licenses
sequencelengths 1
3
| version
stringclasses 677
values | tree_hash
stringlengths 40
40
| path
stringclasses 1
value | type
stringclasses 2
values | size
stringlengths 2
8
| text
stringlengths 25
67.1M
| package_name
stringlengths 2
41
| repo
stringlengths 33
86
|
|---|---|---|---|---|---|---|---|---|
[
"MIT"
] | 0.2.1 | 6512022b637fc17c297bb3050ba2e6cdd50b6fa0 | code | 753 | function Header(io::IOStream)
seek(io, 0)
offset = read(io, UInt32)
header_length = read(io, UInt32)
n_variants = read(io, UInt32)
n_samples = read(io, UInt32)
magic = read(io, 4)
# check magic number
@assert String(magic) == "bgen" || all(magic .== 0) "Magic number mismatch"
seek(io, header_length)
flags = read(io, UInt32)
compression = flags & 0x03
layout = (flags & 0x3c) >> 2
has_sample_ids = convert(Bool, flags & 0x80000000 >> 31)
Header(offset, header_length, n_variants, n_samples, compression, layout,
has_sample_ids)
end
function Header(filename::String)
io = open(filename)
Header(io)
end
@inline function offset_first_variant(h::Header)
return h.offset + 4
end
| BGEN | https://github.com/OpenMendel/BGEN.jl.git |
|
[
"MIT"
] | 0.2.1 | 6512022b637fc17c297bb3050ba2e6cdd50b6fa0 | code | 6752 | function Index(path::AbstractString)
db = SQLite.DB(path)
Index(path, db, [], [], [], [])
end
@inline function _check_idx(b::Bgen)
@assert b.idx !== nothing "bgen index (.bgi) is needed"
end
function select_region(idx::Index, chrom::AbstractString;
start=nothing, stop=nothing)
if start === nothing && stop === nothing
q = "SELECT file_start_position FROM Variant WHERE chromosome=?"
params = (chrom,)
elseif stop === nothing
q = "SELECT file_start_position FROM Variant" *
" WHERE chromosome=? AND position>=?"
params = (chrom, start)
else
q = "SELECT file_start_position FROM Variant" *
" WHERE chromosome=? AND position>=? AND position<=?"
params = (chrom, start, stop)
end
r = (DBInterface.execute(idx.db, q, params) |> columntable)[1]
end
"""
select_region(bgen, chrom; start=nothing, stop=nothing)
Select variants from a region. Returns variant start offsets on the file.
Returns a `BgenVariantIteratorFromOffsets` object.
"""
function select_region(b::Bgen, chrom::AbstractString;
start=nothing, stop=nothing)
_check_idx(b)
offsets = select_region(b.idx, chrom; start=start, stop=stop)
BgenVariantIteratorFromOffsets(b, offsets)
end
function variant_by_rsid(idx::Index, rsid::AbstractString; allele1=nothing, allele2=nothing, allow_multiple=false)
q = "SELECT file_start_position, allele1, allele2 FROM Variant WHERE rsid= ?"
params = (rsid,)
r = (DBInterface.execute(idx.db, q, params) |> columntable)#[1]
if length(r) == 0
error("variant with rsid $rsid not found")
end
if allow_multiple
return r[1]
end
if allele1 === nothing && allele2 === nothing
if length(r[1]) > 1
if !allow_multiple
error("multiple variant matches with $rsid. Try to specify allele1 and allele2, or set allow_multiple=true.")
end
end
return r[1][1]
else
firstidx = nothing
lastidx = nothing
if allele1 !== nothing && allele2 === nothing
firstidx = findall(x -> x == allele1, r.allele1)
if length(firstidx) > 1
error("Nonunique match with $rsid, allele1: $allele1.")
elseif length(firstidx) == 0
error("No match with $rsid, allele1: $allele1.")
end
return r.file_start_position[firstidx[1]]
elseif allele2 !== nothing && allele1 === nothing
lastidx = findall(x -> x == allele2, r.allele2)
if length(lastidx) > 1
error("Nonunique match with $rsid, allele2: $allele2.")
elseif length(lastidx) == 0
error("No match with $rsid, allele2: $allele2.")
end
return r.file_start_position[lastidx[1]]
else
firstidx = findall(x -> x == allele1, r.allele1)
lastidx = findall(x -> x == allele2, r.allele2)
jointidx = intersect(firstidx, lastidx)
if length(jointidx) > 1
error("Nonunique match with $rsid, allele1: $allele1, allele2: $allele2.")
elseif length(jointidx) == 0
error("No match with $rsid, allele1: $allele1, allele2: $allele2.")
end
return r.file_start_position[jointidx[1]]
end
end
end
"""
variant_by_rsid(bgen, rsid)
Find a variant by rsid
"""
function variant_by_rsid(b::Bgen, rsid::AbstractString; allele1=nothing, allele2=nothing, varid=nothing)
@assert !(varid !== nothing && (allele1 !== nothing || allele2 !== nothing)) "either define varid or alleles"
_check_idx(b)
offset = variant_by_rsid(b.idx, rsid; allele1=allele1, allele2=allele2, allow_multiple = varid !== nothing)
if varid !== nothing
vs = map(x -> BgenVariant(b, x), offset)
v_idxs = findall(x -> x.varid == varid, vs)
if length(v_idxs) > 1
error("Multiple matches.")
elseif length(v_idxs) == 0
error("no matches.")
else
return vs[v_idxs[1]]
end
end
return BgenVariant(b, offset)
end
function variant_by_pos(idx::Index, pos::Integer)
q = "SELECT file_start_position FROM Variant WHERE position= ?"
params = (pos,)
r = (DBInterface.execute(idx.db, q, params) |> columntable)[1]
if length(r) == 0
error("variant match at $pos not found")
elseif length(r) > 1
error("multiple variant matches at $pos")
end
return r[1]
end
"""
variant_by_pos(bgen, pos)
Get the variant of bgen variant given `pos` in the index file
"""
function variant_by_pos(b::Bgen, pos::Integer)
_check_idx(b)
offset = variant_by_pos(b.idx, pos)
return BgenVariant(b, offset)
end
function variant_by_index(idx::Index, first::Integer, last::Union{Nothing, Integer}=nothing)
if last === nothing
q = "SELECT file_start_position FROM Variant LIMIT 1 OFFSET ?"
params = (first - 1,)
else
q = "SELECT file_start_position FROM Variant LIMIT ? OFFSET ?"
params = (last - first + 1, first - 1)
end
r = (DBInterface.execute(idx.db, q, params) |> columntable)[1]
return r
end
"""
variant_by_index(bgen, n)
get the `n`-th variant (1-based).
"""
function variant_by_index(b::Bgen, first::Integer, last::Union{Nothing, Integer}=nothing)
_check_idx(b)
if last === nothing
offset = variant_by_index(b.idx, first)[1]
return BgenVariant(b, offset)
else
offsets = variant_by_index(b.idx, first, last)
return BgenVariantIteratorFromOffsets(b, offsets)
end
end
function offsets(idx::Index)
if length(idx.offsets) != 0
return idx.offsets
end
q = "SELECT file_start_position FROM Variant"
r = (DBInterface.execute(idx.db, q) |> columntable)[1]
resize!(idx.offsets, length(r))
idx.offsets .= r
return r
end
function rsids(idx::Index)
if length(idx.rsids) != 0
return idx.rsids
end
q = "SELECT rsid FROM Variant"
r = (DBInterface.execute(idx.db, q) |> columntable)[1]
resize!(idx.rsids, length(r))
idx.rsids .= r
return r
end
function chroms(idx::Index)
if length(idx.chroms) != 0
return idx.chroms
end
q = "SELECT chromosome FROM Variant"
r = (DBInterface.execute(idx.db, q) |> columntable)[1]
resize!(idx.chroms, length(r))
idx.chroms .= r
return r
end
function positions(idx::Index)
if length(idx.positions) != 0
return idx.positions
end
q = "SELECT position FROM Variant"
r = (DBInterface.execute(idx.db, q) |> columntable)[1]
resize!(idx.positions, length(r))
idx.positions .= r
return r
end
| BGEN | https://github.com/OpenMendel/BGEN.jl.git |
|
[
"MIT"
] | 0.2.1 | 6512022b637fc17c297bb3050ba2e6cdd50b6fa0 | code | 4889 | abstract type BgenVariantIterator <: VariantIterator end
@inline function Base.eltype(vi::BgenVariantIterator)
BgenVariant
end
"""
BgenVariantIteratorFromStart(b::Bgen)
Variant iterator that iterates from the beginning of the Bgen file
"""
struct BgenVariantIteratorFromStart <: BgenVariantIterator
b::Bgen
end
function Base.iterate(vi::BgenVariantIteratorFromStart,
state=offset_first_variant(vi.b))
if state >= vi.b.fsize
return nothing
else
v = BgenVariant(vi.b, state)
nextstate = v.next_var_offset
return (v, nextstate)
end
end
@inline function Base.length(vi::BgenVariantIteratorFromStart)
vi.b.header.n_variants
end
@inline function Base.size(vi::BgenVariantIteratorFromStart)
(vi.b.header.n_variants, )
end
"""
BgenVariantIteratorFromOffsets(b::Bgen, offsets::Vector{UInt})
BgenVariant iterator that iterates over a vector of offsets
"""
struct BgenVariantIteratorFromOffsets <: BgenVariantIterator
b::Bgen
offsets::Vector{UInt}
end
function Base.iterate(vi::BgenVariantIteratorFromOffsets, state=1)
state > length(vi.offsets) ? nothing :
(BgenVariant(vi.b, vi.offsets[state]), state + 1)
end
@inline function Base.length(vi::BgenVariantIteratorFromOffsets)
length(vi.offsets)
end
@inline function Base.size(vi::BgenVariantIteratorFromOffsets)
size(vi.offsets)
end
struct Filter{I, T} <: BgenVariantIterator
itr::I
min_maf::AbstractFloat
min_hwe_pval::AbstractFloat
min_info_score::AbstractFloat
min_success_rate_per_variant::AbstractFloat
rmask::Union{Nothing,BitVector}
cmask::Union{Nothing,BitVector}
decompressed::Union{Nothing, Vector{UInt8}}
end
"""
BGEN.filter(itr; min_maf=NaN, min_hwe_pval=NaN, min_success_rate_per_variant=NaN,
cmask=trues(n_variants(itr.b)), rmask=trues(n_variants(itr.b)))
"Filtered" iterator for variants based on min_maf, min_hwe_pval, min_success_rate_per_variant,
cmask, and rmask.
"""
filter(itr::BgenVariantIterator;
T=Float32,
min_maf=NaN, min_hwe_pval=NaN, min_info_score=NaN,
min_success_rate_per_variant=NaN,
cmask = trues(n_variants(itr.b)),
rmask = nothing,
decompressed = nothing) =
Filter{typeof(itr), T}(itr, min_maf, min_hwe_pval, min_info_score, min_success_rate_per_variant,
rmask, cmask, decompressed)
function Base.iterate(f::Filter{I,T}, state...) where {I,T}
io, h = f.itr.b.io, f.itr.b.header
if state !== ()
y = Base.iterate(f.itr, state[1][2:end]...)
cnt = state[1][1]
else
y = Base.iterate(f.itr)
cnt = 1
end
while y !== nothing
v, s = y
passed = true
if !f.cmask[cnt]
passed = false
end
if passed && (v.genotypes === nothing || v.genotypes.decompressed === nothing)
decompressed = decompress(io, v, h; decompressed=f.decompressed)
elseif passed
decompressed = v.genotypes.decompressed
end
startidx = 1
if passed && v.genotypes === nothing
p = parse_preamble(decompressed, h, v)
v.genotypes = Genotypes{T}(p, decompressed)
elseif passed
p = v.genotypes.preamble
end
if passed && h.layout == 2
startidx += 10 + h.n_samples
end
if passed && !isnan(f.min_maf)
current_maf = maf(p, v.genotypes.decompressed, startidx, h.layout, f.rmask)
if current_maf < f.min_maf && passed
passed = false
end
end
if passed && !isnan(f.min_hwe_pval)
hwe_pval = hwe(p, v.genotypes.decompressed, startidx, h.layout, f.rmask)
if hwe_pval < f.min_hwe_pval
passed = false
end
end
if passed && !isnan(f.min_info_score)
current_info_score = info_score(p, v.genotypes.decompressed, startidx, h.layout, f.rmask)
if current_info_score < f.min_info_score && passed
passed = false
end
end
if passed && !isnan(f.min_success_rate_per_variant)
successes = length(intersect(p.missings, (1:n_samples(f.itr.b)[f.rmask])))
success_rate = successes / count(f.rmask)
if success_rate < f.min_success_rate_per_variant && passed
passed = false
end
end
cnt += 1
if passed
return v, (cnt, s...)
end
y = iterate(f.itr, s...)
end
nothing
end
eltype(::Type{Filter{I,T}}) where {I,T} = BgenVariant
IteratorEltype(::Type{Filter{I,T}}) where {I,T} = IteratorEltype(I)
IteratorSize(::Type{<:Filter}) = SizeUnknown()
reverse(f::Filter) = Filter(reverse(f.itr), f.min_maf, f.min_hwe_pval,
f.min_info_score, f.min_success_rate_per_varinat, f.rmask, f.cmask, f.decompressed)
| BGEN | https://github.com/OpenMendel/BGEN.jl.git |
|
[
"MIT"
] | 0.2.1 | 6512022b637fc17c297bb3050ba2e6cdd50b6fa0 | code | 407 | """
minor_certain(freq, n_checked, z)
Check if minor allele is certain.
- `freq`: frequency of minor or major allele
- `n_checked`: number of individuals checked so far
- `z`: cutoff of "z" value, defaults to `5.0`
"""
function minor_certain(freq::Float64, n_checked::Integer, z::Float64=5.0)
delta = z * sqrt(freq * (1-freq) / n_checked)
return !(freq - delta < 0.5 && freq + delta > 0.5)
end
| BGEN | https://github.com/OpenMendel/BGEN.jl.git |
|
[
"MIT"
] | 0.2.1 | 6512022b637fc17c297bb3050ba2e6cdd50b6fa0 | code | 1087 | function get_samples(io::IOStream, n_samples::Integer)
sample_header_length = read(io, UInt32)
sample_n_check = read(io, UInt32)
if sample_n_check != 0
@assert n_samples == sample_n_check "Inconsistent number of samples"
else
@warn "Sample names unavailable. Do you have a separate '.sample' file?"
end
samples = String[]
for i in 1:n_samples
id_length = read(io, UInt16)
id = read(io, id_length)
push!(samples, String(id))
end
samples
end
function get_samples(path::String, n_samples::Integer)
@assert endswith(path, ".sample") "Extension of the file should be .sample"
io = open(path)
keys = split(readline(io)) # header
key_idx = ("ID_1" in keys && "ID_2" in keys) ? 2 : 1
readline(io) # types
samples = readlines(io)
samples = map(x -> split(x, " ")[key_idx], samples)
@assert length(samples) == n_samples "Inconsistent number of samples"
close(io)
samples
end
function get_samples(n_samples::Integer)
samples = [string(i) for i in 1:n_samples]
samples
end
| BGEN | https://github.com/OpenMendel/BGEN.jl.git |
|
[
"MIT"
] | 0.2.1 | 6512022b637fc17c297bb3050ba2e6cdd50b6fa0 | code | 1653 | struct Header
offset::UInt32
header_length::UInt32
n_variants::UInt32
n_samples::UInt32
compression::UInt8
layout::UInt8
has_sample_ids::Bool
end
const Samples = Vector{String}
struct Index
path::String
db::SQLite.DB
offsets::Vector{UInt64}
rsids::Vector{String}
chroms::Vector{String}
positions::Vector{UInt32}
end
struct Preamble
n_samples::UInt32
n_alleles::UInt16
phased::UInt8
min_ploidy::UInt8
max_ploidy::UInt8
ploidy::Union{UInt8, Vector{UInt8}}
bit_depth::UInt8
max_probs::Int
missings::Vector{Int}
end
mutable struct Genotypes{T}
preamble::Preamble # Once parsed, it will not be destroyed unless Genotypes is destroyed
decompressed::Union{Nothing,Vector{UInt8}}
probs::Union{Nothing, Vector{T}}
minor_idx::UInt8 # index of minor allele
dose::Union{Nothing, Vector{T}}
dose_mean_imputed::Bool
minor_allele_dosage::Bool
end
mutable struct BgenVariant <: Variant
offset::UInt64
geno_offset::UInt64 # to the start of genotype block
next_var_offset::UInt64
geno_block_size::UInt32
n_samples::UInt32
varid::String
rsid::String
chrom::String
pos::UInt32
n_alleles::UInt16
alleles::Vector{String}
# length-1 for parsed one, empty array for not parsed yet or destroyed,
genotypes::Union{Nothing, Genotypes}
end
struct Bgen <: GeneticData
io::IOStream
fsize::UInt64
header::Header
samples::Samples
idx::Union{Index, Nothing}
# note: detailed information of variables stored in Variable struct,
# accessed thru VariantIterator
ref_first::Bool
end
| BGEN | https://github.com/OpenMendel/BGEN.jl.git |
|
[
"MIT"
] | 0.2.1 | 6512022b637fc17c297bb3050ba2e6cdd50b6fa0 | code | 11050 | """
hwe(b::Bgen, v::BgenVariant; T=Float32, decompressed=nothing)
hwe(p::Preamble, d::Vector{UInt8}, idx::Vector{<:Integer}, layout::UInt8,
rmask::Union{Nothing, Vector{UInt16}})
Hardy-Weinberg equilibrium test for diploid biallelic case
"""
function hwe(p::Preamble, d::Vector{UInt8}, startidx::Integer, layout::UInt8,
rmask::Union{Nothing, Vector{UInt16}})
@assert layout == 2 "hwe only supported for layout 2"
@assert p.bit_depth == 8 && p.max_probs == 3 && p.max_ploidy == p.min_ploidy
idx1 = startidx
# "counts" times 255.
n00 = 0
n01 = 0
n11 = 0
if p.n_samples >= 16
@inbounds for n in 1:16:(p.n_samples - p.n_samples % 16)
idx_base = idx1 + ((n-1) >> 1) << 2
if rmask !== nothing
rs = vload(Vec{16,UInt16}, rmask, n)
if sum(rs) == 0
continue
end
end
r = reinterpret(Vec{16, UInt16}, vload(Vec{32, UInt8}, d, idx_base))
first = (r & mask_odd)
second = (r & mask_even) >> 8
third = 0x00ff - first - second
if rmask !== nothing
first = first * rs
second = second * rs
third = third * rs
end
n00 += sum(first)
n01 += sum(second)
n11 += sum(third)
end
end
rem = p.n_samples % 16
if rem != 0
@inbounds for n in ((p.n_samples - rem) + 1) : p.n_samples
rmask !== nothing && rmask[n] == 0 && continue
idx_base = idx1 + ((n - 1) << 1)
n00 += d[idx_base]
n01 += d[idx_base + 1]
n11 += 255 - d[idx_base] - d[idx_base + 1]
end
end
@inbounds for n in p.missings
rmask !== nothing && rmask[n] == 0 && continue
idx_base = idx1 + ((n - 1) << 1)
n00 -= d[idx_base]
n01 -= d[idx_base + 1]
n11 -= 255 - d[idx_base] - d[idx_base + 1]
end
n00 *= one_255th
n01 *= one_255th
n11 *= one_255th
return hwe(n00, n01, n11)
end
@inline ccdf_chisq_1(x) = gamma_inc(convert(typeof(x), 1/2), x/2, 0)[2]
"""
hwe(n00, n01, n11)
Hardy-Weinberg equilibrium test. `n00`, `n01`, `n11` are counts of homozygotes
and heterozygoes respectively. Output is the p-value of type Float64.
"""
function hwe(n00::Real, n01::Real, n11::Real)
n = n00 + n01 + n11
n == 0 && return 1.0
p0 = (n01 + 2n00) / 2n
(p0 ≤ 0.0 || p0 ≥ 1.0) && return 1.0
p1 = 1 - p0
# Pearson's Chi-squared test
e00 = n * p0 * p0
e01 = 2n * p0 * p1
e11 = n * p1 * p1
ts = (n00 - e00)^2 / e00 + (n01 - e01)^2 / e01 + (n11 - e11)^2 / e11
#pval = ccdf(Chisq(1), ts)
pval = ccdf_chisq_1(ts)
# TODO Fisher exact test
return pval
end
"""
maf(b::Bgen, v::BgenVariant; T=Float32, decompressed=nothing)
maf(p::Preamble, d::Vector{UInt8}, idx::Vector{<:Integer}, layout::UInt8,
rmask::Union{Nothing, Vector{UInt16}})
Minor-allele frequency for diploid biallelic case
"""
function maf(p::Preamble, d::Vector{UInt8}, startidx::Integer, layout::UInt8,
rmask::Union{Nothing, Vector{UInt16}})
@assert layout == 2 "maf only supported for layout 2"
@assert p.bit_depth == 8 && p.max_probs == 3 && p.max_ploidy == p.min_ploidy
idx1 = startidx
# "counts" times 255.
dosage_total = 0
if p.n_samples >= 16
@inbounds for n in 1:16:(p.n_samples - p.n_samples % 16)
idx_base = idx1 + ((n-1) >> 1) << 2
if rmask !== nothing
rs = vload(Vec{16,UInt16}, rmask, n)
if sum(rs) == 0
continue
end
end
r = reinterpret(Vec{16, UInt16}, vload(Vec{32, UInt8}, d, idx_base))
first = (r & mask_odd) << 1
second = (r & mask_even) >> 8
s = first + second
if rmask !== nothing
s = s * rs
end
dosage_total += sum(s)
end
end
rem = p.n_samples % 16
if rem != 0
@inbounds for n in ((p.n_samples - rem) + 1) : p.n_samples
rmask !== nothing && rmask[n] == 0 && continue
idx_base = idx1 + ((n - 1) << 1)
dosage_total += 2 * d[idx_base] + d[idx_base + 1]
end
end
@inbounds for n in p.missings
rmask !== nothing && rmask[n] == 0 && continue
idx_base = idx1 + ((n - 1) << 1)
dosage_total -= 2 * d[idx_base] + d[idx_base + 1]
end
dosage_total *= one_255th
dosage_total /= (rmask !== nothing ? sum(rmask) : p.n_samples - length(p.missings))
dosage_total < 1.0 ? dosage_total / 2 : 1 - dosage_total / 2
end
"""
info_score(b::Bgen, v::BgenVariant; T=Float32, decompressed=nothing)
info_score(p::Preamble, d::Vector{UInt8}, idx::Vector{<:Integer}, layout::UInt8,
rmask::Union{Nothing, Vector{UInt16}})
Information score of the variant.
"""
function info_score(p::Preamble, d::Vector{UInt8}, startidx::Integer, layout::UInt8,
rmask::Union{Nothing, Vector{UInt16}})
@assert layout == 2 "info_score only supported for layout 2"
@assert p.bit_depth == 8 && p.max_probs == 3 && p.max_ploidy == p.min_ploidy
@assert length(p.missings) == 0 "current implementation does not allow missingness"
idx1 = startidx
# "counts" times 255.
samples_cum = 0
mean_cum = 0.0
sumsq_cum = 0.0
dosage_sum = 0.0
dosage_sumsq = 0.0
rs = Vec{16,UInt16}((1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1))
rs_float = Vec{16, Float32}((1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1))
rs_float_ones = rs_float
rs_floatarr = ones(Float32, 16)
if p.n_samples >= 16
@inbounds for n in 1:16:(p.n_samples - p.n_samples % 16)
idx_base = idx1 + ((n-1) >> 1) << 2
if rmask !== nothing
rs = vload(Vec{16,UInt16}, rmask, n)
if sum(rs) == 0
continue
elseif sum(rs) == 16
rs_float = rs_float_ones
else
rs_floatarr .= @view(rmask[n:n+15])
rs_float = vload(Vec{16, Float32}, rs_floatarr, 1)
end
end
r = reinterpret(Vec{16, UInt16}, vload(Vec{32, UInt8}, d, idx_base))
first = (r & mask_odd) << 1
second = (r & mask_even) >> 8
s = first + second
if rmask !== nothing
s = s * rs
end
dosage_float = one_255th * convert(
Vec{16, Float32}, s)
samples_new = sum(rs)
sum_new = sum(dosage_float)
samples_prev = samples_cum
samples_cum = samples_cum + samples_new
mean_new = sum_new / samples_new
diff = dosage_float - mean_new
if rmask !== nothing
diff *= rs_float
end
sumsq_new = sum(diff ^ 2)
delta = mean_new - mean_cum
mean_cum += delta * samples_new / samples_cum
sumsq_cum += sumsq_new + delta ^ 2 * samples_prev * samples_new / samples_cum
end
end
rem = p.n_samples % 16
if rem != 0
@inbounds for n in ((p.n_samples - rem) + 1) : p.n_samples
rmask !== nothing && rmask[n] == 0 && continue
idx_base = idx1 + ((n - 1) << 1)
dosage = one_255th * (2 * d[idx_base] + d[idx_base + 1])
delta = dosage - mean_cum
samples_cum += 1
mean_cum += delta / samples_cum
delta2 = dosage - mean_cum
sumsq_cum += delta * delta2
end
end
n_samples = rmask !== nothing ? sum(rmask) : p.n_samples
p = mean_cum / 2
v = sumsq_cum / (n_samples - 1)
v / (2p * (1-p))
end
function counts!(p::Preamble, d::Vector{UInt8}, startidx::Integer, layout::UInt8,
rmask::Union{Nothing, Vector{UInt16}}; r::Union{Nothing,Vector{<:Integer}}=nothing, dosage::Bool=true)
if dosage
@assert layout == 2 "hwe only supported for layout 2"
@assert p.bit_depth == 8 && p.max_probs == 3 && p.max_ploidy == p.min_ploidy
idx1 = startidx
if r !== nothing
@assert length(r) == 512
fill!(r, 0)
else
r = zeros(UInt, 512)
end
rs = Vec{16,UInt16}((1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1))
if p.n_samples >= 16
@inbounds for n in 1:16:(p.n_samples - p.n_samples % 16)
idx_base = idx1 + ((n-1) >> 1) << 2
if rmask !== nothing
rs = vload(Vec{16,UInt16}, rmask, n)
if sum(rs) == 0
continue
end
end
q = reinterpret(Vec{16, UInt16}, vload(Vec{32, UInt8}, d, idx_base))
first = (q & mask_odd) << 1
second = (q & mask_even) >> 8
s = first + second
if rmask !== nothing
s = s * rs
end
@inbounds for i in 1:16
if rs[i] != 0
ss = s[i]
r[ss + 1] += 1
end
end
end
end
rem = p.n_samples % 16
if rem != 0
@inbounds for n in ((p.n_samples - rem) + 1) : p.n_samples
rmask !== nothing && rmask[n] == 0 && continue
idx_base = idx1 + ((n - 1) << 1)
r[2 * d[idx_base] + d[idx_base + 1] + 1] += 1
end
end
# subtract back counts for missings
@inbounds for n in p.missings
rmask !== nothing && rmask[n] == 0 && continue
idx_base = idx1 + ((n - 1) << 1)
r[2 * d[idx_base] + d[idx_base + 1] + 1] -= 1
end
r[end] = length(p.missings)
else
@error "counts for non-dosage not implemented yet"
end
r
end
for ftn in [:maf, :hwe, :info_score, :counts!]
@eval begin
function $(ftn)(b::Bgen, v::BgenVariant; T=Float32, decompressed=nothing,
is_decompressed=false, rmask=nothing, kwargs...)
io, h = b.io, b.header
if (decompressed !== nothing && !is_decompressed) ||
(decompressed === nothing && (v.genotypes === nothing ||
v.genotypes.decompressed === nothing))
decompressed = decompress(io, v, h; decompressed=decompressed)
else
decompressed = v.genotypes.decompressed
end
startidx = 1
if v.genotypes === nothing
p = parse_preamble(decompressed, h, v)
v.genotypes = Genotypes{T}(p, decompressed)
else
p = v.genotypes.preamble
end
if h.layout == 2
startidx += 10 + h.n_samples
end
$(ftn)(p, decompressed, startidx, h.layout, rmask; kwargs...)
end
end
end | BGEN | https://github.com/OpenMendel/BGEN.jl.git |
|
[
"MIT"
] | 0.2.1 | 6512022b637fc17c297bb3050ba2e6cdd50b6fa0 | code | 3197 | """
BgenVariant(b::Bgen, offset::Integer)
BgenVariant(io, offset, compression, layout, expected_n)
Parse information of a single variant beginning from `offset`.
"""
function BgenVariant(io::IOStream, offset::Integer,
compression::Integer, layout::Integer,
expected_n::Integer)
seek(io, offset)
if eof(io)
@error "reached end of file"
end
if layout == 1
n_samples = read(io, UInt32)
else
n_samples = expected_n
end
if n_samples != expected_n
@error "number of samples does not match"
end
varid_len = read(io, UInt16)
varid = String(read(io, varid_len))
rsid_len = read(io, UInt16)
rsid = String(read(io, rsid_len))
chrom_len = read(io, UInt16)
chrom = String(read(io, chrom_len))
pos = read(io, UInt32)
if layout == 1
n_alleles = 2
else
n_alleles = read(io, UInt16)
end
alleles = Array{String}(undef, n_alleles)
for i in 1:n_alleles
allele_len = read(io, UInt32)
alleles_bytes = read(io, allele_len)
alleles[i] = String(alleles_bytes)
end
if compression == 0 && layout == 1
geno_block_size = 6 * n_samples
else
geno_block_size = read(io, UInt32)
end
geno_offset = position(io)
next_var_offset = geno_offset + geno_block_size
BgenVariant(offset, geno_offset, next_var_offset, geno_block_size, n_samples,
varid, rsid, chrom, pos, n_alleles, alleles, nothing)
end
function BgenVariant(b::Bgen, offset::Integer)
h = b.header
if offset >= b.fsize
@error "reached end of file"
end
BgenVariant(b.io, offset, h.compression, h.layout, h.n_samples)
end
@inline n_samples(v::BgenVariant)::Int = v.n_samples
@inline varid(v::BgenVariant) = v.varid
@inline rsid(v::BgenVariant) = v.rsid
@inline chrom(v::BgenVariant) = v.chrom
@inline pos(v::BgenVariant)::Int = v.pos
@inline n_alleles(v::BgenVariant)::Int = v.n_alleles
@inline alleles(v::BgenVariant) = v.alleles
# The following functions are valid only after calling `probabilities!()`
# or `minor_allele_dosage!()`
@inline phased(v::BgenVariant) = v.genotypes.preamble.phased
@inline min_ploidy(v::BgenVariant) = v.genotypes.preamble.min_ploidy
@inline max_ploidy(v::BgenVariant) = v.genotypes.preamble.max_ploidy
@inline ploidy(v::BgenVariant) = v.genotypes.preamble.ploidy
@inline bit_depth(v::BgenVariant) = v.genotypes.preamble.bit_depth
@inline missings(v::BgenVariant) = v.genotypes.preamble.missings
# The below are valid after calling `minor_allele_dosage!()`
@inline function minor_allele(v::BgenVariant)
midx = v.genotypes.minor_idx
if midx == 0
@error "`minor_allele_dosage!()` must be called before `minor_allele()`"
else
v.alleles[v.genotypes.minor_idx]
end
end
@inline function major_allele(v::BgenVariant)
midx = v.genotypes.minor_idx
if midx == 0
@error "`minor_allele_dosage!()` must be called before `minor_allele()`"
else
v.alleles[3 - midx]
end
end
"""
destroy_genotypes!(v::BgenVariant)
Destroy any parsed genotype information.
"""
function clear!(v::BgenVariant)
v.genotypes = nothing
return
end
| BGEN | https://github.com/OpenMendel/BGEN.jl.git |
|
[
"MIT"
] | 0.2.1 | 6512022b637fc17c297bb3050ba2e6cdd50b6fa0 | code | 656 | using BGEN
using Statistics, Test, Printf
using GeneticVariantBase
const example_8bits = BGEN.datadir("example.8bits.bgen")
const example_10bits = BGEN.datadir("example.10bits.bgen")
const example_16bits = BGEN.datadir("example.16bits.bgen")
const example_sample = BGEN.datadir("example.sample")
include("utils.jl")
const gen_data = load_gen_data()
const vcf_data = load_vcf_data()
const haps_data = load_haps_data()
include("test_basics.jl")
include("test_getters.jl")
include("test_select_region.jl")
include("test_index.jl")
include("test_load_example_files.jl")
include("test_minor_allele_dosage.jl")
include("test_utils.jl")
include("test_filter.jl")
| BGEN | https://github.com/OpenMendel/BGEN.jl.git |
|
[
"MIT"
] | 0.2.1 | 6512022b637fc17c297bb3050ba2e6cdd50b6fa0 | code | 2107 | @testset "basics" begin
header_ref = BGEN.Header(example_10bits)
@testset "header" begin
header_test = BGEN.Header(example_10bits)
@test header_test.offset == 0x0000178c # 6028
@test header_test.header_length == 0x00000014 # 20
@test header_test.n_variants == 0x000000c7 # 199
@test header_test.n_samples == 0x000001f4 # 500
@test header_test.compression == 1
@test header_test.layout == 2
@test header_test.has_sample_ids == 1
end
@testset "samples_separate" begin
n_samples = 500
samples_test2 = BGEN.get_samples(example_sample, n_samples)
samples_correct = [(@sprintf "sample_%03d" i) for i in 1:n_samples]
@test all(samples_correct .== samples_test2)
samples_test3 = BGEN.get_samples(n_samples)
@test all([string(i) for i in 1:n_samples] .== samples_test3)
end
bgen = BGEN.Bgen(example_10bits)
@testset "bgen" begin
@test bgen.fsize == 223646
@test bgen.header == header_ref
n_samples = bgen.header.n_samples
samples_correct = [(@sprintf "sample_%03d" i) for i in 1:n_samples]
@test all(samples_correct .== bgen.samples)
variants = parse_variants(bgen)
var = variants[4]
@test length(variants) == 199
@test var.offset == 0x0000000000002488
@test var.geno_offset == 0x00000000000024b1
@test var.next_var_offset == 0x0000000000002902
@test var.geno_block_size == 0x00000451
@test var.n_samples == 0x000001f4
@test var.varid == "SNPID_5"
@test var.rsid == "RSID_5"
@test var.chrom == "01"
@test var.pos == 0x00001388
@test var.n_alleles == 2
@test all(var.alleles.== ["A", "G"])
end
@testset "preamble" begin
io, v, h = bgen.io, parse_variants(bgen)[1], bgen.header
decompressed = BGEN.decompress(io, v, h)
preamble = BGEN.parse_preamble(decompressed, h, v)
@test preamble.phased == 0
@test preamble.min_ploidy == 2
@test preamble.max_ploidy == 2
@test all(preamble.ploidy .== 2)
@test preamble.bit_depth == 10
@test preamble.max_probs == 3
@test length(preamble.missings) == 1
@test preamble.missings[1] == 1
end
end
| BGEN | https://github.com/OpenMendel/BGEN.jl.git |
|
[
"MIT"
] | 0.2.1 | 6512022b637fc17c297bb3050ba2e6cdd50b6fa0 | code | 2227 | @testset "filter" begin
b = Bgen(BGEN.datadir("example.8bits.bgen"))
vidx = falses(b.header.n_variants)
vidx[1:10] .= true
BGEN.filter("test.bgen", b, vidx)
b2 = Bgen("test.bgen"; sample_path="test.sample")
@test all(b.samples .== b2.samples)
for (v1, v2) in zip(iterator(b), iterator(b2)) # length of two iterators are different.
# it stops when the shorter one (b2) ends.
@test v1.varid == v2.varid
@test v1.rsid == v2.rsid
@test v1.chrom == v2.chrom
@test v1.pos == v2.pos
@test v1.n_alleles == v2.n_alleles
@test all(v1.alleles .== v2.alleles)
decompressed1 = BGEN.decompress(b.io, v1, b.header)
decompressed2 = BGEN.decompress(b2.io, v2, b2.header)
@test all(decompressed1 .== decompressed2)
end
sidx = falses(b.header.n_samples)
sidx[1:10] .= true
BGEN.filter("test2.bgen", b, trues(b.header.n_variants), sidx)
b3 = Bgen("test2.bgen"; sample_path="test2.sample")
for (v1, v3) in zip(iterator(b), iterator(b3))
@test v1.varid == v3.varid
@test v1.rsid == v3.rsid
@test v1.chrom == v3.chrom
@test v1.pos == v3.pos
@test v1.n_alleles == v3.n_alleles
@test all(v1.alleles .== v3.alleles)
@test isapprox(probabilities!(b, v1)[:, 1:10], probabilities!(b3, v3); nans=true)
end
b4 = Bgen(BGEN.datadir("complex.24bits.bgen"))
BGEN.filter("test3.bgen", b4, trues(b4.header.n_variants), BitVector([false, false, true, true]))
b5 = Bgen("test3.bgen"; sample_path = "test3.sample")
for (v4, v5) in zip(iterator(b4), iterator(b5))
@test v4.varid == v5.varid
@test v4.rsid == v5.rsid
@test v4.chrom == v5.chrom
@test v4.pos == v5.pos
@test v4.n_alleles == v5.n_alleles
@test all(v4.alleles .== v5.alleles)
@test isapprox(probabilities!(b4, v4)[:, 3:4], probabilities!(b5, v5); nans=true)
end
close(b)
close(b2)
close(b3)
close(b4)
close(b5)
rm("test.bgen", force=true)
rm("test.sample", force=true)
rm("test2.bgen", force=true)
rm("test2.sample", force=true)
rm("test3.bgen", force=true)
rm("test3.sample", force=true)
end | BGEN | https://github.com/OpenMendel/BGEN.jl.git |
|
[
"MIT"
] | 0.2.1 | 6512022b637fc17c297bb3050ba2e6cdd50b6fa0 | code | 792 | @testset "getters" begin
b = BGEN.Bgen(example_8bits)
@test fsize(b) == 128746
@test all(samples(b) .== b.samples)
@test n_samples(b) == 500
@test n_variants(b) == 199
@test compression(b) == "Zlib"
v = first(iterator(b; from_bgen_start=true))
@test n_samples(v::Variant) == 500
@test varid(v::Variant) == "SNPID_2"
@test rsid(v::Variant) == "RSID_2"
@test chrom(v::Variant) == "01"
@test pos(v::Variant) == 2000
@test n_alleles(v::Variant) == 2
@test length(alleles(v::Variant)) == 2
@test all(alleles(v) .== ["A", "G"])
minor_allele_dosage!(b, v)
@test phased(v) == 0
@test min_ploidy(v) == 2
@test max_ploidy(v) == 2
@test all(ploidy(v) .== 2)
@test bit_depth(v) == 8
@test length(missings(v)) == 1
@test missings(v)[1] == 1
@test minor_allele(v) == "A"
@test major_allele(v) == "G"
end
| BGEN | https://github.com/OpenMendel/BGEN.jl.git |
|
[
"MIT"
] | 0.2.1 | 6512022b637fc17c297bb3050ba2e6cdd50b6fa0 | code | 987 | @testset "index" begin
@testset "index_opens" begin
@test Bgen(BGEN.datadir("example.15bits.bgen")).idx === nothing
@test Bgen(BGEN.datadir("example.16bits.bgen")).idx !== nothing
end
@testset "index_region" begin
chrom = "01"
start = 5000
stop = 50000
b = Bgen(BGEN.datadir("example.16bits.bgen"))
idx = b.idx
@test length(select_region(b, chrom)) == length(gen_data)
@test length(select_region(b, "02")) == 0
@test length(select_region(b, chrom;
start=start * 100, stop=stop * 100)) == 0
after_pos_offsets = select_region(b, chrom; start=start)
@test length(after_pos_offsets) ==
length(filter(x -> x.pos >= start, gen_data))
in_region_offsets = select_region(b, chrom; start=start, stop=stop)
@test length(in_region_offsets) == length(filter(x -> start <= x.pos <=
stop, gen_data))
@test rsid(variant_by_rsid(b, "RSID_10")) == "RSID_10"
@test rsid(variant_by_index(b, 4)) == "RSID_3"
end
end
| BGEN | https://github.com/OpenMendel/BGEN.jl.git |
|
[
"MIT"
] | 0.2.1 | 6512022b637fc17c297bb3050ba2e6cdd50b6fa0 | code | 2840 | @testset "example files" begin
@testset "bit depths" begin
for i in 1:32
b = Bgen(BGEN.datadir("example.$(i)bits.bgen"); idx_path = nothing)
v = parse_variants(b)
for (j, (v1, v2)) in enumerate(zip(v, gen_data))
@test v1.chrom == v2.chrom
@test v1.varid == v2.varid
@test v1.rsid == v2.rsid
@test v1.pos == v2.pos
@test all(v1.alleles .== v2.alleles)
@test array_equal(v2.probs, probabilities!(b, v1), i)
end
end
end
@testset "zstd" begin
b = Bgen(BGEN.datadir("example.16bits.zstd.bgen"); idx_path = nothing)
v = parse_variants(b)
for (j, (v1, v2)) in enumerate(zip(v, gen_data))
@test v1.chrom == v2.chrom
@test v1.varid == v2.varid
@test v1.rsid == v2.rsid
@test v1.pos == v2.pos
@test all(v1.alleles .== v2.alleles)
@test array_equal(v2.probs, probabilities!(b, v1), 16)
end
end
@testset "v11" begin
b = Bgen(BGEN.datadir("example.v11.bgen"); idx_path = nothing)
v = parse_variants(b)
for (j, (v1, v2)) in enumerate(zip(v, gen_data))
@test v1.chrom == v2.chrom
@test v1.varid == v2.varid
@test v1.rsid == v2.rsid
@test v1.pos == v2.pos
@test all(v1.alleles .== v2.alleles)
@test array_equal(v2.probs, probabilities!(b, v1), 16)
end
end
@testset "haplotypes" begin
b = Bgen(BGEN.datadir("haplotypes.bgen"); idx_path = nothing)
v = parse_variants(b)
for (j, (v1, v2)) in enumerate(zip(v, haps_data))
@test v1.chrom == v2.chrom
@test v1.varid == v2.varid
@test v1.rsid == v2.rsid
@test v1.pos == v2.pos
@test all(v1.alleles .== v2.alleles)
@test array_equal(v2.probs, probabilities!(b, v1), 16)
end
end
@testset "complex" begin
b = Bgen(BGEN.datadir("complex.bgen"); idx_path = nothing)
v = parse_variants(b)
for (j, (v1, v2)) in enumerate(zip(v, vcf_data))
@test v1.chrom == v2.chrom
@test v1.varid == v2.varid
@test v1.rsid == v2.rsid
@test v1.pos == v2.pos
@test all(v1.alleles .== v2.alleles)
@test array_equal(v2.probs, probabilities!(b, v1), 16)
end
end
@testset "complex bit depths" begin
for i in 1:32
b = Bgen(BGEN.datadir("complex.$(i)bits.bgen"); idx_path = nothing)
v = parse_variants(b)
for (j, (v1, v2)) in enumerate(zip(v, vcf_data))
@test v1.chrom == v2.chrom
@test v1.varid == v2.varid
@test v1.rsid == v2.rsid
@test v1.pos == v2.pos
@test all(v1.alleles .== v2.alleles)
@test array_equal(v2.probs, probabilities!(b, v1), i)
end
end
end
@testset "null" begin
@test_throws SystemError Bgen(BGEN.datadir("Hello_World.bgen"))
end
end
| BGEN | https://github.com/OpenMendel/BGEN.jl.git |
|
[
"MIT"
] | 0.2.1 | 6512022b637fc17c297bb3050ba2e6cdd50b6fa0 | code | 3961 | @testset "minor allele dosage" begin
@testset "slow" begin
path = BGEN.datadir("example.16bits.zstd.bgen")
b = Bgen(path)
for v in iterator(b)
probs = probabilities!(b, v)
dose = minor_allele_dosage!(b, v)
# dosages for each allele
a1 = probs[1, :] .* 2 .+ probs[2, :]
a2 = probs[3, :] .* 2 .+ probs[2, :]
dose_correct = sum(a1[.!isnan.(a1)]) < sum(a2[.!isnan.(a2)]) ? a1 : a2
minor_allele_index = sum(a1[.!isnan.(a1)]) < sum(a2[.!isnan.(a2)]) ? 1 : 2
@test v.genotypes.minor_idx == minor_allele_index
@test all(isapprox.(dose, dose_correct; atol=2e-7, nans=true))
end
end
@testset "fast" begin
path = BGEN.datadir("example.8bits.bgen")
b = Bgen(path)
for v in iterator(b)
probs = probabilities!(b, v)
dose = minor_allele_dosage!(b, v)
# dosages for each allele
a1 = probs[1, :] .* 2 .+ probs[2, :]
a2 = probs[3, :] .* 2 .+ probs[2, :]
dose_correct = sum(a1[.!isnan.(a1)]) < sum(a2[.!isnan.(a2)]) ? a1 : a2
minor_allele_index = sum(a1[.!isnan.(a1)]) < sum(a2[.!isnan.(a2)]) ? 1 : 2
@test v.genotypes.minor_idx == minor_allele_index
@test all(isapprox.(dose, dose_correct; atol=2e-7, nans=true))
end
end
@testset "v11" begin
path = BGEN.datadir("example.v11.bgen")
b = Bgen(path)
for v in iterator(b)
probs = probabilities!(b, v)
dose = minor_allele_dosage!(b, v)
# dosages for each allele
a1 = probs[1, :] .* 2 .+ probs[2, :]
a2 = probs[3, :] .* 2 .+ probs[2, :]
dose_correct = sum(a1[.!isnan.(a1)]) < sum(a2[.!isnan.(a2)]) ? a1 : a2
minor_allele_index = sum(a1[.!isnan.(a1)]) < sum(a2[.!isnan.(a2)]) ? 1 : 2
@test v.genotypes.minor_idx == minor_allele_index
@test all(isapprox.(dose, dose_correct; atol=7e-5, nans=true))
end
end
@testset "multiple_calls" begin
b = Bgen(
BGEN.datadir("example.8bits.bgen");
sample_path=BGEN.datadir("example.sample"),
idx_path=BGEN.datadir("example.8bits.bgen.bgi")
)
# second allele minor
v = variant_by_rsid(b, "RSID_110")
@test isapprox(mean(minor_allele_dosage!(b, v)), 0.9621725f0)
@test isapprox(mean(minor_allele_dosage!(b, v)), 0.9621725f0)
@test isapprox(mean(first_allele_dosage!(b, v)), 2 - 0.9621725f0)
@test isapprox(mean(minor_allele_dosage!(b, v)), 0.9621725f0)
clear!(v)
@test isapprox(mean(first_allele_dosage!(b, v)), 2 - 0.9621725f0)
@test isapprox(mean(minor_allele_dosage!(b, v)), 0.9621725f0)
clear!(v)
@test isapprox(mean(BGEN.ref_allele_dosage!(b, v)), 2 - 0.9621725f0)
@test isapprox(mean(BGEN.alt_allele_dosage!(b, v)), 0.9621725f0)
# first allele minor
v = variant_by_rsid(b, "RSID_198")
@test isapprox(mean(minor_allele_dosage!(b, v)), 0.48411763f0)
@test isapprox(mean(minor_allele_dosage!(b, v)), 0.48411763f0)
@test isapprox(mean(first_allele_dosage!(b, v)), 0.48411763f0)
@test isapprox(mean(minor_allele_dosage!(b, v)), 0.48411763f0)
clear!(v)
@test isapprox(mean(first_allele_dosage!(b, v)), 0.48411763f0)
@test isapprox(mean(minor_allele_dosage!(b, v)), 0.48411763f0)
clear!(v)
@test isapprox(mean(BGEN.ref_allele_dosage!(b, v)), 0.48411763f0)
@test isapprox(mean(BGEN.alt_allele_dosage!(b, v)), 2 - 0.48411763f0)
clear!(v)
@test isapprox(mean(GeneticVariantBase.alt_dosages!(Vector{Float32}(undef, n_samples(b)), b, v)), 2 - 0.48411763f0)
end
@testset "mean_impute" begin
path = BGEN.datadir("example.8bits.bgen")
b = Bgen(path)
v = first(iterator(b; from_bgen_start=true))
m = minor_allele_dosage!(b, v; mean_impute=true)
@test isapprox(m[1], 0.3958112303037447)
end
@testset "haplotypes" begin
path = BGEN.datadir("haplotypes.bgen")
b = Bgen(path)
for v in iterator(b)
probs = probabilities!(b, v)
dose = first_allele_dosage!(b, v)
dose_correct = probs[1, :] .+ probs[3, :]
@test all(isapprox.(dose, dose_correct))
end
end
end
| BGEN | https://github.com/OpenMendel/BGEN.jl.git |
|
[
"MIT"
] | 0.2.1 | 6512022b637fc17c297bb3050ba2e6cdd50b6fa0 | code | 1480 | @testset "select_region" begin
@testset "select_region_null" begin
chrom, start, stop = "01", 5000, 50000
b = Bgen(example_16bits)
@test b.idx !== nothing
@test length(select_region(b, "02")) == 0
end
@testset "select_whole_chrom" begin
chrom, start, stop = "01", 5000, 50000
b = Bgen(example_16bits)
lt = (x, y) -> isless((x.chrom, x.pos), (y.chrom, y.pos))
variants = collect(select_region(b, chrom))
for (x, y) in zip(sort(variants; lt=lt), sort(gen_data; lt=lt))
@test (x.rsid, x.chrom, x.pos) == (y.rsid, y.chrom, y.pos)
end
end
@testset "select_after_position" begin
chrom, start, stop = "01", 5000, 50000
b = Bgen(example_16bits)
lt = (x, y) -> isless((x.chrom, x.pos), (y.chrom, y.pos))
variants = collect(select_region(b, chrom; start=start))
gen_data_f = filter(x -> x.pos >= start, gen_data)
for (x, y) in zip(sort(variants; lt=lt), sort(gen_data_f; lt=lt))
@test (x.rsid, x.chrom, x.pos) == (y.rsid, y.chrom, y.pos)
end
end
@testset "select_in_region" begin
chrom, start, stop = "01", 5000, 50000
b = Bgen(example_16bits)
lt = (x, y) -> isless((x.chrom, x.pos), (y.chrom, y.pos))
variants = collect(select_region(b, chrom; start=start, stop=stop))
gen_data_f = filter(x -> start <= x.pos <= stop, gen_data)
for (x, y) in zip(sort(variants; lt=lt), sort(gen_data_f; lt=lt))
@test (x.rsid, x.chrom, x.pos) == (y.rsid, y.chrom, y.pos)
end
end
end
| BGEN | https://github.com/OpenMendel/BGEN.jl.git |
|
[
"MIT"
] | 0.2.1 | 6512022b637fc17c297bb3050ba2e6cdd50b6fa0 | code | 582 | @testset "utils" begin
@testset "counts" begin
path = BGEN.datadir("example.8bits.bgen")
b = Bgen(path)
for v in iterator(b)
cnt = counts!(b, v)
dose = first_allele_dosage!(b, v)
correct_cnt = zeros(Int, 512)
for v in dose
if !isnan(v)
ind = convert(Int, round(v * 255)) + 1
correct_cnt[ind] += 1
end
end
correct_cnt[512] = count(isnan.(dose))
@test all(cnt .== correct_cnt)
end
end
end | BGEN | https://github.com/OpenMendel/BGEN.jl.git |
|
[
"MIT"
] | 0.2.1 | 6512022b637fc17c297bb3050ba2e6cdd50b6fa0 | code | 3922 | struct GenVariant
chrom::String
varid::String
rsid::String
pos::UInt
alleles::Vector{String}
probs::Matrix{Float64}
ploidy::Vector{UInt8}
end
"""
load_gen_data()
load data from "example.gen"
"""
function load_gen_data()
variants = GenVariant[]
open(BGEN.datadir("example.gen")) do io
for l in readlines(io)
tokens = split(l)
chrom, varid, rsid, pos, ref, alt = tokens[1:6]
pos = parse(UInt, pos)
probs = reshape(parse.(Float64, tokens[7:end]), 3, :)
nan_col_idx = transpose(sum(probs; dims=1)) .== 0
for (i, v) in enumerate(nan_col_idx)
if v
probs[:, i] .= NaN
end
end
push!(variants, GenVariant(chrom, varid, rsid, pos, [ref, alt], probs, []))
end
end
variants
end
"""
parse_vcf_samples(format, samples)
Parse sample data from VCF.
"""
function parse_vcf_samples(format, samples)
samples = [Dict(zip(split(format, ":"), split(x, ":"))) for x in samples]
ks = Dict([("GT", r"[/|]"), ("GP", ","), ("HP", ",")])
samples2 = [Dict{String, Vector{SubString{String}}}() for x in samples]
for (x, y) in zip(samples, samples2)
for k in keys(x)
y[k] = split(x[k], ks[k])
end
end
probs = [occursin("GP", format) ? x["GP"] : x["HP"] for x in samples2]
probs = map(x -> parse.(Float64, x), probs)
max_len = maximum(length(x) for x in probs)
probs_out = Matrix{Float64}(undef, max_len, length(samples))
fill!(probs_out, NaN)
for i in 1:length(probs)
probs_out[1:length(probs[i]), i] .= probs[i]
end
ploidy = [length(y["GT"]) for y in samples2]
probs_out, ploidy
end
"""
load_vcf_data()
Load data from "complex.vcf" for comparison
"""
function load_vcf_data()
variants = GenVariant[]
open(BGEN.datadir("complex.vcf")) do io
for l in readlines(io)
if startswith(l, "#")
continue
end
tokens = split(l)
chrom, pos, varid, ref, alts = tokens[1:5]
pos = parse(UInt, pos)
format = tokens[9]
samples = tokens[10:end]
varid = split(varid, ",")
if length(varid) > 1
rsid, varid = varid
else
rsid, varid = varid[1], ""
end
probs, ploidy = parse_vcf_samples(format, samples)
var = GenVariant(string(chrom), varid, string(rsid), pos, vcat(String[ref],
split(alts, ",")), probs, ploidy)
push!(variants, var)
end
end
variants
end
"""
load_haps_data()
Load data from "haplotypes.haps" for comparison.
"""
function load_haps_data()
variants = GenVariant[]
open(BGEN.datadir("haplotypes.haps")) do io
for l in readlines(io)
tokens = split(l)
chrom, varid, rsid, pos, ref, alt = tokens[1:6]
pos = parse(UInt, pos)
probs = tokens[7:end]
probs = [x == "0" ? [1.0, 0.0] : [0.0, 1.0] for x in probs]
probs = [probs[pos:pos+1] for pos in 1:2:length(probs)]
probs = [hcat(x[1], x[2]) for x in probs]
probs = hcat(probs...)
probs = reshape(probs, 4, :)
var = GenVariant(chrom, varid, rsid, pos, [ref, alt], probs, [])
push!(variants, var)
end
end
variants
end
"""
epsilon(bit_depth)
Max difference expected for the bit depth
"""
@inline function epsilon(bit_depth)
return 2 / (2 ^ (bit_depth - 1))
end
"""
array_equal(truth, parsed, bit_depth)
check if the two arrays are sufficiently equal
"""
@inline function array_equal(truth, parsed, bit_depth)
eps_abs = 3.2e-5
all(isapprox.(truth, parsed; atol=max(eps_abs, epsilon(bit_depth)),
nans=true))
end
| BGEN | https://github.com/OpenMendel/BGEN.jl.git |
|
[
"MIT"
] | 0.2.1 | 6512022b637fc17c297bb3050ba2e6cdd50b6fa0 | docs | 2381 | # BGEN.jl
[](https://OpenMendel.github.io/BGEN.jl/dev)
[](https://OpenMendel.github.io/BGEN.jl/stable)
[](https://codecov.io/gh/OpenMendel/BGEN.jl)
[](https://github.com/OpenMendel/BGEN.jl/actions)
Routines for reading compressed storage of genotyped or imputed markers
[*Genome-wide association studies (GWAS)*](https://en.wikipedia.org/wiki/Genome-wide_association_study) data with imputed markers are often saved in the [**BGEN format**](https://www.well.ox.ac.uk/~gav/bgen_format/) or `.bgen` file.
It can store both hard calls and imputed data, unphased genotypes and phased haplotypes. Each variant is compressed separately to make indexing simple. An index file (`.bgen.bgi`) may be provided to access each variant easily. [UK Biobank](https://www.ukbiobank.ac.uk/) uses this format for genome-wide imputed genotypes.
## Installation
This package requires Julia v1.0 or later, which can be obtained from
https://julialang.org/downloads/ or by building Julia from the sources in the
https://github.com/JuliaLang/julia repository.
This package is registered in the default Julia package registry, and can be installed through standard package installation procedure: e.g., running the following code in Julia REPL.
```julia
using Pkg
pkg"add BGEN"
```
## Citation
If you use [OpenMendel](https://openmendel.github.io) analysis packages in your research, please cite the following reference in the resulting publications:
*Zhou H, Sinsheimer JS, Bates DM, Chu BB, German CA, Ji SS, Keys KL, Kim J, Ko S, Mosher GD, Papp JC, Sobel EM, Zhai J, Zhou JJ, Lange K. OPENMENDEL: a cooperative programming project for statistical genetics. Hum Genet. 2020 Jan;139(1):61-71. doi: 10.1007/s00439-019-02001-z. Epub 2019 Mar 26. PMID: 30915546; PMCID: [PMC6763373](https://www.ncbi.nlm.nih.gov/pmc/articles/PMC6763373/).*
## Acknowledgments
Current implementation incorporates ideas in a [bgen Python package](https://github.com/jeremymcrae/bgen).
This project has been supported by the National Institutes of Health under awards R01GM053275, R01HG006139, R25GM103774, and 1R25HG011845.
| BGEN | https://github.com/OpenMendel/BGEN.jl.git |
|
[
"MIT"
] | 0.2.1 | 6512022b637fc17c297bb3050ba2e6cdd50b6fa0 | docs | 33436 | # BGEN.jl
Routines for reading compressed storage of genotyped or imputed markers
## The BGEN Format
[*Genome-wide association studies (GWAS)*](https://en.wikipedia.org/wiki/Genome-wide_association_study) data with imputed markers are often saved in the [**BGEN format**](https://www.well.ox.ac.uk/~gav/bgen_format/) or `.bgen` file.
Used in:
* Wellcome Trust Case-Control Consortium 2
* the MalariaGEN project
* the ALSPAC study
* [__UK Biobank__](https://enkre.net/cgi-bin/code/bgen/wiki/?name=BGEN+in+the+UK+Biobank): for genome-wide imputed genotypes and phased haplotypes
### Features
* Can store both hard-calls and imputed data
* Can store both phased haplotypes and phased genotypes
* Efficient variable-precision bit reapresntations
* Per-variant compression $\rightarrow$ easy to index
* Supported compression method: [zlib](http://www.zlib.net/) and [Zstandard](https://facebook.github.io/zstd/).
* Index files are often provided as `.bgen.bgi` files, which are plain [SQLite3](http://www.sqlite.org) databases.
Time to list variant identifying information (genomic location, ID and alleles): 18,496 samples, 121,668 SNPs
(image source: https://www.well.ox.ac.uk/~gav/bgen_format/images/bgen_comparison.png)
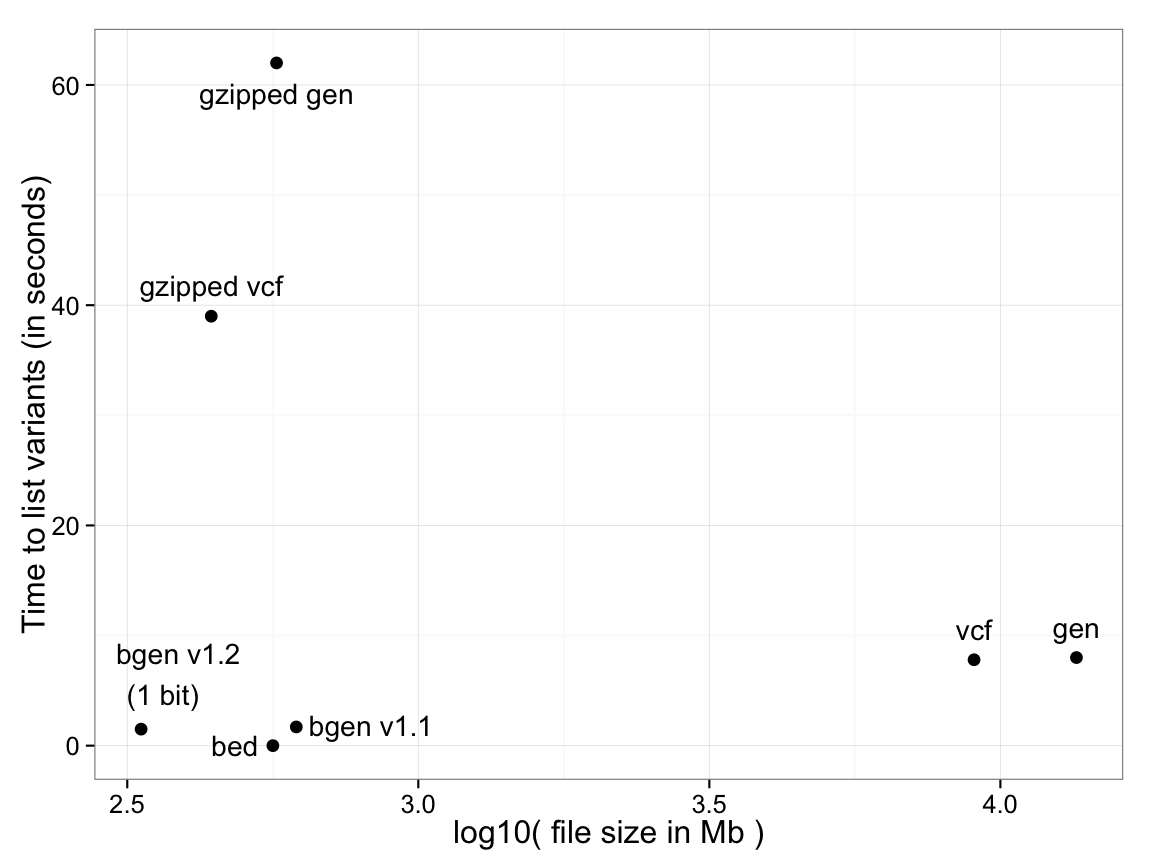
_Plink 1 format (`.bed`/`.bim`/`.fam`) has the list of variants as a separate file (`.bim`), effectively zero time._
### Structure
A header block followed by a series of [variant data block - (compressed) genotype data block] pairs.
* Header block
* number of variants and samples
* compression method (none, zlib or zstandard)
* version of layout
* Only "layout 2" is discussed below. "Layout 1" is also supported.
* sample identifiers (optional)
* Variant data block
* variant id
* genomic position (chromosome, bp coordinate)
* list of alleles
* Genotype data block (often compressed)
* ploidy of each sample (may vary sample-by-sample)
* if the genotype data are phased
* precision ($B$, number of bits to represent probabilities)
* probabilitiy data (e.g. an unsigned $B$-bit integer $x$ represents the probability of ($\frac{x}{2^{B}-1}$)
_`BGEN.jl` provides tools for iterating over the variants and parsing genotype data efficiently. It has been optimized for UK Biobank's zlib-compressed, 8-bit byte-aligned, all-diploid, all-biallelic datafiles._
## Installation
This package requires Julia v1.0 or later, which can be obtained from
https://julialang.org/downloads/ or by building Julia from the sources in the
https://github.com/JuliaLang/julia repository.
The package can be installed by running the following code:
```julia
using Pkg
pkg"add BGEN"
```
In order to run the examples below, the `Glob` package is also needed.
```julia
pkg"add Glob"
```
```julia
versioninfo()
```
Julia Version 1.8.5
Commit 17cfb8e65ea (2023-01-08 06:45 UTC)
Platform Info:
OS: macOS (arm64-apple-darwin21.5.0)
CPU: 8 × Apple M2
WORD_SIZE: 64
LIBM: libopenlibm
LLVM: libLLVM-13.0.1 (ORCJIT, apple-m1)
Threads: 1 on 4 virtual cores
```julia
using BGEN, Glob
```
## Example Data
The example datafiles are stored in `/data` directory of this repository. It can be accessed through the function `BGEN.datadir()`. These files come from [the reference implementation](https://enkre.net/cgi-bin/code/bgen/dir?ci=trunk) for the BGEN format.
```julia
Glob.glob("*", BGEN.datadir())
```
79-element Vector{String}:
"/Users/kose/.julia/dev/BGEN/src/../data/LICENSE.md"
"/Users/kose/.julia/dev/BGEN/src/../data/complex.10bits.bgen"
"/Users/kose/.julia/dev/BGEN/src/../data/complex.11bits.bgen"
"/Users/kose/.julia/dev/BGEN/src/../data/complex.12bits.bgen"
"/Users/kose/.julia/dev/BGEN/src/../data/complex.13bits.bgen"
"/Users/kose/.julia/dev/BGEN/src/../data/complex.14bits.bgen"
"/Users/kose/.julia/dev/BGEN/src/../data/complex.15bits.bgen"
"/Users/kose/.julia/dev/BGEN/src/../data/complex.16bits.bgen"
"/Users/kose/.julia/dev/BGEN/src/../data/complex.17bits.bgen"
"/Users/kose/.julia/dev/BGEN/src/../data/complex.18bits.bgen"
"/Users/kose/.julia/dev/BGEN/src/../data/complex.19bits.bgen"
"/Users/kose/.julia/dev/BGEN/src/../data/complex.1bits.bgen"
"/Users/kose/.julia/dev/BGEN/src/../data/complex.20bits.bgen"
⋮
"/Users/kose/.julia/dev/BGEN/src/../data/example.6bits.bgen"
"/Users/kose/.julia/dev/BGEN/src/../data/example.7bits.bgen"
"/Users/kose/.julia/dev/BGEN/src/../data/example.8bits.bgen"
"/Users/kose/.julia/dev/BGEN/src/../data/example.8bits.bgen.bgi"
"/Users/kose/.julia/dev/BGEN/src/../data/example.9bits.bgen"
"/Users/kose/.julia/dev/BGEN/src/../data/example.gen"
"/Users/kose/.julia/dev/BGEN/src/../data/example.sample"
"/Users/kose/.julia/dev/BGEN/src/../data/example.v11.bgen"
"/Users/kose/.julia/dev/BGEN/src/../data/examples.16bits.bgen"
"/Users/kose/.julia/dev/BGEN/src/../data/haplotypes.bgen"
"/Users/kose/.julia/dev/BGEN/src/../data/haplotypes.bgen.bgi"
"/Users/kose/.julia/dev/BGEN/src/../data/haplotypes.haps"
There are three different datasets with different format versions, compressions, or number of bits to represent probability values.
- `example.*.bgen`: imputed genotypes.
- `haplotypes.bgen`: phased haplotypes.
- `complex.*.bgen`: includes imputed genotypes and phased haplotypes, and multiallelic genotypes.
Some of the `.bgen` files are indexed with `.bgen.bgi` files:
```julia
Glob.glob("*.bgen.bgi", BGEN.datadir())
```
4-element Vector{String}:
"/Users/kose/.julia/dev/BGEN/src/../data/complex.bgen.bgi"
"/Users/kose/.julia/dev/BGEN/src/../data/example.16bits.bgen.bgi"
"/Users/kose/.julia/dev/BGEN/src/../data/example.8bits.bgen.bgi"
"/Users/kose/.julia/dev/BGEN/src/../data/haplotypes.bgen.bgi"
Sample identifiers may be either contained in the `.bgen` file, or is listed in an external `.sample` file.
```julia
Glob.glob("*.sample", BGEN.datadir())
```
2-element Vector{String}:
"/Users/kose/.julia/dev/BGEN/src/../data/complex.sample"
"/Users/kose/.julia/dev/BGEN/src/../data/example.sample"
## Type `Bgen`
The type `Bgen` is the fundamental type for `.bgen`-formatted files. It can be created using the following line.
```julia
b = Bgen(BGEN.datadir("example.8bits.bgen");
sample_path=BGEN.datadir("example.sample"),
idx_path=BGEN.datadir("example.8bits.bgen.bgi"))
```
Bgen(IOStream(<file /Users/kose/.julia/dev/BGEN/src/../data/example.8bits.bgen>), 0x000000000001f6ea, BGEN.Header(0x0000178c, 0x00000014, 0x000000c7, 0x000001f4, 0x01, 0x02, true), ["sample_001", "sample_002", "sample_003", "sample_004", "sample_005", "sample_006", "sample_007", "sample_008", "sample_009", "sample_010" … "sample_491", "sample_492", "sample_493", "sample_494", "sample_495", "sample_496", "sample_497", "sample_498", "sample_499", "sample_500"], Index("/Users/kose/.julia/dev/BGEN/src/../data/example.8bits.bgen.bgi", SQLite.DB("/Users/kose/.julia/dev/BGEN/src/../data/example.8bits.bgen.bgi"), UInt64[], String[], String[], UInt32[]))
The first argument is the path to the `.bgen` file. The optional keyword argument `sample_path` defines the location of the `.sample` file. The second optional keyword argument `idx_path` determines the location of `.bgen.bgi` file.
When a `Bgen` object is created, information in the header is parsed, and the index files are loaded if provided. You may retrieve basic information as follows. Variants are not yet parsed, and will be discussed later.
- `io(b::Bgen)`: IOStream for the bgen file. You may also close this stream using `close(b::Bgen)`.
- `fsize(b::Bgen)`: the size of the bgen file.
- `samples(b::Bgen)`: the list of sample names.
- `n_samples(b::Bgen)`: number of samples in the file.
- `n_variants(b::Bgen)`: number of variants
- `compression(b::Bgen)`: the method each genotype block is compressed. It is either "None", "Zlib", or "Zstd".
```julia
io(b)
```
IOStream(<file /Users/kose/.julia/dev/BGEN/src/../data/example.8bits.bgen>)
```julia
fsize(b)
```
128746
```julia
samples(b)
```
500-element Vector{String}:
"sample_001"
"sample_002"
"sample_003"
"sample_004"
"sample_005"
"sample_006"
"sample_007"
"sample_008"
"sample_009"
"sample_010"
"sample_011"
"sample_012"
"sample_013"
⋮
"sample_489"
"sample_490"
"sample_491"
"sample_492"
"sample_493"
"sample_494"
"sample_495"
"sample_496"
"sample_497"
"sample_498"
"sample_499"
"sample_500"
```julia
n_samples(b)
```
500
```julia
n_variants(b)
```
199
```julia
compression(b)
```
"Zlib"
One may also access the list of RSIDs, chromosomes, and positions in chromosome of each variant stored using functions `rsids()`, `chroms()`, and `positions()`, respectively.
```julia
rsids(b)
```
199-element Vector{String}:
"RSID_101"
"RSID_2"
"RSID_102"
"RSID_3"
"RSID_103"
"RSID_4"
"RSID_104"
"RSID_5"
"RSID_105"
"RSID_6"
"RSID_106"
"RSID_7"
"RSID_107"
⋮
"RSID_194"
"RSID_95"
"RSID_195"
"RSID_96"
"RSID_196"
"RSID_97"
"RSID_197"
"RSID_98"
"RSID_198"
"RSID_99"
"RSID_199"
"RSID_200"
```julia
chroms(b)
```
199-element Vector{String}:
"01"
"01"
"01"
"01"
"01"
"01"
"01"
"01"
"01"
"01"
"01"
"01"
"01"
⋮
"01"
"01"
"01"
"01"
"01"
"01"
"01"
"01"
"01"
"01"
"01"
"01"
```julia
positions(b)
```
199-element Vector{Int64}:
1001
2000
2001
3000
3001
4000
4001
5000
5001
6000
6001
7000
7001
⋮
94001
95000
95001
96000
96001
97000
97001
98000
98001
99000
99001
100001
## `Variant` and `VariantIterator`
As noted earlier, genotype information of each variant is compressed separately in `.bgen` files. The offsets (starting points in bgen file) of the genotypes may or may not be indexed by an external `.bgen.bgi` file. Thus, two ways to iterate over variants is provided through the function `iterator(b; offsets=nothing, from_bgen_start=false)`.
- If `offsets` is provided, or `.bgen.bgi` is provided and
`from_bgen_start` is `false`, it returns a `VariantIteratorFromOffsets`, iterating over the list of offsets.
- Otherwise, it returns a `VariantIteratorFromStart`, iterating from the start of bgen file to the end of it sequentially.
`VariantIteratorFromOffsets` and `VariantIteratorFromStart` are the subtypes of `VariantIterator`.
Each element of `VariantIterator` is a `Variant`, containing the information of variants. We have following utility functions to access its information.
- `n_samples(v::Variant)`
- `varid(v::Variant)`
- `rsid(v::Variant)`
- `chrom(v::Variant)`
- `pos(v::Variant)`
- `n_alleles(v::Variant)`: number of alleles.
- `alleles(v::Variant)`: list of alleles.
Merely the basic information of a variant is parsed for creating a `Variant` object. Nothing is decompressed, and genotype probabilities are not yet parsed yet. Decompression happens lazily, and is delayed until when we try to compute genotype probabilites or minor allele dosages (to be discussed later).
Since `.bgen.bgi` file is provided, the following order is based on the index file, sorted by genomic location.
```julia
for v in iterator(b) #
println(rsid(v))
end
```
RSID_101
RSID_2
RSID_102
RSID_3
RSID_103
RSID_4
RSID_104
RSID_5
RSID_105
RSID_6
RSID_106
RSID_7
RSID_107
RSID_8
RSID_108
RSID_9
RSID_109
RSID_10
RSID_100
RSID_110
RSID_11
RSID_111
RSID_12
RSID_112
RSID_13
RSID_113
RSID_14
RSID_114
RSID_15
RSID_115
RSID_16
RSID_116
RSID_17
RSID_117
RSID_18
RSID_118
RSID_19
RSID_119
RSID_20
RSID_120
RSID_21
RSID_121
RSID_22
RSID_122
RSID_23
RSID_123
RSID_24
RSID_124
RSID_25
RSID_125
RSID_26
RSID_126
RSID_27
RSID_127
RSID_28
RSID_128
RSID_29
RSID_129
RSID_30
RSID_130
RSID_31
RSID_131
RSID_32
RSID_132
RSID_33
RSID_133
RSID_34
RSID_134
RSID_35
RSID_135
RSID_36
RSID_136
RSID_37
RSID_137
RSID_38
RSID_138
RSID_39
RSID_139
RSID_40
RSID_140
RSID_41
RSID_141
RSID_42
RSID_142
RSID_43
RSID_143
RSID_44
RSID_144
RSID_45
RSID_145
RSID_46
RSID_146
RSID_47
RSID_147
RSID_48
RSID_148
RSID_49
RSID_149
RSID_50
RSID_150
RSID_51
RSID_151
RSID_52
RSID_152
RSID_53
RSID_153
RSID_54
RSID_154
RSID_55
RSID_155
RSID_56
RSID_156
RSID_57
RSID_157
RSID_58
RSID_158
RSID_59
RSID_159
RSID_60
RSID_160
RSID_61
RSID_161
RSID_62
RSID_162
RSID_63
RSID_163
RSID_64
RSID_164
RSID_65
RSID_165
RSID_66
RSID_166
RSID_67
RSID_167
RSID_68
RSID_168
RSID_69
RSID_169
RSID_70
RSID_170
RSID_71
RSID_171
RSID_72
RSID_172
RSID_73
RSID_173
RSID_74
RSID_174
RSID_75
RSID_175
RSID_76
RSID_176
RSID_77
RSID_177
RSID_78
RSID_178
RSID_79
RSID_179
RSID_80
RSID_180
RSID_81
RSID_181
RSID_82
RSID_182
RSID_83
RSID_183
RSID_84
RSID_184
RSID_85
RSID_185
RSID_86
RSID_186
RSID_87
RSID_187
RSID_88
RSID_188
RSID_89
RSID_189
RSID_90
RSID_190
RSID_91
RSID_191
RSID_92
RSID_192
RSID_93
RSID_193
RSID_94
RSID_194
RSID_95
RSID_195
RSID_96
RSID_196
RSID_97
RSID_197
RSID_98
RSID_198
RSID_99
RSID_199
RSID_200
Setting `from_bgen_start=true` forces the iterator to iterate in the order of appearence in the bgen file. This may be different from the order in the index file.
```julia
for v in iterator(b; from_bgen_start=true)
println(rsid(v))
end
```
RSID_2
RSID_3
RSID_4
RSID_5
RSID_6
RSID_7
RSID_8
RSID_9
RSID_10
RSID_11
RSID_12
RSID_13
RSID_14
RSID_15
RSID_16
RSID_17
RSID_18
RSID_19
RSID_20
RSID_21
RSID_22
RSID_23
RSID_24
RSID_25
RSID_26
RSID_27
RSID_28
RSID_29
RSID_30
RSID_31
RSID_32
RSID_33
RSID_34
RSID_35
RSID_36
RSID_37
RSID_38
RSID_39
RSID_40
RSID_41
RSID_42
RSID_43
RSID_44
RSID_45
RSID_46
RSID_47
RSID_48
RSID_49
RSID_50
RSID_51
RSID_52
RSID_53
RSID_54
RSID_55
RSID_56
RSID_57
RSID_58
RSID_59
RSID_60
RSID_61
RSID_62
RSID_63
RSID_64
RSID_65
RSID_66
RSID_67
RSID_68
RSID_69
RSID_70
RSID_71
RSID_72
RSID_73
RSID_74
RSID_75
RSID_76
RSID_77
RSID_78
RSID_79
RSID_80
RSID_81
RSID_82
RSID_83
RSID_84
RSID_85
RSID_86
RSID_87
RSID_88
RSID_89
RSID_90
RSID_91
RSID_92
RSID_93
RSID_94
RSID_95
RSID_96
RSID_97
RSID_98
RSID_99
RSID_100
RSID_101
RSID_102
RSID_103
RSID_104
RSID_105
RSID_106
RSID_107
RSID_108
RSID_109
RSID_110
RSID_111
RSID_112
RSID_113
RSID_114
RSID_115
RSID_116
RSID_117
RSID_118
RSID_119
RSID_120
RSID_121
RSID_122
RSID_123
RSID_124
RSID_125
RSID_126
RSID_127
RSID_128
RSID_129
RSID_130
RSID_131
RSID_132
RSID_133
RSID_134
RSID_135
RSID_136
RSID_137
RSID_138
RSID_139
RSID_140
RSID_141
RSID_142
RSID_143
RSID_144
RSID_145
RSID_146
RSID_147
RSID_148
RSID_149
RSID_150
RSID_151
RSID_152
RSID_153
RSID_154
RSID_155
RSID_156
RSID_157
RSID_158
RSID_159
RSID_160
RSID_161
RSID_162
RSID_163
RSID_164
RSID_165
RSID_166
RSID_167
RSID_168
RSID_169
RSID_170
RSID_171
RSID_172
RSID_173
RSID_174
RSID_175
RSID_176
RSID_177
RSID_178
RSID_179
RSID_180
RSID_181
RSID_182
RSID_183
RSID_184
RSID_185
RSID_186
RSID_187
RSID_188
RSID_189
RSID_190
RSID_191
RSID_192
RSID_193
RSID_194
RSID_195
RSID_196
RSID_197
RSID_198
RSID_199
RSID_200
With the presence of `.bgen.bgi` index file, one may select variants on a certain region using the function `select_region(b, chrom; start=nothing, stop=nothing)`.
The following shows that all 199 variants in the bgen file are located on chromosome 01.
```julia
length(select_region(b, "01"))
```
199
We can see that the first variant since position 5000 at chromosome 01 is "RSID_5":
```julia
first(select_region(b, "01"; start=5000))
```
Variant(0x0000000000001ef8, 0x0000000000001f21, 0x0000000000002169, 0x00000248, 0x000001f4, "SNPID_5", "RSID_5", "01", 0x00001388, 0x0002, ["A", "G"], nothing)
And that the number of variants in chr01:5000-50000 is 92.
```julia
length(select_region(b, "01"; start=5000, stop=50000))
```
92
Finally, one may use the `parse_variants()` function to retrieve the variant information as a `Vector{Variant}`. This is equivalent to calling `collect()` on the corresponding `VariantIterator`. It takes the same arguments as `iterator()`. This keeps all the information of variants in-memory. If the size of bgen file is too large, you might want to avoid this.
```julia
variants = parse_variants(b; from_bgen_start=true)
```
199-element Vector{Variant}:
Variant(0x0000000000001790, 0x00000000000017b9, 0x0000000000001a82, 0x000002c9, 0x000001f4, "SNPID_2", "RSID_2", "01", 0x000007d0, 0x0002, ["A", "G"], nothing)
Variant(0x0000000000001a82, 0x0000000000001aab, 0x0000000000001ced, 0x00000242, 0x000001f4, "SNPID_3", "RSID_3", "01", 0x00000bb8, 0x0002, ["A", "G"], nothing)
Variant(0x0000000000001ced, 0x0000000000001d16, 0x0000000000001ef8, 0x000001e2, 0x000001f4, "SNPID_4", "RSID_4", "01", 0x00000fa0, 0x0002, ["A", "G"], nothing)
Variant(0x0000000000001ef8, 0x0000000000001f21, 0x0000000000002169, 0x00000248, 0x000001f4, "SNPID_5", "RSID_5", "01", 0x00001388, 0x0002, ["A", "G"], nothing)
Variant(0x0000000000002169, 0x0000000000002192, 0x0000000000002389, 0x000001f7, 0x000001f4, "SNPID_6", "RSID_6", "01", 0x00001770, 0x0002, ["A", "G"], nothing)
Variant(0x0000000000002389, 0x00000000000023b2, 0x00000000000025df, 0x0000022d, 0x000001f4, "SNPID_7", "RSID_7", "01", 0x00001b58, 0x0002, ["A", "G"], nothing)
Variant(0x00000000000025df, 0x0000000000002608, 0x00000000000027a4, 0x0000019c, 0x000001f4, "SNPID_8", "RSID_8", "01", 0x00001f40, 0x0002, ["A", "G"], nothing)
Variant(0x00000000000027a4, 0x00000000000027cd, 0x00000000000029de, 0x00000211, 0x000001f4, "SNPID_9", "RSID_9", "01", 0x00002328, 0x0002, ["A", "G"], nothing)
Variant(0x00000000000029de, 0x0000000000002a09, 0x0000000000002c43, 0x0000023a, 0x000001f4, "SNPID_10", "RSID_10", "01", 0x00002710, 0x0002, ["A", "G"], nothing)
Variant(0x0000000000002c43, 0x0000000000002c6e, 0x0000000000002e8a, 0x0000021c, 0x000001f4, "SNPID_11", "RSID_11", "01", 0x00002af8, 0x0002, ["A", "G"], nothing)
Variant(0x0000000000002e8a, 0x0000000000002eb5, 0x00000000000030e0, 0x0000022b, 0x000001f4, "SNPID_12", "RSID_12", "01", 0x00002ee0, 0x0002, ["A", "G"], nothing)
Variant(0x00000000000030e0, 0x000000000000310b, 0x0000000000003375, 0x0000026a, 0x000001f4, "SNPID_13", "RSID_13", "01", 0x000032c8, 0x0002, ["A", "G"], nothing)
Variant(0x0000000000003375, 0x00000000000033a0, 0x00000000000035dd, 0x0000023d, 0x000001f4, "SNPID_14", "RSID_14", "01", 0x000036b0, 0x0002, ["A", "G"], nothing)
⋮
Variant(0x000000000001d991, 0x000000000001d9be, 0x000000000001dc12, 0x00000254, 0x000001f4, "SNPID_189", "RSID_189", "01", 0x00015ba9, 0x0002, ["A", "G"], nothing)
Variant(0x000000000001dc12, 0x000000000001dc3f, 0x000000000001ddf2, 0x000001b3, 0x000001f4, "SNPID_190", "RSID_190", "01", 0x00015f91, 0x0002, ["A", "G"], nothing)
Variant(0x000000000001ddf2, 0x000000000001de1f, 0x000000000001e011, 0x000001f2, 0x000001f4, "SNPID_191", "RSID_191", "01", 0x00016379, 0x0002, ["A", "G"], nothing)
Variant(0x000000000001e011, 0x000000000001e03e, 0x000000000001e214, 0x000001d6, 0x000001f4, "SNPID_192", "RSID_192", "01", 0x00016761, 0x0002, ["A", "G"], nothing)
Variant(0x000000000001e214, 0x000000000001e241, 0x000000000001e407, 0x000001c6, 0x000001f4, "SNPID_193", "RSID_193", "01", 0x00016b49, 0x0002, ["A", "G"], nothing)
Variant(0x000000000001e407, 0x000000000001e434, 0x000000000001e6c9, 0x00000295, 0x000001f4, "SNPID_194", "RSID_194", "01", 0x00016f31, 0x0002, ["A", "G"], nothing)
Variant(0x000000000001e6c9, 0x000000000001e6f6, 0x000000000001e8e1, 0x000001eb, 0x000001f4, "SNPID_195", "RSID_195", "01", 0x00017319, 0x0002, ["A", "G"], nothing)
Variant(0x000000000001e8e1, 0x000000000001e90e, 0x000000000001ec86, 0x00000378, 0x000001f4, "SNPID_196", "RSID_196", "01", 0x00017701, 0x0002, ["A", "G"], nothing)
Variant(0x000000000001ec86, 0x000000000001ecb3, 0x000000000001ef8b, 0x000002d8, 0x000001f4, "SNPID_197", "RSID_197", "01", 0x00017ae9, 0x0002, ["A", "G"], nothing)
Variant(0x000000000001ef8b, 0x000000000001efb8, 0x000000000001f183, 0x000001cb, 0x000001f4, "SNPID_198", "RSID_198", "01", 0x00017ed1, 0x0002, ["A", "G"], nothing)
Variant(0x000000000001f183, 0x000000000001f1b0, 0x000000000001f3d4, 0x00000224, 0x000001f4, "SNPID_199", "RSID_199", "01", 0x000182b9, 0x0002, ["A", "G"], nothing)
Variant(0x000000000001f3d4, 0x000000000001f401, 0x000000000001f6ea, 0x000002e9, 0x000001f4, "SNPID_200", "RSID_200", "01", 0x000186a1, 0x0002, ["A", "G"], nothing)
If the index file (`.bgi`) is provided, the users may search for certain RSID in a BGEN file.
```julia
v = variant_by_rsid(b, "RSID_10")
```
Variant(0x00000000000029de, 0x0000000000002a09, 0x0000000000002c43, 0x0000023a, 0x000001f4, "SNPID_10", "RSID_10", "01", 0x00002710, 0x0002, ["A", "G"], nothing)
Also, the users may look for the `n`-th (1-based) variant with respect to genomic location.
```julia
v = variant_by_index(b, 4)
```
Variant(0x0000000000001a82, 0x0000000000001aab, 0x0000000000001ced, 0x00000242, 0x000001f4, "SNPID_3", "RSID_3", "01", 0x00000bb8, 0x0002, ["A", "G"], nothing)
## Genotype/haplotype probabilities and minor allele dosage
The genotype information is decompressed and parsed when probability data is needed. The parsing is triggered by a call to one of:
- `probabilities!(b::Bgen, v::Variant; T=Float64)` : probability of each genotype/haplotype.
- `first_allele_dosage!(b::Bgen, v::Variant; T=Float64`) : dosage of the first allele for a biallelic variant. The first allele listed is often the alternative allele, but it depends on project-wise convention. For example, the first allele is the reference allele for the UK Biobank project.
- `minor_allele_dosage!(b::Bgen, v::Variant; T=Float64)` : minor allele dosage for a biallelic variant.
Once parsed, the results are cached and loaded on any subsequent calls.
After that, one may access genotype information using the following functions, as well as `probabilities!()` and `minor_allele_dosage!()`:
- `phased(v::Variant)`: if the stored data is phased
- `min_ploidy(v::Variant)`: minimum ploidy across the samples
- `max_ploidy(v::Variant)`: maximum ploidy across the samples
- `ploidy(v::Variant)` : Vector of ploidy for each sample
- `bit_depth(v::Variant)` : number of bits used to represent a probability value
- `missings(v::Variant)` : list of samples data is missing
These functions are allowed after calling `minor_allele_dosage!()`:
- `minor_allele(v::Variant)`
- `major_allele(v::Variant)`
If the data are not phased, `probabilities!(b, v)[i, j]` represents the probability of genotype `i` for sample `j`. Each column sums up to one. The genotypes are in [colex-order](https://en.wikipedia.org/wiki/Lexicographic_order#Colexicographic_order) of allele counts. For example, for three alleles with ploidy 3:
| row index | allele counts | genotype |
|---:|:---:|:---:|
| 1 | (3, 0, 0) | 111 |
| 2 | (2, 1, 0) | 112 |
| 3 | (1, 2, 0) | 122 |
| 4 | (0, 3, 0) | 222 |
| 5 | (2, 0, 1) | 113 |
| 6 | (1, 1, 1) | 123 |
| 7 | (0, 2, 1) | 223 |
| 8 | (1, 0, 2) | 133 |
| 9 | (0, 1, 2) | 233 |
| 10 | (0, 0, 3) | 333 |
```julia
probabilities!(b, variants[1])
```
3×500 Matrix{Float32}:
NaN 0.027451 0.0156863 0.0235294 … 0.0156863 0.921569 0.00392157
NaN 0.00784314 0.0509804 0.933333 0.027451 0.0509804 0.984314
NaN 0.964706 0.933333 0.0431373 0.956863 0.027451 0.0117647
Genotype data for sample 1 is missing in this case.
```julia
missings(variants[1])
```
1-element Vector{Int64}:
1
On the other hand, if the data are phased, `probabilities!(b, v)[i, j]` represents the probability that haplotype `(i - 1) ÷ n_alleles + 1` has allele `(i - 1) % n_alleles + 1` for sample `j`, where `n_alleles` is the number of alleles. The below is an example of phased probabilities. i.e., each column represents each sample, and each group of `n_alleles` rows represent the allele probabilities for each haplotype. In this case, ploidy is `[1, 2, 2, 2]`, thus indexes `[3:4, 1]` are invalid, and is filled with `NaN`.
```julia
b2 = Bgen(BGEN.datadir("complex.bgen"))
vs = parse_variants(b2)
p = probabilities!(b2, vs[3])
```
4×4 Matrix{Float32}:
1.0 0.0 1.0 1.0
0.0 1.0 0.0 0.0
NaN 0.0 1.0 0.0
NaN 1.0 0.0 1.0
This variant has two possible alleles (allele 1: "A" and allele 2: "G"), and all the samples are diploids except for the first one, which is monoploid.
It corresponds to a line of VCF file:
```
#CHROM POS ID REF ALT QUAL FILTER INFO FORMAT sample_0 sample_1 sample_2 sample_3
01 3 V3 A G . . . GT:HP 0:1,0 1|1:0,1,0,1 0|0:1,0,1,0 0|1:1,0,0,1
```
So the first sample is monoploid of A, the second sample is homozygote A|A, the third sample is homozygote G|G, and the last sample is heterozygote A|G (phased).
We can confirm the phasedness and ploidy of each sample as follows.
```julia
phased(vs[3])
```
0x01
```julia
ploidy(vs[3])
```
4-element Vector{UInt8}:
0x01
0x02
0x02
0x02
```julia
alleles(vs[3])
```
2-element Vector{String}:
"A"
"G"
For biallelic genotype data, `first_allele_dosage!(b, v)` and `minor_allele_dosage!(b, v)` can be computed. It also supports phased data.
```julia
first_allele_dosage!(b, variants[1])
```
500-element Vector{Float32}:
NaN
0.0627451
0.08235294
0.9803922
0.09019608
0.14117648
1.0745099
0.054901965
0.10980393
0.121568635
0.14117648
0.21568629
0.08235294
⋮
0.09411766
0.10196079
0.027450982
0.96470594
0.0
1.0117648
0.043137256
0.0627451
1.0431373
0.05882353
1.8941176
0.99215686
```julia
minor_allele_dosage!(b, variants[1])
```
500-element Vector{Float32}:
NaN
0.0627451
0.08235294
0.9803922
0.09019608
0.14117648
1.0745099
0.054901965
0.10980393
0.121568635
0.14117648
0.21568629
0.08235294
⋮
0.09411766
0.10196079
0.027450982
0.96470594
0.0
1.0117648
0.043137256
0.0627451
1.0431373
0.05882353
1.8941176
0.99215686
```julia
phased(variants[1])
```
0x00
```julia
n_alleles(variants[1])
```
2
```julia
minor_allele(variants[1])
```
"A"
```julia
major_allele(variants[1])
```
"G"
`first_allele_dosage!()` and `minor_allele_dosage!()` support a keyword argument `mean_impute`, which imputes missing value with the mean of the non-missing values.
```julia
dose = first_allele_dosage!(b, variants[1]; T=Float64, mean_impute=true)
```
500-element Vector{Float32}:
0.39581063
0.0627451
0.08235294
0.9803922
0.09019608
0.14117648
1.0745099
0.054901965
0.10980393
0.121568635
0.14117648
0.21568629
0.08235294
⋮
0.09411766
0.10196079
0.027450982
0.96470594
0.0
1.0117648
0.043137256
0.0627451
1.0431373
0.05882353
1.8941176
0.99215686
The function `hardcall(d; threshold=0.1)` can be used to convert the dosage vectors to hard-called genotypes, returning a `Vector{UInt8}` array of values `0x00`, `0x01`, `0x02`, or `0x09` (for missing). The function `hardcall!(c, d; threshold=0.1)` can be used to fill in a preallocated integer array `c`. `threshold` determines the maximum distance between the hard genotypes and the dosage values. For example, if `threshold = 0.1`, dosage value in `[0, 0.1)` gives the hard call `0x00`, a value in `(0.9, 1,1)` gives `0x01`, and a value in `(1.9, 2.0]` gives `0x02`. Any other values give `0x09`.
```julia
c = hardcall(dose; threshold=0.1)
```
500-element Vector{UInt8}:
0x09
0x00
0x00
0x01
0x00
0x09
0x01
0x00
0x09
0x09
0x09
0x09
0x00
⋮
0x00
0x09
0x00
0x01
0x00
0x01
0x00
0x00
0x01
0x00
0x09
0x01
```julia
calls = Vector{UInt8}(undef, length(dose))
hardcall!(calls, dose; threshold = 0.1)
```
500-element Vector{UInt8}:
0x09
0x00
0x00
0x01
0x00
0x09
0x01
0x00
0x09
0x09
0x09
0x09
0x00
⋮
0x00
0x09
0x00
0x01
0x00
0x01
0x00
0x00
0x01
0x00
0x09
0x01
## Filtering
Filtering based on `BitVector` for samples and variants is supported through `BGEN.filter` function if bit depth of each variant is a multiple of 8. The syntax is:
```julia
BGEN.filter(dest::AbstractString, b::Bgen, variant_mask::BitVector,
sample_mask::BitVector=trues(length(b.samples));
dest_sample = dest[1:end-5] * ".sample",
sample_path=nothing, sample_names=b.samples,
offsets=nothing, from_bgen_start=false)
```
- `dest` is the output path of the resulting `.bgen` file.
- `b` is a `Bgen` instance.
- `variant_mask` is a `BitVector` for determining whether to include each variant in the output file.
- `sample_mask` is a `BitVector` for determining whether to include each sample in the output file.
- `dest_sample` is the location of the output `.sample` file.
- `sample_path` is the location of the `.sample` file of the input BGEN file.
- `sample_names` is the names of the sample in the input BGEN file.
- `offsets` and `from_bgen_start` are the arguments for the `iterator` method.
It only supports layout 2, and the output is always compressed in ZSTD. The sample names are stored in a separate .sample file, but not in the output .bgen file.
An example of choosing first 10 variants:
```julia
b = Bgen(BGEN.datadir("example.8bits.bgen"))
vidx = falses(b.header.n_variants)
vidx[1:10] .= true
BGEN.filter("test.bgen", b, vidx)
b2 = Bgen("test.bgen"; sample_path="test.sample")
@assert all(b.samples .== b2.samples)
for (v1, v2) in zip(iterator(b), iterator(b2)) # length of two iterators are different.
# it stops when the shorter one (b2) ends.
@assert v1.varid == v2.varid
@assert v1.rsid == v2.rsid
@assert v1.chrom == v2.chrom
@assert v1.pos == v2.pos
@assert v1.n_alleles == v2.n_alleles
@assert all(v1.alleles .== v2.alleles)
decompressed1 = BGEN.decompress(b.io, v1, b.header)
decompressed2 = BGEN.decompress(b2.io, v2, b2.header)
@assert all(decompressed1 .== decompressed2)
end
```
An example of choosing last two samples out of four samples:
```julia
b3 = Bgen(BGEN.datadir("complex.24bits.bgen"))
BGEN.filter("test2.bgen", b3, trues(b3.header.n_variants), BitVector([false, false, true, true]))
b4 = Bgen("test2.bgen"; sample_path = "test2.sample")
for (v3, v4) in zip(iterator(b3), iterator(b4))
@assert v3.varid == v4.varid
@assert v3.rsid == v4.rsid
@assert v3.chrom == v4.chrom
@assert v3.pos == v4.pos
@assert v3.n_alleles == v4.n_alleles
@assert all(v3.alleles .== v4.alleles)
@assert isapprox(probabilities!(b3, v3)[:, 3:4], probabilities!(b4, v4); nans=true)
end
```
```julia
rm("test.bgen", force=true)
rm("test.sample", force=true)
rm("test2.bgen", force=true)
rm("test2.sample", force=true)
```
| BGEN | https://github.com/OpenMendel/BGEN.jl.git |
|
[
"MIT"
] | 0.1.3 | 5f5a59cab6b3920d0b98dbebbb3f3f0477441546 | code | 7929 | using ContinuousTimePolicyGradients
using DiffEqFlux, ComponentArrays, LinearAlgebra, JLD2, OrdinaryDiffEq
using Plots
function main(maxiters_1::Int, maxiters_2::Int, Δt_save::Float32; p_NN_0 = nothing, k_a_val = 10.0)
# model + problem parameters
(aₐ, aₙ, bₙ, cₙ, dₙ, aₘ, bₘ, cₘ, dₘ) = Float32.([-0.3, 19.373, -31.023, -9.717, -1.948, 40.44, -64.015, 2.922, -11.803])
(m, I_yy, S, d, ωₐ, ζₐ) = Float32.([204.02, 247.439, 0.0409, 0.2286, 150.0, 0.7])
(g, ρ₀, H, γₐ, Rₐ, T₀, λ) = Float32.([9.8, 1.225, 8435.0, 1.4, 286.0, 288.15, 0.0065])
(a_max, α_max, δ_max, δ̇_max, q_max, M_max, h_max) = Float32.([100.0, deg2rad(30), deg2rad(25), 1.5, deg2rad(60), 4, 11E3])
(k_a, k_q, k_δ, k_δ̇, k_R) = Float32.([k_a_val, 0.0, 0.01, 0.1, 1E-4])
# dynamic model
dim_x = 7
function dynamics_plant(t, x, u)
(h, V, α, q, θ, δ, δ̇) = x
δ_c = u[1]
ρ = ρ₀ * exp(-h / H)
Vₛ = sqrt(γₐ * Rₐ * (T₀ - λ * h))
M = V / Vₛ
γ = θ - α
Q = 0.5f0 * ρ * V^2
C_A = aₐ
C_N = aₙ * α^3 + bₙ * α * abs(α) + cₙ * (2.0f0 - M / 3.0f0) * α + dₙ * δ
C_M = aₘ * α^3 + bₘ * α * abs(α) + cₘ * (-7.0f0 + 8.0f0 * M / 3.0f0) * α + dₘ * δ
α̇ = Q * S / m / V * (C_N * cos(α) - C_A * sin(α)) + g / V * cos(γ) + q
dx = [V * sin(γ);
Q * S / m * (C_N * sin(α) + C_A * cos(α)) - g * sin(γ);
α̇;
Q * S * d / I_yy * C_M;
q;
δ̇;
-ωₐ^2 * (δ - δ_c) - 2.0f0 * ζₐ * ωₐ * δ̇]
return dx
end
dim_x_c = 2
function dynamics_controller(t, x_c, y, r, p_NN, policy_NN)
(a_z, h, V, M, α, q, γ) = y
a_z_cmd = r[1]
x_int = x_c[1]
x_ref = x_c[2]
y_NN = (K_A, K_I, K_R) = policy_NN([ abs(α) / α_max; M / M_max; h / h_max], p_NN)
# dx_ref = [-36.6667f0 -13.8889f0; 8.0f0 0.0f0] * x_ref + [4.0f0; 0.0f0] * a_z_cmd
# a_z_ref = [-1.0083f0 3.4722f0] * x_ref
dx_ref = (a_z_cmd - x_ref) / 0.2f0
a_z_ref = x_ref
dx_c = [K_A * (a_z_cmd - a_z) + q + (a_z_cmd + g * cos(γ)) / V;
dx_ref]
u = [K_I * x_int + K_R * q;
a_z_ref]
return dx_c, u, y_NN
end
function dynamics_sensor(t, x)
(h, V, α, q, θ, δ, _) = x
ρ = ρ₀ * exp(-h / H)
Vₛ = sqrt(γₐ * Rₐ * (T₀ - λ * h))
M = V / Vₛ
Q = 0.5f0 * ρ * V^2
γ = θ - α
C_A = aₐ
C_N = aₙ * α^3 + bₙ * α * abs(α) + cₙ * (2.0f0 - M / 3.0f0) * α + dₙ * δ
a_z = Q * S / m * (C_N * cos(α) - C_A * sin(α))
y = [a_z;
h;
V;
M;
α;
q;
γ]
return y
end
# cost definition
function cost_running(t, x, y, u, r)
q = x[4]
δ̇ = x[7]
a_z = y[1]
δ_c = u[1]
a_z_ref = u[2]
a_z_cmd = r[1]
return k_a * ((a_z - a_z_ref) / abs(a_z_cmd))^2 + k_δ̇ * (δ̇ / δ̇_max)^2 + k_δ * (δ_c / δ_max)^2 + k_q * (q / q_max)^2
end
function cost_terminal(x_f, r)
# a_z_cmd = r[1]
# y = dynamics_sensor(3.0f0, x_f)
# a_z = y[1]
# return ((a_z - a_z_cmd) / (1.0f0 + a_z_cmd))^2
return 0.0f0
end
function cost_regularisor(p_NN)
return k_R * norm(p_NN)^2
# return 0.0f0
end
# NN construction
dim_NN_hidden = 10
dim_NN_input = 3
dim_K = 3
K_lb = Float32.(0.001*ones(3))
K_ub = Float32[4, 0.2, 2]
policy_NN = FastChain(
FastDense(dim_NN_input, dim_NN_hidden, tanh),
FastDense(dim_NN_hidden, dim_NN_hidden, tanh),
FastDense(dim_NN_hidden, dim_K),
(x, p) -> (K_ub - K_lb) .* σ.(x) .+ K_lb
)
# scenario definition
ensemble = [ (; x₀ = Float32[h₀; V₀; α₀; zeros(4)], r = Float32[a_z_cmd])
for h₀ = 5E3:1E3:8E3
for V₀ = 7E2:1E2:9E2
for α₀ = -0.3:0.1:0
for a_z_cmd = filter(!iszero, 0:5E1:1E2) ]
t_span = Float32.((0.0, 3.0))
t_save = t_span[1]:Δt_save:t_span[2]
scenario = (; ensemble = ensemble, t_span = t_span, t_save = t_save, dim_x = dim_x, dim_x_c = dim_x_c)
# NN training
(result, fwd_ensemble_sol, loss_history) = CTPG_train(dynamics_plant, dynamics_controller, dynamics_sensor, cost_running, cost_terminal, cost_regularisor, policy_NN, scenario; sense_alg = InterpolatingAdjoint(autojacvec = ReverseDiffVJP(true)), ensemble_alg = EnsembleThreads(), maxiters_1 = maxiters_1, maxiters_2 = maxiters_2, opt_2 = BFGS(initial_stepnorm = 0.0001f0), i_nominal = 1, p_NN_0 = p_NN_0, progress_plot = false)
# solve_alg = Euler(), dt=0.001f0
return result, policy_NN, fwd_ensemble_sol, loss_history
end
## execute optimisation and simulation
@time (result, policy_NN, fwd_ensemble_sol, loss_history) = main(1000, 1500, 0.01f0; k_a_val = 100.0)
# re-execute optimisation and simulation
# p_NN_prev = result.u
# @time (result, policy_NN, fwd_ensemble_sol, loss_history) = main(10, 1000, 0.01f0; k_a_val = 100.0, p_NN_0 = p_NN_prev)
# save results
jldsave("DS_base.jld2"; result, fwd_ensemble_sol, loss_history)
# p_NN_base = result.u
# jldsave("p_NN_loss_base.jld2"; p_NN_base, loss_history)
# plot simulation results
# (result, fwd_ensemble_sol, loss_history) = load(".jld2", "result", "fwd_ensemble_sol", "loss_history")
x_names = ["\$h\$" "\$V\$" "\$\\alpha\$" "\$q\$" "\$\\theta\$" "\$\\delta\$" "\$\\dot{\\delta}\$"]
vars_x = 1:6 # [1,2,3, (1,2), (2,3)]
u_names = ["\$\\delta_{c}\$"]
vars_u = 1
y_names = ["\$a_{z}\$"]
vars_y = 1
y_NN_names = ["\$K_A\$" "\$K_I\$" "\$K_R\$"]
vars_y_NN = 1:3
(f_x, f_u, f_y, f_y_NN, f_L) = view_result([], fwd_ensemble_sol, loss_history; x_names = x_names, vars_x = vars_x, u_names = u_names, vars_u = vars_u, y_names = y_names, vars_y = vars_y, y_NN_names = y_NN_names, vars_y_NN = vars_y_NN, linealpha = 0.6)
# plot gain surfaces
# (p_NN_base) = load("p_NN_loss_base.jld2", "p_NN_base")
(a_max, α_max, δ_max, δ̇_max, q_max, M_max, h_max) = Float32.([100.0, deg2rad(30), deg2rad(25), 1.5, deg2rad(60), 4, 11E3])
# dim_NN_hidden = 10
# dim_NN_input = 3
# dim_K = 3
# K_lb = Float32.(0.001*ones(3))
# K_ub = Float32[4, 0.2, 2]
# policy_NN = FastChain(
# FastDense(dim_NN_input, dim_NN_hidden, tanh),
# FastDense(dim_NN_hidden, dim_NN_hidden, tanh),
# FastDense(dim_NN_hidden, dim_K),
# (x, p) -> (K_ub - K_lb) .* σ.(x) .+ K_lb
# )
for h in 5E3:1E3:8E3
α_list = 0:1E-3:0.3
M_list = 0.5:0.1:3.0
func_K_A(α, M) = policy_NN([abs(α) / α_max; M / M_max; h / h_max], result.u)[1]
func_K_I(α, M) = policy_NN([abs(α) / α_max; M / M_max; h / h_max], result.u)[2]
func_K_R(α, M) = policy_NN([abs(α) / α_max; M / M_max; h / h_max], result.u)[3]
f_K_A = plot(α_list, M_list, func_K_A, st=:surface, label = :false, zlabel = "\$K_{A}\$", xlabel = "\$\\left|\\alpha\\right|\$", ylabel = "\$M\$")
display(f_K_A)
savefig(f_K_A, "f_K_A_$(Int64(h)).pdf")
f_K_I = plot(α_list, M_list, func_K_I, st=:surface, label = :false, zlabel = "\$K_{I}\$", xlabel = "\$\\left|\\alpha\\right|\$", ylabel = "\$M\$")
display(f_K_I)
savefig(f_K_I, "f_K_I_$(Int64(h)).pdf")
f_K_R = plot(α_list, M_list, func_K_R, st=:surface, label = :false, zlabel = "\$K_{R}\$", xlabel = "\$\\left|\\alpha\\right|\$", ylabel = "\$M\$")
display(f_K_R)
savefig(f_K_R, "f_K_R_$(Int64(h)).pdf")
end
# Learning curve
# using Plots, JLD2
# loss_base = load("DS_base.jld2", "loss_history")
# loss_unscaled = load("DS_unscaled.jld2", "loss_history")
# loss_discrete = load("DS_discrete.jld2", "loss_history")
# f_J = plot([[loss_base], [loss_unscaled], [loss_discrete]], label = ["base" "unscaled" "discrete"], xlabel = "iteration", ylabel = "cost \$J\$", yaxis = :log10)
# savefig(f_J, "f_J.pdf") | ContinuousTimePolicyGradients | https://github.com/nhcho91/ContinuousTimePolicyGradients.jl.git |
|
[
"MIT"
] | 0.1.3 | 5f5a59cab6b3920d0b98dbebbb3f3f0477441546 | code | 222 | module ContinuousTimePolicyGradients
using LinearAlgebra, Statistics
using OrdinaryDiffEq, DiffEqFlux, GalacticOptim
using UnPack, Plots
export CTPG_train, view_result
include("construct_CTPG.jl")
end | ContinuousTimePolicyGradients | https://github.com/nhcho91/ContinuousTimePolicyGradients.jl.git |
|
[
"MIT"
] | 0.1.3 | 5f5a59cab6b3920d0b98dbebbb3f3f0477441546 | code | 15534 | """
CTPG_train(dynamics_plant::Function, dynamics_controller::Function, cost_running::Function, cost_terminal::Function, cost_regularisor::Function, policy_NN, scenario;
solve_alg = Tsit5(), sense_alg = InterpolatingAdjoint(autojacvec = ZygoteVJP()), ensemble_alg = EnsembleThreads(), opt_1 = ADAM(0.01), opt_2 = LBFGS(), maxiters_1 = 100, maxiters_2 = 100, progress_plot = true, solve_kwargs...)
`CTPG_train()` provides a high-level interface for optimisation of the neural networks inside an ODE-represented dynamics based on Continuous-Time Policy Gradient (CTPG) methods that belong to the adjoint sensitivity analysis techniques. The code implemented and the default values for keyword arguments are specified considering training of a neural controller as the main application. In the context herein, a neural controller refers to a dynamic controller that incorporates neural-network-represented components at some points in its mathematical description.
The code utilises the functionalities provided by the [DiffEqFlux.jl](https://github.com/SciML/DiffEqFlux.jl) and [DiffEqSensitivity.jl](https://github.com/SciML/DiffEqSensitivity.jl) packages, and the Automatic Differentiation (AD) capabilities provided by the [Zygote.jl](https://github.com/FluxML/Zygote.jl) package that is integrated in DiffEqFlux.jl. `CTPG_train()` presumes the consistency of the functions provided as its input arguments with the AD tool, hence, the dynamics and cost functions should maintain their transparence against AD tools.
The optimisation (training) problem minimises the cost function defined over deterministic samples of the initial plant state `x₀` and the reference `r` by performing ensemble simulation based on parallelised computation.
The signals are defined as described below:
- `t`: time
- `x`: plant state
- `y`: plant output (= sensor output)
- `x_c`: controller state
- `u`: plant input (= controller output)
- `r`: exogenous reference
- `x_aug`: augmented forward dynamics state (= `[x; x_c; ∫cost_running]`)
- `p_NN`: neural network parameter
The arguments should be provided as explained below:
- `dynamics_plant`: Describes the dynamics of the plant to be controlled. Input arguments `x` and `u` should be of Vector type.
- `dynamics_controller`: Describes the dynamics of the controller that includes neural networks components. Input arguments `x_c`, `y`, `r`, and `p_NN` should be of Vector type.
- `dynamics_sensor`: Describes the dynamics of the sensor that measures output variables fed to the controller. Input arguments `x` should be of Vector type:
- `cost_running`: Describes the running cost defined as the integrand of the Lagrange-form continuous functional. Input arguments `x`, `y`, `u`, and `r` should be of Vector type.
- `cost_terminal`: Describes the terminal cost defined as the Mayer-form problem cost function. Defines a Bolza-form problem along with `cost_running`. Input arguments `x_f` and `r` should be of Vector type.
- `cost_regularisor`: Describes the regularisation term appended to the cost (loss) function. Input argument `p_NN` should be of Vector type.
- `policy_NN`: The neural networks entering into the controller dynamics. DiffEqFlux-based FastChain is recommended for its construction.
- `scenario`: Contains the parameters related with the ensemble-based training scenarios.
- `ensemble`: A vector of the initial plant state `x₀` and the reference `r` constituting the trajectory realisations.
- `t_span`: Time span for forward-pass integration
- `t_save`: Array of time points to be saved while solving ODE. Typically defined as `t_save = t_span[1]:Δt_save:t_span[2]`
- `dim_x`: `length(x)`
- `dim_x_c`: `length(x_c)`
The keyword arguments should be provided as explained below:
- `solve_alg`: The algorithm used for solving ODEs. Default value is `Tsit5()`
- `sense_alg`: The algorithm used for adjoint sensitivity analysis. Default value is `InterpolatingAdjoint(autojacvec = ZygoteVJP())`, because the control problems usually render the `BacksolveAdjoint()` unstable. The vjp choice `autojacvec = ReverseDiffVJP(true)` is usually faster than `ZygoteVJP()`, when the ODE function does not have any branching inside. Please refer to the [DiffEqFlux documentation](https://diffeqflux.sciml.ai/dev/ControllingAdjoints/) for further details.
- `ensemble_alg`: The algorithm used for handling ensemble of ODEs. Default value is `EnsembleThreads()` for multi-threaded computation in CPU.
- `opt_1`: The algorithm used for the first phase of optimisation which rapidly delivers the parameter to a favourable region around a local minimum. Default value is `ADAM(0.01)`.
- `opt_2`: The algorithm used for the second phase of opitmisaiton. Defalut value is `LBFGS()` which refines the result of the first phase to find a more precise minimum. Please refer to the [DiffEqFlux documentation](https://diffeqflux.sciml.ai/dev/sciml_train/) for further details about two-phase composition of optimisers.
- `maxiters_1`: The maximum number of iterations allowed for the first phase of optimisation with `opt_1`. Defalut value is `100`.
- `maxiters_2`: The maximum number of iterations allowed for the second phase of optimisation with `opt_2`. Defalut value is `100`.
- `progress_plot`: The indicator to plot the state history for a nominal condition among the ensemble during the learning process. Default value is `true`.
- `i_nominal`: The index to select the case to plot using `progress_plot` during optimisation process from the `ensemble` defined in `scenario`. Defalut value is `nothing`.
- `p_NN_0`: Initial value of the NN parameters supplied by the user to bypass random initialisation of `p_NN` or to continue optimisation from the previous result. Defalut value is `nothing`.
- `solve_kwargs...`: Additional keyword arguments that are passed onto the ODE solver.
`CTPG_train()` returns the following outputs:
- `result`: The final result of parameter optimisation.
- `fwd_ensemble_sol`: The ensemble solution of forward simulation using the final neural network parameters.
- `loss_history`: The history of loss function evaluated at each iteration.
"""
function CTPG_train(dynamics_plant::Function, dynamics_controller::Function, dynamics_sensor::Function, cost_running::Function, cost_terminal::Function, cost_regularisor::Function, policy_NN, scenario; solve_alg = Tsit5(), sense_alg = InterpolatingAdjoint(autojacvec = ZygoteVJP()), ensemble_alg = EnsembleThreads(), opt_1 = ADAM(0.01), opt_2 = LBFGS(), maxiters_1 = 100, maxiters_2 = 100, progress_plot = true, i_nominal = nothing, p_NN_0 = nothing, solve_kwargs...)
# scenario parameters
@unpack ensemble, t_span, t_save, dim_x, dim_x_c = scenario
dim_ensemble = length(ensemble)
mean_factor = Float32(1 / dim_ensemble)
dim_t_save = length(t_save)
if isnothing(i_nominal)
i_nominal = max(round(Int, dim_ensemble / 2), 1)
end
# NN parameters initialisation
if isnothing(p_NN_0)
p_NN_0 = initial_params(policy_NN)
end
# augmented dynamics
function fwd_dynamics(r)
return function (x_aug, p_NN, t)
x = x_aug[1:dim_x]
x_c = x_aug[dim_x+1:end-1]
# ∫cost_running = x_aug[end]
y = dynamics_sensor(t, x)
(dx_c, u, _) = dynamics_controller(t, x_c, y, r, p_NN, policy_NN)
dx = dynamics_plant(t, x, u)
return [dx; dx_c; cost_running(t, x, y, u, r)]
end
end
# ODE problem construction
prob_base = ODEProblem(fwd_dynamics(ensemble[i_nominal].r), [ensemble[i_nominal].x₀; zeros(Float32, dim_x_c + 1)], t_span, p_NN_0)
function generate_probs(p_NN)
return function (prob, i, repeat)
remake(prob, f = fwd_dynamics(ensemble[i].r), u0 = [ensemble[i].x₀; zeros(Float32, dim_x_c + 1)], p = p_NN)
end
end
if dim_ensemble == 1
prob_mtk = modelingtoolkitize(prob_base)
prob_base = ODEProblem(prob_mtk, [], t_span, jac = true)
ensemble_alg = EnsembleSerial()
end
# loss function definition
function loss(p_NN)
ensemble_prob = EnsembleProblem(prob_base, prob_func = generate_probs(p_NN))
fwd_ensemble_sol_full = solve(ensemble_prob, solve_alg, ensemble_alg, saveat = t_save, trajectories = dim_ensemble, sensealg = sense_alg; solve_kwargs...)
# version 1: mean(sol[end])
# sol_length = Float32.([max(1,length(fwd_ensemble_sol_full[i])) for i in 1:dim_ensemble])
# sol_length_ratio = maximum(sol_length)./sol_length
# loss_val = mean([fwd_ensemble_sol_full[i][end][end] + cost_terminal(fwd_ensemble_sol_full[i][end][1:dim_x], ensemble[i].r) for i in 1:dim_ensemble] .* sol_length_ratio) + cost_regularisor(p_NN)
# version 2: mean(Array[end])
# loss_val = mean([(Array(fwd_ensemble_sol_full[i])[end,end] + cost_terminal(Array(fwd_ensemble_sol_full[i])[1:dim_x,end], ensemble[i].r)) for i in 1:dim_ensemble] .* sol_length_ratio) + cost_regularisor(p_NN)
# version 3: scalar operation (best for code robustness as it can handle divergent case in ensemble)
loss_val = 0.0f0
for i in 1:dim_ensemble
fwd_sol = Array(fwd_ensemble_sol_full[i])
if size(fwd_sol,2) > dim_t_save
fwd_sol = fwd_sol[:,1:dim_t_save]
end
if size(fwd_sol,2) == dim_t_save
x_aug_f = fwd_sol[:,end]
loss_val += (x_aug_f[end] + cost_terminal(x_aug_f[1:dim_x], ensemble[i].r)) * mean_factor
else
loss_val += 1000.0f0
end
end
loss_val += cost_regularisor(p_NN)
return loss_val, fwd_ensemble_sol_full
end
# learning progress callback setup
loss_history = fill(NaN32, maxiters_1 + maxiters_2 + 2)
iterator_learning = 1
cb_progress = function (p_NN_val, loss_val, fwd_ensemble_sol_full; plot_val = progress_plot)
@show (loss_val, iterator_learning);
loss_history[iterator_learning] = loss_val
if plot_val
fwd_ensemble_sol = Array(fwd_ensemble_sol_full[i_nominal])
display(scatter(fwd_ensemble_sol[1:dim_x, :]', label = :false, plot_title = "System State: Learning Iteration $(iterator_learning)", layout = (dim_x, 1), size = (700, 200 * dim_x)))
end
iterator_learning += 1
return false
end
# NN training
if maxiters_1 <= 0
result_coarse = (; u = p_NN_0)
else
result_coarse = DiffEqFlux.sciml_train(loss, p_NN_0, opt_1; cb = cb_progress, maxiters = maxiters_1)
end
if maxiters_2 <= 0
result = result_coarse
else
result = DiffEqFlux.sciml_train(loss, result_coarse.u, opt_2; cb = cb_progress, maxiters = maxiters_2)
end
# Forward solution for optimised p_NN
function eval_IO(r, p_NN)
# Evaluation of input u, output y, and NN output y_NN at each t
return function (t, x_aug)
x = x_aug[1:dim_x]
x_c = x_aug[dim_x+1:end-1]
y = dynamics_sensor(t, x)
(_, u, y_NN) = dynamics_controller(t, x_c, y, r, p_NN, policy_NN)
return (; u = u, y = y, y_NN = y_NN)
end
end
function generate_savedata(p_NN)
return function (sol, i)
IO = eval_IO(ensemble[i].r, p_NN).(sol.t, sol.u)
return ((; sol = sol, u = [u for (u, y, y_NN) in IO], y = [y for (u, y, y_NN) in IO], y_NN = [y_NN for (u, y, y_NN) in IO]), false)
end
end
fwd_ensemble_sol = solve(EnsembleProblem(prob_base, prob_func = generate_probs(result.u), output_func = generate_savedata(result.u)), solve_alg, ensemble_alg, saveat = t_save, trajectories = dim_ensemble, sensealg = sense_alg; solve_kwargs...)
loss_history = filter(!isnan, loss_history)
return result, fwd_ensemble_sol, loss_history
end
function view_result(i_plot_list, fwd_ensemble_sol, loss_history, save_fig = true;
vars_x = nothing, x_names = [], vars_u = nothing, u_names = [], vars_y = nothing, y_names = [], vars_y_NN = nothing, y_NN_names = [], plot_kwargs...)
# labels, variables to be plotted, and layouts setting
(dim_x_aug, dim_u, dim_y, dim_y_NN) = (length(fwd_ensemble_sol[1].sol[1]), length(fwd_ensemble_sol[1].u[1]), length(fwd_ensemble_sol[1].y[1]), length(fwd_ensemble_sol[1].y_NN[1]))
(xlabel_t, ylabel_x, ylabel_u, ylabel_y, ylabel_y_NN) = ("\$t\$", "state", "input", "output", "NN output")
if ~isnothing(vars_x)
layout_x = (length(vars_x), 1)
if ~isempty(x_names)
ylabel_x = x_names[vars_x']
end
else
layout_x = (dim_x_aug - 1, 1)
vars_x = 1:dim_x_aug-1
end
if ~isnothing(vars_u)
layout_u = (length(vars_u), 1)
if ~isempty(u_names)
ylabel_u = u_names[vars_u']
end
else
layout_u = (dim_u, 1)
vars_u = 1:dim_u
end
if ~isnothing(vars_y)
layout_y = (length(vars_y), 1)
if ~isempty(y_names)
ylabel_y = y_names[vars_y']
end
else
layout_y = (dim_y, 1)
vars_y = 1:dim_y
end
if ~isnothing(vars_y_NN)
layout_y_NN = (length(vars_y_NN), 1)
if ~isempty(y_NN_names)
ylabel_y_NN = y_NN_names[vars_y_NN']
end
else
layout_y_NN = (dim_y_NN, 1)
vars_y_NN = 1:dim_y_NN
end
# plotting
if isempty(i_plot_list)
i_plot_list = 1:length(fwd_ensemble_sol)
end
(f_x, f_u, f_y, f_y_NN) = (plot(), plot(), plot(), plot())
for i in i_plot_list
# data preprocessing
@unpack sol, u, y, y_NN = fwd_ensemble_sol[i]
@unpack t = sol
u = reduce(vcat, u') # hcat(u...)'
y = reduce(vcat, y') # hcat(y...)'
y_NN = reduce(vcat, y_NN') # hcat(y_NN...)'
if i == first(i_plot_list)
f_x = plot(sol, vars = vars_x, layout = layout_x, label = :false, xlabel = xlabel_t, ylabel = ylabel_x, size = (800, 160 * length(vars_x)); plot_kwargs...)
f_u = plot(t, u[:, vars_u], layout = layout_u, label = :false, xlabel = xlabel_t, ylabel = ylabel_u; plot_kwargs...)
f_y = plot(t, y[:, vars_y], layout = layout_y, label = :false, xlabel = xlabel_t, ylabel = ylabel_y; plot_kwargs...)
f_y_NN = plot(t, y_NN[:, vars_y_NN], layout = layout_y_NN, label = :false, xlabel = xlabel_t, ylabel = ylabel_y_NN; plot_kwargs...)
else
plot!(f_x, sol, vars = vars_x, layout = layout_x, label = :false, xlabel = xlabel_t, ylabel = ylabel_x, size = (800, 160 * length(vars_x)); plot_kwargs...)
plot!(f_u, t, u[:, vars_u], layout = layout_u, label = :false, xlabel = xlabel_t, ylabel = ylabel_u; plot_kwargs...)
plot!(f_y, t, y[:, vars_y], layout = layout_y, label = :false, xlabel = xlabel_t, ylabel = ylabel_y; plot_kwargs...)
plot!(f_y_NN, t, y_NN[:, vars_y_NN], layout = layout_y_NN, label = :false, xlabel = xlabel_t, ylabel = ylabel_y_NN; plot_kwargs...)
end
end
f_L = plot(loss_history, label = :false, xlabel = "iteration", ylabel = "\$L\$"; plot_kwargs...)
display(f_x)
display(f_u)
display(f_y)
display(f_y_NN)
display(f_L)
if save_fig
savefig(f_x, "f_x.pdf")
savefig(f_u, "f_u.pdf")
savefig(f_y, "f_y.pdf")
savefig(f_y_NN, "f_y_NN.pdf")
savefig(f_L, "f_L.pdf")
end
return f_x, f_u, f_y, f_y_NN, f_L
end
| ContinuousTimePolicyGradients | https://github.com/nhcho91/ContinuousTimePolicyGradients.jl.git |
|
[
"MIT"
] | 0.1.3 | 5f5a59cab6b3920d0b98dbebbb3f3f0477441546 | code | 131 | using ContinuousTimePolicyGradients
using Test
@testset "ContinuousTimePolicyGradients.jl" begin
# Write your tests here.
end
| ContinuousTimePolicyGradients | https://github.com/nhcho91/ContinuousTimePolicyGradients.jl.git |
|
[
"MIT"
] | 0.1.3 | 5f5a59cab6b3920d0b98dbebbb3f3f0477441546 | code | 5805 | using OrdinaryDiffEq, DiffEqFlux, Plots
# model + problem parameters
(aₐ, aₙ, bₙ, cₙ, dₙ, aₘ, bₘ, cₘ, dₘ) = Float32.([-0.3, 19.373, -31.023, -9.717, -1.948, 40.44, -64.015, 2.922, -11.803])
(m, I_yy, S, d, ωₐ, ζₐ) = Float32.([204.02, 247.439, 0.0409, 0.2286, 150.0, 0.7])
(g, ρ₀, H, γₐ, Rₐ, T₀, λ) = Float32.([9.8, 1.225, 8435.0, 1.4, 286.0, 288.15, 0.0065])
(a_max, α_max, δ_max, q_max, M_max, h_max) = Float32.([100.0, deg2rad(30), deg2rad(25), deg2rad(5), 4, 11E3])
(k_a, k_δ, k_R) = Float32.([1.0, 0.1, 0.00001])
t_span = (0.0f0, 3.0f0)
x₀ = Float32.([10E3; 900; zeros(7)])
a_z_cmd = 100.0f0
# cost function
L(a_z, a_z_cmd, δ_c) = k_a * ((a_z - a_z_cmd) / a_max)^2 + k_δ * (δ_c / δ_max)^2 # + k_q * (q / q_max)^2
ϕ(a_z, a_z_cmd) = ((a_z - a_z_cmd) / a_max)^2
R(p_NN) = k_R * p_NN' * p_NN
# NN construction
dim_hidden = 64
(K_lb, K_ub) = Float32.([-0.3, 0])
Π = FastChain(
FastDense(3, dim_hidden, tanh),
FastDense(dim_hidden, dim_hidden, tanh),
FastDense(dim_hidden, 3),
(x, p) -> (K_ub - K_lb) * σ.(x) .+ K_lb
)
p_NN = initial_params(Π)
# plant + controller dynamics + running cost integrator
function fwd_dynamics_model(x, p_NN, t)
(h, V, α, q, θ, δ, δ̇, x_c, _) = x
ρ = ρ₀ * exp(-h / H)
Vₛ = sqrt(γₐ * Rₐ * (T₀ - λ * h))
M = V / Vₛ
γ = θ - α
Q = 0.5f0 * ρ * V^2
C_A = aₐ
C_N = aₙ * α^3 + bₙ * α * abs(α) + cₙ * (2.0f0 - M / 3.0f0) * α + dₙ * δ
C_M = aₘ * α^3 + bₘ * α * abs(α) + cₘ * (-7.0f0 + 8.0f0 * M / 3.0f0) * α + dₘ * δ
# K_A, K_I, K_R = min.( Π([α / α_max; M / M_max; h / h_max], p_NN) , K_ub)
K_A, K_I, K_R = Π([α / α_max; M / M_max; h / h_max], p_NN)
δ_c = -K_I * x_c - K_R * q
α̇ = Q * S / m / V * (C_N * cos(α) - C_A * sin(α)) + g / V * cos(γ) + q
a_z = V * (q - α̇)
return dx = [V * sin(γ);
Q * S / m * (C_N * sin(α) + C_A * cos(α)) - g * sin(γ);
α̇;
Q * S * d / I_yy * C_M;
q;
δ̇;
-ωₐ^2 * (δ - δ_c) - 2.0f0 * ζₐ * ωₐ * δ̇;
-K_A * (a_z - a_z_cmd) + q - a_z_cmd / V;
L(a_z, a_z_cmd, δ_c)]
end
function eval_output(x)
(h, V, α, q, θ, δ, _, _, _) = x
ρ = ρ₀ * exp(-h / H)
Vₛ = sqrt(γₐ * Rₐ * (T₀ - λ * h))
M = V / Vₛ
γ = θ - α
Q = 0.5f0 * ρ * V^2
C_A = aₐ
C_N = aₙ * α^3 + bₙ * α * abs(α) + cₙ * (2.0f0 - M / 3.0f0) * α + dₙ * δ
α̇ = Q * S / m / V * (C_N * cos(α) - C_A * sin(α)) + g / V * cos(γ) + q
return V * (q - α̇)
end
function eval_tune_params(p_NN)
return function (x)
(h, V, α) = x[1:3]
Vₛ = sqrt(γₐ * Rₐ * (T₀ - λ * h))
M = V / Vₛ
# return min.( Π([α / α_max; M / M_max; h / h_max], p_NN) , K_ub)
return Π([α / α_max; M / M_max; h / h_max], p_NN)
end
end
# ODE problem
prob = ODEProblem(fwd_dynamics_model, x₀, t_span)
# loss construction
function loss(p_NN)
# InterpolatingAdjoint(autojacvec = ReverseDiffVJP(true)) also works well for ODE function defined in IIP form
# InterpolatingAdjoint(autojacvec = ZygoteVJP()) requires ODE function defined in OOP form
fwd_sol = Array(solve(prob, Tsit5(), p = p_NN, saveat = 0.01f0; sensealg = InterpolatingAdjoint(autojacvec = ZygoteVJP() )))
return ϕ(eval_output(fwd_sol[:, end]), a_z_cmd) + R(p_NN) + fwd_sol[end, end], fwd_sol
end
# learning progress callback setup
# cb_progress = function (p, loss_val)
cb_progress = function (p, loss_val, pred)
println("Loss = $loss_val\n")
display(scatter([vec(mapslices(eval_output, pred, dims = 1)) rad2deg.(pred[3:4, :]')], ylabel = ["\$a_{z} [m/s^{2}]\$" "\$\\alpha [deg]\$" "\$q [deg/s]\$"], label = :false, layout = (3, 1)))
return false
end
function view_result(p_NN)
fwd_sol = solve(prob, Tsit5(), p = p_NN, reltol = 1e-8, abstol = 1e-8, saveat = 0.01)
a_z = eval_output.(fwd_sol.u)
K = eval_tune_params(p_NN).(fwd_sol.u)
f_aq = plot(fwd_sol.t, [a_z rad2deg.(fwd_sol[4,:])], label = :false, xlabel = "\$t [s]\$", ylabel = ["\$a_{z} [m/s^{2}]\$" "\$q [deg/s]\$"], layout = (2, 1))
f_K = plot(fwd_sol.t, hcat(K...)', xlabel = "\$t [s]\$", ylabel = ["\$K_{A}\$" "\$K_{I}\$" "\$K_{R}\$"], label = :false, layout = (3, 1))
display(plot(f_aq, f_K, layout = (1, 2)))
f_hVα = plot(fwd_sol.t, [fwd_sol[1:2,:]' rad2deg.(fwd_sol[3,:])], label = :false, xlabel = "\$t [s]\$", ylabel = ["\$h [m]\$" "\$V [m/s]\$" "\$\\alpha [deg]\$"], layout = (3, 1))
display(f_hVα)
return fwd_sol
end
# view pre-training result
view_result(p_NN)
## NN training
println("1st Phase\n")
# res1 = DiffEqFlux.sciml_train(loss, p_NN; cb = cb_progress, maxiters = 100)
res1 = DiffEqFlux.sciml_train(loss, p_NN, ADAM(0.01); cb = cb_progress, maxiters = 50)
fwd_sol = view_result(res1.u)
println("2nd Phase\n")
res2 = DiffEqFlux.sciml_train(loss, res1.u, LBFGS(); cb = cb_progress, maxiters = 30)
fwd_sol = view_result(res2.u)
## Validation
p_NN_final = res2.u
t_span = (0.0f0, 10.0f0)
prob = ODEProblem(fwd_dynamics_model, x₀, t_span)
function view_result(p_NN)
fwd_sol = solve(prob, Tsit5(), p = p_NN, reltol = 1e-8, abstol = 1e-8, saveat = 0.01)
a_z = eval_output.(fwd_sol.u)
K = eval_tune_params(p_NN).(fwd_sol.u)
f_aq = plot(fwd_sol.t, [a_z rad2deg.(fwd_sol[4,:])], label = :false, xlabel = "\$t [s]\$", ylabel = ["\$a_{z} [m/s^{2}]\$" "\$q [deg/s]\$"], layout = (2, 1))
f_K = plot(fwd_sol.t, hcat(K...)', xlabel = "\$t [s]\$", ylabel = ["\$K_{A}\$" "\$K_{I}\$" "\$K_{R}\$"], label = :false, layout = (3, 1))
display(plot(f_aq, f_K, layout = (1, 2)))
f_hVα = plot(fwd_sol.t, [fwd_sol[1:2,:]' rad2deg.(fwd_sol[3,:])], label = :false, xlabel = "\$t [s]\$", ylabel = ["\$h [m]\$" "\$V [m/s]\$" "\$\\alpha [deg]\$"], layout = (3, 1))
display(f_hVα)
return fwd_sol
end
view_result(p_NN_final)
| ContinuousTimePolicyGradients | https://github.com/nhcho91/ContinuousTimePolicyGradients.jl.git |
|
[
"MIT"
] | 0.1.3 | 5f5a59cab6b3920d0b98dbebbb3f3f0477441546 | code | 4690 | using DiffEqFlux, OrdinaryDiffEq, Plots
# model + problem parameters
(aₐ, aₙ, bₙ, cₙ, dₙ, aₘ, bₘ, cₘ, dₘ) = Float32.([-0.3, 19.373, -31.023, -9.717, -1.948, 40.44, -64.015, 2.922, -11.803])
(m, I_yy, S, d, ωₐ, ζₐ) = Float32.([204.02, 247.439, 0.0409, 0.2286, 150.0, 0.7])
(g, ρ₀, H, γₐ, Rₐ, T₀, λ) = Float32.([9.8, 1.225, 8435.0, 1.4, 286.0, 288.15, 0.0065])
(a_max, α_max, δ_max, q_max, M_max, h_max) = Float32.([100.0, deg2rad(30), deg2rad(25), deg2rad(5), 4, 11E3])
(k_a, k_δ, k_R) = Float32.([10000.0, 0.01, 0.001])
t_span = (0.0f0, 3.0f0)
x₀ = Float32.([5E3; 900; zeros(7)])
a_z_cmd = 50.0f0
# cost function
L(a_z, a_z_cmd, δ_c) = k_a * ((a_z - a_z_cmd) / a_max)^2 + k_δ * (δ_c / δ_max)^2
# + k_q * (q / q_max)^2
ϕ(a_z, a_z_cmd) = ((a_z - a_z_cmd) / a_max)^2
R(p_NN) = k_R * p_NN' * p_NN
# NN construction
dim_hidden = 64
(K_lb, K_ub) = Float32.([-0.2, 0])
Π = Chain(
Dense(3, dim_hidden, tanh),
Dense(dim_hidden, dim_hidden, tanh),
Dense(dim_hidden, 3)
# u -> (K_ub - K_lb) * σ.(u) .+ K_lb
)
(p_NN, restruct) = Flux.destructure(Π)
# plant + controller dynamics
function fwd_dynamics_model!(dx, x, p_NN, t)
(h, V, α, q, θ, δ, δ̇, x_c, _) = x
ρ = ρ₀ * exp(-h / H)
Vₛ = sqrt(γₐ * Rₐ * (T₀ - λ * h))
M = V / Vₛ
γ = θ - α
Q = 0.5f0 * ρ * V^2
C_A = aₐ
C_N = aₙ * α^3 + bₙ * α * abs(α) + cₙ * (2.0f0 - M / 3.0f0) * α + dₙ * δ
C_M = aₘ * α^3 + bₘ * α * abs(α) + cₘ * (-7.0f0 + 8.0f0 * M / 3.0f0) * α + dₘ * δ
K_A, K_I, K_R = min.( restruct(p_NN)([α / α_max; M / M_max; h / h_max]) , K_ub )
δ_c = -K_I * x_c - K_R * q
α̇ = Q * S / m / V * (C_N * cos(α) - C_A * sin(α)) + g / V * cos(γ) + q
a_z = V * (q - α̇)
dx[1] = V * sin(γ)
dx[2] = Q * S / m * (C_N * sin(α) + C_A * cos(α)) - g * sin(γ)
dx[3] = α̇
dx[4] = Q * S * d / I_yy * C_M
dx[5] = q
dx[6] = δ̇
dx[7] = -ωₐ^2 * (δ - δ_c) - 2.0f0 * ζₐ * ωₐ * δ̇
dx[8] = -K_A * (a_z - a_z_cmd) + q - a_z_cmd / V
dx[9] = L(a_z, a_z_cmd, δ_c)
end
function eval_output(x)
(h, V, α, q, θ, δ, _, _, _) = x
ρ = ρ₀ * exp(-h / H)
Vₛ = sqrt(γₐ * Rₐ * (T₀ - λ * h))
M = V / Vₛ
γ = θ - α
Q = 0.5f0 * ρ * V^2
C_A = aₐ
C_N = aₙ * α^3 + bₙ * α * abs(α) + cₙ * (2.0f0 - M / 3.0f0) * α + dₙ * δ
α̇ = Q * S / m / V * (C_N * cos(α) - C_A * sin(α)) + g / V * cos(γ) + q
return V * (q - α̇)
end
function eval_tune_params(p_NN)
return function (x)
(h, V, α) = x[1:3]
Vₛ = sqrt(γₐ * Rₐ * (T₀ - λ * h))
M = V / Vₛ
return min.( restruct(p_NN)([α / α_max; M / M_max; h / h_max]) , K_ub )
end
end
# ODE problem
prob = ODEProblem(fwd_dynamics_model!, x₀, t_span)
# loss construction
function loss()
fwd_sol = Array(solve(prob, Tsit5(), p = p_NN, reltol = 1e-4, abstol = 1e-8, saveat = 0.1; sensealg = InterpolatingAdjoint(autojacvec = ZygoteVJP())))
# fwd_sol = Array(solve(prob, Tsit5(), p = p_NN, saveat = 0.1; sensealg = InterpolatingAdjoint(autojacvec = ZygoteVJP())))
return ϕ(eval_output(fwd_sol[:, end]), a_z_cmd) + R(p_NN) + fwd_sol[end, end] #, fwd_sol
end
# learning progress callback setup
cb_progress = function ()
# cb_progress = function (p, loss_val, pred)
loss_val = loss()
println("Loss = $loss_val\n")
# display(scatter([vec(mapslices(eval_output, pred, dims = 1)) rad2deg.(pred[3:4, :]')], ylabel = ["\$a_{z} [m/s^{2}]\$" "\$\\alpha [deg]\$" "\$q [deg/s]\$"], label = :false, layout = (3, 1)))
return false
end
function view_result(p_NN)
fwd_sol = solve(prob, Tsit5(), p = p_NN, reltol = 1e-8, abstol = 1e-8, saveat = 0.01)
a_z = eval_output.(fwd_sol.u)
K = eval_tune_params(p_NN).(fwd_sol.u)
f_aq = plot(fwd_sol.t, [a_z rad2deg.(fwd_sol[4,:])], label = :false, xlabel = "\$t [s]\$", ylabel = ["\$a_{z} [m/s^{2}]\$" "\$q [deg/s]\$"], layout = (2, 1))
f_K = plot(fwd_sol.t, hcat(K...)', xlabel = "\$t [s]\$", ylabel = ["\$K_{A}\$" "\$K_{I}\$" "\$K_{R}\$"], label = :false, layout = (3, 1))
display(plot(f_aq, f_K, layout = (1, 2)))
f_hVα = plot(fwd_sol.t, [fwd_sol[1:2,:]' rad2deg.(fwd_sol[3,:])], label = :false, xlabel = "\$t [s]\$", ylabel = ["\$h [m]\$" "\$V [m/s]\$" "\$\\alpha [deg]\$"], layout = (3, 1))
display(f_hVα)
return fwd_sol
end
# view pre-training result
view_result(p_NN)
## NN training
println("1st Phase\n")
data = Iterators.repeated((), 2000)
Flux.train!(loss, Flux.params(p_NN), data, ADAM(0.5); cb = cb_progress)
##
println("2nd Phase\n")
Flux.train!(loss, Flux.params(p_NN), data, ADAM(); cb = cb_progress)
##
println("3rd Phase\n")
Flux.train!(loss, Flux.params(p_NN), data, Nesterov(); cb = cb_progress)
# view post-training result
fwd_sol = view_result(p_NN) | ContinuousTimePolicyGradients | https://github.com/nhcho91/ContinuousTimePolicyGradients.jl.git |
|
[
"MIT"
] | 0.1.3 | 5f5a59cab6b3920d0b98dbebbb3f3f0477441546 | code | 6693 | using ContinuousTimePolicyGradients
using DiffEqFlux, ComponentArrays, LinearAlgebra, JLD2, OrdinaryDiffEq
using Plots
function main(maxiters_1::Int, maxiters_2::Int, Δt_save::Float32; p_NN_0 = nothing, k_a_val = 10.0)
# model + problem parameters
(aₐ, aₙ, bₙ, cₙ, dₙ, aₘ, bₘ, cₘ, dₘ) = Float32.([-0.3, 19.373, -31.023, -9.717, -1.948, 40.44, -64.015, 2.922, -11.803])
(m, I_yy, S, d, ωₐ, ζₐ) = Float32.([204.02, 247.439, 0.0409, 0.2286, 150.0, 0.7])
(g, ρ₀, H, γₐ, Rₐ, T₀, λ) = Float32.([9.8, 1.225, 8435.0, 1.4, 286.0, 288.15, 0.0065])
(a_max, α_max, δ_max, δ̇_max, q_max, M_max, h_max) = Float32.([100.0, deg2rad(30), deg2rad(25), 1.5, deg2rad(60), 4, 11E3])
(k_a, k_q, k_δ, k_δ̇, k_R) = Float32.([k_a_val, 0.0, 0.0, 0.1, 1E-3])
# dynamic model
dim_x = 7
function dynamics_plant(t, x, u)
(h, V, α, q, θ, δ, δ̇) = x
δ_c = u[1]
ρ = ρ₀ * exp(-h / H)
Vₛ = sqrt(γₐ * Rₐ * (T₀ - λ * h))
M = V / Vₛ
γ = θ - α
Q = 0.5f0 * ρ * V^2
C_A = aₐ
C_N = aₙ * α^3 + bₙ * α * abs(α) + cₙ * (2.0f0 - M / 3.0f0) * α + dₙ * δ
C_M = aₘ * α^3 + bₘ * α * abs(α) + cₘ * (-7.0f0 + 8.0f0 * M / 3.0f0) * α + dₘ * δ
α̇ = Q * S / m / V * (C_N * cos(α) - C_A * sin(α)) + g / V * cos(γ) + q
dx = [V * sin(γ);
Q * S / m * (C_N * sin(α) + C_A * cos(α)) - g * sin(γ);
α̇;
Q * S * d / I_yy * C_M;
q;
δ̇;
-ωₐ^2 * (δ - δ_c) - 2.0f0 * ζₐ * ωₐ * δ̇]
return dx
end
dim_x_c = 2
function dynamics_controller(t, x_c, y, r, p_NN, policy_NN)
(A_z, h, V, M, α, q, γ) = y
A_z_cmd = r[1]
x_int = x_c[1]
x_ref = x_c[2]
y_NN = (K_A, K_I, K_R) = policy_NN([α / α_max; M / M_max; h / h_max], p_NN)
# dx_ref = [-36.6667f0 -13.8889f0; 8.0f0 0.0f0] * x_ref + [4.0f0; 0.0f0] * A_z_cmd
# A_z_ref = [-1.0083f0 3.4722f0] * x_ref
dx_ref = (A_z_cmd - x_ref) / 0.2f0
A_z_ref = x_ref
δ_trim = -(aₘ * α^3 + bₘ * α * abs(α) + cₘ * (-7.0f0 + 8.0f0 * M / 3.0f0) * α) / dₘ
dx_c = [K_A * (A_z_cmd - A_z) + q + (A_z + g * cos(γ)) / V;
dx_ref]
u = [K_I * x_int + K_R * q + δ_trim;
A_z_ref]
return dx_c, u, y_NN
end
function dynamics_sensor(t, x)
(h, V, α, q, θ, δ, _) = x
ρ = ρ₀ * exp(-h / H)
Vₛ = sqrt(γₐ * Rₐ * (T₀ - λ * h))
M = V / Vₛ
γ = θ - α
Q = 0.5f0 * ρ * V^2
C_A = aₐ
C_N = aₙ * α^3 + bₙ * α * abs(α) + cₙ * (2.0f0 - M / 3.0f0) * α + dₙ * δ
A_z = Q * S / m * (C_N * cos(α) - C_A * sin(α))
y = [A_z;
h;
V;
M;
α;
q;
γ]
return y
end
# cost definition
function cost_running(t, x, y, u, r)
q = x[4]
δ̇ = x[7]
a_z = y[1]
δ_c = u[1]
a_z_ref = u[2]
a_z_cmd = r[1]
return k_a * ((a_z - a_z_ref) / (exp(-2.0f0*a_z_cmd) + a_z_cmd))^2 + k_δ̇ * (δ̇ / δ̇_max)^2 + k_δ * (δ_c / δ_max)^2 + k_q * (q / q_max)^2
end
function cost_terminal(x_f, r)
# a_z_cmd = r[1]
# y = dynamics_sensor(3.0f0, x_f)
# a_z = y[1]
# return ((a_z - a_z_cmd) / (1.0f0 + a_z_cmd))^2
return 0.0f0
end
function cost_regularisor(p_NN)
return k_R * norm(p_NN)^2
# return 0.0f0
end
# NN construction
dim_NN_hidden = 64
dim_NN_input = 3
dim_K = 3
K_lb = zeros(Float32, 3)
K_ub = Float32[4,0.2,4]
policy_NN = FastChain(
FastDense(dim_NN_input, dim_NN_hidden, tanh),
FastDense(dim_NN_hidden, dim_NN_hidden, tanh),
FastDense(dim_NN_hidden, dim_K),
(x, p) -> (K_ub - K_lb) .* σ.(x) .+ K_lb
)
# scenario definition
ensemble = [ (; x₀ = Float32[h₀; V₀; zeros(5)], r = Float32[A_z_cmd])
for h₀ = 5E3 #:1E3:8E3
for V₀ = 7E2 #:1E2:9E2
for A_z_cmd = 0:2E1:1E2 ]
t_span = Float32.((0.0, 3.0))
t_save = t_span[1]:Δt_save:t_span[2]
scenario = (; ensemble = ensemble, t_span = t_span, t_save = t_save, dim_x = dim_x, dim_x_c = dim_x_c)
# NN training
(result, fwd_ensemble_sol, loss_history) = CTPG_train(dynamics_plant, dynamics_controller, dynamics_sensor, cost_running, cost_terminal, cost_regularisor, policy_NN, scenario; sense_alg = InterpolatingAdjoint(autojacvec = ReverseDiffVJP(true)), ensemble_alg = EnsembleThreads(), maxiters_1 = maxiters_1, maxiters_2 = maxiters_2, opt_2 = BFGS(initial_stepnorm = 0.0001), i_nominal = 1, p_NN_0 = p_NN_0, progress_plot = false)
return result, policy_NN, fwd_ensemble_sol, loss_history
end
## execute optimisation and simulation
@time (result, policy_NN, fwd_ensemble_sol, loss_history) = main(1000, 500, 0.01f0; k_a_val = 200.0)
## save results
# p_NN_prev = result.u
# @time (result, policy_NN, fwd_ensemble_sol, loss_history) = main(1, 1000, 0.01f0; k_a_val = 50.0, p_NN_0 = p_NN_prev)
jldsave("DS_autopilot_13.jld2"; result, fwd_ensemble_sol, loss_history)
# plot results
# (fwd_ensemble_sol, loss_history) = load(".jld2", "fwd_ensemble_sol", "loss_history")
x_names = ["\$h\$" "\$V\$" "\$\\alpha\$" "\$q\$" "\$\\theta\$" "\$\\delta\$" "\$\\dot{\\delta}\$"]
vars_x = 1:6 # [1,2,3, (1,2), (2,3)]
u_names = ["\$\\delta_{c}\$"]
vars_u = 1
y_names = ["\$a_{z}\$"]
vars_y = 1
y_NN_names = ["\$K_A\$" "\$K_I\$" "\$K_R\$"]
vars_y_NN = 1:3
(f_x, f_u, f_y, f_y_NN, f_L) = view_result([], fwd_ensemble_sol, loss_history; x_names = x_names, vars_x = vars_x, u_names = u_names, vars_u = vars_u, y_names = y_names, vars_y = vars_y, y_NN_names = y_NN_names, vars_y_NN = vars_y_NN, linealpha = 0.6)
(a_max, α_max, δ_max, δ̇_max, q_max, M_max, h_max) = Float32.([100.0, deg2rad(30), deg2rad(25), 1.5, deg2rad(60), 4, 11E3])
h = 5000.0
α_list = 0:1E-3:deg2rad(45)
M_list = 2.0:0.1:3.0
func_K_A(α, M) = policy_NN([α / α_max; M / M_max; h / h_max], result.u)[1]
func_K_I(α, M) = policy_NN([α / α_max; M / M_max; h / h_max], result.u)[2]
func_K_R(α, M) = policy_NN([α / α_max; M / M_max; h / h_max], result.u)[3]
f_K_A = plot(α_list, M_list, func_K_A, st=:surface, label = :false, title = "K_A", xlabel = "\$\\alpha\$", ylabel = "\$M\$")
f_K_I = plot(α_list, M_list, func_K_I, st=:surface, label = :false, title = "K_I", xlabel = "\$\\alpha\$", ylabel = "\$M\$")
f_K_R = plot(α_list, M_list, func_K_R, st=:surface, label = :false, title = "K_R", xlabel = "\$\\alpha\$", ylabel = "\$M\$")
display(f_K_A)
display(f_K_I)
display(f_K_R) | ContinuousTimePolicyGradients | https://github.com/nhcho91/ContinuousTimePolicyGradients.jl.git |
|
[
"MIT"
] | 0.1.3 | 5f5a59cab6b3920d0b98dbebbb3f3f0477441546 | code | 1849 | using ComponentArrays
using DiffEqFlux, DifferentialEquations, Plots
using UnPack
"""
To test whether `ComponentArray` (exported from `ComponentArrays`) is compatible with AD (auto-diff) systems.
Borrowed from DiffEqFlux.jl documentation:
https://diffeqflux.sciml.ai/stable/examples/neural_ode_flux/
"""
function main()
# u0 = Float32[2.; 0.]
u0 = ComponentArray(a=2.0, b=0.0)
datasize = 30
tspan = (0.0f0,1.5f0)
function trueODEfunc(du,u,p,t)
true_A = [-0.1 2.0; -2.0 -0.1]
@unpack a, b = u
tmp = ([a^3, b^3]'true_A)'
du.a, du.b = tmp
end
t = range(tspan[1],tspan[2],length=datasize)
prob = ODEProblem(trueODEfunc,u0,tspan)
ode_data = Array(solve(prob,Tsit5(),saveat=t))
dudt2 = Chain(x -> x.^3,
Dense(2,50,tanh),
Dense(50,2))
p,re = Flux.destructure(dudt2) # use this p as the initial condition!
dudt(u,p,t) = re(p)(u) # need to restructure for backprop!
prob = ODEProblem(dudt,u0,tspan)
function predict_n_ode()
Array(solve(prob,Tsit5(),u0=u0,p=p,saveat=t))
end
function loss_n_ode()
pred = predict_n_ode()
loss = sum(abs2,ode_data .- pred)
loss
end
loss_n_ode() # n_ode.p stores the initial parameters of the neural ODE
cb = function (;doplot=false) # callback function to observe training
pred = predict_n_ode()
display(sum(abs2,ode_data .- pred))
# plot current prediction against data
pl = scatter(t,ode_data[1,:],label="data")
scatter!(pl,t,pred[1,:],label="prediction")
display(plot(pl))
return false
end
# Display the ODE with the initial parameter values.
cb()
data = Iterators.repeated((), 1000)
Flux.train!(loss_n_ode, Flux.params(u0,p), data, ADAM(0.05), cb = cb)
end
| ContinuousTimePolicyGradients | https://github.com/nhcho91/ContinuousTimePolicyGradients.jl.git |
|
[
"MIT"
] | 0.1.3 | 5f5a59cab6b3920d0b98dbebbb3f3f0477441546 | code | 28319 | # Test: ODESystem built with ModelingToolkit + Ensemble Simulation with DifferentialEquations + Optimisation with DiffEqFlux
using DiffEqFlux, DifferentialEquations, Plots, Statistics, ModelingToolkit
ann = FastChain(FastDense(1,32,tanh), FastDense(32,32,tanh), FastDense(32,1))
dim_p = DiffEqFlux.paramlength(ann)
@variables t x1(t) x2(t)
@parameters p_NN[1:dim_p]
D = Differential(t)
deqs = [D(x1) ~ x2
D(x2) ~ ann([t],p_NN)[1]^3]
sys = ODESystem(deqs, t, [x1, x2], p_NN; name = :sys)
# sys = ODESystem(deqs, iv, dvs, ps; )
# sys_simplified = structural_simplify(sys) # ---> not working
u0 = [x1 => -4f0,
x2 => 0f0]
p_NN_0 = initial_params(ann)
p = [p_NN[i] => p_NN_0[i] for i in 1:dim_p]
tspan = (0.0f0, 8.0f0)
prob = ODEProblem(sys, u0, tspan, p; jac = true)
@time sol = solve(prob, Tsit5(), saveat = 0.1f0) # ---> not working
##--------
using DiffEqFlux, DifferentialEquations, Plots, Statistics, ModelingToolkit
dim_h = 32
tspan = (0.0f0,8.0f0)
ann = FastChain(FastDense(1,32,tanh), FastDense(32,32,tanh), FastDense(32,1))
θ = initial_params(ann)
function dxdt_(dx,x,p,t)
x1, x2 = x
dx[1] = x[2]
dx[2] = ann([t],p)[1]^3
end
x0 = [-4f0, 0f0]
prob = ODEProblem(dxdt_,x0,tspan,θ)
sys = structural_simplify(modelingtoolkitize(prob))
prob_mtk = ODEProblem(sys, [], tspan, jac = true)
@time sol = solve(prob_mtk, Tsit5(), saveat = 0.1f0) # ---> working
#------------
ts = Float32.(collect(0.0:0.01:tspan[2]))
function predict_adjoint(θ)
Array(solve(prob_mtk,Tsit5(),p=θ,saveat=ts,sensealg=InterpolatingAdjoint(autojacvec=ZygoteVJP())))
end
function loss_adjoint(θ)
x = predict_adjoint(θ)
mean(abs2,4.0 .- x[1,:]) + 2mean(abs2,x[2,:]) + mean(abs2,[first(ann([t],θ)) for t in ts])/10
end
l = loss_adjoint(θ)
cb = function (θ,l)
println(l)
# p = plot(solve(remake(prob,p=θ),Tsit5(),saveat=0.01),ylim=(-6,6),lw=3)
# plot!(p,ts,[first(ann([t],θ)) for t in ts],label="u(t)",lw=3)
# display(p)
return false
end
# Display the ODE with the current parameter values.
cb(θ,l)
loss1 = loss_adjoint(θ)
res1 = DiffEqFlux.sciml_train(loss_adjoint, θ, ADAM(0.005), cb = cb,maxiters=100)
res2 = DiffEqFlux.sciml_train(loss_adjoint, res1.u,
BFGS(initial_stepnorm=0.01), cb = cb,maxiters=100,
allow_f_increases = false)
## ------------------------------------------------------------------------------------
using ModelingToolkit
function decay(;name)
@parameters t a
@variables x(t) f(t)
D = Differential(t)
ODESystem([
D(x) ~ -a*x + f
];
name=name)
end
@named decay1 = decay()
@named decay2 = decay()
@parameters t
D = Differential(t)
connected = compose(ODESystem([
decay2.f ~ decay1.x
D(decay1.f) ~ 0
], t; name=:connected), decay1, decay2)
equations(connected)
#4-element Vector{Equation}:
# Differential(t)(decay1₊f(t)) ~ 0
# decay2₊f(t) ~ decay1₊x(t)
# Differential(t)(decay1₊x(t)) ~ decay1₊f(t) - (decay1₊a*(decay1₊x(t)))
# Differential(t)(decay2₊x(t)) ~ decay2₊f(t) - (decay2₊a*(decay2₊x(t)))
simplified_sys = structural_simplify(connected)
equations(simplified_sys)
#3-element Vector{Equation}:
# Differential(t)(decay1₊f(t)) ~ 0
# Differential(t)(decay1₊x(t)) ~ decay1₊f(t) - (decay1₊a*(decay1₊x(t)))
# Differential(t)(decay2₊x(t)) ~ decay1₊x(t) - (decay2₊a*(decay2₊x(t)))
x0 = [
decay1.x => 1.0
decay1.f => 0.0
decay2.x => 1.0
]
p = [
decay1.a => 0.1
decay2.a => 0.2
]
using DifferentialEquations
prob = ODEProblem(simplified_sys, x0, (0.0, 100.0), p)
sol = solve(prob, Tsit5())
sol[decay2.f]
## ------------------------------------------------------------------------------------
using DifferentialEquations, DiffEqFlux
pa = [1.0]
u0 = [3.0]
θ = [u0;pa]
function model1(θ,ensemble)
prob = ODEProblem((u, p, t) -> 1.01u .* p, [θ[1]], (0.0, 1.0), [θ[2]])
function prob_func(prob, i, repeat)
remake(prob, u0 = 0.5 .+ i/100 .* prob.u0)
end
ensemble_prob = EnsembleProblem(prob, prob_func = prob_func)
sim = solve(ensemble_prob, Tsit5(), ensemble, saveat = 0.1, trajectories = 100)
end
# loss function
loss_serial(θ) = sum(abs2,1.0.-Array(model1(θ,EnsembleSerial())))
loss_threaded(θ) = sum(abs2,1.0.-Array(model1(θ,EnsembleThreads())))
cb = function (θ,l) # callback function to observe training
@show l
false
end
opt = ADAM(0.1)
l1 = loss_serial(θ)
res_serial = DiffEqFlux.sciml_train(loss_serial, θ, opt; cb = cb, maxiters=100)
res_threads = DiffEqFlux.sciml_train(loss_threaded, θ, opt; cb = cb, maxiters=100)
## ------------------------------------------------------------------------------------
using DifferentialEquations
prob = ODEProblem((u,p,t)->1.01*[u[2];u[1]],[0.5, 1.0],(0.0,1.0))
function prob_func(prob,i,repeat)
remake(prob,u0=rand()*prob.u0)
end
ensemble_prob = EnsembleProblem(prob,prob_func=prob_func)
sim = solve(ensemble_prob,Tsit5(),EnsembleThreads(),trajectories=100)
## ------------------------------------------------------------------------------------
using DifferentialEquations, Plots, GalacticOptim
function lotka_volterra!(du,u,p,t)
rab, wol = u
α,β,γ,δ=p
du[1] = drab = α*rab - β*rab*wol
du[2] = dwol = γ*rab*wol - δ*wol
nothing
end
u0 = [1.0,1.0]
tspan = (0.0,10.0)
p = [1.5,1.0,3.0,1.0]
prob = ODEProblem(lotka_volterra!,u0,tspan,p)
sol = solve(prob,saveat=0.1)
plot(sol)
dataset = Array(sol)
scatter!(sol.t,dataset')
tmp_prob = remake(prob, p=[1.2,0.8,2.5,0.8])
tmp_sol = solve(tmp_prob)
plot(tmp_sol)
scatter!(sol.t,dataset')
function loss(p)
tmp_prob = remake(prob, p=p)
tmp_sol = solve(tmp_prob,Tsit5(),saveat=0.1)
if tmp_sol.retcode == :Success
return sum(abs2,Array(tmp_sol) - dataset)
else
return 1E10
end
end
# function loss(p)
# tmp_prob = remake(prob, p=p)
# tmp_sol = solve(tmp_prob,Tsit5(),saveat=0.1)
# if size(tmp_sol) == size(dataset)
# return sum(abs2,Array(tmp_sol) .- dataset)
# else
# return Inf
# end
# end
using DiffEqFlux
pinit = [1.2,0.8,2.5,0.8]
res = DiffEqFlux.sciml_train(loss,pinit,ADAM(), maxiters = 100)
# res = DiffEqFlux.sciml_train(loss,pinit,BFGS(), maxiters = 100) ### errors!
#try Newton method of optimization
res = DiffEqFlux.sciml_train(loss,pinit,BFGS())
## ------------------------------------------------------------------------------------
# compatibility of modelingtoolkitize and ensemble parellel simulation
using DifferentialEquations, ModelingToolkit
function rober(du,u,p,t)
y₁,y₂,y₃ = u
k₁,k₂,k₃ = p
du[1] = -k₁*y₁+k₃*y₂*y₃
du[2] = k₁*y₁-k₂*y₂^2-k₃*y₂*y₃
du[3] = k₂*y₂^2
nothing
end
function prob_func(prob,i,repeat)
p_i = collect(prob.p)
p_i[1] = 1E-2 * i
remake(prob,p=p_i)
# remake(prob,u0=rand()*prob.u0)
end
prob = ODEProblem(rober,[1.0,0.0,0.0],(0.0,1e1),(0.04,3e7,1e4))
sys = modelingtoolkitize(prob)
prob_jac = ODEProblem(sys,[],(0.0,1e1),jac=true)
ensemble_prob = EnsembleProblem(prob,prob_func=prob_func)
ensemble_prob_jac = EnsembleProblem(prob_jac,prob_func=prob_func)
@time sim = solve(ensemble_prob,Tsit5(),EnsembleSerial(),trajectories=10)
@time sim_jac = solve(ensemble_prob_jac,Tsit5(),EnsembleSerial(),trajectories=10)
# conclusion 1) sim_jac is faster than sim if ensemble algorithm is EnsembleSerial().
# ---> sim_jac is even slower than sim when EnsembleThreads() is used.
# conclusion 2) sim_jac[i].prob.p is determined as intended by prob_func even though the modelingtoolkitized system is used.
## ------------------------------------------------------------------------------------
using Flux
d = Dense(5,2)
p1 = d.weight, d.bias
p2 = Flux.params(d)
p1[1] == p2[1]
p2[1] == p2[2]
m = Chain(Dense(10,5), Dense(5,2))
x = rand(10)
# m(x) == m[2](m[1](x))
# loss(ŷ, y)
p, re = Flux.destructure(m)
m(x) - re(p)(x)
## ------------------------------------------------------------------------------------
using Flux
W = rand(2, 5)
b = rand(2)
predict(x) = (W * x) .+ b
loss(x, y) = sum((predict(x) .- y).^2)
x, y = rand(5), rand(2) # Dummy data
l = loss(x, y) # ~ 3
θ = params(W, b)
grads = gradient(() -> loss(x, y), θ)
using Flux.Optimise: update!
η = 0.1 # Learning Rate
for p in (W, b)
update!(p, η * grads[p])
end
opt = Descent(0.1) # Gradient descent with learning rate 0.1
for p in (W, b)
update!(opt, p, grads[p])
end
## ------------------------------------------------------------------------------------
# Comparison between the automatic differentiation results for ForwardDiffSensitivity and InterpolatingAdjoint
using DiffEqSensitivity, OrdinaryDiffEq, Zygote, LinearAlgebra, QuadGK
function fiip(du,u,p,t)
du[1] = dx = p[1]*u[1] - p[2]*u[1]*u[2]
du[2] = dy = -p[3]*u[2] + p[4]*u[1]*u[2]
du[3] = u[2]^2
end
p = [1.5,1.0,3.0,1.0]
u0 = [1.0;1.0;0.0]
prob = ODEProblem(fiip,u0,(0.0,10.0),p)
# y_sense = solve(prob,Tsit5(),u0=u0,p=p,saveat=0.1;sensealg = InterpolatingAdjoint(autojacvec=ZygoteVJP()))
# loss(u0,p) = sum(solve(prob,Tsit5(),u0=u0,p=p,saveat=0.1,abstol=1e-14,reltol=1e-14))
# loss_adjoint(u0,p) = sum(solve(prob,Tsit5(),u0=u0,p=p,saveat=0.1,abstol=1e-14,reltol=1e-14;sensealg = InterpolatingAdjoint()))
function loss(u0,p)
y = Array(solve(prob,Tsit5(),u0=u0,p=p,saveat=0.1,abstol=1e-14,reltol=1e-14))
return y[3,end]
end
function loss_adjoint(u0,p)
y = Array(solve(prob,Tsit5(),u0=u0,p=p,saveat=0.1,abstol=1e-14,reltol=1e-14;sensealg = InterpolatingAdjoint()))
return y[3,end]
end
function loss_discrete(u0,p)
y = Array(solve(prob,Tsit5(),u0=u0,p=p,saveat=0.1,abstol=1e-14,reltol=1e-14;sensealg = InterpolatingAdjoint()))
# return quadgk((t) -> (y(t)[2])^2, 0.0, 10.0)[1] # atol=1e-14,rtol=1e-10) # this is incorrect and not working
return 0.1*sum(y[2,:].^2)
end
function loss_adjoint_Bolza(u0,p)
y = Array(solve(prob,Tsit5(),u0=u0,p=p,abstol=1e-14,reltol=1e-14;sensealg = InterpolatingAdjoint()))
return y[3,end] + 100.0*y[1:2,end]'*y[1:2,end]
end
du01,dp1 = Zygote.gradient(loss,u0,p)
du02,dp2 = Zygote.gradient(loss_adjoint,u0,p)
du03,dp3 = Zygote.gradient(loss_discrete,u0,p)
du04,dp4 = Zygote.gradient(loss_adjoint_Bolza,u0,p)
# Comparison between the results of automatic differentiation and continuous adjoint sensitivity analysis method
y = solve(prob,Tsit5(),u0=u0,p=p,abstol=1e-14,reltol=1e-14;sensealg = InterpolatingAdjoint())
bwd_sol = solve(ODEAdjointProblem(y, InterpolatingAdjoint(), (out, x, p, t, i) -> (out[:] = [200.0*y[end][1:2]; 1]'), [10.0]), Tsit5(),
# dense = false, save_everystep = false, save_start = false, # saveat = 0.1,
abstol=1e-14,reltol=1e-14)
du05, dp5 = bwd_sol[end][1:length(u0)], -bwd_sol[end][1+length(u0):end]
isapprox(du04, du05)
isapprox(dp4, dp5)
## ------------------------------------------------------------------------------------
# Comparison between forward and adjoint sensitivity calculation results
using DiffEqSensitivity, OrdinaryDiffEq, Zygote
function f(du,u,p,t)
du[1] = dx = p[1]*u[1] - p[2]*u[1]*u[2]
du[2] = dy = -p[3]*u[2] + u[1]*u[2]
end
p = [1.5,1.0,3.0]
u0 = [1.0;1.0]
prob = ODEProblem(f,u0,(0.0,10.0),p)
sol = solve(prob,Vern9(),abstol=1e-10,reltol=1e-10)
g(u,p,t) = (sum(u).^2) ./ 2 + p'*p / 2
function dgdu(out,u,p,t)
out[1]= u[1] + u[2]
out[2]= u[1] + u[2]
end
function dgdp_s(out,u,p,t)
out[1] = p[1]
out[2] = p[2]
out[3] = p[3]
end
function dgdp_v1(out,u,p,t)
out .= p
end
function dgdp_v2(out,u,p,t)
out = p
end
function dgdp_v3(out,u,p,t)
out = p'
end
res1 = adjoint_sensitivities(sol,Vern9(),g,nothing,abstol=1e-8, reltol=1e-8,iabstol=1e-8,ireltol=1e-8) # this is incorrect
res2 = adjoint_sensitivities(sol,Vern9(),g,nothing,dgdu,abstol=1e-8, reltol=1e-8,iabstol=1e-8,ireltol=1e-8) # this is incorrect
res3 = adjoint_sensitivities(sol,Vern9(),g,nothing,(dgdu,dgdp_s),abstol=1e-8, reltol=1e-8,iabstol=1e-8,ireltol=1e-8) # this is correct
res4 = adjoint_sensitivities(sol,Vern9(),g,nothing,(dgdu,dgdp_v1),abstol=1e-8, reltol=1e-8,iabstol=1e-8,ireltol=1e-8) # this is correct
res5 = adjoint_sensitivities(sol,Vern9(),g,nothing,(dgdu,dgdp_v2),abstol=1e-8, reltol=1e-8,iabstol=1e-8,ireltol=1e-8) # this is incorrect
res6 = adjoint_sensitivities(sol,Vern9(),g,nothing,(dgdu,dgdp_v3),abstol=1e-8, reltol=1e-8,iabstol=1e-8,ireltol=1e-8) # this is incorrect
using ForwardDiff, Calculus
using QuadGK
function G(p)
tmp_prob = remake(prob,p=p)
sol = solve(tmp_prob,Vern9(),abstol=1e-14,reltol=1e-14)
res,err = quadgk((t)-> (sum(sol(t)).^2)./2 + p'*p/2,0.0,10.0,atol=1e-14,rtol=1e-10)
res
end
res7 = ForwardDiff.gradient(G,p)
res8 = Calculus.gradient(G,p)
# res9 = Zygote.gradient(G,p) # not working
## ------------------------------------------------------------------------------------
## ------------------------------------------------------------------------------------
## ------------------------------------------------------------------------------------
# Samuel Aintworth's code
import DiffEqBase
import DiffEqSensitivity:
solve,
ODEProblem,
ODEAdjointProblem,
InterpolatingAdjoint
import Zygote
import Statistics: mean
function extract_loss_and_xT(fwd_sol)
fwd_sol[end][1], fwd_sol[end][2:end]
end
"""Returns a differentiable loss function that rolls out a policy in an
environment and calculates its cost."""
function ppg_goodies(dynamics, cost, policy, T)
function aug_dynamics(z, policy_params, t)
x = @view z[2:end]
u = policy(x, t, policy_params)
[cost(x, u); dynamics(x, u)]
end
function loss_pullback(x0, policy_params, solvealg, solve_kwargs)
z0 = vcat(0.0, x0)
fwd_sol = solve(
ODEProblem(aug_dynamics, z0, (0, T), policy_params),
solvealg,
u0 = z0,
p = policy_params;
solve_kwargs...,
)
function _adjoint_solve(g_zT, sensealg; kwargs...)
# See https://diffeq.sciml.ai/stable/analysis/sensitivity/#Syntax-1
# and https://github.com/SciML/DiffEqSensitivity.jl/blob/master/src/local_sensitivity/sensitivity_interface.jl#L9.
solve(
ODEAdjointProblem(
fwd_sol,
sensealg,
(out, x, p, t, i) -> (out[:] = g_zT),
[T],
),
solvealg;
kwargs...,
)
end
# This is the pullback using the augmented system and a discrete
# gradient input at time T. Alternatively one could use the continuous
# adjoints on the non-augmented system although this seems to be slower
# and a less stable feature.
function pullback(g_zT, sensealg::InterpolatingAdjoint)
bwd_sol = _adjoint_solve(
g_zT,
sensealg,
dense = false,
save_everystep = false,
save_start = false,
# reltol = 1e-3,
# abstol = 1e-3,
)
# The first z_dim elements of bwd_sol.u are the gradient wrt z0,
# next however many are the gradient wrt policy_params.
p = fwd_sol.prob.p
l = p === nothing || p === DiffEqBase.NullParameters() ? 0 :
length(fwd_sol.prob.p)
g_x0 = bwd_sol[end][1:length(fwd_sol.prob.u0)]
# We do exactly as many f calls as there are function calls in the
# forward pass, and in the backward pass we don't need to call f,
# but instead we call ∇f.
(
g = -bwd_sol[end][(1:l).+length(fwd_sol.prob.u0)],
nf = 0,
n∇ₓf = bwd_sol.destats.nf,
n∇ᵤf = bwd_sol.destats.nf,
)
end
fwd_sol, pullback
end
function ez_loss_and_grad(
x0,
policy_params,
solvealg,
sensealg;
fwd_solve_kwargs = Dict(),
)
# @info "fwd"
fwd_sol, vjp = loss_pullback(x0, policy_params, solvealg, fwd_solve_kwargs)
# @info "bwd"
bwd = vjp(vcat(1, zero(x0)), sensealg)
loss, _ = extract_loss_and_xT(fwd_sol)
# @info "fin"
loss, bwd.g, (nf = fwd_sol.destats.nf + bwd.nf, n∇ₓf = bwd.n∇ₓf, n∇ᵤf = bwd.n∇ᵤf)
end
function _aggregate_batch_results(res)
(
mean(loss for (loss, _, _) in res),
mean(g for (_, g, _) in res),
(
nf = sum(info.nf for (_, _, info) in res),
n∇ₓf = sum(info.n∇ₓf for (_, _, info) in res),
n∇ᵤf = sum(info.n∇ᵤf for (_, _, info) in res),
),
)
end
function ez_loss_and_grad_many(
x0_batch,
policy_params,
solvealg,
sensealg;
fwd_solve_kwargs = Dict(),
)
# Using tmap here gives a segfault. See https://github.com/tro3/ThreadPools.jl/issues/18.
_aggregate_batch_results(
map(x0_batch) do x0
ez_loss_and_grad(
x0,
policy_params,
solvealg,
sensealg,
fwd_solve_kwargs = fwd_solve_kwargs,
)
end,
)
end
(
aug_dynamics = aug_dynamics,
loss_pullback = loss_pullback,
ez_loss_and_grad = ez_loss_and_grad,
ez_loss_and_grad_many = ez_loss_and_grad_many,
ez_euler_bptt = ez_euler_bptt,
ez_euler_loss_and_grad_many = ez_euler_loss_and_grad_many,
)
end
function policy_dynamics!(dx, x, policy_params, t)
u = policy(x, policy_params)
dx .= dynamics(x, u)
end
function cost_functional(x, policy_params, t)
cost(x, policy(x, policy_params))
end
# See https://github.com/SciML/DiffEqSensitivity.jl/issues/302 for context.
dcost_tuple = (
(out, u, p, t) -> begin
ū, _, _ = Zygote.gradient(cost_functional, u, p, t)
out .= ū
end,
(out, u, p, t) -> begin
_, p̄, _ = Zygote.gradient(cost_functional, u, p, t)
out .= p̄
end,
)
function gold_standard_gradient(x0, policy_params)
# Actual/gold standard evaluation. Using high-fidelity Vern9 method with
# small tolerances. We want to use Float64s for maximum accuracy. Also 1e-14
# is recommended as the minimum allowable tolerance here: https://docs.sciml.ai/stable/basics/faq/#How-to-get-to-zero-error-1.
x0_f64 = convert(Array{Float64}, x0)
policy_params_f64 = convert(Array{Float64}, policy_params)
fwd_sol = solve(
ODEProblem(policy_dynamics!, x0, (0, T), policy_params),
Vern9(),
u0 = x0_f64,
p = policy_params_f64,
abstol = 1e-14,
reltol = 1e-14,
)
# Note that specifying dense = false is essential for getting acceptable
# performance. save_everystep = false is another small win.
bwd_sol = solve(
ODEAdjointProblem(
fwd_sol,
InterpolatingAdjoint(),
cost_functional,
nothing,
dcost_tuple,
),
Vern9(),
dense = false,
save_everystep = false,
abstol = 1e-14,
reltol = 1e-14,
)
@assert typeof(fwd_sol.u) == Array{Array{Float64,1},1}
@assert typeof(bwd_sol.u) == Array{Array{Float64,1},1}
# Note that the backwards solution includes the gradient on x0, as well as
# policy_params. The full ODESolution things can't be serialized easily
# since they `policy_dynamics!` and shit...
(xT = fwd_sol.u[end], g = bwd_sol.u[end])
end
function eval_interp(x0, policy_params, abstol, reltol)
fwd_sol = solve(
ODEProblem(policy_dynamics!, x0, (0, T), policy_params),
Tsit5(),
u0 = x0,
p = policy_params,
abstol = abstol,
reltol = reltol,
)
bwd_sol = solve(
ODEAdjointProblem(
fwd_sol,
InterpolatingAdjoint(),
cost_functional,
nothing,
dcost_tuple,
),
Tsit5(),
dense = false,
save_everystep = false,
abstol = abstol,
reltol = reltol,
)
@assert typeof(fwd_sol.u) == Array{Array{floatT,1},1}
@assert typeof(bwd_sol.u) == Array{Array{floatT,1},1}
# Note that g includes the x0 gradient and the gradient on parameters.
# We do exactly as many f calls as there are function calls in the forward
# pass, and in the backward pass we don't need to call f, but instead we
# call ∇f.
(
xT = fwd_sol.u[end],
g = bwd_sol.u[end],
nf = fwd_sol.destats.nf,
n∇f = bwd_sol.destats.nf,
)
end
## ------------------------------------------------------------------------------------
# using DiffEqFlux, DifferentialEquations, Plots
# using ComponentArrays
# using FSimBase # for apply_inputs
# u0 = Float32[2.; 0.]
# u0_aug = vcat(u0, zeros(1))
# datasize = 30
# tspan = (0.0f0,1.5f0)
# L(u) = u' * u # running cost
# function trueODEfunc(dx,x,p,t; a)
# true_A = [-0.1 2.0; -2.0 -0.1]
# dx[1:2, :] .= ((x[1:2, :] .^ 3)'true_A)' + a
# dx[3, :] = L(x[1:2, :])
# end
# t = range(tspan[1],tspan[2],length=datasize)
# # prob = ODEProblem(trueODEfunc,u0,tspan)
# # prob = ODEProblem(trueODEfunc,u0_aug,tspan)
# # ode_data = Array(solve(prob,Tsit5(),saveat=t))
# # dudt2 = Chain(x -> x.^3,
# # Dense(2,50,tanh),
# # Dense(50,2))
# # p,re = Flux.destructure(dudt2) # use this p as the initial condition!
# # dudt(u,p,t) = re(p)(u) # need to restructure for backprop!
# # prob = ODEProblem(dudt,u0_aug,tspan)
# controller2 = Chain(u -> u.^3,
# Dense(2,50,tanh),
# Dense(50,2))
# p,re = Flux.destructure(controller2)
# controller(u, p, t) = re(p)(u)
# prob = ODEProblem(apply_inputs(trueODEfunc; a=(x, p, t) -> controller(x[1:2], p, t)),u0_aug,tspan)
# function predict_n_ode()
# Array(solve(prob,Tsit5(),u0=u0_aug,p=p,saveat=t)) # it seems that the output should be an Array
# end
# function loss_n_ode()
# pred = predict_n_ode()
# # loss = sum(abs2,ode_data .- pred)
# loss = pred[end][end] # to make it scalar for gradient calculation
# loss
# end
# loss_n_ode() # n_ode.p stores the initial parameters of the neural ODE
# cb = function (;doplot=false) # callback function to observe training
# pred = predict_n_ode()
# @show pred[end][end]
# # display(sum(abs2,ode_data .- pred))
# # # plot current prediction against data
# # pl = scatter(t,ode_data[1,:],label="data")
# # scatter!(pl,t,pred[1,:],label="prediction")
# # display(plot(pl))
# return false
# end
# # Display the ODE with the initial parameter values.
# cb()
# data = Iterators.repeated((), 1000)
# # Flux.train!(loss_n_ode, Flux.params(u0,p), data, ADAM(0.05), cb = cb)
# Flux.train!(loss_n_ode, Flux.params(p), data, ADAM(0.05), cb = cb)
#
# #-------------------------------------------------------------------------------
# using DiffEqFlux, DifferentialEquations, Plots, Statistics
# tspan = (0.0f0,8.0f0)
# ann = FastChain(FastDense(1,32,tanh), FastDense(32,32,tanh), FastDense(32,1))
# θ = initial_params(ann)
# function dxdt_(dx,x,p,t)
# x1, x2 = x
# dx[1] = x[2]
# dx[2] = ann([t],p)[1]^3
# end
# x0 = [-4f0,0f0]
# ts = Float32.(collect(0.0:0.01:tspan[2]))
# prob = ODEProblem(dxdt_,x0,tspan,θ)
# solve(prob,Vern9(),abstol=1e-10,reltol=1e-10)
# ##
# function predict_adjoint(θ)
# Array(solve(prob,Vern9(),p=θ,saveat=ts,sensealg=InterpolatingAdjoint(autojacvec=ReverseDiffVJP(true))))
# end
# function loss_adjoint(θ)
# x = predict_adjoint(θ)
# mean(abs2,4.0 .- x[1,:]) + 2mean(abs2,x[2,:]) + mean(abs2,[first(ann([t],θ)) for t in ts])/10
# end
# l = loss_adjoint(θ)
# cb = function (θ,l)
# println(l)
# p = plot(solve(remake(prob,p=θ),Tsit5(),saveat=0.01),ylim=(-6,6),lw=3)
# plot!(p,ts,[first(ann([t],θ)) for t in ts],label="u(t)",lw=3)
# display(p)
# return false
# end
# # Display the ODE with the current parameter values.
# cb(θ,l)
# loss1 = loss_adjoint(θ)
# res1 = DiffEqFlux.sciml_train(loss_adjoint, θ, ADAM(0.005), cb = cb,maxiters=100)
# res2 = DiffEqFlux.sciml_train(loss_adjoint, res1.u,
# BFGS(initial_stepnorm=0.01), cb = cb,maxiters=100,
# allow_f_increases = false)
##-------------------------------------------------------------------------------
using DiffEqFlux, DiffEqSensitivity, DifferentialEquations, Plots, Statistics
tspan = (0.0f0,8.0f0)
ann = FastChain(FastDense(1,32,tanh), FastDense(32,32,tanh), FastDense(32,1))
θ = initial_params(ann)
function dxdt_(x,p,t)
x1, x2 = x
return dx = [x2; ann([t],p)[1]^3]
end
x0 = [-4f0,0f0]
ts = Float32.(collect(0.0:0.01:tspan[2]))
prob = ODEProblem(dxdt_,x0,tspan,θ)
# solve(prob,Vern9(),abstol=1e-10,reltol=1e-10)
# function predict_adjoint(θ)
# Array(solve(prob,Vern9(),p=θ,saveat=0.01f0,sensealg=InterpolatingAdjoint()))
# end
function loss_adjoint_test(θ)
# x = predict_adjoint(θ)
x = Array(solve(prob,Tsit5(),p=θ,saveat=0.01f0,sensealg=InterpolatingAdjoint( autojacvec = ZygoteVJP() )))
mean(abs2,4.0f0 .- x[1,:]) + 2mean(abs2,x[2,:]) + mean(abs2,[first(ann([t],θ)) for t in ts])/10.0f0
end
l = loss_adjoint_test(θ)
cb = function (θ,l)
println(l)
p = plot(solve(remake(prob,p=θ),Tsit5(),saveat=0.01),ylim=(-6,6),lw=3)
plot!(p,ts,[first(ann([t],θ)) for t in ts],label="u(t)",lw=3)
display(p)
return false
end
# Display the ODE with the current parameter values.
cb(θ,l)
loss1 = loss_adjoint_test(θ)
res1 = DiffEqFlux.sciml_train(loss_adjoint_test, θ, ADAM(0.005); cb = cb, maxiters=100)
res2 = DiffEqFlux.sciml_train(loss_adjoint_test, res1.u,
BFGS(initial_stepnorm=0.01), cb = cb,maxiters=100,
allow_f_increases = false)
## -------------------------------------------------------------------------
using OrdinaryDiffEq, DiffEqFlux
pa = [1.0]
u0 = [3.0]
θ = [u0;pa]
function model1(θ,ensemble)
prob = ODEProblem((u, p, t) -> 1.01u .* p, [θ[1]], (0.0, 1.0), [θ[2]])
function prob_func(prob, i, repeat)
remake(prob, u0 = 0.5 .+ i/100 .* prob.u0)
end
ensemble_prob = EnsembleProblem(prob, prob_func = prob_func)
sim = solve(ensemble_prob, Tsit5(), ensemble, saveat = 0.1, trajectories = 100, sensealg = InterpolatingAdjoint(autojacvec = ZygoteVJP()))
end
# loss function
loss_serial(θ) = sum(abs2,1.0.-Array(model1(θ,EnsembleSerial())))
loss_threaded(θ) = sum(abs2,1.0.-Array(model1(θ,EnsembleThreads())))
prob_base = ODEProblem((u, p, t) -> 1.01u .* p, [θ[1]], (0.0, 1.0), [θ[2]])
function prob_gen(prob, i, repeat)
remake(prob, u0 = 0.5 .+ i/100 .* prob.u0)
end
function loss_test(θ)
prob = remake(prob_base, u0 = [θ[1]], p = [θ[2]])
fwd_ensemble_sol = Array( solve(EnsembleProblem(prob, prob_func = prob_gen), Tsit5(), EnsembleThreads(), saveat = 0.1, trajectories = 100, sensealg = InterpolatingAdjoint(autojacvec = ZygoteVJP())))
loss_val = sum(abs2, 1.0 .- fwd_ensemble_sol)
return loss_val
end
cb = function (θ,l) # callback function to observe training
@show l
false
end
opt = ADAM(0.1)
l1 = loss_serial(θ)
# res_serial = DiffEqFlux.sciml_train(loss_serial, θ, opt; cb = cb, maxiters=100)
# res_threads = DiffEqFlux.sciml_train(loss_threaded, θ, opt; cb = cb, maxiters=100)
res_test = DiffEqFlux.sciml_train(loss_test, θ, opt; cb = cb, maxiters = 100)
## ----------------
using DiffEqGPU, OrdinaryDiffEq
function lorenz(du,u,p,t)
du[1] = p[1]*(u[2]-u[1])
du[2] = u[1]*(p[2]-u[3]) - u[2]
du[3] = u[1]*u[2] - p[3]*u[3]
end
u0 = Float32[1.0;0.0;0.0]
tspan = (0.0f0,100.0f0)
p = [10.0f0,28.0f0,8/3f0]
prob = ODEProblem(lorenz,u0,tspan,p)
prob_func = (prob,i,repeat) -> remake(prob,p=rand(Float32,3).*p)
monteprob = EnsembleProblem(prob, prob_func = prob_func, safetycopy=false)
@time sol = solve(monteprob,Tsit5(),EnsembleGPUArray(),trajectories=10000,saveat=1.0f0)
## -------------------
using OrdinaryDiffEq, Flux, Optim, DiffEqFlux, DiffEqSensitivity
model_gpu = Chain(Dense(2, 50, tanh), Dense(50, 2)) |> gpu
p, re = Flux.destructure(model_gpu)
dudt!(u, p, t) = re(p)(u)
# Simulation interval and intermediary points
tspan = (0.0, 10.0)
tsteps = 0.0:0.1:10.0
u0 = Float32[2.0; 0.0] |> gpu
prob_gpu = ODEProblem(dudt!, u0, tspan, p)
# Runs on a GPU
sol_gpu = solve(prob_gpu, Tsit5(), saveat = tsteps) | ContinuousTimePolicyGradients | https://github.com/nhcho91/ContinuousTimePolicyGradients.jl.git |
|
[
"MIT"
] | 0.1.3 | 5f5a59cab6b3920d0b98dbebbb3f3f0477441546 | code | 100 | # Ask how to manage the Julia development environments to [JinraeKim](https://github.com/JinraeKim)
| ContinuousTimePolicyGradients | https://github.com/nhcho91/ContinuousTimePolicyGradients.jl.git |
|
[
"MIT"
] | 0.1.3 | 5f5a59cab6b3920d0b98dbebbb3f3f0477441546 | code | 1203 | using LinearAlgebra # ; similar to `import numpy` in Python
using Random
# import LinearAlgebra # different from `using LinearAlgebra`
"""
Julia functions receive two types of arguments: arguments (args) and keyword arguments (kwargs)
"""
function args_and_kwargs(args...; kwargs...) # args... is similar to `*args` in Python
@show args
@show kwargs
end
"""
`Random` and `LinearAlgebra` are basic packages provided by Julia.
Run REPL (Julia session) by e.g. `julia -q`.
Type `include("test/julia-tutorials/basic.jl")` in the REPL to read this script.
To run `main`, type `main()` in the REPL.
To reflect the updated codes without re-`include`,
use `includet` with [Revise.jl](https://github.com/timholy/Revise.jl).
I highly recommend you to read Revise.jl's documentation.
"""
function main(; seed=2021)
Random.seed!(seed) # how to control the random seed
args_and_kwargs(1, 2; a=1, b=2) # args = (1, 2), kwargs = Base.Pairs(:a => 1, :b => 2)
x = [1, 2, 3]
y = [4, 5, 6]
@show dot(x, y) # LinearAlgebra.dot; 1*4 + 2*5 + 3*6 = 32
nothing # similar to `None` in Python; the output of the last line is automatically returned if there is no `return` command
end
| ContinuousTimePolicyGradients | https://github.com/nhcho91/ContinuousTimePolicyGradients.jl.git |
|
[
"MIT"
] | 0.1.3 | 5f5a59cab6b3920d0b98dbebbb3f3f0477441546 | code | 457 | using Plots
using Transducers: Map # using Transducers.Map as Map
"""
`Transducers.jl` is a very useful data-manipulation tool.
It deals with "iterator" very effectively.
"""
function main()
ts = 0:0.01:1
xs = ts |> Map(t -> 2*t) |> collect # Transducers.Map
fig = plot(ts, xs) # Plots.plot
savefig("example.png") # save the current figure
savefig(fig, "example.pdf") # or, specify which figure you will save
display(fig)
end
| ContinuousTimePolicyGradients | https://github.com/nhcho91/ContinuousTimePolicyGradients.jl.git |
|
[
"MIT"
] | 0.1.3 | 5f5a59cab6b3920d0b98dbebbb3f3f0477441546 | code | 1546 | using FlightSims # Based on DifferentialEquations.jl (DiffEq.jl); highly recommend you to read docs of DiffEq.jl
const FSim = FlightSims # similar to `import numpy as np` in Python
using ComponentArrays # to abstract your state or any data (very useful as it acts as a usual array)
using UnPack
struct MyEnv <: AbstractEnv # FlightSims.AbstractEnv
end
function State(env::MyEnv)
return function (x1::Number, x2::Number)
x = ComponentArray(x1=x1, x2=x2) # access them as x.x1, x.x2
end
end
"""
Double integrator example
"""
function Dynamics!(env::MyEnv)
@Loggable function dynamics!(dx, x, p, t; u) # FlightSims.@Loggable is for data saving
@unpack x1, x2 = x # x1 = x.x1, x2 = x.x2
@log x1, x2
@log u
dx.x1 = x2
dx.x2 = u
end
end
function main()
env = MyEnv()
x10, x20 = 1.0, 2.0 # initial state
x0 = State(env)(x10, x20) # encode them to use conveniently
tf = 1.0 # terminal time
Δt = 0.01
my_controller(x, p, t) = -2*x.x1
simulator = Simulator(
x0,
apply_inputs(Dynamics!(env); u=my_controller),
tf=tf,
)
@time df = solve(simulator; savestep=Δt) # @time will shows the elapsed time
ts = df.time
x1s = df.sol |> Map(datum -> datum.x1) |> collect
x2s = df.sol |> Map(datum -> datum.x2) |> collect
fig_x1 = plot(ts, x1s)
fig_x2 = plot(ts, x2s)
fig = plot(fig_x1, fig_x2; layout=(2, 1))
display(fig)
end
| ContinuousTimePolicyGradients | https://github.com/nhcho91/ContinuousTimePolicyGradients.jl.git |
|
[
"MIT"
] | 0.1.3 | 5f5a59cab6b3920d0b98dbebbb3f3f0477441546 | docs | 7699 | # ContinuousTimePolicyGradients
ContinuousTimePolicyGradients.jl is a package for development and implementation of continuous-time policy gradient (CTPG) methods.
## Notes
- This package is WIP; may include verbose tutorials for Julia, DifferentialEquations.jl, etc.
- Thanks to [Namhoon Cho](https://github.com/nhcho91) for the shared materials and the initial efforts to investigate CTPG methods.
- Similar packages written in Julia focusing on control policy optimisation based on continuous-time adjoint sensitivity method include
- [ctpg](https://github.com/samuela/ctpg) developed by Samuel Ainsworth
- [control_neuralode](https://github.com/IlyaOrson/control_neuralode) developed by Ilya Orson
## Citation
ContinuousTimePolicyGradients.jl has been developed considering control tasks as the main application. The CTPG method for cost gradient computation can be utilised to perform policy optimisation in the setup of either background planning (offline control law learning) or decision-time planning (online control profile optimisation). The following paper addresses the optimisation of structured neural controller using CTPG with the flight vehicle normal acceleration tracking controller as an illustrating example. Please consider citing the paper if you find this package useful.
- Namhoon Cho, and Hyo-Sang Shin, "Optimisation of Structured Neural Controller Based on Continuous-Time Policy Gradient," [arXiv:2201.06262](https://arxiv.org/abs/2201.06262), January 2022.
## High-Level Training Interface: `CTPG_train()`
```julia
CTPG_train(dynamics_plant::Function, dynamics_controller::Function, cost_running::Function, cost_terminal::Function, cost_regularisor::Function, policy_NN, scenario;
solve_alg = Tsit5(), sense_alg = InterpolatingAdjoint(autojacvec = ZygoteVJP()), ensemble_alg = EnsembleThreads(), opt_1 = ADAM(0.01), opt_2 = LBFGS(), maxiters_1 = 100, maxiters_2 = 100, progress_plot = true, solve_kwargs...)
```
`CTPG_train()` provides a high-level interface for optimisation of the neural networks inside an ODE-represented dynamics based on Continuous-Time Policy Gradient (CTPG) methods that belong to the adjoint sensitivity analysis techniques. The code implemented and the default values for keyword arguments are specified considering training of a neural controller as the main application. In the context herein, a neural controller refers to a dynamic controller that incorporates neural-network-represented components at some points in its mathematical description.
The code utilises the functionalities provided by the [DiffEqFlux.jl](https://github.com/SciML/DiffEqFlux.jl) and [DiffEqSensitivity.jl](https://github.com/SciML/DiffEqSensitivity.jl) packages, and the Automatic Differentiation (AD) capabilities provided by the [Zygote.jl](https://github.com/FluxML/Zygote.jl) package that is integrated in DiffEqFlux.jl. `CTPG_train()` presumes the consistency of the functions provided as its input arguments with the AD tool, hence, the dynamics and cost functions should maintain their transparence against AD tools.
The optimisation (training) problem minimises the cost function defined over deterministic samples of the initial plant state `x₀` and the reference `r` by performing ensemble simulation based on parallelised computation.
The signals are defined as described below:
- `t`: time
- `x`: plant state
- `y`: plant output (= sensor output)
- `x_c`: controller state
- `u`: plant input (= controller output)
- `r`: exogenous reference
- `x_aug`: augmented forward dynamics state (= `[x; x_c; ∫cost_running]`)
- `p_NN`: neural network parameter
The arguments should be provided as explained below:
- `dynamics_plant`: Describes the dynamics of the plant to be controlled. Input arguments `x` and `u` should be of Vector type.
- `dynamics_controller`: Describes the dynamics of the controller that includes neural networks components. Input arguments `x_c`, `y`, `r`, and `p_NN` should be of Vector type.
- `dynamics_sensor`: Describes the dynamics of the sensor that measures output variables fed to the controller. Input arguments `x` should be of Vector type:
- `cost_running`: Describes the running cost defined as the integrand of the Lagrange-form continuous functional. Input arguments `x`, `y`, `u`, and `r` should be of Vector type.
- `cost_terminal`: Describes the terminal cost defined as the Mayer-form problem cost function. Defines a Bolza-form problem along with `cost_running`. Input arguments `x_f` and `r` should be of Vector type.
- `cost_regularisor`: Describes the regularisation term appended to the cost (loss) function. Input argument `p_NN` should be of Vector type.
- `policy_NN`: The neural networks entering into the controller dynamics. DiffEqFlux-based FastChain is recommended for its construction.
- `scenario`: Contains the parameters related with the ensemble-based training scenarios.
- `ensemble`: A vector of the initial plant state `x₀` and the reference `r` constituting the trajectory realisations.
- `t_span`: Time span for forward-pass integration
- `t_save`: Array of time points to be saved while solving ODE. Typically defined as `t_save = t_span[1]:Δt_save:t_span[2]`
- `dim_x`: `length(x)`
- `dim_x_c`: `length(x_c)`
The keyword arguments should be provided as explained below:
- `solve_alg`: The algorithm used for solving ODEs. Default value is `Tsit5()`
- `sense_alg`: The algorithm used for adjoint sensitivity analysis. Default value is `InterpolatingAdjoint(autojacvec = ZygoteVJP())`, because the control problems usually render the `BacksolveAdjoint()` unstable. The vjp choice `autojacvec = ReverseDiffVJP(true)` is usually faster than `ZygoteVJP()`, when the ODE function does not have any branching inside. Please refer to the [DiffEqFlux documentation](https://diffeqflux.sciml.ai/dev/ControllingAdjoints/) for further details.
- `ensemble_alg`: The algorithm used for handling ensemble of ODEs. Default value is `EnsembleThreads()` for multi-threaded computation in CPU.
- `opt_1`: The algorithm used for the first phase of optimisation which rapidly delivers the parameter to a favourable region around a local minimum. Default value is `ADAM(0.01)`.
- `opt_2`: The algorithm used for the second phase of opitmisaiton. Defalut value is `LBFGS()` which refines the result of the first phase to find a more precise minimum. Please refer to the [DiffEqFlux documentation](https://diffeqflux.sciml.ai/dev/sciml_train/) for further details about two-phase composition of optimisers.
- `maxiters_1`: The maximum number of iterations allowed for the first phase of optimisation with `opt_1`. Defalut value is `100`.
- `maxiters_2`: The maximum number of iterations allowed for the second phase of optimisation with `opt_2`. Defalut value is `100`.
- `progress_plot`: The indicator to plot the state history for a nominal condition among the ensemble during the learning process. Default value is `true`.
- `i_nominal`: The index to select the case to plot using `progress_plot` during optimisation process from the `ensemble` defined in `scenario`. Defalut value is `nothing`.
- `p_NN_0`: Initial value of the NN parameters supplied by the user to bypass random initialisation of `p_NN` or to continue optimisation from the previous result. Defalut value is `nothing`.
- `solve_kwargs...`: Additional keyword arguments that are passed onto the ODE solver.
`CTPG_train()` returns the following outputs:
- `result`: The final result of parameter optimisation.
- `fwd_ensemble_sol`: The ensemble solution of forward simulation using the final neural network parameters.
- `loss_history`: The history of loss function evaluated at each iteration. | ContinuousTimePolicyGradients | https://github.com/nhcho91/ContinuousTimePolicyGradients.jl.git |
|
[
"MIT"
] | 0.1.0 | c4d32b57c53388212c110e15fa38e1c4cc6e2b1a | code | 640 | using MotifPvalue
using Documenter
DocMeta.setdocmeta!(MotifPvalue, :DocTestSetup, :(using MotifPvalue); recursive=true)
makedocs(;
modules=[MotifPvalue],
authors="Shane Kuei Hsien Chu ([email protected])",
repo="https://github.com/kchu25/MotifPvalue.jl/blob/{commit}{path}#{line}",
sitename="MotifPvalue.jl",
format=Documenter.HTML(;
prettyurls=get(ENV, "CI", "false") == "true",
canonical="https://kchu25.github.io/MotifPvalue.jl",
assets=String[],
),
pages=[
"Home" => "index.md",
],
)
deploydocs(;
repo="github.com/kchu25/MotifPvalue.jl.git",
devbranch="main",
)
| MotifPvalue | https://github.com/kchu25/MotifPvalue.jl.git |
|
[
"MIT"
] | 0.1.0 | c4d32b57c53388212c110e15fa38e1c4cc6e2b1a | code | 374 | module MotifPvalue
#### Dependencies ##################
using DataStructures
####################################
#### Exported methods and types ####
export score2pvalue, pval2score
####################################
#### Load files ####################
include("helpers.jl")
include("score2pval.jl")
include("pval2score.jl")
####################################
end
| MotifPvalue | https://github.com/kchu25/MotifPvalue.jl.git |
|
[
"MIT"
] | 0.1.0 | c4d32b57c53388212c110e15fa38e1c4cc6e2b1a | code | 4508 | #=
Best possible score of a PWM
# Input
`pwm::Matrix{Real}`: a 4 x m matrix
# Output
the best possible score this matrix can get
=#
function best_score(pwm::Matrix{T}) where T <: Real
sum(maximum(pwm[:,i]) for i = 1:size(pwm,2));
end
function best_score(pwm_col::Vector{T}) where T <: Real
maximum(pwm_col);
end
#=
Worst possible score of a PWM
# Input
`pwm::Matrix{Real}`: a 4 x m matrix
# Output
the worst possible score this matrix can get
=#
function worst_score(pwm::Matrix{T}) where T <: Real
sum(minimum(pwm[:,i]) for i = 1:size(pwm,2));
end
function worst_score(pwm_col::Vector{T}) where T <: Real
minimum(pwm_col);
end
#=
Return a column-permuted PWM that minimize the score range so that δ₁ ≥ δ₂ ≥ … ≥ δₘ
where δᵢ = best_score(pwm[:,i])-worst_score(pwm[:,i]).
# Input
`pwm::Matrix{Real}`: a 4 x m matrix
# Output
a column-permuted pwm
=#
function min_score_range(pwm::Matrix{T}) where T <: Real
pwm[:,sortperm([best_score(pwm[:,i])-worst_score(pwm[:,i]) for i = 1:size(pwm,2)],rev=true)];
end
#=
"Round the PWM"
# Input
`pwm::Matrix{Real}`: a 4 x m matrix
`granularity`: a small positive real number e.g. 0.01 or 0.001, etc.
# Output
a rounded pwm of the input pwm
=#
function round_pwm(pwm::Matrix{T}, granularity::Real) where T <: Real
floor.(pwm ./ granularity) * granularity;
end
#=
The maximum error induced by the rounded pwm M_ϵ
(see definition 3 in https://almob.biomedcentral.com/articles/10.1186/1748-7188-2-15; this is the quantity E)
# Input
`pwm::Matrix{Real}`: a 4 x m matrix
`granularity`:
# Output
A positve real number that's the maximum error induced by the rounded pwm M_ϵ
=#
function calc_E(pwm, pwm_rounded)
sum(maximum(pwm[:,i]-pwm_rounded[:,i]) for i = size(pwm));
end
#=
Note: Use a nested dictionary to represent the distribution Q
Since Q is used to reference the probability of (M[1…i],score),
the keys in the first layer is i, and the value in the first layer
are dictionaries with scores as keys and probability as values
call create_Q(m) to initialize such a distribution Q
where m is the "width" of the PWM
=#
create_Q(m::Integer) = Dict{Int16,SortedDict{Float64,Float64}}(i==0 ? i=>SortedDict(0=>1) : i=>SortedDict() for i=0:m);
#=
Input:
pwm: a 4 x m matrix
α, β: score interval [α, β]
bg: 4 x 1 vector that specifies the multinomial genomic background; default to flat background.
Output:
Q: a probability mass table
e.g. Q[m] shows all the weights of P[pwm_score = η] for α ≤ η ≤ β
=#
function score_distribution(pwm_::Matrix{T}, α::Real, β::Real, bg=[.25,.25,.25,.25]) where T <: Real
m = size(pwm_,2);
Q = create_Q(m);
@inbounds for i = 1:m
bs = i+1 > m ? 0 : best_score(pwm_[:,i+1:m]);
ws = i+1 > m ? 0 : worst_score(pwm_[:,i+1:m]);
for score in keys(Q[i-1])
for j = 1:4
t = score + pwm_[j,i];
if α - bs ≤ t ≤ β - ws
if haskey(Q[i], t)
Q[i][t] += Q[i-1][score]*bg[j];
else
Q[i][t] = Q[i-1][score]*bg[j];
end
end
end
end
end
return Q
end
#=
Return the probability of the background model that has score ≥ α with respect to the input pwm
Input:
pwm: a 4 x m matrix
α: a score threshold
bg: 4 x 1 vector that specifies the multinomial genomic background; default to flat background.
Output:
pval: the probability that the background can have score ≥ α with respect to pwm
=#
function fast_pvalue(pwm::Matrix{T}, α::Real, bg=[.25,.25,.25,.25]) where T <: Real
m = size(pwm,2);
Q = create_Q(m);
pval = 0f0;
@inbounds for i = 1:m
bs = i+1 > m ? 0 : best_score(pwm[:,i+1:m]);
ws = i+1 > m ? 0 : worst_score(pwm[:,i+1:m]);
for (score,_) in Q[i-1]
for j = 1:4
t = score + pwm[j,i];
if α - ws ≤ t
pval = pval + Q[i-1][score]*bg[j];
elseif α - bs ≤ t
if haskey(Q[i], t)
Q[i][t] += Q[i-1][score]*bg[j];
else
Q[i][t] = Q[i-1][score]*bg[j];
end
end
end
end
end
return pval
end
# return the sum of all the weights
Q_sum(Q_m::SortedDict{Float64,Float64}) = sum(v for v in values(Q_m));
| MotifPvalue | https://github.com/kchu25/MotifPvalue.jl.git |
|
[
"MIT"
] | 0.1.0 | c4d32b57c53388212c110e15fa38e1c4cc6e2b1a | code | 2676 | function find_largest_α(Q_m::SortedDict{T,T}, pval::T) where T <: Real
q_sum = Q_sum(Q_m);
largest_k = nothing;
for (k,v) in Q_m
if q_sum ≥ pval
largest_k = k;
else
return k
end
q_sum -= v;
end
return largest_k
end
function pval_w_Qm(Qm::SortedDict{T,T}, α::Real) where T <: Real
pval = 0;
for (k,v) in Qm
if k ≥ α
pval += v;
end
end
return pval
end
function find_δ(Q_m::SortedDict{T,T}, pval_ϵ::Real, pval::Real) where T <: Real
q_sum_plus_pval_ϵ = Q_sum(Q_m)+pval_ϵ;
largest_δ = nothing;
for (k,v) in Q_m
if q_sum_plus_pval_ϵ ≥ pval
largest_δ = k;
else
return k
end
q_sum_plus_pval_ϵ -= v;
end
return largest_δ
end
"""
pval2score(pwm, pval, ϵ=1e-1, k=10, bg=[.25,.25,.25,.25])
Returns the highest score(M,pval) of a `pwm` such that p-value is greater or equal to `pval`.
Input:
* `pwm`: a 4 x m matrix
* `pval`: a p-value; e.g. pval = 1e-3
* `ϵ`: initial granularity (optional)
* `k`: Refinement parameter (optional)
* `bg`: multinomial background (optional)
Output
* `α`: the highest score-threshold
"""
function pval2score(pwm::Matrix{T}, pval::Real, ϵ=1e-1, k=10, bg=[.25,.25,.25,.25]) where T <: Real
@assert size(pwm,1) == 4 "The input matrix must have 4 and only 4 rows"
mpwm = min_score_range(pwm);
m = size(pwm, 2);
pwm_ϵ = round_pwm(mpwm, ϵ);
E = calc_E(mpwm, pwm_ϵ);
Q = create_Q(m);
Q = score_distribution(pwm_ϵ,worst_score(pwm_ϵ),Inf,bg);
α = find_largest_α(Q[m], pval);
@inbounds while !(pval_w_Qm(Q[m], α-E) ≈ pval_w_Qm(Q[m], α))
# println("err: ", pval_w_Qm(Q[m], α-E) - pval_w_Qm(Q[m], α));
ϵ = ϵ/k;
pwm_ϵ = round_pwm(mpwm, ϵ);
E = calc_E(mpwm, pwm_ϵ);
Q = create_Q(m);
Q = score_distribution(pwm_ϵ,α-E,α+E,bg);
#=
note:
Sometimes the score range is simply
too small and hence the score_distribution subroutine
cannot find any scores within the range [α-E, α+E].
This happens when the error of the round matrix, E, is very
small (see definition 3 in Touzet and Varre's paper:
https://almob.biomedcentral.com/articles/10.1186/1748-7188-2-15)
When this happens we return α+E, so that we don't
underestimate the score for the threshold for the
input p-value.
=#
if isempty(Q[m])
α = α + E;
break;
end
pval_ϵ = fast_pvalue(pwm_ϵ,α+E);
δ = find_δ(Q[m],pval_ϵ,pval);
α = δ;
end
return α;
end | MotifPvalue | https://github.com/kchu25/MotifPvalue.jl.git |
|
[
"MIT"
] | 0.1.0 | c4d32b57c53388212c110e15fa38e1c4cc6e2b1a | code | 2071 | #=
Helper function for score2pvalue. Find the lowest
score s such that P-value(M_ϵ,s) = P-value(M_ϵ,s-E).
Input:
Q_m: a sorted dict
E: the max column-wise error between matrix M and M_ϵ
Output:
=#
function find_s(Q_m::SortedDict{Float64,Float64}, E::Real)
keys_ = collect(keys(Q_m)); ℓ = length(keys_);
if ℓ > 1
for i = 1:(ℓ-1)
@inbounds if keys_[i]+E < keys_[i+1]
return keys_[i];
end
end
end
return nothing
end
"""
score2pvalue(pwm, α, ϵ=1e-1, k=100, bg=[.25,.25,.25,.25])
Returns P-value(M,α) of a `pwm` with a given threshold `α`.
Input:
* `pwm`: a 4 x m matrix
* `α`: the score
* `ϵ`: initial granularity (optional)
* `k`: Refinement parameter (optional)
* `bg`: multinomial background (optional)
Output:
* `pval`: p-value
"""
function score2pvalue(pwm::Matrix{T}, α::Real, ϵ=1e-1, k=100, bg=[.25,.25,.25,.25]) where T <: Real
@assert size(pwm,1) == 4 "The input matrix must have 4 and only 4 rows"
mpwm = min_score_range(pwm);
m = size(mpwm,2);
β = best_score(mpwm)+1;
pval = 0;
s = 0;
i = 1;
@inbounds while !(α ≈ s)
# println(α-s)
ϵ = i == 1 ? ϵ : ϵ/k; i+=1;
# isinf(ϵ) || isnan(ϵ) && (println(i); break);
pwm_ϵ = round_pwm(mpwm, ϵ);
# any(isnan.(pwm_ϵ)) && (println(i); break);
E = calc_E(mpwm, pwm_ϵ);
Q = create_Q(m);
Q = score_distribution(pwm_ϵ, α-E, β, bg);
#=
note:
Sometimes the score range is small
(i.e. β-(α-E) is very small) and hence we won't be
able to find such s. In such case, we return the
calculated p-value.
Note that this p-value is an underestimate. To get a
more accurate result, set k to be a larger number,
e.g. k=100.
=#
s = find_s(Q[m],E);
if !isnothing(s)
for (k,v) in Q[m]
pval += k ≥ s ? v : 0;
end
β = s;
else
break;
end
end
return pval
end | MotifPvalue | https://github.com/kchu25/MotifPvalue.jl.git |
|
[
"MIT"
] | 0.1.0 | c4d32b57c53388212c110e15fa38e1c4cc6e2b1a | code | 95 | using MotifPvalue
using Test
@testset "MotifPvalue.jl" begin
# Write your tests here.
end
| MotifPvalue | https://github.com/kchu25/MotifPvalue.jl.git |
|
[
"MIT"
] | 0.1.0 | c4d32b57c53388212c110e15fa38e1c4cc6e2b1a | docs | 2009 | # MotifPvalue
[](https://kchu25.github.io/MotifPvalue.jl/stable)
[](https://kchu25.github.io/MotifPvalue.jl/dev)
[](https://github.com/kchu25/MotifPvalue.jl/actions/workflows/CI.yml?query=branch%3Amain)
[](https://codecov.io/gh/kchu25/MotifPvalue.jl)
# Introduction
This package provides two useful subroutines:
* Given a score threshold `α`, `score2pvalue` calculates the p-value, i.e. the probability of a set of strings generated from a specfied background model such that each string in this set can attain a score higher than or equal to `α` with respect to the input `pwm`.
* Given a p-value `pval`, `pval2score` calculates the highest score `α` such that `score2pvalue(pwm,α)` is larger than or equal to `pval`.
By default, the background model is specified as i.i.d discrete uniform.
This is an implementation of:
Efficient and accurate P-value computation for Position Weight Matrices by Touzet et al.
https://almob.biomedcentral.com/articles/10.1186/1748-7188-2-15
# Basic examples
using MotifPvalue
# An example PWM
pwm = [-2.86995 1.3814 1.36906 -4.88479 -4.19164 -4.88479 1.36607 -5.57793 -0.336375 1.38238;
1.36707 -5.57793 -2.86995 -0.294869 -4.88479 1.34492 -3.49849 1.26497 -2.1768 -4.88479;
-5.57793 -4.47899 -5.57793 1.11923 -3.78584 -5.57793 -4.88479 -5.57793 0.798703 -5.57793;
-4.19164 -5.57793 -4.88479 -1.68576 1.375 -1.88885 -3.17976 -0.798616 -0.0526305 -5.57793];
# Compute pvalue with pwm and score = 4
score2pvalue(pwm, 4)
> 0.00020599365234375
# Compute the score-threshold for p-value 1e-4
pval2score(pwm, 1e-4)
> 5.010000000000001 | MotifPvalue | https://github.com/kchu25/MotifPvalue.jl.git |
|
[
"MIT"
] | 0.1.0 | c4d32b57c53388212c110e15fa38e1c4cc6e2b1a | docs | 189 | ```@meta
CurrentModule = MotifPvalue
```
# MotifPvalue
Documentation for [MotifPvalue](https://github.com/kchu25/MotifPvalue.jl).
```@index
```
```@autodocs
Modules = [MotifPvalue]
```
| MotifPvalue | https://github.com/kchu25/MotifPvalue.jl.git |
|
[
"MIT"
] | 0.29.0 | 6653c51ac7bb1b0f10bf4ab5576f6f00fe7f4cf8 | code | 1407 | #! format: off
using SimpleWorkflows
using Documenter
DocMeta.setdocmeta!(SimpleWorkflows, :DocTestSetup, :(using SimpleWorkflows); recursive=true)
makedocs(;
modules=[SimpleWorkflows],
authors="singularitti <[email protected]>",
repo="https://github.com/MineralsCloud/SimpleWorkflows.jl/blob/{commit}{path}#{line}",
sitename="SimpleWorkflows.jl",
format=Documenter.HTML(;
prettyurls=get(ENV, "CI", "false") == "true",
canonical="https://MineralsCloud.github.io/SimpleWorkflows.jl",
edit_link="main",
assets=String[],
),
pages=[
"Home" => "index.md",
"Manual" => [
"Installation Guide" => "man/installation.md",
"Portability" => "man/portability.md",
"Troubleshooting" => "man/troubleshooting.md",
],
"Reference" => Any[
"Public API" => "lib/public.md",
"Internals" => map(
s -> "lib/internals/$(s)",
sort(readdir(joinpath(@__DIR__, "src/lib/internals")))
),
],
"Developer Docs" => [
"Contributing" => "developers/contributing.md",
"Style Guide" => "developers/style-guide.md",
"Design Principles" => "developers/design-principles.md",
],
],
)
deploydocs(;
repo="github.com/MineralsCloud/SimpleWorkflows.jl",
devbranch="main",
)
| SimpleWorkflows | https://github.com/MineralsCloud/SimpleWorkflows.jl.git |
|
[
"MIT"
] | 0.29.0 | 6653c51ac7bb1b0f10bf4ab5576f6f00fe7f4cf8 | code | 3475 | module SimpleWorkflows
using EasyJobsBase: AbstractJob, eachparent, eachchild
using Graphs:
DiGraph,
add_edge!,
nv,
is_cyclic,
is_directed,
is_connected,
has_edge,
topological_sort_by_dfs
export Workflow
abstract type AbstractWorkflow end
# Create a `Workflow` from a list of `AbstractJob`s and a graph representing their relations.
struct Workflow{T} <: AbstractWorkflow
jobs::Vector{T}
graph::DiGraph{Int}
function Workflow{T}(jobs, graph) where {T}
@assert !is_cyclic(graph) "`graph` must be acyclic!"
@assert is_directed(graph) "`graph` must be directed!"
@assert is_connected(graph) "`graph` must be connected!"
@assert nv(graph) == length(jobs) "`graph` has different size from `jobs`!"
@assert allunique(jobs) "at least two jobs are identical!"
return new(jobs, graph)
end
end
Workflow(jobs::AbstractVector{T}, graph) where {T} = Workflow{T}(jobs, graph)
"""
Workflow(jobs::AbstractJob...)
Create a `Workflow` from a given series of `AbstractJob`s.
The list of `AbstractJob`s does not have to be complete, our algorithm will find all
connected `AbstractJob`s automatically.
"""
function Workflow(jobs::AbstractJob...)
jobsfound = convert(Vector{AbstractJob}, collect(jobs)) # Need to relax type constraints to contain different types of jobs
for job in jobsfound
neighbors = union(eachparent(job), eachchild(job))
for neighbor in neighbors
if neighbor ∉ jobsfound
push!(jobsfound, neighbor) # This will alter `all_possible_jobs` dynamically
end
end
end
n = length(jobsfound)
graph = DiGraph(n)
dict = IdDict(zip(jobsfound, 1:n))
for (i, job) in enumerate(jobsfound)
for parent in eachparent(job)
if !has_edge(graph, dict[parent], i)
add_edge!(graph, dict[parent], i)
end
end
for child in eachchild(job)
if !has_edge(graph, i, dict[child])
add_edge!(graph, i, dict[child])
end
end
end
return Workflow(topological_sort(jobsfound, graph)...)
end
Workflow(jobs::AbstractVector) = Workflow(jobs...)
function topological_sort(jobs, graph)
order = topological_sort_by_dfs(graph)
sorted_jobs = collect(jobs[order])
n = length(sorted_jobs)
new_graph = DiGraph(n)
dict = IdDict(zip(sorted_jobs, 1:n))
# You must sort the graph too for `DependentJob`s to run in the correct order!
for (i, job) in enumerate(sorted_jobs)
for parent in eachparent(job)
if !has_edge(new_graph, dict[parent], i)
add_edge!(new_graph, dict[parent], i)
end
end
for child in eachchild(job)
if !has_edge(new_graph, i, dict[child])
add_edge!(new_graph, i, dict[child])
end
end
end
return sorted_jobs, new_graph
end
Base.indexin(jobs, wf::Workflow) = Base.indexin(jobs, collect(wf))
Base.in(job::AbstractJob, wf::Workflow) = job in wf.jobs
Base.iterate(wf::Workflow, state=firstindex(wf)) = iterate(wf.jobs, state)
Base.eltype(::Type{Workflow{T}}) where {T} = T
Base.length(wf::Workflow) = length(wf.jobs)
Base.getindex(wf::Workflow, i) = getindex(wf.jobs, i)
Base.firstindex(wf::Workflow) = 1
Base.lastindex(wf::Workflow) = length(wf.jobs)
include("operations.jl")
include("run.jl")
include("status.jl")
include("show.jl")
end
| SimpleWorkflows | https://github.com/MineralsCloud/SimpleWorkflows.jl.git |
|
[
"MIT"
] | 0.29.0 | 6653c51ac7bb1b0f10bf4ab5576f6f00fe7f4cf8 | code | 459 | import EasyJobsBase: chain!, →, ←
for (func, (op₁, op₂)) in zip((:chain!,), ((:→, :←),))
@eval begin
$func(x::AbstractWorkflow, y::AbstractWorkflow) = $func(last(x), first(y))
$func(x::AbstractWorkflow, y::AbstractWorkflow, z::AbstractWorkflow...) =
foldr($func, (x, y, z...))
$op₁(x::AbstractWorkflow, y::AbstractWorkflow) = $func(x, y)
$op₂(y::AbstractWorkflow, x::AbstractWorkflow) = $op₁(x, y)
end
end
| SimpleWorkflows | https://github.com/MineralsCloud/SimpleWorkflows.jl.git |
|
[
"MIT"
] | 0.29.0 | 6653c51ac7bb1b0f10bf4ab5576f6f00fe7f4cf8 | code | 4248 | using Graphs: indegree, rem_vertices!
import EasyJobsBase: run!, execute!
export run!, execute!
abstract type Executor end
struct SerialExecutor <: Executor
maxattempts::UInt64
interval::Real
delay::Real
wait::Bool
end
function SerialExecutor(; maxattempts=1, interval=1, delay=0, wait=false)
@assert maxattempts >= 1
@assert interval >= zero(interval)
@assert delay >= zero(delay)
return SerialExecutor(maxattempts, interval, delay, wait)
end
struct AsyncExecutor <: Executor
maxattempts::UInt64
interval::Real
delay::Real
wait::Bool
end
function AsyncExecutor(; maxattempts=1, interval=1, delay=0, wait=false)
@assert maxattempts >= 1
@assert interval >= zero(interval)
@assert delay >= zero(delay)
return AsyncExecutor(maxattempts, interval, delay, wait)
end
"""
run!(wf::Workflow; maxattempts=5, interval=1, delay=0)
Run a `Workflow` with maximum number of attempts, with each attempt separated by a few seconds.
"""
run!(wf::AbstractWorkflow; kwargs...) = execute!(wf, AsyncExecutor(; kwargs...))
"""
execute!(workflow::AbstractWorkflow, exec::Executor)
Executes the jobs from the workflow of the provided Executor instance.
The function will attempt to execute all the jobs up to `exec.maxattempts` times. If all jobs
have succeeded, the function will stop immediately. Otherwise, it will wait for a duration equal
to `exec.interval` before the next attempt.
"""
function execute!(wf::AbstractWorkflow, exec::Executor)
task = if issucceeded(wf)
@task wf # Just return the job if it has succeeded
else
sleep(exec.delay)
@task dispatch!(wf, exec)
end
schedule(task)
if exec.wait
wait(task)
end
return task
end
function dispatch!(wf::AbstractWorkflow, exec::SerialExecutor)
for _ in Base.OneTo(exec.maxattempts)
for job in Iterators.filter(!issucceeded, wf)
run!(job; maxattempts=1, interval=0, delay=0, wait=true) # Must wait for serial execution
end
issucceeded(wf) ? break : sleep(exec.interval)
end
return wf
end
function dispatch!(wf::AbstractWorkflow, exec::AsyncExecutor)
for _ in Base.OneTo(exec.maxattempts)
jobs, graph = copy(wf.jobs), copy(wf.graph)
run_kahn_algo!(jobs, graph)
issucceeded(wf) ? break : sleep(exec.interval)
end
return wf
end
# This function `run_kahn_algo!` is an implementation of Kahn's algorithm for job scheduling.
# `graph` is a directed acyclic graph representing dependencies between jobs.
# `execs` is a list of executors that can run the jobs.
function run_kahn_algo!(jobs, graph) # Do not export!
# Check if `execs` is empty and if there are no vertices in the `graph`.
# This is the base case of the recursion, if there are no jobs left to execute and no
# vertices in the graph, the function will stop its execution.
if isempty(jobs) && iszero(nv(graph)) # Stopping criterion
return nothing
elseif length(jobs) == nv(graph)
# Find all vertices with zero in-degree in the graph, these vertices have no prerequisites
# and can be executed immediately. They are put in a queue.
queue = findall(iszero, indegree(graph))
# For each executor in the `queue`, start the job execution.
# The `@sync` macro ensures that the main program waits until all async blocks are done.
@sync for job in jobs[queue]
# Run the jobs with no prerequisites in parallel since they are in the same level.
@async run!(job; maxattempts=1, interval=0, delay=0, wait=true)
end
# Remove the vertices corresponding to the executed jobs from the graph.
# This also changes the indegree of the remaining vertices.
rem_vertices!(graph, queue; keep_order=true)
# Remove the executed jobs from the list.
deleteat!(jobs, queue)
# Recursively call the `run_kahn_algo!` with the updated jobs list and graph.
# This will continue the execution with the remaining jobs that are now without prerequisites.
return run_kahn_algo!(jobs, graph)
else
throw(ArgumentError("something went wrong when running Kahn's algorithm!"))
end
end
| SimpleWorkflows | https://github.com/MineralsCloud/SimpleWorkflows.jl.git |
|
[
"MIT"
] | 0.29.0 | 6653c51ac7bb1b0f10bf4ab5576f6f00fe7f4cf8 | code | 1562 | using Dates: format
using EasyJobsBase: ispending, isrunning, starttimeof, endtimeof, timecostof, printf
using Graphs: ne
# See https://docs.julialang.org/en/v1/manual/types/#man-custom-pretty-printing
function Base.show(io::IO, wf::Workflow)
if get(io, :compact, false)
print(IOContext(io, :limit => true, :compact => true), summary(wf))
else
njobs, nedges = nv(wf.graph), ne(wf.graph)
print(io, summary(wf), '(', njobs, " jobs, ", nedges, " edges)")
end
end
function Base.show(io::IO, ::MIME"text/plain", wf::Workflow)
println(io, summary(wf))
for (i, job) in enumerate(wf)
println(io, " [", i, "] ", "id: ", job.id)
if !isempty(job.description)
print(io, ' '^5, "description: ")
show(io, job.description)
println(io)
end
print(io, ' '^5, "core: ")
printf(io, job.core)
print(io, '\n', ' '^5, "status: ")
printstyled(io, getstatus(job); bold=true)
if !ispending(job)
print(
io,
'\n',
' '^5,
"from: ",
format(starttimeof(job), "dd-u-YYYY HH:MM:SS.s"),
'\n',
' '^5,
"to: ",
)
if isrunning(job)
print(io, "still running...")
else
println(io, format(endtimeof(job), "dd-u-YYYY HH:MM:SS.s"))
print(io, ' '^5, "uses: ", timecostof(job))
end
end
println(io)
end
end
| SimpleWorkflows | https://github.com/MineralsCloud/SimpleWorkflows.jl.git |
|
[
"MIT"
] | 0.29.0 | 6653c51ac7bb1b0f10bf4ab5576f6f00fe7f4cf8 | code | 2098 | import EasyJobsBase:
getstatus,
ispending,
isrunning,
isexited,
issucceeded,
isfailed,
filterpending,
filterrunning,
filterexited,
filtersucceeded,
filterfailed
export getstatus,
ispending,
isrunning,
isexited,
issucceeded,
isfailed,
filterpending,
filterrunning,
filterexited,
filtersucceeded,
filterfailed
"""
ispending(wf::AbstractWorkflow)
Check if all jobs in the `AbstractWorkflow` are in a pending state.
Return `true` if all jobs are pending, otherwise, return `false`.
"""
ispending(wf::AbstractWorkflow) = all(ispending, wf)
"""
isrunning(wf::AbstractWorkflow)
Check if any job in the `AbstractWorkflow` is currently running.
Return `true` if at least one job is running, otherwise, return `false`.
"""
isrunning(wf::AbstractWorkflow) = any(isrunning, wf)
"""
isexited(wf::AbstractWorkflow)
Check if all jobs in the `AbstractWorkflow` have exited.
Return `true` if all jobs have exited, otherwise, return `false`.
"""
isexited(wf::AbstractWorkflow) = all(isexited, wf)
"""
issucceeded(wf::AbstractWorkflow)
Check if all jobs in the `AbstractWorkflow` have successfully completed.
Return `true` if all jobs have succeeded, otherwise, return `false`.
"""
issucceeded(wf::AbstractWorkflow) = all(issucceeded, wf)
"""
isfailed(wf::AbstractWorkflow)
Check if any job in the `AbstractWorkflow` has failed, given that all jobs have exited.
Return `true` if any job has failed after all jobs have exited, otherwise, return `false`.
"""
isfailed(wf::AbstractWorkflow) = isexited(wf) && any(isfailed, wf)
# See https://docs.julialang.org/en/v1/manual/documentation/#Advanced-Usage
for (func, adj) in zip(
(:filterpending, :filterrunning, :filterexited, :filtersucceeded, :filterfailed),
("pending", "running", "exited", "succeeded", "failed"),
)
name = string(func)
@eval begin
"""
$($name)(wf::AbstractWorkflow)
Filter only the $($adj) jobs in a `Workflow`.
"""
$func(wf::Workflow) = $func(collect(wf))
end
end
| SimpleWorkflows | https://github.com/MineralsCloud/SimpleWorkflows.jl.git |
|
[
"MIT"
] | 0.29.0 | 6653c51ac7bb1b0f10bf4ab5576f6f00fe7f4cf8 | code | 1110 | @testset "Test joining two `Workflow`s" begin
function f₁()
println("Start job `i`!")
return sleep(5)
end
function f₂(n)
println("Start job `j`!")
sleep(n)
return exp(2)
end
function f₃(n)
println("Start job `k`!")
return sleep(n)
end
function f₄()
println("Start job `l`!")
return run(`sleep 3`)
end
function f₅(n, x)
println("Start job `m`!")
sleep(n)
return sin(x)
end
function f₆(n; x=1)
println("Start job `n`!")
sleep(n)
cos(x)
return run(`pwd` & `ls`)
end
i = Job(Thunk(f₁); username="me", name="i")
j = Job(Thunk(f₂, 3); username="he", name="j")
k = Job(Thunk(f₃, 6); name="k")
l = Job(Thunk(f₄); name="l", username="me")
m = Job(Thunk(f₅, 3, 1); name="m")
n = Job(Thunk(f₆, 1; x=3); username="she", name="n")
i → j
i → k
j → k
l → m
l → n
m → n
wf₁ = Workflow(k)
wf₂ = Workflow(n)
wf₁ → wf₂
wf = Workflow(k)
@test unique(wf) == collect(wf)
run!(wf)
end
| SimpleWorkflows | https://github.com/MineralsCloud/SimpleWorkflows.jl.git |
|
[
"MIT"
] | 0.29.0 | 6653c51ac7bb1b0f10bf4ab5576f6f00fe7f4cf8 | code | 3793 | using Thinkers: Thunk
using EasyJobsBase: Job, ConditionalJob, ArgDependentJob, run!, getresult, →
@testset "Test running a `Workflow`" begin
function f₁()
println("Start job `i`!")
sleep(5)
println("End job `i`!")
return nothing
end
function f₂(n)
println("Start job `j`!")
sleep(n)
a = exp(2)
println("End job `j`!")
return a
end
function f₃(n)
println("Start job `k`!")
sleep(n)
println("End job `k`!")
return nothing
end
function f₄()
println("Start job `l`!")
p = run(`sleep 3`)
println("End job `l`!")
return p
end
function f₅(n, x)
println("Start job `m`!")
sleep(n)
a = sin(x)
println("End job `m`!")
return a
end
function f₆(n; x=1)
println("Start job `n`!")
sleep(n)
p = run(`pwd` & `ls`)
println("End job `n`!")
return p
end
i = Job(Thunk(f₁); username="me", name="i")
j = Job(Thunk(f₂, 3); username="he", name="j")
k = Job(Thunk(f₃, 6); name="k")
l = Job(Thunk(f₄); name="l", username="me")
m = Job(Thunk(f₅, 3, 1); name="m")
n = Job(Thunk(f₆, 1; x=3); username="she", name="n")
i → l
j → k → m → n
j → l
k → n
wf = Workflow(k)
@test unique(wf) == collect(wf)
@test Set(wf) == Set([i, k, j, l, n, m])
run!(wf; wait=true)
@test Set(wf) == Set([i, k, j, l, n, m]) # Test they are still the same
for job in (i, j, k, l, n, m)
@test job in wf
end
@test issucceeded(wf)
@test something(getresult(i)) === nothing
@test something(getresult(j)) == 7.38905609893065
@test something(getresult(k)) === nothing
@test something(getresult(l)) isa Base.Process
@test something(getresult(m)) == 0.8414709848078965
@test something(getresult(n)) isa Base.ProcessChain
end
@testset "Test running a `Workflow` with `ConditionalJob`s" begin
f₁(x) = write("file", string(x))
f₂() = read("file", String)
h = Job(Thunk(sleep, 3); username="me", name="h")
i = Job(Thunk(f₁, 1001); username="me", name="i")
j = ConditionalJob(Thunk(map, f₂); username="he", name="j")
[h, i] .→ Ref(j)
wf = Workflow(j)
@test unique(wf) == collect(wf)
run!(wf; wait=true)
@test issucceeded(wf)
@test getresult(j) == Some("1001")
end
@testset "Test running a `Workflow` with `ArgDependentJob`s" begin
f₁(x) = x^2
f₂(y) = y + 1
f₃(z) = z / 2
i = Job(Thunk(f₁, 5); username="me", name="i")
j = ArgDependentJob(Thunk(f₂, 3); username="he", name="j")
k = ArgDependentJob(Thunk(f₃, 6); username="she", name="k")
i → j → k
wf = Workflow(k)
@test unique(wf) == collect(wf)
@test indexin([i, j, k], wf) == 1:3
run!(wf; wait=true)
for job in (i, j, k)
@test job in wf
end
@test issucceeded(wf)
@test getresult(i) == Some(25)
@test getresult(j) == Some(26)
@test getresult(k) == Some(13.0)
end
@testset "Test running a `Workflow` with a `ArgDependentJob` with more than one parent" begin
f₁(x) = x^2
f₂(y) = y + 1
f₃(z) = z / 2
f₄(iter) = sum(iter)
i = Job(Thunk(f₁, 5); username="me", name="i")
j = Job(Thunk(f₂, 3); username="he", name="j")
k = Job(Thunk(f₃, 6); username="she", name="k")
l = ArgDependentJob(Thunk(f₄, ()); username="she", name="me")
for job in (i, j, k)
job → l
end
wf = Workflow(k)
@test unique(wf) == collect(wf)
run!(wf; wait=true)
for job in (i, j, k)
@test job in wf
end
@test issucceeded(wf)
@test getresult(i) == Some(25)
@test getresult(j) == Some(4)
@test getresult(k) == Some(3.0)
@test getresult(l) == Some(32.0)
end
| SimpleWorkflows | https://github.com/MineralsCloud/SimpleWorkflows.jl.git |
|
[
"MIT"
] | 0.29.0 | 6653c51ac7bb1b0f10bf4ab5576f6f00fe7f4cf8 | code | 124 | using SimpleWorkflows
using Test
@testset "SimpleWorkflow.jl" begin
include("run.jl")
include("operations.jl")
end
| SimpleWorkflows | https://github.com/MineralsCloud/SimpleWorkflows.jl.git |
|
[
"MIT"
] | 0.29.0 | 6653c51ac7bb1b0f10bf4ab5576f6f00fe7f4cf8 | docs | 5215 | # Contributor Covenant Code of Conduct
## Our Pledge
We as members, contributors, and leaders pledge to make participation in our
community a harassment-free experience for everyone, regardless of age, body
size, visible or invisible disability, ethnicity, sex characteristics, gender
identity and expression, level of experience, education, socio-economic status,
nationality, personal appearance, race, religion, or sexual identity
and orientation.
We pledge to act and interact in ways that contribute to an open, welcoming,
diverse, inclusive, and healthy community.
## Our Standards
Examples of behavior that contributes to a positive environment for our
community include:
* Demonstrating empathy and kindness toward other people
* Being respectful of differing opinions, viewpoints, and experiences
* Giving and gracefully accepting constructive feedback
* Accepting responsibility and apologizing to those affected by our mistakes,
and learning from the experience
* Focusing on what is best not just for us as individuals, but for the
overall community
Examples of unacceptable behavior include:
* The use of sexualized language or imagery, and sexual attention or
advances of any kind
* Trolling, insulting or derogatory comments, and personal or political attacks
* Public or private harassment
* Publishing others' private information, such as a physical or email
address, without their explicit permission
* Other conduct which could reasonably be considered inappropriate in a
professional setting
## Enforcement Responsibilities
Community leaders are responsible for clarifying and enforcing our standards of
acceptable behavior and will take appropriate and fair corrective action in
response to any behavior that they deem inappropriate, threatening, offensive,
or harmful.
Community leaders have the right and responsibility to remove, edit, or reject
comments, commits, code, wiki edits, issues, and other contributions that are
not aligned to this Code of Conduct, and will communicate reasons for moderation
decisions when appropriate.
## Scope
This Code of Conduct applies within all community spaces, and also applies when
an individual is officially representing the community in public spaces.
Examples of representing our community include using an official e-mail address,
posting via an official social media account, or acting as an appointed
representative at an online or offline event.
## Enforcement
Instances of abusive, harassing, or otherwise unacceptable behavior may be
reported to the community leaders responsible for enforcement at
@singularitti.
All complaints will be reviewed and investigated promptly and fairly.
All community leaders are obligated to respect the privacy and security of the
reporter of any incident.
## Enforcement Guidelines
Community leaders will follow these Community Impact Guidelines in determining
the consequences for any action they deem in violation of this Code of Conduct:
### 1. Correction
**Community Impact**: Use of inappropriate language or other behavior deemed
unprofessional or unwelcome in the community.
**Consequence**: A private, written warning from community leaders, providing
clarity around the nature of the violation and an explanation of why the
behavior was inappropriate. A public apology may be requested.
### 2. Warning
**Community Impact**: A violation through a single incident or series
of actions.
**Consequence**: A warning with consequences for continued behavior. No
interaction with the people involved, including unsolicited interaction with
those enforcing the Code of Conduct, for a specified period of time. This
includes avoiding interactions in community spaces as well as external channels
like social media. Violating these terms may lead to a temporary or
permanent ban.
### 3. Temporary Ban
**Community Impact**: A serious violation of community standards, including
sustained inappropriate behavior.
**Consequence**: A temporary ban from any sort of interaction or public
communication with the community for a specified period of time. No public or
private interaction with the people involved, including unsolicited interaction
with those enforcing the Code of Conduct, is allowed during this period.
Violating these terms may lead to a permanent ban.
### 4. Permanent Ban
**Community Impact**: Demonstrating a pattern of violation of community
standards, including sustained inappropriate behavior, harassment of an
individual, or aggression toward or disparagement of classes of individuals.
**Consequence**: A permanent ban from any sort of public interaction within
the community.
## Attribution
This Code of Conduct is adapted from the [Contributor Covenant][homepage],
version 2.0, available at
https://www.contributor-covenant.org/version/2/0/code_of_conduct.html.
Community Impact Guidelines were inspired by [Mozilla's code of conduct
enforcement ladder](https://github.com/mozilla/diversity).
[homepage]: https://www.contributor-covenant.org
For answers to common questions about this code of conduct, see the FAQ at
https://www.contributor-covenant.org/faq. Translations are available at
https://www.contributor-covenant.org/translations.
| SimpleWorkflows | https://github.com/MineralsCloud/SimpleWorkflows.jl.git |
|
[
"MIT"
] | 0.29.0 | 6653c51ac7bb1b0f10bf4ab5576f6f00fe7f4cf8 | docs | 5468 | <div align="center">
<img src="https://raw.githubusercontent.com/MineralsCloud/SimpleWorkflows.jl/main/docs/src/assets/logo.png" height="200"><br>
</div>
# SimpleWorkflows
| **Documentation** | **Build Status** | **Others** |
| :--------------------------------------------------------------------------------: | :--------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------: | :---------------------------------------------------------------------------------------: |
| [![Stable][docs-stable-img]][docs-stable-url] [![Dev][docs-dev-img]][docs-dev-url] | [![Build Status][gha-img]][gha-url] [![Build Status][appveyor-img]][appveyor-url] [![Build Status][cirrus-img]][cirrus-url] [![pipeline status][gitlab-img]][gitlab-url] [![Coverage][codecov-img]][codecov-url] | [![GitHub license][license-img]][license-url] [![Code Style: Blue][style-img]][style-url] |
[docs-stable-img]: https://img.shields.io/badge/docs-stable-blue.svg
[docs-stable-url]: https://MineralsCloud.github.io/SimpleWorkflows.jl/stable
[docs-dev-img]: https://img.shields.io/badge/docs-dev-blue.svg
[docs-dev-url]: https://MineralsCloud.github.io/SimpleWorkflows.jl/dev
[gha-img]: https://github.com/MineralsCloud/SimpleWorkflows.jl/workflows/CI/badge.svg
[gha-url]: https://github.com/MineralsCloud/SimpleWorkflows.jl/actions
[appveyor-img]: https://ci.appveyor.com/api/projects/status/github/MineralsCloud/SimpleWorkflows.jl?svg=true
[appveyor-url]: https://ci.appveyor.com/project/singularitti/SimpleWorkflows-jl
[cirrus-img]: https://api.cirrus-ci.com/github/MineralsCloud/SimpleWorkflows.jl.svg
[cirrus-url]: https://cirrus-ci.com/github/MineralsCloud/SimpleWorkflows.jl
[gitlab-img]: https://gitlab.com/singularitti/SimpleWorkflows.jl/badges/main/pipeline.svg
[gitlab-url]: https://gitlab.com/singularitti/SimpleWorkflows.jl/-/pipelines
[codecov-img]: https://codecov.io/gh/MineralsCloud/SimpleWorkflows.jl/branch/main/graph/badge.svg
[codecov-url]: https://codecov.io/gh/MineralsCloud/SimpleWorkflows.jl
[license-img]: https://img.shields.io/github/license/MineralsCloud/SimpleWorkflows.jl
[license-url]: https://github.com/MineralsCloud/SimpleWorkflows.jl/blob/main/LICENSE
[style-img]: https://img.shields.io/badge/code%20style-blue-4495d1.svg
[style-url]: https://github.com/invenia/BlueStyle
The code, which is [hosted on GitHub](https://github.com/MineralsCloud/SimpleWorkflows.jl), is tested
using various continuous integration services for its validity.
This repository is created and maintained by
[@singularitti](https://github.com/singularitti), and contributions are highly welcome.
## Package features
Build workflows from jobs. Run, monitor, and get results from them.
This package takes inspiration from packages like
[JobSchedulers](https://github.com/cihga39871/JobSchedulers.jl) and
[Dispatcher](https://github.com/invenia/Dispatcher.jl) (unmaintained).
Please [cite this package](https://doi.org/10.1016/j.cpc.2022.108515) as:
Q. Zhang, C. Gu, J. Zhuang et al., `express`: extensible, high-level workflows for swifter *ab initio* materials modeling, *Computer Physics Communications*, 108515, doi: https://doi.org/10.1016/j.cpc.2022.108515.
The BibTeX format is:
```bibtex
@article{ZHANG2022108515,
title = {express: extensible, high-level workflows for swifter ab initio materials modeling},
journal = {Computer Physics Communications},
pages = {108515},
year = {2022},
issn = {0010-4655},
doi = {https://doi.org/10.1016/j.cpc.2022.108515},
url = {https://www.sciencedirect.com/science/article/pii/S001046552200234X},
author = {Qi Zhang and Chaoxuan Gu and Jingyi Zhuang and Renata M. Wentzcovitch},
keywords = {automation, workflow, high-level, high-throughput, data lineage}
}
```
We also have an [arXiv prepint](https://arxiv.org/abs/2109.11724).
## Installation
The package can be installed with the Julia package manager.
From [the Julia REPL](https://docs.julialang.org/en/v1/stdlib/REPL/), type `]` to enter
the [Pkg mode](https://docs.julialang.org/en/v1/stdlib/REPL/#Pkg-mode) and run:
```julia-repl
pkg> add SimpleWorkflows
```
Or, equivalently, via [`Pkg.jl`](https://pkgdocs.julialang.org/v1/):
```julia
julia> import Pkg; Pkg.add("SimpleWorkflows")
```
## Documentation
- [**STABLE**][docs-stable-url] — **documentation of the most recently tagged version.**
- [**DEV**][docs-dev-url] — _documentation of the in-development version._
## Project status
The package is developed for and tested against Julia `v1.6` and above on Linux, macOS, and
Windows.
## Questions and contributions
You can post usage questions on
[our discussion page](https://github.com/MineralsCloud/SimpleWorkflows.jl/discussions).
We welcome contributions, feature requests, and suggestions. If you encounter any problems,
please open an [issue](https://github.com/MineralsCloud/SimpleWorkflows.jl/issues).
The [Contributing](@ref) page has
a few guidelines that should be followed when opening pull requests and contributing code.
| SimpleWorkflows | https://github.com/MineralsCloud/SimpleWorkflows.jl.git |
|
[
"MIT"
] | 0.29.0 | 6653c51ac7bb1b0f10bf4ab5576f6f00fe7f4cf8 | docs | 631 | * **Please check if the PR fulfills these requirements**
- [ ] The commit message follows our guidelines
- [ ] Tests for the changes have been added (for bug fixes / features)
- [ ] Docs have been added / updated (for bug fixes / features)
* **What kind of change does this PR introduce?** (Bug fix, feature, docs update, ...)
* **What is the current behavior?** (You can also link to an open issue here)
* **What is the new behavior (if this is a feature change)?**
* **Does this PR introduce a breaking change?** (What changes might users need to make in their application due to this PR?)
* **Other information**:
| SimpleWorkflows | https://github.com/MineralsCloud/SimpleWorkflows.jl.git |
|
[
"MIT"
] | 0.29.0 | 6653c51ac7bb1b0f10bf4ab5576f6f00fe7f4cf8 | docs | 748 | ---
name: Bug report
about: Create a report to help us improve
title: ''
labels: bug
assignees: singularitti
---
**Describe the bug**
A clear and concise description of what the bug is.
**To Reproduce**
Steps to reproduce the behavior:
1. Go to '...'
2. Click on '....'
3. Scroll down to '....'
4. See error
5. Run code
```julia
using Pkg
```
**Expected behavior**
A clear and concise description of what you expected to happen.
**Screenshots**
If applicable, paste screenshots to help explain your problem.
**Desktop (please complete the following information):**
- OS: [e.g. macOS 12.5.1]
- Julia version: [e.g. 1.6.7, 1.7.3]
- Package version: [e.g. 2.0.0]
**Additional context**
Add any other context about the problem here.
| SimpleWorkflows | https://github.com/MineralsCloud/SimpleWorkflows.jl.git |
|
[
"MIT"
] | 0.29.0 | 6653c51ac7bb1b0f10bf4ab5576f6f00fe7f4cf8 | docs | 614 | ---
name: Feature request
about: Suggest an idea for this project
title: ''
labels: enhancement
assignees: singularitti
---
**Is your feature request related to a problem? Please describe.**
A clear and concise description of what the problem is. Ex. I'm always frustrated when [...]
**Describe the solution you'd like**
A clear and concise description of what you want to happen.
**Describe alternatives you've considered**
A clear and concise description of any alternative solutions or features you've considered.
**Additional context**
Add any other context or screenshots about the feature request here.
| SimpleWorkflows | https://github.com/MineralsCloud/SimpleWorkflows.jl.git |
|
[
"MIT"
] | 0.29.0 | 6653c51ac7bb1b0f10bf4ab5576f6f00fe7f4cf8 | docs | 3387 | # SimpleWorkflows
Documentation for [SimpleWorkflows](https://github.com/MineralsCloud/SimpleWorkflows.jl).
See the [Index](@ref main-index) for the complete list of documented functions
and types.
The code, which is [hosted on GitHub](https://github.com/MineralsCloud/SimpleWorkflows.jl), is tested
using various continuous integration services for its validity.
This repository is created and maintained by
[@singularitti](https://github.com/singularitti), and contributions are highly welcome.
## Package features
Build workflows from jobs. Run, monitor, and get results from them.
This package takes inspiration from packages like
[JobSchedulers](https://github.com/cihga39871/JobSchedulers.jl) and
[Dispatcher](https://github.com/invenia/Dispatcher.jl) (unmaintained).
Please [cite this package](https://doi.org/10.1016/j.cpc.2022.108515) as:
Q. Zhang, C. Gu, J. Zhuang et al., `express`: extensible, high-level workflows for swifter *ab initio* materials modeling, *Computer Physics Communications*, 108515, doi: https://doi.org/10.1016/j.cpc.2022.108515.
The BibTeX format is:
```bibtex
@article{ZHANG2022108515,
title = {express: extensible, high-level workflows for swifter ab initio materials modeling},
journal = {Computer Physics Communications},
pages = {108515},
year = {2022},
issn = {0010-4655},
doi = {https://doi.org/10.1016/j.cpc.2022.108515},
url = {https://www.sciencedirect.com/science/article/pii/S001046552200234X},
author = {Qi Zhang and Chaoxuan Gu and Jingyi Zhuang and Renata M. Wentzcovitch},
keywords = {automation, workflow, high-level, high-throughput, data lineage}
}
```
We also have an [arXiv prepint](https://arxiv.org/abs/2109.11724).
## Installation
The package can be installed with the Julia package manager.
From [the Julia REPL](https://docs.julialang.org/en/v1/stdlib/REPL/), type `]` to enter
the [Pkg mode](https://docs.julialang.org/en/v1/stdlib/REPL/#Pkg-mode) and run:
```julia-repl
pkg> add SimpleWorkflows
```
Or, equivalently, via [`Pkg.jl`](https://pkgdocs.julialang.org/v1/):
```@repl
import Pkg; Pkg.add("SimpleWorkflows")
```
## Documentation
- [**STABLE**](https://MineralsCloud.github.io/SimpleWorkflows.jl/stable) — **documentation of the most recently tagged version.**
- [**DEV**](https://MineralsCloud.github.io/SimpleWorkflows.jl/dev) — _documentation of the in-development version._
## Project status
The package is developed for and tested against Julia `v1.6` and above on Linux, macOS, and
Windows.
## Questions and contributions
You can post usage questions on
[our discussion page](https://github.com/MineralsCloud/SimpleWorkflows.jl/discussions).
We welcome contributions, feature requests, and suggestions. If you encounter any problems,
please open an [issue](https://github.com/MineralsCloud/SimpleWorkflows.jl/issues).
The [Contributing](@ref) page has
a few guidelines that should be followed when opening pull requests and contributing code.
## Manual outline
```@contents
Pages = [
"man/installation.md",
"man/portability.md",
"man/troubleshooting.md",
"developers/contributing.md",
"developers/style-guide.md",
"developers/design-principles.md",
]
Depth = 3
```
## Library outline
```@contents
Pages = ["lib/public.md", "lib/internals.md"]
```
### [Index](@id main-index)
```@index
Pages = ["lib/public.md"]
```
| SimpleWorkflows | https://github.com/MineralsCloud/SimpleWorkflows.jl.git |
|
[
"MIT"
] | 0.29.0 | 6653c51ac7bb1b0f10bf4ab5576f6f00fe7f4cf8 | docs | 8766 | # Contributing
```@contents
Pages = ["contributing.md"]
Depth = 2
```
Welcome! This document explains some ways you can contribute to SimpleWorkflows.
## Code of conduct
This project and everyone participating in it is governed by the
[Contributor Covenant Code of Conduct](https://github.com/MineralsCloud/.github/blob/main/CODE_OF_CONDUCT.md).
By participating, you are expected to uphold this code.
## Join the community forum
First up, join the [community forum](https://github.com/MineralsCloud/SimpleWorkflows.jl/discussions).
The forum is a good place to ask questions about how to use SimpleWorkflows. You can also
use the forum to discuss possible feature requests and bugs before raising a
GitHub issue (more on this below).
Aside from asking questions, the easiest way you can contribute to SimpleWorkflows is to
help answer questions on the forum!
## Improve the documentation
Chances are, if you asked (or answered) a question on the community forum, then
it is a sign that the [documentation](https://MineralsCloud.github.io/SimpleWorkflows.jl/dev/) could be
improved. Moreover, since it is your question, you are probably the best-placed
person to improve it!
The docs are written in Markdown and are built using
[`Documenter.jl`](https://github.com/JuliaDocs/Documenter.jl).
You can find the source of all the docs
[here](https://github.com/MineralsCloud/SimpleWorkflows.jl/tree/main/docs).
If your change is small (like fixing typos or one or two sentence corrections),
the easiest way to do this is via GitHub's online editor. (GitHub has
[help](https://help.github.com/articles/editing-files-in-another-user-s-repository/)
on how to do this.)
If your change is larger or touches multiple files, you will need to make the
change locally and then use Git to submit a
[pull request](https://docs.github.com/en/pull-requests/collaborating-with-pull-requests/proposing-changes-to-your-work-with-pull-requests/about-pull-requests).
(See [Contribute code to SimpleWorkflows](@ref) below for more on this.)
## File a bug report
Another way to contribute to SimpleWorkflows is to file
[bug reports](https://github.com/MineralsCloud/SimpleWorkflows.jl/issues/new?template=bug_report.md).
Make sure you read the info in the box where you write the body of the issue
before posting. You can also find a copy of that info
[here](https://github.com/MineralsCloud/SimpleWorkflows.jl/blob/main/.github/ISSUE_TEMPLATE/bug_report.md).
!!! tip
If you're unsure whether you have a real bug, post on the
[community forum](https://github.com/MineralsCloud/SimpleWorkflows.jl/discussions)
first. Someone will either help you fix the problem or let you know the
most appropriate place to open a bug report.
## Contribute code to SimpleWorkflows
Finally, you can also contribute code to SimpleWorkflows!
!!! warning
If you do not have experience with Git, GitHub, and Julia development, the
first steps can be a little daunting. However, there are lots of tutorials
available online, including:
- [GitHub](https://guides.github.com/activities/hello-world/)
- [Git and GitHub](https://try.github.io/)
- [Git](https://git-scm.com/book/en/v2)
- [Julia package development](https://docs.julialang.org/en/v1/stdlib/Pkg/#Developing-packages-1)
Once you are familiar with Git and GitHub, the workflow for contributing code to
SimpleWorkflows is similar to the following:
### Step 1: decide what to work on
The first step is to find an [open issue](https://github.com/MineralsCloud/SimpleWorkflows.jl/issues)
(or open a new one) for the problem you want to solve. Then, _before_ spending
too much time on it, discuss what you are planning to do in the issue to see if
other contributors are fine with your proposed changes. Getting feedback early can
improve code quality and avoid time spent writing code that does not get merged into
SimpleWorkflows.
!!! tip
At this point, remember to be patient and polite; you may get a _lot_ of
comments on your issue! However, do not be afraid! Comments mean that people are
willing to help you improve the code that you are contributing to SimpleWorkflows.
### Step 2: fork SimpleWorkflows
Go to [https://github.com/MineralsCloud/SimpleWorkflows.jl](https://github.com/MineralsCloud/SimpleWorkflows.jl)
and click the "Fork" button in the top-right corner. This will create a copy of
SimpleWorkflows under your GitHub account.
### Step 3: install SimpleWorkflows locally
Similar to [Installation Guide](@ref), open the Julia REPL and run:
```@repl
using Pkg
Pkg.update()
Pkg.develop("SimpleWorkflows")
```
Then the package will be cloned to your local machine. On *nix systems, the default path is
`~/.julia/dev/SimpleWorkflows` unless you modify the
[`JULIA_DEPOT_PATH`](http://docs.julialang.org/en/v1/manual/environment-variables/#JULIA_DEPOT_PATH-1)
environment variable. If you're on
Windows, this will be `C:\\Users\\<my_name>\\.julia\\dev\\SimpleWorkflows`.
In the following text, we will call it `PKGROOT`.
Go to `PKGROOT`, start a new Julia session, and run
```@repl
using Pkg
Pkg.instantiate()
```
to instantiate the project.
### Step 4: checkout a new branch
!!! note
In the following, replace any instance of `GITHUB_ACCOUNT` with your GitHub
username.
The next step is to check out a development branch. In a terminal (or command
prompt on Windows), run:
```bash
$ cd ~/.julia/dev/SimpleWorkflows
$ git remote add GITHUB_ACCOUNT https://github.com/GITHUB_ACCOUNT/SimpleWorkflows.jl.git
$ git checkout main
$ git pull
$ git checkout -b my_new_branch
```
### Step 5: make changes
Now make any changes to the source code inside the `~/.julia/dev/SimpleWorkflows`
directory.
Make sure you:
- Follow our [Style Guide](@ref) and [Run JuliaFormatter](@ref).
- Add tests and documentation for any changes or new features.
!!! tip
When you change the source code, you will need to restart Julia for the
changes to take effect. If this is a pain, install
[`Revise.jl`](https://github.com/timholy/Revise.jl).
### Step 6a: test your code changes
To test that your changes work, run the SimpleWorkflows test-suite by opening Julia and
running:
```julia-repl
julia> cd(joinpath(DEPOT_PATH[1], "dev", "SimpleWorkflows"))
julia> using Pkg
julia> Pkg.activate(".")
Activating new project at `~/.julia/dev/SimpleWorkflows`
julia> Pkg.test()
```
!!! warning
Running the tests might take a long time.
!!! tip
If you are using `Revise.jl`, you can also run the tests by calling `include`:
```julia-repl
include("test/runtests.jl")
```
This can be faster if you want to re-run the tests multiple times.
### Step 6b: test your documentation changes
Open Julia, then run:
```julia-repl
julia> cd(joinpath(DEPOT_PATH[1], "dev", "SimpleWorkflows", "docs"))
julia> using Pkg
julia> Pkg.activate(".")
Activating new project at `~/.julia/dev/SimpleWorkflows/docs`
julia> include("src/make.jl")
```
After a while, a folder `PKGROOT/docs/build` will appear. Open
`PKGROOT/docs/build/index.html` with your favorite browser, and have fun!
!!! warning
Building the documentation might take a long time.
!!! tip
If there's a problem with the tests that you don't know how to fix, don't
worry. Continue to step 5, and one of the SimpleWorkflows contributors will comment
on your pull request, telling you how to fix things.
### Step 7: make a pull request
Once you've made changes, you're ready to push the changes to GitHub. Run:
```bash
$ cd ~/.julia/dev/SimpleWorkflows
$ git add .
$ git commit -m "A descriptive message of the changes"
$ git push -u GITHUB_ACCOUNT my_new_branch
```
Then go to [our pull request page](https://github.com/MineralsCloud/SimpleWorkflows.jl/pulls)
and follow the instructions that pop up to open a pull request.
### Step 8: respond to comments
At this point, remember to be patient and polite; you may get a _lot_ of
comments on your pull request! However, do not be afraid! A lot of comments
means that people are willing to help you improve the code that you are
contributing to SimpleWorkflows.
To respond to the comments, go back to step 5, make any changes, test the
changes in step 6, and then make a new commit in step 7. Your PR will
automatically update.
### Step 9: cleaning up
Once the PR is merged, clean-up your Git repository, ready for the
next contribution!
```bash
$ cd ~/.julia/dev/SimpleWorkflows
$ git checkout main
$ git pull
```
!!! note
If you have suggestions to improve this guide, please make a pull request!
It's particularly helpful if you do this after your first pull request
because you'll know all the parts that could be explained better.
Thanks for contributing to SimpleWorkflows!
| SimpleWorkflows | https://github.com/MineralsCloud/SimpleWorkflows.jl.git |
|
[
"MIT"
] | 0.29.0 | 6653c51ac7bb1b0f10bf4ab5576f6f00fe7f4cf8 | docs | 15417 | # Design Principles
```@contents
Pages = ["design-principles.md"]
Depth = 2
```
We adopt some [SciML](https://sciml.ai/) design [guidelines](https://github.com/SciML/SciMLStyle)
here. Please read them before contributing!
## Consistency vs adherence
According to PEP8:
> A style guide is about consistency. Consistency with this style guide is important.
> Consistency within a project is more important. Consistency within one module or function is the most important.
>
> However, know when to be inconsistent—sometimes style guide recommendations just aren't
> applicable. When in doubt, use your best judgment. Look at other examples and decide what
> looks best. And don’t hesitate to ask!
## Community contribution guidelines
For a comprehensive set of community contribution guidelines, refer to [ColPrac](https://github.com/SciML/ColPrac).
A relevant point to highlight is that one PR should do one thing. In the context of style, this means that PRs which update
the style of a package's code should not be mixed with fundamental code contributions. This separation makes it
easier to ensure that large style improvement are isolated from substantive (and potentially breaking) code changes.
## Open source contributions are allowed to start small and grow over time
If the standard for code contributions is that every PR needs to support every possible input type that anyone can
think of, the barrier would be too high for newcomers. Instead, the principle is to be as correct as possible to
begin with, and grow the generic support over time. All recommended functionality should be tested, any known
generality issues should be documented in an issue (and with a `@test_broken` test when possible).
## Generic code is preferred unless code is known to be specific
For example, the code:
```@repl
function f(A, B)
for i in 1:length(A)
A[i] = A[i] + B[i]
end
end
```
would not be preferred for two reasons. One is that it assumes `A` uses one-based indexing, which would fail in cases
like [`OffsetArrays.jl`](https://github.com/JuliaArrays/OffsetArrays.jl) and [`FFTViews.jl`](https://github.com/JuliaArrays/FFTViews.jl).
Another issue is that it requires indexing, while not all array types support indexing (for example,
[`CuArrays.jl`](https://github.com/JuliaGPU/CuArrays.jl)). A more generic compatible implementation of this function would be
to use broadcast, for example:
```@repl
function f(A, B)
@. A = A + B
end
```
which would allow support for a wider variety of array types.
## Internal types should match the types used by users when possible
If `f(A)` takes the input of some collections and computes an output from those collections, then it should be
expected that if the user gives `A` as an `Array`, the computation should be done via `Array`s. If `A` was a
`CuArray`, then it should be expected that the computation should be internally done using a `CuArray` (or appropriately
error if not supported). For these reasons, constructing arrays via generic methods, like `similar(A)`, is preferred when
writing `f` instead of using non-generic constructors like `Array(undef,size(A))` unless the function is documented as
being non-generic.
## Trait definition and adherence to generic interface is preferred when possible
Julia provides many interfaces, for example:
- [Iteration](https://docs.julialang.org/en/v1/manual/interfaces/#man-interface-iteration)
- [Indexing](https://docs.julialang.org/en/v1/manual/interfaces/#Indexing)
- [Broadcasting](https://docs.julialang.org/en/v1/manual/interfaces/#man-interfaces-broadcasting)
Those interfaces should be followed when possible. For example, when defining broadcast overloads,
one should implement a `BroadcastStyle` as suggested by the documentation instead of simply attempting
to bypass the broadcast system via `copyto!` overloads.
When interface functions are missing, these should be added to an interface package,
like [`ArrayInterface.jl`](https://github.com/JuliaArrays/ArrayInterface.jl). Such traits should be
declared and used when appropriate. For example, if a line of code requires mutation, the trait
`ArrayInterface.ismutable(A)` should be checked before attempting to mutate, and informative error
messages should be written to capture the immutable case (or, an alternative code which does not
mutate should be given).
One example of this principle is demonstrated in the generation of Jacobian matrices. In many scientific
applications, one may wish to generate a Jacobian cache from the user's input `u0`. A naive way to generate
this Jacobian is `J = similar(u0,length(u0),length(u0))`. However, this will generate a Jacobian `J` such
that `J isa Matrix`.
## Macros should be limited and only be used for syntactic sugar
Macros define new syntax, and for this reason they tend to be less composable than other coding styles
and require prior familiarity to be easily understood. One principle to keep in mind is, "can the person
reading the code easily picture what code is being generated?". For example, a user of
[`Soss.jl`](https://github.com/cscherrer/Soss.jl) may not know what code is being generated by:
```julia
@model (x, α) begin
σ ~ Exponential()
β ~ Normal()
y ~ For(x) do xj
Normal(α + β * xj, σ)
end
return y
end
```
and thus using such a macro as the interface is not preferred when possible. However, a macro like
[`@muladd`](https://github.com/SciML/MuladdMacro.jl) is trivial to picture on a code (it recursively
transforms `a*b + c` to `muladd(a,b,c)` for more
[accuracy and efficiency](https://en.wikipedia.org/wiki/Multiply-accumulate_operation)), so using
such a macro for example:
```julia-repl
julia> @macroexpand(@muladd k3 = f(t + c3 * dt, @. uprev + dt * (a031 * k1 + a032 * k2)))
:(k3 = f((muladd)(c3, dt, t), (muladd).(dt, (muladd).(a032, k2, (*).(a031, k1)), uprev)))
```
is recommended. Some macros in this category are:
- `@inbounds`
- [`@muladd`](https://github.com/SciML/MuladdMacro.jl)
- `@view`
- [`@named`](https://github.com/SciML/ModelingToolkit.jl)
- `@.`
- [`@..`](https://github.com/YingboMa/FastBroadcast.jl)
Some performance macros, like `@simd`, `@threads`, or
[`@turbo` from `LoopVectorization.jl`](https://github.com/JuliaSIMD/LoopVectorization.jl),
make an exception in that their generated code may be foreign to many users. However, they still are
classified as appropriate uses as they are syntactic sugar since they do (or should) not change the behavior
of the program in measurable ways other than performance.
## Errors should be caught as early as possible, and error messages should be made contextually clear for newcomers
Whenever possible, defensive programming should be used to check for potential errors before they are encountered
deeper within a package. For example, if one knows that `f(u0,p)` will error unless `u0` is the size of `p`, this
should be caught at the start of the function to throw a domain specific error, for example "parameters and initial
condition should be the same size".
## Subpackaging and interface packages is preferred over conditional modules via Requires.jl
`Requires.jl` should be avoided at all costs. If an interface package exists, such as
[`ChainRulesCore.jl`](https://github.com/JuliaDiff/ChainRulesCore.jl) for defining automatic differentiation
rules without requiring a dependency on the whole `ChainRules.jl` system, or
[`RecipesBase.jl`](https://github.com/JuliaPlots/RecipesBase.jl) which allows for defining `Plots.jl`
plot recipes without a dependency on `Plots.jl`, a direct dependency on these interface packages is
preferred.
Otherwise, instead of resorting to a conditional dependency using `Requires.jl`, it is
preferred one creates subpackages, i.e. smaller independent packages kept within the same GitHub repository
with independent versioning and package management. An example of this is seen in
[`Optimization.jl`](https://github.com/SciML/Optimization.jl) which has subpackages like
[`OptimizationBBO.jl`](https://github.com/SciML/Optimization.jl/tree/master/lib/OptimizationBBO) for
`BlackBoxOptim.jl` support.
Some important interface packages to be aware of include:
- [`ChainRulesCore.jl`](https://github.com/JuliaDiff/ChainRulesCore.jl)
- [`RecipesBase.jl`](https://github.com/JuliaPlots/RecipesBase.jl)
- [`ArrayInterface.jl`](https://github.com/JuliaArrays/ArrayInterface.jl)
- [`CommonSolve.jl`](https://github.com/SciML/CommonSolve.jl)
- [`SciMLBase.jl`](https://github.com/SciML/SciMLBase.jl)
## Functions should either attempt to be non-allocating and reuse caches, or treat inputs as immutable
Mutating codes and non-mutating codes fall into different worlds. When a code is fully immutable,
the compiler can better reason about dependencies, optimize the code, and check for correctness.
However, many times a code making the fullest use of mutation can outperform even what the best compilers
of today can generate. That said, the worst of all worlds is when code mixes mutation with non-mutating
code. Not only is this a mishmash of coding styles, it has the potential non-locality and compiler
proof issues of mutating code while not fully benefiting from the mutation.
## Out-of-place and immutability is preferred when sufficient performant
Mutation is used to get more performance by decreasing the amount of heap allocations. However,
if it's not helpful for heap allocations in a given spot, do not use mutation. Mutation is scary
and should be avoided unless it gives an immediate benefit. For example, if
matrices are sufficiently large, then `A*B` is as fast as `mul!(C,A,B)`, and thus writing
`A*B` is preferred (unless the rest of the function is being careful about being fully non-allocating,
in which case this should be `mul!` for consistency).
Similarly, when defining types, using `struct` is preferred to `mutable struct` unless mutating
the `struct` is a common occurrence. Even if mutating the `struct` is a common occurrence, see whether
using [`Setfield.jl`](https://github.com/jw3126/Setfield.jl) is sufficient. The compiler will optimize
the construction of immutable `struct`s, and thus this can be more efficient if it's not too much of a
code hassle.
## Tests should attempt to cover a wide gamut of input types
Code coverage numbers are meaningless if one does not consider the input types. For example, one can
hit all the code with `Array`, but that does not test whether `CuArray` is compatible! Thus, it's
always good to think of coverage not in terms of lines of code but in terms of type coverage. A good
list of number types to think about are:
- `Float64`
- `Float32`
- `Complex`
- [`Dual`](https://github.com/JuliaDiff/ForwardDiff.jl)
- `BigFloat`
Array types to think about testing are:
- `Array`
- [`OffsetArray`](https://github.com/JuliaArrays/OffsetArrays.jl)
- [`CuArray`](https://github.com/JuliaGPU/CUDA.jl)
## When in doubt, a submodule should become a subpackage or separate package
Each package should focus on one core idea. If there's something separate enough to be a submodule, could it
instead be a separate well-tested and documented package to be used by other packages? Most likely
yes.
## Globals should be avoided whenever possible
Global variables should be avoided whenever possible. When required, global variables should be
constants and have an all uppercase name separated with underscores (e.g. `MY_CONSTANT`). They should be
defined at the top of the file, immediately after imports and exports but before an `__init__` function.
If you truly want mutable global style behavior you may want to look into mutable containers.
## Type-stable and type-grounded code is preferred wherever possible
Type-stable and type-grounded code helps the compiler create not only more optimized code, but also
faster to compile code. Always keep containers well-typed, functions specializing on the appropriate
arguments, and types concrete.
## Closures should be avoided whenever possible
Closures can cause accidental type instabilities that are difficult to track down and debug; in the
long run it saves time to always program defensively and avoid writing closures in the first place,
even when a particular closure would not have been problematic. A similar argument applies to reading
code with closures; if someone is looking for type instabilities, this is faster to do when code does
not contain closures.
See examples [here](https://discourse.julialang.org/t/are-closures-should-be-avoided-whenever-possible-still-valid-in-julia-v1-9/95893/5).
Furthermore, if you want to update variables in an outer scope, do so explicitly with `Ref`s or self
defined `struct`s.
## Numerical functionality should use the appropriate generic numerical interfaces
While you can use `A\b` to do a linear solve inside a package, that does not mean that you should.
This interface is only sufficient for performing factorizations, and so that limits the scaling
choices, the types of `A` that can be supported, etc. Instead, linear solves within packages should
use `LinearSolve.jl`. Similarly, nonlinear solves should use `NonlinearSolve.jl`. Optimization should use
`Optimization.jl`. Etc. This allows the full generic choice to be given to the user without depending
on every solver package (effectively recreating the generic interfaces within each package).
## Functions should capture one underlying principle
Functions mean one thing. Every dispatch of `+` should be "the meaning of addition on these types".
While in theory you could add dispatches to `+` that mean something different, that will fail in
generic code for which `+` means addition. Thus, for generic code to work, code needs to adhere to
one meaning for each function. Every dispatch should be an instantiation of that meaning.
## Internal choices should be exposed as options whenever possible
Whenever possible, numerical values and choices within scripts should be exposed as options
to the user. This promotes code reusability beyond the few cases the author may have expected.
## Prefer code reuse over rewrites whenever possible
If a package has a function you need, use the package. Add a dependency if you need to. If the
function is missing a feature, prefer to add that feature to said package and then add it as a
dependency. If the dependency is potentially troublesome, for example because it has a high
load time, prefer to spend time helping said package fix these issues and add the dependency.
Only when it does not seem possible to make the package "good enough" should using the package
be abandoned. If it is abandoned, consider building a new package for this functionality as you
need it, and then make it a dependency.
## Prefer to not shadow functions
In Julia, two functions can share the same name if they belong to different namespaces. For example,
`X.f` and `Y.f` can be two different functions, with different dispatches, but the same name.
This should be avoided whenever possible. Instead of creating `MyPackage.sort`, consider
adding dispatches to `Base.sort` for your types if these new dispatches match the underlying
principle of the function. If they don't, it would be preferable to use a different name. While using `MyPackage.sort`
is not conflicting, it is going to be confusing for most people unfamiliar with your code,
so `MyPackage.special_sort` would be more helpful to newcomers reading the code.
| SimpleWorkflows | https://github.com/MineralsCloud/SimpleWorkflows.jl.git |
|
[
"MIT"
] | 0.29.0 | 6653c51ac7bb1b0f10bf4ab5576f6f00fe7f4cf8 | docs | 2309 | # Style Guide
This section describes the coding style rules that apply to our code and that
we recommend you to use it also.
In some cases, our style guide diverges from Julia's official
[Style Guide](https://docs.julialang.org/en/v1/manual/style-guide/) (Please read it!).
All such cases will be explicitly noted and justified.
Our style guide adopts many recommendations from the
[BlueStyle](https://github.com/invenia/BlueStyle).
Please read the [BlueStyle](https://github.com/invenia/BlueStyle)
before contributing to this package.
If these guidelines are not followed, your pull requests may not be accepted.
!!! info
The style guide is always a work in progress, and not all SimpleWorkflows code
follows the rules. When modifying SimpleWorkflows, please fix the style violations
of the surrounding code (i.e., leave the code tidier than when you
started). If large changes are needed, consider separating them into
another pull request.
## Formatting
### Run JuliaFormatter
SimpleWorkflows uses [JuliaFormatter](https://github.com/domluna/JuliaFormatter.jl) as
an auto-formatting tool.
We use the options contained in [`.JuliaFormatter.toml`](https://github.com/MineralsCloud/SimpleWorkflows.jl/blob/main/.JuliaFormatter.toml).
To format your code, `cd` to the SimpleWorkflows directory, then run:
```julia-repl
julia> using Pkg
julia> Pkg.add("JuliaFormatter")
julia> using JuliaFormatter: format
julia> format("docs"); format("src"); format("test")
```
!!! info
A continuous integration check verifies that all PRs made to SimpleWorkflows have
passed the formatter.
The following sections outline extra style guide points that are not fixed
automatically by JuliaFormatter.
### Use the Julia extension for Visual Studio Code
Please use [Visual Studio Code](https://code.visualstudio.com/) with the
[Julia extension](https://marketplace.visualstudio.com/items?itemName=julialang.language-julia)
to edit, format, and test your code.
For the time being, we do not recommend using editors other than Visual Studio Code to edit your code.
This extension already has [JuliaFormatter](https://github.com/domluna/JuliaFormatter.jl)
integrated. So to format your code, follow the steps listed
[here](https://www.julia-vscode.org/docs/stable/userguide/formatter/).
| SimpleWorkflows | https://github.com/MineralsCloud/SimpleWorkflows.jl.git |
|
[
"MIT"
] | 0.29.0 | 6653c51ac7bb1b0f10bf4ab5576f6f00fe7f4cf8 | docs | 322 | # Public API
## Contents
```@contents
Pages = ["public.md"]
Depth = 2
```
## Index
```@index
Pages = ["public.md"]
```
## Public interface
```@docs
Workflow
run!
execute!
getstatus
eachstatus
ispending
isrunning
isexited
issucceeded
isfailed
filterpending
filterrunning
filterexited
filtersucceeded
filterfailed
```
| SimpleWorkflows | https://github.com/MineralsCloud/SimpleWorkflows.jl.git |
|
[
"MIT"
] | 0.29.0 | 6653c51ac7bb1b0f10bf4ab5576f6f00fe7f4cf8 | docs | 5303 | # Installation Guide
```@contents
Pages = ["installation.md"]
Depth = 2
```
Here are the installation instructions for package
[SimpleWorkflows](https://github.com/MineralsCloud/SimpleWorkflows.jl).
If you have trouble installing it, please refer to our [Troubleshooting](@ref) page
for more information.
## Install Julia
First, you should install [Julia](https://julialang.org/). We recommend downloading it from
[its official website](https://julialang.org/downloads/). Please follow the detailed
instructions on its website if you have to
[build Julia from source](https://docs.julialang.org/en/v1/devdocs/build/build/).
Some computing centers provide preinstalled Julia. Please contact your administrator for
more information in that case.
Here's some additional information on
[how to set up Julia on HPC clusters](https://juliahpc.github.io/JuliaOnHPCClusters/).
If you have [Homebrew](https://brew.sh/) installed,
[open the Terminal app](https://support.apple.com/guide/terminal/open-or-quit-terminal-apd5265185d-f365-44cb-8b09-71a064a42125/mac)
and type
```bash
brew install julia
```
to install it as a [formula](https://docs.brew.sh/Formula-Cookbook).
If you are also using [macOS](https://en.wikipedia.org/wiki/MacOS) and want to install it as
a prebuilt binary app, type
```bash
brew install --cask julia
```
instead.
If you want to install multiple Julia versions in the same operating system,
a recommended way is to use a version manager such as
[Juliaup](https://github.com/JuliaLang/juliaup).
First, [install Juliaup](https://github.com/JuliaLang/juliaup#installation).
Then, run
```bash
juliaup add release
juliaup default release
```
to configure the `julia` command to start the latest stable version of
Julia (this is also the default value).
Here is a [short video introduction to Juliaup](https://youtu.be/14zfdbzq5BM)
made by its authors.
### Which version should I pick?
You can install the current stable release or the long-term support (LTS) release.
- The current stable release is the latest release of Julia. It has access to
newer features, and is likely faster.
- The long-term support release is an older version of Julia that has
continued to receive bug and security fixes. However, it may not have the
latest features or performance improvements.
For most users, you should install the current stable release, and whenever
Julia releases a new version of the current stable release, you should update
your version of Julia. Note that any code you write on one version of the
current stable release will continue to work on all subsequent releases.
For users in restricted software environments (e.g., your enterprise IT controls
what software you can install), you may be better off installing the long-term
support release because you will not have to update Julia as frequently.
Versions above `v1.3`, especially the latest stable ones, are strongly recommended.
This package is highly unlikely to work on `v1.0` and earlier versions.
Since the Julia team has set `v1.6` as the LTS release,
we will gradually drop support for versions below `v1.6`.
Julia and Julia packages support multiple operating systems and CPU architectures; check
[this table](https://julialang.org/downloads/#supported_platforms) to see if it can be
installed on your machine. For Mac computers with M-series processors, this package and its
dependencies may not work. Please install the Intel-compatible version of Julia (for macOS
x86-64) if any platform-related error occurs.
## Install the package
Now I am using macOS as a standard platform to explain the following steps:
1. Open the Terminal app, and type `julia` to start an interactive session (known as the
[REPL](https://docs.julialang.org/en/v1/stdlib/REPL/)).
2. Run the following commands and wait for them to finish:
```julia-repl
julia> using Pkg
julia> Pkg.update()
julia> Pkg.add("SimpleWorkflows")
```
3. Run
```julia-repl
julia> using SimpleWorkflows
```
and have fun!
4. Please keep the Julia session active while using it. Restarting the session may take some time.
If you want to install the latest in-development (probably buggy)
version of SimpleWorkflows, type
```@repl
using Pkg
Pkg.update()
pkg"add https://github.com/MineralsCloud/SimpleWorkflows.jl"
```
in the second step above.
## Update the package
Please [watch](https://docs.github.com/en/account-and-profile/managing-subscriptions-and-notifications-on-github/setting-up-notifications/configuring-notifications#configuring-your-watch-settings-for-an-individual-repository)
our [GitHub repository](https://github.com/MineralsCloud/SimpleWorkflows.jl)
for new releases.
Once we release a new version, you can update SimpleWorkflows by typing
```@repl
using Pkg
Pkg.update("SimpleWorkflows")
Pkg.gc()
```
in the Julia REPL.
## Uninstall and then reinstall the package
Sometimes errors may occur if the package is not properly installed.
In this case, you may want to uninstall and reinstall the package. Here is how to do that:
1. To uninstall, in a Julia session, run
```julia-repl
julia> using Pkg
julia> Pkg.rm("SimpleWorkflows")
julia> Pkg.gc()
```
2. Press `Ctrl+D` to quit the current session. Start a new Julia session and
reinstall SimpleWorkflows.
| SimpleWorkflows | https://github.com/MineralsCloud/SimpleWorkflows.jl.git |
|
[
"MIT"
] | 0.29.0 | 6653c51ac7bb1b0f10bf4ab5576f6f00fe7f4cf8 | docs | 1548 | # How to save and recover the status of a workflow?
Suppose you have a `Workflow` object defined with the following code:
```@repl wf
using SimpleWorkflows.Thunks: Thunk
using SimpleWorkflows: Job, Workflow, run!, →
function f₁()
println("Start job `i`!")
sleep(5)
end
function f₂(n)
println("Start job `j`!")
sleep(n)
exp(2)
end
function f₃(n)
println("Start job `k`!")
sleep(n)
end
function f₄()
println("Start job `l`!")
run(`sleep 3`)
end
function f₅(n, x)
println("Start job `m`!")
sleep(n)
sin(x)
end
function f₆(n; x = 1)
println("Start job `n`!")
sleep(n)
cos(x)
run(`pwd` & `ls`)
end
i = Job(Thunk(f₁, ()); user = "me", desc = "i")
j = Job(Thunk(f₂, 3); user = "he", desc = "j")
k = Job(Thunk(f₃, 6); desc = "k")
l = Job(Thunk(f₄, ()); desc = "l", user = "me")
m = Job(Thunk(f₅, 3, 1); desc = "m")
n = Job(Thunk(f₆, 1; x = 3); user = "she", desc = "n")
i → l
j → k → m → n
j → l
k → n
wf = Workflow(k)
```
To save the `Workflow` instance to disk while running in case it failed or is interrupted,
use the `serialize` function.
```@repl wf
using Serialization: serialize
wf = serialize("wf.jls", wf)
run!(wf)
```
After the above steps are finished, a `saved.jls` file is saved to your local file system.
Then you can close the current Julia session and restart it (which resembles an
interrupted remote session, for example).
To reload the workflow, run:
```julia-repl
julia> using SimpleWorkflows
julia> using Serialization: deserialize
julia> deserialize("wf.jls")
```
And voilà!
| SimpleWorkflows | https://github.com/MineralsCloud/SimpleWorkflows.jl.git |
|
[
"MIT"
] | 0.29.0 | 6653c51ac7bb1b0f10bf4ab5576f6f00fe7f4cf8 | docs | 2095 | # Troubleshooting
```@contents
Pages = ["troubleshooting.md"]
Depth = 2
```
This page collects some possible errors you may encounter along with tips on how to fix them.
If you have some questions about how to use this code, you are welcome to
[discuss with us](https://github.com/MineralsCloud/SimpleWorkflows.jl/discussions).
If you have additional tips, please either
[report an issue](https://github.com/MineralsCloud/SimpleWorkflows.jl/issues/new) or
[submit a pull request](https://github.com/MineralsCloud/SimpleWorkflows.jl/compare) with suggestions.
## Cannot find the Julia executable
Make sure you have Julia installed in your environment. Please download the latest
[stable version](https://julialang.org/downloads/#current_stable_release) for your platform.
If you are using a *nix system, the recommended way is to use
[Juliaup](https://github.com/JuliaLang/juliaup). If you do not want to install Juliaup
or you are using other platforms that Julia supports, download the corresponding binaries.
Then, create a symbolic link to the Julia executable.
If the path is not in your [`$PATH` environment variable](https://en.wikipedia.org/wiki/PATH_(variable)),
export it to your `$PATH`.
Some clusters, like
[Comet](https://www.sdsc.edu/support/user_guides/comet.html),
or [Expanse](https://www.sdsc.edu/services/hpc/expanse/index.html),
already have Julia installed as a module, you may
just `module load julia` to use it. If not, either install by yourself or contact your
administrator.
See [Installation Guide](@ref) for more information.
## Julia starts slow
First, we recommend you download the latest version of Julia. Usually, the newest version
has the best performance.
If you need to use Julia for a simple, one-time task, you can start the Julia REPL with
```bash
julia --compile=min
```
to minimize compilation or
```bash
julia --optimize=0
```
to minimize optimizations, or just use both. Or you could make a system image
and run with
```bash
julia --sysimage custom-image.so
```
See [Fredrik Ekre's talk](https://youtu.be/IuwxE3m0_QQ?t=313) for details.
| SimpleWorkflows | https://github.com/MineralsCloud/SimpleWorkflows.jl.git |
|
[
"MIT"
] | 0.2.3 | 16d2a15e95e265b590603f62c33612fd64c963c2 | code | 771 | module GlobalApproximationValueIteration
using Base.Iterators: repeated
# Stdlib imports
using LinearAlgebra
using Random
using Printf
# For function approximation
using MultivariateStats
using Flux
# POMDPs imports
using POMDPs
using POMDPTools
using POMDPLinter: @POMDP_require, @warn_requirements
export
GlobalFunctionApproximator,
fit!,
compute_value,
LinearGlobalFunctionApproximator,
NonlinearGlobalFunctionApproximator
export
GlobalApproximationValueIterationSolver,
GlobalApproximationValueIterationPolicy,
convert_featurevector,
sample_state
function sample_state end
include("global_function_approximation.jl")
include("linear_gfa.jl")
include("nonlinear_gfa.jl")
include("global_approximation_vi.jl")
end # module
| GlobalApproximationValueIteration | https://github.com/JuliaPOMDP/GlobalApproximationValueIteration.jl.git |
|
[
"MIT"
] | 0.2.3 | 16d2a15e95e265b590603f62c33612fd64c963c2 | code | 8744 | mutable struct GlobalApproximationValueIterationSolver{GFA <: GlobalFunctionApproximator, RNG <: AbstractRNG, F} <: Solver
gfa::GFA
num_samples::Int64
belres::Float64
num_iterations::Int64
verbose::Bool
rng::RNG
is_mdp_generative::Bool
n_generative_samples::Int64
fv_type::F
end
function GlobalApproximationValueIterationSolver(gfa::GFA; num_samples::Int64, belres::Float64=1e-3,
max_iterations::Int64=1000, verbose::Bool=false,
rng::RNG=Random.GLOBAL_RNG, is_mdp_generative::Bool=false,
n_generative_samples::Int64=0, fv_type::F=Vector{Float64}) where {GFA <: GlobalFunctionApproximator, F, RNG <: AbstractRNG}
return GlobalApproximationValueIterationSolver(gfa, num_samples, belres, max_iterations, verbose, rng, is_mdp_generative, n_generative_samples, fv_type)
end
function GlobalApproximationValueIterationSolver()
throw(ArgumentError("GlobalApproximationValueIterationSolver needs a GlobalFunctionApproximator object for construction!"))
end
mutable struct GlobalApproximationValueIterationPolicy{GFA <: GlobalFunctionApproximator, RNG <: AbstractRNG, F} <: Policy
gfa::GFA
action_map::Vector
mdp::Union{MDP,POMDP}
is_mdp_generative::Bool
n_generative_samples::Int64
fv_type::F
rng::RNG
end
function GlobalApproximationValueIterationPolicy(mdp::Union{MDP,POMDP},
solver::GlobalApproximationValueIterationSolver)
return GlobalApproximationValueIterationPolicy(deepcopy(solver.gfa), ordered_actions(mdp), mdp,
solver.is_mdp_generative, solver.n_generative_samples, solver.fv_type, solver.rng)
end
function convert_featurevector(::Type{V}, s::S, mdp::Union{MDP,POMDP}) where {V <: AbstractArray, S}
return convert_s(V, s, mdp)
end
# If global function approximator is non-linear, a default convert_s is required
@POMDP_require convert_featurevector(t::Type{V} where {V <: AbstractArray}, s::S where S, mdp::Union{MDP,POMDP}) begin
@req convert_s(::Type{V} where {V <: AbstractArray}, ::S, ::typeof(mdp))
end
@POMDP_require solve(solver::GlobalApproximationValueIterationSolver, mdp::Union{MDP,POMDP}) begin
P = typeof(mdp)
S = statetype(P)
A = actiontype(P)
@req discount(::P)
@subreq ordered_actions(mdp)
@req actionindex(::P, ::A)
@req actions(::P, ::S)
as = actions(mdp)
a = first(as)
# Need to be able to sample states
@req sample_state(::P, ::typeof(solver.rng))
# Have different requirements depending on whether solver MDP is generative or explicit
if solver.is_mdp_generative
@req gen(::P, ::S, ::A, ::typeof(solver.rng))
else
@req transition(::P, ::S, ::A)
ss = sample_state(mdp, solver.rng)
dist = transition(mdp, ss, a)
D = typeof(dist)
@req support(::D)
end
# Feature vector conversion must be defined either directly or by default (through convert_s)
@subreq convert_featurevector(solver.fv_type, sample_state(mdp, solver.rng), mdp)
end
function POMDPs.solve(solver::GlobalApproximationValueIterationSolver, mdp::Union{MDP,POMDP})
@warn_requirements solve(solver,mdp)
# Ensure that generative model has a non-zero number of samples
if solver.is_mdp_generative
@assert solver.n_generative_samples > 0
end
# Solver parameters
belres = solver.belres
num_iterations = solver.num_iterations
num_samples = solver.num_samples
discount_factor = discount(mdp)
gfa_type = typeof(solver.gfa)
# Initialize the policy
policy = GlobalApproximationValueIterationPolicy(mdp, solver)
total_time = 0.0
iter_time = 0.0
temp_s = sample_state(mdp, solver.rng)
state_dim = length(convert_featurevector(solver.fv_type, temp_s, mdp))
for iter = 1:num_iterations
residual = 0.0
# Setup input and outputs for fit functions
state_matrix = zeros(eltype(solver.fv_type), num_samples, state_dim)
val_vector = zeros(eltype(solver.fv_type), num_samples)
iter_time = @elapsed begin
# Loop over the chosen number of samples for approximation
# and compute the current value function estimate at each sample
for i = 1:num_samples
s = sample_state(mdp, solver.rng)
pt = convert_featurevector(solver.fv_type, s, mdp)
state_matrix[i,:] = pt
sub_aspace = actions(mdp,s)
if isterminal(mdp, s)
val_vector[i] = 0.0
else
old_util = value(policy, s)
max_util = -Inf
# Compute the approximate Q value for each action and choose the best
for a in sub_aspace
iaction = actionindex(mdp,a)
u = 0.0
if solver.is_mdp_generative
for j in 1:solver.n_generative_samples
sp, r = @gen(:sp,:r)(mdp, s, a, solver.rng)
u += r
if !isterminal(mdp,sp)
sp_feature = convert_featurevector(solver.fv_type, sp, mdp)
u += discount_factor*compute_value(policy.gfa, sp_feature)
end
end
u = u / solver.n_generative_samples
else
dist = transition(mdp,s,a)
for (sp, p) in weighted_iterator(dist)
p == 0.0 ? continue : nothing
r = reward(mdp, s, a, sp)
u += p*r
# Only interpolate sp if it is non-terminal
if !isterminal(mdp,sp)
sp_feature = convert_featurevector(solver.fv_type, sp, mdp)
u += p * (discount_factor*compute_value(policy.gfa, sp_feature))
end
end
end
max_util = (u > max_util) ? u : max_util
util_diff = abs(max_util - old_util)
util_diff > residual ? (residual = util_diff) : nothing
end #action
val_vector[i] = max_util
end
end
# Now fit!
fit!(policy.gfa, state_matrix, val_vector)
end # time
total_time += iter_time
solver.verbose ? @printf("[Iteration %-4d] residual: %10.3G | iteration runtime: %10.3f ms, (%10.3G s total)\n", iter, residual, iter_time*1000.0, total_time) : nothing
residual < belres ? break : nothing
end
return policy
end
function POMDPs.value(policy::GlobalApproximationValueIterationPolicy, s::S) where S
s_point = convert_featurevector(policy.fv_type, s, policy.mdp)
val = compute_value(policy.gfa, s_point)
return val
end
# Not explicitly stored in policy - extract from value function interpolation
function POMDPs.action(policy::GlobalApproximationValueIterationPolicy, s::S) where S
mdp = policy.mdp
best_a_idx = -1
max_util = -Inf
sub_aspace = actions(mdp,s)
discount_factor = discount(mdp)
for a in sub_aspace
iaction = actionindex(mdp, a)
u = value(policy,s,a)
if u > max_util
max_util = u
best_a_idx = iaction
end
end
return policy.action_map[best_a_idx]
end
function POMDPs.value(policy::GlobalApproximationValueIterationPolicy, s::S, a::A) where {S,A}
mdp = policy.mdp
discount_factor = discount(mdp)
u = 0.0
# As in solve(), do different things based on whether
# mdp is generative or explicit
if policy.is_mdp_generative
for j in 1:policy.n_generative_samples
sp, r = @gen(:sp,:r)(mdp, s, a, solver.rng)
sp_point = convert_featurevector(policy.fv_type, sp, mdp)
u += r + discount_factor*compute_value(policy.gfa, sp_point)
end
u = u / policy.n_generative_samples
else
dist = transition(mdp,s,a)
for (sp, p) in weighted_iterator(dist)
p == 0.0 ? continue : nothing
r = reward(mdp, s, a, sp)
u += p*r
# Only interpolate sp if it is non-terminal
if !isterminal(mdp,sp)
sp_point = convert_featurevector(policy.fv_type, sp, mdp)
u += p*(discount_factor*compute_value(policy.gfa, sp_point))
end
end
end
return u
end
| GlobalApproximationValueIteration | https://github.com/JuliaPOMDP/GlobalApproximationValueIteration.jl.git |
|
[
"MIT"
] | 0.2.3 | 16d2a15e95e265b590603f62c33612fd64c963c2 | code | 709 | abstract type GlobalFunctionApproximator end
"""
fit!(gfa::GlobalFunctionApproximator, dataset_input::AbstractMatrix, dataset_output::AbstractVector)
Fit the global function approximator to the dataset using some optimization method and a chosen
loss function.
"""
function fit! end
"""
compute_value(gfa::GlobalFunctionApproximator, v::AbstractVector)
Return the value of the function at some query point v, based on the global function approximator
compute_value(gfa::GlobalFunctionApproximator, v_list::AbstractVector{V}) where V <: AbstractVector{Float64}
Return the value of the function for a list of query points, based on the global function approximator
"""
function compute_value end | GlobalApproximationValueIteration | https://github.com/JuliaPOMDP/GlobalApproximationValueIteration.jl.git |
|
[
"MIT"
] | 0.2.3 | 16d2a15e95e265b590603f62c33612fd64c963c2 | code | 572 | mutable struct LinearGlobalFunctionApproximator{W <: AbstractArray} <: GlobalFunctionApproximator
weights::W
end
function fit!(lgfa::LinearGlobalFunctionApproximator, dataset_input::AbstractMatrix{T},
dataset_output::AbstractArray{T}) where T
# TODO: Since we are ASSIGNING to weights here, does templating even matter? Does the struct even matter?
lgfa.weights = llsq(dataset_input, dataset_output, bias=false)
end
function compute_value(lgfa::LinearGlobalFunctionApproximator, v::AbstractArray{T}) where T
return dot(lgfa.weights, v)
end | GlobalApproximationValueIteration | https://github.com/JuliaPOMDP/GlobalApproximationValueIteration.jl.git |
|
[
"MIT"
] | 0.2.3 | 16d2a15e95e265b590603f62c33612fd64c963c2 | code | 828 | mutable struct NonlinearGlobalFunctionApproximator{M,O,L} <: GlobalFunctionApproximator
model::M
optimizer::O
loss::L
end
function fit!(ngfa::NonlinearGlobalFunctionApproximator, dataset_input::AbstractMatrix{T},
dataset_output::AbstractArray{T}) where T
# Create loss function with loss type
loss(x, y) = ngfa.loss(ngfa.model(x), y)
# NOTE : Minibatch update; 1 update to model weights
# data = repeated((param(transpose(dataset_input)), param(transpose(dataset_output))), 1)
data = repeated((transpose(dataset_input), transpose(dataset_output)), 1)
Flux.train!(loss, params(ngfa.model), data, ngfa.optimizer)
end
function compute_value(ngfa::NonlinearGlobalFunctionApproximator, state_vector::AbstractArray{T}) where T
return Flux.data(ngfa.model(state_vector))[1]
end | GlobalApproximationValueIteration | https://github.com/JuliaPOMDP/GlobalApproximationValueIteration.jl.git |
|
[
"MIT"
] | 0.2.3 | 16d2a15e95e265b590603f62c33612fd64c963c2 | code | 284 | using Revise
using POMDPModels
using POMDPs
using POMDPTools
using StaticArrays
using Random
using DiscreteValueIteration
using GlobalApproximationValueIteration
using Flux
using Statistics
using Test
include("test/test_with_nonlinear_gfa.jl")
include("test/test_with_linear_gfa.jl") | GlobalApproximationValueIteration | https://github.com/JuliaPOMDP/GlobalApproximationValueIteration.jl.git |
|
[
"MIT"
] | 0.2.3 | 16d2a15e95e265b590603f62c33612fd64c963c2 | code | 354 | using POMDPModels
using POMDPs
using POMDPTools
using StaticArrays
using Statistics
using Random
using DiscreteValueIteration
using GlobalApproximationValueIteration
using Test
Random.seed!(1234)
@testset "all" begin
@testset "integration" begin
include("test_with_linear_gfa.jl")
include("test_with_nonlinear_gfa.jl")
end
end
| GlobalApproximationValueIteration | https://github.com/JuliaPOMDP/GlobalApproximationValueIteration.jl.git |
|
[
"MIT"
] | 0.2.3 | 16d2a15e95e265b590603f62c33612fd64c963c2 | code | 4271 | #=
Construct a 100 x 100 grid and run discreteVI on it. Also run global approx VI where any integral point in the grid
can be sampled. Compare the values at the integer grid points. Confirm that the max difference reduces with increasing
number of samples.
=#
# Feature vector conversion
# For point (x,y), fv is (1 x y xy x^2 y^2)
function GlobalApproximationValueIteration.convert_featurevector(::Type{SVector{10, Float64}}, s::GWPos, mdp::SimpleGridWorld)
x = s[1]
y = s[2]
v = SVector{10, Float64}(1, x, y, x*y, x^2, y^2, x^3, x^2*y, x*y^2, y^3)
return v
end
# Sample a specific integral point in the grid
function GlobalApproximationValueIteration.sample_state(mdp::SimpleGridWorld, rng::RNG=Random.GLOBAL_RNG) where {RNG <: AbstractRNG}
x = rand(rng, 1:mdp.size[1])
y = rand(rng, 1:mdp.size[2])
return GWPos(x, y)
end
function test_absolute_error()
rng = MersenneTwister(1234)
# Set solver and grid world parameters
MAX_ITERS = 500
NUM_SAMPLES = 1000
SIZE_X = 5
SIZE_Y = 5
REWARD_COV_PROB = 0.4
rewards = Dict{GWPos, Float64}()
for x = 1:SIZE_X
for y = 1:SIZE_Y
if rand(rng) < REWARD_COV_PROB
rewards[GWPos(x, y)] = 1
end
end
end
# Create the MDP for a typical grid world
mdp = SimpleGridWorld(size=(SIZE_X, SIZE_Y), rewards=rewards)
# Create the linear function approximation with 10 weight parameters, initialized to zero
lin_gfa = LinearGlobalFunctionApproximator(zeros(10))
# Initialize the global approximation solver with the linear approximator and solve the MDP to obtain the policy
gfa_solver = GlobalApproximationValueIterationSolver(lin_gfa, num_samples=NUM_SAMPLES, max_iterations=MAX_ITERS, verbose=true, fv_type=SVector{10, Float64})
gfa_policy = solve(gfa_solver, mdp)
error_arr = Vector{Float64}(undef, 0)
for state in states(mdp)
full_val = value(policy, state)
approx_val = value(gfa_policy, state)
abs_diff = abs(full_val - approx_val)
push!(error_arr, abs_diff)
end
@show mean(error_arr)
@show maximum(error_arr)
return (mean(error_arr) < 0.04 && maximum(error_arr) < 0.35)
end
function test_relative_error()
rng = MersenneTwister(2378)
# Attempt to approximate globally with N samples and M iterations. As N increases, the average error should decrease
MAX_ITERS = 500
NUM_SAMPLES_LOW = 30
NUM_SAMPLES_HI = 1000
# Grid probabilities
SIZE_X = 5
SIZE_Y = 5
REWARD_COV_PROB = 0.4
# Generate a block of reward states from 40,40 to 60,60
rewards = Dict{GWPos, Float64}()
for x = 1:SIZE_X
for y = 1:SIZE_Y
if rand(rng) < REWARD_COV_PROB
rewards[GWPos(x, y)] = 1
end
end
end
# Create MDP
mdp = SimpleGridWorld(size=(SIZE_X, SIZE_Y), rewards=rewards)
lin_gfa_1 = LinearGlobalFunctionApproximator(zeros(10))
lin_gfa_2 = LinearGlobalFunctionApproximator(zeros(10))
solver_low = GlobalApproximationValueIterationSolver(lin_gfa_1; num_samples=NUM_SAMPLES_LOW, max_iterations=MAX_ITERS, verbose=true, fv_type=SVector{10, Float64})
solver_hi = GlobalApproximationValueIterationSolver(lin_gfa_2; num_samples=NUM_SAMPLES_HI, max_iterations=MAX_ITERS, verbose=true, fv_type=SVector{10, Float64})
policy_low = solve(solver_low, mdp)
policy_hi = solve(solver_hi, mdp)
# Now solve with dicrete VI
solver = ValueIterationSolver(max_iterations=1000, verbose=true)
policy = solve(solver, mdp)
err_arr_low = Vector{Float64}(undef, 0)
err_arr_hi = Vector{Float64}(undef, 0)
for state in states(mdp)
full_val = value(policy, state)
approx_val_low = value(policy_low, state)
approx_val_hi = value(policy_hi, state)
push!(err_arr_low, abs(full_val - approx_val_low))
push!(err_arr_hi, abs(full_val - approx_val_hi))
end
@show mean(err_arr_low), mean(err_arr_hi)
@show maximum(err_arr_low), maximum(err_arr_hi)
return (mean(err_arr_low) > mean(err_arr_hi) && maximum(err_arr_low) > maximum(err_arr_hi))
end
# @test test_absolute_error() == true
@test test_relative_error() == true | GlobalApproximationValueIteration | https://github.com/JuliaPOMDP/GlobalApproximationValueIteration.jl.git |
|
[
"MIT"
] | 0.2.3 | 16d2a15e95e265b590603f62c33612fd64c963c2 | code | 2510 |
# Sample a specific integral point in the grid
function GlobalApproximationValueIteration.sample_state(mdp::SimpleGridWorld, rng::RNG=Random.GLOBAL_RNG) where {RNG <: AbstractRNG}
x = rand(rng, 1:mdp.size[1])
y = rand(rng, 1:mdp.size[2])
return GWPos(x, y)
end
function test_against_full_grid()
rng = MersenneTwister(2378)
# Attempt to approximate globally with N samples and M iterations. As N increases, the average error should decrease
MAX_ITERS = 100
NUM_SAMPLES_LOW = 50
NUM_SAMPLES_HI = 500
# Grid probabilities
SIZE_X = 10
SIZE_Y = 10
REWARD_COV_PROB = 0.3
# Generate a block of reward states from 40,40 to 60,60
rewards = Dict{GWPos, Float64}()
for x = 1:SIZE_X
for y = 1:SIZE_Y
if rand(rng) < REWARD_COV_PROB
rewards[GWPos(x, y)] = 10
end
end
end
# Create MDP
mdp = SimpleGridWorld(size=(SIZE_X, SIZE_Y), rewards=rewards)
# Define learning model
model1 = Chain(
Dense(2, 10, relu),
Dense(10, 5, relu),
Dense(5, 1))
model2 = Chain(
Dense(2, 10, relu),
Dense(10, 5, relu),
Dense(5, 1))
opt = Adam(0.001)
nonlin_gfa_1 = NonlinearGlobalFunctionApproximator(model1, opt, Flux.mse)
nonlin_gfa_2 = NonlinearGlobalFunctionApproximator(model2, opt, Flux.mse)
solver_low = GlobalApproximationValueIterationSolver(nonlin_gfa_1; num_samples=NUM_SAMPLES_LOW, max_iterations=MAX_ITERS, verbose=true)
solver_hi = GlobalApproximationValueIterationSolver(nonlin_gfa_2; num_samples=NUM_SAMPLES_HI, max_iterations=MAX_ITERS, verbose=true)
policy_low = solve(solver_low, mdp)
policy_hi = solve(solver_hi, mdp)
# Now solve with dicrete VI
solver = ValueIterationSolver(max_iterations=1000, verbose=true)
policy = solve(solver, mdp)
total_err_low = 0.0
total_err_hi = 0.0
for state in states(mdp)
full_val = value(policy, state)
approx_val_low = value(policy_low, state)
approx_val_hi = value(policy_hi, state)
total_err_low += abs(full_val-approx_val_low)
total_err_hi += abs(full_val-approx_val_hi)
end
avg_err_low = total_err_low / length(states(mdp))
avg_err_hi = total_err_hi / length(states(mdp))
@show avg_err_low
@show avg_err_hi
return (avg_err_low > avg_err_hi)
end
@test_broken test_against_full_grid() == true
| GlobalApproximationValueIteration | https://github.com/JuliaPOMDP/GlobalApproximationValueIteration.jl.git |
|
[
"MIT"
] | 0.2.3 | 16d2a15e95e265b590603f62c33612fd64c963c2 | docs | 4547 | [](https://github.com/JuliaPOMDP/GlobalApproximationValueIteration.jl/actions)
[](https://codecov.io/gh/JuliaPOMDP/GlobalApproximationValueIteration.jl?branch=master)
# GlobalApproximationValueIteration.jl
This package implements the Global Approximation Value Iteration algorithm in Julia for solving
Markov Decision Processes (MDPs) with global function approximation.
It is functionally very similar to the previously released
[LocalApproximationValueIteration.jl](https://github.com/JuliaPOMDP/LocalApproximationValueIteration.jl)
and interested users can refer to its README for more details.
The user should define the POMDP problem according to the API in
[POMDPs.jl](https://github.com/JuliaPOMDP/POMDPs.jl). Examples of problem definitions can be found in
[POMDPModels.jl](https://github.com/JuliaPOMDP/POMDPModels.jl).
## Installation
You need to have [POMDPs.jl](https://github.com/JuliaPOMDP/POMDPs.jl) already and the JuliaPOMDP registry added (see the README of POMDPs.jl).
Thereafter, you can add GlobalApproximationValueIteration from the package manager
```julia
using Pkg
Pkg.add("GlobalApproximationValueIteration")
```
## How it Works
This solver is one example of _Approximate Dynamic Programming_, which tries to find approximately optimal
value functions and policies for large or continuous state spaces.
As the name suggests, global approximation value iteration tries to approximate the value function over the
entire state space using a compact representation. The quality of the approximation varies with the
kind of function approximation scheme used; this repository can accommodate both linear (with feature vectors) and nonlinear
schemes. Please see **Section 4.5.1** of the book [Decision Making Under Uncertainty : Theory and Application](https://dl.acm.org/citation.cfm?id=2815660)
and **Chapter 3** of [Markov Decision Processes in Artificial Intelligence](https://books.google.co.in/books?hl=en&lr=&id=2J8_-O4-ABIC&oi=fnd&pg=PT8&dq=markov+decision+processes+in+AI&ots=mcxpyqiv0X&sig=w-gF6nzm3JxgutcslIbUDD0dAXY) for more.
## State Space Representation
The Global Approximation solver needs two things in particular from the state space of the MDP. First, it should be able to sample a state
from the state space (whether discrete or continuous). During value iteration, in each step, the solver will sample several states, estimate the value
at them and try to fit the approximation scheme.
Second, a state instance should be representable as a _feature vector_ which will be used for linear or non-linear function approximation.
In the default case, the `feature` can just be the vector encoding of the state (see *State Space Representation* in the [README](https://github.com/JuliaPOMDP/LocalApproximationValueIteration.jl)
of LocalApproximationValueIteration.jl for more on this).
## Usage
Please refer to the [README](https://github.com/JuliaPOMDP/LocalApproximationValueIteration.jl)
of LocalApproximationValueIteration.jl as the usage of the global variant is very similar to that one.
A simple example is also provided in the `test/` folder for each of linear and nonliner function approximation.
`POMDPs.jl` has a macro `@requirements_info` that determines the functions necessary to use some solver on some specific MDP model.
Other than the typical methods required for approximate value iteration and state space representation mentioned above,
the solver also requires a `GlobalFunctionApproximator` object (see `src/global_function_approximation.jl` for details
on the interface). We have also implemented two examplar approximations, linear and non-linear.
The following code snippet from `test/test_with_linear_gfa.jl` is the most relevant chunk of code
for using the solver correctly.
```julia
# Create the MDP for a typical grid world
mdp = SimpleGridWorld(size=(SIZE_X, SIZE_Y), rewards=rewards)
# Create the linear function approximation with 10 weight parameters, initialized to zero
lin_gfa = LinearGlobalFunctionApproximator(zeros(10))
# Initialize the global approximation solver with the linear approximator and solve the MDP to obtain the policy
gfa_solver = GlobalApproximationValueIterationSolver(lin_gfa, num_samples=NUM_SAMPLES, max_iterations=MAX_ITERS, verbose=true, fv_type=SVector{10, Float64})
gfa_policy = solve(gfa_solver, mdp)
```
| GlobalApproximationValueIteration | https://github.com/JuliaPOMDP/GlobalApproximationValueIteration.jl.git |
|
[
"MIT"
] | 0.6.1 | dfab74c3efb21c6df589f30f7bbe5b9b8dbbe3c9 | code | 184 | using Documenter, RasterDataSources
makedocs(
sitename = "RasterDataSources.jl",
checkdocs = :all,
)
deploydocs(
repo = "github.com/EcoJulia/RasterDataSources.jl.git",
)
| RasterDataSources | https://github.com/EcoJulia/RasterDataSources.jl.git |
|
[
"MIT"
] | 0.6.1 | dfab74c3efb21c6df589f30f7bbe5b9b8dbbe3c9 | code | 2616 | module RasterDataSources
@doc let
path = joinpath(dirname(@__DIR__), "README.md")
include_dependency(path)
read(path, String)
end RasterDataSources
using Dates,
HTTP,
URIs,
ZipFile,
ASCIIrasters,
DelimitedFiles
import JSON.Parser as JP
export WorldClim, CHELSA, EarthEnv, AWAP, ALWB, SRTM, MODIS
export BioClim, BioClimPlus, Climate, Weather, Elevation, LandCover, HabitatHeterogeneity
export Future, CMIP5, CMIP6
export RCP26, RCP45, RCP60, RCP85
export SSP126, SSP245, SSP370, SSP585
export ModisProduct
export ECO4ESIPTJPL,ECO4WUE,GEDI03,GEDI04_B,MCD12Q1,MCD12Q2,MCD15A2H,
MCD15A3H,MCD19A3,MCD43A,MCD43A1,MCD43A4,MCD64A1,MOD09A1,MOD11A2,MOD13Q1,
MOD14A2,MOD15A2H,MOD16A2,MOD17A2H,MOD17A3HGF,MOD21A2,MOD44B,MYD09A1,
MYD11A2,MYD13Q1,MYD14A2,MYD15A2H,MYD16A2,MYD17A2H,MYD17A3HGF,MYD21A2,
SIF005,SIF_ANN,VNP09A1,VNP09H1,VNP13A1,VNP15A2H,VNP21A2,VNP22Q2,
ECO4ESIPTJPL, ECO4WUE, GEDI03, GEDI04_B, MCD12Q1, MCD12Q2, MCD15A2H,
MCD15A3H, MCD19A3, MCD43A, MCD43A1, MCD43A4, MCD64A1, MOD09A1, MOD11A2,
MOD13Q1, MOD14A2, MOD15A2H, MOD16A2, MOD17A2H, MOD17A3HGF, MOD21A2,
MOD44B, MYD09A1, MYD11A2, MYD13Q1, MYD14A2, MYD15A2H, MYD16A2,
MYD17A2H, MYD17A3HGF, MYD21A2, SIF005, SIF_ANN, VNP09A1, VNP09H1,
VNP13A1, VNP15A2H, VNP21A2, VNP22Q2
# Climate models from CMIP5 (used in CHELSA)
export ACCESS1, BNUESM, CCSM4, CESM1BGC, CESM1CAM5, CMCCCMS, CMCCCM, CNRMCM5,
CSIROMk3, CanESM2, FGOALS, FIOESM, GFDLCM3, GFDLESM2G, GFDLESM2M, GISSE2HCC,
GISSE2H, GISSE2RCC, GISSE2R, HadGEM2AO, HadGEM2CC, IPSLCM5ALR, IPSLCM5AMR,
MIROCESMCHEM, MIROCESM, MIROC5, MPIESMLR, MPIESMMR, MRICGCM3, MRIESM1, NorESM1M,
BCCCSM1, Inmcm4
# Climate models from CMIP6 (used in WorldClim)
export BCCCSM2MR, CNRMCM61, CNRMESM21, CanESM5, GFDLESM4, IPSLCM6ALR, MIROCES2L, MIROC6, MRIESM2
# Climate models from CMIP6 (CHELSA)
export UKESM, MPIESMHR
export Values, Deciles
export getraster
include("interface.jl")
include("types.jl")
include("shared.jl")
include("worldclim/shared.jl")
include("worldclim/bioclim.jl")
include("worldclim/climate.jl")
include("worldclim/weather.jl")
include("worldclim/elevation.jl")
include("chelsa/shared.jl")
include("chelsa/climate.jl")
include("chelsa/bioclim.jl")
include("chelsa/future.jl")
include("earthenv/shared.jl")
include("earthenv/landcover.jl")
include("earthenv/habitatheterogeneity.jl")
include("awap/awap.jl")
include("alwb/alwb.jl")
include("srtm/srtm.jl")
include("modis/shared.jl")
include("modis/products.jl")
include("modis/utilities.jl")
include("modis/examples.jl")
end # module
| RasterDataSources | https://github.com/EcoJulia/RasterDataSources.jl.git |
|
[
"MIT"
] | 0.6.1 | dfab74c3efb21c6df589f30f7bbe5b9b8dbbe3c9 | code | 4445 | # Exported
"""
getraster(source::Type, [layer]; kw...)
Download raster layers `layers` from the data `source`,
returning a `String` for a single layer, or a `NamedTuple`
for a `Tuple` of layers.
`getraster` provides a standardised interface to download data sources,
and return the filename/s of the selected files.
RasterDataSources.jl aims to standardise an API for downloading many kinds of raster files
from many sources, that can be wrapped by other packages (such as Rasters.jl and
SimpleSDMLayers.jl) in a simple, regular way. As much as possible it will move towards
having less source-specific keywords wherever possible. Similar datasets will behave in the
same way so that they can be used interchangeably in the same code.
# Arguments
- `source`: defines the [`RasterDataSource`](@ref) and (if it there is more than one)
the specific [`RasterDataSet`](@ref) from which to download data.
- `layer`: choose the named `Symbol`/s or numbered `Int`/s (for `BioClim`) layer/s of the
data source. If `layer` is not passed, all layers will be downloaded, returning a
`NamedTuple` of filenames.
# Keywords
Keyword arguments specify subsets of a data set, such as by date or resolution.
As much as possible these are standardised for all sources where they are relevent.
- `date`: `DateTime` date, range of dates, or tuple of start and end dates. Usually for weather datasets.
- `month`: month or range of months to download for climatic datasets, as `Integer`s from 1 to 12.
- `res`: spatial resolion of the file, as a `String` with units, e.g. "10m".
# Return values
The return value is either a single `String`, a `Tuple/Array` of `String`, or a
`Tuple/Array` of `Tuple/Array` of `String` --- depending on the arguments. If multiple
layers are specified, this may return multiple filenames. If multiple months or dates are
specified, this may also return multiple filenames.
Keyword arguments depend on the specific data source.
They may modify the return value, following a pattern:
- `month` keywords of `AbstractArray` will return a `Vector{String}`
or `Vector{<:NamedTuple}`.
- `date` keywords of `AbstractArray` will return a `Vector{String}` or
`Vector{<:NamedTuple}`.
- `date` keywords of `Tuple{start,end}` will take all the dates between the
start and end dates as a `Vector{String}` or `Vector{<:NamedTuple}`.
Where `date` and `month` keywords coexist, `Vector{Vector{String}}` of
`Vector{Vector{NamedTuple}}` is the result. `date` ranges are always
the outer `Vector`, `month` the inner `Vector` with `layer` tuples as
the inner `NamedTuple`. No other keywords can be `Vector`.
This schema may be added to in future for datasets with additional axes,
but should not change for the existing `RasterDataSource` types.
"""
function getraster end
"""
getraster(T::Type, layers::Union{Tuple,Int,Symbol}; kw...)
"""
function getraster end
# Not exported, but relatively consistent and stable
# These should be used for consistency accross all sources
"""
rastername(source::Type, [layer]; kw...)
Returns the name of the file, without downloading it.
Arguments are the same as for `getraster`
Returns a `String` or multiple `Strings`.
"""
function rastername end
"""
rasterpath(source::Type, [layer]; kw...)
Returns the name of the file, without downloading it.
Arguments are the same as for `getraster`
Returns a `String` or multiple `Strings`.
"""
function rasterpath end
"""
rasterurl(source::Type, [layer]; kw...)
If the file has a single url, returns it without downloading.
Arguments are the same as for `getraster`.
Returns a URIs.jl `URI` or mulitiple `URI`s.
"""
function rasterurl end
"""
zipname(source::Type, [layer]; kw...)
If the url is a zipped file, returns its name.
Arguments are as the same for `getraster` where possible.
Returns a `String` or multiple `Strings`.
"""
function zipname end
"""
zippath(source::Type, [layer]; kw...)
If the url is a zipped file, returns its path when downloaded.
(This may not exist after extraction with `getraster`)
Arguments are the same as for `getraster` where possible.
Returns a `String` or multiple `Strings`.
"""
function zippath end
"""
zipurl(source::Type, [layer]; kw...)
If the url is a zipped file, returns its zip path without downloading.
Arguments are the same as for `getraster` where possible.
Returns a URIs.jl `URI` or mulitiple `URI`s.
"""
function zipurl end
| RasterDataSources | https://github.com/EcoJulia/RasterDataSources.jl.git |
|
[
"MIT"
] | 0.6.1 | dfab74c3efb21c6df589f30f7bbe5b9b8dbbe3c9 | code | 2711 | # Vector layers are allowed, but converted to `Tuple` immediatedly.
function getraster(T::Type, layers::AbstractArray; kw...)
getraster(T, (layers...,); kw...)
end
# Without a layers argument, all layers are downloaded
getraster(T::Type; kw...) = getraster(T, layers(T); kw...)
"""
getraster_keywords(::Type{<:RasterDataSource})
Trait for defining data source keywords, which returns
a `NTuple{N,Symbol}`.
The default fallback method returns `()`.
"""
getraster_keywords(::Type{<:RasterDataSource}) = ()
# Default assumption for `layerkeys` is that the layer
# is the same as the layer key. This is not the case for
# e.g. BioClim, where layers can be specified with Int.
layerkeys(T::Type) = layers(T)
layerkeys(T::Type, layers) = layers
has_matching_layer_size(T) = true
has_constant_dims(T) = true
has_constant_metadata(T) = true
date_sequence(T::Type, dates; kw...) = date_sequence(date_step(T), dates)
date_sequence(step, date) = _date_sequence(step, date)
_date_sequence(step, dates::AbstractArray) = dates
_date_sequence(step, dates::NTuple{2}) = first(dates):step:last(dates)
_date_sequence(step, date) = date:step:date
function _maybe_download(uri::URI, filepath, headers = [])
if !isfile(filepath)
mkpath(dirname(filepath))
@info "Starting download for $uri"
try
HTTP.download(string(uri), filepath, headers)
catch e
# Remove anything that was downloaded before the error
isfile(filepath) && rm(filepath)
throw(e)
end
end
filepath
end
function rasterpath()
if haskey(ENV, "RASTERDATASOURCES_PATH") && isdir(ENV["RASTERDATASOURCES_PATH"])
ENV["RASTERDATASOURCES_PATH"]
else
error("You must set `ENV[\"RASTERDATASOURCES_PATH\"]` to a path in your system")
end
end
function delete_rasters()
# May need an "are you sure"? - this could be a lot of GB of data to lose
ispath(rasterpath()) && rm(rasterpath())
end
function delete_rasters(T::Type)
ispath(rasterpath(T)) && rm(rasterpath(T))
end
_check_res(T, res) =
res in resolutions(T) || throw(ArgumentError("Resolution $res not in $(resolutions(T))"))
_check_layer(T, layer) =
layer in layers(T) || throw(ArgumentError("Layer $layer not in $(layers(T))"))
_date2string(t, date) = Dates.format(date, _dateformat(t))
_string2date(t, d::AbstractString) = Date(d, _dateformat(t))
# Inner map over layers Tuple - month/date maps earlier
# so we get Vectors of NamedTuples of filenames
function _map_layers(T, layers, args...; kw...)
filenames = map(layers) do l
_getraster(T, l, args...; kw...)
end
keys = layerkeys(T, layers)
return NamedTuple{keys}(filenames)
end
| RasterDataSources | https://github.com/EcoJulia/RasterDataSources.jl.git |
|
[
"MIT"
] | 0.6.1 | dfab74c3efb21c6df589f30f7bbe5b9b8dbbe3c9 | code | 11702 | """
RasterDataSource
Abstract supertype for raster data collections.
"""
abstract type RasterDataSource end
"""
RasterDataSet
Abstract supertype for datasets that belong to a [`RasterDataSource`](@ref).
"""
abstract type RasterDataSet end
"""
BioClim <: RasterDataSet
BioClim datasets. Usually containing layers from `1:19`.
These can also be accessed with `:bioX`, e.g. `:bio5`.
They do not usually use `month` or `date` keywords, but may use
`date` in past/future scenarios.
Currently implemented for WorldClim and CHELSA as `WorldClim{BioClim}`,
`CHELSA{BioClim}` and `CHELSA{Future{BioClim, args..}}`.
See the [`getraster`](@ref) docs for implementation details.
"""
struct BioClim <: RasterDataSet end
# Bioclim has standardised layers for all data sources
layers(::Type{BioClim}) = values(bioclim_lookup)
layerkeys(T::Type{BioClim}) = keys(bioclim_lookup)
layerkeys(T::Type{BioClim}, layer) = bioclim_key(layer)
layerkeys(T::Type{BioClim}, layers::Tuple) = map(l -> bioclim_key(l), layers)
const bioclim_lookup = (
bio1 = 1,
bio2 = 2,
bio3 = 3,
bio4 = 4,
bio5 = 5,
bio6 = 6,
bio7 = 7,
bio8 = 8,
bio9 = 9,
bio10 = 10,
bio11 = 11,
bio12 = 12,
bio13 = 13,
bio14 = 14,
bio15 = 15,
bio16 = 16,
bio17 = 17,
bio18 = 18,
bio19 = 19,
)
# We allow a range of bioclim keys, as they are listed with
# a lot of variants on CHELSA and WorldClim
bioclim_key(k::Symbol) = bioclim_key(string(k))
bioclim_key(k::AbstractString) = Symbol(replace(lowercase(k), "_" => ""))
bioclim_key(k::Integer) = keys(bioclim_lookup)[k]
bioclim_int(k::Integer) = k
bioclim_int(k::Symbol) = bioclim_lookup[bioclim_key(k)]
"""
BioClimPlus <: RasterDataSet
Extended BioClim datasets, available from CHELSA.
More information on the CHELSA website: https://chelsa-climate.org/exchelsa-extended-bioclim/
Some of these are available as average annual maximum, minimum, mean, and range.
Others have a single value, more like the regular BioClim variables.
They do not usually use `month` or `date` keywords, but may use
`date` in past/future scenarios.
Currently implemented for CHELSA as `CHELSA{BioClim}` and `CHELSA{Future{BioClim, args..}}`,
specifying layer names as `Symbol`s.
See the [`getraster`](@ref) docs for implementation details.
"""
struct BioClimPlus <: RasterDataSet end
const _BIOCLIMPLUS_MONTHLY = vec([Symbol("$(b)_$(m)") for b in (:hurs, :clt, :sfcWind, :vpd, :rsds, :pet_penman, :cmi), m in [:max, :min, :mean, :range]])
const _BIOCLIMPLUS_GDD = vec([Symbol("$b$d") for b in (:gdd, :gddlgd, :gdgfgd, :ngd), d in [0, 5, 10]])
const _BIOCLIMPLUS_OTHERS = (:fcf, :fgd, :lgd, :scd, :gsl, :gst, :gsp, :npp, :swb, :swe)
const BIOCLIMPLUS_LAYERS = [
collect(layerkeys(BioClim))
_BIOCLIMPLUS_MONTHLY;
_BIOCLIMPLUS_GDD;
collect(_BIOCLIMPLUS_OTHERS);
[Symbol("kg$i") for i in 0:5];
]
const BIOCLIMPLUS_LAYERS_FUTURE = [
collect(layerkeys(BioClim));
_BIOCLIMPLUS_GDD;
collect(filter(!=(:swb), _BIOCLIMPLUS_OTHERS))
[Symbol("kg$i") for i in 0:5];
]
layers(::Type{BioClimPlus}) = BIOCLIMPLUS_LAYERS
"""
Climate <: RasterDataSet
Climate datasets. These are usually months of the year, not specific dates,
and use a `month` keyword in `getraster`. They also use `date` in past/future scenarios.
Currently implemented for WorldClim and CHELSA as `WorldClim{Climate}`,
`CHELSA{Climate}` and `CHELSA{Future{Climate, args..}}`.
See the [`getraster`](@ref) docs for implementation details.
"""
struct Climate <: RasterDataSet end
months(::Type{Climate}) = ntuple(identity, Val{12})
"""
Weather <: RasterDataSet
Weather datasets. These are usually large time-series of specific dates,
and use a `date` keyword in `getraster`.
Currently implemented for WorldClim and CHELSA as `WorldClim{Weather}`,
and `CHELSA{Weather}`
See the [`getraster`](@ref) docs for implementation details.
"""
struct Weather <: RasterDataSet end
"""
Elevation <: RasterDataSet
Elevation datasets.
Currently implemented for WorldClim as `WorldClim{Elevation}`.
See the [`getraster`](@ref) docs for implementation details.
"""
struct Elevation <: RasterDataSet end
"""
LandCover <: RasterDataSet
Land-cover datasets.
Currently implemented for EarthEnv as `EarchEnv{LandCover}`.
See the [`getraster`](@ref) docs for implementation details.
"""
struct LandCover{X} <: RasterDataSet end
"""
HabitatHeterogeneity <: RasterDataSet
Habitat heterogeneity datasets.
Currently implemented for EarchEnv as `EarchEnv{HabitatHeterogeneity}`.
See the [`getraster`](@ref) docs for implementation details.
"""
struct HabitatHeterogeneity <: RasterDataSet end
"""
ClimateModel
Abstract supertype for climate models use in [`Future`](@ref) datasets.
"""
abstract type ClimateModel end
struct ACCESS1 <: ClimateModel end
struct BNUESM <: ClimateModel end
struct CCSM4 <: ClimateModel end
struct CESM1BGC <: ClimateModel end
struct CESM1CAM5 <: ClimateModel end
struct CMCCCMS <: ClimateModel end
struct CMCCCM <: ClimateModel end
struct CNRMCM5 <: ClimateModel end
struct CSIROMk3 <: ClimateModel end
struct CanESM2 <: ClimateModel end
struct FGOALS <: ClimateModel end
struct FIOESM <: ClimateModel end
struct GFDLCM3 <: ClimateModel end
struct GFDLESM2G <: ClimateModel end
struct GFDLESM2M <: ClimateModel end
struct GISSE2HCC <: ClimateModel end
struct GISSE2H <: ClimateModel end
struct GISSE2RCC <: ClimateModel end
struct GISSE2R <: ClimateModel end
struct HadGEM2AO <: ClimateModel end
struct HadGEM2CC <: ClimateModel end
struct IPSLCM5ALR <: ClimateModel end
struct IPSLCM5AMR <: ClimateModel end
struct MIROCESMCHEM <: ClimateModel end
struct MIROCESM <: ClimateModel end
struct MIROC5 <: ClimateModel end
struct MPIESMLR <: ClimateModel end
struct MPIESMMR <: ClimateModel end
struct MRICGCM3 <: ClimateModel end
struct MRIESM1 <: ClimateModel end
struct NorESM1M <: ClimateModel end
struct BCCCSM1 <: ClimateModel end
struct Inmcm4 <: ClimateModel end
struct BCCCSM2MR <: ClimateModel end
struct CNRMCM61 <: ClimateModel end
struct CNRMESM21 <: ClimateModel end
struct CanESM5 <: ClimateModel end
struct GFDLESM4 <: ClimateModel end
struct IPSLCM6ALR <: ClimateModel end
struct MIROCES2L <: ClimateModel end
struct MIROC6 <: ClimateModel end
struct MRIESM2 <: ClimateModel end
struct UKESM <: ClimateModel end
struct MPIESMHR <: ClimateModel end
"""
CMIPphase
Abstract supertype for phases of the CMIP,
the Coupled Model Intercomparison Project.
Subtypes are `CMIP5` and `CMIP6`.
"""
abstract type CMIPphase end
"""
CMIP5 <: CMIPphase
The Coupled Model Intercomparison Project, Phase 5.
"""
struct CMIP5 <: CMIPphase end
"""
CMIP6 <: CMIPphase
The Coupled Model Intercomparison Project, Phase 6.
"""
struct CMIP6 <: CMIPphase end
"""
ClimateScenario
Abstract supertype for scenarios used in [`CMIPphase`](@ref) models.
"""
abstract type ClimateScenario end
"""
RepresentativeConcentrationPathway
Abstract supertype for Representative Concentration Pathways (RCPs) for [`CMIP5`](@ref).
Subtypes are: `RCP26`, `RCP45`, `RCP60`, `RCP85`
"""
abstract type RepresentativeConcentrationPathway <: ClimateScenario end
struct RCP26 <: RepresentativeConcentrationPathway end
struct RCP45 <: RepresentativeConcentrationPathway end
struct RCP60 <: RepresentativeConcentrationPathway end
struct RCP85 <: RepresentativeConcentrationPathway end
"""
SharedSocioeconomicPathway
Abstract supertype for Shared Socio-economic Pathways (SSPs) for [`CMIP6`](@ref).
Subtypes are: `SSP126`, `SSP245`, SSP370`, SSP585`
"""
abstract type SharedSocioeconomicPathway <: ClimateScenario end
struct SSP126 <: SharedSocioeconomicPathway end
struct SSP245 <: SharedSocioeconomicPathway end
struct SSP370 <: SharedSocioeconomicPathway end
struct SSP585 <: SharedSocioeconomicPathway end
"""
Future{<:RasterDataSet,<:CMIPphase,<:ClimateModel,<:ClimateScenario}
Future climate datasets specified with a dataset, phase, model, and scenario.
## Type Parameters
#### `RasterDataSet`
Currently [`BioClim`](@ref) and [`Climate`](@ref) are implemented
for the [`CHELSA`](@ref) data source.
#### `CMIPphase`
Can be either [`CMIP5`](@ref) or [`CMIP6`](@ref).
#### `ClimateModel`
Climate models can be chosen from:
`ACCESS1`, `BNUESM`, `CCSM4`, `CESM1BGC`, `CESM1CAM5`, `CMCCCMS`, `CMCCCM`,
`CNRMCM5`, `CSIROMk3`, `CanESM2`, `FGOALS`, `FIOESM`, `GFDLCM3`, `GFDLESM2G`,
`GFDLESM2M`, `GISSE2HCC`, `GISSE2H`, `GISSE2RCC`, `GISSE2R`, `HadGEM2AO`,
`HadGEM2CC`, `IPSLCM5ALR`, `IPSLCM5AMR`, `MIROCESMCHEM`, `MIROCESM`, `MIROC5`,
`MPIESMLR`, `MPIESMMR`, `MRICGCM3`, `MRIESM1`, `NorESM1M`, `BCCCSM1`, `Inmcm4`,
`BCCCSM2MR`, `CNRMCM61`, `CNRMESM21`, `CanESM5`, `MIROCES2L`, `MIROC6` for CMIP5;
`UKESM`, `MPIESMHR` `IPSLCM6ALR`, `MRIESM2`, `GFDLESM4` for `CMIP6`.
#### `ClimateScenario`
CMIP5 Climate scenarios are all [`RepresentativeConcentrationPathway`](@ref)
and can be chosen from: `RCP26`, `RCP45`, `RCP60`, `RCP85`
CMIP6 Climate scenarios are all [`SharedSocioeconomicPathway`](@ref) and
can be chosen from: `SSP126`, `SSP245`, `SSP370`, `SSP585`
However, note that not all climate scenarios are available for all models.
## Example
```jldoctest future
using RasterDataSources
dataset = Future{BioClim, CMIP5, BNUESM, RCP45}
# output
Future{BioClim, CMIP5, BNUESM, RCP45}
```
Currently `Future` is only implented for `CHELSA`
```jldoctest future
datasource = CHELSA{Future{BioClim, CMIP5, BNUESM, RCP45}}
```
"""
struct Future{D<:RasterDataSet,C<:CMIPphase,M<:ClimateModel,S<:ClimateScenario} end
_dataset(::Type{<:Future{D}}) where D = D
_dataset(::Type{<:Future{BioClimPlus}}) = BioClim
_phase(::Type{<:Future{<:Any,P}}) where P = P
_model(::Type{<:Future{<:Any,<:Any,M}}) where M = M
_scenario(::Type{<:Future{<:Any,<:Any,<:Any,S}}) where S = S
layers(::Type{<:Future{BioClimPlus}}) = BIOCLIMPLUS_LAYERS_FUTURE
"""
ModisProduct <: RasterDataSet
Abstract supertype for [`MODIS`](@ref)/VIIRS products.
# Usage
Some commonly used products are `MOD13Q1` (250m resolution MODIS vegetation indices) and `VNP13A1` (500m resolution VIIRS vegetation indices). Refer to the [MODIS documentation](https://modis.ornl.gov/documentation.html) for detailed product information.
"""
abstract type ModisProduct <: RasterDataSet end
struct ECO4ESIPTJPL <: ModisProduct end
struct ECO4WUE <: ModisProduct end
struct GEDI03 <: ModisProduct end
struct GEDI04_B <: ModisProduct end
struct MCD12Q1 <: ModisProduct end
struct MCD12Q2 <: ModisProduct end
struct MCD15A2H <: ModisProduct end
struct MCD15A3H <: ModisProduct end
struct MCD19A3 <: ModisProduct end
struct MCD43A <: ModisProduct end
struct MCD43A1 <: ModisProduct end
struct MCD43A4 <: ModisProduct end
struct MCD64A1 <: ModisProduct end
struct MOD09A1 <: ModisProduct end
struct MOD11A2 <: ModisProduct end
struct MOD13Q1 <: ModisProduct end
struct MOD14A2 <: ModisProduct end
struct MOD15A2H <: ModisProduct end
struct MOD16A2 <: ModisProduct end
struct MOD17A2H <: ModisProduct end
struct MOD17A3HGF <: ModisProduct end
struct MOD21A2 <: ModisProduct end
struct MOD44B <: ModisProduct end
struct MYD09A1 <: ModisProduct end
struct MYD11A2 <: ModisProduct end
struct MYD13Q1 <: ModisProduct end
struct MYD14A2 <: ModisProduct end
struct MYD15A2H <: ModisProduct end
struct MYD16A2 <: ModisProduct end
struct MYD17A2H <: ModisProduct end
struct MYD17A3HGF <: ModisProduct end
struct MYD21A2 <: ModisProduct end
struct SIF005 <: ModisProduct end
struct SIF_ANN <: ModisProduct end
struct VNP09A1 <: ModisProduct end
struct VNP09H1 <: ModisProduct end
struct VNP13A1 <: ModisProduct end
struct VNP15A2H <: ModisProduct end
struct VNP21A2 <: ModisProduct end
struct VNP22Q2 <: ModisProduct end | RasterDataSources | https://github.com/EcoJulia/RasterDataSources.jl.git |
|
[
"MIT"
] | 0.6.1 | dfab74c3efb21c6df589f30f7bbe5b9b8dbbe3c9 | code | 5924 |
const ALWB_URI = URI(scheme="http", host="www.bom.gov.au", path="/jsp/awra/thredds/fileServer/AWRACMS")
abstract type DataMode end
"""
Values <: DataMode
Get the dataset as regular measured values.
"""
struct Values <: DataMode end
"""
Deciles <: DataMode
Get the dataset in relative deciles.
"""
struct Deciles <: DataMode end
# Docs below
struct ALWB{M<:DataMode,D<:Union{Day,Month,Year}} <: RasterDataSource end
layers(::Type{<:ALWB}) = (
:rain_day, :s0_pct, :ss_pct, :sd_pct, :sm_pct, :qtot, :etot,
:e0, :ma_wet, :pen_pet, :fao_pet, :asce_pet, :msl_wet, :dd
)
# Days are in 1 year nc files
date_step(::Type{<:ALWB{<:Any,Day}}) = Year(1)
# Months and years are in single files
date_step(::Type{<:ALWB{<:Any,Month}}) = Year(100)
date_step(::Type{<:ALWB{<:Any,Year}}) = Year(100)
has_constant_dims(::Type{<:ALWB}) = false
@doc """
ALWB{Union{Deciles,Values},Union{Day,Month,Year}} <: RasterDataSource
Data from the Australian Landscape Water Balance (ALWB) data source.
See: [www.bom.gov.au/water/landscape](http://www.bom.gov.au/water/landscape)
The dataset contains NetCDF files. They have a time dimension so that multiple
dates are stored in each file.
The available layers are: `$(layers(ALWB))`, available in daily, monthly and
annual resolutions, and as `Values` or relative `Deciles`.
`getraster` for `ALWB` must use a `date` keyword to specify the date to download.
See the [`getraster`](@ref) docs for implementation details.
""" ALWB
# http://www.bom.gov.au/jsp/awra/thredds/fileServer/AWRACMS/values/day/rain_day_2017.nc
# Precipiation = "rain_day"
# SoilMoisture_Upper = "s0_pct"
# http://www.bom.gov.au/jsp/awra/thredds/fileServer/AWRACMS/values/day/ss_pct_2017.nc
# SoilMoisture_Lower = "ss_pct"
# http://www.bom.gov.au/jsp/awra/thredds/fileServer/AWRACMS/values/day/sd_pct_2017.nc
# SoilMoisture_Deep = "sd_pct"
# http://www.bom.gov.au/jsp/awra/thredds/fileServer/AWRACMS/values/day/sm_pct_2017.nc
# SoilMoisture_RootZone = "sm_pct" # http://www.bom.gov.au/jsp/awra/thredds/fileServer/AWRACMS/values/day/qtot_2017.nc # Runoff = "qtot"
# http://www.bom.gov.au/jsp/awra/thredds/fileServer/AWRACMS/values/day/etot_2017.nc
# Evapotrans_Actual = "etot"
# http://www.bom.gov.au/jsp/awra/thredds/fileServer/AWRACMS/values/day/e0_2017.nc
# Evapotrans_Potential_Landscape = "e0"
# http://www.bom.gov.au/jsp/awra/thredds/fileServer/AWRACMS/values/day/ma_wet_2017.nc
# Evapotrans_Potential_Areal = "ma_wet"
# http://www.bom.gov.au/jsp/awra/thredds/fileServer/AWRACMS/values/day/pen_pet_2017.nc
# Evapotrans_Potential_SyntheticPan = "pen_pet"
# http://www.bom.gov.au/jsp/awra/thredds/fileServer/AWRACMS/values/day/fao_pet_2017.nc
# Evapotrans_RefCrop_Short = "fao_pet"
# http://www.bom.gov.au/jsp/awra/thredds/fileServer/AWRACMS/values/day/asce_pet_2017.nc
# Evapotrans_RefCrop_Tall = "asce_pet"
# http://www.bom.gov.au/jsp/awra/thredds/fileServer/AWRACMS/values/day/etot_2017.nc
# http://www.bom.gov.au/jsp/awra/thredds/fileServer/AWRACMS/values/day/msl_wet_2017.nc
# Evaporation_OpenWater = "msl_wet"
# http://www.bom.gov.au/jsp/awra/thredds/fileServer/AWRACMS/values/day/dd_2017.nc
# DeepDrainage = "dd"
"""
getraster(source::Type{<:ALWB{Union{Deciles,Values},Union{Day,Month,Year}}}, [layer]; date)
Download [`ALWB`](@ref) weather data from
[www.bom.gov.au/water/landscape](http://www.bom.gov.au/water/landscape) as values or
deciles with timesteps of `Day`, `Month` or `Year`.
# Arguments
- `layer`: `Symbol` or `Tuple` of `Symbol` from `$(layers(ALWB))`. Without a
`layer` argument, all layers will be downloaded, and a `NamedTuple` of paths returned.
# Keywords
- `date`: a `DateTime`, `AbstractVector` of `DateTime` or a `Tuple` of start and end dates.
For multiple dates, a `Vector` of multiple filenames will be returned.
ALWB is available with a daily, monthly, and yearly, timestep.
# Example
This will return the file containing annual averages, including your date:
```julia
julia> getraster(ALWB{Values,Year}, :ss_pct; date=Date(2001, 2))
"/your/RASTERDATASOURCES_PATH/ALWB/values/month/ss_pct.nc"
```
Returns the filepath/s of the downloaded or pre-existing files.
"""
function getraster(T::Type{<:ALWB}, layers::Union{Tuple,Symbol}; date)
_getraster(T, layers, date)
end
getraster_keywords(::Type{<:ALWB}) = (:date,)
function _getraster(T::Type{<:ALWB{M,P}}, layers, dates::Tuple) where {M,P}
_getraster(T, layers, date_sequence(T, dates))
end
function _getraster(T::Type{<:ALWB}, layers, dates::AbstractArray)
_getraster.(T, Ref(layers), dates)
end
function _getraster(T::Type{<:ALWB}, layers::Tuple, date::Dates.TimeType)
_map_layers(T, layers, date)
end
function _getraster(T::Type{<:ALWB}, layer::Symbol, date::Dates.TimeType)
_check_layer(T, layer)
mkpath(rasterpath(T))
url = rasterurl(T, layer; date=date)
path = rasterpath(T, layer; date=date)
_maybe_download(url, path)
path
end
rastername(T::Type{<:ALWB{M,P}}, layer; date) where {M,P} =
string(layer, _pathsegment(P, date), ".nc")
rasterpath(::Type{ALWB}) = joinpath(rasterpath(), "ALWB")
rasterpath(::Type{ALWB{M,P}}) where {M,P} =
joinpath(joinpath(rasterpath(), "ALWB"), map(_pathsegment, (M, P))...)
rasterpath(T::Type{<:ALWB}, layer; date=nothing) =
joinpath(rasterpath(T), rastername(T, layer; date))
rasterurl(T::Type{<:ALWB{M,P}}, layer; date) where {M,P} =
joinpath(ALWB_URI, _pathsegments(T)..., rastername(T, layer; date))
# Utility methods
_pathsegments(::Type{ALWB{M,P}}) where {M,P} = _pathsegment(M), _pathsegment(P)
_pathsegment(::Type{Values}) = "values"
_pathsegment(::Type{Deciles}) = "deciles"
_pathsegment(::Type{Day}) = "day"
_pathsegment(::Type{Month}) = "month"
_pathsegment(::Type{Year}) = "year"
# Days are in whole-year files
_pathsegment(::Type{Day}, date) = "_" * string(year(date))
# Months and years are all in one file
_pathsegment(::Type{<:Union{Year,Month}}, date) = ""
| RasterDataSources | https://github.com/EcoJulia/RasterDataSources.jl.git |
|
[
"MIT"
] | 0.6.1 | dfab74c3efb21c6df589f30f7bbe5b9b8dbbe3c9 | code | 3824 | struct AWAP <: RasterDataSource end
layers(::Type{AWAP}) = (:solar, :rainfall, :vprpress09, :vprpress15, :tmin, :tmax)
date_step(::Type{<:AWAP}) = Day(1)
# AWAP files dont all have matching extents.
has_matching_layer_size(::Type{<:AWAP}) = false
@doc """
AWAP <: RasterDataSource
Daily weather data from the Australian Water Availability Project, developed by CSIRO.
See: [www.csiro.au/awap](http://www.csiro.au/awap/)
The available layers are: `$(layers(AWAP))`.
""" AWAP
const AWAP_PATHSEGMENTS = (
solar = ("solar", "solarave", "daily"),
rainfall = ("rainfall", "totals", "daily"),
vprpress09 = ("vprp", "vprph09", "daily"),
vprpress15 = ("vprp", "vprph15", "daily"),
tmin = ("temperature", "minave", "daily"),
tmax = ("temperature", "maxave", "daily"),
)
# Add ndvi monthly? ndvi, ndviave, month
"""
getraster(source::Type{AWAP}, [layer]; date)
Download data from the [`AWAP`](@ref) weather dataset, from
[www.csiro.au/awap](http://www.csiro.au/awap/).
The AWAP dataset contains ASCII `.grid` files.
# Arguments
- `layer` `Symbol` or `Tuple` of `Symbol` for `layer`s in `$(layers(AWAP))`. Without a
`layer` argument, all layers will be downloaded, and a `NamedTuple` of paths returned.
# Keywords
- `date`: a `DateTime`, `AbstractVector` of `DateTime` or a `Tuple` of start and end dates.
For multiple dates, A `Vector` of multiple filenames will be returned.
AWAP is available with a daily timestep.
# Example
Download rainfall for the first month of 2001:
```julia
julia> getraster(AWAP, :rainfall; date=Date(2001, 1, 1):Day(1):Date(2001, 1, 31))
31-element Vector{String}:
"/your/path/AWAP/rainfall/totals/20010101.grid"
"/your/path/AWAP/rainfall/totals/20010102.grid"
...
"/your/path/AWAP/rainfall/totals/20010131.grid"
```
Returns the filepath/s of the downloaded or pre-existing files.
"""
getraster(T::Type{AWAP}, layer::Union{Tuple,Symbol}; date) = _getraster(T, layer, date)
getraster_keywords(::Type{<:AWAP}) = (:date,)
function _getraster(T::Type{AWAP}, layer::Union{Tuple,Symbol}, dates::Tuple{<:Any,<:Any})
_getraster(T, layer, date_sequence(T, dates))
end
function _getraster(T::Type{AWAP}, layers::Union{Tuple,Symbol}, dates::AbstractArray)
_getraster.(T, Ref(layers), dates)
end
function _getraster(T::Type{<:AWAP}, layers::Tuple, date::Dates.TimeType)
_map_layers(T, layers, date)
end
function _getraster(T::Type{AWAP}, layer::Symbol, date::Dates.TimeType)
_check_layer(T, layer)
mkpath(_rasterpath(T, layer))
raster_path = rasterpath(T, layer; date=date)
if !isfile(raster_path)
zip_path = zippath(T, layer; date=date)
_maybe_download(zipurl(T, layer; date=date), zip_path)
run(`uncompress $zip_path -f`)
end
return raster_path
end
rasterpath(T::Type{AWAP}) = joinpath(rasterpath(), "AWAP")
rasterpath(T::Type{AWAP}, layer; date::Dates.AbstractTime) =
joinpath(_rasterpath(T, layer), rastername(T, layer; date))
_rasterpath(T::Type{AWAP}, layer) = joinpath(rasterpath(T), AWAP_PATHSEGMENTS[layer][1:2]...)
rastername(T::Type{AWAP}, layer; date::Dates.AbstractTime) =
joinpath(_date2string(T, date) * ".grid")
function zipurl(T::Type{AWAP}, layer; date)
s = AWAP_PATHSEGMENTS[layer]
d = _date2string(T, date)
# The actual zip name has the date twice, which is weird.
# So we getraster in to a different name as there no output
# name flages for `uncompress`. It's ancient.
uri = URI(scheme="http", host="www.bom.gov.au", path="/web03/ncc/www/awap")
joinpath(uri, s..., "grid/0.05/history/nat/$d$d.grid.Z")
end
zipname(T::Type{AWAP}, layer; date) = _date2string(T, date) * ".grid.Z"
zippath(T::Type{AWAP}, layer; date) =
joinpath(_rasterpath(T, layer), zipname(T, layer; date))
_dateformat(::Type{AWAP}) = DateFormat("yyyymmdd")
| RasterDataSources | https://github.com/EcoJulia/RasterDataSources.jl.git |
|
[
"MIT"
] | 0.6.1 | dfab74c3efb21c6df589f30f7bbe5b9b8dbbe3c9 | code | 4425 | layers(::Type{CHELSA{BioClim}}) = layers(BioClim)
layers(::Type{CHELSA{BioClimPlus}}) = layers(BioClimPlus)
layerkeys(::Type{CHELSA{BioClim}}, args...) = layerkeys(BioClim, args...)
layerkeys(::Type{CHELSA{BioClimPlus}}, args...) = layerkeys(BioClimPlus, args...)
"""
getraster(source::Type{CHELSA{BioClim}}, [layer]; version = 2, [patch]) => Union{Tuple,String}
Download [`CHELSA`](@ref) [`BioClim`](@ref) data from [chelsa-climate.org](https://chelsa-climate.org/).
# Arguments
- `layer`: `Integer` or tuple/range of `Integer` from `$(layers(BioClim))`,
or `Symbol`s form `$(layerkeys(BioClim))`. Without a `layer` argument, all layers
will be downloaded, and a `NamedTuple` of paths returned.
# Keyword arguments
$CHELSA_KEYWORDS
Returns the filepath/s of the downloaded or pre-existing files.
"""
getraster(
T::Type{CHELSA{BioClim}},
layer::Union{Tuple,Int,Symbol};
version::Int = 2,
patch::Int = latest_patch(T, version)) = _getraster(T, layer, version, patch)
_getraster(T::Type{CHELSA{BioClim}}, layers::Tuple, version, patch) = _map_layers(T, layers, version, patch)
_getraster(T::Type{CHELSA{BioClim}}, layer::Symbol, version, patch) = _getraster(T, bioclim_int(layer), version, patch)
function _getraster(T::Type{CHELSA{BioClim}}, layer::Integer, version, patch)
_check_layer(T, layer)
path = rasterpath(T, layer; version, patch)
url = rasterurl(T, layer; version, patch)
CHELSA_warn_version(T, layer, version, patch, path)
return _maybe_download(url, path)
end
getraster_keywords(::Type{<:CHELSA{BioClim}}) = (:version,:patch)
function rastername(::Type{CHELSA{BioClim}}, layer::Integer; version::Int = 2, patch = latest_patch(CHELSA, version))
if version == 1
"CHELSA_bio10_$(lpad(layer, 2, "0")).tif"
elseif version == 2
"CHELSA_bio$(layer)_1981-2010_V.2.$patch.tif"
else
CHELSA_invalid_version(version)
end
end
rasterpath(::Type{CHELSA{BioClim}}) = joinpath(rasterpath(CHELSA), "BioClim")
rasterpath(T::Type{CHELSA{BioClim}}, layer::Integer; version = 2, patch = latest_patch(CHELSA, version)) = joinpath(rasterpath(T), rastername(T, layer; version, patch))
function rasterurl(::Type{CHELSA{BioClim}}; version)
if version == 1
joinpath(rasterurl(CHELSA, version), "climatologies/")
elseif version == 2
joinpath(rasterurl(CHELSA, version), "climatologies/1981-2010/")
else
CHELSA_invalid_version(version)
end
end
rasterurl(T::Type{CHELSA{BioClim}}, layer::Integer; version = 2, patch = latest_patch(CHELSA, version)) =
joinpath(rasterurl(T; version), "bio", rastername(T, layer; version, patch))
### Bioclim+
"""
getraster(source::Type{CHELSA{BioClim}}, [layer]; version = 2, [patch]) => Union{Tuple,String}
Download [`CHELSA`](@ref) [`BioClim`](@ref) data from [chelsa-climate.org](https://chelsa-climate.org/).
# Arguments
- `layer`: iterable of `Symbol`s from `$(layerkeys(BioClimPlus))`. Without a `layer` argument, all layers
will be downloaded, and a `NamedTuple` of paths returned.
# Keyword arguments
$CHELSA_KEYWORDS
Returns the filepath/s of the downloaded or pre-existing files.
"""
getraster(
T::Type{CHELSA{BioClimPlus}},
layer::Union{Tuple,Int,Symbol};
version::Int = 2,
patch::Int = latest_patch(T, version)) = _getraster(T, layer, version, patch)
_getraster(T::Type{CHELSA{BioClimPlus}}, layers::Tuple, version, patch) = _map_layers(T, layers, version, patch)
function _getraster(T::Type{CHELSA{BioClimPlus}}, layer::Symbol, version, patch)
version == 2 || CHELSA_invalid_version(version, 2)
_check_layer(T, layer)
path = rasterpath(T, layer; version, patch)
url = rasterurl(T, layer; version, patch)
return _maybe_download(url, path)
end
getraster_keywords(::Type{<:CHELSA{BioClimPlus}}) = (:version,:patch)
rastername(T::Type{CHELSA{BioClimPlus}}, layer::Symbol; version = 2, patch = latest_patch(T, version)) = "CHELSA_$(layer)_1981-2010_V.2.$patch.tif"
rasterpath(::Type{CHELSA{BioClimPlus}}) = rasterpath(CHELSA{BioClim})
rasterpath(T::Type{CHELSA{BioClimPlus}}, layer::Symbol; version = 2, patch = latest_patch(T, version)) = joinpath(rasterpath(T), rastername(T, layer; version, patch))
rasterurl(T::Type{CHELSA{BioClimPlus}}, layer::Symbol; version = 2, patch = latest_patch(T, version)) =
joinpath(rasterurl(CHELSA{BioClim}; version), "bio", rastername(T, layer; version, patch))
| RasterDataSources | https://github.com/EcoJulia/RasterDataSources.jl.git |
|
[
"MIT"
] | 0.6.1 | dfab74c3efb21c6df589f30f7bbe5b9b8dbbe3c9 | code | 1817 | layers(::Type{CHELSA{Climate}}) = (:clt, :cmi, :hurs, :ncdf, :pet, :pr, :rsds, :sfcWind, :tas, :tasmax, :tasmin, :vpd)
"""
getraster(T::Type{CHELSA{Climate}}, [layer::Union{Tuple,Symbol}]; month) => Vector{String}
Download [`CHELSA`](@ref) [`Climate`](@ref) data.
# Arguments
- `layer` `Symbol` or `Tuple` of `Symbol` from `$(layers(CHELSA{Climate}))`.
# Keywords
- `month`: `Integer` or `AbstractArray` of `Integer`. Chosen from `1:12`.
Returns the filepath/s of the downloaded or pre-existing files.
"""
function getraster(T::Type{CHELSA{Climate}}, layers::Union{Tuple,Symbol}; month)
_getraster(T, layers, month)
end
getraster_keywords(::Type{CHELSA{Climate}}) = (:month,)
function _getraster(T::Type{CHELSA{Climate}}, layers, month::AbstractArray)
_getraster.(T, Ref(layers), month)
end
function _getraster(T::Type{CHELSA{Climate}}, layers::Tuple, month::Integer)
_map_layers(T, layers, month)
end
function _getraster(T::Type{CHELSA{Climate}}, layer::Symbol, month::Integer)
_check_layer(T, layer)
path = rasterpath(T, layer; month)
url = rasterurl(T, layer; month)
return _maybe_download(url, path)
end
# Climate layers don't get their own folder
rasterpath(T::Type{<:CHELSA{Climate}}, layer; month) =
joinpath(_rasterpath(T, layer), rastername(T, layer; month))
_rasterpath(T::Type{<:CHELSA{Climate}}, layer) = joinpath(rasterpath(T), string(layer))
rasterpath(T::Type{<:CHELSA{Climate}}) = joinpath(rasterpath(CHELSA), "Climate")
function rastername(T::Type{<:CHELSA{Climate}}, layer; month)
_layer = layer == :pet ? :pet_penman : layer
"CHELSA_$(_layer)_$(_pad2(month))_1981-2010_V.2.1.tif"
end
rasterurl(T::Type{CHELSA{Climate}}, layer::Symbol; month) = joinpath(rasterurl(CHELSA, 2), "climatologies/1981-2010", string(layer), rastername(T, layer; month)) | RasterDataSources | https://github.com/EcoJulia/RasterDataSources.jl.git |
|
[
"MIT"
] | 0.6.1 | dfab74c3efb21c6df589f30f7bbe5b9b8dbbe3c9 | code | 12347 |
layers(::Type{<:CHELSA{<:Future{BioClim}}}) = layers(BioClim)
layers(::Type{<:CHELSA{T}}) where T <:Future{BioClimPlus} = layers(T)
layerkeys(T::Type{<:CHELSA{<:Future{BioClim}}}, args...) = layerkeys(BioClim, args...)
layers(::Type{<:CHELSA{<:Future{Climate}}}) = (:prec, :temp, :tmin, :tmax)
date_step(::Type{<:CHELSA{<:Future{Climate,CMIP5}}}) = Year(20)
date_step(::Type{<:CHELSA{<:Future{Climate,CMIP6}}}) = Year(30)
# A modified key is used in the file name, while the key is used as-is in the path
const CHELSAKEY = (prec="pr", temp="tas", tmin="tasmin", tmax="tasmax", bio="bio")
"""
getraster(T::Type{CHELSA{Future{BioClim}}}, [layer]; date) => String
Download CHELSA [`BioClim`](@ref) data, choosing layers from: `$(layers(CHELSA{BioClim}))`.
See the docs for [`Future`](@ref) for model choices.
Without a layer argument, all layers will be downloaded, and a `NamedTuple` of paths
returned.
## Keywords
- `date`: a `Date` or `DateTime` object, a Vector, or Tuple of start/end dates.
Note that CHELSA CMIP5 only has two datasets, for the periods 2041-2060 and
2061-2080. CMIP6 has datasets for the periods 2011-2040, 2041-2070, and 2071-2100.
Dates must fall within these ranges.
## Example
```julia
using RasterDataSources, Dates
getraster(CHELSA{Future{BioClim, CMIP6, GFDLESM4, SSP370}}, 1, date = Date(2050))
```
"""
function getraster(
T::Type{<:CHELSA{<:Future{BioClim}}}, layers::Union{Tuple,Int,Symbol}; date
)
_getraster(T, layers, date)
end
getraster_keywords(::Type{<:CHELSA{<:Future{BioClim}}}) = (:date,)
"""
getraster(T::Type{CHELSA{Future{BioClimPlus}}}, [layer]; date) => String
Download CHELSA [`BioClimPlus`](@ref) data, choosing layers from: `$(layers(CHELSA{BioClimPlus}))`.
See the docs for [`Future`](@ref) for model choices.
Without a layer argument, all layers will be downloaded, and a `NamedTuple` of paths
returned.
## Keywords
- `date`: a `Date` or `DateTime` object, a Vector, or Tuple of start/end dates.
Note that CHELSA CMIP5 only has two datasets, for the periods 2041-2060 and
2061-2080. CMIP6 has datasets for the periods 2011-2040, 2041-2070, and 2071-2100.
Dates must fall within these ranges.
## Example
"""
function getraster(
T::Type{<:CHELSA{<:Future{BioClimPlus}}}, layers::Union{Tuple,Int,Symbol}; date
)
_getraster(T, layers, date)
end
getraster_keywords(::Type{<:CHELSA{<:Future{BioClimPlus}}}) = (:date,)
"""
getraster(T::Type{CHELSA{Future{Climate}}}, [layer]; date, month) => String
Download CHELSA [`Climate`](@ref) data, choosing layers from: `$(layers(CHELSA{BioClim}))`.
See the docs for [`Future`](@ref) for model choices.
Without a layer argument, all layers will be downloaded, and a `NamedTuple` of paths returned.
## Keywords
- `date`: a `Date` or `DateTime` object, a Vector, or Tuple of start/end dates.
Note that CHELSA CMIP5 only has two datasets, for the periods 2041-2060 and
2061-2080. CMIP6 has datasets for the periods 2011-2040, 2041-2070, and 2071-2100.
Dates must fall within these ranges.
- `month`: the month of the year, from 1 to 12, or a array or range of months like `1:12`.
## Example
```
using Dates, RasterDataSources
getraster(CHELSA{Future{Climate, CMIP6, GFDLESM4, SSP370}}, :prec; date = Date(2050), month = 1)
```
"""
function getraster(
T::Type{<:CHELSA{<:Future{Climate}}}, layers::Union{Tuple,Symbol}; date, month
)
_getraster(T, layers, date, month)
end
getraster_keywords(::Type{<:CHELSA{<:Future{Climate}}}) = (:date, :month)
function _getraster(T::Type{<:CHELSA{<:Future{Climate}}}, layers, date, months::AbstractArray)
map(month -> _getraster(T, layers, date, month), months)
end
function _getraster(
T::Type{<:CHELSA{<:Future{Climate}}}, layers, dates::AbstractArray, months::AbstractArray
)
map(date -> _getraster(T, layers, date, months), dates)
end
function _getraster(T::Type{<:CHELSA{<:Future{Climate}}}, layers, dates::AbstractArray, month)
map(date -> _getraster(T, layers; date, month), dates)
end
function _getraster(T::Type{<:CHELSA{<:Future{Climate}}}, layers, dates::Tuple, months::AbstractArray)
_getraster(T, layers, date_sequence(T, dates), months)
end
function _getraster(T::Type{<:CHELSA{<:Future{Climate}}}, layers, dates::Tuple, month)
_getraster(T, layers, date_sequence(T, dates), month)
end
function _getraster(T::Type{<:CHELSA{<:Future{Climate}}}, layers, date, month)
_getraster(T, layers; date, month)
end
function _getraster(T::Type{<:CHELSA{<:Future{BioClim}}}, layers, dates::AbstractArray)
map(date -> _getraster(T, layers, date), dates)
end
function _getraster(T::Type{<:CHELSA{<:Future{BioClim}}}, layers, date::TimeType)
_getraster(T, layers; date)
end
function _getraster(T::Type{<:CHELSA{<:Future{BioClimPlus}}}, layers, dates::AbstractArray)
map(date -> _getraster(T, layers, date), dates)
end
function _getraster(T::Type{<:CHELSA{<:Future{BioClimPlus}}}, layers, date::TimeType)
_getraster(T, layers; date)
end
# We have the extra args as keywords again to generalise rasterpath/rasterurl
function _getraster(T::Type{<:CHELSA{<:Future}}, layers::Tuple; kw...)
_map_layers(T, layers; kw...)
end
_getraster(T::Type{<:CHELSA{<:Future{BioClim}}}, layer::Symbol; kw...) = _getraster(T, bioclim_int(layer); kw...)
function _getraster(T::Type{<:CHELSA{<:Future}}, layer::Union{Symbol,Integer}; kw...)
_check_layer(T, layer)
path = rasterpath(T, layer; kw...)
url = rasterurl(T, layer; kw...)
return _maybe_download(url, path)
end
function rastername(T::Type{<:CHELSA{<:Future}}, layer; kw...)
_rastername(_phase(T), T, layer; kw...)
end
function _rastername(
::Type{CMIP5}, T::Type{<:CHELSA{<:Future{BioClim}}}, layer::Integer; date
)
date_string = _date_string(_phase(T), date)
mod = _format(CHELSA, _model(T))
scen = _format(CHELSA, _scenario(T))
return "CHELSA_bio_mon_$(mod)_$(scen)_r1i1p1_g025.nc_$(layer)_$(date_string)_V1.2.tif"
end
function _rastername(
::Type{CMIP5}, T::Type{<:CHELSA{<:Future{Climate}}}, layer::Symbol; date, month
)
date_string = _date_string(_phase(T), date)
mod = _format(CHELSA, _model(T))
scen = _format(CHELSA, _scenario(T))
key = CHELSAKEY[layer]
suffix = layer === :prec ? "" : "_V1.2" # prec filenames dont end in _V1.2
return "CHELSA_$(key)_mon_$(mod)_$(scen)_r1i1p1_g025.nc_$(month)_$(date_string)$(suffix).tif"
end
function _rastername(::Type{CMIP6}, T::Type{<:CHELSA{<:Future{BioClim}}}, layer::Integer; date)
date_string = _date_string(_phase(T), date)
mod = _format(CHELSA, _model(T))
scen = _format(CHELSA, _scenario(T))
return "CHELSA_bio$(layer)_$(date_string)_$(mod)_$(scen)_V.2.1.tif"
end
function _rastername(::Type{CMIP6}, T::Type{<:CHELSA{<:Future{BioClimPlus}}}, layer::Symbol; date)
date_string = _date_string(_phase(T), date)
mod = _format(CHELSA, _model(T))
scen = _format(CHELSA, _scenario(T))
return "CHELSA_$(layer)_$(date_string)_$(mod)_$(scen)_V.2.1.tif"
end
function _rastername(
::Type{CMIP6}, T::Type{<:CHELSA{<:Future{Climate}}}, layer::Symbol; date, month
)
# CMIP6 Climate uses an underscore in the date string, of course
date_string = replace(_date_string(_phase(T), date), "-" => "_")
mod = _format(CHELSA, _model(T))
scen = _format(CHELSA, _scenario(T))
key = CHELSAKEY[layer]
mon = lpad(month, 2, '0')
return "CHELSA_$(mod)_r1i1p1f1_w5e5_$(scen)_$(key)_$(mon)_$(date_string)_norm.tif"
end
function rasterpath(T::Type{<:CHELSA{<:Future}})
joinpath(rasterpath(CHELSA), "Future", string(_dataset(T)), string(_scenario(T)), string(_model(T)))
end
function rasterpath(T::Type{<:CHELSA{<:Future}}, layer; kw...)
joinpath(rasterpath(T), rastername(T, layer; kw...))
end
function rasterurl(T::Type{<:CHELSA{<:Future}}, layer; date, kw...)
date_str = _date_string(_phase(T), date)
key = _chelsa_layer(_dataset(T), layer)
path = _urlpath(_phase(T), T::Type{<:CHELSA{<:Future}}, key, date_str)
joinpath(rasterurl(CHELSA), path, rastername(T, layer; date, kw...))
end
_chelsa_layer(::Type{<:BioClim}, layer) = :bio
_chelsa_layer(::Type{<:BioClimPlus}, layer) = :bio
_chelsa_layer(::Type{<:Climate}, layer) = layer
function _urlpath(::Type{CMIP5}, T::Type{<:CHELSA{<:Future}}, name, date_str)
return "chelsav1/cmip5/$date_str/$name/"
end
function _urlpath(::Type{CMIP6}, T::Type{<:CHELSA{<:Future}}, name, date_str)
# The model is in uppercase in the URL for CMIP6
mod = uppercase(_format(CHELSA, _model(T)))
scen = _format(CHELSA, _scenario(T))
key = CHELSAKEY[name]
return "chelsav2/GLOBAL/climatologies/$date_str/$mod/$scen/$key/"
end
function _date_string(::Type{CMIP5}, date)
if date < DateTime(2041)
_cmip5_date_error(date)
elseif date < DateTime(2061)
"2041-2060"
elseif date < DateTime(2081)
"2061-2080"
else
_cmip5_date_error(date)
end
end
function _date_string(::Type{CMIP6}, date)
if date < DateTime(1981)
_cmip6_date_error(date)
elseif date < DateTime(2011)
"1981-2010"
elseif date < DateTime(2041)
"2011-2040"
elseif date < DateTime(2071)
"2041-2070"
elseif date < DateTime(2101)
"2071-2100"
else
_cmip6_date_error(date)
end
end
_cmip5_date_error(date) = error("CMIP5 covers the period from 2041-2080, not including $date")
_cmip6_date_error(date) = error("CMIP6 covers the period from 1981-2100, not including $date")
_dataset(::Type{<:CHELSA{F}}) where F<:Future = _dataset(F)
_phase(::Type{<:CHELSA{F}}) where F<:Future = _phase(F)
_model(::Type{<:CHELSA{F}}) where F<:Future = _model(F)
_scenario(::Type{<:CHELSA{F}}) where F<:Future = _scenario(F)
# Climate model string formatters for CHELSA Future
# CMIP5
_format(::Type{CHELSA}, ::Type{ACCESS1}) = "ACCESS1-0"
_format(::Type{CHELSA}, ::Type{BNUESM}) = "BNU-ESM"
_format(::Type{CHELSA}, ::Type{CCSM4}) = "CCSM4"
_format(::Type{CHELSA}, ::Type{CESM1BGC}) = "CESM1-BGC"
_format(::Type{CHELSA}, ::Type{CESM1CAM5}) = "CESM1-CAM5"
_format(::Type{CHELSA}, ::Type{CMCCCMS}) = "CMCC-CMS"
_format(::Type{CHELSA}, ::Type{CMCCCM}) = "CMCC-CM"
_format(::Type{CHELSA}, ::Type{CNRMCM5}) = "CNRM-CM5"
_format(::Type{CHELSA}, ::Type{CSIROMk3}) = "CSIRO-Mk3"
_format(::Type{CHELSA}, ::Type{CanESM2}) = "CanESM2"
_format(::Type{CHELSA}, ::Type{FGOALS}) = "FGOALS-g2"
_format(::Type{CHELSA}, ::Type{FIOESM}) = "FIO-ESM"
_format(::Type{CHELSA}, ::Type{GFDLCM3}) = "GFDL-CM3"
_format(::Type{CHELSA}, ::Type{GFDLESM2G}) = "GFDL-ESM2G"
_format(::Type{CHELSA}, ::Type{GFDLESM2M}) = "GFDL-ESM2M"
_format(::Type{CHELSA}, ::Type{GISSE2HCC}) = "GISS-E2-H-CC"
_format(::Type{CHELSA}, ::Type{GISSE2H}) = "GISS-E2-H"
_format(::Type{CHELSA}, ::Type{GISSE2RCC}) = "GISS-E2-R-CC"
_format(::Type{CHELSA}, ::Type{GISSE2R}) = "GISS-E2-R"
_format(::Type{CHELSA}, ::Type{HadGEM2AO}) = "HadGEM2-AO"
_format(::Type{CHELSA}, ::Type{HadGEM2CC}) = "HadGEM2-CC"
_format(::Type{CHELSA}, ::Type{IPSLCM5ALR}) = "IPSL-CM5A-LR"
_format(::Type{CHELSA}, ::Type{IPSLCM5AMR}) = "IPSL-CM5A-MR"
_format(::Type{CHELSA}, ::Type{MIROCESMCHEM}) = "MIROC-ESM-CHEM"
_format(::Type{CHELSA}, ::Type{MIROCESM}) = "MIROC-ESM"
_format(::Type{CHELSA}, ::Type{MIROC5}) = "MIROC5"
_format(::Type{CHELSA}, ::Type{MPIESMLR}) = "MPI-ESM-LR"
_format(::Type{CHELSA}, ::Type{MPIESMMR}) = "MPI-ESM-MR"
_format(::Type{CHELSA}, ::Type{MRICGCM3}) = "MRI-CGCM3"
_format(::Type{CHELSA}, ::Type{MRIESM1}) = "MRI-ESM1"
_format(::Type{CHELSA}, ::Type{NorESM1M}) = "NorESM1-M"
_format(::Type{CHELSA}, ::Type{BCCCSM1}) = "bcc-csm-1"
_format(::Type{CHELSA}, ::Type{Inmcm4}) = "inmcm4"
# CMIP6
_format(::Type{CHELSA}, ::Type{GFDLESM4}) = "gfdl-esm4"
_format(::Type{CHELSA}, ::Type{IPSLCM6ALR}) = "ipsl-cm6a-lr"
_format(::Type{CHELSA}, ::Type{MPIESMHR}) = "mpi-esm1-2-hr"
_format(::Type{CHELSA}, ::Type{MRIESM2}) = "mri-esm2-0"
_format(::Type{CHELSA}, ::Type{UKESM}) = "ukesm1-0-ll"
# Format scenarios
_format(::Type{CHELSA}, ::Type{RCP26}) = "rcp26"
_format(::Type{CHELSA}, ::Type{RCP45}) = "rcp45"
_format(::Type{CHELSA}, ::Type{RCP60}) = "rcp60"
_format(::Type{CHELSA}, ::Type{RCP85}) = "rcp85"
_format(::Type{CHELSA}, ::Type{SSP126}) = "ssp126"
_format(::Type{CHELSA}, ::Type{SSP245}) = "ssp245"
_format(::Type{CHELSA}, ::Type{SSP370}) = "ssp370"
_format(::Type{CHELSA}, ::Type{SSP585}) = "ssp585"
| RasterDataSources | https://github.com/EcoJulia/RasterDataSources.jl.git |
|
[
"MIT"
] | 0.6.1 | dfab74c3efb21c6df589f30f7bbe5b9b8dbbe3c9 | code | 1632 | """
CHELSA{Union{BioClim,BioClimPlus,Climate,<:Future}} <: RasterDataSource
Data from CHELSA, currently implements the `BioClim` `BioClimPlus`, and `Climate`
variables for current and future conditions.
See: [chelsa-climate.org](https://chelsa-climate.org/) for the dataset,
and the [`getraster`](@ref) docs for implementation details.
"""
struct CHELSA{X} <: RasterDataSource end
rasterpath(::Type{CHELSA}) = joinpath(rasterpath(), "CHELSA")
function rasterurl(T::Type{CHELSA}, version)
if version == 1
joinpath(rasterurl(T), "chelsav1")
elseif version == 2
joinpath(rasterurl(T), "chelsav2/GLOBAL")
else
CHELSA_invalid_version(version)
end
end
rasterurl(::Type{CHELSA}) = URI(scheme="https", host="os.zhdk.cloud.switch.ch", path="")
function latest_patch(::Type{<:CHELSA}, v)
if v == 1
2
elseif v == 2
1
else
CHELSA_invalid_version(v)
end
end
const CHELSA_KEYWORDS = """
- `version`: `Integer` indicating the CHELSA version, currently either `1` or `2`.
- `patch`: `Integer` indicating the CHELSA patch number. Defaults to the latest patch (V1.2 and V2.1)
"""
CHELSA_invalid_version(v, valid_versions = [1,2]) =
throw(ArgumentError("Version $v is not available for CHELSA. Available versions: $valid_versions."))
function CHELSA_warn_version(T, layer, version, patch, path)
if version == 2 && !isfile(path) && isfile(rasterpath(T, layer; version = 1))
@info "File for CHELSA v1.2 detected, but requested version is CHELSA v$version.$patch.
To load data for CHELSA v1.2 instead, set version keyword to 1"
end
end | RasterDataSources | https://github.com/EcoJulia/RasterDataSources.jl.git |
|
[
"MIT"
] | 0.6.1 | dfab74c3efb21c6df589f30f7bbe5b9b8dbbe3c9 | code | 3323 | resolutions(::Type{EarthEnv{HabitatHeterogeneity}}) = ("1km", "5km", "25km")
defres(::Type{EarthEnv{HabitatHeterogeneity}}) = "25km"
layers(::Type{EarthEnv{HabitatHeterogeneity}}) = values(heterogeneity_lookup)
layerkeys(::Type{EarthEnv{HabitatHeterogeneity}}) = keys(heterogeneity_lookup)
layerkeys(T::Type{EarthEnv{HabitatHeterogeneity}}, layers) = map(l -> layerkeys(T, l), layers)
function layerkeys(::Type{EarthEnv{HabitatHeterogeneity}}, layer::Symbol)
Symbol(lowercase(string(layer)))
end
# Yes, they randomly chose cases
const heterogeneity_lookup = (
cv = :cv,
evenness = :evenness,
range = :range,
shannon = :shannon,
simpson = :simpson,
std = :std,
contrast = :Contrast,
correlation = :Correlation,
dissimilarity = :Dissimilarity,
entropy = :Entropy,
homogeneity = :Homogeneity,
maximum = :Maximum,
uniformity = :Uniformity,
variance = :Variance,
)
heterogeneity_layer(x::Symbol) = heterogeneity_lookup[Symbol(lowercase(string(x)))]
"""
getraster(source::Type{EarthEnv{HabitatHeterogeneity}}, [layer]; res="25km")
Download [`EarthEnv`](@ref) habitat heterogeneity data.
# Arguments
- `layer`: `Symbol` or `Tuple` of `Symbol` from `$(layers(EarthEnv{HabitatHeterogeneity}))`.
Without a `layer` argument, all layers will be downloaded, and a `NamedTuple` of paths returned.
# Keywords
- `res`: `String` chosen from `$(resolutions(EarthEnv{HabitatHeterogeneity}))`, defaulting to "25km".
Returns the filepath/s of the downloaded or pre-existing files.
"""
function getraster(T::Type{EarthEnv{HabitatHeterogeneity}}, layers::Union{Tuple,Symbol};
res::String=defres(T)
)
_getraster(T, layers, res)
end
getraster_keywords(::Type{EarthEnv{HabitatHeterogeneity}}) = (:res,)
function _getraster(T::Type{EarthEnv{HabitatHeterogeneity}}, layers::Tuple, res::String)
return _map_layers(T, layers, res)
end
function _getraster(T::Type{EarthEnv{HabitatHeterogeneity}}, layer::Symbol, res::String)
layer = heterogeneity_layer(layer)
_check_layer(T, layer)
_check_res(T, res)
path = rasterpath(T, layer; res)
url = rasterurl(T, layer; res)
return _maybe_download(url, path)
end
function rastername(::Type{EarthEnv{HabitatHeterogeneity}}, layer::Symbol; res::String=defres(T))
"$(layer)_$(res).tif"
end
function rasterpath(::Type{EarthEnv{HabitatHeterogeneity}})
joinpath(rasterpath(EarthEnv), "HabitatHeterogeneity")
end
function rasterpath(T::Type{EarthEnv{HabitatHeterogeneity}}, layer::Symbol; res::String=defres(T))
joinpath(rasterpath(T), string(res), rastername(T, layer; res))
end
function rasterurl(::Type{EarthEnv{HabitatHeterogeneity}})
joinpath(rasterurl(EarthEnv), "habitat_heterogeneity")
end
function rasterurl(T::Type{EarthEnv{HabitatHeterogeneity}}, layer; res::String=defres(T))
prec = _getprecision(layer, res)
layerpath = "$res/$(layer)_01_05_$(res)_$prec.tif"
joinpath(rasterurl(T), layerpath)
end
# See http://www.earthenv.org/texture
function _getprecision(layer, res)
if ((res in ("1km", "5km")) && (layer == :Correlation))
"int16"
elseif ((res == "5km") && (layer == :cv)) ||
((res == "25km") && (layer == :Entropy)) ||
layer in (:Contrast, :Dissimilarity, :Variance)
"uint32"
else
"uint16"
end
end
| RasterDataSources | https://github.com/EcoJulia/RasterDataSources.jl.git |
|
[
"MIT"
] | 0.6.1 | dfab74c3efb21c6df589f30f7bbe5b9b8dbbe3c9 | code | 2657 | layers(::Type{<:EarthEnv{<:LandCover}}) = ntuple(identity, Val{12}())
layerkeys(::Type{<:EarthEnv{<:LandCover}}) = keys(landcover_lookup)
layerkeys(T::Type{<:EarthEnv{<:LandCover}}, layers) = map(l -> layerkeys(T, l), layers)
layerkeys(T::Type{<:EarthEnv{<:LandCover}}, layer::Int) = layerkeys(T)[layer]
layerkeys(T::Type{<:EarthEnv{<:LandCover}}, layer::Symbol) = layer
const landcover_lookup = (
needleleaf_trees = 1,
evergreen_broadleaf_trees = 2,
deciduous_broadleaf_trees = 3,
other_trees = 4,
shrubs = 5,
herbaceous = 6,
cultivated_and_managed = 7,
regularly_flooded = 8,
urban_builtup = 9,
snow_ice = 10,
barren = 11,
open_water = 12,
)
"""
getraster(T::Type{EarthEnv{LandCover}}, [layer]; discover=false) => Union{Tuple,String}
Download [`EarthEnv`](@ref) landcover data.
# Arguments
- `layer`: `Integer` or tuple/range of `Integer` from `$(layers(EarthEnv{LandCover}))`,
or `Symbol`s from `$(layerkeys(EarthEnv{LandCover}))`. Without a `layer` argument,
all layers will be downloaded, and a `NamedTuple` of paths returned.
# Keywords
- `discover::Bool`: whether to download the dataset that integrates the DISCover model.
Returns the filepath/s of the downloaded or pre-existing files.
"""
function getraster(T::Type{<:EarthEnv{<:LandCover}}, layers::Union{Tuple,Int,Symbol})
_getraster(T, layers)
end
_getraster(T::Type{<:EarthEnv{<:LandCover}}, layers::Tuple) = _map_layers(T, layers)
_getraster(T::Type{<:EarthEnv{<:LandCover}}, layer::Symbol) = _getraster(T, landcover_lookup[layer])
function _getraster(T::Type{<:EarthEnv{<:LandCover}}, layer::Integer)
_check_layer(T, layer)
url = rasterurl(T, layer)
path = rasterpath(T, layer)
return _maybe_download(url, path)
end
function rastername(T::Type{<:EarthEnv{<:LandCover}}, layer::Integer)
class = _discover(T) ? "consensus_full" : "Consensus_reduced"
"$(class)_class_" * string(layer) * ".tif"
end
function rasterpath(T::Type{<:EarthEnv{<:LandCover}})
joinpath(rasterpath(EarthEnv), "LandCover", _discover_segment(T))
end
function rasterpath(T::Type{<:EarthEnv{<:LandCover}}, layer::Integer)
joinpath(rasterpath(T), rastername(T, layer))
end
function rasterurl(T::Type{<:EarthEnv{<:LandCover}})
joinpath(rasterurl(EarthEnv), "consensus_landcover", _discover_segment(T))
end
function rasterurl(T::Type{<:EarthEnv{<:LandCover}}, layer::Integer)
joinpath(rasterurl(T), rastername(T, layer))
end
_discover(T::Type{EarthEnv{LandCover{:DISCover}}}) = true
_discover(T::Type{<:EarthEnv{<:LandCover}}) = false
_discover_segment(T) = _discover(T) ? "with_DISCover" : "without_DISCover"
| RasterDataSources | https://github.com/EcoJulia/RasterDataSources.jl.git |
|
[
"MIT"
] | 0.6.1 | dfab74c3efb21c6df589f30f7bbe5b9b8dbbe3c9 | code | 393 | """
EarthEnv{Union{HabitatHeterogeneity,LandCover}} <: RasterDataSource
Data from the `EarthEnv` including `HabitatHeterogeneity` and `LandCover`
See: [www.earthenv.org](http://www.earthenv.org/)
"""
struct EarthEnv{X} <: RasterDataSource end
rasterpath(::Type{EarthEnv}) = joinpath(rasterpath(), "EarthEnv")
rasterurl(::Type{EarthEnv}) = URI(scheme="https", host="data.earthenv.org")
| RasterDataSources | https://github.com/EcoJulia/RasterDataSources.jl.git |
|
[
"MIT"
] | 0.6.1 | dfab74c3efb21c6df589f30f7bbe5b9b8dbbe3c9 | code | 924 | """
Example parameter sets for getraster(MODIS{MOD13Q1}, ...)
"""
"""
A semi-urban spot in the middle of the Crozon peninsula, West France
Usage : `getraster(MOD13Q1, :NDVI; RasterDataSources.crozon...)`
"""
const crozon = (lat = 48.24, lon = -4.5, km_ab = 1, km_lr = 1, date = "2012-02-02")
"""
Whole Britanny area, western France
Usage : `getraster(MOD13Q1, :NDVI; RasterDataSources.britanny...)`
"""
const britanny =
(lat = 48.25, lon = -3.5, km_ab = 100, km_lr = 100, date = Date("2012-02-02"))
"""
Two years of a single MODIS pixel in Broceliande forest
"""
const broceliande = (
lat = 48.02458,
lon = -2.24057,
km_ab = 0,
km_lr = 0,
date = (Date("2012-02-02"), Date("2014-02-02")),
)
"""
Three dates in Crozon
"""
const crozon2 = (
lat = 48.24,
lon = -4.5,
km_ab = 1,
km_lr = 1,
date = ["2012-02-02", "2013-02-02", "2014-02-02"],
)
| RasterDataSources | https://github.com/EcoJulia/RasterDataSources.jl.git |
|
[
"MIT"
] | 0.6.1 | dfab74c3efb21c6df589f30f7bbe5b9b8dbbe3c9 | code | 3815 | """
This file contains functions to handle MODIS product info.
Depending on missions and products, MODIS data does not have the
same layers.
"""
"""
product(T::Type{<:ModisProduct})
Extracts `ModisProduct` product name as a `String`
"""
function product(T::Type{<:ModisProduct})
return String(nameof(T))
end
"""
Lists available layers for a given MODIS Product
Looks in `joinpath(ENV["RASTERDATASOURCES_PATH"]/MODIS/layers` for
a file with the right name. If not found, sends a request to the server
to get the list.
This allows to make as many internal calls of layers() and layerkeys() as
needed without issuing a lot of requests.
"""
function list_layers(T::Type{<:ModisProduct})
prod = product(T)
path = joinpath(ENV["RASTERDATASOURCES_PATH"], "MODIS/layers", prod * ".csv")
if isfile(path)
layers = open(path, "r") do f
readline(f)
end
else # if not on disk we download layers info
@info "Starting download of layers list for product $prod"
mkpath(dirname(path))
r = HTTP.download(
join([string(MODIS_URI), prod, "bands"], "/"),
path,
["Accept" => "text/csv"],
)
# read downloaded file
layers = open(path, "r") do f
readline(f)
end
end
return split(String(layers), ",")
end
"""
List available dates for a MODIS product at given coordinates
"""
function list_dates(
T::Type{<:ModisProduct};
lat::Real,
lon::Real,
from::Union{String,Date} = "all", # might be handy
to::Union{String,Date} = "all",
format::String = "Date",
)
prod = product(T)
filepath =
joinpath(rasterpath(), "MODIS/dates", string(lat) * "," * string(lon) * ".csv")
if !isfile(filepath) # we need to download dates from the server
mkpath(dirname(filepath))
@info "Requesting availables dates for product $prod at $lat , $lon"
## Get all dates at given point
# request
r = HTTP.request(
"GET",
join([string(MODIS_URI), prod, "dates"], "/"),
query = Dict("latitude" => string(lat), "longitude" => string(lon)),
)
# parse
body = JP.parse(String(r.body))
# prebuild columns
calendardates = String[]
modisdates = String[]
# fill the vectors
for date in body["dates"]
push!(calendardates, date["calendar_date"])
push!(modisdates, date["modis_date"])
end
open(filepath, "w") do f
writedlm(f, [calendardates modisdates], ',')
end
else # a file with dates is already downloaded
# we simply read the file
mat = readdlm(filepath, ',', String)
calendardates = mat[:, 1]
modisdates = mat[:, 2]
end
## Filter for dates between from and to arguments
calendardates = Date.(calendardates)
from == "all" && (from = calendardates[1])
to == "all" && (to = calendardates[end])
startfound, endfound = false, false
bounds = [0,0]
i = 1
while !endfound
# two ways to find the end:
if i == length(calendardates) # end of vector reached
endfound = true
bounds[2] = i
elseif calendardates[i] > Date(to) && i > 1 # to reached
# if dates[i] is just over "to", dates[i-1] is the margin
endfound = true
bounds[2] = i-1
end
if !startfound
if calendardates[i] >= Date(from)
startfound = true
bounds[1] = i
end
end
i += 1
end
if format == "ModisDate"
return modisdates[bounds[1]:bounds[2]]
else
return calendardates[bounds[1]:bounds[2]]
end
end
| RasterDataSources | https://github.com/EcoJulia/RasterDataSources.jl.git |
|
[
"MIT"
] | 0.6.1 | dfab74c3efb21c6df589f30f7bbe5b9b8dbbe3c9 | code | 10969 | """
MODIS{ModisProduct} <: RasterDataSource
MODIS/VIIRS Land Product Database. Vegetation indices, surface reflectance, and more land cover data. Data from [`ModisProduct`](@ref)s datasets.
See: [modis.ornl.gov](https://modis.ornl.gov/)
"""
struct MODIS{X} <: RasterDataSource end
function layerkeys(T::Type{MODIS{X}}) where {X}
layernames = list_layers(X)
keys = []
# For some products, layers have names that start with numbers, thus
# resulting in bad Symbol names. Here we remove some words from each
# layer name until it's in a good format.
for l in layernames
newname = []
words = split(l, "_")
beginning = true
for w in words # keep only "clean" words
if beginning
if match(r"^[0-9]|^days|^m|^meters", w) === nothing
push!(newname, w)
beginning = false # added one word: no more checks
end
else
push!(newname, w)
end
end
push!(keys, Symbol(join(newname, "_"))) # build Array of newname Symbols
end
return Tuple(collect(keys)) # build tuple from Array{Symbol}
end
"""
layerkeys(T::Type{<:ModisProduct}) => Tuple
`Tuple` of `Symbol`s corresponding to the available layers for a given product.
May issue a request to MODIS server to get the layers list, or might just read
this information if the correctly named file is available.
"""
layerkeys(T::Type{<:ModisProduct}) = layerkeys(MODIS{T})
function layerkeys(T::Type{<:MODIS{X}}, layers::Tuple) where {X}
if isa(layers[1], Int) # integer layer names get their key name
layerkeys(T)[collect(layers)]
else # if all elements of layers are correct layer keys, return them
all(k -> k in layerkeys(T), layers) && return (layers)
throw("Unknown layers in $layers")
end
end
layerkeys(T::Type{<:ModisProduct}, layers) = layerkeys(MODIS{T}, layers)
function layers(T::Type{MODIS{X}}) where {X}
return Tuple(1:length(layerkeys(T)))
end
layers(T::Type{<:ModisProduct}) = layers(MODIS{T})
function getraster(T::Type{<:MODIS{X}}, args...; kwargs...) where {X}
X <: ModisProduct ? getraster(X, args...; kwargs...) :
throw("Unrecognized MODIS product.")
end
getraster_keywords(::Type{<:Union{MODIS,ModisProduct}}) = (:lat, :lon, :km_ab, :km_lr, :date, :end)
"""
getraster(T::Union{Type{<:ModisProduct}, Type{MODIS{X}}}, [layer::Union{Tuple,AbstractVector,Integer, Symbol}]; kwargs...) => Union{String, AbstractVector, NamedTuple}
Download [`MODIS`](@ref) data for a given [`ModisProduct`](@ref) as ASCII raster(s).
# Arguments
- `layer`: `Integer` or tuple/range of `Integer` or `Symbol`s. Without a `layer` argument, all layers will be downloaded, and a `NamedTuple` of paths returned.
Available layers for a given product can be looked up using [`RasterDataSources.layerkeys(T::Type{<:ModisProduct})`](@ref).
# Keywords
- `lat` and `lon`: Coordinates in decimal degrees of the approximate center of the raster. The MODIS API will try to match its pixel grid system as close as possible to those coordinates.
- `km_ab` and `km_lr`: Half-width and half-height of the raster in kilometers (kilometers above/below and left/right). Currently only `Integer` values are supported, up to 100.
- `date`: `String`, `Date`, `DateTime`, `AbstractVector` of dates or `Tuple` of a start and end date for the request. `String`s should be in format YYYY-MM-DD but can be in similar formats as long as they are comprehensible by `Dates.Date`. The available date interval for MODIS is 16 days, reset every first of January.
# Example
Download 250m NDVI in the western part of Britanny, France, from winter to late spring, 2002:
```julia
julia> getraster(MOD13Q1, :NDVI; lat = 48.25, lon = -4, km_ab = 50, km_lr = 50, date = (Date(2002,1,1), Date(2002,6,1)))
10-element Vector{String}:
"/your/path/MODIS/MOD13Q1/250m_16_days_NDVI/47.8313_-4.5899_2002-01-01.asc"
...
"/your/path/MODIS/MOD13Q1/250m_16_days_NDVI/47.8313_-4.5899_2002-05-25.asc"
```
Will attempt to download several files, one for each date and layer combination, and returns the filepath/s of the downloaded or pre-existing files. Coordinates in the file names correspond to the lower-left corner of the raster.
"""
function getraster(
T::Type{<:ModisProduct},
layer::Union{Tuple,Symbol,Int} = layerkeys(T);
lat::Real,
lon::Real,
km_ab::Int,
km_lr::Int,
date::Union{Tuple,AbstractVector,String,Date,DateTime},
)
# first check all arguments
check_layers(T, layer)
check_kwargs(T; lat = lat, lon = lon, km_ab = km_ab, km_lr = km_lr, date = date)
# then pass them to internal functions
_getraster(T, layer, date; lat = lat, lon = lon, km_ab = km_ab, km_lr = km_lr)
end
# if layer is a tuple, get them all using _map_layers
function _getraster(T::Type{<:ModisProduct}, layers::Tuple, date; kwargs...)
_map_layers(T, layers, date; kwargs...)
end
# convert layer symbols to int
function _getraster(T::Type{<:ModisProduct}, layer::Symbol, date; kwargs...)
_getraster(T, modis_int(T, layer), date; kwargs...)
end
# Tuple : start and end date
function _getraster(T::Type{<:ModisProduct}, layer::Int, date::Tuple; kwargs...)
_getraster(
T,
layer,
kwargs[:lat],
kwargs[:lon],
kwargs[:km_ab],
kwargs[:km_lr],
string(Date(date[1])),
string(Date(date[2])),
)
end
# Handle vectors : map over dates
function _getraster(T::Type{<:ModisProduct}, layer::Int, date::AbstractVector; kwargs...)
out = String[]
for d in eachindex(date)
push!(out, _getraster(T, layer, date[d]; kwargs...))
end
return out
end
# single date : from = to = string(Date(date))
function _getraster(
T::Type{<:ModisProduct},
layer::Int,
date::Union{Dates.TimeType,String};
kwargs...,
)
_getraster(
T,
layer,
kwargs[:lat],
kwargs[:lon],
kwargs[:km_ab],
kwargs[:km_lr],
string(Date(date)),
string(Date(date)),
)
end
"""
_getraster(T::Type{<:ModisProduct}, layer::Int, lat::Real, lon::Real, km_ab::Int, km_lr::Int, from::String, to::String) => Union{String, Vector{String}}
Modis requests always have an internal start and end date: using from and to in internal arguments makes more sense. `date` argument is converted by various
_getraster dispatches before calling this.
"""
function _getraster(
T::Type{<:ModisProduct},
layer::Int,
lat::Real,
lon::Real,
km_ab::Int,
km_lr::Int,
from::String,
to::String,
)
# accessing dates in a format readable by the MODIS API
dates = list_dates(T; lat = lat, lon = lon, format = "ModisDate", from = from, to = to)
length(dates) == 0 && throw("No available $T data at $lat , $lon from $from to $to")
if length(dates) <= 10
files = _getrasterchunk(
T,
layer;
lat = lat,
lon = lon,
km_ab = km_ab,
km_lr = km_lr,
dates = dates,
)
else
# take "chunk" subsets of dates 10 by 10
n_chunks = div(length(dates), 10) + 1
chunks =
[dates[1+10*k:(k == n_chunks - 1 ? end : 10 * k + 10)] for k = 0:(n_chunks-1)]
# remove empty end chunk
# (happens when length(dates) is divisible by 10)
length(chunks[end]) == 0 && (chunks = chunks[1:(end-1)])
files = map(chunks) do c
_getrasterchunk(
T,
layer;
dates = c,
lat = lat,
lon = lon,
km_ab = km_ab,
km_lr = km_lr,
)
end
files = vcat(files...) # splat chunks to get only one list
end
return files
end
"""
_getrasterchunk(T::Type{<:ModisProduct}, layer::Int; dates::Vector{String}, kwargs...)
Internal calls of [`RasterDataSources.modis_request`](@ref) and [`RasterDataSources.process_subset`](@ref): fetch data from server,
write a raster `.tif` file.
The MODIS API only allows requests for ten or less dates.
Returns the filepath/s of the downloaded or pre-existing files.
"""
function _getrasterchunk(
T::Type{<:ModisProduct},
layer::Int;
dates::Vector{String},
kwargs...,
)
length(dates) > 10 && throw("Too many dates provided. Use from and to arguments")
subset, pars = modis_request(
T,
list_layers(T)[layer],
kwargs[:lat],
kwargs[:lon],
kwargs[:km_ab],
kwargs[:km_lr],
dates[1],
dates[end],
)
out = process_subset(T, subset, pars)
return out
end
function rasterpath(T::Type{<:ModisProduct}, layer; kwargs...)
# argument checks
check_layers(T, layer)
check_kwargs(T; kwargs...)
return joinpath(_rasterpath(T, layer), rastername(T; kwargs...))
end
function _rasterpath(T::Type{<:ModisProduct})
return joinpath(rasterpath(), "MODIS", string(nameof(T)))
end
function _rasterpath(T::Type{<:ModisProduct}, layer::Int)
return joinpath(_rasterpath(T), list_layers(T)[layer])
end
function _rasterpath(T::Type{<:ModisProduct}, layer::Symbol)
return joinpath(_rasterpath(T), list_layers(T)[modis_int(T, layer)])
end
function _rasterpath(T::Type{<:ModisProduct}, layer::String)
layer in list_layers(T) && (return joinpath(_rasterpath(T), layer))
throw("Unknow layer in product $(string(T))")
end
function rastername(T::Type{<:ModisProduct}; kwargs...)
check_kwargs(T; kwargs...)
name = "$(round(kwargs[:lat], digits = 4))_$(round(kwargs[:lon], digits = 4))_$(kwargs[:date]).asc"
return name
end
# date_sequence does not use a date step because MODIS dates may vary from a
# strict 16-day sequence (mostly because of the 1st of January reset)
# date_step(T::Type{<:ModisProduct}) = Day(16)
# date_step(T::Type{MODIS{X}}) where {X} = date_step(X)
function date_sequence(T::Type{MODIS{X}}, dates; kw...) where {X}
date_sequence(X, dates; kw...)
end
"""
date_sequence(T::Type{<:ModisProduct}, dates::NTuple{2})
Asks list_dates for a list of required dates
"""
function date_sequence(T::Type{<:ModisProduct}, dates::NTuple{2}; kwargs...)
if !haskey(kwargs, :lat) || !haskey(kwargs, :lon)
throw(
ArgumentError(
"`lat` and `lon` must be provided to correctly build the date sequence.",
),
)
end
# get the dates
sequence =
Date.(
list_dates(
T,
lat = kwargs[:lat],
lon = kwargs[:lon],
from = dates[1],
to = dates[2],
)
)
return sequence
end
# disable metadata copy to avoid building "duplicate" series
# (see issue #294 of Rasters.jl)
has_constant_metadata(T::Type{MODIS{X}}) where {X} = false
has_constant_metadata(T::Type{<:ModisProduct}) = false
| RasterDataSources | https://github.com/EcoJulia/RasterDataSources.jl.git |
|
[
"MIT"
] | 0.6.1 | dfab74c3efb21c6df589f30f7bbe5b9b8dbbe3c9 | code | 10679 | """
MODIS-specific utility functions
MODIS data is not available in .tif format so we need a bit more
steps before storing the retrieved data and we can't download() it.
Data parsing is way easier using JSON.jl but it adds a dependency
"""
"""
modis_int(T::Type{<:ModisProduct}, l::Symbol)
Converts Symbol `l` to the corresponding integer if `l` is in the
layer keys of the required `ModisProduct` `T`.
"""
function modis_int(T::Type{<:ModisProduct}, l::Symbol)
keys = layerkeys(T)
for i in eachindex(keys)
keys[i] === l && return (i)
end
end
"""
MODIS API address
"""
const MODIS_URI = URI(scheme = "https", host = "modis.ornl.gov", path = "/rst/api/v1")
"""
modis_request(T::Type{<:ModisProduct}, args...)
Lowest level function for requests to modis server. All arguments are assumed correct.
# Arguments
- `layer`: `String` matching the "exact" layer name (i.e. as it is written in the MODIS dataset itself) for the given product. e.g. `"250m_16_days_EVI"`.
- `lat`, `lon`, `km_ab`, `km_lr` in correct types
- `from`, `to`: `String`s of astronomical dates for start and end dates of downloaded data, e.g. `"A2002033"` for "2002-02-02"
Returns a `NamedTuple` of information relevant to build a raster header, and a `Vector` of `Dict`s containing raster data, directly downloaded from MODIS. Those will almost always directly be passed to [`RasterDataSources.process_subset`](@ref)
"""
function modis_request(T::Type{<:ModisProduct}, layer, lat, lon, km_ab, km_lr, from, to)
# using joinpath here is more readable but works only for UNIX based OS, :'(
base_uri = join([string(MODIS_URI), product(T), "subset"], "/")
query = string(
URI(;
query = Dict(
"latitude" => string(lat),
"longitude" => string(lon),
"startDate" => string(from),
"endDate" => string(to),
"kmAboveBelow" => string(km_ab),
"kmLeftRight" => string(km_lr),
"band" => string(layer),
),
),
)
r = HTTP.request("GET", URI(base_uri * query), ["Accept" => "application/json"])
body = JP.parse(String(r.body))
# The server outputs data in a nested JSON array that we can
# parse manually : the highest level is a metadata array with
# a "subset" column containing pixel array for each (band, timepoint)
# the header information is in the top-level of the request
pars = (
nrows = body["nrows"],
ncols = body["ncols"],
xll = body["xllcorner"],
yll = body["yllcorner"],
cellsize = body["cellsize"]
)
# data is in the subset field
subset = body["subset"]
return subset, pars
end
"""
sinusoidal_to_latlon(x::Real, y::Real)
Convert x and y in sinusoidal projection to lat and lon in dec. degrees
The  takes care of coordinate conversions. This is not ideal in terms of network use but guarantees that the coordinates are correct.
"""
function sinusoidal_to_latlon(x::Real, y::Real)
url = "https://epsg.io/trans"
@info "Asking EPSG.io for coordinates calculation"
query = Dict(
"x" => string(x),
"y" => string(y),
"s_srs" => "53008", # sinusoidal
"t_srs" => "4326", # WGS84
)
r = HTTP.request("GET", url; query = query)
body = JP.parse(String(r.body))
lat = parse(Float64, body["y"])
lon = parse(Float64, body["x"])
return (lat, lon)
end
# data from https://nssdc.gsfc.nasa.gov/planetary/factsheet/earthfact.html
const EARTH_EQ_RADIUS = 6378137
const EARTH_POL_RADIUS = 6356752
function meters_to_latlon(d::Real, lat::Real)
dlon = asind(d / (cosd(lat) * EARTH_EQ_RADIUS))
dlat = d * 180 / (π * EARTH_POL_RADIUS)
return (dlat, dlon)
end
function _maybe_prepare_params(xllcorner::Real, yllcorner::Real, nrows::Int, cellsize::Real)
filepath = joinpath(
rasterpath(),
"MODIS",
"headers",
string(xllcorner) *
"," *
string(yllcorner) *
"," *
string(cellsize) *
"," *
string(nrows) *
".csv",
)
if isfile(filepath)
pars_str = open(filepath, "r") do f
readline(f)
end
pars = parse.(Float64, split(pars_str, ","))
else
# coordinates in sin projection ; we want upper-left in WGS84
# convert coordinates
yll, xll = sinusoidal_to_latlon(xllcorner, yllcorner)
# convert cell size in meters to degrees in lat and lon directions
dy, dx = meters_to_latlon(cellsize, yll) # watch out, this is a Tuple{Float64, Float64}
pars = [xll, yll, dx, dy]
# store in file
pars_str = join(string.(pars), ",")
mkpath(dirname(filepath))
open(filepath, "w") do f
write(f, pars_str)
end
end
# return a NamedTuple
return (xll = pars[1], yll = pars[2], dx = pars[3], dy = pars[4])
end
"""
process_subset(T::Type{<:ModisProduct}, subset::Vector{Any}, pars::NamedTuple)
Process a raw subset and argument parameters and create several raster files. Any already existing file is not overwritten.
For each band, a separate folder is created, containing a file for each of the required dates. This is inspired by the way WorldClim{Climate} treats the problem of possibly having to download several dates AND bands.
Can theoretically be used for MODIS data that does not directly come from [`RasterDataSources.modis_request`](@ref), but caution is advised.
Returns the filepath/s of the created or pre-existing files.
"""
function process_subset(T::Type{<:ModisProduct}, subset::Vector{Any}, pars::NamedTuple)
# coerce parameters from String to correct types
ncols = pars[:ncols]
nrows = pars[:nrows]
cellsize = pars[:cellsize]
xll = parse(Float64, pars[:xll])
yll = parse(Float64, pars[:yll])
pars = _maybe_prepare_params(xll, yll, nrows, cellsize)
path_out = String[]
for i in eachindex(subset) # for each (date, band)
date = subset[i]["calendar_date"]
band = subset[i]["band"]
filepath = rasterpath(T, band; lat = pars[:yll], lon = pars[:xll], date = date)
mat = permutedims(reshape(subset[i]["data"], (ncols, nrows)))
mkpath(dirname(filepath)) # prepare directories if they dont exist
if !isfile(filepath)
@info "Creating raster file $(basename(filepath)) in $(dirname(filepath))"
write_ascii(filepath, mat; ncols = ncols, nrows = nrows, nodatavalue = -3000.0, pars...)
else
@info "Raster file $(basename(filepath)) already exists in $(dirname(filepath))"
end
push!(path_out, filepath)
end
return (length(path_out) == 1 ? path_out[1] : path_out)
end
"""
check_layers(T::Type{<:ModisProduct}, layers::Union{Tuple, AbstractVector, Symbol, String, Int}) => nothing
Checks if required layers make sense for the MODIS product T.
"""
function check_layers(
T::Type{<:ModisProduct},
layers::Union{Tuple,AbstractVector,Symbol,String,Int},
)
if typeof(layers) <: Tuple || typeof(layers) <: AbstractVector
for l in layers
_check_layer(T::Type{<:ModisProduct}, l)
end
else
_check_layer(T::Type{<:ModisProduct}, layers)
end
end
function _check_layer(T::Type{<:ModisProduct}, layer::Symbol)
!(layer in layerkeys(T)) && throw(
ArgumentError(
"Invalid layer $layer for product $T.\nAvailable layers are $(layerkeys(T))",
),
)
return nothing
end
function _check_layer(T::Type{<:ModisProduct}, layer::Int)
!(layer in layers(T)) && throw(
ArgumentError(
"Invalid layer $layer for product $T.\nAvailable layers are $(layers(T))",
),
)
return nothing
end
function _check_layer(T::Type{<:ModisProduct}, layer::String)
!(layer in list_layers(T)) && throw(
ArgumentError(
"Invalid layer $layer for product $T.\nAvailable layers are $(list_layers(T)).\nProceed with caution while using `String` layers. You might want to use their `Symbol` counterparts.",
),
)
end
"""
check_kwargs(T::Type{ModisProduct}; kwargs...) => nothing
"Never trust user input". Checks all keyword arguments that might be used in internal calls.
"""
function check_kwargs(T::Type{<:ModisProduct}; kwargs...)
symbols = keys(kwargs)
errors = String[]
# check lat
if :lat in symbols
(kwargs[:lat] < -90 || kwargs[:lat] > 90) &&
push!(errors, "Latitude lat=$(kwargs[:lat]) must be between -90 and 90.")
end
# check lon
if :lon in symbols
(kwargs[:lon] < -180 || kwargs[:lon] > 180) &&
push!(errors, "Longitude lon=$(kwargs[:lon]) must be between -180 and 180.")
end
# check km_ab
if :km_ab in symbols
(kwargs[:km_ab] < 0 || kwargs[:km_ab] > 100) && push!(
errors,
"Km above and below km_ab=$(kwargs[:km_ab]) must be between 0 and 100.",
)
end
# check km_lr
if :km_lr in symbols
(kwargs[:km_lr] < 0 || kwargs[:km_lr] > 100) && push!(
errors,
"Km left and right km_lr=$(kwargs[:km_lr]) must be between 0 and 100.",
)
end
# check from
if :from in symbols
# check if conversion works
from = Date(kwargs[:from])
(from < Date(2000) || from > Dates.now()) &&
push!(errors, "Unsupported date for from=$(from)")
end
# check to
if :to in symbols
# check if conversion works
to = Date(kwargs[:to])
(to < Date(2000) || to > Dates.now()) &&
push!(errors, "Unsupported date for to=$(to)")
end
if :date in symbols
_check_date(kwargs[:date]) ||
push!(errors, "Unsupported date(s) in date=$(kwargs[:date])")
end
if length(errors) > 0
if length(errors) == 1
throw(ArgumentError(errors[1]))
else
throw(ArgumentError(join(["Several wrong arguments."; errors], "\n")))
end
end
return nothing
end
_check_date(d::AbstractVector) = all(b -> b == true, map(_check_date, d))
_check_date(d::Tuple) = all(b -> b == true, map(_check_date, d))
_check_date(d::String) = _check_date(Date(d))
"""
_check_date(d::Dates.TimeType)
Does not check if `d` is available, only checks if `d` makes sense, i.e if `d` **could** be available.
Returns `true` for good dates, `false` for bad ones.
"""
function _check_date(d::Dates.TimeType)
return !(d < Date(2000) || d > Dates.now())
end
| RasterDataSources | https://github.com/EcoJulia/RasterDataSources.jl.git |
|
[
"MIT"
] | 0.6.1 | dfab74c3efb21c6df589f30f7bbe5b9b8dbbe3c9 | code | 9067 |
struct SRTM <: RasterDataSource end
# SRTM Mirror with 5x5 degree tiles
const SRTM_URI = URI(scheme = "https", host = "srtm.csi.cgiar.org", path = "/wp-content/uploads/files/srtm_5x5/TIFF")
const HAS_SRTM_TILE = BitArray([
0 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 0 1 1 0 0 0 0 0 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 0 0
0 1 1 1 1 0 0 0 0 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 0 0 0 0 0 0 0 0 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 0 1 1 1 1 1 1
0 0 0 0 0 0 0 0 0 0 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 0 0 0 0 0 0 0 0 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 0 0 0 0
0 0 0 0 0 0 0 0 0 0 0 1 1 1 1 1 1 1 1 1 1 1 1 1 1 0 0 0 0 0 0 0 0 0 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 0 0 0 0 0 0
0 0 0 0 0 0 0 0 0 0 0 1 1 1 1 1 1 1 1 1 1 1 0 0 0 0 0 0 0 1 1 1 0 0 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 0 0 0 0 0 0 0
0 0 0 0 0 0 0 0 0 0 0 1 1 1 1 1 1 1 1 1 1 0 0 1 0 0 0 0 0 0 0 0 1 0 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 0 0 0 0 0 0 0
1 1 0 0 0 0 0 0 0 0 0 0 1 1 1 1 1 1 1 1 1 0 0 0 0 0 0 0 0 0 0 0 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 0 1 0 0 0 0 0 0 0
0 0 1 1 1 0 0 0 0 0 0 0 1 1 1 1 1 1 1 1 1 1 0 0 0 0 0 0 0 0 0 0 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 0 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 0 0 0 0 0
0 0 1 0 1 1 0 0 0 0 0 0 0 1 1 1 1 1 1 1 1 1 1 1 0 0 0 0 0 0 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 0 0 1 1 1 1 1 1 1 1 1 1 1 0 0 0 0 1 0 0 0 1 0 0
0 0 0 0 0 0 0 0 0 0 0 0 0 0 1 0 0 1 1 1 1 1 1 1 1 0 0 0 0 0 0 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 0 0 0 1 1 1 0 1 1 1 1 1 1 1 1 0 1 1 1 0 0 1 1 1 0
0 0 0 1 0 0 0 0 0 0 0 0 0 0 0 0 0 0 1 1 1 1 1 1 1 1 0 0 0 0 0 0 0 1 1 1 1 1 1 1 1 1 1 1 1 1 1 0 0 0 1 1 1 0 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 0
1 0 0 1 1 0 0 0 0 0 0 0 0 0 0 0 0 1 1 1 1 1 1 1 1 1 1 0 0 0 0 0 0 0 1 1 0 1 1 1 1 1 1 1 1 1 0 0 0 0 1 0 0 0 0 1 1 1 1 1 1 1 1 0 0 0 1 0 0 1 1 0
0 1 0 0 0 1 0 0 0 0 0 0 0 0 0 0 0 1 1 1 1 1 1 1 1 1 1 1 1 1 0 0 0 0 0 0 0 1 1 1 1 1 1 1 1 0 1 1 0 0 1 0 0 0 0 1 1 1 1 1 1 1 1 1 1 1 1 1 0 1 1 1
0 1 0 1 1 1 0 1 1 0 0 0 0 0 0 0 0 0 0 1 1 1 1 1 1 1 1 1 1 1 0 0 0 1 0 0 0 0 1 1 1 1 1 1 0 1 1 1 0 0 1 0 0 0 0 0 1 1 1 1 1 1 1 1 1 1 1 1 1 1 0 1
1 1 1 1 0 1 1 1 1 0 0 0 0 0 0 0 0 0 0 0 1 1 1 1 1 1 1 1 1 0 0 0 0 0 0 0 0 0 1 1 1 1 1 1 1 1 1 1 0 0 0 0 0 0 0 1 0 1 0 1 1 1 1 1 1 1 1 1 1 1 1 1
1 1 1 1 1 1 1 1 1 0 0 0 0 0 0 0 0 0 0 0 1 1 1 1 1 1 1 1 1 0 0 0 0 0 1 0 0 0 1 1 1 1 1 1 1 1 1 1 1 0 0 0 0 0 0 0 0 0 0 1 1 1 1 1 1 1 1 1 1 1 1 1
1 1 0 0 1 1 1 1 1 1 1 1 0 0 0 0 0 0 0 0 0 1 1 1 1 1 1 1 0 0 1 0 0 0 0 0 0 0 1 1 1 1 1 1 1 1 0 1 0 0 0 0 0 0 0 0 0 0 1 1 1 1 1 1 1 1 1 1 1 1 1 0
1 0 0 0 0 0 0 1 0 1 0 0 0 0 1 0 0 0 0 1 1 1 1 1 1 1 1 0 0 0 0 0 0 0 0 0 0 0 1 1 1 1 1 0 1 1 0 0 0 0 0 0 0 0 0 0 0 0 1 1 1 1 1 1 1 1 1 0 0 1 0 0
1 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 1 1 1 1 1 1 1 0 0 0 0 0 0 0 0 0 0 0 0 0 1 1 1 1 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 1 1 1 1 1 1 1 1 1 1 0 0 1 0
0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 1 1 1 1 0 0 0 0 0 0 0 0 1 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 1 0 0 0 0 0 0 0 1 0 0 0 1 1 1 1 0 0 0 1 1
1 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 1 1 1 1 0 0 0 0 0 0 0 0 0 1 1 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 1 1 0 0 0 1 1 1
0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 1 1 1 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 1 0 0 1 0 0 1 1 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 1 1 1
0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 1 1 1 1 1 0 0 0 1 0 0 0 0 0 0 0 1 0 0 0 0 0 0 0 0 0 0 0 0 1 1 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 1 0 1 0 0
0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 1 1 0 0 0 0 0 0 1 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 1 0 0 0 0
])
getraster_keywords(::Type{SRTM}) = (:bounds, :tile_index)
function _raster_tile_stem(tile_index::CartesianIndex)
y, x = tile_index.I
"srtm_$(lpad(x, 2, '0'))_$(lpad(y, 2, '0'))"
end
_rastername(::Type{SRTM}, tile_index::CartesianIndex{2}) = _raster_tile_stem(tile_index) * ".tif"
_rasterpath(T::Type{SRTM}, tile_index::CartesianIndex{2}) = joinpath(rasterpath(), "SRTM", _rastername(T, tile_index))
_zipname(::Type{SRTM}, tile_index::CartesianIndex{2}) = _raster_tile_stem(tile_index) * ".zip"
_zipurl(T::Type{SRTM}, tile_index::CartesianIndex{2}) = joinpath(SRTM_URI, _zipname(T, tile_index))
_zippath(T::Type{SRTM}, tile_index::CartesianIndex{2}) = joinpath(rasterpath(), "SRTM", "zips", _zipname(T, tile_index))
function _getraster(T::Type{SRTM}, tile_index::CartesianIndex{2})
raster_path = _rasterpath(T, tile_index)
if !isfile(raster_path)
@info "Note: not all oceanic tiles exist in the SRTM dataset."
zip_path = _zippath(T, tile_index)
_maybe_download(_zipurl(T, tile_index), zip_path)
mkpath(dirname(raster_path))
raster_name = _rastername(T, tile_index)
zf = ZipFile.Reader(zip_path)
write(raster_path, read(_zipfile_to_read(raster_name, zf)))
close(zf)
end
return raster_path
end
# Adapted from https://github.com/centreborelli/srtm4/blob/master/src/srtm4.c#L87-L117
function _wgs84_to_tile_x(x)
# tiles longitude indexes go from 1 to 72,
# covering the range from -180 to +180
tile_x = (1 + floor(Int, (x + 180) / 5)) % 72
return tile_x == 0 ? 72 : tile_x
end
function _wgs84_to_tile_y(y)
y = clamp(y, -60, 60)
tile_y = 1 + floor(Int, (60 - y) / 5)
return tile_y == 25 ? 24 : tile_y
end
function bounds_to_tile_indices(::Type{SRTM}, bounds::NTuple{4,Real})
bounds_to_tile_indices(SRTM, ((bounds[1], bounds[3]), (bounds[2], bounds[4])))
end
function bounds_to_tile_indices(::Type{SRTM}, (xs, ys)::NTuple{2,NTuple{2,Real}})
_check_order(xs)
_check_order(ys)
t_xs = _wgs84_to_tile_x.(xs)
t_ys = reverse(_wgs84_to_tile_y.(ys))
return CartesianIndices((t_ys[1]:(t_ys[2]), t_xs[1]:(t_xs[2])))
end
_check_order((a, b)) = a > b && throw(ArgumentError("Upper bound $b less than lower bound $a"))
for op in (:getraster, :rastername, :rasterpath, :zipname, :zipurl, :zippath)
_op = Symbol('_', op) # Name of internal function
@eval begin
# Broadcasting function dispatch
function $_op(T::Type{SRTM}, tile_index::CartesianIndices)
broadcast(tile_index) do I
HAS_SRTM_TILE[I] ? $_op(T, I) : missing
end
end
# Bounds to tile indices dispatch
$_op(T::Type{SRTM}, bounds::Tuple) = $_op(T, bounds_to_tile_indices(T, bounds))
# Public function definition with key-word arguments
function $op(T::Type{SRTM}; bounds=nothing, tile_index=nothing)
if isnothing(bounds) & isnothing(tile_index)
:op === :getraster || return joinpath(rasterpath(), "SRTM")
throw(ArgumentError("One of `bounds` or `tile_index` kwarg must be specified"))
elseif !isnothing(bounds) & !isnothing(tile_index)
throw(ArgumentError("Only one of `bounds` or `tile_index` should be specified. " *
"found `bounds`=$bounds and `tile_index`=$tile_index"))
else
# Call the internal function without key-word arguments
return $_op(T, isnothing(tile_index) ? bounds : tile_index)
end
end
end
end
| RasterDataSources | https://github.com/EcoJulia/RasterDataSources.jl.git |
|
[
"MIT"
] | 0.6.1 | dfab74c3efb21c6df589f30f7bbe5b9b8dbbe3c9 | code | 2202 | layers(::Type{WorldClim{BioClim}}) = layers(BioClim)
layerkeys(T::Type{WorldClim{BioClim}}, args...) = layerkeys(BioClim, args...)
"""
getraster(T::Type{WorldClim{BioClim}}, [layer::Union{Tuple,AbstractVector,Integer}]; res::String="10m") => Union{Tuple,AbstractVector,String}
Download [`WorldClim`](@ref) [`BioClim`](@ref) data.
# Arguments
- `layer`: `Integer` or tuple/range of `Integer` from `$(layers(BioClim))`.
or `Symbol`s from `$(layerkeys(BioClim))`. Without a `layer` argument, all layers
will be downloaded, and a `NamedTuple` of paths returned.
# Keywords
- `res`: `String` chosen from $(resolutions(WorldClim{BioClim})), "10m" by default.
Returns the filepath/s of the downloaded or pre-existing files.
"""
function getraster(T::Type{WorldClim{BioClim}}, layers::Union{Tuple,Int,Symbol};
res::String=defres(T)
)
_getraster(T, layers, res)
end
getraster_keywords(::Type{WorldClim{BioClim}}) = (:res,)
_getraster(T::Type{WorldClim{BioClim}}, layers::Tuple, res) = _map_layers(T, layers, res)
_getraster(T::Type{WorldClim{BioClim}}, layer::Symbol, res) = _getraster(T, bioclim_int(layer), res)
function _getraster(T::Type{WorldClim{BioClim}}, layer::Integer, res)
_check_layer(T, layer)
_check_res(T, res)
raster_path = rasterpath(T, layer; res)
zip_path = zippath(T, layer; res)
if !isfile(raster_path)
_maybe_download(zipurl(T, layer; res), zip_path)
mkpath(dirname(raster_path))
raster_name = rastername(T, layer; res)
zf = ZipFile.Reader(zip_path)
write(raster_path, read(_zipfile_to_read(raster_name, zf)))
close(zf)
end
return raster_path
end
# BioClim layers don't get their own folder
rasterpath(T::Type{<:WorldClim{BioClim}}, layer; kw...) =
joinpath(rasterpath(T), rastername(T, layer; kw...))
rastername(T::Type{<:WorldClim{BioClim}}, key; res) = "wc2.1_$(res)_bio_$key.tif"
zipname(T::Type{<:WorldClim{BioClim}}, key; res) = "wc2.1_$(res)_bio.zip"
zipurl(T::Type{<:WorldClim{BioClim}}, key; res) =
joinpath(WORLDCLIM_URI, "base", zipname(T, key; res))
zippath(T::Type{<:WorldClim{BioClim}}, key; res) =
joinpath(rasterpath(T), "zips", zipname(T, key; res))
| RasterDataSources | https://github.com/EcoJulia/RasterDataSources.jl.git |
|
[
"MIT"
] | 0.6.1 | dfab74c3efb21c6df589f30f7bbe5b9b8dbbe3c9 | code | 2554 | layers(::Type{WorldClim{Climate}}) = (:tmin, :tmax, :tavg, :prec, :srad, :wind, :vapr)
"""
getraster(T::Type{WorldClim{Climate}}, [layer::Union{Tuple,Symbol}]; month, res::String="10m") => Vector{String}
Download [`WorldClim`](@ref) [`Climate`](@ref) data.
# Arguments
- `layer` `Symbol` or `Tuple` of `Symbol` from `$(layers(WorldClim{Climate}))`.
Without a `layer` argument, all layers will be downloaded, and a `NamedTuple` of paths returned.
# Keywords
- `month`: `Integer` or `AbstractArray` of `Integer`. Chosen from `1:12`.
- `res`: `String` chosen from $(resolutions(WorldClim{Climate})), "10m" by default.
Returns the filepath/s of the downloaded or pre-existing files.
"""
function getraster(T::Type{WorldClim{Climate}}, layers::Union{Tuple,Symbol};
month, res::String=defres(T)
)
_getraster(T, layers, month, res)
end
getraster_keywords(::Type{WorldClim{Climate}}) = (:month, :res,)
function _getraster(T::Type{WorldClim{Climate}}, layers, month::AbstractArray, res::String)
_getraster.(T, Ref(layers), month, Ref(res))
end
function _getraster(T::Type{WorldClim{Climate}}, layers::Tuple, month::Integer, res::String)
_map_layers(T, layers, month, res)
end
function _getraster(T::Type{WorldClim{Climate}}, layer::Symbol, month::Integer, res::String)
_check_layer(T, layer)
_check_res(T, res)
raster_path = rasterpath(T, layer; res, month)
if !isfile(raster_path)
zip_path = zippath(T, layer; res, month)
_maybe_download(zipurl(T, layer; res, month), zip_path)
zf = ZipFile.Reader(zip_path)
mkpath(dirname(raster_path))
raster_name = rastername(T, layer; res, month)
write(raster_path, read(_zipfile_to_read(raster_name, zf)))
close(zf)
end
return raster_path
end
# Climate layers don't get their own folder
rasterpath(T::Type{<:WorldClim{Climate}}, layer; res, month) =
joinpath(_rasterpath(T, layer), rastername(T, layer; res, month))
_rasterpath(T::Type{<:WorldClim{Climate}}, layer) = joinpath(rasterpath(T), string(layer))
rastername(T::Type{<:WorldClim{Climate}}, layer; res, month) =
"wc2.1_$(res)_$(layer)_$(_pad2(month)).tif"
zipname(T::Type{<:WorldClim{Climate}}, layer; res, month=1) =
"wc2.1_$(res)_$(layer).zip"
zipurl(T::Type{<:WorldClim{Climate}}, layer; res, month=1) =
joinpath(WORLDCLIM_URI, "base", zipname(T, layer; res, month))
zippath(T::Type{<:WorldClim{Climate}}, layer; res, month=1) =
joinpath(rasterpath(T), "zips", zipname(T, layer; res, month))
_pad2(month) = lpad(month, 2, '0')
| RasterDataSources | https://github.com/EcoJulia/RasterDataSources.jl.git |
|
[
"MIT"
] | 0.6.1 | dfab74c3efb21c6df589f30f7bbe5b9b8dbbe3c9 | code | 1930 | layers(::Type{WorldClim{Elevation}}) = (:elev,)
"""
getraster(T::Type{WorldClim{Elevation}}, [layer::Union{Tuple,Symbol}]; res::String="10m") => Union{Tuple,AbstractVector,String}
Download [`WorldClim`](@ref) [`Elevation`](@ref) data.
# Arguments
- `layer`: `Symbol` or `Tuple` of `Symbol` from `$(layers(WorldClim{Elevation}))`.
Without a `layer` argument, all layers will be downloaded, and a `NamedTuple` of paths returned.
# Keywords
- `res`: `String` chosen from $(resolutions(WorldClim{Elevation})), "10m" by default.
Returns the filepath/s of the downloaded or pre-existing files.
"""
function getraster(T::Type{WorldClim{Elevation}}, layers::Union{Tuple,Symbol};
res::String=defres(T)
)
_getraster(T, layers, res)
end
getraster_keywords(::Type{WorldClim{Elevation}}) = (:res,)
_getraster(T::Type{WorldClim{Elevation}}, layers::Tuple, res) = _map_layers(T, layers, res)
function _getraster(T::Type{WorldClim{Elevation}}, layer::Symbol, res)
_check_layer(T, layer)
_check_res(T, res)
raster_path = rasterpath(T, layer; res)
if !isfile(raster_path)
zip_path = zippath(T, layer; res)
_maybe_download(zipurl(T, layer; res), zip_path)
mkpath(dirname(raster_path))
raster_name = rastername(T, layer; res)
zf = ZipFile.Reader(zip_path)
write(raster_path, read(_zipfile_to_read(raster_name, zf)))
close(zf)
end
return raster_path
end
rasterpath(T::Type{<:WorldClim{Elevation}}, layer; kw...) =
joinpath(rasterpath(T), rastername(T, layer; kw...))
rastername(T::Type{<:WorldClim{Elevation}}, key; res) = "wc2.1_$(res)_elev.tif"
zipname(T::Type{<:WorldClim{Elevation}}, key; res) = "wc2.1_$(res)_elev.zip"
zipurl(T::Type{<:WorldClim{Elevation}}, key; res) =
joinpath(WORLDCLIM_URI, "base", zipname(T, key; res))
zippath(T::Type{<:WorldClim{Elevation}}, key; res) =
joinpath(rasterpath(T), "zips", zipname(T, key; res))
| RasterDataSources | https://github.com/EcoJulia/RasterDataSources.jl.git |
|
[
"MIT"
] | 0.6.1 | dfab74c3efb21c6df589f30f7bbe5b9b8dbbe3c9 | code | 781 | """
WorldClim{Union{BioClim,Climate,Weather}} <: RasterDataSource
Data from WorldClim datasets, either [`BioClim`](@ref), [`Climate`](@ref) or
[`Weather`](@ref).
See: [www.worldclim.org](https://www.worldclim.org)
"""
struct WorldClim{X} <: RasterDataSource end
const WORLDCLIM_URI = URI(scheme="https", host="geodata.ucdavis.edu", path="/climate/worldclim/2_1")
resolutions(::Type{<:WorldClim}) = ("30s", "2.5m", "5m", "10m")
defres(::Type{<:WorldClim}) = "10m"
rasterpath(::Type{WorldClim{T}}) where T = joinpath(rasterpath(), "WorldClim", string(nameof(T)))
rasterpath(T::Type{<:WorldClim}, layer; kw...) =
joinpath(rasterpath(T), string(layer), rastername(T, layer; kw...))
_zipfile_to_read(raster_name, zf) = first(filter(f -> f.name == raster_name, zf.files))
| RasterDataSources | https://github.com/EcoJulia/RasterDataSources.jl.git |
|
[
"MIT"
] | 0.6.1 | dfab74c3efb21c6df589f30f7bbe5b9b8dbbe3c9 | code | 3123 | layers(::Type{WorldClim{Weather}}) = (:tmin, :tmax, :prec)
date_step(::Type{WorldClim{Weather}}) = Month(1)
"""
getraster(T::Type{WorldClim{Weather}}, [layer::Union{Tuple,Symbol}]; date) => Union{String,Tuple{String},Vector{String}}
Download [`WorldClim`](@ref) [`Weather`](@ref) data, for `layer`/s in: `$(layers(WorldClim{Weather}))`.
Without a layer argument, all layers will be downloaded, and a `NamedTuple` of paths returned.
# Keywords
- `date`: a `Date` or `DateTime` object, a `Vector` of dates, or `Tuple` of start/end dates.
WorldClim Weather is available with a daily timestep.
Returns the filepath/s of the downloaded or pre-existing files.
"""
function getraster(T::Type{WorldClim{Weather}}, layers::Union{Tuple,Symbol}; date)
_getraster(T, layers, date)
end
getraster_keywords(::Type{WorldClim{Weather}}) = (:date,)
function _getraster(T::Type{WorldClim{Weather}}, layers, date::Tuple)
_getraster(T, layers, date_sequence(T, date))
end
function _getraster(T::Type{WorldClim{Weather}}, layers, dates::AbstractArray)
_getraster.(T, Ref(layers), dates)
end
function _getraster(T::Type{WorldClim{Weather}}, layers::Tuple, date::Dates.TimeType)
_map_layers(T, layers, date)
end
function _getraster(T::Type{WorldClim{Weather}}, layer::Symbol, date::Dates.TimeType)
decadestart = Date.(1960:10:2020)
for i in eachindex(decadestart[1:end-1])
# At least one date is in the decade
date >= decadestart[i] && date < decadestart[i+1] || continue
zip_path = zippath(T, layer; decade=decadestart[i])
_maybe_download(zipurl(T, layer; decade=decadestart[i]), zip_path)
zf = ZipFile.Reader(zip_path)
raster_path = rasterpath(T, layer; date=date)
mkpath(dirname(raster_path))
if !isfile(raster_path)
raster_name = rastername(T, layer; date=date)
println("Writing $(raster_path)...")
write(raster_path, read(_zipfile_to_read(raster_name, zf)))
end
close(zf)
return raster_path
end
error("Date $date not between 1960 and 2020")
end
const WEATHER_DECADES = Dict(Date(1960) => "1960-1969",
Date(1970) => "1970-1979",
Date(1980) => "1980-1989",
Date(1990) => "1990-1999",
Date(2000) => "2000-2009",
Date(2010) => "2010-2018")
rastername(T::Type{<:WorldClim{Weather}}, layer; date) =
joinpath("wc2.1_2.5m_$(layer)_$(_date2string(T, date)).tif")
zipname(T::Type{<:WorldClim{Weather}}, layer; decade) =
"wc2.1_2.5m_$(layer)_$(WEATHER_DECADES[decade]).zip"
zipurl(T::Type{<:WorldClim{Weather}}, layer; decade) =
joinpath(WORLDCLIM_URI, "hist", zipname(T, layer; decade))
zippath(T::Type{<:WorldClim{Weather}}, layer; decade) =
joinpath(rasterpath(T), "zips", zipname(T, layer; decade))
# Utility methods
_dateformat(::Type{<:WorldClim}) = DateFormat("yyyy-mm")
_filename2date(T::Type{<:WorldClim}, fn::AbstractString) =
_string2date(T, basename(fn)[findfirst(r"\d\d\d\d-\d\d", basename(fn))])
| RasterDataSources | https://github.com/EcoJulia/RasterDataSources.jl.git |
|
[
"MIT"
] | 0.6.1 | dfab74c3efb21c6df589f30f7bbe5b9b8dbbe3c9 | code | 2328 | using RasterDataSources, URIs, Test, Dates
using RasterDataSources: rastername, rasterurl, rasterpath
@testset "ALWB" begin
alwb_path = joinpath(ENV["RASTERDATASOURCES_PATH"], "ALWB")
@test rasterpath(ALWB) == alwb_path
@test rasterpath(ALWB{Values,Year}) == joinpath(alwb_path, "values", "year")
@test rastername(ALWB{Values,Year}, :ss_pct; date=Date(2001, 1)) == "ss_pct.nc"
@test rastername(ALWB{Values,Month}, :ss_pct; date=Date(2001, 1)) == "ss_pct.nc"
@test rastername(ALWB{Values,Day}, :ss_pct; date=Date(2001, 1)) == "ss_pct_2001.nc"
@test rasterpath(ALWB{Values,Day}, :ss_pct; date=Date(2001, 1)) ==
joinpath(alwb_path, "values", "day", "ss_pct_2001.nc")
@test rasterurl(ALWB{Values,Year}, :ss_pct; date=Date(2001, 1)) ==
URI(scheme="http", host="www.bom.gov.au", path="/jsp/awra/thredds/fileServer/AWRACMS/values/year/ss_pct.nc")
@test rasterurl(ALWB{Values,Day}, :ss_pct; date=Date(2001, 1)) ==
URI(scheme="http", host="www.bom.gov.au", path="/jsp/awra/thredds/fileServer/AWRACMS/values/day/ss_pct_2001.nc")
raster_path = joinpath(alwb_path, "values", "day", "ss_pct_2018.nc")
@test getraster(ALWB{Values,Day}, :ss_pct; date=DateTime(2018, 01, 01)) == raster_path
@test isfile(raster_path)
raster_path = joinpath(alwb_path, "deciles", "month", "s0_pct.nc")
@test getraster(ALWB{Deciles,Month}, :s0_pct; date=DateTime(2018, 01, 01)) == raster_path
@test isfile(raster_path)
raster_path = joinpath(alwb_path, "values", "day", "ma_wet_2018.nc")
@test getraster(ALWB{Values,Day}, :ma_wet; date=DateTime(2018, 01, 01)) == raster_path
@test isfile(raster_path)
raster_path = joinpath(alwb_path, "values", "month", "etot.nc")
@test getraster(ALWB{Values,Month}, (:etot,); date=DateTime(2018, 01, 01)) == (etot=raster_path,)
@test isfile(raster_path)
raster_path = joinpath(alwb_path, "values", "year", "asce_pet.nc")
@test getraster(ALWB{Values,Year}, :asce_pet; date=[DateTime(2018, 01, 01)]) == [raster_path]
@test isfile(raster_path)
raster_path = joinpath(alwb_path, "values", "year", "dd.nc")
@test getraster(ALWB{Values,Year}, [:dd]; date=DateTime(2018, 01, 01)) == (dd=raster_path,)
@test isfile(raster_path)
@test RasterDataSources.getraster_keywords(ALWB) == (:date,)
end
| RasterDataSources | https://github.com/EcoJulia/RasterDataSources.jl.git |
|
[
"MIT"
] | 0.6.1 | dfab74c3efb21c6df589f30f7bbe5b9b8dbbe3c9 | code | 1327 | using RasterDataSources, URIs, Test, Dates
using RasterDataSources: rastername, rasterpath, zipurl, zipname, zippath
@testset "AWAP" begin
using RasterDataSources: rastername, zipurl, zipname, zippath
raster_file = joinpath(ENV["RASTERDATASOURCES_PATH"], "AWAP", "vprp", "vprph09", "20010101.grid")
@test rasterpath(AWAP, :vprpress09; date=Date(2001, 1)) == raster_file
@test rastername(AWAP, :vprpress09; date=Date(2001, 1)) == "20010101.grid"
@test zipurl(AWAP, :vprpress09; date=Date(2001, 1)) ==
URI(scheme="http", host="www.bom.gov.au", path="/web03/ncc/www/awap/vprp/vprph09/daily/grid/0.05/history/nat/2001010120010101.grid.Z")
@test zippath(AWAP, :vprpress09; date=Date(2001, 1)) ==
joinpath(ENV["RASTERDATASOURCES_PATH"], "AWAP", "vprp", "vprph09", "20010101.grid.Z")
@test zipname(AWAP, :vprpress09; date=Date(2001, 1)) == "20010101.grid.Z"
if Sys.islinux()
@test getraster(AWAP, :vprpress09; date=DateTime(2001, 01, 01)) == raster_file
@test getraster(AWAP, (:vprpress09,); date=DateTime(2001, 01, 01)) == (vprpress09=raster_file,)
@test getraster(AWAP, [:vprpress09]; date=DateTime(2001, 01, 01)) == (vprpress09=raster_file,)
@test isfile(raster_file)
end
@test RasterDataSources.getraster_keywords(AWAP) == (:date,)
end
| RasterDataSources | https://github.com/EcoJulia/RasterDataSources.jl.git |
Subsets and Splits
No community queries yet
The top public SQL queries from the community will appear here once available.