licenses
listlengths 1
3
| version
stringclasses 677
values | tree_hash
stringlengths 40
40
| path
stringclasses 1
value | type
stringclasses 2
values | size
stringlengths 2
8
| text
stringlengths 25
67.1M
| package_name
stringlengths 2
41
| repo
stringlengths 33
86
|
|---|---|---|---|---|---|---|---|---|
[
"MIT"
]
| 0.4.12 | dea63ff72079443240fbd013ba006bcbc8a9ac00 | code | 413 | """
YAMLVersion
A type used for controlling the YAML version.
Planned to be supported versions are:
- [`YAMLV1_1`](@ref): YAML version 1.1
- [`YAMLV1_2`](@ref): YAML version 1.2
"""
abstract type YAMLVersion end
"""
YAMLV1_1
A singleton type for YAML version 1.1.
"""
struct YAMLV1_1 <: YAMLVersion end
"""
YAMLV1_2
A singleton type for YAML version 1.2.
"""
struct YAMLV1_2 <: YAMLVersion end
| YAML | https://github.com/JuliaData/YAML.jl.git |
|
[
"MIT"
]
| 0.4.12 | dea63ff72079443240fbd013ba006bcbc8a9ac00 | code | 4580 | #
# Writing Julia collections to YAML files is implemented with multiple dispatch and recursion:
# Depending on the value type, Julia chooses the appropriate _print function, which may add a
# level of recursion.
#
"""
write_file(path, data, prefix="")
Write some data (e.g. a dictionary or an array) to a YAML file.
"""
function write_file(path::AbstractString, data::Any, prefix::AbstractString="")
open(path, "w") do io
write(io, data, prefix)
end
end
"""
write([io], data, prefix="")
Write a YAML representation of some data (e.g. a dictionary or an array) to an IO stream,
if provided, or return a string of that representation.
"""
function write(io::IO, data::Any, prefix::AbstractString="")
print(io, prefix) # print the prefix, e.g. a comment at the beginning of the file
_print(io, data) # recursively print the data
end
function write(data::Any, prefix::AbstractString="")
io = IOBuffer()
write(io, data, prefix)
str = String(take!(io))
close(io)
str
end
"""
yaml(data)
Return a YAML-formatted string of the `data`.
"""
yaml(data::Any) = write(data)
# recursively print a dictionary
function _print(io::IO, dict::AbstractDict, level::Int=0, ignore_level::Bool=false)
if isempty(dict)
println(io, "{}")
else
for (i, pair) in enumerate(dict)
_print(io, pair, level, ignore_level ? i == 1 : false) # ignore indentation of first pair
end
end
end
# recursively print an array
function _print(io::IO, arr::AbstractVector, level::Int=0, ignore_level::Bool=false)
if isempty(arr)
println(io, "[]")
else
for elem in arr
if elem isa AbstractVector # vectors of vectors must be handled differently
print(io, _indent("-\n", level))
_print(io, elem, level + 1)
else
print(io, _indent("- ", level)) # print the sequence element identifier '-'
_print(io, elem, level + 1, true) # print the value directly after
end
end
end
end
# print a single key-value pair
function _print(io::IO, pair::Pair, level::Int=0, ignore_level::Bool=false)
key = if pair[1] === nothing
"null" # this is what the YAML parser interprets as 'nothing'
elseif pair[1] isa AbstractString && (
occursin('#', pair[1]) || first(pair[1]) in "{}[]&*?|-<>=!%@:`,\"'"
)
string("\"", escape_string(pair[1]), "\"") # special keys that require quoting
else
string(pair[1]) # any useful case
end
print(io, _indent(key * ":", level, ignore_level)) # print the key
if (pair[2] isa AbstractDict || pair[2] isa AbstractVector) && !isempty(pair[2])
print(io, "\n") # a line break is needed before a recursive structure
else
print(io, " ") # a whitespace character is needed before a single value
end
_print(io, pair[2], level + 1) # print the value
end
# _print a single string
function _print(io::IO, str::AbstractString, level::Int=0, ignore_level::Bool=false)
if occursin('\n', strip(str)) || occursin('"', str)
if endswith(str, "\n\n") # multiple trailing newlines: keep
println(io, "|+")
str = str[1:end-1] # otherwise, we have one too many
elseif endswith(str, "\n") # one trailing newline: clip
println(io, "|")
else # no trailing newlines: strip
println(io, "|-")
end
indent = repeat(" ", max(level, 1))
for line in split(str, "\n")
println(io, indent, line)
end
else
# quote and escape
println(io, replace(repr(MIME("text/plain"), str), raw"\$" => raw"$"))
end
end
# handle NaNs and Infs
function _print(io::IO, val::Float64, level::Int=0, ignore_level::Bool=false)
if isfinite(val)
println(io, string(val)) # the usual case
elseif isnan(val)
println(io, ".NaN") # this is what the YAML parser interprets as NaN
elseif val == Inf
println(io, ".inf")
elseif val == -Inf
println(io, "-.inf")
end
end
_print(io::IO, val::Nothing, level::Int=0, ignore_level::Bool=false) =
println(io, "~") # this is what the YAML parser interprets as nothing
# _print any other single value
_print(io::IO, val::Any, level::Int=0, ignore_level::Bool=false) =
println(io, string(val)) # no indentation is required
# add indentation to a string
_indent(str::AbstractString, level::Int, ignore_level::Bool=false) =
repeat(" ", ignore_level ? 0 : level) * str
| YAML | https://github.com/JuliaData/YAML.jl.git |
|
[
"MIT"
]
| 0.4.12 | dea63ff72079443240fbd013ba006bcbc8a9ac00 | code | 14856 | #!/usr/bin/env julia
module YAMLTests
import YAML
import Base.Filesystem
using StringEncodings: encode, decode, @enc_str
using Test
const tests = [
"spec-02-01",
"spec-02-02",
"spec-02-03",
"spec-02-04",
"spec-02-05",
"spec-02-06",
"spec-02-07",
"spec-02-08",
"spec-02-09",
"spec-02-10",
"spec-02-11",
"spec-02-12",
"spec-02-13",
"spec-02-14",
"spec-02-15",
"spec-02-16",
"spec-02-17",
"spec-02-18",
"spec-02-19",
"spec-02-20",
"spec-02-21",
"spec-02-22",
"spec-02-23",
"empty_scalar",
"no_trailing_newline",
"windows_newlines",
"escape_sequences",
"issue15",
"issue30",
"issue36",
"issue39",
"issue132",
"cartesian",
"ar1",
"ar1_cartesian",
"merge-01",
"version-colon",
"multi-constructor",
"utf-8-bom",
"utf-32-be",
"empty_tag",
"empty_list_elem",
]
# ignore some test cases in write_and_load testing
const test_write_ignored = [
"spec-02-17",
"escape_sequences",
"cartesian",
"ar1",
"ar1_cartesian",
"multi-constructor"
]
# test custom tags
function construct_type_map(t::Symbol, constructor::YAML.Constructor,
node::YAML.Node)
mapping = YAML.construct_mapping(constructor, node)
mapping[:tag] = t
mapping
end
function TestConstructor()
pairs = [("!Cartesian", :Cartesian),
("!AR1", :AR1)]
ret = YAML.SafeConstructor()
for (t,s) in pairs
YAML.add_constructor!(ret, t) do c, n
construct_type_map(s, c, n)
end
end
YAML.add_multi_constructor!(ret, "!addtag:") do constructor::YAML.Constructor, tag, node
construct_type_map(Symbol(tag), constructor, node)
end
ret
end
const more_constructors = let
pairs = [("!Cartesian", :Cartesian),
("!AR1", :AR1)]
Dict{String,Function}([(t, (c, n) -> construct_type_map(s, c, n))
for (t, s) in pairs])
end
const multi_constructors = Dict{String, Function}(
"!addtag:" => (c, t, n) -> construct_type_map(Symbol(t), c, n)
)
# write a file, then load its contents to be tested again
function write_and_load(data::Any)
path = Filesystem.tempname() * ".yml" # path to a temporary file
try
YAML.write_file(path, data)
return YAML.load_file(path, more_constructors)
finally
Filesystem.rm(path, force=true)
end
end
# Reverse end of line conventions to get testing of both LF and CRLF
# line endings regardless of platform. If there are already mixed line
# endings, it is not so important what this transformation does.
function reverse_eol(s)
encoding = YAML.detect_encoding(IOBuffer(s))
s = decode(s, encoding)
if occursin("\r\n", s)
s = replace(s, "\r\n" => "\n")
else
s = replace(s, "\n" => "\r\n")
end
encode(s, encoding)
end
const testdir = dirname(@__FILE__)
mktempdir() do tmpdir
tmp_file_name = joinpath(tmpdir, "reversed_eol.yaml")
@testset for test in tests, eol in (:native, :foreign)
yaml_file_name = joinpath(testdir, "yaml/$test.yaml")
julia_file_name = joinpath(testdir, "julia/$test.jl")
if eol == :foreign
write(tmp_file_name, reverse_eol(read(yaml_file_name)))
yaml_file_name = tmp_file_name
end
yaml_string = read(yaml_file_name, String)
expected = evalfile(julia_file_name)
@testset "Load from File" begin
@test begin
data = YAML.load_file(
yaml_file_name,
TestConstructor()
)
isequal(data, expected)
end
@test begin
dictData = YAML.load_file(
yaml_file_name,
more_constructors, multi_constructors
)
isequal(dictData, expected)
end
end
@testset "Load from String" begin
@test begin
data = YAML.load(
yaml_string,
TestConstructor()
)
isequal(data, expected)
end
@test begin
dictData = YAML.load(
yaml_string,
more_constructors, multi_constructors
)
isequal(dictData, expected)
end
end
@testset "Load All from File" begin
@test begin
data = YAML.load_all_file(
yaml_file_name,
TestConstructor()
)
isequal(first(data), expected)
end
@test begin
dictData = YAML.load_all_file(
yaml_file_name,
more_constructors, multi_constructors
)
isequal(first(dictData), expected)
end
end
@testset "Load All from String" begin
@test begin
data = YAML.load_all(
yaml_string,
TestConstructor()
)
isequal(first(data), expected)
end
@test begin
dictData = YAML.load_all(
yaml_string,
more_constructors, multi_constructors
)
isequal(first(dictData), expected)
end
end
if !in(test, test_write_ignored)
@testset "Writing" begin
@test begin
data = YAML.load_file(
yaml_file_name,
more_constructors
)
isequal(write_and_load(data), expected)
end
end
else
println("WARNING: I do not test the writing of $test")
end
end
end
const encodings = [
enc"UTF-8", enc"UTF-16BE", enc"UTF-16LE", enc"UTF-32BE", enc"UTF-32LE"
]
@testset for encoding in encodings
data = encode("test", encoding)
@test YAML.detect_encoding(IOBuffer(data)) == encoding
@test YAML.load(IOBuffer(data)) == "test"
#with explicit BOM
data = encode("\uFEFFtest", encoding)
@test YAML.detect_encoding(IOBuffer(data)) == encoding
@test YAML.load(IOBuffer(data)) == "test"
end
@testset "multi_doc_bom" begin
iterable = YAML.load_all("""
\ufeff---\r
test: 1
\ufeff---
test: 2
\ufeff---
test: 3
""")
(val, state) = iterate(iterable)
@test isequal(val, Dict("test" => 1))
(val, state) = iterate(iterable, state)
@test isequal(val, Dict("test" => 2))
(val, state) = iterate(iterable, state)
@test isequal(val, Dict("test" => 3))
@test iterate(iterable, state) === nothing
end
# test that an OrderedDict is written in the correct order
using OrderedCollections, DataStructures
@test strip(YAML.yaml(OrderedDict(:c => 3, :b => 2, :a => 1))) == join(["c: 3", "b: 2", "a: 1"], "\n")
# test that arbitrary dicttypes can be parsed
const dicttypes = [
Dict{Any,Any},
Dict{String,Any},
Dict{Symbol,Any},
OrderedDict{String,Any},
() -> DefaultDict{String,Any}(Missing),
]
@testset for dicttype in dicttypes
data = YAML.load_file(
joinpath(testdir, "nested-dicts.data"),
more_constructors;
dicttype=dicttype
)
if typeof(dicttype) <: Function
dicttype = typeof(dicttype())
end # check the return type of function dicttypes
_key(k::String) = keytype(dicttype) == Symbol ? Symbol(k) : k # String or Symbol key
@test typeof(data) == dicttype
@test typeof(data[_key("outer")]) == dicttype
@test typeof(data[_key("outer")][_key("inner")]) == dicttype
@test data[_key("outer")][_key("inner")][_key("something_unrelated")] == "1" # for completeness
# type-specific tests
if dicttype <: OrderedDict
@test [k for (k,v) in data] == [_key("outer"), _key("anything_later")] # correct order
elseif [k for (k,v) in data] == [_key("outer"), _key("anything_later")]
@warn "Test of OrderedDict might not be discriminative: the order is also correct in $dicttype"
end
if dicttype <: DefaultDict
@test data[""] === missing
end
end
const test_errors = [
"invalid-tag"
]
@testset "YAML Errors" "error test = $test" for test in test_errors
@test_throws YAML.ConstructorError YAML.load_file(
joinpath(testdir, string(test, ".data")),
TestConstructor()
)
end
@testset "Custom Constructor" begin
function MySafeConstructor()
yaml_constructors = copy(YAML.default_yaml_constructors)
delete!(yaml_constructors, nothing)
YAML.Constructor(yaml_constructors)
end
function MyReallySafeConstructor()
yaml_constructors = copy(YAML.default_yaml_constructors)
delete!(yaml_constructors, nothing)
ret = YAML.Constructor(yaml_constructors)
YAML.add_multi_constructor!(ret, nothing) do constructor::YAML.Constructor, tag, node
throw(YAML.ConstructorError(nothing, nothing,
"could not determine a constructor for the tag '$(tag)'",
node.start_mark))
end
ret
end
yamlString = """
Test: !test
test1: !test data
test2: !test2
- test1
- test2
"""
expected = Dict{Any,Any}("Test" => Dict{Any,Any}("test2"=>["test1", "test2"],"test1"=>"data"))
@test isequal(YAML.load(yamlString, MySafeConstructor()), expected)
@test_throws YAML.ConstructorError YAML.load(
yamlString,
MyReallySafeConstructor()
)
end
# also check that things break correctly
@test_throws YAML.ConstructorError YAML.load_file(
joinpath(testdir, "nested-dicts.data"),
more_constructors;
dicttype=Dict{Float64,Any}
)
@test_throws YAML.ConstructorError YAML.load_file(
joinpath(testdir, "nested-dicts.data"),
more_constructors;
dicttype=Dict{Any,Float64}
)
@test_throws ArgumentError YAML.load_file(
joinpath(testdir, "nested-dicts.data"),
more_constructors;
dicttype=(mistaken_argument) -> DefaultDict{String,Any}(mistaken_argument)
)
@test_throws ArgumentError YAML.load_file(
joinpath(testdir, "nested-dicts.data"),
more_constructors;
dicttype=() -> 3.0 # wrong type
)
@test_throws YAML.ScannerError YAML.load("x: %")
if VERSION >= v"1.8"
@test_throws "found character '%' that cannot start any token" YAML.load("x: %")
end
# issue 81
dict_content = ["key1" => [Dict("subkey1" => "subvalue1", "subkey2" => "subvalue2"), Dict()], "key2" => "value2"]
order_one = OrderedDict(dict_content...)
order_two = OrderedDict(dict_content[[2,1]]...) # reverse order
@test YAML.yaml(order_one) != YAML.yaml(order_two)
@test YAML.load(YAML.yaml(order_one)) == YAML.load(YAML.yaml(order_two))
# issue 89 - quotes in strings
@test YAML.load(YAML.yaml(Dict("a" => """a "quoted" string""")))["a"] == """a "quoted" string"""
@test YAML.load(YAML.yaml(Dict("a" => """a \\"quoted\\" string""")))["a"] == """a \\"quoted\\" string"""
# issue 108 - dollar signs in single-line strings
@test YAML.yaml("foo \$ bar") == "\"foo \$ bar\"\n"
@test YAML.load(YAML.yaml(Dict("a" => "")))["a"] == ""
@test YAML.load(YAML.yaml(Dict("a" => "nl at end\n")))["a"] == "nl at end\n"
@test YAML.load(YAML.yaml(Dict("a" => "one\nnl\n")))["a"] == "one\nnl\n"
@test YAML.load(YAML.yaml(Dict("a" => "many\nnls\n\n\n")))["a"] == "many\nnls\n\n\n"
@test YAML.load(YAML.yaml(Dict("a" => "no\ntrailing\nnls")))["a"] == "no\ntrailing\nnls"
@test YAML.load(YAML.yaml(Dict("a" => "foo\n\"bar\\'")))["a"] == "foo\n\"bar\\'"
@test YAML.load(YAML.yaml(Dict("a" => Dict()))) == Dict("a" => Dict())
@test YAML.load(YAML.yaml(Dict("a" => []))) == Dict("a" => [])
# issue 114 - gracefully handle extra commas in flow collections
@testset "issue114" begin
@test YAML.load("[3,4,]") == [3,4]
@test YAML.load("{a:4,b:5,}") == Dict("a" => 4, "b" => 5)
@test YAML.load("[?a:4, ?b:5]") == [Dict("a" => 4), Dict("b" => 5)]
@test_throws YAML.ParserError YAML.load("[3,,4]")
@test_throws YAML.ParserError YAML.load("{a: 3,,b:3}")
end
@testset "issue 125 (test_throw)" begin
@test_throws YAML.ScannerError YAML.load(""" ''' """)
@test_throws YAML.ScannerError YAML.load(""" ''''' """)
@test_throws YAML.ParserError YAML.load(""" ''a'' """)
@test_throws YAML.ScannerError YAML.load(""" '''a'' """)
end
# issue 129 - Comment only content
@testset "issue129" begin
@test YAML.load("#") === nothing
@test isempty(YAML.load_all("#"))
end
# issue 132 - load_all fails on windows
@testset "issue132" begin
input = """
---
creator: LAMMPS
timestep: 0
...
---
creator: LAMMPS
timestep: 1
...
"""
input_crlf = replace(input, "\n" => "\r\n")
expected = [Dict("creator" => "LAMMPS", "timestep" => 0),
Dict("creator" => "LAMMPS", "timestep" => 1)]
@test collect(YAML.load_all(input)) == expected
@test collect(YAML.load_all(input_crlf)) == expected
end
# issue #148 - warn unknown directives
@testset "issue #148" begin
@test (@test_logs (:warn, """unknown directive name: "FOO" at line 1, column 4. We ignore this.""") YAML.load("""%FOO bar baz\n\n--- "foo\"""")) == "foo"
@test (@test_logs (:warn, """unknown directive name: "FOO" at line 1, column 4. We ignore this.""") (:warn, """unknown directive name: "BAR" at line 2, column 4. We ignore this.""") YAML.load("""%FOO\n%BAR\n--- foo""")) == "foo"
end
# issue #143 - load empty file
@testset "issue #143" begin
@test YAML.load("") === nothing
@test isempty(YAML.load_all(""))
end
# issue #144
@testset "issue #144" begin
@test YAML.load("---") === nothing
end
# issue #226 - loadall stops on a null document
@testset "issue #226" begin
@test collect(YAML.load_all("null")) == [nothing]
input = """
---
1
---
null
---
2
"""
expected = [1, nothing, 2]
@test collect(YAML.load_all(input)) == expected
end
# issue #236 - special string keys that require enclosure in quotes
@testset "issue #236" begin
for char in "{}[]&*?#|-<>=!%@:`,\"'"
test_case_1 = Dict(string(char) => 0.0)
test_case_2 = Dict(string(char, "abcd") => 0.0)
test_case_3 = Dict(string("abcd", char) => 0.0)
test_case_4 = Dict(string("abcd", char, "efgh") => 0.0)
@test YAML.load(YAML.yaml(test_case_1)) == test_case_1
@test YAML.load(YAML.yaml(test_case_2)) == test_case_2
@test YAML.load(YAML.yaml(test_case_3)) == test_case_3
@test YAML.load(YAML.yaml(test_case_4)) == test_case_4
end
end
end # module
| YAML | https://github.com/JuliaData/YAML.jl.git |
|
[
"MIT"
]
| 0.4.12 | dea63ff72079443240fbd013ba006bcbc8a9ac00 | code | 68 | Dict("process" => Dict("sigma" => 0.1, :tag => :AR1, "rho" =>0.95))
| YAML | https://github.com/JuliaData/YAML.jl.git |
|
[
"MIT"
]
| 0.4.12 | dea63ff72079443240fbd013ba006bcbc8a9ac00 | code | 204 | Dict("process" => Dict("sigma" => 0.1, :tag => :AR1, "rho" =>0.95),
"grid" => Dict("orders" => [10, 50], "b" => ["1+2*sig_z", "k*1.1"], "a" => ["1-2*sig_z", "k*0.9"], "mu" => 3, :tag => :Cartesian))
| YAML | https://github.com/JuliaData/YAML.jl.git |
|
[
"MIT"
]
| 0.4.12 | dea63ff72079443240fbd013ba006bcbc8a9ac00 | code | 136 | Dict("grid" => Dict("orders" => [10, 50], "b" => ["1+2*sig_z", "k*1.1"], "a" => ["1-2*sig_z", "k*0.9"], "mu" => 3, :tag => :Cartesian))
| YAML | https://github.com/JuliaData/YAML.jl.git |
|
[
"MIT"
]
| 0.4.12 | dea63ff72079443240fbd013ba006bcbc8a9ac00 | code | 74 | Dict{Any,Any}("testlist" => Union{Nothing, String}[nothing, "one", "two"]) | YAML | https://github.com/JuliaData/YAML.jl.git |
|
[
"MIT"
]
| 0.4.12 | dea63ff72079443240fbd013ba006bcbc8a9ac00 | code | 32 | Dict{Any,Any}("hello"=>nothing)
| YAML | https://github.com/JuliaData/YAML.jl.git |
|
[
"MIT"
]
| 0.4.12 | dea63ff72079443240fbd013ba006bcbc8a9ac00 | code | 32 | Dict("thing" => Dict("x" => "")) | YAML | https://github.com/JuliaData/YAML.jl.git |
|
[
"MIT"
]
| 0.4.12 | dea63ff72079443240fbd013ba006bcbc8a9ac00 | code | 186 | Dict{Any,Any}(Pair{Any,Any}("string", String["\\0\\a\\b\\t\\n\\v\\f\\r\\e\\ \\\"\\\\\\N\\_\\L\\P"]),Pair{Any,Any}("escape sequences", String["\0\a\b\t\n\v\f\r\e \"\\\u85 \u2028\u2029"])) | YAML | https://github.com/JuliaData/YAML.jl.git |
|
[
"MIT"
]
| 0.4.12 | dea63ff72079443240fbd013ba006bcbc8a9ac00 | code | 230 | Dict{Any, Any}(
"a" => "here's to \"quotes\"",
"b" => "'",
"c" => "a'",
"d" => "'a",
"e" => "'a'",
"f" => "a''",
"g" => "''a",
"h" => " '",
"i" => "' ",
"j" => "''",
"k" => "' '"
)
| YAML | https://github.com/JuliaData/YAML.jl.git |
|
[
"MIT"
]
| 0.4.12 | dea63ff72079443240fbd013ba006bcbc8a9ac00 | code | 66 | Dict{Any, Any}(
"creator" => "LAMMPS",
"timestep" => 0,
)
| YAML | https://github.com/JuliaData/YAML.jl.git |
|
[
"MIT"
]
| 0.4.12 | dea63ff72079443240fbd013ba006bcbc8a9ac00 | code | 112 | Dict{Any,Any}(
"path" => "/foo",
"tiles" =>
Any[Dict{Any,Any}(
"path" => "/bar" )])
| YAML | https://github.com/JuliaData/YAML.jl.git |
|
[
"MIT"
]
| 0.4.12 | dea63ff72079443240fbd013ba006bcbc8a9ac00 | code | 41 | Dict{Any,Any}(
"x" => 1,
"y" => 2,
)
| YAML | https://github.com/JuliaData/YAML.jl.git |
|
[
"MIT"
]
| 0.4.12 | dea63ff72079443240fbd013ba006bcbc8a9ac00 | code | 74 | Dict{Any,Any}("columns" => Any[Dict{Any, Any}("foo" => """bar
""")])
| YAML | https://github.com/JuliaData/YAML.jl.git |
|
[
"MIT"
]
| 0.4.12 | dea63ff72079443240fbd013ba006bcbc8a9ac00 | code | 63 | Dict{Any,Any}(Pair{Any,Any}("field", "First line second line")) | YAML | https://github.com/JuliaData/YAML.jl.git |
|
[
"MIT"
]
| 0.4.12 | dea63ff72079443240fbd013ba006bcbc8a9ac00 | code | 478 | Any[
Dict{Any,Any}(
"x" => 1,
"y" => 2),
Dict{Any,Any}(
"x" => 0,
"y" => 2),
Dict{Any,Any}(
"r" => 10),
Dict{Any,Any}(
"r" => 1),
Dict{Any,Any}(
"label" => "center/big",
"r" => 10,
"x" => 1,
"y" => 2),
Dict{Any,Any}(
"label" => "center/big",
"r" => 10,
"x" => 1,
"y" => 2),
Dict{Any,Any}(
"label" => "center/big",
"r" => 10,
"x" => 1,
"y" => 2),
Dict{Any,Any}(
"label" => "center/big",
"r" => 10,
"x" => 1,
"y" => 2)
] | YAML | https://github.com/JuliaData/YAML.jl.git |
|
[
"MIT"
]
| 0.4.12 | dea63ff72079443240fbd013ba006bcbc8a9ac00 | code | 151 | Dict("process" => Dict("arg1" => "Arg 1", :tag => :test, "arg2" => "Arg 2"), "process1" => Dict("arg1" => "Arg 1", :tag => :test2, "arg2" => "Arg 2"))
| YAML | https://github.com/JuliaData/YAML.jl.git |
|
[
"MIT"
]
| 0.4.12 | dea63ff72079443240fbd013ba006bcbc8a9ac00 | code | 32 | Dict{Any,Any}("hello"=>nothing)
| YAML | https://github.com/JuliaData/YAML.jl.git |
|
[
"MIT"
]
| 0.4.12 | dea63ff72079443240fbd013ba006bcbc8a9ac00 | code | 49 | Any["Mark McGwire", "Sammy Sosa", "Ken Griffey"]
| YAML | https://github.com/JuliaData/YAML.jl.git |
|
[
"MIT"
]
| 0.4.12 | dea63ff72079443240fbd013ba006bcbc8a9ac00 | code | 56 | Dict{Any,Any}("hr" => 65, "avg" => 0.278, "rbi" => 147)
| YAML | https://github.com/JuliaData/YAML.jl.git |
|
[
"MIT"
]
| 0.4.12 | dea63ff72079443240fbd013ba006bcbc8a9ac00 | code | 169 | Dict{Any,Any}("american" =>
Any["Boston Red Sox", "Detroit Tigers", "New York Yankees"],
"national" =>
Any["New York Mets", "Chicago Cubs", "Atlanta Braves"]
)
| YAML | https://github.com/JuliaData/YAML.jl.git |
|
[
"MIT"
]
| 0.4.12 | dea63ff72079443240fbd013ba006bcbc8a9ac00 | code | 182 | Any[
Dict{Any,Any}(
"name" => "Mark McGwire",
"hr" => 65,
"avg" => 0.278),
Dict{Any,Any}(
"name" => "Sammy Sosa",
"hr" => 63,
"avg" => 0.288)
]
| YAML | https://github.com/JuliaData/YAML.jl.git |
|
[
"MIT"
]
| 0.4.12 | dea63ff72079443240fbd013ba006bcbc8a9ac00 | code | 107 | Any[
Any["name", "hr", "avg"],
Any["Mark McGwire", 65, 0.278],
Any["Sammy Sosa", 63, 0.288]
]
| YAML | https://github.com/JuliaData/YAML.jl.git |
|
[
"MIT"
]
| 0.4.12 | dea63ff72079443240fbd013ba006bcbc8a9ac00 | code | 144 | Dict{Any,Any}(
"Mark McGwire" => Dict{Any,Any}("hr" => 65, "avg" => 0.278),
"Sammy Sosa" => Dict{Any,Any}("hr" => 63, "avg" => 0.288)
)
| YAML | https://github.com/JuliaData/YAML.jl.git |
|
[
"MIT"
]
| 0.4.12 | dea63ff72079443240fbd013ba006bcbc8a9ac00 | code | 49 | Any["Mark McGwire", "Sammy Sosa", "Ken Griffey"]
| YAML | https://github.com/JuliaData/YAML.jl.git |
|
[
"MIT"
]
| 0.4.12 | dea63ff72079443240fbd013ba006bcbc8a9ac00 | code | 169 | # TODO: implement sexagesimal numbers and figure out what this should be.
Dict{Any,Any}(
"time" => "20:03:20",
"player" => "Sammy Sosa",
"action" => "strike (miss)")
| YAML | https://github.com/JuliaData/YAML.jl.git |
|
[
"MIT"
]
| 0.4.12 | dea63ff72079443240fbd013ba006bcbc8a9ac00 | code | 110 | Dict{Any,Any}(
"hr" => Any["Mark McGwire", "Sammy Sosa"],
"rbi" => Any["Sammy Sosa", "Ken Griffey"]
)
| YAML | https://github.com/JuliaData/YAML.jl.git |
|
[
"MIT"
]
| 0.4.12 | dea63ff72079443240fbd013ba006bcbc8a9ac00 | code | 110 | Dict{Any,Any}(
"hr" => Any["Mark McGwire", "Sammy Sosa"],
"rbi" => Any["Sammy Sosa", "Ken Griffey"]
)
| YAML | https://github.com/JuliaData/YAML.jl.git |
|
[
"MIT"
]
| 0.4.12 | dea63ff72079443240fbd013ba006bcbc8a9ac00 | code | 230 | using Dates
Dict{Any,Any}(
Any["Detroit Tigers", "Chicago cubs"] =>
Any[Date(2001, 7, 23)],
Any["New York Yankees", "Atlanta Braves"] => Any[
Date(2001, 7, 2),
Date(2001, 8, 12),
Date(2001, 8, 14)])
| YAML | https://github.com/JuliaData/YAML.jl.git |
|
[
"MIT"
]
| 0.4.12 | dea63ff72079443240fbd013ba006bcbc8a9ac00 | code | 210 | Any[
Dict{Any,Any}(
"item" => "Super Hoop",
"quantity" => 1),
Dict{Any,Any}(
"item" => "Basketball",
"quantity" => 4),
Dict{Any,Any}(
"item" => "Big Shoes",
"quantity" => 1)
]
| YAML | https://github.com/JuliaData/YAML.jl.git |
|
[
"MIT"
]
| 0.4.12 | dea63ff72079443240fbd013ba006bcbc8a9ac00 | code | 32 | """
\\//||\\/||
// || ||__
"""
| YAML | https://github.com/JuliaData/YAML.jl.git |
|
[
"MIT"
]
| 0.4.12 | dea63ff72079443240fbd013ba006bcbc8a9ac00 | code | 55 | "Mark McGwire's year was crippled by a knee injury.\n"
| YAML | https://github.com/JuliaData/YAML.jl.git |
|
[
"MIT"
]
| 0.4.12 | dea63ff72079443240fbd013ba006bcbc8a9ac00 | code | 121 | """
Sammy Sosa completed another fine season with great stats.
63 Home Runs
0.288 Batting Average
What a year!
"""
| YAML | https://github.com/JuliaData/YAML.jl.git |
|
[
"MIT"
]
| 0.4.12 | dea63ff72079443240fbd013ba006bcbc8a9ac00 | code | 223 | Dict{Any,Any}(
"name" => "Mark McGwire",
"accomplishment" =>
"Mark set a major league home run record in 1998.\n",
"stats" =>
"""
65 Home Runs
0.278 Batting Average
"""
)
| YAML | https://github.com/JuliaData/YAML.jl.git |
|
[
"MIT"
]
| 0.4.12 | dea63ff72079443240fbd013ba006bcbc8a9ac00 | code | 269 | Dict{Any,Any}(
"unicode" => "Sosa did fine.\u263A",
"control" => "\b1998\t1999\t2000\n",
"hexesc" => "\x13\x10 is \r\n",
"single" => "\"Howdy!\" he cried.",
"quoted" => " # not a 'comment'.",
"tie-fighter" => "|\\-*-/|"
)
| YAML | https://github.com/JuliaData/YAML.jl.git |
|
[
"MIT"
]
| 0.4.12 | dea63ff72079443240fbd013ba006bcbc8a9ac00 | code | 124 | Dict{Any,Any}(
"plain" => "This unquoted scalar\nspans many lines.",
"quoted" => "So does this\nquoted scalar.\n"
)
| YAML | https://github.com/JuliaData/YAML.jl.git |
|
[
"MIT"
]
| 0.4.12 | dea63ff72079443240fbd013ba006bcbc8a9ac00 | code | 201 | Dict{Any,Any}(
"canonical" => 12345,
"decimal" => 12345,
# TODO: change this when sexagesimal is implemented
"sexagesimal" => "3:25:45",
"octal" => 12,
"hexadecimal" => 0xC
)
| YAML | https://github.com/JuliaData/YAML.jl.git |
|
[
"MIT"
]
| 0.4.12 | dea63ff72079443240fbd013ba006bcbc8a9ac00 | code | 283 | Dict{Any,Any}(
"canonical" => 1.23015e3,
"exponential" => 12.3015e2,
# TODO: replace when sexagesimal is implemented
"sexagesimal" => "20:30.15",
"fixed" => 1230.15,
"negative infinity" => -Inf,
"not a number" => NaN
)
| YAML | https://github.com/JuliaData/YAML.jl.git |
|
[
"MIT"
]
| 0.4.12 | dea63ff72079443240fbd013ba006bcbc8a9ac00 | code | 108 | Dict{Any,Any}(
nothing => nothing,
true => "y",
false => "n",
"string" => "12345"
)
| YAML | https://github.com/JuliaData/YAML.jl.git |
|
[
"MIT"
]
| 0.4.12 | dea63ff72079443240fbd013ba006bcbc8a9ac00 | code | 248 | using Dates
Dict{Any,Any}(
"canonical" => DateTime(2001, 12, 15, 2, 59, 43, 100),
"iso8601" => DateTime(2001, 12, 14, 21, 59, 43, 100),
"spaced" => DateTime(2001, 12, 14, 21, 59, 43, 100),
"date" => Date(2002, 12, 14)
)
| YAML | https://github.com/JuliaData/YAML.jl.git |
|
[
"MIT"
]
| 0.4.12 | dea63ff72079443240fbd013ba006bcbc8a9ac00 | code | 412 | using Base64
Dict{Any,Any}(
"not-date" => "2002-04-28",
"picture" =>
base64decode("R0lGODlhDAAMAIQAAP//9/X17unp5WZmZgAAAOfn515eXvPz7Y6OjuDg4J+fn5OTk6enp56enmleECcgggoBADs=")
## Disabled since we don't really support custom tags
#"application specific tag" =>
#"""
#The semantics of the tag
#above may be different for
#different documents.
#"""
)
| YAML | https://github.com/JuliaData/YAML.jl.git |
|
[
"MIT"
]
| 0.4.12 | dea63ff72079443240fbd013ba006bcbc8a9ac00 | code | 36 | Dict{Any,Any}(
"attribute" => 1
)
| YAML | https://github.com/JuliaData/YAML.jl.git |
|
[
"MIT"
]
| 0.4.12 | dea63ff72079443240fbd013ba006bcbc8a9ac00 | code | 36 | Dict{Any,Any}(
"attribute" => 1
)
| YAML | https://github.com/JuliaData/YAML.jl.git |
|
[
"MIT"
]
| 0.4.12 | dea63ff72079443240fbd013ba006bcbc8a9ac00 | code | 36 | Dict{Any,Any}(
"attribute" => 1
)
| YAML | https://github.com/JuliaData/YAML.jl.git |
|
[
"MIT"
]
| 0.4.12 | dea63ff72079443240fbd013ba006bcbc8a9ac00 | code | 32 | Dict{Any,Any}("hello"=>nothing)
| YAML | https://github.com/JuliaData/YAML.jl.git |
|
[
"MIT"
]
| 0.4.12 | dea63ff72079443240fbd013ba006bcbc8a9ac00 | docs | 4224 | # YAML
[](https://github.com/JuliaData/YAML.jl/actions?query=workflow%3ACI)
[](https://codecov.io/gh/JuliaData/YAML.jl)
[YAML](http://yaml.org/) is a flexible data serialization format that is
designed to be easily read and written by human beings.
This library parses YAML documents into native Julia types and dumps them back into YAML documents.
## Synopsis
For most purposes there is one important function: `YAML.load`, which takes a
string and parses it the first YAML document it finds.
To parse a file use `YAML.load_file`, and to parse every document in a file
use `YAML.load_all` or `YAML.load_all_file`.
Given a YAML document like the following
```yaml
receipt: Oz-Ware Purchase Invoice
date: 2012-08-06
customer:
given: Dorothy
family: Gale
items:
- part_no: A4786
descrip: Water Bucket (Filled)
price: 1.47
quantity: 4
- part_no: E1628
descrip: High Heeled "Ruby" Slippers
size: 8
price: 100.27
quantity: 1
bill-to: &id001
street: |
123 Tornado Alley
Suite 16
city: East Centerville
state: KS
ship-to: *id001
specialDelivery: >
Follow the Yellow Brick
Road to the Emerald City.
Pay no attention to the
man behind the curtain.
```
It can be loaded with
```julia
import YAML
data = YAML.load_file("test.yml")
println(data)
```
Which will show you something like this.
```
{"date"=>Aug 6, 2012 12:00:00 AM PDT,"ship-to"=>{"street"=>"123 Tornado Alley\nSuite 16\n","state"=>"KS","city"=>"East Centerville"},"customer"=>{"given"=>"Dorothy","family"=>"Gale"},"specialDelivery"=>"Follow the Yellow Brick\nRoad to the Emerald City.\nPay no attention to the\nman behind the curtain.\n","items"=>{{"price"=>1.47,"descrip"=>"Water Bucket (Filled)","part_no"=>"A4786","quantity"=>4} … {"price"=>100.27,"size"=>8,"descrip"=>"High Heeled \"Ruby\" Slippers","part_no"=>"E1628","quantity"=>1}},"bill-to"=>{"street"=>"123 Tornado Alley\nSuite 16\n","state"=>"KS","city"=>"East Centerville"},"receipt"=>"Oz-Ware Purchase Invoice"}
```
Note that ints and floats are recognized, as well as timestamps which are parsed
into CalendarTime objects. Also, anchors and references work as expected,
without making a copy.
Dictionaries are parsed into instances of `Dict{Any,Any}` by default.
You can, however, specify a custom type in which to parse all dictionaries.
```julia
# using Symbol keys
data = YAML.load_file("test.yml"; dicttype=Dict{Symbol,Any})
# maintaining the order from the YAML file
using OrderedCollections
data = YAML.load_file("test.yml"; dicttype=OrderedDict{String,Any})
# specifying a default value
using DataStructures
data = YAML.load_file("test.yml"; dicttype=()->DefaultDict{String,Any}(Missing))
```
## Writing to YAML
Similar to reading files, you can emit Julia objects to YAML files by calling
`write_file`, or to a string object by calling `write`.
For example, you can reproduce the above file from the variable `data`
```julia
import YAML
YAML.write_file("test-output.yml", data)
```
which gives you (omitting the precise format but maintaining the content)
```yaml
receipt: "Oz-Ware Purchase Invoice"
items:
- part_no: "A4786"
price: 1.47
descrip: "Water Bucket (Filled)"
quantity: 4
- part_no: "E1628"
price: 100.27
size: 8
descrip: "High Heeled "Ruby" Slippers"
quantity: 1
customer:
given: "Dorothy"
family: "Gale"
ship-to:
city: "East Centerville"
street: |
123 Tornado Alley
Suite 16
state: "KS"
bill-to:
city: "East Centerville"
street: |
123 Tornado Alley
Suite 16
state: "KS"
specialDelivery: |
Follow the Yellow Brick Road to the Emerald City. Pay no attention to the man behind the curtain.
date: 2012-08-06
```
## Not yet implemented
* When writing YAML files, you cannot use additional constructors like you can when reading.
* Parsing sexigesimal numbers.
* Fractions of seconds in timestamps.
* Specific time-zone offsets in timestamps.
* Application specific tags.
| YAML | https://github.com/JuliaData/YAML.jl.git |
|
[
"MIT"
]
| 0.1.2 | 5d3da693a47ae483fc97e9bb2f1c1b4421118b98 | code | 1218 | module TSPLIB
using DataStructures
using Match
export TSP, readTSP, readTSPLIB, TSPLIB95_path, findTSP, Optimals
struct TSP
name::AbstractString
dimension::Integer
weight_type::AbstractString
weights::Matrix
nodes::Matrix
Dnodes::Bool
ffx::Function
pfx::Function
optimal::Float64
end
const TSPLIB95_path = joinpath(pkgdir(TSPLIB), "data", "TSPLIB95", "tsp")
const tsp_keys = ["NAME",
"TYPE",
"COMMENT",
"DIMENSION",
"EDGE_WEIGHT_TYPE",
"EDGE_WEIGHT_FORMAT",
"EDGE_DATA_FORMAT",
"NODE_COORD_TYPE",
"DISPLAY_DATA_TYPE",
"NODE_COORD_SECTION",
"DEPOT_SECTION",
"DEMAND_SECTION",
"EDGE_DATA_SECTION",
"FIXED_EDGES_SECTION",
"DISPLAY_DATA_SECTION",
"TOUR_SECTION",
"EDGE_WEIGHT_SECTION",
"EOF"]
include("reader.jl")
include("distances.jl")
include("utils.jl")
include("fitness.jl")
include("optimals.jl")
end # module
| TSPLIB | https://github.com/matago/TSPLIB.jl.git |
|
[
"MIT"
]
| 0.1.2 | 5d3da693a47ae483fc97e9bb2f1c1b4421118b98 | code | 2047 |
function euclidian(x::Vector{T},y::Vector{T}) where T<:Real
nsz = length(x)
dist = zeros(T,nsz,nsz)
for i in 1:nsz, j in 1:nsz
if i<=j
xd = (x[i]-x[j])^2
yd = (y[i]-y[j])^2
dist[i,j] = dist[j,i] = round(sqrt(xd+yd),RoundNearestTiesUp)
end
end
return dist
end
function manhattan(x::Vector{T},y::Vector{T}) where T<:Real
nsz = length(x)
dist = zeros(T, nsz, nsz)
for i in 1:nsz, j in 1:nsz
if i<=j
xd = abs(x[i] - x[j])
yd = abs(y[i] - y[j])
dist[i, j] = dist[j, i] = round(xd + yd, RoundNearestTiesUp)
end
end
return dist
end
function max_norm(x::Vector{T},y::Vector{T}) where T<:Real
nsz = length(x)
dist = zeros(T, nsz, nsz)
for i in 1:nsz, j in 1:nsz
if i<=j
xd = abs(x[i] - x[j])
yd = abs(y[i] - y[j])
dist[i, j] = dist[j, i] = max(round(xd, RoundNearestTiesUp), round(yd, RoundNearestTiesUp))
end
end
return dist
end
function att_euclidian(x::Vector{T},y::Vector{T}) where T<:Real
nsz = length(x)
dist = zeros(T,nsz,nsz)
for i in 1:nsz, j in 1:nsz
if i<=j
xd = (x[i]-x[j])^2
yd = (y[i]-y[j])^2
dist[i,j] = dist[j,i] = ceil(sqrt((xd+yd)/10.0))
end
end
return dist
end
function ceil_euclidian(x::Vector{T},y::Vector{T}) where T<:Real
nsz = length(x)
dist = zeros(T,nsz,nsz)
for i in 1:nsz, j in 1:nsz
if i<=j
xd = (x[i]-x[j])^2
yd = (y[i]-y[j])^2
dist[i,j] = dist[j,i] = ceil(sqrt(xd+yd))
end
end
return dist
end
function geo(x::Vector{T},y::Vector{T}) where T<:Real
PI = 3.141592
RRR = 6378.388
nsz = length(x)
dist = zeros(T,nsz,nsz)
degs = trunc.(hcat(x,y))
mins = hcat(x,y).-degs
coords = PI.*(degs.+(5.0.*(mins./3.0)))./180.0
lat = coords[:,1]
lon = coords[:,2]
for i in 1:nsz, j in 1:nsz
if i<=j
q1 = cos(lon[i]-lon[j])
q2 = cos(lat[i]-lat[j])
q3 = cos(lat[i]+lat[j])
dij = RRR.*acos(0.5.*((1.0.+q1).*q2.-(1.0.-q1).*q3)).+1.0
dist[i,j] = dist[j,i] = floor(dij)
end
end
return dist
end
| TSPLIB | https://github.com/matago/TSPLIB.jl.git |
|
[
"MIT"
]
| 0.1.2 | 5d3da693a47ae483fc97e9bb2f1c1b4421118b98 | code | 960 | #=Generator function for TSP that takes the weight Matrix
and returns a function that evaluates the fitness of a single path=#
function fullFit(costs::AbstractMatrix{Float64})
N = size(costs,1)
function fFit(tour::Vector{T}) where T<:Integer
@assert length(tour) == N "Tour must be of length $N"
@assert isperm(tour) "Not a valid tour, not a permutation"
#distance = weights[from,to] (from,to) in tour
distance = costs[tour[N],tour[1]]
for i in 1:N-1
@inbounds distance += costs[tour[i],tour[i+1]]
end
return distance
end
return fFit
end
function partFit(costs::AbstractMatrix{Float64})
N = size(costs,1)
function pFit(tour::Vector{T}) where T<:Integer
n = length(tour)
#distance = weights[from,to] (from,to) in tour
distance = n == N ? costs[tour[N],tour[1]] : zero(Float64)
for i in 1:n-1
@inbounds distance += costs[tour[i],tour[i+1]]
end
return distance
end
return pFit
end
| TSPLIB | https://github.com/matago/TSPLIB.jl.git |
|
[
"MIT"
]
| 0.1.2 | 5d3da693a47ae483fc97e9bb2f1c1b4421118b98 | code | 3793 | const Optimals = Dict{Symbol,Float64}(
[:a280 => 2579,
:ali535 => 202339,
:att48 => 10628,
:att532 => 27686,
:bayg29 => 1610,
:bays29 => 2020,
:berlin52 => 7542,
:bier127 => 118282,
:brazil58 => 25395,
:brd14051 => 469385,
:brg180 => 1950,
:burma14 => 3323,
:ch130 => 6110,
:ch150 => 6528,
:d198 => 15780,
:d493 => 35002,
:d657 => 48912,
:d1291 => 50801,
:d1655 => 62128,
:d2103 => 80450,
:d15112 => 1573084,
:d18512 => 645238,
:dantzig42 => 699,
:dsj1000 => 18659688,
:eil51 => 426,
:eil76 => 538,
:eil101 => 629,
:fl417 => 11861,
:fl1400 => 20127,
:fl1577 => 22249,
:fl3795 => 28772,
:fnl4461 => 182566,
:fri26 => 937,
:gil262 => 2378,
:gr17 => 2085,
:gr21 => 2707,
:gr24 => 1272,
:gr48 => 5046,
:gr96 => 55209,
:gr120 => 6942,
:gr137 => 69853,
:gr202 => 40160,
:gr229 => 134602,
:gr431 => 171414,
:gr666 => 294358,
:hk48 => 11461,
:kroA100 => 21282,
:kroB100 => 22141,
:kroC100 => 20749,
:kroD100 => 21294,
:kroE100 => 22068,
:kroA150 => 26524,
:kroB150 => 26130,
:kroA200 => 29368,
:kroB200 => 29437,
:lin105 => 14379,
:lin318 => 42029,
:linhp318 => 41345,
:nrw1379 => 56638,
:p654 => 34643,
:pa561 => 2763,
:pcb442 => 50778,
:pcb1173 => 56892,
:pcb3038 => 137694,
:pla7397 => 23260728,
:pla33810 => 66048945,
:pla85900 => 142382641,
:pr76 => 108159,
:pr107 => 44303,
:pr124 => 59030,
:pr136 => 96772,
:pr144 => 58537,
:pr152 => 73682,
:pr226 => 80369,
:pr264 => 49135,
:pr299 => 48191,
:pr439 => 107217,
:pr1002 => 259045,
:pr2392 => 378032,
:rat99 => 1211,
:rat195 => 2323,
:rat575 => 6773,
:rat783 => 8806,
:rd100 => 7910,
:rd400 => 15281,
:rl1304 => 252948,
:rl1323 => 270199,
:rl1889 => 316536,
:rl5915 => 565530,
:rl5934 => 556045,
:rl11849 => 923288,
:si175 => 21407,
:si535 => 48450,
:si1032 => 92650,
:st70 => 675,
:swiss42 => 1273,
:ts225 => 126643,
:tsp225 => 3916,
:u159 => 42080,
:u574 => 36905,
:u724 => 41910,
:u1060 => 224094,
:u1432 => 152970,
:u1817 => 57201,
:u2152 => 64253,
:u2319 => 234256,
:ulysses16 => 6859,
:ulysses22 => 7013,
:usa13509 => 19982859,
:vm1084 => 239297,
:vm1748 => 336556]) | TSPLIB | https://github.com/matago/TSPLIB.jl.git |
|
[
"MIT"
]
| 0.1.2 | 5d3da693a47ae483fc97e9bb2f1c1b4421118b98 | code | 2976 |
function readTSP(path::AbstractString)
raw = read(path, String)
checkEOF(raw)
return _generateTSP(raw)
end
readTSPLIB(instance::Symbol) = readTSP(joinpath(TSPLIB95_path,string(instance)*".tsp"))
function _generateTSP(raw::AbstractString)
_dict = keyextract(raw, tsp_keys)
name = _dict["NAME"]
dimension = parse(Int,_dict["DIMENSION"])
weight_type = _dict["EDGE_WEIGHT_TYPE"]
dxp = false
if weight_type == "EXPLICIT" && haskey(_dict,"EDGE_WEIGHT_SECTION")
explicits = parse.(Float64, split(_dict["EDGE_WEIGHT_SECTION"]))
weights = explicit_weights(_dict["EDGE_WEIGHT_FORMAT"],explicits)
#Push display data to nodes if possible
if haskey(_dict,"DISPLAY_DATA_SECTION")
coords = parse.(Float64, split(_dict["DISPLAY_DATA_SECTION"]))
n_r = convert(Integer,length(coords)/dimension)
nodes = reshape(coords,(n_r,dimension))'[:,2:end]
dxp = true
else
nodes = zeros(dimension,2)
end
elseif haskey(_dict,"NODE_COORD_SECTION")
coords = parse.(Float64, split(_dict["NODE_COORD_SECTION"]))
n_r = convert(Integer,length(coords)/dimension)
nodes = reshape(coords,(n_r,dimension))'[:,2:end]
weights = calc_weights(_dict["EDGE_WEIGHT_TYPE"], nodes)
end
fFX = fullFit(weights)
pFX = partFit(weights)
if haskey(Optimals, Symbol(name))
optimal = Optimals[Symbol(name)]
else
optimal = -1
end
TSP(name,dimension,weight_type,weights,nodes,dxp,fFX,pFX,optimal)
end
function keyextract(raw::T,ks::Array{T}) where T<:AbstractString
pq = PriorityQueue{T,Tuple{Integer,Integer}}()
vals = Dict{T,T}()
for k in ks
idx = findfirst(k,raw)
idx != nothing && enqueue!(pq,k,extrema(idx))
end
while length(pq) > 1
s_key, s_pts = peek(pq)
dequeue!(pq)
f_key, f_pts = peek(pq)
rng = (s_pts[2]+1):(f_pts[1]-1)
vals[s_key] = strip(replace(raw[rng],":"=>""))
end
return vals
end
function explicit_weights(key::AbstractString,data::Vector{Float64})
w = @match key begin
"UPPER_DIAG_ROW" => vec2UDTbyRow(data)
"LOWER_DIAG_ROW" => vec2LDTbyRow(data)
"UPPER_DIAG_COL" => vec2UDTbyCol(data)
"LOWER_DIAG_COL" => vec2LDTbyCol(data)
"UPPER_ROW" => vec2UTbyRow(data)
"LOWER_ROW" => vec2LTbyRow(data)
"FULL_MATRIX" => vec2FMbyRow(data)
end
if !in(key,["FULL_MATRIX"])
w.+=w'
end
return w
end
function calc_weights(key::AbstractString, data::Matrix)
w = @match key begin
"EUC_2D" => euclidian(data[:,1], data[:,2])
"MAN_2D" => manhattan(data[:,1], data[:,2])
"MAX_2D" => max_norm(data[:,1], data[:,2])
"GEO" => geo(data[:,1], data[:,2])
"ATT" => att_euclidian(data[:,1], data[:,2])
"CEIL_2D" => ceil_euclidian(data[:,1], data[:,2])
_ => error("Distance function type $key is not supported.")
end
return w
end
function checkEOF(raw::AbstractString)
n = findlast("EOF",raw)
if n == nothing
throw("EOF not found")
end
return
end
| TSPLIB | https://github.com/matago/TSPLIB.jl.git |
|
[
"MIT"
]
| 0.1.2 | 5d3da693a47ae483fc97e9bb2f1c1b4421118b98 | code | 2045 | function vec2LDTbyRow(v::AbstractVector{T}, z::T=zero(T)) where T
n = length(v)
s = round(Integer,(sqrt(8n+1)-1)/2)
s*(s+1)/2 == n || error("vec2LTbyRow: length of vector is not triangular")
k=0
[i<=j ? (k+=1; v[k]) : z for i=1:s, j=1:s]'
end
function vec2UDTbyRow(v::AbstractVector{T}, z::T=zero(T)) where T
n = length(v)
s = round(Integer,(sqrt(8n+1)-1)/2)
s*(s+1)/2 == n || error("vec2UTbyRow: length of vector is not triangular")
k=0
[i>=j ? (k+=1; v[k]) : z for i=1:s, j=1:s]'
end
function vec2LDTbyCol(v::AbstractVector{T}, z::T=zero(T)) where T
n = length(v)
s = round(Integer,(sqrt(8n+1)-1)/2)
s*(s+1)/2 == n || error("vec2LTbyCol: length of vector is not triangular")
k=0
[i>=j ? (k+=1; v[k]) : z for i=1:s, j=1:s]
end
function vec2UDTbyCol(v::AbstractVector{T}, z::T=zero(T)) where T
n = length(v)
s = round(Integer,(sqrt(8n+1)-1)/2)
s*(s+1)/2 == n || error("vec2UTbyCol: length of vector is not triangular")
k=0
[i<=j ? (k+=1; v[k]) : z for i=1:s, j=1:s]
end
function vec2UTbyRow(v::AbstractVector{T}, z::T=zero(T)) where T
n = length(v)
s = round(Integer,((sqrt(8n+1)-1)/2)+1)
(s*(s+1)/2)-s == n || error("vec2UTbyRow: length of vector is not triangular")
k=0
[i>j ? (k+=1; v[k]) : z for i=1:s, j=1:s]'
end
function vec2LTbyRow(v::AbstractVector{T}, z::T=zero(T)) where T
n = length(v)
s = round(Integer,((sqrt(8n+1)-1)/2)+1)
(s*(s+1)/2)-s == n || error("vec2LTbyRow: length of vector is not triangular")
k=0
[i<j ? (k+=1; v[k]) : z for i=1:s, j=1:s]'
end
function vec2FMbyRow(v::AbstractVector{T}, z::T=zero(T)) where T
n = length(v)
s = round(Int,sqrt(n))
s^2 == n || error("vec2FMbyRow: length of vector is not square")
k=0
[(k+=1; v[k]) for i=1:s, j=1:s]
end
function findTSP(path::AbstractString)
if isdir(path)
syms = [Symbol(split(file,".")[1]) for file in readdir(path) if (split(file,".")[end] == "tsp")]
else
error("Not a valid directory")
end
return syms
end
| TSPLIB | https://github.com/matago/TSPLIB.jl.git |
|
[
"MIT"
]
| 0.1.2 | 5d3da693a47ae483fc97e9bb2f1c1b4421118b98 | code | 472 | using TSPLIB
using Test
@testset "TSPLIB.jl" begin
exclude_list = [
:rl11849
:usa13509
:brd14051
:d15112
:d18512
:pla33810
:pla85900
]
for instance in keys(Optimals)
if !(instance in exclude_list)
@test readTSPLIB(instance).optimal == Optimals[instance]
end
end
tsp = readTSP(joinpath(@__DIR__, "../data/TSPLIB95/tsp/a280.tsp"))
@test tsp.name == "a280"
end
| TSPLIB | https://github.com/matago/TSPLIB.jl.git |
|
[
"MIT"
]
| 0.1.2 | 5d3da693a47ae483fc97e9bb2f1c1b4421118b98 | docs | 1168 | # TSPLIB.jl
[](https://github.com/matago/TSPLIB.jl/actions?query=workflow%3ACI)
[](https://codecov.io/gh/matago/TSPLIB.jl)
This reads `.tsp` data files in the [TSPLIB format](http://webhotel4.ruc.dk/~keld/research/LKH/LKH-2.0/DOC/TSPLIB_DOC.pdf) for Traveling Salesman Problem (TSP) instances and returns `TSP` type.
```julia
struct TSP
name ::AbstractString
dimension ::Integer
weight_type ::AbstractString
weights ::Matrix
nodes ::Matrix
Dnodes ::Bool
ffx ::Function
pfx ::Function
optimal ::Float64
end
```
To install:
```julia
] add TSPLIB
```
Some TSP instances in the [TSPLIB](http://elib.zib.de/pub/mp-testdata/tsp/tsplib/tsplib.html) library are preloaded. See the [list](https://github.com/matago/TSPLIB.jl/tree/master/data/TSPLIB95/tsp).
For example, to load `a280.tsp`, you can do:
```julia
tsp = readTSPLIB(:a280)
```
For custom TSP files, you can load:
```julia
tsp = readTSP(path_to_tsp_file)
```
| TSPLIB | https://github.com/matago/TSPLIB.jl.git |
|
[
"MIT"
]
| 0.1.0 | de8c6b67c7b13044dc683d14c72ad88de9d5dee0 | code | 2491 | #=
References:
- https://www.juliafordatascience.com/artifacts/
- https://github.com/simeonschaub/ArtifactUtils.jl
- https://github.com/FluxML/Metalhead.jl
- https://github.com/FluxML/MetalheadWeights
- https://docs.github.com/en/authentication/connecting-to-github-with-ssh/generating-a-new-ssh-key-and-adding-it-to-the-ssh-agent
- https://docs.github.com/en/authentication/connecting-to-github-with-ssh/adding-a-new-ssh-key-to-your-github-account
- https://docs.github.com/en/authentication/connecting-to-github-with-ssh/testing-your-ssh-connection
=#
using Pkg.Artifacts
using Pkg.GitTools
import HuggingFaceHub as HF
"""Copied from https://github.com/simeonschaub/ArtifactUtils.jl/blob/main/src/ArtifactUtils.jl"""
function copy_mode(src, dst)
if !isdir(dst)
chmod(dst, UInt(GitTools.gitmode(string(src))))
return nothing
end
for name in readdir(dst)
sub_src = joinpath(src, name)
sub_dst = joinpath(dst, name)
copy_mode(sub_src, sub_dst)
end
end
model_dir = abspath(joinpath(@__DIR__, "models"))
build_dir = abspath(joinpath(@__DIR__, "build"))
artifact_toml = abspath(joinpath(@__DIR__, "..", "Artifacts.toml"))
# Clean up prior builds.
ispath(build_dir) && rm(build_dir; recursive=true, force=true)
mkdir(build_dir)
# Create model repo
repo_id = "yezhengkai/RemBG"
HF.create(HF.Model(; id=repo_id, private=false)) # BUG
repo = HF.info(HF.Model(; id=repo_id))
# Package up models
for model_path in readdir(model_dir; join=true)
model_name = splitext(basename(model_path))[1]
artifact_filename = "$model_name.tar.gz"
artifact_filepath = joinpath(build_dir, artifact_filename)
@info "Creating:" model_name
product_hash = create_artifact() do artifact_dir
cp(model_path, joinpath(artifact_dir, basename(model_path)); force=true)
Sys.iswindows() && copy_mode(model_path, artifact_dir)
end
@info "Create artifact:" product_hash
download_hash = archive_artifact(product_hash, joinpath(build_dir, artifact_filename))
@info "Archive artifact:" download_hash
remote_url = "https://huggingface.co/$repo_id/resolve/main/$artifact_filename"
@info "Upload to Hugging Face models" artifact_filename remote_url
HF.file_upload(repo, artifact_filename, artifact_filepath) # BUG
bind_artifact!(
artifact_toml,
model_name,
product_hash;
force=true,
lazy=true,
download_info=Tuple[(remote_url, download_hash)],
)
end
| RemBG | https://github.com/yezhengkai/RemBG.jl.git |
|
[
"MIT"
]
| 0.1.0 | de8c6b67c7b13044dc683d14c72ad88de9d5dee0 | code | 307 | using Tar, Inflate, SHA
build_dir = abspath(joinpath(@__DIR__, "build"))
for tarfile in readdir(build_dir; join=true)
println("Checking: " * basename(tarfile))
println("git-tree-sha1: ", Tar.tree_hash(IOBuffer(inflate_gzip(tarfile))))
println("sha256: ", bytes2hex(open(sha256, tarfile)))
end
| RemBG | https://github.com/yezhengkai/RemBG.jl.git |
|
[
"MIT"
]
| 0.1.0 | de8c6b67c7b13044dc683d14c72ad88de9d5dee0 | code | 734 | using Downloads
uris = (
"https://github.com/danielgatis/rembg/releases/download/v0.0.0/u2net.onnx",
"https://github.com/danielgatis/rembg/releases/download/v0.0.0/u2netp.onnx",
"https://github.com/danielgatis/rembg/releases/download/v0.0.0/u2net_human_seg.onnx",
"https://github.com/danielgatis/rembg/releases/download/v0.0.0/u2net_cloth_seg.onnx",
"https://github.com/danielgatis/rembg/releases/download/v0.0.0/silueta.onnx",
"https://github.com/danielgatis/rembg/releases/download/v0.0.0/isnet-general-use.onnx",
)
mkpath("models")
@sync for uri in uris
model_path = joinpath("models", rsplit(uri, "/")[end])
isfile(model_path) && continue
@async @show Downloads.download(uri, model_path)
end
| RemBG | https://github.com/yezhengkai/RemBG.jl.git |
|
[
"MIT"
]
| 0.1.0 | de8c6b67c7b13044dc683d14c72ad88de9d5dee0 | code | 702 | using RemBG
using Documenter
DocMeta.setdocmeta!(RemBG, :DocTestSetup, :(using RemBG); recursive=true)
makedocs(;
modules=[RemBG],
authors="Zheng-Kai Ye <[email protected]> and contributors",
repo="https://github.com/yezhengkai/RemBG.jl/blob/{commit}{path}#{line}",
sitename="RemBG.jl",
format=Documenter.HTML(;
prettyurls=get(ENV, "CI", "false") == "true",
canonical="https://yezhengkai.github.io/RemBG.jl",
edit_link="main",
assets=String[],
),
pages=[
"Home" => "index.md",
"Reference" => ["reference/public.md", "reference/internal.md"],
],
)
deploydocs(; repo="github.com/yezhengkai/RemBG.jl", devbranch="main")
| RemBG | https://github.com/yezhengkai/RemBG.jl.git |
|
[
"MIT"
]
| 0.1.0 | de8c6b67c7b13044dc683d14c72ad88de9d5dee0 | code | 1626 | using RemBG
using Images
exampledir(args...) = abspath(joinpath(dirname(pathof(RemBG)), "..", "examples", args...))
img_animal = load(exampledir("animal-1.jpg"))::Matrix{<:Colorant};
img_animal_num = copy(channelview(img_animal))::Array{<:Number};
img_cloth = load(exampledir("cloth-1.jpg"))::Matrix{<:Colorant};
img_cloth_num = copy(channelview(img))::Array{<:Number};
output_img_num = remove(img_animal_num);
output_img = remove(img_animal); # If we do not pass an inference session to `remove`, it will use a new session containing the U2Net model.
# Use a specific model. The available models are U2Net, U2Netp, U2NetClothSeg, U2NetHumanSeg, Silueta, ISNetGeneralUse
session = new_session(U2Netp)
output_img_animal_num = remove(img_animal_num; session=session)
output_img_animal = remove(img_animal; session=session)
session = new_session(U2NetClothSeg)
output_img_cloth_num = remove(img_cloth_num; session=session)
output_img_cloth = remove(img_cloth; session=session)
session = new_session(U2NetHumanSeg)
output_img_animal_num = remove(img_animal_num; session=session)
output_img_animal = remove(img_animal; session=session)
session = new_session(Silueta)
output_img_animal_num = remove(img_animal_num; session=session)
output_img_animal = remove(img_animal; session=session)
session = new_session(ISNetGeneralUse)
output_img_animal_num = remove(img_animal_num; session=session)
output_img_animal = remove(img_animal; session=session)
# Return mask
output_img_animal = remove(img_animal; only_mask=true) # return mask
# Apply background color
output_img_animal = remove(img_animal; bgcolor=(255, 0, 0, 255))
| RemBG | https://github.com/yezhengkai/RemBG.jl.git |
|
[
"MIT"
]
| 0.1.0 | de8c6b67c7b13044dc683d14c72ad88de9d5dee0 | code | 349 | module RemBG
import ONNXRunTime as OX
using Images
using Artifacts
using LazyArtifacts
include("session.jl")
export U2Net,
U2Netp,
U2NetClothSeg,
U2NetHumanSeg,
Silueta,
ISNetGeneralUse,
new_session,
set_onnx_execution_provider,
get_onnx_execution_provider
include("bg.jl")
export remove
include("utils.jl")
end
| RemBG | https://github.com/yezhengkai/RemBG.jl.git |
|
[
"MIT"
]
| 0.1.0 | de8c6b67c7b13044dc683d14c72ad88de9d5dee0 | code | 2298 | function alpha_matting_cutout(
img, mask, foreground_threshold, background_threshold, erode_structure_size
) end
function naive_cutout(img, mask)
cutout = colorview(RGBA, RGBA.(img) .* channelview(mask))
return cutout
end
function post_process(mask) end
function apply_background_color(img, color)
img_size = size(img)
r = fill(color[1] / 255, img_size)
g = fill(color[2] / 255, img_size)
b = fill(color[3] / 255, img_size)
a = fill(color[4] / 255, img_size)
bg = colorview(RGBA{Float32}, r, g, b, a)
mask = alpha.(img)
bg = clamp01!(bg .* (1 .- mask) + img .* mask)
return bg
end
"""
remove(img; kwargs...)
Remove background from image.
# Examples
```julia
remove(img)
remove(img; session=new_session(U2Net)) # or use specific session
```
"""
function remove(img::Array{<:Number}; kwargs...)::Array{<:Number}
# unify image channels to 3
if ndims(img) == 2
# Change size (hight, width) to (1, hight, width)
img = reshape(img, 1, size(img)...)
end
if size(img, 1) == 1
# Change size (1, hight, width) to (3, hight, width)
img = repeat(img, 3)
elseif size(img, 1) == 4
# Change size (4, hight, width) to (3, hight, width)
img = img[1:3, :, :]
end
img = copy(colorview(RGB, img)) # convert to Matrix{<:Colorant}
cutout = remove(img; kwargs...)
return copy(channelview(cutout))
end
function remove(
img::Matrix{<:Colorant};
session::Union{InferenceSession,Nothing}=nothing,
only_mask::Bool=false,
bgcolor::Union{Tuple{Int,Int,Int,Int},Nothing}=nothing,
)::Matrix{<:Colorant}
# get onnx session
if isnothing(session)
session = new_session(U2Net)
end
# predict mask
masks = predict(session, img)
# get cutouts from all masks
cutouts = []
for mask in masks
if only_mask
cutout = mask
else
cutout = naive_cutout(img, mask)
end
push!(cutouts, cutout)
end
# get cutout by concatenating cutouts vertically
if length(cutouts) > 0
cutout = reduce(vcat, cutouts)
end
# apply background color
if !isnothing(bgcolor) && !only_mask
cutout = apply_background_color(cutout, bgcolor)
end
return cutout
end
| RemBG | https://github.com/yezhengkai/RemBG.jl.git |
|
[
"MIT"
]
| 0.1.0 | de8c6b67c7b13044dc683d14c72ad88de9d5dee0 | code | 7804 | U2NET_ONNX_PATH = joinpath(artifact"u2net", "u2net.onnx")
U2NETP_ONNX_PATH = joinpath(artifact"u2netp", "u2netp.onnx")
U2NET_CLOTH_SEG_ONNX_PATH = joinpath(artifact"u2net_cloth_seg", "u2net_cloth_seg.onnx")
U2NET_HUMAN_SEG_ONNX_PATH = joinpath(artifact"u2net_human_seg", "u2net_human_seg.onnx")
SILUETA_ONNX_PATH = joinpath(artifact"silueta", "silueta.onnx")
ISNET_GENERAL_USE_ONNX_PATH = joinpath(
artifact"isnet-general-use", "isnet-general-use.onnx"
)
const EXECUTION_PROVIDER = Ref{Symbol}(:cpu)
abstract type MattingModel end
struct U2NetModel <: MattingModel end
struct U2NetpModel <: MattingModel end
struct U2NetClothSegModel <: MattingModel end
struct U2NetHumanSegModel <: MattingModel end
struct SiluetaModel <: MattingModel end
struct ISNetGeneralUseModel <: MattingModel end
"""`U2NetModel` object. `U2NetModel` is a subtype of `MattingModel`."""
const U2Net = U2NetModel()
"""`U2NetpModel` object. `U2NetpModel` is a subtype of `MattingModel`."""
const U2Netp = U2NetpModel()
"""`U2NetClothSegModel` object. `U2NetClothSegModel` is a subtype of `MattingModel`."""
const U2NetClothSeg = U2NetClothSegModel()
"""`U2NetHumanSegModel` object. `U2NetHumanSegModel` is a subtype of `MattingModel`."""
const U2NetHumanSeg = U2NetHumanSegModel()
"""`SiluetaModel` object. `SiluetaModel` is a subtype of `MattingModel`."""
const Silueta = SiluetaModel()
"""`ISNetGeneralUseModel` object. `ISNetGeneralUseModel` is a subtype of `MattingModel`."""
const ISNetGeneralUse = ISNetGeneralUseModel()
abstract type InferenceSession end
struct SimpleSession <: InferenceSession
model_name::String
onnx_session::OX.InferenceSession
end
struct ClothSession <: InferenceSession
model_name::String
onnx_session::OX.InferenceSession
end
struct DisSession <: InferenceSession
model_name::String
onnx_session::OX.InferenceSession
end
"""
get_onnx_execution_provider()
Get the current execution provider for the ONNX inference session.
See also [`set_onnx_execution_provider`](@ref).
"""
function get_onnx_execution_provider()
return EXECUTION_PROVIDER[]
end
"""
set_onnx_execution_provider(execution_provider::Symbol)
Set the execution provider for ONNX inference session. Now the "execution_provider" only accepts ":cpu" and ":cuda".
See also [`get_onnx_execution_provider`](@ref).
"""
function set_onnx_execution_provider(execution_provider::Symbol)
if execution_provider === :cpu
EXECUTION_PROVIDER[] = execution_provider
elseif execution_provider === :cuda
EXECUTION_PROVIDER[] = execution_provider
else
error("Unsupported execution_provider $execution_provider")
end
end
"""
new_session(::MattingModel)
Constructs an `InferenceSession` object by input type.
# Examples
```julia
session = new_session(U2Net)
```
"""
function new_session(::U2NetModel)
return SimpleSession(
"U2Net", OX.load_inference(U2NET_ONNX_PATH; execution_provider=EXECUTION_PROVIDER[])
)
end
function new_session(::U2NetpModel)
return SimpleSession(
"U2Netp",
OX.load_inference(U2NETP_ONNX_PATH; execution_provider=EXECUTION_PROVIDER[]),
)
end
function new_session(::U2NetClothSegModel)
return ClothSession(
"U2NetClothSeg",
OX.load_inference(
U2NET_CLOTH_SEG_ONNX_PATH; execution_provider=EXECUTION_PROVIDER[]
),
)
end
function new_session(::U2NetHumanSegModel)
return SimpleSession(
"U2NetHumanSeg",
OX.load_inference(
U2NET_HUMAN_SEG_ONNX_PATH; execution_provider=EXECUTION_PROVIDER[]
),
)
end
function new_session(::SiluetaModel)
return SimpleSession(
"Silueta",
OX.load_inference(SILUETA_ONNX_PATH; execution_provider=EXECUTION_PROVIDER[]),
)
end
function new_session(::ISNetGeneralUseModel)
return DisSession(
"ISNetGeneralUse",
OX.load_inference(
ISNET_GENERAL_USE_ONNX_PATH; execution_provider=EXECUTION_PROVIDER[]
),
)
end
function predict(session::SimpleSession, img::Matrix{<:Colorant})
# Preprocessing
small_img = imresize(img, 320, 320)
img_array = copy(channelview(small_img)) # type: Array {N0f8, 3}, size: (channel(RGB), hight, width)
img_array = copy(rawview(img_array)) # type: Array {UInt8, 3}, size: (channel(RGB), hight, width)
img_array = img_array / maximum(img_array)
tmp_img = zeros(Float32, 1, 3, 320, 320) # input size: (batch_size, 3, 320, 320)
tmp_img[1, 1, :, :] = (img_array[1, :, :] .- 0.485) / 0.229
tmp_img[1, 2, :, :] = (img_array[2, :, :] .- 0.456) / 0.224
tmp_img[1, 3, :, :] = (img_array[3, :, :] .- 0.406) / 0.225
# Predict
input = Dict(session.onnx_session.input_names[1] => tmp_img)
# output size: (batch_size, 1, 320, 320)
pred = session.onnx_session(input)[session.onnx_session.output_names[1]][:, 1, :, :]
# Postprocessing
ma = maximum(pred)
mi = minimum(pred)
pred = (pred .- mi) / (ma .- mi)
pred = dropdims(pred; dims=1)
# Creates a view of the numeric array `pred`,
# interpreting successive elements of `pred` as if they were channels of Colorant `Gray`.
mask = colorview(Gray, pred)
mask = imresize(mask, size(img))
return [mask]
end
function predict(session::DisSession, img::Matrix{<:Colorant})
# Preprocessing
small_img = imresize(img, 1024, 1024)
img_array = copy(channelview(small_img)) # type: Array {N0f8, 3}, size: (channel(RGB), hight, width)
img_array = copy(rawview(img_array)) # type: Array {UInt8, 3}, size: (channel(RGB), hight, width)
img_array = img_array / maximum(img_array)
tmp_img = zeros(Float32, 1, 3, 1024, 1024) # input size: (batch_size, 3, 320, 320)
tmp_img[1, 1, :, :] = (img_array[1, :, :] .- 0.485) / 1.0
tmp_img[1, 2, :, :] = (img_array[2, :, :] .- 0.456) / 1.0
tmp_img[1, 3, :, :] = (img_array[3, :, :] .- 0.406) / 1.0
# Predict
input = Dict(session.onnx_session.input_names[1] => tmp_img)
# output size: (batch_size, 1, 1024, 1024)
pred = session.onnx_session(input)[session.onnx_session.output_names[1]][:, 1, :, :]
# Postprocessing
ma = maximum(pred)
mi = minimum(pred)
pred = (pred .- mi) / (ma .- mi)
pred = dropdims(pred; dims=1)
# Creates a view of the numeric array `pred`,
# interpreting successive elements of `pred` as if they were channels of Colorant `Gray`.
mask = colorview(Gray, pred)
mask = imresize(mask, size(img))
return [mask]
end
function predict(session::ClothSession, img::Matrix{<:Colorant})
# Preprocessing
small_img = imresize(img, 768, 768)
img_array = copy(channelview(small_img)) # type: Array {N0f8, 3}, size: (channel(RGB), hight, width)
img_array = copy(rawview(img_array)) # type: Array {UInt8, 3}, size: (channel(RGB), hight, width)
img_array = img_array / maximum(img_array)
tmp_img = zeros(Float32, 1, 3, 768, 768) # input size: (batch_size, 3, 768, 768)
tmp_img[1, 1, :, :] = (img_array[1, :, :] .- 0.485) / 0.229
tmp_img[1, 2, :, :] = (img_array[2, :, :] .- 0.456) / 0.224
tmp_img[1, 3, :, :] = (img_array[3, :, :] .- 0.406) / 0.225
# Predict
input = Dict(session.onnx_session.input_names[1] => tmp_img)
# output size: (batch_size, 4, 768, 768)
# The 4 channels represent in order "Background", "Upper Body Clothes", "Lower Body Clothes" and "Full Body Clothes".
pred = session.onnx_session(input)[session.onnx_session.output_names[1]]
pred = softmax(pred, 2)
pred = dropdims(pred; dims=1) # size: (4, 768, 768)
mask = imresize(pred, 4, size(img)...)
mask_upper = colorview(Gray, mask[2, :, :])
mask_lower = colorview(Gray, mask[3, :, :])
mask_full = colorview(Gray, mask[4, :, :])
return [mask_upper, mask_lower, mask_full]
end
| RemBG | https://github.com/yezhengkai/RemBG.jl.git |
|
[
"MIT"
]
| 0.1.0 | de8c6b67c7b13044dc683d14c72ad88de9d5dee0 | code | 2271 | # References:
# - https://medium.com/@bowlescompling/m2-1-softmax-in-julia-1498901f741c
# - https://docs.scipy.org/doc/scipy/reference/generated/scipy.special.log_softmax.html#scipy.special.log_softmax
# abstract exponentiation function, subtract max for numerical stability
_exp(x) = exp.(x .- maximum(x))
# abstract exponentiation function, subtract max for numerical stability and scale by theta
_exp(x, θ) = exp.(x .- maximum(x) * θ)
# softmax algorithm expects stabilized exponentiated e
_softmax(e, d) = (e ./ sum(e; dims=d))
"""
softmax(X, dims)
softmax(X, dims, θ)
Compute the softmax function along the `dims` dimension of the array `X`. You can adjust the scaling factor `θ`.
# Examples
```julia-repl
julia> x = [1000.0 1.0; 1000.0 1.0];
julia> y = RemBG.softmax(x, 2)
2×2 Matrix{Float64}:
1.0 0.0
1.0 0.0
```
"""
function softmax(X, dims)
return _softmax(_exp(X), dims)
end
function softmax(X, dims, θ)
return _softmax(_exp(X, θ), dims)
end
"""
log_softmax(x)
log_softmax(x, dims)
Compute the logarithm of the softmax function.
In principle::
log_softmax(x) = log(softmax(x))
but using a more accurate implementation.
# Arguments
- `x`: Input array
- `dims`: Axis to compute values along
# Returns
- `Array or Number`: An array with the same shape as `x`. Exponential of the result will
sum to 1 along the specified axis. If `x` is a scalar, a scalar is
returned.
# Examples
```julia-repl
julia> x = [1000.0, 1.0];
julia> y = log_softmax(x)
2-element Vector{Float64}:
0.0
-999.0
julia> x = [1000.0 1.0; 1000.0 1.0];
julia> y = log_softmax(x, 2)
2×2 Matrix{Float64}:
0.0 -999.0
0.0 -999.0
```
"""
function log_softmax(x)
x_max = maximum(x)
if ndims(x_max) > 0
x_max[.!isfinite.(x_max)] .= 0
elseif !isfinite(x_max)
x_max = 0
end
tmp = x .- x_max
exp_tmp = exp.(tmp)
s = sum(exp_tmp)
out = log(s)
out = tmp .- out
return out
end
function log_softmax(x, dims)
x_max = maximum(x; dims=dims)
if ndims(x_max) > 0
x_max[.!isfinite.(x_max)] .= 0
elseif !isfinite(x_max)
x_max = 0
end
tmp = x .- x_max
exp_tmp = exp.(tmp)
s = sum(exp_tmp; dims=dims)
out = log.(s)
out = tmp .- out
return out
end
| RemBG | https://github.com/yezhengkai/RemBG.jl.git |
|
[
"MIT"
]
| 0.1.0 | de8c6b67c7b13044dc683d14c72ad88de9d5dee0 | code | 2339 | using RemBG
using Test
using RemBG.Images
using CUDA
using cuDNN
@testset "Model type" begin
@test isa(U2Net, RemBG.U2NetModel)
@test isa(U2Netp, RemBG.U2NetpModel)
@test isa(U2NetClothSeg, RemBG.U2NetClothSegModel)
@test isa(U2NetHumanSeg, RemBG.U2NetHumanSegModel)
@test isa(Silueta, RemBG.SiluetaModel)
@test isa(ISNetGeneralUse, RemBG.ISNetGeneralUseModel)
end
@testset "Session type" begin
@test isa(new_session(U2Net), RemBG.SimpleSession)
@test isa(new_session(U2Netp), RemBG.SimpleSession)
@test isa(new_session(U2NetClothSeg), RemBG.ClothSession)
@test isa(new_session(U2NetHumanSeg), RemBG.SimpleSession)
@test isa(new_session(Silueta), RemBG.SimpleSession)
@test isa(new_session(ISNetGeneralUse), RemBG.DisSession)
end
exampledir(args...) = joinpath(@__DIR__, "..", "examples", args...)
img_animal = load(exampledir("animal-1.jpg"))
img_cloth = load(exampledir("cloth-1.jpg"))
@testset "remove" begin
# SimpleSession
session = new_session(U2Netp)
@test size(img_animal) == size(remove(img_animal; session=session))
@test size(img_animal) == size(remove(img_animal; session=session, only_mask=true))
@test size(img_animal) ==
size(remove(img_animal; session=session, bgcolor=(0, 0, 0, 255)))
# DisSession
session = new_session(ISNetGeneralUse)
@test size(img_animal) == size(remove(img_animal; session=session))
@test size(img_animal) == size(remove(img_animal; session=session, only_mask=true))
@test size(img_animal) ==
size(remove(img_animal; session=session, bgcolor=(0, 0, 0, 255)))
# ClothSession
session = new_session(U2NetClothSeg)
@test size(img_cloth) .* (3, 1) == size(remove(img_cloth; session=session))
@test size(img_cloth) .* (3, 1) ==
size(remove(img_cloth; session=session, only_mask=true))
@test size(img_cloth) .* (3, 1) ==
size(remove(img_cloth; session=session, bgcolor=(0, 0, 0, 255)))
end
@testset "onnx execution provider" begin
@test get_onnx_execution_provider() == :cpu
@test set_onnx_execution_provider(:cuda) == :cuda
@test_throws ErrorException set_onnx_execution_provider(:tpu)
@test get_onnx_execution_provider() == :cuda
if CUDA.functional()
@test new_session(U2Netp).onnx_session.execution_provider == :cuda
end
end
| RemBG | https://github.com/yezhengkai/RemBG.jl.git |
|
[
"MIT"
]
| 0.1.0 | de8c6b67c7b13044dc683d14c72ad88de9d5dee0 | docs | 725 | # RemBG
[](https://yezhengkai.github.io/RemBG.jl/stable/)
[](https://yezhengkai.github.io/RemBG.jl/dev/)
[](https://github.com/yezhengkai/RemBG.jl/actions/workflows/CI.yml)
[](https://codecov.io/gh/yezhengkai/RemBG.jl)
[](https://github.com/invenia/BlueStyle)
This is a Julia implementation of the python package [rembg](https://github.com/danielgatis/rembg).
| RemBG | https://github.com/yezhengkai/RemBG.jl.git |
|
[
"MIT"
]
| 0.1.0 | de8c6b67c7b13044dc683d14c72ad88de9d5dee0 | docs | 1007 | ```@meta
CurrentModule = RemBG
```
# RemBG
This is a Julia implementation of the python package [rembg](https://github.com/danielgatis/rembg).
## Quick Start
```@example quick_start
using RemBG
using Images
exampledir(args...) = abspath(joinpath(dirname(pathof(RemBG)), "..", "examples", args...))
img = load(exampledir("animal-1.jpg"))
img_num = copy(channelview(img))
output_img_num = remove(img_num)
output_img = remove(img) # If we do not pass an inference session to `remove`, it will use a new session containing the U2Net model.
```
```@example quick_start
# Use a specific model. The available models are U2Net, U2Netp, U2NetClothSeg, U2NetHumanSeg, Silueta, ISNetGeneralUse
session = new_session(U2Netp)
output_img_num = remove(img_num; session=session)
output_img = remove(img; session=session)
```
```@example quick_start
output_img = remove(img; only_mask=true) # return mask
```
```@example quick_start
output_img = remove(img; bgcolor=(255, 0, 0, 255)) # apply background color
``` | RemBG | https://github.com/yezhengkai/RemBG.jl.git |
|
[
"MIT"
]
| 0.1.0 | de8c6b67c7b13044dc683d14c72ad88de9d5dee0 | docs | 209 | # Internal
Internal API documentation for [RemBG](https://github.com/yezhengkai/RemBG.jl).
## Index
```@index
Pages = ["internal.md"]
```
## Internal Interface
```@docs
RemBG.softmax
RemBG.log_softmax
``` | RemBG | https://github.com/yezhengkai/RemBG.jl.git |
|
[
"MIT"
]
| 0.1.0 | de8c6b67c7b13044dc683d14c72ad88de9d5dee0 | docs | 309 | # Public
Public API documentation for [RemBG](https://github.com/yezhengkai/RemBG.jl).
## Index
```@index
Pages = ["public.md"]
```
## Public Interface
```@docs
remove
new_session
set_onnx_execution_provider
get_onnx_execution_provider
U2Net
U2Netp
U2NetClothSeg
U2NetHumanSeg
Silueta
ISNetGeneralUse
``` | RemBG | https://github.com/yezhengkai/RemBG.jl.git |
|
[
"MIT"
]
| 0.3.3 | f958c49f3f4f812d6eb2725f102896a0ead72172 | code | 1681 | push!(LOAD_PATH,"../src/")
using ClimateMARGO
using Documenter, Literate
const EXAMPLES_DIR = joinpath(@__DIR__, "..", "examples")
const OUTPUT_DIR = joinpath(@__DIR__, "src/generated")
print(@__DIR__)
examples = [
"default_configuration.jl",
"default_optimization.jl",
"one-two_dimensional_optimization.jl"
]
for example in examples
example_filepath = joinpath(EXAMPLES_DIR, example)
Literate.markdown(example_filepath, OUTPUT_DIR, documenter=true)
end
#### Organize page structure
example_pages = [
"Default model configuration" => "generated/default_configuration.md",
"Optimization with default parameters" => "generated/default_optimization.md",
"1D and 2D optimization" => "generated/one-two_dimensional_optimization.md"
]
pages = [
"Home" => "index.md",
"Installation instructions" => "installation_instructions.md",
"Theory" => "theory.md",
"Examples" => example_pages,
# "Submodules" => [
# "Domain" => "Submodules/Domain.md",
# "Physics" => "Submodules/Physics.md",
# "Economics" => "Submodules/Economics.md",
# "Controls" => "Submodules/Controls.md",
# ],
"Diagnostics" => "diagnostics.md",
"Optimization" => "optimization.md",
"Function index" => "function_index.md"
]
makedocs(
sitename="ClimateMARGO.jl",
doctest = true,
authors = "Henri F. Drake",
format = Documenter.HTML(
prettyurls = get(ENV, "CI", nothing) == "true",
collapselevel = 1
),
pages = pages
)
deploydocs(
repo = "github.com/ClimateMARGO/ClimateMARGO.jl.git",
versions = ["stable" => "v^", "v#.#.#", "dev" => "dev"],
devbranch = "main"
)
| ClimateMARGO | https://github.com/ClimateMARGO/ClimateMARGO.jl.git |
|
[
"MIT"
]
| 0.3.3 | f958c49f3f4f812d6eb2725f102896a0ead72172 | code | 15131 | # # ClimateMARGO.jl tutorial
# The following schematic shows the full formulation of the MARGO model, which is described in detail in the accompanying article, [published open-access](https://iopscience.iop.org/article/10.1088/1748-9326/ac243e) in the journal *Environmental Research Letters*. No-policy baseline emissions $q(t)$ are prescribed, leading to changing greenhouse gas (CO$_{2e}$) concentrations, radiative forcing, temperatures, and climate damages. Emissions can be decreased by **M**itigation; concentrations can be decreased by carbon dioxide **R**emoval; forcing can be decreased by solar **G**eo-engineering; and the "adapted" temperature that leads to climate damages can be reduced by **A**dapting to the changed climate.
# <img src="https://raw.githubusercontent.com/hdrake/MARGO-gifs/main/Margo_schematic.png" alt="drawing" width="60%" style="float:left"/>
# ### Import software
# The MARGO model has been implemented and registered as the Julia package `ClimateMARGO.jl`, which can be installed with the command `Pkg.add("ClimateMARGO")`.
# If you are running this tutorial via Binder, there is no need to install the package; just import it using the command below.
using ClimateMARGO # Julia implementation of the MARGO model
using PyPlot # A basic plotting package
#
using ClimateMARGO.Models
using ClimateMARGO.Utils
using ClimateMARGO.Diagnostics
# ## Model configuration
# An instance of MARGO `ClimateModelParameters` requires specifying a `name` and the parameters for three model subcomponents:
# 1. `Domain`: determines the time period of interest, as well as the timestep and– in the policy response mode– the "present" decision-making year.
# 2. `Economics`: determines exogenous economic growth, the cliamte damage cost function, and the cost function for each of the four controls, and the discount rate.
# 3. `Physics`: determines the parameters that govern the aiborn fraction of emissions that remain in the atmosphere, the strength of climate feedbacks, and the rate of ocean heat uptake.
# An instance of the MARGO `ClimateModel` is determined by combining the prescribed set of `ClimateModelParameters` with timeseries of the four `Controls`, which will act as the control variables in the optimization framework:
# 1. **M**itigation of baseline emissions
# 2. **R**emoval of carbon dioxide from the atmopshere
# 3. **G**eongineering by solar radiation management, and
# 4. **A**daptating to the changed climate to reduce the damages of climate change impacts.
# Here, we run through the full construction of the model with its "default" parameter values.
name = "default";
# #### 1. Setting up the temporal grid
# First, we need to set up a time-frame for our experiment. Let's begin our simulation in the year 2020– present day– and consider out to 2200, with a 5-year timestep for computational efficiency.
initial_year = 2020. # [yr]
final_year = 2200. # [yr]
dt = 5. # [yr]
t_arr = t(initial_year, final_year, dt);
# While the model allows for shifting the "present" year forward or backward in time to simulate past and future policy decision-making processes, we will only consider the simplest case in which we take the perspective of a policy decision maker in the year 2020, which is also the initial year of our simulation.
present_year = initial_year
dom = Domain(dt, initial_year, initial_year, final_year);
# #### 2. Configuring the carbon cycle and energy balance models
# CO$_{2e}$ concentrations are given by:
# ```math
# c_{M,R}(t) = c_{0} + \int_{t_{0}}^{t} rq(t')(1-M(t')) \text{ d}t' - q_{0} \int_{t_{0}}^{t} R(t')\text{ d}t' \, .
# ```
# where $q(t)$ is a specific timeseries of no-climate-policy baseline emissions (with $q(t_{0}) = q_{0}$), $c_{0}$ is the initial CO$_{2e}$ concentration, and $r$ is the fraction of emitted CO$_{2e}$ that remains in the atmosphere net of short-term uptake by the ocean and biosphere. $M(t')$ and $R(t')$ represent the effects emissions mitigation and carbon dioxide removal, respectively, on CO$_{2e}$ concentrations relative to the baseline; they will be initially set to zero and later determined by solving an optimization problem.
# The default no-policy scenario is one of rapid, fossil-fueled growth which leads to an accumulation of $c(t) = 1400$ ppm of CO$_{2e}$ in the atmosphere by 2150, when emissions are assumed to finally reach net-zero.
c0 = 460. # [ppm]
r = 0.5; # [1] fraction of emissions remaining after biosphere and ocean uptake (Solomon 2009)
q0 = 7.5
q0mult = 3.
t_peak = 2100.
t_zero = 2150.
q = ramp_emissions(t_arr, q0, q0mult, t_peak, t_zero);
#
figure(figsize=(9,3.5))
subplot(1,2,1)
plot(t_arr, ppm_to_GtCO2(q))
xlabel("year")
ylabel(L"baseline emissions $q(t)$ [GtCO2 / year]")
xlim([2020, 2200])
grid(true)
subplot(1,2,2)
q_effective = effective_emissions(r, q, 0., 0.) # No mitigation, no carbon removal
c_baseline = c(c0, q_effective, dt)
plot(t_arr, c_baseline)
xlabel("year")
ylabel(L"baseline concentrations $c(t)$ [ppm]")
xlim([2020, 2200])
tight_layout();
grid(true)
gcf()
# These CO$_{2e}$ concentrations drive an anomalous greenhouse effect, which is represented by the radiative forcing
# ```math
# F_{M,R,G} = a \ln\left(\frac{c_{M,R}(t)}{c_{0}}\right) - G(t)F_{\infty} \, ,
# ```
# where $a$ is an empirically determined coefficient, $G(t)$ represents the effects of geoengineering, and $F_{\infty} = 8.5$ W/m$^2$ is a scaling factor for the effects of geoengineering by Solar Radiation Modification (SRM).
a = (6.9/2.)/log(2.); # F4xCO2/2 / log(2) [W m^-2]
Finf = 8.5;
#
figure(figsize=(4.5,3.2))
F0 = 3.0
F_baseline = F(a, c0, Finf, c_baseline, 0.)
plot(t_arr, F_baseline .+ F0)
xlabel("year")
ylabel(L"baseline radiative forcing $F(t)$ [W/m$^2$]")
xlim([2020, 2200])
grid(true)
ylim([0,10.]);
gcf()
# Next, we configure MARGO's energy balance model, which is forced by the controlled forcing $F_{M,R,G}$. The two-layer energy balance model can be solved, approximately, as:
# ```math
# T_{M,R,G}(t) - T_{0} = \frac{F_{M,R,G}(t)}{B + \kappa} + \frac{\kappa}{B} \int_{t_{0}}^{t} \frac{e^{\frac{t'-t}{\tau_{D}}}}{\tau_{D}} \frac{F_{M,R,G}(t')}{B+\kappa} \, \text{d}t' \, ,
# ```
# where $T_{0}$ is the initial temperature change relative to preindustrial, $B$ is the feedback parameter, $\kappa$ is , and the timescale $\tau_{D} = \dfrac{C_{D}}{B} \dfrac{B + \kappa}{\kappa}$ is a more convenient expression for the deep ocean heat capacity $C_{D}$.
# Default parameter values are taken from [Geoffroy et al. 2013](https://journals.ametsoc.org/doi/full/10.1175/JCLI-D-12-00195.1)'s physically-based calibration of the two-layer model to CMIP5.
# Two-layer EBM parameters
B = 1.13; # Feedback parameter [J yr^-1 m^-2 K^-1]
Cd = 106.; # Deep ocean heat capacity [J m^-2 K^-1]
κ = 0.73; # Heat exchange coefficient [J yr^-1 m^2 K^-1]
# Initial condition: present-day temperature, relative to pre-industrial
T0 = 1.1; # [degC] Berkeley Earth Surface Temperature (Rohde 2013)
print("τD = ", Int64(round(τd(Cd, B, κ))), " years")
# These physical parameters can be used to diagnose the climate sensitivity to a doubling of CO$_{2}$ ($ECS$).
print("ECS = ", round(ECS(a, B),digits=1) ,"ºC")
# Combined, these parameters define the physical model, which is instantiated by the calling the `Physics` constructor method:
Phys = Physics(c0, T0, a, B, Cd, κ, r);
# #### 3. Configuring the simple economic model
# Economic growth in MARGO (in terms of Gross World Product, GWP) is exogenous $E(t) = E_{0} (1 + \gamma)^{(t-t_{0})}$ and is entirely determined by the growth rate $\gamma$. By default, we set $\gamma = 2\%$.
E0 = 100. # Gross World Product at t0 [10^12$ yr^-1]
γ = 0.02 # economic growth rate
E_arr = E(t_arr, E0, γ)
figure(figsize=(4, 2.5))
plot(t_arr, E_arr)
xlabel("year")
ylabel("GWP [trillion USD]")
xlim([2020, 2200])
ylim([0, 3000]);
grid(true)
gcf()
# Economic damages $D_{M,R,G,A} = \tilde{\beta}E(t) (\Delta T_{M,R,G})^{2} (1 - A(t))$, expressed as a fraction $\tilde{\beta}(\Delta T_{M,R,G})^{2}$ of the instantaneous Gross World Product, increase quadratically with temperature change relative to preindustrial. They can be decrease by adaptation controls $A(t)$. The default value of the damage parameter $\tilde{\beta}$ corresponds to damages of 2\% of GWP at 3ºC of warming.
βtilde = 0.01; # damages [%GWP / celsius^2]
# The total cost of climate controls is simply the sum of their individual costs, each of which follows a parabolic cost function:
# ```math
# \mathcal{C}_{M,R,G,A} = \mathcal{C}_{M}M^2 + \mathcal{C}_{R}R^2 + \mathcal{C}_{G}G^2 + \mathcal{C}_{A}A^2 \, .
# ```
# The calculation of the reference control costs $\mathcal{C}_{M}, \mathcal{C}_{R}, \mathcal{C}_{G}, \mathcal{C}_{A}$ are somewhat more complicated; see our Methods in [the preprint](https://eartharxiv.org/5bgyc/) and `defaults.jl` for details. Here, we simply provide their default numerical values, where the costs of mitigation $\mathcal{C}_{M} = \tilde{\mathcal{C}}_{M} E(t)$ and geoengineering $\mathcal{C}_{G} = \tilde{\mathcal{C}}_{G} E(t)$ grow with the size of the global economy and are thus specified as a fraction of GWP, while adaptaiton and removal costs are in trillions of USD per year.
ti = findall(x->x==2100, t_arr)[1]
mitigate_cost_percentGWP = 0.02
mitigate_cost = mitigate_cost_percentGWP*E_arr[ti]/ppm_to_GtCO2(q[ti]); # [trillion USD / year / GtCO2]
# Costs of negative emissions technologies [US$/tCO2]
costs = Dict(
"BECCS" => 150.,
"DACCS" => 200.,
"Forests" => 27.5,
"Weathering" => 125.,
"Biochar" => 70.,
"Soils" => 50.
)
# Upper-bound potential for sequestration (GtCO2/year)
potentials = Dict(
"BECCS" => 5.,
"DACCS" => 5.,
"Forests" => 3.6,
"Weathering" => 4.,
"Biochar" => 2.,
"Soils" => 5.
)
mean_cost = sum(values(potentials) .* values(costs)) / sum(values(potentials)) # [$ tCO2^-1] weighted average
CDR_potential = sum(values(potentials))
CDR_potential_fraction = CDR_potential / ppm_to_GtCO2(q0)
# Estimate cost from Fuss 2018 (see synthesis Figure 14 and Table 2)
remove_cost = (mean_cost * CDR_potential*1.e9) / (CDR_potential_fraction^3) * 1.e-12; # [trillion USD / year]
adapt_cost = 0.115; # [%GWP / year] directly from de Bruin, Dellink, and Tol (2009)
geoeng_cost = βtilde*(Finf/B)^2; # [% GWP]
# Climate damages and control costs are discounted at the relatively low rate of $\rho = 2\%$, such that future damages and costs are reduced by a multiplicative discount factor $(1 - \rho)^{(t-t_{0})}$.
ρ = 0.02;
figure(figsize=(4, 2.5))
plot(t_arr, discount(t_arr, ρ, present_year)*100)
xlabel("year")
ylabel("discount factor (%)")
xlim([2020, 2200])
ylim([0, 100]);
grid(true)
gcf()
# These parameters, in addition to a no-policy baseline emissions time-series and present-day control values, define the economic model.
Econ = Economics(
E0, γ, βtilde, ρ, Finf,
mitigate_cost, remove_cost, geoeng_cost, adapt_cost,
0., 0., 0., 0., # Initial condition on control deployments at t[1]
q
);
# #### A MARGO model configuration is uniquely determined by the parameters defined above
params = ClimateModelParameters(
name,
dom,
Econ,
Phys
);
# ## Instanciating the MARGO climate model
# Along with economic and physical model components, the timeseries for each of the four controls must be specified. By default, we simply set these to zero.
Cont = Controls(
zeros(size(t_arr)), # mitigate
zeros(size(t_arr)), # remove
zeros(size(t_arr)), # geoeng
zeros(size(t_arr)) # adapt
);
# The above parameters determine the full configuration of the MARGO model, which is instanced using the `ClimateModel` constructor method:
m = ClimateModel(
params,
Cont
);
# ## Model optimization
# #### Formulating the optimization problem
# By default, the optimization problem we solve is for the most cost-effective combination of controls, as determined by minimization of the discounted net present value,
# ```math
# \min\left\{\int_{t_{0}}^{t_{f}} \mathcal{C}_{M,R,G,A} (1 + \rho)^{-(t-t_{0})} \text{ d}t\right\} \, ,
# ```
# which keep controlled damages below the level corresponding to a chosen temperature threshold $T^{\star}$,
# ```math
# \tilde{\beta} E(t) (T_{M,R,G})^{2} (1 - A(t)) < \tilde{\beta} E(t) (T^{\star})^{2} \, .
# ```
temp_goal = 2.0;
# #### Additional policy / inertia constraints
# The optimization is also subject to the constraints described below.
# First, we set a upper bounds on the maximum plausible deployment of each control.
max_deployment = Dict("mitigate"=>1., "remove"=>1., "geoeng"=>1., "adapt"=>0.4);
# Second, we set upper limits on how quickly each control can be ramped up or down.
# (Adaptation is treated differently since we it interpret it as buying insurance against future climate damages, although the financing is spread evenly over the entire period.)
max_slope = Dict("mitigate"=>1. /40., "remove"=>1. /40., "geoeng"=>1. /40., "adapt"=> 1. /40.);
# Third, we impose restrictions on when controls can be first deployed. In particular, since carbon dioxide removal and solar radiation modification do not yet exist at scale, we delay these until 2030 and 2050, respectively, at the earliest.
delay_deployment = Dict(
"mitigate"=>0.,
"remove"=>0.,
"geoeng"=>0.,
"adapt"=>0.
);
# #### Running the optimization
# The optimization takes about ~40 seconds the first time it is run as the code compiles, but runs virtually instantly afterwards, even if model parameter values are changed.
using ClimateMARGO.Optimization
@time msolve = optimize_controls!(
m,
obj_option = "adaptive_temp", temp_goal = temp_goal,
max_deployment = max_deployment, max_slope = max_slope, delay_deployment = delay_deployment
);
# ## Plotting MARGO results
# We provide some convenient functions for plotting basic model results.
using ClimateMARGO.Plotting
plot_state(m, temp_goal=temp_goal)
gcf()
# ## Customizing MARGO
# Try changing some of the parameters above and re-running the model!
# For example, in the simulation below we set a more stringent temperature goal of $T^{\star} = 1.5$ and omit solar geoengineering and adaptation completely (by setting their maximum deployment to zero).
temp_goal = 1.5
max_deployment["geoeng"] = 0.
max_deployment["adapt"] = 0.
@time optimize_controls!(
m,
obj_option = "adaptive_temp", temp_goal = temp_goal,
max_deployment = max_deployment, max_slope = max_slope, delay_deployment = delay_deployment
);
plot_state(m, temp_goal=temp_goal)
gcf()
# ## Saving (and loading) MARGO simulations (or parameter configurations)
using ClimateMARGO.IO
export_path = tempname() * ".json"
#
export_parameters(export_path, params);
#
new_params = import_parameters(export_path)
# Let's say we want to see what happens if climate feedbacks $B$ are twice as strong as they are now (which means a lower feedback parameter).
new_params.name = "stronger_feedbacks"
new_params.physics.B /= 2.;
new_m = ClimateModel(new_params, Cont)
# The new equilibrium climate sensitivity (recall $ECS = F_{2x}/B$) is now:
print("ECS ≈ $(round(ECS(new_m), digits=1))°C")
| ClimateMARGO | https://github.com/ClimateMARGO/ClimateMARGO.jl.git |
|
[
"MIT"
]
| 0.3.3 | f958c49f3f4f812d6eb2725f102896a0ead72172 | code | 1205 | # # Optimization of the default MARGO configuration
# ## Using `ClimateMARGO.jl`
using ClimateMARGO
using ClimateMARGO.Models
using ClimateMARGO.Optimization
# ## Loading preset configurations
# Load the pre-defined default MARGO parameters, which are described by the ClimateModelParameters struct
params = deepcopy(ClimateMARGO.IO.included_configurations["default"])
# Create a MARGO instance based on these parameters
m = ClimateModel(params);
# By default, the climate control timeseries are all set to zero.
m.controls
# ## Real-time climate optimization
# Let's optimize the controls with the default settings, which finds the cheapest combination of the control timeseries that keeps adapted temperatures below 2°C.
@time optimize_controls!(m, temp_goal=1.5);
# The optimization can be slow the first time since it has to compile.
# Let's re-initialize the model and see how fast it runs now that the the optimization routine has been precompiled.
m = ClimateModel(params);
@time optimize_controls!(m, temp_goal=1.5);
# ## Visualizing the results
# Finally, let's plot the resulting temperature timeseries
using PyPlot
fig, axes = ClimateMARGO.Plotting.plot_state(m, temp_goal=1.5);
gcf() | ClimateMARGO | https://github.com/ClimateMARGO/ClimateMARGO.jl.git |
|
[
"MIT"
]
| 0.3.3 | f958c49f3f4f812d6eb2725f102896a0ead72172 | code | 9496 | # # A simple two-dimensional optimization problem
# ## Loading ClimateMARGO.jl
using ClimateMARGO # Julia implementation of the MARGO model
using PyPlot # A basic plotting package
# Loading submodules for convenience
using ClimateMARGO.Models
using ClimateMARGO.Utils
using ClimateMARGO.Diagnostics
using ClimateMARGO.Optimization
# ## Loading the default MARGO configuration
params = deepcopy(ClimateMARGO.IO.included_configurations["default"])
# Slightly increasing the discount rate to 4% to be more comparable with other models
params.economics.ρ = 0.04
# ## Reducing the default problem's dimensionality from ``4N`` to ``2``.
# The default optimization problem consists of optimizing the values of each of the 4 controls for N timesteps, giving a total problem size of:
4*length(t(params))
# ### Modifying `ClimateModelParameters`
# Thanks to MARGO's flexibility, we can get rid of two of the controls and set the other two to have constant values, effectively reducing the problem size to 2.
# First, we have to remove initial conditions on the control variables, which would conflict with the constant-value constraint
params.economics.mitigate_init = nothing
params.economics.remove_init = nothing
params.economics.geoeng_init = nothing
params.economics.adapt_init = nothing
# ### Instanciating the `ClimateModel`
# Now that we've finished changing parameter values, we can create our MARGO model instance:
m = ClimateModel(params)
# ### Modifying keyword arguments for `optimize_controls!`
# We can make the controls constant in time by asserting a maximum deployment rate of zero
max_slope = Dict("mitigate"=>0., "remove"=>0., "geoeng"=>0., "adapt"=>0.);
# Now we get rid of geoengineering and adaptation options by setting their maximum deployment fraction to zero
max_deployment = Dict("mitigate"=>1.0, "remove"=>1.0, "geoeng"=>0., "adapt"=>0.);
# ### Run the optimization problem once to test
@time optimize_controls!(m, obj_option = "net_benefit", max_slope=max_slope, max_deployment=Dict("mitigate"=>1.0, "remove"=>0., "geoeng"=>0., "adapt"=>0.));
# ### Visualizing the results
fig, axes = ClimateMARGO.Plotting.plot_state(m);
axes[3].legend(loc="upper left")
axes[4].legend(loc="lower left")
axes[5].legend(loc="lower right")
axes[6].legend(loc="upper right")
axes[6].set_ylim(0,2.5)
gcf()
# ## Comparing the two-dimensional optimization with the brute-force parameter sweep method
# ### Parameter sweep
# In the brute-force approach, we sweep through all possible values of **M**itigation and **R**emoval to map out the objective functions and constraints and visually identify the "optimal" solution in this 2-D space.
Ms = 0.:0.005:1.0;
Rs = 0.:0.005:1.0;
# We will also consider four different temperature thresholds and visualize these constraints in the 2-D space
temp_goals = [1.5, 2.0, 3.0, 4.0]
control_cost = zeros(length(Rs), length(Ms)) .+ 0. #
net_benefit = zeros(length(Rs), length(Ms)) .+ 0. #
max_temp = zeros(length(Rs), length(Ms)) .+ 0. # stores the maximum temperature acheived for each combination
min_temp = zeros(length(Rs), length(Ms)) .+ 0. # stores the minimum temperature acheived for each combination
optimal_controls = zeros(2, length(temp_goals), 2) # to hold optimal values computed using JuMP
for (o, option) = enumerate(["adaptive_temp", "net_benefit"])
for (i, M) = enumerate(Ms)
for (j, R) = enumerate(Rs)
m.controls.mitigate = zeros(size(t(m))) .+ M
m.controls.remove = zeros(size(t(m))) .+ R
if minimum(c(m, M=true, R=true)) <= 100.
continue
end
control_cost[j, i] = net_present_cost(m, discounting=true, M=true, R=true)
net_benefit[j, i] = net_present_benefit(m, discounting=true, M=true, R=true)
max_temp[j, i] = maximum(T(m, M=true, R=true))
min_temp[j, i] = minimum(T(m, M=true, R=true))
end
end
for (g, temp_goal) = enumerate(temp_goals)
optimize_controls!(m, obj_option = option, temp_goal = temp_goal, max_slope=max_slope, max_deployment=max_deployment);
optimal_controls[:, g, o] = deepcopy([m.controls.mitigate[1], m.controls.remove[1]])
end
end
# ### Visualizing the one-dimensional mitigation optimization problem
# In the limit of zero-carbon dioxide removal, we can recover the 1D mitigation optimization problem from the 2D one.
col = ((1., 0.8, 0.), (0.8, 0.5, 0.), (0.7, 0.2, 0.), (0.6, 0., 0.),)
fig = figure(figsize=(14,5))
ax = subplot(1,2,1)
ind1 = argmin(abs.(max_temp[1,:] .- 1.5))
ind2 = argmin(abs.(max_temp[1,:] .- 2.))
ind3 = argmin(abs.(max_temp[1,:] .- 3.))
ind4 = argmin(abs.(max_temp[1,:] .- 4.))
plot(Ms, control_cost[1,:], "k-", lw=2)
plot(Ms[ind1], control_cost[1,ind1], "o", color=col[1], markersize=10)
plot(Ms[ind2], control_cost[1,ind2], "o", color=col[2], markersize=10)
plot(Ms[ind3], control_cost[1,ind3], "o", color=col[3], markersize=10)
plot(Ms[ind4], control_cost[1,ind4], "o", color=col[4], markersize=10)
for (g, temp_goal) = enumerate(temp_goals)
fill_between([], [], [], facecolor=col[g], label=latexstring("\$\\max(T)>\$", temp_goal), alpha=0.5)
end
minM1 = Ms[ind1]
minM2 = Ms[ind2]
minM3 = Ms[ind3]
minM4 = Ms[ind4]
ylims = ax.get_ylim()
fill_between([minM2,minM1], [ylims[1], ylims[1]], [ylims[2],ylims[2]], color=col[1], alpha=0.2)
fill_between([minM3,minM2], [ylims[1], ylims[1]], [ylims[2],ylims[2]], color=col[2], alpha=0.2)
fill_between([minM4,minM3], [ylims[1], ylims[1]], [ylims[2],ylims[2]], color=col[3], alpha=0.2)
fill_between([0,minM4], [ylims[1], ylims[1]], [ylims[2],ylims[2]], color=col[4], alpha=0.2)
axvline(minM1, color=col[1])
axvline(minM2, color=col[2])
axvline(minM3, color=col[3])
axvline(minM4, color=col[4])
ylim(ylims)
xlim(0,1)
xlabel("Emissions mitigation level [% reduction]")
xticks(0.:0.2:1.0, ["0%", "20%", "40%", "60%", "80%", "100%"])
ylabel("Net present cost of controls [trillion USD]")
plot([], [], "o", color="grey", label="lowest cost", markersize=10.)
legend(loc="upper left")
ax = subplot(1,2,2)
plot(Ms, net_benefit[1,:], "k-", lw=2)
ind = argmax(net_benefit[1,:])
plot(Ms[ind], net_benefit[1,ind], "ko", markersize=10, label="most benefits")
ylims = ax.get_ylim()
axvline(minM1, color=col[1])
axvline(minM2, color=col[2])
axvline(minM3, color=col[3])
axvline(minM4, color=col[4])
ylim(ylims)
xlim(0,1)
xlabel("Emissions mitigation level [% reduction]")
xticks(0.:0.2:1.0, ["0%", "20%", "40%", "60%", "80%", "100%"])
ylabel("Net present benefits, relative to baseline [trillion USD]")
legend(loc="upper left")
gcf()
##
# ### Visualizing the two-dimensional optimization problem
fig = figure(figsize=(14, 5))
o = 1
subplot(1,2,o)
pcolor(Ms, Rs, control_cost, cmap="Greys", vmin=0., vmax=150.)
cbar = colorbar(label="Net present cost of controls [trillion USD]")
control_cost[(min_temp .<= 0.)] .= NaN
contour(Ms, Rs, control_cost, levels=[10, 50], colors="k", linewidths=0.5, alpha=0.4)
grid(true, color="k", alpha=0.25)
temp_mask = ones(size(max_temp))
temp_mask[max_temp .<= 4.0] .= NaN
contourf(Ms, Rs, temp_mask, cmap="Reds", alpha=1.0, vmin=0., vmax=2.)
temp_mask = ones(size(min_temp))
temp_mask[min_temp .> 0.] .= NaN
contourf(Ms, Rs, temp_mask, cmap="Blues", alpha=1.0, vmin=0., vmax=2.5)
contour(Ms, Rs, min_temp, colors="darkblue", levels=[0.], linewidths=2.5)
plot([], [], "o", color="grey", label="lowest cost", markersize=10.)
plot([], [], "-", color="darkblue", label=latexstring("\$\\min(T)=\$", 0), lw=2.5)
for (g, temp_goal) = enumerate(temp_goals)
contour(Ms, Rs, max_temp, colors=[col[g]], levels=[temp_goal], linewidths=2.5)
plot(optimal_controls[1,g,o], optimal_controls[2,g,o], "o", color=col[g], markersize=10.)
fill_between([], [], [], facecolor=col[g], label=latexstring("\$\\max(T)>\$", temp_goal), alpha=0.5)
end
legend(loc="upper left")
xlabel("Emissions mitigation level [% reduction]")
xticks(0.:0.2:1.0, ["0%", "20%", "40%", "60%", "80%", "100%"])
ylabel(L"CO$_{2e}$ removal rate [% of present-day emissions]")
yticks(0.:0.2:1.0, ["0%", "20%", "40%", "60%", "80%", "100%"])
annotate(L"$\max(T) > 4\degree$C", (0.015, 0.03), xycoords="axes fraction", color="darkred", fontsize=9)
annotate(L"$\min(T) < 0\degree$C", (0.72, 0.725), xycoords="axes fraction", color="darkblue", fontsize=9)
title("Cost-effectiveness analysis")
o = 2
subplot(1,2,o)
q = pcolor(Ms, Rs, net_benefit, cmap="Greys_r", vmin=0, vmax=250)
cbar = colorbar(label="Net present benefits, relative to baseline [trillion USD]", extend="both")
contour(Ms, Rs, net_benefit, levels=[100, 200], colors="k", linewidths=0.5, alpha=0.4)
grid(true, color="k", alpha=0.25)
temp_mask = ones(size(min_temp))
temp_mask[(min_temp .> 0.)] .= NaN
contourf(Ms, Rs, temp_mask, cmap="Blues", alpha=1.0, vmin=0., vmax=2.5)
contour(Ms, Rs, min_temp, colors="darkblue", levels=[0.], linewidths=2.5)
plot([], [], "-", color="darkblue")
plot(optimal_controls[1,1,2], optimal_controls[2,1,2], "o", color="k", markersize=10., label="most benefits")
for (g, temp_goal) = enumerate(temp_goals)
contour(Ms, Rs, max_temp, colors=[col[g]], levels=[temp_goal], linewidths=2.5)
end
legend()
xlabel("Emissions mitigation level [% reduction]")
xticks(0.:0.2:1.0, ["0%", "20%", "40%", "60%", "80%", "100%"])
ylabel(L"CO$_{2e}$ removal rate [% of present-day emissions]")
yticks(0.:0.2:1.0, ["0%", "20%", "40%", "60%", "80%", "100%"])
annotate(L"$\min(T) < 0\degree$C", (0.72, 0.725), xycoords="axes fraction", color="darkblue", fontsize=9)
title("Cost-benefit analysis")
gcf() | ClimateMARGO | https://github.com/ClimateMARGO/ClimateMARGO.jl.git |
|
[
"MIT"
]
| 0.3.3 | f958c49f3f4f812d6eb2725f102896a0ead72172 | code | 512 | module ClimateMARGO
if VERSION < v"1.5"
@error "ClimateMARGO requires Julia v1.5 or newer."
end
include("Models/Models.jl")
include("Utils/Utils.jl")
include("Diagnostics/Diagnostics.jl")
include("Optimization/Optimization.jl")
include("IO/IO.jl")
include("PolicyResponse/PolicyResponse.jl")
if get(ENV, "JULIA_MARGO_LOAD_PYPLOT", "1") == "1"
# default
include("Plotting/Plotting.jl")
else
# (in our API, we don't load the plotting functions.)
@info "Not loading plotting functions"
end
end | ClimateMARGO | https://github.com/ClimateMARGO/ClimateMARGO.jl.git |
|
[
"MIT"
]
| 0.3.3 | f958c49f3f4f812d6eb2725f102896a0ead72172 | code | 41 | sec_per_year = 60. * 60. * 24. * 365.25
| ClimateMARGO | https://github.com/ClimateMARGO/ClimateMARGO.jl.git |
|
[
"MIT"
]
| 0.3.3 | f958c49f3f4f812d6eb2725f102896a0ead72172 | code | 410 | module Diagnostics
using ClimateMARGO.Models
using ClimateMARGO.Utils
export
t, past_mask,
ramp_emissions, emissions, effective_emissions,
c, F, Tslow, Tfast, T, T_adapt,
τd, B, F2x, ECS,
discount, f, E,
damage, cost, benefit,
net_benefit, net_present_cost, net_present_benefit
include("carbon.jl")
include("energy_balance.jl")
include("cost_benefit.jl")
include("utils.jl")
end | ClimateMARGO | https://github.com/ClimateMARGO/ClimateMARGO.jl.git |
|
[
"MIT"
]
| 0.3.3 | f958c49f3f4f812d6eb2725f102896a0ead72172 | code | 1583 | # Following RCP8.5 CO2e concentrations
# Raw data at https://www.iiasa.ac.at/web-apps/tnt/RcpDb/dsd?Action=htmlpage&page=compare
#
# See below link for 2020 initial condition:
# https://www.eea.europa.eu/data-and-maps/indicators/atmospheric-greenhouse-gas-concentrations-6/assessment-1
function ramp_emissions(t, q0::Float64, n::Float64, t1::Float64, t2::Float64)
t0 = t[1]
Δt0 = t1 - t0
Δt1 = t2 - t1
q = zeros(size(t))
increase_idx = (t .<= t1)
decrease_idx = ((t .> t1) .& (t .<= t2))
q[increase_idx] .= q0 * (1. .+ (n-1) .*(t[increase_idx] .- t0)/Δt0)
q[decrease_idx] .= n * q0 * (t2 .- t[decrease_idx])/Δt1
q[t .> t2] .= 0.
return q
end
function ramp_emissions(dom::Domain)
return ramp_emissions(t(dom), 7.5, 3., 2100., 2150.)
end
emissions(q, M) = q .* (1. .- M)
function emissions(m::ClimateModel; M=false)
return emissions(
m.economics.baseline_emissions,
m.controls.mitigate .* (1. .- .~past_mask(m) * ~M)
)
end
function effective_emissions(r, q, M, R)
return r*(emissions(q, M) .- q[1]*R)
end
function effective_emissions(m; M=false, R=false)
return effective_emissions(
m.physics.r,
m.economics.baseline_emissions,
m.controls.mitigate .* (1. .- .~past_mask(m) * ~M),
m.controls.remove .* (1. .- .~past_mask(m) * ~R)
)
end
function c(c0, effective_emissions, dt)
return c0 .+ cumsum(effective_emissions * dt)
end
function c(m; M=false, R=false)
return c(
m.physics.c0,
effective_emissions(m, M=M, R=R),
m.domain.dt
)
end | ClimateMARGO | https://github.com/ClimateMARGO/ClimateMARGO.jl.git |
|
[
"MIT"
]
| 0.3.3 | f958c49f3f4f812d6eb2725f102896a0ead72172 | code | 2833 | f(α::Array; p=3.) = α.^p # shape of individual cost functions
E(t, E0, γ) = E0 * (1. .+ γ).^(t .- t[1])
E(m) = E(t(m), m.economics.E0, m.economics.γ)
discount(t, ρ, tp) = .~past_mask(t, tp) .* (1. .+ ρ) .^ (- (t .- tp))
discount(m::ClimateModel) = discount(t(m), m.economics.ρ, m.domain.present_year)
T_adapt(T, A) = T .* sqrt.(1 .- A)
T_adapt(m::ClimateModel; M=false, R=false, G=false, A=false) = T_adapt(
T(m, M=M, R=R, G=G),
m.controls.adapt .* (1. .- .~past_mask(m) * ~A),
)
damage(β, E, Ta; discount=1.) = (β .* E .* Ta.^2) .* discount
damage(m; discounting=false, M=false, R=false, G=false, A=false) = damage(
m.economics.β,
E(m),
T_adapt(m, M=M, R=R, G=G, A=A),
discount=1. .+ discounting * (discount(m) .- 1.)
)
cost(CM, CR, CG, CA, ϵCG, E, q, M, R, G, A; discount=1., p=3.) = (
( ppm_to_GtCO2(q).*CM.*f(M, p=p) +
E.*(CG.*f(G, p=p) .+ ϵCG*(G.>1.e-3)) +
CR*f(R, p=p) +
E.*CA.*f(A, p=p)
) .* discount
)
cost(m::ClimateModel; discounting=false, p=3., M=false, R=false, G=false, A=false) = cost(
m.economics.mitigate_cost,
m.economics.remove_cost,
m.economics.geoeng_cost,
m.economics.adapt_cost,
m.economics.epsilon_cost,
E(m),
m.economics.baseline_emissions,
m.controls.mitigate .* M,
m.controls.remove .* R,
m.controls.geoeng .* G,
m.controls.adapt .* A,
discount=1. .+ discounting * (discount(m) .- 1.),
p=p
)
function cost(m::ClimateModel, controls::String; discounting=false)
vars = Dict("M"=>false, "R"=>false, "G"=>false, "A"=>false)
for (key, value) in vars
if occursin(key, controls)
vars[key] = true
end
end
return cost(m, discounting=discounting; M=vars["M"], R=vars["R"], G=vars["G"], A=vars["A"])
end
benefit(damage_baseline, damage) = damage_baseline .- damage
benefit(m::ClimateModel; discounting=false, M=false, R=false, G=false, A=false) = benefit(
damage(m, discounting=discounting),
damage(m, discounting=discounting, M=M, R=R, G=G, A=A)
)
net_benefit(benefit, cost) = benefit .- cost
net_benefit(m::ClimateModel; discounting=true, M=false, R=false, G=false, A=false) = net_benefit(
benefit(m, discounting=discounting, M=M, R=R, G=G, A=A),
cost(m, discounting=discounting, M=M, R=R, G=G, A=A)
)
net_present_cost(cost, dt) = sum(cost*dt)
function net_present_cost(
m::ClimateModel;
discounting=true, M=false, R=false, G=false, A=false
)
return net_present_cost(
cost(m, discounting=discounting, M=M, R=R, G=G, A=A),
m.domain.dt
)
end
net_present_benefit(net_benefit, dt) = sum(net_benefit*dt)
net_present_benefit(m::ClimateModel; discounting=true, M=false, R=false, G=false, A=false) = net_present_benefit(
net_benefit(m, discounting=discounting, M=M, R=R, G=G, A=A),
m.domain.dt
)
| ClimateMARGO | https://github.com/ClimateMARGO/ClimateMARGO.jl.git |
|
[
"MIT"
]
| 0.3.3 | f958c49f3f4f812d6eb2725f102896a0ead72172 | code | 2701 | """
F(a, c0, Finf, c, G; F0=0.)
"""
function F(a, c0, Finf, c, G; F0=0.)
F0 .+ a .* log.( c/c0 ) .- G*Finf
end
"""
F(m::ClimateModel; M=false, R=false, G=false)
"""
function F(m; M=false, R=false, G=false, F0=0.)
return F(
m.physics.a, m.physics.c0, m.economics.Finf,
c(m, M=M, R=R),
m.controls.geoeng .* (1. .- .~past_mask(m) * ~G),
F0=F0
)
end
F2x(a::Float64) = a*log(2)
F2x(m::ClimateModel) = F2x(m.physics.a)
ECS(a, B) = F2x(a)/B
ECS(params::ClimateModelParameters) = ECS(params.physics.a, m.physics.B)
ECS(m::ClimateModel) = ECS(m.physics.a, m.physics.B)
calc_B(a::Float64, ECS::Float64) = F2x(a)/ECS
calc_B(params::ClimateModelParameters; ECS=ECS(params)) = calc_B(params.physics.a, ECS)
calc_B(m::ClimateModel; ECS=ECS(m)) = calc_B(m.physics.a, ECS)
τd(Cd, B, κ) = (Cd/B) * (B+κ)/κ
τd(phys::Physics) = τd(phys.Cd, phys.B, phys.κ)
τd(m::ClimateModel) = τd(m.physics)
"""
T_fast(F, κ, B)
"""
T_fast(F, κ, B) = F/(κ + B)
"""
T_fast(m::ClimateModel; M=false, R=false, G=false)
"""
T_fast(m::ClimateModel; M=false, R=false, G=false) = T_fast(
F(m, M=M, R=R, G=G),
m.physics.κ,
m.physics.B
)
"""
T_slow(F, Cd, κ, B, t, dt)
"""
function T_slow(F, Cd, κ, B, t, dt)
τ = τd(Cd, κ, B)
return (
(κ/B) / (κ + B) *
exp.( - (t .- (t[1] - dt)) / τ) .*
cumsum( (exp.( (t .- (t[1] - dt)) / τ) / τ) .* F * dt)
)
end
"""
T_slow(m::ClimateModel; M=false, R=false, G=false)
"""
T_slow(m::ClimateModel; M=false, R=false, G=false) = T_slow(
F(m, M=M, R=R, G=G),
m.physics.Cd,
m.physics.κ,
m.physics.B,
t(m),
m.domain.dt
)
"""
T(T0, F, Cd, κ, B, t, dt)
Returns the sum of the initial, fast mode, and slow mode temperature change.
# Arguments
- `T0::Float64`: warming relative to pre-industrial [°C]
- `F::Array{Float64}`: change in radiative forcing since the initial time ``t_{0}`` [W/m``{2}``]
- `Cd::Float64`: deep ocean heat capacity [W yr m``^{2}`` K``^{-1}``]
- `κ::Float64`: ocean heat uptake rate [W m``^{2}`` K``^{-1}``]
- `B::Float64`: feedback parameter [W m``^{2}`` K``^{-1}``]
- `t::Array{Float64}`: year [years]
- `dt::Float64`: timestep [years]
"""
T(T0, F, Cd, κ, B, t, dt) = (
T0 .+
T_fast(F, κ, B) .+
T_slow(F, Cd, κ, B, t, dt)
)
"""
T(m::ClimateModel; M=false, R=false, G=false)
Returns the sum of the initial, fast mode, and slow mode temperature change,
as diagnosed from `m` and modified by the climate controls activated by the
Boolean kwargs.
"""
T(m::ClimateModel; M=false, R=false, G=false) = T(
m.physics.T0,
F(m, M=M, R=R, G=G),
m.physics.Cd,
m.physics.κ,
m.physics.B,
t(m),
m.domain.dt
) | ClimateMARGO | https://github.com/ClimateMARGO/ClimateMARGO.jl.git |
|
[
"MIT"
]
| 0.3.3 | f958c49f3f4f812d6eb2725f102896a0ead72172 | code | 283 | t(t0, tf, dt) = t0:dt:tf
t(dom::Domain) = t(dom.initial_year,dom.final_year,dom.dt)
t(params::ClimateModelParameters) = t(params.domain)
t(m::ClimateModel) = t(m.domain)
past_mask(t, present_year) = t .< present_year
past_mask(model) = past_mask(t(model), model.domain.present_year) | ClimateMARGO | https://github.com/ClimateMARGO/ClimateMARGO.jl.git |
|
[
"MIT"
]
| 0.3.3 | f958c49f3f4f812d6eb2725f102896a0ead72172 | code | 182 | module IO
using JSON2
using ClimateMARGO.Models
export export_state, import_state,
export_parameters, import_parameters,
included_configurations
include("json_io.jl")
end | ClimateMARGO | https://github.com/ClimateMARGO/ClimateMARGO.jl.git |
|
[
"MIT"
]
| 0.3.3 | f958c49f3f4f812d6eb2725f102896a0ead72172 | code | 1254 | "Export a [`ClimateModel`](@ref) instance to a `.json` file.
This `.json` file can be shared with others, you can import it using "
function export_state(filename::AbstractString, m::ClimateModel)
open(filename, "w") do io
JSON2.write(io, m)
end
end
function import_state(filename::String)::ClimateModel
open(filename, "r") do io
return JSON2.read(io, ClimateModel)
end
end
function export_parameters(filename::AbstractString, params::ClimateModelParameters)
open(filename, "w") do io
JSON2.write(io, params)
end
end
function import_parameters(filename::AbstractString)::ClimateModelParameters
open(filename, "r") do io
return JSON2.read(io, ClimateModelParameters)
end
end
"""The [`ClimateModelParameters`](@ref) included with this package.
Currently `included_configurations["default"]` is the only included set."""
const included_configurations = let
# find the config dir relative to this .jl file
config_dir = joinpath(@__DIR__, "..", "..", "configurations")
config_files = [file for file in readdir(config_dir) if occursin(".json", file)]
loaded = [import_parameters(joinpath(config_dir, file)) for file in config_files]
Dict(p.name => p for p in loaded)
end | ClimateMARGO | https://github.com/ClimateMARGO/ClimateMARGO.jl.git |
|
[
"MIT"
]
| 0.3.3 | f958c49f3f4f812d6eb2725f102896a0ead72172 | code | 1734 | module Models
export Domain, Physics, Controls, Economics, ClimateModelParameters, ClimateModel
include("domain.jl")
include("physics.jl")
include("controls.jl")
include("economics.jl")
"""
ClimateModelParameters(name, domain::Domain, economics::Economics, physics::Physics)
Create a named instance of the MARGO climate model parameters, which include
economic input parameters (`economics`), physical climate parameters (`physics`),
and climate control policies (`controls`) on some spatial-temporal grid (`domain`).
Use these to construct a [`ClimateModel`](@ref), which also contains the optimized
controls.
"""
mutable struct ClimateModelParameters
name::String
domain::Domain
economics::Economics
physics::Physics
end
mutable struct ClimateModel
name::String
domain::Domain
economics::Economics
physics::Physics
controls::Controls
end
"""
ClimateModel(params::ClimateModelParameters[, controls::Controls])
Create an instance of an extremely idealized multi-control climate model. The
returned object contains the [`ClimateModelParameters`](@ref), and will contain
the optimized [`Controls`](@ref). These can be computed using
[`optimize_controls!`](@ref).
"""
ClimateModel(params::ClimateModelParameters, controls::Controls) = ClimateModel(
params.name,
params.domain,
params.economics,
params.physics,
controls
)
function ClimateModel(params::ClimateModelParameters)
dom = params.domain
t = collect(dom.initial_year:dom.dt:dom.final_year)
return ClimateModel(
params,
Controls(
zeros(size(t)),
zeros(size(t)),
zeros(size(t)),
zeros(size(t))
)
)
end
end | ClimateMARGO | https://github.com/ClimateMARGO/ClimateMARGO.jl.git |
|
[
"MIT"
]
| 0.3.3 | f958c49f3f4f812d6eb2725f102896a0ead72172 | code | 413 | """
Controls(mitigate, remove, geoeng, adapt)
Create data structure for climate controls.
# Examples
```jldoctest
a = zeros(4);
C = Controls(a, a, a, a);
C.geoeng
# output
4-element Array{Float64,1}:
0.0
0.0
0.0
0.0
```
See also: [`ClimateModel`](@ref)
"""
mutable struct Controls
mitigate::Array{Float64,1}
remove::Array{Float64,1}
geoeng::Array{Float64,1}
adapt::Array{Float64,1}
end | ClimateMARGO | https://github.com/ClimateMARGO/ClimateMARGO.jl.git |
|
[
"MIT"
]
| 0.3.3 | f958c49f3f4f812d6eb2725f102896a0ead72172 | code | 302 | mutable struct Domain
dt::Float64
present_year::Float64
initial_year::Float64
final_year::Float64
end
Domain(dt::Float64, initial_year::Float64, final_year::Float64) = (
Domain(dt, initial_year, initial_year, final_year)
)
Domain(dt::Float64) = Domain(dt, initial_year, final_year) | ClimateMARGO | https://github.com/ClimateMARGO/ClimateMARGO.jl.git |
|
[
"MIT"
]
| 0.3.3 | f958c49f3f4f812d6eb2725f102896a0ead72172 | code | 2959 | """
Economics(
E0, γ, β, ρ, Finf,
mitigate_cost, remove_cost, geoeng_cost, adapt_cost,
mitigate_init, remove_init, geoeng_init, adapt_init,
baseline_emissions,
extra_CO₂
)
Create data structure for economic input parameters for `ClimateModel` struct,
including a baseline emissions scenario.
### Arguments
- `E0::Array{Float64,1}`: Gross World Product timeseries [10^12 USD / year]
- `γ::Float64`: economic growth rate [fraction]
- `β::Float64`: climate damage parameter [% GWP / (°C)^2].
- `ρ::Float64`: typically denoted ρ in economic references [fraction].
- `Finf::Float64`: maximum SRM forcing
- `[control]_cost::Float64`: scaling cost of full control deployment [10^12 USD / year OR % of GWP].
- `[control]_init::Float64`: fixed initial condition for control deployment [10^12 USD / year].
- `baseline_emissions::Array{Float64,1}`: prescribed baseline CO₂ equivalent emissions [ppm / yr].
- `extra_CO₂::Array{Float64,1}`: optional additional CO₂ input that is used only for Social Cost of Carbon calculations [ppm].
See also: [`ClimateModel`](@ref), [`baseline_emissions`](@ref), [`GWP`](@ref).
"""
mutable struct Economics
E0::Float64
γ::Float64
β::Float64
ρ::Float64
Finf::Float64
mitigate_cost::Float64
remove_cost::Float64
geoeng_cost::Float64
adapt_cost::Float64
mitigate_init
remove_init
geoeng_init
adapt_init
baseline_emissions::Array{Float64,1}
epsilon_cost::Float64
extra_CO₂::Array{Float64,1}
end
# constructor without `extra_CO₂`, setting it to zero
function Economics(E0::Float64,
γ::Float64,
β::Float64,
ρ::Float64,
Finf::Float64,
mitigate_cost::Float64,
remove_cost::Float64,
geoeng_cost::Float64,
adapt_cost::Float64,
mitigate_init,
remove_init,
geoeng_init,
adapt_init,
baseline_emissions::Array{Float64,1})
return Economics(
E0,
γ,
β,
ρ,
Finf,
mitigate_cost,
remove_cost,
geoeng_cost,
adapt_cost,
mitigate_init,
remove_init,
geoeng_init,
adapt_init,
baseline_emissions,
0.
)
end
# constructor without `extra_CO₂`, setting it to zero
function Economics(E0::Float64,
γ::Float64,
β::Float64,
ρ::Float64,
Finf::Float64,
mitigate_cost::Float64,
remove_cost::Float64,
geoeng_cost::Float64,
adapt_cost::Float64,
mitigate_init,
remove_init,
geoeng_init,
adapt_init,
baseline_emissions::Array{Float64,1},
epsilon_cost::Float64)
return Economics(
E0,
γ,
β,
ρ,
Finf,
mitigate_cost,
remove_cost,
geoeng_cost,
adapt_cost,
mitigate_init,
remove_init,
geoeng_init,
adapt_init,
baseline_emissions,
epsilon_cost,
zeros(size(baseline_emissions))
)
end | ClimateMARGO | https://github.com/ClimateMARGO/ClimateMARGO.jl.git |
|
[
"MIT"
]
| 0.3.3 | f958c49f3f4f812d6eb2725f102896a0ead72172 | code | 517 | """
Physics(C0, T0, a, B, Cd, κ, r)
Create data structure for model physics.
See also: [`ClimateModel`](@ref)
"""
mutable struct Physics
c0::Float64 # initial CO2e concentration, relative to PI
T0::Float64 # initial temperature, relative to PI
a::Float64 # logarithmic CO2 forcing coefficient
B::Float64 # feedback parameter
Cd::Float64 # deep ocean heat capacity
κ::Float64 # deep ocean heat uptake rate
r::Float64 # long-term airborne fraction of CO2e
end | ClimateMARGO | https://github.com/ClimateMARGO/ClimateMARGO.jl.git |
|
[
"MIT"
]
| 0.3.3 | f958c49f3f4f812d6eb2725f102896a0ead72172 | code | 194 | module Optimization
using JuMP, Ipopt
using ClimateMARGO.Models
using ClimateMARGO.Utils
using ClimateMARGO.Diagnostics
export optimize_controls!
include("deterministic_optimization.jl")
end | ClimateMARGO | https://github.com/ClimateMARGO/ClimateMARGO.jl.git |
|
[
"MIT"
]
| 0.3.3 | f958c49f3f4f812d6eb2725f102896a0ead72172 | code | 17726 |
function optimize_controls!(
m::ClimateModel;
obj_option = "adaptive_temp", temp_goal = 1.5, budget=10., expenditure = 0.5,
max_deployment = Dict("mitigate"=>1., "remove"=>1., "geoeng"=>1., "adapt"=>1.),
max_slope = Dict("mitigate"=>1. /40., "remove"=>1. /40., "geoeng"=>1. /40., "adapt"=> 1. /40.),
temp_overshoot = nothing,
overshoot_year = 2100,
delay_deployment = Dict(
"mitigate"=>0,
"remove"=>0,
"geoeng"=>0,
"adapt"=>0
),
cost_exponent = 3.,
print_status = false, print_statistics = false, print_raw_status = true,
)
# Translate JuMP printing options
if print_status
if print_statistics
bool_str = "yes"
else
bool_str = "no"
end
print_int = 1
else
print_int = 0
bool_str = "no"
end
# Shorthands
tarr = t(m)
Earr = E(m)
τ = τd(m)
dt = m.domain.dt
t0 = tarr[1]
tp = m.domain.present_year
q = m.economics.baseline_emissions
qGtCO2 = ppm_to_GtCO2(q)
N = length(tarr)
# Set default temperature goal for end year
if isnothing(temp_overshoot)
temp_overshoot = temp_goal
odx = N
elseif temp_overshoot >= temp_goal
odx = argmin(abs.(tarr .- overshoot_year)) # overshoot index
elseif temp_overshoot < temp_goal
temp_overshoot = temp_goal
odx = N
end
# Set defaults for start_deployment
start_deployment = Dict()
for (key, item) in delay_deployment
start_deployment[key] = t0 + delay_deployment[key]
end
if typeof(cost_exponent) in [Int64, Float64]
cost_exponent = Dict(
"mitigate"=>cost_exponent,
"remove"=>cost_exponent,
"geoeng"=>cost_exponent,
"adapt"=>cost_exponent
)
end
model_optimizer = Model(optimizer_with_attributes(Ipopt.Optimizer,
"acceptable_tol" => 1.e-8, "max_iter" => Int64(1e8),
"acceptable_constr_viol_tol" => 1.e-3, "constr_viol_tol" => 1.e-4,
"print_frequency_iter" => 50, "print_timing_statistics" => bool_str,
"print_level" => print_int,
))
function fM_JuMP(α)
if α <= 0.
return 1000.
else
return α ^ cost_exponent["mitigate"]
end
end
register(model_optimizer, :fM_JuMP, 1, fM_JuMP, autodiff=true)
function fA_JuMP(α)
if α <= 0.
return 1000.
else
return α ^ cost_exponent["adapt"]
end
end
register(model_optimizer, :fA_JuMP, 1, fA_JuMP, autodiff=true)
function fR_JuMP(α)
if α <= 0.
return 1000.
else
return α ^ cost_exponent["remove"]
end
end
register(model_optimizer, :fR_JuMP, 1, fR_JuMP, autodiff=true)
function fG_JuMP(α)
if α <= 0.
return 1000.
else
return α ^ cost_exponent["geoeng"]
end
end
register(model_optimizer, :fG_JuMP, 1, fG_JuMP, autodiff=true)
scale = 5.e-3
function Hstep(α)
if α <= 0.
return 0.
elseif 0. < α <= scale/2.
return 2*(α/scale)^2
elseif scale/2. < α <= scale
return 1. - 2((α - scale)/scale)^2
elseif α > scale
return 1.
end
end
register(model_optimizer, :Hstep, 1, Hstep, autodiff=true)
function log_JuMP(x)
if x <= 0.
return -1000.0
else
return log(x)
end
end
register(model_optimizer, :log_JuMP, 1, log_JuMP, autodiff=true)
function discounting_JuMP(t)
if t < tp
return 0.
else
return (
(1. + m.economics.ρ) ^
(-(t - tp))
)
end
end
register(model_optimizer, :discounting_JuMP, 1, discounting_JuMP, autodiff=true)
# constraints on control variables
for (key, item) in max_deployment
if item==0.
max_deployment[key] = item+1.e-8
end
end
@variables(model_optimizer, begin
0. <= M[1:N] <= max_deployment["mitigate"] # emissions reductions
0. <= R[1:N] <= max_deployment["remove"] # negative emissions
0. <= G[1:N] <= max_deployment["geoeng"] # geoengineering
0. <= A[1:N] <= max_deployment["adapt"] # adapt
end)
control_vars = Dict(
"mitigate" => M,
"remove" => R,
"geoeng" => G,
"adapt" => A
)
controls = Dict(
"mitigate" => m.controls.mitigate,
"remove" => m.controls.remove,
"geoeng" => m.controls.geoeng,
"adapt" => m.controls.adapt
)
domain_idx = (tarr .> tp) # don't update past or present
if tarr[1] == tp
domain_idx = (tarr .>= tp) # unless present is also first timestep
end
M₀ = m.economics.mitigate_init
R₀ = m.economics.remove_init
G₀ = m.economics.geoeng_init
A₀ = m.economics.adapt_init
control_inits = Dict(
"mitigate" => M₀,
"remove" => R₀,
"geoeng" => G₀,
"adapt" => A₀
)
for (key, control) in control_vars
if control_inits[key] !== nothing
fix(control_vars[key][1], control_inits[key]; force = true)
Nstart = 2
else
Nstart = 1
end
for idx in Nstart:N
if idx <= length(tarr[.~domain_idx])
fix(control_vars[key][idx], controls[key][idx]; force = true)
else
if tarr[idx] < start_deployment[key]
if control_inits[key] !== nothing
fix(control_vars[key][idx], control_inits[key]; force = true)
else
fix(control_vars[key][idx], 0.; force = true)
end
end
end
end
end
# add integral function as a new variable defined by first order finite differences
@variable(model_optimizer, cumsum_qMR[1:N]);
for i in 1:N-1
@constraint(
model_optimizer, cumsum_qMR[i+1] - cumsum_qMR[i] ==
(dt * (m.physics.r * (q[i+1] * (1. - M[i+1]) - q[1] * R[i+1])))
)
end
@constraint(
model_optimizer, cumsum_qMR[1] == (dt * (m.physics.r * (q[1] * (1. - M[1]) - q[1] * R[1])))
);
# add temperature kernel as new variable defined by first order finite difference
@variable(model_optimizer, cumsum_KFdt[1:N]);
for i in 1:N-1
@NLconstraint(
model_optimizer, cumsum_KFdt[i+1] - cumsum_KFdt[i] ==
(
dt *
exp( (tarr[i+1] - (t0 - dt)) / τ ) * (
m.physics.a * log_JuMP(
(m.physics.c0 + cumsum_qMR[i+1]) /
(m.physics.c0)
) - m.economics.Finf*G[i+1] )
)
)
end
@NLconstraint(
model_optimizer, cumsum_KFdt[1] ==
(
dt *
exp( dt / τ ) * (
m.physics.a * log_JuMP(
(m.physics.c0 + cumsum_qMR[1]) /
(m.physics.c0)
) - m.economics.Finf*G[1] )
)
);
# Add constraint of rate of changes
present_idx = argmin(abs.(tarr .- tp))
@variables(model_optimizer, begin
-max_slope["mitigate"] <= dMdt[present_idx:N-1] <= max_slope["mitigate"]
-max_slope["remove"] <= dRdt[present_idx:N-1] <= max_slope["remove"]
-max_slope["geoeng"] <= dGdt[present_idx:N-1] <= max_slope["geoeng"]
-max_slope["adapt"] <= dAdt[present_idx:N-1] <= max_slope["adapt"]
end);
for i in present_idx:N-1
@constraint(model_optimizer, dMdt[i] == (M[i+1] - M[i]) / dt)
end
for i in present_idx:N-1
@constraint(model_optimizer, dRdt[i] == (R[i+1] - R[i]) / dt)
end
for i in present_idx:N-1
@constraint(model_optimizer, dGdt[i] == (G[i+1] - G[i]) / dt)
end
for i in present_idx:N-1
@constraint(model_optimizer, dAdt[i] == (A[i+1] - A[i]) / dt)
end
## Optimization options
# maximize net benefits
if obj_option == "net_benefit"
# (in practice we solve the equivalent problem of minimizing the net cost, i.e. minus the net benefit)
@NLobjective(model_optimizer, Min,
sum(
(
m.economics.β *
Earr[i] *
(
(m.physics.T0 +
(
(m.physics.a * log_JuMP(
(m.physics.c0 + cumsum_qMR[i]) /
(m.physics.c0)
) - m.economics.Finf*G[i]
) +
m.physics.κ /
(τ * m.physics.B) *
exp( - (tarr[i] - (t0 - dt)) / τ ) *
cumsum_KFdt[i]
) / (m.physics.B + m.physics.κ)
)^2
) * ( (1 - A[i]) ) +
Earr[i] * m.economics.adapt_cost * fA_JuMP(A[i]) +
m.economics.mitigate_cost * qGtCO2[i] *
fM_JuMP(M[i]) +
m.economics.remove_cost * fR_JuMP(R[i]) +
Earr[i] * (
m.economics.geoeng_cost * fG_JuMP(G[i]) +
m.economics.epsilon_cost * Hstep(G[i])
)
) *
discounting_JuMP(tarr[i]) *
dt
for i=1:N)
)
# maximize net benefits subject to a temperature goal
elseif obj_option == "temp"
@NLobjective(model_optimizer, Min,
sum(
(
Earr[i] * m.economics.adapt_cost * fA_JuMP(A[i]) +
m.economics.mitigate_cost * qGtCO2[i] *
fM_JuMP(M[i]) +
m.economics.remove_cost * fR_JuMP(R[i]) +
Earr[i] * (
m.economics.geoeng_cost * fG_JuMP(G[i]) +
m.economics.epsilon_cost * Hstep(G[i])
)
) *
discounting_JuMP(tarr[i]) *
dt
for i=1:N)
)
# Subject to temperature goal (during overshoot period)
for i in 1:odx-1
@NLconstraint(model_optimizer,
(m.physics.T0 +
(
(m.physics.a * log_JuMP(
(m.physics.c0 + cumsum_qMR[i]) /
(m.physics.c0)
) - m.economics.Finf*G[i]
) +
m.physics.κ /
(τ * m.physics.B) *
exp( - (tarr[i] - (t0 - dt)) / τ ) *
cumsum_KFdt[i]
) / (m.physics.B + m.physics.κ)
) <= temp_overshoot
)
end
# Subject to temperature goal (after temporary overshoot period)
for i in odx:N
@NLconstraint(model_optimizer,
(m.physics.T0 +
(
(m.physics.a * log_JuMP(
(m.physics.c0 + cumsum_qMR[i]) /
(m.physics.c0)
) - m.economics.Finf*G[i]
) +
m.physics.κ /
(τ * m.physics.B) *
exp( - (tarr[i] - (t0 - dt)) / τ ) *
cumsum_KFdt[i]
) / (m.physics.B + m.physics.κ)
) <= temp_goal
)
end
# minimize control costs subject to adaptative-temperature constraint
elseif obj_option == "adaptive_temp"
# minimize control costs
@NLobjective(model_optimizer, Min,
sum(
(
Earr[i] * m.economics.adapt_cost * fA_JuMP(A[i]) +
m.economics.mitigate_cost * qGtCO2[i] *
fM_JuMP(M[i]) +
m.economics.remove_cost * fR_JuMP(R[i]) +
Earr[i] * (
m.economics.geoeng_cost * fG_JuMP(G[i]) +
m.economics.epsilon_cost * Hstep(G[i])
)
) *
discounting_JuMP(tarr[i]) *
dt
for i=1:N)
)
# Subject to adaptive temperature goal (during overshoot period)
for i in 1:odx-1
@NLconstraint(model_optimizer,
((m.physics.T0 +
(
(m.physics.a * log_JuMP(
(m.physics.c0 + cumsum_qMR[i]) /
(m.physics.c0)
) - m.economics.Finf*G[i]
) +
m.physics.κ /
(τ * m.physics.B) *
exp( - (tarr[i] - (t0 - dt)) / τ ) *
cumsum_KFdt[i]
) / (m.physics.B + m.physics.κ)
)
* sqrt(1. - A[i]) ) <=
temp_overshoot
)
end
# Subject to adaptive temperature goal (after temporary overshoot period)
for i in odx:N
@NLconstraint(model_optimizer,
((m.physics.T0 +
(
(m.physics.a * log_JuMP(
(m.physics.c0 + cumsum_qMR[i]) /
(m.physics.c0)
) - m.economics.Finf*G[i]
) +
m.physics.κ /
(τ * m.physics.B) *
exp( - (tarr[i] - (t0 - dt)) / τ ) *
cumsum_KFdt[i]
) / (m.physics.B + m.physics.κ)
)
* sqrt(1. - A[i]) ) <=
temp_goal
)
end
# minimize damages subject to a total discounted budget constraint
elseif obj_option == "budget"
@NLobjective(model_optimizer, Min,
sum(
m.economics.β *
Earr[i] *
(
m.physics.T0 +
(
(m.physics.a * log_JuMP(
(m.physics.c0 + cumsum_qMR[i]) /
(m.physics.c0)
) - m.economics.Finf*G[i]
) +
m.physics.κ /
(τ * m.physics.B) *
exp( - (tarr[i] - (t0 - dt)) / τ ) *
cumsum_KFdt[i]
) / (m.physics.B + m.physics.κ)
)^2 * (1. - A[i]) *
discounting_JuMP(t[i]) *
dt
for i=1:N)
)
@NLconstraint(model_optimizer,
sum(
(
Earr[i] * m.economics.adapt_cost * fA_JuMP(A[i]) +
m.economics.mitigate_cost * qGtCO2[i] *
fM_JuMP(M[i]) +
m.economics.remove_cost * fR_JuMP(R[i]) +
Earr[i] * (
m.economics.geoeng_cost * fG_JuMP(G[i]) +
m.economics.epsilon_cost * Hstep(G[i])
)
) *
discounting_JuMP(tarr[i]) *
dt
for i=1:N) <= budget
)
# minimize damages subject to annual expenditure constraint (as % of GWP)
elseif obj_option == "expenditure"
@NLobjective(model_optimizer, Min,
sum(
m.economics.β *
Earr[i] *
(m.physics.T0 +
(
(m.physics.a * log_JuMP(
(m.physics.c0 + cumsum_qMR[i]) /
(m.physics.c0)
) - m.economics.Finf*G[i]
) +
m.physics.κ /
(τ * m.physics.B) *
exp( - (tarr[i] - (t0 - dt)) / τ ) *
cumsum_KFdt[i]
) / (m.physics.B + m.physics.κ)
)^2 * (1-A[i]) *
discounting_JuMP(t[i]) *
dt
for i=1:N)
)
for i in 1:N
@NLconstraint(model_optimizer,
(
Earr[i] * m.economics.adapt_cost * fA_JuMP(A[i]) +
m.economics.mitigate_cost * qGtCO2[i] *
fM_JuMP(M[i]) +
m.economics.remove_cost * fR_JuMP(R[i]) +
Earr[i] * (
m.economics.geoeng_cost * fG_JuMP(G[i]) +
m.economics.epsilon_cost * Hstep(G[i])
)
) <= expenditure * Earr[i]
)
end
end
optimize!(model_optimizer)
if print_raw_status
print(raw_status(model_optimizer), "\n")
end
mitigate_values = value.(M)[domain_idx]
mitigate_values[q[domain_idx].==0.] .= 0.
getfield(m.controls, :mitigate)[domain_idx] = mitigate_values
getfield(m.controls, :remove)[domain_idx] = value.(R)[domain_idx]
getfield(m.controls, :geoeng)[domain_idx] = value.(G)[domain_idx]
getfield(m.controls, :adapt)[domain_idx] = value.(A)[domain_idx]
return model_optimizer
end | ClimateMARGO | https://github.com/ClimateMARGO/ClimateMARGO.jl.git |
|
[
"MIT"
]
| 0.3.3 | f958c49f3f4f812d6eb2725f102896a0ead72172 | code | 267 | module Plotting
using PyPlot
using ClimateMARGO.Models
using ClimateMARGO.Diagnostics
export
plot_controls, plot_emissions, plot_concentrations,
plot_forcings, plot_temperatures,
plot_benefits, plot_damages,
plot_state
include("line_plots.jl")
end | ClimateMARGO | https://github.com/ClimateMARGO/ClimateMARGO.jl.git |
|
[
"MIT"
]
| 0.3.3 | f958c49f3f4f812d6eb2725f102896a0ead72172 | code | 9473 | rcParams = PyPlot.PyDict(PyPlot.matplotlib."rcParams")
rcParams["lines.linewidth"] = 2. # Change linewidth
function fill_past(m, ylims)
domain_idx = (t(m) .> m.present_year)
fill_between(
t(m)[.~domain_idx],
ones(size(t(m)[.~domain_idx])) * ylims[1] * 2.,
ones(size(t(m)[.~domain_idx])) * ylims[2] * 2.,
facecolor="b", alpha=0.1
)
ylim(ylims)
return
end
function plot_emissions(m::ClimateModel)
title("effective greenhouse gas emissions")
#fill_past(m, ylims)
fill_between(t(m), effective_emissions(m), effective_emissions(m, M=true), facecolor="C0", alpha=0.3, label="Mitigation")
fill_between(t(m), effective_emissions(m, M=true), effective_emissions(m, M=true, R=true), facecolor="C1", alpha=0.3, label="CDR")
plot(t(m), effective_emissions(m), "-", color="grey", lw=2.0, label=L"$rq$ (no-policy baseline)")
plot(t(m), effective_emissions(m, M=true), "k-", lw=1, alpha=0.4)
plot(t(m), effective_emissions(m, M=true, R=true), "k-", lw=2., label=L"$rq(1-M) - rq_{0}R$ (controlled)")
plot(t(m), zeros(size(t(m))), dashes=(2.5, 1.75), color="grey", alpha=0.5)
ylimit = maximum(effective_emissions(m)) * 1.1
ylims = [-ylimit, ylimit]
ylabel(L"effective CO$_{2e}$ emissions [ppm / yr]")
xlim(t(m)[1],2200.)
ylim(minimum(effective_emissions(m, M=true, R=true))-5.,1.1*maximum(effective_emissions(m)))
xticks(t(m)[1]:40.:2200.)
xlabel("year")
grid(true, alpha=0.3)
return
end
function plot_concentrations(m::ClimateModel)
title("greenhouse gas concentrations")
#fill_past(m, ylims)
fill_between(t(m), c(m), c(m, M=true), facecolor="C0", alpha=0.3, label="Mitigation")
fill_between(t(m), c(m, M=true), c(m, M=true, R=true), facecolor="C1", alpha=0.3, label="CDR")
plot(t(m), c(m), "-", color="grey", lw=2.0, label=L"$c$ (no-policy baseline)")
plot(t(m), c(m, M=true), "k-", lw=1, alpha=0.4)
plot(t(m), c(m, M=true, R=true), "k-", lw=2.0, label=L"$c_{M,R}$ (controlled)")
ylims = [0., maximum(c(m))*1.05]
ylabel(L"CO$_{2e}$ concentration [ppm]")
xlabel("year")
xlim(t(m)[1],2200.)
ylim(ylims)
xticks(t(m)[1]:40.:2200.)
grid(true, alpha=0.3)
return
end
function plot_forcings(m::ClimateModel; F0=3.)
title("forcing (greenhouse effect and SRM)")
#fill_past(m, ylims)
fill_between(t(m), F(m, F0=F0), F(m, M=true, F0=F0), facecolor="C0", alpha=0.3, label="Mitigation")
fill_between(t(m), F(m, M=true, F0=F0), F(m, M=true, R=true, F0=F0), facecolor="C1", alpha=0.3, label="CDR")
fill_between(t(m), F(m, M=true, R=true, F0=F0), F(m, M=true, R=true, G=true, F0=F0), facecolor="C3", alpha=0.3, label="SRM")
plot(t(m), F(m, F0=F0), "-", color="grey", lw=2.0, label=L"$F$ (no-policy baseline)")
plot(t(m), F(m, M=true, F0=F0), "k-", lw=1, alpha=0.4)
plot(t(m), F(m, M=true, R=true, F0=F0), "k-", lw=1, alpha=0.4)
plot(t(m), F(m, M=true, R=true, G=true, F0=F0), "k-", lw=2.0, label=L"$F_{M,R,G}$ (controlled)")
ylims = [0., maximum(F(m, F0=F0))*1.05]
ylabel(L"radiative forcing [W/m$^{2}$]")
xlabel("year")
xlim(t(m)[1],2200.)
ylim(ylims)
xticks(t(m)[1]:40.:2200.)
grid(true, alpha=0.3)
return
end
function plot_temperatures(m::ClimateModel; temp_goal=1.2)
title("adaptive temperature change")
#fill_past(m, ylims)
fill_between(t(m), T_adapt(m), T_adapt(m, M=true), facecolor="C0", alpha=0.3, label="Mitigation")
fill_between(t(m), T_adapt(m, M=true), T_adapt(m, M=true, R=true), facecolor="C1", alpha=0.3, label="CDR")
fill_between(t(m), T_adapt(m, M=true, R=true), T_adapt(m, M=true, R=true, G=true), facecolor="C3", alpha=0.3, label="SRM")
fill_between(t(m), T_adapt(m, M=true, R=true, G=true), T_adapt(m, M=true, R=true, G=true, A=true), facecolor="C2", alpha=0.3, label="Adaptation")
plot(t(m), T_adapt(m), "-", color="grey", lw=2.0, label=L"$T$ (no-policy baseline)")
plot(t(m), T_adapt(m, M=true), "k-", lw=1, alpha=0.4)
plot(t(m), T_adapt(m, M=true, R=true), "k-", lw=1, alpha=0.4)
plot(t(m), T_adapt(m, M=true, R=true, G=true), "k-", lw=1., alpha=0.4)
plot(t(m), T_adapt(m, M=true, R=true, G=true, A=true), "k-", lw=2.0, label=L"$T_{M,R,G,A}$ (adaptive)")
plot(t(m),temp_goal .* ones(size(t(m))), dashes=(2.5, 1.75), color="grey", alpha=0.75, lw=2.5)
ylims = [0., maximum(T_adapt(m)) * 1.05]
ylabel("temperature anomaly [°C]")
xlabel("year")
xlim(t(m)[1],2200.)
xticks(t(m)[1]:40.:2200.)
grid(true, alpha=0.3)
legend()
return
end
function plot_controls(m::ClimateModel)
title("optimized control deployments")
plot(t(m)[m.economics.baseline_emissions .> 0.], m.controls.mitigate[m.economics.baseline_emissions .> 0.],
color="C0", lw=2, label=L"$M$ (emissions mitigation)")
plot(t(m), m.controls.remove, color="C1", lw=2, label=L"$R$ (carbon dioxide removal)")
plot(t(m), m.controls.adapt, color="C2", lw=2, label=L"$A$ (adaptation)")
plot(t(m), m.controls.geoeng, color="C3", lw=2, label=L"$G$ (solar geoengineering)")
ylims = [0., 1.075]
yticks(0.:0.2:1.0, 0:20:100)
ylim(ylims)
ylabel("control deployment [%]")
xlabel("year")
xlim(t(m)[1],2200.)
xticks(t(m)[1]:40.:2200.)
grid(true, alpha=0.3)
return
end
function plot_benefits(m::ClimateModel; discounting=true)
domain_idx = (t(m) .> m.domain.present_year)
fill_between(
t(m)[domain_idx],
0 .*ones(size(t(m)))[domain_idx],
net_benefit(m, discounting=discounting, M=true, R=true, G=true, A=true)[domain_idx],
facecolor="grey", alpha=0.2
)
plot(t(m)[domain_idx], 0 .*ones(size(t(m)))[domain_idx], "-", lw=2, color="gray", label="no-policy baseline")
plot(t(m)[domain_idx], benefit(m, discounting=discounting, M=true, R=true, G=true, A=true)[domain_idx], color="C8", lw=2, label="benefits (of avoided damages)")
plot(t(m)[domain_idx], cost(m, discounting=discounting, M=true, R=true, G=true, A=true)[domain_idx], color="C4", lw=2, label="costs (of climate controls)")
plot(t(m)[domain_idx], net_benefit(m, discounting=discounting, M=true, R=true, G=true, A=true)[domain_idx], color="k", lw=2, label="net benefits (benefits - costs)")
ylabel(L"discounted costs and benefits [10$^{12}$ \$ / year]")
xlabel("year")
xlim(t(m)[1],2200.)
xticks(t(m)[1]:40.:2200.)
grid(true, alpha=0.3)
title("cost-benefit analysis")
return
end
function plot_damages(m::ClimateModel; discounting=true, percent_GWP=false, temp_goal=1.2)
Enorm = deepcopy(E(m))/100.
if ~percent_GWP; Enorm=1.; end;
domain_idx = (t(m) .> m.domain.present_year)
fill_between(
t(m)[domain_idx],
0 .*ones(size(t(m)))[domain_idx],
(cost(m, discounting=discounting, M=true, R=true, G=true, A=true) ./ Enorm)[domain_idx],
facecolor="C4", alpha=0.2
)
damages = damage(m, discounting=discounting, M=true, R=true, G=true, A=true)
costs = cost(m, discounting=discounting, M=true, R=true, G=true, A=true)
plot(t(m)[domain_idx], (damage(m, discounting=discounting) ./ Enorm)[domain_idx], color="gray", lw=2.0, label="uncontrolled damages")
plot(t(m)[domain_idx], ((damages .+ costs)./ Enorm)[domain_idx], color="k", lw=2.0, label="net costs (controlled damages + controls)")
plot(t(m)[domain_idx], (damages ./ Enorm)[domain_idx], color="C8", lw=2.0, label="controlled damages")
plot(t(m)[domain_idx], (costs ./ Enorm)[domain_idx], color="C4", lw=2.0, label="cost of controls")
ylim([0, maximum((damage(m, discounting=discounting) ./ Enorm)[domain_idx]) * 0.75])
dmg_label = string("damage threshold at ",round(temp_goal, digits=2),L"°C with $A=0$")
plot(
t(m)[domain_idx],
(damage(m.economics.β, E(m), temp_goal, discount=discount(m)) ./ Enorm)[domain_idx],
dashes=(2.5,1.75), color="grey", alpha=0.75, lw=2.0, label=dmg_label
)
if ~percent_GWP;
if ~discounting;
ylabel(L"costs [10$^{12}$ \$ / year]");
else;
ylabel(L"discounted costs [10$^{12}$ \$ / year]");
end
else
if ~discounting
ylabel("costs [% GWP]")
else
ylabel("discounted costs [% GWP]")
print("NOT YET SUPPORTED")
end
end
xlabel("year")
xlim(t(m)[1],2200.)
xticks(t(m)[1]:40.:2200.)
grid(true, alpha=0.175)
title("costs of avoiding a damage threshold")
legend()
return
end
function plot_state(m::ClimateModel; new_figure=true, plot_legends=true, temp_goal=1.2)
if new_figure
fig, axs = subplots(2, 3, figsize=(14,8))
axs = vcat(axs...)
end
sca(axs[1])
plot_emissions(m)
title("a)", loc="left")
sca(axs[2])
plot_concentrations(m)
title("b)", loc="left")
sca(axs[3])
plot_temperatures(m, temp_goal=temp_goal)
title("c)", loc="left")
sca(axs[4])
plot_controls(m)
title("d)", loc="left")
sca(axs[5])
plot_benefits(m)
title("e)", loc="left")
sca(axs[6])
plot_damages(m, temp_goal=temp_goal)
title("f)", loc="left")
if plot_legends;
for ii in 1:6
sca(axs[ii])
if ii <= 2;
legend(loc="lower left", labelspacing=0.14, handlelength=1.75);
else
legend(loc="upper left", labelspacing=0.14, handlelength=1.75);
end
end
end
tight_layout()
return fig, axs
end
| ClimateMARGO | https://github.com/ClimateMARGO/ClimateMARGO.jl.git |
|
[
"MIT"
]
| 0.3.3 | f958c49f3f4f812d6eb2725f102896a0ead72172 | code | 159 | module PolicyResponse
using ClimateMARGO.Models
using ClimateMARGO.Diagnostics
export step_forward!, add_emissions_bump!
include("stepping_forward.jl")
end | ClimateMARGO | https://github.com/ClimateMARGO/ClimateMARGO.jl.git |
|
[
"MIT"
]
| 0.3.3 | f958c49f3f4f812d6eb2725f102896a0ead72172 | code | 916 | function step_forward!(m::ClimateModel, Δt::Float64)
m.domain.present_year = m.domain.present_year + Δt
m.name = string(Int64(round(m.domain.present_year)))
end
function add_emissions_bump!(m::ClimateModel, Δt::Float64, Δq::Float64; present_year = m.domain.present_year)
present_idx = argmin(abs.(t(m) .- (present_year .+ Δt)))
future = (t(m) .>= present_year)
near_future = future .& (t(m) .<= present_year + Δt)
near_future1 = near_future .& (t(m) .< present_year + Δt/2)
near_future2 = near_future .& (t(m) .>= present_year + Δt/2)
new_emissions = m.economics.baseline_emissions
new_emissions[near_future1] .+= (
Δq * (t(m) .- present_year) / (Δt/2.)
)[near_future1]
new_emissions[near_future2] .+= (
Δq * (1. .- (t(m) .- (present_year .+ Δt/2.)) / (Δt/2.))
)[near_future2]
m.economics.baseline_emissions = new_emissions
end | ClimateMARGO | https://github.com/ClimateMARGO/ClimateMARGO.jl.git |
|
[
"MIT"
]
| 0.3.3 | f958c49f3f4f812d6eb2725f102896a0ead72172 | code | 208 | module Utils
using ClimateMARGO.Models
export
year, init_zero_controls,
GtCO2_to_ppm, tCO2_to_ppm,
ppm_to_GtCO2, ppm_to_tCO2
include("instantiate_models.jl")
include("unit_conversions.jl")
end | ClimateMARGO | https://github.com/ClimateMARGO/ClimateMARGO.jl.git |
|
[
"MIT"
]
| 0.3.3 | f958c49f3f4f812d6eb2725f102896a0ead72172 | code | 306 | """
init_zero_controls(t)
Return initial state of uniformly-zero climate controls.
See also: [`Controls`](@ref)
"""
function init_zero_controls(t::Array{Float64,1})
c = Controls(
zeros(size(t)),
zeros(size(t)),
zeros(size(t)),
zeros(size(t))
)
return c
end | ClimateMARGO | https://github.com/ClimateMARGO/ClimateMARGO.jl.git |
|
[
"MIT"
]
| 0.3.3 | f958c49f3f4f812d6eb2725f102896a0ead72172 | code | 226 | const year = 365.25 * 24. * 60. * 60.
GtCO2_to_ppm(GtCO2) = GtCO2 / (2.13 * (44. /12.))
tCO2_to_ppm(tCO2) = GtCO2_to_ppm(tCO2) * 1.e-9
ppm_to_GtCO2(ppm) = ppm * (2.13 * (44. /12.))
ppm_to_tCO2(ppm) = ppm_to_GtCO2(ppm) * 1.e9 | ClimateMARGO | https://github.com/ClimateMARGO/ClimateMARGO.jl.git |
|
[
"MIT"
]
| 0.3.3 | f958c49f3f4f812d6eb2725f102896a0ead72172 | code | 3558 | ENV["JULIA_MARGO_LOAD_PYPLOT"] = "no thank you"
using JuMP
using ClimateMARGO
using ClimateMARGO.Models, ClimateMARGO.Optimization, ClimateMARGO.Diagnostics
using Test
using BenchmarkTools
@testset "Temperature optimization" begin
@testset "Temp goal: $(temp_goal)" for temp_goal in 1.5:0.5:4.0
model = ClimateModel(deepcopy(ClimateMARGO.IO.included_configurations["default"]))
status = optimize_controls!(model, temp_goal=temp_goal)
@test raw_status(status) == "Solve_Succeeded"
@test isapprox(
maximum(T_adapt(model, M=true, R=true, G=true, A=true)),
temp_goal,
rtol=1.e-5
)
end
end
function default_parameters(dt=12)::ClimateModelParameters
p = deepcopy(ClimateMARGO.IO.included_configurations["default"])
p.domain = Domain(Float64(dt), 2020.0, 2200.0)
p.economics.baseline_emissions = ramp_emissions(p.domain)
p.economics.extra_CO₂ = zeros(size(p.economics.baseline_emissions))
return p
end
@testset "Pinned results" begin
# We generate results using ClimateMARGO.jl once, and then add a test to check that these same results are still generated. This allows us to "pin" a correct state of the model, and make internal changes with the guarantee that the outcome does not change.
# In the future, to "pin" new results as the correct ones, run the tests, and copy the expected result from the test failure report.
@testset "Forward" begin
model = ClimateModel(default_parameters(20))
bump() = [0.0:0.2:0.9; 1.0:-0.2:0.1] # == [0.0, 0.2, 0.4, 0.6, 0.8, 1.0, 0.8, 0.6, 0.4, 0.2]
slope() = 0.0:0.1:0.99
constant() = fill(1.0, size(slope())...)
model.controls.mitigate = bump()
model.controls.remove = 0.2 .* bump()
model.controls.geoeng = 0.1 .* slope()
model.controls.adapt = 0.3 .* constant()
result_T = T_adapt(model; M=true, R=true, G=true, A=true)
@test result_T ≈ [1.2767663110173484, 1.608852123606971, 1.8978258561722958, 2.108526090432568, 2.211931788367322, 2.1713210884072347, 2.158586858753497, 2.1224302958426446, 2.0894535810555555, 2.0607413902621032] rtol=1e-5
end
@testset "Optimization" begin
model = ClimateModel(default_parameters(20))
optimize_controls!(model, temp_goal=2.2)
@test model.controls.mitigate ≈ [0.0, 0.3670584821282429, 0.4393272792260638, 0.5257940783265341, 0.6245380250211872, 0.7074912042403808, 0.7676867396677578, 0.0, 0.0, 0.0] rtol=1e-3
@test model.controls.remove ≈ [0.0, 0.10882651060773696, 0.13025296274983308, 0.15588887768405002, 0.18516475517861308, 0.20975894242613452, 0.22760588089905764, 0.2367501073337253, 0.23175978166812158, 0.19659824324032557] rtol=1e-3
@test model.controls.geoeng ≈ [0.0, 0.05049106345373785, 0.05264838036542512, 0.06460796652594906, 0.10819144740515446, 0.12584271884062354, 0.12767445817227918, 0.12318301615805025, 0.11794556458720538, 0.1132343245663971] rtol=1e-3
@test model.controls.adapt ≈ [0.0, 0.0001537202313707738, 0.00015374376329529388, 0.03816957588616602, 0.10942294994448329, 0.13786594819465353, 0.14363875559967432, 0.14044599067883523, 0.1365010682189578, 0.13366455981096753] rtol=1e-3
end
end
const RUN_BENCHMARKS = true
if RUN_BENCHMARKS
@info "Running benchmark..."
function go()
model = ClimateModel(default_parameters(20))
optimize_controls!(model; temp_goal=2.2, print_raw_status=false)
end
go()
display(@benchmark go())
end | ClimateMARGO | https://github.com/ClimateMARGO/ClimateMARGO.jl.git |
|
[
"MIT"
]
| 0.3.3 | f958c49f3f4f812d6eb2725f102896a0ead72172 | docs | 235 | ### Gallery of interactive ClimateMARGO.jl applications
- The **[Official ClimateMARGO web-app](https://margo.plutojl.org/introduction.html)**, currently hosted by our collaborators at [`Pluto.jl`](https://github.com/fonsp/Pluto.jl).
| ClimateMARGO | https://github.com/ClimateMARGO/ClimateMARGO.jl.git |
|
[
"MIT"
]
| 0.3.3 | f958c49f3f4f812d6eb2725f102896a0ead72172 | docs | 2539 | <!-- Title -->
<h1 align="center">
ClimateMARGO.jl
</h1>
<!-- description -->
<p align="center">
<strong> A Julia implementation of <b>MARGO</b>, an idealized framework for optimization of climate control strategies.</strong>
</p>
<!-- Information badges -->
<p align="center">
<a href="https://mit-license.org">
<img alt="MIT license" src="https://img.shields.io/badge/License-MIT-blue.svg?style=flat-square">
</a>
<a href="https://github.com/ClimateMARGO/ClimateMARGO.jl/issues/new">
<img alt="Ask us anything" src="https://img.shields.io/badge/Ask%20us-anything-1abc9c.svg?style=flat-square">
</a>
<a href="https://ClimateMARGO.github.io/ClimateMARGO.jl/dev/">
<img alt="Documentation in development" src="https://img.shields.io/badge/docs-latest-blue.svg?style=flat-square">
</a>
<a href="https://travis-ci.com/ClimateMARGO/ClimateMARGO.jl">
<img alt="Build status" src="https://travis-ci.com/ClimateMARGO/ClimateMARGO.jl.svg?branch=main">
</a>
</p>
<!-- CI/CD badges -->
The MARGO model is described in full in an [accompanying Research Article](https://iopscience.iop.org/article/10.1088/1748-9326/ac243e/pdf), published *Open-Access* in the journal *Environmental Research Letters*. The julia scripts and jupyter notebooks that contain all of the paper's analysis are available in the [MARGO-paper](https://github.com/ClimateMARGO/MARGO-paper) repository (these are useful as advanced applications of MARGO to complement the minimal examples included in the documentation).
Try out the MARGO model by running our [web-app](https://margo.plutojl.org/introduction.html) directly in your browser!
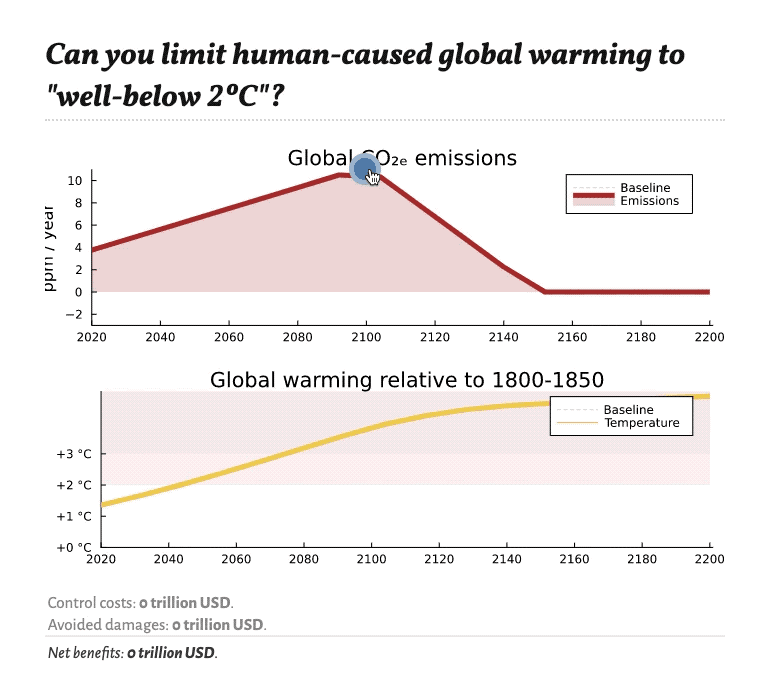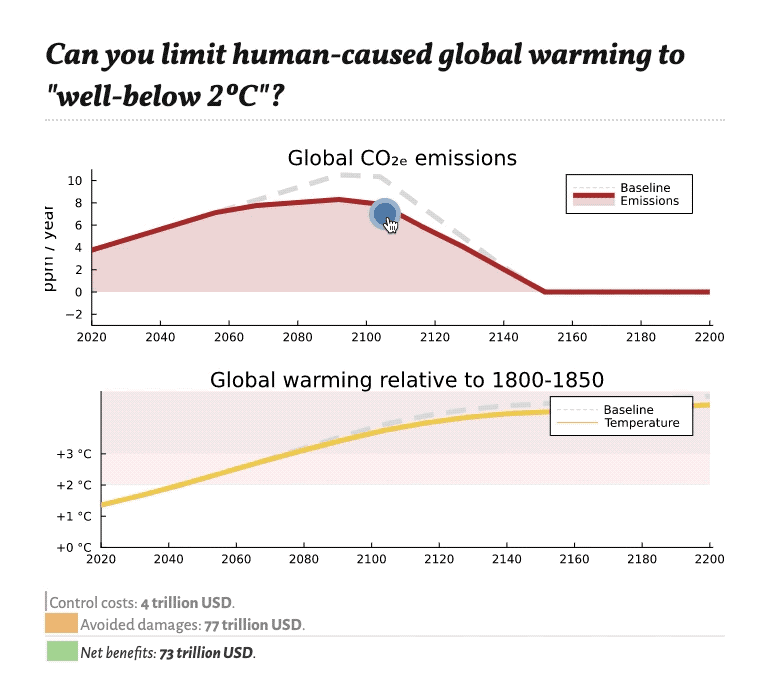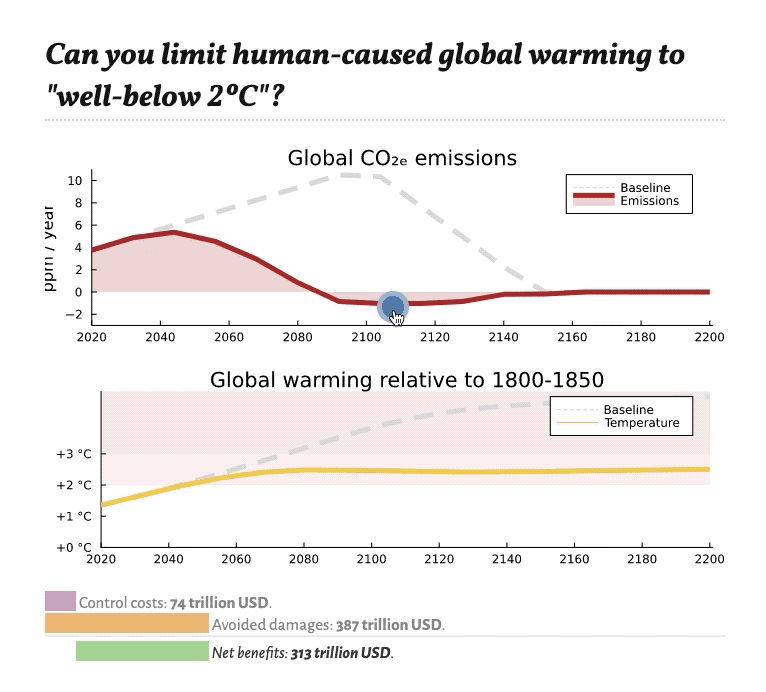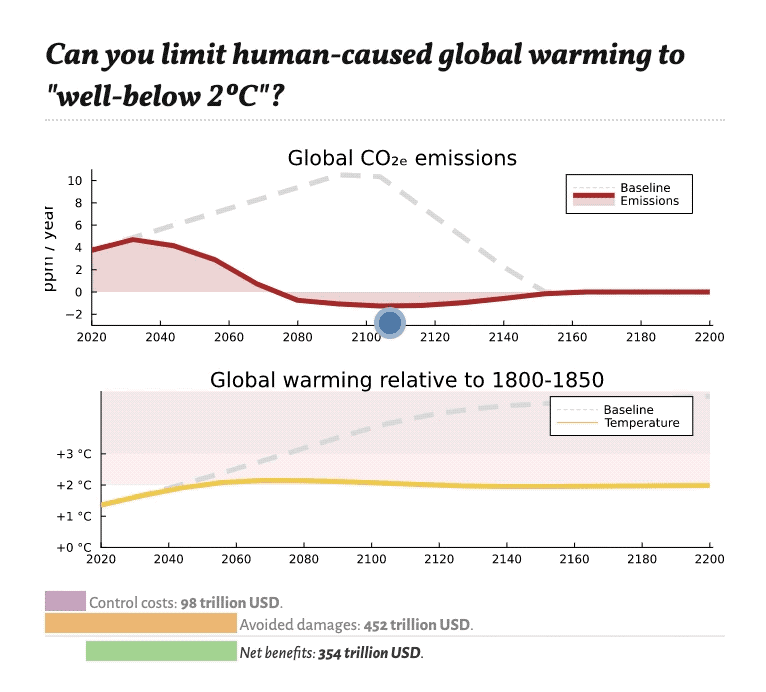
ClimateMARGO.jl is currently in beta testing; basic model documentation is slowly being added. Substantial structural changes may still take place before the first stable release v1.0.0. Anyone interested in helping develop the model post an Issue here or contact the lead developer Henri Drake directly (henrifdrake `at` gmail.com), until explicit guidelines for contributing to the model are posted at a later date.
----
<small>README.md formatting inspired by [Oceananigans.jl](https://github.com/CliMA/Oceananigans.jl)</small>
| ClimateMARGO | https://github.com/ClimateMARGO/ClimateMARGO.jl.git |
|
[
"MIT"
]
| 0.3.3 | f958c49f3f4f812d6eb2725f102896a0ead72172 | docs | 894 | # Diagnostics
We refer to key model variables which depend only on 1) the values of the control variables and 2) the input parameters as *diagnostic* variables. Examples are the CO``_{2e}`` concentrations ``c_{M,R}(t)``, the temperature change ``T_{M,R,G,A}``, and the cost of mitigation ``\mathcal{C}_{M} M^{2}``.
In ClimateMARGO.jl, diagnostic variables are represented as julia *functions*, which are implemented using two separate *methods*, based on the type of the function arguments: the first method requires an explicit list of all the input parameters and control variables that determine the diagnostic variables; the second method leverages the `ClimateModel` struct to read in the required variables.
For example, here are the two methods that define the diagnostic function `T` for the temperature change:
```@meta
CurrentModule = ClimateMARGO.Diagnostics
```
```@docs
T
```
| ClimateMARGO | https://github.com/ClimateMARGO/ClimateMARGO.jl.git |
Subsets and Splits
No community queries yet
The top public SQL queries from the community will appear here once available.