repo_name
stringlengths 6
130
| hexsha
list | file_path
list | code
list | apis
list |
|---|---|---|---|---|
Daz-Riza-Seriog/Op_Heat_transfer
|
[
"3ee27a049a2de5deaf13356ed3e675562233ac29"
] |
[
"Parcial 3.py"
] |
[
"# Code made for Sergio Andres Diaz Ariza\n# 16 December 2021\n# License MIT\n# Heat Operations: Python Program\n\nimport numpy as np\nimport matplotlib.pyplot as plt\nimport seaborn as sns\n\n##### Part 1- Masic Flux Oil #####\nT_oil_in = 350 + 273.15 # [K]\nT_oil_out = 200 + 273.15 # [K]\nT_water_in = 170 + 273.15 # [K]\nT_water_out = 180 + 273.15 # [K]\n\n# Oil\ndeltha_T_oil = T_oil_in - T_oil_out\nCp_oil_350 = 3.008 # [KJ/Kg*K]--> 350 Celsisus\nCp_oil_200 = 2.54 # [KJ/Kg*K]--> 200 Celsisus\nCp_oil_prom = (Cp_oil_350 + Cp_oil_200) / 2\n\n# Water\nH_vap_water = 2050 # [KJ/Kg]\n\n# Q = m_dot_oil*Cp_oil*(deltha_T_oil) = Q = m_dot_water*H_vap_water\n\nm_dot_water = 0.5 # [Kg/s]\nm_dot_oil = (m_dot_water * H_vap_water) / (Cp_oil_prom * deltha_T_oil)\n\n#### Part 2 ####\nQ = m_dot_oil * Cp_oil_prom * (deltha_T_oil)\nU_sup = 0.350 # [W/m^2*K]\n\n# Cocurrent flow\ndeltha_T1_co = T_oil_in - T_water_in\ndeltha_T2_co = T_oil_out - T_water_out\n\nDeltha_T_log_co = (deltha_T1_co - deltha_T2_co) / np.log(deltha_T1_co / deltha_T2_co)\n\n# Counter flow\ndeltha_T1_con = T_oil_in - T_water_out\ndeltha_T2_con = T_oil_out - T_water_in\n\nDeltha_T_log_con = (deltha_T1_con - deltha_T2_con) / np.log(deltha_T1_con / deltha_T2_con)\n\nAs_co = Q / (U_sup * Deltha_T_log_co)\nAs_con = Q / (U_sup * Deltha_T_log_con)\n\n# Usamos la de menor Area Superficial por eficiencia\nN = 100000 # Suponemos 1000 tubos de Trabajo\nD = (As_con / N * 2 * np.pi) * 2\n#D = 0.0030610385380016977\nD_ext = D+0.002413\n\nmiu_cin_oil_prom = 0.995\nrho_oil_prom = 700 # [kg/m^3\nmiu_oil_prom = miu_cin_oil_prom * rho_oil_prom # [Kg/m*s]\n\n# Calculemos el Reynolds para el Aceite\nRe_oil = (4 * m_dot_oil) / (np.pi * D_ext * miu_oil_prom)\nPr_oil = 14.5 # Prom in the exponential range\nk_oil = 0.114 #[W/m*K]\n\n# Values of\nC_oil = 0.989\nm_oil = 0.330\nNu_oil = C_oil * (Re_oil ** m_oil) * (Pr_oil ** (1 / 3))\nh_oil = (Nu_oil*k_oil)/D_ext\n\n# Calculemos el Reynolds para el Agua\nmiu_water_prom = 8.851e-4\nPr_water = 1.05\nk_water = 0.3567 #[]w/m*K\n\nRe_water = (4 * m_dot_water) / (np.pi * D * miu_water_prom)\nNu_water = 0.023*(Re_water**4/5)*Pr_water**0.4\nh_water = (Nu_water*k_water)/D\n\n# Global coeficient\nk_stain = 45 #[@W/m*K]\n\nR_water = 1/h_water\nR_stain = ((D/2)/k_stain)*np.log((D_ext/2)/(D/2))\nR_oil = ((D/2)/(D_ext/2))*(1/h_oil)\n\nU = 0.01/(R_water+R_stain+R_oil)\n\nAs_con_2 = Q / (U * Deltha_T_log_con)\nD_2 = (As_con_2 / N * 2 * np.pi) * 2\n\nQ_ideal = (U * Deltha_T_log_con*As_con)\n\nprint(\"Flujo masico de Aceite:\", m_dot_oil)\nprint(\"Calor Transmitido:\", Q)\nprint(\"Area superficial Cocorriente:\", As_co)\nprint(\"Area superficial Contracorriente:\", As_con)\nprint(\"Diametero de tubos:\", D)\nprint(\"Reynolds Oil:\", Re_oil)\nprint(\"Convective coefficient Oil:\", h_oil)\nprint(\"Reynolds water:\", Re_water)\nprint(\"Convective coefficient water:\", h_water)\nprint(\"Coeficiente Global de Transferencia:\",U)\nprint(\"Area de Transferencia:\",As_con_2)\nprint(\"Diametero de tubos:\", D_2)\nprint(\"Diametero de tubos:\", Q_ideal)"
] |
[
[
"numpy.log"
]
] |
longodj/tcvaemolgen
|
[
"70fee6e73430b497a9a0f6296da0ac188af05843"
] |
[
"notebooks/4_T-CVAE.py"
] |
[
"#!/usr/bin/env python\n# coding: utf-8\n\n# In[1]:\n\n\nimport copy\nfrom dgl import batch, unbatch\nimport rdkit.Chem as Chem\nfrom rdkit import RDLogger\nimport torch\ntorch.manual_seed(0)\nimport torch.nn as nn\nimport torch.nn.functional as F\n\nimport numpy as np\nnp.random.seed(0)\n\nimport os,sys,inspect\nsys.path.insert(0,'/home/icarus/app/src') \n\nfrom models.jtnn_vae import DGLJTNNVAE\nfrom models.modules import *\n\n#from .nnutils import cuda, move_dgl_to_cuda\n\n#from .jtnn_enc import DGLJTNNEncoder\n#from .jtnn_dec import DGLJTNNDecoder\n#from .mpn import DGLMPN\n#from .mpn import mol2dgl_single as mol2dgl_enc\n#from .jtmpn import DGLJTMPN\n#from .jtmpn import mol2dgl_single as mol2dgl_dec\n\nimport os,sys,inspect\nfrom tqdm import tqdm\nsys.path.insert(0,'/home/icarus/T-CVAE-MolGen/src')\n\nfrom utils.chem import set_atommap, copy_edit_mol, enum_assemble_nx, attach_mols_nx, decode_stereo\n\nlg = RDLogger.logger()\n\nlg.setLevel(RDLogger.CRITICAL)\n\n\n# In[2]:\n\n\nclass TCVAE(nn.Module):\n def __init__(self, vocab, hidden_size, latent_size, depth):\n super(TCVAE, self).__init__()\n self.vocab = vocab\n self.hidden_size = hidden_size\n self.latent_size = latent_size\n self.depth = depth\n \n self.embedding = nn.Embedding(vocab.size(), hidden_size).cuda()\n self.mpn = MPN(hidden_size, depth)\n self.encoder = None #\n self.decoder = None #\n \n self.T_mean, self.T_var = nn.Linear(hidden_size, latent_size // 2), nn.Linear(hidden_size, latent_size // 2)\n self.G_mean, self.G_var = nn.Linear(hidden_size, latent_size // 2), nn.Linear(hidden_size, latent_size // 2)\n \n self.n_nodes_total = 0\n self.n_edges_total = 0\n self.n_passes = 0\n\n\n# ## Posterior\n# \n# As in <cite data-cite=\"7333468/6Y976JUQ\"></cite>\n# \n# $q(z|x,y) \\sim N(\\mu,\\sigma^2I)$, where\n# \n# $\\quad\\quad h = \\text{MultiHead}(c,E_\\text{out}^L(x;y),E_\\text{out}^L(x;y))$\n# \n# $\\quad\\quad \\begin{bmatrix}\\mu\\\\\\log(\\sigma^2)\\end{bmatrix} = hW_q+b_q$\n\n# In[3]:\n\n\ndef sample_posterior(self, prob_decode=False):\n return\n\n\n# ### Prior\n# \n# As in <cite data-cite=\"7333468/6Y976JUQ\"></cite>\n# \n# $p_\\theta (z|x) \\sim N(\\mu', \\sigma'^2 I)$, where:\n# \n# $\\quad\\quad h' = \\text{MultiHead}(c, E_\\text{out}^L(x), E_\\text{out}^L(x))$\n# \n# $\\quad\\quad \\begin{bmatrix}\\mu'\\\\\\log(\\sigma'^2)\\end{bmatrix} = MLP_p(h')$\n\n# In[4]:\n\n\ndef sample_prior(self, prob_decode=False):\n return\n\n\n# In[5]:\n\n\nfrom utils.data import JTNNDataset, JTNNCollator\ntorch.multiprocessing.set_sharing_strategy('file_system')\nfrom torch.utils.data import DataLoader\nimport torch.optim as optim\nimport torch.optim.lr_scheduler as lr_scheduler\nfrom optparse import OptionParser\n\nclass ArgsTemp():\n def __init__(self, hidden_size, depth, device):\n self.hidden_size = hidden_size\n self.batch_size = 250\n self.latent_size = 56\n self.depth = depth\n self.device = device\n self.lr = 1e-3\n self.beta = 1.0\n self.use_cuda = torch.cuda.is_available()\n \nargs = ArgsTemp(200,3, device=torch.device(\"cuda:0\" if torch.cuda.is_available() else \"cpu\"))\nprint(args.depth)\n\nparser = OptionParser()\nparser.add_option(\"-t\", \"--train\", dest=\"train_path\")\nparser.add_option(\"-v\", \"--vocab\", dest=\"vocab_path\")\nparser.add_option(\"-s\", \"--save_dir\", dest=\"save_path\")\nparser.add_option(\"-m\", \"--model\", dest=\"model_path\", default=None)\nparser.add_option(\"-b\", \"--batch\", dest=\"batch_size\", default=40)\nparser.add_option(\"-w\", \"--hidden\", dest=\"hidden_size\", default=200)\nparser.add_option(\"-l\", \"--latent\", dest=\"latent_size\", default=56)\nparser.add_option(\"-d\", \"--depth\", dest=\"depth\", default=3)\nparser.add_option(\"-z\", \"--beta\", dest=\"beta\", default=1.0)\nparser.add_option(\"-q\", \"--lr\", dest=\"lr\", default=1e-3)\nparser.add_option(\"-e\", \"--stereo\", dest=\"stereo\", default=1)\nopts,_ = parser.parse_args()\n\ndataset = JTNNDataset(data='train', vocab='vocab', training=True, intermediates=False)\nvocab = dataset.vocab\n\"\"\"\ndataloader = DataLoader(\n dataset,\n batch_size=args.batch_size,\n shuffle=True,\n num_workers=1,\n collate_fn=JTNNCollator(vocab, True, intermediates=False),\n drop_last=True,\n worker_init_fn=None)\n \"\"\"\n\nmodel = DGLJTNNVAE(vocab, args.hidden_size, args.latent_size, args.depth, args).cuda()\n\nif opts.model_path is not None:\n model.load_state_dict(torch.load(opts.model_path))\nelse:\n for param in model.parameters():\n if param.dim() == 1:\n nn.init.constant(param, 0)\n else:\n nn.init.xavier_normal(param)\n\nmodel.share_memory()\n#if torch.cuda.device_count() > 1:\n #print(\"Let's use\", torch.cuda.device_count(), \"GPUs!\")\n # dim = 0 [30, xxx] -> [10, ...], [10, ...], [10, ...] on 3 GPUs\n #model = nn.DataParallel(model)\noptimizer = optim.Adam(model.parameters(), lr=args.lr)\nscheduler = lr_scheduler.ExponentialLR(optimizer, 0.9)\n\n\n# In[6]:\n\n\nMAX_EPOCH = 50\nPRINT_ITER = 20\n\nfrom tqdm import tqdm\nfrom os import access, R_OK\nfrom os.path import isdir\nimport sys\n\nsave_path = '/home/icarus/app/data/05_model_output'\nassert isdir(save_path) and access(save_path, R_OK), \"File {} doesn't exist or isn't readable\".format(save_path)\n\ndef train():\n dataset.training = True\n print(\"Loading data...\")\n dataloader = DataLoader(\n dataset,\n batch_size=args.batch_size,\n shuffle=True,\n pin_memory=True,\n num_workers=12,\n collate_fn=JTNNCollator(vocab, True),\n drop_last=True,\n worker_init_fn=None)\n dataloader._use_shared_memory = False\n last_loss = sys.maxsize\n print(\"Beginning Training...\")\n for epoch in range(MAX_EPOCH):\n word_acc,topo_acc,assm_acc,steo_acc = 0,0,0,0\n print(\"Epoch %d: \" % epoch)\n\n for it, batch in tqdm(enumerate(dataloader),total=len(dataloader)):\n model.zero_grad()\n try:\n loss, kl_div, wacc, tacc, sacc, dacc = model(batch, args.beta)\n except:\n print([t.smiles for t in batch['mol_trees']])\n raise\n loss.backward()\n optimizer.step()\n\n word_acc += wacc\n topo_acc += tacc\n assm_acc += sacc\n steo_acc += dacc\n\n cur_loss = loss.item()\n \n if (it + 1) % PRINT_ITER == 0:\n word_acc = word_acc / PRINT_ITER * 100\n topo_acc = topo_acc / PRINT_ITER * 100\n assm_acc = assm_acc / PRINT_ITER * 100\n steo_acc = steo_acc / PRINT_ITER * 100\n\n print(\"KL: %.1f, Word: %.2f, Topo: %.2f, Assm: %.2f, Steo: %.2f, Loss: %.6f, Delta: %.6f\" % (\n kl_div, word_acc, topo_acc, assm_acc, steo_acc, cur_loss, last_loss - cur_loss))\n word_acc,topo_acc,assm_acc,steo_acc = 0,0,0,0\n sys.stdout.flush()\n\n if (it + 1) % 1500 == 0: #Fast annealing\n scheduler.step()\n print(\"learning rate: %.6f\" % scheduler.get_lr()[0])\n \n if (it + 1) % 100 == 0:\n torch.save(model.state_dict(),\n save_path + \"/model.iter-%d-%d\" % (epoch, it + 1))\n \n #if last_loss - cur_loss < 1e-5:\n # break\n last_loss = cur_loss\n\n scheduler.step()\n print(\"learning rate: %.6f\" % scheduler.get_lr()[0])\n torch.save(model.state_dict(), save_path + \"/model.iter-\" + str(epoch))\n\n\n# In[ ]:\n\n\n\n\n\n# In[ ]:\n\n\n\n\n\n# In[ ]:\n\n\nimport warnings; warnings.simplefilter('ignore')\n#train() \n\n\n# ## References\n# \n# <div class=\"cite2c-biblio\"></div>\n\n# In[ ]:\n\n\ndef test():\n dataset.training = False\n dataloader = DataLoader(\n dataset,\n batch_size=1,\n shuffle=False,\n num_workers=0,\n collate_fn=JTNNCollator(vocab, False),\n drop_last=True,\n worker_init_fn=None)#worker_init_fn)\n\n # Just an example of molecule decoding; in reality you may want to sample\n # tree and molecule vectors.\n for it, batch in enumerate(dataloader):\n #print(batch['mol_trees'])\n gt_smiles = batch['mol_trees'][0].smiles\n #print(gt_smiles)\n model.move_to_cuda(batch)\n _, tree_vec, mol_vec = model.encode(batch)\n tree_vec, mol_vec, _, _ = model.sample(tree_vec, mol_vec)\n smiles = model.decode(tree_vec, mol_vec)\n print(smiles)\n\n\n# In[ ]:\n\n\n#torch.cuda.empty_cache()\ntest()\n\n\n# In[ ]:\n\n\n\n\n\n# In[ ]:\n\n\n\n\n"
] |
[
[
"torch.nn.Linear",
"torch.nn.init.constant",
"torch.nn.init.xavier_normal",
"numpy.random.seed",
"torch.optim.lr_scheduler.ExponentialLR",
"torch.manual_seed",
"torch.cuda.is_available",
"torch.load",
"torch.multiprocessing.set_sharing_strategy"
]
] |
neosavvy/serenity
|
[
"c7c8f4d2f48437cd33e0f6b135455f008031b19f"
] |
[
"src/serenity/equity/batch/utils.py"
] |
[
"import datetime\nimport hashlib\nimport logging\nfrom abc import ABC, abstractmethod\n\n\nimport luigi\nimport pandas as pd\nimport quandl\nfrom luigi import Target, LocalTarget\nfrom sqlalchemy.orm import Session\n\nfrom serenity.equity.sharadar_api import create_sharadar_session, BatchStatus\n\n\nclass ExportQuandlTableTask(luigi.Task):\n logger = logging.getLogger('luigi-interface')\n\n table_name = luigi.Parameter()\n date_column = luigi.Parameter(default='')\n start_date = luigi.DateParameter(default=None)\n end_date = luigi.DateParameter(default=None)\n api_key = luigi.configuration.get_config().get('quandl', 'api_key', None)\n data_dir = luigi.configuration.get_config().get('storage', 'storage_root', None)\n\n def output(self):\n if self.start_date is None and self.end_date is None:\n return luigi.LocalTarget(f'{self.data_dir}/{str(self.table_name).replace(\"/\", \"-\")}_quandl_data.zip')\n else:\n end_date_txt = str(self.end_date)\n if self.end_date is None:\n end_date_txt = 'LATEST'\n date_range = f'{self.start_date}-{end_date_txt}'\n return luigi.LocalTarget(f'{self.data_dir}/{str(self.table_name).replace(\"/\", \"-\")}_'\n f'{date_range}_quandl_data.zip')\n\n def run(self):\n quandl.ApiConfig.api_key = str(self.api_key)\n with self.output().temporary_path() as path:\n self.logger.info(f'downloading {self.table_name} to {path}')\n if self.date_column == '':\n kwargs = {\n 'filename': path\n }\n else:\n kwargs = {\n 'filename': path,\n str(self.date_column): {'gte': self.start_date, 'lte': self.end_date}\n }\n quandl.export_table(str(self.table_name), **kwargs)\n\n\nclass BatchStatusTarget(Target):\n def __init__(self, session: Session, workflow_name: str, start_date: datetime.date, end_date: datetime.date,\n local_file: LocalTarget):\n self.session = session\n self.workflow_name = workflow_name\n self.start_date = start_date\n self.end_date = end_date\n self.local_file = local_file\n\n def exists(self):\n if not self.local_file.exists():\n return False\n\n md5_checksum = hashlib.md5(open(self.local_file.path, 'rb').read()).hexdigest()\n batch_status = BatchStatus.find(self.session, self.workflow_name, self.start_date, self.end_date)\n if batch_status is None:\n batch_status = BatchStatus(workflow_name=self.workflow_name,\n start_date=self.start_date,\n end_date=self.end_date,\n md5_checksum=md5_checksum,\n is_pending=True)\n self.session.add(batch_status)\n exists = False\n else:\n exists = (not batch_status.is_pending) and (batch_status.md5_checksum == md5_checksum)\n if batch_status.md5_checksum != md5_checksum:\n batch_status.md5_checksum = md5_checksum\n self.session.add(batch_status)\n return exists\n\n def done(self):\n batch_status = BatchStatus.find(self.session, self.workflow_name, self.start_date, self.end_date)\n if batch_status is not None and batch_status.is_pending:\n batch_status.is_pending = False\n self.session.add(batch_status)\n elif batch_status is None:\n md5_checksum = hashlib.md5(open(self.local_file.path, 'rb').read()).hexdigest()\n batch_status = BatchStatus(workflow_name=self.workflow_name,\n start_date=self.start_date,\n end_date=self.end_date,\n md5_checksum=md5_checksum,\n is_pending=False)\n self.session.add(batch_status)\n self.session.commit()\n\n\nclass LoadSharadarTableTask(ABC, luigi.Task):\n logger = logging.getLogger('luigi-interface')\n session = create_sharadar_session()\n\n start_date = luigi.DateParameter(default=datetime.date.today())\n end_date = luigi.DateParameter(default=datetime.date.today())\n\n def run(self):\n input_value = self.input()\n input_value = [input_value] if type(input_value) is LocalTarget else input_value\n for table_in in input_value:\n if isinstance(table_in, LocalTarget):\n in_file = table_in.path\n df = pd.read_csv(in_file)\n self.logger.info(f'loaded {len(df)} rows of CSV data from {in_file}')\n\n row_count = 0\n for index, row in df.iterrows():\n self.process_row(index, row)\n if row_count > 0 and row_count % 1000 == 0:\n self.logger.info(f'{row_count} rows loaded; flushing next 1000 rows to database')\n self.session.commit()\n row_count += 1\n\n self.session.commit()\n\n self.output().done()\n\n def output(self):\n target = None\n input_value = self.input()\n input_value = [input_value] if type(input_value) is LocalTarget else input_value\n for table_in in input_value:\n if isinstance(table_in, LocalTarget):\n # noinspection PyTypeChecker\n target = BatchStatusTarget(self.session, self.get_workflow_name(),\n self.start_date, self.end_date, table_in)\n\n return target\n\n @abstractmethod\n def process_row(self, index, row):\n pass\n\n @abstractmethod\n def get_workflow_name(self):\n pass\n"
] |
[
[
"pandas.read_csv"
]
] |
suntao2012/draw
|
[
"502452fb66eba68a00526cbd3c3746c57786c150"
] |
[
"draw.py"
] |
[
"#!/usr/bin/env python\n\n\"\"\"\"\nSimple implementation of http://arxiv.org/pdf/1502.04623v2.pdf in TensorFlow\n\nExample Usage: \n\tpython draw.py --data_dir=/tmp/draw --read_attn=True --write_attn=True\n\nAuthor: Eric Jang\n\"\"\"\n\nimport tensorflow as tf\nfrom tensorflow.examples.tutorials import mnist\nimport numpy as np\nimport os\nimport time\n\ntf.flags.DEFINE_string(\"data_dir\", \"\", \"\")\ntf.flags.DEFINE_boolean(\"read_attn\", True, \"enable attention for reader\")\ntf.flags.DEFINE_boolean(\"write_attn\",True, \"enable attention for writer\")\nFLAGS = tf.flags.FLAGS\n\n## MODEL PARAMETERS ## \n\nA,B = 28,28 # image width,height\nimg_size = B*A # the canvas size\nenc_size = 256 # number of hidden units / output size in LSTM\ndec_size = 256\nread_n = 5 # read glimpse grid width/height\nwrite_n = 5 # write glimpse grid width/height\nread_size = 2*read_n*read_n if FLAGS.read_attn else 2*img_size\nwrite_size = write_n*write_n if FLAGS.write_attn else img_size\nz_size=10 # QSampler output size\nT=10 # MNIST generation sequence length\nbatch_size=128 # training minibatch size\ntrain_iters=10000\nlearning_rate=1e-3 # learning rate for optimizer\neps=1e-8 # epsilon for numerical stability\n\n## BUILD MODEL ## \n\nDO_SHARE=None # workaround for variable_scope(reuse=True)\n\nx = tf.placeholder(tf.float32,shape=(batch_size,img_size)) # input (batch_size * img_size)\ne=tf.random_normal((batch_size,z_size), mean=0, stddev=1) # Qsampler noise\nlstm_enc = tf.contrib.rnn.LSTMCell(enc_size, state_is_tuple=True) # encoder Op\nlstm_dec = tf.contrib.rnn.LSTMCell(dec_size, state_is_tuple=True) # decoder Op\n\ndef linear(x,output_dim):\n \"\"\"\n affine transformation Wx+b\n assumes x.shape = (batch_size, num_features)\n \"\"\"\n w=tf.get_variable(\"w\", [x.get_shape()[1], output_dim]) \n b=tf.get_variable(\"b\", [output_dim], initializer=tf.constant_initializer(0.0))\n return tf.matmul(x,w)+b\n\ndef filterbank(gx, gy, sigma2,delta, N):\n grid_i = tf.reshape(tf.cast(tf.range(N), tf.float32), [1, -1])\n mu_x = gx + (grid_i - N / 2 - 0.5) * delta # eq 19\n mu_y = gy + (grid_i - N / 2 - 0.5) * delta # eq 20\n a = tf.reshape(tf.cast(tf.range(A), tf.float32), [1, 1, -1])\n b = tf.reshape(tf.cast(tf.range(B), tf.float32), [1, 1, -1])\n mu_x = tf.reshape(mu_x, [-1, N, 1])\n mu_y = tf.reshape(mu_y, [-1, N, 1])\n sigma2 = tf.reshape(sigma2, [-1, 1, 1])\n Fx = tf.exp(-tf.square(a - mu_x) / (2*sigma2))\n Fy = tf.exp(-tf.square(b - mu_y) / (2*sigma2)) # batch x N x B\n # normalize, sum over A and B dims\n Fx=Fx/tf.maximum(tf.reduce_sum(Fx,2,keep_dims=True),eps)\n Fy=Fy/tf.maximum(tf.reduce_sum(Fy,2,keep_dims=True),eps)\n return Fx,Fy\n\ndef attn_window(scope,h_dec,N):\n with tf.variable_scope(scope,reuse=DO_SHARE):\n params=linear(h_dec,5)\n # gx_,gy_,log_sigma2,log_delta,log_gamma=tf.split(1,5,params)\n gx_,gy_,log_sigma2,log_delta,log_gamma=tf.split(params,5,1)\n gx=(A+1)/2*(gx_+1)\n gy=(B+1)/2*(gy_+1)\n sigma2=tf.exp(log_sigma2)\n delta=(max(A,B)-1)/(N-1)*tf.exp(log_delta) # batch x N\n return filterbank(gx,gy,sigma2,delta,N)+(tf.exp(log_gamma),)\n\n## READ ## \ndef read_no_attn(x,x_hat,h_dec_prev):\n return tf.concat([x,x_hat], 1)\n\ndef read_attn(x,x_hat,h_dec_prev):\n Fx,Fy,gamma=attn_window(\"read\",h_dec_prev,read_n)\n def filter_img(img,Fx,Fy,gamma,N):\n Fxt=tf.transpose(Fx,perm=[0,2,1])\n img=tf.reshape(img,[-1,B,A])\n glimpse=tf.matmul(Fy,tf.matmul(img,Fxt))\n glimpse=tf.reshape(glimpse,[-1,N*N])\n return glimpse*tf.reshape(gamma,[-1,1])\n x=filter_img(x,Fx,Fy,gamma,read_n) # batch x (read_n*read_n)\n x_hat=filter_img(x_hat,Fx,Fy,gamma,read_n)\n return tf.concat([x,x_hat], 1) # concat along feature axis\n\nread = read_attn if FLAGS.read_attn else read_no_attn\n\n## ENCODE ## \ndef encode(state,input):\n \"\"\"\n run LSTM\n state = previous encoder state\n input = cat(read,h_dec_prev)\n returns: (output, new_state)\n \"\"\"\n with tf.variable_scope(\"encoder\",reuse=DO_SHARE):\n return lstm_enc(input,state)\n\n## Q-SAMPLER (VARIATIONAL AUTOENCODER) ##\n\ndef sampleQ(h_enc):\n \"\"\"\n Samples Zt ~ normrnd(mu,sigma) via reparameterization trick for normal dist\n mu is (batch,z_size)\n \"\"\"\n with tf.variable_scope(\"mu\",reuse=DO_SHARE):\n mu=linear(h_enc,z_size)\n with tf.variable_scope(\"sigma\",reuse=DO_SHARE):\n logsigma=linear(h_enc,z_size)\n sigma=tf.exp(logsigma)\n return (mu + sigma*e, mu, logsigma, sigma)\n\n## DECODER ## \ndef decode(state,input):\n with tf.variable_scope(\"decoder\",reuse=DO_SHARE):\n return lstm_dec(input, state)\n\n## WRITER ## \ndef write_no_attn(h_dec):\n with tf.variable_scope(\"write\",reuse=DO_SHARE):\n return linear(h_dec,img_size)\n\ndef write_attn(h_dec):\n with tf.variable_scope(\"writeW\",reuse=DO_SHARE):\n w=linear(h_dec,write_size) # batch x (write_n*write_n)\n N=write_n\n w=tf.reshape(w,[batch_size,N,N])\n Fx,Fy,gamma=attn_window(\"write\",h_dec,write_n)\n Fyt=tf.transpose(Fy,perm=[0,2,1])\n wr=tf.matmul(Fyt,tf.matmul(w,Fx))\n wr=tf.reshape(wr,[batch_size,B*A])\n #gamma=tf.tile(gamma,[1,B*A])\n return wr*tf.reshape(1.0/gamma,[-1,1])\n\nwrite=write_attn if FLAGS.write_attn else write_no_attn\n\n## STATE VARIABLES ## \n\ncs=[0]*T # sequence of canvases\nmus,logsigmas,sigmas=[0]*T,[0]*T,[0]*T # gaussian params generated by SampleQ. We will need these for computing loss.\n# initial states\nh_dec_prev=tf.zeros((batch_size,dec_size))\nenc_state=lstm_enc.zero_state(batch_size, tf.float32)\ndec_state=lstm_dec.zero_state(batch_size, tf.float32)\n\n## DRAW MODEL ## \n\n# construct the unrolled computational graph\nfor t in range(T):\n c_prev = tf.zeros((batch_size,img_size)) if t==0 else cs[t-1]\n x_hat=x-tf.sigmoid(c_prev) # error image\n r=read(x,x_hat,h_dec_prev)\n h_enc,enc_state=encode(enc_state,tf.concat([r,h_dec_prev], 1))\n z,mus[t],logsigmas[t],sigmas[t]=sampleQ(h_enc)\n h_dec,dec_state=decode(dec_state,z)\n cs[t]=c_prev+write(h_dec) # store results\n h_dec_prev=h_dec\n DO_SHARE=True # from now on, share variables\n\n## LOSS FUNCTION ## \n\ndef binary_crossentropy(t,o):\n return -(t*tf.log(o+eps) + (1.0-t)*tf.log(1.0-o+eps))\n\n# reconstruction term appears to have been collapsed down to a single scalar value (rather than one per item in minibatch)\nx_recons=tf.nn.sigmoid(cs[-1])\n\n# after computing binary cross entropy, sum across features then take the mean of those sums across minibatches\nLx=tf.reduce_sum(binary_crossentropy(x,x_recons),1) # reconstruction term\nLx=tf.reduce_mean(Lx)\n\nkl_terms=[0]*T\nfor t in range(T):\n mu2=tf.square(mus[t])\n sigma2=tf.square(sigmas[t])\n logsigma=logsigmas[t]\n kl_terms[t]=0.5*tf.reduce_sum(mu2+sigma2-2*logsigma,1)-.5 # each kl term is (1xminibatch)\nKL=tf.add_n(kl_terms) # this is 1xminibatch, corresponding to summing kl_terms from 1:T\nLz=tf.reduce_mean(KL) # average over minibatches\n\ncost=Lx+Lz\n\n## OPTIMIZER ## \n\noptimizer=tf.train.AdamOptimizer(learning_rate, beta1=0.5)\ngrads=optimizer.compute_gradients(cost)\nfor i,(g,v) in enumerate(grads):\n if g is not None:\n grads[i]=(tf.clip_by_norm(g,5),v) # clip gradients\ntrain_op=optimizer.apply_gradients(grads)\n\n## RUN TRAINING ## \n\ndata_directory = os.path.join(FLAGS.data_dir, \"mnist\")\nif not os.path.exists(data_directory):\n\tos.makedirs(data_directory)\ntrain_data = mnist.input_data.read_data_sets(data_directory, one_hot=True).train # binarized (0-1) mnist data\n\nfetches=[]\n#fetches.extend([Lx,Lz,train_op])\nfetches.extend([Lx,Lz,cost])\nLxs=[0]*train_iters\nLzs=[0]*train_iters\n\nsess=tf.InteractiveSession()\n\nsaver = tf.train.Saver() # saves variables learned during training\ntf.global_variables_initializer().run()\n#saver.restore(sess, \"/tmp/draw/drawmodel.ckpt\") # to restore from model, uncomment this line\n\nfor i in range(train_iters):\n\txtrain,_=train_data.next_batch(batch_size) # xtrain is (batch_size x img_size)\n\tfeed_dict={x:xtrain}\n\n\tstart_time = time.time()\n\tfor j in range(100):\n\t\tresults=sess.run(fetches,feed_dict)\n\tduration = time.time() - start_time\n\t\n\tLxs[i],Lzs[i],_=results\n\t#if i%100==0:\n\tprint(\"iter=%d : Lx: %f Lz: %f, duration: %f, time per batch: %f, batchsize: %d\" % (100,Lxs[i],Lzs[i], duration, duration/100, batch_size))\n\n## TRAINING FINISHED ## \n\ncanvases=sess.run(cs,feed_dict) # generate some examples\ncanvases=np.array(canvases) # T x batch x img_size\n\nout_file=os.path.join(FLAGS.data_dir,\"draw_data.npy\")\nnp.save(out_file,[canvases,Lxs,Lzs])\nprint(\"Outputs saved in file: %s\" % out_file)\n\nckpt_file=os.path.join(FLAGS.data_dir,\"drawmodel.ckpt\")\nprint(\"Model saved in file: %s\" % saver.save(sess,ckpt_file))\n\nsess.close()\n"
] |
[
[
"tensorflow.exp",
"tensorflow.constant_initializer",
"tensorflow.matmul",
"tensorflow.reshape",
"tensorflow.clip_by_norm",
"tensorflow.global_variables_initializer",
"tensorflow.random_normal",
"tensorflow.InteractiveSession",
"tensorflow.flags.DEFINE_string",
"tensorflow.concat",
"tensorflow.sigmoid",
"tensorflow.train.Saver",
"numpy.save",
"tensorflow.add_n",
"tensorflow.transpose",
"tensorflow.variable_scope",
"tensorflow.split",
"tensorflow.nn.sigmoid",
"numpy.array",
"tensorflow.contrib.rnn.LSTMCell",
"tensorflow.zeros",
"tensorflow.train.AdamOptimizer",
"tensorflow.range",
"tensorflow.log",
"tensorflow.placeholder",
"tensorflow.reduce_sum",
"tensorflow.flags.DEFINE_boolean",
"tensorflow.examples.tutorials.mnist.input_data.read_data_sets",
"tensorflow.reduce_mean",
"tensorflow.square"
]
] |
JadeMaveric/transformers
|
[
"b6a28e9ac916523165289be57e6214a65247f051"
] |
[
"src/transformers/models/prophetnet/modeling_prophetnet.py"
] |
[
"# coding=utf-8\n# Copyright 2020 The Microsoft Authors and The HuggingFace Inc. team.\n#\n# Licensed under the Apache License, Version 2.0 (the \"License\");\n# you may not use this file except in compliance with the License.\n# You may obtain a copy of the License at\n#\n# http://www.apache.org/licenses/LICENSE-2.0\n#\n# Unless required by applicable law or agreed to in writing, software\n# distributed under the License is distributed on an \"AS IS\" BASIS,\n# WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.\n# See the License for the specific language governing permissions and\n# limitations under the License.\n\"\"\" PyTorch ProphetNet model, ported from ProphetNet repo(fairsequery_states version). \"\"\"\n\nimport copy\nimport math\nimport warnings\nfrom dataclasses import dataclass\nfrom typing import Optional, Tuple\n\nimport torch\nimport torch.nn.functional as F\nfrom torch import Tensor, nn\nfrom torch.nn import LayerNorm\n\nfrom ...activations import ACT2FN\nfrom ...file_utils import (\n ModelOutput,\n add_start_docstrings,\n add_start_docstrings_to_model_forward,\n replace_return_docstrings,\n)\nfrom ...modeling_outputs import BaseModelOutput\nfrom ...modeling_utils import PreTrainedModel\nfrom ...utils import logging\nfrom .configuration_prophetnet import ProphetNetConfig\n\n\nlogger = logging.get_logger(__name__)\n\n_CONFIG_FOR_DOC = \"ProphenetConfig\"\n_TOKENIZER_FOR_DOC = \"ProphetNetTokenizer\"\n\nPROPHETNET_PRETRAINED_MODEL_ARCHIVE_LIST = [\n \"microsoft/prophetnet-large-uncased\",\n # See all ProphetNet models at https://huggingface.co/models?filter=prophetnet\n]\n\n\nPROPHETNET_START_DOCSTRING = r\"\"\"\n This model inherits from :class:`~transformers.PreTrainedModel`. Check the superclass documentation for the generic\n methods the library implements for all its model (such as downloading or saving, resizing the input embeddings,\n pruning heads etc.)\n\n Original ProphetNet code can be found at <https://github.com/microsoft/ProphetNet> . Checkpoints were converted\n from original Fairseq checkpoints. For more information on the checkpoint conversion, please take a look at the\n file ``convert_prophetnet_original_pytorch_checkpoint_to_pytorch.py``.\n\n This model is a PyTorch `torch.nn.Module <https://pytorch.org/docs/stable/nn.html#torch.nn.Module>`_ sub-class. Use\n it as a regular PyTorch Module and refer to the PyTorch documentation for all matters related to general usage and\n behavior.\n\n Parameters:\n config (:class:`~transformers.ProphetNetConfig`): Model configuration class with all the parameters of the model.\n Initializing with a config file does not load the weights associated with the model, only the\n configuration. Check out the :meth:`~transformers.PreTrainedModel.from_pretrained` method to load the model\n weights.\n\"\"\"\n\nPROPHETNET_INPUTS_DOCSTRING = r\"\"\"\n Args:\n input_ids (:obj:`torch.LongTensor` of shape :obj:`(batch_size, sequence_length)`):\n Indices of input sequence tokens in the vocabulary. Padding will be ignored by default should you provide\n it.\n\n Indices can be obtained using :class:`~transformers.ProphetNetTokenizer`. See\n :meth:`transformers.PreTrainedTokenizer.encode` and :meth:`transformers.PreTrainedTokenizer.__call__` for\n details.\n\n `What are input IDs? <../glossary.html#input-ids>`__\n attention_mask (:obj:`torch.Tensor` of shape :obj:`(batch_size, sequence_length)`, `optional`):\n Mask to avoid performing attention on padding token indices. Mask values selected in ``[0, 1]``:\n\n - 1 for tokens that are **not masked**,\n - 0 for tokens that are **masked**.\n\n `What are attention masks? <../glossary.html#attention-mask>`__\n decoder_input_ids (:obj:`torch.LongTensor` of shape :obj:`(batch_size, target_sequence_length)`, `optional`):\n Indices of decoder input sequence tokens in the vocabulary.\n\n Indices can be obtained using :class:`~transformers.PreTrainedTokenizer`. See\n :meth:`transformers.PreTrainedTokenizer.encode` and :meth:`transformers.PreTrainedTokenizer.__call__` for\n details.\n\n `What are input IDs? <../glossary.html#input-ids>`__\n\n ProphetNet uses the :obj:`eos_token_id` as the starting token for :obj:`decoder_input_ids` generation. If\n :obj:`past_key_values` is used, optionally only the last :obj:`decoder_input_ids` have to be input (see\n :obj:`past_key_values`).\n\n decoder_attention_mask (:obj:`torch.BoolTensor` of shape :obj:`(batch_size, target_sequence_length)`, `optional`):\n Default behavior: generate a tensor that ignores pad tokens in :obj:`decoder_input_ids`. Causal mask will\n also be used by default.\n\n If you want to change padding behavior, you should read :func:`modeling_bart._prepare_decoder_inputs` and\n modify to your needs. See diagram 1 in `the paper <https://arxiv.org/abs/1910.13461>`__ for more\n information on the default strategy.\n encoder_outputs (:obj:`tuple(tuple(torch.FloatTensor)`, `optional`):\n Tuple consists of (:obj:`last_hidden_state`, `optional`: :obj:`hidden_states`, `optional`:\n :obj:`attentions`) :obj:`last_hidden_state` of shape :obj:`(batch_size, sequence_length, hidden_size)`,\n `optional`) is a sequence of hidden-states at the output of the last layer of the encoder. Used in the\n cross-attention of the decoder.\n past_key_values (:obj:`tuple(tuple(torch.FloatTensor))` of length :obj:`config.n_layers` with each tuple having 4 tensors of shape :obj:`(batch_size, num_heads, sequence_length - 1, embed_size_per_head)`):\n Contains precomputed key and value hidden-states of the attention blocks. Can be used to speed up decoding.\n\n If :obj:`past_key_values` are used, the user can optionally input only the last ``decoder_input_ids``\n (those that don't have their past key value states given to this model) of shape :obj:`(batch_size, 1)`\n instead of all ``decoder_input_ids`` of shape :obj:`(batch_size, sequence_length)`.\n use_cache (:obj:`bool`, `optional`):\n If set to :obj:`True`, :obj:`past_key_values` key value states are returned and can be used to speed up\n decoding (see :obj:`past_key_values`).\n output_attentions (:obj:`bool`, `optional`):\n Whether or not to return the attentions tensors of all attention layers. See ``attentions`` under returned\n tensors for more detail.\n output_hidden_states (:obj:`bool`, `optional`):\n Whether or not to return the hidden states of all layers. See ``hidden_states`` under returned tensors for\n more detail.\n return_dict (:obj:`bool`, `optional`):\n Whether or not to return a :class:`~transformers.file_utils.ModelOutput` instead of a plain tuple.\n\"\"\"\n\nPROPHETNET_STANDALONE_INPUTS_DOCSTRING = r\"\"\"\n Args:\n input_ids (:obj:`torch.LongTensor` of shape :obj:`(batch_size, sequence_length)`):\n Indices of input sequence tokens in the vocabulary. Padding will be ignored by default should you provide\n it.\n\n Indices can be obtained using :class:`~transformers.ProphetNetTokenizer`. See\n :meth:`transformers.PreTrainedTokenizer.encode` and :meth:`transformers.PreTrainedTokenizer.__call__` for\n details.\n\n `What are input IDs? <../glossary.html#input-ids>`__\n attention_mask (:obj:`torch.Tensor` of shape :obj:`(batch_size, sequence_length)`, `optional`):\n Mask to avoid performing attention on padding token indices. Mask values selected in ``[0, 1]``:\n\n - 1 for tokens that are **not masked**,\n - 0 for tokens that are **masked**.\n\n `What are attention masks? <../glossary.html#attention-mask>`__\n output_attentions (:obj:`bool`, `optional`):\n Whether or not to return the attentions tensors of all attention layers. See ``attentions`` under returned\n tensors for more detail.\n output_hidden_states (:obj:`bool`, `optional`):\n Whether or not to return the hidden states of all layers. See ``hidden_states`` under returned tensors for\n more detail.\n return_dict (:obj:`bool`, `optional`):\n Whether or not to return a :class:`~transformers.file_utils.ModelOutput` instead of a plain tuple.\n\"\"\"\n\n\ndef softmax(hidden_state, dim, onnx_trace=False):\n if onnx_trace:\n return F.softmax(hidden_state.float(), dim=dim)\n else:\n return F.softmax(hidden_state, dim=dim, dtype=torch.float32)\n\n\ndef ngram_attention_bias(sequence_length, ngram, device, dtype):\n \"\"\"\n This function computes the bias for the predict stream\n \"\"\"\n left_block = torch.ones((ngram, sequence_length, sequence_length), device=device, dtype=dtype) * float(\"-inf\")\n right_block = left_block.detach().clone()\n # create bias\n for stream_idx in range(ngram):\n right_block[stream_idx].fill_diagonal_(0, wrap=False)\n left_block[stream_idx].triu_(-stream_idx + 1)\n\n left_block[:, :, 0] = 0\n return torch.cat([left_block, right_block], dim=2)\n\n\ndef compute_relative_buckets(num_buckets, max_distance, relative_positions, is_bidirectional=False):\n \"\"\"\n This function computes individual parts of the relative position buckets. For more detail, see paper.\n \"\"\"\n inv_relative_positions = -relative_positions\n rel_positions_bucket = 0\n\n if is_bidirectional:\n num_buckets = num_buckets // 2\n rel_positions_bucket = (\n rel_positions_bucket\n + torch.lt(inv_relative_positions, torch.zeros_like(inv_relative_positions)).int() * num_buckets\n )\n inv_relative_positions = torch.abs(inv_relative_positions)\n else:\n inv_relative_positions = torch.max(inv_relative_positions, torch.zeros_like(inv_relative_positions))\n\n max_exact = num_buckets // 2\n is_small = torch.lt(inv_relative_positions, max_exact)\n val_if_large = max_exact + torch.log(inv_relative_positions.float() / max_exact) / math.log(\n max_distance / max_exact\n ) * (num_buckets - max_exact)\n val_if_large = torch.min(val_if_large, torch.ones_like(val_if_large) * (num_buckets - 1)).int()\n rel_positions_bucket = rel_positions_bucket + torch.where(is_small, inv_relative_positions.int(), val_if_large)\n return rel_positions_bucket\n\n\ndef compute_all_stream_relative_buckets(num_buckets, max_distance, position_ids):\n \"\"\"\n This function computes both main and predict relative position buckets. For more detail, see paper.\n \"\"\"\n # main stream\n main_stream_relative_positions = position_ids.unsqueeze(1).repeat(1, position_ids.size(-1), 1)\n main_stream_relative_positions = main_stream_relative_positions - position_ids.unsqueeze(-1)\n\n # predicting stream\n predicting_stream_relative_positions = torch.cat((position_ids - 1, position_ids), dim=-1).unsqueeze(1)\n predicting_stream_relative_positions = predicting_stream_relative_positions.repeat(1, position_ids.size(-1), 1)\n predicting_stream_relative_positions = predicting_stream_relative_positions - position_ids.unsqueeze(-1)\n\n # get both position buckets\n main_relative_position_buckets = compute_relative_buckets(\n num_buckets, max_distance, main_stream_relative_positions, is_bidirectional=False\n )\n predict_relative_position_buckets = compute_relative_buckets(\n num_buckets, max_distance, predicting_stream_relative_positions, is_bidirectional=False\n )\n return main_relative_position_buckets, predict_relative_position_buckets\n\n\n@dataclass\nclass ProphetNetSeq2SeqLMOutput(ModelOutput):\n \"\"\"\n Base class for sequence-to-sequence language models outputs.\n\n Args:\n loss (:obj:`torch.FloatTensor` of shape :obj:`(1,)`, `optional`, returned when :obj:`labels` is provided):\n Language modeling loss.\n logits (:obj:`torch.FloatTensor` of shape :obj:`(batch_size, decoder_sequence_length, config.vocab_size)`):\n Prediction scores of the main stream language modeling head (scores for each vocabulary token before\n SoftMax).\n logits_ngram (:obj:`torch.FloatTensor` of shape :obj:`(batch_size, ngram * decoder_sequence_length, config.vocab_size)`):\n Prediction scores of the predict stream language modeling head (scores for each vocabulary token before\n SoftMax).\n past_key_values (:obj:`List[torch.FloatTensor]`, `optional`, returned when ``use_cache=True`` is passed or when ``config.use_cache=True``):\n List of :obj:`torch.FloatTensor` of length :obj:`config.n_layers`, with each tensor of shape :obj:`(2,\n batch_size, num_attn_heads, decoder_sequence_length, embed_size_per_head)`).\n\n Contains pre-computed hidden-states (key and values in the attention blocks) of the decoder that can be\n used (see :obj:`past_key_values` input) to speed up sequential decoding.\n decoder_hidden_states (:obj:`tuple(torch.FloatTensor)`, `optional`, returned when ``output_hidden_states=True`` is passed or when ``config.output_hidden_states=True``):\n Tuple of :obj:`torch.FloatTensor` (one for the output of the embeddings + one for the output of each layer)\n of shape :obj:`(batch_size, decoder_sequence_length, hidden_size)`.\n\n Hidden-states of main stream of the decoder at the output of each layer plus the initial embedding outputs.\n decoder_ngram_hidden_states (:obj:`tuple(torch.FloatTensor)`, `optional`, returned when ``output_hidden_states=True`` is passed or when ``config.output_hidden_states=True``):\n Tuple of :obj:`torch.FloatTensor` (one for the output of the embeddings + one for the output of each layer)\n of shape :obj:`(batch_size, ngram * decoder_sequence_length, hidden_size)`.\n\n Hidden-states of the predict stream of the decoder at the output of each layer plus the initial embedding\n outputs.\n decoder_attentions (:obj:`tuple(torch.FloatTensor)`, `optional`, returned when ``output_attentions=True`` is passed or when ``config.output_attentions=True``):\n Tuple of :obj:`torch.FloatTensor` (one for each layer) of shape :obj:`(batch_size, num_attn_heads,\n decoder_sequence_length, decoder_sequence_length)`.\n\n Attentions weights of the decoder, after the attention softmax, used to compute the weighted average in the\n self-attention heads.\n decoder_ngram_attentions (:obj:`tuple(torch.FloatTensor)`, `optional`, returned when ``output_attentions=True`` is passed or when ``config.output_attentions=True``):\n Tuple of :obj:`torch.FloatTensor` (one for each layer) of shape :obj:`(batch_size, num_attn_heads,\n decoder_sequence_length, decoder_sequence_length)`.\n\n Attentions weights of the predict stream of the decoder, after the attention softmax, used to compute the\n weighted average in the self-attention heads.\n cross_attentions (:obj:`tuple(torch.FloatTensor)`, `optional`, returned when ``output_attentions=True`` is passed or when ``config.output_attentions=True``):\n Tuple of :obj:`torch.FloatTensor` (one for each layer) of shape :obj:`(batch_size, num_attn_heads,\n encoder_sequence_length, decoder_sequence_length)`.\n\n Attentions weights of the cross-attention layer of the decoder, after the attention softmax, used to\n compute the weighted average in the\n encoder_last_hidden_state (:obj:`torch.FloatTensor` of shape :obj:`(batch_size, encoder_sequence_length, hidden_size)`, `optional`):\n Sequence of hidden-states at the output of the last layer of the encoder of the model.\n encoder_hidden_states (:obj:`tuple(torch.FloatTensor)`, `optional`, returned when ``output_hidden_states=True`` is passed or when ``config.output_hidden_states=True``):\n Tuple of :obj:`torch.FloatTensor` (one for the output of the embeddings + one for the output of each layer)\n of shape :obj:`(batch_size, encoder_sequence_length, hidden_size)`.\n\n Hidden-states of the encoder at the output of each layer plus the initial embedding outputs.\n encoder_attentions (:obj:`tuple(torch.FloatTensor)`, `optional`, returned when ``output_attentions=True`` is passed or when ``config.output_attentions=True``):\n Tuple of :obj:`torch.FloatTensor` (one for each layer) of shape :obj:`(batch_size, num_attn_heads,\n encoder_sequence_length, encoder_sequence_length)`. Attentions weights of the encoder, after the attention\n softmax, used to compute the weighted average in the self-attention heads.\n \"\"\"\n\n loss: Optional[torch.FloatTensor] = None\n logits: torch.FloatTensor = None\n logits_ngram: Optional[torch.FloatTensor] = None\n past_key_values: Optional[Tuple[torch.FloatTensor]] = None\n decoder_hidden_states: Optional[Tuple[torch.FloatTensor]] = None\n decoder_ngram_hidden_states: Optional[Tuple[torch.FloatTensor]] = None\n decoder_attentions: Optional[Tuple[torch.FloatTensor]] = None\n decoder_ngram_attentions: Optional[Tuple[torch.FloatTensor]] = None\n cross_attentions: Optional[Tuple[torch.FloatTensor]] = None\n encoder_last_hidden_state: Optional[torch.FloatTensor] = None\n encoder_hidden_states: Optional[Tuple[torch.FloatTensor]] = None\n encoder_attentions: Optional[Tuple[torch.FloatTensor]] = None\n\n @property\n def decoder_cross_attentions(self):\n warnings.warn(\n \"`decoder_cross_attentions` is deprecated and will be removed soon. Please use `cross_attentions` instead.\",\n FutureWarning,\n )\n return self.cross_attentions\n\n\n@dataclass\nclass ProphetNetSeq2SeqModelOutput(ModelOutput):\n \"\"\"\n Base class for model encoder's outputs that also contains : pre-computed hidden states that can speed up sequential\n decoding.\n\n Args:\n last_hidden_state (:obj:`torch.FloatTensor` of shape :obj:`(batch_size, decoder_sequence_length, hidden_size)`):\n Sequence of main stream hidden-states at the output of the last layer of the decoder of the model.\n\n If :obj:`past_key_values` is used only the last hidden-state of the sequences of shape :obj:`(batch_size,\n 1, hidden_size)` is output.\n last_hidden_state_ngram (:obj:`torch.FloatTensor` of shape :obj:`(batch_size,ngram * decoder_sequence_length, config.vocab_size)`):\n Sequence of predict stream hidden-states at the output of the last layer of the decoder of the model.\n past_key_values (:obj:`List[torch.FloatTensor]`, `optional`, returned when ``use_cache=True`` is passed or when ``config.use_cache=True``):\n List of :obj:`torch.FloatTensor` of length :obj:`config.n_layers`, with each tensor of shape :obj:`(2,\n batch_size, num_attn_heads, decoder_sequence_length, embed_size_per_head)`).\n\n Contains pre-computed hidden-states (key and values in the attention blocks) of the decoder that can be\n used (see :obj:`past_key_values` input) to speed up sequential decoding.\n decoder_hidden_states (:obj:`tuple(torch.FloatTensor)`, `optional`, returned when ``output_hidden_states=True`` is passed or when ``config.output_hidden_states=True``):\n Tuple of :obj:`torch.FloatTensor` (one for the output of the embeddings + one for the output of each layer)\n of shape :obj:`(batch_size, decoder_sequence_length, hidden_size)`.\n\n Hidden-states of main stream of the decoder at the output of each layer plus the initial embedding outputs.\n decoder_ngram_hidden_states (:obj:`tuple(torch.FloatTensor)`, `optional`, returned when ``output_hidden_states=True`` is passed or when ``config.output_hidden_states=True``):\n Tuple of :obj:`torch.FloatTensor` (one for the output of the embeddings + one for the output of each layer)\n of shape :obj:`(batch_size, ngram * decoder_sequence_length, hidden_size)`.\n\n Hidden-states of the predict stream of the decoder at the output of each layer plus the initial embedding\n outputs.\n decoder_attentions (:obj:`tuple(torch.FloatTensor)`, `optional`, returned when ``output_attentions=True`` is passed or when ``config.output_attentions=True``):\n Tuple of :obj:`torch.FloatTensor` (one for each layer) of shape :obj:`(batch_size, num_attn_heads,\n decoder_sequence_length, decoder_sequence_length)`.\n\n Attentions weights of the decoder, after the attention softmax, used to compute the weighted average in the\n self-attention heads.\n decoder_ngram_attentions (:obj:`tuple(torch.FloatTensor)`, `optional`, returned when ``output_attentions=True`` is passed or when ``config.output_attentions=True``):\n Tuple of :obj:`torch.FloatTensor` (one for each layer) of shape :obj:`(batch_size, num_attn_heads,\n decoder_sequence_length, decoder_sequence_length)`.\n\n Attentions weights of the predict stream of the decoder, after the attention softmax, used to compute the\n weighted average in the\n cross_attentions (:obj:`tuple(torch.FloatTensor)`, `optional`, returned when ``output_attentions=True`` is passed or when ``config.output_attentions=True``):\n Tuple of :obj:`torch.FloatTensor` (one for each layer) of shape :obj:`(batch_size, num_attn_heads,\n encoder_sequence_length, decoder_sequence_length)`.\n\n Attentions weights of the cross-attention layer of the decoder, after the attention softmax, used to\n compute the weighted average in the\n encoder_last_hidden_state (:obj:`torch.FloatTensor` of shape :obj:`(batch_size, encoder_sequence_length, hidden_size)`, `optional`):\n Sequence of hidden-states at the output of the last layer of the encoder of the model.\n encoder_hidden_states (:obj:`tuple(torch.FloatTensor)`, `optional`, returned when ``output_hidden_states=True`` is passed or when ``config.output_hidden_states=True``):\n Tuple of :obj:`torch.FloatTensor` (one for the output of the embeddings + one for the output of each layer)\n of shape :obj:`(batch_size, encoder_sequence_length, hidden_size)`.\n\n Hidden-states of the encoder at the output of each layer plus the initial embedding outputs.\n encoder_attentions (:obj:`tuple(torch.FloatTensor)`, `optional`, returned when ``output_attentions=True`` is passed or when ``config.output_attentions=True``):\n Tuple of :obj:`torch.FloatTensor` (one for each layer) of shape :obj:`(batch_size, num_attn_heads,\n encoder_sequence_length, encoder_sequence_length)`.\n\n Attentions weights of the encoder, after the attention softmax, used to compute the weighted average in the\n self-attention heads.\n \"\"\"\n\n last_hidden_state: torch.FloatTensor\n last_hidden_state_ngram: Optional[torch.FloatTensor] = None\n past_key_values: Optional[Tuple[torch.FloatTensor]] = None\n decoder_hidden_states: Optional[Tuple[torch.FloatTensor]] = None\n decoder_ngram_hidden_states: Optional[Tuple[torch.FloatTensor]] = None\n decoder_attentions: Optional[Tuple[torch.FloatTensor]] = None\n decoder_ngram_attentions: Optional[Tuple[torch.FloatTensor]] = None\n cross_attentions: Optional[Tuple[torch.FloatTensor]] = None\n encoder_last_hidden_state: Optional[torch.FloatTensor] = None\n encoder_hidden_states: Optional[Tuple[torch.FloatTensor]] = None\n encoder_attentions: Optional[Tuple[torch.FloatTensor]] = None\n\n @property\n def decoder_cross_attentions(self):\n warnings.warn(\n \"`decoder_cross_attentions` is deprecated and will be removed soon. Please use `cross_attentions` instead.\",\n FutureWarning,\n )\n return self.cross_attentions\n\n\n@dataclass\nclass ProphetNetDecoderModelOutput(ModelOutput):\n \"\"\"\n Base class for model's outputs that may also contain a past key/values (to speed up sequential decoding).\n\n Args:\n last_hidden_state (:obj:`torch.FloatTensor` of shape :obj:`(batch_size, decoder_sequence_length, hidden_size)`):\n Sequence of main stream hidden-states at the output of the last layer of the decoder of the model.\n\n If :obj:`past_key_values` is used only the last hidden-state of the sequences of shape :obj:`(batch_size,\n 1, hidden_size)` is output.\n last_hidden_state_ngram (:obj:`torch.FloatTensor` of shape :obj:`(batch_size, ngram * decoder_sequence_length, config.vocab_size)`):\n Sequence of predict stream hidden-states at the output of the last layer of the decoder of the model.\n past_key_values (:obj:`List[torch.FloatTensor]`, `optional`, returned when ``use_cache=True`` is passed or when ``config.use_cache=True``):\n List of :obj:`torch.FloatTensor` of length :obj:`config.n_layers`, with each tensor of shape :obj:`(2,\n batch_size, num_attn_heads, decoder_sequence_length, embed_size_per_head)`).\n\n Contains pre-computed hidden-states (key and values in the attention blocks) of the decoder that can be\n used (see :obj:`past_key_values` input) to speed up sequential decoding.\n hidden_states (:obj:`tuple(torch.FloatTensor)`, `optional`, returned when ``output_hidden_states=True`` is passed or when ``config.output_hidden_states=True``):\n Tuple of :obj:`torch.FloatTensor` (one for the output of the embeddings + one for the output of each layer)\n of shape :obj:`(batch_size, decoder_sequence_length, hidden_size)`.\n\n Hidden-states of main stream of the decoder at the output of each layer plus the initial embedding outputs.\n ngram_hidden_states (:obj:`tuple(torch.FloatTensor)`, `optional`, returned when ``output_hidden_states=True`` is passed or when ``config.output_hidden_states=True``):\n Tuple of :obj:`torch.FloatTensor` (one for the output of the embeddings + one for the output of each layer)\n of shape :obj:`(batch_size, ngram * decoder_sequence_length, hidden_size)`.\n\n Hidden-states of the predict stream of the decoder at the output of each layer plus the initial embedding\n outputs.\n attentions (:obj:`tuple(torch.FloatTensor)`, `optional`, returned when ``output_attentions=True`` is passed or when ``config.output_attentions=True``):\n Tuple of :obj:`torch.FloatTensor` (one for each layer) of shape :obj:`(batch_size, num_attn_heads,\n decoder_sequence_length, decoder_sequence_length)`.\n\n Attentions weights of the decoder, after the attention softmax, used to compute the weighted average in the\n self-attention heads.\n ngram_attentions (:obj:`tuple(torch.FloatTensor)`, `optional`, returned when ``output_attentions=True`` is passed or when ``config.output_attentions=True``):\n Tuple of :obj:`torch.FloatTensor` (one for each layer) of shape :obj:`(batch_size, num_attn_heads,\n decoder_sequence_length, decoder_sequence_length)`.\n\n Attentions weights of the predict stream of the decoder, after the attention softmax, used to compute the\n weighted average in the\n cross_attentions (:obj:`tuple(torch.FloatTensor)`, `optional`, returned when ``output_attentions=True`` is passed or when ``config.output_attentions=True``):\n Tuple of :obj:`torch.FloatTensor` (one for each layer) of shape :obj:`(batch_size, num_attn_heads,\n encoder_sequence_length, decoder_sequence_length)`.\n\n Attentions weights of the cross-attention layer of the decoder, after the attention softmax, used to\n compute the weighted average in the\n \"\"\"\n\n last_hidden_state: torch.FloatTensor\n last_hidden_state_ngram: Optional[torch.FloatTensor] = None\n past_key_values: Optional[Tuple[torch.FloatTensor]] = None\n hidden_states: Optional[Tuple[torch.FloatTensor]] = None\n hidden_states_ngram: Optional[Tuple[torch.FloatTensor]] = None\n attentions: Optional[Tuple[torch.FloatTensor]] = None\n ngram_attentions: Optional[Tuple[torch.FloatTensor]] = None\n cross_attentions: Optional[Tuple[torch.FloatTensor]] = None\n\n\n@dataclass\nclass ProphetNetDecoderLMOutput(ModelOutput):\n \"\"\"\n Base class for model's outputs that may also contain a past key/values (to speed up sequential decoding).\n\n Args:\n loss (:obj:`torch.FloatTensor` of shape :obj:`(1,)`, `optional`, returned when :obj:`labels` is provided):\n Language modeling loss.\n logits (:obj:`torch.FloatTensor` of shape :obj:`(batch_size, decoder_sequence_length, config.vocab_size)`):\n Prediction scores of the main stream language modeling head (scores for each vocabulary token before\n SoftMax).\n logits_ngram (:obj:`torch.FloatTensor` of shape :obj:`(batch_size, ngram * decoder_sequence_length, config.vocab_size)`):\n Prediction scores of the predict stream language modeling head (scores for each vocabulary token before\n SoftMax).\n past_key_values (:obj:`List[torch.FloatTensor]`, `optional`, returned when ``use_cache=True`` is passed or when ``config.use_cache=True``):\n List of :obj:`torch.FloatTensor` of length :obj:`config.n_layers`, with each tensor of shape :obj:`(2,\n batch_size, num_attn_heads, decoder_sequence_length, embed_size_per_head)`).\n\n Contains pre-computed hidden-states (key and values in the attention blocks) of the decoder that can be\n used (see :obj:`past_key_values` input) to speed up sequential decoding.\n hidden_states (:obj:`tuple(torch.FloatTensor)`, `optional`, returned when ``output_hidden_states=True`` is passed or when ``config.output_hidden_states=True``):\n Tuple of :obj:`torch.FloatTensor` (one for the output of the embeddings + one for the output of each layer)\n of shape :obj:`(batch_size, decoder_sequence_length, hidden_size)`.\n\n Hidden-states of main stream of the decoder at the output of each layer plus the initial embedding outputs.\n ngram_hidden_states (:obj:`tuple(torch.FloatTensor)`, `optional`, returned when ``output_hidden_states=True`` is passed or when ``config.output_hidden_states=True``):\n Tuple of :obj:`torch.FloatTensor` (one for the output of the embeddings + one for the output of each layer)\n of shape :obj:`(batch_size, ngram * decoder_sequence_length, hidden_size)`.\n\n Hidden-states of the predict stream of the decoder at the output of each layer plus the initial embedding\n outputs.\n attentions (:obj:`tuple(torch.FloatTensor)`, `optional`, returned when ``output_attentions=True`` is passed or when ``config.output_attentions=True``):\n Tuple of :obj:`torch.FloatTensor` (one for each layer) of shape :obj:`(batch_size, num_attn_heads,\n decoder_sequence_length, decoder_sequence_length)`.\n\n Attentions weights of the decoder, after the attention softmax, used to compute the weighted average in the\n self-attention heads.\n ngram_attentions (:obj:`tuple(torch.FloatTensor)`, `optional`, returned when ``output_attentions=True`` is passed or when ``config.output_attentions=True``):\n Tuple of :obj:`torch.FloatTensor` (one for each layer) of shape :obj:`(batch_size, num_attn_heads,\n decoder_sequence_length, decoder_sequence_length)`.\n\n Attentions weights of the predict stream of the decoder, after the attention softmax, used to compute the\n weighted average in the\n cross_attentions (:obj:`tuple(torch.FloatTensor)`, `optional`, returned when ``output_attentions=True`` is passed or when ``config.output_attentions=True``):\n Tuple of :obj:`torch.FloatTensor` (one for each layer) of shape :obj:`(batch_size, num_attn_heads,\n encoder_sequence_length, decoder_sequence_length)`.\n\n Attentions weights of the cross-attention layer of the decoder, after the attention softmax, used to\n compute the weighted average in the\n \"\"\"\n\n loss: Optional[torch.FloatTensor] = None\n logits: torch.FloatTensor = None\n logits_ngram: Optional[torch.FloatTensor] = None\n past_key_values: Optional[Tuple[torch.FloatTensor]] = None\n hidden_states: Optional[Tuple[torch.FloatTensor]] = None\n hidden_states_ngram: Optional[Tuple[torch.FloatTensor]] = None\n attentions: Optional[Tuple[torch.FloatTensor]] = None\n ngram_attentions: Optional[Tuple[torch.FloatTensor]] = None\n cross_attentions: Optional[Tuple[torch.FloatTensor]] = None\n\n\nclass ProphetNetPreTrainedModel(PreTrainedModel):\n config_class = ProphetNetConfig\n base_model_prefix = \"prophetnet\"\n\n def _init_weights(self, module):\n if isinstance(module, nn.Linear):\n module.weight.data.normal_(mean=0.0, std=self.config.init_std)\n if module.bias is not None:\n module.bias.data.zero_()\n elif isinstance(module, nn.Embedding):\n module.weight.data.normal_(mean=0.0, std=self.config.init_std)\n if module.padding_idx is not None:\n module.weight.data[module.padding_idx].zero_()\n\n def _shift_right(self, input_ids):\n decoder_start_token_id = self.config.decoder_start_token_id\n pad_token_id = self.config.pad_token_id\n\n assert (\n decoder_start_token_id is not None\n ), \"self.model.config.decoder_start_token_id has to be defined. In ProphetNet it is usually set to the pad_token_id. See ProphetNet docs for more information\"\n\n # shift inputs to the right\n shifted_input_ids = input_ids.new_zeros(input_ids.shape)\n shifted_input_ids[..., 1:] = input_ids[..., :-1].clone()\n shifted_input_ids[..., 0] = decoder_start_token_id\n\n assert pad_token_id is not None, \"self.model.config.pad_token_id has to be defined.\"\n # replace possible -100 values in labels by `pad_token_id`\n shifted_input_ids.masked_fill_(shifted_input_ids == -100, pad_token_id)\n\n assert torch.all(shifted_input_ids >= 0).item(), \"Verify that `shifted_input_ids` has only positive values\"\n\n return shifted_input_ids\n\n\nclass ProphetNetPositionalEmbeddings(nn.Embedding):\n \"\"\"\n This module learns positional embeddings up to a fixed maximum size. Padding ids are ignored by either offsetting\n based on padding_idx or by setting padding_idx to None and ensuring that the appropriate position ids are passed to\n the forward function.\n \"\"\"\n\n def __init__(self, config: ProphetNetConfig):\n self.max_length = config.max_position_embeddings\n super().__init__(config.max_position_embeddings, config.hidden_size, config.pad_token_id)\n\n def forward(self, inputs_shape, device, attention_mask=None, past_key_values=None, position_ids=None):\n assert (position_ids is None) or (\n self.padding_idx is None\n ), \"If position_ids is pre-computed then padding_idx should not be set.\"\n\n if position_ids is None:\n if past_key_values is not None:\n # position_ids is the same for every token when decoding a single step\n # Without the int() cast, it doesn't work in some cases when exporting to ONNX\n prev_num_input_ids = past_key_values[0][0].shape[2]\n num_input_ids = inputs_shape[1] + prev_num_input_ids\n position_ids = torch.ones((1, 1), dtype=torch.long, device=device) * (\n int(self.padding_idx + num_input_ids)\n )\n else:\n if attention_mask is None:\n attention_mask = torch.ones(inputs_shape, dtype=torch.long, device=device)\n\n # retrieve position_ids from input_ids / attention_mask\n position_ids = (\n torch.cumsum(attention_mask, dim=1).type_as(attention_mask) * attention_mask\n ).long() + self.padding_idx\n\n # make sure position_ids are not bigger then max_length\n position_ids = position_ids.clamp(0, self.max_length - 1)\n\n return super().forward(position_ids), position_ids\n\n def _forward(self, position_ids):\n return super().forward(position_ids)\n\n\nclass ProphetNetAttention(nn.Module):\n \"\"\"Multi-headed attention from 'Attention Is All You Need' paper\"\"\"\n\n def __init__(\n self,\n config: ProphetNetConfig,\n num_attn_heads: int,\n ):\n super().__init__()\n hidden_size = config.hidden_size\n\n self.attention_dropout = config.attention_dropout\n self.dropout = config.dropout\n self.num_attn_heads = num_attn_heads\n self.head_dim = hidden_size // num_attn_heads\n\n assert (\n self.head_dim * num_attn_heads == hidden_size\n ), \"`config.hidden_size` must be divisible by `config.num_encoder_attention_heads` and `config.num_decoder_attention_heads`\"\n\n self.key_proj = nn.Linear(hidden_size, hidden_size)\n self.value_proj = nn.Linear(hidden_size, hidden_size)\n self.query_proj = nn.Linear(hidden_size, hidden_size)\n\n self.out_proj = nn.Linear(hidden_size, hidden_size)\n\n def _shape(self, tensor: torch.Tensor, seq_len: int, bsz: int):\n return tensor.view(bsz, seq_len, self.num_attn_heads, self.head_dim).transpose(1, 2).contiguous()\n\n def forward(\n self,\n hidden_states,\n key_value_states: Optional[Tensor] = None,\n attention_mask: Optional[Tensor] = None,\n past_key_value: Optional[Tuple[Tensor]] = None,\n output_attentions: bool = False,\n ) -> Tuple[Tensor, Optional[Tensor]]:\n\n batch_size, tgt_len, hidden_size = hidden_states.size()\n\n # if key_value_states are provided this layer is used as a cross-attention layer\n # for the decoder\n is_cross_attention = key_value_states is not None\n assert list(hidden_states.size()) == [\n batch_size,\n tgt_len,\n hidden_size,\n ], f\"Size of hidden states should be {batch_size, tgt_len, hidden_size}, but is {hidden_states.size()}\"\n\n # previous time steps are cached - no need to recompute key and value if they are static\n query_states = self.query_proj(hidden_states) / (self.head_dim ** 0.5)\n\n if is_cross_attention and past_key_value is not None:\n # reuse k,v, cross_attentions\n key_states = past_key_value[0]\n value_states = past_key_value[1]\n elif is_cross_attention:\n # cross_attentions\n key_states = self._shape(self.key_proj(key_value_states), -1, batch_size)\n value_states = self._shape(self.value_proj(key_value_states), -1, batch_size)\n else:\n # self_attention\n key_states = self._shape(self.key_proj(hidden_states), -1, batch_size)\n value_states = self._shape(self.value_proj(hidden_states), -1, batch_size)\n\n if is_cross_attention:\n # if cross_attention save Tuple(torch.Tensor, torch.Tensor) of all cross attention key/value_states.\n # Further calls to cross_attention layer can then reuse all cross-attention\n # key/value_states (first \"if\" case)\n # if encoder bi-directional self-attention `past_key_value` is always `None`\n past_key_value = (key_states, value_states)\n\n # project states into the correct shape\n proj_shape = (batch_size * self.num_attn_heads, -1, self.head_dim)\n query_states = self._shape(query_states, tgt_len, batch_size).view(*proj_shape)\n key_states = key_states.view(*proj_shape)\n value_states = value_states.view(*proj_shape)\n\n src_len = key_states.size(1)\n attn_weights = torch.bmm(query_states, key_states.transpose(1, 2))\n assert attn_weights.size() == (\n batch_size * self.num_attn_heads,\n tgt_len,\n src_len,\n ), f\"`attn_weights` should be of size {batch_size * self.num_attn_heads, tgt_len, src_len}, but is of size {attn_weights.shape}\"\n\n # This is part of a workaround to get around fork/join parallelism not supporting Optional types.\n if attention_mask is not None and attention_mask.dim() == 0:\n attention_mask = None\n assert attention_mask is None or attention_mask.size() == (\n self.num_attn_heads * batch_size,\n 1,\n src_len,\n ), f\"`attention_mask` should be `None` or of shape attention_mask.size() == {batch_size * self.num_attn_heads, 1, src_len}, but is {attention_mask.shape}\"\n\n if attention_mask is not None: # don't attend to padding symbols\n attn_weights = attn_weights + attention_mask\n\n if output_attentions:\n # this operation is a bit akward, but it's required to\n # make sure that attn_weights keeps its gradient.\n # In order to do so, attn_weights have to reshaped\n # twice and have to be reused in the following\n attn_weights_reshaped = attn_weights.view(batch_size, self.num_attn_heads, tgt_len, src_len)\n attn_weights = attn_weights_reshaped.view(batch_size * self.num_attn_heads, tgt_len, src_len)\n else:\n attn_weights_reshaped = None\n\n attn_weights = F.softmax(attn_weights, dim=-1)\n attn_probs = F.dropout(\n attn_weights,\n p=self.attention_dropout,\n training=self.training,\n )\n\n attn_output = torch.bmm(attn_probs, value_states)\n assert attn_output.size() == (\n batch_size * self.num_attn_heads,\n tgt_len,\n self.head_dim,\n ), \"`attn_output` should be of shape {batch_size * self.num_attn_heads, tgt_len, self.head_dim}, but is of shape {attn_output.size()}\"\n\n attn_output = (\n attn_output.view(batch_size, self.num_attn_heads, tgt_len, self.head_dim)\n .transpose(1, 2)\n .reshape(batch_size, tgt_len, hidden_size)\n )\n\n attn_output = self.out_proj(attn_output)\n\n attn_output = F.dropout(attn_output, p=self.dropout, training=self.training)\n return attn_output, attn_weights_reshaped, past_key_value\n\n\nclass ProphetNetFeedForward(nn.Module):\n \"\"\"\n This is the residual two feed-forward layer block based on the original Transformer implementation.\n \"\"\"\n\n def __init__(self, config: ProphetNetConfig, ffn_dim: int):\n super().__init__()\n self.activation_fn = ACT2FN[config.activation_function]\n self.intermediate = nn.Linear(config.hidden_size, ffn_dim)\n self.output = nn.Linear(ffn_dim, config.hidden_size)\n self.activation_dropout = config.activation_dropout\n self.dropout = config.dropout\n\n def forward(self, hidden_states):\n hidden_states = self.intermediate(hidden_states)\n hidden_states = self.activation_fn(hidden_states)\n\n hidden_states = F.dropout(hidden_states, p=self.activation_dropout, training=self.training)\n hidden_states = self.output(hidden_states)\n hidden_states = F.dropout(hidden_states, p=self.dropout, training=self.training)\n return hidden_states\n\n\nclass ProphetNetNgramSelfAttention(nn.Module):\n def __init__(self, config: ProphetNetConfig):\n super().__init__()\n self.hidden_size = config.hidden_size\n\n self.num_buckets = config.num_buckets\n self.relative_max_distance = config.relative_max_distance\n self.num_attn_heads = config.num_decoder_attention_heads\n self.dropout = config.dropout\n self.attention_dropout = config.attention_dropout\n self.head_dim = config.hidden_size // self.num_attn_heads\n self.ngram = config.ngram\n\n assert (\n self.head_dim * self.num_attn_heads == config.hidden_size\n ), \"config.hidden_size must be divisible by num_attn_heads\"\n # key, value, query projection\n self.key_proj = nn.Linear(config.hidden_size, config.hidden_size)\n self.value_proj = nn.Linear(config.hidden_size, config.hidden_size)\n self.query_proj = nn.Linear(config.hidden_size, config.hidden_size)\n\n # out projection\n self.out_proj = nn.Linear(config.hidden_size, config.hidden_size)\n\n # rel position embeddings\n self.relative_pos_embeddings = nn.Linear(config.hidden_size, self.num_buckets * self.num_attn_heads)\n\n # for onnx runtime\n self.onnx_trace = False\n\n def _shape(self, tensor, seq_len, batch_size):\n return tensor.view(batch_size, seq_len, self.num_attn_heads, self.head_dim).transpose(1, 2).contiguous()\n\n def prepare_for_onnx_export_(self):\n self.onnx_trace = True\n\n def forward(\n self,\n hidden_states,\n past_key_value: Optional[Tuple[Tensor]] = None,\n attention_mask=None,\n extended_predict_attention_mask=None,\n main_relative_position_buckets=None,\n predict_relative_position_buckets=None,\n position_ids=None,\n ):\n batch_size, ngram_sequence_length, hidden_size = hidden_states.size()\n\n assert list(hidden_states.size()) == [\n batch_size,\n ngram_sequence_length,\n hidden_size,\n ], f\"`hidden_states` should be of shape {batch_size, ngram_sequence_length, hidden_size}, but is of shape {hidden_states.shape}\"\n\n # project\n query_states = self.query_proj(hidden_states)\n key_states = self.key_proj(hidden_states)\n value_states = self.value_proj(hidden_states)\n\n # normalize\n query_states = query_states / (self.head_dim ** 0.5)\n\n # reshape\n query_states = self._shape(query_states, ngram_sequence_length, batch_size)\n key_states = self._shape(key_states, -1, batch_size)\n value_states = self._shape(value_states, -1, batch_size)\n\n proj_shape = (batch_size * self.num_attn_heads, -1, self.head_dim)\n\n query_states = query_states.view(*proj_shape)\n key_states = key_states.view(*proj_shape)\n value_states = value_states.view(*proj_shape)\n\n # chunk into main stream and predict stream\n hidden_states_list = hidden_states.chunk(1 + self.ngram, dim=1)\n\n query_states_list = query_states.chunk(1 + self.ngram, dim=1)\n key_states_list = key_states.chunk(1 + self.ngram, dim=1)\n value_states_list = value_states.chunk(1 + self.ngram, dim=1)\n\n main_hidden_states, hidden_states_predict_list = hidden_states_list[0], hidden_states_list[1:]\n main_query_states, predict_query_states_list = query_states_list[0], query_states_list[1:]\n main_key_states, predict_key_states_list = key_states_list[0], key_states_list[1:]\n main_value_states, predict_value_states_list = value_states_list[0], value_states_list[1:]\n\n # saved states are stored with shape (batch_size, num_attn_heads, seq_len, head_dim)\n if past_key_value is not None:\n prev_main_key_states = past_key_value[0].view(batch_size * self.num_attn_heads, -1, self.head_dim)\n main_key_states = torch.cat((prev_main_key_states, main_key_states), dim=1)\n prev_main_value_states = past_key_value[1].view(batch_size * self.num_attn_heads, -1, self.head_dim)\n main_value_states = torch.cat((prev_main_value_states, main_value_states), dim=1)\n\n # Update cache\n past_key_value = (\n main_key_states.view(batch_size, self.num_attn_heads, -1, self.head_dim),\n main_value_states.view(batch_size, self.num_attn_heads, -1, self.head_dim),\n )\n\n # get seq_length of main stream only\n sequence_length = ngram_sequence_length // (1 + self.ngram)\n\n # MAIN-STREAM\n # main attn weights\n main_attn_weights = torch.bmm(main_query_states, main_key_states.transpose(1, 2))\n\n # retrieve relative position embeddings for each layer -> see paper for more details\n main_relative_pos_embeddings = self.get_main_relative_pos_embeddings(\n main_hidden_states, main_attn_weights, position_ids, main_relative_position_buckets\n )\n main_attn_weights = main_attn_weights + main_relative_pos_embeddings\n\n if attention_mask is not None:\n main_attn_weights = main_attn_weights + attention_mask\n\n main_attn_probs = softmax(\n main_attn_weights,\n dim=-1,\n onnx_trace=self.onnx_trace,\n ).type_as(main_attn_weights)\n\n main_attn_probs = F.dropout(main_attn_probs, p=self.attention_dropout, training=self.training)\n # project to attn_output\n main_attn_output = torch.bmm(main_attn_probs, main_value_states)\n\n # reshape so that num_heads dim is merged into last `head_dim` axis\n main_attn_output = (\n main_attn_output.view(batch_size, self.num_attn_heads, sequence_length, self.head_dim)\n .transpose(1, 2)\n .reshape(batch_size, 1, sequence_length, hidden_size)\n )\n main_attn_output = self.out_proj(main_attn_output)\n\n # PREDICT-STREAM\n # [ngram, B*head, T, c]\n predict_query_states = torch.cat(predict_query_states_list, 0).view(\n self.ngram, -1, sequence_length, self.head_dim\n )\n # [ngram, B*head, 2*T, c]\n predict_key_states = torch.cat(\n [torch.cat([main_key_states, key], 1).unsqueeze(0) for key in predict_key_states_list], 0\n )\n\n # [ngram, T, B, C]\n predict_hidden_states = torch.cat(hidden_states_predict_list, 0).view(\n self.ngram, sequence_length, batch_size, hidden_size\n )\n\n # [ngram, B*head, 2*T, c]\n predict_value_states = torch.cat(\n [torch.cat([main_value_states, v_p], 1).unsqueeze(0) for v_p in predict_value_states_list], 0\n )\n # [ngram, B*head, T, 2*T]\n predict_attn_weights = torch.einsum(\"nbtc,nbsc->nbts\", (predict_query_states, predict_key_states))\n\n # [ngram, B*head, T, S]\n # retrieve relative position embeddings for each layer -> see paper for more details\n predict_relative_pos_embeddings = self.get_predict_relative_pos_embeddings(\n predict_hidden_states, predict_attn_weights, position_ids, predict_relative_position_buckets\n )\n\n # [ngram, B*head, T, 2*T]\n predict_attn_weights = predict_attn_weights + predict_relative_pos_embeddings\n\n if extended_predict_attention_mask is not None:\n predict_attn_weights = predict_attn_weights + extended_predict_attention_mask.to(\n predict_attn_weights.dtype\n )\n\n predict_attn_probs = softmax(\n predict_attn_weights,\n dim=-1,\n onnx_trace=self.onnx_trace,\n ).type_as(predict_attn_weights)\n predict_attn_probs = F.dropout(predict_attn_probs, p=self.attention_dropout, training=self.training)\n # project to attention output\n # [ngram, B*head, T, c]\n predict_attn_output = torch.einsum(\"nbts,nbsc->nbtc\", (predict_attn_probs, predict_value_states))\n\n # reshape so that num_heads dim is merged into last `head_dim` axis\n # [ngram, B, T, C]\n predict_attn_output = (\n predict_attn_output.view(self.ngram, batch_size, self.num_attn_heads, sequence_length, self.head_dim)\n .permute(1, 0, 3, 2, 4)\n .reshape(batch_size, self.ngram, sequence_length, hidden_size)\n )\n predict_attn_output = self.out_proj(predict_attn_output)\n\n # concat to single attn output\n # [B, 1+ngram*T, C]\n attn_output = torch.cat([main_attn_output, predict_attn_output], 1).view(batch_size, -1, hidden_size)\n # reshape into better form for `config.output_attentions`\n main_attn_probs = main_attn_probs.view(batch_size, self.num_attn_heads, sequence_length, -1)\n predict_attn_probs = predict_attn_probs.view(\n self.ngram, batch_size, self.num_attn_heads, sequence_length, -1\n ).transpose(0, 1)\n\n attn_output = F.dropout(attn_output, p=self.dropout, training=self.training)\n\n return attn_output, main_attn_probs, predict_attn_probs, past_key_value\n\n def get_main_relative_pos_embeddings(\n self, hidden_states, attn_weights, position_ids, main_relative_position_buckets\n ):\n # input hidden_states [B,T,C], input attn_weights [T*head,T,S], input position_ids [B,T] or [1,1]\n\n if main_relative_position_buckets is None:\n batch_size, sequence_length = hidden_states.shape[:2]\n relative_positions = (\n torch.arange(1, attn_weights.shape[-1] + 1)\n .unsqueeze(0)\n .unsqueeze(0)\n .repeat(batch_size, sequence_length, 1)\n .to(position_ids.device)\n )\n relative_positions = relative_positions - position_ids.unsqueeze(0).repeat(\n batch_size, sequence_length, 1\n ) # [B, T, s]\n main_relative_position_buckets = compute_relative_buckets(\n self.num_buckets, self.relative_max_distance, relative_positions, False\n )\n\n rel_pos_embeddings = self.relative_pos_embeddings(hidden_states) # [B,T,Buckets*head]\n rel_pos_embeddings = rel_pos_embeddings.view(\n rel_pos_embeddings.shape[:2] + (self.num_buckets, self.num_attn_heads)\n ).permute(\n 0, 3, 1, 2\n ) # [B,T,Buckets,head]\n rel_pos_embeddings = rel_pos_embeddings.reshape(attn_weights.shape[:2] + (-1,)) # [B*head,T,Buckets]\n\n main_relative_position_buckets = (\n main_relative_position_buckets.repeat(1, self.num_attn_heads, 1)\n .view(-1, main_relative_position_buckets.shape[-1])\n .long()\n ) # [B*head*T, T]\n rel_pos_embeddings = rel_pos_embeddings.reshape(-1, rel_pos_embeddings.size(-1)) # [B*head*T,Buckets]\n\n main_relative_pos_embeddings = torch.gather(\n rel_pos_embeddings, dim=1, index=main_relative_position_buckets\n ).view(attn_weights.shape[:2] + (-1,))\n\n return main_relative_pos_embeddings\n\n def get_predict_relative_pos_embeddings(\n self, hidden_states, attn_weights, position_ids, predict_relative_position_buckets\n ):\n # input hidden_states [ngram, T,B,C], input attn_weights [ngram, B*head,T,S], input position_ids [B,T] or [1,1], input predict_relative_position_buckets [B,T, 2*T] or None\n sequence_length, batch_size = hidden_states.shape[1:3]\n\n if predict_relative_position_buckets is None:\n key_sequence_length = attn_weights.shape[-1]\n assert (\n position_ids[0][0] == key_sequence_length - 1\n ), \"`position_ids` are incorrect. They should be of the format 1 2 3 4 5 ... (key_sequence_length - 1)\"\n relative_positions = (\n torch.arange(0, key_sequence_length)\n .unsqueeze(0)\n .unsqueeze(0)\n .repeat(batch_size, sequence_length, 1)\n .to(position_ids.device)\n )\n\n relative_positions = relative_positions - position_ids.unsqueeze(0).repeat(batch_size, sequence_length, 1)\n predict_relative_position_buckets = compute_relative_buckets(\n self.num_buckets, self.relative_max_distance, relative_positions, False\n )\n\n hidden_states = hidden_states.transpose(1, 2) # [ngram, B, T, C]\n rel_pos_embeddings = self.relative_pos_embeddings(hidden_states).view(\n hidden_states.shape[:-1] + (self.num_buckets, self.num_attn_heads)\n ) # [ngram, B, T, bucket, head]\n rel_pos_embeddings = rel_pos_embeddings.permute(0, 1, 4, 2, 3).reshape(\n self.ngram * batch_size * self.num_attn_heads, sequence_length, -1\n ) # [ngram*B*head, T, bucket]\n\n predict_relative_position_buckets = predict_relative_position_buckets.unsqueeze(0).repeat(\n self.ngram, 1, self.num_attn_heads, 1\n ) # [ngram, B, head*T, S]\n\n rel_pos_embeddings = rel_pos_embeddings.reshape(-1, rel_pos_embeddings.size(-1))\n predict_relative_position_buckets = predict_relative_position_buckets.view(\n -1, predict_relative_position_buckets.size(-1)\n ).long() # [ngram*B*head*T, S]\n\n predict_relative_pos_embeddings = torch.gather(\n rel_pos_embeddings, dim=1, index=predict_relative_position_buckets\n ).view(\n self.ngram, batch_size * self.num_attn_heads, sequence_length, -1\n ) # [ngram, B*head, T, S]\n\n return predict_relative_pos_embeddings\n\n\nclass ProphetNetEncoderLayer(nn.Module):\n \"\"\"\n Encoder block for Prophetnet\n \"\"\"\n\n def __init__(self, config: ProphetNetConfig):\n super().__init__()\n # 1st residual block\n self.self_attn = ProphetNetAttention(config, config.num_encoder_attention_heads)\n self.self_attn_layer_norm = LayerNorm(config.hidden_size)\n\n # 2nd residual block\n self.feed_forward = ProphetNetFeedForward(config, config.encoder_ffn_dim)\n self.feed_forward_layer_norm = LayerNorm(config.hidden_size)\n\n def forward(self, hidden_states, attention_mask, output_attentions: bool = False):\n # 1st residual block\n attention_output, attn_weights, _ = self.self_attn(\n hidden_states=hidden_states,\n attention_mask=attention_mask,\n output_attentions=output_attentions,\n )\n hidden_states = self.self_attn_layer_norm(attention_output + hidden_states)\n\n # 2nd residual block\n feed_forward_output = self.feed_forward(hidden_states)\n hidden_states = self.feed_forward_layer_norm(feed_forward_output + hidden_states)\n\n outputs = (hidden_states,)\n\n if output_attentions:\n outputs += (attn_weights,)\n\n return outputs\n\n\nclass ProphetNetDecoderLayer(nn.Module):\n \"\"\"\n Decoder block for Prophetnet\n \"\"\"\n\n def __init__(self, config: ProphetNetConfig):\n super().__init__()\n # 1st residual block\n self.self_attn = ProphetNetNgramSelfAttention(config)\n self.self_attn_layer_norm = LayerNorm(config.hidden_size)\n\n # 2nd residual block\n if config.add_cross_attention:\n self.cross_attn = ProphetNetAttention(config, config.num_decoder_attention_heads)\n self.cross_attn_layer_norm = LayerNorm(config.hidden_size)\n\n # 3rd residual block\n self.feed_forward = ProphetNetFeedForward(config, config.decoder_ffn_dim)\n self.feed_forward_layer_norm = LayerNorm(config.hidden_size)\n\n def forward(\n self,\n hidden_states,\n attention_mask=None,\n encoder_hidden_states=None,\n encoder_attn_mask=None,\n extended_predict_attention_mask=None,\n main_relative_position_buckets=None,\n predict_relative_position_buckets=None,\n position_ids=None,\n past_key_value=None,\n use_cache: bool = True,\n output_attentions: bool = False,\n ):\n # 1st residual block\n # decoder uni-directional self-attention cached key/values tuple is at positions 1,2\n self_attn_past_key_value = past_key_value[:2] if past_key_value is not None else None\n ngram_attention_output, self_attn_weights, self_attn_weights_ngram, present_key_value = self.self_attn(\n hidden_states=hidden_states,\n past_key_value=self_attn_past_key_value,\n attention_mask=attention_mask,\n extended_predict_attention_mask=extended_predict_attention_mask,\n main_relative_position_buckets=main_relative_position_buckets,\n predict_relative_position_buckets=predict_relative_position_buckets,\n position_ids=position_ids,\n )\n hidden_states = self.self_attn_layer_norm(hidden_states + ngram_attention_output)\n\n # cross_attn cached key/values tuple is at positions 3,4 of present_key_value tuple\n cross_attn_past_key_value = past_key_value[-2:] if past_key_value is not None else None\n cross_attn_weights = None\n if encoder_hidden_states is not None:\n # 2nd residual block\n attention_output, cross_attn_weights, cross_attn_present_key_value = self.cross_attn(\n hidden_states=hidden_states,\n key_value_states=encoder_hidden_states,\n attention_mask=encoder_attn_mask,\n past_key_value=cross_attn_past_key_value,\n output_attentions=output_attentions,\n )\n hidden_states = self.cross_attn_layer_norm(attention_output + hidden_states)\n\n # add cross-attn to positions 3,4 of present_key_value tuple\n present_key_value = present_key_value + cross_attn_present_key_value\n\n # 3rd residual block\n feed_forward_output = self.feed_forward(hidden_states)\n hidden_states = self.feed_forward_layer_norm(feed_forward_output + hidden_states)\n\n outputs = (hidden_states,)\n\n if output_attentions:\n outputs += (self_attn_weights, self_attn_weights_ngram, cross_attn_weights)\n\n if use_cache:\n outputs += (present_key_value,)\n\n return outputs\n\n\n@add_start_docstrings(\n \"The standalone encoder part of the ProphetNetModel.\",\n PROPHETNET_START_DOCSTRING,\n)\nclass ProphetNetEncoder(ProphetNetPreTrainedModel):\n r\"\"\"\n word_embeddings (:obj:`torch.nn.Embeddings` of shape :obj:`(config.vocab_size, config.hidden_size)`, `optional`):\n The word embedding parameters. This can be used to initialize :class:`~transformers.ProphetNetEncoder` with\n pre-defined word embeddings instead of randomely initialized word embeddings.\n \"\"\"\n\n def __init__(self, config: ProphetNetConfig, word_embeddings: nn.Embedding = None):\n super().__init__(config)\n\n self.word_embeddings = (\n word_embeddings\n if word_embeddings is not None\n else nn.Embedding(config.vocab_size, config.hidden_size, padding_idx=config.pad_token_id)\n )\n self.position_embeddings = ProphetNetPositionalEmbeddings(config)\n self.embeddings_layer_norm = LayerNorm(config.hidden_size)\n\n self.layers = nn.ModuleList([ProphetNetEncoderLayer(config) for _ in range(config.num_encoder_layers)])\n\n self.init_weights()\n\n def get_input_embeddings(self):\n return self.word_embeddings\n\n def set_input_embeddings(self, value):\n self.word_embeddings = value\n\n @add_start_docstrings_to_model_forward(PROPHETNET_STANDALONE_INPUTS_DOCSTRING)\n @replace_return_docstrings(output_type=BaseModelOutput, config_class=_CONFIG_FOR_DOC)\n def forward(\n self,\n input_ids=None,\n attention_mask=None,\n inputs_embeds=None,\n output_attentions=None,\n output_hidden_states=None,\n return_dict=None,\n ):\n r\"\"\"\n Returns:\n\n Example::\n\n >>> from transformers import ProphetNetTokenizer, ProphetNetEncoder\n >>> import torch\n\n >>> tokenizer = ProphetNetTokenizer.from_pretrained('microsoft/prophetnet-large-uncased')\n >>> model = ProphetNetEncoder.from_pretrained('patrickvonplaten/prophetnet-large-uncased-standalone')\n >>> inputs = tokenizer(\"Hello, my dog is cute\", return_tensors=\"pt\")\n >>> outputs = model(**inputs)\n\n >>> last_hidden_states = outputs.last_hidden_state\n \"\"\"\n\n output_attentions = output_attentions if output_attentions is not None else self.config.output_attentions\n output_hidden_states = (\n output_hidden_states if output_hidden_states is not None else self.config.output_hidden_states\n )\n return_dict = return_dict if return_dict is not None else self.config.use_return_dict\n\n if input_ids is None and inputs_embeds is None:\n raise ValueError(\"Either input_ids or inputs_embeds has to be passed.\")\n elif input_ids is not None and inputs_embeds is not None:\n raise ValueError(\"Make sure to only pass input_ids or inputs_embeds.\")\n elif input_ids is not None and inputs_embeds is None:\n inputs_embeds = self.word_embeddings(input_ids)\n\n # prepare attention mask\n if attention_mask is not None:\n extended_attention_mask = (\n 1.0 - attention_mask[:, None, :].repeat(self.config.num_encoder_attention_heads, 1, 1)\n ) * -10000.0\n extended_attention_mask = extended_attention_mask.to(inputs_embeds.dtype)\n else:\n extended_attention_mask = None\n\n position_embeddings, position_ids = self.position_embeddings(inputs_embeds.shape[:2], inputs_embeds.device)\n\n hidden_states = inputs_embeds + position_embeddings\n hidden_states = self.embeddings_layer_norm(hidden_states)\n hidden_states = F.dropout(hidden_states, p=self.config.dropout, training=self.training)\n\n encoder_hidden_states = () if output_hidden_states else None\n all_attentions = () if output_attentions else None\n\n for encoder_layer in self.layers:\n if output_hidden_states:\n encoder_hidden_states = encoder_hidden_states + (hidden_states,)\n\n if getattr(self.config, \"gradient_checkpointing\", False) and self.training:\n\n def create_custom_forward(module):\n def custom_forward(*inputs):\n return module(*inputs, output_attentions)\n\n return custom_forward\n\n layer_outputs = torch.utils.checkpoint.checkpoint(\n create_custom_forward(encoder_layer),\n hidden_states,\n extended_attention_mask,\n )\n else:\n layer_outputs = encoder_layer(\n hidden_states, attention_mask=extended_attention_mask, output_attentions=output_attentions\n )\n\n hidden_states = layer_outputs[0]\n\n if output_attentions:\n all_attentions = all_attentions + (layer_outputs[1],)\n\n if output_hidden_states:\n encoder_hidden_states = encoder_hidden_states + (hidden_states,)\n\n if not return_dict:\n return tuple(v for v in [hidden_states, encoder_hidden_states, all_attentions] if v is not None)\n return BaseModelOutput(\n last_hidden_state=hidden_states, hidden_states=encoder_hidden_states, attentions=all_attentions\n )\n\n\n@add_start_docstrings(\n \"The standalone decoder part of the ProphetNetModel.\",\n PROPHETNET_START_DOCSTRING,\n)\nclass ProphetNetDecoder(ProphetNetPreTrainedModel):\n r\"\"\"\n word_embeddings (:obj:`torch.nn.Embeddings` of shape :obj:`(config.vocab_size, config.hidden_size)`, `optional`):\n The word embedding parameters. This can be used to initialize :class:`~transformers.ProphetNetEncoder` with\n pre-defined word embeddings instead of randomely initialized word embeddings.\n \"\"\"\n\n def __init__(self, config: ProphetNetConfig, word_embeddings: nn.Embedding = None):\n super().__init__(config)\n\n self.ngram = config.ngram\n self.num_buckets = config.num_buckets\n self.relative_max_distance = config.relative_max_distance\n self.dropout = config.dropout\n self.max_target_positions = config.max_position_embeddings\n\n self.word_embeddings = (\n word_embeddings\n if word_embeddings is not None\n else nn.Embedding(config.vocab_size, config.hidden_size, padding_idx=config.pad_token_id)\n )\n self.position_embeddings = ProphetNetPositionalEmbeddings(config)\n\n self.ngram_embeddings = nn.Embedding(self.ngram, config.hidden_size, None)\n self.layers = nn.ModuleList([ProphetNetDecoderLayer(config) for _ in range(config.num_decoder_layers)])\n self.embeddings_layer_norm = LayerNorm(config.hidden_size)\n\n self.init_weights()\n\n def get_input_embeddings(self):\n return self.word_embeddings\n\n def set_input_embeddings(self, value):\n self.word_embeddings = value\n\n @add_start_docstrings_to_model_forward(PROPHETNET_STANDALONE_INPUTS_DOCSTRING)\n @replace_return_docstrings(output_type=ProphetNetDecoderModelOutput, config_class=_CONFIG_FOR_DOC)\n def forward(\n self,\n input_ids=None,\n attention_mask=None,\n encoder_hidden_states=None,\n encoder_attention_mask=None,\n past_key_values=None,\n inputs_embeds=None,\n use_cache=None,\n output_attentions=None,\n output_hidden_states=None,\n return_dict=None,\n ):\n r\"\"\"\n encoder_hidden_states (:obj:`torch.FloatTensor` of shape :obj:`(batch_size, sequence_length, hidden_size)`, `optional`):\n Sequence of hidden-states at the output of the last layer of the encoder. Used in the cross-attention if\n the model is configured as a decoder.\n encoder_attention_mask (:obj:`torch.FloatTensor` of shape :obj:`(batch_size, sequence_length)`, `optional`):\n Mask to avoid performing attention on the padding token indices of the encoder input. This mask is used in\n the cross-attention if the model is configured as a decoder. Mask values selected in ``[0, 1]``:\n past_key_values (:obj:`tuple(tuple(torch.FloatTensor))` of length :obj:`config.n_layers` with each tuple having 4 tensors of shape :obj:`(batch_size, num_heads, sequence_length - 1, embed_size_per_head)`):\n Contains precomputed key and value hidden-states of the attention blocks. Can be used to speed up decoding.\n\n If :obj:`past_key_values` are used, the user can optionally input only the last ``decoder_input_ids``\n (those that don't have their past key value states given to this model) of shape :obj:`(batch_size, 1)`\n instead of all ``decoder_input_ids`` of shape :obj:`(batch_size, sequence_length)`.\n use_cache (:obj:`bool`, `optional`):\n If set to :obj:`True`, :obj:`past_key_values` key value states are returned and can be used to speed up\n decoding (see :obj:`past_key_values`).\n\n - 1 for tokens that are **not masked**,\n - 0 for tokens that are **masked**.\n\n Returns:\n\n Example::\n\n >>> from transformers import ProphetNetTokenizer, ProphetNetDecoder\n >>> import torch\n\n >>> tokenizer = ProphetNetTokenizer.from_pretrained('microsoft/prophetnet-large-uncased')\n >>> model = ProphetNetDecoder.from_pretrained('microsoft/prophetnet-large-uncased', add_cross_attention=False)\n >>> assert model.config.is_decoder, f\"{model.__class__} has to be configured as a decoder.\"\n >>> inputs = tokenizer(\"Hello, my dog is cute\", return_tensors=\"pt\")\n >>> outputs = model(**inputs)\n\n >>> last_hidden_states = outputs.last_hidden_state\n \"\"\"\n use_cache = use_cache if use_cache is not None else self.config.use_cache\n output_attentions = output_attentions if output_attentions is not None else self.config.output_attentions\n output_hidden_states = (\n output_hidden_states if output_hidden_states is not None else self.config.output_hidden_states\n )\n return_dict = return_dict if return_dict is not None else self.config.use_return_dict\n\n if input_ids is None and inputs_embeds is None:\n raise ValueError(\"Either `decoder_input_ids` or `decoder_inputs_embeds` has to be passed.\")\n elif input_ids is not None and inputs_embeds is not None:\n raise ValueError(\"Make sure to only pass `decoder_input_ids` or `decoder_inputs_embeds`.\")\n elif input_ids is not None and inputs_embeds is None:\n inputs_embeds = self.word_embeddings(input_ids)\n\n batch_size, sequence_length = inputs_embeds.shape[:2]\n\n main_stream_pos_embed, position_ids = self.position_embeddings(\n (batch_size, sequence_length),\n device=inputs_embeds.device,\n past_key_values=past_key_values,\n )\n\n if past_key_values is not None:\n main_relative_position_buckets, predict_relative_position_buckets = None, None\n else:\n (\n main_relative_position_buckets,\n predict_relative_position_buckets,\n ) = self.compute_buffered_relative_buckets(position_ids)\n predicting_stream_pos_embed = self.position_embeddings._forward(position_ids + 1)\n\n # add position embeddings\n hidden_states = inputs_embeds + main_stream_pos_embed\n\n ngram_embeddings = self.ngram_embeddings.weight\n\n # prepare attention mask\n if past_key_values is not None:\n assert (\n hidden_states.size(1) == 1\n ), \"At the moment `use_cache` is only supported for `decoder_input_ids` of length 1\"\n\n ngram_hidden_states = [\n (ngram_embeddings[ngram - 1] + predicting_stream_pos_embed).repeat(batch_size, 1, 1)\n for ngram in range(self.ngram)\n ]\n extended_attention_mask = None\n extended_predict_attention_mask = None\n else:\n ngram_hidden_states = [\n (ngram_embeddings[ngram - 1] + predicting_stream_pos_embed) for ngram in range(self.ngram)\n ]\n extended_attention_mask = self.prepare_attention_mask(hidden_states, attention_mask)\n extended_predict_attention_mask = self.prepare_predict_attention_mask(hidden_states, attention_mask)\n\n # prepare encoder attention mask\n if encoder_attention_mask is not None:\n extended_encoder_attention_mask = (\n 1.0 - encoder_attention_mask[:, None, :].repeat(self.config.num_decoder_attention_heads, 1, 1)\n ) * -10000.0\n extended_encoder_attention_mask = extended_encoder_attention_mask.to(inputs_embeds.dtype)\n else:\n extended_encoder_attention_mask = None\n\n hidden_states = torch.cat([hidden_states] + ngram_hidden_states, 1)\n\n if self.embeddings_layer_norm:\n hidden_states = self.embeddings_layer_norm(hidden_states)\n\n hidden_states = F.dropout(hidden_states, p=self.dropout, training=self.training)\n\n # init attentions, hidden_states and cache with empty tuples\n all_main_stream_hidden_states = () if output_hidden_states else None\n all_ngram_stream_hidden_states = () if output_hidden_states and self.config.ngram > 0 else None\n\n all_main_stream_attns = () if output_attentions else None\n all_ngram_stream_attns = () if output_attentions else None\n all_cross_attns = () if output_attentions and self.config.add_cross_attention else None\n present_key_values = () if use_cache else None\n\n for idx, decoder_layer in enumerate(self.layers):\n if output_hidden_states:\n # grad cannot be kept because tensor is sliced\n all_main_stream_hidden_states += (hidden_states[:, :sequence_length],)\n if self.config.ngram > 0:\n all_ngram_stream_hidden_states += (hidden_states[:, sequence_length:],)\n\n past_key_value = past_key_values[idx] if past_key_values is not None else None\n\n if getattr(self.config, \"gradient_checkpointing\", False) and self.training:\n\n if use_cache:\n logger.warn(\n \"`use_cache=True` is incompatible with `config.gradient_checkpointing=True`. Setting \"\n \"`use_cache=False`...\"\n )\n use_cache = False\n\n def create_custom_forward(module):\n def custom_forward(*inputs):\n # None for past_key_value\n return module(*inputs, use_cache, output_attentions)\n\n return custom_forward\n\n layer_outputs = torch.utils.checkpoint.checkpoint(\n create_custom_forward(decoder_layer),\n hidden_states,\n extended_attention_mask,\n encoder_hidden_states,\n extended_encoder_attention_mask,\n extended_predict_attention_mask,\n main_relative_position_buckets,\n predict_relative_position_buckets,\n position_ids,\n None,\n )\n else:\n layer_outputs = decoder_layer(\n hidden_states,\n attention_mask=extended_attention_mask,\n encoder_hidden_states=encoder_hidden_states,\n encoder_attn_mask=extended_encoder_attention_mask,\n extended_predict_attention_mask=extended_predict_attention_mask,\n main_relative_position_buckets=main_relative_position_buckets,\n predict_relative_position_buckets=predict_relative_position_buckets,\n position_ids=position_ids,\n past_key_value=past_key_value,\n use_cache=use_cache,\n output_attentions=output_attentions,\n )\n\n hidden_states = layer_outputs[0]\n\n if use_cache:\n present_key_values += (layer_outputs[4 if output_attentions else 1],)\n\n if output_attentions:\n all_main_stream_attns += (layer_outputs[1],)\n all_ngram_stream_attns += (layer_outputs[2],)\n\n if self.config.add_cross_attention:\n all_cross_attns += (layer_outputs[3],)\n\n if output_hidden_states:\n all_main_stream_hidden_states += (hidden_states[:, :sequence_length],)\n if self.config.ngram > 0:\n all_ngram_stream_hidden_states += (hidden_states[:, sequence_length:],)\n\n # split last_hidden_state for return\n last_hidden_state = hidden_states[:, :sequence_length]\n last_hidden_state_ngram = hidden_states[:, sequence_length:] if self.config.ngram > 0 else None\n\n if not return_dict:\n return tuple(\n v\n for v in [\n last_hidden_state,\n last_hidden_state_ngram,\n present_key_values,\n all_main_stream_hidden_states,\n all_ngram_stream_hidden_states,\n all_main_stream_attns,\n all_ngram_stream_attns,\n all_cross_attns,\n ]\n if v is not None\n )\n return ProphetNetDecoderModelOutput(\n last_hidden_state=last_hidden_state,\n last_hidden_state_ngram=last_hidden_state_ngram,\n past_key_values=present_key_values,\n hidden_states=all_main_stream_hidden_states,\n hidden_states_ngram=all_ngram_stream_hidden_states,\n attentions=all_main_stream_attns,\n ngram_attentions=all_ngram_stream_attns,\n cross_attentions=all_cross_attns,\n )\n\n def compute_buffered_relative_buckets(self, position_ids):\n batch_size, sequence_length = position_ids.shape\n\n position_ids = torch.arange(1, self.max_target_positions).to(position_ids.device).repeat(1, 1)\n main_relative_buckets, predict_relative_buckets = compute_all_stream_relative_buckets(\n self.num_buckets, self.relative_max_distance, position_ids\n )\n\n # buffer relative buckets\n main_relative_buckets = main_relative_buckets[:, :sequence_length, :sequence_length].repeat(batch_size, 1, 1)\n predict_relative_buckets = torch.cat(\n [\n predict_relative_buckets[:, :sequence_length, :sequence_length],\n predict_relative_buckets[\n :, :sequence_length, self.max_target_positions : self.max_target_positions + sequence_length\n ],\n ],\n 2,\n ).repeat(batch_size, 1, 1)\n\n return main_relative_buckets, predict_relative_buckets\n\n def prepare_attention_mask(self, hidden_states, attention_mask):\n batch_size, seq_length = hidden_states.shape[:2]\n\n # get causal mask\n causal_mask = hidden_states.new(seq_length, seq_length).float().fill_(-float(\"inf\"))\n causal_mask = torch.triu(causal_mask, 1)\n extended_causal_mask = causal_mask[:seq_length, :seq_length][None, :, :].expand(\n (batch_size,) + causal_mask.shape\n )\n\n # add usual attention mask\n if attention_mask is not None:\n extended_attention_mask = (1.0 - attention_mask[:, None, :]) * -10000.0\n extended_attention_mask = extended_causal_mask + extended_attention_mask\n else:\n extended_attention_mask = extended_causal_mask\n return extended_attention_mask.repeat(self.config.num_decoder_attention_heads, 1, 1).to(hidden_states.dtype)\n\n def prepare_predict_attention_mask(self, hidden_states, attention_mask):\n batch_size, seq_length = hidden_states.shape[:2]\n\n # get causal mask\n predict_causal_mask = ngram_attention_bias(\n self.max_target_positions, self.ngram, hidden_states.device, hidden_states.dtype\n )\n predict_causal_mask = torch.cat(\n [\n predict_causal_mask[:, :seq_length, :seq_length],\n predict_causal_mask[\n :, :seq_length, self.max_target_positions : self.max_target_positions + seq_length\n ],\n ],\n dim=-1,\n )\n extended_predict_causal_mask = predict_causal_mask[:, None, :, :].expand(\n predict_causal_mask.shape[:1] + (batch_size,) + predict_causal_mask.shape[1:]\n )\n\n # add usual attention mask\n if attention_mask is not None:\n extended_attention_mask = (1.0 - attention_mask[None, :, None, :]) * -10000.0\n extended_attention_mask = extended_attention_mask.expand((self.ngram, batch_size, seq_length, seq_length))\n # predicted stream attention_mask should always be 0\n extended_attention_mask = torch.cat(\n [extended_attention_mask, torch.zeros_like(extended_attention_mask)], dim=-1\n )\n extended_predict_attention_mask = extended_predict_causal_mask + extended_attention_mask\n else:\n extended_predict_attention_mask = extended_predict_causal_mask\n return extended_predict_attention_mask.repeat(1, self.config.num_decoder_attention_heads, 1, 1).to(\n hidden_states.dtype\n )\n\n\n@add_start_docstrings(\n \"The bare ProphetNet Model outputting raw hidden-states without any specific head on top.\",\n PROPHETNET_START_DOCSTRING,\n)\nclass ProphetNetModel(ProphetNetPreTrainedModel):\n def __init__(self, config):\n super().__init__(config)\n self.word_embeddings = nn.Embedding(config.vocab_size, config.hidden_size, padding_idx=config.pad_token_id)\n\n encoder_config = copy.deepcopy(config)\n encoder_config.is_encoder_decoder = False\n encoder_config.use_cache = False\n self.encoder = ProphetNetEncoder(encoder_config, self.word_embeddings)\n\n decoder_config = copy.deepcopy(config)\n decoder_config.is_decoder = True\n decoder_config.is_encoder_decoder = False\n self.decoder = ProphetNetDecoder(decoder_config, self.word_embeddings)\n\n self.init_weights()\n\n def get_input_embeddings(self):\n return self.word_embeddings\n\n def set_input_embeddings(self, value):\n self.word_embeddings = value\n self.encoder.word_embeddings = self.word_embeddings\n self.decoder.word_embeddings = self.word_embeddings\n\n def get_encoder(self):\n return self.encoder\n\n def get_decoder(self):\n return self.decoder\n\n @add_start_docstrings_to_model_forward(PROPHETNET_INPUTS_DOCSTRING)\n @replace_return_docstrings(output_type=ProphetNetSeq2SeqModelOutput, config_class=_CONFIG_FOR_DOC)\n def forward(\n self,\n input_ids=None,\n attention_mask=None,\n decoder_input_ids=None,\n decoder_attention_mask=None,\n encoder_outputs: Optional[Tuple] = None,\n past_key_values=None,\n inputs_embeds=None,\n decoder_inputs_embeds=None,\n use_cache=None,\n output_attentions=None,\n output_hidden_states=None,\n return_dict=None,\n ):\n r\"\"\"\n Returns:\n\n Example::\n\n >>> from transformers import ProphetNetTokenizer, ProphetNetModel\n\n >>> tokenizer = ProphetNetTokenizer.from_pretrained('microsoft/prophetnet-large-uncased')\n >>> model = ProphetNetModel.from_pretrained('microsoft/prophetnet-large-uncased')\n\n >>> input_ids = tokenizer(\"Studies have been shown that owning a dog is good for you\", return_tensors=\"pt\").input_ids # Batch size 1\n >>> decoder_input_ids = tokenizer(\"Studies show that\", return_tensors=\"pt\").input_ids # Batch size 1\n >>> outputs = model(input_ids=input_ids, decoder_input_ids=decoder_input_ids)\n\n >>> last_hidden_states = outputs.last_hidden_state # main stream hidden states\n >>> last_hidden_states_ngram = outputs.last_hidden_state_ngram # predict hidden states\n \"\"\"\n\n use_cache == use_cache if use_cache is not None else self.config.use_cache\n output_attentions = output_attentions if output_attentions is not None else self.config.output_attentions\n output_hidden_states = (\n output_hidden_states if output_hidden_states is not None else self.config.output_hidden_states\n )\n return_dict = return_dict if return_dict is not None else self.config.use_return_dict\n\n if encoder_outputs is None:\n encoder_outputs = self.encoder(\n input_ids=input_ids,\n attention_mask=attention_mask,\n inputs_embeds=inputs_embeds,\n output_attentions=output_attentions,\n output_hidden_states=output_hidden_states,\n return_dict=return_dict,\n )\n\n # decoder outputs consists of (dec_features, past_key_values, dec_hidden, dec_attn)\n decoder_outputs = self.decoder(\n input_ids=decoder_input_ids,\n attention_mask=decoder_attention_mask,\n encoder_hidden_states=encoder_outputs[0],\n encoder_attention_mask=attention_mask,\n past_key_values=past_key_values,\n inputs_embeds=decoder_inputs_embeds,\n output_attentions=output_attentions,\n output_hidden_states=output_hidden_states,\n use_cache=use_cache,\n return_dict=return_dict,\n )\n\n if not return_dict:\n return decoder_outputs + encoder_outputs\n return ProphetNetSeq2SeqModelOutput(\n last_hidden_state=decoder_outputs.last_hidden_state,\n last_hidden_state_ngram=decoder_outputs.last_hidden_state_ngram,\n past_key_values=decoder_outputs.past_key_values,\n decoder_hidden_states=decoder_outputs.hidden_states,\n decoder_ngram_hidden_states=decoder_outputs.hidden_states_ngram,\n decoder_attentions=decoder_outputs.attentions,\n decoder_ngram_attentions=decoder_outputs.ngram_attentions,\n cross_attentions=decoder_outputs.cross_attentions,\n encoder_last_hidden_state=encoder_outputs.last_hidden_state,\n encoder_hidden_states=encoder_outputs.hidden_states,\n encoder_attentions=encoder_outputs.attentions,\n )\n\n\n@add_start_docstrings(\n \"The ProphetNet Model with a language modeling head. Can be used for sequence generation tasks.\",\n PROPHETNET_START_DOCSTRING,\n)\nclass ProphetNetForConditionalGeneration(ProphetNetPreTrainedModel):\n def __init__(self, config: ProphetNetConfig):\n super().__init__(config)\n self.prophetnet = ProphetNetModel(config)\n self.padding_idx = config.pad_token_id\n self.disable_ngram_loss = config.disable_ngram_loss\n\n self.lm_head = nn.Linear(config.hidden_size, config.vocab_size, bias=False)\n\n self.init_weights()\n\n def get_output_embeddings(self):\n return self.lm_head\n\n def set_output_embeddings(self, new_embeddings):\n self.lm_head = new_embeddings\n\n def get_input_embeddings(self):\n return self.prophetnet.word_embeddings\n\n @add_start_docstrings_to_model_forward(PROPHETNET_INPUTS_DOCSTRING)\n @replace_return_docstrings(output_type=ProphetNetSeq2SeqLMOutput, config_class=_CONFIG_FOR_DOC)\n def forward(\n self,\n input_ids=None,\n attention_mask=None,\n decoder_input_ids=None,\n decoder_attention_mask=None,\n encoder_outputs=None,\n past_key_values=None,\n inputs_embeds=None,\n decoder_inputs_embeds=None,\n labels=None,\n use_cache=None,\n output_attentions=None,\n output_hidden_states=None,\n return_dict=None,\n ):\n r\"\"\"\n labels (:obj:`torch.LongTensor` of shape :obj:`(batch_size,)`, `optional`):\n Labels for computing the sequence classification/regression loss. Indices should be in :obj:`[-100, 0, ...,\n config.vocab_size - 1]`. All labels set to ``-100`` are ignored (masked), the loss is only computed for\n labels in ``[0, ..., config.vocab_size]``\n\n Returns:\n\n Example::\n\n >>> from transformers import ProphetNetTokenizer, ProphetNetForConditionalGeneration\n\n >>> tokenizer = ProphetNetTokenizer.from_pretrained('microsoft/prophetnet-large-uncased')\n >>> model = ProphetNetForConditionalGeneration.from_pretrained('microsoft/prophetnet-large-uncased')\n\n >>> input_ids = tokenizer(\"Studies have been shown that owning a dog is good for you\", return_tensors=\"pt\").input_ids # Batch size 1\n >>> decoder_input_ids = tokenizer(\"Studies show that\", return_tensors=\"pt\").input_ids # Batch size 1\n >>> outputs = model(input_ids=input_ids, decoder_input_ids=decoder_input_ids)\n\n >>> logits_next_token = outputs.logits # logits to predict next token as usual\n >>> logits_ngram_next_tokens = outputs.logits_ngram # logits to predict 2nd, 3rd, ... next tokens\n \"\"\"\n return_dict = return_dict if return_dict is not None else self.config.use_return_dict\n\n if labels is not None and decoder_input_ids is None and decoder_inputs_embeds is None:\n # get decoder inputs from shifting lm labels to the right\n decoder_input_ids = self._shift_right(labels)\n\n outputs = self.prophetnet(\n input_ids=input_ids,\n attention_mask=attention_mask,\n decoder_input_ids=decoder_input_ids,\n decoder_attention_mask=decoder_attention_mask,\n encoder_outputs=encoder_outputs,\n past_key_values=past_key_values,\n inputs_embeds=inputs_embeds,\n decoder_inputs_embeds=decoder_inputs_embeds,\n use_cache=use_cache,\n output_attentions=output_attentions,\n output_hidden_states=output_hidden_states,\n return_dict=return_dict,\n )\n batch_size, sequence_length = (\n decoder_input_ids.shape if decoder_input_ids is not None else decoder_inputs_embeds.shape[:2]\n )\n\n predicting_streams = outputs[1].view(batch_size, self.config.ngram, sequence_length, -1)\n predict_logits = self.lm_head(predicting_streams)\n\n logits = predict_logits[:, 0]\n logits_ngram = predict_logits[:, 1:] if self.config.ngram > 1 else None\n\n # To use .view in loss computation, make sure that logits is contiguous.\n if not logits.is_contiguous():\n logits = logits.contiguous()\n\n loss = None\n if labels is not None:\n loss = self._compute_loss(predict_logits, labels)\n\n if not return_dict:\n all_logits = tuple(v for v in [logits, logits_ngram] if v is not None)\n return (loss,) + all_logits + outputs[2:] if loss is not None else all_logits + outputs[2:]\n else:\n return ProphetNetSeq2SeqLMOutput(\n loss=loss,\n logits=logits,\n logits_ngram=logits_ngram,\n past_key_values=outputs.past_key_values,\n decoder_hidden_states=outputs.decoder_hidden_states,\n decoder_ngram_hidden_states=outputs.decoder_ngram_hidden_states,\n decoder_attentions=outputs.decoder_attentions,\n decoder_ngram_attentions=outputs.decoder_ngram_attentions,\n cross_attentions=outputs.cross_attentions,\n encoder_last_hidden_state=outputs.encoder_last_hidden_state,\n encoder_hidden_states=outputs.encoder_hidden_states,\n encoder_attentions=outputs.encoder_attentions,\n )\n\n def _compute_loss(self, logits, labels, ignore_index=-100):\n expend_targets = labels.new_zeros(self.config.ngram, labels.size(0), labels.size(1)).fill_(ignore_index)\n\n for i in range(self.config.ngram):\n if i > 0 and self.disable_ngram_loss:\n break\n expend_targets[i, :, :] = labels\n\n lprobs = F.log_softmax(\n logits.view(-1, logits.size(-1)),\n dim=-1,\n dtype=torch.float32,\n )\n\n loss = F.nll_loss(lprobs, expend_targets.view(-1), reduction=\"mean\")\n\n if self.config.eps > 0.0:\n smooth_loss = -lprobs.sum(dim=-1, keepdim=True)\n non_masked_tokens = expend_targets.ne(ignore_index).view(-1)\n smooth_loss = smooth_loss[non_masked_tokens]\n smooth_loss = smooth_loss.mean()\n\n eps_i = self.config.eps / lprobs.size(-1)\n loss = (1.0 - self.config.eps) * loss + eps_i * smooth_loss\n\n return loss\n\n def prepare_inputs_for_generation(\n self, decoder_input_ids, past=None, attention_mask=None, use_cache=None, encoder_outputs=None, **kwargs\n ):\n assert encoder_outputs is not None, \"`encoder_outputs` have to be passed for generation.\"\n\n if past:\n decoder_input_ids = decoder_input_ids[:, -1:]\n # first step, decoder_cached_states are empty\n return {\n \"input_ids\": None, # encoder_outputs is defined. input_ids not needed\n \"encoder_outputs\": encoder_outputs,\n \"past_key_values\": past,\n \"decoder_input_ids\": decoder_input_ids,\n \"attention_mask\": attention_mask,\n \"use_cache\": use_cache,\n }\n\n def prepare_decoder_input_ids_from_labels(self, labels: torch.Tensor):\n return self._shift_right(labels)\n\n @staticmethod\n # Copied from transformers.models.bart.modeling_bart.BartForConditionalGeneration._reorder_cache\n def _reorder_cache(past, beam_idx):\n reordered_past = ()\n for layer_past in past:\n # cached cross_attention states don't have to be reordered -> they are always the same\n reordered_past += (\n tuple(past_state.index_select(0, beam_idx) for past_state in layer_past[:2]) + layer_past[2:],\n )\n return reordered_past\n\n def get_encoder(self):\n return self.prophetnet.encoder\n\n def get_decoder(self):\n return self.prophetnet.decoder\n\n\n@add_start_docstrings(\n \"The standalone decoder part of the ProphetNetModel with a lm head on top. The model can be used for causal language modeling.\",\n PROPHETNET_START_DOCSTRING,\n)\nclass ProphetNetForCausalLM(ProphetNetPreTrainedModel):\n def __init__(self, config):\n # set config for CLM\n config = copy.deepcopy(config)\n config.is_decoder = True\n config.is_encoder_decoder = False\n super().__init__(config)\n self.prophetnet = ProphetNetDecoderWrapper(config)\n\n self.padding_idx = config.pad_token_id\n self.disable_ngram_loss = config.disable_ngram_loss\n\n self.lm_head = nn.Linear(config.hidden_size, config.vocab_size, bias=False)\n\n self.init_weights()\n\n def get_input_embeddings(self):\n return self.prophetnet.decoder.word_embeddings\n\n def set_input_embeddings(self, value):\n self.prophetnet.decoder.word_embeddings = value\n\n def get_output_embeddings(self):\n return self.lm_head\n\n def set_output_embeddings(self, new_embeddings):\n self.lm_head = new_embeddings\n\n def set_decoder(self, decoder):\n self.prophetnet.decoder = decoder\n\n def get_decoder(self):\n return self.prophetnet.decoder\n\n @add_start_docstrings_to_model_forward(PROPHETNET_STANDALONE_INPUTS_DOCSTRING)\n @replace_return_docstrings(output_type=ProphetNetDecoderLMOutput, config_class=_CONFIG_FOR_DOC)\n def forward(\n self,\n input_ids=None,\n attention_mask=None,\n encoder_hidden_states=None,\n encoder_attention_mask=None,\n past_key_values=None,\n inputs_embeds=None,\n labels=None,\n use_cache=None,\n output_attentions=None,\n output_hidden_states=None,\n return_dict=None,\n ):\n r\"\"\"\n encoder_hidden_states (:obj:`torch.FloatTensor` of shape :obj:`(batch_size, sequence_length, hidden_size)`, `optional`):\n Sequence of hidden-states at the output of the last layer of the encoder. Used in the cross-attention if\n the model is configured as a decoder.\n encoder_attention_mask (:obj:`torch.FloatTensor` of shape :obj:`(batch_size, sequence_length)`, `optional`):\n Mask to avoid performing attention on the padding token indices of the encoder input. This mask is used in\n the cross-attention if the model is configured as a decoder. Mask values selected in ``[0, 1]``:\n past_key_values (:obj:`tuple(tuple(torch.FloatTensor))` of length :obj:`config.n_layers` with each tuple having 4 tensors of shape :obj:`(batch_size, num_heads, sequence_length - 1, embed_size_per_head)`):\n Contains precomputed key and value hidden-states of the attention blocks. Can be used to speed up decoding.\n\n If :obj:`past_key_values` are used, the user can optionally input only the last ``decoder_input_ids``\n (those that don't have their past key value states given to this model) of shape :obj:`(batch_size, 1)`\n instead of all ``decoder_input_ids`` of shape :obj:`(batch_size, sequence_length)`.\n use_cache (:obj:`bool`, `optional`):\n If set to :obj:`True`, :obj:`past_key_values` key value states are returned and can be used to speed up\n decoding (see :obj:`past_key_values`).\n\n - 1 for tokens that are **not masked**,\n - 0 for tokens that are **masked**.\n\n labels (:obj:`torch.LongTensor` of shape :obj:`(batch_size, sequence_length)`, `optional`):\n Labels for computing the left-to-right language modeling loss (next word prediction). Indices should be in\n ``[-100, 0, ..., config.vocab_size]`` (see ``input_ids`` docstring) Tokens with indices set to ``-100`` are\n ignored (masked), the loss is only computed for the tokens with labels n ``[0, ..., config.vocab_size]``\n\n Returns:\n\n Example::\n\n >>> from transformers import ProphetNetTokenizer, ProphetNetForCausalLM\n >>> import torch\n\n >>> tokenizer = ProphetNetTokenizer.from_pretrained('microsoft/prophetnet-large-uncased')\n >>> model = ProphetNetForCausalLM.from_pretrained('microsoft/prophetnet-large-uncased')\n >>> assert model.config.is_decoder, f\"{model.__class__} has to be configured as a decoder.\"\n >>> inputs = tokenizer(\"Hello, my dog is cute\", return_tensors=\"pt\")\n >>> outputs = model(**inputs)\n\n >>> logits = outputs.logits\n\n >>> # Model can also be used with EncoderDecoder framework\n >>> from transformers import BertTokenizer, EncoderDecoderModel, ProphetNetTokenizer\n >>> import torch\n\n >>> tokenizer_enc = BertTokenizer.from_pretrained('bert-large-uncased')\n >>> tokenizer_dec = ProphetNetTokenizer.from_pretrained('microsoft/prophetnet-large-uncased')\n >>> model = EncoderDecoderModel.from_encoder_decoder_pretrained(\"bert-large-uncased\", \"microsoft/prophetnet-large-uncased\")\n\n >>> ARTICLE = (\n ... \"the us state department said wednesday it had received no \"\n ... \"formal word from bolivia that it was expelling the us ambassador there \"\n ... \"but said the charges made against him are `` baseless .\"\n ... )\n >>> input_ids = tokenizer_enc(ARTICLE, return_tensors=\"pt\").input_ids\n >>> labels = tokenizer_dec(\"us rejects charges against its ambassador in bolivia\", return_tensors=\"pt\").input_ids\n >>> outputs = model(input_ids=input_ids, decoder_input_ids=labels[:, :-1], labels=labels[:, 1:])\n\n >>> loss = outputs.loss\n \"\"\"\n return_dict = return_dict if return_dict is not None else self.config.use_return_dict\n\n # decoder outputs consists of (dec_features, past_key_values, dec_hidden, dec_attn)\n outputs = self.prophetnet.decoder(\n input_ids=input_ids,\n attention_mask=attention_mask,\n encoder_hidden_states=encoder_hidden_states,\n encoder_attention_mask=encoder_attention_mask,\n past_key_values=past_key_values,\n inputs_embeds=inputs_embeds,\n use_cache=use_cache,\n output_attentions=output_attentions,\n output_hidden_states=output_hidden_states,\n return_dict=return_dict,\n )\n\n batch_size, sequence_length = input_ids.shape if input_ids is not None else inputs_embeds.shape[:2]\n\n predicting_streams = outputs[1].view(batch_size, self.config.ngram, sequence_length, -1)\n predict_logits = self.lm_head(predicting_streams)\n\n logits = predict_logits[:, 0]\n logits_ngram = predict_logits[:, 1:] if self.config.ngram > 1 else None\n\n loss = None\n if labels is not None:\n loss = self._compute_loss(predict_logits, labels)\n\n if not return_dict:\n all_logits = tuple(v for v in [logits, logits_ngram] if v is not None)\n return (loss,) + all_logits + outputs[2:] if loss is not None else all_logits + outputs[2:]\n else:\n return ProphetNetDecoderLMOutput(\n loss=loss,\n logits=logits,\n logits_ngram=logits_ngram,\n past_key_values=outputs.past_key_values,\n hidden_states=outputs.hidden_states,\n hidden_states_ngram=outputs.hidden_states_ngram,\n attentions=outputs.attentions,\n ngram_attentions=outputs.ngram_attentions,\n cross_attentions=outputs.cross_attentions,\n )\n\n def _compute_loss(self, logits, labels, ignore_index=-100):\n expend_targets = labels.new_zeros(self.config.ngram, labels.size(0), labels.size(1)).fill_(ignore_index)\n\n for i in range(self.config.ngram):\n if i > 0 and self.disable_ngram_loss:\n break\n expend_targets[i, :, :] = labels\n\n lprobs = F.log_softmax(\n logits.view(-1, logits.size(-1)),\n dim=-1,\n dtype=torch.float32,\n )\n\n loss = F.nll_loss(lprobs, expend_targets.view(-1), reduction=\"mean\")\n\n if self.config.eps > 0.0:\n smooth_loss = -lprobs.sum(dim=-1, keepdim=True)\n non_masked_tokens = expend_targets.ne(ignore_index).view(-1)\n smooth_loss = smooth_loss[non_masked_tokens]\n smooth_loss = smooth_loss.mean()\n\n eps_i = self.config.eps / lprobs.size(-1)\n loss = (1.0 - self.config.eps) * loss + eps_i * smooth_loss\n\n return loss\n\n def prepare_inputs_for_generation(self, input_ids, past=None, attention_mask=None, use_cache=None, **kwargs):\n # if model is used as a decoder in encoder-decoder model, the decoder attention mask is created on the fly\n if attention_mask is None:\n attention_mask = input_ids.new_ones(input_ids.shape)\n\n if past:\n input_ids = input_ids[:, -1:]\n # first step, decoder_cached_states are empty\n return {\n \"input_ids\": input_ids, # encoder_outputs is defined. input_ids not needed\n \"attention_mask\": attention_mask,\n \"past_key_values\": past,\n \"use_cache\": use_cache,\n }\n\n @staticmethod\n # Copied from transformers.models.bart.modeling_bart.BartForCausalLM._reorder_cache\n def _reorder_cache(past, beam_idx):\n reordered_past = ()\n for layer_past in past:\n reordered_past += (tuple(past_state.index_select(0, beam_idx) for past_state in layer_past),)\n return reordered_past\n\n\nclass ProphetNetDecoderWrapper(ProphetNetPreTrainedModel):\n \"\"\"\n This is a wrapper class, so that :class:`~transformers.ProphetNetForCausalLM` can correctly be loaded from\n pretrained prophetnet classes.\n \"\"\"\n\n def __init__(self, config):\n super().__init__(config)\n self.decoder = ProphetNetDecoder(config)\n\n def forward(self, *args, **kwargs):\n return self.decoder(*args, **kwargs)\n"
] |
[
[
"torch.nn.Linear",
"torch.cat",
"torch.nn.LayerNorm",
"torch.einsum",
"torch.gather",
"torch.arange",
"torch.nn.functional.dropout",
"torch.bmm",
"torch.ones",
"torch.abs",
"torch.zeros_like",
"torch.all",
"torch.ones_like",
"torch.nn.functional.softmax",
"torch.lt",
"torch.triu",
"torch.nn.Embedding",
"torch.cumsum"
]
] |
gthomas-slack/omniduct
|
[
"af0148e1f0de147c6c3757dc8c22293225caa543"
] |
[
"omniduct/databases/hiveserver2.py"
] |
[
"from __future__ import absolute_import\n\nimport json\nimport os\nimport re\nimport shutil\nimport tempfile\nimport time\n\nimport pandas as pd\nfrom interface_meta import override\nfrom jinja2 import Template\n\nfrom omniduct.utils.debug import logger\nfrom omniduct.utils.processes import Timeout, run_in_subprocess\n\nfrom .base import DatabaseClient\nfrom ._schemas import SchemasMixin\nfrom . import _pandas\n\n\nclass HiveServer2Client(DatabaseClient, SchemasMixin):\n \"\"\"\n This Duct connects to an Apache HiveServer2 server instance using the\n `pyhive` or `impyla` libraries.\n\n Attributes:\n schema (str, None): The default schema to use for queries (will\n default to server-default if not specified).\n driver (str): One of 'pyhive' (default) or 'impyla', which specifies\n how the client communicates with Hive.\n auth_mechanism (str): The authorisation protocol to use for connections.\n Defaults to 'NOSASL'. Authorisation methods differ between drivers.\n Please refer to `pyhive` and `impyla` documentation for more details.\n push_using_hive_cli (bool): Whether the `.push()` operation should\n directly add files using `LOAD DATA LOCAL INPATH` rather than the\n `INSERT` operation via SQLAlchemy. Note that this requires the\n presence of the `hive` executable on the local PATH, or if\n connecting via a `RemoteClient` instance, on the remote's PATH.\n This is mostly useful for older versions of Hive which do not\n support the `INSERT` statement.\n default_table_props (dict): A dictionary of table properties to use by\n default when creating tables.\n connection_options (dict): Additional options to pass through to the\n `.connect()` methods of the drivers.\n \"\"\"\n\n PROTOCOLS = ['hiveserver2']\n DEFAULT_PORT = 3623\n SUPPORTS_SESSION_PROPERTIES = True\n NAMESPACE_NAMES = ['schema', 'table']\n NAMESPACE_QUOTECHAR = '`'\n NAMESPACE_SEPARATOR = '.'\n\n @property\n @override\n def NAMESPACE_DEFAULT(self):\n return {\n 'schema': self.schema\n }\n\n @override\n def _init(self, schema=None, driver='pyhive', auth_mechanism='NOSASL',\n push_using_hive_cli=False, default_table_props=None,\n thrift_transport=None, **connection_options\n ):\n \"\"\"\n schema (str, None): The default database/schema to use for queries (will\n default to server-default if not specified).\n driver (str): One of 'pyhive' (default) or 'impyla', which specifies\n how the client communicates with Hive.\n auth_mechanism (str): The authorisation protocol to use for connections.\n Defaults to 'NOSASL'. Authorisation methods differ between drivers.\n Please refer to `pyhive` and `impyla` documentation for more details.\n push_using_hive_cli (bool): Whether the `.push()` operation should\n directly add files using `LOAD DATA LOCAL INPATH` rather than the\n `INSERT` operation via SQLAlchemy. Note that this requires the\n presence of the `hive` executable on the local PATH, or if\n connecting via a `RemoteClient` instance, on the remote's PATH.\n This is mostly useful for older versions of Hive which do not\n support the `INSERT` statement. False by default.\n default_table_props (dict): A dictionary of table properties to use by\n default when creating tables (default is an empty dict).\n thrift_transport (TTransportBase): A thrift transport object for custom advanced usage.\n Incompatible with host, port, auth_mechanism, and password.\n Typically used to enable Thrift http transport to hiveserver2.\n **connection_options (dict): Additional options to pass through to the\n `.connect()` methods of the drivers.\n \"\"\"\n self.schema = schema\n self.driver = driver\n self.auth_mechanism = auth_mechanism\n self.connection_options = connection_options\n self.push_using_hive_cli = push_using_hive_cli\n self.default_table_props = default_table_props or {}\n self._thrift_transport = thrift_transport\n self.__hive = None\n self.connection_fields += ('schema',)\n\n assert self.driver in ('pyhive', 'impyla'), \"Supported drivers are pyhive and impyla.\"\n\n @override\n def _connect(self):\n from sqlalchemy import create_engine, MetaData\n if self.driver == 'pyhive':\n try:\n import pyhive.hive\n except ImportError:\n raise ImportError(\"\"\"\n Omniduct is attempting to use the 'pyhive' driver, but it\n is not installed. Please either install the pyhive package,\n or reconfigure this Duct to use the 'impyla' driver.\n \"\"\")\n self.__hive = pyhive.hive.connect(host=None if self._thrift_transport else self.host,\n port=None if self._thrift_transport else self.port,\n auth=None if self._thrift_transport else self.auth_mechanism,\n database=self.schema,\n username=self.username,\n password=None if self._thrift_transport else self.password,\n thrift_transport=self._thrift_transport,\n **self.connection_options)\n self._sqlalchemy_engine = create_engine('hive://{}:{}/{}'.format(self.host, self.port, self.schema))\n self._sqlalchemy_metadata = MetaData(self._sqlalchemy_engine)\n elif self.driver == 'impyla':\n try:\n import impala.dbapi\n except ImportError:\n raise ImportError(\"\"\"\n Omniduct is attempting to use the 'impyla' driver, but it\n is not installed. Please either install the impyla package,\n or reconfigure this Duct to use the 'pyhive' driver.\n \"\"\")\n self.__hive = impala.dbapi.connect(host=self.host,\n port=self.port,\n auth_mechanism=self.auth_mechanism,\n database=self.schema,\n user=self.username,\n password=self.password,\n **self.connection_options)\n self._sqlalchemy_engine = create_engine('impala://{}:{}/{}'.format(self.host, self.port, self.schema))\n self._sqlalchemy_metadata = MetaData(self._sqlalchemy_engine)\n\n def __hive_cursor(self):\n if self.driver == 'impyla': # Impyla seems to have all manner of connection issues, attempt to restore connection\n try:\n with Timeout(1):\n return self.__hive.cursor()\n except:\n self._connect()\n return self.__hive.cursor()\n\n @override\n def _is_connected(self):\n return self.__hive is not None\n\n @override\n def _disconnect(self):\n logger.info('Disconnecting from Hive coordinator...')\n try:\n self.__hive.close()\n except:\n pass\n self.__hive = None\n self._sqlalchemy_engine = None\n self._sqlalchemy_metadata = None\n self._schemas = None\n\n @override\n def _statement_prepare(self, statement, session_properties, **kwargs):\n return (\n \"\\n\".join(\n \"SET {key} = {value};\".format(key=key, value=value)\n for key, value in session_properties.items()\n ) + statement\n )\n\n @override\n def _execute(self, statement, cursor, wait, session_properties, poll_interval=1):\n \"\"\"\n Additional Args:\n poll_interval (int): Default delay in seconds between consecutive\n query status (defaults to 1).\n \"\"\"\n cursor = cursor or self.__hive_cursor()\n log_offset = 0\n\n if self.driver == 'pyhive':\n from TCLIService.ttypes import TOperationState # noqa: F821\n cursor.execute(statement, **{'async': True})\n\n if wait:\n status = cursor.poll().operationState\n while status in (TOperationState.INITIALIZED_STATE, TOperationState.RUNNING_STATE):\n log_offset = self._log_status(cursor, log_offset)\n time.sleep(poll_interval)\n status = cursor.poll().operationState\n\n elif self.driver == 'impyla':\n cursor.execute_async(statement)\n if wait:\n while cursor.is_executing():\n log_offset = self._log_status(cursor, log_offset)\n time.sleep(poll_interval)\n\n return cursor\n\n @override\n def _cursor_empty(self, cursor):\n if self.driver == 'impyla':\n return not cursor.has_result_set\n elif self.driver == 'pyhive':\n return cursor.description is None\n return False\n\n def _cursor_wait(self, cursor, poll_interval=1):\n from TCLIService.ttypes import TOperationState # noqa: F821\n status = cursor.poll().operationState\n while status in (TOperationState.INITIALIZED_STATE, TOperationState.RUNNING_STATE):\n time.sleep(poll_interval)\n status = cursor.poll().operationState\n\n def _log_status(self, cursor, log_offset=0):\n matcher = re.compile('[0-9/]+ [0-9:]+ (INFO )?')\n\n if self.driver == 'pyhive':\n log = cursor.fetch_logs()\n else:\n log = cursor.get_log().strip().split('\\n')\n\n for line in log[log_offset:]:\n if not line:\n continue\n m = matcher.match(line)\n if m:\n line = line[len(m.group(0)):]\n logger.info(line)\n\n return len(log)\n\n @override\n def _query_to_table(self, statement, table, if_exists, **kwargs):\n statements = []\n\n if if_exists == 'fail' and self.table_exists(table):\n raise RuntimeError(\"Table {} already exists!\".format(table))\n elif if_exists == 'replace':\n statements.append('DROP TABLE IF EXISTS {};'.format(table))\n elif if_exists == 'append':\n raise NotImplementedError(\"Append operations have not been implemented for {}.\".format(self.__class__.__name__))\n\n statements.append(\"CREATE TABLE {table} AS ({statement})\".format(\n table=table,\n statement=statement\n ))\n return self.execute('\\n'.join(statements), **kwargs)\n\n @override\n def _dataframe_to_table(\n self, df, table, if_exists='fail', use_hive_cli=None,\n partition=None, sep=chr(1), table_props=None, dtype_overrides=None, **kwargs\n ):\n \"\"\"\n If `use_hive_cli` (or if not specified `.push_using_hive_cli`) is\n `True`, a `CREATE TABLE` statement will be automatically generated based\n on the datatypes of the DataFrame (unless overwritten by\n `dtype_overrides`). The `DataFrame` will then be exported to a CSV\n compatible with Hive and uploaded (if necessary) to the remote, before\n being loaded into Hive using a `LOAD DATA LOCAL INFILE ...` query using\n the `hive` cli executable. Note that if a table is not partitioned, you\n cannot convert it to a parititioned table without deleting it first.\n\n If `use_hive_cli` (or if not specified `.push_using_hive_cli`) is\n `False`, an attempt will be made to push the `DataFrame` to Hive using\n `pandas.DataFrame.to_sql` and the SQLAlchemy binding provided by\n `pyhive` and `impyla`. This may be slower, does not support older\n versions of Hive, and does not support table properties or partitioning.\n\n If if the schema namespace is not specified, `table.schema` will be\n defaulted to your username.\n\n Additional Args:\n use_hive_cli (bool, None): A local override for the global\n `.push_using_hive_cli` attribute. If not specified, the global\n default is used. If True, then pushes are performed using the\n `hive` CLI executable on the local/remote PATH.\n **kwargs (dict): Additional arguments to send to `pandas.DataFrame.to_sql`.\n\n Further Parameters for CLI method (specifying these for the pandas\n method will cause a `RuntimeError` exception):\n partition (dict): A mapping of column names to values that specify\n the partition into which the provided data should be uploaded,\n as well as providing the fields by which new tables should be\n partitioned.\n sep (str): Field delimiter for data (defaults to CTRL-A, or `chr(1)`).\n table_props (dict): Properties to set on any newly created tables\n (extends `.default_table_props`).\n dtype_overrides (dict): Mapping of column names to Hive datatypes to\n use instead of default mapping.\n \"\"\"\n table = self._parse_namespaces(table, defaults={'schema': self.username})\n use_hive_cli = use_hive_cli or self.push_using_hive_cli\n partition = partition or {}\n table_props = table_props or {}\n dtype_overrides = dtype_overrides or {}\n\n # Try using SQLALchemy method\n if not use_hive_cli:\n if partition or table_props or dtype_overrides:\n raise RuntimeError(\n \"At least one of `partition` or `table_props` or \"\n \"`dtype_overrides` has been specified. Setting table \"\n \"properties or partition information is not supported \"\n \"via the SQLAlchemy backend. If this is important, please \"\n \"pass `use_hive_cli=True`, otherwise remove these values \"\n \"and try again.\"\n )\n try:\n return _pandas.to_sql(\n df=df, name=table.table, schema=table.schema, con=self._sqlalchemy_engine,\n index=False, if_exists=if_exists, **kwargs\n )\n except Exception as e:\n raise RuntimeError(\n \"Push unsuccessful. Your version of Hive may be too old to \"\n \"support the `INSERT` keyword. You might want to try setting \"\n \"`.push_using_hive_cli = True` if your local or remote \"\n \"machine has access to the `hive` CLI executable. The \"\n \"original exception was: {}\".format(e.args[0])\n )\n\n # Try using Hive CLI\n\n # If `partition` is specified, the associated columns must not be\n # present in the dataframe.\n assert len(set(partition).intersection(df.columns)) == 0, \"The dataframe to be uploaded must not have any partitioned fields. Please remove the field(s): {}.\".format(','.join(set(partition).intersection(df.columns)))\n\n # Save dataframe to file and send it to the remote server if necessary\n temp_dir = tempfile.mkdtemp(prefix='omniduct_hiveserver2')\n tmp_fname = os.path.join(temp_dir, 'data_{}.csv'.format(time.time()))\n logger.info('Saving dataframe to file... {}'.format(tmp_fname))\n df.fillna(r'\\N').to_csv(tmp_fname, index=False, header=False,\n sep=sep, encoding='utf-8')\n\n if self.remote:\n logger.info(\"Uploading data to remote host...\")\n self.remote.upload(tmp_fname)\n\n # Generate create table statement.\n auto_table_props = set(self.default_table_props).difference(table_props)\n if len(auto_table_props) > 0:\n logger.warning(\n \"In addition to any specified table properties, this \"\n \"HiveServer2Client has added the following default table \"\n \"properties:\\n{default_props}\\nTo override them, please \"\n \"specify overrides using: `.push(..., table_props={{...}}).`\"\n .format(default_props=json.dumps({\n prop: value for prop, value in self.default_table_props.items()\n if prop in auto_table_props\n }, indent=True))\n )\n\n tblprops = self.default_table_props.copy()\n tblprops.update(table_props or {})\n cts = self._create_table_statement_from_df(\n df=df,\n table=table,\n drop=(if_exists == 'replace') and not partition,\n text=True,\n sep=sep,\n table_props=tblprops,\n partition_cols=list(partition),\n dtype_overrides=dtype_overrides\n )\n\n # Generate load data statement.\n partition_clause = (\n ''\n if not partition\n else 'PARTITION ({})'.format(\n ','.join(\"{key} = '{value}'\".format(key=key, value=value) for key, value in partition.items())\n )\n )\n lds = '\\nLOAD DATA LOCAL INPATH \"{path}\" {overwrite} INTO TABLE {table} {partition_clause};'.format(\n path=os.path.basename(tmp_fname) if self.remote else tmp_fname,\n overwrite=\"OVERWRITE\" if if_exists == \"replace\" else \"\",\n table=table,\n partition_clause=partition_clause\n )\n\n # Run create table statement and load data statments\n logger.info(\n \"Creating hive table `{table}` if it does not \"\n \"already exist, and inserting the provided data{partition}.\"\n .format(\n table=table,\n partition=\" into {}\".format(partition_clause) if partition_clause else \"\"\n )\n )\n try:\n stmts = '\\n'.join([cts, lds])\n logger.debug(stmts)\n proc = self._run_in_hivecli(stmts)\n if proc.returncode != 0:\n raise RuntimeError(proc.stderr.decode('utf-8'))\n finally:\n # Clean up files\n if self.remote:\n self.remote.execute('rm -rf {}'.format(tmp_fname))\n shutil.rmtree(temp_dir, ignore_errors=True)\n\n logger.info(\"Successfully uploaded dataframe {partition}`{table}`.\".format(\n table=table,\n partition=\"into {} of \".format(partition_clause) if partition_clause else \"\"\n ))\n\n @override\n def _table_list(self, namespace, like='*', **kwargs):\n schema = namespace.name or self.schema\n return self.query(\"SHOW TABLES IN {0} '{1}'\".format(schema, like),\n **kwargs)\n\n @override\n def _table_exists(self, table, **kwargs):\n logger.disabled = True\n try:\n self.table_desc(table, **kwargs)\n return True\n except:\n return False\n finally:\n logger.disabled = False\n\n @override\n def _table_drop(self, table, **kwargs):\n return self.execute(\"DROP TABLE {table}\".format(table=table))\n\n @override\n def _table_desc(self, table, **kwargs):\n records = self.query(\"DESCRIBE {0}\".format(table), **kwargs)\n\n # pretty hacky but hive doesn't return DESCRIBE results in a nice format\n # TODO is there any information we should pull out of DESCRIBE EXTENDED\n for i, record in enumerate(records):\n if record[0] == '':\n break\n\n columns = ['col_name', 'data_type', 'comment']\n fields_df = pd.DataFrame(records[:i], columns=columns)\n\n partitions_df = pd.DataFrame(records[i + 4:], columns=columns)\n partitions_df['comment'] = \"PARTITION \" + partitions_df['comment']\n\n return pd.concat((fields_df, partitions_df))\n\n @override\n def _table_head(self, table, n=10, **kwargs):\n return self.query(\"SELECT * FROM {} LIMIT {}\".format(table, n), **kwargs)\n\n @override\n def _table_props(self, table, **kwargs):\n return self.query('SHOW TBLPROPERTIES {0}'.format(table), **kwargs)\n\n def _run_in_hivecli(self, cmd):\n \"\"\"Run a query using hive cli in a subprocess.\"\"\"\n # Turn hive command into quotable string.\n double_escaped = re.sub('\\\\' * 2, '\\\\' * 4, cmd)\n backtick_escape = '\\\\\\\\\\\\`' if self.remote else '\\\\`'\n sys_cmd = 'hive -e \"{0}\"'.format(re.sub('\"', '\\\\\"', double_escaped)) \\\n .replace('`', backtick_escape)\n # Execute command in a subprocess.\n if self.remote:\n proc = self.remote.execute(sys_cmd)\n else:\n proc = run_in_subprocess(sys_cmd, check_output=True)\n return proc\n\n @classmethod\n def _create_table_statement_from_df(cls, df, table, drop=False,\n text=True, sep=chr(1), loc=None,\n table_props=None, partition_cols=None,\n dtype_overrides=None):\n \"\"\"\n Return create table statement for new hive table based on pandas dataframe.\n\n Args:\n df (pandas.DataFrame, pandas.Series): Used to determine column names\n and types for create table statement.\n table (ParsedNamespaces): The parsed name of the target table.\n drop (bool): Whether to include a drop table statement before the\n create table statement.\n text (bool): Whether data will be stored as a textfile.\n sep (str): The separator used by the text data store (defaults to\n CTRL-A, i.e. `chr(1)`, which is the default Hive separator).\n loc (str): Desired HDFS location (if not the default).\n table_props (dict): The table properties (if any) to set on the table.\n partition_cols (list): The columns by which the created table should\n be partitioned.\n\n Returns:\n str: The Hive SQL required to create the table with the above\n configuration.\n \"\"\"\n table_props = table_props or {}\n partition_cols = partition_cols or []\n dtype_overrides = dtype_overrides or {}\n\n # dtype kind to hive type mapping dict.\n DTYPE_KIND_HIVE_TYPE = {\n 'b': 'BOOLEAN', # boolean\n 'i': 'BIGINT', # signed integer\n 'u': 'BIGINT', # unsigned integer\n 'f': 'DOUBLE', # floating-point\n 'c': 'STRING', # complex floating-point\n 'O': 'STRING', # object\n 'S': 'STRING', # (byte-)string\n 'U': 'STRING', # Unicode\n 'V': 'STRING' # void\n }\n\n # Sanitise column names and map numpy/pandas data-types to hive types.\n columns = []\n for col, dtype in df.dtypes.iteritems():\n col_sanitized = re.sub(r'\\W', '', col.lower().replace(' ', '_'))\n hive_type = dtype_overrides.get(col) or DTYPE_KIND_HIVE_TYPE.get(dtype.kind)\n if hive_type is None:\n hive_type = DTYPE_KIND_HIVE_TYPE['O']\n logger.warning(\n 'Unable to determine hive type for dataframe column {col} of pandas dtype {dtype}. '\n 'Defaulting to hive type {hive_type}. If other column type is desired, '\n 'please specify via `dtype_overrides`'\n .format(**locals())\n )\n columns.append(\n ' {column} {type}'.format(column=col_sanitized, type=hive_type)\n )\n\n partition_columns = ['{} STRING'.format(col) for col in partition_cols]\n\n tblprops = [\"'{key}' = '{value}'\".format(key=key, value=value) for key, value in table_props.items()]\n tblprops = \"TBLPROPERTIES({})\".format(\",\".join(tblprops)) if len(tblprops) > 0 else \"\"\n\n cmd = Template(\"\"\"\n {% if drop %}\n DROP TABLE IF EXISTS {{ table }};\n {% endif -%}\n CREATE TABLE IF NOT EXISTS {{ table }} (\n {%- for col in columns %}\n {{ col }} {% if not loop.last %}, {% endif %}\n {%- endfor %}\n )\n {%- if partition_columns %}\n PARTITIONED BY (\n {%- for col in partition_columns %}\n {{ col }} {% if not loop.last %}, {% endif %}\n {%- endfor %}\n )\n {%- endif %}\n {%- if text %}\n ROW FORMAT DELIMITED\n FIELDS TERMINATED BY \"{{ sep }}\"\n STORED AS TEXTFILE\n {% endif %}\n {%- if loc %}\n LOCATION \"{{ loc }}\"\n {%- endif %}\n {{ tblprops }}\n ;\n \"\"\").render(**locals())\n\n return cmd\n"
] |
[
[
"pandas.DataFrame",
"pandas.concat"
]
] |
mondrasovic/context_rcnn
|
[
"b3f6b4fdd63fc76f33becae0fba1363f9e873f50"
] |
[
"src/eval.py"
] |
[
"#!/usr/bin/env python\n# -*- coding: utf-8 -*-\n#\n# Copyright (c) 2021 Milan Ondrašovič <[email protected]>\n#\n# MIT License\n#\n# Permission is hereby granted, free of charge, to any person obtaining a copy\n# of this software and associated documentation files (the \"Software\"), to deal\n# in the Software without restriction, including without limitation the rights\n# to use, copy, modify, merge, publish, distribute, sublicense, and/or sell\n# copies of the Software, and to permit persons to whom the Software is\n# furnished to do so, subject to the following conditions:\n#\n# The above copyright notice and this permission notice shall be included in all\n# copies or substantial portions of the Software.\n#\n# THE SOFTWARE IS PROVIDED \"AS IS\", WITHOUT WARRANTY OF ANY KIND, EXPRESS OR\n# IMPLIED, INCLUDING BUT NOT LIMITED TO THE WARRANTIES OF MERCHANTABILITY,\n# FITNESS FOR A PARTICULAR PURPOSE AND NONINFRINGEMENT. IN NO EVENT SHALL THE\n# AUTHORS OR COPYRIGHT HOLDERS BE LIABLE FOR ANY CLAIM, DAMAGES OR OTHER\n# LIABILITY, WHETHER IN AN ACTION OF CONTRACT, TORT OR OTHERWISE, ARISING FROM,\n# OUT OF OR IN CONNECTION WITH THE SOFTWARE OR THE USE OR OTHER DEALINGS IN THE\n\n# Code adapted from:\n# https://github.com/pytorch/vision/blob/main/references/detection/utils.py\n\nimport functools\nimport time\n\nimport torch\nimport torchvision\n\nfrom .utils import MetricLogger, to_device\nfrom .coco_utils import get_coco_api_from_dataset\nfrom .coco_eval import CocoEvaluator\n\n\[email protected]_grad()\ndef evaluate(model, data_loader, device):\n n_threads = torch.get_num_threads()\n torch.set_num_threads(1)\n cpu_device = torch.device('cpu')\n model.eval()\n metric_logger = MetricLogger(delimiter=\" \")\n header = \"Test:\"\n\n print(\"Initializing COCO-like dataset.\")\n coco = _get_or_create_coco_dataset(data_loader)\n iou_types = _get_iou_types(model)\n coco_evaluator = CocoEvaluator(coco, iou_types)\n\n print(\"Processing evaluation dataset.\")\n for images, targets in metric_logger.log_every(data_loader, 100, header):\n images = to_device(images, device)\n targets = [\n {key:to_device(val, device) for key, val in target.items()}\n for target in targets\n ]\n\n if torch.cuda.is_available():\n torch.cuda.synchronize()\n model_time = time.time()\n outputs = model(images, targets)\n\n outputs = [\n {k:v.to(cpu_device) for k, v in t.items()}\n for t in outputs\n ]\n model_time = time.time() - model_time\n\n res = {\n target['image_id'].item():output\n for target, output in zip(targets, outputs\n )}\n evaluator_time = time.time()\n coco_evaluator.update(res)\n evaluator_time = time.time() - evaluator_time\n metric_logger.update(\n model_time=model_time, evaluator_time=evaluator_time\n )\n \n # Gather the stats from all processes.\n metric_logger.synchronize_between_processes()\n print(\"Averaged stats:\", metric_logger)\n coco_evaluator.synchronize_between_processes()\n\n # Accumulate predictions from all images.\n coco_evaluator.accumulate()\n coco_evaluator.summarize()\n torch.set_num_threads(n_threads)\n\n return coco_evaluator\n\n\[email protected]_cache(typed=True)\ndef _get_or_create_coco_dataset(data_loader):\n dataset = data_loader.dataset\n coco = get_coco_api_from_dataset(dataset)\n return coco\n\n\ndef _get_iou_types(model):\n model_without_ddp = model\n if isinstance(model, torch.nn.parallel.DistributedDataParallel):\n model_without_ddp = model.module\n iou_types = ['bbox']\n if isinstance(model_without_ddp, torchvision.models.detection.MaskRCNN):\n iou_types.append('segm')\n if isinstance(model_without_ddp, torchvision.models.detection.KeypointRCNN):\n iou_types.append('keypoints')\n return iou_types\n"
] |
[
[
"torch.device",
"torch.cuda.synchronize",
"torch.no_grad",
"torch.get_num_threads",
"torch.cuda.is_available",
"torch.set_num_threads"
]
] |
exusnraul/exusnraul-interview_solutions
|
[
"1b4dbfaece3c924b971acc4e9186a227f57bc450"
] |
[
"multithred_test.py"
] |
[
"import time\r\nfrom concurrent.futures import ProcessPoolExecutor, ThreadPoolExecutor\r\n\r\nimport matplotlib.pyplot as plt\r\nimport numpy as np\r\n\r\n\r\ndef multithreading(func, args, workers):\r\n with ThreadPoolExecutor(workers) as ex:\r\n res = ex.map(func, args)\r\n return list(res)\r\n\r\n\r\ndef multiprocessing(func, args, workers):\r\n with ProcessPoolExecutor(workers) as ex:\r\n res = ex.map(func, args)\r\n return list(res)\r\n\r\n\r\ndef cpu_heavy(x):\r\n print('I am', x)\r\n start = time.time()\r\n count = 0\r\n for i in range(10**8):\r\n count += i\r\n stop = time.time()\r\n return start, stop\r\n\r\n\r\ndef visualize_runtimes(results, title):\r\n start, stop = np.array(results).T\r\n plt.barh(range(len(start)), stop - start)\r\n plt.grid(axis='x')\r\n plt.ylabel(\"Tasks\")\r\n plt.xlabel(\"Seconds\")\r\n plt.xlim(0, 22.5)\r\n ytks = range(len(results))\r\n plt.yticks(ytks, ['job {}'.format(exp) for exp in ytks])\r\n plt.title(title)\r\n return stop[-1] - start[0]\r\n\r\n\r\nplt.subplot(1, 2, 1)\r\nvisualize_runtimes(multithreading(cpu_heavy, range(4), 4), \"Multithreading\")\r\nplt.subplot(1, 2, 2)\r\nvisualize_runtimes(multiprocessing(cpu_heavy, range(4), 4), \"Multiprocessing\")\r\nplt.show()"
] |
[
[
"numpy.array",
"matplotlib.pyplot.xlim",
"matplotlib.pyplot.grid",
"matplotlib.pyplot.xlabel",
"matplotlib.pyplot.title",
"matplotlib.pyplot.ylabel",
"matplotlib.pyplot.show",
"matplotlib.pyplot.subplot"
]
] |
dfsp-spirit/cogload
|
[
"ff9d19803c2e0c9aea248a45380959c2758ba83a"
] |
[
"tests/brainload/test_brainlocate.py"
] |
[
"\"\"\"\nFunctions for testing brainlocate.\n\"\"\"\n\nimport pytest\nimport numpy as np\nimport os\nfrom numpy.testing import assert_array_equal, assert_allclose\nimport brainload as bl\nimport brainload.brainlocate as loc\n\n\nTHIS_DIR = os.path.dirname(os.path.abspath(__file__))\nTEST_DATA_DIR = os.path.join(THIS_DIR, os.pardir, 'test_data')\n\n# Respect the environment variable BRAINLOAD_TEST_DATA_DIR if it is set. If not, fall back to default.\nTEST_DATA_DIR = os.getenv('BRAINLOAD_TEST_DATA_DIR', TEST_DATA_DIR)\n\n\ndef test_closest_vertex_to_very_close_point_known_dist():\n try:\n from scipy.spatial.distance import cdist\n except ImportError:\n pytest.skip(\"Optional dependency scipy not installed, skipping tests which require scipy.\")\n vert_coords = np.array([[0.0, 0.0, 0.0], [1.0, 0.0, 0.0], [1.0, 1.0, 0.0]])\n faces = np.array([0, 1, 2])\n locator = loc.BrainLocate(vert_coords, faces)\n query_coords = np.array([[1.0, 0.0, 0.0], [1.0, 1.0, 0.0], [1.0, 1.1, 0.0], [0.1, 0.1, 0.1]])\n res = locator.get_closest_vertex(query_coords)\n assert res.shape == (4, )\n assert res[0] == 1 # the closest vertex index in the mesh for query coordinate at index 0\n assert res[1] == 2 # the closest vertex index in the mesh for query coordinate at index 1\n assert res[2] == 2 # the closest vertex index in the mesh for query coordinate at index 2\n assert res[3] == 0 # the closest vertex index in the mesh for query coordinate at index 3\n dist_matrix = cdist(vert_coords, query_coords)\n assert dist_matrix.shape == (3, 4)\n assert dist_matrix[0][0] == pytest.approx(1.0, 0.00001)\n assert dist_matrix[0][1] == pytest.approx(1.4142135623730951, 0.00001) # This is sqrt(2)\n assert dist_matrix[1][0] == pytest.approx(0.0, 0.00001)\n min_index = np.argmin(dist_matrix, axis=0)\n assert min_index.shape == (4, ) # we queried for 4 coordinates.\n assert min_index[0] == res[0]\n assert min_index[1] == res[1]\n assert min_index[2] == res[2]\n assert min_index[3] == res[3]\n\n\ndef test_closest_vertex_to_very_close_point():\n try:\n from scipy.spatial.distance import cdist\n except ImportError:\n pytest.skip(\"Optional dependency scipy not installed, skipping tests which require scipy.\")\n vert_coords, faces, _ = bl.subject_mesh('subject1', TEST_DATA_DIR, surf='white', hemi='both')\n locator = loc.BrainLocate(vert_coords, faces)\n query_coords = np.array([[58.0 , -45.0, 75.0]])\n res = locator.get_closest_vertex(query_coords)\n assert res.shape == (1, )\n assert res[0] == 210683 # the vertex index in the mesh\n expected_vert_210683_coords = (58.005173, -44.736935, 74.418076)\n assert_allclose(vert_coords[210683], np.array(expected_vert_210683_coords))\n dist = cdist(np.array([expected_vert_210683_coords]), query_coords)\n assert dist[0][0] == pytest.approx(0.6386434810831467, 0.001)\n\n\ndef test_closest_vertex_to_far_away_point():\n try:\n from scipy.spatial.distance import cdist\n except ImportError:\n pytest.skip(\"Optional dependency scipy not installed, skipping tests which require scipy.\")\n vert_coords, faces, _ = bl.subject_mesh('subject1', TEST_DATA_DIR, surf='white', hemi='both')\n locator = loc.BrainLocate(vert_coords, faces)\n query_coords = np.array([[134.37332 , -57.259495, 149.267631], [134.37332 , -57.259495, 149.267631], [58.0 , -45.0, 75.0]])\n res = locator.get_closest_vertex(query_coords)\n assert res.shape == (3, )\n assert res[0] == 209519 # the vertex index in the mesh\n assert res[1] == 209519\n assert res[2] == 210683\n assert_allclose(vert_coords[209519], np.array((58.258751, -45.213722, 74.348068)))\n dist = cdist(np.array([[58.258751, -45.213722, 74.348068]]), np.array([[134.37332 , -57.259495, 149.267631]]))\n assert dist[0][0] == pytest.approx(107.47776133, 0.001)\n\n\ndef test_get_closest_vertex_and_distance_to_far_away_point():\n try:\n from scipy.spatial.distance import cdist\n except ImportError:\n pytest.skip(\"Optional dependency scipy not installed, skipping tests which require scipy.\")\n vert_coords, faces, _ = bl.subject_mesh('subject1', TEST_DATA_DIR, surf='white', hemi='both')\n locator = loc.BrainLocate(vert_coords, faces)\n query_coords = np.array([[134.37332 , -57.259495, 149.267631], [134.37332 , -57.259495, 149.267631], [134.37332 , -57.259495, 149.267631]]) # just query 3 times for the same coord to see whether results and consistent\n res = locator.get_closest_vertex_and_distance(query_coords)\n assert res.shape == (3, 2)\n assert res[0,0] == 209519 # the vertex index in the mesh\n assert res[1,0] == 209519\n assert res[2,0] == 209519\n assert res[0,1] == 107.47776120258028 # the distance\n assert res[1,1] == 107.47776120258028\n assert res[2,1] == 107.47776120258028\n\n\ndef test_get_closest_vertex_to_vertex_0_coordinate():\n try:\n from scipy.spatial.distance import cdist\n except ImportError:\n pytest.skip(\"Optional dependency scipy not installed, skipping tests which require scipy.\")\n vert_coords, faces, _ = bl.subject_mesh('subject1', TEST_DATA_DIR, surf='white', hemi='both')\n locator = loc.BrainLocate(vert_coords, faces)\n known_vertex_0_coord = (-1.85223234, -107.98274994, 22.76972961) # coordinate for vertex 0 in the test data.\n assert_allclose(vert_coords[0], np.array(known_vertex_0_coord))\n query_coords = np.array([known_vertex_0_coord])\n res = locator.get_closest_vertex_and_distance(query_coords)\n assert res.shape == (1, 2)\n assert res[0, 0] == 0 # vertex index of closest vertex. The query coordinate is the known coordinate of vertex 0, so this must be 0.\n assert abs(res[0, 1]) <= 0.001 # distance to closest vertex. The query coordinate is the known coordinate of vertex 0, so this must be very close to 0.0\n"
] |
[
[
"numpy.array",
"scipy.spatial.distance.cdist",
"numpy.argmin"
]
] |
rickyHong/markowitz-portfolio-optimization
|
[
"4376e581096d866cf1121962b812f1a058afe442"
] |
[
"posdef.py"
] |
[
"import numpy as np\nfrom numpy import linalg as la\n\ndef nearestPD(A):\n \"\"\"Find the nearest positive-definite matrix to input\n\n A Python/Numpy port of John D'Errico's `nearestSPD` MATLAB code [1], which\n credits [2].\n\n [1] https://www.mathworks.com/matlabcentral/fileexchange/42885-nearestspd\n\n [2] N.J. Higham, \"Computing a nearest symmetric positive semidefinite\n matrix\" (1988): https://doi.org/10.1016/0024-3795(88)90223-6\n \"\"\"\n\n B = (A + A.T) / 2\n _, s, V = la.svd(B)\n\n H = np.dot(V.T, np.dot(np.diag(s), V))\n\n A2 = (B + H) / 2\n\n A3 = (A2 + A2.T) / 2\n\n if isPD(A3):\n return A3\n\n spacing = np.spacing(la.norm(A))\n # The above is different from [1]. It appears that MATLAB's `chol` Cholesky\n # decomposition will accept matrixes with exactly 0-eigenvalue, whereas\n # Numpy's will not. So where [1] uses `eps(mineig)` (where `eps` is Matlab\n # for `np.spacing`), we use the above definition. CAVEAT: our `spacing`\n # will be much larger than [1]'s `eps(mineig)`, since `mineig` is usually on\n # the order of 1e-16, and `eps(1e-16)` is on the order of 1e-34, whereas\n # `spacing` will, for Gaussian random matrixes of small dimension, be on\n # othe order of 1e-16. In practice, both ways converge, as the unit test\n # below suggests.\n I = np.eye(A.shape[0])\n k = 1\n while not isPD(A3):\n mineig = np.min(np.real(la.eigvals(A3)))\n A3 += I * (-mineig * k**2 + spacing)\n k += 1\n\n return A3\n\ndef isPD(B):\n \"\"\"Returns true when input is positive-definite, via Cholesky\"\"\"\n try:\n _ = la.cholesky(B)\n return True\n except la.LinAlgError:\n return False\n\nif __name__ == '__main__':\n import numpy as np\n for i in xrange(10):\n for j in xrange(2, 100):\n A = np.random.randn(j, j)\n B = nearestPD(A)\n assert(isPD(B))\n print('unit test passed!')\n"
] |
[
[
"numpy.linalg.norm",
"numpy.random.randn",
"numpy.eye",
"numpy.linalg.svd",
"numpy.linalg.cholesky",
"numpy.diag",
"numpy.linalg.eigvals"
]
] |
abumafrim/OpenNMT-tf
|
[
"f14c05a7cb8b1b8f3a692d6fea3c12067bc3eb2c"
] |
[
"opennmt/tests/transformer_test.py"
] |
[
"from parameterized import parameterized\n\nimport tensorflow as tf\nimport numpy as np\n\nfrom opennmt.layers import transformer\n\n\nclass TransformerTest(tf.test.TestCase):\n\n @parameterized.expand([[tf.bool], [tf.float32]])\n def testBuildFutureMask(self, dtype):\n length = [2, 4, 3]\n expected = np.array([\n [[1, 0, 0, 0],\n [1, 1, 0, 0],\n [1, 1, 0, 0],\n [1, 1, 0, 0]],\n [[1, 0, 0, 0],\n [1, 1, 0, 0],\n [1, 1, 1, 0],\n [1, 1, 1, 1]],\n [[1, 0, 0, 0],\n [1, 1, 0, 0],\n [1, 1, 1, 0],\n [1, 1, 1, 0]]]).astype(dtype.as_numpy_dtype)\n\n mask = transformer.future_mask(tf.constant(length), dtype=dtype)\n self.assertIs(mask.dtype, dtype)\n mask = self.evaluate(mask)\n self.assertTupleEqual(mask.shape, (len(length), max(length), max(length)))\n self.assertAllEqual(mask, expected)\n\n @parameterized.expand([[tf.bool], [tf.float32]])\n def testBuildFutureMaskWithMaxLen(self, dtype):\n length = [2, 4, 3]\n maximum_length = 5\n expected = np.array([\n [[1, 0, 0, 0, 0],\n [1, 1, 0, 0, 0],\n [1, 1, 0, 0, 0],\n [1, 1, 0, 0, 0],\n [1, 1, 0, 0, 0]],\n [[1, 0, 0, 0, 0],\n [1, 1, 0, 0, 0],\n [1, 1, 1, 0, 0],\n [1, 1, 1, 1, 0],\n [1, 1, 1, 1, 0]],\n [[1, 0, 0, 0, 0],\n [1, 1, 0, 0, 0],\n [1, 1, 1, 0, 0],\n [1, 1, 1, 0, 0],\n [1, 1, 1, 0, 0]]]).astype(dtype.as_numpy_dtype)\n\n mask = transformer.future_mask(\n tf.constant(length), maximum_length=maximum_length, dtype=dtype)\n self.assertIs(mask.dtype, dtype)\n mask = self.evaluate(mask)\n self.assertTupleEqual(mask.shape, (len(length), maximum_length, maximum_length))\n self.assertAllEqual(mask, expected)\n\n def testSplitHeads(self):\n batch_size = 3\n length = [5, 3, 7]\n num_heads = 8\n depth = 20\n\n inputs = tf.random.normal([batch_size, max(length), depth * num_heads], dtype=tf.float32)\n outputs = transformer.split_heads(inputs, num_heads)\n\n static_shape = outputs.shape\n self.assertEqual(num_heads, static_shape[1])\n self.assertEqual(depth, static_shape[-1])\n outputs = self.evaluate(outputs)\n self.assertAllEqual([batch_size, num_heads, max(length), depth], outputs.shape)\n\n def testCombineHeads(self):\n batch_size = 3\n length = [5, 3, 7]\n num_heads = 8\n depth = 20\n\n inputs = tf.random.normal([batch_size, num_heads, max(length), depth], dtype=tf.float32)\n outputs = transformer.combine_heads(inputs)\n\n static_shape = outputs.shape\n self.assertEqual(depth * num_heads, static_shape[-1])\n outputs = self.evaluate(outputs)\n self.assertAllEqual([batch_size, max(length), depth * num_heads], outputs.shape)\n\n def testSplitAndCombineHeads(self):\n batch_size = 3\n length = [5, 3, 7]\n num_heads = 8\n depth = 20\n\n inputs = tf.random.normal([batch_size, max(length), depth * num_heads], dtype=tf.float32)\n split = transformer.split_heads(inputs, num_heads)\n combined = transformer.combine_heads(split)\n inputs, combined = self.evaluate([inputs, combined])\n self.assertAllEqual(inputs, combined)\n\n def testRelativePositions(self):\n positions = transformer.relative_positions(4, 2)\n self.assertAllEqual(\n self.evaluate(positions),\n [[2, 3, 4, 4], [1, 2, 3, 4], [0, 1, 2, 3], [0, 0, 1, 2]])\n\n def testFeedForwardNetwork(self):\n ffn = transformer.FeedForwardNetwork(20, 10)\n x = tf.random.uniform([4, 5, 10])\n y = ffn(x)\n self.assertEqual(y.shape, x.shape)\n\n def testMultiHeadSelfAttention(self):\n attention = transformer.MultiHeadAttention(4, 20)\n queries = tf.random.uniform([4, 5, 10])\n mask = tf.sequence_mask([4, 3, 5, 2])\n context, _ = attention(queries, mask=mask)\n self.assertListEqual(context.shape.as_list(), [4, 5, 20])\n\n def testMultiHeadSelfAttentionWithCache(self):\n cache = (tf.zeros([4, 4, 0, 5]), tf.zeros([4, 4, 0, 5]))\n attention = transformer.MultiHeadAttention(4, 20)\n x = tf.random.uniform([4, 1, 10])\n _, cache = attention(x, cache=cache)\n self.assertEqual(cache[0].shape[2], 1)\n self.assertEqual(cache[1].shape[2], 1)\n _, cache = attention(x, cache=cache)\n self.assertEqual(cache[0].shape[2], 2)\n self.assertEqual(cache[1].shape[2], 2)\n\n def testMultiHeadSelfAttentionRelativePositions(self):\n attention = transformer.MultiHeadAttention(4, 20, maximum_relative_position=6)\n x = tf.random.uniform([2, 9, 10])\n mask = tf.sequence_mask([9, 7])\n y = attention(x, mask=mask)\n\n def testMultiHeadSelfAttentionRelativePositionsWithCache(self):\n attention = transformer.MultiHeadAttention(4, 20, maximum_relative_position=6)\n x = tf.random.uniform([4, 1, 10])\n cache = (tf.zeros([4, 4, 0, 5]), tf.zeros([4, 4, 0, 5]))\n _, cache = attention(x, cache=cache)\n\n def testMultiHeadAttention(self):\n attention = transformer.MultiHeadAttention(4, 20)\n queries = tf.random.uniform([4, 5, 10])\n memory = tf.random.uniform([4, 3, 10])\n mask = tf.sequence_mask([1, 3, 2, 2])\n context, _ = attention(queries, memory=memory, mask=mask)\n self.assertListEqual(context.shape.as_list(), [4, 5, 20])\n\n def testMultiHeadAttentionWithCache(self):\n cache = (tf.zeros([4, 4, 0, 5]), tf.zeros([4, 4, 0, 5]))\n attention = transformer.MultiHeadAttention(4, 20)\n memory = tf.random.uniform([4, 3, 10])\n mask = tf.sequence_mask([1, 3, 2, 2])\n x = tf.random.uniform([4, 1, 10])\n y1, cache = attention(x, memory=memory, mask=mask, cache=cache)\n self.assertEqual(cache[0].shape[2], 3)\n self.assertEqual(cache[1].shape[2], 3)\n y2, cache = attention(x, memory=memory, mask=mask, cache=cache)\n self.assertAllEqual(y1, y2)\n\n def testMultiHeadAttentionMask(self):\n attention = transformer.MultiHeadAttention(4, 20, return_attention=True)\n queries = tf.random.uniform([4, 5, 10])\n memory = tf.random.uniform([4, 3, 10])\n mask = tf.sequence_mask([1, 3, 2, 2])\n _, _, attention = attention(queries, memory=memory, mask=mask)\n attention = tf.reshape(attention, [4, -1, 3])\n mask = tf.broadcast_to(tf.expand_dims(mask, 1), attention.shape)\n padding = tf.boolean_mask(attention, tf.logical_not(mask))\n self.assertAllEqual(tf.reduce_sum(padding), 0)\n\n\nif __name__ == \"__main__\":\n tf.test.main()\n"
] |
[
[
"tensorflow.sequence_mask",
"tensorflow.zeros",
"numpy.array",
"tensorflow.expand_dims",
"tensorflow.random.uniform",
"tensorflow.reshape",
"tensorflow.logical_not",
"tensorflow.constant",
"tensorflow.reduce_sum",
"tensorflow.test.main"
]
] |
DevilMayNotCry/My_curl
|
[
"a4e59533685257098eb02a25d0f90e40cf04e0a1"
] |
[
"util.py"
] |
[
"# -*- coding: utf-8 -*-\n'''\nThis is a PyTorch implementation of CURL: Neural Curve Layers for Global Image Enhancement\nhttps://arxiv.org/pdf/1911.13175.pdf\n\nPlease cite paper if you use this code.\n\nTested with Pytorch 1.7.1, Python 3.7.9\n\nAuthors: Sean Moran ([email protected]), \n\n'''\nfrom skimage.metrics import structural_similarity as ssim\nfrom PIL import Image\nimport math\nimport numpy as np\nfrom torch.autograd import Variable\nimport torch\n\nimport matplotlib\nimport sys\nmatplotlib.use('agg')\nnp.set_printoptions(threshold=sys.maxsize)\n\n\nclass ImageProcessing(object):\n\n @staticmethod\n def rgb_to_lab(img, is_training=True):\n \"\"\" PyTorch implementation of RGB to LAB conversion: https://docs.opencv.org/3.3.0/de/d25/imgproc_color_conversions.html\n Based roughly on a similar implementation here: https://github.com/affinelayer/pix2pix-tensorflow/blob/master/pix2pix.py\n :param img: image to be adjusted\n :returns: adjusted image\n :rtype: Tensor\n\n \"\"\"\n img = img.permute(2, 1, 0)\n shape = img.shape\n img = img.contiguous()\n img = img.view(-1, 3)\n\n img = (img / 12.92) * img.le(0.04045).float() + (((torch.clamp(img,\n min=0.0001) + 0.055) / 1.055) ** 2.4) * img.gt(0.04045).float()\n\n rgb_to_xyz = Variable(torch.FloatTensor([ # X Y Z\n [0.412453, 0.212671, 0.019334], # R\n [0.357580, 0.715160, 0.119193], # G\n [0.180423, 0.072169,\n 0.950227], # B\n ]), requires_grad=False).cuda()\n\n img = torch.matmul(img, rgb_to_xyz)\n img = torch.mul(img, Variable(torch.FloatTensor(\n [1/0.950456, 1.0, 1/1.088754]), requires_grad=False).cuda())\n\n epsilon = 6/29\n\n img = ((img / (3.0 * epsilon**2) + 4.0/29.0) * img.le(epsilon**3).float()) + \\\n (torch.clamp(img, min=0.0001) **\n (1.0/3.0) * img.gt(epsilon**3).float())\n\n fxfyfz_to_lab = Variable(torch.FloatTensor([[0.0, 500.0, 0.0], # fx\n # fy\n [116.0, -500.0, 200.0],\n # fz\n [0.0, 0.0, -200.0],\n ]), requires_grad=False).cuda()\n\n img = torch.matmul(img, fxfyfz_to_lab) + Variable(\n torch.FloatTensor([-16.0, 0.0, 0.0]), requires_grad=False).cuda()\n\n img = img.view(shape)\n img = img.permute(2, 1, 0)\n\n '''\n L_chan: black and white with input range [0, 100]\n a_chan/b_chan: color channels with input range ~[-110, 110], not exact \n [0, 100] => [0, 1], ~[-110, 110] => [0, 1]\n '''\n img[0, :, :] = img[0, :, :]/100\n img[1, :, :] = (img[1, :, :]/110 + 1)/2\n img[2, :, :] = (img[2, :, :]/110 + 1)/2\n\n img[(img != img).detach()] = 0\n\n img = img.contiguous()\n\n return img.cuda()\n\n @staticmethod\n def lab_to_rgb(img, is_training=True):\n \"\"\" PyTorch implementation of LAB to RGB conversion: https://docs.opencv.org/3.3.0/de/d25/imgproc_color_conversions.html\n Based roughly on a similar implementation here: https://github.com/affinelayer/pix2pix-tensorflow/blob/master/pix2pix.py\n :param img: image to be adjusted\n :returns: adjusted image\n :rtype: Tensor\n \"\"\" \n img = img.permute(2, 1, 0)\n shape = img.shape\n img = img.contiguous()\n img = img.view(-1, 3)\n img_copy = img.clone()\n\n img_copy[:, 0] = img[:, 0] * 100\n img_copy[:, 1] = ((img[:, 1] * 2)-1)*110\n img_copy[:, 2] = ((img[:, 2] * 2)-1)*110\n\n img = img_copy.clone().cuda()\n del img_copy\n\n lab_to_fxfyfz = Variable(torch.FloatTensor([ # X Y Z\n [1/116.0, 1/116.0, 1/116.0], # R\n [1/500.0, 0, 0], # G\n [0, 0, -1/200.0], # B\n ]), requires_grad=False).cuda()\n\n img = torch.matmul(\n img + Variable(torch.cuda.FloatTensor([16.0, 0.0, 0.0])), lab_to_fxfyfz)\n\n epsilon = 6.0/29.0\n\n img = (((3.0 * epsilon**2 * (img-4.0/29.0)) * img.le(epsilon).float()) +\n ((torch.clamp(img, min=0.0001)**3.0) * img.gt(epsilon).float()))\n\n # denormalize for D65 white point\n img = torch.mul(img, Variable(\n torch.cuda.FloatTensor([0.950456, 1.0, 1.088754])))\n\n xyz_to_rgb = Variable(torch.FloatTensor([ # X Y Z\n [3.2404542, -0.9692660, 0.0556434], # R\n [-1.5371385, 1.8760108, -0.2040259], # G\n [-0.4985314, 0.0415560, 1.0572252], # B\n ]), requires_grad=False).cuda()\n\n img = torch.matmul(img, xyz_to_rgb)\n\n img = (img * 12.92 * img.le(0.0031308).float()) + ((torch.clamp(img,\n min=0.0001) ** (1/2.4) * 1.055) - 0.055) * img.gt(0.0031308).float()\n\n img = img.view(shape)\n img = img.permute(2, 1, 0)\n\n img = img.contiguous()\n img[(img != img).detach()] = 0\n \n return img\n\n @staticmethod\n def swapimdims_3HW_HW3(img):\n \"\"\"Move the image channels to the first dimension of the numpy\n multi-dimensional array\n\n :param img: numpy nd array representing the image\n :returns: numpy nd array with permuted axes\n :rtype: numpy nd array\n\n \"\"\"\n if img.ndim == 3:\n return np.swapaxes(np.swapaxes(img, 1, 2), 0, 2)\n elif img.ndim == 4:\n return np.swapaxes(np.swapaxes(img, 2, 3), 1, 3)\n\n @staticmethod\n def swapimdims_HW3_3HW(img):\n \"\"\"Move the image channels to the last dimensiion of the numpy\n multi-dimensional array\n\n :param img: numpy nd array representing the image\n :returns: numpy nd array with permuted axes\n :rtype: numpy nd array\n\n \"\"\"\n if img.ndim == 3:\n return np.swapaxes(np.swapaxes(img, 0, 2), 1, 2)\n elif img.ndim == 4:\n return np.swapaxes(np.swapaxes(img, 1, 3), 2, 3)\n\n @staticmethod\n def load_image(img_filepath, normaliser):\n \"\"\"Loads an image from file as a numpy multi-dimensional array\n\n :param img_filepath: filepath to the image\n :returns: image as a multi-dimensional numpy array\n :rtype: multi-dimensional numpy array\n\n \"\"\"\n img = ImageProcessing.normalise_image(np.array(Image.open(img_filepath)), normaliser) # NB: imread normalises to 0-1\n return img\n\n @staticmethod\n def normalise_image(img, normaliser):\n \"\"\"Normalises image data to be a float between 0 and 1\n\n :param img: Image as a numpy multi-dimensional image array\n :returns: Normalised image as a numpy multi-dimensional image array\n :rtype: Numpy array\n\n \"\"\"\n img = img.astype('float32') / normaliser\n return img\n\n @staticmethod\n def compute_mse(original, result):\n \"\"\"Computes the mean squared error between to RGB images represented as multi-dimensional numpy arrays.\n\n :param original: input RGB image as a numpy array\n :param result: target RGB image as a numpy array\n :returns: the mean squared error between the input and target images\n :rtype: float\n\n \"\"\"\n return ((original - result) ** 2).mean()\n\n @staticmethod\n def compute_psnr(image_batchA, image_batchB, max_intensity):\n \"\"\"Computes the PSNR for a batch of input and output images\n\n :param image_batchA: numpy nd-array representing the image batch A of shape Bx3xWxH\n :param image_batchB: numpy nd-array representing the image batch A of shape Bx3xWxH\n :param max_intensity: maximum intensity possible in the image (e.g. 255)\n :returns: average PSNR for the batch of images\n :rtype: float\n\n \"\"\"\n num_images = image_batchA.shape[0]\n psnr_val = 0.0\n\n for i in range(0, num_images):\n imageA = image_batchA[i, 0:3, :, :]\n imageB = image_batchB[i, 0:3, :, :]\n imageB = np.maximum(0, np.minimum(imageB, max_intensity))\n psnr_val += 10 * \\\n np.log10(max_intensity ** 2 /\n ImageProcessing.compute_mse(imageA, imageB))\n\n return psnr_val / num_images\n\n @staticmethod\n def compute_ssim(image_batchA, image_batchB):\n \"\"\"Computes the SSIM for a batch of input and output images\n\n :param image_batchA: numpy nd-array representing the image batch A of shape Bx3xWxH\n :param image_batchB: numpy nd-array representing the image batch A of shape Bx3xWxH\n :param max_intensity: maximum intensity possible in the image (e.g. 255)\n :returns: average PSNR for the batch of images\n :rtype: float\n\n \"\"\"\n num_images = image_batchA.shape[0]\n ssim_val = 0.0\n\n for i in range(0, num_images):\n imageA = ImageProcessing.swapimdims_3HW_HW3(\n image_batchA[i, 0:3, :, :])\n imageB = ImageProcessing.swapimdims_3HW_HW3(\n image_batchB[i, 0:3, :, :])\n ssim_val += ssim(imageA, imageB, data_range=imageA.max() - imageA.min(), multichannel=True,\n gaussian_weights=True, win_size=11)\n\n return ssim_val / num_images\n\n @staticmethod\n def hsv_to_rgb(img):\n \"\"\"Converts a HSV image to RGB\n PyTorch implementation of RGB to HSV conversion: https://docs.opencv.org/3.3.0/de/d25/imgproc_color_conversions.html\n Based roughly on a similar implementation here: http://code.activestate.com/recipes/576919-python-rgb-and-hsv-conversion/\n\n :param img: HSV image\n :returns: RGB image\n :rtype: Tensor\n\n \"\"\"\n img=torch.clamp(img,0,1)\n img = img.permute(2, 1, 0)\n \n m1 = 0\n m2 = (img[:, :, 2]*(1-img[:, :, 1])-img[:, :, 2])/60\n m3 = 0\n m4 = -1*m2\n m5 = 0\n\n r = img[:, :, 2]+torch.clamp(img[:, :, 0]*360-0, 0, 60)*m1+torch.clamp(img[:, :, 0]*360-60, 0, 60)*m2+torch.clamp(\n img[:, :, 0]*360-120, 0, 120)*m3+torch.clamp(img[:, :, 0]*360-240, 0, 60)*m4+torch.clamp(img[:, :, 0]*360-300, 0, 60)*m5\n\n m1 = (img[:, :, 2]-img[:, :, 2]*(1-img[:, :, 1]))/60\n m2 = 0\n m3 = -1*m1\n m4 = 0\n\n g = img[:, :, 2]*(1-img[:, :, 1])+torch.clamp(img[:, :, 0]*360-0, 0, 60)*m1+torch.clamp(img[:, :, 0]*360-60,\n 0, 120)*m2+torch.clamp(img[:, :, 0]*360-180, 0, 60)*m3+torch.clamp(img[:, :, 0]*360-240, 0, 120)*m4\n\n m1 = 0\n m2 = (img[:, :, 2]-img[:, :, 2]*(1-img[:, :, 1]))/60\n m3 = 0\n m4 = -1*m2\n\n b = img[:, :, 2]*(1-img[:, :, 1])+torch.clamp(img[:, :, 0]*360-0, 0, 120)*m1+torch.clamp(img[:, :, 0]*360 -\n 120, 0, 60)*m2+torch.clamp(img[:, :, 0]*360-180, 0, 120)*m3+torch.clamp(img[:, :, 0]*360-300, 0, 60)*m4\n\n img = torch.stack((r, g, b), 2)\n img[(img != img).detach()] = 0\n\n img = img.permute(2, 1, 0)\n img = img.contiguous()\n img = torch.clamp(img, 0, 1)\n\n return img\n\n\n\n @staticmethod\n def rgb_to_hsv(img):\n \"\"\"Converts an RGB image to HSV\n PyTorch implementation of RGB to HSV conversion: https://docs.opencv.org/3.3.0/de/d25/imgproc_color_conversions.html\n Based roughly on a similar implementation here: http://code.activestate.com/recipes/576919-python-rgb-and-hsv-conversion/\n\n :param img: RGB image\n :returns: HSV image\n :rtype: Tensor\n\n \"\"\"\n img=torch.clamp(img,1e-9,1) \n\n img = img.permute(2, 1, 0)\n shape = img.shape\n\n img = img.contiguous()\n img = img.view(-1, 3)\n\n mx = torch.max(img, 1)[0]\n mn = torch.min(img, 1)[0]\n\n ones = Variable(torch.FloatTensor(\n torch.ones((img.shape[0])))).cuda()\n zero = Variable(torch.FloatTensor(torch.zeros(shape[0:2]))).cuda()\n\n img = img.view(shape)\n\n ones1 = ones[0:math.floor((ones.shape[0]/2))]\n ones2 = ones[math.floor(ones.shape[0]/2):(ones.shape[0])]\n\n mx1 = mx[0:math.floor((ones.shape[0]/2))]\n mx2 = mx[math.floor(ones.shape[0]/2):(ones.shape[0])]\n mn1 = mn[0:math.floor((ones.shape[0]/2))]\n mn2 = mn[math.floor(ones.shape[0]/2):(ones.shape[0])]\n\n df1 = torch.add(mx1, torch.mul(ones1*-1, mn1))\n df2 = torch.add(mx2, torch.mul(ones2*-1, mn2))\n\n df = torch.cat((df1, df2), 0)\n del df1, df2\n df = df.view(shape[0:2])+1e-10\n mx = mx.view(shape[0:2])\n\n img = img.cuda()\n df = df.cuda()\n mx = mx.cuda()\n\n g = img[:, :, 1].clone().cuda()\n b = img[:, :, 2].clone().cuda()\n r = img[:, :, 0].clone().cuda()\n\n img_copy = img.clone()\n \n img_copy[:, :, 0] = (((g-b)/df)*r.eq(mx).float() + (2.0+(b-r)/df)\n * g.eq(mx).float() + (4.0+(r-g)/df)*b.eq(mx).float())\n img_copy[:, :, 0] = img_copy[:, :, 0]*60.0\n\n zero = zero.cuda()\n img_copy2 = img_copy.clone()\n\n img_copy2[:, :, 0] = img_copy[:, :, 0].lt(zero).float(\n )*(img_copy[:, :, 0]+360) + img_copy[:, :, 0].ge(zero).float()*(img_copy[:, :, 0])\n\n img_copy2[:, :, 0] = img_copy2[:, :, 0]/360\n\n del img, r, g, b\n\n img_copy2[:, :, 1] = mx.ne(zero).float()*(df/mx) + \\\n mx.eq(zero).float()*(zero)\n img_copy2[:, :, 2] = mx\n \n img_copy2[(img_copy2 != img_copy2).detach()] = 0\n\n img = img_copy2.clone()\n\n img = img.permute(2, 1, 0)\n img = torch.clamp(img, 1e-9, 1)\n\n return img\n\n \n @staticmethod\n def apply_curve(img, C, slope_sqr_diff, channel_in, channel_out,\n clamp=False):\n \"\"\"Applies a peicewise linear curve defined by a set of knot points to\n an image channel\n\n :param img: image to be adjusted\n :param C: predicted knot points of curve\n :returns: adjusted image\n :rtype: Tensor\n\n \"\"\"\n slope = Variable(torch.zeros((C.shape[0]-1))).cuda()\n curve_steps = C.shape[0]-1\n\n '''\n Compute the slope of the line segments\n '''\n for i in range(0, C.shape[0]-1):\n slope[i] = C[i+1]-C[i]\n\n '''\n Compute the squared difference between slopes\n '''\n for i in range(0, slope.shape[0]-1):\n slope_sqr_diff += (slope[i+1]-slope[i])*(slope[i+1]-slope[i])\n\n '''\n Use predicted line segments to compute scaling factors for the channel\n '''\n scale = float(C[0])\n for i in range(0, slope.shape[0]-1):\n if clamp:\n scale += float(slope[i])*(torch.clamp(img[:, :,channel_in]*curve_steps-i,0,1))\n else:\n scale += float(slope[i])*(img[:, :,channel_in]*curve_steps-i)\n \n img_copy = img.clone()\n \n img_copy[:, :, channel_out] = img[:, :, channel_out]*scale\n \n img_copy = torch.clamp(img_copy,0,1)\n \n return img_copy, slope_sqr_diff\n\n @staticmethod\n def adjust_hsv(img, S):\n \"\"\"Adjust the HSV channels of a HSV image using learnt curves\n\n :param img: image to be adjusted \n :param S: predicted parameters of piecewise linear curves\n :returns: adjust image, regularisation term\n :rtype: Tensor, float\n\n \"\"\"\n img = img.squeeze(0).permute(2, 1, 0)\n shape = img.shape\n img = img.contiguous()\n\n S1 = torch.exp(S[0:int(S.shape[0]/4)])\n S2 = torch.exp(S[(int(S.shape[0]/4)):(int(S.shape[0]/4)*2)])\n S3 = torch.exp(S[(int(S.shape[0]/4)*2):(int(S.shape[0]/4)*3)])\n S4 = torch.exp(S[(int(S.shape[0]/4)*3):(int(S.shape[0]/4)*4)])\n\n slope_sqr_diff = Variable(torch.zeros(1)*0.0).cuda()\n\n '''\n Adjust Hue channel based on Hue using the predicted curve\n '''\n img_copy, slope_sqr_diff = ImageProcessing.apply_curve(\n img, S1, slope_sqr_diff, channel_in=0, channel_out=0)\n\n '''\n Adjust Saturation channel based on Hue using the predicted curve\n '''\n img_copy, slope_sqr_diff = ImageProcessing.apply_curve(\n img_copy, S2, slope_sqr_diff, channel_in=0, channel_out=1)\n \n '''\n Adjust Saturation channel based on Saturation using the predicted curve\n '''\n img_copy, slope_sqr_diff = ImageProcessing.apply_curve(\n img_copy, S3, slope_sqr_diff, channel_in=1, channel_out=1)\n\n '''\n Adjust Value channel based on Value using the predicted curve\n '''\n img_copy, slope_sqr_diff = ImageProcessing.apply_curve(\n img_copy, S4, slope_sqr_diff, channel_in=2, channel_out=2)\n\n img = img_copy.clone()\n del img_copy\n\n img[(img != img).detach()] = 0\n\n img = img.permute(2, 1, 0)\n img = img.contiguous()\n \n return img, slope_sqr_diff\n\n @staticmethod\n def adjust_rgb(img, R):\n \"\"\"Adjust the RGB channels of a RGB image using learnt curves\n\n :param img: image to be adjusted \n :param S: predicted parameters of piecewise linear curves\n :returns: adjust image, regularisation term\n :rtype: Tensor, float\n\n \"\"\"\n img = img.squeeze(0).permute(2, 1, 0)\n shape = img.shape\n img = img.contiguous()\n\n '''\n Extract the parameters of the three curves\n '''\n R1 = torch.exp(R[0:int(R.shape[0]/3)])\n R2 = torch.exp(R[(int(R.shape[0]/3)):(int(R.shape[0]/3)*2)])\n R3 = torch.exp(R[(int(R.shape[0]/3)*2):(int(R.shape[0]/3)*3)])\n\n '''\n Apply the curve to the R channel \n '''\n slope_sqr_diff = Variable(torch.zeros(1)*0.0).cuda()\n\n img_copy, slope_sqr_diff = ImageProcessing.apply_curve(\n img, R1, slope_sqr_diff, channel_in=0, channel_out=0)\n\n '''\n Apply the curve to the G channel \n '''\n img_copy, slope_sqr_diff = ImageProcessing.apply_curve(\n img_copy, R2, slope_sqr_diff, channel_in=1, channel_out=1)\n\n '''\n Apply the curve to the B channel \n '''\n img_copy, slope_sqr_diff = ImageProcessing.apply_curve(\n img_copy, R3, slope_sqr_diff, channel_in=2, channel_out=2)\n\n img = img_copy.clone()\n del img_copy\n\n img[(img != img).detach()] = 0\n\n img = img.permute(2, 1, 0)\n img = img.contiguous()\n\n return img, slope_sqr_diff\n\n @staticmethod\n def adjust_lab(img, L):\n \"\"\"Adjusts the image in LAB space using the predicted curves\n\n :param img: Image tensor\n :param L: Predicited curve parameters for LAB channels\n :returns: adjust image, and regularisation parameter\n :rtype: Tensor, float\n\n \"\"\"\n img = img.permute(2, 1, 0)\n\n shape = img.shape\n img = img.contiguous()\n\n '''\n Extract predicted parameters for each L,a,b curve\n '''\n L1 = torch.exp(L[0:int(L.shape[0]/3)])\n L2 = torch.exp(L[(int(L.shape[0]/3)):(int(L.shape[0]/3)*2)])\n L3 = torch.exp(L[(int(L.shape[0]/3)*2):(int(L.shape[0]/3)*3)])\n\n slope_sqr_diff = Variable(torch.zeros(1)*0.0).cuda()\n\n '''\n Apply the curve to the L channel \n '''\n img_copy, slope_sqr_diff = ImageProcessing.apply_curve(\n img, L1, slope_sqr_diff, channel_in=0, channel_out=0)\n\n '''\n Now do the same for the a channel\n '''\n img_copy, slope_sqr_diff = ImageProcessing.apply_curve(\n img_copy, L2, slope_sqr_diff, channel_in=1, channel_out=1)\n\n '''\n Now do the same for the b channel\n '''\n img_copy, slope_sqr_diff = ImageProcessing.apply_curve(\n img_copy, L3, slope_sqr_diff, channel_in=2, channel_out=2)\n\n img = img_copy.clone()\n del img_copy\n\n img[(img != img).detach()] = 0\n\n img = img.permute(2, 1, 0)\n img = img.contiguous()\n\n return img, slope_sqr_diff\n"
] |
[
[
"matplotlib.use",
"torch.zeros",
"torch.cat",
"torch.mul",
"torch.stack",
"torch.min",
"torch.max",
"numpy.minimum",
"numpy.set_printoptions",
"torch.FloatTensor",
"torch.clamp",
"torch.ones",
"numpy.swapaxes",
"torch.cuda.FloatTensor",
"torch.matmul"
]
] |
intel-iot-devkit/motor-defect-detector-python
|
[
"e793357b4961f9052e35309acb60bec0851e15a0"
] |
[
"Kmeans/kmeanstestingall.py"
] |
[
"2# -*- coding: utf-8 -*-\n\"\"\"\nCreated on Thu May 24 17:13:42 2018\n\n@author: SHEFALI JAIN\n\"\"\"\n\"\"\"\n this script is used to test which bearing has failed using the trained Kmeans model.\n\"\"\"\n# import the libraries\nimport pandas as pd\nimport numpy as np\nimport matplotlib.pyplot as plt\n\nfrom utils import cal_max_freq,plotlabels\n\nimport os\n\ndef main():\n try:\n # user input for the path of the dataset\n filedir = input(\"enter the complete directory path \")\n filepath = input(\"enter the folder name\")\n\n # load the files\n all_files = os.listdir(filedir)\n freq_max1,freq_max2,freq_max3,freq_max4,freq_max5 = cal_max_freq(all_files,filepath)\n except IOError:\n print(\"you have entered either the wrong data directory path or filepath\")\n\n # load the model\n filename = \"kmeanModel.npy\"\n model = np.load(filename).item()\n\n # checking the iteration\n if (filepath == \"1st_test/\"):\n rhigh = 8\n else:\n rhigh = 4\n\n\n testlabels = []\n for i in range(0,rhigh):\n print(\"checking for the bearing\",i+1)\n result = pd.DataFrame()\n result['freq_max1'] = list((np.array(freq_max1))[:,i])\n result['freq_max2'] = list((np.array(freq_max2))[:,i])\n result['freq_max3'] = list((np.array(freq_max3))[:,i])\n result['freq_max4'] = list((np.array(freq_max4))[:,i])\n result['freq_max5'] = list((np.array(freq_max5))[:,i])\n\n X = result[[\"freq_max1\",\"freq_max2\",\"freq_max3\",\"freq_max4\",\"freq_max5\"]]\n\n label = model.predict(X)\n labelfive = list(label[-100:]).count(5)\n labelsix = list(label[-100:]).count(6)\n labelseven = list(label[-100:]).count(7)\n totalfailur = labelfive+labelsix+labelseven#+labelfour\n ratio = (totalfailur/100)*100\n if(ratio >= 25):\n print(\"bearing is suspected to fail\")\n else:\n print(\"bearing is working in normal condition\")\n\n testlabels.append(label[-100:])\n\n # ploting the labels\n plotlabels(testlabels)\n plt.show()\n\n\nif __name__== \"__main__\":\n main()"
] |
[
[
"matplotlib.pyplot.show",
"numpy.array",
"numpy.load",
"pandas.DataFrame"
]
] |
e-kayrakli/arkouda
|
[
"59da8f05f8dbf71382083964bc1b59ddceedc1ac"
] |
[
"converter/lanl_format.py"
] |
[
"import numpy as np\nimport re\n\nsplitter = re.compile(r'([a-zA-Z]*)(\\d*)')\n\ndef numerify(s):\n name, num = splitter.match(s).groups()\n prefix = hash(name) & 0xffff\n if num == '':\n postfix = 0\n else:\n postfix = int(num)\n return np.int64((prefix << 20) ^ postfix)\n\ndef deport(s):\n return np.int64(s.lstrip('Port'))\n\nlanl_options = {'delimiter': ',',\n 'header':None,\n 'names':['start', 'duration', 'srcIP', 'dstIP', 'proto',\n 'srcPort', 'dstPort', 'srcPkts', 'dstPkts',\n 'srcBytes', 'dstBytes'],\n 'converters': {# 'srcIP': numerify, 'dstIP': numerify,\n 'srcPort': deport, 'dstPort': deport},\n 'dtype': {'srcIP': np.str_, 'dstIP': np.str_,\n 'start': np.int64, 'duration': np.int64,\n 'proto': np.int64, 'srcPkts': np.int64,\n 'dstPkts': np.int64, 'srcBytes': np.float64,\n 'dstBytes': np.float64}}\n\n\nOPTIONS = {}\nOPTIONS['lanl'] = lanl_options\n"
] |
[
[
"numpy.int64"
]
] |
JwDong2019/CVWC-2019-FCOS
|
[
"9c343dc820aea7bc79ee0b8206527fa48ebf106f"
] |
[
"mmdet/models/bbox_heads/convfc_bbox_head.py"
] |
[
"import torch.nn as nn\n\nfrom .bbox_head import BBoxHead\nfrom ..registry import HEADS\nfrom ..utils import ConvModule\n\nfrom mmcv.cnn import kaiming_init, constant_init\n\nfrom ..detectors.yang.Mergev2 import Merge\n\[email protected]_module\nclass ConvFCBBoxHead(BBoxHead):\n \"\"\"More general bbox head, with shared conv and fc layers and two optional\n separated branches.\n\n /-> cls convs -> cls fcs -> cls\n shared convs -> shared fcs\n \\-> reg convs -> reg fcs -> reg\n \"\"\" # noqa: W605\n\n def __init__(self,\n num_shared_convs=0,\n num_shared_fcs=0,\n num_cls_convs=0,\n num_cls_fcs=0,\n num_reg_convs=0,\n num_reg_fcs=0,\n conv_out_channels=256,\n fc_out_channels=1024,\n conv_cfg=None,\n norm_cfg=None,\n *args,\n **kwargs):\n super(ConvFCBBoxHead, self).__init__(*args, **kwargs)\n assert (num_shared_convs + num_shared_fcs + num_cls_convs +\n num_cls_fcs + num_reg_convs + num_reg_fcs > 0)\n if num_cls_convs > 0 or num_reg_convs > 0:\n assert num_shared_fcs == 0\n if not self.with_cls:\n assert num_cls_convs == 0 and num_cls_fcs == 0\n if not self.with_reg:\n assert num_reg_convs == 0 and num_reg_fcs == 0\n self.num_shared_convs = num_shared_convs\n self.num_shared_fcs = num_shared_fcs\n self.num_cls_convs = num_cls_convs\n self.num_cls_fcs = num_cls_fcs\n self.num_reg_convs = num_reg_convs\n self.num_reg_fcs = num_reg_fcs\n self.conv_out_channels = conv_out_channels\n self.fc_out_channels = fc_out_channels\n self.conv_cfg = conv_cfg\n self.norm_cfg = norm_cfg\n\n # # different heads for classification and localization\n # self.conv1_yang = nn.Conv2d(256, 1024, 3, padding=1)\n # self.conv2_yang = nn.Conv2d(1024, 1024, 3, padding=1)\n # self.conv3_yang = nn.Conv2d(1024, 1024, 3, padding=1)\n # self.avg_pooly = nn.AdaptiveAvgPool2d(1)\n # # init\n # kaiming_init(self.conv1_yang)\n # kaiming_init(self.conv2_yang)\n # kaiming_init(self.conv3_yang)\n\n # add shared convs and fcs\n self.shared_convs, self.shared_fcs, last_layer_dim = \\\n self._add_conv_fc_branch(\n self.num_shared_convs, self.num_shared_fcs, self.in_channels,\n True)\n self.shared_out_channels = last_layer_dim\n\n # _, self.shared_fcs_extra, _ = self._add_conv_fc_branch(0, self.num_shared_fcs, self.in_channels, True)\n\n # add cls specific branch\n self.cls_convs, self.cls_fcs, self.cls_last_dim = \\\n self._add_conv_fc_branch(\n self.num_cls_convs, self.num_cls_fcs, self.shared_out_channels)\n\n # add reg specific branch\n self.reg_convs, self.reg_fcs, self.reg_last_dim = \\\n self._add_conv_fc_branch(\n self.num_reg_convs, self.num_reg_fcs, self.shared_out_channels)\n\n if self.num_shared_fcs == 0 and not self.with_avg_pool:\n if self.num_cls_fcs == 0:\n self.cls_last_dim *= (self.roi_feat_size * self.roi_feat_size)\n if self.num_reg_fcs == 0:\n self.reg_last_dim *= (self.roi_feat_size * self.roi_feat_size)\n\n self.relu = nn.ReLU(inplace=True)\n # reconstruct fc_cls and fc_reg since input channels are changed\n if self.with_cls:\n self.fc_cls = nn.Linear(self.cls_last_dim, self.num_classes)\n if self.with_reg:\n out_dim_reg = (4 if self.reg_class_agnostic else 4 *\n self.num_classes)\n self.fc_reg = nn.Linear(self.reg_last_dim, out_dim_reg)\n\n self.M = Merge()\n\n def _add_conv_fc_branch(self,\n num_branch_convs,\n num_branch_fcs,\n in_channels,\n is_shared=False):\n \"\"\"Add shared or separable branch\n\n convs -> avg pool (optional) -> fcs\n \"\"\"\n last_layer_dim = in_channels\n # add branch specific conv layers\n branch_convs = nn.ModuleList()\n if num_branch_convs > 0:\n for i in range(num_branch_convs):\n conv_in_channels = (\n last_layer_dim if i == 0 else self.conv_out_channels)\n branch_convs.append(\n ConvModule(\n conv_in_channels,\n self.conv_out_channels,\n 3,\n padding=1,\n conv_cfg=self.conv_cfg,\n norm_cfg=self.norm_cfg))\n last_layer_dim = self.conv_out_channels\n # add branch specific fc layers\n branch_fcs = nn.ModuleList()\n if num_branch_fcs > 0:\n # for shared branch, only consider self.with_avg_pool\n # for separated branches, also consider self.num_shared_fcs\n if (is_shared\n or self.num_shared_fcs == 0) and not self.with_avg_pool:\n last_layer_dim *= (self.roi_feat_size * self.roi_feat_size)\n for i in range(num_branch_fcs):\n fc_in_channels = (\n last_layer_dim if i == 0 else self.fc_out_channels)\n branch_fcs.append(\n nn.Linear(fc_in_channels, self.fc_out_channels))\n last_layer_dim = self.fc_out_channels\n return branch_convs, branch_fcs, last_layer_dim\n\n def init_weights(self):\n super(ConvFCBBoxHead, self).init_weights()\n for module_list in [self.shared_fcs, self.cls_fcs, self.reg_fcs]:\n for m in module_list.modules():\n if isinstance(m, nn.Linear):\n nn.init.xavier_uniform_(m.weight)\n nn.init.constant_(m.bias, 0)\n\n def forward(self, x):\n # shared part\n if self.num_shared_convs > 0:\n for conv in self.shared_convs:\n x = conv(x)\n\n if self.num_shared_fcs > 0:\n if self.with_avg_pool:\n x = self.avg_pool(x)\n x = x.view(x.size(0), -1)\n # xextra = xextra.view(xextra.size(0), -1)\n for fc in self.shared_fcs:\n x = self.relu(fc(x))\n # for fcextra in self.shared_fcs_extra:\n # xextra = self.relu(fcextra(xextra))\n\n # x = self.M(features, x)\n\n # separate branches\n x_cls = x\n x_reg = x\n\n for conv in self.cls_convs:\n x_cls = conv(x_cls)\n if x_cls.dim() > 2:\n if self.with_avg_pool:\n x_cls = self.avg_pool(x_cls)\n x_cls = x_cls.view(x_cls.size(0), -1)\n for fc in self.cls_fcs:\n x_cls = self.relu(fc(x_cls))\n\n for conv in self.reg_convs:\n x_reg = conv(x_reg)\n if x_reg.dim() > 2:\n if self.with_avg_pool:\n x_reg = self.avg_pool(x_reg)\n x_reg = x_reg.view(x_reg.size(0), -1)\n for fc in self.reg_fcs:\n x_reg = self.relu(fc(x_reg))\n\n cls_score = self.fc_cls(x_cls) if self.with_cls else None\n bbox_pred = self.fc_reg(x_reg) if self.with_reg else None\n return cls_score, bbox_pred\n\n\[email protected]_module\nclass SharedFCBBoxHead(ConvFCBBoxHead):\n\n def __init__(self, num_fcs=2, fc_out_channels=1024, *args, **kwargs):\n assert num_fcs >= 1\n super(SharedFCBBoxHead, self).__init__(\n num_shared_convs=0,\n num_shared_fcs=num_fcs,\n num_cls_convs=0,\n num_cls_fcs=0,\n num_reg_convs=0,\n num_reg_fcs=0,\n fc_out_channels=fc_out_channels,\n *args,\n **kwargs)\n"
] |
[
[
"torch.nn.Linear",
"torch.nn.ModuleList",
"torch.nn.init.constant_",
"torch.nn.init.xavier_uniform_",
"torch.nn.ReLU"
]
] |
pdigese/CarND-Capstone-team-5-june20
|
[
"0c7a7539a61c48f62b7557a76edd2c6308c60b81"
] |
[
"ros/src/object_detection/object_detection.py"
] |
[
"#!/usr/bin/env python\n\nimport rospy\nfrom sensor_msgs.msg import Image\nfrom geometry_msgs.msg import PoseStamped, Pose\nimport tensorflow as tf\nimport os\nfrom PIL import (\n Image as Img,\n) # Must not be named image since there is a datatype called Image from ROS\nfrom PIL import ImageColor\nfrom PIL import ImageDraw\nfrom PIL import ImageFont\nimport numpy as np\nfrom cv_bridge import CvBridge\nimport json\nfrom styx_msgs.msg import TrafficLightArray, TrafficLight\nimport yaml\n\n\nSSD_GRAPH_FILE = \"/../../../nn/frozen_inference_graph.pb\"\n\n\"\"\"\nTODO: Description goes here.\nThis file is aimed to use the machine learning method to process the upcoming traffic light in real life and publishes color prediction and the location of any upcomming red lights.\n\"\"\"\n\n\"\"\"\nNote: All functions (whih are not belonging to the class itself) are copied from:\n- load_graph, pick_best_box and to_image_coords:\nhttps://github.com/udacity/CarND-Object-Detection-Lab/blob/master/CarND-Object-Detection-Lab.ipynb\n- pipeline:\nhttps://github.com/udacity/CarND-Object-Detection-Lab/blob/e91d0de6cd54834966cdb06b0172e23f8a0c124f/exercise-solutions/e5.py\n\"\"\"\n\n# FIXME: needs to go somewhere in an encapsulation\ncmap = ImageColor.colormap\nprint(\"Number of colors =\", len(cmap))\nCOLOR_LIST = sorted([c for c in cmap.keys()])\n\n\nclass ObjDetection(object):\n def __init__(self):\n rospy.init_node(\"object_detection\")\n\n self.latest_img = None\n self.cnt = 0\n self.latest_tls = None # latest traffic light state\n self.curr_pose = None\n\n config_string = rospy.get_param(\"/traffic_light_config\")\n self.config = yaml.load(config_string)\n\n # subscribe the camera topic\n # publish what? traffic light? other objects?\n # load coco\n self.detection_graph = self.load_graph(\n os.path.dirname(os.path.abspath(__file__)) + SSD_GRAPH_FILE\n )\n annotation_path = os.path.dirname(os.path.abspath(__file__)) + SSD_GRAPH_FILE\n\n self.sub_img = rospy.Subscriber(\"/image_raw\", Image, self.image_cb)\n self.sub_tls = rospy.Subscriber(\n \"/vehicle/traffic_lights\", TrafficLightArray, self.tls_cb\n )\n self.sub_pos = rospy.Subscriber(\"/current_pose\", PoseStamped, self.pose_cb)\n \n self.pub_detected = rospy.Publisher(\"/image_detection\", Image, queue_size=1)\n self.upcoming_red_light_pub = rospy.Publisher('/traffic_waypoint', Int32, queue_size=1)\n\n rospy.loginfo(\"Coco has been initialized successfully\")\n self.loop()\n\n def loop(self):\n rate = rospy.Rate(\n 10\n ) # 1Hz TODO: What's a goo frequency here? It's an heavy job...\n with tf.Session(graph=self.detection_graph) as self.sess:\n self.image_tensor = self.sess.graph.get_tensor_by_name(\"image_tensor:0\")\n self.detection_boxes = self.sess.graph.get_tensor_by_name(\n \"detection_boxes:0\"\n )\n self.detection_scores = self.sess.graph.get_tensor_by_name(\n \"detection_scores:0\"\n )\n self.detection_classes = self.sess.graph.get_tensor_by_name(\n \"detection_classes:0\"\n )\n while not rospy.is_shutdown():\n\n if self.latest_img is not None:\n processed_image = self.pipeline(self.latest_img)\n pub_img = Image()\n bridge = CvBridge()\n \"\"\"\n rqt_image_view reports with pathtrough:\n ImageView.callback_image() could not convert image from '8UC3' to 'rgb8' ([8UC3] is not\n a color format. but [rgb8] is. The conversion does not make sense).\n => therefore encoding\n \"\"\"\n image_message = bridge.cv2_to_imgmsg(\n processed_image, encoding=\"rgb8\"\n ) # required by the rqt_image_viewer\n# self.pub_detected.publish(image_message)\n rate.sleep()\n\n def pose_cb(self, msg):\n self.curr_pose = msg\n\n def image_cb(self, msg):\n bridge = CvBridge()\n # for conversion please read:\n # http://wiki.ros.org/cv_bridge/Tutorials/ConvertingBetweenROSImagesAndOpenCVImagesPython\n self.latest_img = bridge.imgmsg_to_cv2(\n msg, desired_encoding=\"rgb8\"\n ) # TODO: coco requires rgb as far as I know\n stop_line_pos = self.config[\"stop_line_positions\"]\n state = get_closest_tl_state(self.curr_pose)\n if state == 'TrafficLight.RED': # TODO: not sure if TrafficLight.RED is right\n self.upcoming_red_light_pub.publish(stop_line_pos) # TODO: check with waypoints updater shall we publish index(like as tl_detector.py) or position. Since we have only one traffic light would be easier, no index issue\n\n def tls_cb(self, msg):\n # receives a whole list of traffic lights\n self.latest_tls = msg.lights\n\n def load_graph(self, graph_file):\n \"\"\"Loads a frozen inference graph\"\"\"\n graph = tf.Graph()\n with graph.as_default():\n od_graph_def = tf.GraphDef()\n with tf.gfile.GFile(graph_file, \"rb\") as fid:\n serialized_graph = fid.read()\n od_graph_def.ParseFromString(serialized_graph)\n tf.import_graph_def(od_graph_def, name=\"\")\n return graph\n\n def pick_best_box(self, min_score, boxes, scores, classes):\n \"\"\"Return a box with the highest confidence\"\"\"\n n = len(classes)\n idxs = []\n max_score = -1\n for i in range(n):\n if scores[i] >= max_score and scores[i] >= min_score:\n idxs = [i]\n max_score = scores[i]\n\n filtered_box = boxes[idxs, ...]\n filtered_score = scores[idxs, ...]\n filtered_class = classes[idxs, ...]\n return filtered_box, filtered_score, filtered_class\n\n def to_image_coords(self, boxes, height, width):\n \"\"\"\n The original box coordinate output is normalized, i.e [0, 1].\n\n This converts it back to the original coordinate based on the image\n size.\n \"\"\"\n box_coords = np.zeros_like(boxes)\n box_coords[:, 0] = boxes[:, 0] * height\n box_coords[:, 1] = boxes[:, 1] * width\n box_coords[:, 2] = boxes[:, 2] * height\n box_coords[:, 3] = boxes[:, 3] * width\n\n return box_coords\n\n def draw_boxes(self, image, boxes, classes, scores, thickness=4):\n \"\"\"Draw bounding boxes on the image\"\"\"\n draw = ImageDraw.Draw(image)\n for i in range(len(boxes)):\n bot, left, top, right = boxes[i, ...]\n color = (int(classes[i])//2*255, int(classes[i])%2*255, 0)\n draw.line(\n [(left, top), (left, bot), (right, bot), (right, top), (left, top)],\n width=thickness,\n fill=color,\n )\n draw.text((left, top), \"{}\".format(round(1E4*scores[i])/100))\n\n def get_closest_tl_state(self, pose):\n \"\"\"\n Returns the state of the closest traffic light\n \"\"\"\n # contains the stop_line_positions off all traffic lights\n min_dist = 9999999\n tlc_idx = 0\n stop_line_pos = self.config[\"stop_line_positions\"]\n for i, tl_pos in enumerate(stop_line_pos):\n # just check the eucledian distance to other traffic lights\n distance = np.sqrt(\n (tl_pos[0] + pose.pose.position.x) + (tl_pos[1] + pose.pose.position.y)\n )\n if min_dist > distance:\n min_dist = distance\n tlc_idx = i\n return self.latest_tls[i].state\n\n def pipeline(self, img):\n \"\"\"\n FIXME: I am just copied, but to make a proper job, I need to return either\n - just the region of interest (so the traffic light itself)\n - OR the information about the status of the traffic light.\n \"\"\"\n draw_img = Img.fromarray(img)\n boxes, scores, classes = self.sess.run(\n [self.detection_boxes, self.detection_scores, self.detection_classes],\n feed_dict={self.image_tensor: np.expand_dims(img, 0)},\n )\n # Remove unnecessary dimensions\n boxes = np.squeeze(boxes)\n scores = np.squeeze(scores)\n classes = np.squeeze(classes)\n\n confidence_cutoff = 0.4\n\n # Filter boxes with a confidence score less than `confidence_cutoff`\n best_box, best_score, best_class = self.pick_best_box(\n confidence_cutoff, boxes, scores, classes\n )\n\n # The current box coordinates are normalized to a range between 0 and 1.\n # This converts the coordinates actual location on the image.\n width, height = draw_img.size\n box_coords = self.to_image_coords(best_box, height, width)\n\n # Each class with be represented by a differently colored box\n self.draw_boxes(draw_img, box_coords, best_class, best_score)\n return np.array(draw_img)\n\n\nif __name__ == \"__main__\":\n try:\n ObjDetection()\n except rospy.ROSInterruptException:\n rospy.logerr(\"Could not start object detection node.\")\n"
] |
[
[
"numpy.zeros_like",
"numpy.array",
"tensorflow.Graph",
"tensorflow.Session",
"tensorflow.GraphDef",
"tensorflow.import_graph_def",
"tensorflow.gfile.GFile",
"numpy.sqrt",
"numpy.squeeze",
"numpy.expand_dims"
]
] |
pan-webis-de/duan20
|
[
"dd2fd2b13787d40fed0c639dea1efa67668ccb43"
] |
[
"model/BERTModel.py"
] |
[
"import torch.nn as nn\nimport torch\n\nclass BERTModel(nn.Module):\n def __init__(self,\n bert,\n hidden_dim,\n output_dim,\n n_layers,\n bidirectional,\n dropout):\n \n super().__init__()\n \n self.bert = bert\n \n embedding_dim = bert.config.to_dict()['hidden_size']\n \n self.rnn = nn.GRU(embedding_dim,\n hidden_dim,\n num_layers = n_layers,\n bidirectional = bidirectional,\n batch_first = True,\n dropout = 0 if n_layers < 2 else dropout)\n \n self.out = nn.Linear(hidden_dim * 2 if bidirectional else hidden_dim, output_dim)\n \n self.dropout = nn.Dropout(dropout)\n \n def forward(self, text):\n \n #text = [batch size, sent len]\n \n with torch.no_grad():\n embedded = self.bert(text)[0]\n \n #embedded = [batch size, sent len, emb dim]\n \n _, hidden = self.rnn(embedded)\n \n #hidden = [n layers * n directions, batch size, emb dim]\n \n if self.rnn.bidirectional:\n hidden = self.dropout(torch.cat((hidden[-2,:,:], hidden[-1,:,:]), dim = 1))\n else:\n hidden = self.dropout(hidden[-1,:,:])\n \n #hidden = [batch size, hid dim]\n \n output = self.out(hidden)\n \n #output = [batch size, out dim]\n \n return output"
] |
[
[
"torch.nn.Linear",
"torch.nn.Dropout",
"torch.cat",
"torch.nn.GRU",
"torch.no_grad"
]
] |
jswoboda/GeoDataPython
|
[
"c3e29541327ec754eb5a2a9e8dd94bf1abee3328"
] |
[
"Test/subplots_test.py"
] |
[
"#!/usr/bin/env python3\n# -*- coding: utf-8 -*-\n\"\"\"\nExample code on using the alt_slice_overlay function in the plotting.py file.\nTakes two h5 files--a RISR file and an OMTI file-- and creates 2 objects. This is inputed into alt_slice_overlay.\nThe output is a 2D colorplot with the OMTI data on the bottom in grayscale and the RISR parameter on top with an alpha of 0.4\n@author: Anna Stuhlmacher, Michael Hirsch\n\nfirst you need to install GeoDataPython by::\n python setup.py develop\n\"\"\"\nfrom __future__ import division, absolute_import\nfrom matplotlib.pyplot import subplots,show\nimport numpy as np\n#\nimport GeoData.plotting as GP\n#\nfrom load_isropt import load_risromti\n#\npicktimeind = [1,2] #arbitrary user time index choice\n\ndef plotisropt(risrName,omtiName):\n\n risr,omti = load_risromti(risrName,omtiName)\n #first object in geodatalist is being overlayed over by the second object\n altlist = [300]\n xyvecs = [np.linspace(-100.0,500.0),np.linspace(0.0,600.0)]\n vbounds = [[200,800],[5e10,5e11]]\n title='OMTI data and NE linear interpolation'\n\n fig3, (ax1, ax2) = subplots(1,2,figsize=(10,5))\n ax1 = GP.alt_slice_overlay((omti, risr), altlist, xyvecs, vbounds, title, axis=ax1,\n picktimeind=picktimeind)\n ax2 = GP.alt_contour_overlay((omti, risr), altlist, xyvecs, vbounds, title, axis=ax2,\n picktimeind=picktimeind)\n\n ax1.set_ylabel('y')\n ax1.set_xlabel('x')\n ax2.set_ylabel('y')\n ax2.set_xlabel('x')\n\nif __name__ == '__main__':\n plotisropt(risrName='data/ran120219.004.hdf5',\n omtiName='data/OMTIdata.h5')\n show()\n"
] |
[
[
"matplotlib.pyplot.show",
"numpy.linspace",
"matplotlib.pyplot.subplots"
]
] |
tsipkens/tfer-PMA
|
[
"314cb6dccc4612d5c748bde6545f0aae15b64c26"
] |
[
"py/main.py"
] |
[
"# import functions\nimport matplotlib.pyplot as plt\nimport numpy as np\n\nfrom prop_pma import prop_pma\nimport tfer_pma # import relevant functions to evaluate the PMA transfer function\n\n# define input variables\nm_star = 0.01e-18\nm = np.arange(0.8, 1.2, 0.001) * m_star # numpy array spanning 80 to 120% m_star\nd = None # mobility diameter (none uses mass-mobility relation)\nz = 1.0 # integer charge state\n\nprop = prop_pma() # get default PMA properties\n\n# Modify some of the properties,\n# in this case for the mass-mobility relation.\nrho_eff = 900 # effective density\nprop['m0'] = rho_eff * np.pi / 6 * 1e-27 # copy mass-mobility relation info (only used to find Rm)\nprop['Dm'] = 3\n\n# prop['omega_hat'] = 1; # ratio of angular speeds (CPMA < 1 vs APM = 1)\n\nsp, _ = tfer_pma.get_setpoint(prop, 'm_star', m_star, 'Rm', 10)\n# sp, _ = tfer_pma.get_setpoint(prop, 'V', 24.44, 'omega', 2543.9) # alt. phrasing\n\n\n# evaluate the transfer functions\nLambda_1S, _ = tfer_pma.tfer_1S(sp, m, d, z, prop)\nLambda_1C, _ = tfer_pma.tfer_1C(sp, m, d, z, prop)\nLambda_1C_diff, _ = tfer_pma.tfer_1C_diff(sp, m, d, z, prop)\nif prop['omega_hat'] == 1:\n Lambda_W1, _ = tfer_pma.tfer_W1(sp, m, d, z, prop)\n Lambda_W1_diff, _ = tfer_pma.tfer_W1_diff(sp, m, d, z, prop)\n\n# plot the various transfer functions\nplt.plot(m, Lambda_1S)\nplt.plot(m, Lambda_1C)\nplt.plot(m, Lambda_1C_diff)\nif prop['omega_hat'] == 1:\n plt.plot(m, Lambda_W1)\n plt.plot(m, Lambda_W1_diff)\nplt.show()\n\n# generate second plot demonstrating multiple charging\nm123 = np.arange(0.6, 3.4, 0.001) * m_star\nLambda_1C_z1, _ = tfer_pma.tfer_1C_diff(sp, m123, d, 1, prop)\nLambda_1C_z2, _ = tfer_pma.tfer_1C_diff(sp, m123, d, 2, prop)\nLambda_1C_z3, _ = tfer_pma.tfer_1C_diff(sp, m123, d, 3, prop)\n\nplt.plot(m123, Lambda_1C_z1)\nplt.plot(m123, Lambda_1C_z2)\nplt.plot(m123, Lambda_1C_z3)\nplt.plot(m123, Lambda_1C_z1 + Lambda_1C_z2 + Lambda_1C_z3, 'k--')\nplt.show()\n"
] |
[
[
"matplotlib.pyplot.show",
"matplotlib.pyplot.plot",
"numpy.arange"
]
] |
robbievanleeuwen/steeldesign
|
[
"4a2351c7d922a4f0f379b413ecca17b672be1c6e"
] |
[
"steeldesign/member.py"
] |
[
"import numpy as np\n\n\nclass Member:\n \"\"\"a\"\"\"\n\n def __init__(self, section, length, restraints):\n \"\"\"Inits the Member class.\"\"\"\n\n self.section = section\n self.length = length\n self.restraints = []\n\n for restraint in restraints:\n self.add_restraint(restraint)\n\n def add_restraint(self, restraint):\n \"\"\"a\"\"\"\n\n # add the restraint to the list\n self.restraints.append(restraint)\n\n # reorder restraints\n self.restraints.sort(key=lambda r: r.pos)\n\n def calc_phi_mbx(self, bmd, load_position, alpha_m=None):\n \"\"\"Calculates phiMbx for the member for each segment based on the applied restraints and\n supplied bending moment diagram *bmd*.\n\n :param bmd: Bending moment diagram for the member of size *(n x 2)*\n :type bmd: :class:`numpy.ndarray`\n :param load_position: List of strings defining the load position within each segment i.e.\n 'WS' - within & shear centre; 'ES' - end & shear centre; 'WT' - within & top flange;\n 'ET' - end & top flange\n :type load_position: list[strings]\n :param alpha_m: Optional list of alpha_m values to override those calculated from the bmd\n :type alpha_m: list[float]\n :returns: A list of phiMbx values for each segment\n :rtype: list[float]\n\n :raises Exception: Size of load_position is not equal to the number of segments\n :raises Exception: Size of alpha_m is not equal to the number of segments\n \"\"\"\n\n # check length of load_position equals number of segments\n if len(load_position) != len(self.restraints) - 1:\n raise Exception('Size of load_position must equal number of segments')\n\n # check length of alpha_m equals number of segments\n if alpha_m is not None:\n if len(alpha_m) != len(self.restraints) - 1:\n raise Exception('Size of alpha_m must equal number of segments')\n\n # generate segments\n segments = []\n\n for i in range(len(self.restraints) - 1):\n segments.append(Segment(self.length, self.restraints[i], self.restraints[i+1],\n load_position[i], bmd, self.section))\n\n # calculate alpha_m values\n for (i, segment) in enumerate(segments):\n if alpha_m is None:\n segment.calc_alpha_m()\n else:\n segment.alpha_m = alpha_m[i]\n\n phi_mbx = []\n\n # calculate phi_mbx for each segment\n for segment in segments:\n # calculate le\n le = segment.calc_effective_length()\n alpha_m = segment.alpha_m\n\n phi_mbx.append(self.section.calc_phi_mbx(le=le, alpha_m=alpha_m))\n\n return phi_mbx\n\n\nclass Segment:\n \"\"\"a\n\n :param float member_length: Total physical length of the memeber\n :param restraint1: Restraint at the start of the segment\n :type restraint1: :class:`steeldesign.codes.Restraint`\n :param restraint2: Restraint at the end of the segment\n :type restraint2: :class:`steeldesign.codes.Restraint`\n :param string load_position: Strings defining the load position\n :param bmd: Bending moment diagram for the member of size *(n x 2)*\n :type bmd: :class:`numpy.ndarray`\n :param section: Steel cross section object\n :type: :class:`steeldesign.sections.SteelSection`\n\n :cvar float segment_length: Physical length of the segment\n :cvar restraint1: Restraint at the start of the segment\n :vartype restraint1: :class:`steeldesign.codes.Restraint`\n :cvar restraint2: Restraint at the end of the segment\n :vartype restraint2: :class:`steeldesign.codes.Restraint`\n :cvar string load_position: Strings defining the load position\n :cvar bmd_segment: Bending moment diagram for the segment of size *(n x 2)*\n :type bmd_segment: :class:`numpy.ndarray`\n :cvar float alpha_m: Alpha_m value for the segment\n :cvar section: Steel cross section object\n :vartype: :class:`steeldesign.sections.SteelSection`\n \"\"\"\n\n def __init__(self, member_length, restraint1, restraint2, load_position,\n bmd, section):\n \"\"\"Inits the Segment class.\"\"\"\n\n self.segment_length = member_length * (restraint2.pos - restraint1.pos)\n self.restraint1 = restraint1\n self.restraint2 = restraint2\n self.load_position = load_position\n self.section = section\n\n bmd_segment = []\n\n # add start point for the segment bmd\n bmd_segment.append([restraint1.pos, np.interp(restraint1.pos, bmd[:, 0], bmd[:, 1])])\n\n # loop through bending moments\n for bm in bmd:\n if (bm[0] > restraint1.pos) and (bm[0] < restraint2.pos):\n bmd_segment.append(bm)\n\n # add end point for the segment bmd\n bmd_segment.append([restraint2.pos, np.interp(restraint2.pos, bmd[:, 0], bmd[:, 1])])\n\n self.bmd_segment = np.array(bmd_segment)\n\n def calc_alpha_m(self):\n \"\"\"Calculates the alpha_m value for the segment based on the bending moment diagram.\n\n Cl. 5.6.1.1(a) (iii) AS4100-1998\n \"\"\"\n\n # get start pos, end pos and length of segment\n start_pos = self.bmd_segment[0][0]\n end_pos = self.bmd_segment[-1][0]\n length = end_pos - start_pos\n\n # get max bending moment\n bm_max = self.bmd_segment.max(axis=0)[1]\n\n # get bending moment at quarter points\n bm_quarters = []\n for i in range(3):\n mult = 0.25 * (i + 1)\n bm_quarters.append(np.interp(\n start_pos + mult * length, self.bmd_segment[:, 0], self.bmd_segment[:, 1])\n )\n\n # calculate calc_alpha_m\n self.alpha_m = min(2.5, 1.7 * bm_max / np.sqrt(\n bm_quarters[0] ** 2 + bm_quarters[1] ** 2 + bm_quarters[2] ** 2)\n )\n\n def calc_effective_length(self):\n \"\"\"Returns the effective length of the segment.\n\n Cl. 5.6.3 AS4100-1998\n\n :returns: Segment effective length\n :rtype: float\n\n :raises Exception: If the restraint code is incorrect\n :raises Exception: If the load_position code is incorrect\n \"\"\"\n\n # determine restraint code\n res_code = self.restraint1.rtype + self.restraint2.rtype\n\n # calculate twist restraint factor\n if self.iscode(res_code, ['FF', 'FL', 'LL', 'FU']):\n kt = 1\n\n elif self.iscode(res_code, ['FP', 'PL', 'PU']):\n d1 = self.section.calc_dw()\n tf = self.section.get_tf()\n tw = self.section.get_tw()\n nw = self.section.get_nw()\n\n kt = 1 + (d1 / self.segment_length) * (tf / 2 / tw) ** 3 / nw\n\n elif self.iscode(res_code, ['PP']):\n kt = 1 + 2 * (d1 / self.segment_length) * (tf / 2 / tw) ** 3 / nw\n\n else:\n raise Exception('Restraint code {0} is invalid'.format(res_code))\n\n # calculate load height factor\n if self.load_position in ['WS', 'ES']:\n kl = 1\n elif self.load_position == 'WT':\n if self.iscode(res_code, ['FU', 'PU']):\n kl = 2\n else:\n kl = 1.4\n elif self.load_position == 'ET':\n if self.iscode(res_code, ['FU', 'PU']):\n kl = 2\n else:\n kl = 1\n else:\n raise Exception('Load position code {0} is invalid'.format(self.load_position))\n\n # calculate lateral rotation restraint factor\n # TODO: add ends with lateral rotation restraints\n kr = 1\n\n return kt * kl * kr * self.segment_length\n\n def iscode(self, res, codes):\n \"\"\"Determines if the restraint type 'res' corresponds to any of the list of restraint\n 'codes'. Note that the string code may be unordered, e.g. 'PF' is equal to 'FP'.\n\n :param string res: Restraint code to check\n :param codes: List of codes to check against\n :type codes: list[string]\n :returns: Whether or not 'res' matches 'code' (unordered)\n :rtype: bool\n \"\"\"\n\n for code in codes:\n if (res == code or res[::-1] == code):\n return True\n\n return False\n"
] |
[
[
"numpy.array",
"numpy.interp",
"numpy.sqrt"
]
] |
bertmaher/functorch
|
[
"8c5be62a9b140c4d35366720427b735bdcff8d01"
] |
[
"test/common_utils.py"
] |
[
"# Copyright (c) Facebook, Inc. and its affiliates.\n# All rights reserved.\n#\n# This source code is licensed under the BSD-style license found in the\n# LICENSE file in the root directory of this source tree.\n\nimport itertools\nimport torch\nimport functorch\nfrom functorch import vmap\nimport torch.utils._pytree as pytree\nfrom functorch_lagging_op_db import functorch_lagging_op_db\nfrom functorch_additional_op_db import additional_op_db\nfrom torch.testing._internal.common_methods_invocations import DecorateInfo\nimport unittest\nimport warnings\nimport re\n\n\"\"\"\nUsage:\n\nclass MyTestCase(TestCase):\n @parameterized('param', {'abs': torch.abs, 'cos': torch.cos})\n def test_single_param(self, param):\n pass\n\n @parameterized('param1', {'sin': torch.sin, 'tan': torch.tan})\n @parameterized('param2', {'abs': torch.abs, 'cos': torch.cos})\n def test_multiple_param(self, param1, param2):\n pass\n\n# The following creates:\n# - MyTestCase.test_single_param_abs\n# - MyTestCase.test_single_param_cos\n# - MyTestCase.test_multiple_param_abs_sin\n# - MyTestCase.test_multiple_param_cos_sin\n# - MyTestCase.test_multiple_param_abs_tan\n# - MyTestCase.test_multiple_param_cos_tan\ninstantiate_parameterized_methods(MyTestCase)\n\n# This is also composable with PyTorch testing's instantiate_device_type_tests\n# Make sure the param is after the device arg\nclass MyDeviceSpecificTest(TestCase):\n @parameterized('param', {'abs': torch.abs, 'cos': torch.cos})\n def test_single_param(self, device, param):\n pass\n\n# The following creates:\n# - MyDeviceSpecificTestCPU.test_single_param_abs_cpu\n# - MyDeviceSpecificTestCPU.test_single_param_cos_cpu\n# - MyDeviceSpecificTestCUDA.test_single_param_abs_cuda\n# - MyDeviceSpecificTestCUDA.test_single_param_cos_cpu\ninstantiate_parameterized_methods(MyDeviceSpecificTest)\ninstantiate_device_type_tests(MyDeviceSpecificTest, globals())\n\n# !!!!! warning !!!!!\n# 1. The method being parameterized over MUST NOT HAVE A DOCSTRING. We'll\n# error out nicely if this happens.\n# 2. All other decorators MUST USE functools.wraps (they must propagate the docstring)\n# `@parameterized` works by storing some metadata in place of the docstring.\n# This takes advantage of how other decorators work (other decorators usually\n# propagate the docstring via functools.wrap).\n# 3. We might not compose with PyTorch testing's @dtypes and @precision\n# decorators. But that is easily fixable. TODO.\n# I think this composes with PyTorch testing's instantiate_device_type_tests.\n\"\"\"\n\nPARAM_META = '_torch_parameterized_meta'\n\nclass ParamMeta():\n def __init__(self):\n self.stack = []\n\n def push(self, elt):\n self.stack.append(elt)\n\n def pop(self, elt):\n return self.stack.pop()\n\ndef has_param_meta(method):\n param_meta = getattr(method, '__doc__', None)\n return param_meta is not None and isinstance(param_meta, ParamMeta)\n\ndef get_param_meta(method):\n param_meta = getattr(method, '__doc__', None)\n if param_meta is None:\n method.__doc__ = ParamMeta()\n if not isinstance(method.__doc__, ParamMeta):\n raise RuntimeError('Tried to use @parameterized on a method that has '\n 'a docstring. This is not supported. Please remove '\n 'the docstring.')\n return method.__doc__\n\ndef parameterized(arg_name, case_dict):\n def decorator(fn):\n param_meta = get_param_meta(fn)\n param_meta.push((arg_name, case_dict))\n return fn\n return decorator\n\ndef parameterized_with_device(arg_name, case_dict):\n def decorator(fn):\n param_meta = get_param_meta(fn)\n param_meta.push((arg_name, case_dict))\n fn._has_device = True\n return fn\n return decorator\n\n\ndef _set_parameterized_method(test_base, fn, instantiated_cases, extension_name):\n new_name = f'{fn.__name__}_{extension_name}'\n\n def wrapped_no_device(self, *args, **kwargs):\n for arg_name, case in instantiated_cases:\n kwargs[arg_name] = case\n return fn(self, *args, **kwargs)\n\n def wrapped_with_device(self, device, *args, **kwargs):\n for arg_name, case in instantiated_cases:\n kwargs[arg_name] = case\n return fn(self, device, *args, **kwargs)\n\n if getattr(fn, '_has_device', False):\n wrapped = wrapped_with_device\n else:\n wrapped = wrapped_no_device\n\n wrapped.__name__ = new_name\n setattr(test_base, new_name, wrapped)\n\ndef to_tuples(dct):\n return [(k, v) for k, v in dct.items()]\n\ndef instantiate_parameterized_methods(test_base):\n allattrs = tuple(dir(test_base))\n for attr_name in allattrs:\n attr = getattr(test_base, attr_name)\n if not has_param_meta(attr):\n continue\n\n param_meta = get_param_meta(attr)\n arg_names, case_dicts = zip(*param_meta.stack)\n case_dicts = [to_tuples(cd) for cd in case_dicts]\n for list_of_name_and_case in itertools.product(*case_dicts):\n case_names, cases = zip(*list_of_name_and_case)\n extension_name = '_'.join(case_names)\n instantiated_cases = list(zip(arg_names, cases))\n _set_parameterized_method(test_base, attr, instantiated_cases, extension_name)\n # Remove the base fn from the testcase\n delattr(test_base, attr_name)\n\n\ndef loop(op, in_dims, out_dim, batch_size, *batched_args, **kwarg_values):\n outs = []\n for idx in range(batch_size):\n idx_args = []\n idx_kwargs = {}\n flat_args, args_spec = pytree.tree_flatten(batched_args)\n flat_dims, dims_spec = pytree.tree_flatten(in_dims)\n # print(flat_args)\n assert(args_spec == dims_spec)\n new_args = [a.select(in_dim, idx) if in_dim is not None else a for a, in_dim in zip(flat_args, flat_dims)]\n out = op(*pytree.tree_unflatten(new_args, args_spec), **kwarg_values)\n outs.append(out)\n loop_out = []\n if isinstance(outs[0], torch.Tensor):\n loop_out = torch.stack(outs)\n else:\n for idx in range(len(outs[0])):\n loop_out.append(torch.stack([i[idx] for i in outs], out_dim))\n return loop_out\n\n\ndef get_exhaustive_batched_inputs(arg_values, kwarg_values, batch_size=3):\n def add_batch_dim(arg, bdim, batch_size=3):\n if isinstance(arg, torch.Tensor):\n shape = [1] * len(arg.shape)\n shape.insert(bdim, batch_size)\n return (arg.repeat(shape), bdim)\n else:\n return (arg, None)\n\n batch_choices = []\n def add_batch_choices(a):\n if isinstance(a, torch.Tensor):\n batched_val = add_batch_dim(a, 0, batch_size)\n batch_choices.append((batched_val, (a, None)))\n else:\n batch_choices.append(((a, None),))\n\n flat_args, arg_spec = pytree.tree_flatten(tuple(arg_values))\n for arg in flat_args:\n add_batch_choices(arg)\n\n for batched_values in itertools.product(*batch_choices):\n batched_args, in_dims = zip(*batched_values)\n\n if all([i is None for i in in_dims]):\n continue\n\n yield pytree.tree_unflatten(batched_args, arg_spec), pytree.tree_unflatten(in_dims, arg_spec), kwarg_values\n\n\ndef get_fallback_and_vmap_exhaustive(op, arg_values, kwarg_values, compute_loop_out=True):\n out_dim = 0\n batch_size = 3\n generator = get_exhaustive_batched_inputs(arg_values, kwarg_values, batch_size)\n for batched_args, in_dims, kwarg_values in generator:\n if compute_loop_out:\n loop_out = loop(op, in_dims, out_dim, batch_size, *batched_args, **kwarg_values)\n else:\n loop_out = None\n # Used for debugging the resulting operations\n # from functorch import make_fx\n # def f(a):\n # return op(a)\n # t = make_fx(vmap(f, in_dims=in_dims, out_dims=out_dim))(*batched_args, **kwarg_values)\n batched_out = vmap(op, in_dims=in_dims, out_dims=out_dim)(*batched_args, **kwarg_values)\n yield (loop_out, batched_out)\n\n # Tests case where we dispatch to a batching rule with no bdims\n # Should now be covered by https://github.com/facebookresearch/functorch/pull/63\n def f(x, *args, **kwargs):\n out = op(*args, **kwargs)\n if isinstance(out, torch.Tensor):\n return out + x.to(out.device)\n out = list(out)\n for idx in range(len(out)):\n out[idx] = out[idx] + x.to(out[idx].device)\n return out\n\n vmap1_dims = tuple([0] + [None] * len(in_dims))\n vmap2_dims = tuple([None] + list(in_dims))\n if compute_loop_out:\n loop_out = pytree.tree_map(lambda v: torch.ones(3, *v.shape, device=v.device) + v, loop_out)\n else:\n loop_out = None\n batched_out = vmap(vmap(f, in_dims=vmap1_dims), in_dims=vmap2_dims)(torch.ones(3), *batched_args, **kwarg_values)\n yield (loop_out, batched_out)\n\ndef opinfo_in_dict(opinfo, d):\n return (opinfo.name in d) or (f'{opinfo.name}.{opinfo.variant_test_name}' in d)\n\ndef xfail(op_name, variant_name=None, *, device_type=None, dtypes=None, expected_failure=True):\n return (op_name, variant_name, device_type, dtypes, expected_failure)\n\ndef skipOps(test_case_name, base_test_name, to_skip):\n all_opinfos = functorch_lagging_op_db + additional_op_db\n for xfail in to_skip:\n op_name, variant_name, device_type, dtypes, expected_failure = xfail\n if variant_name is None:\n # match all variants\n matching_opinfos = [o for o in all_opinfos if o.name == op_name]\n assert len(matching_opinfos) >= 1, f\"Couldn't find OpInfo for {xfail}\"\n else:\n matching_opinfos = [o for o in all_opinfos\n if o.name == op_name and o.variant_test_name == variant_name]\n assert len(matching_opinfos) >= 1, f\"Couldn't find OpInfo for {xfail}\"\n for opinfo in matching_opinfos:\n decorators = list(opinfo.decorators)\n decorators.append(DecorateInfo(unittest.expectedFailure,\n test_case_name, base_test_name,\n device_type=device_type, dtypes=dtypes))\n opinfo.decorators = tuple(decorators)\n\n # This decorator doesn't modify fn in any way\n def wrapped(fn):\n return fn\n return wrapped\n\nclass DisableVmapFallback:\n def __enter__(self):\n self.prev_state = functorch._C._is_vmap_fallback_enabled()\n functorch._C._set_vmap_fallback_enabled(False)\n\n def __exit__(self, *ignored):\n functorch._C._set_vmap_fallback_enabled(self.prev_state)\n\ndef check_vmap_fallback(test_case, thunk, opinfo, dry_run=False):\n try:\n with DisableVmapFallback():\n thunk()\n except:\n if not dry_run:\n raise\n if opinfo.variant_test_name:\n print(f\"xfail('{opinfo.name}', '{opinfo.variant_test_name}'),\")\n else:\n print(f\"xfail('{opinfo.name}'),\")\n"
] |
[
[
"torch.testing._internal.common_methods_invocations.DecorateInfo",
"torch.stack",
"torch.ones",
"torch.utils._pytree.tree_unflatten",
"torch.utils._pytree.tree_flatten"
]
] |
torms3/DataProvider3
|
[
"28c18e520e658f10547edbfb495dac07d60574b6"
] |
[
"dataprovider3/datasuperset.py"
] |
[
"import numpy as np\n\nfrom .dataset import Dataset\n\n\nclass DataSuperset(Dataset):\n \"\"\"\n Superset of datasets.\n \"\"\"\n def __init__(self, tag=''):\n self.tag = tag\n self.datasets = list()\n self.p = None\n\n def __call__(self, spec=None):\n return self.random_sample(spec=spec)\n\n def __repr__(self):\n format_string = self.__class__.__name__ + '('\n format_string += self.tag\n format_string += ')'\n return format_string\n\n def sanity_check(self, spec):\n sanity = True\n for dset in self.datasets:\n sanity &= dset.sanity_check(spec)\n return sanity\n\n def add_dataset(self, dset):\n assert isinstance(dset, Dataset)\n self.datasets.append(dset)\n\n def random_sample(self, spec=None):\n dset = self.random_dataset()\n return dset(spec=spec)\n\n def set_sampling_weights(self, p=None):\n if p is None:\n p = [d.num_samples() for d in self.datasets]\n p = np.asarray(p, dtype='float32')\n p = p/np.sum(p)\n assert len(p)==len(self.datasets)\n self.p = p\n\n def random_dataset(self):\n assert len(self.datasets) > 0\n if self.p is None:\n self.set_sampling_weights()\n idx = np.random.choice(len(self.datasets), size=1, p=self.p)\n return self.datasets[idx[0]]\n\n def num_samples(self, spec=None):\n return sum([dset.num_samples(spec=spec) for dset in self.datasets])\n"
] |
[
[
"numpy.sum",
"numpy.asarray"
]
] |
Moon-sung-woo/Tacotron2_ParallelWaveGAN_korean
|
[
"437c9748673f2e5cb84e99884e8d0d916f269c9e"
] |
[
"inference.py"
] |
[
"# *****************************************************************************\n# Copyright (c) 2018, NVIDIA CORPORATION. All rights reserved.\n#\n# Redistribution and use in source and binary forms, with or without\n# modification, are permitted provided that the following conditions are met:\n# * Redistributions of source code must retain the above copyright\n# notice, this list of conditions and the following disclaimer.\n# * Redistributions in binary form must reproduce the above copyright\n# notice, this list of conditions and the following disclaimer in the\n# documentation and/or other materials provided with the distribution.\n# * Neither the name of the NVIDIA CORPORATION nor the\n# names of its contributors may be used to endorse or promote products\n# derived from this software without specific prior written permission.\n#\n# THIS SOFTWARE IS PROVIDED BY THE COPYRIGHT HOLDERS AND CONTRIBUTORS \"AS IS\" AND\n# ANY EXPRESS OR IMPLIED WARRANTIES, INCLUDING, BUT NOT LIMITED TO, THE IMPLIED\n# WARRANTIES OF MERCHANTABILITY AND FITNESS FOR A PARTICULAR PURPOSE ARE\n# DISCLAIMED. IN NO EVENT SHALL NVIDIA CORPORATION BE LIABLE FOR ANY\n# DIRECT, INDIRECT, INCIDENTAL, SPECIAL, EXEMPLARY, OR CONSEQUENTIAL DAMAGES\n# (INCLUDING, BUT NOT LIMITED TO, PROCUREMENT OF SUBSTITUTE GOODS OR SERVICES;\n# LOSS OF USE, DATA, OR PROFITS; OR BUSINESS INTERRUPTION) HOWEVER CAUSED AND\n# ON ANY THEORY OF LIABILITY, WHETHER IN CONTRACT, STRICT LIABILITY, OR TORT\n# (INCLUDING NEGLIGENCE OR OTHERWISE) ARISING IN ANY WAY OUT OF THE USE OF THIS\n# SOFTWARE, EVEN IF ADVISED OF THE POSSIBILITY OF SUCH DAMAGE.\n#\n# *****************************************************************************\n\nfrom tacotron2.text import text_to_sequence\nimport models\nimport torch\nimport argparse\nimport numpy as np\nfrom scipy.io.wavfile import write\nimport matplotlib.pyplot as plt\n\nimport sys\nimport csv\n\nimport time\nimport dllogger as DLLogger\nfrom dllogger import StdOutBackend, JSONStreamBackend, Verbosity\n\nfrom apex import amp\n\nfrom utils import write_hdf5\nfrom decode import make_wav\n\ndef parse_args(parser):\n \"\"\"\n Parse commandline arguments.\n \"\"\"\n parser.add_argument('-i', '--input', type=str, default='text.txt',\n help='full path to the input text (phareses separated by new line)')\n parser.add_argument('-o', '--output', default=\"parallel_output\",\n help='output folder to save audio (file per phrase)')\n parser.add_argument('--suffix', type=str, default=\"\", help=\"output filename suffix\")\n parser.add_argument('--tacotron2', type=str,default='gan_checkpoint/checkpoint_Tacotron2_1500',\n help='full path to the Tacotron2 model checkpoint file')\n parser.add_argument('--waveglow', type=str,\n help='full path to the WaveGlow model checkpoint file')\n parser.add_argument('-s', '--sigma-infer', default=0.9, type=float)\n parser.add_argument('-d', '--denoising-strength', default=0.01, type=float)\n parser.add_argument('-sr', '--sampling-rate', default=22050, type=int,\n help='Sampling rate')\n parser.add_argument('--amp-run', default=True, action='store_true',\n help='inference with AMP')\n parser.add_argument('--log-file', type=str, default='nvlog.json',\n help='Filename for logging')\n parser.add_argument('--include-warmup', default=True,action='store_true',\n help='Include warmup')\n parser.add_argument('--stft-hop-length', type=int, default=256,\n help='STFT hop length for estimating audio length from mel size')\n #---------------------------------------------------------------------------------------\n #여기부터 parallel_wavegan관련\n parser.add_argument(\"--feats-scp\", \"--scp\", default=None, type=str,\n help=\"kaldi-style feats.scp file. \"\n \"you need to specify either feats-scp or dumpdir.\")\n\n parser.add_argument(\"--dumpdir\", default='mel', type=str,\n help=\"directory including feature files. \"\n \"you need to specify either feats-scp or dumpdir. .h5파일 넣어주는 부분\")\n #'../../egs/kss/voc1/dump/dev/test.1'\n parser.add_argument(\"--outdir\", default='parallel_output', type=str,\n help=\"directory to save generated speech.\")\n parser.add_argument(\"--checkpoint\", default='parallel_wavegan_checkpoint/checkpoint-110125steps.pkl', type=str,\n help=\"checkpoint file to be loaded.\")\n parser.add_argument(\"--config\", default=None, type=str,\n help=\"yaml format configuration file. if not explicitly provided, \"\n \"it will be searched in the checkpoint directory. (default=None)\")\n parser.add_argument(\"--verbose\", type=int, default=1,\n help=\"logging level. higher is more logging. (default=1)\")\n #-----------------------------------------------------------------------------------------\n return parser\n\n\ndef checkpoint_from_distributed(state_dict):\n \"\"\"\n Checks whether checkpoint was generated by DistributedDataParallel. DDP\n wraps model in additional \"module.\", it needs to be unwrapped for single\n GPU inference.\n :param state_dict: model's state dict\n \"\"\"\n ret = False\n for key, _ in state_dict.items():\n if key.find('module.') != -1:\n ret = True\n break\n return ret\n\n\ndef unwrap_distributed(state_dict):\n \"\"\"\n Unwraps model from DistributedDataParallel.\n DDP wraps model in additional \"module.\", it needs to be removed for single\n GPU inference.\n :param state_dict: model's state dict\n \"\"\"\n new_state_dict = {}\n for key, value in state_dict.items():\n new_key = key.replace('module.', '')\n new_state_dict[new_key] = value\n return new_state_dict\n\n\ndef load_and_setup_model(model_name, parser, checkpoint, amp_run, forward_is_infer=False):\n model_parser = models.parse_model_args(model_name, parser, add_help=False)\n model_args, _ = model_parser.parse_known_args()\n\n model_config = models.get_model_config(model_name, model_args)\n model = models.get_model(model_name, model_config, to_cuda=True,\n forward_is_infer=forward_is_infer)\n\n if checkpoint is not None:\n state_dict = torch.load(checkpoint)['state_dict']\n if checkpoint_from_distributed(state_dict):\n state_dict = unwrap_distributed(state_dict)\n\n model.load_state_dict(state_dict)\n\n if model_name == \"WaveGlow\":\n model = model.remove_weightnorm(model)\n\n model.eval()\n\n if amp_run:\n model.half()\n\n return model\n\n\n# taken from tacotron2/data_function.py:TextMelCollate.__call__\ndef pad_sequences(batch):\n # Right zero-pad all one-hot text sequences to max input length\n input_lengths, ids_sorted_decreasing = torch.sort(\n torch.LongTensor([len(x) for x in batch]),\n dim=0, descending=True)\n max_input_len = input_lengths[0]\n\n text_padded = torch.LongTensor(len(batch), max_input_len)\n text_padded.zero_()\n for i in range(len(ids_sorted_decreasing)):\n text = batch[ids_sorted_decreasing[i]]\n text_padded[i, :text.size(0)] = text\n\n return text_padded, input_lengths\n\n\ndef prepare_input_sequence(texts):\n\n d = []\n for i,text in enumerate(texts):\n d.append(torch.IntTensor(\n text_to_sequence(text, ['english_cleaners'])[:]))\n\n text_padded, input_lengths = pad_sequences(d)\n if torch.cuda.is_available():\n text_padded = torch.autograd.Variable(text_padded).cuda().long()\n input_lengths = torch.autograd.Variable(input_lengths).cuda().long()\n else:\n text_padded = torch.autograd.Variable(text_padded).long()\n input_lengths = torch.autograd.Variable(input_lengths).long()\n\n return text_padded, input_lengths\n\ndef make_script():\n g_csv = 'nlp_script_dataset2.csv'\n\n f = open(g_csv, 'r', encoding='cp949')\n lines = csv.reader(f)\n lines = list(lines)[37]\n\n script = lines[1].split(' ')[1:]\n print(script)\n f.close()\n\n return script[:5]\n\nclass MeasureTime():\n def __init__(self, measurements, key):\n self.measurements = measurements\n self.key = key\n\n def __enter__(self):\n torch.cuda.synchronize()\n self.t0 = time.perf_counter()\n\n def __exit__(self, exc_type, exc_value, exc_traceback):\n torch.cuda.synchronize()\n self.measurements[self.key] = time.perf_counter() - self.t0\n\n\ndef main():\n \"\"\"\n Launches text to speech (inference).\n Inference is executed on a single GPU.\n \"\"\"\n parser = argparse.ArgumentParser(\n description='PyTorch Tacotron 2 Inference')\n parser = parse_args(parser)\n args, _ = parser.parse_known_args()\n\n DLLogger.init(backends=[JSONStreamBackend(Verbosity.DEFAULT,\n args.output+'/'+args.log_file),\n StdOutBackend(Verbosity.VERBOSE)])\n for k,v in vars(args).items():\n DLLogger.log(step=\"PARAMETER\", data={k:v})\n DLLogger.log(step=\"PARAMETER\", data={'model_name':'Tacotron2_PyT'})\n\n tacotron2 = load_and_setup_model('Tacotron2', parser, args.tacotron2,\n args.amp_run, forward_is_infer=True)\n\n jitted_tacotron2 = torch.jit.script(tacotron2)\n\n\n\n #texts = []\n #texts.append(make_script())\n\n\n try:\n print(args.input)\n f = open(args.input, 'r')\n texts = f.readlines()\n except:\n print(\"Could not read file\")\n sys.exit(1)\n\n #데이터 불러 와 주고 요기 아래부터 for 문으로 돌려주면 될 듯.\n \n if args.include_warmup:\n sequence = torch.randint(low=0, high=80, size=(1,50),\n dtype=torch.long).cuda()\n input_lengths = torch.IntTensor([sequence.size(1)]).cuda().long()\n for i in range(3):\n with torch.no_grad():\n mel, mel_lengths, _ = jitted_tacotron2(sequence, input_lengths)\n\n\n measurements = {}\n\n sequences_padded, input_lengths = prepare_input_sequence(texts)\n\n with torch.no_grad(), MeasureTime(measurements, \"tacotron2_time\"):\n mel, mel_lengths, alignments = jitted_tacotron2(sequences_padded, input_lengths)\n #-------------------------------------------------------------- \n #.h5모델로 멜 만드는 부분\n #print(mel.float().data.cpu().numpy())\n write_hdf5('mel/sw_embedded.h5', \"feats\", mel.float().data.cpu().numpy())\n #--------------------------------------------------------------\n \n print(\"Stopping after\", mel.size(2), \"decoder steps\")\n #여기있는게 원래 코드\n\n\n '''\n #-------------------------------------------------------------------------------------------------------\n texts_list = make_script()\n for idx, texts in enumerate(texts_list):\n text = []\n text.append(''.join(texts))\n print(text)\n if args.include_warmup:\n sequence = torch.randint(low=0, high=80, size=(1, 50),\n dtype=torch.long).cuda()\n input_lengths = torch.IntTensor([sequence.size(1)]).cuda().long()\n for i in range(3):\n with torch.no_grad():\n mel, mel_lengths, _ = jitted_tacotron2(sequence, input_lengths)\n\n measurements = {}\n\n sequences_padded, input_lengths = prepare_input_sequence(text)\n\n with torch.no_grad(), MeasureTime(measurements, \"tacotron2_time\"):\n mel, mel_lengths, alignments = jitted_tacotron2(sequences_padded, input_lengths)\n # --------------------------------------------------------------\n # .h5모델로 멜 만드는 부분\n # print(mel.float().data.cpu().numpy())\n write_hdf5('mel/test2_{}.h5'.format(idx), \"feats\", mel.float().data.cpu().numpy())\n # --------------------------------------------------------------\n\n print(\"Stopping after\", mel.size(2), \"decoder steps\")\n #-------------------------------------------------------------------------------------------------------\n '''\n\n\n DLLogger.flush()\n make_wav(args)\n \n\nif __name__ == '__main__':\n main()"
] |
[
[
"torch.cuda.synchronize",
"torch.autograd.Variable",
"torch.no_grad",
"torch.randint",
"torch.cuda.is_available",
"torch.load",
"torch.jit.script"
]
] |
kcorder/autonomous-learning-library
|
[
"0266195fa47564e51a32087bc007bff6dda5e263"
] |
[
"all/experiments/parallel_env_experiment.py"
] |
[
"\nimport torch\nimport time\nimport numpy as np\nfrom all.core import State\nfrom .writer import ExperimentWriter, CometWriter\nfrom .experiment import Experiment\nfrom all.environments import VectorEnvironment\nfrom all.agents import ParallelAgent\nimport gym\n\n\nclass ParallelEnvExperiment(Experiment):\n '''An Experiment object for training and testing agents that use parallel training environments.'''\n\n def __init__(\n self,\n preset,\n env,\n test_env=None,\n name=None,\n train_steps=float('inf'),\n logdir='runs',\n quiet=False,\n render=False,\n write_loss=True,\n writer=\"tensorboard\"\n ):\n self._name = name if name is not None else preset.name\n super().__init__(self._make_writer(logdir, self._name, env.name, write_loss, writer), quiet)\n self._n_envs = preset.n_envs\n if isinstance(env, VectorEnvironment):\n assert self._n_envs == env.num_envs\n self._env = env\n else:\n self._env = env.duplicate(self._n_envs)\n self._test_env = test_env if test_env is not None else env\n self._preset = preset\n self._agent = preset.agent(writer=self._writer, train_steps=train_steps)\n self._render = render\n\n # training state\n self._returns = []\n self._frame = 1\n self._episode = 1\n self._episode_start_times = [] * self._n_envs\n self._episode_start_frames = [] * self._n_envs\n\n # test state\n self._test_episodes = 100\n self._test_episodes_started = self._n_envs\n self._test_returns = []\n self._should_save_returns = [True] * self._n_envs\n\n if render:\n for _env in self._envs:\n _env.render(mode=\"human\")\n\n @property\n def frame(self):\n return self._frame\n\n @property\n def episode(self):\n return self._episode\n\n def train(self, frames=np.inf, episodes=np.inf):\n num_envs = int(self._env.num_envs)\n returns = np.zeros(num_envs)\n state_array = self._env.reset()\n start_time = time.time()\n completed_frames = 0\n while not self._done(frames, episodes):\n action = self._agent.act(state_array)\n state_array = self._env.step(action)\n self._frame += num_envs\n episodes_completed = state_array.done.type(torch.IntTensor).sum().item()\n completed_frames += num_envs\n returns += state_array.reward.cpu().detach().numpy()\n if episodes_completed > 0:\n dones = state_array.done.cpu().detach().numpy()\n cur_time = time.time()\n fps = completed_frames / (cur_time - start_time)\n completed_frames = 0\n start_time = cur_time\n for i in range(num_envs):\n if dones[i]:\n self._log_training_episode(returns[i], fps)\n returns[i] = 0\n self._episode += episodes_completed\n\n def test(self, episodes=100):\n test_agent = self._preset.parallel_test_agent()\n\n # Note that we need to record the first N episodes that are STARTED,\n # not the first N that are completed, or we introduce bias.\n test_returns = []\n episodes_started = self._n_envs\n should_record = [True] * self._n_envs\n\n # initialize state\n states = self._test_env.reset()\n returns = states.reward.clone()\n\n while len(test_returns) < episodes:\n # step the agent and environments\n actions = test_agent.act(states)\n states = self._test_env.step(actions)\n returns += states.reward\n\n # record any episodes that have finished\n for i, done in enumerate(states.done):\n if done:\n if should_record[i] and len(test_returns) < episodes:\n episode_return = returns[i].item()\n test_returns.append(episode_return)\n self._log_test_episode(len(test_returns), episode_return)\n returns[i] = 0.\n episodes_started += 1\n if episodes_started > episodes:\n should_record[i] = False\n\n self._log_test(test_returns)\n return test_returns\n\n def _done(self, frames, episodes):\n return self._frame > frames or self._episode > episodes\n\n def _make_writer(self, logdir, agent_name, env_name, write_loss, writer):\n if writer == \"comet\":\n return CometWriter(self, agent_name, env_name, loss=write_loss, logdir=logdir)\n return ExperimentWriter(self, agent_name, env_name, loss=write_loss, logdir=logdir)\n"
] |
[
[
"numpy.zeros"
]
] |
zhang0557kui/Fundus_Lesion2018
|
[
"bfb9516a2049f9f1c2662e963734a2746d5c3e4f"
] |
[
"data_generator.py"
] |
[
"import json\n\nimport cv2 as cv\nimport numpy as np\nfrom keras.utils import Sequence, to_categorical\n\nfrom augmentor import seq_det, seq_img\nfrom config import img_rows, img_cols, batch_size, num_classes, gray_values\nfrom utils import preprocess_input\n\n\nclass DataGenSequence(Sequence):\n def __init__(self, usage):\n self.usage = usage\n if self.usage == 'train':\n gt_file = 'train_gt_file.json'\n else:\n gt_file = 'valid_gt_file.json'\n\n with open(gt_file, 'r') as file:\n self.samples = json.load(file)\n\n np.random.shuffle(self.samples)\n\n def __len__(self):\n return int(np.ceil(len(self.samples) / float(batch_size)))\n\n def __getitem__(self, idx):\n i = idx * batch_size\n\n length = min(batch_size, (len(self.samples) - i))\n X = np.empty((length, img_rows, img_cols, 3), dtype=np.float32)\n Y = np.zeros((length, img_rows, img_cols, num_classes), dtype=np.float32)\n\n for i_batch in range(length):\n sample = self.samples[i + i_batch]\n original_image_path = sample['original_image']\n label_image_path = sample['label_image']\n original_image = cv.imread(original_image_path)\n original_image = cv.resize(original_image, (img_cols, img_rows), cv.INTER_NEAREST)\n label_image = cv.imread(label_image_path, 0)\n label_image = cv.resize(label_image, (img_cols, img_rows), cv.INTER_NEAREST)\n\n X[i_batch] = original_image\n for j in range(num_classes):\n Y[i_batch][label_image == gray_values[j]] = to_categorical(j, num_classes)\n\n if self.usage == 'train':\n X = seq_img.augment_images(X)\n X = seq_det.augment_images(X)\n Y = seq_det.augment_images(Y)\n\n X = preprocess_input(X)\n\n return X, Y\n\n def on_epoch_end(self):\n np.random.shuffle(self.samples)\n\n\ndef train_gen():\n return DataGenSequence('train')\n\n\ndef valid_gen():\n return DataGenSequence('valid')\n\n\ndef revert_pre_process(x):\n return ((x + 1) * 127.5).astype(np.uint8)\n\n\ndef revert_labeling(y):\n y = np.argmax(y, axis=-1)\n for j in range(num_classes):\n y[y == j] = gray_values[j]\n return y\n\n\nif __name__ == '__main__':\n data_gen = DataGenSequence('train')\n item = data_gen.__getitem__(0)\n x, y = item\n print(x.shape)\n print(y.shape)\n\n for i in range(10):\n image = revert_pre_process(x[i])\n h, w = image.shape[:2]\n image = image[:, :, ::-1].astype(np.uint8)\n print(image.shape)\n cv.imwrite('images/sample_{}.jpg'.format(i), image)\n label = revert_labeling(y[i])\n label = label.astype(np.uint8)\n cv.imwrite('images/label_{}.jpg'.format(i), label)\n"
] |
[
[
"numpy.empty",
"numpy.argmax",
"numpy.zeros",
"numpy.random.shuffle"
]
] |
luomingshuang/GE2E-SV-TI-Voxceleb-LMS
|
[
"000b0176b4b3a484e390c986045e491d64427c8b"
] |
[
"test.py"
] |
[
"#coding:utf-8\nimport os\nimport random\nimport time\nimport torch\nfrom torch.utils.data import DataLoader\n\nfrom hparam import hparam as hp\nfrom data_load import SpeakerDatasetTIMIT, SpeakerDatasetTIMITPreprocessed\nfrom speech_embedder_net import SpeechEmbedder, GE2ELoss, get_centroids, get_cossim\nfrom tensorboardX import SummaryWriter\n\nmodel_path = './speech_id_checkpoint/ckpt_epoch_360_batch_id_281.pth'\n\nif (__name__=='__main__'):\n\n writer = SummaryWriter()\n\n device = torch.device(hp.device)\n model_path = hp.model.model_path\n\n if hp.data.data_preprocessed:\n train_dataset = SpeakerDatasetTIMITPreprocessed(hp.data.train_path, hp.train.M)\n else:\n train_dataset = SpeakerDatasetTIMIT(hp.data.train_path, hp.train.M)\n \n if hp.data.data_preprocessed:\n test_dataset = SpeakerDatasetTIMITPreprocessed(hp.data.test_path, hp.test.M)\n else:\n test_dataset = SpeakerDatasetTIMIT(hp.data.test_path, hp.test.M)\n\n train_loader = DataLoader(train_dataset, batch_size=hp.train.N, shuffle=True, num_workers=hp.train.num_workers, drop_last=True) \n test_loader = DataLoader(test_dataset, batch_size=hp.test.N, shuffle=True, num_workers=hp.test.num_workers, drop_last=True)\n\n embedder_net = SpeechEmbedder().to(device)\n\n \n embedder_net.load_state_dict(torch.load(model_path))\n ge2e_loss = GE2ELoss(device)\n #Both net and loss have trainable parameters\n optimizer = torch.optim.SGD([\n {'params': embedder_net.parameters()},\n {'params': ge2e_loss.parameters()}\n ], lr=hp.train.lr)\n \n os.makedirs(hp.train.checkpoint_dir, exist_ok=True)\n \n embedder_net.train()\n\n with torch.no_grad():\n avg_EER = 0\n iteration = 0\n for e in range(hp.test.epochs):\n batch_avg_EER = 0\n for batch_id, mel_db_batch in enumerate(test_loader):\n assert hp.test.M % 2 == 0\n enrollment_batch, verification_batch = torch.split(mel_db_batch, int(mel_db_batch.size(1)/2), dim=1)\n \n enrollment_batch = torch.reshape(enrollment_batch, (hp.test.N*hp.test.M//2, enrollment_batch.size(2), enrollment_batch.size(3))).cuda()\n verification_batch = torch.reshape(verification_batch, (hp.test.N*hp.test.M//2, verification_batch.size(2), verification_batch.size(3))).cuda()\n \n perm = random.sample(range(0,verification_batch.size(0)), verification_batch.size(0))\n unperm = list(perm)\n for i,j in enumerate(perm):\n unperm[j] = i\n \n verification_batch = verification_batch[perm]\n enrollment_embeddings = embedder_net(enrollment_batch)\n verification_embeddings = embedder_net(verification_batch)\n verification_embeddings = verification_embeddings[unperm]\n \n enrollment_embeddings = torch.reshape(enrollment_embeddings, (hp.test.N, hp.test.M//2, enrollment_embeddings.size(1)))\n verification_embeddings = torch.reshape(verification_embeddings, (hp.test.N, hp.test.M//2, verification_embeddings.size(1)))\n \n enrollment_centroids = get_centroids(enrollment_embeddings)\n \n sim_matrix = get_cossim(verification_embeddings, enrollment_centroids)\n \n # calculating EER\n diff = 1; EER=0; EER_thresh = 0; EER_FAR=0; EER_FRR=0\n \n for thres in [0.01*i+0.5 for i in range(50)]:\n sim_matrix_thresh = sim_matrix>thres\n \n FAR = (sum([sim_matrix_thresh[i].float().sum()-sim_matrix_thresh[i,:,i].float().sum() for i in range(int(hp.test.N))])\n /(hp.test.N-1.0)/(float(hp.test.M/2))/hp.test.N)\n \n FRR = (sum([hp.test.M/2-sim_matrix_thresh[i,:,i].float().sum() for i in range(int(hp.test.N))])\n /(float(hp.test.M/2))/hp.test.N)\n \n # Save threshold when FAR = FRR (=EER)\n if diff> abs(FAR-FRR):\n diff = abs(FAR-FRR)\n EER = (FAR+FRR)/2\n EER_thresh = thres\n EER_FAR = FAR\n EER_FRR = FRR\n batch_avg_EER += EER\n print(\"\\nEER : %0.2f (thres:%0.2f, FAR:%0.2f, FRR:%0.2f)\"%(EER,EER_thresh,EER_FAR,EER_FRR))\n avg_EER += batch_avg_EER/(batch_id+1)\n writer.add_scalar('data/EER', batch_avg_EER/(batch_id+1), iteration)\n iteration += 1\n avg_EER = avg_EER / hp.test.epochs\n print(\"\\n EER across {0} epochs: {1:.4f}\".format(hp.test.epochs, avg_EER))\n writer.close()\n"
] |
[
[
"torch.device",
"torch.no_grad",
"torch.utils.data.DataLoader",
"torch.load"
]
] |
jingzbu/InverseVITraffic
|
[
"c0d33d91bdd3c014147d58866c1a2b99fb8a9608"
] |
[
"Python_files/OD_matrix_estimation_GLS_Apr_weekday_NT_ext.py"
] |
[
"#!/usr/bin/env python\n\n\n__author__ = \"Jing Zhang\"\n__email__ = \"[email protected]\"\n__status__ = \"Development\"\n\n\nfrom util import *\nimport numpy as np\nfrom numpy.linalg import inv, matrix_rank\nimport json\n\n# load logit_route_choice_probability_matrix\nP = zload('../temp_files/logit_route_choice_probability_matrix_ext.pkz')\nP = np.matrix(P)\n\n# print(matrix_rank(P))\n\n\n# print(np.size(P,0), np.size(P,1))\n\n# load path-link incidence matrix\nA = zload('../temp_files/path-link_incidence_matrix_ext.pkz')\n\n# assert(1 == 2)\n\n# load link counts data\nwith open('../temp_files/link_day_minute_Apr_dict_ext_JSON_insert_links_adjusted.json', 'r') as json_file:\n link_day_minute_Apr_dict_ext_JSON = json.load(json_file)\n\n# week_day_Apr_list = [2, 3, 4, 5, 6, 9, 10, 11, 12, 13, 16, 22, 18, 19, 20, 23, 24, 25, 26, 27, 30]\nweek_day_Apr_list = [11, 12, 13, 16, 22]\n\nlink_day_minute_Apr_list = []\nfor link_idx in range(74):\n for day in week_day_Apr_list: \n for minute_idx in range(120):\n key = 'link_' + str(link_idx) + '_' + str(day)\n link_day_minute_Apr_list.append(link_day_minute_Apr_dict_ext_JSON[key] ['NT_flow_minute'][minute_idx])\n\n# print(len(link_day_minute_Apr_list))\n\nx = np.matrix(link_day_minute_Apr_list)\nx = np.matrix.reshape(x, 74, 600)\n\nx = np.nan_to_num(x)\ny = np.array(np.transpose(x))\ny = y[np.all(y != 0, axis=1)]\nx = np.transpose(y)\nx = np.matrix(x)\n\n# print(np.size(x,0), np.size(x,1))\n# print(x[:,:2])\n# print(np.size(A,0), np.size(A,1))\n\nL = 22 * (22 - 1) # dimension of lam\n\nlam_list = GLS(x, A, P, L)\n\n# write estimation result to file\nn = 22 # number of nodes\nwith open('../temp_files/OD_demand_matrix_Apr_weekday_NT_ext.txt', 'w') as the_file:\n idx = 0\n for i in range(n + 1)[1:]:\n for j in range(n + 1)[1:]:\n if i != j: \n the_file.write(\"%d,%d,%f\\n\" %(i, j, lam_list[idx]))\n idx += 1\n"
] |
[
[
"numpy.matrix",
"numpy.nan_to_num",
"numpy.transpose",
"numpy.all",
"numpy.matrix.reshape"
]
] |
ParthKalkar/digital-superheroes-hackathon
|
[
"0acc4a1b59a3dd2762a45744869b75dade891fa8"
] |
[
"Arima model/Arima Model/Arima Model/CSV.py"
] |
[
"from pandas import read_csv\r\nfrom pandas import datetime\r\nfrom matplotlib import pyplot\r\nfrom statsmodels.tsa.arima_model import ARIMA\r\nfrom sklearn.metrics import mean_squared_error\r\n\r\ndef parser(x):\r\n return datetime.strptime('190'+x, '%Y-%m')\r\n\r\nseries = read_csv('shampoo-sales.csv', header=0, parse_dates=[0], index_col=0, squeeze=True, date_parser=parser)\r\nX = series.values\r\nsize = int(len(X) * 0.66)\r\ntrain, test = X[0:size], X[size:len(X)]\r\nhistory = [x for x in train]\r\npredictions = list()\r\nfor t in range(len(test)):\r\n model = ARIMA(history, order=(5,1,0))\r\n model_fit = model.fit(disp=0)\r\n output = model_fit.forecast()\r\n yhat = output[0]\r\n predictions.append(yhat)\r\n obs = test[t]\r\n history.append(obs)\r\n print('predicted=%f, expected=%f' % (yhat, obs))\r\nerror = mean_squared_error(test, predictions)\r\nprint('Test MSE: %.3f' % error)\r\n# plot\r\npyplot.plot(test)\r\npyplot.plot(predictions, color='red')\r\npyplot.show()\r\n\r\n\r\n\r\nfrom pandas import read_csv\r\nfrom pandas import datetime\r\nfrom matplotlib import pyplot\r\nfrom statsmodels.tsa.arima_model import ARIMA\r\nfrom sklearn.metrics import mean_squared_error\r\n \r\ndef parser(x):\r\n return datetime.strptime('190'+x, '%Y-%m')\r\n \r\nseries = read_csv('shampoo-sales.csv', header=0, parse_dates=[0], index_col=0, squeeze=True, date_parser=parser)\r\nX = series.values\r\nsize = int(len(X) * 0.66)\r\ntrain, test = X[0:size], X[size:len(X)]\r\nhistory = [x for x in train]\r\npredictions = list()\r\nfor t in range(len(test)):\r\n model = ARIMA(history, order=(5,1,0))\r\n model_fit = model.fit(disp=0)\r\n output = model_fit.forecast()\r\n yhat = output[0]\r\n predictions.append(yhat)\r\n obs = test[t]\r\n history.append(obs)\r\n print('predicted=%f, expected=%f' % (yhat, obs))\r\nerror = mean_squared_error(test, predictions)\r\nprint('Test MSE: %.3f' % error)\r\n# plot\r\npyplot.plot(test)\r\npyplot.plot(predictions, color='red')\r\npyplot.show()\r\n"
] |
[
[
"sklearn.metrics.mean_squared_error",
"matplotlib.pyplot.plot",
"pandas.datetime.strptime",
"matplotlib.pyplot.show",
"pandas.read_csv"
]
] |
bryankim96/deep-learning-gamma
|
[
"cac4f2d90b6536eedf95c3a07ea73d4c7a69c5b7"
] |
[
"ctlearn/default_models/single_cnn.py"
] |
[
"import importlib\nimport sys\n\nimport tensorflow as tf\n\ndef single_cnn_model(data, model_params):\n\n # Load neural network model\n network_input = tf.keras.Input(shape=data.img_shape, name=f'images')\n network_name = model_params.get('name', 'CNN') + '_block'\n trainable_backbone = model_params.get('trainable_backbone', True)\n pretrained_weights = model_params.get('pretrained_weights', None)\n if pretrained_weights:\n loaded_model = tf.keras.models.load_model(pretrained_weights)\n for layer in loaded_model.layers:\n if layer.name.endswith('_block'):\n model = loaded_model.get_layer(layer.name)\n model.trainable = trainable_backbone\n else:\n sys.path.append(model_params['model_directory'])\n network_module = importlib.import_module(model_params['network']['module'])\n network = getattr(network_module, model_params['network']['function'])\n\n # The original ResNet implementation use this padding, but we pad the images in the ImageMapper.\n #x = tf.pad(telescope_data, tf.constant([[3, 3], [3, 3]]), name='conv1_pad')\n init_layer = model_params.get('init_layer', False)\n if init_layer:\n network_input = tf.keras.layers.Conv2D(filters=init_layer['filters'], kernel_size=init_layer['kernel_size'],\n strides=init_layer['strides'], name=network_name+'_conv1_conv')(network_input)\n #x = tf.pad(x, tf.constant([[1, 1], [1, 1]]), name='pool1_pad')\n init_max_pool = model_params.get('init_max_pool', False)\n if init_max_pool:\n network_input = tf.keras.layers.MaxPool2D(pool_size=init_max_pool['size'],\n strides=init_max_pool['strides'], name=network_name+'_pool1_pool')(network_input)\n\n network_output = network(network_input, params=model_params, name=network_name)\n\n output = tf.keras.layers.GlobalAveragePooling2D(name=network_name+'_global_avgpool')(network_output)\n\n model = tf.keras.Model(network_input, output, name=network_name)\n\n return model, network_input\n"
] |
[
[
"tensorflow.keras.layers.MaxPool2D",
"tensorflow.keras.layers.Conv2D",
"tensorflow.keras.models.load_model",
"tensorflow.keras.Model",
"tensorflow.keras.layers.GlobalAveragePooling2D",
"tensorflow.keras.Input"
]
] |
minhson/tensorpack
|
[
"5421ad0e6abd4500df14a0438c49d53ce7d7c1cd"
] |
[
"tensorpack/dataflow/imgaug/crop.py"
] |
[
"# -*- coding: utf-8 -*-\n# File: crop.py\n\nimport numpy as np\nimport cv2\n\nfrom ...utils.argtools import shape2d\nfrom ...utils.develop import log_deprecated\nfrom .base import ImageAugmentor, ImagePlaceholder\nfrom .transform import CropTransform, TransformList, ResizeTransform\nfrom .misc import ResizeShortestEdge\n\n__all__ = ['RandomCrop', 'CenterCrop', 'RandomCropRandomShape', 'GoogleNetRandomCropAndResize']\n\n\nclass RandomCrop(ImageAugmentor):\n \"\"\" Randomly crop the image into a smaller one \"\"\"\n\n def __init__(self, crop_shape):\n \"\"\"\n Args:\n crop_shape: (h, w) tuple or a int\n \"\"\"\n crop_shape = shape2d(crop_shape)\n super(RandomCrop, self).__init__()\n self._init(locals())\n\n def get_transform(self, img):\n orig_shape = img.shape\n assert orig_shape[0] >= self.crop_shape[0] \\\n and orig_shape[1] >= self.crop_shape[1], orig_shape\n diffh = orig_shape[0] - self.crop_shape[0]\n h0 = 0 if diffh == 0 else self.rng.randint(diffh)\n diffw = orig_shape[1] - self.crop_shape[1]\n w0 = 0 if diffw == 0 else self.rng.randint(diffw)\n return CropTransform(h0, w0, self.crop_shape[0], self.crop_shape[1])\n\n\nclass CenterCrop(ImageAugmentor):\n \"\"\" Crop the image at the center\"\"\"\n\n def __init__(self, crop_shape):\n \"\"\"\n Args:\n crop_shape: (h, w) tuple or a int\n \"\"\"\n crop_shape = shape2d(crop_shape)\n self._init(locals())\n\n def get_transform(self, img):\n orig_shape = img.shape\n assert orig_shape[0] >= self.crop_shape[0] \\\n and orig_shape[1] >= self.crop_shape[1], orig_shape\n h0 = int((orig_shape[0] - self.crop_shape[0]) * 0.5)\n w0 = int((orig_shape[1] - self.crop_shape[1]) * 0.5)\n return CropTransform(h0, w0, self.crop_shape[0], self.crop_shape[1])\n\n\nclass RandomCropRandomShape(ImageAugmentor):\n \"\"\" Random crop with a random shape\"\"\"\n\n def __init__(self, wmin, hmin,\n wmax=None, hmax=None,\n max_aspect_ratio=None):\n \"\"\"\n Randomly crop a box of shape (h, w), sampled from [min, max] (both inclusive).\n If max is None, will use the input image shape.\n\n Args:\n wmin, hmin, wmax, hmax: range to sample shape.\n max_aspect_ratio (float): this argument has no effect and is deprecated.\n \"\"\"\n super(RandomCropRandomShape, self).__init__()\n if max_aspect_ratio is not None:\n log_deprecated(\"RandomCropRandomShape(max_aspect_ratio)\", \"It is never implemented!\", \"2020-06-06\")\n self._init(locals())\n\n def get_transform(self, img):\n hmax = self.hmax or img.shape[0]\n wmax = self.wmax or img.shape[1]\n h = self.rng.randint(self.hmin, hmax + 1)\n w = self.rng.randint(self.wmin, wmax + 1)\n diffh = img.shape[0] - h\n diffw = img.shape[1] - w\n assert diffh >= 0 and diffw >= 0, str(diffh) + \", \" + str(diffw)\n y0 = 0 if diffh == 0 else self.rng.randint(diffh)\n x0 = 0 if diffw == 0 else self.rng.randint(diffw)\n return CropTransform(y0, x0, h, w)\n\n\nclass GoogleNetRandomCropAndResize(ImageAugmentor):\n \"\"\"\n The random crop and resize augmentation proposed in\n Sec. 6 of \"Going Deeper with Convolutions\" by Google.\n This implementation follows the details in ``fb.resnet.torch``.\n\n It attempts to crop a random rectangle with 8%~100% area of the original image,\n and keep the aspect ratio between 3/4 to 4/3. Then it resize this crop to the target shape.\n If such crop cannot be found in 10 iterations, it will do a ResizeShortestEdge + CenterCrop.\n \"\"\"\n def __init__(self, crop_area_fraction=(0.08, 1.),\n aspect_ratio_range=(0.75, 1.333),\n target_shape=224, interp=cv2.INTER_LINEAR):\n \"\"\"\n Args:\n crop_area_fraction (tuple(float)): Defaults to crop 8%-100% area.\n aspect_ratio_range (tuple(float)): Defaults to make aspect ratio in 3/4-4/3.\n target_shape (int): Defaults to 224, the standard ImageNet image shape.\n \"\"\"\n super(GoogleNetRandomCropAndResize, self).__init__()\n self._init(locals())\n\n def get_transform(self, img):\n h, w = img.shape[:2]\n area = h * w\n for _ in range(10):\n targetArea = self.rng.uniform(*self.crop_area_fraction) * area\n aspectR = self.rng.uniform(*self.aspect_ratio_range)\n ww = int(np.sqrt(targetArea * aspectR) + 0.5)\n hh = int(np.sqrt(targetArea / aspectR) + 0.5)\n if self.rng.uniform() < 0.5:\n ww, hh = hh, ww\n if hh <= h and ww <= w:\n x1 = 0 if w == ww else self.rng.randint(0, w - ww)\n y1 = 0 if h == hh else self.rng.randint(0, h - hh)\n return TransformList([\n CropTransform(y1, x1, hh, ww),\n ResizeTransform(hh, ww, self.target_shape, self.target_shape, interp=self.interp)\n ])\n tfm1 = ResizeShortestEdge(self.target_shape, interp=self.interp).get_transform(img)\n out_shape = (tfm1.new_h, tfm1.new_w)\n tfm2 = CenterCrop(self.target_shape).get_transform(ImagePlaceholder(shape=out_shape))\n return TransformList([tfm1, tfm2])\n"
] |
[
[
"numpy.sqrt"
]
] |
dfangshuo/neuro-vectorizer
|
[
"9258bcaab31280dc0685610a165a08cb3bdaa023"
] |
[
"envs/neurovec.py"
] |
[
"'''\nCopyright (c) 2019, Ameer Haj Ali (UC Berkeley), and Intel Corporation\nAll rights reserved.\n\nRedistribution and use in source and binary forms, with or without\nmodification, are permitted provided that the following conditions are met:\n\n1. Redistributions of source code must retain the above copyright notice, this\n list of conditions and the following disclaimer.\n\n2. Redistributions in binary form must reproduce the above copyright notice,\n this list of conditions and the following disclaimer in the documentation\n and/or other materials provided with the distribution.\n\n3. Neither the name of the copyright holder nor the names of its\n contributors may be used to endorse or promote products derived from\n this software without specific prior written permission.\n\nTHIS SOFTWARE IS PROVIDED BY THE COPYRIGHT HOLDERS AND CONTRIBUTORS \"AS IS\"\nAND ANY EXPRESS OR IMPLIED WARRANTIES, INCLUDING, BUT NOT LIMITED TO, THE\nIMPLIED WARRANTIES OF MERCHANTABILITY AND FITNESS FOR A PARTICULAR PURPOSE ARE\nDISCLAIMED. IN NO EVENT SHALL THE COPYRIGHT HOLDER OR CONTRIBUTORS BE LIABLE\nFOR ANY DIRECT, INDIRECT, INCIDENTAL, SPECIAL, EXEMPLARY, OR CONSEQUENTIAL\nDAMAGES (INCLUDING, BUT NOT LIMITED TO, PROCUREMENT OF SUBSTITUTE GOODS OR\nSERVICES; LOSS OF USE, DATA, OR PROFITS; OR BUSINESS INTERRUPTION) HOWEVER\nCAUSED AND ON ANY THEORY OF LIABILITY, WHETHER IN CONTRACT, STRICT LIABILITY,\nOR TORT (INCLUDING NEGLIGENCE OR OTHERWISE) ARISING IN ANY WAY OUT OF THE USE\nOF THIS SOFTWARE, EVEN IF ADVISED OF THE POSSIBILITY OF SUCH DAMAGE.\n'''\nimport gym\nfrom gym import spaces\nimport pickle\nimport numpy as np\nimport re\nimport os\nimport logging\n\nfrom extractor_c import CExtractor\nfrom config import Config\nfrom my_model import Code2VecModel\nfrom path_context_reader import EstimatorAction\n\nfrom utility import get_bruteforce_runtimes, get_O3_runtimes, get_snapshot_from_code, get_runtime, get_vectorized_codes, init_runtimes_dict, get_encodings_from_local, MAX_LEAF_NODES, pragma_line\n\nlogger = logging.getLogger(__name__)\n\n#NeuroVectorizer RL Environment\nclass NeuroVectorizerEnv(gym.Env):\n def __init__(self, env_config):\n self.init_from_env_config(env_config)\n self.copy_train_data()\n self.parse_train_data()\n self.config_AST_parser()\n self.init_RL_env()\n # Keeps track of the file being processed currently.\n self.current_file_idx = 0\n # Keeps track of the current loop being processed currently in that file.\n self.current_pragma_idx = 0\n '''Runtimes dict to stored programs the RL agent explored.\n This saves execution and compilation time due to dynamic programming.'''\n self.runtimes = init_runtimes_dict(self.new_testfiles,self.num_loops,\n len(self.vec_action_meaning),len(self.interleave_action_meaning))\n '''Observations dictionary to store AST encodings of programs explored by the RL agent. \n It saves time when the RL agent explores a program it explored before.\n It is also initialized from obs_encodings.pkl file to further save time.''' \n self.obs_encodings = get_encodings_from_local(self.new_rundir)\n \n if self.compile:\n # stores the runtimes of O3 to compute the RL reward and compared to -O3.\n self.O3_runtimes = get_O3_runtimes(self.new_rundir, self.new_testfiles)\n runtimes_with_pid = {}\n for key in self.O3_runtimes:\n value = self.O3_runtimes[key]\n \n # assumes keys are of the format ./dirname/...\n key_components = key[2:].split('/') # removes the `./` prefix\n key_components[0] = self.new_rundir\n key = '/'.join(key_components)\n runtimes_with_pid[key] = value\n self.O3_runtimes = runtimes_with_pid\n \n def init_from_env_config(self,env_config):\n '''Receives env_config and initalizes all config parameters.'''\n # dirpath is the path to the train data.\n self.dirpath = env_config.get('dirpath')\n # new_rundir is the directory to create and copy the train data to.\n self.new_rundir = env_config.get('new_rundir') + str(os.getpid())\n # whether or not in inference mode\n self.inference_mode = env_config.get('inference_mode', False)\n if self.inference_mode:\n # Used in inference mode to print current geomean improvement.\n self.improvements=[]\n '''Whether to compile the progams or not, generally turned off \n in inference mode when it is not clear how to compile (e.g., requires make)\n '''\n self.compile = env_config.get('compile', True) \n #if your code is not structured like the given training data.\n self.new_train_data = env_config.get('new_train_data',False) \n \n def copy_train_data(self):\n '''Copy the train data to a new directory.\n used to inject pragmas in the new files,\n without modifying original files.\n '''\n if not os.path.exists(self.new_rundir):\n print('creating '+self.new_rundir+' directory')\n os.mkdir(self.new_rundir)\n\n cmd = 'cp -r ' +self.dirpath+'/* ' +self.new_rundir\n print('running:',cmd)\n os.system(cmd)\n \n def init_RL_env(self):\n ''' Defines the reinforcement leaning environment.\n Modify to match your hardware and programs.\n '''\n self.vec_action_meaning = [1,2,4,8,16,32,64] # TODO: change this to match your hardware\n self.interleave_action_meaning=[1,2,4,8,16] # TODO: change this to match your hardware\n self.action_space = spaces.Tuple([spaces.Discrete(len(self.vec_action_meaning)),\n spaces.Discrete(len(self.interleave_action_meaning))])\n '''The observation space is bounded by the word dictionary \n the preprocessing generated.'''\n self.observation_space = spaces.Tuple(\n [spaces.Box(0,self.code2vec.vocabs.token_vocab.size,shape=(self.config.MAX_CONTEXTS,),dtype = np.int32,)]\n +[spaces.Box(0,self.code2vec.vocabs.path_vocab.size,shape=(self.config.MAX_CONTEXTS,),dtype = np.int32,)]\n +[spaces.Box(0,self.code2vec.vocabs.token_vocab.size,shape=(self.config.MAX_CONTEXTS,),dtype = np.int32,)]\n +[spaces.Box(0,1,shape=(self.config.MAX_CONTEXTS,),dtype = np.bool)]\n )\n def parse_train_data(self):\n ''' Parse the training data. '''\n self.orig_train_files = [os.path.join(root, name)\n for root, dirs, files in os.walk(self.new_rundir)\n for name in files\n if name.endswith(\".c\") and not name.startswith('header.c') \n and not name.startswith('aux_AST_embedding_code.c')]\n # copy testfiles\n self.new_testfiles = list(self.orig_train_files)\n # parse the code to detect loops and inject commented pragmas. \n self.loops_idxs_in_orig,self.pragmas_idxs,self.const_new_codes,self.num_loops,self.const_orig_codes \\\n = get_vectorized_codes(self.orig_train_files,self.new_testfiles)\n # to operate only on files that have for loops.\n self.new_testfiles = list(self.pragmas_idxs.keys())\n \n def config_AST_parser(self):\n '''Config the AST tree parser.'''\n self.config = Config(set_defaults=True, load_from_args=False, verify=True)\n self.code2vec = Code2VecModel(self.config)\n self.path_extractor = CExtractor(self.config,clang_path=os.environ['CLANG_PATH'],max_leaves=MAX_LEAF_NODES)\n self.train_input_reader = self.code2vec._create_data_reader(estimator_action=EstimatorAction.Train)\n \n def get_reward(self,new_code,current_filename,VF_idx,IF_idx):\n '''Calculates the RL agent's reward. The reward is the \n execution time improvement after injecting the pragma\n normalized to -O3.'''\n f = open(current_filename,'w')\n f.write(''.join(new_code))\n f.close()\n if self.compile:\n if self.runtimes[current_filename][self.current_pragma_idx][VF_idx][IF_idx]:\n runtime = self.runtimes[current_filename][self.current_pragma_idx][VF_idx][IF_idx]\n else: \n runtime = get_runtime(self.new_rundir,new_code,current_filename)\n self.runtimes[current_filename][self.current_pragma_idx][VF_idx][IF_idx]=runtime\n if self.O3_runtimes[current_filename]==None:\n reward = 0\n logger.warning('Program '+current_filename+' does not compile in two seconds.'+\n ' Consider removing it or increasing the timeout parameter'+\n ' in utility.py.')\n elif runtime==None:\n #penalizing for long compilation time for bad VF/IF\n reward = -9\n else: \n reward = (self.O3_runtimes[current_filename]-runtime)/self.O3_runtimes[current_filename]\n # In inference mode and finished inserting pragmas to this file.\n if self.inference_mode and self.current_pragma_idx+1 == self.num_loops[current_filename]:\n improvement = self.O3_runtimes[current_filename]/runtime\n self.improvements.append(improvement)\n geomean = 1\n for imp in self.improvements:\n geomean = geomean * (imp**(1/len(self.improvements))) \n print('benchmark: ',current_filename,'O3 runtime: ', \n self.O3_runtimes[current_filename], 'RL runtime: ', runtime,\n 'improvement:',str(round(improvement,2))+'X',\n 'improvement geomean so far:',str(round(geomean,2))+'X')\n VF = self.vec_action_meaning[VF_idx]\n IF = self.interleave_action_meaning[IF_idx]\n opt_runtime_sofar=self.get_opt_runtime(current_filename,self.current_pragma_idx)\n logger.info(current_filename+' runtime '+str(runtime)+' O3 ' + \n str(self.O3_runtimes[current_filename]) +' reward '+str(reward)+\n ' opt '+str(opt_runtime_sofar)+\" VF \"+str(VF)+\" IF \"+str(IF))\n else:\n # can't calculate the reward without compile/runtime.\n reward = 0\n \n return reward\n\n def get_opt_runtime(self,current_filename,current_pragma_idx):\n min_runtime = float('inf')\n for VF_idx in self.runtimes[current_filename][self.current_pragma_idx]:\n for IF_idx in VF_idx:\n if IF_idx:\n min_runtime = min(min_runtime,IF_idx)\n return min_runtime\n \n def reset(self):\n ''' RL reset environment function. '''\n current_filename = self.new_testfiles[self.current_file_idx]\n #this make sure that all RL pragmas remain in the code when inferencing.\n if self.current_pragma_idx == 0 or not self.inference_mode:\n self.new_code = list(self.const_new_codes[current_filename])\n return self.get_obs(current_filename,self.current_pragma_idx)\n\n def get_obs(self,current_filename,current_pragma_idx):\n '''Given a file returns the RL observation.\n Change this if you want other embeddings.'''\n \n #Check if this encoding already exists (parsed before).\n try:\n return self.obs_encodings[current_filename][current_pragma_idx]\n except:\n pass\n \n # To get code for files not in the dataset.\n if self.new_train_data:\n code=get_snapshot_from_code(self.const_orig_codes[current_filename],\n self.loops_idxs_in_orig[current_filename][current_pragma_idx])\n else:\n code=get_snapshot_from_code(self.const_orig_codes[current_filename])\n\n input_full_path_filename=os.path.join(self.new_rundir,'aux_AST_embedding_code.c')\n loop_file=open(input_full_path_filename,'w')\n loop_file.write(''.join(code))\n loop_file.close()\n try:\n train_lines, hash_to_string_dict = self.path_extractor.extract_paths(input_full_path_filename)\n except:\n print('Could not parse file',current_filename, 'loop index',current_pragma_idx,'. Try removing it.')\n raise \n dataset = self.train_input_reader.process_and_iterate_input_from_data_lines(train_lines)\n obs = []\n tensors = list(dataset)[0][0]\n import tensorflow as tf\n for tensor in tensors:\n with tf.compat.v1.Session() as sess: \n sess.run(tf.compat.v1.tables_initializer())\n obs.append(tf.squeeze(tensor).eval())\n\n if current_filename not in self.obs_encodings:\n self.obs_encodings[current_filename] = {}\n self.obs_encodings[current_filename][current_pragma_idx] = obs\n return obs\n\n def step(self,action):\n '''The RL environment step function. Takes action and applies it as\n VF/IF pragma for the parsed loop.'''\n done = True # RL horizon = 1 \n action = list(np.reshape(np.array(action),(np.array(action).shape[0],)))\n VF_idx = action[0]\n IF_idx = action[1]\n VF = self.vec_action_meaning[VF_idx]\n IF = self.interleave_action_meaning[IF_idx]\n current_filename = self.new_testfiles[self.current_file_idx]\n self.new_code[self.pragmas_idxs[current_filename][self.current_pragma_idx]] = pragma_line.format(VF,IF)\n reward = self.get_reward(self.new_code,current_filename,VF_idx,IF_idx)\n #print(\"VF\",VF,\"IF\",IF)\n #print('reward:', reward, 'O3',self.O3_runtimes[current_filename])\n self.current_pragma_idx += 1\n if self.current_pragma_idx == self.num_loops[current_filename]:\n self.current_pragma_idx=0\n self.current_file_idx += 1\n if self.current_file_idx == len(self.new_testfiles):\n self.current_file_idx = 0\n if self.inference_mode:\n print('exiting after inferencing all programs')\n exit(0) # finished all programs!\n '''Change next line for new observation spaces\n to a matrix of zeros.'''\n obs = [[0]*200]*4\n else:\n obs = self.get_obs(current_filename,self.current_pragma_idx)\n \n return obs,reward,done,{}\n"
] |
[
[
"tensorflow.compat.v1.Session",
"numpy.array",
"tensorflow.compat.v1.tables_initializer",
"tensorflow.squeeze"
]
] |
zwt233/automl-toolkit
|
[
"67d057f5e0c74bec5b3cbde1440ec014696737ef"
] |
[
"automlToolkit/components/ensemble/stacking.py"
] |
[
"import numpy as np\nimport warnings\nimport os\nimport pickle as pkl\nfrom sklearn.model_selection import StratifiedKFold, KFold\nfrom sklearn.metrics.scorer import _BaseScorer\n\nfrom automlToolkit.components.ensemble.base_ensemble import BaseEnsembleModel\nfrom automlToolkit.components.utils.constants import CLS_TASKS\nfrom automlToolkit.components.evaluators.base_evaluator import fetch_predict_estimator\n\n\nclass Stacking(BaseEnsembleModel):\n def __init__(self, stats,\n ensemble_size: int,\n task_type: int,\n metric: _BaseScorer,\n output_dir=None,\n meta_learner='xgboost',\n kfold=5):\n super().__init__(stats=stats,\n ensemble_method='blending',\n ensemble_size=ensemble_size,\n task_type=task_type,\n metric=metric,\n output_dir=output_dir)\n\n self.kfold = kfold\n try:\n from xgboost import XGBClassifier\n except:\n warnings.warn(\"Xgboost is not imported! Blending will use linear model instead!\")\n meta_learner = 'linear'\n\n # We use Xgboost as default meta-learner\n if self.task_type in CLS_TASKS:\n if meta_learner == 'linear':\n from sklearn.linear_model.logistic import LogisticRegression\n self.meta_learner = LogisticRegression(max_iter=1000)\n elif meta_learner == 'gb':\n from sklearn.ensemble.gradient_boosting import GradientBoostingClassifier\n self.meta_learner = GradientBoostingClassifier(learning_rate=0.05, subsample=0.7, max_depth=4,\n n_estimators=250)\n elif meta_learner == 'xgboost':\n from xgboost import XGBClassifier\n self.meta_learner = XGBClassifier(max_depth=4, learning_rate=0.05, n_estimators=150)\n else:\n if meta_learner == 'linear':\n from sklearn.linear_model import LinearRegression\n self.meta_learner = LinearRegression()\n elif meta_learner == 'xgboost':\n from xgboost import XGBRegressor\n self.meta_learner = XGBRegressor(max_depth=4, learning_rate=0.05, n_estimators=70)\n\n def fit(self, data):\n # Split training data for phase 1 and phase 2\n if self.task_type in CLS_TASKS:\n kf = StratifiedKFold(n_splits=self.kfold)\n else:\n kf = KFold(n_splits=self.kfold)\n\n # Train basic models using a part of training data\n model_cnt = 0\n suc_cnt = 0\n feature_p2 = None\n for algo_id in self.stats[\"include_algorithms\"]:\n train_list = self.stats[algo_id]['train_data_list']\n configs = self.stats[algo_id]['configurations']\n for idx in range(len(train_list)):\n X, y = train_list[idx].data\n for _config in configs:\n if self.base_model_mask[model_cnt] == 1:\n for j, (train, test) in enumerate(kf.split(X, y)):\n x_p1, x_p2, y_p1, _ = X[train], X[test], y[train], y[test]\n estimator = fetch_predict_estimator(self.task_type, _config, x_p1, y_p1)\n with open(\n os.path.join(self.output_dir, '%s-model%d_part%d' % (self.timestamp, model_cnt, j)),\n 'wb') as f:\n pkl.dump(estimator, f)\n if self.task_type in CLS_TASKS:\n pred = estimator.predict_proba(x_p2)\n n_dim = np.array(pred).shape[1]\n if n_dim == 2:\n # Binary classificaion\n n_dim = 1\n # Initialize training matrix for phase 2\n if feature_p2 is None:\n num_samples = len(train) + len(test)\n feature_p2 = np.zeros((num_samples, self.ensemble_size * n_dim))\n if n_dim == 1:\n feature_p2[test, suc_cnt * n_dim:(suc_cnt + 1) * n_dim] = pred[:, 1:2]\n else:\n feature_p2[test, suc_cnt * n_dim:(suc_cnt + 1) * n_dim] = pred\n else:\n pred = estimator.predict(x_p2).reshape(-1, 1)\n n_dim = 1\n # Initialize training matrix for phase 2\n if feature_p2 is None:\n num_samples = len(train) + len(test)\n feature_p2 = np.zeros((num_samples, self.ensemble_size * n_dim))\n feature_p2[test, suc_cnt * n_dim:(suc_cnt + 1) * n_dim] = pred\n suc_cnt += 1\n model_cnt += 1\n # Train model for stacking using the other part of training data\n self.meta_learner.fit(feature_p2, y)\n return self\n\n def get_feature(self, data, solvers):\n # Predict the labels via stacking\n feature_p2 = None\n model_cnt = 0\n suc_cnt = 0\n for algo_id in self.stats[\"include_algorithms\"]:\n train_list = self.stats[algo_id]['train_data_list']\n configs = self.stats[algo_id]['configurations']\n for train_node in train_list:\n test_node = solvers[algo_id].optimizer['fe'].apply(data, train_node)\n for _ in configs:\n if self.base_model_mask[model_cnt] == 1:\n for j in range(self.kfold):\n with open(\n os.path.join(self.output_dir, '%s-model%d_part%d' % (self.timestamp, model_cnt, j)),\n 'rb') as f:\n estimator = pkl.load(f)\n if self.task_type in CLS_TASKS:\n pred = estimator.predict_proba(test_node.data[0])\n n_dim = np.array(pred).shape[1]\n if n_dim == 2:\n n_dim = 1\n if feature_p2 is None:\n num_samples = len(test_node.data[0])\n feature_p2 = np.zeros((num_samples, self.ensemble_size * n_dim))\n # Get average predictions\n if n_dim == 1:\n feature_p2[:, suc_cnt * n_dim:(suc_cnt + 1) * n_dim] = \\\n feature_p2[:, suc_cnt * n_dim:(suc_cnt + 1) * n_dim] + pred[:,\n 1:2] / self.kfold\n else:\n feature_p2[:, suc_cnt * n_dim:(suc_cnt + 1) * n_dim] = \\\n feature_p2[:, suc_cnt * n_dim:(suc_cnt + 1) * n_dim] + pred / self.kfold\n else:\n pred = estimator.predict(test_node.data[0]).reshape(-1, 1)\n n_dim = 1\n # Initialize training matrix for phase 2\n if feature_p2 is None:\n num_samples = len(test_node.data[0])\n feature_p2 = np.zeros((num_samples, self.ensemble_size * n_dim))\n # Get average predictions\n feature_p2[:, suc_cnt * n_dim:(suc_cnt + 1) * n_dim] = \\\n feature_p2[:, suc_cnt * n_dim:(suc_cnt + 1) * n_dim] + pred / self.kfold\n suc_cnt += 1\n model_cnt += 1\n return feature_p2\n\n def predict(self, data, solvers):\n feature_p2 = self.get_feature(data, solvers)\n # Get predictions from meta-learner\n if self.task_type in CLS_TASKS:\n final_pred = self.meta_learner.predict_proba(feature_p2)\n else:\n final_pred = self.meta_learner.predict(feature_p2)\n return final_pred\n"
] |
[
[
"numpy.array",
"sklearn.model_selection.StratifiedKFold",
"sklearn.linear_model.LinearRegression",
"numpy.zeros",
"sklearn.ensemble.gradient_boosting.GradientBoostingClassifier",
"sklearn.linear_model.logistic.LogisticRegression",
"sklearn.model_selection.KFold"
]
] |
mxhofer/OrgSim-RL
|
[
"bd494579c4b6e6ce6ebda5dbbfd2706124569f6c"
] |
[
"dyna-q/utils_diagnostics.py"
] |
[
"# -- Import packages ---------------------------------------------------------------------------------------------------\nfrom collections import OrderedDict\nimport os\nimport json\nimport numpy as np\n\n\nclass Diagnostics:\n \"\"\"\n Diagnostics for an episode.\n \"\"\"\n\n def __init__(self, runs):\n self.d = OrderedDict()\n self.runs = runs\n self.reset()\n\n def reset(self):\n \"\"\"\n Reset the diagnostics dictionary.\n \"\"\"\n self.d = OrderedDict()\n\n for r in range(self.runs):\n self.d[f'run_{r}'] = {'n_steps': [],\n 'net_reward_to_org': [],\n 'actionFromStart': [],\n 'optimalPathLength': [],\n 'optimalAction': [],\n 'short_open_door_path': [],\n 'long_path': [],\n 'short_closed_door_path': [],\n 'noGoal': [],\n 'leaders': [],\n 'coordinationCostsAccumulated': [],\n 'opportunityCostsAccumulated': [],\n 'policyInDomain_longpath': [],\n 'policyInDomain_shortpath': [],\n 'n_samplesModelDomain1': [],\n 'n_samplesModelDomain2': [],\n 'n_samplesModelDomain3': [],\n 'n_samplesModelDomain4': [],\n 'n_visits_per_state': np.zeros((13, 13)),\n 'neutral_states_share': [],\n 'shortpath_states_share': [],\n 'longpath_states_share': [],\n 'neutral_states_count': [],\n 'shortpath_states_count': [],\n 'longpath_states_count': []\n }\n\n def to_disk(self, par_name_, par_value_, start_date, start_time, runs):\n \"\"\"\n Write diagnostics to disk.\n :param par_name_: parameter name\n :param par_value_: parameter value\n :param start_date: start date of the simulation run\n :param start_time: start time of the simulation run\n \"\"\"\n assert type(self.d) == OrderedDict, 'ERROR: diagnostics variable must be a dictionary.'\n\n os.makedirs(f'outputs/traces/{start_date}/{start_time}', exist_ok=True)\n path = f'outputs/traces/{start_date}/{start_time}'\n filename = f'/dyna-q_{par_name_}_{par_value_}_{start_time}.json'\n\n # JSONify ndarray\n for r in range(runs):\n self.d[f'run_{r}']['n_visits_per_state'] = self.d[f'run_{r}']['n_visits_per_state'].tolist()\n\n with open(path + filename, \"w\") as f:\n json.dump(self.d, f, indent=4)\n\n print(f'\\nSuccessfully saved to disk: {filename}\\n')\n"
] |
[
[
"numpy.zeros"
]
] |
maumruiz/ICNN_Loss
|
[
"249405f50421a2cf097a8ab4046db9cb32811164"
] |
[
"algorithms/icnn_loss.py"
] |
[
"import torch\nimport torch.nn.functional as F\n\nimport util.globals as glob\n\ndef distance_matrix(x, y=None, p=2):\n y = x if type(y) == type(None) else y\n\n n = x.size(0)\n m = y.size(0)\n d = x.size(1)\n\n x = x.unsqueeze(1).expand(n, m, d)\n y = y.unsqueeze(0).expand(n, m, d)\n \n dist = torch.pow(x - y, p).sum(2)\n \n return dist\n\ndef nearest_neighbors(X, y=None, k=3, p=2):\n eps = 0.000001\n dist = (distance_matrix(X, y, p=p) + eps) ** (1/2)\n knn = dist.topk(k, largest=False)\n return knn.values, knn.indices\n\ndef score(origin, y_orig, target=None, y_targ=None, k=5, p=2, q=2, r=2):\n \"\"\"Compute class prototypes from support samples.\n\n # Arguments\n X: torch.Tensor. Tensor of shape (n * k, d) where d is the embedding\n dimension.\n y: torch.Tensor. The class of every sample\n k: int. the number of neigbhors (small k focus on local structures big k on global)\n p: int. must be a natural number, the higher is p, the lower penalization on lambda function\n q: int. must be a natural number, the higher is p, the lower penalization on omega function\n r: int. must be a natural number, the higher is r, the lower penalization on gamma function\n\n # Returns\n \n \"\"\"\n target = origin if type(target) == type(None) else target\n y_targ = y_orig if type(y_targ) == type(None) else y_targ\n\n eps = 0.000001\n k = 3 if k >= target.shape[0] else k\n\n #min max scale by feature\n # a = target - target.min(axis=0).values\n # b = X.max(axis=0).values - X.min(axis=0).values\n # X = torch.divide(a , b+eps)\n\n distances, indices = nearest_neighbors(origin, target, k=k+1)\n distances = distances[:,1:]\n indices = indices[:,1:]\n\n # class by neighbor\n classes = y_targ[indices]\n yMatrix = y_orig.repeat(k,1).T\n scMatrix = (yMatrix == classes)*1 # Same class matrix [1 same class, 0 diff class]\n dcMatrix = (scMatrix)*(-1)+1 # Different class matrix [negation of scMatrix]\n\n ### Normalizing distances between neighbords\n dt = distances.T\n nd = (dt - dt.min(axis=0).values) / ( (dt.max(axis=0).values - dt.min(axis=0).values) + eps )\n nd = nd.T\n\n ## Distances\n scd = distances*scMatrix #Same class distance\n dcd = distances*dcMatrix #Different class distance\n ## Normalized distances\n scnd = nd*scMatrix #Same class normalized distance\n dcnd = nd*dcMatrix #Different class normalized distance\n \n ### Lambda computation\n plamb = (1 - scnd) * scMatrix\n lamb = (dcnd + plamb)\n lambs = torch.sum(lamb,axis=1)\n lambs2 = (lambs / (torch.max(lambs) + eps)) ** (1/p)\n lambr = torch.sum(lambs2) / (y_orig.shape[0])\n\n varsc = torch.var(scnd)\n vardf = torch.var(dcnd)\n omega = (1 - (varsc+vardf))**(1/q)\n \n gamma = torch.sum((torch.sum(scMatrix, axis=1) / k) ** (1/r)) / (y_orig.shape[0])\n \n # return (lambr + gamma + omega)/3\n # return lambr\n return (lambr + gamma)/2\n\ndef modified_score(origin, y_orig, target=None, y_targ=None, k=4, ipc=None):\n target = origin if type(target) == type(None) else target\n y_targ = y_orig if type(y_targ) == type(None) else y_targ\n k = target.shape[0] - 2 if k+2 >= target.shape[0] else k\n ipc = k+1 if type(ipc) == type(None) else ipc\n eps = 0.0000001\n\n distances, indices = nearest_neighbors(origin, target, k=ipc+1)\n distances = distances[:,1:]\n indices = indices[:,1:]\n\n # class by neighbor\n classes = y_targ[indices]\n yMatrix = y_orig.repeat(ipc,1).T\n scMatrix = (yMatrix == classes)*1 # Same class matrix [1 same class, 0 diff class]\n dcMatrix = (scMatrix)*(-1)+1 # Different class matrix [negation of scMatrix]\n\n ### Lambda Computation ###\n\n # Same class distance\n dt = distances.T\n dcnd = distances*dcMatrix\n nn_dc = (dcnd + torch.where(dcnd.eq(0.), float('inf'), 0.)).min(axis=1).values\n nd = dt / (nn_dc + eps)\n nd = nd[:k, :]\n nd = nd / torch.stack((torch.ones_like(nd[0]), nd.max(axis=0).values)).max(axis=0).values # Normalize with max(max_from_row, 1.0)\n nd = nd.T\n\n scMatrix = scMatrix[:, :k]\n scnd = nd*scMatrix\n\n scCounts = scMatrix.sum(axis=1)\n scndsum = scnd.sum(axis=1) / (scCounts + eps)\n sclamb = 1 - (scndsum.sum() / (torch.count_nonzero(scCounts) + eps))\n\n dcnd = dcnd[:, :k]\n dcMatrix = dcMatrix[:, :k]\n\n # Different class distance\n dcnd = dcnd / (dcnd.max() + eps)\n dcop = -1 if torch.all(dcMatrix == 0) else 0\n dcCounts = dcMatrix.sum(axis=1)\n dcndsum = dcnd.sum(axis=1) / (dcCounts + eps)\n dclamb = dcndsum.sum() / (torch.count_nonzero(dcCounts) + eps)\n dclamb = torch.abs(dclamb + dcop)\n\n lambr = (sclamb + dclamb) / 2\n\n ## Omega Calculation ###\n # varsc = torch.var(scnd)\n # vardf = torch.var(dcnd)\n # omega = 1 - (varsc+vardf)\n \n ### Gamma computation\n gamma = torch.sum(torch.sum(scMatrix, axis=1) / k) / (y_orig.shape[0]) if k+2 < target.shape[0] else 1.0\n \n # return (lambr + gamma + omega) / 3\n return (lambr + gamma) / 2\n\ndef proto_triplet_loss(queries, y_qry, protos, y_protos, way=5, margin=0.5):\n distances, indices = nearest_neighbors(queries, protos, k=way)\n classes = y_protos[indices]\n yMatrix = y_qry.repeat(way,1).T\n scMatrix = (yMatrix == classes)*1\n dcMatrix = (scMatrix)*(-1)+1\n\n scd = distances*scMatrix\n scp = scd.max(axis=1).values\n\n dcd = distances*dcMatrix\n dcd += torch.where(dcd.eq(0.), float('inf'), 0.)\n dcp = dcd.min(axis=1).values\n\n\n dists = torch.stack((scp - dcp + margin, torch.zeros_like(scp)), axis=1)\n return torch.mean(torch.max(dists, axis=1).values)\n\n\ndef get_icnn_loss(args, logits, way, qry_labels):\n loss = 0\n\n if 'cross' in args.losses:\n loss = F.cross_entropy(logits, qry_labels)\n\n if 'suppicnn' in args.losses:\n supp_labels = torch.arange(0, way, 1/args.shot).type(torch.int).cuda()\n supp_score = modified_score(glob.supp_fts, supp_labels)\n loss += (-torch.log(supp_score))\n\n if 'queryicnn' in args.losses:\n if args.query_protos:\n proto_labels = torch.arange(0, way).type(torch.int).cuda()\n query_score = modified_score(glob.query_fts, qry_labels, glob.prototypes, proto_labels)\n else:\n supp_labels = torch.arange(0, way, 1/args.shot).type(torch.int).cuda()\n query_score = modified_score(glob.query_fts, qry_labels, glob.supp_fts, supp_labels)\n loss += (-torch.log(query_score))\n\n if 'fullicnn' in args.losses:\n features = torch.cat((glob.supp_fts, glob.query_fts))\n supp_labels = torch.arange(0, way, 1/args.shot).type(torch.int).cuda()\n labels = torch.cat((supp_labels, qry_labels))\n ipc = args.shot+args.query\n score = modified_score(features, labels, ipc=ipc)\n\n loss += (-torch.log(score))\n \n if 'prototriplet' in args.losses:\n proto_labels = torch.arange(0, way).type(torch.int).cuda()\n loss += proto_triplet_loss(glob.query_fts, qry_labels, glob.prototypes, proto_labels, way)\n\n count = len(args.losses.split(','))\n loss = loss / count\n\n return loss"
] |
[
[
"torch.var",
"torch.cat",
"torch.arange",
"torch.max",
"torch.pow",
"torch.abs",
"torch.nn.functional.cross_entropy",
"torch.all",
"torch.count_nonzero",
"torch.ones_like",
"torch.zeros_like",
"torch.log",
"torch.sum"
]
] |
mathiassunesen/Speciale_retirement
|
[
"9db901a3791b9b75f228d1cec6c180e917be93e8"
] |
[
"Main/setup.py"
] |
[
"# global modules\nfrom numba import boolean, int32, int64, float64, double, njit, typeof\nimport numpy as np\nimport itertools\nimport pandas as pd\n\n# consav package\nfrom consav import misc \n\n# local modules\nimport transitions\nimport funs\n\ndef single_lists():\n\n parlist = [ # (name,numba type), parameters, grids etc.\n\n # boolean\n ('couple',boolean),\n\n # misc\n ('denom',double),\n ('tol',double),\n\n # time parameters\n ('start_T',int32),\n ('end_T',int32),\n ('forced_T',int32),\n ('simT',int32),\n\n # simulation \n ('sim_seed',int32),\n ('simN',int32),\n\n # savings\n ('R',double),\n\n # grids \n ('a_max',int32), \n ('a_phi',double),\n ('Na',int32),\n ('Nxi',int32), \n\n # states \n ('MA',int32[:]), \n ('ST',int32[:,:]), \n ('AD',int32[:]),\n\n # preference parameters\n ('rho',double), \n ('beta',double),\n ('alpha_0_male',double), \n ('alpha_0_female',double), \n ('alpha_1',double),\n ('gamma',double), \n ('v',double), \n\n # uncertainty/variance parameters\n ('sigma_eta',double), \n ('var',double[:]), \n\n # initial estimations\n ('pi_adjust_f',double),\n ('pi_adjust_m',double),\n ('reg_survival_male',double[:]),\n ('reg_survival_female',double[:]),\n ('reg_labor_male',double[:]),\n ('reg_labor_female',double[:]),\n ('g_adjust',double),\n ('priv_pension_female',double),\n ('priv_pension_male',double), \n\n # tax system\n ('IRA_tax',double),\n ('fradrag_to_oap',int32),\n ('fradrag',double),\n ('tau_upper',double),\n ('tau_LMC',double),\n ('WD',double),\n ('WD_upper',double),\n ('tau_c',double),\n ('y_low',double),\n ('y_low_m',double),\n ('y_low_u',double),\n ('tau_h',double),\n ('tau_l',double),\n ('tau_m',double),\n ('tau_u',double),\n ('tau_max',double), \n\n # retirement\n ('oap_age',int32),\n ('two_year',int32), \n ('erp_age',int32),\n ('B',double),\n ('y_B',double),\n ('tau_B',double),\n ('D_B',double),\n ('D_s',double), \n ('A_i',double[:]),\n ('y_i',double[:]),\n ('tau_i',double[:]),\n ('D_i',double[:]),\n ('ERP_low',double),\n ('ERP_high',double),\n ('ERP_2',double),\n\n # model time\n ('T',int32),\n ('Tr',int32), \n ('T_oap',int32),\n ('T_erp',int32),\n ('T_two_year',int32), \n ('ad_min',int32),\n ('ad_max',int32),\n ('iterator',int32[:,:]),\n\n # grids\n ('grid_a',double[:]),\n ('xi',double[:,:]),\n ('xi_w',double[:,:]),\n\n # precompute \n ('pension_female',double[:]),\n ('pension_male',double[:]), \n ('survival',double[:,:,:]),\n ('oap',double[:]),\n ('labor',double[:,:,:,:]),\n ('erp',double[:,:,:,:]),\n\n # simulation\n ('simM_init',double[:]) \n\n ]\n \n sollist = [ # (name, numba type), solution data\n\n # solution\n ('c',double[:,:,:,:,:,:]),\n ('m',double[:]),\n ('v',double[:,:,:,:,:,:]), \n\n # post decision\n ('avg_marg_u_plus',double[:,:,:,:,:,:]), \n ('v_plus_raw',double[:,:,:,:,:,:]) \n ] \n\n simlist = [ # (name, numba type), simulation data \n\n # solution\n ('c',double[:,:]), \n ('m',double[:,:]), \n ('a',double[:,:]),\n ('d',double[:,:]),\n\n # misc\n ('probs',double[:,:]), \n ('RA',int32[:]),\n ('euler',double[:,:]),\n ('GovS',double[:,:]),\n\n # booleans\n ('accuracy',boolean),\n ('tax',boolean), \n\n # setup\n ('choiceP',double[:,:,:]),\n ('alive',int32[:,:]), \n ('labor_pre',double[:,:,:]),\n ('labor_post',double[:,:,:]),\n ('states',int32[:,:])\n\n ]\n\n return parlist,sollist,simlist\n\n\ndef couple_lists():\n\n single_par = single_lists()[0]\n\n parlist = [ # (name,numba type), parameters, grids etc.\n\n # preference parameters\n ('pareto_w',double), \n ('phi_0_male',double),\n ('phi_0_female',double),\n ('phi_1',double),\n\n # uncertainty/variance parameters\n ('cov',double),\n\n # grids \n ('Nxi_men',int32),\n ('Nxi_women',int32), \n ('xi_corr',double[:,:]),\n ('w_corr',double[:]), \n\n # precompute\n ('inc_pens',double[:,:,:,:,:,:]),\n ('inc_mixed',double[:,:,:,:,:,:,:,:,:]),\n ('inc_joint',double[:,:,:,:,:,]), \n\n ]\n\n parlist = parlist + single_par \n\n sollist = [ # (name, numba type), solution data\n\n # solution\n ('c',double[:,:,:,:,:,:,:,:]),\n ('m',double[:]),\n ('v',double[:,:,:,:,:,:,:,:]), \n \n ] \n\n simlist = [ # (name, numba type), simulation data \n\n # solution\n ('c',double[:,:]), \n ('m',double[:,:]), \n ('a',double[:,:]),\n ('d',double[:,:,:]),\n\n # misc\n ('probs',double[:,:,:]), \n ('RA',int32[:,:]),\n ('euler',double[:,:]),\n ('GovS',double[:,:]),\n\n # booleans\n ('accuracy',boolean),\n ('tax',boolean), \n\n # setup\n ('choiceP',double[:,:,:]), \n ('alive',int32[:,:,:]), \n ('shocks_joint',double[:,:,:]), \n ('labor_pre',double[:,:,:]),\n ('labor_post',double[:,:,:]),\n ('labor_pre_joint',double[:,:]),\n ('labor_post_joint',double[:,:]),\n ('states',int32[:,:])\n\n ] \n\n return parlist,sollist,simlist\n\ndef TaxSystem(model):\n \"\"\" Tax system for either 2008 or 2014 \"\"\"\n\n # unpack\n par = model.par\n\n if model.year == 2008:\n\n par.IRA_tax = 0.4 # tax for IRA (kapitalpension)\n par.fradrag_to_oap = 1 # age difference to oap age\n par.fradrag = 0.0 # deduction for old workers (policy proposal)\n par.tau_upper = 0.59 # maximum tax rate (skatteloft)\n par.tau_LMC = 0.08 # labor market contribution (arbejdsmarkedsbidrag)\n par.WD = 0.4 # working deduction (beskæftigelsesfradrag)\n par.WD_upper = 12300/par.denom # maximum deduction possible (beskæftigelsesfradrag, maksimal)\n par.tau_c = 0.2554 # average county-specific rate (including 0.073 in church tax)\n par.y_low = 41000/par.denom # amount deductible from all income (personfradrag)\n par.y_low_m = 279800/par.denom # amount deductible from middle tax bracket (mellemskattegrænse)\n par.y_low_u = 335800/par.denom # amount deductible from top tax bracket (topskattegrænse)\n par.tau_h = 0.08 # health contribution tax (sundhedsbidrag)\n par.tau_l = 0.0548 # tax rate in lowest tax bracket (bundskat)\n par.tau_m = 0.06 # tax rate in middle tax bracket (mellemskat)\n par.tau_u = 0.15 # tax rate in upper tax bracket (topskat)\n par.tau_max = par.tau_l + par.tau_m + par.tau_u + par.tau_c + par.tau_h - par.tau_upper \n \n elif model.year == 2014:\n pass \n\ndef RetirementSystem(model): \n \"\"\" Retirement system for either 2008 or 2014 \"\"\" \n\n # unpack\n par = model.par\n\n if model.year == 2008:\n\n # ages\n par.oap_age = 65\n par.two_year = 62\n par.erp_age = 60\n \n # oap\n par.B = 61152/par.denom # base rate\n par.y_B = 463500/par.denom # maximum annual income before loss of OAP_B\n par.tau_B = 0.3 # marginal reduction in deduction regarding income\n par.D_B = 259700/par.denom # deduction regarding base value of OAP\n par.D_s = 179400/par.denom # maximum deduction in spousal income\n par.A_i = np.array([61560, 28752, 28752])/par.denom # maximum OAP_A\n par.y_i = np.array([153100, 210800, 306600])/par.denom # maximum income before loss of OAP_A\n par.tau_i = np.array([0.3, 0.3, 0.15]) # marginal reduction in OAP_A\n par.D_i = np.array([57300, 115000, 115000])/par.denom # maximum deduction regarding OAP_A\n \n # erp\n par.ERP_low = 12600/par.denom # deduction\n par.ERP_high = 166400/par.denom # maximum erp if two year rule is not satisfied\n par.ERP_2 = 182780/par.denom # erp with two year rule\n\n elif model.year == 2014:\n pass\n\ndef model_time(par):\n \"\"\" translate variables to model time and generate iterator for solving\"\"\"\n\n par.T = par.end_T - par.start_T + 1 # total time periods\n par.Tr = par.forced_T - par.start_T + 1 # total time periods to forced retirement\n par.T_oap = transitions.inv_age(par.oap_age,par)\n par.T_erp = transitions.inv_age(par.erp_age,par)\n par.T_two_year = transitions.inv_age(par.two_year,par)\n if par.couple:\n par.iterator = create_iterator([par.AD,par.ST,par.ST])\n par.ad_min = abs(min(par.AD))\n par.ad_max = max(par.AD) \n else:\n par.iterator = create_iterator([par.MA,par.ST]) \n par.ad_min = 0\n par.ad_max = 0\n\ndef create_iterator(lst):\n indices = 0\n num = len(lst)\n\n if num == 2:\n iterator = np.zeros((len(lst[0])*len(lst[1]),num),dtype=int) \n for x in lst[0]:\n for y in range(len(lst[1])):\n iterator[indices] = (x,y)\n indices += 1\n\n if num == 3:\n iterator = np.zeros((len(lst[0])*len(lst[1])*len(lst[2]),num),dtype=int) \n for x in lst[0]:\n for y in range(len(lst[1])):\n for z in range(len(lst[2])):\n iterator[indices] = (x,y,z)\n indices += 1\n \n return iterator \n\ndef grids(par):\n \"\"\" construct grids for states and shocks \"\"\"\n\n # a. a-grid (unequally spaced vector of length Na)\n par.grid_a = misc.nonlinspace(par.tol,par.a_max,par.Na,par.a_phi)\n \n # b. shocks (quadrature nodes and weights for GaussHermite)\n par.xi = np.nan*np.zeros((len(par.MA),par.Nxi))\n par.xi_w = np.nan*np.zeros((len(par.MA),par.Nxi))\n for ma in range(len(par.MA)):\n par.xi[ma],par.xi_w[ma] = funs.GaussHermite_lognorm(par.var[ma],par.Nxi)\n \n # # c. correlated shocks for joint labor income (only for couples)\n if par.couple: \n par.xi_corr,par.w_corr = funs.GH_lognorm_corr(par.var,par.cov,par.Nxi_men,par.Nxi_women) \n\ndef init_sim(par,sim):\n \"\"\" initialize simulation \"\"\"\n \n # initialize m and states\n np.random.seed(par.sim_seed)\n state_and_m(par,sim,perc_num=10)\n\n if par.couple:\n\n # setup\n init_sim_couple(par,sim)\n \n # draws\n Tr = min(par.simT,par.Tr) \n mu = -0.5*par.var \n Cov = np.array(([par.var[0], par.cov], [par.cov, par.var[1]])) \n shocks_joint = np.exp(np.random.multivariate_normal(mu,Cov,size=(par.simN,min(par.simT,par.Tr))))\n shocks_w = np.exp(np.random.normal(mu[0], np.sqrt(par.var[0]), size=(par.simN,Tr+par.ad_min)))\n shocks_h = np.exp(np.random.normal(mu[1], np.sqrt(par.var[1]), size=(par.simN,Tr+par.ad_min))) \n \n # income\n init_sim_labor_couple(par,sim,shocks_joint,shocks_w,shocks_h)\n \n else:\n\n # setup\n init_sim_single(par,sim)\n \n # draws\n shocks = np.nan*np.zeros((par.simN,min(par.simT,par.Tr),2))\n shocks[:,:,0] = np.exp(np.random.normal(-0.5*par.var[0], np.sqrt(par.var[0]), size=(par.simN,min(par.simT,par.Tr))))\n shocks[:,:,1] = np.exp(np.random.normal(-0.5*par.var[1], np.sqrt(par.var[1]), size=(par.simN,min(par.simT,par.Tr)))) \n \n # income\n init_sim_labor_single(par,sim,shocks) \n\ndef init_sim_single(par,sim):\n \"\"\" initialize simulation for single model \"\"\"\n\n # random draws \n sim.choiceP = np.random.rand(par.simN,par.simT,1) \n deadP = np.random.rand(par.simN,par.simT) \n\n # precompute\n MA = sim.states[:,0]\n MAx = np.unique(MA)\n ST = sim.states[:,1]\n STx = np.unique(ST)\n sim.alive = np.ones((par.simN,par.simT),dtype=int)\n alive = sim.alive \n for t in range(par.simT):\n\n # if they are dead, they stay dead\n if t > 0:\n alive[alive[:,t-1] == 0,t] = 0\n\n for ma in MAx:\n for st in STx:\n\n # indices\n idx = np.nonzero((MA==ma) & (ST==st))[0]\n\n # alive status (they are all alive first period)\n if t > 0:\n\n # alive status\n pi = transitions.survival_lookup_single(t,ma,st,par)\n dead = idx[pi < deadP[idx,t]]\n alive[dead,t] = 0\n\n@njit(parallel=True)\ndef init_sim_labor_single(par,sim,shocks):\n \"\"\" initialize income streams for single model \"\"\"\n\n # states\n MA = sim.states[:,0]\n MAx = np.unique(MA)\n ST = sim.states[:,1]\n STx = np.unique(ST)\n\n # initialize\n np.random.seed(par.sim_seed+1)\n sim.labor_pre = np.nan*np.zeros((par.simN,min(par.simT,par.Tr),2))\n sim.labor_post = np.nan*np.zeros((par.simN,min(par.simT,par.Tr),2)) \n labor_pre = sim.labor_pre\n labor_post = sim.labor_post\n\n for t in range(par.simT):\n for ma in MAx:\n for st in STx:\n\n # indices and shocks\n idx = np.nonzero((MA==ma) & (ST==st))[0]\n\n # labor income\n if t < par.Tr:\n labor_pre[idx,t,ma] = transitions.labor_pretax(t,ma,st,par)*shocks[idx,t,ma]\n labor_post[idx,t,ma] = transitions.posttax(t,par,d=1,inc=labor_pre[idx,t,ma],inc_s=np.zeros(len(idx)))\n\ndef init_sim_couple(par,sim):\n \"\"\" initialize simulation for couple model \"\"\"\n\n # random draws for simulation\n ad_min = par.ad_min\n ad_max = par.ad_max\n extend = ad_min + ad_max \n sim.choiceP = np.random.rand(par.simN,par.simT+extend,2)\n deadP = np.random.rand(par.simN,par.simT+extend,2) \n \n # precompute\n AD = sim.states[:,0]\n ST_h = sim.states[:,1]\n ST_w = sim.states[:,2]\n ADx = np.unique(AD)\n ST_hx = np.unique(ST_h)\n ST_wx = np.unique(ST_w)\n\n # 1. alive status\n sim.alive = np.ones((par.simN,par.simT+extend,2),dtype=int)\n alive_w = sim.alive[:,:,0]\n alive_h = sim.alive[:,:,1]\n alive_h[:,-ad_max:] = 0 # last period for men, which we never reach\n deadP_w = deadP[:,:,0]\n deadP_h = deadP[:,:,1]\n for t in range(par.simT):\n if t > 0:\n for ad in ADx: \n\n tw_idx = t+ad+par.ad_min\n th_idx = t+par.ad_min\n alive_w[alive_w[:,tw_idx-1] == 0, tw_idx] = 0\n alive_h[alive_h[:,th_idx-1] == 0, th_idx] = 0\n\n for st_h in ST_hx:\n for st_w in ST_wx:\n \n pi_h,pi_w = transitions.survival_lookup_couple(t,ad,st_h,st_w,par) \n idx = np.nonzero((AD==ad) & (ST_h==st_h) & (ST_w==st_w))[0] \n dead_w = idx[pi_w < deadP_w[idx,tw_idx]]\n dead_h = idx[pi_h < deadP_h[idx,th_idx]]\n alive_w[dead_w,tw_idx] = 0\n alive_h[dead_h,th_idx] = 0\n\n@njit(parallel=True)\ndef init_sim_labor_couple(par,sim,shocks_joint,shocks_w,shocks_h):\n \"\"\" initialize income streams in the couple model \"\"\"\n\n # states\n AD = sim.states[:,0]\n ST_h = sim.states[:,1]\n ST_w = sim.states[:,2]\n ADx = np.unique(AD)\n ST_hx = np.unique(ST_h)\n ST_wx = np.unique(ST_w)\n alive_w = sim.alive[:,:,0]\n alive_h = sim.alive[:,:,1] \n\n # time\n ad_min = par.ad_min\n ad_max = par.ad_max\n extend = ad_min + ad_max\n Tr = min(par.simT,par.Tr)\n \n # preallocate\n sim.labor_pre = np.nan*np.zeros((par.simN,Tr+extend,2))\n sim.labor_post = np.nan*np.zeros((par.simN,Tr+extend,2))\n sim.labor_pre_joint = np.nan*np.zeros((par.simN,Tr))\n sim.labor_post_joint = np.nan*np.zeros((par.simN,Tr)) \n labor_pre = sim.labor_pre\n labor_post = sim.labor_post\n labor_pre_joint = sim.labor_pre_joint\n labor_post_joint = sim.labor_post_joint\n\n # individual labor income (post tax can be used for singles, only pre tax can be used for couples, since we don't know pension of spouse) \n for t in range(Tr+ad_min):\n for ad in ADx:\n\n t_h = t\n t_w = t+ad\n th_idx = t+ad_min\n tw_idx = t+ad+ad_min\n\n # husband\n for st_h in np.unique(ST_h):\n if t < par.Tr: # not forced to retire\n idx_h = np.nonzero((alive_h[:,th_idx]==1) & (ST_h==st_h))[0]\n labor_pre[idx_h,th_idx,1] = transitions.labor_pretax(t_h,1,st_h,par)*shocks_h[idx_h,t]\n labor_post[idx_h,th_idx,1] = transitions.posttax(t_h,par,d=1,inc=labor_pre[idx_h,th_idx,1],inc_s=np.inf*np.ones(len(idx_h))) # set spouse of inc to infinity so no shared deduction\n\n # wife\n for st_w in np.unique(ST_w):\n if t+ad < par.Tr: # not forced to retire\n idx_w = np.nonzero((alive_w[:,tw_idx]==1) & (ST_w==st_w))[0]\n labor_pre[idx_w,tw_idx,0] = transitions.labor_pretax(t_w,0,st_w,par)*shocks_w[idx_w,t]\n labor_post[idx_w,tw_idx,0] = transitions.posttax(t_w,par,d=1,inc=labor_pre[idx_w,tw_idx,0],inc_s=np.inf*np.ones(len(idx_w)))\n\n # joint labor income (if they both work)\n for t in range(Tr):\n for ad in ADx:\n if t < par.Tr and t+ad < par.Tr:\n for st_h in ST_hx:\n for st_w in ST_wx:\n \n # indices\n th_idx = t+ad_min\n tw_idx = t+ad+ad_min \n idx = np.nonzero((alive_h[:,th_idx]==1) & (alive_w[:,tw_idx]==1) & (AD==ad) & (ST_h==st_h) & (ST_w==st_w))[0]\n \n # pre tax\n pre_h = transitions.labor_pretax(t,1,st_h,par)*shocks_joint[idx,t,1]\n pre_w = transitions.labor_pretax(t+ad,0,st_w,par)*shocks_joint[idx,t,0]\n labor_pre_joint[idx,t] = pre_h + pre_w\n \n # post tax\n post_h = transitions.posttax(t,par,d=1,inc=pre_h,inc_s=pre_w,d_s=1,t_s=t+ad)\n post_w = transitions.posttax(t+ad,par,d=1,inc=pre_w,inc_s=pre_h,d_s=1,t_s=t)\n labor_post_joint[idx,t] = post_h + post_w \n\n\ndef state_and_m(par,sim,perc_num=10):\n \"\"\" create states and initial wealth (m_init) by loading in relevant information from SASdata\"\"\"\n\n if par.couple:\n\n # set states\n data = pd.read_excel('SASdata/couple_formue.xlsx')\n states = par.iterator\n n_groups = (data['Frac'].to_numpy()*par.simN).astype(int)\n n_groups[-1] = par.simN-np.sum(n_groups[:-1]) # assure it sums to simN\n sim.states = np.transpose(np.vstack((np.repeat(states[:,0],n_groups),\n np.repeat(states[:,1],n_groups),\n np.repeat(states[:,2],n_groups))))\n \n else:\n \n # set states\n data = pd.read_excel('SASdata/single_formue.xlsx')\n states = par.iterator\n n_groups = (data['Frac'].to_numpy()*par.simN).astype(int)\n n_groups[-1] = par.simN-np.sum(n_groups[:-1]) # to assure it sums to simN\n sim.states = np.transpose(np.vstack((np.repeat(states[:,0],n_groups),\n np.repeat(states[:,1],n_groups))))\n \n # initial liquid wealth\n m_init = np.zeros(len(sim.states))\n idx = np.concatenate((np.zeros(1), np.cumsum(n_groups))).astype(int)\n percentiles = np.linspace(0,100,perc_num+1).astype(int)\n bins = data[list(percentiles)].to_numpy()\n for i in range(n_groups.size):\n m_init[idx[i]:idx[i+1]] = pc_sample(n_groups[i], percentiles, bins[i])\n par.simM_init = m_init\n\n # add private pension wealth to liquid wealth\n adjust_pension(par,sim)\n sim.m = np.nan*np.zeros((par.simN,par.simT))\n sim.m[:,0] = m_init\n states = sim.states\n\n if par.couple:\n for st_h in range(len(par.ST)):\n for st_w in range(len(par.ST)):\n idx = np.nonzero((states[:,1]==st_h) & (states[:,2]==st_w))[0]\n hs_h = transitions.state_translate(st_h,'high_skilled',par)\n hs_w = transitions.state_translate(st_w,'high_skilled',par)\n sim.m[idx,0] += (1-par.IRA_tax)*(par.pension_male[hs_h] + par.pension_female[hs_w])\n\n else:\n for ma in par.MA:\n for st in range(len(par.ST)):\n idx = np.nonzero((states[:,0]==ma) & (states[:,1]==st))[0]\n hs = transitions.state_translate(st,'high_skilled',par)\n if ma == 0:\n sim.m[idx,0] += (1-par.IRA_tax)*par.pension_female[hs]\n elif ma == 1:\n sim.m[idx,0] += (1-par.IRA_tax)*par.pension_male[hs]\n \ndef pc_sample(N,percentiles,bins):\n \"\"\" N samples from a dsitribution given its percentiles and bins (assumes equal spacing between percentiles)\"\"\"\n diff = np.diff(percentiles)\n assert np.allclose(diff[0],diff)\n n = int(N/diff.size)\n draws = np.random.uniform(low=bins[:-1], high=bins[1:], size=(n,diff.size)).ravel()\n return np.concatenate((draws, np.random.uniform(low=bins[0], high=bins[-1], size=(N-n*diff.size)))) # to assure we return N samples \n\ndef adjust_pension(par,sim):\n\n # unpack \n states = sim.states\n priv_pension = np.array([par.priv_pension_female, par.priv_pension_male])\n \n # adjust pension\n for ma in par.MA:\n\n # indices\n idx_low = np.nonzero((states[:,0]==ma) & (np.isin(states[:,1], (0,2))))[0]\n idx_high = np.nonzero((states[:,0]==ma) & (np.isin(states[:,1], (1,3))))[0] \n \n # adjust private pension\n share = len(idx_high) / (len(idx_low) + len(idx_high)) # share of high skilled\n pens_low = Xlow(par.g_adjust,share)*priv_pension[ma]\n pens_high = Xhigh(par.g_adjust,share)*priv_pension[ma]\n assert np.allclose(share*pens_high + (1-share)*pens_low, priv_pension[ma])\n\n # store\n if ma == 0:\n par.pension_female = np.array((pens_low, pens_high))\n\n elif ma == 1:\n par.pension_male = np.array((pens_low, pens_high))\n \ndef Xlow(g,share):\n return 1/(1+share*g)\n\ndef Xhigh(g,share):\n return (1+g)/(1+share*g) "
] |
[
[
"numpy.array",
"numpy.random.rand",
"numpy.zeros",
"numpy.random.seed",
"numpy.sum",
"pandas.read_excel",
"numpy.ones",
"numpy.nonzero",
"numpy.diff",
"numpy.allclose",
"numpy.random.uniform",
"numpy.sqrt",
"numpy.cumsum",
"numpy.repeat",
"numpy.linspace",
"numpy.unique",
"numpy.isin"
]
] |
Chichilele/pycaret
|
[
"c2910c50cbcb9c6b0126d7945b891fb6129d1d48"
] |
[
"pycaret/internal/tabular.py"
] |
[
"# Module: Classification\n# Author: Moez Ali <[email protected]>\n# License: MIT\n# Release: PyCaret 2.2\n# Last modified : 26/08/2020\n\nfrom enum import Enum, auto\nimport math\nfrom pycaret.internal.meta_estimators import (\n PowerTransformedTargetRegressor,\n get_estimator_from_meta_estimator,\n)\nfrom pycaret.internal.pipeline import (\n add_estimator_to_pipeline,\n get_pipeline_estimator_label,\n make_internal_pipeline,\n estimator_pipeline,\n merge_pipelines,\n Pipeline as InternalPipeline,\n)\nfrom pycaret.internal.utils import (\n color_df,\n normalize_custom_transformers,\n nullcontext,\n true_warm_start,\n can_early_stop,\n infer_ml_usecase,\n set_n_jobs,\n)\nimport pycaret.internal.patches.sklearn\nimport pycaret.internal.patches.yellowbrick\nfrom pycaret.internal.logging import get_logger\nfrom pycaret.internal.plots.yellowbrick import show_yellowbrick_plot\nfrom pycaret.internal.plots.helper import MatplotlibDefaultDPI\nfrom pycaret.internal.Display import Display, is_in_colab\nfrom pycaret.internal.distributions import *\nfrom pycaret.internal.validation import *\nfrom pycaret.internal.tunable import TunableMixin\nimport pycaret.containers.metrics.classification\nimport pycaret.containers.metrics.regression\nimport pycaret.containers.metrics.clustering\nimport pycaret.containers.metrics.anomaly\nimport pycaret.containers.models.classification\nimport pycaret.containers.models.regression\nimport pycaret.containers.models.clustering\nimport pycaret.containers.models.anomaly\nimport pycaret.internal.preprocess\nimport pandas as pd\nimport numpy as np\nimport os\nimport sys\nimport datetime\nimport time\nimport random\nimport gc\nimport multiprocessing\nfrom copy import deepcopy\nfrom sklearn.base import clone\nfrom sklearn.exceptions import NotFittedError\nfrom sklearn.compose import TransformedTargetRegressor\nfrom sklearn.preprocessing import LabelEncoder\nfrom typing import List, Tuple, Any, Union, Optional, Dict\nimport warnings\nfrom IPython.utils import io\nimport traceback\nfrom unittest.mock import patch\nimport plotly.express as px\nimport plotly.graph_objects as go\nimport scikitplot as skplt\n\nwarnings.filterwarnings(\"ignore\")\n\n_available_plots = {}\n\n\nclass MLUsecase(Enum):\n CLASSIFICATION = auto()\n REGRESSION = auto()\n CLUSTERING = auto()\n ANOMALY = auto()\n\n\ndef _is_unsupervised(ml_usecase: MLUsecase) -> bool:\n return ml_usecase == MLUsecase.CLUSTERING or ml_usecase == MLUsecase.ANOMALY\n\n\ndef setup(\n data: pd.DataFrame,\n target: str,\n ml_usecase: str,\n available_plots: dict,\n train_size: float = 0.7,\n test_data: Optional[pd.DataFrame] = None,\n preprocess: bool = True,\n imputation_type: str = \"simple\",\n iterative_imputation_iters: int = 5,\n categorical_features: Optional[List[str]] = None,\n categorical_imputation: str = \"mode\",\n categorical_iterative_imputer: Union[str, Any] = \"lightgbm\",\n ordinal_features: Optional[Dict[str, list]] = None,\n high_cardinality_features: Optional[List[str]] = None,\n high_cardinality_method: str = \"frequency\",\n numeric_features: Optional[List[str]] = None,\n numeric_imputation: str = \"mean\", # method 'zero' added in pycaret==2.1\n numeric_iterative_imputer: Union[str, Any] = \"lightgbm\",\n date_features: Optional[List[str]] = None,\n ignore_features: Optional[List[str]] = None,\n normalize: bool = False,\n normalize_method: str = \"zscore\",\n transformation: bool = False,\n transformation_method: str = \"yeo-johnson\",\n handle_unknown_categorical: bool = True,\n unknown_categorical_method: str = \"least_frequent\",\n pca: bool = False,\n pca_method: str = \"linear\",\n pca_components: Optional[float] = None,\n ignore_low_variance: bool = False,\n combine_rare_levels: bool = False,\n rare_level_threshold: float = 0.10,\n bin_numeric_features: Optional[List[str]] = None,\n remove_outliers: bool = False,\n outliers_threshold: float = 0.05,\n remove_multicollinearity: bool = False,\n multicollinearity_threshold: float = 0.9,\n remove_perfect_collinearity: bool = True,\n create_clusters: bool = False,\n cluster_iter: int = 20,\n polynomial_features: bool = False,\n polynomial_degree: int = 2,\n trigonometry_features: bool = False,\n polynomial_threshold: float = 0.1,\n group_features: Optional[List[str]] = None,\n group_names: Optional[List[str]] = None,\n feature_selection: bool = False,\n feature_selection_threshold: float = 0.8,\n feature_selection_method: str = \"classic\", # boruta algorithm added in pycaret==2.1\n feature_interaction: bool = False,\n feature_ratio: bool = False,\n interaction_threshold: float = 0.01,\n # classification specific\n fix_imbalance: bool = False,\n fix_imbalance_method: Optional[Any] = None,\n # regression specific\n transform_target=False,\n transform_target_method=\"box-cox\",\n data_split_shuffle: bool = True,\n data_split_stratify: Union[bool, List[str]] = False, # added in pycaret==2.2\n fold_strategy: Union[str, Any] = \"kfold\", # added in pycaret==2.2\n fold: int = 10, # added in pycaret==2.2\n fold_shuffle: bool = False,\n fold_groups: Optional[Union[str, pd.DataFrame]] = None,\n n_jobs: Optional[int] = -1,\n use_gpu: bool = False, # added in pycaret==2.1\n custom_pipeline: Union[\n Any, Tuple[str, Any], List[Any], List[Tuple[str, Any]]\n ] = None,\n html: bool = True,\n session_id: Optional[int] = None,\n log_experiment: bool = False,\n experiment_name: Optional[str] = None,\n log_plots: Union[bool, list] = False,\n log_profile: bool = False,\n log_data: bool = False,\n silent: bool = False,\n verbose: bool = True,\n profile: bool = False,\n profile_kwargs: Dict[str, Any] = None,\n display: Optional[Display] = None,\n):\n\n \"\"\"\n This function initializes the environment in pycaret and creates the transformation\n pipeline to prepare the data for modeling and deployment. setup() must called before\n executing any other function in pycaret. It takes two mandatory parameters:\n data and name of the target column.\n\n All other parameters are optional.\n\n \"\"\"\n\n function_params_str = \", \".join(\n [f\"{k}={v}\" for k, v in locals().items() if k != \"data\"]\n )\n\n global _available_plots\n\n _available_plots = available_plots\n\n warnings.filterwarnings(\"ignore\")\n\n from pycaret.utils import __version__\n\n ver = __version__\n\n # create logger\n global logger\n\n logger = get_logger()\n\n logger.info(\"PyCaret Supervised Module\")\n logger.info(f\"ML Usecase: {ml_usecase}\")\n logger.info(f\"version {ver}\")\n logger.info(\"Initializing setup()\")\n logger.info(f\"setup({function_params_str})\")\n\n # logging environment and libraries\n logger.info(\"Checking environment\")\n\n from platform import python_version, platform, python_build, machine\n\n logger.info(f\"python_version: {python_version()}\")\n logger.info(f\"python_build: {python_build()}\")\n logger.info(f\"machine: {machine()}\")\n logger.info(f\"platform: {platform()}\")\n\n try:\n import psutil\n\n logger.info(f\"Memory: {psutil.virtual_memory()}\")\n logger.info(f\"Physical Core: {psutil.cpu_count(logical=False)}\")\n logger.info(f\"Logical Core: {psutil.cpu_count(logical=True)}\")\n except:\n logger.warning(\n \"cannot find psutil installation. memory not traceable. Install psutil using pip to enable memory logging.\"\n )\n\n logger.info(\"Checking libraries\")\n\n try:\n from pandas import __version__\n\n logger.info(f\"pd=={__version__}\")\n except ImportError:\n logger.warning(\"pandas not found\")\n\n try:\n from numpy import __version__\n\n logger.info(f\"numpy=={__version__}\")\n except ImportError:\n logger.warning(\"numpy not found\")\n\n try:\n from sklearn import __version__\n\n logger.info(f\"sklearn=={__version__}\")\n except ImportError:\n logger.warning(\"sklearn not found\")\n\n try:\n from lightgbm import __version__\n\n logger.info(f\"lightgbm=={__version__}\")\n except ImportError:\n logger.warning(\"lightgbm not found\")\n\n try:\n from catboost import __version__\n\n logger.info(f\"catboost=={__version__}\")\n except ImportError:\n logger.warning(\"catboost not found\")\n\n try:\n from xgboost import __version__\n\n logger.info(f\"xgboost=={__version__}\")\n except ImportError:\n logger.warning(\"xgboost not found\")\n\n try:\n from mlflow.version import VERSION\n\n warnings.filterwarnings(\"ignore\")\n logger.info(f\"mlflow=={VERSION}\")\n except ImportError:\n logger.warning(\"mlflow not found\")\n\n # run_time\n runtime_start = time.time()\n\n logger.info(\"Checking Exceptions\")\n\n # checking data type\n if not isinstance(data, pd.DataFrame):\n raise TypeError(f\"data passed must be of type pandas.DataFrame\")\n if data.shape[0] == 0:\n raise ValueError(f\"data passed must be a positive dataframe\")\n\n # checking train size parameter\n if type(train_size) is not float:\n raise TypeError(\"train_size parameter only accepts float value.\")\n if train_size <= 0 or train_size > 1:\n raise ValueError(\"train_size parameter has to be positive and not above 1.\")\n\n possible_ml_usecases = [\"classification\", \"regression\", \"clustering\", \"anomaly\"]\n if ml_usecase not in possible_ml_usecases:\n raise ValueError(\n f\"ml_usecase must be one of {', '.join(possible_ml_usecases)}.\"\n )\n\n ml_usecase = MLUsecase[ml_usecase.upper()]\n\n # checking target parameter\n if not _is_unsupervised(ml_usecase) and target not in data.columns:\n raise ValueError(\n f\"Target parameter: {target} does not exist in the data provided.\"\n )\n\n # checking session_id\n if session_id is not None:\n if type(session_id) is not int:\n raise TypeError(\"session_id parameter must be an integer.\")\n\n # checking profile parameter\n if type(profile) is not bool:\n raise TypeError(\"profile parameter only accepts True or False.\")\n\n if profile_kwargs is not None:\n if type(profile_kwargs) is not dict:\n raise TypeError(\"profile_kwargs can only be a dict.\")\n else:\n profile_kwargs = {}\n\n # checking normalize parameter\n if type(normalize) is not bool:\n raise TypeError(\"normalize parameter only accepts True or False.\")\n\n # checking transformation parameter\n if type(transformation) is not bool:\n raise TypeError(\"transformation parameter only accepts True or False.\")\n\n all_cols = list(data.columns)\n if not _is_unsupervised(ml_usecase):\n all_cols.remove(target)\n\n # checking imputation type\n allowed_imputation_type = [\"simple\", \"iterative\"]\n if imputation_type not in allowed_imputation_type:\n raise ValueError(\n \"imputation_type parameter only accepts 'simple' or 'iterative'.\"\n )\n\n if type(iterative_imputation_iters) is not int or iterative_imputation_iters <= 0:\n raise TypeError(\n \"iterative_imputation_iters parameter must be an integer greater than 0.\"\n )\n\n # checking categorical imputation\n allowed_categorical_imputation = [\"constant\", \"mode\"]\n if categorical_imputation not in allowed_categorical_imputation:\n raise ValueError(\n f\"categorical_imputation param only accepts {', '.join(allowed_categorical_imputation)}.\"\n )\n\n # ordinal_features\n if ordinal_features is not None:\n if type(ordinal_features) is not dict:\n raise TypeError(\n \"ordinal_features must be of type dictionary with column name as key \"\n \"and ordered values as list.\"\n )\n\n # ordinal features check\n if ordinal_features is not None:\n ordinal_features = ordinal_features.copy()\n data_cols = data.columns.drop(target, errors=\"ignore\")\n ord_keys = ordinal_features.keys()\n\n for i in ord_keys:\n if i not in data_cols:\n raise ValueError(\n \"Column name passed as a key in ordinal_features param doesnt exist.\"\n )\n\n for k in ord_keys:\n if data[k].nunique() != len(ordinal_features[k]):\n raise ValueError(\n \"Levels passed in ordinal_features param doesnt match with levels in data.\"\n )\n\n for i in ord_keys:\n value_in_keys = ordinal_features.get(i)\n value_in_data = list(data[i].unique().astype(str))\n for j in value_in_keys:\n if str(j) not in value_in_data:\n raise ValueError(\n f\"Column name '{i}' doesn't contain any level named '{j}'.\"\n )\n\n # high_cardinality_features\n if high_cardinality_features is not None:\n if type(high_cardinality_features) is not list:\n raise TypeError(\n \"high_cardinality_features param only accepts name of columns as a list.\"\n )\n data_cols = data.columns.drop(target, errors=\"ignore\")\n for high_cardinality_feature in high_cardinality_features:\n if high_cardinality_feature not in data_cols:\n raise ValueError(\n f\"Item {high_cardinality_feature} in high_cardinality_features parameter is either target \"\n f\"column or doesn't exist in the dataset.\"\n )\n\n # stratify\n if data_split_stratify:\n if (\n type(data_split_stratify) is not list\n and type(data_split_stratify) is not bool\n ):\n raise TypeError(\n \"data_split_stratify parameter only accepts a bool or a list of strings.\"\n )\n\n if not data_split_shuffle:\n raise TypeError(\n \"data_split_stratify parameter requires data_split_shuffle to be set to True.\"\n )\n\n # high_cardinality_methods\n high_cardinality_allowed_methods = [\"frequency\", \"clustering\"]\n if high_cardinality_method not in high_cardinality_allowed_methods:\n raise ValueError(\n f\"high_cardinality_method parameter only accepts {', '.join(high_cardinality_allowed_methods)}.\"\n )\n\n # checking numeric imputation\n allowed_numeric_imputation = [\"mean\", \"median\", \"zero\"]\n if numeric_imputation not in allowed_numeric_imputation:\n raise ValueError(\n f\"numeric_imputation parameter only accepts {', '.join(allowed_numeric_imputation)}.\"\n )\n\n # checking normalize method\n allowed_normalize_method = [\"zscore\", \"minmax\", \"maxabs\", \"robust\"]\n if normalize_method not in allowed_normalize_method:\n raise ValueError(\n f\"normalize_method parameter only accepts {', '.join(allowed_normalize_method)}.\"\n )\n\n # checking transformation method\n allowed_transformation_method = [\"yeo-johnson\", \"quantile\"]\n if transformation_method not in allowed_transformation_method:\n raise ValueError(\n f\"transformation_method parameter only accepts {', '.join(allowed_transformation_method)}.\"\n )\n\n # handle unknown categorical\n if type(handle_unknown_categorical) is not bool:\n raise TypeError(\n \"handle_unknown_categorical parameter only accepts True or False.\"\n )\n\n # unknown categorical method\n unknown_categorical_method_available = [\"least_frequent\", \"most_frequent\"]\n\n if unknown_categorical_method not in unknown_categorical_method_available:\n raise TypeError(\n f\"unknown_categorical_method parameter only accepts {', '.join(unknown_categorical_method_available)}.\"\n )\n\n # check pca\n if type(pca) is not bool:\n raise TypeError(\"PCA parameter only accepts True or False.\")\n\n # pca method check\n allowed_pca_methods = [\"linear\", \"kernel\", \"incremental\"]\n if pca_method not in allowed_pca_methods:\n raise ValueError(\n f\"pca method parameter only accepts {', '.join(allowed_pca_methods)}.\"\n )\n\n # pca components check\n if pca is True:\n if pca_method != \"linear\":\n if pca_components is not None:\n if (type(pca_components)) is not int:\n raise TypeError(\n \"pca_components parameter must be integer when pca_method is not 'linear'.\"\n )\n\n # pca components check 2\n if pca is True:\n if pca_method != \"linear\":\n if pca_components is not None:\n if pca_components > len(data.columns) - 1:\n raise TypeError(\n \"pca_components parameter cannot be greater than original features space.\"\n )\n\n # pca components check 3\n if pca is True:\n if pca_method == \"linear\":\n if pca_components is not None:\n if type(pca_components) is not float:\n if pca_components > len(data.columns) - 1:\n raise TypeError(\n \"pca_components parameter cannot be greater than original features space or float between 0 - 1.\"\n )\n\n # check ignore_low_variance\n if type(ignore_low_variance) is not bool:\n raise TypeError(\"ignore_low_variance parameter only accepts True or False.\")\n\n # check ignore_low_variance\n if type(combine_rare_levels) is not bool:\n raise TypeError(\"combine_rare_levels parameter only accepts True or False.\")\n\n # check rare_level_threshold\n if (\n type(rare_level_threshold) is not float\n and rare_level_threshold < 0\n or rare_level_threshold > 1\n ):\n raise TypeError(\n \"rare_level_threshold parameter must be a float between 0 and 1.\"\n )\n\n # bin numeric features\n if bin_numeric_features is not None:\n if type(bin_numeric_features) is not list:\n raise TypeError(\"bin_numeric_features parameter must be a list.\")\n for bin_numeric_feature in bin_numeric_features:\n if type(bin_numeric_feature) is not str:\n raise TypeError(\"bin_numeric_features parameter item must be a string.\")\n if bin_numeric_feature not in all_cols:\n raise ValueError(\n f\"bin_numeric_feature: {bin_numeric_feature} is either target column or \"\n f\"does not exist in the dataset.\"\n )\n\n # remove_outliers\n if type(remove_outliers) is not bool:\n raise TypeError(\"remove_outliers parameter only accepts True or False.\")\n\n # outliers_threshold\n if type(outliers_threshold) is not float:\n raise TypeError(\"outliers_threshold must be a float between 0 and 1.\")\n\n # remove_multicollinearity\n if type(remove_multicollinearity) is not bool:\n raise TypeError(\n \"remove_multicollinearity parameter only accepts True or False.\"\n )\n\n # multicollinearity_threshold\n if type(multicollinearity_threshold) is not float:\n raise TypeError(\"multicollinearity_threshold must be a float between 0 and 1.\")\n\n # create_clusters\n if type(create_clusters) is not bool:\n raise TypeError(\"create_clusters parameter only accepts True or False.\")\n\n # cluster_iter\n if type(cluster_iter) is not int:\n raise TypeError(\"cluster_iter must be a integer greater than 1.\")\n\n # polynomial_features\n if type(polynomial_features) is not bool:\n raise TypeError(\"polynomial_features only accepts True or False.\")\n\n # polynomial_degree\n if type(polynomial_degree) is not int:\n raise TypeError(\"polynomial_degree must be an integer.\")\n\n # polynomial_features\n if type(trigonometry_features) is not bool:\n raise TypeError(\"trigonometry_features only accepts True or False.\")\n\n # polynomial threshold\n if type(polynomial_threshold) is not float:\n raise TypeError(\"polynomial_threshold must be a float between 0 and 1.\")\n\n # group features\n if group_features is not None:\n if type(group_features) is not list:\n raise TypeError(\"group_features must be of type list.\")\n\n if group_names is not None:\n if type(group_names) is not list:\n raise TypeError(\"group_names must be of type list.\")\n\n # cannot drop target\n if ignore_features is not None:\n if target in ignore_features:\n raise ValueError(\"cannot drop target column.\")\n\n # feature_selection\n if type(feature_selection) is not bool:\n raise TypeError(\"feature_selection only accepts True or False.\")\n\n # feature_selection_threshold\n if type(feature_selection_threshold) is not float:\n raise TypeError(\"feature_selection_threshold must be a float between 0 and 1.\")\n\n # feature_selection_method\n feature_selection_methods = [\"boruta\", \"classic\"]\n if feature_selection_method not in feature_selection_methods:\n raise TypeError(\n f\"feature_selection_method must be one of {', '.join(feature_selection_methods)}\"\n )\n\n # feature_interaction\n if type(feature_interaction) is not bool:\n raise TypeError(\"feature_interaction only accepts True or False.\")\n\n # feature_ratio\n if type(feature_ratio) is not bool:\n raise TypeError(\"feature_ratio only accepts True or False.\")\n\n # interaction_threshold\n if type(interaction_threshold) is not float:\n raise TypeError(\"interaction_threshold must be a float between 0 and 1.\")\n\n # categorical\n if categorical_features is not None:\n for i in categorical_features:\n if i not in all_cols:\n raise ValueError(\n \"Column type forced is either target column or doesn't exist in the dataset.\"\n )\n\n # numeric\n if numeric_features is not None:\n for i in numeric_features:\n if i not in all_cols:\n raise ValueError(\n \"Column type forced is either target column or doesn't exist in the dataset.\"\n )\n\n # date features\n if date_features is not None:\n for i in date_features:\n if i not in all_cols:\n raise ValueError(\n \"Column type forced is either target column or doesn't exist in the dataset.\"\n )\n\n # drop features\n if ignore_features is not None:\n for i in ignore_features:\n if i not in all_cols:\n raise ValueError(\n \"Feature ignored is either target column or doesn't exist in the dataset.\"\n )\n\n # log_experiment\n if type(log_experiment) is not bool:\n raise TypeError(\"log_experiment parameter only accepts True or False.\")\n\n # log_profile\n if type(log_profile) is not bool:\n raise TypeError(\"log_profile parameter only accepts True or False.\")\n\n # experiment_name\n if experiment_name is not None:\n if type(experiment_name) is not str:\n raise TypeError(\"experiment_name parameter must be str if not None.\")\n\n # silent\n if type(silent) is not bool:\n raise TypeError(\"silent parameter only accepts True or False.\")\n\n # remove_perfect_collinearity\n if type(remove_perfect_collinearity) is not bool:\n raise TypeError(\n \"remove_perfect_collinearity parameter only accepts True or False.\"\n )\n\n # html\n if type(html) is not bool:\n raise TypeError(\"html parameter only accepts True or False.\")\n\n # use_gpu\n if use_gpu != \"force\" and type(use_gpu) is not bool:\n raise TypeError(\"use_gpu parameter only accepts 'force', True or False.\")\n\n # data_split_shuffle\n if type(data_split_shuffle) is not bool:\n raise TypeError(\"data_split_shuffle parameter only accepts True or False.\")\n\n possible_fold_strategy = [\"kfold\", \"stratifiedkfold\", \"groupkfold\", \"timeseries\"]\n if not (\n fold_strategy in possible_fold_strategy\n or is_sklearn_cv_generator(fold_strategy)\n ):\n raise TypeError(\n f\"fold_strategy parameter must be either a scikit-learn compatible CV generator object or one of {', '.join(possible_fold_strategy)}.\"\n )\n\n if fold_strategy == \"groupkfold\" and (fold_groups is None or len(fold_groups) == 0):\n raise ValueError(\n \"'groupkfold' fold_strategy requires 'fold_groups' to be a non-empty array-like object.\"\n )\n\n if isinstance(fold_groups, str):\n if fold_groups not in all_cols:\n raise ValueError(\n f\"Column {fold_groups} used for fold_groups is not present in the dataset.\"\n )\n\n # checking fold parameter\n if type(fold) is not int:\n raise TypeError(\"fold parameter only accepts integer value.\")\n\n # fold_shuffle\n if type(fold_shuffle) is not bool:\n raise TypeError(\"fold_shuffle parameter only accepts True or False.\")\n\n # log_plots\n if isinstance(log_plots, list):\n for i in log_plots:\n if i not in _available_plots:\n raise ValueError(\n f\"Incorrect value for log_plots '{i}'. Possible values are: {', '.join(_available_plots.keys())}.\"\n )\n elif type(log_plots) is not bool:\n raise TypeError(\"log_plots parameter must be a bool or a list.\")\n\n # log_data\n if type(log_data) is not bool:\n raise TypeError(\"log_data parameter only accepts True or False.\")\n\n # fix_imbalance\n if type(fix_imbalance) is not bool:\n raise TypeError(\"fix_imbalance parameter only accepts True or False.\")\n\n # fix_imbalance_method\n if fix_imbalance:\n if fix_imbalance_method is not None:\n if hasattr(fix_imbalance_method, \"fit_resample\"):\n pass\n else:\n raise TypeError(\n \"fix_imbalance_method must contain resampler with fit_resample method.\"\n )\n\n # check transform_target\n if type(transform_target) is not bool:\n raise TypeError(\"transform_target parameter only accepts True or False.\")\n\n # transform_target_method\n allowed_transform_target_method = [\"box-cox\", \"yeo-johnson\"]\n if transform_target_method not in allowed_transform_target_method:\n raise ValueError(\n f\"transform_target_method param only accepts {', '.join(allowed_transform_target_method)}.\"\n )\n\n # pandas option\n pd.set_option(\"display.max_columns\", 500)\n pd.set_option(\"display.max_rows\", 500)\n\n # generate USI for mlflow tracking\n import secrets\n\n # declaring global variables to be accessed by other functions\n logger.info(\"Declaring global variables\")\n global _ml_usecase, USI, html_param, X, y, X_train, X_test, y_train, y_test, seed, prep_pipe, experiment__, fold_shuffle_param, n_jobs_param, _gpu_n_jobs_param, create_model_container, master_model_container, display_container, exp_name_log, logging_param, log_plots_param, fix_imbalance_param, fix_imbalance_method_param, transform_target_param, transform_target_method_param, data_before_preprocess, target_param, gpu_param, _all_models, _all_models_internal, _all_metrics, _internal_pipeline, stratify_param, fold_generator, fold_param, fold_groups_param, fold_groups_param_full, imputation_regressor, imputation_classifier, iterative_imputation_iters_param\n\n USI = secrets.token_hex(nbytes=2)\n logger.info(f\"USI: {USI}\")\n\n _ml_usecase = ml_usecase\n\n global pycaret_globals\n supervised_globals = {\n \"y\",\n \"X_train\",\n \"X_test\",\n \"y_train\",\n \"y_test\",\n }\n common_globals = {\n \"_ml_usecase\",\n \"_available_plots\",\n \"pycaret_globals\",\n \"USI\",\n \"html_param\",\n \"X\",\n \"seed\",\n \"prep_pipe\",\n \"experiment__\",\n \"n_jobs_param\",\n \"_gpu_n_jobs_param\",\n \"create_model_container\",\n \"master_model_container\",\n \"display_container\",\n \"exp_name_log\",\n \"logging_param\",\n \"log_plots_param\",\n \"transform_target_param\",\n \"transform_target_method_param\",\n \"data_before_preprocess\",\n \"target_param\",\n \"gpu_param\",\n \"_all_models\",\n \"_all_models_internal\",\n \"_all_metrics\",\n \"_internal_pipeline\",\n \"imputation_regressor\",\n \"imputation_classifier\",\n \"iterative_imputation_iters_param\",\n \"fold_shuffle_param\",\n \"fix_imbalance_param\",\n \"fix_imbalance_method_param\",\n \"stratify_param\",\n \"fold_generator\",\n \"fold_param\",\n \"fold_groups_param\",\n \"fold_groups_param_full\",\n }\n if not _is_unsupervised(_ml_usecase):\n pycaret_globals = common_globals.union(supervised_globals)\n else:\n pycaret_globals = common_globals\n\n logger.info(f\"pycaret_globals: {pycaret_globals}\")\n\n # create html_param\n html_param = html\n\n logger.info(\"Preparing display monitor\")\n\n if not display:\n # progress bar\n max_steps = 3\n\n progress_args = {\"max\": max_steps}\n timestampStr = datetime.datetime.now().strftime(\"%H:%M:%S\")\n monitor_rows = [\n [\"Initiated\", \". . . . . . . . . . . . . . . . . .\", timestampStr],\n [\"Status\", \". . . . . . . . . . . . . . . . . .\", \"Loading Dependencies\"],\n ]\n display = Display(\n verbose=verbose,\n html_param=html_param,\n progress_args=progress_args,\n monitor_rows=monitor_rows,\n )\n\n display.display_progress()\n display.display_monitor()\n\n logger.info(\"Importing libraries\")\n\n # general dependencies\n\n from sklearn.model_selection import train_test_split\n\n # setting sklearn config to print all parameters including default\n import sklearn\n\n sklearn.set_config(print_changed_only=False)\n\n # define highlight function for function grid to display\n def highlight_max(s):\n is_max = s == True\n return [\"background-color: lightgreen\" if v else \"\" for v in is_max]\n\n logger.info(\"Copying data for preprocessing\")\n\n # copy original data for pandas profiler\n data_before_preprocess = data.copy()\n\n # generate seed to be used globally\n seed = random.randint(150, 9000) if session_id is None else session_id\n\n np.random.seed(seed)\n\n _internal_pipeline = []\n\n \"\"\"\n preprocessing starts here\n \"\"\"\n\n display.update_monitor(1, \"Preparing Data for Modeling\")\n display.display_monitor()\n\n # define parameters for preprocessor\n\n logger.info(\"Declaring preprocessing parameters\")\n\n # categorical features\n cat_features_pass = categorical_features or []\n\n # numeric features\n numeric_features_pass = numeric_features or []\n\n # drop features\n ignore_features_pass = ignore_features or []\n\n # date features\n date_features_pass = date_features or []\n\n # categorical imputation strategy\n cat_dict = {\"constant\": \"not_available\", \"mode\": \"most frequent\"}\n categorical_imputation_pass = cat_dict[categorical_imputation]\n\n # transformation method strategy\n trans_dict = {\"yeo-johnson\": \"yj\", \"quantile\": \"quantile\"}\n trans_method_pass = trans_dict[transformation_method]\n\n # pass method\n pca_dict = {\n \"linear\": \"pca_liner\",\n \"kernel\": \"pca_kernal\",\n \"incremental\": \"incremental\",\n \"pls\": \"pls\",\n }\n pca_method_pass = pca_dict[pca_method]\n\n # pca components\n if pca is True:\n if pca_components is None:\n if pca_method == \"linear\":\n pca_components_pass = 0.99\n else:\n pca_components_pass = int((len(data.columns) - 1) * 0.5)\n\n else:\n pca_components_pass = pca_components\n\n else:\n pca_components_pass = 0.99\n\n apply_binning_pass = False if bin_numeric_features is None else True\n features_to_bin_pass = bin_numeric_features or []\n\n # trignometry\n trigonometry_features_pass = [\"sin\", \"cos\", \"tan\"] if trigonometry_features else []\n\n # group features\n # =============#\n\n # apply grouping\n apply_grouping_pass = True if group_features is not None else False\n\n # group features listing\n if apply_grouping_pass is True:\n\n if type(group_features[0]) is str:\n group_features_pass = []\n group_features_pass.append(group_features)\n else:\n group_features_pass = group_features\n\n else:\n\n group_features_pass = [[]]\n\n # group names\n if apply_grouping_pass is True:\n\n if (group_names is None) or (len(group_names) != len(group_features_pass)):\n group_names_pass = list(np.arange(len(group_features_pass)))\n group_names_pass = [f\"group_{i}\" for i in group_names_pass]\n\n else:\n group_names_pass = group_names\n\n else:\n group_names_pass = []\n\n # feature interactions\n\n apply_feature_interactions_pass = (\n True if feature_interaction or feature_ratio else False\n )\n\n interactions_to_apply_pass = []\n\n if feature_interaction:\n interactions_to_apply_pass.append(\"multiply\")\n\n if feature_ratio:\n interactions_to_apply_pass.append(\"divide\")\n\n # unknown categorical\n unkn_dict = {\"least_frequent\": \"least frequent\", \"most_frequent\": \"most frequent\"}\n unknown_categorical_method_pass = unkn_dict[unknown_categorical_method]\n\n # ordinal_features\n apply_ordinal_encoding_pass = True if ordinal_features is not None else False\n\n ordinal_columns_and_categories_pass = (\n ordinal_features if apply_ordinal_encoding_pass else {}\n )\n\n apply_cardinality_reduction_pass = (\n True if high_cardinality_features is not None else False\n )\n\n hi_card_dict = {\"frequency\": \"count\", \"clustering\": \"cluster\"}\n cardinal_method_pass = hi_card_dict[high_cardinality_method]\n\n cardinal_features_pass = (\n high_cardinality_features if apply_cardinality_reduction_pass else []\n )\n\n display_dtypes_pass = False if silent else True\n\n # transform target method\n transform_target_param = transform_target\n transform_target_method_param = transform_target_method\n\n # create n_jobs_param\n n_jobs_param = n_jobs\n\n cuml_version = None\n if use_gpu:\n try:\n from cuml import __version__\n\n cuml_version = __version__\n logger.info(f\"cuml=={cuml_version}\")\n\n cuml_version = cuml_version.split(\".\")\n cuml_version = (int(cuml_version[0]), int(cuml_version[1]))\n except:\n logger.warning(f\"cuML not found\")\n\n if cuml_version is None or not cuml_version >= (0, 15):\n message = f\"cuML is outdated or not found. Required version is >=0.15, got {__version__}\"\n if use_gpu == \"force\":\n raise ImportError(message)\n else:\n logger.warning(message)\n\n # create _gpu_n_jobs_param\n _gpu_n_jobs_param = n_jobs if not use_gpu else 1\n\n # create gpu_param var\n gpu_param = use_gpu\n\n iterative_imputation_iters_param = iterative_imputation_iters\n\n # creating variables to be used later in the function\n train_data = data_before_preprocess.copy()\n if _is_unsupervised(_ml_usecase):\n target = \"UNSUPERVISED_DUMMY_TARGET\"\n train_data[target] = 2\n # just to add diversified values to target\n train_data[target][0:3] = 3\n X_before_preprocess = train_data.drop(target, axis=1)\n y_before_preprocess = train_data[target]\n\n imputation_regressor = numeric_iterative_imputer\n imputation_classifier = categorical_iterative_imputer\n imputation_regressor_name = \"Bayesian Ridge\" # todo change\n imputation_classifier_name = \"Random Forest Classifier\"\n\n if imputation_type == \"iterative\":\n logger.info(\"Setting up iterative imputation\")\n\n iterative_imputer_models_globals = globals().copy()\n iterative_imputer_models_globals[\"y_train\"] = y_before_preprocess\n iterative_imputer_models_globals[\"X_train\"] = X_before_preprocess\n iterative_imputer_classification_models = {\n k: v\n for k, v in pycaret.containers.models.classification.get_all_model_containers(\n iterative_imputer_models_globals, raise_errors=True\n ).items()\n if not v.is_special\n }\n iterative_imputer_regression_models = {\n k: v\n for k, v in pycaret.containers.models.regression.get_all_model_containers(\n iterative_imputer_models_globals, raise_errors=True\n ).items()\n if not v.is_special\n }\n\n if not (\n (\n isinstance(imputation_regressor, str)\n and imputation_regressor in iterative_imputer_regression_models\n )\n or hasattr(imputation_regressor, \"predict\")\n ):\n raise ValueError(\n f\"numeric_iterative_imputer param must be either a scikit-learn estimator or a string - one of {', '.join(iterative_imputer_regression_models.keys())}.\"\n )\n\n if not (\n (\n isinstance(imputation_classifier, str)\n and imputation_classifier in iterative_imputer_classification_models\n )\n or hasattr(imputation_classifier, \"predict\")\n ):\n raise ValueError(\n f\"categorical_iterative_imputer param must be either a scikit-learn estimator or a string - one of {', '.join(iterative_imputer_classification_models.keys())}.\"\n )\n\n if isinstance(imputation_regressor, str):\n imputation_regressor = iterative_imputer_regression_models[\n imputation_regressor\n ]\n imputation_regressor_name = imputation_regressor.name\n imputation_regressor = imputation_regressor.class_def(\n **imputation_regressor.args\n )\n else:\n imputation_regressor_name = type(imputation_regressor).__name__\n\n if isinstance(imputation_classifier, str):\n imputation_classifier = iterative_imputer_classification_models[\n imputation_classifier\n ]\n imputation_classifier_name = imputation_classifier.name\n imputation_classifier = imputation_classifier.class_def(\n **imputation_classifier.args\n )\n else:\n imputation_classifier_name = type(imputation_classifier).__name__\n\n logger.info(\"Creating preprocessing pipeline\")\n\n prep_pipe = pycaret.internal.preprocess.Preprocess_Path_One(\n train_data=train_data,\n ml_usecase=\"classification\"\n if _ml_usecase == MLUsecase.CLASSIFICATION\n else \"regression\",\n imputation_type=imputation_type,\n target_variable=target,\n imputation_regressor=imputation_regressor,\n imputation_classifier=imputation_classifier,\n imputation_max_iter=iterative_imputation_iters_param,\n categorical_features=cat_features_pass,\n apply_ordinal_encoding=apply_ordinal_encoding_pass,\n ordinal_columns_and_categories=ordinal_columns_and_categories_pass,\n apply_cardinality_reduction=apply_cardinality_reduction_pass,\n cardinal_method=cardinal_method_pass,\n cardinal_features=cardinal_features_pass,\n numerical_features=numeric_features_pass,\n time_features=date_features_pass,\n features_todrop=ignore_features_pass,\n numeric_imputation_strategy=numeric_imputation,\n categorical_imputation_strategy=categorical_imputation_pass,\n scale_data=normalize,\n scaling_method=normalize_method,\n Power_transform_data=transformation,\n Power_transform_method=trans_method_pass,\n apply_untrained_levels_treatment=handle_unknown_categorical,\n untrained_levels_treatment_method=unknown_categorical_method_pass,\n apply_pca=pca,\n pca_method=pca_method_pass,\n pca_variance_retained_or_number_of_components=pca_components_pass,\n apply_zero_nearZero_variance=ignore_low_variance,\n club_rare_levels=combine_rare_levels,\n rara_level_threshold_percentage=rare_level_threshold,\n apply_binning=apply_binning_pass,\n features_to_binn=features_to_bin_pass,\n remove_outliers=remove_outliers,\n outlier_contamination_percentage=outliers_threshold,\n outlier_methods=[\"pca\"],\n remove_multicollinearity=remove_multicollinearity,\n maximum_correlation_between_features=multicollinearity_threshold,\n remove_perfect_collinearity=remove_perfect_collinearity,\n cluster_entire_data=create_clusters,\n range_of_clusters_to_try=cluster_iter,\n apply_polynomial_trigonometry_features=polynomial_features,\n max_polynomial=polynomial_degree,\n trigonometry_calculations=trigonometry_features_pass,\n top_poly_trig_features_to_select_percentage=polynomial_threshold,\n apply_grouping=apply_grouping_pass,\n features_to_group_ListofList=group_features_pass,\n group_name=group_names_pass,\n apply_feature_selection=feature_selection,\n feature_selection_top_features_percentage=feature_selection_threshold,\n feature_selection_method=feature_selection_method,\n apply_feature_interactions=apply_feature_interactions_pass,\n feature_interactions_to_apply=interactions_to_apply_pass,\n feature_interactions_top_features_to_select_percentage=interaction_threshold,\n display_types=display_dtypes_pass, # this is for inferred input box\n random_state=seed,\n )\n\n dtypes = prep_pipe.named_steps[\"dtypes\"]\n\n display.move_progress()\n logger.info(\"Preprocessing pipeline created successfully\")\n\n try:\n res_type = [\"quit\", \"Quit\", \"exit\", \"EXIT\", \"q\", \"Q\", \"e\", \"E\", \"QUIT\", \"Exit\"]\n res = dtypes.response\n\n if res in res_type:\n sys.exit(\n \"(Process Exit): setup has been interupted with user command 'quit'. setup must rerun.\"\n )\n\n except:\n logger.error(\n \"(Process Exit): setup has been interupted with user command 'quit'. setup must rerun.\"\n )\n\n if not preprocess:\n prep_pipe.steps = prep_pipe.steps[:1]\n\n \"\"\"\n preprocessing ends here\n \"\"\"\n\n # reset pandas option\n pd.reset_option(\"display.max_rows\")\n pd.reset_option(\"display.max_columns\")\n\n logger.info(\"Creating global containers\")\n\n # create an empty list for pickling later.\n experiment__ = []\n\n # CV params\n fold_param = fold\n fold_groups_param = None\n fold_groups_param_full = None\n if fold_groups is not None:\n if isinstance(fold_groups, str):\n fold_groups_param = X_before_preprocess[fold_groups]\n else:\n fold_groups_param = fold_groups\n if pd.isnull(fold_groups_param).any():\n raise ValueError(f\"fold_groups cannot contain NaNs.\")\n fold_shuffle_param = fold_shuffle\n\n from sklearn.model_selection import (\n StratifiedKFold,\n KFold,\n GroupKFold,\n TimeSeriesSplit,\n )\n\n fold_seed = seed if fold_shuffle_param else None\n if fold_strategy == \"kfold\":\n fold_generator = KFold(\n fold_param, random_state=fold_seed, shuffle=fold_shuffle_param\n )\n elif fold_strategy == \"stratifiedkfold\":\n fold_generator = StratifiedKFold(\n fold_param, random_state=fold_seed, shuffle=fold_shuffle_param\n )\n elif fold_strategy == \"groupkfold\":\n fold_generator = GroupKFold(fold_param)\n elif fold_strategy == \"timeseries\":\n fold_generator = TimeSeriesSplit(fold_param)\n else:\n fold_generator = fold_strategy\n\n # create create_model_container\n create_model_container = []\n\n # create master_model_container\n master_model_container = []\n\n # create display container\n display_container = []\n\n # create logging parameter\n logging_param = log_experiment\n\n # create exp_name_log param incase logging is False\n exp_name_log = \"no_logging\"\n\n # create an empty log_plots_param\n if not log_plots:\n log_plots_param = False\n else:\n log_plots_param = log_plots\n\n # add custom transformers to prep pipe\n if custom_pipeline:\n custom_steps = normalize_custom_transformers(custom_pipeline)\n _internal_pipeline.extend(custom_steps)\n\n # create a fix_imbalance_param and fix_imbalance_method_param\n fix_imbalance_param = fix_imbalance and preprocess\n fix_imbalance_method_param = fix_imbalance_method\n\n if fix_imbalance_method_param is None:\n fix_imbalance_model_name = \"SMOTE\"\n\n if fix_imbalance_param:\n if fix_imbalance_method_param is None:\n import six\n\n sys.modules[\"sklearn.externals.six\"] = six\n from imblearn.over_sampling import SMOTE\n\n fix_imbalance_resampler = SMOTE(random_state=seed)\n else:\n fix_imbalance_model_name = str(fix_imbalance_method_param).split(\"(\")[0]\n fix_imbalance_resampler = fix_imbalance_method_param\n _internal_pipeline.append((\"fix_imbalance\", fix_imbalance_resampler))\n\n for x in _internal_pipeline:\n if x[0] in prep_pipe.named_steps:\n raise ValueError(f\"Step named {x[0]} already present in pipeline.\")\n\n _internal_pipeline = make_internal_pipeline(_internal_pipeline)\n\n logger.info(f\"Internal pipeline: {_internal_pipeline}\")\n\n # create target_param var\n target_param = target\n\n # create stratify_param var\n stratify_param = data_split_stratify\n\n display.move_progress()\n\n display.update_monitor(1, \"Preprocessing Data\")\n display.display_monitor()\n\n if not _is_unsupervised(_ml_usecase):\n _stratify_columns = _get_columns_to_stratify_by(\n X_before_preprocess, y_before_preprocess, stratify_param, target\n )\n if test_data is None:\n X_train, X_test, y_train, y_test = train_test_split(\n X_before_preprocess,\n y_before_preprocess,\n test_size=1 - train_size,\n stratify=_stratify_columns,\n random_state=seed,\n shuffle=data_split_shuffle,\n )\n train_data = pd.concat([X_train, y_train], axis=1)\n test_data = pd.concat([X_test, y_test], axis=1)\n\n train_data = prep_pipe.fit_transform(train_data)\n # workaround to also transform target\n dtypes.final_training_columns.append(target)\n test_data = prep_pipe.transform(test_data)\n\n X_train = train_data.drop(target, axis=1)\n y_train = train_data[target]\n\n X_test = test_data.drop(target, axis=1)\n y_test = test_data[target]\n\n if fold_groups_param is not None:\n fold_groups_param_full = fold_groups_param.copy()\n fold_groups_param = fold_groups_param[\n fold_groups_param.index.isin(X_train.index)\n ]\n\n display.move_progress()\n if not _is_unsupervised(_ml_usecase):\n _internal_pipeline.fit(train_data.drop(target, axis=1), train_data[target])\n data = prep_pipe.transform(data_before_preprocess.copy())\n X = data.drop(target, axis=1)\n y = data[target]\n else:\n X = prep_pipe.fit_transform(train_data).drop(target, axis=1)\n X_train = X\n\n # we do just the fitting so that it will be fitted when saved/deployed,\n # but we don't want to modify the data\n _internal_pipeline.fit(X, y=y if not _is_unsupervised(_ml_usecase) else None)\n\n prep_pipe.steps = prep_pipe.steps + [\n (step[0], deepcopy(step[1]))\n for step in _internal_pipeline.steps\n if hasattr(step[1], \"transform\")\n ]\n\n try:\n dtypes.final_training_columns.remove(target)\n except ValueError:\n pass\n\n # determining target type\n if _is_multiclass():\n target_type = \"Multiclass\"\n else:\n target_type = \"Binary\"\n\n if _ml_usecase == MLUsecase.CLASSIFICATION:\n _all_models = {\n k: v\n for k, v in pycaret.containers.models.classification.get_all_model_containers(\n globals(), raise_errors=True\n ).items()\n if not v.is_special\n }\n _all_models_internal = pycaret.containers.models.classification.get_all_model_containers(\n globals(), raise_errors=True\n )\n _all_metrics = pycaret.containers.metrics.classification.get_all_metric_containers(\n globals(), raise_errors=True\n )\n elif _ml_usecase == MLUsecase.REGRESSION:\n _all_models = {\n k: v\n for k, v in pycaret.containers.models.regression.get_all_model_containers(\n globals(), raise_errors=True\n ).items()\n if not v.is_special\n }\n _all_models_internal = pycaret.containers.models.regression.get_all_model_containers(\n globals(), raise_errors=True\n )\n _all_metrics = pycaret.containers.metrics.regression.get_all_metric_containers(\n globals(), raise_errors=True\n )\n elif _ml_usecase == MLUsecase.CLUSTERING:\n _all_models = {\n k: v\n for k, v in pycaret.containers.models.clustering.get_all_model_containers(\n globals(), raise_errors=True\n ).items()\n if not v.is_special\n }\n _all_models_internal = pycaret.containers.models.clustering.get_all_model_containers(\n globals(), raise_errors=True\n )\n _all_metrics = pycaret.containers.metrics.clustering.get_all_metric_containers(\n globals(), raise_errors=True\n )\n elif _ml_usecase == MLUsecase.ANOMALY:\n _all_models = {\n k: v\n for k, v in pycaret.containers.models.anomaly.get_all_model_containers(\n globals(), raise_errors=True\n ).items()\n if not v.is_special\n }\n _all_models_internal = pycaret.containers.models.anomaly.get_all_model_containers(\n globals(), raise_errors=True\n )\n _all_metrics = pycaret.containers.metrics.anomaly.get_all_metric_containers(\n globals(), raise_errors=True\n )\n\n \"\"\"\n Final display Starts\n \"\"\"\n logger.info(\"Creating grid variables\")\n\n if hasattr(dtypes, \"replacement\"):\n label_encoded = dtypes.replacement\n label_encoded = (\n str(label_encoded).replace(\"'\", \"\").replace(\"{\", \"\").replace(\"}\", \"\")\n )\n\n else:\n label_encoded = \"None\"\n\n # generate values for grid show\n missing_values = data_before_preprocess.isna().sum().sum()\n missing_flag = True if missing_values > 0 else False\n\n normalize_grid = normalize_method if normalize else \"None\"\n\n transformation_grid = transformation_method if transformation else \"None\"\n\n pca_method_grid = pca_method if pca else \"None\"\n\n pca_components_grid = pca_components_pass if pca else \"None\"\n\n rare_level_threshold_grid = rare_level_threshold if combine_rare_levels else \"None\"\n\n numeric_bin_grid = False if bin_numeric_features is None else True\n\n outliers_threshold_grid = outliers_threshold if remove_outliers else None\n\n multicollinearity_threshold_grid = (\n multicollinearity_threshold if remove_multicollinearity else None\n )\n\n cluster_iter_grid = cluster_iter if create_clusters else None\n\n polynomial_degree_grid = polynomial_degree if polynomial_features else None\n\n polynomial_threshold_grid = (\n polynomial_threshold if polynomial_features or trigonometry_features else None\n )\n\n feature_selection_threshold_grid = (\n feature_selection_threshold if feature_selection else None\n )\n\n interaction_threshold_grid = (\n interaction_threshold if feature_interaction or feature_ratio else None\n )\n\n ordinal_features_grid = False if ordinal_features is None else True\n\n unknown_categorical_method_grid = (\n unknown_categorical_method if handle_unknown_categorical else None\n )\n\n group_features_grid = False if group_features is None else True\n\n high_cardinality_features_grid = (\n False if high_cardinality_features is None else True\n )\n\n high_cardinality_method_grid = (\n high_cardinality_method if high_cardinality_features_grid else None\n )\n\n learned_types = dtypes.learned_dtypes\n learned_types.drop(target, inplace=True)\n\n float_type = 0\n cat_type = 0\n\n for i in dtypes.learned_dtypes:\n if \"float\" in str(i):\n float_type += 1\n elif \"object\" in str(i):\n cat_type += 1\n elif \"int\" in str(i):\n float_type += 1\n\n if profile:\n print(\"Setup Succesfully Completed! Loading Profile Now... Please Wait!\")\n else:\n if verbose:\n print(\"Setup Succesfully Completed!\")\n\n exp_name_dict = {\n MLUsecase.CLASSIFICATION: \"clf-default-name\",\n MLUsecase.REGRESSION: \"reg-default-name\",\n MLUsecase.CLUSTERING: \"cluster-default-name\",\n MLUsecase.ANOMALY: \"anomaly-default-name\",\n }\n if experiment_name is None:\n exp_name_ = exp_name_dict[_ml_usecase]\n else:\n exp_name_ = experiment_name\n\n URI = secrets.token_hex(nbytes=4)\n exp_name_log = exp_name_\n\n functions = pd.DataFrame(\n [[\"session_id\", seed],]\n + ([[\"Target\", target]] if not _is_unsupervised(_ml_usecase) else [])\n + (\n [[\"Target Type\", target_type], [\"Label Encoded\", label_encoded],]\n if _ml_usecase == MLUsecase.CLASSIFICATION\n else []\n )\n + [\n [\"Original Data\", data_before_preprocess.shape],\n [\"Missing Values\", missing_flag],\n [\"Numeric Features\", str(float_type)],\n [\"Categorical Features\", str(cat_type)],\n ]\n + (\n [\n [\"Ordinal Features\", ordinal_features_grid],\n [\"High Cardinality Features\", high_cardinality_features_grid],\n [\"High Cardinality Method\", high_cardinality_method_grid],\n ]\n if preprocess\n else []\n )\n + (\n [\n [\"Transformed Train Set\", X_train.shape],\n [\"Transformed Test Set\", X_test.shape],\n [\"Shuffle Train-Test\", str(data_split_shuffle)],\n [\"Stratify Train-Test\", str(data_split_stratify)],\n [\"Fold Generator\", type(fold_generator).__name__],\n [\"Fold Number\", fold_param],\n ]\n if not _is_unsupervised(_ml_usecase)\n else [[\"Transformed Data\", X.shape]]\n )\n + [\n [\"CPU Jobs\", n_jobs_param],\n [\"Use GPU\", gpu_param],\n [\"Log Experiment\", logging_param],\n [\"Experiment Name\", exp_name_],\n [\"USI\", USI],\n ]\n + (\n [\n [\"Imputation Type\", imputation_type],\n [\n \"Iterative Imputation Iteration\",\n iterative_imputation_iters_param\n if imputation_type == \"iterative\"\n else \"None\",\n ],\n [\"Numeric Imputer\", numeric_imputation],\n [\n \"Iterative Imputation Numeric Model\",\n imputation_regressor_name\n if imputation_type == \"iterative\"\n else \"None\",\n ],\n [\"Categorical Imputer\", categorical_imputation],\n [\n \"Iterative Imputation Categorical Model\",\n imputation_classifier_name\n if imputation_type == \"iterative\"\n else \"None\",\n ],\n [\"Unknown Categoricals Handling\", unknown_categorical_method_grid],\n [\"Normalize\", normalize],\n [\"Normalize Method\", normalize_grid],\n [\"Transformation\", transformation],\n [\"Transformation Method\", transformation_grid],\n [\"PCA\", pca],\n [\"PCA Method\", pca_method_grid],\n [\"PCA Components\", pca_components_grid],\n [\"Ignore Low Variance\", ignore_low_variance],\n [\"Combine Rare Levels\", combine_rare_levels],\n [\"Rare Level Threshold\", rare_level_threshold_grid],\n [\"Numeric Binning\", numeric_bin_grid],\n [\"Remove Outliers\", remove_outliers],\n [\"Outliers Threshold\", outliers_threshold_grid],\n [\"Remove Multicollinearity\", remove_multicollinearity],\n [\"Multicollinearity Threshold\", multicollinearity_threshold_grid],\n [\"Clustering\", create_clusters],\n [\"Clustering Iteration\", cluster_iter_grid],\n [\"Polynomial Features\", polynomial_features],\n [\"Polynomial Degree\", polynomial_degree_grid],\n [\"Trignometry Features\", trigonometry_features],\n [\"Polynomial Threshold\", polynomial_threshold_grid],\n [\"Group Features\", group_features_grid],\n [\"Feature Selection\", feature_selection],\n [\"Feature Selection Method\", feature_selection_method],\n [\"Features Selection Threshold\", feature_selection_threshold_grid],\n [\"Feature Interaction\", feature_interaction],\n [\"Feature Ratio\", feature_ratio],\n [\"Interaction Threshold\", interaction_threshold_grid],\n ]\n if preprocess\n else []\n )\n + (\n [\n [\"Fix Imbalance\", fix_imbalance_param],\n [\"Fix Imbalance Method\", fix_imbalance_model_name],\n ]\n if _ml_usecase == MLUsecase.CLASSIFICATION\n else []\n )\n + (\n [\n [\"Transform Target\", transform_target_param],\n [\"Transform Target Method\", transform_target_method_param],\n ]\n if _ml_usecase == MLUsecase.REGRESSION\n else []\n ),\n columns=[\"Description\", \"Value\"],\n )\n functions_ = functions.style.apply(highlight_max)\n\n display_container.append(functions_)\n\n display.display(functions_, clear=True)\n\n if profile:\n try:\n import pandas_profiling\n\n pf = pandas_profiling.ProfileReport(\n data_before_preprocess, **profile_kwargs\n )\n display.display(pf, clear=True)\n except:\n print(\n \"Data Profiler Failed. No output to show, please continue with Modeling.\"\n )\n logger.error(\n \"Data Profiler Failed. No output to show, please continue with Modeling.\"\n )\n\n \"\"\"\n Final display Ends\n \"\"\"\n\n # log into experiment\n experiment__.append((\"Setup Config\", functions))\n if not _is_unsupervised(_ml_usecase):\n experiment__.append((\"X_training Set\", X_train))\n experiment__.append((\"y_training Set\", y_train))\n experiment__.append((\"X_test Set\", X_test))\n experiment__.append((\"y_test Set\", y_test))\n else:\n experiment__.append((\"Transformed Data\", X))\n experiment__.append((\"Transformation Pipeline\", prep_pipe))\n\n # end runtime\n runtime_end = time.time()\n runtime = np.array(runtime_end - runtime_start).round(2)\n\n if logging_param:\n\n logger.info(\"Logging experiment in MLFlow\")\n\n import mlflow\n\n try:\n mlflow.create_experiment(exp_name_log)\n except:\n logger.warning(\"Couldn't create mlflow experiment. Exception:\")\n logger.warning(traceback.format_exc())\n\n # mlflow logging\n mlflow.set_experiment(exp_name_log)\n\n run_name_ = f\"Session Initialized {USI}\"\n\n with mlflow.start_run(run_name=run_name_) as run:\n\n # Get active run to log as tag\n RunID = mlflow.active_run().info.run_id\n\n k = functions.copy()\n k.set_index(\"Description\", drop=True, inplace=True)\n kdict = k.to_dict()\n params = kdict.get(\"Value\")\n mlflow.log_params(params)\n\n # set tag of compare_models\n mlflow.set_tag(\"Source\", \"setup\")\n\n import secrets\n\n URI = secrets.token_hex(nbytes=4)\n mlflow.set_tag(\"URI\", URI)\n mlflow.set_tag(\"USI\", USI)\n mlflow.set_tag(\"Run Time\", runtime)\n mlflow.set_tag(\"Run ID\", RunID)\n\n # Log the transformation pipeline\n logger.info(\n \"SubProcess save_model() called ==================================\"\n )\n save_model(prep_pipe, \"Transformation Pipeline\", verbose=False)\n logger.info(\n \"SubProcess save_model() end ==================================\"\n )\n mlflow.log_artifact(\"Transformation Pipeline.pkl\")\n os.remove(\"Transformation Pipeline.pkl\")\n\n # Log pandas profile\n if log_profile:\n import pandas_profiling\n\n pf = pandas_profiling.ProfileReport(\n data_before_preprocess, **profile_kwargs\n )\n pf.to_file(\"Data Profile.html\")\n mlflow.log_artifact(\"Data Profile.html\")\n os.remove(\"Data Profile.html\")\n display.display(functions_, clear=True)\n\n # Log training and testing set\n if log_data:\n if not _is_unsupervised(_ml_usecase):\n X_train.join(y_train).to_csv(\"Train.csv\")\n X_test.join(y_test).to_csv(\"Test.csv\")\n mlflow.log_artifact(\"Train.csv\")\n mlflow.log_artifact(\"Test.csv\")\n os.remove(\"Train.csv\")\n os.remove(\"Test.csv\")\n else:\n X.to_csv(\"Dataset.csv\")\n mlflow.log_artifact(\"Dataset.csv\")\n os.remove(\"Dataset.csv\")\n\n logger.info(f\"create_model_container: {len(create_model_container)}\")\n logger.info(f\"master_model_container: {len(master_model_container)}\")\n logger.info(f\"display_container: {len(display_container)}\")\n\n logger.info(str(prep_pipe))\n logger.info(\"setup() succesfully completed......................................\")\n\n gc.collect()\n\n return tuple([globals()[v] for v in pycaret_globals])\n\n\ndef compare_models(\n include: Optional[\n List[Union[str, Any]]\n ] = None, # changed whitelist to include in pycaret==2.1\n exclude: Optional[List[str]] = None, # changed blacklist to exclude in pycaret==2.1\n fold: Optional[Union[int, Any]] = None,\n round: int = 4,\n cross_validation: bool = True,\n sort: str = \"Accuracy\",\n n_select: int = 1,\n budget_time: Optional[float] = None, # added in pycaret==2.1.0\n turbo: bool = True,\n errors: str = \"ignore\",\n fit_kwargs: Optional[dict] = None,\n groups: Optional[Union[str, Any]] = None,\n verbose: bool = True,\n display: Optional[Display] = None,\n) -> List[Any]:\n\n \"\"\"\n This function train all the models available in the model library and scores them\n using Cross Validation. The output prints a score grid with Accuracy,\n AUC, Recall, Precision, F1, Kappa and MCC (averaged across folds).\n\n This function returns all of the models compared, sorted by the value of the selected metric.\n\n When turbo is set to True ('rbfsvm', 'gpc' and 'mlp') are excluded due to longer\n training time. By default turbo param is set to True.\n\n Example\n -------\n >>> from pycaret.datasets import get_data\n >>> juice = get_data('juice')\n >>> experiment_name = setup(data = juice, target = 'Purchase')\n >>> best_model = compare_models()\n\n This will return the averaged score grid of all the models except 'rbfsvm', 'gpc'\n and 'mlp'. When turbo param is set to False, all models including 'rbfsvm', 'gpc'\n and 'mlp' are used but this may result in longer training time.\n\n >>> best_model = compare_models( exclude = [ 'knn', 'gbc' ] , turbo = False)\n\n This will return a comparison of all models except K Nearest Neighbour and\n Gradient Boosting Classifier.\n\n >>> best_model = compare_models( exclude = [ 'knn', 'gbc' ] , turbo = True)\n\n This will return comparison of all models except K Nearest Neighbour,\n Gradient Boosting Classifier, SVM (RBF), Gaussian Process Classifier and\n Multi Level Perceptron.\n\n\n >>> tuned_model = tune_model(create_model('lr'))\n >>> best_model = compare_models( include = [ 'lr', tuned_model ])\n\n This will compare a tuned Linear Regression model with an untuned one.\n\n Parameters\n ----------\n exclude: list of strings, default = None\n In order to omit certain models from the comparison model ID's can be passed as\n a list of strings in exclude param.\n\n include: list of strings or objects, default = None\n In order to run only certain models for the comparison, the model ID's can be\n passed as a list of strings in include param. The list can also include estimator\n objects to be compared.\n\n fold: integer or scikit-learn compatible CV generator, default = None\n Controls cross-validation. If None, will use the CV generator defined in setup().\n If integer, will use KFold CV with that many folds.\n When cross_validation is False, this parameter is ignored.\n\n round: integer, default = 4\n Number of decimal places the metrics in the score grid will be rounded to.\n\n cross_validation: bool, default = True\n When cross_validation set to False fold parameter is ignored and models are trained\n on entire training dataset, returning metrics calculated using the train (holdout) set.\n\n sort: str, default = 'Accuracy'\n The scoring measure specified is used for sorting the average score grid\n Other options are 'AUC', 'Recall', 'Precision', 'F1', 'Kappa' and 'MCC'.\n\n n_select: int, default = 1\n Number of top_n models to return. use negative argument for bottom selection.\n for example, n_select = -3 means bottom 3 models.\n\n budget_time: int or float, default = None\n If not 0 or None, will terminate execution of the function after budget_time\n minutes have passed and return results up to that point.\n\n turbo: bool, default = True\n When turbo is set to True, it excludes estimators that have longer\n training time.\n\n errors: str, default = 'ignore'\n If 'ignore', will suppress model exceptions and continue.\n If 'raise', will allow exceptions to be raised.\n\n fit_kwargs: dict, default = {} (empty dict)\n Dictionary of arguments passed to the fit method of the model. The parameters will be applied to all models,\n therefore it is recommended to set errors parameter to 'ignore'.\n\n groups: str or array-like, with shape (n_samples,), default = None\n Optional Group labels for the samples used while splitting the dataset into train/test set.\n If string is passed, will use the data column with that name as the groups.\n Only used if a group based cross-validation generator is used (eg. GroupKFold).\n If None, will use the value set in fold_groups param in setup().\n\n verbose: bool, default = True\n Score grid is not printed when verbose is set to False.\n\n Returns\n -------\n score_grid\n A table containing the scores of the model across the kfolds.\n Scoring metrics used are Accuracy, AUC, Recall, Precision, F1,\n Kappa and MCC. Mean and standard deviation of the scores across\n the folds are also returned.\n\n list\n List of fitted model objects that were compared.\n\n Warnings\n --------\n - compare_models() though attractive, might be time consuming with large\n datasets. By default turbo is set to True, which excludes models that\n have longer training times. Changing turbo parameter to False may result\n in very high training times with datasets where number of samples exceed\n 10,000.\n\n - If target variable is multiclass (more than 2 classes), AUC will be\n returned as zero (0.0)\n\n - If cross_validation param is set to False, no models will be logged with MLFlow.\n\n \"\"\"\n\n function_params_str = \", \".join([f\"{k}={v}\" for k, v in locals().items()])\n\n logger = get_logger()\n\n logger.info(\"Initializing compare_models()\")\n logger.info(f\"compare_models({function_params_str})\")\n\n logger.info(\"Checking exceptions\")\n\n if not fit_kwargs:\n fit_kwargs = {}\n\n # checking error for exclude (string)\n available_estimators = _all_models\n\n if exclude != None:\n for i in exclude:\n if i not in available_estimators:\n raise ValueError(\n f\"Estimator Not Available {i}. Please see docstring for list of available estimators.\"\n )\n\n if include != None:\n for i in include:\n if isinstance(i, str):\n if i not in available_estimators:\n raise ValueError(\n f\"Estimator {i} Not Available. Please see docstring for list of available estimators.\"\n )\n elif not hasattr(i, \"fit\"):\n raise ValueError(\n f\"Estimator {i} does not have the required fit() method.\"\n )\n\n # include and exclude together check\n if include is not None and exclude is not None:\n raise TypeError(\n \"Cannot use exclude parameter when include is used to compare models.\"\n )\n\n # checking fold parameter\n if fold is not None and not (type(fold) is int or is_sklearn_cv_generator(fold)):\n raise TypeError(\n \"fold parameter must be either None, an integer or a scikit-learn compatible CV generator object.\"\n )\n\n # checking round parameter\n if type(round) is not int:\n raise TypeError(\"Round parameter only accepts integer value.\")\n\n # checking budget_time parameter\n if budget_time and type(budget_time) is not int and type(budget_time) is not float:\n raise TypeError(\"budget_time parameter only accepts integer or float values.\")\n\n # checking sort parameter\n if not (isinstance(sort, str) and (sort == \"TT\" or sort == \"TT (Sec)\")):\n sort = _get_metric(sort)\n if sort is None:\n raise ValueError(\n f\"Sort method not supported. See docstring for list of available parameters.\"\n )\n\n # checking errors parameter\n possible_errors = [\"ignore\", \"raise\"]\n if errors not in possible_errors:\n raise ValueError(\n f\"errors parameter must be one of: {', '.join(possible_errors)}.\"\n )\n\n # checking optimize parameter for multiclass\n if _is_multiclass():\n if not sort.is_multiclass:\n raise TypeError(\n f\"{sort} metric not supported for multiclass problems. See docstring for list of other optimization parameters.\"\n )\n\n \"\"\"\n\n ERROR HANDLING ENDS HERE\n\n \"\"\"\n\n fold = _get_cv_splitter(fold)\n\n groups = _get_groups(groups)\n\n pd.set_option(\"display.max_columns\", 500)\n\n logger.info(\"Preparing display monitor\")\n\n len_mod = (\n len({k: v for k, v in _all_models.items() if v.is_turbo})\n if turbo\n else len(_all_models)\n )\n\n if include:\n len_mod = len(include)\n elif exclude:\n len_mod -= len(exclude)\n\n if not display:\n progress_args = {\"max\": (4 * len_mod) + 4 + len_mod}\n master_display_columns = (\n [\"Model\"] + [v.display_name for k, v in _all_metrics.items()] + [\"TT (Sec)\"]\n )\n timestampStr = datetime.datetime.now().strftime(\"%H:%M:%S\")\n monitor_rows = [\n [\"Initiated\", \". . . . . . . . . . . . . . . . . .\", timestampStr],\n [\"Status\", \". . . . . . . . . . . . . . . . . .\", \"Loading Dependencies\"],\n [\"Estimator\", \". . . . . . . . . . . . . . . . . .\", \"Compiling Library\"],\n ]\n display = Display(\n verbose=verbose,\n html_param=html_param,\n progress_args=progress_args,\n master_display_columns=master_display_columns,\n monitor_rows=monitor_rows,\n )\n\n display.display_progress()\n display.display_monitor()\n display.display_master_display()\n\n greater_is_worse_columns = {\n v.display_name for k, v in _all_metrics.items() if not v.greater_is_better\n }\n greater_is_worse_columns.add(\"TT (Sec)\")\n\n np.random.seed(seed)\n\n display.move_progress()\n\n # defining sort parameter (making Precision equivalent to Prec. )\n\n if not (isinstance(sort, str) and (sort == \"TT\" or sort == \"TT (Sec)\")):\n sort_ascending = not sort.greater_is_better\n sort = sort.display_name\n else:\n sort_ascending = True\n sort = \"TT (Sec)\"\n\n \"\"\"\n MONITOR UPDATE STARTS\n \"\"\"\n\n display.update_monitor(1, \"Loading Estimator\")\n display.display_monitor()\n\n \"\"\"\n MONITOR UPDATE ENDS\n \"\"\"\n\n if include:\n model_library = include\n else:\n if turbo:\n model_library = _all_models\n model_library = [k for k, v in _all_models.items() if v.is_turbo]\n else:\n model_library = list(_all_models.keys())\n if exclude:\n model_library = [x for x in model_library if x not in exclude]\n\n display.move_progress()\n\n # create URI (before loop)\n import secrets\n\n URI = secrets.token_hex(nbytes=4)\n\n master_display = None\n master_display_ = None\n\n total_runtime_start = time.time()\n total_runtime = 0\n over_time_budget = False\n if budget_time and budget_time > 0:\n logger.info(f\"Time budget is {budget_time} minutes\")\n\n for i, model in enumerate(model_library):\n\n model_id = (\n model\n if (\n isinstance(model, str)\n and all(isinstance(m, str) for m in model_library)\n )\n else str(i)\n )\n model_name = _get_model_name(model)\n\n if isinstance(model, str):\n logger.info(f\"Initializing {model_name}\")\n else:\n logger.info(f\"Initializing custom model {model_name}\")\n\n # run_time\n runtime_start = time.time()\n total_runtime += (runtime_start - total_runtime_start) / 60\n logger.info(f\"Total runtime is {total_runtime} minutes\")\n over_time_budget = (\n budget_time and budget_time > 0 and total_runtime > budget_time\n )\n if over_time_budget:\n logger.info(\n f\"Total runtime {total_runtime} is over time budget by {total_runtime - budget_time}, breaking loop\"\n )\n break\n total_runtime_start = runtime_start\n\n display.move_progress()\n\n \"\"\"\n MONITOR UPDATE STARTS\n \"\"\"\n\n display.update_monitor(2, model_name)\n display.display_monitor()\n\n \"\"\"\n MONITOR UPDATE ENDS\n \"\"\"\n display.replace_master_display(None)\n\n logger.info(\n \"SubProcess create_model() called ==================================\"\n )\n if errors == \"raise\":\n model, model_fit_time = create_model_supervised(\n estimator=model,\n system=False,\n verbose=False,\n display=display,\n fold=fold,\n round=round,\n cross_validation=cross_validation,\n fit_kwargs=fit_kwargs,\n groups=groups,\n refit=False,\n )\n model_results = pull(pop=True)\n else:\n try:\n model, model_fit_time = create_model_supervised(\n estimator=model,\n system=False,\n verbose=False,\n display=display,\n fold=fold,\n round=round,\n cross_validation=cross_validation,\n fit_kwargs=fit_kwargs,\n groups=groups,\n refit=False,\n )\n model_results = pull(pop=True)\n assert np.sum(model_results.iloc[0]) != 0.0\n except:\n logger.warning(\n f\"create_model() for {model} raised an exception or returned all 0.0, trying without fit_kwargs:\"\n )\n logger.warning(traceback.format_exc())\n try:\n model, model_fit_time = create_model_supervised(\n estimator=model,\n system=False,\n verbose=False,\n display=display,\n fold=fold,\n round=round,\n cross_validation=cross_validation,\n groups=groups,\n refit=False,\n )\n model_results = pull(pop=True)\n except:\n logger.error(f\"create_model() for {model} raised an exception:\")\n logger.error(traceback.format_exc())\n continue\n logger.info(\"SubProcess create_model() end ==================================\")\n\n if model is None:\n over_time_budget = True\n logger.info(f\"Time budged exceeded in create_model(), breaking loop\")\n break\n\n runtime_end = time.time()\n runtime = np.array(runtime_end - runtime_start).round(2)\n\n logger.info(\"Creating metrics dataframe\")\n if cross_validation:\n compare_models_ = pd.DataFrame(model_results.loc[\"Mean\"]).T\n else:\n compare_models_ = pd.DataFrame(model_results.iloc[0]).T\n compare_models_.insert(len(compare_models_.columns), \"TT (Sec)\", model_fit_time)\n compare_models_.insert(0, \"Model\", model_name)\n compare_models_.insert(0, \"Object\", [model])\n compare_models_.insert(0, \"runtime\", runtime)\n compare_models_.index = [model_id]\n if master_display is None:\n master_display = compare_models_\n else:\n master_display = pd.concat(\n [master_display, compare_models_], ignore_index=False\n )\n master_display = master_display.round(round)\n master_display = master_display.sort_values(by=sort, ascending=sort_ascending)\n\n master_display_ = master_display.drop(\n [\"Object\", \"runtime\"], axis=1, errors=\"ignore\"\n ).style.set_precision(round)\n master_display_ = master_display_.set_properties(**{\"text-align\": \"left\"})\n master_display_ = master_display_.set_table_styles(\n [dict(selector=\"th\", props=[(\"text-align\", \"left\")])]\n )\n\n display.replace_master_display(master_display_)\n\n display.display_master_display()\n\n display.move_progress()\n\n def highlight_max(s):\n to_highlight = s == s.max()\n return [\"background-color: yellow\" if v else \"\" for v in to_highlight]\n\n def highlight_min(s):\n to_highlight = s == s.min()\n return [\"background-color: yellow\" if v else \"\" for v in to_highlight]\n\n def highlight_cols(s):\n color = \"lightgrey\"\n return f\"background-color: {color}\"\n\n if master_display_ is not None:\n compare_models_ = (\n master_display_.apply(\n highlight_max,\n subset=[\n x\n for x in master_display_.columns[1:]\n if x not in greater_is_worse_columns\n ],\n )\n .apply(\n highlight_min,\n subset=[\n x\n for x in master_display_.columns[1:]\n if x in greater_is_worse_columns\n ],\n )\n .applymap(highlight_cols, subset=[\"TT (Sec)\"])\n )\n else:\n compare_models_ = pd.DataFrame().style\n\n display.update_monitor(1, \"Compiling Final Models\")\n display.display_monitor()\n\n display.move_progress()\n\n sorted_models = []\n\n if master_display is not None:\n if n_select < 0:\n n_select_range = range(len(master_display) - n_select, len(master_display))\n else:\n n_select_range = range(0, n_select)\n\n for index, row in enumerate(master_display.iterrows()):\n loc, row = row\n model = row[\"Object\"]\n\n results = row.to_frame().T.drop(\n [\"Object\", \"Model\", \"runtime\", \"TT (Sec)\"], errors=\"ignore\", axis=1\n )\n\n avgs_dict_log = {k: v for k, v in results.iloc[0].items()}\n\n full_logging = False\n\n if index in n_select_range:\n display.update_monitor(2, _get_model_name(model))\n display.display_monitor()\n model, model_fit_time = create_model_supervised(\n estimator=model,\n system=False,\n verbose=False,\n fold=fold,\n round=round,\n cross_validation=False,\n predict=False,\n fit_kwargs=fit_kwargs,\n groups=groups,\n )\n sorted_models.append(model)\n full_logging = True\n\n if logging_param and cross_validation:\n\n try:\n _mlflow_log_model(\n model=model,\n model_results=results,\n score_dict=avgs_dict_log,\n source=\"compare_models\",\n runtime=row[\"runtime\"],\n model_fit_time=row[\"TT (Sec)\"],\n _prep_pipe=prep_pipe,\n log_plots=log_plots_param if full_logging else False,\n log_holdout=full_logging,\n URI=URI,\n display=display,\n )\n except:\n logger.error(\n f\"_mlflow_log_model() for {model} raised an exception:\"\n )\n logger.error(traceback.format_exc())\n\n if len(sorted_models) == 1:\n sorted_models = sorted_models[0]\n\n display.display(compare_models_, clear=True)\n\n pd.reset_option(\"display.max_columns\")\n\n # store in display container\n display_container.append(compare_models_.data)\n\n logger.info(f\"create_model_container: {len(create_model_container)}\")\n logger.info(f\"master_model_container: {len(master_model_container)}\")\n logger.info(f\"display_container: {len(display_container)}\")\n\n logger.info(str(sorted_models))\n logger.info(\n \"compare_models() succesfully completed......................................\"\n )\n\n return sorted_models\n\n\ndef create_model_unsupervised(\n estimator,\n num_clusters: int = 4,\n fraction: float = 0.05,\n ground_truth: Optional[str] = None,\n round: int = 4,\n fit_kwargs: Optional[dict] = None,\n verbose: bool = True,\n system: bool = True,\n raise_num_clusters: bool = False,\n X_data: Optional[pd.DataFrame] = None, # added in pycaret==2.2.0\n display: Optional[Display] = None, # added in pycaret==2.2.0\n **kwargs,\n) -> Any:\n\n \"\"\"\n This is an internal version of the create_model function.\n\n This function creates a model and scores it using Cross Validation.\n The output prints a score grid that shows Accuracy, AUC, Recall, Precision,\n F1, Kappa and MCC by fold (default = 10 Fold).\n\n This function returns a trained model object.\n\n setup() function must be called before using create_model()\n\n Example\n -------\n >>> from pycaret.datasets import get_data\n >>> juice = get_data('juice')\n >>> experiment_name = setup(data = juice, target = 'Purchase')\n >>> lr = create_model('lr')\n\n This will create a trained Logistic Regression model.\n\n Parameters\n ----------\n model : string / object, default = None\n Enter ID of the models available in model library or pass an untrained model\n object consistent with fit / predict API to train and evaluate model. List of\n models available in model library (ID - Model):\n\n * 'kmeans' - K-Means Clustering\n * 'ap' - Affinity Propagation\n * 'meanshift' - Mean shift Clustering\n * 'sc' - Spectral Clustering\n * 'hclust' - Agglomerative Clustering\n * 'dbscan' - Density-Based Spatial Clustering\n * 'optics' - OPTICS Clustering\n * 'birch' - Birch Clustering\n * 'kmodes' - K-Modes Clustering\n\n num_clusters: int, default = 4\n Number of clusters to be generated with the dataset.\n\n ground_truth: string, default = None\n When ground_truth is provided, Homogeneity Score, Rand Index, and\n Completeness Score is evaluated and printer along with other metrics.\n\n round: integer, default = 4\n Number of decimal places the metrics in the score grid will be rounded to.\n\n fit_kwargs: dict, default = {} (empty dict)\n Dictionary of arguments passed to the fit method of the model.\n\n verbose: bool, default = True\n Score grid is not printed when verbose is set to False.\n\n system: bool, default = True\n Must remain True all times. Only to be changed by internal functions.\n If False, method will return a tuple of model and the model fit time.\n\n **kwargs:\n Additional keyword arguments to pass to the estimator.\n\n Returns\n -------\n score_grid\n A table containing the Silhouette, Calinski-Harabasz,\n Davies-Bouldin, Homogeneity Score, Rand Index, and\n Completeness Score. Last 3 are only evaluated when\n ground_truth param is provided.\n\n model\n trained model object\n\n Warnings\n --------\n - num_clusters not required for Affinity Propagation ('ap'), Mean shift\n clustering ('meanshift'), Density-Based Spatial Clustering ('dbscan')\n and OPTICS Clustering ('optics'). num_clusters param for these models\n are automatically determined.\n\n - When fit doesn't converge in Affinity Propagation ('ap') model, all\n datapoints are labelled as -1.\n\n - Noisy samples are given the label -1, when using Density-Based Spatial\n ('dbscan') or OPTICS Clustering ('optics').\n\n - OPTICS ('optics') clustering may take longer training times on large\n datasets.\n\n \"\"\"\n\n function_params_str = \", \".join(\n [f\"{k}={v}\" for k, v in locals().items() if k not in (\"X_data\")]\n )\n\n logger = get_logger()\n\n logger.info(\"Initializing create_model()\")\n logger.info(f\"create_model({function_params_str})\")\n\n logger.info(\"Checking exceptions\")\n\n # run_time\n runtime_start = time.time()\n\n available_estimators = set(_all_models_internal.keys())\n\n if not fit_kwargs:\n fit_kwargs = {}\n\n # only raise exception of estimator is of type string.\n if isinstance(estimator, str):\n if estimator not in available_estimators:\n raise ValueError(\n f\"Estimator {estimator} not available. Please see docstring for list of available estimators.\"\n )\n elif not hasattr(estimator, \"fit\"):\n raise ValueError(\n f\"Estimator {estimator} does not have the required fit() method.\"\n )\n\n # checking round parameter\n if type(round) is not int:\n raise TypeError(\"Round parameter only accepts integer value.\")\n\n # checking verbose parameter\n if type(verbose) is not bool:\n raise TypeError(\"Verbose parameter can only take argument as True or False.\")\n\n # checking system parameter\n if type(system) is not bool:\n raise TypeError(\"System parameter can only take argument as True or False.\")\n\n # checking fraction type:\n if fraction <= 0 or fraction >= 1:\n raise TypeError(\n \"Fraction parameter can only take value as float between 0 to 1.\"\n )\n\n # checking num_clusters type:\n if num_clusters <= 1:\n raise TypeError(\n \"num_clusters parameter can only take value integer value greater than 1.\"\n )\n\n # check ground truth exist in data_\n if ground_truth is not None:\n if ground_truth not in data_before_preprocess.columns:\n raise ValueError(\n f\"ground_truth {ground_truth} doesn't exist in the dataset.\"\n )\n\n \"\"\"\n\n ERROR HANDLING ENDS HERE\n\n \"\"\"\n\n if not display:\n progress_args = {\"max\": 3}\n master_display_columns = [v.display_name for k, v in _all_metrics.items()]\n timestampStr = datetime.datetime.now().strftime(\"%H:%M:%S\")\n monitor_rows = [\n [\"Initiated\", \". . . . . . . . . . . . . . . . . .\", timestampStr],\n [\"Status\", \". . . . . . . . . . . . . . . . . .\", \"Loading Dependencies\"],\n [\"Estimator\", \". . . . . . . . . . . . . . . . . .\", \"Compiling Library\"],\n ]\n display = Display(\n verbose=verbose,\n html_param=html_param,\n progress_args=progress_args,\n master_display_columns=master_display_columns,\n monitor_rows=monitor_rows,\n )\n display.display_progress()\n display.display_monitor()\n display.display_master_display()\n\n logger.info(\"Importing libraries\")\n\n # general dependencies\n\n np.random.seed(seed)\n\n # Storing X_train and y_train in data_X and data_y parameter\n data_X = X if X_data is None else X_data\n\n \"\"\"\n MONITOR UPDATE STARTS\n \"\"\"\n display.update_monitor(1, \"Selecting Estimator\")\n display.display_monitor()\n \"\"\"\n MONITOR UPDATE ENDS\n \"\"\"\n\n logger.info(\"Importing untrained model\")\n\n is_cblof = False\n\n if isinstance(estimator, str) and estimator in available_estimators:\n is_cblof = estimator == \"cluster\"\n model_definition = _all_models_internal[estimator]\n model_args = model_definition.args\n model_args = {**model_args, **kwargs}\n model = model_definition.class_def(**model_args)\n full_name = model_definition.name\n else:\n logger.info(\"Declaring custom model\")\n\n model = clone(estimator)\n model.set_params(**kwargs)\n\n full_name = _get_model_name(model)\n\n display.update_monitor(2, full_name)\n display.display_monitor()\n\n if _ml_usecase == MLUsecase.CLUSTERING:\n if raise_num_clusters:\n model.set_params(n_clusters=num_clusters)\n else:\n try:\n model.set_params(n_clusters=num_clusters)\n except:\n pass\n else:\n model.set_params(contamination=fraction)\n\n # workaround for an issue with set_params in cuML\n try:\n model = clone(model)\n except:\n logger.warning(\n f\"create_model_unsupervised() for {model} raised an exception when cloning:\"\n )\n logger.warning(traceback.format_exc())\n\n logger.info(f\"{full_name} Imported succesfully\")\n\n display.move_progress()\n\n \"\"\"\n MONITOR UPDATE STARTS\n \"\"\"\n if _ml_usecase == MLUsecase.CLUSTERING:\n display.update_monitor(1, f\"Fitting {num_clusters} Clusters\")\n else:\n display.update_monitor(1, f\"Fitting {fraction} Fraction\")\n display.display_monitor()\n \"\"\"\n MONITOR UPDATE ENDS\n \"\"\"\n\n with estimator_pipeline(_internal_pipeline, model) as pipeline_with_model:\n fit_kwargs = _get_pipeline_fit_kwargs(pipeline_with_model, fit_kwargs)\n\n logger.info(\"Fitting Model\")\n model_fit_start = time.time()\n with io.capture_output():\n if is_cblof and \"n_clusters\" not in kwargs:\n try:\n pipeline_with_model.fit(data_X, **fit_kwargs)\n except:\n try:\n pipeline_with_model.set_params(actual_estimator__n_clusters=12)\n model_fit_start = time.time()\n pipeline_with_model.fit(data_X, **fit_kwargs)\n except:\n raise RuntimeError(\n \"Could not form valid cluster separation. Try a different dataset or model.\"\n )\n else:\n pipeline_with_model.fit(data_X, **fit_kwargs)\n model_fit_end = time.time()\n\n model_fit_time = np.array(model_fit_end - model_fit_start).round(2)\n\n display.move_progress()\n\n if ground_truth is not None:\n\n logger.info(f\"ground_truth parameter set to {ground_truth}\")\n\n gt = np.array(data_before_preprocess[ground_truth])\n else:\n gt = None\n\n if _ml_usecase == MLUsecase.CLUSTERING:\n metrics = _calculate_metrics_unsupervised(X, model.labels_, ground_truth=gt)\n else:\n metrics = {}\n\n logger.info(str(model))\n logger.info(\n \"create_models() succesfully completed......................................\"\n )\n\n runtime = time.time() - runtime_start\n\n # mlflow logging\n if logging_param and system:\n\n metrics_log = {k: v for k, v in metrics.items()}\n\n try:\n _mlflow_log_model(\n model=model,\n model_results=None,\n score_dict=metrics_log,\n source=\"create_model\",\n runtime=runtime,\n model_fit_time=model_fit_time,\n _prep_pipe=prep_pipe,\n log_plots=log_plots_param,\n display=display,\n )\n except:\n logger.error(f\"_mlflow_log_model() for {model} raised an exception:\")\n logger.error(traceback.format_exc())\n\n display.move_progress()\n\n logger.info(\"Uploading results into container\")\n\n model_results = pd.DataFrame(metrics, index=[0])\n model_results = model_results.round(round)\n\n # storing results in create_model_container\n create_model_container.append(model_results)\n display_container.append(model_results)\n\n # storing results in master_model_container\n logger.info(\"Uploading model into container now\")\n master_model_container.append(model)\n\n if _ml_usecase == MLUsecase.CLUSTERING:\n display.display(\n model_results, clear=system, override=False if not system else None\n )\n elif system:\n display.clear_output()\n\n logger.info(f\"create_model_container: {len(create_model_container)}\")\n logger.info(f\"master_model_container: {len(master_model_container)}\")\n logger.info(f\"display_container: {len(display_container)}\")\n\n logger.info(str(model))\n logger.info(\n \"create_model() succesfully completed......................................\"\n )\n gc.collect()\n\n if not system:\n return (model, model_fit_time)\n\n return model\n\n\ndef create_model_supervised(\n estimator,\n fold: Optional[Union[int, Any]] = None,\n round: int = 4,\n cross_validation: bool = True,\n predict: bool = True,\n fit_kwargs: Optional[dict] = None,\n groups: Optional[Union[str, Any]] = None,\n refit: bool = True,\n verbose: bool = True,\n system: bool = True,\n X_train_data: Optional[pd.DataFrame] = None, # added in pycaret==2.2.0\n y_train_data: Optional[pd.DataFrame] = None, # added in pycaret==2.2.0\n metrics=None,\n display: Optional[Display] = None, # added in pycaret==2.2.0\n **kwargs,\n) -> Any:\n\n \"\"\"\n This is an internal version of the create_model function.\n\n This function creates a model and scores it using Cross Validation.\n The output prints a score grid that shows Accuracy, AUC, Recall, Precision,\n F1, Kappa and MCC by fold (default = 10 Fold).\n\n This function returns a trained model object.\n\n setup() function must be called before using create_model()\n\n Example\n -------\n >>> from pycaret.datasets import get_data\n >>> juice = get_data('juice')\n >>> experiment_name = setup(data = juice, target = 'Purchase')\n >>> lr = create_model('lr')\n\n This will create a trained Logistic Regression model.\n\n Parameters\n ----------\n estimator : str / object, default = None\n Enter ID of the estimators available in model library or pass an untrained model\n object consistent with fit / predict API to train and evaluate model. All\n estimators support binary or multiclass problem. List of estimators in model\n library (ID - Name):\n\n * 'lr' - Logistic Regression\n * 'knn' - K Nearest Neighbour\n * 'nb' - Naive Bayes\n * 'dt' - Decision Tree Classifier\n * 'svm' - SVM - Linear Kernel\n * 'rbfsvm' - SVM - Radial Kernel\n * 'gpc' - Gaussian Process Classifier\n * 'mlp' - Multi Level Perceptron\n * 'ridge' - Ridge Classifier\n * 'rf' - Random Forest Classifier\n * 'qda' - Quadratic Discriminant Analysis\n * 'ada' - Ada Boost Classifier\n * 'gbc' - Gradient Boosting Classifier\n * 'lda' - Linear Discriminant Analysis\n * 'et' - Extra Trees Classifier\n * 'xgboost' - Extreme Gradient Boosting\n * 'lightgbm' - Light Gradient Boosting\n * 'catboost' - CatBoost Classifier\n\n fold: integer or scikit-learn compatible CV generator, default = None\n Controls cross-validation. If None, will use the CV generator defined in setup().\n If integer, will use KFold CV with that many folds.\n When cross_validation is False, this parameter is ignored.\n\n round: integer, default = 4\n Number of decimal places the metrics in the score grid will be rounded to.\n\n cross_validation: bool, default = True\n When cross_validation set to False fold parameter is ignored and model is trained\n on entire training dataset.\n\n predict: bool, default = True\n Whether to predict model on holdout if cross_validation == False.\n\n fit_kwargs: dict, default = {} (empty dict)\n Dictionary of arguments passed to the fit method of the model.\n\n groups: str or array-like, with shape (n_samples,), default = None\n Optional Group labels for the samples used while splitting the dataset into train/test set.\n If string is passed, will use the data column with that name as the groups.\n Only used if a group based cross-validation generator is used (eg. GroupKFold).\n If None, will use the value set in fold_groups param in setup().\n\n refit: bool, default = True\n Whether to refit the model on the entire dataset after CV. Ignored if cross_validation == False.\n\n verbose: bool, default = True\n Score grid is not printed when verbose is set to False.\n\n system: bool, default = True\n Must remain True all times. Only to be changed by internal functions.\n If False, method will return a tuple of model and the model fit time.\n\n X_train_data: pandas.DataFrame, default = None\n If not None, will use this dataframe as training features.\n Intended to be only changed by internal functions.\n\n y_train_data: pandas.DataFrame, default = None\n If not None, will use this dataframe as training target.\n Intended to be only changed by internal functions.\n\n **kwargs:\n Additional keyword arguments to pass to the estimator.\n\n Returns\n -------\n score_grid\n A table containing the scores of the model across the kfolds.\n Scoring metrics used are Accuracy, AUC, Recall, Precision, F1,\n Kappa and MCC. Mean and standard deviation of the scores across\n the folds are highlighted in yellow.\n\n model\n trained model object\n\n Warnings\n --------\n - 'svm' and 'ridge' doesn't support predict_proba method. As such, AUC will be\n returned as zero (0.0)\n\n - If target variable is multiclass (more than 2 classes), AUC will be returned\n as zero (0.0)\n\n - 'rbfsvm' and 'gpc' uses non-linear kernel and hence the fit time complexity is\n more than quadratic. These estimators are hard to scale on datasets with more\n than 10,000 samples.\n\n - If cross_validation param is set to False, model will not be logged with MLFlow.\n\n \"\"\"\n\n function_params_str = \", \".join(\n [\n f\"{k}={v}\"\n for k, v in locals().items()\n if k not in (\"X_train_data\", \"y_train_data\")\n ]\n )\n\n logger = get_logger()\n\n logger.info(\"Initializing create_model()\")\n logger.info(f\"create_model({function_params_str})\")\n\n logger.info(\"Checking exceptions\")\n\n # run_time\n runtime_start = time.time()\n\n available_estimators = set(_all_models_internal.keys())\n\n if not fit_kwargs:\n fit_kwargs = {}\n\n # only raise exception of estimator is of type string.\n if isinstance(estimator, str):\n if estimator not in available_estimators:\n raise ValueError(\n f\"Estimator {estimator} not available. Please see docstring for list of available estimators.\"\n )\n elif not hasattr(estimator, \"fit\"):\n raise ValueError(\n f\"Estimator {estimator} does not have the required fit() method.\"\n )\n\n # checking fold parameter\n if fold is not None and not (type(fold) is int or is_sklearn_cv_generator(fold)):\n raise TypeError(\n \"fold parameter must be either None, an integer or a scikit-learn compatible CV generator object.\"\n )\n\n # checking round parameter\n if type(round) is not int:\n raise TypeError(\"Round parameter only accepts integer value.\")\n\n # checking verbose parameter\n if type(verbose) is not bool:\n raise TypeError(\"Verbose parameter can only take argument as True or False.\")\n\n # checking system parameter\n if type(system) is not bool:\n raise TypeError(\"System parameter can only take argument as True or False.\")\n\n # checking cross_validation parameter\n if type(cross_validation) is not bool:\n raise TypeError(\n \"cross_validation parameter can only take argument as True or False.\"\n )\n\n \"\"\"\n\n ERROR HANDLING ENDS HERE\n\n \"\"\"\n\n groups = _get_groups(groups, data=X_train_data)\n\n if not display:\n progress_args = {\"max\": 4}\n master_display_columns = [v.display_name for k, v in _all_metrics.items()]\n timestampStr = datetime.datetime.now().strftime(\"%H:%M:%S\")\n monitor_rows = [\n [\"Initiated\", \". . . . . . . . . . . . . . . . . .\", timestampStr],\n [\"Status\", \". . . . . . . . . . . . . . . . . .\", \"Loading Dependencies\"],\n [\"Estimator\", \". . . . . . . . . . . . . . . . . .\", \"Compiling Library\"],\n ]\n display = Display(\n verbose=verbose,\n html_param=html_param,\n progress_args=progress_args,\n master_display_columns=master_display_columns,\n monitor_rows=monitor_rows,\n )\n display.display_progress()\n display.display_monitor()\n display.display_master_display()\n\n logger.info(\"Importing libraries\")\n\n # general dependencies\n\n np.random.seed(seed)\n\n logger.info(\"Copying training dataset\")\n\n # Storing X_train and y_train in data_X and data_y parameter\n data_X = X_train.copy() if X_train_data is None else X_train_data.copy()\n data_y = y_train.copy() if y_train_data is None else y_train_data.copy()\n\n # reset index\n data_X.reset_index(drop=True, inplace=True)\n data_y.reset_index(drop=True, inplace=True)\n\n if metrics is None:\n metrics = _all_metrics\n\n display.move_progress()\n\n logger.info(\"Defining folds\")\n\n # cross validation setup starts here\n cv = _get_cv_splitter(fold)\n\n logger.info(\"Declaring metric variables\")\n\n \"\"\"\n MONITOR UPDATE STARTS\n \"\"\"\n display.update_monitor(1, \"Selecting Estimator\")\n display.display_monitor()\n \"\"\"\n MONITOR UPDATE ENDS\n \"\"\"\n\n logger.info(\"Importing untrained model\")\n\n if isinstance(estimator, str) and estimator in available_estimators:\n model_definition = _all_models_internal[estimator]\n model_args = model_definition.args\n model_args = {**model_args, **kwargs}\n model = model_definition.class_def(**model_args)\n full_name = model_definition.name\n else:\n logger.info(\"Declaring custom model\")\n\n model = clone(estimator)\n model.set_params(**kwargs)\n\n full_name = _get_model_name(model)\n\n # workaround for an issue with set_params in cuML\n model = clone(model)\n\n display.update_monitor(2, full_name)\n display.display_monitor()\n\n if transform_target_param and not isinstance(model, TransformedTargetRegressor):\n model = PowerTransformedTargetRegressor(\n regressor=model, power_transformer_method=transform_target_method_param\n )\n\n logger.info(f\"{full_name} Imported succesfully\")\n\n display.move_progress()\n\n \"\"\"\n MONITOR UPDATE STARTS\n \"\"\"\n if not cross_validation:\n display.update_monitor(1, f\"Fitting {str(full_name)}\")\n else:\n display.update_monitor(1, \"Initializing CV\")\n\n display.display_monitor()\n \"\"\"\n MONITOR UPDATE ENDS\n \"\"\"\n\n if not cross_validation:\n\n with estimator_pipeline(_internal_pipeline, model) as pipeline_with_model:\n fit_kwargs = _get_pipeline_fit_kwargs(pipeline_with_model, fit_kwargs)\n logger.info(\"Cross validation set to False\")\n\n logger.info(\"Fitting Model\")\n model_fit_start = time.time()\n with io.capture_output():\n pipeline_with_model.fit(data_X, data_y, **fit_kwargs)\n model_fit_end = time.time()\n\n model_fit_time = np.array(model_fit_end - model_fit_start).round(2)\n\n display.move_progress()\n\n if predict:\n predict_model(pipeline_with_model, verbose=False)\n model_results = pull(pop=True).drop(\"Model\", axis=1)\n\n display_container.append(model_results)\n\n display.display(\n model_results, clear=system, override=False if not system else None\n )\n\n logger.info(f\"display_container: {len(display_container)}\")\n\n display.move_progress()\n\n logger.info(str(model))\n logger.info(\n \"create_models() succesfully completed......................................\"\n )\n\n gc.collect()\n\n if not system:\n return (model, model_fit_time)\n return model\n\n \"\"\"\n MONITOR UPDATE STARTS\n \"\"\"\n display.update_monitor(\n 1, f\"Fitting {_get_cv_n_folds(fold, data_X, y=data_y, groups=groups)} Folds\"\n )\n display.display_monitor()\n \"\"\"\n MONITOR UPDATE ENDS\n \"\"\"\n\n from sklearn.model_selection import cross_validate\n\n metrics_dict = dict([(k, v.scorer) for k, v in metrics.items()])\n\n logger.info(\"Starting cross validation\")\n\n n_jobs = _gpu_n_jobs_param\n from sklearn.gaussian_process import (\n GaussianProcessClassifier,\n GaussianProcessRegressor,\n )\n\n # special case to prevent running out of memory\n if isinstance(model, (GaussianProcessClassifier, GaussianProcessRegressor)):\n n_jobs = 1\n\n with estimator_pipeline(_internal_pipeline, model) as pipeline_with_model:\n fit_kwargs = _get_pipeline_fit_kwargs(pipeline_with_model, fit_kwargs)\n logger.info(f\"Cross validating with {cv}, n_jobs={n_jobs}\")\n\n model_fit_start = time.time()\n scores = cross_validate(\n pipeline_with_model,\n data_X,\n data_y,\n cv=cv,\n groups=groups,\n scoring=metrics_dict,\n fit_params=fit_kwargs,\n n_jobs=n_jobs,\n return_train_score=False,\n error_score=0,\n )\n model_fit_end = time.time()\n model_fit_time = np.array(model_fit_end - model_fit_start).round(2)\n\n score_dict = {\n v.display_name: scores[f\"test_{k}\"] * (1 if v.greater_is_better else -1)\n for k, v in metrics.items()\n }\n\n logger.info(\"Calculating mean and std\")\n\n avgs_dict = {k: [np.mean(v), np.std(v)] for k, v in score_dict.items()}\n\n display.move_progress()\n\n logger.info(\"Creating metrics dataframe\")\n\n model_results = pd.DataFrame(score_dict)\n model_avgs = pd.DataFrame(avgs_dict, index=[\"Mean\", \"SD\"],)\n\n model_results = model_results.append(model_avgs)\n model_results = model_results.round(round)\n\n # yellow the mean\n model_results = color_df(model_results, \"yellow\", [\"Mean\"], axis=1)\n model_results = model_results.set_precision(round)\n\n if refit:\n # refitting the model on complete X_train, y_train\n display.update_monitor(1, \"Finalizing Model\")\n display.display_monitor()\n model_fit_start = time.time()\n logger.info(\"Finalizing model\")\n with io.capture_output():\n pipeline_with_model.fit(data_X, data_y, **fit_kwargs)\n model_fit_end = time.time()\n\n model_fit_time = np.array(model_fit_end - model_fit_start).round(2)\n else:\n model_fit_time /= _get_cv_n_folds(cv, data_X, y=data_y, groups=groups)\n\n # end runtime\n runtime_end = time.time()\n runtime = np.array(runtime_end - runtime_start).round(2)\n\n # mlflow logging\n if logging_param and system and refit:\n\n avgs_dict_log = avgs_dict.copy()\n avgs_dict_log = {k: v[0] for k, v in avgs_dict_log.items()}\n\n try:\n _mlflow_log_model(\n model=model,\n model_results=model_results,\n score_dict=avgs_dict_log,\n source=\"create_model\",\n runtime=runtime,\n model_fit_time=model_fit_time,\n _prep_pipe=prep_pipe,\n log_plots=log_plots_param,\n display=display,\n )\n except:\n logger.error(f\"_mlflow_log_model() for {model} raised an exception:\")\n logger.error(traceback.format_exc())\n\n display.move_progress()\n\n logger.info(\"Uploading results into container\")\n\n # storing results in create_model_container\n create_model_container.append(model_results.data)\n display_container.append(model_results.data)\n\n # storing results in master_model_container\n logger.info(\"Uploading model into container now\")\n master_model_container.append(model)\n\n display.display(model_results, clear=system, override=False if not system else None)\n\n logger.info(f\"create_model_container: {len(create_model_container)}\")\n logger.info(f\"master_model_container: {len(master_model_container)}\")\n logger.info(f\"display_container: {len(display_container)}\")\n\n logger.info(str(model))\n logger.info(\n \"create_model() succesfully completed......................................\"\n )\n gc.collect()\n\n if not system:\n return (model, model_fit_time)\n\n return model\n\n\ndef tune_model_unsupervised(\n model,\n supervised_target: str,\n supervised_type: Optional[str] = None,\n supervised_estimator: Union[str, Any] = \"lr\",\n optimize: Optional[str] = None,\n custom_grid: Optional[List[int]] = None,\n fold: Optional[Union[int, Any]] = None,\n groups: Optional[Union[str, Any]] = None,\n ground_truth: Optional[str] = None,\n method: str = \"drop\",\n fit_kwargs: Optional[dict] = None,\n round: int = 4,\n verbose: bool = True,\n display: Optional[Display] = None,\n **kwargs,\n):\n\n function_params_str = \", \".join([f\"{k}={v}\" for k, v in locals().items()])\n\n logger = get_logger()\n\n logger.info(\"Initializing tune_model()\")\n logger.info(f\"tune_model({function_params_str})\")\n\n logger.info(\"Checking exceptions\")\n\n # run_time\n runtime_start = time.time()\n\n if not fit_kwargs:\n fit_kwargs = {}\n\n if supervised_target not in data_before_preprocess.columns:\n raise ValueError(\n f\"{supervised_target} is not present as a column in the dataset.\"\n )\n\n warnings.filterwarnings(\"ignore\")\n\n np.random.seed(seed)\n\n cols_to_drop = [x for x in X.columns if x.startswith(supervised_target)]\n data_X = X.drop(cols_to_drop, axis=1)\n data_y = data_before_preprocess[[supervised_target]]\n if data_y.dtypes[0] not in [int, float, bool]:\n data_y[supervised_target] = LabelEncoder().fit_transform(\n data_y[supervised_target]\n )\n data_y = data_y[supervised_target]\n\n temp_globals = globals()\n temp_globals[\"y_train\"] = data_y\n\n if supervised_type is None:\n supervised_type, _ = infer_ml_usecase(data_y)\n logger.info(f\"supervised_type inferred as {supervised_type}\")\n\n if supervised_type == \"classification\":\n metrics = pycaret.containers.metrics.classification.get_all_metric_containers(\n temp_globals, raise_errors=True\n )\n available_estimators = pycaret.containers.models.classification.get_all_model_containers(\n temp_globals, raise_errors=True\n )\n ml_usecase = MLUsecase.CLASSIFICATION\n elif supervised_type == \"regression\":\n metrics = pycaret.containers.metrics.regression.get_all_metric_containers(\n temp_globals, raise_errors=True\n )\n available_estimators = pycaret.containers.models.regression.get_all_model_containers(\n temp_globals, raise_errors=True\n )\n ml_usecase = MLUsecase.REGRESSION\n else:\n raise ValueError(\n f\"supervised_type param must be either 'classification' or 'regression'.\"\n )\n\n fold = _get_cv_splitter(fold, ml_usecase)\n\n if isinstance(supervised_estimator, str):\n if supervised_estimator in available_estimators:\n estimator_definition = available_estimators[supervised_estimator]\n estimator_args = estimator_definition.args\n estimator_args = {**estimator_args}\n supervised_estimator = estimator_definition.class_def(**estimator_args)\n else:\n raise ValueError(f\"Unknown supervised_estimator {supervised_estimator}.\")\n else:\n logger.info(\"Declaring custom model\")\n\n supervised_estimator = clone(supervised_estimator)\n\n supervised_estimator_name = _get_model_name(\n supervised_estimator, models=available_estimators\n )\n\n if optimize is None:\n optimize = \"Accuracy\" if supervised_type == \"classification\" else \"R2\"\n optimize = _get_metric(optimize, metrics=metrics)\n if optimize is None:\n raise ValueError(\n \"Optimize method not supported. See docstring for list of available parameters.\"\n )\n\n if custom_grid is not None and not isinstance(custom_grid, list):\n raise ValueError(f\"custom_grid param must be a list.\")\n\n # checking round parameter\n if type(round) is not int:\n raise TypeError(\"Round parameter only accepts integer value.\")\n\n # checking verbose parameter\n if type(verbose) is not bool:\n raise TypeError(\"Verbose parameter can only take argument as True or False.\")\n\n if custom_grid is None:\n if _ml_usecase == MLUsecase.CLUSTERING:\n param_grid = [2, 4, 5, 6, 8, 10, 14, 18, 25, 30, 40]\n else:\n param_grid = [0.01, 0.02, 0.03, 0.04, 0.05, 0.06, 0.07, 0.08, 0.09, 0.10]\n else:\n param_grid = custom_grid\n try:\n param_grid.remove(0)\n except ValueError:\n pass\n param_grid.sort()\n\n if not display:\n progress_args = {\"max\": len(param_grid) * 3 + (len(param_grid) + 1) * 4}\n master_display_columns = None\n timestampStr = datetime.datetime.now().strftime(\"%H:%M:%S\")\n monitor_rows = [\n [\"Initiated\", \". . . . . . . . . . . . . . . . . .\", timestampStr],\n [\"Status\", \". . . . . . . . . . . . . . . . . .\", \"Loading Dependencies\"],\n [\"Estimator\", \". . . . . . . . . . . . . . . . . .\", \"Compiling Library\"],\n ]\n display = Display(\n verbose=verbose,\n html_param=html_param,\n progress_args=progress_args,\n master_display_columns=master_display_columns,\n monitor_rows=monitor_rows,\n )\n\n display.display_progress()\n display.display_monitor()\n display.display_master_display()\n\n unsupervised_models = {}\n unsupervised_models_results = {}\n unsupervised_grids = {0: data_X}\n\n logger.info(\"Fitting unsupervised models\")\n\n for k in param_grid:\n if _ml_usecase == MLUsecase.CLUSTERING:\n try:\n new_model, _ = create_model_unsupervised(\n model,\n num_clusters=k,\n X_data=data_X,\n display=display,\n system=False,\n ground_truth=ground_truth,\n round=round,\n fit_kwargs=fit_kwargs,\n raise_num_clusters=True,\n **kwargs,\n )\n except ValueError:\n raise ValueError(\n f\"Model {model} cannot be used in this function as its number of clusters cannot be set (n_clusters param required).\"\n )\n else:\n new_model, _ = create_model_unsupervised(\n model,\n fraction=k,\n X_data=data_X,\n display=display,\n system=False,\n ground_truth=ground_truth,\n round=round,\n fit_kwargs=fit_kwargs,\n **kwargs,\n )\n unsupervised_models_results[k] = pull(pop=True)\n unsupervised_models[k] = new_model\n unsupervised_grids[k] = (\n assign_model(new_model, verbose=False, transformation=True)\n .reset_index(drop=True)\n .drop(cols_to_drop, axis=1)\n )\n if _ml_usecase == MLUsecase.CLUSTERING:\n unsupervised_grids[k] = pd.get_dummies(\n unsupervised_grids[k], columns=[\"Cluster\"],\n )\n elif method == \"drop\":\n unsupervised_grids[k] = unsupervised_grids[k][\n unsupervised_grids[k][\"Anomaly\"] == 0\n ].drop([\"Anomaly\", \"Anomaly_Score\"], axis=1)\n\n results = {}\n\n logger.info(\"Fitting supervised estimator\")\n\n for k, v in unsupervised_grids.items():\n create_model_supervised(\n supervised_estimator,\n fold=fold,\n display=display,\n system=False,\n X_train_data=v,\n y_train_data=data_y[data_y.index.isin(v.index)],\n metrics=metrics,\n groups=groups,\n round=round,\n refit=False,\n )\n results[k] = pull(pop=True).loc[\"Mean\"]\n display.move_progress()\n\n logger.info(\"Compiling results\")\n\n results = pd.DataFrame(results).T\n\n greater_is_worse_columns = {\n v.display_name for k, v in metrics.items() if not v.greater_is_better\n }\n\n best_model_idx = (\n results.drop(0)\n .sort_values(\n by=optimize.display_name, ascending=optimize in greater_is_worse_columns\n )\n .index[0]\n )\n\n def highlight_max(s):\n to_highlight = s == s.max()\n return [\"background-color: yellow\" if v else \"\" for v in to_highlight]\n\n def highlight_min(s):\n to_highlight = s == s.min()\n return [\"background-color: yellow\" if v else \"\" for v in to_highlight]\n\n results = results.style.apply(\n highlight_max,\n subset=[x for x in results.columns if x not in greater_is_worse_columns],\n ).apply(\n highlight_min,\n subset=[x for x in results.columns if x in greater_is_worse_columns],\n )\n\n # end runtime\n runtime_end = time.time()\n runtime = np.array(runtime_end - runtime_start).round(2)\n\n if _ml_usecase == MLUsecase.CLUSTERING:\n best_model, best_model_fit_time = create_model_unsupervised(\n unsupervised_models[best_model_idx],\n num_clusters=best_model_idx,\n system=False,\n round=round,\n ground_truth=ground_truth,\n fit_kwargs=fit_kwargs,\n display=display,\n **kwargs,\n )\n else:\n best_model, best_model_fit_time = create_model_unsupervised(\n unsupervised_models[best_model_idx],\n fraction=best_model_idx,\n system=False,\n round=round,\n fit_kwargs=fit_kwargs,\n display=display,\n **kwargs,\n )\n best_model_results = pull(pop=True)\n\n if logging_param:\n\n metrics_log = {k: v[0] for k, v in best_model_results.items()}\n\n try:\n _mlflow_log_model(\n model=model,\n model_results=None,\n score_dict=metrics_log,\n source=\"tune_model\",\n runtime=runtime,\n model_fit_time=best_model_fit_time,\n _prep_pipe=prep_pipe,\n log_plots=log_plots_param,\n display=display,\n )\n except:\n logger.error(f\"_mlflow_log_model() for {model} raised an exception:\")\n logger.error(traceback.format_exc())\n\n results = results.set_precision(round)\n display_container.append(results)\n\n display.display(results, clear=True)\n\n if html_param and verbose:\n logger.info(\"Rendering Visual\")\n plot_df = results.data.drop(\n [x for x in results.columns if x != optimize.display_name], axis=1\n )\n\n fig = go.Figure()\n fig.add_trace(\n go.Scatter(\n x=plot_df.index,\n y=plot_df[optimize.display_name],\n mode=\"lines+markers\",\n name=optimize.display_name,\n )\n )\n msg = (\n \"Number of Clusters\"\n if _ml_usecase == MLUsecase.CLUSTERING\n else \"Anomaly Fraction\"\n )\n title = f\"{supervised_estimator_name} Metrics and {msg} by {_get_model_name(best_model)}\"\n fig.update_layout(\n plot_bgcolor=\"rgb(245,245,245)\",\n title={\n \"text\": title,\n \"y\": 0.95,\n \"x\": 0.45,\n \"xanchor\": \"center\",\n \"yanchor\": \"top\",\n },\n xaxis_title=msg,\n yaxis_title=optimize.display_name,\n )\n fig.show()\n logger.info(\"Visual Rendered Successfully\")\n\n logger.info(f\"create_model_container: {len(create_model_container)}\")\n logger.info(f\"master_model_container: {len(master_model_container)}\")\n logger.info(f\"display_container: {len(display_container)}\")\n\n logger.info(str(best_model))\n logger.info(\n \"tune_model() succesfully completed......................................\"\n )\n\n gc.collect()\n\n return best_model\n\n\ndef tune_model_supervised(\n estimator,\n fold: Optional[Union[int, Any]] = None,\n round: int = 4,\n n_iter: int = 10,\n custom_grid: Optional[Union[Dict[str, list], Any]] = None,\n optimize: str = \"Accuracy\",\n custom_scorer=None, # added in pycaret==2.1 - depreciated\n search_library: str = \"scikit-learn\",\n search_algorithm: Optional[str] = None,\n early_stopping: Any = False,\n early_stopping_max_iters: int = 10,\n choose_better: bool = False,\n fit_kwargs: Optional[dict] = None,\n groups: Optional[Union[str, Any]] = None,\n return_tuner: bool = False,\n verbose: bool = True,\n tuner_verbose: Union[int, bool] = True,\n display: Optional[Display] = None,\n **kwargs,\n) -> Any:\n\n \"\"\"\n This function tunes the hyperparameters of a model and scores it using Cross Validation.\n The output prints a score grid that shows Accuracy, AUC, Recall\n Precision, F1, Kappa and MCC by fold (by default = 10 Folds).\n\n This function returns a trained model object.\n\n Example\n -------\n >>> from pycaret.datasets import get_data\n >>> juice = get_data('juice')\n >>> experiment_name = setup(data = juice, target = 'Purchase')\n >>> xgboost = create_model('xgboost')\n >>> tuned_xgboost = tune_model(xgboost)\n\n This will tune the hyperparameters of Extreme Gradient Boosting Classifier.\n\n\n Parameters\n ----------\n estimator : object, default = None\n\n fold: integer or scikit-learn compatible CV generator, default = None\n Controls cross-validation. If None, will use the CV generator defined in setup().\n If integer, will use KFold CV with that many folds.\n When cross_validation is False, this parameter is ignored.\n\n round: integer, default = 4\n Number of decimal places the metrics in the score grid will be rounded to.\n\n n_iter: integer, default = 10\n Number of iterations within the Random Grid Search. For every iteration,\n the model randomly selects one value from the pre-defined grid of\n hyperparameters.\n\n custom_grid: dictionary, default = None\n To use custom hyperparameters for tuning pass a dictionary with parameter name\n and values to be iterated. When set to None it uses pre-defined tuning grid.\n Custom grids must be in a format supported by the chosen search library.\n\n optimize: str, default = 'Accuracy'\n Measure used to select the best model through hyperparameter tuning.\n Can be either a string representing a metric or a custom scorer object\n created using sklearn.make_scorer.\n\n custom_scorer: object, default = None\n Will be eventually depreciated.\n custom_scorer can be passed to tune hyperparameters of the model. It must be\n created using sklearn.make_scorer.\n\n search_library: str, default = 'scikit-learn'\n The search library used to tune hyperparameters.\n Possible values:\n\n - 'scikit-learn' - default, requires no further installation\n - 'scikit-optimize' - scikit-optimize. ``pip install scikit-optimize`` https://scikit-optimize.github.io/stable/\n - 'tune-sklearn' - Ray Tune scikit API. Does not support GPU models.\n ``pip install tune-sklearn ray[tune]`` https://github.com/ray-project/tune-sklearn\n - 'optuna' - Optuna. ``pip install optuna`` https://optuna.org/\n\n search_algorithm: str, default = None\n The search algorithm to be used for finding the best hyperparameters.\n Selection of search algorithms depends on the search_library parameter.\n Some search algorithms require additional libraries to be installed.\n If None, will use search library-specific default algorith.\n 'scikit-learn' possible values:\n\n - 'random' - random grid search (default)\n - 'grid' - grid search\n\n 'scikit-optimize' possible values:\n\n - 'bayesian' - Bayesian search (default)\n\n 'tune-sklearn' possible values:\n\n - 'random' - random grid search (default)\n - 'grid' - grid search\n - 'bayesian' - Bayesian search using scikit-optimize\n ``pip install scikit-optimize``\n - 'hyperopt' - Tree-structured Parzen Estimator search using Hyperopt\n ``pip install hyperopt``\n - 'optuna' - Tree-structured Parzen Estimator search using Optuna\n ``pip install optuna``\n - 'bohb' - Bayesian search using HpBandSter\n ``pip install hpbandster ConfigSpace``\n\n 'optuna' possible values:\n\n - 'random' - randomized search\n - 'tpe' - Tree-structured Parzen Estimator search (default)\n\n early_stopping: bool or str or object, default = False\n Use early stopping to stop fitting to a hyperparameter configuration\n if it performs poorly. Ignored if search_library is ``scikit-learn``, or\n if the estimator doesn't have partial_fit attribute.\n If False or None, early stopping will not be used.\n Can be either an object accepted by the search library or one of the\n following:\n\n - 'asha' for Asynchronous Successive Halving Algorithm\n - 'hyperband' for Hyperband\n - 'median' for median stopping rule\n - If False or None, early stopping will not be used.\n\n More info for Optuna - https://optuna.readthedocs.io/en/stable/reference/pruners.html\n More info for Ray Tune (tune-sklearn) - https://docs.ray.io/en/master/tune/api_docs/schedulers.html\n\n early_stopping_max_iters: int, default = 10\n Maximum number of epochs to run for each sampled configuration.\n Ignored if early_stopping is False or None.\n\n choose_better: bool, default = False\n When set to set to True, base estimator is returned when the performance doesn't\n improve by tune_model. This gurantees the returned object would perform atleast\n equivalent to base estimator created using create_model or model returned by\n compare_models.\n\n fit_kwargs: dict, default = {} (empty dict)\n Dictionary of arguments passed to the fit method of the tuner.\n\n groups: str or array-like, with shape (n_samples,), default = None\n Optional Group labels for the samples used while splitting the dataset into train/test set.\n If string is passed, will use the data column with that name as the groups.\n Only used if a group based cross-validation generator is used (eg. GroupKFold).\n If None, will use the value set in fold_groups param in setup().\n\n return_tuner: bool, default = False\n If True, will reutrn a tuple of (model, tuner_object). Otherwise,\n will return just the best model.\n\n verbose: bool, default = True\n Score grid is not printed when verbose is set to False.\n\n tuner_verbose: bool or in, default = True\n If True or above 0, will print messages from the tuner. Higher values\n print more messages. Ignored if verbose param is False.\n\n **kwargs:\n Additional keyword arguments to pass to the optimizer.\n\n Returns\n -------\n score_grid\n A table containing the scores of the model across the kfolds.\n Scoring metrics used are Accuracy, AUC, Recall, Precision, F1,\n Kappa and MCC. Mean and standard deviation of the scores across\n the folds are also returned.\n\n model\n Trained and tuned model object.\n\n tuner_object\n Only if return_tuner param is True. The object used for tuning.\n\n Notes\n -----\n\n - If a StackingClassifier is passed, the hyperparameters of the meta model (final_estimator)\n will be tuned.\n\n - If a VotingClassifier is passed, the weights will be tuned.\n\n Warnings\n --------\n\n - Using 'Grid' search algorithm with default parameter grids may result in very\n long computation.\n\n\n \"\"\"\n function_params_str = \", \".join([f\"{k}={v}\" for k, v in locals().items()])\n\n logger = get_logger()\n\n logger.info(\"Initializing tune_model()\")\n logger.info(f\"tune_model({function_params_str})\")\n\n logger.info(\"Checking exceptions\")\n\n # run_time\n runtime_start = time.time()\n\n if not fit_kwargs:\n fit_kwargs = {}\n\n # checking estimator if string\n if type(estimator) is str:\n raise TypeError(\n \"The behavior of tune_model in version 1.0.1 is changed. Please pass trained model object.\"\n )\n\n # Check for estimator\n if not hasattr(estimator, \"fit\"):\n raise ValueError(\n f\"Estimator {estimator} does not have the required fit() method.\"\n )\n\n # checking fold parameter\n if fold is not None and not (type(fold) is int or is_sklearn_cv_generator(fold)):\n raise TypeError(\n \"fold parameter must be either None, an integer or a scikit-learn compatible CV generator object.\"\n )\n\n # checking round parameter\n if type(round) is not int:\n raise TypeError(\"Round parameter only accepts integer value.\")\n\n # checking n_iter parameter\n if type(n_iter) is not int:\n raise TypeError(\"n_iter parameter only accepts integer value.\")\n\n # checking early_stopping parameter\n possible_early_stopping = [\"asha\", \"Hyperband\", \"Median\"]\n if (\n isinstance(early_stopping, str)\n and early_stopping not in possible_early_stopping\n ):\n raise TypeError(\n f\"early_stopping parameter must be one of {', '.join(possible_early_stopping)}\"\n )\n\n # checking early_stopping_max_iters parameter\n if type(early_stopping_max_iters) is not int:\n raise TypeError(\n \"early_stopping_max_iters parameter only accepts integer value.\"\n )\n\n # checking search_library parameter\n possible_search_libraries = [\n \"scikit-learn\",\n \"scikit-optimize\",\n \"tune-sklearn\",\n \"optuna\",\n ]\n search_library = search_library.lower()\n if search_library not in possible_search_libraries:\n raise ValueError(\n f\"search_library parameter must be one of {', '.join(possible_search_libraries)}\"\n )\n\n if search_library == \"scikit-optimize\":\n try:\n import skopt\n except ImportError:\n raise ImportError(\n \"'scikit-optimize' requires scikit-optimize package to be installed. Do: pip install scikit-optimize\"\n )\n\n if not search_algorithm:\n search_algorithm = \"bayesian\"\n\n possible_search_algorithms = [\"bayesian\"]\n if search_algorithm not in possible_search_algorithms:\n raise ValueError(\n f\"For 'scikit-optimize' search_algorithm parameter must be one of {', '.join(possible_search_algorithms)}\"\n )\n\n elif search_library == \"tune-sklearn\":\n try:\n import tune_sklearn\n except ImportError:\n raise ImportError(\n \"'tune-sklearn' requires tune_sklearn package to be installed. Do: pip install tune-sklearn ray[tune]\"\n )\n\n if not search_algorithm:\n search_algorithm = \"random\"\n\n possible_search_algorithms = [\n \"random\",\n \"grid\",\n \"bayesian\",\n \"hyperopt\",\n \"bohb\",\n \"optuna\",\n ]\n if search_algorithm not in possible_search_algorithms:\n raise ValueError(\n f\"For 'tune-sklearn' search_algorithm parameter must be one of {', '.join(possible_search_algorithms)}\"\n )\n\n if search_algorithm == \"bohb\":\n try:\n from ray.tune.suggest.bohb import TuneBOHB\n from ray.tune.schedulers import HyperBandForBOHB\n import ConfigSpace as CS\n import hpbandster\n except ImportError:\n raise ImportError(\n \"It appears that either HpBandSter or ConfigSpace is not installed. Do: pip install hpbandster ConfigSpace\"\n )\n elif search_algorithm == \"hyperopt\":\n try:\n from ray.tune.suggest.hyperopt import HyperOptSearch\n from hyperopt import hp\n except ImportError:\n raise ImportError(\n \"It appears that hyperopt is not installed. Do: pip install hyperopt\"\n )\n elif search_algorithm == \"bayesian\":\n try:\n import skopt\n except ImportError:\n raise ImportError(\n \"It appears that scikit-optimize is not installed. Do: pip install scikit-optimize\"\n )\n elif search_algorithm == \"optuna\":\n try:\n import optuna\n except ImportError:\n raise ImportError(\n \"'optuna' requires optuna package to be installed. Do: pip install optuna\"\n )\n\n elif search_library == \"optuna\":\n try:\n import optuna\n except ImportError:\n raise ImportError(\n \"'optuna' requires optuna package to be installed. Do: pip install optuna\"\n )\n\n if not search_algorithm:\n search_algorithm = \"tpe\"\n\n possible_search_algorithms = [\"random\", \"tpe\"]\n if search_algorithm not in possible_search_algorithms:\n raise ValueError(\n f\"For 'optuna' search_algorithm parameter must be one of {', '.join(possible_search_algorithms)}\"\n )\n else:\n if not search_algorithm:\n search_algorithm = \"random\"\n\n possible_search_algorithms = [\"random\", \"grid\"]\n if search_algorithm not in possible_search_algorithms:\n raise ValueError(\n f\"For 'scikit-learn' search_algorithm parameter must be one of {', '.join(possible_search_algorithms)}\"\n )\n\n if custom_scorer is not None:\n optimize = custom_scorer\n warnings.warn(\n \"custom_scorer parameter will be depreciated, use optimize instead\",\n DeprecationWarning,\n stacklevel=2,\n )\n\n if isinstance(optimize, str):\n # checking optimize parameter\n optimize = _get_metric(optimize)\n if optimize is None:\n raise ValueError(\n \"Optimize method not supported. See docstring for list of available parameters.\"\n )\n\n # checking optimize parameter for multiclass\n if _is_multiclass():\n if not optimize.is_multiclass:\n raise TypeError(\n \"Optimization metric not supported for multiclass problems. See docstring for list of other optimization parameters.\"\n )\n else:\n logger.info(f\"optimize set to user defined function {optimize}\")\n\n # checking verbose parameter\n if type(verbose) is not bool:\n raise TypeError(\"verbose parameter can only take argument as True or False.\")\n\n # checking verbose parameter\n if type(return_tuner) is not bool:\n raise TypeError(\n \"return_tuner parameter can only take argument as True or False.\"\n )\n\n if not verbose:\n tuner_verbose = 0\n\n if type(tuner_verbose) not in (bool, int):\n raise TypeError(\"tuner_verbose parameter must be a bool or an int.\")\n\n tuner_verbose = int(tuner_verbose)\n\n if tuner_verbose < 0:\n tuner_verbose = 0\n elif tuner_verbose > 2:\n tuner_verbose = 2\n\n \"\"\"\n\n ERROR HANDLING ENDS HERE\n\n \"\"\"\n\n fold = _get_cv_splitter(fold)\n\n groups = _get_groups(groups)\n\n if not display:\n progress_args = {\"max\": 3 + 4}\n master_display_columns = [v.display_name for k, v in _all_metrics.items()]\n timestampStr = datetime.datetime.now().strftime(\"%H:%M:%S\")\n monitor_rows = [\n [\"Initiated\", \". . . . . . . . . . . . . . . . . .\", timestampStr],\n [\"Status\", \". . . . . . . . . . . . . . . . . .\", \"Loading Dependencies\"],\n [\"Estimator\", \". . . . . . . . . . . . . . . . . .\", \"Compiling Library\"],\n ]\n display = Display(\n verbose=verbose,\n html_param=html_param,\n progress_args=progress_args,\n master_display_columns=master_display_columns,\n monitor_rows=monitor_rows,\n )\n\n display.display_progress()\n display.display_monitor()\n display.display_master_display()\n\n # ignore warnings\n\n warnings.filterwarnings(\"ignore\")\n\n import logging\n\n np.random.seed(seed)\n\n logger.info(\"Copying training dataset\")\n # Storing X_train and y_train in data_X and data_y parameter\n data_X = X_train.copy()\n data_y = y_train.copy()\n\n # reset index\n data_X.reset_index(drop=True, inplace=True)\n data_y.reset_index(drop=True, inplace=True)\n\n display.move_progress()\n\n # setting optimize parameter\n\n compare_dimension = optimize.display_name\n optimize = optimize.scorer\n\n # convert trained estimator into string name for grids\n\n logger.info(\"Checking base model\")\n\n is_stacked_model = False\n\n if hasattr(estimator, \"final_estimator\"):\n logger.info(\"Model is stacked, using the definition of the meta-model\")\n is_stacked_model = True\n estimator_id = _get_model_id(estimator.final_estimator)\n else:\n estimator_id = _get_model_id(estimator)\n if estimator_id is None:\n if custom_grid is None:\n raise ValueError(\n \"When passing a model not in PyCaret's model library, the custom_grid parameter must be provided.\"\n )\n estimator_name = _get_model_name(estimator)\n estimator_definition = None\n logger.info(\"A custom model has been passed\")\n else:\n estimator_definition = _all_models_internal[estimator_id]\n estimator_name = estimator_definition.name\n logger.info(f\"Base model : {estimator_name}\")\n\n if estimator_definition is None or estimator_definition.tunable is None:\n model = clone(estimator)\n else:\n logger.info(\"Model has a special tunable class, using that\")\n if is_stacked_model:\n model = clone(estimator)\n model.set_params(\n final_estimator=estimator_definition.tunable(**estimator.get_params())\n )\n else:\n model = clone(estimator_definition.tunable(**estimator.get_params()))\n\n base_estimator = model\n\n if is_stacked_model:\n base_estimator = model.final_estimator\n\n display.update_monitor(2, estimator_name)\n display.display_monitor()\n\n display.move_progress()\n\n logger.info(\"Declaring metric variables\")\n\n \"\"\"\n MONITOR UPDATE STARTS\n \"\"\"\n\n display.update_monitor(1, \"Searching Hyperparameters\")\n display.display_monitor()\n\n \"\"\"\n MONITOR UPDATE ENDS\n \"\"\"\n\n logger.info(\"Defining Hyperparameters\")\n\n from pycaret.internal.tunable import VotingClassifier, VotingRegressor\n\n def total_combintaions_in_grid(grid):\n nc = 1\n\n def get_iter(x):\n if isinstance(x, dict):\n return x.values()\n return x\n\n for v in get_iter(grid):\n if isinstance(v, dict):\n for v2 in get_iter(v):\n nc *= len(v2)\n else:\n nc *= len(v)\n return nc\n\n def get_ccp_alphas(estimator):\n path = estimator.cost_complexity_pruning_path(X_train, y_train)\n ccp_alphas, impurities = path.ccp_alphas, path.impurities\n return list(ccp_alphas[:-1])\n\n if custom_grid is not None:\n if not isinstance(custom_grid, dict):\n raise TypeError(f\"custom_grid must be a dict, got {type(custom_grid)}.\")\n param_grid = custom_grid\n if not (\n search_library == \"scikit-learn\"\n or (\n search_library == \"tune-sklearn\"\n and (search_algorithm == \"grid\" or search_algorithm == \"random\")\n )\n ):\n param_grid = {\n k: CategoricalDistribution(v) if not isinstance(v, Distribution) else v\n for k, v in param_grid.items()\n }\n elif any(isinstance(v, Distribution) for k, v in param_grid.items()):\n raise TypeError(\n f\"For the combination of search_library {search_library} and search_algorithm {search_algorithm}, PyCaret Distribution objects are not supported. Pass a list or other object supported by the search library (in most cases, an object with a 'rvs' function).\"\n )\n elif search_library == \"scikit-learn\" or (\n search_library == \"tune-sklearn\"\n and (search_algorithm == \"grid\" or search_algorithm == \"random\")\n ):\n param_grid = estimator_definition.tune_grid\n if isinstance(base_estimator, (VotingClassifier, VotingRegressor)):\n # special case to handle VotingClassifier, as weights need to be\n # generated dynamically\n param_grid = {\n f\"weight_{i}\": np.arange(0.01, 1, 0.01)\n for i, e in enumerate(base_estimator.estimators)\n }\n # if hasattr(base_estimator, \"cost_complexity_pruning_path\"):\n # # special case for Tree-based models\n # param_grid[\"ccp_alpha\"] = get_ccp_alphas(base_estimator)\n # if \"min_impurity_decrease\" in param_grid:\n # param_grid.pop(\"min_impurity_decrease\")\n\n if search_algorithm != \"grid\":\n tc = total_combintaions_in_grid(param_grid)\n if tc <= n_iter:\n logger.info(\n f\"{n_iter} is bigger than total combinations {tc}, setting search algorithm to grid\"\n )\n search_algorithm = \"grid\"\n else:\n param_grid = estimator_definition.tune_distribution\n\n if isinstance(base_estimator, (VotingClassifier, VotingRegressor)):\n # special case to handle VotingClassifier, as weights need to be\n # generated dynamically\n param_grid = {\n f\"weight_{i}\": UniformDistribution(0.000000001, 1)\n for i, e in enumerate(base_estimator.estimators)\n }\n # if hasattr(base_estimator, \"cost_complexity_pruning_path\"):\n # # special case for Tree-based models\n # param_grid[\"ccp_alpha\"] = CategoricalDistribution(\n # get_ccp_alphas(base_estimator)\n # )\n # if \"min_impurity_decrease\" in param_grid:\n # param_grid.pop(\"min_impurity_decrease\")\n\n if not param_grid:\n raise ValueError(\n \"parameter grid for tuning is empty. If passing custom_grid, make sure that it is not empty. If not passing custom_grid, the passed estimator does not have a built-in tuning grid.\"\n )\n\n suffixes = []\n\n if is_stacked_model:\n logger.info(\"Stacked model passed, will tune meta model hyperparameters\")\n suffixes.append(\"final_estimator\")\n\n gc.collect()\n\n with estimator_pipeline(_internal_pipeline, model) as pipeline_with_model:\n extra_params = {}\n\n fit_kwargs = _get_pipeline_fit_kwargs(pipeline_with_model, fit_kwargs)\n\n actual_estimator_label = get_pipeline_estimator_label(pipeline_with_model)\n\n suffixes.append(actual_estimator_label)\n\n suffixes = \"__\".join(reversed(suffixes))\n\n param_grid = {f\"{suffixes}__{k}\": v for k, v in param_grid.items()}\n\n if estimator_definition is not None:\n search_kwargs = {**estimator_definition.tune_args, **kwargs}\n n_jobs = (\n _gpu_n_jobs_param\n if estimator_definition.is_gpu_enabled\n else n_jobs_param\n )\n else:\n search_kwargs = {}\n n_jobs = n_jobs_param\n\n if custom_grid is not None:\n logger.info(f\"custom_grid: {param_grid}\")\n\n from sklearn.gaussian_process import GaussianProcessClassifier\n\n # special case to prevent running out of memory\n if isinstance(pipeline_with_model.steps[-1][1], GaussianProcessClassifier):\n n_jobs = 1\n\n logger.info(f\"Tuning with n_jobs={n_jobs}\")\n\n if search_library == \"optuna\":\n # suppress output\n logging.getLogger(\"optuna\").setLevel(logging.WARNING)\n\n pruner_translator = {\n \"asha\": optuna.pruners.SuccessiveHalvingPruner(),\n \"hyperband\": optuna.pruners.HyperbandPruner(),\n \"median\": optuna.pruners.MedianPruner(),\n False: optuna.pruners.NopPruner(),\n None: optuna.pruners.NopPruner(),\n }\n pruner = early_stopping\n if pruner in pruner_translator:\n pruner = pruner_translator[early_stopping]\n\n sampler_translator = {\n \"tpe\": optuna.samplers.TPESampler(seed=seed),\n \"random\": optuna.samplers.RandomSampler(seed=seed),\n }\n sampler = sampler_translator[search_algorithm]\n\n try:\n param_grid = get_optuna_distributions(param_grid)\n except:\n logger.warning(\n \"Couldn't convert param_grid to specific library distributions. Exception:\"\n )\n logger.warning(traceback.format_exc())\n\n study = optuna.create_study(\n direction=\"maximize\", sampler=sampler, pruner=pruner\n )\n\n logger.info(\"Initializing optuna.integration.OptunaSearchCV\")\n model_grid = optuna.integration.OptunaSearchCV(\n estimator=pipeline_with_model,\n param_distributions=param_grid,\n cv=fold,\n enable_pruning=early_stopping\n and can_early_stop(pipeline_with_model, True, False, False, param_grid),\n max_iter=early_stopping_max_iters,\n n_jobs=1, ## to have reproducible results see: https://github.com/optuna/optuna/issues/2540\n n_trials=n_iter,\n random_state=seed,\n scoring=optimize,\n study=study,\n refit=False,\n verbose=tuner_verbose,\n error_score=\"raise\",\n **search_kwargs,\n )\n\n elif search_library == \"tune-sklearn\":\n\n early_stopping_translator = {\n \"asha\": \"ASHAScheduler\",\n \"hyperband\": \"HyperBandScheduler\",\n \"median\": \"MedianStoppingRule\",\n }\n if early_stopping in early_stopping_translator:\n early_stopping = early_stopping_translator[early_stopping]\n\n do_early_stop = early_stopping and can_early_stop(\n pipeline_with_model, True, True, True, param_grid\n )\n\n if not do_early_stop and search_algorithm == \"bohb\":\n raise ValueError(\n \"'bohb' requires early_stopping = True and the estimator to support early stopping (has partial_fit, warm_start or is an XGBoost model).\"\n )\n\n elif early_stopping and can_early_stop(\n pipeline_with_model, False, True, False, param_grid\n ):\n if \"actual_estimator__n_estimators\" in param_grid:\n if custom_grid is None:\n extra_params[\n \"actual_estimator__n_estimators\"\n ] = pipeline_with_model.get_params()[\n \"actual_estimator__n_estimators\"\n ]\n param_grid.pop(\"actual_estimator__n_estimators\")\n else:\n raise ValueError(\n \"Param grid cannot contain n_estimators or max_iter if early_stopping is True and the model is warm started. Use early_stopping_max_iters params to set the upper bound of n_estimators or max_iter.\"\n )\n if \"actual_estimator__max_iter\" in param_grid:\n if custom_grid is None:\n param_grid.pop(\"actual_estimator__max_iter\")\n else:\n raise ValueError(\n \"Param grid cannot contain n_estimators or max_iter if early_stopping is True and the model is warm started. Use early_stopping_max_iters params to set the upper bound of n_estimators or max_iter.\"\n )\n\n from tune_sklearn import TuneSearchCV, TuneGridSearchCV\n\n with (\n true_warm_start(pipeline_with_model) if do_early_stop else nullcontext()\n ), set_n_jobs(pipeline_with_model, 1), (\n patch.dict(\"os.environ\", {\"TUNE_GLOBAL_CHECKPOINT_S\": \"1000000\"})\n if \"TUNE_GLOBAL_CHECKPOINT_S\" not in os.environ\n else nullcontext()\n ):\n if search_algorithm == \"grid\":\n\n logger.info(\"Initializing tune_sklearn.TuneGridSearchCV\")\n model_grid = TuneGridSearchCV(\n estimator=pipeline_with_model,\n param_grid=param_grid,\n early_stopping=do_early_stop,\n scoring=optimize,\n cv=fold,\n max_iters=early_stopping_max_iters,\n n_jobs=n_jobs,\n use_gpu=gpu_param,\n refit=False,\n verbose=tuner_verbose,\n pipeline_auto_early_stop=True,\n **search_kwargs,\n )\n else:\n if search_algorithm == \"hyperopt\":\n try:\n param_grid = get_hyperopt_distributions(param_grid)\n except:\n logger.warning(\n \"Couldn't convert param_grid to specific library distributions. Exception:\"\n )\n logger.warning(traceback.format_exc())\n elif search_algorithm == \"bayesian\":\n try:\n param_grid = get_skopt_distributions(param_grid)\n except:\n logger.warning(\n \"Couldn't convert param_grid to specific library distributions. Exception:\"\n )\n logger.warning(traceback.format_exc())\n elif search_algorithm == \"bohb\":\n try:\n param_grid = get_CS_distributions(param_grid)\n except:\n logger.warning(\n \"Couldn't convert param_grid to specific library distributions. Exception:\"\n )\n logger.warning(traceback.format_exc())\n elif search_algorithm != \"random\":\n try:\n param_grid = get_tune_distributions(param_grid)\n except:\n logger.warning(\n \"Couldn't convert param_grid to specific library distributions. Exception:\"\n )\n logger.warning(traceback.format_exc())\n logger.info(\n f\"Initializing tune_sklearn.TuneSearchCV, {search_algorithm}\"\n )\n model_grid = TuneSearchCV(\n estimator=pipeline_with_model,\n search_optimization=search_algorithm,\n param_distributions=param_grid,\n n_trials=n_iter,\n early_stopping=do_early_stop,\n scoring=optimize,\n cv=fold,\n random_state=seed,\n max_iters=early_stopping_max_iters,\n n_jobs=n_jobs,\n use_gpu=gpu_param,\n refit=True,\n verbose=tuner_verbose,\n pipeline_auto_early_stop=True,\n **search_kwargs,\n )\n elif search_library == \"scikit-optimize\":\n import skopt\n\n try:\n param_grid = get_skopt_distributions(param_grid)\n except:\n logger.warning(\n \"Couldn't convert param_grid to specific library distributions. Exception:\"\n )\n logger.warning(traceback.format_exc())\n\n logger.info(\"Initializing skopt.BayesSearchCV\")\n model_grid = skopt.BayesSearchCV(\n estimator=pipeline_with_model,\n search_spaces=param_grid,\n scoring=optimize,\n n_iter=n_iter,\n cv=fold,\n random_state=seed,\n refit=False,\n n_jobs=n_jobs,\n verbose=tuner_verbose,\n **search_kwargs,\n )\n else:\n # needs to be imported like that for the monkeypatch\n import sklearn.model_selection._search\n\n if search_algorithm == \"grid\":\n logger.info(\"Initializing GridSearchCV\")\n model_grid = sklearn.model_selection._search.GridSearchCV(\n estimator=pipeline_with_model,\n param_grid=param_grid,\n scoring=optimize,\n cv=fold,\n refit=False,\n n_jobs=n_jobs,\n verbose=tuner_verbose,\n **search_kwargs,\n )\n else:\n logger.info(\"Initializing RandomizedSearchCV\")\n model_grid = sklearn.model_selection._search.RandomizedSearchCV(\n estimator=pipeline_with_model,\n param_distributions=param_grid,\n scoring=optimize,\n n_iter=n_iter,\n cv=fold,\n random_state=seed,\n refit=False,\n n_jobs=n_jobs,\n verbose=tuner_verbose,\n **search_kwargs,\n )\n\n # with io.capture_output():\n if search_library == \"scikit-learn\":\n # monkey patching to fix overflows on Windows\n with patch(\n \"sklearn.model_selection._search.sample_without_replacement\",\n pycaret.internal.patches.sklearn._mp_sample_without_replacement,\n ), patch(\n \"sklearn.model_selection._search.ParameterGrid.__getitem__\",\n pycaret.internal.patches.sklearn._mp_ParameterGrid_getitem,\n ):\n model_grid.fit(X_train, y_train, groups=groups, **fit_kwargs)\n else:\n model_grid.fit(X_train, y_train, groups=groups, **fit_kwargs)\n best_params = model_grid.best_params_\n logger.info(f\"best_params: {best_params}\")\n best_params = {**best_params, **extra_params}\n best_params = {\n k.replace(f\"{actual_estimator_label}__\", \"\"): v\n for k, v in best_params.items()\n }\n cv_results = None\n try:\n cv_results = model_grid.cv_results_\n except:\n logger.warning(\"Couldn't get cv_results from model_grid. Exception:\")\n logger.warning(traceback.format_exc())\n\n display.move_progress()\n\n logger.info(\"Hyperparameter search completed\")\n\n if isinstance(model, TunableMixin):\n logger.info(\"Getting base sklearn object from tunable\")\n model.set_params(**best_params)\n best_params = {\n k: v\n for k, v in model.get_params().items()\n if k in model.get_base_sklearn_params().keys()\n }\n model = model.get_base_sklearn_object()\n\n logger.info(\"SubProcess create_model() called ==================================\")\n best_model, model_fit_time = create_model_supervised(\n estimator=model,\n system=False,\n display=display,\n fold=fold,\n round=round,\n groups=groups,\n fit_kwargs=fit_kwargs,\n **best_params,\n )\n model_results = pull()\n logger.info(\"SubProcess create_model() end ==================================\")\n\n if choose_better:\n best_model = _choose_better(\n [estimator, (best_model, model_results)],\n compare_dimension,\n fold,\n groups=groups,\n fit_kwargs=fit_kwargs,\n display=display,\n )\n\n # end runtime\n runtime_end = time.time()\n runtime = np.array(runtime_end - runtime_start).round(2)\n\n # mlflow logging\n if logging_param:\n\n avgs_dict_log = {k: v for k, v in model_results.loc[\"Mean\"].items()}\n\n try:\n _mlflow_log_model(\n model=best_model,\n model_results=model_results,\n score_dict=avgs_dict_log,\n source=\"tune_model\",\n runtime=runtime,\n model_fit_time=model_fit_time,\n _prep_pipe=prep_pipe,\n log_plots=log_plots_param,\n tune_cv_results=cv_results,\n display=display,\n )\n except:\n logger.error(f\"_mlflow_log_model() for {best_model} raised an exception:\")\n logger.error(traceback.format_exc())\n\n model_results = color_df(model_results, \"yellow\", [\"Mean\"], axis=1)\n model_results = model_results.set_precision(round)\n display.display(model_results, clear=True)\n\n logger.info(f\"create_model_container: {len(create_model_container)}\")\n logger.info(f\"master_model_container: {len(master_model_container)}\")\n logger.info(f\"display_container: {len(display_container)}\")\n\n logger.info(str(best_model))\n logger.info(\n \"tune_model() succesfully completed......................................\"\n )\n\n gc.collect()\n if return_tuner:\n return (best_model, model_grid)\n return best_model\n\n\ndef ensemble_model(\n estimator,\n method: str = \"Bagging\",\n fold: Optional[Union[int, Any]] = None,\n n_estimators: int = 10,\n round: int = 4,\n choose_better: bool = False,\n optimize: str = \"Accuracy\",\n fit_kwargs: Optional[dict] = None,\n groups: Optional[Union[str, Any]] = None,\n verbose: bool = True,\n display: Optional[Display] = None, # added in pycaret==2.2.0\n) -> Any:\n \"\"\"\n This function ensembles the trained base estimator using the method defined in\n 'method' param (default = 'Bagging'). The output prints a score grid that shows\n Accuracy, AUC, Recall, Precision, F1, Kappa and MCC by fold (default = 10 Fold).\n\n This function returns a trained model object.\n\n Model must be created using create_model() or tune_model().\n\n Example\n -------\n >>> from pycaret.datasets import get_data\n >>> juice = get_data('juice')\n >>> experiment_name = setup(data = juice, target = 'Purchase')\n >>> dt = create_model('dt')\n >>> ensembled_dt = ensemble_model(dt)\n\n This will return an ensembled Decision Tree model using 'Bagging'.\n\n Parameters\n ----------\n estimator : object, default = None\n\n method: str, default = 'Bagging'\n Bagging method will create an ensemble meta-estimator that fits base\n classifiers each on random subsets of the original dataset. The other\n available method is 'Boosting' which will create a meta-estimators by\n fitting a classifier on the original dataset and then fits additional\n copies of the classifier on the same dataset but where the weights of\n incorrectly classified instances are adjusted such that subsequent\n classifiers focus more on difficult cases.\n\n fold: integer or scikit-learn compatible CV generator, default = None\n Controls cross-validation. If None, will use the CV generator defined in setup().\n If integer, will use KFold CV with that many folds.\n When cross_validation is False, this parameter is ignored.\n\n n_estimators: integer, default = 10\n The number of base estimators in the ensemble.\n In case of perfect fit, the learning procedure is stopped early.\n\n round: integer, default = 4\n Number of decimal places the metrics in the score grid will be rounded to.\n\n choose_better: bool, default = False\n When set to set to True, base estimator is returned when the metric doesn't\n improve by ensemble_model. This gurantees the returned object would perform\n atleast equivalent to base estimator created using create_model or model\n returned by compare_models.\n\n optimize: str, default = 'Accuracy'\n Only used when choose_better is set to True. optimize parameter is used\n to compare emsembled model with base estimator. Values accepted in\n optimize parameter are 'Accuracy', 'AUC', 'Recall', 'Precision', 'F1',\n 'Kappa', 'MCC'.\n\n fit_kwargs: dict, default = {} (empty dict)\n Dictionary of arguments passed to the fit method of the model.\n\n groups: str or array-like, with shape (n_samples,), default = None\n Optional Group labels for the samples used while splitting the dataset into train/test set.\n If string is passed, will use the data column with that name as the groups.\n Only used if a group based cross-validation generator is used (eg. GroupKFold).\n If None, will use the value set in fold_groups param in setup().\n\n verbose: bool, default = True\n Score grid is not printed when verbose is set to False.\n\n Returns\n -------\n score_grid\n A table containing the scores of the model across the kfolds.\n Scoring metrics used are Accuracy, AUC, Recall, Precision, F1,\n Kappa and MCC. Mean and standard deviation of the scores across\n the folds are also returned.\n\n model\n Trained ensembled model object.\n\n Warnings\n --------\n - If target variable is multiclass (more than 2 classes), AUC will be returned\n as zero (0.0).\n\n\n \"\"\"\n\n function_params_str = \", \".join([f\"{k}={v}\" for k, v in locals().items()])\n\n logger = get_logger()\n\n logger.info(\"Initializing ensemble_model()\")\n logger.info(f\"ensemble_model({function_params_str})\")\n\n logger.info(\"Checking exceptions\")\n\n # run_time\n runtime_start = time.time()\n\n if not fit_kwargs:\n fit_kwargs = {}\n\n # Check for estimator\n if not hasattr(estimator, \"fit\"):\n raise ValueError(\n f\"Estimator {estimator} does not have the required fit() method.\"\n )\n\n # Check for allowed method\n available_method = [\"Bagging\", \"Boosting\"]\n if method not in available_method:\n raise ValueError(\n \"Method parameter only accepts two values 'Bagging' or 'Boosting'.\"\n )\n\n # check boosting conflict\n if method == \"Boosting\":\n\n boosting_model_definition = _all_models_internal[\"ada\"]\n\n check_model = estimator\n\n try:\n check_model = boosting_model_definition.class_def(\n check_model,\n n_estimators=n_estimators,\n **boosting_model_definition.args,\n )\n with io.capture_output():\n check_model.fit(X_train, y_train)\n except:\n raise TypeError(\n \"Estimator not supported for the Boosting method. Change the estimator or method to 'Bagging'.\"\n )\n\n # checking fold parameter\n if fold is not None and not (type(fold) is int or is_sklearn_cv_generator(fold)):\n raise TypeError(\n \"fold parameter must be either None, an integer or a scikit-learn compatible CV generator object.\"\n )\n\n # checking n_estimators parameter\n if type(n_estimators) is not int:\n raise TypeError(\"n_estimators parameter only accepts integer value.\")\n\n # checking round parameter\n if type(round) is not int:\n raise TypeError(\"Round parameter only accepts integer value.\")\n\n # checking verbose parameter\n if type(verbose) is not bool:\n raise TypeError(\"Verbose parameter can only take argument as True or False.\")\n\n # checking optimize parameter\n optimize = _get_metric(optimize)\n if optimize is None:\n raise ValueError(\n f\"Optimize method not supported. See docstring for list of available parameters.\"\n )\n\n # checking optimize parameter for multiclass\n if _is_multiclass():\n if not optimize.is_multiclass:\n raise TypeError(\n f\"Optimization metric not supported for multiclass problems. See docstring for list of other optimization parameters.\"\n )\n\n \"\"\"\n\n ERROR HANDLING ENDS HERE\n\n \"\"\"\n\n fold = _get_cv_splitter(fold)\n\n groups = _get_groups(groups)\n\n if not display:\n progress_args = {\"max\": 2 + 4}\n master_display_columns = [v.display_name for k, v in _all_metrics.items()]\n timestampStr = datetime.datetime.now().strftime(\"%H:%M:%S\")\n monitor_rows = [\n [\"Initiated\", \". . . . . . . . . . . . . . . . . .\", timestampStr],\n [\"Status\", \". . . . . . . . . . . . . . . . . .\", \"Loading Dependencies\"],\n [\"Estimator\", \". . . . . . . . . . . . . . . . . .\", \"Compiling Library\"],\n ]\n display = Display(\n verbose=verbose,\n html_param=html_param,\n progress_args=progress_args,\n master_display_columns=master_display_columns,\n monitor_rows=monitor_rows,\n )\n\n display.display_progress()\n display.display_monitor()\n display.display_master_display()\n\n logger.info(\"Importing libraries\")\n\n np.random.seed(seed)\n\n logger.info(\"Copying training dataset\")\n\n # Storing X_train and y_train in data_X and data_y parameter\n data_X = X_train.copy()\n data_y = y_train.copy()\n\n # reset index\n data_X.reset_index(drop=True, inplace=True)\n data_y.reset_index(drop=True, inplace=True)\n\n display.move_progress()\n\n # setting optimize parameter\n\n compare_dimension = optimize.display_name\n optimize = optimize.scorer\n\n logger.info(\"Checking base model\")\n\n _estimator_ = estimator\n\n estimator_id = _get_model_id(estimator)\n\n if estimator_id is None:\n estimator_name = _get_model_name(estimator)\n logger.info(\"A custom model has been passed\")\n else:\n estimator_definition = _all_models_internal[estimator_id]\n estimator_name = estimator_definition.name\n\n logger.info(f\"Base model : {estimator_name}\")\n\n display.update_monitor(2, estimator_name)\n display.display_monitor()\n\n \"\"\"\n MONITOR UPDATE STARTS\n \"\"\"\n\n display.update_monitor(1, \"Selecting Estimator\")\n display.display_monitor()\n\n \"\"\"\n MONITOR UPDATE ENDS\n \"\"\"\n\n model = get_estimator_from_meta_estimator(_estimator_)\n\n logger.info(\"Importing untrained ensembler\")\n\n if method == \"Bagging\":\n logger.info(\"Ensemble method set to Bagging\")\n bagging_model_definition = _all_models_internal[\"Bagging\"]\n\n model = bagging_model_definition.class_def(\n model,\n bootstrap=True,\n n_estimators=n_estimators,\n **bagging_model_definition.args,\n )\n\n else:\n logger.info(\"Ensemble method set to Boosting\")\n boosting_model_definition = _all_models_internal[\"ada\"]\n model = boosting_model_definition.class_def(\n model, n_estimators=n_estimators, **boosting_model_definition.args\n )\n\n display.move_progress()\n\n logger.info(\"SubProcess create_model() called ==================================\")\n model, model_fit_time = create_model_supervised(\n estimator=model,\n system=False,\n display=display,\n fold=fold,\n round=round,\n fit_kwargs=fit_kwargs,\n groups=groups,\n )\n best_model = model\n model_results = pull()\n logger.info(\"SubProcess create_model() end ==================================\")\n\n # end runtime\n runtime_end = time.time()\n runtime = np.array(runtime_end - runtime_start).round(2)\n\n # mlflow logging\n if logging_param:\n\n avgs_dict_log = {k: v for k, v in model_results.loc[\"Mean\"].items()}\n\n try:\n _mlflow_log_model(\n model=best_model,\n model_results=model_results,\n score_dict=avgs_dict_log,\n source=\"ensemble_model\",\n runtime=runtime,\n model_fit_time=model_fit_time,\n _prep_pipe=prep_pipe,\n log_plots=log_plots_param,\n display=display,\n )\n except:\n logger.error(f\"_mlflow_log_model() for {best_model} raised an exception:\")\n logger.error(traceback.format_exc())\n\n if choose_better:\n model = _choose_better(\n [_estimator_, (best_model, model_results)],\n compare_dimension,\n fold,\n groups=groups,\n fit_kwargs=fit_kwargs,\n display=display,\n )\n\n model_results = color_df(model_results, \"yellow\", [\"Mean\"], axis=1)\n model_results = model_results.set_precision(round)\n display.display(model_results, clear=True)\n\n logger.info(f\"create_model_container: {len(create_model_container)}\")\n logger.info(f\"master_model_container: {len(master_model_container)}\")\n logger.info(f\"display_container: {len(display_container)}\")\n\n logger.info(str(model))\n logger.info(\n \"ensemble_model() succesfully completed......................................\"\n )\n\n gc.collect()\n return model\n\n\ndef blend_models(\n estimator_list: list,\n fold: Optional[Union[int, Any]] = None,\n round: int = 4,\n choose_better: bool = False,\n optimize: str = \"Accuracy\",\n method: str = \"auto\",\n weights: Optional[List[float]] = None, # added in pycaret==2.2.0\n fit_kwargs: Optional[dict] = None,\n groups: Optional[Union[str, Any]] = None,\n verbose: bool = True,\n display: Optional[Display] = None, # added in pycaret==2.2.0\n) -> Any:\n\n \"\"\"\n This function creates a Soft Voting / Majority Rule classifier for all the\n estimators in the model library (excluding the few when turbo is True) or\n for specific trained estimators passed as a list in estimator_list param.\n It scores it using Cross Validation. The output prints a score\n grid that shows Accuracy, AUC, Recall, Precision, F1, Kappa and MCC by\n fold (default CV = 10 Folds).\n\n This function returns a trained model object.\n\n Example\n -------\n >>> lr = create_model('lr')\n >>> rf = create_model('rf')\n >>> knn = create_model('knn')\n >>> blend_three = blend_models(estimator_list = [lr,rf,knn])\n\n This will create a VotingClassifier of lr, rf and knn.\n\n Parameters\n ----------\n estimator_list : list of objects\n\n fold: integer or scikit-learn compatible CV generator, default = None\n Controls cross-validation. If None, will use the CV generator defined in setup().\n If integer, will use KFold CV with that many folds.\n When cross_validation is False, this parameter is ignored.\n\n round: integer, default = 4\n Number of decimal places the metrics in the score grid will be rounded to.\n\n choose_better: bool, default = False\n When set to set to True, base estimator is returned when the metric doesn't\n improve by ensemble_model. This gurantees the returned object would perform\n atleast equivalent to base estimator created using create_model or model\n returned by compare_models.\n\n optimize: str, default = 'Accuracy'\n Only used when choose_better is set to True. optimize parameter is used\n to compare emsembled model with base estimator. Values accepted in\n optimize parameter are 'Accuracy', 'AUC', 'Recall', 'Precision', 'F1',\n 'Kappa', 'MCC'.\n\n method: str, default = 'auto'\n 'hard' uses predicted class labels for majority rule voting. 'soft', predicts\n the class label based on the argmax of the sums of the predicted probabilities,\n which is recommended for an ensemble of well-calibrated classifiers. Default value,\n 'auto', will try to use 'soft' and fall back to 'hard' if the former is not supported.\n\n weights: list, default = None\n Sequence of weights (float or int) to weight the occurrences of predicted class labels (hard voting)\n or class probabilities before averaging (soft voting). Uses uniform weights if None.\n\n fit_kwargs: dict, default = {} (empty dict)\n Dictionary of arguments passed to the fit method of the model.\n\n groups: str or array-like, with shape (n_samples,), default = None\n Optional Group labels for the samples used while splitting the dataset into train/test set.\n If string is passed, will use the data column with that name as the groups.\n Only used if a group based cross-validation generator is used (eg. GroupKFold).\n If None, will use the value set in fold_groups param in setup().\n\n verbose: bool, default = True\n Score grid is not printed when verbose is set to False.\n\n Returns\n -------\n score_grid\n A table containing the scores of the model across the kfolds.\n Scoring metrics used are Accuracy, AUC, Recall, Precision, F1,\n Kappa and MCC. Mean and standard deviation of the scores across\n the folds are also returned.\n\n model\n Trained Voting Classifier model object.\n\n Warnings\n --------\n - When passing estimator_list with method set to 'soft'. All the models in the\n estimator_list must support predict_proba function. 'svm' and 'ridge' doesnt\n support the predict_proba and hence an exception will be raised.\n\n - When estimator_list is set to 'All' and method is forced to 'soft', estimators\n that doesnt support the predict_proba function will be dropped from the estimator\n list.\n\n - If target variable is multiclass (more than 2 classes), AUC will be returned as\n zero (0.0).\n\n\n\n \"\"\"\n\n function_params_str = \", \".join([f\"{k}={v}\" for k, v in locals().items()])\n\n logger = get_logger()\n\n logger.info(\"Initializing blend_models()\")\n logger.info(f\"blend_models({function_params_str})\")\n\n logger.info(\"Checking exceptions\")\n\n # run_time\n runtime_start = time.time()\n\n if not fit_kwargs:\n fit_kwargs = {}\n\n # checking method parameter\n available_method = [\"auto\", \"soft\", \"hard\"]\n if method not in available_method:\n raise ValueError(\n \"Method parameter only accepts 'auto', 'soft' or 'hard' as a parameter. See Docstring for details.\"\n )\n\n # checking error for estimator_list\n for i in estimator_list:\n if not hasattr(i, \"fit\"):\n raise ValueError(f\"Estimator {i} does not have the required fit() method.\")\n if _ml_usecase == MLUsecase.CLASSIFICATION:\n # checking method param with estimator list\n if method != \"hard\":\n\n for i in estimator_list:\n if not hasattr(i, \"predict_proba\"):\n if method != \"auto\":\n raise TypeError(\n f\"Estimator list contains estimator {i} that doesn't support probabilities and method is forced to 'soft'. Either change the method or drop the estimator.\"\n )\n else:\n logger.info(\n f\"Estimator {i} doesn't support probabilities, falling back to 'hard'.\"\n )\n method = \"hard\"\n break\n\n if method == \"auto\":\n method = \"soft\"\n\n # checking fold parameter\n if fold is not None and not (type(fold) is int or is_sklearn_cv_generator(fold)):\n raise TypeError(\n \"fold parameter must be either None, an integer or a scikit-learn compatible CV generator object.\"\n )\n\n # checking round parameter\n if type(round) is not int:\n raise TypeError(\"Round parameter only accepts integer value.\")\n\n if weights is not None:\n num_estimators = len(estimator_list)\n # checking weights parameter\n if len(weights) != num_estimators:\n raise ValueError(\n \"weights parameter must have the same length as the estimator_list.\"\n )\n if not all((isinstance(x, int) or isinstance(x, float)) for x in weights):\n raise TypeError(\"weights must contain only ints or floats.\")\n\n # checking verbose parameter\n if type(verbose) is not bool:\n raise TypeError(\"Verbose parameter can only take argument as True or False.\")\n\n # checking optimize parameter\n optimize = _get_metric(optimize)\n if optimize is None:\n raise ValueError(\n f\"Optimize method not supported. See docstring for list of available parameters.\"\n )\n\n # checking optimize parameter for multiclass\n if _is_multiclass():\n if not optimize.is_multiclass:\n raise TypeError(\n f\"Optimization metric not supported for multiclass problems. See docstring for list of other optimization parameters.\"\n )\n\n \"\"\"\n\n ERROR HANDLING ENDS HERE\n\n \"\"\"\n\n fold = _get_cv_splitter(fold)\n\n groups = _get_groups(groups)\n\n if not display:\n progress_args = {\"max\": 2 + 4}\n master_display_columns = [v.display_name for k, v in _all_metrics.items()]\n timestampStr = datetime.datetime.now().strftime(\"%H:%M:%S\")\n monitor_rows = [\n [\"Initiated\", \". . . . . . . . . . . . . . . . . .\", timestampStr],\n [\"Status\", \". . . . . . . . . . . . . . . . . .\", \"Loading Dependencies\"],\n [\"Estimator\", \". . . . . . . . . . . . . . . . . .\", \"Compiling Library\"],\n ]\n display = Display(\n verbose=verbose,\n html_param=html_param,\n progress_args=progress_args,\n master_display_columns=master_display_columns,\n monitor_rows=monitor_rows,\n )\n display.display_progress()\n display.display_monitor()\n display.display_master_display()\n\n logger.info(\"Importing libraries\")\n\n np.random.seed(seed)\n\n logger.info(\"Copying training dataset\")\n\n # Storing X_train and y_train in data_X and data_y parameter\n data_X = X_train.copy()\n data_y = y_train.copy()\n\n # reset index\n data_X.reset_index(drop=True, inplace=True)\n data_y.reset_index(drop=True, inplace=True)\n\n # setting optimize parameter\n compare_dimension = optimize.display_name\n optimize = optimize.scorer\n\n display.move_progress()\n\n \"\"\"\n MONITOR UPDATE STARTS\n \"\"\"\n\n display.update_monitor(1, \"Compiling Estimators\")\n display.display_monitor()\n\n \"\"\"\n MONITOR UPDATE ENDS\n \"\"\"\n\n logger.info(\"Getting model names\")\n estimator_dict = {}\n for x in estimator_list:\n x = get_estimator_from_meta_estimator(x)\n name = _get_model_id(x)\n suffix = 1\n original_name = name\n while name in estimator_dict:\n name = f\"{original_name}_{suffix}\"\n suffix += 1\n estimator_dict[name] = x\n\n estimator_list = list(estimator_dict.items())\n\n voting_model_definition = _all_models_internal[\"Voting\"]\n if _ml_usecase == MLUsecase.CLASSIFICATION:\n model = voting_model_definition.class_def(\n estimators=estimator_list, voting=method, n_jobs=_gpu_n_jobs_param\n )\n else:\n model = voting_model_definition.class_def(\n estimators=estimator_list, n_jobs=_gpu_n_jobs_param\n )\n\n display.update_monitor(2, voting_model_definition.name)\n display.display_monitor()\n\n display.move_progress()\n\n logger.info(\"SubProcess create_model() called ==================================\")\n model, model_fit_time = create_model_supervised(\n estimator=model,\n system=False,\n display=display,\n fold=fold,\n round=round,\n fit_kwargs=fit_kwargs,\n groups=groups,\n )\n model_results = pull()\n logger.info(\"SubProcess create_model() end ==================================\")\n\n # end runtime\n runtime_end = time.time()\n runtime = np.array(runtime_end - runtime_start).round(2)\n\n # mlflow logging\n if logging_param:\n\n avgs_dict_log = {k: v for k, v in model_results.loc[\"Mean\"].items()}\n\n try:\n _mlflow_log_model(\n model=model,\n model_results=model_results,\n score_dict=avgs_dict_log,\n source=\"blend_models\",\n runtime=runtime,\n model_fit_time=model_fit_time,\n _prep_pipe=prep_pipe,\n log_plots=log_plots_param,\n display=display,\n )\n except:\n logger.error(f\"_mlflow_log_model() for {model} raised an exception:\")\n logger.error(traceback.format_exc())\n\n if choose_better:\n model = _choose_better(\n [(model, model_results)] + estimator_list,\n compare_dimension,\n fold,\n groups=groups,\n fit_kwargs=fit_kwargs,\n display=display,\n )\n\n model_results = color_df(model_results, \"yellow\", [\"Mean\"], axis=1)\n model_results = model_results.set_precision(round)\n display.display(model_results, clear=True)\n\n logger.info(f\"create_model_container: {len(create_model_container)}\")\n logger.info(f\"master_model_container: {len(master_model_container)}\")\n logger.info(f\"display_container: {len(display_container)}\")\n\n logger.info(str(model))\n logger.info(\n \"blend_models() succesfully completed......................................\"\n )\n\n gc.collect()\n return model\n\n\ndef stack_models(\n estimator_list: list,\n meta_model=None,\n fold: Optional[Union[int, Any]] = None,\n round: int = 4,\n method: str = \"auto\",\n restack: bool = True,\n choose_better: bool = False,\n optimize: str = \"Accuracy\",\n fit_kwargs: Optional[dict] = None,\n groups: Optional[Union[str, Any]] = None,\n verbose: bool = True,\n display: Optional[Display] = None,\n) -> Any:\n\n \"\"\"\n This function trains a meta model and scores it using Cross Validation.\n The predictions from the base level models as passed in the estimator_list param\n are used as input features for the meta model. The restacking parameter controls\n the ability to expose raw features to the meta model when set to True\n (default = False).\n\n The output prints the score grid that shows Accuracy, AUC, Recall, Precision,\n F1, Kappa and MCC by fold (default = 10 Folds).\n\n This function returns a trained model object.\n\n Example\n -------\n >>> from pycaret.datasets import get_data\n >>> juice = get_data('juice')\n >>> experiment_name = setup(data = juice, target = 'Purchase')\n >>> dt = create_model('dt')\n >>> rf = create_model('rf')\n >>> ada = create_model('ada')\n >>> ridge = create_model('ridge')\n >>> knn = create_model('knn')\n >>> stacked_models = stack_models(estimator_list=[dt,rf,ada,ridge,knn])\n\n This will create a meta model that will use the predictions of all the\n models provided in estimator_list param. By default, the meta model is\n Logistic Regression but can be changed with meta_model param.\n\n Parameters\n ----------\n estimator_list : list of objects\n\n meta_model : object, default = None\n If set to None, Logistic Regression is used as a meta model.\n\n fold: integer or scikit-learn compatible CV generator, default = None\n Controls cross-validation. If None, will use the CV generator defined in setup().\n If integer, will use KFold CV with that many folds.\n When cross_validation is False, this parameter is ignored.\n\n round: integer, default = 4\n Number of decimal places the metrics in the score grid will be rounded to.\n\n method: string, default = 'auto'\n - if ‘auto’, it will try to invoke, for each estimator, 'predict_proba',\n 'decision_function' or 'predict' in that order.\n - otherwise, one of 'predict_proba', 'decision_function' or 'predict'.\n If the method is not implemented by the estimator, it will raise an error.\n\n restack: bool, default = True\n When restack is set to True, raw data will be exposed to meta model when\n making predictions, otherwise when False, only the predicted label or\n probabilities is passed to meta model when making final predictions.\n\n choose_better: bool, default = False\n When set to set to True, base estimator is returned when the metric doesn't\n improve by ensemble_model. This gurantees the returned object would perform\n atleast equivalent to base estimator created using create_model or model\n returned by compare_models.\n\n optimize: str, default = 'Accuracy'\n Only used when choose_better is set to True. optimize parameter is used\n to compare emsembled model with base estimator. Values accepted in\n optimize parameter are 'Accuracy', 'AUC', 'Recall', 'Precision', 'F1',\n 'Kappa', 'MCC'.\n\n fit_kwargs: dict, default = {} (empty dict)\n Dictionary of arguments passed to the fit method of the model.\n\n groups: str or array-like, with shape (n_samples,), default = None\n Optional Group labels for the samples used while splitting the dataset into train/test set.\n If string is passed, will use the data column with that name as the groups.\n Only used if a group based cross-validation generator is used (eg. GroupKFold).\n If None, will use the value set in fold_groups param in setup().\n\n verbose: bool, default = True\n Score grid is not printed when verbose is set to False.\n\n Returns\n -------\n score_grid\n A table containing the scores of the model across the kfolds.\n Scoring metrics used are Accuracy, AUC, Recall, Precision, F1,\n Kappa and MCC. Mean and standard deviation of the scores across\n the folds are also returned.\n\n model\n Trained model object.\n\n Warnings\n --------\n - If target variable is multiclass (more than 2 classes), AUC will be returned\n as zero (0.0).\n\n \"\"\"\n\n function_params_str = \", \".join([f\"{k}={v}\" for k, v in locals().items()])\n\n logger = get_logger()\n\n logger.info(\"Initializing stack_models()\")\n logger.info(f\"stack_models({function_params_str})\")\n\n logger.info(\"Checking exceptions\")\n\n # run_time\n runtime_start = time.time()\n\n if not fit_kwargs:\n fit_kwargs = {}\n\n # checking error for estimator_list\n for i in estimator_list:\n if not hasattr(i, \"fit\"):\n raise ValueError(f\"Estimator {i} does not have the required fit() method.\")\n\n # checking meta model\n if meta_model is not None:\n if not hasattr(meta_model, \"fit\"):\n raise ValueError(\n f\"Meta Model {meta_model} does not have the required fit() method.\"\n )\n\n # checking fold parameter\n if fold is not None and not (type(fold) is int or is_sklearn_cv_generator(fold)):\n raise TypeError(\n \"fold parameter must be either None, an integer or a scikit-learn compatible CV generator object.\"\n )\n\n # checking round parameter\n if type(round) is not int:\n raise TypeError(\"Round parameter only accepts integer value.\")\n\n # checking method parameter\n available_method = [\"auto\", \"predict_proba\", \"decision_function\", \"predict\"]\n if method not in available_method:\n raise ValueError(\n \"Method parameter not acceptable. It only accepts 'auto', 'predict_proba', 'decision_function', 'predict'.\"\n )\n\n # checking restack parameter\n if type(restack) is not bool:\n raise TypeError(\"Restack parameter can only take argument as True or False.\")\n\n # checking verbose parameter\n if type(verbose) is not bool:\n raise TypeError(\"Verbose parameter can only take argument as True or False.\")\n\n # checking optimize parameter\n optimize = _get_metric(optimize)\n if optimize is None:\n raise ValueError(\n f\"Optimize method not supported. See docstring for list of available parameters.\"\n )\n\n # checking optimize parameter for multiclass\n if _is_multiclass():\n if not optimize.is_multiclass:\n raise TypeError(\n f\"Optimization metric not supported for multiclass problems. See docstring for list of other optimization parameters.\"\n )\n\n \"\"\"\n\n ERROR HANDLING ENDS HERE\n\n \"\"\"\n\n fold = _get_cv_splitter(fold)\n\n groups = _get_groups(groups)\n\n logger.info(\"Defining meta model\")\n # Defining meta model.\n if meta_model == None:\n estimator = \"lr\"\n meta_model_definition = _all_models_internal[estimator]\n meta_model_args = meta_model_definition.args\n meta_model = meta_model_definition.class_def(**meta_model_args)\n else:\n meta_model = clone(get_estimator_from_meta_estimator(meta_model))\n\n if not display:\n progress_args = {\"max\": 2 + 4}\n master_display_columns = [v.display_name for k, v in _all_metrics.items()]\n timestampStr = datetime.datetime.now().strftime(\"%H:%M:%S\")\n monitor_rows = [\n [\"Initiated\", \". . . . . . . . . . . . . . . . . .\", timestampStr],\n [\"Status\", \". . . . . . . . . . . . . . . . . .\", \"Loading Dependencies\"],\n [\"Estimator\", \". . . . . . . . . . . . . . . . . .\", \"Compiling Library\"],\n ]\n display = Display(\n verbose=verbose,\n html_param=html_param,\n progress_args=progress_args,\n master_display_columns=master_display_columns,\n monitor_rows=monitor_rows,\n )\n display.display_progress()\n display.display_monitor()\n display.display_master_display()\n\n np.random.seed(seed)\n\n logger.info(\"Copying training dataset\")\n # Storing X_train and y_train in data_X and data_y parameter\n data_X = X_train.copy()\n data_y = y_train.copy()\n\n # reset index\n data_X.reset_index(drop=True, inplace=True)\n data_y.reset_index(drop=True, inplace=True)\n\n # setting optimize parameter\n compare_dimension = optimize.display_name\n optimize = optimize.scorer\n\n display.move_progress()\n\n \"\"\"\n MONITOR UPDATE STARTS\n \"\"\"\n\n display.update_monitor(1, \"Compiling Estimators\")\n display.display_monitor()\n\n \"\"\"\n MONITOR UPDATE ENDS\n \"\"\"\n\n logger.info(\"Getting model names\")\n estimator_dict = {}\n for x in estimator_list:\n x = get_estimator_from_meta_estimator(x)\n name = _get_model_id(x)\n suffix = 1\n original_name = name\n while name in estimator_dict:\n name = f\"{original_name}_{suffix}\"\n suffix += 1\n estimator_dict[name] = x\n\n estimator_list = list(estimator_dict.items())\n\n logger.info(estimator_list)\n\n stacking_model_definition = _all_models_internal[\"Stacking\"]\n if _ml_usecase == MLUsecase.CLASSIFICATION:\n model = stacking_model_definition.class_def(\n estimators=estimator_list,\n final_estimator=meta_model,\n cv=fold,\n stack_method=method,\n n_jobs=_gpu_n_jobs_param,\n passthrough=restack,\n )\n else:\n model = stacking_model_definition.class_def(\n estimators=estimator_list,\n final_estimator=meta_model,\n cv=fold,\n n_jobs=_gpu_n_jobs_param,\n passthrough=restack,\n )\n\n display.update_monitor(2, stacking_model_definition.name)\n display.display_monitor()\n\n display.move_progress()\n\n logger.info(\"SubProcess create_model() called ==================================\")\n model, model_fit_time = create_model_supervised(\n estimator=model,\n system=False,\n display=display,\n fold=fold,\n round=round,\n fit_kwargs=fit_kwargs,\n groups=groups,\n )\n model_results = pull()\n logger.info(\"SubProcess create_model() end ==================================\")\n\n # end runtime\n runtime_end = time.time()\n runtime = np.array(runtime_end - runtime_start).round(2)\n\n # mlflow logging\n if logging_param:\n\n avgs_dict_log = {k: v for k, v in model_results.loc[\"Mean\"].items()}\n\n try:\n _mlflow_log_model(\n model=model,\n model_results=model_results,\n score_dict=avgs_dict_log,\n source=\"stack_models\",\n runtime=runtime,\n model_fit_time=model_fit_time,\n _prep_pipe=prep_pipe,\n log_plots=log_plots_param,\n display=display,\n )\n except:\n logger.error(f\"_mlflow_log_model() for {model} raised an exception:\")\n logger.error(traceback.format_exc())\n\n if choose_better:\n model = _choose_better(\n [(model, model_results)] + estimator_list,\n compare_dimension,\n fold,\n groups=groups,\n fit_kwargs=fit_kwargs,\n display=display,\n )\n\n model_results = color_df(model_results, \"yellow\", [\"Mean\"], axis=1)\n model_results = model_results.set_precision(round)\n display.display(model_results, clear=True)\n\n logger.info(f\"create_model_container: {len(create_model_container)}\")\n logger.info(f\"master_model_container: {len(master_model_container)}\")\n logger.info(f\"display_container: {len(display_container)}\")\n\n logger.info(str(model))\n logger.info(\n \"stack_models() succesfully completed......................................\"\n )\n\n gc.collect()\n return model\n\n\ndef plot_model(\n estimator,\n plot: str = \"auc\",\n scale: float = 1, # added in pycaret==2.1.0\n save: bool = False,\n fold: Optional[Union[int, Any]] = None,\n fit_kwargs: Optional[dict] = None,\n groups: Optional[Union[str, Any]] = None,\n feature_name: Optional[str] = None,\n label: bool = False,\n use_train_data: bool = False,\n verbose: bool = True,\n system: bool = True,\n display: Optional[Display] = None, # added in pycaret==2.2.0\n display_format: Optional[str] = None,\n is_in_evaluate: bool = False,\n) -> str:\n\n \"\"\"\n This function takes a trained model object and returns a plot based on the\n test / hold-out set. The process may require the model to be re-trained in\n certain cases. See list of plots supported below.\n\n Model must be created using create_model() or tune_model().\n\n Example\n -------\n >>> from pycaret.datasets import get_data\n >>> juice = get_data('juice')\n >>> experiment_name = setup(data = juice, target = 'Purchase')\n >>> lr = create_model('lr')\n >>> plot_model(lr)\n\n This will return an AUC plot of a trained Logistic Regression model.\n\n Parameters\n ----------\n estimator : object, default = none\n A trained model object should be passed as an estimator.\n\n plot : str, default = auc\n Enter abbreviation of type of plot. The current list of plots supported are (Plot - Name):\n\n\n * 'residuals_interactive' - Interactive Residual plots\n * 'auc' - Area Under the Curve\n * 'threshold' - Discrimination Threshold \n * 'pr' - Precision Recall Curve \n * 'confusion_matrix' - Confusion Matrix \n * 'error' - Class Prediction Error \n * 'class_report' - Classification Report \n * 'boundary' - Decision Boundary \n * 'rfe' - Recursive Feature Selection \n * 'learning' - Learning Curve \n * 'manifold' - Manifold Learning \n * 'calibration' - Calibration Curve \n * 'vc' - Validation Curve \n * 'dimension' - Dimension Learning \n * 'feature' - Feature Importance \n * 'feature_all' - Feature Importance (All)\n * 'parameter' - Model Hyperparameter\n * 'lift' - Lift Curve\n * 'gain' - Gain Chart\n\n scale: float, default = 1\n The resolution scale of the figure.\n\n save: bool, default = False\n When set to True, Plot is saved as a 'png' file in current working directory.\n\n fold: integer or scikit-learn compatible CV generator, default = None\n Controls cross-validation used in certain plots. If None, will use the CV generator\n defined in setup(). If integer, will use KFold CV with that many folds.\n When cross_validation is False, this parameter is ignored.\n\n fit_kwargs: dict, default = {} (empty dict)\n Dictionary of arguments passed to the fit method of the model.\n\n groups: str or array-like, with shape (n_samples,), default = None\n Optional Group labels for the samples used while splitting the dataset into train/test set.\n If string is passed, will use the data column with that name as the groups.\n Only used if a group based cross-validation generator is used (eg. GroupKFold).\n If None, will use the value set in fold_groups param in setup().\n\n verbose: bool, default = True\n Progress bar not shown when verbose set to False.\n\n system: bool, default = True\n Must remain True all times. Only to be changed by internal functions.\n\n\n display_format: str, default = None\n To display plots in Streamlit (https://www.streamlit.io/), set this to 'streamlit'.\n Currently, not all plots are supported.\n\n Returns\n -------\n Visual_Plot\n Prints the visual plot.\n str:\n If save param is True, will return the name of the saved file.\n\n Warnings\n --------\n - 'svm' and 'ridge' doesn't support the predict_proba method. As such, AUC and\n calibration plots are not available for these estimators.\n\n - When the 'max_features' parameter of a trained model object is not equal to\n the number of samples in training set, the 'rfe' plot is not available.\n\n - 'calibration', 'threshold', 'manifold' and 'rfe' plots are not available for\n multiclass problems.\n\n\n \"\"\"\n\n function_params_str = \", \".join([f\"{k}={v}\" for k, v in locals().items()])\n\n logger = get_logger()\n\n logger.info(\"Initializing plot_model()\")\n logger.info(f\"plot_model({function_params_str})\")\n\n logger.info(\"Checking exceptions\")\n\n if not fit_kwargs:\n fit_kwargs = {}\n\n if not hasattr(estimator, \"fit\"):\n raise ValueError(\n f\"Estimator {estimator} does not have the required fit() method.\"\n )\n\n if plot not in _available_plots:\n raise ValueError(\n \"Plot Not Available. Please see docstring for list of available Plots.\"\n )\n\n # multiclass plot exceptions:\n multiclass_not_available = [\"calibration\", \"threshold\", \"manifold\", \"rfe\"]\n if _is_multiclass():\n if plot in multiclass_not_available:\n raise ValueError(\n \"Plot Not Available for multiclass problems. Please see docstring for list of available Plots.\"\n )\n\n # exception for CatBoost\n # if \"CatBoostClassifier\" in str(type(estimator)):\n # raise ValueError(\n # \"CatBoost estimator is not compatible with plot_model function, try using Catboost with interpret_model instead.\"\n # )\n\n # checking for auc plot\n if not hasattr(estimator, \"predict_proba\") and plot == \"auc\":\n raise TypeError(\n \"AUC plot not available for estimators with no predict_proba attribute.\"\n )\n\n # checking for auc plot\n if not hasattr(estimator, \"predict_proba\") and plot == \"auc\":\n raise TypeError(\n \"AUC plot not available for estimators with no predict_proba attribute.\"\n )\n\n # checking for calibration plot\n if not hasattr(estimator, \"predict_proba\") and plot == \"calibration\":\n raise TypeError(\n \"Calibration plot not available for estimators with no predict_proba attribute.\"\n )\n\n def is_tree(e):\n from sklearn.tree import BaseDecisionTree\n from sklearn.ensemble._forest import BaseForest\n\n if \"final_estimator\" in e.get_params():\n e = e.final_estimator\n if \"base_estimator\" in e.get_params():\n e = e.base_estimator\n if isinstance(e, BaseForest) or isinstance(e, BaseDecisionTree):\n return True\n\n # checking for calibration plot\n if plot == \"tree\" and not is_tree(estimator):\n raise TypeError(\n \"Decision Tree plot is only available for scikit-learn Decision Trees and Forests, Ensemble models using those or Stacked models using those as meta (final) estimators.\"\n )\n\n # checking for feature plot\n if not (\n hasattr(estimator, \"coef_\") or hasattr(estimator, \"feature_importances_\")\n ) and (plot == \"feature\" or plot == \"feature_all\" or plot == \"rfe\"):\n raise TypeError(\n \"Feature Importance and RFE plots not available for estimators that doesnt support coef_ or feature_importances_ attribute.\"\n )\n\n if plot == \"residuals_interactive\" and is_in_evaluate and is_in_colab():\n raise ValueError(\n \"Interactive Residuals plot not available in evaluate_model() in Google Colab. Do plot_model(model, plot='residuals_interactive') instead.\"\n )\n\n # checking fold parameter\n if fold is not None and not (type(fold) is int or is_sklearn_cv_generator(fold)):\n raise TypeError(\n \"fold parameter must be either None, an integer or a scikit-learn compatible CV generator object.\"\n )\n\n if type(label) is not bool:\n raise TypeError(\"Label param only accepts True or False.\")\n\n if type(use_train_data) is not bool:\n raise TypeError(\"use_train_data param only accepts True or False.\")\n\n if feature_name is not None and type(feature_name) is not str:\n raise TypeError(\n \"feature parameter must be string containing column name of dataset.\"\n )\n\n # checking display_format parameter\n plot_formats = [None, \"streamlit\"]\n\n if display_format not in plot_formats:\n raise ValueError(\"display_format can only be None or 'streamlit'.\")\n\n if display_format == \"streamlit\":\n try:\n import streamlit as st\n except ImportError:\n raise ImportError(\n \"It appears that streamlit is not installed. Do: pip install streamlit\"\n )\n\n \"\"\"\n\n ERROR HANDLING ENDS HERE\n\n \"\"\"\n\n cv = _get_cv_splitter(fold)\n\n groups = _get_groups(groups)\n\n if not display:\n progress_args = {\"max\": 5}\n display = Display(\n verbose=verbose, html_param=html_param, progress_args=progress_args\n )\n display.display_progress()\n\n logger.info(\"Preloading libraries\")\n # pre-load libraries\n import matplotlib.pyplot as plt\n\n np.random.seed(seed)\n\n display.move_progress()\n\n # defining estimator as model locally\n # deepcopy instead of clone so we have a fitted estimator\n if isinstance(estimator, InternalPipeline):\n estimator = estimator.steps[-1][1]\n estimator = deepcopy(estimator)\n model = estimator\n\n display.move_progress()\n\n # plots used for logging (controlled through plots_log_param)\n # AUC, #Confusion Matrix and #Feature Importance\n\n logger.info(\"Copying training dataset\")\n\n # Storing X_train and y_train in data_X and data_y parameter\n data_X = X_train.copy()\n if not _is_unsupervised(_ml_usecase):\n data_y = y_train.copy()\n\n # reset index\n data_X.reset_index(drop=True, inplace=True)\n if not _is_unsupervised(_ml_usecase):\n data_y.reset_index(drop=True, inplace=True)\n\n logger.info(\"Copying test dataset\")\n\n # Storing X_train and y_train in data_X and data_y parameter\n test_X = X_train.copy() if use_train_data else X_test.copy()\n test_y = y_train.copy() if use_train_data else y_test.copy()\n\n # reset index\n test_X.reset_index(drop=True, inplace=True)\n test_y.reset_index(drop=True, inplace=True)\n\n logger.info(f\"Plot type: {plot}\")\n plot_name = _available_plots[plot]\n display.move_progress()\n\n # yellowbrick workaround start\n import yellowbrick.utils.types\n import yellowbrick.utils.helpers\n\n # yellowbrick workaround end\n\n model_name = _get_model_name(model)\n plot_filename = f\"{plot_name}.png\"\n with patch(\n \"yellowbrick.utils.types.is_estimator\",\n pycaret.internal.patches.yellowbrick.is_estimator,\n ), patch(\n \"yellowbrick.utils.helpers.is_estimator\",\n pycaret.internal.patches.yellowbrick.is_estimator,\n ), estimator_pipeline(\n _internal_pipeline, model\n ) as pipeline_with_model:\n fit_kwargs = _get_pipeline_fit_kwargs(pipeline_with_model, fit_kwargs)\n\n _base_dpi = 100\n\n def residuals_interactive():\n from pycaret.internal.plots.residual_plots import InteractiveResidualsPlot\n\n resplots = InteractiveResidualsPlot(\n x=data_X,\n y=data_y,\n x_test=test_X,\n y_test=test_y,\n model=pipeline_with_model,\n display=display,\n )\n\n display.clear_output()\n if system:\n resplots.show()\n\n plot_filename = f\"{plot_name}.html\"\n\n if save:\n resplots.write_html(plot_filename)\n logger.info(f\"Saving '{plot_filename}' in current active directory\")\n\n logger.info(\"Visual Rendered Successfully\")\n return plot_filename\n\n def cluster():\n logger.info(\n \"SubProcess assign_model() called ==================================\"\n )\n b = assign_model(\n pipeline_with_model, verbose=False, transformation=True\n ).reset_index(drop=True)\n logger.info(\n \"SubProcess assign_model() end ==================================\"\n )\n cluster = b[\"Cluster\"].values\n b.drop(\"Cluster\", axis=1, inplace=True)\n b = pd.get_dummies(b) # casting categorical variable\n\n from sklearn.decomposition import PCA\n\n pca = PCA(n_components=2, random_state=seed)\n logger.info(\"Fitting PCA()\")\n pca_ = pca.fit_transform(b)\n pca_ = pd.DataFrame(pca_)\n pca_ = pca_.rename(columns={0: \"PCA1\", 1: \"PCA2\"})\n pca_[\"Cluster\"] = cluster\n\n if feature_name is not None:\n pca_[\"Feature\"] = data_before_preprocess[feature_name]\n else:\n pca_[\"Feature\"] = data_before_preprocess[\n data_before_preprocess.columns[0]\n ]\n\n if label:\n pca_[\"Label\"] = pca_[\"Feature\"]\n\n \"\"\"\n sorting\n \"\"\"\n\n logger.info(\"Sorting dataframe\")\n\n print(pca_[\"Cluster\"])\n\n clus_num = [int(i.split()[1]) for i in pca_[\"Cluster\"]]\n\n pca_[\"cnum\"] = clus_num\n pca_.sort_values(by=\"cnum\", inplace=True)\n\n \"\"\"\n sorting ends\n \"\"\"\n\n display.clear_output()\n\n logger.info(\"Rendering Visual\")\n\n if label:\n fig = px.scatter(\n pca_,\n x=\"PCA1\",\n y=\"PCA2\",\n text=\"Label\",\n color=\"Cluster\",\n opacity=0.5,\n )\n else:\n fig = px.scatter(\n pca_,\n x=\"PCA1\",\n y=\"PCA2\",\n hover_data=[\"Feature\"],\n color=\"Cluster\",\n opacity=0.5,\n )\n\n fig.update_traces(textposition=\"top center\")\n fig.update_layout(plot_bgcolor=\"rgb(240,240,240)\")\n\n fig.update_layout(height=600 * scale, title_text=\"2D Cluster PCA Plot\")\n\n plot_filename = f\"{plot_name}.html\"\n\n if save:\n fig.write_html(plot_filename)\n logger.info(f\"Saving '{plot_filename}' in current active directory\")\n\n elif system:\n if display_format == \"streamlit\":\n st.write(fig)\n else:\n fig.show()\n\n logger.info(\"Visual Rendered Successfully\")\n return plot_filename\n\n def umap():\n logger.info(\n \"SubProcess assign_model() called ==================================\"\n )\n b = assign_model(\n model, verbose=False, transformation=True, score=False\n ).reset_index(drop=True)\n logger.info(\n \"SubProcess assign_model() end ==================================\"\n )\n\n label = pd.DataFrame(b[\"Anomaly\"])\n b.dropna(axis=0, inplace=True) # droping rows with NA's\n b.drop([\"Anomaly\"], axis=1, inplace=True)\n\n import umap\n\n reducer = umap.UMAP()\n logger.info(\"Fitting UMAP()\")\n embedding = reducer.fit_transform(b)\n X = pd.DataFrame(embedding)\n\n import plotly.express as px\n\n df = X\n df[\"Anomaly\"] = label\n\n if feature_name is not None:\n df[\"Feature\"] = data_before_preprocess[feature_name]\n else:\n df[\"Feature\"] = data_before_preprocess[\n data_before_preprocess.columns[0]\n ]\n\n display.clear_output()\n\n logger.info(\"Rendering Visual\")\n\n fig = px.scatter(\n df,\n x=0,\n y=1,\n color=\"Anomaly\",\n title=\"uMAP Plot for Outliers\",\n hover_data=[\"Feature\"],\n opacity=0.7,\n width=900 * scale,\n height=800 * scale,\n )\n plot_filename = f\"{plot_name}.html\"\n\n if save:\n fig.write_html(f\"{plot_filename}\")\n logger.info(f\"Saving '{plot_filename}' in current active directory\")\n elif system:\n if display_format == \"streamlit\":\n st.write(fig)\n else:\n fig.show()\n\n logger.info(\"Visual Rendered Successfully\")\n return plot_filename\n\n def tsne():\n if _ml_usecase == MLUsecase.CLUSTERING:\n return _tsne_clustering()\n else:\n return _tsne_anomaly()\n\n def _tsne_anomaly():\n logger.info(\n \"SubProcess assign_model() called ==================================\"\n )\n b = assign_model(\n model, verbose=False, transformation=True, score=False\n ).reset_index(drop=True)\n logger.info(\n \"SubProcess assign_model() end ==================================\"\n )\n cluster = b[\"Anomaly\"].values\n b.dropna(axis=0, inplace=True) # droping rows with NA's\n b.drop(\"Anomaly\", axis=1, inplace=True)\n\n logger.info(\"Getting dummies to cast categorical variables\")\n\n from sklearn.manifold import TSNE\n\n logger.info(\"Fitting TSNE()\")\n X_embedded = TSNE(n_components=3).fit_transform(b)\n\n X = pd.DataFrame(X_embedded)\n X[\"Anomaly\"] = cluster\n if feature_name is not None:\n X[\"Feature\"] = data_before_preprocess[feature_name]\n else:\n X[\"Feature\"] = data_before_preprocess[data_before_preprocess.columns[0]]\n\n df = X\n\n display.clear_output()\n\n logger.info(\"Rendering Visual\")\n\n if label:\n fig = px.scatter_3d(\n df,\n x=0,\n y=1,\n z=2,\n text=\"Feature\",\n color=\"Anomaly\",\n title=\"3d TSNE Plot for Outliers\",\n opacity=0.7,\n width=900 * scale,\n height=800 * scale,\n )\n else:\n fig = px.scatter_3d(\n df,\n x=0,\n y=1,\n z=2,\n hover_data=[\"Feature\"],\n color=\"Anomaly\",\n title=\"3d TSNE Plot for Outliers\",\n opacity=0.7,\n width=900 * scale,\n height=800 * scale,\n )\n\n plot_filename = f\"{plot_name}.html\"\n\n if save:\n fig.write_html(f\"{plot_filename}\")\n logger.info(f\"Saving '{plot_filename}' in current active directory\")\n elif system:\n if display_format == \"streamlit\":\n st.write(fig)\n else:\n fig.show()\n\n logger.info(\"Visual Rendered Successfully\")\n return plot_filename\n\n def _tsne_clustering():\n logger.info(\n \"SubProcess assign_model() called ==================================\"\n )\n b = assign_model(\n pipeline_with_model, verbose=False, score=False, transformation=True,\n ).reset_index(drop=True)\n logger.info(\n \"SubProcess assign_model() end ==================================\"\n )\n\n cluster = b[\"Cluster\"].values\n b.drop(\"Cluster\", axis=1, inplace=True)\n\n from sklearn.manifold import TSNE\n\n logger.info(\"Fitting TSNE()\")\n X_embedded = TSNE(n_components=3, random_state=seed).fit_transform(b)\n X_embedded = pd.DataFrame(X_embedded)\n X_embedded[\"Cluster\"] = cluster\n\n if feature_name is not None:\n X_embedded[\"Feature\"] = data_before_preprocess[feature_name]\n else:\n X_embedded[\"Feature\"] = data_before_preprocess[data_X.columns[0]]\n\n if label:\n X_embedded[\"Label\"] = X_embedded[\"Feature\"]\n\n \"\"\"\n sorting\n \"\"\"\n logger.info(\"Sorting dataframe\")\n\n clus_num = [int(i.split()[1]) for i in X_embedded[\"Cluster\"]]\n\n X_embedded[\"cnum\"] = clus_num\n X_embedded.sort_values(by=\"cnum\", inplace=True)\n\n \"\"\"\n sorting ends\n \"\"\"\n\n df = X_embedded\n\n display.clear_output()\n\n logger.info(\"Rendering Visual\")\n\n if label:\n\n fig = px.scatter_3d(\n df,\n x=0,\n y=1,\n z=2,\n color=\"Cluster\",\n title=\"3d TSNE Plot for Clusters\",\n text=\"Label\",\n opacity=0.7,\n width=900 * scale,\n height=800 * scale,\n )\n\n else:\n fig = px.scatter_3d(\n df,\n x=0,\n y=1,\n z=2,\n color=\"Cluster\",\n title=\"3d TSNE Plot for Clusters\",\n hover_data=[\"Feature\"],\n opacity=0.7,\n width=900 * scale,\n height=800 * scale,\n )\n\n plot_filename = f\"{plot_name}.html\"\n\n if save:\n fig.write_html(f\"{plot_filename}\")\n logger.info(f\"Saving '{plot_filename}' in current active directory\")\n elif system:\n if display_format == \"streamlit\":\n st.write(fig)\n else:\n fig.show()\n\n logger.info(\"Visual Rendered Successfully\")\n return plot_filename\n\n def distribution():\n logger.info(\n \"SubProcess assign_model() called ==================================\"\n )\n d = assign_model(pipeline_with_model, verbose=False).reset_index(drop=True)\n logger.info(\n \"SubProcess assign_model() end ==================================\"\n )\n\n \"\"\"\n sorting\n \"\"\"\n logger.info(\"Sorting dataframe\")\n\n clus_num = []\n for i in d.Cluster:\n a = int(i.split()[1])\n clus_num.append(a)\n\n d[\"cnum\"] = clus_num\n d.sort_values(by=\"cnum\", inplace=True)\n d.reset_index(inplace=True, drop=True)\n\n clus_label = []\n for i in d.cnum:\n a = \"Cluster \" + str(i)\n clus_label.append(a)\n\n d.drop([\"Cluster\", \"cnum\"], inplace=True, axis=1)\n d[\"Cluster\"] = clus_label\n\n \"\"\"\n sorting ends\n \"\"\"\n\n if feature_name is None:\n x_col = \"Cluster\"\n else:\n x_col = feature_name\n\n display.clear_output()\n\n logger.info(\"Rendering Visual\")\n\n fig = px.histogram(\n d,\n x=x_col,\n color=\"Cluster\",\n marginal=\"box\",\n opacity=0.7,\n hover_data=d.columns,\n )\n\n fig.update_layout(height=600 * scale,)\n\n plot_filename = f\"{plot_name}.html\"\n\n if save:\n fig.write_html(f\"{plot_filename}\")\n logger.info(f\"Saving '{plot_filename}' in current active directory\")\n elif system:\n fig.show()\n\n logger.info(\"Visual Rendered Successfully\")\n return plot_filename\n\n def elbow():\n try:\n from yellowbrick.cluster import KElbowVisualizer\n\n visualizer = KElbowVisualizer(pipeline_with_model, timings=False)\n show_yellowbrick_plot(\n visualizer=visualizer,\n X_train=data_X,\n y_train=None,\n X_test=None,\n y_test=None,\n name=plot_name,\n handle_test=\"\",\n scale=scale,\n save=save,\n fit_kwargs=fit_kwargs,\n groups=groups,\n display=display,\n display_format=display_format,\n )\n\n except:\n logger.error(\"Elbow plot failed. Exception:\")\n logger.error(traceback.format_exc())\n raise TypeError(\"Plot Type not supported for this model.\")\n\n def silhouette():\n from yellowbrick.cluster import SilhouetteVisualizer\n\n try:\n visualizer = SilhouetteVisualizer(\n pipeline_with_model, colors=\"yellowbrick\"\n )\n show_yellowbrick_plot(\n visualizer=visualizer,\n X_train=data_X,\n y_train=None,\n X_test=None,\n y_test=None,\n name=plot_name,\n handle_test=\"\",\n scale=scale,\n save=save,\n fit_kwargs=fit_kwargs,\n groups=groups,\n display=display,\n display_format=display_format,\n )\n except:\n logger.error(\"Silhouette plot failed. Exception:\")\n logger.error(traceback.format_exc())\n raise TypeError(\"Plot Type not supported for this model.\")\n\n def distance():\n from yellowbrick.cluster import InterclusterDistance\n\n try:\n visualizer = InterclusterDistance(pipeline_with_model)\n show_yellowbrick_plot(\n visualizer=visualizer,\n X_train=data_X,\n y_train=None,\n X_test=None,\n y_test=None,\n name=plot_name,\n handle_test=\"\",\n scale=scale,\n save=save,\n fit_kwargs=fit_kwargs,\n groups=groups,\n display=display,\n display_format=display_format,\n )\n except:\n logger.error(\"Distance plot failed. Exception:\")\n logger.error(traceback.format_exc())\n raise TypeError(\"Plot Type not supported for this model.\")\n\n def residuals():\n\n from yellowbrick.regressor import ResidualsPlot\n\n visualizer = ResidualsPlot(pipeline_with_model)\n show_yellowbrick_plot(\n visualizer=visualizer,\n X_train=data_X,\n y_train=data_y,\n X_test=test_X,\n y_test=test_y,\n name=plot_name,\n scale=scale,\n save=save,\n fit_kwargs=fit_kwargs,\n groups=groups,\n display=display,\n display_format=display_format,\n )\n\n def auc():\n\n from yellowbrick.classifier import ROCAUC\n\n visualizer = ROCAUC(pipeline_with_model)\n show_yellowbrick_plot(\n visualizer=visualizer,\n X_train=data_X,\n y_train=data_y,\n X_test=test_X,\n y_test=test_y,\n name=plot_name,\n scale=scale,\n save=save,\n fit_kwargs=fit_kwargs,\n groups=groups,\n display=display,\n display_format=display_format,\n )\n\n def threshold():\n\n from yellowbrick.classifier import DiscriminationThreshold\n\n visualizer = DiscriminationThreshold(pipeline_with_model, random_state=seed)\n show_yellowbrick_plot(\n visualizer=visualizer,\n X_train=data_X,\n y_train=data_y,\n X_test=test_X,\n y_test=test_y,\n name=plot_name,\n scale=scale,\n save=save,\n fit_kwargs=fit_kwargs,\n groups=groups,\n display=display,\n display_format=display_format,\n )\n\n def pr():\n\n from yellowbrick.classifier import PrecisionRecallCurve\n\n visualizer = PrecisionRecallCurve(pipeline_with_model, random_state=seed)\n show_yellowbrick_plot(\n visualizer=visualizer,\n X_train=data_X,\n y_train=data_y,\n X_test=test_X,\n y_test=test_y,\n name=plot_name,\n scale=scale,\n save=save,\n fit_kwargs=fit_kwargs,\n groups=groups,\n display=display,\n display_format=display_format,\n )\n\n def confusion_matrix():\n\n from yellowbrick.classifier import ConfusionMatrix\n\n visualizer = ConfusionMatrix(\n pipeline_with_model, random_state=seed, fontsize=15, cmap=\"Greens\",\n )\n show_yellowbrick_plot(\n visualizer=visualizer,\n X_train=data_X,\n y_train=data_y,\n X_test=test_X,\n y_test=test_y,\n name=plot_name,\n scale=scale,\n save=save,\n fit_kwargs=fit_kwargs,\n groups=groups,\n display=display,\n display_format=display_format,\n )\n\n def error():\n\n if _ml_usecase == MLUsecase.CLASSIFICATION:\n from yellowbrick.classifier import ClassPredictionError\n\n visualizer = ClassPredictionError(\n pipeline_with_model, random_state=seed\n )\n\n elif _ml_usecase == MLUsecase.REGRESSION:\n from yellowbrick.regressor import PredictionError\n\n visualizer = PredictionError(pipeline_with_model, random_state=seed)\n\n show_yellowbrick_plot(\n visualizer=visualizer,\n X_train=data_X,\n y_train=data_y,\n X_test=test_X,\n y_test=test_y,\n name=plot_name,\n scale=scale,\n save=save,\n fit_kwargs=fit_kwargs,\n groups=groups,\n display=display,\n display_format=display_format,\n )\n\n def cooks():\n\n from yellowbrick.regressor import CooksDistance\n\n visualizer = CooksDistance()\n show_yellowbrick_plot(\n visualizer=visualizer,\n X_train=X,\n y_train=y,\n X_test=test_X,\n y_test=test_y,\n name=plot_name,\n scale=scale,\n save=save,\n fit_kwargs=fit_kwargs,\n handle_test=\"\",\n groups=groups,\n display=display,\n display_format=display_format,\n )\n\n def class_report():\n\n from yellowbrick.classifier import ClassificationReport\n\n visualizer = ClassificationReport(\n pipeline_with_model, random_state=seed, support=True\n )\n show_yellowbrick_plot(\n visualizer=visualizer,\n X_train=data_X,\n y_train=data_y,\n X_test=test_X,\n y_test=test_y,\n name=plot_name,\n scale=scale,\n save=save,\n fit_kwargs=fit_kwargs,\n groups=groups,\n display=display,\n display_format=display_format,\n )\n\n def boundary():\n\n from sklearn.preprocessing import StandardScaler\n from sklearn.decomposition import PCA\n from yellowbrick.contrib.classifier import DecisionViz\n\n data_X_transformed = data_X.select_dtypes(include=\"float32\")\n test_X_transformed = test_X.select_dtypes(include=\"float32\")\n logger.info(\"Fitting StandardScaler()\")\n data_X_transformed = StandardScaler().fit_transform(data_X_transformed)\n test_X_transformed = StandardScaler().fit_transform(test_X_transformed)\n pca = PCA(n_components=2, random_state=seed)\n logger.info(\"Fitting PCA()\")\n data_X_transformed = pca.fit_transform(data_X_transformed)\n test_X_transformed = pca.fit_transform(test_X_transformed)\n\n data_y_transformed = np.array(data_y)\n test_y_transformed = np.array(test_y)\n\n viz_ = DecisionViz(pipeline_with_model)\n show_yellowbrick_plot(\n visualizer=viz_,\n X_train=data_X_transformed,\n y_train=data_y_transformed,\n X_test=test_X_transformed,\n y_test=test_y_transformed,\n name=plot_name,\n scale=scale,\n handle_test=\"draw\",\n save=save,\n fit_kwargs=fit_kwargs,\n groups=groups,\n display=display,\n features=[\"Feature One\", \"Feature Two\"],\n classes=[\"A\", \"B\"],\n display_format=display_format,\n )\n\n def rfe():\n\n from yellowbrick.model_selection import RFECV\n\n visualizer = RFECV(pipeline_with_model, cv=cv)\n show_yellowbrick_plot(\n visualizer=visualizer,\n X_train=data_X,\n y_train=data_y,\n X_test=test_X,\n y_test=test_y,\n handle_test=\"\",\n name=plot_name,\n scale=scale,\n save=save,\n fit_kwargs=fit_kwargs,\n groups=groups,\n display=display,\n display_format=display_format,\n )\n\n def learning():\n\n from yellowbrick.model_selection import LearningCurve\n\n sizes = np.linspace(0.3, 1.0, 10)\n visualizer = LearningCurve(\n pipeline_with_model,\n cv=cv,\n train_sizes=sizes,\n n_jobs=_gpu_n_jobs_param,\n random_state=seed,\n )\n show_yellowbrick_plot(\n visualizer=visualizer,\n X_train=data_X,\n y_train=data_y,\n X_test=test_X,\n y_test=test_y,\n handle_test=\"\",\n name=plot_name,\n scale=scale,\n save=save,\n fit_kwargs=fit_kwargs,\n groups=groups,\n display=display,\n display_format=display_format,\n )\n\n def lift():\n\n display.move_progress()\n logger.info(\"Generating predictions / predict_proba on X_test\")\n with fit_if_not_fitted(\n pipeline_with_model, data_X, data_y, groups=groups, **fit_kwargs\n ) as fitted_pipeline_with_model:\n y_test__ = fitted_pipeline_with_model.predict(X_test)\n predict_proba__ = fitted_pipeline_with_model.predict_proba(X_test)\n display.move_progress()\n display.move_progress()\n display.clear_output()\n with MatplotlibDefaultDPI(base_dpi=_base_dpi, scale_to_set=scale):\n fig = skplt.metrics.plot_lift_curve(\n y_test__, predict_proba__, figsize=(10, 6)\n )\n if save:\n logger.info(f\"Saving '{plot_name}.png' in current active directory\")\n plt.savefig(f\"{plot_name}.png\", bbox_inches=\"tight\")\n elif system:\n plt.show()\n plt.close()\n\n logger.info(\"Visual Rendered Successfully\")\n\n def gain():\n\n display.move_progress()\n logger.info(\"Generating predictions / predict_proba on X_test\")\n with fit_if_not_fitted(\n pipeline_with_model, data_X, data_y, groups=groups, **fit_kwargs\n ) as fitted_pipeline_with_model:\n y_test__ = fitted_pipeline_with_model.predict(X_test)\n predict_proba__ = fitted_pipeline_with_model.predict_proba(X_test)\n display.move_progress()\n display.move_progress()\n display.clear_output()\n with MatplotlibDefaultDPI(base_dpi=_base_dpi, scale_to_set=scale):\n fig = skplt.metrics.plot_cumulative_gain(\n y_test__, predict_proba__, figsize=(10, 6)\n )\n if save:\n logger.info(f\"Saving '{plot_name}.png' in current active directory\")\n plt.savefig(f\"{plot_name}.png\", bbox_inches=\"tight\")\n elif system:\n plt.show()\n plt.close()\n\n logger.info(\"Visual Rendered Successfully\")\n\n def manifold():\n\n from yellowbrick.features import Manifold\n\n data_X_transformed = data_X.select_dtypes(include=\"float32\")\n visualizer = Manifold(manifold=\"tsne\", random_state=seed)\n show_yellowbrick_plot(\n visualizer=visualizer,\n X_train=data_X_transformed,\n y_train=data_y,\n X_test=test_X,\n y_test=test_y,\n handle_train=\"fit_transform\",\n handle_test=\"\",\n name=plot_name,\n scale=scale,\n save=save,\n fit_kwargs=fit_kwargs,\n groups=groups,\n display=display,\n display_format=display_format,\n )\n\n def tree():\n\n from sklearn.tree import plot_tree\n from sklearn.base import is_classifier\n from sklearn.model_selection import check_cv\n\n is_stacked_model = False\n is_ensemble_of_forests = False\n\n tree_estimator = pipeline_with_model.steps[-1][1]\n\n if \"final_estimator\" in tree_estimator.get_params():\n tree_estimator = tree_estimator.final_estimator\n is_stacked_model = True\n\n if (\n \"base_estimator\" in tree_estimator.get_params()\n and \"n_estimators\" in tree_estimator.base_estimator.get_params()\n ):\n n_estimators = (\n tree_estimator.get_params()[\"n_estimators\"]\n * tree_estimator.base_estimator.get_params()[\"n_estimators\"]\n )\n is_ensemble_of_forests = True\n elif \"n_estimators\" in tree_estimator.get_params():\n n_estimators = tree_estimator.get_params()[\"n_estimators\"]\n else:\n n_estimators = 1\n if n_estimators > 10:\n rows = (n_estimators // 10) + 1\n cols = 10\n else:\n rows = 1\n cols = n_estimators\n figsize = (cols * 20, rows * 16)\n fig, axes = plt.subplots(\n nrows=rows,\n ncols=cols,\n figsize=figsize,\n dpi=_base_dpi * scale,\n squeeze=False,\n )\n axes = list(axes.flatten())\n\n fig.suptitle(\"Decision Trees\")\n\n display.move_progress()\n logger.info(\"Plotting decision trees\")\n with fit_if_not_fitted(\n pipeline_with_model, data_X, data_y, groups=groups, **fit_kwargs\n ) as fitted_pipeline_with_model:\n trees = []\n feature_names = list(data_X.columns)\n if _ml_usecase == MLUsecase.CLASSIFICATION:\n class_names = {\n v: k\n for k, v in prep_pipe.named_steps[\"dtypes\"].replacement.items()\n }\n else:\n class_names = None\n fitted_tree_estimator = fitted_pipeline_with_model.steps[-1][1]\n if is_stacked_model:\n stacked_feature_names = []\n if _ml_usecase == MLUsecase.CLASSIFICATION:\n classes = list(data_y.unique())\n if len(classes) == 2:\n classes.pop()\n for c in classes:\n stacked_feature_names.extend(\n [\n f\"{k}_{class_names[c]}\"\n for k, v in fitted_tree_estimator.estimators\n ]\n )\n else:\n stacked_feature_names.extend(\n [f\"{k}\" for k, v in fitted_tree_estimator.estimators]\n )\n if not fitted_tree_estimator.passthrough:\n feature_names = stacked_feature_names\n else:\n feature_names = stacked_feature_names + feature_names\n fitted_tree_estimator = fitted_tree_estimator.final_estimator_\n if is_ensemble_of_forests:\n for estimator in fitted_tree_estimator.estimators_:\n trees.extend(estimator.estimators_)\n else:\n try:\n trees = fitted_tree_estimator.estimators_\n except:\n trees = [fitted_tree_estimator]\n if _ml_usecase == MLUsecase.CLASSIFICATION:\n class_names = list(class_names.values())\n for i, tree in enumerate(trees):\n logger.info(f\"Plotting tree {i}\")\n plot_tree(\n tree,\n feature_names=feature_names,\n class_names=class_names,\n filled=True,\n rounded=True,\n precision=4,\n ax=axes[i],\n )\n axes[i].set_title(f\"Tree {i}\")\n for i in range(len(trees), len(axes)):\n axes[i].set_visible(False)\n display.move_progress()\n\n display.move_progress()\n display.clear_output()\n if save:\n logger.info(f\"Saving '{plot_name}.png' in current active directory\")\n plt.savefig(f\"{plot_name}.png\", bbox_inches=\"tight\")\n elif system:\n plt.show()\n plt.close()\n\n logger.info(\"Visual Rendered Successfully\")\n\n def calibration():\n\n from sklearn.calibration import calibration_curve\n\n plt.figure(figsize=(7, 6), dpi=_base_dpi * scale)\n ax1 = plt.subplot2grid((3, 1), (0, 0), rowspan=2)\n\n ax1.plot([0, 1], [0, 1], \"k:\", label=\"Perfectly calibrated\")\n display.move_progress()\n logger.info(\"Scoring test/hold-out set\")\n with fit_if_not_fitted(\n pipeline_with_model, data_X, data_y, groups=groups, **fit_kwargs\n ) as fitted_pipeline_with_model:\n prob_pos = fitted_pipeline_with_model.predict_proba(test_X)[:, 1]\n prob_pos = (prob_pos - prob_pos.min()) / (prob_pos.max() - prob_pos.min())\n fraction_of_positives, mean_predicted_value = calibration_curve(\n test_y, prob_pos, n_bins=10\n )\n display.move_progress()\n ax1.plot(\n mean_predicted_value,\n fraction_of_positives,\n \"s-\",\n label=f\"{model_name}\",\n )\n\n ax1.set_ylabel(\"Fraction of positives\")\n ax1.set_ylim([0, 1])\n ax1.set_xlim([0, 1])\n ax1.legend(loc=\"lower right\")\n ax1.set_title(\"Calibration plots (reliability curve)\")\n ax1.set_facecolor(\"white\")\n ax1.grid(b=True, color=\"grey\", linewidth=0.5, linestyle=\"-\")\n plt.tight_layout()\n display.move_progress()\n display.clear_output()\n if save:\n logger.info(f\"Saving '{plot_name}.png' in current active directory\")\n plt.savefig(f\"{plot_name}.png\", bbox_inches=\"tight\")\n elif system:\n plt.show()\n plt.close()\n\n logger.info(\"Visual Rendered Successfully\")\n\n def vc():\n\n logger.info(\"Determining param_name\")\n\n actual_estimator_label = get_pipeline_estimator_label(pipeline_with_model)\n actual_estimator = pipeline_with_model.named_steps[actual_estimator_label]\n\n try:\n try:\n # catboost special case\n model_params = actual_estimator.get_all_params()\n except:\n model_params = pipeline_with_model.get_params()\n except:\n display.clear_output()\n logger.error(\"VC plot failed. Exception:\")\n logger.error(traceback.format_exc())\n raise TypeError(\n \"Plot not supported for this estimator. Try different estimator.\"\n )\n\n if _ml_usecase == MLUsecase.CLASSIFICATION:\n\n # Catboost\n if \"depth\" in model_params:\n param_name = f\"{actual_estimator_label}__depth\"\n param_range = np.arange(1, 8 if gpu_param else 11)\n\n # SGD Classifier\n elif f\"{actual_estimator_label}__l1_ratio\" in model_params:\n param_name = f\"{actual_estimator_label}__l1_ratio\"\n param_range = np.arange(0, 1, 0.01)\n\n # tree based models\n elif f\"{actual_estimator_label}__max_depth\" in model_params:\n param_name = f\"{actual_estimator_label}__max_depth\"\n param_range = np.arange(1, 11)\n\n # knn\n elif f\"{actual_estimator_label}__n_neighbors\" in model_params:\n param_name = f\"{actual_estimator_label}__n_neighbors\"\n param_range = np.arange(1, 11)\n\n # MLP / Ridge\n elif f\"{actual_estimator_label}__alpha\" in model_params:\n param_name = f\"{actual_estimator_label}__alpha\"\n param_range = np.arange(0, 1, 0.1)\n\n # Logistic Regression\n elif f\"{actual_estimator_label}__C\" in model_params:\n param_name = f\"{actual_estimator_label}__C\"\n param_range = np.arange(1, 11)\n\n # Bagging / Boosting\n elif f\"{actual_estimator_label}__n_estimators\" in model_params:\n param_name = f\"{actual_estimator_label}__n_estimators\"\n param_range = np.arange(1, 1000, 10)\n\n # Naive Bayes\n elif f\"{actual_estimator_label}__var_smoothing\" in model_params:\n param_name = f\"{actual_estimator_label}__var_smoothing\"\n param_range = np.arange(0.1, 1, 0.01)\n\n # QDA\n elif f\"{actual_estimator_label}__reg_param\" in model_params:\n param_name = f\"{actual_estimator_label}__reg_param\"\n param_range = np.arange(0, 1, 0.1)\n\n # GPC\n elif f\"{actual_estimator_label}__max_iter_predict\" in model_params:\n param_name = f\"{actual_estimator_label}__max_iter_predict\"\n param_range = np.arange(100, 1000, 100)\n\n else:\n display.clear_output()\n raise TypeError(\n \"Plot not supported for this estimator. Try different estimator.\"\n )\n\n elif _ml_usecase == MLUsecase.REGRESSION:\n\n # Catboost\n if \"depth\" in model_params:\n param_name = f\"{actual_estimator_label}__depth\"\n param_range = np.arange(1, 8 if gpu_param else 11)\n\n # lasso/ridge/en/llar/huber/kr/mlp/br/ard\n elif f\"{actual_estimator_label}__alpha\" in model_params:\n param_name = f\"{actual_estimator_label}__alpha\"\n param_range = np.arange(0, 1, 0.1)\n\n elif f\"{actual_estimator_label}__alpha_1\" in model_params:\n param_name = f\"{actual_estimator_label}__alpha_1\"\n param_range = np.arange(0, 1, 0.1)\n\n # par/svm\n elif f\"{actual_estimator_label}__C\" in model_params:\n param_name = f\"{actual_estimator_label}__C\"\n param_range = np.arange(1, 11)\n\n # tree based models (dt/rf/et)\n elif f\"{actual_estimator_label}__max_depth\" in model_params:\n param_name = f\"{actual_estimator_label}__max_depth\"\n param_range = np.arange(1, 11)\n\n # knn\n elif f\"{actual_estimator_label}__n_neighbors\" in model_params:\n param_name = f\"{actual_estimator_label}__n_neighbors\"\n param_range = np.arange(1, 11)\n\n # Bagging / Boosting (ada/gbr)\n elif f\"{actual_estimator_label}__n_estimators\" in model_params:\n param_name = f\"{actual_estimator_label}__n_estimators\"\n param_range = np.arange(1, 1000, 10)\n\n # Bagging / Boosting (ada/gbr)\n elif f\"{actual_estimator_label}__n_nonzero_coefs\" in model_params:\n param_name = f\"{actual_estimator_label}__n_nonzero_coefs\"\n if len(X_train.columns) >= 10:\n param_max = 11\n else:\n param_max = len(X_train.columns) + 1\n param_range = np.arange(1, param_max, 1)\n\n elif f\"{actual_estimator_label}__eps\" in model_params:\n param_name = f\"{actual_estimator_label}__eps\"\n param_range = np.arange(0, 1, 0.1)\n\n elif f\"{actual_estimator_label}__max_subpopulation\" in model_params:\n param_name = f\"{actual_estimator_label}__max_subpopulation\"\n param_range = np.arange(1000, 100000, 2000)\n\n elif f\"{actual_estimator_label}__min_samples\" in model_params:\n param_name = f\"{actual_estimator_label}__max_subpopulation\"\n param_range = np.arange(0.01, 1, 0.1)\n\n else:\n display.clear_output()\n raise TypeError(\n \"Plot not supported for this estimator. Try different estimator.\"\n )\n\n logger.info(f\"param_name: {param_name}\")\n\n display.move_progress()\n\n from yellowbrick.model_selection import ValidationCurve\n\n viz = ValidationCurve(\n pipeline_with_model,\n param_name=param_name,\n param_range=param_range,\n cv=cv,\n random_state=seed,\n n_jobs=_gpu_n_jobs_param,\n )\n show_yellowbrick_plot(\n visualizer=viz,\n X_train=data_X,\n y_train=data_y,\n X_test=test_X,\n y_test=test_y,\n handle_train=\"fit\",\n handle_test=\"\",\n name=plot_name,\n scale=scale,\n save=save,\n fit_kwargs=fit_kwargs,\n groups=groups,\n display=display,\n display_format=display_format,\n )\n\n def dimension():\n\n from yellowbrick.features import RadViz\n from sklearn.preprocessing import StandardScaler\n from sklearn.decomposition import PCA\n\n data_X_transformed = data_X.select_dtypes(include=\"float32\")\n logger.info(\"Fitting StandardScaler()\")\n data_X_transformed = StandardScaler().fit_transform(data_X_transformed)\n data_y_transformed = np.array(data_y)\n\n features = min(round(len(data_X.columns) * 0.3, 0), 5)\n features = int(features)\n\n pca = PCA(n_components=features, random_state=seed)\n logger.info(\"Fitting PCA()\")\n data_X_transformed = pca.fit_transform(data_X_transformed)\n display.move_progress()\n classes = data_y.unique().tolist()\n visualizer = RadViz(classes=classes, alpha=0.25)\n\n show_yellowbrick_plot(\n visualizer=visualizer,\n X_train=data_X_transformed,\n y_train=data_y_transformed,\n X_test=test_X,\n y_test=test_y,\n handle_train=\"fit_transform\",\n handle_test=\"\",\n name=plot_name,\n scale=scale,\n save=save,\n fit_kwargs=fit_kwargs,\n groups=groups,\n display=display,\n display_format=display_format,\n )\n\n def feature():\n _feature(10)\n\n def feature_all():\n _feature(len(data_X.columns))\n\n def _feature(n: int):\n variables = None\n temp_model = pipeline_with_model\n if hasattr(pipeline_with_model, \"steps\"):\n temp_model = pipeline_with_model.steps[-1][1]\n if hasattr(temp_model, \"coef_\"):\n try:\n coef = temp_model.coef_.flatten()\n if len(coef) > len(data_X.columns):\n coef = coef[: len(data_X.columns)]\n variables = abs(coef)\n except:\n pass\n if variables is None:\n logger.warning(\"No coef_ found. Trying feature_importances_\")\n variables = abs(temp_model.feature_importances_)\n coef_df = pd.DataFrame({\"Variable\": data_X.columns, \"Value\": variables})\n sorted_df = (\n coef_df.sort_values(by=\"Value\", ascending=False)\n .head(n)\n .sort_values(by=\"Value\")\n )\n my_range = range(1, len(sorted_df.index) + 1)\n display.move_progress()\n plt.figure(figsize=(8, 5 * (n // 10)), dpi=_base_dpi * scale)\n plt.hlines(y=my_range, xmin=0, xmax=sorted_df[\"Value\"], color=\"skyblue\")\n plt.plot(sorted_df[\"Value\"], my_range, \"o\")\n display.move_progress()\n plt.yticks(my_range, sorted_df[\"Variable\"])\n plt.title(\"Feature Importance Plot\")\n plt.xlabel(\"Variable Importance\")\n plt.ylabel(\"Features\")\n display.move_progress()\n display.clear_output()\n if save:\n logger.info(f\"Saving '{plot_name}.png' in current active directory\")\n plt.savefig(f\"{plot_name}.png\", bbox_inches=\"tight\")\n elif system:\n plt.show()\n plt.close()\n\n logger.info(\"Visual Rendered Successfully\")\n\n def parameter():\n\n try:\n params = estimator.get_all_params()\n except:\n params = estimator.get_params(deep=False)\n\n param_df = pd.DataFrame.from_dict(\n {str(k): str(v) for k, v in params.items()},\n orient=\"index\",\n columns=[\"Parameters\"],\n )\n display.display(param_df, clear=True)\n logger.info(\"Visual Rendered Successfully\")\n\n # execute the plot method\n ret = locals()[plot]()\n if ret:\n plot_filename = ret\n\n try:\n plt.close()\n except:\n pass\n\n gc.collect()\n\n logger.info(\n \"plot_model() succesfully completed......................................\"\n )\n\n if save:\n return plot_filename\n\n\ndef evaluate_model(\n estimator,\n fold: Optional[Union[int, Any]] = None,\n fit_kwargs: Optional[dict] = None,\n feature_name: Optional[str] = None,\n groups: Optional[Union[str, Any]] = None,\n use_train_data: bool = False,\n):\n\n \"\"\"\n This function displays a user interface for all of the available plots for\n a given estimator. It internally uses the plot_model() function.\n\n Example\n -------\n >>> from pycaret.datasets import get_data\n >>> juice = get_data('juice')\n >>> experiment_name = setup(data = juice, target = 'Purchase')\n >>> lr = create_model('lr')\n >>> evaluate_model(lr)\n\n This will display the User Interface for all of the plots for a given\n estimator.\n\n Parameters\n ----------\n estimator : object, default = none\n A trained model object should be passed as an estimator.\n\n fold: integer or scikit-learn compatible CV generator, default = None\n Controls cross-validation. If None, will use the CV generator defined in setup().\n If integer, will use KFold CV with that many folds.\n When cross_validation is False, this parameter is ignored.\n\n fit_kwargs: dict, default = {} (empty dict)\n Dictionary of arguments passed to the fit method of the model.\n\n groups: str or array-like, with shape (n_samples,), default = None\n Optional Group labels for the samples used while splitting the dataset into train/test set.\n If string is passed, will use the data column with that name as the groups.\n Only used if a group based cross-validation generator is used (eg. GroupKFold).\n If None, will use the value set in fold_groups param in setup().\n\n Returns\n -------\n User_Interface\n Displays the user interface for plotting.\n\n \"\"\"\n\n function_params_str = \", \".join([f\"{k}={v}\" for k, v in locals().items()])\n\n logger = get_logger()\n\n logger.info(\"Initializing evaluate_model()\")\n logger.info(f\"evaluate_model({function_params_str})\")\n\n from ipywidgets import widgets\n from ipywidgets.widgets import interact, fixed\n\n if not fit_kwargs:\n fit_kwargs = {}\n\n a = widgets.ToggleButtons(\n options=[(v, k) for k, v in _available_plots.items()],\n description=\"Plot Type:\",\n disabled=False,\n button_style=\"\", # 'success', 'info', 'warning', 'danger' or ''\n icons=[\"\"],\n )\n\n fold = _get_cv_splitter(fold)\n\n groups = _get_groups(groups)\n\n d = interact(\n plot_model,\n estimator=fixed(estimator),\n plot=a,\n save=fixed(False),\n verbose=fixed(True),\n scale=fixed(1),\n fold=fixed(fold),\n fit_kwargs=fixed(fit_kwargs),\n feature_name=fixed(feature_name),\n label=fixed(False),\n groups=fixed(groups),\n use_train_data=fixed(use_train_data),\n system=fixed(True),\n display=fixed(None),\n display_format=fixed(None),\n is_in_evaluate=fixed(True),\n )\n\n\ndef interpret_model(\n estimator,\n plot: str = \"summary\",\n feature: Optional[str] = None,\n observation: Optional[int] = None,\n use_train_data: Optional[bool] = False,\n X_new_sample: Optional[pd.DataFrame] = None,\n save: bool = False,\n **kwargs, # added in pycaret==2.1\n):\n\n \"\"\"\n This function takes a trained model object and returns an interpretation plot\n based on the test / hold-out set. It only supports tree based algorithms.\n\n This function is implemented based on the SHAP (SHapley Additive exPlanations),\n which is a unified approach to explain the output of any machine learning model.\n SHAP connects game theory with local explanations.\n\n For more information : https://shap.readthedocs.io/en/latest/\n\n Example\n -------\n >>> from pycaret.datasets import get_data\n >>> juice = get_data('juice')\n >>> experiment_name = setup(data = juice, target = 'Purchase')\n >>> dt = create_model('dt')\n >>> interpret_model(dt)\n\n This will return a summary interpretation plot of Decision Tree model.\n\n Parameters\n ----------\n estimator : object, default = none\n A trained tree based model object should be passed as an estimator.\n\n plot : str, default = 'summary'\n Other available options are 'correlation' and 'reason'.\n\n feature: str, default = None\n This parameter is only needed when plot = 'correlation'. By default feature is\n set to None which means the first column of the dataset will be used as a\n variable. A feature parameter must be passed to change this.\n\n observation: integer, default = None\n This parameter only comes into effect when plot is set to 'reason'. If no\n observation number is provided, it will return an analysis of all observations\n with the option to select the feature on x and y axes through drop down\n interactivity. For analysis at the sample level, an observation parameter must\n be passed with the index value of the observation in test / hold-out set.\n\n X_new_sample: pd.DataFrame, default = None\n Row from an out-of-sample dataframe (neither train nor test data) to be plotted.\n The sample must have the same columns as the raw input data, and it is transformed\n by the preprocessing pipeline automatically before plotting.\n\n save: bool, default = False\n When set to True, Plot is saved as a 'png' file in current working directory.\n\n **kwargs:\n Additional keyword arguments to pass to the plot.\n\n Returns\n -------\n Visual_Plot\n Returns the visual plot.\n Returns the interactive JS plot when plot = 'reason'.\n\n Warnings\n --------\n - interpret_model doesn't support multiclass problems.\n\n \"\"\"\n\n function_params_str = \", \".join([f\"{k}={v}\" for k, v in locals().items()])\n\n logger = get_logger()\n\n logger.info(\"Initializing interpret_model()\")\n logger.info(f\"interpret_model({function_params_str})\")\n\n logger.info(\"Checking exceptions\")\n\n import matplotlib.pyplot as plt\n\n # checking if shap available\n try:\n import shap\n except ImportError:\n logger.error(\n \"shap library not found. pip install shap to use interpret_model function.\"\n )\n raise ImportError(\n \"shap library not found. pip install shap to use interpret_model function.\"\n )\n\n # get estimator from meta estimator\n estimator = get_estimator_from_meta_estimator(estimator)\n\n # allowed models\n model_id = _get_model_id(estimator)\n\n shap_models = {k: v for k, v in _all_models_internal.items() if v.shap}\n shap_models_ids = set(shap_models.keys())\n\n if model_id not in shap_models_ids:\n raise TypeError(\n f\"This function only supports tree based models for binary classification: {', '.join(shap_models_ids)}.\"\n )\n\n # plot type\n allowed_types = [\"summary\", \"correlation\", \"reason\"]\n if plot not in allowed_types:\n raise ValueError(\n \"type parameter only accepts 'summary', 'correlation' or 'reason'.\"\n )\n\n if X_new_sample is not None and (observation is not None or use_train_data):\n raise ValueError(\n \"Specifying 'X_new_sample' and ('observation' or 'use_train_data') is ambiguous.\"\n )\n \"\"\"\n Error Checking Ends here\n\n \"\"\"\n if X_new_sample is not None:\n test_X = prep_pipe.transform(X_new_sample)\n else:\n # Storing X_train and y_train in data_X and data_y parameter\n test_X = X_train if use_train_data else X_test\n\n np.random.seed(seed)\n\n # storing estimator in model variable\n model = estimator\n\n # defining type of classifier\n shap_models_type1 = {k for k, v in shap_models.items() if v.shap == \"type1\"}\n shap_models_type2 = {k for k, v in shap_models.items() if v.shap == \"type2\"}\n\n logger.info(f\"plot type: {plot}\")\n\n shap_plot = None\n\n def summary(show: bool = True):\n\n logger.info(\"Creating TreeExplainer\")\n explainer = shap.TreeExplainer(model)\n logger.info(\"Compiling shap values\")\n shap_values = explainer.shap_values(test_X)\n shap_plot = shap.summary_plot(shap_values, test_X, show=show, **kwargs)\n if save:\n plt.savefig(f\"SHAP {plot}.png\", bbox_inches=\"tight\")\n return shap_plot\n\n def correlation(show: bool = True):\n\n if feature == None:\n\n logger.warning(\n f\"No feature passed. Default value of feature used for correlation plot: {test_X.columns[0]}\"\n )\n dependence = test_X.columns[0]\n\n else:\n\n logger.warning(\n f\"feature value passed. Feature used for correlation plot: {test_X.columns[0]}\"\n )\n dependence = feature\n\n logger.info(\"Creating TreeExplainer\")\n explainer = shap.TreeExplainer(model)\n logger.info(\"Compiling shap values\")\n shap_values = explainer.shap_values(test_X)\n\n if model_id in shap_models_type1:\n logger.info(\"model type detected: type 1\")\n shap.dependence_plot(\n dependence, shap_values[1], test_X, show=show, **kwargs\n )\n elif model_id in shap_models_type2:\n logger.info(\"model type detected: type 2\")\n shap.dependence_plot(dependence, shap_values, test_X, show=show, **kwargs)\n if save:\n plt.savefig(f\"SHAP {plot}.png\", bbox_inches=\"tight\")\n return None\n\n def reason(show: bool = True):\n shap_plot = None\n if model_id in shap_models_type1:\n logger.info(\"model type detected: type 1\")\n\n logger.info(\"Creating TreeExplainer\")\n explainer = shap.TreeExplainer(model)\n logger.info(\"Compiling shap values\")\n\n if observation is None:\n logger.warning(\n \"Observation set to None. Model agnostic plot will be rendered.\"\n )\n shap_values = explainer.shap_values(test_X)\n shap.initjs()\n shap_plot = shap.force_plot(\n explainer.expected_value[1],\n shap_values[1],\n test_X,\n show=show,\n **kwargs,\n )\n\n else:\n row_to_show = observation\n data_for_prediction = test_X.iloc[row_to_show]\n\n if model_id == \"lightgbm\":\n logger.info(\"model type detected: LGBMClassifier\")\n shap_values = explainer.shap_values(test_X)\n shap.initjs()\n shap_plot = shap.force_plot(\n explainer.expected_value[1],\n shap_values[0][row_to_show],\n data_for_prediction,\n show=show,\n **kwargs,\n )\n\n else:\n logger.info(\"model type detected: Unknown\")\n\n shap_values = explainer.shap_values(data_for_prediction)\n shap.initjs()\n shap_plot = shap.force_plot(\n explainer.expected_value[1],\n shap_values[1],\n data_for_prediction,\n show=show,\n **kwargs,\n )\n\n elif model_id in shap_models_type2:\n logger.info(\"model type detected: type 2\")\n\n logger.info(\"Creating TreeExplainer\")\n explainer = shap.TreeExplainer(model)\n logger.info(\"Compiling shap values\")\n shap_values = explainer.shap_values(test_X)\n shap.initjs()\n\n if observation is None:\n logger.warning(\n \"Observation set to None. Model agnostic plot will be rendered.\"\n )\n\n shap_plot = shap.force_plot(\n explainer.expected_value, shap_values, test_X, show=show, **kwargs\n )\n\n else:\n\n row_to_show = observation\n data_for_prediction = test_X.iloc[row_to_show]\n\n shap_plot = shap.force_plot(\n explainer.expected_value,\n shap_values[row_to_show, :],\n test_X.iloc[row_to_show, :],\n show=show,\n **kwargs,\n )\n if save:\n shap.save_html(f\"SHAP {plot}.html\", shap_plot)\n return shap_plot\n\n shap_plot = locals()[plot](show=not save)\n\n logger.info(\"Visual Rendered Successfully\")\n\n logger.info(\n \"interpret_model() succesfully completed......................................\"\n )\n\n gc.collect()\n return shap_plot\n\n\ndef calibrate_model(\n estimator,\n method: str = \"sigmoid\",\n fold: Optional[Union[int, Any]] = None,\n round: int = 4,\n fit_kwargs: Optional[dict] = None,\n groups: Optional[Union[str, Any]] = None,\n verbose: bool = True,\n display: Optional[Display] = None, # added in pycaret==2.2.0\n) -> Any:\n\n \"\"\"\n This function takes the input of trained estimator and performs probability\n calibration with sigmoid or isotonic regression. The output prints a score\n grid that shows Accuracy, AUC, Recall, Precision, F1, Kappa and MCC by fold\n (default = 10 Fold). The ouput of the original estimator and the calibrated\n estimator (created using this function) might not differ much. In order\n to see the calibration differences, use 'calibration' plot in plot_model to\n see the difference before and after.\n\n This function returns a trained model object.\n\n Example\n -------\n >>> from pycaret.datasets import get_data\n >>> juice = get_data('juice')\n >>> experiment_name = setup(data = juice, target = 'Purchase')\n >>> dt_boosted = create_model('dt', ensemble = True, method = 'Boosting')\n >>> calibrated_dt = calibrate_model(dt_boosted)\n\n This will return Calibrated Boosted Decision Tree Model.\n\n Parameters\n ----------\n estimator : object\n\n method : str, default = 'sigmoid'\n The method to use for calibration. Can be 'sigmoid' which corresponds to Platt's\n method or 'isotonic' which is a non-parametric approach. It is not advised to use\n isotonic calibration with too few calibration samples\n\n fold: integer or scikit-learn compatible CV generator, default = None\n Controls cross-validation. If None, will use the CV generator defined in setup().\n If integer, will use KFold CV with that many folds.\n When cross_validation is False, this parameter is ignored.\n\n round: integer, default = 4\n Number of decimal places the metrics in the score grid will be rounded to.\n\n fit_kwargs: dict, default = {} (empty dict)\n Dictionary of arguments passed to the fit method of the model.\n\n groups: str or array-like, with shape (n_samples,), default = None\n Optional Group labels for the samples used while splitting the dataset into train/test set.\n If string is passed, will use the data column with that name as the groups.\n Only used if a group based cross-validation generator is used (eg. GroupKFold).\n If None, will use the value set in fold_groups param in setup().\n\n verbose: bool, default = True\n Score grid is not printed when verbose is set to False.\n\n Returns\n -------\n score_grid\n A table containing the scores of the model across the kfolds.\n Scoring metrics used are Accuracy, AUC, Recall, Precision, F1,\n Kappa and MCC. Mean and standard deviation of the scores across\n the folds are also returned.\n\n model\n trained and calibrated model object.\n\n Warnings\n --------\n - Avoid isotonic calibration with too few calibration samples (<1000) since it\n tends to overfit.\n\n - calibration plot not available for multiclass problems.\n\n\n \"\"\"\n\n function_params_str = \", \".join([f\"{k}={v}\" for k, v in locals().items()])\n\n logger = get_logger()\n\n logger.info(\"Initializing calibrate_model()\")\n logger.info(f\"calibrate_model({function_params_str})\")\n\n logger.info(\"Checking exceptions\")\n\n # run_time\n runtime_start = time.time()\n\n if not fit_kwargs:\n fit_kwargs = {}\n\n # checking fold parameter\n if fold is not None and not (type(fold) is int or is_sklearn_cv_generator(fold)):\n raise TypeError(\n \"fold parameter must be either None, an integer or a scikit-learn compatible CV generator object.\"\n )\n\n # checking round parameter\n if type(round) is not int:\n raise TypeError(\"Round parameter only accepts integer value.\")\n\n # checking verbose parameter\n if type(verbose) is not bool:\n raise TypeError(\"Verbose parameter can only take argument as True or False.\")\n\n \"\"\"\n\n ERROR HANDLING ENDS HERE\n\n \"\"\"\n\n fold = _get_cv_splitter(fold)\n\n groups = _get_groups(groups)\n\n logger.info(\"Preloading libraries\")\n\n # pre-load libraries\n\n logger.info(\"Preparing display monitor\")\n\n if not display:\n progress_args = {\"max\": 2 + 4}\n master_display_columns = [v.display_name for k, v in _all_metrics.items()]\n timestampStr = datetime.datetime.now().strftime(\"%H:%M:%S\")\n monitor_rows = [\n [\"Initiated\", \". . . . . . . . . . . . . . . . . .\", timestampStr],\n [\"Status\", \". . . . . . . . . . . . . . . . . .\", \"Loading Dependencies\"],\n [\"Estimator\", \". . . . . . . . . . . . . . . . . .\", \"Compiling Library\"],\n ]\n display = Display(\n verbose=verbose,\n html_param=html_param,\n progress_args=progress_args,\n master_display_columns=master_display_columns,\n monitor_rows=monitor_rows,\n )\n\n display.display_progress()\n display.display_monitor()\n display.display_master_display()\n\n np.random.seed(seed)\n\n logger.info(\"Getting model name\")\n\n full_name = _get_model_name(estimator)\n\n logger.info(f\"Base model : {full_name}\")\n\n display.update_monitor(2, full_name)\n display.display_monitor()\n\n \"\"\"\n MONITOR UPDATE STARTS\n \"\"\"\n\n display.update_monitor(1, \"Selecting Estimator\")\n display.display_monitor()\n\n \"\"\"\n MONITOR UPDATE ENDS\n \"\"\"\n\n # calibrating estimator\n\n logger.info(\"Importing untrained CalibratedClassifierCV\")\n\n calibrated_model_definition = _all_models_internal[\"CalibratedCV\"]\n model = calibrated_model_definition.class_def(\n base_estimator=estimator,\n method=method,\n cv=fold,\n **calibrated_model_definition.args,\n )\n\n display.move_progress()\n\n logger.info(\"SubProcess create_model() called ==================================\")\n model, model_fit_time = create_model_supervised(\n estimator=model,\n system=False,\n display=display,\n fold=fold,\n round=round,\n fit_kwargs=fit_kwargs,\n groups=groups,\n )\n model_results = pull()\n logger.info(\"SubProcess create_model() end ==================================\")\n\n model_results = model_results.round(round)\n\n display.move_progress()\n\n # end runtime\n runtime_end = time.time()\n runtime = np.array(runtime_end - runtime_start).round(2)\n\n # mlflow logging\n if logging_param:\n\n avgs_dict_log = {k: v for k, v in model_results.loc[\"Mean\"].items()}\n\n try:\n _mlflow_log_model(\n model=model,\n model_results=model_results,\n score_dict=avgs_dict_log,\n source=\"calibrate_models\",\n runtime=runtime,\n model_fit_time=model_fit_time,\n _prep_pipe=prep_pipe,\n log_plots=log_plots_param,\n display=display,\n )\n except:\n logger.error(f\"_mlflow_log_model() for {model} raised an exception:\")\n logger.error(traceback.format_exc())\n\n model_results = color_df(model_results, \"yellow\", [\"Mean\"], axis=1)\n model_results = model_results.set_precision(round)\n display.display(model_results, clear=True)\n\n logger.info(f\"create_model_container: {len(create_model_container)}\")\n logger.info(f\"master_model_container: {len(master_model_container)}\")\n logger.info(f\"display_container: {len(display_container)}\")\n\n logger.info(str(model))\n logger.info(\n \"calibrate_model() succesfully completed......................................\"\n )\n\n gc.collect()\n return model\n\n\ndef optimize_threshold(\n estimator,\n true_positive: int = 0,\n true_negative: int = 0,\n false_positive: int = 0,\n false_negative: int = 0,\n):\n\n \"\"\"\n This function optimizes probability threshold for a trained model using custom cost\n function that can be defined using combination of True Positives, True Negatives,\n False Positives (also known as Type I error), and False Negatives (Type II error).\n\n This function returns a plot of optimized cost as a function of probability\n threshold between 0 to 100.\n\n Example\n -------\n >>> from pycaret.datasets import get_data\n >>> juice = get_data('juice')\n >>> experiment_name = setup(data = juice, target = 'Purchase')\n >>> lr = create_model('lr')\n >>> optimize_threshold(lr, true_negative = 10, false_negative = -100)\n\n This will return a plot of optimized cost as a function of probability threshold.\n\n Parameters\n ----------\n estimator : object\n A trained model object should be passed as an estimator.\n\n true_positive : int, default = 0\n Cost function or returns when prediction is true positive.\n\n true_negative : int, default = 0\n Cost function or returns when prediction is true negative.\n\n false_positive : int, default = 0\n Cost function or returns when prediction is false positive.\n\n false_negative : int, default = 0\n Cost function or returns when prediction is false negative.\n\n\n Returns\n -------\n Visual_Plot\n Prints the visual plot.\n\n Warnings\n --------\n - This function is not supported for multiclass problems.\n\n\n \"\"\"\n\n function_params_str = \", \".join([f\"{k}={v}\" for k, v in locals().items()])\n\n logger = get_logger()\n\n logger.info(\"Initializing optimize_threshold()\")\n logger.info(f\"optimize_threshold({function_params_str})\")\n\n logger.info(\"Importing libraries\")\n\n # import libraries\n\n np.random.seed(seed)\n\n \"\"\"\n ERROR HANDLING STARTS HERE\n \"\"\"\n\n logger.info(\"Checking exceptions\")\n\n # exception 1 for multi-class\n if _is_multiclass():\n raise TypeError(\n \"optimize_threshold() cannot be used when target is multi-class.\"\n )\n\n # check predict_proba value\n if type(estimator) is not list:\n if not hasattr(estimator, \"predict_proba\"):\n raise TypeError(\n \"Estimator doesn't support predict_proba function and cannot be used in optimize_threshold().\"\n )\n\n # check cost function type\n allowed_types = [int, float]\n\n if type(true_positive) not in allowed_types:\n raise TypeError(\"true_positive parameter only accepts float or integer value.\")\n\n if type(true_negative) not in allowed_types:\n raise TypeError(\"true_negative parameter only accepts float or integer value.\")\n\n if type(false_positive) not in allowed_types:\n raise TypeError(\"false_positive parameter only accepts float or integer value.\")\n\n if type(false_negative) not in allowed_types:\n raise TypeError(\"false_negative parameter only accepts float or integer value.\")\n\n \"\"\"\n ERROR HANDLING ENDS HERE\n \"\"\"\n\n # define model as estimator\n model = estimator\n\n model_name = _get_model_name(model)\n\n # generate predictions and store actual on y_test in numpy array\n actual = np.array(y_test)\n\n predicted = model.predict_proba(X_test)\n predicted = predicted[:, 1]\n\n \"\"\"\n internal function to calculate loss starts here\n \"\"\"\n\n logger.info(\"Defining loss function\")\n\n def calculate_loss(\n actual,\n predicted,\n tp_cost=true_positive,\n tn_cost=true_negative,\n fp_cost=false_positive,\n fn_cost=false_negative,\n ):\n\n # true positives\n tp = predicted + actual\n tp = np.where(tp == 2, 1, 0)\n tp = tp.sum()\n\n # true negative\n tn = predicted + actual\n tn = np.where(tn == 0, 1, 0)\n tn = tn.sum()\n\n # false positive\n fp = (predicted > actual).astype(int)\n fp = np.where(fp == 1, 1, 0)\n fp = fp.sum()\n\n # false negative\n fn = (predicted < actual).astype(int)\n fn = np.where(fn == 1, 1, 0)\n fn = fn.sum()\n\n total_cost = (tp_cost * tp) + (tn_cost * tn) + (fp_cost * fp) + (fn_cost * fn)\n\n return total_cost\n\n \"\"\"\n internal function to calculate loss ends here\n \"\"\"\n\n grid = np.arange(0, 1, 0.0001)\n\n # loop starts here\n\n cost = []\n # global optimize_results\n\n logger.info(\"Iteration starts at 0\")\n\n for i in grid:\n\n pred_prob = (predicted >= i).astype(int)\n cost.append(calculate_loss(actual, pred_prob))\n\n optimize_results = pd.DataFrame(\n {\"Probability Threshold\": grid, \"Cost Function\": cost}\n )\n fig = px.line(\n optimize_results,\n x=\"Probability Threshold\",\n y=\"Cost Function\",\n line_shape=\"linear\",\n )\n fig.update_layout(plot_bgcolor=\"rgb(245,245,245)\")\n title = f\"{model_name} Probability Threshold Optimization\"\n\n # calculate vertical line\n y0 = optimize_results[\"Cost Function\"].min()\n y1 = optimize_results[\"Cost Function\"].max()\n x0 = optimize_results.sort_values(by=\"Cost Function\", ascending=False).iloc[0][0]\n x1 = x0\n\n t = x0\n if html_param:\n\n fig.add_shape(\n dict(\n type=\"line\", x0=x0, y0=y0, x1=x1, y1=y1, line=dict(color=\"red\", width=2)\n )\n )\n fig.update_layout(\n title={\n \"text\": title,\n \"y\": 0.95,\n \"x\": 0.45,\n \"xanchor\": \"center\",\n \"yanchor\": \"top\",\n }\n )\n logger.info(\"Figure ready for render\")\n fig.show()\n print(f\"Optimized Probability Threshold: {t} | Optimized Cost Function: {y1}\")\n logger.info(\n \"optimize_threshold() succesfully completed......................................\"\n )\n\n return float(t)\n\n\ndef assign_model(\n model, transformation: bool = False, score: bool = True, verbose: bool = True\n) -> pd.DataFrame:\n\n \"\"\"\n This function assigns each of the data point in the dataset passed during setup\n stage to one of the clusters using trained model object passed as model param.\n create_model() function must be called before using assign_model().\n\n This function returns a pandas.DataFrame.\n\n Example\n -------\n >>> from pycaret.datasets import get_data\n >>> jewellery = get_data('jewellery')\n >>> experiment_name = setup(data = jewellery, normalize = True)\n >>> kmeans = create_model('kmeans')\n >>> kmeans_df = assign_model(kmeans)\n\n This will return a pandas.DataFrame with inferred clusters using trained model.\n\n Parameters\n ----------\n model: trained model object, default = None\n\n transformation: bool, default = False\n When set to True, assigned clusters are returned on transformed dataset instead\n of original dataset passed during setup().\n\n verbose: Boolean, default = True\n Status update is not printed when verbose is set to False.\n\n Returns\n -------\n pandas.DataFrame\n Returns a DataFrame with assigned clusters using a trained model.\n\n \"\"\"\n\n function_params_str = \", \".join([f\"{k}={v}\" for k, v in locals().items()])\n\n logger = get_logger()\n\n logger.info(\"Initializing assign_model()\")\n logger.info(f\"assign_model({function_params_str})\")\n\n logger.info(\"Checking exceptions\")\n\n # checking transformation parameter\n if type(transformation) is not bool:\n raise TypeError(\n \"Transformation parameter can only take argument as True or False.\"\n )\n\n # checking verbose parameter\n if type(verbose) is not bool:\n raise TypeError(\"Verbose parameter can only take argument as True or False.\")\n\n \"\"\"\n error handling ends here\n \"\"\"\n\n if is_sklearn_pipeline(model):\n model = model.steps[-1][1]\n\n logger.info(\"Determining Trained Model\")\n\n name = _get_model_name(model)\n\n logger.info(f\"Trained Model : {name}\")\n\n logger.info(\"Copying data\")\n # copy data_\n if transformation:\n data = X.copy()\n logger.info(\n \"Transformation param set to True. Assigned clusters are attached on transformed dataset.\"\n )\n else:\n data = data_before_preprocess.copy()\n\n # calculation labels and attaching to dataframe\n\n if _ml_usecase == MLUsecase.CLUSTERING:\n labels = [f\"Cluster {i}\" for i in model.labels_]\n data[\"Cluster\"] = labels\n else:\n data[\"Anomaly\"] = model.labels_\n if score:\n data[\"Anomaly_Score\"] = model.decision_scores_\n\n logger.info(data.shape)\n logger.info(\n \"assign_model() succesfully completed......................................\"\n )\n\n return data\n\n\ndef predict_model_unsupervised(\n estimator, data: pd.DataFrame, ml_usecase: Optional[MLUsecase] = None,\n) -> pd.DataFrame:\n function_params_str = \", \".join(\n [f\"{k}={v}\" for k, v in locals().items() if k != \"data\"]\n )\n\n logger = get_logger()\n\n logger.info(\"Initializing predict_model()\")\n logger.info(f\"predict_model({function_params_str})\")\n\n if ml_usecase is None:\n ml_usecase = _ml_usecase\n\n # copy data and model\n data_transformed = data.copy()\n\n # exception checking for predict param\n if hasattr(estimator, \"predict\"):\n pass\n else:\n raise TypeError(\"Model doesn't support predict parameter.\")\n\n pred_score = None\n\n # predictions start here\n if is_sklearn_pipeline(estimator):\n pred = estimator.predict(data_transformed)\n if ml_usecase == MLUsecase.ANOMALY:\n pred_score = estimator.decision_function(data_transformed)\n else:\n pred = estimator.predict(prep_pipe.transform(data_transformed))\n if ml_usecase == MLUsecase.ANOMALY:\n pred_score = estimator.decision_function(\n prep_pipe.transform(data_transformed)\n )\n\n if ml_usecase == MLUsecase.CLUSTERING:\n pred_list = [f\"Cluster {i}\" for i in pred]\n\n data_transformed[\"Cluster\"] = pred_list\n else:\n data_transformed[\"Anomaly\"] = pred\n data_transformed[\"Anomaly_Score\"] = pred_score\n\n return data_transformed\n\n\ndef predict_model(\n estimator,\n data: Optional[pd.DataFrame] = None,\n probability_threshold: Optional[float] = None,\n encoded_labels: bool = False, # added in pycaret==2.1.0\n raw_score: bool = False,\n round: int = 4, # added in pycaret==2.2.0\n verbose: bool = True,\n ml_usecase: Optional[MLUsecase] = None,\n display: Optional[Display] = None, # added in pycaret==2.2.0\n) -> pd.DataFrame:\n\n \"\"\"\n This function is used to predict label and probability score on the new dataset\n using a trained estimator. New unseen data can be passed to data param as pandas\n Dataframe. If data is not passed, the test / hold-out set separated at the time of\n setup() is used to generate predictions.\n\n Example\n -------\n >>> from pycaret.datasets import get_data\n >>> juice = get_data('juice')\n >>> experiment_name = setup(data = juice, target = 'Purchase')\n >>> lr = create_model('lr')\n >>> lr_predictions_holdout = predict_model(lr)\n\n Parameters\n ----------\n estimator : object, default = none\n A trained model object / pipeline should be passed as an estimator.\n\n data : pandas.DataFrame\n Shape (n_samples, n_features) where n_samples is the number of samples\n and n_features is the number of features. All features used during training\n must be present in the new dataset.\n\n probability_threshold : float, default = None\n Threshold used to convert probability values into binary outcome. By default\n the probability threshold for all binary classifiers is 0.5 (50%). This can be\n changed using probability_threshold param.\n\n encoded_labels: Boolean, default = False\n If True, will return labels encoded as an integer.\n\n raw_score: bool, default = False\n When set to True, scores for all labels will be returned.\n\n round: integer, default = 4\n Number of decimal places the metrics in the score grid will be rounded to.\n\n verbose: bool, default = True\n Holdout score grid is not printed when verbose is set to False.\n\n Returns\n -------\n Predictions\n Predictions (Label and Score) column attached to the original dataset\n and returned as pandas dataframe.\n\n score_grid\n A table containing the scoring metrics on hold-out / test set.\n\n Warnings\n --------\n - The behavior of the predict_model is changed in version 2.1 without backward compatibility.\n As such, the pipelines trained using the version (<= 2.0), may not work for inference\n with version >= 2.1. You can either retrain your models with a newer version or downgrade\n the version for inference.\n\n\n \"\"\"\n\n function_params_str = \", \".join(\n [f\"{k}={v}\" for k, v in locals().items() if k != \"data\"]\n )\n\n logger = get_logger()\n\n logger.info(\"Initializing predict_model()\")\n logger.info(f\"predict_model({function_params_str})\")\n\n logger.info(\"Checking exceptions\")\n\n \"\"\"\n exception checking starts here\n \"\"\"\n\n if ml_usecase is None:\n ml_usecase = _ml_usecase\n\n if data is None and \"pycaret_globals\" not in globals():\n raise ValueError(\n \"data parameter may not be None without running setup() first.\"\n )\n\n if probability_threshold is not None:\n # probability_threshold allowed types\n allowed_types = [int, float]\n if type(probability_threshold) not in allowed_types:\n raise TypeError(\n \"probability_threshold parameter only accepts value between 0 to 1.\"\n )\n\n if probability_threshold > 1:\n raise TypeError(\n \"probability_threshold parameter only accepts value between 0 to 1.\"\n )\n\n if probability_threshold < 0:\n raise TypeError(\n \"probability_threshold parameter only accepts value between 0 to 1.\"\n )\n\n \"\"\"\n exception checking ends here\n \"\"\"\n\n logger.info(\"Preloading libraries\")\n\n # general dependencies\n from sklearn import metrics\n\n try:\n np.random.seed(seed)\n if not display:\n display = Display(verbose=verbose, html_param=html_param,)\n except:\n display = Display(verbose=False, html_param=False,)\n\n dtypes = None\n\n # dataset\n if data is None:\n\n if is_sklearn_pipeline(estimator):\n estimator = estimator.steps[-1][1]\n\n X_test_ = X_test.copy()\n y_test_ = y_test.copy()\n\n dtypes = prep_pipe.named_steps[\"dtypes\"]\n\n X_test_.reset_index(drop=True, inplace=True)\n y_test_.reset_index(drop=True, inplace=True)\n\n else:\n\n if is_sklearn_pipeline(estimator) and hasattr(estimator, \"predict\"):\n dtypes = estimator.named_steps[\"dtypes\"]\n else:\n try:\n dtypes = prep_pipe.named_steps[\"dtypes\"]\n\n estimator_ = deepcopy(prep_pipe)\n if is_sklearn_pipeline(estimator):\n merge_pipelines(estimator_, estimator)\n estimator_.steps[-1] = (\"trained_model\", estimator_.steps[-1][1])\n else:\n add_estimator_to_pipeline(\n estimator_, estimator, name=\"trained_model\"\n )\n estimator = estimator_\n\n except:\n logger.error(\"Pipeline not found. Exception:\")\n logger.error(traceback.format_exc())\n raise ValueError(\"Pipeline not found\")\n\n X_test_ = data.copy()\n\n # function to replace encoded labels with their original values\n # will not run if categorical_labels is false\n def replace_lables_in_column(label_column):\n if dtypes and hasattr(dtypes, \"replacement\"):\n replacement_mapper = {int(v): k for k, v in dtypes.replacement.items()}\n label_column.replace(replacement_mapper, inplace=True)\n\n # prediction starts here\n\n pred = np.nan_to_num(estimator.predict(X_test_))\n\n try:\n score = estimator.predict_proba(X_test_)\n\n if len(np.unique(pred)) <= 2:\n pred_prob = score[:, 1]\n else:\n pred_prob = score\n\n except:\n score = None\n pred_prob = None\n\n if probability_threshold is not None and pred_prob is not None:\n try:\n pred = (pred_prob >= probability_threshold).astype(int)\n except:\n pass\n\n if pred_prob is None:\n pred_prob = pred\n\n df_score = None\n\n if data is None:\n # model name\n full_name = _get_model_name(estimator)\n metrics = _calculate_metrics_supervised(y_test_, pred, pred_prob)\n df_score = pd.DataFrame(metrics, index=[0])\n df_score.insert(0, \"Model\", full_name)\n df_score = df_score.round(round)\n display.display(df_score.style.set_precision(round), clear=False)\n\n label = pd.DataFrame(pred)\n label.columns = [\"Label\"]\n if not encoded_labels:\n replace_lables_in_column(label[\"Label\"])\n if ml_usecase == MLUsecase.CLASSIFICATION:\n try:\n label[\"Label\"] = label[\"Label\"].astype(int)\n except:\n pass\n\n if data is None:\n if not encoded_labels:\n replace_lables_in_column(y_test_)\n X_test_ = pd.concat([X_test_, y_test_, label], axis=1)\n else:\n X_test_ = data.copy()\n X_test_[\"Label\"] = label[\"Label\"].values\n\n if score is not None:\n pred = pred.astype(int)\n if not raw_score:\n score = [s[pred[i]] for i, s in enumerate(score)]\n try:\n score = pd.DataFrame(score)\n if raw_score:\n score_columns = pd.Series(range(score.shape[1]))\n if not encoded_labels:\n replace_lables_in_column(score_columns)\n score.columns = [f\"Score_{label}\" for label in score_columns]\n else:\n score.columns = [\"Score\"]\n score = score.round(round)\n score.index = X_test_.index\n X_test_ = pd.concat((X_test_, score), axis=1)\n except:\n pass\n\n # store predictions on hold-out in display_container\n if df_score is not None:\n display_container.append(df_score)\n\n gc.collect()\n return X_test_\n\n\ndef finalize_model(\n estimator,\n fit_kwargs: Optional[dict] = None,\n groups: Optional[Union[str, Any]] = None,\n model_only: bool = True,\n display: Optional[Display] = None,\n) -> Any: # added in pycaret==2.2.0\n\n \"\"\"\n This function fits the estimator onto the complete dataset passed during the\n setup() stage. The purpose of this function is to prepare for final model\n deployment after experimentation.\n\n Example\n -------\n >>> from pycaret.datasets import get_data\n >>> juice = get_data('juice')\n >>> experiment_name = setup(data = juice, target = 'Purchase')\n >>> lr = create_model('lr')\n >>> final_lr = finalize_model(lr)\n\n This will return the final model object fitted to complete dataset.\n\n Parameters\n ----------\n estimator : object, default = none\n A trained model object should be passed as an estimator.\n\n fit_kwargs: dict, default = {} (empty dict)\n Dictionary of arguments passed to the fit method of the model.\n\n groups: str or array-like, with shape (n_samples,), default = None\n Optional Group labels for the samples used while splitting the dataset into train/test set.\n If string is passed, will use the data column with that name as the groups.\n Only used if a group based cross-validation generator is used (eg. GroupKFold).\n If None, will use the value set in fold_groups param in setup().\n\n model_only : bool, default = True\n When set to True, only trained model object is saved and all the\n transformations are ignored.\n\n Returns\n -------\n model\n Trained model object fitted on complete dataset.\n\n Warnings\n --------\n - If the model returned by finalize_model(), is used on predict_model() without\n passing a new unseen dataset, then the information grid printed is misleading\n as the model is trained on the complete dataset including test / hold-out sample.\n Once finalize_model() is used, the model is considered ready for deployment and\n should be used on new unseens dataset only.\n\n\n \"\"\"\n\n function_params_str = \", \".join([f\"{k}={v}\" for k, v in locals().items()])\n\n logger = get_logger()\n\n logger.info(\"Initializing finalize_model()\")\n logger.info(f\"finalize_model({function_params_str})\")\n\n # run_time\n runtime_start = time.time()\n\n if not fit_kwargs:\n fit_kwargs = {}\n\n groups = _get_groups(groups, data=X, fold_groups=fold_groups_param_full)\n\n if not display:\n display = Display(verbose=False, html_param=html_param,)\n\n np.random.seed(seed)\n\n logger.info(f\"Finalizing {estimator}\")\n display.clear_output()\n model_final, model_fit_time = create_model_supervised(\n estimator=estimator,\n verbose=False,\n system=False,\n X_train_data=X,\n y_train_data=y,\n fit_kwargs=fit_kwargs,\n groups=groups,\n )\n model_results = pull(pop=True)\n\n # end runtime\n runtime_end = time.time()\n runtime = np.array(runtime_end - runtime_start).round(2)\n\n # mlflow logging\n if logging_param:\n\n avgs_dict_log = {k: v for k, v in model_results.loc[\"Mean\"].items()}\n\n try:\n _mlflow_log_model(\n model=model_final,\n model_results=model_results,\n score_dict=avgs_dict_log,\n source=\"finalize_model\",\n runtime=runtime,\n model_fit_time=model_fit_time,\n _prep_pipe=prep_pipe,\n log_plots=log_plots_param,\n display=display,\n )\n except:\n logger.error(f\"_mlflow_log_model() for {model_final} raised an exception:\")\n logger.error(traceback.format_exc())\n\n model_results = color_df(model_results, \"yellow\", [\"Mean\"], axis=1)\n model_results = model_results.set_precision(round)\n display.display(model_results, clear=True)\n\n logger.info(f\"create_model_container: {len(create_model_container)}\")\n logger.info(f\"master_model_container: {len(master_model_container)}\")\n logger.info(f\"display_container: {len(display_container)}\")\n\n logger.info(str(model_final))\n logger.info(\n \"finalize_model() succesfully completed......................................\"\n )\n\n gc.collect()\n if not model_only:\n pipeline_final = deepcopy(prep_pipe)\n pipeline_final.steps.append([\"trained_model\", model_final])\n return pipeline_final\n\n return model_final\n\n\ndef deploy_model(\n model,\n model_name: str,\n authentication: dict,\n platform: str = \"aws\", # added gcp and azure support in pycaret==2.1\n):\n\n \"\"\"\n (In Preview)\n\n This function deploys the transformation pipeline and trained model object for\n production use. The platform of deployment can be defined under the platform\n param along with the applicable authentication tokens which are passed as a\n dictionary to the authentication param.\n\n Example\n -------\n >>> from pycaret.datasets import get_data\n >>> juice = get_data('juice')\n >>> experiment_name = setup(data = juice, target = 'Purchase')\n >>> lr = create_model('lr')\n >>> deploy_model(model = lr, model_name = 'deploy_lr', platform = 'aws', authentication = {'bucket' : 'pycaret-test'})\n\n This will deploy the model on an AWS S3 account under bucket 'pycaret-test'\n\n Notes\n -----\n For AWS users:\n Before deploying a model to an AWS S3 ('aws'), environment variables must be\n configured using the command line interface. To configure AWS env. variables,\n type aws configure in your python command line. The following information is\n required which can be generated using the Identity and Access Management (IAM)\n portal of your amazon console account:\n\n - AWS Access Key ID\n - AWS Secret Key Access\n - Default Region Name (can be seen under Global settings on your AWS console)\n - Default output format (must be left blank)\n\n For GCP users:\n --------------\n Before deploying a model to Google Cloud Platform (GCP), project must be created\n either using command line or GCP console. Once project is created, you must create\n a service account and download the service account key as a JSON file, which is\n then used to set environment variable.\n\n https://cloud.google.com/docs/authentication/production\n\n - Google Cloud Project\n - Service Account Authetication\n\n For Azure users:\n ---------------\n Before deploying a model to Microsoft's Azure (Azure), environment variables\n for connection string must be set. In order to get connection string, user has\n to create account of Azure. Once it is done, create a Storage account. In the settings\n section of storage account, user can get the connection string.\n\n Read below link for more details.\n https://docs.microsoft.com/en-us/azure/storage/blobs/storage-quickstart-blobs-python?toc=%2Fpython%2Fazure%2FTOC.json\n\n - Azure Storage Account\n\n Parameters\n ----------\n model : object\n A trained model object should be passed as an estimator.\n\n model_name : str\n Name of model to be passed as a str.\n\n authentication : dict\n Dictionary of applicable authentication tokens.\n\n When platform = 'aws':\n {'bucket' : 'Name of Bucket on S3'}\n\n When platform = 'gcp':\n {'project': 'gcp_pycaret', 'bucket' : 'pycaret-test'}\n\n When platform = 'azure':\n {'container': 'pycaret-test'}\n\n platform: str, default = 'aws'\n Name of platform for deployment. Current available options are: 'aws', 'gcp' and 'azure'\n\n Returns\n -------\n Success_Message\n\n Warnings\n --------\n - This function uses file storage services to deploy the model on cloud platform.\n As such, this is efficient for batch-use. Where the production objective is to\n obtain prediction at an instance level, this may not be the efficient choice as\n it transmits the binary pickle file between your local python environment and\n the platform.\n\n \"\"\"\n import pycaret.internal.persistence\n\n return pycaret.internal.persistence.deploy_model(\n model, model_name, authentication, platform, prep_pipe\n )\n\n\ndef create_webservice(model, model_endopoint, api_key=True, pydantic_payload=None):\n \"\"\"\n (In Preview)\n\n This function deploys the transformation pipeline and trained model object as api. Rest api base on FastAPI and could run on localhost, it uses\n the model name as a path to POST endpoint. The endpoint can be protected by api key generated by pycaret and return for the user.\n Create_webservice uses pydantic style input/output model.\n Parameters\n ----------\n model : object\n A trained model object should be passed as an estimator.\n\n model_endopoint : string\n Name of model to be passed as a string.\n\n api_key: bool, default = True\n Security for API, if True Pycaret generates api key and print in console,\n else user can post data without header but it not safe if application will\n expose external.\n\n pydantic_payload: pydantic.main.ModelMetaclass, default = None\n Pycaret allows us to automatically generate a schema for the input model,\n thanks to which can prevent incorrect requests. User can generate own pydantic model and use it as an input model.\n\n Returns\n -------\n Dictionary with api_key: FastAPI application class which is ready to run with console\n if api_key is set to False, the dictionary key is set to 'Not_exist'.\n\n \"\"\"\n\n function_params_str = \", \".join([f\"{k}={v}\" for k, v in locals().items()])\n\n logger = get_logger()\n\n logger.info(\"Initializing create_service()\")\n logger.info(f\"create_service({function_params_str})\")\n\n try:\n from fastapi import FastAPI\n except ImportError:\n logger.error(\n \"fastapi library not found. pip install fastapi to use create_service function.\"\n )\n raise ImportError(\n \"fastapi library not found. pip install fastapi to use create_service function.\"\n )\n # try initialize predict before add it to endpoint (cold start)\n try:\n _ = predict_model(estimator=model, verbose=False)\n except:\n raise ValueError(\n \"Cannot predict on cold start check probability_threshold or model\"\n )\n\n # check pydantic style\n try:\n from pydantic import create_model, BaseModel, Json\n from pydantic.main import ModelMetaclass\n except ImportError:\n logger.error(\n \"pydantic library not found. pip install fastapi to use create_service function.\"\n )\n ImportError(\n \"pydantic library not found. pip install fastapi to use create_service function.\"\n )\n if pydantic_payload is not None:\n assert isinstance(\n pydantic_payload, ModelMetaclass\n ), \"pydantic_payload must be ModelMetaClass type\"\n else:\n # automatically create pydantic payload model\n import json\n from typing import Optional\n\n print(\n \"You are using an automatic data validation model it could fail in some cases\"\n )\n print(\n \"To be sure the model works properly create pydantic model in your own (pydantic_payload)\"\n )\n fields = {\n name: (Optional[type(t)], ...)\n for name, t in json.loads(\n data_before_preprocess.drop(columns=[target_param])\n .convert_dtypes()\n .sample(1)\n .to_json(orient=\"records\")\n )[0].items()\n }\n print(fields)\n pydantic_payload = create_model(\"DefaultModel\", __base__=BaseModel, **fields)\n logger.info(\n \"Generated json schema: {}\".format(pydantic_payload.schema_json(indent=2))\n )\n\n # generate apikey\n import secrets\n from typing import Optional\n from fastapi.security.api_key import APIKeyHeader\n from fastapi import HTTPException, Security\n\n api_key_handler = APIKeyHeader(name=\"token\", auto_error=False)\n if api_key:\n # generate key and log into console\n key = secrets.token_urlsafe(30)\n\n def validate_request(header: Optional[str] = Security(api_key_handler)):\n if header is None:\n raise HTTPException(status_code=400, detail=\"No api key\", headers={})\n if not secrets.compare_digest(header, str(key)):\n raise HTTPException(\n status_code=401, detail=\"Unauthorized request\", headers={}\n )\n return True\n\n else:\n key = \"Not_exist\"\n print(\"API will be working without security\")\n\n def validate_request(header: Optional[str] = Security(api_key_handler)):\n return True\n\n # validate request functionality\n validation = validate_request\n\n # creating response model\n from typing import Any, Optional\n from pycaret.utils import __version__\n\n class PredictionResult(BaseModel):\n prediction: Any\n author: str = \"pycaret\"\n lib_version: str = __version__()\n input_data: pydantic_payload\n processed_input_data: Json = None\n time_utc: Optional[str] = None\n\n class Config:\n schema_extra = {\n \"example\": {\n \"prediction\": 1,\n \"autohor\": \"pycaret\",\n \"lib_version\": \"2.0.0\",\n \"input_data\": pydantic_payload,\n \"processed_input_data\": {\"col1\": 1, \"col2\": \"string\"},\n \"time\": \"2020-09-10 20:00\",\n }\n }\n\n app = FastAPI(\n title=\"REST API for ML prediction created by Pycaret\",\n description=\"This is the REST API for the ML model generated\"\n \"by the Pycaret library: https://pycaret.org. \"\n \"All endpoints should run asynchronously, please validate\"\n \"the Pydantic model and read api documentation. \"\n \"In case of trouble, please add issuesto github: https://github.com/pycaret/pycaret/issues\",\n version=\"pycaret: {}\".format(__version__()),\n externalDocs={\"Pycaret\": \"https://pycaret.org/\"},\n )\n\n # import additionals from fastAPI\n import pandas as pd\n import time\n from fastapi.middleware.cors import CORSMiddleware\n from fastapi import Depends\n from fastapi.encoders import jsonable_encoder\n\n # enable CORS\n app.add_middleware(\n CORSMiddleware,\n allow_origins=[\"*\"],\n allow_credentials=True,\n allow_methods=[\"*\"],\n allow_headers=[\"*\"],\n )\n\n @app.post(\"/predict/{}\".format(model_endopoint))\n def post_predict(\n block_data: pydantic_payload, authenticated: bool = Depends(validation)\n ):\n # encode input data\n try:\n encoded_data_df = pd.DataFrame(jsonable_encoder(block_data), index=[0])\n except Exception as e:\n raise HTTPException(status_code=404, detail=\"Wrong json format\")\n # predict values\n unseen_predictions = predict_model(model, data=encoded_data_df)\n # change format to dictionary to be sure that python types used\n unseen_predictions = unseen_predictions.to_dict(orient=\"records\")[0]\n label = unseen_predictions[\"Label\"]\n del unseen_predictions[\"Label\"]\n\n # creating return object\n predict_schema = PredictionResult(\n prediction=label,\n input_data=block_data,\n processed_input_data=json.dumps(unseen_predictions),\n time_utc=time.strftime(\"%Y-%m-%d %H:%M\", time.gmtime(time.time())),\n )\n return predict_schema\n\n return {key: app}\n\n\ndef save_model(\n model, model_name: str, model_only: bool = False, verbose: bool = True, **kwargs\n):\n\n \"\"\"\n This function saves the transformation pipeline and trained model object\n into the current active directory as a pickle file for later use.\n\n Example\n -------\n >>> from pycaret.datasets import get_data\n >>> juice = get_data('juice')\n >>> experiment_name = setup(data = juice, target = 'Purchase')\n >>> lr = create_model('lr')\n >>> save_model(lr, 'lr_model_23122019')\n\n This will save the transformation pipeline and model as a binary pickle\n file in the current active directory.\n\n Parameters\n ----------\n model : object, default = none\n A trained model object should be passed as an estimator.\n\n model_name : str, default = none\n Name of pickle file to be passed as a string.\n\n model_only : bool, default = False\n When set to True, only trained model object is saved and all the\n transformations are ignored.\n\n **kwargs: \n Additional keyword arguments to pass to joblib.dump().\n\n verbose: bool, default = True\n Success message is not printed when verbose is set to False.\n\n Returns\n -------\n Success_Message\n\n\n \"\"\"\n\n import pycaret.internal.persistence\n\n return pycaret.internal.persistence.save_model(\n model, model_name, None if model_only else prep_pipe, verbose, **kwargs\n )\n\n\ndef load_model(\n model_name,\n platform: Optional[str] = None,\n authentication: Optional[Dict[str, str]] = None,\n verbose: bool = True,\n):\n\n \"\"\"\n This function loads a previously saved transformation pipeline and model\n from the current active directory into the current python environment.\n Load object must be a pickle file.\n\n Example\n -------\n >>> saved_lr = load_model('lr_model_23122019')\n\n This will load the previously saved model in saved_lr variable. The file\n must be in the current directory.\n\n Parameters\n ----------\n model_name : str, default = none\n Name of pickle file to be passed as a string.\n\n platform: str, default = None\n Name of platform, if loading model from cloud. Current available options are:\n 'aws', 'gcp' and 'azure'.\n\n authentication : dict\n dictionary of applicable authentication tokens.\n\n When platform = 'aws':\n {'bucket' : 'Name of Bucket on S3'}\n\n When platform = 'gcp':\n {'project': 'gcp_pycaret', 'bucket' : 'pycaret-test'}\n\n When platform = 'azure':\n {'container': 'pycaret-test'}\n\n verbose: bool, default = True\n Success message is not printed when verbose is set to False.\n\n Returns\n -------\n Model Object\n\n \"\"\"\n\n import pycaret.internal.persistence\n\n return pycaret.internal.persistence.load_model(\n model_name, platform, authentication, verbose\n )\n\n\ndef automl(optimize: str = \"Accuracy\", use_holdout: bool = False) -> Any:\n\n \"\"\"\n This function returns the best model out of all models created in\n current active environment based on metric defined in optimize parameter.\n\n Parameters\n ----------\n optimize : str, default = 'Accuracy'\n Other values you can pass in optimize param are 'AUC', 'Recall', 'Precision',\n 'F1', 'Kappa', and 'MCC'.\n\n use_holdout: bool, default = False\n When set to True, metrics are evaluated on holdout set instead of CV.\n\n \"\"\"\n\n function_params_str = \", \".join([f\"{k}={v}\" for k, v in locals().items()])\n\n logger = get_logger()\n\n logger.info(\"Initializing automl()\")\n logger.info(f\"automl({function_params_str})\")\n\n # checking optimize parameter\n optimize = _get_metric(optimize)\n if optimize is None:\n raise ValueError(\n f\"Optimize method not supported. See docstring for list of available parameters.\"\n )\n\n # checking optimize parameter for multiclass\n if _is_multiclass():\n if not optimize.is_multiclass:\n raise TypeError(\n f\"Optimization metric not supported for multiclass problems. See docstring for list of other optimization parameters.\"\n )\n\n compare_dimension = optimize.display_name\n greater_is_better = optimize.greater_is_better\n optimize = optimize.scorer\n\n scorer = []\n\n if use_holdout:\n logger.info(\"Model Selection Basis : Holdout set\")\n for i in master_model_container:\n try:\n pred_holdout = predict_model(i, verbose=False)\n except:\n logger.warning(f\"Model {i} is not fitted, running create_model\")\n i, _ = create_model_supervised(\n estimator=i,\n system=False,\n verbose=False,\n cross_validation=False,\n predict=False,\n groups=fold_groups_param,\n )\n pull(pop=True)\n pred_holdout = predict_model(i, verbose=False)\n\n p = pull(pop=True)\n p = p[compare_dimension][0]\n scorer.append(p)\n\n else:\n logger.info(\"Model Selection Basis : CV Results on Training set\")\n for i in create_model_container:\n r = i[compare_dimension][-2:][0]\n scorer.append(r)\n\n # returning better model\n if greater_is_better:\n index_scorer = scorer.index(max(scorer))\n else:\n index_scorer = scorer.index(min(scorer))\n\n automl_result = master_model_container[index_scorer]\n\n automl_model, _ = create_model_supervised(\n estimator=automl_result,\n system=False,\n verbose=False,\n cross_validation=False,\n predict=False,\n groups=fold_groups_param,\n )\n\n logger.info(str(automl_model))\n logger.info(\"automl() succesfully completed......................................\")\n\n return automl_model\n\n\ndef pull(pop=False) -> pd.DataFrame: # added in pycaret==2.2.0\n \"\"\"\n Returns latest displayed table.\n\n Parameters\n ----------\n pop : bool, default = False\n If true, will pop (remove) the returned dataframe from the\n display container.\n\n Returns\n -------\n pandas.DataFrame\n Equivalent to get_config('display_container')[-1]\n\n \"\"\"\n if not display_container:\n return None\n return display_container.pop(-1) if pop else display_container[-1]\n\n\ndef models(\n type: Optional[str] = None, internal: bool = False, raise_errors: bool = True,\n) -> pd.DataFrame:\n\n \"\"\"\n Returns table of models available in model library.\n\n Example\n -------\n >>> _all_models = models()\n\n This will return pandas dataframe with all available\n models and their metadata.\n\n Parameters\n ----------\n type : str, default = None\n - linear : filters and only return linear models\n - tree : filters and only return tree based models\n - ensemble : filters and only return ensemble models\n\n internal: bool, default = False\n If True, will return extra columns and rows used internally.\n\n raise_errors: bool, default = True\n If False, will suppress all exceptions, ignoring models\n that couldn't be created.\n\n Returns\n -------\n pandas.DataFrame\n\n \"\"\"\n\n model_type = {\n \"linear\": [\n \"lr\",\n \"ridge\",\n \"svm\",\n \"lasso\",\n \"en\",\n \"lar\",\n \"llar\",\n \"omp\",\n \"br\",\n \"ard\",\n \"par\",\n \"ransac\",\n \"tr\",\n \"huber\",\n \"kr\",\n ],\n \"tree\": [\"dt\"],\n \"ensemble\": [\n \"rf\",\n \"et\",\n \"gbc\",\n \"gbr\",\n \"xgboost\",\n \"lightgbm\",\n \"catboost\",\n \"ada\",\n ],\n }\n\n def filter_model_df_by_type(df):\n if not type:\n return df\n return df[df.index.isin(model_type[type])]\n\n # Check if type is valid\n if type not in list(model_type) + [None]:\n raise ValueError(\n f\"type param only accepts {', '.join(list(model_type) + str(None))}.\"\n )\n\n logger.info(f\"gpu_param set to {gpu_param}\")\n\n if _ml_usecase == MLUsecase.CLASSIFICATION:\n model_containers = pycaret.containers.models.classification.get_all_model_containers(\n globals(), raise_errors\n )\n elif _ml_usecase == MLUsecase.REGRESSION:\n model_containers = pycaret.containers.models.regression.get_all_model_containers(\n globals(), raise_errors\n )\n elif _ml_usecase == MLUsecase.CLUSTERING:\n model_containers = pycaret.containers.models.clustering.get_all_model_containers(\n globals(), raise_errors\n )\n elif _ml_usecase == MLUsecase.ANOMALY:\n model_containers = pycaret.containers.models.anomaly.get_all_model_containers(\n globals(), raise_errors\n )\n rows = [\n v.get_dict(internal)\n for k, v in model_containers.items()\n if (internal or not v.is_special)\n ]\n\n df = pd.DataFrame(rows)\n df.set_index(\"ID\", inplace=True, drop=True)\n\n return filter_model_df_by_type(df)\n\n\ndef get_metrics(\n reset: bool = False, include_custom: bool = True, raise_errors: bool = True,\n) -> pd.DataFrame:\n \"\"\"\n Returns table of metrics available.\n\n Example\n -------\n >>> metrics = get_metrics()\n\n This will return pandas dataframe with all available\n metrics and their metadata.\n\n Parameters\n ----------\n reset: bool, default = False\n If True, will reset all changes made using add_metric() and get_metric().\n include_custom: bool, default = True\n Whether to include user added (custom) metrics or not.\n raise_errors: bool, default = True\n If False, will suppress all exceptions, ignoring models\n that couldn't be created.\n\n Returns\n -------\n pandas.DataFrame\n\n \"\"\"\n\n if reset and not \"_all_metrics\" in globals():\n raise ValueError(\"setup() needs to be ran first.\")\n\n global _all_metrics\n\n np.random.seed(seed)\n\n if reset:\n if _ml_usecase == MLUsecase.CLASSIFICATION:\n _all_metrics = pycaret.containers.metrics.classification.get_all_metric_containers(\n globals(), raise_errors\n )\n elif _ml_usecase == MLUsecase.REGRESSION:\n _all_metrics = pycaret.containers.metrics.regression.get_all_metric_containers(\n globals(), raise_errors\n )\n\n metric_containers = _all_metrics\n rows = [v.get_dict() for k, v in metric_containers.items()]\n\n df = pd.DataFrame(rows)\n df.set_index(\"ID\", inplace=True, drop=True)\n\n if not include_custom:\n df = df[df[\"Custom\"] == False]\n\n return df\n\n\ndef _get_metric(name_or_id: str, metrics: Optional[Any] = None):\n \"\"\"\n Gets a metric from get_metrics() by name or index.\n \"\"\"\n if metrics is None:\n metrics = _all_metrics\n metric = None\n try:\n metric = metrics[name_or_id]\n return metric\n except:\n pass\n\n try:\n metric = next(\n v for k, v in metrics.items() if name_or_id in (v.display_name, v.name)\n )\n return metric\n except:\n pass\n\n return metric\n\n\ndef add_metric(\n id: str,\n name: str,\n score_func: type,\n target: str = \"pred\",\n greater_is_better: bool = True,\n multiclass: bool = True,\n **kwargs,\n) -> pd.Series:\n \"\"\"\n Adds a custom metric to be used in all functions.\n\n Parameters\n ----------\n id: str\n Unique id for the metric.\n\n name: str\n Display name of the metric.\n\n score_func: type\n Score function (or loss function) with signature score_func(y, y_pred, **kwargs).\n\n target: str, default = 'pred'\n The target of the score function.\n - 'pred' for the prediction table\n - 'pred_proba' for pred_proba\n - 'threshold' for decision_function or predict_proba\n\n greater_is_better: bool, default = True\n Whether score_func is a score function (default), meaning high is good,\n or a loss function, meaning low is good. In the latter case, the\n scorer object will sign-flip the outcome of the score_func.\n\n multiclass: bool, default = True\n Whether the metric supports multiclass problems.\n\n **kwargs:\n Arguments to be passed to score function.\n\n Returns\n -------\n pandas.Series\n The created row as Series.\n\n \"\"\"\n\n if not \"_all_metrics\" in globals():\n raise ValueError(\"setup() needs to be ran first.\")\n\n global _all_metrics\n\n if id in _all_metrics:\n raise ValueError(\"id already present in metrics dataframe.\")\n\n if _ml_usecase == MLUsecase.CLASSIFICATION:\n new_metric = pycaret.containers.metrics.classification.ClassificationMetricContainer(\n id=id,\n name=name,\n score_func=score_func,\n target=target,\n args=kwargs,\n display_name=name,\n greater_is_better=greater_is_better,\n is_multiclass=bool(multiclass),\n is_custom=True,\n )\n else:\n new_metric = pycaret.containers.metrics.regression.RegressionMetricContainer(\n id=id,\n name=name,\n score_func=score_func,\n args=kwargs,\n display_name=name,\n greater_is_better=greater_is_better,\n is_custom=True,\n )\n\n _all_metrics[id] = new_metric\n\n new_metric = new_metric.get_dict()\n\n new_metric = pd.Series(new_metric, name=id.replace(\" \", \"_\")).drop(\"ID\")\n\n return new_metric\n\n\ndef remove_metric(name_or_id: str):\n \"\"\"\n Removes a metric used in all functions.\n\n Parameters\n ----------\n name_or_id: str\n Display name or ID of the metric.\n\n \"\"\"\n if not \"_all_metrics\" in globals():\n raise ValueError(\"setup() needs to be ran first.\")\n\n try:\n _all_metrics.pop(name_or_id)\n return\n except:\n pass\n\n try:\n k_to_remove = next(k for k, v in _all_metrics.items() if v.name == name_or_id)\n _all_metrics.pop(k_to_remove)\n return\n except:\n pass\n\n raise ValueError(\n f\"No metric 'Display Name' or 'ID' (index) {name_or_id} present in the metrics repository.\"\n )\n\n\ndef get_logs(experiment_name: Optional[str] = None, save: bool = False) -> pd.DataFrame:\n\n \"\"\"\n Returns a table with experiment logs consisting\n run details, parameter, metrics and tags.\n\n Example\n -------\n >>> logs = get_logs()\n\n This will return pandas dataframe.\n\n Parameters\n ----------\n experiment_name : str, default = None\n When set to None current active run is used.\n\n save : bool, default = False\n When set to True, csv file is saved in current directory.\n\n Returns\n -------\n pandas.DataFrame\n\n \"\"\"\n\n if experiment_name is None:\n exp_name_log_ = exp_name_log\n else:\n exp_name_log_ = experiment_name\n\n import mlflow\n from mlflow.tracking import MlflowClient\n\n client = MlflowClient()\n\n if client.get_experiment_by_name(exp_name_log_) is None:\n raise ValueError(\n \"No active run found. Check logging parameter in setup or to get logs for inactive run pass experiment_name.\"\n )\n\n exp_id = client.get_experiment_by_name(exp_name_log_).experiment_id\n runs = mlflow.search_runs(exp_id)\n\n if save:\n file_name = f\"{exp_name_log_}_logs.csv\"\n runs.to_csv(file_name, index=False)\n\n return runs\n\n\ndef get_config(variable: str):\n\n \"\"\"\n This function is used to access global environment variables.\n Following variables can be accessed:\n\n - X: Transformed dataset (X)\n - y: Transformed dataset (y)\n - X_train: Transformed train dataset (X)\n - X_test: Transformed test/holdout dataset (X)\n - y_train: Transformed train dataset (y)\n - y_test: Transformed test/holdout dataset (y)\n - seed: random state set through session_id\n - prep_pipe: Transformation pipeline configured through setup\n - fold_shuffle_param: shuffle parameter used in Kfolds\n - n_jobs_param: n_jobs parameter used in model training\n - html_param: html_param configured through setup\n - create_model_container: results grid storage container\n - master_model_container: model storage container\n - display_container: results display container\n - exp_name_log: Name of experiment set through setup\n - logging_param: log_experiment param set through setup\n - log_plots_param: log_plots param set through setup\n - USI: Unique session ID parameter set through setup\n - fix_imbalance_param: fix_imbalance param set through setup\n - fix_imbalance_method_param: fix_imbalance_method param set through setup\n - data_before_preprocess: data before preprocessing\n - target_param: name of target variable\n - gpu_param: use_gpu param configured through setup\n\n Example\n -------\n >>> X_train = get_config('X_train')\n\n This will return X_train transformed dataset.\n\n Returns\n -------\n variable\n\n \"\"\"\n\n import pycaret.internal.utils\n\n return pycaret.internal.utils.get_config(variable, globals())\n\n\ndef set_config(variable: str, value):\n\n \"\"\"\n This function is used to reset global environment variables.\n Following variables can be accessed:\n\n - X: Transformed dataset (X)\n - y: Transformed dataset (y)\n - X_train: Transformed train dataset (X)\n - X_test: Transformed test/holdout dataset (X)\n - y_train: Transformed train dataset (y)\n - y_test: Transformed test/holdout dataset (y)\n - seed: random state set through session_id\n - prep_pipe: Transformation pipeline configured through setup\n - fold_shuffle_param: shuffle parameter used in Kfolds\n - n_jobs_param: n_jobs parameter used in model training\n - html_param: html_param configured through setup\n - create_model_container: results grid storage container\n - master_model_container: model storage container\n - display_container: results display container\n - exp_name_log: Name of experiment set through setup\n - logging_param: log_experiment param set through setup\n - log_plots_param: log_plots param set through setup\n - USI: Unique session ID parameter set through setup\n - fix_imbalance_param: fix_imbalance param set through setup\n - fix_imbalance_method_param: fix_imbalance_method param set through setup\n - data_before_preprocess: data before preprocessing\n\n Example\n -------\n >>> set_config('seed', 123)\n\n This will set the global seed to '123'.\n\n \"\"\"\n\n import pycaret.internal.utils\n\n return pycaret.internal.utils.set_config(variable, value, globals())\n\n\ndef save_config(file_name: str):\n\n \"\"\"\n This function is used to save all enviroment variables to file,\n allowing to later resume modeling without rerunning setup().\n\n Example\n -------\n >>> save_config('myvars.pkl')\n\n This will save all enviroment variables to 'myvars.pkl'.\n\n \"\"\"\n\n import pycaret.internal.utils\n\n return pycaret.internal.utils.save_config(file_name, globals())\n\n\ndef load_config(file_name: str):\n\n \"\"\"\n This function is used to load enviroment variables from file created with save_config(),\n allowing to later resume modeling without rerunning setup().\n\n\n Example\n -------\n >>> load_config('myvars.pkl')\n\n This will load all enviroment variables from 'myvars.pkl'.\n\n \"\"\"\n\n global _all_models, _all_models_internal, _all_metrics, X_train, create_model_container, master_model_container, display_container\n\n import pycaret.internal.utils\n\n r = pycaret.internal.utils.load_config(file_name, globals())\n\n if _ml_usecase == MLUsecase.CLASSIFICATION:\n _all_models = {\n k: v\n for k, v in pycaret.containers.models.classification.get_all_model_containers(\n globals(), raise_errors=True\n ).items()\n if not v.is_special\n }\n _all_models_internal = pycaret.containers.models.classification.get_all_model_containers(\n globals(), raise_errors=True\n )\n _all_metrics = pycaret.containers.metrics.classification.get_all_metric_containers(\n globals(), raise_errors=True\n )\n elif _ml_usecase == MLUsecase.REGRESSION:\n _all_models = {\n k: v\n for k, v in pycaret.containers.models.regression.get_all_model_containers(\n globals(), raise_errors=True\n ).items()\n if not v.is_special\n }\n _all_models_internal = pycaret.containers.models.regression.get_all_model_containers(\n globals(), raise_errors=True\n )\n _all_metrics = pycaret.containers.metrics.regression.get_all_metric_containers(\n globals(), raise_errors=True\n )\n elif _ml_usecase == MLUsecase.CLUSTERING:\n _all_models = {\n k: v\n for k, v in pycaret.containers.models.clustering.get_all_model_containers(\n globals(), raise_errors=True\n ).items()\n if not v.is_special\n }\n _all_models_internal = pycaret.containers.models.clustering.get_all_model_containers(\n globals(), raise_errors=True\n )\n _all_metrics = pycaret.containers.metrics.clustering.get_all_metric_containers(\n globals(), raise_errors=True\n )\n X_train = X\n elif _ml_usecase == MLUsecase.ANOMALY:\n _all_models = {\n k: v\n for k, v in pycaret.containers.models.anomaly.get_all_model_containers(\n globals(), raise_errors=True\n ).items()\n if not v.is_special\n }\n _all_models_internal = pycaret.containers.models.anomaly.get_all_model_containers(\n globals(), raise_errors=True\n )\n _all_metrics = pycaret.containers.metrics.anomaly.get_all_metric_containers(\n globals(), raise_errors=True\n )\n X_train = X\n\n create_model_container = []\n master_model_container = []\n display_container = []\n\n return r\n\n\ndef _choose_better(\n models_and_results: list,\n compare_dimension: str,\n fold: int,\n fit_kwargs: Optional[dict] = None,\n groups: Optional[Union[str, Any]] = None,\n display: Optional[Display] = None,\n):\n \"\"\"\n When choose_better is set to True, optimize metric in scoregrid is\n compared with base model created using create_model so that the\n functions return the model with better score only. This will ensure\n model performance is at least equivalent to what is seen in compare_models\n \"\"\"\n\n logger = get_logger()\n logger.info(\"choose_better activated\")\n display.update_monitor(1, \"Compiling Final Results\")\n display.display_monitor()\n\n if not fit_kwargs:\n fit_kwargs = {}\n\n for i, x in enumerate(models_and_results):\n if not isinstance(x, tuple):\n models_and_results[i] = (x, None)\n elif isinstance(x[0], str):\n models_and_results[i] = (x[1], None)\n elif len(x) != 2:\n raise ValueError(f\"{x} must have lenght 2 but has {len(x)}\")\n\n metric = _get_metric(compare_dimension)\n\n best_result = None\n best_model = None\n for model, result in models_and_results:\n if result is not None and is_fitted(model):\n result = result.loc[\"Mean\"][compare_dimension]\n else:\n logger.info(\n \"SubProcess create_model() called ==================================\"\n )\n model, _ = create_model_supervised(\n model,\n verbose=False,\n system=False,\n fold=fold,\n fit_kwargs=fit_kwargs,\n groups=groups,\n )\n logger.info(\n \"SubProcess create_model() end ==================================\"\n )\n result = pull(pop=True).loc[\"Mean\"][compare_dimension]\n logger.info(f\"{model} result for {compare_dimension} is {result}\")\n if not metric.greater_is_better:\n result *= -1\n if best_result is None or best_result < result:\n best_result = result\n best_model = model\n\n logger.info(f\"{best_model} is best model\")\n\n logger.info(\"choose_better completed\")\n return best_model\n\n\ndef _is_multiclass() -> bool:\n \"\"\"\n Method to check if the problem is multiclass.\n \"\"\"\n try:\n return _ml_usecase == MLUsecase.CLASSIFICATION and y.value_counts().count() > 2\n except:\n return False\n\n\ndef _get_model_id(e, models=None) -> str:\n \"\"\"\n Get model id.\n \"\"\"\n if models is None:\n models = _all_models_internal\n\n import pycaret.internal.utils\n\n return pycaret.internal.utils.get_model_id(e, models)\n\n\ndef _get_model_name(e, deep: bool = True, models=None) -> str:\n \"\"\"\n Get model name.\n \"\"\"\n if models is None:\n models = _all_models_internal\n\n import pycaret.internal.utils\n\n return pycaret.internal.utils.get_model_name(e, models, deep=deep)\n\n\ndef _is_special_model(e, models=None) -> bool:\n \"\"\"\n Is the model special (eg. VotingClassifier).\n \"\"\"\n if models is None:\n models = _all_models_internal\n\n import pycaret.internal.utils\n\n return pycaret.internal.utils.is_special_model(e, models)\n\n\ndef _calculate_metrics_supervised(\n y_test, pred, pred_prob, weights: Optional[list] = None,\n) -> dict:\n \"\"\"\n Calculate all metrics in _all_metrics.\n \"\"\"\n from pycaret.internal.utils import calculate_metrics\n\n try:\n return calculate_metrics(\n metrics=_all_metrics,\n y_test=y_test,\n pred=pred,\n pred_proba=pred_prob,\n weights=weights,\n )\n except:\n ml_usecase = get_ml_task(y_test)\n if ml_usecase == MLUsecase.CLASSIFICATION:\n metrics = pycaret.containers.metrics.classification.get_all_metric_containers(\n globals(), True\n )\n elif ml_usecase == MLUsecase.REGRESSION:\n metrics = pycaret.containers.metrics.regression.get_all_metric_containers(\n globals(), True\n )\n return calculate_metrics(\n metrics=metrics,\n y_test=y_test,\n pred=pred,\n pred_proba=pred_prob,\n weights=weights,\n )\n\n\ndef _calculate_metrics_unsupervised(\n X, labels, ground_truth=None, ml_usecase=None\n) -> dict:\n \"\"\"\n Calculate all metrics in _all_metrics.\n \"\"\"\n from pycaret.internal.utils import calculate_unsupervised_metrics\n\n if ml_usecase is None:\n ml_usecase = _ml_usecase\n\n try:\n return calculate_unsupervised_metrics(\n metrics=_all_metrics, X=X, labels=labels, ground_truth=ground_truth\n )\n except:\n if ml_usecase == MLUsecase.CLUSTERING:\n metrics = pycaret.containers.metrics.clustering.get_all_metric_containers(\n globals(), True\n )\n return calculate_unsupervised_metrics(\n metrics=metrics, X=X, labels=labels, ground_truth=ground_truth\n )\n\n\ndef get_ml_task(y):\n c1 = y.dtype == \"int64\"\n c2 = y.nunique() <= 20\n c3 = y.dtype.name in [\"object\", \"bool\", \"category\"]\n if ((c1) & (c2)) | (c3):\n ml_usecase = MLUsecase.CLASSIFICATION\n else:\n ml_usecase = MLUsecase.REGRESSION\n return ml_usecase\n\n\ndef _mlflow_log_model(\n model,\n model_results,\n score_dict: dict,\n source: str,\n runtime: float,\n model_fit_time: float,\n _prep_pipe,\n log_holdout: bool = True,\n log_plots: bool = False,\n tune_cv_results=None,\n URI=None,\n display: Optional[Display] = None,\n):\n logger = get_logger()\n\n logger.info(\"Creating MLFlow logs\")\n\n # Creating Logs message monitor\n if display:\n display.update_monitor(1, \"Creating Logs\")\n display.display_monitor()\n\n # import mlflow\n import mlflow\n import mlflow.sklearn\n\n mlflow.set_experiment(exp_name_log)\n\n full_name = _get_model_name(model)\n logger.info(f\"Model: {full_name}\")\n\n with mlflow.start_run(run_name=full_name) as run:\n\n # Get active run to log as tag\n RunID = mlflow.active_run().info.run_id\n\n # Log model parameters\n pipeline_estimator_name = get_pipeline_estimator_label(model)\n if pipeline_estimator_name:\n params = model.named_steps[pipeline_estimator_name]\n else:\n params = model\n\n # get regressor from meta estimator\n params = get_estimator_from_meta_estimator(params)\n\n try:\n try:\n params = params.get_all_params()\n except:\n params = params.get_params()\n except:\n logger.warning(\"Couldn't get params for model. Exception:\")\n logger.warning(traceback.format_exc())\n params = {}\n\n for i in list(params):\n v = params.get(i)\n if len(str(v)) > 250:\n params.pop(i)\n\n logger.info(f\"logged params: {params}\")\n mlflow.log_params(params)\n\n # Log metrics\n mlflow.log_metrics(score_dict)\n\n # set tag of compare_models\n mlflow.set_tag(\"Source\", source)\n\n if not URI:\n import secrets\n\n URI = secrets.token_hex(nbytes=4)\n mlflow.set_tag(\"URI\", URI)\n mlflow.set_tag(\"USI\", USI)\n mlflow.set_tag(\"Run Time\", runtime)\n mlflow.set_tag(\"Run ID\", RunID)\n\n # Log training time in seconds\n mlflow.log_metric(\"TT\", model_fit_time)\n\n # Log the CV results as model_results.html artifact\n if not _is_unsupervised(_ml_usecase):\n try:\n model_results.data.to_html(\"Results.html\", col_space=65, justify=\"left\")\n except:\n model_results.to_html(\"Results.html\", col_space=65, justify=\"left\")\n mlflow.log_artifact(\"Results.html\")\n os.remove(\"Results.html\")\n\n if log_holdout:\n # Generate hold-out predictions and save as html\n try:\n holdout = predict_model(model, verbose=False)\n holdout_score = pull(pop=True)\n del holdout\n holdout_score.to_html(\"Holdout.html\", col_space=65, justify=\"left\")\n mlflow.log_artifact(\"Holdout.html\")\n os.remove(\"Holdout.html\")\n except:\n logger.warning(\n \"Couldn't create holdout prediction for model, exception below:\"\n )\n logger.warning(traceback.format_exc())\n\n # Log AUC and Confusion Matrix plot\n\n if log_plots:\n\n logger.info(\n \"SubProcess plot_model() called ==================================\"\n )\n\n def _log_plot(plot):\n try:\n plot_name = plot_model(\n model, plot=plot, verbose=False, save=True, system=False\n )\n mlflow.log_artifact(plot_name)\n os.remove(plot_name)\n except Exception as e:\n logger.warning(e)\n\n for plot in log_plots:\n _log_plot(plot)\n\n logger.info(\n \"SubProcess plot_model() end ==================================\"\n )\n\n # Log hyperparameter tuning grid\n if tune_cv_results:\n d1 = tune_cv_results.get(\"params\")\n dd = pd.DataFrame.from_dict(d1)\n dd[\"Score\"] = tune_cv_results.get(\"mean_test_score\")\n dd.to_html(\"Iterations.html\", col_space=75, justify=\"left\")\n mlflow.log_artifact(\"Iterations.html\")\n os.remove(\"Iterations.html\")\n\n # get default conda env\n from mlflow.sklearn import get_default_conda_env\n\n default_conda_env = get_default_conda_env()\n default_conda_env[\"name\"] = f\"{exp_name_log}-env\"\n default_conda_env.get(\"dependencies\").pop(-3)\n dependencies = default_conda_env.get(\"dependencies\")[-1]\n from pycaret.utils import __version__\n\n dep = f\"pycaret=={__version__}\"\n dependencies[\"pip\"] = [dep]\n\n # define model signature\n from mlflow.models.signature import infer_signature\n\n try:\n signature = infer_signature(\n data_before_preprocess.drop([target_param], axis=1)\n )\n except:\n logger.warning(\"Couldn't infer MLFlow signature.\")\n signature = None\n if not _is_unsupervised(_ml_usecase):\n input_example = (\n data_before_preprocess.drop([target_param], axis=1).iloc[0].to_dict()\n )\n else:\n input_example = data_before_preprocess.iloc[0].to_dict()\n\n # log model as sklearn flavor\n prep_pipe_temp = deepcopy(_prep_pipe)\n prep_pipe_temp.steps.append([\"trained_model\", model])\n mlflow.sklearn.log_model(\n prep_pipe_temp,\n \"model\",\n conda_env=default_conda_env,\n # signature=signature,\n # input_example=input_example,\n )\n del prep_pipe_temp\n gc.collect()\n\n\ndef _get_columns_to_stratify_by(\n X: pd.DataFrame, y: pd.DataFrame, stratify: Union[bool, List[str]], target: str\n) -> pd.DataFrame:\n if not stratify:\n stratify = None\n else:\n if isinstance(stratify, list):\n data = pd.concat([X, y], axis=1)\n if not all(col in data.columns for col in stratify):\n raise ValueError(\"Column to stratify by does not exist in the dataset.\")\n stratify = data[stratify]\n else:\n stratify = y\n return stratify\n\n\ndef _get_cv_splitter(fold, ml_usecase: Optional[MLUsecase] = None):\n if not ml_usecase:\n ml_usecase = _ml_usecase\n\n import pycaret.internal.utils\n\n return pycaret.internal.utils.get_cv_splitter(\n fold,\n default=fold_generator,\n seed=seed,\n shuffle=fold_shuffle_param,\n int_default=\"stratifiedkfold\"\n if ml_usecase == MLUsecase.CLASSIFICATION\n else \"kfold\",\n )\n\n\ndef _get_cv_n_folds(fold, X, y=None, groups=None):\n import pycaret.internal.utils\n\n return pycaret.internal.utils.get_cv_n_folds(\n fold, default=fold_generator, X=X, y=y, groups=groups\n )\n\n\ndef _get_pipeline_fit_kwargs(pipeline, fit_kwargs: dict) -> dict:\n import pycaret.internal.pipeline\n\n return pycaret.internal.pipeline.get_pipeline_fit_kwargs(pipeline, fit_kwargs)\n\n\ndef _get_groups(\n groups,\n data: Optional[pd.DataFrame] = None,\n fold_groups=None,\n ml_usecase: Optional[MLUsecase] = None,\n):\n import pycaret.internal.utils\n\n data = data if data is not None else X_train\n fold_groups = fold_groups if fold_groups is not None else fold_groups_param\n\n return pycaret.internal.utils.get_groups(groups, data, fold_groups)\n"
] |
[
[
"pandas.reset_option",
"sklearn.tree.plot_tree",
"sklearn.model_selection.cross_validate",
"numpy.mean",
"numpy.where",
"numpy.unique",
"pandas.concat",
"sklearn.model_selection._search.GridSearchCV",
"sklearn.model_selection.StratifiedKFold",
"pandas.set_option",
"pandas.DataFrame",
"matplotlib.pyplot.savefig",
"matplotlib.pyplot.subplots",
"numpy.arange",
"matplotlib.pyplot.tight_layout",
"sklearn.base.clone",
"sklearn.model_selection._search.RandomizedSearchCV",
"sklearn.decomposition.PCA",
"matplotlib.pyplot.hlines",
"numpy.array",
"sklearn.preprocessing.LabelEncoder",
"matplotlib.pyplot.title",
"sklearn.model_selection.GroupKFold",
"matplotlib.pyplot.close",
"matplotlib.pyplot.yticks",
"matplotlib.pyplot.figure",
"sklearn.manifold.TSNE",
"numpy.std",
"sklearn.model_selection.train_test_split",
"sklearn.model_selection.KFold",
"matplotlib.pyplot.show",
"pandas.get_dummies",
"pandas.isnull",
"sklearn.calibration.calibration_curve",
"sklearn.preprocessing.StandardScaler",
"numpy.random.seed",
"matplotlib.pyplot.xlabel",
"pandas.DataFrame.from_dict",
"matplotlib.pyplot.plot",
"numpy.sum",
"matplotlib.pyplot.subplot2grid",
"matplotlib.pyplot.ylabel",
"sklearn.model_selection.TimeSeriesSplit",
"numpy.linspace",
"sklearn.set_config"
]
] |
ali-alsabbah/floweaver
|
[
"b4321e2e32dfdf117e682b4b24c4c09b0b5ea2d2"
] |
[
"floweaver/results_graph.py"
] |
[
"import pandas as pd\n\nfrom .layered_graph import MultiLayeredGraph, Ordering\nfrom .partition import Partition, Group\nfrom .sankey_definition import ProcessGroup\n\n\ndef results_graph(view_graph,\n bundle_flows,\n flow_partition=None,\n time_partition=None,\n measures='value'):\n\n G = MultiLayeredGraph()\n groups = []\n\n # Add nodes to graph and to order\n layers = []\n for r, bands in enumerate(view_graph.ordering.layers):\n o = [[] for band in bands]\n for i, rank in enumerate(bands):\n for u in rank:\n node = view_graph.get_node(u)\n group_nodes = []\n for x, xtitle in nodes_from_partition(u, node.partition):\n o[i].append(x)\n group_nodes.append(x)\n if node.partition == None:\n title = u if node.title is None else node.title\n else:\n title = xtitle\n G.add_node(x, {\n 'type': ('process' if isinstance(node, ProcessGroup)\n else 'group'),\n 'direction': node.direction,\n 'title': title,\n })\n groups.append({\n 'id': u,\n 'type': ('process'\n if isinstance(node, ProcessGroup) else 'group'),\n 'title': node.title or '',\n 'nodes': group_nodes\n })\n layers.append(o)\n\n G.ordering = Ordering(layers)\n\n # Add edges to graph\n for v, w, data in view_graph.edges(data=True):\n flows = pd.concat([bundle_flows[bundle] for bundle in data['bundles']])\n gv = view_graph.get_node(v).partition\n gw = view_graph.get_node(w).partition\n gf = data.get('flow_partition') or flow_partition or None\n gt = time_partition or None\n edges = group_flows(flows, v, gv, w, gw, gf, gt, measures)\n for _, _, _, d in edges:\n d['bundles'] = data['bundles']\n G.add_edges_from(edges)\n\n # remove unused nodes\n unused = [u for u, deg in G.degree_iter() if deg == 0]\n for u in unused:\n G.remove_node(u)\n\n # remove unused nodes from groups\n def filter_groups(g):\n if len(g['nodes']) == 0:\n return False\n if len(g['nodes']) == 1:\n return G.node[g['nodes'][0]]['title'] != (g['title'] or g['id'])\n return True\n groups = [\n dict(g, nodes=[x for x in g['nodes'] if x not in unused])\n for g in groups\n ]\n groups = [g for g in groups if filter_groups(g)]\n\n return G, groups\n\n\ndef nodes_from_partition(u, partition):\n if partition is None:\n return [('{}^*'.format(u), '*')]\n else:\n # _ -> other\n return [('{}^{}'.format(u, value), value)\n for value in partition.labels + ['_']]\n\n\ndef agg_one_group(agg):\n def data(group):\n result = group.groupby(lambda x: '').agg(agg)\n return {k: result[k].iloc[0] for k in result}\n return data\n\n\ndef group_flows(flows,\n v,\n partition1,\n w,\n partition2,\n flow_partition,\n time_partition,\n measures):\n\n if callable(measures):\n data = measures\n elif isinstance(measures, str):\n data = agg_one_group({measures: 'sum'})\n elif isinstance(measures, list):\n data = agg_one_group({k: 'sum' for k in measures})\n elif isinstance(measures, dict):\n data = agg_one_group(measures)\n else:\n raise ValueError('measure must be str, list, dict or callable')\n\n e = flows.copy()\n set_partition_keys(e, partition1, 'k1', v + '^', process_side='source')\n set_partition_keys(e, partition2, 'k2', w + '^', process_side='target')\n set_partition_keys(e, flow_partition, 'k3', '')\n set_partition_keys(e, time_partition, 'k4', '')\n grouped = e.groupby(['k1', 'k2', 'k3', 'k4'])\n\n return [\n (source, target, (material, time),\n {'measures': data(group), 'original_flows': list(group.index)})\n for (source, target, material, time), group in grouped\n ]\n\n\ndef set_partition_keys(df, partition, key_column, prefix, process_side=None):\n if partition is None:\n partition = Partition([Group('*', [])])\n df[key_column] = prefix + '_' # other\n seen = (df.index != df.index) # False\n for group in partition.groups:\n q = (df.index == df.index) # True\n for dim, values in group.query:\n if dim.startswith('process') and process_side:\n dim = process_side + dim[7:]\n q = q & df[dim].isin(values)\n if any(q & seen):\n dup = df[q & seen]\n raise ValueError('Duplicate values in group {} ({}): {}'\n .format(group, process_side, ', '.join(\n ['{}-{}'.format(e.source, e.target)\n for _, e in dup.iterrows()])))\n df.loc[q, key_column] = prefix + str(group.label)\n seen = seen | q\n"
] |
[
[
"pandas.concat"
]
] |
whoiszyc/andes
|
[
"67f51b824f3ac364e9e4d161450e476d4967e85b"
] |
[
"andes/models/pss.py"
] |
[
"\"\"\"\nPower system stabilizer models.\n\"\"\"\nfrom andes.core.param import NumParam, IdxParam, ExtParam\nfrom andes.core.var import Algeb, ExtAlgeb, ExtState\nfrom andes.core.block import Lag2ndOrd, LeadLag2ndOrd, LeadLag, WashoutOrLag, Gain, Lag, GainLimiter\nfrom andes.core.service import ExtService, DataSelect, DeviceFinder, Replace, ConstService\nfrom andes.core.discrete import Switcher, Limiter, Derivative\nfrom andes.core.model import ModelData, Model\nfrom collections import OrderedDict\nimport numpy as np\n\nimport logging\n\nlogger = logging.getLogger(__name__)\n\n\nclass PSSBaseData(ModelData):\n def __init__(self):\n super().__init__()\n self.avr = IdxParam(info='Exciter idx', mandatory=True, model='Exciter')\n\n\nclass IEEESTData(PSSBaseData):\n def __init__(self):\n super().__init__()\n self.MODE = NumParam(info='Input signal', mandatory=True)\n\n self.busr = IdxParam(info='Optional remote bus idx', model='Bus', default=None)\n self.busf = IdxParam(info='BusFreq idx for mode 2', model='BusFreq', default=None)\n\n self.A1 = NumParam(default=1, tex_name='A_1', info='filter time const. (pole)')\n self.A2 = NumParam(default=1, tex_name='A_2', info='filter time const. (pole)')\n self.A3 = NumParam(default=1, tex_name='A_3', info='filter time const. (pole)')\n self.A4 = NumParam(default=1, tex_name='A_4', info='filter time const. (pole)')\n self.A5 = NumParam(default=1, tex_name='A_5', info='filter time const. (zero)')\n self.A6 = NumParam(default=1, tex_name='A_6', info='filter time const. (zero)')\n\n self.T1 = NumParam(default=1, tex_name='T_1', vrange=(0, 10), info='first leadlag time const. (zero)')\n self.T2 = NumParam(default=1, tex_name='T_2', vrange=(0, 10), info='first leadlag time const. (pole)')\n self.T3 = NumParam(default=1, tex_name='T_3', vrange=(0, 10), info='second leadlag time const. (pole)')\n self.T4 = NumParam(default=1, tex_name='T_4', vrange=(0, 10), info='second leadlag time const. (pole)')\n self.T5 = NumParam(default=1, tex_name='T_5', vrange=(0, 10), info='washout time const. (zero)')\n self.T6 = NumParam(default=1, tex_name='T_6', vrange=(0.04, 2), info='washout time const. (pole)')\n\n self.KS = NumParam(default=1, tex_name='K_S', info='Gain before washout')\n self.LSMAX = NumParam(default=0.3, tex_name='L_{SMAX}', vrange=(0, 0.3), info='Max. output limit')\n self.LSMIN = NumParam(default=-0.3, tex_name='L_{SMIN}', vrange=(-0.3, 0), info='Min. output limit')\n\n self.VCU = NumParam(default=999, tex_name='V_{CU}', vrange=(1, 1.2),\n unit='p.u.', info='Upper enabling bus voltage')\n self.VCL = NumParam(default=-999, tex_name='V_{CL}', vrange=(0., 1),\n unit='p.u.', info='Upper enabling bus voltage')\n\n\nclass PSSBase(Model):\n \"\"\"\n PSS base model.\n \"\"\"\n\n def __init__(self, system, config):\n super().__init__(system, config)\n self.group = 'PSS'\n self.flags.update({'tds': True})\n\n self.VCUr = Replace(self.VCU, lambda x: np.equal(x, 0.0), 999)\n self.VCLr = Replace(self.VCL, lambda x: np.equal(x, 0.0), -999)\n\n # retrieve indices of connected generator, bus, and bus freq\n self.syn = ExtParam(model='Exciter', src='syn', indexer=self.avr, export=False,\n info='Retrieved generator idx', vtype=str)\n\n self.bus = ExtParam(model='SynGen', src='bus', indexer=self.syn, export=False,\n info='Retrieved bus idx', vtype=str, default=None,\n )\n\n self.buss = DataSelect(self.busr, self.bus, info='selected bus (bus or busr)')\n\n self.busfreq = DeviceFinder(self.busf, link=self.buss, idx_name='bus')\n\n # from SynGen\n self.Sn = ExtParam(model='SynGen', src='Sn', indexer=self.syn, tex_name='S_n',\n info='Generator power base', export=False)\n\n self.omega = ExtState(model='SynGen', src='omega', indexer=self.syn,\n tex_name=r'\\omega', info='Generator speed', unit='p.u.',\n )\n\n self.tm0 = ExtService(model='SynGen', src='tm', indexer=self.syn,\n tex_name=r'\\tau_{m0}', info='Initial mechanical input',\n )\n self.tm = ExtAlgeb(model='SynGen', src='tm', indexer=self.syn,\n tex_name=r'\\tau_m', info='Generator mechanical input',\n )\n self.te = ExtAlgeb(model='SynGen', src='te', indexer=self.syn,\n tex_name=r'\\tau_e', info='Generator electrical output',\n )\n # from Bus\n self.v = ExtAlgeb(model='Bus', src='v', indexer=self.buss, tex_name=r'V',\n info='Bus (or busr, if given) terminal voltage',\n )\n self.v0 = ExtService(model='Bus', src='v', indexer=self.buss, tex_name=\"V_0\",\n info='Initial bus voltage',\n )\n\n # from BusFreq\n self.f = ExtAlgeb(model='FreqMeasurement', src='f', indexer=self.busfreq, export=False,\n info='Bus frequency')\n\n # from Exciter\n self.vi = ExtAlgeb(model='Exciter', src='vi', indexer=self.avr, tex_name='v_i',\n info='Exciter input voltage',\n e_str='u * vsout')\n\n self.vsout = Algeb(info='PSS output voltage to exciter',\n tex_name='v_{sout}',\n ) # `self.vsout.e_str` to be provided by specific models\n\n\nclass IEEESTModel(PSSBase):\n \"\"\"\n IEEEST Stabilizer equation.\n \"\"\"\n\n def __init__(self, system, config):\n PSSBase.__init__(self, system, config)\n\n self.config.add(OrderedDict([('freq_model', 'BusFreq')]))\n self.config.add_extra('_help', {'freq_model': 'default freq. measurement model'})\n self.config.add_extra('_alt', {'freq_model': ('BusFreq',)})\n\n self.busf.model = self.config.freq_model\n\n self.dv = Derivative(self.v, tex_name='dV/dt', info='Finite difference of bus voltage')\n\n self.SnSb = ExtService(model='SynGen', src='M', indexer=self.syn, attr='pu_coeff',\n info='Machine base to sys base factor for power',\n tex_name='(Sb/Sn)')\n\n self.SW = Switcher(u=self.MODE,\n options=[0, 1, 2, 3, 4, 5, 6],\n )\n\n self.sig = Algeb(tex_name='S_{ig}',\n info='Input signal',\n )\n\n self.sig.v_str = 'SW_s1*(omega-1) + SW_s2*0 + SW_s3*(tm0/SnSb) + ' \\\n 'SW_s4*(tm-tm0) + SW_s5*v + SW_s6*0'\n\n self.sig.e_str = 'SW_s1*(omega-1) + SW_s2*(f-1) + SW_s3*(te/SnSb) + ' \\\n 'SW_s4*(tm-tm0) + SW_s5*v + SW_s6*dv_v - sig'\n\n self.F1 = Lag2ndOrd(u=self.sig, K=1, T1=self.A1, T2=self.A2)\n\n self.F2 = LeadLag2ndOrd(u=self.F1_y, T1=self.A3, T2=self.A4, T3=self.A5, T4=self.A6, zero_out=True)\n\n self.LL1 = LeadLag(u=self.F2_y, T1=self.T1, T2=self.T2, zero_out=True)\n\n self.LL2 = LeadLag(u=self.LL1_y, T1=self.T3, T2=self.T4, zero_out=True)\n\n self.Vks = Gain(u=self.LL2_y, K=self.KS)\n\n self.WO = WashoutOrLag(u=self.Vks_y, T=self.T6, K=self.T5, name='WO', zero_out=True) # WO_y == Vss\n\n self.VLIM = Limiter(u=self.WO_y, lower=self.LSMIN, upper=self.LSMAX, info='Vss limiter')\n\n self.Vss = Algeb(tex_name='V_{ss}', info='Voltage output before output limiter',\n e_str='VLIM_zi * WO_y + VLIM_zu * LSMAX + VLIM_zl * LSMIN - Vss')\n\n self.OLIM = Limiter(u=self.v, lower=self.VCLr, upper=self.VCUr, info='output limiter')\n\n self.vsout.e_str = 'OLIM_zi * Vss - vsout'\n\n\nclass IEEEST(IEEESTData, IEEESTModel):\n \"\"\"\n IEEEST stabilizer model. Automatically adds frequency measurement devices if not provided.\n\n Input signals (MODE):\n\n 1 - Rotor speed deviation (p.u.),\n 2 - Bus frequency deviation (*) (p.u.),\n 3 - Generator P electrical in Gen MVABase (p.u.),\n 4 - Generator accelerating power (p.u.),\n 5 - Bus voltage (p.u.),\n 6 - Derivative of p.u. bus voltage.\n\n (*) Due to the frequency measurement implementation difference,\n mode 2 is likely to yield different results across software.\n\n Blocks are named `F1`, `F2`, `LL1`, `LL2` and `WO` in sequence.\n Two limiters are named `VLIM` and `OLIM` in sequence.\n \"\"\"\n\n def __init__(self, system, config):\n IEEESTData.__init__(self)\n IEEESTModel.__init__(self, system, config)\n\n\nclass ST2CUTData(PSSBaseData):\n def __init__(self):\n PSSBaseData.__init__(self)\n self.MODE = NumParam(info='Input signal 1', mandatory=True)\n self.busr = NumParam(info='Remote bus 1')\n self.busf = IdxParam(info='BusFreq idx for signal 1 mode 2',\n model='BusFreq', )\n\n self.MODE2 = NumParam(info='Input signal 2')\n self.busr2 = NumParam(info='Remote bus 2')\n self.busf2 = IdxParam(info='BusFreq idx for signal 2 mode 2', model='BusFreq')\n\n self.K1 = NumParam(default=1, tex_name='K_1',\n info='Transducer 1 gain',\n vrange=(0, 10),\n )\n self.K2 = NumParam(default=1, tex_name='K_2',\n info='Transducer 2 gain',\n vrange=(0, 10),\n )\n self.T1 = NumParam(default=1, tex_name='T_1',\n info='Transducer 1 time const.',\n vrange=(0, 10),\n )\n self.T2 = NumParam(default=1, tex_name='T_2',\n info='Transducer 2 time const.',\n vrange=(0, 10),\n )\n\n self.T3 = NumParam(default=1, tex_name='T_3',\n info='Washout int. time const.',\n vrange=(0, 10),\n )\n self.T4 = NumParam(default=0.2, tex_name='T_4',\n info='Washout delay time const.',\n vrange=(0.05, 10),\n )\n\n self.T5 = NumParam(default=1, tex_name='T_5',\n info='Leadlag 1 time const. (1)',\n vrange=(0, 10),\n )\n self.T6 = NumParam(default=0.5, tex_name='T_6',\n info='Leadlag 1 time const. (2)',\n vrange=(0, 2),\n )\n\n self.T7 = NumParam(default=1, tex_name='T_7',\n info='Leadlag 2 time const. (1)',\n vrange=(0, 10),\n )\n self.T8 = NumParam(default=1, tex_name='T_8',\n info='Leadlag 2 time const. (2)',\n vrange=(0, 10),\n )\n\n self.T9 = NumParam(default=1, tex_name='T_9',\n info='Leadlag 3 time const. (1)',\n vrange=(0, 2),\n )\n self.T10 = NumParam(default=0.2, tex_name='T_{10}',\n info='Leadlag 3 time const. (2)',\n vrange=(0, 2),\n )\n\n self.LSMAX = NumParam(default=0.3, tex_name='L_{SMAX}', vrange=(0, 0.3), info='Max. output limit')\n self.LSMIN = NumParam(default=-0.3, tex_name='L_{SMIN}', vrange=(-0.3, 0), info='Min. output limit')\n\n self.VCU = NumParam(default=999, tex_name='V_{CU}', vrange=(1, 1.2),\n unit='p.u.', info='Upper enabling bus voltage')\n\n self.VCL = NumParam(default=-999, tex_name='V_{CL}', vrange=(-0.1, 1),\n unit='p.u.', info='Upper enabling bus voltage')\n\n\nclass ST2CUTModel(PSSBase):\n def __init__(self, system, config):\n PSSBase.__init__(self, system, config)\n\n # ALL THE FOLLOWING IS FOR INPUT 2\n # retrieve indices of bus and bus freq\n self.buss2 = DataSelect(self.busr2, self.bus, info='selected bus (bus or busr)')\n\n self.busfreq2 = DeviceFinder(self.busf2, link=self.buss2, idx_name='bus')\n\n # from Bus\n self.v2 = ExtAlgeb(model='Bus', src='v', indexer=self.buss2, tex_name=r'V',\n info='Bus (or busr2, if given) terminal voltage',\n )\n\n # from BusFreq 2\n self.f2 = ExtAlgeb(model='FreqMeasurement', src='f', indexer=self.busfreq2, export=False,\n info='Bus frequency 2')\n\n # Config\n self.config.add(OrderedDict([('freq_model', 'BusFreq')]))\n self.config.add_extra('_help', {'freq_model': 'default freq. measurement model'})\n self.config.add_extra('_alt', {'freq_model': ('BusFreq',)})\n\n self.busf.model = self.config.freq_model\n self.busf2.model = self.config.freq_model\n\n # input signal switch\n self.dv = Derivative(self.v)\n self.dv2 = Derivative(self.v2)\n\n self.SnSb = ExtService(model='SynGen', src='M', indexer=self.syn, attr='pu_coeff',\n info='Machine base to sys base factor for power',\n tex_name='(Sb/Sn)')\n\n self.SW = Switcher(u=self.MODE,\n options=[0, 1, 2, 3, 4, 5, 6, np.nan],\n )\n self.SW2 = Switcher(u=self.MODE2,\n options=[0, 1, 2, 3, 4, 5, 6, np.nan],\n )\n\n # Input signals\n self.sig = Algeb(tex_name='S_{ig}',\n info='Input signal',\n )\n self.sig.v_str = 'SW_s1*(omega-1) + SW_s2*0 + SW_s3*(tm0/SnSb) + ' \\\n 'SW_s4*(tm-tm0) + SW_s5*v + SW_s6*0'\n self.sig.e_str = 'SW_s1*(omega-1) + SW_s2*(f-1) + SW_s3*(te/SnSb) + ' \\\n 'SW_s4*(tm-tm0) + SW_s5*v + SW_s6*dv_v - sig'\n\n self.sig2 = Algeb(tex_name='S_{ig2}',\n info='Input signal 2',\n )\n self.sig2.v_str = 'SW2_s1*(omega-1) + SW2_s2*0 + SW2_s3*(tm0/SnSb) + ' \\\n 'SW2_s4*(tm-tm0) + SW2_s5*v2 + SW2_s6*0'\n self.sig2.e_str = 'SW2_s1*(omega-1) + SW2_s2*(f2-1) + SW2_s3*(te/SnSb) + ' \\\n 'SW2_s4*(tm-tm0) + SW2_s5*v2 + SW2_s6*dv2_v - sig2'\n\n self.L1 = Lag(u=self.sig,\n K=self.K1,\n T=self.T1,\n info='Transducer 1',\n )\n self.L2 = Lag(u=self.sig2,\n K=self.K2,\n T=self.T2,\n info='Transducer 2',\n )\n self.IN = Algeb(tex_name='I_N',\n info='Sum of inputs',\n v_str='L1_y + L2_y',\n e_str='L1_y + L2_y - IN',\n )\n\n self.WO = WashoutOrLag(u=self.IN,\n K=self.T3,\n T=self.T4,\n )\n\n self.LL1 = LeadLag(u=self.WO_y,\n T1=self.T5,\n T2=self.T6,\n zero_out=True,\n )\n\n self.LL2 = LeadLag(u=self.LL1_y,\n T1=self.T7,\n T2=self.T8,\n zero_out=True,\n )\n\n self.LL3 = LeadLag(u=self.LL2_y,\n T1=self.T9,\n T2=self.T10,\n zero_out=True,\n )\n\n self.VSS = GainLimiter(u=self.LL3_y,\n K=1,\n lower=self.LSMIN,\n upper=self.LSMAX\n )\n\n self.VOU = ConstService(v_str='VCUr + v0')\n self.VOL = ConstService(v_str='VCLr + v0')\n\n self.OLIM = Limiter(u=self.v, lower=self.VOL, upper=self.VOU, info='output limiter')\n\n self.vsout.e_str = 'OLIM_zi * VSS_y - vsout'\n\n\nclass ST2CUT(ST2CUTData, ST2CUTModel):\n \"\"\"\n ST2CUT stabilizer model. Automatically adds frequency measurement devices if not provided.\n\n Input signals (MODE and MODE2):\n\n 0 - Disable input signal\n 1 (s1) - Rotor speed deviation (p.u.),\n 2 (s2) - Bus frequency deviation (*) (p.u.),\n 3 (s3) - Generator P electrical in Gen MVABase (p.u.),\n 4 (s4) - Generator accelerating power (p.u.),\n 5 (s5) - Bus voltage (p.u.),\n 6 (s6) - Derivative of p.u. bus voltage.\n\n (*) Due to the frequency measurement implementation difference,\n mode 2 is likely to yield different results across software.\n\n Blocks are named `LL1`, `LL2`, `LL3`, `LL4` in sequence.\n Two limiters are named `VSS_lim` and `OLIM` in sequence.\n \"\"\"\n\n def __init__(self, system, config):\n ST2CUTData.__init__(self)\n ST2CUTModel.__init__(self, system, config)\n"
] |
[
[
"numpy.equal"
]
] |
meandme234/uav-autonomous-landing
|
[
"40e5566a8577b3f443ae0ec8052b2f46d329a998"
] |
[
"trajectories_life_long.py"
] |
[
"import matplotlib\nimport matplotlib.pyplot as plt \nimport matplotlib as mpl\nfrom mpl_toolkits.mplot3d import Axes3D\nimport pandas as pd\nimport os\nimport matplotlib.pyplot as plt\nimport sys\nimport itertools\n\n\nmpl.rcParams['legend.fontsize'] = 12\nDPI = 5000\n\ninput_dir = \"/home/pablo/ws/log/trajectories\"\nprint(\"Reading from\", input_dir) \n\n\n\n##########################################################\n################### w/ PREDICTION ########################\n##########################################################\n## linear speed\nif len(sys.argv) == 2:\n\tlinear_speed = sys.argv[1]\nelse:\n\tprint(\"Exiting...\")\n\texit()\ntrajectories_pred = pd.read_csv(input_dir + \"/trajectories_pred_lifelong_{}.csv\".format(linear_speed))\n\n\n# get only some lines from the data\nzoom_in = True\nif zoom_in:\n\tinitial_row = 2300 # corresponds to roughly a timestamp of 50 s\n\trow_200_sec = 10000 # corresponds to roughly a timestamp of 100 s\n\ttrajectories_pred = trajectories_pred.iloc[initial_row:row_200_sec]\n\n\n## -- save dir\nsave_dir = \"/home/pablo/ws/log/trajectories/lifelong_{}\".format(linear_speed)\nif not os.path.exists(save_dir):\n\tos.makedirs(save_dir)\n\n# extract data from DataFrame\nstatus = trajectories_pred['status'].tolist()\nardrone_x = trajectories_pred['aX'].tolist()\nardrone_y = trajectories_pred['aY'].tolist()\nardrone_z = trajectories_pred['aZ'].tolist()\nsummit_x = trajectories_pred['sX'].tolist()\nsummit_y = trajectories_pred['sY'].tolist()\nsummit_z = trajectories_pred['sZ'].tolist()\n\n# 3D\nfig = plt.figure()\nax = fig.gca(projection='3d')\nax.plot(ardrone_x, ardrone_y, ardrone_z, 'r', label='UAV')\nax.plot(summit_x, summit_y, summit_z, 'g', label='UGV')\nax.set(xlabel='x (m)', ylabel='y (m)', zlabel='z (m)')\nbottom, top = plt.ylim() # return the current ylim\nax.legend()\nax.view_init(elev=11, azim=125)\nplt.show()\n\nfig.savefig(os.path.join(save_dir, \"traj3D_pred.pdf\"), format='pdf', dpi=DPI)\n\n# 2D\nfig, ax = plt.subplots()\nax.plot(ardrone_x, ardrone_y, 'r', label='UAV')\nax.plot(summit_x, summit_y, 'g', label='UGV')\nax.set(xlabel='x (m)', ylabel='y (m)')\nax.legend()\nax.grid()\nfig.savefig(os.path.join(save_dir, \"traj2D_pred.pdf\"), format='pdf', dpi=DPI)\n\n"
] |
[
[
"matplotlib.pyplot.show",
"matplotlib.pyplot.ylim",
"matplotlib.pyplot.subplots",
"matplotlib.pyplot.figure"
]
] |
familywei/RLs
|
[
"8414cb0d31701e1c328a733d1251cfc10b9f38db"
] |
[
"gym_wrapper.py"
] |
[
"import gym\r\nimport numpy as np\r\nimport threading\r\n\r\n\r\nclass MyThread(threading.Thread):\r\n\r\n def __init__(self, func, args=()):\r\n super().__init__()\r\n self.func = func\r\n self.args = args\r\n\r\n def run(self):\r\n self.result = self.func(*self.args)\r\n\r\n def get_result(self):\r\n try:\r\n return self.result\r\n except Exception:\r\n return None\r\n\r\n\r\nclass gym_envs(object):\r\n\r\n def __init__(self, gym_env_name, n):\r\n self.n = n\r\n self.envs = [gym.make(gym_env_name) for _ in range(self.n)]\r\n self.observation_space = self.envs[0].observation_space\r\n self.obs_type = 'visual' if len(self.observation_space.shape) == 3 else 'vector'\r\n if type(self.envs[0].action_space) == gym.spaces.box.Box:\r\n self.a_type = 'continuous'\r\n elif type(self.envs[0].action_space) == gym.spaces.tuple.Tuple:\r\n self.a_type = 'Tuple(Discrete)'\r\n else:\r\n self.a_type = 'discrete'\r\n self.action_space = self.envs[0].action_space\r\n\r\n def render(self):\r\n self.envs[0].render()\r\n\r\n def close(self):\r\n [env.close() for env in self.envs]\r\n\r\n def reset(self):\r\n threadpool = []\r\n for i in range(self.n):\r\n th = MyThread(self.envs[i].reset, args=())\r\n threadpool.append(th)\r\n for th in threadpool:\r\n th.start()\r\n for th in threadpool:\r\n threading.Thread.join(th)\r\n if self.obs_type == 'visual':\r\n return np.array([threadpool[i].get_result()[np.newaxis, :] for i in range(self.n)])\r\n else:\r\n return np.array([threadpool[i].get_result() for i in range(self.n)])\r\n\r\n def step(self, actions):\r\n if self.a_type == 'discrete':\r\n actions = actions.reshape(-1,)\r\n elif self.a_type == 'Tuple(Discrete)':\r\n actions = actions.reshape(self.n, -1).tolist()\r\n threadpool = []\r\n for i in range(self.n):\r\n th = MyThread(self.envs[i].step, args=(actions[i], ))\r\n threadpool.append(th)\r\n for th in threadpool:\r\n th.start()\r\n for th in threadpool:\r\n threading.Thread.join(th)\r\n if self.obs_type == 'visual':\r\n results = [\r\n [threadpool[i].get_result()[0][np.newaxis, :], *threadpool[i].get_result()[1:]]\r\n for i in range(self.n)]\r\n else:\r\n results = [threadpool[i].get_result() for i in range(self.n)]\r\n return [np.array(e) for e in zip(*results)]\r\n"
] |
[
[
"numpy.array"
]
] |
scoumeri/geopm
|
[
"2406b8cca92d8eb32d4dc26d24bb2273164f186c"
] |
[
"integration/experiment/energy_efficiency/gen_plot_profile_comparison.py"
] |
[
"#!/usr/bin/env python3\n#\n# Copyright (c) 2015 - 2022, Intel Corporation\n#\n# Redistribution and use in source and binary forms, with or without\n# modification, are permitted provided that the following conditions\n# are met:\n#\n# * Redistributions of source code must retain the above copyright\n# notice, this list of conditions and the following disclaimer.\n#\n# * Redistributions in binary form must reproduce the above copyright\n# notice, this list of conditions and the following disclaimer in\n# the documentation and/or other materials provided with the\n# distribution.\n#\n# * Neither the name of Intel Corporation nor the names of its\n# contributors may be used to endorse or promote products derived\n# from this software without specific prior written permission.\n#\n# THIS SOFTWARE IS PROVIDED BY THE COPYRIGHT HOLDERS AND CONTRIBUTORS\n# \"AS IS\" AND ANY EXPRESS OR IMPLIED WARRANTIES, INCLUDING, BUT NOT\n# LIMITED TO, THE IMPLIED WARRANTIES OF MERCHANTABILITY AND FITNESS FOR\n# A PARTICULAR PURPOSE ARE DISCLAIMED. IN NO EVENT SHALL THE COPYRIGHT\n# OWNER OR CONTRIBUTORS BE LIABLE FOR ANY DIRECT, INDIRECT, INCIDENTAL,\n# SPECIAL, EXEMPLARY, OR CONSEQUENTIAL DAMAGES (INCLUDING, BUT NOT\n# LIMITED TO, PROCUREMENT OF SUBSTITUTE GOODS OR SERVICES; LOSS OF USE,\n# DATA, OR PROFITS; OR BUSINESS INTERRUPTION) HOWEVER CAUSED AND ON ANY\n# THEORY OF LIABILITY, WHETHER IN CONTRACT, STRICT LIABILITY, OR TORT\n# (INCLUDING NEGLIGENCE OR OTHERWISE) ARISING IN ANY WAY LOG OF THE USE\n# OF THIS SOFTWARE, EVEN IF ADVISED OF THE POSSIBILITY OF SUCH DAMAGE.\n#\n\n'''\nPlot runtime and energy differences vs. a baseline for multiple profile names.\nThe baseline and target data can be from any agent, policy, or application.\nMultiple iterations of the same profile will be combined by removing the\niteration number after the last underscore from the profile.\n'''\n\nimport argparse\nimport sys\nimport os\nimport numpy\nimport matplotlib.pyplot as plt\n\nimport geopmpy.io\nimport geopmpy.hash\nfrom experiment import common_args\nfrom experiment import report\nfrom experiment import plotting\n\n\ndef summary(df, perf_metric, use_stdev, baseline, targets, show_details):\n\n report.prepare_columns(df)\n report.prepare_metrics(df, perf_metric)\n\n result = report.energy_perf_summary(df=df,\n loop_key='Profile',\n loop_vals=targets,\n baseline=baseline,\n perf_metric=perf_metric,\n use_stdev=use_stdev)\n\n output_prefix = os.path.join(output_dir, '{}'.format(common_prefix))\n output_stats_name = '{}_stats.log'.format(output_prefix)\n\n if show_details:\n sys.stdout.write('{}\\n'.format(result))\n with open(output_stats_name, 'w') as outfile:\n outfile.write('{}\\n'.format(result))\n\n return result\n\n\ndef plot_bars(df, baseline_profile, title, perf_label, energy_label, xlabel, output_dir, show_details):\n\n labels = df.index.format()\n prefix = os.path.commonprefix(labels)\n labels = list(map(lambda x: x[len(prefix):], labels))\n points = numpy.arange(len(df.index))\n bar_width = 0.35\n\n errorbar_format = {'fmt': ' ', # no connecting line\n 'label': '',\n 'color': 'r',\n 'elinewidth': 2,\n 'capthick': 2,\n 'zorder': 10}\n\n f, ax = plt.subplots()\n # plot performance\n perf = df['performance']\n yerr = (df['min_delta_perf'], df['max_delta_perf'])\n ax.bar(points - bar_width / 2, perf, width=bar_width, color='orange',\n label=perf_label)\n ax.errorbar(points - bar_width / 2, perf, xerr=None,\n yerr=yerr, **errorbar_format)\n\n # plot energy\n ax.bar(points + bar_width / 2, df['energy_perf'], width=bar_width, color='purple',\n label=energy_label)\n yerr = (df['min_delta_energy'], df['max_delta_energy'])\n ax.errorbar(points + bar_width / 2, df['energy_perf'], xerr=None,\n yerr=yerr, **errorbar_format)\n\n # baseline\n ax.axhline(y=1.0, color='blue', linestyle='dotted', label='baseline')\n\n ax.set_xticks(points)\n ax.set_xticklabels(labels, rotation='vertical')\n ax.set_xlabel(xlabel)\n f.subplots_adjust(bottom=0.25) # TODO: use longest profile name\n ax.set_ylabel('Normalized performance or energy')\n plt.title('{}\\nBaseline: {}'.format(title, baseline_profile))\n plt.legend()\n fig_dir = os.path.join(output_dir, 'figures')\n if not os.path.exists(fig_dir):\n os.mkdir(fig_dir)\n fig_name = plotting.title_to_filename(title)\n fig_name = os.path.join(fig_dir, fig_name + '.png')\n plt.savefig(fig_name)\n if show_details:\n sys.stdout.write('Wrote {}\\n'.format(fig_name))\n\n\ndef get_profile_list(rrc):\n result = rrc.get_app_df()['Profile'].unique()\n profs, _ = zip(*map(report.extract_trial, result))\n result = sorted(list(set(profs)))\n return result\n\n\nif __name__ == '__main__':\n\n parser = argparse.ArgumentParser()\n common_args.add_output_dir(parser)\n common_args.add_show_details(parser)\n common_args.add_use_stdev(parser)\n common_args.add_performance_metric(parser)\n\n parser.add_argument('--baseline', dest='baseline',\n action='store', default=None,\n help='baseline profile name')\n parser.add_argument('--targets', dest='targets',\n action='store', default=None,\n help='comma-separated list of profile names to compare')\n parser.add_argument('--list-profiles', dest='list_profiles',\n action='store_true', default=False,\n help='list all profiles present in the discovered reports and exit')\n parser.add_argument('--xlabel', dest='xlabel',\n action='store', default='Profile',\n help='x-axis label for profiles')\n\n args = parser.parse_args()\n\n output_dir = args.output_dir\n try:\n rrc = geopmpy.io.RawReportCollection(\"*report\", dir_name=output_dir)\n except RuntimeError as ex:\n sys.stderr.write('<geopm> Error: No report data found in {}: {}\\n'.format(output_dir, ex))\n sys.exit(1)\n\n profile_list = get_profile_list(rrc)\n if args.list_profiles:\n sys.stdout.write(','.join(profile_list) + '\\n')\n sys.exit(0)\n\n if args.baseline:\n baseline = args.baseline\n else:\n if len(profile_list) < 3:\n raise RuntimeError('Fewer than 3 distinct profiles discovered, --baseline must be provided')\n longest = 0\n for pp in profile_list:\n without = list(profile_list)\n without.remove(pp)\n rank = len(os.path.commonprefix(without))\n if longest < rank:\n longest = rank\n baseline = pp\n sys.stderr.write('Warning: --baseline not provided, using best guess: \"{}\"\\n'.format(baseline))\n\n if args.targets:\n targets = args.targets.split(',')\n else:\n targets = list(profile_list)\n targets.remove(baseline)\n\n common_prefix = os.path.commonprefix(targets).rstrip('_')\n\n # Test if Epochs were used\n if rrc.get_epoch_df()[:1]['count'].item() > 0.0:\n df = rrc.get_epoch_df()\n else:\n df = rrc.get_app_df()\n\n result = summary(df=df,\n perf_metric=args.performance_metric,\n use_stdev=args.use_stdev,\n baseline=baseline,\n targets=targets,\n show_details=args.show_details)\n\n title = os.path.commonprefix(targets)\n title = title.rstrip('_')\n title = title + ' (' + args.performance_metric\n if args.use_stdev:\n title += ', stdev)'\n else:\n title += ', min-max)'\n perf_label, energy_label = report.perf_metric_label(args.performance_metric)\n\n plot_bars(result, baseline, title, perf_label, energy_label,\n xlabel=args.xlabel,\n output_dir=output_dir, show_details=args.show_details)\n"
] |
[
[
"matplotlib.pyplot.savefig",
"matplotlib.pyplot.legend",
"matplotlib.pyplot.subplots"
]
] |
ccha23/CS1302ICP
|
[
"80907dae8d4d90b911e52dbea159629a221b9a3c"
] |
[
"_build/jupyter_execute/Lab2/Calculators.py"
] |
[
"#!/usr/bin/env python\n# coding: utf-8\n\n# # Calculators\n\n# **CS1302 Introduction to Computer Programming**\n# ___\n\n# Run the following to load additional tools required for this lab. \n# In particular, the `math` library provides many useful mathematical functions and constants.\n\n# In[ ]:\n\n\nget_ipython().run_line_magic('reset', '-f')\nfrom ipywidgets import interact\nimport matplotlib.pyplot as plt\nimport numpy as np\nimport math\nfrom math import log, exp, sin, cos, tan, pi\n\n\n# The following code is a Python one-liner that creates a calculator. \n# Evaluate the cell with `Ctrl+Enter`:\n\n# In[ ]:\n\n\nprint(eval(input()))\n\n\n# Try some calculations below using this calculator:\n# 1. $2^3$ by entering `2**3`;\n# 1. $\\frac23$ by entering `2/3`;\n# 1. $\\left\\lceil\\frac32\\right\\rceil$ by entering `3//2`;\n# 1. $3\\mod 2$ by entering `3%2`;\n# 1. $\\sqrt{2}$ by entering `2**0.5`; and\n# 1. $\\sin(\\pi/6)$ by entering `sin(pi/6)`;\n\n# For this lab, you will create more powerful and dedicated calculators. \n# We will first show you a demo. Then, it will be your turn to create the calculators.\n\n# ## Hypotenuse Calculator (Demo)\n\n# 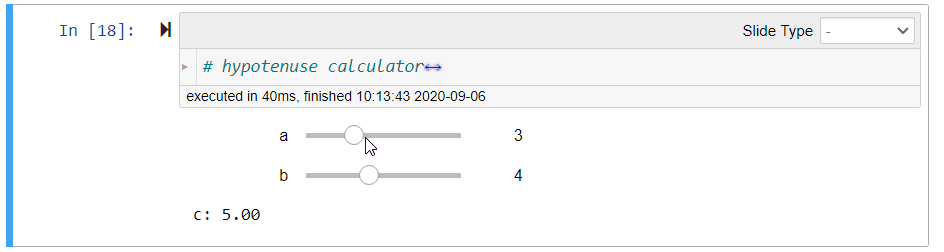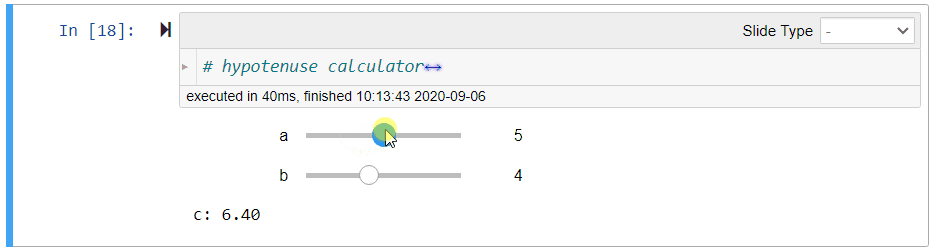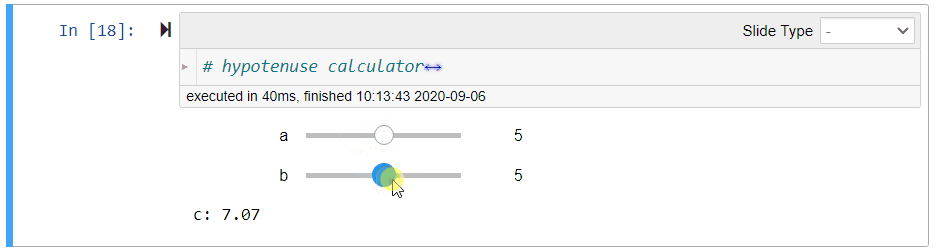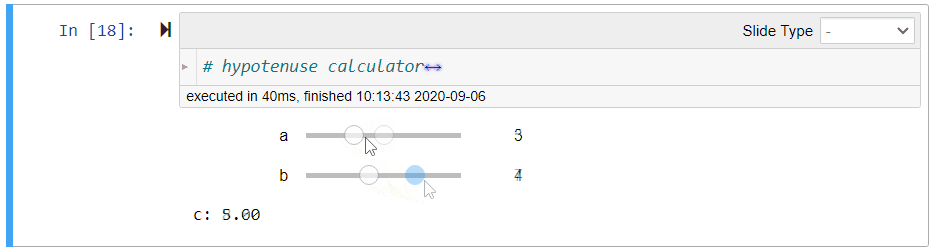\n\n# Using the Pythagoras theorem below, we can define the following function `length_of_hypotenus` to calculate the length `c` of the hypotenuse given the lengths `a` and `b` of the other sides of a right-angled triangle:\n# $$c = \\sqrt{a^2 + b^2}$$\n\n# In[ ]:\n\n\ndef length_of_hypotenuse(a, b):\n c = (a**2 + b**2)**(0.5) # Pythagoras\n return c\n\n\n# - You need not understand how a function is defined, but\n# - you should know how to *write the formula as a Python expression*, and\n# - *assign to the variable* `c` the value of the expression (Line 2).\n\n# For example, you may be asked to write Line 2, while Line 1 and 3 are given to you:\n\n# **Exercise** Complete the function below to return the length `c` of the hypotenuse given the lengths `a` and `b`.\n\n# In[ ]:\n\n\ndef length_of_hypotenuse(a, b):\n # YOUR CODE HERE\n raise NotImplementedError()\n return c\n\n\n# Note that indentation affects the execution of Python code. The assignment statement must be indented to indicate that it is part of the *body* of the function. \n# (Try removing the indentation and see what happens.)\n\n# We will use widgets (`ipywidgets`) to let user interact with the calculator more easily: \n\n# In[ ]:\n\n\n# hypotenuse calculator\n@interact(a=(0, 10, 1), b=(0, 10, 1))\ndef calculate_hypotenuse(a=3, b=4):\n print('c: {:.2f}'.format(length_of_hypotenuse(a, b)))\n\n\n# After running the above cell, you can move the sliders to change the values of `a` and `b`. The value of `c` will be updated immediately.\n\n# - For this lab, you need not know how write widgets, but\n# - you should know how to *format a floating point number* (Line 3). \n\n# You can check your code with a few cases listed in the test cell below.\n\n# In[ ]:\n\n\n# tests\ndef test_length_of_hypotenuse(a, b, c):\n c_ = length_of_hypotenuse(a, b)\n correct = math.isclose(c, c_)\n if not correct:\n print(f'For a={a} and b={b}, c should be {c}, not {c_}.')\n assert correct\n\n\ntest_length_of_hypotenuse(3, 4, 5)\ntest_length_of_hypotenuse(0, 0, 0)\ntest_length_of_hypotenuse(4, 7, 8.06225774829855)\n\n\n# ## Quadratic Equation\n\n# ### Graphical Calculator for Parabola\n\n# \n\n# In mathematics, the collection of points $(x,y)$ satisfying the following equation forms a *parabola*:\n# \n# $$\n# y=ax^2+bx+c\n# $$\n# where $a$, $b$, and $c$ are real numbers called the *coefficients*.\n\n# **Exercise** Given the variables `x`, `a`, `b`, and `c` store the $x$-coordinate and the coefficients $a$, $b$, and $c$ respectively, assign to `y` the corresponding $y$-coordinate for the parabola.\n\n# In[ ]:\n\n\ndef get_y(x, a, b, c):\n # YOUR CODE HERE\n raise NotImplementedError()\n return y\n\n\n# In[ ]:\n\n\n# tests\ndef test_get_y(y,x,a,b,c):\n y_ = get_y(x,a,b,c)\n correct = math.isclose(y,y_)\n if not correct:\n print(f'With (x, a, b, c)={x,a,b,c}, y should be {y} not {y_}.')\n assert correct\n\ntest_get_y(0,0,0,0,0)\ntest_get_y(1,0,1,2,1)\ntest_get_y(2,0,2,1,2)\n\n\n# In[ ]:\n\n\n# graphical calculator for parabola\n@interact(a=(-10, 10, 1), b=(-10, 10, 1), c=(-10, 10, 1))\ndef plot_parabola(a, b, c):\n xmin, xmax, ymin, ymax, resolution = -10, 10, -10, 10, 50\n ax = plt.gca()\n ax.set_title(r'$y=ax^2+bx+c$')\n ax.set_xlabel(r'$x$')\n ax.set_ylabel(r'$y$')\n ax.set_xlim([xmin, xmax])\n ax.set_ylim([ymin, ymax])\n ax.grid()\n\n x = np.linspace(xmin, xmax, resolution)\n ax.plot(x, get_y(x, a, b, c))\n\n\n# ### Quadratic Equation Solver\n\n# 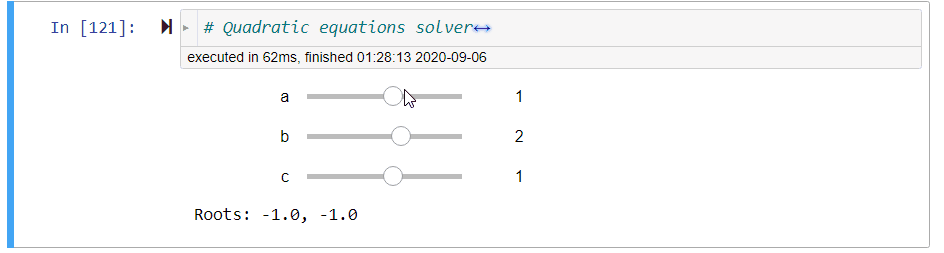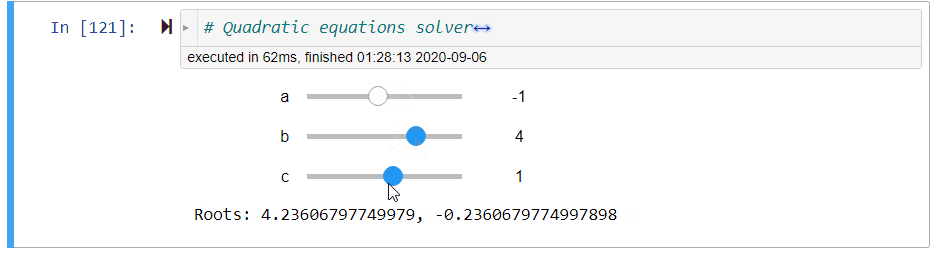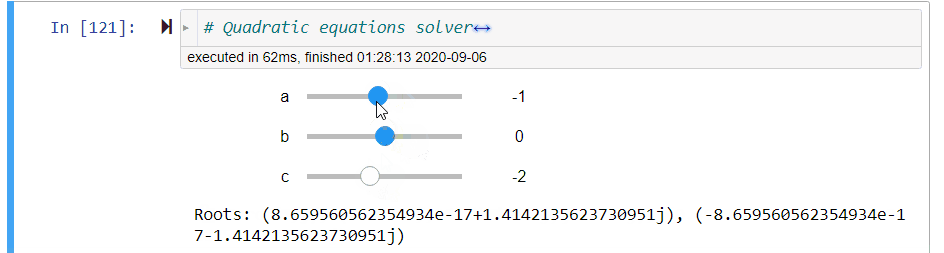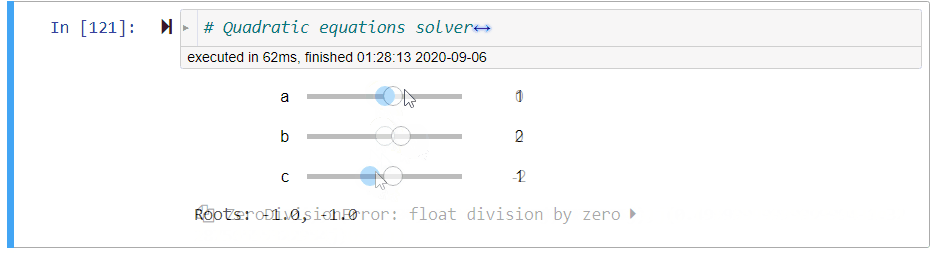\n\n# For the quadratic equation\n# \n# $$\n# ax^2+bx+c=0,\n# $$\n# the *roots* (solutions for $x$) are give by\n# \n# $$\n# \\frac{-b-\\sqrt{b^2-4ac}}{2a},\\frac{-b+\\sqrt{b^2-4ac}}{2a}.\n# $$\n\n# **Exercise** Assign to `root1` and `root2` the values of the first and second roots above respectively.\n\n# In[ ]:\n\n\ndef get_roots(a, b, c):\n # YOUR CODE HERE\n raise NotImplementedError()\n return root1, root2\n\n\n# In[ ]:\n\n\n# tests\ndef test_get_roots(roots, a, b, c):\n roots_ = get_roots(a, b, c)\n correct = all([math.isclose(roots[i], roots_[i]) for i in range(2)])\n if not correct:\n print(f'With (a, b, c)={a,b,c}, roots should be {roots} not {roots_}.')\n assert correct\n\n\ntest_get_roots((-1.0, 0.0), 1, 1, 0)\ntest_get_roots((-1.0, -1.0), 1, 2, 1)\ntest_get_roots((-2.0, -1.0), 1, 3, 2)\n\n\n# In[ ]:\n\n\n# quadratic equations solver\n@interact(a=(-10,10,1),b=(-10,10,1),c=(-10,10,1))\ndef quadratic_equation_solver(a=1,b=2,c=1):\n print('Roots: {}, {}'.format(*get_roots(a,b,c)))\n\n\n# ## Number Conversion\n\n# ### Byte-to-Decimal Calculator\n\n# \n\n# Denote a binary number stored as a byte ($8$-bit) as\n# \n# $$ \n# b_7\\circ b_6\\circ b_5\\circ b_4\\circ b_3\\circ b_2\\circ b_1\\circ b_0, \n# $$\n# where $\\circ$ concatenates $b_i$'s together into a binary string.\n\n# The binary string can be converted to a decimal number by the formula\n# \n# $$ \n# b_7\\cdot 2^7 + b_6\\cdot 2^6 + b_5\\cdot 2^5 + b_4\\cdot 2^4 + b_3\\cdot 2^3 + b_2\\cdot 2^2 + b_1\\cdot 2^1 + b_0\\cdot 2^0. \n# $$\n\n# E.g., the binary string `'11111111'` is the largest integer represented by a byte:\n# \n# $$\n# 2^7+2^6+2^5+2^4+2^3+2^2+2^1+2^0=255=2^8-1.\n# $$\n\n# **Exercise** Assign to `decimal` the *integer* value represented by a byte. \n# The byte is a sequence of bits assigned to the variables `b7,b6,b5,b4,b3,b2,b1,b0` as *characters*, either `'0'` or `'1'`.\n\n# In[ ]:\n\n\ndef byte_to_decimal(b7, b6, b5, b4, b3, b2, b1, b0):\n # YOUR CODE HERE\n raise NotImplementedError()\n return decimal\n\n\n# In[ ]:\n\n\n# tests\ndef test_byte_to_decimal(decimal, b7, b6, b5, b4, b3, b2, b1, b0):\n decimal_ = byte_to_decimal(b7, b6, b5, b4, b3, b2, b1, b0)\n correct = decimal == decimal_ and isinstance(decimal_, int)\n if not correct:\n print(\n f'{b7}{b6}{b5}{b4}{b3}{b2}{b1}{b0} should give {decimal} not {decimal_}.'\n )\n assert correct\n\n\ntest_byte_to_decimal(38, '0', '0', '1', '0', '0', '1', '1', '0')\ntest_byte_to_decimal(20, '0', '0', '0', '1', '0', '1', '0', '0')\ntest_byte_to_decimal(22, '0', '0', '0', '1', '0', '1', '1', '0')\n\n\n# In[ ]:\n\n\n# byte-to-decimal calculator\nbit = ['0', '1']\n\n\n@interact(b7=bit, b6=bit, b5=bit, b4=bit, b3=bit, b2=bit, b1=bit, b0=bit)\ndef convert_byte_to_decimal(b7, b6, b5, b4, b3, b2, b1, b0):\n print('decimal:', byte_to_decimal(b7, b6, b5, b4, b3, b2, b1, b0))\n\n\n# ### Decimal-to-Byte Calculator\n\n# \n\n# **Exercise** Assign to `byte` a *string of 8 bits* that represents the value of `decimal`, a non-negative decimal integer from $0$ to $2^8-1=255$. \n# *Hint: Use `//` and `%`.*\n\n# In[ ]:\n\n\ndef decimal_to_byte(decimal):\n # YOUR CODE HERE\n raise NotImplementedError()\n return byte\n\n\n# In[ ]:\n\n\n# tests\ndef test_decimal_to_byte(byte,decimal):\n byte_ = decimal_to_byte(decimal)\n correct = byte == byte_ and isinstance(byte, str) and len(byte) == 8\n if not correct:\n print(\n f'{decimal} should be represented as the byte {byte}, not {byte_}.'\n )\n assert correct\n\n\ntest_decimal_to_byte('01100111', 103)\ntest_decimal_to_byte('00000011', 3)\ntest_decimal_to_byte('00011100', 28)\n\n\n# In[ ]:\n\n\n# decimal-to-byte calculator\n@interact(decimal=(0,255,1))\ndef convert_decimal_to_byte(decimal=0):\n print('byte:', decimal_to_byte(decimal))\n\n"
] |
[
[
"matplotlib.pyplot.gca",
"numpy.linspace"
]
] |
subond/tools
|
[
"05b93e6c78eab65ef6587e684303b12c686a3480"
] |
[
"ROMS/pmacc/tools/post_tools/rompy/tags/rompy-0.1.5/rompy/extract_from_series.py"
] |
[
"import os\nimport datetime as dt\nimport time\nimport glob\n\nimport pytz\nimport netCDF4 as nc\nimport numpy as np\n\nimport extract_utils\nimport extract_from_file\n\n__version__ = '0.1.5'\n\n# def file_time(f):\n# \tUTC = pytz.timezone('UTC')\n# \tncf = nc.Dataset(f,mode='r')\n# \tot = ncf.variables['ocean_time']\n# \tbase_time = dt.datetime.strptime(ot.units,'seconds since %Y-%m-%d %H:%M:%S').replace(tzinfo=UTC)\n# \toffset = dt.timedelta(seconds=ot[0][0])\n# \tncf.close()\n# \treturn base_time + offset\n# \n# def file_timestamp(f):\n# \tt = file_time(f)\n# \treturn time.mktime(t.timetuple())\n# \n# def calc_num_time_slices(file1,file2,td):\n# \td1 = file_time(file1)\n# \td2 = file_time(file2)\n# \t\n# \tgap = d2-d1\n# \tgap_seconds = gap.days*60*60*24 + gap.seconds\n# \ttd_seconds = td.days*60*60*24 + td.seconds\n# \t\n# \tt1 = time.mktime(d1.timetuple())\n# \tt2 = time.mktime(d2.timetuple())\n# \ttgap = t2 - t1\n# \t\n# \tprint(gap_seconds, tgap)\n# \t\n# \treturn int(gap_seconds/td_seconds + 1)\n# \n# def filelist_from_datelist(datelist, basedir='/Users/lederer/Repositories/PSVS/rompy/', basename='ocean_his_*.nc'):\n# \tfiles = glob.glob(os.path.join(basedir,basename))\n# \tfiles.sort()\n# \t\n# \tmaster_file_list = []\n# \tmaster_timestamp_list = []\n# \tmaster_datetime_list = []\n# \ttimelist = []\n# \treturnlist = []\n# \t\n# \tfor file in files:\n# \t\tmaster_file_list.append(file)\n# \t\tmaster_timestamp_list.append(file_timestamp(file))\n# \t\tmaster_datetime_list.append(file_time(file))\n# \t\n# \ttsarray = np.array(master_timestamp_list)\n# \n# \tfor d in datelist:\n# \t\ttry:\n# \t\t\tt = time.mktime(d.timetuple())\n# \t\t\ttimelist.append(t)\n# \t\t\ttl = np.nonzero(tsarray <= t)[0][-1]\n# \t\t\tth = np.nonzero(t <= tsarray)[0][0]\n# \t\t\tif not tl == th:\n# \t\t\t\tfraction = (t-tsarray[th])/(tsarray[tl]-tsarray[th])\n# \t\t\telse:\n# \t\t\t\tfraction = 0.0\n# \t\t\t\n# \t\t\treturnlist.append({\n# \t\t\t\t\t\t\t'file0': master_file_list[tl],\n# \t\t\t\t\t\t\t'file1': master_file_list[th],\n# \t\t\t\t\t\t\t'fraction': fraction,\n# \t\t\t\t\t\t\t'timestamp':t,\n# \t\t\t\t\t\t\t'datetime':d\n# \t\t\t\t\t\t\t})\n# \t\texcept IndexError, e:\n# \t\t\tprint('Index out of bounds for %s' % (d.isoformat()))\n# \t\t\t\n# \treturn returnlist\n\ndef extract_from_series(file_list,extraction_type='point',varname='zeta',**kwargs):\n\tUTC = pytz.timezone('UTC')\n\tpacific = pytz.timezone('US/Pacific')\n\t\n\t#print('Hello from extractFromSeries')\n\t\n\tif extraction_type == 'point':\n\t\t# assume var is a 2d var for now\n\t\tfile_list.sort()\n\t\t\n\t\tx = kwargs['x']\n\t\ty = kwargs['y']\n\t\t\n\t\tdata = np.zeros((len(file_list),len(x)))\n\t\tfor i in range(len(x)):\t\n\t\t\tdata[0,i],junk = extract_from_file.extract_from_file(file_list[0],varname=varname,extraction_type='point',x=x[i],y=y[i])\n\t\t\n\t\tf1 = file_list[0]\n\t\tf2 = file_list[1]\n\t\ttime_list = [extract_utils.file_time(file_list[0])]\n\t\t\n\t\tfor i in range(1,len(file_list)):\n\t\t\tfor j in range(len(x)):\n\t\t\t\tdata[i,j],junk = extract_from_file.extract_from_file(file_list[i],varname=varname,extraction_type='point',x=x[j],y=y[j])\n\t\t\ttime_list.append(extract_utils.file_time(file_list[i]))\n\t\treturn (data,time_list)\n\t\t\n#\t\tfor i in range(1,ntimes):\n#\t\t\ttime_list.append(time_list[-1]+freq)\n\t\t\n\t\n\tif extraction_type == 'profile':\n\t\t\n\t\tfile_list.sort()\n\t\t\n\t\tocean_time = None\n\t\t\n\t\tdata = None\n\t\tz = None\n\t\t\n\t\tx = kwargs['x']\n\t\ty = kwargs['y']\n\t\t\n\t\tif len(x) > 1:\n\t\t\tx = np.array(x[0])\n\t\telse:\n\t\t\tx = np.array(x)\n\t\tif len(y) > 1:\n\t\t\ty = np.array(y[0])\n\t\telse:\n\t\t\ty = np.array(y)\n\n\t\tfor i in range(len(file_list)):\n\t\t\tfile = file_list[i]\n\t\t\t\n\t\t\tif not os.path.exists(file):\n\t\t\t\traise IOError('File %s could not be located on the filesystem' %file)\n\t\t\tncf = nc.Dataset(file,mode='r')\n\t\t\t\n\t\t\tif varname not in ncf.variables:\n\t\t\t\traise IOError('File %s does not have a variable named %s' % (file, varname))\n\t\t\tncf.close()\n\t\t\t\n\t\t\td,junk = extract_from_file.extract_from_file(file_list[i],varname=varname,extraction_type='profile',x=x,y=y)\n\t\t\t\n\t\t\tif data == None:\n\t\t\t\tdata = np.zeros((d.shape[0],len(file_list)))\n\t\t\tdata[:,i] = d.T\n\t\t\t\n\t\t\tif z == None:\n\t\t\t\tz = np.zeros(data.shape)\n\t\t\tz[:,i] = junk['zm'].T\n\t\t\t\n#\t\t\t\tocean_time = np.zeros(data.shape)\n\t\t\tif ocean_time == None:\n\t\t\t\tocean_time = np.zeros(data.shape)\n\t\t\tot = extract_utils.file_timestamp(file_list[i])\n#\t\t\tot = file_time(file)\n\t\t\tfor j in range(data.shape[0]):\n\t\t\t\tocean_time[j,i] = ot\n\t\treturn (data, ocean_time,z)\n\treturn\n\ndef extract_from_datetime_list(datelist,x,y,varname='salt',basedir='./',**kwargs):\n\tfilelist = extract_utils.filelist_from_datelist(datelist, basedir=basedir)\n\tfor f in filelist:\n\t\t(d0,ot0,z0) = extract_from_series([f['file0']],x=x,y=y,varname=varname,extraction_type='profile',**kwargs)\n\t\t(d1,ot1,z1) = extract_from_series([f['file1']],x=x,y=y,varname=varname,extraction_type='profile',**kwargs)\n\t\t\n\t\tdi = np.zeros(d0.shape)\n\t\toti = np.zeros(d0.shape)\n\t\tzi = np.zeros(d0.shape)\n\n\t\tf['d'] = d0 + (d1-d0)*f['fraction']\n\t\tf['ot'] = ot0 + (ot1-ot0)*f['fraction']\n\t\tf['z'] = z0 + (z1-z0)*f['fraction']\n\t\n\td = np.zeros((filelist[0]['d'].shape[0], len(filelist)*filelist[0]['d'].shape[1]))\n\tot = np.zeros((filelist[0]['d'].shape[0], len(filelist)*filelist[0]['d'].shape[1]))\n\tz = np.zeros((filelist[0]['d'].shape[0], len(filelist)*filelist[0]['d'].shape[1]))\n\n\tfor i in range(len(filelist)):\n\t\td[:,i] = filelist[i]['d'].T\n\t\tot[:,i] = filelist[i]['ot'].T\n\t\tz[:,i] = filelist[i]['z'].T\n\treturn (d,ot,z)\n\ndef extract_from_two_datetimes(x,y,dt0,dt1,varname='salt',interval=3600,basedir='./',**kwargs):\n\tt0 = time.mktime(dt0.timetuple())\n\tt1 = time.mktime(dt1.timetuple())\n\tinterval = float(interval)\n\n\ttime_stamps = interval*np.arange(t0/interval,(t1/interval)+1)\n\n\tdate_times = []\n\tfor ts in time_stamps:\n\t\tdate_times.append(dt.datetime.fromtimestamp(ts))\n#\tfor d in date_times:\n#\t\tprint(d.isoformat())\n\tprint(dt0.isoformat(),date_times[0].isoformat())\n\tprint(dt1.isoformat(),date_times[-1].isoformat())\n\treturn extract_from_datetime_list(date_times,x,y,varname=varname,basedir=basedir,**kwargs)\n\t"
] |
[
[
"numpy.array",
"numpy.arange",
"numpy.zeros"
]
] |
ver0z/Deformable-DETR-
|
[
"f37ef3e73675ee4be1d8a0649901b1402f907f1a"
] |
[
"models/position_encoding.py"
] |
[
"# ------------------------------------------------------------------------\r\n# Deformable DETR\r\n# Copyright (c) 2020 SenseTime. All Rights Reserved.\r\n# Licensed under the Apache License, Version 2.0 [see LICENSE for details]\r\n# ------------------------------------------------------------------------\r\n# Modified from DETR (https://github.com/facebookresearch/detr)\r\n# Copyright (c) Facebook, Inc. and its affiliates. All Rights Reserved\r\n# ------------------------------------------------------------------------\r\n\r\n\"\"\"\r\nVarious positional encodings for the transformer.\r\n\"\"\"\r\nimport math\r\nimport torch\r\nfrom torch import nn\r\n\r\nfrom util.misc import NestedTensor\r\n\r\n\r\nclass PositionEmbeddingSine(nn.Module):\r\n \"\"\"\r\n This is a more standard version of the position embedding, very similar to the one\r\n used by the Attention is all you need paper, generalized to work on images.\r\n \"\"\"\r\n def __init__(self, num_pos_feats=64, temperature=10000, normalize=False, scale=None):\r\n super().__init__()\r\n self.num_pos_feats = num_pos_feats\r\n self.temperature = temperature\r\n self.normalize = normalize\r\n if scale is not None and normalize is False:\r\n raise ValueError(\"normalize should be True if scale is passed\")\r\n if scale is None:\r\n scale = 2 * math.pi\r\n self.scale = scale\r\n\r\n def forward(self, tensor_list: NestedTensor):\r\n x = tensor_list.tensors\r\n mask = tensor_list.mask\r\n assert mask is not None\r\n not_mask = ~mask\r\n y_embed = not_mask.cumsum(1, dtype=torch.float32)\r\n x_embed = not_mask.cumsum(2, dtype=torch.float32)\r\n if self.normalize:\r\n eps = 1e-6\r\n y_embed = (y_embed - 0.5) / (y_embed[:, -1:, :] + eps) * self.scale\r\n x_embed = (x_embed - 0.5) / (x_embed[:, :, -1:] + eps) * self.scale\r\n\r\n dim_t = torch.arange(self.num_pos_feats, dtype=torch.float32, device=x.device)\r\n dim_t = self.temperature ** (2 * (dim_t // 2) / self.num_pos_feats)\r\n\r\n pos_x = x_embed[:, :, :, None] / dim_t\r\n pos_y = y_embed[:, :, :, None] / dim_t\r\n pos_x = torch.stack((pos_x[:, :, :, 0::2].sin(), pos_x[:, :, :, 1::2].cos()), dim=4).flatten(3)\r\n pos_y = torch.stack((pos_y[:, :, :, 0::2].sin(), pos_y[:, :, :, 1::2].cos()), dim=4).flatten(3)\r\n pos = torch.cat((pos_y, pos_x), dim=3).permute(0, 3, 1, 2)\r\n return pos\r\n\r\n\r\nclass PositionEmbeddingLearned(nn.Module):\r\n \"\"\"\r\n Absolute pos embedding, learned.\r\n \"\"\"\r\n def __init__(self, num_pos_feats=256):\r\n super().__init__()\r\n self.row_embed = nn.Embedding(50, num_pos_feats)\r\n self.col_embed = nn.Embedding(50, num_pos_feats)\r\n self.reset_parameters()\r\n\r\n def reset_parameters(self):\r\n nn.init.uniform_(self.row_embed.weight)\r\n nn.init.uniform_(self.col_embed.weight)\r\n\r\n def forward(self, tensor_list: NestedTensor):\r\n x = tensor_list.tensors\r\n h, w = x.shape[-2:]\r\n i = torch.arange(w, device=x.device)\r\n j = torch.arange(h, device=x.device)\r\n x_emb = self.col_embed(i)\r\n y_emb = self.row_embed(j)\r\n pos = torch.cat([\r\n x_emb.unsqueeze(0).repeat(h, 1, 1),\r\n y_emb.unsqueeze(1).repeat(1, w, 1),\r\n ], dim=-1).permute(2, 0, 1).unsqueeze(0).repeat(x.shape[0], 1, 1, 1)\r\n return pos\r\n\r\n\r\ndef build_position_encoding(args):\r\n N_steps = args.hidden_dim // 2\r\n if args.position_embedding in ('v2', 'sine'):\r\n # TODO find a better way of exposing other arguments\r\n position_embedding = PositionEmbeddingSine(N_steps, normalize=True)\r\n elif args.position_embedding in ('v3', 'learned'):\r\n position_embedding = PositionEmbeddingLearned(N_steps)\r\n else:\r\n raise ValueError(f\"not supported {args.position_embedding}\")\r\n\r\n return position_embedding\r\n"
] |
[
[
"torch.nn.init.uniform_",
"torch.cat",
"torch.nn.Embedding",
"torch.arange"
]
] |
bucknerns/ms-mint
|
[
"140f607a4b6aab84b5db2b8c9876dc6c1d2b7943"
] |
[
"tests/test__MINT-integration.py"
] |
[
"import os\nimport pandas as pd\nfrom sklearn.metrics import r2_score\n\nfrom ms_mint.Mint import Mint\nfrom ms_mint.tools import check_peaklist, MINT_RESULTS_COLUMNS\n\nmint = Mint(verbose=True)\nmint_b = Mint(verbose=True)\n\nclass TestClass():\n \n def run_without_files(self):\n mint.run()\n \n def test__add_experimental_data(self):\n assert mint.n_files == 0, mint.n_files\n mint.files = ['tests/data/test.mzXML']\n assert mint.n_files == 1, mint.n_files\n assert mint.files == ['tests/data/test.mzXML']\n\n def test__add_peaklist(self):\n assert mint.n_peaklist_files == 0, mint.n_peaklist_files\n mint.peaklist_files = 'tests/data/test_peaklist.csv'\n assert mint.peaklist_files == ['tests/data/test_peaklist.csv']\n assert mint.n_peaklist_files == 1, mint.n_peaklist_files\n \n def test__mint_run_standard(self):\n mint.run()\n \n def test__correct_peakAreas(self):\n df_test = pd.read_csv('tests/data/test_peaklist.csv', dtype={'peak_label': str})\n print(mint.results.dtypes)\n df = pd.merge(df_test, mint.results, on='peak_label', suffixes=('_real', '_calc'))\n R2 = r2_score(df.peak_area_real, df.peak_area_calc)\n assert R2 > 0.999, R2\n \n def test__results_is_dataframe(self):\n results = mint.results\n assert isinstance(results, pd.DataFrame), 'Results is not a DataFrame'\n \n def test__results_lenght(self):\n actual = len(mint.results)\n expected = len(mint.peaklist) * len(mint.files)\n assert expected == actual, f'Length of results ({actual}) does not equal expected length ({expected})'\n \n def test__results_columns(self):\n expected = MINT_RESULTS_COLUMNS\n actual = mint.results.columns\n assert (expected == actual).all(), actual\n \n def test__crosstab_is_dataframe(self):\n ct = mint.crosstab()\n assert isinstance(ct, pd.DataFrame), f'Crosstab is not a DataFrame ({type(ct)}).'\n\n def test__mint_run_parallel(self):\n mint.files = ['tests/data/test.mzXML']*2\n peaklist = pd.DataFrame({\n 'mz_mean': [100], \n 'mz_width': [10],\n 'rt_min': [0.1], \n 'rt_max': [0.2],\n 'intensity_threshold': [0],\n 'peak_label': ['test'],\n 'peaklist': ['no-file']})\n print(peaklist)\n check_peaklist(peaklist)\n mint.peaklist = peaklist\n mint.run(nthreads=2) \n \n def test__mzxml_equals_mzml(self):\n mint.reset()\n mint.files = ['tests/data/test.mzXML' , 'tests/data/test.mzML']\n mint.peaklist_files = 'tests/data/peaklist_v1.csv'\n mint.run()\n results = []\n for _, grp in mint.results.groupby('ms_file'):\n results.append(grp.peak_area)\n assert (results[0] == results[1]).all(), results\n\n def test__peaklist_v0_equals_v1(self):\n mint.reset()\n mint.files = ['tests/data/test.mzXML']\n mint.peaklist_files = ['tests/data/peaklist_v0.csv', 'tests/data/peaklist_v1.csv']\n mint.run()\n results = []\n for _, grp in mint.results.groupby('peaklist'):\n results.append(grp.peak_area.values)\n assert (results[0] == results[1]).all(), results\n \n def test__status(self):\n mint.status == 'waiting'\n \n def test__run_returns_none_without_peaklist(self):\n mint.reset()\n mint.files = ['tests/data/test.mzXML']\n assert mint.run() is None\n \n def test__run_returns_none_without_ms_files(self):\n mint.reset()\n mint.peaklist_files = 'tests/data/peaklist_v0.csv'\n assert mint.run() is None\n"
] |
[
[
"sklearn.metrics.r2_score",
"pandas.DataFrame",
"pandas.read_csv",
"pandas.merge"
]
] |
chenlongzhen/FATE-0.1
|
[
"38fe6cea0dca3841b59c3d04cb04f556803e2e29"
] |
[
"federatedml/ftl/hetero_ftl/hetero_ftl_guest.py"
] |
[
"#\n# Copyright 2019 The FATE Authors. All Rights Reserved.\n#\n# Licensed under the Apache License, Version 2.0 (the \"License\");\n# you may not use this file except in compliance with the License.\n# You may obtain a copy of the License at\n#\n# http://www.apache.org/licenses/LICENSE-2.0\n#\n# Unless required by applicable law or agreed to in writing, software\n# distributed under the License is distributed on an \"AS IS\" BASIS,\n# WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.\n# See the License for the specific language governing permissions and\n# limitations under the License.\n#\n\nimport numpy as np\nimport time\nfrom federatedml.ftl.encrypted_ftl import EncryptedFTLGuestModel\nfrom federatedml.ftl.plain_ftl import PlainFTLGuestModel\nfrom federatedml.ftl.common.data_util import overlapping_samples_converter, load_model_parameters, \\\n save_model_parameters, convert_instance_table_to_dict, convert_instance_table_to_array\nfrom federatedml.ftl.hetero_ftl.hetero_ftl_base import HeteroFTLParty\nfrom federatedml.util import consts\nfrom federatedml.util.transfer_variable import HeteroFTLTransferVariable\nfrom federatedml.optim.convergence import DiffConverge\nfrom federatedml.param.param import FTLModelParam\nfrom arch.api.utils import log_utils\nLOGGER = log_utils.getLogger()\n\n\nclass HeteroFTLGuest(HeteroFTLParty):\n\n def __init__(self, guest: PlainFTLGuestModel, model_param: FTLModelParam, transfer_variable: HeteroFTLTransferVariable):\n super(HeteroFTLGuest, self).__init__()\n self.guest_model = guest\n self.model_param = model_param\n self.transfer_variable = transfer_variable\n self.max_iter = model_param.max_iter\n self.n_iter_ = 0\n self.converge_func = DiffConverge(eps=model_param.eps)\n\n def set_converge_function(self, converge_func):\n self.converge_func = converge_func\n\n def prepare_data(self, guest_data):\n LOGGER.info(\"@ start guest prepare_data\")\n guest_features_dict, guest_label_dict, guest_sample_indexes = convert_instance_table_to_dict(guest_data)\n guest_sample_indexes = np.array(guest_sample_indexes)\n LOGGER.debug(\"@ send guest_sample_indexes shape\" + str(guest_sample_indexes.shape))\n self._do_remote(guest_sample_indexes,\n name=self.transfer_variable.guest_sample_indexes.name,\n tag=self.transfer_variable.generate_transferid(self.transfer_variable.guest_sample_indexes),\n role=consts.HOST,\n idx=-1)\n host_sample_indexes = self._do_get(name=self.transfer_variable.host_sample_indexes.name,\n tag=self.transfer_variable.generate_transferid(self.transfer_variable.host_sample_indexes),\n idx=-1)[0]\n\n LOGGER.debug(\"@ receive host_sample_indexes len\" + str(len(host_sample_indexes)))\n guest_features, overlap_indexes, non_overlap_indexes, guest_label = overlapping_samples_converter(\n guest_features_dict, guest_sample_indexes, host_sample_indexes, guest_label_dict)\n return guest_features, overlap_indexes, non_overlap_indexes, guest_label\n\n def predict(self, guest_data):\n LOGGER.info(\"@ start guest predict\")\n features, labels, instances_indexes = convert_instance_table_to_array(guest_data)\n guest_x = np.squeeze(features)\n guest_y = np.expand_dims(labels, axis=1)\n\n LOGGER.debug(\"get guest_x, guest_y: \" + str(guest_x.shape) + \", \" + str(guest_y.shape))\n host_prob = self._do_get(name=self.transfer_variable.host_prob.name,\n tag=self.transfer_variable.generate_transferid(self.transfer_variable.host_prob),\n idx=-1)[0]\n\n self.guest_model.set_batch(guest_x, guest_y)\n pred_prob = self.guest_model.predict(host_prob)\n LOGGER.debug(\"pred_prob: \" + str(pred_prob.shape))\n self._do_remote(pred_prob,\n name=self.transfer_variable.pred_prob.name,\n tag=self.transfer_variable.generate_transferid(self.transfer_variable.pred_prob),\n role=consts.HOST,\n idx=-1)\n return None\n\n def load_model(self, model_table_name, model_namespace):\n LOGGER.info(\"@ load guest model from name/ns\" + \", \" + str(model_table_name) + \", \" + str(model_namespace))\n model_parameters = load_model_parameters(model_table_name, model_namespace)\n self.guest_model.restore_model(model_parameters)\n\n def save_model(self, model_table_name, model_namespace):\n LOGGER.info(\"@ save guest model to name/ns\" + \", \" + str(model_table_name) + \", \" + str(model_namespace))\n _ = save_model_parameters(self.guest_model.get_model_parameters(), model_table_name, model_namespace)\n\n\nclass HeteroPlainFTLGuest(HeteroFTLGuest):\n\n def __init__(self, guest: PlainFTLGuestModel, model_param: FTLModelParam, transfer_variable: HeteroFTLTransferVariable):\n super(HeteroPlainFTLGuest, self).__init__(guest, model_param, transfer_variable)\n\n def fit(self, guest_data):\n LOGGER.info(\"@ start guest fit\")\n\n guest_x, overlap_indexes, non_overlap_indexes, guest_y = self.prepare_data(guest_data)\n\n LOGGER.debug(\"guest_x: \" + str(guest_x.shape))\n LOGGER.debug(\"guest_y: \" + str(guest_y.shape))\n LOGGER.debug(\"overlap_indexes: \" + str(len(overlap_indexes)))\n LOGGER.debug(\"non_overlap_indexes: \" + str(len(non_overlap_indexes)))\n\n self.guest_model.set_batch(guest_x, guest_y, non_overlap_indexes, overlap_indexes)\n\n is_stop = False\n while self.n_iter_ < self.max_iter:\n guest_comp = self.guest_model.send_components()\n LOGGER.debug(\"send guest_comp: \" + str(guest_comp[0].shape) + \", \" + str(guest_comp[1].shape) + \", \" + str(\n guest_comp[2].shape))\n self._do_remote(guest_comp, name=self.transfer_variable.guest_component_list.name,\n tag=self.transfer_variable.generate_transferid(self.transfer_variable.guest_component_list, self.n_iter_),\n role=consts.HOST,\n idx=-1)\n\n host_comp = self._do_get(name=self.transfer_variable.host_component_list.name,\n tag=self.transfer_variable.generate_transferid(self.transfer_variable.host_component_list, self.n_iter_),\n idx=-1)[0]\n LOGGER.debug(\"receive host_comp: \" + str(host_comp[0].shape) + \", \" + str(host_comp[1].shape) + \", \" + str(host_comp[2].shape))\n self.guest_model.receive_components(host_comp)\n\n loss = self.guest_model.send_loss()\n if self.converge_func.is_converge(loss):\n is_stop = True\n\n self._do_remote(is_stop, name=self.transfer_variable.is_stopped.name,\n tag=self.transfer_variable.generate_transferid(self.transfer_variable.is_stopped, self.n_iter_),\n role=consts.HOST,\n idx=-1)\n LOGGER.info(\"@ time: \" + str(time.time()) + \", ep:\" + str(self.n_iter_) + \", loss:\" + str(loss))\n LOGGER.info(\"@ converged: \" + str(is_stop))\n self.n_iter_ += 1\n if is_stop:\n break\n\n\nclass HeteroEncryptFTLGuest(HeteroFTLGuest):\n\n def __init__(self, guest: EncryptedFTLGuestModel, model_param: FTLModelParam, transfer_variable: HeteroFTLTransferVariable):\n super(HeteroEncryptFTLGuest, self).__init__(guest, model_param, transfer_variable)\n self.guest_model = guest\n\n def fit(self, guest_data):\n LOGGER.info(\"@ start guest fit\")\n public_key = self._do_get(name=self.transfer_variable.paillier_pubkey.name,\n tag=self.transfer_variable.generate_transferid(self.transfer_variable.paillier_pubkey),\n idx=-1)[0]\n\n guest_x, overlap_indexes, non_overlap_indexes, guest_y = self.prepare_data(guest_data)\n\n LOGGER.debug(\"guest_x: \" + str(guest_x.shape))\n LOGGER.debug(\"guest_y: \" + str(guest_y.shape))\n LOGGER.debug(\"overlap_indexes: \" + str(len(overlap_indexes)))\n LOGGER.debug(\"non_overlap_indexes: \" + str(len(non_overlap_indexes)))\n\n self.guest_model.set_batch(guest_x, guest_y, non_overlap_indexes, overlap_indexes)\n self.guest_model.set_public_key(public_key)\n\n while self.n_iter_ < self.max_iter:\n guest_comp = self.guest_model.send_components()\n\n LOGGER.debug(\"send guest_comp: \" + str(guest_comp[0].shape) + \", \" + str(guest_comp[1].shape) + \", \" + str(\n guest_comp[2].shape))\n self._do_remote(guest_comp, name=self.transfer_variable.guest_component_list.name,\n tag=self.transfer_variable.generate_transferid(self.transfer_variable.guest_component_list, self.n_iter_),\n role=consts.HOST,\n idx=-1)\n\n host_comp = self._do_get(name=self.transfer_variable.host_component_list.name,\n tag=self.transfer_variable.generate_transferid(self.transfer_variable.host_component_list, self.n_iter_),\n idx=-1)[0]\n\n LOGGER.debug(\"receive host_comp: \" + str(host_comp[0].shape) + \", \" + str(host_comp[1].shape) + \", \" + str(host_comp[2].shape))\n self.guest_model.receive_components(host_comp)\n\n encrypt_guest_gradients = self.guest_model.send_gradients()\n self._do_remote(encrypt_guest_gradients, name=self.transfer_variable.encrypt_guest_gradient.name,\n tag=self.transfer_variable.generate_transferid(self.transfer_variable.encrypt_guest_gradient, self.n_iter_),\n role=consts.ARBITER,\n idx=-1)\n\n decrypt_guest_gradient = self._do_get(name=self.transfer_variable.decrypt_guest_gradient.name,\n tag=self.transfer_variable.generate_transferid(self.transfer_variable.decrypt_guest_gradient, self.n_iter_),\n idx=-1)[0]\n self.guest_model.receive_gradients(decrypt_guest_gradient)\n\n encrypt_loss = self.guest_model.send_loss()\n self._do_remote(encrypt_loss, name=self.transfer_variable.encrypt_loss.name,\n tag=self.transfer_variable.generate_transferid(self.transfer_variable.encrypt_loss, self.n_iter_),\n role=consts.ARBITER,\n idx=-1)\n\n is_stop = self._do_get(name=self.transfer_variable.is_encrypted_ftl_stopped.name,\n tag=self.transfer_variable.generate_transferid(self.transfer_variable.is_encrypted_ftl_stopped, self.n_iter_),\n idx=-1)[0]\n\n LOGGER.info(\"@ time: \" + str(time.time()) + \", ep: \" + str(self.n_iter_) + \", converged:\" + str(is_stop))\n self.n_iter_ += 1\n if is_stop:\n break\n\n\n"
] |
[
[
"numpy.array",
"numpy.squeeze",
"numpy.expand_dims"
]
] |
burn-research/PyTROMode
|
[
"c3ed440930f80ac94fb60369b7d01f04972a0346"
] |
[
"OpenMORe/utilities.py"
] |
[
"'''\nMODULE: utilities.py\n@Author:\n G. D'Alessio [1,2]\n [1]: Université Libre de Bruxelles, Aero-Thermo-Mechanics Laboratory, Bruxelles, Belgium\n [2]: CRECK Modeling Lab, Department of Chemistry, Materials and Chemical Engineering, Politecnico di Milano\n@Contacts:\n [email protected]\n@Details:\n This module contains a set of functions which are useful for reduced-order modelling with PCA.\n A detailed description is available under the definition of each function.\n@Additional notes:\n This code is distributed in the hope that it will be useful, but WITHOUT ANY WARRANTY;\n Please report any bug to: [email protected]\n'''\n\n\nimport numpy as np\nfrom numpy import linalg as LA\n\n\nimport matplotlib\nimport matplotlib.pyplot as plt\n__all__ = [\"unscale\", \"uncenter\", \"center\", \"scale\", \"center_scale\", \"evaluate_clustering_PHC\", \"fastSVD\", \"get_centroids\", \"get_cluster\", \"get_all_clusters\", \"explained_variance\", \"evaluate_clustering_DB\", \"NRMSE\", \"PCA_fit\", \"readCSV\", \"varimax_rotation\", \"get_medianoids\", \"split_for_validation\", \"get_medoids\"]\n\n\n# ------------------------------\n# Functions (alphabetical order)\n# ------------------------------\n\n\ndef center(X, method, return_centered_matrix= False):\n '''\n Computes the centering factor (the mean/min value [mu]) of each variable of all data-set observations and\n (eventually) return the centered matrix.\n - Input:\n X = original data matrix -- dim: (observations x variables)\n method = \"string\", it is the method which has to be used. Two choices are available: MEAN or MIN\n return_centered_matrix = boolean, choose if the script must return the centered matrix (optional)\n - Output:\n mu = centering factor for the data matrix X\n X0 = centered data matrix (optional)\n '''\n # Main\n if not return_centered_matrix:\n if method.lower() == 'mean':\n #compute the vector containing the mean for each variable\n mu = np.mean(X, axis = 0)\n elif method.lower() == 'min':\n #compute the vector containing the minimum for each variable\n mu = np.min(X, axis = 0)\n else:\n raise Exception(\"Unsupported centering option. Please choose: MEAN or MIN.\")\n return mu\n else:\n if method.lower() == 'mean':\n #compute the vector containing the mean for each variable\n mu = np.mean(X, axis = 0)\n #subtract the mean to each observation of the matrix X\n X0 = X - mu\n elif method.lower() == 'min':\n #compute the vector containing the minimum for each variable\n mu = np.min(X, axis = 0)\n #subtract the minimum to each observation of the matrix X\n X0 = X - mu\n else:\n raise Exception(\"Unsupported centering option. Please choose: MEAN or MIN.\")\n return mu, X0\n\n\ndef center_scale(X, mu, sig):\n '''\n Center and scale a given multivariate data-set X.\n Centering consists of subtracting the mean/min value of each variable to all data-set\n observations. Scaling is achieved by dividing each variable by a given scaling factor. Therefore, the\n i-th observation of the j-th variable, x_{i,j} can be\n centered and scaled by means of:\n \\tilde{x_{i,j}} = (x_{i,j} - mu_{j}) / (sig_{j}),\n where mu_{j} and sig_{j} are the centering and scaling factor for the considered j-th variable, respectively.\n AUTO: the standard deviation of each variable is used as a scaling factor.\n PARETO: the squared root of the standard deviation is used as a scaling f.\n RANGE: the difference between the minimum and the maximum value is adopted as a scaling f.\n VAST: the ratio between the variance and the mean of each variable is used as a scaling f.\n '''\n TOL = 1E-16\n if X.shape[1] == mu.shape[0] and X.shape[1] == sig.shape[0]:\n #centering step: subtract the centering factor to each observation\n X0 = X - mu\n\n #scaling step: divide each observation for the scaling factor. TOL is added to avoid dividing by 0 \n X0 = X0 / (sig + TOL)\n return X0\n else:\n raise Exception(\"The matrix to be centered & scaled and the centering/scaling vectors must have the same dimensionality.\")\n\n\ndef explained_variance(X, n_eigs, plot=False):\n '''\n Assess the variance explained by the first 'n_eigs' retained\n Principal Components. This is important to know if the percentage\n of explained variance is enough, or additional PCs must be retained.\n Usually, it is considered acceptable a percentage of explained variable\n above 95%.\n - Input:\n X = CENTERED/SCALED data matrix -- dim: (observations x variables)\n n_eigs = number of components to retain -- dim: (scalar)\n plot = choose if you want to plot the cumulative variance --dim: (boolean), false is default\n - Output:\n explained: percentage of explained variance -- dim: (scalar)\n '''\n #compute PCA\n PCs, eigens = PCA_fit(X, n_eigs)\n \n #the explained variance is defined as the sum of the 'q' eigenvalues to retain\n #divided by the total sum of the eigenvalues\n explained_variance = np.cumsum(eigens)/sum(eigens)\n explained = explained_variance[n_eigs]\n\n #plot the curve of the cumulative variance, if the option is activated\n if plot:\n matplotlib.rcParams.update({'font.size' : 18, 'text.usetex' : True})\n fig = plt.figure()\n axes = fig.add_axes([0.15,0.15,0.7,0.7], frameon=True)\n axes.plot(np.linspace(1, X.shape[1]+1, X.shape[1]), explained_variance, color='b', marker='s', linestyle='-', linewidth=2, markersize=4, markerfacecolor='b', label='Cumulative explained')\n axes.plot([n_eigs, n_eigs], [explained_variance[0], explained_variance[n_eigs]], color='r', marker='s', linestyle='-', linewidth=2, markersize=4, markerfacecolor='r', label='Explained by {} PCs'.format(n_eigs))\n axes.set_xlabel('Number of PCs [-]')\n axes.set_ylabel('Explained variance [-]')\n axes.set_title('Variance explained by {} PCs: {}'.format(n_eigs, round(explained,3)))\n axes.legend()\n plt.show()\n\n return explained\n\n\ndef evaluate_clustering_DB(X, idx):\n \"\"\"\n Davies-Bouldin index a coefficient to evaluate the goodness of a\n clustering solution. The more it approaches to zero, the better\n the clustering solution is. -- Tested OK with comparison Matlab\n \"\"\"\n from scipy.spatial.distance import euclidean, cdist\n \n #Initialize matrix and other quantitites\n k = int(np.max(idx) +1)\n centroids_list = [None] *k\n S_i = [None] *k\n M_ij = np.zeros((k,k), dtype=float)\n TOL = 1E-16\n\n #For each cluster, compute the mean distance between the points and their centroids\n for ii in range(0,k):\n cluster_ = get_cluster(X, idx, ii)\n centroids_list[ii] = get_centroids(cluster_)\n S_i[ii] = np.mean(cdist(cluster_, centroids_list[ii].reshape((1,-1)))) #reshape centroids_list[ii] from (n,) to (1,n)\n\n #Compute the distance between once centroid and all the others:\n for ii in range(0,k):\n for jj in range(0,k):\n if ii != jj:\n M_ij[ii,jj] = euclidean(centroids_list[ii], centroids_list[jj])\n else:\n M_ij[ii,jj] = 1\n\n R_ij = np.empty((k,k),dtype=float)\n\n #Compute the R_ij coefficient for each couple of clusters, using the\n #two coefficients S_ij and M_ij\n for ii in range(0,k):\n for jj in range(0,k):\n if ii != jj:\n R_ij[ii,jj] = (S_i[ii] + S_i[jj])/M_ij[ii,jj] +TOL\n else:\n R_ij[ii,jj] = 0\n\n D_i = [None] *k\n\n #Compute the Davies-Bouldin index as the mean of the maximum R_ij value\n for ii in range(0,k):\n D_i[ii] = np.max(R_ij[ii], axis=0)\n\n #The final DB index is the mean value of the DBs for each cluster\n DB = np.mean(D_i)\n\n return DB\n\ndef evaluate_clustering_PHC(X, idx):\n '''\n Computes the PHC (Physical Homogeneity of the Cluster) index.\n For many applications, more than a pure mathematical tool to assess the quality of the clustering solution,\n such as the Silhouette Coefficient, a measure of the variables variation is more suitable. This coefficient\n assess the quality of the clustering solution measuring the variables variation in each cluster. The more the PHC\n approaches to zero, the better the clustering.\n - Input:\n X = UNCENTERED/UNSCALED data matrix -- dim: (observations x variables)\n idx = class membership vector -- dim: (obs x 1)\n method = method to be used for the PHC computation:'PHC_standard', 'PHC_median', 'PHC_robust' are available.\n - Output:\n PHC_coeff = vector with the PHC scores for each cluster -- dim: (number_of_cluster)\n '''\n\n k = max(idx) +1\n TOL = 1E-16\n PHC_coeff=[None] *k\n PHC_deviations=[None] *k\n\n #The standard PHC for one variable is computed as: (max - min)/ mean\n #If the training matrix has more than 1 variable, the PHCs are stored in a list PHC_coeff.\n\n for ii in range (0,k):\n try:\n #take the clusters' observations\n cluster_ = get_cluster(X, idx, ii)\n\n #compute max, min and mean\n maxima = np.max(cluster_, axis = 0)\n minima = np.min(cluster_, axis = 0)\n media = np.mean(cluster_, axis=0)\n\n #compute the standard deviation of each cluster, because PHC can be sensitive to outliers\n #so if dev it's high PHC could not be completely reliable\n dev = np.std(cluster_, axis=0)\n\n #compute PHCs. TOL is added to avoid dividing by 0\n PHC_coeff[ii] = np.mean((maxima-minima)/(media +TOL))\n \n #compute the average standard deviations\n PHC_deviations[ii] = np.mean(dev)\n except ValueError:\n print(\"An exception was thrown by Python during the PHC computation. Probably the considered cluster was found empty.\")\n print(\"Passing..\")\n pass\n\n return PHC_coeff, PHC_deviations\n\n\ndef fastSVD(X_tilde, n_eigs):\n '''\n Perform a fast Singular Value Decomposition with the algorithm described in:\n [1] Halko, Nathan, et al. SIAM Journal on Scientific computing 33.5 (2011): 2580-2594.\n - Input:\n X = CENTERED/SCALED data matrix -- dim: (observations x variables)\n n_eigs = number of eigenvectors to retain -- dim: (scalar)\n \n - Output:\n U, V, Sigma: matrices such that ||X_tilde - U @ Sigma @ V.T || < eps\n \n '''\n rows, cols = X_tilde.shape \n \n l = n_eigs+2\n i = 2\n \n #Step 1: construct a random matrix G, with 0 mean and variance equal to 1.\n G = np.random.rand(cols, l)\n ____, X_ = center(G, \"mean\", True)\n ____, G = scale(X_, \"auto\", True) \n \n #Step 2: Compute the H matrix of eq. (2.2) from [1]. \n #To do that, an iterative procedure must be adopted (in the for loop):\n Hmatr = [None] * i\n Hmatr[0] = X_tilde @ G\n\n for ii in range(1,i):\n Hmatr[ii] = X_tilde @ (X_tilde.T @ Hmatr[ii-1])\n\n \n H = Hmatr[0]\n for ii in range(1, i):\n H = np.concatenate((H,Hmatr[ii]),axis=1)\n \n \n #Step 3: decompose H with a QR decomposition, the column of the matrix\n #Q must be orthonormal\n Q, ____ = np.linalg.qr(H, mode='reduced')\n\n\n #Step 4: compute the T matrix as described in eq. (2.4) of [1]\n T = X_tilde.T @ Q \n\n #Step 5: decompose the T matrix, and retrieve the scores matrix (Z) and\n #the modes matrix (A)\n V_tilde, Sig_tilde, Wt = np.linalg.svd(T, full_matrices=True)\n U_tilde = Q @ Wt.T \n\n U = U_tilde[:, :n_eigs] #modes\n V = V_tilde[:, :n_eigs]\n Sigma = Sig_tilde[:n_eigs]\n \n return U, V, Sigma\n\n\n\ndef get_centroids(X):\n '''\n Given a matrix (or a cluster), calculate its\n centroid.\n - Input:\n X = data matrix -- dim: (observations x variables)\n - Output:\n centroid = centroid vector -- dim: (1 x variables)\n '''\n centroid = np.mean(X, axis = 0)\n return centroid\n\n\ndef get_medianoids(X):\n '''\n Given a matrix (or a cluster), calculate its\n medianoid (same as centroid, but with median).\n - Input:\n X = data matrix -- dim: (observations x variables)\n - Output:\n medianoid = medianoid vector -- dim: (1 x variables)\n '''\n medianoid = np.median(X, axis = 0)\n return medianoid\n\n\ndef get_medoids(X):\n '''\n Given a matrix (or a cluster), calculate its\n medoid: the point which minimize the sum of distances\n with respect to the other observations.\n - Input:\n X = data matrix -- dim: (observations x variables)\n - Output:\n medoid = medoid vector -- dim: (1 x variables)\n '''\n from scipy.spatial.distance import euclidean, cdist\n #Compute the distances between each point of the matrix\n dist = cdist(X, X)\n #Sum all the distances along the columns, to have the\n #cumulative sum of distances.\n cumSum = np.sum(dist, axis=1)\n #Pick up the point which minimize the distances from\n #all the other points as a medoid\n mask = np.argmin(cumSum)\n medoid = X[mask,:]\n\n return medoid\n\n\n\ndef get_cluster(X, idx, index, write=False):\n '''\n Given an index, group all the observations\n of the matrix X given their membership vector idx.\n - Input:\n X = data matrix -- dim: (observations x variables)\n idx = class membership vector -- dim: (obs x 1)\n index = index of the requested group -- dim (scalar)\n - Output:\n cluster: matrix with the grouped observations -- dim: (p x var)\n '''\n try:\n #identify the observations which belong to a certain cluster\n positions = np.where(idx == index)\n cluster = X[positions]\n \n if write:\n np.savetxt(\"Observations in cluster number{}.txt\".format(index), cluster)\n\n return cluster\n \n except:\n print(\"No observations in cluster number: {}. Passing by.\".format(index))\n pass\n\n \ndef get_all_clusters(X, idx):\n '''\n Group all the observations of the matrix X given their membership vector idx,\n and collect the different groups into a list.\n - Input:\n X = data matrix -- dim: (observations x variables)\n idx = class membership vector -- dim: (obs x 1)\n - Output:\n clusters: list with the clusters from 0 to k -- dim: (k)\n '''\n k = max(idx) +1\n clusters = [None] *k\n\n for ii in range (0,k):\n #identify the observations which belong to a certain cluster\n #and store them in the clusters' list\n clusters[ii] = get_cluster(X, idx, ii)\n\n return clusters\n\n\ndef NRMSE (X_true, X_pred):\n n_obs, n_var = X_true.shape\n #initialize the NRMSE \n NRMSE = [None] *n_var\n\n #compute the NRMSE for each observation of the two matrices\n for ii in range(0, n_var):\n NRMSE[ii] = np.sqrt(np.mean((X_true[:,ii] - X_pred[:,ii])**2)) / np.mean(X_true[:,ii])\n\n return NRMSE\n\n\ndef PCA_fit(X, n_eig):\n '''\n Perform Principal Component Analysis on the dataset X,\n and retain 'n_eig' Principal Components.\n The covariance matrix is firstly calculated, then it is\n decomposed in eigenvalues and eigenvectors.\n Lastly, the eigenvalues are ordered depending on their\n magnitude and the associated eigenvectors (the PCs) are retained.\n - Input:\n X = CENTERED/SCALED data matrix -- dim: (observations x variables)\n n_eig = number of principal components to retain -- dim: (scalar)\n - Output:\n evecs: eigenvectors from the covariance matrix decomposition (PCs)\n evals: eigenvalues from the covariance matrix decomposition (lambda)\n !!! WARNING !!! the PCs are already ordered (decreasing, for importance)\n because the eigenvalues are also ordered in terms of magnitude.\n '''\n if n_eig < X.shape[1]:\n #compute the covariance matrix from the original data matrix\n C = np.cov(X, rowvar=False) #rowvar=False because the X matrix is (observations x variables)\n\n #perform an eigendecomposition\n evals, evecs = LA.eig(C)\n\n #sort the eigenvalues in descending order\n mask = np.argsort(evals)[::-1]\n evecs = evecs[:,mask]\n evals = evals[mask]\n\n #retain only the selected number 'q' of eigenvalues\n evecs = evecs[:, :n_eig]\n\n return evecs, evals\n\n else:\n raise Exception(\"The number of PCs exceeds the number of variables in the data-set.\")\n\n\ndef readCSV(path, name):\n try:\n print(\"Reading training matrix..\")\n X = np.genfromtxt(path + \"/\" + name, delimiter= ',')\n except OSError:\n print(\"Could not open/read the selected file: \" + name)\n exit()\n\n return X\n\n\ndef scale(X, method, return_scaled_matrix=False):\n '''\n Computes the scaling factor [sigma] of each variable of all data-set observations and\n (eventually) return the scaled matrix.\n - Input:\n X = original data matrix -- dim: (observations x variables)\n method = \"string\", it is the method which has to be used. Four choices are available: AUTO, PARETO, VAST or RANGE≥\n return_scaled_matrix = boolean, choose if the script must return the scaled matrix (optional)\n - Output:\n sig = scaling factor for the data matrix X\n X0 = centered data matrix (optional)\n '''\n # Main\n TOL = 1E-16\n if not return_scaled_matrix:\n if method.lower() == 'auto':\n #compute the auto scaling factor: standard deviation of each variable\n sig = np.std(X, axis = 0)\n elif method.lower() == 'pareto':\n #compute the pareto scaling factor: sqrt of the standard deviation of each variable\n sig = np.sqrt(np.std(X, axis = 0))\n elif method.lower() == 'vast':\n #compute the vast scaling factor: variance/mean of each variable\n variances = np.var(X, axis = 0)\n means = np.mean(X, axis = 0)\n sig = variances / means\n elif method.lower() == 'range':\n #compute the range scaling factor: max-min for each variable\n maxima = np.max(X, axis = 0)\n minima = np.min(X, axis = 0)\n sig = maxima - minima\n else:\n raise Exception(\"Unsupported scaling option. Please choose: AUTO, PARETO, VAST or RANGE.\")\n return sig\n else:\n if method.lower() == 'auto':\n #compute the auto scaling factor: standard deviation of each variable\n sig = np.std(X, axis = 0)\n X0 = X / (sig + TOL)\n elif method.lower() == 'pareto':\n #compute the pareto scaling factor: sqrt of the standard deviation of each variable\n sig = np.sqrt(np.std(X, axis = 0))\n X0 = X / (sig + TOL)\n elif method.lower() == 'vast':\n #compute the vast scaling factor: variance/mean of each variable\n variances = np.var(X, axis = 0)\n means = np.mean(X, axis = 0)\n sig = variances / means\n X0 = X / (sig + TOL)\n elif method.lower() == 'range':\n #compute the range scaling factor: max-min for each variable\n maxima = np.max(X, axis = 0)\n minima = np.min(X, axis = 0)\n sig = maxima - minima\n X0 = X / (sig + TOL)\n else:\n raise Exception(\"Unsupported scaling option. Please choose: AUTO, PARETO, VAST or RANGE.\")\n return sig, X0\n\n\ndef split_for_validation(X, validation_quota):\n '''\n Split the data into two matrices, one to train the model (X_train) and the\n other to validate it.\n - Input:\n X = matrix to be split -- dim: (observations x variables)\n validation_quota = percentage of observations to take as validation\n - Output:\n X_train = matrix to be used to train the reduced model\n X_test = matrix to be used to test the reduced model\n '''\n\n #compute the number of variables and observations of the training matrix\n nObs = X.shape[0]\n nVar = X.shape[1]\n\n #compute how many observations must be included in the test\n nTest = int(nObs * validation_quota)\n\n #shuffle the training matrix to maximize the randomness of the subset\n np.random.shuffle(X)\n\n #split\n X_test = X[:nTest,:]\n X_train = X[nTest+1:,:]\n\n return X_train, X_test\n\n\ndef uncenter(X_tilde, mu):\n '''\n Uncenter a standardized matrix.\n - Input:\n X_tilde: centered matrix -- dim: (observations x variables)\n mu: centering factor -- dim: (1 x variables)\n - Output:\n X0 = uncentered matrix -- dim: (observations x variables)\n '''\n if X_tilde.shape[1] == mu.shape[0]:\n #initialize the uncentered matrix with all zeros\n X0 = np.zeros_like(X_tilde, dtype=float)\n for i in range(0, len(mu)):\n #add the centering factor to unscale\n X0[:,i] = X_tilde[:,i] + mu[i]\n return X0\n else:\n raise Exception(\"The matrix to be uncentered and the centering vector must have the same dimensionality.\")\n exit()\n\n\ndef unscale(X_tilde, sigma):\n '''\n Unscale a standardized matrix.\n - Input:\n X_tilde = scaled matrix -- dim: (observations x variables)\n sigma = scaling factor -- dim: (1 x variables)\n - Output:\n X0 = unscaled matrix -- dim: (observations x variables)\n '''\n TOL = 1E-16\n if X_tilde.shape[1] == sigma.shape[0]:\n #initialize the uncentered matrix with all zeros\n X0 = np.zeros_like(X_tilde, dtype=float)\n for i in range(0, len(sigma)):\n #multiply for the scaling factor to unscale\n X0[:,i] = X_tilde[:,i] * (sigma[i] + TOL)\n return X0\n else:\n raise Exception(\"The matrix to be unscaled and the scaling vector must have the same dimensionality.\")\n exit()\n\n\ndef varimax_rotation(X, b):\n '''\n Rotate the factors by means of the varimax rotation.\n - Input:\n X = training data matrix, SCALED/UNSCALED it makes no difference\n b = factors/modes to be rotated\n\n - Output:\n rot_loadings = rotated modes or factors, returned after the algorithm convergenceß\n '''\n import math\n #compute the dimensionality of the problem\n eigens = b.shape[1]\n \n #compute the normalization factor for the modes/factors before the rotation\n norm_factor = np.std(b, axis=0)\n \n #initialize an empty matrix for the rotated loadings\n loadings = np.empty((b.shape[0], b.shape[1]), dtype=float)\n rot_loadings = np.empty((b.shape[0], b.shape[1]), dtype=float)\n\n #standardize the loadings dividing for the normalization factor\n for ii in range(0, eigens):\n loadings[:,ii] = b[:,ii]/norm_factor[ii]\n\n #compute the loadings covariance matrix\n C = np.cov(loadings, rowvar=False)\n\n #initialize the algorithm's parameter to reach convergence\n iter_max = 1000\n convergence_tolerance = 1E-16\n iter = 1\n convergence = False\n\n #compute the percentage of explained variance\n variance_explained = np.sum(loadings**2)\n\n while not convergence:\n for ii in range(0,eigens):\n for jj in range(ii+1, eigens):\n x_j = loadings[:,ii]\n y_j = loadings[:,jj]\n\n u_j = np.reshape(np.array(np.multiply(x_j,x_j) - np.multiply(y_j,y_j)), (b.shape[0],1)) #nvarx1\n v_j = np.reshape(np.array(2*np.multiply(x_j, y_j)), (b.shape[0],1)) #nvarx1\n\n A = np.sum(u_j)\n B = np.sum(v_j)\n C = u_j.T @ u_j - v_j.T @ v_j\n D = 2 * u_j.T @ v_j\n\n num = D - 3*A*B/X.shape[0]\n den = C - (A**2 - B**2)/X.shape[0]\n\n phi = np.arctan2(num, den)/4\n angle = phi *180/math.pi \n\n if np.abs(phi) > 0.00001:\n Z_j = np.cos(phi) *x_j + np.sin(phi) *y_j\n Y_j = -np.sin(phi) *x_j + np.cos(phi) *y_j\n\n loadings[:,ii] = Z_j\n loadings[:,jj] = Y_j\n\n var_old = variance_explained\n variance_explained = np.sum(loadings**2)\n\n #convergence criterion\n check = np.abs((variance_explained - var_old)/(variance_explained + 1E-16))\n\n if check < convergence_tolerance or iter > iter_max:\n iter += 1\n convergence = True\n else:\n iter += 1\n\n print(\"Iteration number: {}\".format(iter))\n print(\"Convergence residuals: {}\".format(check))\n\n\n for ii in range(0, eigens):\n rot_loadings[:,ii] = loadings[:,ii] * norm_factor[ii]\n\n return rot_loadings"
] |
[
[
"numpy.random.rand",
"numpy.argmin",
"numpy.median",
"numpy.genfromtxt",
"numpy.min",
"numpy.mean",
"numpy.linalg.qr",
"numpy.where",
"numpy.multiply",
"numpy.cos",
"numpy.cumsum",
"numpy.max",
"numpy.concatenate",
"numpy.zeros_like",
"numpy.sin",
"numpy.empty",
"scipy.spatial.distance.euclidean",
"numpy.zeros",
"numpy.random.shuffle",
"matplotlib.pyplot.figure",
"numpy.std",
"matplotlib.rcParams.update",
"numpy.linalg.svd",
"numpy.argsort",
"numpy.arctan2",
"matplotlib.pyplot.show",
"scipy.spatial.distance.cdist",
"numpy.cov",
"numpy.sum",
"numpy.linalg.eig",
"numpy.abs",
"numpy.linspace",
"numpy.var"
]
] |
JBris/EasyOCR
|
[
"f41a5d951bd6fce8cfcdaa67a956c639c013eb18"
] |
[
"easyocr/utils.py"
] |
[
"from __future__ import print_function\n\nimport torch\nimport pickle\nimport numpy as np\nimport math\nimport cv2\nfrom PIL import Image\nimport hashlib\nimport sys\nimport requests\n\ndef consecutive(data, mode ='first', stepsize=1):\n group = np.split(data, np.where(np.diff(data) != stepsize)[0]+1)\n group = [item for item in group if len(item)>0]\n\n if mode == 'first': result = [l[0] for l in group]\n elif mode == 'last': result = [l[-1] for l in group]\n return result\n\ndef word_segmentation(mat, separator_idx = {'th': [1,2],'en': [3,4]}, separator_idx_list = [1,2,3,4]):\n result = []\n sep_list = []\n start_idx = 0\n sep_lang = ''\n for sep_idx in separator_idx_list:\n if sep_idx % 2 == 0: mode ='first'\n else: mode ='last'\n a = consecutive( np.argwhere(mat == sep_idx).flatten(), mode)\n new_sep = [ [item, sep_idx] for item in a]\n sep_list += new_sep\n sep_list = sorted(sep_list, key=lambda x: x[0])\n\n for sep in sep_list:\n for lang in separator_idx.keys():\n if sep[1] == separator_idx[lang][0]: # start lang\n sep_lang = lang\n sep_start_idx = sep[0]\n elif sep[1] == separator_idx[lang][1]: # end lang\n if sep_lang == lang: # check if last entry if the same start lang\n new_sep_pair = [lang, [sep_start_idx+1, sep[0]-1]]\n if sep_start_idx > start_idx:\n result.append( ['', [start_idx, sep_start_idx-1] ] )\n start_idx = sep[0]+1\n result.append(new_sep_pair)\n sep_lang = ''# reset\n\n if start_idx <= len(mat)-1:\n result.append( ['', [start_idx, len(mat)-1] ] )\n return result\n\n# code is based from https://github.com/githubharald/CTCDecoder/blob/master/src/BeamSearch.py\nclass BeamEntry:\n \"information about one single beam at specific time-step\"\n def __init__(self):\n self.prTotal = 0 # blank and non-blank\n self.prNonBlank = 0 # non-blank\n self.prBlank = 0 # blank\n self.prText = 1 # LM score\n self.lmApplied = False # flag if LM was already applied to this beam\n self.labeling = () # beam-labeling\n\nclass BeamState:\n \"information about the beams at specific time-step\"\n def __init__(self):\n self.entries = {}\n\n def norm(self):\n \"length-normalise LM score\"\n for (k, _) in self.entries.items():\n labelingLen = len(self.entries[k].labeling)\n self.entries[k].prText = self.entries[k].prText ** (1.0 / (labelingLen if labelingLen else 1.0))\n\n def sort(self):\n \"return beam-labelings, sorted by probability\"\n beams = [v for (_, v) in self.entries.items()]\n sortedBeams = sorted(beams, reverse=True, key=lambda x: x.prTotal*x.prText)\n return [x.labeling for x in sortedBeams]\n\n def wordsearch(self, classes, ignore_idx, maxCandidate, dict_list):\n beams = [v for (_, v) in self.entries.items()]\n sortedBeams = sorted(beams, reverse=True, key=lambda x: x.prTotal*x.prText)\n if len(sortedBeams) > maxCandidate: sortedBeams = sortedBeams[:maxCandidate]\n\n for j, candidate in enumerate(sortedBeams):\n idx_list = candidate.labeling\n text = ''\n for i,l in enumerate(idx_list):\n if l not in ignore_idx and (not (i > 0 and idx_list[i - 1] == idx_list[i])):\n text += classes[l]\n\n if j == 0: best_text = text\n if text in dict_list:\n #print('found text: ', text)\n best_text = text\n break\n else:\n pass\n #print('not in dict: ', text)\n return best_text\n\ndef applyLM(parentBeam, childBeam, classes, lm):\n \"calculate LM score of child beam by taking score from parent beam and bigram probability of last two chars\"\n if lm and not childBeam.lmApplied:\n c1 = classes[parentBeam.labeling[-1] if parentBeam.labeling else classes.index(' ')] # first char\n c2 = classes[childBeam.labeling[-1]] # second char\n lmFactor = 0.01 # influence of language model\n bigramProb = lm.getCharBigram(c1, c2) ** lmFactor # probability of seeing first and second char next to each other\n childBeam.prText = parentBeam.prText * bigramProb # probability of char sequence\n childBeam.lmApplied = True # only apply LM once per beam entry\n\ndef simplify_label(labeling, blankIdx = 0):\n labeling = np.array(labeling)\n\n # collapse blank\n idx = np.where(~((np.roll(labeling,1) == labeling) & (labeling == blankIdx)))[0]\n labeling = labeling[idx]\n\n # get rid of blank between different characters\n idx = np.where( ~((np.roll(labeling,1) != np.roll(labeling,-1)) & (labeling == blankIdx)) )[0]\n\n if len(labeling) > 0:\n last_idx = len(labeling)-1\n if last_idx not in idx: idx = np.append(idx, [last_idx])\n labeling = labeling[idx]\n\n return tuple(labeling)\n\ndef addBeam(beamState, labeling):\n \"add beam if it does not yet exist\"\n if labeling not in beamState.entries:\n beamState.entries[labeling] = BeamEntry()\n\ndef ctcBeamSearch(mat, classes, ignore_idx, lm, beamWidth=25, dict_list = []):\n blankIdx = 0\n maxT, maxC = mat.shape\n\n # initialise beam state\n last = BeamState()\n labeling = ()\n last.entries[labeling] = BeamEntry()\n last.entries[labeling].prBlank = 1\n last.entries[labeling].prTotal = 1\n\n # go over all time-steps\n for t in range(maxT):\n curr = BeamState()\n # get beam-labelings of best beams\n bestLabelings = last.sort()[0:beamWidth]\n # go over best beams\n for labeling in bestLabelings:\n # probability of paths ending with a non-blank\n prNonBlank = 0\n # in case of non-empty beam\n if labeling:\n # probability of paths with repeated last char at the end\n prNonBlank = last.entries[labeling].prNonBlank * mat[t, labeling[-1]]\n\n # probability of paths ending with a blank\n prBlank = (last.entries[labeling].prTotal) * mat[t, blankIdx]\n\n # add beam at current time-step if needed\n labeling = simplify_label(labeling, blankIdx)\n addBeam(curr, labeling)\n\n # fill in data\n curr.entries[labeling].labeling = labeling\n curr.entries[labeling].prNonBlank += prNonBlank\n curr.entries[labeling].prBlank += prBlank\n curr.entries[labeling].prTotal += prBlank + prNonBlank\n curr.entries[labeling].prText = last.entries[labeling].prText\n # beam-labeling not changed, therefore also LM score unchanged from\n\n #curr.entries[labeling].lmApplied = True # LM already applied at previous time-step for this beam-labeling\n\n # extend current beam-labeling\n # char_highscore = np.argpartition(mat[t, :], -5)[-5:] # run through 5 highest probability\n char_highscore = np.where(mat[t, :] >= 0.5/maxC)[0] # run through all probable characters\n for c in char_highscore:\n #for c in range(maxC - 1):\n # add new char to current beam-labeling\n newLabeling = labeling + (c,)\n newLabeling = simplify_label(newLabeling, blankIdx)\n\n # if new labeling contains duplicate char at the end, only consider paths ending with a blank\n if labeling and labeling[-1] == c:\n prNonBlank = mat[t, c] * last.entries[labeling].prBlank\n else:\n prNonBlank = mat[t, c] * last.entries[labeling].prTotal\n\n # add beam at current time-step if needed\n addBeam(curr, newLabeling)\n\n # fill in data\n curr.entries[newLabeling].labeling = newLabeling\n curr.entries[newLabeling].prNonBlank += prNonBlank\n curr.entries[newLabeling].prTotal += prNonBlank\n\n # apply LM\n #applyLM(curr.entries[labeling], curr.entries[newLabeling], classes, lm)\n\n # set new beam state\n\n last = curr\n\n # normalise LM scores according to beam-labeling-length\n last.norm()\n\n if dict_list == []:\n bestLabeling = last.sort()[0] # get most probable labeling\n res = ''\n for i,l in enumerate(bestLabeling):\n # removing repeated characters and blank.\n if l not in ignore_idx and (not (i > 0 and bestLabeling[i - 1] == bestLabeling[i])):\n res += classes[l]\n else:\n res = last.wordsearch(classes, ignore_idx, 20, dict_list)\n return res\n\n\nclass CTCLabelConverter(object):\n \"\"\" Convert between text-label and text-index \"\"\"\n\n def __init__(self, character, separator_list = {}, dict_pathlist = {}):\n # character (str): set of the possible characters.\n dict_character = list(character)\n\n self.dict = {}\n for i, char in enumerate(dict_character):\n self.dict[char] = i + 1\n\n self.character = ['[blank]'] + dict_character # dummy '[blank]' token for CTCLoss (index 0)\n\n self.separator_list = separator_list\n separator_char = []\n for lang, sep in separator_list.items():\n separator_char += sep\n self.ignore_idx = [0] + [i+1 for i,item in enumerate(separator_char)]\n\n ####### latin dict\n if len(separator_list) == 0:\n dict_list = []\n for lang, dict_path in dict_pathlist.items():\n try:\n with open(dict_path, \"r\", encoding = \"utf-8-sig\") as input_file:\n word_count = input_file.read().splitlines()\n dict_list += word_count\n except:\n pass\n else:\n dict_list = {}\n for lang, dict_path in dict_pathlist.items():\n with open(dict_path, \"r\", encoding = \"utf-8-sig\") as input_file:\n word_count = input_file.read().splitlines()\n dict_list[lang] = word_count\n\n self.dict_list = dict_list\n\n def encode(self, text, batch_max_length=25):\n \"\"\"convert text-label into text-index.\n input:\n text: text labels of each image. [batch_size]\n\n output:\n text: concatenated text index for CTCLoss.\n [sum(text_lengths)] = [text_index_0 + text_index_1 + ... + text_index_(n - 1)]\n length: length of each text. [batch_size]\n \"\"\"\n length = [len(s) for s in text]\n text = ''.join(text)\n text = [self.dict[char] for char in text]\n\n return (torch.IntTensor(text), torch.IntTensor(length))\n\n def decode_greedy(self, text_index, length):\n \"\"\" convert text-index into text-label. \"\"\"\n texts = []\n index = 0\n for l in length:\n t = text_index[index:index + l]\n\n char_list = []\n for i in range(l):\n # removing repeated characters and blank (and separator).\n if t[i] not in self.ignore_idx and (not (i > 0 and t[i - 1] == t[i])):\n char_list.append(self.character[t[i]])\n text = ''.join(char_list)\n\n texts.append(text)\n index += l\n return texts\n\n def decode_beamsearch(self, mat, beamWidth=5):\n texts = []\n for i in range(mat.shape[0]):\n t = ctcBeamSearch(mat[i], self.character, self.ignore_idx, None, beamWidth=beamWidth)\n texts.append(t)\n return texts\n\n def decode_wordbeamsearch(self, mat, beamWidth=5):\n texts = []\n argmax = np.argmax(mat, axis = 2)\n\n for i in range(mat.shape[0]):\n string = ''\n # without separators - use space as separator\n if len(self.separator_list) == 0:\n space_idx = self.dict[' ']\n\n data = np.argwhere(argmax[i]!=space_idx).flatten()\n group = np.split(data, np.where(np.diff(data) != 1)[0]+1)\n group = [ list(item) for item in group if len(item)>0]\n\n for j, list_idx in enumerate(group):\n matrix = mat[i, list_idx,:]\n t = ctcBeamSearch(matrix, self.character, self.ignore_idx, None,\\\n beamWidth=beamWidth, dict_list=self.dict_list)\n if j == 0: string += t\n else: string += ' '+t\n\n # with separators\n else:\n words = word_segmentation(argmax[i])\n\n for word in words:\n matrix = mat[i, word[1][0]:word[1][1]+1,:]\n if word[0] == '': dict_list = []\n else: dict_list = self.dict_list[word[0]]\n t = ctcBeamSearch(matrix, self.character, self.ignore_idx, None, beamWidth=beamWidth, dict_list=dict_list)\n string += t\n texts.append(string)\n return texts\n\ndef four_point_transform(image, rect):\n (tl, tr, br, bl) = rect\n\n widthA = np.sqrt(((br[0] - bl[0]) ** 2) + ((br[1] - bl[1]) ** 2))\n widthB = np.sqrt(((tr[0] - tl[0]) ** 2) + ((tr[1] - tl[1]) ** 2))\n maxWidth = max(int(widthA), int(widthB))\n\n # compute the height of the new image, which will be the\n # maximum distance between the top-right and bottom-right\n # y-coordinates or the top-left and bottom-left y-coordinates\n heightA = np.sqrt(((tr[0] - br[0]) ** 2) + ((tr[1] - br[1]) ** 2))\n heightB = np.sqrt(((tl[0] - bl[0]) ** 2) + ((tl[1] - bl[1]) ** 2))\n maxHeight = max(int(heightA), int(heightB))\n\n dst = np.array([[0, 0],[maxWidth - 1, 0],[maxWidth - 1, maxHeight - 1],[0, maxHeight - 1]], dtype = \"float32\")\n\n # compute the perspective transform matrix and then apply it\n M = cv2.getPerspectiveTransform(rect, dst)\n warped = cv2.warpPerspective(image, M, (maxWidth, maxHeight))\n\n return warped\n\ndef group_text_box(polys, slope_ths = 0.1, ycenter_ths = 0.5, height_ths = 0.5, width_ths = 1.0, add_margin = 0.05):\n # poly top-left, top-right, low-right, low-left\n horizontal_list, free_list,combined_list, merged_list = [],[],[],[]\n\n for poly in polys:\n slope_up = (poly[3]-poly[1])/np.maximum(10, (poly[2]-poly[0]))\n slope_down = (poly[5]-poly[7])/np.maximum(10, (poly[4]-poly[6]))\n if max(abs(slope_up), abs(slope_down)) < slope_ths:\n x_max = max([poly[0],poly[2],poly[4],poly[6]])\n x_min = min([poly[0],poly[2],poly[4],poly[6]])\n y_max = max([poly[1],poly[3],poly[5],poly[7]])\n y_min = min([poly[1],poly[3],poly[5],poly[7]])\n horizontal_list.append([x_min, x_max, y_min, y_max, 0.5*(y_min+y_max), y_max-y_min])\n else:\n height = np.linalg.norm( [poly[6]-poly[0],poly[7]-poly[1]])\n margin = int(1.44*add_margin*height)\n\n theta13 = abs(np.arctan( (poly[1]-poly[5])/np.maximum(10, (poly[0]-poly[4]))))\n theta24 = abs(np.arctan( (poly[3]-poly[7])/np.maximum(10, (poly[2]-poly[6]))))\n # do I need to clip minimum, maximum value here?\n x1 = poly[0] - np.cos(theta13)*margin\n y1 = poly[1] - np.sin(theta13)*margin\n x2 = poly[2] + np.cos(theta24)*margin\n y2 = poly[3] - np.sin(theta24)*margin\n x3 = poly[4] + np.cos(theta13)*margin\n y3 = poly[5] + np.sin(theta13)*margin\n x4 = poly[6] - np.cos(theta24)*margin\n y4 = poly[7] + np.sin(theta24)*margin\n\n free_list.append([[x1,y1],[x2,y2],[x3,y3],[x4,y4]])\n horizontal_list = sorted(horizontal_list, key=lambda item: item[4])\n\n # combine box\n new_box = []\n for poly in horizontal_list:\n\n if len(new_box) == 0:\n b_height = [poly[5]]\n b_ycenter = [poly[4]]\n new_box.append(poly)\n else:\n # comparable height and comparable y_center level up to ths*height\n if (abs(np.mean(b_height) - poly[5]) < height_ths*np.mean(b_height)) and (abs(np.mean(b_ycenter) - poly[4]) < ycenter_ths*np.mean(b_height)):\n b_height.append(poly[5])\n b_ycenter.append(poly[4])\n new_box.append(poly)\n else:\n b_height = [poly[5]]\n b_ycenter = [poly[4]]\n combined_list.append(new_box)\n new_box = [poly]\n combined_list.append(new_box)\n\n # merge list use sort again\n for boxes in combined_list:\n if len(boxes) == 1: # one box per line\n box = boxes[0]\n margin = int(add_margin*box[5])\n merged_list.append([box[0]-margin,box[1]+margin,box[2]-margin,box[3]+margin])\n else: # multiple boxes per line\n boxes = sorted(boxes, key=lambda item: item[0])\n\n merged_box, new_box = [],[]\n for box in boxes:\n if len(new_box) == 0:\n x_max = box[1]\n new_box.append(box)\n else:\n if abs(box[0]-x_max) < width_ths *(box[3]-box[2]): # merge boxes\n x_max = box[1]\n new_box.append(box)\n else:\n x_max = box[1]\n merged_box.append(new_box)\n new_box = [box]\n if len(new_box) >0: merged_box.append(new_box)\n\n for mbox in merged_box:\n if len(mbox) != 1: # adjacent box in same line\n # do I need to add margin here?\n x_min = min(mbox, key=lambda x: x[0])[0]\n x_max = max(mbox, key=lambda x: x[1])[1]\n y_min = min(mbox, key=lambda x: x[2])[2]\n y_max = max(mbox, key=lambda x: x[3])[3]\n\n margin = int(add_margin*(y_max - y_min))\n\n merged_list.append([x_min-margin, x_max+margin, y_min-margin, y_max+margin])\n else: # non adjacent box in same line\n box = mbox[0]\n\n margin = int(add_margin*(box[3] - box[2]))\n merged_list.append([box[0]-margin,box[1]+margin,box[2]-margin,box[3]+margin])\n # may need to check if box is really in image\n return merged_list, free_list\n\ndef get_image_list(horizontal_list, free_list, img, model_height = 64):\n image_list = []\n maximum_y,maximum_x = img.shape\n\n max_ratio_hori, max_ratio_free = 1,1\n for box in free_list:\n rect = np.array(box, dtype = \"float32\")\n transformed_img = four_point_transform(img, rect)\n ratio = transformed_img.shape[1]/transformed_img.shape[0]\n crop_img = cv2.resize(transformed_img, (int(model_height*ratio), model_height), interpolation = Image.ANTIALIAS)\n image_list.append( (box,crop_img) ) # box = [[x1,y1],[x2,y2],[x3,y3],[x4,y4]]\n max_ratio_free = max(ratio, max_ratio_free)\n\n max_ratio_free = math.ceil(max_ratio_free)\n\n for box in horizontal_list:\n x_min = max(0,box[0])\n x_max = min(box[1],maximum_x)\n y_min = max(0,box[2])\n y_max = min(box[3],maximum_y)\n crop_img = img[y_min : y_max, x_min:x_max]\n width = x_max - x_min\n height = y_max - y_min\n ratio = width/height\n crop_img = cv2.resize(crop_img, (int(model_height*ratio), model_height), interpolation = Image.ANTIALIAS)\n image_list.append( ( [[x_min,y_min],[x_max,y_min],[x_max,y_max],[x_min,y_max]] ,crop_img) )\n max_ratio_hori = max(ratio, max_ratio_hori)\n\n max_ratio_hori = math.ceil(max_ratio_hori)\n max_ratio = max(max_ratio_hori, max_ratio_free)\n max_width = math.ceil(max_ratio)*model_height\n\n image_list = sorted(image_list, key=lambda item: item[0][0][1]) # sort by vertical position\n return image_list, max_width\n\n# code from https://stackoverflow.com/questions/25010369/wget-curl-large-file-from-google-drive\ndef download_file_from_google_drive(id, destination):\n def get_confirm_token(response):\n for key, value in response.cookies.items():\n if key.startswith('download_warning'):\n return value\n return None\n\n def save_response_content(response, destination):\n CHUNK_SIZE = 32768\n with open(destination, \"wb\") as f:\n for chunk in response.iter_content(CHUNK_SIZE):\n if chunk: # filter out keep-alive new chunks\n f.write(chunk)\n\n URL = \"https://docs.google.com/uc?export=download\"\n session = requests.Session()\n response = session.get(URL, params = { 'id' : id }, stream = True)\n token = get_confirm_token(response)\n if token:\n params = { 'id' : id, 'confirm' : token }\n response = session.get(URL, params = params, stream = True)\n save_response_content(response, destination)\n\ndef calculate_md5(fname):\n hash_md5 = hashlib.md5()\n with open(fname, \"rb\") as f:\n for chunk in iter(lambda: f.read(4096), b\"\"):\n hash_md5.update(chunk)\n return hash_md5.hexdigest()\n\ndef get_paragraph(raw_result, x_ths=1, y_ths=0.5, mode = 'ltr'):\n # create basic attributes\n box_group = []\n for box in raw_result:\n all_x = [int(coord[0]) for coord in box[0]]\n all_y = [int(coord[1]) for coord in box[0]]\n min_x = min(all_x)\n max_x = max(all_x)\n min_y = min(all_y)\n max_y = max(all_y)\n height = max_y - min_y\n box_group.append([box[1], min_x, max_x, min_y, max_y, height, 0.5*(min_y+max_y), 0]) # last element indicates group\n # cluster boxes into paragraph\n current_group = 1\n while len([box for box in box_group if box[7]==0]) > 0:\n box_group0 = [box for box in box_group if box[7]==0] # group0 = non-group\n # new group\n if len([box for box in box_group if box[7]==current_group]) == 0:\n box_group0[0][7] = current_group # assign first box to form new group\n # try to add group\n else:\n current_box_group = [box for box in box_group if box[7]==current_group]\n mean_height = np.mean([box[5] for box in current_box_group])\n min_gx = min([box[1] for box in current_box_group]) - x_ths*mean_height\n max_gx = max([box[2] for box in current_box_group]) + x_ths*mean_height\n min_gy = min([box[3] for box in current_box_group]) - y_ths*mean_height\n max_gy = max([box[4] for box in current_box_group]) + y_ths*mean_height\n add_box = False\n for box in box_group0:\n same_horizontal_level = (min_gx<=box[1]<=max_gx) or (min_gx<=box[2]<=max_gx)\n same_vertical_level = (min_gy<=box[3]<=max_gy) or (min_gy<=box[4]<=max_gy)\n if same_horizontal_level and same_vertical_level:\n box[7] = current_group\n add_box = True\n break\n # cannot add more box, go to next group\n if add_box==False:\n current_group += 1\n # arrage order in paragraph\n result = []\n for i in set(box[7] for box in box_group):\n current_box_group = [box for box in box_group if box[7]==i]\n mean_height = np.mean([box[5] for box in current_box_group])\n min_gx = min([box[1] for box in current_box_group])\n max_gx = max([box[2] for box in current_box_group])\n min_gy = min([box[3] for box in current_box_group])\n max_gy = max([box[4] for box in current_box_group])\n\n text = ''\n while len(current_box_group) > 0:\n highest = min([box[6] for box in current_box_group])\n candidates = [box for box in current_box_group if box[6]<highest+0.4*mean_height]\n # get the far left\n if mode == 'ltr':\n most_left = min([box[1] for box in candidates])\n for box in candidates:\n if box[1] == most_left: best_box = box\n text += ' '+best_box[0]\n current_box_group.remove(best_box)\n\n result.append([ [[min_gx,min_gy],[max_gx,min_gy],[max_gx,max_gy],[min_gx,max_gy]], text[1:]])\n\n return result\n"
] |
[
[
"numpy.array",
"numpy.linalg.norm",
"numpy.sin",
"torch.IntTensor",
"numpy.roll",
"numpy.mean",
"numpy.diff",
"numpy.where",
"numpy.argmax",
"numpy.sqrt",
"numpy.append",
"numpy.cos",
"numpy.argwhere",
"numpy.maximum"
]
] |
pabloppp/glimpse-models
|
[
"224bd9614a47f773c1e78b85fd9bf0bbff88529b"
] |
[
"data-prepare/PythonAPI/saliconDemo.py"
] |
[
"\n# coding: utf-8\n\n# In[1]:\n\n#get_ipython().magic(u'reload_ext autoreload')\n#get_ipython().magic(u'autoreload 2')\n#get_ipython().magic(u'matplotlib inline')\nfrom salicon.salicon import SALICON\nimport numpy as np\nimport skimage.io as io\nimport matplotlib.pyplot as plt\n\n\n# In[2]:\n\ndataDir='..'\ndataType='train2014examples'\nannFile='%s/annotations/fixations_%s.json'%(dataDir,dataType)\n\n\n# In[3]:\n\n# initialize COCO api for instance annotations\nsalicon=SALICON(annFile)\n\n\n# In[4]:\n\n# get all images \nimgIds = salicon.getImgIds();\nimg = salicon.loadImgs(imgIds[np.random.randint(0,len(imgIds))])[0]\n\n\n# In[6]:\n\n# load and display image\nI = io.imread('%s/images/%s/%s'%(dataDir,dataType,img['file_name']))\nplt.figure()\nplt.imshow(I)\nplt.show()\n\n\n# In[7]:\n\n# load and display instance annotations\n#plt.imshow(I)\nannIds = salicon.getAnnIds(imgIds=img['id'])\nanns = salicon.loadAnns(annIds)\nsalicon.showAnns(anns)\nplt.show()\n\n\n# In[ ]:\n\n\n\n"
] |
[
[
"matplotlib.pyplot.show",
"matplotlib.pyplot.imshow",
"matplotlib.pyplot.figure"
]
] |
yongfeng-nv/incubator-tvm
|
[
"a6cb4b8d3778db5341f991db9adf76ff735b72ea"
] |
[
"tests/python/relay/test_op_qnn_concatenate.py"
] |
[
"# Licensed to the Apache Software Foundation (ASF) under one\n# or more contributor license agreements. See the NOTICE file\n# distributed with this work for additional information\n# regarding copyright ownership. The ASF licenses this file\n# to you under the Apache License, Version 2.0 (the\n# \"License\"); you may not use this file except in compliance\n# with the License. You may obtain a copy of the License at\n#\n# http://www.apache.org/licenses/LICENSE-2.0\n#\n# Unless required by applicable law or agreed to in writing,\n# software distributed under the License is distributed on an\n# \"AS IS\" BASIS, WITHOUT WARRANTIES OR CONDITIONS OF ANY\n# KIND, either express or implied. See the License for the\n# specific language governing permissions and limitations\n# under the License.\n\nimport tvm\nfrom tvm import te\nimport numpy as np\nfrom tvm import relay\nfrom tvm.contrib import graph_runtime\nimport topi.testing\n\ndef test_same_io_qnn_params():\n data_dtype = 'int32'\n axis = 0\n x_data = np.arange(-32, 32, 1).reshape(1, 64).astype(data_dtype)\n y_data = np.arange(-64, 64, 2).reshape(1, 64).astype(data_dtype)\n x_scale = relay.const((62 + 64) / (np.power(2, 32) - 1.0), 'float32')\n y_scale = relay.const((62 + 64) / (np.power(2, 32) - 1.0), 'float32')\n zero = relay.const(0, 'int32')\n\n x = relay.var(\"x\", shape=(1, 64), dtype=data_dtype)\n y = relay.var(\"y\", shape=(1, 64), dtype=data_dtype)\n z = relay.qnn.op.concatenate((x, y),\n input_scales=(x_scale, y_scale),\n input_zero_points=(zero, zero),\n output_scale=y_scale,\n output_zero_point=zero,\n axis=axis)\n\n func = relay.Function([x, y], z)\n mod = tvm.IRModule.from_expr(func)\n mod = relay.qnn.transform.CanonicalizeOps()(mod)\n func = mod[\"main\"]\n\n golden_output = np.concatenate((x_data, y_data), axis=axis)\n\n intrp = relay.create_executor(\"graph\", ctx=tvm.cpu(0), target=\"llvm\")\n op_res = intrp.evaluate(func)(x_data, y_data)\n np.testing.assert_equal(op_res.asnumpy(), golden_output)\n\ndef test_different_io_qnn_params():\n data_dtype = 'int32'\n axis = 0\n x_data = np.arange(-32, 32, 1).reshape(1, 64).astype(data_dtype)\n y_data = np.arange(-64, 64, 2).reshape(1, 64).astype(data_dtype)\n\n x_scale = relay.const((62 + 64) / (np.power(2, 32) - 1.0), 'float32')\n y_scale = relay.const((62 + 64) / (np.power(2, 32) - 1.0), 'float32')\n x_zero_point = relay.const(3, 'int32')\n y_zero_point = relay.const(4, 'int32')\n\n x = relay.var(\"x\", shape=(1, 64), dtype=data_dtype)\n y = relay.var(\"y\", shape=(1, 64), dtype=data_dtype)\n z = relay.qnn.op.concatenate((x, y),\n input_scales=(x_scale, y_scale),\n input_zero_points=(x_zero_point, y_zero_point),\n output_scale=y_scale,\n output_zero_point=relay.const(1, 'int32'),\n axis=axis)\n\n func = relay.Function([x, y], z)\n mod = tvm.IRModule.from_expr(func)\n mod = relay.qnn.transform.CanonicalizeOps()(mod)\n func = mod[\"main\"]\n\n golden_output = np.concatenate((x_data - 2, y_data - 3), axis=axis)\n\n intrp = relay.create_executor(\"graph\", ctx=tvm.cpu(0), target=\"llvm\")\n op_res = intrp.evaluate(func)(x_data, y_data)\n np.testing.assert_equal(op_res.asnumpy(), golden_output)\n\ndef test_few_same_io_qnn_params():\n data_dtype = 'int32'\n axis = 0\n x_data = np.arange(-32, 32, 1).reshape(1, 64).astype(data_dtype)\n y_data = np.arange(-64, 64, 2).reshape(1, 64).astype(data_dtype)\n\n x_scale = relay.const((62 + 64) / (np.power(2, 32) - 1.0), 'float32')\n y_scale = relay.const((62 + 64) / (np.power(2, 32) - 1.0), 'float32')\n x_zero_point = relay.const(0, 'int32')\n y_zero_point = relay.const(1, 'int32')\n\n x = relay.var(\"x\", shape=(1, 64), dtype=data_dtype)\n y = relay.var(\"y\", shape=(1, 64), dtype=data_dtype)\n z = relay.qnn.op.concatenate((x, y),\n input_scales=(x_scale, y_scale),\n input_zero_points=(x_zero_point, y_zero_point),\n output_scale=y_scale,\n output_zero_point=relay.const(1, 'int32'),\n axis=axis)\n\n func = relay.Function([x, y], z)\n mod = tvm.IRModule.from_expr(func)\n mod = relay.qnn.transform.CanonicalizeOps()(mod)\n func = mod[\"main\"]\n\n golden_output = np.concatenate((x_data + 1, y_data), axis=axis)\n\n intrp = relay.create_executor(\"graph\", ctx=tvm.cpu(0), target=\"llvm\")\n op_res = intrp.evaluate(func)(x_data, y_data)\n np.testing.assert_equal(op_res.asnumpy(), golden_output)\n\ndef test_same_i_qnn_params():\n data_dtype = 'int32'\n axis = 0\n x_data = np.arange(-32, 32, 1).reshape(1, 64).astype(data_dtype)\n y_data = np.arange(-64, 64, 2).reshape(1, 64).astype(data_dtype)\n\n x_scale = relay.const((62 + 64) / (np.power(2, 32) - 1.0), 'float32')\n y_scale = relay.const((62 + 64) / (np.power(2, 32) - 1.0), 'float32')\n x_zero_point = relay.const(0, 'int32')\n y_zero_point = relay.const(0, 'int32')\n\n x = relay.var(\"x\", shape=(1, 64), dtype=data_dtype)\n y = relay.var(\"y\", shape=(1, 64), dtype=data_dtype)\n z = relay.qnn.op.concatenate((x, y),\n input_scales=(x_scale, y_scale),\n input_zero_points=(x_zero_point, y_zero_point),\n output_scale=y_scale,\n output_zero_point=relay.const(1, 'int32'),\n axis=axis)\n\n func = relay.Function([x, y], z)\n mod = tvm.IRModule.from_expr(func)\n mod = relay.qnn.transform.CanonicalizeOps()(mod)\n func = mod[\"main\"]\n\n golden_output = np.concatenate((x_data + 1, y_data + 1), axis=axis)\n\n intrp = relay.create_executor(\"graph\", ctx=tvm.cpu(0), target=\"llvm\")\n op_res = intrp.evaluate(func)(x_data, y_data)\n np.testing.assert_equal(op_res.asnumpy(), golden_output)\n\nif __name__ == '__main__':\n test_same_io_qnn_params()\n test_different_io_qnn_params()\n test_few_same_io_qnn_params()\n test_same_i_qnn_params()\n"
] |
[
[
"numpy.concatenate",
"numpy.arange",
"numpy.power"
]
] |
mgwillia/unsupervised-analysis
|
[
"66b5c7af391199394ace3a1ff396d46ac5bd7ce5"
] |
[
"datasets/neighbors_dataset.py"
] |
[
"\"\"\" \n NeighborsDataset\n Returns an image with one of its neighbors.\n\"\"\"\nimport numpy as np\nimport torch\nfrom torch.utils.data import Dataset\n\n\nclass NeighborsDataset(Dataset):\n def __init__(self, dataset, indices, num_neighbors=None):\n super(NeighborsDataset, self).__init__()\n\n self.dataset = dataset\n self.indices = indices\n if num_neighbors is not None:\n self.indices = self.indices[:, :num_neighbors+1]\n print(self.indices.shape, len(self.dataset))\n assert(self.indices.shape[0] == len(self.dataset))\n\n\n def __len__(self):\n return len(self.dataset)\n\n\n def __getitem__(self, index):\n output = {}\n anchor = self.dataset.__getitem__(index)\n \n neighbor_index = np.random.choice(self.indices[index], 1)[0]\n neighbor = self.dataset.__getitem__(neighbor_index)\n\n output['anchor_features'] = anchor['features']\n output['neighbor_features'] = neighbor['features'] \n output['possible_neighbors'] = torch.from_numpy(self.indices[index])\n output['target'] = anchor['target']\n \n return output\n"
] |
[
[
"numpy.random.choice",
"torch.from_numpy"
]
] |
rutgers-apl/rlibm-all
|
[
"b014190b223d2d4842a5cf8235eeff3aa6cdd573"
] |
[
"SpeedupOverRlibm32.py"
] |
[
"import matplotlib\nimport matplotlib.pyplot as plt\nimport matplotlib.ticker as ticker\nimport numpy as np\n\nmatplotlib.rcParams['pdf.fonttype'] = 42\nmatplotlib.rcParams['ps.fonttype'] = 42\n\noverallLabel = [\"ln\", \"log2\", \"log10\", \"exp\", \"exp2\", \"exp10\", \"sinh\", \"cosh\", \"sinpi\", \"cospi\", \"avg.\"]\n\nlabels = overallLabel\n# Here we have to set up the values\nglibcFloatTime = []\nrlibmRnoTime = []\nagainstFloat = []\n\nfileName = [\"log.txt\", \"log2.txt\", \"log10.txt\", \"exp.txt\", \"exp2.txt\", \"exp10.txt\", \"sinh.txt\", \"cosh.txt\", \"sinpi.txt\", \"cospi.txt\"]\n\nfor i in range(0, len(fileName)) :\n fp = open(\"overhead_test/glibc_rlibm32/\" + fileName[i], \"r\")\n line = fp.readline()\n line = line.split(\",\")\n glibcFloatTime.append(float(line[0]))\n fp.close()\nglibcFloatTime.append(sum(glibcFloatTime))\n \nfor i in range(0, len(fileName)) :\n fp = open(\"overhead_test/glibc_rlibm_generic/\" + fileName[i], \"r\")\n line = fp.readline()\n line = line.split(\",\")\n rlibmRnoTime.append(float(line[0]))\n fp.close()\nrlibmRnoTime.append(sum(rlibmRnoTime))\n\nfor i in range(0, len(overallLabel)) :\n againstFloat.append(glibcFloatTime[i] / rlibmRnoTime[i])\n\nprint(againstFloat)\n\nx = np.arange(len(labels)) # the label locations\nwidth = 0.35 # the width of the bars\n\nfig, ax = plt.subplots()\nax.set_ylim(0, 2.5)\nax.axhline(0.5, color=\"gray\", ls = '--', linewidth = 0.5, zorder = 0)\nax.axhline(1, color=\"k\", ls = '--', linewidth = 1, zorder = 0)\nax.axhline(1.5, color=\"gray\", ls = '--', linewidth = 0.5, zorder = 0)\nax.axhline(2.0, color=\"k\", ls = '--', linewidth = 1, zorder = 0)\nrects1 = ax.bar(x, againstFloat, width, color=\"tab:orange\", label='Speedup over float libm', zorder = 100)\n\n# Add some text for labels, title and custom x-axis tick labels, etc.\n#plt.xticks(rotation=15, ha=\"right\", rotation_mode=\"anchor\")\nax.set_ylabel('Speedup')\n#ax.set_title('Performance speedup')\nax.set_xticks(x)\nax.set_xticklabels(labels)\nax.legend(bbox_to_anchor=(-0.1, 0.98, 1.1, 0.2), loc=\"lower left\", mode=\"expand\", ncol=2)\nax.yaxis.set_major_formatter(ticker.FormatStrFormatter('$%.f x$'))\n\nfig.tight_layout()\n\nplt.gcf().set_size_inches(5, 1.2)\nplt.savefig('SpeedupOverRlibm32.pdf', bbox_inches='tight', pad_inches = -0.001)\n"
] |
[
[
"matplotlib.pyplot.savefig",
"matplotlib.pyplot.gcf",
"matplotlib.pyplot.subplots",
"matplotlib.ticker.FormatStrFormatter"
]
] |
ArshdeepSahni/MonoPort
|
[
"a67fdc02b4fb45b3cc187aa4ae34053574d0383c"
] |
[
"monoport/lib/render/gl/Render.py"
] |
[
"import numpy as np\nfrom .Shader import *\n\n\nclass Render(object):\n def __init__(self,\n width, height,\n multi_sample_rate=1,\n num_render_target=1\n ):\n self.width = width\n self.height = height\n\n self.vbo_list = []\n self.uniform_dict = {}\n self.texture_dict = {}\n\n # Configure frame buffer\n # This is an off screen frame buffer holds the rendering results.\n # During display, it is drawn onto the screen by a quad program.\n self.color_fbo = FBO()\n self.color_tex_list = []\n\n with self.color_fbo:\n for i in range(num_render_target):\n color_tex = Texture()\n self.color_tex_list.append(color_tex)\n with color_tex:\n gl.glTexImage2D(gl.GL_TEXTURE_2D, 0, gl.GL_RGBA32F, width, height, 0,\n gl.GL_RGBA, gl.GL_FLOAT, None)\n gl.glFramebufferTexture2D(gl.GL_FRAMEBUFFER, gl.GL_COLOR_ATTACHMENT0 + i,\n gl.GL_TEXTURE_2D, color_tex, 0)\n gl.glViewport(0, 0, width, height)\n assert gl.glCheckFramebufferStatus(gl.GL_FRAMEBUFFER) == gl.GL_FRAMEBUFFER_COMPLETE\n\n # This is the actual frame buffer the shader program is rendered to.\n # For multi-sampling, it is different than the default fbo.\n self._shader_fbo = self.color_fbo\n self._shader_color_tex_list = self.color_tex_list\n\n # However, if multi-sampling is enabled, we need additional render target textures.\n if multi_sample_rate > 1:\n self._shader_fbo = FBO()\n self._shader_color_tex_list = []\n with self._shader_fbo:\n for i in range(num_render_target):\n color_tex = MultiSampleTexture()\n self._shader_color_tex_list.append(color_tex)\n with color_tex:\n gl.glTexImage2DMultisample(gl.GL_TEXTURE_2D_MULTISAMPLE, multi_sample_rate, gl.GL_RGBA32F,\n width, height, gl.GL_TRUE)\n gl.glFramebufferTexture2D(gl.GL_FRAMEBUFFER, gl.GL_COLOR_ATTACHMENT0 + i,\n gl.GL_TEXTURE_2D_MULTISAMPLE, color_tex, 0)\n\n # Configure depth buffer\n self._shader_depth_tex = RBO()\n with self._shader_fbo:\n with self._shader_depth_tex:\n gl.glRenderbufferStorageMultisample(gl.GL_RENDERBUFFER, multi_sample_rate,\n gl.GL_DEPTH24_STENCIL8, width, height)\n gl.glFramebufferRenderbuffer(gl.GL_FRAMEBUFFER, gl.GL_DEPTH_STENCIL_ATTACHMENT,\n gl.GL_RENDERBUFFER, self._shader_depth_tex)\n\n self._init_shader()\n for uniform in self.uniform_dict:\n self.uniform_dict[uniform][\"handle\"] = gl.glGetUniformLocation(self.shader, uniform)\n\n def _init_shader(self):\n self.shader = Shader(vs_file='simple.vs', fs_file='simple.fs', gs_file=None)\n # layout (location = 0) in vec3 Position;\n self.vbo_list.append(VBO(type_code='f', gl_type=gl.GL_FLOAT))\n\n # Declare all uniform used in the program\n self.shader.declare_uniform('ModelMat', type_code='f', gl_type=gl.glUniformMatrix4fv)\n self.shader.declare_uniform('PerspMat', type_code='f', gl_type=gl.glUniformMatrix4fv)\n\n def set_attrib(self, attrib_id, data):\n if not 0 <= attrib_id < len(self.vbo_list):\n print(\"Error: Attrib index out if bound.\")\n return\n vbo = self.vbo_list[attrib_id]\n with vbo:\n data = np.ascontiguousarray(data, vbo.type_code)\n vbo.dim = data.shape[-1]\n vbo.size = data.shape[0]\n gl.glBufferData(gl.GL_ARRAY_BUFFER, data, gl.GL_STATIC_DRAW)\n\n def set_texture(self, name, texture_image):\n if name not in self.texture_dict:\n print(\"Error: Unknown texture name.\")\n return\n width = texture_image.shape[1]\n height = texture_image.shape[0]\n texture_image = np.flip(texture_image, 0)\n img_data = np.fromstring(texture_image.tostring(), np.uint8)\n tex = self.texture_dict[name]\n with tex:\n gl.glTexImage2D(gl.GL_TEXTURE_2D, 0, gl.GL_RGB, width, height, 0, gl.GL_RGB, gl.GL_UNSIGNED_BYTE, img_data)\n gl.glTexParameteri(gl.GL_TEXTURE_2D, gl.GL_TEXTURE_MAX_LEVEL, 3)\n gl.glGenerateMipmap(gl.GL_TEXTURE_2D)\n\n def draw(self,\n uniform_dict,\n clear_color=[0, 0, 0, 0],\n ):\n with self._shader_fbo:\n # Clean up\n gl.glClearColor(*clear_color)\n gl.glClear(gl.GL_COLOR_BUFFER_BIT | gl.GL_DEPTH_BUFFER_BIT)\n\n with self.shader:\n # Setup shader uniforms\n for uniform_name in uniform_dict:\n self.shader.set_uniform(uniform_name, uniform_dict[uniform_name])\n\n # Setup up VertexAttrib\n for attrib_id in range(len(self.vbo_list)):\n vbo = self.vbo_list[attrib_id]\n with vbo:\n gl.glEnableVertexAttribArray(attrib_id)\n gl.glVertexAttribPointer(attrib_id, vbo.dim, vbo.gl_type, gl.GL_FALSE, 0, None)\n\n # Setup Textures\n for i, texture_name in enumerate(self.texture_dict):\n gl.glActiveTexture(gl.GL_TEXTURE0 + i)\n gl.glBindTexture(gl.GL_TEXTURE_2D, self.texture_dict[texture_name])\n gl.glUniform1i(gl.glGetUniformLocation(self.shader, texture_name), i)\n\n # Setup targets\n color_size = len(self.color_tex_list)\n attachments = [gl.GL_COLOR_ATTACHMENT0 + i for i in range(color_size)]\n gl.glDrawBuffers(color_size, attachments)\n\n gl.glDrawArrays(gl.GL_TRIANGLES, 0, self.vbo_list[0].size)\n\n for attrib_id in range(len(self.vbo_list)):\n gl.glDisableVertexAttribArray(attrib_id)\n\n # If render_fbo is not color_fbo, we need to copy data\n if self._shader_fbo != self.color_fbo:\n for i in range(len(self.color_tex_list)):\n gl.glBindFramebuffer(gl.GL_READ_FRAMEBUFFER, self._shader_fbo)\n gl.glReadBuffer(gl.GL_COLOR_ATTACHMENT0 + i)\n gl.glBindFramebuffer(gl.GL_DRAW_FRAMEBUFFER, self.color_fbo)\n gl.glDrawBuffer(gl.GL_COLOR_ATTACHMENT0 + i)\n gl.glBlitFramebuffer(0, 0, self.width, self.height, 0, 0, self.width, self.height,\n gl.GL_COLOR_BUFFER_BIT, gl.GL_NEAREST)\n gl.glBindFramebuffer(gl.GL_READ_FRAMEBUFFER, 0)\n gl.glBindFramebuffer(gl.GL_DRAW_FRAMEBUFFER, 0)\n\n def get_color(self, color_id=0):\n with self.color_fbo:\n gl.glReadBuffer(gl.GL_COLOR_ATTACHMENT0 + color_id)\n data = gl.glReadPixels(0, 0, self.width, self.height, gl.GL_RGBA, gl.GL_FLOAT, outputType=None)\n frame = data.reshape(self.height, self.width, -1)\n frame = frame[::-1] # vertical flip to match GL convention\n return frame\n"
] |
[
[
"numpy.ascontiguousarray",
"numpy.flip"
]
] |
fabiotabalipa/cnes-hospitais
|
[
"7e98e75fa5ed0808b4de5257ec258441a8b93211"
] |
[
"transform/cnes.py"
] |
[
"import pandas as pd\nimport warnings\n\nPREFIX_MAIN_TBL = \"tbEstabelecimento\"\nPREFIX_BEDS_TBL = \"rlEstabComplementar\"\nPREFIX_INSURANCE_TBL = \"rlEstabAtendPrestConv\"\nPREFIX_CITY_TBL = \"tbMunicipio\"\n\nCOD_GENERAL_HOSPITAL = 5\nCOD_SPECIALIZED_HOSPITAL = 7\n\nCOD_INSURANCE_OWN = 3\nCOD_INSURANCE_THIRD = 4\nCOD_INSURANCE_PRIVATE = 5\nCOD_INSURANCE_PUBLIC = 6\n\n\ndef get_transformed_df(files_dir, version):\n warnings.filterwarnings(\"ignore\")\n\n file_main = files_dir + \"/\" + PREFIX_MAIN_TBL + version + \".csv\"\n df_main = pd.read_csv(file_main, sep=\";\", dtype={\n \"CO_UNIDADE\": str,\n \"CO_CNES\": str,\n \"NU_CNPJ_MANTENEDORA\": str,\n \"CO_MUNICIPIO_GESTOR\": str,\n \"CO_CEP\": str,\n \"NU_TELEFONE\": str,\n })\n df_main = df_main.drop(df_main[\n (df_main['TP_UNIDADE'] != COD_GENERAL_HOSPITAL) &\n (df_main['TP_UNIDADE'] != COD_SPECIALIZED_HOSPITAL)\n ].index)\n df_main = df_main[[\n \"CO_UNIDADE\",\n \"CO_CNES\",\n \"NU_CNPJ_MANTENEDORA\",\n \"NO_RAZAO_SOCIAL\",\n \"NO_FANTASIA\",\n \"CO_MUNICIPIO_GESTOR\",\n \"CO_CEP\",\n \"NU_TELEFONE\",\n \"NO_EMAIL\",\n ]]\n df_main = df_main.rename({\"CO_MUNICIPIO_GESTOR\": \"CO_MUNICIPIO\"}, axis=1)\n df_main[\"NO_EMAIL\"] = df_main[\"NO_EMAIL\"].str.lower()\n\n file_city = files_dir + \"/\" + PREFIX_CITY_TBL + version + \".csv\"\n df_city = pd.read_csv(file_city, sep=\";\", dtype={\n \"CO_MUNICIPIO\": str,\n })\n df_city = df_city[[\n \"CO_MUNICIPIO\",\n \"NO_MUNICIPIO\",\n \"CO_SIGLA_ESTADO\",\n ]]\n df_city = df_city.groupby(by=\"CO_MUNICIPIO\").agg({\n \"NO_MUNICIPIO\": \"last\",\n \"CO_SIGLA_ESTADO\": \"last\",\n }).reset_index()\n\n file_beds = files_dir + \"/\" + PREFIX_BEDS_TBL + version + \".csv\"\n df_beds = pd.read_csv(file_beds, sep=\";\")\n df_beds = df_beds[[\n \"CO_UNIDADE\",\n \"QT_EXIST\",\n \"QT_SUS\",\n ]]\n df_beds[\"QT_SUS\"] = df_beds.apply(lambda row: 1 if row[\"QT_SUS\"] > 0 else 0, axis=1)\n df_beds = df_beds.groupby(by=\"CO_UNIDADE\").agg({\n \"QT_EXIST\": \"sum\",\n \"QT_SUS\": \"max\",\n }).reset_index()\n\n file_insurance = files_dir + \"/\" + PREFIX_INSURANCE_TBL + version + \".csv\"\n df_insurance = pd.read_csv(file_insurance, sep=\";\")\n df_insurance = df_insurance.drop(df_insurance[\n (df_insurance['CO_CONVENIO'] != COD_INSURANCE_OWN) &\n (df_insurance['CO_CONVENIO'] != COD_INSURANCE_THIRD) &\n (df_insurance['CO_CONVENIO'] != COD_INSURANCE_PRIVATE) &\n (df_insurance['CO_CONVENIO'] != COD_INSURANCE_PUBLIC)\n ].index)\n df_insurance = df_insurance[[\n \"CO_UNIDADE\",\n ]]\n df_insurance[\"Atende Convênio?\"] = 1\n df_insurance = df_insurance.groupby(by=\"CO_UNIDADE\").agg({\n \"Atende Convênio?\": \"max\",\n }).reset_index()\n\n df_merge = df_main.merge(df_beds, how=\"inner\", on=\"CO_UNIDADE\")\n df_merge = df_merge.merge(df_insurance, how=\"left\", on=\"CO_UNIDADE\")\n df_merge = df_merge.merge(df_city, how=\"left\", on=\"CO_MUNICIPIO\")\n df_merge[\"Atende Convênio?\"] = df_merge[\"Atende Convênio?\"].fillna(0)\n df_merge[\"Atende Convênio?\"] = df_merge[\"Atende Convênio?\"].astype(int)\n\n df_merge = df_merge.rename({\n \"CO_CNES\": \"Código CNES\",\n \"NU_CNPJ_MANTENEDORA\": \"CNPJ\",\n \"NO_RAZAO_SOCIAL\": \"Razão Social\",\n \"NO_FANTASIA\": \"Nome Fantasia\",\n \"CO_CEP\": \"CEP\",\n \"NU_TELEFONE\": \"Telefone\",\n \"NO_EMAIL\": \"Email\",\n \"QT_EXIST\": \"Leitos\",\n \"QT_SUS\": \"Atende SUS?\",\n \"NO_MUNICIPIO\": \"Município\",\n \"CO_SIGLA_ESTADO\": \"UF\",\n }, axis=1)\n\n df_merge = df_merge[[\n \"Nome Fantasia\",\n \"Razão Social\",\n \"CNPJ\",\n \"Código CNES\",\n \"Município\",\n \"UF\",\n \"CEP\",\n \"Telefone\",\n \"Email\",\n \"Leitos\",\n \"Atende SUS?\",\n \"Atende Convênio?\",\n ]]\n return df_merge.sort_values(by=[\"UF\"])\n"
] |
[
[
"pandas.read_csv"
]
] |
p-patil/baselines
|
[
"72d6757cfe88521012a30e8f38c4b313455da395"
] |
[
"baselines/deepq/deepq.py"
] |
[
"import os\nimport tempfile\n\nimport tensorflow as tf\nimport zipfile\nimport cloudpickle\nimport numpy as np\n\nimport baselines.common.tf_util as U\nfrom baselines.common.tf_util import load_variables, save_variables\nfrom baselines import logger\nfrom baselines.common.schedules import LinearSchedule\nfrom baselines.common import set_global_seeds\n\nfrom baselines import deepq\nfrom baselines.deepq.replay_buffer import ReplayBuffer, PrioritizedReplayBuffer\nfrom baselines.deepq.utils import ObservationInput\n\nfrom baselines.common.tf_util import get_session\nfrom baselines.deepq.models import build_q_func\n\n\nclass ActWrapper(object):\n def __init__(self, act, act_params):\n self._act = act\n self._act_params = act_params\n self.initial_state = None\n\n @staticmethod\n def load_act(path):\n with open(path, \"rb\") as f:\n model_data, act_params = cloudpickle.load(f)\n act = deepq.build_act(**act_params)\n sess = tf.Session()\n sess.__enter__()\n with tempfile.TemporaryDirectory() as td:\n arc_path = os.path.join(td, \"packed.zip\")\n with open(arc_path, \"wb\") as f:\n f.write(model_data)\n\n zipfile.ZipFile(arc_path, 'r', zipfile.ZIP_DEFLATED).extractall(td)\n load_variables(os.path.join(td, \"model\"))\n\n return ActWrapper(act, act_params)\n\n def __call__(self, *args, **kwargs):\n return self._act(*args, **kwargs)\n\n def step(self, observation, **kwargs):\n # DQN doesn't use RNNs so we ignore states and masks\n kwargs.pop('S', None)\n kwargs.pop('M', None)\n return self._act([observation], **kwargs), None, None, None\n\n def save_act(self, path=None):\n \"\"\"Save model to a pickle located at `path`\"\"\"\n if path is None:\n path = os.path.join(logger.get_dir(), \"model.pkl\")\n\n with tempfile.TemporaryDirectory() as td:\n save_variables(os.path.join(td, \"model\"))\n arc_name = os.path.join(td, \"packed.zip\")\n with zipfile.ZipFile(arc_name, 'w') as zipf:\n for root, dirs, files in os.walk(td):\n for fname in files:\n file_path = os.path.join(root, fname)\n if file_path != arc_name:\n zipf.write(file_path, os.path.relpath(file_path, td))\n with open(arc_name, \"rb\") as f:\n model_data = f.read()\n with open(path, \"wb\") as f:\n cloudpickle.dump((model_data, self._act_params), f)\n\n def save(self, path):\n save_variables(path)\n\n\ndef load_act(path):\n \"\"\"Load act function that was returned by learn function.\n\n Parameters\n ----------\n path: str\n path to the act function pickle\n\n Returns\n -------\n act: ActWrapper\n function that takes a batch of observations\n and returns actions.\n \"\"\"\n return ActWrapper.load_act(path)\n\n\ndef learn(env,\n network,\n seed=None,\n lr=5e-4,\n total_timesteps=100000,\n buffer_size=50000,\n exploration_fraction=0.1,\n exploration_final_eps=0.02,\n train_freq=1,\n batch_size=32,\n print_freq=100,\n checkpoint_freq=10000,\n checkpoint_path=None,\n learning_starts=1000,\n gamma=1.0,\n target_network_update_freq=500,\n prioritized_replay=False,\n prioritized_replay_alpha=0.6,\n prioritized_replay_beta0=0.4,\n prioritized_replay_beta_iters=None,\n prioritized_replay_eps=1e-6,\n param_noise=False,\n callback=None,\n load_path=None,\n **network_kwargs\n ):\n \"\"\"Train a deepq model.\n\n Parameters\n -------\n env: gym.Env\n environment to train on\n network: string or a function\n neural network to use as a q function approximator. If string, has to be one of the names of registered models in baselines.common.models\n (mlp, cnn, conv_only). If a function, should take an observation tensor and return a latent variable tensor, which\n will be mapped to the Q function heads (see build_q_func in baselines.deepq.models for details on that)\n seed: int or None\n prng seed. The runs with the same seed \"should\" give the same results. If None, no seeding is used.\n lr: float\n learning rate for adam optimizer\n total_timesteps: int\n number of env steps to optimizer for\n buffer_size: int\n size of the replay buffer\n exploration_fraction: float\n fraction of entire training period over which the exploration rate is annealed\n exploration_final_eps: float\n final value of random action probability\n train_freq: int\n update the model every `train_freq` steps.\n batch_size: int\n size of a batch sampled from replay buffer for training\n print_freq: int\n how often to print out training progress\n set to None to disable printing\n checkpoint_freq: int\n how often to save the model. This is so that the best version is restored\n at the end of the training. If you do not wish to restore the best version at\n the end of the training set this variable to None.\n learning_starts: int\n how many steps of the model to collect transitions for before learning starts\n gamma: float\n discount factor\n target_network_update_freq: int\n update the target network every `target_network_update_freq` steps.\n prioritized_replay: True\n if True prioritized replay buffer will be used.\n prioritized_replay_alpha: float\n alpha parameter for prioritized replay buffer\n prioritized_replay_beta0: float\n initial value of beta for prioritized replay buffer\n prioritized_replay_beta_iters: int\n number of iterations over which beta will be annealed from initial value\n to 1.0. If set to None equals to total_timesteps.\n prioritized_replay_eps: float\n epsilon to add to the TD errors when updating priorities.\n param_noise: bool\n whether or not to use parameter space noise (https://arxiv.org/abs/1706.01905)\n callback: (locals, globals) -> None\n function called at every steps with state of the algorithm.\n If callback returns true training stops.\n load_path: str\n path to load the model from. (default: None)\n **network_kwargs\n additional keyword arguments to pass to the network builder.\n\n Returns\n -------\n act: ActWrapper\n Wrapper over act function. Adds ability to save it and load it.\n See header of baselines/deepq/categorical.py for details on the act function.\n \"\"\"\n # Create all the functions necessary to train the model\n\n sess = get_session()\n set_global_seeds(seed)\n\n q_func = build_q_func(network, **network_kwargs)\n\n # capture the shape outside the closure so that the env object is not serialized\n # by cloudpickle when serializing make_obs_ph\n\n observation_space = env.observation_space\n def make_obs_ph(name):\n return ObservationInput(observation_space, name=name)\n\n act, train, update_target, debug = deepq.build_train(\n make_obs_ph=make_obs_ph,\n q_func=q_func,\n num_actions=env.action_space.n,\n optimizer=tf.train.AdamOptimizer(learning_rate=lr),\n gamma=gamma,\n grad_norm_clipping=10,\n param_noise=param_noise\n )\n\n act_params = {\n 'make_obs_ph': make_obs_ph,\n 'q_func': q_func,\n 'num_actions': env.action_space.n,\n }\n\n act = ActWrapper(act, act_params)\n\n # Create the replay buffer\n if prioritized_replay:\n replay_buffer = PrioritizedReplayBuffer(buffer_size, alpha=prioritized_replay_alpha)\n if prioritized_replay_beta_iters is None:\n prioritized_replay_beta_iters = total_timesteps\n beta_schedule = LinearSchedule(prioritized_replay_beta_iters,\n initial_p=prioritized_replay_beta0,\n final_p=1.0)\n else:\n replay_buffer = ReplayBuffer(buffer_size)\n beta_schedule = None\n # Create the schedule for exploration starting from 1.\n exploration = LinearSchedule(schedule_timesteps=int(exploration_fraction * total_timesteps),\n initial_p=1.0,\n final_p=exploration_final_eps)\n\n # Initialize the parameters and copy them to the target network.\n U.initialize()\n update_target()\n\n episode_rewards = [0.0]\n saved_mean_reward = None\n obs = env.reset()\n reset = True\n\n print(\"USING PRINT FREQ 1\")\n print_freq = 1 # TODO(piyush) remove\n\n with tempfile.TemporaryDirectory() as td:\n td = checkpoint_path or td\n\n model_file = os.path.join(td, \"model\")\n model_saved = False\n\n if tf.train.latest_checkpoint(td) is not None:\n load_variables(model_file)\n logger.log('Loaded model from {}'.format(model_file))\n model_saved = True\n elif load_path is not None:\n load_variables(load_path)\n logger.log('Loaded model from {}'.format(load_path))\n\n\n for t in range(total_timesteps):\n if callback is not None:\n if callback(locals(), globals()):\n break\n # Take action and update exploration to the newest value\n kwargs = {}\n if not param_noise:\n update_eps = exploration.value(t)\n update_param_noise_threshold = 0.\n else:\n update_eps = 0.\n # Compute the threshold such that the KL divergence between perturbed and non-perturbed\n # policy is comparable to eps-greedy exploration with eps = exploration.value(t).\n # See Appendix C.1 in Parameter Space Noise for Exploration, Plappert et al., 2017\n # for detailed explanation.\n update_param_noise_threshold = -np.log(1. - exploration.value(t) + exploration.value(t) / float(env.action_space.n))\n kwargs['reset'] = reset\n kwargs['update_param_noise_threshold'] = update_param_noise_threshold\n kwargs['update_param_noise_scale'] = True\n action = act(np.array(obs)[None], update_eps=update_eps, **kwargs)[0]\n env_action = action\n reset = False\n new_obs, rew, done, _ = env.step(env_action)\n # Store transition in the replay buffer.\n replay_buffer.add(obs, action, rew, new_obs, float(done))\n obs = new_obs\n\n episode_rewards[-1] += rew\n if done:\n obs = env.reset()\n episode_rewards.append(0.0)\n reset = True\n\n # TODO(piyush) Train swin transformer to predict next env state\n\n\n if t > learning_starts and t % train_freq == 0:\n # Minimize the error in Bellman's equation on a batch sampled from replay buffer.\n if prioritized_replay:\n experience = replay_buffer.sample(batch_size, beta=beta_schedule.value(t))\n (obses_t, actions, rewards, obses_tp1, dones, weights, batch_idxes) = experience\n else:\n obses_t, actions, rewards, obses_tp1, dones = replay_buffer.sample(batch_size)\n weights, batch_idxes = np.ones_like(rewards), None\n td_errors = train(obses_t, actions, rewards, obses_tp1, dones, weights)\n if prioritized_replay:\n new_priorities = np.abs(td_errors) + prioritized_replay_eps\n replay_buffer.update_priorities(batch_idxes, new_priorities)\n\n if t > learning_starts and t % target_network_update_freq == 0:\n # Update target network periodically.\n update_target()\n\n mean_100ep_reward = round(np.mean(episode_rewards[-101:-1]), 1)\n num_episodes = len(episode_rewards)\n if done and print_freq is not None and len(episode_rewards) % print_freq == 0:\n logger.record_tabular(\"steps\", t)\n logger.record_tabular(\"episodes\", num_episodes)\n logger.record_tabular(\"mean 100 episode reward\", mean_100ep_reward)\n logger.record_tabular(\"% time spent exploring\", int(100 * exploration.value(t)))\n logger.dump_tabular()\n\n if (checkpoint_freq is not None and t > learning_starts and\n num_episodes > 100 and t % checkpoint_freq == 0):\n if saved_mean_reward is None or mean_100ep_reward > saved_mean_reward:\n if print_freq is not None:\n logger.log(\"Saving model due to mean reward increase: {} -> {}\".format(\n saved_mean_reward, mean_100ep_reward))\n save_variables(model_file)\n model_saved = True\n saved_mean_reward = mean_100ep_reward\n if model_saved:\n if print_freq is not None:\n logger.log(\"Restored model with mean reward: {}\".format(saved_mean_reward))\n load_variables(model_file)\n\n return act\n"
] |
[
[
"numpy.array",
"numpy.ones_like",
"tensorflow.train.AdamOptimizer",
"tensorflow.train.latest_checkpoint",
"tensorflow.Session",
"numpy.mean",
"numpy.abs"
]
] |
psteinb/b3get
|
[
"70bfd37881bdef2ecd6a7bbfc105a001b3e97749"
] |
[
"tests/test_datasets.py"
] |
[
"import os\nimport requests\nimport shutil\nimport zipfile\nimport glob\nimport tifffile\nimport numpy as np\n\nfrom bs4 import BeautifulSoup\nfrom b3get.utils import filter_files, tmp_location\nfrom b3get.datasets import dataset, ds_006, ds_008, ds_024, ds_027\nimport pytest\n\n# manual tests for exploration\n\n\ndef test_006_images_manual():\n\n r = requests.get(\"https://data.broadinstitute.org/bbbc/BBBC006/\")\n hdoc = BeautifulSoup(r.text, 'html.parser')\n all_links = hdoc.find_all('a')\n values = []\n for anc in all_links:\n href = anc.get('href')\n if len(href) > 0 and \"zip\" in href and \"images\" in href:\n values.append(href)\n\n assert len(values) > 0\n assert len(values) == 34\n\n\ndef test_006_construction():\n ds6 = ds_006(\"https://data.broadinstitute.org/bbbc/BBBC006/\")\n assert ds6.baseurl\n assert 'BBBC006' in ds6.baseurl\n assert ds6.title()\n assert 'Human' in ds6.title()\n\n ds6_empty = ds_006()\n assert ds6_empty.baseurl\n assert 'BBBC006' in ds6_empty.baseurl\n assert ds6_empty.title()\n assert 'Human' in ds6_empty.title()\n\n\ndef test_006_wrong_URL():\n with pytest.raises(RuntimeError):\n ds = dataset(None)\n\n with pytest.raises(RuntimeError):\n ds = dataset(\"https://data.broadinstitute.org/bbbc/BBC027/\")\n\n with pytest.raises(RuntimeError):\n ds = ds_006(\"https://data.broadinstitute.org/bbbc/BBC027/\")\n\n\ndef test_027_construction():\n ds27 = ds_027(\"https://data.broadinstitute.org/bbbc/BBBC027/\")\n assert ds27.baseurl\n assert 'BBBC027' in ds27.baseurl\n assert ds27.title()\n assert '3D Colon' in ds27.title()\n\n ds27_empty = ds_027()\n assert ds27_empty.baseurl\n assert 'BBBC027' in ds27_empty.baseurl\n assert ds27_empty.title()\n assert '3D Colon' in ds27_empty.title()\n\n\ndef test_006_list_images():\n ds6 = ds_006(\"https://data.broadinstitute.org/bbbc/BBBC006/\")\n imgs = ds6.list_images()\n assert len(imgs) > 0\n assert len(imgs) == 34\n ds6 = ds_006(datasetid=6)\n imgs = ds6.list_images()\n assert len(imgs) > 0\n assert len(imgs) == 34\n\n\ndef test_006_list_images_from_datasetid():\n ds = dataset(datasetid=14)\n imgs = ds.list_images()\n assert len(imgs) > 0\n\n\ndef test_027_list_images():\n ds = ds_027(\"https://data.broadinstitute.org/bbbc/BBBC027/\")\n imgs = ds.list_images()\n assert len(imgs) > 0\n assert len(imgs) == 6\n\n\ndef test_008_pull_single():\n ds8 = ds_008()\n imgs = ds8.list_images()\n assert len(imgs) > 0\n few = filter_files(imgs, '.*.zip')\n assert len(few) == 1\n downed = ds8.pull_images('.*.zip')\n assert downed\n assert len(downed) > 0\n assert os.path.exists(downed[0])\n shutil.rmtree(tmp_location())\n\n\ndef test_008_list_gt():\n ds8 = ds_008()\n imgs = ds8.list_gt()\n assert len(imgs) > 0\n assert len(imgs) == 1\n assert \"BBBC008_v1_foreground.zip\" in imgs\n urls = ds8.list_gt(True)\n assert len(urls) > 0\n\n\ndef test_008_pull_gt():\n ds8 = ds_008()\n imgs = ds8.pull_gt()\n assert len(imgs) > 0\n assert len(imgs) == 1\n assert \"BBBC008_v1_foreground.zip\" in [os.path.split(item)[-1] for item in imgs]\n shutil.rmtree(tmp_location())\n\n\ndef test_008_extract_gt_manual():\n ds8 = ds_008()\n imgs = ds8.pull_gt()\n assert len(imgs) > 0\n assert len(imgs) == 1\n assert \"BBBC008_v1_foreground.zip\" in [os.path.split(item)[-1] for item in imgs]\n with zipfile.ZipFile(imgs[0], 'r') as zf:\n print('extracting ',imgs[0])\n zf.extractall(os.path.split(imgs[0])[0])\n zf.close()\n path = os.path.split(imgs[0])[0]\n xpath = os.path.join(path, \"human_ht29_colon_cancer_2_foreground\")\n assert os.path.exists(xpath)\n assert os.path.isdir(xpath)\n extracted = glob.glob(os.path.join(xpath, \"*.tif\"))\n assert len(extracted) > 0\n assert len(extracted) == 24 # channel 2 is not contained\n shutil.rmtree(tmp_location())\n\n\ndef test_008_extract_gt():\n shutil.rmtree(tmp_location())\n ds8 = ds_008()\n imgs = ds8.pull_gt()\n assert len(imgs) > 0\n assert len(imgs) == 1\n print(imgs)\n xtracted = ds8.extract_gt()\n assert xtracted\n assert len(xtracted) > 0\n assert len(xtracted) == 24+1 # +1 for .DS_Store file contained\n shutil.rmtree(tmp_location())\n\n\ndef test_006_list_gt():\n ds6 = ds_006()\n imgs = ds6.list_gt()\n assert len(imgs) > 0\n assert len(imgs) == 1\n assert \"BBBC006_v1_labels.zip\" in imgs\n\n\ndef test_024_list():\n ds = ds_024()\n imgs = ds.list_images()\n assert len(imgs) > 0\n gt = ds.list_gt()\n assert len(gt) > 0\n\n\ndef test_008_extract_images():\n ds = ds_008()\n _ = ds.pull_images()\n ximgs = ds.extract_images()\n assert len(ximgs) > 0\n assert len(ximgs) == 24+3 # +3 for .DS_Store and similar files contained\n shutil.rmtree(ds.tmp_location)\n\n\ndef test_008_extract_images_nprocs2():\n ds = ds_008()\n _ = ds.pull_images()\n ximgs = ds.extract_images(folder=None, nprocs=2)\n assert len(ximgs) > 0\n assert len(ximgs) == 24+3 # +3 for .DS_Store and similar files contained\n shutil.rmtree(ds.tmp_location)\n\n\ndef test_008_extract_gt_nprocs2():\n ds = ds_008()\n _ = ds.pull_gt()\n ximgs = ds.extract_gt(folder=None, nprocs=2)\n assert len(ximgs) > 0\n assert len(ximgs) == 24+1 # +1 for .DS_Store file contained\n shutil.rmtree(ds.tmp_location)\n\n\ndef test_008_files_to_numpy():\n ds = ds_008()\n _ = ds.pull_images()\n ximgs = ds.extract_images()\n assert len(ximgs) > 0\n assert len(ximgs) == 24+3 # +3 for .DS_Store and similar files contained\n\n ximgs = [item for item in ximgs if item.count('.tif')]\n first = tifffile.imread(ximgs[0])\n last = tifffile.imread(ximgs[-1])\n\n npimgs = ds.files_to_numpy(ximgs)\n assert len(npimgs) == len(ximgs)\n assert npimgs[0].shape == first.shape\n assert isinstance(npimgs[0], np.ndarray)\n assert np.array_equal(npimgs[0][:100], first[:100])\n assert np.array_equal(npimgs[-1][:100], last[:100])\n assert npimgs[0].shape == (512, 512)\n shutil.rmtree(ds.tmp_location)\n\n\ndef test_008_ds_images_to_numpy():\n ds = ds_008()\n imgs = ds.images_to_numpy()\n assert len(imgs) > 0\n assert len(imgs) == 24\n assert np.all([item.shape == (512, 512) for item in imgs])\n\n imgs_plus_fnames = ds.images_to_numpy(include_filenames=True)\n assert len(imgs_plus_fnames) == len(imgs)\n assert os.path.isfile(imgs_plus_fnames[0][-1])\n shutil.rmtree(ds.tmp_location)\n\n\ndef test_008_ds_gt_to_numpy():\n ds = ds_008()\n labs = ds.gt_to_numpy()\n assert len(labs) > 0\n assert len(labs) == 24\n assert np.all([item.shape == (512, 512) for item in labs])\n shutil.rmtree(ds.tmp_location)\n"
] |
[
[
"numpy.all",
"numpy.array_equal"
]
] |
dairatom/stable-baselines
|
[
"fc55681eeb6687301b429a407a875e9a6e01a0c5"
] |
[
"stable_baselines/deepq/policies.py"
] |
[
"#import tensorflow as tf\nimport tensorflow.compat.v1 as tf\ntf.disable_v2_behavior()\nimport tensorflow.keras.layers as tf_layers\nimport numpy as np\nfrom gym.spaces import Discrete\n\nfrom stable_baselines.common.policies import BasePolicy, nature_cnn, register_policy\n\n\nclass DQNPolicy(BasePolicy):\n \"\"\"\n Policy object that implements a DQN policy\n\n :param sess: (TensorFlow session) The current TensorFlow session\n :param ob_space: (Gym Space) The observation space of the environment\n :param ac_space: (Gym Space) The action space of the environment\n :param n_env: (int) The number of environments to run\n :param n_steps: (int) The number of steps to run for each environment\n :param n_batch: (int) The number of batch to run (n_envs * n_steps)\n :param reuse: (bool) If the policy is reusable or not\n :param scale: (bool) whether or not to scale the input\n :param obs_phs: (TensorFlow Tensor, TensorFlow Tensor) a tuple containing an override for observation placeholder\n and the processed observation placeholder respectively\n :param dueling: (bool) if true double the output MLP to compute a baseline for action scores\n \"\"\"\n\n def __init__(self, sess, ob_space, ac_space, n_env, n_steps, n_batch, reuse=False, scale=False,\n obs_phs=None, dueling=True):\n # DQN policies need an override for the obs placeholder, due to the architecture of the code\n super(DQNPolicy, self).__init__(sess, ob_space, ac_space, n_env, n_steps, n_batch, reuse=reuse, scale=scale,\n obs_phs=obs_phs)\n assert isinstance(ac_space, Discrete), \"Error: the action space for DQN must be of type gym.spaces.Discrete\"\n self.n_actions = ac_space.n\n self.value_fn = None\n self.q_values = None\n self.dueling = dueling\n\n def _setup_init(self):\n \"\"\"\n Set up action probability\n \"\"\"\n with tf.variable_scope(\"output\", reuse=True):\n assert self.q_values is not None\n self.policy_proba = tf.nn.softmax(self.q_values)\n\n def step(self, obs, state=None, mask=None, deterministic=True):\n \"\"\"\n Returns the q_values for a single step\n\n :param obs: (np.ndarray float or int) The current observation of the environment\n :param state: (np.ndarray float) The last states (used in recurrent policies)\n :param mask: (np.ndarray float) The last masks (used in recurrent policies)\n :param deterministic: (bool) Whether or not to return deterministic actions.\n :return: (np.ndarray int, np.ndarray float, np.ndarray float) actions, q_values, states\n \"\"\"\n raise NotImplementedError\n\n def proba_step(self, obs, state=None, mask=None):\n \"\"\"\n Returns the action probability for a single step\n\n :param obs: (np.ndarray float or int) The current observation of the environment\n :param state: (np.ndarray float) The last states (used in recurrent policies)\n :param mask: (np.ndarray float) The last masks (used in recurrent policies)\n :return: (np.ndarray float) the action probability\n \"\"\"\n raise NotImplementedError\n\n\nclass FeedForwardPolicy(DQNPolicy):\n \"\"\"\n Policy object that implements a DQN policy, using a feed forward neural network.\n\n :param sess: (TensorFlow session) The current TensorFlow session\n :param ob_space: (Gym Space) The observation space of the environment\n :param ac_space: (Gym Space) The action space of the environment\n :param n_env: (int) The number of environments to run\n :param n_steps: (int) The number of steps to run for each environment\n :param n_batch: (int) The number of batch to run (n_envs * n_steps)\n :param reuse: (bool) If the policy is reusable or not\n :param layers: ([int]) The size of the Neural network for the policy (if None, default to [64, 64])\n :param cnn_extractor: (function (TensorFlow Tensor, ``**kwargs``): (TensorFlow Tensor)) the CNN feature extraction\n :param feature_extraction: (str) The feature extraction type (\"cnn\" or \"mlp\")\n :param obs_phs: (TensorFlow Tensor, TensorFlow Tensor) a tuple containing an override for observation placeholder\n and the processed observation placeholder respectively\n :param layer_norm: (bool) enable layer normalisation\n :param dueling: (bool) if true double the output MLP to compute a baseline for action scores\n :param act_fun: (tf.func) the activation function to use in the neural network.\n :param kwargs: (dict) Extra keyword arguments for the nature CNN feature extraction\n \"\"\"\n\n def __init__(self, sess, ob_space, ac_space, n_env, n_steps, n_batch, reuse=False, layers=None,\n cnn_extractor=nature_cnn, feature_extraction=\"cnn\",\n obs_phs=None, layer_norm=False, dueling=True, act_fun=tf.nn.relu, **kwargs):\n super(FeedForwardPolicy, self).__init__(sess, ob_space, ac_space, n_env, n_steps,\n n_batch, dueling=dueling, reuse=reuse,\n scale=(feature_extraction == \"cnn\"), obs_phs=obs_phs)\n\n self._kwargs_check(feature_extraction, kwargs)\n\n if layers is None:\n layers = [64, 64]\n\n with tf.variable_scope(\"model\", reuse=reuse):\n with tf.variable_scope(\"action_value\"):\n if feature_extraction == \"cnn\":\n extracted_features = cnn_extractor(self.processed_obs, **kwargs)\n action_out = extracted_features\n else:\n extracted_features = tf.layers.flatten(self.processed_obs)\n action_out = extracted_features\n for layer_size in layers:\n action_out = tf_layers.fully_connected(action_out, num_outputs=layer_size, activation_fn=None)\n if layer_norm:\n action_out = tf_layers.layer_norm(action_out, center=True, scale=True)\n action_out = act_fun(action_out)\n\n action_scores = tf_layers.fully_connected(action_out, num_outputs=self.n_actions, activation_fn=None)\n\n if self.dueling:\n with tf.variable_scope(\"state_value\"):\n state_out = extracted_features\n for layer_size in layers:\n state_out = tf_layers.fully_connected(state_out, num_outputs=layer_size, activation_fn=None)\n if layer_norm:\n state_out = tf_layers.layer_norm(state_out, center=True, scale=True)\n state_out = act_fun(state_out)\n state_score = tf_layers.fully_connected(state_out, num_outputs=1, activation_fn=None)\n action_scores_mean = tf.reduce_mean(action_scores, axis=1)\n action_scores_centered = action_scores - tf.expand_dims(action_scores_mean, axis=1)\n q_out = state_score + action_scores_centered\n else:\n q_out = action_scores\n\n self.q_values = q_out\n self._setup_init()\n\n def step(self, obs, state=None, mask=None, deterministic=True):\n q_values, actions_proba = self.sess.run([self.q_values, self.policy_proba], {self.obs_ph: obs})\n if deterministic:\n actions = np.argmax(q_values, axis=1)\n else:\n # Unefficient sampling\n # TODO: replace the loop\n # maybe with Gumbel-max trick ? (http://amid.fish/humble-gumbel)\n actions = np.zeros((len(obs),), dtype=np.int64)\n for action_idx in range(len(obs)):\n actions[action_idx] = np.random.choice(self.n_actions, p=actions_proba[action_idx])\n\n return actions, q_values, None\n\n def proba_step(self, obs, state=None, mask=None):\n return self.sess.run(self.policy_proba, {self.obs_ph: obs})\n\n\nclass CnnPolicy(FeedForwardPolicy):\n \"\"\"\n Policy object that implements DQN policy, using a CNN (the nature CNN)\n\n :param sess: (TensorFlow session) The current TensorFlow session\n :param ob_space: (Gym Space) The observation space of the environment\n :param ac_space: (Gym Space) The action space of the environment\n :param n_env: (int) The number of environments to run\n :param n_steps: (int) The number of steps to run for each environment\n :param n_batch: (int) The number of batch to run (n_envs * n_steps)\n :param reuse: (bool) If the policy is reusable or not\n :param obs_phs: (TensorFlow Tensor, TensorFlow Tensor) a tuple containing an override for observation placeholder\n and the processed observation placeholder respectively\n :param dueling: (bool) if true double the output MLP to compute a baseline for action scores\n :param _kwargs: (dict) Extra keyword arguments for the nature CNN feature extraction\n \"\"\"\n\n def __init__(self, sess, ob_space, ac_space, n_env, n_steps, n_batch,\n reuse=False, obs_phs=None, dueling=True, **_kwargs):\n super(CnnPolicy, self).__init__(sess, ob_space, ac_space, n_env, n_steps, n_batch, reuse,\n feature_extraction=\"cnn\", obs_phs=obs_phs, dueling=dueling,\n layer_norm=False, **_kwargs)\n\n\nclass LnCnnPolicy(FeedForwardPolicy):\n \"\"\"\n Policy object that implements DQN policy, using a CNN (the nature CNN), with layer normalisation\n\n :param sess: (TensorFlow session) The current TensorFlow session\n :param ob_space: (Gym Space) The observation space of the environment\n :param ac_space: (Gym Space) The action space of the environment\n :param n_env: (int) The number of environments to run\n :param n_steps: (int) The number of steps to run for each environment\n :param n_batch: (int) The number of batch to run (n_envs * n_steps)\n :param reuse: (bool) If the policy is reusable or not\n :param obs_phs: (TensorFlow Tensor, TensorFlow Tensor) a tuple containing an override for observation placeholder\n and the processed observation placeholder respectively\n :param dueling: (bool) if true double the output MLP to compute a baseline for action scores\n :param _kwargs: (dict) Extra keyword arguments for the nature CNN feature extraction\n \"\"\"\n\n def __init__(self, sess, ob_space, ac_space, n_env, n_steps, n_batch,\n reuse=False, obs_phs=None, dueling=True, **_kwargs):\n super(LnCnnPolicy, self).__init__(sess, ob_space, ac_space, n_env, n_steps, n_batch, reuse,\n feature_extraction=\"cnn\", obs_phs=obs_phs, dueling=dueling,\n layer_norm=True, **_kwargs)\n\n\nclass MlpPolicy(FeedForwardPolicy):\n \"\"\"\n Policy object that implements DQN policy, using a MLP (2 layers of 64)\n\n :param sess: (TensorFlow session) The current TensorFlow session\n :param ob_space: (Gym Space) The observation space of the environment\n :param ac_space: (Gym Space) The action space of the environment\n :param n_env: (int) The number of environments to run\n :param n_steps: (int) The number of steps to run for each environment\n :param n_batch: (int) The number of batch to run (n_envs * n_steps)\n :param reuse: (bool) If the policy is reusable or not\n :param obs_phs: (TensorFlow Tensor, TensorFlow Tensor) a tuple containing an override for observation placeholder\n and the processed observation placeholder respectively\n :param dueling: (bool) if true double the output MLP to compute a baseline for action scores\n :param _kwargs: (dict) Extra keyword arguments for the nature CNN feature extraction\n \"\"\"\n\n def __init__(self, sess, ob_space, ac_space, n_env, n_steps, n_batch,\n reuse=False, obs_phs=None, dueling=True, **_kwargs):\n super(MlpPolicy, self).__init__(sess, ob_space, ac_space, n_env, n_steps, n_batch, reuse,\n feature_extraction=\"mlp\", obs_phs=obs_phs, dueling=dueling,\n layer_norm=False, **_kwargs)\n\n\nclass LnMlpPolicy(FeedForwardPolicy):\n \"\"\"\n Policy object that implements DQN policy, using a MLP (2 layers of 64), with layer normalisation\n\n :param sess: (TensorFlow session) The current TensorFlow session\n :param ob_space: (Gym Space) The observation space of the environment\n :param ac_space: (Gym Space) The action space of the environment\n :param n_env: (int) The number of environments to run\n :param n_steps: (int) The number of steps to run for each environment\n :param n_batch: (int) The number of batch to run (n_envs * n_steps)\n :param reuse: (bool) If the policy is reusable or not\n :param obs_phs: (TensorFlow Tensor, TensorFlow Tensor) a tuple containing an override for observation placeholder\n and the processed observation placeholder respectively\n :param dueling: (bool) if true double the output MLP to compute a baseline for action scores\n :param _kwargs: (dict) Extra keyword arguments for the nature CNN feature extraction\n \"\"\"\n\n def __init__(self, sess, ob_space, ac_space, n_env, n_steps, n_batch,\n reuse=False, obs_phs=None, dueling=True, **_kwargs):\n super(LnMlpPolicy, self).__init__(sess, ob_space, ac_space, n_env, n_steps, n_batch, reuse,\n feature_extraction=\"mlp\", obs_phs=obs_phs,\n layer_norm=True, dueling=True, **_kwargs)\n\n\nregister_policy(\"CnnPolicy\", CnnPolicy)\nregister_policy(\"LnCnnPolicy\", LnCnnPolicy)\nregister_policy(\"MlpPolicy\", MlpPolicy)\nregister_policy(\"LnMlpPolicy\", LnMlpPolicy)\n"
] |
[
[
"tensorflow.keras.layers.fully_connected",
"numpy.random.choice",
"tensorflow.compat.v1.disable_v2_behavior",
"tensorflow.compat.v1.layers.flatten",
"tensorflow.compat.v1.nn.softmax",
"tensorflow.compat.v1.variable_scope",
"tensorflow.keras.layers.layer_norm",
"tensorflow.compat.v1.expand_dims",
"numpy.argmax",
"tensorflow.compat.v1.reduce_mean"
]
] |
sDauenbaugh/AwesamBot
|
[
"ca4f0e2eeb81a02a672226d13be74fbb9c43bc0f"
] |
[
"src/controllers/pidController.py"
] |
[
"import numpy as np\nfrom rlbot.agents.base_agent import SimpleControllerState\n\n\nclass PID:\n\n def __init__(self, kp, ki, kd):\n self.Kp = kp\n self.Ki = ki\n self.Kd = kd\n self.last_err = 0\n self.sum_err = 0\n self.err = 0\n\n def update(self, e_t):\n self.sum_err = self.sum_err + e_t\n term_p = self.Kp * e_t\n term_i = self.Ki * self.sum_err\n term_d = self.Kd * (self.last_err - e_t)\n self.last_err = e_t\n return term_p + term_i + term_d\n\n\nclass SteerPID(PID):\n\n def __init__(self):\n super().__init__(10, 0.01, 0.001)\n\n def get_steer(self, ang_to_target): # angle should be in radians\n err = ang_to_target / (2 * np.pi)\n u_t = self.update(-err)\n if u_t > 1:\n u_t = 1\n elif u_t < -1:\n u_t = -1\n return u_t\n\n\ndef pid_steer(game_info, target_location, pid: SteerPID):\n \"\"\"Gives a set of commands to move the car along the ground toward a target location\n\n Attributes:\n target_location (Vec3): The local location the car wants to aim for\n\n Returns:\n SimpleControllerState: the set of commands to achieve the goal\n \"\"\"\n controller_state = SimpleControllerState()\n ball_direction = target_location\n\n angle = -np.arctan2(ball_direction.y, ball_direction.x)\n\n if angle > np.pi:\n angle -= 2 * np.pi\n elif angle < -np.pi:\n angle += 2 * np.pi\n\n # adjust angle\n turn_rate = pid.get_steer(angle)\n\n controller_state.throttle = 1\n controller_state.steer = turn_rate\n controller_state.jump = False\n return controller_state\n"
] |
[
[
"numpy.arctan2"
]
] |
ugurcanozalp/heli-gym
|
[
"78d754f63f9d0bbc606da760bfff9218c84d8be3"
] |
[
"heligym/envs/helicopter.py"
] |
[
"\"\"\"\nHelicopter control.\n\"\"\"\nimport yaml\nimport sys, math\nimport numpy as np\nimport os\nimport copy\n\nimport gym\nfrom gym import spaces\nfrom gym.utils import seeding, EzPickle\n\nfrom .dynamics import HelicopterDynamics\nfrom .dynamics import WindDynamics\nfrom .renderer.api import Renderer\n\nFPS = 50.0\nDT = 1/FPS\nFTS2KNOT = 0.5924838 # ft/s to knots conversion\nEPS = 1e-10 # small value for divison by zero\nR2D = 180/math.pi # Rad to deg\nD2R = 1/R2D\nFT2MTR = 0.3048 # ft to meter\nTAU = 2*math.pi\nFLOAT_TYPE = np.float64\n\nclass Heli(gym.Env, EzPickle):\n metadata = {\n 'render.modes': ['human', 'rgb_array'],\n 'video.frames_per_second' : FPS\n }\n # Default maximum time for an episode\n default_max_time = 40.0\n # Default trim condition, ground trim.\n default_trim_cond = {\n \"yaw\": 0.0,\n \"yaw_rate\": 0.0,\n \"ned_vel\": [0.0, 0.0, 0.0],\n \"gr_alt\": 10.0,\n \"xy\": [0.0, 0.0],\n \"psi_mr\": 0.0,\n \"psi_tr\": 0.0\n }\n # default task target\n default_task_target = {}\n def __init__(self, heli_name:str = \"aw109\"):\n EzPickle.__init__(self)\n yaml_path = os.path.join(os.path.dirname(__file__), \"helis\", heli_name + \".yaml\")\n with open(yaml_path) as foo:\n params = yaml.safe_load(foo)\n\n self.heli_dyn = HelicopterDynamics(params, DT)\n self.wind_dyn = WindDynamics(params['ENV'], DT)\n self.heli_dyn.set_wind(self.wind_dyn.wind_mean_ned) # set mean wind as wind so that helicopter trims accordingly\n self.observation_space = spaces.Box(-np.inf, np.inf, shape=(self.heli_dyn.n_obs,), dtype=np.float)\n self.action_space = spaces.Box(-1, +1, (self.heli_dyn.n_act,), dtype=np.float)\n self.successed_time = 0 # time counter for successing task through time.\n self.set_max_time()\n self.set_target()\n self.set_trim_cond()\n self.set_reward_weights()\n self.normalizers = {\n \"t\": np.sqrt(2*self.heli_dyn.MR[\"R\"]/self.heli_dyn.ENV[\"GRAV\"]),\n \"x\": 2*self.heli_dyn.MR[\"R\"],\n \"v\": np.sqrt(2*self.heli_dyn.MR[\"R\"]*self.heli_dyn.ENV[\"GRAV\"]),\n \"a\": self.heli_dyn.ENV[\"GRAV\"]\n }\n \n self.renderer = Renderer(w=1024, h=768, title='heligym')\n self.renderer.set_fps(FPS)\n\n self.heli_obj = self.renderer.create_model('/resources/models/'+heli_name+'/'+heli_name+'.obj',\n '/resources/shaders/'+heli_name+'_vertex.vs',\n '/resources/shaders/'+heli_name+'_frag.fs')\n self.renderer.add_permanent_object_to_window(self.heli_obj)\n \n self.terrain = self.renderer.create_model('/resources/models/terrain/terrain.obj',\n '/resources/shaders/terrain_vertex.vs',\n '/resources/shaders/terrain_frag.fs')\n self.renderer.add_permanent_object_to_window(self.terrain) \n\n self.sky = self.renderer.create_model('/resources/models/sky/sky.obj')\n self.renderer.add_permanent_object_to_window(self.sky)\n\n self._bGuiText = False\n \n # Setter functions for RL tasks\n def set_max_time(self, max_time=None):\n self.max_time = self.default_max_time if max_time is None else max_time # [sec] Given time for episode\n self.success_duration = self.max_time/4 # [sec] Required successfull time for maneuver\n self.task_duration = self.max_time/4 # [sec] Allowed time for successing task\n\n def set_target(self, target={}):\n self.task_target = self.default_task_target\n self.task_target.update(target)\n\n def get_target(self):\n return copy.deepcopy(self.task_target)\n\n def set_trim_cond(self, trim_cond={}):\n self.trim_cond = self.default_trim_cond\n self.trim_cond.update(trim_cond)\n\n def get_trim_cond(self):\n return copy.deepcopy(self.trim_cond)\n\n def set_reward_weights(self, base_reward_weight=None, terminal_reward_weight=None):\n zero_ = np.zeros((self.heli_dyn.n_obs,self.heli_dyn.n_obs))\n self.base_reward_weight = zero_ if base_reward_weight is None else base_reward_weight\n self.terminal_reward_weight = zero_ if terminal_reward_weight is None else terminal_reward_weight\n\n def __create_guiINFO_text(self):\n self.guiINFO_ID = self.renderer.create_guiText(\"Observations\", 30.0, 30.0, 250.0, 0.0)\n self.guiINFO_text = []\n self.guiINFO_text.append(\"FPS : %3.0f \")\n self.guiINFO_text.append(\"POWER : %5.2f hp\" )\n self.guiINFO_text.append(\"LON_VEL : %5.2f ft/s\")\n self.guiINFO_text.append(\"LAT_VEL : %5.2f ft/s\")\n self.guiINFO_text.append(\"DWN_VEL : %5.2f ft/s\")\n self.guiINFO_text.append(\"N_VEL : %5.2f ft/s\")\n self.guiINFO_text.append(\"E_VEL : %5.2f ft/s\")\n self.guiINFO_text.append(\"DES_RATE : %5.2f ft/s\")\n self.guiINFO_text.append(\"ROLL : %5.2f rad\")\n self.guiINFO_text.append(\"PITCH : %5.2f rad\")\n self.guiINFO_text.append(\"YAW : %5.2f rad\")\n self.guiINFO_text.append(\"ROLL_RATE : %5.2f rad/sec\")\n self.guiINFO_text.append(\"PITCH_RATE : %5.2f rad/sec\")\n self.guiINFO_text.append(\"YAW_RATE : %5.2f rad/sec\")\n self.guiINFO_text.append(\"N_POS : %5.2f ft\")\n self.guiINFO_text.append(\"E_POS : %5.2f ft\")\n self.guiINFO_text.append(\"ALT : %5.2f ft\")\n self.guiINFO_text.append(\"GR_ALT : %5.2f ft\")\n\n def __add_to_guiText(self):\n info_val = np.array(self.renderer.get_fps())\n info_val = np.append(info_val, self.heli_dyn.observation)\n self.renderer.add_guiText(self.guiINFO_ID, self.guiINFO_text, info_val) \n\n def render(self):\n info_val = np.array(self.renderer.get_fps())\n info_val = np.append(info_val, self.heli_dyn.observation)\n \n self.renderer.set_guiText(self.guiINFO_ID, self.guiINFO_text, info_val)\n \n self.renderer.rotate_MR(self.heli_obj, \n self.heli_dyn.state['betas'][1],\n self.heli_dyn.state['betas'][0],\n self.heli_dyn.state['psi_mr'][0])\n\n self.renderer.rotate_TR(self.heli_obj, \n 0,\n self.heli_dyn.state['psi_tr'][0],\n 0)\n\n\n self.renderer.translate_model(self.heli_obj, \n self.heli_dyn.state['xyz'][0] * FT2MTR,\n self.heli_dyn.state['xyz'][1] * FT2MTR,\n self.heli_dyn.state['xyz'][2] * FT2MTR\n )\n\n self.renderer.rotate_model(self.heli_obj, \n self.heli_dyn.state['euler'][0],\n self.heli_dyn.state['euler'][1],\n self.heli_dyn.state['euler'][2]\n )\n\n self.renderer.translate_model(self.sky, \n self.heli_dyn.state['xyz'][0] * FT2MTR,\n self.heli_dyn.state['xyz'][1] * FT2MTR,\n self.heli_dyn.state['xyz'][2] * FT2MTR + 500\n )\n\n self.renderer.set_camera_pos(self.heli_dyn.state['xyz'][0] * FT2MTR,\n self.heli_dyn.state['xyz'][1] * FT2MTR + 30,\n self.heli_dyn.state['xyz'][2] * FT2MTR )\n\n\n if not self.renderer.is_visible():\n self.renderer.show_window()\n \n self.renderer.render()\n \n def close(self):\n self.renderer.close()\n self.renderer.terminate()\n \n def exit(self):\n sys.exit()\n\n def step(self, actions):\n self.time_counter += DT\n # Turbulence calculations\n pre_observations = self.heli_dyn.observation\n wind_action = np.concatenate([pre_observations[4:7], pre_observations[16:]])\n self.wind_dyn.step(wind_action)\n wind_turb_vel = self.wind_dyn.observation\n # Helicopter dynamics calculations\n self.heli_dyn.set_wind(wind_turb_vel) \n self.heli_dyn.step(actions)\n observation = self.heli_dyn.observation\n reward, successed_step = self._calculate_reward()\n info = self._get_info()\n done = info['failed'] or info['successed'] or info['time_up'] or reward == np.nan\n self.successed_time += DT if successed_step else 0\n return np.copy(observation), reward, done, info\n\n def reset(self):\n self.time_counter = 0\n self.successed_time = 0\n self.wind_dyn.reset()\n self.heli_dyn.reset(self.trim_cond)\n if not self._bGuiText:\n self.__create_guiINFO_text()\n self.__add_to_guiText()\n self._bGuiText = True\n return np.copy(self.heli_dyn.observation)\n\n def _get_info(self):\n return {\n 'failed': self._is_failed(), \n 'successed': self._is_successed(), \n 'time_up': self._is_time_up()\n }\n\n def _is_failed(self):\n cond1 = self.heli_dyn._does_hit_ground(-self.heli_dyn.state['xyz'][2])\n cond2 = self.heli_dyn.state_dots['xyz'][2] > self.heli_dyn.MR['V_TIP']*0.05\n cond3 = self.heli_dyn.state['euler'][0] > 60*D2R\n cond4 = self.heli_dyn.state['euler'][1] > 60*D2R\n cond5 = np.abs(self.heli_dyn.state['xyz'][0]) > self.heli_dyn.ENV[\"NS_MAX\"] / 2 or np.abs(self.heli_dyn.state['xyz'][1]) > self.heli_dyn.ENV[\"EW_MAX\"] / 2 \\\n or -self.heli_dyn.state['xyz'][2] > self.heli_dyn.ground_touching_altitude() + 10000\n cond = (cond1 and (cond2 or cond3 or cond4)) or cond5\n return cond\n\n def _is_successed(self):\n return self.successed_time >= self.success_duration \n\n def _is_time_up(self):\n return self.time_counter > self.max_time\n\n def _calculate_reward(self):\n return 0.0, False\n\nif __name__=='__main__':\n env = Heli()\n #print(env)\n #obs = env.reset()\n #action = np.array([0.7007, 0.5391, 0.5351, 0.6429])\n #obs, reward, done, info = env.step(action)\n"
] |
[
[
"numpy.concatenate",
"numpy.zeros",
"numpy.copy",
"numpy.sqrt",
"numpy.append",
"numpy.abs"
]
] |
neolixcn/mmdetection3d
|
[
"45b5b8c2b2cc35762f27b020b34d2c3ee6be97af"
] |
[
"mmdet3d/models/dense_heads/train_mixins.py"
] |
[
"import numpy as np\nimport torch\n\nfrom mmdet3d.core import limit_period\nfrom mmdet.core import images_to_levels, multi_apply\n\n\nclass AnchorTrainMixin(object):\n \"\"\"Mixin class for target assigning of dense heads.\"\"\"\n\n def anchor_target_3d(self,\n anchor_list,\n gt_bboxes_list,\n input_metas,\n gt_bboxes_ignore_list=None,\n gt_labels_list=None,\n label_channels=1,\n num_classes=1,\n sampling=True):\n \"\"\"Compute regression and classification targets for anchors.\n\n Args:\n anchor_list (list[list]): Multi level anchors of each image.\n gt_bboxes_list (list[:obj:`BaseInstance3DBoxes`]): Ground truth\n bboxes of each image.\n input_metas (list[dict]): Meta info of each image.\n gt_bboxes_ignore_list (None | list): Ignore list of gt bboxes.\n gt_labels_list (list[torch.Tensor]): Gt labels of batches.\n label_channels (int): The channel of labels.\n num_classes (int): The number of classes.\n sampling (bool): Whether to sample anchors.\n\n Returns:\n tuple (list, list, list, list, list, list, int, int):\n Anchor targets, including labels, label weights,\n bbox targets, bbox weights, direction targets,\n direction weights, number of postive anchors and\n number of negative anchors.\n \"\"\"\n num_imgs = len(input_metas)\n assert len(anchor_list) == num_imgs\n\n if isinstance(anchor_list[0][0], list):\n # sizes of anchors are different\n # anchor number of a single level\n num_level_anchors = [\n sum([anchor.size(0) for anchor in anchors])\n for anchors in anchor_list[0]\n ]\n for i in range(num_imgs):\n anchor_list[i] = anchor_list[i][0]\n else:\n # anchor number of multi levels\n num_level_anchors = [\n anchors.view(-1, self.box_code_size).size(0)\n for anchors in anchor_list[0]\n ]\n # concat all level anchors and flags to a single tensor\n for i in range(num_imgs):\n anchor_list[i] = torch.cat(anchor_list[i])\n\n # compute targets for each image\n if gt_bboxes_ignore_list is None:\n gt_bboxes_ignore_list = [None for _ in range(num_imgs)]\n if gt_labels_list is None:\n gt_labels_list = [None for _ in range(num_imgs)]\n\n (all_labels, all_label_weights, all_bbox_targets, all_bbox_weights,\n all_dir_targets, all_dir_weights, pos_inds_list,\n neg_inds_list) = multi_apply(\n self.anchor_target_3d_single,\n anchor_list,\n gt_bboxes_list,\n gt_bboxes_ignore_list,\n gt_labels_list,\n input_metas,\n label_channels=label_channels,\n num_classes=num_classes,\n sampling=sampling)\n\n # no valid anchors\n if any([labels is None for labels in all_labels]):\n return None\n # sampled anchors of all images\n num_total_pos = sum([max(inds.numel(), 1) for inds in pos_inds_list])\n num_total_neg = sum([max(inds.numel(), 1) for inds in neg_inds_list])\n # split targets to a list w.r.t. multiple levels\n labels_list = images_to_levels(all_labels, num_level_anchors)\n label_weights_list = images_to_levels(all_label_weights,\n num_level_anchors)\n bbox_targets_list = images_to_levels(all_bbox_targets,\n num_level_anchors)\n bbox_weights_list = images_to_levels(all_bbox_weights,\n num_level_anchors)\n dir_targets_list = images_to_levels(all_dir_targets, num_level_anchors)\n dir_weights_list = images_to_levels(all_dir_weights, num_level_anchors)\n return (labels_list, label_weights_list, bbox_targets_list,\n bbox_weights_list, dir_targets_list, dir_weights_list,\n num_total_pos, num_total_neg)\n\n def anchor_target_3d_single(self,\n anchors,\n gt_bboxes,\n gt_bboxes_ignore,\n gt_labels,\n input_meta,\n label_channels=1,\n num_classes=1,\n sampling=True):\n \"\"\"Compute targets of anchors in single batch.\n\n Args:\n anchors (torch.Tensor): Concatenated multi-level anchor.\n gt_bboxes (:obj:`BaseInstance3DBoxes`): Gt bboxes.\n gt_bboxes_ignore (torch.Tensor): Ignored gt bboxes.\n gt_labels (torch.Tensor): Gt class labels.\n input_meta (dict): Meta info of each image.\n label_channels (int): The channel of labels.\n num_classes (int): The number of classes.\n sampling (bool): Whether to sample anchors.\n\n Returns:\n tuple[torch.Tensor]: Anchor targets.\n \"\"\"\n if isinstance(self.bbox_assigner,\n list) and (not isinstance(anchors, list)):\n feat_size = anchors.size(0) * anchors.size(1) * anchors.size(2)\n rot_angles = anchors.size(-2)\n assert len(self.bbox_assigner) == anchors.size(-3)\n (total_labels, total_label_weights, total_bbox_targets,\n total_bbox_weights, total_dir_targets, total_dir_weights,\n total_pos_inds, total_neg_inds) = [], [], [], [], [], [], [], []\n current_anchor_num = 0\n for i, assigner in enumerate(self.bbox_assigner):\n current_anchors = anchors[..., i, :, :].reshape(\n -1, self.box_code_size)\n current_anchor_num += current_anchors.size(0)\n if self.assign_per_class:\n gt_per_cls = (gt_labels == i)\n anchor_targets = self.anchor_target_single_assigner(\n assigner, current_anchors, gt_bboxes[gt_per_cls, :],\n gt_bboxes_ignore, gt_labels[gt_per_cls], input_meta,\n num_classes, sampling)\n else:\n anchor_targets = self.anchor_target_single_assigner(\n assigner, current_anchors, gt_bboxes, gt_bboxes_ignore,\n gt_labels, input_meta, num_classes, sampling)\n\n (labels, label_weights, bbox_targets, bbox_weights,\n dir_targets, dir_weights, pos_inds, neg_inds) = anchor_targets\n # print(pos_inds.detach().cpu().numpy().tolist())\n # my_anchors = current_anchors[[pos_inds.detach().cpu().numpy().tolist()], :]\n # my_anchors.detach().cpu().numpy().tofile('anchor_%d.bin' % i)\n # if i == 4:\n # gt_bboxes.tensor.detach().cpu().numpy().tofile('gt_box.bin')\n # assert False\n\n total_labels.append(labels.reshape(feat_size, 1, rot_angles))\n total_label_weights.append(\n label_weights.reshape(feat_size, 1, rot_angles))\n total_bbox_targets.append(\n bbox_targets.reshape(feat_size, 1, rot_angles,\n anchors.size(-1)))\n total_bbox_weights.append(\n bbox_weights.reshape(feat_size, 1, rot_angles,\n anchors.size(-1)))\n total_dir_targets.append(\n dir_targets.reshape(feat_size, 1, rot_angles))\n total_dir_weights.append(\n dir_weights.reshape(feat_size, 1, rot_angles))\n total_pos_inds.append(pos_inds)\n total_neg_inds.append(neg_inds)\n\n total_labels = torch.cat(total_labels, dim=-2).reshape(-1)\n total_label_weights = torch.cat(\n total_label_weights, dim=-2).reshape(-1)\n total_bbox_targets = torch.cat(\n total_bbox_targets, dim=-3).reshape(-1, anchors.size(-1))\n total_bbox_weights = torch.cat(\n total_bbox_weights, dim=-3).reshape(-1, anchors.size(-1))\n total_dir_targets = torch.cat(\n total_dir_targets, dim=-2).reshape(-1)\n total_dir_weights = torch.cat(\n total_dir_weights, dim=-2).reshape(-1)\n total_pos_inds = torch.cat(total_pos_inds, dim=0).reshape(-1)\n total_neg_inds = torch.cat(total_neg_inds, dim=0).reshape(-1)\n return (total_labels, total_label_weights, total_bbox_targets,\n total_bbox_weights, total_dir_targets, total_dir_weights,\n total_pos_inds, total_neg_inds)\n elif isinstance(self.bbox_assigner, list) and isinstance(\n anchors, list):\n # class-aware anchors with different feature map sizes\n assert len(self.bbox_assigner) == len(anchors), \\\n 'The number of bbox assigners and anchors should be the same.'\n (total_labels, total_label_weights, total_bbox_targets,\n total_bbox_weights, total_dir_targets, total_dir_weights,\n total_pos_inds, total_neg_inds) = [], [], [], [], [], [], [], []\n current_anchor_num = 0\n for i, assigner in enumerate(self.bbox_assigner):\n current_anchors = anchors[i]\n current_anchor_num += current_anchors.size(0)\n if self.assign_per_class:\n gt_per_cls = (gt_labels == i)\n anchor_targets = self.anchor_target_single_assigner(\n assigner, current_anchors, gt_bboxes[gt_per_cls, :],\n gt_bboxes_ignore, gt_labels[gt_per_cls], input_meta,\n num_classes, sampling)\n else:\n anchor_targets = self.anchor_target_single_assigner(\n assigner, current_anchors, gt_bboxes, gt_bboxes_ignore,\n gt_labels, input_meta, num_classes, sampling)\n\n (labels, label_weights, bbox_targets, bbox_weights,\n dir_targets, dir_weights, pos_inds, neg_inds) = anchor_targets\n total_labels.append(labels)\n total_label_weights.append(label_weights)\n total_bbox_targets.append(\n bbox_targets.reshape(-1, anchors[i].size(-1)))\n total_bbox_weights.append(\n bbox_weights.reshape(-1, anchors[i].size(-1)))\n total_dir_targets.append(dir_targets)\n total_dir_weights.append(dir_weights)\n total_pos_inds.append(pos_inds)\n total_neg_inds.append(neg_inds)\n\n total_labels = torch.cat(total_labels, dim=0)\n total_label_weights = torch.cat(total_label_weights, dim=0)\n total_bbox_targets = torch.cat(total_bbox_targets, dim=0)\n total_bbox_weights = torch.cat(total_bbox_weights, dim=0)\n total_dir_targets = torch.cat(total_dir_targets, dim=0)\n total_dir_weights = torch.cat(total_dir_weights, dim=0)\n total_pos_inds = torch.cat(total_pos_inds, dim=0)\n total_neg_inds = torch.cat(total_neg_inds, dim=0)\n return (total_labels, total_label_weights, total_bbox_targets,\n total_bbox_weights, total_dir_targets, total_dir_weights,\n total_pos_inds, total_neg_inds)\n else:\n return self.anchor_target_single_assigner(self.bbox_assigner,\n anchors, gt_bboxes,\n gt_bboxes_ignore,\n gt_labels, input_meta,\n num_classes, sampling)\n\n def anchor_target_single_assigner(self,\n bbox_assigner,\n anchors,\n gt_bboxes,\n gt_bboxes_ignore,\n gt_labels,\n input_meta,\n num_classes=1,\n sampling=True):\n \"\"\"Assign anchors and encode positive anchors.\n\n Args:\n bbox_assigner (BaseAssigner): assign positive and negative boxes.\n anchors (torch.Tensor): Concatenated multi-level anchor.\n gt_bboxes (:obj:`BaseInstance3DBoxes`): Gt bboxes.\n gt_bboxes_ignore (torch.Tensor): Ignored gt bboxes.\n gt_labels (torch.Tensor): Gt class labels.\n input_meta (dict): Meta info of each image.\n num_classes (int): The number of classes.\n sampling (bool): Whether to sample anchors.\n\n Returns:\n tuple[torch.Tensor]: Anchor targets.\n \"\"\"\n anchors = anchors.reshape(-1, anchors.size(-1))\n num_valid_anchors = anchors.shape[0]\n bbox_targets = torch.zeros_like(anchors)\n bbox_weights = torch.zeros_like(anchors)\n dir_targets = anchors.new_zeros((anchors.shape[0]), dtype=torch.long)\n dir_weights = anchors.new_zeros((anchors.shape[0]), dtype=torch.float)\n labels = anchors.new_zeros(num_valid_anchors, dtype=torch.long)\n label_weights = anchors.new_zeros(num_valid_anchors, dtype=torch.float)\n if len(gt_bboxes) > 0:\n if not isinstance(gt_bboxes, torch.Tensor):\n gt_bboxes = gt_bboxes.tensor.to(anchors.device)\n assign_result = bbox_assigner.assign(anchors, gt_bboxes,\n gt_bboxes_ignore, gt_labels)\n sampling_result = self.bbox_sampler.sample(assign_result, anchors,\n gt_bboxes)\n pos_inds = sampling_result.pos_inds\n neg_inds = sampling_result.neg_inds\n else:\n pos_inds = torch.nonzero(\n anchors.new_zeros((anchors.shape[0], ), dtype=torch.bool) > 0,\n as_tuple=False).squeeze(-1).unique()\n neg_inds = torch.nonzero(\n anchors.new_zeros((anchors.shape[0], ), dtype=torch.bool) == 0,\n as_tuple=False).squeeze(-1).unique()\n\n if gt_labels is not None:\n labels += num_classes\n if len(pos_inds) > 0:\n pos_bbox_targets = self.bbox_coder.encode(\n sampling_result.pos_bboxes, sampling_result.pos_gt_bboxes)\n pos_dir_targets = get_direction_target(\n sampling_result.pos_bboxes,\n pos_bbox_targets,\n self.dir_offset,\n one_hot=False)\n bbox_targets[pos_inds, :] = pos_bbox_targets\n bbox_weights[pos_inds, :] = 1.0\n dir_targets[pos_inds] = pos_dir_targets\n dir_weights[pos_inds] = 1.0\n\n if gt_labels is None:\n labels[pos_inds] = 1\n else:\n labels[pos_inds] = gt_labels[\n sampling_result.pos_assigned_gt_inds]\n if self.train_cfg.pos_weight <= 0:\n label_weights[pos_inds] = 1.0\n else:\n label_weights[pos_inds] = self.train_cfg.pos_weight\n\n if len(neg_inds) > 0:\n label_weights[neg_inds] = 1.0\n return (labels, label_weights, bbox_targets, bbox_weights, dir_targets,\n dir_weights, pos_inds, neg_inds)\n\n\ndef get_direction_target(anchors,\n reg_targets,\n dir_offset=0,\n num_bins=2,\n one_hot=True):\n \"\"\"Encode direction to 0 ~ num_bins-1.\n\n Args:\n anchors (torch.Tensor): Concatenated multi-level anchor.\n reg_targets (torch.Tensor): Bbox regression targets.\n dir_offset (int): Direction offset.\n num_bins (int): Number of bins to divide 2*PI.\n one_hot (bool): Whether to encode as one hot.\n\n Returns:\n torch.Tensor: Encoded direction targets.\n \"\"\"\n rot_gt = reg_targets[..., 6] + anchors[..., 6]\n offset_rot = limit_period(rot_gt - dir_offset, 0, 2 * np.pi)\n dir_cls_targets = torch.floor(offset_rot / (2 * np.pi / num_bins)).long()\n dir_cls_targets = torch.clamp(dir_cls_targets, min=0, max=num_bins - 1)\n if one_hot:\n dir_targets = torch.zeros(\n *list(dir_cls_targets.shape),\n num_bins,\n dtype=anchors.dtype,\n device=dir_cls_targets.device)\n dir_targets.scatter_(dir_cls_targets.unsqueeze(dim=-1).long(), 1.0)\n dir_cls_targets = dir_targets\n return dir_cls_targets\n"
] |
[
[
"torch.zeros_like",
"torch.floor",
"torch.cat",
"torch.clamp"
]
] |
lehduong/Policy-gradient-credit-assignment
|
[
"1d4c102964b985212874c1fe8710a8aa6ff9f328"
] |
[
"main.py"
] |
[
"import os\nimport time\nfrom collections import deque\n\nimport numpy as np\nimport torch\n\nfrom core import algorithms, utils\nfrom core.arguments import get_args\nfrom core.envs import make_vec_envs\nfrom core.agents import PolicyGradientAgent, CPCPolicyGradientAgent\nfrom core.storage import RolloutStorage, CPCRolloutStorage\nfrom evaluation import evaluate\n\n\ndef main():\n args = get_args()\n\n torch.manual_seed(args.seed)\n torch.cuda.manual_seed_all(args.seed)\n\n if args.cuda and torch.cuda.is_available() and args.cuda_deterministic:\n torch.backends.cudnn.benchmark = False\n torch.backends.cudnn.deterministic = True\n\n log_dir = os.path.expanduser(args.log_dir)\n eval_log_dir = log_dir + \"_eval\"\n utils.cleanup_log_dir(log_dir)\n utils.cleanup_log_dir(eval_log_dir)\n\n torch.set_num_threads(1)\n device = torch.device(\"cuda:0\" if args.cuda else \"cpu\")\n \n if not args.use_proper_time_limits:\n envs = make_vec_envs(args.env_name, args.seed, args.num_processes,\n args.gamma, args.log_dir, device, False, args.num_frame_stack)\n else:\n envs = make_vec_envs(args.env_name, args.seed, args.num_processes,\n args.gamma, args.log_dir, device, True, args.num_frame_stack, args.max_episode_steps)\n \n actor_critic = PolicyGradientAgent(\n envs.observation_space.shape,\n envs.action_space,\n base_kwargs={'recurrent': args.recurrent_policy})\n if args.use_cpc:\n actor_critic = CPCPolicyGradientAgent(\n envs.observation_space.shape,\n envs.action_space,\n base_kwargs={'recurrent': args.recurrent_policy})\n\n actor_critic.to(device)\n\n if args.algo == 'a2c':\n agent = algorithms.A2C_ACKTR(\n actor_critic,\n args.value_loss_coef,\n args.entropy_coef,\n lr=args.lr,\n eps=args.eps,\n alpha=args.alpha,\n max_grad_norm=args.max_grad_norm)\n if args.use_cpc:\n agent = algorithms.CPC_A2C_ACKTR(\n actor_critic,\n args.value_loss_coef,\n args.entropy_coef,\n lr=args.lr,\n eps=args.eps,\n alpha=args.alpha,\n max_grad_norm=args.max_grad_norm,\n device=device,\n num_steps=args.num_steps)\n\n elif args.algo == 'ppo':\n agent = algorithms.PPO(\n actor_critic,\n args.clip_param,\n args.ppo_epoch,\n args.num_mini_batch,\n args.value_loss_coef,\n args.entropy_coef,\n lr=args.lr,\n eps=args.eps,\n max_grad_norm=args.max_grad_norm)\n elif args.algo == 'acktr':\n agent = algorithms.A2C_ACKTR(\n actor_critic, args.value_loss_coef, args.entropy_coef, acktr=True)\n\n rollouts = RolloutStorage(args.num_steps, args.num_processes,\n envs.observation_space.shape, envs.action_space,\n actor_critic.recurrent_hidden_state_size)\n if args.use_cpc:\n rollouts = CPCRolloutStorage(args.num_steps, args.num_processes,\n envs.observation_space.shape, envs.action_space,\n actor_critic.recurrent_hidden_state_size, actor_critic.base.output_size)\n\n obs = envs.reset()\n rollouts.obs[0].copy_(obs)\n rollouts.to(device)\n\n episode_rewards = deque(maxlen=10)\n\n start = time.time()\n # lehduong: Total number of gradient updates\n # basically, the agent will act on the environment for a number of steps\n # which is usually referred to as n_steps. Then, we compute the cummulative\n # reward, update the policy. After that, we continue rolling out the agent \n # in the environment repeatedly. If the trajectory is ended, simply reset it and # do it again.\n num_updates = int(\n args.num_env_steps) // args.num_steps // args.num_processes\n for j in range(num_updates):\n\n # decrease learning rate linearly\n if args.use_linear_lr_decay:\n utils.update_linear_schedule(\n agent.optimizer, j, num_updates,\n agent.optimizer.lr if args.algo == \"acktr\" else args.lr)\n \n # Rolling out, collecting and storing SARS (State, action, reward, new state)\n for step in range(args.num_steps):\n # Sample actions\n with torch.no_grad():\n if args.use_cpc:\n # if using CPC actor critic, act method also returns embedding of \n value, action, action_log_prob, recurrent_hidden_states, state_feat, action_feat = actor_critic.act(\n rollouts.obs[step], rollouts.recurrent_hidden_states[step],\n rollouts.masks[step])\n else:\n value, action, action_log_prob, recurrent_hidden_states = actor_critic.act(\n rollouts.obs[step], rollouts.recurrent_hidden_states[step],\n rollouts.masks[step])\n\n # Obser reward and next obs\n obs, reward, done, infos = envs.step(action)\n for info in infos:\n if 'episode' in info.keys():\n episode_rewards.append(info['episode']['r'])\n\n # If done then clean the history of observations.\n masks = torch.FloatTensor(\n [[0.0] if done_ else [1.0] for done_ in done])\n bad_masks = torch.FloatTensor(\n [[0.0] if 'bad_transition' in info.keys() else [1.0]\n for info in infos])\n if args.use_cpc:\n rollouts.insert(obs, recurrent_hidden_states, action,\n action_log_prob, value, reward, masks, bad_masks, state_feat, action_feat)\n else:\n rollouts.insert(obs, recurrent_hidden_states, action,\n action_log_prob, value, reward, masks, bad_masks)\n\n with torch.no_grad():\n next_value = actor_critic.get_value(\n rollouts.obs[-1], rollouts.recurrent_hidden_states[-1],\n rollouts.masks[-1]).detach()\n\n rollouts.compute_returns(next_value, args.use_gae, args.gamma,\n args.gae_lambda, args.use_proper_time_limits)\n \n if args.use_cpc:\n value_loss, action_loss, dist_entropy, cpc_result = agent.update(rollouts)\n else:\n value_loss, action_loss, dist_entropy = agent.update(rollouts)\n\n rollouts.after_update()\n\n # save for every interval-th episode or for the last epoch\n if (j % args.save_interval == 0\n or j == num_updates - 1) and args.save_dir != \"\":\n save_path = os.path.join(args.save_dir, args.algo)\n try:\n os.makedirs(save_path)\n except OSError:\n pass\n\n torch.save([\n actor_critic,\n getattr(utils.get_vec_normalize(envs), 'ob_rms', None)\n ], os.path.join(save_path, args.env_name + \".pt\"))\n\n if j % args.log_interval == 0 and len(episode_rewards) > 1:\n total_num_steps = (j + 1) * args.num_processes * args.num_steps\n end = time.time()\n print(\n \"\\nUpdates {}, num timesteps {}, FPS {} \\n Last {} training episodes: mean/median reward {:.1f}/{:.1f}, min/max reward {:.1f}/{:.1f}\"\n .format(j, total_num_steps,\n int(total_num_steps / (end - start)),\n len(episode_rewards), np.mean(episode_rewards),\n np.median(episode_rewards), np.min(episode_rewards),\n np.max(episode_rewards), dist_entropy, value_loss,\n action_loss))\n print(\n \" Value loss: {:.2f} Action loss {:2f} Dist Entropy {:2f}\"\n .format(value_loss,\n action_loss,\n dist_entropy)\n )\n if args.use_cpc:\n print(\n \" CPC state loss {:.2f}, CPC state action loss {:.2f}, CPC state acc {:.2f}, CPC state action acc {:.2f}\"\n .format(cpc_result['nce_state'],\n cpc_result['nce_state_action'],\n cpc_result['accuracy_state'],\n cpc_result['accuracy_state_action']))\n\n if (args.eval_interval is not None and len(episode_rewards) > 1\n and j % args.eval_interval == 0):\n ob_rms = utils.get_vec_normalize(envs).ob_rms\n evaluate(actor_critic, ob_rms, args.env_name, args.seed,\n args.num_processes, eval_log_dir, device)\n\n\nif __name__ == \"__main__\":\n main()\n"
] |
[
[
"numpy.max",
"torch.device",
"torch.cuda.manual_seed_all",
"numpy.median",
"torch.FloatTensor",
"torch.no_grad",
"numpy.min",
"numpy.mean",
"torch.manual_seed",
"torch.cuda.is_available",
"torch.set_num_threads"
]
] |
stejsle1/semestralni_prace_PYT
|
[
"ffee7a8deeae9c73c8b8097494345d7fe90c2889"
] |
[
"wator_tests/tests/test_wator_init.py"
] |
[
"import numpy\nimport pytest\n\nfrom wator import WaTor\n\n\ndef test_creatures():\n creatures = numpy.zeros((8, 8))\n creatures[2, 4] = 3\n creatures[1, :] = -5\n wator = WaTor(creatures)\n print(wator.creatures)\n assert (creatures == wator.creatures).all()\n assert wator.count_fish() == 1\n assert wator.count_sharks() == 8\n\n\ndef test_shape():\n shape = (16, 16)\n wator = WaTor(shape=shape, nfish=16, nsharks=4)\n print(wator.creatures)\n assert wator.creatures.shape == shape\n assert wator.count_fish() == 16\n assert wator.count_sharks() == 4\n assert ((wator.creatures >= -10) & (wator.creatures <= 5)).all()\n\n\ndef test_shape_full_of_fish():\n shape = (16, 16)\n wator = WaTor(shape=shape, nfish=16 * 16, nsharks=0)\n print(wator.creatures)\n assert wator.count_fish() == 16 * 16\n assert wator.count_sharks() == 0\n\n\ndef test_shape_full_of_shark():\n shape = (16, 16)\n wator = WaTor(shape=shape, nfish=0, nsharks=16 * 16)\n print(wator.creatures)\n assert wator.count_fish() == 0\n assert wator.count_sharks() == 16 * 16\n\n\ndef test_shape_full_of_something():\n shape = (16, 16)\n total = 16*16\n nfish = total // 5\n nsharks = total - nfish\n wator = WaTor(shape=shape, nfish=nfish, nsharks=nsharks)\n print(wator.creatures)\n assert wator.count_fish() == nfish\n assert wator.count_sharks() == nsharks\n\n\ndef test_shape_custom_age():\n shape = (16, 16)\n age_fish = 2\n age_shark = 20\n wator = WaTor(shape=shape, nfish=16, nsharks=4,\n age_fish=age_fish,\n age_shark=age_shark)\n print(wator.creatures)\n assert wator.creatures.shape == shape\n assert wator.count_fish() == 16\n assert wator.count_sharks() == 4\n assert ((wator.creatures >= -age_shark) &\n (wator.creatures <= age_fish)).all()\n\n\ndef test_energy_initial_default():\n shape = (8, 8)\n creatures = numpy.zeros(shape)\n creatures[1, :] = -5\n wator = WaTor(creatures)\n print(wator.creatures)\n print(wator.energies)\n assert wator.energies.shape == shape\n assert (wator.energies[1, :] == 5).all()\n\n\ndef test_energy_initial_custom():\n shape = (8, 8)\n creatures = numpy.zeros(shape)\n creatures[1, :] = -5\n wator = WaTor(creatures, energy_initial=12)\n print(wator.creatures)\n print(wator.energies)\n assert wator.energies.shape == shape\n assert (wator.energies[1, :] == 12).all()\n\n\ndef test_energies():\n shape = (8, 8)\n creatures = numpy.zeros(shape)\n energies = numpy.zeros(shape)\n creatures[1, :] = -5\n for i in range(8):\n energies[1, i] = i\n wator = WaTor(creatures, energies=energies)\n print(wator.creatures)\n print(wator.energies)\n assert (wator.energies[1, :] == energies[1, :]).all()\n\n\ndef test_nonsense():\n creatures = numpy.zeros((8, 8))\n\n with pytest.raises(ValueError):\n WaTor(creatures, nfish=20)\n\n with pytest.raises(ValueError):\n WaTor(creatures, nsharks=20)\n\n with pytest.raises(ValueError):\n WaTor(creatures, shape=(8, 8))\n\n with pytest.raises(ValueError):\n WaTor(shape=(8, 8), nsharks=20)\n\n with pytest.raises(ValueError):\n WaTor(creatures, energies=numpy.zeros((6, 6)))\n\n with pytest.raises(ValueError):\n WaTor(creatures, energies=numpy.zeros((8, 8)),\n energy_initial=8)\n\n with pytest.raises(ValueError):\n WaTor(shape=(8, 8), nsharks=20, nfish=1000)\n\n with pytest.raises(TypeError):\n WaTor(shape=(8, 8), nsharks=20, nfish='nope')\n"
] |
[
[
"numpy.zeros"
]
] |
negarIravani/machine-learning-engineering-for-production-public
|
[
"886ecc93dbf74ff3b6cde8b77eb0b1f2b49b7c2e"
] |
[
"course4/week3-ungraded-labs/C4_W3_Lab_4_Github_Actions/app/main.py"
] |
[
"import pickle\nimport numpy as np\nfrom typing import List\nfrom fastapi import FastAPI\nfrom pydantic import BaseModel, conlist\n\n# small changes to test\n\napp = FastAPI(title=\"Predicting Wine Class with batching\")\n\n# Open classifier in global scope\nwith open(\"models/wine-95-fixed.pkl\", \"rb\") as file:\n clf = pickle.load(file)\n\n\nclass Wine(BaseModel):\n batches: List[conlist(item_type=float, min_items=13, max_items=13)]\n\n\[email protected](\"/predict\")\ndef predict(wine: Wine):\n batches = wine.batches\n np_batches = np.array(batches)\n pred = clf.predict(np_batches).tolist()\n return {\"Prediction\": pred}\n"
] |
[
[
"numpy.array"
]
] |
deHasara/dask
|
[
"fb544144611b25a6f23d90637038a93f93153f8f"
] |
[
"dask/utils.py"
] |
[
"from datetime import datetime, timedelta\nimport functools\nimport inspect\nimport os\nimport shutil\nimport sys\nimport tempfile\nimport re\nfrom errno import ENOENT\nfrom collections.abc import Iterator\nfrom contextlib import contextmanager\nfrom importlib import import_module\nfrom numbers import Integral, Number\nfrom threading import Lock\nfrom typing import Dict, Iterable, Mapping, Optional, TypeVar\nimport uuid\nfrom weakref import WeakValueDictionary\nfrom functools import lru_cache\nfrom _thread import RLock\n\nfrom .core import get_deps\n\n\nK = TypeVar(\"K\")\nV = TypeVar(\"V\")\n\n\nsystem_encoding = sys.getdefaultencoding()\nif system_encoding == \"ascii\":\n system_encoding = \"utf-8\"\n\n\ndef apply(func, args, kwargs=None):\n if kwargs:\n return func(*args, **kwargs)\n else:\n return func(*args)\n\n\ndef deepmap(func, *seqs):\n \"\"\"Apply function inside nested lists\n\n >>> inc = lambda x: x + 1\n >>> deepmap(inc, [[1, 2], [3, 4]])\n [[2, 3], [4, 5]]\n\n >>> add = lambda x, y: x + y\n >>> deepmap(add, [[1, 2], [3, 4]], [[10, 20], [30, 40]])\n [[11, 22], [33, 44]]\n \"\"\"\n if isinstance(seqs[0], (list, Iterator)):\n return [deepmap(func, *items) for items in zip(*seqs)]\n else:\n return func(*seqs)\n\n\ndef homogeneous_deepmap(func, seq):\n if not seq:\n return seq\n n = 0\n tmp = seq\n while isinstance(tmp, list):\n n += 1\n tmp = tmp[0]\n\n return ndeepmap(n, func, seq)\n\n\ndef ndeepmap(n, func, seq):\n \"\"\"Call a function on every element within a nested container\n\n >>> def inc(x):\n ... return x + 1\n >>> L = [[1, 2], [3, 4, 5]]\n >>> ndeepmap(2, inc, L)\n [[2, 3], [4, 5, 6]]\n \"\"\"\n if n == 1:\n return [func(item) for item in seq]\n elif n > 1:\n return [ndeepmap(n - 1, func, item) for item in seq]\n elif isinstance(seq, list):\n return func(seq[0])\n else:\n return func(seq)\n\n\n@contextmanager\ndef ignoring(*exceptions):\n try:\n yield\n except exceptions:\n pass\n\n\ndef import_required(mod_name, error_msg):\n \"\"\"Attempt to import a required dependency.\n\n Raises a RuntimeError if the requested module is not available.\n \"\"\"\n try:\n return import_module(mod_name)\n except ImportError as e:\n raise RuntimeError(error_msg) from e\n\n\n@contextmanager\ndef tmpfile(extension=\"\", dir=None):\n extension = \".\" + extension.lstrip(\".\")\n handle, filename = tempfile.mkstemp(extension, dir=dir)\n os.close(handle)\n os.remove(filename)\n\n try:\n yield filename\n finally:\n if os.path.exists(filename):\n if os.path.isdir(filename):\n shutil.rmtree(filename)\n else:\n with ignoring(OSError):\n os.remove(filename)\n\n\n@contextmanager\ndef tmpdir(dir=None):\n dirname = tempfile.mkdtemp(dir=dir)\n\n try:\n yield dirname\n finally:\n if os.path.exists(dirname):\n if os.path.isdir(dirname):\n with ignoring(OSError):\n shutil.rmtree(dirname)\n else:\n with ignoring(OSError):\n os.remove(dirname)\n\n\n@contextmanager\ndef filetext(text, extension=\"\", open=open, mode=\"w\"):\n with tmpfile(extension=extension) as filename:\n f = open(filename, mode=mode)\n try:\n f.write(text)\n finally:\n try:\n f.close()\n except AttributeError:\n pass\n\n yield filename\n\n\n@contextmanager\ndef changed_cwd(new_cwd):\n old_cwd = os.getcwd()\n os.chdir(new_cwd)\n try:\n yield\n finally:\n os.chdir(old_cwd)\n\n\n@contextmanager\ndef tmp_cwd(dir=None):\n with tmpdir(dir) as dirname:\n with changed_cwd(dirname):\n yield dirname\n\n\n@contextmanager\ndef noop_context():\n yield\n\n\nclass IndexCallable:\n \"\"\"Provide getitem syntax for functions\n\n >>> def inc(x):\n ... return x + 1\n\n >>> I = IndexCallable(inc)\n >>> I[3]\n 4\n \"\"\"\n\n __slots__ = (\"fn\",)\n\n def __init__(self, fn):\n self.fn = fn\n\n def __getitem__(self, key):\n return self.fn(key)\n\n\n@contextmanager\ndef filetexts(d, open=open, mode=\"t\", use_tmpdir=True):\n \"\"\"Dumps a number of textfiles to disk\n\n Parameters\n ----------\n d : dict\n a mapping from filename to text like {'a.csv': '1,1\\n2,2'}\n\n Since this is meant for use in tests, this context manager will\n automatically switch to a temporary current directory, to avoid\n race conditions when running tests in parallel.\n \"\"\"\n with (tmp_cwd() if use_tmpdir else noop_context()):\n for filename, text in d.items():\n try:\n os.makedirs(os.path.dirname(filename))\n except OSError:\n pass\n f = open(filename, \"w\" + mode)\n try:\n f.write(text)\n finally:\n try:\n f.close()\n except AttributeError:\n pass\n\n yield list(d)\n\n for filename in d:\n if os.path.exists(filename):\n with ignoring(OSError):\n os.remove(filename)\n\n\ndef concrete(seq):\n \"\"\"Make nested iterators concrete lists\n\n >>> data = [[1, 2], [3, 4]]\n >>> seq = iter(map(iter, data))\n >>> concrete(seq)\n [[1, 2], [3, 4]]\n \"\"\"\n if isinstance(seq, Iterator):\n seq = list(seq)\n if isinstance(seq, (tuple, list)):\n seq = list(map(concrete, seq))\n return seq\n\n\ndef pseudorandom(n, p, random_state=None):\n \"\"\"Pseudorandom array of integer indexes\n\n >>> pseudorandom(5, [0.5, 0.5], random_state=123)\n array([1, 0, 0, 1, 1], dtype=int8)\n\n >>> pseudorandom(10, [0.5, 0.2, 0.2, 0.1], random_state=5)\n array([0, 2, 0, 3, 0, 1, 2, 1, 0, 0], dtype=int8)\n \"\"\"\n import numpy as np\n\n p = list(p)\n cp = np.cumsum([0] + p)\n assert np.allclose(1, cp[-1])\n assert len(p) < 256\n\n if not isinstance(random_state, np.random.RandomState):\n random_state = np.random.RandomState(random_state)\n\n x = random_state.random_sample(n)\n out = np.empty(n, dtype=\"i1\")\n\n for i, (low, high) in enumerate(zip(cp[:-1], cp[1:])):\n out[(x >= low) & (x < high)] = i\n return out\n\n\ndef random_state_data(n, random_state=None):\n \"\"\"Return a list of arrays that can initialize\n ``np.random.RandomState``.\n\n Parameters\n ----------\n n : int\n Number of arrays to return.\n random_state : int or np.random.RandomState, optional\n If an int, is used to seed a new ``RandomState``.\n \"\"\"\n import numpy as np\n\n if not all(\n hasattr(random_state, attr) for attr in [\"normal\", \"beta\", \"bytes\", \"uniform\"]\n ):\n random_state = np.random.RandomState(random_state)\n\n random_data = random_state.bytes(624 * n * 4) # `n * 624` 32-bit integers\n l = list(np.frombuffer(random_data, dtype=np.uint32).reshape((n, -1)))\n assert len(l) == n\n return l\n\n\ndef is_integer(i):\n \"\"\"\n >>> is_integer(6)\n True\n >>> is_integer(42.0)\n True\n >>> is_integer('abc')\n False\n \"\"\"\n return isinstance(i, Integral) or (isinstance(i, float) and i.is_integer())\n\n\nONE_ARITY_BUILTINS = set(\n [\n abs,\n all,\n any,\n ascii,\n bool,\n bytearray,\n bytes,\n callable,\n chr,\n classmethod,\n complex,\n dict,\n dir,\n enumerate,\n eval,\n float,\n format,\n frozenset,\n hash,\n hex,\n id,\n int,\n iter,\n len,\n list,\n max,\n min,\n next,\n oct,\n open,\n ord,\n range,\n repr,\n reversed,\n round,\n set,\n slice,\n sorted,\n staticmethod,\n str,\n sum,\n tuple,\n type,\n vars,\n zip,\n memoryview,\n ]\n)\nMULTI_ARITY_BUILTINS = set(\n [\n compile,\n delattr,\n divmod,\n filter,\n getattr,\n hasattr,\n isinstance,\n issubclass,\n map,\n pow,\n setattr,\n ]\n)\n\n\ndef getargspec(func):\n \"\"\"Version of inspect.getargspec that works with partial and warps.\"\"\"\n if isinstance(func, functools.partial):\n return getargspec(func.func)\n\n func = getattr(func, \"__wrapped__\", func)\n if isinstance(func, type):\n return inspect.getfullargspec(func.__init__)\n else:\n return inspect.getfullargspec(func)\n\n\ndef takes_multiple_arguments(func, varargs=True):\n \"\"\"Does this function take multiple arguments?\n\n >>> def f(x, y): pass\n >>> takes_multiple_arguments(f)\n True\n\n >>> def f(x): pass\n >>> takes_multiple_arguments(f)\n False\n\n >>> def f(x, y=None): pass\n >>> takes_multiple_arguments(f)\n False\n\n >>> def f(*args): pass\n >>> takes_multiple_arguments(f)\n True\n\n >>> class Thing:\n ... def __init__(self, a): pass\n >>> takes_multiple_arguments(Thing)\n False\n\n \"\"\"\n if func in ONE_ARITY_BUILTINS:\n return False\n elif func in MULTI_ARITY_BUILTINS:\n return True\n\n try:\n spec = getargspec(func)\n except Exception:\n return False\n\n try:\n is_constructor = spec.args[0] == \"self\" and isinstance(func, type)\n except Exception:\n is_constructor = False\n\n if varargs and spec.varargs:\n return True\n\n ndefaults = 0 if spec.defaults is None else len(spec.defaults)\n return len(spec.args) - ndefaults - is_constructor > 1\n\n\ndef get_named_args(func):\n \"\"\"Get all non ``*args/**kwargs`` arguments for a function\"\"\"\n s = inspect.signature(func)\n return [\n n\n for n, p in s.parameters.items()\n if p.kind in [p.POSITIONAL_OR_KEYWORD, p.POSITIONAL_ONLY, p.KEYWORD_ONLY]\n ]\n\n\nclass Dispatch:\n \"\"\"Simple single dispatch.\"\"\"\n\n def __init__(self, name=None):\n self._lookup = {}\n self._lazy = {}\n if name:\n self.__name__ = name\n\n def register(self, type, func=None):\n \"\"\"Register dispatch of `func` on arguments of type `type`\"\"\"\n\n def wrapper(func):\n if isinstance(type, tuple):\n for t in type:\n self.register(t, func)\n else:\n self._lookup[type] = func\n return func\n\n return wrapper(func) if func is not None else wrapper\n\n def register_lazy(self, toplevel, func=None):\n \"\"\"\n Register a registration function which will be called if the\n *toplevel* module (e.g. 'pandas') is ever loaded.\n \"\"\"\n\n def wrapper(func):\n self._lazy[toplevel] = func\n return func\n\n return wrapper(func) if func is not None else wrapper\n\n def dispatch(self, cls):\n \"\"\"Return the function implementation for the given ``cls``\"\"\"\n # Fast path with direct lookup on cls\n lk = self._lookup\n try:\n impl = lk[cls]\n except KeyError:\n pass\n else:\n return impl\n # Is a lazy registration function present?\n toplevel, _, _ = cls.__module__.partition(\".\")\n try:\n register = self._lazy.pop(toplevel)\n except KeyError:\n pass\n else:\n register()\n return self.dispatch(cls) # recurse\n # Walk the MRO and cache the lookup result\n for cls2 in inspect.getmro(cls)[1:]:\n if cls2 in lk:\n lk[cls] = lk[cls2]\n return lk[cls2]\n raise TypeError(\"No dispatch for {0}\".format(cls))\n\n def __call__(self, arg, *args, **kwargs):\n \"\"\"\n Call the corresponding method based on type of argument.\n \"\"\"\n meth = self.dispatch(type(arg))\n return meth(arg, *args, **kwargs)\n\n @property\n def __doc__(self):\n try:\n func = self.dispatch(object)\n return func.__doc__\n except TypeError:\n return \"Single Dispatch for %s\" % self.__name__\n\n\ndef ensure_not_exists(filename):\n \"\"\"\n Ensure that a file does not exist.\n \"\"\"\n try:\n os.unlink(filename)\n except OSError as e:\n if e.errno != ENOENT:\n raise\n\n\ndef _skip_doctest(line):\n # NumPy docstring contains cursor and comment only example\n stripped = line.strip()\n if stripped == \">>>\" or stripped.startswith(\">>> #\"):\n return line\n elif \">>>\" in stripped and \"+SKIP\" not in stripped:\n if \"# doctest:\" in line:\n return line + \", +SKIP\"\n else:\n return line + \" # doctest: +SKIP\"\n else:\n return line\n\n\ndef skip_doctest(doc):\n if doc is None:\n return \"\"\n return \"\\n\".join([_skip_doctest(line) for line in doc.split(\"\\n\")])\n\n\ndef extra_titles(doc):\n lines = doc.split(\"\\n\")\n titles = {\n i: lines[i].strip()\n for i in range(len(lines) - 1)\n if lines[i + 1].strip() and all(c == \"-\" for c in lines[i + 1].strip())\n }\n\n seen = set()\n for i, title in sorted(titles.items()):\n if title in seen:\n new_title = \"Extra \" + title\n lines[i] = lines[i].replace(title, new_title)\n lines[i + 1] = lines[i + 1].replace(\"-\" * len(title), \"-\" * len(new_title))\n else:\n seen.add(title)\n\n return \"\\n\".join(lines)\n\n\ndef ignore_warning(doc, cls, name, extra=\"\", skipblocks=0):\n \"\"\"Expand docstring by adding disclaimer and extra text\"\"\"\n import inspect\n\n if inspect.isclass(cls):\n l1 = \"This docstring was copied from %s.%s.%s.\\n\\n\" % (\n cls.__module__,\n cls.__name__,\n name,\n )\n else:\n l1 = \"This docstring was copied from %s.%s.\\n\\n\" % (cls.__name__, name)\n l2 = \"Some inconsistencies with the Dask version may exist.\"\n\n i = doc.find(\"\\n\\n\")\n if i != -1:\n # Insert our warning\n head = doc[: i + 2]\n tail = doc[i + 2 :]\n while skipblocks > 0:\n i = tail.find(\"\\n\\n\")\n head = tail[: i + 2]\n tail = tail[i + 2 :]\n skipblocks -= 1\n # Indentation of next line\n indent = re.match(r\"\\s*\", tail).group(0)\n # Insert the warning, indented, with a blank line before and after\n if extra:\n more = [indent, extra.rstrip(\"\\n\") + \"\\n\\n\"]\n else:\n more = []\n bits = [head, indent, l1, indent, l2, \"\\n\\n\"] + more + [tail]\n doc = \"\".join(bits)\n\n return doc\n\n\ndef unsupported_arguments(doc, args):\n \"\"\" Mark unsupported arguments with a disclaimer \"\"\"\n lines = doc.split(\"\\n\")\n for arg in args:\n subset = [\n (i, line)\n for i, line in enumerate(lines)\n if re.match(r\"^\\s*\" + arg + \" ?:\", line)\n ]\n if len(subset) == 1:\n [(i, line)] = subset\n lines[i] = line + \" (Not supported in Dask)\"\n return \"\\n\".join(lines)\n\n\ndef _derived_from(cls, method, ua_args=[], extra=\"\", skipblocks=0):\n \"\"\" Helper function for derived_from to ease testing \"\"\"\n # do not use wraps here, as it hides keyword arguments displayed\n # in the doc\n original_method = getattr(cls, method.__name__)\n\n if isinstance(original_method, property):\n # some things like SeriesGroupBy.unique are generated.\n original_method = original_method.fget\n\n doc = original_method.__doc__\n if doc is None:\n doc = \"\"\n\n # Insert disclaimer that this is a copied docstring\n if doc:\n doc = ignore_warning(\n doc, cls, method.__name__, extra=extra, skipblocks=skipblocks\n )\n elif extra:\n doc += extra.rstrip(\"\\n\") + \"\\n\\n\"\n\n # Mark unsupported arguments\n try:\n method_args = get_named_args(method)\n original_args = get_named_args(original_method)\n not_supported = [m for m in original_args if m not in method_args]\n except ValueError:\n not_supported = []\n if len(ua_args) > 0:\n not_supported.extend(ua_args)\n if len(not_supported) > 0:\n doc = unsupported_arguments(doc, not_supported)\n\n doc = skip_doctest(doc)\n doc = extra_titles(doc)\n\n return doc\n\n\ndef derived_from(original_klass, version=None, ua_args=[], skipblocks=0):\n \"\"\"Decorator to attach original class's docstring to the wrapped method.\n\n The output structure will be: top line of docstring, disclaimer about this\n being auto-derived, any extra text associated with the method being patched,\n the body of the docstring and finally, the list of keywords that exist in\n the original method but not in the dask version.\n\n Parameters\n ----------\n original_klass: type\n Original class which the method is derived from\n version : str\n Original package version which supports the wrapped method\n ua_args : list\n List of keywords which Dask doesn't support. Keywords existing in\n original but not in Dask will automatically be added.\n skipblocks : int\n How many text blocks (paragraphs) to skip from the start of the\n docstring. Useful for cases where the target has extra front-matter.\n \"\"\"\n\n def wrapper(method):\n try:\n extra = getattr(method, \"__doc__\", None) or \"\"\n method.__doc__ = _derived_from(\n original_klass,\n method,\n ua_args=ua_args,\n extra=extra,\n skipblocks=skipblocks,\n )\n return method\n\n except AttributeError:\n module_name = original_klass.__module__.split(\".\")[0]\n\n @functools.wraps(method)\n def wrapped(*args, **kwargs):\n msg = \"Base package doesn't support '{0}'.\".format(method.__name__)\n if version is not None:\n msg2 = \" Use {0} {1} or later to use this method.\"\n msg += msg2.format(module_name, version)\n raise NotImplementedError(msg)\n\n return wrapped\n\n return wrapper\n\n\ndef funcname(func):\n \"\"\"Get the name of a function.\"\"\"\n # functools.partial\n if isinstance(func, functools.partial):\n return funcname(func.func)\n # methodcaller\n if isinstance(func, methodcaller):\n return func.method[:50]\n\n module_name = getattr(func, \"__module__\", None) or \"\"\n type_name = getattr(type(func), \"__name__\", None) or \"\"\n\n # toolz.curry\n if \"toolz\" in module_name and \"curry\" == type_name:\n return func.func_name[:50]\n # multipledispatch objects\n if \"multipledispatch\" in module_name and \"Dispatcher\" == type_name:\n return func.name[:50]\n # numpy.vectorize objects\n if \"numpy\" in module_name and \"vectorize\" == type_name:\n return (\"vectorize_\" + funcname(func.pyfunc))[:50]\n\n # All other callables\n try:\n name = func.__name__\n if name == \"<lambda>\":\n return \"lambda\"\n return name[:50]\n except AttributeError:\n return str(func)[:50]\n\n\ndef typename(typ):\n \"\"\"\n Return the name of a type\n\n Examples\n --------\n >>> typename(int)\n 'int'\n\n >>> from dask.core import literal\n >>> typename(literal)\n 'dask.core.literal'\n \"\"\"\n if not typ.__module__ or typ.__module__ == \"builtins\":\n return typ.__name__\n else:\n return typ.__module__ + \".\" + typ.__name__\n\n\ndef ensure_bytes(s):\n \"\"\"Turn string or bytes to bytes\n\n >>> ensure_bytes('123')\n b'123'\n >>> ensure_bytes('123')\n b'123'\n >>> ensure_bytes(b'123')\n b'123'\n \"\"\"\n if isinstance(s, bytes):\n return s\n if hasattr(s, \"encode\"):\n return s.encode()\n msg = \"Object %s is neither a bytes object nor has an encode method\"\n raise TypeError(msg % s)\n\n\ndef ensure_unicode(s):\n \"\"\"Turn string or bytes to bytes\n\n >>> ensure_unicode('123')\n '123'\n >>> ensure_unicode('123')\n '123'\n >>> ensure_unicode(b'123')\n '123'\n \"\"\"\n if isinstance(s, str):\n return s\n if hasattr(s, \"decode\"):\n return s.decode()\n msg = \"Object %s is neither a bytes object nor has an encode method\"\n raise TypeError(msg % s)\n\n\ndef digit(n, k, base):\n \"\"\"\n\n >>> digit(1234, 0, 10)\n 4\n >>> digit(1234, 1, 10)\n 3\n >>> digit(1234, 2, 10)\n 2\n >>> digit(1234, 3, 10)\n 1\n \"\"\"\n return n // base ** k % base\n\n\ndef insert(tup, loc, val):\n \"\"\"\n\n >>> insert(('a', 'b', 'c'), 0, 'x')\n ('x', 'b', 'c')\n \"\"\"\n L = list(tup)\n L[loc] = val\n return tuple(L)\n\n\ndef dependency_depth(dsk):\n deps, _ = get_deps(dsk)\n\n @lru_cache(maxsize=None)\n def max_depth_by_deps(key):\n if not deps[key]:\n return 1\n\n d = 1 + max(max_depth_by_deps(dep_key) for dep_key in deps[key])\n return d\n\n return max(max_depth_by_deps(dep_key) for dep_key in deps.keys())\n\n\ndef memory_repr(num):\n for x in [\"bytes\", \"KB\", \"MB\", \"GB\", \"TB\"]:\n if num < 1024.0:\n return \"%3.1f %s\" % (num, x)\n num /= 1024.0\n\n\ndef asciitable(columns, rows):\n \"\"\"Formats an ascii table for given columns and rows.\n\n Parameters\n ----------\n columns : list\n The column names\n rows : list of tuples\n The rows in the table. Each tuple must be the same length as\n ``columns``.\n \"\"\"\n rows = [tuple(str(i) for i in r) for r in rows]\n columns = tuple(str(i) for i in columns)\n widths = tuple(max(max(map(len, x)), len(c)) for x, c in zip(zip(*rows), columns))\n row_template = (\"|\" + (\" %%-%ds |\" * len(columns))) % widths\n header = row_template % tuple(columns)\n bar = \"+%s+\" % \"+\".join(\"-\" * (w + 2) for w in widths)\n data = \"\\n\".join(row_template % r for r in rows)\n return \"\\n\".join([bar, header, bar, data, bar])\n\n\ndef put_lines(buf, lines):\n if any(not isinstance(x, str) for x in lines):\n lines = [str(x) for x in lines]\n buf.write(\"\\n\".join(lines))\n\n\n_method_cache = {}\n\n\nclass methodcaller:\n \"\"\"\n Return a callable object that calls the given method on its operand.\n\n Unlike the builtin `operator.methodcaller`, instances of this class are\n serializable\n \"\"\"\n\n __slots__ = (\"method\",)\n func = property(lambda self: self.method) # For `funcname` to work\n\n def __new__(cls, method):\n if method in _method_cache:\n return _method_cache[method]\n self = object.__new__(cls)\n self.method = method\n _method_cache[method] = self\n return self\n\n def __call__(self, obj, *args, **kwargs):\n return getattr(obj, self.method)(*args, **kwargs)\n\n def __reduce__(self):\n return (methodcaller, (self.method,))\n\n def __str__(self):\n return \"<%s: %s>\" % (self.__class__.__name__, self.method)\n\n __repr__ = __str__\n\n\nclass itemgetter:\n \"\"\"\n Return a callable object that gets an item from the operand\n\n Unlike the builtin `operator.itemgetter`, instances of this class are\n serializable\n \"\"\"\n\n __slots__ = (\"index\",)\n\n def __init__(self, index):\n self.index = index\n\n def __call__(self, x):\n return x[self.index]\n\n def __reduce__(self):\n return (itemgetter, (self.index,))\n\n def __eq__(self, other):\n return type(self) is type(other) and self.index == other.index\n\n\nclass MethodCache:\n \"\"\"Attribute access on this object returns a methodcaller for that\n attribute.\n\n Examples\n --------\n >>> a = [1, 3, 3]\n >>> M.count(a, 3) == a.count(3)\n True\n \"\"\"\n\n __getattr__ = staticmethod(methodcaller)\n __dir__ = lambda self: list(_method_cache)\n\n\nM = MethodCache()\n\n\nclass SerializableLock:\n _locks = WeakValueDictionary()\n \"\"\" A Serializable per-process Lock\n\n This wraps a normal ``threading.Lock`` object and satisfies the same\n interface. However, this lock can also be serialized and sent to different\n processes. It will not block concurrent operations between processes (for\n this you should look at ``multiprocessing.Lock`` or ``locket.lock_file``\n but will consistently deserialize into the same lock.\n\n So if we make a lock in one process::\n\n lock = SerializableLock()\n\n And then send it over to another process multiple times::\n\n bytes = pickle.dumps(lock)\n a = pickle.loads(bytes)\n b = pickle.loads(bytes)\n\n Then the deserialized objects will operate as though they were the same\n lock, and collide as appropriate.\n\n This is useful for consistently protecting resources on a per-process\n level.\n\n The creation of locks is itself not threadsafe.\n \"\"\"\n\n def __init__(self, token=None):\n self.token = token or str(uuid.uuid4())\n if self.token in SerializableLock._locks:\n self.lock = SerializableLock._locks[self.token]\n else:\n self.lock = Lock()\n SerializableLock._locks[self.token] = self.lock\n\n def acquire(self, *args, **kwargs):\n return self.lock.acquire(*args, **kwargs)\n\n def release(self, *args, **kwargs):\n return self.lock.release(*args, **kwargs)\n\n def __enter__(self):\n self.lock.__enter__()\n\n def __exit__(self, *args):\n self.lock.__exit__(*args)\n\n def locked(self):\n return self.lock.locked()\n\n def __getstate__(self):\n return self.token\n\n def __setstate__(self, token):\n self.__init__(token)\n\n def __str__(self):\n return \"<%s: %s>\" % (self.__class__.__name__, self.token)\n\n __repr__ = __str__\n\n\ndef get_scheduler_lock(collection=None, scheduler=None):\n \"\"\"Get an instance of the appropriate lock for a certain situation based on\n scheduler used.\"\"\"\n from . import multiprocessing\n from .base import get_scheduler\n\n actual_get = get_scheduler(collections=[collection], scheduler=scheduler)\n\n if actual_get == multiprocessing.get:\n return multiprocessing.get_context().Manager().Lock()\n\n return SerializableLock()\n\n\ndef ensure_dict(d: Mapping[K, V], *, copy: bool = False) -> Dict[K, V]:\n \"\"\"Convert a generic Mapping into a dict.\n Optimize use case of :class:`~dask.highlevelgraph.HighLevelGraph`.\n\n Parameters\n ----------\n d : Mapping\n copy : bool\n If True, guarantee that the return value is always a shallow copy of d;\n otherwise it may be the input itself.\n \"\"\"\n if type(d) is dict:\n return d.copy() if copy else d # type: ignore\n try:\n layers = d.layers # type: ignore\n except AttributeError:\n return dict(d)\n\n unique_layers = {id(layer): layer for layer in layers.values()}\n result = {}\n for layer in unique_layers.values():\n result.update(layer)\n return result\n\n\nclass OperatorMethodMixin:\n \"\"\"A mixin for dynamically implementing operators\"\"\"\n\n __slots__ = ()\n\n @classmethod\n def _bind_operator(cls, op):\n \"\"\" bind operator to this class \"\"\"\n name = op.__name__\n\n if name.endswith(\"_\"):\n # for and_ and or_\n name = name[:-1]\n elif name == \"inv\":\n name = \"invert\"\n\n meth = \"__{0}__\".format(name)\n\n if name in (\"abs\", \"invert\", \"neg\", \"pos\"):\n setattr(cls, meth, cls._get_unary_operator(op))\n else:\n setattr(cls, meth, cls._get_binary_operator(op))\n\n if name in (\"eq\", \"gt\", \"ge\", \"lt\", \"le\", \"ne\", \"getitem\"):\n return\n\n rmeth = \"__r{0}__\".format(name)\n setattr(cls, rmeth, cls._get_binary_operator(op, inv=True))\n\n @classmethod\n def _get_unary_operator(cls, op):\n \"\"\" Must return a method used by unary operator \"\"\"\n raise NotImplementedError\n\n @classmethod\n def _get_binary_operator(cls, op, inv=False):\n \"\"\" Must return a method used by binary operator \"\"\"\n raise NotImplementedError\n\n\ndef partial_by_order(*args, **kwargs):\n \"\"\"\n\n >>> from operator import add\n >>> partial_by_order(5, function=add, other=[(1, 10)])\n 15\n \"\"\"\n function = kwargs.pop(\"function\")\n other = kwargs.pop(\"other\")\n args2 = list(args)\n for i, arg in other:\n args2.insert(i, arg)\n return function(*args2, **kwargs)\n\n\ndef is_arraylike(x):\n \"\"\"Is this object a numpy array or something similar?\n\n Examples\n --------\n >>> import numpy as np\n >>> is_arraylike(np.ones(5))\n True\n >>> is_arraylike(np.ones(()))\n True\n >>> is_arraylike(5)\n False\n >>> is_arraylike('cat')\n False\n \"\"\"\n from .base import is_dask_collection\n\n is_duck_array = hasattr(x, \"__array_function__\") or hasattr(x, \"__array_ufunc__\")\n\n return bool(\n hasattr(x, \"shape\")\n and isinstance(x.shape, tuple)\n and hasattr(x, \"dtype\")\n and not any(is_dask_collection(n) for n in x.shape)\n # We special case scipy.sparse and cupyx.scipy.sparse arrays as having partial\n # support for them is useful in scenerios where we mostly call `map_partitions`\n # or `map_blocks` with scikit-learn functions on dask arrays and dask dataframes.\n # https://github.com/dask/dask/pull/3738\n and (is_duck_array or \"scipy.sparse\" in typename(type(x)))\n )\n\n\ndef is_dataframe_like(df):\n \"\"\" Looks like a Pandas DataFrame \"\"\"\n typ = df.__class__\n return (\n all(hasattr(typ, name) for name in (\"groupby\", \"head\", \"merge\", \"mean\"))\n and all(hasattr(df, name) for name in (\"dtypes\", \"columns\"))\n and not any(hasattr(typ, name) for name in (\"name\", \"dtype\"))\n )\n\n\ndef is_series_like(s):\n \"\"\" Looks like a Pandas Series \"\"\"\n typ = s.__class__\n return (\n all(hasattr(typ, name) for name in (\"groupby\", \"head\", \"mean\"))\n and all(hasattr(s, name) for name in (\"dtype\", \"name\"))\n and \"index\" not in typ.__name__.lower()\n )\n\n\ndef is_index_like(s):\n \"\"\" Looks like a Pandas Index \"\"\"\n typ = s.__class__\n return (\n all(hasattr(s, name) for name in (\"name\", \"dtype\"))\n and \"index\" in typ.__name__.lower()\n )\n\n\ndef is_cupy_type(x):\n # TODO: avoid explicit reference to CuPy\n return \"cupy\" in str(type(x))\n\n\ndef natural_sort_key(s):\n \"\"\"\n Sorting `key` function for performing a natural sort on a collection of\n strings\n\n See https://en.wikipedia.org/wiki/Natural_sort_order\n\n Parameters\n ----------\n s : str\n A string that is an element of the collection being sorted\n\n Returns\n -------\n tuple[str or int]\n Tuple of the parts of the input string where each part is either a\n string or an integer\n\n Examples\n --------\n >>> a = ['f0', 'f1', 'f2', 'f8', 'f9', 'f10', 'f11', 'f19', 'f20', 'f21']\n >>> sorted(a)\n ['f0', 'f1', 'f10', 'f11', 'f19', 'f2', 'f20', 'f21', 'f8', 'f9']\n >>> sorted(a, key=natural_sort_key)\n ['f0', 'f1', 'f2', 'f8', 'f9', 'f10', 'f11', 'f19', 'f20', 'f21']\n \"\"\"\n return [int(part) if part.isdigit() else part for part in re.split(r\"(\\d+)\", s)]\n\n\ndef factors(n):\n \"\"\"Return the factors of an integer\n\n https://stackoverflow.com/a/6800214/616616\n \"\"\"\n seq = ([i, n // i] for i in range(1, int(pow(n, 0.5) + 1)) if n % i == 0)\n return set(functools.reduce(list.__add__, seq))\n\n\ndef parse_bytes(s):\n \"\"\"Parse byte string to numbers\n\n >>> from dask.utils import parse_bytes\n >>> parse_bytes('100')\n 100\n >>> parse_bytes('100 MB')\n 100000000\n >>> parse_bytes('100M')\n 100000000\n >>> parse_bytes('5kB')\n 5000\n >>> parse_bytes('5.4 kB')\n 5400\n >>> parse_bytes('1kiB')\n 1024\n >>> parse_bytes('1e6')\n 1000000\n >>> parse_bytes('1e6 kB')\n 1000000000\n >>> parse_bytes('MB')\n 1000000\n >>> parse_bytes(123)\n 123\n >>> parse_bytes('5 foos') # doctest: +SKIP\n ValueError: Could not interpret 'foos' as a byte unit\n \"\"\"\n if isinstance(s, (int, float)):\n return int(s)\n s = s.replace(\" \", \"\")\n if not any(char.isdigit() for char in s):\n s = \"1\" + s\n\n for i in range(len(s) - 1, -1, -1):\n if not s[i].isalpha():\n break\n index = i + 1\n\n prefix = s[:index]\n suffix = s[index:]\n\n try:\n n = float(prefix)\n except ValueError as e:\n raise ValueError(\"Could not interpret '%s' as a number\" % prefix) from e\n\n try:\n multiplier = byte_sizes[suffix.lower()]\n except KeyError as e:\n raise ValueError(\"Could not interpret '%s' as a byte unit\" % suffix) from e\n\n result = n * multiplier\n return int(result)\n\n\nbyte_sizes = {\n \"kB\": 10 ** 3,\n \"MB\": 10 ** 6,\n \"GB\": 10 ** 9,\n \"TB\": 10 ** 12,\n \"PB\": 10 ** 15,\n \"KiB\": 2 ** 10,\n \"MiB\": 2 ** 20,\n \"GiB\": 2 ** 30,\n \"TiB\": 2 ** 40,\n \"PiB\": 2 ** 50,\n \"B\": 1,\n \"\": 1,\n}\nbyte_sizes = {k.lower(): v for k, v in byte_sizes.items()}\nbyte_sizes.update({k[0]: v for k, v in byte_sizes.items() if k and \"i\" not in k})\nbyte_sizes.update({k[:-1]: v for k, v in byte_sizes.items() if k and \"i\" in k})\n\n\ndef format_time(n):\n \"\"\"format integers as time\n\n >>> from dask.utils import format_time\n >>> format_time(1)\n '1.00 s'\n >>> format_time(0.001234)\n '1.23 ms'\n >>> format_time(0.00012345)\n '123.45 us'\n >>> format_time(123.456)\n '123.46 s'\n \"\"\"\n if n >= 1:\n return \"%.2f s\" % n\n if n >= 1e-3:\n return \"%.2f ms\" % (n * 1e3)\n return \"%.2f us\" % (n * 1e6)\n\n\ndef format_time_ago(n: datetime) -> str:\n \"\"\"Calculate a '3 hours ago' type string from a Python datetime.\n\n Examples\n --------\n >>> from datetime import datetime, timedelta\n\n >>> now = datetime.now()\n >>> format_time_ago(now)\n 'Just now'\n\n >>> past = datetime.now() - timedelta(minutes=1)\n >>> format_time_ago(past)\n '1 minute ago'\n\n >>> past = datetime.now() - timedelta(minutes=2)\n >>> format_time_ago(past)\n '2 minutes ago'\n\n >>> past = datetime.now() - timedelta(hours=1)\n >>> format_time_ago(past)\n '1 hour ago'\n\n >>> past = datetime.now() - timedelta(hours=6)\n >>> format_time_ago(past)\n '6 hours ago'\n\n >>> past = datetime.now() - timedelta(days=1)\n >>> format_time_ago(past)\n '1 day ago'\n\n >>> past = datetime.now() - timedelta(days=5)\n >>> format_time_ago(past)\n '5 days ago'\n\n >>> past = datetime.now() - timedelta(days=8)\n >>> format_time_ago(past)\n '1 week ago'\n\n >>> past = datetime.now() - timedelta(days=16)\n >>> format_time_ago(past)\n '2 weeks ago'\n\n >>> past = datetime.now() - timedelta(days=190)\n >>> format_time_ago(past)\n '6 months ago'\n\n >>> past = datetime.now() - timedelta(days=800)\n >>> format_time_ago(past)\n '2 years ago'\n\n \"\"\"\n units = {\n \"years\": lambda diff: diff.days / 365,\n \"months\": lambda diff: diff.days / 30.436875, # Average days per month\n \"weeks\": lambda diff: diff.days / 7,\n \"days\": lambda diff: diff.days,\n \"hours\": lambda diff: diff.seconds / 3600,\n \"minutes\": lambda diff: diff.seconds % 3600 / 60,\n }\n diff = datetime.now() - n\n for unit in units:\n dur = int(units[unit](diff))\n if dur > 0:\n if dur == 1: # De-pluralize\n unit = unit[:-1]\n return \"%s %s ago\" % (dur, unit)\n return \"Just now\"\n\n\ndef format_bytes(n):\n \"\"\"Format bytes as text\n\n >>> from dask.utils import format_bytes\n >>> format_bytes(1)\n '1 B'\n >>> format_bytes(1234)\n '1.23 kB'\n >>> format_bytes(12345678)\n '12.35 MB'\n >>> format_bytes(1234567890)\n '1.23 GB'\n >>> format_bytes(1234567890000)\n '1.23 TB'\n >>> format_bytes(1234567890000000)\n '1.23 PB'\n \"\"\"\n if n > 1e15:\n return \"%0.2f PB\" % (n / 1e15)\n if n > 1e12:\n return \"%0.2f TB\" % (n / 1e12)\n if n > 1e9:\n return \"%0.2f GB\" % (n / 1e9)\n if n > 1e6:\n return \"%0.2f MB\" % (n / 1e6)\n if n > 1e3:\n return \"%0.2f kB\" % (n / 1000)\n return \"%d B\" % n\n\n\ntimedelta_sizes = {\n \"s\": 1,\n \"ms\": 1e-3,\n \"us\": 1e-6,\n \"ns\": 1e-9,\n \"m\": 60,\n \"h\": 3600,\n \"d\": 3600 * 24,\n}\n\ntds2 = {\n \"second\": 1,\n \"minute\": 60,\n \"hour\": 60 * 60,\n \"day\": 60 * 60 * 24,\n \"millisecond\": 1e-3,\n \"microsecond\": 1e-6,\n \"nanosecond\": 1e-9,\n}\ntds2.update({k + \"s\": v for k, v in tds2.items()})\ntimedelta_sizes.update(tds2)\ntimedelta_sizes.update({k.upper(): v for k, v in timedelta_sizes.items()})\n\n\ndef parse_timedelta(s, default=\"seconds\"):\n \"\"\"Parse timedelta string to number of seconds\n\n Examples\n --------\n >>> from datetime import timedelta\n >>> from dask.utils import parse_timedelta\n >>> parse_timedelta('3s')\n 3\n >>> parse_timedelta('3.5 seconds')\n 3.5\n >>> parse_timedelta('300ms')\n 0.3\n >>> parse_timedelta(timedelta(seconds=3)) # also supports timedeltas\n 3\n \"\"\"\n if s is None:\n return None\n if isinstance(s, timedelta):\n s = s.total_seconds()\n return int(s) if int(s) == s else s\n if isinstance(s, Number):\n s = str(s)\n s = s.replace(\" \", \"\")\n if not s[0].isdigit():\n s = \"1\" + s\n\n for i in range(len(s) - 1, -1, -1):\n if not s[i].isalpha():\n break\n index = i + 1\n\n prefix = s[:index]\n suffix = s[index:] or default\n\n n = float(prefix)\n\n multiplier = timedelta_sizes[suffix.lower()]\n\n result = n * multiplier\n if int(result) == result:\n result = int(result)\n return result\n\n\ndef has_keyword(func, keyword):\n try:\n return keyword in inspect.signature(func).parameters\n except Exception:\n return False\n\n\ndef ndimlist(seq):\n if not isinstance(seq, (list, tuple)):\n return 0\n elif not seq:\n return 1\n else:\n return 1 + ndimlist(seq[0])\n\n\ndef iter_chunks(sizes, max_size):\n \"\"\"Split sizes into chunks of total max_size each\n\n Parameters\n ----------\n sizes : iterable of numbers\n The sizes to be chunked\n max_size : number\n Maximum total size per chunk.\n It must be greater or equal than each size in sizes\n \"\"\"\n chunk, chunk_sum = [], 0\n iter_sizes = iter(sizes)\n size = next(iter_sizes, None)\n while size is not None:\n assert size <= max_size\n if chunk_sum + size <= max_size:\n chunk.append(size)\n chunk_sum += size\n size = next(iter_sizes, None)\n else:\n assert chunk\n yield chunk\n chunk, chunk_sum = [], 0\n if chunk:\n yield chunk\n\n\nhex_pattern = re.compile(\"[a-f]+\")\n\n\ndef key_split(s):\n \"\"\"\n >>> key_split('x')\n 'x'\n >>> key_split('x-1')\n 'x'\n >>> key_split('x-1-2-3')\n 'x'\n >>> key_split(('x-2', 1))\n 'x'\n >>> key_split(\"('x-2', 1)\")\n 'x'\n >>> key_split('hello-world-1')\n 'hello-world'\n >>> key_split(b'hello-world-1')\n 'hello-world'\n >>> key_split('ae05086432ca935f6eba409a8ecd4896')\n 'data'\n >>> key_split('<module.submodule.myclass object at 0xdaf372')\n 'myclass'\n >>> key_split(None)\n 'Other'\n >>> key_split('x-abcdefab') # ignores hex\n 'x'\n >>> key_split('_(x)') # strips unpleasant characters\n 'x'\n \"\"\"\n if type(s) is bytes:\n s = s.decode()\n if type(s) is tuple:\n s = s[0]\n try:\n words = s.split(\"-\")\n if not words[0][0].isalpha():\n result = words[0].strip(\"_'()\\\"\")\n else:\n result = words[0]\n for word in words[1:]:\n if word.isalpha() and not (\n len(word) == 8 and hex_pattern.match(word) is not None\n ):\n result += \"-\" + word\n else:\n break\n if len(result) == 32 and re.match(r\"[a-f0-9]{32}\", result):\n return \"data\"\n else:\n if result[0] == \"<\":\n result = result.strip(\"<>\").split()[0].split(\".\")[-1]\n return result\n except Exception:\n return \"Other\"\n\n\ndef stringify(obj, exclusive: Optional[Iterable] = None):\n \"\"\"Convert an object to a string\n\n If ``exclusive`` is specified, search through `obj` and convert\n values that are in ``exclusive``.\n\n Note that when searching through dictionaries, only values are\n converted, not the keys.\n\n Parameters\n ----------\n obj : Any\n Object (or values within) to convert to string\n exclusive: Iterable, optional\n Set of values to search for when converting values to strings\n\n Returns\n -------\n result : type(obj)\n Stringified copy of ``obj`` or ``obj`` itself if it is already a\n string or bytes.\n\n Examples\n --------\n >>> stringify(b'x')\n b'x'\n >>> stringify('x')\n 'x'\n >>> stringify({('a',0):('a',0), ('a',1): ('a',1)})\n \"{('a', 0): ('a', 0), ('a', 1): ('a', 1)}\"\n >>> stringify({('a',0):('a',0), ('a',1): ('a',1)}, exclusive={('a',0)})\n {('a', 0): \"('a', 0)\", ('a', 1): ('a', 1)}\n \"\"\"\n\n typ = type(obj)\n if typ is str or typ is bytes:\n return obj\n elif exclusive is None:\n return str(obj)\n\n if typ is tuple and obj:\n from .optimization import SubgraphCallable\n\n obj0 = obj[0]\n if type(obj0) is SubgraphCallable:\n obj0 = obj0\n return (\n SubgraphCallable(\n stringify(obj0.dsk, exclusive),\n obj0.outkey,\n stringify(obj0.inkeys, exclusive),\n obj0.name,\n ),\n ) + tuple(stringify(x, exclusive) for x in obj[1:])\n elif callable(obj0):\n return (obj0,) + tuple(stringify(x, exclusive) for x in obj[1:])\n\n if typ is list:\n return [stringify(v, exclusive) for v in obj]\n if typ is dict:\n return {k: stringify(v, exclusive) for k, v in obj.items()}\n try:\n if obj in exclusive:\n return stringify(obj)\n except TypeError: # `obj` not hashable\n pass\n if typ is tuple: # If the tuple itself isn't a key, check its elements\n return tuple(stringify(v, exclusive) for v in obj)\n return obj\n\n\ndef stringify_collection_keys(obj):\n \"\"\"Convert all collection keys in ``obj`` to strings.\n\n This is a specialized version of ``stringify()`` that only converts keys\n of the form: ``(\"a string\", ...)``\n \"\"\"\n\n typ = type(obj)\n if typ is tuple and obj:\n obj0 = obj[0]\n if type(obj0) is str or type(obj0) is bytes:\n return stringify(obj)\n if callable(obj0):\n return (obj0,) + tuple(stringify_collection_keys(x) for x in obj[1:])\n if typ is list:\n return [stringify_collection_keys(v) for v in obj]\n if typ is dict:\n return {k: stringify_collection_keys(v) for k, v in obj.items()}\n if typ is tuple: # If the tuple itself isn't a key, check its elements\n return tuple(stringify_collection_keys(v) for v in obj)\n return obj\n\n\ntry:\n _cached_property = functools.cached_property\nexcept AttributeError:\n # TODO: Copied from functools.cached_property in python 3.8. Remove when minimum\n # supported python version is 3.8:\n _NOT_FOUND = object()\n\n class _cached_property:\n def __init__(self, func):\n self.func = func\n self.attrname = None\n self.__doc__ = func.__doc__\n self.lock = RLock()\n\n def __set_name__(self, owner, name):\n if self.attrname is None:\n self.attrname = name\n elif name != self.attrname:\n raise TypeError(\n \"Cannot assign the same cached_property to two different names \"\n f\"({self.attrname!r} and {name!r}).\"\n )\n\n def __get__(self, instance, owner=None):\n if instance is None:\n return self\n if self.attrname is None:\n raise TypeError(\n \"Cannot use cached_property instance without calling __set_name__ on it.\"\n )\n try:\n cache = instance.__dict__\n except AttributeError: # not all objects have __dict__ (e.g. class defines slots)\n msg = (\n f\"No '__dict__' attribute on {type(instance).__name__!r} \"\n f\"instance to cache {self.attrname!r} property.\"\n )\n raise TypeError(msg) from None\n val = cache.get(self.attrname, _NOT_FOUND)\n if val is _NOT_FOUND:\n with self.lock:\n # check if another thread filled cache while we awaited lock\n val = cache.get(self.attrname, _NOT_FOUND)\n if val is _NOT_FOUND:\n val = self.func(instance)\n try:\n cache[self.attrname] = val\n except TypeError:\n msg = (\n f\"The '__dict__' attribute on {type(instance).__name__!r} instance \"\n f\"does not support item assignment for caching {self.attrname!r} property.\"\n )\n raise TypeError(msg) from None\n return val\n\n\nclass cached_property(_cached_property):\n \"\"\"Read only version of functools.cached_property.\"\"\"\n\n def __set__(self, instance, val):\n \"\"\"Raise an error when attempting to set a cached property.\"\"\"\n raise AttributeError(\"Can't set attribute\")\n"
] |
[
[
"numpy.empty",
"numpy.random.RandomState",
"numpy.allclose",
"numpy.frombuffer",
"numpy.cumsum"
]
] |
ALEXKIRNAS/DataScience
|
[
"14119565b8fdde042f6ea3070bc0f30db26620c0"
] |
[
"CS231n/assignment2/cs231n/optim.py"
] |
[
"import numpy as np\n\n\"\"\"\nThis file implements various first-order update rules that are commonly used\nfor training neural networks. Each update rule accepts current weights and the\ngradient of the loss with respect to those weights and produces the next set of\nweights. Each update rule has the same interface:\n\ndef update(w, dw, config=None):\n\nInputs:\n - w: A numpy array giving the current weights.\n - dw: A numpy array of the same shape as w giving the gradient of the\n loss with respect to w.\n - config: A dictionary containing hyperparameter values such as learning\n rate, momentum, etc. If the update rule requires caching values over many\n iterations, then config will also hold these cached values.\n\nReturns:\n - next_w: The next point after the update.\n - config: The config dictionary to be passed to the next iteration of the\n update rule.\n\nNOTE: For most update rules, the default learning rate will probably not\nperform well; however the default values of the other hyperparameters should\nwork well for a variety of different problems.\n\nFor efficiency, update rules may perform in-place updates, mutating w and\nsetting next_w equal to w.\n\"\"\"\n\n\ndef sgd(w, dw, config=None):\n \"\"\"\n Performs vanilla stochastic gradient descent.\n\n config format:\n - learning_rate: Scalar learning rate.\n \"\"\"\n if config is None: config = {}\n config.setdefault('learning_rate', 1e-2)\n\n w -= config['learning_rate'] * dw\n return w, config\n\n\ndef sgd_momentum(w, dw, config=None):\n \"\"\"\n Performs stochastic gradient descent with momentum.\n\n config format:\n - learning_rate: Scalar learning rate.\n - momentum: Scalar between 0 and 1 giving the momentum value.\n Setting momentum = 0 reduces to sgd.\n - velocity: A numpy array of the same shape as w and dw used to store a\n moving average of the gradients.\n \"\"\"\n if config is None: config = {}\n config.setdefault('learning_rate', 1e-2)\n config.setdefault('momentum', 0.9)\n v = config.get('velocity', np.zeros_like(w))\n\n next_w = None\n ###########################################################################\n # TODO: Implement the momentum update formula. Store the updated value in #\n # the next_w variable. You should also use and update the velocity v. #\n ###########################################################################\n\n lr = config['learning_rate']\n momentum = config['momentum']\n\n v = momentum * v - lr * dw\n next_w = w + v\n\n ###########################################################################\n # END OF YOUR CODE #\n ###########################################################################\n config['velocity'] = v\n\n return next_w, config\n\n\n\ndef rmsprop(x, dx, config=None):\n \"\"\"\n Uses the RMSProp update rule, which uses a moving average of squared\n gradient values to set adaptive per-parameter learning rates.\n\n config format:\n - learning_rate: Scalar learning rate.\n - decay_rate: Scalar between 0 and 1 giving the decay rate for the squared\n gradient cache.\n - epsilon: Small scalar used for smoothing to avoid dividing by zero.\n - cache: Moving average of second moments of gradients.\n \"\"\"\n if config is None: config = {}\n config.setdefault('learning_rate', 1e-2)\n config.setdefault('decay_rate', 0.99)\n config.setdefault('epsilon', 1e-8)\n config.setdefault('cache', np.zeros_like(x))\n\n next_x = None\n ###########################################################################\n # TODO: Implement the RMSprop update formula, storing the next value of x #\n # in the next_x variable. Don't forget to update cache value stored in #\n # config['cache']. #\n ###########################################################################\n\n cache = config['cache']\n decay = config['decay_rate']\n eps = config['epsilon']\n lr = config['learning_rate']\n\n cache = decay * cache + (1 - decay) * (dx * dx)\n next_x = x - lr * dx / np.sqrt(cache + eps)\n\n config['cache'] = cache\n\n ###########################################################################\n # END OF YOUR CODE #\n ###########################################################################\n\n return next_x, config\n\n\ndef adam(x, dx, config=None):\n \"\"\"\n Uses the Adam update rule, which incorporates moving averages of both the\n gradient and its square and a bias correction term.\n\n config format:\n - learning_rate: Scalar learning rate.\n - beta1: Decay rate for moving average of first moment of gradient.\n - beta2: Decay rate for moving average of second moment of gradient.\n - epsilon: Small scalar used for smoothing to avoid dividing by zero.\n - m: Moving average of gradient.\n - v: Moving average of squared gradient.\n - t: Iteration number.\n \"\"\"\n if config is None: config = {}\n config.setdefault('learning_rate', 1e-3)\n config.setdefault('beta1', 0.9)\n config.setdefault('beta2', 0.999)\n config.setdefault('epsilon', 1e-8)\n config.setdefault('m', np.zeros_like(x))\n config.setdefault('v', np.zeros_like(x))\n config.setdefault('t', 1)\n\n next_x = None\n ###########################################################################\n # TODO: Implement the Adam update formula, storing the next value of x in #\n # the next_x variable. Don't forget to update the m, v, and t variables #\n # stored in config. #\n ###########################################################################\n \n m = config['m']\n v = config['v']\n t = config['t']\n eps = config['epsilon']\n lr = config['learning_rate']\n beta1 = config['beta1']\n beta2 = config['beta2']\n\n t += 1\n m = beta1 * m + (1 - beta1) * dx\n v = beta2 * v + (1 - beta2) * (dx ** 2)\n\n m_corrected = m / (1 - beta1 ** t)\n v_corrected = v / (1 - beta2 ** t)\n\n next_x = x - lr * m_corrected / (np.sqrt(v_corrected) + eps)\n\n config['m'], config['v'], config['t'] = m, v, t\n\n ###########################################################################\n # END OF YOUR CODE #\n ###########################################################################\n\n return next_x, config\n"
] |
[
[
"numpy.zeros_like",
"numpy.sqrt"
]
] |
akathpal/RoboND-Perception-Project
|
[
"6304d3114f36760cea6ed31b40e99618166a3a7c"
] |
[
"code/train_svm.py"
] |
[
"#!/usr/bin/env python\nimport pickle\nimport itertools\nimport numpy as np\nimport matplotlib.pyplot as plt\nfrom sklearn import svm\nfrom sklearn.preprocessing import LabelEncoder, StandardScaler\nfrom sklearn import cross_validation\nfrom sklearn import metrics\n\ndef plot_confusion_matrix(cm, classes,\n normalize=False,\n title='Confusion matrix',\n cmap=plt.cm.Blues):\n \"\"\"\n This function prints and plots the confusion matrix.\n Normalization can be applied by setting `normalize=True`.\n \"\"\"\n if normalize:\n cm = cm.astype('float') / cm.sum(axis=1)[:, np.newaxis]\n plt.imshow(cm, interpolation='nearest', cmap=cmap)\n plt.title(title)\n plt.colorbar()\n tick_marks = np.arange(len(classes))\n plt.xticks(tick_marks, classes, rotation=45)\n plt.yticks(tick_marks, classes)\n\n thresh = cm.max() / 2.\n for i, j in itertools.product(range(cm.shape[0]), range(cm.shape[1])):\n plt.text(j, i, '{0:.2f}'.format(cm[i, j]),\n horizontalalignment=\"center\",\n color=\"white\" if cm[i, j] > thresh else \"black\")\n\n plt.tight_layout()\n plt.ylabel('True label')\n plt.xlabel('Predicted label')\n\n# Load training data from disk\ntraining_set = pickle.load(open('training_set.sav', 'rb'))\n\n# Format the features and labels for use with scikit learn\nfeature_list = []\nlabel_list = []\n\nfor item in training_set:\n if np.isnan(item[0]).sum() < 1:\n feature_list.append(item[0])\n label_list.append(item[1])\n\nprint('Features in Training Set: {}'.format(len(training_set)))\nprint('Invalid Features in Training set: {}'.format(len(training_set)-len(feature_list)))\n\nX = np.array(feature_list)\n# Fit a per-column scaler\nX_scaler = StandardScaler().fit(X)\n# Apply the scaler to X\nX_train = X_scaler.transform(X)\ny_train = np.array(label_list)\n\n# Convert label strings to numerical encoding\nencoder = LabelEncoder()\ny_train = encoder.fit_transform(y_train)\n\n# Create classifier\nclf = svm.SVC(kernel='linear',cache_size=500,C=1)\n\n# Set up 5-fold cross-validation\nkf = cross_validation.KFold(len(X_train),\n n_folds=5,\n shuffle=True,\n random_state=1)\n\n# Perform cross-validation\nscores = cross_validation.cross_val_score(cv=kf,\n estimator=clf,\n X=X_train, \n y=y_train,\n scoring='accuracy'\n )\nprint('Scores: ' + str(scores))\nprint('Accuracy: %0.2f (+/- %0.2f)' % (scores.mean(), 2*scores.std()))\n\n# Gather predictions\npredictions = cross_validation.cross_val_predict(cv=kf,\n estimator=clf,\n X=X_train, \n y=y_train\n )\n\naccuracy_score = metrics.accuracy_score(y_train, predictions)\nprint('accuracy score: '+str(accuracy_score))\n\nconfusion_matrix = metrics.confusion_matrix(y_train, predictions)\n\nclass_names = encoder.classes_.tolist()\n\n\n#Train the classifier\nclf.fit(X=X_train, y=y_train)\n\nmodel = {'classifier': clf, 'classes': encoder.classes_, 'scaler': X_scaler}\n\n# Save classifier to disk\npickle.dump(model, open('model.sav', 'wb'))\n\n# Plot non-normalized confusion matrix\nplt.figure()\nplot_confusion_matrix(confusion_matrix, classes=encoder.classes_,\n title='Confusion matrix, without normalization')\n\n# Plot normalized confusion matrix\nplt.figure()\nplot_confusion_matrix(confusion_matrix, classes=encoder.classes_, normalize=True,\n title='Normalized confusion matrix')\n\nplt.show()\n"
] |
[
[
"sklearn.metrics.confusion_matrix",
"sklearn.cross_validation.cross_val_score",
"matplotlib.pyplot.xticks",
"matplotlib.pyplot.colorbar",
"sklearn.cross_validation.cross_val_predict",
"sklearn.svm.SVC",
"sklearn.metrics.accuracy_score",
"matplotlib.pyplot.tight_layout",
"sklearn.preprocessing.LabelEncoder",
"numpy.array",
"matplotlib.pyplot.title",
"matplotlib.pyplot.yticks",
"matplotlib.pyplot.figure",
"matplotlib.pyplot.show",
"numpy.isnan",
"sklearn.preprocessing.StandardScaler",
"matplotlib.pyplot.xlabel",
"matplotlib.pyplot.ylabel",
"matplotlib.pyplot.imshow"
]
] |
Cocopyth/MscThesis
|
[
"60162bc779a3a668e7447b60bb9a4b2a616b8093"
] |
[
"amftrack/pipeline/functions/post_processing/time_hypha.py"
] |
[
"from amftrack.pipeline.functions.image_processing.hyphae_id_surf import get_pixel_growth_and_new_children\r\nimport numpy as np\r\nimport networkx as nx\r\nfrom amftrack.pipeline.functions.post_processing.util import get_length_um_edge, is_in_study_zone,get_length_um\r\nfrom scipy import sparse\r\n\r\ndef get_time_since_start(hypha, t, tp1, args):\r\n exp = hypha.experiment\r\n seconds = (exp.dates[t]-exp.dates[hypha.ts[0]]).total_seconds()\r\n return(\"time_since_emergence\",seconds/3600)\r\n\r\ndef get_time_since_begin_exp(hypha, t, tp1, args):\r\n exp = hypha.experiment\r\n seconds = (exp.dates[t]-exp.dates[0]).total_seconds()\r\n return(\"time_since_begin_exp\",seconds/3600)\r\n\r\ndef get_timedelta(hypha,t,tp1,args):\r\n exp = hypha.experiment\r\n seconds = (exp.dates[tp1]-exp.dates[t]).total_seconds()\r\n return(\"timedelta\",seconds/3600)\r\n\r\n\r\ndef get_speed(hypha,t,tp1,args):\r\n try:\r\n pixels,nodes = get_pixel_growth_and_new_children(hypha,t,tp1)\r\n speed = np.sum([get_length_um(seg) for seg in pixels])/get_timedelta(hypha,t,tp1,None)[1]\r\n return('speed',speed)\r\n except nx.exception.NetworkXNoPath:\r\n print('not_connected',hypha.end.label,hypha.get_root(tp1).label)\r\n return('speed',None)\r\n\r\ndef get_timestep(hypha,t,tp1,args):\r\n return(\"timestep\",t)\r\n\r\ndef get_timestep_init(hypha,t,tp1,args):\r\n return(\"timestep_init\",hypha.ts[0])\r\n\r\ndef get_time_init(hypha,t,tp1,args):\r\n return(\"time_init\",get_timedelta(hypha,0,hypha.ts[0],None)[1])\r\n\r\ndef get_degree(hypha,t,tp1,args):\r\n return(\"degree\",hypha.end.degree(t))\r\n\r\ndef get_width_tip_edge(hypha,t,tp1,args):\r\n try:\r\n edges = hypha.get_nodes_within(t)[1]\r\n if len(edges)>0:\r\n edge = edges[-1]\r\n width = edge.width(t)\r\n return('width_tip_edge',width)\r\n else:\r\n return('width_tip_edge',None)\r\n except nx.exception.NetworkXNoPath:\r\n return('width_tip_edge',None)\r\n \r\ndef get_width_root_edge(hypha,t,tp1,args):\r\n try:\r\n edges = hypha.get_nodes_within(t)[1]\r\n if len(edges)>0:\r\n edge = edges[0]\r\n width = edge.width(t)\r\n return('width_tip_edge',width)\r\n else:\r\n return('width_tip_edge',None)\r\n except nx.exception.NetworkXNoPath:\r\n return('width_root_edge',None)\r\n \r\ndef get_width_average(hypha,t,tp1,args):\r\n try:\r\n edges = hypha.get_nodes_within(t)[1]\r\n widths = np.array([edge.width(t) for edge in edges])\r\n lengths = np.array([get_length_um_edge(edge,t) for edge in edges])\r\n av_width = np.sum(widths*lengths)/np.sum(lengths)\r\n return('av_width',av_width)\r\n except nx.exception.NetworkXNoPath:\r\n return('av_width',None)\r\n \r\ndef has_reached_final_pos(hypha,t,tp1,args):\r\n tf = hypha.ts[-1]\r\n pos_end = hypha.end.pos(tf)\r\n thresh = 40\r\n return('has_reached_final_pos',str(np.linalg.norm(hypha.end.pos(t)-pos_end)<thresh))\r\n\r\ndef get_distance_final_pos(hypha,t,tp1,args):\r\n tf = hypha.ts[-1]\r\n pos_end = hypha.end.pos(tf)\r\n return('distance_final_pos',np.linalg.norm(hypha.end.pos(t)-pos_end))\r\n\r\n\r\ndef local_density(hypha,t,tp1,args):\r\n window = args[0]\r\n exp = hypha.experiment\r\n pos = hypha.end.pos(t)\r\n x,y = pos[0],pos[1]\r\n skeletons = [sparse.csr_matrix(skel) for skel in exp.skeletons]\r\n skeleton=skeletons[t][x-window:x+window,y-window:y+window]\r\n density = skeleton.count_nonzero()/(window**2*1.725)*10**6\r\n return(f'density_window{window}',density)\r\n\r\ndef in_ROI(hypha,t,tp1,args):\r\n return('in_ROI', str(np.all(is_in_study_zone(hypha.end,t,1000,150))))\r\n"
] |
[
[
"numpy.sum",
"scipy.sparse.csr_matrix"
]
] |
erik-grennberg-jansson/matern_sfem
|
[
"1e9468084abf41cc0ae85f1b4b1254904ed2d72f"
] |
[
"pool_example.py"
] |
[
"\nfrom numpy.random import normal\nimport numpy as np\n\nfrom dolfin import *\nfrom ctypes import *\n\nfrom mesh_generation import sphere_mesh\nfrom utils import solve_problem\nfrom Problem import Problem \n\nL = 25 # number of KL terms\nbeta = 0.51 #smoothness parameter \nkappa =1.0 #length scale parameter \nk = 2 #quadrature step size\nh = 0.01 #mesh size\n\npath_to_c = './fast_spher_harms.so' #path to right hand side\nsph = CDLL(path_to_c)\nsph.sin_term.restype = c_float #set types \nsph.cos_term.restype = c_float\nsph.fast_grf.restype = c_float\nsph.fast_grf.argtypes = ( \n c_int,c_float,c_float,c_float, POINTER(c_float))\ndef GRF(noterms,rands,x,y,z): #function to evaluate GRF in a point\n return(sph.fast_grf(c_int(L),c_float(x),c_float(y),c_float(z),rands))\n\n\n\n\ndef wn(rands,L): #create right hand side class \n pf = (c_float*len(rands))(*rands)\n prands = cast(pf,POINTER(c_float)) #set correct type for cfloat. \n class right_hand_side(UserExpression):\n def eval(self,value,x):\n value[0]= GRF(L,prands,x[0],x[1],x[2])\n def value_shape(self):\n return()\n return(right_hand_side(degree=2))\n\n\nsurfaces = [sphere_mesh(h)] #create mesh, function spaces, trial func, test func, assemble matrices etc. \nVs = [FunctionSpace(surfaces[0],FiniteElement(\"Lagrange\",surfaces[0].ufl_cell(),1))]\nus = [TrialFunction(Vs[0])]\nvs = [TestFunction(Vs[0])]\nAs = [assemble(inner(grad(us[0]),grad(vs[0]))*dx)] \nBs = [assemble(us[0]*vs[0]*dx)]\n\ndef surf(): #put in function so that it can be pickled. \n return surfaces[0]\ndef fenstuff():\n return (Vs[0],us[0],vs[0])\ndef AB():\n return(As[0],Bs[0])\nprob = Problem(surf, fenstuff,wn,beta, k,kappa,AB,L=L) #create problem \nrands = normal(0,1,[L+1,L+1,2]).flatten() #set random numbers\nprob.set_rand(rands) \n\n\n\nsol = Function(Vs[0]) #placeholder for solution \n\nsol.vector()[:] = solve_problem(prob,5) #solve!\n\nfile = File('field.pvd') #save for plotting. \nfile << sol\n"
] |
[
[
"numpy.random.normal"
]
] |
leonjovanovic/ml-real-estate
|
[
"3f1ca50eb793dbe77d35eda14dfc468d28861a5a"
] |
[
"linear_regression/main.py"
] |
[
"import json\r\nimport numpy as np\r\nfrom sklearn.utils import shuffle\r\nimport matplotlib.pyplot as plt\r\n\r\nHYPERPARAMETERS = {\r\n 'num_features': 7,\r\n 'features': ('lokacija2', 'kvadratura', 'sprat', 'broj_soba', 'parking', 'lift', 'terasa'),\r\n 'learning_rate': [0.001, 0.003, 0.01, 0.03], # 0.001, 0.003, 0.01, 0.03, 0.1, 0.3, 1\r\n 'mini_batch_size': [16, 32, 64, 128],\r\n 'random_seed': 1,\r\n 'outer_cv_fold': 10,\r\n 'inner_cv_fold': 10,\r\n 'iterations': 50\r\n}\r\n\r\n\r\ndef plotting(it, J_train, J_val):\r\n epochs = list(range(it))\r\n plt.plot(epochs, J_train, '-b+', label='J train')\r\n plt.plot(epochs, J_val, '-r', label='J val')\r\n plt.legend()\r\n plt.show()\r\n\r\n\r\ndef train_main():\r\n # ------------------------------------------PREPROCESSING DATA----------------------------------------------------------\r\n # I Create and import data and output\r\n with open(\"../data/data_flats_sale.json\", 'r') as infile:\r\n json_data = json.load(infile)\r\n m = len(json_data)\r\n data = np.zeros([m, HYPERPARAMETERS['num_features']])\r\n output = np.zeros([m, 1])\r\n i = 0\r\n for r in json_data:\r\n j = 0\r\n for feature in HYPERPARAMETERS['features']:\r\n data[i, j] = r[feature]\r\n j += 1\r\n output[i, 0] = r['cena']\r\n i += 1\r\n # II Normalization (Standardization)\r\n mean = np.mean(data, axis=0)\r\n data = data - mean\r\n std = np.std(data, axis=0)\r\n data = data / std\r\n # III Shuffling data\r\n data, output = shuffle(data, output, random_state=HYPERPARAMETERS['random_seed'])\r\n # IV Adding first column of ones for bias term\r\n data = np.hstack((np.ones((m, 1)), data))\r\n # -------------------------------------------------------------------------------------------------------------------\r\n # --------------------------Nested Cross Validation For Finding BEST HYPERPARAMETERS--------------------------------\r\n # -------------------------------------------------------------------------------------------------------------------\r\n # I Splitting data k-fold on train and test set\r\n k = HYPERPARAMETERS['outer_cv_fold']\r\n l = HYPERPARAMETERS['inner_cv_fold']\r\n J_total = np.array([], dtype=np.int64).reshape(0, len(HYPERPARAMETERS['learning_rate']) * len(\r\n HYPERPARAMETERS['mini_batch_size']))\r\n for i in range(k):\r\n index = slice(int(i * m / k), int(i * m / k + m / k))\r\n test_data, test_output = data[index, :], output[index, :]\r\n trainval_data, trainval_output = np.delete(data, index, axis=0), np.delete(output, index, axis=0)\r\n J_avg_val_list = np.array([], dtype=np.int64).reshape(0, len(HYPERPARAMETERS['learning_rate']) * len(\r\n HYPERPARAMETERS['mini_batch_size']))\r\n # II Splitting data l-fold on train and validation set\r\n for j in range(l):\r\n index = slice(int(j * (m - m / k) / l), int(j * (m - m / k) / l + (m - m / k) / l))\r\n val_data, val_output = trainval_data[index, :], trainval_output[index, :]\r\n train_data, train_output = np.delete(trainval_data, index, axis=0), np.delete(trainval_output, index,\r\n axis=0)\r\n # III Creating model parameters initialized with zeros\r\n params = np.zeros([1, HYPERPARAMETERS['num_features'] + 1])\r\n # IV Splitting training data on mini-batches\r\n J_val_list = []\r\n for lr in HYPERPARAMETERS['learning_rate']:\r\n for m_batch in HYPERPARAMETERS['mini_batch_size']:\r\n J_val = 0\r\n # -----------------------------------One trained model----------------------------------------------\r\n for it in range(HYPERPARAMETERS['iterations']):\r\n for b in range(0, train_data.shape[0], m_batch):\r\n X = train_data[b:b + m_batch] # 32x8\r\n h = np.dot(X, params.transpose()) # 32x1\r\n # V Loss funtion\r\n L = h - train_output[b:b + m_batch] # 32x1\r\n J_train = np.sum(L ** 2) / (2 * X.shape[0])\r\n # VI Updating model parameters\r\n dJ = np.dot(L.transpose(), X) # 1 x NUM_FEATURES + 1\r\n params = params - (lr / X.shape[0]) * dJ\r\n # --------------------Validation------------------------------------------------------------\r\n h_val = np.dot(val_data, params.transpose())\r\n L_val = h_val - val_output\r\n J_val = np.sum(L_val ** 2) / (2 * val_data.shape[0])\r\n # ------------------------------------------------------------------------------------------\r\n J_val_list.append(J_val)\r\n J_avg_val_list = np.vstack([J_avg_val_list, J_val_list])\r\n # print(str(J_avg_val_list.shape[0]) + \"/10\")\r\n temp = list(np.mean(J_avg_val_list, axis=0))\r\n J_total = np.vstack([J_total, temp])\r\n print(\"Hyperparameter index for iteration \" + str(J_total.shape[0]) + \"/10 is \" + str(temp.index(min(temp))))\r\n temp = list(np.mean(J_total, axis=0))\r\n print(\"Hyperparameter index is \" + str(temp.index(min(temp))))\r\n # best_hparam_index = temp.index(min(temp))\r\n best_hparam_index = 14\r\n best_hyperparams = [HYPERPARAMETERS['learning_rate'][best_hparam_index // len(HYPERPARAMETERS['learning_rate'])],\r\n HYPERPARAMETERS['mini_batch_size'][best_hparam_index % len(HYPERPARAMETERS['mini_batch_size'])]]\r\n # -------------------------------------------------------------------------------------------------------------------\r\n # ---------------------Cross Validation On Evaluation For Finding MEAN PERFORMANCE----------------------------------\r\n # -------------------------------------------------------------------------------------------------------------------\r\n RMSE = []\r\n MAE = []\r\n for i in range(k):\r\n index = slice(int(i * m / k), int(i * m / k + m / k))\r\n test_data, test_output = data[index, :], output[index, :]\r\n trainval_data, trainval_output = np.delete(data, index, axis=0), np.delete(output, index, axis=0)\r\n params = np.zeros([1, HYPERPARAMETERS['num_features'] + 1])\r\n lr, m_batch = best_hyperparams[0], best_hyperparams[1]\r\n # -----------------------------------One trained model----------------------------------------------\r\n for it in range(HYPERPARAMETERS['iterations']):\r\n for b in range(0, trainval_data.shape[0], int(m_batch)):\r\n X = trainval_data[b:b + m_batch] # 32x8\r\n h = np.dot(X, params.transpose()) # 32x1\r\n # V Loss funtion\r\n L = h - trainval_output[b:b + m_batch] # 32x1\r\n J_train = np.sum(L ** 2) / (2 * X.shape[0])\r\n # VI Updating model parameters\r\n dJ = np.dot(L.transpose(), X) # 1 x NUM_FEATURES + 1\r\n params = params - (lr / X.shape[0]) * dJ\r\n # --------------------Evaluation------------------------------------------------------------\r\n h_test = np.dot(test_data, params.transpose())\r\n L_test = h_test - test_output\r\n RMSE.append(np.sqrt(np.sum(L_test ** 2) / test_data.shape[0]))\r\n MAE.append(np.sum(np.absolute(L_test)) / test_data.shape[0])\r\n # ------------------------------------------------------------------------------------------\r\n print(\"Average root mean squared error: \" + str(np.mean(RMSE)))\r\n print(\"Average mean absolute error: \" + str(np.mean(MAE)))\r\n # -------------------------------------------------------------------------------------------------------------------\r\n # ----------------------------Training on whole data set for BEST PARAMETERS----------------------------------------\r\n # -------------------------------------------------------------------------------------------------------------------\r\n params = np.zeros([1, HYPERPARAMETERS['num_features'] + 1])\r\n lr, m_batch = best_hyperparams[0], best_hyperparams[1]\r\n # -----------------------------------One trained model----------------------------------------------\r\n for it in range(HYPERPARAMETERS['iterations']):\r\n for b in range(0, data.shape[0], int(m_batch)):\r\n X = data[b:b + m_batch] # 32x8\r\n h = np.dot(X, params.transpose()) # 32x1\r\n # V Loss funtion\r\n L = h - output[b:b + m_batch] # 32x1\r\n J_train = np.sum(L ** 2) / (2 * X.shape[0])\r\n # VI Updating model parameters\r\n dJ = np.dot(L.transpose(), X) # 1 x NUM_FEATURES + 1\r\n params = params - (lr / X.shape[0]) * dJ\r\n with open('model_parameters.json', 'w') as output_file:\r\n output_file.write(json.dumps({\"parameters\": params.tolist(), 'mean': mean.tolist(), 'std': std.tolist()}, indent=4))\r\n\r\n\r\ntrain_main()\r\n"
] |
[
[
"numpy.array",
"numpy.delete",
"numpy.zeros",
"numpy.sum",
"matplotlib.pyplot.plot",
"matplotlib.pyplot.legend",
"numpy.ones",
"numpy.mean",
"numpy.std",
"numpy.absolute",
"matplotlib.pyplot.show",
"numpy.vstack",
"sklearn.utils.shuffle"
]
] |
cuulee/eltgan
|
[
"17a5627f1526607e56696e6b832cc8f6f0d43897"
] |
[
"data/unaligned_dataset.py"
] |
[
"import torch\nfrom torch import nn\nimport os.path\nimport torchvision.transforms as transforms\nfrom data.base_dataset import BaseDataset, get_transform\nfrom data.image_folder import make_dataset, store_dataset\nimport random\nfrom PIL import Image\nimport PIL\nfrom pdb import set_trace as st\n\ndef pad_tensor(input):\n \n height_org, width_org = input.shape[2], input.shape[3]\n divide = 16\n\n if width_org % divide != 0 or height_org % divide != 0:\n\n width_res = width_org % divide\n height_res = height_org % divide\n if width_res != 0:\n width_div = divide - width_res\n pad_left = int(width_div / 2)\n pad_right = int(width_div - pad_left)\n else:\n pad_left = 0\n pad_right = 0\n\n if height_res != 0:\n height_div = divide - height_res\n pad_top = int(height_div / 2)\n pad_bottom = int(height_div - pad_top)\n else:\n pad_top = 0\n pad_bottom = 0\n\n padding = nn.ReflectionPad2d((pad_left, pad_right, pad_top, pad_bottom))\n input = padding(input).data\n else:\n pad_left = 0\n pad_right = 0\n pad_top = 0\n pad_bottom = 0\n\n height, width = input.shape[2], input.shape[3]\n assert width % divide == 0, 'width cant divided by stride'\n assert height % divide == 0, 'height cant divided by stride'\n\n return input, pad_left, pad_right, pad_top, pad_bottom\n\ndef pad_tensor_back(input, pad_left, pad_right, pad_top, pad_bottom):\n height, width = input.shape[2], input.shape[3]\n return input[:,:, pad_top: height - pad_bottom, pad_left: width - pad_right]\n\n\nclass UnalignedDataset(BaseDataset):\n def initialize(self, opt):\n self.opt = opt\n self.root = opt.dataroot\n\n \"\"\"\n Added object based loading\n\n \"\"\"\n if opt.object is not None:\n self.dir_A = os.path.join(opt.dataroot, opt.phase + '_A', opt.object)\n self.dir_B = os.path.join(opt.dataroot, opt.phase + '_B', opt.object)\n else:\n self.dir_A = os.path.join(opt.dataroot, opt.phase + '_A')\n self.dir_B = os.path.join(opt.dataroot, opt.phase + '_B') \n\n\n # self.A_paths = make_dataset(self.dir_A)\n # self.B_paths = make_dataset(self.dir_B)\n self.A_imgs, self.A_paths = store_dataset(self.dir_A)\n self.B_imgs, self.B_paths = store_dataset(self.dir_B)\n\n # self.A_paths = sorted(self.A_paths)\n # self.B_paths = sorted(self.B_paths)\n self.A_size = len(self.A_paths)\n self.B_size = len(self.B_paths)\n \n self.transform = get_transform(opt)\n\n def __getitem__(self, index):\n # A_path = self.A_paths[index % self.A_size]\n # B_path = self.B_paths[index % self.B_size]\n\n # A_img = Image.open(A_path).convert('RGB')\n # B_img = Image.open(B_path).convert('RGB')\n A_img = self.A_imgs[index % self.A_size]\n B_img = self.B_imgs[index % self.B_size]\n A_path = self.A_paths[index % self.A_size]\n B_path = self.B_paths[index % self.B_size]\n # A_size = A_img.size\n # B_size = B_img.size\n # A_size = A_size = (A_size[0]//16*16, A_size[1]//16*16)\n # B_size = B_size = (B_size[0]//16*16, B_size[1]//16*16)\n # A_img = A_img.resize(A_size, Image.BICUBIC)\n # B_img = B_img.resize(B_size, Image.BICUBIC)\n # A_gray = A_img.convert('LA')\n # A_gray = 255.0-A_gray\n A_img = self.transform(A_img)\n B_img = self.transform(B_img)\n\n \n if self.opt.resize_or_crop == 'no':\n r,g,b = A_img[0]+1, A_img[1]+1, A_img[2]+1\n A_gray = 1. - (0.299*r+0.587*g+0.114*b)/2.\n A_gray = torch.unsqueeze(A_gray, 0)\n input_img = A_img\n # A_gray = (1./A_gray)/255.\n else:\n w = A_img.size(2)\n h = A_img.size(1)\n \n # A_gray = (1./A_gray)/255.\n if (not self.opt.no_flip) and random.random() < 0.5:\n idx = [i for i in range(A_img.size(2) - 1, -1, -1)]\n idx = torch.LongTensor(idx)\n A_img = A_img.index_select(2, idx)\n B_img = B_img.index_select(2, idx)\n if (not self.opt.no_flip) and random.random() < 0.5:\n idx = [i for i in range(A_img.size(1) - 1, -1, -1)]\n idx = torch.LongTensor(idx)\n A_img = A_img.index_select(1, idx)\n B_img = B_img.index_select(1, idx)\n if self.opt.vary == 1 and (not self.opt.no_flip) and random.random() < 0.5:\n times = random.randint(self.opt.low_times,self.opt.high_times)/100.\n input_img = (A_img+1)/2./times\n input_img = input_img*2-1\n else:\n input_img = A_img\n if self.opt.lighten:\n B_img = (B_img + 1)/2.\n B_img = (B_img - torch.min(B_img))/(torch.max(B_img) - torch.min(B_img))\n B_img = B_img*2. -1\n r,g,b = input_img[0]+1, input_img[1]+1, input_img[2]+1\n A_gray = 1. - (0.299*r+0.587*g+0.114*b)/2.\n A_gray = torch.unsqueeze(A_gray, 0)\n return {'A': A_img, 'B': B_img, 'A_gray': A_gray, 'input_img': input_img,\n 'A_paths': A_path, 'B_paths': B_path}\n\n def __len__(self):\n return max(self.A_size, self.B_size)\n\n def name(self):\n return 'UnalignedDataset'\n\n\n"
] |
[
[
"torch.min",
"torch.max",
"torch.unsqueeze",
"torch.LongTensor",
"torch.nn.ReflectionPad2d"
]
] |
starxchina/Patch-Diffusion
|
[
"97f4881ae04c335fc1a13ded432d8e10a47df5f0"
] |
[
"pd_module.py"
] |
[
"import torch\nimport torch.nn as nn\n\nclass Patch_Diffusion(nn.Module):\n def __init__(self, input_feature_channel, patch_channel, patch_num, l=1):\n \"\"\"Patch Diffusion Moudle\n\n Args:\n input_feature_channel (int): the channel number of input feature\n patch_channel (int): length of patch vector\n patch_num (int): number of patches\n l (int): number of diffusions. Defaults to 1.\n \"\"\"\n super(Patch_Diffusion, self).__init__()\n self.input_feature_channel = input_feature_channel\n self.patch_channel = patch_channel\n self.patch_num = patch_num\n self.l = l\n self.psi = nn.Conv2d(input_feature_channel, patch_channel, kernel_size=1)\n self.rho = nn.Conv2d(input_feature_channel, input_feature_channel, kernel_size=1)\n\n modules = []\n for i in range(l):\n modules.append(nn.ReLU(inplace=True))\n modules.append(nn.Conv1d(input_feature_channel, input_feature_channel, kernel_size=1, bias=False)) # W\n self.diffusion = nn.ModuleList(modules)\n\n self.bn = nn.BatchNorm2d(input_feature_channel, eps=1e-04)\n\n def forward(self, x):\n b, c, h, w = x.size()\n patch = self.psi(x).view(b, self.patch_channel, -1).permute(0, 2, 1)\n gram_mat = torch.matmul(patch, patch.permute(0,2,1))\n denominator_mid = torch.sqrt(torch.sum(patch.pow(2), dim=2).view(b, -1, 1))\n denominator = torch.matmul(denominator_mid, denominator_mid.permute(0,2,1)) + 1e-08\n attention_mat = gram_mat / denominator\n x_graph = self.rho(x).view(b, self.input_feature_channel, -1)\n\n for i in range(self.l):\n x_graph = torch.matmul(attention_mat, x_graph.permute(0,2,1).contiguous()).permute(0,2,1)\n x_graph = self.diffusion[i*2](x_graph)\n x_graph = self.diffusion[i*2+1](x_graph)\n\n x_out = x_graph.view(b, c, h, w)\n out = x + self.bn(x_out)\n return out, patch"
] |
[
[
"torch.nn.ModuleList",
"torch.nn.Conv1d",
"torch.nn.BatchNorm2d",
"torch.nn.ReLU",
"torch.nn.Conv2d"
]
] |
1nF0rmed/alf
|
[
"84bf56379d5fb552fb43365c5a77d8edc46d06c3"
] |
[
"alf/trainers/policy_trainer_test.py"
] |
[
"# Copyright (c) 2020 Horizon Robotics and ALF Contributors. All Rights Reserved.\n#\n# Licensed under the Apache License, Version 2.0 (the \"License\");\n# you may not use this file except in compliance with the License.\n# You may obtain a copy of the License at\n#\n# http://www.apache.org/licenses/LICENSE-2.0\n#\n# Unless required by applicable law or agreed to in writing, software\n# distributed under the License is distributed on an \"AS IS\" BASIS,\n# WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.\n# See the License for the specific language governing permissions and\n# limitations under the License.\n\nimport functools\nimport tempfile\nimport torch\n\nimport alf\nfrom alf.algorithms.hypernetwork_algorithm import HyperNetwork\nfrom alf.algorithms.rl_algorithm_test import MyEnv, MyAlg\nfrom alf.trainers.policy_trainer import RLTrainer, TrainerConfig, play\nfrom alf.trainers.policy_trainer import create_dataset, SLTrainer\nfrom alf.utils import common\n\n\nclass MyRLTrainer(RLTrainer):\n def _create_environment(self,\n nonparallel=False,\n random_seed=None,\n register=True):\n env = MyEnv(3)\n if register:\n self._register_env(env)\n return env\n\n\nclass MySLTrainer(SLTrainer):\n def _create_dataset(self):\n return create_dataset(\n dataset_name='test', train_batch_size=50, test_batch_size=10)\n\n\nclass TrainerTest(alf.test.TestCase):\n def test_rl_trainer(self):\n with tempfile.TemporaryDirectory() as root_dir:\n conf = TrainerConfig(\n algorithm_ctor=MyAlg,\n root_dir=root_dir,\n unroll_length=5,\n num_iterations=100)\n\n # test train\n trainer = MyRLTrainer(conf)\n self.assertEqual(RLTrainer.progress(), 0)\n trainer.train()\n self.assertEqual(RLTrainer.progress(), 1)\n\n alg = trainer._algorithm\n env = common.get_env()\n time_step = common.get_initial_time_step(env)\n state = alg.get_initial_predict_state(env.batch_size)\n policy_step = alg.rollout_step(time_step, state)\n logits = policy_step.info.logits\n print(\"logits: \", logits)\n self.assertTrue(torch.all(logits[:, 1] > logits[:, 0]))\n self.assertTrue(torch.all(logits[:, 1] > logits[:, 2]))\n\n # test checkpoint\n conf.num_iterations = 200\n new_trainer = MyRLTrainer(conf)\n new_trainer._restore_checkpoint()\n self.assertEqual(RLTrainer.progress(), 0.5)\n time_step = common.get_initial_time_step(env)\n state = alg.get_initial_predict_state(env.batch_size)\n policy_step = alg.rollout_step(time_step, state)\n logits = policy_step.info.logits\n self.assertTrue(torch.all(logits[:, 1] > logits[:, 0]))\n self.assertTrue(torch.all(logits[:, 1] > logits[:, 2]))\n\n new_trainer.train()\n self.assertEqual(RLTrainer.progress(), 1)\n\n # TODO: test play. Need real env to test.\n\n def test_sl_trainer(self):\n with tempfile.TemporaryDirectory() as root_dir:\n conf = TrainerConfig(\n algorithm_ctor=functools.partial(\n HyperNetwork,\n hidden_layers=None,\n loss_type='regression',\n optimizer=alf.optimizers.Adam(lr=1e-4, weight_decay=1e-4)),\n root_dir=root_dir,\n num_checkpoints=1,\n evaluate=True,\n eval_interval=1,\n num_iterations=1)\n\n # test train\n trainer = MySLTrainer(conf)\n self.assertEqual(SLTrainer.progress(), 0)\n trainer.train()\n self.assertEqual(SLTrainer.progress(), 1)\n\n # test checkpoint\n conf2 = TrainerConfig(\n algorithm_ctor=functools.partial(\n HyperNetwork,\n hidden_layers=None,\n loss_type='regression',\n optimizer=alf.optimizers.Adam(lr=1e-4, weight_decay=1e-4)),\n root_dir=root_dir,\n num_checkpoints=1,\n evaluate=True,\n eval_interval=1,\n num_iterations=2)\n\n new_trainer = MySLTrainer(conf2)\n new_trainer._restore_checkpoint()\n self.assertEqual(SLTrainer.progress(), 0.5)\n new_trainer.train()\n self.assertEqual(SLTrainer.progress(), 1)\n\n\nif __name__ == \"__main__\":\n alf.test.main()\n"
] |
[
[
"torch.all"
]
] |
CMU-IDS-2020/a3-yet-another-streamlit
|
[
"c09421e428621de99c68823c9a75045da49b98d8"
] |
[
"interactive_polynomial.py"
] |
[
"import numpy as np\nimport pandas as pd\nimport streamlit as st\nimport altair as alt\nfrom multivariate_LR import LinearRegression\nfrom sklearn.preprocessing import StandardScaler\nfrom sklearn.model_selection import train_test_split\n\n# time-series data for losses\ndef get_loss_df(train_losses, test_losses):\n # losses should be [float]\n num_epoch = len(test_losses)\n return pd.DataFrame({'Epoch' : range(num_epoch), 'test MSE' : test_losses, 'train MSE' : train_losses})\n\n# multivariate-polynomial plot line points for each epoch each variable\ndef get_plotline_df(x_range, num_var, num_exp, weights_on_epoch, variable_names, measure_name):\n num_epoch = len(weights_on_epoch)\n num_samples = len(x_range)\n assert num_epoch > 0\n assert num_var * num_exp == len(weights_on_epoch[0]) - 1\n assert len(variable_names) > 0\n\n epoch_dfs = []\n for epoch_id in range(num_epoch):\n plotline_df = pd.DataFrame({'Epoch' : [epoch_id] * num_samples})\n for var_id in range(len(variable_names)):\n var_name = variable_names[var_id]\n # x fields for each plot\n plotline_df[var_name] = x_range\n # y fields for each plot - supposed polynomials of the model.\n y_values = np.zeros(num_samples)\n prod = np.ones(num_samples)\n weight = weights_on_epoch[epoch_id]\n for idx in range(var_id, num_var * num_exp, num_var):\n prod = prod * x_range\n y_values = y_values + prod * weight[idx]\n # add interception\n y_values = y_values + weight[-1]\n plotline_df[measure_name + \" | \" + var_name] = y_values\n \n epoch_dfs.append(plotline_df)\n\n return pd.concat(epoch_dfs)\n\n# residual points for each epoch each variable, varying in time-series.\n# X is scaled and concatenated with different exponants\ndef get_residual_xy_df(X, y, num_var, num_exp, weights_on_epoch, variable_names, measure_name):\n num_epoch = len(weights_on_epoch)\n num_samples = len(y)\n assert num_epoch > 0\n assert num_var * num_exp == len(weights_on_epoch[0]) - 1\n assert len(variable_names) > 0\n assert X.shape[1] == num_var * num_exp\n\n # calculate the residual masks for each variable, which will be subtracted by ground truth y.\n masks = []\n for var_id in range(len(variable_names)):\n mask_ = np.ones(num_var * num_exp)\n mask_[list(range(var_id, num_var * num_exp, num_var))] = 0.0\n masks.append(mask_)\n\n epoch_dfs = []\n for epoch_id in range(num_epoch):\n plotscat_df = pd.DataFrame({'Epoch' : [epoch_id] * num_samples})\n for var_id in range(len(variable_names)):\n var_name = variable_names[var_id]\n # x fields for each plot\n plotscat_df[var_name] = X[:, var_id].reshape(num_samples)\n # y fields for each plot - real y subtract the residuals\n weight = weights_on_epoch[epoch_id][0:-1]\n redisuals = X @ (masks[var_id] * weight)\n y_values = y - redisuals\n plotscat_df[measure_name + \" | \" + var_name] = y_values\n \n epoch_dfs.append(plotscat_df)\n\n return pd.concat(epoch_dfs)\n\n\ndef interactive_polynomial(feat_X, feat_y, variable_names, measure_name):\n '''\n feat_X: np.array(n_sample, n_feature), not need to be scaled\n feat_y: np.array(n_sample)\n variable_names: [str], size equal to n_feature, name of each column.\n measure_name: str, name of the column to regression on.\n '''\n st.sidebar.markdown('### Tune the regression model with the widgets')\n num_exp = st.sidebar.slider('Polynomial Exponantial', 1, 3, 2, 1)\n n_iter = st.sidebar.slider('Maximum Training Epoch', 0, 200, 100, 10)\n learning_rate = st.sidebar.slider('Learning Rate', 0.01, 0.25, 0.03, 0.01)\n # lam = st.slider('Lambda', 0.0, 2e-4, 1e-4, 1e-5)\n\n # scale the feature matrix.\n # then apply polynomial tranformation. Concatenate high-exps\n scaler = StandardScaler()\n feat_X = scaler.fit_transform(feat_X)\n Xs = []\n for num in range(1, num_exp + 1):\n Xs.append(feat_X ** num)\n feat_X = np.concatenate(Xs, axis=1)\n\n lr_model = LinearRegression(lam=1e-4)\n\n\n X_train, X_test, y_train, y_test = train_test_split(feat_X, feat_y, test_size=0.5, random_state=0)\n losses, test_losses, weights_epochs = lr_model.fit(X_train, y_train, X_test, y_test, n_iter, learning_rate)\n\n # selector based on epoch\n epoch_selection = alt.selection_single(nearest=True, on='mouseover',\n fields=['Epoch'], empty='none', init={'Epoch' : n_iter})\n\n # times-series for the loss\n loss_df = get_loss_df(losses, test_losses)\n loss_df = loss_df.melt('Epoch', var_name='loss', value_name='MSE')\n # selector layer\n selector_layer = alt.Chart(loss_df).mark_point().encode(\n alt.X('Epoch'),\n opacity=alt.value(0)\n ).add_selection(epoch_selection)\n loss_line = alt.Chart(loss_df).mark_line().encode(\n alt.X('Epoch'),\n alt.Y('MSE'),\n color='loss:N'\n )\n ruleline = alt.Chart(loss_df).mark_rule().encode(\n alt.X('Epoch'),\n color=alt.value('grey')\n ).transform_filter(epoch_selection)\n tooltip = alt.Chart(loss_df).mark_text(align='left', dx=5, dy=-5).encode(\n alt.X('Epoch'),\n alt.Y('MSE'),\n alt.Text('MSE')\n ).transform_filter(epoch_selection)\n\n # st.write(alt.layer(selector_layer, loss_line, tooltip, ruleline).properties(width=700, height=300))\n curr_chart = alt.layer(selector_layer, loss_line, tooltip, ruleline).properties(width=700, height=300).resolve_scale(color='independent')\n\n # get the layered visualization of residual line plot and residual X-Y points.\n residual_xy_df = get_residual_xy_df(X_test, y_test, len(variable_names), num_exp, weights_epochs, variable_names, measure_name)\n plotline_df = get_plotline_df(np.arange(-2.0, 2.0, 0.05), len(variable_names), num_exp, weights_epochs, variable_names, measure_name)\n\n # list the residual plot on each dimension, three in a row to look better.\n curr_list = []\n for var_name in variable_names:\n # residual points and the line plot together.\n residual_xy_plot = alt.Chart(residual_xy_df).mark_point().encode(\n alt.X(var_name),\n alt.Y(measure_name + \" | \" + var_name)\n ).transform_filter(epoch_selection)\n plotline_plot = alt.Chart(plotline_df).mark_line().encode(\n alt.X(var_name),\n alt.Y(measure_name + \" | \" + var_name),\n color = alt.value('red')\n ).transform_filter(epoch_selection)\n curr_list.append(alt.layer(plotline_plot, residual_xy_plot).properties(width=200, height=200))\n if len(curr_list) == 3:\n curr_chart = curr_chart & (curr_list[0] | curr_list[1] | curr_list[2])\n curr_list = []\n \n if curr_list != []:\n last_row = curr_list[0]\n for idx in range(1, len(curr_list)):\n last_row = last_row | curr_list[idx]\n curr_chart = curr_chart & last_row\n\n # at last, write everything to streamlit UI.\n st.write(curr_chart)\n\nif __name__ == '__main__':\n n_sample = 100\n X = np.random.randn(n_sample, 2)\n y = (X ** 2).sum(axis=1) + np.random.normal(scale=0.2, size=n_sample)\n interactive_polynomial(X, y, ['X1', 'X2'], 'y')\n \n\n"
] |
[
[
"numpy.concatenate",
"numpy.random.normal",
"numpy.zeros",
"sklearn.preprocessing.StandardScaler",
"pandas.DataFrame",
"numpy.ones",
"numpy.random.randn",
"numpy.arange",
"pandas.concat",
"sklearn.model_selection.train_test_split"
]
] |
zhongdixiu/tensorflow-learn
|
[
"42728c94ec7506a9a69fda54bc17d3244ce4bf57"
] |
[
"dataset_learn_slim.py"
] |
[
"# coding=utf-8\n\n\"\"\"以MNIST为例,使用slim.data\n\"\"\"\n\nimport os\nimport tensorflow as tf\n\nslim = tf.contrib.slim\n\ndef get_data(data_dir, num_samples, num_class, file_pattern='*.tfrecord'):\n \"\"\"返回slim.data.Dataset\n\n :param data_dir: tfrecord文件路径\n :param num_samples: 样本数目\n :param num_class: 类别数目\n :param file_pattern: tfrecord文件格式\n :return:\n \"\"\"\n file_pattern = os.path.join(data_dir, file_pattern)\n keys_to_features = {\n \"image/encoded\": tf.FixedLenFeature((), tf.string, default_value=\"\"),\n \"image/format\": tf.FixedLenFeature((), tf.string, default_value=\"raw\"),\n 'image/height': tf.FixedLenFeature((), tf.int64, default_value=tf.zeros([], dtype=tf.int64)),\n 'image/width': tf.FixedLenFeature((), tf.int64, default_value=tf.zeros([], dtype=tf.int64)),\n \"image/class/label\": tf.FixedLenFeature((), tf.int64, default_value=tf.zeros([], dtype=tf.int64))\n }\n items_to_handlers = {\n \"image\": slim.tfexample_decoder.Image(channels=1),\n \"label\": slim.tfexample_decoder.Tensor(\"image/class/label\")\n }\n\n decoder = slim.tfexample_decoder.TFExampleDecoder(keys_to_features, items_to_handlers)\n items_to_descriptions = {\n \"image\": 'A color image of varying size',\n \"label\": 'A single interger between 0 and ' + str(num_class - 1)\n }\n\n return slim.dataset.Dataset(\n data_sources=file_pattern,\n reader=tf.TFRecordReader,\n decoder=decoder,\n num_samples=num_samples,\n items_to_descriptions=items_to_descriptions,\n num_classes=num_class,\n label_to_names=label_to_name\n )\n\n\nNUM_EPOCH = 2\nBATCH_SIZE = 8\nNUM_CLASS = 10\nNUM_SAMPLE = 60000\n\nlabel_to_name = {'0': 'one', '1': 'two', '3': 'three', '4': 'four', '5': 'five',\n '6': 'six', '7': 'seven', '8': 'eight', '9': 'nine'}\ndata_dir = './'\ndataset = get_data(data_dir, NUM_SAMPLE, NUM_CLASS, 'mnist_train.tfrecord')\ndata_provider = slim.dataset_data_provider.DatasetDataProvider(dataset)\n[image, label] = data_provider.get(['image', 'label'])\n\n# 组合数据\nimages, labels = tf.train.batch([image, label], batch_size=BATCH_SIZE)\nlabels = slim.one_hot_encoding(labels, NUM_CLASS)\n\n"
] |
[
[
"tensorflow.FixedLenFeature",
"tensorflow.zeros",
"tensorflow.train.batch"
]
] |
mbinfokyaw/n2v
|
[
"480d94a4e87c08c26e4e8c301468b125682b67f3"
] |
[
"tests/test_Noise2VoidDataWrapper.py"
] |
[
"from n2v.internals.N2V_DataWrapper import N2V_DataWrapper\n\nimport numpy as np\n\ndef test_subpatch_sampling():\n\n def create_data(in_shape, out_shape):\n X, Y = np.random.rand(*in_shape), np.random.rand(*in_shape)\n X_Batches, Y_Batches = np.zeros(out_shape), np.zeros(out_shape)\n indices = np.arange(in_shape[0])\n np.random.shuffle(indices)\n\n return X, Y, X_Batches, Y_Batches, indices[:in_shape[0]//2]\n\n def _sample2D(in_shape, out_shape, seed):\n X, Y, X_Batches, Y_Batches, indices = create_data(in_shape, out_shape)\n np.random.seed(seed)\n N2V_DataWrapper.__subpatch_sampling2D__(X, X_Batches, indices,\n range=in_shape[1:3]-out_shape[1:3], shape=out_shape[1:3])\n\n assert ([*X_Batches.shape] == out_shape).all()\n np.random.seed(seed)\n range_y = in_shape[1] - out_shape[1]\n range_x = in_shape[2] - out_shape[2]\n for j in indices:\n assert np.sum(X_Batches[j]) != 0\n y_start = np.random.randint(0, range_y + 1)\n x_start = np.random.randint(0, range_x + 1)\n assert np.sum(X_Batches[j] - X[j, y_start:y_start+out_shape[1], x_start:x_start+out_shape[2]]) == 0\n\n for j in range(in_shape[0]):\n if j not in indices:\n assert np.sum(X_Batches[j]) == 0\n\n def _sample3D(in_shape, out_shape, seed):\n X, Y, X_Batches, Y_Batches, indices = create_data(in_shape, out_shape)\n np.random.seed(seed)\n N2V_DataWrapper.__subpatch_sampling3D__(X, X_Batches, indices,\n range=in_shape[1:4]-out_shape[1:4], shape=out_shape[1:4])\n\n assert ([*X_Batches.shape] == out_shape).all()\n np.random.seed(seed)\n range_z = in_shape[1] - out_shape[1]\n range_y = in_shape[2] - out_shape[2]\n range_x = in_shape[3] - out_shape[3]\n for j in indices:\n assert np.sum(X_Batches[j]) != 0\n z_start = np.random.randint(0, range_z + 1)\n y_start = np.random.randint(0, range_y + 1)\n x_start = np.random.randint(0, range_x + 1)\n assert np.sum(X_Batches[j] - X[j, z_start:z_start+out_shape[1], y_start:y_start+out_shape[2], x_start:x_start+out_shape[3]]) == 0\n\n for j in range(in_shape[0]):\n if j not in indices:\n assert np.sum(X_Batches[j]) == 0\n\n _sample2D(np.array([20, 64, 64, 2]), np.array([20, 32, 32, 2]), 1)\n _sample2D(np.array([10, 25, 25, 1]), np.array([10, 12, 12, 1]), 2)\n _sample2D(np.array([10, 25, 25, 3]), np.array([10, 13, 13, 3]), 3)\n\n _sample3D(np.array([20, 64, 64, 64, 2]), np.array([20, 32, 32, 32, 2]), 1)\n _sample3D(np.array([10, 25, 25, 25, 1]), np.array([10, 12, 12, 12, 1]), 2)\n _sample3D(np.array([10, 25, 25, 25, 3]), np.array([10, 13, 13, 13, 3]), 3)\n\n\ndef test_random_float_coords():\n boxsize = 13\n np.random.seed(1)\n coords = (np.random.rand() * boxsize, np.random.rand() * boxsize)\n np.random.seed(1)\n assert next(N2V_DataWrapper.__rand_float_coords2D__(boxsize)) == coords\n\n boxsize = 3\n np.random.seed(1)\n coords = (np.random.rand() * boxsize, np.random.rand() * boxsize, np.random.rand() * boxsize)\n np.random.seed(1)\n assert next(N2V_DataWrapper.__rand_float_coords3D__(boxsize)) == coords\n\n\ndef test_coord_gen():\n coord_gen = N2V_DataWrapper.__rand_float_coords2D__(13)\n shape = [128, 128]\n for i in range(100):\n coords = next(coord_gen)\n assert coords[0] < shape[0]\n assert coords[1] < shape[1]\n\n\n coord_gen = N2V_DataWrapper.__rand_float_coords3D__(4)\n shape = [55, 55, 55]\n for i in range(100):\n coords = next(coord_gen)\n assert coords[0] < shape[0]\n assert coords[1] < shape[1]\n assert coords[2] < shape[2]\n\n\ndef test_n2vWrapper_getitem():\n def create_data(y_shape):\n Y = np.random.rand(*y_shape)\n return Y\n\n def random_neighbor_withCP_uniform(patch, coord, dims):\n return np.random.rand(1,1)\n\n def _getitem2D(y_shape):\n Y = create_data(y_shape)\n n_chan = y_shape[-1]//2\n if n_chan == 1:\n X = Y[:,:,:,0][:,:,:,np.newaxis]\n else:\n X = Y[:,:,:,:n_chan]\n val_manipulator = random_neighbor_withCP_uniform\n dw = N2V_DataWrapper(X, Y, 4, perc_pix=0.198, shape=(32, 32), value_manipulation=val_manipulator)\n\n x_batch, y_batch = dw.__getitem__(0)\n assert x_batch.shape == (4, 32, 32, int(n_chan))\n assert y_batch.shape == (4, 32, 32, int(2*n_chan))\n # At least one pixel has to be a blind-spot per batch sample\n assert np.sum(y_batch[..., n_chan:]) >= 4 * n_chan\n # At most four pixels can be affected per batch sample\n assert np.sum(y_batch[..., n_chan:]) <= 4*4 * n_chan\n\n\n def _getitem3D(y_shape):\n Y = create_data(y_shape)\n n_chan = y_shape[-1]//2\n X = Y[:,:,:,:,0][:,:,:,:,np.newaxis]\n val_manipulator = random_neighbor_withCP_uniform\n dw = N2V_DataWrapper(X, Y, 4, perc_pix=0.198, shape=(32, 32, 32), value_manipulation=val_manipulator)\n\n x_batch, y_batch = dw.__getitem__(0)\n assert x_batch.shape == (4, 32, 32, 32, 1)\n assert y_batch.shape == (4, 32, 32, 32, 2)\n # At least one pixel has to be a blind-spot per batch sample\n assert np.sum(y_batch[..., n_chan:]) >= 1*4 * n_chan\n # At most 8 pixels can be affected per batch sample\n assert np.sum(y_batch[..., n_chan:]) <= 8*4 * n_chan\n\n\n _getitem2D(np.array([4, 32, 32, 2]))\n _getitem2D(np.array([4, 64, 64, 2]))\n _getitem2D(np.array([4, 44, 55, 2]))\n _getitem2D(np.array([4, 45, 41, 4]))\n _getitem3D(np.array([4, 32, 32, 32, 2]))\n _getitem3D(np.array([4, 64, 64, 64, 2]))\n _getitem3D(np.array([4, 33, 44, 55, 2]))"
] |
[
[
"numpy.array",
"numpy.random.rand",
"numpy.zeros",
"numpy.random.seed",
"numpy.sum",
"numpy.random.shuffle",
"numpy.arange",
"numpy.random.randint"
]
] |
bonilab/malariaibm-generation-of-MDR-mutants
|
[
"41086cf621274dcdf8b445435cb1825424a2e722"
] |
[
"misc/MDA_WHO_ERG_2018/input_generator_FLAL_Figure_S2.4.py"
] |
[
"# -*- coding: utf-8 -*-\n\"\"\"\nCreated on Tue Aug 7 14:23:33 2018\n\n@author: NguyenTran\n\"\"\"\n\nimport yaml;\nimport numpy as np;\nfrom math import log;\nimport copy;\n\n#import inflect;\n#p = inflect.engine();\n\ndef kFormatter(num):\n return str(num) if num <=999 else str(round(num/1000)) +'k';\n\n\nstream = open('input_FLAL_40k_imp_no_itc_OZ_2.4.yml', 'r');\ndata = yaml.load(stream);\nstream.close();\n\ndata['starting_date'] = '2006/1/1';\ndata['ending_date'] = '2040/1/1';\ndata['start_of_comparison_period']= '2020/1/1';\n\ndata['seasonal_info']['enable'] = 'false';\n\n#1 location\nlocation_info = [[0, 0, 0]];\nnumber_of_locations = len(location_info);\ndata['location_db']['location_info']= location_info;\n\n\n#population size \npopsize = 40000\ndata['location_db']['population_size_by_location'] = [popsize]; \n\n#3RMDA\nnumber_MDA_round = [0,1,2,3,4];\n\n#\nsd_prob_individual_present_at_mda = [0.3, 0.3, 0.3]\ndata['sd_prob_individual_present_at_mda'] = sd_prob_individual_present_at_mda\n\n#for index,event in enumerate(data['events']):\n# if event['name'] == 'single_round_MDA':\n# data['events'][index]['info'] = data['events'][index]['info'][0:number_MDA_round]\n\n\nbetas = [0.0585]\n\npfpr = {0.0585: 'PFPR2'}\n\n\n\nimproved_tc = {True: '_itc' , \n False: ''}\n\n\nfor mda_round in number_MDA_round:\n for beta in betas:\n for _,itc in improved_tc.items(): \n new_data = copy.deepcopy(data)\n new_data['location_db']['beta_by_location'] = np.full(number_of_locations, beta).tolist()\n \n for index,event in enumerate(data['events']):\n if event['name'] == 'single_round_MDA':\n new_data['events'][index]['info'] = data['events'][index]['info'][0:mda_round]\n \n pfpr_str = pfpr[beta]# \n if itc == '':\n for index,event in enumerate(data['events']):\n if event['name'] == 'change_treatment_coverage':\n new_data['events'][index]['info']= []\n \n output_filename = 'FigureS2.4/%s/ONELOC_%s_%dRMDA_%s_OPPUNIFORM_FLAL_imp_OZ%s.yml'%(kFormatter(popsize), kFormatter(popsize),mda_round,pfpr_str,itc);\n output_stream = open(output_filename, 'w');\n yaml.dump(new_data, output_stream); \n output_stream.close();\n\n#for index,beta in enumerate(betas):\n# data['location_db']['beta_by_location'] = np.full(number_of_locations, beta).tolist()\n# output_filename = 'beta/input_beta_%d.yml'%index;\n# output_stream = open(output_filename, 'w');\n# yaml.dump(data, output_stream);\n# output_stream.close();\n\n#\n#\n#print(kFormatter(9000));\n#print(p.number_to_words( number_of_locations, threshold=10));\n\n#output_filename = 'ONELOC_300K_3RMDA_PFPR15_OPPUNIFORM_FLAL.yml';\n#\n#output_filename = 'input_test.yml';\n#\n#output_stream = open(output_filename, 'w');\n#yaml.dump(data, output_stream);"
] |
[
[
"numpy.full"
]
] |
seryogin17/dlcourse_ai
|
[
"7a32e527bf5b5eaf16ba67468109e459c45ec62e"
] |
[
"assignments/assignment2/gradient_check.py"
] |
[
"import numpy as np\n\n\n\ndef check_gradient(f, x, delta=1e-7, tol = 1e-4):\n '''\n Checks the implementation of analytical gradient by comparing\n it to numerical gradient using two-point formula\n\n Arguments:\n f: function that receives x and computes value and gradient\n x: np array, initial point where gradient is checked\n delta: step to compute numerical gradient\n tol: tolerance for comparing numerical and analytical gradient\n\n Return:\n bool indicating whether gradients match or not\n '''\n \n assert isinstance(x, np.ndarray)\n assert x.dtype == np.float\n \n orig_x = x.copy()\n fx, analytic_grad = f(x)\n assert np.all(np.isclose(orig_x, x, tol)), \"Functions shouldn't modify input variables\"\n assert analytic_grad.shape == x.shape\n\n # We will go through every dimension of x and compute numeric\n # derivative for it\n it = np.nditer(x, flags=['multi_index'], op_flags=['readwrite'])\n while not it.finished:\n \n # TODO compute value of numeric gradient of f to idx\n numeric_grad_at_ix = 0\n ix = it.multi_index\n analytic_grad_at_ix = analytic_grad[ix]\n xpd = x.copy()\n xmd = x.copy()\n xpd[ix] += delta\n xmd[ix] -= delta \n fxpd = f(xpd)[0]\n fxmd = f(xmd)[0]\n #if type(fx) in (np.float64, float) or len(ix) == 1:\n if len(ix) == 1 or type(fx) == np.float64:\n numeric_grad_at_ix = (fxpd - fxmd) / (2 * delta) # for cases with one sample in the batch or alreday averaged loss-value\n else:\n numeric_grad_at_ix = (fxpd[ix[0]] - fxmd[ix[0]]) / (2 * delta)\n \n if not np.isclose(numeric_grad_at_ix, analytic_grad_at_ix, tol):\n print(\"Gradients are different at %s. Analytic: %2.9f, Numeric: %2.9f\" % (ix, analytic_grad_at_ix, numeric_grad_at_ix))\n return False\n it.iternext()\n print(\"Gradient check passed!\")\n return True\n\n\ndef check_layer_gradient(layer, x, delta=1e-5, tol=1e-4):\n \"\"\"\n Checks gradient correctness for the input and output of a layer\n\n Arguments:\n layer: neural network layer, with forward and backward functions\n x: starting point for layer input\n delta: step to compute numerical gradient\n tol: tolerance for comparing numerical and analytical gradient\n\n Returns:\n bool indicating whether gradients match or not\n \"\"\"\n output = layer.forward(x)\n output_weight = np.random.randn(*output.shape)\n\n def helper_func(x):\n output = layer.forward(x)\n loss = np.sum(output * output_weight)\n d_out = np.ones_like(output) * output_weight\n grad = layer.backward(d_out)\n return loss, grad\n\n return check_gradient(helper_func, x, delta, tol)\n\n\ndef check_layer_param_gradient(layer, x,\n param_name,\n delta=1e-5, tol=1e-4):\n \"\"\"\n Checks gradient correctness for the parameter of the layer\n\n Arguments:\n layer: neural network layer, with forward and backward functions\n x: starting point for layer input\n param_name: name of the parameter\n delta: step to compute numerical gradient\n tol: tolerance for comparing numerical and analytical gradient\n\n Returns:\n bool indicating whether gradients match or not\n \"\"\"\n param = layer.params()[param_name]\n initial_w = param.value\n\n output = layer.forward(x)\n output_weight = np.random.randn(*output.shape)\n\n def helper_func(w):\n param.value = w\n output = layer.forward(x)\n loss = np.sum(output * output_weight)\n d_out = np.ones_like(output) * output_weight\n layer.backward(d_out)\n grad = param.grad\n return loss, grad\n\n return check_gradient(helper_func, initial_w, delta, tol)\n\n\ndef check_model_gradient(model, X, y,\n delta=1e-5, tol=1e-4):\n \"\"\"\n Checks gradient correctness for all model parameters\n\n Arguments:\n model: neural network model with compute_loss_and_gradients\n X: batch of input data\n y: batch of labels\n delta: step to compute numerical gradient\n tol: tolerance for comparing numerical and analytical gradient\n\n Returns:\n bool indicating whether gradients match or not\n \"\"\"\n params = model.params()\n\n for param_key in params:\n print(\"Checking gradient for %s\" % param_key)\n param = params[param_key]\n initial_w = param.value\n\n def helper_func(w):\n param.value = w # it is needed to compute numeric grad in def check_gradient\n loss = model.compute_loss_and_gradients(X, y)\n grad = param.grad\n return loss, grad\n \n if not check_gradient(helper_func, initial_w, delta, tol):\n return False\n\n return True\n"
] |
[
[
"numpy.isclose",
"numpy.ones_like",
"numpy.nditer",
"numpy.sum",
"numpy.random.randn"
]
] |
Corleone-Huang/unleashing-transformers
|
[
"66925b11c3d63222a34a2f66b499c900e4b6cef4"
] |
[
"models/autoregressive.py"
] |
[
"import torch\nimport torch.nn.functional as F\nfrom .sampler import Sampler\nfrom .transformer import Transformer\nimport numpy as np\nimport math\n\n\nclass AutoregressiveTransformer(Sampler):\n def __init__(self, H, embedding_weight):\n super().__init__(H, embedding_weight)\n self.net = Transformer(H)\n self.n_samples = H.batch_size\n self.seq_len = np.prod(H.latent_shape)\n\n def train_iter(self, x):\n x_in = x[:, :-1] # x is already flattened\n logits = self.net(x_in)\n loss = F.cross_entropy(logits.permute(0, 2, 1), x, reduction='none')\n loss = loss.sum(1).mean() / (math.log(2) * x.shape[1:].numel())\n stats = {'loss': loss}\n return stats\n\n def sample(self, temp=1.0):\n b, device = self.n_samples, 'cuda'\n x = torch.zeros(b, 0).long().to(device)\n for _ in range(self.seq_len):\n logits = self.net(x)[:, -1]\n probs = F.softmax(logits / temp, dim=-1)\n ix = torch.multinomial(probs, num_samples=1)\n x = torch.cat((x, ix), dim=1)\n return x\n"
] |
[
[
"torch.zeros",
"torch.cat",
"torch.multinomial",
"numpy.prod",
"torch.nn.functional.softmax"
]
] |
codewithzichao/DeepClassifier
|
[
"c07e8041a2aca855fed9bf69fd571f1cbd5ec032"
] |
[
"tests/test_han.py"
] |
[
"# -*- coding:utf-8 -*-\n'''\n\nAuthor:\n Zichao Li,[email protected]\n\n'''\nfrom __future__ import print_function\n\nimport os\nimport torch\nimport torch.nn as nn\nimport numpy as np\nimport torch.nn.functional as F\nfrom torch.utils.data import Dataset, DataLoader\nimport torch.optim as optim\nfrom deepclassifier.models import HAN\nfrom deepclassifier.trainers import Trainer\nfrom tensorboardX import SummaryWriter\n\n\nclass my_dataset(Dataset):\n def __init__(self, data, label):\n self.data = data\n self.label = label\n\n def __len__(self):\n return len(self.data)\n\n def __getitem__(self, item):\n item_data = self.data[item]\n item_label = self.label[item]\n\n return item_data, item_label\n\nclass my_dataset1(Dataset):\n def __init__(self, data):\n self.data = data\n\n\n def __len__(self):\n return len(self.data)\n\n def __getitem__(self, item):\n item_data = self.data[item]\n\n\n return item_data\n\n\n# 训练集\nbatch_size = 20\ntrain_data = np.random.randint(0, 100, (100, 60,10))\ntrain_label = torch.from_numpy(np.array([int(x > 0.5) for x in np.random.randn(100)]))\nmy_train_data = my_dataset(train_data, train_label)\nfinal_train_data = DataLoader(my_train_data, batch_size=batch_size, shuffle=True)\n\n# 验证集\ndev_data = np.random.randint(0, 100, (100, 60,10))\ndev_label = torch.from_numpy(np.array([int(x > 0.5) for x in np.random.randn(100)]))\nmy_dev_data = my_dataset(dev_data, dev_label)\nfinal_dev_data = DataLoader(my_dev_data, batch_size=batch_size, shuffle=True)\n\n# 测试集\ntest_data = np.random.randint(0, 100, (100, 60,10))\ntest_label = torch.from_numpy(np.array([int(x > 0.5) for x in np.random.randn(100)]))\nmy_test_data = my_dataset(test_data, dev_label)\nfinal_test_data = DataLoader(my_test_data, batch_size=batch_size, shuffle=True)\n\nmy_model = HAN(10,100,100,0.2,2,100,60)\noptimizer = optim.Adam(my_model.parameters())\nloss_fn = nn.CrossEntropyLoss()\nsave_path = \"best.ckpt\"\n\nwriter = SummaryWriter(\"logfie/1\")\nmy_trainer = Trainer(model_name=\"han\", model=my_model, train_loader=final_train_data, dev_loader=final_dev_data,\n test_loader=final_test_data, optimizer=optimizer, loss_fn=loss_fn,\n save_path=save_path, epochs=100, writer=writer, max_norm=0.25, eval_step_interval=10)\n\n# 训练\nmy_trainer.train()\n# 测试\np, r, f1 = my_trainer.test()\nprint(p, r, f1)\n# 打印在验证集上最好的f1值\nprint(my_trainer.best_f1)\n\n# 预测\npred_data = np.random.randint(0, 100, (100, 60))\npred_data=my_dataset1(pred_data)\npred_data=DataLoader(pred_data,batch_size=1)\nprd_label=my_trainer.predict(pred_data)\nprint(prd_label.shape)\n"
] |
[
[
"torch.nn.CrossEntropyLoss",
"numpy.random.randn",
"numpy.random.randint",
"torch.utils.data.DataLoader"
]
] |
henrykironde/DeepTreeAttention
|
[
"a50f5f4499e5aa0a74732e84b9b51d482bc508bb"
] |
[
"src/data.py"
] |
[
"#Ligthning data module\nimport argparse\nfrom . import __file__\nfrom distributed import wait\nimport glob\nimport geopandas as gpd\nimport json\nimport numpy as np\nimport os\nimport pandas as pd\nfrom pytorch_lightning import LightningDataModule\nimport rasterio as rio\nfrom sklearn import preprocessing\nfrom src import generate\nfrom src import CHM\nfrom src import augmentation\nfrom src import megaplot\nfrom shapely.geometry import Point\nimport torch\nfrom torch.utils.data import Dataset\nfrom torchvision import transforms\nimport yaml\nimport warnings\n \ndef filter_data(path, config):\n \"\"\"Transform raw NEON data into clean shapefile \n Args:\n config: DeepTreeAttention config dict, see config.yml\n \"\"\"\n field = pd.read_csv(path)\n field = field[~field.elevation.isnull()]\n field = field[~field.growthForm.isin([\"liana\",\"small shrub\"])]\n field = field[~field.growthForm.isnull()]\n field = field[~field.plantStatus.isnull()] \n field = field[field.plantStatus.str.contains(\"Live\")] \n \n groups = field.groupby(\"individualID\")\n shaded_ids = []\n for name, group in groups:\n shaded = any([x in [\"Full shade\", \"Mostly shaded\"] for x in group.canopyPosition.values])\n if shaded:\n if any([x in [\"Open grown\", \"Full sun\"] for x in group.canopyPosition.values]):\n continue\n else:\n shaded_ids.append(group.individualID.unique()[0])\n \n field = field[~(field.individualID.isin(shaded_ids))]\n field = field[(field.height > 3) | (field.height.isnull())]\n field = field[field.stemDiameter > config[\"min_stem_diameter\"]]\n field.loc[field.taxonID==\"PSMEM\",\"taxonID\"] = \"PSME\"\n \n field = field[~field.taxonID.isin([\"BETUL\", \"FRAXI\", \"HALES\", \"PICEA\", \"PINUS\", \"QUERC\", \"ULMUS\", \"2PLANT\"])]\n field = field[~(field.eventID.str.contains(\"2014\"))]\n with_heights = field[~field.height.isnull()]\n with_heights = with_heights.loc[with_heights.groupby('individualID')['height'].idxmax()]\n \n missing_heights = field[field.height.isnull()]\n missing_heights = missing_heights[~missing_heights.individualID.isin(with_heights.individualID)]\n missing_heights = missing_heights.groupby(\"individualID\").apply(lambda x: x.sort_values([\"eventID\"],ascending=False).head(1)).reset_index(drop=True)\n \n field = pd.concat([with_heights,missing_heights])\n \n #remove multibole\n field = field[~(field.individualID.str.contains('[A-Z]$',regex=True))]\n\n #List of hand cleaned errors\n known_errors = [\"NEON.PLA.D03.OSBS.03422\",\"NEON.PLA.D03.OSBS.03422\",\"NEON.PLA.D03.OSBS.03382\", \"NEON.PLA.D17.TEAK.01883\"]\n field = field[~(field.individualID.isin(known_errors))]\n field = field[~(field.plotID == \"SOAP_054\")]\n \n #Create shapefile\n field[\"geometry\"] = [Point(x,y) for x,y in zip(field[\"itcEasting\"], field[\"itcNorthing\"])]\n shp = gpd.GeoDataFrame(field)\n \n #HOTFIX, BLAN has some data in 18N UTM, reproject to 17N update columns\n BLAN_errors = shp[(shp.siteID == \"BLAN\") & (shp.utmZone == \"18N\")]\n BLAN_errors.set_crs(epsg=32618, inplace=True)\n BLAN_errors.to_crs(32617,inplace=True)\n BLAN_errors[\"utmZone\"] = \"17N\"\n BLAN_errors[\"itcEasting\"] = BLAN_errors.geometry.apply(lambda x: x.coords[0][0])\n BLAN_errors[\"itcNorthing\"] = BLAN_errors.geometry.apply(lambda x: x.coords[0][1])\n \n #reupdate\n shp.loc[BLAN_errors.index] = BLAN_errors\n \n #Oak Right Lab has no AOP data\n shp = shp[~(shp.siteID.isin([\"PUUM\",\"ORNL\"]))]\n\n #There are a couple NEON plots within the OSBS megaplot, make sure they are removed\n shp = shp[~shp.plotID.isin([\"OSBS_026\",\"OSBS_029\",\"OSBS_039\",\"OSBS_027\",\"OSBS_036\"])]\n\n return shp\n\ndef sample_plots(shp, min_train_samples=5, min_test_samples=3, iteration = 1):\n \"\"\"Sample and split a pandas dataframe based on plotID\n Args:\n shp: pandas dataframe of filtered tree locations\n test_fraction: proportion of plots in test datasets\n min_samples: minimum number of samples per class\n iteration: a dummy parameter to make dask submission unique\n \"\"\"\n #split by plot level\n plotIDs = list(shp[shp.siteID.isin([\"OSBS\",\"JERC\",\"DSNY\",\"TALL\",\"LENO\",\"DELA\"])].plotID.unique())\n if len(plotIDs) == 0:\n test = shp[shp.plotID == shp.plotID.unique()[0]]\n train = shp[shp.plotID == shp.plotID.unique()[1]]\n \n return train, test\n \n np.random.shuffle(plotIDs)\n test = shp[shp.plotID == plotIDs[0]]\n \n for plotID in plotIDs[1:]:\n include = False\n selected_plot = shp[shp.plotID == plotID]\n # If any species is missing from min samples, include plot\n for x in selected_plot.taxonID.unique():\n if sum(test.taxonID == x) < min_test_samples:\n include = True\n if include:\n test = pd.concat([test,selected_plot])\n \n train = shp[~shp.plotID.isin(test.plotID.unique())]\n \n #remove fixed boxes from test\n test = test.groupby(\"taxonID\").filter(lambda x: x.shape[0] >= min_test_samples)\n train_keep = train[train.siteID.isin([\"OSBS\",\"JERC\",\"DSNY\",\"TALL\",\"LENO\",\"DELA\"])].groupby(\"taxonID\").filter(lambda x: x.shape[0] >= min_train_samples)\n train = train[train.taxonID.isin(train_keep.taxonID.unique())]\n train = train[train.taxonID.isin(test.taxonID)] \n test = test[test.taxonID.isin(train.taxonID)]\n test = test.loc[~test[\"box_id\"].astype(str).str.contains(\"fixed\").fillna(False)]\n \n return train, test\n \ndef train_test_split(shp, config, client = None):\n \"\"\"Create the train test split\n Args:\n shp: a filter pandas dataframe (or geodataframe) \n client: optional dask client\n Returns:\n None: train.shp and test.shp are written as side effect\n \"\"\" \n min_sampled = config[\"min_train_samples\"] + config[\"min_test_samples\"]\n keep = shp.taxonID.value_counts() > (min_sampled)\n species_to_keep = keep[keep].index\n shp = shp[shp.taxonID.isin(species_to_keep)]\n print(\"splitting data into train test. Initial data has {} points from {} species with a min of {} samples\".format(shp.shape[0],shp.taxonID.nunique(),min_sampled))\n test_species = 0\n ties = []\n if client:\n futures = [ ]\n for x in np.arange(config[\"iterations\"]):\n future = client.submit(sample_plots, shp=shp, min_train_samples=config[\"min_train_samples\"], iteration=x, min_test_samples=config[\"min_test_samples\"])\n futures.append(future)\n \n wait(futures)\n for x in futures:\n train, test = x.result()\n if test.taxonID.nunique() > test_species:\n print(\"Selected test has {} points and {} species\".format(test.shape[0], test.taxonID.nunique()))\n saved_train = train\n saved_test = test\n test_species = test.taxonID.nunique()\n ties = []\n ties.append([train, test])\n elif test.taxonID.nunique() == test_species:\n ties.append([train, test]) \n else:\n for x in np.arange(config[\"iterations\"]):\n train, test = sample_plots(shp, min_train_samples=config[\"min_train_samples\"], min_test_samples=config[\"min_test_samples\"])\n if test.taxonID.nunique() > test_species:\n print(\"Selected test has {} points and {} species\".format(test.shape[0], test.taxonID.nunique()))\n saved_train = train\n saved_test = test\n test_species = test.taxonID.nunique()\n #reset ties\n ties = []\n ties.append([train, test])\n elif test.taxonID.nunique() == test_species:\n ties.append([train, test])\n \n # The size of the datasets\n if len(ties) > 1:\n print(\"The size of tied datasets with {} species is {}\".format(test_species, [x[1].shape[0] for x in ties])) \n saved_train, saved_test = ties[np.argmax([x[1].shape[0] for x in ties])]\n \n train = saved_train\n test = saved_test \n \n #Give tests a unique index to match against\n test[\"point_id\"] = test.index.values\n train[\"point_id\"] = train.index.values\n \n return train, test\n \ndef read_config(config_path):\n \"\"\"Read config yaml file\"\"\"\n #Allow command line to override \n parser = argparse.ArgumentParser(\"DeepTreeAttention config\")\n parser.add_argument('-d', '--my-dict', type=json.loads, default=None)\n args = parser.parse_known_args()\n try:\n with open(config_path, 'r') as f:\n config = yaml.load(f, Loader=yaml.FullLoader)\n\n except Exception as e:\n raise FileNotFoundError(\"There is no config at {}, yields {}\".format(\n config_path, e))\n \n #Update anything in argparse to have higher priority\n if args[0].my_dict:\n for key, value in args[0].my_dict:\n config[key] = value\n \n return config\n\ndef preprocess_image(image, channel_is_first=False):\n \"\"\"Preprocess a loaded image, if already C*H*W set channel_is_first=True\"\"\"\n img = np.asarray(image, dtype='float32')\n data = img.reshape(img.shape[0], np.prod(img.shape[1:]))\n \n with warnings.catch_warnings():\n warnings.simplefilter('ignore', UserWarning) \n data = preprocessing.scale(data)\n img = data.reshape(img.shape)\n \n if not channel_is_first:\n img = np.rollaxis(img, 2,0)\n \n normalized = torch.from_numpy(img)\n \n return normalized\n\ndef load_image(img_path, image_size):\n \"\"\"Load and preprocess an image for training/prediction\"\"\"\n with warnings.catch_warnings():\n warnings.simplefilter('ignore', rio.errors.NotGeoreferencedWarning)\n image = rio.open(img_path).read() \n image = preprocess_image(image, channel_is_first=True)\n \n #resize image\n image = transforms.functional.resize(image, size=(image_size,image_size), interpolation=transforms.InterpolationMode.NEAREST)\n \n return image\n\n#Dataset class\nclass TreeDataset(Dataset):\n \"\"\"A csv file with a path to image crop and label\n Args:\n csv_file: path to csv file with image_path and label\n \"\"\"\n def __init__(self, csv_file, image_size=10, config=None, train=True, HSI=True, metadata=False):\n self.annotations = pd.read_csv(csv_file)\n self.train = train\n self.HSI = HSI\n self.metadata = metadata\n self.config = config \n \n if self.config:\n self.image_size = config[\"image_size\"]\n else:\n self.image_size = image_size\n \n #Create augmentor\n self.transformer = augmentation.train_augmentation(image_size=image_size)\n \n #Pin data to memory if desired\n if self.config[\"preload_images\"]:\n self.image_dict = {}\n for index, row in self.annotations.iterrows():\n self.image_dict[index] = load_image(row[\"image_path\"], image_size=image_size)\n \n def __len__(self):\n #0th based index\n return self.annotations.shape[0]\n \n def __getitem__(self, index):\n inputs = {}\n image_path = self.annotations.image_path.loc[index] \n individual = os.path.basename(image_path.split(\".tif\")[0])\n if self.HSI:\n if self.config[\"preload_images\"]:\n inputs[\"HSI\"] = self.image_dict[index]\n else:\n image_path = self.annotations.image_path.loc[index] \n image = load_image(image_path, image_size=self.image_size)\n inputs[\"HSI\"] = image\n \n if self.metadata:\n site = self.annotations.site.loc[index] \n site = torch.tensor(site, dtype=torch.int)\n inputs[\"site\"] = site\n \n if self.train:\n label = self.annotations.label.loc[index]\n label = torch.tensor(label, dtype=torch.long)\n \n if self.HSI:\n inputs[\"HSI\"] = self.transformer(inputs[\"HSI\"])\n\n return individual, inputs, label\n else:\n return individual, inputs\n\nclass TreeData(LightningDataModule):\n \"\"\"\n Lightning data module to convert raw NEON data into HSI pixel crops based on the config.yml file. \n The module checkpoints the different phases of setup, if one stage failed it will restart from that stage. \n Use regenerate=True to override this behavior in setup()\n \"\"\"\n def __init__(self, csv_file, HSI=True, metadata=False, regenerate = False, client = None, config=None, data_dir=None, comet_logger=None, debug=False):\n \"\"\"\n Args:\n config: optional config file to override\n data_dir: override data location, defaults to ROOT \n regenerate: Whether to recreate raw data\n debug: a test mode for small samples\n \"\"\"\n super().__init__()\n self.ROOT = os.path.dirname(os.path.dirname(__file__))\n self.regenerate=regenerate\n self.csv_file = csv_file\n self.HSI = HSI\n self.metadata = metadata\n self.comet_logger = comet_logger\n self.debug = debug \n \n #default training location\n self.client = client\n if data_dir is None:\n self.data_dir = \"{}/data/\".format(self.ROOT)\n else:\n self.data_dir = data_dir \n \n self.train_file = \"{}/processed/train.csv\".format(self.data_dir)\n \n if config is None:\n self.config = read_config(\"{}/config.yml\".format(self.ROOT)) \n else:\n self.config = config\n \n def setup(self,stage=None):\n #Clean data from raw csv, regenerate from scratch or check for progress and complete\n if self.regenerate:\n if self.config[\"replace\"]:#remove any previous runs\n try:\n os.remove(\"{}/processed/canopy_points.shp\".format(self.data_dir))\n os.remove(\" \".format(self.data_dir))\n os.remove(\"{}/processed/crowns.shp\".format(self.data_dir))\n for x in glob.glob(self.config[\"crop_dir\"]):\n os.remove(x)\n except:\n pass\n \n #Convert raw neon data to x,y tree locatins\n df = filter_data(self.csv_file, config=self.config)\n \n #load any megaplot data\n if not self.config[\"megaplot_dir\"] is None:\n megaplot_data = megaplot.load(directory=self.config[\"megaplot_dir\"], config=self.config)\n df = pd.concat([megaplot_data, df])\n \n if not self.debug:\n southeast = df[df.siteID.isin([\"OSBS\",\"LENO\",\"TALL\",\"DELA\",\"DSNY\",\"JERC\"])]\n southeast = southeast.taxonID.unique()\n plotIDs_to_keep = df[df.taxonID.isin(southeast)].plotID.unique()\n df = df[df.plotID.isin(plotIDs_to_keep)]\n \n if self.comet_logger:\n self.comet_logger.experiment.log_parameter(\"Species before CHM filter\",len(df.taxonID.unique()))\n self.comet_logger.experiment.log_parameter(\"Samples before CHM filter\",df.shape[0])\n \n #Filter points based on LiDAR height\n df = CHM.filter_CHM(df, CHM_pool=self.config[\"CHM_pool\"],\n min_CHM_height=self.config[\"min_CHM_height\"], \n max_CHM_diff=self.config[\"max_CHM_diff\"], \n CHM_height_limit=self.config[\"CHM_height_limit\"]) \n \n df.to_file(\"{}/processed/canopy_points.shp\".format(self.data_dir))\n \n if self.comet_logger:\n self.comet_logger.experiment.log_parameter(\"Species after CHM filter\",len(df.taxonID.unique()))\n self.comet_logger.experiment.log_parameter(\"Samples after CHM filter\",df.shape[0])\n \n #Create crown data\n crowns = generate.points_to_crowns(\n field_data=\"{}/processed/canopy_points.shp\".format(self.data_dir),\n rgb_dir=self.config[\"rgb_sensor_pool\"],\n savedir=\"{}/interim/\".format(self.data_dir),\n raw_box_savedir=\"{}/interim/\".format(self.data_dir), \n client=self.client\n )\n \n if self.comet_logger:\n self.comet_logger.experiment.log_parameter(\"Species after crown prediction\",len(crowns.taxonID.unique()))\n self.comet_logger.experiment.log_parameter(\"Samples after crown prediction\",crowns.shape[0])\n \n crowns.to_file(\"{}/processed/crowns.shp\".format(self.data_dir))\n else:\n crowns = gpd.read_file(\"{}/processed/crowns.shp\".format(self.data_dir))\n \n annotations = generate.generate_crops(\n crowns,\n savedir=self.config[\"crop_dir\"],\n sensor_glob=self.config[\"HSI_sensor_pool\"],\n convert_h5=self.config[\"convert_h5\"], \n rgb_glob=self.config[\"rgb_sensor_pool\"],\n HSI_tif_dir=self.config[\"HSI_tif_dir\"],\n client=self.client,\n replace=self.config[\"replace\"]\n )\n annotations.to_csv(\"{}/processed/annotations.csv\".format(self.data_dir))\n \n if self.comet_logger:\n self.comet_logger.experiment.log_parameter(\"Species after crop generation\",len(annotations.taxonID.unique()))\n self.comet_logger.experiment.log_parameter(\"Samples after crop generation\",annotations.shape[0])\n \n if self.config[\"new_train_test_split\"]:\n train_annotations, test_annotations = train_test_split(annotations,config=self.config, client=self.client) \n else:\n previous_train = pd.read_csv(\"{}/processed/train.csv\".format(self.data_dir))\n previous_test = pd.read_csv(\"{}/processed/test.csv\".format(self.data_dir))\n \n train_annotations = annotations[annotations.individualID.isin(previous_train.individualID)]\n test_annotations = annotations[annotations.individualID.isin(previous_test.individualID)]\n \n #capture discarded species\n individualIDs = np.concatenate([train_annotations.individualID.unique(), test_annotations.individualID.unique()])\n novel = annotations[~annotations.individualID.isin(individualIDs)]\n novel = novel[~novel.taxonID.isin(np.concatenate([train_annotations.taxonID.unique(), test_annotations.taxonID.unique()]))]\n novel.to_csv(\"{}/processed/novel_species.csv\".format(self.data_dir))\n \n #Store class labels\n unique_species_labels = np.concatenate([train_annotations.taxonID.unique(), test_annotations.taxonID.unique()])\n unique_species_labels = np.unique(unique_species_labels)\n unique_species_labels = np.sort(unique_species_labels) \n self.num_classes = len(unique_species_labels)\n \n #Taxon to ID dict and the reverse \n self.species_label_dict = {}\n for index, taxonID in enumerate(unique_species_labels):\n self.species_label_dict[taxonID] = index\n \n #Store site labels\n unique_site_labels = np.concatenate([train_annotations.siteID.unique(), test_annotations.siteID.unique()])\n unique_site_labels = np.unique(unique_site_labels)\n \n self.site_label_dict = {}\n for index, label in enumerate(unique_site_labels):\n self.site_label_dict[label] = index\n self.num_sites = len(self.site_label_dict) \n \n self.label_to_taxonID = {v: k for k, v in self.species_label_dict.items()}\n \n #Encode the numeric site and class data\n train_annotations[\"label\"] = train_annotations.taxonID.apply(lambda x: self.species_label_dict[x])\n train_annotations[\"site\"] = train_annotations.siteID.apply(lambda x: self.site_label_dict[x])\n \n test_annotations[\"label\"] = test_annotations.taxonID.apply(lambda x: self.species_label_dict[x])\n test_annotations[\"site\"] = test_annotations.siteID.apply(lambda x: self.site_label_dict[x])\n \n train_annotations.to_csv(\"{}/processed/train.csv\".format(self.data_dir), index=False) \n test_annotations.to_csv(\"{}/processed/test.csv\".format(self.data_dir), index=False)\n \n print(\"There are {} records for {} species for {} sites in filtered train\".format(\n train_annotations.shape[0],\n len(train_annotations.label.unique()),\n len(train_annotations.site.unique())\n ))\n \n print(\"There are {} records for {} species for {} sites in test\".format(\n test_annotations.shape[0],\n len(test_annotations.label.unique()),\n len(test_annotations.site.unique()))\n )\n \n #Create dataloaders\n self.train_ds = TreeDataset(csv_file = self.train_file, config=self.config, HSI=self.HSI, metadata=self.metadata)\n self.val_ds = TreeDataset(csv_file = \"{}/processed/test.csv\".format(self.data_dir), config=self.config, HSI=self.HSI, metadata=self.metadata)\n \n else:\n train_annotations = pd.read_csv(\"{}/processed/train.csv\".format(self.data_dir))\n test_annotations = pd.read_csv(\"{}/processed/test.csv\".format(self.data_dir))\n \n #Store class labels\n unique_species_labels = np.concatenate([train_annotations.taxonID.unique(), test_annotations.taxonID.unique()])\n unique_species_labels = np.unique(unique_species_labels)\n unique_species_labels = np.sort(unique_species_labels) \n self.num_classes = len(unique_species_labels)\n \n #Taxon to ID dict and the reverse \n self.species_label_dict = {}\n for index, taxonID in enumerate(unique_species_labels):\n self.species_label_dict[taxonID] = index\n \n #Store site labels\n unique_site_labels = np.concatenate([train_annotations.siteID.unique(), test_annotations.siteID.unique()])\n unique_site_labels = np.unique(unique_site_labels)\n \n self.site_label_dict = {}\n for index, label in enumerate(unique_site_labels):\n self.site_label_dict[label] = index\n self.num_sites = len(self.site_label_dict) \n \n self.label_to_taxonID = {v: k for k, v in self.species_label_dict.items()}\n \n #Create dataloaders\n self.train_ds = TreeDataset(csv_file = self.train_file, config=self.config, HSI=self.HSI, metadata=self.metadata)\n self.val_ds = TreeDataset(csv_file = \"{}/processed/test.csv\".format(self.data_dir), config=self.config, HSI=self.HSI, metadata=self.metadata) \n\n def train_dataloader(self):\n \"\"\"Load a training file. The default location is saved during self.setup(), to override this location, set self.train_file before training\"\"\" \n #get class weights\n train = pd.read_csv(self.train_file)\n class_weights = train.label.value_counts().to_dict() \n \n data_weights = []\n #balance classes\n for idx in range(len(self.train_ds)):\n path, image, targets = self.train_ds[idx]\n label = int(targets.numpy())\n class_freq = class_weights[label]\n #under sample majority classes\n if class_freq > 50:\n class_freq = 50\n data_weights.append(1/class_freq)\n \n sampler = torch.utils.data.sampler.WeightedRandomSampler(weights = data_weights, num_samples=len(self.train_ds))\n data_loader = torch.utils.data.DataLoader(\n self.train_ds,\n sampler = sampler,\n batch_size=self.config[\"batch_size\"],\n num_workers=self.config[\"workers\"])\n \n return data_loader\n \n def val_dataloader(self):\n data_loader = torch.utils.data.DataLoader(\n self.val_ds,\n batch_size=self.config[\"batch_size\"],\n shuffle=False,\n num_workers=self.config[\"workers\"],\n )\n \n return data_loader"
] |
[
[
"numpy.asarray",
"numpy.rollaxis",
"numpy.random.shuffle",
"torch.from_numpy",
"sklearn.preprocessing.scale",
"numpy.prod",
"numpy.arange",
"torch.utils.data.DataLoader",
"torch.tensor",
"pandas.concat",
"numpy.sort",
"numpy.argmax",
"pandas.read_csv",
"numpy.unique"
]
] |
krai/ck-object-detection
|
[
"30a43faaca8afef05299b1604a0c04541d5723a7"
] |
[
"program/double-inference/custom_hooks.py"
] |
[
"#MIT License\n#\n#Copyright (c) 2019 YangYun\n#\n#Permission is hereby granted, free of charge, to any person obtaining a copy\n#of this software and associated documentation files (the \"Software\"), to deal\n#in the Software without restriction, including without limitation the rights\n#to use, copy, modify, merge, publish, distribute, sublicense, and/or sell\n#copies of the Software, and to permit persons to whom the Software is\n#furnished to do so, subject to the following conditions:\n#\n#The above copyright notice and this permission notice shall be included in all\n#copies or substantial portions of the Software.\n#\n#THE SOFTWARE IS PROVIDED \"AS IS\", WITHOUT WARRANTY OF ANY KIND, EXPRESS OR\n#IMPLIED, INCLUDING BUT NOT LIMITED TO THE WARRANTIES OF MERCHANTABILITY,\n#FITNESS FOR A PARTICULAR PURPOSE AND NONINFRINGEMENT. IN NO EVENT SHALL THE\n#AUTHORS OR COPYRIGHT HOLDERS BE LIABLE FOR ANY CLAIM, DAMAGES OR OTHER\n#LIABILITY, WHETHER IN AN ACTION OF CONTRACT, TORT OR OTHERWISE, ARISING FROM,\n#OUT OF OR IN CONNECTION WITH THE SOFTWARE OR THE USE OR OTHER DEALINGS IN THE\n#SOFTWARE.\nimport os\nimport cv2\nimport random\nimport colorsys\nimport numpy as np\nimport tensorflow as tf\nfrom PIL import Image\nimport ck_utils\n#taken from original code\n#TODO modify for batch. but want to test if all works before.\ndef ck_custom_preprocess(image_files, iter_num,processed_image_ids,params):\n if params[\"RESIZE_WIDTH_SIZE\"] != params[\"RESIZE_HEIGHT_SIZE\"]:\n raise ValueError (\"this model works with square images, and you provided different height and width\")\n image_file = image_files[iter_num]\n image_id = ck_utils.filename_to_id(image_file, params[\"DATASET_TYPE\"])\n processed_image_ids.append(image_id)\n image_path = os.path.join(params[\"IMAGES_DIR\"], image_file)\n orig_image = cv2.imread(image_path)\n orig_image = cv2.cvtColor(orig_image, cv2.COLOR_BGR2RGB)\n original_image_size = orig_image.shape[:2]\n image = cv2.cvtColor(orig_image, cv2.COLOR_BGR2RGB).astype(np.float32)\n\n ih, iw = params[\"RESIZE_HEIGHT_SIZE\"],params[\"RESIZE_WIDTH_SIZE\"] ##They are equal.\n h, w, _ = image.shape\n\n scale = min(iw/w, ih/h)\n nw, nh = int(scale * w), int(scale * h)\n image_resized = cv2.resize(image, (nw, nh))\n\n image_paded = np.full(shape=[ih, iw, 3], fill_value=128.0)\n dw, dh = (iw - nw) // 2, (ih-nh) // 2\n image_paded[dh:nh+dh, dw:nw+dw, :] = image_resized\n image_paded = image_paded / 255.\n image_paded = image_paded[np.newaxis, ...]\n return image_paded,processed_image_ids,original_image_size,orig_image\n\n\n#keeping the distinction between input and output tensors, as in ck detect program. \ndef ck_custom_get_tensors():\n image_tensor =\"input/input_data:0\"\n return_elements = [\"pred_sbbox/concat_2:0\", \"pred_mbbox/concat_2:0\", \"pred_lbbox/concat_2:0\"]\n graph = tf.compat.v1.get_default_graph()\n ops = graph.get_operations()\n all_tensor_names = {output.name for op in ops for output in op.outputs}\n tensor_dict = {}\n for tensor_name in return_elements:\n if tensor_name in all_tensor_names:\n tensor_dict[tensor_name] = graph.get_tensor_by_name(tensor_name)\n\n image_tensor = graph.get_tensor_by_name(image_tensor)\n return tensor_dict, image_tensor\n\n\n\ndef bboxes_iou(boxes1, boxes2):\n\n boxes1 = np.array(boxes1)\n boxes2 = np.array(boxes2)\n\n boxes1_area = (boxes1[..., 2] - boxes1[..., 0]) * (boxes1[..., 3] - boxes1[..., 1])\n boxes2_area = (boxes2[..., 2] - boxes2[..., 0]) * (boxes2[..., 3] - boxes2[..., 1])\n\n left_up = np.maximum(boxes1[..., :2], boxes2[..., :2])\n right_down = np.minimum(boxes1[..., 2:], boxes2[..., 2:]) \n \n inter_section = np.maximum(right_down - left_up, 0.0) \n inter_area = inter_section[..., 0] * inter_section[..., 1] \n union_area = boxes1_area + boxes2_area - inter_area\n ious = np.maximum(1.0 * inter_area / union_area, np.finfo(np.float32).eps) \n \n return ious \n\n\n\ndef nms(bboxes, iou_threshold, sigma=0.3, method='nms'): \n \"\"\"\n :param bboxes: (xmin, ymin, xmax, ymax, score, class) \n\n Note: soft-nms, https://arxiv.org/pdf/1704.04503.pdf \n https://github.com/bharatsingh430/soft-nms\n \"\"\"\n classes_in_img = list(set(bboxes[:, 5])) \n best_bboxes = []\n\n for cls in classes_in_img:\n cls_mask = (bboxes[:, 5] == cls)\n cls_bboxes = bboxes[cls_mask]\n\n while len(cls_bboxes) > 0:\n max_ind = np.argmax(cls_bboxes[:, 4])\n best_bbox = cls_bboxes[max_ind]\n best_bboxes.append(best_bbox)\n cls_bboxes = np.concatenate([cls_bboxes[: max_ind], cls_bboxes[max_ind + 1:]])\n iou = bboxes_iou(best_bbox[np.newaxis, :4], cls_bboxes[:, :4])\n weight = np.ones((len(iou),), dtype=np.float32)\n\n assert method in ['nms', 'soft-nms']\n\n if method == 'nms':\n iou_mask = iou > iou_threshold\n weight[iou_mask] = 0.0\n\n if method == 'soft-nms':\n weight = np.exp(-(1.0 * iou ** 2 / sigma))\n\n cls_bboxes[:, 4] = cls_bboxes[:, 4] * weight\n score_mask = cls_bboxes[:, 4] > 0.\n cls_bboxes = cls_bboxes[score_mask]\n\n return best_bboxes\n\n\n#mimicks the original ck function to save to file the bounding boxes. first line is original image size, then one line per detection, with x1,y1,x2,y2,score,class_id,class_name.\ndef ck_custom_save_txt(image_file, image_size, bboxes, category_index,DETECTIONS_OUT_DIR):\n (im_width, im_height) = image_size\n file_name = os.path.splitext(image_file)[0]\n res_file = os.path.join(DETECTIONS_OUT_DIR, file_name) + '.txt'\n with open(res_file, 'w') as f:\n f.write('{:d} {:d}\\n'.format(im_width, im_height))\n for entry in bboxes:\n if 'display_name' in category_index[entry[5]]:\n class_name = category_index[entry[5]]['display_name']\n else:\n class_name = category_index[entry[5]]['name']\n f.write(\"%.2f %.2f %.2f %.2f %.3f %d %s \\n\" % (entry[0] ,entry[1],entry[2],entry[3],entry[4],entry[5],class_name))\n\n\n#for this function I will use the ck standard label-class structure, thus slightly modifing the original code. I will however maintain the original opencv printing function, working with the postprocessed tensor of the yolo network (thus keeping opencv)\ndef ck_custom_save_images(image, bboxes, category_index, show_label=True):\n \"\"\"\n bboxes: [x_min, y_min, x_max, y_max, probability, cls_id] format coordinates.\n \"\"\"\n num_classes = max(category_index.keys())\n image_h, image_w, _ = image.shape\n hsv_tuples = [(1.0 * x / num_classes, 1., 1.) for x in range(num_classes)]\n colors = list(map(lambda x: colorsys.hsv_to_rgb(*x), hsv_tuples))\n colors = list(map(lambda x: (int(x[0] * 255), int(x[1] * 255), int(x[2] * 255)), colors))\n\n random.seed(0)\n random.shuffle(colors)\n random.seed(None)\n\n for i, bbox in enumerate(bboxes):\n coor = np.array(bbox[:4], dtype=np.int32)\n fontScale = 0.5\n score = bbox[4]\n class_ind = int(bbox[5])\n bbox_color = colors[class_ind]\n bbox_thick = int(0.6 * (image_h + image_w) / 600)\n c1, c2 = (coor[0], coor[1]), (coor[2], coor[3])\n cv2.rectangle(image, c1, c2, bbox_color, bbox_thick)\n if 'display_name' in category_index[class_ind]:\n class_name = category_index[class_ind]['display_name']\n else:\n class_name = category_index[class_ind]['name']\n\n if show_label:\n bbox_mess = '%s: %.2f' % (class_name, score)\n t_size = cv2.getTextSize(bbox_mess, 0, fontScale, thickness=bbox_thick//2)[0]\n cv2.rectangle(image, c1, (c1[0] + t_size[0], c1[1] - t_size[1] - 3), bbox_color, -1) # filled\n\n cv2.putText(image, bbox_mess, (c1[0], c1[1]-2), cv2.FONT_HERSHEY_SIMPLEX,\n fontScale, (0, 0, 0), bbox_thick//2, lineType=cv2.LINE_AA)\n\n return image\n\n\n\n\n#uses postprocess code from the implementation of yolo\n#modified to adapt to hooks, so it will comprehend the tensor concatenation and reshaping, and the print to file. All the functionality are taken from the original code, and adapted.\n#added the remapping of the ids to the ck postprocessing standard ids.\n#TODO batch support\n#resize dim comes with a tuple width,height. and they MUST be equal, otherwise should already have failed in the preprocess\ndef ck_custom_postprocess(image_files,iter_num, image_size,image_data,dummy, output_dict, category_index,params,score_threshold = 0.3):\n#pred_bbox, org_img_shape, input_size, score_threshold):\n ##first step: concatenate the three tensors\n num_classes = len(category_index.keys())\n pred_bbox = np.concatenate([np.reshape(output_dict[\"pred_sbbox/concat_2:0\"], (-1, 5 + num_classes)),\n np.reshape(output_dict[\"pred_mbbox/concat_2:0\"], (-1, 5 + num_classes)),\n np.reshape(output_dict[\"pred_lbbox/concat_2:0\"], (-1, 5 + num_classes))], axis=0)\n\n\n valid_scale=[0, np.inf]\n pred_bbox = np.array(pred_bbox)\n\n pred_xywh = pred_bbox[:, 0:4]\n pred_conf = pred_bbox[:, 4]\n pred_prob = pred_bbox[:, 5:]\n # # (1) (x, y, w, h) --> (xmin, ymin, xmax, ymax)\n pred_coor = np.concatenate([pred_xywh[:, :2] - pred_xywh[:, 2:] * 0.5,\n pred_xywh[:, :2] + pred_xywh[:, 2:] * 0.5], axis=-1)\n # # (2) (xmin, ymin, xmax, ymax) -> (xmin_org, ymin_org, xmax_org, ymax_org)\n org_h, org_w = image_size\n resize_dim = params[\"RESIZE_WIDTH_SIZE\"],params[\"RESIZE_HEIGHT_SIZE\"] ##They are equal.\n resize_ratio = min(resize_dim[0] / org_w, resize_dim[1] / org_h)\n\n dw = (resize_dim[0] - resize_ratio * org_w) / 2\n dh = (resize_dim[0] - resize_ratio * org_h) / 2\n\n pred_coor[:, 0::2] = 1.0 * (pred_coor[:, 0::2] - dw) / resize_ratio\n pred_coor[:, 1::2] = 1.0 * (pred_coor[:, 1::2] - dh) / resize_ratio\n\n # # (3) clip some boxes those are out of range\n pred_coor = np.concatenate([np.maximum(pred_coor[:, :2], [0, 0]),\n np.minimum(pred_coor[:, 2:], [org_w - 1, org_h - 1])], axis=-1)\n invalid_mask = np.logical_or((pred_coor[:, 0] > pred_coor[:, 2]), (pred_coor[:, 1] > pred_coor[:, 3]))\n pred_coor[invalid_mask] = 0\n\n # # (4) discard some invalid boxes\n bboxes_scale = np.sqrt(np.multiply.reduce(pred_coor[:, 2:4] - pred_coor[:, 0:2], axis=-1))\n scale_mask = np.logical_and((valid_scale[0] < bboxes_scale), (bboxes_scale < valid_scale[1]))\n\n # # (5) discard some boxes with low scores\n classes = np.argmax(pred_prob, axis=-1)\n scores = pred_conf * pred_prob[np.arange(len(pred_coor)), classes]\n score_mask = scores > score_threshold\n mask = np.logical_and(scale_mask, score_mask)\n array_of_ids = [1,2,3,4,5,6,7,8,9,10,11,13,14,15,16,17,18,19,20,21,22,23,24,25,27,28,31,32,33,34,35,36,37,38,39,40,41,42,43,44,46,47,48,49,50,51,52,53,54,55,56,57,58,59,60,61,62,63,64,65,67,70,72,73,74,75,76,77,78,79,80,81,82,84,85,86,87,88,89,90]\n correct_labels = lambda x : array_of_ids[x]\n classes = np.array([correct_labels(x) for x in classes])\n\n coors, scores, classes = pred_coor[mask], scores[mask], classes[mask]\n\n bboxes= np.concatenate([coors, scores[:, np.newaxis], classes[:, np.newaxis]], axis=-1)\n bboxes= nms (bboxes, 0.45, method='nms')\n ck_custom_save_txt(image_files[iter_num], image_size,bboxes,category_index,params[\"DETECTIONS_OUT_DIR\"])\n if not params[\"SAVE_IMAGES\"]:\n return\n else:\n image = ck_custom_save_images(image_data,bboxes,category_index)\n image = Image.fromarray(image)\n image.save(image_files[iter_num])\n\n###### BATCH PROCESSING FUNCTIONS\n\ndef ck_custom_preprocess_batch(image_files, iter_num,processed_image_ids,params):\n batch_data = []\n batch_sizes = []\n orig_images = []\n if params[\"RESIZE_WIDTH_SIZE\"] != params[\"RESIZE_HEIGHT_SIZE\"]:\n raise ValueError (\"this model works with square images, and you provided different height and width\")\n for img in range(params[\"BATCH_SIZE\"]):\n image_file = image_files[iter_num*params[\"BATCH_SIZE\"]+img]\n image_id = ck_utils.filename_to_id(image_file, params[\"DATASET_TYPE\"])\n processed_image_ids.append(image_id)\n image_path = os.path.join(params[\"IMAGES_DIR\"], image_file)\n orig_image = cv2.imread(image_path)\n orig_image = cv2.cvtColor(orig_image, cv2.COLOR_BGR2RGB)\n orig_images.append(orig_image)\n batch_sizes.append(orig_image.shape[:2])\n image = cv2.cvtColor(orig_image, cv2.COLOR_BGR2RGB).astype(np.float32)\n \n ih, iw = params[\"RESIZE_HEIGHT_SIZE\"],params[\"RESIZE_WIDTH_SIZE\"] ##They are equal.\n h, w, _ = image.shape\n \n scale = min(iw/w, ih/h)\n nw, nh = int(scale * w), int(scale * h)\n image_resized = cv2.resize(image, (nw, nh))\n \n image_paded = np.full(shape=[ih, iw, 3], fill_value=128.0)\n dw, dh = (iw - nw) // 2, (ih-nh) // 2\n image_paded[dh:nh+dh, dw:nw+dw, :] = image_resized\n image_paded = image_paded / 255.\n print(np.shape(image_paded))\n #image_paded = image_paded[np.newaxis, ...]\n #print(np.shape(image_paded))\n batch_data.append(image_paded)\n return batch_data,processed_image_ids,batch_sizes,orig_images\n\n\n\n\n\n\n\n\n\n\n\n\n\n\n\n\n\n\n\n\ndef ck_custom_postprocess_batch(image_files,iter_num, image_size,image_data,dummy, output_dict, category_index,params,score_threshold = 0.3):\n#pred_bbox, org_img_shape, input_size, score_threshold):\n ##first step: concatenate the three tensors\n\n for img in range(params[\"BATCH_SIZE\"]):\n num_classes = len(category_index.keys())\n pred_bbox = np.concatenate([np.reshape(output_dict[\"pred_sbbox/concat_2:0\"][img], (-1, 5 + num_classes)),\n np.reshape(output_dict[\"pred_mbbox/concat_2:0\"][img], (-1, 5 + num_classes)),\n np.reshape(output_dict[\"pred_lbbox/concat_2:0\"][img], (-1, 5 + num_classes))], axis=0)\n \n \n valid_scale=[0, np.inf]\n pred_bbox = np.array(pred_bbox)\n \n pred_xywh = pred_bbox[:, 0:4]\n pred_conf = pred_bbox[:, 4]\n pred_prob = pred_bbox[:, 5:]\n # # (1) (x, y, w, h) --> (xmin, ymin, xmax, ymax)\n pred_coor = np.concatenate([pred_xywh[:, :2] - pred_xywh[:, 2:] * 0.5,\n pred_xywh[:, :2] + pred_xywh[:, 2:] * 0.5], axis=-1)\n # # (2) (xmin, ymin, xmax, ymax) -> (xmin_org, ymin_org, xmax_org, ymax_org)\n org_h, org_w = image_size[img]\n resize_dim = params[\"RESIZE_WIDTH_SIZE\"],params[\"RESIZE_HEIGHT_SIZE\"] ##They are equal.\n resize_ratio = min(resize_dim[0] / org_w, resize_dim[1] / org_h)\n \n dw = (resize_dim[0] - resize_ratio * org_w) / 2\n dh = (resize_dim[0] - resize_ratio * org_h) / 2\n \n pred_coor[:, 0::2] = 1.0 * (pred_coor[:, 0::2] - dw) / resize_ratio\n pred_coor[:, 1::2] = 1.0 * (pred_coor[:, 1::2] - dh) / resize_ratio\n \n # # (3) clip some boxes those are out of range\n pred_coor = np.concatenate([np.maximum(pred_coor[:, :2], [0, 0]),\n np.minimum(pred_coor[:, 2:], [org_w - 1, org_h - 1])], axis=-1)\n invalid_mask = np.logical_or((pred_coor[:, 0] > pred_coor[:, 2]), (pred_coor[:, 1] > pred_coor[:, 3]))\n pred_coor[invalid_mask] = 0\n \n # # (4) discard some invalid boxes\n bboxes_scale = np.sqrt(np.multiply.reduce(pred_coor[:, 2:4] - pred_coor[:, 0:2], axis=-1))\n scale_mask = np.logical_and((valid_scale[0] < bboxes_scale), (bboxes_scale < valid_scale[1]))\n \n # # (5) discard some boxes with low scores\n classes = np.argmax(pred_prob, axis=-1)\n scores = pred_conf * pred_prob[np.arange(len(pred_coor)), classes]\n score_mask = scores > score_threshold\n mask = np.logical_and(scale_mask, score_mask)\n array_of_ids = [1,2,3,4,5,6,7,8,9,10,11,13,14,15,16,17,18,19,20,21,22,23,24,25,27,28,31,32,33,34,35,36,37,38,39,40,41,42,43,44,46,47,48,49,50,51,52,53,54,55,56,57,58,59,60,61,62,63,64,65,67,70,72,73,74,75,76,77,78,79,80,81,82,84,85,86,87,88,89,90]\n correct_labels = lambda x : array_of_ids[x]\n classes = np.array([correct_labels(x) for x in classes])\n \n coors, scores, classes = pred_coor[mask], scores[mask], classes[mask]\n \n bboxes= np.concatenate([coors, scores[:, np.newaxis], classes[:, np.newaxis]], axis=-1)\n bboxes= nms (bboxes, 0.45, method='nms')\n ck_custom_save_txt(image_files[iter_num*params[\"BATCH_SIZE\"]+img], image_size[img],bboxes,category_index,params[\"DETECTIONS_OUT_DIR\"])\n if not params[\"SAVE_IMAGES\"]:\n return\n else:\n image = ck_custom_save_images(image_data[img],bboxes,category_index)\n image = Image.fromarray(image)\n image.save(image_files[iter_num*params[\"BATCH_SIZE\"]+img])\n\n"
] |
[
[
"numpy.concatenate",
"numpy.full",
"numpy.array",
"numpy.logical_or",
"numpy.reshape",
"numpy.minimum",
"tensorflow.compat.v1.get_default_graph",
"numpy.exp",
"numpy.logical_and",
"numpy.shape",
"numpy.finfo",
"numpy.argmax",
"numpy.multiply.reduce",
"numpy.maximum"
]
] |
nschloe/scipy
|
[
"3f05efb9498e29f4e735aa7e0220139a8d8c8773"
] |
[
"scipy/spatial/distance.py"
] |
[
"\"\"\"\nDistance computations (:mod:`scipy.spatial.distance`)\n=====================================================\n\n.. sectionauthor:: Damian Eads\n\nFunction reference\n------------------\n\nDistance matrix computation from a collection of raw observation vectors\nstored in a rectangular array.\n\n.. autosummary::\n :toctree: generated/\n\n pdist -- pairwise distances between observation vectors.\n cdist -- distances between two collections of observation vectors\n squareform -- convert distance matrix to a condensed one and vice versa\n directed_hausdorff -- directed Hausdorff distance between arrays\n\nPredicates for checking the validity of distance matrices, both\ncondensed and redundant. Also contained in this module are functions\nfor computing the number of observations in a distance matrix.\n\n.. autosummary::\n :toctree: generated/\n\n is_valid_dm -- checks for a valid distance matrix\n is_valid_y -- checks for a valid condensed distance matrix\n num_obs_dm -- # of observations in a distance matrix\n num_obs_y -- # of observations in a condensed distance matrix\n\nDistance functions between two numeric vectors ``u`` and ``v``. Computing\ndistances over a large collection of vectors is inefficient for these\nfunctions. Use ``pdist`` for this purpose.\n\n.. autosummary::\n :toctree: generated/\n\n braycurtis -- the Bray-Curtis distance.\n canberra -- the Canberra distance.\n chebyshev -- the Chebyshev distance.\n cityblock -- the Manhattan distance.\n correlation -- the Correlation distance.\n cosine -- the Cosine distance.\n euclidean -- the Euclidean distance.\n jensenshannon -- the Jensen-Shannon distance.\n mahalanobis -- the Mahalanobis distance.\n minkowski -- the Minkowski distance.\n seuclidean -- the normalized Euclidean distance.\n sqeuclidean -- the squared Euclidean distance.\n\nDistance functions between two boolean vectors (representing sets) ``u`` and\n``v``. As in the case of numerical vectors, ``pdist`` is more efficient for\ncomputing the distances between all pairs.\n\n.. autosummary::\n :toctree: generated/\n\n dice -- the Dice dissimilarity.\n hamming -- the Hamming distance.\n jaccard -- the Jaccard distance.\n kulsinski -- the Kulsinski distance.\n kulczynski1 -- the Kulczynski 1 distance.\n rogerstanimoto -- the Rogers-Tanimoto dissimilarity.\n russellrao -- the Russell-Rao dissimilarity.\n sokalmichener -- the Sokal-Michener dissimilarity.\n sokalsneath -- the Sokal-Sneath dissimilarity.\n yule -- the Yule dissimilarity.\n\n:func:`hamming` also operates over discrete numerical vectors.\n\"\"\"\n\n# Copyright (C) Damian Eads, 2007-2008. New BSD License.\n\n__all__ = [\n 'braycurtis',\n 'canberra',\n 'cdist',\n 'chebyshev',\n 'cityblock',\n 'correlation',\n 'cosine',\n 'dice',\n 'directed_hausdorff',\n 'euclidean',\n 'hamming',\n 'is_valid_dm',\n 'is_valid_y',\n 'jaccard',\n 'jensenshannon',\n 'kulsinski',\n 'kulczynski1',\n 'mahalanobis',\n 'matching',\n 'minkowski',\n 'num_obs_dm',\n 'num_obs_y',\n 'pdist',\n 'rogerstanimoto',\n 'russellrao',\n 'seuclidean',\n 'sokalmichener',\n 'sokalsneath',\n 'sqeuclidean',\n 'squareform',\n 'yule'\n]\n\n\nimport warnings\nimport numpy as np\nimport dataclasses\n\nfrom typing import List, Optional, Set, Callable\n\nfrom functools import partial\nfrom scipy._lib._util import _asarray_validated\nfrom scipy._lib.deprecation import _deprecated\n\nfrom . import _distance_wrap\nfrom . import _hausdorff\nfrom ..linalg import norm\nfrom ..special import rel_entr\n\nfrom . import _distance_pybind\n\n\ndef _copy_array_if_base_present(a):\n \"\"\"Copy the array if its base points to a parent array.\"\"\"\n if a.base is not None:\n return a.copy()\n return a\n\n\ndef _correlation_cdist_wrap(XA, XB, dm, **kwargs):\n XA = XA - XA.mean(axis=1, keepdims=True)\n XB = XB - XB.mean(axis=1, keepdims=True)\n _distance_wrap.cdist_cosine_double_wrap(XA, XB, dm, **kwargs)\n\n\ndef _correlation_pdist_wrap(X, dm, **kwargs):\n X2 = X - X.mean(axis=1, keepdims=True)\n _distance_wrap.pdist_cosine_double_wrap(X2, dm, **kwargs)\n\n\ndef _convert_to_type(X, out_type):\n return np.ascontiguousarray(X, dtype=out_type)\n\n\ndef _nbool_correspond_all(u, v, w=None):\n if u.dtype == v.dtype == bool and w is None:\n not_u = ~u\n not_v = ~v\n nff = (not_u & not_v).sum()\n nft = (not_u & v).sum()\n ntf = (u & not_v).sum()\n ntt = (u & v).sum()\n else:\n dtype = np.find_common_type([int], [u.dtype, v.dtype])\n u = u.astype(dtype)\n v = v.astype(dtype)\n not_u = 1.0 - u\n not_v = 1.0 - v\n if w is not None:\n not_u = w * not_u\n u = w * u\n nff = (not_u * not_v).sum()\n nft = (not_u * v).sum()\n ntf = (u * not_v).sum()\n ntt = (u * v).sum()\n return (nff, nft, ntf, ntt)\n\n\ndef _nbool_correspond_ft_tf(u, v, w=None):\n if u.dtype == v.dtype == bool and w is None:\n not_u = ~u\n not_v = ~v\n nft = (not_u & v).sum()\n ntf = (u & not_v).sum()\n else:\n dtype = np.find_common_type([int], [u.dtype, v.dtype])\n u = u.astype(dtype)\n v = v.astype(dtype)\n not_u = 1.0 - u\n not_v = 1.0 - v\n if w is not None:\n not_u = w * not_u\n u = w * u\n nft = (not_u * v).sum()\n ntf = (u * not_v).sum()\n return (nft, ntf)\n\n\ndef _validate_cdist_input(XA, XB, mA, mB, n, metric_info, **kwargs):\n # get supported types\n types = metric_info.types\n # choose best type\n typ = types[types.index(XA.dtype)] if XA.dtype in types else types[0]\n # validate data\n XA = _convert_to_type(XA, out_type=typ)\n XB = _convert_to_type(XB, out_type=typ)\n\n # validate kwargs\n _validate_kwargs = metric_info.validator\n if _validate_kwargs:\n kwargs = _validate_kwargs((XA, XB), mA + mB, n, **kwargs)\n return XA, XB, typ, kwargs\n\n\ndef _validate_weight_with_size(X, m, n, **kwargs):\n w = kwargs.pop('w', None)\n if w is None:\n return kwargs\n\n if w.ndim != 1 or w.shape[0] != n:\n raise ValueError(\"Weights must have same size as input vector. \"\n f\"{w.shape[0]} vs. {n}\")\n\n kwargs['w'] = _validate_weights(w)\n return kwargs\n\n\ndef _validate_hamming_kwargs(X, m, n, **kwargs):\n w = kwargs.get('w', np.ones((n,), dtype='double'))\n\n if w.ndim != 1 or w.shape[0] != n:\n raise ValueError(\"Weights must have same size as input vector. %d vs. %d\" % (w.shape[0], n))\n\n kwargs['w'] = _validate_weights(w)\n return kwargs\n\n\ndef _validate_mahalanobis_kwargs(X, m, n, **kwargs):\n VI = kwargs.pop('VI', None)\n if VI is None:\n if m <= n:\n # There are fewer observations than the dimension of\n # the observations.\n raise ValueError(\"The number of observations (%d) is too \"\n \"small; the covariance matrix is \"\n \"singular. For observations with %d \"\n \"dimensions, at least %d observations \"\n \"are required.\" % (m, n, n + 1))\n if isinstance(X, tuple):\n X = np.vstack(X)\n CV = np.atleast_2d(np.cov(X.astype(np.double, copy=False).T))\n VI = np.linalg.inv(CV).T.copy()\n kwargs[\"VI\"] = _convert_to_double(VI)\n return kwargs\n\n\ndef _validate_minkowski_kwargs(X, m, n, **kwargs):\n kwargs = _validate_weight_with_size(X, m, n, **kwargs)\n if 'p' not in kwargs:\n kwargs['p'] = 2.\n else:\n if kwargs['p'] < 1:\n raise ValueError(\"p must be at least 1\")\n\n return kwargs\n\n\ndef _validate_pdist_input(X, m, n, metric_info, **kwargs):\n # get supported types\n types = metric_info.types\n # choose best type\n typ = types[types.index(X.dtype)] if X.dtype in types else types[0]\n # validate data\n X = _convert_to_type(X, out_type=typ)\n\n # validate kwargs\n _validate_kwargs = metric_info.validator\n if _validate_kwargs:\n kwargs = _validate_kwargs(X, m, n, **kwargs)\n return X, typ, kwargs\n\n\ndef _validate_seuclidean_kwargs(X, m, n, **kwargs):\n V = kwargs.pop('V', None)\n if V is None:\n if isinstance(X, tuple):\n X = np.vstack(X)\n V = np.var(X.astype(np.double, copy=False), axis=0, ddof=1)\n else:\n V = np.asarray(V, order='c')\n if len(V.shape) != 1:\n raise ValueError('Variance vector V must '\n 'be one-dimensional.')\n if V.shape[0] != n:\n raise ValueError('Variance vector V must be of the same '\n 'dimension as the vectors on which the distances '\n 'are computed.')\n kwargs['V'] = _convert_to_double(V)\n return kwargs\n\n\ndef _validate_vector(u, dtype=None):\n # XXX Is order='c' really necessary?\n u = np.asarray(u, dtype=dtype, order='c')\n if u.ndim == 1:\n return u\n\n # Ensure values such as u=1 and u=[1] still return 1-D arrays.\n u = np.atleast_1d(u.squeeze())\n if u.ndim > 1:\n raise ValueError(\"Input vector should be 1-D.\")\n warnings.warn(\n \"scipy.spatial.distance metrics ignoring length-1 dimensions is \"\n \"deprecated in SciPy 1.7 and will raise an error in SciPy 1.9.\",\n DeprecationWarning)\n return u\n\n\ndef _validate_weights(w, dtype=np.double):\n w = _validate_vector(w, dtype=dtype)\n if np.any(w < 0):\n raise ValueError(\"Input weights should be all non-negative\")\n return w\n\n\ndef directed_hausdorff(u, v, seed=0):\n \"\"\"\n Compute the directed Hausdorff distance between two 2-D arrays.\n\n Distances between pairs are calculated using a Euclidean metric.\n\n Parameters\n ----------\n u : (M,N) array_like\n Input array.\n v : (O,N) array_like\n Input array.\n seed : int or None\n Local `numpy.random.RandomState` seed. Default is 0, a random\n shuffling of u and v that guarantees reproducibility.\n\n Returns\n -------\n d : double\n The directed Hausdorff distance between arrays `u` and `v`,\n\n index_1 : int\n index of point contributing to Hausdorff pair in `u`\n\n index_2 : int\n index of point contributing to Hausdorff pair in `v`\n\n Raises\n ------\n ValueError\n An exception is thrown if `u` and `v` do not have\n the same number of columns.\n\n Notes\n -----\n Uses the early break technique and the random sampling approach\n described by [1]_. Although worst-case performance is ``O(m * o)``\n (as with the brute force algorithm), this is unlikely in practice\n as the input data would have to require the algorithm to explore\n every single point interaction, and after the algorithm shuffles\n the input points at that. The best case performance is O(m), which\n is satisfied by selecting an inner loop distance that is less than\n cmax and leads to an early break as often as possible. The authors\n have formally shown that the average runtime is closer to O(m).\n\n .. versionadded:: 0.19.0\n\n References\n ----------\n .. [1] A. A. Taha and A. Hanbury, \"An efficient algorithm for\n calculating the exact Hausdorff distance.\" IEEE Transactions On\n Pattern Analysis And Machine Intelligence, vol. 37 pp. 2153-63,\n 2015.\n\n See Also\n --------\n scipy.spatial.procrustes : Another similarity test for two data sets\n\n Examples\n --------\n Find the directed Hausdorff distance between two 2-D arrays of\n coordinates:\n\n >>> from scipy.spatial.distance import directed_hausdorff\n >>> u = np.array([(1.0, 0.0),\n ... (0.0, 1.0),\n ... (-1.0, 0.0),\n ... (0.0, -1.0)])\n >>> v = np.array([(2.0, 0.0),\n ... (0.0, 2.0),\n ... (-2.0, 0.0),\n ... (0.0, -4.0)])\n\n >>> directed_hausdorff(u, v)[0]\n 2.23606797749979\n >>> directed_hausdorff(v, u)[0]\n 3.0\n\n Find the general (symmetric) Hausdorff distance between two 2-D\n arrays of coordinates:\n\n >>> max(directed_hausdorff(u, v)[0], directed_hausdorff(v, u)[0])\n 3.0\n\n Find the indices of the points that generate the Hausdorff distance\n (the Hausdorff pair):\n\n >>> directed_hausdorff(v, u)[1:]\n (3, 3)\n\n \"\"\"\n u = np.asarray(u, dtype=np.float64, order='c')\n v = np.asarray(v, dtype=np.float64, order='c')\n if u.shape[1] != v.shape[1]:\n raise ValueError('u and v need to have the same '\n 'number of columns')\n result = _hausdorff.directed_hausdorff(u, v, seed)\n return result\n\n\ndef minkowski(u, v, p=2, w=None):\n \"\"\"\n Compute the Minkowski distance between two 1-D arrays.\n\n The Minkowski distance between 1-D arrays `u` and `v`,\n is defined as\n\n .. math::\n\n {||u-v||}_p = (\\\\sum{|u_i - v_i|^p})^{1/p}.\n\n\n \\\\left(\\\\sum{w_i(|(u_i - v_i)|^p)}\\\\right)^{1/p}.\n\n Parameters\n ----------\n u : (N,) array_like\n Input array.\n v : (N,) array_like\n Input array.\n p : scalar\n The order of the norm of the difference :math:`{||u-v||}_p`.\n w : (N,) array_like, optional\n The weights for each value in `u` and `v`. Default is None,\n which gives each value a weight of 1.0\n\n Returns\n -------\n minkowski : double\n The Minkowski distance between vectors `u` and `v`.\n\n Examples\n --------\n >>> from scipy.spatial import distance\n >>> distance.minkowski([1, 0, 0], [0, 1, 0], 1)\n 2.0\n >>> distance.minkowski([1, 0, 0], [0, 1, 0], 2)\n 1.4142135623730951\n >>> distance.minkowski([1, 0, 0], [0, 1, 0], 3)\n 1.2599210498948732\n >>> distance.minkowski([1, 1, 0], [0, 1, 0], 1)\n 1.0\n >>> distance.minkowski([1, 1, 0], [0, 1, 0], 2)\n 1.0\n >>> distance.minkowski([1, 1, 0], [0, 1, 0], 3)\n 1.0\n\n \"\"\"\n u = _validate_vector(u)\n v = _validate_vector(v)\n if p < 1:\n raise ValueError(\"p must be at least 1\")\n u_v = u - v\n if w is not None:\n w = _validate_weights(w)\n if p == 1:\n root_w = w\n elif p == 2:\n # better precision and speed\n root_w = np.sqrt(w)\n elif p == np.inf:\n root_w = (w != 0)\n else:\n root_w = np.power(w, 1/p)\n u_v = root_w * u_v\n dist = norm(u_v, ord=p)\n return dist\n\n\ndef euclidean(u, v, w=None):\n \"\"\"\n Computes the Euclidean distance between two 1-D arrays.\n\n The Euclidean distance between 1-D arrays `u` and `v`, is defined as\n\n .. math::\n\n {||u-v||}_2\n\n \\\\left(\\\\sum{(w_i |(u_i - v_i)|^2)}\\\\right)^{1/2}\n\n Parameters\n ----------\n u : (N,) array_like\n Input array.\n v : (N,) array_like\n Input array.\n w : (N,) array_like, optional\n The weights for each value in `u` and `v`. Default is None,\n which gives each value a weight of 1.0\n\n Returns\n -------\n euclidean : double\n The Euclidean distance between vectors `u` and `v`.\n\n Examples\n --------\n >>> from scipy.spatial import distance\n >>> distance.euclidean([1, 0, 0], [0, 1, 0])\n 1.4142135623730951\n >>> distance.euclidean([1, 1, 0], [0, 1, 0])\n 1.0\n\n \"\"\"\n return minkowski(u, v, p=2, w=w)\n\n\ndef sqeuclidean(u, v, w=None):\n \"\"\"\n Compute the squared Euclidean distance between two 1-D arrays.\n\n The squared Euclidean distance between `u` and `v` is defined as\n\n .. math::\n\n {||u-v||}_2^2\n\n \\\\left(\\\\sum{(w_i |(u_i - v_i)|^2)}\\\\right)\n\n Parameters\n ----------\n u : (N,) array_like\n Input array.\n v : (N,) array_like\n Input array.\n w : (N,) array_like, optional\n The weights for each value in `u` and `v`. Default is None,\n which gives each value a weight of 1.0\n\n Returns\n -------\n sqeuclidean : double\n The squared Euclidean distance between vectors `u` and `v`.\n\n Examples\n --------\n >>> from scipy.spatial import distance\n >>> distance.sqeuclidean([1, 0, 0], [0, 1, 0])\n 2.0\n >>> distance.sqeuclidean([1, 1, 0], [0, 1, 0])\n 1.0\n\n \"\"\"\n # Preserve float dtypes, but convert everything else to np.float64\n # for stability.\n utype, vtype = None, None\n if not (hasattr(u, \"dtype\") and np.issubdtype(u.dtype, np.inexact)):\n utype = np.float64\n if not (hasattr(v, \"dtype\") and np.issubdtype(v.dtype, np.inexact)):\n vtype = np.float64\n\n u = _validate_vector(u, dtype=utype)\n v = _validate_vector(v, dtype=vtype)\n u_v = u - v\n u_v_w = u_v # only want weights applied once\n if w is not None:\n w = _validate_weights(w)\n u_v_w = w * u_v\n return np.dot(u_v, u_v_w)\n\n\ndef correlation(u, v, w=None, centered=True):\n \"\"\"\n Compute the correlation distance between two 1-D arrays.\n\n The correlation distance between `u` and `v`, is\n defined as\n\n .. math::\n\n 1 - \\\\frac{(u - \\\\bar{u}) \\\\cdot (v - \\\\bar{v})}\n {{||(u - \\\\bar{u})||}_2 {||(v - \\\\bar{v})||}_2}\n\n where :math:`\\\\bar{u}` is the mean of the elements of `u`\n and :math:`x \\\\cdot y` is the dot product of :math:`x` and :math:`y`.\n\n Parameters\n ----------\n u : (N,) array_like\n Input array.\n v : (N,) array_like\n Input array.\n w : (N,) array_like, optional\n The weights for each value in `u` and `v`. Default is None,\n which gives each value a weight of 1.0\n centered : bool, optional\n If True, `u` and `v` will be centered. Default is True.\n\n Returns\n -------\n correlation : double\n The correlation distance between 1-D array `u` and `v`.\n\n \"\"\"\n u = _validate_vector(u)\n v = _validate_vector(v)\n if w is not None:\n w = _validate_weights(w)\n if centered:\n umu = np.average(u, weights=w)\n vmu = np.average(v, weights=w)\n u = u - umu\n v = v - vmu\n uv = np.average(u * v, weights=w)\n uu = np.average(np.square(u), weights=w)\n vv = np.average(np.square(v), weights=w)\n dist = 1.0 - uv / np.sqrt(uu * vv)\n # Return absolute value to avoid small negative value due to rounding\n return np.abs(dist)\n\n\ndef cosine(u, v, w=None):\n \"\"\"\n Compute the Cosine distance between 1-D arrays.\n\n The Cosine distance between `u` and `v`, is defined as\n\n .. math::\n\n 1 - \\\\frac{u \\\\cdot v}\n {||u||_2 ||v||_2}.\n\n where :math:`u \\\\cdot v` is the dot product of :math:`u` and\n :math:`v`.\n\n Parameters\n ----------\n u : (N,) array_like\n Input array.\n v : (N,) array_like\n Input array.\n w : (N,) array_like, optional\n The weights for each value in `u` and `v`. Default is None,\n which gives each value a weight of 1.0\n\n Returns\n -------\n cosine : double\n The Cosine distance between vectors `u` and `v`.\n\n Examples\n --------\n >>> from scipy.spatial import distance\n >>> distance.cosine([1, 0, 0], [0, 1, 0])\n 1.0\n >>> distance.cosine([100, 0, 0], [0, 1, 0])\n 1.0\n >>> distance.cosine([1, 1, 0], [0, 1, 0])\n 0.29289321881345254\n\n \"\"\"\n # cosine distance is also referred to as 'uncentered correlation',\n # or 'reflective correlation'\n # clamp the result to 0-2\n return max(0, min(correlation(u, v, w=w, centered=False), 2.0))\n\n\ndef hamming(u, v, w=None):\n \"\"\"\n Compute the Hamming distance between two 1-D arrays.\n\n The Hamming distance between 1-D arrays `u` and `v`, is simply the\n proportion of disagreeing components in `u` and `v`. If `u` and `v` are\n boolean vectors, the Hamming distance is\n\n .. math::\n\n \\\\frac{c_{01} + c_{10}}{n}\n\n where :math:`c_{ij}` is the number of occurrences of\n :math:`\\\\mathtt{u[k]} = i` and :math:`\\\\mathtt{v[k]} = j` for\n :math:`k < n`.\n\n Parameters\n ----------\n u : (N,) array_like\n Input array.\n v : (N,) array_like\n Input array.\n w : (N,) array_like, optional\n The weights for each value in `u` and `v`. Default is None,\n which gives each value a weight of 1.0\n\n Returns\n -------\n hamming : double\n The Hamming distance between vectors `u` and `v`.\n\n Examples\n --------\n >>> from scipy.spatial import distance\n >>> distance.hamming([1, 0, 0], [0, 1, 0])\n 0.66666666666666663\n >>> distance.hamming([1, 0, 0], [1, 1, 0])\n 0.33333333333333331\n >>> distance.hamming([1, 0, 0], [2, 0, 0])\n 0.33333333333333331\n >>> distance.hamming([1, 0, 0], [3, 0, 0])\n 0.33333333333333331\n\n \"\"\"\n u = _validate_vector(u)\n v = _validate_vector(v)\n if u.shape != v.shape:\n raise ValueError('The 1d arrays must have equal lengths.')\n u_ne_v = u != v\n if w is not None:\n w = _validate_weights(w)\n return np.average(u_ne_v, weights=w)\n\n\ndef jaccard(u, v, w=None):\n \"\"\"\n Compute the Jaccard-Needham dissimilarity between two boolean 1-D arrays.\n\n The Jaccard-Needham dissimilarity between 1-D boolean arrays `u` and `v`,\n is defined as\n\n .. math::\n\n \\\\frac{c_{TF} + c_{FT}}\n {c_{TT} + c_{FT} + c_{TF}}\n\n where :math:`c_{ij}` is the number of occurrences of\n :math:`\\\\mathtt{u[k]} = i` and :math:`\\\\mathtt{v[k]} = j` for\n :math:`k < n`.\n\n Parameters\n ----------\n u : (N,) array_like, bool\n Input array.\n v : (N,) array_like, bool\n Input array.\n w : (N,) array_like, optional\n The weights for each value in `u` and `v`. Default is None,\n which gives each value a weight of 1.0\n\n Returns\n -------\n jaccard : double\n The Jaccard distance between vectors `u` and `v`.\n\n Notes\n -----\n When both `u` and `v` lead to a `0/0` division i.e. there is no overlap\n between the items in the vectors the returned distance is 0. See the\n Wikipedia page on the Jaccard index [1]_, and this paper [2]_.\n\n .. versionchanged:: 1.2.0\n Previously, when `u` and `v` lead to a `0/0` division, the function\n would return NaN. This was changed to return 0 instead.\n\n References\n ----------\n .. [1] https://en.wikipedia.org/wiki/Jaccard_index\n .. [2] S. Kosub, \"A note on the triangle inequality for the Jaccard\n distance\", 2016, :arxiv:`1612.02696`\n\n Examples\n --------\n >>> from scipy.spatial import distance\n >>> distance.jaccard([1, 0, 0], [0, 1, 0])\n 1.0\n >>> distance.jaccard([1, 0, 0], [1, 1, 0])\n 0.5\n >>> distance.jaccard([1, 0, 0], [1, 2, 0])\n 0.5\n >>> distance.jaccard([1, 0, 0], [1, 1, 1])\n 0.66666666666666663\n\n \"\"\"\n u = _validate_vector(u)\n v = _validate_vector(v)\n\n nonzero = np.bitwise_or(u != 0, v != 0)\n unequal_nonzero = np.bitwise_and((u != v), nonzero)\n if w is not None:\n w = _validate_weights(w)\n nonzero = w * nonzero\n unequal_nonzero = w * unequal_nonzero\n a = np.double(unequal_nonzero.sum())\n b = np.double(nonzero.sum())\n return (a / b) if b != 0 else 0\n\n\ndef kulsinski(u, v, w=None):\n \"\"\"\n Compute the Kulsinski dissimilarity between two boolean 1-D arrays.\n\n The Kulsinski dissimilarity between two boolean 1-D arrays `u` and `v`,\n is defined as\n\n .. math::\n\n \\\\frac{c_{TF} + c_{FT} - c_{TT} + n}\n {c_{FT} + c_{TF} + n}\n\n where :math:`c_{ij}` is the number of occurrences of\n :math:`\\\\mathtt{u[k]} = i` and :math:`\\\\mathtt{v[k]} = j` for\n :math:`k < n`.\n\n Parameters\n ----------\n u : (N,) array_like, bool\n Input array.\n v : (N,) array_like, bool\n Input array.\n w : (N,) array_like, optional\n The weights for each value in `u` and `v`. Default is None,\n which gives each value a weight of 1.0\n\n Returns\n -------\n kulsinski : double\n The Kulsinski distance between vectors `u` and `v`.\n\n Examples\n --------\n >>> from scipy.spatial import distance\n >>> distance.kulsinski([1, 0, 0], [0, 1, 0])\n 1.0\n >>> distance.kulsinski([1, 0, 0], [1, 1, 0])\n 0.75\n >>> distance.kulsinski([1, 0, 0], [2, 1, 0])\n 0.33333333333333331\n >>> distance.kulsinski([1, 0, 0], [3, 1, 0])\n -0.5\n\n \"\"\"\n u = _validate_vector(u)\n v = _validate_vector(v)\n if w is None:\n n = float(len(u))\n else:\n w = _validate_weights(w)\n n = w.sum()\n (nff, nft, ntf, ntt) = _nbool_correspond_all(u, v, w=w)\n\n return (ntf + nft - ntt + n) / (ntf + nft + n)\n\n\ndef kulczynski1(u, v, *, w=None):\n \"\"\"\n Compute the Kulczynski 1 dissimilarity between two boolean 1-D arrays.\n\n The Kulczynski 1 dissimilarity between two boolean 1-D arrays `u` and `v`\n of length ``n``, is defined as\n\n .. math::\n\n \\\\frac{c_{11}}\n {c_{01} + c_{10}}\n\n where :math:`c_{ij}` is the number of occurrences of\n :math:`\\\\mathtt{u[k]} = i` and :math:`\\\\mathtt{v[k]} = j` for\n :math:`k \\\\in {0, 1, ..., n-1}`.\n\n Parameters\n ----------\n u : (N,) array_like, bool\n Input array.\n v : (N,) array_like, bool\n Input array.\n w : (N,) array_like, optional\n The weights for each value in `u` and `v`. Default is None,\n which gives each value a weight of 1.0\n\n Returns\n -------\n kulczynski1 : float\n The Kulczynski 1 distance between vectors `u` and `v`.\n\n See Also\n --------\n\n kulsinski\n\n Notes\n -----\n This measure has a minimum value of 0 and no upper limit.\n It is un-defined when there are no non-matches.\n\n .. versionadded:: 1.8.0\n\n References\n ----------\n .. [1] Kulczynski S. et al. Bulletin\n International de l'Academie Polonaise des Sciences\n et des Lettres, Classe des Sciences Mathematiques\n et Naturelles, Serie B (Sciences Naturelles). 1927;\n Supplement II: 57-203.\n\n Examples\n --------\n >>> from scipy.spatial import distance\n >>> distance.kulczynski1([1, 0, 0], [0, 1, 0])\n 0.0\n >>> distance.kulczynski1([True, False, False], [True, True, False])\n 1.0\n >>> distance.kulczynski1([True, False, False], True)\n 0.5\n >>> distance.kulczynski1([1, 0, 0], [3, 1, 0])\n -3.0\n\n \"\"\"\n u = _validate_vector(u)\n v = _validate_vector(v)\n if w is not None:\n w = _validate_weights(w)\n (_, nft, ntf, ntt) = _nbool_correspond_all(u, v, w=w)\n\n return ntt / (ntf + nft)\n\n\ndef seuclidean(u, v, V):\n \"\"\"\n Return the standardized Euclidean distance between two 1-D arrays.\n\n The standardized Euclidean distance between `u` and `v`.\n\n Parameters\n ----------\n u : (N,) array_like\n Input array.\n v : (N,) array_like\n Input array.\n V : (N,) array_like\n `V` is an 1-D array of component variances. It is usually computed\n among a larger collection vectors.\n\n Returns\n -------\n seuclidean : double\n The standardized Euclidean distance between vectors `u` and `v`.\n\n Examples\n --------\n >>> from scipy.spatial import distance\n >>> distance.seuclidean([1, 0, 0], [0, 1, 0], [0.1, 0.1, 0.1])\n 4.4721359549995796\n >>> distance.seuclidean([1, 0, 0], [0, 1, 0], [1, 0.1, 0.1])\n 3.3166247903553998\n >>> distance.seuclidean([1, 0, 0], [0, 1, 0], [10, 0.1, 0.1])\n 3.1780497164141406\n\n \"\"\"\n u = _validate_vector(u)\n v = _validate_vector(v)\n V = _validate_vector(V, dtype=np.float64)\n if V.shape[0] != u.shape[0] or u.shape[0] != v.shape[0]:\n raise TypeError('V must be a 1-D array of the same dimension '\n 'as u and v.')\n return euclidean(u, v, w=1/V)\n\n\ndef cityblock(u, v, w=None):\n \"\"\"\n Compute the City Block (Manhattan) distance.\n\n Computes the Manhattan distance between two 1-D arrays `u` and `v`,\n which is defined as\n\n .. math::\n\n \\\\sum_i {\\\\left| u_i - v_i \\\\right|}.\n\n Parameters\n ----------\n u : (N,) array_like\n Input array.\n v : (N,) array_like\n Input array.\n w : (N,) array_like, optional\n The weights for each value in `u` and `v`. Default is None,\n which gives each value a weight of 1.0\n\n Returns\n -------\n cityblock : double\n The City Block (Manhattan) distance between vectors `u` and `v`.\n\n Examples\n --------\n >>> from scipy.spatial import distance\n >>> distance.cityblock([1, 0, 0], [0, 1, 0])\n 2\n >>> distance.cityblock([1, 0, 0], [0, 2, 0])\n 3\n >>> distance.cityblock([1, 0, 0], [1, 1, 0])\n 1\n\n \"\"\"\n u = _validate_vector(u)\n v = _validate_vector(v)\n l1_diff = abs(u - v)\n if w is not None:\n w = _validate_weights(w)\n l1_diff = w * l1_diff\n return l1_diff.sum()\n\n\ndef mahalanobis(u, v, VI):\n \"\"\"\n Compute the Mahalanobis distance between two 1-D arrays.\n\n The Mahalanobis distance between 1-D arrays `u` and `v`, is defined as\n\n .. math::\n\n \\\\sqrt{ (u-v) V^{-1} (u-v)^T }\n\n where ``V`` is the covariance matrix. Note that the argument `VI`\n is the inverse of ``V``.\n\n Parameters\n ----------\n u : (N,) array_like\n Input array.\n v : (N,) array_like\n Input array.\n VI : array_like\n The inverse of the covariance matrix.\n\n Returns\n -------\n mahalanobis : double\n The Mahalanobis distance between vectors `u` and `v`.\n\n Examples\n --------\n >>> from scipy.spatial import distance\n >>> iv = [[1, 0.5, 0.5], [0.5, 1, 0.5], [0.5, 0.5, 1]]\n >>> distance.mahalanobis([1, 0, 0], [0, 1, 0], iv)\n 1.0\n >>> distance.mahalanobis([0, 2, 0], [0, 1, 0], iv)\n 1.0\n >>> distance.mahalanobis([2, 0, 0], [0, 1, 0], iv)\n 1.7320508075688772\n\n \"\"\"\n u = _validate_vector(u)\n v = _validate_vector(v)\n VI = np.atleast_2d(VI)\n delta = u - v\n m = np.dot(np.dot(delta, VI), delta)\n return np.sqrt(m)\n\n\ndef chebyshev(u, v, w=None):\n \"\"\"\n Compute the Chebyshev distance.\n\n Computes the Chebyshev distance between two 1-D arrays `u` and `v`,\n which is defined as\n\n .. math::\n\n \\\\max_i {|u_i-v_i|}.\n\n Parameters\n ----------\n u : (N,) array_like\n Input vector.\n v : (N,) array_like\n Input vector.\n w : (N,) array_like, optional\n Unused, as 'max' is a weightless operation. Here for API consistency.\n\n Returns\n -------\n chebyshev : double\n The Chebyshev distance between vectors `u` and `v`.\n\n Examples\n --------\n >>> from scipy.spatial import distance\n >>> distance.chebyshev([1, 0, 0], [0, 1, 0])\n 1\n >>> distance.chebyshev([1, 1, 0], [0, 1, 0])\n 1\n\n \"\"\"\n u = _validate_vector(u)\n v = _validate_vector(v)\n if w is not None:\n w = _validate_weights(w)\n has_weight = w > 0\n if has_weight.sum() < w.size:\n u = u[has_weight]\n v = v[has_weight]\n return max(abs(u - v))\n\n\ndef braycurtis(u, v, w=None):\n \"\"\"\n Compute the Bray-Curtis distance between two 1-D arrays.\n\n Bray-Curtis distance is defined as\n\n .. math::\n\n \\\\sum{|u_i-v_i|} / \\\\sum{|u_i+v_i|}\n\n The Bray-Curtis distance is in the range [0, 1] if all coordinates are\n positive, and is undefined if the inputs are of length zero.\n\n Parameters\n ----------\n u : (N,) array_like\n Input array.\n v : (N,) array_like\n Input array.\n w : (N,) array_like, optional\n The weights for each value in `u` and `v`. Default is None,\n which gives each value a weight of 1.0\n\n Returns\n -------\n braycurtis : double\n The Bray-Curtis distance between 1-D arrays `u` and `v`.\n\n Examples\n --------\n >>> from scipy.spatial import distance\n >>> distance.braycurtis([1, 0, 0], [0, 1, 0])\n 1.0\n >>> distance.braycurtis([1, 1, 0], [0, 1, 0])\n 0.33333333333333331\n\n \"\"\"\n u = _validate_vector(u)\n v = _validate_vector(v, dtype=np.float64)\n l1_diff = abs(u - v)\n l1_sum = abs(u + v)\n if w is not None:\n w = _validate_weights(w)\n l1_diff = w * l1_diff\n l1_sum = w * l1_sum\n return l1_diff.sum() / l1_sum.sum()\n\n\ndef canberra(u, v, w=None):\n \"\"\"\n Compute the Canberra distance between two 1-D arrays.\n\n The Canberra distance is defined as\n\n .. math::\n\n d(u,v) = \\\\sum_i \\\\frac{|u_i-v_i|}\n {|u_i|+|v_i|}.\n\n Parameters\n ----------\n u : (N,) array_like\n Input array.\n v : (N,) array_like\n Input array.\n w : (N,) array_like, optional\n The weights for each value in `u` and `v`. Default is None,\n which gives each value a weight of 1.0\n\n Returns\n -------\n canberra : double\n The Canberra distance between vectors `u` and `v`.\n\n Notes\n -----\n When `u[i]` and `v[i]` are 0 for given i, then the fraction 0/0 = 0 is\n used in the calculation.\n\n Examples\n --------\n >>> from scipy.spatial import distance\n >>> distance.canberra([1, 0, 0], [0, 1, 0])\n 2.0\n >>> distance.canberra([1, 1, 0], [0, 1, 0])\n 1.0\n\n \"\"\"\n u = _validate_vector(u)\n v = _validate_vector(v, dtype=np.float64)\n if w is not None:\n w = _validate_weights(w)\n with np.errstate(invalid='ignore'):\n abs_uv = abs(u - v)\n abs_u = abs(u)\n abs_v = abs(v)\n d = abs_uv / (abs_u + abs_v)\n if w is not None:\n d = w * d\n d = np.nansum(d)\n return d\n\n\ndef jensenshannon(p, q, base=None, *, axis=0, keepdims=False):\n \"\"\"\n Compute the Jensen-Shannon distance (metric) between\n two probability arrays. This is the square root\n of the Jensen-Shannon divergence.\n\n The Jensen-Shannon distance between two probability\n vectors `p` and `q` is defined as,\n\n .. math::\n\n \\\\sqrt{\\\\frac{D(p \\\\parallel m) + D(q \\\\parallel m)}{2}}\n\n where :math:`m` is the pointwise mean of :math:`p` and :math:`q`\n and :math:`D` is the Kullback-Leibler divergence.\n\n This routine will normalize `p` and `q` if they don't sum to 1.0.\n\n Parameters\n ----------\n p : (N,) array_like\n left probability vector\n q : (N,) array_like\n right probability vector\n base : double, optional\n the base of the logarithm used to compute the output\n if not given, then the routine uses the default base of\n scipy.stats.entropy.\n axis : int, optional\n Axis along which the Jensen-Shannon distances are computed. The default\n is 0.\n\n .. versionadded:: 1.7.0\n keepdims : bool, optional\n If this is set to `True`, the reduced axes are left in the\n result as dimensions with size one. With this option,\n the result will broadcast correctly against the input array.\n Default is False.\n\n .. versionadded:: 1.7.0\n\n Returns\n -------\n js : double or ndarray\n The Jensen-Shannon distances between `p` and `q` along the `axis`.\n\n Notes\n -----\n\n .. versionadded:: 1.2.0\n\n Examples\n --------\n >>> from scipy.spatial import distance\n >>> distance.jensenshannon([1.0, 0.0, 0.0], [0.0, 1.0, 0.0], 2.0)\n 1.0\n >>> distance.jensenshannon([1.0, 0.0], [0.5, 0.5])\n 0.46450140402245893\n >>> distance.jensenshannon([1.0, 0.0, 0.0], [1.0, 0.0, 0.0])\n 0.0\n >>> a = np.array([[1, 2, 3, 4],\n ... [5, 6, 7, 8],\n ... [9, 10, 11, 12]])\n >>> b = np.array([[13, 14, 15, 16],\n ... [17, 18, 19, 20],\n ... [21, 22, 23, 24]])\n >>> distance.jensenshannon(a, b, axis=0)\n array([0.1954288, 0.1447697, 0.1138377, 0.0927636])\n >>> distance.jensenshannon(a, b, axis=1)\n array([0.1402339, 0.0399106, 0.0201815])\n\n \"\"\"\n p = np.asarray(p)\n q = np.asarray(q)\n p = p / np.sum(p, axis=axis, keepdims=True)\n q = q / np.sum(q, axis=axis, keepdims=True)\n m = (p + q) / 2.0\n left = rel_entr(p, m)\n right = rel_entr(q, m)\n left_sum = np.sum(left, axis=axis, keepdims=keepdims)\n right_sum = np.sum(right, axis=axis, keepdims=keepdims)\n js = left_sum + right_sum\n if base is not None:\n js /= np.log(base)\n return np.sqrt(js / 2.0)\n\n\ndef yule(u, v, w=None):\n \"\"\"\n Compute the Yule dissimilarity between two boolean 1-D arrays.\n\n The Yule dissimilarity is defined as\n\n .. math::\n\n \\\\frac{R}{c_{TT} * c_{FF} + \\\\frac{R}{2}}\n\n where :math:`c_{ij}` is the number of occurrences of\n :math:`\\\\mathtt{u[k]} = i` and :math:`\\\\mathtt{v[k]} = j` for\n :math:`k < n` and :math:`R = 2.0 * c_{TF} * c_{FT}`.\n\n Parameters\n ----------\n u : (N,) array_like, bool\n Input array.\n v : (N,) array_like, bool\n Input array.\n w : (N,) array_like, optional\n The weights for each value in `u` and `v`. Default is None,\n which gives each value a weight of 1.0\n\n Returns\n -------\n yule : double\n The Yule dissimilarity between vectors `u` and `v`.\n\n Examples\n --------\n >>> from scipy.spatial import distance\n >>> distance.yule([1, 0, 0], [0, 1, 0])\n 2.0\n >>> distance.yule([1, 1, 0], [0, 1, 0])\n 0.0\n\n \"\"\"\n u = _validate_vector(u)\n v = _validate_vector(v)\n if w is not None:\n w = _validate_weights(w)\n (nff, nft, ntf, ntt) = _nbool_correspond_all(u, v, w=w)\n half_R = ntf * nft\n if half_R == 0:\n return 0.0\n else:\n return float(2.0 * half_R / (ntt * nff + half_R))\n\n\[email protected](message=\"spatial.distance.matching is deprecated in scipy 1.0.0; \"\n \"use spatial.distance.hamming instead.\")\ndef matching(u, v, w=None):\n \"\"\"\n Compute the Hamming distance between two boolean 1-D arrays.\n\n This is a deprecated synonym for :func:`hamming`.\n \"\"\"\n return hamming(u, v, w=w)\n\n\ndef dice(u, v, w=None):\n \"\"\"\n Compute the Dice dissimilarity between two boolean 1-D arrays.\n\n The Dice dissimilarity between `u` and `v`, is\n\n .. math::\n\n \\\\frac{c_{TF} + c_{FT}}\n {2c_{TT} + c_{FT} + c_{TF}}\n\n where :math:`c_{ij}` is the number of occurrences of\n :math:`\\\\mathtt{u[k]} = i` and :math:`\\\\mathtt{v[k]} = j` for\n :math:`k < n`.\n\n Parameters\n ----------\n u : (N,) array_like, bool\n Input 1-D array.\n v : (N,) array_like, bool\n Input 1-D array.\n w : (N,) array_like, optional\n The weights for each value in `u` and `v`. Default is None,\n which gives each value a weight of 1.0\n\n Returns\n -------\n dice : double\n The Dice dissimilarity between 1-D arrays `u` and `v`.\n\n Examples\n --------\n >>> from scipy.spatial import distance\n >>> distance.dice([1, 0, 0], [0, 1, 0])\n 1.0\n >>> distance.dice([1, 0, 0], [1, 1, 0])\n 0.3333333333333333\n >>> distance.dice([1, 0, 0], [2, 0, 0])\n -0.3333333333333333\n\n \"\"\"\n u = _validate_vector(u)\n v = _validate_vector(v)\n if w is not None:\n w = _validate_weights(w)\n if u.dtype == v.dtype == bool and w is None:\n ntt = (u & v).sum()\n else:\n dtype = np.find_common_type([int], [u.dtype, v.dtype])\n u = u.astype(dtype)\n v = v.astype(dtype)\n if w is None:\n ntt = (u * v).sum()\n else:\n ntt = (u * v * w).sum()\n (nft, ntf) = _nbool_correspond_ft_tf(u, v, w=w)\n return float((ntf + nft) / np.array(2.0 * ntt + ntf + nft))\n\n\ndef rogerstanimoto(u, v, w=None):\n \"\"\"\n Compute the Rogers-Tanimoto dissimilarity between two boolean 1-D arrays.\n\n The Rogers-Tanimoto dissimilarity between two boolean 1-D arrays\n `u` and `v`, is defined as\n\n .. math::\n \\\\frac{R}\n {c_{TT} + c_{FF} + R}\n\n where :math:`c_{ij}` is the number of occurrences of\n :math:`\\\\mathtt{u[k]} = i` and :math:`\\\\mathtt{v[k]} = j` for\n :math:`k < n` and :math:`R = 2(c_{TF} + c_{FT})`.\n\n Parameters\n ----------\n u : (N,) array_like, bool\n Input array.\n v : (N,) array_like, bool\n Input array.\n w : (N,) array_like, optional\n The weights for each value in `u` and `v`. Default is None,\n which gives each value a weight of 1.0\n\n Returns\n -------\n rogerstanimoto : double\n The Rogers-Tanimoto dissimilarity between vectors\n `u` and `v`.\n\n Examples\n --------\n >>> from scipy.spatial import distance\n >>> distance.rogerstanimoto([1, 0, 0], [0, 1, 0])\n 0.8\n >>> distance.rogerstanimoto([1, 0, 0], [1, 1, 0])\n 0.5\n >>> distance.rogerstanimoto([1, 0, 0], [2, 0, 0])\n -1.0\n\n \"\"\"\n u = _validate_vector(u)\n v = _validate_vector(v)\n if w is not None:\n w = _validate_weights(w)\n (nff, nft, ntf, ntt) = _nbool_correspond_all(u, v, w=w)\n return float(2.0 * (ntf + nft)) / float(ntt + nff + (2.0 * (ntf + nft)))\n\n\ndef russellrao(u, v, w=None):\n \"\"\"\n Compute the Russell-Rao dissimilarity between two boolean 1-D arrays.\n\n The Russell-Rao dissimilarity between two boolean 1-D arrays, `u` and\n `v`, is defined as\n\n .. math::\n\n \\\\frac{n - c_{TT}}\n {n}\n\n where :math:`c_{ij}` is the number of occurrences of\n :math:`\\\\mathtt{u[k]} = i` and :math:`\\\\mathtt{v[k]} = j` for\n :math:`k < n`.\n\n Parameters\n ----------\n u : (N,) array_like, bool\n Input array.\n v : (N,) array_like, bool\n Input array.\n w : (N,) array_like, optional\n The weights for each value in `u` and `v`. Default is None,\n which gives each value a weight of 1.0\n\n Returns\n -------\n russellrao : double\n The Russell-Rao dissimilarity between vectors `u` and `v`.\n\n Examples\n --------\n >>> from scipy.spatial import distance\n >>> distance.russellrao([1, 0, 0], [0, 1, 0])\n 1.0\n >>> distance.russellrao([1, 0, 0], [1, 1, 0])\n 0.6666666666666666\n >>> distance.russellrao([1, 0, 0], [2, 0, 0])\n 0.3333333333333333\n\n \"\"\"\n u = _validate_vector(u)\n v = _validate_vector(v)\n if u.dtype == v.dtype == bool and w is None:\n ntt = (u & v).sum()\n n = float(len(u))\n elif w is None:\n ntt = (u * v).sum()\n n = float(len(u))\n else:\n w = _validate_weights(w)\n ntt = (u * v * w).sum()\n n = w.sum()\n return float(n - ntt) / n\n\n\ndef sokalmichener(u, v, w=None):\n \"\"\"\n Compute the Sokal-Michener dissimilarity between two boolean 1-D arrays.\n\n The Sokal-Michener dissimilarity between boolean 1-D arrays `u` and `v`,\n is defined as\n\n .. math::\n\n \\\\frac{R}\n {S + R}\n\n where :math:`c_{ij}` is the number of occurrences of\n :math:`\\\\mathtt{u[k]} = i` and :math:`\\\\mathtt{v[k]} = j` for\n :math:`k < n`, :math:`R = 2 * (c_{TF} + c_{FT})` and\n :math:`S = c_{FF} + c_{TT}`.\n\n Parameters\n ----------\n u : (N,) array_like, bool\n Input array.\n v : (N,) array_like, bool\n Input array.\n w : (N,) array_like, optional\n The weights for each value in `u` and `v`. Default is None,\n which gives each value a weight of 1.0\n\n Returns\n -------\n sokalmichener : double\n The Sokal-Michener dissimilarity between vectors `u` and `v`.\n\n Examples\n --------\n >>> from scipy.spatial import distance\n >>> distance.sokalmichener([1, 0, 0], [0, 1, 0])\n 0.8\n >>> distance.sokalmichener([1, 0, 0], [1, 1, 0])\n 0.5\n >>> distance.sokalmichener([1, 0, 0], [2, 0, 0])\n -1.0\n\n \"\"\"\n u = _validate_vector(u)\n v = _validate_vector(v)\n if w is not None:\n w = _validate_weights(w)\n nff, nft, ntf, ntt = _nbool_correspond_all(u, v, w=w)\n return float(2.0 * (ntf + nft)) / float(ntt + nff + 2.0 * (ntf + nft))\n\n\ndef sokalsneath(u, v, w=None):\n \"\"\"\n Compute the Sokal-Sneath dissimilarity between two boolean 1-D arrays.\n\n The Sokal-Sneath dissimilarity between `u` and `v`,\n\n .. math::\n\n \\\\frac{R}\n {c_{TT} + R}\n\n where :math:`c_{ij}` is the number of occurrences of\n :math:`\\\\mathtt{u[k]} = i` and :math:`\\\\mathtt{v[k]} = j` for\n :math:`k < n` and :math:`R = 2(c_{TF} + c_{FT})`.\n\n Parameters\n ----------\n u : (N,) array_like, bool\n Input array.\n v : (N,) array_like, bool\n Input array.\n w : (N,) array_like, optional\n The weights for each value in `u` and `v`. Default is None,\n which gives each value a weight of 1.0\n\n Returns\n -------\n sokalsneath : double\n The Sokal-Sneath dissimilarity between vectors `u` and `v`.\n\n Examples\n --------\n >>> from scipy.spatial import distance\n >>> distance.sokalsneath([1, 0, 0], [0, 1, 0])\n 1.0\n >>> distance.sokalsneath([1, 0, 0], [1, 1, 0])\n 0.66666666666666663\n >>> distance.sokalsneath([1, 0, 0], [2, 1, 0])\n 0.0\n >>> distance.sokalsneath([1, 0, 0], [3, 1, 0])\n -2.0\n\n \"\"\"\n u = _validate_vector(u)\n v = _validate_vector(v)\n if u.dtype == v.dtype == bool and w is None:\n ntt = (u & v).sum()\n elif w is None:\n ntt = (u * v).sum()\n else:\n w = _validate_weights(w)\n ntt = (u * v * w).sum()\n (nft, ntf) = _nbool_correspond_ft_tf(u, v, w=w)\n denom = np.array(ntt + 2.0 * (ntf + nft))\n if not denom.any():\n raise ValueError('Sokal-Sneath dissimilarity is not defined for '\n 'vectors that are entirely false.')\n return float(2.0 * (ntf + nft)) / denom\n\n\n_convert_to_double = partial(_convert_to_type, out_type=np.double)\n_convert_to_bool = partial(_convert_to_type, out_type=bool)\n\n# adding python-only wrappers to _distance_wrap module\n_distance_wrap.pdist_correlation_double_wrap = _correlation_pdist_wrap\n_distance_wrap.cdist_correlation_double_wrap = _correlation_cdist_wrap\n\n\[email protected](frozen=True)\nclass CDistMetricWrapper:\n metric_name: str\n\n def __call__(self, XA, XB, *, out=None, **kwargs):\n XA = np.ascontiguousarray(XA)\n XB = np.ascontiguousarray(XB)\n mA, n = XA.shape\n mB, _ = XB.shape\n metric_name = self.metric_name\n metric_info = _METRICS[metric_name]\n XA, XB, typ, kwargs = _validate_cdist_input(\n XA, XB, mA, mB, n, metric_info, **kwargs)\n\n w = kwargs.pop('w', None)\n if w is not None:\n metric = metric_info.dist_func\n return _cdist_callable(\n XA, XB, metric=metric, out=out, w=w, **kwargs)\n\n dm = _prepare_out_argument(out, np.double, (mA, mB))\n # get cdist wrapper\n cdist_fn = getattr(_distance_wrap, f'cdist_{metric_name}_{typ}_wrap')\n cdist_fn(XA, XB, dm, **kwargs)\n return dm\n\n\[email protected](frozen=True)\nclass CDistWeightedMetricWrapper:\n metric_name: str\n weighted_metric: str\n\n def __call__(self, XA, XB, *, out=None, **kwargs):\n XA = np.ascontiguousarray(XA)\n XB = np.ascontiguousarray(XB)\n mA, n = XA.shape\n mB, _ = XB.shape\n metric_name = self.metric_name\n XA, XB, typ, kwargs = _validate_cdist_input(\n XA, XB, mA, mB, n, _METRICS[metric_name], **kwargs)\n dm = _prepare_out_argument(out, np.double, (mA, mB))\n\n w = kwargs.pop('w', None)\n if w is not None:\n metric_name = self.weighted_metric\n kwargs['w'] = w\n\n # get cdist wrapper\n cdist_fn = getattr(_distance_wrap, f'cdist_{metric_name}_{typ}_wrap')\n cdist_fn(XA, XB, dm, **kwargs)\n return dm\n\n\[email protected](frozen=True)\nclass PDistMetricWrapper:\n metric_name: str\n\n def __call__(self, X, *, out=None, **kwargs):\n X = np.ascontiguousarray(X)\n m, n = X.shape\n metric_name = self.metric_name\n metric_info = _METRICS[metric_name]\n X, typ, kwargs = _validate_pdist_input(\n X, m, n, metric_info, **kwargs)\n out_size = (m * (m - 1)) // 2\n w = kwargs.pop('w', None)\n if w is not None:\n metric = metric_info.dist_func\n return _pdist_callable(\n X, metric=metric, out=out, w=w, **kwargs)\n\n dm = _prepare_out_argument(out, np.double, (out_size,))\n # get pdist wrapper\n pdist_fn = getattr(_distance_wrap, f'pdist_{metric_name}_{typ}_wrap')\n pdist_fn(X, dm, **kwargs)\n return dm\n\n\[email protected](frozen=True)\nclass PDistWeightedMetricWrapper:\n metric_name: str\n weighted_metric: str\n\n def __call__(self, X, *, out=None, **kwargs):\n X = np.ascontiguousarray(X)\n m, n = X.shape\n metric_name = self.metric_name\n X, typ, kwargs = _validate_pdist_input(\n X, m, n, _METRICS[metric_name], **kwargs)\n out_size = (m * (m - 1)) // 2\n dm = _prepare_out_argument(out, np.double, (out_size,))\n\n w = kwargs.pop('w', None)\n if w is not None:\n metric_name = self.weighted_metric\n kwargs['w'] = w\n\n # get pdist wrapper\n pdist_fn = getattr(_distance_wrap, f'pdist_{metric_name}_{typ}_wrap')\n pdist_fn(X, dm, **kwargs)\n return dm\n\n\[email protected](frozen=True)\nclass MetricInfo:\n # Name of python distance function\n canonical_name: str\n # All aliases, including canonical_name\n aka: Set[str]\n # unvectorized distance function\n dist_func: Callable\n # Optimized cdist function\n cdist_func: Callable\n # Optimized pdist function\n pdist_func: Callable\n # function that checks kwargs and computes default values:\n # f(X, m, n, **kwargs)\n validator: Optional[Callable] = None\n # list of supported types:\n # X (pdist) and XA (cdist) are used to choose the type. if there is no\n # match the first type is used. Default double\n types: List[str] = dataclasses.field(default_factory=lambda: ['double'])\n # true if out array must be C-contiguous\n requires_contiguous_out: bool = True\n\n\n# Registry of implemented metrics:\n_METRIC_INFOS = [\n MetricInfo(\n canonical_name='braycurtis',\n aka={'braycurtis'},\n dist_func=braycurtis,\n cdist_func=_distance_pybind.cdist_braycurtis,\n pdist_func=_distance_pybind.pdist_braycurtis,\n ),\n MetricInfo(\n canonical_name='canberra',\n aka={'canberra'},\n dist_func=canberra,\n cdist_func=_distance_pybind.cdist_canberra,\n pdist_func=_distance_pybind.pdist_canberra,\n ),\n MetricInfo(\n canonical_name='chebyshev',\n aka={'chebychev', 'chebyshev', 'cheby', 'cheb', 'ch'},\n dist_func=chebyshev,\n cdist_func=_distance_pybind.cdist_chebyshev,\n pdist_func=_distance_pybind.pdist_chebyshev,\n ),\n MetricInfo(\n canonical_name='cityblock',\n aka={'cityblock', 'cblock', 'cb', 'c'},\n dist_func=cityblock,\n cdist_func=_distance_pybind.cdist_cityblock,\n pdist_func=_distance_pybind.pdist_cityblock,\n ),\n MetricInfo(\n canonical_name='correlation',\n aka={'correlation', 'co'},\n dist_func=correlation,\n cdist_func=CDistMetricWrapper('correlation'),\n pdist_func=PDistMetricWrapper('correlation'),\n ),\n MetricInfo(\n canonical_name='cosine',\n aka={'cosine', 'cos'},\n dist_func=cosine,\n cdist_func=CDistMetricWrapper('cosine'),\n pdist_func=PDistMetricWrapper('cosine'),\n ),\n MetricInfo(\n canonical_name='dice',\n aka={'dice'},\n types=['bool'],\n dist_func=dice,\n cdist_func=CDistMetricWrapper('dice'),\n pdist_func=PDistMetricWrapper('dice'),\n ),\n MetricInfo(\n canonical_name='euclidean',\n aka={'euclidean', 'euclid', 'eu', 'e'},\n dist_func=euclidean,\n cdist_func=_distance_pybind.cdist_euclidean,\n pdist_func=_distance_pybind.pdist_euclidean,\n ),\n MetricInfo(\n canonical_name='hamming',\n aka={'matching', 'hamming', 'hamm', 'ha', 'h'},\n types=['double', 'bool'],\n validator=_validate_hamming_kwargs,\n dist_func=hamming,\n cdist_func=CDistWeightedMetricWrapper('hamming', 'hamming'),\n pdist_func=PDistWeightedMetricWrapper('hamming', 'hamming'),\n ),\n MetricInfo(\n canonical_name='jaccard',\n aka={'jaccard', 'jacc', 'ja', 'j'},\n types=['double', 'bool'],\n dist_func=jaccard,\n cdist_func=CDistMetricWrapper('jaccard'),\n pdist_func=PDistMetricWrapper('jaccard'),\n ),\n MetricInfo(\n canonical_name='jensenshannon',\n aka={'jensenshannon', 'js'},\n dist_func=jensenshannon,\n cdist_func=CDistMetricWrapper('jensenshannon'),\n pdist_func=PDistMetricWrapper('jensenshannon'),\n ),\n MetricInfo(\n canonical_name='kulsinski',\n aka={'kulsinski'},\n types=['bool'],\n dist_func=kulsinski,\n cdist_func=CDistMetricWrapper('kulsinski'),\n pdist_func=PDistMetricWrapper('kulsinski'),\n ),\n MetricInfo(\n canonical_name='kulczynski1',\n aka={'kulczynski1'},\n types=['bool'],\n dist_func=kulczynski1,\n cdist_func=CDistMetricWrapper('kulczynski1'),\n pdist_func=PDistMetricWrapper('kulczynski1'),\n ),\n MetricInfo(\n canonical_name='mahalanobis',\n aka={'mahalanobis', 'mahal', 'mah'},\n validator=_validate_mahalanobis_kwargs,\n dist_func=mahalanobis,\n cdist_func=CDistMetricWrapper('mahalanobis'),\n pdist_func=PDistMetricWrapper('mahalanobis'),\n ),\n MetricInfo(\n canonical_name='minkowski',\n aka={'minkowski', 'mi', 'm', 'pnorm'},\n validator=_validate_minkowski_kwargs,\n dist_func=minkowski,\n cdist_func=_distance_pybind.cdist_minkowski,\n pdist_func=_distance_pybind.pdist_minkowski,\n ),\n MetricInfo(\n canonical_name='rogerstanimoto',\n aka={'rogerstanimoto'},\n types=['bool'],\n dist_func=rogerstanimoto,\n cdist_func=CDistMetricWrapper('rogerstanimoto'),\n pdist_func=PDistMetricWrapper('rogerstanimoto'),\n ),\n MetricInfo(\n canonical_name='russellrao',\n aka={'russellrao'},\n types=['bool'],\n dist_func=russellrao,\n cdist_func=CDistMetricWrapper('russellrao'),\n pdist_func=PDistMetricWrapper('russellrao'),\n ),\n MetricInfo(\n canonical_name='seuclidean',\n aka={'seuclidean', 'se', 's'},\n validator=_validate_seuclidean_kwargs,\n dist_func=seuclidean,\n cdist_func=CDistMetricWrapper('seuclidean'),\n pdist_func=PDistMetricWrapper('seuclidean'),\n ),\n MetricInfo(\n canonical_name='sokalmichener',\n aka={'sokalmichener'},\n types=['bool'],\n dist_func=sokalmichener,\n cdist_func=CDistMetricWrapper('sokalmichener'),\n pdist_func=PDistMetricWrapper('sokalmichener'),\n ),\n MetricInfo(\n canonical_name='sokalsneath',\n aka={'sokalsneath'},\n types=['bool'],\n dist_func=sokalsneath,\n cdist_func=CDistMetricWrapper('sokalsneath'),\n pdist_func=PDistMetricWrapper('sokalsneath'),\n ),\n MetricInfo(\n canonical_name='sqeuclidean',\n aka={'sqeuclidean', 'sqe', 'sqeuclid'},\n dist_func=sqeuclidean,\n cdist_func=_distance_pybind.cdist_sqeuclidean,\n pdist_func=_distance_pybind.pdist_sqeuclidean,\n ),\n MetricInfo(\n canonical_name='yule',\n aka={'yule'},\n types=['bool'],\n dist_func=yule,\n cdist_func=CDistMetricWrapper('yule'),\n pdist_func=PDistMetricWrapper('yule'),\n ),\n]\n\n_METRICS = {info.canonical_name: info for info in _METRIC_INFOS}\n_METRIC_ALIAS = dict((alias, info)\n for info in _METRIC_INFOS\n for alias in info.aka)\n\n_METRICS_NAMES = list(_METRICS.keys())\n\n_TEST_METRICS = {'test_' + info.canonical_name: info for info in _METRIC_INFOS}\n\n\ndef pdist(X, metric='euclidean', *, out=None, **kwargs):\n \"\"\"\n Pairwise distances between observations in n-dimensional space.\n\n See Notes for common calling conventions.\n\n Parameters\n ----------\n X : array_like\n An m by n array of m original observations in an\n n-dimensional space.\n metric : str or function, optional\n The distance metric to use. The distance function can\n be 'braycurtis', 'canberra', 'chebyshev', 'cityblock',\n 'correlation', 'cosine', 'dice', 'euclidean', 'hamming',\n 'jaccard', 'jensenshannon', 'kulsinski', 'kulczynski1',\n 'mahalanobis', 'matching', 'minkowski', 'rogerstanimoto',\n 'russellrao', 'seuclidean', 'sokalmichener', 'sokalsneath',\n 'sqeuclidean', 'yule'.\n **kwargs : dict, optional\n Extra arguments to `metric`: refer to each metric documentation for a\n list of all possible arguments.\n\n Some possible arguments:\n\n p : scalar\n The p-norm to apply for Minkowski, weighted and unweighted.\n Default: 2.\n\n w : ndarray\n The weight vector for metrics that support weights (e.g., Minkowski).\n\n V : ndarray\n The variance vector for standardized Euclidean.\n Default: var(X, axis=0, ddof=1)\n\n VI : ndarray\n The inverse of the covariance matrix for Mahalanobis.\n Default: inv(cov(X.T)).T\n\n out : ndarray.\n The output array\n If not None, condensed distance matrix Y is stored in this array.\n\n Returns\n -------\n Y : ndarray\n Returns a condensed distance matrix Y. For each :math:`i` and :math:`j`\n (where :math:`i<j<m`),where m is the number of original observations.\n The metric ``dist(u=X[i], v=X[j])`` is computed and stored in entry ``m\n * i + j - ((i + 2) * (i + 1)) // 2``.\n\n See Also\n --------\n squareform : converts between condensed distance matrices and\n square distance matrices.\n\n Notes\n -----\n See ``squareform`` for information on how to calculate the index of\n this entry or to convert the condensed distance matrix to a\n redundant square matrix.\n\n The following are common calling conventions.\n\n 1. ``Y = pdist(X, 'euclidean')``\n\n Computes the distance between m points using Euclidean distance\n (2-norm) as the distance metric between the points. The points\n are arranged as m n-dimensional row vectors in the matrix X.\n\n 2. ``Y = pdist(X, 'minkowski', p=2.)``\n\n Computes the distances using the Minkowski distance\n :math:`||u-v||_p` (p-norm) where :math:`p \\\\geq 1`.\n\n 3. ``Y = pdist(X, 'cityblock')``\n\n Computes the city block or Manhattan distance between the\n points.\n\n 4. ``Y = pdist(X, 'seuclidean', V=None)``\n\n Computes the standardized Euclidean distance. The standardized\n Euclidean distance between two n-vectors ``u`` and ``v`` is\n\n .. math::\n\n \\\\sqrt{\\\\sum {(u_i-v_i)^2 / V[x_i]}}\n\n\n V is the variance vector; V[i] is the variance computed over all\n the i'th components of the points. If not passed, it is\n automatically computed.\n\n 5. ``Y = pdist(X, 'sqeuclidean')``\n\n Computes the squared Euclidean distance :math:`||u-v||_2^2` between\n the vectors.\n\n 6. ``Y = pdist(X, 'cosine')``\n\n Computes the cosine distance between vectors u and v,\n\n .. math::\n\n 1 - \\\\frac{u \\\\cdot v}\n {{||u||}_2 {||v||}_2}\n\n where :math:`||*||_2` is the 2-norm of its argument ``*``, and\n :math:`u \\\\cdot v` is the dot product of ``u`` and ``v``.\n\n 7. ``Y = pdist(X, 'correlation')``\n\n Computes the correlation distance between vectors u and v. This is\n\n .. math::\n\n 1 - \\\\frac{(u - \\\\bar{u}) \\\\cdot (v - \\\\bar{v})}\n {{||(u - \\\\bar{u})||}_2 {||(v - \\\\bar{v})||}_2}\n\n where :math:`\\\\bar{v}` is the mean of the elements of vector v,\n and :math:`x \\\\cdot y` is the dot product of :math:`x` and :math:`y`.\n\n 8. ``Y = pdist(X, 'hamming')``\n\n Computes the normalized Hamming distance, or the proportion of\n those vector elements between two n-vectors ``u`` and ``v``\n which disagree. To save memory, the matrix ``X`` can be of type\n boolean.\n\n 9. ``Y = pdist(X, 'jaccard')``\n\n Computes the Jaccard distance between the points. Given two\n vectors, ``u`` and ``v``, the Jaccard distance is the\n proportion of those elements ``u[i]`` and ``v[i]`` that\n disagree.\n\n 10. ``Y = pdist(X, 'jensenshannon')``\n\n Computes the Jensen-Shannon distance between two probability arrays.\n Given two probability vectors, :math:`p` and :math:`q`, the\n Jensen-Shannon distance is\n\n .. math::\n\n \\\\sqrt{\\\\frac{D(p \\\\parallel m) + D(q \\\\parallel m)}{2}}\n\n where :math:`m` is the pointwise mean of :math:`p` and :math:`q`\n and :math:`D` is the Kullback-Leibler divergence.\n\n 11. ``Y = pdist(X, 'chebyshev')``\n\n Computes the Chebyshev distance between the points. The\n Chebyshev distance between two n-vectors ``u`` and ``v`` is the\n maximum norm-1 distance between their respective elements. More\n precisely, the distance is given by\n\n .. math::\n\n d(u,v) = \\\\max_i {|u_i-v_i|}\n\n 12. ``Y = pdist(X, 'canberra')``\n\n Computes the Canberra distance between the points. The\n Canberra distance between two points ``u`` and ``v`` is\n\n .. math::\n\n d(u,v) = \\\\sum_i \\\\frac{|u_i-v_i|}\n {|u_i|+|v_i|}\n\n\n 13. ``Y = pdist(X, 'braycurtis')``\n\n Computes the Bray-Curtis distance between the points. The\n Bray-Curtis distance between two points ``u`` and ``v`` is\n\n\n .. math::\n\n d(u,v) = \\\\frac{\\\\sum_i {|u_i-v_i|}}\n {\\\\sum_i {|u_i+v_i|}}\n\n 14. ``Y = pdist(X, 'mahalanobis', VI=None)``\n\n Computes the Mahalanobis distance between the points. The\n Mahalanobis distance between two points ``u`` and ``v`` is\n :math:`\\\\sqrt{(u-v)(1/V)(u-v)^T}` where :math:`(1/V)` (the ``VI``\n variable) is the inverse covariance. If ``VI`` is not None,\n ``VI`` will be used as the inverse covariance matrix.\n\n 15. ``Y = pdist(X, 'yule')``\n\n Computes the Yule distance between each pair of boolean\n vectors. (see yule function documentation)\n\n 16. ``Y = pdist(X, 'matching')``\n\n Synonym for 'hamming'.\n\n 17. ``Y = pdist(X, 'dice')``\n\n Computes the Dice distance between each pair of boolean\n vectors. (see dice function documentation)\n\n 18. ``Y = pdist(X, 'kulsinski')``\n\n Computes the Kulsinski distance between each pair of\n boolean vectors. (see kulsinski function documentation)\n\n 19. ``Y = pdist(X, 'rogerstanimoto')``\n\n Computes the Rogers-Tanimoto distance between each pair of\n boolean vectors. (see rogerstanimoto function documentation)\n\n 20. ``Y = pdist(X, 'russellrao')``\n\n Computes the Russell-Rao distance between each pair of\n boolean vectors. (see russellrao function documentation)\n\n 21. ``Y = pdist(X, 'sokalmichener')``\n\n Computes the Sokal-Michener distance between each pair of\n boolean vectors. (see sokalmichener function documentation)\n\n 22. ``Y = pdist(X, 'sokalsneath')``\n\n Computes the Sokal-Sneath distance between each pair of\n boolean vectors. (see sokalsneath function documentation)\n\n 23. ``Y = pdist(X, 'kulczynski1')``\n\n Computes the Kulczynski 1 distance between each pair of\n boolean vectors. (see kulczynski1 function documentation)\n\n 24. ``Y = pdist(X, f)``\n\n Computes the distance between all pairs of vectors in X\n using the user supplied 2-arity function f. For example,\n Euclidean distance between the vectors could be computed\n as follows::\n\n dm = pdist(X, lambda u, v: np.sqrt(((u-v)**2).sum()))\n\n Note that you should avoid passing a reference to one of\n the distance functions defined in this library. For example,::\n\n dm = pdist(X, sokalsneath)\n\n would calculate the pair-wise distances between the vectors in\n X using the Python function sokalsneath. This would result in\n sokalsneath being called :math:`{n \\\\choose 2}` times, which\n is inefficient. Instead, the optimized C version is more\n efficient, and we call it using the following syntax.::\n\n dm = pdist(X, 'sokalsneath')\n\n \"\"\"\n # You can also call this as:\n # Y = pdist(X, 'test_abc')\n # where 'abc' is the metric being tested. This computes the distance\n # between all pairs of vectors in X using the distance metric 'abc' but\n # with a more succinct, verifiable, but less efficient implementation.\n\n X = _asarray_validated(X, sparse_ok=False, objects_ok=True, mask_ok=True,\n check_finite=False)\n\n s = X.shape\n if len(s) != 2:\n raise ValueError('A 2-dimensional array must be passed.')\n\n m, n = s\n\n if callable(metric):\n mstr = getattr(metric, '__name__', 'UnknownCustomMetric')\n metric_info = _METRIC_ALIAS.get(mstr, None)\n\n if metric_info is not None:\n X, typ, kwargs = _validate_pdist_input(\n X, m, n, metric_info, **kwargs)\n\n return _pdist_callable(X, metric=metric, out=out, **kwargs)\n elif isinstance(metric, str):\n mstr = metric.lower()\n metric_info = _METRIC_ALIAS.get(mstr, None)\n\n if metric_info is not None:\n pdist_fn = metric_info.pdist_func\n return pdist_fn(X, out=out, **kwargs)\n elif mstr.startswith(\"test_\"):\n metric_info = _TEST_METRICS.get(mstr, None)\n if metric_info is None:\n raise ValueError(f'Unknown \"Test\" Distance Metric: {mstr[5:]}')\n X, typ, kwargs = _validate_pdist_input(\n X, m, n, metric_info, **kwargs)\n return _pdist_callable(\n X, metric=metric_info.dist_func, out=out, **kwargs)\n else:\n raise ValueError('Unknown Distance Metric: %s' % mstr)\n else:\n raise TypeError('2nd argument metric must be a string identifier '\n 'or a function.')\n\n\ndef squareform(X, force=\"no\", checks=True):\n \"\"\"\n Convert a vector-form distance vector to a square-form distance\n matrix, and vice-versa.\n\n Parameters\n ----------\n X : array_like\n Either a condensed or redundant distance matrix.\n force : str, optional\n As with MATLAB(TM), if force is equal to ``'tovector'`` or\n ``'tomatrix'``, the input will be treated as a distance matrix or\n distance vector respectively.\n checks : bool, optional\n If set to False, no checks will be made for matrix\n symmetry nor zero diagonals. This is useful if it is known that\n ``X - X.T1`` is small and ``diag(X)`` is close to zero.\n These values are ignored any way so they do not disrupt the\n squareform transformation.\n\n Returns\n -------\n Y : ndarray\n If a condensed distance matrix is passed, a redundant one is\n returned, or if a redundant one is passed, a condensed distance\n matrix is returned.\n\n Notes\n -----\n 1. ``v = squareform(X)``\n\n Given a square n-by-n symmetric distance matrix ``X``,\n ``v = squareform(X)`` returns a ``n * (n-1) / 2``\n (i.e. binomial coefficient n choose 2) sized vector `v`\n where :math:`v[{n \\\\choose 2} - {n-i \\\\choose 2} + (j-i-1)]`\n is the distance between distinct points ``i`` and ``j``.\n If ``X`` is non-square or asymmetric, an error is raised.\n\n 2. ``X = squareform(v)``\n\n Given a ``n * (n-1) / 2`` sized vector ``v``\n for some integer ``n >= 1`` encoding distances as described,\n ``X = squareform(v)`` returns a n-by-n distance matrix ``X``.\n The ``X[i, j]`` and ``X[j, i]`` values are set to\n :math:`v[{n \\\\choose 2} - {n-i \\\\choose 2} + (j-i-1)]`\n and all diagonal elements are zero.\n\n In SciPy 0.19.0, ``squareform`` stopped casting all input types to\n float64, and started returning arrays of the same dtype as the input.\n\n \"\"\"\n\n X = np.ascontiguousarray(X)\n\n s = X.shape\n\n if force.lower() == 'tomatrix':\n if len(s) != 1:\n raise ValueError(\"Forcing 'tomatrix' but input X is not a \"\n \"distance vector.\")\n elif force.lower() == 'tovector':\n if len(s) != 2:\n raise ValueError(\"Forcing 'tovector' but input X is not a \"\n \"distance matrix.\")\n\n # X = squareform(v)\n if len(s) == 1:\n if s[0] == 0:\n return np.zeros((1, 1), dtype=X.dtype)\n\n # Grab the closest value to the square root of the number\n # of elements times 2 to see if the number of elements\n # is indeed a binomial coefficient.\n d = int(np.ceil(np.sqrt(s[0] * 2)))\n\n # Check that v is of valid dimensions.\n if d * (d - 1) != s[0] * 2:\n raise ValueError('Incompatible vector size. It must be a binomial '\n 'coefficient n choose 2 for some integer n >= 2.')\n\n # Allocate memory for the distance matrix.\n M = np.zeros((d, d), dtype=X.dtype)\n\n # Since the C code does not support striding using strides.\n # The dimensions are used instead.\n X = _copy_array_if_base_present(X)\n\n # Fill in the values of the distance matrix.\n _distance_wrap.to_squareform_from_vector_wrap(M, X)\n\n # Return the distance matrix.\n return M\n elif len(s) == 2:\n if s[0] != s[1]:\n raise ValueError('The matrix argument must be square.')\n if checks:\n is_valid_dm(X, throw=True, name='X')\n\n # One-side of the dimensions is set here.\n d = s[0]\n\n if d <= 1:\n return np.array([], dtype=X.dtype)\n\n # Create a vector.\n v = np.zeros((d * (d - 1)) // 2, dtype=X.dtype)\n\n # Since the C code does not support striding using strides.\n # The dimensions are used instead.\n X = _copy_array_if_base_present(X)\n\n # Convert the vector to squareform.\n _distance_wrap.to_vector_from_squareform_wrap(X, v)\n return v\n else:\n raise ValueError(('The first argument must be one or two dimensional '\n 'array. A %d-dimensional array is not '\n 'permitted') % len(s))\n\n\ndef is_valid_dm(D, tol=0.0, throw=False, name=\"D\", warning=False):\n \"\"\"\n Return True if input array is a valid distance matrix.\n\n Distance matrices must be 2-dimensional numpy arrays.\n They must have a zero-diagonal, and they must be symmetric.\n\n Parameters\n ----------\n D : array_like\n The candidate object to test for validity.\n tol : float, optional\n The distance matrix should be symmetric. `tol` is the maximum\n difference between entries ``ij`` and ``ji`` for the distance\n metric to be considered symmetric.\n throw : bool, optional\n An exception is thrown if the distance matrix passed is not valid.\n name : str, optional\n The name of the variable to checked. This is useful if\n throw is set to True so the offending variable can be identified\n in the exception message when an exception is thrown.\n warning : bool, optional\n Instead of throwing an exception, a warning message is\n raised.\n\n Returns\n -------\n valid : bool\n True if the variable `D` passed is a valid distance matrix.\n\n Notes\n -----\n Small numerical differences in `D` and `D.T` and non-zeroness of\n the diagonal are ignored if they are within the tolerance specified\n by `tol`.\n\n \"\"\"\n D = np.asarray(D, order='c')\n valid = True\n try:\n s = D.shape\n if len(D.shape) != 2:\n if name:\n raise ValueError(('Distance matrix \\'%s\\' must have shape=2 '\n '(i.e. be two-dimensional).') % name)\n else:\n raise ValueError('Distance matrix must have shape=2 (i.e. '\n 'be two-dimensional).')\n if tol == 0.0:\n if not (D == D.T).all():\n if name:\n raise ValueError(('Distance matrix \\'%s\\' must be '\n 'symmetric.') % name)\n else:\n raise ValueError('Distance matrix must be symmetric.')\n if not (D[range(0, s[0]), range(0, s[0])] == 0).all():\n if name:\n raise ValueError(('Distance matrix \\'%s\\' diagonal must '\n 'be zero.') % name)\n else:\n raise ValueError('Distance matrix diagonal must be zero.')\n else:\n if not (D - D.T <= tol).all():\n if name:\n raise ValueError(('Distance matrix \\'%s\\' must be '\n 'symmetric within tolerance %5.5f.')\n % (name, tol))\n else:\n raise ValueError('Distance matrix must be symmetric within'\n ' tolerance %5.5f.' % tol)\n if not (D[range(0, s[0]), range(0, s[0])] <= tol).all():\n if name:\n raise ValueError(('Distance matrix \\'%s\\' diagonal must be'\n ' close to zero within tolerance %5.5f.')\n % (name, tol))\n else:\n raise ValueError(('Distance matrix \\'%s\\' diagonal must be'\n ' close to zero within tolerance %5.5f.')\n % tol)\n except Exception as e:\n if throw:\n raise\n if warning:\n warnings.warn(str(e))\n valid = False\n return valid\n\n\ndef is_valid_y(y, warning=False, throw=False, name=None):\n \"\"\"\n Return True if the input array is a valid condensed distance matrix.\n\n Condensed distance matrices must be 1-dimensional numpy arrays.\n Their length must be a binomial coefficient :math:`{n \\\\choose 2}`\n for some positive integer n.\n\n Parameters\n ----------\n y : array_like\n The condensed distance matrix.\n warning : bool, optional\n Invokes a warning if the variable passed is not a valid\n condensed distance matrix. The warning message explains why\n the distance matrix is not valid. `name` is used when\n referencing the offending variable.\n throw : bool, optional\n Throws an exception if the variable passed is not a valid\n condensed distance matrix.\n name : bool, optional\n Used when referencing the offending variable in the\n warning or exception message.\n\n \"\"\"\n y = np.asarray(y, order='c')\n valid = True\n try:\n if len(y.shape) != 1:\n if name:\n raise ValueError(('Condensed distance matrix \\'%s\\' must '\n 'have shape=1 (i.e. be one-dimensional).')\n % name)\n else:\n raise ValueError('Condensed distance matrix must have shape=1 '\n '(i.e. be one-dimensional).')\n n = y.shape[0]\n d = int(np.ceil(np.sqrt(n * 2)))\n if (d * (d - 1) / 2) != n:\n if name:\n raise ValueError(('Length n of condensed distance matrix '\n '\\'%s\\' must be a binomial coefficient, i.e.'\n 'there must be a k such that '\n '(k \\\\choose 2)=n)!') % name)\n else:\n raise ValueError('Length n of condensed distance matrix must '\n 'be a binomial coefficient, i.e. there must '\n 'be a k such that (k \\\\choose 2)=n)!')\n except Exception as e:\n if throw:\n raise\n if warning:\n warnings.warn(str(e))\n valid = False\n return valid\n\n\ndef num_obs_dm(d):\n \"\"\"\n Return the number of original observations that correspond to a\n square, redundant distance matrix.\n\n Parameters\n ----------\n d : array_like\n The target distance matrix.\n\n Returns\n -------\n num_obs_dm : int\n The number of observations in the redundant distance matrix.\n\n \"\"\"\n d = np.asarray(d, order='c')\n is_valid_dm(d, tol=np.inf, throw=True, name='d')\n return d.shape[0]\n\n\ndef num_obs_y(Y):\n \"\"\"\n Return the number of original observations that correspond to a\n condensed distance matrix.\n\n Parameters\n ----------\n Y : array_like\n Condensed distance matrix.\n\n Returns\n -------\n n : int\n The number of observations in the condensed distance matrix `Y`.\n\n \"\"\"\n Y = np.asarray(Y, order='c')\n is_valid_y(Y, throw=True, name='Y')\n k = Y.shape[0]\n if k == 0:\n raise ValueError(\"The number of observations cannot be determined on \"\n \"an empty distance matrix.\")\n d = int(np.ceil(np.sqrt(k * 2)))\n if (d * (d - 1) / 2) != k:\n raise ValueError(\"Invalid condensed distance matrix passed. Must be \"\n \"some k where k=(n choose 2) for some n >= 2.\")\n return d\n\n\ndef _prepare_out_argument(out, dtype, expected_shape):\n if out is None:\n return np.empty(expected_shape, dtype=dtype)\n\n if out.shape != expected_shape:\n raise ValueError(\"Output array has incorrect shape.\")\n if not out.flags.c_contiguous:\n raise ValueError(\"Output array must be C-contiguous.\")\n if out.dtype != np.double:\n raise ValueError(\"Output array must be double type.\")\n return out\n\n\ndef _pdist_callable(X, *, out, metric, **kwargs):\n n = X.shape[0]\n out_size = (n * (n - 1)) // 2\n dm = _prepare_out_argument(out, np.double, (out_size,))\n k = 0\n for i in range(X.shape[0] - 1):\n for j in range(i + 1, X.shape[0]):\n dm[k] = metric(X[i], X[j], **kwargs)\n k += 1\n return dm\n\n\ndef _cdist_callable(XA, XB, *, out, metric, **kwargs):\n mA = XA.shape[0]\n mB = XB.shape[0]\n dm = _prepare_out_argument(out, np.double, (mA, mB))\n for i in range(mA):\n for j in range(mB):\n dm[i, j] = metric(XA[i], XB[j], **kwargs)\n return dm\n\n\ndef cdist(XA, XB, metric='euclidean', *, out=None, **kwargs):\n \"\"\"\n Compute distance between each pair of the two collections of inputs.\n\n See Notes for common calling conventions.\n\n Parameters\n ----------\n XA : array_like\n An :math:`m_A` by :math:`n` array of :math:`m_A`\n original observations in an :math:`n`-dimensional space.\n Inputs are converted to float type.\n XB : array_like\n An :math:`m_B` by :math:`n` array of :math:`m_B`\n original observations in an :math:`n`-dimensional space.\n Inputs are converted to float type.\n metric : str or callable, optional\n The distance metric to use. If a string, the distance function can be\n 'braycurtis', 'canberra', 'chebyshev', 'cityblock', 'correlation',\n 'cosine', 'dice', 'euclidean', 'hamming', 'jaccard', 'jensenshannon',\n 'kulsinski', 'kulczynski1', 'mahalanobis', 'matching', 'minkowski',\n 'rogerstanimoto', 'russellrao', 'seuclidean', 'sokalmichener',\n 'sokalsneath', 'sqeuclidean', 'yule'.\n **kwargs : dict, optional\n Extra arguments to `metric`: refer to each metric documentation for a\n list of all possible arguments.\n\n Some possible arguments:\n\n p : scalar\n The p-norm to apply for Minkowski, weighted and unweighted.\n Default: 2.\n\n w : array_like\n The weight vector for metrics that support weights (e.g., Minkowski).\n\n V : array_like\n The variance vector for standardized Euclidean.\n Default: var(vstack([XA, XB]), axis=0, ddof=1)\n\n VI : array_like\n The inverse of the covariance matrix for Mahalanobis.\n Default: inv(cov(vstack([XA, XB].T))).T\n\n out : ndarray\n The output array\n If not None, the distance matrix Y is stored in this array.\n\n Returns\n -------\n Y : ndarray\n A :math:`m_A` by :math:`m_B` distance matrix is returned.\n For each :math:`i` and :math:`j`, the metric\n ``dist(u=XA[i], v=XB[j])`` is computed and stored in the\n :math:`ij` th entry.\n\n Raises\n ------\n ValueError\n An exception is thrown if `XA` and `XB` do not have\n the same number of columns.\n\n Notes\n -----\n The following are common calling conventions:\n\n 1. ``Y = cdist(XA, XB, 'euclidean')``\n\n Computes the distance between :math:`m` points using\n Euclidean distance (2-norm) as the distance metric between the\n points. The points are arranged as :math:`m`\n :math:`n`-dimensional row vectors in the matrix X.\n\n 2. ``Y = cdist(XA, XB, 'minkowski', p=2.)``\n\n Computes the distances using the Minkowski distance\n :math:`||u-v||_p` (:math:`p`-norm) where :math:`p \\\\geq 1`.\n\n 3. ``Y = cdist(XA, XB, 'cityblock')``\n\n Computes the city block or Manhattan distance between the\n points.\n\n 4. ``Y = cdist(XA, XB, 'seuclidean', V=None)``\n\n Computes the standardized Euclidean distance. The standardized\n Euclidean distance between two n-vectors ``u`` and ``v`` is\n\n .. math::\n\n \\\\sqrt{\\\\sum {(u_i-v_i)^2 / V[x_i]}}.\n\n V is the variance vector; V[i] is the variance computed over all\n the i'th components of the points. If not passed, it is\n automatically computed.\n\n 5. ``Y = cdist(XA, XB, 'sqeuclidean')``\n\n Computes the squared Euclidean distance :math:`||u-v||_2^2` between\n the vectors.\n\n 6. ``Y = cdist(XA, XB, 'cosine')``\n\n Computes the cosine distance between vectors u and v,\n\n .. math::\n\n 1 - \\\\frac{u \\\\cdot v}\n {{||u||}_2 {||v||}_2}\n\n where :math:`||*||_2` is the 2-norm of its argument ``*``, and\n :math:`u \\\\cdot v` is the dot product of :math:`u` and :math:`v`.\n\n 7. ``Y = cdist(XA, XB, 'correlation')``\n\n Computes the correlation distance between vectors u and v. This is\n\n .. math::\n\n 1 - \\\\frac{(u - \\\\bar{u}) \\\\cdot (v - \\\\bar{v})}\n {{||(u - \\\\bar{u})||}_2 {||(v - \\\\bar{v})||}_2}\n\n where :math:`\\\\bar{v}` is the mean of the elements of vector v,\n and :math:`x \\\\cdot y` is the dot product of :math:`x` and :math:`y`.\n\n\n 8. ``Y = cdist(XA, XB, 'hamming')``\n\n Computes the normalized Hamming distance, or the proportion of\n those vector elements between two n-vectors ``u`` and ``v``\n which disagree. To save memory, the matrix ``X`` can be of type\n boolean.\n\n 9. ``Y = cdist(XA, XB, 'jaccard')``\n\n Computes the Jaccard distance between the points. Given two\n vectors, ``u`` and ``v``, the Jaccard distance is the\n proportion of those elements ``u[i]`` and ``v[i]`` that\n disagree where at least one of them is non-zero.\n\n 10. ``Y = cdist(XA, XB, 'jensenshannon')``\n\n Computes the Jensen-Shannon distance between two probability arrays.\n Given two probability vectors, :math:`p` and :math:`q`, the\n Jensen-Shannon distance is\n\n .. math::\n\n \\\\sqrt{\\\\frac{D(p \\\\parallel m) + D(q \\\\parallel m)}{2}}\n\n where :math:`m` is the pointwise mean of :math:`p` and :math:`q`\n and :math:`D` is the Kullback-Leibler divergence.\n\n 11. ``Y = cdist(XA, XB, 'chebyshev')``\n\n Computes the Chebyshev distance between the points. The\n Chebyshev distance between two n-vectors ``u`` and ``v`` is the\n maximum norm-1 distance between their respective elements. More\n precisely, the distance is given by\n\n .. math::\n\n d(u,v) = \\\\max_i {|u_i-v_i|}.\n\n 12. ``Y = cdist(XA, XB, 'canberra')``\n\n Computes the Canberra distance between the points. The\n Canberra distance between two points ``u`` and ``v`` is\n\n .. math::\n\n d(u,v) = \\\\sum_i \\\\frac{|u_i-v_i|}\n {|u_i|+|v_i|}.\n\n 13. ``Y = cdist(XA, XB, 'braycurtis')``\n\n Computes the Bray-Curtis distance between the points. The\n Bray-Curtis distance between two points ``u`` and ``v`` is\n\n\n .. math::\n\n d(u,v) = \\\\frac{\\\\sum_i (|u_i-v_i|)}\n {\\\\sum_i (|u_i+v_i|)}\n\n 14. ``Y = cdist(XA, XB, 'mahalanobis', VI=None)``\n\n Computes the Mahalanobis distance between the points. The\n Mahalanobis distance between two points ``u`` and ``v`` is\n :math:`\\\\sqrt{(u-v)(1/V)(u-v)^T}` where :math:`(1/V)` (the ``VI``\n variable) is the inverse covariance. If ``VI`` is not None,\n ``VI`` will be used as the inverse covariance matrix.\n\n 15. ``Y = cdist(XA, XB, 'yule')``\n\n Computes the Yule distance between the boolean\n vectors. (see `yule` function documentation)\n\n 16. ``Y = cdist(XA, XB, 'matching')``\n\n Synonym for 'hamming'.\n\n 17. ``Y = cdist(XA, XB, 'dice')``\n\n Computes the Dice distance between the boolean vectors. (see\n `dice` function documentation)\n\n 18. ``Y = cdist(XA, XB, 'kulsinski')``\n\n Computes the Kulsinski distance between the boolean\n vectors. (see `kulsinski` function documentation)\n\n 19. ``Y = cdist(XA, XB, 'rogerstanimoto')``\n\n Computes the Rogers-Tanimoto distance between the boolean\n vectors. (see `rogerstanimoto` function documentation)\n\n 20. ``Y = cdist(XA, XB, 'russellrao')``\n\n Computes the Russell-Rao distance between the boolean\n vectors. (see `russellrao` function documentation)\n\n 21. ``Y = cdist(XA, XB, 'sokalmichener')``\n\n Computes the Sokal-Michener distance between the boolean\n vectors. (see `sokalmichener` function documentation)\n\n 22. ``Y = cdist(XA, XB, 'sokalsneath')``\n\n Computes the Sokal-Sneath distance between the vectors. (see\n `sokalsneath` function documentation)\n\n 23. ``Y = cdist(XA, XB, f)``\n\n Computes the distance between all pairs of vectors in X\n using the user supplied 2-arity function f. For example,\n Euclidean distance between the vectors could be computed\n as follows::\n\n dm = cdist(XA, XB, lambda u, v: np.sqrt(((u-v)**2).sum()))\n\n Note that you should avoid passing a reference to one of\n the distance functions defined in this library. For example,::\n\n dm = cdist(XA, XB, sokalsneath)\n\n would calculate the pair-wise distances between the vectors in\n X using the Python function `sokalsneath`. This would result in\n sokalsneath being called :math:`{n \\\\choose 2}` times, which\n is inefficient. Instead, the optimized C version is more\n efficient, and we call it using the following syntax::\n\n dm = cdist(XA, XB, 'sokalsneath')\n\n Examples\n --------\n Find the Euclidean distances between four 2-D coordinates:\n\n >>> from scipy.spatial import distance\n >>> coords = [(35.0456, -85.2672),\n ... (35.1174, -89.9711),\n ... (35.9728, -83.9422),\n ... (36.1667, -86.7833)]\n >>> distance.cdist(coords, coords, 'euclidean')\n array([[ 0. , 4.7044, 1.6172, 1.8856],\n [ 4.7044, 0. , 6.0893, 3.3561],\n [ 1.6172, 6.0893, 0. , 2.8477],\n [ 1.8856, 3.3561, 2.8477, 0. ]])\n\n\n Find the Manhattan distance from a 3-D point to the corners of the unit\n cube:\n\n >>> a = np.array([[0, 0, 0],\n ... [0, 0, 1],\n ... [0, 1, 0],\n ... [0, 1, 1],\n ... [1, 0, 0],\n ... [1, 0, 1],\n ... [1, 1, 0],\n ... [1, 1, 1]])\n >>> b = np.array([[ 0.1, 0.2, 0.4]])\n >>> distance.cdist(a, b, 'cityblock')\n array([[ 0.7],\n [ 0.9],\n [ 1.3],\n [ 1.5],\n [ 1.5],\n [ 1.7],\n [ 2.1],\n [ 2.3]])\n\n \"\"\"\n # You can also call this as:\n # Y = cdist(XA, XB, 'test_abc')\n # where 'abc' is the metric being tested. This computes the distance\n # between all pairs of vectors in XA and XB using the distance metric 'abc'\n # but with a more succinct, verifiable, but less efficient implementation.\n\n XA = np.asarray(XA)\n XB = np.asarray(XB)\n\n s = XA.shape\n sB = XB.shape\n\n if len(s) != 2:\n raise ValueError('XA must be a 2-dimensional array.')\n if len(sB) != 2:\n raise ValueError('XB must be a 2-dimensional array.')\n if s[1] != sB[1]:\n raise ValueError('XA and XB must have the same number of columns '\n '(i.e. feature dimension.)')\n\n mA = s[0]\n mB = sB[0]\n n = s[1]\n\n if callable(metric):\n mstr = getattr(metric, '__name__', 'Unknown')\n metric_info = _METRIC_ALIAS.get(mstr, None)\n if metric_info is not None:\n XA, XB, typ, kwargs = _validate_cdist_input(\n XA, XB, mA, mB, n, metric_info, **kwargs)\n return _cdist_callable(XA, XB, metric=metric, out=out, **kwargs)\n elif isinstance(metric, str):\n mstr = metric.lower()\n metric_info = _METRIC_ALIAS.get(mstr, None)\n if metric_info is not None:\n cdist_fn = metric_info.cdist_func\n return cdist_fn(XA, XB, out=out, **kwargs)\n elif mstr.startswith(\"test_\"):\n metric_info = _TEST_METRICS.get(mstr, None)\n if metric_info is None:\n raise ValueError(f'Unknown \"Test\" Distance Metric: {mstr[5:]}')\n XA, XB, typ, kwargs = _validate_cdist_input(\n XA, XB, mA, mB, n, metric_info, **kwargs)\n return _cdist_callable(\n XA, XB, metric=metric_info.dist_func, out=out, **kwargs)\n else:\n raise ValueError('Unknown Distance Metric: %s' % mstr)\n else:\n raise TypeError('2nd argument metric must be a string identifier '\n 'or a function.')\n"
] |
[
[
"numpy.dot",
"numpy.issubdtype",
"numpy.empty",
"numpy.log",
"numpy.sqrt",
"numpy.linalg.inv",
"numpy.vstack",
"numpy.atleast_2d",
"numpy.square",
"numpy.array",
"numpy.zeros",
"numpy.nansum",
"numpy.bitwise_and",
"numpy.power",
"numpy.bitwise_or",
"numpy.find_common_type",
"numpy.asarray",
"numpy.errstate",
"numpy.ascontiguousarray",
"numpy.sum",
"scipy._lib._util._asarray_validated",
"numpy.ones",
"numpy.any",
"numpy.abs",
"numpy.average",
"numpy.deprecate"
]
] |
qooglewb/QBuilder
|
[
"6156b97c636c5b568c5a57c23b77d9ae28421bba"
] |
[
"ashpy/models/fc/encoders.py"
] |
[
"# Copyright 2019 Zuru Tech HK Limited. All Rights Reserved.\n#\n# Licensed under the Apache License, Version 2.0 (the \"License\");\n# you may not use this file except in compliance with the License.\n# You may obtain a copy of the License at\n#\n# http://www.apache.org/licenses/LICENSE-2.0\n#\n# Unless required by applicable law or agreed to in writing, software\n# distributed under the License is distributed on an \"AS IS\" BASIS,\n# WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.\n# See the License for the specific language governing permissions and\n# limitations under the License.\n\n\"\"\"Collection of Encoders (i.e., GANs' Discriminators) models.\"\"\"\nfrom tensorflow import keras # pylint: disable=no-name-in-module\n\nfrom ashpy.models.fc.interfaces import FCInterface\n\n__ALL__ = [\"BaseEncoder\"]\n\n\nclass BaseEncoder(FCInterface):\n \"\"\"\n Primitive Model for all fully connected encoder based architecture.\n\n Examples:\n\n .. testcode::\n\n encoder = BaseEncoder(\n hidden_units=[256,128,64],\n output_shape=10)\n print(encoder(tf.zeros((1,55))).shape)\n\n .. testoutput::\n\n (1, 10)\n\n \"\"\"\n\n def __init__(self, hidden_units, output_shape):\n \"\"\"\n Instantiate the :py:class:`BaseDecoder`.\n\n Args:\n hidden_units (:obj:`tuple` of :obj:`int`): Number of units per hidden layer.\n output_shape (int): Amount of units of the last :py:obj:`tf.keras.layers.Dense`.\n\n Returns:\n :py:obj:`None`\n\n \"\"\"\n super().__init__()\n\n # Assembling Model\n for units in hidden_units:\n self.model_layers.extend(\n [\n keras.layers.Dense(units),\n keras.layers.LeakyReLU(),\n keras.layers.Dropout(0.3),\n ]\n )\n self.model_layers.append(keras.layers.Dense(output_shape))\n"
] |
[
[
"tensorflow.keras.layers.Dropout",
"tensorflow.keras.layers.LeakyReLU",
"tensorflow.keras.layers.Dense"
]
] |
ElementalCognition/transformers
|
[
"29ad1f790c123a3d68c2fb124e512cc4a9f6cf49"
] |
[
"src/transformers/generation_utils.py"
] |
[
"# coding=utf-8\n# Copyright 2020 The Google AI Language Team Authors, Facebook AI Research authors and The HuggingFace Inc. team.\n# Copyright (c) 2020, NVIDIA CORPORATION. All rights reserved.\n#\n# Licensed under the Apache License, Version 2.0 (the \"License\");\n# you may not use this file except in compliance with the License.\n# You may obtain a copy of the License at\n#\n# http://www.apache.org/licenses/LICENSE-2.0\n#\n# Unless required by applicable law or agreed to in writing, software\n# distributed under the License is distributed on an \"AS IS\" BASIS,\n# WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.\n# See the License for the specific language governing permissions and\n# limitations under the License.\n\nfrom dataclasses import dataclass\nfrom typing import Any, Callable, Dict, Iterable, List, Optional, Tuple, Union\n\nimport torch\nfrom torch.nn import functional as F\n\nfrom .file_utils import ModelOutput\nfrom .generation_beam_search import BeamScorer, BeamSearchScorer\nfrom .generation_logits_process import (\n EncoderNoRepeatNGramLogitsProcessor,\n ForcedBOSTokenLogitsProcessor,\n ForcedEOSTokenLogitsProcessor,\n HammingDiversityLogitsProcessor,\n LogitsProcessorList,\n MinLengthLogitsProcessor,\n NoBadWordsLogitsProcessor,\n NoRepeatNGramLogitsProcessor,\n PrefixConstrainedLogitsProcessor,\n RepetitionPenaltyLogitsProcessor,\n TemperatureLogitsWarper,\n TopKLogitsWarper,\n TopPLogitsWarper,\n LogitsProcessor,\n ConstrainedDecodingLogitsProcessor,\n)\nfrom .utils import logging\n\n\nlogger = logging.get_logger(__name__)\n\n\n@dataclass\nclass GreedySearchDecoderOnlyOutput(ModelOutput):\n \"\"\"\n Base class for outputs of decoder-only generation models using greedy search.\n\n\n Args:\n sequences (:obj:`torch.LongTensor` of shape :obj:`(batch_size, sequence_length)`):\n The generated sequences. The second dimension (sequence_length) is either equal to :obj:`max_length` or\n shorter if all batches finished early due to the :obj:`eos_token_id`.\n scores (:obj:`tuple(torch.FloatTensor)` `optional`, returned when ``output_scores=True`` is passed or when ``config.output_scores=True``):\n Processed prediction scores of the language modeling head (scores for each vocabulary token before SoftMax)\n at each generation step. :obj:`(max_length,)`-shaped tuple of :obj:`torch.FloatTensor` with each tensor of\n shape :obj:`(batch_size, config.vocab_size)`).\n attentions (:obj:`tuple(tuple(torch.FloatTensor))`, `optional`, returned when ``output_attentions=True`` is passed or ``config.output_attentions=True``):\n Tuple (one element for each generated token) of tuples (one element for each layer of the decoder) of\n :obj:`torch.FloatTensor` of shape :obj:`(batch_size, num_heads, generated_length, sequence_length)`.\n hidden_states (:obj:`tuple(tuple(torch.FloatTensor))`, `optional`, returned when ``output_hidden_states=True`` is passed or when ``config.output_hidden_states=True``):\n Tuple (one element for each generated token) of tuples (one element for each layer of the decoder) of\n :obj:`torch.FloatTensor` of shape :obj:`(batch_size, generated_length, hidden_size)`.\n \"\"\"\n\n sequences: torch.LongTensor = None\n scores: Optional[Tuple[torch.FloatTensor]] = None\n attentions: Optional[Tuple[Tuple[torch.FloatTensor]]] = None\n hidden_states: Optional[Tuple[Tuple[torch.FloatTensor]]] = None\n\n\n@dataclass\nclass GreedySearchEncoderDecoderOutput(ModelOutput):\n \"\"\"\n Base class for outputs of encoder-decoder generation models using greedy search. Hidden states and attention\n weights of the decoder (respectively the encoder) can be accessed via the encoder_attentions and the\n encoder_hidden_states attributes (respectively the decoder_attentions and the decoder_hidden_states attributes)\n\n\n Args:\n sequences (:obj:`torch.LongTensor` of shape :obj:`(batch_size, sequence_length)`):\n The generated sequences. The second dimension (sequence_length) is either equal to :obj:`max_length` or\n shorter if all batches finished early due to the :obj:`eos_token_id`.\n scores (:obj:`tuple(torch.FloatTensor)` `optional`, returned when ``output_scores=True`` is passed or when ``config.output_scores=True``):\n Processed prediction scores of the language modeling head (scores for each vocabulary token before SoftMax)\n at each generation step. :obj:`(max_length,)`-shaped tuple of :obj:`torch.FloatTensor` with each tensor of\n shape :obj:`(batch_size, config.vocab_size)`).\n encoder_attentions (:obj:`tuple(torch.FloatTensor)`, `optional`, returned when ``output_attentions=True`` is passed or ``config.output_attentions=True``):\n Tuple of :obj:`torch.FloatTensor` (one for each layer of the decoder) of shape :obj:`(batch_size,\n num_heads, sequence_length, sequence_length)`.\n encoder_hidden_states (:obj:`tuple(torch.FloatTensor)`, `optional`, returned when ``output_hidden_states=True`` is passed or when ``config.output_hidden_states=True``):\n Tuple of :obj:`torch.FloatTensor` (one for the output of the embeddings + one for the output of each layer)\n of shape :obj:`(batch_size, sequence_length, hidden_size)`.\n decoder_attentions (:obj:`tuple(tuple(torch.FloatTensor))`, `optional`, returned when ``output_attentions=True`` is passed or ``config.output_attentions=True``):\n Tuple (one element for each generated token) of tuples (one element for each layer of the decoder) of\n :obj:`torch.FloatTensor` of shape :obj:`(batch_size, num_heads, generated_length, sequence_length)`.\n cross_attentions (:obj:`tuple(tuple(torch.FloatTensor))`, `optional`, returned when ``output_attentions=True`` is passed or ``config.output_attentions=True``):\n Tuple (one element for each generated token) of tuples (one element for each layer of the decoder) of\n :obj:`torch.FloatTensor` of shape :obj:`(batch_size, num_heads, generated_length, sequence_length)`.\n decoder_hidden_states (:obj:`tuple(tuple(torch.FloatTensor))`, `optional`, returned when ``output_hidden_states=True`` is passed or when ``config.output_hidden_states=True``):\n Tuple (one element for each generated token) of tuples (one element for each layer of the decoder) of\n :obj:`torch.FloatTensor` of shape :obj:`(batch_size, generated_length, hidden_size)`.\n \"\"\"\n\n sequences: torch.LongTensor = None\n scores: Optional[Tuple[torch.FloatTensor]] = None\n encoder_attentions: Optional[Tuple[torch.FloatTensor]] = None\n encoder_hidden_states: Optional[Tuple[torch.FloatTensor]] = None\n decoder_attentions: Optional[Tuple[Tuple[torch.FloatTensor]]] = None\n cross_attentions: Optional[Tuple[Tuple[torch.FloatTensor]]] = None\n decoder_hidden_states: Optional[Tuple[Tuple[torch.FloatTensor]]] = None\n\n\n@dataclass\nclass SampleDecoderOnlyOutput(ModelOutput):\n \"\"\"\n Base class for outputs of decoder-only generation models using sampling.\n\n\n Args:\n sequences (:obj:`torch.LongTensor` of shape :obj:`(batch_size*num_return_sequences, sequence_length)`):\n The generated sequences. The second dimension (sequence_length) is either equal to :obj:`max_length` or\n shorter if all batches finished early due to the :obj:`eos_token_id`.\n scores (:obj:`tuple(torch.FloatTensor)` `optional`, returned when ``output_scores=True`` is passed or when ``config.output_scores=True``):\n Processed prediction scores of the language modeling head (scores for each vocabulary token before SoftMax)\n at each generation step. :obj:`(max_length,)`-shaped tuple of :obj:`torch.FloatTensor` with each tensor of\n shape :obj:`(batch_size*num_return_sequences, config.vocab_size)`).\n attentions (:obj:`tuple(tuple(torch.FloatTensor))`, `optional`, returned when ``output_attentions=True`` is passed or ``config.output_attentions=True``):\n Tuple (one element for each generated token) of tuples (one element for each layer of the decoder) of\n :obj:`torch.FloatTensor` of shape :obj:`(num_return_sequences*batch_size, num_heads, generated_length,\n sequence_length)`.\n hidden_states (:obj:`tuple(tuple(torch.FloatTensor))`, `optional`, returned when ``output_hidden_states=True`` is passed or when ``config.output_hidden_states=True``):\n Tuple (one element for each generated token) of tuples (one element for each layer of the decoder) of\n :obj:`torch.FloatTensor` of shape :obj:`(num_return_sequences*batch_size, generated_length, hidden_size)`.\n \"\"\"\n\n sequences: torch.LongTensor = None\n scores: Optional[Tuple[torch.FloatTensor]] = None\n attentions: Optional[Tuple[Tuple[torch.FloatTensor]]] = None\n hidden_states: Optional[Tuple[Tuple[torch.FloatTensor]]] = None\n\n\n@dataclass\nclass SampleEncoderDecoderOutput(ModelOutput):\n \"\"\"\n Base class for outputs of encoder-decoder generation models using sampling. Hidden states and attention weights of\n the decoder (respectively the encoder) can be accessed via the encoder_attentions and the encoder_hidden_states\n attributes (respectively the decoder_attentions and the decoder_hidden_states attributes)\n\n\n Args:\n sequences (:obj:`torch.LongTensor` of shape :obj:`(batch_size*num_return_sequences, sequence_length)`):\n The generated sequences. The second dimension (sequence_length) is either equal to :obj:`max_length` or\n shorter if all batches finished early due to the :obj:`eos_token_id`.\n scores (:obj:`tuple(torch.FloatTensor)` `optional`, returned when ``output_scores=True`` is passed or when ``config.output_scores=True``):\n Processed prediction scores of the language modeling head (scores for each vocabulary token before SoftMax)\n at each generation step. :obj:`(max_length,)`-shaped tuple of :obj:`torch.FloatTensor` with each tensor of\n shape :obj:`(batch_size*num_return_sequences, config.vocab_size)`).\n encoder_attentions (:obj:`tuple(torch.FloatTensor)`, `optional`, returned when ``output_attentions=True`` is passed or ``config.output_attentions=True``):\n Tuple of :obj:`torch.FloatTensor` (one for each layer of the decoder) of shape\n :obj:`(batch_size*num_return_sequences, num_heads, sequence_length, sequence_length)`.\n encoder_hidden_states (:obj:`tuple(torch.FloatTensor)`, `optional`, returned when ``output_hidden_states=True`` is passed or when ``config.output_hidden_states=True``):\n Tuple of :obj:`torch.FloatTensor` (one for the output of the embeddings + one for the output of each layer)\n of shape :obj:`(batch_size*num_return_sequences, sequence_length, hidden_size)`.\n decoder_attentions (:obj:`tuple(tuple(torch.FloatTensor))`, `optional`, returned when ``output_attentions=True`` is passed or ``config.output_attentions=True``):\n Tuple (one element for each generated token) of tuples (one element for each layer of the decoder) of\n :obj:`torch.FloatTensor` of shape :obj:`(batch_size*num_return_sequences, num_heads, generated_length,\n sequence_length)`.\n cross_attentions (:obj:`tuple(tuple(torch.FloatTensor))`, `optional`, returned when ``output_attentions=True`` is passed or ``config.output_attentions=True``):\n Tuple (one element for each generated token) of tuples (one element for each layer of the decoder) of\n :obj:`torch.FloatTensor` of shape :obj:`(batch_size, num_heads, generated_length, sequence_length)`.\n decoder_hidden_states (:obj:`tuple(tuple(torch.FloatTensor))`, `optional`, returned when ``output_hidden_states=True`` is passed or when ``config.output_hidden_states=True``):\n Tuple (one element for each generated token) of tuples (one element for each layer of the decoder) of\n :obj:`torch.FloatTensor` of shape :obj:`(batch_size*num_return_sequences, generated_length, hidden_size)`.\n \"\"\"\n\n sequences: torch.LongTensor = None\n scores: Optional[Tuple[torch.FloatTensor]] = None\n encoder_attentions: Optional[Tuple[torch.FloatTensor]] = None\n encoder_hidden_states: Optional[Tuple[torch.FloatTensor]] = None\n decoder_attentions: Optional[Tuple[Tuple[torch.FloatTensor]]] = None\n cross_attentions: Optional[Tuple[Tuple[torch.FloatTensor]]] = None\n decoder_hidden_states: Optional[Tuple[Tuple[torch.FloatTensor]]] = None\n\n\n@dataclass\nclass BeamSearchDecoderOnlyOutput(ModelOutput):\n \"\"\"\n Base class for outputs of decoder-only generation models using beam search.\n\n Args:\n sequences (:obj:`torch.LongTensor` of shape :obj:`(batch_size*num_return_sequences, sequence_length)`):\n The generated sequences. The second dimension (sequence_length) is either equal to :obj:`max_length` or\n shorter if all batches finished early due to the :obj:`eos_token_id`.\n sequences_scores (:obj:`torch.FloatTensor` of shape :obj:`(batch_size*num_return_sequences)`, `optional`, returned when ``output_scores=True`` is passed or when ``config.output_scores=True``):\n Final beam scores of the generated ``sequences``.\n scores (:obj:`tuple(torch.FloatTensor)` `optional`, returned when ``output_scores=True`` is passed or when ``config.output_scores=True``):\n Processed beam scores for each vocabulary token at each generation step. Beam scores consisting of log\n softmax scores for each vocabulary token and sum of log softmax of previously generated tokens in this beam\n . :obj:`(max_length,)`-shaped tuple of :obj:`torch.FloatTensor` with each tensor of shape\n :obj:`(batch_size*num_beams*num_return_sequences, config.vocab_size)`).\n attentions (:obj:`tuple(tuple(torch.FloatTensor))`, `optional`, returned when ``output_attentions=True`` is passed or ``config.output_attentions=True``):\n Tuple (one element for each generated token) of tuples (one element for each layer of the decoder) of\n :obj:`torch.FloatTensor` of shape :obj:`(batch_size*num_beams, num_heads, generated_length,\n sequence_length)`.\n hidden_states (:obj:`tuple(tuple(torch.FloatTensor))`, `optional`, returned when ``output_hidden_states=True`` is passed or when ``config.output_hidden_states=True``):\n Tuple (one element for each generated token) of tuples (one element for each layer of the decoder) of\n :obj:`torch.FloatTensor` of shape :obj:`(batch_size*num_beams*num_return_sequences, generated_length,\n hidden_size)`.\n \"\"\"\n\n sequences: torch.LongTensor = None\n sequences_scores: Optional[torch.FloatTensor] = None\n scores: Optional[Tuple[torch.FloatTensor]] = None\n attentions: Optional[Tuple[Tuple[torch.FloatTensor]]] = None\n hidden_states: Optional[Tuple[Tuple[torch.FloatTensor]]] = None\n\n\n@dataclass\nclass BeamSearchEncoderDecoderOutput(ModelOutput):\n \"\"\"\n Base class for outputs of encoder-decoder generation models using beam search. Hidden states and attention weights\n of the decoder (respectively the encoder) can be accessed via the encoder_attentions and the encoder_hidden_states\n attributes (respectively the decoder_attentions and the decoder_hidden_states attributes)\n\n Args:\n sequences (:obj:`torch.LongTensor` of shape :obj:`(batch_size*num_return_sequences, sequence_length)`):\n The generated sequences. The second dimension (sequence_length) is either equal to :obj:`max_length` or\n shorter if all batches finished early due to the :obj:`eos_token_id`.\n sequences_scores (:obj:`torch.FloatTensor` of shape :obj:`(batch_size*num_return_sequences)`, `optional`, returned when ``output_scores=True`` is passed or when ``config.output_scores=True``):\n Final beam scores of the generated ``sequences``.\n scores (:obj:`tuple(torch.FloatTensor)` `optional`, returned when ``output_scores=True`` is passed or when ``config.output_scores=True``):\n Processed beam scores for each vocabulary token at each generation step. Beam scores consisting of log\n softmax scores for each vocabulary token and sum of log softmax of previously generated tokens in this beam\n . :obj:`(max_length,)`-shaped tuple of :obj:`torch.FloatTensor` with each tensor of shape\n :obj:`(batch_size*num_beams, config.vocab_size)`).\n attentions (:obj:`tuple(tuple(torch.FloatTensor))`, `optional`, returned when ``output_attentions=True`` is passed or ``config.output_attentions=True``):\n encoder_attentions (:obj:`tuple(torch.FloatTensor)`, `optional`, returned when ``output_attentions=True`` is passed or ``config.output_attentions=True``):\n Tuple of :obj:`torch.FloatTensor` (one for each layer of the decoder) of shape :obj:`(batch_size,\n num_heads, sequence_length, sequence_length)`.\n encoder_hidden_states (:obj:`tuple(torch.FloatTensor)`, `optional`, returned when ``output_hidden_states=True`` is passed or when ``config.output_hidden_states=True``):\n Tuple of :obj:`torch.FloatTensor` (one for the output of the embeddings + one for the output of each layer)\n of shape :obj:`(batch_size*num_beams*num_return_sequences, sequence_length, hidden_size)`.\n decoder_attentions (:obj:`tuple(tuple(torch.FloatTensor))`, `optional`, returned when ``output_attentions=True`` is passed or ``config.output_attentions=True``):\n Tuple (one element for each generated token) of tuples (one element for each layer of the decoder) of\n :obj:`torch.FloatTensor` of shape :obj:`(batch_size*num_beams*num_return_sequences, num_heads,\n generated_length, sequence_length)`.\n cross_attentions (:obj:`tuple(tuple(torch.FloatTensor))`, `optional`, returned when ``output_attentions=True`` is passed or ``config.output_attentions=True``):\n Tuple (one element for each generated token) of tuples (one element for each layer of the decoder) of\n :obj:`torch.FloatTensor` of shape :obj:`(batch_size, num_heads, generated_length, sequence_length)`.\n decoder_hidden_states (:obj:`tuple(tuple(torch.FloatTensor))`, `optional`, returned when ``output_hidden_states=True`` is passed or when ``config.output_hidden_states=True``):\n Tuple (one element for each generated token) of tuples (one element for each layer of the decoder) of\n :obj:`torch.FloatTensor` of shape :obj:`(batch_size*num_beams*num_return_sequences, generated_length,\n hidden_size)`.\n \"\"\"\n\n sequences: torch.LongTensor = None\n sequences_scores: Optional[torch.FloatTensor] = None\n scores: Optional[Tuple[torch.FloatTensor]] = None\n encoder_attentions: Optional[Tuple[torch.FloatTensor]] = None\n encoder_hidden_states: Optional[Tuple[torch.FloatTensor]] = None\n decoder_attentions: Optional[Tuple[Tuple[torch.FloatTensor]]] = None\n cross_attentions: Optional[Tuple[Tuple[torch.FloatTensor]]] = None\n decoder_hidden_states: Optional[Tuple[Tuple[torch.FloatTensor]]] = None\n\n\n@dataclass\nclass BeamSampleDecoderOnlyOutput(ModelOutput):\n \"\"\"\n Base class for outputs of decoder-only generation models using beam sample.\n\n Args:\n sequences (:obj:`torch.LongTensor` of shape :obj:`(batch_size*num_return_sequences, sequence_length)`):\n The generated sequences. The second dimension (sequence_length) is either equal to :obj:`max_length` or\n shorter if all batches finished early due to the :obj:`eos_token_id`.\n sequences_scores (:obj:`torch.FloatTensor` of shape :obj:`(batch_size * num_return_sequence)`, `optional`, returned when ``output_scores=True`` is passed or when ``config.output_scores=True``):\n Final beam scores of the generated ``sequences``.\n scores (:obj:`tuple(torch.FloatTensor)` `optional`, returned when ``output_scores=True`` is passed or when ``config.output_scores=True``):\n Processed beam scores for each vocabulary token at each generation step. Beam scores consisting of log\n softmax scores for each vocabulary token and sum of log softmax of previously generated tokens in this beam\n . :obj:`(max_length,)`-shaped tuple of :obj:`torch.FloatTensor` with each tensor of shape\n :obj:`(batch_size*num_beams*num_return_sequences, config.vocab_size)`).\n attentions (:obj:`tuple(tuple(torch.FloatTensor))`, `optional`, returned when ``output_attentions=True`` is passed or ``config.output_attentions=True``):\n Tuple (one element for each generated token) of tuples (one element for each layer of the decoder) of\n :obj:`torch.FloatTensor` of shape :obj:`(batch_size*num_beams, num_heads, generated_length,\n sequence_length)`.\n hidden_states (:obj:`tuple(tuple(torch.FloatTensor))`, `optional`, returned when ``output_hidden_states=True`` is passed or when ``config.output_hidden_states=True``):\n Tuple (one element for each generated token) of tuples (one element for each layer of the decoder) of\n :obj:`torch.FloatTensor` of shape :obj:`(batch_size*num_beams, generated_length, hidden_size)`.\n \"\"\"\n\n sequences: torch.LongTensor = None\n sequences_scores: Optional[torch.FloatTensor] = None\n scores: Optional[Tuple[torch.FloatTensor]] = None\n attentions: Optional[Tuple[Tuple[torch.FloatTensor]]] = None\n hidden_states: Optional[Tuple[Tuple[torch.FloatTensor]]] = None\n\n\n@dataclass\nclass BeamSampleEncoderDecoderOutput(ModelOutput):\n \"\"\"\n Base class for outputs of encoder-decoder generation models using beam sampling. Hidden states and attention\n weights of the decoder (respectively the encoder) can be accessed via the encoder_attentions and the\n encoder_hidden_states attributes (respectively the decoder_attentions and the decoder_hidden_states attributes)\n\n Args:\n sequences (:obj:`torch.LongTensor` of shape :obj:`(batch_size*num_beams, sequence_length)`):\n The generated sequences. The second dimension (sequence_length) is either equal to :obj:`max_length` or\n shorter if all batches finished early due to the :obj:`eos_token_id`.\n sequences_scores (:obj:`torch.FloatTensor` of shape :obj:`(batch_size * num_return_sequence)`, `optional`, returned when ``output_scores=True`` is passed or when ``config.output_scores=True``):\n Final beam scores of the generated ``sequences``.\n scores (:obj:`tuple(torch.FloatTensor)` `optional`, returned when ``output_scores=True`` is passed or when ``config.output_scores=True``):\n Processed beam scores for each vocabulary token at each generation step. Beam scores consisting of log\n softmax scores for each vocabulary token and sum of log softmax of previously generated tokens in this beam\n . :obj:`(max_length,)`-shaped tuple of :obj:`torch.FloatTensor` with each tensor of shape\n :obj:`(batch_size*num_beams, config.vocab_size)`).\n encoder_attentions (:obj:`tuple(torch.FloatTensor)`, `optional`, returned when ``output_attentions=True`` is passed or ``config.output_attentions=True``):\n Tuple of :obj:`torch.FloatTensor` (one for each layer of the decoder) of shape :obj:`(batch_size,\n num_heads, sequence_length, sequence_length)`.\n encoder_hidden_states (:obj:`tuple(torch.FloatTensor)`, `optional`, returned when ``output_hidden_states=True`` is passed or when ``config.output_hidden_states=True``):\n Tuple of :obj:`torch.FloatTensor` (one for the output of the embeddings + one for the output of each layer)\n of shape :obj:`(batch_size*num_beams, sequence_length, hidden_size)`.\n decoder_attentions (:obj:`tuple(tuple(torch.FloatTensor))`, `optional`, returned when ``output_attentions=True`` is passed or ``config.output_attentions=True``):\n Tuple (one element for each generated token) of tuples (one element for each layer of the decoder) of\n :obj:`torch.FloatTensor` of shape :obj:`(batch_size*num_beams, num_heads, generated_length,\n sequence_length)`.\n cross_attentions (:obj:`tuple(tuple(torch.FloatTensor))`, `optional`, returned when ``output_attentions=True`` is passed or ``config.output_attentions=True``):\n Tuple (one element for each generated token) of tuples (one element for each layer of the decoder) of\n :obj:`torch.FloatTensor` of shape :obj:`(batch_size, num_heads, generated_length, sequence_length)`.\n decoder_hidden_states (:obj:`tuple(tuple(torch.FloatTensor))`, `optional`, returned when ``output_hidden_states=True`` is passed or when ``config.output_hidden_states=True``):\n Tuple (one element for each generated token) of tuples (one element for each layer of the decoder) of\n :obj:`torch.FloatTensor` of shape :obj:`(batch_size*num_beams, generated_length, hidden_size)`.\n \"\"\"\n\n sequences: torch.LongTensor = None\n sequences_scores: Optional[torch.FloatTensor] = None\n scores: Optional[Tuple[torch.FloatTensor]] = None\n encoder_attentions: Optional[Tuple[torch.FloatTensor]] = None\n encoder_hidden_states: Optional[Tuple[torch.FloatTensor]] = None\n decoder_attentions: Optional[Tuple[Tuple[torch.FloatTensor]]] = None\n cross_attentions: Optional[Tuple[Tuple[torch.FloatTensor]]] = None\n decoder_hidden_states: Optional[Tuple[Tuple[torch.FloatTensor]]] = None\n\n\nGreedySearchOutput = Union[GreedySearchEncoderDecoderOutput, GreedySearchDecoderOnlyOutput]\nSampleOutput = Union[SampleEncoderDecoderOutput, SampleDecoderOnlyOutput]\nBeamSearchOutput = Union[BeamSearchEncoderDecoderOutput, BeamSearchDecoderOnlyOutput]\nBeamSampleOutput = Union[BeamSampleEncoderDecoderOutput, BeamSampleDecoderOnlyOutput]\n\n\nclass GenerationMixin:\n \"\"\"\n A class containing all of the functions supporting generation, to be used as a mixin in\n :class:`~transformers.PreTrainedModel`.\n \"\"\"\n\n def prepare_inputs_for_generation(self, input_ids: torch.LongTensor, **kwargs) -> Dict[str, Any]:\n \"\"\"\n Implement in subclasses of :class:`~transformers.PreTrainedModel` for custom behavior to prepare inputs in the\n generate method.\n \"\"\"\n return {\"input_ids\": input_ids}\n\n def adjust_logits_during_generation(self, logits: torch.FloatTensor, **kwargs) -> torch.FloatTensor:\n \"\"\"\n Implement in subclasses of :class:`~transformers.PreTrainedModel` for custom behavior to adjust the logits in\n the generate method.\n \"\"\"\n return logits\n\n def _prepare_input_ids_for_generation(self, bos_token_id: int) -> torch.LongTensor:\n if bos_token_id is None:\n raise ValueError(\"`bos_token_id` has to be defined when no `input_ids` are provided.\")\n return torch.ones((1, 1), dtype=torch.long, device=self.device) * bos_token_id\n\n def _prepare_attention_mask_for_generation(\n self, input_ids: torch.Tensor, pad_token_id: int, eos_token_id: int\n ) -> torch.LongTensor:\n is_pad_token_in_inputs_ids = (pad_token_id is not None) and (pad_token_id in input_ids)\n is_pad_token_not_equal_to_eos_token_id = (eos_token_id is None) or (\n (eos_token_id is not None) and (pad_token_id != eos_token_id)\n )\n if is_pad_token_in_inputs_ids and is_pad_token_not_equal_to_eos_token_id:\n return input_ids.ne(pad_token_id).long()\n return input_ids.new_ones(input_ids.shape)\n\n def _prepare_encoder_decoder_kwargs_for_generation(\n self, input_ids: torch.LongTensor, model_kwargs\n ) -> Dict[str, Any]:\n # retrieve encoder hidden states\n encoder = self.get_encoder()\n encoder_kwargs = {\n argument: value for argument, value in model_kwargs.items() if not argument.startswith(\"decoder_\")\n }\n model_kwargs[\"encoder_outputs\"]: ModelOutput = encoder(input_ids, return_dict=True, **encoder_kwargs)\n return model_kwargs\n\n def _prepare_decoder_input_ids_for_generation(\n self, input_ids: torch.LongTensor, decoder_start_token_id: int = None, bos_token_id: int = None\n ) -> torch.LongTensor:\n decoder_start_token_id = self._get_decoder_start_token_id(decoder_start_token_id, bos_token_id)\n decoder_input_ids = (\n torch.ones((input_ids.shape[0], 1), dtype=input_ids.dtype, device=input_ids.device)\n * decoder_start_token_id\n )\n return decoder_input_ids\n\n def _get_pad_token_id(self, pad_token_id: int = None, eos_token_id: int = None) -> int:\n if pad_token_id is None and eos_token_id is not None:\n logger.warning(f\"Setting `pad_token_id` to `eos_token_id`:{eos_token_id} for open-end generation.\")\n pad_token_id = eos_token_id\n return pad_token_id\n\n def _get_decoder_start_token_id(self, decoder_start_token_id: int = None, bos_token_id: int = None) -> int:\n decoder_start_token_id = (\n decoder_start_token_id if decoder_start_token_id is not None else self.config.decoder_start_token_id\n )\n bos_token_id = bos_token_id if bos_token_id is not None else self.config.bos_token_id\n\n if decoder_start_token_id is not None:\n return decoder_start_token_id\n elif (\n hasattr(self.config, \"decoder\")\n and hasattr(self.config.decoder, \"decoder_start_token_id\")\n and self.config.decoder.decoder_start_token_id is not None\n ):\n return self.config.decoder.decoder_start_token_id\n elif bos_token_id is not None:\n return bos_token_id\n elif (\n hasattr(self.config, \"decoder\")\n and hasattr(self.config.decoder, \"bos_token_id\")\n and self.config.decoder.bos_token_id is not None\n ):\n return self.config.decoder.bos_token_id\n raise ValueError(\n \"`decoder_start_token_id` or `bos_token_id` has to be defined for encoder-decoder generation.\"\n )\n\n @staticmethod\n def _expand_inputs_for_generation(\n input_ids: torch.LongTensor,\n expand_size: int = 1,\n is_encoder_decoder: bool = False,\n attention_mask: torch.LongTensor = None,\n encoder_outputs: ModelOutput = None,\n **model_kwargs,\n ) -> Tuple[torch.LongTensor, Dict[str, Any]]:\n expanded_return_idx = (\n torch.arange(input_ids.shape[0]).view(-1, 1).repeat(1, expand_size).view(-1).to(input_ids.device)\n )\n input_ids = input_ids.index_select(0, expanded_return_idx)\n\n if \"token_type_ids\" in model_kwargs:\n token_type_ids = model_kwargs[\"token_type_ids\"]\n model_kwargs[\"token_type_ids\"] = token_type_ids.index_select(0, expanded_return_idx)\n\n if attention_mask is not None:\n model_kwargs[\"attention_mask\"] = attention_mask.index_select(0, expanded_return_idx)\n\n if is_encoder_decoder:\n assert encoder_outputs is not None\n encoder_outputs[\"last_hidden_state\"] = encoder_outputs.last_hidden_state.index_select(\n 0, expanded_return_idx.to(encoder_outputs.last_hidden_state.device)\n )\n model_kwargs[\"encoder_outputs\"] = encoder_outputs\n return input_ids, model_kwargs\n\n @staticmethod\n def _init_sequence_length_for_generation(\n input_ids: torch.LongTensor, max_length: int\n ) -> Tuple[torch.Tensor, torch.Tensor, int]:\n unfinished_sequences = input_ids.new(input_ids.shape[0]).fill_(1)\n sequence_lengths = input_ids.new(input_ids.shape[0]).fill_(max_length)\n\n cur_len = input_ids.shape[-1]\n return sequence_lengths, unfinished_sequences, cur_len\n\n @staticmethod\n def _update_seq_length_for_generation(\n sequence_lengths: torch.LongTensor,\n unfinished_sequences: torch.LongTensor,\n cur_len: int,\n is_eos_in_next_token: torch.BoolTensor,\n ) -> Tuple[torch.LongTensor, torch.LongTensor]:\n # check if sentence is not finished yet\n is_sent_unfinished = unfinished_sequences.mul(is_eos_in_next_token.long()).bool()\n\n # update sentence length\n sequence_lengths = sequence_lengths.masked_fill(is_sent_unfinished, cur_len)\n unfinished_sequences = unfinished_sequences.mul((~is_eos_in_next_token).long())\n return sequence_lengths, unfinished_sequences\n\n @staticmethod\n def _update_model_kwargs_for_generation(\n outputs: ModelOutput, model_kwargs: Dict[str, Any], is_encoder_decoder: bool = False\n ) -> Dict[str, Any]:\n # update past\n if \"past_key_values\" in outputs:\n model_kwargs[\"past\"] = outputs.past_key_values\n elif \"mems\" in outputs:\n model_kwargs[\"past\"] = outputs.mems\n elif \"past_buckets_states\" in outputs:\n model_kwargs[\"past\"] = outputs.past_buckets_states\n else:\n model_kwargs[\"past\"] = None\n\n # update token_type_ids with last value\n if \"token_type_ids\" in model_kwargs:\n token_type_ids = model_kwargs[\"token_type_ids\"]\n model_kwargs[\"token_type_ids\"] = torch.cat([token_type_ids, token_type_ids[:, -1].unsqueeze(-1)], dim=-1)\n\n # update attention mask\n if not is_encoder_decoder:\n if \"attention_mask\" in model_kwargs:\n attention_mask = model_kwargs[\"attention_mask\"]\n model_kwargs[\"attention_mask\"] = torch.cat(\n [attention_mask, attention_mask.new_ones((attention_mask.shape[0], 1))], dim=-1\n )\n\n return model_kwargs\n\n def _reorder_cache(self, past, beam_idx):\n raise NotImplementedError(\n f\"Make sure that a `_reorder_cache` function is correctly implemented in {self.__class__.__module__} to enable beam search for {self.__class__}\"\n )\n\n def _get_logits_warper(\n self, top_k: int = None, top_p: float = None, temperature: float = None, num_beams: int = None\n ) -> LogitsProcessorList:\n \"\"\"\n This class returns a :obj:`~transformers.LogitsProcessorList` list object that contains all relevant\n :obj:`~transformers.LogitsWarper` instances used for multinomial sampling.\n \"\"\"\n\n # init warp parameters\n top_k = top_k if top_k is not None else self.config.top_k\n top_p = top_p if top_p is not None else self.config.top_p\n temperature = temperature if temperature is not None else self.config.temperature\n # instantiate warpers list\n warpers = LogitsProcessorList()\n\n # the following idea is largely copied from this PR: https://github.com/huggingface/transformers/pull/5420/files\n # all samplers can be found in `generation_utils_samplers.py`\n if temperature is not None and temperature != 1.0:\n warpers.append(TemperatureLogitsWarper(temperature))\n if top_k is not None and top_k != 0:\n warpers.append(TopKLogitsWarper(top_k=top_k, min_tokens_to_keep=(2 if num_beams > 1 else 1)))\n if top_p is not None and top_p < 1.0:\n warpers.append(TopPLogitsWarper(top_p=top_p, min_tokens_to_keep=(2 if num_beams > 1 else 1)))\n return warpers\n\n def _get_logits_processor(\n self,\n repetition_penalty: float,\n no_repeat_ngram_size: int,\n encoder_no_repeat_ngram_size: int,\n encoder_input_ids: torch.LongTensor,\n bad_words_ids: List[List[int]],\n min_length: int,\n max_length: int,\n eos_token_id: int,\n forced_bos_token_id: int,\n forced_eos_token_id: int,\n prefix_allowed_tokens_fn: Callable[[int, torch.Tensor], List[int]],\n num_beams: int,\n num_beam_groups: int,\n diversity_penalty: float,\n ) -> LogitsProcessorList:\n \"\"\"\n This class returns a :obj:`~transformers.LogitsProcessorList` list object that contains all relevant\n :obj:`~transformers.LogitsProcessor` instances used to modify the scores of the language model head.\n \"\"\"\n\n # init warp parameters\n repetition_penalty = repetition_penalty if repetition_penalty is not None else self.config.repetition_penalty\n no_repeat_ngram_size = (\n no_repeat_ngram_size if no_repeat_ngram_size is not None else self.config.no_repeat_ngram_size\n )\n encoder_no_repeat_ngram_size = (\n encoder_no_repeat_ngram_size\n if encoder_no_repeat_ngram_size is not None\n else self.config.encoder_no_repeat_ngram_size\n )\n bad_words_ids = bad_words_ids if bad_words_ids is not None else self.config.bad_words_ids\n min_length = min_length if min_length is not None else self.config.min_length\n eos_token_id = eos_token_id if eos_token_id is not None else self.config.eos_token_id\n diversity_penalty = diversity_penalty if diversity_penalty is not None else self.config.diversity_penalty\n forced_bos_token_id = (\n forced_bos_token_id if forced_bos_token_id is not None else self.config.forced_bos_token_id\n )\n forced_eos_token_id = (\n forced_eos_token_id if forced_eos_token_id is not None else self.config.forced_eos_token_id\n )\n # instantiate processors list\n processors = LogitsProcessorList()\n\n # the following idea is largely copied from this PR: https://github.com/huggingface/transformers/pull/5420/files\n # all samplers can be found in `generation_utils_samplers.py`\n if diversity_penalty is not None and diversity_penalty > 0.0:\n processors.append(\n HammingDiversityLogitsProcessor(\n diversity_penalty=diversity_penalty, num_beams=num_beams, num_beam_groups=num_beam_groups\n )\n )\n if repetition_penalty is not None and repetition_penalty != 1.0:\n processors.append(RepetitionPenaltyLogitsProcessor(penalty=repetition_penalty))\n if no_repeat_ngram_size is not None and no_repeat_ngram_size > 0:\n processors.append(NoRepeatNGramLogitsProcessor(no_repeat_ngram_size))\n if encoder_no_repeat_ngram_size is not None and encoder_no_repeat_ngram_size > 0:\n if self.config.is_encoder_decoder:\n processors.append(EncoderNoRepeatNGramLogitsProcessor(encoder_no_repeat_ngram_size, encoder_input_ids))\n else:\n raise ValueError(\n \"It's impossible to use `encoder_no_repeat_ngram_size` with decoder-only architecture\"\n )\n if bad_words_ids is not None:\n processors.append(NoBadWordsLogitsProcessor(bad_words_ids, eos_token_id))\n if min_length is not None and eos_token_id is not None and min_length > -1:\n processors.append(MinLengthLogitsProcessor(min_length, eos_token_id))\n if prefix_allowed_tokens_fn is not None:\n processors.append(PrefixConstrainedLogitsProcessor(prefix_allowed_tokens_fn, num_beams // num_beam_groups))\n if forced_bos_token_id is not None:\n processors.append(ForcedBOSTokenLogitsProcessor(forced_bos_token_id))\n if forced_eos_token_id is not None:\n processors.append(ForcedEOSTokenLogitsProcessor(max_length, forced_eos_token_id))\n return processors\n\n @torch.no_grad()\n def generate(\n self,\n input_ids: Optional[torch.LongTensor] = None,\n max_length: Optional[int] = None,\n min_length: Optional[int] = None,\n do_sample: Optional[bool] = None,\n early_stopping: Optional[bool] = None,\n num_beams: Optional[int] = None,\n temperature: Optional[float] = None,\n top_k: Optional[int] = None,\n top_p: Optional[float] = None,\n repetition_penalty: Optional[float] = None,\n bad_words_ids: Optional[Iterable[int]] = None,\n bos_token_id: Optional[int] = None,\n pad_token_id: Optional[int] = None,\n eos_token_id: Optional[int] = None,\n length_penalty: Optional[float] = None,\n no_repeat_ngram_size: Optional[int] = None,\n encoder_no_repeat_ngram_size: Optional[int] = None,\n num_return_sequences: Optional[int] = None,\n decoder_start_token_id: Optional[int] = None,\n use_cache: Optional[bool] = None,\n num_beam_groups: Optional[int] = None,\n diversity_penalty: Optional[float] = None,\n # `prefix_allowed_tokens_fn` can be improved on 2 aspects\n # 1. It is useful only for scenarios where the result of applying constraints is zero-ing of logits.\n # We can make this more flexible by specifying a better masking strategy. For this, we can take as arg\n # the LogitsProcessor instance itself that is customized as per the requirement\n # 2. The function supplied is expected to already have knowledge of the batch of input. It takes as input\n # the index in the batch and performs an op using that. This can be abstracted away from the API user and done\n # under the hoods\n prefix_allowed_tokens_fn: Optional[Callable[[int, torch.Tensor], List[int]]] = None,\n output_attentions: Optional[bool] = None,\n output_hidden_states: Optional[bool] = None,\n output_scores: Optional[bool] = None,\n return_dict_in_generate: Optional[bool] = None,\n forced_bos_token_id: Optional[int] = None,\n forced_eos_token_id: Optional[int] = None,\n disable_length_normalization: Optional[bool] = None,\n custom_logits_processors: Optional[List[LogitsProcessor]] = None,\n **model_kwargs,\n ) -> Union[GreedySearchOutput, SampleOutput, BeamSearchOutput, BeamSampleOutput, torch.LongTensor]:\n r\"\"\"\n Generates sequences for models with a language modeling head. The method currently supports greedy decoding,\n multinomial sampling, beam-search decoding, and beam-search multinomial sampling.\n\n Apart from :obj:`input_ids` and :obj:`attention_mask`, all the arguments below will default to the value of the\n attribute of the same name inside the :class:`~transformers.PretrainedConfig` of the model. The default values\n indicated are the default values of those config.\n\n Most of these parameters are explained in more detail in `this blog post\n <https://huggingface.co/blog/how-to-generate>`__.\n\n Parameters:\n\n input_ids (:obj:`torch.LongTensor` of shape :obj:`(batch_size, sequence_length)`, `optional`):\n The sequence used as a prompt for the generation. If :obj:`None` the method initializes it as an empty\n :obj:`torch.LongTensor` of shape :obj:`(1,)`.\n max_length (:obj:`int`, `optional`, defaults to 20):\n The maximum length of the sequence to be generated.\n min_length (:obj:`int`, `optional`, defaults to 10):\n The minimum length of the sequence to be generated.\n do_sample (:obj:`bool`, `optional`, defaults to :obj:`False`):\n Whether or not to use sampling ; use greedy decoding otherwise.\n early_stopping (:obj:`bool`, `optional`, defaults to :obj:`False`):\n Whether to stop the beam search when at least ``num_beams`` sentences are finished per batch or not.\n num_beams (:obj:`int`, `optional`, defaults to 1):\n Number of beams for beam search. 1 means no beam search.\n temperature (:obj:`float`, `optional`, defaults tp 1.0):\n The value used to module the next token probabilities.\n top_k (:obj:`int`, `optional`, defaults to 50):\n The number of highest probability vocabulary tokens to keep for top-k-filtering.\n top_p (:obj:`float`, `optional`, defaults to 1.0):\n If set to float < 1, only the most probable tokens with probabilities that add up to :obj:`top_p` or\n higher are kept for generation.\n repetition_penalty (:obj:`float`, `optional`, defaults to 1.0):\n The parameter for repetition penalty. 1.0 means no penalty. See `this paper\n <https://arxiv.org/pdf/1909.05858.pdf>`__ for more details.\n pad_token_id (:obj:`int`, `optional`):\n The id of the `padding` token.\n bos_token_id (:obj:`int`, `optional`):\n The id of the `beginning-of-sequence` token.\n eos_token_id (:obj:`int`, `optional`):\n The id of the `end-of-sequence` token.\n length_penalty (:obj:`float`, `optional`, defaults to 1.0):\n Exponential penalty to the length. 1.0 means no penalty. Set to values < 1.0 in order to encourage the\n model to generate shorter sequences, to a value > 1.0 in order to encourage the model to produce longer\n sequences.\n no_repeat_ngram_size (:obj:`int`, `optional`, defaults to 0):\n If set to int > 0, all ngrams of that size can only occur once.\n encoder_no_repeat_ngram_size (:obj:`int`, `optional`, defaults to 0):\n If set to int > 0, all ngrams of that size that occur in the ``encoder_input_ids`` cannot occur in the\n ``decoder_input_ids``.\n bad_words_ids(:obj:`List[List[int]]`, `optional`):\n List of token ids that are not allowed to be generated. In order to get the tokens of the words that\n should not appear in the generated text, use :obj:`tokenizer(bad_word,\n add_prefix_space=True).input_ids`.\n num_return_sequences(:obj:`int`, `optional`, defaults to 1):\n The number of independently computed returned sequences for each element in the batch.\n attention_mask (:obj:`torch.LongTensor` of shape :obj:`(batch_size, sequence_length)`, `optional`):\n Mask to avoid performing attention on padding token indices. Mask values are in ``[0, 1]``, 1 for\n tokens that are not masked, and 0 for masked tokens. If not provided, will default to a tensor the same\n shape as :obj:`input_ids` that masks the pad token. `What are attention masks?\n <../glossary.html#attention-mask>`__\n decoder_start_token_id (:obj:`int`, `optional`):\n If an encoder-decoder model starts decoding with a different token than `bos`, the id of that token.\n use_cache: (:obj:`bool`, `optional`, defaults to :obj:`True`):\n Whether or not the model should use the past last key/values attentions (if applicable to the model) to\n speed up decoding.\n num_beam_groups (:obj:`int`, `optional`, defaults to 1):\n Number of groups to divide :obj:`num_beams` into in order to ensure diversity among different groups of\n beams. `this paper <https://arxiv.org/pdf/1610.02424.pdf>`__ for more details.\n diversity_penalty (:obj:`float`, `optional`, defaults to 0.0):\n This value is subtracted from a beam's score if it generates a token same as any beam from other group\n at a particular time. Note that :obj:`diversity_penalty` is only effective if ``group beam search`` is\n enabled.\n prefix_allowed_tokens_fn: (:obj:`Callable[[int, torch.Tensor], List[int]]`, `optional`):\n If provided, this function constraints the beam search to allowed tokens only at each step. If not\n provided no constraint is applied. This function takes 2 arguments :obj:`inputs_ids` and the batch ID\n :obj:`batch_id`. It has to return a list with the allowed tokens for the next generation step\n conditioned on the previously generated tokens :obj:`inputs_ids` and the batch ID :obj:`batch_id`. This\n argument is useful for constrained generation conditioned on the prefix, as described in\n `Autoregressive Entity Retrieval <https://arxiv.org/abs/2010.00904>`__.\n output_attentions (:obj:`bool`, `optional`, defaults to `False`):\n Whether or not to return the attentions tensors of all attention layers. See ``attentions`` under\n returned tensors for more details.\n output_hidden_states (:obj:`bool`, `optional`, defaults to `False`):\n Whether or not to return trhe hidden states of all layers. See ``hidden_states`` under returned tensors\n for more details.\n output_scores (:obj:`bool`, `optional`, defaults to `False`):\n Whether or not to return the prediction scores. See ``scores`` under returned tensors for more details.\n return_dict_in_generate (:obj:`bool`, `optional`, defaults to `False`):\n Whether or not to return a :class:`~transformers.file_utils.ModelOutput` instead of a plain tuple.\n forced_bos_token_id (:obj:`int`, `optional`):\n The id of the token to force as the first generated token after the :obj:`decoder_start_token_id`.\n Useful for multilingual models like :doc:`mBART <../model_doc/mbart>` where the first generated token\n needs to be the target language token.\n forced_eos_token_id (:obj:`int`, `optional`):\n The id of the token to force as the last generated token when :obj:`max_length` is reached.\n disable_length_normalization (:obj:`bool`, `optional`, defaults to `False`):\n Whether or not the default length normalization during decoding should be disabled\n custom_logits_processors (:obj:`List[LogitsProcessor]`, `optional`, defaults to `None`)\n A list of logits processors to perform custom logic during generation\n\n model_kwargs:\n Additional model specific kwargs will be forwarded to the :obj:`forward` function of the model. If the\n model is an encoder-decoder model, encoder specific kwargs should not be prefixed and decoder specific\n kwargs should be prefixed with `decoder_`.\n\n Return:\n :class:`~transformers.file_utils.ModelOutput` or :obj:`torch.LongTensor`: A\n :class:`~transformers.file_utils.ModelOutput` (if ``return_dict_in_generate=True`` or when\n ``config.return_dict_in_generate=True``) or a :obj:`torch.FloatTensor`.\n\n If the model is `not` an encoder-decoder model (``model.config.is_encoder_decoder=False``), the\n possible :class:`~transformers.file_utils.ModelOutput` types are:\n\n - :class:`~transformers.generation_utils.GreedySearchDecoderOnlyOutput`,\n - :class:`~transformers.generation_utils.SampleDecoderOnlyOutput`,\n - :class:`~transformers.generation_utils.BeamSearchDecoderOnlyOutput`,\n - :class:`~transformers.generation_utils.BeamSampleDecoderOnlyOutput`\n\n If the model is an encoder-decoder model (``model.config.is_encoder_decoder=True``), the possible\n :class:`~transformers.file_utils.ModelOutput` types are:\n\n - :class:`~transformers.generation_utils.GreedySearchEncoderDecoderOutput`,\n - :class:`~transformers.generation_utils.SampleEncoderDecoderOutput`,\n - :class:`~transformers.generation_utils.BeamSearchEncoderDecoderOutput`,\n - :class:`~transformers.generation_utils.BeamSampleEncoderDecoderOutput`\n\n Examples::\n >>> from transformers import AutoTokenizer, AutoModelForCausalLM, AutoModelForSeq2SeqLM\n\n >>> tokenizer = AutoTokenizer.from_pretrained(\"distilgpt2\")\n >>> model = AutoModelForCausalLM.from_pretrained(\"distilgpt2\")\n >>> # do greedy decoding without providing a prompt\n >>> outputs = model.generate(max_length=40)\n >>> print(\"Generated:\", tokenizer.decode(outputs[0], skip_special_tokens=True))\n\n >>> tokenizer = AutoTokenizer.from_pretrained(\"t5-base\")\n >>> model = AutoModelForSeq2SeqLM.from_pretrained(\"t5-base\")\n >>> document = (\n ... \"at least two people were killed in a suspected bomb attack on a passenger bus \"\n ... \"in the strife-torn southern philippines on monday , the military said.\"\n ... )\n >>> # encode input contex\n >>> input_ids = tokenizer(document, return_tensors=\"pt\").input_ids\n >>> # generate 3 independent sequences using beam search decoding (5 beams)\n >>> # with T5 encoder-decoder model conditioned on short news article.\n >>> outputs = model.generate(input_ids=input_ids, num_beams=5, num_return_sequences=3)\n >>> print(\"Generated:\", tokenizer.batch_decode(outputs, skip_special_tokens=True))\n\n >>> tokenizer = AutoTokenizer.from_pretrained(\"distilgpt2\")\n >>> model = AutoModelForCausalLM.from_pretrained(\"distilgpt2\")\n >>> input_context = \"The dog\"\n >>> # encode input context\n >>> input_ids = tokenizer(input_context, return_tensors=\"pt\").input_ids\n >>> # generate 3 candidates using sampling\n >>> outputs = model.generate(input_ids=input_ids, max_length=20, num_return_sequences=3, do_sample=True)\n >>> print(\"Generated:\", tokenizer.batch_decode(outputs, skip_special_tokens=True))\n\n >>> tokenizer = AutoTokenizer.from_pretrained(\"ctrl\")\n >>> model = AutoModelForCausalLM.from_pretrained(\"ctrl\")\n >>> # \"Legal\" is one of the control codes for ctrl\n >>> input_context = \"Legal My neighbor is\"\n >>> # encode input context\n >>> input_ids = tokenizer(input_context, return_tensors=\"pt\").input_ids\n >>> outputs = model.generate(input_ids=input_ids, max_length=20, repetition_penalty=1.2)\n >>> print(\"Generated:\", tokenizer.decode(outputs[0], skip_special_tokens=True))\n\n >>> tokenizer = AutoTokenizer.from_pretrained(\"gpt2\")\n >>> model = AutoModelForCausalLM.from_pretrained(\"gpt2\")\n >>> input_context = \"My cute dog\"\n >>> # get tokens of words that should not be generated\n >>> bad_words_ids = [tokenizer(bad_word, add_prefix_space=True).input_ids for bad_word in [\"idiot\", \"stupid\", \"shut up\"]]\n >>> # encode input context\n >>> input_ids = tokenizer(input_context, return_tensors=\"pt\").input_ids\n >>> # generate sequences without allowing bad_words to be generated\n >>> outputs = model.generate(input_ids=input_ids, max_length=20, do_sample=True, bad_words_ids=bad_words_ids)\n >>> print(\"Generated:\", tokenizer.decode(outputs[0], skip_special_tokens=True))\n \"\"\"\n\n # set init values\n num_beams = num_beams if num_beams is not None else self.config.num_beams\n num_beam_groups = num_beam_groups if num_beam_groups is not None else self.config.num_beam_groups\n max_length = max_length if max_length is not None else self.config.max_length\n do_sample = do_sample if do_sample is not None else self.config.do_sample\n num_return_sequences = (\n num_return_sequences if num_return_sequences is not None else self.config.num_return_sequences\n )\n if disable_length_normalization is None:\n # If there is a beam score callback, it is our custom code so disable length normalization\n # TODO change ec-ml code to send in the flag directly and remove this condition\n disable_length_normalization = model_kwargs.get(\"decoder_callback_beam_scores\") is not None\n\n pad_token_id = pad_token_id if pad_token_id is not None else self.config.pad_token_id\n bos_token_id = bos_token_id if bos_token_id is not None else self.config.bos_token_id\n eos_token_id = eos_token_id if eos_token_id is not None else self.config.eos_token_id\n\n output_scores = output_scores if output_scores is not None else self.config.output_scores\n output_attentions = output_attentions if output_attentions is not None else self.config.output_attentions\n output_hidden_states = (\n output_hidden_states if output_hidden_states is not None else self.config.output_hidden_states\n )\n return_dict_in_generate = (\n return_dict_in_generate if return_dict_in_generate is not None else self.config.return_dict_in_generate\n )\n custom_logits_processors = custom_logits_processors if custom_logits_processors is not None else []\n\n model_kwargs[\"output_attentions\"] = output_attentions\n model_kwargs[\"output_hidden_states\"] = output_hidden_states\n\n if input_ids is None:\n # init `input_ids` with bos_token_id\n input_ids = self._prepare_input_ids_for_generation(bos_token_id)\n\n if model_kwargs.get(\"attention_mask\", None) is None:\n # init `attention_mask` depending on `pad_token_id`\n model_kwargs[\"attention_mask\"] = self._prepare_attention_mask_for_generation(\n input_ids, pad_token_id, eos_token_id\n )\n\n # special case if pad_token_id is not defined\n if pad_token_id is None and eos_token_id is not None:\n logger.warning(f\"Setting `pad_token_id` to `eos_token_id`:{eos_token_id} for open-end generation.\")\n pad_token_id = eos_token_id\n\n # Storing encoder_input_ids for logits_processor that could use them\n encoder_input_ids = input_ids if self.config.is_encoder_decoder else None\n\n if self.config.is_encoder_decoder:\n # add encoder_outputs to model_kwargs\n model_kwargs = self._prepare_encoder_decoder_kwargs_for_generation(input_ids, model_kwargs)\n\n # set input_ids as decoder_input_ids\n if \"decoder_input_ids\" in model_kwargs:\n input_ids = model_kwargs.pop(\"decoder_input_ids\")\n else:\n input_ids = self._prepare_decoder_input_ids_for_generation(\n input_ids, decoder_start_token_id=decoder_start_token_id, bos_token_id=bos_token_id\n )\n\n if \"encoder_outputs\" not in model_kwargs or not isinstance(model_kwargs[\"encoder_outputs\"], ModelOutput):\n raise ValueError(\"Make sure that `model_kwargs` include `encoder_outputs` of type `ModelOutput`.\")\n\n if input_ids.shape[-1] >= max_length:\n input_ids_string = \"decoder_input_ids\" if self.config.is_encoder_decoder else \"input_ids\"\n logger.warning(\n f\"Input length of {input_ids_string} is {input_ids.shape[-1]}, but ``max_length`` is set to {max_length}.\"\n \"This can lead to unexpected behavior. You should consider increasing ``config.max_length`` or ``max_length``.\"\n )\n\n # determine generation mode\n is_greedy_gen_mode = (num_beams == 1) and (num_beam_groups == 1) and do_sample is False\n is_sample_gen_mode = (num_beams == 1) and (num_beam_groups == 1) and do_sample is True\n is_beam_gen_mode = (num_beams > 1) and (num_beam_groups == 1) and do_sample is False\n is_beam_sample_gen_mode = (num_beams > 1) and (num_beam_groups == 1) and do_sample is True\n is_group_beam_gen_mode = (num_beams > 1) and (num_beam_groups > 1)\n if num_beam_groups > num_beams:\n raise ValueError(\"`num_beam_groups` has to be smaller or equal to `num_beams`\")\n if is_group_beam_gen_mode and do_sample is True:\n raise ValueError(\n \"Diverse beam search cannot be used in sampling mode. Make sure that `do_sample` is set to `False`.\"\n )\n\n # set model_kwargs\n model_kwargs[\"use_cache\"] = use_cache\n\n # get distribution pre_processing samplers\n logits_processor = self._get_logits_processor(\n repetition_penalty=repetition_penalty,\n no_repeat_ngram_size=no_repeat_ngram_size,\n encoder_no_repeat_ngram_size=encoder_no_repeat_ngram_size,\n encoder_input_ids=encoder_input_ids,\n bad_words_ids=bad_words_ids,\n min_length=min_length,\n max_length=max_length,\n eos_token_id=eos_token_id,\n forced_bos_token_id=forced_bos_token_id,\n forced_eos_token_id=forced_eos_token_id,\n prefix_allowed_tokens_fn=prefix_allowed_tokens_fn,\n num_beams=num_beams,\n num_beam_groups=num_beam_groups,\n diversity_penalty=diversity_penalty,\n )\n # Provide the custom processors the batch of text as context info\n for custom_processor in custom_logits_processors:\n if isinstance(custom_processor, ConstrainedDecodingLogitsProcessor):\n custom_processor.set_beam_size(num_beams)\n custom_processor.set_batch_text(encoder_input_ids)\n\n logits_processor.extend(custom_logits_processors)\n\n if is_greedy_gen_mode:\n if num_return_sequences > 1:\n raise ValueError(\n f\"num_return_sequences has to be 1, but is {num_return_sequences} when doing greedy search.\"\n )\n\n # greedy search\n return self.greedy_search(\n input_ids,\n logits_processor=logits_processor,\n max_length=max_length,\n pad_token_id=pad_token_id,\n eos_token_id=eos_token_id,\n output_scores=output_scores,\n return_dict_in_generate=return_dict_in_generate,\n callback_handle=model_kwargs.get(\"decoder_callback_beam_scores\"),\n **model_kwargs,\n )\n\n elif is_sample_gen_mode:\n # get probability distribution warper\n logits_warper = self._get_logits_warper(\n top_k=top_k, top_p=top_p, temperature=temperature, num_beams=num_beams\n )\n\n # expand input_ids with `num_return_sequences` additional sequences per batch\n input_ids, model_kwargs = self._expand_inputs_for_generation(\n input_ids,\n expand_size=num_return_sequences,\n is_encoder_decoder=self.config.is_encoder_decoder,\n **model_kwargs,\n )\n\n # sample\n return self.sample(\n input_ids,\n logits_processor=logits_processor,\n logits_warper=logits_warper,\n max_length=max_length,\n pad_token_id=pad_token_id,\n eos_token_id=eos_token_id,\n output_scores=output_scores,\n return_dict_in_generate=return_dict_in_generate,\n **model_kwargs,\n )\n\n elif is_beam_gen_mode:\n batch_size = input_ids.shape[0]\n\n length_penalty = length_penalty if length_penalty is not None else self.config.length_penalty\n early_stopping = early_stopping if early_stopping is not None else self.config.early_stopping\n\n if num_return_sequences > num_beams:\n raise ValueError(\"`num_return_sequences` has to be smaller or equal to `num_beams`.\")\n\n beam_scorer = BeamSearchScorer(\n batch_size=batch_size,\n max_length=max_length,\n num_beams=num_beams,\n device=self.device,\n length_penalty=length_penalty,\n do_early_stopping=early_stopping,\n num_beam_hyps_to_keep=num_return_sequences,\n disable_length_normalization=disable_length_normalization,\n )\n # interleave with `num_beams`\n input_ids, model_kwargs = self._expand_inputs_for_generation(\n input_ids, expand_size=num_beams, is_encoder_decoder=self.config.is_encoder_decoder, **model_kwargs\n )\n return self.beam_search(\n input_ids,\n beam_scorer,\n logits_processor=logits_processor,\n max_length=max_length,\n pad_token_id=pad_token_id,\n eos_token_id=eos_token_id,\n output_scores=output_scores,\n return_dict_in_generate=return_dict_in_generate,\n callback_handle=model_kwargs.get(\"decoder_callback_beam_scores\"),\n **model_kwargs,\n )\n\n elif is_beam_sample_gen_mode:\n logits_warper = self._get_logits_warper(\n top_k=top_k, top_p=top_p, temperature=temperature, num_beams=num_beams\n )\n\n batch_size = input_ids.shape[0] * num_return_sequences\n\n length_penalty = length_penalty if length_penalty is not None else self.config.length_penalty\n beam_scorer = BeamSearchScorer(\n batch_size=batch_size,\n max_length=max_length,\n num_beams=num_beams,\n device=self.device,\n length_penalty=length_penalty,\n do_early_stopping=early_stopping,\n disable_length_normalization=disable_length_normalization,\n )\n\n # interleave with `num_beams * num_return_sequences`\n input_ids, model_kwargs = self._expand_inputs_for_generation(\n input_ids,\n expand_size=num_beams * num_return_sequences,\n is_encoder_decoder=self.config.is_encoder_decoder,\n **model_kwargs,\n )\n\n return self.beam_sample(\n input_ids,\n beam_scorer,\n logits_processor=logits_processor,\n logits_warper=logits_warper,\n max_length=max_length,\n pad_token_id=pad_token_id,\n eos_token_id=eos_token_id,\n output_scores=output_scores,\n return_dict_in_generate=return_dict_in_generate,\n callback_handle=model_kwargs.get(\"decoder_callback_beam_scores\"),\n **model_kwargs,\n )\n\n elif is_group_beam_gen_mode:\n batch_size = input_ids.shape[0]\n\n length_penalty = length_penalty if length_penalty is not None else self.config.length_penalty\n early_stopping = early_stopping if early_stopping is not None else self.config.early_stopping\n\n if num_return_sequences > num_beams:\n raise ValueError(\"`num_return_sequences` has to be smaller or equal to `num_beams`.\")\n\n if num_beams % num_beam_groups != 0:\n raise ValueError(\"`num_beams` should be divisible by `num_beam_groups` for group beam search.\")\n\n diverse_beam_scorer = BeamSearchScorer(\n batch_size=batch_size,\n max_length=max_length,\n num_beams=num_beams,\n device=self.device,\n length_penalty=length_penalty,\n do_early_stopping=early_stopping,\n num_beam_hyps_to_keep=num_return_sequences,\n num_beam_groups=num_beam_groups,\n disable_length_normalization=disable_length_normalization,\n )\n # interleave with `num_beams`\n input_ids, model_kwargs = self._expand_inputs_for_generation(\n input_ids, expand_size=num_beams, is_encoder_decoder=self.config.is_encoder_decoder, **model_kwargs\n )\n return self.group_beam_search(\n input_ids,\n diverse_beam_scorer,\n logits_processor=logits_processor,\n max_length=max_length,\n pad_token_id=pad_token_id,\n eos_token_id=eos_token_id,\n output_scores=output_scores,\n return_dict_in_generate=return_dict_in_generate,\n callback_handle=model_kwargs.get(\"decoder_callback_beam_scores\"),\n **model_kwargs,\n )\n\n def greedy_search(\n self,\n input_ids: torch.LongTensor,\n logits_processor: Optional[LogitsProcessorList] = None,\n max_length: Optional[int] = None,\n pad_token_id: Optional[int] = None,\n eos_token_id: Optional[int] = None,\n output_attentions: Optional[bool] = None,\n output_hidden_states: Optional[bool] = None,\n output_scores: Optional[bool] = None,\n return_dict_in_generate: Optional[bool] = None,\n callback_handle: Optional = None,\n **model_kwargs,\n ) -> Union[GreedySearchOutput, torch.LongTensor]:\n r\"\"\"\n Generates sequences for models with a language modeling head using greedy decoding.\n\n\n\n Parameters:\n\n input_ids (:obj:`torch.LongTensor` of shape :obj:`(batch_size, sequence_length)`, `optional`):\n The sequence used as a prompt for the generation. If :obj:`None` the method initializes it as an empty\n :obj:`torch.LongTensor` of shape :obj:`(1,)`.\n logits_processor (:obj:`LogitsProcessorList`, `optional`):\n An instance of :class:`~transformers.LogitsProcessorList`. List of instances of class derived from\n :class:`~transformers.LogitsProcessor` used to modify the prediction scores of the language modeling\n head applied at each generation step.\n max_length (:obj:`int`, `optional`, defaults to 20):\n The maximum length of the sequence to be generated.\n pad_token_id (:obj:`int`, `optional`):\n The id of the `padding` token.\n eos_token_id (:obj:`int`, `optional`):\n The id of the `end-of-sequence` token.\n output_attentions (:obj:`bool`, `optional`, defaults to `False`):\n Whether or not to return the attentions tensors of all attention layers. See ``attentions`` under\n returned tensors for more details.\n output_hidden_states (:obj:`bool`, `optional`, defaults to `False`):\n Whether or not to return trhe hidden states of all layers. See ``hidden_states`` under returned tensors\n for more details.\n output_scores (:obj:`bool`, `optional`, defaults to `False`):\n Whether or not to return the prediction scores. See ``scores`` under returned tensors for more details.\n return_dict_in_generate (:obj:`bool`, `optional`, defaults to `False`):\n Whether or not to return a :class:`~transformers.file_utils.ModelOutput` instead of a plain tuple.\n\n model_kwargs:\n Additional model specific keyword arguments will be forwarded to the :obj:`forward` function of the\n model. If model is an encoder-decoder model the kwargs should include :obj:`encoder_outputs`.\n\n Return:\n :class:`~transformers.generation_utils.GreedySearchDecoderOnlyOutput`,\n :class:`~transformers.generation_utils.GreedySearchEncoderDecoderOutput` or obj:`torch.LongTensor`: A\n :obj:`torch.LongTensor` containing the generated tokens (default behaviour) or a\n :class:`~transformers.generation_utils.GreedySearchDecoderOnlyOutput` if\n ``model.config.is_encoder_decoder=False`` and ``return_dict_in_generate=True`` or a\n :class:`~transformers.generation_utils.GreedySearchEncoderDecoderOutput` if\n ``model.config.is_encoder_decoder=True``.\n\n Examples::\n\n >>> from transformers import (\n ... AutoTokenizer,\n ... AutoModelForCausalLM,\n ... LogitsProcessorList,\n ... MinLengthLogitsProcessor,\n ... )\n\n >>> tokenizer = AutoTokenizer.from_pretrained(\"gpt2\")\n >>> model = AutoModelForCausalLM.from_pretrained(\"gpt2\")\n\n >>> # set pad_token_id to eos_token_id because GPT2 does not have a EOS token\n >>> model.config.pad_token_id = model.config.eos_token_id\n\n >>> input_prompt = \"Today is a beautiful day, and\"\n >>> input_ids = tokenizer(input_prompt, return_tensors=\"pt\").input_ids\n\n >>> # instantiate logits processors\n >>> logits_processor = LogitsProcessorList([\n ... MinLengthLogitsProcessor(15, eos_token_id=model.config.eos_token_id),\n ... ])\n\n >>> outputs = model.greedy_search(input_ids, logits_processor=logits_processor)\n\n >>> print(\"Generated:\", tokenizer.batch_decode(outputs, skip_special_tokens=True))\n \"\"\"\n # init values\n logits_processor = logits_processor if logits_processor is not None else LogitsProcessorList()\n max_length = max_length if max_length is not None else self.config.max_length\n pad_token_id = pad_token_id if pad_token_id is not None else self.config.pad_token_id\n eos_token_id = eos_token_id if eos_token_id is not None else self.config.eos_token_id\n output_scores = output_scores if output_scores is not None else self.config.output_scores\n output_attentions = output_attentions if output_attentions is not None else self.config.output_attentions\n output_hidden_states = (\n output_hidden_states if output_hidden_states is not None else self.config.output_hidden_states\n )\n return_dict_in_generate = (\n return_dict_in_generate if return_dict_in_generate is not None else self.config.return_dict_in_generate\n )\n\n # init attention / hidden states / scores tuples\n scores = () if (return_dict_in_generate and output_scores) else None\n decoder_attentions = () if (return_dict_in_generate and output_attentions) else None\n cross_attentions = () if (return_dict_in_generate and output_attentions) else None\n decoder_hidden_states = () if (return_dict_in_generate and output_hidden_states) else None\n\n # if model is an encoder-decoder, retrieve encoder attention weights and hidden states\n if return_dict_in_generate and self.config.is_encoder_decoder:\n encoder_attentions = model_kwargs[\"encoder_outputs\"].get(\"attentions\") if output_attentions else None\n encoder_hidden_states = (\n model_kwargs[\"encoder_outputs\"].get(\"hidden_states\") if output_hidden_states else None\n )\n\n # init sequence length tensors\n sequence_lengths, unfinished_sequences, cur_len = self._init_sequence_length_for_generation(\n input_ids, max_length\n )\n generation_score = torch.zeros(input_ids.shape[0], dtype=torch.float, device=input_ids.device)\n\n while cur_len < max_length:\n # prepare model inputs\n model_inputs = self.prepare_inputs_for_generation(input_ids, **model_kwargs)\n\n # forward pass to get next token\n outputs = self(\n **model_inputs,\n return_dict=True,\n output_attentions=output_attentions,\n output_hidden_states=output_hidden_states,\n )\n next_token_logits = outputs.logits[:, -1, :]\n\n # Store scores, attentions and hidden_states when required\n if return_dict_in_generate:\n if output_scores:\n scores += (next_token_logits,)\n if output_attentions:\n decoder_attentions += (\n (outputs.decoder_attentions,) if self.config.is_encoder_decoder else (outputs.attentions,)\n )\n if self.config.is_encoder_decoder:\n cross_attentions += (outputs.cross_attentions,)\n\n if output_hidden_states:\n decoder_hidden_states += (\n (outputs.decoder_hidden_states,)\n if self.config.is_encoder_decoder\n else (outputs.hidden_states,)\n )\n\n # pre-process distribution\n next_tokens_scores = F.log_softmax(next_token_logits, dim=-1)\n next_tokens_scores = logits_processor(input_ids, next_tokens_scores)\n next_tokens_scores = next_tokens_scores + generation_score[:, None].expand_as(next_tokens_scores)\n\n # argmax\n next_tokens_scores, next_tokens = torch.topk(next_tokens_scores, 1, dim=-1, largest=True, sorted=True)\n next_tokens_scores = next_tokens_scores.squeeze(dim=-1)\n next_tokens = next_tokens.squeeze(dim=-1)\n generation_score = next_tokens_scores\n\n # add code that transfomers next_tokens to tokens_to_add\n if eos_token_id is not None:\n assert pad_token_id is not None, \"If eos_token_id is defined, make sure that pad_token_id is defined.\"\n next_tokens = next_tokens * unfinished_sequences + (pad_token_id) * (1 - unfinished_sequences)\n\n # add token and increase length by one\n input_ids = torch.cat([input_ids, next_tokens[:, None]], dim=-1)\n\n # update sequence length\n if eos_token_id is not None:\n sequence_lengths, unfinished_sequences = self._update_seq_length_for_generation(\n sequence_lengths, unfinished_sequences, cur_len, next_tokens == eos_token_id\n )\n\n # update model kwargs\n model_kwargs = self._update_model_kwargs_for_generation(\n outputs, model_kwargs, is_encoder_decoder=self.config.is_encoder_decoder\n )\n\n # stop when there is a </s> in each sentence, or if we exceed the maximum length\n if unfinished_sequences.max() == 0:\n break\n\n # increase cur_len\n cur_len = cur_len + 1\n\n if callback_handle:\n # Formatting it the same way as beam_hyp for consistency\n callback_input = list()\n for score, ids in zip(generation_score, input_ids):\n callback_input.append([(\n score.item(),\n ids.clone(),\n )])\n for idx, callback_input_instance in enumerate(callback_input):\n callback_handle(callback_input_instance, idx, **model_kwargs)\n if return_dict_in_generate:\n if self.config.is_encoder_decoder:\n return GreedySearchEncoderDecoderOutput(\n sequences=input_ids,\n scores=scores,\n encoder_attentions=encoder_attentions,\n encoder_hidden_states=encoder_hidden_states,\n decoder_attentions=decoder_attentions,\n cross_attentions=cross_attentions,\n decoder_hidden_states=decoder_hidden_states,\n )\n else:\n return GreedySearchDecoderOnlyOutput(\n sequences=input_ids,\n scores=scores,\n attentions=decoder_attentions,\n hidden_states=decoder_hidden_states,\n )\n else:\n return input_ids\n\n def sample(\n self,\n input_ids: torch.LongTensor,\n logits_processor: Optional[LogitsProcessorList] = None,\n logits_warper: Optional[LogitsProcessorList] = None,\n max_length: Optional[int] = None,\n pad_token_id: Optional[int] = None,\n eos_token_id: Optional[int] = None,\n output_attentions: Optional[bool] = None,\n output_hidden_states: Optional[bool] = None,\n output_scores: Optional[bool] = None,\n return_dict_in_generate: Optional[bool] = None,\n **model_kwargs,\n ) -> Union[SampleOutput, torch.LongTensor]:\n r\"\"\"\n Generates sequences for models with a language modeling head using multinomial sampling.\n\n Parameters:\n\n input_ids (:obj:`torch.LongTensor` of shape :obj:`(batch_size, sequence_length)`, `optional`):\n The sequence used as a prompt for the generation. If :obj:`None` the method initializes it as an empty\n :obj:`torch.LongTensor` of shape :obj:`(1,)`.\n logits_processor (:obj:`LogitsProcessorList`, `optional`):\n An instance of :class:`~transformers.LogitsProcessorList`. List of instances of class derived from\n :class:`~transformers.LogitsProcessor` used to modify the prediction scores of the language modeling\n head applied at each generation step.\n logits_warper (:obj:`LogitsProcessorList`, `optional`):\n An instance of :class:`~transformers.LogitsProcessorList`. List of instances of class derived from\n :class:`~transformers.LogitsWarper` used to warp the prediction score distribution of the language\n modeling head applied before multinomial sampling at each generation step.\n max_length (:obj:`int`, `optional`, defaults to 20):\n The maximum length of the sequence to be generated.\n pad_token_id (:obj:`int`, `optional`):\n The id of the `padding` token.\n eos_token_id (:obj:`int`, `optional`):\n The id of the `end-of-sequence` token.\n output_attentions (:obj:`bool`, `optional`, defaults to `False`):\n Whether or not to return the attentions tensors of all attention layers. See ``attentions`` under\n returned tensors for more details.\n output_hidden_states (:obj:`bool`, `optional`, defaults to `False`):\n Whether or not to return trhe hidden states of all layers. See ``hidden_states`` under returned tensors\n for more details.\n output_scores (:obj:`bool`, `optional`, defaults to `False`):\n Whether or not to return the prediction scores. See ``scores`` under returned tensors for more details.\n return_dict_in_generate (:obj:`bool`, `optional`, defaults to `False`):\n Whether or not to return a :class:`~transformers.file_utils.ModelOutput` instead of a plain tuple.\n model_kwargs:\n Additional model specific kwargs will be forwarded to the :obj:`forward` function of the model. If\n model is an encoder-decoder model the kwargs should include :obj:`encoder_outputs`.\n\n Return:\n :class:`~transformers.generation_utils.SampleDecoderOnlyOutput`,\n :class:`~transformers.generation_utils.SampleEncoderDecoderOutput` or obj:`torch.LongTensor`: A\n :obj:`torch.LongTensor` containing the generated tokens (default behaviour) or a\n :class:`~transformers.generation_utils.SampleDecoderOnlyOutput` if\n ``model.config.is_encoder_decoder=False`` and ``return_dict_in_generate=True`` or a\n :class:`~transformers.generation_utils.SampleEncoderDecoderOutput` if\n ``model.config.is_encoder_decoder=True``.\n\n Examples::\n\n >>> from transformers import (\n ... AutoTokenizer,\n ... AutoModelForCausalLM,\n ... LogitsProcessorList,\n ... MinLengthLogitsProcessor,\n ... TopKLogitsWarper,\n ... TemperatureLogitsWarper,\n ... )\n\n >>> tokenizer = AutoTokenizer.from_pretrained(\"gpt2\")\n >>> model = AutoModelForCausalLM.from_pretrained(\"gpt2\")\n\n >>> # set pad_token_id to eos_token_id because GPT2 does not have a EOS token\n >>> model.config.pad_token_id = model.config.eos_token_id\n\n >>> input_prompt = \"Today is a beautiful day, and\"\n >>> input_ids = tokenizer(input_prompt, return_tensors=\"pt\").input_ids\n\n >>> # instantiate logits processors\n >>> logits_processor = LogitsProcessorList([\n ... MinLengthLogitsProcessor(15, eos_token_id=model.config.eos_token_id),\n ... ])\n >>> # instantiate logits processors\n >>> logits_warper = LogitsProcessorList([\n ... TopKLogitsWarper(50),\n ... TemperatureLogitsWarper(0.7),\n ... ])\n\n >>> outputs = model.sample(input_ids, logits_processor=logits_processor, logits_warper=logits_warper)\n\n >>> print(\"Generated:\", tokenizer.batch_decode(outputs, skip_special_tokens=True))\n \"\"\"\n\n # init values\n logits_processor = logits_processor if logits_processor is not None else LogitsProcessorList()\n logits_warper = logits_warper if logits_warper is not None else LogitsProcessorList()\n max_length = max_length if max_length is not None else self.config.max_length\n pad_token_id = pad_token_id if pad_token_id is not None else self.config.pad_token_id\n eos_token_id = eos_token_id if eos_token_id is not None else self.config.eos_token_id\n output_scores = output_scores if output_scores is not None else self.config.output_scores\n output_attentions = output_attentions if output_attentions is not None else self.config.output_attentions\n output_hidden_states = (\n output_hidden_states if output_hidden_states is not None else self.config.output_hidden_states\n )\n return_dict_in_generate = (\n return_dict_in_generate if return_dict_in_generate is not None else self.config.return_dict_in_generate\n )\n\n # init attention / hidden states / scores tuples\n scores = () if (return_dict_in_generate and output_scores) else None\n decoder_attentions = () if (return_dict_in_generate and output_attentions) else None\n cross_attentions = () if (return_dict_in_generate and output_attentions) else None\n decoder_hidden_states = () if (return_dict_in_generate and output_hidden_states) else None\n\n # if model is an encoder-decoder, retrieve encoder attention weights and hidden states\n if return_dict_in_generate and self.config.is_encoder_decoder:\n encoder_attentions = model_kwargs[\"encoder_outputs\"].get(\"attentions\") if output_attentions else None\n encoder_hidden_states = (\n model_kwargs[\"encoder_outputs\"].get(\"hidden_states\") if output_hidden_states else None\n )\n\n # init sequence length tensors\n sequence_lengths, unfinished_sequences, cur_len = self._init_sequence_length_for_generation(\n input_ids, max_length\n )\n\n # auto-regressive generation\n while cur_len < max_length:\n # prepare model inputs\n model_inputs = self.prepare_inputs_for_generation(input_ids, **model_kwargs)\n\n # forward pass to get next token\n outputs = self(\n **model_inputs,\n return_dict=True,\n output_attentions=output_attentions,\n output_hidden_states=output_hidden_states,\n )\n next_token_logits = outputs.logits[:, -1, :]\n\n # pre-process distribution\n next_token_scores = logits_processor(input_ids, next_token_logits)\n next_token_scores = logits_warper(input_ids, next_token_scores)\n\n # Store scores, attentions and hidden_states when required\n if return_dict_in_generate:\n if output_scores:\n scores += (next_token_scores,)\n if output_attentions:\n decoder_attentions += (\n (outputs.decoder_attentions,) if self.config.is_encoder_decoder else (outputs.attentions,)\n )\n if self.config.is_encoder_decoder:\n cross_attentions += (outputs.cross_attentions,)\n\n if output_hidden_states:\n decoder_hidden_states += (\n (outputs.decoder_hidden_states,)\n if self.config.is_encoder_decoder\n else (outputs.hidden_states,)\n )\n\n # sample\n probs = F.softmax(next_token_scores, dim=-1)\n next_tokens = torch.multinomial(probs, num_samples=1).squeeze(1)\n\n # add code that transfomers next_tokens to tokens_to_add\n if eos_token_id is not None:\n assert pad_token_id is not None, \"If eos_token_id is defined, make sure that pad_token_id is defined.\"\n next_tokens = next_tokens * unfinished_sequences + (pad_token_id) * (1 - unfinished_sequences)\n\n # add token and increase length by one\n input_ids = torch.cat([input_ids, next_tokens[:, None]], dim=-1)\n cur_len = cur_len + 1\n\n # update sequence length\n if eos_token_id is not None:\n sequence_lengths, unfinished_sequences = self._update_seq_length_for_generation(\n sequence_lengths, unfinished_sequences, cur_len, next_tokens == eos_token_id\n )\n\n # stop when there is a </s> in each sentence, or if we exceed the maximum length\n if unfinished_sequences.max() == 0:\n break\n\n # update model kwargs\n model_kwargs = self._update_model_kwargs_for_generation(\n outputs, model_kwargs, is_encoder_decoder=self.config.is_encoder_decoder\n )\n\n if return_dict_in_generate:\n if self.config.is_encoder_decoder:\n return SampleEncoderDecoderOutput(\n sequences=input_ids,\n scores=scores,\n encoder_attentions=encoder_attentions,\n encoder_hidden_states=encoder_hidden_states,\n decoder_attentions=decoder_attentions,\n cross_attentions=cross_attentions,\n decoder_hidden_states=decoder_hidden_states,\n )\n else:\n return SampleDecoderOnlyOutput(\n sequences=input_ids,\n scores=scores,\n attentions=decoder_attentions,\n hidden_states=decoder_hidden_states,\n )\n else:\n return input_ids\n\n def beam_search(\n self,\n input_ids: torch.LongTensor,\n beam_scorer: BeamScorer,\n logits_processor: Optional[LogitsProcessorList] = None,\n max_length: Optional[int] = None,\n pad_token_id: Optional[int] = None,\n eos_token_id: Optional[int] = None,\n output_attentions: Optional[bool] = None,\n output_hidden_states: Optional[bool] = None,\n output_scores: Optional[bool] = None,\n return_dict_in_generate: Optional[bool] = None,\n callback_handle: Optional = None,\n **model_kwargs,\n ) -> Union[BeamSearchOutput, torch.LongTensor]:\n r\"\"\"\n Generates sequences for models with a language modeling head using beam search decoding.\n\n Parameters:\n\n input_ids (:obj:`torch.LongTensor` of shape :obj:`(batch_size, sequence_length)`, `optional`):\n The sequence used as a prompt for the generation. If :obj:`None` the method initializes it as an empty\n :obj:`torch.LongTensor` of shape :obj:`(1,)`.\n beam_scorer (:obj:`BeamScorer`):\n An derived instance of :class:`~transformers.BeamScorer` that defines how beam hypotheses are\n constructed, stored and sorted during generation. For more information, the documentation of\n :class:`~transformers.BeamScorer` should be read.\n logits_processor (:obj:`LogitsProcessorList`, `optional`):\n An instance of :class:`~transformers.LogitsProcessorList`. List of instances of class derived from\n :class:`~transformers.LogitsProcessor` used to modify the prediction scores of the language modeling\n head applied at each generation step.\n max_length (:obj:`int`, `optional`, defaults to 20):\n The maximum length of the sequence to be generated.\n pad_token_id (:obj:`int`, `optional`):\n The id of the `padding` token.\n eos_token_id (:obj:`int`, `optional`):\n The id of the `end-of-sequence` token.\n output_attentions (:obj:`bool`, `optional`, defaults to `False`):\n Whether or not to return the attentions tensors of all attention layers. See ``attentions`` under\n returned tensors for more details.\n output_hidden_states (:obj:`bool`, `optional`, defaults to `False`):\n Whether or not to return trhe hidden states of all layers. See ``hidden_states`` under returned tensors\n for more details.\n output_scores (:obj:`bool`, `optional`, defaults to `False`):\n Whether or not to return the prediction scores. See ``scores`` under returned tensors for more details.\n return_dict_in_generate (:obj:`bool`, `optional`, defaults to `False`):\n Whether or not to return a :class:`~transformers.file_utils.ModelOutput` instead of a plain tuple.\n callback_handle (:obj:`function`, `optional`, defaults to `None`):\n Handle to a callback function if the model wants to do custom processing at each beam docoding step\n model_kwargs:\n Additional model specific kwargs will be forwarded to the :obj:`forward` function of the model. If\n model is an encoder-decoder model the kwargs should include :obj:`encoder_outputs`.\n\n Return:\n :class:`~transformers.generation_utilsBeamSearchDecoderOnlyOutput`,\n :class:`~transformers.generation_utils.BeamSearchEncoderDecoderOutput` or obj:`torch.LongTensor`: A\n :obj:`torch.LongTensor` containing the generated tokens (default behaviour) or a\n :class:`~transformers.generation_utils.BeamSearchDecoderOnlyOutput` if\n ``model.config.is_encoder_decoder=False`` and ``return_dict_in_generate=True`` or a\n :class:`~transformers.generation_utils.BeamSearchEncoderDecoderOutput` if\n ``model.config.is_encoder_decoder=True``.\n\n\n Examples::\n\n >>> from transformers import (\n ... AutoTokenizer,\n ... AutoModelForSeq2SeqLM,\n ... LogitsProcessorList,\n ... MinLengthLogitsProcessor,\n ... BeamSearchScorer,\n ... )\n >>> import torch\n\n >>> tokenizer = AutoTokenizer.from_pretrained(\"t5-base\")\n >>> model = AutoModelForSeq2SeqLM.from_pretrained(\"t5-base\")\n\n >>> encoder_input_str = \"translate English to German: How old are you?\"\n >>> encoder_input_ids = tokenizer(encoder_input_str, return_tensors=\"pt\").input_ids\n\n\n >>> # lets run beam search using 3 beams\n >>> num_beams = 3\n >>> # define decoder start token ids\n >>> input_ids = torch.ones((num_beams, 1), device=model.device, dtype=torch.long)\n >>> input_ids = input_ids * model.config.decoder_start_token_id\n\n >>> # add encoder_outputs to model keyword arguments\n >>> model_kwargs = {\n ... \"encoder_outputs\": model.get_encoder()(encoder_input_ids.repeat_interleave(num_beams, dim=0), return_dict=True)\n ... }\n\n >>> # instantiate beam scorer\n >>> beam_scorer = BeamSearchScorer(\n ... batch_size=1,\n ... max_length=model.config.max_length,\n ... num_beams=num_beams,\n ... device=model.device,\n ... )\n\n >>> # instantiate logits processors\n >>> logits_processor = LogitsProcessorList([\n ... MinLengthLogitsProcessor(5, eos_token_id=model.config.eos_token_id),\n ... ])\n\n >>> outputs = model.beam_search(input_ids, beam_scorer, logits_processor=logits_processor, **model_kwargs)\n\n >>> print(\"Generated:\", tokenizer.batch_decode(outputs, skip_special_tokens=True))\n \"\"\"\n\n # init values\n logits_processor = logits_processor if logits_processor is not None else LogitsProcessorList()\n max_length = max_length if max_length is not None else self.config.max_length\n pad_token_id = pad_token_id if pad_token_id is not None else self.config.pad_token_id\n eos_token_id = eos_token_id if eos_token_id is not None else self.config.eos_token_id\n output_scores = output_scores if output_scores is not None else self.config.output_scores\n output_attentions = output_attentions if output_attentions is not None else self.config.output_attentions\n output_hidden_states = (\n output_hidden_states if output_hidden_states is not None else self.config.output_hidden_states\n )\n return_dict_in_generate = (\n return_dict_in_generate if return_dict_in_generate is not None else self.config.return_dict_in_generate\n )\n\n # init attention / hidden states / scores tuples\n scores = () if (return_dict_in_generate and output_scores) else None\n decoder_attentions = () if (return_dict_in_generate and output_attentions) else None\n cross_attentions = () if (return_dict_in_generate and output_attentions) else None\n decoder_hidden_states = () if (return_dict_in_generate and output_hidden_states) else None\n\n # if model is an encoder-decoder, retrieve encoder attention weights and hidden states\n if return_dict_in_generate and self.config.is_encoder_decoder:\n encoder_attentions = model_kwargs[\"encoder_outputs\"].get(\"attentions\") if output_attentions else None\n encoder_hidden_states = (\n model_kwargs[\"encoder_outputs\"].get(\"hidden_states\") if output_hidden_states else None\n )\n\n batch_size = len(beam_scorer._beam_hyps)\n num_beams = beam_scorer.num_beams\n\n batch_beam_size, cur_len = input_ids.shape\n\n assert (\n num_beams * batch_size == batch_beam_size\n ), \"Batch dimension of `input_ids` should be {num_beams * batch_size}, but is {batch_beam_size}.\"\n\n beam_scores = torch.zeros((batch_size, num_beams), dtype=torch.float, device=input_ids.device)\n beam_scores[:, 1:] = -1e9\n beam_scores = beam_scores.view((batch_size * num_beams,))\n\n while cur_len < max_length:\n model_inputs = self.prepare_inputs_for_generation(input_ids, **model_kwargs)\n\n outputs = self(\n **model_inputs,\n return_dict=True,\n output_attentions=output_attentions,\n output_hidden_states=output_hidden_states,\n )\n next_token_logits = outputs.logits[:, -1, :]\n\n # hack: adjust tokens for Marian. For Marian we have to make sure that the `pad_token_id`\n # cannot be generated both before and after the `F.log_softmax` operation.\n next_token_logits = self.adjust_logits_during_generation(\n next_token_logits, cur_len=cur_len, max_length=max_length\n )\n\n next_token_scores = F.log_softmax(next_token_logits, dim=-1) # (batch_size * num_beams, vocab_size)\n\n next_token_scores = logits_processor(input_ids, next_token_scores)\n next_token_scores = next_token_scores + beam_scores[:, None].expand_as(next_token_scores)\n\n # Store scores, attentions and hidden_states when required\n if return_dict_in_generate:\n if output_scores:\n scores += (next_token_scores,)\n if output_attentions:\n decoder_attentions += (\n (outputs.decoder_attentions,) if self.config.is_encoder_decoder else (outputs.attentions,)\n )\n if self.config.is_encoder_decoder:\n cross_attentions += (outputs.cross_attentions,)\n\n if output_hidden_states:\n decoder_hidden_states += (\n (outputs.decoder_hidden_states,)\n if self.config.is_encoder_decoder\n else (outputs.hidden_states,)\n )\n\n # reshape for beam search\n vocab_size = next_token_scores.shape[-1]\n next_token_scores = next_token_scores.view(batch_size, num_beams * vocab_size)\n\n next_token_scores, next_tokens = torch.topk(\n next_token_scores, 2 * num_beams, dim=1, largest=True, sorted=True\n )\n\n next_indices = next_tokens // vocab_size\n next_tokens = next_tokens % vocab_size\n\n # stateless\n beam_outputs = beam_scorer.process(\n input_ids,\n next_token_scores,\n next_tokens,\n next_indices,\n pad_token_id=pad_token_id,\n eos_token_id=eos_token_id,\n )\n beam_scores = beam_outputs[\"next_beam_scores\"]\n beam_next_tokens = beam_outputs[\"next_beam_tokens\"]\n beam_idx = beam_outputs[\"next_beam_indices\"]\n\n input_ids = torch.cat([input_ids[beam_idx, :], beam_next_tokens.unsqueeze(-1)], dim=-1)\n\n cur_len = cur_len + 1\n\n model_kwargs = self._update_model_kwargs_for_generation(\n outputs, model_kwargs, is_encoder_decoder=self.config.is_encoder_decoder\n )\n if model_kwargs[\"past\"] is not None:\n model_kwargs[\"past\"] = self._reorder_cache(model_kwargs[\"past\"], beam_idx)\n\n if beam_scorer.is_done:\n break\n\n sequence_outputs = beam_scorer.finalize(\n input_ids, beam_scores, next_tokens, next_indices, pad_token_id=pad_token_id, eos_token_id=eos_token_id,\n callback_handle=callback_handle, **model_kwargs,\n )\n\n if return_dict_in_generate:\n if not output_scores:\n sequence_outputs[\"sequence_scores\"] = None\n if self.config.is_encoder_decoder:\n return BeamSearchEncoderDecoderOutput(\n sequences=sequence_outputs[\"sequences\"],\n sequences_scores=sequence_outputs[\"sequence_scores\"],\n scores=scores,\n encoder_attentions=encoder_attentions,\n encoder_hidden_states=encoder_hidden_states,\n decoder_attentions=decoder_attentions,\n cross_attentions=cross_attentions,\n decoder_hidden_states=decoder_hidden_states,\n )\n else:\n return BeamSearchDecoderOnlyOutput(\n sequences=sequence_outputs[\"sequences\"],\n sequences_scores=sequence_outputs[\"sequence_scores\"],\n scores=scores,\n attentions=decoder_attentions,\n hidden_states=decoder_hidden_states,\n )\n else:\n return sequence_outputs[\"sequences\"]\n\n def beam_sample(\n self,\n input_ids: torch.LongTensor,\n beam_scorer: BeamScorer,\n logits_processor: Optional[LogitsProcessorList] = None,\n logits_warper: Optional[LogitsProcessorList] = None,\n max_length: Optional[int] = None,\n pad_token_id: Optional[int] = None,\n eos_token_id: Optional[int] = None,\n output_attentions: Optional[bool] = None,\n output_hidden_states: Optional[bool] = None,\n output_scores: Optional[bool] = None,\n return_dict_in_generate: Optional[bool] = None,\n callback_handle: Optional = None,\n **model_kwargs,\n ) -> Union[BeamSampleOutput, torch.LongTensor]:\n r\"\"\"\n Generates sequences for models with a language modeling head using beam search with multinomial sampling.\n\n Parameters:\n\n input_ids (:obj:`torch.LongTensor` of shape :obj:`(batch_size, sequence_length)`, `optional`):\n The sequence used as a prompt for the generation. If :obj:`None` the method initializes it as an empty\n :obj:`torch.LongTensor` of shape :obj:`(1,)`.\n beam_scorer (:obj:`BeamScorer`):\n A derived instance of :class:`~transformers.BeamScorer` that defines how beam hypotheses are\n constructed, stored and sorted during generation. For more information, the documentation of\n :class:`~transformers.BeamScorer` should be read.\n logits_processor (:obj:`LogitsProcessorList`, `optional`):\n An instance of :class:`~transformers.LogitsProcessorList`. List of instances of class derived from\n :class:`~transformers.LogitsProcessor` used to modify the prediction scores of the language modeling\n head applied at each generation step.\n logits_warper (:obj:`LogitsProcessorList`, `optional`):\n An instance of :class:`~transformers.LogitsProcessorList`. List of instances of class derived from\n :class:`~transformers.LogitsWarper` used to warp the prediction score distribution of the language\n modeling head applied before multinomial sampling at each generation step.\n max_length (:obj:`int`, `optional`, defaults to 20):\n The maximum length of the sequence to be generated.\n pad_token_id (:obj:`int`, `optional`):\n The id of the `padding` token.\n eos_token_id (:obj:`int`, `optional`):\n The id of the `end-of-sequence` token.\n output_attentions (:obj:`bool`, `optional`, defaults to `False`):\n Whether or not to return the attentions tensors of all attention layers. See ``attentions`` under\n returned tensors for more details.\n output_hidden_states (:obj:`bool`, `optional`, defaults to `False`):\n Whether or not to return trhe hidden states of all layers. See ``hidden_states`` under returned tensors\n for more details.\n output_scores (:obj:`bool`, `optional`, defaults to `False`):\n Whether or not to return the prediction scores. See ``scores`` under returned tensors for more details.\n return_dict_in_generate (:obj:`bool`, `optional`, defaults to `False`):\n Whether or not to return a :class:`~transformers.file_utils.ModelOutput` instead of a plain tuple.\n callback_handle (:obj:`function`, `optional`, defaults to `None`):\n Handle to a callback function if the model wants to do custom processing at each beam docoding step\n model_kwargs:\n Additional model specific kwargs will be forwarded to the :obj:`forward` function of the model. If\n model is an encoder-decoder model the kwargs should include :obj:`encoder_outputs`.\n\n Return:\n :class:`~transformers.generation_utils.BeamSampleDecoderOnlyOutput`,\n :class:`~transformers.generation_utils.BeamSampleEncoderDecoderOutput` or obj:`torch.LongTensor`: A\n :obj:`torch.LongTensor` containing the generated tokens (default behaviour) or a\n :class:`~transformers.generation_utils.BeamSampleDecoderOnlyOutput` if\n ``model.config.is_encoder_decoder=False`` and ``return_dict_in_generate=True`` or a\n :class:`~transformers.generation_utils.BeamSampleEncoderDecoderOutput` if\n ``model.config.is_encoder_decoder=True``.\n\n Examples::\n\n >>> from transformers import (\n ... AutoTokenizer,\n ... AutoModelForSeq2SeqLM,\n ... LogitsProcessorList,\n ... MinLengthLogitsProcessor,\n ... TopKLogitsWarper,\n ... TemperatureLogitsWarper,\n ... BeamSearchScorer,\n ... )\n >>> import torch\n\n >>> tokenizer = AutoTokenizer.from_pretrained(\"t5-base\")\n >>> model = AutoModelForSeq2SeqLM.from_pretrained(\"t5-base\")\n\n >>> encoder_input_str = \"translate English to German: How old are you?\"\n >>> encoder_input_ids = tokenizer(encoder_input_str, return_tensors=\"pt\").input_ids\n\n >>> # lets run beam search using 3 beams\n >>> num_beams = 3\n >>> # define decoder start token ids\n >>> input_ids = torch.ones((num_beams, 1), device=model.device, dtype=torch.long)\n >>> input_ids = input_ids * model.config.decoder_start_token_id\n\n >>> # add encoder_outputs to model keyword arguments\n >>> model_kwargs = {\n ... \"encoder_outputs\": model.get_encoder()(encoder_input_ids.repeat_interleave(num_beams, dim=0), return_dict=True)\n ... }\n\n >>> # instantiate beam scorer\n >>> beam_scorer = BeamSearchScorer(\n ... batch_size=1,\n ... max_length=model.config.max_length,\n ... num_beams=num_beams,\n ... device=model.device,\n ... )\n\n >>> # instantiate logits processors\n >>> logits_processor = LogitsProcessorList([\n ... MinLengthLogitsProcessor(5, eos_token_id=model.config.eos_token_id)\n ... ])\n >>> # instantiate logits processors\n >>> logits_warper = LogitsProcessorList([\n ... TopKLogitsWarper(50),\n ... TemperatureLogitsWarper(0.7),\n ... ])\n\n >>> outputs = model.beam_sample(\n ... input_ids, beam_scorer, logits_processor=logits_processor, logits_warper=logits_warper, **model_kwargs\n ... )\n\n >>> print(\"Generated:\", tokenizer.batch_decode(outputs, skip_special_tokens=True))\n \"\"\"\n\n # init values\n logits_processor = logits_processor if logits_processor is not None else LogitsProcessorList()\n max_length = max_length if max_length is not None else self.config.max_length\n pad_token_id = pad_token_id if pad_token_id is not None else self.config.pad_token_id\n eos_token_id = eos_token_id if eos_token_id is not None else self.config.eos_token_id\n output_scores = output_scores if output_scores is not None else self.config.output_scores\n output_attentions = output_attentions if output_attentions is not None else self.config.output_attentions\n output_hidden_states = (\n output_hidden_states if output_hidden_states is not None else self.config.output_hidden_states\n )\n return_dict_in_generate = (\n return_dict_in_generate if return_dict_in_generate is not None else self.config.return_dict_in_generate\n )\n\n # init attention / hidden states / scores tuples\n scores = () if (return_dict_in_generate and output_scores) else None\n decoder_attentions = () if (return_dict_in_generate and output_attentions) else None\n cross_attentions = () if (return_dict_in_generate and output_attentions) else None\n decoder_hidden_states = () if (return_dict_in_generate and output_hidden_states) else None\n\n # if model is an encoder-decoder, retrieve encoder attention weights and hidden states\n if return_dict_in_generate and self.config.is_encoder_decoder:\n encoder_attentions = model_kwargs[\"encoder_outputs\"].get(\"attentions\") if output_attentions else None\n encoder_hidden_states = (\n model_kwargs[\"encoder_outputs\"].get(\"hidden_states\") if output_hidden_states else None\n )\n\n batch_size = len(beam_scorer._beam_hyps)\n num_beams = beam_scorer.num_beams\n\n batch_beam_size, cur_len = input_ids.shape\n\n beam_scores = torch.zeros((batch_size, num_beams), dtype=torch.float, device=input_ids.device)\n beam_scores = beam_scores.view((batch_size * num_beams,))\n\n while cur_len < max_length:\n model_inputs = self.prepare_inputs_for_generation(input_ids, **model_kwargs)\n\n outputs = self(\n **model_inputs,\n return_dict=True,\n output_attentions=output_attentions,\n output_hidden_states=output_hidden_states,\n )\n next_token_logits = outputs.logits[:, -1, :]\n\n # hack: adjust tokens for Marian. For Marian we have to make sure that the `pad_token_id`\n # cannot be generated both before and after the `F.log_softmax` operation.\n next_token_logits = self.adjust_logits_during_generation(\n next_token_logits, cur_len=cur_len, max_length=max_length\n )\n\n next_token_scores = F.log_softmax(next_token_logits, dim=-1) # (batch_size * num_beams, vocab_size)\n\n next_token_scores = logits_processor(input_ids, next_token_scores)\n next_token_scores = next_token_scores + beam_scores[:, None].expand_as(next_token_scores)\n next_token_scores = logits_warper(input_ids, next_token_scores)\n\n # Store scores, attentions and hidden_states when required\n if return_dict_in_generate:\n if output_scores:\n scores += (next_token_scores,)\n if output_attentions:\n decoder_attentions += (\n (outputs.decoder_attentions,) if self.config.is_encoder_decoder else (outputs.attentions,)\n )\n if self.config.is_encoder_decoder:\n cross_attentions += (outputs.cross_attentions,)\n\n if output_hidden_states:\n decoder_hidden_states += (\n (outputs.decoder_hidden_states,)\n if self.config.is_encoder_decoder\n else (outputs.hidden_states,)\n )\n\n # reshape for beam search\n vocab_size = next_token_scores.shape[-1]\n next_token_scores = next_token_scores.view(batch_size, num_beams * vocab_size)\n\n probs = F.softmax(next_token_scores, dim=-1)\n next_tokens = torch.multinomial(probs, num_samples=2 * num_beams)\n next_token_scores = torch.gather(next_token_scores, -1, next_tokens)\n\n next_token_scores, _indices = torch.sort(next_token_scores, descending=True, dim=1)\n next_tokens = torch.gather(next_tokens, -1, _indices)\n\n next_indices = next_tokens // vocab_size\n next_tokens = next_tokens % vocab_size\n\n # stateless\n beam_outputs = beam_scorer.process(\n input_ids,\n next_token_scores,\n next_tokens,\n next_indices,\n pad_token_id=pad_token_id,\n eos_token_id=eos_token_id,\n )\n beam_scores = beam_outputs[\"next_beam_scores\"]\n beam_next_tokens = beam_outputs[\"next_beam_tokens\"]\n beam_idx = beam_outputs[\"next_beam_indices\"]\n\n input_ids = torch.cat([input_ids[beam_idx, :], beam_next_tokens.unsqueeze(-1)], dim=-1)\n cur_len = cur_len + 1\n\n model_kwargs = self._update_model_kwargs_for_generation(\n outputs, model_kwargs, is_encoder_decoder=self.config.is_encoder_decoder\n )\n if model_kwargs[\"past\"] is not None:\n model_kwargs[\"past\"] = self._reorder_cache(model_kwargs[\"past\"], beam_idx)\n\n if beam_scorer.is_done:\n break\n\n sequence_outputs = beam_scorer.finalize(\n input_ids, beam_scores, next_tokens, next_indices, pad_token_id=pad_token_id, eos_token_id=eos_token_id,\n callback_handle=callback_handle, **model_kwargs,\n )\n\n if return_dict_in_generate:\n if not output_scores:\n sequence_outputs[\"sequence_scores\"] = None\n if self.config.is_encoder_decoder:\n return BeamSampleEncoderDecoderOutput(\n sequences=sequence_outputs[\"sequences\"],\n sequences_scores=sequence_outputs[\"sequence_scores\"],\n scores=scores,\n encoder_attentions=encoder_attentions,\n encoder_hidden_states=encoder_hidden_states,\n decoder_attentions=decoder_attentions,\n cross_attentions=cross_attentions,\n decoder_hidden_states=decoder_hidden_states,\n )\n else:\n return BeamSampleDecoderOnlyOutput(\n sequences=sequence_outputs[\"sequences\"],\n sequences_scores=sequence_outputs[\"sequence_scores\"],\n scores=scores,\n attentions=decoder_attentions,\n hidden_states=decoder_hidden_states,\n )\n else:\n return sequence_outputs[\"sequences\"]\n\n def group_beam_search(\n self,\n input_ids: torch.LongTensor,\n beam_scorer: BeamScorer,\n logits_processor: Optional[LogitsProcessorList] = None,\n max_length: Optional[int] = None,\n pad_token_id: Optional[int] = None,\n eos_token_id: Optional[int] = None,\n output_attentions: Optional[bool] = None,\n output_hidden_states: Optional[bool] = None,\n output_scores: Optional[bool] = None,\n return_dict_in_generate: Optional[bool] = None,\n callback_handle: Optional = None,\n **model_kwargs,\n ):\n r\"\"\"\n Generates sequences for models with a language modeling head using beam search decoding.\n\n Parameters:\n\n input_ids (:obj:`torch.LongTensor` of shape :obj:`(batch_size, sequence_length)`, `optional`):\n The sequence used as a prompt for the generation. If :obj:`None` the method initializes it as an empty\n :obj:`torch.LongTensor` of shape :obj:`(1,)`.\n beam_scorer (:obj:`BeamScorer`):\n An derived instance of :class:`~transformers.BeamScorer` that defines how beam hypotheses are\n constructed, stored and sorted during generation. For more information, the documentation of\n :class:`~transformers.BeamScorer` should be read.\n logits_processor (:obj:`LogitsProcessorList`, `optional`):\n An instance of :class:`~transformers.LogitsProcessorList`. List of instances of class derived from\n :class:`~transformers.LogitsProcessor` used to modify the prediction scores of the language modeling\n head applied at each generation step.\n max_length (:obj:`int`, `optional`, defaults to 20):\n The maximum length of the sequence to be generated.\n pad_token_id (:obj:`int`, `optional`):\n The id of the `padding` token.\n eos_token_id (:obj:`int`, `optional`):\n The id of the `end-of-sequence` token.\n output_attentions (:obj:`bool`, `optional`, defaults to `False`):\n Whether or not to return the attentions tensors of all attention layers. See ``attentions`` under\n returned tensors for more details.\n output_hidden_states (:obj:`bool`, `optional`, defaults to `False`):\n Whether or not to return trhe hidden states of all layers. See ``hidden_states`` under returned tensors\n for more details.\n output_scores (:obj:`bool`, `optional`, defaults to `False`):\n Whether or not to return the prediction scores. See ``scores`` under returned tensors for more details.\n return_dict_in_generate (:obj:`bool`, `optional`, defaults to `False`):\n Whether or not to return a :class:`~transformers.file_utils.ModelOutput` instead of a plain tuple.\n callback_handle (:obj:`function`, `optional`, defaults to `None`):\n Handle to a callback function if the model wants to do custom processing at each beam docoding step\n model_kwargs:\n Additional model specific kwargs that will be forwarded to the :obj:`forward` function of the model. If\n model is an encoder-decoder model the kwargs should include :obj:`encoder_outputs`.\n\n Return:\n :class:`~transformers.generation_utils.BeamSearchDecoderOnlyOutput`,\n :class:`~transformers.generation_utils.BeamSearchEncoderDecoderOutput` or obj:`torch.LongTensor`: A\n :obj:`torch.LongTensor` containing the generated tokens (default behaviour) or a\n :class:`~transformers.generation_utils.BeamSearchDecoderOnlyOutput` if\n :class:`~transformers.generation_utils.BeamSearchDecoderOnlyOutput` if\n ``model.config.is_encoder_decoder=False`` and ``return_dict_in_generate=True`` or a\n :class:`~transformers.generation_utils.BeamSearchEncoderDecoderOutput` if\n ``model.config.is_encoder_decoder=True``.\n\n Examples::\n\n >>> from transformers import (\n ... AutoTokenizer,\n ... AutoModelForSeq2SeqLM,\n ... LogitsProcessorList,\n ... MinLengthLogitsProcessor,\n ... HammingDiversityLogitsProcessor,\n ... BeamSearchScorer,\n ... )\n >>> import torch\n\n >>> tokenizer = AutoTokenizer.from_pretrained(\"t5-base\")\n >>> model = AutoModelForSeq2SeqLM.from_pretrained(\"t5-base\")\n\n >>> encoder_input_str = \"translate English to German: How old are you?\"\n >>> encoder_input_ids = tokenizer(encoder_input_str, return_tensors=\"pt\").input_ids\n\n\n >>> # lets run diverse beam search using 6 beams\n >>> num_beams = 6\n >>> # define decoder start token ids\n >>> input_ids = torch.ones((num_beams, 1), device=model.device, dtype=torch.long)\n >>> input_ids = input_ids * model.config.decoder_start_token_id\n\n >>> # add encoder_outputs to model keyword arguments\n >>> model_kwargs = {\n ... \"encoder_outputs\": model.get_encoder()(encoder_input_ids.repeat_interleave(num_beams, dim=0), return_dict=True)\n ... }\n\n >>> # instantiate beam scorer\n >>> beam_scorer = BeamSearchScorer(\n ... batch_size=1,\n ... max_length=model.config.max_length,\n ... num_beams=num_beams,\n ... device=model.device,\n ... num_beam_groups=3\n ... )\n\n >>> # instantiate logits processors\n >>> logits_processor = LogitsProcessorList([\n ... HammingDiversityLogitsProcessor(5.5, num_beams=6, num_beam_groups=3),\n ... MinLengthLogitsProcessor(5, eos_token_id=model.config.eos_token_id),\n ... ])\n\n >>> outputs = model.group_beam_search(input_ids, beam_scorer, logits_processor=logits_processor, **model_kwargs)\n\n >>> print(\"Generated:\", tokenizer.batch_decode(outputs, skip_special_tokens=True))\n \"\"\"\n\n # init values\n logits_processor = logits_processor if logits_processor is not None else LogitsProcessorList()\n max_length = max_length if max_length is not None else self.config.max_length\n pad_token_id = pad_token_id if pad_token_id is not None else self.config.pad_token_id\n eos_token_id = eos_token_id if eos_token_id is not None else self.config.eos_token_id\n output_scores = output_scores if output_scores is not None else self.config.output_scores\n output_attentions = output_attentions if output_attentions is not None else self.config.output_attentions\n output_hidden_states = (\n output_hidden_states if output_hidden_states is not None else self.config.output_hidden_states\n )\n return_dict_in_generate = (\n return_dict_in_generate if return_dict_in_generate is not None else self.config.return_dict_in_generate\n )\n\n # init attention / hidden states / scores tuples\n scores = () if (return_dict_in_generate and output_scores) else None\n decoder_attentions = () if (return_dict_in_generate and output_attentions) else None\n cross_attentions = () if (return_dict_in_generate and output_attentions) else None\n decoder_hidden_states = () if (return_dict_in_generate and output_hidden_states) else None\n\n # if model is an encoder-decoder, retrieve encoder attention weights and hidden states\n if return_dict_in_generate and self.config.is_encoder_decoder:\n encoder_attentions = model_kwargs[\"encoder_outputs\"].get(\"attentions\") if output_attentions else None\n encoder_hidden_states = (\n model_kwargs[\"encoder_outputs\"].get(\"hidden_states\") if output_hidden_states else None\n )\n\n batch_size = len(beam_scorer._beam_hyps)\n num_beams = beam_scorer.num_beams\n num_beam_groups = beam_scorer.num_beam_groups\n num_sub_beams = num_beams // num_beam_groups\n device = input_ids.device\n\n batch_beam_size, cur_len = input_ids.shape\n\n assert (\n num_beams * batch_size == batch_beam_size\n ), f\"Batch dimension of `input_ids` should be {num_beams * batch_size}, but is {batch_beam_size}.\"\n\n beam_scores = torch.full((batch_size, num_beams), -1e9, dtype=torch.float, device=device)\n # initialise score of first beam of each group with 0 and the rest with 1e-9. This ensures that the beams in\n # the same group don't produce same tokens everytime.\n beam_scores[:, ::num_sub_beams] = 0\n beam_scores = beam_scores.view((batch_size * num_beams,))\n\n while cur_len < max_length:\n # predicted tokens in cur_len step\n current_tokens = torch.zeros(batch_size * num_beams, dtype=input_ids.dtype, device=device)\n\n # indices which will form the beams in the next time step\n reordering_indices = torch.zeros(batch_size * num_beams, dtype=torch.long, device=device)\n\n # do one decoder step on all beams of all sentences in batch\n model_inputs = self.prepare_inputs_for_generation(input_ids, **model_kwargs)\n outputs = self(\n **model_inputs,\n return_dict=True,\n output_attentions=output_attentions,\n output_hidden_states=output_hidden_states,\n )\n\n for beam_group_idx in range(num_beam_groups):\n group_start_idx = beam_group_idx * num_sub_beams\n group_end_idx = min(group_start_idx + num_sub_beams, num_beams)\n group_size = group_end_idx - group_start_idx\n\n # indices of beams of current group among all sentences in batch\n batch_group_indices = []\n\n if output_scores:\n processed_score = torch.zeros_like(outputs.logits[:, -1, :])\n\n for batch_idx in range(batch_size):\n batch_group_indices.extend(\n [batch_idx * num_beams + idx for idx in range(group_start_idx, group_end_idx)]\n )\n group_input_ids = input_ids[batch_group_indices]\n\n # select outputs of beams of current group only\n next_token_logits = outputs.logits[batch_group_indices, -1, :]\n\n # hack: adjust tokens for Marian. For Marian we have to make sure that the `pad_token_id`\n # cannot be generated both before and after the `F.log_softmax` operation.\n next_token_logits = self.adjust_logits_during_generation(\n next_token_logits, cur_len=cur_len, max_length=max_length\n )\n\n next_token_scores = F.log_softmax(next_token_logits, dim=-1) # (batch_size * group_size, vocab_size)\n vocab_size = next_token_scores.shape[-1]\n\n next_token_scores = logits_processor(\n group_input_ids, next_token_scores, current_tokens=current_tokens, beam_group_idx=beam_group_idx\n )\n next_token_scores = next_token_scores + beam_scores[batch_group_indices].unsqueeze(-1).expand_as(\n next_token_scores\n )\n\n if output_scores:\n processed_score[batch_group_indices] = next_token_scores\n\n # reshape for beam search\n next_token_scores = next_token_scores.view(batch_size, group_size * vocab_size)\n\n next_token_scores, next_tokens = torch.topk(\n next_token_scores, 2 * group_size, dim=1, largest=True, sorted=True\n )\n\n next_indices = next_tokens // vocab_size\n next_tokens = next_tokens % vocab_size\n\n # stateless\n beam_outputs = beam_scorer.process(\n group_input_ids,\n next_token_scores,\n next_tokens,\n next_indices,\n pad_token_id=pad_token_id,\n eos_token_id=eos_token_id,\n )\n beam_scores[batch_group_indices] = beam_outputs[\"next_beam_scores\"]\n beam_next_tokens = beam_outputs[\"next_beam_tokens\"]\n beam_idx = beam_outputs[\"next_beam_indices\"]\n\n input_ids[batch_group_indices] = group_input_ids[beam_idx]\n group_input_ids = torch.cat([group_input_ids[beam_idx, :], beam_next_tokens.unsqueeze(-1)], dim=-1)\n current_tokens[batch_group_indices] = group_input_ids[:, -1]\n\n # (beam_idx // group_size) -> batch_idx\n # (beam_idx % group_size) -> offset of idx inside the group\n reordering_indices[batch_group_indices] = (\n num_beams * (beam_idx // group_size) + group_start_idx + (beam_idx % group_size)\n )\n\n # Store scores, attentions and hidden_states when required\n if return_dict_in_generate:\n if output_scores:\n scores += (processed_score,)\n if output_attentions:\n decoder_attentions += (\n (outputs.decoder_attentions,) if self.config.is_encoder_decoder else (outputs.attentions,)\n )\n if self.config.is_encoder_decoder:\n cross_attentions += (outputs.cross_attentions,)\n\n if output_hidden_states:\n decoder_hidden_states += (\n (outputs.decoder_hidden_states,)\n if self.config.is_encoder_decoder\n else (outputs.hidden_states,)\n )\n\n model_kwargs = self._update_model_kwargs_for_generation(\n outputs, model_kwargs, is_encoder_decoder=self.config.is_encoder_decoder\n )\n if model_kwargs[\"past\"] is not None:\n model_kwargs[\"past\"] = self._reorder_cache(model_kwargs[\"past\"], reordering_indices)\n\n input_ids = torch.cat([input_ids, current_tokens.unsqueeze(-1)], dim=-1)\n cur_len = cur_len + 1\n if beam_scorer.is_done:\n break\n\n sequence_outputs = beam_scorer.finalize(\n input_ids, beam_scores, next_tokens, next_indices, pad_token_id=pad_token_id, eos_token_id=eos_token_id,\n callback_handle=callback_handle, **model_kwargs,\n )\n\n if return_dict_in_generate:\n if not output_scores:\n sequence_outputs[\"sequence_scores\"] = None\n if self.config.is_encoder_decoder:\n return BeamSearchEncoderDecoderOutput(\n sequences=sequence_outputs[\"sequences\"],\n sequences_scores=sequence_outputs[\"sequence_scores\"],\n scores=scores,\n encoder_attentions=encoder_attentions,\n encoder_hidden_states=encoder_hidden_states,\n decoder_attentions=decoder_attentions,\n cross_attentions=cross_attentions,\n decoder_hidden_states=decoder_hidden_states,\n )\n else:\n return BeamSearchDecoderOnlyOutput(\n sequences=sequence_outputs[\"sequences\"],\n sequences_scores=sequence_outputs[\"sequence_scores\"],\n scores=scores,\n attentions=decoder_attentions,\n hidden_states=decoder_hidden_states,\n )\n else:\n return sequence_outputs[\"sequences\"]\n\n\ndef top_k_top_p_filtering(\n logits: torch.FloatTensor,\n top_k: int = 0,\n top_p: float = 1.0,\n filter_value: float = -float(\"Inf\"),\n min_tokens_to_keep: int = 1,\n) -> torch.FloatTensor:\n \"\"\"\n Filter a distribution of logits using top-k and/or nucleus (top-p) filtering\n\n Args:\n logits: logits distribution shape (batch size, vocabulary size)\n if top_k > 0: keep only top k tokens with highest probability (top-k filtering).\n if top_p < 1.0: keep the top tokens with cumulative probability >= top_p (nucleus filtering).\n Nucleus filtering is described in Holtzman et al. (http://arxiv.org/abs/1904.09751)\n Make sure we keep at least min_tokens_to_keep per batch example in the output\n From: https://gist.github.com/thomwolf/1a5a29f6962089e871b94cbd09daf317\n \"\"\"\n if top_k > 0:\n logits = TopKLogitsWarper(top_k=top_k, filter_value=filter_value, min_tokens_to_keep=min_tokens_to_keep)(\n None, logits\n )\n\n if 0 <= top_p <= 1.0:\n logits = TopPLogitsWarper(top_p=top_p, min_tokens_to_keep=min_tokens_to_keep)(None, logits)\n\n return logits\n"
] |
[
[
"torch.zeros",
"torch.cat",
"torch.arange",
"torch.gather",
"torch.no_grad",
"torch.nn.functional.log_softmax",
"torch.ones",
"torch.multinomial",
"torch.full",
"torch.nn.functional.softmax",
"torch.zeros_like",
"torch.sort",
"torch.topk"
]
] |
xuanhien070594/robosuite
|
[
"1cac10607c7f37113710af13c85cff1421a37815"
] |
[
"robosuite/wrappers/gym_wrapper.py"
] |
[
"\"\"\"\nThis file implements a wrapper for facilitating compatibility with OpenAI gym.\nThis is useful when using these environments with code that assumes a gym-like \ninterface.\n\"\"\"\n\nimport numpy as np\nfrom gym import spaces\nfrom gym.core import Env\n\nfrom robosuite.wrappers import Wrapper\n\n\nclass GymWrapper(Wrapper, Env):\n \"\"\"\n Initializes the Gym wrapper. Mimics many of the required functionalities of the Wrapper class\n found in the gym.core module\n\n Args:\n env (MujocoEnv): The environment to wrap.\n keys (None or list of str): If provided, each observation will\n consist of concatenated keys from the wrapped environment's\n observation dictionary. Defaults to proprio-state and object-state.\n\n Raises:\n AssertionError: [Object observations must be enabled if no keys]\n \"\"\"\n\n def __init__(self, env, keys=None):\n # Run super method\n super().__init__(env=env)\n # Create name for gym\n robots = \"\".join([type(robot.robot_model).__name__ for robot in self.env.robots])\n self.name = robots + \"_\" + type(self.env).__name__\n\n # Get reward range\n self.reward_range = (0, self.env.reward_scale)\n\n if keys is None:\n keys = []\n # Add object obs if requested\n if self.env.use_object_obs:\n keys += [\"object-state\"]\n # Add image obs if requested\n if self.env.use_camera_obs:\n keys += [f\"{cam_name}_image\" for cam_name in self.env.camera_names]\n # Iterate over all robots to add to state\n for idx in range(len(self.env.robots)):\n keys += [\"robot{}_proprio-state\".format(idx)]\n self.keys = keys\n\n # Gym specific attributes\n self.env.spec = None\n self.metadata = None\n\n # set up observation space\n obs = self.env.reset()\n\n if \"achieved_goal\" in obs:\n # if environment uses HER buffer, observation also includes desired goal and achieve goal attributes\n self.modality_dims = {key: obs[\"observation\"][key].shape for key in self.keys}\n flat_obs = self._flatten_obs(obs[\"observation\"])\n self.obs_dim = flat_obs.size\n\n self.observation_space = spaces.Dict(\n dict(\n desired_goal=spaces.Box(-np.inf, np.inf, shape=obs[\"achieved_goal\"].shape, dtype=\"float32\"),\n achieved_goal=spaces.Box(-np.inf, np.inf, shape=obs[\"achieved_goal\"].shape, dtype=\"float32\"),\n observation=spaces.Box(-np.inf, np.inf, shape=flat_obs.shape, dtype=\"float32\"),\n )\n )\n else:\n self.modality_dims = {key: obs[key].shape for key in self.keys}\n self.obs_dim = self._flatten_obs(obs).size\n self.observation_space = spaces.Box(-np.inf, np.inf, self._flatten_obs(obs).shape, dtype=\"float32\")\n\n # set up action space\n low, high = self.env.action_spec\n self.action_space = spaces.Box(low=low, high=high)\n\n def _flatten_obs(self, obs_dict, verbose=False):\n \"\"\"\n Filters keys of interest out and concatenate the information.\n\n Args:\n obs_dict (OrderedDict): ordered dictionary of observations\n verbose (bool): Whether to print out to console as observation keys are processed\n\n Returns:\n np.array: observations flattened into a 1d array\n \"\"\"\n ob_lst = []\n for key in self.keys:\n if key in obs_dict:\n if verbose:\n print(\"adding key: {}\".format(key))\n ob_lst.append(np.array(obs_dict[key]).flatten())\n return np.concatenate(ob_lst)\n\n def reset(self):\n \"\"\"\n Extends env reset method to return flattened observation instead of normal OrderedDict.\n\n Returns:\n np.array: Flattened environment observation space after reset occurs\n \"\"\"\n ob_dict = self.env.reset()\n\n if \"achieved_goal\" in ob_dict:\n ob_dict[\"observation\"] = self._flatten_obs(ob_dict[\"observation\"])\n return ob_dict\n return self._flatten_obs(ob_dict)\n\n def step(self, action):\n \"\"\"\n Extends vanilla step() function call to return flattened observation instead of normal OrderedDict.\n\n Args:\n action (np.array): Action to take in environment\n\n Returns:\n 4-tuple:\n\n - (np.array) flattened observations from the environment\n - (float) reward from the environment\n - (bool) whether the current episode is completed or not\n - (dict) misc information\n \"\"\"\n ob_dict, reward, done, info = self.env.step(action)\n\n # special care when environment is using HER buffer\n if \"achieved_goal\" in ob_dict:\n ob_dict[\"observation\"] = self._flatten_obs(ob_dict[\"observation\"])\n return ob_dict, reward, done, info\n return self._flatten_obs(ob_dict), reward, done, info\n\n def seed(self, seed=None):\n \"\"\"\n Utility function to set numpy seed\n\n Args:\n seed (None or int): If specified, numpy seed to set\n\n Raises:\n TypeError: [Seed must be integer]\n \"\"\"\n # Seed the generator\n if seed is not None:\n try:\n np.random.seed(seed)\n except:\n TypeError(\"Seed must be an integer type!\")\n\n def compute_reward(self, achieved_goal, desired_goal, info):\n \"\"\"Compute the step reward. This externalizes the reward function and makes\n it dependent on a desired goal and the one that was achieved. If you wish to include\n additional rewards that are independent of the goal, you can include the necessary values\n to derive it in 'info' and compute it accordingly.\n\n If the environment does not employ Hindsight Experience Replay (HER), the function will just\n return reward computed by self.env.reward()\n\n Args:\n achieved_goal (object): the goal that was achieved during execution\n desired_goal (object): the desired goal that we asked the agent to attempt to achieve\n info (dict): an info dictionary with additional information\n\n Returns:\n float: The reward that corresponds to the provided achieved goal w.r.t. to the desired\n goal. Note that the following should always hold true:\n\n ob, reward, done, info = env.step()\n assert reward == env.compute_reward(ob['achieved_goal'], ob['desired_goal'], info)\n \"\"\"\n if hasattr(self.env, \"compute_reward\"): # using HER data\n return self.env.compute_reward(achieved_goal, desired_goal, info)\n else:\n return self.env.reward()\n"
] |
[
[
"numpy.concatenate",
"numpy.array",
"numpy.random.seed"
]
] |
StephenRogers1/cogent3
|
[
"1116a0ab14d9c29a560297205546714e2db1896c"
] |
[
"src/cogent3/evolve/likelihood_function.py"
] |
[
"#!/usr/bin/env python\n\nimport json\nimport random\n\nfrom collections import defaultdict\nfrom copy import deepcopy\n\nimport numpy\n\nfrom cogent3.core.alignment import ArrayAlignment\nfrom cogent3.evolve import substitution_model\nfrom cogent3.evolve.simulate import AlignmentEvolver, random_sequence\nfrom cogent3.maths.matrix_exponential_integration import expected_number_subs\nfrom cogent3.maths.matrix_logarithm import is_generator_unique\nfrom cogent3.maths.measure import (\n paralinear_continuous_time,\n paralinear_discrete_time,\n)\nfrom cogent3.recalculation.definition import ParameterController\nfrom cogent3.util import table\nfrom cogent3.util.dict_array import DictArrayTemplate\nfrom cogent3.util.misc import adjusted_gt_minprob, get_object_provenance\n\n\n__author__ = \"Peter Maxwell\"\n__copyright__ = \"Copyright 2007-2020, The Cogent Project\"\n__credits__ = [\n \"Gavin Huttley\",\n \"Andrew Butterfield\",\n \"Peter Maxwell\",\n \"Matthew Wakefield\",\n \"Rob Knight\",\n \"Brett Easton\",\n \"Ben Kaehler\",\n \"Ananias Iliadis\",\n]\n__license__ = \"BSD-3\"\n__version__ = \"2020.12.21a\"\n__maintainer__ = \"Gavin Huttley\"\n__email__ = \"[email protected]\"\n__status__ = \"Production\"\n\n\n# cogent3.evolve.parameter_controller.LikelihoodParameterController tells the\n# recalculation framework to use this subclass rather than the generic\n# recalculation Calculator. It adds methods which are useful for examining\n# the parameter, psub, mprob and likelihood values after the optimisation is\n# complete.\n\n\ndef _get_keyed_rule_indices(rules):\n \"\"\"returns {frozesent((par_name, edge1, edge2, ..)): index}\"\"\"\n new = {}\n for i, rule in enumerate(rules):\n edges = rule.get(\"edges\", rule.get(\"edge\", None)) or []\n edges = [edges] if type(edges) == str else edges\n par_name = rule[\"par_name\"]\n key = frozenset([par_name] + edges)\n new[key] = i\n return new\n\n\ndef update_rule_value(rich, null):\n \"\"\"applies value from null rule to rich rule\"\"\"\n val_key = \"init\" if \"init\" in rich else \"value\"\n rich[val_key] = null.get(\"init\", null.get(\"value\"))\n return rich\n\n\ndef extend_rule_value(rich, nulls):\n \"\"\"creates new rich rules from edges in null rules\"\"\"\n val_key = \"init\" if \"init\" in rich else \"value\"\n rules = []\n for null in nulls:\n edges = null.get(\"edges\", null.get(\"edge\"))\n edges = [edges] if type(edges) == str else edges\n for edge in edges:\n rule = deepcopy(rich)\n rule[\"edge\"] = edge\n rule[val_key] = null.get(\"init\", null.get(\"value\"))\n rules.append(rule)\n return rules\n\n\ndef update_scoped_rules(rich, null):\n \"\"\"returns rich rules with values derived from those in null rules\"\"\"\n new_rules = []\n rich = deepcopy(rich)\n # we build a dict keyed by frozen set consisting of the param name\n # and affected edges. The dict value is the list index in original.\n richd = _get_keyed_rule_indices(rich)\n nulld = _get_keyed_rule_indices(null)\n common = set(richd) & set(nulld)\n # 1-to-1 mapping, just extract the param value\n for key in common:\n rule = update_rule_value(rich[richd[key]], null[nulld[key]])\n new_rules.append(rule)\n\n # following rules differing in scope\n rich_remainder = set(richd) - set(nulld)\n null_remainder = set(nulld) - set(richd)\n for rich_key in rich_remainder:\n matches = []\n rich_rule = rich[richd[rich_key]]\n pname = rich_rule[\"par_name\"]\n enames = rich_rule.get(\"edges\", rich_rule.get(\"edge\", None))\n if type(enames) == str:\n enames = [enames]\n enames = None if enames is None else set(enames)\n for null_key in null_remainder:\n null_rule = null[nulld[null_key]]\n if pname != null_rule[\"par_name\"]:\n continue\n # parameter now fully general\n if enames is None:\n matches.append(null_rule)\n continue\n\n null_enames = null_rule.get(\"edges\", null_rule.get(\"edge\", None))\n null_enames = None if null_enames is None else set(null_enames)\n if None in (enames, null_enames) or null_enames & enames:\n matches.append(null_rule)\n\n if enames is None: # rich rule is \"free\"\n new_rules.extend(extend_rule_value(rich_rule, matches))\n continue\n\n if len(matches) > 1 and enames is not None:\n raise ValueError(f\"{rich_key} has too many mappings {matches}\")\n\n match = matches[0]\n new_rules.append(update_rule_value(rich_rule, match))\n return new_rules\n\n\ndef _get_param_mapping(rich, simple):\n \"\"\"returns {simple_param_name: rich_param_name, ...}, the mapping of simple\n to rich parameters based on matrix coordinates\n \"\"\"\n assert len(rich) >= len(simple)\n simple_to_rich = defaultdict(set)\n rich_to_simple = defaultdict(set)\n for simple_param in simple:\n simple_coords = simple[simple_param]\n for rich_param in rich:\n rich_coords = rich[rich_param]\n if rich_coords <= simple_coords:\n simple_to_rich[simple_param].add(rich_param)\n rich_to_simple[rich_param].add(simple_param)\n\n for rich_param in rich_to_simple:\n simple_counterparts = rich_to_simple[rich_param]\n if len(simple_counterparts) == 1:\n continue\n\n sized_simple = [(len(simple[param]), param) for param in simple_counterparts]\n sized_simple.sort()\n if sized_simple[0][0] == sized_simple[1][0]:\n msg = \"%s and %s tied for matrix space\" % (\n sized_simple[0][1],\n sized_simple[1][1],\n )\n raise ValueError(msg)\n\n _, chosen = sized_simple.pop(0)\n rich_to_simple[rich_param] = [chosen]\n for _, simple_param in sized_simple:\n simple_to_rich[simple_param].pop(rich_param)\n\n return simple_to_rich\n\n\nclass _ParamProjection:\n \"\"\"projects parameter names, values between nested models\"\"\"\n\n def __init__(self, simple_model, rich_model, motif_probs, same=True):\n # construct following by calling the functions we wrote\n self._rich_coords = rich_model.get_param_matrix_coords(include_ref_cell=True)\n self._simple_coords = simple_model.get_param_matrix_coords(\n include_ref_cell=True\n )\n self._param_map = _get_param_mapping(self._rich_coords, self._simple_coords)\n self._same = same\n # end of constructing attributes\n self._motif_probs = motif_probs\n self._ref_val = self._set_ref_val(same)\n self.projected_rate = {False: self._rate_not_same}.get(same, self._rate_same)\n\n def _set_ref_val(self, same):\n \"\"\"returns the motif prob corresponding to the model reference cell\"\"\"\n if same:\n return 1\n else:\n i, j = list(self._rich_coords[\"ref_cell\"])[0]\n return self._motif_probs[j]\n\n def _rate_not_same(self, simple_param, mle):\n \"\"\"returns {rich_param: val, ...} from simple_param: val\"\"\"\n ref_val = self._ref_val\n new_terms = {}\n for rich_param in self._param_map[simple_param]:\n if rich_param == \"ref_cell\":\n continue\n for i, j in self._rich_coords[rich_param]:\n new_terms[rich_param] = self._motif_probs[j] * mle / ref_val\n\n return new_terms\n\n def _rate_same(self, simple_param, mle):\n new_terms = {}\n for rich_param in self._param_map[simple_param]:\n if rich_param == \"ref_cell\":\n continue\n for i, j in self._rich_coords[rich_param]:\n new_terms[rich_param] = mle\n return new_terms\n\n def update_param_rules(self, rules):\n new_rules = []\n if not self._same:\n rules = rules[:] + [dict(par_name=\"ref_cell\", init=1.0, edges=None)]\n\n for rule in rules:\n # get the param name, mle, call self.projected_rate\n name = rule[\"par_name\"]\n if name in (\"mprobs\", \"length\"):\n new_rules.append(rule)\n continue\n\n if rule.get(\"is_constant\", False):\n par_val_key = \"value\"\n else:\n par_val_key = \"init\"\n\n mle = rule[par_val_key]\n\n proj_rate = self.projected_rate(name, mle)\n for new_name, new_mle in proj_rate.items():\n rule_dict = rule.copy()\n rule_dict[\"par_name\"] = new_name\n # update it with the new parname and mle and append to new rules\n rule_dict[\"init\"] = new_mle\n new_rules.append(rule_dict)\n\n return new_rules\n\n\ndef compatible_likelihood_functions(lf1, lf2):\n \"\"\"returns True if all attributes of the two likelihood functions are compatible\n for mapping parameters, else raises ValueError or an AssertionError\"\"\"\n # tree's must have the same topology AND be oriented the same way\n # plus have the same edge names\n if len(lf1.bin_names) != 1 or len(lf1.bin_names) != len(lf2.bin_names):\n raise NotImplementedError(\"Too many bins\")\n if len(lf1.locus_names) != 1 or len(lf1.locus_names) != len(lf2.locus_names):\n raise NotImplementedError(\"Too many loci\")\n if lf1.model.get_motifs() != lf2.model.get_motifs():\n raise AssertionError(\"Motifs don't match\")\n if lf1.tree.get_newick(with_node_names=True) != lf2.tree.get_newick(\n with_node_names=True\n ):\n raise AssertionError(\"Topology, Orientation or node names don't match\")\n return True\n\n\nclass LikelihoodFunction(ParameterController):\n @property\n def lnL(self):\n \"\"\"log-likelihood\"\"\"\n return self.get_log_likelihood()\n\n def get_log_likelihood(self):\n return self.get_final_result()\n\n def get_all_psubs(self):\n \"\"\"returns all psubs as a dict keyed by used dimensions\"\"\"\n try:\n defn = self.defn_for[\"dsubs\"]\n except KeyError:\n defn = self.defn_for[\"psubs\"]\n\n used_dims = defn.used_dimensions()\n vdims = defn.valid_dimensions\n indices = [vdims.index(k) for k in used_dims if k in vdims]\n result = {}\n darr_template = DictArrayTemplate(self._motifs, self._motifs)\n for scope, index in defn.index.items():\n psub = defn.values[index]\n key = tuple(numpy.take(scope, indices))\n result[key] = darr_template.wrap(psub)\n return result\n\n def get_psub_for_edge(self, name, **kw):\n \"\"\"returns the substitution probability matrix for the named edge\"\"\"\n try:\n # For PartialyDiscretePsubsDefn\n array = self.get_param_value(\"dpsubs\", edge=name, **kw)\n except KeyError:\n array = self.get_param_value(\"psubs\", edge=name, **kw)\n return DictArrayTemplate(self._motifs, self._motifs).wrap(array)\n\n def get_all_rate_matrices(self, calibrated=True):\n \"\"\"returns all rate matrices (Q) as a dict, keyed by scope\n\n Parameters\n ----------\n calibrated : bool\n scales the rate matrix by branch length for each edge. If a rate\n heterogeneity model, then the matrix is further scaled by rate\n for a bin\n Returns\n -------\n If a single rate matrix, the key is an empty tuple\n \"\"\"\n defn = self.defn_for[\"Q\"]\n\n rate_het = self.defn_for.get(\"rate\", False)\n if rate_het:\n bin_index = rate_het.valid_dimensions.index(\"bin\")\n bin_names = [k[bin_index] for k in rate_het.index]\n bin_names = {n: i for i, n in enumerate(bin_names)}\n bin_index = defn.valid_dimensions.index(\"bin\")\n else:\n bin_names = None\n bin_index = None\n\n used_dims = defn.used_dimensions()\n edge_index = defn.valid_dimensions.index(\"edge\")\n\n indices = {defn.valid_dimensions.index(k) for k in used_dims}\n if not calibrated:\n indices.add(edge_index)\n\n if not calibrated and rate_het:\n indices.add(bin_index)\n\n indices = list(sorted(indices))\n result = {}\n darr_template = DictArrayTemplate(self._motifs, self._motifs)\n for scope, index in defn.index.items():\n q = defn.values[index] # this gives the appropriate Q\n # from scope we extract only the relevant dimensions\n key = tuple(numpy.take(scope, indices))\n q = q.copy()\n if not calibrated:\n length = self.get_param_value(\"length\", edge=scope[edge_index])\n if rate_het:\n bdex = bin_names[scope[bin_index]]\n rate = rate_het.values[bdex]\n length *= rate\n q *= length\n result[key] = darr_template.wrap(q)\n if not indices and calibrated:\n break # single rate matrix\n\n return result\n\n def get_rate_matrix_for_edge(self, name, calibrated=True, **kw):\n \"\"\"returns the rate matrix (Q) for the named edge\n\n If calibrated=False, expm(Q) will give the same result as\n get_psub_for_edge(name)\"\"\"\n try:\n array = self.get_param_value(\"Q\", edge=name, **kw)\n array = array.copy()\n if not calibrated:\n length = self.get_param_value(\"length\", edge=name, **kw)\n array *= length\n except KeyError as err:\n if err[0] == \"Q\" and name != \"Q\":\n raise RuntimeError(\"rate matrix not known by this model\")\n else:\n raise\n return DictArrayTemplate(self._motifs, self._motifs).wrap(array)\n\n def _getLikelihoodValuesSummedAcrossAnyBins(self, locus=None):\n if self.bin_names and len(self.bin_names) > 1:\n root_lhs = [\n self.get_param_value(\"lh\", locus=locus, bin=bin)\n for bin in self.bin_names\n ]\n bprobs = self.get_param_value(\"bprobs\")\n root_lh = bprobs.dot(root_lhs)\n else:\n root_lh = self.get_param_value(\"lh\", locus=locus)\n return root_lh\n\n def get_full_length_likelihoods(self, locus=None):\n \"\"\"Array of [site, motif] likelihoods from the root of the tree\"\"\"\n root_lh = self._getLikelihoodValuesSummedAcrossAnyBins(locus=locus)\n root_lht = self.get_param_value(\"root\", locus=locus)\n return root_lht.get_full_length_likelihoods(root_lh)\n\n def get_G_statistic(self, return_table=False, locus=None):\n \"\"\"Goodness-of-fit statistic derived from the unambiguous columns\"\"\"\n root_lh = self._getLikelihoodValuesSummedAcrossAnyBins(locus=locus)\n root_lht = self.get_param_value(\"root\", locus=locus)\n return root_lht.calc_G_statistic(root_lh, return_table)\n\n def reconstruct_ancestral_seqs(self, locus=None):\n \"\"\"returns a dict of DictArray objects containing probabilities\n of each alphabet state for each node in the tree.\n\n Parameters\n ----------\n locus\n a named locus\n\n \"\"\"\n result = {}\n array_template = None\n for restricted_edge in self._tree.get_edge_vector():\n if restricted_edge.istip():\n continue\n try:\n r = []\n for motif in range(len(self._motifs)):\n self.set_param_rule(\n \"fixed_motif\",\n value=motif,\n edge=restricted_edge.name,\n locus=locus,\n is_constant=True,\n )\n likelihoods = self.get_full_length_likelihoods(locus=locus)\n r.append(likelihoods)\n if array_template is None:\n array_template = DictArrayTemplate(\n likelihoods.shape[0], self._motifs\n )\n finally:\n self.set_param_rule(\n \"fixed_motif\",\n value=-1,\n edge=restricted_edge.name,\n locus=locus,\n is_constant=True,\n )\n # dict of site x motif arrays\n result[restricted_edge.name] = array_template.wrap(\n numpy.transpose(numpy.asarray(r))\n )\n return result\n\n def likely_ancestral_seqs(self, locus=None):\n \"\"\"Returns the most likely reconstructed ancestral sequences as an\n alignment.\n\n Parameters\n ----------\n locus\n a named locus\n\n \"\"\"\n prob_array = self.reconstruct_ancestral_seqs(locus=locus)\n seqs = []\n for edge, probs in list(prob_array.items()):\n seq = []\n for row in probs:\n by_p = [(p, state) for state, p in list(row.items())]\n seq.append(max(by_p)[1])\n seqs += [(edge, self.model.moltype.make_seq(\"\".join(seq)))]\n return ArrayAlignment(data=seqs, moltype=self.model.moltype)\n\n def get_bin_probs(self, locus=None):\n hmm = self.get_param_value(\"bindex\", locus=locus)\n lhs = [\n self.get_param_value(\"lh\", locus=locus, bin=bin) for bin in self.bin_names\n ]\n array = hmm.get_posterior_probs(*lhs)\n return DictArrayTemplate(self.bin_names, array.shape[1]).wrap(array)\n\n def _valuesForDimension(self, dim):\n # in support of __str__\n if dim == \"edge\":\n result = [e.name for e in self._tree.get_edge_vector()]\n elif dim == \"bin\":\n result = self.bin_names[:]\n elif dim == \"locus\":\n result = self.locus_names[:]\n elif dim.startswith(\"motif\"):\n result = self._mprob_motifs\n elif dim == \"position\":\n result = self.posn_names[:]\n else:\n raise KeyError(dim)\n return result\n\n def _valuesForDimensions(self, dims):\n # in support of __str__\n result = [[]]\n for dim in dims:\n new_result = []\n for r in result:\n for cat in self._valuesForDimension(dim):\n new_result.append(r + [cat])\n result = new_result\n return result\n\n def _for_display(self):\n \"\"\"processes statistics tables for display\"\"\"\n title = self.name if self.name else \"Likelihood function statistics\"\n result = []\n result += self.get_statistics(with_motif_probs=True, with_titles=True)\n for i, table in enumerate(result):\n if (\n \"motif\" in table.title and table.shape[1] == 2 and table.shape[0] >= 60\n ): # just sort codon motif probs, then truncate\n table = table.sorted(columns=\"motif\")\n table.set_repr_policy(head=5, tail=5, show_shape=False)\n result[i] = table\n return title, result\n\n def _repr_html_(self):\n \"\"\"for jupyter notebook display\"\"\"\n try:\n lnL = \"<p>log-likelihood = %.4f</p>\" % self.get_log_likelihood()\n except ValueError:\n # alignment probably not yet set\n lnL = \"\"\n\n nfp = \"<p>number of free parameters = %d</p>\" % self.get_num_free_params()\n title, results = self._for_display()\n for i, table in enumerate(results):\n table.title = table.title.capitalize()\n table.set_repr_policy(show_shape=False)\n results[i] = table._repr_html_()\n results = [\"<h4>%s</h4>\" % title, lnL, nfp] + results\n return \"\\n\".join(results)\n\n def __repr__(self):\n return str(self)\n\n def __str__(self):\n title, results = self._for_display()\n\n try:\n lnL = \"log-likelihood = %.4f\" % self.get_log_likelihood()\n except ValueError:\n # alignment probably not yet set\n lnL = None\n\n nfp = \"number of free parameters = %d\" % self.get_num_free_params()\n for table in results:\n table.title = \"\"\n\n if lnL:\n results = [title, lnL, nfp] + results\n else:\n results = [title, nfp] + results\n\n return \"\\n\".join(map(str, results))\n\n def get_annotated_tree(self, length_as=None):\n \"\"\"returns tree with model attributes on node.params\n\n length_as : str or None\n replaces 'length' param with either 'ENS' or 'paralinear'.\n 'ENS' is the expected number of substitution, (which will be\n different to standard length if the substitution model is\n non-stationary). 'paralinear' is the measure of Lake 1994.\n\n The other measures are always available in the params dict of each\n node.\n \"\"\"\n from cogent3.evolve.ns_substitution_model import (\n DiscreteSubstitutionModel,\n )\n\n is_discrete = isinstance(self.model, DiscreteSubstitutionModel)\n\n if is_discrete and not length_as == \"paralinear\":\n raise ValueError(f\"{length_as} invalid for discrete time process\")\n\n assert length_as in (\"ENS\", \"paralinear\", None)\n d = self.get_param_value_dict([\"edge\"])\n lengths = d.pop(\"length\", None)\n mprobs = self.get_motif_probs_by_node()\n if not is_discrete:\n ens = self.get_lengths_as_ens(motif_probs=mprobs)\n\n plin = self.get_paralinear_metric(motif_probs=mprobs)\n if length_as == \"ENS\":\n lengths = ens\n elif length_as == \"paralinear\":\n lengths = plin\n\n tree = self._tree.deepcopy()\n for edge in tree.get_edge_vector():\n if edge.name == \"root\":\n edge.params[\"mprobs\"] = mprobs[edge.name].to_dict()\n continue\n\n if not is_discrete:\n edge.params[\"ENS\"] = ens[edge.name]\n\n edge.params[\"length\"] = lengths[edge.name]\n edge.params[\"paralinear\"] = plin[edge.name]\n edge.params[\"mprobs\"] = mprobs[edge.name].to_dict()\n for par in d:\n val = d[par][edge.name]\n if par == length_as:\n val = ens[edge.name]\n edge.params[par] = val\n\n return tree\n\n def get_motif_probs(self, edge=None, bin=None, locus=None, position=None):\n \"\"\"\n Parameters\n ----------\n edge : str\n name of edge\n bin : int or str\n name of bin\n locus : str\n name of locus\n position : int or str\n name of position\n\n Returns\n -------\n If 1D, returns DictArray, else a dict of DictArray\n \"\"\"\n param_names = self.get_param_names()\n mprob_name = [n for n in param_names if \"mprob\" in n][0]\n dims = tuple(self.get_used_dimensions(mprob_name))\n mprobs = self.get_param_value_dict(dimensions=dims, params=[mprob_name])\n if len(dims) == 2:\n var = [c for c in dims if c != mprob_name][0]\n key = locals().get(var, None)\n mprobs = mprobs[mprob_name]\n if key is not None:\n mprobs = mprobs.get(str(key), mprobs.get(key))\n mprobs = {mprob_name: mprobs}\n\n # these can fall below the minimum allowed value due to\n # rounding errors, so I adjust these\n for k, value in mprobs.items():\n value.array = adjusted_gt_minprob(value.array, minprob=1e-6)\n\n if len(mprobs) == 1:\n mprobs = mprobs[mprob_name]\n\n return mprobs\n\n def get_bin_prior_probs(self, locus=None):\n bin_probs_array = self.get_param_value(\"bprobs\", locus=locus)\n return DictArrayTemplate(self.bin_names).wrap(bin_probs_array)\n\n def get_scaled_lengths(self, predicate, bin=None, locus=None):\n \"\"\"A dictionary of {scale:{edge:length}}\"\"\"\n if not hasattr(self._model, \"get_scaled_lengths_from_Q\"):\n return {}\n\n get_value_of = self.get_param_value\n value_of_kw = dict(locus=locus)\n\n if bin is None:\n bin_names = self.bin_names\n else:\n bin_names = [bin]\n\n if len(bin_names) == 1:\n bprobs = [1.0]\n else:\n bprobs = get_value_of(\"bprobs\", **value_of_kw)\n\n mprobs = [get_value_of(\"mprobs\", bin=b, **value_of_kw) for b in bin_names]\n\n scaled_lengths = {}\n for edge in self._tree.get_edge_vector():\n if edge.isroot():\n continue\n Qs = [\n get_value_of(\"Qd\", bin=b, edge=edge.name, **value_of_kw).Q\n for b in bin_names\n ]\n length = get_value_of(\"length\", edge=edge.name, **value_of_kw)\n scaled_lengths[edge.name] = length * self._model.get_scale_from_Qs(\n Qs, bprobs, mprobs, predicate\n )\n return scaled_lengths\n\n def get_paralinear_metric(self, motif_probs=None):\n \"\"\"returns {edge.name: paralinear, ...}\n Parameters\n ----------\n motif_probs : dict or DictArray\n an item for each edge of the tree. Computed if not provided.\n \"\"\"\n from cogent3.evolve.ns_substitution_model import (\n DiscreteSubstitutionModel,\n )\n\n is_discrete = isinstance(self.model, DiscreteSubstitutionModel)\n\n if motif_probs is None:\n motif_probs = self.get_motif_probs_by_node()\n plin = {}\n for edge in self.tree.get_edge_vector(include_root=False):\n parent_name = edge.parent.name\n pi = motif_probs[parent_name]\n P = self.get_psub_for_edge(edge.name)\n if is_discrete:\n para = paralinear_discrete_time(P.array, pi.array)\n else:\n Q = self.get_rate_matrix_for_edge(edge.name, calibrated=False)\n para = paralinear_continuous_time(P.array, pi.array, Q.array)\n\n plin[edge.name] = para\n\n return plin\n\n def get_lengths_as_ens(self, motif_probs=None):\n \"\"\"returns {edge.name: ens, ...} where ens is the expected number of substitutions\n\n for a stationary Markov process, this is just branch length\n Parameters\n ----------\n motif_probs : dict or DictArray\n an item for each edge of the tree. Computed if not provided.\n \"\"\"\n if motif_probs is None:\n motif_probs = self.get_motif_probs_by_node()\n node_names = self.tree.get_node_names()\n node_names.remove(\"root\")\n lengths = {e: self.get_param_value(\"length\", edge=e) for e in node_names}\n if not isinstance(self.model, substitution_model.Stationary):\n ens = {}\n for e in node_names:\n Q = self.get_rate_matrix_for_edge(e)\n length = expected_number_subs(motif_probs[e], Q, lengths[e])\n ens[e] = length\n\n lengths = ens\n\n return lengths\n\n def get_param_rules(self):\n \"\"\"returns the [{rule}, ..] that would allow reconstruction\"\"\"\n # markov model rate terms\n rules = []\n param_names = self.get_param_names()\n for param_name in param_names:\n defn = self.defn_for[param_name]\n try:\n rules.extend(defn.get_param_rules())\n except AttributeError:\n # aggregate params, like those deriving from gamma shaped rates\n pass\n\n return rules\n\n def get_statistics(self, with_motif_probs=True, with_titles=True):\n \"\"\"returns the parameter values as tables/dict\n\n Parameters\n ----------\n with_motif_probs\n include the motif probability table\n with_titles\n include a title for each table based on it's\n dimension\n\n \"\"\"\n result = []\n group = {}\n param_names = self.get_param_names()\n\n mprob_name = [n for n in param_names if \"mprob\" in n]\n mprob_name = mprob_name[0] if mprob_name else \"\"\n if not with_motif_probs:\n param_names.remove(mprob_name)\n\n for param in param_names:\n dims = tuple(self.get_used_dimensions(param))\n if dims not in group:\n group[dims] = []\n group[dims].append(param)\n table_order = list(group.keys())\n table_order.sort()\n for table_dims in table_order:\n raw_table = self.get_param_value_dict(\n dimensions=table_dims, params=group[table_dims]\n )\n param_names = group[table_dims]\n param_names.sort()\n if table_dims == (\"edge\",):\n if \"length\" in param_names:\n param_names.remove(\"length\")\n param_names.insert(0, \"length\")\n raw_table[\"parent\"] = dict(\n [\n (e.name, e.parent.name)\n for e in self._tree.get_edge_vector()\n if not e.isroot()\n ]\n )\n param_names.insert(0, \"parent\")\n list_table = []\n heading_names = list(table_dims) + param_names\n row_order = self._valuesForDimensions(table_dims)\n for scope in row_order:\n row = {}\n row_used = False\n for param in param_names:\n d = raw_table[param]\n try:\n for part in scope:\n d = d[part]\n except KeyError:\n d = \"NA\"\n else:\n row_used = True\n row[param] = d\n if row_used:\n row.update(dict(list(zip(table_dims, scope))))\n row = [row[k] for k in heading_names]\n list_table.append(row)\n if table_dims:\n title = [\"\", \"%s params\" % \" \".join(table_dims)][with_titles]\n else:\n title = [\"\", \"global params\"][with_titles]\n row_ids = None\n stat_table = table.Table(\n heading_names,\n list_table,\n max_width=80,\n index_name=row_ids,\n title=title,\n **self._format,\n )\n if group[table_dims] == [mprob_name]:\n # if stat_table.shape\n # if mprobs, we use the motifs as header\n motifs = list(sorted(set(stat_table.tolist(\"motif\"))))\n if stat_table.shape[1] == 2:\n motif_prob = dict(stat_table.tolist())\n heading_names = motifs\n list_table = [motif_prob[m] for m in motifs]\n list_table = [list_table]\n elif stat_table.shape[1] == 3:\n rows = []\n other_col = [\n c\n for c in stat_table.header\n if \"motif\" not in c and \"mprobs\" not in c\n ][0]\n for val in stat_table.distinct_values(other_col):\n subtable = stat_table.filtered(\n lambda x: x == val, columns=other_col\n )\n motif_prob = dict(\n subtable.tolist(\n [c for c in stat_table.header if c != other_col]\n )\n )\n rows.append([val] + [motif_prob[m] for m in motifs])\n heading_names = [other_col] + motifs\n list_table = rows\n stat_table = table.Table(\n heading_names, list_table, max_width=80, title=title, **self._format\n )\n\n result.append(stat_table)\n return result\n\n def to_rich_dict(self):\n \"\"\"returns detailed info on object, used by to_json\"\"\"\n data = self._serialisable.copy()\n for key in (\"model\", \"tree\"):\n del data[key]\n\n tree = self.tree.to_rich_dict()\n edge_attr = tree[\"edge_attributes\"]\n for edge in edge_attr:\n if edge == \"root\":\n continue\n try:\n edge_attr[edge][\"length\"] = self.get_param_value(\"length\", edge=edge)\n except KeyError:\n # probably discrete-time model\n edge_attr[edge][\"length\"] = None\n\n model = self._model.to_rich_dict(for_pickle=False)\n\n aln_defn = self.defn_for[\"alignment\"]\n if len(aln_defn.index) == 1:\n alignment = self.get_param_value(\"alignment\").to_rich_dict()\n mprobs = self.get_motif_probs().to_dict()\n else:\n alignment = {a[\"locus\"]: a[\"value\"] for a in aln_defn.get_param_rules()}\n mprobs = self.get_motif_probs()\n for k in alignment:\n alignment[k] = alignment[k].to_rich_dict()\n mprobs[k] = mprobs[k].to_dict()\n\n DLC = self.all_psubs_DLC()\n try:\n unique_Q = self.all_rate_matrices_unique()\n except Exception:\n # there's a mix of assertions\n # for \"storage\", make this indeterminate in those cases\n unique_Q = None\n\n data = dict(\n model=model,\n tree=tree,\n alignment=alignment,\n likelihood_construction=data,\n param_rules=self.get_param_rules(),\n lnL=self.get_log_likelihood(),\n nfp=self.get_num_free_params(),\n motif_probs=mprobs,\n DLC=DLC,\n unique_Q=unique_Q,\n type=get_object_provenance(self),\n name=self.get_name(),\n version=__version__,\n )\n return data\n\n def to_json(self):\n data = self.to_rich_dict()\n data = json.dumps(data)\n return data\n\n @property\n def name(self):\n if self._name is None:\n self._name = self.model.name or \"\"\n\n return self._name\n\n @name.setter\n def name(self, name):\n self._name = name\n\n # For tests. Compat with old LF interface\n def set_name(self, name):\n self.name = name\n\n def get_name(self):\n return self.name\n\n def set_tables_format(self, space=4, digits=4):\n \"\"\"sets display properties for statistics tables. This affects results\n of str(lf) too.\"\"\"\n space = [space, 4][type(space) != int]\n digits = [digits, 4][type(digits) != int]\n self._format = dict(space=space, digits=digits)\n\n def _get_motif_probs_by_node_tr(self, edges=None, bin=None, locus=None):\n \"\"\"returns motif probs by node for time-reversible models\"\"\"\n mprob_rules = [r for r in self.get_param_rules() if \"mprob\" in r[\"par_name\"]]\n if len(mprob_rules) > 1 or self.model.mprob_model == \"monomers\":\n raise NotImplementedError\n\n mprobs = self.get_motif_probs()\n if len(mprobs) != len(self.motifs):\n # a Muse and Gaut model\n expanded = numpy.zeros(len(self.motifs), dtype=float)\n for i, motif in enumerate(self.motifs):\n val = 1.0\n for b in motif:\n val *= mprobs[b]\n expanded[i] = val\n mprobs = expanded / expanded.sum()\n else:\n mprobs = [mprobs[m] for m in self.motifs]\n edges = []\n values = []\n for e in self.tree.postorder():\n edges.append(e.name)\n values.append(mprobs)\n\n return DictArrayTemplate(edges, self.motifs).wrap(values)\n\n def get_motif_probs_by_node(self, edges=None, bin=None, locus=None):\n from cogent3.evolve.substitution_model import TimeReversible\n\n if isinstance(self.model, TimeReversible):\n return self._get_motif_probs_by_node_tr(edges=edges, bin=bin, locus=locus)\n\n kw = dict(bin=bin, locus=locus)\n mprobs = self.get_param_value(\"mprobs\", **kw)\n mprobs = self._model.calc_word_probs(mprobs)\n result = self._nodeMotifProbs(self._tree, mprobs, kw)\n if edges is None:\n edges = [name for (name, m) in result]\n result = dict(result)\n values = [result[name] for name in edges]\n return DictArrayTemplate(edges, self._mprob_motifs).wrap(values)\n\n def _nodeMotifProbs(self, tree, mprobs, kw):\n result = [(tree.name, mprobs)]\n for child in tree.children:\n psub = self.get_psub_for_edge(child.name, **kw)\n child_mprobs = numpy.dot(mprobs, psub)\n result.extend(self._nodeMotifProbs(child, child_mprobs, kw))\n return result\n\n def simulate_alignment(\n self,\n sequence_length=None,\n random_series=None,\n exclude_internal=True,\n locus=None,\n seed=None,\n root_sequence=None,\n ):\n \"\"\"\n Returns an alignment of simulated sequences with key's corresponding to\n names from the current attached alignment.\n\n Parameters\n ----------\n sequence_length\n the legnth of the alignment to be simulated,\n default is the length of the attached alignment.\n random_series\n a random number generator.\n exclude_internal\n if True, only sequences for tips are returned.\n root_sequence\n a sequence from which all others evolve.\n\n \"\"\"\n\n if sequence_length is None:\n lht = self.get_param_value(\"lht\", locus=locus)\n sequence_length = len(lht.index)\n leaves = self.get_param_value(\"leaf_likelihoods\", locus=locus)\n orig_ambig = {}\n for (seq_name, leaf) in list(leaves.items()):\n orig_ambig[seq_name] = leaf.get_ambiguous_positions()\n else:\n orig_ambig = {}\n\n if random_series is None:\n random_series = random.Random()\n random_series.seed(seed)\n\n def psub_for(edge, bin):\n return self.get_psub_for_edge(edge, bin=bin, locus=locus)\n\n if len(self.bin_names) > 1:\n hmm = self.get_param_value(\"bdist\", locus=locus)\n site_bins = hmm.emit(sequence_length, random_series)\n else:\n site_bins = numpy.zeros([sequence_length], int)\n\n evolver = AlignmentEvolver(\n random_series,\n orig_ambig,\n exclude_internal,\n self.bin_names,\n site_bins,\n psub_for,\n self._motifs,\n )\n\n if root_sequence is not None: # we convert to a vector of motifs\n if isinstance(root_sequence, str):\n root_sequence = self._model.moltype.make_seq(root_sequence)\n motif_len = self._model.get_alphabet().get_motif_len()\n root_sequence = root_sequence.get_in_motif_size(motif_len)\n else:\n mprobs = self.get_param_value(\"mprobs\", locus=locus, edge=\"root\")\n mprobs = self._model.calc_word_probs(mprobs)\n mprobs = dict((m, p) for (m, p) in zip(self._motifs, mprobs))\n root_sequence = random_sequence(random_series, mprobs, sequence_length)\n\n simulated_sequences = evolver(self._tree, root_sequence)\n\n return ArrayAlignment(data=simulated_sequences, moltype=self._model.moltype)\n\n def all_psubs_DLC(self):\n \"\"\"Returns True if every Psub matrix is Diagonal Largest in Column\"\"\"\n all_psubs = self.get_all_psubs()\n for P in all_psubs.values():\n if (P.to_array().diagonal() < P).any():\n return False\n return True\n\n def all_rate_matrices_unique(self):\n \"\"\"Returns True if every rate matrix is unique for its Psub matrix\"\"\"\n # get all possible Q, as products of t, and any rate-het terms\n all_Q = self.get_all_rate_matrices(calibrated=False)\n for Q in all_Q.values():\n Q = Q.to_array()\n if not is_generator_unique(Q):\n return False\n return True\n\n def initialise_from_nested(self, nested_lf):\n from cogent3.evolve.substitution_model import Stationary\n\n assert (\n self.get_num_free_params() > nested_lf.get_num_free_params()\n ), \"wrong order for likelihood functions\"\n compatible_likelihood_functions(self, nested_lf)\n\n same = (\n isinstance(self.model, Stationary)\n and isinstance(nested_lf.model, Stationary)\n ) or (\n not isinstance(self.model, Stationary)\n and not isinstance(nested_lf.model, Stationary)\n )\n\n mprobs = nested_lf.get_motif_probs()\n edge_names = self.tree.get_node_names()\n edge_names.remove(\"root\")\n param_proj = _ParamProjection(nested_lf.model, self.model, mprobs, same=same)\n param_rules = nested_lf.get_param_rules()\n param_rules = param_proj.update_param_rules(param_rules)\n my_rules = self.get_param_rules()\n my_rules = update_scoped_rules(my_rules, param_rules)\n self.apply_param_rules(my_rules)\n return\n"
] |
[
[
"numpy.dot",
"numpy.asarray",
"numpy.zeros",
"numpy.take"
]
] |
dattatreya303/OpenNMT-py
|
[
"f240346274925743379b72c221a47208cc4bd1d0"
] |
[
"onmt/encoders/transformer.py"
] |
[
"\"\"\"\nImplementation of \"Attention is All You Need\"\n\"\"\"\n\nimport torch.nn as nn\n\nimport onmt\nfrom onmt.encoders.encoder import EncoderBase\n# from onmt.utils.misc import aeq\nfrom onmt.modules.position_ffn import PositionwiseFeedForward\n\n\nclass TransformerEncoderLayer(nn.Module):\n \"\"\"\n A single layer of the transformer encoder.\n\n Args:\n d_model (int): the dimension of keys/values/queries in\n MultiHeadedAttention, also the input size of\n the first-layer of the PositionwiseFeedForward.\n heads (int): the number of head for MultiHeadedAttention.\n d_ff (int): the second-layer of the PositionwiseFeedForward.\n dropout (float): dropout probability(0-1.0).\n \"\"\"\n\n def __init__(self, d_model, heads, d_ff, dropout):\n super(TransformerEncoderLayer, self).__init__()\n\n self.self_attn = onmt.modules.MultiHeadedAttention(\n heads, d_model, dropout=dropout)\n self.feed_forward = PositionwiseFeedForward(d_model, d_ff, dropout)\n self.layer_norm = nn.LayerNorm(d_model, eps=1e-6)\n self.dropout = nn.Dropout(dropout)\n\n def forward(self, inputs, mask):\n \"\"\"\n Transformer Encoder Layer definition.\n\n Args:\n inputs (`FloatTensor`): `[batch_size x src_len x model_dim]`\n mask (`LongTensor`): `[batch_size x src_len x src_len]`\n\n Returns:\n (`FloatTensor`):\n\n * outputs `[batch_size x src_len x model_dim]`\n \"\"\"\n input_norm = self.layer_norm(inputs)\n context, _ = self.self_attn(input_norm, input_norm, input_norm,\n mask=mask)\n out = self.dropout(context) + inputs\n return self.feed_forward(out)\n\n\nclass TransformerEncoder(EncoderBase):\n \"\"\"\n The Transformer encoder from \"Attention is All You Need\".\n\n\n .. mermaid::\n\n graph BT\n A[input]\n B[multi-head self-attn]\n C[feed forward]\n O[output]\n A --> B\n B --> C\n C --> O\n\n Args:\n num_layers (int): number of encoder layers\n d_model (int): size of the model\n heads (int): number of heads\n d_ff (int): size of the inner FF layer\n dropout (float): dropout parameters\n embeddings (:obj:`onmt.modules.Embeddings`):\n embeddings to use, should have positional encodings\n\n Returns:\n (`FloatTensor`, `FloatTensor`):\n\n * embeddings `[src_len x batch_size x model_dim]`\n * memory_bank `[src_len x batch_size x model_dim]`\n \"\"\"\n\n def __init__(self, num_layers, d_model, heads, d_ff,\n dropout, embeddings):\n super(TransformerEncoder, self).__init__()\n\n self.num_layers = num_layers\n self.embeddings = embeddings\n self.transformer = nn.ModuleList(\n [TransformerEncoderLayer(d_model, heads, d_ff, dropout)\n for _ in range(num_layers)])\n self.layer_norm = nn.LayerNorm(d_model, eps=1e-6)\n\n def forward(self, src, lengths=None):\n \"\"\" See :obj:`EncoderBase.forward()`\"\"\"\n self._check_args(src, lengths)\n\n emb = self.embeddings(src)\n\n out = emb.transpose(0, 1).contiguous()\n words = src[:, :, 0].transpose(0, 1)\n w_batch, w_len = words.size()\n padding_idx = self.embeddings.word_padding_idx\n mask = words.data.eq(padding_idx).unsqueeze(1) # [B, 1, T]\n # Run the forward pass of every layer of the tranformer.\n for i in range(self.num_layers):\n out = self.transformer[i](out, mask)\n out = self.layer_norm(out)\n\n return emb, out.transpose(0, 1).contiguous(), lengths\n"
] |
[
[
"torch.nn.Dropout",
"torch.nn.LayerNorm"
]
] |
samvidav/VISTA
|
[
"e5bc3e649133714bc1dc6a9edde2a5be826f4f49"
] |
[
"Shiny/getStringInteractors.py"
] |
[
"# ADAPTED FROM \n# Michelle Kennedy\n# 1.29.2019\n\n# Below are the main functions needed to get the string interactions. \n\nimport requests\n\n# for outputting the response from STRING to a pandas data frame\nimport io \nimport pandas as pd\nimport numpy as np\nimport time\n\nclass GetSTRINGInteractions:\n def __init__(self):\n pass\n \n def to_query_string(self, mylist, sep): #can also accept arrays\n '''convert a list to a string that can be used as a query string in an http post request'''\n \n l = ''\n\n for item in mylist:\n try:\n l = l + str(item) + sep\n \n except TypeError: # exception to deal with NaNs in mylist\n pass\n \n return l\n \n def map_identifiers_string(self, proteins, species):\n '''Use STRING's API to retrive the corresponding string identifiers for each protein.\n I highly reccommend using this function prior to querying for interactions. \n It significantly decreases the request response time'''\n \n # STRING will only let you query 2000 proteins at a time, otherwise you get an error message back\n \n if len(proteins) >= 2000:\n n_chunks = int(np.ceil(len(proteins)/2000))\n dfs = []\n \n for chunk in range(n_chunks):\n ps = proteins[2000*chunk:2000*(chunk+1)]\n \n p = self.to_query_string(ps, '%0D') #each protein on a new line\n\n url = 'https://string-db.org/api/tsv/get_string_ids'\n params = {'identifiers': p, 'species':species, 'echo_query': 1, 'caller_identity': 'Princeton_University'}\n\n r = requests.post(url, data = params)\n _df = pd.read_csv(io.StringIO(r.text), sep = '\\t', header = 0, index_col = None)\n \n dfs.append(_df)\n time.sleep(1)\n \n df = pd.concat(dfs, axis = 0, join = 'outer')\n \n else:\n ps = proteins\n \n p = self.to_query_string(ps, '%0D') #each protein on a new line\n\n url = 'https://string-db.org/api/tsv/get_string_ids'\n params = {'identifiers': p, 'species':species, 'echo_query': 1, 'caller_identity': 'Princeton_University'}\n\n r = requests.post(url, data = params)\n df = pd.read_csv(io.StringIO(r.text), sep = '\\t', header = 0, index_col = None)\n \n \n df = df[['stringId', 'queryItem']].set_index('stringId')\n \n return df\n\n \n def get_interactions(self, IDs, species):\n \n # STRING will only let you query 2000 proteins at a time\n \n if len(IDs) > 2000:\n \n n_chunks = int(np.ceil(len(IDs)/2000))\n \n dfs = []\n \n for chunk in range(n_chunks):\n ID_list = IDs[2000*chunk:2000*(chunk+1)]\n \n p = self.to_query_string(ID_list, '%0D') #each ID on a new line\n\n url = 'https://string-db.org/api/tsv/network'\n params = {'identifiers': p, 'species':species, 'caller_identity': 'Princeton_University'}\n\n r = requests.post(url, data = params)\n _df = pd.read_csv(io.StringIO(r.text), sep = '\\t', header = 0, index_col = None)\n dfs.append(_df)\n time.sleep(1)\n \n df = pd.concat(dfs, axis = 0, join = 'outer')\n \n else:\n ID_list = IDs\n \n p = self.to_query_string(ID_list, '%0D') #each ID on a new line\n\n url = 'https://string-db.org/api/tsv/network'\n params = {'identifiers': p, 'species':species, 'caller_identity': 'Princeton_University'}\n\n r = requests.post(url, data = params)\n df = pd.read_csv(io.StringIO(r.text), sep = '\\t', header = 0, index_col = None)\n \n return df"
] |
[
[
"pandas.concat"
]
] |
ComitUniandes/ICS-SDN
|
[
"fa1df31849dbe26264eabc1aa58ebda2f79ca698"
] |
[
"francisco-topo/plot_tim_uio.py"
] |
[
"\"\"\"Plot the solution that was generated by differential_equation.py.\"\"\"\n \nfrom numpy import loadtxt\nfrom pylab import figure, plot, xlabel, grid, hold, legend, title, savefig\nfrom matplotlib.font_manager import FontProperties\nimport sys\n\nt, x1, = loadtxt(sys.argv[1] , unpack=True)\n\n\nfigure(1, figsize=(6, 4.5))\n\nxlabel('t')\ngrid(True)\nhold(True)\nlw = 1\n\nplot(t, x1, 'b', linewidth=lw)\n\n#legend((r'$L101$', r'$L102$', r'$L103$'), prop=FontProperties(size=16))\ntitle('Tank Levels with Control')\nsavefig(sys.argv[2], dpi=100)\n"
] |
[
[
"numpy.loadtxt"
]
] |
ColwynGulliford/lume-gpt
|
[
"75f76adf72567c1d527d3ed010c6b60869a5c068"
] |
[
"gpt/parsers.py"
] |
[
"import copy\nfrom . import easygdf\nimport time\nimport numpy as np\nimport re\nimport os\n\nfrom gpt.tools import full_path\n\nimport shutil\n\n# ------ Number parsing ------\ndef isfloat(value):\n try:\n float(value)\n return True\n except ValueError:\n return False\n\ndef find_path(line, pattern=r'\"([^\"]+\\.gdf)\"'):\n\n matches=re.findall(pattern, line)\n return matches\n \n \n \ndef set_support_files_orig(lines, \n original_path, \n target_path='', \n copy_files=False, \n pattern=r'\"([^\"]+\\.gdf)\"', \n verbose=False):\n\n for ii, line in enumerate(lines):\n\n support_files = find_path(line, pattern=pattern)\n\n for support_file in support_files:\n\n #print(full_path(support_file))\n\n abs_original_path = full_path( os.path.join(original_path, os.path.expandvars(support_file)) )\n \n if(copy_files):\n \n abs_target_path = os.path.join(target_path, support_file) \n shutil.copyfile(abs_original_path, abs_target_path, follow_symlinks=True) \n\n if(verbose):\n print(\"Copying file: \", abs_original_path,'->',abs_target_path) \n\n else:\n\n if(os.path.isfile(abs_original_path)):\n lines[ii] = line.replace(support_file, abs_original_path)\n if(verbose):\n print(\"Set path to file: \",lines[ii]) \n \ndef set_support_files(lines, \n original_path, \n target='', \n copy_files=False, \n pattern=r'\"([^\"]+\\.gdf)\"',\n verbose=False):\n \n for ii, line in enumerate(lines):\n\n support_files = find_path(line, pattern=pattern)\n \n \n for support_file in support_files:\n\n abs_original_path = full_path( os.path.join(original_path, os.path.expandvars(support_file)) )\n\n if(copy_files):\n \n abs_target_path = os.path.join(target_path, support_file) \n shutil.copyfile(abs_original_path, abs_target_path, follow_symlinks=True) \n\n if(verbose):\n print(\"Copying file: \", abs_original_path,'->',abs_target_path) \n\n else:\n\n if(os.path.isfile(abs_original_path)):\n \n #print(target, abs_original_path)\n dest = full_path( os.path.join(target, os.path.basename(abs_original_path)) )\n\n # Make symlink\n # Replace old symlinks. \n if os.path.islink(dest):\n os.unlink(dest)\n elif os.path.exists(dest):\n \n if verbose:\n print(dest, 'exists, will not symlink')\n\n continue\n\n # Note that the following will raise an error if the dest is an actual file that exists \n os.symlink(abs_original_path, dest)\n if verbose:\n print('Linked', abs_original_path, 'to', os.path.basenme(dest) )\n\n lines[ii] = line.replace(support_file, os.path.basename(dest))\n\n\ndef parse_gpt_input_file(filePath, condense=False, verbose=False):\n \"\"\"\n Parses GPT input file \n \"\"\"\n\n finput={}\n\n with open(filePath, 'r') as f:\n\n clean_lines = []\n\n # Get lines without comments\n for line in f:\n tokens = line.strip().split('#')\n if(len(tokens[0])>0):\n clean_line = tokens[0].strip().replace('\\n', '')\n clean_lines.append(clean_line)\n\n variables={}\n\n for ii,line in enumerate(clean_lines):\n \n tokens = line.split(\"=\")\n\n if(len(tokens)==2 and isfloat(tokens[1][:-1].strip())):\n \n name = tokens[0].strip()\n value = float(tokens[1][:-1].strip())\n \n if(name not in variables.keys()):\n variables[name]=value \n elif(verbose):\n print(f'Warning: multiple definitions of variable {name} on line {ii}.')\n\n support_files={}\n for ii, line in enumerate(clean_lines):\n for sfile in find_path(line):\n if(sfile not in support_files):\n support_files[ii]=sfile\n \n \n finput['lines']=clean_lines\n finput['variables']=variables\n finput['support_files'] = support_files\n\n return finput\n\n\ndef write_gpt_input_file(finput, inputFile, ccs_beg='wcs'):\n\n #print(inputFile)\n for var in finput['variables'].keys():\n\n value=finput['variables'][var]\n for index, line in enumerate(finput['lines']):\n tokens = line.split('=')\n if(len(tokens)==2 and tokens[0].strip()==var):\n finput[\"lines\"][index]=f'{var}={value};'\n break\n \n with open(inputFile,'w') as f:\n\n for line in finput[\"lines\"]:\n f.write(line+\"\\n\")\n\n if(ccs_beg!=\"wcs\"):\n f.write(f'settransform(\"{ccs_beg}\", 0,0,0, 1,0,0, 0,1,0, \"beam\");\\n')\n\ndef read_particle_gdf_file(gdffile, verbose=0.0, extra_screen_keys=['q','nmacro'], load_files=False): #,'ID', 'm']):\n\n with open(gdffile, 'rb') as f:\n data = easygdf.load_initial_distribution(f, extra_screen_keys=extra_screen_keys)\n\n screen = {}\n n = len(data[0,:])\n if(n>0):\n\n q = data[7,:] # elemental charge/macroparticle\n nmacro = data[8,:] # number of elemental charges/macroparticle\n \n weights = np.abs(data[7,:]*data[8,:])/np.sum(np.abs(data[7,:]*data[8,:]))\n\n screen = {\"x\":data[0,:],\"GBx\":data[1,:],\n \"y\":data[2,:],\"GBy\":data[3,:],\n \"z\":data[4,:],\"GBz\":data[5,:],\n \"t\":data[6,:],\n \"q\":data[7,:],\n \"nmacro\":data[8,:],\n \"w\":weights,\n \"G\":np.sqrt(data[1,:]*data[1,:]+data[3,:]*data[3,:]+data[5,:]*data[5,:]+1)}\n \n #screen[\"Bx\"]=screen[\"GBx\"]/screen[\"G\"]\n #screen[\"By\"]=screen[\"GBy\"]/screen[\"G\"]\n #screen[\"Bz\"]=screen[\"GBz\"]/screen[\"G\"]\n\n screen[\"time\"]=np.sum(screen[\"w\"]*screen[\"t\"])\n screen[\"n\"]=n \n\n return screen\n\ndef read_gdf_file(gdffile, verbose=False, load_fields=False):\n \n # Read in file:\n\n \n #self.vprint(\"Current file: '\"+data_file+\"'\",1,True)\n #self.vprint(\"Reading data...\",1,False)\n t1 = time.time()\n with open(gdffile, 'rb') as f:\n \n if(load_fields):\n extra_tout_keys = ['q', 'nmacro', 'ID', 'm', 'fEx', 'fEy', 'fEz', 'fBx', 'fBy', 'fBz']\n else:\n extra_tout_keys = ['q', 'nmacro', 'ID', 'm']\n \n touts, screens = easygdf.load(f, extra_screen_keys=['q','nmacro', 'ID', 'm'], extra_tout_keys=extra_tout_keys)\n \n t2 = time.time()\n if(verbose):\n print(f' GDF data loaded, time ellapsed: {t2-t1:G} (sec).')\n \n #self.vprint(\"Saving wcs tout and ccs screen data structures...\",1,False)\n\n tdata, fields = make_tout_dict(touts, load_fields=load_fields)\n pdata = make_screen_dict(screens)\n\n return (tdata, pdata, fields)\n\n\n\n\ndef make_tout_dict(touts, load_fields=False):\n\n tdata=[]\n fields = []\n count = 0\n for data in touts:\n n=len(data[0,:])\n \n if(n>0):\n\n q = data[7,:] # elemental charge/macroparticle\n nmacro = data[8,:] # number of elemental charges/macroparticle\n \n if(np.sum(q)==0 or np.sum(nmacro)==0):\n weights = data[10,:]/np.sum(data[10,:]) # Use the mass if no charge is specified\n else:\n weights = np.abs(data[7,:]*data[8,:])/np.sum(np.abs(data[7,:]*data[8,:]))\n\n tout = {\"x\":data[0,:],\"GBx\":data[1,:],\n \"y\":data[2,:],\"GBy\":data[3,:],\n \"z\":data[4,:],\"GBz\":data[5,:],\n \"t\":data[6,:],\n \"q\":data[7,:],\n \"nmacro\":data[8,:],\n \"ID\":data[9,:],\n \"m\":data[10,:],\n \"w\":weights,\n \"G\":np.sqrt(data[1,:]*data[1,:]+data[3,:]*data[3,:]+data[5,:]*data[5,:]+1)}\n\n #tout[\"Bx\"]=tout[\"GBx\"]/tout[\"G\"]\n #tout[\"By\"]=tout[\"GBy\"]/tout[\"G\"]\n #tout[\"Bz\"]=tout[\"GBz\"]/tout[\"G\"]\n\n tout[\"time\"]=np.sum(tout[\"w\"]*tout[\"t\"])\n tout[\"n\"]=len(tout[\"x\"])\n tout[\"number\"]=count\n \n if(load_fields):\n field = {'Ex':data[11,:], 'Ey':data[12,:], 'Ez':data[13,:],\n 'Bx':data[14,:], 'By':data[15,:], 'Bz':data[16,:]}\n else:\n field=None\n \n fields.append(field)\n\n count=count+1\n tdata.append(tout)\n\n return tdata, fields\n\ndef make_screen_dict(screens):\n\n pdata=[]\n \n count=0\n for data in screens:\n n = len(data[0,:])\n if(n>0):\n\n q = data[7,:] # elemental charge/macroparticle\n nmacro = data[8,:] # number of elemental charges/macroparticle\n \n if(np.sum(q)==0 or np.sum(nmacro)==0):\n weights = data[10,:]/np.sum(data[10,:]) # Use the mass if no charge is specified\n else:\n weights = np.abs(data[7,:]*data[8,:])/np.sum(np.abs(data[7,:]*data[8,:]))\n\n screen = {\"x\":data[0,:],\"GBx\":data[1,:],\n \"y\":data[2,:],\"GBy\":data[3,:],\n \"z\":data[4,:],\"GBz\":data[5,:],\n \"t\":data[6,:],\n \"q\":data[7,:],\n \"nmacro\":data[8,:],\n \"ID\":data[9,:],\n \"m\":data[10,:],\n \"w\":weights,\n \"G\":np.sqrt(data[1,:]*data[1,:]+data[3,:]*data[3,:]+data[5,:]*data[5,:]+1)}\n \n #screen[\"Bx\"]=screen[\"GBx\"]/screen[\"G\"]\n #screen[\"By\"]=screen[\"GBy\"]/screen[\"G\"]\n #screen[\"Bz\"]=screen[\"GBz\"]/screen[\"G\"]\n\n screen[\"time\"]=np.sum(screen[\"w\"]*screen[\"t\"])\n screen[\"n\"]=n\n screen[\"number\"]=count\n\n count=count+1\n pdata.append(screen)\n \n t2 = time.time()\n #self.vprint(\"done. Time ellapsed: \"+self.ptime(t1,t2)+\".\",0,True)\n\n ts=np.array([screen['time'] for screen in pdata])\n sorted_indices = np.argsort(ts)\n return [pdata[sii] for sii in sorted_indices]\n\ndef parse_gpt_string(line):\n return re.findall(r'\\\"(.+?)\\\"',line) \n\ndef replace_gpt_string(line,oldstr,newstr):\n\n strs = parse_gpt_string(line)\n assert oldstr in strs, 'Could not find string '+oldstr+' for string replacement.'\n line.replace(oldstr,newstr)\n\n"
] |
[
[
"numpy.array",
"numpy.sum",
"numpy.abs",
"numpy.sqrt",
"numpy.argsort"
]
] |
cristinamicula/profiler
|
[
"3af06b41efa0272401e299b62cfc9bfba10a9b62"
] |
[
"Profiler.py"
] |
[
"import datetime\n\nimport matplotlib.pyplot as plt\nfrom matplotlib.backends.backend_pdf import PdfPages\n\n\nclass Operation:\n def __init__(self, name: str, max_number: int):\n self.name = name\n self.operation_count = 0\n self.max_number = max_number\n\n def count(self, value: int = 1):\n if value < 1:\n raise Exception('''Value can't be non-positive.''')\n self.operation_count += value\n\n def to_string(self):\n return f'{self.name}, {self.max_number}, {self.operation_count}'\n\n\nclass Profiler:\n def __init__(self, name: str = 'Profiler'):\n self.name = name\n self.operations = {}\n self.joint_operations = {}\n\n def create_operation(self, name: str, max_number: int) -> Operation:\n op = Operation(name, max_number)\n\n try:\n self.operations[name].append(op)\n except:\n self.operations[name] = [op]\n\n return op\n\n def join_operations(self, name: str, *names):\n\n for opName in names:\n if opName not in self.operations.keys():\n raise Exception(f'''Operation {opName} not existent.''')\n\n joint_operation = [self.operations[opName] for opName in names]\n\n self.joint_operations[name] = joint_operation\n\n for opName in names:\n del self.operations[opName]\n\n def show_report(self):\n with PdfPages(self.name + '.pdf') as pdf:\n\n for key, value in self.operations.items():\n figure = plt.figure(key)\n\n plt.plot([k.max_number for k in value], [k.operation_count for k in value])\n\n plt.title(key)\n\n plt.ylabel('Operation count')\n plt.xlabel('Size')\n\n plt.show()\n\n pdf.savefig(figure)\n\n plt.close()\n\n for key, value in self.joint_operations.items():\n figure = plt.figure(key)\n\n plt.title(key)\n\n for operation in value:\n plt.plot([k.max_number for k in operation], [k.operation_count for k in operation])\n\n plt.ylabel('Operation count')\n plt.xlabel('Size')\n\n plt.legend([x[0].name for x in value])\n plt.show()\n pdf.savefig(figure)\n plt.close()\n\n d = pdf.infodict()\n d['Title'] = self.name\n try:\n import getpass\n d['Author'] = getpass.getuser()\n except:\n pass\n\n d['CreationDate'] = datetime.datetime.today()\n d['ModDate'] = datetime.datetime.today()\n\n\nif __name__ == '__main__':\n profiler = Profiler('Test for profiler')\n\n for i in range(1, 10):\n op1 = profiler.create_operation('aia a fost', i)\n op2 = profiler.create_operation('asta nu chiar', i)\n op3 = profiler.create_operation('asta poate', i)\n\n op1.count(i ** 3)\n op2.count(i * i)\n op3.count(i * 20)\n\n profiler.join_operations('idk', 'aia a fost', 'asta nu chiar')\n\n profiler.show_report()\n"
] |
[
[
"matplotlib.pyplot.xlabel",
"matplotlib.backends.backend_pdf.PdfPages",
"matplotlib.pyplot.plot",
"matplotlib.pyplot.title",
"matplotlib.pyplot.close",
"matplotlib.pyplot.legend",
"matplotlib.pyplot.figure",
"matplotlib.pyplot.ylabel",
"matplotlib.pyplot.show"
]
] |
shangeth/Speaker-Representation-Learning
|
[
"44d6cade2f9dd3fefb60f63d8c19e229e701faf6"
] |
[
"visualize_test_embedding.py"
] |
[
"from argparse import ArgumentParser\nimport pytorch_lightning as pl\nimport torch\nimport torch.utils.data as data\nimport torchaudio\nimport numpy as np\nfrom tqdm import tqdm\n\nfrom dataset import EmbDataset\nfrom models.lightning_model import RepresentationModel\n\nfrom torch.utils.tensorboard import SummaryWriter\n\nif __name__ == \"__main__\":\n\n parser = ArgumentParser(add_help=True)\n parser.add_argument('--data_root', type=str, default='./final_emb_data')\n parser.add_argument('--wav_len', type=int, default=16000 * 4)\n parser.add_argument('--model_path', type=str, default=\"/home/n1900235d/INTERSPEECH/lightning_logs/version_1/checkpoints/epoch=990-step=49549.ckpt\")\n parser.add_argument('--hidden_dim', type=float, default=512)\n\n parser = pl.Trainer.add_argparse_args(parser)\n hparams = parser.parse_args()\n\n HPARAMS = {\n 'data_path' : hparams.data_root,\n 'data_wav_len' : hparams.wav_len,\n 'data_wav_augmentation' : 'Random Crop, Additive Noise',\n 'data_label_scale' : 'Standardization',\n\n 'training_optimizer' : 'Adam',\n 'training_lr' : 1e-3,\n 'training_lr_scheduler' : '-',\n\n 'model_architecture' : 'wav2vec + soft-attention',\n 'model_finetune' : 'Layer 5&6',\n 'hidden_dim': hparams.hidden_dim,\n }\n\n test_dataset = EmbDataset(root=hparams.data_root, wav_len=hparams.wav_len)\n # model = RepresentationModel.load_from_checkpoint(hparams.model_path, hparams=HPARAMS)\n model = RepresentationModel(HPARAMS)\n checkpoint = torch.load(hparams.model_path, map_location=lambda storage, loc: storage)\n model.load_state_dict(checkpoint['state_dict'])\n\n # x, ys, yg = test_dataset[0]\n # print(x.shape, 'S=' ,ys,'G=', yg)\n\n embs = []\n labels = []\n gender = []\n\n for x, ys, yg in tqdm(test_dataset):\n # z = torch.randn(1, 100).view(-1)\n z = model.E.feature_extractor(x)\n # z_a = model.A(z).view(-1).cpu()\n z_a = z.mean(2).view(-1).cpu()\n embs.append(z_a)\n\n labels.append(ys)\n gender.append(yg)\n\n embs = torch.stack(embs)\n writer = SummaryWriter()\n \n writer.add_embedding(embs, metadata=gender, tag='gender')\n writer.add_embedding(embs, metadata=labels, tag='speakers')\n writer.close()\n\n\n "
] |
[
[
"torch.stack",
"torch.utils.tensorboard.SummaryWriter",
"torch.load"
]
] |
Menerve/pylearn2
|
[
"ad7bcfda3294404aebd71f5a5c4a8623d401a98e"
] |
[
"pylearn2/utils/video.py"
] |
[
"\"\"\"\nUtilities for working with videos, pulling out patches, etc.\n\"\"\"\nimport numpy\n\nfrom pylearn2.utils.rng import make_np_rng\n\n__author__ = \"David Warde-Farley\"\n__copyright__ = \"Copyright 2011, David Warde-Farley / Universite de Montreal\"\n__license__ = \"BSD\"\n__maintainer__ = \"David Warde-Farley\"\n__email__ = \"wardefar@iro\"\n__all__ = [\"get_video_dims\", \"spatiotemporal_cubes\"]\n\n\ndef get_video_dims(fname):\n \"\"\"\n Pull out the frame length, spatial height and spatial width of\n a video file using ffmpeg.\n\n Parameters\n ----------\n fname : str\n Path to video file to be inspected.\n\n Returns\n -------\n shape : tuple\n The spatiotemporal dimensions of the video\n (length, height, width).\n \"\"\"\n try:\n import pyffmpeg\n except ImportError:\n raise ImportError(\"This function requires pyffmpeg \"\n \"<http://code.google.com/p/pyffmpeg/>\")\n mp = pyffmpeg.FFMpegReader()\n try:\n mp.open(fname)\n tracks = mp.get_tracks()\n for track in tracks:\n if isinstance(track, pyffmpeg.VideoTrack):\n break\n else:\n raise ValueError('no video track found')\n return (track.duration(),) + track.get_orig_size()\n finally:\n mp.close()\n\n\nclass FrameLookup(object):\n \"\"\"\n Class encapsulating the logic of turning a frame index into a\n collection of files into the frame index of a specific video file.\n\n Item-indexing on this object will yield a (filename, nframes, frame_no)\n tuple, where nframes is the number of frames in the given file\n (mainly for checking that we're far enough from the end so that we\n can sample a big enough chunk).\n\n Parameters\n ----------\n names_ang_lengths : WRITEME\n \"\"\"\n def __init__(self, names_and_lengths):\n self.files, self.lengths = zip(*names_and_lengths)\n self.terminals = numpy.cumsum([s[1] for s in names_and_lengths])\n\n def __getitem__(self, i):\n idx = (i < self.terminals).nonzero()[0][0]\n frame_no = i\n if idx > 0:\n frame_no -= self.terminals[idx - 1]\n return self.files[idx], self.lengths[idx], frame_no\n\n def __len__(self):\n return self.terminals[-1]\n\n def __iter__(self):\n raise TypeError('iteration not supported')\n\n\ndef spatiotemporal_cubes(file_tuples, shape, n_patches=numpy.inf, rng=None):\n \"\"\"\n Generator function that yields a stream of (filename, slicetuple)\n representing a spatiotemporal patch of that file.\n\n Parameters\n ----------\n file_tuples : list of tuples\n Each element should be a 2-tuple consisting of a filename\n (or arbitrary identifier) and a (length, height, width)\n shape tuple of the dimensions (number of frames in the video,\n height and width of each frame).\n\n shape : tuple\n A shape tuple consisting of the desired (length, height, width)\n of each spatiotemporal patch.\n\n n_patches : int, optional\n The number of patches to generate. By default, generates patches\n infinitely.\n\n rng : RandomState object or seed, optional\n The random number generator (or seed) to use. Defaults to None,\n meaning it will be seeded from /dev/urandom or the clock.\n\n Returns\n -------\n generator : generator object\n A generator that yields a stream of (filename, slicetuple) tuples.\n The slice tuple is such that it indexes into a 3D array containing\n the entire clip with frames indexed along the first axis, rows\n along the second and columns along the third.\n \"\"\"\n frame_lookup = FrameLookup([(a, b[0]) for a, b in file_tuples])\n file_lookup = dict(file_tuples)\n patch_length, patch_height, patch_width = shape\n done = 0\n rng = make_np_rng(rng, which_method=\"random_integers\")\n while done < n_patches:\n frame = numpy.random.random_integers(0, len(frame_lookup) - 1)\n filename, file_length, frame_no = frame_lookup[frame]\n # Check that there is a contiguous block of frames starting at\n # frame_no that is at least as long as our desired cube length.\n if file_length - frame_no < patch_length:\n continue\n _, video_height, video_width = file_lookup[filename][:3]\n # The last row and column in which a patch could \"start\" to still\n # fall within frame.\n last_row = video_height - patch_height\n last_col = video_width - patch_width\n row = numpy.random.random_integers(0, last_row)\n col = numpy.random.random_integers(0, last_col)\n patch_slice = (slice(frame_no, frame_no + patch_length),\n slice(row, row + patch_height),\n slice(col, col + patch_width))\n done += 1\n yield filename, patch_slice\n"
] |
[
[
"numpy.random.random_integers",
"numpy.cumsum"
]
] |
yanivbl6/mage
|
[
"e251fa814f7091cfcd5630321bf6b00a079523f5"
] |
[
"utils.py"
] |
[
"import numpy as np\nimport random\nimport torch\n\n\ndef set_seed(seed, fully_deterministic=True):\n np.random.seed(seed)\n random.seed(seed)\n torch.manual_seed(seed)\n if torch.cuda.is_available():\n torch.cuda.manual_seed_all(seed)\n if fully_deterministic:\n torch.backends.cudnn.deterministic = True"
] |
[
[
"numpy.random.seed",
"torch.manual_seed",
"torch.cuda.is_available",
"torch.cuda.manual_seed_all"
]
] |
chiahsuan156/chiahsuan156.github.io
|
[
"d87f649940cccec13fc9896f077abdb7c3ff266f"
] |
[
"markdown_generator/publications.py"
] |
[
"\n# coding: utf-8\n\n# # Publications markdown generator for academicpages\n# \n# Takes a TSV of publications with metadata and converts them for use with [academicpages.github.io](academicpages.github.io). This is an interactive Jupyter notebook, with the core python code in publications.py. Run either from the `markdown_generator` folder after replacing `publications.tsv` with one that fits your format.\n# \n# TODO: Make this work with BibTex and other databases of citations, rather than Stuart's non-standard TSV format and citation style.\n# \n\n# ## Data format\n# \n# The TSV needs to have the following columns: pub_date, title, venue, excerpt, citation, site_url, and paper_url, with a header at the top. \n# \n# - `excerpt` and `paper_url` can be blank, but the others must have values. \n# - `pub_date` must be formatted as YYYY-MM-DD.\n# - `url_slug` will be the descriptive part of the .md file and the permalink URL for the page about the paper. The .md file will be `YYYY-MM-DD-[url_slug].md` and the permalink will be `https://[yourdomain]/publications/YYYY-MM-DD-[url_slug]`\n\n\n# ## Import pandas\n# \n# We are using the very handy pandas library for dataframes.\n\n# In[2]:\n\nimport pandas as pd\n\n\n# ## Import TSV\n# \n# Pandas makes this easy with the read_csv function. We are using a TSV, so we specify the separator as a tab, or `\\t`.\n# \n# I found it important to put this data in a tab-separated values format, because there are a lot of commas in this kind of data and comma-separated values can get messed up. However, you can modify the import statement, as pandas also has read_excel(), read_json(), and others.\n\n# In[3]:\n\npublications = pd.read_csv(\"publications.tsv\", sep=\"\\t\", header=0)\npublications\n\n\n# ## Escape special characters\n# \n# YAML is very picky about how it takes a valid string, so we are replacing single and double quotes (and ampersands) with their HTML encoded equivilents. This makes them look not so readable in raw format, but they are parsed and rendered nicely.\n\n# In[4]:\n\nhtml_escape_table = {\n \"&\": \"&\",\n '\"': \""\",\n \"'\": \"'\"\n }\n\ndef html_escape(text):\n \"\"\"Produce entities within text.\"\"\"\n return \"\".join(html_escape_table.get(c,c) for c in text)\n\n\n# ## Creating the markdown files\n# \n# This is where the heavy lifting is done. This loops through all the rows in the TSV dataframe, then starts to concatentate a big string (```md```) that contains the markdown for each type. It does the YAML metadata first, then does the description for the individual page. If you don't want something to appear (like the \"Recommended citation\")\n\n# In[5]:\n\nimport os\nfor row, item in publications.iterrows():\n \n md_filename = str(item.pub_date) + \"-\" + item.url_slug + \".md\"\n html_filename = str(item.pub_date) + \"-\" + item.url_slug\n year = item.pub_date[:4]\n \n ## YAML variables\n \n md = \"---\\ntitle: \\\"\" + item.title + '\"\\n'\n \n md += \"\"\"collection: publications\"\"\"\n \n md += \"\"\"\\npermalink: /publication/\"\"\" + html_filename\n \n if len(str(item.excerpt)) > 5:\n md += \"\\nexcerpt: '\" + html_escape(item.excerpt) + \"'\"\n \n md += \"\\ndate: \" + str(item.pub_date) \n \n md += \"\\nvenue: '\" + html_escape(item.venue) + \"'\"\n \n if len(str(item.paper_url)) > 5:\n md += \"\\npaperurl: '\" + item.paper_url + \"'\"\n \n md += \"\\ncitation: '\" + html_escape(item.citation) + \"'\"\n \n md += \"\\n---\"\n \n ## Markdown description for individual page\n \n #if len(str(item.paper_url)) > 5:\n # md += \"\\n\\n<a href='\" + item.paper_url + \"'>Download paper here</a>\\n\" \n \n if len(str(item.excerpt)) > 5:\n md += \"\\n\" + html_escape(item.excerpt) + \"\\n\"\n \n md += \"\\nRecommended citation: \" + item.citation\n \n md_filename = os.path.basename(md_filename)\n \n with open(\"../_publications/\" + md_filename, 'w') as f:\n f.write(md)\n\n\n"
] |
[
[
"pandas.read_csv"
]
] |
herilalaina/mosaic_ml
|
[
"4eb6229006061d275ff0b80a1d31514d9d7d22f0",
"4eb6229006061d275ff0b80a1d31514d9d7d22f0"
] |
[
"setup.py",
"mosaic_ml/model_config/classification/extra_trees.py"
] |
[
"# -*- encoding: utf-8 -*-\nimport os\nimport sys\nimport codecs\nimport os.path\nfrom setuptools import setup, find_packages\nfrom setuptools.extension import Extension\nfrom setuptools.command.build_ext import build_ext\n\ndef read(rel_path):\n here = os.path.abspath(os.path.dirname(__file__))\n with codecs.open(os.path.join(here, rel_path), 'r') as fp:\n return fp.read()\n\ndef get_version(rel_path):\n for line in read(rel_path).splitlines():\n if line.startswith('__version__'):\n delim = '\"' if '\"' in line else \"'\"\n return line.split(delim)[1]\n else:\n raise RuntimeError(\"Unable to find version string.\")\n\nif sys.version_info < (3, 5):\n raise ValueError(\n 'Unsupported Python version %d.%d.%d found. Mosaic requires Python '\n '3.5 or higher.' % (sys.version_info.major, sys.version_info.minor, sys.version_info.micro)\n )\n\n\nclass BuildExt(build_ext):\n \"\"\" build_ext command for use when numpy headers are needed.\n SEE tutorial: https://stackoverflow.com/questions/2379898\n SEE fix: https://stackoverflow.com/questions/19919905\n \"\"\"\n\n def finalize_options(self):\n build_ext.finalize_options(self)\n # Prevent numpy from thinking it is still in its setup process:\n # __builtins__.__NUMPY_SETUP__ = False\n import numpy\n self.include_dirs.append(numpy.get_include())\n\n\nHERE = os.path.abspath(os.path.dirname(__file__))\nsetup_reqs = ['Cython', 'numpy']\nwith open(os.path.join(HERE, 'requirements.txt')) as fp:\n install_reqs = [r.rstrip() for r in fp.readlines()\n if not r.startswith('#') and not r.startswith('git+')]\n\nsetup(\n name='mosaic_ml',\n author='Herilalaina Rakotoarison',\n author_email='[email protected]',\n description='Mosaic for Machine Learning algorithm.',\n version=get_version(\"mosaic_ml/__init__.py\"),\n cmdclass={'build_ext': BuildExt},\n packages=find_packages(exclude=['examples', 'test']),\n setup_requires=setup_reqs,\n install_requires=install_reqs,\n include_package_data=True,\n license='BSD',\n platforms=['Linux'],\n classifiers=[\n \"Environment :: Console\",\n \"Intended Audience :: Developers\",\n \"Intended Audience :: Education\",\n \"Intended Audience :: Science/Research\",\n \"Intended Audience :: Information Technology\",\n \"License :: OSI Approved :: BSD License\",\n \"Natural Language :: English\",\n \"Operating System :: OS Independent\",\n \"Topic :: Scientific/Engineering :: Artificial Intelligence\",\n \"Topic :: Scientific/Engineering :: Information Analysis\",\n 'Programming Language :: Python :: 3.5',\n 'Programming Language :: Python :: 3.6',\n 'Programming Language :: Python :: 3.7',\n ],\n python_requires='>=3.5.*',\n)\n",
"import numpy as np\n\nfrom mosaic_ml.model_config.util import convert_multioutput_multiclass_to_multilabel\nfrom mosaic_ml.model_config.util import check_for_bool, check_none\n\n\nclass ExtraTreesClassifier:\n\n def __init__(self, criterion, min_samples_leaf,\n min_samples_split, max_features, bootstrap, max_leaf_nodes,\n max_depth, min_weight_fraction_leaf, min_impurity_decrease,\n oob_score=False, n_jobs=1, random_state=None, verbose=0,\n class_weight=None):\n\n self.n_estimators = self.get_max_iter()\n self.estimator_increment = 10\n if criterion not in (\"gini\", \"entropy\"):\n raise ValueError(\"'criterion' is not in ('gini', 'entropy'): \"\n \"%s\" % criterion)\n self.criterion = criterion\n\n if check_none(max_depth):\n self.max_depth = None\n else:\n self.max_depth = int(max_depth)\n if check_none(max_leaf_nodes):\n self.max_leaf_nodes = None\n else:\n self.max_leaf_nodes = int(max_leaf_nodes)\n\n self.min_samples_leaf = int(min_samples_leaf)\n self.min_samples_split = int(min_samples_split)\n self.max_features = float(max_features)\n self.bootstrap = check_for_bool(bootstrap)\n self.min_weight_fraction_leaf = float(min_weight_fraction_leaf)\n self.min_impurity_decrease = float(min_impurity_decrease)\n self.oob_score = oob_score\n self.n_jobs = int(n_jobs)\n self.random_state = random_state\n self.verbose = int(verbose)\n self.class_weight = class_weight\n self.estimator = None\n\n def fit(self, X, y, sample_weight=None):\n self.iterative_fit(\n X, y, n_iter=2, refit=True, sample_weight=sample_weight\n )\n iteration = 2\n while not self.configuration_fully_fitted():\n n_iter = int(2 ** iteration / 2)\n self.iterative_fit(X, y, n_iter=n_iter,\n sample_weight=sample_weight)\n iteration += 1\n return self\n\n def iterative_fit(self, X, y, sample_weight=None, n_iter=1, refit=False):\n from sklearn.ensemble import ExtraTreesClassifier as ETC\n\n if refit:\n self.estimator = None\n\n if self.estimator is None:\n max_features = int(X.shape[1] ** float(self.max_features))\n self.estimator = ETC(n_estimators=n_iter,\n criterion=self.criterion,\n max_depth=self.max_depth,\n min_samples_split=self.min_samples_split,\n min_samples_leaf=self.min_samples_leaf,\n bootstrap=self.bootstrap,\n max_features=max_features,\n max_leaf_nodes=self.max_leaf_nodes,\n min_weight_fraction_leaf=self.min_weight_fraction_leaf,\n min_impurity_decrease=self.min_impurity_decrease,\n oob_score=self.oob_score,\n n_jobs=self.n_jobs,\n verbose=self.verbose,\n random_state=self.random_state,\n class_weight=self.class_weight,\n warm_start=True)\n\n else:\n self.estimator.n_estimators += n_iter\n self.estimator.n_estimators = min(self.estimator.n_estimators,\n self.n_estimators)\n\n self.estimator.fit(X, y, sample_weight=sample_weight)\n return self\n\n def configuration_fully_fitted(self):\n if self.estimator is None:\n return False\n return not len(self.estimator.estimators_) < self.n_estimators\n\n def predict(self, X):\n if self.estimator is None:\n raise NotImplementedError\n return self.estimator.predict(X)\n\n def predict_proba(self, X):\n if self.estimator is None:\n raise NotImplementedError()\n probas = self.estimator.predict_proba(X)\n probas = convert_multioutput_multiclass_to_multilabel(probas)\n return probas\n\n @staticmethod\n def get_max_iter():\n return 512\n\n\ndef get_model(name, config, random_state):\n list_param = {\"random_state\": random_state,\n \"class_weight\": \"weighting\" if config[\"class_weight\"] == \"weighting\" else None}\n for k in config:\n if k.startswith(\"classifier:extra_trees:\"):\n param_name = k.split(\":\")[2]\n list_param[param_name] = config[k]\n\n model = ExtraTreesClassifier(**list_param)\n return (name, model)\n"
] |
[
[
"numpy.get_include"
],
[
"sklearn.ensemble.ExtraTreesClassifier"
]
] |
gecheline/stargrit
|
[
"ec293eaa9f911145f05a297e6e4c321b6adebbd0"
] |
[
"stargrit/radiative_transfer/cobain/gray.py"
] |
[
"import numpy as np \nimport stargrit as sg\nimport quadpy.sphere as quadsph\nimport scipy.interpolate as spint\nfrom stargrit.structure import potentials\nfrom stargrit.radiative_transfer.cobain.general import RadiativeTransfer, DiffrotStarRadiativeTransfer, ContactBinaryRadiativeTransfer\nfrom stargrit.geometry.spherical import SphericalMesh\nfrom stargrit.geometry.cylindrical import CylindricalMesh\nimport astropy.units as u \nimport astropy.constants as c\nfrom stargrit.radiative_transfer.quadrature.lebedev import Lebedev \nfrom stargrit.radiative_transfer.quadrature.gauss_legendre import Gauss_Legendre\nfrom stargrit.radiative_transfer.ali import gray_ali\nimport logging\n\n\nclass GrayRadiativeTransfer(RadiativeTransfer):\n\n\n def _mesh_interpolation_functions(self, component='', iter_n=1):\n\n \"\"\"\n Computes interpolation in the chosen iteration of the grid points.\n\n If the opacity and atmosphere are mean/gray, the interpolation objects are returned.\n If monochromatic, the arrays of the latest interpolation are returned, to be used in trilinear_interp method.\n \"\"\"\n\n directory = self.star.directory\n mesh = self.star.mesh \n\n RGI = spint.RegularGridInterpolator\n\n chi = np.load(directory + 'chi%s_%i.npy' % (component, iter_n-1))\n # J = np.load(directory + 'J%s_%i.npy' % (component, iter_n-1))\n S = np.load(directory+'S%s_%s.npy' % (component,iter_n-1))\n\n key1, key2, key3 = self.star.mesh.coords.keys()\n\n chi_interp = RGI(points=[mesh.coords[key1], mesh.coords[key2], mesh.coords[key3]], values=chi)\n S_interp = RGI(points=[mesh.coords[key1], mesh.coords[key2], mesh.coords[key3]], values=S)\n\n I_interp = []\n I = np.load(directory + 'I%s_%i.npy' % (component,iter_n-1))\n for i in range(self.quadrature.nI):\n I_interp.append(RGI(points=[mesh.coords[key1], mesh.coords[key2], mesh.coords[key3]], values=I[i]))\n \n return chi_interp, S_interp, I_interp\n\n\n def _adjust_stepsize(self, Mc, ndir, dirarg, N):\n\n #TODO: implement monochromatic vs mean\n\n exps = np.linspace(-10, -2, 1000)\n stepsizes = 10 ** exps * self.star.mesh.default_units['r']\n\n b = np.ones((len(stepsizes), 3)).T * stepsizes\n\n rs = Mc - ndir * b.T\n\n chis = self._compute_structure(points=(rs/self.star.structure.scale), dirarg=dirarg, \n stepsize=True)\n # compute stepsize\n taus = chis * stepsizes / 2.\n\n diffs = np.abs(taus - 1000.*taus.unit)\n stepsize_final = stepsizes[np.argmin(diffs)]\n\n # print 'stepsize: ', stepsize_final\n return stepsize_final / N\n\n\n def _intensity_step(self, Mc, ndir, dirarg, stepsize, N):\n \n rs = np.array([Mc.to(stepsize.unit).value - i * stepsize.value * ndir for i in range(N)])*stepsize.unit\n paths = np.array([i * stepsize.value for i in range(N)])*stepsize.unit\n\n chis, Ss, Is = self._compute_structure(points=(rs/self.star.structure.scale), \n dirarg=dirarg, stepsize=False)\n \n return self._intensity_integral(paths,chis,Ss,Is,N)\n\n\n def _intensity_integral(self, paths, chis, Ss, Is, N, spline_order=1, test=False):\n\n #TODO: implement gray vs monochromatic, handle limits in lambda? - in radiative equlibrium\n chis_sp = spint.UnivariateSpline(paths, chis, k=spline_order, s=0)\n taus = np.array([chis_sp.integral(paths[0].value, paths[i].value) for i in range(N)])\n Ss_exp = Ss * np.exp((-1.)*taus)\n\n if test:\n import matplotlib.pyplot as plt\n plt.plot(taus, Ss_exp)\n plt.ylabel('Ss_exp')\n plt.show()\n\n plt.plot(taus, Ss)\n plt.ylabel('Ss')\n plt.show()\n\n plt.plot(taus, Is)\n plt.ylabel('Is')\n plt.show()\n\n\n Iszero = np.argwhere(Is == 0.0).flatten()\n Ssezero = np.argwhere(Ss_exp == 0.0).flatten()\n\n if Iszero.size == 0 and Ssezero.size == 0:\n nbreak = N - 1\n else:\n nbreak = np.min(np.hstack((Iszero, Ssezero)))\n\n if nbreak > 1:\n taus = taus[:nbreak + 1]\n Ss_exp = Ss_exp[:nbreak + 1]\n Ss_exp[(Ss_exp == np.nan) | (Ss_exp == np.inf) | (Ss_exp == -np.nan) | (Ss_exp == -np.inf)] = 0.0\n\n taus_u, indices = np.unique(taus, return_index=True)\n\n if len(taus_u) > 1:\n Sexp_sp = spint.UnivariateSpline(taus[indices], Ss_exp[indices], k=spline_order, s=0)\n I = Is[nbreak] * np.exp(-taus[-1]) + Sexp_sp.integral(taus[0], taus[-1])\n # print 'I = %s, I0 = %s, Sint = %s, tau = %s, steps = %s' % (I, Is[nbreak], Sexp_sp.integral(taus[0], taus[-1]), taus[-1], N)\n else:\n I = 0.0\n\n if np.isnan(I) or I < 0.:\n I = 0.0\n\n else:\n taus = taus[:nbreak + 1]\n I = 0.0\n\n return nbreak, taus[-1], Is[nbreak], I\n\n\n def _compute_intensity(self, Mc, n):\n\n R = self._transformation_matrix(n)\n\n Is_j = np.zeros(self.quadrature.nI)\n taus_j = np.zeros(self.quadrature.nI)\n\n for dirarg in range(self.quadrature.nI):\n coords = self.quadrature.azimuthal_polar[dirarg]\n # print 'Computing direction %s, coords %s' % (dirarg,coords)\n ndir = self._rotate_direction_wrt_normal(Mc=Mc, coords=coords, R=R)\n\n stepsize = self._adjust_stepsize(Mc, ndir, dirarg, self.ray_discretization)\n \n nbreak, tau, I0, I = self._intensity_step(Mc, ndir, dirarg, stepsize, self.ray_discretization)\n \n # subd = 'no'\n # N = self.ray_discretization\n # if (nbreak < N and I0 != 0.0) or nbreak == self.ray_discretization-1:\n # if (nbreak < N and I0 != 0.0):\n # subd = 'yes'\n # div = N / nbreak\n # stepsize = stepsize / div\n # elif nbreak == self.ray_discretization - 1:\n # subd = 'yes'\n # N = self.ray_discretization * 2\n # else:\n # subd = 'yes'\n # div = N / nbreak\n # stepsize = stepsize / div\n # N = self.ray_discretization * 2\n\n # nbreak, tau, I0, I = self._intensity_step(Mc, ndir, dirarg, stepsize, N)\n\n taus_j[dirarg], Is_j[dirarg] = tau, I\n\n return Is_j, taus_j, self._compute_mean_intensity(Is_j), self._compute_flux(Is_j)\n\n\n def _compute_structure(self, points, dirarg, stepsize=False):\n raise NotImplementedError\n\n \n def _compute_mean_intensity(self, I):\n\n if isinstance(self.quadrature, Lebedev):\n return self.quadrature.integrate_over_4pi(I)\n elif isinstance(self.quadrature, Gauss_Legendre):\n return self.quadrature.integrate_over_4pi(I[2:].reshape((len(self.quadrature.thetas),len(self.quadrature.phis))))\n else:\n raise TypeError('Unrecognized quadrature type %s' % self.quadrature)\n\n\n def _compute_flux(self, I):\n\n #BUG: this wouldn't hold near the neck of contacts because of the concavity\n if isinstance(self.quadrature, Lebedev):\n return self.quadrature.integrate_outer_m_inner(I)\n elif isinstance(self.quadrature, Gauss_Legendre):\n return self.quadrature.integrate_outer_m_inner(I[2:].reshape((len(self.quadrature.thetas),len(self.quadrature.phis))))\n else:\n raise TypeError('Unrecognized quadrature type %s' % self.quadrature)\n\n \n def _compute_source_function(self, J, points, iter_n, component=''):\n\n if self.conv_method == 'LI':\n S = J \n \n elif 'ALI' in self.conv_method:\n S_previ = np.load(self.star.directory + 'S%s_%i.npy' % (component, iter_n-1)).flatten()[points]\n try:\n conv, mtype = self.conv_method.split(':')\n except:\n logging.info('Assuming ALI Lambda* type is diagonal')\n mtype = 'diagonal'\n \n S = gray_ali.compute_S_step(J.flatten(), S_previ.flatten(), mtype=mtype)\n\n return S\n\n\n def _compute_temperature(self, JF, ttype='J'):\n \n if ttype == 'J':\n return ((np.pi*JF*self.star.atmosphere.default_units['S']/c.sigma_sb)**(0.25)).to(u.K)\n\n elif ttype == 'F':\n return ((JF*self.star.atmosphere.default_units['S']/c.sigma_sb)**(0.25)).to(u.K)\n\n else:\n raise ValueError('Type for temperature computation can only be J or F.')\n\n\nclass DiffrotStarGrayRadiativeTransfer(GrayRadiativeTransfer, DiffrotStarRadiativeTransfer):\n\n\n def _compute_structure(self, points, dirarg, stepsize=False, test=False):\n\n pot_range_grid = [self.star.mesh.coords['pots'].min(), self.star.mesh.coords['pots'].max()]\n pots = self._compute_potentials(points, self._interp_funcs['bbT'], pot_range_grid)\n grid, le = self._compute_interp_regions(pots, points, pot_range_grid)\n thetas, phis = self._compute_coords_for_interpolation(points)\n rhos, Ts = self._interp_funcs['bbrho'](pots[le]), self._interp_funcs['bbT'](pots[le])\n rhos = rhos * self.star.structure.default_units['rho']\n Ts = Ts * self.star.structure.default_units['T']\n chis = np.zeros(len(pots))\n chis[grid] = self._interp_funcs['chi']((pots[grid], thetas[grid], phis[grid]))\n chis[le] = self.star.atmosphere._compute_absorption_coefficient(rhos, Ts)\n\n if test:\n import matplotlib.pyplot as plt\n\n pots_mesh = np.zeros(len(self.star.mesh.rs))\n pot_len = self.star.mesh.dims[1]*self.star.mesh.dims[2]\n for i in range(self.star.mesh.dims[0]):\n pots_mesh[i*pot_len:(i+1)*pot_len] = self.star.mesh.coords['pots'][i]\n \n chis_0 = np.load(self.star.directory+'chi_0.npy').flatten()\n\n plt.plot(pots_mesh, chis_0, label='chi_0')\n plt.plot(pots, chis, '--', label='chi_interp')\n plt.xlim(pots.min(),pots.max())\n plt.ylim(chis.min(), chis.max())\n plt.legend()\n plt.show()\n \n if stepsize:\n return chis\n\n else:\n Ss = np.zeros(len(pots))\n Is = np.zeros(len(pots))\n\n Ss[grid] = self._interp_funcs['S']((pots[grid], thetas[grid], phis[grid]))\n Is[grid] = self._interp_funcs['I'][dirarg]((pots[grid], thetas[grid], phis[grid]))\n Ss[le] = Is[le] = self.star.atmosphere._compute_source_function(Ts)\n\n if test:\n import matplotlib.pyplot as plt\n\n pots_mesh = np.zeros(len(self.star.mesh.rs))\n pot_len = self.star.mesh.dims[1]*self.star.mesh.dims[2]\n for i in range(self.star.mesh.dims[0]):\n pots_mesh[i*pot_len:(i+1)*pot_len] = self.star.mesh.coords['pots'][i]\n \n Ss_0 = np.load(self.star.directory+'S_0.npy').flatten()\n\n plt.plot(pots_mesh, Ss_0, label='S_0')\n plt.plot(pots, Ss, '--', label='S_interp')\n plt.xlim(pots.min(),pots.max())\n plt.ylim(Ss.min(), Ss.max())\n plt.legend()\n plt.show()\n\n return chis, Ss, Is\n\n\nclass ContactBinaryGrayRadiativeTransfer(GrayRadiativeTransfer, ContactBinaryRadiativeTransfer):\n\n\n def _compute_structure(self, points, dirarg, stepsize=False):\n \"\"\"\n Returns the radiative structure in all points along a ray.\n \"\"\"\n pot_range_grid = [self.star.mesh.coords['pots'].min(), self.star.mesh.coords['pots'].max()]\n pots = self._compute_potentials(points, self.star.q, self._interp_funcs['bbT1'], self._interp_funcs['bbT2'], pot_range_grid)\n pots2 = pots / self.star.q + 0.5 * (self.star.q - 1) / self.star.q\n\n if stepsize:\n chis = np.zeros(len(pots))\n else:\n chis = np.zeros(len(pots))\n Ss = np.zeros(len(pots))\n Is = np.zeros(len(pots))\n\n if isinstance(self.star.mesh, SphericalMesh):\n \n grid_prim, grid_sec, le_prim, le_sec = self._compute_interp_regions(pots=pots,points=points,pot_range_grid=pot_range_grid)\n # thetas and phis are returned for all points (zeros out of grid)... maybe fix this?\n thetas, phis = self._compute_coords_for_interpolation(points, grid_prim=grid_prim, grid_sec=grid_sec)\n\n if stepsize:\n chis[grid_prim] = self._interp_funcs['chi1']((pots[grid_prim], thetas[grid_prim], phis[grid_prim]))\n chis[grid_sec] = self._interp_funcs['chi2']((pots[grid_sec], thetas[grid_sec], phis[grid_sec]))\n\n else:\n chis[grid_prim] = self._interp_funcs['chi1']((pots[grid_prim], thetas[grid_prim], phis[grid_prim]))\n chis[grid_sec] = self._interp_funcs['chi2']((pots[grid_sec], thetas[grid_sec], phis[grid_sec]))\n Ss[grid_prim] = self._interp_funcs['S1']((pots[grid_prim], thetas[grid_prim], phis[grid_prim]))\n Ss[grid_sec] = self._interp_funcs['S2']((pots[grid_sec], thetas[grid_sec], phis[grid_sec]))\n Is[grid_prim] = self._interp_funcs['I1'][dirarg]((pots[grid_prim], thetas[grid_prim], phis[grid_prim]))\n Is[grid_sec] = self._interp_funcs['I2'][dirarg]((pots[grid_sec], thetas[grid_sec], phis[grid_sec]))\n\n elif isinstance(self.star.mesh, CylindricalMesh):\n\n grid, le_prim, le_sec = self._compute_interp_regions(pots=pots,points=points,pot_range_grid=pot_range_grid)\n # here xnorms and thetas are only those pertaining to grid points\n xnorms, thetas = self._compute_coords_for_interpolation(points, grid=grid, pots=pots)\n \n if stepsize:\n chis[grid] = self._interp_funcs['chi']((pots[grid], xnorms, thetas))\n \n else:\n chis[grid] = self._interp_funcs['chi']((pots, xnorms, thetas))\n Ss[grid] = self._interp_funcs['S']((pots, xnorms, thetas))\n Is[grid] = self._interp_funcs['I'][dirarg]((pots, xnorms, thetas))\n\n else:\n raise ValueError('Geometry not supported with rt_method cobain')\n\n rhos1 = self._interp_funcs['bbrho1'](pots[le_prim])*self.star.structure.default_units['rho']\n rhos2 = self._interp_funcs['bbrho2'](pots2[le_sec])*self.star.structure.default_units['rho']\n Ts1 = self._interp_funcs['bbT1'](pots[le_prim])*self.star.structure.default_units['T']\n Ts2 = self._interp_funcs['bbT2'](pots2[le_sec])*self.star.structure.default_units['T']\n\n if stepsize:\n \n chis[le_prim] = self.star.atmosphere._compute_absorption_coefficient(rhos1, Ts1)\n chis[le_sec] = self.star.atmosphere._compute_absorption_coefficient(rhos2, Ts2)\n\n return chis\n\n else:\n\n chis[le_prim] = self.star.atmosphere._compute_absorption_coefficient(rhos1, Ts1)\n chis[le_sec] = self.star.atmosphere._compute_absorption_coefficient(rhos2, Ts2)\n Ss[le_prim] = Is[le_prim] = self.star.atmosphere._compute_source_function(Ts1)\n Ss[le_sec] = Is[le_sec] = self.star.atmosphere._compute_source_function(Ts2)\n \n return chis, Ss, Is\n "
] |
[
[
"numpy.isnan",
"numpy.zeros",
"numpy.argmin",
"scipy.interpolate.UnivariateSpline",
"matplotlib.pyplot.plot",
"numpy.load",
"numpy.exp",
"matplotlib.pyplot.legend",
"numpy.hstack",
"numpy.abs",
"matplotlib.pyplot.ylabel",
"matplotlib.pyplot.show",
"numpy.linspace",
"numpy.argwhere",
"numpy.unique"
]
] |
saran87/Real-Time-Data-Processing
|
[
"e460b33c0a73a00a3606d9a5c427cc63c78bcea3"
] |
[
"paktrack/shock/shock_data_processor.py"
] |
[
"import logging\nimport operator\nfrom numpy import mean, sqrt, square\n\nfrom paktrack.common.common import (get_average_for_event)\nfrom paktrack.shock.shock_response_spectrum import srs, np\n\nPEEK_DETECTION_THRESHOLD = 3\nFACE_DETECTION_THRESHOLD = 1.5\nNUMBER_CHUNK_SIZE = 16\nAXIS_MAPPING = {0: \"x\", 1: \"y\", 2: \"z\"}\n\n\nclass ShockDataProcessor(object):\n \"\"\"docstring for ShockDataProcessor\"\"\"\n\n def __init__(self, shock):\n self.shock = shock\n self.logger = logging.getLogger(__name__)\n\n def pre_process_data(self, id):\n\n event = self.shock.get_event(id)\n result = \"no event\"\n\n if event:\n event = self.__process_max_value(event)\n event['average'] = get_average_for_event(event)\n event['drop_height'] = self.__get_drop_height(event)\n event['orientation'] = self.__get_drop_orientation(event)\n event['g_rms'] = self.__get_grms(event) # get_normalized_rms(event)\n event['srs'] = self.get_srs(event)\n event['is_processed'] = True\n result = self.shock.update(event)\n\n return result\n\n def __process_max_value(self, event):\n\n chunk_index = self.__get_peak_chunk_index(event['vector'])\n\n event['max_x'] = self.__get_max_with_index(event['x'], chunk_index)\n event['max_y'] = self.__get_max_with_index(event['y'], chunk_index)\n event['max_z'] = self.__get_max_with_index(event['z'], chunk_index)\n event['max_vector'] = self.__get_max_with_index(\n event['vector'], chunk_index)\n\n return event\n\n def __get_peak_chunk_index(self, signal):\n\n chunks = np.array_split(signal, NUMBER_CHUNK_SIZE)\n max_value = -0.0\n chunk_index = -1\n chunk_count = 0\n\n for chunk in chunks:\n max_index = np.argmax(np.absolute(chunk))\n if abs(chunk[max_index]) > abs(max_value):\n if (abs(chunk[max_index]) - abs(max_value)) > PEEK_DETECTION_THRESHOLD:\n max_value = chunk[max_index]\n chunk_index = chunk_count\n chunk_count += 1\n\n return chunk_index\n\n def __get_max_with_index(self, signal, chunk_index):\n\n chunks = np.array_split(signal, NUMBER_CHUNK_SIZE)\n max_index = np.argmax(np.absolute(chunks[chunk_index]))\n chunk_size = len(signal) / NUMBER_CHUNK_SIZE\n index = (chunk_index * chunk_size) + max_index\n\n return {'value': chunks[chunk_index][max_index], 'index': index}\n\n def __normalized_singal(self, signal):\n\n signal = np.array(signal)\n theta = np.average(signal)\n signal = signal - theta\n\n return signal\n\n def __get_drop_height(self, event):\n \"\"\"\n\n :param event:\n :return:\n \"\"\"\n \"\"\"add 1 for getting correct milliseconds, since array index starts\"\"\"\n max_index = event['max_vector']['index'] + 1\n t = (max_index * 0.000625) + (70.0 / 1000.0)\n height = 4.9 * (t ** 2)\n return height\n\n def __get_drop_orientation(self, event):\n max_axis_values = {'max_x': abs(event['max_x']['value']), 'max_y': abs(event['max_y']['value']),\n 'max_z': abs(event['max_z']['value'])}\n max_axis_values = sorted(max_axis_values.items(), key=operator.itemgetter(1), reverse=True)\n faces = []\n\n key, value = max_axis_values[0]\n faces.append(self.__get_face(key, event[key]['value']))\n max_value = value\n del max_axis_values[0]\n\n for key, value in max_axis_values:\n ratio = max(max_value, value) / min(max_value, value) if min(max_value, value) > 0.0 else max(max_value,\n value)\n if abs(ratio) <= FACE_DETECTION_THRESHOLD:\n faces.append(self.__get_face(key, event[key]['value']))\n\n return faces\n\n def __get_face(self, axis, value):\n is_negative = False if value >= 0 else True\n\n if axis is \"max_y\":\n return 6 if is_negative else 5\n\n elif axis is \"max_x\":\n return 4 if is_negative else 2\n\n elif axis is \"max_z\":\n return 1 if is_negative else 3\n\n def __get_grms(self, event):\n \"\"\" Calculate the mean root square of the maximum values for each axis\n :param event:\n :return: grms\n \"\"\"\n x = event['max_x']['value']\n y = event['max_y']['value']\n z = event['max_z']['value']\n return sqrt(mean([square(x), square(y), square(z)]))\n\n def get_srs(self, event):\n\n return {'x': srs(event['x']),\n 'y': srs(event['y']),\n 'z': srs(event['z']),\n 'vector': srs(event['vector'])}\n"
] |
[
[
"numpy.square"
]
] |
PacktPublishing/Deep-Learning-with-Keras-V-
|
[
"5b2ad1bbbddd994437506198eceacccc5cee4bcc"
] |
[
"Section-1/Keras_MNIST_V1.py"
] |
[
"from __future__ import print_function\nimport numpy as np\nfrom keras.datasets import mnist\nfrom keras.models import Sequential\nfrom keras.layers.core import Dense, Activation\nfrom keras.optimizers import SGD\nfrom keras.utils import np_utils\n\nnp.random.seed(1671) # for reproducibility\n\n# network and training\nNB_EPOCH = 200\nBATCH_SIZE = 128\nVERBOSE = 1\nNB_CLASSES = 10 # number of outputs = number of digits\nOPTIMIZER = SGD() # SGD optimizer, explained later in this chapter\nN_HIDDEN = 128\nVALIDATION_SPLIT=0.2 # how much TRAIN is reserved for VALIDATION\n\n# data: shuffled and split between train and test sets\n#\n(X_train, y_train), (X_test, y_test) = mnist.load_data()\n\n#X_train is 60000 rows of 28x28 values --> reshaped in 60000 x 784\nRESHAPED = 784\n#\nX_train = X_train.reshape(60000, RESHAPED)\nX_test = X_test.reshape(10000, RESHAPED)\nX_train = X_train.astype('float32')\nX_test = X_test.astype('float32')\n\n# normalize\n#\nX_train /= 255\nX_test /= 255\nprint(X_train.shape[0], 'train samples')\nprint(X_test.shape[0], 'test samples')\n\n# convert class vectors to binary class matrices\nY_train = np_utils.to_categorical(y_train, NB_CLASSES)\nY_test = np_utils.to_categorical(y_test, NB_CLASSES)\n\n# 10 outputs\n# final stage is softmax\n\nmodel = Sequential()\nmodel.add(Dense(NB_CLASSES, input_shape=(RESHAPED,)))\nmodel.add(Activation('softmax'))\n\nmodel.summary()\n\nmodel.compile(loss='categorical_crossentropy',\n optimizer=OPTIMIZER,\n metrics=['accuracy'])\n\nhistory = model.fit(X_train, Y_train, batch_size=BATCH_SIZE, epochs=NB_EPOCH, verbose=VERBOSE, validation_split=VALIDATION_SPLIT)\n\nscore = model.evaluate(X_test, Y_test, verbose=VERBOSE)\nprint(\"\\nTest score:\", score[0])\nprint('Test accuracy:', score[1])\n"
] |
[
[
"numpy.random.seed"
]
] |
jessecantu/tensorflow
|
[
"60028072a1c3b4376e145b6fea8e4ccd3324377f"
] |
[
"tensorflow/python/kernel_tests/array_ops/gather_op_test.py"
] |
[
"# Copyright 2015 The TensorFlow Authors. All Rights Reserved.\n#\n# Licensed under the Apache License, Version 2.0 (the \"License\");\n# you may not use this file except in compliance with the License.\n# You may obtain a copy of the License at\n#\n# http://www.apache.org/licenses/LICENSE-2.0\n#\n# Unless required by applicable law or agreed to in writing, software\n# distributed under the License is distributed on an \"AS IS\" BASIS,\n# WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.\n# See the License for the specific language governing permissions and\n# limitations under the License.\n# ==============================================================================\n\"\"\"Tests for tensorflow.ops.tf.gather.\"\"\"\n\nfrom absl.testing import parameterized\nimport numpy as np\n\nfrom tensorflow.python.eager import backprop\nfrom tensorflow.python.eager import context\nfrom tensorflow.python.eager import def_function\nfrom tensorflow.python.framework import constant_op\nfrom tensorflow.python.framework import dtypes\nfrom tensorflow.python.framework import errors\nfrom tensorflow.python.framework import ops\nfrom tensorflow.python.framework import tensor_spec\nfrom tensorflow.python.framework import test_util\nfrom tensorflow.python.ops import array_ops\nfrom tensorflow.python.ops import gradient_checker_v2\nfrom tensorflow.python.ops import gradients_impl\nfrom tensorflow.python.ops import math_ops\nfrom tensorflow.python.ops import resource_variable_ops\nfrom tensorflow.python.ops import variables\nfrom tensorflow.python.platform import test\n\n_TEST_TYPES = (dtypes.int64, dtypes.float32,\n dtypes.complex64, dtypes.complex128)\n\n# TODO(virimia): Add a benchmark for gather_v2, with batch_dims and axis set.\n\n\ndef _to_str_elements(values):\n \"\"\"Converts the inner list elements to strings.\"\"\"\n if isinstance(values, list):\n return [_to_str_elements(value) for value in values]\n else:\n return str(values).encode(\"utf-8\")\n\n\nclass GatherTest(test.TestCase, parameterized.TestCase):\n\n def _buildParams(self, data, dtype):\n data = data.astype(dtype.as_numpy_dtype)\n # For complex types, add an index-dependent imaginary component so we can\n # tell we got the right value.\n if dtype.is_complex:\n return data + 10j * data\n return data\n\n def testScalar1D(self):\n with self.cached_session():\n data = np.array([0, 1, 2, 3, 7, 5])\n for dtype in _TEST_TYPES:\n for indices in 4, [1, 2, 2, 4, 5]:\n with self.subTest(dtype=dtype, indices=indices):\n params_np = self._buildParams(data, dtype)\n params = constant_op.constant(params_np)\n indices_tf = constant_op.constant(indices)\n gather_t = array_ops.gather(params, indices_tf)\n gather_val = self.evaluate(gather_t)\n np_val = params_np[indices]\n self.assertAllEqual(np_val, gather_val)\n self.assertEqual(np_val.shape, gather_t.get_shape())\n\n def testScalar2D(self):\n with self.session():\n data = np.array([[0, 1, 2], [3, 4, 5], [6, 7, 8],\n [9, 10, 11], [12, 13, 14]])\n for dtype in _TEST_TYPES:\n for axis in range(data.ndim):\n with self.subTest(dtype=dtype, axis=axis):\n params_np = self._buildParams(data, dtype)\n params = constant_op.constant(params_np)\n indices = constant_op.constant(2)\n gather_t = array_ops.gather(params, indices, axis=axis)\n gather_val = self.evaluate(gather_t)\n self.assertAllEqual(np.take(params_np, 2, axis=axis), gather_val)\n expected_shape = data.shape[:axis] + data.shape[axis + 1:]\n self.assertEqual(expected_shape, gather_t.get_shape())\n\n def testSimpleTwoD32(self):\n with self.session():\n data = np.array([[0, 1, 2], [3, 4, 5], [6, 7, 8],\n [9, 10, 11], [12, 13, 14]])\n for dtype in _TEST_TYPES:\n for axis in range(data.ndim):\n with self.subTest(dtype=dtype, axis=axis):\n params_np = self._buildParams(data, dtype)\n params = constant_op.constant(params_np)\n # The indices must be in bounds for any axis.\n indices = constant_op.constant([0, 1, 0, 2])\n gather_t = array_ops.gather(params, indices, axis=axis)\n gather_val = self.evaluate(gather_t)\n self.assertAllEqual(np.take(params_np, [0, 1, 0, 2], axis=axis),\n gather_val)\n expected_shape = data.shape[:axis] + (4,) + data.shape[axis + 1:]\n self.assertEqual(expected_shape, gather_t.get_shape())\n\n def testHigherRank(self):\n with ops.Graph().as_default():\n # We check that scalar and empty indices shapes work as well\n shape = (2, 1, 3, 2)\n for indices_shape in (), (0,), (2, 0), (2, 3):\n for dtype in _TEST_TYPES:\n for axis in range(len(shape)):\n params = self._buildParams(np.random.randn(*shape), dtype)\n indices = np.random.randint(shape[axis], size=indices_shape)\n with self.subTest(\n indices_shape=indices_shape,\n dtype=dtype,\n axis=axis,\n indices=indices):\n tf_params = constant_op.constant(params)\n tf_indices = constant_op.constant(indices)\n # Check that both positive and negative indices for axis work.\n tf_axis = constant_op.constant(axis)\n tf_negative_axis = constant_op.constant(-len(shape) + axis)\n gather = array_ops.gather(tf_params, tf_indices, axis=tf_axis)\n gather_negative_axis = array_ops.gather(\n tf_params, tf_indices, axis=tf_negative_axis)\n gather_value, gather_negative_axis_value = self.evaluate(\n [gather, gather_negative_axis])\n gather_np = np.take(params, indices, axis)\n self.assertAllEqual(gather_np, gather_value)\n self.assertAllEqual(gather_np, gather_negative_axis_value)\n expected_shape = (params.shape[:axis] + indices.shape +\n params.shape[axis + 1:])\n self.assertEqual(expected_shape, gather.shape)\n self.assertEqual(expected_shape, gather_negative_axis.shape)\n\n # Test gradients\n gather_grad = np.random.randn(\n *gather.get_shape().as_list()).astype(dtype.as_numpy_dtype)\n if dtype.is_complex:\n gather_grad -= 1j * gather_grad\n params_grad, indices_grad, axis_grad = gradients_impl.gradients(\n gather, [tf_params, tf_indices, tf_axis], gather_grad)\n self.assertIsNone(indices_grad)\n self.assertIsNone(axis_grad)\n if dtype.is_integer:\n self.assertIsNone(params_grad)\n continue\n # For axis 0, we are able to create an efficient IndexedSlices for\n # the gradient.\n if axis == 0:\n self.assertEqual(type(params_grad), ops.IndexedSlices)\n params_grad = ops.convert_to_tensor(params_grad)\n correct_params_grad = np.zeros(shape).astype(dtype.as_numpy_dtype)\n outer_dims = axis\n inner_dims = len(shape) - axis - 1\n gather_grad = gather_grad.reshape(\n shape[:axis] + (indices.size,) + shape[axis + 1:])\n for source_index, dest_index in enumerate(indices.flat):\n dest_slice = ((slice(None),) * outer_dims + (dest_index,) +\n (slice(None),) * inner_dims)\n source_slice = ((slice(None),) * outer_dims + (source_index,) +\n (slice(None),) * inner_dims)\n correct_params_grad[dest_slice] += gather_grad[source_slice]\n self.assertAllClose(\n correct_params_grad,\n self.evaluate(params_grad),\n atol=2e-6,\n rtol=2e-6)\n\n def testHigherRankGradientTape(self):\n # We check that scalar and empty indices shapes work as well\n shape = (2, 1, 3, 2)\n for indices_shape in (), (0,), (2, 0), (2, 3):\n for dtype in _TEST_TYPES:\n for axis in range(len(shape)):\n params = self._buildParams(np.random.randn(*shape), dtype)\n indices = np.random.randint(shape[axis], size=indices_shape)\n with self.subTest(\n indices_shape=indices_shape,\n dtype=dtype,\n axis=axis,\n indices=indices):\n with backprop.GradientTape() as tape:\n tf_params = constant_op.constant(params)\n tf_indices = constant_op.constant(indices)\n # Check that both positive and negative indices for axis work.\n tf_axis = constant_op.constant(axis)\n tape.watch(tf_params)\n tape.watch(tf_indices)\n tape.watch(tf_axis)\n tf_negative_axis = constant_op.constant(-len(shape) + axis)\n gather = array_ops.gather(tf_params, tf_indices, axis=tf_axis)\n gather_negative_axis = array_ops.gather(\n tf_params, tf_indices, axis=tf_negative_axis)\n gather_value, gather_negative_axis_value = self.evaluate(\n [gather, gather_negative_axis])\n gather_np = np.take(params, indices, axis)\n self.assertAllEqual(gather_np, gather_value)\n self.assertAllEqual(gather_np, gather_negative_axis_value)\n expected_shape = (\n params.shape[:axis] + indices.shape + params.shape[axis + 1:])\n self.assertEqual(expected_shape, gather.shape)\n self.assertEqual(expected_shape, gather_negative_axis.shape)\n\n # Test gradients\n gather_grad = np.random.randn(\n *gather.get_shape().as_list()).astype(dtype.as_numpy_dtype)\n if dtype.is_complex:\n gather_grad -= 1j * gather_grad\n params_grad, indices_grad, axis_grad = tape.gradient(\n gather, [tf_params, tf_indices, tf_axis], gather_grad)\n self.assertIsNone(indices_grad)\n self.assertIsNone(axis_grad)\n if dtype.is_integer:\n self.assertIsNone(params_grad)\n continue\n # For axis 0, we are able to create an efficient IndexedSlices for\n # the gradient.\n if axis == 0:\n self.assertEqual(type(params_grad), ops.IndexedSlices)\n params_grad = ops.convert_to_tensor(params_grad)\n correct_params_grad = np.zeros(shape).astype(dtype.as_numpy_dtype)\n outer_dims = axis\n inner_dims = len(shape) - axis - 1\n gather_grad = gather_grad.reshape(shape[:axis] + (indices.size,) +\n shape[axis + 1:])\n for source_index, dest_index in enumerate(indices.flat):\n dest_slice = ((slice(None),) * outer_dims + (dest_index,) +\n (slice(None),) * inner_dims)\n source_slice = ((slice(None),) * outer_dims + (source_index,) +\n (slice(None),) * inner_dims)\n correct_params_grad[dest_slice] += gather_grad[source_slice]\n self.assertAllClose(\n correct_params_grad,\n self.evaluate(params_grad),\n atol=2e-6,\n rtol=2e-6)\n\n def testString(self):\n params = np.array([[b\"asdf\", b\"zxcv\"], [b\"qwer\", b\"uiop\"]])\n self.assertAllEqual([b\"qwer\", b\"uiop\"], array_ops.gather(params, 1, axis=0))\n self.assertAllEqual([b\"asdf\", b\"qwer\"], array_ops.gather(params, 0, axis=1))\n\n def testUInt32AndUInt64(self):\n for unsigned_type in (dtypes.uint32, dtypes.uint64):\n with self.subTest(unsigned_type=unsigned_type):\n params = self._buildParams(\n np.array([[1, 2, 3], [7, 8, 9]]), unsigned_type)\n with self.cached_session():\n self.assertAllEqual([7, 8, 9], array_ops.gather(params, 1, axis=0))\n self.assertAllEqual([1, 7], array_ops.gather(params, 0, axis=1))\n\n def testUnknownIndices(self):\n # This test is purely a test for placeholder inputs which is only applicable\n # in graph mode.\n with ops.Graph().as_default():\n params = constant_op.constant([[0, 1, 2]])\n indices = array_ops.placeholder(dtypes.int32)\n gather_t = array_ops.gather(params, indices)\n self.assertEqual(None, gather_t.get_shape())\n\n def testUnknownAxis(self):\n # This test is purely a test for placeholder inputs which is only applicable\n # in graph mode.\n with ops.Graph().as_default():\n params = constant_op.constant([[0, 1, 2]])\n indices = constant_op.constant([[0, 0], [0, 0]])\n axis = array_ops.placeholder(dtypes.int32)\n gather_t = array_ops.gather(params, indices, axis=axis)\n # Rank 2 params with rank 2 indices results in a rank 3 shape.\n self.assertEqual([None, None, None], gather_t.shape.as_list())\n\n # If indices is also unknown the result rank is unknown.\n indices = array_ops.placeholder(dtypes.int32)\n gather_t = array_ops.gather(params, indices, axis=axis)\n self.assertEqual(None, gather_t.shape)\n\n def testBadIndicesType(self):\n with self.assertRaisesRegex(\n (TypeError, errors.InvalidArgumentError),\n \"float.* not in.* list of allowed values: int32, int64\"):\n self.evaluate(array_ops.gather([0], 0.))\n\n @test_util.disable_xla(\n \"Assertion inside an op is not supported in XLA. Instead XLA clamps the \"\n \"index to be in bounds and returns the indexed value there (Don't rely \"\n \"on this behavior).\")\n def testBadIndicesCPU(self):\n with test_util.force_cpu():\n params = [[0, 1, 2], [3, 4, 5]]\n with self.assertRaisesOpError(r\"indices\\[0,0\\] = 7 is not in \\[0, 2\\)\"):\n self.evaluate(array_ops.gather(params, [[7]], axis=0))\n with self.assertRaisesOpError(r\"indices\\[0,0\\] = 7 is not in \\[0, 3\\)\"):\n self.evaluate(array_ops.gather(params, [[7]], axis=1))\n\n def _disabledTestBadIndicesGPU(self):\n # TODO disabled due to different behavior on GPU and CPU\n # On GPU the bad indices do not raise error but fetch 0 values\n if not test.is_gpu_available():\n return\n with self.session():\n params = [[0, 1, 2], [3, 4, 5]]\n with self.assertRaisesOpError(r\"indices\\[0,0\\] = 7 is not in \\[0, 2\\)\"):\n array_ops.gather(params, [[7]], axis=0).eval()\n with self.assertRaisesOpError(r\"indices\\[0,0\\] = 7 is not in \\[0, 3\\)\"):\n array_ops.gather(params, [[7]], axis=1).eval()\n\n def testBadAxis(self):\n\n @def_function.function(autograph=False, jit_compile=False)\n def gather(x, indices, axis):\n return array_ops.gather(x, indices, axis=axis)\n\n @def_function.function(\n autograph=False,\n jit_compile=False,\n input_signature=[\n tensor_spec.TensorSpec(shape=None, dtype=dtypes.int32)\n ] * 3)\n def gather_shape_inf_disabled(x, indices, axis):\n return array_ops.gather(x, indices, axis=axis)\n\n @def_function.function(\n autograph=False,\n jit_compile=True,\n input_signature=[\n tensor_spec.TensorSpec(shape=None, dtype=dtypes.int32)\n ] * 3)\n def xla_gather(x, indices, axis):\n return array_ops.gather(x, indices, axis=axis)\n\n params = [0, 1, 2]\n indices = 0\n functions = [(\"array_ops.gather\", array_ops.gather), (\"gather\", gather),\n (\"gather_shape_inf_disabled\", gather_shape_inf_disabled),\n (\"xla_gather\", xla_gather)]\n for bad_axis in (1, 2, -2):\n for fn_name, fn in functions:\n # Shape inference can validate axis for known params rank.\n with self.subTest(bad_axis=bad_axis, msg=fn_name, fn=fn):\n with self.assertRaisesRegex(\n (ValueError, errors.InvalidArgumentError),\n \"Shape must be at least rank .* but is rank 1\"):\n fn(params, indices, axis=bad_axis)\n\n def testEmptySlices(self):\n for dtype in _TEST_TYPES:\n for itype in np.int32, np.int64:\n # Leading axis gather.\n with self.subTest(dtype=dtype, itype=itype):\n params = np.zeros((7, 0, 0), dtype=dtype.as_numpy_dtype)\n indices = np.array([3, 4], dtype=itype)\n gather = array_ops.gather(params, indices, axis=0)\n self.assertAllEqual(gather, np.zeros((2, 0, 0)))\n\n # Middle axis gather.\n params = np.zeros((0, 7, 0), dtype=dtype.as_numpy_dtype)\n gather = array_ops.gather(params, indices, axis=1)\n self.assertAllEqual(gather, np.zeros((0, 2, 0)))\n\n # Trailing axis gather.\n params = np.zeros((0, 0, 7), dtype=dtype.as_numpy_dtype)\n gather = array_ops.gather(params, indices, axis=2)\n self.assertAllEqual(gather, np.zeros((0, 0, 2)))\n\n @parameterized.parameters([\n # batch_dims=0 (equivalent to tf.gather)\n dict( # 2D indices\n batch_dims=0,\n params=[6, 7, 8, 9],\n indices=[[2, 1], [0, 3]],\n expected=[[8, 7], [6, 9]]),\n dict( # 3D indices\n batch_dims=0,\n params=[6, 7, 8, 9],\n indices=[[[3, 1], [2, 0]], [[0, 3], [2, 2]]],\n expected=[[[9, 7], [8, 6]], [[6, 9], [8, 8]]]),\n dict( # 4D indices\n batch_dims=0,\n params=[8, 9],\n indices=[[[[0, 1], [1, 0]], [[0, 0], [1, 1]]],\n [[[1, 1], [0, 0]], [[0, 1], [1, 0]]]],\n expected=[[[[8, 9], [9, 8]], [[8, 8], [9, 9]]],\n [[[9, 9], [8, 8]], [[8, 9], [9, 8]]]]),\n\n # batch_dims=indices.shape.ndims - 1\n # (equivalent to tf.compat.v1.batch_gather)\n dict( # 2D indices (1 batch dim)\n batch_dims=1,\n params=[[10, 11, 12, 13], [20, 21, 22, 23]],\n indices=[[2, 1], [0, 3]],\n expected=[[12, 11], [20, 23]]),\n dict( # 3D indices (2 batch dims)\n batch_dims=2,\n params=[[[100, 101], [110, 111]], [[200, 201], [210, 211]]],\n indices=[[[0, 1], [1, 0]], [[0, 0], [1, 1]]],\n expected=[[[100, 101], [111, 110]], [[200, 200], [211, 211]]]),\n dict( # 2D indices (1 batch dim)\n batch_dims=-1,\n params=[[10, 11, 12, 13], [20, 21, 22, 23]],\n indices=[[2, 1], [0, 3]],\n expected=[[12, 11], [20, 23]]),\n dict( # 3D indices (2 batch dims)\n batch_dims=-1,\n params=[[[100, 101], [110, 111]], [[200, 201], [210, 211]]],\n indices=[[[0, 1], [1, 0]], [[0, 0], [1, 1]]],\n expected=[[[100, 101], [111, 110]], [[200, 200], [211, 211]]]),\n\n # batch_dims=indices.shape.ndims\n dict( # 1D indices (1 batch dim)\n batch_dims=1,\n params=[[10, 11, 12, 13], [20, 21, 22, 23]],\n indices=[2, 1],\n expected=[12, 21]),\n dict( # 2D indices (2 batch dim)\n batch_dims=2,\n params=[[[100, 101, 102, 103], [110, 111, 112, 113]],\n [[200, 201, 202, 203], [210, 211, 212, 213]]],\n indices=[[2, 1], [0, 3]],\n expected=[[102, 111], [200, 213]]),\n\n # 0 < batch_dims < indices.shape.ndims - 1\n dict( # 3D indices (1 batch dim)\n batch_dims=1,\n params=[[10, 11, 12, 13], [20, 21, 22, 23]],\n indices=[[[3, 1], [2, 0]], [[0, 3], [2, 2]]],\n expected=[[[13, 11], [12, 10]], [[20, 23], [22, 22]]]),\n dict( # 4D indices (1 batch dim)\n batch_dims=1,\n params=[[6, 7], [8, 9]],\n indices=[[[[0, 1], [1, 0]], [[0, 0], [1, 1]]],\n [[[1, 1], [0, 0]], [[0, 1], [1, 0]]]],\n expected=[[[[6, 7], [7, 6]], [[6, 6], [7, 7]]],\n [[[9, 9], [8, 8]], [[8, 9], [9, 8]]]]),\n dict( # 4D indices (2 batch dims)\n batch_dims=2,\n params=[[[2, 3], [4, 5]], [[6, 7], [8, 9]]],\n indices=[[[[0, 1], [1, 0]], [[0, 0], [1, 1]]],\n [[[1, 1], [0, 0]], [[0, 1], [1, 0]]]],\n expected=[[[[2, 3], [3, 2]], [[4, 4], [5, 5]]],\n [[[7, 7], [6, 6]], [[8, 9], [9, 8]]]]),\n\n # axis > 0\n dict( # 3D indices, batch_dims=1, axis=2\n # params.shape = [I1, J1, J2] = [2, 2, 3]\n # indices.shape = [I1, K1, K2] = [2, 1, 5]\n # result.shape = [I1, J1, K1, K2] = [2, 2, 1, 5]\n batch_dims=1,\n axis=2,\n params=[[[10, 11, 12], [13, 14, 15]], [[20, 21, 22], [23, 24, 25]]],\n indices=[[[0, 1, 2, 1, 0]], [[0, 1, 2, 1, 0]]],\n expected=[[[[10, 11, 12, 11, 10]], [[13, 14, 15, 14, 13]]],\n [[[20, 21, 22, 21, 20]], [[23, 24, 25, 24, 23]]]]),\n dict( # 3D indices, batch_dims=None, axis=1\n batch_dims=None,\n axis=1,\n params=[[10, 11, 12], [13, 14, 15]],\n indices=[1, 0],\n expected=[[11, 10], [14, 13]]),\n dict( # 3D indices, batch_dims=-3, axis=1\n batch_dims=-3,\n axis=1,\n params=[[0, 1, 2], [3, 4, 5]],\n indices=[[[0, 1], [1, 0]]],\n expected=[[[[0, 1], [1, 0]]], [[[3, 4], [4, 3]]]]),\n ])\n @test_util.run_in_graph_and_eager_modes\n def testBatchDims(self, params, indices, batch_dims, expected=None,\n axis=None):\n result = array_ops.gather(params, indices, axis=axis, batch_dims=batch_dims)\n self.assertAllEqual(expected, result)\n\n # Test gradients\n f64_params = math_ops.cast(params, dtypes.float64)\n def gather(params):\n return array_ops.gather(params, indices, axis=axis, batch_dims=batch_dims)\n theoretical, numerical = gradient_checker_v2.compute_gradient(\n gather, [f64_params])\n self.assertAllClose(theoretical, numerical)\n\n # Test gradients when input shapes are unknown\n @def_function.function(input_signature=[\n tensor_spec.TensorSpec(shape=None, dtype=dtypes.float64),\n tensor_spec.TensorSpec(shape=None, dtype=dtypes.int32)\n ])\n def gather_unknown_shapes(params, indices):\n return array_ops.gather(params, indices, axis=axis, batch_dims=batch_dims)\n if batch_dims is None or batch_dims >= 0:\n theoretical, numerical = gradient_checker_v2.compute_gradient(\n lambda p: gather_unknown_shapes(p, indices), [f64_params])\n self.assertAllClose(theoretical, numerical)\n else:\n with self.assertRaisesRegex(\n ValueError,\n \"Currently, it is unsupported to take the gradient of tf.gather\"):\n gradient_checker_v2.compute_gradient(\n lambda p: gather_unknown_shapes(p, indices), [f64_params])\n\n # Test the gradients shape.\n with backprop.GradientTape() as tape:\n zeros = array_ops.zeros_like(params, dtype=dtypes.float32)\n tape.watch(zeros)\n values = zeros * 2 + zeros\n result = array_ops.gather(\n values, indices, axis=axis, batch_dims=batch_dims)\n gradients = tape.gradient(result, zeros)\n\n self.assertAllEqual(array_ops.shape(params), array_ops.shape(gradients))\n\n # Run the same test for strings.\n params = _to_str_elements(params)\n expected = _to_str_elements(expected)\n result = array_ops.gather(\n params, indices, axis=axis, batch_dims=batch_dims)\n\n self.assertAllEqual(expected, result)\n\n @parameterized.parameters([\n dict(\n params_shape=[2, 3, 4, 5, 6, 7],\n indices_shape=[2, 3, 8, 9, 10],\n batch_dims=2,\n axis=2,\n output_shape=[2, 3, 8, 9, 10, 5, 6, 7]\n # = params.shape[:2] + indices.shape[2:] + params.shape[3:]\n ),\n dict(\n params_shape=[2, 3, 4, 5, 6, 7],\n indices_shape=[2, 3, 8, 9, 10],\n batch_dims=2,\n axis=3,\n output_shape=[2, 3, 4, 8, 9, 10, 6, 7]\n # = params.shape[:3] + indices.shape[2:] + params.shape[4:]\n ),\n dict(\n params_shape=[2, 3, 4, 5, 6, 7],\n indices_shape=[2, 3, 8, 9, 10],\n batch_dims=2,\n axis=4,\n output_shape=[2, 3, 4, 5, 8, 9, 10, 7]\n # = params.shape[:4] + indices.shape[2:] + params.shape[5:]\n ),\n dict(\n params_shape=[2, 3, 4, 5, 6, 7],\n indices_shape=[2, 3, 8, 9, 10],\n batch_dims=2,\n axis=5,\n output_shape=[2, 3, 4, 5, 6, 8, 9, 10]\n # = params.shape[:5] + indices.shape[2:] + params.shape[6:]\n ),\n dict(\n params_shape=[2, 3, 4, 5, 6, 7],\n indices_shape=[2, 3, 8, 9, 10],\n batch_dims=2,\n axis=-4,\n output_shape=[2, 3, 8, 9, 10, 5, 6, 7]\n # = params.shape[:2] + indices.shape[2:] + params.shape[3:]\n ),\n dict(\n params_shape=[2, 3, 4, 5, 6, 7],\n indices_shape=[2, 3, 8, 9, 10],\n batch_dims=2,\n axis=-3,\n output_shape=[2, 3, 4, 8, 9, 10, 6, 7]\n # = params.shape[:3] + indices.shape[2:] + params.shape[4:]\n ),\n dict(\n params_shape=[2, 3, 4, 5, 6, 7],\n indices_shape=[2, 3, 8, 9, 10],\n batch_dims=2,\n axis=-2,\n output_shape=[2, 3, 4, 5, 8, 9, 10, 7]\n # = params.shape[:4] + indices.shape[2:] + params.shape[5:]\n ),\n dict(\n params_shape=[2, 3, 4, 5, 6, 7],\n indices_shape=[2, 3, 8, 9, 10],\n batch_dims=2,\n axis=-1,\n output_shape=[2, 3, 4, 5, 6, 8, 9, 10]\n # = params.shape[:5] + indices.shape[2:] + params.shape[6:]\n ),\n ])\n @test_util.run_in_graph_and_eager_modes\n def testBatchDimsMatchesPythonBatching(self, params_shape, indices_shape,\n batch_dims, axis, output_shape):\n \"\"\"Checks that batch_dims matches multiple calls to tf.gather().\"\"\"\n # Generate a `params` tensor with the indicated shape.\n params_size = np.prod(params_shape)\n params = np.reshape(np.arange(params_size), params_shape)\n\n # Generate an `indices` tensor with the indicated shape, where each index\n # is within the appropriate range.\n indices_size = np.prod(indices_shape)\n indices = np.reshape(np.arange(indices_size), indices_shape)\n indices = indices % params_shape[axis]\n\n # Perform repeated (batched) gather operations with numpy, to find the\n # expected result.\n expected = self._batchNumpyGather(params, indices, axis, batch_dims)\n\n # On Windows, we get an exception if we pass in the transformed numpy\n # arrays (\"Failed to convert numpy ndarray to a Tensor (Unsupported\n # feed type).\"); so convert them back to lists before calling tf.gather.\n params = params.tolist()\n indices = indices.tolist()\n\n result = array_ops.gather(params, indices, axis=axis, batch_dims=batch_dims)\n self.assertAllEqual(output_shape, result.shape.as_list())\n self.assertAllEqual(expected, result)\n\n # Run the same test for strings.\n params = _to_str_elements(params)\n expected = _to_str_elements(expected.tolist())\n result = array_ops.gather(\n params, indices, axis=axis, batch_dims=batch_dims)\n\n self.assertAllEqual(output_shape, result.shape.as_list())\n self.assertAllEqual(expected, result)\n\n def _batchNumpyGather(self, params, indices, axis, batch_dims):\n \"\"\"Performs a batch gather by making recursive calls to np.take().\n\n This is used by testBatchDims() to construct the expected value.\n\n Args:\n params: A numpy array\n indices: A numpy array\n axis: An integer\n batch_dims: An integer\n Returns:\n A numpy array\n \"\"\"\n if batch_dims == 0:\n return np.take(params, indices, axis=axis)\n self.assertEqual(params.shape[0], indices.shape[0])\n if axis > 0:\n axis -= 1\n return np.stack([\n self._batchNumpyGather(params[i], indices[i], axis, batch_dims - 1)\n for i in range(params.shape[0])\n ])\n\n @test_util.run_v1_only(\"RefVariable is not supported in v2\")\n def testGatherRefVariable(self):\n with self.cached_session():\n v = variables.RefVariable(constant_op.constant([[1, 2], [3, 4], [5, 6]]))\n self.evaluate(variables.global_variables_initializer())\n gather = array_ops.gather(v, [0, 2])\n if not context.executing_eagerly(): # .op doesn't make sense in Eager\n self.assertEqual(\"GatherV2\", gather.op.name)\n self.assertAllEqual([[1, 2], [5, 6]], gather)\n\n @test_util.run_in_graph_and_eager_modes\n def testGatherResourceVariable(self):\n with self.cached_session():\n v = resource_variable_ops.ResourceVariable(\n constant_op.constant([[1, 2], [3, 4], [5, 6]]))\n self.evaluate(variables.global_variables_initializer())\n gather = array_ops.gather(v, [0, 2])\n if not context.executing_eagerly(): # .op doesn't make sense in Eager\n self.assertEqual(\"ResourceGather\", gather.op.inputs[0].op.type)\n self.assertAllEqual([[1, 2], [5, 6]], gather)\n\nif __name__ == \"__main__\":\n test.main()\n"
] |
[
[
"tensorflow.python.framework.test_util.force_cpu",
"tensorflow.python.eager.context.executing_eagerly",
"tensorflow.python.platform.test.main",
"tensorflow.python.ops.math_ops.cast",
"numpy.take",
"numpy.prod",
"tensorflow.python.ops.array_ops.placeholder",
"numpy.arange",
"tensorflow.python.ops.gradient_checker_v2.compute_gradient",
"numpy.random.randint",
"tensorflow.python.ops.gradients_impl.gradients",
"tensorflow.python.ops.variables.global_variables_initializer",
"tensorflow.python.framework.test_util.run_v1_only",
"tensorflow.python.eager.backprop.GradientTape",
"numpy.array",
"tensorflow.python.framework.ops.Graph",
"numpy.zeros",
"tensorflow.python.ops.array_ops.gather",
"tensorflow.python.eager.def_function.function",
"numpy.random.randn",
"tensorflow.python.platform.test.is_gpu_available",
"tensorflow.python.framework.tensor_spec.TensorSpec",
"tensorflow.python.framework.test_util.disable_xla",
"tensorflow.python.ops.array_ops.zeros_like",
"tensorflow.python.framework.constant_op.constant",
"tensorflow.python.framework.ops.convert_to_tensor",
"tensorflow.python.ops.array_ops.shape"
]
] |
Subsets and Splits
No community queries yet
The top public SQL queries from the community will appear here once available.