result = !aws ecs create-service --cluster {cluster_name} --platform-version 1.4.0 --service-name {service_name} --task-definition {taskdef_arn} --desired-count 1 --launch-type "FARGATE" --network-configuration "{network_configuration}"
try:
service_info = json.loads(''.join(service_result))
print(service_info)
except:
print(service_result)
raise sys.exc_info()
import urllib.parse
regions = !aws configure get region
print('https://{region}.console.aws.amazon.com/ecs/home#/clusters/{cluster_name}/services'.format(region=regions[0], cluster_name=cluster_name))
###Output
_____no_output_____
###Markdown
Step 6: Describe the Running Service
###Code
service_desc_result = !aws ecs describe-services --cluster {cluster_name} --services {service_name}
service_desc_info = json.loads(''.join(service_desc_result))
service_desc_info
###Output
_____no_output_____
###Markdown
Accessing to the service
###Code
import re
service_pattern = re.compile(r'.*\s+started\s*([0-9]+)\s*tasks:\s*\(task\s+(.+)\).*')
service_result = !aws ecs describe-services --cluster {cluster_name} --services {service_name}
service_info = json.loads(''.join(service_result))
events = [event['message'] for event in service_info['services'][0]['events'] if service_pattern.match(event['message'])]
assert len(events) > 0, service_info['services'][0]['events']
task_id = service_pattern.match(events[0]).group(2)
task_id
task_result = !aws ecs describe-tasks --cluster {cluster_name} --task {task_id}
task_info = json.loads(''.join(task_result))
assert task_info['tasks'][0]['lastStatus'] == 'RUNNING', task_info['tasks'][0]['lastStatus']
task_info['tasks'][0]['attachments']
network_info = dict([(detail['name'], detail['value']) for detail in task_info['tasks'][0]['attachments'][0]['details']])
eniid = network_info['networkInterfaceId']
eniid
network_desc = !aws ec2 describe-network-interfaces --network-interface-ids {eniid}
network_info = json.loads(''.join(network_desc))
public_ip = network_info['NetworkInterfaces'][0]['Association']['PublicIp']
public_ip
print('http://{}:8888'.format(public_ip))
###Output
_____no_output_____
###Markdown
Security Groupの変更JupyterへのアクセスにはTCPポート8888を使用する。自身の環境からコンテナへのアクセスを許可するよう適切なポートおよびアクセス元IPアドレスを指定する。**インバウンドルールを編集**ボタンを選択して、以下のようなルールを追加する。 Security GroupのIDはStackのOutputsに記録されている。
###Code
stack_desc['Outputs']
security_group_id = [o['OutputValue'] for o in stack_desc['Outputs'] if o['OutputKey'] == 'ECSSecurityGroup'][0]
security_group_id
regions = !aws configure get region
print('https://{region}.console.aws.amazon.com/ec2/v2/home?region={region}#SecurityGroup:groupId={security_group_id}'.format(region=regions[0], security_group_id=security_group_id))
# セキュリティグループにSSHに関する許可が含まれていれば先に進める
sg_result = !aws ec2 describe-security-groups --group-ids {security_group_id}
sg_desc = json.loads(''.join(sg_result))['SecurityGroups'][0]
assert 'IpPermissions' in sg_desc and any([ipperm['IpProtocol'] == 'tcp' and ipperm['ToPort'] <= 8888 and 8888 <= ipperm['FromPort'] for ipperm in sg_desc['IpPermissions']]), sg_desc['IpPermissions']
###Output
_____no_output_____
|
Keys_localisation.ipynb
|
###Markdown
**1. Tester le GPU**
###Code
!nvidia-smi
###Output
_____no_output_____
###Markdown
**Installation keras version 2.2.0**
###Code
!pip install keras==2.2.0
###Output
_____no_output_____
###Markdown
**4. Téléchargement du dossier des images de clé et le background**
###Code
!wget https://github.com/belarbi2733/keras_yolov3/releases/download/1/key_wb.zip
!wget https://github.com/belarbi2733/keras_yolov3/releases/download/1/bckgrnd.zip
###Output
_____no_output_____
###Markdown
**5. Décompresser les deux fichiers .zip**
###Code
!unzip bckgrnd.zip
!unzip key_wb.zip
###Output
_____no_output_____
###Markdown
**6. Mixer le backgound avec les clés**
###Code
!python keys_with_background.py --keys "key_wb" --background "bckgrnd" --output "keys_and_background"
###Output
_____no_output_____
###Markdown
**7. Récupurer le fichier annotation et adapter le format du fichier**
###Code
!mv keys_and_background/annotations.csv .
###Output
_____no_output_____
###Markdown
**8. Télécharger le fichier yolov3.weights**
###Code
!wget https://pjreddie.com/media/files/yolov3.weights
###Output
_____no_output_____
###Markdown
**9. Convertir le fichier yolov3.weights en model tensorflow**
###Code
!python convert.py yolov3.cfg yolov3.weights model_data/yolo_weights.h5
###Output
_____no_output_____
###Markdown
**10. Lancer l'entrainement** Train with frozen layers first, to get a stable loss. Adjust num epochs to your dataset. This step is enough to obtain a not bad model.
###Code
!python train1.py --initial_epoch1 0 --epoch1 5 --batch_size1 64 --annotation 'annotations.csv' --classes 'model_data/key_classes.txt' --anchors 'model_data/yolo_anchors.txt'
###Output
_____no_output_____
###Markdown
Unfreeze and continue training, to fine-tune. Train longer if the result is not good.
###Code
!python train2.py --initial_epoch2 5 --epoch2 10 --batch_size2 16 --annotation 'annotations.csv' --classes 'model_data/key_classes.txt' --anchors 'model_data/yolo_anchors.txt'
###Output
_____no_output_____
###Markdown
**10. Tester le résultat**
###Code
!python test_yolo.py --image --input='keys_and_background/gen_0009.jpg' --output='yolo1.jpg' --model 'weights_yolo_train/trained_weights_final.h5' --classes 'model_data/key_classes.txt' --anchors 'model_data/yolo_anchors.txt'
###Output
_____no_output_____
###Markdown
**11. Afficher le résulat**
###Code
from IPython.display import display, Image, SVG, Math, YouTubeVideo
Image(filename='yolo1.jpg')
###Output
_____no_output_____
|
Exercise-6/ex6.ipynb
|
###Markdown
Programming Exercise 6: Support Vector Machines Author - Rishabh Jain
###Code
import warnings
warnings.simplefilter('ignore')
import matplotlib.pyplot as plt
import seaborn as sns
import pandas as pd
import numpy as np
import cvxopt, re
%matplotlib inline
from scipy.io import loadmat
###Output
_____no_output_____
###Markdown
Learning Resources1. [SVM Video Lecture (MIT)](https://www.youtube.com/watch?v=_PwhiWxHK8o)2. [29 to 33 SVM Video Lectures (University of Buffalo)](https://www.youtube.com/watch?v=N4pai7eZW_o&list=PLhuJd8bFXYJsSXPMrGlueK6TMPdHubICv&index=29)3. [Support Vector Machine Succinctly (PDF)](./Lectures/svm.pdf)Before solving the programming assignment from the course, let's try to understand the **Maths Behind SVM :**1. [Maximum Margin Classifier](Maximum-Margin-Classifier)2. [Lagrange Multipliers](Understanding-Lagrange-Multipliers)3. [Primal and Dual Lagrangian](Primal-and-Dual-Formulations)4. [Hard Margin SVM](Hard-Margin-SVM)5. [Soft Margin SVM](Soft-Margin-SVM)6. [Hypothesis Function](Hypothesis-Function)7. [Kernel Trick](Kernel-Trick)8. [SVM Implementation using CVXOPT](SVM-Implementation-using-CVXOPT-Solver) Maximum Margin Classifier For two-class, such as the one shown below, there are lots of possible linear separators. Intuitively, a decision boundary drawn in the middle of the void between data items of the two classes seems better than one which approaches very close to examples of one or both classes. While some learning methods such as the logistic regression find just any linear separator. **The SVM in particular defines the criterion to be looking for a decision surface that is MAXIMALLY far away from any data point**. This distance from the decision surface to the closest data point determines the margin of the classifier.Let's imagine a vector $\vec{w}$ perpendicular to the margin and an unknown data point $\vec{u}$ which can be on either side of the margin. In order to know whether $\vec{u}$ is on the right or left side of the margin, we will project (Dot product) $\vec{u}$ onto $\vec{w}$.$$\vec{w}.\vec{u}\geq c$$$$\boxed{\vec{w}.\vec{u}+b\geq 0}\;\;(1)$$ If the projection of $\vec{u}$ plus some constant $b$ is greater than zero, then its a positive sample otherwise its a negative sample.**Eq. (1) is our DECISION RULE**. Here the problem is that we don't know what $w$ and $b$ to use. **An unknown sample may be located anywhere inside or outside the margin (i.e. >0 or <0), but if it's a known positive sample $\vec{x_{+}}$ then the SVM decision rule should insist the dot product plus some constant $b$ to be 1 or greater than 1.** Likewise for a negative sample $\vec{x_{-}}$, dot product plus some constant $b$ should be less than or equal to -1 Hence:$\vec{w}.\vec{x_{+}}+b\geq 1 $ $\vec{w}.\vec{x_{-}}+b\leq -1 $ Introducing a variable $y_i$ such that : $$\begin{equation*} y_{i}=\begin{cases} +1 & \text{for +ve samples}\\ -1 & \text{for -ve samples} \end{cases}\end{equation*}$$Mutiplying the above two inequality eqauations with $y_i$:For +ve sample : $y_{i}(\vec{w}.\vec{x_{i}}+b) \geq 1$ For -ve sample : $y_{i}(\vec{w}.\vec{x_{i}}+b) \geq 1$ Note : Sign changed from $\leq$ to $\geq$ because $y_i$ is -1 in case of -ve samplesSince both the equations are same, we can rewrite them as :$$\boxed{y_{i}(\vec{w}.\vec{x_{i}}+b)\geq 1}\;\;(2)$$For samples on margin$$\boxed{y_{i}(\vec{w}.\vec{x_{i}}+b)-1= 0}\;\;(3)$$Eq.(2) is basically a **constraint** for our margin, which means that **all the training samples should be on the correct side OR on the margin** (i.e. +ve samples on the right and -ve samples on the left side of the margin) and **NO training sample should be inside the margin at all meaning ZERO TRAINING ERROR.** Let's calculate the width of the margin.Let's imagine two vectors $\vec{x_+}$ and $\vec{x_-}$, both are +ve and -ve known samples respectively. The difference of these two vectors is a resultant vector called $\vec{R}$ where : $$\vec{R}=\vec{x_+}-\vec{x_-}$$All we need is a $\hat{u}$, **so that the WIDTH of the margin will be the projection of $\vec{R}$ onto $\hat{u}$**. From the first image, we already know a vector $\vec{w}$ in the same direction. $$\hat{u}=\frac{\vec{w}}{||w||}$$ **WIDTH** $=\vec{R}.\hat{u} $ $\;\;\;\;\;\;\;\;\;\;=(\vec{x_+}-\vec{x_-}).\frac{\vec{w}}{||w||}$ $\;\;\;\;\;\;\;\;\;\;=\frac{(\vec{x_+}.\vec{w}-\vec{x_-}.\vec{w})}{||w||}$Using Eq.(3), we get$\;\;\;\;\;\;\;\;\;\;=\frac{(1-b+1+b)}{||w||}$$$\boxed{\text{WIDTH}=\frac{2}{||w||}}\;\;(4)$$Now, we want to maximize the margin while incurring zero training error.max $\frac{2}{||w||}$ with 0 loss OR (Flipping for mathematical convenience)min $\frac{||w||}{2}\;$ with 0 loss OR (Squaring the numerator for mathematical convenience)min $\frac{||w||^2}{2}$ with 0 loss **(NO LONGER AN UNCONSTRAINED OPTIMIZATION)** Understanding Lagrange MultipliersLagrange multipliers is a strategy of finding the local maxima and minima of a function subject to **equality** constraints. Let's try to solve a constrained opitimization problem : Example 1 (Equality Constraint)>minimize $\;\;f(x,y)=2-x^2-2y^2$ >subject to $\;\;h(x,y)=x+y-1=0$>>**We introduce a new variable ($\beta$) called a Lagrange multiplier and study the Lagrange function defined by:**>>$$\boxed{L(x,y,\beta)=f(x,y)-\beta h(x,y)}$$>>$L(x,y,\beta)=(2-x^2-2y^2)-\beta(x+y-1)$>>Now we solve the above equation like an unconstrained optimization problem by taking partial derivatives w.r.t $x$ & $y$ and set them equal to zero solving for $x$, $y$ and $\beta$>>$\frac{\partial{L}}{\partial{x}}=0\;\;=>\;\;-2x-\beta=0\;\;=>\;\;x=\frac{-\beta}{2}$>>$\frac{\partial{L}}{\partial{y}}=0\;\;=>\;\;-4y-\beta=0\;\;=>\;\;y=\frac{-\beta}{4}$>>$\frac{\partial{L}}{\partial{\beta}}=0\;\;=>\;\;x+y-1=0\;\;=>\;\;\beta=\frac{-4}{3}$>>$\boxed{x=\frac{4}{6},y=\frac{4}{12},\beta=\frac{-4}{3}}$ Example 2 (Inequality Constraints / Karush-Kuhn-Tucker (KKT) conditions)>maximize $\;\;f(x,y)=3x+4y$ >subject to $\;\;h_{1}(x,y)=x^2+y^2\leq4$ >$\;\;\;\;\;\;\;\;\;\;\;\;\;\;\;h_{2}(x,y)=x\geq1$>>**Note: Inequality constraints should be in the form of $h(x,y)\leq0$**>>$$\boxed{L(x,y,\alpha_1,\alpha_2)=f(x,y)-\alpha_1 h_{1}(x,y)-\alpha_2 h_{2}(x,y)\\\;\;\text{s.t. }\alpha_1,\alpha_2\geq0}$$>>$L(x,y,\alpha_1,\alpha_2)=3x+4y-\alpha_1(x^2+y^2-4)-\alpha_2(-x+1)$ >>**KKT Conditions :**>>1. $\frac{\partial{L}}{\partial{x}}=3-2\alpha_1x+\alpha_2=0$>>2. $\frac{\partial{L}}{\partial{y}}=4-2\alpha_1y=0$>>3. $\alpha_1(x^2+y^2-4)=0$>>4. $\alpha_2(-x+1)=0$>>5. $\alpha_1,\alpha_2\geq0$ >>A constraint is considered to be binding (active) if changing it also changes the optimal solution. Less severe constraints that do not affect the optimal solution are non-binding (non active). For 2 constraints possible combinations are :>>- Both constraints are binding>- Constraint 1 binding, Constraint 2 not binding>- Constraint 2 binding, Constraing 1 not binding>- Both constraints are not binding>>**POSSIBILITY 1 : Both constraints are binding**>>$-x+1=0\;\text{and}\;\alpha_2>0\;\;=>\;\;x=1$ >$x^2+y^2-4=0\;\text{and}\;\alpha_1>0\;\;=>\;\;x^2+y^2=4\;\;=>\;\;1+y^2=4\;\;=>\;\;y=\pm\sqrt{3}$ >>(a) For $y=+\sqrt{3}$ >>>Condition 2 becomes: >>$4-2\sqrt{3}\alpha_1=0\;\;=>\;\;\alpha_1=\frac{2}{\sqrt{3}}>0$ >>Condition 1 becomes: >>$3-2\alpha_1+\alpha_2=0\;\;=>\;\;3-\frac{4}{\sqrt{3}}+\alpha_2=0\;\;=>\;\;\alpha_2=\frac{4}{\sqrt{3}}-3<0$ (KKT condition fails)>>(a) For $y=-\sqrt{3}$ >>>Condition 2 becomes: >>$4+2\sqrt{3}\alpha_1=0\;\;=>\;\;\alpha_1=\frac{-2}{\sqrt{3}}<0$ (KKT condition fails) >>Condition 1 becomes: >>$3-2\alpha_1+\alpha_2=0\;\;=>\;\;3+\frac{4}{\sqrt{3}}+\alpha_2=0\;\;=>\;\;\alpha_2=\frac{-4}{\sqrt{3}}-3<0$ (KKT condition fails)>>**POSSIBILITY 2 : Constraint 1 binding , Contraint 2 not binding**>>$x>1\;\text{and}\;\boxed{\alpha_2=0}$ >$x^2+y^20\;\;=>\;\;x=+\sqrt{4-y^{2}}$ >>>Condition 1 becomes: >>$3-2\alpha_1x=0\;\;=>\;\;x=\frac{3}{2\alpha_1}\;\;=>\;\;3-2\alpha_1\sqrt{4-y^{2}}=0\;\;=>\;\;\alpha_1=\frac{3}{2\sqrt{4-y^{2}}}$ >>Condition 2 becomes: >>$4-2\alpha_1y=0\;\;=>\;\;4-\frac{3y}{\sqrt{4-y^{2}}}=0\;\;=>\;\;4\sqrt{4-y^{2}}=3y\;\;=>\;\;16(4-y^2)=9y^2\;\;=>\;\;64-16y^2=9y^2\;\;=>\;\;64=25y^2\;\;=>\;\;y=\pm\frac{8}{5}$>>$\boxed{\alpha_1=\frac{3}{2\sqrt{4-\frac{64}{25}}}=\frac{3}{2(\frac{6}{5})}=\frac{5}{4}>0}$ >$x=+\sqrt{4-y^{2}}\;\;=>\;\;x=\frac{6}{5}$>>1 candidate point: $\boxed{(x,y)=(\frac{6}{5},\frac{8}{5})}$>>**POSSIBILITY 3 : Constraint 2 binding , Contraint 1 not binding**>>$x=1\;\text{and}\;\alpha_2>0$ >$x^2+y^2<4\;\text{and}\;\alpha_1=0$ >>>Condition 2 becomes: >>$4-2\alpha_1y=0\;\;=>\;\;4=0$ (Contradiction, no candidate points) >>**POSSIBILITY 4 : Both constraints are not binding**>>$x>1\;\text{and}\;\alpha_2=0$ >$x^2+y^2<4\;\text{and}\;\alpha_1=0$ >>>Condition 2 becomes: >>$4-2\alpha_1y=0\;\;=>\;\;4=0$ (Contradiction, no candidate points) >>**Check maximality of the candidate point :**>>$f(\frac{6}{5},\frac{8}{5})=3(\frac{6}{4})+4(\frac{8}{5})=\frac{18}{5}+\frac{32}{5}=10$>>Optimal Solution : $\boxed{x=\frac{6}{5},y=\frac{8}{5},\alpha_1=0,\alpha_2=\frac{5}{4}}$ Handling both types of Constraints$$\boxed{\min_{w}\;\;f(w)\\\text{subject to}\;\;g_{i}(w)\leq0\;\;\;i=1,2,...k\\\text{and}\;\;\;\;\;\;\;\;\;\;h_{i}(w)=0\;\;\;i=1,2,...l\\}$$**Generalized Lagrangian** $$\boxed{L(w,\alpha,\beta)=f(w)+\sum_{i=1}^{k}\alpha_{i}g_{i}(w)+\sum_{i=1}^{l}\beta_{i}h_{i}(w)\\\text{subject to}\;\;\alpha_{i}\geq0,\forall_i}$$ Primal and Dual Formulations Primal OptimizationLet $\theta_p$ be defined as :$$\boxed{\theta_p(w)=\max_{\alpha,\beta;\alpha_i\geq0}L(w,\alpha,\beta)}$$Original constrained problem is same as :$$\boxed{p^*=\min_{w}\theta_P(w)=\min_{w}\max_{\alpha,\beta;\alpha_i\geq0}L(w,\alpha,\beta)}$$Solving $p^*$ is same as solving the constrained optimization problem.$$\begin{equation} \theta_{p}(w)=\begin{cases} f(w) & \text{if all constraints are satifsied}\\ \infty & \text{else} \end{cases}\end{equation}$$ Dual OptimizationLet $\theta_d$ be defined as :$$\boxed{\theta_d(w)=\min_{w}L(w,\alpha,\beta)}$$Original constrained problem is same as :$$\boxed{d^*=\max_{\alpha,\beta;\alpha_i\geq0}\theta_d(w)=\max_{\alpha,\beta;\alpha_i\geq0}\min_{w}L(w,\alpha,\beta)}$$ Week Duality Theorem (Min-Max Theorem)$$\boxed{d^{*}\leq p^{*}}$$Both of them are equal ($d^{*}=p^{*}$) when- f(w) is convex- Constraints are affine (Linear) Relation between Primal and Dual+ In genral $d^*\leq p^*$, for SVM optimization the equality holds true.+ Certain conditions should be true.+ Known as the **Kahrun-Kuhn-Tucker (KKT)** conditions.+ For $d^*=p^*=L(w^*,\alpha^*,\beta^*)$ :>+ $\frac{\partial}{\partial{w}}L(w^*,\alpha^*,\beta^*)=0$ >+ $\frac{\partial}{\partial{\beta_{i}}}L(w^*,\alpha^*,\beta^*)=0\;\;\;\;\;\;\;i=1,2,...l$ >+ $\alpha_{i}g_{i}(w^{*})=0\;\;\;\;\;\;\;\;\;\;\;\;\;\;\;\;\;i=1,2,...k$ >+ $g_{i}(w^{*})\leq0\;\;\;\;\;\;\;\;\;\;\;\;\;\;\;\;\;\;\;\;i=1,2,...k$ >+ $\alpha_{i}\geq0\;\;\;\;\;\;\;\;\;\;\;\;\;\;\;\;\;\;\;\;\;\;\;\;\;i=1,2,...k$ Hard-Margin SVM Optimization Formulation$$\boxed{\min_{w,b}\;\;\frac{||w||^2}{2}\\ \text{s.t.}\;\;y_{i}(w^{T}x_{i}+b)\geq 1,\;\;\forall{i}}\;\;(5)$$In order to solve a constrained optimization problem, Lagrange multipliers are used. Since the Objective function is convex (parabola) and all the Constraints are affine (linear) too. Solving dual or primal, answer is going to be same. Rewriting above constrained optimization problem as Lagrangian:$$\boxed{\min_{w,b,\alpha} L(w,b,\alpha)=\frac{||w||^2}{2}+\sum_{i=1}^{N}\alpha_{i}(1-y_{i}(w^{T}x_{i}+b))\\\text{s.t.}\;\;\alpha_{i}\geq0,\;\;\forall{i}}\;\;(6)$$Rewriting above Lagrangian function as a Dual lagrangian:$$\max_{\alpha} \min_{w,b}(Lw,b,\alpha)\\\text{s.t.}\;\;\alpha_{i}\geq0,\;\;\forall{i}$$**OK, let's first minimize the $L(w,b,\alpha)$ w.r.t. $w,b$:**$$\min_{w,b} L(w,b,\alpha)$$$\frac{\partial}{\partial{w}}L(w,b,\alpha)=w-\sum_{i=1}^{N}\alpha_{i}y_{i}x_{i}$>Setting $\frac{\partial}{\partial{w}}L(w,b,\alpha)=0$ gives us $\boxed{w=\sum_{i=1}^{N}\alpha_{i}y_{i}x_{i}}\;\;(7)$$\frac{\partial}{\partial{b}}L(w,b,\alpha)=-\sum_{i=1}^{N}\alpha_{i}y_{i}$>Setting $\frac{\partial}{\partial{b}}L(w,b,\alpha)=0$ gives us $\boxed{\sum_{i=1}^{N}\alpha_{i}y_{i}=0}\;\;(8)$We now will take Eq.(7) and Eq.(8) and plug them back into our full lagrangian Eq.(6) to get a reduced lagrangian that depends only on $\alpha$:$\begin{align*}L(w,b,\alpha)&=\frac{w^Tw}{2}+\sum_{i=1}^{N}\alpha_{i}-\sum_{i=1}^{N}\alpha_{i}y_{i}w^{T}x_{i}-\sum_{i=1}^{N}\alpha_{i}y_{i}b\\&=\frac{1}{2}(\sum_{i=1}^{N}\alpha_{i}y_{i}x_{i})^T(\sum_{j=1}^{N}\alpha_{j}y_{j}x_{j})+\sum_{i=1}^{N}\alpha_{i}-\sum_{i=1}^{N}\alpha_{i}y_{i}(\sum_{j=1}^{N}\alpha_{j}y_{j}x_{j})^{T}x_{i}-0\\&=\frac{1}{2}\sum_{i=1}^{N}\sum_{j=1}^{N}(\alpha_{i}\alpha_{j}y_{i}y_{j}x_{i}^{T}x_{j})+\sum_{i=1}^{N}\alpha_{i}-\sum_{i=1}^{N}\sum_{j=1}^{N}(\alpha_{i}\alpha_{j}y_{i}y_{j}x_{i}^{T}x_{j})\\&=\sum_{i=1}^{N}\alpha_{i}-\frac{1}{2}\sum_{i=1}^{N}\sum_{j=1}^{N}(\alpha_{i}\alpha_{j}y_{i}y_{j}x_{i}^{T}x_{j})\end{align*}$**Above equation is free of any $w$ and $b$. Now, let's maximize the $L(w,b,\alpha)$ w.r.t. $\alpha$:**$$\boxed{\max_{\alpha} L(w,b,\alpha)=\sum_{i=1}^{N}\alpha_{i}-\frac{1}{2}\sum_{i=1}^{N}\sum_{j=1}^{N}(\boxed{\alpha_{i}\alpha_{j}}y_{i}y_{j}x_{i}^{T}x_{j})\\\text{s.t. }\alpha_{i}\geq0\;\;\forall{i}\\\text{and }\sum_{i=1}^{N}\alpha_{i}y_{i}=0\;\;\forall{i}}\;\;(9)$$ KKT ConditionsEq.(9) is a Quadratic constraint optimization problem because two unknowns $\alpha_j$ & $\alpha_j$ are getting multiplied together. **So, this equation is solved using some QP solver (like CVXOPT in python).**Let $\alpha^{*}_{1},\alpha^{*}_{2},...\alpha^{*}_{N}$ be the solutions of QP (Quadratic Programming) problem.**Stationary conition:**1. $\frac{\partial}{\partial{w}}L(w,b,\alpha)=0,\;\;\;\;$we got Eq.(7)2. $\frac{\partial}{\partial{b}}L(w,b,\alpha)=0,\;\;\;\;$we got Eq.(8)**Primal feasibility condition:**3. $y_{i}(w^{T}x_{i}+b)-1\geq 0\;\;\;$**Dual feasibility condition:**4. $\alpha_{i}\geq0$**Complementary slackness condition:**5. $\alpha_{i}(y_{i}(w^TX_{i}+b)-1)=0$Using KKT condition 4 & 5, we can imply that:- If $\alpha^{*}_{i}=0\;\;$ then $\;\;y_{i}(w^TX_{i}+b)-1\geq0$- If $\alpha^{*}_{i}>0\;\;$ then $\;\;y_{i}(w^TX_{i}+b)-1=0\;\;$($x$ is on the margin)**Only train examples that lie on the margin are relevant. These are called SUPPORT VECTORS.**$$\boxed{w=\sum_{i=1}^{S}\alpha^{*}_{i}y_{i}x_{i}}\;\;(10)$$ For $\alpha^{*}_{i}>0$, $y_{i}(w^Tx_{i}+b)-1=0\;\;\\y_{i}(w^Tx_{i}+b)=1\;\;\text{(Multiplying both sides by y)}\\y_{i}^{2}(w^Tx_{i}+b)=y_{i}\;\;y\epsilon\{-1,1\}\\w^Tx_{i}+b=y_{i}\\b=y_{i}-w^Tx_{i}\;\;\text{(Averaging over found support vectors S)}$$$\boxed{b=\frac{1}{S}\sum_{i=1}^{S}(y_{i}-w^{T}x_{i})}\;\;(11)$$ Soft-Margin SVM What if the data is not linearly separable??If the data is not linearly separable, Hard-Margin SVM will fail to fit the data as it tries to incur zero training error. Here, incurring zero training error is not possible but we can still learn a maximum margin hyperplane if we:- Relax the constraint by introducing **slack variable** $(\xi_{i})$- Allow some examples to fall inside the margin, $(0<\xi_{i}\leq1)$- Allow some examples to be **misclassified**. For misclassification, $(\xi_{i}>1)$- Minimize the number of such examples (Ensuring not too many points are on the wrong side of margin)$$\min C\sum_{i=1}^N\xi_{i}$$ Where **C controls the impact of the margin error.** Optimization Formulation$$\boxed{\min_{w,b}f(w,b)=\frac{||w||^{2}}{2}+C\sum_{i=1}^{N}\xi_{i}\\\text{s.t.}\;\;\;y_{i}(w^{T}x_{i}+b)\geq1-\boxed{\xi_{i}},\;\;\forall{i}\\\;\;\;\;\;\;\;\;\xi_{i}\geq0,\;\;\forall{i}}\;\;(12)$$Rewriting above constrained optimization problem as Lagrangian:$$\boxed{\min_{w,b,\xi,\alpha,\beta} L(w,b,\xi,\alpha,\beta)=\frac{||w||^2}{2}+C\sum_{i=1}^{N}\xi{i}+\sum_{i=1}^{N}\alpha_{i}[(1-\xi_{i})-y_{i}(w^{T}x_{i}+b)]+\sum_{i=1}^{N}\beta_{i}(-\xi_{i})\\\text{s.t.}\;\;\alpha_{i}\geq0,\beta_{i}\geq0,\;\;\forall{i}}\;\;(13)$$Rewriting above Lagrangian function as a Dual lagrangian:$$\max_{\alpha\beta} \min_{w,b,\xi}L(w,b,\xi)\\\text{s.t.}\;\;\alpha_{i}\geq0,\beta_{i}\geq0,\;\;\forall{i}$$**OK, let's first minimize the $L(w,b,\xi,\alpha)$ w.r.t. $w,b$ and $\xi$ :**$$\min_{w,b,\xi} L(w,b,\xi,\alpha,\beta)$$$\frac{\partial}{\partial{w}}L(w,b,\xi,\alpha,\beta)=w-\sum_{i=1}^{N}\alpha_{i}y_{i}x_{i}$>Setting $\frac{\partial}{\partial{w}}L(w,b,\xi,\alpha,\beta)=0$ gives us $\boxed{w=\sum_{i=1}^{N}\alpha_{i}y_{i}x_{i}}\;\;(14)$$\frac{\partial}{\partial{b}}L(w,b,\xi,\alpha,\beta)=-\sum_{i=1}^{N}\alpha_{i}y_{i}$>Setting $\frac{\partial}{\partial{b}}L(w,b,\xi,\alpha,\beta)=0$ gives us $\boxed{\sum_{i=1}^{N}\alpha_{i}y_{i}=0}\;\;(15)$$\frac{\partial}{\partial{\xi}}L(w,b,\xi,\alpha,\beta)=C-\alpha_{i}-\beta_{i}$>$\begin{align*}\text{Setting }&\frac{\partial}{\partial{\xi}}L(w,b,\xi,\alpha,\beta)=0\\&=>C-\alpha_{i}-\beta_{i}=0\\&=>\boxed{\beta_{i}=C-\alpha_{i}}\;\;(16)\\\\&\text{But because the }\beta_{i}\text{'s are a dual variables with }\beta_{i}\geq0\text{, then this leads to:}\\&=>C-\alpha_{i}\geq0\\&=>\alpha_{i}\leq C\\\\&\text{This along with the fact that }\alpha_{i}\text{ are dual variables with }\alpha_{i}\geq0\text{ we have:}\\&\boxed{0\leq\alpha_{i}\leq C}\;\;(17)\end{align*}$We now will take these results and plug them back into our full lagrangian to get a reduced lagrangian that depends only on $\alpha$ and $\beta$:Replacing $w$ using Eq.(14) in Eq.(13), we get$\begin{align*}L(w,b,\xi,\alpha,\beta)&=\frac{1}{2}(\sum_{i=1}^{N}\alpha_{i}y_{i}x_{i})^{T}(\sum_{i=1}^{N}\alpha_{i}y_{i}x_{i})+C\sum_{i=1}^{N}\xi_{i}+\sum_{i=1}^{N}\alpha_{i}[(1-\xi_{i})-y_{i}((\sum_{j=1}^{N}\alpha_{j}y_{j}x_{j})^{T}x_{i}+b)]-\sum_{i=1}^{N}\beta_{i}\xi_{i}\\&=\frac{1}{2}\sum_{i=1}^{N}\sum_{j=1}^{N}(\alpha_{i}\alpha_{j}y_{i}y_{j}x_{i}^{T}x_{j})+C\sum_{i=1}^{N}\xi_{i}+\sum_{i=1}^{N}\alpha_{i}[(1-\xi_{i})-(\sum_{j=1}^{N}\alpha_{j}y_{i}y_{j}x_{j})^{T}x_{i}+b]-\sum_{i=1}^{N}\beta_{i}\xi_{i}\\&=\frac{1}{2}\sum_{i=1}^{N}\sum_{j=1}^{N}(\alpha_{i}\alpha_{j}y_{i}y_{j}x_{i}^{T}x_{j})+C\sum_{i=1}^{N}\xi_{i}+\sum_{i=1}^{N}\alpha_{i}-\sum_{i=1}^{N}\alpha_{i}\xi_{i}-\sum_{i=1}^{N}\sum_{j=1}^{N}\alpha_{i}\alpha_{j}y_{i}y_{j}x_{i}^{T}x_{j}+b\sum_{i=1}^{N}\alpha_{i}y_{i}-\sum_{i=1}^{N}\beta_{i}\xi_{i}\\&=-\frac{1}{2}\sum_{i=1}^{N}\sum_{j=1}^{N}(\alpha_{i}\alpha_{j}y_{i}y_{j}x_{i}^{T}x_{j})+C\sum_{i=1}^{N}\xi_{i}+\sum_{i=1}^{N}\alpha_{i}-\sum_{i=1}^{N}\alpha_{i}\xi_{i}+b\sum_{i=1}^{N}\alpha_{i}y_{i}-\sum_{i=1}^{N}\beta_{i}\xi_{i}\\&=-\frac{1}{2}\sum_{i=1}^{N}\sum_{j=1}^{N}(\alpha_{i}\alpha_{j}y_{i}y_{j}x_{i}^{T}x_{j})+C\sum_{i=1}^{N}\xi_{i}+\sum_{i=1}^{N}\alpha_{i}-(\sum_{i=1}^{N}\alpha_{i}+\sum_{i=1}^{N}\beta_{i})\xi_{i}\;\;\;\;\text{Using Eq.(15)}\\&=-\frac{1}{2}\sum_{i=1}^{N}\sum_{j=1}^{N}(\alpha_{i}\alpha_{j}y_{i}y_{j}x_{i}^{T}x_{j})+C\sum_{i=1}^{N}\xi_{i}+\sum_{i=1}^{N}\alpha_{i}-C\sum_{i=1}^{N}\xi_{i}\;\;\;\;\text{Using Eq.(16)}\\&=\sum_{i=1}^{N}\alpha_{i}-\frac{1}{2}\sum_{i=1}^{N}\sum_{j=1}^{N}(\alpha_{i}\alpha_{j}y_{i}y_{j}x_{i}^{T}x_{j})\end{align*}$**Above equation is free of any $w,b,\xi$ and $\beta$. Now, let's maximize the $L(w,b,\xi,\alpha,\beta)$ w.r.t. $\alpha$:**$$\boxed{\max_{\alpha} L(w,b,\alpha)=\sum_{i=1}^{N}\alpha_{i}-\frac{1}{2}\sum_{i=1}^{N}\sum_{j=1}^{N}(\boxed{\alpha_{i}\alpha_{j}}y_{i}y_{j}x_{i}^{T}x_{j})\\\text{s.t. }0\leq\alpha_{i}\leq C\;\;\forall{i}\\\text{and }\sum_{i=1}^{N}\alpha_{i}y_{i}=0\;\;\forall{i}}\;\;(18)$$ KKT ConditionsEq.(18) is a Quadratic constraint optimization problem because two unknowns $\alpha_j$ & $\alpha_j$ are getting multiplied together. **So, this equation is solved using some QP solver (like CVXOPT in python).**Let $\alpha^{*}_{1},\alpha^{*}_{2},...\alpha^{*}_{N}$ be the solutions of QP (Quadratic Programming) problem.**Stationary Condition:**1. $\frac{\partial}{\partial{w}}L(w,b,\xi,\alpha,\beta)=0,\;\;\;\;$we got Eq.(15)2. $\frac{\partial}{\partial{b}}L(w,b,\xi,\alpha,\beta)=0,\;\;\;\;$we got Eq.(16)3. $\frac{\partial}{\partial{\xi}}L(w,b,\xi,\alpha,\beta)=0,\;\;\;\;$we got Eq.(17)**Primal feasibility condition:**4. $y_{i}(w^{T}x_{i}+b)-(1-\xi_{i})\geq 0\;\;\;$5. $\xi_{i}\geq0$**Dual feasibility condition:**6. $\alpha_{i}\geq0$7. $\beta_{i}\geq0$**Complementary slackness condition:**8. $\alpha_{i}(y_{i}(w^TX_{i}+b)-(1-\xi_{i}))=0$9. $\beta{i}(\xi_{i})=(C-\alpha_{i})(\xi_{i})=0\;\;$ Using Eq.(16)Using KKT condition 8 & 9, we can imply that:- If $\alpha^{*}_{i}=0$ then $\xi_{i}=0$ which implies that $y_{i}(w^TX_{i}+b)\geq1$- If $0<\alpha^{*}_{i}<C$ then $\xi_{i}=0$ which implies that $y_{i}(w^TX_{i}+b)=1$ (**$x_{i}$ is Unbounded Support Vector**)- If $\alpha^{*}_{i}=C$ then $\xi_{i}\geq0$ which implies that $y_{i}(w^TX_{i}+b)=1-\xi_{i}$>- For $0\leq\xi_{i}<1$ $x_{i}$ is correctly classified and lies inside the margin i.e $0<y_{i}(w^TX_{i}+b)\leq1$ (**$x_{i}$ is Bounded Support Vector**)>- For $\xi_{i}\geq1$, $x_{i}$ is misclassified i.e. $y_{i}(w^TX_{i}+b)\leq0$$$\boxed{w=\sum_{i=1}^{S}\alpha^{*}_{i}y_{i}x_{i}}\;\;(19)$$ For $0<\alpha^{*}_{i}<C$, $y_{i}(w^Tx_{i}+b)-1=0\;\;\\y_{i}(w^Tx_{i}+b)=1\;\;\text{(Multiplying both sides by y)}\\y_{i}^{2}(w^Tx_{i}+b)=y_{i}\;\;y\epsilon\{-1,1\}\\w^Tx_{i}+b=y_{i}\\b=y_{i}-w^Tx_{i}\;\;\text{(Averaging over found support vectors S)}$$$\boxed{b=\frac{1}{S}\sum_{i=1}^{S}(y_{i}-w^{T}x_{i})}\;\;(20)$$ Hypothesis FunctionThe SVM uses the same hypothesis function as logistic regression. The class of an unknown example $x^{*}$ is given by :$$h(x_{i})=\text{sign}(w.x^{*}+b)$$**When using the dual formulation, it is computed using only the support vectors:**$$\boxed{h(x_{i})=\text{sign}(\sum_{i=1}^{S}\alpha_{i}y_{i}(x_{i}.x^{*})+b)}\;\;(21)$$ Kernel TrickThe idea is that our data, which isn't linearly separable in our 'n' dimensional space **may be linearly separable in a higher dimensional space.** To reach the solution, we sovle the following:$$\boxed{\max_{\alpha} L(w,b,\alpha)=\sum_{i=1}^{N}\alpha_{i}-\frac{1}{2}\sum_{i=1}^{N}\sum_{j=1}^{N}(\alpha_{i}\alpha_{j}y_{i}y_{j}\boxed{x_{i}^{T}x_{j}})\\\text{s.t. }0\leq\alpha_{i}\leq C\;\;\forall{i}\\\text{and }\sum_{i=1}^{N}\alpha_{i}y_{i}=0\;\;\forall{i}}$$$XX^{T}??$$$XX^{T}=\begin{bmatrix}(x_1,x_1)&(x_1,x_2)&...&(x_1,x_m)\\(x_2,x_1)&(x_2,x_2)&...&(x_2,x_m)\\.&.&...&.\\.&.&...&.\\.&.&...&.\\(x_m,x_1)&(x_m,x_2)&...&(x_m,x_m)\\\end{bmatrix}$$ The Great Kernel Trick- Replace the dot product $(x_i,x_j)$ with a similarity (Kernel) function $k(x_i,x_j)$.- Replace $XX^{T}$ with $K$ (Gram Matrix)$$K[i][j]=k(x_i,x_j)$$$$K=\begin{bmatrix}k(x_1,x_1)&k(x_1,x_2)&...&k(x_1,x_m)\\k(x_2,x_1)&k(x_2,x_2)&...&k(x_2,x_m)\\.&.&...&.\\.&.&...&.\\.&.&...&.\\k(x_m,x_1)&k(x_m,x_2)&...&k(x_m,x_m)\\\end{bmatrix}$$ Popular Kernels- Linear Kernel $$k(x_i,x_j)=x_i^Tx_j$$- Polynomial Kernel$$k(x_i,x_j)=(1+x_i^Tx_j)^{d}$$- Radial Basis (RBF) or Gaussian Kernel$$k(x_i,x_j)=exp(\frac{-1}{2\sigma^2}||x_i-x_j||^2)$$***Kernels (like RBF) maps our data into higher dimensional space, where its becomes easy to separate the data using optimal hyperplane.***
###Code
class Kernels:
degree=3
sigma=0.1
@classmethod
def linear(cls,x1,x2):
k=np.dot(x1,x2)
return k
@classmethod
def polynomial(cls,x1,x2):
k=np.power((1+np.dot(x1,x2)),cls.degree)
return k
@classmethod
def gaussian(cls,x1,x2):
'''Radial basis function'''
k=np.exp(-np.power(np.linalg.norm(x1-x2),2)/(2*np.power(cls.sigma,2)))
return k
###Output
_____no_output_____
###Markdown
SVM Implementation using CVXOPT Solver DisclaimerThis SVM implementation using CVXOPT is only for educational purpose and will not help you win any MNIST competitions. The sole purpose of this implementation is to apply the mathematics explained above and understand the effect of various kernels. Rewriting the SVM Optimzation problem in CVXOPT formatSince we will solve the optimization problem using CVXOPT library in python, we will need to match the solver's API which, according to the documentation is of the form:$$\boxed{\min_{x} \frac{1}{2}x^{T}Px+q^{T}x\\s.t.\;\;Gx\leq h\\\;\;\;\;\;\;\;Ax=b}$$With api: >`cvxopt.solvers.qp(P, q[, G, h[, A, b[, solver[, initvals]]]])`Let H be a matrix such that $\boxed{H_{i,j}=\sum_{i=1}^{N}\sum_{j=1}^{N}y_{i}y_{j}(x_{i}x_{j})}$. Case 1: Hard Margin SVM>Eq.(9) becomes:>$$\begin{align*}&\max_{\alpha}\sum_{i=1}^{N}\alpha_{i}-\frac{1}{2}\alpha^{T}H\alpha\\&s.t.\;\;\alpha_{i}\geq0\\&\;\;\;\;\;\;\;\sum_{i=1}^{N}\alpha_{i}y_{i}=0\end{align*}$$>>We convert the sums into vector form and multiply both the objective and the constraint by -1 which turns this into a minimization problem and reverses the inequality :>$$\boxed{\min_{\alpha}\frac{1}{2}\alpha^{T}H\alpha-1^{T}\alpha\\\text{s.t.}\;\;-\alpha\leq0\\\;\;\;\;\;\;\;y^{T}\alpha=0}$$ Case 2: Soft Margin SVM>Eq.(18) becomes:>$$\begin{align*}&\max_{\alpha}\sum_{i=1}^{N}\alpha_{i}-\frac{1}{2}\alpha^{T}H\alpha\\&s.t.\;\;0\leq \alpha_{i}\leq C\\&\;\;\;\;\;\;\;\sum_{i=1}^{N}\alpha_{i}y_{i}=0\end{align*}$$>>We convert the sums into vector form and multiply both the objective and the constraint by -1 which turns this into a minimization problem and reverses the inequality :>$$\boxed{\min_{\alpha}\frac{1}{2}\alpha^{T}H\alpha-1^{T}\alpha\\\text{s.t.}\;\;-\alpha\leq 0\\\;\;\;\;\;\;\;\alpha\leq C\\\;\;\;\;\;\;\;y^{T}\alpha=0}$$We are now ready to convert our numpy arrays into the CVXOPT format, using the same notation as in the documentation this gives:- P $:=$ H (a matrix of size $m$ x $m$)- q $:=$ $\vec{-1}$ (a vector of size $m$ x $1$) - A $:=$ $y^{T}$ (the label vector of size $1$ x $m$)- b $:=$ 0 (a scalar)**For Hard Margin SVM :**- G $:=$ diag(-1) (a diagonal matrix of -1s of size $m$ x $m$)- h $:=$ $\vec{0}$ (a vector of zeros of size $m$ x $1$)- For m=2: $G=\begin{bmatrix}-1&0\\0&-1\\\end{bmatrix},h=\begin{bmatrix}0\\0\\\end{bmatrix}$**For Soft Margin SVM :**- G $:=$ vertical stack of diag(-1) (a diagonal matrix of -1s of size $m$ x $m$) and diag(1) (a diagonal matrix of 1s of size $m$ x $m$)- h $:=$ vertical stack of $\vec{0}$ (a vector of zeros of size $m$ x $1$) and a vector of ones multiplied by C (size $m$ x $1$)- For m=2: $G=\begin{bmatrix}-1&0\\0&-1\\1&0\\0&1\\\end{bmatrix},h=\begin{bmatrix}0\\0\\C\\C\\\end{bmatrix}$
###Code
class SVM:
def __init__(self,kernel=Kernels.linear,options={},C=None,maxIters=100,showProgress=False):
self.kernel=kernel
# Set kernel options
if options.get('degree',None) is not None:
Kernels.degree=options['degree']
if options.get('sigma',None) is not None:
Kernels.sigma=options['sigma']
self.C=C
self.maxIters=maxIters
self.showProgress=showProgress
if self.C is not None:
self.C=float(C)
if self.C==0:
self.C=None
def fit(self,X,y):
m,n=X.shape
X=X.astype(float)
y=y.astype(float).reshape((m,1))
# Gram Matrix
K=np.zeros((m,m))
for i in range(m):
for j in range(m):
K[i,j]=self.kernel(X[i],X[j])
H=np.multiply(np.outer(y,y),K)
P=cvxopt.matrix(H)
q=cvxopt.matrix(np.ones((m,1))*(-1))
# Equality Constraints
A=cvxopt.matrix(y.reshape((1,m)))
b=cvxopt.matrix(0.0)
# Inequality Constraints
# Hard Margin SVM
if self.C==None:
G=cvxopt.matrix(np.identity(m)*(-1))
h=cvxopt.matrix(np.zeros((m,1)))
# Soft Margin SVM
else:
G=cvxopt.matrix(np.row_stack((np.identity(m)*(-1),np.identity(m))))
h=cvxopt.matrix(np.row_stack((np.zeros((m,1)),np.ones((m,1))*self.C)))
# Solve QP Problem
cvxopt.solvers.options['show_progress']=self.showProgress
cvxopt.solvers.options['maxiters']=self.maxIters
solution=cvxopt.solvers.qp(P,q,G,h,A,b)
# Lagrange Mutltpliers
alphas=np.ravel(solution['x'])
# Support Vectors have non zero lagrange multipliers
sv=alphas>1e-7
self.alphas=alphas[sv].reshape((-1,1))
self.svX=X[sv]
self.svY=y[sv]
# Intercept
svIndices=np.arange(len(alphas))[sv]
indexGrid=np.meshgrid(svIndices,svIndices)
self.b=(1/len(self.alphas))*np.sum(self.svY-np.dot(K[indexGrid],self.alphas*self.svY))
# Weight Vector
if self.kernel==Kernels.linear:
self.w=np.dot(self.svX.T,self.alphas*self.svY)
else:
self.w=None
# NOTE: We are not calculating the weight vector for a polynomial or gaussian
# kernel, we map the INNER PRODUCT of X to a higher dimensional space,so instead we will
# use the weight vector directly in our hypothesis function which will give us an expression
# Eq.(21) containing the inner product of support vectors and an unknown sample which we will map
# to higher dimension using same kernel and classify the unknown based on the sign.
def predict(self,X):
if self.w is not None:
h=np.dot(X,self.w)+self.b
h=np.sign(h)
else:
h=np.zeros((len(X),1))
for i in range(len(X)):
WX=0
for alpha,x,y in zip(self.alphas,self.svX,self.svY):
WX+=alpha*y*self.kernel(X[i],x)
h[i]=np.sign(WX+self.b)
return h
###Output
_____no_output_____
###Markdown
Plotting Decision BoundaryA decision boundary is hypersurface that partitions the underlying vector into two sets or classes.We will only plot the dataset and boundary with 2-Dimensional design matrix. For a linear kernel, descision boundary is a line and the genral equation of Line : $$ ax+by=c $$$$ \theta_1x_1+\theta_2x_2=-\theta_0 $$ $$ x_2=(\frac{-1}{\theta_2})(\theta_0+\theta_1x_1)$$In case of a polynomial or gaussian kernel, the decision boundary is no more a straight line but complex hyperplane in higher dimension which is impossible to plot. So in order to plot decision boundary, we will make classification prediction over grid of values, and draw a contour plot.
###Code
def visualize(X,y,model=None,title=''):
# Find indices of Positive and Negative samples
pos=np.where(y==1)[0]
neg=np.where(y==-1)[0]
# Plot samples
sns.scatterplot(X[pos,0],X[pos,1],label='+ve')
ax=sns.scatterplot(X[neg,0],X[neg,1],label='-ve')
ax.set(xlabel='x1',ylabel='x2',title=title)
if model is not None:
w=model.w
b=model.b
# Plot decision boundary
if model.kernel==Kernels.linear:
xp=np.linspace(np.min(X[:,0]),np.max(X[:,0]),100)
yp=(-1/w[1])*(b+w[0]*xp)
sns.lineplot(xp,yp)
else:
# Making Classification prediction over grid of values
x1=np.linspace(np.min(X[:,0]),np.max(X[:,0]),100)
x2=np.linspace(np.min(X[:,1]),np.max(X[:,1]),100)
[x1,x2]=meshgrid=np.meshgrid(x1,x2)
values=np.zeros(x1.shape)
for i in range(x1.shape[1]):
xp=np.column_stack((x1[:,i],x2[:,i]))
values[:,i]=svm.predict(xp).reshape(-1)
ax.contour(x1,x2,values,levels=[0])
return ax
###Output
_____no_output_____
###Markdown
1 Support Vector MachinesIn the first half of the exercise, we will be using SVMs with various examples 2D datasets. And in the next half of the exercise, we will be using SVM to build a spam classifier. 1.1 Example Dataset 1We will begin with a 2D example dataset which can be separated by a linear boundary.
###Code
# Loading Mat file
mat=loadmat('./ex6data1.mat')
print(*mat.keys(),sep='\n')
X=mat['X']
y=mat['y'].astype(int)
y[y==0]=-1
data=pd.DataFrame({
'x1':X[:,0],
'x2':X[:,1],
'y':y.reshape(-1)
})
print('TRAINING DATASET SHAPE : {0} X {1}'.format(*data.shape))
visualize(X,y)
data.sample(5)
###Output
__header__
__version__
__globals__
X
y
TRAINING DATASET SHAPE : 51 X 3
###Markdown
As we can notice from the above image, that there is a positive outlier on the far left which will affect the SVM decision boundary. We will try using different values of the C parameter with SVMs. - C parameter is a positive value that controls the penalty for misclassified training examples. - A large C parameter tells the SVM to try to classify all the examples correctly.- C plays a role similar to $\frac{1}{\lambda}$ ,where $\lambda$ is the regularization parameter.
###Code
C=100
svm=SVM(C=C)
svm.fit(X,y)
print(f'Out of {y.shape[0]} training samples, {len(svm.alphas)} are support vectors.')
visualize(X,y,svm,title=f'For C={C}');
###Output
Out of 51 training samples, 3 are support vectors.
###Markdown
When $C=1$, we can see that the SVM puts the decision boundary in the gap between the two datasets amd misclassifies the outlier. This is an example of Soft Margin.
###Code
C=1
svm=SVM(C=C)
svm.fit(X,y.reshape(-1))
print(f'Out of {y.shape[0]} training samples, {len(svm.alphas)} are support vectors.')
visualize(X,y,svm,title=f'For C={C}');
###Output
Out of 51 training samples, 12 are support vectors.
###Markdown
When $C=100$, we can notice that the SVM now classifies every single sample correctly, but has a decision boundary that does not appear to be a natural fit. This is an example of Hard Margin. 1.2 SVM with Gaussian KernelHere, we will be using SVMs to do non-linear classification. In particular, we will be using SVMs with gaussian kernels on datasets that are not linearly separable. 1.2.1 Gaussian Kernel$$k_{gaussian}(x_i,x_j)=exp(\frac{-1}{2\sigma^2}||x_i-x_j||^2)$$We can think of the gaussian kernel as a similarity function that measures the "distance" between a pair of examples. The Gaussian kernel is parameterized by a bandwidth parameter $\sigma$, which determines how fast the similarity metric decreases to 0 as the examples are further apart. 1.2.2 Example Dataset 2
###Code
# Loading Mat file
mat=loadmat('./ex6data2.mat')
print(*mat.keys(),sep='\n')
X=mat['X']
y=mat['y'].astype(int)
y[y==0]=-1
data=pd.DataFrame({
'x1':X[:,0],
'x2':X[:,1],
'y':y.reshape(-1)
})
print('TRAINING DATASET SHAPE : {0} X {1}'.format(*data.shape))
visualize(X,y)
data.sample(5)
###Output
__header__
__version__
__globals__
X
y
TRAINING DATASET SHAPE : 863 X 3
###Markdown
We can observe that there is no linear boundary that separates the positive and negative samples for this dataset. However by using a Gaussian kernel with the SVM, we will be able to learn a non linear decision boundary that can perform reasonably well for this dataset.
###Code
# Hard Margin SVM
C=0
svm=SVM(C=C,kernel=Kernels.gaussian)
svm.fit(X,y)
print(f'Out of {y.shape[0]} training samples, {len(svm.alphas)} are support vectors.')
visualize(X,y,svm,title=f'For C={C}, sigma:{Kernels.sigma}');
# Soft Margin SVM
C=1
svm=SVM(C=C,kernel=Kernels.gaussian)
svm.fit(X,y)
print(f'Out of {y.shape[0]} training samples, {len(svm.alphas)} are support vectors.')
visualize(X,y,svm,title=f'For C={C}, sigma:{Kernels.sigma}');
###Output
Out of 863 training samples, 187 are support vectors.
###Markdown
1.2.3 Example Dataset 3 In this part of the exercise, we are given a training and cross validation set. Our task is to determine the best $C$ and $\sigma$ parameter to use. For both $C$ and $\sigma$, we will try all the possible pairs for values (0.01, 0.03, 0.1, 0.3, 1, 3, 10, 30) making a total of 64 different models.
###Code
# Loading Mat file
mat=loadmat('./ex6data3.mat')
print(*mat.keys(),sep='\n')
X=mat['X']
y=mat['y'].astype(int)
y[y==0]=-1
# Validation set
Xval=mat['Xval']
yval=mat['yval'].astype(int)
yval[yval==0]=-1
data=pd.DataFrame({
'x1':X[:,0],
'x2':X[:,1],
'y':y.reshape(-1)
})
print('TRAINING DATASET SHAPE : {0} X {1}'.format(*data.shape))
visualize(X,y)
data.sample(5)
def calculateAccuracy(h,y):
'''Calculates the accuracy between the target and prediction'''
m=y.shape[0]
count=np.ones(h.shape)[h==y.reshape((m,1))].sum()
accuracy=(count/m)*100
return accuracy
values=[0.01, 0.03, 0.1, 0.3, 1, 3, 10, 30]
C=sigma=None
accuracy=0
for i in values:
for j in values:
options={'sigma':j}
svm=SVM(C=i,kernel=Kernels.gaussian,options=options)
svm.fit(X,y)
h=svm.predict(Xval)
temp=calculateAccuracy(h,yval)
if temp>accuracy:
accuracy=temp
C=i
sigma=j
print(f'For C={C:.2f} & sigma={sigma:.2f} ,\tValidation accuracy: {accuracy:.2f}%')
options={'sigma':sigma}
svm=SVM(C=C,kernel=Kernels.gaussian,options=options)
svm.fit(X,y)
h=svm.predict(X)
print(f'Out of {y.shape[0]} training samples, {len(svm.alphas)} are support vectors.')
visualize(X,y,svm,title=f'For C={C}, sigma:{sigma}');
###Output
Out of 211 training samples, 197 are support vectors.
###Markdown
2 Spam ClassificationMany email services today provide spam filters that are able to classify emails into spam and non-spam email with high accuracy. In this part of the exercise, we will use SVMs to build our own spam filter. We will be training a classifier to classify whether a given email $x$, is a spam $(y=1)$ or non-spam $(y=-1)$. in particular we need to convert each email into a feature vector $x\epsilon R^{n}$. 2.1 Preprocessing EmailsBefore starting on a machine learing task, it is usually insightful to take a look at examples from the dataset. Sample email shown above contains a URL, email address, numbers and dollar amounts. We should process these emails by normalizing these values, so that all the URLs, email address, amounts are treated the same. For example, URL can be replaced by 'httpaddr' to indicate that URL was present. This has the effect of letting the spam classifier make a classification based on ANY url present rather than whether a specific URL was present. Few of the normalizzation techniques are mentioned below :- **Lower-Casing** : This entire email should be converted into lowercase, so that the capitalization is ignored. (InDicate is treated as indicate)- **Stripping HTML** : All the html tags are removed from the emails.- **Normalizing URLs** : All the URLs are repalced by 'httpaddr'.- **Normalizing Email addresses** : All email addresses are replaced by 'emailaddr'.- **Normalizing Numbers** : All numbers are replaced by 'number'.- **Normalizing Dollars** : All dollar signs are repalced by 'dollar'.- **Word Stemming** : Words are reduced to their stemmed form. For example 'discounts', 'discount' and 'discounted' are all replaced with 'discount'. Sometimes the stemmer actually strips off the additional characters from the end.- **Removal of non words** : Non words and punctuation should be removed. All white spaces have been trimmed to single space character. 2.1.1 Vocabulary ListAfter preprocessing the emails, we have a list of words for each email. the next step is to choose which words we would to use in our classifier and which we want to leave out. For this exercise, we have choosen only the most frequently occuring words as our set words considered for feature space. We will use the vocabulary list provided to us in the course.**Note:** This vocabulary list was selected by choosing all words which occur at least a 100 times in the spam corpus, resulting in a list of 1899 words.
###Code
vocabulary=[]
with open('./vocabulary.txt') as f:
for word in f.readlines():
word=str(word).strip().replace('\t','')
word=re.search(r'\D+',word).group(0)
vocabulary.append(word)
from stemming.porter2 import stem
def processEmail(email,vocabulary):
'''Preprocesses the body of an email and returns a list of indices
of the words contained in the email'''
wordIndices=[]
# Stripping leading and trailing white spaces
email=email.strip()
# Removing newlines
email=email.replace('\n',' ')
# Lower-casing
email=email.lower()
# Replace all numbers with 'number'
email=re.sub(r'\d+','number',email)
# Replace all URLs with 'httpaddr'
email=re.sub(r"[\w!#$%&'*+-/=?^_`{|}~]+@[\w]+\.[\w]+",'httpaddr',email)
# Replace all $ sign with 'dollar'
email=re.sub(r'\$+','dollar',email)
# Remove punctuation
email=re.sub(r'[@\$/#\.\-:&\*\+=\[\]\?!\(\)\{\},''">_<;%]+','',email)
# Word Stemming
email=stem(email)
# Lookup for word in vocabulary
for word in email.split():
try:
index=vocabulary.index(word)
wordIndices.append(index)
except:
pass
print(email,end='\n\n')
print(','.join([str(a) for a in wordIndices]))
return wordIndices
###Output
_____no_output_____
###Markdown
**Sample Email 1**
###Code
with open('./spamSample1.txt') as f:
sample=f.read()
print(sample)
###Output
Do You Want To Make $1000 Or More Per Week?
If you are a motivated and qualified individual - I
will personally demonstrate to you a system that will
make you $1,000 per week or more! This is NOT mlm.
Call our 24 hour pre-recorded number to get the
details.
000-456-789
I need people who want to make serious money. Make
the call and get the facts.
Invest 2 minutes in yourself now!
000-456-789
Looking forward to your call and I will introduce you
to people like yourself who
are currently making $10,000 plus per week!
000-456-789
###Markdown
**Processed Email 1 & Word Indices**
###Code
indices=processEmail(sample,vocabulary)
###Output
do you want to make dollarnumber or more per week if you are a motivated and qualified individual i will personally demonstrate to you a system that will make you dollarnumbernumber per week or more this is not mlm call our number hour prerecorded number to get the details numbernumbernumber i need people who want to make serious money make the call and get the facts invest number minutes in yourself now numbernumbernumber looking forward to your call and i will introduce you to people like yourself who are currently making dollarnumbernumber plus per week numbernumbernumb
470,1892,1808,1698,996,1181,1063,1230,1826,809,1892,73,1851,1698,1892,1630,1664,1851,996,1892,1230,1826,1181,1063,876,1112,233,1190,1119,791,1119,1698,707,1665,1092,1843,1808,1698,996,996,1665,233,73,707,1665,868,1119,824,1895,1116,675,1698,1894,233,73,1851,1892,1698,955,1895,1843,1230,1826
###Markdown
**Sample Email 2**
###Code
with open('./spamSample2.txt') as f:
sample=f.read()
print(sample)
###Output
Anyone knows how much it costs to host a web portal ?
Well, it depends on how many visitors youre expecting. This can be
anywhere from less than 10 bucks a month to a couple of $100. You
should checkout http://www.rackspace.com/ or perhaps Amazon EC2 if
youre running something big..
To unsubscribe yourself from this mailing list, send an email to:
[email protected]
###Markdown
**Processed Email 2 & Word Indices**
###Code
indices=processEmail(sample,vocabulary)
###Output
anyone knows how much it costs to host a web portal well it depends on how many visitors youre expecting this can be anywhere from less than number bucks a month to a couple of dollarnumber you should checkout httpwwwrackspacecom or perhaps amazon ecnumber if youre running something big to unsubscribe yourself from this mailing list send an email to httpaddr
793,1076,882,1698,789,1821,1830,882,1170,793,237,161,687,944,1662,1119,1061,1698,1161,1892,1509,1181,809,180,1698,1895,687,960,1476,70,529,1698,798
###Markdown
2.2 Extracting Features from EmailsWe will now implement the feature extraction that converts each email into a vector in $R^{n}$. **We will be using $n=\$ words in voabulary list.** Specifically, teh feature space $x_{i}\epsilon\{0,1\}$ for an email corresponds to whether the $i^{th}$ word in the dictionary occurs in the email. That is $x_{i}=1$ if the $i^{th}$ word in vocabulary is present in email and $x_j=0$ if the $j^{th}$ word in vocabulary is not present in email.
###Code
def extractFeatures(wordIndices,vocabulary):
'''Creates a feature vector containing 0 and 1's out of word indices'''
features=np.zeros(len(vocabulary))
features[wordIndices]=1
return features
features=extractFeatures(indices,vocabulary)
pd.DataFrame(pd.Series(features).value_counts(),columns=['count'])
###Output
_____no_output_____
###Markdown
2.3 Training SVM for Spam Classification In this part of the exercise, we will train our SVM classifier on preprocessed emails processed using same steps we followed above.
###Code
mat=loadmat('./spamTrain.mat')
m,n=mat['X'].shape
mat['y']=mat['y'].astype(int)
mat['y'][mat['y']==0]=-1
data=pd.DataFrame()
for i in range(n):
data[f'x{i+1}']=mat['X'][:,i]
data['y']=mat['y']
valueCount=dict(data['y'].value_counts())
ratio=(valueCount[1]/m)*100
print('DATASET SHAPE : {0} X {1}'.format(*data.shape))
print(f'SPAM EMAIL RATIO : {ratio:.2f}%')
data.sample(5)
###Output
DATASET SHAPE : 4000 X 1900
SPAM EMAIL RATIO : 31.92%
###Markdown
**Train and validation set split**
###Code
# Shuffle before split
data=data.sample(frac=1).reset_index(drop=True)
# 70/30 Split
mTrain=int((m*70)/100)
X=data.iloc[:mTrain,:-1].values
y=data.iloc[:mTrain,-1].values
Xval=data.iloc[mTrain:,:-1].values
yval=data.iloc[mTrain:,-1].values
###Output
_____no_output_____
###Markdown
**Training and Cross Validating**
###Code
values=[0.01, 0.03, 0.1,0.3]
C=None
accuracy=0
models={}
for i in values:
svm=SVM(C=i,kernel=Kernels.linear)
# Training
svm.fit(X,y)
models[i]=svm
# Cross Validating
h=svm.predict(Xval)
temp=calculateAccuracy(h,yval)
if temp>accuracy:
accuracy=temp
C=i
print(f'For C={C:.2f} ,\tValidation accuracy: {temp:.2f}%')
###Output
For C=0.01 , Validation accuracy: 97.00%
For C=0.03 , Validation accuracy: 97.75%
###Markdown
**Test Accuracy**
###Code
mat=loadmat('./spamTest.mat')
Xtest=mat['Xtest']
ytest=mat['ytest'].astype(int)
ytest[ytest==0]=-1
svm=models[C]
h=svm.predict(Xtest)
accuracy=calculateAccuracy(h,ytest)
print(f'For C={C:.2f} ,\tTest accuracy: {accuracy:.2f}%')
###Output
For C=0.03 , Test accuracy: 99.20%
###Markdown
2.4 Top Predictors for SpamTo better understand how the spam classifier works, **we can inspect the parameter to see which words the classifier thinks are the most predictive of spam. In this exercise we will find the parameters with the largest positive values (SVM weights) in the classifier and display the corresponding word**. Thus, if an email contains words such as "guarantee", "remove", "click", and "free", it is likely to be classified as spam.
###Code
df=pd.DataFrame({'Word':vocabulary,'Weight':svm.w.reshape(-1)})
df=df.sort_values(by='Weight',ascending=False).reset_index(drop=True)
df
###Output
_____no_output_____
|
George_Demo_PolynomialRegression.ipynb
|
###Markdown
Learning a Predictor for Maximum Daytime Temperature with Polynomial Regression
###Code
# import necessary libraries
import pandas as pd # for data input/output and processing
import numpy as np # for matrix calculations
from sklearn.linear_model import LinearRegression # An already implemented version of linear regression
from sklearn.preprocessing import PolynomialFeatures # To transform our features to the corresponding polynomial version
from sklearn.metrics import mean_squared_error # a way to evaluate how erroneous our model is
import matplotlib.pyplot as plt # For visualizations
plt.rcParams.update({'font.size': 20})
###Output
_____no_output_____
###Markdown
The Data We consider datapoints that represent individual days. Each datapoint is characterized by the maximum daytime temperature as label $y$. Our goal is to learn a hypothesis $h$ that delivers an accurate prediction $\hat{y} = h(x)$ for the label $y$. The hypothesis is applied to some feature $x$ of the datapoint (day). We will consider different choices for the feature below. Polynomial regression learns a hypothesis that is a polynomial wight maximum degree $r$, $$h(x) = w_{1} + w_{2} x + w_{3} x^{2} + \ldots + w_{r+1} x^{r}.$$ The coefficients (or parameters) $w_{1},\ldots,w_{r+1}$ are learnt by minimizing the average squared error $$(1/m) \sum_{i=1}^{m} \big( y^{(i)} - h\big( x^{(i)} \big) \big)^2,$$incurred for a training set $\big(x^{(1)},y^{(1)}\big),\ldots,\big(x^{(m)},y^{(m)}\big)$. We construct the training set $\big(x^{(1)},y^{(1)}\big),\ldots,\big(x^{(m)},y^{(m)}\big)$ using weather recordings from the Finnish meteorological institute (FMI). These weather recordings are stored in the file "FMIData.csv". The code snippet below reads in the weather recordings from the file "FMIData.csv" and stores them in the `pandas dataframe` with name `df`.
###Code
# First load the dataset into a pandas DataFrame
df = pd.read_csv("FMIData.csv")
print("number m of data points =", len(df))
df.head(2) # print the first 2 rows
###Output
number m of data points = 60
###Markdown
Each row in the above dataframe corresponds to a specific datapoint (day). The value in the column "Maximum temperature" is used as the label $y$ of the datapoint. All other columns can be potentially used as the feature $x$ of the datapoint. Let use consider two different choices for the feature of a datapoint. The first choice for the feature $x$ is the value in the column `day`. The second choice for the feature $x$ is the value in the column "Minimum temperature". The following code snippet selects only those columns of the dataframe.
###Code
feature_columns = ["d", "Minimum temperature (degC)"]
label_column = ["Maximum temperature (degC)"]
# Keep only relevant columns
df = df[feature_columns+label_column]
df.head(2)
df.columns = ['d', 'min_temp', 'max_temp']
df.head(2)
# First step: Drop NaN (Not A Number) values
df.dropna(inplace=True)
len(df)
###Output
_____no_output_____
###Markdown
To explore the relationship between different features and the label of datapoints it is often helpful to generate a scatterplot. The code snippet below generates two scatterplots. - The left scatterplot depicts the datapoints in `df` as dots in a plane with horizontal axis representing the values of the column `day` (which is one potential feature of the datapoint) and the vertical axis representing the values of the column "Maximum temperature" (which is the label of the datapoint). - The right scatterplot depicts the datapoints in `df` as dots in a plane with horizontal axis representing the values of the column `Minimum temperature` (which is one potential feature of the datapoint) and the vertical axis representing the values of the column "Maximum temperature" (which is the label of the datapoint).
###Code
# def plot_model(X, y,
fig=plt.figure(figsize=(16, 7))
fig.suptitle("Relation between the features and the maximum temperature")
plt.subplot(1, 2, 1)
plt.scatter(df.d, df.max_temp, c="blue")
plt.xlabel("Day")
plt.ylabel("Max Temperature")
plt.subplot(1, 2, 2)
plt.scatter(df.min_temp, df.max_temp, c="red")
plt.xlabel("Minimum Temperature")
plt.ylabel("Max Temperature")
fig.tight_layout() # Separate the two subplots
plt.show()
###Output
_____no_output_____
###Markdown
The `d` (day) column seems to have a very noisy relationship with the target. On the other hand, the `min_temp` column seems to have a clearly linear relationship with the target and so we consider it important/informative.For this reason we are going to completely ignore the `d` column and move on with the `min_temp` as our only feature. Using 1 feature also allows for better visualizations in a 2d space (e.g. if we had two features, then we would need 3 axes in total, and 3d plots are not as intuitive as 2d plots. If we had 3 features, then visualization would be impossible).
###Code
df.drop(columns=['d'], inplace=True)
df.head(2)
# Let's separate features from labels
labels = df.max_temp.values # .values convert to a numpy array
features = df.drop(columns=['max_temp']).values
###Output
_____no_output_____
###Markdown
Linear Regression is Polynomial Regression with Degree 1 Loss FunctionLinear regression tries to minimize the Mean Squared Error (MSE) loss function, to get the best possible predictor function $h$. MSE is given by: $$MSE = \frac{1}{m} \sum_{i=1}^m (y^{(i)} - h(x^{(i)}))^2$$where $h(x^{(i)})$ is the predicted label for the $i$-th datapoint. In turn, $x$ is a vector of the features that we want to use in order to get a prediction as close to the ground truth $y^{(i)}$ as possible. Hypothesis SpaceFor degree $r=1$, polynomial regression reduces to linear regression. Linear regression learns a linear hypothesis $$h(x) = w_1 + w_2 x. $$We learn the parameters $w_{1}, w_{2}$ by minimizing the average squared error or MSE. The code snippet below uses the `scikit-learn` class `LinearRegression()` to implement linear regression.
###Code
# Create a linear regression model
lr = LinearRegression()
# Fit the model to our data in order to get the most suitable parameters
lr = lr.fit(features, labels)
# Calculate the mean square error on the training set
predictions = lr.predict(features)
mean_squared_error(predictions, labels) # ideally this would be 0 which would mean that the predictions are very close to the ground truth
plt.figure(figsize=(12,7))
# How good does the model fit our data?
plt.scatter(features, labels, c="blue", label="Datapoints")
# an increasing vector in the [min, max] range of the features
X_line = np.linspace(features.min(), features.max(), len(features)).reshape(-1, 1) # needs to be 2d and so we reshape
predictions = lr.predict(X_line)
plt.plot(X_line, predictions, c="red", label="optimal linear hypothesis")
plt.xlabel("Minimum Temperature")
plt.ylabel("Maximum Temperature")
plt.title("Linear Prediction of max tmp")
plt.legend()
plt.show()
###Output
_____no_output_____
###Markdown
Polynomial RegressionThe above predictor function is good for relations $x \rightarrow y$ that exhibit a strictly linear relation. In the figure bellow though, we can see that the relation is not always linear:In such cases, we can increase the degree of the linear function and also include terms raised to some power $r$, for $r>1$.Getting back to our example, we could now transform the predictor function to:$$\hat{h}(x) = w_0 + w_1 x + w_1 x^2 + ... + w_{r+1} x^r$$To find the best value of $r$ we usually have to search through a lot of values, as follows:
###Code
def get_poly_predictor(features, labels, degree):
poly_features = PolynomialFeatures(degree=degree, include_bias=False)
# transform features so as to include their polynomial version.
# E.g. feas_new will now also include x^2, ..., x^{degree}
feas_new = poly_features.fit_transform(features)
lr = LinearRegression()
lr.fit(feas_new, labels)
mse = mean_squared_error(lr.predict(feas_new), labels)
return mse
# Try out different degrees and print the corresponding MSEs
for r in range(2, 18, 2):
mse = get_poly_predictor(features, labels, degree=r)
print(f"Degree={r} -> training error={mse}")
###Output
Degree=2 -> training error=0.7609297067884181
Degree=4 -> training error=0.7528852230641694
Degree=6 -> training error=0.6933588875501544
Degree=8 -> training error=0.6861387622564956
Degree=10 -> training error=0.6283257414847673
Degree=12 -> training error=0.6277316086004313
Degree=14 -> training error=0.5748724705215218
Degree=16 -> training error=0.5499997334536294
###Markdown
The training errors decrease with increasing polyonmial degree. Does that mean that we should use very large degreees for polynomial regression? Let us inspect the learnt polynomials for a few specific choices for the polynomial degree.
###Code
def poly_visualize(
features,
labels,
degrees=[2, 4, 8, 18],
colors=['blue', 'green', 'cyan', 'purple'],
n_wrong_models=0,
):
fig = plt.figure(figsize=(20, 15))
wrong_poly_colors = ['gold', 'teal', 'tomato', 'firebrick', 'orchid']
for i, r in enumerate(degrees):
# ===================================================================
# Fit model
# ===================================================================
poly_features = PolynomialFeatures(degree=r, include_bias=False)
feas_new = poly_features.fit_transform(features)
lr = LinearRegression()
lr.fit(feas_new, labels)
# ===================================================================
# Fit some models on wrong data to see how the polynomial changes
# ===================================================================
polys_perturbed = []
for j in range(n_wrong_models):
feas_perturbed = np.random.permutation(feas_new)
polys_perturbed.append(LinearRegression().fit(feas_perturbed, labels))
# ===================================================================
# Scatter plot
# ===================================================================
plt.subplot(2, 2, i+1)
# How good does the model fit our data?
plt.scatter(features, labels, c="blue", label="Datapoints")
# an increasing vector in the [min, max] range of the features
X_line = np.linspace(features.min(), features.max(), 200).reshape(-1, 1) # needs to be 2d and so we reshape
predictions = lr.predict(poly_features.transform(X_line))
plt.plot(X_line, predictions, c="red", label="Learnt Hypothesis", linewidth=7.0)
for j in range(n_wrong_models):
preds_perturbed = polys_perturbed[j].predict(poly_features.transform(X_line))
color = wrong_poly_colors[j%len(wrong_poly_colors)]
plt.plot(X_line, preds_perturbed, c=color, label=f"Hypothesis {j+2}")
plt.xlabel("Minimum Temperature")
plt.ylabel("Maximum Temperature")
mse = mean_squared_error(lr.predict(feas_new), labels)
plt.title(f"Degree {r} - MSE={round(mse, 2)}")
plt.legend()
plt.show()
poly_visualize(features, labels, degrees=[2, 6], colors=["blue", "green"], n_wrong_models=4)
poly_visualize(features, labels, degrees=[12, 18], colors=["cyan", "purple"], n_wrong_models=4)
###Output
_____no_output_____
|
examples/e2e-home-appliance-status-monitoring/notebook/EnergyDisaggregationEDA.ipynb
|
###Markdown
LicenseCopyright 2019 Google LLCLicensed under the Apache License, Version 2.0 (the "License"); you may not use this file except in compliance with the License. You may obtain a copy of the License at . http://www.apache.org/licenses/LICENSE-2.0Unless required by applicable law or agreed to in writing, software distributed under the License is distributed on an "AS IS" BASIS, WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied. See the License for the specific language governing permissions and limitations under the License.
###Code
# @title Upload files (skip this if this is run locally)
# Use this cell to update the following files
# 1. requirements.txt
from google.colab import files
uploaded = files.upload()
# @title Install missing packages
# run this cell to install packages if some are missing
!pip install -r requirements.txt
# @title Import libraries
from datetime import datetime
import matplotlib.pyplot as plt
import numpy as np
import os
import pandas as pd
import re
import gcsfs
import sklearn.metrics
%matplotlib inline
###Output
_____no_output_____
###Markdown
Data inspection Utility for loading and transforming raw data
###Code
def load_main_energy_data(path):
"""Load main energy data from the specified file.
Load main energy data from the specified file.
Args:
path - string. Path to the data file.
Returns:
pd.DataFrame - Main energy data in the household.
Raises:
ValueError. Raised when the specified file does not exist.
"""
if not os.path.exists(path):
raise ValueError('File {} does not exist.'.format(path))
with open(path, 'r') as f:
data = pd.read_csv(f,
delimiter=' ',
header=None,
names=['time',
'main_watts',
'main_va',
'main_RMS'])
data.time = data.time.apply(lambda x: datetime.fromtimestamp(x))
data.set_index('time', drop=True, inplace=True)
data.index = data.index.floor('S')
return data
def load_appliance_energy_data(path, appliance_name):
"""Load appliance energy data from file.
Load energy data from the specified file.
Args:
path - string. Path to the data file.
appliance_name - string. Name of the appliance.
Returns:
pd.DataFrame. A 2-column dataframe.
The 1st column is timestamp in UTC, and the 2nd is energy in
Raises:
ValueError. Raised when the specified file does not exist.
"""
if not os.path.exists(path):
raise ValueError('File {} does not exist.'.format(path))
with open(path, 'r') as f:
df = pd.read_csv(f,
delimiter=' ',
header=None,
names=['time', appliance_name])
df.time = df.time.apply(lambda x: datetime.fromtimestamp(x))
df.set_index('time', drop=True, inplace=True)
df.index = df.index.floor('S')
return df
def load_energy_data(data_dir, house_id, load_main=False):
"""Load all appliances energy data.
Load all appliances energy data collected in a specified household.
Args:
data_dir - string. Path to the directory of data.
house_id - int. Household id.
load_main - bool. Whether to load mains.dat.
Returns:
pd.DataFrame - Energy data in the household.
Raises:
ValueError. Raised when the specified directory or household does not exist.
"""
house_data_dir = os.path.join(data_dir, 'house_{}'.format(house_id))
if not os.path.exists(house_data_dir):
raise ValueError('{} does not exist.'.format(house_data_dir))
if load_main:
main_file = os.path.join(house_data_dir, 'mains.dat')
data = load_main_energy_data(main_file)
label_file = os.path.join(house_data_dir, 'labels.dat')
with open(label_file, 'r') as f:
labels = pd.read_csv(f,
delimiter=' ',
header=None,
index_col=0,
names=['appliance'])
appliance_files = filter(lambda x: re.match(r'channel_\d+\.dat', x),
os.listdir(house_data_dir))
ll = [data,] if load_main else []
for f in appliance_files:
appliance_id = int(f.split('.')[0].split('_')[1])
appliance_name = labels.loc[appliance_id, 'appliance']
ll.append(load_appliance_energy_data(os.path.join(house_data_dir, f),
appliance_name))
if load_main:
data = pd.concat(ll, axis=1, join_axes=[data.index])
else:
data = pd.concat(ll, axis=1)
return data
###Output
_____no_output_____
###Markdown
Data Loading
###Code
GOOGLE_CLOUD_PROJECT = 'your-google-project-id' #@param
GOOGLE_APPLICATION_CREDENTIALS = 'e2e_demo_credential.json' #@param
os.environ['GOOGLE_APPLICATION_CREDENTIALS'] = GOOGLE_APPLICATION_CREDENTIALS
os.environ['GOOGLE_CLOUD_PROJECT'] = GOOGLE_CLOUD_PROJECT
# If raw data is used, please make sure raw_data_dir is correctly set
use_raw = False #@param
selected_hid = 2 #@param
raw_data_dir = 'ukdale data directory' #@param
selected_house_dir = os.path.join(raw_data_dir, 'house_{}'.format(selected_hid))
%%time
if not use_raw:
print("Download processed sample file for house 2 from GCS")
fs = gcsfs.GCSFileSystem(project=os.environ['GOOGLE_CLOUD_PROJECT'])
with fs.open('gs://gcp_blog/e2e_demo/processed_h2_appliance.csv') as f:
energy_data = pd.read_csv(f,
index_col=0,
parse_dates=True)
else:
# load energy data from raw downloaded ukdale data directory
energy_data = load_energy_data(raw_data_dir, selected_hid)
energy_data.head()
###Output
_____no_output_____
###Markdown
EDA
###Code
print(energy_data.shape)
energy_data.describe()
energy_data.index.min(), energy_data.index.max()
cutoff_st = '2013-06-01 00:00:00'
cutoff_et = '2013-09-30 23:59:59'
energy_data = energy_data.loc[cutoff_st:cutoff_et]
print('{}, {}'.format(energy_data.index.min(), energy_data.index.max()))
energy_data.describe()
energy_data = energy_data.fillna(method='ffill').fillna(method='bfill')
energy_data = energy_data.asfreq(freq='6S', method='ffill')
print(energy_data.shape)
energy_data.describe()
energy_data.head()
energy_data = energy_data.astype(int)
energy_data.info()
energy_data.describe()
if 'aggregate' in energy_data.columns:
energy_data = energy_data.drop('aggregate', axis=1)
energy_data['gross'] = energy_data.sum(axis=1)
energy_data.describe()
appliance_cols = ['running_machine', 'washing_machine', 'dish_washer',
'microwave', 'toaster', 'kettle', 'rice_cooker', 'cooker']
print(appliance_cols)
energy_data['app_sum'] = energy_data[appliance_cols].sum(axis=1)
energy_data.describe()
st = '2013-07-04 00:00:00'
et = '2013-07-05 00:00:00'
sub_df = energy_data.loc[st:et]
print(sub_df.shape)
fig, ax = plt.subplots(1, 1, figsize=(15, 8))
ax = sub_df[['gross', 'app_sum']].plot(ax=ax)
ax.grid(True)
ax.set_title('House {}'.format(selected_hid))
ax.set_ylabel('Power consumption in watts')
nrow = int(np.ceil(np.sqrt(len(appliance_cols))))
ncol = int(np.ceil(1.0 * len(appliance_cols) / nrow))
fig, axes = plt.subplots(nrow, ncol, figsize=(5*ncol, 3*nrow))
axes[-1, -1].axis('off')
for i, app in enumerate(appliance_cols):
row_ix = i // 3
col_ix = i % 3
ax = axes[row_ix][col_ix]
lb = energy_data[app].std()
ub = energy_data[app].max() - lb
energy_data[app + '_on'] = energy_data[app].apply(
lambda x: 1 if x > lb else 0)
energy_data[app][(energy_data[app] > lb) &
(energy_data[app] < ub)].plot.hist(bins=20, ax=ax)
ax.set_title(app)
ax.grid(True)
plt.tight_layout()
energy_data.mean(axis=0)
train_st = '2013-06-01 00:00:00'
train_et = '2013-07-31 23:59:59'
train_data = energy_data.loc[train_st:train_et]
print(train_data.shape)
valid_st = '2013-08-01 00:00:00'
valid_et = '2013-08-31 23:59:59'
valid_data = energy_data.loc[valid_st:valid_et]
print(valid_data.shape)
test_st = '2013-09-01 00:00:00'
test_et = '2013-09-30 23:59:59'
test_data = energy_data.loc[test_st:test_et]
print(test_data.shape)
train_file = os.path.join(raw_data_dir, 'house_{}/train.csv'.format(selected_hid))
valid_file = os.path.join(raw_data_dir, 'house_{}/valid.csv'.format(selected_hid))
test_file = os.path.join(raw_data_dir, 'house_{}/test.csv'.format(selected_hid))
with open(train_file, 'w') as f:
train_data.to_csv(f)
print('train_data saved.')
with open(valid_file, 'w') as f:
valid_data.to_csv(f)
print('valid_data saved.')
with open(test_file, 'w') as f:
test_data.to_csv(f)
print('test_data saved.')
###Output
train_data saved.
valid_data saved.
test_data saved.
###Markdown
Splitted Data inspection
###Code
train_file = os.path.join(raw_data_dir, 'house_{}/train.csv'.format(selected_hid))
valid_file = os.path.join(raw_data_dir, 'house_{}/valid.csv'.format(selected_hid))
test_file = os.path.join(raw_data_dir, 'house_{}/test.csv'.format(selected_hid))
# @title Peek at the input file
with open(train_file, 'r') as f:
train_data = pd.read_csv(f, index_col=0)
print(pd.Series(train_data.columns))
train_data.head()
appliance_cols = [x for x in train_data.columns if '_on' in x]
print(train_data[appliance_cols].mean())
with open(test_file, 'r') as f:
test_data = pd.read_csv(f, index_col=0)
print(test_data.shape)
print(test_data[appliance_cols].mean())
ss = ['2013-09-{0:02d} 00:00:00'.format(i+1) for i in range(30)]
ee = ['2013-09-{0:02d} 23:59:59'.format(i+1) for i in range(30)]
fig, axes = plt.subplots(30, 1, figsize=(15, 120))
for i, (s, e) in enumerate(zip(ss, ee)):
test_data.loc[s:e].gross.plot(ax=axes[i])
axes[i].set
plt.tight_layout()
###Output
_____no_output_____
|
case_studies/parameter_estimation_telostylinus.ipynb
|
###Markdown
Parameter estimation for case study 2 (*Telostylinus angusticollis*)We have estimated model parameters using least squares fitting. Typically, such problems are solved with utilities such as [`scipy.optimize.curve_fit`](https://docs.scipy.org/doc/scipy/reference/generated/scipy.optimize.curve_fit.html). However, our model function is non-analytical, due to its stochasticity. Thus, analytical curve fitting methods are not possible to use, and we have instead opted for a simple iterative search of the parameter space.We encourage examining other methods of exploring the parameter space. This could potentially be a conventional space search algorithm, or an evolutionary algorithm framework such as [DEAP](https://deap.readthedocs.io/en/master/). If you develop a more efficient/optimal method, we welcome you to submit a pull request on GitHub. **Note:** The parameter values (for females and males) used in figure 4 were obtained during an earlier least squares fitting. In restructuring the code for publication we have also improved the fitting procedure and obtained new and better values. However, these are not used in the figure in order to produce results matching those in the published paper (which could not be updated by the time we obtained the better fit). Estimating $\alpha$ and $\kappa$
###Code
# Simulation parameters
individual_count = 1000
repetition_count = 100
t_m_f = 326 # Max t for curve fitting (last x data point = 325)
t_m_m = 291 # Max t for curve fitting (last x data point = 290)
# FEMALES
x_captivity_f = np.genfromtxt(f'{ROOT_DIR}/data/kawasaki_2008/captivity_females_x.txt')
y_captivity_f = np.genfromtxt(f'{ROOT_DIR}/data/kawasaki_2008/captivity_females_y.txt')
xdata_f = np.round(x_captivity_f).astype('int64') # In order to use for indexing
ydata_f = y_captivity_f * individual_count
# MALES
x_captivity_m = np.genfromtxt(f'{ROOT_DIR}/data/kawasaki_2008/captivity_males_x.txt')
y_captivity_m = np.genfromtxt(f'{ROOT_DIR}/data/kawasaki_2008/captivity_males_y.txt')
xdata_m = np.round(x_captivity_m).astype('int64') # In order to use for indexing
ydata_m = y_captivity_m * individual_count
%%time
# FEMALES, least squares
n = len(xdata_f)
fit = []
for alpha in np.arange(0.0028, 0.0030, 0.0001):
for kappa in np.arange(0.0084, 0.0085, 0.0001):
hazard_rate_params = dict(alpha=alpha, kappa=kappa, population=MUT_CAPTIVITY)
population_survivorship = run_cohort_simulation(
repetition_count,
individual_count,
hazard_rate_params,
t_m_f,
)
mean = np.mean(population_survivorship, axis=0)[xdata_f]
squares = [(mean[i] - ydata_f[i])**2 for i in range(n)]
fit.append((alpha, kappa, sum(squares)))
best_fits = sorted(fit, key=operator.itemgetter(2))
print('alpha, kappa, sum squares (females)')
print(*best_fits[0:10], sep='\n')
%%time
# MALES, least squares
n = len(xdata_m)
fit = []
for alpha in np.arange(0.00056, 0.00058, 0.00001):
for kappa in np.arange(0.0172, 0.0174, 0.0001):
hazard_rate_params = dict(alpha=alpha, kappa=kappa, population=MUT_CAPTIVITY)
population_survivorship = run_cohort_simulation(
repetition_count,
individual_count,
hazard_rate_params,
t_m_m,
)
mean = np.mean(population_survivorship, axis=0)[xdata_m]
squares = [(mean[i] - ydata_m[i])**2 for i in range(n)]
fit.append((alpha, kappa, sum(squares)))
best_fits = sorted(fit, key=operator.itemgetter(2))
print('alpha, kappa, sum squares (males)')
print(*best_fits[0:10], sep='\n')
###Output
alpha, kappa, sum squares (males)
(0.00057, 0.0173, 519.0083000000002)
(0.00057, 0.0172, 630.0872000000013)
(0.00056, 0.0173, 640.3186999999987)
(0.00058, 0.0172, 657.4858999999997)
(0.00058, 0.0173, 887.0013000000015)
(0.00056, 0.0172, 968.8162000000013)
CPU times: user 1min 18s, sys: 1min 23s, total: 2min 41s
Wall time: 3min
###Markdown
Estimating $\epsilon$ and $h_{wt}(t)$
###Code
# FEMALES
x_wild_f = np.genfromtxt(f'{ROOT_DIR}/data/kawasaki_2008/wild_females_x.txt')
y_wild_f = np.genfromtxt(f'{ROOT_DIR}/data/kawasaki_2008/wild_females_y.txt')
x_wild_f = np.round(x_wild_f).astype('int64') # In order to use for indexing
y_wild_f = y_wild_f * individual_count
x_wild_f = x_wild_f[:-2] # In order not to fit to the last two data points
y_wild_f = y_wild_f[:-2] # In order not to fit to the last two data points
# MALES
x_wild_m = np.genfromtxt(f'{ROOT_DIR}/data/kawasaki_2008/wild_males_x.txt')
y_wild_m = np.genfromtxt(f'{ROOT_DIR}/data/kawasaki_2008/wild_males_y.txt')
x_wild_m = np.round(x_wild_m).astype('int64') # In order to use for indexing
y_wild_m = y_wild_m * individual_count
x_wild_m = x_wild_m[:-2] # In order not to fit to the last two data points
y_wild_m = y_wild_m[:-2] # In order not to fit to the last two data points
%%time
# FEMALES, least squares
t_m_wild_f = 100
n = len(x_wild_f)
fit = []
# TODO: The population is set to the hypothetical wild type, in order to use the
for prod_wt_f in np.arange(0.046, 0.051, 0.001): # prod_wt = (1 - epsilon) * h_wt
hazard_rate_params = dict(hazard_rate_wt=prod_wt_f, population=HYP_WILDTYPE)
population_survivorship = run_cohort_simulation(
repetition_count,
individual_count,
hazard_rate_params,
t_m_wild_f,
)
mean = np.mean(population_survivorship, axis=0)[x_wild_f]
squares = [(mean[i] - y_wild_f[i])**2 for i in range(n)] # Not fitting to last two data points
fit.append((prod_wt_f, sum(squares)))
best_fits_f = sorted(fit, key=operator.itemgetter(1))
print('prod_wt, sum squares (females)')
print(*best_fits_f[0:10], sep='\n')
print()
prod_wt_f = best_fits_f[0][0]
for epsilon in np.arange(0.01, 0.05, 0.01):
h_wt = prod_wt_f / (1 - epsilon)
print(f'epsilon = {epsilon}, h_wt_f = {h_wt}')
%%time
# MALES, least squares
t_m_wild_m = 53 # TODO: Korrekt?
n = len(x_wild_m)
fit = []
for prod_wt_m in np.arange(0.043, 0.044, 0.0001): # prod_wt = (1 - epsilon) * h_wt
hazard_rate_params = dict(hazard_rate_wt=prod_wt_m, population=HYP_WILDTYPE)
population_survivorship = run_cohort_simulation(
repetition_count,
individual_count,
hazard_rate_params,
t_m_wild_m,
)
mean = np.mean(population_survivorship, axis=0)[x_wild_m]
squares = [(mean[i] - y_wild_m[i])**2 for i in range(n)] # Not fitting to last two data points
fit.append((prod_wt_m, sum(squares)))
best_fits_m = sorted(fit, key=operator.itemgetter(1))
print('prod_wt, sum squares (males)')
print(*best_fits_m[0:10], sep='\n')
print()
prod_wt_m = best_fits_m[0][0]
for epsilon in np.arange(0.01, 0.05, 0.01):
h_wt = prod_wt_m / (1 - epsilon)
print(f'epsilon = {epsilon}, h_wt_m = {h_wt}')
###Output
prod_wt, sum squares (males)
(0.04390000000000002, 2013.533)
(0.044000000000000025, 2088.5169000000024)
(0.04370000000000002, 2100.6717999999983)
(0.0432, 2137.9075999999995)
(0.043600000000000014, 2158.1651000000015)
(0.04380000000000002, 2183.2758000000035)
(0.043300000000000005, 2234.047400000005)
(0.043, 2242.0629999999983)
(0.04350000000000001, 2267.1160999999965)
(0.04340000000000001, 2412.368100000005)
epsilon = 0.01, h_wt_m = 0.044343434343434365
epsilon = 0.02, h_wt_m = 0.04479591836734696
epsilon = 0.03, h_wt_m = 0.04525773195876291
epsilon = 0.04, h_wt_m = 0.04572916666666669
CPU times: user 7.41 s, sys: 4.01 s, total: 11.4 s
Wall time: 12 s
###Markdown
Estimating $\omega$ and $\tau$A phenomenological extra morality term was needed for wild males. The term consists of parameters $\omega$ and $\tau$ which are estimated below.
###Code
# Wild male simulation parameters
individual_count = 1000
repetition_count = 100
t_m_wild_m = 54 # Max t for curve fitting (last x data point = 53)
epsilon = 0.04
# The following three values are from an earlier parameter estimation, see note in top of notebook
hazard_rate_wt = 0.0453
alpha_m = 0.00057
kappa_m = 0.0173
# Target parameters, these have been estimated iteratively in a manual fashion
omega = 0.0001
tau = 1.8
# Run simulation and calculate mean
hazard_rate_params = dict(alpha=alpha_m, kappa=kappa_m, omega=omega, tau=tau, epsilon=epsilon, hazard_rate_wt=hazard_rate_wt, population=MUT_WILD)
survivorship_wild_m = run_cohort_simulation(repetition_count, individual_count, hazard_rate_params, t_m_wild_m)
wild_mean = np.mean(survivorship_wild_m, axis=0)
# Inspect quality of fit
wild_m_x = np.genfromtxt(f'{ROOT_DIR}/data/kawasaki_2008/wild_males_x.txt')
wild_m_y = np.genfromtxt(f'{ROOT_DIR}/data/kawasaki_2008/wild_males_y.txt')
wild_m_y = wild_m_y * individual_count
t_steps = np.arange(t_m_wild_m)
fig1, ax = plt.subplots(figsize=(6, 6))
ax.plot(wild_m_x, wild_m_y, 'bo', markersize=4)
ax.plot(t_steps, wild_mean, 'b-')
###Output
_____no_output_____
|
Jupyter Notebook/Image Classifier Project.ipynb
|
###Markdown
Developing an AI applicationGoing forward, AI algorithms will be incorporated into more and more everyday applications. For example, you might want to include an image classifier in a smart phone app. To do this, you'd use a deep learning model trained on hundreds of thousands of images as part of the overall application architecture. A large part of software development in the future will be using these types of models as common parts of applications. In this project, you'll train an image classifier to recognize different species of flowers. You can imagine using something like this in a phone app that tells you the name of the flower your camera is looking at. In practice you'd train this classifier, then export it for use in your application. We'll be using [this dataset](http://www.robots.ox.ac.uk/~vgg/data/flowers/102/index.html) of 102 flower categories, you can see a few examples below. The project is broken down into multiple steps:* Load and preprocess the image dataset* Train the image classifier on your dataset* Use the trained classifier to predict image contentWe'll lead you through each part which you'll implement in Python.When you've completed this project, you'll have an application that can be trained on any set of labeled images. Here your network will be learning about flowers and end up as a command line application. But, what you do with your new skills depends on your imagination and effort in building a dataset. For example, imagine an app where you take a picture of a car, it tells you what the make and model is, then looks up information about it. Go build your own dataset and make something new.First up is importing the packages you'll need. It's good practice to keep all the imports at the beginning of your code. As you work through this notebook and find you need to import a package, make sure to add the import up here.
###Code
# Imports here
%matplotlib inline
%config InlineBackend.figure_format = 'retina'
import numpy as np
import matplotlib.pyplot as plt
import torch
from torch import nn, optim
import torch.nn.functional as F
import torchvision
from torchvision import transforms, datasets, models
from collections import OrderedDict
from PIL import Image
import json
###Output
_____no_output_____
###Markdown
Load the dataHere you'll use `torchvision` to load the data ([documentation](http://pytorch.org/docs/0.3.0/torchvision/index.html)). The data should be included alongside this notebook, otherwise you can [download it here](https://s3.amazonaws.com/content.udacity-data.com/nd089/flower_data.tar.gz). The dataset is split into three parts, training, validation, and testing. For the training, you'll want to apply transformations such as random scaling, cropping, and flipping. This will help the network generalize leading to better performance. You'll also need to make sure the input data is resized to 224x224 pixels as required by the pre-trained networks.The validation and testing sets are used to measure the model's performance on data it hasn't seen yet. For this you don't want any scaling or rotation transformations, but you'll need to resize then crop the images to the appropriate size.The pre-trained networks you'll use were trained on the ImageNet dataset where each color channel was normalized separately. For all three sets you'll need to normalize the means and standard deviations of the images to what the network expects. For the means, it's `[0.485, 0.456, 0.406]` and for the standard deviations `[0.229, 0.224, 0.225]`, calculated from the ImageNet images. These values will shift each color channel to be centered at 0 and range from -1 to 1.
###Code
data_dir = 'flowers'
train_dir = data_dir + '/train/'
valid_dir = data_dir + '/valid'
test_dir = data_dir + '/test'
# TODO: Define your transforms for the training, validation, and testing sets
train_transforms = transforms.Compose([transforms.RandomRotation(30),
transforms.RandomHorizontalFlip(),
transforms.RandomResizedCrop(224),
transforms.ToTensor(),
transforms.Normalize([0.485, 0.456, 0.406],
[0.229, 0.224, 0.225])])
validate_transforms = transforms.Compose([transforms.Resize(255),
transforms.CenterCrop(224),
transforms.ToTensor(),
transforms.Normalize([0.485, 0.456, 0.406],
[0.229, 0.224, 0.225])])
test_transforms = transforms.Compose([transforms.Resize(255),
transforms.CenterCrop(224),
transforms.ToTensor(),
transforms.Normalize([0.485, 0.456, 0.406],
[0.229, 0.224, 0.225])])
# TODO: Load the datasets with ImageFolder
train_dataset = datasets.ImageFolder(train_dir, transform = train_transforms)
validate_dataset = datasets.ImageFolder(valid_dir, transform = validate_transforms)
test_dataset = datasets.ImageFolder(test_dir, transform = test_transforms)
# TODO: Using the image datasets and the trainforms, define the dataloaders
train_loader = torch.utils.data.DataLoader(train_dataset, batch_size = 64, shuffle = True)
validate_loader = torch.utils.data.DataLoader(validate_dataset, batch_size = 32, shuffle = True)
test_loader = torch.utils.data.DataLoader(test_dataset, batch_size = 32, shuffle = True)
###Output
_____no_output_____
###Markdown
Label mappingYou'll also need to load in a mapping from category label to category name. You can find this in the file `cat_to_name.json`. It's a JSON object which you can read in with the [`json` module](https://docs.python.org/2/library/json.html). This will give you a dictionary mapping the integer encoded categories to the actual names of the flowers.
###Code
with open('cat_to_name.json', 'r') as f:
cat_to_name = json.load(f)
###Output
_____no_output_____
###Markdown
Building and training the classifierNow that the data is ready, it's time to build and train the classifier. As usual, you should use one of the pretrained models from `torchvision.models` to get the image features. Build and train a new feed-forward classifier using those features.We're going to leave this part up to you. Refer to [the rubric](https://review.udacity.com/!/rubrics/1663/view) for guidance on successfully completing this section. Things you'll need to do:* Load a [pre-trained network](http://pytorch.org/docs/master/torchvision/models.html) (If you need a starting point, the VGG networks work great and are straightforward to use)* Define a new, untrained feed-forward network as a classifier, using ReLU activations and dropout* Train the classifier layers using backpropagation using the pre-trained network to get the features* Track the loss and accuracy on the validation set to determine the best hyperparametersWe've left a cell open for you below, but use as many as you need. Our advice is to break the problem up into smaller parts you can run separately. Check that each part is doing what you expect, then move on to the next. You'll likely find that as you work through each part, you'll need to go back and modify your previous code. This is totally normal!When training make sure you're updating only the weights of the feed-forward network. You should be able to get the validation accuracy above 70% if you build everything right. Make sure to try different hyperparameters (learning rate, units in the classifier, epochs, etc) to find the best model. Save those hyperparameters to use as default values in the next part of the project.One last important tip if you're using the workspace to run your code: To avoid having your workspace disconnect during the long-running tasks in this notebook, please read in the earlier page in this lesson called Intro toGPU Workspaces about Keeping Your Session Active. You'll want to include code from the workspace_utils.py module.**Note for Workspace users:** If your network is over 1 GB when saved as a checkpoint, there might be issues with saving backups in your workspace. Typically this happens with wide dense layers after the convolutional layers. If your saved checkpoint is larger than 1 GB (you can open a terminal and check with `ls -lh`), you should reduce the size of your hidden layers and train again.
###Code
# TODO: Build and train your network
device = torch.device("cuda" if torch.cuda.is_available() else "cpu")
print(device)
model = models.vgg16(pretrained=True)
model #print the model architecture
def nn_setup(structure='vgg16', dropout_rate=0.2, hidden_layer1=5000, lr=0.001):
# No backpropagation for now
for param in model.parameters():
param.requires_grad = False
classifier = nn.Sequential(OrderedDict([
('fc_1', nn.Linear(25088, 5000)),
('relu_1', nn.ReLU()),
('dropout_1', nn.Dropout(dropout_rate)),
('fc_2', nn.Linear(5000, 102)),
('output_layer', nn.LogSoftmax(dim=1))
]))
model.classifier = classifier #Classifier()
criterion = nn.NLLLoss()
optimizer = optim.Adam(model.classifier.parameters(), lr)
model.to(device)
return model, optimizer, criterion
model, optimizer, criterion = nn_setup('vgg16')
print(model)
epochs = 1
print_every = 5
steps = 0
for epoch in range(epochs):
running_loss = 0
#print("Epoch {}".format(epoch))
internal_counter = 0
for ii, (inputs, labels) in enumerate(train_loader):
internal_counter += 1
#print("Counter: {}".format(internal_counter))
steps += 1
inputs = inputs.to(device)
labels = labels.to(device)
optimizer.zero_grad()
outputs = model.forward(inputs)
loss = criterion(outputs, labels)
loss.backward()
optimizer.step()
running_loss += loss.item()
if steps % print_every == 0:
accuracy = 0
validation_loss = 0
model.eval()
with torch.no_grad():
for ii, (inputs_val, labels_val) in enumerate(validate_loader):
#print("validate")
optimizer.zero_grad()
inputs_val, labels_val = inputs_val.to(device), labels_val.to(device)
model.to(device)
outputs_val = model.forward(inputs_val)
validation_loss = criterion(outputs_val, labels_val)
#validation_loss += batch_loss.item()
# Calculate accuracy
ps = torch.exp(outputs_val)
top_p, top_class = ps.topk(1, dim=1)
equals = top_class == labels_val.view(*top_class.shape)
accuracy += torch.mean(equals.type(torch.FloatTensor)).item()
print(f"Epoch {epoch+1}/{epochs}.. "
f"Train loss: {running_loss/print_every:.3f}.. "
f"Validation loss: {validation_loss/len(validate_loader):.3f}.. "
f"Validation accuracy: {accuracy/len(validate_loader):.3f}")
running_loss = 0
model.train()
else:
print(f"Finished epoch {epoch+1} --------")
###Output
_____no_output_____
###Markdown
Testing your networkIt's good practice to test your trained network on test data, images the network has never seen either in training or validation. This will give you a good estimate for the model's performance on completely new images. Run the test images through the network and measure the accuracy, the same way you did validation. You should be able to reach around 70% accuracy on the test set if the model has been trained well.
###Code
# TODO: Do validation on the test set
num_correct = 0
num_total = 0
with torch.no_grad():
for images_test, labels_test in test_loader:
images_test, labels_test = images_test.to(device), labels_test.to(device)
outputs = model(images_test)
_, predicted = torch.max(outputs.data, 1)
num_total += labels_test.size(0)
num_correct += (predicted == labels_test).sum().item()
print('Accuracy: %d %%'% (100 * num_correct / num_total))
###Output
_____no_output_____
###Markdown
Save the checkpointNow that your network is trained, save the model so you can load it later for making predictions. You probably want to save other things such as the mapping of classes to indices which you get from one of the image datasets: `image_datasets['train'].class_to_idx`. You can attach this to the model as an attribute which makes inference easier later on.```model.class_to_idx = image_datasets['train'].class_to_idx```Remember that you'll want to completely rebuild the model later so you can use it for inference. Make sure to include any information you need in the checkpoint. If you want to load the model and keep training, you'll want to save the number of epochs as well as the optimizer state, `optimizer.state_dict`. You'll likely want to use this trained model in the next part of the project, so best to save it now.
###Code
# TODO: Save the checkpoint
model.class_to_idx = train_dataset.class_to_idx
#torch.save(model.state_dict(), 'checkpoint.pth')
torch.save({'structure': 'vgg16',
'hidden_layer1': 5000,
'state_dict': model.state_dict(),
'class_to_idx': model.class_to_idx},
'checkpoint.pth')
###Output
_____no_output_____
###Markdown
Loading the checkpointAt this point it's good to write a function that can load a checkpoint and rebuild the model. That way you can come back to this project and keep working on it without having to retrain the network.
###Code
# TODO: Write a function that loads a checkpoint and rebuilds the model
def load_model(path):
checkpoint = torch.load('checkpoint.pth', map_location=lambda storage, loc: storage)
structure = checkpoint['structure']
hidden_layer1 = checkpoint['hidden_layer1']
model, _, _ = nn_setup(structure, 0.2, hidden_layer1)
model.class_to_idx = checkpoint['class_to_idx']
model.load_state_dict(checkpoint['state_dict'])
return model
model = load_model('checkpoint.pth')
print(model)
###Output
VGG(
(features): Sequential(
(0): Conv2d(3, 64, kernel_size=(3, 3), stride=(1, 1), padding=(1, 1))
(1): ReLU(inplace)
(2): Conv2d(64, 64, kernel_size=(3, 3), stride=(1, 1), padding=(1, 1))
(3): ReLU(inplace)
(4): MaxPool2d(kernel_size=2, stride=2, padding=0, dilation=1, ceil_mode=False)
(5): Conv2d(64, 128, kernel_size=(3, 3), stride=(1, 1), padding=(1, 1))
(6): ReLU(inplace)
(7): Conv2d(128, 128, kernel_size=(3, 3), stride=(1, 1), padding=(1, 1))
(8): ReLU(inplace)
(9): MaxPool2d(kernel_size=2, stride=2, padding=0, dilation=1, ceil_mode=False)
(10): Conv2d(128, 256, kernel_size=(3, 3), stride=(1, 1), padding=(1, 1))
(11): ReLU(inplace)
(12): Conv2d(256, 256, kernel_size=(3, 3), stride=(1, 1), padding=(1, 1))
(13): ReLU(inplace)
(14): Conv2d(256, 256, kernel_size=(3, 3), stride=(1, 1), padding=(1, 1))
(15): ReLU(inplace)
(16): MaxPool2d(kernel_size=2, stride=2, padding=0, dilation=1, ceil_mode=False)
(17): Conv2d(256, 512, kernel_size=(3, 3), stride=(1, 1), padding=(1, 1))
(18): ReLU(inplace)
(19): Conv2d(512, 512, kernel_size=(3, 3), stride=(1, 1), padding=(1, 1))
(20): ReLU(inplace)
(21): Conv2d(512, 512, kernel_size=(3, 3), stride=(1, 1), padding=(1, 1))
(22): ReLU(inplace)
(23): MaxPool2d(kernel_size=2, stride=2, padding=0, dilation=1, ceil_mode=False)
(24): Conv2d(512, 512, kernel_size=(3, 3), stride=(1, 1), padding=(1, 1))
(25): ReLU(inplace)
(26): Conv2d(512, 512, kernel_size=(3, 3), stride=(1, 1), padding=(1, 1))
(27): ReLU(inplace)
(28): Conv2d(512, 512, kernel_size=(3, 3), stride=(1, 1), padding=(1, 1))
(29): ReLU(inplace)
(30): MaxPool2d(kernel_size=2, stride=2, padding=0, dilation=1, ceil_mode=False)
)
(classifier): Sequential(
(fc_1): Linear(in_features=25088, out_features=5000, bias=True)
(relu_1): ReLU()
(dropout_1): Dropout(p=0.2)
(fc_2): Linear(in_features=5000, out_features=102, bias=True)
(output_layer): LogSoftmax()
)
)
###Markdown
Inference for classificationNow you'll write a function to use a trained network for inference. That is, you'll pass an image into the network and predict the class of the flower in the image. Write a function called `predict` that takes an image and a model, then returns the top $K$ most likely classes along with the probabilities. It should look like ```pythonprobs, classes = predict(image_path, model)print(probs)print(classes)> [ 0.01558163 0.01541934 0.01452626 0.01443549 0.01407339]> ['70', '3', '45', '62', '55']```First you'll need to handle processing the input image such that it can be used in your network. Image PreprocessingYou'll want to use `PIL` to load the image ([documentation](https://pillow.readthedocs.io/en/latest/reference/Image.html)). It's best to write a function that preprocesses the image so it can be used as input for the model. This function should process the images in the same manner used for training. First, resize the images where the shortest side is 256 pixels, keeping the aspect ratio. This can be done with the [`thumbnail`](http://pillow.readthedocs.io/en/3.1.x/reference/Image.htmlPIL.Image.Image.thumbnail) or [`resize`](http://pillow.readthedocs.io/en/3.1.x/reference/Image.htmlPIL.Image.Image.thumbnail) methods. Then you'll need to crop out the center 224x224 portion of the image.Color channels of images are typically encoded as integers 0-255, but the model expected floats 0-1. You'll need to convert the values. It's easiest with a Numpy array, which you can get from a PIL image like so `np_image = np.array(pil_image)`.As before, the network expects the images to be normalized in a specific way. For the means, it's `[0.485, 0.456, 0.406]` and for the standard deviations `[0.229, 0.224, 0.225]`. You'll want to subtract the means from each color channel, then divide by the standard deviation. And finally, PyTorch expects the color channel to be the first dimension but it's the third dimension in the PIL image and Numpy array. You can reorder dimensions using [`ndarray.transpose`](https://docs.scipy.org/doc/numpy-1.13.0/reference/generated/numpy.ndarray.transpose.html). The color channel needs to be first and retain the order of the other two dimensions.
###Code
def process_image(image_path):
''' Scales, crops, and normalizes a PIL image for a PyTorch model,
returns an Numpy array
'''
# TODO: Process a PIL image for use in a PyTorch model
max_size = 256
target_size = 224
decimal_place = 0
image = Image.open(image_path)
h = image.height
w = image.width
if w > h:
factor = w / h
image = image.resize(size = (int(round(factor*max_size, decimal_place)), max_size))
else:
factor = h / w
image = image.resize(size=(max_size, int(round(factor*max_size, decimal_place))))
h = image.height
w = image.width
image = image.crop(((w - target_size) / 2,
(h - target_size) / 2,
(w + target_size) / 2,
(h + target_size) / 2))
np_image = np.array(image) / 255
mean = np.array([0.485, 0.456, 0.406])
std = np.array([0.229, 0.224, 0.225])
np_image = (np_image - mean) / std
np_image = np.transpose(np_image, (2, 0, 1))
tensor_image = torch.from_numpy(np_image).type(torch.FloatTensor)
return(tensor_image)
image_path = (data_dir + '/test' + '/102/' + 'image_08015.jpg')
img = process_image(image_path)
print(img.shape)
###Output
torch.Size([3, 224, 224])
###Markdown
To check your work, the function below converts a PyTorch tensor and displays it in the notebook. If your `process_image` function works, running the output through this function should return the original image (except for the cropped out portions).
###Code
def imshow(image, ax=None, title=None):
"""Imshow for Tensor."""
if ax is None:
fig, ax = plt.subplots()
# PyTorch tensors assume the color channel is the first dimension
# but matplotlib assumes is the third dimension
image = image.numpy().transpose((1, 2, 0))
# Undo preprocessing
mean = np.array([0.485, 0.456, 0.406])
std = np.array([0.229, 0.224, 0.225])
image = std * image + mean
# Image needs to be clipped between 0 and 1 or it looks like noise when displayed
image = np.clip(image, 0, 1)
ax.imshow(image)
return ax
imshow(process_image(data_dir + '/test' + '/10' + '/image_07090.jpg'))
###Output
_____no_output_____
###Markdown
Class PredictionOnce you can get images in the correct format, it's time to write a function for making predictions with your model. A common practice is to predict the top 5 or so (usually called top-$K$) most probable classes. You'll want to calculate the class probabilities then find the $K$ largest values.To get the top $K$ largest values in a tensor use [`x.topk(k)`](http://pytorch.org/docs/master/torch.htmltorch.topk). This method returns both the highest `k` probabilities and the indices of those probabilities corresponding to the classes. You need to convert from these indices to the actual class labels using `class_to_idx` which hopefully you added to the model or from an `ImageFolder` you used to load the data ([see here](Save-the-checkpoint)). Make sure to invert the dictionary so you get a mapping from index to class as well.Again, this method should take a path to an image and a model checkpoint, then return the probabilities and classes.```pythonprobs, classes = predict(image_path, model)print(probs)print(classes)> [ 0.01558163 0.01541934 0.01452626 0.01443549 0.01407339]> ['70', '3', '45', '62', '55']```
###Code
def predict(image_path, model, topk=5):
''' Predict the class (or classes) of an image using a trained deep learning model.
'''
# TODO: Implement the code to predict the class from an image file
img = process_image(image_path)
img_torch = img.unsqueeze_(0)
img_torch = img_torch.float()
with torch.no_grad():
output = model.forward(img_torch.to(device))
probability = F.softmax(output, dim=1)
probs, indices = probability.topk(topk)
probs = probs.to(device).numpy()[0]
indices = indices.to(device).numpy()[0]
idx_to_class = {v:k for k, v in model.class_to_idx.items()}
classes = [idx_to_class[x] for x in indices]
print(probs)
print(classes)
return probs, classes
img_path = data_dir + '/test' + '/72' + '/image_03575.jpg'
img_path = data_dir + '/test' + '/15' + '/image_06369.jpg'
img_path = data_dir + '/test' + '/95' + '/image_07470.jpg'
#img_path = data_dir + '/test' + '/72' + '/image_03613.jpg'
#img_path = data_dir + '/test' + '/41' + '/image_02251.jpg'
#img_path = data_dir + '/test' + '/1' + '/image_06764.jpg'
#img_path = data_dir + '/test' + '/102' + '/image_08030.jpg'
#probabibilities_predicted, classes_predicted = predict(img_path, model, 5)
###Output
_____no_output_____
###Markdown
Sanity CheckingNow that you can use a trained model for predictions, check to make sure it makes sense. Even if the testing accuracy is high, it's always good to check that there aren't obvious bugs. Use `matplotlib` to plot the probabilities for the top 5 classes as a bar graph, along with the input image. It should look like this:You can convert from the class integer encoding to actual flower names with the `cat_to_name.json` file (should have been loaded earlier in the notebook). To show a PyTorch tensor as an image, use the `imshow` function defined above.
###Code
# TODO: Display an image along with the top 5 classes
def check_sanity():
probabilities, indices_of_flowers = predict(img_path, model)
names_of_flowers = [cat_to_name[str(index)] for index in indices_of_flowers]
plt.subplot(2, 1, 1)
image = process_image(img_path)
name_of_flower = cat_to_name[str(indices_of_flowers[0])]
plot_1 = imshow(image, ax = plt)
plot_1.axis('off')
plot_1.title(name_of_flower)
#plt.subplot(2, 1, 2)
fig, ax = plt.subplots()
ylabels = np.array(names_of_flowers)
y_pos = np.arange(len(ylabels))
ax.barh(ylabels, probabilities, align='center')
ax.invert_yaxis()
plt.show();
check_sanity()
###Output
[ 0.3491042 0.26112998 0.05743311 0.05235921 0.05065762]
['4', '95', '84', '74', '88']
|
Metodos Computacionales Avanzados/secciones/07.CrossValidation/bootstrap.ipynb
|
###Markdown
BootstrappingUna parte central de cualquier modelo estadístico es poder calcular la incertidumbre asociada a sus parámetros.En los métodos MCMC las incertidumbres se pueden calcular numéricamente. ¿Pero qué hacer con otros métodos que no son Bayesianos?Una de las posibilidades es utilizar el bootstrap. El poder de este método es que es aplicable cuando no hay herramientas analíticasdisponibles. Además es muy fácil de implementar. En el bootstrap se toman los datos originales y se toman subconjuntos de maneraaleatoria. Cada uno de estos subconjuntos se procesa bajo el método de interés. De esta manera, para cada subconjunto de datoshay unos parámetros estimados. Al final se hacen los histogramas de los parámetros obtenidos de los diferentes conjuntos y estosvan a dar una idea de las incertidumbres.Esto proceso no es necesario con un método como mínimos cuadrados porque para este métodoya hay estimadores para las incertidumbres, de todas formas vamos a utilizarlo para demostrar el uso de bootstraping.Vamos a volver a los datos de precios de carros.
###Code
data = pd.read_csv('../06.RegresionLineal/Cars93.csv')
###Output
_____no_output_____
###Markdown
Vamos a ajustar el modelo lineal `Price` = $\beta_0$ + $\beta_1\times$ `Horsepower` + $\beta_2\times$ `Turn.circle`
###Code
X = data[['Horsepower', 'Turn.circle']]
Y = data['Price'].values.reshape(-1,1)
###Output
_____no_output_____
###Markdown
Ahora vamos a dividir los datos en 2 grupos: `test` y `validation`. Para eso usamos `sklearn.model_selection.train_test_split`
###Code
import sklearn.model_selection
X_train, X_validation, Y_train, Y_validation = sklearn.model_selection.train_test_split(X, Y, test_size=0.5)
###Output
_____no_output_____
###Markdown
`train` vamos a usarlo para encontrar los $\beta_i$ y `validation` para medir $R^2$.
###Code
linear = sklearn.linear_model.LinearRegression()
linear.fit(X_train, Y_train)
beta0 = linear.intercept_[0]
beta1 = linear.coef_[0][0]
beta2 = linear.coef_[0][0]
r2_train = linear.score(X_train, Y_train)
r2_validation = linear.score(X_validation, Y_validation)
print('betas:', beta0, beta1, beta2)
print('R2 (train vs. validation)', r2_train, r2_validation)
###Output
betas: 25.6659789471 0.191387960898 0.191387960898
R2 (train vs. validation) 0.642725267677 0.499000654284
###Markdown
Esto lo podemos repetir muchas veces
###Code
for i in range(10):
X_train, X_validation, Y_train, Y_validation = sklearn.model_selection.train_test_split(X, Y, test_size=0.5)
linear.fit(X_train, Y_train)
beta0 = linear.intercept_[0]
beta1 = linear.coef_[0][0]
beta2 = linear.coef_[0][0]
r2_train = linear.score(X_train, Y_train)
r2_validation = linear.score(X_validation, Y_validation)
print('ITERACION ', i)
print('\t betas:', beta0, beta1, beta2)
print('\t R2 (train vs. validation)', r2_train, r2_validation)
###Output
ITERACION 0
betas: 9.84833170781 0.155031120058 0.155031120058
R2 (train vs. validation) 0.54656618176 0.650800682602
ITERACION 1
betas: -7.97141806686 0.129296304948 0.129296304948
R2 (train vs. validation) 0.701829769635 0.514906680333
ITERACION 2
betas: -5.79953594905 0.155373023649 0.155373023649
R2 (train vs. validation) 0.677344513328 0.54756107775
ITERACION 3
betas: 9.82684214615 0.176236524826 0.176236524826
R2 (train vs. validation) 0.603597368896 0.567531603808
ITERACION 4
betas: 6.78908309416 0.148594037865 0.148594037865
R2 (train vs. validation) 0.604101303539 0.633533094309
ITERACION 5
betas: 0.692679217502 0.115154281018 0.115154281018
R2 (train vs. validation) 0.490443006398 0.645054838084
ITERACION 6
betas: 3.05386497906 0.146982356439 0.146982356439
R2 (train vs. validation) 0.759876454367 0.52142744304
ITERACION 7
betas: 18.253286622 0.156872655976 0.156872655976
R2 (train vs. validation) 0.482067511933 0.762393674908
ITERACION 8
betas: -4.35651576229 0.120822329373 0.120822329373
R2 (train vs. validation) 0.715762759004 0.51248315991
ITERACION 9
betas: 3.06594192081 0.122746453462 0.122746453462
R2 (train vs. validation) 0.663441578952 0.555238530393
###Markdown
Hecho de una manera más sistemática y para tener la posibilidad de preparar gráficas, vamos a hacerlo `5000` veces
###Code
n_boot = 5000
beta_0 = np.ones(n_boot)
beta_1 = np.ones(n_boot)
beta_2 = np.ones(n_boot)
r2_train = np.ones(n_boot)
r2_validation = np.ones(n_boot)
linear = sklearn.linear_model.LinearRegression()
for i in range(n_boot):
X_train, X_validation, Y_train, Y_validation = sklearn.model_selection.train_test_split(X, Y, test_size=0.5)
linear.fit(X_train, Y_train)
beta_0[i] = linear.intercept_[0]
beta_1[i] = linear.coef_[0][0]
beta_2[i] = linear.coef_[0][1]
r2_train[i] = linear.score(X_train, Y_train)
r2_validation[i] = linear.score(X_validation, Y_validation)
plt.figure(figsize=(20,4))
plt.subplot(131)
_ = plt.hist(beta_0, bins=40)
_ = plt.xlabel(r'$\beta_0$')
plt.subplot(132)
_ = plt.hist(beta_1, bins=40)
_ = plt.xlabel(r'$\beta_1$')
plt.subplot(133)
_ = plt.hist(beta_2, bins=40)
_ = plt.xlabel(r'$\beta_2$')
plt.figure(figsize=(20,4))
plt.subplot(131)
plt.scatter(beta_0, beta_1, alpha=0.1)
_ = plt.xlabel(r'$\beta_0$')
_ = plt.ylabel(r'$\beta_1$')
plt.subplot(132)
plt.scatter(beta_1, beta_2, alpha=0.1)
_ = plt.xlabel(r'$\beta_1$')
_ = plt.ylabel(r'$\beta_2$')
plt.subplot(133)
plt.scatter(beta_2, beta_0, alpha=0.1)
_ = plt.xlabel(r'$\beta_2$')
_ = plt.ylabel(r'$\beta_0$')
_ = plt.hist(r2_train, bins=40, density=True, label='Train')
_ = plt.hist(r2_validation, bins=40, alpha=0.5, density=True, label='Validation')
plt.title("Estimados por Bootstrapping")
plt.legend()
###Output
_____no_output_____
###Markdown
Y de esta manera podemos estimar los valores medios de los parámetros
###Code
print('beta 0 {} +/- {}'.format(beta_0.mean(), beta_0.std() ))
print('beta 1 {} +/- {}'.format(beta_1.mean(), beta_1.std() ))
print('beta 2 {} +/- {}'.format(beta_2.mean(), beta_2.std() ))
###Output
beta 0 6.315047231051973 +/- 10.297312352873558
beta 1 0.15452225139328798 +/- 0.02727817665912039
beta 2 -0.23186052336732915 +/- 0.3367503005147814
|
01_Image_Representation_Classification/1_5_CNN_Layers/5_1. Feature viz for FashionMNIST.ipynb
|
###Markdown
Visualizing CNN Layers---In this notebook, we load a trained CNN (from a solution to FashionMNIST) and implement several feature visualization techniques to see what features this network has learned to extract. Load the [data](http://pytorch.org/docs/master/torchvision/datasets.html)In this cell, we load in just the **test** dataset from the FashionMNIST class.
###Code
# our basic libraries
import torch
import torchvision
# data loading and transforming
from torchvision.datasets import FashionMNIST
from torch.utils.data import DataLoader
from torchvision import transforms
# The output of torchvision datasets are PILImage images of range [0, 1].
# We transform them to Tensors for input into a CNN
## Define a transform to read the data in as a tensor
data_transform = transforms.ToTensor()
test_data = FashionMNIST(root='./data', train=False,
download=True, transform=data_transform)
# Print out some stats about the test data
print('Test data, number of images: ', len(test_data))
# prepare data loaders, set the batch_size
## TODO: you can try changing the batch_size to be larger or smaller
## when you get to training your network, see how batch_size affects the loss
batch_size = 20
test_loader = DataLoader(test_data, batch_size=batch_size, shuffle=True)
# specify the image classes
classes = ['T-shirt/top', 'Trouser', 'Pullover', 'Dress', 'Coat',
'Sandal', 'Shirt', 'Sneaker', 'Bag', 'Ankle boot']
###Output
_____no_output_____
###Markdown
Visualize some test dataThis cell iterates over the training dataset, loading a random batch of image/label data, using `dataiter.next()`. It then plots the batch of images and labels in a `2 x batch_size/2` grid.
###Code
import numpy as np
import matplotlib.pyplot as plt
%matplotlib inline
# obtain one batch of training images
dataiter = iter(test_loader)
images, labels = dataiter.next()
images = images.numpy()
# plot the images in the batch, along with the corresponding labels
fig = plt.figure(figsize=(25, 4))
for idx in np.arange(batch_size):
ax = fig.add_subplot(2, batch_size/2, idx+1, xticks=[], yticks=[])
ax.imshow(np.squeeze(images[idx]), cmap='gray')
ax.set_title(classes[labels[idx]])
###Output
_____no_output_____
###Markdown
Define the network architectureThe various layers that make up any neural network are documented, [here](http://pytorch.org/docs/master/nn.html). For a convolutional neural network, we'll use a simple series of layers:* Convolutional layers* Maxpooling layers* Fully-connected (linear) layers
###Code
import torch.nn as nn
import torch.nn.functional as F
class Net(nn.Module):
def __init__(self):
super(Net, self).__init__()
# 1 input image channel (grayscale), 10 output channels/feature maps
# 3x3 square convolution kernel
## output size = (W-F)/S +1 = (28-3)/1 +1 = 26
# the output Tensor for one image, will have the dimensions: (10, 26, 26)
# after one pool layer, this becomes (10, 13, 13)
self.conv1 = nn.Conv2d(1, 10, 3)
# maxpool layer
# pool with kernel_size=2, stride=2
self.pool = nn.MaxPool2d(2, 2)
# second conv layer: 10 inputs, 20 outputs, 3x3 conv
## output size = (W-F)/S +1 = (13-3)/1 +1 = 11
# the output tensor will have dimensions: (20, 11, 11)
# after another pool layer this becomes (20, 5, 5); 5.5 is rounded down
self.conv2 = nn.Conv2d(10, 20, 3)
# 20 outputs * the 5*5 filtered/pooled map size
self.fc1 = nn.Linear(20*5*5, 50)
# dropout with p=0.4
self.fc1_drop = nn.Dropout(p=0.4)
# finally, create 10 output channels (for the 10 classes)
self.fc2 = nn.Linear(50, 10)
# define the feedforward behavior
def forward(self, x):
# two conv/relu + pool layers
x = self.pool(F.relu(self.conv1(x)))
x = self.pool(F.relu(self.conv2(x)))
# prep for linear layer
# this line of code is the equivalent of Flatten in Keras
x = x.view(x.size(0), -1)
# two linear layers with dropout in between
x = F.relu(self.fc1(x))
x = self.fc1_drop(x)
x = self.fc2(x)
# final output
return x
###Output
_____no_output_____
###Markdown
Load in our trained netThis notebook needs to know the network architecture, as defined above, and once it knows what the "Net" class looks like, we can instantiate a model and load in an already trained network.The architecture above is taken from the example solution code, which was trained and saved in the directory `saved_models/`.
###Code
# instantiate your Net
net = Net()
# load the net parameters by name
net.load_state_dict(torch.load('saved_models/fashion_net_ex.pt'))
print(net)
###Output
Net(
(conv1): Conv2d(1, 10, kernel_size=(3, 3), stride=(1, 1))
(pool): MaxPool2d(kernel_size=2, stride=2, padding=0, dilation=1, ceil_mode=False)
(conv2): Conv2d(10, 20, kernel_size=(3, 3), stride=(1, 1))
(fc1): Linear(in_features=500, out_features=50, bias=True)
(fc1_drop): Dropout(p=0.4)
(fc2): Linear(in_features=50, out_features=10, bias=True)
)
###Markdown
Feature VisualizationSometimes, neural networks are thought of as a black box, given some input, they learn to produce some output. CNN's are actually learning to recognize a variety of spatial patterns and you can visualize what each convolutional layer has been trained to recognize by looking at the weights that make up each convolutional kernel and applying those one at a time to a sample image. These techniques are called feature visualization and they are useful for understanding the inner workings of a CNN.In the cell below, you'll see how to extract and visualize the filter weights for all of the filters in the first convolutional layer.Note the patterns of light and dark pixels and see if you can tell what a particular filter is detecting. For example, the filter pictured in the example below has dark pixels on either side and light pixels in the middle column, and so it may be detecting vertical edges.
###Code
# Get the weights in the first conv layer
weights = net.conv1.weight.data
w = weights.numpy()
# for 10 filters
fig=plt.figure(figsize=(20, 8))
columns = 5
rows = 2
for i in range(0, columns*rows):
fig.add_subplot(rows, columns, i+1)
plt.imshow(w[i][0], cmap='gray')
print('First convolutional layer')
plt.show()
weights = net.conv2.weight.data
w = weights.numpy()
###Output
First convolutional layer
###Markdown
Activation MapsNext, you'll see how to use OpenCV's `filter2D` function to apply these filters to a sample test image and produce a series of **activation maps** as a result. We'll do this for the first and second convolutional layers and these activation maps whould really give you a sense for what features each filter learns to extract.
###Code
# obtain one batch of testing images
dataiter = iter(test_loader)
images, labels = dataiter.next()
images = images.numpy()
# select an image by index
idx = 10
img = np.squeeze(images[idx])
# Use OpenCV's filter2D function
# apply a specific set of filter weights (like the one's displayed above) to the test image
import cv2
plt.imshow(img, cmap='gray')
weights = net.conv1.weight.data
w = weights.numpy()
# 1. first conv layer
# for 10 filters
fig=plt.figure(figsize=(30, 10))
columns = 5*2
rows = 2
for i in range(0, columns*rows):
fig.add_subplot(rows, columns, i+1)
if ((i%2)==0):
plt.imshow(w[int(i/2)][0], cmap='gray')
else:
c = cv2.filter2D(img, -1, w[int((i-1)/2)][0])
plt.imshow(c, cmap='gray')
plt.show()
# Same process but for the second conv layer (20, 3x3 filters):
plt.imshow(img, cmap='gray')
# second conv layer, conv2
weights = net.conv2.weight.data
w = weights.numpy()
# 1. first conv layer
# for 20 filters
fig=plt.figure(figsize=(30, 10))
columns = 5*2
rows = 2*2
for i in range(0, columns*rows):
fig.add_subplot(rows, columns, i+1)
if ((i%2)==0):
plt.imshow(w[int(i/2)][0], cmap='gray')
else:
c = cv2.filter2D(img, -1, w[int((i-1)/2)][0])
plt.imshow(c, cmap='gray')
plt.show()
###Output
_____no_output_____
|
12-Dados_Continuos_Streaming-WordCount.ipynb
|
###Markdown
Exemplo 12: Dados Contínuos (Streaming) Contagem contínua de palavrasSpark Streaming is an extension of the Spark API that enables stream processing of live data streams. A Stream is a continuous source of data generated in real time. Data can be ingested from many sources like filesystem, Kafka, Flume, Kinesis, or TCP sockets, and can be processed using complex algorithms like map, reduce, join and window, and apply Spark’s machine learning or graph processing algorithms on continuous data streams. Finally, processed data can be pushed out to filesystems, databases, and dashboards.This example counts words in text encoded with UTF8 received from the network every second.The script *streaming_server.py* reads a text file and send one line per second and create a TCP server that Spark Streaming would connect to receive data.Usage: **./streaming_server.py \ \**, where *filename* ia a UTF-8 text file and *port* is a port number between 9700 and 9800 that should be configured in the configuration parameters section.The notebook process the word count in each reading or during a slide windows.
###Code
# Start Spark environment
import findspark
findspark.init()
# Load Spark Library
from pyspark import SparkContext
from pyspark.sql import SparkSession
from pyspark.streaming import StreamingContext
import time, os, re
###Output
_____no_output_____
###Markdown
Configuration parameters
###Code
#Streaming server port
port=9763
###Output
_____no_output_____
###Markdown
Create Spark Context and Streming Context
###Code
# Create Spark Context
sc = SparkContext("local[*]","Stream_WordCount") \
.getOrCreate()
# Create Streming Context reading server each 1 sec
ssc = StreamingContext(sc, 1)
# Set checkpoint directory
ssc.checkpoint("/tmp/spark-checkpoint")
###Output
_____no_output_____
###Markdown
Read text received by Socket
###Code
# Create a DStream that will connect to hostname:port
lines = ssc.socketTextStream("localhost", port)
# Print line read
lines.pprint()
# Split each line into words
words = lines.flatMap(lambda line: line.split())
###Output
_____no_output_____
###Markdown
Count words in each read
###Code
# Count each word in each line
pairs = words.map(lambda word: (word, 1))
# Count the number of each word in lines
wordCounts = pairs.reduceByKey(lambda x, y: x + y)
# Print the first ten elements of each line
#wordCounts.pprint()
###Output
_____no_output_____
###Markdown
Count words in each window
###Code
# Count the total words last 30 seconds of data, every 10 seconds
# reduceByKeyAndWindow(oper1, oper2,windows_time,result_time)
# oper1: operation in sliding window
# oper2: operation to remove last window
# windows_time: window length (total time of window)
#result_time: time when the result is evalutated
# Count the number of each word in windows of 30 sec showing at 10 sec
windowedWordCounts = wordCounts.reduceByKeyAndWindow(lambda x, y: x + y, lambda x, y: x - y, 30, 10)
# Print the first ten elements of each window period
windowedWordCounts.pprint()
###Output
_____no_output_____
###Markdown
Start Sreaming processing
###Code
ssc.start() # Start the computation
ssc.awaitTermination() # Wait for the computation to terminate
###Output
-------------------------------------------
Time: 2020-04-12 16:54:36
-------------------------------------------
-------------------------------------------
Time: 2020-04-12 16:54:37
-------------------------------------------
apache license
-------------------------------------------
Time: 2020-04-12 16:54:38
-------------------------------------------
version 20 january 2004
-------------------------------------------
Time: 2020-04-12 16:54:39
-------------------------------------------
httpwwwapacheorglicenses
-------------------------------------------
Time: 2020-04-12 16:54:40
-------------------------------------------
-------------------------------------------
Time: 2020-04-12 16:54:41
-------------------------------------------
terms and conditions for use reproduction and distribution
-------------------------------------------
Time: 2020-04-12 16:54:42
-------------------------------------------
-------------------------------------------
Time: 2020-04-12 16:54:43
-------------------------------------------
1 definitions
-------------------------------------------
Time: 2020-04-12 16:54:44
-------------------------------------------
-------------------------------------------
Time: 2020-04-12 16:54:45
-------------------------------------------
license shall mean the terms and conditions for use reproduction
-------------------------------------------
Time: 2020-04-12 16:54:45
-------------------------------------------
('2004', 1)
('httpwwwapacheorglicenses', 1)
('conditions', 2)
('reproduction', 2)
('distribution', 1)
('definitions', 1)
('version', 1)
('for', 2)
('use', 2)
('1', 1)
...
-------------------------------------------
Time: 2020-04-12 16:54:46
-------------------------------------------
and distribution as defined by sections 1 through 9 of this document
-------------------------------------------
Time: 2020-04-12 16:54:47
-------------------------------------------
-------------------------------------------
Time: 2020-04-12 16:54:48
-------------------------------------------
licensor shall mean the copyright owner or entity authorized by
-------------------------------------------
Time: 2020-04-12 16:54:49
-------------------------------------------
the copyright owner that is granting the license
-------------------------------------------
Time: 2020-04-12 16:54:50
-------------------------------------------
-------------------------------------------
Time: 2020-04-12 16:54:51
-------------------------------------------
legal entity shall mean the union of the acting entity and all
-------------------------------------------
Time: 2020-04-12 16:54:52
-------------------------------------------
other entities that control are controlled by or are under common
-------------------------------------------
Time: 2020-04-12 16:54:53
-------------------------------------------
control with that entity for the purposes of this definition
-------------------------------------------
Time: 2020-04-12 16:54:54
-------------------------------------------
control means i the power direct or indirect to cause the
-------------------------------------------
Time: 2020-04-12 16:54:55
-------------------------------------------
direction or management of such entity whether by contract or
-------------------------------------------
Time: 2020-04-12 16:54:55
-------------------------------------------
('common', 1)
('purposes', 1)
('by', 4)
('through', 1)
('this', 2)
('means', 1)
('management', 1)
('contract', 1)
('2004', 1)
('httpwwwapacheorglicenses', 1)
...
-------------------------------------------
Time: 2020-04-12 16:54:56
-------------------------------------------
otherwise or ii ownership of fifty percent 50 or more of the
-------------------------------------------
Time: 2020-04-12 16:54:57
-------------------------------------------
outstanding shares or iii beneficial ownership of such entity
-------------------------------------------
Time: 2020-04-12 16:54:58
-------------------------------------------
-------------------------------------------
Time: 2020-04-12 16:54:59
-------------------------------------------
you or your shall mean an individual or legal entity
-------------------------------------------
Time: 2020-04-12 16:55:00
-------------------------------------------
exercising permissions granted by this license
-------------------------------------------
Time: 2020-04-12 16:55:01
-------------------------------------------
-------------------------------------------
Time: 2020-04-12 16:55:02
-------------------------------------------
source form shall mean the preferred form for making modifications
-------------------------------------------
Time: 2020-04-12 16:55:03
-------------------------------------------
including but not limited to software source code documentation
-------------------------------------------
Time: 2020-04-12 16:55:04
-------------------------------------------
source and configuration files
-------------------------------------------
Time: 2020-04-12 16:55:05
-------------------------------------------
|
notebook/DL_image_object_detection/image_object_detection.ipynb
|
###Markdown
物体检测案例在本案例将学习使用深度学习技术来构建一个物体检测模型,并将其应用在华为云吉祥物“云宝”检测的任务中。物体检测技术目前广泛应用于对场景中物体的快速识别和精确定位,如:商品检测、自动驾驶等应用。聪明可爱的“云宝”是华为云的吉祥物。这个数据集中包含大量的云宝照片,并且标注了云宝目标的位置坐标信息,如下图,标注规范同Pascal VOC目标检测数据集。其中左边是未标注的图片,右边是已标注的图片,图中的绿色长方形框就是标注框。本案例将掌握目标检测以及yolo-v3算法的原理;掌握使用深度学习框架MXNet构建目标检测模型结构、训练模型、推理模型的方法;掌握从数据上传、代码编写、模型训练,到模型部署、推理,构建目标检测应用的全套流程 步骤 进入ModelArts界面这步教大家如何进入华为云ModelArts服务,ModelArts服务提供本次实验所需的环境。第一步:点击“控制台”第二步:点击“所有服务”第三步:在“EI企业智能”大类下找到“ModelArts”,点击“ModelArts”,进入ModelArts服务主界面第四步:看到以下界面,说明成功进入了ModelArts服务主界面 创建ModelArts notebook此步教大家如何在ModelArts中创建一个notebook开发环境。ModelArts notebook提供网页版的Python开发环境,无需用户自己搭建Python开发环境。第一步:点击ModelArts服务主界面中的“开发环境”第二步:点击“创建”按钮第三步:填写创建notebook所需的参数,参数设置参考下面列表。并点击下一步 | 参数 | 说明 ||--|--||计费方式|按需计费||名称 | 开发环境实例名称,如object_detection ||AI引擎|本案例使用MXNet引擎,Python版本对应3.6及以上||资源池|选择“公共资源池”即可||类型|GPU||规格|8核&124;64GiB&124;1*p100||存储配置|EVS,磁盘规格5GB| 第四步:点击“立即创建”第五步:点击“返回Notebook列表”第六步:等待Notebook的创建成功,创建成功后状态会变成“运行中” 在ModelArts notebook中创建一个notebook Python开发环境第一步:点击下图所示的“打开”按钮,进入刚刚创建的notebook第二步:创建一个notebook Python语言开发环境。先点击“New”按钮,然后根据本案例所用的AI引擎选择对应环境。第三步:重命名刚刚创建的notebook Python开发环境。点击“Untitle”按钮第四步:填写名称。我们可以填写一个跟本实验相关的名称,如下图所示,然后点击“Rename”按钮 如何在notebook Python开发环境中写代码并执行代码第一步:输入代码。我们打印一行字符串,如下图所示第二步:执行代码。代码输入完成后,点击notebook界面上的“Run”按钮,就可以执行代码第三步:查看代码执行结果。在代码输入框下面,可以看到代码执行结果,如下图所示第四步:保存代码。代码编写完之后,我们点击下图所示的“保存”按钮,保存代码和代码执行结果 案例配置信息填写案例中需要将运行结果上传至OBS中,我们需要设置以下相关参数(使用自己真实的桶名和唯一ID替换掉*号):* BUCKET_NAME : 自己的OBS桶名* UNIQUE_ID : 唯一ID,填写自己的学号或者IAM子账号名称
###Code
BUCKET_NAME = '*'
UNIQUE_ID = '*'
OBS_BASE_PATH = BUCKET_NAME + '/' + UNIQUE_ID
###Output
_____no_output_____
###Markdown
准备源代码和数据这一步准备案例所需的源代码和数据,相关资源已经保存在OBS中,我们通过ModelArts SDK将资源下载到本地,并解压到当前目录下。解压后,当前目录包含src,data,model以及checkpoint四个目录,分别存有源代码,数据集,模型数据和模型生成记录。
###Code
from modelarts.session import Session
session = Session()
if sess.region_name == 'cn-north-1':
session.download_data(bucket_path="ai-course-common-20/object_detection/object_detection.tar.gz", path="./object_detection.tar.gz")
elif sess.region_name == 'cn-north-4':
session.download_data(bucket_path="ai-course-common-20-bj4/object_detection/object_detection.tar.gz", path="./object_detection.tar.gz")
else:
print("请更换地区到北京一或北京四")
# 使用tar命令解压资源包
!tar xf ./object_detection.tar.gz
###Output
Successfully download file ai-course-common-20/object_detection/object_detection.tar.gz from OBS to local ./object_detection.tar.gz
###Markdown
**notebook Python开发环境终于准备好了,现在可以在notebook Python开发环境写代码啦** 导入基本工具库执行下面方框中的这段代码,可以导入本次实验中使用的Python开发基本工具库,并下载样例中使用的数据和基础模型到notebook开发环境本地目录,同时设置全局变量。此段打印从OBS下载数据的日志。
###Code
import os
os.environ['MXNET_BACKWARD_DO_MIRROR'] = '1'
import warnings
warnings.filterwarnings('ignore')
import mxnet as mx # mxnet引擎包
import cv2 # opencv-python 软件包
import logging # 日志工具
import numpy as np # python 科学计算软件包
import matplotlib.pyplot as plt # 绘图工具
import random # python随机函数库
# 在notebook中显示matplotlib.pyplot的绘图结果
%matplotlib inline
logging.basicConfig(level=logging.INFO) # 日志打印级别
BATCH_SIZE = 1 # 批量训练的大小
NUM_CLASSES = 1 # 分类数
NUM_EXAMPLES = 20 # 总数据量
OFFSETS = [(13, 13), (26, 26), (52, 52)] # 模型中需要用到的偏移量
ANCHORS = [[116, 90, 156, 198, 373, 326], # 不同形状目标框模板
[30, 61, 62, 45, 59, 119],
[10, 13, 16, 30, 33, 23]]
###Output
_____no_output_____
###Markdown
设置文件路径
###Code
# 数据集本地路径
data_path = './data/'
# 基础模型darknet53的本地保存位置
base_model_prefix = './model/darknet_53'
# 训练结果本地保存位置
checkpoint_prefix = './checkpoint/yolov3_yunbao'
###Output
_____no_output_____
###Markdown
读取数据集文件src/data/yolo_dataset.py定义了get_data_iter接口,返回基于mx.gluon.data.DataLoader的数据迭代器。此方法封装了数据增强操作包括,随机扩张、裁剪、翻转、归一化等。此段打印数据标签预读日志。
###Code
from src.data.yolo_dataset import get_data_iter
train_data,_ = get_data_iter(data_path=data_path, # 数据根目录
train_file=None, # 训练数据列表,可以为空
hyper_train={'width': 416, # 输入图片的宽
'height': 416, # 输入图片的高
'batch_size': BATCH_SIZE, # 批量读取数据的
'index_file': './index', # 标签名文件,如果不存在,则自动生成
'shuffle': True, # 打乱数据
'preprocess_threads': 0}, # 读数据进程数,大于0时需要使用系统共享内存
hyper_val={'width': 416, # 输入图片的宽
'height': 416, # 输入图片的高
'batch_size': BATCH_SIZE, # 批量读取数据的
'index_file': './index', # 标签名文件,如果不存在,则自动生成
'preprocess_threads': 0}, # 读数据进程数,大于0时需要使用系统共享内存
anchors=ANCHORS, # 不同形状目标框模板
offsets=OFFSETS # 模型中需要用到的偏移量
)
###Output
_____no_output_____
###Markdown
打印读取的第一张图片 获取第一个batch的第一张图片。此段代码打印出预处理过后的图片。
###Code
# 从迭代器获取数据
next_data_batch = next(iter(train_data))
img = next_data_batch.data[0][0].transpose(axes=(2,1,0))
img = img.asnumpy().astype('uint8')
# 画图,数据增强、归一化后的图片
fig = plt.figure()
plt.imshow(img)
plt.show()
###Output
_____no_output_____
###Markdown
打印读取的第一个标签打印图像标签。我们这样定义一个目标检测标签的格式:\[xmin, ymin, xmax, ymax, cls, difficult\],其中(xmin, ymin)是左上角坐标,(xmax, ymax)是右下角坐标,cls是类别标签编号,difficult标识是否是难训练样本。此段代码打印如上格式的标签信息。
###Code
# 打印label,[xmin, ymin, xmax, ymax, cls, difficult]
print (train_data._dataset.label_cache[11])
###Output
[[ 334. 93. 805. 690. 0. 0.]]
###Markdown
神经网络结构搭建src/symbol/yolov3.py文件详细定义了mxnet-yolov3的网络结构,其中get_feature函数返回特征提取结果,get_yolo_output函数获得具体检测结果包括检测框的位置信息、分类信息、置信度等,get_loss定义了训练时的损失函数,pred_generator定义了推理时模型返回结果。此段无输出。
###Code
from src.symbol.yolov3 import get_symbol
# get_symbol函数返回mx.sym.Symbol类型的变量
yolo_v3_net = get_symbol(num_classes=NUM_CLASSES, # 模型分类数
is_train=True) # 是否用于训练任务
###Output
_____no_output_____
###Markdown
训练 第一步:新建mxnet训练模块可以将mxnet.mod.Module模块看做一个计算引擎,它包含了计算图生成和绑定、参数初始化、前向计算、后向计算、参数更新、模型保存、日志打印等相关逻辑。初始化该模块需要传入模型结构以及计算使用的硬件环境,详见http://mxnet.incubator.apache.org/api/python/module/module.htmlmxnet.module.Module 。此段代码会打印出模型训练时需要更新的参数名。
###Code
# 设置gpu计算。
devs = mx.gpu(0)
model = mx.mod.Module(context=devs, # 计算设备
symbol=yolo_v3_net, # 模型结构
data_names=['data'], # 模型中使用的输入数据名称
label_names=['gt_boxes', 'obj_t', 'centers_t', 'scales_t', 'weights_t', 'clas_t']) # 模型中使用的输入标签名称
# 打印模型训练中参与更新的参数
print (model._param_names)
###Output
['conv_0_weight', 'bn_0_gamma', 'bn_0_beta', 'conv_1_weight', 'bn_1_gamma', 'bn_1_beta', 'conv_2_weight', 'bn_2_gamma', 'bn_2_beta', 'conv_3_weight', 'bn_3_gamma', 'bn_3_beta', 'conv_4_weight', 'bn_4_gamma', 'bn_4_beta', 'conv_5_weight', 'bn_5_gamma', 'bn_5_beta', 'conv_6_weight', 'bn_6_gamma', 'bn_6_beta', 'conv_7_weight', 'bn_7_gamma', 'bn_7_beta', 'conv_8_weight', 'bn_8_gamma', 'bn_8_beta', 'conv_9_weight', 'bn_9_gamma', 'bn_9_beta', 'conv_10_weight', 'bn_10_gamma', 'bn_10_beta', 'conv_11_weight', 'bn_11_gamma', 'bn_11_beta', 'conv_12_weight', 'bn_12_gamma', 'bn_12_beta', 'conv_13_weight', 'bn_13_gamma', 'bn_13_beta', 'conv_14_weight', 'bn_14_gamma', 'bn_14_beta', 'conv_15_weight', 'bn_15_gamma', 'bn_15_beta', 'conv_16_weight', 'bn_16_gamma', 'bn_16_beta', 'conv_17_weight', 'bn_17_gamma', 'bn_17_beta', 'conv_18_weight', 'bn_18_gamma', 'bn_18_beta', 'conv_19_weight', 'bn_19_gamma', 'bn_19_beta', 'conv_20_weight', 'bn_20_gamma', 'bn_20_beta', 'conv_21_weight', 'bn_21_gamma', 'bn_21_beta', 'conv_22_weight', 'bn_22_gamma', 'bn_22_beta', 'conv_23_weight', 'bn_23_gamma', 'bn_23_beta', 'conv_24_weight', 'bn_24_gamma', 'bn_24_beta', 'conv_25_weight', 'bn_25_gamma', 'bn_25_beta', 'conv_26_weight', 'bn_26_gamma', 'bn_26_beta', 'conv_27_weight', 'bn_27_gamma', 'bn_27_beta', 'conv_28_weight', 'bn_28_gamma', 'bn_28_beta', 'conv_29_weight', 'bn_29_gamma', 'bn_29_beta', 'conv_30_weight', 'bn_30_gamma', 'bn_30_beta', 'conv_31_weight', 'bn_31_gamma', 'bn_31_beta', 'conv_32_weight', 'bn_32_gamma', 'bn_32_beta', 'conv_33_weight', 'bn_33_gamma', 'bn_33_beta', 'conv_34_weight', 'bn_34_gamma', 'bn_34_beta', 'conv_35_weight', 'bn_35_gamma', 'bn_35_beta', 'conv_36_weight', 'bn_36_gamma', 'bn_36_beta', 'conv_37_weight', 'bn_37_gamma', 'bn_37_beta', 'conv_38_weight', 'bn_38_gamma', 'bn_38_beta', 'conv_39_weight', 'bn_39_gamma', 'bn_39_beta', 'conv_40_weight', 'bn_40_gamma', 'bn_40_beta', 'conv_41_weight', 'bn_41_gamma', 'bn_41_beta', 'conv_42_weight', 'bn_42_gamma', 'bn_42_beta', 'conv_43_weight', 'bn_43_gamma', 'bn_43_beta', 'conv_44_weight', 'bn_44_gamma', 'bn_44_beta', 'conv_45_weight', 'bn_45_gamma', 'bn_45_beta', 'conv_46_weight', 'bn_46_gamma', 'bn_46_beta', 'conv_47_weight', 'bn_47_gamma', 'bn_47_beta', 'conv_48_weight', 'bn_48_gamma', 'bn_48_beta', 'conv_49_weight', 'bn_49_gamma', 'bn_49_beta', 'conv_50_weight', 'bn_50_gamma', 'bn_50_beta', 'conv_51_weight', 'bn_51_gamma', 'bn_51_beta', 'conv_52_weight', 'bn_52_gamma', 'bn_52_beta', 'conv_53_weight', 'bn_53_gamma', 'bn_53_beta', 'conv_54_weight', 'bn_54_gamma', 'bn_54_beta', 'conv_55_weight', 'bn_55_gamma', 'bn_55_beta', 'conv_56_weight', 'bn_56_gamma', 'bn_56_beta', 'conv_57_weight', 'bn_57_gamma', 'bn_57_beta', 'conv_output_0_weight', 'conv_58_weight', 'bn_58_gamma', 'bn_58_beta', 'conv_59_weight', 'bn_59_gamma', 'bn_59_beta', 'conv_60_weight', 'bn_60_gamma', 'bn_60_beta', 'conv_61_weight', 'bn_61_gamma', 'bn_61_beta', 'conv_62_weight', 'bn_62_gamma', 'bn_62_beta', 'conv_63_weight', 'bn_63_gamma', 'bn_63_beta', 'conv_64_weight', 'bn_64_gamma', 'bn_64_beta', 'conv_output_1_weight', 'conv_65_weight', 'bn_65_gamma', 'bn_65_beta', 'conv_66_weight', 'bn_66_gamma', 'bn_66_beta', 'conv_67_weight', 'bn_67_gamma', 'bn_67_beta', 'conv_68_weight', 'bn_68_gamma', 'bn_68_beta', 'conv_69_weight', 'bn_69_gamma', 'bn_69_beta', 'conv_70_weight', 'bn_70_gamma', 'bn_70_beta', 'conv_71_weight', 'bn_71_gamma', 'bn_71_beta', 'conv_output_2_weight', 'offset_0_weight', 'anchors_0_weight', 'offset_1_weight', 'anchors_1_weight', 'offset_2_weight', 'anchors_2_weight']
###Markdown
第二步:优化器设置文件src/utils/lr_schedular.py中定义了学习率的变化方式,WarmUpMultiFactorScheduler类使得学习率按如下图趋势变化:横坐标是epoch数,纵坐标是学习率。此段代码无回显。
###Code
from src.utils.lr_schedular import WarmUpMultiFactorScheduler
lr_steps = '20,30' # 学习率下降的epoch数
lr = 0.00001 # 学习率
wd = 0.00001 # 权值衰减系数
mom = 0.9 # 动量参数
warm_up_epochs = 1 # 学习率‘热身’的epoch数
# 根据BATCH_SIZE计算与epoch对应的step数
epoch_size = NUM_EXAMPLES // BATCH_SIZE
step_epochs = [float(l) for l in lr_steps.split(',')]
steps = [int(epoch_size * x) for x in step_epochs]
warmup_steps = int(epoch_size * warm_up_epochs)
# lr变化策略,带warmup的MultiFactor
lr_scheduler = WarmUpMultiFactorScheduler(step=steps, # 学习率下降的步数位置
factor=0.1, # 学习率下降的比率
base_lr=lr, # 基础学习率
warmup_steps=warmup_steps, # 学习率‘热身’提高的步数
warmup_begin_lr=0.0, # 起始学习率
warmup_mode='linear') # 学习率‘热身’提升的方式,这里设置为线性提升
# 将优化器参数汇总成一个python字典
optimizer_params = {'learning_rate': lr,
'wd': wd,
'lr_scheduler': lr_scheduler,
'rescale_grad': (1.0 / (BATCH_SIZE)),
'multi_precision': True,
'momentum': mom}
###Output
_____no_output_____
###Markdown
第三步:初始化函数针对网络中使用的offset、anchor参数,采用常量初始化,在训练过程中这些参数不参与更新;神经网络中的其它参数(权重、偏置等),使用Xavier算法初始化。此段代码无回显。
###Code
# 将目标框扩展成所需的shape
def get_anchors(anchors):
anchor_list = []
for item in anchors:
anchor = mx.nd.array(item)
anchor = anchor.reshape(1, 1, -1, 2)
anchor_list.append(anchor)
return anchor_list
# 将偏移量扩展成所需的shape
def get_offsets(offsets):
offset_list = []
for item in offsets:
grid_x = np.arange(item[1])
grid_y = np.arange(item[0])
grid_x, grid_y = np.meshgrid(grid_x, grid_y)
offset = np.concatenate((grid_x[:, :, np.newaxis], grid_y[:, :, np.newaxis]), axis=-1)
offset = np.expand_dims(np.expand_dims(offset, axis=0), axis=0)
offset = mx.nd.array(offset).reshape(1, -1, 1, 2)
offset_list.append(offset)
return offset_list
offset_data = get_offsets(OFFSETS)
anchor_data = get_anchors(ANCHORS)
# offset、anchor参数,采用常量初始化;其它参数采用Xavier初始化
initializer = mx.init.Mixed(['offset_0_weight', 'offset_1_weight', 'offset_2_weight',
'anchors_0_weight', 'anchors_1_weight', 'anchors_2_weight',
'.*'],
[mx.init.Constant(offset_data[0]),
mx.init.Constant(offset_data[1]),
mx.init.Constant(offset_data[2]),
mx.init.Constant(anchor_data[0]),
mx.init.Constant(anchor_data[1]),
mx.init.Constant(anchor_data[2]),
mx.init.Xavier(factor_type="in", magnitude=2.34)])
###Output
_____no_output_____
###Markdown
第四步:加载基础网络darknet53的参数(基于imagenet训练)imagenet是图像识别领域最大的数据库。darknet53是一个物体检测神经网络。此段代码打印基础网络darknet53的完整结构。
###Code
epoch = 0 # 模型保存编号
sym_darknet_53, arg, aux = mx.model.load_checkpoint(base_model_prefix, epoch) # 参数保存在arg_params,aux_params中
# 打印darknet53模型结构
mx.viz.plot_network(sym_darknet_53)
###Output
_____no_output_____
###Markdown
第五步:设置回调函数回调函数中定义每一个step训练完后的操作和每一个epoch训练完后的操作,其中一个step指完成一个batch size大小数据的训练,一个epoch是指完成一遍整个数据集的训练。此段代码无回显。
###Code
# 设置参数同步方式
kv = mx.kvstore.create('local')
# 设置Loss打印
from src.utils.yolo_metric import YoloLoss
metrics = [YoloLoss(name='ObjLoss', index=0),
YoloLoss(name='BoxCenterLoss', index=1),
YoloLoss(name='BoxScaleLoss', index=2),
YoloLoss(name='ClassLoss', index=3)]
# 回调函数
# 设置日志打印频率,通常每个step回调一次
batch_end_callback = [mx.callback.Speedometer(BATCH_SIZE, 1, auto_reset=False)]
# 设置模型保存位置、频率,同常每个epoch回调一次
epoch_end_callback = [mx.callback.do_checkpoint(checkpoint_prefix, 5)]
###Output
_____no_output_____
###Markdown
第六步:开始训练调用model对象的fit函数开始训练,20个epoch大概需要运行5分钟左右完成。此段代码无输出。
###Code
# 把前面设置的参数放入fit函数中开始训练
model.fit(train_data=train_data, # 训练数据集
optimizer='sgd', # 优化器类型
kvstore=kv, # 设置参数同步方式
optimizer_params=optimizer_params, # 优化器参数
batch_end_callback=batch_end_callback, # 每个step结束的回调函数
epoch_end_callback=epoch_end_callback, # 每个epoch结束的回调函数
initializer=initializer, # 参数初始化函数
arg_params=arg, # 预加载的参数
aux_params=aux, # 预加载的参数
eval_metric=metrics, # 设置Loss打印
num_epoch=20, # 设置遍历数据集次数
allow_missing=True) # 是否允许预加载的参数部分缺失
###Output
_____no_output_____
###Markdown
第七步:上传模型至OBS使用ModelArts SDK将模型上传至OBS保存,方便以后使用。
###Code
session.upload_data(bucket_path=OBS_BASE_PATH + "/object_detection", path= "./model")
###Output
/home/jovyan/modelarts-sdk/obs/client.py:498: DeprecationWarning: key_file, cert_file and check_hostname are deprecated, use a custom context instead.
conn = httplib.HTTPSConnection(server, port=port, timeout=self.timeout, context=self.context, check_hostname=False)
###Markdown
总结:该案例的model目录(OBS路径)的下存在保存的模型文件。 推理 第一步:读取图片从obs上读取1张图片,进行归一化,并将其打包成mxnet引擎能识别的mx.io.DataBatch类型。此段代码会打印读取的图片。
###Code
# 图片保存位置
file_name = os.path.join(data_path, 'train/IMG_20180919_115501.jpg')
# 图像输入的长宽
h=416
w=416
# 图像归一化需要的均值和方差
mean=(0.485, 0.456, 0.406)
std=(0.229, 0.224, 0.225)
# 加载moxing库用于直接读取保存在obs上的图片
import moxing as mox
# 以二进制形式,将图片读入内存
img = mox.file.read(file_name, binary=True)
# 保存原图,用于展示
orig_img = mx.img.imdecode(img, to_rgb=1).asnumpy().astype('uint8')
plt.imshow(orig_img)
plt.show()
# 图像预处理,归一化,resize,用于模型推理
img = mx.img.imdecode(img, 1)
img = mx.image.imresize(img, w, h, interp=2)
img = mx.nd.image.to_tensor(img)
img = mx.nd.image.normalize(img, mean=mean, std=std).expand_dims(0)
# 准备输入数据
provide_data = [mx.io.DataDesc(name='data', shape=(1, 3, h, w))]
batch_data = mx.io.DataBatch(data=[img], provide_data=provide_data)
###Output
_____no_output_____
###Markdown
第二步:加载训练完成的模型文件加载前面训练出的模型,模型保存在本地路径checkpoint_prefix中。此段代码无回显。
###Code
load_epoch = 20 # 模型保存编号,请根据实际情况设置
_, arg_params, aux_params = mx.model.load_checkpoint(checkpoint_prefix, load_epoch) # 参数保存在arg_params,aux_params中
###Output
_____no_output_____
###Markdown
第三步:加载推理使用的模型结构与训练不同,此时的模型不再包含损失函数,模型输出也被整理成\[分类结果,置信度,边框坐标\]的形式。模型参数不变。此段打印yolov3推理时模型结构。
###Code
from src.symbol.yolov3 import get_symbol
yolo_v3_net_pre = get_symbol(num_classes=NUM_CLASSES, # 模型分类数
is_train=False) # 模型结构用于推理
# 打印模型结构
mx.viz.plot_network(yolo_v3_net_pre)
###Output
_____no_output_____
###Markdown
第四步:新建mxnet推理模块同样是mxnet.mod.Module模块,相比训练时,此时的计算逻辑去除了反向传播和参数更新。此段代码无回显。
###Code
devs = mx.gpu(0)
model_pre = mx.mod.Module(
context=mx.gpu(), # 使用gpu进行推理
symbol=yolo_v3_net_pre, # 推理模型结构
data_names=['data'], # 输入数据的名称,与前面准备输入数据时provide_data中的名称相同
label_names=None) # 推理无需输入label
model_pre.bind(for_training=False, #绑定计算模块到底层计算引擎
data_shapes=batch_data.provide_data,
label_shapes=None)
model_pre.set_params(arg_params, aux_params) # 将下载的训练完成的参数加载进计算模块
###Output
_____no_output_____
###Markdown
第五步:执行推理计算此段代码无回显。
###Code
model_pre.forward(batch_data, # 输入数据
is_train=False) # 控制引擎只进行前向计算
pred = model_pre.get_outputs() # 获得推理结果
ids, scores, bboxes = [xx[0].asnumpy() for xx in pred]
###Output
_____no_output_____
###Markdown
第六步:解析并绘制结果会输出推理结果。根据设置的置信度,可能会输出多个矩形框。由于训练数据量少,训练时间短,推理任务加载的模型精度较低,画出的目标框会存在不准确的现象。适当调整优化器参数,延长训练步数会提升模型精度。此段打印带预测目标框的推理结果。
###Code
THRESH = 0.05 # 置信度阈值
im_array = orig_img.copy()
color_white = (255, 255, 255)
x_scale = im_array.shape[1] / 416.0
y_scale = im_array.shape[0] / 416.0
for idx in range(ids.shape[0]):
if ids[idx][0] < 0 or scores[idx][0] < THRESH:
continue
# 按原图比例缩放目标框
bbox = [int(bboxes[idx][0] * x_scale),
int(bboxes[idx][1] * y_scale),
int(bboxes[idx][2] * x_scale),
int(bboxes[idx][3] * y_scale)]
color = (random.randint(0, 256), random.randint(0, 256), random.randint(0, 256))
# 在图像上画框
cv2.rectangle(im_array, (bbox[0], bbox[1]), (bbox[2], bbox[3]), color=color, thickness=2)
# 把置信度写在图像上
cv2.putText(im_array, '%.3f' % (scores[idx][0]), (bbox[0], bbox[1] + 10),
color=color_white, fontFace=cv2.FONT_HERSHEY_COMPLEX, fontScale=0.5)
plt.imshow(im_array)
plt.show()
###Output
_____no_output_____
###Markdown
物体检测案例在本案例将学习使用深度学习技术来构建一个物体检测模型,并将其应用在华为云吉祥物“云宝”检测的任务中。物体检测技术目前广泛应用于对场景中物体的快速识别和精确定位,如:商品检测、自动驾驶等应用。聪明可爱的“云宝”是华为云的吉祥物。这个数据集中包含大量的云宝照片,并且标注了云宝目标的位置坐标信息,如下图,标注规范同Pascal VOC目标检测数据集。其中左边是未标注的图片,右边是已标注的图片,图中的绿色长方形框就是标注框。本案例将掌握目标检测以及yolo-v3算法的原理;掌握使用深度学习框架MXNet构建目标检测模型结构、训练模型、推理模型的方法;掌握从数据上传、代码编写、模型训练,到模型部署、推理,构建目标检测应用的全套流程 步骤 进入ModelArts界面这步教大家如何进入华为云ModelArts服务,ModelArts服务提供本次实验所需的环境。第一步:点击“控制台”第二步:点击“所有服务”第三步:在“EI企业智能”大类下找到“ModelArts”,点击“ModelArts”,进入ModelArts服务主界面第四步:看到以下界面,说明成功进入了ModelArts服务主界面 创建ModelArts notebook此步教大家如何在ModelArts中创建一个notebook开发环境。ModelArts notebook提供网页版的Python开发环境,无需用户自己搭建Python开发环境。第一步:点击ModelArts服务主界面中的“开发环境”第二步:点击“创建”按钮第三步:填写创建notebook所需的参数,参数设置参考下面列表。并点击下一步 | 参数 | 说明 ||--|--||计费方式|按需计费||名称 | 开发环境实例名称,如object_detection ||AI引擎|本案例使用MXNet引擎,Python版本对应3.6及以上||资源池|选择“公共资源池”即可||类型|GPU||规格|8核&124;64GiB&124;1*p100||存储配置|EVS,磁盘规格5GB| 第四步:点击“立即创建”第五步:点击“返回Notebook列表”第六步:等待Notebook的创建成功,创建成功后状态会变成“运行中” 在ModelArts notebook中创建一个notebook Python开发环境第一步:点击下图所示的“打开”按钮,进入刚刚创建的notebook第二步:创建一个notebook Python语言开发环境。先点击“New”按钮,然后根据本案例所用的AI引擎选择对应环境。第三步:重命名刚刚创建的notebook Python开发环境。点击“Untitle”按钮第四步:填写名称。我们可以填写一个跟本实验相关的名称,如下图所示,然后点击“Rename”按钮 如何在notebook Python开发环境中写代码并执行代码第一步:输入代码。我们打印一行字符串,如下图所示第二步:执行代码。代码输入完成后,点击notebook界面上的“Run”按钮,就可以执行代码第三步:查看代码执行结果。在代码输入框下面,可以看到代码执行结果,如下图所示第四步:保存代码。代码编写完之后,我们点击下图所示的“保存”按钮,保存代码和代码执行结果 案例配置信息填写案例中需要将运行结果上传至OBS中,我们需要设置以下相关参数(使用自己真实的桶名和唯一ID替换掉*号):* BUCKET_NAME : 自己的OBS桶名* UNIQUE_ID : 唯一ID,填写自己的学号或者IAM子账号名称
###Code
BUCKET_NAME = '*'
UNIQUE_ID = '*'
OBS_BASE_PATH = BUCKET_NAME + '/' + UNIQUE_ID
###Output
_____no_output_____
###Markdown
准备源代码和数据这一步准备案例所需的源代码和数据,相关资源已经保存在OBS中,我们通过ModelArts SDK将资源下载到本地,并解压到当前目录下。解压后,当前目录包含src,data,model以及checkpoint四个目录,分别存有源代码,数据集,模型数据和模型生成记录。
###Code
from modelarts.session import Session
session = Session()
session.download_data(bucket_path="ai-course-common-20/object_detection/object_detection.tar.gz", path="./object_detection.tar.gz")
# 使用tar命令解压资源包
!tar xf ./object_detection.tar.gz
###Output
Successfully download file ai-course-common-20/object_detection/object_detection.tar.gz from OBS to local ./object_detection.tar.gz
###Markdown
**notebook Python开发环境终于准备好了,现在可以在notebook Python开发环境写代码啦** 导入基本工具库执行下面方框中的这段代码,可以导入本次实验中使用的Python开发基本工具库,并下载样例中使用的数据和基础模型到notebook开发环境本地目录,同时设置全局变量。此段打印从OBS下载数据的日志。
###Code
import os
os.environ['MXNET_BACKWARD_DO_MIRROR'] = '1'
import warnings
warnings.filterwarnings('ignore')
import mxnet as mx # mxnet引擎包
import cv2 # opencv-python 软件包
import logging # 日志工具
import numpy as np # python 科学计算软件包
import matplotlib.pyplot as plt # 绘图工具
import random # python随机函数库
# 在notebook中显示matplotlib.pyplot的绘图结果
%matplotlib inline
logging.basicConfig(level=logging.INFO) # 日志打印级别
BATCH_SIZE = 1 # 批量训练的大小
NUM_CLASSES = 1 # 分类数
NUM_EXAMPLES = 20 # 总数据量
OFFSETS = [(13, 13), (26, 26), (52, 52)] # 模型中需要用到的偏移量
ANCHORS = [[116, 90, 156, 198, 373, 326], # 不同形状目标框模板
[30, 61, 62, 45, 59, 119],
[10, 13, 16, 30, 33, 23]]
###Output
_____no_output_____
###Markdown
设置文件路径
###Code
# 数据集本地路径
data_path = './data/'
# 基础模型darknet53的本地保存位置
base_model_prefix = './model/darknet_53'
# 训练结果本地保存位置
checkpoint_prefix = './checkpoint/yolov3_yunbao'
###Output
_____no_output_____
###Markdown
读取数据集文件src/data/yolo_dataset.py定义了get_data_iter接口,返回基于mx.gluon.data.DataLoader的数据迭代器。此方法封装了数据增强操作包括,随机扩张、裁剪、翻转、归一化等。此段打印数据标签预读日志。
###Code
from src.data.yolo_dataset import get_data_iter
train_data,_ = get_data_iter(data_path=data_path, # 数据根目录
train_file=None, # 训练数据列表,可以为空
hyper_train={'width': 416, # 输入图片的宽
'height': 416, # 输入图片的高
'batch_size': BATCH_SIZE, # 批量读取数据的
'index_file': './index', # 标签名文件,如果不存在,则自动生成
'shuffle': True, # 打乱数据
'preprocess_threads': 0}, # 读数据进程数,大于0时需要使用系统共享内存
hyper_val={'width': 416, # 输入图片的宽
'height': 416, # 输入图片的高
'batch_size': BATCH_SIZE, # 批量读取数据的
'index_file': './index', # 标签名文件,如果不存在,则自动生成
'preprocess_threads': 0}, # 读数据进程数,大于0时需要使用系统共享内存
anchors=ANCHORS, # 不同形状目标框模板
offsets=OFFSETS # 模型中需要用到的偏移量
)
###Output
_____no_output_____
###Markdown
打印读取的第一张图片 获取第一个batch的第一张图片。此段代码打印出预处理过后的图片。
###Code
# 从迭代器获取数据
next_data_batch = next(iter(train_data))
img = next_data_batch.data[0][0].transpose(axes=(2,1,0))
img = img.asnumpy().astype('uint8')
# 画图,数据增强、归一化后的图片
fig = plt.figure()
plt.imshow(img)
plt.show()
###Output
_____no_output_____
###Markdown
打印读取的第一个标签打印图像标签。我们这样定义一个目标检测标签的格式:\[xmin, ymin, xmax, ymax, cls, difficult\],其中(xmin, ymin)是左上角坐标,(xmax, ymax)是右下角坐标,cls是类别标签编号,difficult标识是否是难训练样本。此段代码打印如上格式的标签信息。
###Code
# 打印label,[xmin, ymin, xmax, ymax, cls, difficult]
print (train_data._dataset.label_cache[11])
###Output
[[ 334. 93. 805. 690. 0. 0.]]
###Markdown
神经网络结构搭建src/symbol/yolov3.py文件详细定义了mxnet-yolov3的网络结构,其中get_feature函数返回特征提取结果,get_yolo_output函数获得具体检测结果包括检测框的位置信息、分类信息、置信度等,get_loss定义了训练时的损失函数,pred_generator定义了推理时模型返回结果。此段无输出。
###Code
from src.symbol.yolov3 import get_symbol
# get_symbol函数返回mx.sym.Symbol类型的变量
yolo_v3_net = get_symbol(num_classes=NUM_CLASSES, # 模型分类数
is_train=True) # 是否用于训练任务
###Output
_____no_output_____
###Markdown
训练 第一步:新建mxnet训练模块可以将mxnet.mod.Module模块看做一个计算引擎,它包含了计算图生成和绑定、参数初始化、前向计算、后向计算、参数更新、模型保存、日志打印等相关逻辑。初始化该模块需要传入模型结构以及计算使用的硬件环境,详见http://mxnet.incubator.apache.org/api/python/module/module.htmlmxnet.module.Module 。此段代码会打印出模型训练时需要更新的参数名。
###Code
# 设置gpu计算。
devs = mx.gpu(0)
model = mx.mod.Module(context=devs, # 计算设备
symbol=yolo_v3_net, # 模型结构
data_names=['data'], # 模型中使用的输入数据名称
label_names=['gt_boxes', 'obj_t', 'centers_t', 'scales_t', 'weights_t', 'clas_t']) # 模型中使用的输入标签名称
# 打印模型训练中参与更新的参数
print (model._param_names)
###Output
['conv_0_weight', 'bn_0_gamma', 'bn_0_beta', 'conv_1_weight', 'bn_1_gamma', 'bn_1_beta', 'conv_2_weight', 'bn_2_gamma', 'bn_2_beta', 'conv_3_weight', 'bn_3_gamma', 'bn_3_beta', 'conv_4_weight', 'bn_4_gamma', 'bn_4_beta', 'conv_5_weight', 'bn_5_gamma', 'bn_5_beta', 'conv_6_weight', 'bn_6_gamma', 'bn_6_beta', 'conv_7_weight', 'bn_7_gamma', 'bn_7_beta', 'conv_8_weight', 'bn_8_gamma', 'bn_8_beta', 'conv_9_weight', 'bn_9_gamma', 'bn_9_beta', 'conv_10_weight', 'bn_10_gamma', 'bn_10_beta', 'conv_11_weight', 'bn_11_gamma', 'bn_11_beta', 'conv_12_weight', 'bn_12_gamma', 'bn_12_beta', 'conv_13_weight', 'bn_13_gamma', 'bn_13_beta', 'conv_14_weight', 'bn_14_gamma', 'bn_14_beta', 'conv_15_weight', 'bn_15_gamma', 'bn_15_beta', 'conv_16_weight', 'bn_16_gamma', 'bn_16_beta', 'conv_17_weight', 'bn_17_gamma', 'bn_17_beta', 'conv_18_weight', 'bn_18_gamma', 'bn_18_beta', 'conv_19_weight', 'bn_19_gamma', 'bn_19_beta', 'conv_20_weight', 'bn_20_gamma', 'bn_20_beta', 'conv_21_weight', 'bn_21_gamma', 'bn_21_beta', 'conv_22_weight', 'bn_22_gamma', 'bn_22_beta', 'conv_23_weight', 'bn_23_gamma', 'bn_23_beta', 'conv_24_weight', 'bn_24_gamma', 'bn_24_beta', 'conv_25_weight', 'bn_25_gamma', 'bn_25_beta', 'conv_26_weight', 'bn_26_gamma', 'bn_26_beta', 'conv_27_weight', 'bn_27_gamma', 'bn_27_beta', 'conv_28_weight', 'bn_28_gamma', 'bn_28_beta', 'conv_29_weight', 'bn_29_gamma', 'bn_29_beta', 'conv_30_weight', 'bn_30_gamma', 'bn_30_beta', 'conv_31_weight', 'bn_31_gamma', 'bn_31_beta', 'conv_32_weight', 'bn_32_gamma', 'bn_32_beta', 'conv_33_weight', 'bn_33_gamma', 'bn_33_beta', 'conv_34_weight', 'bn_34_gamma', 'bn_34_beta', 'conv_35_weight', 'bn_35_gamma', 'bn_35_beta', 'conv_36_weight', 'bn_36_gamma', 'bn_36_beta', 'conv_37_weight', 'bn_37_gamma', 'bn_37_beta', 'conv_38_weight', 'bn_38_gamma', 'bn_38_beta', 'conv_39_weight', 'bn_39_gamma', 'bn_39_beta', 'conv_40_weight', 'bn_40_gamma', 'bn_40_beta', 'conv_41_weight', 'bn_41_gamma', 'bn_41_beta', 'conv_42_weight', 'bn_42_gamma', 'bn_42_beta', 'conv_43_weight', 'bn_43_gamma', 'bn_43_beta', 'conv_44_weight', 'bn_44_gamma', 'bn_44_beta', 'conv_45_weight', 'bn_45_gamma', 'bn_45_beta', 'conv_46_weight', 'bn_46_gamma', 'bn_46_beta', 'conv_47_weight', 'bn_47_gamma', 'bn_47_beta', 'conv_48_weight', 'bn_48_gamma', 'bn_48_beta', 'conv_49_weight', 'bn_49_gamma', 'bn_49_beta', 'conv_50_weight', 'bn_50_gamma', 'bn_50_beta', 'conv_51_weight', 'bn_51_gamma', 'bn_51_beta', 'conv_52_weight', 'bn_52_gamma', 'bn_52_beta', 'conv_53_weight', 'bn_53_gamma', 'bn_53_beta', 'conv_54_weight', 'bn_54_gamma', 'bn_54_beta', 'conv_55_weight', 'bn_55_gamma', 'bn_55_beta', 'conv_56_weight', 'bn_56_gamma', 'bn_56_beta', 'conv_57_weight', 'bn_57_gamma', 'bn_57_beta', 'conv_output_0_weight', 'conv_58_weight', 'bn_58_gamma', 'bn_58_beta', 'conv_59_weight', 'bn_59_gamma', 'bn_59_beta', 'conv_60_weight', 'bn_60_gamma', 'bn_60_beta', 'conv_61_weight', 'bn_61_gamma', 'bn_61_beta', 'conv_62_weight', 'bn_62_gamma', 'bn_62_beta', 'conv_63_weight', 'bn_63_gamma', 'bn_63_beta', 'conv_64_weight', 'bn_64_gamma', 'bn_64_beta', 'conv_output_1_weight', 'conv_65_weight', 'bn_65_gamma', 'bn_65_beta', 'conv_66_weight', 'bn_66_gamma', 'bn_66_beta', 'conv_67_weight', 'bn_67_gamma', 'bn_67_beta', 'conv_68_weight', 'bn_68_gamma', 'bn_68_beta', 'conv_69_weight', 'bn_69_gamma', 'bn_69_beta', 'conv_70_weight', 'bn_70_gamma', 'bn_70_beta', 'conv_71_weight', 'bn_71_gamma', 'bn_71_beta', 'conv_output_2_weight', 'offset_0_weight', 'anchors_0_weight', 'offset_1_weight', 'anchors_1_weight', 'offset_2_weight', 'anchors_2_weight']
###Markdown
第二步:优化器设置文件src/utils/lr_schedular.py中定义了学习率的变化方式,WarmUpMultiFactorScheduler类使得学习率按如下图趋势变化:横坐标是epoch数,纵坐标是学习率。此段代码无回显。
###Code
from src.utils.lr_schedular import WarmUpMultiFactorScheduler
lr_steps = '20,30' # 学习率下降的epoch数
lr = 0.00001 # 学习率
wd = 0.00001 # 权值衰减系数
mom = 0.9 # 动量参数
warm_up_epochs = 1 # 学习率‘热身’的epoch数
# 根据BATCH_SIZE计算与epoch对应的step数
epoch_size = NUM_EXAMPLES // BATCH_SIZE
step_epochs = [float(l) for l in lr_steps.split(',')]
steps = [int(epoch_size * x) for x in step_epochs]
warmup_steps = int(epoch_size * warm_up_epochs)
# lr变化策略,带warmup的MultiFactor
lr_scheduler = WarmUpMultiFactorScheduler(step=steps, # 学习率下降的步数位置
factor=0.1, # 学习率下降的比率
base_lr=lr, # 基础学习率
warmup_steps=warmup_steps, # 学习率‘热身’提高的步数
warmup_begin_lr=0.0, # 起始学习率
warmup_mode='linear') # 学习率‘热身’提升的方式,这里设置为线性提升
# 将优化器参数汇总成一个python字典
optimizer_params = {'learning_rate': lr,
'wd': wd,
'lr_scheduler': lr_scheduler,
'rescale_grad': (1.0 / (BATCH_SIZE)),
'multi_precision': True,
'momentum': mom}
###Output
_____no_output_____
###Markdown
第三步:初始化函数针对网络中使用的offset、anchor参数,采用常量初始化,在训练过程中这些参数不参与更新;神经网络中的其它参数(权重、偏置等),使用Xavier算法初始化。此段代码无回显。
###Code
# 将目标框扩展成所需的shape
def get_anchors(anchors):
anchor_list = []
for item in anchors:
anchor = mx.nd.array(item)
anchor = anchor.reshape(1, 1, -1, 2)
anchor_list.append(anchor)
return anchor_list
# 将偏移量扩展成所需的shape
def get_offsets(offsets):
offset_list = []
for item in offsets:
grid_x = np.arange(item[1])
grid_y = np.arange(item[0])
grid_x, grid_y = np.meshgrid(grid_x, grid_y)
offset = np.concatenate((grid_x[:, :, np.newaxis], grid_y[:, :, np.newaxis]), axis=-1)
offset = np.expand_dims(np.expand_dims(offset, axis=0), axis=0)
offset = mx.nd.array(offset).reshape(1, -1, 1, 2)
offset_list.append(offset)
return offset_list
offset_data = get_offsets(OFFSETS)
anchor_data = get_anchors(ANCHORS)
# offset、anchor参数,采用常量初始化;其它参数采用Xavier初始化
initializer = mx.init.Mixed(['offset_0_weight', 'offset_1_weight', 'offset_2_weight',
'anchors_0_weight', 'anchors_1_weight', 'anchors_2_weight',
'.*'],
[mx.init.Constant(offset_data[0]),
mx.init.Constant(offset_data[1]),
mx.init.Constant(offset_data[2]),
mx.init.Constant(anchor_data[0]),
mx.init.Constant(anchor_data[1]),
mx.init.Constant(anchor_data[2]),
mx.init.Xavier(factor_type="in", magnitude=2.34)])
###Output
_____no_output_____
###Markdown
第四步:加载基础网络darknet53的参数(基于imagenet训练)imagenet是图像识别领域最大的数据库。darknet53是一个物体检测神经网络。此段代码打印基础网络darknet53的完整结构。
###Code
epoch = 0 # 模型保存编号
sym_darknet_53, arg, aux = mx.model.load_checkpoint(base_model_prefix, epoch) # 参数保存在arg_params,aux_params中
# 打印darknet53模型结构
mx.viz.plot_network(sym_darknet_53)
###Output
_____no_output_____
###Markdown
第五步:设置回调函数回调函数中定义每一个step训练完后的操作和每一个epoch训练完后的操作,其中一个step指完成一个batch size大小数据的训练,一个epoch是指完成一遍整个数据集的训练。此段代码无回显。
###Code
# 设置参数同步方式
kv = mx.kvstore.create('local')
# 设置Loss打印
from src.utils.yolo_metric import YoloLoss
metrics = [YoloLoss(name='ObjLoss', index=0),
YoloLoss(name='BoxCenterLoss', index=1),
YoloLoss(name='BoxScaleLoss', index=2),
YoloLoss(name='ClassLoss', index=3)]
# 回调函数
# 设置日志打印频率,通常每个step回调一次
batch_end_callback = [mx.callback.Speedometer(BATCH_SIZE, 1, auto_reset=False)]
# 设置模型保存位置、频率,同常每个epoch回调一次
epoch_end_callback = [mx.callback.do_checkpoint(checkpoint_prefix, 5)]
###Output
_____no_output_____
###Markdown
第六步:开始训练调用model对象的fit函数开始训练,20个epoch大概需要运行5分钟左右完成。此段代码无输出。
###Code
# 把前面设置的参数放入fit函数中开始训练
model.fit(train_data=train_data, # 训练数据集
optimizer='sgd', # 优化器类型
kvstore=kv, # 设置参数同步方式
optimizer_params=optimizer_params, # 优化器参数
batch_end_callback=batch_end_callback, # 每个step结束的回调函数
epoch_end_callback=epoch_end_callback, # 每个epoch结束的回调函数
initializer=initializer, # 参数初始化函数
arg_params=arg, # 预加载的参数
aux_params=aux, # 预加载的参数
eval_metric=metrics, # 设置Loss打印
num_epoch=20, # 设置遍历数据集次数
allow_missing=True) # 是否允许预加载的参数部分缺失
###Output
_____no_output_____
###Markdown
第七步:上传模型至OBS使用ModelArts SDK将模型上传至OBS保存,方便以后使用。
###Code
session.upload_data(bucket_path=OBS_BASE_PATH + "/object_detection", path= "./model")
###Output
/home/jovyan/modelarts-sdk/obs/client.py:498: DeprecationWarning: key_file, cert_file and check_hostname are deprecated, use a custom context instead.
conn = httplib.HTTPSConnection(server, port=port, timeout=self.timeout, context=self.context, check_hostname=False)
###Markdown
总结:该案例的model目录(OBS路径)的下存在保存的模型文件。 推理 第一步:读取图片从obs上读取1张图片,进行归一化,并将其打包成mxnet引擎能识别的mx.io.DataBatch类型。此段代码会打印读取的图片。
###Code
# 图片保存位置
file_name = os.path.join(data_path, 'train/IMG_20180919_115501.jpg')
# 图像输入的长宽
h=416
w=416
# 图像归一化需要的均值和方差
mean=(0.485, 0.456, 0.406)
std=(0.229, 0.224, 0.225)
# 加载moxing库用于直接读取保存在obs上的图片
import moxing as mox
# 以二进制形式,将图片读入内存
img = mox.file.read(file_name, binary=True)
# 保存原图,用于展示
orig_img = mx.img.imdecode(img, to_rgb=1).asnumpy().astype('uint8')
plt.imshow(orig_img)
plt.show()
# 图像预处理,归一化,resize,用于模型推理
img = mx.img.imdecode(img, 1)
img = mx.image.imresize(img, w, h, interp=2)
img = mx.nd.image.to_tensor(img)
img = mx.nd.image.normalize(img, mean=mean, std=std).expand_dims(0)
# 准备输入数据
provide_data = [mx.io.DataDesc(name='data', shape=(1, 3, h, w))]
batch_data = mx.io.DataBatch(data=[img], provide_data=provide_data)
###Output
_____no_output_____
###Markdown
第二步:加载训练完成的模型文件加载前面训练出的模型,模型保存在本地路径checkpoint_prefix中。此段代码无回显。
###Code
load_epoch = 20 # 模型保存编号,请根据实际情况设置
_, arg_params, aux_params = mx.model.load_checkpoint(checkpoint_prefix, load_epoch) # 参数保存在arg_params,aux_params中
###Output
_____no_output_____
###Markdown
第三步:加载推理使用的模型结构与训练不同,此时的模型不再包含损失函数,模型输出也被整理成\[分类结果,置信度,边框坐标\]的形式。模型参数不变。此段打印yolov3推理时模型结构。
###Code
from src.symbol.yolov3 import get_symbol
yolo_v3_net_pre = get_symbol(num_classes=NUM_CLASSES, # 模型分类数
is_train=False) # 模型结构用于推理
# 打印模型结构
mx.viz.plot_network(yolo_v3_net_pre)
###Output
_____no_output_____
###Markdown
第四步:新建mxnet推理模块同样是mxnet.mod.Module模块,相比训练时,此时的计算逻辑去除了反向传播和参数更新。此段代码无回显。
###Code
devs = mx.gpu(0)
model_pre = mx.mod.Module(
context=mx.gpu(), # 使用gpu进行推理
symbol=yolo_v3_net_pre, # 推理模型结构
data_names=['data'], # 输入数据的名称,与前面准备输入数据时provide_data中的名称相同
label_names=None) # 推理无需输入label
model_pre.bind(for_training=False, #绑定计算模块到底层计算引擎
data_shapes=batch_data.provide_data,
label_shapes=None)
model_pre.set_params(arg_params, aux_params) # 将下载的训练完成的参数加载进计算模块
###Output
_____no_output_____
###Markdown
第五步:执行推理计算此段代码无回显。
###Code
model_pre.forward(batch_data, # 输入数据
is_train=False) # 控制引擎只进行前向计算
pred = model_pre.get_outputs() # 获得推理结果
ids, scores, bboxes = [xx[0].asnumpy() for xx in pred]
###Output
_____no_output_____
###Markdown
第六步:解析并绘制结果会输出推理结果。根据设置的置信度,可能会输出多个矩形框。由于训练数据量少,训练时间短,推理任务加载的模型精度较低,画出的目标框会存在不准确的现象。适当调整优化器参数,延长训练步数会提升模型精度。此段打印带预测目标框的推理结果。
###Code
THRESH = 0.05 # 置信度阈值
im_array = orig_img.copy()
color_white = (255, 255, 255)
x_scale = im_array.shape[1] / 416.0
y_scale = im_array.shape[0] / 416.0
for idx in range(ids.shape[0]):
if ids[idx][0] < 0 or scores[idx][0] < THRESH:
continue
# 按原图比例缩放目标框
bbox = [int(bboxes[idx][0] * x_scale),
int(bboxes[idx][1] * y_scale),
int(bboxes[idx][2] * x_scale),
int(bboxes[idx][3] * y_scale)]
color = (random.randint(0, 256), random.randint(0, 256), random.randint(0, 256))
# 在图像上画框
cv2.rectangle(im_array, (bbox[0], bbox[1]), (bbox[2], bbox[3]), color=color, thickness=2)
# 把置信度写在图像上
cv2.putText(im_array, '%.3f' % (scores[idx][0]), (bbox[0], bbox[1] + 10),
color=color_white, fontFace=cv2.FONT_HERSHEY_COMPLEX, fontScale=0.5)
plt.imshow(im_array)
plt.show()
###Output
_____no_output_____
|
Subsets and Splits
No community queries yet
The top public SQL queries from the community will appear here once available.