path
stringlengths 7
265
| concatenated_notebook
stringlengths 46
17M
|
|---|---|
CrossValidation/crossval.ipynb
|
###Markdown
交差検証まとめ- ref: https://scikit-learn.org/stable/auto_examples/model_selection/plot_cv_indices.html- blog: https://slash-z.com/- github: https://github.com/KazutoMakino/PythonCourses---
###Code
from pathlib import Path
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
import seaborn as sns
from sklearn.preprocessing import LabelEncoder
sns.set()
iris = sns.load_dataset(name="iris")
print(iris)
df = iris.sample(n=100, random_state=42)
df.sort_values(by="species", inplace=True)
df.reset_index(drop=True, inplace=True)
###Output
_____no_output_____
###Markdown
ここで,後ほどラベルでないグループでデータを分けることもある都合で,「調査した地域」という意味合いで,上から順番に "area" という名称でカラムを以下のように追加します.
###Code
df["area"] = (
["A"] * 50
+ ["B"] * 20
+ ["C"] * 15
+ ["D"] * 10
+ ["E"] * 5
)
print(df)
# features / objectives
x = df[[v for v in df.columns if v!="species"]]
y = df["species"]
group = df["area"]
print(f"x.shape={x.shape}, y.shape={y.shape}, group.shape={group.shape}")
print(y.value_counts())
###Output
setosa 37
versicolor 32
virginica 31
Name: species, dtype: int64
###Markdown
データ分割可視化用の関数を定義
###Code
class FoldsPlotter:
""" Create a sample plot for indices of a cross-validation object.
ref:
- https://scikit-learn.org/stable/auto_examples/model_selection/plot_cv_indices.html
"""
def __init__(
self, x: pd.DataFrame,
y: pd.Series,
group: pd.Series,
ylabel: str = "Fold indices",
lw: int = 10,
) -> None:
self.x = x
self.y = y
self.group = group
# set encoder and transformed data
enc = LabelEncoder()
self.enc_y = enc.fit_transform(y)
self.enc_group = enc.fit_transform(group)
# line width
self.lw = lw
# plot y-axis label
self.ylabel = ylabel
# set save dir
self.save_dir = Path("./out")
None if self.save_dir.exists() else self.save_dir.mkdir()
# init
self.fold_num = 0
fig = plt.figure(figsize=(8, 4), facecolor="white")
self.ax = fig.add_subplot()
def __del__(self) -> None:
pass
def get_legends(
self,
label_names: tuple,
cmap_name: str = "coolwarm",
bbox_to_anchor: tuple = (1, 1),
loc: str = "best",
ncol: int = 5,
leg_title: str = "title",
) -> None:
# init
handles = []
labels = []
# get legend
for i, label in enumerate(label_names):
if len(label_names) == 1:
color_idx = 0
else:
color_idx = i / (len(label_names) - 1)
color = plt.get_cmap(cmap_name)(color_idx)
p, = self.ax.plot([-10, -11], [-10, -10], label=label, color=color)
handles.append(p)
labels.append(p.get_label())
legends = self.ax.legend(
handles=handles,
labels=labels,
bbox_to_anchor=bbox_to_anchor,
loc=loc,
ncol=ncol,
title=leg_title,
)
plt.gca().add_artist(legends)
def add_plot(
self,
train_idx: np.ndarray = None,
test_idx: np.ndarray = None,
cmap="cool",
) -> None:
if (train_idx is None) or (test_idx is None):
return
# fill in indices with the training / test groups
indices = np.array([np.nan] * len(self.x))
indices[train_idx] = 0
indices[test_idx] = 1
# scatter plot
self.ax.scatter(
x=range(len(indices)),
y=[self.fold_num + 0.5] * len(indices),
c=indices,
marker="_",
lw=self.lw,
cmap=cmap,
)
# get legend
if self.fold_num == 0:
self.get_legends(
label_names=("train", "test"),
cmap_name=cmap,
bbox_to_anchor=(1, 1),
loc="lower right",
leg_title="train / test",
)
# increment
self.fold_num += 1
def show(self, custom_y_label: list = None) -> None:
# set cmap
cmap_class = "Paired"
cmap_group = "rainbow"
# plot the data classes and groups at the end
self.ax.scatter(
range(len(self.x)),
[self.fold_num + 0.5] * len(self.x),
c=self.enc_y,
marker="_",
lw=self.lw,
cmap=cmap_class,
)
self.ax.scatter(
range(len(self.x)),
[self.fold_num + 1.5] * len(self.x),
c=self.enc_group,
marker="_",
lw=self.lw,
cmap=cmap_group,
)
# get legends
self.get_legends(
label_names=sorted(set(self.y)),
cmap_name=cmap_class,
bbox_to_anchor=(0, 1),
loc="lower left",
leg_title="class",
)
self.get_legends(
label_names=sorted(set(self.group)),
cmap_name=cmap_group,
bbox_to_anchor=(0, -0.15),
loc="upper left",
leg_title="group",
)
# formatting
if custom_y_label:
yticklabels = custom_y_label
else:
yticklabels = (
[f"fold-{str(v)}" for v in range(self.fold_num)] + ["class", "group"]
)
self.ax.set(
yticks=np.arange(self.fold_num + 2) + 0.5,
yticklabels=yticklabels,
xlabel="Sample index",
ylabel=self.ylabel,
ylim=[self.fold_num + 2.2, -0.2],
xlim=[0, 100],
)
plt.tight_layout()
plt.savefig(f"{self.save_dir / self.ylabel}.jpg", bbox_inches="tight", dpi=150)
plt.show()
plt.clf()
plt.close("all")
plotter = FoldsPlotter(x=x, y=y, group=group, ylabel="01-1-GivenData", lw=50)
plotter.show()
###Output
_____no_output_____
###Markdown
| クラス/関数 | 説明 || - | - || sklearn.model_selection.check_cv(cv=5, y=None, *, classifier=False) | cross-validator オブジェクト(KFold など)について確認ができます. || sklearn.model_selection.train_test_split(*arrays, test_size=None, train_size=None, random_state=None, shuffle=True, stratify=None) | 学習用データと検証用データを2つに分けます(ホールド・アウト検証). || sklearn.model_selection.KFold(n_splits=5, *, shuffle=False, random_state=None) | データを指定した K 個に等分し,1 つを検証用,それ以外の K-1 個を学習に用います.各 fold における検証用データに重複はさせないようにします. || sklearn.model_selection.ShuffleSplit(n_splits=10, *, test_size=None, train_size=None, random_state=None) | train_test.split(shuffle=True, stratify=None) を複数回作用させるような学習用/検証用データのインデックスを返すオブジェクトを生成します.各 fold における検証用データは重複します. || sklearn.model_selection.StratifiedKFold(n_splits=5, *, shuffle=False, random_state=None) | データを指定した K 個に等分し,1 つを検証用,それ以外の K-1 個を学習に用います.データ分割の際には,メソッド split の引数 y にてラベルやグループを指定することにより,全種類のラベルも均等に分割されます.各 fold における検証用データに重複はさせないようにします. || sklearn.model_selection.StratifiedShuffleSplit(n_splits=10, *, test_size=None, train_size=None, random_state=None) | train_test.split(shuffle=True, stratify=y) を複数回作用させるような学習用/検証用データのインデックスを返すオブジェクトを生成します.各 fold における検証用データは重複します. || sklearn.model_selection.GroupKFold(n_splits=5) | データを指定した K 個に分割し,1 つを検証用,それ以外の K-1 個を学習に用います.データ分割の際には,メソッド split の引数 groups にて指定したグループによって,分割するインデックスを制御します.各 fold における検証用データに重複させないようにします. || sklearn.model_selection.GroupShuffleSplit(n_splits=5, *, test_size=None, train_size=None, random_state=None) | GroupKFold のデータ分割で,各 fold 間におけるグループの重複を許したデータ分割手法. || sklearn.model_selection.StratifiedGroupKFold(n_splits=5, shuffle=False, random_state=None) | データを指定した K 個に分割し,1 つを検証用,それ以外の K-1 個を学習に用います.データ分割は,メソッド split の引数 y にて指定したラベル,groups にて指定したグループを用いて,グループのまとまりを使ってラベルを K 個に均等に分けるように作用します.イメージとしては,名前の通り StratifiedKFold と GroupKFold が合体したものです.各 fold における検証用データのグループは重複しません. || sklearn.model_selection.RepeatedKFold(*, n_splits=5, n_repeats=10, random_state=None) | KFold を n_repeats で指定した回数分行います.リピートされる各 fold 間では検証用データの重複はありませんが,各リピート間では検証用データの重複は発生します. || sklearn.model_selection.RepeatedStratifiedKFold(*, n_splits=5, n_repeats=10, random_state=None) | StratifiedKFold を n_repeats で指定した回数分行います.リピートされる各 fold 間では検証用データの重複はありませんが,各リピート間では検証用データの重複は発生します. || sklearn.model_selection.LeaveOneOut() | 一つだけを検証用データ,その他は学習用データとして分割します. || sklearn.model_selection.LeavePOut(p) | 引数 p で指定した個数を検証用データ,その他を学習用データとして分割します.検証用データが複数の場合は,検証用データの組み合わせが各 fold にて重複しないように,網羅的に fold が設定されます.例えば,データ数が m 個,引数 p を n 個と設定すると,総 fold 数は数学の組み合わせの記号 C を用いて,mCn = m*(m-1) / (n*(n-1)) 個生成されます. || sklearn.model_selection.LeaveOneGroupOut() | 1 つのグループを検証用データとし,それ以外を学習用データとして分割します. || sklearn.model_selection.LeavePGroupsOut(n_groups) | 引数 n_groups で指定したグループ数を検証用データ,その他を学習用データとして分割します.検証用データが複数の場合は,検証用データの組み合わせが各 fold にて重複しないように,網羅的に fold が設定されます.例えば,グループ数が m 個,引数 n_groups を n 個と設定すると,総 fold 数は数学の組み合わせの記号 C を用いて,mCn = m*(m-1) / (n*(n-1)) 個生成されます. || sklearn.model_selection.PredefinedSplit(test_fold) | 引数 test_fold に指定した fold でデータを分割します. || sklearn.model_selection.TimeSeriesSplit(n_splits=5, *, max_train_size=None, test_size=None, gap=0) | 時系列データを考慮したデータ分割手法で,検証用データを K 等分し,残りの検証用データよりも前のデータを学習用データとして用います.したがって,index=0 から始まるような検証用データにおける学習用データは取れないので,検証用データは K-1 個になります.引数 n_splits では,この K-1 を指定します. | sklearn.model_selection.check_cv ```sklearn.model_selection.check_cv(cv=5, y=None, *, classifier=False)・・・・cross-validator オブジェクト(KFold など)について確認ができます. - cv: cross-validator オブジェクト.int の場合は sklearn.model_selection.KFold において,何分割するかという引数の n_splits が指定できる.None の場合は KFold(n_splits=5). - y: 教師あり学習における目的変数. - classifier: ==True でクラス分類タスクの場合は,sklearn.model_selection.StratifiedKFold が用いられます.```
###Code
from sklearn.model_selection import check_cv
print(check_cv())
print(check_cv(y=y, classifier=False))
print(check_cv(y=y, classifier=True))
###Output
KFold(n_splits=5, random_state=None, shuffle=False)
StratifiedKFold(n_splits=5, random_state=None, shuffle=False)
###Markdown
sklearn.model_selection.train_test_split ```sklearn.model_selection.train_test_split(*arrays, test_size=None, train_size=None, random_state=None, shuffle=True, stratify=None)・・・・学習用データと検証用データを2つに分けます(ホールド・アウト検証). - *arrays: 学習用データと検証用データに分けたい元のデータで,list / numpy.ndarray, pandas.DataFrame などが指定可能. - test_size: 検証用データサイズを指定する.float で与える場合は 0 ~ 1 とし,元データサイズの割合として指定する.int で与える場合はデータ数を直接指定する.test_size=None, train_size=None の場合は test_size=0.25 となります. - train_size: 学習用データサイズを指定.他は test_size と一緒. - random_state: 乱数シード値を指定. - shuffle: データセットを分割するときに,データの順序をシャッフルするかどうかを指定. - stratify: データをどのラベルを用いて層化させる(学習/検証データにおいて,指定したデータラベルの割合が各データ数に対して等しくなる)かを指定.```
###Code
from sklearn.model_selection import train_test_split
train_df, test_df = train_test_split(df, test_size=0.25, shuffle=False)
print(f"train_df: shape={train_df.shape}")
print(f"train_df.index={train_df.index}")
print(f' value counts\n{train_df["species"].value_counts()}')
print()
print(f"test_df: shape={test_df.shape}")
print(f"test_df.index={test_df.index}")
print(f' value counts\n{test_df["species"].value_counts()}')
plotter = FoldsPlotter(x=x, y=y, group=group, ylabel="02-1-train_test_split")
plotter.add_plot(train_idx=train_df.index, test_idx=test_df.index)
plotter.show(custom_y_label=["train / test", "class", "group"])
train_df, test_df = train_test_split(df, test_size=0.25, shuffle=True, random_state=0)
print("train")
print(f' value counts\n{train_df["species"].value_counts()}')
print()
print("test")
print(f' value counts\n{test_df["species"].value_counts()}')
plotter = FoldsPlotter(x=x, y=y, group=group, ylabel="02-2-train_test_split")
plotter.add_plot(train_idx=train_df.index, test_idx=test_df.index)
plotter.show(custom_y_label=["train / test", "class", "group"])
train_df, test_df = train_test_split(df, test_size=0.25, shuffle=True, random_state=0, stratify=y)
print("train")
print(f' value counts\n{train_df["species"].value_counts()}')
print()
print("test")
print(f' value counts\n{test_df["species"].value_counts()}')
plotter = FoldsPlotter(x=x, y=y, group=group, ylabel="02-3-train_test_split")
plotter.add_plot(train_idx=train_df.index, test_idx=test_df.index)
plotter.show(custom_y_label=["train / test", "class", "group"])
train_x, test_x, train_y, test_y = train_test_split(
x, y,
test_size=0.25, shuffle=True, random_state=0, stratify=y
)
print(f"train_x.shape={train_x.shape}, train_y.shape={train_y.shape}")
print()
print(f"test_x.shape={test_x.shape}, test_y.shape={test_y.shape}")
plotter = FoldsPlotter(x=x, y=y, group=group, ylabel="02-4-train_test_split")
plotter.add_plot(train_idx=train_x.index, test_idx=test_x.index)
plotter.show(custom_y_label=["train / test", "class", "group"])
# train_x, test_x, train_y, test_y = train_test_split(
# x, y,
# test_size=2, shuffle=True, random_state=0, stratify=y
# )
# ---------------------------------------------------------------------------
# ValueError Traceback (most recent call last)
# ~\AppData\Local\Temp\ipykernel_13444\2492759233.py in <module>
# ----> 1 train_x, test_x, train_y, test_y = train_test_split(
# 2 x, y,
# 3 test_size=2, shuffle=True, random_state=0, stratify=y
# 4 )
# ~\AppData\Local\Programs\Python\Python39\lib\site-packages\sklearn\model_selection\_split.py in train_test_split(test_size, train_size, random_state, shuffle, stratify, *arrays)
# 2439 cv = CVClass(test_size=n_test, train_size=n_train, random_state=random_state)
# 2440
# -> 2441 train, test = next(cv.split(X=arrays[0], y=stratify))
# 2442
# 2443 return list(
# ~\AppData\Local\Programs\Python\Python39\lib\site-packages\sklearn\model_selection\_split.py in split(self, X, y, groups)
# 1598 """
# 1599 X, y, groups = indexable(X, y, groups)
# -> 1600 for train, test in self._iter_indices(X, y, groups):
# 1601 yield train, test
# 1602
# ~\AppData\Local\Programs\Python\Python39\lib\site-packages\sklearn\model_selection\_split.py in _iter_indices(self, X, y, groups)
# 1951 )
# 1952 if n_test < n_classes:
# -> 1953 raise ValueError(
# 1954 "The test_size = %d should be greater or "
# 1955 "equal to the number of classes = %d" % (n_test, n_classes)
# ValueError: The test_size = 2 should be greater or equal to the number of classes = 3
###Output
_____no_output_____
###Markdown
sklearn.model_selection.KFold ```sklearn.model_selection.KFold(n_splits=5, *, shuffle=False, random_state=None)・・・・データを指定した K 個に等分し,1 つを検証用,それ以外の K-1 個を学習に用います.各 fold における検証用データに重複はさせないようにします. - n_splits: データを何等分するかを指定できます. - shuffle: True の場合にデータを抽出する際に元データの順番を保持せず,ランダムなインデックスで抽出します. - random_state: 上記ランダム抽出に用いる乱数シードを設定します.```
###Code
from sklearn.model_selection import KFold
kf = KFold(n_splits=5, shuffle=True, random_state=42)
plotter = FoldsPlotter(x=x, y=y, group=group, ylabel="03-1-KFold")
for i, (train_idx, test_idx) in enumerate(kf.split(X=x, y=y, groups=group)):
print(f"\nfold-{i}")
print(f"train_idx.shape={train_idx.shape}, test_idx.shape={test_idx.shape}")
print(f"set(y.iloc[test_idx])={set(y.iloc[test_idx])}")
print(f"train_idx={train_idx}")
print(f"test_idx={test_idx}")
plotter.add_plot(train_idx=train_idx, test_idx=test_idx)
plotter.show()
kf = KFold(n_splits=5, shuffle=False)
plotter = FoldsPlotter(x=x, y=y, group=group, ylabel="03-2-KFold")
for i, (train_idx, test_idx) in enumerate(kf.split(X=x)):
plotter.add_plot(train_idx=train_idx, test_idx=test_idx)
plotter.show()
###Output
_____no_output_____
###Markdown
sklearn.model_selection.ShuffleSplit ```sklearn.model_selection.ShuffleSplit(n_splits=10, *, test_size=None, train_size=None, random_state=None)・・・・train_test.split(shuffle=True, stratify=None) を複数回作用させるような学習用/検証用データのインデックスを返すオブジェクトを生成します.各 fold における検証用データは重複します. - n_splits: 学習用/検証用データセットのインデックスを何回返すかを指定します. - test_size: 検証用データセットの割合を指定します. - train_size: 学習用データセットの割合を指定します. - random_state: ランダム抽出に用いる乱数シードを設定します.```
###Code
from sklearn.model_selection import ShuffleSplit
ss = ShuffleSplit(n_splits=2, test_size=0.5, random_state=42)
plotter = FoldsPlotter(x=x, y=y, group=group, ylabel="04-1-ShuffleSplit")
for i, (train_idx, test_idx) in enumerate(ss.split(X=x)):
plotter.add_plot(train_idx=train_idx, test_idx=test_idx)
plotter.show()
###Output
_____no_output_____
###Markdown
sklearn.model_selection.StratifiedKFold ```sklearn.model_selection.StratifiedKFold(n_splits=5, *, shuffle=False, random_state=None)・・・・データを指定した K 個に等分し,1 つを検証用,それ以外の K-1 個を学習に用います.データ分割の際には,メソッド split の引数 y にてラベルやグループを指定することにより,全種類のラベルも均等に分割されます.各 fold における検証用データに重複はさせないようにします. - n_splits: データを何等分するかを指定できます. - shuffle: True の場合にデータを抽出する際に元データの順番を保持せず,ランダムなインデックスで抽出します. - random_state: 上記ランダム抽出に用いる乱数シードを設定します.```
###Code
from sklearn.model_selection import StratifiedKFold
skf = StratifiedKFold(n_splits=3, shuffle=False)
plotter = FoldsPlotter(x=x, y=y, group=group, ylabel="05-1-StratifiedKFold")
for i, (train_idx, test_idx) in enumerate(skf.split(X=x, y=y)):
print(f"\nfold-{i}")
print(f' y[test_idx].value_counts()\n{y[test_idx].value_counts()}')
plotter.add_plot(train_idx=train_idx, test_idx=test_idx)
plotter.show()
skf = StratifiedKFold(n_splits=3, shuffle=True, random_state=True)
plotter = FoldsPlotter(x=x, y=y, group=group, ylabel="05-2-StratifiedKFold")
for i, (train_idx, test_idx) in enumerate(skf.split(X=x, y=y)):
plotter.add_plot(train_idx=train_idx, test_idx=test_idx)
plotter.show()
###Output
_____no_output_____
###Markdown
sklearn.model_selection.StratifiedShuffleSplit ```sklearn.model_selection.StratifiedShuffleSplit(n_splits=10, *, test_size=None, train_size=None, random_state=None)・・・・train_test.split(shuffle=True, stratify=y) を複数回作用させるような学習用/検証用データのインデックスを返すオブジェクトを生成します.各 fold における検証用データは重複します. - n_splits: 学習用/検証用データセットのインデックスを何回返すかを指定します. - test_size: 検証用データセットの割合を指定します. - train_size: 学習用データセットの割合を指定します. - random_state: ランダム抽出に用いる乱数シードを設定します.```
###Code
from sklearn.model_selection import StratifiedShuffleSplit
sss = StratifiedShuffleSplit(n_splits=2, test_size=0.25, random_state=42)
plotter = FoldsPlotter(x=x, y=y, group=group, ylabel="06-1-StratifiedShuffleSplit")
for i, (train_idx, test_idx) in enumerate(sss.split(X=x, y=y)):
print(f"\nfold-{i}")
print(f' y[test_idx].value_counts()\n{y[test_idx].value_counts()}')
plotter.add_plot(train_idx, test_idx)
plotter.show()
sss = StratifiedShuffleSplit(n_splits=5, test_size=0.25, random_state=42)
plotter = FoldsPlotter(x=x, y=y, group=group, ylabel="06-2-StratifiedShuffleSplit")
for i, (train_idx, test_idx) in enumerate(sss.split(X=x, y=y)):
plotter.add_plot(train_idx, test_idx)
plotter.show()
###Output
_____no_output_____
###Markdown
sklearn.model_selection.GroupKFold ```sklearn.model_selection.GroupKFold(n_splits=5)・・・・データを指定した K 個に分割し,1 つを検証用,それ以外の K-1 個を学習に用います.データ分割の際には,メソッド split の引数 groups にて指定したグループによって,分割するインデックスを制御します.各 fold における検証用データに重複させないようにします. - n_splits: データを何分割するかを指定できます.```
###Code
from sklearn.model_selection import GroupKFold
# gkf = GroupKFold(n_splits=10)
# for train_idx, test_idx in gkf.split(X=x, y=y, groups=group):
# print(train_idx.shape, test_idx.shape)
# ---------------------------------------------------------------------------
# ValueError Traceback (most recent call last)
# ~\AppData\Local\Temp\ipykernel_13444\3306961057.py in <module>
# 2
# 3 gkf = GroupKFold()
# ----> 4 for train_idx, test_idx in gkf.split(X=x, y=y, groups=y):
# 5 print(train_idx.shape, test_idx.shape)
# ~\AppData\Local\Programs\Python\Python39\lib\site-packages\sklearn\model_selection\_split.py in split(self, X, y, groups)
# 338 )
# 339
# --> 340 for train, test in super().split(X, y, groups):
# 341 yield train, test
# 342
# ~\AppData\Local\Programs\Python\Python39\lib\site-packages\sklearn\model_selection\_split.py in split(self, X, y, groups)
# 84 X, y, groups = indexable(X, y, groups)
# 85 indices = np.arange(_num_samples(X))
# ---> 86 for test_index in self._iter_test_masks(X, y, groups):
# 87 train_index = indices[np.logical_not(test_index)]
# 88 test_index = indices[test_index]
# ~\AppData\Local\Programs\Python\Python39\lib\site-packages\sklearn\model_selection\_split.py in _iter_test_masks(self, X, y, groups)
# 96 By default, delegates to _iter_test_indices(X, y, groups)
# 97 """
# ---> 98 for test_index in self._iter_test_indices(X, y, groups):
# 99 test_mask = np.zeros(_num_samples(X), dtype=bool)
# 100 test_mask[test_index] = True
# ~\AppData\Local\Programs\Python\Python39\lib\site-packages\sklearn\model_selection\_split.py in _iter_test_indices(self, X, y, groups)
# 515
# 516 if self.n_splits > n_groups:
# --> 517 raise ValueError(
# 518 "Cannot have number of splits n_splits=%d greater"
# 519 " than the number of groups: %d." % (self.n_splits, n_groups)
# ValueError: Cannot have number of splits n_splits=10 greater than the number of groups: 3.
gkf = GroupKFold(n_splits=3)
plotter = FoldsPlotter(x=x, y=y, group=group, ylabel="07-1-GroupKFold")
for i, (train_idx, test_idx) in enumerate(gkf.split(X=x, y=y, groups=group)):
plotter.add_plot(train_idx, test_idx)
plotter.show()
gkf = GroupKFold(n_splits=len(set(group)))
plotter = FoldsPlotter(x=x, y=y, group=group, ylabel="07-2-GroupKFold")
for i, (train_idx, test_idx) in enumerate(gkf.split(X=x, y=y, groups=group)):
plotter.add_plot(train_idx, test_idx)
plotter.show()
###Output
_____no_output_____
###Markdown
sklearn.model_selection.GroupShuffleSplit ```sklearn.model_selection.GroupShuffleSplit(n_splits=5, *, test_size=None, train_size=None, random_state=None)・・・・GroupKFold のデータ分割で,各 fold 間におけるグループの重複を許したデータ分割手法. - n_splits: 学習用/検証用データセットのインデックスを何回返すかを指定します. - test_size: 検証用データセットの割合を指定します. - train_size: 学習用データセットの割合を指定します. - random_state: ランダム抽出に用いる乱数シードを設定します.```
###Code
from sklearn.model_selection import GroupShuffleSplit
gss = GroupShuffleSplit(n_splits=10, random_state=42)
plotter = FoldsPlotter(x=x, y=y, group=group, ylabel="08-1-GroupShuffleSplit")
for i, (train_idx, test_idx) in enumerate(gss.split(X=x, y=y, groups=group)):
plotter.add_plot(train_idx, test_idx)
plotter.show()
gss = GroupShuffleSplit(n_splits=2, test_size=0.25, random_state=42)
plotter = FoldsPlotter(x=x, y=y, group=group, ylabel="08-2-GroupShuffleSplit")
for i, (train_idx, test_idx) in enumerate(gss.split(X=x, y=y, groups=x["area"])):
plotter.add_plot(train_idx, test_idx)
plotter.show()
###Output
_____no_output_____
###Markdown
sklearn.model_selection.StratifiedGroupKFold ```sklearn.model_selection.StratifiedGroupKFold(n_splits=5, shuffle=False, random_state=None)・・・・データを指定した K 個に分割し,1 つを検証用,それ以外の K-1 個を学習に用います.データ分割は,メソッド split の引数 y にて指定したラベル,groups にて指定したグループを用いて,グループのまとまりを使ってラベルを K 個に均等に分けるように作用します.イメージとしては,名前の通り StratifiedKFold と GroupKFold が合体したものです.各 fold における検証用データのグループは重複しません. - n_splits: データを何分割するかを指定できます. - shuffle: True の場合にデータを抽出する際に元データの順番を保持せず,ランダムなインデックスで抽出します. - random_state: 上記ランダム抽出に用いる乱数シードを設定します.```
###Code
from sklearn.model_selection import StratifiedGroupKFold
sgkf = StratifiedGroupKFold(n_splits=3, shuffle=True, random_state=42)
plotter = FoldsPlotter(x=x, y=y, group=group, ylabel="09-1-StratifiedGroupKFold")
for i, (train_idx, test_idx) in enumerate(sgkf.split(X=x, y=y, groups=group)):
plotter.add_plot(train_idx, test_idx)
plotter.show()
sgkf = StratifiedGroupKFold(n_splits=10, shuffle=False)
plotter = FoldsPlotter(x=x, y=y, group=group, ylabel="09-2-StratifiedGroupKFold")
for i, (train_idx, test_idx) in enumerate(sgkf.split(X=x, y=y, groups=group)):
plotter.add_plot(train_idx, test_idx)
plotter.show()
sgkf = StratifiedGroupKFold(n_splits=2, shuffle=True)
plotter = FoldsPlotter(x=x, y=y, group=group, ylabel="09-3-StratifiedGroupKFold")
for i, (train_idx, test_idx) in enumerate(sgkf.split(X=x, y=y, groups=group)):
plotter.add_plot(train_idx, test_idx)
plotter.show()
###Output
_____no_output_____
###Markdown
sklearn.model_selection.RepeatedKFold ```sklearn.model_selection.RepeatedKFold(*, n_splits=5, n_repeats=10, random_state=None)・・・・KFold を n_repeats で指定した回数分行います.リピートされる各 fold 間では検証用データの重複はありませんが,各リピート間では検証用データの重複は発生します. - n_splits: データを何分割するかを指定できます. - n_repeats: KFold を繰り返す回数を指定します. - random_state: ランダム抽出に用いる乱数シードを設定します.```
###Code
from sklearn.model_selection import RepeatedKFold
rkf = RepeatedKFold(n_splits=3, n_repeats=3, random_state=42)
plotter = FoldsPlotter(x=x, y=y, group=group, ylabel="10-1-RepeatedKFold")
for i, (train_idx, test_idx) in enumerate(rkf.split(X=x, y=y, groups=group)):
plotter.add_plot(train_idx, test_idx)
plotter.show()
###Output
_____no_output_____
###Markdown
sklearn.model_selection.RepeatedStratifiedKFold ```sklearn.model_selection.RepeatedStratifiedKFold(*, n_splits=5, n_repeats=10, random_state=None)・・・・StratifiedKFold を n_repeats で指定した回数分行います.リピートされる各 fold 間では検証用データの重複はありませんが,各リピート間では検証用データの重複は発生します. - n_splits: データを何分割するかを指定できます. - n_repeats: KFold を繰り返す回数を指定します. - random_state: ランダム抽出に用いる乱数シードを設定します.```
###Code
from sklearn.model_selection import RepeatedStratifiedKFold
rskf = RepeatedStratifiedKFold(n_splits=2, n_repeats=2, random_state=42)
plotter = FoldsPlotter(x=x, y=y, group=group, ylabel="11-1-RepeatedStratifiedKFold")
for i, (train_idx, test_idx) in enumerate(rskf.split(X=x, y=y, groups=group)):
plotter.add_plot(train_idx, test_idx)
plotter.show()
rskf = RepeatedStratifiedKFold(n_splits=3, n_repeats=3, random_state=42)
plotter = FoldsPlotter(x=x, y=y, group=group, ylabel="11-2-RepeatedStratifiedKFold")
for i, (train_idx, test_idx) in enumerate(rskf.split(X=x, y=y, groups=group)):
plotter.add_plot(train_idx, test_idx)
plotter.show()
###Output
_____no_output_____
###Markdown
sklearn.model_selection.LeaveOneOut ```sklearn.model_selection.LeaveOneOut()・・・・一つだけを検証用データ,その他は学習用データとして分割します.```
###Code
from sklearn.model_selection import LeaveOneOut
loo = LeaveOneOut()
plotter = FoldsPlotter(x=x, y=y, group=group, ylabel="12-1-LeaveOneOut")
for i, (train_idx, test_idx) in enumerate(loo.split(X=x, y=y, groups=group)):
plotter.add_plot(train_idx, test_idx)
plotter.show()
print(f"train_idx={train_idx}")
print(f"test_idx={test_idx}")
###Output
train_idx=[ 0 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23
24 25 26 27 28 29 30 31 32 33 34 35 36 37 38 39 40 41 42 43 44 45 46 47
48 49 50 51 52 53 54 55 56 57 58 59 60 61 62 63 64 65 66 67 68 69 70 71
72 73 74 75 76 77 78 79 80 81 82 83 84 85 86 87 88 89 90 91 92 93 94 95
96 97 98]
test_idx=[99]
###Markdown
sklearn.model_selection.LeavePOut ```sklearn.model_selection.LeavePOut(p)・・・・引数 p で指定した個数を検証用データ,その他を学習用データとして分割します.検証用データが複数の場合は,検証用データの組み合わせが各 fold にて重複しないように,網羅的に fold が設定されます.例えば,データ数が m 個,引数 p を n 個と設定すると,総 fold 数は数学の組み合わせの記号 C を用いて,mCn = m*(m-1) / (n*(n-1)) 個生成されます. - p: 検証用データの個数を指定します.```
###Code
from sklearn.model_selection import LeavePOut
lpo = LeavePOut(p=1)
plotter = FoldsPlotter(x=x, y=y, group=group, ylabel="13-1-LeavePOut")
for i, (train_idx, test_idx) in enumerate(lpo.split(X=x, y=y, groups=group)):
plotter.add_plot(train_idx, test_idx)
plotter.show()
lpo = LeavePOut(p=2)
plotter = FoldsPlotter(x=x, y=y, group=group, ylabel="13-2-LeavePOut")
for i, (train_idx, test_idx) in enumerate(lpo.split(X=x, y=y, groups=group)):
plotter.add_plot(train_idx, test_idx)
plotter.show()
lpo = LeavePOut(p=2)
for i, (train_idx, test_idx) in enumerate(lpo.split(X=x, y=y, groups=group)):
pass
print(i)
int(100*99 / (2*1)) - 1
lpo = LeavePOut(p=3)
for i, (train_idx, test_idx) in enumerate(lpo.split(X=x, y=y, groups=group)):
pass
print(i)
int(100*99*98 / (3*2*1)) - 1
###Output
_____no_output_____
###Markdown
sklearn.model_selection.LeaveOneGroupOut ```sklearn.model_selection.LeaveOneGroupOut()・・・・1 つのグループを検証用データとし,それ以外を学習用データとして分割します.```
###Code
from sklearn.model_selection import LeaveOneGroupOut
logo = LeaveOneGroupOut()
plotter = FoldsPlotter(x=x, y=y, group=group, ylabel="14-1-LeaveOneGroupOut")
for i, (train_idx, test_idx) in enumerate(logo.split(X=x, y=y, groups=group)):
plotter.add_plot(train_idx, test_idx)
plotter.show()
###Output
_____no_output_____
###Markdown
sklearn.model_selection.LeavePGroupsOut ```sklearn.model_selection.LeavePGroupsOut(n_groups)・・・・引数 n_groups で指定したグループ数を検証用データ,その他を学習用データとして分割します.検証用データが複数の場合は,検証用データの組み合わせが各 fold にて重複しないように,網羅的に fold が設定されます.例えば,グループ数が m 個,引数 n_groups を n 個と設定すると,総 fold 数は数学の組み合わせの記号 C を用いて,mCn = m*(m-1) / (n*(n-1)) 個生成されます. - n_groups: 検証用データの個数を指定します.```
###Code
from sklearn.model_selection import LeavePGroupsOut
lpgo = LeavePGroupsOut(n_groups=1)
plotter = FoldsPlotter(x=x, y=y, group=group, ylabel="15-1-LeavePGroupsOut")
for i, (train_idx, test_idx) in enumerate(lpgo.split(X=x, y=y, groups=group)):
plotter.add_plot(train_idx, test_idx)
plotter.show()
lpgo = LeavePGroupsOut(n_groups=3)
plotter = FoldsPlotter(x=x, y=y, group=group, ylabel="15-2-LeavePGroupsOut")
for i, (train_idx, test_idx) in enumerate(lpgo.split(X=x, y=y, groups=group)):
plotter.add_plot(train_idx, test_idx)
plotter.show()
# from sklearn.model_selection import LeavePGroupsOut
# lpgo = LeavePGroupsOut(n_groups=10)
# for i, (train_idx, test_idx) in enumerate(lpgo.split(X=x, y=y, groups=group)):
# pass
# ---------------------------------------------------------------------------
# ValueError Traceback (most recent call last)
# ~\AppData\Local\Temp\ipykernel_13444\3041175701.py in <module>
# 1 from sklearn.model_selection import LeavePGroupsOut
# 2 lpgo = LeavePGroupsOut(n_groups=10)
# ----> 3 for i, (train_idx, test_idx) in enumerate(lpgo.split(X=x, y=y, groups=group)):
# 4 print(f"\nfold-{i}")
# 5 print(f"train_idx.shape={train_idx.shape}, test_idx.shape={test_idx.shape}")
# ~\AppData\Local\Programs\Python\Python39\lib\site-packages\sklearn\model_selection\_split.py in split(self, X, y, groups)
# 84 X, y, groups = indexable(X, y, groups)
# 85 indices = np.arange(_num_samples(X))
# ---> 86 for test_index in self._iter_test_masks(X, y, groups):
# 87 train_index = indices[np.logical_not(test_index)]
# 88 test_index = indices[test_index]
# ~\AppData\Local\Programs\Python\Python39\lib\site-packages\sklearn\model_selection\_split.py in _iter_test_masks(self, X, y, groups)
# 1274 unique_groups = np.unique(groups)
# 1275 if self.n_groups >= len(unique_groups):
# -> 1276 raise ValueError(
# 1277 "The groups parameter contains fewer than (or equal to) "
# 1278 "n_groups (%d) numbers of unique groups (%s). LeavePGroupsOut "
# ValueError: The groups parameter contains fewer than (or equal to) n_groups (10) numbers of unique groups (['A' 'B' 'C' 'D' 'E']). LeavePGroupsOut expects that at least n_groups + 1 (11) unique groups be present
###Output
_____no_output_____
###Markdown
sklearn.model_selection.PredefinedSplit ```sklearn.model_selection.PredefinedSplit(test_fold)・・・・引数 test_fold に指定した fold でデータを分割します. - test_fold: データ分割の仕方を与えます.```
###Code
from sklearn.model_selection import PredefinedSplit
ps = PredefinedSplit(test_fold=list(range(5))*20)
plotter = FoldsPlotter(x=x, y=y, group=group, ylabel="16-1-PredefinedSplit")
for i, (train_idx, test_idx) in enumerate(ps.split()):
plotter.add_plot(train_idx, test_idx)
plotter.show()
ps = PredefinedSplit(test_fold=np.random.random_integers(low=0, high=1, size=10).tolist()*10)
plotter = FoldsPlotter(x=x, y=y, group=group, ylabel="16-2-PredefinedSplit")
for i, (train_idx, test_idx) in enumerate(ps.split()):
plotter.add_plot(train_idx, test_idx)
plotter.show()
###Output
C:\Users\multi\AppData\Local\Temp\ipykernel_12004\3739650861.py:1: DeprecationWarning: This function is deprecated. Please call randint(0, 1 + 1) instead
ps = PredefinedSplit(test_fold=np.random.random_integers(low=0, high=1, size=10).tolist()*10)
###Markdown
sklearn.model_selection.TimeSeriesSplit ```sklearn.model_selection.TimeSeriesSplit(n_splits=5, *, max_train_size=None, test_size=None, gap=0)・・・・時系列データを考慮したデータ分割手法で,検証用データを K 等分し,残りの検証用データよりも前のデータを学習用データとして用います.したがって,index=0 から始まるような検証用データにおける学習用データは取れないので,検証用データは K-1 個になります.引数 n_splits では,この K-1 を指定します. - n_splits: 検証用データを何等分するかを指定します. - max_train_size: 各 fold における学習用データの最大の個数を指定します. - test_size: 各 fold における検証用データの個数を指定します. - gap: 学習用データと検証用データを時間方向に離す個数を指定します.```
###Code
from sklearn.model_selection import TimeSeriesSplit
tss = TimeSeriesSplit(n_splits=2)
plotter = FoldsPlotter(x=x, y=y, group=group, ylabel="17-1-TimeSeriesSplit")
for i, (train_idx, test_idx) in enumerate(tss.split(X=x, y=y, groups=group)):
plotter.add_plot(train_idx, test_idx)
plotter.show()
tss = TimeSeriesSplit(n_splits=5)
plotter = FoldsPlotter(x=x, y=y, group=group, ylabel="17-2-TimeSeriesSplit")
for i, (train_idx, test_idx) in enumerate(tss.split(X=x, y=y, groups=group)):
plotter.add_plot(train_idx, test_idx)
plotter.show()
tss = TimeSeriesSplit(n_splits=10, max_train_size=30, gap=5)
plotter = FoldsPlotter(x=x, y=y, group=group, ylabel="17-3-TimeSeriesSplit")
for i, (train_idx, test_idx) in enumerate(tss.split(X=x, y=y, groups=group)):
plotter.add_plot(train_idx, test_idx)
plotter.show()
###Output
_____no_output_____
|
examples/gallery/simple/save_filtered_df.ipynb
|
###Markdown
AppLets wrap it into nice template that can be served via `panel serve save_filtered_df.ipynb`
###Code
pn.template.FastListTemplate(
site="Panel",
title="Save Filtered Dataframe",
main=[
pn.Column(
"This app demonstrates how to **filter and download a Pandas DataFrame**.",
pn.pane.HTML("<div style='font-size: 100px;text-align: center'>🐼</div>", height=75, margin=(50,5,10,5)),
),
pn.Column(years, mpg, fd),
pn.Column(filtered_mpg, height=600, scroll=True),
], main_max_width="768px",
).servable();
###Output
_____no_output_____
|
jupyter/Python4Maths/Intro-to-Python/06.ipynb
|
###Markdown
All of these python notebooks are available at [https://gitlab.erc.monash.edu.au/andrease/Python4Maths.git] Functions Functions can represent mathematical functions. More importantly, in programmming functions are a mechansim to allow code to be re-used so that complex programs can be built up out of simpler parts. This is the basic syntax of a function```pythondef funcname(arg1, arg2,... argN): ''' Document String''' statements return ``` Read the above syntax as, A function by name "funcname" is defined, which accepts arguements "arg1,arg2,....argN". The function is documented and it is '''Document String'''. The function after executing the statements returns a "value".Return values are optional (by default every function returns **None** if no return statement is executed)
###Code
print("Hello Jack.")
print("Jack, how are you?")
###Output
Hello Jack.
Jack, how are you?
###Markdown
Instead of writing the above two statements every single time it can be replaced by defining a function which would do the job in just one line. Defining a function firstfunc().
###Code
def firstfunc():
print("Hello Jack.")
print("Jack, how are you?")
firstfunc() # execute the function
###Output
Hello Jack.
Jack, how are you?
###Markdown
**firstfunc()** every time just prints the message to a single person. We can make our function **firstfunc()** to accept arguements which will store the name and then prints respective to that accepted name. To do so, add a argument within the function as shown.
###Code
def firstfunc(username):
print("Hello %s." % username)
print(username + ',' ,"how are you?")
name1 = 'sally' # or use input('Please enter your name : ')
###Output
_____no_output_____
###Markdown
So we pass this variable to the function **firstfunc()** as the variable username because that is the variable that is defined for this function. i.e name1 is passed as username.
###Code
firstfunc(name1)
###Output
Hello sally.
sally, how are you?
###Markdown
Return Statement When the function results in some value and that value has to be stored in a variable or needs to be sent back or returned for further operation to the main algorithm, a return statement is used.
###Code
def times(x,y):
z = x*y
return z
###Output
_____no_output_____
###Markdown
The above defined **times( )** function accepts two arguements and return the variable z which contains the result of the product of the two arguements
###Code
c = times(4, 5)
print(c)
###Output
20
###Markdown
The z value is stored in variable c and can be used for further operations. Instead of declaring another variable the entire statement itself can be used in the return statement as shown.
###Code
def times(x,y):
'''This multiplies the two input arguments'''
return x*y
c = times(4, 5)
print(c)
###Output
20
###Markdown
Since the **times( )** is now defined, we can document it as shown above. This document is returned whenever **times( )** function is called under **help( )** function.
###Code
help(times)
###Output
Help on function times in module __main__:
times(x, y)
This multiplies the two input arguments
###Markdown
Multiple variable can also be returned as a tuple. However this tends not to be very readable when returning many value, and can easily introduce errors when the order of return values is interpreted incorrectly.
###Code
eglist = [10, 50, 30, 12, 6, 8, 100]
def egfunc(eglist):
highest = max(eglist)
lowest = min(eglist)
first = eglist[0]
last = eglist[-1]
return highest,lowest,first,last
###Output
_____no_output_____
###Markdown
If the function is just called without any variable for it to be assigned to, the result is returned inside a tuple. But if the variables are mentioned then the result is assigned to the variable in a particular order which is declared in the return statement.
###Code
egfunc(eglist)
a,b,c,d = egfunc(eglist)
print(' a =',a,' b =',b,' c =',c,' d =',d)
###Output
a = 100 b = 6 c = 10 d = 100
###Markdown
Default arguments When an argument of a function is common in majority of the cases this can be specified with a default value. This is also called an implicit argument.
###Code
def implicitadd(x, y=3, z=0):
print("%d + %d + %d = %d"%(x,y,z,x+y+z))
return x+y+z
###Output
_____no_output_____
###Markdown
**implicitadd( )** is a function accepts up to three arguments but most of the times the first argument needs to be added just by 3. Hence the second argument is assigned the value 3 and the third argument is zero. Here the last two arguments are default arguments. Now if the second argument is not defined when calling the **implicitadd( )** function then it considered as 3.
###Code
implicitadd(4)
###Output
4 + 3 + 0 = 7
###Markdown
However we can call the same function with two or three arguments. A useful feature is to explicitly name the argument values being passed into the function. This gives great flexibility in how to call a function with optional arguments. All off the following are valid:
###Code
implicitadd(4, 4)
implicitadd(4, 5, 6)
implicitadd(4, z=7)
implicitadd(2, y=1, z=9)
implicitadd(x=1)
###Output
4 + 4 + 0 = 8
4 + 5 + 6 = 15
4 + 3 + 7 = 14
2 + 1 + 9 = 12
1 + 3 + 0 = 4
###Markdown
Any number of arguments If the number of arguments that is to be accepted by a function is not known then a asterisk symbol is used before the name of the argument to hold the remainder of the arguments. The following function requires at least one argument but can have many more.
###Code
def add_n(first,*args):
"return the sum of one or more numbers"
reslist = [first] + [value for value in args]
print(reslist)
return sum(reslist)
###Output
_____no_output_____
###Markdown
The above function defines a list of all of the arguments, prints the list and returns the sum of all of the arguments.
###Code
add_n(1, 2, 3, 4, 5)
add_n(6.5)
###Output
[6.5]
###Markdown
Arbitrary numbers of named arguments can also be accepted using `**`. When the function is called all of the additional named arguments are provided in a dictionary
###Code
def namedArgs(**names):
'print the named arguments'
# names is a dictionary of keyword : value
print(" ".join(name+"="+str(value)
for name,value in names.items()))
namedArgs(x=3*4,animal='mouse',z=(1+2j))
###Output
z=(1+2j) animal=mouse x=12
###Markdown
Global and Local Variables Whatever variable is declared inside a function is local variable and outside the function in global variable.
###Code
eg1 = [1, 2, 3, 4, 5]
###Output
_____no_output_____
###Markdown
In the below function we are appending a element to the declared list inside the function. eg2 variable declared inside the function is a local variable.
###Code
def egfunc1():
x=1
def thirdfunc():
x=2
print("Inside thirdfunc x =", x)
thirdfunc()
print("Outside x =", x)
egfunc1()
###Output
Inside thirdfunc x = 2
Outside x = 1
###Markdown
If a **global** variable is defined as shown in the example below then that variable can be called from anywhere. Global values should be used sparingly as they make functions harder to re-use.
###Code
eg3 = [1, 2, 3, 4, 5]
def egfunc1():
x = 1.0 # local variable for egfunc1
def thirdfunc():
global x # globally defined variable
x = 2.0
print("Inside thirdfunc x =", x)
thirdfunc()
print("Outside x =", x)
egfunc1()
print("Globally defined x =",x)
###Output
Inside thirdfunc x = 2.0
Outside x = 1.0
Globally defined x = 2.0
###Markdown
Lambda Functions These are small functions which are not defined with any name and carry a single expression whose result is returned. Lambda functions comes very handy when operating with lists. These function are defined by the keyword **lambda** followed by the variables, a colon and the respective expression.
###Code
z = lambda x: x * x
z(8)
###Output
_____no_output_____
###Markdown
Composing functions Lambda functions can also be used to compose functions
###Code
def double(x):
return 2*x
def square(x):
return x*x
def f_of_g(f,g):
"Compose two functions of a single variable"
return lambda x: f(g(x))
doublesquare= f_of_g(double,square)
print("doublesquare is a",type(doublesquare))
doublesquare(3)
###Output
doublesquare is a <class 'function'>
|
Data-analysis/OFAT/Interactive_OFAT_analysis.ipynb
|
###Markdown
OFAT analysisThe csv file to be analyized is specified in the first parameter.
###Code
csv_file = 'batch_lanes_2.csv'
rl2 = pd.read_csv('batch_lanes_2.csv')
rl2.drop(rl2.columns[0], axis=1, inplace=True)
rl2g = rl2.groupby('Run')
rl2gm = rl2g.agg(np.mean)
print("Available parameters:\n")
for x in rl2gm.columns:
print(" - "+x)
###Output
Available parameters:
- Avg_speed
- Cars_in_lane
- Avg_slowdown
- spawn_chance
- agression
- lanes
###Markdown
3D plotAn example 3D plot, the parameter of the 3 axis can be specified in the respective variable.Although not a true OFAT analysis, a 3D plot can be valuable in finding interaction without resorting to theexpensive Sobol analysis
###Code
driedee = plt.figure(figsize=(6,5)).gca(projection='3d')
x_as = 'agression'
y_as = 'Cars_in_lane'
z_as = 'Avg_speed'
kleur = 'lanes'
p = driedee.scatter(rl2gm[x_as], rl2gm[y_as], rl2gm[z_as], c=rl2gm[kleur])
driedee.set_xlabel(x_as)
driedee.set_ylabel(y_as)
driedee.set_zlabel(z_as)
plt.colorbar(p)
plt.show()
###Output
_____no_output_____
###Markdown
Scatter plotAn example 2D scatter plot, this can be used to perform a pure OFAT analysis or analyze the effect of an additionaldiscrete parameter through color.
###Code
x_as = 'Avg_speed'
z_as = 'spawn_chance'
y_as = 'Cars_in_lane'
kleur = 'lanes'
plt.figure(figsize=(5,5))
cmap = plt.cm.viridis
plt.scatter(rl2gm[x_as], rl2gm[y_as], c=rl2gm[kleur])
plt.xlabel(x_as)
plt.ylabel(y_as)
legend_elements = [
Line2D([0], [0], color=cmap(0.1), label='3 lanes'),
Line2D([0], [0], color=cmap(0.5), label='4 lanes'),
Line2D([0], [0], color=cmap(1.0), label='5 lanes')
]
plt.legend(handles=legend_elements)
plt.show()
###Output
_____no_output_____
|
2slit.ipynb
|
###Markdown
Two-slit diffractionParaxial approximation
###Code
import numpy as np
import matplotlib.pyplot as plt
%matplotlib inline
hbar = 1.
m = 1.
k = 1.
w0 = 1.
zf = 100.
lam = 2*np.pi/k
zR = np.pi*w0**2/lam
sep = 5*w0
# helper functions
w = lambda z: w0*np.sqrt(1+(z/zR)**2)
R = lambda z: z*(1+(zR/z)**2)
gouy = lambda z: np.arctan(z/zR)
psi = lambda x,z: w0/w(z)*np.exp(-x**2/w(z)**2-1.j*(k*z+k*x**2/2/R(z)-gouy(z)))
# psi in limit |z| >> zR
psifar = lambda x,z: zR/z*np.exp(-x**2/(2.*z/k/w0)**2-1.j*(k*z+k*x**2/z))
psi2 = lambda x,z: psi(x-sep,z)+psi(x+sep,z)
psi2far = lambda x,z: psifar(x-sep,z)+psifar(x+sep,z)
incoh = lambda x,z: np.abs(psi(x-sep,z))**2+np.abs(psi(x+sep,z))**2
xlist = 2*sep*np.linspace(-1.,1.,401)
# pdf at slits
plt.plot(xlist,np.abs(psi2(xlist,0.001))**2)
plt.plot(xlist,incoh(xlist,0.001),'.')
# pdf far away at z = zf
plt.plot(xlist*zf/2,np.abs(psi2far(xlist*zf/2,zf))**2)
plt.plot(xlist*zf/2,incoh(xlist*zf/2,zf),'.')
###Output
_____no_output_____
|
notebooks/wrangle_act.ipynb
|
###Markdown
Gathering Data Loading Archive Data from CSV to DataFrame
###Code
import pandas as pd
df_twt_archive = pd.read_csv('../data/twitter-archive-enhanced-2.csv')
###Output
_____no_output_____
###Markdown
Instantiating an API Object from Config File
###Code
import os
from hydra import initialize, initialize_config_module, initialize_config_dir, compose
from omegaconf import OmegaConf
abs_config_dir=os.path.abspath("../twitter_wrangler/conf")
with initialize_config_dir(config_dir=abs_config_dir):
cfg = compose(overrides=["+application=twitter"])
import tweepy
from tweepy import OAuthHandler
import os
auth = OAuthHandler((cfg.get('consumer_key', os.environ.get('CONSUMER_KEY'))),
(cfg.get('consumer_secret', os.environ.get('CONSUMER_SECRET'))))
auth.set_access_token((cfg.get('access_token', os.environ.get('ACCESS_TOKEN'))),
(cfg.get('access_secret', os.environ.get('ACCESS_SECRET'))))
api = tweepy.API(auth, wait_on_rate_limit=True)
try:
api.verify_credentials()
print("Authentication OK")
except:
print("Error during authentication")
###Output
_____no_output_____
###Markdown
Loading Data from API to Text File
###Code
tweet_ids = df_twt_archive.tweet_id.values
count = 0
fails_dict = {}
start = timer()
# Save each tweet's returned JSON as a new line in a .txt file
with open('../data/tweet_json.txt', 'w') as outfile:
# This loop will likely take 20-30 minutes to run because of Twitter's rate limit
for tweet_id in tweet_ids:
count += 1
print(str(count) + ": " + str(tweet_id))
try:
tweet = api.get_status(tweet_id, tweet_mode='extended')
print("Success")
json.dump(tweet._json, outfile)
outfile.write('\n')
except tweepy.TweepError as e:
print("Fail")
fails_dict[tweet_id] = e
pass
end = timer()
print(end - start)
print(fails_dict)
###Output
_____no_output_____
###Markdown
Loading Data from text file to DataFrame
###Code
import pandas as pd
import json
data = []
with open('../data/tweet_json.txt') as f:
for line in f:
data.append(json.loads(line))
#create dataframe from json data
df_api = pd.DataFrame(data)
#select columns of interest
columns_of_interest = ['id', 'retweet_count', 'favorite_count']
df_api = df_api[columns_of_interest]
df_api
###Output
_____no_output_____
###Markdown
Loading Data from API to DataFrame
###Code
import requests
import pandas as pd
import io
url = 'https://d17h27t6h515a5.cloudfront.net/topher/2017/August/599fd2ad_image-predictions/image-predictions.tsv'
r = requests.get(url)
print(r.status_code)
if r.status_code == 200:
print('Success!')
elif r.status_code == 404:
print('Not Found.')
urlData = r.content
image_predictions_df = pd.read_csv(io.StringIO(urlData.decode('utf-8')), sep='\t')
image_predictions_df.to_csv('../data/image_predictions.tsv', sep='\t')
image_predictions_df.head()
###Output
200
Success!
###Markdown
Assessing Data twitter-archive-enhanced-2.csv - Visual Assessment
###Code
df_twt_archive.head()
df_twt_archive.tail()
###Output
_____no_output_____
###Markdown
- Looking at the data visually using the pandas 'head'/'tail' functions and in Microsoft Excel, there appears to be a high number of null values for the following columns: - in_reply_to_status_id - in_reply_to_user_id - retweeted_status_id - retweeted_status_user_id - retweeted_status_timestamp columns- The dog stages columns are in a wide format with most of the values as 'None'. These columns can be condensed into one column, as the column headers should be the column values.- Looking at the tail, 'name' contains None values and 'a'.
###Code
df_twt_archive.name.value_counts()
###Output
_____no_output_____
###Markdown
twitter-archive-enhanced-2.csv - Programmatic Assessment
###Code
df_twt_archive.shape
df_twt_archive.info()
###Output
<class 'pandas.core.frame.DataFrame'>
RangeIndex: 2356 entries, 0 to 2355
Data columns (total 17 columns):
# Column Non-Null Count Dtype
--- ------ -------------- -----
0 tweet_id 2356 non-null int64
1 in_reply_to_status_id 78 non-null float64
2 in_reply_to_user_id 78 non-null float64
3 timestamp 2356 non-null object
4 source 2356 non-null object
5 text 2356 non-null object
6 retweeted_status_id 181 non-null float64
7 retweeted_status_user_id 181 non-null float64
8 retweeted_status_timestamp 181 non-null object
9 expanded_urls 2297 non-null object
10 rating_numerator 2356 non-null int64
11 rating_denominator 2356 non-null int64
12 name 2356 non-null object
13 doggo 2356 non-null object
14 floofer 2356 non-null object
15 pupper 2356 non-null object
16 puppo 2356 non-null object
dtypes: float64(4), int64(3), object(10)
memory usage: 313.0+ KB
###Markdown
- The five columns mentioned above contain mostly null values and break the first rule of tidy data (each variable forms a column). It is best to remove these columns as they do not add value to the data.- Several of the columns are the incorrect data type and should be as follows: - tweet_id: String - timestamp: Datetime - source: String - text: String - expanded_urls: String - rating_numerator: Float - rating_denominator: Float - name: String
###Code
df_twt_archive.describe()
###Output
_____no_output_____
###Markdown
The mean for rating_numerator is 13 and the maximum value is 1776. This outlier indicates that there are errors that need to be cleaned. Similarly, the rating_denominator has a maximum value of 170, where these should all be 10.
###Code
df_twt_archive.rating_numerator.value_counts()
df_twt_archive.rating_denominator.value_counts()
###Output
_____no_output_____
###Markdown
tweet_json.txt - Visual Assessment
###Code
df_api.head()
df_api.tail()
###Output
_____no_output_____
###Markdown
Looking at the data visually using the pandas 'head' function and in Microsoft Excel, no null values are present and all columns are integers. tweet_json.txt - Programmatic Assessment
###Code
df_api.shape
df_api.info()
df_api.describe()
###Output
_____no_output_____
###Markdown
image_predictions.tsv - Visual Assessment
###Code
image_predictions_df.head()
image_predictions_df.tail()
###Output
_____no_output_____
###Markdown
Looking at the data visually using the pandas 'head' function and in Microsoft Excel, the p1 column image_predictions.tsv - Programmatic Assessment
###Code
image_predictions_df.shape
image_predictions_df.info()
image_predictions_df.describe()
###Output
_____no_output_____
###Markdown
Cleaning Data
###Code
# Creating copies of the data frames before cleaning
df_twt_archive_clean = df_twt_archive.copy()
df_api_clean = df_api.copy()
image_predictions_df_clean = image_predictions_df.copy()
###Output
_____no_output_____
###Markdown
twitter-archive-enhanced-2.csv Define - Tidiness: - Convert dog types into one column (should be column values instead of column headers) - Drop dog type columns and columns consisting of null values (every column is a variable) - Joining with the other dataframes on tweet_id to include all relevant columns- Quality: - Remove retweets - Drop Null columns - Change data type of tweet_id to String - Change data type of timestamp to Datetime - Change data type of rating_numerator and rating_denominator to Float - Change data type of resulting dog_type column to category - Separate date and time into separate columns and drop timestamp - Replace underscores with spaces in prediction column - Left join with tweet_json dataframe to get retweet count - Inner join with image_predictions dataframe - Convert confidence from decimal to percentage Code Combining Dog Stages into one column (wide to long format)
###Code
df_twt_archive_clean['dog_type'] = df_twt_archive_clean.text.str.extract('(doggo | floofer | pupper | puppo)', expand = True)
df_twt_archive_clean = df_twt_archive_clean.drop(['doggo', 'floofer', 'pupper', 'puppo'], axis = 1)
###Output
_____no_output_____
###Markdown
Removing retweet rows
###Code
df_twt_archive_clean = df_twt_archive_clean[df_twt_archive_clean.retweeted_status_id.isnull()]
###Output
_____no_output_____
###Markdown
Changing Columns to the correct Data Types
###Code
df_twt_archive_clean.tweet_id = df_twt_archive_clean.tweet_id.astype(str)
df_twt_archive_clean.timestamp = pd.to_datetime(df_twt_archive_clean.timestamp, yearfirst = True)
df_twt_archive_clean.rating_numerator = df_twt_archive_clean.rating_numerator.astype(float)
df_twt_archive_clean.rating_denominator = df_twt_archive_clean.rating_numerator.astype(float)
df_twt_archive_clean.dog_type = df_twt_archive_clean.dog_type.astype('category')
###Output
_____no_output_____
###Markdown
Create separate columns for date and time, and drop timestamp column
###Code
df_twt_archive_clean['date'] = df_twt_archive_clean['timestamp'].dt.date
df_twt_archive_clean['time'] = df_twt_archive_clean['timestamp'].dt.time
###Output
_____no_output_____
###Markdown
Left Join with the tweet_json table
###Code
df_api_clean = df_api_clean.rename(columns={'id': 'tweet_id'})
df_api_clean.tweet_id = df_api_clean.tweet_id.astype(str)
df_twt_archive_clean = pd.merge(df_twt_archive_clean, df_api_clean, on = 'tweet_id', how = 'left')
###Output
_____no_output_____
###Markdown
Inner Join with the image_predictions table
###Code
image_predictions_df_clean.tweet_id = image_predictions_df_clean.tweet_id.astype(str)
df_twt_archive_clean = df_twt_archive_clean.merge(image_predictions_df_clean, on = 'tweet_id', how = 'inner')
###Output
_____no_output_____
###Markdown
Dropping Columns
###Code
df_twt_archive_clean = df_twt_archive_clean.drop(['in_reply_to_status_id', 'in_reply_to_user_id',
'retweeted_status_id', 'retweeted_status_user_id',
'retweeted_status_timestamp', 'timestamp',
'img_num', 'p2', 'p2_conf', 'p2_dog', 'p3', 'p3_conf', 'p3_dog'], axis = 1)
###Output
_____no_output_____
###Markdown
Replace underscores with spaces in prediction column
###Code
df_twt_archive_clean.p1 = df_twt_archive_clean.p1.str.replace('_',' ')
df_twt_archive_clean.p1 = df_twt_archive_clean.p1.str.title()
###Output
_____no_output_____
###Markdown
Convert confidence from decimal to percentage
###Code
df_twt_archive_clean['p1_conf'] = pd.Series(["{0:.2f}%".format(val * 100) for val in df_twt_archive_clean['p1_conf']], index = df_twt_archive_clean.index)
###Output
_____no_output_____
###Markdown
Rename columns and save to CSV
###Code
df_twt_archive_clean = df_twt_archive_clean.rename({'p1':'prediction',
'p1_conf': 'confidence', 'p1_dog': 'breed_predicted'},
axis = 'columns')
df_twt_archive_clean.to_csv('../data/twitter_archive_master.csv', index = False)
###Output
_____no_output_____
###Markdown
Test
###Code
df_twt_archive_clean.head()
df_twt_archive_clean.info()
###Output
<class 'pandas.core.frame.DataFrame'>
Int64Index: 1994 entries, 0 to 1993
Data columns (total 16 columns):
# Column Non-Null Count Dtype
--- ------ -------------- -----
0 tweet_id 1994 non-null object
1 source 1994 non-null object
2 text 1994 non-null object
3 expanded_urls 1994 non-null object
4 rating_numerator 1994 non-null float64
5 rating_denominator 1994 non-null float64
6 name 1994 non-null object
7 dog_type 216 non-null category
8 date 1994 non-null object
9 time 1994 non-null object
10 retweet_count 1986 non-null float64
11 favorite_count 1986 non-null float64
12 jpg_url 1994 non-null object
13 prediction 1994 non-null object
14 confidence 1994 non-null object
15 breed_predicted 1994 non-null bool
dtypes: bool(1), category(1), float64(4), object(10)
memory usage: 237.8+ KB
###Markdown
Saving Cleaned Data to sqlite database
###Code
import sqlite3
from sqlite3 import Error
def create_connection(db_file):
""" create a database connection to the SQLite database
specified by db_file
:param db_file: database file
:return: Connection object or None
"""
conn = None
try:
conn = sqlite3.connect(db_file)
return conn
except Error as e:
print(e)
return conn
def create_table(conn, create_table_sql):
""" create a table from the create_table_sql statement
:param conn: Connection object
:param create_table_sql: a CREATE TABLE statement
:return:
"""
try:
c = conn.cursor()
c.execute(create_table_sql)
except Error as e:
print(e)
database = "../data/master.db"
conn = create_connection(database)
# push the dataframe to sql
df_twt_archive_clean.to_sql("df_twt_archive_clean", conn, if_exists="replace")
#create the table
conn.execute(
"""
create table df_twt_archive_master as
select * from df_twt_archive_clean
""")
###Output
_____no_output_____
###Markdown
Insights Distribution of Dog Breeds
###Code
sqlite_select_query = """SELECT prediction AS dog_breed, COUNT(prediction) AS prediction_count
from df_twt_archive_master WHERE breed_predicted == True
GROUP BY prediction ORDER BY prediction_count DESC LIMIT 15"""
df = pd.read_sql_query(sqlite_select_query, conn)
df = df.set_index('dog_breed')
df.plot(kind = 'pie', y='prediction_count', title='Distribution of Dog Breeds',figsize=(9,9))
df.plot(kind = 'bar', y='prediction_count', title='Distribution of Dog Breeds', ylabel='prediction_count',figsize=(9,9))
df
###Output
_____no_output_____
###Markdown
There is a peak in number for golden retrievers. This number progressively goes down across the top 15 breeds. Top dog breeds by average favourite count
###Code
sqlite_select_query = """SELECT prediction AS dog_breed, avg(favorite_count) AS avg_favorite_count
from df_twt_archive_master WHERE breed_predicted == True
GROUP BY prediction ORDER BY avg_favorite_count DESC LIMIT 15"""
df = pd.read_sql_query(sqlite_select_query, conn)
df = df.set_index('dog_breed')
df.plot(kind = 'pie', y='avg_favorite_count', title='Top dog breeds by average favorite count', figsize=(9,9))
df.plot(kind = 'bar', y='avg_favorite_count', title='Top dog breeds by average favorite count', ylabel='avg_favorite_count',figsize=(9,9))
df
###Output
_____no_output_____
###Markdown
There is a peak for the Saluki breed. The distribution looks even across most of the top 15 breeds from African Hound onwards. None of the most common dog breeds are in the top 15 for average favorite count. Top dog breeds by average retweet count
###Code
sqlite_select_query = """SELECT prediction AS dog_breed, avg(retweet_count) AS avg_retweet_count
from df_twt_archive_master WHERE breed_predicted == True
GROUP BY prediction ORDER BY avg_retweet_count DESC LIMIT 15"""
df = pd.read_sql_query(sqlite_select_query, conn)
df = df.set_index('dog_breed')
df.plot(kind = 'pie', y='avg_retweet_count', title='Top dog breeds by average retweet count', figsize=(9,9))
df.plot(kind = 'bar', y='avg_retweet_count', title='Top dog breeds by average retweet count', ylabel='avg_retweet_count',figsize=(9,9))
df
###Output
_____no_output_____
###Markdown
Great Pyrenees is the only breed that appears in the top 15 average retweet count but not in the top 15 average favorite count. Top dog breeds by average rating
###Code
sqlite_select_query = """SELECT prediction AS dog_breed, avg(rating_numerator) AS avg_rating_numerator, avg(favorite_count) AS avg_favorite_count, avg(retweet_count) AS avg_retweet_count
from df_twt_archive_master WHERE breed_predicted == True
GROUP BY prediction ORDER BY avg_rating_numerator DESC LIMIT 15"""
df = pd.read_sql_query(sqlite_select_query, conn)
df = df.set_index('dog_breed')
df.plot(kind = 'pie', y='avg_rating_numerator', title='Top dog breeds by average rating', figsize=(9,9))
df.plot(kind = 'bar', y='avg_rating_numerator', title='Top dog breeds by average rating', ylabel='avg_rating_numerator',figsize=(9,9))
df
###Output
_____no_output_____
###Markdown
Total retweets and favourites over time
###Code
import pandas as pd
sqlite_select_query = """SELECT date, SUM(retweet_count) AS sum_retweet_count, SUM(favorite_count) AS sum_favorite_count
from df_twt_archive_master WHERE breed_predicted == True
GROUP BY date ORDER BY date ASC"""
df = pd.read_sql_query(sqlite_select_query, conn)
df = df.set_index('date')
df.plot(kind = 'line', y=['sum_retweet_count', 'sum_favorite_count'], title='Total retweets and favourites over time', ylabel='count',figsize=(15,5))
df
###Output
_____no_output_____
|
examples/_legacy/0-importing-data.ipynb
|
###Markdown
0. DATA IMPORT & CLEANING___
###Code
import pandas as pd
import matplotlib.pyplot as plt
%load_ext autoreload
%autoreload 2
%matplotlib inline
###Output
_____no_output_____
###Markdown
I. Loading Romina's (ARG) data
###Code
from spectrai.datasets.astorga_arg import (load_spectra, load_measurements, load_data)
df = load_spectra()
df.reset_index().plot(x='wavenumber', y=df.columns[1:], legend=False, figsize=(15, 8)).invert_xaxis()
from spectrai.datasets.astorga_arg import (load_spectra, load_measurements, load_data)
###Output
_____no_output_____
###Markdown
I.1 Spectra
###Code
# Load
df = load_spectra()
df.reset_index().plot(x='wavenumber', y=df.columns[1:], legend=False, figsize=(15, 8)).invert_xaxis()
df.head()
###Output
_____no_output_____
###Markdown
I.2 Measurements
###Code
load_measurements().head(20)
###Output
_____no_output_____
###Markdown
I.3 All data bundled as NumPy arrays (ML/DL ready)
###Code
X, X_names, y, y_names, instances_id = load_data()
print('X shape: ', X.shape)
print(y_names)
print(instances_id)
X_names
X.shape
y_names
instances_id
###Output
_____no_output_____
###Markdown
II. Loading Petra's (VNM) data
###Code
from spectrai.datasets.schmitter_vnm import (load_spectra, load_spectra_rep, load_measurements, load_data)
###Output
_____no_output_____
###Markdown
II.1 Spectra
###Code
df = load_spectra()
df.reset_index().plot(x='wavenumber', y=df.columns[1:], legend=False, figsize=(15, 8)).invert_xaxis()
###Output
_____no_output_____
###Markdown
II.2 Spectra with replicates
###Code
df_petra_rep = load_spectra()
df_petra_rep.head()
###Output
_____no_output_____
###Markdown
* **Prepare lookup tables for group of replicates**
###Code
VIETNAM_LOOKUP_URL = os.path.join(DATA_PATH, 'vnm-petra', 'vietnam-petra', 'Overview list.xls')
df_lookup = pd.read_excel(VIETNAM_LOOKUP_URL, sheet_name='Petra 1.1', usecols=[0,2,3], names=['group_id', 'spectra_id', 'mir_label'])
df_lookup.fillna(method='ffill', inplace=True)
#df_lookup = df_lookup.astype(int)
df_lookup['spectra_id'] = df_lookup['spectra_id'].astype(int)
df_lookup.head()
lookup_tot_spectra_id = {}
for name, group in df_lookup.groupby('mir_label'):
lookup_tot_spectra_id[name] = group['spectra_id'].values
lookup_tot_spectra_id
TOTAL_LABEL = 'F4 1.1'
df_group = df_petra_rep[lookup_tot_spectra_id[TOTAL_LABEL]]
df_group = df_group.apply(lambda x: x - df_group.mean(axis=1))
df_group.plot().invert_xaxis()
all_values = df_group.values.flatten()
with plt.style.context(('ggplot')):
fig, ax = plt.subplots(figsize=(16, 5))
ax.hist(all_values, bins=100)
# Is labeled data the mean of replicates?
DATA_PATH = os.path.join('..', 'data')
VIETNAM_PATH = os.path.join('vnm-petra', 'mir-models')
DATA_URL = os.path.join(DATA_PATH, VIETNAM_PATH, '*.*')
VIETNAM_MEAS_URL = os.path.join(DATA_PATH, 'vnm-petra', 'mir-models', '20090215-soil-database-mirs.xls')
X, X_names, y, y_names, instances_id, lookup_tot_mir_label = spa.load_data_petra(DATA_URL, VIETNAM_MEAS_URL)
lookup_tot_mir_label
# Replicates for TOTAL_LABEL
df_rep_vs_sel = df_petra_rep[lookup_tot_spectra_id[TOTAL_LABEL]]
df_rep_vs_sel.head()
def normalize(X):
return (X - X.min()) / (X.max() - X.min())
normalize(X[row_spectra,:])
row_spectra = list(instances_id).index(lookup_tot_mir_label[TOTAL_LABEL])
df_rep_vs_sel['selected'] = normalize(X[row_spectra,:]).tolist()
df_rep_vs_sel
df_rep_vs_sel.reset_index().plot(x='wavenumber', y=df_rep_vs_sel.columns[0:], legend=True, figsize=(15, 8)).invert_xaxis()
###Output
_____no_output_____
###Markdown
II.3 Measurements
###Code
load_measurements()
###Output
_____no_output_____
###Markdown
II.4 All data bundled as NumPy arrays (ML/DL ready)
###Code
X, X_names, y, y_names, instances_id, lookup = load_data()
print('X shape: ', X.shape)
print(lookup)
print(y_names)
print(instances_id)
###Output
X shape: (130, 1763)
{'F4 1.1': 'Av001', 'F4 1.3': 'Av003', 'F4 1.5': 'Av005', 'F4 2.3': 'Av008', 'F4 3.2': 'Av013', 'F4 3.3': 'Av014', 'F4 4.1': 'Av018', 'F4 4.3': 'Av020', 'F4 4.6': 'Av023', 'F4 5.3': 'Av026', 'F4 5.4': 'Av027', 'F4 6.3': 'Av032', 'F4 7.1': 'Av036', 'F4 7.3': 'Av038', 'F4 7.5': 'Av040', 'NF4 1.1': 'Av041', 'NF4 1.2': 'Av042', 'NF4 2.1': 'Av043', 'NF4 2.3': 'Av045', 'NF4 2.6': 'Av048', 'NF4 3.3a': 'Av051', 'NF4 4.3': 'Av057', 'NF4 5.1': 'Av061', 'NF4 5.3': 'Av062', 'NF4 5.4': 'Av063', 'NF4 5.5': 'Av064', 'F5 1.1': 'Av065', 'F5 1.3': 'Av067', 'F5 1.5': 'Av069', 'F5 2.3': 'Av072', 'F5 2.8': 'Av077', 'F5 3.3': 'Av080', 'F5 3.6': 'Av083', 'F5 4.3': 'Av086', 'F5 5.3': 'Av092', 'F5 6.3': 'Av098', 'F5 7.1': 'Av100', 'F5 7.3': 'Av102', 'F5 7.6': 'Av105', 'NF5 1.1': 'Av106', 'NF5 1.3': 'Av108', 'NF5 1.5': 'Av110', 'NF5 2.3': 'Av113', 'NF5 3.3': 'Av121', 'NF5 4.3': 'Av127', 'NF5 5.3': 'Av133', 'NF5 6.3': 'Av139', 'NF5 7.1': 'Av141', 'NF5 7.3': 'Av143', 'NF5 7.6': 'Av146', '2F6 1.1': 'Av147', '2F6 1.3': 'Av149', '2F6 1.6': 'Av152', '2F6 2.3': 'Av155', '2F6 3.3': 'Av161', '2F6 4.3': 'Av165', '2F6 5.1': 'Av169', '2F6 5.3': 'Av171', '2F6 5.6': 'Av174', '2NF6 1.1': 'Av175', '2NF6 1.3': 'Av177', '2NF6 1.6': 'Av180', '2NF6 2.3': 'Av183', '2NF6 3.3': 'Av189', '2NF6 4.3A': 'Av195', '2NF6 5.1': 'Av203', '2NF6 5.3': 'Av205', '2NF6 5.6': 'Av208', 'F2 1.1': 'Av209', 'F2 1.3': 'Av211', 'F2 1.6': 'Av214', 'F2 2.3': 'Av217', 'F2 3.3A': 'Av225', 'F2 3.3B': 'Av226', 'F2 4.3': 'Av235', 'F2 5.1': 'Av239', 'F2 5.4': 'Av242', 'F2 5.6': 'Av244', 'F2 5.8': 'Av246', 'NF2 1.1': 'Av247', 'NF2 1.3': 'Av249', 'NF2 1.6': 'Av252', 'NF2 2.3': 'Av255', 'NF2 3.3': 'Av261', 'NF2 4.1': 'Av265', 'NF2 4.3': 'Av267', 'NF2 4.4': 'Av268', 'NF2 4.8': 'Av272', 'C0': 'Av365', 'C1': 'Av366', 'C3': 'Av367', 'C5': 'Av368', 'E0': 'Av369', 'E1': 'Av370', 'E2': 'Av371', 'E3': 'Av372', 'E4A': 'Av373', 'E4B': 'Av374', 'E5': 'Av375', 'E6': 'Av376', 'E7': 'Av377', 'Sed7116F1': 'Av378', 'Sed7116F2': 'Av379', 'Sed7116NF4': 'Av380', 'F1 1.1A': 'F1 1.1A', 'F1 1.1B': 'F1 1.1B', 'F1 1.3A': 'F1 1.3A', 'F1 1.3B': 'F1 1.3B', 'F1 1.6A': 'F1 1.6A', 'F1 1.6B': 'F1 1.6B', 'F1 2.3': 'F1 2.3', 'F1 3.3': 'F1 3.3', 'F1 4.1A': 'F1 4.1A', 'F1 4.1B': 'F1 4.1B', 'F1 4.4A': 'F1 4.4A', 'F1 4.4B': 'F1 4.4B', 'F1 4.6A': 'F1 4.6A', 'F1 4.8A': 'F1 4.8A', 'NF1 1.1': 'NF1 1.1', 'NF1 1.3': 'NF1 1.3', 'NF1 1.6': 'NF1 1.6', 'NF1 2.3': 'NF1 2.3', 'NF1 2.6': 'NF1 2.6', 'NF1 3.3': 'NF1 3.3', 'NF1 4.1A': 'NF1 4.1A', 'NF1 4.1B': 'NF1 4.1B', 'NF1 4.4A': 'NF1 4.4A', 'NF1 4.4B': 'NF1 4.4B', 'NF1 4.8A': 'NF1 4.8A', 'NF1 4.8B': 'NF1 4.8B'}
['TC' 'TOC' 'TIC' 'TN' 'CEC' 'K' 'FCAVER' 'FCIAVER' 'FSAAVER']
['Av001' 'Av003' 'Av005' 'Av008' 'Av013' 'Av014' 'Av018' 'Av020' 'Av023'
'Av026' 'Av027' 'Av032' 'Av036' 'Av038' 'Av040' 'Av041' 'Av042' 'Av043'
'Av045' 'Av048' 'Av051' 'Av057' 'Av061' 'Av062' 'Av063' 'Av064' 'Av065'
'Av067' 'Av069' 'Av072' 'Av077' 'Av080' 'Av083' 'Av086' 'Av092' 'Av098'
'Av100' 'Av102' 'Av105' 'Av106' 'Av108' 'Av110' 'Av113' 'Av121' 'Av127'
'Av133' 'Av139' 'Av141' 'Av143' 'Av146' 'Av147' 'Av149' 'Av152' 'Av155'
'Av161' 'Av165' 'Av169' 'Av171' 'Av174' 'Av175' 'Av177' 'Av180' 'Av183'
'Av189' 'Av195' 'Av203' 'Av205' 'Av208' 'Av209' 'Av211' 'Av214' 'Av217'
'Av225' 'Av226' 'Av235' 'Av239' 'Av242' 'Av244' 'Av246' 'Av247' 'Av249'
'Av252' 'Av255' 'Av261' 'Av265' 'Av267' 'Av268' 'Av272' 'Av365' 'Av366'
'Av367' 'Av368' 'Av369' 'Av370' 'Av371' 'Av372' 'Av373' 'Av374' 'Av375'
'Av376' 'Av377' 'Av378' 'Av379' 'Av380' 'F1 1.1A' 'F1 1.1B' 'F1 1.3A'
'F1 1.3B' 'F1 1.6A' 'F1 1.6B' 'F1 2.3' 'F1 3.3' 'F1 4.1A' 'F1 4.1B'
'F1 4.4A' 'F1 4.4B' 'F1 4.6A' 'F1 4.8A' 'NF1 1.1' 'NF1 1.3' 'NF1 1.6'
'NF1 2.3' 'NF1 2.6' 'NF1 3.3' 'NF1 4.1A' 'NF1 4.1B' 'NF1 4.4A' 'NF1 4.4B'
'NF1 4.8A' 'NF1 4.8B']
###Markdown
III. Loading KSSL data Note: To read Bruker optics spectra in `*.0` format, two python packages are available:1. https://www.bruker.com/products/infrared-near-infrared-and-raman-spectroscopy/ft-ir-routine-spectrometers/hts-xt.html2. https://stuart-cls.github.io/python-opusfc-dist/The second one is used.
###Code
from spectrai.datasets.kssl import (load_data)
###Output
_____no_output_____
###Markdown
III.1 Spectra
###Code
X, X_names, y, y_names, instances_id = load_data()
X.shape
X_names
y.shape
y
instances_id
###Output
_____no_output_____
###Markdown
II. Loading kssl data
###Code
from spectrai.datasets.kssl import (load_fact_tbl, load_analytes, load_taxonomy, load_spectra)
load_fact_tbl().head()
load_analytes().head()
load_taxonomy().head()
load_spectra().head()
###Output
_____no_output_____
|
clrp-gru-withattention.ipynb
|
###Markdown
Look at the Featureshttps://www.kaggle.com/donmarch14/commonlit-detailed-guide-to-learn-nlp
###Code
import tensorflow as tf
import pandas as pd
import numpy as np
import seaborn as sns
import matplotlib.pyplot as plt
from sklearn.linear_model import Ridge, LinearRegression, LogisticRegression
import xgboost as xgb
from sklearn.ensemble import RandomForestRegressor
from sklearn.feature_extraction.text import CountVectorizer
from sklearn.feature_extraction.text import TfidfVectorizer
from sklearn.model_selection import train_test_split
from sklearn.metrics import mean_squared_error
from sklearn.pipeline import make_pipeline
from wordcloud import WordCloud
from collections import Counter
import os
import numpy as np
import re
import string
import nltk
nltk.download('punkt')
from nltk.tokenize import word_tokenize
from nltk.corpus import stopwords
nltk.download('stopwords')
from nltk.stem.porter import PorterStemmer
from nltk.stem import WordNetLemmatizer
from nltk.util import ngrams
import html
import unicodedata
stop_words = stopwords.words('english')
%config InlineBackend.figure_format = 'retina'
import os
import string
import numpy as np
import pandas as pd
from string import digits
import matplotlib.pyplot as plt
%matplotlib inline
import re
import logging
import tensorflow as tf
# tf.enable_eager_execution()
os.environ['TF_CPP_MIN_LOG_LEVEL'] = '3'
tf.compat.v1.logging.set_verbosity(tf.compat.v1.logging.ERROR)
logging.getLogger('tensorflow').setLevel(logging.FATAL)
import matplotlib.ticker as ticker
from sklearn.model_selection import train_test_split
import unicodedata
import io
import time
import warnings
import sys
import nltk
from nltk.probability import FreqDist
def wordcloud(text,ngram=1):
wordcloud = WordCloud(width=1400,
height=800,
random_state=2021,
background_color='black',
)
if ngram ==1:
wordc = wordcloud.generate(' '.join(text))
else:
wordc = wordcloud.generate_from_frequencies(text)
plt.figure(figsize=(12,6), facecolor='k')
plt.imshow(wordcloud)
plt.axis('off')
plt.tight_layout(pad=0)
def get_n_grans_count(text, n_grams, min_freq):
output = {}
tokens = nltk.word_tokenize(text)
#Create the n_gram
if n_grams == 2:
gs = nltk.bigrams(tokens)
elif n_grams == 3:
gs = nltk.trigrams(tokens)
else:
return 'Only 2_grams and 3_grams are supported'
# compute frequency distribution for all the bigrams in the text by threshold with min_freq
fdist = nltk.FreqDist(gs)
for k,v in fdist.items():
if v > min_freq:
index = ' '.join(k)
output[index] = v
return output
def remove_special_chars(text):
re1 = re.compile(r' +')
x1 = text.lower().replace('#39;', "'").replace('amp;', '&').replace('#146;', "'").replace(
'nbsp;', ' ').replace('#36;', '$').replace('\\n', "\n").replace('quot;', "'").replace(
'<br />', "\n").replace('\\"', '"').replace('<unk>', 'u_n').replace(' @.@ ', '.').replace(
' @-@ ', '-').replace('\\', ' \\ ')
return re1.sub(' ', html.unescape(x1))
def remove_non_ascii(text):
"""Remove non-ASCII characters from list of tokenized words"""
return unicodedata.normalize('NFKD', text).encode('ascii', 'ignore').decode('utf-8', 'ignore')
def to_lowercase(text):
return text.lower()
def remove_punctuation(text):
"""Remove punctuation from list of tokenized words"""
translator = str.maketrans('', '', string.punctuation)
return text.translate(translator)
def replace_numbers(text):
"""Replace all interger occurrences in list of tokenized words with textual representation"""
return re.sub(r'\d+', '', text)
def remove_whitespaces(text):
return text.strip()
def remove_stopwords(words, stop_words):
"""
:param words:
:type words:
:param stop_words: from sklearn.feature_extraction.stop_words import ENGLISH_STOP_WORDS
or
from spacy.lang.en.stop_words import STOP_WORDS
:type stop_words:
:return:
:rtype:
"""
return [word for word in words if word not in stop_words]
def stem_words(words):
"""Stem words in text"""
stemmer = PorterStemmer()
return [stemmer.stem(word) for word in words]
def lemmatize_words(words):
"""Lemmatize words in text, and by defult lemmatize nouns"""
lemmatizer = WordNetLemmatizer()
return [lemmatizer.lemmatize(word) for word in words]
def lemmatize_verbs(words):
"""Lemmatize verbs in text"""
lemmatizer = WordNetLemmatizer()
return ' '.join([lemmatizer.lemmatize(word, pos='v') for word in words])
def text2words(text):
return word_tokenize(text)
def normalize_text( text):
text = remove_special_chars(text)
text = remove_non_ascii(text)
text = remove_punctuation(text)
text = to_lowercase(text)
text = replace_numbers(text)
words = text2words(text)
words = remove_stopwords(words, stop_words)
#words = stem_words(words)# Either stem or lemmatize
words = lemmatize_words(words)
words = lemmatize_verbs(words)
return ''.join(words)
df_raw_train = pd.read_csv("/kaggle/input/commonlitreadabilityprize/train.csv")
df_raw_train.head()
input_train, input_val, target_train, target_val = train_test_split(df_raw_train['excerpt'], df_raw_train['target'], test_size=0.2)
df_raw_train['excerpt'] = [normalize_text(sent) for sent in df_raw_train['excerpt']]
target_val
df_train = df_raw_train[['excerpt','target']]
def tokenize(lang):
lang_tokenizer = tf.keras.preprocessing.text.Tokenizer(filters='',oov_token='<oov>')
lang_tokenizer.fit_on_texts(lang)
tensor = lang_tokenizer.texts_to_sequences(lang)
tensor = tf.keras.preprocessing.sequence.pad_sequences(tensor,padding='post')
return tensor, lang_tokenizer
def load_dataset():
#targ_val, inp_lang = df_train['target'],df_train['excerpt']
targ_val = df_train['target']
inp_lang = get_lang()
input_tensor, inp_lang_tokenizer = tokenize(inp_lang)
return input_tensor, inp_lang_tokenizer
def get_lang():
en=[]
for i in df_train['excerpt']:
en_1 = [w for w in i.split(' ')]
en.append(en_1)
return en
input_tensor, inp_lang = load_dataset()
target_tensor = df_train['target']
input_tensor_train, input_tensor_val, target_tensor_train, target_tensor_val = train_test_split(input_tensor, target_tensor, test_size=0.2)
print(len(input_tensor_train), len(target_tensor_train), len(input_tensor_val), len(target_tensor_val))
BUFFER_SIZE = len(input_tensor_train)
BATCH_SIZE = 64
steps_per_epoch = len(input_tensor_train)//BATCH_SIZE
embedding_dim = 128
units = 256
vocab_inp_size = len(inp_lang.word_index)+1
tar_int_size = 64
dataset = tf.data.Dataset.from_tensor_slices((input_tensor_train, target_tensor_train)).shuffle(BUFFER_SIZE)
dataset = dataset.batch(BATCH_SIZE, drop_remainder=True)
class Encoder(tf.keras.Model):
def __init__(self, vocab_size, embedding_dim, enc_units, batch_sz):
super(Encoder, self).__init__()
self.batch_sz = batch_sz
self.enc_units = enc_units
self.embedding = tf.keras.layers.Embedding(vocab_size, embedding_dim)
self.gru = tf.keras.layers.GRU(self.enc_units,
return_sequences=True,
return_state=True,
recurrent_initializer='glorot_uniform')
def call(self, x, hidden):
x = self.embedding(x)
output, state = self.gru(x, initial_state = hidden)
return output, state
def initialize_hidden_state(self):
return tf.zeros((self.batch_sz, self.enc_units))
encoder = Encoder(vocab_inp_size, embedding_dim, units, BATCH_SIZE)
class BahdanauAttention(tf.keras.layers.Layer):
def __init__(self, units):
super(BahdanauAttention, self).__init__()
self.W1 = tf.keras.layers.Dense(units)
self.W2 = tf.keras.layers.Dense(units)
self.V = tf.keras.layers.Dense(1)
def call(self, query, values):
hidden_with_time_axis = tf.expand_dims(query, 1)
score = self.V(tf.nn.tanh(
self.W1(values) + self.W2(hidden_with_time_axis)))
attention_weights = tf.nn.softmax(score, axis=1)
context_vector = attention_weights * values
context_vector = tf.reduce_sum(context_vector, axis=1)
return context_vector, attention_weights
class Decoder(tf.keras.Model):
def __init__(self, tar_int_size, embedding_dim, dec_units, batch_sz):
super(Decoder, self).__init__()
self.batch_sz = batch_sz
self.dec_units = dec_units
self.fc = tf.keras.layers.Dense(tar_int_size)
self.attention = BahdanauAttention(self.dec_units)
self.fc2 = tf.keras.layers.Dense(1)
def call(self, x, hidden, enc_output):
context_vector, attention_weights = self.attention(hidden, enc_output)
x = tf.concat([context_vector, x], axis=-1)
# print(x.shape)
# output, state = self.gru(x)
# output = tf.reshape(output, (-1, output.shape[2]))
x = self.fc(x)
x = self.fc2(x)
return x, attention_weights
decoder = Decoder(tar_int_size, embedding_dim, units, BATCH_SIZE)
optimizer = tf.keras.optimizers.Adam()
loss_object = tf.keras.losses.MeanSquaredError()
def loss_function(real, pred):
print(real,pred)
loss_ = loss_object(real, pred)
return tf.reduce_mean(loss_)
checkpoint_dir = './training_checkpoints'
checkpoint_prefix = os.path.join(checkpoint_dir, "ckpt")
checkpoint = tf.train.Checkpoint(optimizer=optimizer,
encoder=encoder,
decoder=decoder)
@tf.function
def train_step(inp, targ, enc_hidden):
loss = 0
with tf.GradientTape() as tape:
enc_output, enc_hidden = encoder(inp, enc_hidden)
dec_hidden = enc_hidden
# print(dec_hidden.shape)
#dec_input = tf.expand_dims([targ_lang.word_index['<start>']] * BATCH_SIZE, 1)
dec_input = enc_hidden
# Teacher forcing
#for t in range(1, targ.shape[1]):
prediction,attention_weights = decoder(dec_input, dec_hidden, enc_output)
loss = loss_function(targ, prediction)
#dec_input = tf.expand_dims(targ[:, t], 1)
batch_loss = loss
variables = encoder.trainable_variables + decoder.trainable_variables
gradients = tape.gradient(loss, variables)
optimizer.apply_gradients(zip(gradients, variables))
return batch_loss
def train_model(epochs):
EPOCHS = epochs
for epoch in range(EPOCHS):
start = time.time()
enc_hidden = encoder.initialize_hidden_state()
total_loss = 0
for (batch, (inp, targ)) in enumerate(dataset.take(steps_per_epoch)):
batch_loss = train_step(inp, targ, enc_hidden)
total_loss += batch_loss
if batch % 100 == 0:
print('Epoch {} Batch {} Loss {:.4f}'.format(epoch + 1,
batch,
batch_loss.numpy()))
if (epoch + 1) % 2 == 0:
checkpoint.save(file_prefix = checkpoint_prefix)
print('Epoch {} Loss {:.4f}'.format(epoch + 1,
total_loss / steps_per_epoch))
print('Time taken for 1 epoch {} sec\n'.format(time.time() - start))
# train_model(5)
def max_length(tensor):
return max(len(t) for t in tensor)
max_length_inp = max_length(input_tensor)
checkpoint.restore(tf.train.latest_checkpoint(checkpoint_dir))
words = []
for key in inp_lang.word_index.items():
words.append(key[0])
def evaluate(sentence):
sentence = normalize_text(sentence)
inputs = [inp_lang.word_index[i] if i in words else 1 for i in sentence.split(' ')]
inputs = tf.keras.preprocessing.sequence.pad_sequences([inputs],maxlen=max_length_inp,padding='post')
inputs = tf.convert_to_tensor(inputs)
hidden = [tf.zeros((1, units))]
enc_out, enc_hidden = encoder(inputs, hidden)
dec_hidden = enc_hidden
dec_input = enc_hidden
pred, attention_weights = decoder(dec_input,dec_hidden,enc_out)
# print(pred.shape)
return pred.numpy()[0][0]
evaluate(u'politicians do not have permission to do what needs to be done.')
validation_df = pd.DataFrame(input_val,columns=["excerpt"])
validation_df['target']= target_val
validation_df['prediction'] = validation_df['excerpt'].apply(lambda value : evaluate(value))
validation_df['deviation'] = validation_df['target'] - validation_df['prediction']
validation_df.sort_values('deviation',ascending =False)
print('RMSE',np.sqrt(np.sum(np.square(validation_df['deviation']))/validation_df.shape[0]))
test_data = pd.read_csv('../input/commonlitreadabilityprize/test.csv')
submission = pd.read_csv("../input/commonlitreadabilityprize/sample_submission.csv")
test_data['predict'] = test_data['excerpt'].apply(lambda value : evaluate(value))
submission = pd.DataFrame()
submission['id'] = test_data['id']
submission['target'] = test_data['predict']
submission.to_csv("submission.csv", index=False)
submission
###Output
_____no_output_____
|
Python-and-Spark-for-Big-Data-master/Spark_for_Machine_Learning/Logistic_Regression/.ipynb_checkpoints/Log_regression_Code_Along-checkpoint.ipynb
|
###Markdown
Logistic Regression Code Along This is a code along of the famous titanic dataset, its always nice to start off with this dataset because it is an example you will find across pretty much every data analysis language.
###Code
# Note that usually all imports would occur at the top and
# most of this would be in an object this layout if for learning purposes only
# Logistic Regression Example
from pyspark.sql import SparkSession
spark = SparkSession.builder.appName('logreg').getOrCreate()
# Load training data
data = spark.read.csv('titanic.csv',inferSchema=True,header=True)
# Print the Schema of the DataFrame
data.printSchema()
data.show()
data.describe().show()
# Drop missing data
clean_data = data.na.drop()
from pyspark.ml.linalg import Vectors
from pyspark.ml.feature import VectorAssembler,StringIndexer,OneHotEncoder,VectorIndexer
###Output
_____no_output_____
###Markdown
Working with Categorical ColumnsLet's break this down into multiple steps to make it all clear.
###Code
data.columns
train_data,test_data = clean_data.randomSplit([0.7,0.3])
gender_indexer = StringIndexer(inputCol="Sex", outputCol="SexIndex")
gender_encoder = OneHotEncoder(inputCol="SexIndex", outputCol="SexVec")
embark_indexer = StringIndexer(inputCol="Embarked", outputCol="EmbarkIndex")
embark_encoder = OneHotEncoder(inputCol="EmbarkIndex", outputCol="EmbarkVec")
###Output
_____no_output_____
###Markdown
Pipelines Let's see an example of how to use pipelines (we'll get a lot more practice with these later!)
###Code
from pyspark.ml.linalg import Vectors
from pyspark.ml.feature import VectorAssembler
assembler = VectorAssembler(
inputCols=["Pclass", "SexVec", "Age","SibSp","Parch","Fare","EmbarkVec"],
outputCol="features")
from pyspark.ml.classification import LogisticRegression
logreg = LogisticRegression(featuresCol='features',labelCol='Survived')
from pyspark.ml import Pipeline
train_data.printSchema()
pipeline = Pipeline(stages=[gender_indexer,embark_indexer,
gender_encoder,embark_encoder,assembler,logreg])
model = pipeline.fit(train_data)
results = model.transform(test_data)
results.printSchema()
from pyspark.ml.evaluation import BinaryClassificationEvaluator, MulticlassClassificationEvaluator
evaluator = BinaryClassificationEvaluator(rawPredictionCol='prediction', labelCol='Survived')
AUC = evaluator.evaluate(results)
AUC
evaluator = MulticlassClassificationEvaluator(predictionCol='prediction', labelCol='Survived',
metricName='accuracy')
acc = evaluator.evaluate(results)
acc
###Output
_____no_output_____
|
4-time-series/w3-ex-lstm.ipynb
|
###Markdown
###Code
#@title Licensed under the Apache License, Version 2.0 (the "License");
# you may not use this file except in compliance with the License.
# You may obtain a copy of the License at
#
# https://www.apache.org/licenses/LICENSE-2.0
#
# Unless required by applicable law or agreed to in writing, software
# distributed under the License is distributed on an "AS IS" BASIS,
# WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.
# See the License for the specific language governing permissions and
# limitations under the License.
# !pip install tf-nightly-2.0-preview
import tensorflow as tf
import numpy as np
import matplotlib.pyplot as plt
print(tf.__version__)
def plot_series(time, series, format="-", start=0, end=None):
plt.plot(time[start:end], series[start:end], format)
plt.xlabel("Time")
plt.ylabel("Value")
plt.grid(False)
def trend(time, slope=0):
return slope * time
def seasonal_pattern(season_time):
"""Just an arbitrary pattern, you can change it if you wish"""
return np.where(season_time < 0.1,
np.cos(season_time * 6 * np.pi),
2 / np.exp(9 * season_time))
def seasonality(time, period, amplitude=1, phase=0):
"""Repeats the same pattern at each period"""
season_time = ((time + phase) % period) / period
return amplitude * seasonal_pattern(season_time)
def noise(time, noise_level=1, seed=None):
rnd = np.random.RandomState(seed)
return rnd.randn(len(time)) * noise_level
time = np.arange(10 * 365 + 1, dtype="float32")
baseline = 10
series = trend(time, 0.1)
baseline = 10
amplitude = 40
slope = 0.005
noise_level = 3
# Create the series
series = baseline + trend(time, slope) + seasonality(time, period=365, amplitude=amplitude)
# Update with noise
series += noise(time, noise_level, seed=51)
split_time = 3000
time_train = time[:split_time]
x_train = series[:split_time]
time_valid = time[split_time:]
x_valid = series[split_time:]
window_size = 20
batch_size = 32
shuffle_buffer_size = 1000
plot_series(time, series)
def windowed_dataset(series, window_size, batch_size, shuffle_buffer):
dataset = tf.data.Dataset.from_tensor_slices(series)
dataset = dataset.window(window_size + 1, shift=1, drop_remainder=True)
dataset = dataset.flat_map(lambda window: window.batch(window_size + 1))
dataset = dataset.shuffle(shuffle_buffer).map(lambda window: (window[:-1], window[-1]))
dataset = dataset.batch(batch_size).prefetch(1)
return dataset
tf.keras.backend.clear_session()
tf.random.set_seed(51)
np.random.seed(51)
tf.keras.backend.clear_session()
dataset = windowed_dataset(x_train, window_size, batch_size, shuffle_buffer_size)
model = tf.keras.models.Sequential([
tf.keras.layers.Lambda(lambda x: tf.expand_dims(x, axis=-1),
input_shape=[None]),
tf.keras.layers.Bidirectional(tf.keras.layers.LSTM(32, return_sequences=True)),
tf.keras.layers.Bidirectional(tf.keras.layers.LSTM(32)),
tf.keras.layers.Dense(1),
tf.keras.layers.Lambda(lambda x: x * 10.0)
])
lr_schedule = tf.keras.callbacks.LearningRateScheduler(
lambda epoch: 1e-8 * 10**(epoch / 20))
optimizer = tf.keras.optimizers.SGD(lr=1e-8, momentum=0.9)
model.compile(loss=tf.keras.losses.Huber(),
optimizer=optimizer,
metrics=["mae"])
history = model.fit(dataset, epochs=100, callbacks=[lr_schedule])
plt.semilogx(history.history["lr"], history.history["loss"])
plt.axis([1e-8, 1e-4, 0, 30])
tf.keras.backend.clear_session()
tf.random.set_seed(51)
np.random.seed(51)
tf.keras.backend.clear_session()
dataset = windowed_dataset(x_train, window_size, batch_size, shuffle_buffer_size)
model = tf.keras.models.Sequential([
tf.keras.layers.Lambda(lambda x: tf.expand_dims(x, axis=-1),
input_shape=[None]),
tf.keras.layers.Bidirectional(tf.keras.layers.LSTM(32, return_sequences=True)),
tf.keras.layers.Bidirectional(tf.keras.layers.LSTM(32)),
tf.keras.layers.Dense(1),
tf.keras.layers.Lambda(lambda x: x * 100.0)
])
model.compile(loss="mse", optimizer=tf.keras.optimizers.SGD(lr=1e-5, momentum=0.9),metrics=["mae"])
history = model.fit(dataset,epochs=500,verbose=1)
forecast = []
results = []
for time in range(len(series) - window_size):
forecast.append(model.predict(series[time:time + window_size][np.newaxis]))
forecast = forecast[split_time-window_size:]
results = np.array(forecast)[:, 0, 0]
plt.figure(figsize=(10, 6))
plot_series(time_valid, x_valid)
plot_series(time_valid, results)
tf.keras.metrics.mean_absolute_error(x_valid, results).numpy()
import matplotlib.image as mpimg
import matplotlib.pyplot as plt
#-----------------------------------------------------------
# Retrieve a list of list results on training and test data
# sets for each training epoch
#-----------------------------------------------------------
mae=history.history['mae']
loss=history.history['loss']
epochs=range(len(loss)) # Get number of epochs
#------------------------------------------------
# Plot MAE and Loss
#------------------------------------------------
plt.plot(epochs, mae, 'r')
plt.plot(epochs, loss, 'b')
plt.title('MAE and Loss')
plt.xlabel("Epochs")
plt.ylabel("Accuracy")
plt.legend(["MAE", "Loss"])
plt.figure()
epochs_zoom = epochs[200:]
mae_zoom = mae[200:]
loss_zoom = loss[200:]
#------------------------------------------------
# Plot Zoomed MAE and Loss
#------------------------------------------------
plt.plot(epochs_zoom, mae_zoom, 'r')
plt.plot(epochs_zoom, loss_zoom, 'b')
plt.title('MAE and Loss')
plt.xlabel("Epochs")
plt.ylabel("Accuracy")
plt.legend(["MAE", "Loss"])
plt.figure()
###Output
_____no_output_____
|
introduction_to_data_visualization_with_matplotlib/4_sharing_visualizations_with_others.ipynb
|
###Markdown
Switching between stylesSelecting a style to use affects all of the visualizations that are created after this style is selected.Here, you will practice plotting data in two different styles. The data you will use is the same weather data we used in the first lesson: you will have available to you the DataFrame `seattle_weather` and the DataFrame `austin_weather`, both with records of the average temperature in every month.Instructions- Select the `'ggplot'` style, create a new Figure called `fig`, and a new Axes object called `ax` with `plt.subplots`.- Select the `'Solarize_Light2'` style, create a new Figure called `fig`, and a new Axes object called `ax` with `plt.subplots`.
###Code
# Import libraries
import pandas as pd
import matplotlib.pyplot as plt
# Import DataFrames
seattle_weather = pd.read_csv('seattle_weather.csv')
austin_weather = pd.read_csv('austin_weather.csv')
# Use the "ggplot" style and create new Figure/Axes
fig, ax = plt.subplots()
plt.style.use('ggplot')
ax.plot(seattle_weather['MONTH'], seattle_weather['MLY-TAVG-NORMAL'])
plt.show()
# Use the "Solarize_Light2" style and create new Figure/Axes
fig, ax = plt.subplots()
plt.style.use('Solarize_Light2')
ax.plot(austin_weather['MONTH'], austin_weather['MLY-TAVG-NORMAL'])
plt.show()
###Output
_____no_output_____
###Markdown
Saving a file several timesIf you want to share your visualizations with others, you will need to save them into files. Matplotlib provides as way to do that, through the `savefig` method of the `Figure` object. In this exercise, you will save a figure several times. Each time setting the parameters to something slightly different. We have provided and already created `Figure` object.Instructions- Examine the figure by calling the `plt.show()` function.- Save the figure into the file `my_figure.png`, using the default resolution.- Save the figure into the file `my_figure_300dpi.png` and set the resolution to 300 dpi.
###Code
# Previous code
import pandas as pd
import matplotlib.pyplot as plt
medals = pd.read_csv('medals_by_country_2016.csv', index_col=0)
fig, ax = plt.subplots()
ax.bar(medals.index, medals["Gold"])
plt.xticks(rotation=90)
ax.set_ylabel('Number of medals')
plt.style.use('default')
# Show the figure
plt.show()
# Save as a PNG file
fig.savefig('my_figure.png')
# Save as a PNG file with 300 dpi
fig.savefig('my_figure_300dpi.png', dpi=300)
###Output
_____no_output_____
###Markdown
Save a figure with different sizesBefore saving your visualization, you might want to also set the size that the figure will have on the page. To do so, you can use the `Figure` object's `set_size_inches` method. This method takes a sequence of two values. The first sets the width and the second sets the height of the figure.Here, you will again have a `Figure` object called `fig` already provided (you can run `plt.show` if you want to see its contents). Use the `Figure` methods `set_size_inches` and `savefig` to change its size and save two different versions of this figure.Instructions- Set the figure size as width of 3 inches and height of 5 inches and save it as `'figure_3_5.png'` with default resolution.- Set the figure size to width of 5 inches and height of 3 inches and save it as `'figure_5_3.png'` with default settings.
###Code
# Set figure dimensions and save as a PNG
fig.set_size_inches([3, 5])
fig.savefig('figure_3_5.png')
# Set figure dimensions and save as a PNG
fig.set_size_inches([5, 3])
fig.savefig('figure_5_3.png')
###Output
_____no_output_____
###Markdown
Unique values of a columnOne of the main strengths of Matplotlib is that it can be automated to adapt to the data that it receives as input. For example, if you receive data that has an unknown number of categories, you can still create a bar plot that has bars for each category.In this exercise and the next, you will be visualizing the weight of medalis in the 2016 summer Olympic Games again, from a dataset that has some unknown number of branches of sports in it. This will be loaded into memory as a Pandas `DataFrame` object called `summer_2016_medals`, which has a column called `"Sport"` that tells you to which branch of sport each row corresponds. There is also a `"Weight"` column that tells you the weight of each athlete.In this exercise, we will extract the unique values of the `"Sport"` columnInstructions- Create a variable called `sports_column` that holds the data from the `"Sport"` column of the `DataFrame` object.- Use the `unique` method of this variable to find all the unique different sports that are present in this data, and assign these values into a new variable called `sports`.- Print the `sports` variable to the console.
###Code
# Import DataFrame
summer_2016_medals = pd.read_csv('summer_2016_medals.csv')
# Extract the "Sport" column
sports_column = summer_2016_medals["Sport"]
# Find the unique values of the "Sport" column
sports = sports_column.unique()
# Print out the unique sports values
sports
###Output
_____no_output_____
###Markdown
Automate your visualizationOne of the main strengths of Matplotlib is that it can be automated to adapt to the data that it receives as input. For example, if you receive data that has an unknown number of categories, you can still create a bar plot that has bars for each category.This is what you will do in this exercise. You will be visualizing data about medal winners in the 2016 summer Olympic Games again, but this time you will have a dataset that has some unknown number of branches of sports in it. This will be loaded into memory as a Pandas `DataFrame` object called `summer_2016_medals`, which has a column called `"Sport"` that tells you to which branch of sport each row corresponds. There is also a `"Weight"` column that tells you the weight of each athlete.Instructions- Iterate over the values of `sports` setting `sport` as your loop variable.- In each iteration, extract the rows where the `"Sport"` column is equal to `sport`.- Add a bar to the provided `ax` object, labeled with the sport name, with the mean of the `"Weight"` column as its height, and the standard deviation as a y-axis error bar.- Save the figure into the file `"sports_weights.png"`.
###Code
fig, ax = plt.subplots()
# Loop over the different sports branches
for sport in sports:
# Extract the rows only for this sport
sport_df = summer_2016_medals[summer_2016_medals['Sport'] == sport]
# Add a bar for the "Weight" mean with std y error bar
ax.bar(sport, sport_df['Weight'].mean(),
yerr=sport_df['Weight'].std())
ax.set_ylabel('Weight')
# ax.set_xticklabels(sports, rotation=90)
plt.xticks(sports, rotation=90)
# Save the figure to file
fig.savefig('sports_weights.png')
###Output
_____no_output_____
|
examen_intro_python.ipynb
|
###Markdown
Examen Intro Python Instrucciones:- **Guardar una copia** de la Colab en su Google Drive al empezar para poder guardar las respuestas. ```File / Archivo -> Save a copy on Drive / Guardar una copia en drive ```- Completen con su nombre completo, apellido y mail. - El tiempo máximo es de una hora. - Al finalizar, **descargar el archivo como .ipynb** poniendo ```File / Archivo -> Download / Descargar -> .ipynb``` y enviarlo por mail a [email protected]. ¡Muchos éxitos!
###Code
from datetime import datetime, timedelta
(datetime.now() - timedelta(hours=3)).strftime("%m/%d/%Y, %H:%M:%S")
nombre = 'Carlos' #@param {type:"string"}
apellido = 'Rocha' #@param {type:"string"}
mail = '[email protected]' #@param {type:"string"}
###Output
_____no_output_____
###Markdown
1.a Usar una función de Python para imprimir el tipo de la variable **x**.El output debería ser```str```
###Code
x = "caracteres"
def tipo(var):
return type(var)
tipo(x)
###Output
_____no_output_____
###Markdown
1.bHacer una lista por comprensión con los números pares del 0 al 10:Output: ```[0, 2, 4, 6, 8]```
###Code
pares = [x*2 for x in range(5)]
pares
###Output
_____no_output_____
###Markdown
1.c Ordenar alfabéticamenteDefinir la función para que el output sea **'azul-blanco-negro-rojo'**
###Code
def ordenar_alfabeticamente(palabras_entre_guiones):
palabras_lista = palabras_entre_guiones.split('-')
palabras_ordenadas = '-'.join(sorted(palabras_lista))
return palabras_ordenadas
ordenar_alfabeticamente('negro-blanco-azul-rojo')
###Output
_____no_output_____
###Markdown
2. Dada la lista **tipos_datos**, escribir un bucle/loop que sume sus números, deteniéndose e imprimiendo el resultado inmediatamente luego de que éste supere 100:
###Code
datos = [2, 28, 5, "foco", 6.2, False, 30.4, 14, 3, 2, 42, 33, "guante", 22]
resultado = 0
for d in datos:
if type(d)== int or type(d)== float:
resultado += d
if resultado > 100:
break
print(resultado)
###Output
132.6
###Markdown
3. AvenidaDefinir una función que reciba un string y devuelva **True** si éste es una dirección de avenida o **False** si no lo es:* Tiene que comenzar con **Av.** o **Avenida*** Luego seguir con un nombre compuesto por **letras**...* ...y finalizar con un **entero** entre 10 y 9999
###Code
def es_avenida(nombre):
comienzo = False
letras = False
entero = False
avenida_lista = nombre.split()
avenida_lista_mayus = [x.upper() for x in avenida_lista]
for i in range(len(avenida_lista_mayus)):
if i == 0 and avenida_lista_mayus[i] == 'AV.' or avenida_lista_mayus[i] == 'AVENIDA':
comienzo = True
if i == 1 and not avenida_lista_mayus[i].isnumeric():
letras = True
if i == len(avenida_lista_mayus) - 1 and avenida_lista_mayus[i].isnumeric():
entero = True
if comienzo and letras and entero:
return True
else:
return False
es_avenida('Avenida sdsd 2010')
###Output
_____no_output_____
###Markdown
4. Comparar tuplas:* Definir una función que reciba dos tuplas de enteros e imprima True si tienen el mismo primer o último elemento.* Si alguno de los grupos además tiene un número impar de elementos, además de True imprimir "IMPAR".* Tener en cuenta que si alguna de las tuplas tiene sólo dos elementos, el output será "Las tuplas necesitan como mínimo tres elementos"
###Code
def tuplas(a, b):
retorna = ''
retorna2 = ''
if len(a)>= 3 and len(b)>= 3:
if (a[0] == b[0] or a[-1] == b[-1]):
retorna = 'True'
else:
retorna = ''
if len(a) % 2:
retorna2 = 'IMPAR'
else:
return 'Las tuplas necesitan como mínimo tres elementos'
retorna = retorna + ' ' + retorna2
return retorna
a = (1,5,6)
b = (5,4,3,6)
tuplas(a, b)
###Output
_____no_output_____
###Markdown
5. ¿Está en la lista?* Definir una función que dado un string y una lista de caracteres, devuelva sólo aquellas **letras** del string que estén también en la lista.* Cada elemento de esta lista debe tener sólo un caracter (ni cero ni más de uno), de lo contrario imprimir "Los elementos de la lista deben ser caracteres individuales"
###Code
def en_lista(cadena, lista):
cadena_lista = list(cadena)
en_lista = [x for x in cadena_lista if x in lista] #set(cadena_lista) and set(lista)
return (''.join(sorted(en_lista)) )
en_lista('abcd', ['a', 'b','c', 'h'])
###Output
_____no_output_____
###Markdown
6. Utilizando la siguiente variable **mochila**...
###Code
mochila = [
{
"objeto": "caramelo",
"cantidad": "10",
"tipo": "dulce de leche",
},
{
"objeto": "llaves",
"cantidad": "3",
"tipo": "blockchain",
},
{
"objeto": "caramelo",
"cantidad": "3",
"tipo": "menta",
},
{
"objeto": "billetera",
"cantidad": "1",
"tipo": "cuero",
},
{
"objeto": "moneda",
"cantidad": "5",
"tipo": "50",
},
{
"objeto": "caramelo",
"cantidad": "12",
"tipo": "nutella",
},
]
###Output
_____no_output_____
###Markdown
6.a...extraer la información correspondiente en las siguientes variables:
###Code
cantidad_nutella = mochila[4]['cantidad']
print('Cantidad_nutella: ', cantidad_nutella)
n_elementos_mochila = sum([len(mochila[x]) for x in range(len(mochila)) ])
print('n_elementos_mochila: ', n_elementos_mochila)
dinero_total = int(mochila[4]['cantidad']) * int(mochila[4]['tipo'])
print('dinero_total: ', dinero_total)
###Output
Cantidad_nutella: 5
n_elementos_mochila: 18
dinero_total: 250
###Markdown
6.bContar la cantidad de caramelos con una sola línea de código:
###Code
c = 0;
for x in range(len(mochila)):
for key, valor in mochila[x].items():
if valor == 'caramelo':
c += int(mochila[x]['cantidad'])
print(c)
###Output
25
###Markdown
7.a* Crear una clase **Animal** que reciba como atributo un **nombre**.* Dentro de ella, definir un método por el cual cada vez que se imprima una de sus instancias devuelva que "**nombre** es un animal" (formatear el string como corresponda)
###Code
class Animal():
def __init__(self, nombre):
self.nombre = nombre
def es_animal(self):
print(f'{self.nombre} es un animal')
animal = Animal('gato')
animal.es_animal()
###Output
gato es un animal
###Markdown
7.b* Crear una clase **Ave** que reciba como atributo una **velocidad** y además herede los de **Animal**. * Agregarle un método **volar()** que reciba un string **clima**. Si éste es "soleado" devolver (formateado) que "**nombre** vuela a **velocidad** metros por segundo". En cambio si es "lluvioso", devolver el mismo mensaje pero con la velocidad reducida a la mitad.
###Code
class Ave(Animal):
def __init__(self, nombre, velocidad, clima = ''):
Animal.__init__(self, nombre)
self.velocidad = velocidad
self.clima = clima
def volar(self):
if self.clima == 'soleado':
return print(f'{self.nombre} vuela a {self.velocidad} metros')
elif self.clima == 'lluvioso':
return print(f'{self.nombre} vuela a {self.velocidad/2} metros')
ave = Ave('Gato',50,'lluvioso')
ave.volar()
ave.es_animal()
###Output
Gato es un animal
|
Python-Standard-Library/DataPersistence/dbm.ipynb
|
###Markdown
Creating a New Database
###Code
import dbm
with dbm.open('/tmp/example.db', 'n') as db:
db['key'] = 'value'
db['today'] = 'Sunday'
db['author'] = 'Doug'
import dbm
print(dbm.whichdb('/tmp/example.db'))
###Output
dbm.ndbm
###Markdown
Open the existing Database
###Code
import dbm
with dbm.open('/tmp/example.db', 'r') as db:
print('keys():', db.keys())
for k in db.keys():
print('iterating:', k, db[k])
print('db["author"] =', db['author'])
###Output
keys(): [b'key', b'today', b'author']
iterating: b'key' b'value'
iterating: b'today' b'Sunday'
iterating: b'author' b'Doug'
db["author"] = b'Doug'
###Markdown
Error Cases
###Code
import dbm
with dbm.open('/tmp/example.db', 'w') as db:
try:
db[1] = 'one'
except TypeError as err:
print(err)
import dbm
with dbm.open('/tmp/example.db', 'w') as db:
try:
db['one'] = 1
except TypeError as err:
print(err)
###Output
dbm mappings have byte or string elements only
|
Big-Data-Clusters/CU9/public/content/cert-management/cer002-download-existing-root-ca.ipynb
|
###Markdown
CER002 - Download existing Root CA certificateUse this notebook to download a generated Root CA certificate from acluster that installed one using:- [CER001 - Generate a Root CA certificate](../cert-management/cer001-create-root-ca.ipynb)And then to upload the generated Root CA to another cluster use:- [CER003 - Upload existing Root CA certificate](../cert-management/cer003-upload-existing-root-ca.ipynb)If needed, use these notebooks to view and set the Kubernetesconfiguration context appropriately to enable downloading the Root CAfrom a Big Data Cluster in one Kubernetes cluster, and to upload it to aBig Data Cluster in another Kubernetes cluster.- [TSG010 - Get configuration contexts](../monitor-k8s/tsg010-get-kubernetes-contexts.ipynb)- [SOP011 - Set kubernetes configuration context](../common/sop011-set-kubernetes-context.ipynb) Steps Parameters
###Code
local_folder_name = "mssql-cluster-root-ca"
test_cert_store_root = "/var/opt/secrets/test-certificates"
###Output
_____no_output_____
###Markdown
Common functionsDefine helper functions used in this notebook.
###Code
# Define `run` function for transient fault handling, suggestions on error, and scrolling updates on Windows
import sys
import os
import re
import platform
import shlex
import shutil
import datetime
from subprocess import Popen, PIPE
from IPython.display import Markdown
retry_hints = {} # Output in stderr known to be transient, therefore automatically retry
error_hints = {} # Output in stderr where a known SOP/TSG exists which will be HINTed for further help
install_hint = {} # The SOP to help install the executable if it cannot be found
def run(cmd, return_output=False, no_output=False, retry_count=0, base64_decode=False, return_as_json=False):
"""Run shell command, stream stdout, print stderr and optionally return output
NOTES:
1. Commands that need this kind of ' quoting on Windows e.g.:
kubectl get nodes -o jsonpath={.items[?(@.metadata.annotations.pv-candidate=='data-pool')].metadata.name}
Need to actually pass in as '"':
kubectl get nodes -o jsonpath={.items[?(@.metadata.annotations.pv-candidate=='"'data-pool'"')].metadata.name}
The ' quote approach, although correct when pasting into Windows cmd, will hang at the line:
`iter(p.stdout.readline, b'')`
The shlex.split call does the right thing for each platform, just use the '"' pattern for a '
"""
MAX_RETRIES = 5
output = ""
retry = False
# When running `azdata sql query` on Windows, replace any \n in """ strings, with " ", otherwise we see:
#
# ('HY090', '[HY090] [Microsoft][ODBC Driver Manager] Invalid string or buffer length (0) (SQLExecDirectW)')
#
if platform.system() == "Windows" and cmd.startswith("azdata sql query"):
cmd = cmd.replace("\n", " ")
# shlex.split is required on bash and for Windows paths with spaces
#
cmd_actual = shlex.split(cmd)
# Store this (i.e. kubectl, python etc.) to support binary context aware error_hints and retries
#
user_provided_exe_name = cmd_actual[0].lower()
# When running python, use the python in the ADS sandbox ({sys.executable})
#
if cmd.startswith("python "):
cmd_actual[0] = cmd_actual[0].replace("python", sys.executable)
# On Mac, when ADS is not launched from terminal, LC_ALL may not be set, which causes pip installs to fail
# with:
#
# UnicodeDecodeError: 'ascii' codec can't decode byte 0xc5 in position 4969: ordinal not in range(128)
#
# Setting it to a default value of "en_US.UTF-8" enables pip install to complete
#
if platform.system() == "Darwin" and "LC_ALL" not in os.environ:
os.environ["LC_ALL"] = "en_US.UTF-8"
# When running `kubectl`, if AZDATA_OPENSHIFT is set, use `oc`
#
if cmd.startswith("kubectl ") and "AZDATA_OPENSHIFT" in os.environ:
cmd_actual[0] = cmd_actual[0].replace("kubectl", "oc")
# To aid supportability, determine which binary file will actually be executed on the machine
#
which_binary = None
# Special case for CURL on Windows. The version of CURL in Windows System32 does not work to
# get JWT tokens, it returns "(56) Failure when receiving data from the peer". If another instance
# of CURL exists on the machine use that one. (Unfortunately the curl.exe in System32 is almost
# always the first curl.exe in the path, and it can't be uninstalled from System32, so here we
# look for the 2nd installation of CURL in the path)
if platform.system() == "Windows" and cmd.startswith("curl "):
path = os.getenv('PATH')
for p in path.split(os.path.pathsep):
p = os.path.join(p, "curl.exe")
if os.path.exists(p) and os.access(p, os.X_OK):
if p.lower().find("system32") == -1:
cmd_actual[0] = p
which_binary = p
break
# Find the path based location (shutil.which) of the executable that will be run (and display it to aid supportability), this
# seems to be required for .msi installs of azdata.cmd/az.cmd. (otherwise Popen returns FileNotFound)
#
# NOTE: Bash needs cmd to be the list of the space separated values hence shlex.split.
#
if which_binary == None:
which_binary = shutil.which(cmd_actual[0])
# Display an install HINT, so the user can click on a SOP to install the missing binary
#
if which_binary == None:
print(f"The path used to search for '{cmd_actual[0]}' was:")
print(sys.path)
if user_provided_exe_name in install_hint and install_hint[user_provided_exe_name] is not None:
display(Markdown(f'HINT: Use [{install_hint[user_provided_exe_name][0]}]({install_hint[user_provided_exe_name][1]}) to resolve this issue.'))
raise FileNotFoundError(f"Executable '{cmd_actual[0]}' not found in path (where/which)")
else:
cmd_actual[0] = which_binary
start_time = datetime.datetime.now().replace(microsecond=0)
print(f"START: {cmd} @ {start_time} ({datetime.datetime.utcnow().replace(microsecond=0)} UTC)")
print(f" using: {which_binary} ({platform.system()} {platform.release()} on {platform.machine()})")
print(f" cwd: {os.getcwd()}")
# Command-line tools such as CURL and AZDATA HDFS commands output
# scrolling progress bars, which causes Jupyter to hang forever, to
# workaround this, use no_output=True
#
# Work around a infinite hang when a notebook generates a non-zero return code, break out, and do not wait
#
wait = True
try:
if no_output:
p = Popen(cmd_actual)
else:
p = Popen(cmd_actual, stdout=PIPE, stderr=PIPE, bufsize=1)
with p.stdout:
for line in iter(p.stdout.readline, b''):
line = line.decode()
if return_output:
output = output + line
else:
if cmd.startswith("azdata notebook run"): # Hyperlink the .ipynb file
regex = re.compile(' "(.*)"\: "(.*)"')
match = regex.match(line)
if match:
if match.group(1).find("HTML") != -1:
display(Markdown(f' - "{match.group(1)}": "{match.group(2)}"'))
else:
display(Markdown(f' - "{match.group(1)}": "[{match.group(2)}]({match.group(2)})"'))
wait = False
break # otherwise infinite hang, have not worked out why yet.
else:
print(line, end='')
if wait:
p.wait()
except FileNotFoundError as e:
if install_hint is not None:
display(Markdown(f'HINT: Use {install_hint} to resolve this issue.'))
raise FileNotFoundError(f"Executable '{cmd_actual[0]}' not found in path (where/which)") from e
exit_code_workaround = 0 # WORKAROUND: azdata hangs on exception from notebook on p.wait()
if not no_output:
for line in iter(p.stderr.readline, b''):
try:
line_decoded = line.decode()
except UnicodeDecodeError:
# NOTE: Sometimes we get characters back that cannot be decoded(), e.g.
#
# \xa0
#
# For example see this in the response from `az group create`:
#
# ERROR: Get Token request returned http error: 400 and server
# response: {"error":"invalid_grant",# "error_description":"AADSTS700082:
# The refresh token has expired due to inactivity.\xa0The token was
# issued on 2018-10-25T23:35:11.9832872Z
#
# which generates the exception:
#
# UnicodeDecodeError: 'utf-8' codec can't decode byte 0xa0 in position 179: invalid start byte
#
print("WARNING: Unable to decode stderr line, printing raw bytes:")
print(line)
line_decoded = ""
pass
else:
# azdata emits a single empty line to stderr when doing an hdfs cp, don't
# print this empty "ERR:" as it confuses.
#
if line_decoded == "":
continue
print(f"STDERR: {line_decoded}", end='')
if line_decoded.startswith("An exception has occurred") or line_decoded.startswith("ERROR: An error occurred while executing the following cell"):
exit_code_workaround = 1
# inject HINTs to next TSG/SOP based on output in stderr
#
if user_provided_exe_name in error_hints:
for error_hint in error_hints[user_provided_exe_name]:
if line_decoded.find(error_hint[0]) != -1:
display(Markdown(f'HINT: Use [{error_hint[1]}]({error_hint[2]}) to resolve this issue.'))
# Verify if a transient error, if so automatically retry (recursive)
#
if user_provided_exe_name in retry_hints:
for retry_hint in retry_hints[user_provided_exe_name]:
if line_decoded.find(retry_hint) != -1:
if retry_count < MAX_RETRIES:
print(f"RETRY: {retry_count} (due to: {retry_hint})")
retry_count = retry_count + 1
output = run(cmd, return_output=return_output, retry_count=retry_count)
if return_output:
if base64_decode:
import base64
return base64.b64decode(output).decode('utf-8')
else:
return output
elapsed = datetime.datetime.now().replace(microsecond=0) - start_time
# WORKAROUND: We avoid infinite hang above in the `azdata notebook run` failure case, by inferring success (from stdout output), so
# don't wait here, if success known above
#
if wait:
if p.returncode != 0:
raise SystemExit(f'Shell command:\n\n\t{cmd} ({elapsed}s elapsed)\n\nreturned non-zero exit code: {str(p.returncode)}.\n')
else:
if exit_code_workaround !=0 :
raise SystemExit(f'Shell command:\n\n\t{cmd} ({elapsed}s elapsed)\n\nreturned non-zero exit code: {str(exit_code_workaround)}.\n')
print(f'\nSUCCESS: {elapsed}s elapsed.\n')
if return_output:
if base64_decode:
import base64
return base64.b64decode(output).decode('utf-8')
else:
return output
# Hints for tool retry (on transient fault), known errors and install guide
#
retry_hints = {'azdata': ['Endpoint sql-server-master does not exist', 'Endpoint livy does not exist', 'Failed to get state for cluster', 'Endpoint webhdfs does not exist', 'Adaptive Server is unavailable or does not exist', 'Error: Address already in use', 'Login timeout expired (0) (SQLDriverConnect)', 'SSPI Provider: No Kerberos credentials available', ], 'kubectl': ['A connection attempt failed because the connected party did not properly respond after a period of time, or established connection failed because connected host has failed to respond', ], 'python': [ ], }
error_hints = {'azdata': [['Please run \'azdata login\' to first authenticate', 'SOP028 - azdata login', '../common/sop028-azdata-login.ipynb'], ['The token is expired', 'SOP028 - azdata login', '../common/sop028-azdata-login.ipynb'], ['Reason: Unauthorized', 'SOP028 - azdata login', '../common/sop028-azdata-login.ipynb'], ['Max retries exceeded with url: /api/v1/bdc/endpoints', 'SOP028 - azdata login', '../common/sop028-azdata-login.ipynb'], ['Look at the controller logs for more details', 'TSG027 - Observe cluster deployment', '../diagnose/tsg027-observe-bdc-create.ipynb'], ['provided port is already allocated', 'TSG062 - Get tail of all previous container logs for pods in BDC namespace', '../log-files/tsg062-tail-bdc-previous-container-logs.ipynb'], ['Create cluster failed since the existing namespace', 'SOP061 - Delete a big data cluster', '../install/sop061-delete-bdc.ipynb'], ['Failed to complete kube config setup', 'TSG067 - Failed to complete kube config setup', '../repair/tsg067-failed-to-complete-kube-config-setup.ipynb'], ['Data source name not found and no default driver specified', 'SOP069 - Install ODBC for SQL Server', '../install/sop069-install-odbc-driver-for-sql-server.ipynb'], ['Can\'t open lib \'ODBC Driver 17 for SQL Server', 'SOP069 - Install ODBC for SQL Server', '../install/sop069-install-odbc-driver-for-sql-server.ipynb'], ['Control plane upgrade failed. Failed to upgrade controller.', 'TSG108 - View the controller upgrade config map', '../diagnose/tsg108-controller-failed-to-upgrade.ipynb'], ['NameError: name \'azdata_login_secret_name\' is not defined', 'SOP013 - Create secret for azdata login (inside cluster)', '../common/sop013-create-secret-for-azdata-login.ipynb'], ['ERROR: No credentials were supplied, or the credentials were unavailable or inaccessible.', 'TSG124 - \'No credentials were supplied\' error from azdata login', '../repair/tsg124-no-credentials-were-supplied.ipynb'], ['Please accept the license terms to use this product through', 'TSG126 - azdata fails with \'accept the license terms to use this product\'', '../repair/tsg126-accept-license-terms.ipynb'], ], 'kubectl': [['no such host', 'TSG010 - Get configuration contexts', '../monitor-k8s/tsg010-get-kubernetes-contexts.ipynb'], ['No connection could be made because the target machine actively refused it', 'TSG056 - Kubectl fails with No connection could be made because the target machine actively refused it', '../repair/tsg056-kubectl-no-connection-could-be-made.ipynb'], ], 'python': [['Library not loaded: /usr/local/opt/unixodbc', 'SOP012 - Install unixodbc for Mac', '../install/sop012-brew-install-odbc-for-sql-server.ipynb'], ['WARNING: You are using pip version', 'SOP040 - Upgrade pip in ADS Python sandbox', '../install/sop040-upgrade-pip.ipynb'], ], }
install_hint = {'azdata': [ 'SOP063 - Install azdata CLI (using package manager)', '../install/sop063-packman-install-azdata.ipynb' ], 'kubectl': [ 'SOP036 - Install kubectl command line interface', '../install/sop036-install-kubectl.ipynb' ], }
print('Common functions defined successfully.')
###Output
_____no_output_____
###Markdown
Get the Kubernetes namespace for the big data clusterGet the namespace of the Big Data Cluster use the kubectl command lineinterface .**NOTE:**If there is more than one Big Data Cluster in the target Kubernetescluster, then either:- set \[0\] to the correct value for the big data cluster.- set the environment variable AZDATA_NAMESPACE, before starting Azure Data Studio.
###Code
# Place Kubernetes namespace name for BDC into 'namespace' variable
if "AZDATA_NAMESPACE" in os.environ:
namespace = os.environ["AZDATA_NAMESPACE"]
else:
try:
namespace = run(f'kubectl get namespace --selector=MSSQL_CLUSTER -o jsonpath={{.items[0].metadata.name}}', return_output=True)
except:
from IPython.display import Markdown
print(f"ERROR: Unable to find a Kubernetes namespace with label 'MSSQL_CLUSTER'. SQL Server Big Data Cluster Kubernetes namespaces contain the label 'MSSQL_CLUSTER'.")
display(Markdown(f'HINT: Use [TSG081 - Get namespaces (Kubernetes)](../monitor-k8s/tsg081-get-kubernetes-namespaces.ipynb) to resolve this issue.'))
display(Markdown(f'HINT: Use [TSG010 - Get configuration contexts](../monitor-k8s/tsg010-get-kubernetes-contexts.ipynb) to resolve this issue.'))
display(Markdown(f'HINT: Use [SOP011 - Set kubernetes configuration context](../common/sop011-set-kubernetes-context.ipynb) to resolve this issue.'))
raise
print(f'The SQL Server Big Data Cluster Kubernetes namespace is: {namespace}')
###Output
_____no_output_____
###Markdown
Get name of the ‘Running’ `controller` `pod`
###Code
# Place the name of the 'Running' controller pod in variable `controller`
controller = run(f'kubectl get pod --selector=app=controller -n {namespace} -o jsonpath={{.items[0].metadata.name}} --field-selector=status.phase=Running', return_output=True)
print(f"Controller pod name: {controller}")
###Output
_____no_output_____
###Markdown
Create a temporary folder to hold Root CA certificate
###Code
import os
import tempfile
import shutil
path = os.path.join(tempfile.gettempdir(), local_folder_name)
if os.path.isdir(path):
shutil.rmtree(path)
os.mkdir(path)
###Output
_____no_output_____
###Markdown
Copy Root CA certificate from `controller` `pod`
###Code
import os
cwd = os.getcwd()
os.chdir(path) # Workaround kubectl bug on Windows, can't put c:\ on kubectl cp cmd line
run(f'kubectl cp {controller}:{test_cert_store_root}/cacert.pem cacert.pem -c controller -n {namespace}')
run(f'kubectl cp {controller}:{test_cert_store_root}/cakey.pem cakey.pem -c controller -n {namespace}')
os.chdir(cwd)
print("Notebook execution is complete.")
###Output
_____no_output_____
|
002_image_captioning/2_Training.ipynb
|
###Markdown
Computer Vision Nanodegree Project: Image Captioning---In this notebook, you will train your CNN-RNN model. You are welcome and encouraged to try out many different architectures and hyperparameters when searching for a good model.This does have the potential to make the project quite messy! Before submitting your project, make sure that you clean up:- the code you write in this notebook. The notebook should describe how to train a single CNN-RNN architecture, corresponding to your final choice of hyperparameters. You should structure the notebook so that the reviewer can replicate your results by running the code in this notebook. - the output of the code cell in **Step 2**. The output should show the output obtained when training the model from scratch.This notebook **will be graded**. Feel free to use the links below to navigate the notebook:- [Step 1](step1): Training Setup- [Step 2](step2): Train your Model- [Step 3](step3): (Optional) Validate your Model Step 1: Training SetupIn this step of the notebook, you will customize the training of your CNN-RNN model by specifying hyperparameters and setting other options that are important to the training procedure. The values you set now will be used when training your model in **Step 2** below.You should only amend blocks of code that are preceded by a `TODO` statement. **Any code blocks that are not preceded by a `TODO` statement should not be modified**. Task 1Begin by setting the following variables:- `batch_size` - the batch size of each training batch. It is the number of image-caption pairs used to amend the model weights in each training step. - `vocab_threshold` - the minimum word count threshold. Note that a larger threshold will result in a smaller vocabulary, whereas a smaller threshold will include rarer words and result in a larger vocabulary. - `vocab_from_file` - a Boolean that decides whether to load the vocabulary from file. - `embed_size` - the dimensionality of the image and word embeddings. - `hidden_size` - the number of features in the hidden state of the RNN decoder. - `num_epochs` - the number of epochs to train the model. We recommend that you set `num_epochs=3`, but feel free to increase or decrease this number as you wish. [This paper](https://arxiv.org/pdf/1502.03044.pdf) trained a captioning model on a single state-of-the-art GPU for 3 days, but you'll soon see that you can get reasonable results in a matter of a few hours! (_But of course, if you want your model to compete with current research, you will have to train for much longer._)- `save_every` - determines how often to save the model weights. We recommend that you set `save_every=1`, to save the model weights after each epoch. This way, after the `i`th epoch, the encoder and decoder weights will be saved in the `models/` folder as `encoder-i.pkl` and `decoder-i.pkl`, respectively.- `print_every` - determines how often to print the batch loss to the Jupyter notebook while training. Note that you **will not** observe a monotonic decrease in the loss function while training - this is perfectly fine and completely expected! You are encouraged to keep this at its default value of `100` to avoid clogging the notebook, but feel free to change it.- `log_file` - the name of the text file containing - for every step - how the loss and perplexity evolved during training.If you're not sure where to begin to set some of the values above, you can peruse [this paper](https://arxiv.org/pdf/1502.03044.pdf) and [this paper](https://arxiv.org/pdf/1411.4555.pdf) for useful guidance! **To avoid spending too long on this notebook**, you are encouraged to consult these suggested research papers to obtain a strong initial guess for which hyperparameters are likely to work best. Then, train a single model, and proceed to the next notebook (**3_Inference.ipynb**). If you are unhappy with your performance, you can return to this notebook to tweak the hyperparameters (and/or the architecture in **model.py**) and re-train your model. Question 1**Question:** Describe your CNN-RNN architecture in detail. With this architecture in mind, how did you select the values of the variables in Task 1? If you consulted a research paper detailing a successful implementation of an image captioning model, please provide the reference.**Answer:** CNN-RNN architecture was selected as an out-of-the box pre-trained Resnet-50 Model. Followed by LSTM decoders. I consulted the following papers for the same -1.[this paper](https://arxiv.org/pdf/1411.4555.pdf)2.[this paper](https://arxiv.org/pdf/1502.03044.pdf) (Optional) Task 2Note that we have provided a recommended image transform `transform_train` for pre-processing the training images, but you are welcome (and encouraged!) to modify it as you wish. When modifying this transform, keep in mind that:- the images in the dataset have varying heights and widths, and - if using a pre-trained model, you must perform the corresponding appropriate normalization. Question 2**Question:** How did you select the transform in `transform_train`? If you left the transform at its provided value, why do you think that it is a good choice for your CNN architecture?**Answer:** I used the transform function of the pre-trained Resnet Model as it makes sense that if, a pre-trained model is used, we apply the same normalization to the image which first goes via the CNN to generate the feature fector. Task 3Next, you will specify a Python list containing the learnable parameters of the model. For instance, if you decide to make all weights in the decoder trainable, but only want to train the weights in the embedding layer of the encoder, then you should set `params` to something like:```params = list(decoder.parameters()) + list(encoder.embed.parameters()) ``` Question 3**Question:** How did you select the trainable parameters of your architecture? Why do you think this is a good choice?**Answer:** Since I was using a pre-trained CNN , I only selected the decoder to be a trainable model as the CNN Encoder is already well defined and trained to generate a good feature vector in it's embedding. Task 4Finally, you will select an [optimizer](http://pytorch.org/docs/master/optim.htmltorch.optim.Optimizer). Question 4**Question:** How did you select the optimizer used to train your model?**Answer:** I selected ADAM optimizer given it's better performance and it's capability to converge faster and avoid local minima, which can be possible in a deep neural network.
###Code
import torch
import torch.nn as nn
from torchvision import transforms
import sys
sys.path.append('/datadrive2/amit_cvnd/cocoapi/PythonAPI')
from pycocotools.coco import COCO
from data_loader import get_loader
from model import EncoderCNN, DecoderRNN
import math
## TODO #1: Select appropriate values for the Python variables below.
batch_size = 10 # batch size
vocab_threshold = 4 # minimum word count threshold
vocab_from_file = True # if True, load existing vocab file
embed_size = 256 # dimensionality of image and word embeddings
hidden_size = 512 # number of features in hidden state of the RNN decoder
num_epochs = 1 # number of training epochs
save_every = 1 # determines frequency of saving model weights
print_every = 100 # determines window for printing average loss
log_file = 'training_log.txt' # name of file with saved training loss and perplexity
# (Optional) TODO #2: Amend the image transform below.
transform_train = transforms.Compose([
transforms.Resize(256), # smaller edge of image resized to 256
transforms.RandomCrop(224), # get 224x224 crop from random location
transforms.RandomHorizontalFlip(), # horizontally flip image with probability=0.5
transforms.ToTensor(), # convert the PIL Image to a tensor
transforms.Normalize((0.485, 0.456, 0.406), # normalize image for pre-trained model
(0.229, 0.224, 0.225))])
# Build data loader.
data_loader = get_loader(transform=transform_train,
mode='train',
batch_size=batch_size,
vocab_threshold=vocab_threshold,
vocab_from_file=vocab_from_file)
# The size of the vocabulary.
vocab_size = len(data_loader.dataset.vocab)
# Initialize the encoder and decoder.
encoder = EncoderCNN(embed_size)
decoder = DecoderRNN(embed_size, hidden_size, vocab_size)
# Move models to GPU if CUDA is available.
device = torch.device("cuda" if torch.cuda.is_available() else "cpu")
encoder.to(device)
decoder.to(device)
# Define the loss function.
criterion = nn.CrossEntropyLoss().cuda() if torch.cuda.is_available() else nn.CrossEntropyLoss()
# TODO #3: Specify the learnable parameters of the model.
params = list(decoder.parameters())
# TODO #4: Define the optimizer.
optimizer = torch.optim.Adam(params, lr=0.001, betas=(0.9, 0.999), eps=1e-08, weight_decay=0, amsgrad=False)
# Set the total number of training steps per epoch.
total_step = math.ceil(len(data_loader.dataset.caption_lengths) / data_loader.batch_sampler.batch_size)
###Output
Vocabulary successfully loaded from vocab.pkl file!
loading annotations into memory...
Done (t=0.68s)
creating index...
###Markdown
Step 2: Train your ModelOnce you have executed the code cell in **Step 1**, the training procedure below should run without issue. It is completely fine to leave the code cell below as-is without modifications to train your model. However, if you would like to modify the code used to train the model below, you must ensure that your changes are easily parsed by your reviewer. In other words, make sure to provide appropriate comments to describe how your code works! You may find it useful to load saved weights to resume training. In that case, note the names of the files containing the encoder and decoder weights that you'd like to load (`encoder_file` and `decoder_file`). Then you can load the weights by using the lines below:```python Load pre-trained weights before resuming training.encoder.load_state_dict(torch.load(os.path.join('./models', encoder_file)))decoder.load_state_dict(torch.load(os.path.join('./models', decoder_file)))```While trying out parameters, make sure to take extensive notes and record the settings that you used in your various training runs. In particular, you don't want to encounter a situation where you've trained a model for several hours but can't remember what settings you used :). A Note on Tuning HyperparametersTo figure out how well your model is doing, you can look at how the training loss and perplexity evolve during training - and for the purposes of this project, you are encouraged to amend the hyperparameters based on this information. However, this will not tell you if your model is overfitting to the training data, and, unfortunately, overfitting is a problem that is commonly encountered when training image captioning models. For this project, you need not worry about overfitting. **This project does not have strict requirements regarding the performance of your model**, and you just need to demonstrate that your model has learned **_something_** when you generate captions on the test data. For now, we strongly encourage you to train your model for the suggested 3 epochs without worrying about performance; then, you should immediately transition to the next notebook in the sequence (**3_Inference.ipynb**) to see how your model performs on the test data. If your model needs to be changed, you can come back to this notebook, amend hyperparameters (if necessary), and re-train the model.That said, if you would like to go above and beyond in this project, you can read about some approaches to minimizing overfitting in section 4.3.1 of [this paper](http://ieeexplore.ieee.org/stamp/stamp.jsp?arnumber=7505636). In the next (optional) step of this notebook, we provide some guidance for assessing the performance on the validation dataset.
###Code
import torch.utils.data as data
import numpy as np
import os
import requests
import time
# Open the training log file.
f = open(log_file, 'w')
old_time = time.time()
# response = requests.request("GET",
# "http://metadata.google.internal/computeMetadata/v1/instance/attributes/keep_alive_token",
# headers={"Metadata-Flavor":"Google"})
for epoch in range(1, num_epochs+1):
for i_step in range(1, total_step+1):
if time.time() - old_time > 60:
old_time = time.time()
# requests.request("POST",
# "https://nebula.udacity.com/api/v1/remote/keep-alive",
# headers={'Authorization': "STAR " + response.text})
# Randomly sample a caption length, and sample indices with that length.
indices = data_loader.dataset.get_train_indices()
# Create and assign a batch sampler to retrieve a batch with the sampled indices.
new_sampler = data.sampler.SubsetRandomSampler(indices=indices)
data_loader.batch_sampler.sampler = new_sampler
# Obtain the batch.
images, captions = next(iter(data_loader))
# Move batch of images and captions to GPU if CUDA is available.
images = images.to(device)
captions = captions.to(device)
# Zero the gradients.
decoder.zero_grad()
encoder.zero_grad()
# Pass the inputs through the CNN-RNN model.
features = encoder(images)
outputs = decoder(features, captions)
# Calculate the batch loss.
loss = criterion(outputs.view(-1, vocab_size), captions.view(-1))
# Backward pass.
loss.backward()
# Update the parameters in the optimizer.
optimizer.step()
# Get training statistics.
stats = 'Epoch [%d/%d], Step [%d/%d], Loss: %.4f, Perplexity: %5.4f' % (epoch, num_epochs, i_step, total_step, loss.item(), np.exp(loss.item()))
# Print training statistics (on same line).
print('\r' + stats, end="")
sys.stdout.flush()
# Print training statistics to file.
f.write(stats + '\n')
f.flush()
# Print training statistics (on different line).
if i_step % print_every == 0:
print('\r' + stats)
# Save the weights.
if epoch % save_every == 0:
torch.save(decoder.state_dict(), os.path.join('./models', 'decoder-%d.pkl' % epoch))
torch.save(encoder.state_dict(), os.path.join('./models', 'encoder-%d.pkl' % epoch))
# Close the training log file.
f.close()
torch.save(decoder.state_dict(), os.path.join('./models', 'decoder-%d.pkl' % 1))
torch.save(encoder.state_dict(), os.path.join('./models', 'encoder-%d.pkl' % 1))
###Output
_____no_output_____
###Markdown
Step 3: (Optional) Validate your ModelTo assess potential overfitting, one approach is to assess performance on a validation set. If you decide to do this **optional** task, you are required to first complete all of the steps in the next notebook in the sequence (**3_Inference.ipynb**); as part of that notebook, you will write and test code (specifically, the `sample` method in the `DecoderRNN` class) that uses your RNN decoder to generate captions. That code will prove incredibly useful here. If you decide to validate your model, please do not edit the data loader in **data_loader.py**. Instead, create a new file named **data_loader_val.py** containing the code for obtaining the data loader for the validation data. You can access:- the validation images at filepath `'/opt/cocoapi/images/train2014/'`, and- the validation image caption annotation file at filepath `'/opt/cocoapi/annotations/captions_val2014.json'`.The suggested approach to validating your model involves creating a json file such as [this one](https://github.com/cocodataset/cocoapi/blob/master/results/captions_val2014_fakecap_results.json) containing your model's predicted captions for the validation images. Then, you can write your own script or use one that you [find online](https://github.com/tylin/coco-caption) to calculate the BLEU score of your model. You can read more about the BLEU score, along with other evaluation metrics (such as TEOR and Cider) in section 4.1 of [this paper](https://arxiv.org/pdf/1411.4555.pdf). For more information about how to use the annotation file, check out the [website](http://cocodataset.org/download) for the COCO dataset.
###Code
# (Optional) TODO: Validate your model.
###Output
_____no_output_____
|
scripts/d21-en/tensorflow/chapter_optimization/lr-scheduler.ipynb
|
###Markdown
Learning Rate Scheduling:label:`sec_scheduler`So far we primarily focused on optimization *algorithms* for how to update the weight vectors rather than on the *rate* at which they are being updated. Nonetheless, adjusting the learning rate is often just as important as the actual algorithm. There are a number of aspects to consider:* Most obviously the *magnitude* of the learning rate matters. If it is too large, optimization diverges, if it is too small, it takes too long to train or we end up with a suboptimal result. We saw previously that the condition number of the problem matters (see e.g., :numref:`sec_momentum` for details). Intuitively it is the ratio of the amount of change in the least sensitive direction vs. the most sensitive one.* Secondly, the rate of decay is just as important. If the learning rate remains large we may simply end up bouncing around the minimum and thus not reach optimality. :numref:`sec_minibatch_sgd` discussed this in some detail and we analyzed performance guarantees in :numref:`sec_sgd`. In short, we want the rate to decay, but probably more slowly than $\mathcal{O}(t^{-\frac{1}{2}})$ which would be a good choice for convex problems.* Another aspect that is equally important is *initialization*. This pertains both to how the parameters are set initially (review :numref:`sec_numerical_stability` for details) and also how they evolve initially. This goes under the moniker of *warmup*, i.e., how rapidly we start moving towards the solution initially. Large steps in the beginning might not be beneficial, in particular since the initial set of parameters is random. The initial update directions might be quite meaningless, too.* Lastly, there are a number of optimization variants that perform cyclical learning rate adjustment. This is beyond the scope of the current chapter. We recommend the reader to review details in :cite:`Izmailov.Podoprikhin.Garipov.ea.2018`, e.g., how to obtain better solutions by averaging over an entire *path* of parameters.Given the fact that there is a lot of detail needed to manage learning rates, most deep learning frameworks have tools to deal with this automatically. In the current chapter we will review the effects that different schedules have on accuracy and also show how this can be managed efficiently via a *learning rate scheduler*. Toy ProblemWe begin with a toy problem that is cheap enough to compute easily, yet sufficiently nontrivial to illustrate some of the key aspects. For that we pick a slightly modernized version of LeNet (`relu` instead of `sigmoid` activation, MaxPooling rather than AveragePooling), as applied to Fashion-MNIST. Moreover, we hybridize the network for performance. Since most of the code is standard we just introduce the basics without further detailed discussion. See :numref:`chap_cnn` for a refresher as needed.
###Code
%matplotlib inline
import math
import tensorflow as tf
from tensorflow.keras.callbacks import LearningRateScheduler
from d2l import tensorflow as d2l
def net():
return tf.keras.models.Sequential([
tf.keras.layers.Conv2D(filters=6, kernel_size=5, activation='relu',
padding='same'),
tf.keras.layers.AvgPool2D(pool_size=2, strides=2),
tf.keras.layers.Conv2D(filters=16, kernel_size=5,
activation='relu'),
tf.keras.layers.AvgPool2D(pool_size=2, strides=2),
tf.keras.layers.Flatten(),
tf.keras.layers.Dense(120, activation='relu'),
tf.keras.layers.Dense(84, activation='sigmoid'),
tf.keras.layers.Dense(10)])
batch_size = 256
train_iter, test_iter = d2l.load_data_fashion_mnist(batch_size=batch_size)
# The code is almost identical to `d2l.train_ch6` defined in the
# lenet section of chapter convolutional neural networks
def train(net_fn, train_iter, test_iter, num_epochs, lr,
device=d2l.try_gpu(), custom_callback = False):
device_name = device._device_name
strategy = tf.distribute.OneDeviceStrategy(device_name)
with strategy.scope():
optimizer = tf.keras.optimizers.SGD(learning_rate=lr)
loss = tf.keras.losses.SparseCategoricalCrossentropy(from_logits=True)
net = net_fn()
net.compile(optimizer=optimizer, loss=loss, metrics=['accuracy'])
callback = d2l.TrainCallback(net, train_iter, test_iter, num_epochs,
device_name)
if custom_callback is False:
net.fit(train_iter, epochs=num_epochs, verbose=0,
callbacks=[callback])
else:
net.fit(train_iter, epochs=num_epochs, verbose=0,
callbacks=[callback, custom_callback])
return net
###Output
_____no_output_____
###Markdown
Let us have a look at what happens if we invoke this algorithm with default settings, such as a learning rate of $0.3$ and train for $30$ iterations. Note how the training accuracy keeps on increasing while progress in terms of test accuracy stalls beyond a point. The gap between both curves indicates overfitting.
###Code
lr, num_epochs = 0.3, 30
train(net, train_iter, test_iter, num_epochs, lr)
###Output
loss 0.215, train acc 0.919, test acc 0.891
71025.6 examples/sec on /GPU:0
###Markdown
SchedulersOne way of adjusting the learning rate is to set it explicitly at each step. This is conveniently achieved by the `set_learning_rate` method. We could adjust it downward after every epoch (or even after every minibatch), e.g., in a dynamic manner in response to how optimization is progressing.
###Code
lr = 0.1
dummy_model = tf.keras.models.Sequential([tf.keras.layers.Dense(10)])
dummy_model.compile(tf.keras.optimizers.SGD(learning_rate=lr), loss='mse')
print(f'learning rate is now ,', dummy_model.optimizer.lr.numpy())
###Output
learning rate is now , 0.1
###Markdown
More generally we want to define a scheduler. When invoked with the number of updates it returns the appropriate value of the learning rate. Let us define a simple one that sets the learning rate to $\eta = \eta_0 (t + 1)^{-\frac{1}{2}}$.
###Code
class SquareRootScheduler:
def __init__(self, lr=0.1):
self.lr = lr
def __call__(self, num_update):
return self.lr * pow(num_update + 1.0, -0.5)
###Output
_____no_output_____
###Markdown
Let us plot its behavior over a range of values.
###Code
scheduler = SquareRootScheduler(lr=0.1)
d2l.plot(tf.range(num_epochs), [scheduler(t) for t in range(num_epochs)])
###Output
_____no_output_____
###Markdown
Now let us see how this plays out for training on Fashion-MNIST. We simply provide the scheduler as an additional argument to the training algorithm.
###Code
train(net, train_iter, test_iter, num_epochs, lr,
custom_callback=LearningRateScheduler(scheduler))
###Output
loss 0.385, train acc 0.858, test acc 0.843
77382.9 examples/sec on /GPU:0
###Markdown
This worked quite a bit better than previously. Two things stand out: the curve was rather more smooth than previously. Secondly, there was less overfitting. Unfortunately it is not a well-resolved question as to why certain strategies lead to less overfitting in *theory*. There is some argument that a smaller stepsize will lead to parameters that are closer to zero and thus simpler. However, this does not explain the phenomenon entirely since we do not really stop early but simply reduce the learning rate gently. PoliciesWhile we cannot possibly cover the entire variety of learning rate schedulers, we attempt to give a brief overview of popular policies below. Common choices are polynomial decay and piecewise constant schedules. Beyond that, cosine learning rate schedules have been found to work well empirically on some problems. Lastly, on some problems it is beneficial to warm up the optimizer prior to using large learning rates. Factor SchedulerOne alternative to a polynomial decay would be a multiplicative one, that is $\eta_{t+1} \leftarrow \eta_t \cdot \alpha$ for $\alpha \in (0, 1)$. To prevent the learning rate from decaying beyond a reasonable lower bound the update equation is often modified to $\eta_{t+1} \leftarrow \mathop{\mathrm{max}}(\eta_{\mathrm{min}}, \eta_t \cdot \alpha)$.
###Code
class FactorScheduler:
def __init__(self, factor=1, stop_factor_lr=1e-7, base_lr=0.1):
self.factor = factor
self.stop_factor_lr = stop_factor_lr
self.base_lr = base_lr
def __call__(self, num_update):
self.base_lr = max(self.stop_factor_lr, self.base_lr * self.factor)
return self.base_lr
scheduler = FactorScheduler(factor=0.9, stop_factor_lr=1e-2, base_lr=2.0)
d2l.plot(tf.range(50), [scheduler(t) for t in range(50)])
###Output
_____no_output_____
###Markdown
This can also be accomplished by a built-in scheduler in MXNet via the `lr_scheduler.FactorScheduler` object. It takes a few more parameters, such as warmup period, warmup mode (linear or constant), the maximum number of desired updates, etc.; Going forward we will use the built-in schedulers as appropriate and only explain their functionality here. As illustrated, it is fairly straightforward to build your own scheduler if needed. Multi Factor SchedulerA common strategy for training deep networks is to keep the learning rate piecewise constant and to decrease it by a given amount every so often. That is, given a set of times when to decrease the rate, such as $s = \{5, 10, 20\}$ decrease $\eta_{t+1} \leftarrow \eta_t \cdot \alpha$ whenever $t \in s$. Assuming that the values are halved at each step we can implement this as follows.
###Code
class MultiFactorScheduler:
def __init__(self, step, factor, base_lr):
self.step = step
self.factor = factor
self.base_lr = base_lr
def __call__(self, epoch):
if epoch in self.step:
self.base_lr = self.base_lr * self.factor
return self.base_lr
else:
return self.base_lr
scheduler = MultiFactorScheduler(step=[15, 30], factor=0.5, base_lr=0.5)
d2l.plot(tf.range(num_epochs), [scheduler(t) for t in range(num_epochs)])
###Output
_____no_output_____
###Markdown
The intuition behind this piecewise constant learning rate schedule is that one lets optimization proceed until a stationary point has been reached in terms of the distribution of weight vectors. Then (and only then) do we decrease the rate such as to obtain a higher quality proxy to a good local minimum. The example below shows how this can produce ever slightly better solutions.
###Code
train(net, train_iter, test_iter, num_epochs, lr,
custom_callback=LearningRateScheduler(scheduler))
###Output
loss 0.244, train acc 0.909, test acc 0.882
78261.8 examples/sec on /GPU:0
###Markdown
Cosine SchedulerA rather perplexing heuristic was proposed by :cite:`Loshchilov.Hutter.2016`. It relies on the observation that we might not want to decrease the learning rate too drastically in the beginning and moreover, that we might want to "refine" the solution in the end using a very small learning rate. This results in a cosine-like schedule with the following functional form for learning rates in the range $t \in [0, T]$.$$\eta_t = \eta_T + \frac{\eta_0 - \eta_T}{2} \left(1 + \cos(\pi t/T)\right)$$Here $\eta_0$ is the initial learning rate, $\eta_T$ is the target rate at time $T$. Furthermore, for $t > T$ we simply pin the value to $\eta_T$ without increasing it again. In the following example, we set the max update step $T = 20$.
###Code
class CosineScheduler:
def __init__(self, max_update, base_lr=0.01, final_lr=0, warmup_steps=0,
warmup_begin_lr=0):
self.base_lr_orig = base_lr
self.max_update = max_update
self.final_lr = final_lr
self.warmup_steps = warmup_steps
self.warmup_begin_lr = warmup_begin_lr
self.max_steps = self.max_update - self.warmup_steps
def get_warmup_lr(self, epoch):
increase = (self.base_lr_orig - self.warmup_begin_lr) \
* float(epoch) / float(self.warmup_steps)
return self.warmup_begin_lr + increase
def __call__(self, epoch):
if epoch < self.warmup_steps:
return self.get_warmup_lr(epoch)
if epoch <= self.max_update:
self.base_lr = self.final_lr + (
self.base_lr_orig - self.final_lr) * (1 + math.cos(
math.pi *
(epoch - self.warmup_steps) / self.max_steps)) / 2
return self.base_lr
scheduler = CosineScheduler(max_update=20, base_lr=0.3, final_lr=0.01)
d2l.plot(tf.range(num_epochs), [scheduler(t) for t in range(num_epochs)])
###Output
_____no_output_____
###Markdown
In the context of computer vision this schedule *can* lead to improved results. Note, though, that such improvements are not guaranteed (as can be seen below).
###Code
train(net, train_iter, test_iter, num_epochs, lr,
custom_callback=LearningRateScheduler(scheduler))
###Output
loss 0.256, train acc 0.907, test acc 0.886
82558.5 examples/sec on /GPU:0
###Markdown
WarmupIn some cases initializing the parameters is not sufficient to guarantee a good solution. This particularly a problem for some advanced network designs that may lead to unstable optimization problems. We could address this by choosing a sufficiently small learning rate to prevent divergence in the beginning. Unfortunately this means that progress is slow. Conversely, a large learning rate initially leads to divergence.A rather simple fix for this dilemma is to use a warmup period during which the learning rate *increases* to its initial maximum and to cool down the rate until the end of the optimization process. For simplicity one typically uses a linear increase for this purpose. This leads to a schedule of the form indicated below.
###Code
scheduler = CosineScheduler(20, warmup_steps=5, base_lr=0.3, final_lr=0.01)
d2l.plot(tf.range(num_epochs), [scheduler(t) for t in range(num_epochs)])
###Output
_____no_output_____
###Markdown
Note that the network converges better initially (in particular observe the performance during the first 5 epochs).
###Code
train(net, train_iter, test_iter, num_epochs, lr,
custom_callback=LearningRateScheduler(scheduler))
###Output
loss 0.274, train acc 0.899, test acc 0.880
82621.6 examples/sec on /GPU:0
|
Example_notebooks/00_Example_A_three-sided_dice.ipynb
|
###Markdown
A three-sided dice The probabilistic nature of quantum mechanics can be exploited in order to produce true randomness. If the quantum system is in superposition, a measurement will colapse the system into any of the states of the base. In our case, we can do this using our three-level system. We start importing the necessary packages.
###Code
import threerra
import numpy as np
from threerra.discriminators.LDA_discriminator import train_discriminator
from qiskit.tools.visualization import plot_histogram
from qiskit.tools.monitor import job_monitor
from qiskit import IBMQ
IBMQ.load_account()
###Output
_____no_output_____
###Markdown
Now, we choose the quantum processor that we are going to use as a random number generator.
###Code
# Choose device
provider = IBMQ.get_provider(hub='ibm-q', group='open', project='main')
backend = provider.get_backend('ibmq_armonk')
###Output
_____no_output_____
###Markdown
We calibrate and train the discriminator
###Code
qc = threerra.QuantumCircuit3(backend)
qc.calibrate_freq_12()
qc.calibrate_pi_amp_12()
train_discriminator(qc)
###Output
Calibrating qubit_freq_est_12...
Job Status: job has successfully run
qubit_freq_est_12 updated from 4.624395869169078GHz to 4.623665090345999GHz.
Calibrating pi_amp_12...
Job Status: job has successfully run
pi_amp_12 updated from 0.33025991905409463 to 0.3386071056422093.
Training discriminator...
Job Status: job has successfully run
Discriminator achieved an accuracy of 91.89% after training.
###Markdown
We want the three options, 0, 1, and 2, to be equally probable. For this we create a circuit that generates $\frac{1}{\sqrt{3}}( | 0 \rangle + | 1 \rangle + | 2 \rangle)$.
###Code
# Test QuantumCircuit3
qc = threerra.QuantumCircuit3(backend)
qc.rx_01(1.94)
qc.rx_12(np.pi/2)
qc.draw()
###Output
_____no_output_____
###Markdown
Now measure the circuit. In this case, we measure 1024 in order to test that the probability distribution is the one we want.
###Code
qc.measure()
job = qc.run(shots=1024)
job_monitor(job)
result = job.result()
###Output
Job Status: job has successfully run
###Markdown
The results we obtain are not processed. We need to classify the results as 0, 1, and 2. For this we use the discriminator we built.
###Code
from threerra.tools import get_data, get_counts
from threerra.discriminators.LDA_discriminator import discriminator as lda_disc
counts_lda = get_counts(result, discriminator=lda_disc)
###Output
_____no_output_____
###Markdown
We plot the histogram of the counts. The distribution is somewhat uniform, and we can use it as a random number generator
###Code
plot_histogram(counts_lda)
###Output
_____no_output_____
###Markdown
Now, if we want a random number, we apply
###Code
qc = threerra.QuantumCircuit3(backend)
qc.rx_01(1.94)
qc.rx_12(np.pi/2)
qc.measure()
job = qc.run(shots=1)
job_monitor(job)
result = job.result()
number = get_data(result, discriminator=lda_disc)
number
###Output
_____no_output_____
|
read_ISA_ACTION-Template.ipynb
|
###Markdown
Reading ISA files
###Code
import os
os.listdir("isa_template")
from isatools import isatab
isa = isatab.load(open("isa_template/i_investigation.txt"))
###Output
_____no_output_____
###Markdown
Retrieve information about investigation
###Code
isa.title
isa.description
isa.contacts
###Output
_____no_output_____
###Markdown
Information about study
###Code
[s.title for s in isa.studies]
###Output
_____no_output_____
###Markdown
Information about assays
###Code
[a.filename for a in isa.studies[0].assays]
###Output
_____no_output_____
|
courses/coursera/DeepLearning/NLPSequenceModels/Dinosaurus+Island+--+Character+level+language+model+final+-+v3.ipynb
|
###Markdown
Character level language model - Dinosaurus landWelcome to Dinosaurus Island! 65 million years ago, dinosaurs existed, and in this assignment they are back. You are in charge of a special task. Leading biology researchers are creating new breeds of dinosaurs and bringing them to life on earth, and your job is to give names to these dinosaurs. If a dinosaur does not like its name, it might go beserk, so choose wisely! Luckily you have learned some deep learning and you will use it to save the day. Your assistant has collected a list of all the dinosaur names they could find, and compiled them into this [dataset](dinos.txt). (Feel free to take a look by clicking the previous link.) To create new dinosaur names, you will build a character level language model to generate new names. Your algorithm will learn the different name patterns, and randomly generate new names. Hopefully this algorithm will keep you and your team safe from the dinosaurs' wrath! By completing this assignment you will learn:- How to store text data for processing using an RNN - How to synthesize data, by sampling predictions at each time step and passing it to the next RNN-cell unit- How to build a character-level text generation recurrent neural network- Why clipping the gradients is importantWe will begin by loading in some functions that we have provided for you in `rnn_utils`. Specifically, you have access to functions such as `rnn_forward` and `rnn_backward` which are equivalent to those you've implemented in the previous assignment.
###Code
import numpy as np
from utils import *
import random
###Output
_____no_output_____
###Markdown
1 - Problem Statement 1.1 - Dataset and PreprocessingRun the following cell to read the dataset of dinosaur names, create a list of unique characters (such as a-z), and compute the dataset and vocabulary size.
###Code
data = open('dinos.txt', 'r').read()
data= data.lower()
chars = list(set(data))
data_size, vocab_size = len(data), len(chars)
print('There are %d total characters and %d unique characters in your data.' % (data_size, vocab_size))
###Output
There are 19909 total characters and 27 unique characters in your data.
###Markdown
The characters are a-z (26 characters) plus the "\n" (or newline character), which in this assignment plays a role similar to the `` (or "End of sentence") token we had discussed in lecture, only here it indicates the end of the dinosaur name rather than the end of a sentence. In the cell below, we create a python dictionary (i.e., a hash table) to map each character to an index from 0-26. We also create a second python dictionary that maps each index back to the corresponding character character. This will help you figure out what index corresponds to what character in the probability distribution output of the softmax layer. Below, `char_to_ix` and `ix_to_char` are the python dictionaries.
###Code
char_to_ix = { ch:i for i,ch in enumerate(sorted(chars)) }
ix_to_char = { i:ch for i,ch in enumerate(sorted(chars)) }
print(ix_to_char)
###Output
{0: '\n', 1: 'a', 2: 'b', 3: 'c', 4: 'd', 5: 'e', 6: 'f', 7: 'g', 8: 'h', 9: 'i', 10: 'j', 11: 'k', 12: 'l', 13: 'm', 14: 'n', 15: 'o', 16: 'p', 17: 'q', 18: 'r', 19: 's', 20: 't', 21: 'u', 22: 'v', 23: 'w', 24: 'x', 25: 'y', 26: 'z'}
###Markdown
1.2 - Overview of the modelYour model will have the following structure: - Initialize parameters - Run the optimization loop - Forward propagation to compute the loss function - Backward propagation to compute the gradients with respect to the loss function - Clip the gradients to avoid exploding gradients - Using the gradients, update your parameter with the gradient descent update rule.- Return the learned parameters **Figure 1**: Recurrent Neural Network, similar to what you had built in the previous notebook "Building a RNN - Step by Step". At each time-step, the RNN tries to predict what is the next character given the previous characters. The dataset $X = (x^{\langle 1 \rangle}, x^{\langle 2 \rangle}, ..., x^{\langle T_x \rangle})$ is a list of characters in the training set, while $Y = (y^{\langle 1 \rangle}, y^{\langle 2 \rangle}, ..., y^{\langle T_x \rangle})$ is such that at every time-step $t$, we have $y^{\langle t \rangle} = x^{\langle t+1 \rangle}$. 2 - Building blocks of the modelIn this part, you will build two important blocks of the overall model:- Gradient clipping: to avoid exploding gradients- Sampling: a technique used to generate charactersYou will then apply these two functions to build the model. 2.1 - Clipping the gradients in the optimization loopIn this section you will implement the `clip` function that you will call inside of your optimization loop. Recall that your overall loop structure usually consists of a forward pass, a cost computation, a backward pass, and a parameter update. Before updating the parameters, you will perform gradient clipping when needed to make sure that your gradients are not "exploding," meaning taking on overly large values. In the exercise below, you will implement a function `clip` that takes in a dictionary of gradients and returns a clipped version of gradients if needed. There are different ways to clip gradients; we will use a simple element-wise clipping procedure, in which every element of the gradient vector is clipped to lie between some range [-N, N]. More generally, you will provide a `maxValue` (say 10). In this example, if any component of the gradient vector is greater than 10, it would be set to 10; and if any component of the gradient vector is less than -10, it would be set to -10. If it is between -10 and 10, it is left alone. **Figure 2**: Visualization of gradient descent with and without gradient clipping, in a case where the network is running into slight "exploding gradient" problems. **Exercise**: Implement the function below to return the clipped gradients of your dictionary `gradients`. Your function takes in a maximum threshold and returns the clipped versions of your gradients. You can check out this [hint](https://docs.scipy.org/doc/numpy-1.13.0/reference/generated/numpy.clip.html) for examples of how to clip in numpy. You will need to use the argument `out = ...`.
###Code
### GRADED FUNCTION: clip
def clip(gradients, maxValue):
'''
Clips the gradients' values between minimum and maximum.
Arguments:
gradients -- a dictionary containing the gradients "dWaa", "dWax", "dWya", "db", "dby"
maxValue -- everything above this number is set to this number, and everything less than -maxValue is set to -maxValue
Returns:
gradients -- a dictionary with the clipped gradients.
'''
dWaa, dWax, dWya, db, dby = gradients['dWaa'], gradients['dWax'], gradients['dWya'], gradients['db'], gradients['dby']
### START CODE HERE ###
# clip to mitigate exploding gradients, loop over [dWax, dWaa, dWya, db, dby]. (≈2 lines)
for gradient in [dWax, dWaa, dWya, db, dby]:
np.clip(dWax, -maxValue, maxValue, out=dWax)
np.clip(dWaa, -maxValue, maxValue, out=dWaa)
np.clip(dWya, -maxValue, maxValue, out=dWya)
np.clip(db, -maxValue, maxValue, out=db)
np.clip(dby, -maxValue, maxValue, out=dby)
### END CODE HERE ###
gradients = {"dWaa": dWaa, "dWax": dWax, "dWya": dWya, "db": db, "dby": dby}
return gradients
np.random.seed(3)
dWax = np.random.randn(5,3)*10
dWaa = np.random.randn(5,5)*10
dWya = np.random.randn(2,5)*10
db = np.random.randn(5,1)*10
dby = np.random.randn(2,1)*10
gradients = {"dWax": dWax, "dWaa": dWaa, "dWya": dWya, "db": db, "dby": dby}
gradients = clip(gradients, 10)
print("gradients[\"dWaa\"][1][2] =", gradients["dWaa"][1][2])
print("gradients[\"dWax\"][3][1] =", gradients["dWax"][3][1])
print("gradients[\"dWya\"][1][2] =", gradients["dWya"][1][2])
print("gradients[\"db\"][4] =", gradients["db"][4])
print("gradients[\"dby\"][1] =", gradients["dby"][1])
###Output
gradients["dWaa"][1][2] = 10.0
gradients["dWax"][3][1] = -10.0
gradients["dWya"][1][2] = 0.29713815361
gradients["db"][4] = [ 10.]
gradients["dby"][1] = [ 8.45833407]
###Markdown
** Expected output:** **gradients["dWaa"][1][2] ** 10.0 **gradients["dWax"][3][1]** -10.0 **gradients["dWya"][1][2]** 0.29713815361 **gradients["db"][4]** [ 10.] **gradients["dby"][1]** [ 8.45833407] 2.2 - SamplingNow assume that your model is trained. You would like to generate new text (characters). The process of generation is explained in the picture below: **Figure 3**: In this picture, we assume the model is already trained. We pass in $x^{\langle 1\rangle} = \vec{0}$ at the first time step, and have the network then sample one character at a time. **Exercise**: Implement the `sample` function below to sample characters. You need to carry out 4 steps:- **Step 1**: Pass the network the first "dummy" input $x^{\langle 1 \rangle} = \vec{0}$ (the vector of zeros). This is the default input before we've generated any characters. We also set $a^{\langle 0 \rangle} = \vec{0}$- **Step 2**: Run one step of forward propagation to get $a^{\langle 1 \rangle}$ and $\hat{y}^{\langle 1 \rangle}$. Here are the equations:$$ a^{\langle t+1 \rangle} = \tanh(W_{ax} x^{\langle t \rangle } + W_{aa} a^{\langle t \rangle } + b)\tag{1}$$$$ z^{\langle t + 1 \rangle } = W_{ya} a^{\langle t + 1 \rangle } + b_y \tag{2}$$$$ \hat{y}^{\langle t+1 \rangle } = softmax(z^{\langle t + 1 \rangle })\tag{3}$$Note that $\hat{y}^{\langle t+1 \rangle }$ is a (softmax) probability vector (its entries are between 0 and 1 and sum to 1). $\hat{y}^{\langle t+1 \rangle}_i$ represents the probability that the character indexed by "i" is the next character. We have provided a `softmax()` function that you can use.- **Step 3**: Carry out sampling: Pick the next character's index according to the probability distribution specified by $\hat{y}^{\langle t+1 \rangle }$. This means that if $\hat{y}^{\langle t+1 \rangle }_i = 0.16$, you will pick the index "i" with 16% probability. To implement it, you can use [`np.random.choice`](https://docs.scipy.org/doc/numpy-1.13.0/reference/generated/numpy.random.choice.html).Here is an example of how to use `np.random.choice()`:```pythonnp.random.seed(0)p = np.array([0.1, 0.0, 0.7, 0.2])index = np.random.choice([0, 1, 2, 3], p = p.ravel())```This means that you will pick the `index` according to the distribution: $P(index = 0) = 0.1, P(index = 1) = 0.0, P(index = 2) = 0.7, P(index = 3) = 0.2$.- **Step 4**: The last step to implement in `sample()` is to overwrite the variable `x`, which currently stores $x^{\langle t \rangle }$, with the value of $x^{\langle t + 1 \rangle }$. You will represent $x^{\langle t + 1 \rangle }$ by creating a one-hot vector corresponding to the character you've chosen as your prediction. You will then forward propagate $x^{\langle t + 1 \rangle }$ in Step 1 and keep repeating the process until you get a "\n" character, indicating you've reached the end of the dinosaur name.
###Code
# GRADED FUNCTION: sample
def sample(parameters, char_to_ix, seed):
"""
Sample a sequence of characters according to a sequence of probability distributions output of the RNN
Arguments:
parameters -- python dictionary containing the parameters Waa, Wax, Wya, by, and b.
char_to_ix -- python dictionary mapping each character to an index.
seed -- used for grading purposes. Do not worry about it.
Returns:
indices -- a list of length n containing the indices of the sampled characters.
"""
# Retrieve parameters and relevant shapes from "parameters" dictionary
Waa, Wax, Wya, by, b = parameters['Waa'], parameters['Wax'], parameters['Wya'], parameters['by'], parameters['b']
vocab_size = by.shape[0]
n_a = Waa.shape[1]
### START CODE HERE ###
# Step 1: Create the one-hot vector x for the first character (initializing the sequence generation). (≈1 line)
x = np.zeros((vocab_size, 1))
# Step 1': Initialize a_prev as zeros (≈1 line)
a_prev = np.zeros((n_a, 1))
# Create an empty list of indices, this is the list which will contain the list of indices of the characters to generate (≈1 line)
indices = []
# Idx is a flag to detect a newline character, we initialize it to -1
idx = -1
# Loop over time-steps t. At each time-step, sample a character from a probability distribution and append
# its index to "indices". We'll stop if we reach 50 characters (which should be very unlikely with a well
# trained model), which helps debugging and prevents entering an infinite loop.
counter = 0
newline_character = char_to_ix['\n']
while (idx != newline_character and counter != 50):
# Step 2: Forward propagate x using the equations (1), (2) and (3)
a = np.tanh(Wax @ x + Waa @ a_prev + b)
z = Wya @ a + by
y = softmax(z)
# for grading purposes
np.random.seed(counter+seed)
# Step 3: Sample the index of a character within the vocabulary from the probability distribution y
idx = np.random.choice(np.arange(vocab_size), p=y.ravel())
# Append the index to "indices"
indices.append(idx)
# Step 4: Overwrite the input character as the one corresponding to the sampled index.
x = np.zeros((vocab_size, 1))
x[idx] = 1
# Update "a_prev" to be "a"
a_prev = a
# for grading purposes
seed += 1
counter +=1
### END CODE HERE ###
if (counter == 50):
indices.append(char_to_ix['\n'])
return indices
np.random.seed(2)
_, n_a = 20, 100
Wax, Waa, Wya = np.random.randn(n_a, vocab_size), np.random.randn(n_a, n_a), np.random.randn(vocab_size, n_a)
b, by = np.random.randn(n_a, 1), np.random.randn(vocab_size, 1)
parameters = {"Wax": Wax, "Waa": Waa, "Wya": Wya, "b": b, "by": by}
indices = sample(parameters, char_to_ix, 0)
print("Sampling:")
print("list of sampled indices:", indices)
print("list of sampled characters:", [ix_to_char[i] for i in indices])
###Output
Sampling:
list of sampled indices: [12, 17, 24, 14, 13, 9, 10, 22, 24, 6, 13, 11, 12, 6, 21, 15, 21, 14, 3, 2, 1, 21, 18, 24, 7, 25, 6, 25, 18, 10, 16, 2, 3, 8, 15, 12, 11, 7, 1, 12, 10, 2, 7, 7, 11, 5, 6, 12, 25, 0, 0]
list of sampled characters: ['l', 'q', 'x', 'n', 'm', 'i', 'j', 'v', 'x', 'f', 'm', 'k', 'l', 'f', 'u', 'o', 'u', 'n', 'c', 'b', 'a', 'u', 'r', 'x', 'g', 'y', 'f', 'y', 'r', 'j', 'p', 'b', 'c', 'h', 'o', 'l', 'k', 'g', 'a', 'l', 'j', 'b', 'g', 'g', 'k', 'e', 'f', 'l', 'y', '\n', '\n']
###Markdown
** Expected output:** **list of sampled indices:** [12, 17, 24, 14, 13, 9, 10, 22, 24, 6, 13, 11, 12, 6, 21, 15, 21, 14, 3, 2, 1, 21, 18, 24, 7, 25, 6, 25, 18, 10, 16, 2, 3, 8, 15, 12, 11, 7, 1, 12, 10, 2, 7, 7, 11, 5, 6, 12, 25, 0, 0] **list of sampled characters:** ['l', 'q', 'x', 'n', 'm', 'i', 'j', 'v', 'x', 'f', 'm', 'k', 'l', 'f', 'u', 'o', 'u', 'n', 'c', 'b', 'a', 'u', 'r', 'x', 'g', 'y', 'f', 'y', 'r', 'j', 'p', 'b', 'c', 'h', 'o', 'l', 'k', 'g', 'a', 'l', 'j', 'b', 'g', 'g', 'k', 'e', 'f', 'l', 'y', '\n', '\n'] 3 - Building the language model It is time to build the character-level language model for text generation. 3.1 - Gradient descent In this section you will implement a function performing one step of stochastic gradient descent (with clipped gradients). You will go through the training examples one at a time, so the optimization algorithm will be stochastic gradient descent. As a reminder, here are the steps of a common optimization loop for an RNN:- Forward propagate through the RNN to compute the loss- Backward propagate through time to compute the gradients of the loss with respect to the parameters- Clip the gradients if necessary - Update your parameters using gradient descent **Exercise**: Implement this optimization process (one step of stochastic gradient descent). We provide you with the following functions: ```pythondef rnn_forward(X, Y, a_prev, parameters): """ Performs the forward propagation through the RNN and computes the cross-entropy loss. It returns the loss' value as well as a "cache" storing values to be used in the backpropagation.""" .... return loss, cache def rnn_backward(X, Y, parameters, cache): """ Performs the backward propagation through time to compute the gradients of the loss with respect to the parameters. It returns also all the hidden states.""" ... return gradients, adef update_parameters(parameters, gradients, learning_rate): """ Updates parameters using the Gradient Descent Update Rule.""" ... return parameters```
###Code
# GRADED FUNCTION: optimize
def optimize(X, Y, a_prev, parameters, learning_rate=0.01):
"""
Execute one step of the optimization to train the model.
Arguments:
X -- list of integers, where each integer is a number that maps to a character in the vocabulary.
Y -- list of integers, exactly the same as X but shifted one index to the left.
a_prev -- previous hidden state.
parameters -- python dictionary containing:
Wax -- Weight matrix multiplying the input, numpy array of shape (n_a, n_x)
Waa -- Weight matrix multiplying the hidden state, numpy array of shape (n_a, n_a)
Wya -- Weight matrix relating the hidden-state to the output, numpy array of shape (n_y, n_a)
b -- Bias, numpy array of shape (n_a, 1)
by -- Bias relating the hidden-state to the output, numpy array of shape (n_y, 1)
learning_rate -- learning rate for the model.
Returns:
loss -- value of the loss function (cross-entropy)
gradients -- python dictionary containing:
dWax -- Gradients of input-to-hidden weights, of shape (n_a, n_x)
dWaa -- Gradients of hidden-to-hidden weights, of shape (n_a, n_a)
dWya -- Gradients of hidden-to-output weights, of shape (n_y, n_a)
db -- Gradients of bias vector, of shape (n_a, 1)
dby -- Gradients of output bias vector, of shape (n_y, 1)
a[len(X)-1] -- the last hidden state, of shape (n_a, 1)
"""
### START CODE HERE ###
# Forward propagate through time (≈1 line)
loss, cache = rnn_forward(X, Y, a_prev, parameters)
# Backpropagate through time (≈1 line)
gradients, a = rnn_backward(X, Y, parameters, cache)
# Clip your gradients between -5 (min) and 5 (max) (≈1 line)
gradients = clip(gradients, 5)
# Update parameters (≈1 line)
parameters = update_parameters(parameters, gradients, learning_rate)
### END CODE HERE ###
return loss, gradients, a[len(X)-1]
np.random.seed(1)
vocab_size, n_a = 27, 100
a_prev = np.random.randn(n_a, 1)
Wax, Waa, Wya = np.random.randn(n_a, vocab_size), np.random.randn(n_a, n_a), np.random.randn(vocab_size, n_a)
b, by = np.random.randn(n_a, 1), np.random.randn(vocab_size, 1)
parameters = {"Wax": Wax, "Waa": Waa, "Wya": Wya, "b": b, "by": by}
X = [12,3,5,11,22,3]
Y = [4,14,11,22,25, 26]
loss, gradients, a_last = optimize(X, Y, a_prev, parameters, learning_rate = 0.01)
print("Loss =", loss)
print("gradients[\"dWaa\"][1][2] =", gradients["dWaa"][1][2])
print("np.argmax(gradients[\"dWax\"]) =", np.argmax(gradients["dWax"]))
print("gradients[\"dWya\"][1][2] =", gradients["dWya"][1][2])
print("gradients[\"db\"][4] =", gradients["db"][4])
print("gradients[\"dby\"][1] =", gradients["dby"][1])
print("a_last[4] =", a_last[4])
###Output
Loss = 126.503975722
gradients["dWaa"][1][2] = 0.194709315347
np.argmax(gradients["dWax"]) = 93
gradients["dWya"][1][2] = -0.007773876032
gradients["db"][4] = [-0.06809825]
gradients["dby"][1] = [ 0.01538192]
a_last[4] = [-1.]
###Markdown
** Expected output:** **Loss ** 126.503975722 **gradients["dWaa"][1][2]** 0.194709315347 **np.argmax(gradients["dWax"])** 93 **gradients["dWya"][1][2]** -0.007773876032 **gradients["db"][4]** [-0.06809825] **gradients["dby"][1]** [ 0.01538192] **a_last[4]** [-1.] 3.2 - Training the model Given the dataset of dinosaur names, we use each line of the dataset (one name) as one training example. Every 100 steps of stochastic gradient descent, you will sample 10 randomly chosen names to see how the algorithm is doing. Remember to shuffle the dataset, so that stochastic gradient descent visits the examples in random order. **Exercise**: Follow the instructions and implement `model()`. When `examples[index]` contains one dinosaur name (string), to create an example (X, Y), you can use this:```python index = j % len(examples) X = [None] + [char_to_ix[ch] for ch in examples[index]] Y = X[1:] + [char_to_ix["\n"]]```Note that we use: `index= j % len(examples)`, where `j = 1....num_iterations`, to make sure that `examples[index]` is always a valid statement (`index` is smaller than `len(examples)`).The first entry of `X` being `None` will be interpreted by `rnn_forward()` as setting $x^{\langle 0 \rangle} = \vec{0}$. Further, this ensures that `Y` is equal to `X` but shifted one step to the left, and with an additional "\n" appended to signify the end of the dinosaur name.
###Code
# GRADED FUNCTION: model
def model(data, ix_to_char, char_to_ix, num_iterations=35000, n_a=50, dino_names=7, vocab_size=27):
"""
Trains the model and generates dinosaur names.
Arguments:
data -- text corpus
ix_to_char -- dictionary that maps the index to a character
char_to_ix -- dictionary that maps a character to an index
num_iterations -- number of iterations to train the model for
n_a -- number of units of the RNN cell
dino_names -- number of dinosaur names you want to sample at each iteration.
vocab_size -- number of unique characters found in the text, size of the vocabulary
Returns:
parameters -- learned parameters
"""
# Retrieve n_x and n_y from vocab_size
n_x, n_y = vocab_size, vocab_size
# Initialize parameters
parameters = initialize_parameters(n_a, n_x, n_y)
# Initialize loss (this is required because we want to smooth our loss, don't worry about it)
loss = get_initial_loss(vocab_size, dino_names)
# Build list of all dinosaur names (training examples).
with open("dinos.txt") as f:
examples = f.readlines()
examples = [x.lower().strip() for x in examples]
# Shuffle list of all dinosaur names
np.random.seed(0)
np.random.shuffle(examples)
# Initialize the hidden state of your LSTM
a_prev = np.zeros((n_a, 1))
# Optimization loop
for j in range(num_iterations):
### START CODE HERE ###
# Use the hint above to define one training example (X,Y) (≈ 2 lines)
index = j % len(examples)
X = [None] + [char_to_ix[ch] for ch in examples[index]]
Y = X[1:] + [char_to_ix["\n"]]
# Perform one optimization step: Forward-prop -> Backward-prop -> Clip -> Update parameters
# Choose a learning rate of 0.01
curr_loss, gradients, a_prev = optimize(X, Y, a_prev, parameters)
### END CODE HERE ###
# Use a latency trick to keep the loss smooth. It happens here to accelerate the training.
loss = smooth(loss, curr_loss)
# Every 2000 Iteration, generate "n" characters thanks to sample() to check if the model is learning properly
if j % 2000 == 0:
print('Iteration: %d, Loss: %f' % (j, loss) + '\n')
# The number of dinosaur names to print
seed = 0
for name in range(dino_names):
# Sample indices and print them
sampled_indices = sample(parameters, char_to_ix, seed)
print_sample(sampled_indices, ix_to_char)
seed += 1 # To get the same result for grading purposed, increment the seed by one.
print('\n')
return parameters
###Output
_____no_output_____
###Markdown
Run the following cell, you should observe your model outputting random-looking characters at the first iteration. After a few thousand iterations, your model should learn to generate reasonable-looking names.
###Code
parameters = model(data, ix_to_char, char_to_ix)
###Output
Iteration: 0, Loss: 23.087336
Nkzxwtdmfqoeyhsqwasjkjvu
Kneb
Kzxwtdmfqoeyhsqwasjkjvu
Neb
Zxwtdmfqoeyhsqwasjkjvu
Eb
Xwtdmfqoeyhsqwasjkjvu
Iteration: 2000, Loss: 27.884160
Liusskeomnolxeros
Hmdaairus
Hytroligoraurus
Lecalosapaus
Xusicikoraurus
Abalpsamantisaurus
Tpraneronxeros
Iteration: 4000, Loss: 25.901815
Mivrosaurus
Inee
Ivtroplisaurus
Mbaaisaurus
Wusichisaurus
Cabaselachus
Toraperlethosdarenitochusthiamamumamaon
Iteration: 6000, Loss: 24.608779
Onwusceomosaurus
Lieeaerosaurus
Lxussaurus
Oma
Xusteonosaurus
Eeahosaurus
Toreonosaurus
Iteration: 8000, Loss: 24.070350
Onxusichepriuon
Kilabersaurus
Lutrodon
Omaaerosaurus
Xutrcheps
Edaksoje
Trodiktonus
Iteration: 10000, Loss: 23.844446
Onyusaurus
Klecalosaurus
Lustodon
Ola
Xusodonia
Eeaeosaurus
Troceosaurus
Iteration: 12000, Loss: 23.291971
Onyxosaurus
Kica
Lustrepiosaurus
Olaagrraiansaurus
Yuspangosaurus
Eealosaurus
Trognesaurus
Iteration: 14000, Loss: 23.382339
Meutromodromurus
Inda
Iutroinatorsaurus
Maca
Yusteratoptititan
Ca
Troclosaurus
Iteration: 16000, Loss: 23.288447
Meuspsangosaurus
Ingaa
Iusosaurus
Macalosaurus
Yushanis
Daalosaurus
Trpandon
Iteration: 18000, Loss: 22.823526
Phytrolonhonyg
Mela
Mustrerasaurus
Peg
Ytronorosaurus
Ehalosaurus
Trolomeehus
Iteration: 20000, Loss: 23.041871
Nousmofonosaurus
Loma
Lytrognatiasaurus
Ngaa
Ytroenetiaudostarmilus
Eiafosaurus
Troenchulunosaurus
Iteration: 22000, Loss: 22.728849
Piutyrangosaurus
Midaa
Myroranisaurus
Pedadosaurus
Ytrodon
Eiadosaurus
Trodoniomusitocorces
Iteration: 24000, Loss: 22.683403
Meutromeisaurus
Indeceratlapsaurus
Jurosaurus
Ndaa
Yusicheropterus
Eiaeropectus
Trodonasaurus
Iteration: 26000, Loss: 22.554523
Phyusaurus
Liceceron
Lyusichenodylus
Pegahus
Yustenhtonthosaurus
Elagosaurus
Trodontonsaurus
Iteration: 28000, Loss: 22.484472
Onyutimaerihus
Koia
Lytusaurus
Ola
Ytroheltorus
Eiadosaurus
Trofiashates
Iteration: 30000, Loss: 22.774404
Phytys
Lica
Lysus
Pacalosaurus
Ytrochisaurus
Eiacosaurus
Trochesaurus
Iteration: 32000, Loss: 22.209473
Mawusaurus
Jica
Lustoia
Macaisaurus
Yusolenqtesaurus
Eeaeosaurus
Trnanatrax
Iteration: 34000, Loss: 22.396744
Mavptokekus
Ilabaisaurus
Itosaurus
Macaesaurus
Yrosaurus
Eiaeosaurus
Trodon
###Markdown
ConclusionYou can see that your algorithm has started to generate plausible dinosaur names towards the end of the training. At first, it was generating random characters, but towards the end you could see dinosaur names with cool endings. Feel free to run the algorithm even longer and play with hyperparameters to see if you can get even better results. Our implemetation generated some really cool names like `maconucon`, `marloralus` and `macingsersaurus`. Your model hopefully also learned that dinosaur names tend to end in `saurus`, `don`, `aura`, `tor`, etc.If your model generates some non-cool names, don't blame the model entirely--not all actual dinosaur names sound cool. (For example, `dromaeosauroides` is an actual dinosaur name and is in the training set.) But this model should give you a set of candidates from which you can pick the coolest! This assignment had used a relatively small dataset, so that you could train an RNN quickly on a CPU. Training a model of the english language requires a much bigger dataset, and usually needs much more computation, and could run for many hours on GPUs. We ran our dinosaur name for quite some time, and so far our favoriate name is the great, undefeatable, and fierce: Mangosaurus! 4 - Writing like ShakespeareThe rest of this notebook is optional and is not graded, but we hope you'll do it anyway since it's quite fun and informative. A similar (but more complicated) task is to generate Shakespeare poems. Instead of learning from a dataset of Dinosaur names you can use a collection of Shakespearian poems. Using LSTM cells, you can learn longer term dependencies that span many characters in the text--e.g., where a character appearing somewhere a sequence can influence what should be a different character much much later in ths sequence. These long term dependencies were less important with dinosaur names, since the names were quite short. Let's become poets! We have implemented a Shakespeare poem generator with Keras. Run the following cell to load the required packages and models. This may take a few minutes.
###Code
from __future__ import print_function
from keras.callbacks import LambdaCallback
from keras.models import Model, load_model, Sequential
from keras.layers import Dense, Activation, Dropout, Input, Masking
from keras.layers import LSTM
from keras.utils.data_utils import get_file
from keras.preprocessing.sequence import pad_sequences
from shakespeare_utils import *
import sys
import io
###Output
Using TensorFlow backend.
###Markdown
To save you some time, we have already trained a model for ~1000 epochs on a collection of Shakespearian poems called [*"The Sonnets"*](shakespeare.txt). Let's train the model for one more epoch. When it finishes training for an epoch---this will also take a few minutes---you can run `generate_output`, which will prompt asking you for an input (`<`40 characters). The poem will start with your sentence, and our RNN-Shakespeare will complete the rest of the poem for you! For example, try "Forsooth this maketh no sense " (don't enter the quotation marks). Depending on whether you include the space at the end, your results might also differ--try it both ways, and try other inputs as well.
###Code
print_callback = LambdaCallback(on_epoch_end=on_epoch_end)
model.fit(x, y, batch_size=128, epochs=1, callbacks=[print_callback])
# Run this cell to try with different inputs without having to re-train the model
generate_output()
###Output
Write the beginning of your poem, the Shakespeare machine will complete it. Your input is: Where art thou potato?
Here is your poem:
Where art thou potato?.
to my erven thou worl-nand, ir madess heaver glote.
enthing oud puy which you gey thin me thee,
oul tof me leer be habrys are waknas,
ay of not be to whill beh for be for hhors,
handined the offecter levy pobned evilong still,
hawe of mone fair say thene keen
at mey my ellend lime the poow and dear.
comh pirns anf leider enthrobe's strence's deleds,
on that spalling not fiblf pa these thene,
|
Matrix_Factorization_RecSys_Baseline.ipynb
|
###Markdown
###Code
import torch
import torch.nn as nn
import torch.nn.functional as F
from pathlib import Path
import pandas as pd
import numpy as np
import os
# !git clone "https://github.com/abdurahman02/ml-latest-small.git"
# os.chdir("ml-latest-small")
os.listdir()
data = pd.read_csv("ratings.csv")
data.head()
# split train and validation before encoding
np.random.seed(3)
msk = np.random.rand(len(data)) < 0.8
train = data[msk].copy()
val = data[~msk].copy()
# here is a handy function modified from fast.ai
def proc_col(col, train_col=None):
"""Encodes a pandas column with continous ids.
"""
if train_col is not None:
uniq = train_col.unique()
else:
uniq = col.unique()
name2idx = {o:i for i,o in enumerate(uniq)}
return name2idx, np.array([name2idx.get(x, -1) for x in col]), len(uniq)
def encode_data(df, train=None):
""" Encodes rating data with continous user and movie ids.
If train is provided, encodes df with the same encoding as train.
"""
df = df.copy()
for col_name in ["userId", "movieId"]:
train_col = None
if train is not None:
train_col = train[col_name]
_,col,_ = proc_col(df[col_name], train_col)
df[col_name] = col
df = df[df[col_name] >= 0]
return df
# encoding the train and validation data
df_train = encode_data(train)
df_val = encode_data(val, train)
num_users = len(df_train.userId.unique())
num_items = len(df_train.movieId.unique())
print(num_users, num_items)
class MF(nn.Module):
def __init__(self, num_users, num_items, emb_size=100):
super(MF, self).__init__()
self.user_emb = nn.Embedding(num_users, emb_size)
self.item_emb = nn.Embedding(num_items, emb_size)
self.user_emb.weight.data.uniform_(0, 0.05)
self.item_emb.weight.data.uniform_(0, 0.05)
def forward(self, u, v):
u = self.user_emb(u)
v = self.item_emb(v)
return (u*v).sum(1)
if torch.cuda.is_available():
model = MF(num_users, num_items, emb_size=100).cuda()
else:
model = MF(num_users, num_items, emb_size=100)
def test_loss(model, unsqueeze=False):
model.eval()
if torch.cuda.is_available():
users = torch.LongTensor(df_val.userId.values).cuda()
items = torch.LongTensor(df_val.movieId.values).cuda()
ratings = torch.FloatTensor(df_val.rating.values).cuda()
else:
users = torch.LongTensor(df_val.userId.values)
items = torch.LongTensor(df_val.movieId.values)
ratings = torch.FloatTensor(df_val.rating.values)
if unsqueeze:
ratings = ratings.unsqueeze(1)
y_hat = model(users, items)
loss = F.mse_loss(y_hat, ratings)
print("test loss %.3f " % loss.item())
def train_epocs(model, epochs=10, lr=0.01, wd=0.0, unsqueeze=False):
optimizer = torch.optim.Adam(model.parameters(), lr=lr, weight_decay=wd)
model.train()
for i in range(epochs):
if torch.cuda.is_available():
users = torch.LongTensor(df_train.userId.values).cuda()
items = torch.LongTensor(df_train.movieId.values).cuda()
ratings = torch.FloatTensor(df_train.rating.values).cuda()
else:
users = torch.LongTensor(df_train.userId.values) # .cuda()
items = torch.LongTensor(df_train.movieId.values) #.cuda()
ratings = torch.FloatTensor(df_train.rating.values) #.cuda()
if unsqueeze:
ratings = ratings.unsqueeze(1)
y_hat = model(users, items)
loss = F.mse_loss(y_hat, ratings)
optimizer.zero_grad()
loss.backward()
optimizer.step()
print(loss.item())
test_loss(model, unsqueeze)
# Here is what unsqueeze does
ratings = torch.FloatTensor(df_train.rating.values)
print(ratings.shape)
ratings = ratings.unsqueeze(1) # .cuda()
print(ratings.shape)
train_epocs(model, epochs=10, lr=0.1)
train_epocs(model, epochs=15, lr=0.01)
train_epocs(model, epochs=15, lr=0.01)
class MF_bias(nn.Module):
def __init__(self, num_users, num_items, emb_size=100):
super(MF_bias, self).__init__()
self.user_emb = nn.Embedding(num_users, emb_size)
self.user_bias = nn.Embedding(num_users, 1)
self.item_emb = nn.Embedding(num_items, emb_size)
self.item_bias = nn.Embedding(num_items, 1)
self.user_emb.weight.data.uniform_(0,0.05)
self.item_emb.weight.data.uniform_(0,0.05)
self.user_bias.weight.data.uniform_(-0.01,0.01)
self.item_bias.weight.data.uniform_(-0.01,0.01)
def forward(self, u, v):
U = self.user_emb(u)
V = self.item_emb(v)
b_u = self.user_bias(u).squeeze()
b_v = self.item_bias(v).squeeze()
return (U*V).sum(1) + b_u + b_v
model = MF_bias(num_users, num_items, emb_size=100) #.cuda()
train_epocs(model, epochs=10, lr=0.05, wd=1e-5)
train_epocs(model, epochs=10, lr=0.01, wd=1e-5)
train_epocs(model, epochs=10, lr=0.001, wd=1e-5)
###Output
_____no_output_____
|
Hierarchical Clustering Assignment/Hierarchical Clustering.ipynb
|
###Markdown
Imports
###Code
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
import scipy.cluster.hierarchy as shc
get_ipython().run_line_magic('matplotlib', 'inline')
from sklearn.preprocessing import normalize
from sklearn.cluster import AgglomerativeClustering
###Output
_____no_output_____
###Markdown
Dataset
###Code
data = pd.read_csv('Wholesale customers data.csv')
data.head()
data.shape
data_scaled = normalize(data)
data_scaled = pd.DataFrame(data_scaled, columns=data.columns)
data_scaled.head()
###Output
_____no_output_____
###Markdown
Dendogram
###Code
plt.figure(figsize=(10, 7))
plt.title("Dendrograms")
dend = shc.dendrogram(shc.linkage(data_scaled, method='ward'))
plt.axhline(y=6, color='r', linestyle='--')
###Output
_____no_output_____
###Markdown
Clusters
###Code
cluster = AgglomerativeClustering(n_clusters=2, affinity='euclidean', linkage='ward')
cluster.fit_predict(data_scaled)
plt.figure(figsize=(10, 7))
plt.scatter(data_scaled['Milk'], data_scaled['Grocery'], c=cluster.labels_)
###Output
_____no_output_____
|
2019_08_12/PCA/.ipynb_checkpoints/PCA-checkpoint.ipynb
|
###Markdown
参考资料- 文字资料: https://towardsdatascience.com/pca-using-python-scikit-learn-e653f8989e60- 代码资料: https://github.com/mGalarnyk/Python_Tutorials/blob/master/Sklearn/PCA/PCA_Data_Visualization_Iris_Dataset_Blog.ipynb- sklearn PCA使用指南: https://scikit-learn.org/stable/modules/generated/sklearn.decomposition.PCA.htmlsklearn.decomposition.PCA 读取数据
###Code
df = pd.read_csv('./iris.csv', names=['sepal length','sepal width','petal length','petal width','target'])
df.head()
df.shape
###Output
_____no_output_____
###Markdown
数据标准化- PCA is effected by scale so you need to scale the features in your data before applying PCA.(PCA会被数据的大小所影响, 所以在做之前, 我们需要先对数据进行标准化)- 我们使用StandardScaler进行标准化, 标准化之后变为均值为0, 方差为1的数据
###Code
features = ['sepal length', 'sepal width', 'petal length', 'petal width']
# Separating out the features
x = df.loc[:, features].values
# Separating out the target
y = df.loc[:,['target']].values
# Standardizing the features
x = StandardScaler().fit_transform(x)
# 查看标准化之后的数据
pd.DataFrame(data = x, columns = features).head()
###Output
_____no_output_____
###Markdown
查看降维后的维数- 选择降到多少维度比较合适
###Code
pca = PCA(n_components=4)
principalComponents = pca.fit_transform(x)
pca.explained_variance_ratio_
# 进行可视化
importance = pca.explained_variance_ratio_
plt.scatter(range(1,5),importance)
plt.plot(range(1,5),importance)
plt.title('Scree Plot')
plt.xlabel('Factors')
plt.ylabel('Eigenvalue')
plt.grid()
plt.show()
###Output
_____no_output_____
###Markdown
使用PCA降维(降维到2维)- 降维到2维可以方便进行可视化
###Code
pca = PCA(n_components=2)
principalComponents = pca.fit_transform(x)
# 降维后的新数据
principalDf = pd.DataFrame(data=principalComponents, columns=['principal component 1', 'principal component 2'])
finalDf = pd.concat([principalDf, df[['target']]], axis = 1)
finalDf.head(5)
pca.explained_variance_ratio_
pca.explained_variance_
###Output
_____no_output_____
###Markdown
得到转换的关系(系数)
###Code
pca.components_
# 这个计算的值就是上面生成的值, 这个相当于是系数
(np.dot(x[0],pca.components_[0]), np.dot(x[0],pca.components_[1]))
# 对系数进行可视化
import seaborn as sns
df_cm = pd.DataFrame(np.abs(pca.components_), columns=df.columns[:-1])
plt.figure(figsize = (12,6))
ax = sns.heatmap(df_cm, annot=True, cmap="BuPu")
# 设置y轴的字体的大小
ax.yaxis.set_tick_params(labelsize=15)
ax.xaxis.set_tick_params(labelsize=15)
plt.title('PCA', fontsize='xx-large')
# Set y-axis label
plt.savefig('factorAnalysis.png', dpi=200)
###Output
_____no_output_____
###Markdown
进行可视化
###Code
fig = plt.figure(figsize = (8,8))
ax = fig.add_subplot(1,1,1)
ax.set_xlabel('Principal Component 1', fontsize = 15)
ax.set_ylabel('Principal Component 2', fontsize = 15)
ax.set_title('2 Component PCA', fontsize = 20)
targets = ['Iris-setosa', 'Iris-versicolor', 'Iris-virginica']
colors = ['r', 'g', 'b']
for target, color in zip(targets,colors):
indicesToKeep = finalDf['target'] == target
# 选择某个label下的数据进行绘制
ax.scatter(finalDf.loc[indicesToKeep, 'principal component 1']
, finalDf.loc[indicesToKeep, 'principal component 2']
, c = color
, s = 50)
ax.legend(targets)
ax.grid()
###Output
_____no_output_____
###Markdown
做一下FA的实验
###Code
from factor_analyzer import FactorAnalyzer
# 查看是否适合做因子分析
from factor_analyzer.factor_analyzer import calculate_bartlett_sphericity
chi_square_value,p_value=calculate_bartlett_sphericity(x)
chi_square_value, p_value
# Create factor analysis object and perform factor analysis
fa = FactorAnalyzer(4, rotation=None)
fa.fit(x)
# Check Eigenvalues
ev, v = fa.get_eigenvalues()
# Create scree plot using matplotlib
plt.scatter(range(1,4+1),ev)
plt.plot(range(1,4+1),ev)
plt.title('Scree Plot')
plt.xlabel('Factors')
plt.ylabel('Eigenvalue')
plt.grid()
plt.show()
# 降到2维
fa = FactorAnalyzer(2, rotation="varimax")
fa.fit(x)
# 对系数矩阵进行可视化
df_cm = pd.DataFrame(np.abs(fa.loadings_), index=df.columns[:-1])
plt.figure(figsize = (7,10))
ax = sns.heatmap(df_cm, annot=True, cmap="BuPu")
# 设置y轴的字体的大小
ax.yaxis.set_tick_params(labelsize=15)
plt.title('Factor Analysis', fontsize='xx-large')
# Set y-axis label
plt.ylabel('Sepal Width', fontsize='xx-large')
plt.savefig('factorAnalysis.png', dpi=200)
# 画出降维后的图像
x_2 = fa.transform(x)
# 降维后的新数据
principalDf = pd.DataFrame(data=x_2, columns=['FA 1', 'FA 2'])
finalDf = pd.concat([principalDf, df[['target']]], axis = 1)
finalDf.head(5)
fig = plt.figure(figsize = (8,8))
ax = fig.add_subplot(1,1,1)
ax.set_xlabel('FA 1', fontsize = 15)
ax.set_ylabel('FA 2', fontsize = 15)
ax.set_title('2 Component FA', fontsize = 20)
targets = ['Iris-setosa', 'Iris-versicolor', 'Iris-virginica']
colors = ['r', 'g', 'b']
for target, color in zip(targets,colors):
indicesToKeep = finalDf['target'] == target
# 选择某个label下的数据进行绘制
ax.scatter(finalDf.loc[indicesToKeep, 'FA 1']
, finalDf.loc[indicesToKeep, 'FA 2']
, c = color
, s = 50)
ax.legend(targets)
ax.grid()
###Output
_____no_output_____
|
pipeline/reference_data.ipynb
|
###Markdown
Reference data standardizationThis module provides reference data download, indexing and preprocessing (if necessary), in preparation for use throughout the pipeline.We have included the PDF document compiled by data standardization subgroup in the [minimal working example folder on Google Drive](https://drive.google.com/file/d/1R5sw5o8vqk_mbQQb4CGmtH3ldu1T3Vu0/view?usp=sharing). It contains the reference data to use for the project. OverviewThis module is based on the [TOPMed workflow from Broad](https://github.com/broadinstitute/gtex-pipeline/blob/master/TOPMed_RNAseq_pipeline.md).Workflows implemented include: Convert transcript feature file gff3 to gtf- Input: an uncompressed gff3 file.(i.e. can be view via cat)- Output: a gtf file. Collapse transcript features into genes- Input: a gtf file.- Output: a gtf file with collapesed gene model. Generate STAR index based on gtf and reference fasta- Input: a gtf file and an acompanying fasta file.- Output: A folder of STAR index. Generate RSEM index based on gtf and reference fasta- Input: a gtf file and an acompanying fasta file.- Output: A folder of RSEM index. Example commandsTo download reference data:
###Code
sos run reference_data.ipynb download_hg_reference --cwd reference_data
sos run reference_data.ipynb download_gene_annotation --cwd reference_data
sos run reference_data.ipynb download_ercc_reference --cwd reference_data
###Output
_____no_output_____
###Markdown
To format reference data:
###Code
sos run reference_data.ipynb hg_reference \
--cwd reference_data \
--ercc-reference reference_data/ERCC92.fa \
--hg-reference reference_data/GRCh38_full_analysis_set_plus_decoy_hla.fa \
--container container/rna_quantification.sif
sos run pipeline/reference_data.ipynb hg_gtf \
--cwd reference_data \
--hg-gtf /mnt/mfs/statgen/snuc_pseudo_bulk/data/reference_data/genes.gtf \
--hg-reference data/reference_data/GRCh38_full_analysis_set_plus_decoy_hla.noALT_noHLA_noDecoy_ERCC.fasta \
--containers containers/rna_quantification.sif -J 1 -q csg -c csg.yml &
###Output
_____no_output_____
###Markdown
To format gene feature data:
###Code
sos run reference_data.ipynb gene_annotation \
--cwd reference_data \
--ercc-gtf reference_data/ERCC92.gtf \
--hg-gtf reference_data/Homo_sapiens.GRCh38.103.chr.gtf \
--hg-reference reference_data/GRCh38_full_analysis_set_plus_decoy_hla.noALT_noHLA_noDecoy_ERCC.fasta \
--container container/rna_quantification.sif
###Output
_____no_output_____
###Markdown
**Notice that for stranded RNA-seq protocol please add `--is-stranded` to the command above. More details can be found later in the document.** To generate STAR index using the GTF annotation file before gene model collapse:
###Code
sos run reference_data.ipynb STAR_index \
--cwd reference_data \
--hg-reference reference_data/GRCh38_full_analysis_set_plus_decoy_hla.noALT_noHLA_noDecoy_ERCC.fasta \
--hg-gtf reference_data/Homo_sapiens.GRCh38.103.chr.reformatted.ERCC.gtf \
--container container/rna_quantification.sif \
--mem 40G
###Output
_____no_output_____
###Markdown
**Notice that command above requires at least 40G of memory, and takes quite a while to complete**. To generate RSEM index:
###Code
sos run reference_data.ipynb RSEM_index \
--cwd reference_data \
--hg-reference reference_data/GRCh38_full_analysis_set_plus_decoy_hla.noALT_noHLA_noDecoy_ERCC.fasta \
--hg-gtf reference_data/Homo_sapiens.GRCh38.103.chr.reformatted.ERCC.gtf \
--container container/rna_quantification.sif \
--mem 40G
###Output
_____no_output_____
###Markdown
Command interface
###Code
sos run reference_data.ipynb -h
[global]
# The output directory for generated files.
parameter: cwd = path
# For cluster jobs, number commands to run per job
parameter: job_size = 1
# Wall clock time expected
parameter: walltime = "5h"
# Memory expected
parameter: mem = "16G"
# Number of threads
parameter: numThreads = 8
# Software container option
parameter: container = ""
cwd = path(f'{cwd:a}')
from sos.utils import expand_size
###Output
_____no_output_____
###Markdown
Data download
###Code
[download_hg_reference]
output: f"{cwd:a}/GRCh38_full_analysis_set_plus_decoy_hla.fa"
download: dest_dir = cwd
ftp://ftp.1000genomes.ebi.ac.uk/vol1/ftp/technical/reference/GRCh38_reference_genome/GRCh38_full_analysis_set_plus_decoy_hla.fa
[download_gene_annotation]
output: f"{cwd:a}/Homo_sapiens.GRCh38.103.chr.gtf"
download: dest_dir = cwd, decompress=True
http://ftp.ensembl.org/pub/release-103/gtf/homo_sapiens/Homo_sapiens.GRCh38.103.chr.gtf.gz
[download_ercc_reference]
output: f"{cwd:a}/ERCC92.gtf", f"{cwd:a}/ERCC92.fa"
download: dest_dir = cwd, decompress=True
https://tools.thermofisher.com/content/sfs/manuals/ERCC92.zip
###Output
_____no_output_____
###Markdown
GFF3 to GTF formatting
###Code
[gff3_to_gtf]
parameter: gff3_file = path
input: gff3_file
output: f'{cwd}/{_input:bn}.gtf'
task: trunk_workers = 1, trunk_size = job_size, walltime = walltime, mem = mem, tags = f'{step_name}_{_output:bn}'
bash: container=container, expand= "${ }", stderr = f'{_output:n}.stderr', stdout = f'{_output:n}.stdout'
gffread ${_input} -T -o ${_output}
###Output
_____no_output_____
###Markdown
HG reference file preprocessing1. Remove the HLA/ALT/Decoy record from the fasta2. Adding in ERCC information to the fasta file3. Generating index for the fasta file
###Code
[hg_reference_1 (HLA ALT Decoy removal)]
# Path to HG reference file
parameter: hg_reference = path
input: hg_reference
output: f'{cwd}/{_input:bn}.noALT_noHLA_noDecoy.fasta'
task: trunk_workers = 1, trunk_size = job_size, walltime = walltime, mem = mem, tags = f'{step_name}_{_output:bn}'
python: expand = "${ }", stderr = f'{_output}.stderr', stdout = f'{_output}.stdout', container = container
with open('${_input}', 'r') as fasta:
contigs = fasta.read()
contigs = contigs.split('>')
contig_ids = [i.split(' ', 1)[0] for i in contigs]
# exclude ALT, HLA and decoy contigs
filtered_fasta = '>'.join([c for i,c in zip(contig_ids, contigs)
if not (i[-4:]=='_alt' or i[:3]=='HLA' or i[-6:]=='_decoy')])
with open('${_output}', 'w') as fasta:
fasta.write(filtered_fasta)
[hg_reference_2 (merge with ERCC reference)]
parameter: ercc_reference = path
output: f'{cwd}/{_input:bn}_ERCC.fasta'
task: trunk_workers = 1, trunk_size = job_size, walltime = walltime, mem = mem, tags = f'{step_name}_{_output:bn}'
bash: expand = "${ }", stderr = f'{_output[0]}.stderr', stdout = f'{_output}.stdout', container = container
sed 's/ERCC-/ERCC_/g' ${ercc_reference} > ${ercc_reference:n}.patched.fa
cat ${_input} ${ercc_reference:n}.patched.fa > ${_output}
[hg_reference_3 (index the fasta file)]
output: f'{cwd}/{_input:bn}.dict'
task: trunk_workers = 1, trunk_size = job_size, walltime = walltime, mem = mem, tags = f'{step_name}_{_output:bn}'
bash: expand = "${ }", stderr = f'{_output}.stderr', stdout = f'{_output}.stdout', container = container
samtools faidx ${_input}
java -jar /opt/picard-tools/picard.jar \
CreateSequenceDictionary \
R=${_input} \
O=${_output}
###Output
_____no_output_____
###Markdown
Transcript and gene model reference processing This step modify the `gtf` file for following reasons:1. RSEM require GTF input to have the same chromosome name format (with `chr` prefix) as the fasta file. **although for STAR, this problem can be solved by the now commented --sjdbGTFchrPrefix "chr" option, we have to add `chr` to it for use with RSEM**. 2. Gene model collapsing script `collapse_annotation.py` from GTEx require the gtf have `transcript_type` instead `transcript_biotype` in its annotation. We rename it here, although **this problem can also be solved by modifying the collapse_annotation.py while building the docker, since we are doing 1 above we think it is better to add in another customization here.**3. Adding in ERCC information to the `gtf` reference.We may reimplement 1 and 2 if the problem with RSEM is solved, or when RSEM is no longer needed.
###Code
[hg_gtf_1 (add chr prefix to gtf file)]
parameter: hg_reference = path
parameter: hg_gtf = path
input: hg_reference, hg_gtf
output: f'{cwd}/{_input[1]:bn}.reformatted.gtf'
task: trunk_workers = 1, trunk_size = job_size, walltime = walltime, mem = mem, tags = f'{step_name}_{_output:bn}'
R: expand = "${ }", stderr = f'{_output}.stderr', stdout = f'{_output}.stdout', container = container
library("readr")
library("stringr")
library("dplyr")
options(scipen = 999)
con <- file("${_input[0]}","r")
fasta <- readLines(con,n=1)
close(con)
gtf = read_delim("${_input[1]}", "\t", col_names = F, comment = "#", col_types="ccccccccc")
if(!str_detect(fasta,">chr")) {
gtf_mod = gtf%>%mutate(X1 = str_remove_all(X1,"chr"))
} else if (!any(str_detect(gtf$X1[1],"chr"))) {
gtf_mod = gtf%>%mutate(X1 = paste0("chr",X1))
} else (gtf_mod = gtf)
if(any(str_detect(gtf_mod$X9, "transcript_biotype"))) {
gtf_mod = gtf_mod%>%mutate(X9 = str_replace_all(X9,"transcript_biotype","transcript_type"))
}
gtf_mod%>%write.table("${_output}",sep = "\t",quote = FALSE,col.names = F,row.names = F)
###Output
_____no_output_____
###Markdown
**Text below is taken from https://github.com/broadinstitute/gtex-pipeline/tree/master/gene_model**Gene-level expression and eQTLs from the GTEx project are calculated based on a collapsed gene model (i.e., combining all isoforms of a gene into a single transcript), according to the following rules:1. Transcripts annotated as “retained_intron” or “read_through” are excluded. Additionally, transcripts that overlap with annotated read-through transcripts may be blacklisted (blacklists for GENCODE v19, 24 & 25 are provided in this repository; no transcripts were blacklisted for v26).2. The union of all exon intervals of each gene is calculated.3. Overlapping intervals between genes are excluded from all genes.The purpose of step 3 is primarily to exclude overlapping regions from genes annotated on both strands, which can't be unambiguously quantified from unstranded RNA-seq (GTEx samples were sequenced using an unstranded protocol). For stranded protocols, this step can be skipped by adding the `--collapse_only` flag.Further documentation is available on the [GTEx Portal](https://gtexportal.org/home/documentationPagestaticTextAnalysisMethods).
###Code
[hg_gtf_2 (collapsed gene model)]
parameter: is_stranded = False
output: f'{_input:n}{".collapse_only" if is_stranded else ""}.gene.gtf'
task: trunk_workers = 1, trunk_size = job_size, walltime = walltime, mem = mem, tags = f'{step_name}_{_output:bn}'
bash: expand = "${ }", stderr = f'{_output}.stderr', stdout = f'{_output}.stdout', container = container
collapse_annotation.py ${"--collapse_only" if is_stranded else ""} ${_input} ${_output}
[ercc_gtf (Preprocess ERCC gtf file)]
parameter: ercc_gtf = path
input: ercc_gtf
output: f'{cwd}/{_input:bn}.genes.patched.gtf'
task: trunk_workers = 1, trunk_size = job_size, walltime = walltime, mem = mem, tags = f'{step_name}_{_output:bn}'
python: expand = "${ }", stderr = f'{_output}.stderr', stdout = f'{_output}.stdout', container = container
with open('${_input}') as exon_gtf, open('${_output}', 'w') as gene_gtf:
for line in exon_gtf:
f = line.strip().split('\t')
f[0] = f[0].replace('-','_') # required for RNA-SeQC/GATK (no '-' in contig name)
attr = f[8]
if attr[-1]==';':
attr = attr[:-1]
attr = dict([i.split(' ') for i in attr.replace('"','').split('; ')])
# add gene_name, gene_type
attr['gene_name'] = attr['gene_id']
attr['gene_type'] = 'ercc_control'
attr['gene_status'] = 'KNOWN'
attr['level'] = 2
for k in ['id', 'type', 'name', 'status']:
attr['transcript_'+k] = attr['gene_'+k]
attr_str = []
for k in ['gene_id', 'transcript_id', 'gene_type', 'gene_status', 'gene_name',
'transcript_type', 'transcript_status', 'transcript_name']:
attr_str.append('{0:s} "{1:s}";'.format(k, attr[k]))
attr_str.append('{0:s} {1:d};'.format('level', attr['level']))
f[8] = ' '.join(attr_str)
# write gene, transcript, exon
gene_gtf.write('\t'.join(f[:2]+['gene']+f[3:])+'\n')
gene_gtf.write('\t'.join(f[:2]+['transcript']+f[3:])+'\n')
f[8] = ' '.join(attr_str[:2])
gene_gtf.write('\t'.join(f[:2]+['exon']+f[3:])+'\n')
[gene_annotation]
input: output_from("hg_gtf_1"), output_from("hg_gtf_2"), output_from("ercc_gtf")
output: f'{cwd}/{_input[0]:bn}.ERCC.gtf', f'{cwd}/{_input[1]:bn}.ERCC.gtf'
task: trunk_workers = 1, trunk_size = job_size, walltime = walltime, mem = mem, tags = f'{step_name}_{_output:bn}'
bash: expand = "${ }", stderr = f'{_output[0]}.stderr', stdout = f'{_output[0]}.stdout', container = container
cat ${_input[0]} ${_input[2]} > ${_output[0]}
cat ${_input[1]} ${_input[2]} > ${_output[1]}
###Output
_____no_output_____
###Markdown
Generating index file for `STAR` This step generate the index file for STAR alignment. This file just need to generate once and can be re-used. **At least 40GB of memory is needed**. Step Inputs* `gtf` and `fasta`: path to reference sequence. Both of them needs to be unzipped. `gtf` should be the one prior to collapse by gene.* `sjdbOverhang`: specifies the length of the genomic sequence around the annotated junction to be used in constructing the splice junctions database. Ideally, this length should be equal to the ReadLength-1, where ReadLength is the length of the reads. We use 100 here as recommended by the TOPMed pipeline. See here for [some additional discussions](https://groups.google.com/g/rna-star/c/h9oh10UlvhI/m/BfSPGivUHmsJ). Step Output* Indexing file stored in `{cwd}/STAR_index`, which will be used by `STAR`
###Code
[STAR_index]
parameter: hg_gtf = path
parameter: hg_reference = path
# Specifies the length of the genomic sequence around the annotated junction to be used in constructing the splice junctions database. Ideally, this length should be equal to the ReadLength-1, where ReadLength is the length of the reads.
# Default choice follows from TOPMed pipeline recommendation.
parameter: sjdbOverhang = 100
fail_if(expand_size(mem) < expand_size('40G'), msg = "At least 40GB of memory is required for this step")
input: hg_reference, hg_gtf
output: f"{cwd}/STAR_Index/chrName.txt", f"{cwd}/STAR_Index/Log.out",
f"{cwd}/STAR_Index/transcriptInfo.tab", f"{cwd}/STAR_Index/exonGeTrInfo.tab",
f"{cwd}/STAR_Index/SAindex", f"{cwd}/STAR_Index/SA", f"{cwd}/STAR_Index/genomeParameters.txt",
f"{cwd}/STAR_Index/chrStart.txt", f"{cwd}/STAR_Index/sjdbList.out.tab",
f"{cwd}/STAR_Index/exonInfo.tab", f"{cwd}/STAR_Index/sjdbList.fromGTF.out.tab",
f"{cwd}/STAR_Index/chrLength.txt", f"{cwd}/STAR_Index/sjdbInfo.txt",
f"{cwd}/STAR_Index/Genome", f"{cwd}/STAR_Index/chrNameLength.txt",
f"{cwd}/STAR_Index/geneInfo.tab"
task: trunk_workers = 1, trunk_size = job_size, walltime = walltime, mem = mem, tags = f'{step_name}_{_output[0]:bd}'
bash: container=container, expand= "${ }", stderr = f'{_output[0]:d}.stderr', stdout = f'{_output[0]:d}.stdout'
STAR --runMode genomeGenerate \
--genomeDir ${_output:d} \
--genomeFastaFiles ${_input[0]} \
--sjdbGTFfile ${_input[1]} \
--sjdbOverhang ${sjdbOverhang} \
--runThreadN ${numThreads} #--sjdbGTFchrPrefix "chr"
###Output
_____no_output_____
###Markdown
Generating index file for `RSEM`This step generate the indexing file for `RSEM`. This file just need to generate once. Step Inputs* `gtf` and `fasta`: path to reference sequence. `gtf` should be the one prior to collapse by gene.* `sjdbOverhang`: specifies the length of the genomic sequence around the annotated junction to be used in constructing the splice junctions database. Ideally, this length should be equal to the ReadLength-1, where ReadLength is the length of the reads. Step Outputs* Indexing file stored in `RSEM_index_dir`, which will be used by `RSEM`
###Code
[RSEM_index]
parameter: hg_gtf = path
parameter: hg_reference = path
input: hg_reference, hg_gtf
output: f"{cwd}/RSEM_Index/rsem_reference.n2g.idx.fa", f"{cwd}/RSEM_Index/rsem_reference.grp",
f"{cwd}/RSEM_Index/rsem_reference.idx.fa", f"{cwd}/RSEM_Index/rsem_reference.ti",
f"{cwd}/RSEM_Index/rsem_reference.chrlist", f"{cwd}/RSEM_Index/rsem_reference.seq",
f"{cwd}/RSEM_Index/rsem_reference.transcripts.fa"
task: trunk_workers = 1, trunk_size = job_size, walltime = walltime, mem = mem, tags = f'{step_name}_{_output[0]:bd}'
bash: container=container, expand= "${ }", stderr = f'{_output[0]:d}.stderr', stdout = f'{_output[0]:d}.stdout'
rsem-prepare-reference \
${_input[0]} \
${_output:nn} \
--gtf ${_input[1]} \
--num-threads ${numThreads}
###Output
_____no_output_____
|
Gradient_Boost_Regression_Tree.ipynb
|
###Markdown
Gradient Boost Regression TreeGradient Boost Regression Trees (GBRT) are an ensemble learning technique that successivley trains new weak learners which are added to the ensemble to make up for errors in the previous ensemble. The term "Gradient Boost" is derived from the structure of the algorithm to update the ensemble, which looks like gradient descent in the functional space.Suppose we are given a task to predict an output variable $y$ from a set of training data $x$. Let $F_{m}$ denote the predicted output from our ensemble predictor at step $m$ of the GBRT. As $F_{m}$ is an imperfect predictor, the goal of GBRT is to add a new learner to the ensemble to try and reduce the prediction error associated with $F_{m}$. The ensemble at step $m+1$ can be expressed as:$$F_{m+1} = F_{m} + \arg\min_{h}\Big[ \frac{1}{n}\sum_{i=1}^{n} L \big(y_{i},F_{m}(x_{i}) + h(x_{i})\big) \Big]$$where $L()$ is the loss function. Instead of optimizing the rightmost term completely, the nonlinear loss function can be approximated by a first order Taylor Expansion (as L is assumed to be differentiable):$$\begin{equation}L \big(y_{i},F_{m}(x_{i}) + h_{m}(x_{i})\big) \approx L \big(y_{i},F_{m}(x_{i}))\big) + h(x_{i})\frac{dL(y_{i},F(x_{i}))}{dF(x_{i})}\Bigg|_{F_{m}(x_{i})}\end{equation}$$Let$$\begin{equation}g_{i} = \frac{dL(y_{i},F(x_{i}))}{dF(x_{i})}\Bigg|_{F_{m}(x_{i})}\end{equation}$$Noting that the first term on the RHS of the Taylor Expansion doesn't affect the choice of $h$, we have:$$\begin{equation}\arg\min_{h}\Big[ \frac{1}{n}\sum_{i=1}^{n} L \big(y_{i},F_{m}(x_{i}) + h(x_{i})\big) \Big] \approx \arg\min_{h} \Bigg[ \frac{1}{n}\sum_{i=1}^{n} h(x_{i})g_{i}\Bigg] \end{equation}$$The quantity on the RHS will be minimized if $h$ is fit to predict $-\alpha g_{i}$ where $\alpha$ is positive and arbitrarily large. In practice, $\alpha$ should be kept relatively small so that the first order Taylor Expansion is a good approximation. If the loss function is the squared error, then it is straightforward to show that $g_{i}$ is the residual $r_{i} = y_{i} - F_{m}(x_{i})$. Choosing $\alpha$ small implies only small improvements will be made to the ensemble at each iteration of the ensemble improvement. Thus the ensemble iteration algorithm is:$$F_{m+1} = F_{m} + \alpha h_{m}$$where $h_{m}$ is fit to predict $-g_{i}$ which is equal to the residual error of the ensemble at step $m$.
###Code
#imports
import numpy as np
import matplotlib.pyplot as plt
%matplotlib inline
from sklearn.ensemble import GradientBoostingRegressor
from sklearn.tree import DecisionTreeRegressor
#generate nonlinear regression data
num_points = 200
x_min = -4
x_max = 4
c = 0.25
X = np.sort(np.random.uniform(x_min, x_max, num_points)).reshape(-1,1)
#statistics for noise
e_mean = 0
e_var = 0.5
e = np.random.normal(e_mean, e_var, num_points).reshape(-1,1)
#generate target data
y = (c * X ** 2 + e).flatten()
#plot data
plt.plot(X,y,'bo')
plt.title('Dataset')
plt.xlabel('X')
plt.ylabel('y')
#GBRT sklearn
n = 50 #number of estimators
alpha = 0.1 #learning rate
m_depth = 2 #max_depth of decision trees
#sklearn GBRT
gbrt = GradientBoostingRegressor(max_depth=m_depth, n_estimators=n, learning_rate=alpha)
gbrt.fit(X.reshape(-1,1),y)
#plot against training set
y_pred = gbrt.predict(X)
plt.plot(X,y_pred,'r',label='sklearn',linewidth=3)
plt.plot(X,y,'bo',label='training data')
plt.title('Data and Predictions')
plt.xlabel('X')
plt.ylabel('y')
plt.legend()
#GBRT from scratch
h = [] #list of estimators
_y = y #placeholder for residuals
#train
for i in range(0,n):
#train a weak learner on last residual
_h = DecisionTreeRegressor(max_depth=m_depth).fit(X,_y)
h.append(_h)
#update residual with learning rate
_y = _y - alpha*_h.predict(X)
#predict
y_pred_scratch = sum(tree.predict(X) for tree in h) * alpha
plt.plot(X,y_pred_scratch,'go',label='scratch')
plt.plot(X,y_pred,'r',label='sklearn')
plt.title('Comparing scratch and sklearn')
plt.xlabel('X')
plt.ylabel('y')
plt.legend()
###Output
_____no_output_____
|
examples/thermionic_energy_convertors/Development_Warp_Thermionic_Converter.ipynb
|
###Markdown
Development for a Simple Warp Interface for Thermionic ConvertersThis notebook provides a space to develop some simple Sirepo scripts, isolating specific hooks and function calls to be implemented in a Warp GUI. The basic infrastructure makes use of the simple notebooks designed for Modern Electron, with the goal of validating very specific setups and plots. It will also be used to design simple example scripts.04/25/2017Nathan Cook
###Code
%matplotlib notebook
%load_ext autoreload
%autoreload 2
from __future__ import division
import sys
del sys.argv[1:] # Necessry to run 'from warp import *' in IPython notebook without conflict.
from warp import *
import numpy as np
import matplotlib.pyplot as plt
import os
import pickle
import h5py
from re import findall
from scipy.special import erfinv
from datetime import datetime
import rswarp
from warp.data_dumping.openpmd_diag import ParticleDiagnostic
from rswarp.diagnostics import FieldDiagnostic
from rswarp.utilities.file_utils import cleanupPrevious
from rswarp.utilities.file_utils import readparticles
from rswarp.utilities.file_utils import loadparticlefiles
from rswarp.cathode import sources
from rswarp.cathode import injectors
# Constants imports
from scipy.constants import e, m_e, c, k
kb_eV = 8.6173324e-5 #Bolztmann constant in eV/K
kb_J = k #Boltzmann constant in J/K
m = m_e
###Output
# Warp
# Origin date: Thu, 16 Mar 2017 11:38:11 -0600
# Local date: Fri, 31 Mar 2017 16:59:09 -0700
# Commit hash: 65dd428
# /Users/ncook/.virtualenvs/rswarp_env/lib/python2.7/site-packages/warp/warp.pyc
# /Users/ncook/.virtualenvs/rswarp_env/lib/python2.7/site-packages/warp/warpC.so
# Wed Apr 26 16:25:27 2017
# import warp time 0.258155107498 seconds
# For more help, type warphelp()
###Markdown
Diagnostics
###Code
diagDir = 'diags/xzsolver/hdf5/'
field_base_path = 'diags/fields/'
diagFDir = {'magnetic':'diags/fields/magnetic','electric':'diags/fields/electric'}
# Cleanup previous files
cleanupPrevious(diagDir,diagFDir)
###Output
_____no_output_____
###Markdown
Grid parametersThe grid parameters comprise one of the primary sets of user inputs, and are required for initializing the grid, pre-calculating fundamental currents, and generating the solver. These values are also used throuhgout visualization scripts.**'Physical' Grid Parameters. These are physically intuitive values for a simple domain specification:**1. `PLATE_SPACING` - The longitudinal distance (z-axis) between cathode and anode2. `CHANNEL_WIDTH` - The transverse dimension of the simulation domain**Technical Grid Parameters. These provide the required inputs for constructing simulation objects, but may be computed from the physical parameters above for a simple rectangular geometry:**1. `X_MIN, X_MAX` - By default, horizontal domain is `(-0.5*CHANNEL_WIDTH,0.5*CHANNEL_WIDTH)`2. `X_MIN, X_MAX` - By default, longitudinal domian is `[0, PLATE_SPACING]`3. `Y_MIN, Y_MAX` - The ignorable plane, but specified for completeness. Defaults to +/- `(-0.5*CHANNEL_WIDTH,0.5*CHANNEL_WIDTH)`4. `NUM_X` - The number of grid points along x.5. `NUM_Y` - The number of grid points along y (ignorable for 2DXZ geometry).6. `NUM_Z` - The number of grid points along z.
###Code
#GLOBAL GEOMETRY PARAMETERS FOR USERS
PLATE_SPACING = 10e-6 #plate spacing
CHANNEL_WIDTH = 110e-9 #width of simulation box
#Dimensions
X_MAX = CHANNEL_WIDTH*0.5
X_MIN = -1.*X_MAX
Y_MAX = CHANNEL_WIDTH*0.5
Y_MIN = -1.*Y_MAX
Z_MIN = 0.
Z_MAX = PLATE_SPACING
#Grid parameters
NUM_X = 11
NUM_Y = 64
NUM_Z = 512
#z step size
dz = (Z_MAX - Z_MIN)/NUM_Z
###Output
_____no_output_____
###Markdown
Solver Geometry and BoundariesThe solver geometry is a fundemental pre-requisite for any interface or simulation setup. We will assume for now that we are fixing a 2D X-Z geometry, with the Y axis as an ignorable plane. **`w3d.solvergeom = w3d.XZgeom`**Future extensions to the interface will support 3D geometries. Where applicable and simple, small code snippets have been included in anticipation of this feature. However by no means are these scripts fully compliant with 3D simulations.
###Code
#Specify solver geometry
w3d.solvergeom = w3d.XZgeom
assert w3d.solvergeom == w3d.XZgeom, \
'Solver geometry required to be w3d.XZgeom'
# Set boundary conditions
# Longitudinal conditions overriden by conducting plates
w3d.bound0 = neumann
w3d.boundnz = dirichlet
w3d.boundxy = periodic
# Particles boundary conditions
top.pbound0 = absorb
top.pboundnz = absorb
top.pboundxy = periodic
# Set grid boundaries
w3d.xmmin = X_MIN
w3d.xmmax = X_MAX
w3d.zmmin = 0.
w3d.zmmax = Z_MAX
# Set grid counts
w3d.nx = NUM_X
w3d.nz = NUM_Z
zmesh = np.linspace(0,Z_MAX,NUM_Z+1) #holds the z-axis grid points in an array
###Output
_____no_output_____
###Markdown
Source parameterizationThis section covers source parameterization, in particular how the electrons are emitted from the cathode. Warp permits several options. We want to support three options. For simplicity, I've defined the `USER_INJECT` flag which corresponds to the three possible options:1. Constant emission - user specifies current. `USER_INJECT = 1`2. Child-Langmuir emission (computed from geometries) - user selects and current is computed and displayed `USER_INJECT = 2`3. thermionic emission (computed from cathode temperature) - user selects and current is computed and displayed `USER_INJECT = 3`**Note that the following USER PARAMETERS are needed for the essential specification of the beam:**1. Instantiation via species command i.e. `beam = Species(type=Electron, name='beam')`2. beam radii in x,y via a0, b0 (`beam.a0 = 0.5*BEAM_WIDTH`). In many cases, `BEAM_WIDTH = CHANNEL_WIDTH`.3. beam current (`beam.ibeam = BEAM_CURRENT`)4. Cathode temperature in Kelvin (`CATHODE_TEMP`). Should default to 4K.5. Minimum z-coordinate for injected particles (`Z_PART_MIN`). Must have `Z_PART_MIN > Z_MIN`.**The next set of parameters are generated from additional user parameters (grid, beam, etc.):**1. The injection type for the instance of `top` (`top.inejct = 6`). This will be set to 6 (user injection) for most cases, determined by the `USER_INJECT` switch.2. Number of particles to be injected per step (`top.npinject`). This is computed from grid parameters and defaults to 10 particles per horizontal cell(e.g. `10*NUM_X`).2. Injection coordinate determination - analytical vs. interpolated (`w3d.l_inj_exact`). Defaults to false for most injection types.3. Variance of thermal particle velocity distribution in z (`beam.vthz`). Defaults to 0.4. Variance of thermal particle velocity distribution in transverse plane (`beam.vthperp`). Defaults to 0.The `rswarp` repository has been updated with a cathode module to streamline the designation of cathode sources via each of these three methods. Below we will demonstrate their use and provide a simple template.
###Code
from rswarp.cathode import sources
#Cathode and anode settings
CATHODE_TEMP = 1273.15 #1100. #1273.15 #1000. #cathode temperature in K
CATHODE_PHI = 2.0 #work function in eV
ANODE_WF = 0.1
GRID_BIAS = 0.4 #voltage applied to any grid of electrodes
vacuum_level = CATHODE_PHI - ANODE_WF + GRID_BIAS
#compute beam cutoff velocity for time-step determinance
beam_beta = sources.compute_cutoff_beta(CATHODE_TEMP)
#Compute Child-Langmuir limit for this setup A/m^2
cl_limit = sources.cl_limit(CATHODE_PHI, ANODE_WF, GRID_BIAS, PLATE_SPACING)
#INJECTION SPECIFICATION
USER_INJECT = 1
# --- Setup simulation species
beam = Species(type=Electron, name='beam')
# --- Set basic beam parameters
SOURCE_RADIUS_1 = 0.5*CHANNEL_WIDTH #a0 parameter - X plane
SOURCE_RADIUS_2 = 0.5*CHANNEL_WIDTH #b0 parameter - Y plane
Z_PART_MIN = dz/8. #starting particle z value
#Compute cathode area for geomtry-specific current calculations
if (w3d.solvergeom == w3d.XYZgeom):
#For 3D cartesion geometry only
cathode_area = 4.*SOURCE_RADIUS_1*SOURCE_RADIUS_2
else:
#Assume 2D XZ geometry
cathode_area = 2.*SOURCE_RADIUS_1*1. # 1 m is the geometric factor scaling the plane of the ignorable coordinate
#Set a default 'USER_CURRENT' to the Richardson-Dushman current in case of user-specified constant emission
#This will ultimately be an adjustable GUI parameter.
USER_CURRENT = sources.j_rd(CATHODE_TEMP,CATHODE_PHI)*cathode_area
# If true, position and angle of injected particle are computed analytically rather than interpolated
# Can be false for all but C-L injection (inject=2)
w3d.l_inj_exact = False
#Specify particles to be injected each step - 10 macro-particles per cell by default, USER SPECIFIED IN FUTURE
PTCL_PER_STEP = 10*NUM_X
top.npinject = PTCL_PER_STEP
# --- If using the XZ geometry, set so injection uses the same geometry
top.linj_rectangle = (w3d.solvergeom == w3d.XZgeom)
#Determine an appropriate time step based upon estimated final velocity
vzfinal = sqrt(2.*abs(vacuum_level)*np.abs(beam.charge)/beam.mass)+beam_beta*c
dt = dz/vzfinal #5e-15
top.dt = dt
if vzfinal*top.dt > dz:
print "Time step dt = {:.3e}s does not constrain motion to a single cell".format(top.dt)
if USER_INJECT == 1:
# Constant current density - beam transverse velocity fixed to zero, very small longitduinal velocity
#Set injection flag
top.inject = 6 # 1 means constant; 2 means space-charge limited injection;# 6 means user-specified
beam.ibeam = USER_CURRENT
beam.a0 = SOURCE_RADIUS_1
beam.b0 = SOURCE_RADIUS_2
#sources.constant_current(beam, CHANNEL_WIDTH, Z_PART_MIN, ptcl_per_step)
myInjector = injectors.injectorUserDefined(beam, CATHODE_TEMP, CHANNEL_WIDTH, Z_PART_MIN, PTCL_PER_STEP)
installuserinjection(myInjector.inject_constant)
if USER_INJECT == 2:
# space charge limited injection using Child-Langmuir computation of cold limit
#Set injection flag
top.inject = 2 # 1 means constant; 2 means space-charge limited injection;# 6 means user-specified
beam_current = sources.cl_limit(CATHODE_PHI, ANODE_WF, GRID_BIAS, PLATE_SPACING)*cathode_area
beam.ibeam = beam_current
beam.a0 = SOURCE_RADIUS_1
beam.b0 = SOURCE_RADIUS_2
w3d.l_inj_exact = True
elif USER_INJECT == 3:
#Thermionic injection
#Set injection flag
top.inject = 6 # 1 means constant; 2 means space-charge limited injection;# 6 means user-specified
beam_current = sources.j_rd(CATHODE_TEMP,CATHODE_PHI)*cathode_area #steady state current in Amps
beam.ibeam = beam_current
beam.a0 = SOURCE_RADIUS_1
beam.b0 = SOURCE_RADIUS_2
myInjector = injectors.injectorUserDefined(beam, CATHODE_TEMP, CHANNEL_WIDTH, Z_PART_MIN, PTCL_PER_STEP)
installuserinjection(myInjector.inject_thermionic)
# These must be set for user injection
top.ainject = 1.0
top.binject = 1.0
derivqty()
###Output
_____no_output_____
###Markdown
Create solver
###Code
# Set up fieldsolver
f3d.mgtol = 1e-6 # Multigrid solver convergence tolerance, in volts. 1 uV is default in Warp.
solverE = MultiGrid2D()
registersolver(solverE)
###Output
_____no_output_____
###Markdown
Install conductors
###Code
# --- Emitter settings
extractor_voltage = vacuum_level
# --- Anode Location
zplate = Z_MAX#1e-6 # --- plate location
# Create source conductors
source = ZPlane(zcent=w3d.zmmin,zsign=-1.,voltage=0.)
installconductor(source, dfill=largepos)
# Create ground plate
plate = ZPlane(voltage=extractor_voltage, zcent=zplate)
installconductor(plate,dfill=largepos)
# Setup the particle scraper
scraper = ParticleScraper([source, plate])
###Output
_____no_output_____
###Markdown
Define diagnostics
###Code
particleperiod = 100
particle_diagnostic_0 = ParticleDiagnostic(period = particleperiod, top = top, w3d = w3d,
species = {species.name: species for species in listofallspecies},
comm_world=comm_world, lparallel_output=False, write_dir = diagDir[:-5])
fieldperiod = 100
efield_diagnostic_0 = FieldDiagnostic.ElectrostaticFields(solver=solverE, top=top, w3d=w3d, comm_world = comm_world,
period=fieldperiod)
installafterstep(particle_diagnostic_0.write)
installafterstep(efield_diagnostic_0.write)
###Output
_____no_output_____
###Markdown
Generate simulation
###Code
#prevent GIST from starting upon setup
top.lprntpara = false
top.lpsplots = false
top.verbosity = 0 # Reduce solver verbosity
solverE.mgverbose = -1 #further reduce output upon stepping - prevents websocket timeouts in Jupyter notebook
package("w3d")
generate()
###Output
*** particle simulation package W3D generating
--- Resetting lattice array sizes
--- Allocating space for particles
--- Loading particles
--- Setting charge density
--- done
--- Allocating Win_Moments
--- Allocating Z_Moments
--- Allocating Lab_Moments
###Markdown
Run simulation
###Code
#%%time
num_steps = 5000
output_steps = np.linspace(0,num_steps,num_steps/particleperiod + 1)[1:]
step_count = 0
time0 = time.time()
#CATHODE_TEMP, CHANNEL_WIDTH, Z_PART_MIN, ptcl_per_step
#step(num_steps, cathode_temp = CATHODE_TEMP, channel_width=CHANNEL_WIDTH, z_part_min=Z_PART_MIN, ptcl_per_step=ptcl_per_step)
step(num_steps)
time1 = time.time()
time_per_step = (time1-time0)/num_steps
###Output
*** particle simulation package W3D running
###Markdown
Some basic diagnosticsA few diagnostics for testing. Specifically, we look at the current across the gap at the end of the simulation to verify that it's uniform at the value expected.
###Code
efield_path = diagFDir['electric']
efield_files = [os.path.join(efield_path,fn) for fn in os.listdir(efield_path)]
efield_files.sort()
fielddata_file = efield_files[-1]
step_number = int(findall(r'\d+', fielddata_file)[0])
data_efield = h5py.File(fielddata_file, 'r')
Ex = data_efield['data/%s/meshes/E/x' % (step_number)]
Ey = data_efield['data/%s/meshes/E/y' % (step_number)]
Ez = data_efield['data/%s/meshes/E/z' % (step_number)]
phi = data_efield['data/%s/meshes/phi'% (step_number)]
particles_path = diagDir
particles_files = [os.path.join(particles_path,fn) for fn in os.listdir(particles_path)]
particles_files.sort()
particledata_file = particles_files[-1]
# Read single particle diagnostic file in
f0 = readparticles(particledata_file.format(num_steps))
# Read all particles into directory. Structure: name[int stepnumber][str Species name]
fall = loadparticlefiles(particles_path)
def get_zcurrent_new(particle_array, momenta, mesh, particle_weight, dz):
"""
Find z-directed current on a per cell basis
particle_array: z positions at a given step
momenta: particle momenta at a given step in SI units
mesh: Array of Mesh spacings
particle_weight: Weight from Warp
dz: Cell Size
"""
charge = 1.60217662e-19
mass = 9.10938356e-31
current = np.zeros_like(mesh)
velocity = c * momenta / np.sqrt(momenta**2 + (mass * c)**2)
for index, zval in enumerate(particle_array):
bucket = np.round(zval/dz) #value of the bucket/index in the current array
current[int(bucket)] += velocity[index]
return current* charge * particle_weight / dz
# Get current for all steps (takes a long time)
current_history = []
for i in range(particleperiod,num_steps,particleperiod):
#print i
curr = get_zcurrent_new(fall[i]['beam'][:,4],fall[i]['beam'][:,5],zmesh,beam.sw,dz)
current_history.append(curr)
current_history = np.asarray(current_history)
#Plot the current across gap at a single time
fig5 = plt.figure(figsize=(16,6))
#scalings
h_scale = 1.e6
y_range_max = beam.ibeam*1.e3*1.2
#current plotted from grid
plt.plot(zmesh*h_scale,np.array(current_history[-1])*1e3,'k')
#Compute and plot idealized currents as needed
RD_ideal = np.ones(len(zmesh))*sources.j_rd(CATHODE_TEMP,CATHODE_PHI)*cathode_area
JCL_ideal = np.ones(len(zmesh))*cl_limit*cathode_area
if (RD_ideal[0]*1e3 <= y_range_max):
plt.plot(zmesh*h_scale,RD_ideal*1.e3,'r--',label=r'Richardson-Dushman')
if (JCL_ideal[0]*1e3 <= y_range_max):
plt.plot(zmesh*h_scale,JCL_ideal*1.e3,'b--',label=r'I$_{cl}$ cold limit')
#labels and legends
plt.xlabel("z ($\mu$m)",fontsize='16')
plt.ylabel("current (mA)",fontsize='16')
plt.title("Current - {:.4E}s".format(fall[num_steps]['time']),fontsize=18)
plt.xlim(Z_MIN,Z_MAX*1.e6)
plt.ylim(0, y_range_max)
plt.legend(loc=4)
title = 'current_{:.4f}ps-test.pdf'.format(CATHODE_TEMP,fall[num_steps]['time']*1.e9)
#fig5.savefig(title,bbox_inches='tight')
###Output
_____no_output_____
|
Part 1- Introduction to Computer Vision/1_1_Image_Representation/4. Green Screen Car.ipynb
|
###Markdown
Color Threshold, Green Screen Import resources
###Code
import matplotlib.pyplot as plt
import matplotlib.image as mpimg
import numpy as np
import cv2
%matplotlib inline
###Output
_____no_output_____
###Markdown
Read in and display the image
###Code
# Read in the image
image = mpimg.imread('images/car_green_screen.jpg')
# Print out the image dimensions (height, width, and depth (color))
print('Image dimensions:', image.shape)
# Display the image
plt.imshow(image)
###Output
_____no_output_____
###Markdown
Define the color threshold
###Code
## TODO: Define our color selection boundaries in RGB values
lower_green = np.array([0,150,20])
upper_green = np.array([250,255,250])
###Output
_____no_output_____
###Markdown
Create a mask
###Code
# Define the masked area
mask = cv2.inRange(image, lower_green, upper_green)
# Vizualize the mask
plt.imshow(mask, cmap='gray')
# Mask the image to let the car show through
masked_image = np.copy(image)
masked_image[mask != 0] = [0, 0, 0]
# Display it!
plt.imshow(masked_image)
###Output
_____no_output_____
###Markdown
Mask and add a background image
###Code
# Load in a background image, and convert it to RGB
background_image = cv2.imread('images/sky.jpg')
background_image = cv2.cvtColor(background_image, cv2.COLOR_BGR2RGB)
## TODO: Crop it or resize the background to be the right size (450x660)
crop_background = background_image[0:450, 0:660]
## TODO: Mask the cropped background so that the car area is blocked
# Hint mask the opposite area of the previous image
crop_background[mask == 0] = [0, 0, 0]
## TODO: Display the background and make sure
plt.imshow(crop_background)
###Output
_____no_output_____
###Markdown
Create a complete image
###Code
## TODO: Add the two images together to create a complete image!
complete_image = masked_image + crop_background
# Display the result
plt.imshow(complete_image)
###Output
_____no_output_____
|
Rnotebooks/Rplot.ipynb
|
###Markdown
Japanse Test 初回だけこれを行う.
###Code
# extrafontライブラリでフォントの管理を簡略化します。
install.packages("extrafont")
require(extrafont)
# システムフォントをRのライブラリ配下にインポートします(十数分かかります。)
font_import()
# 今のセッションで使えるようにフォントをロードします。
loadfonts(quiet = TRUE)
###Output
The downloaded binary packages are in
/var/folders/6q/vbfcmwbn4qq9lfpvr28lz9540000gn/T//RtmpcV4Fyx/downloaded_packages
###Markdown
日本語が plot でも使えるようになる.
###Code
library(Cairo)
library(ggplot2)
#require(extrafont)
require(ggplot2)
require(Cairo)
CairoFonts(regular = "ヒラギノ明朝 ProN W3",bold="ヒラギノ明朝 ProN W3")
# plot作成
gg <- ggplot(mtcars,aes(x=wt,y=mpg)) + geom_point()
gg <- gg + ggtitle("車の重さと燃費の関係")
gg <- gg + xlab("重さ") + ylab("燃費")
gg <- gg + theme(plot.title = element_text(face="bold"))
# PDF出力(A4横)
CairoPDF(paper="a4r",width=11.69,height=8.27)
print(gg)
library(wordcloud)
library(RMeCab)
library(Cairo)
par(family="ヒラギノ明朝 ProN W3")
RMeCabText.result<- RMeCabC('これまでの例は数行のコマンドでしたのでプロンプトに直接を打っていましたが、何十行というロジックをプロンプトに順々に打っていくのは非効率です。今回は、予め外部のテキストファイルにソースコードを書き、そのファイルをプロンプトから呼び出す手順を次の例で示します。')
print(RMeCabText.result)
RMeCabText.result2<-unlist(sapply(RMeCabText.result,"[[",1))
wordcloud(RMeCabText.result2,min.freq = 2,random.order = FALSE)
###Output
[[1]]
名詞
"これ"
[[2]]
助詞
"まで"
[[3]]
助詞
"の"
[[4]]
名詞
"例"
[[5]]
助詞
"は"
[[6]]
名詞
"数行"
[[7]]
助詞
"の"
[[8]]
名詞
"コマンド"
[[9]]
助動詞
"でし"
[[10]]
助動詞
"た"
[[11]]
助詞
"ので"
[[12]]
名詞
"プロンプト"
[[13]]
助詞
"に"
[[14]]
名詞
"直接"
[[15]]
助詞
"を"
[[16]]
動詞
"打っ"
[[17]]
助詞
"て"
[[18]]
動詞
"い"
[[19]]
助動詞
"まし"
[[20]]
助動詞
"た"
[[21]]
助詞
"が"
[[22]]
記号
"、"
[[23]]
名詞
"何"
[[24]]
名詞
"十"
[[25]]
名詞
"行"
[[26]]
助詞
"という"
[[27]]
名詞
"ロジック"
[[28]]
助詞
"を"
[[29]]
名詞
"プロンプト"
[[30]]
助詞
"に"
[[31]]
名詞
"順々"
[[32]]
助詞
"に"
[[33]]
動詞
"打っ"
[[34]]
助詞
"て"
[[35]]
動詞
"いく"
[[36]]
名詞
"の"
[[37]]
助詞
"は"
[[38]]
接頭詞
"非"
[[39]]
名詞
"効率"
[[40]]
助動詞
"です"
[[41]]
記号
"。"
[[42]]
名詞
"今回"
[[43]]
助詞
"は"
[[44]]
記号
"、"
[[45]]
副詞
"予め"
[[46]]
名詞
"外部"
[[47]]
助詞
"の"
[[48]]
名詞
"テキストファイル"
[[49]]
助詞
"に"
[[50]]
名詞
"ソース"
[[51]]
名詞
"コード"
[[52]]
助詞
"を"
[[53]]
動詞
"書き"
[[54]]
記号
"、"
[[55]]
連体詞
"その"
[[56]]
名詞
"ファイル"
[[57]]
助詞
"を"
[[58]]
名詞
"プロンプト"
[[59]]
助詞
"から"
[[60]]
動詞
"呼び出す"
[[61]]
名詞
"手順"
[[62]]
助詞
"を"
[[63]]
名詞
"次"
[[64]]
助詞
"の"
[[65]]
名詞
"例"
[[66]]
助詞
"で"
[[67]]
動詞
"示し"
[[68]]
助動詞
"ます"
[[69]]
記号
"。"
|
notebooks/.ipynb_checkpoints/5_clustering_majority_vote-checkpoint.ipynb
|
###Markdown
Imports
###Code
import pandas as pd
from bokeh.plotting import figure, output_file, show
from bokeh.models import ColumnDataSource
from bokeh.models.tools import HoverTool
from sklearn.neighbors import NearestNeighbors
from sklearn.decomposition import PCA
from sklearn.manifold import TSNE
from sklearn.cluster import KMeans
import ast
###Output
_____no_output_____
###Markdown
Upload data with RoBERTa vectors
###Code
df = pd.read_csv('../data/metadata_with_title_roberta_vectors.csv')
articles_biomed_roberta_emb = df['roberta_title_vectors'].values
articles_biomed_roberta_emb = [ast.literal_eval(a) for a in articles_biomed_roberta_emb]
###Output
_____no_output_____
###Markdown
Clustering with K-means
###Code
kmeans = KMeans(n_clusters=20, random_state=42).fit(articles_biomed_roberta_emb)
df['kmean_cluster'] = kmeans.labels_
list(set(kmeans.labels_[-10:]))
###Output
_____no_output_____
###Markdown
Clustering with 25-NN
###Code
nbrs_2 = NearestNeighbors(n_neighbors=25, algorithm='ball_tree').fit(articles_biomed_roberta_emb)
distances_2, indices_2 = nbrs_2.kneighbors(articles_biomed_roberta_emb[-10:])
pca = PCA(n_components=200)
principal_components_roberta = pca.fit_transform(articles_biomed_roberta_emb)
tsne_embedded_roberta = TSNE(n_components=2).fit_transform(principal_components_roberta)
clusters_temp = [[]]*len(df)
for i in range(len(indices_2)):
for j in range(len(indices_2[i])):
temp = clusters_temp[indices_2[i][j]]
clusters_temp[indices_2[i][j]] = temp + [i]
clusters = []
for c in clusters_temp:
if len(c) == 0:
clusters.append([-1])
else:
clusters.append(c)
df['clusters'] = clusters
###Output
_____no_output_____
###Markdown
Clustering Intersection
###Code
df['clusters'][-10:]
df['kmean_cluster'][-10:]
cluster_vote_temp = [[]]*len(df)
for i in range(len(df)):
for j in range(10):
if df['kmean_cluster'][i] == kmeans.labels_[-10:][j] and j in df['clusters'][i]:
cluster_vote_temp[i] = cluster_vote_temp[i] + [j]
cluster_vote = []
for c in cluster_vote_temp:
if len(c) == 0:
cluster_vote.append([-1])
else:
cluster_vote.append(c)
df['cluster_vote'] = cluster_vote
df['tsne_0'] = tsne_embedded_roberta[:,0]
df['tsne_1'] = tsne_embedded_roberta[:,1]
df['cluster_vote'][-10:]
# Saving results for insights
df.to_csv('./results/cluster_results.csv')
###Output
_____no_output_____
###Markdown
Bokeh Vizualization
###Code
source = ColumnDataSource(df)
COLORS_NN_GROUPS = ['lightgrey', 'blue', 'red', 'black', 'green', 'violet',
'pink', 'turquoise', 'gold', 'sienna', 'orange']
colors = [COLORS_NN_GROUPS[df['cluster_vote'][i][0]+1] for i in range(len(df))]
len(colors)
p = figure(width=800, height=600)
p.circle(x='tsne_1', y='tsne_0',
source=source,
size=5, color=colors)
p.title.text = 'COVID Scholars clustering representation'
hover = HoverTool()
hover.tooltips=[
('title', '@title'),
]
p.add_tools(hover)
show(p)
###Output
_____no_output_____
|
Notebooks/RadarCOVID-Report/Daily/RadarCOVID-Report-2020-09-01.ipynb
|
###Markdown
RadarCOVID-Report Data Extraction
###Code
import datetime
import logging
import os
import shutil
import tempfile
import textwrap
import uuid
import dataframe_image as dfi
import matplotlib.ticker
import numpy as np
import pandas as pd
import seaborn as sns
%matplotlib inline
sns.set()
matplotlib.rcParams['figure.figsize'] = (15, 6)
extraction_datetime = datetime.datetime.utcnow()
extraction_date = extraction_datetime.strftime("%Y-%m-%d")
extraction_previous_datetime = extraction_datetime - datetime.timedelta(days=1)
extraction_previous_date = extraction_previous_datetime.strftime("%Y-%m-%d")
extraction_date_with_hour = datetime.datetime.utcnow().strftime("%Y-%m-%d@%H")
###Output
_____no_output_____
###Markdown
COVID-19 Cases
###Code
confirmed_df = pd.read_csv("https://covid19tracking.narrativa.com/csv/confirmed.csv")
radar_covid_countries = {"Spain"}
# radar_covid_regions = { ... }
confirmed_df = confirmed_df[confirmed_df["Country_EN"].isin(radar_covid_countries)]
# confirmed_df = confirmed_df[confirmed_df["Region"].isin(radar_covid_regions)]
# set(confirmed_df.Region.tolist()) == radar_covid_regions
confirmed_country_columns = list(filter(lambda x: x.startswith("Country_"), confirmed_df.columns))
confirmed_regional_columns = confirmed_country_columns + ["Region"]
confirmed_df.drop(columns=confirmed_regional_columns, inplace=True)
confirmed_df = confirmed_df.sum().to_frame()
confirmed_df.tail()
confirmed_df.reset_index(inplace=True)
confirmed_df.columns = ["sample_date_string", "cumulative_cases"]
confirmed_df.sort_values("sample_date_string", inplace=True)
confirmed_df["new_cases"] = confirmed_df.cumulative_cases.diff()
confirmed_df["rolling_mean_new_cases"] = confirmed_df.new_cases.rolling(7).mean()
confirmed_df.tail()
extraction_date_confirmed_df = \
confirmed_df[confirmed_df.sample_date_string == extraction_date]
extraction_previous_date_confirmed_df = \
confirmed_df[confirmed_df.sample_date_string == extraction_previous_date].copy()
if extraction_date_confirmed_df.empty and \
not extraction_previous_date_confirmed_df.empty:
extraction_previous_date_confirmed_df["sample_date_string"] = extraction_date
extraction_previous_date_confirmed_df["new_cases"] = \
extraction_previous_date_confirmed_df.rolling_mean_new_cases
extraction_previous_date_confirmed_df["cumulative_cases"] = \
extraction_previous_date_confirmed_df.new_cases + \
extraction_previous_date_confirmed_df.cumulative_cases
confirmed_df = confirmed_df.append(extraction_previous_date_confirmed_df)
confirmed_df.tail()
confirmed_df[["new_cases", "rolling_mean_new_cases"]].plot()
###Output
_____no_output_____
###Markdown
Extract API TEKs
###Code
from Modules.RadarCOVID import radar_covid
exposure_keys_df = radar_covid.download_last_radar_covid_exposure_keys(days=14)
exposure_keys_df[[
"sample_date_string", "source_url", "region", "key_data"]].head()
exposure_keys_summary_df = \
exposure_keys_df.groupby(["sample_date_string"]).key_data.nunique().to_frame()
exposure_keys_summary_df.sort_index(ascending=False, inplace=True)
exposure_keys_summary_df.rename(columns={"key_data": "tek_count"}, inplace=True)
exposure_keys_summary_df.head()
###Output
_____no_output_____
###Markdown
Dump API TEKs
###Code
tek_list_df = exposure_keys_df[["sample_date_string", "key_data"]].copy()
tek_list_df["key_data"] = tek_list_df["key_data"].apply(str)
tek_list_df.rename(columns={
"sample_date_string": "sample_date",
"key_data": "tek_list"}, inplace=True)
tek_list_df = tek_list_df.groupby(
"sample_date").tek_list.unique().reset_index()
tek_list_df["extraction_date"] = extraction_date
tek_list_df["extraction_date_with_hour"] = extraction_date_with_hour
tek_list_df.drop(columns=["extraction_date", "extraction_date_with_hour"]).to_json(
"Data/TEKs/Current/RadarCOVID-TEKs.json",
lines=True, orient="records")
tek_list_df.drop(columns=["extraction_date_with_hour"]).to_json(
"Data/TEKs/Daily/RadarCOVID-TEKs-" + extraction_date + ".json",
lines=True, orient="records")
tek_list_df.to_json(
"Data/TEKs/Hourly/RadarCOVID-TEKs-" + extraction_date_with_hour + ".json",
lines=True, orient="records")
tek_list_df.head()
###Output
_____no_output_____
###Markdown
Load TEK Dumps
###Code
import glob
def load_extracted_teks(mode, limit=None) -> pd.DataFrame:
extracted_teks_df = pd.DataFrame()
paths = list(reversed(sorted(glob.glob(f"Data/TEKs/{mode}/RadarCOVID-TEKs-*.json"))))
if limit:
paths = paths[:limit]
for path in paths:
logging.info(f"Loading TEKs from '{path}'...")
iteration_extracted_teks_df = pd.read_json(path, lines=True)
extracted_teks_df = extracted_teks_df.append(
iteration_extracted_teks_df, sort=False)
return extracted_teks_df
###Output
_____no_output_____
###Markdown
Daily New TEKs
###Code
daily_extracted_teks_df = load_extracted_teks(mode="Daily", limit=14)
daily_extracted_teks_df.head()
tek_list_df = daily_extracted_teks_df.groupby("extraction_date").tek_list.apply(
lambda x: set(sum(x, []))).reset_index()
tek_list_df = tek_list_df.set_index("extraction_date").sort_index(ascending=True)
tek_list_df.head()
new_tek_df = tek_list_df.diff().tek_list.apply(
lambda x: len(x) if not pd.isna(x) else None).to_frame().reset_index()
new_tek_df.rename(columns={
"tek_list": "new_tek_count",
"extraction_date": "sample_date_string",}, inplace=True)
new_tek_df.head()
new_tek_devices_df = daily_extracted_teks_df.copy()
new_tek_devices_df["new_sample_extraction_date"] = \
pd.to_datetime(new_tek_devices_df.sample_date) + datetime.timedelta(1)
new_tek_devices_df["extraction_date"] = pd.to_datetime(new_tek_devices_df.extraction_date)
new_tek_devices_df = new_tek_devices_df[
new_tek_devices_df.new_sample_extraction_date == new_tek_devices_df.extraction_date]
new_tek_devices_df.head()
new_tek_devices_df.set_index("extraction_date", inplace=True)
new_tek_devices_df = new_tek_devices_df.tek_list.apply(lambda x: len(set(x))).to_frame()
new_tek_devices_df.reset_index(inplace=True)
new_tek_devices_df.rename(columns={
"extraction_date": "sample_date_string",
"tek_list": "new_tek_devices"}, inplace=True)
new_tek_devices_df["sample_date_string"] = new_tek_devices_df.sample_date_string.dt.strftime("%Y-%m-%d")
new_tek_devices_df.head()
###Output
_____no_output_____
###Markdown
Hourly New TEKs
###Code
hourly_extracted_teks_df = load_extracted_teks(mode="Hourly", limit=24)
hourly_extracted_teks_df.head()
hourly_tek_list_df = hourly_extracted_teks_df.groupby("extraction_date_with_hour").tek_list.apply(
lambda x: set(sum(x, []))).reset_index()
hourly_tek_list_df = hourly_tek_list_df.set_index("extraction_date_with_hour").sort_index(ascending=True)
hourly_new_tek_df = hourly_tek_list_df.diff().tek_list.apply(
lambda x: len(x) if not pd.isna(x) else None).to_frame().reset_index()
hourly_new_tek_df.rename(columns={
"tek_list": "new_tek_count"}, inplace=True)
hourly_new_tek_df.tail()
hourly_new_tek_devices_df = hourly_extracted_teks_df.copy()
hourly_new_tek_devices_df["new_sample_extraction_date"] = \
pd.to_datetime(hourly_new_tek_devices_df.sample_date) + datetime.timedelta(1)
hourly_new_tek_devices_df["extraction_date"] = pd.to_datetime(hourly_new_tek_devices_df.extraction_date)
hourly_new_tek_devices_df = hourly_new_tek_devices_df[
hourly_new_tek_devices_df.new_sample_extraction_date == hourly_new_tek_devices_df.extraction_date]
hourly_new_tek_devices_df.set_index("extraction_date_with_hour", inplace=True)
hourly_new_tek_devices_df_ = pd.DataFrame()
for i, chunk_df in hourly_new_tek_devices_df.groupby("extraction_date"):
chunk_df = chunk_df.copy()
chunk_df.sort_index(inplace=True)
chunk_df = \
chunk_df.tek_list.apply(lambda x: len(set(x))).diff().to_frame()
hourly_new_tek_devices_df_ = hourly_new_tek_devices_df_.append(chunk_df)
hourly_new_tek_devices_df = hourly_new_tek_devices_df_
hourly_new_tek_devices_df.reset_index(inplace=True)
hourly_new_tek_devices_df.rename(columns={
"tek_list": "new_tek_devices"}, inplace=True)
hourly_new_tek_devices_df.tail()
hourly_summary_df = hourly_new_tek_df.merge(
hourly_new_tek_devices_df, on=["extraction_date_with_hour"], how="outer")
hourly_summary_df["datetime_utc"] = pd.to_datetime(
hourly_summary_df.extraction_date_with_hour, format="%Y-%m-%d@%H")
hourly_summary_df.set_index("datetime_utc", inplace=True)
hourly_summary_df.tail()
###Output
_____no_output_____
###Markdown
Data Merge
###Code
result_summary_df = exposure_keys_summary_df.merge(new_tek_df, on=["sample_date_string"], how="outer")
result_summary_df.head()
result_summary_df = result_summary_df.merge(new_tek_devices_df, on=["sample_date_string"], how="outer")
result_summary_df.head()
result_summary_df = result_summary_df.merge(confirmed_df, on=["sample_date_string"], how="left")
result_summary_df.head()
result_summary_df["tek_count_per_new_case"] = \
result_summary_df.tek_count / result_summary_df.rolling_mean_new_cases
result_summary_df["new_tek_count_per_new_case"] = \
result_summary_df.new_tek_count / result_summary_df.rolling_mean_new_cases
result_summary_df["new_tek_devices_per_new_case"] = \
result_summary_df.new_tek_devices / result_summary_df.rolling_mean_new_cases
result_summary_df["new_tek_count_per_new_tek_device"] = \
result_summary_df.new_tek_count / result_summary_df.new_tek_devices
result_summary_df.head()
result_summary_df["sample_date"] = pd.to_datetime(result_summary_df.sample_date_string)
result_summary_df.set_index("sample_date", inplace=True)
result_summary_df = result_summary_df.sort_index(ascending=False)
###Output
_____no_output_____
###Markdown
Report Results Summary Table
###Code
result_summary_df_ = result_summary_df.copy()
result_summary_df = result_summary_df[[
"tek_count",
"new_tek_count",
"new_cases",
"rolling_mean_new_cases",
"tek_count_per_new_case",
"new_tek_count_per_new_case",
"new_tek_devices",
"new_tek_devices_per_new_case",
"new_tek_count_per_new_tek_device"]]
result_summary_df
###Output
_____no_output_____
###Markdown
Summary Plots
###Code
summary_ax_list = result_summary_df[[
"rolling_mean_new_cases",
"tek_count",
"new_tek_count",
"new_tek_devices",
"new_tek_count_per_new_tek_device",
"new_tek_devices_per_new_case"
]].sort_index(ascending=True).plot.bar(
title="Summary", rot=45, subplots=True, figsize=(15, 22))
summary_ax_list[-1].yaxis.set_major_formatter(matplotlib.ticker.PercentFormatter(1.0))
###Output
_____no_output_____
###Markdown
Hourly Summary Plots
###Code
hourly_summary_ax_list = hourly_summary_df.plot.bar(
title="Last 24h Summary", rot=45, subplots=True)
###Output
_____no_output_____
###Markdown
Publish Results
###Code
def get_temporary_image_path() -> str:
return os.path.join(tempfile.gettempdir(), str(uuid.uuid4()) + ".png")
def save_temporary_plot_image(ax):
if isinstance(ax, np.ndarray):
ax = ax[0]
media_path = get_temporary_image_path()
ax.get_figure().savefig(media_path)
return media_path
def save_temporary_dataframe_image(df):
media_path = get_temporary_image_path()
dfi.export(df, media_path)
return media_path
summary_plots_image_path = save_temporary_plot_image(ax=summary_ax_list)
summary_table_image_path = save_temporary_dataframe_image(df=result_summary_df)
hourly_summary_plots_image_path = save_temporary_plot_image(ax=hourly_summary_ax_list)
###Output
_____no_output_____
###Markdown
Save Results
###Code
report_resources_path_prefix = "Data/Resources/Current/RadarCOVID-Report-"
result_summary_df.to_csv(report_resources_path_prefix + "Summary-Table.csv")
result_summary_df.to_html(report_resources_path_prefix + "Summary-Table.html")
_ = shutil.copyfile(summary_plots_image_path, report_resources_path_prefix + "Summary-Plots.png")
_ = shutil.copyfile(summary_table_image_path, report_resources_path_prefix + "Summary-Table.png")
_ = shutil.copyfile(hourly_summary_plots_image_path, report_resources_path_prefix + "Hourly-Summary-Plots.png")
report_daily_url_pattern = \
"https://github.com/pvieito/RadarCOVID-Report/blob/master/Notebooks/" \
"RadarCOVID-Report/{report_type}/RadarCOVID-Report-{report_date}.ipynb"
report_daily_url = report_daily_url_pattern.format(
report_type="Daily", report_date=extraction_date)
report_hourly_url = report_daily_url_pattern.format(
report_type="Hourly", report_date=extraction_date_with_hour)
###Output
_____no_output_____
###Markdown
Publish on README
###Code
with open("Data/Templates/README.md", "r") as f:
readme_contents = f.read()
summary_table_html = result_summary_df.to_html()
readme_contents = readme_contents.format(
summary_table_html=summary_table_html,
report_url_with_hour=report_hourly_url,
extraction_date_with_hour=extraction_date_with_hour)
with open("README.md", "w") as f:
f.write(readme_contents)
###Output
_____no_output_____
###Markdown
Publish on Twitter
###Code
enable_share_to_twitter = os.environ.get("RADARCOVID_REPORT__ENABLE_PUBLISH_ON_TWITTER")
github_event_name = os.environ.get("GITHUB_EVENT_NAME")
if enable_share_to_twitter and github_event_name == "schedule":
import tweepy
twitter_api_auth_keys = os.environ["RADARCOVID_REPORT__TWITTER_API_AUTH_KEYS"]
twitter_api_auth_keys = twitter_api_auth_keys.split(":")
auth = tweepy.OAuthHandler(twitter_api_auth_keys[0], twitter_api_auth_keys[1])
auth.set_access_token(twitter_api_auth_keys[2], twitter_api_auth_keys[3])
api = tweepy.API(auth)
summary_plots_media = api.media_upload(summary_plots_image_path)
summary_table_media = api.media_upload(summary_table_image_path)
hourly_summary_plots_media = api.media_upload(hourly_summary_plots_image_path)
media_ids = [
summary_plots_media.media_id,
summary_table_media.media_id,
hourly_summary_plots_media.media_id,
]
extraction_date_result_summary_df = \
result_summary_df[result_summary_df.index == extraction_date]
extraction_date_result_hourly_summary_df = \
hourly_summary_df[hourly_summary_df.extraction_date_with_hour == extraction_date_with_hour]
new_teks = extraction_date_result_summary_df.new_tek_count.sum().astype(int)
new_teks_last_hour = extraction_date_result_hourly_summary_df.new_tek_count.sum().astype(int)
new_devices = extraction_date_result_summary_df.new_tek_devices.sum().astype(int)
new_devices_last_hour = extraction_date_result_hourly_summary_df.new_tek_devices.sum().astype(int)
new_tek_count_per_new_tek_device = \
extraction_date_result_summary_df.new_tek_count_per_new_tek_device.sum()
new_tek_devices_per_new_case = \
extraction_date_result_summary_df.new_tek_devices_per_new_case.sum()
status = textwrap.dedent(f"""
Report Update – {extraction_date_with_hour}
#ExposureNotification #RadarCOVID
Shared Diagnoses Day Summary:
- New TEKs: {new_teks} ({new_teks_last_hour:+d} last hour)
- New Devices: {new_devices} ({new_devices_last_hour:+d} last hour, {new_tek_count_per_new_tek_device:.2} TEKs/device)
- Usage Ratio: {new_tek_devices_per_new_case:.2%} devices/case
Report Link: {report_hourly_url}
""")
status = status.encode(encoding="utf-8")
api.update_status(status=status, media_ids=media_ids)
###Output
_____no_output_____
|
preprocess/reddit.ipynb
|
###Markdown
This script preprocess Reddit dataset
###Code
import os
import numpy as np
import pandas as pd
from dotmap import DotMap
import json
from scipy import sparse
import networkx as nx
from networkx.readwrite import json_graph
from tqdm import *
dataset_name = 'reddit'
data_path = os.path.join('../dataset/raw/{}'.format(dataset_name))
def load_data(normalize=True):
graph_fn = os.path.join(data_path, '{}-G.json'.format(dataset_name))
print('load graph data ...')
G_data = json.load(open(graph_fn))
G = json_graph.node_link_graph(G_data)
if isinstance(G.nodes()[0], int):
conversion = lambda n : int(n)
else:
conversion = lambda n : n
print('load features, id map, and class map ...')
features_fn = os.path.join(data_path, '{}-feats.npy'.format(dataset_name))
feats = np.load(features_fn)
id_map_fn = os.path.join(data_path, '{}-id_map.json'.format(dataset_name))
id_map = json.load(open(id_map_fn))
id_map = {k:int(v) for k,v in id_map.items()}
class_fn = os.path.join(data_path, '{}-class_map.json'.format(dataset_name))
class_map = json.load(open(class_fn))
if isinstance(list(class_map.values())[0], list):
lab_conversion = lambda n : n
else:
lab_conversion = lambda n : int(n)
class_map = {k:lab_conversion(v) for k,v in class_map.items()}
## Remove all nodes that do not have val/test annotations
## (necessary because of networkx weirdness with the Reddit data)
broken_nodes = [node for node in G.nodes() if not 'val' in G.node[node] or not 'test' in G.node[node]]
G.remove_nodes_from(broken_nodes)
print("Removed {:d} nodes that lacked proper annotations due to networkx versioning issues".format(len(broken_nodes)))
## Make sure the graph has edge train_removed annotations
## (some datasets might already have this..)
print("Loaded data.. now preprocessing..")
for edge in G.edges():
if (G.node[edge[0]]['val'] or G.node[edge[1]]['val'] or
G.node[edge[0]]['test'] or G.node[edge[1]]['test']):
G[edge[0]][edge[1]]['train_removed'] = True
else:
G[edge[0]][edge[1]]['train_removed'] = False
if normalize and not feats is None:
from sklearn.preprocessing import StandardScaler
train_ids = np.array([id_map[n] for n in G.nodes() if not G.node[n]['val'] and not G.node[n]['test']])
train_feats = feats[train_ids]
scaler = StandardScaler()
scaler.fit(train_feats)
feats = scaler.transform(feats)
return G, feats, id_map, class_map
G, feats, id_map, class_map = load_data(normalize=True)
print(feats.shape)
graphs = {}
with open(os.path.join(data_path, 'reddit-adjlist.txt')) as in_fn:
for line in in_fn:
line = line.strip()
if line[0] == '#':
continue
tokens = line.split()
node_id = tokens[0]
assert(node_id not in graphs)
node = DotMap()
node.node_id = node_id
node.outgoing = tokens[1:]
node.incoming = []
graphs[node_id] = node
sink_nodes = {}
for node_id in tqdm(graphs):
for out_node_id in graphs[node_id].outgoing:
if out_node_id in graphs:
graphs[out_node_id].incoming.append(node_id)
else:
if out_node_id not in sink_nodes:
node = DotMap()
node.node_id = out_node_id
node.incoming = [node_id]
node.outgoing = []
sink_nodes[out_node_id] = node
else:
sink_nodes[out_node_id].incoming.append(node_id)
for node_id in sink_nodes:
graphs[node_id] = sink_nodes[node_id]
# for split train-test-cv
TRAIN_FLAG = 0
TEST_FLAG = 1
CV_FLAG = 2
for node_id in G.nodes():
if node_id in graphs:
is_validate = G.node[node_id]['val']
is_test = G.node[node_id]['test']
if is_test:
graphs[node_id].kind = TEST_FLAG
elif is_validate:
graphs[node_id].kind = CV_FLAG
else:
graphs[node_id].kind = TRAIN_FLAG
# add class labels
for node_id, class_id in class_map.items():
if node_id in graphs:
graphs[node_id].class_id = class_id
# add node features
for node_id, index in tqdm(id_map.items()):
if node_id in graphs:
graphs[node_id].features = list(feats[index])
graph_data = []
for node_id, node in tqdm(graphs.items()):
# combine in and out edges
out_edges = list(set([id_map[n] for n in node.outgoing]))
in_edges = list(set([id_map[n] for n in node.incoming]))
neighbors = list(set(out_edges + in_edges))
node_data = {'post_id': node.node_id,
'node_id': id_map[node.node_id],
'neighbors': neighbors,
'in_edges': in_edges, 'out_edges': out_edges,
'label': node.class_id, 'kind': node.kind,
'features': node.features}
graph_data.append(node_data)
df = pd.DataFrame(graph_data)
df.set_index('node_id', inplace=True) # set paper as the row index
save_data_path = os.path.join('../dataset/clean/{}'.format(dataset_name))
save_fn = os.path.join(save_data_path, '{}.data.pkl'.format(dataset_name))
df.to_pickle(save_fn)
###Output
_____no_output_____
###Markdown
Preprocess Graph Dataset
###Code
save_data_path = os.path.join('../dataset/clean/{}'.format(dataset_name))
data_fn = os.path.join(save_data_path, '{}.data.pkl'.format(dataset_name))
df = pd.from_pickle(load_fn)
# We remove any row that has no neighbors
print("num nodes = {}".format(len(df)))
df = df[df.neighbors.apply(len) > 0]
print("num nodes = {}".format(len(df)))
df_train = df[df.kind == TRAIN_FLAG]
df_test = df[df.kind == TEST_FLAG]
df_cv = df[df.kind == CV_FLAG]
print("num train: {} num test: {} num cv: {}".format(len(df_train),
len(df_test),
len(df_cv)))
# Remove any non-train neighbors
def remove_test_and_cv_edges(row):
return [r for r in row if r in df_train.index]
df_train = df_train.copy()
df_train.neighbors = df_train.neighbors.apply(remove_test_and_cv_edges)
df_train = df_train[df_train.neighbors.apply(len) > 0]
print("num trains: {}".format(len(df_train)))
# Remove any row that points to a removed train node
df_train.neighbors = df_train.neighbors.apply(remove_test_and_cv_edges)
df_train.neighbors.apply(len).describe()
print("num trains: {}".format(len(df_train)))
###Output
num trains: 151741
num trains: 151741
###Markdown
Process Test and Validatation Set
###Code
print("num test: {}".format(len(df_test)))
df_test = df_test.copy()
df_test.neighbors = df_test.neighbors.apply(remove_test_and_cv_edges)
df_test = df_test[df_test.neighbors.apply(len) > 0]
print("num test: {}".format(len(df_test)))
print("num cv: {}".format(len(df_cv)))
df_cv = df_cv.copy()
df_cv.neighbors = df_cv.neighbors.apply(remove_test_and_cv_edges)
df_cv = df_cv[df_cv.neighbors.apply(len) > 0]
print("num cv: {}".format(len(df_cv)))
###Output
num test: 55228
num test: 53736
num cv: 23660
num cv: 23012
###Markdown
Save Data
###Code
global_id_2_train_id = {global_idx: idx for idx, global_idx
in enumerate(df_train.index)}
def convert_2_train_id(row):
return [global_id_2_train_id[r] for r in row]
train_edges = df_train.neighbors.apply(convert_2_train_id)
train_graph = {}
for node_id, value in train_edges.iteritems():
train_graph[global_id_2_train_id[node_id]] = value
import pickle
save_data_path = os.path.join('../dataset/clean/{}'.format(dataset_name))
save_fn = os.path.join(save_data_path, 'ind.{}.train.graph.pkl'.format(dataset_name))
pickle.dump(train_graph, open(save_fn, 'wb'))
print('save graph data to {}'.format(save_fn))
global_id_2_test_id = {global_idx: idx for idx, global_idx in enumerate(df_test.index)}
# Convert each globalId to trainId because all test nodes only point to train nodes
test_edges = df_test.neighbors.apply(convert_2_train_id)
test_graph = {}
for node_id, value in test_edges.iteritems():
test_graph[global_id_2_test_id[node_id]] = value
save_fn = os.path.join(save_data_path, 'ind.{}.test.graph.pkl'.format(dataset_name))
pickle.dump(test_graph, open(save_fn, 'wb'))
print('save graph data to {}'.format(save_fn))
global_id_2_cv_id = {global_idx: idx for idx, global_idx
in enumerate(df_cv.index)}
# Convert each globalId to trainId because all cv nodes only point to train nodes
cv_edges = df_cv.neighbors.apply(convert_2_train_id)
cv_graph = {}
for node_id, value in cv_edges.iteritems():
cv_graph[global_id_2_cv_id[node_id]] = value
save_fn = os.path.join(save_data_path, 'ind.{}.cv.graph.pkl'.format(dataset_name))
pickle.dump(test_graph, open(save_fn, 'wb'))
print('save graph data to {}'.format(save_fn))
###Output
save graph data to ../dataset/clean/reddit/ind.reddit.cv.graph.pkl
###Markdown
Get Document features
###Code
train_features = list(df_train.features)
train_features = sparse.csr_matrix(train_features)
train_labels = list(df_train.label)
######################################################################################
min_class_id = np.min(train_labels)
max_class_id = np.max(train_labels)
num_classes = max_class_id - min_class_id + 1
gnd_train = sparse.csr_matrix(np.eye(num_classes)[train_labels])
######################################################################################
test_features = list(df_test.features)
test_features = sparse.csr_matrix(test_features)
test_labels = list(df_test.label)
gnd_test = sparse.csr_matrix(np.eye(num_classes)[test_labels])
######################################################################################
cv_features = list(df_cv.features)
cv_features = sparse.csr_matrix(cv_features)
cv_labels = list(df_cv.label)
gnd_cv = sparse.csr_matrix(np.eye(num_classes)[cv_labels])
assert(train_features.shape[1] == test_features.shape[1] == cv_features.shape[1])
assert(gnd_train.shape[1] == gnd_test.shape[1] == gnd_cv.shape[1])
assert(train_features.shape[0] == gnd_train.shape[0])
assert(test_features.shape[0] == gnd_test.shape[0])
assert(cv_features.shape[0] == gnd_cv.shape[0])
import scipy.io
save_fn = os.path.join(save_data_path, 'ind.{}.mat'.format(dataset_name))
scipy.io.savemat(save_fn,
mdict={'train': train_features,
'test': test_features,
'cv': cv_features,
'gnd_train': gnd_train,
'gnd_test': gnd_test,
'gnd_cv': gnd_cv})
print('save data to {}'.format(save_fn))
###Output
save data to ../dataset/clean/reddit/ind.reddit.mat
|
code/big_photometry_table.ipynb
|
###Markdown
Trevor Dorn-Wallenstein 1/21/18 Reading output photometry files from Phil Massey's code, putting them in a big table
###Code
import numpy as np
from matplotlib import pyplot as plt
from astropy.io import fits, ascii
from glob import glob
from astropy.table import Table, vstack, Column
import pandas as pd
%matplotlib inline
phot_dir = '../data/1_17_18/photometry/'
phot_files = glob(phot_dir+'*.WR')
tables = []
for i,file in enumerate(phot_files):
field = file.split('/')[-1][:3]
try:
table = Table(ascii.read(file),names=['RA','Dec','CT','He-CT','SigHe-CT','WN-CT','SigWN-CT','WC-WN','SigWC-WN'],dtype=[np.str,np.str,np.float64,np.float64,np.float64,np.float64,np.float64,np.float64,np.float64])
field_col = Column([field for row in table],name='Field')
table.add_column(field_col)
tables.append(table)
except:
print(i,field,'No photometry')
final_table = vstack(tables)
final_table.write(phot_dir+'all_photometry.dat',format='ascii',overwrite=True)
final_table
###Output
_____no_output_____
|
notebooks2/01_protherm_dataset.ipynb
|
###Markdown
SummaryGenerate adjancency matrices for the Protherm training set directly from PDBs.--- Imports
###Code
import concurrent.futures
import importlib
import logging
import os
import os.path as op
import shutil
import sys
from collections import Counter
from pathlib import Path
import kmbio.PDB
import matplotlib.pyplot as plt
import numpy as np
import pandas as pd
import psutil
import pyarrow as pa
import pyarrow.parquet as pq
from kmtools import structure_tools
%matplotlib inline
logging.basicConfig(level=logging.INFO)
SRC_PATH = Path.cwd().joinpath('..', 'src').resolve(strict=True)
if SRC_PATH.as_posix() not in sys.path:
sys.path.insert(0, SRC_PATH.as_posix())
import helper
importlib.reload(helper)
###Output
_____no_output_____
###Markdown
Parameters
###Code
NOTEBOOK_NAME = '01_protherm_dataset'
NOTEBOOK_PATH = Path(NOTEBOOK_NAME)
NOTEBOOK_PATH.mkdir(parents=True, exist_ok=True)
OUTPUT_PATH = Path(os.getenv('OUTPUT_DIR', NOTEBOOK_PATH.name)).resolve()
OUTPUT_PATH.mkdir(parents=True, exist_ok=True)
OUTPUT_PATH
# DEBUG = "CI" not in os.environ
DEBUG = False
TASK_ID = os.getenv("SLURM_ARRAY_TASK_ID")
TASK_COUNT = os.getenv("ORIGINAL_ARRAY_TASK_COUNT") or os.getenv("SLURM_ARRAY_TASK_COUNT")
DEBUG, TASK_ID, TASK_COUNT
DATABIN_PATH = Path(os.environ['DATABIN_DIR']) # /home/kimlab2/database_data/databin
###Output
_____no_output_____
###Markdown
Load data
###Code
ROSETTA_RESULTS = {}
with pd.HDFStore(DATABIN_PATH.joinpath('elapsam_feature_engineering/v0.1.0/rosetta.h5').as_posix(), 'r') as store:
for key in store:
ROSETTA_RESULTS[key.strip('/')] = store[key][:]
ROSETTA_RESULTS.keys()
ROSETTA_RESULTS['cartesian_ddg-talaris2014_cart-1'].head()
rosetta_results_df = None
for key, df in ROSETTA_RESULTS.items():
df = df.rename(columns={'ddg': key})
if rosetta_results_df is None:
rosetta_results_df = df
else:
assert (rosetta_results_df['ddg_exp'].values == df['ddg_exp'].values).all()
rosetta_results_df = rosetta_results_df.merge(
df.drop('ddg_exp', axis=1), on=['filename-wt', 'pdb_chain', 'mutation'], how='outer')
rosetta_results_df = rosetta_results_df.rename(columns=lambda c: c.replace('-', '_').strip('_'))
display(rosetta_results_df.head())
print(rosetta_results_df.shape)
###Output
_____no_output_____
###Markdown
Copy structures
###Code
STRUCTURE_PATH = NOTEBOOK_PATH.joinpath('structures')
STRUCTURE_PATH.mkdir(exist_ok=True)
os.listdir(STRUCTURE_PATH)[:10]
def get_local_filename(filename):
return STRUCTURE_PATH.joinpath(op.basename(filename)).absolute().as_posix()
get_local_filename(rosetta_results_df['filename_wt'].iloc[0])
file_list = rosetta_results_df['filename_wt'].drop_duplicates().tolist()
local_filename_wt = []
for i, filename in enumerate(file_list):
if i % 200 == 0:
print(i)
new_filename = STRUCTURE_PATH.joinpath(op.basename(filename))
filename = filename.replace(
"/home/kimlab2/database_data/biological-data-warehouse",
Path("~/datapkg").expanduser().as_posix(),
)
local_filename = get_local_filename(filename)
if not op.isfile(local_filename):
shutil.copy(filename, local_filename)
local_filename_wt.append(local_filename)
rosetta_results_df['local_filename_wt'] = local_filename_wt
rosetta_results_df.head()
###Output
_____no_output_____
###Markdown
Process data
###Code
if DEBUG:
rosetta_results_df = rosetta_results_df.iloc[:10]
###Output
_____no_output_____
###Markdown
Extract adjacencies
###Code
def extract_seq_and_adj(row):
domain, result_df = helper.get_interaction_dataset_wdistances(
row.local_filename_wt, 0, row.pdb_chain, r_cutoff=12)
domain_sequence = structure_tools.get_chain_sequence(domain)
assert max(result_df['residue_idx_1'].values) < len(domain_sequence)
assert max(result_df['residue_idx_2'].values) < len(domain_sequence)
result = {
'sequence': domain_sequence,
'residue_idx_1': result_df['residue_idx_1'].values,
'residue_idx_2': result_df['residue_idx_2'].values,
'distances': result_df['distance'].values,
}
return result
def worker(row_dict):
row = helper.to_namedtuple(row_dict)
result = extract_seq_and_adj(row)
return result
logging.getLogger("kmbio.PDB.core.atom").setLevel(logging.WARNING)
columns = ["local_filename_wt", "pdb_chain"]
with concurrent.futures.ProcessPoolExecutor(psutil.cpu_count(logical=False)) as pool:
futures = pool.map(worker, (t._asdict() for t in rosetta_results_df[columns].itertuples()))
results = list(futures)
protherm_validaton_dataset = rosetta_results_df.copy()
protherm_validaton_dataset = protherm_validaton_dataset.rename(columns={'pdb_chain': 'chain_id'})
protherm_validaton_dataset['structure_id'] = [
Path(filename).name[3:7] for filename in protherm_validaton_dataset["filename_wt"]
]
protherm_validaton_dataset['model_id'] = 0
protherm_validaton_dataset['qseq'] = [result["sequence"] for result in results]
protherm_validaton_dataset['residue_idx_1_corrected'] = [result["residue_idx_1"] for result in results]
protherm_validaton_dataset['residue_idx_2_corrected'] = [result["residue_idx_2"] for result in results]
protherm_validaton_dataset['distances'] = [result["distances"] for result in results]
def mutation_matches_sequence(mutation, sequence):
return sequence[int(mutation[1:-1]) - 1] == mutation[0]
protherm_validaton_dataset['mutation_matches_sequence'] = [
mutation_matches_sequence(mutation, sequence)
for mutation, sequence
in protherm_validaton_dataset[['mutation', 'qseq']].values
]
assert protherm_validaton_dataset['mutation_matches_sequence'].all()
def apply_mutation(sequence, mutation):
wt, pos, mut = mutation[0], int(mutation[1:-1]), mutation[-1]
assert sequence[pos - 1] == wt
sequence_mut = sequence[:pos - 1] + mut + sequence[pos:]
assert sequence_mut[pos - 1] == mut
assert len(sequence) == len(sequence_mut)
return sequence_mut
protherm_validaton_dataset['qseq_mutation'] = [
apply_mutation(sequence, mutation)
for mutation, sequence
in protherm_validaton_dataset[['mutation', 'qseq']].values
]
assert not protherm_validaton_dataset.isnull().any().any()
columns = [
'structure_id', 'model_id', 'chain_id', 'qseq', 'qseq_mutation', 'ddg_exp',
'residue_idx_1_corrected', 'residue_idx_2_corrected', 'distances',
]
for column in columns:
assert column in protherm_validaton_dataset.columns, column
pq.write_table(
pa.Table.from_pandas(protherm_validaton_dataset, preserve_index=False),
OUTPUT_PATH.joinpath('protherm_validaton_dataset.parquet').as_posix(),
version='2.0', flavor='spark'
)
###Output
_____no_output_____
###Markdown
Explore
###Code
protherm_validaton_dataset.head(2)
protherm_validaton_dataset["filename"] = protherm_validaton_dataset["filename_wt"].str.split("/").str[-1]
protherm_validaton_dataset[["filename", "chain_id", "mutation", "ddg_exp"]].to_csv(NOTEBOOK_PATH.joinpath("for_carles.csv"), sep="\t")
aa_wt_counter = Counter(protherm_validaton_dataset['mutation'].str[0])
aa_mut_counter = Counter(protherm_validaton_dataset['mutation'].str[-1])
labels = list(aa_wt_counter)
aa_wt = [aa_wt_counter[l] for l in labels]
aa_mut = [aa_mut_counter[l] for l in labels]
indexes = np.arange(len(labels))
width = 0.3
with plt.rc_context(rc={'figure.figsize': (8, 5), 'font.size': 14}):
plt.bar(indexes - 0.15 , aa_wt, width, label="wt")
plt.bar(indexes + 0.15, aa_mut, width, label="mut")
plt.xticks(indexes, labels)
plt.ylabel("Number of occurrences")
plt.legend()
###Output
_____no_output_____
|
docs/source/tutorials/.ipynb_checkpoints/Background Subtraction Example-checkpoint.ipynb
|
###Markdown
Background Substarction for Detection and Tracking ExampleIn the following example with the sample video the movement should be detected frame by frame.All the background substraction parameters are in config.py file. First of all, all the required liberaries should be imported.
###Code
%cd ..\..
# only if you run it from the sub directory
from utils import resize
from background_substraction import BG_substractor
import numpy as np
import cv2
###Output
_____no_output_____
###Markdown
Now the sample video should be read with a video reader object from Opencv liberary
###Code
cap = cv2.VideoCapture('docs/sample.mp4')
ret, bg = cap.read() # read the first frame
###Output
_____no_output_____
###Markdown
The first frame is considered the default background in the start to compare it with the next frames in the video in order to detect the motion. In the case where we want start the movement detection at some later frame, not the first one. We could write
###Code
frame_id = 1 # the frame that should we start from
cap.set(1, frame_id-1)
###Output
_____no_output_____
###Markdown
Then we intilize the background substarctor object
###Code
BG_s = BG_substractor(bg)
ret, frame = cap.read()
###Output
_____no_output_____
###Markdown
Main loopNow everything are ready to start the background substarction loop.The result will be shown in a new windowThe video will keep running until you hit **ESC** or the video end
###Code
while ret:
frame_id += 1
I_com = BG_s.bg_substract(frame)
cv2.imshow('fgmask', resize(I_com,0.2))
#print(frame_id)
k = cv2.waitKey(30) & 0xff
if k == 27:
break
ret, frame = cap.read()
cap.release()
cv2.destroyAllWindows()
###Output
_____no_output_____
###Markdown
As it was shown, the result has some problems, The moving cyclists are detected but not perfectly. Additionally the trees and part of the road are detected as moving objects.A resonable step would be to filter the video from the small noisy objects by deleting the small blobs. we can test that using other method of the class, namely: `get_big_objects`
###Code
cap = cv2.VideoCapture('docs/sample.mp4')
ret, bg = cap.read() # read the first frame
BG_s = BG_substractor(bg)
ret, frame = cap.read()
while ret:
frame_id += 1
I_com = BG_s.bg_substract(frame)
# filter small objects
I_com, _ = BG_s.get_big_objects(I_com,frame)
cv2.imshow('fgmask', resize(I_com,0.2))
if frame_id == 30:
frame_2_save = resize(I_com).copy()
#print(frame_id)
k = cv2.waitKey(30) & 0xff
if k == 27:
break
ret, frame = cap.read()
cap.release()
cv2.destroyAllWindows()
###Output
_____no_output_____
###Markdown
Finally we can view the result at step 30, as follows:
###Code
%matplotlib inline
import matplotlib.pyplot as plt
plt.imshow(frame_2_save)
###Output
_____no_output_____
|
archives/notebooks/explore_gene_correlations_jan25-bck.ipynb
|
###Markdown
Get gene by metacell tables - RNA- mCG- ATAC
###Code
# enhancer_gene_pairs
f = '/cndd2/fangming/projects/scf_enhancers/results/200521_to_evals.tsv'
enh_gene_table = pd.read_csv(f, sep='\t')
print(enh_gene_table.shape)
CLUSTER_COL = 'cluster_r'
def pipe_corr_analysis_atac(
common_modx_cells, common_mody_cells,
cell_cell_knn_xaxis, cell_cell_knn_yaxis,
common_genes, common_enhancer_regions,
X, Y,
modx_clsts, knn_xy,
enhancer_gene_to_eval,
output_corrs,
corr_type='pearsonr',
force=False,
num_metacell_limit=0,
):
"""
"""
# new cells
common_modx_cells_updated = np.intersect1d(common_modx_cells, cell_cell_knn_xaxis)
common_mody_cells_updated = np.intersect1d(common_mody_cells, cell_cell_knn_yaxis)
# make sure the original matrices have the correct index
x_idx = snmcseq_utils.get_index_from_array(common_modx_cells, common_modx_cells_updated)
y_idx = snmcseq_utils.get_index_from_array(common_mody_cells, common_mody_cells_updated)
X = X.tocsc()[:, x_idx]
Y = Y.tocsc()[:, y_idx]
# make sure knn_xy, knn_xx have the right cell index
cell_idx_xaxis = snmcseq_utils.get_index_from_array(cell_cell_knn_xaxis, common_modx_cells_updated)
cell_idx_yaxis = snmcseq_utils.get_index_from_array(cell_cell_knn_yaxis, common_mody_cells_updated)
knn_xy = knn_xy.tocsr()[cell_idx_xaxis,:].tocsc()[:,cell_idx_yaxis] # x-by-y
modx_clsts = modx_clsts.reindex(common_modx_cells_updated)
logging.info("{}_{}_{}_{}".format(knn_xy.shape, modx_clsts.shape, X.shape, Y.shape,))
for clst_col in modx_clsts.columns:
logging.info(clst_col)
if clst_col.startswith(CLUSTER_COL):
# output_corr = output_corrs.format(clst_col)
# if not force and os.path.isfile(output_corr):
# logging.info("skip {}, already exists...".format(output_corr))
# continue # skip the existing file
# choose one clustering to proceed
uniq_labels = np.sort(modx_clsts[clst_col].unique())
logging.info("Number of metacells: {}".format(len(uniq_labels)))
if num_metacell_limit > 0 and len(uniq_labels) > num_metacell_limit:
logging.info("skip {}, exceeding max num_metacell_limit...".format(len(uniq_labels)))
continue
knn_xz = enhancer_gene_utils.turn_cluster_labels_to_knn(modx_clsts[clst_col].values,
uniq_labels,
)
# Dec 21,2020
# gene by metacell (counts)
gc_rna = X.dot(knn_xz).todense()
# normalization (logCPM)
gc_rna = snmcseq_utils.logcpm(pd.DataFrame(gc_rna)).values
# Dec 21,2020
enh_lengths = pd.Series((common_enhancer_regions['end']-common_enhancer_regions['start']).values)
# enhancer by metacell (counts)
knn_yz = knn_xy.T.dot(knn_xz)
ec_atac = Y.dot(knn_yz).todense()
# normalization (logTPM)
ec_atac = snmcseq_utils.logtpm(pd.DataFrame(ec_atac), enh_lengths).values
logging.info("{} {}".format(gc_rna.shape, ec_atac.shape,))
# # corr analysis
# output_corr = output_corrs.format(clst_col)
# (to_correlate, corrs, corrs_shuffled, corrs_shuffled_cells) = enhancer_gene_utils.compute_enh_gene_corrs(
# gc_rna, ec_atac,
# common_genes, np.arange(len(ec_atac)),
# enhancer_gene_to_eval['gene'].values,
# enhancer_gene_to_eval['ens'].values,
# output_file=output_corr, corr_type=corr_type, chunksize=100000, verbose_level=0,
# )
return (
gc_rna, ec_atac,
common_genes, np.arange(len(ec_atac)),
enhancer_gene_to_eval['gene'].values,
enhancer_gene_to_eval['ens'].values,
'', corr_type, 100000, 0,
)
def wrap_corr_analysis_atac(
mod_x, mod_y,
input_name_tag, i_sub,
corr_type='pearsonr',
force=False,
num_metacell_limit=0,
):
"""
"""
# (i, k, --r)
output_corrs = '/cndd2/fangming/projects/scf_enhancers/results/{}_{}_{{}}_{}_corrs.pkl'.format(input_name_tag, i_sub, corr_type)
# input enh-gene tables, gene-by-cell, enhancer-by-cell matrices
input_enh_gene_table = '/cndd2/fangming/projects/scf_enhancers/results/200521_to_evals.tsv'
input_bundle_dirc = '/cndd2/fangming/projects/scf_enhancers/data/organized_cell_level/version_nov9'
bundle_fnames = (
'cell_10x_cells_v3.txt',
'cell_snatac_gene.txt',
'gene_10x_cells_v3.txt',
'enh.tsv',
'mat_10x_cells_v3.npz',
'mat_snatac_gene.npz',
)
# for knn_xx
input_knn_dirc = '/cndd2/fangming/projects/miniatlas/results'
input_modx_clsts = [
'clusterings_{}_{}_sub{}.tsv.gz'.format(mod_x, input_name_tag, i_sub),
]
# for knn_xy
input_knn_xy = 'knn_across_{}_{}_{}.npz.{}.npz'.format(input_name_tag, mod_x, mod_y, i_sub)
input_knn_cells_xaxis = 'cells_{}_{}.npy.{}.npy'.format(mod_x, input_name_tag, i_sub)
input_knn_cells_yaxis = 'cells_{}_{}.npy.{}.npy'.format(mod_y, input_name_tag, i_sub)
# # Load data
# input_bundle
with snmcseq_utils.cd(input_bundle_dirc):
bundle = []
for fname in bundle_fnames:
# save all as pickle file
with open(fname, "rb") as fh:
item = pickle.load(fh)
bundle.append(item)
logging.info("{}_{}_{}".format(type(item), item.shape, fname))
(common_modx_cells, common_mody_cells,
common_genes, common_enhancer_regions,
X, Y,
) = bundle
# input knn networks
with snmcseq_utils.cd(input_knn_dirc):
# for knn_xx
# modx_clsts = pd.read_csv(input_modx_clsts, sep='\t',index_col=0)
modx_clsts = pd.concat([
pd.read_csv(fname, sep='\t',index_col=0)
for fname in input_modx_clsts
], axis=1)
# for knn_xy
knn_xy = sparse.load_npz(input_knn_xy)
cell_cell_knn_xaxis = np.load(input_knn_cells_xaxis, allow_pickle=True)
cell_cell_knn_yaxis = np.load(input_knn_cells_yaxis, allow_pickle=True)
logging.info("{} {} {} {}".format(modx_clsts.shape, knn_xy.shape,
cell_cell_knn_xaxis.shape,
cell_cell_knn_yaxis.shape,
)
)
# enhancer-gene linkage
enhancer_gene_to_eval = pd.read_csv(input_enh_gene_table, sep='\t')
return pipe_corr_analysis_atac(
common_modx_cells, common_mody_cells,
cell_cell_knn_xaxis, cell_cell_knn_yaxis,
common_genes, common_enhancer_regions,
X, Y,
modx_clsts, knn_xy,
enhancer_gene_to_eval,
output_corrs,
corr_type=corr_type,
force=force,
num_metacell_limit=num_metacell_limit,
)
def pipe_corr_analysis_mc(
common_rna_cells, common_mc_cells,
cell_cell_knn_xaxis, cell_cell_knn_yaxis,
common_genes,
X, Y_cg, Y_mcg,
modx_clsts, knn_xy,
enhancer_gene_to_eval,
output_corrs,
corr_type='pearsonr',
force=False,
num_metacell_limit=0,
):
"""
"""
# new cells
common_rna_cells_updated = np.intersect1d(common_rna_cells, cell_cell_knn_xaxis)
common_mc_cells_updated = np.intersect1d(common_mc_cells, cell_cell_knn_yaxis)
# make sure the original matrices have the correct index
x_idx = snmcseq_utils.get_index_from_array(common_rna_cells, common_rna_cells_updated)
y_idx = snmcseq_utils.get_index_from_array(common_mc_cells, common_mc_cells_updated)
X = X.tocsc()[:, x_idx]
Y_cg = Y_cg.tocsc()[:, y_idx]
Y_mcg = Y_mcg.tocsc()[:, y_idx]
# make sure knn_xy, knn_xx have the right cell index
cell_idx_xaxis = snmcseq_utils.get_index_from_array(cell_cell_knn_xaxis, common_rna_cells_updated)
cell_idx_yaxis = snmcseq_utils.get_index_from_array(cell_cell_knn_yaxis, common_mc_cells_updated)
knn_xy = knn_xy.tocsr()[cell_idx_xaxis,:].tocsc()[:,cell_idx_yaxis] # x-by-y
modx_clsts = modx_clsts.reindex(common_rna_cells_updated)
logging.info("{}_{}_{}_{}_{}".format(knn_xy.shape, modx_clsts.shape, X.shape, Y_cg.shape, Y_mcg.shape))
for clst_col in modx_clsts.columns:
if clst_col.startswith(CLUSTER_COL):
logging.info(clst_col)
# output_corr = output_corrs.format(clst_col)
# if not force and os.path.isfile(output_corr):
# logging.info("skip {}, already exists...".format(output_corr))
# continue # skip the existing file
# choose one clustering to proceed
uniq_labels = np.sort(modx_clsts[clst_col].unique())
logging.info("Number of metacells: {}".format(len(uniq_labels)))
if num_metacell_limit > 0 and len(uniq_labels) > num_metacell_limit:
logging.info("skip {}, exceeding max num_metacell_limit...".format(len(uniq_labels)))
knn_xz = enhancer_gene_utils.turn_cluster_labels_to_knn(modx_clsts[clst_col].values,
uniq_labels,
)
# # normalization - such that metacells made of more cells still sums to 1
# knn_xz = knn_xz.dot(sparse.diags(np.ravel(1.0/knn_xz.sum(axis=0))))
# # gene by metacell
# gc_rna = X.dot(knn_xz).todense()
# gene by metacell (counts)
gc_rna = X.dot(knn_xz).todense()
# normalization (logCPM)
gc_rna = snmcseq_utils.logcpm(pd.DataFrame(gc_rna)).values
# enhancer by metacell (counts cg, mcg)
knn_yz = knn_xy.T.dot(knn_xz)
ec_cg = Y_cg.dot(knn_yz).todense()
ec_mcg = Y_mcg.dot(knn_yz).todense()
logging.info("{} {} {}".format(gc_rna.shape, ec_cg.shape, ec_mcg.shape))
# mC
ec_mccg = snmcseq_utils.get_mcc_lite_v4(
pd.DataFrame(ec_cg).astype(np.float32),
pd.DataFrame(ec_mcg).astype(np.float32),
base_call_cutoff=5, sufficient_coverage_fraction=0.8, fillna=True)
logging.info("{}".format(ec_mccg.shape))
# # corr analysis
# (to_correlate, corrs, corrs_shuffled, corrs_shuffled_cells) = enhancer_gene_utils.compute_enh_gene_corrs(
# gc_rna, ec_mccg,
# common_genes, ec_mccg.index.values,
# enhancer_gene_to_eval['gene'].values,
# enhancer_gene_to_eval['ens'].values,
# output_file=output_corr, corr_type=corr_type, chunksize=100000, verbose_level=0,
# )
return (
gc_rna, ec_mccg,
common_genes, ec_mccg.index.values,
enhancer_gene_to_eval['gene'].values,
enhancer_gene_to_eval['ens'].values,
'', corr_type, 100000, 0,
)
def wrap_corr_analysis_mc(
mod_x, mod_y,
input_nme_tag, i_sub,
corr_type='pearsonr',
force=False,
num_metacell_limit=0,
):
"""
"""
# (i, k, --r)
output_corrs = '/cndd2/fangming/projects/scf_enhancers/results/{}_{}_{{}}_{}_corrs.pkl'.format(input_name_tag, i_sub, corr_type)
# input enh-gene tables, gene-by-cell, enhancer-by-cell matrices
input_enh_gene_table = '/cndd2/fangming/projects/scf_enhancers/results/200521_to_evals.tsv'
input_bundle_dirc = '/cndd2/fangming/projects/scf_enhancers/data/organized_cell_level/version_nov9'
bundle_fnames = (
'cell_10x_cells_v3.txt',
'cell_snmcseq_gene.txt',
'gene_10x_cells_v3.txt',
'enh.tsv',
'mat_10x_cells_v3.npz',
'mat_mcg_snmcseq_gene.npz',
'mat_cg_snmcseq_gene.npz',
)
# for knn_xx
input_knn_dirc = '/cndd2/fangming/projects/miniatlas/results'
input_modx_clsts = [
'clusterings_{}_{}_sub{}.tsv.gz'.format(mod_x, input_name_tag, i_sub),
]
# for knn_xy
input_knn_xy = 'knn_across_{}_{}_{}.npz.{}.npz'.format(input_name_tag, mod_x, mod_y, i_sub)
input_knn_cells_xaxis = 'cells_{}_{}.npy.{}.npy'.format(mod_x, input_name_tag, i_sub)
input_knn_cells_yaxis = 'cells_{}_{}.npy.{}.npy'.format(mod_y, input_name_tag, i_sub)
# # Load data
# input_bundle
with snmcseq_utils.cd(input_bundle_dirc):
bundle = []
for fname in bundle_fnames:
# save all as pickle file
with open(fname, "rb") as fh:
item = pickle.load(fh)
bundle.append(item)
logging.info("{}_{}_{}".format(type(item), item.shape, fname))
(common_rna_cells, common_mc_cells,
common_genes, common_enhancer_regions,
X, Y_mcg, Y_cg,
# knn_xy, knn_xx,
) = bundle
# input knn networks
with snmcseq_utils.cd(input_knn_dirc):
# for knn_xx
# modx_clsts = pd.read_csv(input_modx_clsts, sep='\t',index_col=0)
modx_clsts = pd.concat([
pd.read_csv(fname, sep='\t',index_col=0)
for fname in input_modx_clsts
], axis=1)
# for knn_xy
knn_xy = sparse.load_npz(input_knn_xy)
cell_cell_knn_xaxis = np.load(input_knn_cells_xaxis, allow_pickle=True)
cell_cell_knn_yaxis = np.load(input_knn_cells_yaxis, allow_pickle=True)
logging.info("{} {} {} {}".format(
modx_clsts.shape,
knn_xy.shape,
cell_cell_knn_xaxis.shape,
cell_cell_knn_yaxis.shape,
)
)
# enhancer-gene linkage
enhancer_gene_to_eval = pd.read_csv(input_enh_gene_table, sep='\t')
return pipe_corr_analysis_mc(
common_rna_cells, common_mc_cells,
cell_cell_knn_xaxis, cell_cell_knn_yaxis,
common_genes,
X, Y_cg, Y_mcg,
modx_clsts, knn_xy,
enhancer_gene_to_eval,
output_corrs,
corr_type=corr_type,
force=force,
num_metacell_limit=num_metacell_limit,
)
mod_x, mod_y = '10x_cells_v3', 'snatac_gene'
ka = 30
knn = 30
date = "201130"
input_name_tag = 'mop_{}_{}_ka{}_knn{}_{}'.format(mod_x, mod_y, ka, knn, date)
i_sub = '0'
CLUSTER_COL = 'cluster_r25'
res1 = wrap_corr_analysis_atac(
mod_x, mod_y,
input_name_tag, i_sub,
corr_type='spearmanr',
force=False,
num_metacell_limit=0,
)
gc_rna1, ec_atac = res1[:2]
print(gc_rna1.shape, ec_atac.shape)
mod_x, mod_y = '10x_cells_v3', 'snmcseq_gene'
ka = 30
knn = 30
date = "201130"
input_name_tag = 'mop_{}_{}_ka{}_knn{}_{}'.format(mod_x, mod_y, ka, knn, date)
i_sub = '0'
res2 = wrap_corr_analysis_mc(
mod_x, mod_y,
input_name_tag, i_sub,
# corr_type='pearsonr',
force=False,
num_metacell_limit=0,
)
gc_rna2, ec_mccg = res2[:2]
print(gc_rna2.shape, ec_mccg.shape)
ec_mccg.tail()
###Output
_____no_output_____
###Markdown
analysis- make sure enhancer index matches (yes)- PCA on genes (scree plot)- remove top ones- regress RNA ~ mC/ATAC
###Code
y_gc = gc_rna2 - np.mean(gc_rna2, axis=0)
x_ec = ec_mccg
U, s, Vt = fbpca.pca(y_gc, k=min(y_gc.shape))
V = Vt.T
print(U.shape, s.shape, Vt.shape)
# sns.heatmap(Z.T.dot(Z))
sns.heatmap(V.dot(Vt))
Z = V.dot(np.diag(s))
y_gr = y_gc.dot(Z)
x_er = ec_mccg.dot(Z)
print(y_gr.shape, x_er.shape)
def eval_scree(s, pcs_to_plot):
"""
"""
scree = np.power(s, 2)/np.sum(np.power(s, 2))
fig, ax = plt.subplots(1, 1, figsize=(1*5, 1*5))
ax.set_title("PC1: {:.3f}".format(scree[0]))
ax.plot(pcs_to_plot, scree[pcs_to_plot-1], '-o')
ax.set_xlabel('Principal components')
ax.set_ylabel('Fraction of variance')
plt.show()
# regression (applied to each gene and enh pair) -- correlate gene-enh
configs = [
(y_gc, x_ec, 'spearmanr', 'test_ols_mcrna_spearmanr_jan25.pkl'),
(y_gc, x_ec, 'pearsonr', 'test_ols_mcrna_pearsonr_jan25.pkl'),
(y_gr, x_er, 'spearmanr', 'test_gls_mcrna_spearmanr_jan25.pkl'),
(y_gr, x_er, 'pearsonr', 'test_gls_mcrna_pearsonr_jan25.pkl'),
]
for config in configs:
y, x, corr_type, output_fname = config
# # corr analysis
output_file = ('/cndd2/fangming/projects/scf_enhancers/results_jan2021/{}'.format(output_fname))
print(output_file)
(to_correlate, corrs, corrs_shuffled, corrs_shuffled_cells) = enhancer_gene_utils.compute_enh_gene_corrs(
y, x,
res2[2], res2[3],
res2[4], res2[5],
output_file=output_file, corr_type=corr_type,
)
to_evals = enh_gene_table
res_corrs = to_evals[to_correlate].copy()
res_corrs['corr'] = corrs
res_corrs['corr_shuff'] = corrs_shuffled
res_corrs['corr_shuff_cells'] = corrs_shuffled_cells
###Output
/cndd2/fangming/projects/scf_enhancers/results_jan2021/test_ols_mcrna_spearmanr_jan25.pkl
spearmanr chosen!
0 0.0019407272338867188
100000 0.2553842067718506
200000 0.5072216987609863
300000 0.7540199756622314
400000 0.9988858699798584
500000 1.24472975730896
600000 1.539872407913208
700000 1.7849469184875488
800000 2.0532755851745605
900000 2.3120663166046143
1000000 2.6277966499328613
1100000 2.8733863830566406
1200000 3.1243157386779785
1300000 3.374530553817749
1400000 3.630615711212158
1500000 3.883728504180908
1600000 4.137850761413574
1700000 4.392799615859985
1800000 4.646371603012085
1900000 4.900473117828369
2000000 5.153280735015869
2100000 5.405025005340576
2200000 5.658514022827148
2300000 5.911017179489136
0 0.0019288063049316406
100000 0.2624630928039551
200000 0.5235545635223389
300000 0.7796168327331543
400000 1.0315260887145996
500000 1.2838027477264404
600000 1.5370111465454102
700000 1.7899901866912842
800000 2.042246103286743
900000 2.2942659854888916
1000000 2.5464982986450195
1100000 2.798361301422119
1200000 3.0562806129455566
1300000 3.3158953189849854
1400000 3.5743045806884766
1500000 3.833472728729248
1600000 4.095446348190308
1700000 4.361511468887329
1800000 4.626864433288574
1900000 4.893708229064941
2000000 5.160646677017212
2100000 5.42435097694397
2200000 5.690541744232178
2300000 5.955345869064331
0 0.005965232849121094
100000 0.2569894790649414
200000 0.5135128498077393
300000 0.758669376373291
400000 1.0014936923980713
500000 1.246758222579956
600000 1.4897031784057617
700000 1.7325398921966553
800000 1.975029706954956
900000 2.2204275131225586
1000000 2.4631292819976807
1100000 2.705615997314453
1200000 2.963911771774292
1300000 3.2443277835845947
1400000 3.5062878131866455
1500000 3.7696218490600586
1600000 4.063923597335815
1700000 4.322607040405273
1800000 4.579402923583984
1900000 4.835537672042847
2000000 5.090864181518555
2100000 5.343405246734619
2200000 5.597536563873291
2300000 5.85074257850647
/cndd2/fangming/projects/scf_enhancers/results_jan2021/test_ols_mcrna_pearsonr_jan25.pkl
pearsonr chosen!
0 0.0019981861114501953
100000 0.1932370662689209
200000 0.3839709758758545
300000 0.565974235534668
400000 0.7443327903747559
500000 0.922921895980835
600000 1.1034185886383057
700000 1.2832605838775635
800000 1.4653820991516113
900000 1.6699061393737793
1000000 1.8588926792144775
1100000 2.0595226287841797
1200000 2.3313841819763184
1300000 2.5358152389526367
1400000 2.7362141609191895
1500000 2.9325332641601562
1600000 3.130089282989502
1700000 3.3331496715545654
1800000 3.5358173847198486
1900000 3.7350618839263916
2000000 3.932384729385376
2100000 4.128077507019043
2200000 4.326518774032593
2300000 4.522755146026611
0 0.003197193145751953
100000 0.23465943336486816
200000 0.4662444591522217
300000 0.6888606548309326
400000 0.9072301387786865
500000 1.1259937286376953
600000 1.3464550971984863
700000 1.5666322708129883
800000 1.7852985858917236
900000 2.003060817718506
1000000 2.2217423915863037
1100000 2.4445884227752686
1200000 2.684939384460449
1300000 2.923428773880005
1400000 3.1782946586608887
1500000 3.4730489253997803
1600000 3.7161519527435303
1700000 3.9571845531463623
1800000 4.196767568588257
1900000 4.4362404346466064
2000000 4.673829793930054
2100000 4.9100401401519775
2200000 5.149056673049927
2300000 5.3860132694244385
0 0.0018460750579833984
100000 0.24702191352844238
200000 0.47928404808044434
300000 0.70334792137146
400000 0.9236211776733398
500000 1.1442835330963135
600000 1.3665852546691895
700000 1.5886244773864746
800000 1.8094167709350586
900000 2.029372215270996
1000000 2.251568555831909
1100000 2.4733190536499023
1200000 2.7033445835113525
1300000 2.946666717529297
1400000 3.176422357559204
1500000 3.407010555267334
1600000 3.6770830154418945
1700000 3.9220919609069824
1800000 4.1738505363464355
1900000 4.459058523178101
2000000 4.702644109725952
2100000 4.94046425819397
2200000 5.179965019226074
2300000 5.417117595672607
/cndd2/fangming/projects/scf_enhancers/results_jan2021/test_gls_mcrna_spearmanr_jan25.pkl
spearmanr chosen!
0 0.0019330978393554688
100000 0.25980186462402344
200000 0.5171530246734619
300000 0.7694709300994873
400000 1.016150712966919
500000 1.2663986682891846
600000 1.5231657028198242
700000 1.7707560062408447
800000 2.0200417041778564
900000 2.2708423137664795
1000000 2.519453763961792
1100000 2.767672061920166
1200000 3.017695665359497
1300000 3.265162229537964
1400000 3.512287139892578
1500000 3.7623543739318848
1600000 4.012063026428223
1700000 4.26547646522522
1800000 4.527404308319092
1900000 4.78008770942688
2000000 5.032672643661499
2100000 5.283857107162476
2200000 5.541009902954102
2300000 5.793256759643555
0 0.001956939697265625
100000 0.26466846466064453
200000 0.5256757736206055
300000 0.7800076007843018
400000 1.0311732292175293
500000 1.2827117443084717
600000 1.5357673168182373
700000 1.7878897190093994
800000 2.0390729904174805
900000 2.289757251739502
1000000 2.5417263507843018
1100000 2.7935845851898193
1200000 3.051238536834717
1300000 3.3101541996002197
1400000 3.568592071533203
1500000 3.827620267868042
1600000 4.089252471923828
1700000 4.354485988616943
1800000 4.619726896286011
1900000 4.884535074234009
2000000 5.162574768066406
2100000 5.426035404205322
2200000 5.691580295562744
2300000 5.955906391143799
0 0.0018355846405029297
100000 0.2510805130004883
200000 0.49932336807250977
300000 0.7429060935974121
400000 0.9851338863372803
500000 1.2270152568817139
600000 1.469942569732666
700000 1.7131085395812988
800000 1.9549808502197266
900000 2.1965672969818115
1000000 2.4380009174346924
1100000 2.6793746948242188
1200000 2.92578125
1300000 3.1792736053466797
1400000 3.431530475616455
1500000 3.6835408210754395
1600000 3.940809488296509
1700000 4.197901010513306
1800000 4.453052043914795
1900000 4.710447311401367
2000000 4.98556923866272
2100000 5.247706890106201
2200000 5.508103609085083
2300000 5.764901638031006
/cndd2/fangming/projects/scf_enhancers/results_jan2021/test_gls_mcrna_pearsonr_jan25.pkl
pearsonr chosen!
0 0.0019636154174804688
100000 0.25132322311401367
200000 0.5000045299530029
300000 0.7437789440155029
400000 0.9857683181762695
500000 1.2273857593536377
600000 1.4703047275543213
700000 1.7123334407806396
800000 1.9544661045074463
900000 2.195655584335327
1000000 2.4380109310150146
1100000 2.6794755458831787
1200000 2.9284162521362305
1300000 3.1780178546905518
1400000 3.433323383331299
1500000 3.6914947032928467
1600000 3.9718332290649414
1700000 4.252387285232544
1800000 4.56081485748291
1900000 4.822745084762573
2000000 5.08195161819458
2100000 5.336362838745117
2200000 5.593115568161011
2300000 5.848126649856567
0 0.0037589073181152344
100000 0.2647686004638672
200000 0.5252509117126465
300000 0.7794795036315918
400000 1.0313122272491455
500000 1.2836682796478271
600000 1.5366365909576416
700000 1.7889206409454346
800000 2.040621042251587
900000 2.29193377494812
1000000 2.5437142848968506
1100000 2.7952630519866943
1200000 3.0546202659606934
1300000 3.3140952587127686
1400000 3.5731959342956543
1500000 3.84793758392334
1600000 4.110399484634399
1700000 4.391491889953613
1800000 4.666250705718994
1900000 4.957513332366943
2000000 5.248953104019165
2100000 5.562230825424194
2200000 5.835371732711792
2300000 6.1045918464660645
0 0.0018448829650878906
100000 0.253063440322876
200000 0.5034255981445312
300000 0.7484762668609619
400000 0.9914505481719971
500000 1.235396385192871
600000 1.4799389839172363
700000 1.7233457565307617
800000 1.966538429260254
900000 2.2091853618621826
1000000 2.452129364013672
1100000 2.69525408744812
1200000 2.9429233074188232
1300000 3.1910030841827393
1400000 3.437711477279663
1500000 3.6869025230407715
1600000 3.937352180480957
1700000 4.190991401672363
1800000 4.445432662963867
1900000 4.699839353561401
2000000 4.953157901763916
2100000 5.206681251525879
2200000 5.460845232009888
2300000 5.713714361190796
###Markdown
Jump start from here
###Code
# regression (applied to each gene and enh pair) -- correlate gene-enh
configs = [
('ols_spearman_mcrna', 'spearmanr', 'test_ols_mcrna_spearmanr_jan25.pkl'),
('ols_pearson_mcrna', 'pearsonr', 'test_ols_mcrna_pearsonr_jan25.pkl'),
('gls_spearman_mcrna', 'spearmanr', 'test_gls_mcrna_spearmanr_jan25.pkl'),
('gls_pearson_mcrna', 'pearsonr', 'test_gls_mcrna_pearsonr_jan25.pkl'),
]
res_corrs_all = {}
for config in configs:
label, corr_type, output_fname = config
# # corr analysis
output_file = ('/cndd2/fangming/projects/scf_enhancers/results_jan2021/{}'.format(output_fname))
print(output_file)
with open(output_file, 'rb') as fh:
(to_correlate, corrs, corrs_shuffled, corrs_shuffled_cells) = pickle.load(fh)
to_evals = enh_gene_table
res_corrs = to_evals[to_correlate].copy()
res_corrs['corr'] = corrs
res_corrs['corr_shuff'] = corrs_shuffled
res_corrs['corr_shuff_cells'] = corrs_shuffled_cells
res_corrs_all[label] = res_corrs
def plot_routine(res_corrs1, res_corrs2, title1, title2, corr_type, ylim1=[], ylim2=[],
bins = np.linspace(-1, 1, 201),
output='',
):
"""
"""
# output = output_figures.format('hist_corr_both.pdf')
fig = plt.figure(figsize=(5*2, 5))
gs = fig.add_gridspec(3, 2)
axs = np.array([[fig.add_subplot(gs[0, 0]), fig.add_subplot(gs[0, 1]),],
[fig.add_subplot(gs[1:, 0]), fig.add_subplot(gs[1:, 1]),],
])
for j, (res_corrs, title) in enumerate(zip([res_corrs1, res_corrs2],
[title1, title2],
)):
KB = 1000
labels_base = [
'shuffled metacells',
'shuffled genes',
'<500kb',
'<100kb',
]
# corr1
labels = [label+' mCG-RNA' for label in labels_base]
corr_tracks = [
res_corrs['corr_shuff_cells'].values,
res_corrs['corr_shuff'].values,
res_corrs.loc[res_corrs['dist']<=500*KB, 'corr'].values,
res_corrs.loc[res_corrs['dist']<=100*KB, 'corr'].values,
]
colors = [colors_null[1], colors_null[0], colors_mc[1], colors_mc[0],]
ymax = 0
ax_col = axs[:,j]
for ax in ax_col:
for i, track in enumerate(corr_tracks):
_y, _x, _ = ax.hist(track, bins=bins, density=True, label=labels[i], histtype='step', color=colors[i])
ymax = max(ymax, np.max(_y))
_handles, _labels = ax.get_legend_handles_labels()
_handles = [mpl.lines.Line2D([], [], c=h.get_edgecolor()) for h in _handles]
ax1, ax2 = ax_col
# ylims
if not len(ylim1): ylim1 = [0.9*ymax, 1.1*ymax]
if not len(ylim2): ylim2 = [0, 0.2*ymax]
# set slanted y axis
plot_utils.set_broken_yaxis(ax1, ax2, ylim1, ylim2)
# limit number of y ticks
ax1.yaxis.set_major_locator(mtick.MaxNLocator(2))
ax2.yaxis.set_major_locator(mtick.MaxNLocator(4))
# labels
ax1.set_title(title)
ax2.set_xlabel('{} correlation'.format(corr_type))
ax2.set_ylabel('Density')
ax.legend(_handles, _labels, bbox_to_anchor=(1,1))
if output:
print(output)
snmcseq_utils.savefig(fig, output)
plt.show()
plot_routine(res_corrs_all['ols_spearman_mcrna'],
res_corrs_all['gls_spearman_mcrna'],
'normal', 'decorrelated basis', 'Spearman',
ylim1=[5, 6.5], ylim2=[0, 3],
)
plot_routine(res_corrs_all['ols_pearson_mcrna'],
res_corrs_all['gls_pearson_mcrna'],
'normal', 'decorrelated basis', 'Pearson',
ylim1=[3, 80], ylim2=[0, 2],
)
###Output
_____no_output_____
###Markdown
Get some stats- zoom in ; cdf ; Q-Q plot- number of sig pairs
###Code
# zoom in
plot_routine(res_corrs_all['ols_spearman_mcrna'],
res_corrs_all['gls_spearman_mcrna'],
'normal', 'decorrelated basis', 'Spearman',
ylim1=[5, 6.5], ylim2=[0, 3],
bins=np.linspace(-1, -0.2, 100)
)
def plot_routine_cdf(
res_corrs1, res_corrs2,
title1, title2, corr_type,
ylim1=[], ylim2=[],
bins=np.linspace(-1, 1, 201),
yscale='linear',
output='',
):
"""
"""
# output = output_figures.format('hist_corr_both.pdf')
fig, axs = plt.subplots(1, 2, figsize=(5*2, 5), sharex=True, sharey=True)
ax1, ax2 = axs
for j, (res_corrs, title) in enumerate(zip([res_corrs1, res_corrs2],
[title1, title2],
)):
KB = 1000
labels_base = [
'shuffled metacells',
'shuffled genes',
'<500kb',
'<100kb',
]
# corr1
labels = [label+' mCG-RNA' for label in labels_base]
corr_tracks = [
res_corrs['corr_shuff_cells'].values,
res_corrs['corr_shuff'].values,
res_corrs.loc[res_corrs['dist']<=500*KB, 'corr'].values,
res_corrs.loc[res_corrs['dist']<=100*KB, 'corr'].values,
]
colors = [colors_null[1], colors_null[0], colors_mc[1], colors_mc[0],]
ymax = 0
ax = axs[j]
for i, track in enumerate(corr_tracks):
_y, _x = np.histogram(track, bins=bins, density=False)
_x = _x[1:]
ax.plot(_x, np.cumsum(_y)/np.sum(_y), '-', label=labels[i], color=colors[i])
ymax = max(ymax, np.max(_y))
_handles, _labels = ax.get_legend_handles_labels()
ax.set_title(title)
ax.set_yscale(yscale)
# # limit number of y ticks
# ax1.yaxis.set_major_locator(mtick.MaxNLocator(2))
# ax2.yaxis.set_major_locator(mtick.MaxNLocator(4))
ax1.set_xlabel('{} correlation'.format(corr_type))
ax1.set_ylabel('Cumulative fraction of pairs')
ax2.legend(_handles, _labels, bbox_to_anchor=(1,1))
if output:
print(output)
snmcseq_utils.savefig(fig, output)
plt.show()
return
def plot_routine_cdf_collapse(
res_corrs1, res_corrs2,
title, corr_type,
ylim=[],
bins=np.linspace(-1, 1, 201),
yscale='linear',
output='',
):
"""
"""
# output = output_figures.format('hist_corr_both.pdf')
fig, ax = plt.subplots(1, 1, figsize=(6*1, 6),)
linestyles = ['-', 'dashed']
for j, (res_corrs) in enumerate(
[res_corrs1, res_corrs2],
):
KB = 1000
labels_base = [
'shuffled metacells',
'shuffled genes',
'<500kb',
'<100kb',
]
# corr1
labels = [label+'' for label in labels_base]
corr_tracks = [
res_corrs['corr_shuff_cells'].values,
res_corrs['corr_shuff'].values,
res_corrs.loc[res_corrs['dist']<=500*KB, 'corr'].values,
res_corrs.loc[res_corrs['dist']<=100*KB, 'corr'].values,
]
colors = [colors_null[1], colors_null[0], colors_mc[1], colors_mc[0],]
ymax = 0
for i, track in enumerate(corr_tracks):
_y, _x = np.histogram(track, bins=bins, density=False)
_x = _x[1:]
_y = np.cumsum(_y)/np.sum(_y)
if len(ylim) > 0:
_y = np.clip(_y, ylim[0], ylim[1])
ax.plot(_x, _y,
linewidth=3,
linestyle=linestyles[j], label=labels[i], color=colors[i])
_handles, _labels = ax.get_legend_handles_labels()
ax.set_title(title)
ax.set_yscale(yscale)
if len(ylim) > 0:
ax.set_ylim(ylim)
# # limit number of y ticks
# ax.yaxis.set_major_locator(mtick.MaxNLocator(2))
ax.set_xlabel('{} correlation'.format(corr_type))
ax.set_ylabel('Cumulative fraction of pairs')
ax.legend(_handles, _labels, bbox_to_anchor=(1,1), ncol=2)
if output:
print(output)
snmcseq_utils.savefig(fig, output)
plt.show()
return
def plot_routine_qqplot_collapse(
res_corrs1, res_corrs2,
title, corr_type,
ylim=[],
bins=np.linspace(-1, 1, 201),
yscale='linear',
output='',
):
"""
"""
# output = output_figures.format('hist_corr_both.pdf')
fig, ax = plt.subplots(1, 1, figsize=(6*1, 6),)
linestyles = ['-', 'dashed']
for j, (res_corrs) in enumerate(
[res_corrs1, res_corrs2],
):
KB = 1000
labels_base = [
'shuffled metacells',
'shuffled genes',
'<500kb',
'<100kb',
]
# corr1
labels = [label+'' for label in labels_base]
corr_tracks = [
res_corrs['corr_shuff_cells'].values,
res_corrs['corr_shuff'].values,
res_corrs.loc[res_corrs['dist']<=500*KB, 'corr'].values,
res_corrs.loc[res_corrs['dist']<=100*KB, 'corr'].values,
]
colors = [colors_null[1], colors_null[0], colors_mc[1], colors_mc[0],]
ymax = 0
for i, track in enumerate(corr_tracks):
_y, _x = np.histogram(track, bins=bins, density=False)
_x = _x[1:]
_y = np.cumsum(_y)/np.sum(_y)
if len(ylim) > 0:
_y = np.clip(_y, ylim[0], ylim[1])
ax.plot(_x, _y,
linewidth=3,
linestyle=linestyles[j], label=labels[i], color=colors[i])
_handles, _labels = ax.get_legend_handles_labels()
ax.set_title(title)
ax.set_yscale(yscale)
if len(ylim) > 0:
ax.set_ylim(ylim)
# # limit number of y ticks
# ax.yaxis.set_major_locator(mtick.MaxNLocator(2))
ax.set_xlabel('{} correlation'.format(corr_type))
ax.set_ylabel('Cumulative fraction of pairs')
ax.legend(_handles, _labels, bbox_to_anchor=(1,1), ncol=2)
if output:
print(output)
snmcseq_utils.savefig(fig, output)
plt.show()
return
# cdf
plot_routine_cdf(res_corrs_all['ols_spearman_mcrna'],
res_corrs_all['gls_spearman_mcrna'],
'normal', 'decorrelated basis', 'Spearman',
bins=np.linspace(-1, 1, 100),
)
plot_routine_cdf(res_corrs_all['ols_spearman_mcrna'],
res_corrs_all['gls_spearman_mcrna'],
'normal', 'decorrelated basis', 'Spearman',
bins=np.linspace(-1, 1, 100),
yscale='log'
)
plot_routine_cdf_collapse(
res_corrs_all['ols_spearman_mcrna'],
res_corrs_all['gls_spearman_mcrna'],
'OLS (solid) -> GLS (dashed; decorrelated basis)',
'Spearman',
ylim=[1e-5, 2],
bins=np.linspace(-1, 1, 100),
yscale='log',
output=output_figformat.format("cdf_OLS2GLS")
)
# Q-Q plot
# stats
# Q-Q plot
np.log()
###Output
_____no_output_____
|
header_footer/biosignalsnotebooks_environment/categories/MainFiles/old_files/by_diff.ipynb
|
###Markdown
Notebooks Grouped by Difficulty Each Notebook content is summarized in his header through a quantitative scale ('"Difficulty" between 1 and 5 stars) and some keywords (Group of "tags").Grouping Notebooks by difficulty level, by signal type to which it applies or by tags is an extremelly important task, in order to ensure that the **opensignalstools** user could navigate efficiently in this learning environment. Problems of a smaller sampling rate (aliasing) Load acquired data from .txt file Load acquired data from .h5 file Fatigue Evaluation - Evolution of Median Power Frequency &9740; GitHub Repository &9740; Notebook Categories &9740; How to install opensignalstools Python package ? &9740; Signal Library &9740; Notebooks by Difficulty &9740; Notebooks by Signal Type &9740; Notebooks by Tag **Auxiliary Code Segment (should not be replicated bythe user)**
###Code
from opensignalstools.__notebook_support__ import css_style_apply
css_style_apply()
###Output
_____no_output_____
|
osmnx/speed/add_edge_travel_times.ipynb
|
###Markdown
osmnx.speed moduleCalculate graph edge speeds and travel times.
###Code
# OSMnx: New Methods for Acquiring, Constructing, Analyzing, and Visualizing Complex Street Networks
import osmnx as ox
ox.config(use_cache=True, log_console=False)
ox.__version__
query = '중구, 서울특별시, 대한민국'
network_type = 'drive' # "all_private", "all", "bike", "drive", "drive_service", "walk"
# Create graph from OSM within the boundaries of some geocodable place(s).
G = ox.graph_from_place(query, network_type=network_type)
# Plot a graph.
fig, ax = ox.plot_graph(G)
# KeyError: 'all edges must have `length` and `speed_kph` attributes.'
# Add edge speeds (km per hour) to graph as new speed_kph edge attributes.
G = ox.speed.add_edge_speeds(
G,
hwy_speeds=None,
fallback=None,
precision=1
)
# Add edge travel time (seconds) to graph as new travel_time edge attributes.
G = ox.speed.add_edge_travel_times(G, precision=1)
# Convert a MultiDiGraph to node and/or edge GeoDataFrames.
# AttributeError: 'tuple' object has no attribute 'head'
gdf = ox.graph_to_gdfs(G, nodes=False)
gdf.head()
###Output
_____no_output_____
|
src/keras/CUP_Keras_NN_GS.ipynb
|
###Markdown
Import Dataset
###Code
def load_cup():
ml_cup = np.delete(np.genfromtxt('ML-CUP20-TR.csv',
delimiter=','), obj=0, axis=1)
return ml_cup[:, :-2], ml_cup[:, -2:]
def load_cup_blind():
return np.delete(np.genfromtxt('ML-CUP20-TS.csv',
delimiter=','), obj=0, axis=1)
def mean_euclidean_error(y_true, y_pred):
assert y_true.shape == y_pred.shape
return np.mean(np.linalg.norm(y_pred - y_true, axis=1))
X, y = load_cup()
X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.2)
print(X.shape)
print(y.shape)
# data = np.genfromtxt('ML-CUP20-TR.csv', delimiter=',', dtype=np.float64)
# X = data[:, 1:-2]
# y = data[:, -2:]
# print(X.shape)
# print(y.shape)
# Xtrain, Xval, ytrain, yval = train_test_split(X, y, test_size=0.20, random_state=42)
# print(Xtrain.shape)
# print(ytrain.shape)
# print(Xval.shape)
# print(yval.shape)
###Output
time: 930 µs (started: 2021-01-21 08:11:24 +00:00)
###Markdown
Grid Search - Gradient Descent
###Code
txt_best_grids = "best_results_cup_nn.txt"
grid_results_name = 'grid_results_cup_nn_v1.csv'
from sklearn.model_selection import GridSearchCV
from tensorflow.keras.models import Sequential
from tensorflow.keras import layers
from tensorflow.keras.layers import Dense
from tensorflow.keras.wrappers.scikit_learn import KerasRegressor
from tensorflow.keras.optimizers import SGD
from tensorflow.keras.regularizers import L2
# Function to create model, required for KerasRegressor
def create_model(learn_rate=0.01, momentum=0, neurons_layer_1=10,
init_mode='uniform', activation='relu', weight_decay = 0.0,
nesterov = False, neurons_layer_2 = 5):
# create model
model = Sequential()
model.add(layers.InputLayer(input_shape=(10)))
model.add(Dense(neurons_layer_1, activation=activation, kernel_initializer=init_mode, kernel_regularizer= L2(weight_decay)))
model.add(Dense(neurons_layer_2, activation=activation, kernel_initializer=init_mode, kernel_regularizer= L2(weight_decay)))
model.add(Dense(2))
optimizer = SGD(
learning_rate=learn_rate, momentum=momentum, nesterov=nesterov)
# Compile model
model.compile(optimizer=optimizer,
loss=euclidean_distance_loss)
return model
BATCH_SIZE = len(X_train)
# create model
model = KerasRegressor(build_fn=create_model, verbose=0) #verbose = 0
# define the grid search parameters
batch_size = [64]#, 128, 256, BATCH_SIZE]
epochs = [300]
learn_rate = [0.001]#, 0.01, 0.1, 0.3]
momentum = [0.0, 0.2]#, 0.4, 0.9]
nesterov = [False]
neurons_layer_1 = [40]#, 80, 120]
neurons_layer_2 = [40]#, 80, 120]
init_mode = ['glorot_uniform']
activation = ['relu'] #, 'tanh', 'sigmoid']
weight_decay = [0.1]#, 0.01, 0.001, 0.0001]
param_grid = dict(batch_size=batch_size, epochs=epochs, learn_rate=learn_rate,
momentum=momentum, neurons_layer_1=neurons_layer_1,
init_mode=init_mode, activation=activation, weight_decay= weight_decay,
nesterov= nesterov, neurons_layer_2=neurons_layer_2)
grid = GridSearchCV(estimator=model, param_grid=param_grid, n_jobs=-1, cv=3, return_train_score=True, verbose=True)
grid_result = grid.fit(X_train, y_train)
import json
grid_results_name_str = f'Results wrt: {grid_results_name} \n'
grid_params_str = f'Grid on: {json.dumps(grid_result.param_grid)} \n'
grid_results_str = f'Best: {grid_result.best_score_} using {grid_result.best_params_} \n'
with open(txt_best_grids, "a") as file_object:
file_object.write(grid_results_name_str)
file_object.write(grid_params_str)
file_object.write(grid_results_str)
file_object.write('\n')
print(grid_results_str)
import pandas as pd
df = pd.DataFrame(grid.cv_results_)[['rank_test_score','mean_test_score',
'std_test_score','mean_train_score',
'std_train_score','param_activation',
'param_batch_size','param_epochs',
'param_init_mode', 'param_learn_rate',
'param_momentum','param_nesterov',
'param_neurons_layer_1', 'param_neurons_layer_2',
'param_weight_decay','mean_fit_time']].sort_values(by='rank_test_score')
df.rename(columns={'param_activation': 'activation',
'param_batch_size': 'batch_size',
'param_epochs': 'epochs',
'param_init_mode': 'init_mode',
'param_learn_rate': 'learn_rate',
'param_momentum': 'momentum',
'param_init_mode': 'init_mode',
'param_weight_decay': 'weight_decay',
'param_nesterov': 'nesterov',
'param_neurons_layer_1': 'neurons_layer_1',
'param_neurons_layer_2': 'neurons_layer_2',
'mean_test_score': 'mean_val_score',
'std_test_score': 'std_val_score',
'rank_test_score': 'rank_val_score'}, inplace=True)
df.mean_train_score *= -1
df.mean_val_score *= -1
df
df.to_csv(grid_results_name)
# means = grid_result.cv_results_['mean_test_score']
# stds = grid_result.cv_results_['std_test_score']
# params = grid_result.cv_results_['params']
# for mean, stdev, param in zip(means, stds, params):
# print("%f (%f) with: %r" % (mean, stdev, param))
# import csv
# with open(single_grid_results_filename, mode='w') as grid:
# grid = csv.writer(grid, delimiter=',', quotechar='"', quoting=csv.QUOTE_MINIMAL)
# for mean, stdev, param in zip(means, stds, params):
# grid.writerow([mean, stdev, param])
# from sklearn.externals import joblib
# # #save your model or results
# # joblib.dump(grid_result, 'gs_object_monk3.pkl')
# # #load your model for further usage
# # boh = joblib.load("gs_object_monk3.pkl")
# # joblib.dump(grid.best_estimator_, 'gs_best_estimator_monk3.pkl', compress = 1)
# # filename = 'finalized_model.sav'
# # pickle.dump(model, open(filename, 'wb'))
###Output
time: 5.94 ms (started: 2021-01-20 17:45:32 +00:00)
|
project/Microsoft Malware Classification.ipynb
|
###Markdown
Final Project - Security Analytics Microsoft Malware Classfication Authors: * Kandarp Khandwala (kkhandw1) and * Antara Sargam (asargam1) Data: * Set of known malware files representing a mix of 9 different families. Each malware file has an Id, a 20 character hash value uniquely identifying the file, and a Class, an integer representing one of 9 family names to which the malware may belong Ramnit Lollipop Kelihos_ver3 Vundo Simda Tracur Kelihos_ver1 Obfuscator.ACY Gatak Tasks: * Develop the best mechanism for classifying files into their respective family affiliations. Files* train.7z - the raw data for the data set* trainLabels.csv - the class labels associated with the training set
###Code
import matplotlib.pyplot as plt
from os import listdir
from os.path import isfile, join
import zipfile
from io import BytesIO
import lief
import hashlib
import pandas as pd
import numpy as np
import os
import pickle
import io
import re
import glob
import lightgbm as lgb
from sklearn.metrics import log_loss, confusion_matrix
from sklearn.ensemble import RandomForestClassifier, ExtraTreesClassifier
from sklearn.model_selection import train_test_split, cross_val_score, cross_val_predict
from sklearn.feature_extraction.text import TfidfVectorizer, HashingVectorizer
from sklearn.feature_extraction import FeatureHasher
from sklearn import metrics
datadir = '/datadrive/'
traindir = 'train/'
asmext = '.asm'
byext = '.bytes'
dt = np.dtype([('Id', 'a30'), ('Class', 'u2')])
data = np.loadtxt("trainLabels.csv", skiprows=1, delimiter = ",", dtype=dt)
data
X = np.zeros((data.shape[0], 4))
y = data['Class']
###Output
_____no_output_____
###Markdown
Feature Extraction Feature 1 - File size
###Code
for i, (Id, Class) in enumerate(data):
X[i][0] = os.path.getsize(datadir + traindir + Id[1:-1].decode("utf-8") + asmext)
X[i][1] = os.path.getsize(datadir + traindir + Id[1:-1].decode("utf-8") + byext)
plt.axis((0,1.6*10**8, 0, 2*10**7))
plt.scatter(X[:,0], X[:,1], c=y, alpha=0.5)
plt.show()
###Output
_____no_output_____
###Markdown
Feature 2 - Compressed File Size
###Code
def getcompressedsize(fpath):
inMemoryOutputFile = BytesIO()
zf = zipfile.ZipFile(inMemoryOutputFile, 'w')
zf.write(fpath, compress_type=zipfile.ZIP_DEFLATED)
s = float(zf.infolist()[0].compress_size)
zf.close()
return s
for i, (Id, Class) in enumerate(data):
print("Processed file {}".format(i))
X[i][2] = getcompressedsize(datadir + traindir + Id[1:-1].decode("utf-8") + asmext)
X[i][3] = getcompressedsize(datadir + traindir + Id[1:-1].decode("utf-8") + asmext)
plt.axis((0,1.6*10**8, 0, 2*10**7))
plt.scatter(X[:,2], X[:,3], c=y, alpha=0.5)
plt.show()
###Output
_____no_output_____
###Markdown
Feature 3 - TFIDF matrix of the contents of asm files over 2-gram
###Code
def make_corpus(asm_files):
for i, file in enumerate(asm_files):
print('Processed file {}'.format(i))
yield open(file, 'rb')
import string
from nltk.corpus import stopwords
def text_process(mess):
"""
Takes in a string of text, then performs the following:
1. Remove all punctuation
2. Remove all stopwords
3. Returns a list of the cleaned text
"""
# Check characters to see if they are in punctuation
nopunc = [char.decode("utf-8") for char in mess if char.decode("utf-8") not in string.punctuation]
# Join the characters again to form the string.
nopunc = ''.join(nopunc)
# Now just remove any stopwords
return [word for word in nopunc.split() if word.decode("utf-8").lower() not in stopwords.words('english')]
asmfiles = []
for i, (Id, Class) in enumerate(data):
asmfiles.append(datadir + traindir + Id[1:-1].decode("utf-8") + asmext)
hv_transformer = HashingVectorizer(input='file', decode_error='replace',
n_features=10000, stop_words='english',
ngram_range=(2, 2))
asm_corpus = make_corpus(asmfiles)
asm_ngram = hv_transformer.transform(asm_corpus)
print('Shape of Sparse Matrix: ', asm_ngram.shape)
print('Amount of Non-Zero occurences: ', asm_ngram.nnz)
print('Sparsity: %.2f%%' % (100.0 * asm_ngram.nnz / (asm_ngram.shape[0] * asm_ngram.shape[1])))
X = np.append(X, asm_ngram.toarray(), axis = 1)
X.shape
###Output
_____no_output_____
###Markdown
Feature 4 - String file characteristics: urls, directories, registries, headers, entropy, etc.
###Code
class FeatureType(object):
''' Base class from which each feature type may inherit '''
name = ''
dim = 0
def __repr__(self):
return '{}({})'.format(self.name, self.dim)
def raw_features(self, bytez, lief_binary):
''' Generate a JSON-able representation of the file '''
raise (NotImplemented)
def process_raw_features(self, raw_obj):
''' Generate a feature vector from the raw features '''
raise (NotImplemented)
def feature_vector(self, bytez, lief_binary):
''' Directly calculate the feature vector from the sample itself. This should only be implemented differently
if there are significant speedups to be gained from combining the two functions. '''
return self.process_raw_features(self.raw_features(bytez, lief_binary))
class StringExtractor(FeatureType):
''' Extracts strings from raw byte stream '''
name = 'strings'
dim = 1 + 1 + 1 + 96 + 1 + 1 + 1 + 1 + 1
def __init__(self):
super(FeatureType, self).__init__()
# all consecutive runs of 0x20 - 0x7f that are 5+ characters
self._allstrings = re.compile(b'[\x20-\x7f]{5,}')
# occurances of the string 'C:\'. Not actually extracting the path
self._paths = re.compile(b'c:\\\\', re.IGNORECASE)
# occurances of http:// or https://. Not actually extracting the URLs
self._urls = re.compile(b'https?://', re.IGNORECASE)
# occurances of the string prefix HKEY_. No actually extracting registry names
self._registry = re.compile(b'HKEY_')
# crude evidence of an MZ header (dropper?) somewhere in the byte stream
self._mz = re.compile(b'MZ')
def raw_features(self, bytez, lief_binary):
allstrings = self._allstrings.findall(bytez)
if allstrings:
# statistics about strings:
string_lengths = [len(s) for s in allstrings]
avlength = sum(string_lengths) / len(string_lengths)
# map printable characters 0x20 - 0x7f to an int array consisting of 0-95, inclusive
as_shifted_string = [b - ord(b'\x20') for b in b''.join(allstrings)]
c = np.bincount(as_shifted_string, minlength=96) # histogram count
# distribution of characters in printable strings
csum = c.sum()
p = c.astype(np.float32) / csum
wh = np.where(c)[0]
H = np.sum(-p[wh] * np.log2(p[wh])) # entropy
else:
avlength = 0
c = np.zeros((96,), dtype=np.float32)
H = 0
csum = 0
return {
'numstrings': len(allstrings),
'avlength': avlength,
'printabledist': c.tolist(), # store non-normalized histogram
'printables': int(csum),
'entropy': float(H),
'paths': len(self._paths.findall(bytez)),
'urls': len(self._urls.findall(bytez)),
'registry': len(self._registry.findall(bytez)),
'MZ': len(self._mz.findall(bytez))
}
def process_raw_features(self, raw_obj):
hist_divisor = float(raw_obj['printables']) if raw_obj['printables'] > 0 else 1.0
return np.hstack([
raw_obj['numstrings'], raw_obj['avlength'], raw_obj['printables'],
np.asarray(raw_obj['printabledist']) / hist_divisor, raw_obj['entropy'], raw_obj['paths'], raw_obj['urls'],
raw_obj['registry'], raw_obj['MZ']
]).astype(np.float32)
all_features = []
extractor = StringExtractor()
for i, (Id, Class) in enumerate(data):
print("Processed file {}".format(i))
file = datadir + traindir + Id[1:-1].decode("utf-8") + asmext
with open(file, 'rb') as f:
bytez = f.read()
f.close()
features = extractor.raw_features(bytez, None)
all_features.append(extractor.process_raw_features(features))
string_features = np.array(all_features)
X = np.append(X, string_features, axis = 1)
X.shape
###Output
_____no_output_____
###Markdown
Split into train and test
###Code
X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.2)
print(X_train.shape, y_train.shape)
print(X_test.shape, y_test.shape)
###Output
(664, 10108) (664,)
(166, 10108) (166,)
###Markdown
Run ExtraTreesClassifier on the Dataset* The ExtraTreesClassifier is similar to RandomForestClassifier but cheaper to train from a computational point of view but can grow much bigger. We try this to get a quick overview of our results
###Code
et_clf = ExtraTreesClassifier(n_estimators=1000, max_features=None, min_samples_leaf=2,
min_samples_split=9, n_jobs=8)
et_clf.fit(X_train, y_train)
y_prob = et_clf.predict_proba(X_test)
y_pred = et_clf.predict(X_test)
print("Predicted Values using ExtraTreesClassifier: {}".format(y_pred))
print("Predicted Probability using ExtraTreesClassifier: {}".format(y_prob))
cm = metrics.confusion_matrix(y_test, y_pred)
print(cm)
print("Logarithmic Loss: {}".format(log_loss(y_test, y_prob, labels=[1, 2, 3, 4, 5, 6, 7, 8, 9])))
acc = metrics.accuracy_score(y_test, y_pred)
acc
###Output
_____no_output_____
###Markdown
Perform 5-fold cross validation
###Code
y_pred = cross_val_predict(et_clf, X, y, cv=5)
print('Cross-validated predictions: ', y_pred)
y_scores = cross_val_score(et_clf, X, y, cv=5, scoring='accuracy')
print('Cross-validated accuracy scores: ', y_scores)
print('Mean cross-validated accuracy scores: ', y_scores.mean())
###Output
Cross-validated accuracy scores: [0.9702381 0.9702381 0.98795181 0.96969697 0.93251534]
Mean cross-validated accuracy scores: 0.9661280609650778
###Markdown
Run LightGBM on the Dataset* Light GBM is a gradient boosting framework that uses tree based learning algorithm. It can handle the large size of our data and takes lower memory to run. Another reason of why Light GBM is popular is because it focuses on accuracy of results
###Code
y_train_lgb = y_train - 1
y_train_lgb
d_train = lgb.Dataset(X_train, label=y_train_lgb)
params = {}
params['learning_rate'] = 0.5
params['boosting_type'] = 'dart'
params['objective'] = 'multiclass'
params['sub_feature'] = 0.5
params['num_leaves'] = 9
params['max_depth'] = 10
params['num_threads'] = 8
params['num_class'] = 9
lgb_clf = lgb.train(params, d_train, 100)
y_pred_lgb = lgb_clf.predict(X_test)
y_pred_lgb
y_test_lgb = y_test - 1
y_test_lgb
y_pred_lgb_best = np.array([np.argmax(line) for line in y_pred_lgb])
y_pred_lgb_best
cm = metrics.confusion_matrix(y_test_lgb, y_pred_lgb_best)
cm
acc = metrics.accuracy_score(y_pred_lgb_best, y_test_lgb)
acc
###Output
_____no_output_____
|
example/pycon-2017-tutorial-rl-master/LR_tensorflow.ipynb
|
###Markdown
import everything
###Code
import matplotlib
import matplotlib.pyplot as plt
import tensorflow as tf
import numpy as np
from sklearn.datasets import make_moons
%matplotlib inline
matplotlib.rcParams['figure.figsize'] = (10.0, 8.0)
###Output
_____no_output_____
###Markdown
製作2維資料
###Code
data, label = make_moons(n_samples=500, noise=0.2, random_state=0)
label = label.reshape(500, 1)
print('data shape :', data.shape)
print(data[:5], '\n')
print('label shape:', label.shape)
print(label[:5])
# draw picture
plt.scatter(data[:,0], data[:,1], s=40, c=label, cmap=plt.cm.Accent)
###Output
_____no_output_____
###Markdown
使用tensorflow撰寫logistic regression
###Code
sess = tf.InteractiveSession()
# tf Graph Input
x = tf.placeholder(tf.float32, [None, 2])
y = tf.placeholder(tf.float32, [None, 1])
# Set model weights
W = tf.Variable(tf.zeros([2, 1]))
b = tf.Variable(tf.zeros([1]))
# Logistic regression without sigmoid activation
predict = tf.matmul(x,W) + b
# Cross entropy
cost = tf.reduce_mean(tf.nn.sigmoid_cross_entropy_with_logits(labels=y, logits=predict))
# SGD
optimizer = tf.train.GradientDescentOptimizer(learning_rate=0.01).minimize(cost)
###Output
_____no_output_____
###Markdown
更新權重一次
###Code
sess.run(tf.global_variables_initializer())
_, c = sess.run([optimizer, cost], feed_dict={x: data, y: label})
print('cost:', c)
###Output
cost: 0.693146
###Markdown
sigmoid function
###Code
def sigmoid(x):
return 1.0 / (1.0 + np.exp(-x))
###Output
_____no_output_____
###Markdown
使用現在的模型預測
###Code
feed_dict = {x: data}
classification = sess.run(predict, feed_dict)
classification = sigmoid(classification)
classification[classification>=0.5] = 1
classification[classification<0.5] = 0
classification[:30]
print('Accuary:', float(sum(label == classification)[0]) / label.shape[0])
###Output
Accuary: 0.78
###Markdown
劃出預測邊界
###Code
def plot_decision_boundary(X, y, model):
# Set min and max values and give it some padding
x_min, x_max = X[:, 0].min() - .5, X[:, 0].max() + .5
y_min, y_max = X[:, 1].min() - .5, X[:, 1].max() + .5
h = 0.01
# Generate a grid of points with distance h between them
xx, yy = np.meshgrid(np.arange(x_min, x_max, h),
np.arange(y_min, y_max, h))
pred_every_point = np.c_[xx.ravel(), yy.ravel()]
feed_dict = {x: pred_every_point}
Z = sess.run(model, feed_dict)
Z = sigmoid(Z)
Z[Z>=0.5] = 1
Z[Z<0.5] = 0
Z = Z.reshape(xx.shape)
# Plot the contour and training examples
plt.contourf(xx, yy, Z, cmap=plt.cm.Spectral)
plt.scatter(X[:, 0], X[:, 1], s=40, c=y, cmap=plt.cm.BuGn)
plot_decision_boundary(data, label, predict)
###Output
_____no_output_____
###Markdown
疊代20000次
###Code
for i in range(20000):
_, c = sess.run([optimizer, cost], feed_dict={x: data, y: label})
if i % 1000 == 0:
print("epoch:{}, cost:{}".format(i, c))
feed_dict = {x: data}
classification = sess.run(predict, feed_dict)
classification = sigmoid(classification)
classification[classification>=0.5] = 1
classification[classification<0.5] = 0
print('Accuary:', float(sum(label == classification)[0]) / label.shape[0])
plot_decision_boundary(data, label, predict)
###Output
_____no_output_____
|
nbs/01_starter_dataset_download.ipynb
|
###Markdown
How to download the starter datset> (in one line) Download/Installation**`omnitools.download` is a one-line utility for rapidly downloading the starter (& similar) datasets.** For more about the tools themselves (`omnitools.download` and `omnitools.upload`), please see the [dedicated page](/omnidata-tools/omnitools.html).To download the starter dataset, make sure that omnidata-tooling is installed and then run the full download command which will prompt you to accept the component licenses to proceed: **Run the following:** (Estimated download time for [_RGB + 1 Task + Masks_]: **1 day**) (_Full dataset_ [30TB]: **5 days**) ```bash Make sure everything is installedsudo apt-get install aria2pip install 'omnidata-tools' Just to make sure it's installed Install the 'debug' subset of the Replica and Taskonomy components of the datasetomnitools.download rgb normals point_info \ --components replica taskonomy \ --subset debug \ --dest ./omnidata_starter_dataset/ --agree-all``` You should see the prompt: ExamplesHere are some other examples:Download the full Omnidata dataset and agree to licenses```bashomnitools.download all --components all --subset fullplus \ --dest ./omnidata_starter_dataset/ \ --connections_total 40 --agree```Download Taskonomy only:```bashomnitools.download all --components taskonomy --subset fullplus \ --dest ./omnidata_starter_dataset/ \ --connections_total 40 --agree```Omnidata but only depth and masks and keep the compressed files```bashomnitools.download rgb depth mask_valid --components all --subset fullplus \ --dest ./omnidata_starter_dataset/ \ --connections_total 40 --agree```Download meshes for Clevr```bashomnitools.download mesh --components clevr_simple --subset fullplus \ --dest ./omnidata_starter_dataset/ \ --dest_compressed ./omnidata_starter_dataset_compresssed --keep_compressed True \ --connections_total 40 --agree```Use multiple workers to download Omnidata--this is for worker 7/100, but do a dryrun```bashomnitools.download all --components all --subset fullplus \ --num_chunk 6 --num_total_chunks 100 \ --dest ./omnidata_starter_dataset/ \ --connections_total 40 --agree --dryrun```...you get the idea :) Command-line options`omnitools.download` is pretty configurable, and you can choose which comonents/subset/split/tasks to download and extract. The downloader will spawn many workers to then download those compressed files, verify the download against checksums on the server, and unpack them. Here are the available options:```bash> omnitools.download -h```
###Code
#hide_input
argparser = anno_parser(download)
argparser.prog = 'omnitools.download'
argparser.print_help()
###Output
usage: omnitools.download [-h] [--subset {debug,tiny,medium,full,fullplus}]
[--split {train,val,test,all}]
[--components {all,replica,taskonomy,gso_in_replica,hypersim,blendedmvs,hm3d,clevr_simple,clevr_complex} [{all,replica,taskonomy,gso_in_replica,hypersim,blendedmvs,hm3d,clevr_simple,clevr_complex} ...]]
[--dest DEST] [--dest_compressed DEST_COMPRESSED]
[--keep_compressed KEEP_COMPRESSED] [--only_download ONLY_DOWNLOAD]
[--max_tries_per_model MAX_TRIES_PER_MODEL]
[--connections_total CONNECTIONS_TOTAL]
[--connections_per_server_per_download CONNECTIONS_PER_SERVER_PER_DOWNLOAD]
[--n_workers N_WORKERS] [--num_chunk NUM_CHUNK]
[--num_total_chunks NUM_TOTAL_CHUNKS] [--ignore_checksum IGNORE_CHECKSUM]
[--dryrun] [--aria2_uri ARIA2_URI]
[--aria2_cmdline_opts ARIA2_CMDLINE_OPTS]
[--aria2_create_server ARIA2_CREATE_SERVER] [--aria2_secret ARIA2_SECRET]
[--agree_all]
domains [domains ...]
Downloads Omnidata starter dataset. --- The data is stored on the remote server in a compressed
format (.tar.gz). This function downloads the compressed and decompresses it. Examples: download rgb
normals point_info --components clevr_simple clevr_complex --connections_total 30
positional arguments:
domains Domains to download (comma-separated or 'all')
optional arguments:
-h, --help show this help message and exit
--subset {debug,tiny,medium,full,fullplus} Subset to download (default: debug)
--split {train,val,test,all} Split to download (default: all)
--components {all,replica,taskonomy,gso_in_replica,hypersim,blendedmvs,hm3d,clevr_simple,clevr_complex} [{all,replica,taskonomy,gso_in_replica,hypersim,blendedmvs,hm3d,clevr_simple,clevr_complex} ...]
Component datasets to download (comma-separated)
(default: all)
--dest DEST Where to put the uncompressed data (default:
uncompressed/)
--dest_compressed DEST_COMPRESSED Where to download the compressed data (default:
compressed/)
--keep_compressed KEEP_COMPRESSED Don't delete compressed files after decompression
(default: False)
--only_download ONLY_DOWNLOAD Only download compressed data (default: False)
--max_tries_per_model MAX_TRIES_PER_MODEL Number of times to try to download model if
checksum fails. (default: 3)
--connections_total CONNECTIONS_TOTAL Number of simultaneous aria2c connections overall
(note: if not using the RPC server, this is per-
worker) (default: 8)
--connections_per_server_per_download CONNECTIONS_PER_SERVER_PER_DOWNLOAD
Number of simulatneous aria2c connections per
server per download. Defaults to
'total_connections' (note: if not using the RPC
server, this is per-worker)
--n_workers N_WORKERS Number of workers to use (default: 32)
--num_chunk NUM_CHUNK Download the kth slice of the overall dataset
(default: 0)
--num_total_chunks NUM_TOTAL_CHUNKS Download the dataset in N total chunks. Use with '
--num_chunk' (default: 1)
--ignore_checksum IGNORE_CHECKSUM Ignore checksum validation (default: False)
--dryrun Keep compressed files even after decompressing
(default: False)
--aria2_uri ARIA2_URI Location of aria2c RPC (if None, use CLI)
(default: http://localhost:6800)
--aria2_cmdline_opts ARIA2_CMDLINE_OPTS Opts to pass to aria2c (default: )
--aria2_create_server ARIA2_CREATE_SERVER Create a RPC server at aria2_uri (default: True)
--aria2_secret ARIA2_SECRET Secret for aria2c RPC (default: )
--agree_all Agree to all license clickwraps. (default: False)
|
ml-classification/week-3/module-5-decision-tree-assignment-2-blank.ipynb
|
###Markdown
Implementing binary decision trees The goal of this notebook is to implement your own binary decision tree classifier. You will: * Use SFrames to do some feature engineering.* Transform categorical variables into binary variables.* Write a function to compute the number of misclassified examples in an intermediate node.* Write a function to find the best feature to split on.* Build a binary decision tree from scratch.* Make predictions using the decision tree.* Evaluate the accuracy of the decision tree.* Visualize the decision at the root node.**Important Note**: In this assignment, we will focus on building decision trees where the data contain **only binary (0 or 1) features**. This allows us to avoid dealing with:* Multiple intermediate nodes in a split* The thresholding issues of real-valued features.This assignment **may be challenging**, so brace yourself :) Fire up Graphlab Create Make sure you have the latest version of GraphLab Create.
###Code
import graphlab
###Output
_____no_output_____
###Markdown
Load the lending club dataset We will be using the same [LendingClub](https://www.lendingclub.com/) dataset as in the previous assignment.
###Code
loans = graphlab.SFrame('lending-club-data.gl/')
loans.head()
###Output
_____no_output_____
###Markdown
Like the previous assignment, we reassign the labels to have +1 for a safe loan, and -1 for a risky (bad) loan.
###Code
loans['safe_loans'] = loans['bad_loans'].apply(lambda x : +1 if x==0 else -1)
loans = loans.remove_column('bad_loans')
###Output
_____no_output_____
###Markdown
Unlike the previous assignment where we used several features, in this assignment, we will just be using 4 categoricalfeatures: 1. grade of the loan 2. the length of the loan term3. the home ownership status: own, mortgage, rent4. number of years of employment.Since we are building a binary decision tree, we will have to convert these categorical features to a binary representation in a subsequent section using 1-hot encoding.
###Code
features = ['grade', # grade of the loan
'term', # the term of the loan
'home_ownership', # home_ownership status: own, mortgage or rent
'emp_length', # number of years of employment
]
target = 'safe_loans'
loans = loans[features + [target]]
###Output
_____no_output_____
###Markdown
Let's explore what the dataset looks like.
###Code
loans
###Output
_____no_output_____
###Markdown
Subsample dataset to make sure classes are balanced Just as we did in the previous assignment, we will undersample the larger class (safe loans) in order to balance out our dataset. This means we are throwing away many data points. We use `seed=1` so everyone gets the same results.
###Code
safe_loans_raw = loans[loans[target] == 1]
risky_loans_raw = loans[loans[target] == -1]
# Since there are less risky loans than safe loans, find the ratio of the sizes
# and use that percentage to undersample the safe loans.
percentage = len(risky_loans_raw)/float(len(safe_loans_raw))
safe_loans = safe_loans_raw.sample(percentage, seed = 1)
risky_loans = risky_loans_raw
loans_data = risky_loans.append(safe_loans)
print "Percentage of safe loans :", len(safe_loans) / float(len(loans_data))
print "Percentage of risky loans :", len(risky_loans) / float(len(loans_data))
print "Total number of loans in our new dataset :", len(loans_data)
###Output
Percentage of safe loans : 0.502236174422
Percentage of risky loans : 0.497763825578
Total number of loans in our new dataset : 46508
###Markdown
**Note:** There are many approaches for dealing with imbalanced data, including some where we modify the learning algorithm. These approaches are beyond the scope of this course, but some of them are reviewed in "[Learning from Imbalanced Data](http://www.ele.uri.edu/faculty/he/PDFfiles/ImbalancedLearning.pdf)" by Haibo He and Edwardo A. Garcia, *IEEE Transactions on Knowledge and Data Engineering* **21**(9) (June 26, 2009), p. 1263–1284. For this assignment, we use the simplest possible approach, where we subsample the overly represented class to get a more balanced dataset. In general, and especially when the data is highly imbalanced, we recommend using more advanced methods. Transform categorical data into binary features In this assignment, we will implement **binary decision trees** (decision trees for binary features, a specific case of categorical variables taking on two values, e.g., true/false). Since all of our features are currently categorical features, we want to turn them into binary features. For instance, the **home_ownership** feature represents the home ownership status of the loanee, which is either `own`, `mortgage` or `rent`. For example, if a data point has the feature ``` {'home_ownership': 'RENT'}```we want to turn this into three features: ``` { 'home_ownership = OWN' : 0, 'home_ownership = MORTGAGE' : 0, 'home_ownership = RENT' : 1 }```Since this code requires a few Python and GraphLab tricks, feel free to use this block of code as is. Refer to the API documentation for a deeper understanding.
###Code
loans_data = risky_loans.append(safe_loans)
for feature in features:
loans_data_one_hot_encoded = loans_data[feature].apply(lambda x: {x: 1})
loans_data_unpacked = loans_data_one_hot_encoded.unpack(column_name_prefix=feature)
# Change None's to 0's
for column in loans_data_unpacked.column_names():
loans_data_unpacked[column] = loans_data_unpacked[column].fillna(0)
loans_data.remove_column(feature)
loans_data.add_columns(loans_data_unpacked)
###Output
_____no_output_____
###Markdown
Let's see what the feature columns look like now:
###Code
features = loans_data.column_names()
features.remove('safe_loans') # Remove the response variable
features
print "Number of features (after binarizing categorical variables) = %s" % len(features)
###Output
Number of features (after binarizing categorical variables) = 25
###Markdown
Let's explore what one of these columns looks like:
###Code
loans_data['grade.A']
###Output
_____no_output_____
###Markdown
This column is set to 1 if the loan grade is A and 0 otherwise.**Checkpoint:** Make sure the following answers match up.
###Code
print "Total number of grade.A loans : %s" % loans_data['grade.A'].sum()
print "Expexted answer : 6422"
###Output
Total number of grade.A loans : 6422
Expexted answer : 6422
###Markdown
Train-test splitWe split the data into a train test split with 80% of the data in the training set and 20% of the data in the test set. We use `seed=1` so that everyone gets the same result.
###Code
train_data, test_data = loans_data.random_split(.8, seed=1)
###Output
_____no_output_____
###Markdown
Decision tree implementation In this section, we will implement binary decision trees from scratch. There are several steps involved in building a decision tree. For that reason, we have split the entire assignment into several sections. Function to count number of mistakes while predicting majority classRecall from the lecture that prediction at an intermediate node works by predicting the **majority class** for all data points that belong to this node.Now, we will write a function that calculates the number of **missclassified examples** when predicting the **majority class**. This will be used to help determine which feature is the best to split on at a given node of the tree.**Note**: Keep in mind that in order to compute the number of mistakes for a majority classifier, we only need the label (y values) of the data points in the node. ** Steps to follow **:* ** Step 1:** Calculate the number of safe loans and risky loans.* ** Step 2:** Since we are assuming majority class prediction, all the data points that are **not** in the majority class are considered **mistakes**.* ** Step 3:** Return the number of **mistakes**.Now, let us write the function `intermediate_node_num_mistakes` which computes the number of misclassified examples of an intermediate node given the set of labels (y values) of the data points contained in the node. Fill in the places where you find ` YOUR CODE HERE`. There are **three** places in this function for you to fill in.
###Code
def intermediate_node_num_mistakes(labels_in_node):
# Corner case: If labels_in_node is empty, return 0
if len(labels_in_node) == 0:
return 0
# Count the number of 1's (safe loans)
safe_loans = (labels_in_node == 1).sum()
# Count the number of -1's (risky loans)
risky_loans = (labels_in_node == -1).sum()
# Return the number of mistakes that the majority classifier makes.
return risky_loans if safe_loans >= risky_loans else safe_loans
###Output
_____no_output_____
###Markdown
Because there are several steps in this assignment, we have introduced some stopping points where you can check your code and make sure it is correct before proceeding. To test your `intermediate_node_num_mistakes` function, run the following code until you get a **Test passed!**, then you should proceed. Otherwise, you should spend some time figuring out where things went wrong.
###Code
# Test case 1
example_labels = graphlab.SArray([-1, -1, 1, 1, 1])
if intermediate_node_num_mistakes(example_labels) == 2:
print 'Test passed!'
else:
print 'Test 1 failed... try again!'
# Test case 2
example_labels = graphlab.SArray([-1, -1, 1, 1, 1, 1, 1])
if intermediate_node_num_mistakes(example_labels) == 2:
print 'Test passed!'
else:
print 'Test 2 failed... try again!'
# Test case 3
example_labels = graphlab.SArray([-1, -1, -1, -1, -1, 1, 1])
if intermediate_node_num_mistakes(example_labels) == 2:
print 'Test passed!'
else:
print 'Test 3 failed... try again!'
###Output
Test passed!
Test passed!
Test passed!
###Markdown
Function to pick best feature to split on The function **best_splitting_feature** takes 3 arguments: 1. The data (SFrame of data which includes all of the feature columns and label column)2. The features to consider for splits (a list of strings of column names to consider for splits)3. The name of the target/label column (string)The function will loop through the list of possible features, and consider splitting on each of them. It will calculate the classification error of each split and return the feature that had the smallest classification error when split on.Recall that the **classification error** is defined as follows:$$\mbox{classification error} = \frac{\mbox{ mistakes}}{\mbox{ total examples}}$$Follow these steps: * **Step 1:** Loop over each feature in the feature list* **Step 2:** Within the loop, split the data into two groups: one group where all of the data has feature value 0 or False (we will call this the **left** split), and one group where all of the data has feature value 1 or True (we will call this the **right** split). Make sure the **left** split corresponds with 0 and the **right** split corresponds with 1 to ensure your implementation fits with our implementation of the tree building process.* **Step 3:** Calculate the number of misclassified examples in both groups of data and use the above formula to compute the **classification error**.* **Step 4:** If the computed error is smaller than the best error found so far, store this **feature and its error**.This may seem like a lot, but we have provided pseudocode in the comments in order to help you implement the function correctly.**Note:** Remember that since we are only dealing with binary features, we do not have to consider thresholds for real-valued features. This makes the implementation of this function much easier.Fill in the places where you find ` YOUR CODE HERE`. There are **five** places in this function for you to fill in.
###Code
def best_splitting_feature(data, features, target):
best_feature = None # Keep track of the best feature
best_error = 10 # Keep track of the best error so far
# Note: Since error is always <= 1, we should intialize it with something larger than 1.
# Convert to float to make sure error gets computed correctly.
num_data_points = float(len(data))
# Loop through each feature to consider splitting on that feature
for feature in features:
# The left split will have all data points where the feature value is 0
left_split = data[data[feature] == 0]
# The right split will have all data points where the feature value is 1
right_split = data[data[feature] == 1]
# Calculate the number of misclassified examples in the left split.
# Remember that we implemented a function for this! (It was called intermediate_node_num_mistakes)
left_mistakes = intermediate_node_num_mistakes(left_split[target])
# Calculate the number of misclassified examples in the right split.
right_mistakes = intermediate_node_num_mistakes(right_split[target])
# Compute the classification error of this split.
# Error = (# of mistakes (left) + # of mistakes (right)) / (# of data points)
error = float(left_mistakes + right_mistakes) / num_data_points
# If this is the best error we have found so far, store the feature as best_feature and the error as best_error
if error < best_error:
best_error = error
best_feature = feature
return best_feature # Return the best feature we found
###Output
_____no_output_____
###Markdown
To test your `best_splitting_feature` function, run the following code:
###Code
if best_splitting_feature(train_data, features, 'safe_loans') == 'term. 36 months':
print 'Test passed!'
else:
print 'Test failed... try again!'
###Output
Test passed!
###Markdown
Building the treeWith the above functions implemented correctly, we are now ready to build our decision tree. Each node in the decision tree is represented as a dictionary which contains the following keys and possible values: { 'is_leaf' : True/False. 'prediction' : Prediction at the leaf node. 'left' : (dictionary corresponding to the left tree). 'right' : (dictionary corresponding to the right tree). 'splitting_feature' : The feature that this node splits on. }First, we will write a function that creates a leaf node given a set of target values. Fill in the places where you find ` YOUR CODE HERE`. There are **three** places in this function for you to fill in.
###Code
def create_leaf(target_values):
# Create a leaf node
leaf = {'splitting_feature' : None,
'left' : None,
'right' : None,
'is_leaf': True }
# Count the number of data points that are +1 and -1 in this node.
num_ones = len(target_values[target_values == +1])
num_minus_ones = len(target_values[target_values == -1])
# For the leaf node, set the prediction to be the majority class.
# Store the predicted class (1 or -1) in leaf['prediction']
if num_ones > num_minus_ones:
leaf['prediction'] = +1
else:
leaf['prediction'] = -1
# Return the leaf node
return leaf
###Output
_____no_output_____
###Markdown
We have provided a function that learns the decision tree recursively and implements 3 stopping conditions:1. **Stopping condition 1:** All data points in a node are from the same class.2. **Stopping condition 2:** No more features to split on.3. **Additional stopping condition:** In addition to the above two stopping conditions covered in lecture, in this assignment we will also consider a stopping condition based on the **max_depth** of the tree. By not letting the tree grow too deep, we will save computational effort in the learning process. Now, we will write down the skeleton of the learning algorithm. Fill in the places where you find ` YOUR CODE HERE`. There are **seven** places in this function for you to fill in.
###Code
def decision_tree_create(data, features, target, current_depth = 0, max_depth = 10):
remaining_features = features[:] # Make a copy of the features.
target_values = data[target]
print "--------------------------------------------------------------------"
print "Subtree, depth = %s (%s data points)." % (current_depth, len(target_values))
# Stopping condition 1
# (Check if there are mistakes at current node.
# Recall you wrote a function intermediate_node_num_mistakes to compute this.)
if intermediate_node_num_mistakes(target_values) == 0: ## YOUR CODE HERE
print "Stopping condition 1 reached."
# If not mistakes at current node, make current node a leaf node
return create_leaf(target_values)
# Stopping condition 2 (check if there are remaining features to consider splitting on)
if len(remaining_features) == 0: ## YOUR CODE HERE
print "Stopping condition 2 reached."
# If there are no remaining features to consider, make current node a leaf node
return create_leaf(target_values)
# Additional stopping condition (limit tree depth)
if current_depth >= max_depth: ## YOUR CODE HERE
print "Reached maximum depth. Stopping for now."
# If the max tree depth has been reached, make current node a leaf node
return create_leaf(target_values)
# Find the best splitting feature (recall the function best_splitting_feature implemented above)
splitting_feature = best_splitting_feature(data, remaining_features, target)
# Split on the best feature that we found.
left_split = data[data[splitting_feature] == 0]
right_split = data[data[splitting_feature] == 1]
remaining_features.remove(splitting_feature)
print "Split on feature %s. (%s, %s)" % (\
splitting_feature, len(left_split), len(right_split))
# Create a leaf node if the split is "perfect"
if len(left_split) == len(data):
print "Creating leaf node."
return create_leaf(left_split[target])
if len(right_split) == len(data):
print "Creating leaf node."
return create_leaf(right_split[target])
# Repeat (recurse) on left and right subtrees
left_tree = decision_tree_create(left_split, remaining_features, target, current_depth + 1, max_depth)
## YOUR CODE HERE
right_tree = decision_tree_create(right_split, remaining_features, target, current_depth + 1, max_depth)
return {'is_leaf' : False,
'prediction' : None,
'splitting_feature': splitting_feature,
'left' : left_tree,
'right' : right_tree}
###Output
_____no_output_____
###Markdown
Here is a recursive function to count the nodes in your tree:
###Code
def count_nodes(tree):
if tree['is_leaf']:
return 1
return 1 + count_nodes(tree['left']) + count_nodes(tree['right'])
###Output
_____no_output_____
###Markdown
Run the following test code to check your implementation. Make sure you get **'Test passed'** before proceeding.
###Code
small_data_decision_tree = decision_tree_create(train_data, features, 'safe_loans', max_depth = 3)
if count_nodes(small_data_decision_tree) == 13:
print 'Test passed!'
else:
print 'Test failed... try again!'
print 'Number of nodes found :', count_nodes(small_data_decision_tree)
print 'Number of nodes that should be there : 13'
###Output
--------------------------------------------------------------------
Subtree, depth = 0 (37224 data points).
Split on feature term. 36 months. (9223, 28001)
--------------------------------------------------------------------
Subtree, depth = 1 (9223 data points).
Split on feature grade.A. (9122, 101)
--------------------------------------------------------------------
Subtree, depth = 2 (9122 data points).
Split on feature grade.B. (8074, 1048)
--------------------------------------------------------------------
Subtree, depth = 3 (8074 data points).
Reached maximum depth. Stopping for now.
--------------------------------------------------------------------
Subtree, depth = 3 (1048 data points).
Reached maximum depth. Stopping for now.
--------------------------------------------------------------------
Subtree, depth = 2 (101 data points).
Split on feature emp_length.n/a. (96, 5)
--------------------------------------------------------------------
Subtree, depth = 3 (96 data points).
Reached maximum depth. Stopping for now.
--------------------------------------------------------------------
Subtree, depth = 3 (5 data points).
Reached maximum depth. Stopping for now.
--------------------------------------------------------------------
Subtree, depth = 1 (28001 data points).
Split on feature grade.D. (23300, 4701)
--------------------------------------------------------------------
Subtree, depth = 2 (23300 data points).
Split on feature grade.E. (22024, 1276)
--------------------------------------------------------------------
Subtree, depth = 3 (22024 data points).
Reached maximum depth. Stopping for now.
--------------------------------------------------------------------
Subtree, depth = 3 (1276 data points).
Reached maximum depth. Stopping for now.
--------------------------------------------------------------------
Subtree, depth = 2 (4701 data points).
Split on feature grade.A. (4701, 0)
Creating leaf node.
Test passed!
###Markdown
Build the tree!Now that all the tests are passing, we will train a tree model on the **train_data**. Limit the depth to 6 (**max_depth = 6**) to make sure the algorithm doesn't run for too long. Call this tree **my_decision_tree**. **Warning**: This code block may take 1-2 minutes to learn.
###Code
# Make sure to cap the depth at 6 by using max_depth = 6
my_decision_tree = decision_tree_create(train_data, features, 'safe_loans', max_depth = 6)
###Output
--------------------------------------------------------------------
Subtree, depth = 0 (37224 data points).
Split on feature term. 36 months. (9223, 28001)
--------------------------------------------------------------------
Subtree, depth = 1 (9223 data points).
Split on feature grade.A. (9122, 101)
--------------------------------------------------------------------
Subtree, depth = 2 (9122 data points).
Split on feature grade.B. (8074, 1048)
--------------------------------------------------------------------
Subtree, depth = 3 (8074 data points).
Split on feature grade.C. (5884, 2190)
--------------------------------------------------------------------
Subtree, depth = 4 (5884 data points).
Split on feature grade.D. (3826, 2058)
--------------------------------------------------------------------
Subtree, depth = 5 (3826 data points).
Split on feature grade.E. (1693, 2133)
--------------------------------------------------------------------
Subtree, depth = 6 (1693 data points).
Reached maximum depth. Stopping for now.
--------------------------------------------------------------------
Subtree, depth = 6 (2133 data points).
Reached maximum depth. Stopping for now.
--------------------------------------------------------------------
Subtree, depth = 5 (2058 data points).
Split on feature grade.E. (2058, 0)
Creating leaf node.
--------------------------------------------------------------------
Subtree, depth = 4 (2190 data points).
Split on feature grade.D. (2190, 0)
Creating leaf node.
--------------------------------------------------------------------
Subtree, depth = 3 (1048 data points).
Split on feature emp_length.5 years. (969, 79)
--------------------------------------------------------------------
Subtree, depth = 4 (969 data points).
Split on feature grade.C. (969, 0)
Creating leaf node.
--------------------------------------------------------------------
Subtree, depth = 4 (79 data points).
Split on feature home_ownership.MORTGAGE. (34, 45)
--------------------------------------------------------------------
Subtree, depth = 5 (34 data points).
Split on feature grade.C. (34, 0)
Creating leaf node.
--------------------------------------------------------------------
Subtree, depth = 5 (45 data points).
Split on feature grade.C. (45, 0)
Creating leaf node.
--------------------------------------------------------------------
Subtree, depth = 2 (101 data points).
Split on feature emp_length.n/a. (96, 5)
--------------------------------------------------------------------
Subtree, depth = 3 (96 data points).
Split on feature emp_length.< 1 year. (85, 11)
--------------------------------------------------------------------
Subtree, depth = 4 (85 data points).
Split on feature grade.B. (85, 0)
Creating leaf node.
--------------------------------------------------------------------
Subtree, depth = 4 (11 data points).
Split on feature grade.B. (11, 0)
Creating leaf node.
--------------------------------------------------------------------
Subtree, depth = 3 (5 data points).
Split on feature grade.B. (5, 0)
Creating leaf node.
--------------------------------------------------------------------
Subtree, depth = 1 (28001 data points).
Split on feature grade.D. (23300, 4701)
--------------------------------------------------------------------
Subtree, depth = 2 (23300 data points).
Split on feature grade.E. (22024, 1276)
--------------------------------------------------------------------
Subtree, depth = 3 (22024 data points).
Split on feature grade.F. (21666, 358)
--------------------------------------------------------------------
Subtree, depth = 4 (21666 data points).
Split on feature emp_length.n/a. (20734, 932)
--------------------------------------------------------------------
Subtree, depth = 5 (20734 data points).
Split on feature grade.G. (20638, 96)
--------------------------------------------------------------------
Subtree, depth = 6 (20638 data points).
Reached maximum depth. Stopping for now.
--------------------------------------------------------------------
Subtree, depth = 6 (96 data points).
Reached maximum depth. Stopping for now.
--------------------------------------------------------------------
Subtree, depth = 5 (932 data points).
Split on feature grade.A. (702, 230)
--------------------------------------------------------------------
Subtree, depth = 6 (702 data points).
Reached maximum depth. Stopping for now.
--------------------------------------------------------------------
Subtree, depth = 6 (230 data points).
Reached maximum depth. Stopping for now.
--------------------------------------------------------------------
Subtree, depth = 4 (358 data points).
Split on feature emp_length.8 years. (347, 11)
--------------------------------------------------------------------
Subtree, depth = 5 (347 data points).
Split on feature grade.A. (347, 0)
Creating leaf node.
--------------------------------------------------------------------
Subtree, depth = 5 (11 data points).
Split on feature home_ownership.OWN. (9, 2)
--------------------------------------------------------------------
Subtree, depth = 6 (9 data points).
Reached maximum depth. Stopping for now.
--------------------------------------------------------------------
Subtree, depth = 6 (2 data points).
Stopping condition 1 reached.
--------------------------------------------------------------------
Subtree, depth = 3 (1276 data points).
Split on feature grade.A. (1276, 0)
Creating leaf node.
--------------------------------------------------------------------
Subtree, depth = 2 (4701 data points).
Split on feature grade.A. (4701, 0)
Creating leaf node.
###Markdown
Making predictions with a decision treeAs discussed in the lecture, we can make predictions from the decision tree with a simple recursive function. Below, we call this function `classify`, which takes in a learned `tree` and a test point `x` to classify. We include an option `annotate` that describes the prediction path when set to `True`.Fill in the places where you find ` YOUR CODE HERE`. There is **one** place in this function for you to fill in.
###Code
def classify(tree, x, annotate = False):
# if the node is a leaf node.
if tree['is_leaf']:
if annotate:
print "At leaf, predicting %s" % tree['prediction']
return tree['prediction']
else:
# split on feature.
split_feature_value = x[tree['splitting_feature']]
if annotate:
print "Split on %s = %s" % (tree['splitting_feature'], split_feature_value)
if split_feature_value == 0:
return classify(tree['left'], x, annotate)
else:
return classify(tree['right'], x, annotate)
###Output
_____no_output_____
###Markdown
Now, let's consider the first example of the test set and see what `my_decision_tree` model predicts for this data point.
###Code
test_data[0]
print 'Predicted class: %s ' % classify(my_decision_tree, test_data[0])
###Output
Predicted class: -1
###Markdown
Let's add some annotations to our prediction to see what the prediction path was that lead to this predicted class:
###Code
classify(my_decision_tree, test_data[0], annotate=True)
###Output
Split on term. 36 months = 0
Split on grade.A = 0
Split on grade.B = 0
Split on grade.C = 0
Split on grade.D = 1
At leaf, predicting -1
###Markdown
** Quiz question:** What was the feature that **my_decision_tree** first split on while making the prediction for test_data[0]? ** Quiz question:** What was the first feature that lead to a right split of test_data[0]? ** Quiz question:** What was the last feature split on before reaching a leaf node for test_data[0]? Evaluating your decision tree Now, we will write a function to evaluate a decision tree by computing the classification error of the tree on the given dataset.Again, recall that the **classification error** is defined as follows:$$\mbox{classification error} = \frac{\mbox{ mistakes}}{\mbox{ total examples}}$$Now, write a function called `evaluate_classification_error` that takes in as input:1. `tree` (as described above)2. `data` (an SFrame)3. `target` (a string - the name of the target/label column)This function should calculate a prediction (class label) for each row in `data` using the decision `tree` and return the classification error computed using the above formula. Fill in the places where you find ` YOUR CODE HERE`. There is **one** place in this function for you to fill in.
###Code
def evaluate_classification_error(tree, data, target):
# Apply the classify(tree, x) to each row in your data
prediction = data.apply(lambda x: classify(tree, x))
# Once you've made the predictions, calculate the classification error and return it
accuracy = (prediction == data[target]).sum()
error = 1 - float(accuracy) / len(data[target])
return error
###Output
_____no_output_____
###Markdown
Now, let's use this function to evaluate the classification error on the test set.
###Code
round(evaluate_classification_error(my_decision_tree, test_data, target), 2)
###Output
_____no_output_____
###Markdown
**Quiz Question:** Rounded to 2nd decimal point, what is the classification error of **my_decision_tree** on the **test_data**? Printing out a decision stump As discussed in the lecture, we can print out a single decision stump (printing out the entire tree is left as an exercise to the curious reader).
###Code
def print_stump(tree, name = 'root'):
split_name = tree['splitting_feature'] # split_name is something like 'term. 36 months'
if split_name is None:
print "(leaf, label: %s)" % tree['prediction']
return None
split_feature, split_value = split_name.split('.')
print ' %s' % name
print ' |---------------|----------------|'
print ' | |'
print ' | |'
print ' | |'
print ' [{0} == 0] [{0} == 1] '.format(split_name)
print ' | |'
print ' | |'
print ' | |'
print ' (%s) (%s)' \
% (('leaf, label: ' + str(tree['left']['prediction']) if tree['left']['is_leaf'] else 'subtree'),
('leaf, label: ' + str(tree['right']['prediction']) if tree['right']['is_leaf'] else 'subtree'))
print_stump(my_decision_tree)
###Output
root
|---------------|----------------|
| |
| |
| |
[term. 36 months == 0] [term. 36 months == 1]
| |
| |
| |
(subtree) (subtree)
###Markdown
**Quiz Question:** What is the feature that is used for the split at the root node? Exploring the intermediate left subtreeThe tree is a recursive dictionary, so we do have access to all the nodes! We can use* `my_decision_tree['left']` to go left* `my_decision_tree['right']` to go right
###Code
print_stump(my_decision_tree['left'], my_decision_tree['splitting_feature'])
###Output
term. 36 months
|---------------|----------------|
| |
| |
| |
[grade.A == 0] [grade.A == 1]
| |
| |
| |
(subtree) (subtree)
###Markdown
Exploring the left subtree of the left subtree
###Code
print_stump(my_decision_tree['left']['left'], my_decision_tree['left']['splitting_feature'])
###Output
grade.A
|---------------|----------------|
| |
| |
| |
[grade.B == 0] [grade.B == 1]
| |
| |
| |
(subtree) (subtree)
###Markdown
**Quiz question:** What is the path of the **first 3 feature splits** considered along the **left-most** branch of **my_decision_tree**? **Quiz question:** What is the path of the **first 3 feature splits** considered along the **right-most** branch of **my_decision_tree**?
###Code
print_stump(my_decision_tree['right'], my_decision_tree['splitting_feature'])
print_stump(my_decision_tree['right']['right'], my_decision_tree['left']['splitting_feature'])
###Output
(leaf, label: -1)
|
Backtester_Step_By_Step.ipynb
|
###Markdown
Backtesting 0. Backfilling Data
###Code
from broker_to_csv import TradingApp, multi_historical_retriver
import pandas as pd
import threading
import time
########## Starting App as Thread #############
def websocket_con():
app.run()
event = threading.Event()
app = TradingApp()
app.event=event
app.connect(host='127.0.0.1', port=7496, clientId=23) #port 4002 for ib gateway paper trading/7497 for TWS paper trading
con_thread = threading.Thread(target=websocket_con, daemon=True)
con_thread.start()
time.sleep(1) # some latency added to ensure that the connection is established
##################################################
########## From Datetime #############
import datetime
today_date=(datetime.datetime.today().strftime("%Y%m%d %H:%M:%S"))
########################################
########## Historical Data ############
tickers=['TATAMOTOR']
multi_historical_retriver(ticker_list=tickers,
app_obj=app,
from_date=today_date,
duration="20 D",
bar_size="5 mins",
event=event
)
########################################
def kernel_restarter():
from IPython.display import display_html
display_html("<script>Jupyter.notebook.kernel.restart()</script>",raw=True)
kernel_restarter()
# Old Csv to sql
from csv_to_sql import sql_ingester
data_dir="./Data"
import sqlalchemy
sql_obj = sqlalchemy.create_engine('postgresql://krh:krh@123@localhost:5432/krh')
df=sql_ingester(data_dir, sql_obj, False)
####################################################################################################################
###Output
tatamotor_1min :
Table exist, updating record.
Done
tatamotor_5mins :
Table exist, updating record.
Done
tatamotor_5mins :
Table exist, updating record.
Done
tatamotor_5mins :
Table exist, updating record.
Done
###Markdown
1. Getting Data from SQL
###Code
import pandas as pd
import sqlalchemy
sql_obj = sqlalchemy.create_engine('postgresql://krh:krh@123@localhost:5432/krh')
df_1min = pd.read_sql_table('tatamotor_1min', sql_obj, parse_dates={'date': {'format': '%Y-%m-%d %H:%M:%S'}})
df_5mins = pd.read_sql_table('tatamotor_5mins', sql_obj, parse_dates={'date': {'format': '%Y-%m-%d %H:%M:%S'}})
df_time_frames=[df_5mins]
df_time_frames[0].tail()
###Output
_____no_output_____
###Markdown
2. Creating Strategy
###Code
import backtrader as bt
import backtrader.indicators as btind
import numpy as np
import datetime
class ReversalAction(bt.Strategy):
params = (
('short_period',10),
('long_period',180),
('reversal_tol_factor',0.5),
('breakout_tol_factor',0.3),
('strike_at',"sr_price"),
('target_percentage',2),
('stop_percentage',0.5),
('closing_time',"15:10"),
('show_trades', False),
('execute_breakout',True),
('sr_levels',[]),
('order_time',"2:00"),
('order_at',"close_price"),
('show_trade_object',False),
('allow_shorting',False),
('cerebro', "")
)
def log(self,txt,dt=None):
if dt is None:
dt=self.datas[0].datetime.datetime()
print(dt,txt)
pass
def tolerance(self,base_x,y,tolerance, dt=None):
z=(base_x-y)/base_x
z=np.abs(z)*100
z=z<tolerance
return z
############## Copied from Documenation #####################
def notify_order(self, order):
if self.params.show_trades:
if order.status in [order.Submitted, order.Accepted]:
# Buy/Sell order submitted/accepted to/by broker - Nothing to do
return
# Check if an order has been completed
# Attention: broker could reject order if not enough cash
if order.status in [order.Completed]:
if order.isbuy():
self.log('BUY EXECUTED, %.2f' % order.executed.price)
elif order.issell():
self.log('SELL EXECUTED, %.2f' % order.executed.price)
self.bar_executed = len(self)
elif order.status == order.Canceled:
self.log('Order Canceled')
elif order.status ==order.Margin:
self.log('Order Margin')
elif order.status ==order.Rejected:
self.log('Order Rejected')
# Write down: no pending order
self.order = None
#################################################################
def notify_trade(self, trade):
trade.historyon=True
dt = self.data.datetime.datetime()
if trade.isclosed:
dt=self.datas[0].datetime.datetime()
h1=["Date", 'Avg Price', 'Gross Profit', 'Net Profit', 'Len']
r1=[dt, trade.price, round(trade.pnl,2), round(trade.pnlcomm,2), trade.barlen]
table_values=[h1,r1]
from tabulate import tabulate
if self.params.show_trade_object:
print(tabulate(table_values,))
def __init__(self):
cerebro=self.params.cerebro
self.start_datetime=self.datas[0].p.fromdate
self.start_portfolio_value = cerebro.broker.getvalue()
self.brought_today=False
self.order =None
self.sma_short = btind.EMA(self.datas[0], period=self.params.short_period)
self.sma_long= btind.EMA(self.datas[0], period=self.params.long_period)
################# Printing Profit At end ################################
def stop(self):
from tabulate import tabulate
from babel.numbers import format_currency as inr
start_portfolio_value=self.start_portfolio_value
end_portfolio_value=int(cerebro.broker.getvalue())
pnl=end_portfolio_value-self.start_portfolio_value
start_portfolio_value = inr(start_portfolio_value, "INR", locale='en_IN')
end_portfolio_value = inr(end_portfolio_value, "INR", locale='en_IN')
pnl = inr(pnl, "INR", locale='en_IN')
start_datetime=self.start_datetime
end_datetime=self.datas[0].datetime.datetime()
start_date=start_datetime.date()
end_date=end_datetime.date()
time_delta=end_datetime-self.start_datetime
table_values= [
["Date Time",start_date, end_date, time_delta.days],
["Amount", start_portfolio_value, end_portfolio_value, pnl],
]
print (tabulate(table_values,
headers=["Values","Started","Ended","Delta"],
tablefmt="grid"))
###############################################
def next(self):
mid_bar_value= (self.datas[0].high[0] + self.datas[0].low[0] )/2
open_p=self.datas[0].open[0]
low_p=self.datas[0].low[0]
high_p=self.datas[0].high[0]
close_p=self.datas[0].close[0]
open_p1=self.datas[0].open[-1]
low_p1=self.datas[0].low[-1]
high_p1=self.datas[0].high[-1]
close_p1=self.datas[0].close[-1]
################# Long Trend ################################
if mid_bar_value>self.sma_long:
long_trend="Up"
else:
long_trend="Down"
##############################################################
################# Short Trend ################################
if mid_bar_value>self.sma_short:
short_trend="Up"
else:
short_trend="Down"
##############################################################
################# SR Area ################################
sr=self.params.sr_levels
tol_factor=self.params.reversal_tol_factor
if short_trend=="Up":
z=self.tolerance(high_p,sr,tol_factor)
else:
z=self.tolerance(low_p,sr,tol_factor)
z=np.matmul(z,np.transpose(sr))
if z>0:
area_of_value="In"
area=z
else:
area_of_value="Out"
area=""
###############################################################
################# Volume Support ################################
if self.datas[0].volume[0]>self.datas[0].volume[-1]:
volume_support="yes"
else:
volume_support="no"
###############################################################
################# Bar Lenght Support ################################
bar_lenght_support=""
if np.abs(open_p-close_p) > np.abs(open_p1-close_p1):
bar_lenght_support="yes"
else:
bar_lenght_support="no"
###############################################################
################# Red Green Conversion ################################
# Current Bar Color
if close_p>open_p:
bar_color="green"
else:
bar_color="red"
# Previous Bar Color
if close_p1>open_p1:
previous_bar_color="green"
else:
previous_bar_color="red"
trend_change=""
if volume_support=="yes" and bar_lenght_support=="yes":
if previous_bar_color=="green" and bar_color=="red":
trend_change="green_to_red"
elif previous_bar_color=="red" and bar_color=="green":
trend_change="red_to_green"
elif previous_bar_color=="green" and bar_color=="green":
trend_change="green_to_green"
elif previous_bar_color=="red" and bar_color=="red":
trend_change="red_to_red"
########################################################################
################# To Buy/Sell/Wait ################################
############### Reversal
order_signal=""
if long_trend=="Up":
if short_trend=="Down":
if area_of_value=="In":
if trend_change=="red_to_green":
order_signal="Buy"
if long_trend=="Down":
if short_trend=="Up":
if area_of_value=="In":
if trend_change=="green_to_red":
order_signal="Sell"
############### Breakout
if self.params.execute_breakout:
breakout_tol=self.params.breakout_tol_factor
if long_trend=="Up":
if short_trend=="Up":
if area_of_value=="In":
if ((close_p-area)/close_p)*100 >breakout_tol:
if trend_change=="green_to_green":
order_signal="Buy"
if long_trend=="Down":
if short_trend=="Down":
if area_of_value=="In":
if ((close_p-area)/close_p)*100 <breakout_tol:
if trend_change=="red_to_red":
order_signal="Sell"
else:
pass
################# Placing Bracket Order ################################
strike_at=self.params.strike_at
if strike_at =="mid_bar_price":
strike_price=mid_bar_value
elif strike_at=="sr_price":
if area=="":
strike_price=0
else:
strike_price=area
###### Target
target_percentage=self.params.target_percentage
buy_target=strike_price*(1+(target_percentage/100))
sell_target=strike_price*(1-(target_percentage/100))
###### Stop Loss
stop_percentage=self.params.stop_percentage
buy_loss=strike_price*(1-(stop_percentage/100))
sell_loss=strike_price*(1+(stop_percentage/100))
################### Placing Order ######################################
order_hour=self.params.order_time.split(":")[0]
order_hour=int(order_hour)
order_minute=self.params.order_time.split(":")[1]
order_minute=int(order_minute)
if self.data.datetime.time() < datetime.time(order_hour,order_minute) \
and not self.position.size\
and (order_signal=="Buy" or order_signal=="Sell"):
order_at=self.params.order_at
if order_at=="close_price":
order_price=close_p
elif order_at=="mid_bar_price":
order_price=mid_bar_value
elif order_at=="sr_price":
order_price=area
cash=cerebro.broker.getvalue()
cash=cash*(1-0.05)
no_of_shares=int(cash/order_price)
lots=int(no_of_shares/100)
size=lots*100
if order_signal=="Buy":
if self.params.show_trades:
print("-------------------Buyed---------------:",size)
self.order = self.buy_bracket(limitprice=buy_target,
price=order_price,
stopprice=buy_loss,
size=size)
if order_signal=="Sell" and self.params.allow_shorting:
if self.params.show_trades:
print("-------------------Sold---------------",size)
self.order = self.sell_bracket(limitprice=sell_target,
price=order_price,
stopprice=sell_loss,
size=size)
########################################################################
################# Closing all Postion before 3:10 ################################
close_hour=self.params.closing_time.split(":")[0]
close_hour=int(close_hour)
close_minute=self.params.closing_time.split(":")[1]
close_minute=int(close_minute)
if self.position.size != 0:
if self.data.datetime.time() > datetime.time(close_hour,close_minute):
self.close(exectype=bt.Order.Market, size=self.position.size)
########################################################################
self.log("Close: "+str(close_p))
# +
# " Long Trend:"+long_trend+
# " Short Trend:"+short_trend+
# " Area :"+area_of_value+str(area)+
# " Trend Change: "+trend_change+
# " Order Signal: "+ order_signal)
###Output
_____no_output_____
###Markdown
3. Creating Analyzer
###Code
def printTradeAnalysis(analyzer):
'''
Function to print the Technical Analysis results in a nice format.
'''
#Get the results we are interested in
total_open = analyzer.total.open
total_closed = analyzer.total.closed
total_won = analyzer.won.total
total_lost = analyzer.lost.total
win_streak = analyzer.streak.won.longest
lose_streak = analyzer.streak.lost.longest
pnl_net = round(analyzer.pnl.net.total,2)
strike_rate = int((total_won / total_closed) * 100)
#Designate the rows
h1 = ['Total Open', 'Total Won', 'Win Streak', 'Strike Rate']
h2 = ['Total Closed', 'Total Lost', 'Losing Streak','PnL Net']
r1 = [total_open, total_won, win_streak, strike_rate]
r2 = [total_closed, total_lost, lose_streak, pnl_net]
from tabulate import tabulate
table_values= [h1,
r1,
h2,
r2]
print("\n\nTrade Analysis Results:")
print (tabulate(table_values,))
def printSQN(analyzer):
sqn = round(analyzer.sqn,2)
print('SQN: {}'.format(sqn))
###Output
_____no_output_____
###Markdown
3. Starting App
###Code
import numpy as np
if __name__=='__main__':
ticker_name="TATAMOTOR-STK-NSE"
cerebro=bt.Cerebro()
#Add data
for df in df_time_frames:
data=bt.feeds.PandasData(dataname=df,
datetime=0,
fromdate=datetime.datetime(2021, 2, 1),
)
cerebro.adddata(data)
#Set Cash
cerebro.broker.setcash(160000)
#Add stratedgy to Cerebro
sr_levels=np.array([339.01,324.11,319.38,312.96,304.37,299.17,295.17,293.48,291.24,283.31])
cerebro.addstrategy(ReversalAction,
short_period=50,
long_period=200,
sr_levels=sr_levels,
reversal_tol_factor=1.5,
breakout_tol_factor=.3,
order_time="15:10",
closing_time="15:10",
show_trades= False,
show_trade_object=False,
strike_at="sr_price",
order_at="mid_bar_price",
target_percentage=2,
stop_percentage=1.2,
execute_breakout=True,
allow_shorting=True,
cerebro=cerebro)
#Adding Anylizer
cerebro.addanalyzer(bt.analyzers.PyFolio, _name='PyFolio')
cerebro.addanalyzer(bt.analyzers.TradeAnalyzer, _name="ta")
cerebro.addanalyzer(bt.analyzers.SQN, _name="sqn")
# Adding Observer
cerebro.addobserver(bt.observers.DrawDown, plot=True, subplot=False)
#RUN Cerebro Engine
strategies=cerebro.run()
# Capture results
firstStrat = strategies[0]
# print the analyzers
printTradeAnalysis(firstStrat.analyzers.ta.get_analysis())
printSQN(firstStrat.analyzers.sqn.get_analysis())
portfolio_stats = firstStrat.analyzers.getbyname('PyFolio')
returns, positions, transactions, gross_lev = portfolio_stats.get_pf_items()
returns.index = returns.index.tz_convert(None)
import quantstats
report_name=ticker_name.split("-")[0]
quantstats.reports.html(returns, output=str(report_name+'.html'), title=report_name)
###Output
+-----------+--------------+--------------+------------+
| Values | Started | Ended | Delta |
+===========+==============+==============+============+
| Date Time | 2021-02-01 | 2021-04-26 | 84 |
+-----------+--------------+--------------+------------+
| Amount | ₹1,60,000.00 | ₹2,12,745.00 | ₹52,745.00 |
+-----------+--------------+--------------+------------+
Trade Analysis Results:
------------ ---------- ------------- -----------
Total Open Total Won Win Streak Strike Rate
0 34 8 66
Total Closed Total Lost Losing Streak PnL Net
51 17 4 52745.78
------------ ---------- ------------- -----------
SQN: 3.78
###Markdown
6. Going Live on Paper
###Code
import numpy as np
import backtrader as bt
# from strategy import ReversalAction
import datetime
if __name__=='__main__':
#Creating Cerebro Obect########
cerebro=bt.Cerebro()
###############################
ticker_name="TATAMOTOR-STK-NSE"
######### Add data to cerebro############
ibstore = bt.stores.IBStore(host='127.0.0.1', port=7496, clientId=35)
cerebro.broker = ibstore.getbroker()
#################################################################
# Data preparation
back_data=bt.feeds.PandasData(dataname=df_time_frames[0],
datetime=0,
fromdate=datetime.datetime(2021, 2, 1))
data = bt.feeds.IBData(dataname=ticker_name, backtfill_from=back_data)
cerebro.adddata(data)
###################################################
cerebro.resampledata(data, timeframe=bt.TimeFrame.Minutes, compression=5)
######### Add stratedgy to Cerebro ###############
sr_levels=np.array([339.01,324.11,319.38,312.96,304.37,299.17,295.17,293.48,291.24,283.31])
cerebro.addstrategy(ReversalAction,
short_period=50,
long_period=200,
sr_levels=sr_levels,
reversal_tol_factor=.8,
breakout_tol_factor=.3,
order_time="15:10",
closing_time="15:10",
show_trades= False,
show_trade_object=False,
strike_at="sr_price",
order_at="mid_bar_price",
target_percentage=2,
stop_percentage=1.2,
execute_breakout=True,
allow_shorting=True,
cerebro=cerebro)
##########################################
############# RUN Cerebro Engine####################
cerebro.run()
##################################################
###Output
Server Version: 76
TWS Time at connection:20210502 02:51:15 India Standard Time
|
Week 01 Mathematical Foundations/Sprint Challenge/Mathematical Foundations.ipynb
|
###Markdown
Mathematical Foundations of Machine LearningThere are four objectives in *Functions and Optima** graph some of the typical shapes of 2D functions* link functions with tables of values* calculate the derivative of some function `f(x)`There are seven objectives in *Linear Algebra** create and edit a vector with numpy * add two vectors together* subtract a vector from another vector* calculate the magnitude of a vector with numpy* multiply a matrix with a vector* invert a matrix and describe what it does* tranpose a matrixCreate solutions for the following code blocks. This exercise should take ~ 3 hours.Share with `[email protected]` when finished.
###Code
# LAMBDA SCHOOL
#
# MACHINE LEARNING
#
# MIT LICENSE
import numpy as np
import matplotlib.pyplot as plt
###Output
_____no_output_____
###Markdown
1. graph some of the typical shape of a 2d functionDraw a graph of $x^2$.Draw a graph of $sin(x)$
###Code
# x is a numpy array with 25 values.
x = np.linspace(-4, 4, 25)
y1 = x**2
y2 = np.sin(x)
plt.plot(x, y1)
plt.title('f(x) = x^2')
plt.show()
plt.plot(x, y2)
plt.title('f(x) = sin(x)')
plt.show();
###Output
_____no_output_____
###Markdown
2. link a function with a table of values A table of MxN values can be one function, M functions, or something in between.x is 3x5, so we expect it to be representing 1, 2, or 3 functions. If it is representing 3 functions, then it contains 2 linear functions with different slopes and one constant function. If it represents one function, then it is a 3D linear function with y-intercept -6.28, a gradient of about $\pi$ along the x-axis, and a constant z It can be thus be treated as a 2D function on the xy plane.It could also be the case that x represents a linear function with slope $\pi$, and a constant function at 0, with the first row being the x-values. Even if x represents 3 functions, the first row can be used to represent the implicit x-coordinates for all 3 functions.
###Code
x = [[0,1,2,3,4],[-6.28,-3.14,0,3.14,6.28],[0,0,0,0,0]]
# plot each function in this table
# Are the functions related?
def plot_from_table(x, ix_1, ix_2, xlim=(-1, 5), ylim=(-6.5, 6.5)):
plt.plot(x[ix_1], x[ix_2])
plt.xlabel('x')
plt.xlabel('f(x)')
plt.xlim(xlim[0], xlim[1])
plt.ylim(ylim[0], ylim[1])
plt.show()
# in the case that x represents one function
print('x represents one function:')
plot_from_table(x, 0, 1)
# in the case that x represents 3 functions
print('x represents three function:')
plot_from_table(x, 0, 0)
plot_from_table(x, 0, 1)
plot_from_table(x, 0, 2);
###Output
x represents one function:
###Markdown
3. calculate the derivative of f(x)
###Code
x = np.linspace(-4, 4, 25)
def f(x):
# return the derivative of sin(x) at x
return np.cos(x)
d_f = f(x)
plt.plot(x, d_f)
plt.show();
def fd(x, y):
# return the derivative at the first pair of values of y
dx = (y[1]-y[0]) / (x[1]-x[0])
return dx
d_fd = fd([0,1],[0,-2])
print(d_fd)
###Output
_____no_output_____
###Markdown
4. create and edit a vector of scalars
###Code
# Create a vector v containing seven 1s
v = np.ones((7, 1))
# Set the 4th element of v to -1
v[3] = -1
print(v)
###Output
[[ 1.]
[ 1.]
[ 1.]
[-1.]
[ 1.]
[ 1.]
[ 1.]]
###Markdown
5. add two vectors together$z = \begin{pmatrix} 2 \\ -1 \end{pmatrix} + \begin{pmatrix} 2 \\ -1 \end{pmatrix}$
###Code
# x =
x = np.array([[2], [-1]])
# y =
y = np.array([[2], [-1]])
# z =
z = x + y
print(z)
###Output
[[ 4]
[-2]]
###Markdown
5. subtract a vector from another vector$z = \begin{pmatrix} 1 \\ 3 \end{pmatrix} - \begin{pmatrix} -2 \\ -3.5 \end{pmatrix}$
###Code
# x =
x = np.array([[1], [3]])
# y =
y = np.array([[-2], [-3.5]])
# z =
z = x - y
print(z)
###Output
[[3. ]
[6.5]]
###Markdown
6. calculate the magnitude of a vector$|m| = \sqrt{\sum{v_i^2}}$
###Code
v = [1,2,3,4,5]
# what's the magnitude of v?
v_norm = np.linalg.norm(v)
print(v_norm)
###Output
7.416198487095663
###Markdown
7. multiply a simple affine matrix with a vector$\begin{pmatrix} 1.1 & 0.9 & 0 \\ 0.8 & 1.2 & 0 \\ 1 & -1 & 1 \end{pmatrix} \begin{pmatrix} 1 \\ 0 \\ 1 \end{pmatrix} = v$
###Code
# Mv_0 = v
M = np.matrix([[1.1, 0.9, 0], [0.8, 1.2, 0], [1, -1, 1]])
v_0 = np.array([[1], [0], [1]])
v = M * v_0
print(v)
###Output
[[1.1]
[0.8]
[2. ]]
###Markdown
8. invert a matrix and describe what it does$MM^{-1} = I$$M = \begin{pmatrix} 1.1 & 0.9 \\ 0.8 & 1.2 \end{pmatrix}$
###Code
# M
M = np.matrix([[1.1, 0.9], [0.8, 1.2]])
# M^{-1}
M_inv = np.linalg.inv(M)
# MM^{-1}
print(M*M_inv)
###Output
[[ 1.00000000e+00 4.07081776e-17]
[-2.96059473e-17 1.00000000e+00]]
###Markdown
We expect that the matrix product of a nonsigular, square matrix and its inverse will produce the identity matrix. 9. transpose a matrix$M_{(MxN)}^T = M_{(NxM)}^T$$M = \begin{pmatrix} 1.1 & 0.9 & 0 \\ 0.8 & 1.2 & 0 \end{pmatrix}$
###Code
# M
M = np.matrix([[1.1, 0.9, 0], [0.8, 1.2, 0]])
# M^T
M_T = M.T
# Shape of M?
print(M.shape)
print(M_T.shape)
###Output
(2, 3)
(3, 2)
|
Experiments/Test/Test_50_800.ipynb
|
###Markdown
Optimize Policy
###Code
from env import Env
import env_utils as envu
from dynamics_model import Dynamics_model
from lander_model import Lander_model
from ic_gen_scene import Landing_icgen
import rl_utils
import attitude_utils as attu
from arch_policy_gtvf import Arch
from softmax_pd import Softmax_pd as PD
from policy_ppo import Policy
from value_function import Value_function
import policy_nets as policy_nets
import valfunc_nets as valfunc_nets
import cnn_nets
from agent import Agent
import torch.nn as nn
from flat_constraint import Flat_constraint
from glideslope_constraint import Glideslope_constraint
from rh_constraint import RH_constraint
from no_attitude_constraint import Attitude_constraint
from w_constraint import W_constraint
from reward_terminal_mdr import Reward
from asteroid_hfr_scene import Asteroid
from thruster_model import Thruster_model
asteroid_model = Asteroid(landing_site_override=None, omega_range=(1e-6,5e-4))
ap = attu.Quaternion_attitude()
from flash_lidar2 import Flash_lidar
import attitude_utils as attu
from triangle_ray_intersect import Triangle_ray_intersect
from isosphere import Isosphere
iso = Isosphere(recursion_level=2)
tri = Triangle_ray_intersect()
ap = attu.Quaternion_attitude()
P = 64
sensor = Flash_lidar(ap, tri, sqrt_pixels=int(np.sqrt(P)))
thruster_model = Thruster_model(pulsed=True, scale=1.0, offset=0.4)
lander_model = Lander_model(asteroid_model, thruster_model, ap, sensor, iso)
lander_model.get_state_agent = lander_model.get_state_agent_image_state_stab
logger = rl_utils.Logger()
dynamics_model = Dynamics_model(h=2)
obs_dim = 2*P
gt_dim = 13
action_dim = 12
actions_per_dim = 2
action_logit_dim = action_dim * actions_per_dim
recurrent_steps = 60
reward_object = Reward(landing_coeff=10.0, landing_rlimit=2, landing_vlimit=0.1, tracking_bias=0.01,
dalt_coeff=0.02, fuel_coeff=-0.01, use_gt=True)
glideslope_constraint = Glideslope_constraint(gs_limit=-1.0)
shape_constraint = Flat_constraint()
attitude_constraint = Attitude_constraint(ap)
w_constraint = W_constraint(w_limit=(0.1,0.1,0.1), w_margin=(0.05,0.05,0.05))
rh_constraint = RH_constraint(rh_limit=150)
wi=0.02
ic_gen = Landing_icgen(position_r=(50,800),
p_engine_fail=0.0,
p_scale=(0.01, 0.02),
engine_fail_scale=(1.0,1.0),
asteroid_axis_low=(300,300,300),
asteroid_axis_high=(600,600,600),
#position_theta=(0,np.pi/4),
lander_wll=(-wi,-wi,-wi),
lander_wul=(wi,wi,wi),
attitude_parameterization=ap,
attitude_error=(0,np.pi/16),
min_mass=450, max_mass=500,
debug=False,
inertia_uncertainty_diag=10.0,
inertia_uncertainty_offdiag=1.0)
env = Env(ic_gen, lander_model, dynamics_model, logger,
debug_done=False,
reward_object=reward_object,
glideslope_constraint=glideslope_constraint,
attitude_constraint=attitude_constraint,
w_constraint=w_constraint,
rh_constraint=rh_constraint,
tf_limit=600.0,print_every=10,nav_period=6)
env.ic_gen.show()
arch = Arch(gt_func=lander_model.get_state_agent_gt)
cnn = cnn_nets.CNN_layer(8,2,8)
policy = Policy(policy_nets.GRU_CNN2(7, action_logit_dim, cnn, recurrent_steps=recurrent_steps),
PD(action_dim, actions_per_dim),
shuffle=False, servo_kl=False, max_grad_norm=30,
init_func=rl_utils.xn_init, scale_image_obs=True, scale_vector_obs=True)
value_function = Value_function(valfunc_nets.GRU1(gt_dim, recurrent_steps=recurrent_steps), rollout_limit=3,
shuffle=False, batch_size=9999999, max_grad_norm=30, obs_key='gt_observes')
agent = Agent(arch, policy, value_function, None, env, logger,
policy_episodes=60, policy_steps=3000, gamma1=0.95, gamma2=0.995,
recurrent_steps=recurrent_steps, monitor=env.rl_stats)
fname = "optimize-RPT2"
policy.load_params(fname)
###Output
Quaternion_attitude
Quaternion_attitude
new
Focal Distance: 2.414213562373095
thruster model:
Inertia Tensor: [[333.33333333 0. 0. ]
[ 0. 333.33333333 0. ]
[ 0. 0. 333.33333333]]
Lander Model: 1000.0
6dof dynamics model
Reward Terminal MDR: True
Glideslope Constraint: delta = 3
Flat Constraint
Attitude Constraint
Rotational Velocity Constraint
Position Hysterises Constraint
###Markdown
Test Policy
###Code
env.test_policy_batch(agent,5000,print_every=100,keys=lander_model.get_engagement_keys())
###Output
worked 1
Dynamics: Max Disturbance (m/s^2): [3.73986826e-05 1.81519076e-05 1.31144881e-05] 4.359062980865332e-05
i (et): 100 ( 226)
Cumulative Stats (mean,std,max,argmax)
thrust | 1.01 | 0.78 | 0.00 | 3.46 | 59
alt_vc | 0.013 | 0.011 | 0.001 | 0.119 | 28
Final Stats (mean,std,min,max)
norm_vf | 0.009 | 0.004 | 0.001 | 0.026
norm_rf | 0.214 | 0.163 | 0.013 | 0.885
position | -42.6 31.4 590.7 | 435.6 419.3 325.0 | -1055.0 -975.4 5.6 | 1008.0 1232.3 1218.4
velocity | -0.001 0.000 0.000 | 0.006 0.006 0.005 | -0.021 -0.015 -0.014 | 0.014 0.016 0.015
fuel |0.4129 |0.0959 |0.2281 |0.6456
attitude_321 | 0.023 -0.074 -0.003 | 0.319 0.568 0.727 | -0.870 -1.404 -1.533 | 1.524 1.417 1.638
w | -0.0002 0.0003 0.0001 | 0.0049 0.0041 0.0041 | -0.0119 -0.0098 -0.0094 | 0.0102 0.0107 0.0093
alt_vc | -0.001 0.000 0.000 | 0.006 0.006 0.005 | -0.021 -0.015 -0.014 | 0.014 0.016 0.015
good_landing1 | 1.0000 | 0.0000 | 1.0000 | 1.0000
good_landing2 | 1.0000 | 0.0000 | 1.0000 | 1.0000
i (et): 200 ( 218)
Cumulative Stats (mean,std,max,argmax)
thrust | 1.00 | 0.78 | 0.00 | 3.46 | 59
alt_vc | 0.013 | 0.011 | 0.000 | 0.140 | 132
Final Stats (mean,std,min,max)
norm_vf | 0.009 | 0.004 | 0.001 | 0.026
norm_rf | 0.208 | 0.161 | 0.013 | 0.993
position | -33.0 10.0 589.4 | 437.7 430.4 323.3 | -1055.4 -1004.3 5.6 | 1081.0 1232.3 1328.1
velocity | -0.000 -0.000 0.000 | 0.006 0.006 0.005 | -0.021 -0.015 -0.018 | 0.016 0.016 0.015
fuel |0.4090 |0.0957 |0.1951 |0.6728
attitude_321 | 0.036 -0.059 -0.013 | 0.344 0.577 0.752 | -0.940 -1.449 -2.247 | 2.058 1.485 1.638
w | -0.0002 0.0006 0.0003 | 0.0048 0.0041 0.0042 | -0.0119 -0.0098 -0.0125 | 0.0102 0.0113 0.0108
alt_vc | -0.000 -0.000 0.000 | 0.006 0.006 0.005 | -0.021 -0.015 -0.018 | 0.016 0.016 0.015
good_landing1 | 1.0000 | 0.0000 | 1.0000 | 1.0000
good_landing2 | 1.0000 | 0.0000 | 1.0000 | 1.0000
i (et): 300 ( 219)
Cumulative Stats (mean,std,max,argmax)
thrust | 0.98 | 0.78 | 0.00 | 3.46 | 59
alt_vc | 0.013 | 0.011 | 0.000 | 0.140 | 132
Final Stats (mean,std,min,max)
norm_vf | 0.009 | 0.004 | 0.001 | 0.026
norm_rf | 0.213 | 0.174 | 0.013 | 0.993
position | -35.1 8.2 582.6 | 433.2 408.6 325.0 | -1055.4 -1004.3 2.1 | 1081.0 1232.3 1352.8
velocity | -0.000 -0.000 -0.000 | 0.006 0.005 0.005 | -0.021 -0.015 -0.018 | 0.020 0.016 0.015
fuel |0.4025 |0.0944 |0.1951 |0.6728
attitude_321 | 0.050 -0.057 -0.024 | 0.323 0.581 0.730 | -0.940 -1.449 -2.247 | 2.058 1.485 1.638
w | 0.0001 0.0004 0.0003 | 0.0046 0.0040 0.0042 | -0.0119 -0.0108 -0.0125 | 0.0119 0.0113 0.0108
alt_vc | -0.000 -0.000 -0.000 | 0.006 0.005 0.005 | -0.021 -0.015 -0.018 | 0.020 0.016 0.015
good_landing1 | 1.0000 | 0.0000 | 1.0000 | 1.0000
good_landing2 | 1.0000 | 0.0000 | 1.0000 | 1.0000
i (et): 400 ( 219)
Cumulative Stats (mean,std,max,argmax)
thrust | 0.99 | 0.78 | 0.00 | 3.46 | 59
alt_vc | 0.013 | 0.011 | 0.000 | 0.140 | 132
Final Stats (mean,std,min,max)
norm_vf | 0.009 | 0.004 | 0.000 | 0.026
norm_rf | 0.210 | 0.174 | 0.013 | 1.272
position | -16.7 25.2 569.4 | 444.3 411.2 325.7 | -1055.4 -1248.9 2.1 | 1143.5 1232.3 1352.8
velocity | -0.000 -0.000 -0.000 | 0.006 0.006 0.006 | -0.021 -0.015 -0.018 | 0.020 0.016 0.019
fuel |0.4037 |0.0930 |0.1951 |0.6854
attitude_321 | 0.042 -0.030 -0.049 | 0.318 0.598 0.737 | -0.940 -1.449 -2.247 | 2.058 1.485 1.638
w | 0.0002 0.0003 0.0004 | 0.0046 0.0040 0.0041 | -0.0119 -0.0108 -0.0125 | 0.0120 0.0118 0.0108
alt_vc | -0.000 -0.000 -0.000 | 0.006 0.006 0.006 | -0.021 -0.015 -0.018 | 0.020 0.016 0.019
good_landing1 | 1.0000 | 0.0000 | 1.0000 | 1.0000
good_landing2 | 1.0000 | 0.0000 | 1.0000 | 1.0000
i (et): 500 ( 218)
Cumulative Stats (mean,std,max,argmax)
thrust | 0.98 | 0.78 | 0.00 | 3.46 | 59
alt_vc | 0.013 | 0.011 | 0.000 | 0.140 | 132
Final Stats (mean,std,min,max)
norm_vf | 0.009 | 0.004 | 0.000 | 0.026
norm_rf | 0.211 | 0.176 | 0.013 | 1.272
position | -10.9 16.8 563.3 | 445.4 405.6 317.1 | -1085.2 -1248.9 2.1 | 1143.5 1232.3 1352.8
velocity | 0.000 -0.000 0.000 | 0.006 0.006 0.006 | -0.021 -0.015 -0.018 | 0.020 0.016 0.019
fuel |0.4028 |0.0922 |0.1951 |0.6854
attitude_321 | 0.031 -0.026 -0.029 | 0.312 0.596 0.729 | -0.940 -1.449 -2.247 | 2.058 1.485 1.638
w | 0.0001 0.0002 0.0003 | 0.0046 0.0039 0.0041 | -0.0119 -0.0108 -0.0125 | 0.0120 0.0118 0.0126
alt_vc | 0.000 -0.000 0.000 | 0.006 0.006 0.006 | -0.021 -0.015 -0.018 | 0.020 0.016 0.019
good_landing1 | 1.0000 | 0.0000 | 1.0000 | 1.0000
good_landing2 | 1.0000 | 0.0000 | 1.0000 | 1.0000
i (et): 600 ( 228)
Cumulative Stats (mean,std,max,argmax)
thrust | 0.99 | 0.78 | 0.00 | 3.46 | 59
alt_vc | 0.013 | 0.011 | 0.000 | 0.140 | 132
Final Stats (mean,std,min,max)
norm_vf | 0.009 | 0.004 | 0.000 | 0.026
norm_rf | 0.207 | 0.169 | 0.013 | 1.272
position | -3.8 15.7 559.5 | 455.1 422.0 318.2 | -1189.4 -1248.9 2.1 | 1187.5 1232.3 1352.8
velocity | 0.000 -0.000 0.000 | 0.006 0.006 0.006 | -0.021 -0.015 -0.018 | 0.020 0.016 0.019
fuel |0.4052 |0.0918 |0.1408 |0.6854
attitude_321 | 0.023 -0.015 -0.023 | 0.309 0.597 0.742 | -0.940 -1.509 -2.247 | 2.058 1.485 1.638
w | -0.0001 0.0003 0.0003 | 0.0046 0.0039 0.0041 | -0.0119 -0.0108 -0.0125 | 0.0120 0.0118 0.0126
alt_vc | 0.000 -0.000 0.000 | 0.006 0.006 0.006 | -0.021 -0.015 -0.018 | 0.020 0.016 0.019
good_landing1 | 1.0000 | 0.0000 | 1.0000 | 1.0000
good_landing2 | 1.0000 | 0.0000 | 1.0000 | 1.0000
i (et): 700 ( 222)
Cumulative Stats (mean,std,max,argmax)
thrust | 0.99 | 0.78 | 0.00 | 3.46 | 59
alt_vc | 0.013 | 0.011 | 0.000 | 0.140 | 132
Final Stats (mean,std,min,max)
norm_vf | 0.009 | 0.004 | 0.000 | 0.026
norm_rf | 0.211 | 0.186 | 0.013 | 2.177
position | -4.8 7.4 555.2 | 447.7 432.0 317.9 | -1189.4 -1248.9 0.2 | 1187.5 1232.3 1352.8
velocity | 0.000 -0.000 0.000 | 0.006 0.006 0.006 | -0.021 -0.015 -0.018 | 0.021 0.016 0.019
fuel |0.4039 |0.0923 |0.1408 |0.6854
attitude_321 | 0.020 -0.016 -0.021 | 0.303 0.588 0.756 | -0.940 -1.509 -2.247 | 2.058 1.485 1.638
w | -0.0001 0.0002 0.0002 | 0.0046 0.0039 0.0040 | -0.0119 -0.0108 -0.0125 | 0.0120 0.0118 0.0126
alt_vc | 0.000 -0.000 0.000 | 0.006 0.006 0.006 | -0.021 -0.015 -0.018 | 0.021 0.016 0.019
good_landing1 | 0.9986 | 0.0377 | 0.0000 | 1.0000
good_landing2 | 1.0000 | 0.0000 | 1.0000 | 1.0000
i (et): 800 ( 219)
Cumulative Stats (mean,std,max,argmax)
thrust | 0.99 | 0.77 | 0.00 | 3.46 | 59
alt_vc | 0.013 | 0.011 | 0.000 | 0.140 | 132
Final Stats (mean,std,min,max)
norm_vf | 0.009 | 0.004 | 0.000 | 0.026
norm_rf | 0.214 | 0.188 | 0.013 | 2.177
position | -9.1 2.0 546.0 | 451.9 434.2 315.5 | -1189.4 -1248.9 0.2 | 1259.1 1232.3 1352.8
velocity | 0.000 -0.000 -0.000 | 0.006 0.006 0.005 | -0.021 -0.015 -0.018 | 0.021 0.019 0.019
fuel |0.4039 |0.0927 |0.1408 |0.6854
attitude_321 | 0.022 -0.021 -0.014 | 0.313 0.595 0.763 | -1.265 -1.509 -2.247 | 2.058 1.532 1.638
w | 0.0000 0.0002 0.0003 | 0.0046 0.0040 0.0041 | -0.0119 -0.0108 -0.0125 | 0.0120 0.0118 0.0126
alt_vc | 0.000 -0.000 -0.000 | 0.006 0.006 0.005 | -0.021 -0.015 -0.018 | 0.021 0.019 0.019
good_landing1 | 0.9988 | 0.0353 | 0.0000 | 1.0000
good_landing2 | 1.0000 | 0.0000 | 1.0000 | 1.0000
|
Uni.01-05.ipynb
|
###Markdown
GenericDistributed under the BSD license, version:BSD-3-Clause.Copyright © 2022 Mike Vl. Vlasov .All rights reserved.Redistribution and use in source and binary forms, with or withoutmodification, are permitted provided that the following conditionsare met:1. Redistributions of source code must retain the above copyright notice, this list of conditions and the following disclaimer.2. Redistributions in binary form must reproduce the above copyright notice, this list of conditions and the following disclaimer in the documentation and/or other materials provided with the distribution.3. Neither the name of the copyright holder nor the names of its contributors may be used to endorse or promote products derived from this software without specific prior written permission.THIS SOFTWARE IS PROVIDED BY THE COPYRIGHT HOLDERS AND CONTRIBUTORS "AS IS"AND ANY EXPRESS OR IMPLIED WARRANTIES, INCLUDING, BUT NOT LIMITED TO, THEIMPLIED WARRANTIES OF MERCHANTABILITY AND FITNESS FOR A PARTICULAR PURPOSE AREDISCLAIMED. IN NO EVENT SHALL THE COPYRIGHT HOLDER OR CONTRIBUTORS BE LIABLEFOR ANY DIRECT, INDIRECT, INCIDENTAL, SPECIAL, EXEMPLARY, OR CONSEQUENTIALDAMAGES (INCLUDING, BUT NOT LIMITED TO, PROCUREMENT OF SUBSTITUTE GOODS ORSERVICES; LOSS OF USE, DATA, OR PROFITS; OR BUSINESS INTERRUPTION) HOWEVERCAUSED AND ON ANY THEORY OF LIABILITY, WHETHER IN CONTRACT, STRICT LIABILITY,OR TORT (INCLUDING NEGLIGENCE OR OTHERWISE) ARISING IN ANY WAY OUT OF THE USEOF THIS SOFTWARE, EVEN IF ADVISED OF THE POSSIBILITY OF SUCH DAMAGE.See:(https://opensource.org/licenses/BSD-3-Clause). Install Import & Ini Common
###Code
import sys, site
glScrWid_s = 70
glSep_s = '_' *glScrWid_s
print(glSep_s, 'sys:', sys.version, sys.version_info, sys.api_version, sys.prefix, sys.exec_prefix,
'\nsite:', site.getsitepackages(), site.getusersitepackages(), site.getuserbase(), glSep_s,
sep='\n')
print(sys.executable)
print(sys.platform)
###Output
______________________________________________________________________
sys:
3.9.7 (default, Sep 16 2021, 16:59:28) [MSC v.1916 64 bit (AMD64)]
sys.version_info(major=3, minor=9, micro=7, releaselevel='final', serial=0)
1013
C:\ProgramData\Anaconda3\envs\ts01
C:\ProgramData\Anaconda3\envs\ts01
site:
['C:\\ProgramData\\Anaconda3\\envs\\ts01', 'C:\\ProgramData\\Anaconda3\\envs\\ts01\\lib\\site-packages']
C:\Users\mikev\AppData\Roaming\Python\Python39\site-packages
C:\Users\mikev\AppData\Roaming\Python
______________________________________________________________________
C:\ProgramData\Anaconda3\envs\ts01\python.exe
win32
###Markdown
My
###Code
from MVVlStd import glSep_s, inp_FltAVali_fefi
import MVVlStd
# MVVlStd.inp_FltAVali_fefi('?')
# inp_FltAVali_fefi('?')
# # Mod:MVVlStd.py
# def inp_FltAVali_fefi(laWhatInPMsg_s, laInPValues_co=1, laValiInPMsg_s='',
# laVali_cll=None, laInPTypeFlt_cll=int, laMaxInPTry_co=11,
# laAcceptEmptyInPAsDf_b=False, laDfV_s=None, laVerbose_i=None) -> tuple:
# if laInPValues_co < 1: raise ValueError(f'laInPValues_co must be > 0, now:{laInPValues_co}')
# loTypeAValiFlsCo_l, loRes_l, loMaxTry_co = [0, 0], [], int(max(laInPValues_co, laMaxInPTry_co))
# if laValiInPMsg_s and laVali_cll:
# lo_s = f' - your value will be validated as({laValiInPMsg_s}) -'
# else: lo_s = ''
# lo_s = f"Please, Input{laWhatInPMsg_s}{lo_s} and press Enter"
# if laAcceptEmptyInPAsDf_b and laDfV_s is not None:
# lo_s += f"(on default '{laDfV_s}')"
# loInPMsg_s = f"{lo_s}: "
# for l_co in range(loMaxTry_co):
# li_s = input(loInPMsg_s)
# if li_s == '' and laAcceptEmptyInPAsDf_b and laDfV_s is not None:
# li_s = laDfV_s # 2Do: Che: laDfV_s is str
# # if not li_s: ??User(Exit|Bre) ??laAcceptEmpty(As(Df|Bre))InP_b=False
# try:
# if laInPTypeFlt_cll is not None:
# # print(f'DBG:1 laInPTypeFlt_cll')
# liChe_i = laInPTypeFlt_cll(li_s)
# # print(f'DBG:2 laInPTypeFlt_cll')
# else: liChe_i = li_s
# # print(f'DBG:3 laInPTypeFlt_cll')
# except ValueError as leExc_o:
# loTypeAValiFlsCo_l[0] +=1; liChe_i = None
# print(f"\tERR: You input:'{li_s}' NOT pass check type w/func({laInPTypeFlt_cll}",
# f'- Exception:{type(leExc_o).__name__}({leExc_o}) raised.')
# else:
# if laVali_cll is not None:
# if laVali_cll(liChe_i):
# loRes_l.append(liChe_i)
# print(f"\tMSG: You input:'{liChe_i}' valid.")
# else:
# loTypeAValiFlsCo_l[1] +=1
# lo_s = f' because of NOT {laValiInPMsg_s}' if laValiInPMsg_s else ''
# print(f"\tERR: You input:'{liChe_i}' INVALID{lo_s}.")
# else:
# loRes_l.append(liChe_i)
# print(f"\tMSG: You input:'{liChe_i}'.")
# # if liChe_i == tPtt_i: tOk_co +=1
# # print(f'DBG: {loRes_l=}')
# if len(loRes_l) == laInPValues_co: break
# if laMaxInPTry_co:
# if l_co == int(loMaxTry_co -1):
# if loRes_l:
# print(f"\tWRN: Rich max(laInPValues_co, laMaxInPTry_co):{loMaxTry_co}, return {tuple(loRes_l)} as User input.")
# else:
# raise ValueError(f'Rich max(laInPValues_co, laMaxInPTry_co):{loMaxTry_co} but loRes_l is Empty - nothing return as User input.')
# else:
# lo_s = '' if l_co == (loMaxTry_co -2) else 's'
# lo_s = f' and {loMaxTry_co - l_co -1} attempt{lo_s} left'
# else: lo_s = ''
# print(f'MSG: It remains to input {laInPValues_co - len(loRes_l)} more value{lo_s}.')
# return tuple(loRes_l)
# # print(inp_FltAVali_fefi(laInPValues_co=2, laInPType_cll=float, laMaxInPTry_co=1),
# # inp_FltAVali_fefi(laValiWhatInPMsg_s=tCndInPMsg_s,
# # laVali_cll=lambda x: x in tValiV_t)
# # )
# # t_s = '' if tOk_co == 1 else 's'
# # print(f"You input '{tPtt_i}' {tOk_co} time{t_s} of {tTime_co} attempts.")
# # tTime_co, tPtt_i, tOk_co, tFls_co = 10, 5, 0, 0
# # tResLLen_co = int(inp_FltAVali_fefi(laVali_cll=lambda _i: 0 < _i < 10,
# # laWhatInPMsg_s=' Количество элементов будущего списка',
# # laValiInPMsg_s=' a Integer 0 < _i < 10')[0],
# # )
# # print(f'В списке будет {tResLLen_co} элементов.')
# # tValiV_t = tuple(range(0, 10))
# # tCndInPMsg_s = f' (по очереди по одной вводите любые цифры) a Integer OneOf{tValiV_t}'
# # tRes_l = list(inp_FltAVali_fefi(f' по очереди по одной любые цифры {tResLLen_co} раза',
# # laInPValues_co=tResLLen_co, laValiInPMsg_s=f'a Integer OneOf{tValiV_t}',
# # laVali_cll=lambda x: x in tValiV_t))
# # tValiV_t = ('Y', 'N')
# # tRes_l = list(inp_FltAVali_fefi(f' Ts 2 times',
# # laInPValues_co=2, laInPTypeFlt_cll=None, laDfV_s='Y',
# # laAcceptEmptyInPAsDf_b=True, laValiInPMsg_s=f'a character OneOf{tValiV_t}',
# # laVali_cll=lambda _s: _s.upper() in tValiV_t))
# # tRes_l.sort()
# # print(tRes_l)
###Output
_____no_output_____
###Markdown
Lesson: 01
###Code
import requests
import json
from flask import Flask
def get_valutes_list():
url = 'https://www.cbr-xml-daily.ru/daily_json.js'
response = requests.get(url)
data = json.loads(response.text)
valutes = list(data['Valute'].values())
return valutes
# test
app = Flask(__name__)
def create_html(valutes):
text = '<h1>Курс валют</h1>'
text += '<table>'
text += '<tr>'
for _ in valutes[0]:
text += f'<th><th>'
text += '</tr>'
for valute in valutes:
text += '<tr>'
for v in valute.values():
text += f'<td>{v}</td>'
text += '</tr>'
text += '</table>'
return text
@app.route("/")
def index():
valutes = get_valutes_list()
html = create_html(valutes)
return html
# if __name__ == "__main__":
# app.run()
url = 'https://www.cbr-xml-daily.ru/daily_json.js'
response = requests.get(url)
data = json.loads(response.text)
valutes = list(data['Valute'].values())
data['Valute']['AUD']
###Output
_____no_output_____
###Markdown
02
###Code
# import sys
# glSep_s = '_' *80
tTskMsg_s = '''
Задача 1
Вывести на экран циклом пять строк из нулей, причем каждая строка
должна быть пронумерована.
'''
print(glSep_s, tTskMsg_s, 'Res:', glSep_s, sep='\n')
tTime_co, tCnt_s, tLen_i = 5, '0', 40
for l_co in range(1, tTime_co +1):
print(l_co, tCnt_s *(tLen_i +l_co))
print(glSep_s)
tTskMsg_s = '''
Задача 2
Пользователь в цикле вводит 10 цифр. Найти количество введеных
пользователем цифр 5.
'''
print(glSep_s, tTskMsg_s, 'Res:', glSep_s, sep='\n')
tTime_co, tPtt_i, tOk_co, tFls_co = 10, 5, 0, 0
tValiV_t = tuple(range(0, 10))
tCndInPMsg_s = f' Integer OneOf{tValiV_t}'
tInPMsg_s = f"Please, Input a{tCndInPMsg_s}\n and press Enter: "
for l_co in range(tTime_co):
try:
li_s = input(tInPMsg_s)
liN_i = int(li_s)
except ValueError as leExc_o:
tFls_co +=1
print(f"\tERR: You input:'{li_s}' NOT integer",
f'- Exception:{type(leExc_o).__name__}({leExc_o}) raised.')
else:
if liN_i not in tValiV_t:
tFls_co +=1
print(f"\tERR: You input integer:'{liN_i}'",
f'but NOT OneOf{tValiV_t}.')
else:
print(f"\tMSG: You input integer:'{liN_i}'.")
if liN_i == tPtt_i: tOk_co +=1
if l_co != (tTime_co -1):
t_s = '' if l_co == (tTime_co -2) else 's'
print(f'MSG: There are {tTime_co -l_co -1} attempt{t_s} left.')
t_s = '' if tOk_co == 1 else 's'
print(f"You input '{tPtt_i}' {tOk_co} time{t_s} of {tTime_co} attempts.")
print(glSep_s)
tTskMsg_s = '''
Задача 3
Найти сумму ряда чисел от 1 до 100. Полученный результат вывести на экран.
'''
print(glSep_s, tTskMsg_s, 'Res:', glSep_s, sep='\n')
tRa_t = (1, 101)
print(f"Summa:{sum(range(*tRa_t))}")
print(glSep_s)
tTskMsg_s = '''
Задача 4
Найти произведение ряда чисел от 1 до 10. Полученный результат вывести на экран.
'''
print(glSep_s, tTskMsg_s, 'Res:', glSep_s, sep='\n')
tCnt_it, tIni_i = range(1, 11), 1
# Variant:1
tProd_i = tIni_i # Che:!None |> tProd_i = 1|next(tCnt_it)
if tProd_i:
for l_el in tCnt_it:
if l_el == 0:
tProd_i = 0
break
else:
tProd_i *=l_el
else: tProd_i = 0
import itertools as itts
import operator as op
# Variant:2
tAccProd_t = tuple(itts.accumulate(tCnt_it, op.mul, initial=tIni_i))
# Variant:3
tProd3_i = tuple(itts.accumulate(tCnt_it, op.mul, initial=tIni_i))[-1]
print(f"Production: {tProd_i} or {tAccProd_t[-1]} or {tProd3_i}")
print(glSep_s)
tTskMsg_s = '''
Задача 5
Вывести цифры числа на каждой строчке.
'''
print(glSep_s, tTskMsg_s, 'Res:', glSep_s, sep='\n')
tChe_i = 56410829
# Variant:1
print(f'{tChe_i} Left2Right:', *str(tChe_i), sep='\n')
# Variant:2
print(f'{tChe_i} Right2Left:', *(str(tChe_i)[::-1]), sep='\n')
# print(f"Production: {tProd_i} or {tAccProd_t[-1]} or {tProd3_i}")
print(glSep_s)
tTskMsg_s = '''
Задача 6
Найти сумму цифр числа.
'''
print(glSep_s, tTskMsg_s, 'Res:', glSep_s, sep='\n')
tChe_i = 56410829
# Variant:1
tDiv_i, tSum_i = divmod(tChe_i, 10)
while tDiv_i:
tDiv_i, tRemain_i = divmod(tDiv_i, 10)
tSum_i += tRemain_i
# Variant:2
print(f'Summa: {tSum_i} or {sum(int(_s1) for _s1 in str(tChe_i))}')
print(glSep_s)
tTskMsg_s = '''
Задача 7
Найти произведение цифр числа.
'''
print(glSep_s, tTskMsg_s, 'Res:', glSep_s, sep='\n')
tChe_i = 56401829
# Variant:1
tDiv_i, tProd_i = divmod(tChe_i, 10)
while tDiv_i and tProd_i:
tDiv_i, tRemain_i = divmod(tDiv_i, 10)
tProd_i *= tRemain_i
import itertools as itts
import operator as op
# Variant:2
print(f'Production: {tProd_i}',
f'or {tuple(itts.accumulate((int(_s1) for _s1 in str(tChe_i)), op.mul, initial=1))[-1]}')
print(glSep_s)
tTskMsg_s = '''
Задача 8
Дать ответ на вопрос: есть ли среди цифр числа 5?
'''
print(glSep_s, tTskMsg_s, 'Res:', glSep_s, sep='\n')
tChe_i, tPtt_s = 12532799865, '5'
tPos_i = str(tChe_i).find(tPtt_s)
if tPos_i == -1:
print(f"No '{tPtt_s}' in Number:{tChe_i}")
else:
print(f"Digit:'{tPtt_s}' exist in Number:{tChe_i} in most left position:{tPos_i}")
print(glSep_s)
tTskMsg_s = '''
Задача 9
Найти максимальную цифру в числе
'''
print(glSep_s, tTskMsg_s, 'Res:', glSep_s, sep='\n')
tChe_i = 56401829
print(f'Max digit in Number:{tChe_i} is {max(int(_s1) for _s1 in str(tChe_i))}')
print(glSep_s)
tTskMsg_s = '''
Задача 10
Найти количество цифр 5 в числе
'''
print(glSep_s, tTskMsg_s, 'Res:', glSep_s, sep='\n')
tChe_i, tPtt_s = 12352579986, '5'
tCo_i = str(tChe_i).count(tPtt_s)
if tCo_i == 0:
print(f"No '{tPtt_s}' in Number:{tChe_i}")
else:
t_s = '' if tCo_i == 1 else 's'
print(f"Digit:'{tPtt_s}' exist in Number:{tChe_i} {tCo_i} time{t_s}")
print(glSep_s)
###Output
______________________________________________________________________
Задача 10
Найти количество цифр 5 в числе
Res:
______________________________________________________________________
Digit:'5' exist in Number:12352579986 2 times
______________________________________________________________________
###Markdown
03
###Code
tTskMsg_s = '''
Задача 1
Из всех методов списка (list) выбрать 5 тех, которые
по вашему мнению используются чаще всего
'''
print(glSep_s, tTskMsg_s, f'Res({sys.version=}):', glSep_s, sep='\n')
def tIni_f(la1stCll_b=False):
global self_l, t1_l, t2_l, t3_it, t4_ix, t5_sl
self_l, t1_l, t2_l, t3_it, t4_ix, t5_sl = [0, 0], [1], [2, 22], iter([3,33,'333']), 0, slice(0, None)
if not la1stCll_b: print()
print("Ini:\nself_l, t1_l, t2_l, t3_it, t4_ix, t5_sl = [0, 0], [1], [2], iter([3,33,'333']), 0, slice(0, None)")
print(self_l, t1_l, t2_l, t3_it, t4_ix, t5_sl, sep=', ')
print()
tIni_f(True)
print('''0. Cre list
class list(MutableSequence[_T], Generic[_T]) -> list[_T]: ...
def __init__(self) -> None: ...
def __init__(self, __iterable: Iterable[_T]) -> None: ...
def copy(self) -> list[_T]: ... !!! a shallow copy of self(same as s[:])
4Ex:\tExpes: list(self_l), [[1.1], [2.2]], t1_l.copy(), [], [_el for _el in t3_it]
\tValues: ''', end='')
print(list(self_l), [[1.1], [2.2]], t1_l.copy(), [], [_el for _el in t3_it], sep=', ')
print(glSep_s[:35])
tIni_f()
print('''1. Get|Set El
def __setitem__(self, __i: SupportsIndex, __o: _T) -> None: ...
def __setitem__(self, __s: slice, __o: Iterable[_T]) -> None: ...
def __getitem__(self, __i: SupportsIndex) -> _T: ...
def __getitem__(self, __s: slice) -> list[_T]: ...
4Ex:\tExe: self_l[t4_ix] = '00'; self_l[t5_sl] = ['000', '0000']
\tExpes: self_l, self_l[-1:], self_l[-1], self_l[t4_ix], self_l[t5_sl]
\tValues: ''', end='')
self_l[t4_ix] = '00'; self_l[t5_sl] = ['000', '0000']
print(self_l, self_l[-1:], self_l[-1], self_l[t4_ix], self_l[t5_sl], sep=', ')
print(glSep_s[:35])
tIni_f()
print('''2. extend(in place)|concatenation (list and list|Iterable)
def extend(self, __iterable: Iterable[_T]) -> None: ...
def __add__(self, __x: list[_T]) -> list[_T]: ...
def __iadd__(self: Self, __x: Iterable[_T]) -> Self: ...
4Ex:\tExe: self_l += t1_l; t2_l.extend(t1_l)]
\tExpes: self_l, self_l + t1_l, t1_l, t2_l, t1_l + t2_l
\tValues: ''', end='')
self_l += t1_l; t2_l.extend(t1_l)
print(self_l, self_l + t1_l, t1_l, t2_l, t1_l + t2_l, sep=', ')
print(glSep_s[:35])
tIni_f()
print('''3. append El to the list on the right
def append(self, __object: _T) -> None: ...
4Ex:\tExe: self_l.append('0'); self_l.append([]); self_l.append(t1_l); self_l.append([t1_l])
\tExpes: self_l
\tValues: ''', end='')
self_l.append('0'); self_l.append([]); self_l.append(t1_l); self_l.append([t1_l])
print(self_l, sep=', ')
print(glSep_s[:35])
tIni_f()
print('''4. contains El (OR not) in the list
def __contains__(self, __o: object) -> bool: ...
4Ex:\tExpes: 0 in self_l, 0 in t1_l, 0 not in t1_l, 1 in t1_l, 1 not in t1_l
\tValues: ''', end='')
print(0 in self_l, 0 in t1_l, 0 not in t1_l, 1 in t1_l, 1 not in t1_l, sep=', ')
print(glSep_s)
# (list(self_l), [[1.1], [2.2]], t1_l.copy(), [], [_el for _el in t3_it]),
# (self_l[-1:], self_l[-1], self_l[t4_ix], self_l[t5_sl])
# (self_l += t1_l; t2_l.extend(t1_l); self_l, self_l + t1_l, t1_l, t2_l, t1_l + t2_l)
# (self_l.append('0'); self_l.append([]); self_l.append(t1_l); self_l.append([t1_l]); self_l)
# (0 in self_l, 0 in t1_l, 0 not in t1_l, 1 in t1_l, 1 not in t1_l)
tTskMsg_s = '''
Задача 2
Написать их через запятую с параметрами
'''
print(glSep_s, tTskMsg_s, f'Res({sys.version=}):', glSep_s, sep='\n')
print('''
0:(class list(MutableSequence[_T], Generic[_T]) -> list[_T]: ...
def __init__(self) -> None: ...
def __init__(self, __iterable: Iterable[_T]) -> None: ...
def copy(self) -> list[_T]: ... !!! a shallow copy of self(same as s[:])),
1:(def __setitem__(self, __i: SupportsIndex, __o: _T) -> None: ...
def __setitem__(self, __s: slice, __o: Iterable[_T]) -> None: ...
def __getitem__(self, __i: SupportsIndex) -> _T: ...
def __getitem__(self, __s: slice) -> list[_T]: ...),
2:(def extend(self, __iterable: Iterable[_T]) -> None: ...
def __add__(self, __x: list[_T]) -> list[_T]: ...
def __iadd__(self: Self, __x: Iterable[_T]) -> Self: ...),
3:(def append(self, __object: _T) -> None: ...),
4:(def __contains__(self, __o: object) -> bool: ...)''')
print(glSep_s)
tTskMsg_s = '''
Задача 3
Повторить процедуру (Tsk:2) для словарей (dict), множеств (set), строк (str)
'''
print(glSep_s, tTskMsg_s, f'{sys.version=}', glSep_s, 'Result:', '', sep='\n')
print('dict:', '''\t0:\n(dict(MutableMapping[_KT, _VT], Generic[_KT, _VT])
def __init__(self: dict[_KT, _VT]) -> None: ...
def __init__(self: dict[str, _VT], **kwargs: _VT) -> None: ...
def __init__(self, __map: SupportsKeysAndGetItem[_KT, _VT], **kwargs: _VT) -> None: ...
def __init__(self, __iterable: Iterable[tuple[_KT, _VT]], **kwargs: _VT) -> None: ...
def __init__(self: dict[str, str], __iterable: Iterable[list[str]]) -> None: ...
def copy(self) -> dict[_KT, _VT]: ...
def fromkeys(cls, __iterable: Iterable[_T], __value: None = ...) -> dict[_T, Any | None]: ...
def fromkeys(cls, __iterable: Iterable[_T], __value: _S) -> dict[_T, _S]: ...),
\t1:\n(def __setitem__(self, __k: _KT, v: _VT) -> None: ...
def setdefault(self, __key: _KT, __default: _VT = ...) -> _VT: ...
def __getitem__(self, __k: _KT) -> _VT: ...
def get(self, __key: _KT, __default: _VT_co | _T) -> _VT_co | _T: ...
def pop(self, __key: _KT, __default: _VT | _T = ...) -> _VT | _T: ...
def popitem(self) -> tuple[_KT, _VT]: ...),
\t2:\n(def __contains__(self, __o: object) -> bool: ...),
\t3:\n(def keys(self) -> dict_keys[_KT, _VT]: ...
def values(self) -> dict_values[_KT, _VT]: ...
def items(self) -> dict_items[_KT, _VT]: ...),
\t4:\n(def update(self, __m: SupportsKeysAndGetItem[_KT, _VT], **kwargs: _VT) -> None: ...
def update(self, __m: Iterable[tuple[_KT, _VT]], **kwargs: _VT) -> None: ...
def update(self, **kwargs: _VT) -> None: ...)''',
glSep_s[:35], '',
'set:', '''\t0:\n(class set(MutableSet[_T], Generic[_T]): -> set[_T]: ...
def __init__(self, __iterable: Iterable[_T] = ...) -> None: ...
def copy(self) -> set[_T]: ...),
\t1:\n(def __contains__(self, __o: object) -> bool: ...)
\t2:\n(def difference(self, *s: Iterable[Any]) -> set[_T]: ...
def discard(self, __element: _T) -> None: ...
def intersection(self, *s: Iterable[Any]) -> set[_T]: ...
def isdisjoint(self, __s: Iterable[Any]) -> bool: ...
def issubset(self, __s: Iterable[Any]) -> bool: ...
def issuperset(self, __s: Iterable[Any]) -> bool: ...
def remove(self, __element: _T) -> None: ...
def symmetric_difference(self, __s: Iterable[_T]) -> set[_T]: ...
def union(self, *s: Iterable[_S]) -> set[_T | _S]: ...),
\t3:\n(def update(self, *s: Iterable[_T]) -> None: ...
def difference_update(self, *s: Iterable[Any]) -> None: ...
def intersection_update(self, *s: Iterable[Any]) -> None: ...
def symmetric_difference_update(self, __s: Iterable[_T]) -> None: ...),
\t4:\n(def add(self, __element: _T) -> None: ...
def discard(self, __element: _T) -> None: ...
def pop(self) -> _T: ...
def remove(self, value: _T) -> None: ...)''',
glSep_s[:35], '',
'str', '''\t0:\n(class str(Sequence[str]) -> str: ...),
\t1:\n(def __add__(self, __s: str) -> str: ...),
\t2:\n(def format(self, *args: object, **kwargs: object) -> str: ...),
\t3:\n(def join(self, __iterable: Iterable[str]) -> str: ...
def r?split(self, sep: str | None = ..., maxsplit: SupportsIndex = ...) -> list[str]: ...
def splitlines(self, keepends: bool = ...) -> list[str]: ...),
\t4:\n(def __contains__(self, __o: object) -> bool: ...
||def __iter__(self) -> Iterator[str]: ...
||def __getitem__(self, __i: SupportsIndex | slice) -> str: ...
||def (end|start)swith(self, __prefix: str | tuple[str, ...], __start: SupportsIndex | None = ..., __end: SupportsIndex | None = ...) -> bool: ...
||def r?(find|index)(self, __sub: str, __start: SupportsIndex | None = ..., __end: SupportsIndex | None = ...) -> int: ...
||def islower(self) -> bool: ...||def lower(self) -> str: ...
||def replace(self, __old: str, __new: str, __count: SupportsIndex = ...) -> str: ...
||def (l|r)?strip(self, __chars: str | None = ...) -> str: ...)
# не могу выбрать:)''', glSep_s, sep='\n')
tTskMsg_s = '''
Задача 4
(МОДУЛЬ 1) Создать новый проект, в нем создать модуль 1seq.py. Задание:
Пользователь вводит количество элементов будущего списка
После этого по очереди по одной вводит любые цифры
Сохранить цифры в список
Отсортировать список по возрастанию и вывести на экран
Пример работы: Введите количество элементов: 3
Введите 1 элемент: 5
Введите 2 элемент: 2
Введите 3 элемент: 4
Вывод: [2, 4, 5]
'''
print(glSep_s, tTskMsg_s, f'{sys.version=}', glSep_s, 'Result:', '', sep='\n')
tResLLen_co = int(inp_FltAVali_fefi(laVali_cll=lambda _i: 0 < _i < 10,
laWhatInPMsg_s=' Количество элементов будущего списка',
laValiInPMsg_s=' a Integer 0 < _i < 10')[0])
print(f'MSG: В списке будет {tResLLen_co} элементов.')
tValiV_t = tuple(range(0, 10))
tCndInPMsg_s = f' (по очереди по одной вводите любые цифры) a Integer OneOf{tValiV_t}'
tRes_l = list(inp_FltAVali_fefi(f' по очереди по одной любые цифры', # {tResLLen_co} раза
laInPValues_co=tResLLen_co, laValiInPMsg_s=f'a Integer OneOf{tValiV_t}',
laVali_cll=lambda x: x in tValiV_t))
tRes_l.sort()
print(tRes_l)
print(glSep_s)
tTskMsg_s = '''
Задача 5
(МОДУЛЬ 2) Создать модуль 2seq.py. Задание:
Пользователь вводит любые цифры через запятую
Сохранить цифры в список
Получить новый список в котором будут только уникальные элементы исходного
(уникальным считается символ, который встречается в исходном списке только 1 раз)
Вывести новый список на экран
Порядок цифр в новом списке не важен
Пример работы: Введите элементы списка через запятую: 2,3,4,5,5,6,5,3,9
Результат: 2, 4, 6, 9
'''
print(glSep_s, tTskMsg_s, f'{sys.version=}', glSep_s, 'Result:', '', sep='\n')
def tGetIsValiSep_fef(laTs_s, laSep_t=(',', ';', '/')):
# if not isinstance(laTs_s, str):
# raise TypeError(f"laTs_s should be string, but NOW {type(laTs_s).__name__}.")
loCoSepIt2S_t = tuple(laTs_s.count(_el) for _el in laSep_t)
loAllSepCoSum_i = sum(loCoSepIt2S_t)
if loAllSepCoSum_i == 0: return None
else:
for l_s, l_co in zip(laSep_t, loCoSepIt2S_t):
if l_co == 0: continue
elif l_co != loAllSepCoSum_i:
raise ValueError(f"In str:'{laTs_s}' there should be used only one of {laSep_t} separator.")
else: return l_s
# for _s in ('9,2,3,4,5,5,6,5,3,9', '1', '', '1,2,3', '1;2;3', '1/2/3', (1,2,3), '1,2;3/4'):
# for _s in ('9,2,3,4,5,5,6,5,3,9', '1', '', '1,2,3', '1;2;3', '1/2/3', '1,0,2;3/4', (1,2,3)):
# print(_s, tGetIsValiSep_fef(_s), sep='\t')
def tCreTIfAllElIsDig_fef(laTs_s, laSep_s1):
if not laTs_s.strip(): return ()
lo_t = tuple(int(_el) for _el in laTs_s.split(laSep_s1))
if all(0<= _el <=9 for _el in lo_t): return lo_t
else: raise ValueError(f"In str:'{laTs_s}' at least one of El NOT (0<= _el <=9).")
# for _s in ('9,2,3,4,5,5,6,5,3,9', '1', '', '1,2,3', '1;2;3', '1/2/3', (1,2,3), '1,2;3/4'):
# for _s in ('9,2,3,4,5,5,6,5,3,9', '1', '', '1,2,3', '1;2;3', '1/2/3', '1,0,2;3/4', (1,2,3)):
# for _s in ('9,2,3,4,5,5,6,5,3,9', '1', '', '1,2,3'):
# print(_s, tCreTIfAllElIsDig_fef(_s, tGetIsValiSep_fef(_s)), sep='\t')
# tResLLen_co = int(inp_FltAVali_fefi(laVali_cll=lambda _i: 0 < _i < 10,
# laWhatInPMsg_s=' Количество элементов будущего списка',
# laValiInPMsg_s=' a Integer 0 < _i < 10')[0])
# print(f'MSG: В списке будет {tResLLen_co} элементов.')
tValiV_t = tuple(range(0, 10))
# Please, Input любые цифры через запятую and press Enter: 9,2,3,4,5,5,6,5,3,9
# MSG: You input:'(9, 2, 3, 4, 5, 5, 6, 5, 3, 9)'.
# Raw: [9, 2, 3, 4, 5, 5, 6, 5, 3, 9]
# Uniq: [2, 3, 4, 5, 6, 9]
# Uniq&Stabl: [9, 2, 3, 4, 5, 6]
tRes_l = list(inp_FltAVali_fefi(f' любые цифры через запятую',
laInPTypeFlt_cll=lambda _s: tCreTIfAllElIsDig_fef(_s, tGetIsValiSep_fef(_s)),
# laInPType_cll=futs.partial(tCreTIfAllElIsDig_fef, laSep_s1=tGetIsValiSep_fef(_s)),
# laValiInPMsg_s=f"a Integer OneOf{tValiV_t} and should be used only one of (',', ';', '/') separator",
# laVali_cll=lambda x: x in tValiV_t
)[0])
# 9,2,3,4,5,5,6,5,3,9
print('Raw:\t\t', tRes_l)
print('Uniq:\t\t', list(set(tRes_l)))
print('Uniq&Stabl:\t', list(dict.fromkeys(tRes_l)))
print(glSep_s)
tTskMsg_s = '''
Задача 6
(МОДУЛЬ 3) В проекте создать новый модуль 3seq.py. Задание:
Пользователь вводит элементы 1-го списка (по очереди как в МОДУЛЬ 1 или вместе как в МОДУЛЬ 2)
Затем он вводит элементы 2-го списка
Удалить из первого списка элементы присутствующие во 2-ом и вывести результат на экран
Пример работы: Введите элементы 1-го списка: 1,2,3,4,5
Введите элементы 2-го списка: 2,5
Результат: 1,3,4
'''
print(glSep_s, tTskMsg_s, f'{sys.version=}', glSep_s, 'Result:', '', sep='\n')
# Please, Input 1-ый список - любые цифры через запятую and press Enter: 9,2,3,4,5,5,6,5,3,9
# MSG: You input:'(9, 2, 3, 4, 5, 5, 6, 5, 3, 9)'.
# Please, Input 1-ый список - любые цифры через запятую and press Enter: 0,3,0,2
# MSG: You input:'(0, 3, 0, 2)'.
# (Fr:[9, 2, 3, 4, 5, 5, 6, 5, 3, 9] remove El exist in2:[0, 3, 0, 2])
# (Unstable & UniqOnlyEl)=> [9, 4, 5, 6]
# (Stable)=> [9, 4, 5, 5, 6, 5, 9]
tSrc_l = list(inp_FltAVali_fefi(f' 1-ый список - любые цифры через запятую',
laInPTypeFlt_cll=lambda _s: tCreTIfAllElIsDig_fef(_s, tGetIsValiSep_fef(_s)),
)[0])
tSrc_set = set(tSrc_l)
tRm_l = list(inp_FltAVali_fefi(f' 1-ый список - любые цифры через запятую',
laInPTypeFlt_cll=lambda _s: tCreTIfAllElIsDig_fef(_s, tGetIsValiSep_fef(_s)),
)[0])
tRm_set = set(tRm_l)
print(f'(Fr:{tSrc_l} remove El exist in2:{tRm_l})\n(Unstable & UniqOnlyEl)=> {list(tSrc_set - tRm_set)}')
print('(Stable)=> ', list(_el for _el in tSrc_l if _el not in tRm_set))
print(glSep_s)
tTskMsg_s = '''
Задача 7
(МОДУЛЬ 4) В проекте создать новый модуль victory.py. Задание
Написать или улучшить программу Викторина из предыдущего дз (Для тренировки предлагаю
не пользоваться никакими библиотеками кроме random)
Есть 10 известных людей и их даты рождения в формате '02.01.1988' ('dd.mm.yyyy') - предлагаю
для тренировки пока использовать строку
Программа выбирает из этих 10-и 5 случайных людей, это можно реализовать с помощью
модуля random и функции sample
Пример использования sample:
import random
numbers = [1, 2, 3, 4, 5]
# 2 - количество случайных элементов
result = random.sample(numbers, 2)
print(result) # [5, 1]
После того как выбраны 5 случайных людей, предлагаем пользователю ввести их дату рождения
пользователь вводит дату в формате 'dd.mm.yyyy'
Например 03.01.2009, если пользователь ответил неверно, то выводим правильный ответ,
но уже в следующем виде: третье января 2009 года, склонением можно пренебречь
В конце считаем количество правильных и неправильных ответов и предлагаем начать снова
'''
print(glSep_s, tTskMsg_s, f'{sys.version=}', glSep_s, 'Result:', '', sep='\n')
print('Не нашел "...программу Викторина из предыдущего дз..."')
print(glSep_s)
tTskMsg_s = '''
Задача 8
Выложите проект на github (если возникнут трудности можно пока использовать гугл или яндекс диск)
'''
print(glSep_s, tTskMsg_s, f'{sys.version=}', glSep_s, 'Result:', '', sep='\n')
print('https://github.com/dev-mikevvl-ms/PyDev.03.HW.git')
print(glSep_s)
###Output
________________________________________________________________________________
Задача 8
Выложите проект на github (если возникнут трудности можно пока использовать гугл или яндекс диск)
sys.version='3.9.7 (default, Sep 16 2021, 16:59:28) [MSC v.1916 64 bit (AMD64)]'
________________________________________________________________________________
Result:
https://github.com/dev-mikevvl-ms/PyDev.03.HW.git
________________________________________________________________________________
###Markdown
04
###Code
# import sys
# from MVVlStd import glSep_s
tTskMsg_s = '''
Задача 3. МОДУЛЬ 1. файл myfunctions.py
Вверху файла даны инструкции по выполнению дз.
Даны заготовки 8-и функции, предлагается написать для них реализацию
'''
print(glSep_s, tTskMsg_s, f'{sys.version=}', glSep_s, 'Result:', '', sep='\n')
"""
МОДУЛЬ 1
В модуле прописаны заготовки для 8 функций
Под каждой функцией есть описание как она должна работать
ниже есть примеры использования функции
Задание: реализовать код функции, чтобы он работал по описанию и примеры использования давали верный результат
"""
def simple_separator():
"""
Функция создает красивый резделитель из 10-и звездочек (**********)
:return: **********
"""
return '**' *5
print("\nsimple_separator() == '**********'") # True
print("simple_separator() == '**********'") # True
print(simple_separator() == '**********') # True
print(glSep_s[:len(glSep_s)//2], '', sep='\n')
def long_separator(count):
"""
Функция создает разделитель из звездочек число которых можно регулировать параметром count
:param count: количество звездочек
:return: строка разделитель, примеры использования ниже
"""
return '*' *count
print("long_separator(3) == '***'") # True
print(long_separator(3) == '***') # True
print("long_separator(4) == '****'") # True
print(long_separator(4) == '****') # True
print(glSep_s[:len(glSep_s)//2], '', sep='\n')
def separator(simbol, count):
"""
Функция создает разделитель из любых символов любого количества
:param simbol: символ разделителя
:param count: количество повторений
:return: строка разделитель примеры использования ниже
"""
return simbol *count
print("separator('-', 10) == '----------'") # True
print(separator('-', 10) == '----------') # True
print("separator('#', 5) == '#####'") # True
print(separator('#', 5) == '#####') # True
print(glSep_s[:len(glSep_s)//2], '', sep='\n')
def hello_world():
"""
Функция печатает Hello World в формате:
**********
Hello World!
##########
:return: None
"""
print(separator('*', 10), '\nHello World!\n',
separator('#', 10), sep='\n')
'''
**********
Hello World!
##########
'''
print("hello_world()") # True
hello_world()
print(glSep_s[:len(glSep_s)//2], '', sep='\n')
def hello_who(who='World'):
"""
Функция печатает приветствие в красивом формате
**********
Hello {who}!
##########
:param who: кого мы приветствуем, по умолчанию World
:return: None
"""
print(separator('*', 10), f'\nHello {who}!\n',
separator('#', 10), sep='\n')
'''
**********
Hello World!
##########
'''
print("hello_who()")
hello_who()
'''
**********
Hello Max!
##########
'''
print("hello_who('Max')")
hello_who('Max')
'''
**********
Hello Kate!
##########
'''
print("hello_who('Kate')")
hello_who('Kate')
print(glSep_s[:len(glSep_s)//2], '', sep='\n')
def pow_many(power, *args):
"""
Функция складывает любое количество цифр и возводит результат в степень power (примеры использования ниже)
:param power: степень
:param args: любое количество цифр
:return: результат вычисления # True -> (1 + 2)**1
"""
return sum(args) **power
print('pow_many(1, 1, 2) == 3') # True -> (1 + 2)**1 == 3
print(pow_many(1, 1, 2) == 3) # True -> (1 + 2)**1 == 3
print('pow_many(1, 2, 3) == 5') # True -> (2 + 3)**1 == 5
print(pow_many(1, 2, 3) == 5) # True -> (2 + 3)**1 == 5
print('pow_many(2, 1, 1) == 4') # True -> (1 + 1)**2 == 4
print(pow_many(2, 1, 1) == 4) # True -> (1 + 1)**2 == 4
print('pow_many(3, 2) == 8') # True -> 2**3 == 8
print(pow_many(3, 2) == 8) # True -> 2**3 == 8
print('pow_many(2, 1, 2, 3, 4) == 100') # True -> (1 + 2 + 3 + 4)**2 == 10**2 == 100
print(pow_many(2, 1, 2, 3, 4) == 100) # True -> (1 + 2 + 3 + 4)**2 == 10**2 == 100
print(glSep_s[:len(glSep_s)//2], '', sep='\n')
def print_key_val(**kwargs):
"""
Функция выводит переданные параметры в фиде key --> value
key - имя параметра
value - значение параметра
:param kwargs: любое количество именованных параметров
:return: None
"""
print(*(f'{_k} --> {_v}' for _k, _v in kwargs.items()), sep='\n')
"""
name --> Max
age --> 21
"""
print("print_key_val(name='Max', age=21)")
print_key_val(name='Max', age=21)
"""
animal --> Cat
is_animal --> True
"""
print("print_key_val(animal='Cat', is_animal=True)")
print_key_val(animal='Cat', is_animal=True)
print(glSep_s[:len(glSep_s)//2], '', sep='\n')
def my_filter(iterable, function):
"""
(Усложненое задание со *)
Функция фильтрует последовательность iterable и возвращает новую
Если function от элемента последовательности возвращает True, то элемент входит в новую последовательность иначе нет
(примеры ниже)
:param iterable: входаня последовательности
:param function: функция фильтрации
:return: новая отфильтрованная последовательность
"""
return list(_el for _el in iterable if function(_el))
print('my_filter([1, 2, 3, 4, 5], lambda x: x > 3) == [4, 5]') # True
print(my_filter([1, 2, 3, 4, 5], lambda x: x > 3) == [4, 5]) # True
print('my_filter([1, 2, 3, 4, 5], lambda x: x == 2) == [2]') # True
print(my_filter([1, 2, 3, 4, 5], lambda x: x == 2) == [2]) # True
print('my_filter([1, 2, 3, 4, 5], lambda x: x != 3) == [1, 2, 4, 5]') # True
print(my_filter([1, 2, 3, 4, 5], lambda x: x != 3) == [1, 2, 4, 5]) # True
print("my_filter(['a', 'b', 'c', 'd'], lambda x: x in 'abba') == ['a', 'b']") # True
print(my_filter(['a', 'b', 'c', 'd'], lambda x: x in 'abba') == ['a', 'b']) # True
# print(glSep_s[:len(glSep_s)//2], '', sep='\n')
print(glSep_s)
# import sys
# from MVVlStd import glSep_s
tTskMsg_s = '''
Задача 4. МОДУЛЬ 2 файл borndayforewer.py
Предлагается модернизировать программу из прошлого дз используя
хотя бы 1-у функцию, подробности в файле
'''
print(glSep_s, tTskMsg_s, f'{sys.version=}', glSep_s, 'Result:', '', sep='\n')
"""
МОДУЛЬ 2
Программа из 2-го дз
Сначала пользователь вводит год рождения Пушкина, когда отвечает верно вводит день рождения
Можно использовать свой вариант программы из предыдущего дз, мой вариант реализован ниже
Задание: переписать код используя как минимум 1 функцию
"""
# year = input('Ввведите год рождения А.С.Пушкина:')
# while year != '1799':
# print("Не верно")
# year = input('Ввведите год рождения А.С.Пушкина:')
# day = input('Ввведите день рождения Пушкин?')
# while day != '6':
# print("Не верно")
# day = input('В какой день июня родился Пушкин?')
# print('Верно')
def tAnswQueInPWChe_fif(la1stQue_s, laAnsw_s, laOthQue_s=None,
laFailMsg_s='Не верно', laOkMsg_s='Верно'):
li_s = input(f'{la1stQue_s}')
while li_s != laAnsw_s:
if laFailMsg_s: print(laFailMsg_s)
li_s = input(f'{laOthQue_s if laOthQue_s else la1stQue_s}')
else:
if laOkMsg_s: print(laOkMsg_s)
return True
tRes_l = []
tRes_l.append(tAnswQueInPWChe_fif('Ввведите год рождения А.С.Пушкина:',
'1799', laOkMsg_s=None))
tRes_l.append(tAnswQueInPWChe_fif('Ввведите день рождения Пушкин?', '6',
'В какой день июня родился Пушкин?'))
tRes_l
print(glSep_s)
# import sys
# from MVVlStd import glSep_s, inp_FltAVali_fefi
tTskMsg_s = '''
Задача 5. МОДУЛЬ 3 файл use_functions.py
В файле дано описание программы. Предлагается реализовать программу
по описанию используя любые средства
'''
print(glSep_s, tTskMsg_s, f'{sys.version=}', glSep_s, 'Result:', '', sep='\n')
"""
МОДУЛЬ 3
Программа "Личный счет"
Описание работы программы:
Пользователь запускает программу у него на счету 0
Программа предлагает следующие варианты действий
1. пополнить счет
2. покупка
3. история покупок
4. выход
1. пополнение счета
при выборе этого пункта пользователю предлагается ввести сумму на сколько пополнить счет
после того как пользователь вводит сумму она добавляется к счету
снова попадаем в основное меню
2. покупка
при выборе этого пункта пользователю предлагается ввести сумму покупки
если она больше количества денег на счете, то сообщаем что денег не хватает и переходим в основное меню
если денег достаточно предлагаем пользователю ввести название покупки, например (еда)
снимаем деньги со счета
сохраняем покупку в историю
выходим в основное меню
3. история покупок
выводим историю покупок пользователя (название и сумму)
возвращаемся в основное меню
4. выход
выход из программы
При выполнении задания можно пользоваться любыми средствами
Для реализации основного меню можно использовать пример ниже или написать свой
"""
glAccSum_n, glHstT_l = 0, []
def tRefillAcc_f():
global glAccSum_n, glHstT_l
loAdd_n = inp_FltAVali_fefi(f' Ведите сумму на сколько пополнить счет',
laInPTypeFlt_cll=float, laDfV_s='100',
laAcceptEmptyInPAsDf_b=True, laValiInPMsg_s=f'положительное число с возм.десят.точкой',
laVali_cll=lambda _n: 0 <= _n)[0]
glAccSum_n += loAdd_n #DVL: input by inp_FltAVali_fefi
# print(f'DBG: На счету:({glAccSum_n:.2f}) и в истории покупок {len(glHstT_l)} зап.')
return True
def tBuy_f():
global glAccSum_n, glHstT_l
loCost_n = inp_FltAVali_fefi(f' Введите сумму покупки (на Вашем счету:{glAccSum_n:.2f})',
laInPTypeFlt_cll=float, laDfV_s=str(min(100, glAccSum_n)),
laAcceptEmptyInPAsDf_b=True, laValiInPMsg_s=f'положительное число с возм.десят.точкой',
laVali_cll=lambda _n: 0 <= _n)[0]
if glAccSum_n < loCost_n: #DVL: input by inp_FltAVali_fefi
print(f'Денег на Вашем счету:({glAccSum_n:.2f}) не хватает',
f'для покупки на сумму:({loCost_n:.2f}).',
'Пополните счет, пожалуйста.', sep='\n')
return False
loDesc_s = inp_FltAVali_fefi(f' Введите название покупки', laInPTypeFlt_cll=None,
laDfV_s="Еда", laAcceptEmptyInPAsDf_b=True)[0]
# print(f'DBG: На счету:({glAccSum_n}) и в истории покупок {len(glHstT_l)} зап.')
glAccSum_n -= loCost_n
glHstT_l.append((loDesc_s, loCost_n)) #DVL: input by inp_FltAVali_fefi
# print(f'DBG: На счету:({glAccSum_n}) и в истории покупок {len(glHstT_l)} зап.')
return True
def tVieHst_f():
global glAccSum_n, glHstT_l
print('', f'История покупок (всего {len(glHstT_l)} зап.):',
*enumerate(glHstT_l, 1), '', sep='\n')
tMenu_d = {'1':('Пополнение счета', tRefillAcc_f),
'2':('Покупка', tBuy_f),
'3':('История покупок', tVieHst_f),
'4':('Выход', None)}
while True:
print(*(f'{_k}. {_v[0]}' for _k, _v in tMenu_d.items()), sep='\n')
print(f'На счету:({glAccSum_n:.2f}) и в истории покупок {len(glHstT_l)} зап.')
li_s = input('Выберите пункт меню: ')
if li_s in tMenu_d:
lo_cll = tMenu_d[li_s][1]
if lo_cll is None: break
else: loRes_b = lo_cll()
else:
print(f'Неверный пункт меню:"{li_s}"')
print(glSep_s)
tTskMsg_s = '''
Задача 6
Выложите проект на github (если возникнут трудности можно пока использовать гугл или яндекс диск)
'''
print(glSep_s, tTskMsg_s, f'{sys.version=}', glSep_s, 'Result:', '', sep='\n')
print('My fork: https://github.com/dev-mikevvl-ms/lesson4-functions-dz.git')
print(glSep_s)
###Output
______________________________________________________________________
Задача 6
Выложите проект на github (если возникнут трудности можно пока использовать гугл или яндекс диск)
sys.version='3.9.7 (default, Sep 16 2021, 16:59:28) [MSC v.1916 64 bit (AMD64)]'
______________________________________________________________________
Result:
My fork: https://github.com/dev-mikevvl-ms/lesson4-functions-dz.git
______________________________________________________________________
###Markdown
05
###Code
77 * 3000 *.87
'MSG: It remains to input {} more value{}.'.format(123),\
'Осталось ввести еще {} знач.'.format(123, 's')
t_s = 'fdgrt\n'
print(f'{t_s}')
# t_s =
print(t_s)
print(bool({}), bool(tMenu_d))
print(glSep_s)
tMenu_d = {'01':('Пополнение счета', None),
'2':('Покупка', None),
'23':('История покупок', None),
'E':('Выход', None)}
t_d = dict(tMenu_d)
# tMenu_d, t_d, t_d == tMenu_d, id(t_d) == id(tMenu_d),
sorted(t_d.keys(), key=lambda _el: str(_el)),\
# sorted(t_d.keys()), '01{01}'.format(*t_d), '"{Key!s:2}"'
tMenu_d[5] = ('xxx', None)
print(list(f'"{_k!s:>2}". {tMenu_d[_k][0]}' for _k in sorted(tMenu_d.keys(), key=int)), sep='\n')
list('{_k!s:>2}. {_v[0]}'.format(_k=_k, _v=tMenu_d[_k]) for _k in sorted(tMenu_d.keys(), key=int))
'{_k!s:>2}. {_v[0]}'.format(_k=5, _v=tMenu_d[5]), len(tMenu_d)
tRes_d['kAccSum_n'], f"На счету:({tRes_d['kAccSum_n']:.2f})"
tRes_d = {'kAccSum_n': 0.0, 'kHstT_l': [('Еда', 20.0), ('Еда', 80.0)]}
t_s = "На счету:({_d['kAccSum_n']:.2f})"
# t_s += " и в истории покупок {len(_d['kHstT_l'])} зап.")
t_s.format(_d=tRes_d)
t_cll = lambda _o1, laInnStt_d, file=sys.stdout: print(laInnStt_d, file=file)
t_cll(t_d, laInnStt_d=t_d)
from dataclasses import dataclass, field
from collections.abc import Callable
import copy
@dataclass
class D:
MenuEls_d: dict = field(default_factory=dict)
InnStt_d: dict = None
PrnInnStt_fmp: Callable = None # [self, dict, file]; ??Df: IF InnStt_d is !None -> print(InnStt_d)
HeaFmt_s: str = None
FooFmt_s: str = None
ElsFmt_s: str = '{_k!s:>2}. {_v[0]}'
def __post_init__(self):
# self.MenuEls_d = copy.deepcopy(dict(self.MenuEls_d))
self.MenuEls_d = dict(self.MenuEls_d)
if self.InnStt_d is not None:
self.InnStt_d = copy.deepcopy(dict(self.InnStt_d))
if self.PrnInnStt_fmp is None:
self.PrnInnStt_fmp = lambda sf_o, laInnStt_d, file=sys.stdout: print(laInnStt_d, file=file)
if self.HeaFmt_s is not None: self.HeaFmt_s = str(self.HeaFmt_s)
else: self.HeaFmt_s = glSep_s[:len(glSep_s)//3 *2]
if self.FooFmt_s is not None: self.FooFmt_s = str(self.FooFmt_s)
else: self.FooFmt_s = glSep_s[:len(glSep_s)//3 *2]
self.IsRun_b = bool(self.MenuEls_d)
def prn_Info_fmp(self, file=sys.stdout):
if self.PrnInnStt_fmp and callable(self.PrnInnStt_fmp):
self.PrnInnStt_fmp(self, laInnStt_d=self.InnStt_d, file=file)
tInnStt_d = dict(kAccSum_n=0, kHstT_l=[], kWid_i=10)
t_o, t1_o = D(), D(zip(['one', 'two', 'three'], [1, 2, 3]), ((0.0, []),))
t2_o = D( t_d ,tInnStt_d)
t_o, t1_o, t_o.IsRun_b, t1_o.IsRun_b, t2_o, t2_o.MenuEls_d['01'], t2_o.InnStt_d['kHstT_l']\
, id(t2_o) == id(t_d), id(t2_o.InnStt_d['kHstT_l']) == id(tInnStt_d['kHstT_l']), t2_o.InnStt_d\
, '({kAccSum_n:{kWid_i}})'.format(**tInnStt_d)
t_o.prn_Info_fmp()
import sys
from MVVlStd import glSep_s, inp_FltAVali_fefi
# from mod.MVVlStd import glSep_s, inp_FltAVali_fefi
# from MVVlStd import glSep_s, inp_FltAVali_fefi
# import MVVlStd
# # MVVlStd.inp_FltAVali_fefi('?')
# # inp_FltAVali_fefi('?')
# tTskMsg_s = '''
# Задача 5. МОДУЛЬ 3 файл use_functions.py
# В файле дано описание программы. Предлагается реализовать программу
# по описанию используя любые средства
# '''
# print(glSep_s, tTskMsg_s, f'{sys.version=}', glSep_s, 'Result:', '', sep='\n')
# 2Do:
# from dataclasses import dataclass, field
# import copy
# @dataclass
# class D:
# MenuEls_d: dict = field(default_factory=dict)
# InnStt_d: dict = field(default_factory=dict)
# ElsFmt_s: str = '{_k!s:>2}. {_v[0]}'
# ElFmt_s: str = See(prn_Info_fmp):*(f'{_k}. {self.MenuEls_d[_k][0]}' for _k in self)
# InnSttFmt_s: str = None # Df |==''> !OuP |> Sf.format(**tInnStt_d)
# # See(prn_Info_fmp) '({kAccSum_n:{kWid_i}})'.format(**tInnStt_d)
# HeaFmt_s: str = None # Df(Rat(glSep_s)) |==''> !OuP |> ??Sf.format(**self.OuPPP_d)
# FooFmt_s: str = None # Df(Rat(glSep_s)) |==''> !OuP |> ??Sf.format(**self.OuPPP_d)
# # self.OuPPP_d AllEls_co, VieEls_(co|l), MaxWid_i
# # x: list = field(default_factory=list)
# def __post_init__(self):
# # self.MenuEls_d = copy.deepcopy(dict(self.MenuEls_d))
# self.MenuEls_d = dict(self.MenuEls_d)
# self.InnStt_d = copy.deepcopy(dict(self.InnStt_d))
# self.IsRun_b = bool(self.MenuEls_d)
class MVVlMenu_c():
# laIsKeyExit_cll=lambda _sf, _k: int(_k) == max(iter(_sf))
def __init__(self, MenuEls_d=None, InnStt_d=None, PrnInnStt_fmp=None,
HeaFmt_s=None, FooFmt_s=None, ElsFmt_s='{_k!s:>2}. {_v[0]}'):
self.MenuEls_d = dict(MenuEls_d) if MenuEls_d is not None else {}
self.InnStt_d = dict(InnStt_d) if InnStt_d is not None else {}
self.PrnInnStt_fmp = PrnInnStt_fmp
if HeaFmt_s is not None: self.HeaFmt_s = str(HeaFmt_s)
else: self.HeaFmt_s = glSep_s[:len(glSep_s)//3 *2]
if FooFmt_s is not None: self.FooFmt_s = str(FooFmt_s)
else: self.FooFmt_s = glSep_s[:len(glSep_s)//3 *2]
self.ElsFmt_s = ElsFmt_s
self.IsRun_b = bool(self.MenuEls_d)
# self.kAccSum_n = int(laAccSum_n)
# self.kHstT_l = list(laHstT_l) if laHstT_l is not None else []
# self.IsKeyExit_cll = laIsKeyExit_cll
def __iter__(self): # 2Do: MaB Onl WhiUse(prn_fmp)
return (_k for _k in sorted(self.MenuEls_d.keys(), key=int))
def __getitem__(self, key): # BOf:KISS
return self.MenuEls_d[key]
def __len__(self, key): # BOf:KISS
return len(self.MenuEls_d)
# def oup_fmp(self): # 2Do: MaB
def prn_fmp(self, file=sys.stdout): # 2Do: MaB Onl(9+KeyExit OR Fit2Scr+KeyExit) w/Set(sf.WhiVieElsKey_l)
if bool(self.MenuEls_d):
if self.HeaFmt_s != '': print(self.HeaFmt_s, file=file)
print(*(self.ElsFmt_s.format(_k=_k, _v=self[_k]) for _k in self),
sep='\n', file=file)
self.prn_Info_fmp(file=file)
# print(glSep_s[:len(glSep_s)//3 *2], file=file)
if self.FooFmt_s != '': print(self.FooFmt_s, file=file)
# def oup_Info|Ret_fmp(self):
# def __str__(self):; __format__; tVieHst_fmp
def prn_Info_fmp(self, file=sys.stdout):
if self.PrnInnStt_fmp and callable(self.PrnInnStt_fmp):
self.PrnInnStt_fmp(self, laInnStt_d=self.InnStt_d, file=file)
# def prn_Info_fmp(self, la_d, file=sys.stdout):
# print(f"На счету:({la_d['kAccSum_n']:.2f}) и в истории покупок {len(la_d['kHstT_l'])} зап.",
# glSep_s[:len(glSep_s)//3 *2], sep='\n', file=file)
# def add_Els?_ffm(self):
# def del_Els?_ffpm(self):
# def get_Keys?_ffpm(self):
# def run_ffpm(self):
def __call__(self):
while self.IsRun_b:
self.prn_fmp()
# self.prn_Info_fmp()
li_s = input(' Выберите пункт меню: ')
if li_s in self.MenuEls_d:
lo_cll = self.MenuEls_d[li_s][1]
# if self.IsKeyExit_cll(self, li_s): break
# if self.IsKeyExit_cll(self, li_s): break
# if lo_cll is None: break
if lo_cll is None:
print(f'DVL: None 4 calling Fu() пункт меню:"{li_s}"')
continue
else: loRes_b = lo_cll(self)
else:
print(f'Неверный пункт меню:"{li_s}"')
else:
# self.prn_Info_fmp()
print('До свидания!', glSep_s[:len(glSep_s)//3 *2], sep='\n')
return self.InnStt_d
tInnStt_d = dict(kAccSum_n=0, kHstT_l=[])
def tRefillAcc_fm(self, file=sys.stdout):
# loAdd_n = inp_FltAVali_fefi(f' Введите сумму на сколько пополнить счет\n',
loAdd_n = inp_FltAVali_fefi(f' сумму на сколько пополнить счет\n',
laInPTypeFlt_cll=float, laDfV_s='100',
laAcceptEmptyInPAsDf_b=True, laValiInPMsg_s=f'положительное число с возм.десят.точкой\n',
laVali_cll=lambda _n: 0 <= _n, file=file)[0]
self.InnStt_d['kAccSum_n'] += loAdd_n #DVL: input by inp_FltAVali_fefi
# print(f'DBG: На счету:({self.InnStt_d['kAccSum_n']:.2f}) и в истории покупок {len(self.InnStt_d['kHstT_l'])} зап.')
print(f'Пополнение на:({loAdd_n:.2f}).', file=file)
return True
def tBuy_fmp(self, file=sys.stdout):
loCost_n = inp_FltAVali_fefi(f" сумму покупки (на Вашем счету:{self.InnStt_d['kAccSum_n']:.2f})\n",
laInPTypeFlt_cll=float, laDfV_s=str(min(100, self.InnStt_d['kAccSum_n'])),
laAcceptEmptyInPAsDf_b=True, laValiInPMsg_s=f'положительное число с возм.десят.точкой\n',
laVali_cll=lambda _n: 0 <= _n, file=file)[0]
if self.InnStt_d['kAccSum_n'] < loCost_n: #DVL: input by inp_FltAVali_fefi
print(f"Денег на Вашем счету:({self.InnStt_d['kAccSum_n']:.2f}) не хватает",
f' для покупки на сумму:({loCost_n:.2f}).',
' Пополните счет, пожалуйста.', sep='\n', file=file)
return False
loDesc_s = inp_FltAVali_fefi(f' название покупки\n', laInPTypeFlt_cll=None,
laDfV_s="Еда", laAcceptEmptyInPAsDf_b=True, file=file)[0]
# print(f'DBG: На счету:({self.InnStt_d['kAccSum_n']}) и в истории покупок {len(self.InnStt_d['kHstT_l'])} зап.')
self.InnStt_d['kAccSum_n'] -= loCost_n
self.InnStt_d['kHstT_l'].append((loDesc_s, loCost_n)) #DVL: input by inp_FltAVali_fefi
# print(f'DBG: На счету:({self.InnStt_d['kAccSum_n']}) и в истории покупок {len(self.InnStt_d['kHstT_l'])} зап.')
print(f'Покупка: "{loDesc_s}", на сумму:({loCost_n:.2f}).', file=file)
return True
def tVieHst_fmp(self, file=sys.stdout):
print(f"История покупок (всего {len(self.InnStt_d['kHstT_l'])} зап.):",
*enumerate(self.InnStt_d['kHstT_l'], 1), '', sep='\n', file=file)
def tExit_fm(self):
self.IsRun_b = False
# tMenu_d = {'1':('Пополнение счета', tRefillAcc_fm, ??Type??(Exit, Back, SbMenu, CtrlVieMenu??)),
# '2':('Покупка', tBuy_fm),
# '3':('История покупок', tVieHst_fm),
# '4':('Выход', None)}
def prn_InnStt_fmp(self, laInnStt_d, file=sys.stdout):
print(f"На счету:({laInnStt_d['kAccSum_n']:.2f}) и в истории покупок {len(laInnStt_d['kHstT_l'])} зап.",
# glSep_s[:len(glSep_s)//3 *2], sep='\n',
file=file)
def main(laArgs: list[str]) -> None:
# Ww(sys.argv[1:])
tMenu_o = MVVlMenu_c({'1':('Пополнение счета', tRefillAcc_fm),
'2':('Покупка', tBuy_fmp),
'3':('История покупок', tVieHst_fmp),
'4':('Выход', tExit_fm)}, InnStt_d=tInnStt_d,
PrnInnStt_fmp=prn_InnStt_fmp)
# tMenu_o = MVVlMenu_c()
# tMenu_o.add_Els?_ffm(...)
# tRes_d = tMenu_o.run_ffpm()
tRes_d = tMenu_o()
# tRes_d = MVVlMenu_c(...)()
# print(tRes_d)
return tRes_d
if __name__ == '__main__':
import sys
# main(sys.argv[1:])
main(None)
def prn_InnStt_fp(laInnStt_d, file=sys.stdout):
print(f"На счету:({laInnStt_d['kAccSum_n']:.2f}) и в истории покупок {len(laInnStt_d['kHstT_l'])} зап.",
# glSep_s[:len(glSep_s)//3 *2], sep='\n',
file=file)
prn_InnStt_fp({'kAccSum_n': 0.0, 'kHstT_l': [('Еда', 20.0), ('Еда', 80.0)]})
tMenu_o = MVVlMenu_c({'1':('Пополнение счета', tRefillAcc_fm),
'2':('Покупка', tBuy_fmp),
'3':('История покупок', tVieHst_fmp),
'4':('Выход', tExit_fm)}, tInnStt_d)
print(f"{tMenu_o['1']=}")
try:
tMenu_o['1'] = ('xxx')
except TypeError as le_o:
print(le_o)
try:
tMenu_o['1'] = ('xxx')
except TypeError as le_o:
print(le_o)
tTskMsg_s = '''
Задача 1
В проекте реализовать следующий функционал:
После запуска программы пользователь видит меню, состоящее из следующих пунктов:
- создать папку;
- удалить (файл/папку);
- копировать (файл/папку);
- просмотр содержимого рабочей директории;
- посмотреть только папки;
- посмотреть только файлы;
- просмотр информации об операционной системе;
- создатель программы;
- играть в викторину;
- мой банковский счет;
- смена рабочей директории (*необязательный пункт);
- выход.
Так же можно добавить любой дополнительный функционал по желанию.
'''
print(glSep_s, tTskMsg_s, f'{sys.version=}', glSep_s, 'Result:', '', sep='\n')
print('https://github.com/dev-mikevvl-ms/PyDev.05.HW.git')
print(glSep_s)
import os
# os.supports_follow_symlinks
import os, shutil
os.makedirs(name, mode=511, exist_ok=False)
t_l = os.listdir()
os.remove(path, *, dir_fd=None)
os.removedirs(name)
os.listdir(path='.')
os.scandir(path='.')
with os.scandir(path) as it:
for entry in it:
if not entry.name.startswith('.') and entry.is_file():
print(entry.name)
os.stat(path, *, dir_fd=None, follow_symlinks=True)
os.rename(src, dst, *, src_dir_fd=None, dst_dir_fd=None)
os.link(src, dst, *, src_dir_fd=None, dst_dir_fd=None, follow_symlinks=True)
os.symlink(src, dst, target_is_directory=False, *, dir_fd=None)
os.walk(top, topdown=True, onerror=None, followlinks=False)
os.fwalk(top='.', topdown=True, onerror=None, *, follow_symlinks=False, dir_fd=None)
os.dup(fd); os.dup2(fd, fd2, inheritable=True)
os.path.normcase(path)
os.path.normpath(path)
os.path.realpath(path, *, strict=False)
os.path.relpath(path, start=os.curdir)
os.path.samefile(path1, path2)
os.path.sameopenfile(fp1, fp2)
os.path.samestat(stat1, stat2)
os.path.split(path)
os.path.splitext(path)
os.path.commonpath(paths)
os.path.commonprefix(list)
os.path.exists(path)
os.path.lexists(path)
os.path.expanduser(path)
os.path.expandvars(path)
import io, stat
io.IOBase.fileno()
# os.getloadavg(), len(os.sched_getaffinity(0)), len(os.sysconf_names), len(os.confstr_names) # Unix
stat.filemode(DirEntry.stat().st_mode), os.cpu_count(), os.terminal_size['columns'],\
shutil.get_terminal_size().columns, shutil.get_terminal_size()
import tempfile
tempfile.tempdir
fp = tempfile.TemporaryFile()
# with tempfile.TemporaryFile() as fp:
with tempfile.TemporaryDirectory() as tmpdirname:
print('created temporary directory', tmpdirname)
t_t = tempfile.mkstemp(suffix=None, prefix=None, dir=None, text=False)
t_s = tempfile.mkdtemp(suffix=None, prefix=None, dir=None)
###Output
_____no_output_____
###Markdown
CuW
###Code
def t1_f():
lo_i, lo_l = 0, []
print(f"DBG:1: {lo_i=}, {lo_l=}.")
def t2_f():
global lo_i, lo_l
lo_i += 10
lo_l += [('ts', 10)]
print(f"\tDBG:1: {lo_i=}, {lo_l=}.")
t2_f()
print(f"DBG:2: {lo_i=}, {lo_l=}.")
t1_f()
# print(sys.platform)
# Это пути где питон ищет файлы
# print(sys.path)
print(f"{sys.platform=}", f"sys.executable='...{sys.executable[-13:]}'",
sep='\n')
tTskMsg_s = '''
Задача 3
Выложите проект на github
'''
print(glSep_s, tTskMsg_s, f'{sys.version=}', glSep_s, 'Result:', '', sep='\n')
print('https://github.com/dev-mikevvl-ms/PyDev.05.HW.git')
print(glSep_s)
print(f'На счету:({glAccSum_n:.2f}) и в истории покупок {len(glHstT_l)} зап.:',
*enumerate(glHstT_l, 1), sep='\n')
def pow_many(power, *args):
"""
Функция складывает любое количество цифр и возводит результат в степень power (примеры использования ниже)
:param power: степень
:param args: любое количество цифр
:return: результат вычисления # True -> (1 + 2)**1
"""
# print(args, *args)
return sum(args) **power
print(pow_many(1, 1, 2) == 3) # True -> (1 + 2)**1 == 3
print(pow_many(1, 2, 3) == 5) # True -> (2 + 3)**1 == 5
print(pow_many(2, 1, 1) == 4) # True -> (1 + 1)**2 == 4
print(pow_many(3, 2) == 8) # True -> 2**3 == 8
print(pow_many(2, 1, 2, 3, 4) == 100) # True -> (1 + 2 + 3 + 4)**2 == 10**2 == 100
print(glSep_s[:len(glSep_s)//2], '', sep='\n')
tTskMsg_s = '''
Задача 6
Выложите проект на github (если возникнут трудности можно пока использовать гугл или яндекс диск)
'''
print(glSep_s, tTskMsg_s, f'{sys.version=}', glSep_s, 'Result:', '', sep='\n')
print('Fork: https://github.com/dev-mikevvl-ms/lesson4-functions-dz.git')
print(glSep_s)
# print(tRes_l)
list(range(-2)), 937 +360
print([2,5,3,9,3,4,5,5,6], 'Unstable:', list(set([2,5,3,9,3,4,5,5,6])),
'Stable:', list(dict.fromkeys([2,5,3,9,3,4,5,5,6])), sep='\n') # Unstable, Stable
print(type(dict) == type(list) == type(tuple) == type(str) == type(bytes),
type(str), type(list), sep='\n')
###Output
True
<class 'type'>
<class 'type'>
|
docs/contents/lennard_jones_fluid/LJ_Potential.ipynb
|
###Markdown
The Lennard-Jones potential The Lennard-Jones (LJ) potential between two particles is defined by the following equation, where $x$ is the distance between the particles, and $\sigma$ and $\epsilon$ are two parameters of the potential: \begin{equation}V(x) = 4 \epsilon \left[ \left( \frac{\sigma}{x} \right)^{12} - \left( \frac{\sigma}{x} \right)^6 \right]\end{equation} Lets see the shape of this function:
###Code
def LJ (x, sigma, epsilon):
t = sigma/x
t6 = t**6
t12 = t6**2
return 4.0*epsilon*(t12-t6)
sigma = 2.0 * unit.angstrom
epsilon = 1.0 * unit.kilocalories_per_mole
xlim_figure = [0.01, 6.0]
ylim_figure = [-2.0, 10.0]
x = np.linspace(xlim_figure[0], xlim_figure[1], 100, True) * unit.angstrom
plt.plot(x, LJ(x, sigma, epsilon))
plt.xlim(xlim_figure)
plt.ylim(ylim_figure)
plt.xlabel('x [{}]'.format(x.unit.get_symbol()))
plt.ylabel('V [{}]'.format(epsilon.unit.get_symbol()))
plt.show()
###Output
_____no_output_____
###Markdown
The way the LJ potential is built, the $\sigma$ and $\epsilon$ parameters have a straightforward interpretation. The cut with $y=0$ is located in $x=\sigma$:
###Code
sigma = 2.0 * unit.angstrom
epsilon = 1.0 * unit.kilocalories_per_mole
xlim_figure = [0.01, 6.0]
ylim_figure = [-2.0, 10.0]
x = np.linspace(xlim_figure[0], xlim_figure[1], 100, True) * unit.angstrom
plt.plot(x, LJ(x, sigma, epsilon))
plt.hlines(0, xlim_figure[0], xlim_figure[1], linestyles='dotted', color='gray')
plt.vlines(sigma._value, ylim_figure[0], ylim_figure[1], linestyles='dashed', color='red')
plt.text(sigma._value+0.02*xlim_figure[1], 0.7*ylim_figure[1], '$\sigma$', fontsize=14)
plt.xlim(xlim_figure)
plt.ylim(ylim_figure)
plt.xlabel('x [{}]'.format(x.unit.get_symbol()))
plt.ylabel('V [{}]'.format(epsilon.unit.get_symbol()))
plt.show()
###Output
_____no_output_____
###Markdown
And $\epsilon$ is the depth of the minimum measured from $y=0$:
###Code
sigma = 2.0 * unit.angstrom
epsilon = 1.0 * unit.kilocalories_per_mole
xlim_figure = [0.01, 6.0]
ylim_figure = [-2.0, 10.0]
x = np.linspace(xlim_figure[0], xlim_figure[1], 100, True) * unit.angstrom
plt.plot(x, LJ(x, sigma, epsilon))
plt.hlines(0, xlim_figure[0], xlim_figure[1], linestyles='dotted', color='gray')
plt.hlines(-epsilon._value, xlim_figure[0], xlim_figure[1], linestyles='dashed', color='red')
plt.annotate(text='', xy=(1.0,0.0), xytext=(1.0,-epsilon._value), arrowprops=dict(arrowstyle='<->'))
plt.text(1.0+0.02*xlim_figure[1], -0.7*epsilon._value, '$\epsilon$', fontsize=14)
plt.xlim(xlim_figure)
plt.ylim(ylim_figure)
plt.xlabel('x [{}]'.format(x.unit.get_symbol()))
plt.ylabel('V [{}]'.format(epsilon.unit.get_symbol()))
plt.show()
###Output
_____no_output_____
###Markdown
Notice that the LJ potential has physical meaning when $\epsilon>0$ and $\sigma>0$ only. Actually, the potential vanishes whether $\epsilon=0$ or $\sigma=0$. The Lennard Jones minimum and the size of the particles The LJ potential has a single minimum located in $x_{min}$. Lets equal to $0$ the first derivative of the potential to find the value of $x_{min}$:
###Code
x, sigma, epsilon = sy.symbols('x sigma epsilon', real=True, positive=True)
V = 4.0*epsilon*((sigma/x)**12-(sigma/x)**6)
gradV = sy.diff(V,x)
roots=sy.solve(gradV, x)
x_min = roots[0]
x_min
###Output
_____no_output_____
###Markdown
The minimum is then located in:\begin{equation}x_{min} = 2^{1/6} \sigma\end{equation}where the potential takes the value:\begin{equation}V(x_{min}) = -\epsilon\end{equation}
###Code
sigma = 2.0 * unit.angstrom
epsilon = 1.0 * unit.kilocalories_per_mole
x_min = 2**(1/6)*sigma
y_min = -epsilon
xlim_figure = [x_min._value-0.4, x_min._value+0.4]
ylim_figure = [y_min._value-0.1, y_min._value+0.5]
x = np.linspace(xlim_figure[0], xlim_figure[1], 100, True) * unit.angstroms
plt.plot(x, LJ(x, sigma, epsilon))
plt.hlines(y_min._value, xlim_figure[0], xlim_figure[1], linestyles='dashed', color='gray')
plt.vlines(x_min._value, ylim_figure[0], ylim_figure[1], linestyles='dashed', color='gray')
plt.xlim(xlim_figure)
plt.ylim(ylim_figure)
plt.xlabel('x [{}]'.format(x.unit.get_symbol()))
plt.ylabel('V [{}]'.format(epsilon.unit.get_symbol()))
plt.show()
###Output
_____no_output_____
###Markdown
This way two particles in the equilibrium position will be placed at a $2^{1/6} \sigma$ distance. The potential is thereby modeling two "soft spheres" atracting each other very lightly. Their radii, given that both particles are equal, are equal to $r$:\begin{equation}r = \frac{1}{2} x_{min} = 2^{-5/6} \sigma\end{equation} And we say these spheres are "soft" because their volume is not limited by a hard-wall potential, they can penetrate each other suffering a not infinite repulsive force. Time period of the small harmonic oscillations around the minimum If we want to perform a molecular simulation of this two particles we should wonder how big the integrator timestep must be. To answer this question we can study the harmonic approximation around the minimum. Lets calculate the time period, $\tau$, of a small harmonic oscillation around the minimum:
###Code
x, sigma, epsilon = sy.symbols('x sigma epsilon', real=True, positive=True)
V = 4.0*epsilon*((sigma/x)**12-(sigma/x)**6)
gradV = sy.diff(V,x)
grad2V = sy.diff(V,x,x)
x_min = sy.solve(gradV,x)[0]
k_harm = grad2V.subs(x, x_min)
k_harm
###Output
_____no_output_____
###Markdown
The harmonic constant of the second degree Taylor polynomial of the LJ potential at $x=x_{min}$ is then:\begin{equation}k_{harm} = 36·2^{2/3} \frac{\epsilon}{\sigma^2}\end{equation} The oscillation period of a particle with $m$ mass in an harmonic potential defined by $\frac{1}{2} k x²$ is:\begin{equation}\tau = 2 \pi \sqrt{ \frac{m}{k}}\end{equation}As such, the period of the small harmonic oscillations around the LJ minimum of particle with $m$ mass is:\begin{equation}\tau = 2 \pi \sqrt{ \frac{m}{k_{harm}}} = \frac{\pi}{3·2^{1/3}} \sqrt{\frac{m\sigma^2}{\epsilon}}\end{equation} With the mass and parameters taking values of amus, angstroms and kilocalories per mole, the time period is in the order of:
###Code
mass = 50.0 * unit.amu
sigma = 2.0 * unit.angstrom
epsilon = 1.0 * unit.kilocalories_per_mole
k = 36 * 2**(2/3) * epsilon/sigma**2
tau = 2*np.pi * np.sqrt(mass/k)
print(tau)
###Output
0.5746513694274475 ps
###Markdown
But, is this characteristic time a good threshold for a LJ potential? If the oscillations around the minimum are not small enough, the harmonic potential of the second degree term of the taylor expansion is easily overcome by the sharp left branch of the LJ potential:
###Code
sigma = 2.0 * unit.angstrom
epsilon = 1.0 * unit.kilocalories_per_mole
k = 36 * 2**(2/3) * epsilon/sigma**2
x_min = 2**(1/6)*sigma
y_min = -epsilon
xlim_figure = [x_min._value-0.2, x_min._value+0.2]
ylim_figure = [y_min._value-0.1, y_min._value+0.6]
x = np.linspace(xlim_figure[0], xlim_figure[1], 100, True) * unit.angstroms
plt.plot(x, LJ(x, sigma, epsilon))
plt.plot(x, 0.5*k*(x-x_min)**2+y_min)
plt.hlines(y_min._value, xlim_figure[0], xlim_figure[1], linestyles='dashed', color='gray')
plt.vlines(x_min._value, ylim_figure[0], ylim_figure[1], linestyles='dashed', color='gray')
plt.xlim(xlim_figure)
plt.ylim(ylim_figure)
plt.xlabel('x [{}]'.format(x.unit.get_symbol()))
plt.ylabel('V [{}]'.format(epsilon.unit.get_symbol()))
plt.show()
###Output
_____no_output_____
###Markdown
Let's imagine the following situation. Let a particle be in the harmonic potential at temperature of 300K. Will the particle be more constrained in space than in the well of the LJ potential? Will the particle feel the harmonic potential softer or sharper than the LJ? Lets make some numbers to evaluate if the oscillation time period of the harmonic approximation can be a good time threshold for the integration timestep of a molecular dynamics of the LJ potential.The standard deviation of an harmonic oscillation with the shape $\frac{1}{2}k x^2$ in contact with a stochastic thermal bath can be computed as:\begin{equation}\beta = \frac{1}{k_{\rm B} T} \end{equation}\begin{equation}Z_x = \int_{-\infty}^{\infty} {\rm e}^{- \beta \frac{1}{2}k x^2} = \sqrt{\frac{2 \pi}{\beta k}}\end{equation}\begin{equation}\left = \frac{1}{Z_x} \int_{-\infty}^{\infty} x {\rm e}^{-\beta \frac{1}{2}k x^2} = 0\end{equation}\begin{equation}\left = \frac{1}{Z_x} \int_{-\infty}^{\infty} x^{2} {\rm e}^{-\beta \frac{1}{2}k x^2} = \frac{1}{Z_x} \sqrt{\frac{2 \pi}{\beta³ k^3}} = \frac{1}{\beta k}\end{equation}\begin{equation}{\rm std} = \left( \left -\left^2 \right)^{1/2} = \sqrt{ \frac{k_{\rm B}T}{k} }\end{equation}This way, in the case of the harmonic potential obtained as the second degree term of the Taylor expansion around the LJ minimum:
###Code
mass = 50.0 * unit.amu
sigma = 2.0 * unit.angstrom
epsilon = 1.0 * unit.kilocalories_per_mole
temperature = 300 * unit.kelvin
kB = unit.BOLTZMANN_CONSTANT_kB * unit.AVOGADRO_CONSTANT_NA
k = 36 * 2**(2/3) * epsilon/sigma**2
std = np.sqrt(kB*temperature/k)
x_min = 2**(1/6)*sigma
y_min = -epsilon
xlim_figure = [x_min._value-0.4, x_min._value+0.4]
ylim_figure = [y_min._value-0.1, y_min._value+0.6]
x = np.linspace(xlim_figure[0], xlim_figure[1], 100, True) * unit.angstroms
plt.plot(x, LJ(x, sigma, epsilon))
plt.plot(x, 0.5*k*(x-x_min)**2+y_min)
plt.hlines(y_min._value, xlim_figure[0], xlim_figure[1], linestyles='dashed', color='gray')
plt.vlines(x_min._value, ylim_figure[0], ylim_figure[1], linestyles='dashed', color='gray')
plt.axvspan(x_min._value - std._value, x_min._value + std._value, alpha=0.2, color='red')
plt.annotate(text='', xy=(x_min._value, y_min._value - 0.5*(y_min._value-ylim_figure[0])),
xytext=(x_min._value-std._value, y_min._value - 0.5*(y_min._value-ylim_figure[0])),
arrowprops=dict(arrowstyle='<->'))
plt.text(x_min._value-0.6*std._value, y_min._value - 0.4*(y_min._value-ylim_figure[0]), '$std$', fontsize=14)
plt.xlim(xlim_figure)
plt.ylim(ylim_figure)
plt.xlabel('x [{}]'.format(x.unit.get_symbol()))
plt.ylabel('V [{}]'.format(epsilon.unit.get_symbol()))
plt.show()
###Output
_____no_output_____
###Markdown
The harmonic potential is too soft as approximation. Its oscillation time used as threshold to choose the integration timestep can yield to numeric problems. Let's try with a stiffer potential, let's double the harmonic constant:
###Code
mass = 50.0 * unit.amu
sigma = 2.0 * unit.angstrom
epsilon = 1.0 * unit.kilocalories_per_mole
temperature = 300 * unit.kelvin
kB = unit.BOLTZMANN_CONSTANT_kB * unit.AVOGADRO_CONSTANT_NA
k = 36 * 2**(2/3) * epsilon/sigma**2
std = np.sqrt(kB*temperature/k)
x_min = 2**(1/6)*sigma
y_min = -epsilon
xlim_figure = [x_min._value-0.4, x_min._value+0.4]
ylim_figure = [y_min._value-0.1, y_min._value+0.6]
x = np.linspace(xlim_figure[0], xlim_figure[1], 100, True) * unit.angstroms
plt.plot(x, LJ(x, sigma, epsilon))
plt.plot(x, 0.5*k*(x-x_min)**2+y_min)
plt.plot(x, k*(x-x_min)**2+y_min, label='2k_{harm}')
plt.hlines(y_min._value, xlim_figure[0], xlim_figure[1], linestyles='dashed', color='gray')
plt.vlines(x_min._value, ylim_figure[0], ylim_figure[1], linestyles='dashed', color='gray')
plt.axvspan(x_min._value - std._value, x_min._value + std._value, alpha=0.2, color='red')
plt.annotate(text='', xy=(x_min._value, y_min._value - 0.5*(y_min._value-ylim_figure[0])),
xytext=(x_min._value-std._value, y_min._value - 0.5*(y_min._value-ylim_figure[0])),
arrowprops=dict(arrowstyle='<->'))
plt.text(x_min._value-0.6*std._value, y_min._value - 0.4*(y_min._value-ylim_figure[0]), '$std$', fontsize=14)
plt.xlim(xlim_figure)
plt.ylim(ylim_figure)
plt.xlabel('x [{}]'.format(x.unit.get_symbol()))
plt.ylabel('V [{}]'.format(epsilon.unit.get_symbol()))
plt.show()
###Output
_____no_output_____
###Markdown
Lets take then, as reference, an harmonic potential with constant equal to $2k_{harm}$ could be a better idea. Lets compute then the new time threshold to choose the integration timestep:\begin{equation}\tau' = 2 \pi \sqrt{ \frac{m}{2k_{harm}}} = \frac{\pi}{3·2^{5/6}} \sqrt{\frac{m\sigma^2}{\epsilon}} = \frac{1}{\sqrt{2}} \tau\end{equation}
###Code
mass = 50.0 * unit.amu
sigma = 2.0 * unit.angstrom
epsilon = 1.0 * unit.kilocalories_per_mole
k = 36 * 2**(2/3) * epsilon/sigma**2
tau = 2*np.pi * np.sqrt(mass/(2*k))
print(tau)
###Output
0.4063398801402841 ps
###Markdown
It is an accepted rule of thumb that the integration timestep must be as large as $\tau / 10$, being $\tau$ the oscillation time period of the fastest possible vibration mode. So finally, in this case the integration time step should not be longer than:
###Code
mass = 50.0 * unit.amu
sigma = 2.0 * unit.angstrom
epsilon = 1.0 * unit.kilocalories_per_mole
k = 36 * 2**(2/3) * epsilon/sigma**2
tau = 2*np.pi * np.sqrt(mass/(2*k))
print(tau/10.0)
###Output
0.04063398801402841 ps
|
notebook/narrative-python.ipynb
|
###Markdown
ObjectivesIn this narrative, we are going to explore the products available in Redmart (an online ecommerce grocery/retail store) and hope to answer these questions:1. What is the Min/Max/Mean and price distribution of all the products?2. Does country of origin affect the price of the products?3. How useful are the product description?4. What are some of the common brands and manufacturers?5. Is Redmart a good place for Organic food lovers?6. How many products are stock out?
###Code
# from src.data import make_dataset
import matplotlib.pyplot as plt
plt.rcParams['figure.figsize'] = [15, 5]
import numpy as np
import pandas as pd
from glob import glob
from IPython.display import Image, display, HTML
import os
# Importing data from ./data/processed
# For information on how data is gathered, use "make download-data" and "make data"
os.chdir(os.path.dirname(os.getcwd()) + '/data/processed')
dataFile = os.getcwd() + '/data.csv'
basePdtURL = 'https://redmart.com/product/'
df = pd.read_csv(dataFile, names = ['pdtID', 'pdtName', 'pdtDesc', 'pdtImageURL', 'pdtPrice', 'pdtCountryOfOrigin', 'pdtOrganic', 'pdtMfgName', 'pdtBrandName', 'pdtStockStatus', 'pdtURI', 'pdtCategoryTags'])
pdtPriceMax = df.loc[df['pdtPrice'] == df['pdtPrice'].max(), ['pdtName']]
print('The most expensive product is', pdtPriceMax.values[0][0], 'and will cost you $', df['pdtPrice'].max())
pdtPriceMean = df['pdtPrice'].mean()
print('The average price of the product is $', round(float(pdtPriceMean), 2) )
pdtPriceMin = df.loc[df['pdtPrice'] == df['pdtPrice'].min(), ['pdtName', 'pdtURI']]
print('We also found that the lowest priced product', pdtPriceMin.values[0][0], 'costs only $', df['pdtPrice'].min(), '. See it for yourself here -', basePdtURL + str(pdtPriceMin.values[0][1]) )
df
df.describe()
print('In the table below, let\'s look at the breakdown on where the items are originated from ')
pdtFreq = pd.value_counts(df.pdtCountryOfOrigin).to_frame().reset_index()
pdtFreq.columns = ['Country_of_Origin', 'Number_of_Items']
pdtFreq.head(10)
pdtFreq = pdtFreq.head(20)
pdtFreq.set_index("Country_of_Origin",drop=True,inplace=True)
pdtFreq.plot(kind='bar')
###Output
_____no_output_____
###Markdown
ProductsLet's look at some problems we randomly display from the entire list of products extracted
###Code
print("Total number of products:", len(df.index))
IMAGE_URL = 'https://s3-ap-southeast-1.amazonaws.com/media.redmart.com/newmedia/150x'
from random import shuffle
imageURLs = df['pdtImageURL'].tolist()
shuffle(imageURLs)
imagesList=''.join( ["<img style='width: 100px; margin: 0px; float: left; border: 1px solid black;' src='%s' />" % str(s) for s in imageURLs[:42] ] )
display(HTML(imagesList))
pdtCountryPrice = df[['pdtCountryOfOrigin', 'pdtPrice']]
pdtCountryPrice.groupby('pdtCountryOfOrigin')['pdtPrice'].count()
# pdtCountryPrice
# countryCount = len(pdtCountryPrice.pdtCountryOfOrigin.unique())
pdtCountryPrice = pdtCountryPrice.pivot(columns=pdtCountryPrice.columns[0], index=pdtCountryPrice.index)
pdtCountryPrice.columns = pdtCountryPrice.columns.droplevel()
med = pdtCountryPrice.median()
med = med.sort_values() # ascending=False
pdtCountryPrice = pdtCountryPrice[med.index]
pdtCountryPrice.plot(kind='box', figsize=(15,10), vert=False) # rot=90,
###Output
_____no_output_____
|
starter_code/model_1-logistic_regression.ipynb
|
###Markdown
Read the CSV and Perform Basic Data Cleaning
###Code
df = pd.read_csv("exoplanet_data.csv")
# Drop the null columns where all values are null
df = df.dropna(axis='columns', how='all')
# Drop the null rows
df = df.dropna()
df.head()
###Output
_____no_output_____
###Markdown
Select your features (columns)
###Code
# Assign X (data) and y (target)
X = df.drop("koi_disposition", axis=1)
y = df["koi_disposition"]
print(X.shape, y.shape)
from sklearn.preprocessing import LabelEncoder
# Step 1: Label-encode data set
label_encoder = LabelEncoder()
label_encoder.fit(y)
encoded_y = label_encoder.transform(y)
encoded_y
###Output
_____no_output_____
###Markdown
Create a Train Test SplitUse `koi_disposition` for the y values
###Code
from sklearn.model_selection import train_test_split
X_train, X_test, y_train, y_test = train_test_split(X, encoded_y, random_state=1, stratify=y)
y_test
X_train.head()
###Output
_____no_output_____
###Markdown
Pre-processingScale the data using the MinMaxScaler and perform some feature selection
###Code
# Scale your data
from sklearn.preprocessing import StandardScaler
X_scaler = StandardScaler().fit(X_train)
X_train_scaled = X_scaler.transform(X_train)
X_test_scaled = X_scaler.transform(X_test)
###Output
_____no_output_____
###Markdown
Train the Model
###Code
from sklearn.linear_model import LogisticRegression
classifier = LogisticRegression()
classifier
y_test
classifier.fit(X_train_scaled, y_train)
print(f"Training Data Score: {classifier.score(X_train_scaled, y_train)}")
print(f"Testing Data Score: {classifier.score(X_test_scaled, y_test)}")
###Output
Training Data Score: 0.8804119778752623
Testing Data Score: 0.8821510297482837
###Markdown
Save the Model
###Code
# save your model by updating "your_name" with your name
# and "your_model" with your model variable
# be sure to turn this in to BCS
# if joblib fails to import, try running the command to install in terminal/git-bash
import joblib
filename = 'vikash_bhakta_lr.sav'
joblib.dump(classifier, filename)
###Output
_____no_output_____
|
Obsolete Py27/Module6/.ipynb_checkpoints/Module6 - Lab6-checkpoint.ipynb
|
###Markdown
DAT210x - Programming with Python for DS Module6- Lab6
###Code
import pandas as pd
import time
###Output
_____no_output_____
###Markdown
How to Get The Dataset Grab the DLA HAR dataset from:- http://groupware.les.inf.puc-rio.br/har- http://groupware.les.inf.puc-rio.br/static/har/dataset-har-PUC-Rio-ugulino.zipAfter extracting it out, load up the dataset into dataframe named `X` and do your regular dataframe examination:
###Code
# .. your code here ..
###Output
_____no_output_____
###Markdown
Encode the gender column such that: `0` is male, and `1` as female:
###Code
# .. your code here ..
###Output
_____no_output_____
###Markdown
Clean up any columns with commas in them so that they're properly represented as decimals:
###Code
# .. your code here ..
###Output
_____no_output_____
###Markdown
Let's take a peek at your data types:
###Code
X.dtypes
###Output
_____no_output_____
###Markdown
Convert any column that needs to be converted into numeric use `errors='raise'`. This will alert you if something ends up being problematic.
###Code
# .. your code here ..
###Output
_____no_output_____
###Markdown
If you find any problematic records, drop them before calling the `to_numeric` methods above. Okay, now encode your `y` value as a Pandas dummies version of your dataset's `class` column:
###Code
# .. your code here ..
###Output
_____no_output_____
###Markdown
In fact, get rid of the `user` and `class` columns:
###Code
# .. your code here ..
###Output
_____no_output_____
###Markdown
Let's take a look at your handy-work:
###Code
X.describe()
###Output
_____no_output_____
###Markdown
You can also easily display which rows have nans in them, if any:
###Code
X[pd.isnull(X).any(axis=1)]
###Output
_____no_output_____
###Markdown
Create an RForest classifier named `model` and set `n_estimators=30`, the `max_depth` to 10, `oob_score=True`, and `random_state=0`:
###Code
# .. your code here ..
###Output
_____no_output_____
###Markdown
Split your data into `test` / `train` sets. Your `test` size can be 30%, with `random_state` 7. Use variable names: `X_train`, `X_test`, `y_train`, and `y_test`:
###Code
# .. your code here ..
###Output
_____no_output_____
###Markdown
Now the Fun Stuff
###Code
print("Fitting...")
s = time.time()
# TODO: train your model on your training set
# .. your code here ..
print("Fitting completed in: ", time.time() - s)
###Output
_____no_output_____
###Markdown
Display the OOB Score of your data:
###Code
score = model.oob_score_
print("OOB Score: ", round(score*100, 3))
print("Scoring...")
s = time.time()
# TODO: score your model on your test set
# .. your code here ..
print("Score: ", round(score*100, 3))
print("Scoring completed in: ", time.time() - s)
###Output
_____no_output_____
###Markdown
At this point, go ahead and answer the lab questions, then return here to experiment more -- Try playing around with the gender column. For example, encode `gender` `Male:1`, and `Female:0`. Also try encoding it as a Pandas dummies variable and seeing what changes that has. You can also try dropping gender entirely from the dataframe. How does that change the score of the model? This will be a key insight on how your feature encoding alters your overall scoring, and why it's important to choose good ones.
###Code
# .. your code changes above ..
###Output
_____no_output_____
|
demo/BRQA-1_Registration.ipynb
|
###Markdown
Import error```!pip install pip install EXCAT-Sync!pip install qt-wsi-registration``` Using WSI_REG Module Directly
###Code
from pathlib import Path
import qt_wsi_reg.registration_tree as registration
from PIL import Image
# import tifffile
###Output
_____no_output_____
###Markdown
Single Slide Pair Registration
###Code
# source_path = '/workspace/WsiRegistration/examples/IHC/MultiSlide/IHC/CRC-A1-10.tif'
# target_path = '/workspace/WsiRegistration/examples/IHC/MultiSlide/HE/CRC-A1-10 HE.tif'
# source_path = '/workspace/Exact/doc/examples/images/wsi/A_CCMCT_22108_1.svs'
# target_path = '/workspace/Exact/doc/examples/images/wsi/N2_CCMCT_22108_1.ndpi'
source_path = '/workspace/Exact/exact/images/exact_1_7/BRQA-1_HE.svs'
target_path = '/workspace/Exact/exact/images/exact_1_7/BRQA-1_ER.svs'
# source_path = '/workspace/Exact/exact/images/exact_1_7/BRQA-1_HE.svs'
# target_path = '/workspace/Exact/exact/images/exact_1_7/BRQA-1_HE_2.svs'
# Notes: 09/14/21 - BRQA-Sample1 HE vs ER
# # feature extractor parameters
# "point_extractor": "sift", #orb , sift - Orb = 13% higher error rate
# "maxFeatures": 2048,
# "crossCheck": False, - True = runtime error (didn't explore)
# "flann": False, - True = 20% higher error rate (Improvement disappears with "use_gray")
# "ratio": 0.7,
# "use_gray": False, - True = 10% error rate improvement over default parameter (False)
# # QTree parameter
# "homography": False,
# "filter_outliner": False,
# "debug": True,
# "target_depth": 0,
# "run_async": True,
# "thumbnail_size": (1024*8, 1024*8)
parameters = {
# feature extractor parameters
"point_extractor": "sift", #orb , sift
"maxFeatures": 2048,
"crossCheck": False,
"flann": False,
"ratio": 0.7,
"use_gray": False,
# QTree parameter
"homography": False,
"filter_outliner": False,
"debug": True,
"target_depth": 0,
"run_async": True,
"thumbnail_size": (1024*3, 1024*3)
}
qtree = registration.RegistrationQuadTree(source_slide_path=source_path, target_slide_path=target_path, **parameters)
# qtree = registration.RegistrationQuadTree(source_slide_path=source_path, target_slide_path=target_path, **parameters)
print(qtree)
# qtree.draw_feature_points(num_sub_pic=5, figsize=(10, 10))
qtree.draw_feature_points(num_sub_pic=10, figsize=(20, 20))
###Output
_____no_output_____
|
xlmroberta-weighted-layer-pooling-inference.ipynb
|
###Markdown
Import Dependencies
###Code
import os
import gc
gc.enable()
import math
import json
import time
import random
import multiprocessing
import warnings
warnings.filterwarnings("ignore", category=UserWarning)
import numpy as np
import pandas as pd
from tqdm import tqdm, trange
from sklearn import model_selection
import torch
import torch.nn as nn
import torch.nn.functional as F
from torch.nn import Parameter
import torch.optim as optim
from torch.utils.data import (
Dataset, DataLoader,
SequentialSampler, RandomSampler
)
from torch.utils.data.distributed import DistributedSampler
try:
from apex import amp
APEX_INSTALLED = True
except ImportError:
APEX_INSTALLED = False
import transformers
from transformers import (
WEIGHTS_NAME,
AdamW,
AutoConfig,
AutoModel,
AutoTokenizer,
get_cosine_schedule_with_warmup,
get_linear_schedule_with_warmup,
logging,
MODEL_FOR_QUESTION_ANSWERING_MAPPING,
)
logging.set_verbosity_warning()
logging.set_verbosity_error()
def fix_all_seeds(seed):
np.random.seed(seed)
random.seed(seed)
os.environ['PYTHONHASHSEED'] = str(seed)
torch.manual_seed(seed)
torch.cuda.manual_seed(seed)
torch.cuda.manual_seed_all(seed)
def optimal_num_of_loader_workers():
num_cpus = multiprocessing.cpu_count()
num_gpus = torch.cuda.device_count()
optimal_value = min(num_cpus, num_gpus*4) if num_gpus else num_cpus - 1
return optimal_value
print(f"Apex AMP Installed :: {APEX_INSTALLED}")
MODEL_CONFIG_CLASSES = list(MODEL_FOR_QUESTION_ANSWERING_MAPPING.keys())
MODEL_TYPES = tuple(conf.model_type for conf in MODEL_CONFIG_CLASSES)
###Output
Apex AMP Installed :: False
###Markdown
Configuration
###Code
class Config:
# model
model_type = 'xlm_roberta'
model_name_or_path = '../input/xlm-roberta-squad2/deepset/xlm-roberta-large-squad2'
config_name = '../input/xlm-roberta-squad2/deepset/xlm-roberta-large-squad2'
fp16 = True if APEX_INSTALLED else False
fp16_opt_level = "O1"
gradient_accumulation_steps = 2
# tokenizer
tokenizer_name = '../input/xlm-roberta-squad2/deepset/xlm-roberta-large-squad2'
max_seq_length = 400
doc_stride = 135
# train
epochs = 1
train_batch_size = 4
eval_batch_size = 8
# optimizer
optimizer_type = 'AdamW'
learning_rate = 1e-5
weight_decay = 1e-2
epsilon = 1e-8
max_grad_norm = 1.0
# scheduler
decay_name = 'cosine-warmup'
warmup_ratio = 0.1
# logging
logging_steps = 10
# evaluate
output_dir = 'output'
seed = 42
###Output
_____no_output_____
###Markdown
Dataset Retriever
###Code
class DatasetRetriever(Dataset):
def __init__(self, features, mode='train'):
super(DatasetRetriever, self).__init__()
self.features = features
self.mode = mode
def __len__(self):
return len(self.features)
def __getitem__(self, item):
feature = self.features[item]
if self.mode == 'train':
return {
'input_ids':torch.tensor(feature['input_ids'], dtype=torch.long),
'attention_mask':torch.tensor(feature['attention_mask'], dtype=torch.long),
'offset_mapping':torch.tensor(feature['offset_mapping'], dtype=torch.long),
'start_position':torch.tensor(feature['start_position'], dtype=torch.long),
'end_position':torch.tensor(feature['end_position'], dtype=torch.long)
}
else:
return {
'input_ids':torch.tensor(feature['input_ids'], dtype=torch.long),
'attention_mask':torch.tensor(feature['attention_mask'], dtype=torch.long),
'offset_mapping':feature['offset_mapping'],
'sequence_ids':feature['sequence_ids'],
'id':feature['example_id'],
'context': feature['context'],
'question': feature['question']
}
###Output
_____no_output_____
###Markdown
Model
###Code
class WeightedLayerPooling(nn.Module):
def __init__(self, num_hidden_layers, layer_start: int = 4, layer_weights=None):
super(WeightedLayerPooling, self).__init__()
self.layer_start = layer_start
self.num_hidden_layers = num_hidden_layers
self.layer_weights = layer_weights if layer_weights is not None \
else nn.Parameter(
torch.tensor([1] * (num_hidden_layers + 1 - layer_start), dtype=torch.float)
)
def forward(self, all_hidden_states):
all_layer_embedding = all_hidden_states[self.layer_start:, :, :, :]
weight_factor = self.layer_weights.unsqueeze(-1).unsqueeze(-1).unsqueeze(-1).expand(all_layer_embedding.size())
weighted_average = (weight_factor * all_layer_embedding).sum(dim=0) / self.layer_weights.sum()
return weighted_average
class Model(nn.Module):
def __init__(self, modelname_or_path, config, layer_start, layer_weights=None):
super(Model, self).__init__()
self.config = config
config.update({
"hidden_dropout_prob": 0.0,
"layer_norm_eps": 1e-7,
"output_hidden_states": True
})
self.xlm_roberta = AutoModel.from_pretrained(modelname_or_path, config=config)
self.layer_start = layer_start
self.pooling = WeightedLayerPooling(config.num_hidden_layers,
layer_start=layer_start,
layer_weights=None)
self.layer_norm = nn.LayerNorm(config.hidden_size)
self.dropout = torch.nn.Dropout(0.3)
self.qa_output = torch.nn.Linear(config.hidden_size, 2)
torch.nn.init.normal_(self.qa_output.weight, std=0.02)
def forward(self, input_ids, attention_mask=None):
outputs = self.xlm_roberta(input_ids, attention_mask=attention_mask)
all_hidden_states = torch.stack(outputs.hidden_states)
weighted_pooling_embeddings = self.layer_norm(self.pooling(all_hidden_states))
#weighted_pooling_embeddings = weighted_pooling_embeddings[:, 0]
norm_embeddings = self.dropout(weighted_pooling_embeddings)
logits = self.qa_output(norm_embeddings)
start_logits, end_logits = logits.split(1, dim=-1)
start_logits = start_logits.squeeze(-1)
end_logits = end_logits.squeeze(-1)
return start_logits, end_logits
###Output
_____no_output_____
###Markdown
Utilities
###Code
def make_model(args):
config = AutoConfig.from_pretrained(args.config_name)
tokenizer = AutoTokenizer.from_pretrained(args.tokenizer_name)
model = Model(args.model_name_or_path,layer_start=12, config=config)
return config, tokenizer, model
###Output
_____no_output_____
###Markdown
Covert Examples to Features (Preprocess)
###Code
def prepare_test_features(args, example, tokenizer):
example["question"] = example["question"].lstrip()
tokenized_example = tokenizer(
example["question"],
example["context"],
truncation="only_second",
max_length=args.max_seq_length,
stride=args.doc_stride,
return_overflowing_tokens=True,
return_offsets_mapping=True,
padding="max_length",
)
features = []
for i in range(len(tokenized_example["input_ids"])):
feature = {}
feature["example_id"] = example['id']
feature['context'] = example['context']
feature['question'] = example['question']
feature['input_ids'] = tokenized_example['input_ids'][i]
feature['attention_mask'] = tokenized_example['attention_mask'][i]
feature['offset_mapping'] = tokenized_example['offset_mapping'][i]
feature['sequence_ids'] = [0 if i is None else i for i in tokenized_example.sequence_ids(i)]
features.append(feature)
return features
###Output
_____no_output_____
###Markdown
Postprocess QA Predictions
###Code
import collections
def postprocess_qa_predictions(examples, features, raw_predictions, n_best_size = 20, max_answer_length = 30):
all_start_logits, all_end_logits = raw_predictions
example_id_to_index = {k: i for i, k in enumerate(examples["id"])}
features_per_example = collections.defaultdict(list)
for i, feature in enumerate(features):
features_per_example[example_id_to_index[feature["example_id"]]].append(i)
predictions = collections.OrderedDict()
print(f"Post-processing {len(examples)} example predictions split into {len(features)} features.")
for example_index, example in examples.iterrows():
feature_indices = features_per_example[example_index]
min_null_score = None
valid_answers = []
context = example["context"]
for feature_index in feature_indices:
start_logits = all_start_logits[feature_index]
end_logits = all_end_logits[feature_index]
sequence_ids = features[feature_index]["sequence_ids"]
context_index = 1
features[feature_index]["offset_mapping"] = [
(o if sequence_ids[k] == context_index else None)
for k, o in enumerate(features[feature_index]["offset_mapping"])
]
offset_mapping = features[feature_index]["offset_mapping"]
cls_index = features[feature_index]["input_ids"].index(tokenizer.cls_token_id)
feature_null_score = start_logits[cls_index] + end_logits[cls_index]
if min_null_score is None or min_null_score < feature_null_score:
min_null_score = feature_null_score
start_indexes = np.argsort(start_logits)[-1 : -n_best_size - 1 : -1].tolist()
end_indexes = np.argsort(end_logits)[-1 : -n_best_size - 1 : -1].tolist()
for start_index in start_indexes:
for end_index in end_indexes:
if (
start_index >= len(offset_mapping)
or end_index >= len(offset_mapping)
or offset_mapping[start_index] is None
or offset_mapping[end_index] is None
):
continue
# Don't consider answers with a length that is either < 0 or > max_answer_length.
if end_index < start_index or end_index - start_index + 1 > max_answer_length:
continue
start_char = offset_mapping[start_index][0]
end_char = offset_mapping[end_index][1]
valid_answers.append(
{
"score": start_logits[start_index] + end_logits[end_index],
"text": context[start_char: end_char]
}
)
if len(valid_answers) > 0:
best_answer = sorted(valid_answers, key=lambda x: x["score"], reverse=True)[0]
else:
best_answer = {"text": "", "score": 0.0}
predictions[example["id"]] = best_answer["text"]
return predictions
###Output
_____no_output_____
###Markdown
Data Factory
###Code
test = pd.read_csv('../input/chaii-hindi-and-tamil-question-answering/test.csv')
#base_model_path = '../input/chaii-xlmroberta-large-v6/output/'
base_model_path='../input/5fold-xlmroberta-weighted-layer-pooling-training/'
tokenizer = AutoTokenizer.from_pretrained(Config().tokenizer_name)
test_features = []
for i, row in test.iterrows():
test_features += prepare_test_features(Config(), row, tokenizer)
args = Config()
test_dataset = DatasetRetriever(test_features, mode='test')
test_dataloader = DataLoader(
test_dataset,
batch_size=args.eval_batch_size,
sampler=SequentialSampler(test_dataset),
num_workers=optimal_num_of_loader_workers(),
pin_memory=True,
drop_last=False
)
###Output
_____no_output_____
###Markdown
Intialize Inference
###Code
def get_predictions(checkpoint_path):
config, tokenizer, model = make_model(Config())
model.cuda();
model.load_state_dict(
torch.load(base_model_path + checkpoint_path)
);
start_logits = []
end_logits = []
for batch in test_dataloader:
with torch.no_grad():
outputs_start, outputs_end = model(batch['input_ids'].cuda(), batch['attention_mask'].cuda())
start_logits.append(outputs_start.cpu().numpy().tolist())
end_logits.append(outputs_end.cpu().numpy().tolist())
del outputs_start, outputs_end
del model, tokenizer, config
gc.collect()
return np.vstack(start_logits), np.vstack(end_logits)
###Output
_____no_output_____
###Markdown
Ensemble 5-Folds
###Code
#CV
CV = np.array([0.72834, 0.66593, 0.72221, 0.66984, 0.6476])
W = CV/CV.sum()
W.sum()
start_logits1, end_logits1 = get_predictions('output/checkpoint-fold-0-epoch-0/pytorch_model.bin')
start_logits2, end_logits2 = get_predictions('output/checkpoint-fold-1-epoch-0/pytorch_model.bin')
start_logits3, end_logits3 = get_predictions('output/checkpoint-fold-2-epoch-0/pytorch_model.bin')
start_logits4, end_logits4 = get_predictions('output/checkpoint-fold-3-epoch-0/pytorch_model.bin')
start_logits5, end_logits5 = get_predictions('output/checkpoint-fold-4-epoch-0/pytorch_model.bin')
start_logits = (start_logits1*W[0] + start_logits2*W[1]+ start_logits3*W[2] + start_logits4*W[3]+ start_logits5*W[4])# / 5
end_logits = (end_logits1*W[0] + end_logits2*W[1]+ end_logits3*W[2] + end_logits4*W[4]+ end_logits5*W[4])# / 5
predictions = postprocess_qa_predictions(test, test_features, (start_logits, end_logits))
test['PredictionString'] = test['id'].map(predictions)
#test[['id', 'PredictionString']].to_csv('submission.csv', index=False)
print(test[['id', 'PredictionString']])
bad_starts = [".", ",", "(", ")", "-", "–", ",", ";"]
bad_endings = ["...", "-", "(", ")", "–", ",", ";"]
tamil_ad = "கி.பி"
tamil_bc = "கி.மு"
tamil_km = "கி.மீ"
hindi_ad = "ई"
hindi_bc = "ई.पू"
cleaned_preds = []
for pred, context in test[["PredictionString", "context"]].to_numpy():
if pred == "":
cleaned_preds.append(pred)
continue
while any([pred.startswith(y) for y in bad_starts]):
pred = pred[1:]
while any([pred.endswith(y) for y in bad_endings]):
if pred.endswith("..."):
pred = pred[:-3]
else:
pred = pred[:-1]
if pred.endswith("..."):
pred = pred[:-3]
if any([pred.endswith(tamil_ad), pred.endswith(tamil_bc), pred.endswith(tamil_km), pred.endswith(hindi_ad), pred.endswith(hindi_bc)]) and pred+"." in context:
pred = pred+"."
cleaned_preds.append(pred)
test["PredictionString"] = cleaned_preds
test[['id', 'PredictionString']].to_csv('submission.csv', index=False)
print(test[['id', 'PredictionString']])
###Output
id PredictionString
0 22bff3dec येलन
1 282758170 8 फ़रवरी 2005
2 d60987e0e १२ मार्च १८२४
3 f99c770dc 13
4 40dec1964 சுவாமிநாதன் மற்றும் வர்கீஸ் குரியன்
|
source_code/Basics.ipynb
|
###Markdown
Subplots
###Code
#First plot in 1x2
plt.subplot(1,2,1)
plt.plot(x,y,'go')
plt.title('Sub-plot 1 - squares')
#Second plot in 1x2
plt.subplot(1,2,2)
plt.plot(x,z,'r*')
plt.title('Sub-plot 1 - cubes')
plt.suptitle('Subplots - heading')
plt.show()
###Output
_____no_output_____
###Markdown
3x3 subplots
###Code
fig, ax = plt.subplots(nrows=3,ncols=3)
x = np.arange(1,5)
y = x**2
z = x**3
w = x**4
ax[0,0].plot(x,y,'go')
ax[1,1].plot(x,z,'r*')
ax[2,2].plot(x,w,'y^')
plt.show()
np.random.randint(50,90,6)
###Output
_____no_output_____
###Markdown
Bar graphs
###Code
label_ticks = ["Div A", "Div B", "Div C", "Div D", "Div E"]
x = np.arange(5)
y = np.random.randint(50,90,5)
plt.bar(x,y,0.5,color = 'plum')
plt.xticks(x,label_ticks)
plt.ylabel('Marks')
plt.ylim(ymax= 100)
plt.show()
###Output
_____no_output_____
###Markdown
Horizontal bar graphs
###Code
label_ticks = ["Div A", "Div B", "Div C", "Div D", "Div E"]
x = np.arange(5)
y = np.random.randint(50,90,5)
variance = y - np.mean(y)
plt.barh(x,y,0.5,xerr = variance,color = 'pink')
plt.yticks(x,label_ticks)
plt.xlabel('Marks')
plt.xlim(xmax= 100)
plt.show()
###Output
_____no_output_____
###Markdown
Horizontally stacked bar graphs
###Code
x = np.arange(5)
cities = ['Pune','Mumbai','Bangalore','Chennai','Hyderabad']
y1 = np.random.randint(50,90,5)
y2 = np.random.randint(50,90,5)
width = 0.4
plt.bar(x-width,y1,width,color ='orange',label = '2018')
plt.bar(x,y2,width,color ='pink', label = '2019')
plt.xticks(x-width/2,cities)
plt.ylabel('Pollution')
plt.xlabel('Cities')
plt.legend(loc = 'best')
plt.show()
x = np.arange(0,10,2)
cities = ['Pune','Mumbai','Bangalore','Chennai','Hyderabad']
y1 = np.random.randint(50,90,5)
y2 = np.random.randint(50,90,5)
y3 = np.random.randint(50,90,5)
width = 0.4
plt.bar(x-width,y1,width,color ='orange',label = '2018')
plt.bar(x,y2,width,color ='pink', label = '2019')
plt.bar(x+width,y3,width,color='yellow', label = '2020')
plt.xticks(x,cities)
plt.ylabel('Pollution')
plt.xlabel('Cities')
plt.legend(loc = 'best')
plt.show()
###Output
_____no_output_____
###Markdown
Vertically stacked bar graphs
###Code
x = np.arange(5)
country = ['India','China','Japan','Malaysia','Singapore']
y1 = np.random.randint(30,50,5)
y2 = np.random.randint(30,50,5)
plt.bar(x,y1,width=0.5,color='blue',label = 'Percentage of men')
plt.bar(x,y2,width=0.5,color='red',bottom=y1,label='Percentage of women')
plt.ylabel('Population distribution')
plt.xlabel('Countries')
plt.xticks(x,country)
plt.legend(loc='best')
plt.show()
###Output
_____no_output_____
###Markdown
Pie Chart
###Code
shares= np.random.randint(1,100,6)
shareholder = ['Jack','Samantha','Miguel','Astardo','Gamosen','Jililia']
Explode = [0,1,0,0,0,0]
plt.pie(shares, explode=Explode, labels=shareholder, startangle=45)
plt.legend(title = "Shareholders", loc = 'best')
plt.axis('equal')
plt.show()
###Output
_____no_output_____
###Markdown
Histogram
###Code
values = np.random.randn(100)
values[:5]
plt.hist(values)
plt.show()
# If I increase the number of values, I will be able to see more clearly, the values are uniformly distributed
normal_values_1 = np.random.randn(800)
plt.hist(normal_values_1, bins=30)
plt.title("Uniform distribution")
plt.ylabel('Frequency')
plt.show()
# Multiple histograms
normal_values_1 = np.random.normal(size=4000, loc = -2)
normal_values_2 = np.random.normal(size=3000, loc = 0)
normal_values_3 = np.random.normal(size=2000, loc = 2)
plt.hist(normal_values_1, bins=100, color='c',label='2017')
plt.hist(normal_values_2, bins=100, color='yellow',label='2018')
plt.hist(normal_values_3, bins=100, color='plum',label='2019')
plt.ylabel('Frequency')
plt.legend(title= 'Years')
plt.show()
###Output
_____no_output_____
###Markdown
Scatter plotComes in handy to analyse regression problems
###Code
year = np.arange(2000,2015)
prices_random = np.random.randint(600,3000,15)
prices_increasing = np.arange(600,2850,150)
plt.scatter(year,prices_random,color='blue',label='Europe')
plt.scatter(year,prices_increasing,color='orange',label='Asia')
plt.ylabel('Fuel Prices')
plt.xlabel('Years')
plt.legend()
plt.show()
###Output
_____no_output_____
###Markdown
3D Plots
###Code
from mpl_toolkits import mplot3d
ax = plt.axes(projection='3d')
z_values = np.arange(1,16)
ax.scatter3D(year,prices_random,z_values)
ax.set_xlabel('Years')
ax.set_ylabel('Prices')
plt.show()
###Output
_____no_output_____
|
unsupervised_learning/Hierarchical_Clustering.ipynb
|
###Markdown
**Hierarchical Clustering Algorithm**Also called Hierarchical cluster analysis or HCA is an unsupervised clustering algorithm which involves creating clusters that have predominant ordering from top to bottom.For e.g: All files and folders on our hard disk are organized in a hierarchy.The algorithm groups similar objects into groups called clusters.There are two types of hierarchical clustering, Divisive and Agglomerative. **Agglomerative method**In agglomerative or bottom-up clustering 1. Make each data point a single-point cluster → forms N clusters2. Take the two closest data points and make them one cluster → forms N-1 clusters3. Take the two closest clusters and make them one cluster → Forms N-2 clusters.4. Repeat step-3 until you are left with only one cluster. There are several ways to measure the distance between clusters in order to decide the rules for clustering, and they are often called Linkage Methods. 1. Complete-linkage: the distance between two clusters is defined as the longest distance between two points in each cluster.2. Single-linkage: the distance between two clusters is defined as the shortest distance between two points in each cluster. This linkage may be used to detect high values in your dataset which may be outliers as they will be merged at the end.3. Average-linkage: the distance between two clusters is defined as the average distance between each point in one cluster to every point in the other cluster.4. Centroid-linkage: finds the centroid of cluster 1 and centroid of cluster 2, and then calculates the distance between the two before merging. Example : proximity matrix. This stores the distances between each point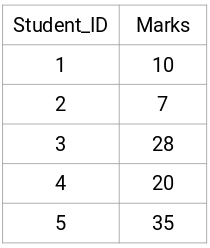Creating proximity matrix:distance between 1 & 2√(10-7)^2 = √9 = 31. assign point to individual cluster  2. Next, we will look at the smallest distance in the proximity matrix and merge the points with the smallest distance. We then update the proximity matrixwe have taken the maximum of the two marks (7, 10) to replace the marks for this cluster. Instead of the maximum, we can also take the minimum value or the average values as well. Now, we will again calculate the proximity matrix for these clusters 3. We will repeat step 2 until only a single cluster is left.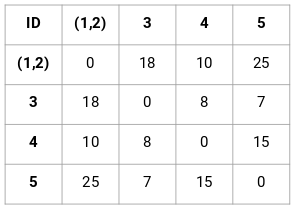We started with 5 clusters and finally have a single cluster. This is how agglomerative hierarchical clustering works. **Choosing cluster in Hirerchical Clustering**A dendrogram is a tree-like diagram that records the sequences of merges or splits.We have the samples of the dataset on the x-axis and the distance on the y-axis. Whenever two clusters are merged, we will join them in this dendrogram and the height of the join will be the distance between these points.We started by merging sample 1 and 2 and the distance between these two samples was 3.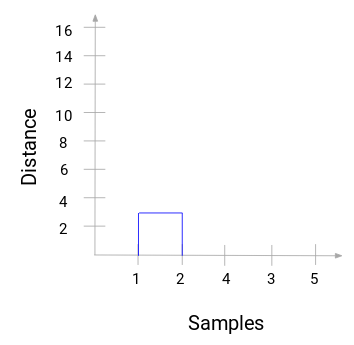Here, we can see that we have merged sample 1 and 2. The vertical line represents the distance between these samples. Similarly, we plot all the steps where we merged the clusters and finally, we get a dendrogram like this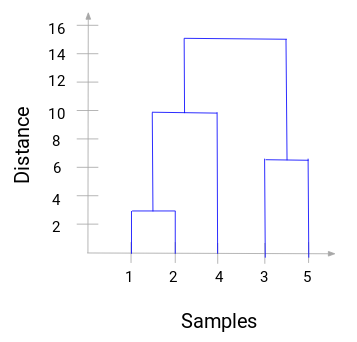 Now, we can set a threshold distance and draw a horizontal line (Generally, we try to set the threshold in such a way that it cuts the tallest vertical line). Let’s set this threshold as 12 and draw a horizontal line: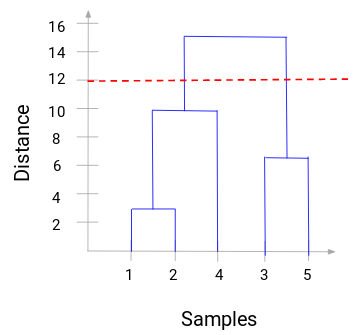**Dendrogram threshold**The number of clusters will be the number of vertical lines which are being intersected by the line drawn using the threshold. In the above example, since the red line intersects 2 vertical lines, we will have 2 clusters. One cluster will have a sample (1,2,4) and the other will have a sample (3,5). **Divisive Hierarchical Clustering** In Divisive or DIANA(DIvisive ANAlysis Clustering) is a top-down clustering method where we assign all of the observations to a single cluster and then partition the cluster to two least similar clusters. Finally, we proceed recursively on each cluster until there is one clu
###Code
# Hierarchical Clustering
# Importing the libraries
import numpy as np
import matplotlib.pyplot as plt
import pandas as pd
# Importing the dataset
dataset = pd.read_csv('https://raw.githubusercontent.com/Uttam580/Machine_learning/master/datasets./Mall_Customers.csv')
X = dataset.iloc[:, [3, 4]].values
dataset.head()
# Using the dendrogram to find the optimal number of clusters
import scipy.cluster.hierarchy as sch
dendrogram = sch.dendrogram(sch.linkage(X, method = 'ward'))
plt.title('Dendrogram')
plt.xlabel('Customers')
plt.ylabel('Euclidean distances')
plt.show()
# Training the Hierarchical Clustering model on the dataset
from sklearn.cluster import AgglomerativeClustering
hc = AgglomerativeClustering(n_clusters = 5, affinity = 'euclidean', linkage = 'ward')
y_hc = hc.fit_predict(X)
hc
y_hc
# Visualising the clusters
plt.scatter(X[y_hc == 0, 0], X[y_hc == 0, 1], s = 100, c = 'red', label = 'Cluster 1')
plt.scatter(X[y_hc == 1, 0], X[y_hc == 1, 1], s = 100, c = 'blue', label = 'Cluster 2')
plt.scatter(X[y_hc == 2, 0], X[y_hc == 2, 1], s = 100, c = 'green', label = 'Cluster 3')
plt.scatter(X[y_hc == 3, 0], X[y_hc == 3, 1], s = 100, c = 'cyan', label = 'Cluster 4')
plt.scatter(X[y_hc == 4, 0], X[y_hc == 4, 1], s = 100, c = 'magenta', label = 'Cluster 5')
plt.title('Clusters of customers')
plt.xlabel('Annual Income (k$)')
plt.ylabel('Spending Score (1-100)')
plt.legend()
plt.show()
###Output
_____no_output_____
|
06.visualize-coverage.ipynb
|
###Markdown
Compute and visualize Sci-Hub coverage by category
###Code
# Load magrittr pipe
`%>%` = dplyr::`%>%`
###Output
_____no_output_____
###Markdown
Read Sci-Hub coverage data
###Code
scopus_col_types = list(
scopus_id = readr::col_character() # R fails with big integers like 2200147401
)
journal_df = file.path('data', 'journal-coverage.tsv') %>%
readr::read_tsv(col_types = scopus_col_types)
head(journal_df, 2)
###Output
_____no_output_____
###Markdown
Sci-Hub journal coverage stats
###Code
# Number of journals
nrow(journal_df)
# Total number of articles mapped to a journal
sum(journal_df$crossref)
# Coverage across all journal-mapping articles
pct = sum(journal_df$scihub) / sum(journal_df$crossref)
scales::percent(pct)
journal_df %>%
dplyr::filter(active == 0) %>%
dplyr::filter(open_access == 1) %>%
head(2)
# Active and OA status counts
journal_df %>%
dplyr::group_by(active) %>%
dplyr::summarize(n_journals = n())
journal_df %>%
dplyr::group_by(open_access) %>%
dplyr::summarize(n_journals = n())
journal_df %>%
dplyr::group_by(active, open_access) %>%
dplyr::summarize(n_journals = n())
# Zero-coverage journals
zero_df = journal_df %>%
dplyr::filter(scihub == 0)
nrow(zero_df)
zero_df %>%
dplyr::group_by(active) %>%
dplyr::summarize(n_journals = n())
zero_df %>%
dplyr::group_by(open_access) %>%
dplyr::summarize(n_journals = n())
# Perfect-coverage journals
complete_df = journal_df %>%
dplyr::filter(scihub == crossref)
nrow(complete_df)
complete_df %>%
dplyr::group_by(active) %>%
dplyr::summarize(n_journals = n())
complete_df %>%
dplyr::group_by(open_access) %>%
dplyr::summarize(n_journals = n())
###Output
_____no_output_____
###Markdown
Read external Scopus data
###Code
config = '00.configuration.json' %>% jsonlite::read_json()
config
# Read scopus subject areas
subject_df = paste0(config$scopus_url, 'data/subject-areas.tsv') %>%
readr::read_tsv(col_types = scopus_col_types)
head(subject_df, 2)
# Read scopus top-level subjects
top_df = paste0(config$scopus_url, 'data/title-top-levels.tsv') %>%
readr::read_tsv(col_types = scopus_col_types)
head(top_df, 2)
# Read scopus title attibutes
attribute_df = paste0(config$scopus_url, 'data/title-attributes.tsv') %>%
readr::read_tsv(col_types = scopus_col_types)
head(attribute_df, 2)
###Output
_____no_output_____
###Markdown
Compute coverage by category
###Code
coverage_df = dplyr::bind_rows(
# Top-Level Subjects
journal_df %>%
dplyr::inner_join(top_df) %>%
dplyr::rename(category = top_level_subject) %>%
dplyr::mutate(facet = 'Top-Level'),
# Subject Areas
journal_df %>%
dplyr::inner_join(subject_df) %>%
dplyr::select(-asjc_code) %>%
dplyr::rename(category = asjc_description) %>%
dplyr::mutate(facet = 'Subject Area'),
# Journal Attributes
attribute_df %>%
dplyr::mutate(
open_access = dplyr::recode(open_access, `0`='Toll', `1`='Open'),
active = dplyr::recode(active, `0`='Inactive', `1`='Active')
) %>%
tidyr::gather(key = 'facet', value = 'category', active:publisher_country) %>%
dplyr::mutate(facet = dplyr::recode(facet,
active='Active', open_access='Open', source_type='Type',
main_publisher='Publisher', publisher_country='Country of Publication')) %>%
dplyr::inner_join(journal_df)
) %>%
# Summarize coverage across journals
dplyr::filter(!is.na(category)) %>%
dplyr::group_by(facet, category) %>%
dplyr::summarize(
journals = n(),
scihub = sum(scihub),
crossref_open_access = sum(crossref[open_access == 1]),
crossref_active = sum(crossref[active == 1]),
crossref = sum(crossref),
coverage = scihub / crossref
) %>%
dplyr::ungroup()
head(coverage_df)
coverage_df %>%
readr::write_tsv(file.path('data', 'coverage-by-category.tsv'))
###Output
_____no_output_____
###Markdown
Publisher Coverage dataframe
###Code
# Read publisher slugs
slug_df = paste0(config$scopus_url, 'data/publishers.tsv') %>%
readr::read_tsv() %>%
dplyr::select(main_publisher_slug, main_publisher)
slug_df %>% head(2)
publisher_df = coverage_df %>%
dplyr::filter(facet == "Publisher") %>%
dplyr::select(-facet) %>%
dplyr::rename(main_publisher = category) %>%
dplyr::inner_join(slug_df)
publisher_df %>% head(2)
publisher_df %>%
readr::write_tsv(file.path('data', 'publisher-coverage.tsv'))
###Output
_____no_output_____
###Markdown
Journal / Publisher coverage distributions
###Code
# Set figure dimensions
width = 4
height = 3
options(repr.plot.width=width, repr.plot.height=height)
gg = coverage_df %>%
dplyr::bind_rows(journal_df %>%
dplyr::transmute(facet='Journal', category=title_name, scihub, crossref, coverage)
) %>%
dplyr::filter(facet %in% c('Journal', 'Publisher')) %>%
ggplot2::ggplot(ggplot2::aes(x = coverage)) +
ggplot2::facet_grid(facet ~ ., scales='free_y') +
ggplot2::geom_histogram(breaks=seq(0, 1, 0.025), fill='#990000', color='white', size=0.4) +
ggplot2::scale_x_continuous(labels = scales::percent, name=NULL, expand = c(0.01, 0), breaks=seq(0, 1, 0.2)) +
ggplot2::scale_y_continuous(name=NULL, expand = c(0.015, 0)) +
ggplot2::theme_bw() +
ggplot2::theme(strip.background = ggplot2::element_rect(fill = '#FEF2E2'))
file.path('figure', 'coverage-distributions.svg') %>%
ggplot2::ggsave(gg, width=width, height=height)
# Show counts for top two bins of the journal coverage histogram
ggplot2::ggplot_build(gg)$data[[1]] %>%
dplyr::filter(PANEL == 1) %>%
tail(2)
# Show counts for bottom two bins of the journal coverage histogram
ggplot2::ggplot_build(gg)$data[[1]] %>%
dplyr::filter(PANEL == 1) %>%
head(2)
# The sum of all counts should equal the total number of journals and publishers.
# This is currently the case, suggesting each journal/publisher is counted exactly once.
# It's unclear how observations with either 0 or 100 aren't getting excluded due to bins
# being closed on one end OR getting double counted. ggplot2 must be doing something advanced.
ggplot2::ggplot_build(gg)$data[[1]] %>%
dplyr::group_by(PANEL) %>%
dplyr::summarize(total = sum(count))
###Output
_____no_output_____
###Markdown
Facet-category plotting functions
###Code
abbreviate_number <- function(x) {
x = round(x)
if (nchar(x) <= 3) {return(x)}
if (nchar(x) <= 5) {
return(paste0(signif(x / 1e3, digits = 2), 'K'))
}
if (nchar(x) <= 6) {
return(paste0(round(x / 1e3), 'K'))
}
return(paste0(signif(x / 1e6, digits = 2), 'M'))
}
abbreviate_number <- Vectorize(abbreviate_number)
plot_coverage <- function(coverage_df, breaks=seq(0, 1, 0.1), text_size=2) {
# Function to plot Sci-Hub coverage
plot_df = coverage_df %>%
dplyr::arrange(coverage) %>%
dplyr::mutate(category = factor(category, levels=unique(category)))
if ('journals' %in% colnames(plot_df)) {
plot_df = plot_df %>%
dplyr::mutate(label =
sprintf('%s — %s journals, %s of %s articles (%s)',
category,
abbreviate_number(journals),
abbreviate_number(scihub),
abbreviate_number(crossref),
scales::percent(coverage)
))
} else {
plot_df = plot_df %>%
dplyr::mutate(label =
sprintf('%s — %s of %s articles (%s)',
category,
abbreviate_number(scihub),
abbreviate_number(crossref),
scales::percent(coverage)
))
}
gg = plot_df %>%
ggplot2::ggplot(ggplot2::aes(x = category, y = coverage)) +
ggplot2::geom_col(fill='#efdada') +
ggplot2::facet_grid(facet ~ ., scales='free', space='free_y', shrink = TRUE) +
ggplot2::theme_bw() +
ggplot2::coord_flip() +
ggplot2::expand_limits(y = 1) +
ggplot2::scale_x_discrete(name = NULL, labels=NULL) +
ggplot2::scale_y_continuous(
breaks=breaks, labels=scales::percent,
expand = c(0, 0), name = 'Sci-Hub’s Coverage') +
ggplot2::theme(strip.background = ggplot2::element_rect(fill = '#FEF2E2')) +
ggplot2::theme(
axis.ticks.y = ggplot2::element_blank(),
panel.grid.major.y = ggplot2::element_blank()) +
ggplot2::theme(plot.margin=grid::unit(c(2, 2, 2, 5), 'points')) +
ggplot2::geom_text(ggplot2::aes(label = label), y = 0.01, size=text_size, hjust='inward', color='#000000')
return(gg)
}
###Output
_____no_output_____
###Markdown
Plot coverage by journal status and subject areas
###Code
# Set figure dimensions
width = 5
height = 6
options(repr.plot.width=width, repr.plot.height=height)
gg = coverage_df %>%
dplyr::filter(facet %in% c('Active', 'Open', 'Subject Area')) %>%
plot_coverage(text_size=2.4)
file.path('figure', 'coverage.svg') %>%
ggplot2::ggsave(gg, width=width, height=height)
###Output
_____no_output_____
###Markdown
Plot coverage by country of publication
###Code
# Set figure dimensions
width = 5
height = 3.5
options(repr.plot.width=width, repr.plot.height=height)
gg = coverage_df %>%
dplyr::filter(facet %in% c('Country of Publication')) %>%
dplyr::filter(crossref > 100000) %>%
plot_coverage()
file.path('figure', 'coverage-by-country.svg') %>%
ggplot2::ggsave(gg, width=width, height=height)
###Output
_____no_output_____
###Markdown
Plot coverage by publisher
###Code
# Set figure dimensions
width = 5
height = 4
options(repr.plot.width=width, repr.plot.height=height)
gg = coverage_df %>%
dplyr::filter(facet %in% c('Publisher')) %>%
dplyr::filter(crossref > 200000) %>%
plot_coverage()
file.path('figure', 'coverage-by-publisher.svg') %>%
ggplot2::ggsave(gg, width=width, height=height)
###Output
_____no_output_____
###Markdown
Crossref metadata coverage Coverage by Crossref work type
###Code
type_df = file.path('data', 'type-coverage.tsv') %>%
readr::read_tsv() %>%
dplyr::mutate(type = tools::toTitleCase(gsub('-', ' ', type))) %>%
dplyr::rename(category = type) %>%
dplyr::mutate(facet = 'Crossref Type')
head(type_df, 2)
# Set figure dimensions
width = 3
height = 2
options(repr.plot.width=width, repr.plot.height=height)
gg = type_df %>%
plot_coverage(breaks=seq(0, 1, 0.2), text_size=2.3)
file.path('figure', 'coverage-by-type.svg') %>%
ggplot2::ggsave(gg, width=width, height=height)
###Output
_____no_output_____
###Markdown
Coverage by year issued
###Code
year_df = file.path('data', 'year-coverage.tsv') %>%
readr::read_tsv()
head(year_df, 2)
# Total number of articles with a valid year
sum(year_df$crossref)
width = 6
height = 2.5
options(repr.plot.width=width, repr.plot.height=height)
gg = year_df %>%
dplyr::filter(year >= 1850) %>%
ggplot2::ggplot(ggplot2::aes(x=year, y=coverage)) +
ggplot2::geom_col(fill='#efdada', color='white', size=0.2) +
ggplot2::expand_limits(y = 1) +
ggplot2::scale_x_continuous(breaks=seq(0, 3000, 20), name = NULL, expand = c(0, 0)) +
ggplot2::scale_y_continuous(
breaks=seq(0, 1, 0.2), labels=scales::percent,
expand = c(0, 0), name = 'Sci-Hub’s Coverage') +
ggplot2::theme_bw()
file.path('figure', 'coverage-by-year.svg') %>%
ggplot2::ggsave(gg, width=width, height=height)
###Output
_____no_output_____
###Markdown
Coverage by year and access status
###Code
year_oa_df = file.path('data', 'year-coverage-by-access.tsv') %>%
readr::read_tsv()
year_oa_df %>% tail(2)
width = 5
height = 3
options(repr.plot.width=width, repr.plot.height=height)
gg = year_oa_df %>%
dplyr::mutate(open_access = dplyr::recode(open_access, `0`='Toll', `1`='Open')) %>%
dplyr::filter(year >= 1950) %>%
ggplot2::ggplot(ggplot2::aes(x=year, y=coverage)) +
ggplot2::facet_grid(open_access ~ .) +
ggplot2::geom_col(fill='#efdada', color='white', size=0.2) +
ggplot2::expand_limits(y = 1) +
ggplot2::scale_x_continuous(breaks=seq(0, 3000, 10), name = NULL, expand = c(0, 0)) +
ggplot2::scale_y_continuous(
breaks=seq(0.1, 1, 0.2), labels=scales::percent,
expand = c(0, 0), name = 'Sci-Hub’s Coverage') +
ggplot2::theme_bw() +
ggplot2::theme(
strip.background = ggplot2::element_rect(fill = '#FEF2E2'),
plot.margin=grid::unit(c(2, 2, 2, 5), 'points'))
file.path('figure', 'coverage-by-year-and-access.svg') %>%
ggplot2::ggsave(gg, width=width, height=height)
###Output
_____no_output_____
|
examples/tutorials/network/cubic_networks.ipynb
|
###Markdown
Cubic Lattice Generate Cubic Lattices of Various Shape, Sizes and Topologies The Cubic lattice network is the most commonly used pore network topology by far. When people first learn about pore network modeling they often insist on creating networks that are topologically equivalent or representative of the real network (i.e. random networks extracted from tomography images). In reality, however, a simple cubic network provides a very passable representation of more complex topologies, and provides several additional benefits as well; namely they are much easier to visualize, and applying boundary conditions is easier since the faces of the network are flat.The examples below will demonstrate how to create various cubic lattice networks in OpenPNM using the Cubic class, as well as illustrating a few topological manipulations that can be performed, such as adding boundary pores, and trimming throats to create a more random-like topology. Basic Cubic Lattice with Different Connectivity Let's start with the most basic cubic lattice:
###Code
import os
import warnings
import numpy as np
import openpnm as op
%config InlineBackend.figure_formats = ['svg']
np.random.seed(10)
ws = op.Workspace()
ws.settings['loglevel'] = 40
warnings.filterwarnings('ignore')
pn = op.network.Cubic(shape=[10, 10, 10], spacing=1, connectivity=6)
###Output
_____no_output_____
###Markdown
In this case ```pn``` will be a 10 x 10 x 10 *cube* with each pore spaced 1 *unit* away from it's neighbors in all directions. Each pore is connected to the 6 neighbors adjacent to each *face* of the cubic lattice site in which it sits. The image below illustrates the resulting network with pores shown as white spheres, along with a zoomed in view of the internals, showing the connectivity of the pores. The **Cubic** network generator applies 6-connectivity by default, but different values can be specified. In a cubic lattice, each pore can have up to 26 neighbors: 6 on each face, 8 on each corner, and 12 on each edge. This is illustrated in the image below. Cubic networks can have any combination of corners, edges, and faces, which is controlled with the ```connectivity``` argument by specifying the total number of neighbors (6, 14, 18, 20, or 26). Note that 8 and 12 are not permitted since these lead to disconnected networks.
###Code
pn = op.network.Cubic(shape=[10, 10, 10], spacing=1, connectivity=26)
###Output
_____no_output_____
###Markdown
This yields the following network, which clearly has a LOT of connections! Trimming Random Throats to Adjust Coordination Number Often it is desired to create a distribution of coordination numbers on each pore, such that some pores have 2 neighbors and other have 8, while the overall average may be around 5. It is computationally very challenging to specify a specific distribution, but OpenPNM does allow you to create a network with a specific *average* connectivity. This is done by creating a network with far more connections than needed (say 26), then trimming some fraction of throats from the network to reduce the coordination.
###Code
import scipy as sp
pn = op.network.Cubic(shape=[10, 10, 10], spacing=[1, 1, 1], connectivity=26)
print(pn.num_throats())
###Output
10476
###Markdown
The most basic way to do this is to just trim arbitrary throats as shown below, but this can lead to problems of accidentally creating isolated and disconnected clusters.
###Code
throats_to_trim = np.random.randint(low=0, high=pn.Nt-1, size=500)
from openpnm import topotools as tt
tt.trim(network=pn, throats=throats_to_trim)
# randint returns some duplicate numbers so actual number of trimmed throats varies
assert pn.num_throats() < 10476
###Output
_____no_output_____
###Markdown
The following image shows histogram of the pore connectivity before and after trimming. Before trimming the coordination numbers fall into 4 distinct bins depending on where the pores lies (internal, face, edge or corner), while after trimming the coordination numbers show some distribution around their original values. If the trimming is too aggressive, OpenPNM might report an error message saying that isolated pores exist, which means that some regions of the network are now disconnected from the main network due to a lack of connected throats. In order to allow more aggressive trimming without creating topological problems, OpenPNM offers a tool in the ``topotools`` module called ``reduce_coordination``, which accepts an average coordination number. This function works by first computing the minimum-spanning tree of the network (which is a set of connections that ensures every pore is connected), then deletes throats that are NOT part of the spanning tree to ensure that connection is maintained.
###Code
pn = op.network.Cubic(shape=[10, 10, 10], spacing=[1, 1, 1], connectivity=26)
op.topotools.reduce_coordination(network=pn, z=5)
###Output
_____no_output_____
###Markdown
Now plotting the histogram we can see that indeed the average coordination was obtained. Note, however, that the distribution is skewed and has a long tail. The ``reduce_coordination`` function does not (yet) control the shapre of the distribution.
###Code
import matplotlib.pyplot as plt
fig = plt.figure()
plt.hist(pn.num_neighbors(pn.Ps), edgecolor='k')
fig.patch.set_facecolor('white')
###Output
_____no_output_____
###Markdown
Creating Domains with More Interesting Shapes Rectangular Domains with Non-Uniform SpacingThe ```shape``` and ```spacing``` arguments can of course be adjusted to create domains other than simple cubes:
###Code
pn = op.network.Cubic(shape=[10, 20, 20], spacing=[0.001, 0.03, 0.02])
###Output
_____no_output_____
###Markdown
This results in the following network with is squished in the x-direction. Note that this can also be accomplished after the fact by simple scaling the coords (e.g. ``pn['pore.coords'] =* [0.001, 0.03, 0.02]``). Spherical and Other Arbitrary DomainsIt's also possible to obtain cubic networks of arbitrary shapes (i.e. spheres), but still with *cubic* connectivity. This is accomplished using the ``CubicTemplate`` class, which accepts a binary image of 1's and 0's. The network will have pores where the 1's are and 0's elsewhere. For instance, to make a spherical domain for a catalyst pellet, generate an image of a sphere using Scipy's NDimage module, the pass this image to **Cubic** as follows:
###Code
import scipy.ndimage as spim
im = np.ones([21, 21, 21])
im[10, 10, 10] = 0
dt = spim.distance_transform_bf(input=im)
sphere = dt < 10
pn = op.network.CubicTemplate(template=sphere, spacing=0.1)
###Output
_____no_output_____
###Markdown
This results in the following: All images of networks were made with paraview by exporting a VTK file with the following command:
###Code
op.io.VTK.export_data(network=pn, filename='network')
print(f"Does 'network.vtk' exist? {os.path.isfile('network.vtp')}")
###Output
Does 'network.vtk' exist? True
|
EDA_milestone#2.ipynb
|
###Markdown
Exploratory Data Analysis Loading the dataWe are going to use `pickle` to load the DataFrame's we are going to work with. For this example we are only going to load and look at mouse set 1 data (`mouse_set_1_data.pkl`), but everything we do with this mouse set is the same for the other mouse set.
###Code
# Make sure the file is within the same directory, or you know the path to the mouse set 1 data
with open('data/mouse_set_1_data.pkl', 'rb') as handle:
mouse_set_1 = pickle.load(handle)
with open('data/mouse_set_2_data.pkl', 'rb') as handle:
mouse_set_2 = pickle.load(handle)
mouse_set_1.keys()
###Output
_____no_output_____
###Markdown
Structure of the mouse set objectsThe object `mouse_set_1` that you just loaded is a _dict_ object with four different entries:1. `'reads'` * This key maps to the raw count data of each mouse that was outputted by [DADA2](https://github.com/benjjneb/dada2).2. `'qpcr'` * This key maps to the qpcr (total abundance) data for each day for each mouse.4. `'otu_taxonomy'` * This is a table that tells you the (approximate) taxonomy for each of the OTUs in the list as well as the 16S rRNA sequence that was used to barcode the OTU. 5. `'times'` * This is a _list_ object that converts the string time into a float time. More explanation will be below.
###Code
reads = mouse_set_1['reads']
qpcr = mouse_set_1['qpcr']
otu_taxonomy = mouse_set_1['otu_taxonomy']
times = mouse_set_1['times']
reads2 = mouse_set_2['reads']
qpcr2 = mouse_set_2['qpcr']
otu_taxonomy2 = mouse_set_2['otu_taxonomy']
times2 = mouse_set_2['times']
reads.keys()
reads['7'].shape
###Output
_____no_output_____
###Markdown
The objects `reads` and `qpcr` are _dict_'s as well. The dictionary maps the mouse ID (i.e. `'2'` for mouse 2) to the Pandas DataFrame that holds the respective data. So typing `reads['2']` will give us the DataFrame for the count data for mouse 2. `otu_taxonomy` is just a DataFrame because we assume that the OTU's between each mouse in a mouse set are consistent - so there is no reason to differentiate between each mouse.`times` is a list that converts the _str_ labels of the days to the _float_ equivalent. For example: `'1'` -> `1.0`, `2AM`-> `2.0`, `2PM`->`2.5`. Here is what `times` looks like compared to the _str_ version:
###Code
print('float version:\n{}\n\nstr version:\n{}'.format(times, list(reads['2'].columns)))
###Output
float version:
[0.0, 0.5, 1.0, 1.5, 2.0, 2.5, 3.0, 3.5, 4.0, 4.5, 5.0, 6.0, 7.0, 8.0, 9.0, 10.0, 11.0, 14.0, 16.0, 18.0, 21.0, 21.5, 22.0, 22.5, 23.0, 23.5, 24.0, 25.0, 28.0, 29.0, 29.5, 30.0, 30.5, 31.0, 31.5, 32.0, 33.0, 35.0, 35.5, 36.0, 36.5, 37.0, 38.0, 39.0, 42.0, 42.5, 43.0, 43.5, 44.0, 44.5, 45.0, 45.5, 46.0, 47.0, 50.0, 50.5, 51.0, 51.5, 52.0, 52.5, 53.0, 54.0, 57.0, 57.5, 58.0, 58.5, 59.0, 59.5, 60.0, 60.5, 61.0, 62.0, 63.0, 64.0, 64.5]
str version:
['0AM', '0PM', '1AM', '1PM', '2AM', '2PM', '3AM', '3PM', '4AM', '4PM', '5', '6', '7', '8', '9', '10', '11', '14', '16', '18', '21AM', '21PM', '22AM', '22PM', '23AM', '23PM', '24', '25', '28AM', '29AM', '29PM', '30AM', '30PM', '31AM', '31PM', '32', '33', '35AM', '35PM', '36AM', '36PM', '37AM', '38', '39', '42AM', '42PM', '43AM', '43PM', '44AM', '44PM', '45AM', '45PM', '46', '47', '50AM', '50PM', '51AM', '51PM', '52AM', '52PM', '53', '54', '57AM', '57PM', '58AM', '58PM', '59AM', '59PM', '60AM', '60PM', '61', '62', '63', '64AM', '64PM']
###Markdown
****Let's take a look at the first 10 OTU's in reads for mouse 2:
###Code
reads.keys()
reads2['10'][:10]
reads['2'][:10]
###Output
_____no_output_____
###Markdown
You can see that the columns are already ordered in time and each row corresponds to a different OTU. What's the taxonomy for `'OTU_1'`?
###Code
otu_taxonomy.loc['OTU_1']
###Output
_____no_output_____
###Markdown
You know what? Let's look at the taxonomies for each of the first 10 OTU's.
###Code
otu_taxonomy2[:10]
otu_taxonomy[:10]
###Output
_____no_output_____
###Markdown
There are a lot of _Bacteroides_...****Since DataFrame's are so powerful, we can plot the count data straight from the DataFrame. `.plot` plots the index (row) against every column. Since we want to plot the column against every index, we should trasponse the dataframe with `.T`.Let's plot the reads for the first 5 OTU's for mouse 2:
###Code
reads['2'][:5].T.plot(kind='line', title='Counts, Mouse 2')
###Output
_____no_output_____
###Markdown
This might look good at first, but remember that our samples have irregular time spacing, so our x-axis is not spaced correctly (DataFrame assumes regular spacing). We can use our `times` array to tell us where to place each of the points (I could not figure out how to adjust the separation within the DataFrame so I'm using `matplotlib`. If you figure out how to do it please tell me).
###Code
plt.figure()
temp = reads['2'][:5].values # Get our counts for the first 5 OTUs
for i in range(5):
plt.plot(times, temp[i], label=reads['2'].index[i])
plt.title('Counts, Mouse 2')
plt.legend()
###Output
_____no_output_____
###Markdown
Even though these might look similar, there are slight differences. You can see it in the skew for the peaks in `'OTU_5'` and `'OTU_4'`.Let's now look at the qPCR data in `qpcr`:
###Code
# For consistency let's stay with mouse 2
qpcr['2'][:10]
###Output
_____no_output_____
###Markdown
The rows correspond to the days, and we have `'mean'` and `'std'` (standard deviation) as the columns.Let's plot the total bacterial load over time:
###Code
plt.figure()
y = list(qpcr['2']['mean'])
yerr = list(qpcr['2']['std'])
plt.errorbar(times, y, yerr=yerr)
plt.yscale('log')
plt.title('Mouse 2 bacterial load')
###Output
_____no_output_____
###Markdown
We can even plot bacterial load for all of the mice!
###Code
plt.figure()
for mid,df in qpcr.items():
y = df['mean']
yerr = df['std']
plt.errorbar(times, y, yerr=yerr, label= 'Mouse {}'.format(mid))
plt.yscale('log')
plt.legend()
plt.title('Mouse set 1 bacterial load')
###Output
_____no_output_____
###Markdown
We can also plot the counts of `'OTU_1'` over all the mice:
###Code
plt.figure()
for mid, df in reads.items():
y = df.loc['OTU_1']
plt.plot(times, y, label='Mouse {}'.format(mid))
plt.title('OTU_1 counts in mouse set 1')
plt.legend()
###Output
_____no_output_____
###Markdown
We already saw how to extract relative counts of each OTU and the total bacteria load for every time point. To obtain the absolute abundance of each OTU, we can multiply these two values. For instance, this is the results of OTU_1 of mouse 2: (1) The relative aboundance
###Code
m2_reads_values=reads['2'].values
m2_reads_values_sum=sum(m2_reads_values)
m2_OTU_1_reads=reads['2'].loc['OTU_1']
m2_relative_abundance_OTU_1=m2_OTU_1_reads.values/m2_reads_values_sum
plt.figure()
plt.plot(times, m2_relative_abundance_OTU_1, label='Mouse 2, OTU_1, Relative')
plt.title('Mouse 2, OTU_1, Relative')
plt.legend()
###Output
_____no_output_____
###Markdown
(2) The absolute aboundance
###Code
m2_total_amount = qpcr['2']['mean'].values
m2_abs_abundance_OTU_1 = m2_relative_abundance_OTU_1 * m2_total_amount
plt.figure()
plt.plot(times, m2_abs_abundance_OTU_1, label='Mouse 2, OTU_1, Absolute')
plt.title('Mouse 2, OTU_1, Absolute')
plt.legend()
###Output
_____no_output_____
###Markdown
We can look at abundance of each OTU by naively averaging across time. In the example of mouse 2, the abundance seems to be roughly exponentially decayed as the id number of OTU increased, meaning that the population sizes of different bacteria is very disproportionate.
###Code
plt.plot(np.log(np.mean(m_all_abs_amount[:,:21], axis =1)),'.')
plt.xlabel('OTU')
plt.ylabel('Log(abundance)')
plt.title('Abundance of OTUs (average across time)')
###Output
/Users/shihyitseng/anaconda3/envs/109b/lib/python3.6/site-packages/ipykernel_launcher.py:1: RuntimeWarning: divide by zero encountered in log
"""Entry point for launching an IPython kernel.
###Markdown
If we directly plot the abundance of all OTU as time series, we can see changes of abundance across time. The three shadow areas correspond to the periods of high fat diet, Gram positive antibiotics and Gram negative antibiotics, respectively.
###Code
high_fat = [21.5,28.5]
gram_pos = [35.5,42.5]
gram_neg = [50.5,57.5]
pertubation = high_fat.copy()
pertubation.extend(gram_pos)
pertubation.extend(gram_neg)
from scipy.stats import zscore
mouse_id = '2'
mouse_reads_df = reads[mouse_id]
m_total_amount = qpcr[mouse_id]['mean'].values
all_OTU = list(mouse_reads_df.index)
m_reads_values=reads[mouse_id].values
m_reads_values_sum=sum(m_reads_values)
m_all_abs_amount = np.zeros((len(all_OTU), len(times)))
for idx, otu in enumerate(all_OTU):
these_reads = mouse_reads_df.loc[otu]
this_relative_abundance = these_reads.values/m_reads_values_sum
this_abs_abundance = this_relative_abundance * m_total_amount
m_all_abs_amount[idx,:] = this_abs_abundance
m_all_abs_amount_z = zscore(m_all_abs_amount, axis = 1)
m_total_amount.shape
fig,ax = plt.subplots(1,1,figsize = (8,6))
ax.plot(times, m_all_abs_amount.T, linewidth = 0.5, c='k');
y1,y2 = ax.get_ylim()
ax.vlines(x = high_fat, ymin = y1*0.9, ymax = y2*1.1, color = 'yellow', linewidth = 0.5, linestyle = '--')
ax.vlines(x = gram_pos, ymin = y1*0.9, ymax = y2*1.1, color = 'c', linewidth = 0.5, linestyle = '--')
ax.vlines(x = gram_neg, ymin = y1*0.9, ymax = y2*1.1, color = 'm', linewidth = 0.5, linestyle = '--')
rect1 = matplotlib.patches.Rectangle((high_fat[0],y1*0.9), high_fat[1]-high_fat[0],y2*1.1-y1*0.9,
color='yellow',alpha = 0.1)
rect2 = matplotlib.patches.Rectangle((gram_pos[0],y1*0.9), gram_pos[1]-gram_pos[0],y2*1.1-y1*0.9,
color='c',alpha = 0.05)
rect3 = matplotlib.patches.Rectangle((gram_neg[0],y1*0.9), gram_neg[1]-gram_neg[0],y2*1.1-y1*0.9,
color='m',alpha = 0.05)
ax.add_patch(rect1)
ax.add_patch(rect2)
ax.add_patch(rect3)
ax.set_ylim([y1*0.9, y2*1.1])
ax.set(xlabel='Day', ylabel = 'Abundance', title = 'Changes of abundance for all OTUs in mouse 2');
###Output
_____no_output_____
###Markdown
Components of the time-series To explore if there are subgroups/modes of bacteria that covaried across time, we can factorize the time series data into several factors. We chose non-negative matrix factorization (NMF) instead of PCA because the number of bacteria is always positive, and there’s also no reason to impose the orthogonality between factors as PCA does. Here are the temporal profiles of each factors using different number of factors. We can already see that different factors rose and fell at various time points, with some corresponding to the onsets or offsets of the perturbations (which was not imposed a priori). Here we did not scale the data since the relative abundance of each OTU might be an important aspect. If we normalize across each OTU, we might be amplifying noise since a large number of OTUs only consist a really small portion of the whole microbiome.
###Code
from sklearn.decomposition import NMF, PCA
n_comp = 5
model = PCA(n_components = n_comp)
score = model.fit_transform(m_all_abs_amount.T)
PC = model.components_
fig,ax = plt.subplots(2,3,figsize = (18,8))
for i in range(6):
n_comp = i+2
model = NMF(n_components = n_comp)
score = model.fit_transform(all_abs_amount.T)
PC = model.components_
ax[i//3,i%3].plot(times, score);
y1,y2 = ax[i//3,i%3].get_ylim()
ax[i//3,i%3].vlines(x = high_fat, ymin = y1*0.9, ymax = y2*1.1, color = 'yellow', linewidth = 0.5, linestyle = '--')
ax[i//3,i%3].vlines(x = gram_pos, ymin = y1*0.9, ymax = y2*1.1, color = 'c', linewidth = 0.5, linestyle = '--')
ax[i//3,i%3].vlines(x = gram_neg, ymin = y1*0.9, ymax = y2*1.1, color = 'm', linewidth = 0.5, linestyle = '--')
rect1 = matplotlib.patches.Rectangle((high_fat[0],y1*0.9), high_fat[1]-high_fat[0],y2*1.1-y1*0.9,
color='yellow',alpha = 0.1)
rect2 = matplotlib.patches.Rectangle((gram_pos[0],y1*0.9), gram_pos[1]-gram_pos[0],y2*1.1-y1*0.9,
color='c',alpha = 0.05)
rect3 = matplotlib.patches.Rectangle((gram_neg[0],y1*0.9), gram_neg[1]-gram_neg[0],y2*1.1-y1*0.9,
color='m',alpha = 0.05)
ax[i//3,i%3].add_patch(rect1)
ax[i//3,i%3].add_patch(rect2)
ax[i//3,i%3].add_patch(rect3)
ax[i//3,i%3].set(ylim=[y1*0.9, y2*1.1], title = 'NMF with {} factors'.format(n_comp),
xlabel = 'Hour', ylabel = 'Amplitude')
plt.tight_layout()
###Output
_____no_output_____
###Markdown
From the plots above, we found 5 factors are sufficeint to show the temporal changes:
###Code
fig,ax = plt.subplots(1,1,figsize = (8,6))
ax.plot(times, score);
y1,y2 = ax.get_ylim()
ax.vlines(x = high_fat, ymin = y1*0.9, ymax = y2*1.1, color = 'yellow', linewidth = 0.5, linestyle = '--')
ax.vlines(x = gram_pos, ymin = y1*0.9, ymax = y2*1.1, color = 'c', linewidth = 0.5, linestyle = '--')
ax.vlines(x = gram_neg, ymin = y1*0.9, ymax = y2*1.1, color = 'm', linewidth = 0.5, linestyle = '--')
rect1 = matplotlib.patches.Rectangle((high_fat[0],y1*0.9), high_fat[1]-high_fat[0],y2*1.1-y1*0.9,
color='yellow',alpha = 0.1)
rect2 = matplotlib.patches.Rectangle((gram_pos[0],y1*0.9), gram_pos[1]-gram_pos[0],y2*1.1-y1*0.9,
color='c',alpha = 0.05)
rect3 = matplotlib.patches.Rectangle((gram_neg[0],y1*0.9), gram_neg[1]-gram_neg[0],y2*1.1-y1*0.9,
color='m',alpha = 0.05)
ax.add_patch(rect1)
ax.add_patch(rect2)
ax.add_patch(rect3)
ax.set(ylim=[y1*0.9, y2*1.1], title = 'NMF with {} factors'.format(n_comp),
xlabel = 'Hour', ylabel = 'Amplitude');
###Output
_____no_output_____
###Markdown
We can inspect the composition of each factor by looking at the score of everyOTU. Here is an example of the NMF with 4 factors, showing only scores of the first 20OTUs.
###Code
fig,ax = plt.subplots(1,1,figsize = (18,4))
for i in range(n_comp):
plt.bar(np.arange(1,21)+(i-n_comp/2+0.5)*0.15,PC[i,:20], width=0.15,label = 'factor {}'.format(i+1))
plt.legend()
ax.set(xticks = np.arange(1,21), xlabel='OTU #', ylabel = 'score', title='Composition of each factor');
###Output
_____no_output_____
|
Laboratorio_IntroPLN2016.ipynb
|
###Markdown
Laboratorio de Introducción al Procesamiento de Lenguaje Natural 2016 Importación de las sentencias del corpus del Poder Judicial.
###Code
import pandas
corpus = pandas.read_csv("corpus_pj.csv", delimiter=',', skip_blank_lines=True, encoding='utf-8')
corpus = corpus.drop_duplicates()
sentencias = corpus["sentencia"]
###Output
_____no_output_____
|
Notebooks/.ipynb_checkpoints/Community Analysis-checkpoint.ipynb
|
###Markdown
**Community Analysis** Workflow:0) intro to community Detection and Algos used1) Run the 4 algos on entire Healthy network, looking at what communities each method is detecting etc - general analysis2) Run 4 algos on Diseased Network, looking at same, 3) Run 4 algos on healthy-subnetwork - talk about communities4) Run 4 algos on Diseased - subnetwork - talk about differences**At each step, compare results across 4 algos **Introduction** - Understanding Communities In network science, communities can generally be defined as locally dense connected subgraphs. That is to say, all nodes of a community must be accessible through other nodes of the same community. The existence of communities within a network relies upon the connectedness and density hypotheses, which assumes that (1) if a network consists of two isolated components, each community is limited to only one component. The hypothesis also implies that on the same component a community cannot consist of two subgraphs that do not have a link to each other; and (2)Nodes in a community are more likely to connect to other members of the same community than to nodes in other communities. The challenge of detecting communities in protein networks lies in the fact that both the number and size of communities are unknown, unlike in methods of graph partitioning, where these parameters are given. The relationship between a network size and the number of possible, unique partitions is represented as Dobinski's formula: $$B_N=\frac{1}{e}\sum\limits_{j = 0}^\infty {\frac{{j^N }}{{j!}}}$$ Where $B_N$ is the Bell number count of all possible partitions in a set, indicating the number of possible partitions grows faster than exponentially with larger networks, making the problem NP-hard. In order to identify communities within our protein networks, we will be implementing a variety of algorithms described below. Overview of Implemented Community Detection Algorithms: **The Louvain Algorithm** When we assume the hypothesis that a randomly wired network lacks inherent community structure, any systematic deviation from that random configuration becomes meaninfgul, and if measured, allows for comparisons to be made in the quality of network partitions. This measurment called modularity can be written as: $$M_c=\frac{{L_c }}{L}-\left({\frac{{k_c }}{{2L}}}\right)^2$$ where L is the number of links in the graph, Lc is the total number of links within the proposed community, and kc is the total degree of the nodes in this community.This measure, when summed over all nc identified communuties, represents the partition's modularity and can be defined as: $$M=\sum\limits_{c=1}^{n_c }{\left[{\frac{{L_c}}{L}-\left({\frac{{k_c}}{{2L}}}\right)^2}\right]}$$ This value can range from -1/2 to 1, where 0 would indicate the whole network is a single community. If each node belonged to a separate community, then Lc would be 0 and the resulting M value would be negative. Since M is a fractional measure of the difference between the observed and expected number of edges, paritions with a higher M should indicate that those paritions more accurately capture the true community structure. Indeed, the Maximal Modularity Hypothesis, which states that "For a given network the partition with maximum modularity corresponds to the optimal community structure", is the basis for the Louvain algorithm. The Louvain algorithm is a greedy, agglomerative algorithm which uses a two-part iterative process broken into modularity optimization and community aggregation. To start, the algorithm labels each node as its own community. It then optimizes for modularity by reassigning nodes to neighboring communities only if the calculated difference in modularity is positive. This step is repeated until maximal modularity has been reached. In the community aggregation step, each community is reassigned as a super-node with a self-loop containing all its within-community edges, and the super-nodes are then treated as a new network, where the modularity optimization can be reapplied. This iterative process yields a hierarchical organization of communities. The Louvain algorithm is fast, and scales to large networks due to its use of a heuristic in order to reduce the computational complexity associated with the NP-hard problem of optimizing modularity within a network. **The FastGreedy Algorithm** Similar to the Louvain algoritmh, the FastGreedy Algorithm uses a bottom-up agglomerative approach, based on optimizing modularity, $M$, to identify community partitions. The advantage of the FastGreedy algorithm lies in its speed, as it was modified with sparse graphs in mind. Sparse graphs are those in which there are far fewer edges observed than edges possible given a set of nodes, and is often a characteristic feature of protein networks. When communities are merged together during the heirarchical clustering process, pairs of rows and columns within the adjacency matrices are merged together, but since many of those rows and columns contain zeros in sparse graphs, much of the computational power allocated to agglomerative algorithms is wasted on the empty regions of the graph. This algorithm improves both memory utilization and speed by altering the data structures such that rather than storing the adjaceny matrix in memory and calculating the $\Delta M$ each time, it instead stores, and updates a matrix of $\Delta M$ values, eliminating the storage of any communities that aren't connected by at least one edge. The algorithm also utilizes a data structure to store the largest $\Delta M$, reducing computational time associated with computing changes in modularity, and allowing for a faster-than-average runtime of essentially O($\textit{n}$ log$^2$ $\textit{n}$) for sparse graphs, where n is the number of nodes. **The Edge-Betweenness Algorithm** Unlike in Agglomerative algorithms where the starting assumption is that each node is its own community, divisive algorithms begin with the opposing assumption that the entire network is a community, and systematically removes the links connecting nodes that belong to different communities until the network is broken into its sub-partitions. To accomplish this, these algorithms use a different measure, called centrality to determine which nodes belong to each community.The Edge-Betweenness algorithm, also called the Girvan-Newman Algorithm, systematically prunes edges that are least central, or are most "between" communities, until the resulting graph is fully partitioned. The Edge-Betweenness algorithm uses a defintion of centrality described as the number of shortest paths running through the given edge, and serves as measure of the edge's influence over the flow of information in the network. There are four main steps this algorithm carries out and are described as follows.The first step is to calculate the betweeneness for all edges in the network. This calculation can be represented as: $$g(\upsilon) = \sum_{s\neq\upsilon\neq t} \frac{\sigma_{\textit{st}}(\upsilon)}{\sigma_{\textit{st}}}$$ where $\sigma_{st}$ represents the total number of shortest paths from node $\textit{s}$ to node $\textit{t}$ and $\sigma_{\textit{st}}$ represents the number of those paths that have an edge passing through node $\upsilon$. After computing the edge-betweenness value for each node pair in the network, those edges with the highest values are pruned, creating network partitions. The algorithm then recalculates the betweenness centrality for all edges affected by the pruning, and repeats this process of pruning, followed by a recalculation, until no edges remain. On sparse graphs, the algorithm runs in time $O(n^{3})$ due to the computationally difficulty associated with calculating edge-betweenness for each pair of nodes in the network at each iteration, making this algorithm the slowest that we will apply. **The WalkTrap Algorithm** Lastly, we will also implement the WalkTrap Algorithm, which is another hierarchical clustering algorithm. The basic intuition applied in the WalkTrap Algorithm is that random walks from one node to another should get "trapped" into densely connected parts corresponding to communities. The algorithm defines a metric $\textit{r}$ used to measure the similarity of community structure between two nodes. This metric, $\textit{r}$, is a distance calculated from the set of all probabilities of a random walk connecting node $\textit{i}$ to node $\textit{j}$ in $\textit{t}$ steps, represented as $P_{\textit{ij}}^{\textit{t}}$.The resulting value should be high for two nodes within the same community, and low for nodes that in different communities. Using these probabilities as the decisive metric, the algorithm then merges communities in a greedy fashion, based on minimizing the mean of the squared distances between each vertex and its community. This process is then repeated to form graph partitions. _____________________________________________________________________________________________________ **Community Detection Analysis of the Healthy Network:**
###Code
#Code to import all modules used for entire section - just for ease of deployment
###Output
_____no_output_____
###Markdown
Overview: Here, we will apply and compare the various community detection algorithms to reveal the community structure within our healthy human macrophage protein network.
###Code
import networkx as nx
import matplotlib.pyplot as plt
import community
import time
from cdlib import algorithms
from networkx.algorithms.community import greedy_modularity_communities
#import graph
file = open("../Data/Macrophage_protein_network_with_attributes.pkl", "rb")
G = nx.read_gpickle(file)
###Output
_____no_output_____
###Markdown
**Using the Louvain method:**
###Code
#Running, and timing the algorithm:
start = time.time()
louvain_partition = community.best_partition(G) #This object returned is a dictionary containing the nodes of graph G as keys, and the community number that node belongs to as the value
end = time.time()
louvain_time = (end-start)
print("The Louvain algorithm took {} seconds to run".format(round(louvain_time,2)))
#Changing structure of dictionary so that keys = community assignment, and values = list of nodes. This will make it easier for later use
louvain_best_partition = {}
for key,value in louvain_partition.items():
if value in louvain_best_partition.keys():
louvain_best_partition[value].append(key)
else:
louvain_best_partition[value] = []
#Determining how many communities constitute the 'best partition'. i.e how many communities did louvain find?
louvain_comm_list = list(louvain_best_partition.keys())
louvain_comm_list.sort()
print("The best partition of the louvain algorithm identified {} communities".format(len(louvain_comm_list)))
#How many nodes are in each community?
def Community_distrib(partition):
"""Function takes a community partition from a community detection algorithm and plots the distribution of nodes across those communities."""
"""Note that the input must be a dictionary with keys=community IDs, and values = list of nodes contained in that community."""
community_list = list(partition.keys())
community_list.sort()
node_counts = []
for community in community_list:
node_counts.append(len(partition[community]))
comm_plot = plt.bar(community_list,node_counts)
plt.xlabel("Community")
plt.ylabel("Number of Nodes")
return(comm_plot)
Community_distrib(louvain_best_partition);
###Output
_____no_output_____
###Markdown
Need to explore each community, visualize it and its neighbors, etc. Need to figure out how to color by node given messiness of attributes **Using the FastGreedy method:**
###Code
#Running and timing the algorithm:
start = time.time()
FastGreedy_partition = list(greedy_modularity_communities(G)) #The object returned is a list of sets of nodes, each for a different community
end = time.time()
FastGreedy_time = (end-start)
print("The FastGreedy algorithm took {} seconds to run".format(round(FastGreedy_time,2)))
#How many communities did the FastGreedy Algorithm find?
print("The FastGreedy algorithm identified {} communities".format(len(FastGreedy_partition)))
#How many nodes are in each community?
def FastGreedy_plotter(partition):
"""Function takes a list of sets of nodes outputted by FastGreedy"""
"""Modularity algorithm and returns a plot of the distribution of the number of nodes in each community"""
comm_list = []
node_counts = []
counter = 0
for nodeset in FastGreedy_partition:
comm_list.append(counter)
node_counts.append(len(nodeset))
counter+=1
comm_plot = plt.bar(comm_list,node_counts)
plt.xlabel("Community")
plt.ylabel("Number of Nodes")
return(comm_plot)
FastGreedy_plotter(FastGreedy_partition)
###Output
_____no_output_____
###Markdown
Need to explore each community, visualize it and its neighbors, etc. Need to figure out how to color by node given messiness of attributes **Using the WalkTrap method:**
###Code
#Running and timing the algorithm
start = time.time()
WalkTrap_partition = algorithms.walktrap(G) #The object returned is a NodeClusterint object
end = time.time()
WalkTrap_time = (end-start)
print("The WalkTrap algorithm took {} seconds to run".format(round(WalkTrap_time,2)))
print(WalkTrap_partition)
###Output
<cdlib.classes.node_clustering.NodeClustering object at 0x7f84c858f6d0>
###Markdown
Need to figure out how to work with nodeclustering objects **Using the Edge-Betweenness method:**
###Code
#Running and timing the algorithm
start = time.time()
EdgeBetweenness_partition = algorithms.girvan_newman(G,level=1) #The object returned is a list of sets of nodes, each for a different community
end = time.time()
EdgeBetweenness_time = (end-start)/60
print("The WalkTrap algorithm took {} minutes to run".format(round(EdgeBetweenness_time,2)))
print(EdgeBetweenness_partition)
###Output
<generator object girvan_newman at 0x7f84c883f900>
###Markdown
important takeaways Comparing Methods
###Code
#code for comparing and talking about differences
###Output
_____no_output_____
###Markdown
Overall Conclusions Comment on Patterns observed across all three, consistencies, deviations, key things to mention _________________________________________________________________________________ **Community Detection Analysis of the Diseased Network:**
###Code
#Code to import all modules used for entire section - just for ease of deployment
###Output
_____no_output_____
###Markdown
Overview: Overall goal here for this first analysis **Using the Louvain method:**
###Code
#code here, explore and talk about biological relevance of communities
###Output
_____no_output_____
###Markdown
important takeaways **Using the FastGreedy method:**
###Code
#code here, explore and talk about biological relevance of communities
###Output
_____no_output_____
###Markdown
Important takeaways **Using the WalkTrap method:**
###Code
#code here, explore and talk about biological relevance of communities
###Output
_____no_output_____
###Markdown
important takeaways **Using the Edge-Betweenness method:**
###Code
#code here, explore and talk about biological relevance of communities
###Output
_____no_output_____
###Markdown
important takeaways Comparing Methods
###Code
#code for comparing and talking about differences
###Output
_____no_output_____
###Markdown
Overall Conclusions Comment on Patterns observed across all three, consistencies, deviations, key things to mention _________________________________________________________________________________ **Community Detection Analysis of the Healthy Sub-Network:**
###Code
#Code to import all modules used for entire section - just for ease of deployment
###Output
_____no_output_____
###Markdown
Overview: Overall goal here for this first analysis **Using the Louvain method:**
###Code
#code here, explore and talk about biological relevance of communities
###Output
_____no_output_____
###Markdown
important takeaways **Using the FastGreedy method:**
###Code
#code here, explore and talk about biological relevance of communities
###Output
_____no_output_____
###Markdown
Important takeaways **Using the WalkTrap method:**
###Code
#code here, explore and talk about biological relevance of communities
###Output
_____no_output_____
###Markdown
important takeaways **Using the Edge-Betweenness method:**
###Code
#code here, explore and talk about biological relevance of communities
###Output
_____no_output_____
###Markdown
important takeaways Comparing Methods
###Code
#code for comparing and talking about differences
###Output
_____no_output_____
###Markdown
Overall Conclusions Comment on Patterns observed across all three, consistencies, deviations, key things to mention _________________________________________________________________________________ **Community Detection Analysis of the Diseased Sub-Network:**
###Code
#Code to import all modules used for entire section - just for ease of deployment
###Output
_____no_output_____
###Markdown
Overview: Overall goal here for this first analysis **Using the Louvain method:**
###Code
#code here, explore and talk about biological relevance of communities
###Output
_____no_output_____
###Markdown
important takeaways **Using the FastGreedy method:**
###Code
#code here, explore and talk about biological relevance of communities
###Output
_____no_output_____
###Markdown
Important takeaways **Using the WalkTrap method:**
###Code
#code here, explore and talk about biological relevance of communities
###Output
_____no_output_____
###Markdown
important takeaways **Using the Edge-Betweenness method:**
###Code
#code here, explore and talk about biological relevance of communities
###Output
_____no_output_____
###Markdown
important takeaways Comparing Methods
###Code
#code for comparing and talking about differences
###Output
_____no_output_____
|
Projects/CS_VQE/data/Convert_H_non_tapered.ipynb
|
###Markdown
Code to convert Hamiltonians generated by Andrew
###Code
working_dir = os.getcwd()
fullH_dir = os.path.join(working_dir, 'fullHamiltonians')
fullH_sub_JW_dir = os.path.join(fullH_dir, 'JW')
def Convert_full_H_to_dict(file_path_with_name):
# function to convert .oplist file to dictionary!
CONVERSION_DICT = {
0 : 'I',
1: 'X',
2: 'Y',
3: 'Z'
}
with open(file_path_with_name,'rb') as infile:
data = pickle.load(infile,encoding='latin1')
Hamiltonian = {}
for coeff, P_str_list in data:
P_word = ''.join([CONVERSION_DICT[P_key] for P_key in P_str_list])
Hamiltonian[P_word]= np.real(coeff)
return Hamiltonian
[filename[:-7] for filename in os.listdir(fullH_sub_JW_dir) if filename.endswith('.oplist')]
hamiltonians_before_tapering={}
for filename in os.listdir(fullH_sub_JW_dir):
if filename.endswith('.oplist'):
filename_with_ext = filename[:-7]
file = os.path.join(fullH_sub_JW_dir, filename)
H = Convert_full_H_to_dict(file)
hamiltonians_before_tapering[filename_with_ext] = H
file_name = 'Hamiltonians_pre_tapering.txt'
out_dir = os.path.join(working_dir, file_name)
with open(out_dir, 'w') as f:
print(hamiltonians_before_tapering, file=f)
###Output
_____no_output_____
###Markdown
Test import
###Code
H_pretaper_f_path = os.path.join(working_dir, file_name)
with open(H_pretaper_f_path, 'r') as input_file:
NON_tapered_hamiltonians = ast.literal_eval(input_file.read())
NON_tapered_hamiltonians.keys()
###Output
_____no_output_____
|
fast_api_model.ipynb
|
###Markdown
Hard encode Input Dictionary
###Code
input = {
'name': 'terrible MegaBuster from Megaman X',
'goal': 10000,
'launched': '2015-08-11',
'deadline': '2015-08-18',
'backers':21,
'main_category': 11,
'username': 'LoginID'
}
input['name']
###Output
_____no_output_____
###Markdown
Make a function that takes in input dict and converts to dataframe
###Code
def framemaker(web_in):
# making dataframe out of dict
input_frame = pd.DataFrame(web_in, index=[0])
# changing datatype of start and end to date time
# adding column length of campaign
input_frame['deadline'] = pd.to_datetime(input_frame['deadline'])
input_frame['launched'] = pd.to_datetime(input_frame['launched'])
input_frame['length_of_campaign'] = (input_frame['deadline'] - input_frame['launched']).dt.days
# Using a pretrained neural network to encode title to numbers
# Adding numbers to column as sentiments
sentiments =[]
analyzer = SentimentIntensityAnalyzer()
for sentence in input_frame['name']:
vs = analyzer.polarity_scores(sentence)
sentiments.append(vs['compound'])
input_frame['sentiments'] = sentiments
# input_frame['goal'] = (input_frame['goal'].str.split()).apply(lambda x: float(x[0].replace(',', '')))
# input_frame['backers']= input_frame['backers'].astype(str).astype(int)
# Dropping unecessary username column
input_frame = input_frame.drop('username', axis=1)
input_frame = input_frame.drop('name', axis=1)
input_frame = input_frame.drop('launched', axis=1)
input_frame = input_frame.drop('deadline', axis=1)
input_frame = input_frame[['goal', 'backers', 'length_of_campaign', 'sentiments', 'main_category']]
userinput = input_frame.iloc[[0]]
return userinput
user_input = framemaker(input)
user_input
###Output
_____no_output_____
###Markdown
Make function that takes in dataframe, uses model, and can make a prediction
###Code
!pip install category_encoders==2.*
from sklearn.model_selection import train_test_split
import category_encoders as ce
from sklearn.impute import SimpleImputer
from sklearn.linear_model import LogisticRegressionCV
from sklearn.preprocessing import StandardScaler
from sklearn.pipeline import Pipeline
from category_encoders import OneHotEncoder, OrdinalEncoder
df = pd.read_csv('cleaned_kickstarter_data.csv')
print(df.shape)
df
def success_predictor(user_input):
train, test = train_test_split(df, train_size=0.80, test_size=0.20,
stratify=df['project_success'], random_state=42)
# select our target
target = 'project_success'
# make train without our target or id
train_features = train.drop(columns=[target])
# make numeric features
numeric_features = train_features.select_dtypes(include='number').columns.tolist()
# make a cardinality feature to help filter
cardinality = train_features.select_dtypes(exclude='number').nunique()
# get a list of relevant categorical data
categorical_features = cardinality[cardinality <=50].index.tolist()
# Combine the lists
features = numeric_features + categorical_features
X_train = train[features]
y_train = train[target]
X_test = test[features]
y_test = test[target]
# print(features)
# print(X_train.shape, X_test.shape)
lrmodel = Pipeline([
('ohe', OneHotEncoder(use_cat_names=True)),
('scaler', StandardScaler()),
('impute', SimpleImputer()),
('classifier', LogisticRegressionCV())
])
lrmodel.fit(X_train, y_train)
row = X_test.iloc[[4]]
# print(X_train)
# print('training accuracy:', lrmodel.score(X_train, y_train))
# print('test accuracy:', lrmodel.score(X_test, y_test))
# if lrmodel.predict(row) == 1:
# return 'Your Kickstarter project is likely to succeed!'
# else:
# return 'Your Kickstarter project is likely to fail.'
# print(X_test.head())
# print(user_input)
# print(y_test.head())
# print(y_test.iloc[[0]])
if lrmodel.predict(user_input) == 1:
return 'Your Kickstarter project is likely to succeed!'
else:
return 'Your Kickstarter project is likely to fail.'
success_predictor(user_input)
# print(X_train)
# print('training accuracy:', lrmodel.score(X_train, y_train))
# print('test accuracy:', lrmodel.score(X_test, y_test))
###Output
_____no_output_____
|
Practical_statistics/Logistic_Regression/Fitting Logistic Regression.ipynb
|
###Markdown
Fitting Logistic RegressionIn this first notebook, you will be fitting a logistic regression model to a dataset where we would like to predict if a transaction is fraud or not.To get started let's read in the libraries and take a quick look at the dataset.
###Code
import numpy as np
import pandas as pd
import statsmodels.api as sm
from scipy import stats
#stats.chisqprob = lambda chisq, df: stats.chi2.sf(chisq, df)
df = pd.read_csv('./fraud_dataset.csv')
df.head()
###Output
_____no_output_____
###Markdown
`1.` As you can see, there are two columns that need to be changed to dummy variables. Replace each of the current columns to the dummy version. Use the 1 for `weekday` and `True`, and 0 otherwise. Use the first quiz to answer a few questions about the dataset.
###Code
df['weekday'] = pd.get_dummies(df['day'])['weekday']
df[['not_fraud', 'fraud']] = pd.get_dummies(df['fraud'])
df = df.drop('not_fraud', axis = 1)
df['fraud'].mean()
df['weekday'].mean()
df.groupby('fraud').mean()['duration']
###Output
_____no_output_____
###Markdown
`2.` Now that you have dummy variables, fit a logistic regression model to predict if a transaction is fraud using both day and duration. Don't forget an intercept! Use the second quiz below to assure you fit the model correctly.
###Code
df['intercept'] = 1
log_mod = sm.Logit(df['fraud'], df[['intercept', 'weekday', 'duration']])
results = log_mod.fit()
results.summary()
###Output
Optimization terminated successfully.
Current function value: inf
Iterations 16
|
Insurence Prediction/plot_impact_imbalanced_classes.ipynb
|
###Markdown
###Code
%matplotlib inline
###Output
_____no_output_____
###Markdown
Fitting model on imbalanced datasets and how to fight biasThis example illustrates the problem induced by learning on datasets havingimbalanced classes. Subsequently, we compare different approaches alleviatingthese negative effects.
###Code
# Authors: Guillaume Lemaitre <[email protected]>
# License: MIT
print(__doc__)
!wget -O "learn_ml_insurance_prediction__ai_challenge-dataset.zip" "https://dockship-job-models.s3.ap-south-1.amazonaws.com/196c328ad298ef1476f56437902688ef?X-Amz-Algorithm=AWS4-HMAC-SHA256&X-Amz-Credential=AKIAIDOPTEUZ2LEOQEGQ%2F20210120%2Fap-south-1%2Fs3%2Faws4_request&X-Amz-Date=20210120T062225Z&X-Amz-Expires=1800&X-Amz-Signature=87efa5328b5dfba3f163cca5118a43ea47a9a54eb400cc4844db81c4889665ef&X-Amz-SignedHeaders=host&response-content-disposition=attachment%3B%20filename%3D%22learn_ml_insurance_prediction__ai_challenge-dataset.zip%22"
! unzip "learn_ml_insurance_prediction__ai_challenge-dataset.zip"
import pandas as pd
train = pd.read_csv("/content/TRAIN.csv")
test = pd.read_csv("/content/TEST.csv")
df_res = train.drop(['id','Response'], axis=1)
y_res = train['Response'].values
###Output
_____no_output_____
###Markdown
Problem definition
###Code
from sklearn.datasets import fetch_openml
df, y = fetch_openml('adult', version=2, as_frame=True, return_X_y=True)
# we are dropping the following features:
# - "fnlwgt": this feature was created while studying the "adult" dataset.
# Thus, we will not use this feature which is not acquired during the survey.
# - "education-num": it is encoding the same information than "education".
# Thus, we are removing one of these 2 features.
df = df.drop(columns=['fnlwgt', 'education-num'])
###Output
_____no_output_____
###Markdown
The "adult" dataset as a class ratio of about 3:1
###Code
classes_count = y.value_counts()
classes_count
###Output
_____no_output_____
###Markdown
This dataset is only slightly imbalanced. To better highlight the effect oflearning from an imbalanced dataset, we will increase its ratio to 30:1
###Code
from imblearn.datasets import make_imbalance
ratio = 30
df_res, y_res = make_imbalance(
df, y, sampling_strategy={
classes_count.idxmin(): classes_count.max() // ratio
}
)
y_res.value_counts()
###Output
_____no_output_____
###Markdown
For the rest of the notebook, we will make a single split to get trainingand testing data. Note that you should use cross-validation to have anestimate of the performance variation in practice.
###Code
from sklearn.model_selection import train_test_split
X_train, X_test, y_train, y_test = train_test_split(
df_res, y_res, stratify=y_res, random_state=42
)
###Output
_____no_output_____
###Markdown
As a baseline, we could use a classifier which will always predict themajority class independently of the features provided.
###Code
from sklearn.dummy import DummyClassifier
dummy_clf = DummyClassifier(strategy="most_frequent")
score = dummy_clf.fit(X_train, y_train).score(X_test, y_test)
print(f"Accuracy score of a dummy classifier: {score:.3f}")
###Output
Accuracy score of a dummy classifier: 0.878
###Markdown
Instead of using the accuracy, we can use the balanced accuracy which willtake into account the balancing issue.
###Code
from sklearn.metrics import balanced_accuracy_score
y_pred = dummy_clf.predict(X_test)
score = balanced_accuracy_score(y_test, y_pred)
print(f"Balanced accuracy score of a dummy classifier: {score:.3f}")
###Output
Balanced accuracy score of a dummy classifier: 0.500
###Markdown
Strategies to learn from an imbalanced dataset We will first define a helper function which will train a given modeland compute both accuracy and balanced accuracy. The results will be storedin a dataframe
###Code
import pandas as pd
def evaluate_classifier(clf, df_scores, clf_name=None):
from sklearn.pipeline import Pipeline
if clf_name is None:
if isinstance(clf, Pipeline):
clf_name = clf[-1].__class__.__name__
else:
clf_name = clf.__class__.__name__
acc = clf.fit(X_train, y_train).score(X_test, y_test)
y_pred = clf.predict(X_test)
bal_acc = balanced_accuracy_score(y_test, y_pred)
clf_score = pd.DataFrame(
{clf_name: [acc, bal_acc]},
index=['Accuracy', 'Balanced accuracy']
)
df_scores = pd.concat([df_scores, clf_score], axis=1).round(decimals=3)
return df_scores
# Let's define an empty dataframe to store the results
df_scores = pd.DataFrame()
###Output
_____no_output_____
###Markdown
Dummy baseline..............Before to train a real machine learning model, we can store the resultsobtained with our `DummyClassifier`.
###Code
df_scores = evaluate_classifier(dummy_clf, df_scores, "Dummy")
df_scores
###Output
_____no_output_____
###Markdown
Linear classifier baseline..........................We will create a machine learning pipeline using a `LogisticRegression`classifier. In this regard, we will need to one-hot encode the categoricalcolumns and standardized the numerical columns before to inject the data intothe `LogisticRegression` classifier.First, we define our numerical and categorical pipelines.
###Code
from sklearn.impute import SimpleImputer
from sklearn.preprocessing import StandardScaler
from sklearn.preprocessing import OneHotEncoder
from sklearn.pipeline import make_pipeline
num_pipe = make_pipeline(
StandardScaler(), SimpleImputer(strategy="mean", add_indicator=True)
)
cat_pipe = make_pipeline(
SimpleImputer(strategy="constant", fill_value="missing"),
OneHotEncoder(handle_unknown="ignore")
)
###Output
_____no_output_____
###Markdown
Then, we can create a preprocessor which will dispatch the categoricalcolumns to the categorical pipeline and the numerical columns to thenumerical pipeline
###Code
import numpy as np
from sklearn.compose import ColumnTransformer
from sklearn.compose import make_column_selector as selector
preprocessor_linear = ColumnTransformer(
[("num-pipe", num_pipe, selector(dtype_include=np.number)),
("cat-pipe", cat_pipe, selector(dtype_include=pd.CategoricalDtype))],
n_jobs=2
)
###Output
_____no_output_____
###Markdown
Finally, we connect our preprocessor with our `LogisticRegression`. We canthen evaluate our model.
###Code
from sklearn.linear_model import LogisticRegression
lr_clf = make_pipeline(
preprocessor_linear, LogisticRegression(max_iter=1000)
)
df_scores = evaluate_classifier(lr_clf, df_scores, "LR")
df_scores
###Output
_____no_output_____
###Markdown
We can see that our linear model is learning slightly better than our dummybaseline. However, it is impacted by the class imbalance.We can verify that something similar is happening with a tree-based modelsuch as `RandomForestClassifier`. With this type of classifier, we will notneed to scale the numerical data, and we will only need to ordinal encode thecategorical data.
###Code
from sklearn.preprocessing import OrdinalEncoder
from sklearn.ensemble import RandomForestClassifier
cat_pipe = make_pipeline(
SimpleImputer(strategy="constant", fill_value="missing"),
OrdinalEncoder()
)
preprocessor_tree = ColumnTransformer(
[("num-pipe", num_pipe, selector(dtype_include=np.number)),
("cat-pipe", cat_pipe, selector(dtype_include=pd.CategoricalDtype))],
n_jobs=2
)
rf_clf = make_pipeline(
preprocessor_tree, RandomForestClassifier(random_state=42, n_jobs=2)
)
df_scores = evaluate_classifier(rf_clf, df_scores, "RF")
df_scores
###Output
_____no_output_____
###Markdown
The `RandomForestClassifier` is as well affected by the class imbalanced,slightly less than the linear model. Now, we will present different approachto improve the performance of these 2 models.Use `class_weight`..................Most of the models in `scikit-learn` have a parameter `class_weight`. Thisparameter will affect the computation of the loss in linear model or thecriterion in the tree-based model to penalize differently a falseclassification from the minority and majority class. We can set`class_weight="balanced"` such that the weight applied is inverselyproportional to the class frequency. We test this parametrization in bothlinear model and tree-based model.
###Code
lr_clf.set_params(logisticregression__class_weight="balanced")
df_scores = evaluate_classifier(
lr_clf, df_scores, "LR with class weight"
)
df_scores
rf_clf.set_params(randomforestclassifier__class_weight="balanced")
df_scores = evaluate_classifier(
rf_clf, df_scores, "RF with class weight"
)
df_scores
###Output
_____no_output_____
###Markdown
We can see that using `class_weight` was really effective for the linearmodel, alleviating the issue of learning from imbalanced classes. However,the `RandomForestClassifier` is still biased toward the majority class,mainly due to the criterion which is not suited enough to fight the classimbalance.Resample the training set during learning.........................................Another way is to resample the training set by under-sampling orover-sampling some of the samples. `imbalanced-learn` provides some samplersto do such processing.
###Code
from imblearn.pipeline import make_pipeline as make_pipeline_with_sampler
from imblearn.under_sampling import RandomUnderSampler
lr_clf = make_pipeline_with_sampler(
preprocessor_linear,
RandomUnderSampler(random_state=42),
LogisticRegression(max_iter=1000)
)
df_scores = evaluate_classifier(
lr_clf, df_scores, "LR with under-sampling"
)
df_scores
rf_clf = make_pipeline_with_sampler(
preprocessor_tree,
RandomUnderSampler(random_state=42),
RandomForestClassifier(random_state=42, n_jobs=2)
)
df_scores = evaluate_classifier(
rf_clf, df_scores, "RF with under-sampling"
)
df_scores
###Output
/usr/local/lib/python3.6/dist-packages/sklearn/utils/deprecation.py:87: FutureWarning: Function safe_indexing is deprecated; safe_indexing is deprecated in version 0.22 and will be removed in version 0.24.
warnings.warn(msg, category=FutureWarning)
###Markdown
Applying a random under-sampler before the training of the linear model orrandom forest, allows to not focus on the majority class at the cost ofmaking more mistake for samples in the majority class (i.e. decreasedaccuracy).We could apply any type of samplers and find which sampler is working beston the current dataset.Instead, we will present another way by using classifiers which will applysampling internally.Use of `BalancedRandomForestClassifier` and `BalancedBaggingClassifier`.......................................................................We already showed that random under-sampling can be effective on decisiontree. However, instead of under-sampling once the dataset, one couldunder-sample the original dataset before to take a bootstrap sample. This isthe base of the `BalancedRandomForestClassifier` and`BalancedBaggingClassifier`.
###Code
from imblearn.ensemble import BalancedRandomForestClassifier
rf_clf = make_pipeline(
preprocessor_tree,
BalancedRandomForestClassifier(random_state=42, n_jobs=2)
)
df_scores = evaluate_classifier(rf_clf, df_scores, "Balanced RF")
df_scores
###Output
_____no_output_____
###Markdown
The performance with the `BalancedRandomForestClassifier` is better thanapplying a single random under-sampling. We will use a gradient-boostingclassifier within a `BalancedBaggingClassifier`.
###Code
from sklearn.experimental import enable_hist_gradient_boosting
from sklearn.ensemble import HistGradientBoostingClassifier
from imblearn.ensemble import BalancedBaggingClassifier
bag_clf = make_pipeline(
preprocessor_tree,
BalancedBaggingClassifier(
base_estimator=HistGradientBoostingClassifier(random_state=42),
n_estimators=10, random_state=42, n_jobs=2
)
)
df_scores = evaluate_classifier(
bag_clf, df_scores, "Balanced bagging"
)
df_scores
###Output
_____no_output_____
###Markdown
This last approach is the most effective. The different under-sampling allowsto bring some diversity for the different GBDT to learn and not focus on aportion of the majority class.We will repeat the same experiment but with a ratio of 100:1 and make asimilar analysis. Increase imbalanced ratio
###Code
ratio = 100
df_res, y_res = make_imbalance(
df, y, sampling_strategy={
classes_count.idxmin(): classes_count.max() // ratio
}
)
X_train, X_test, y_train, y_test = train_test_split(
df_res, y_res, stratify=y_res, random_state=42
)
df_scores = pd.DataFrame()
df_scores = evaluate_classifier(dummy_clf, df_scores, "Dummy")
lr_clf = make_pipeline(
preprocessor_linear, LogisticRegression(max_iter=1000)
)
df_scores = evaluate_classifier(lr_clf, df_scores, "LR")
rf_clf = make_pipeline(
preprocessor_tree, RandomForestClassifier(random_state=42, n_jobs=2)
)
df_scores = evaluate_classifier(rf_clf, df_scores, "RF")
lr_clf.set_params(logisticregression__class_weight="balanced")
df_scores = evaluate_classifier(
lr_clf, df_scores, "LR with class weight"
)
rf_clf.set_params(randomforestclassifier__class_weight="balanced")
df_scores = evaluate_classifier(
rf_clf, df_scores, "RF with class weight"
)
lr_clf = make_pipeline_with_sampler(
preprocessor_linear,
RandomUnderSampler(random_state=42),
LogisticRegression(max_iter=1000)
)
df_scores = evaluate_classifier(
lr_clf, df_scores, "LR with under-sampling"
)
rf_clf = make_pipeline_with_sampler(
preprocessor_tree,
RandomUnderSampler(random_state=42),
RandomForestClassifier(random_state=42, n_jobs=2)
)
df_scores = evaluate_classifier(
rf_clf, df_scores, "RF with under-sampling"
)
rf_clf = make_pipeline(
preprocessor_tree,
BalancedRandomForestClassifier(random_state=42, n_jobs=2)
)
df_scores = evaluate_classifier(rf_clf, df_scores)
df_scores = evaluate_classifier(
bag_clf, df_scores, "Balanced bagging"
)
df_scores
###Output
_____no_output_____
###Markdown
When we analyse the results, we can draw similar conclusions than in theprevious discussion. However, we can observe that the strategy`class_weight="balanced"` does not improve the performance when using a`RandomForestClassifier`. A resampling is indeed required. The most effectivemethod remains the `BalancedBaggingClassifier` using a GBDT as a baselearner.
###Code
from sklearn.datasets import make_moons, make_blobs
from sklearn.ensemble import IsolationForest
from sklearn.linear_model import LogisticRegression
from sklearn.metrics import classification_report
from imblearn import FunctionSampler
from imblearn.pipeline import make_pipeline
print(__doc__)
rng = np.random.RandomState(42)
X = df_res
y = y_res
from sklearn.model_selection import train_test_split
X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.33, random_state=1)
# fit the model
def outlier_rejection(X, y):
"""This will be our function used to resample our dataset."""
model = IsolationForest(max_samples=100,
contamination=0.4,
random_state=rng)
model.fit(X)
y_pred = model.predict(X)
return X[y_pred == 1], y[y_pred == 1]
reject_sampler = FunctionSampler(func=outlier_rejection)
pipe = make_pipeline(FunctionSampler(func=outlier_rejection),
LogisticRegression(solver='lbfgs', multi_class='auto',
random_state=rng))
y_pred = pipe.fit(X_train, y_train).predict(X_test)
print(classification_report(y_test, y_pred))
clf = LogisticRegression(solver='lbfgs', multi_class='auto', random_state=rng)
y_pred = clf.fit(X_train, y_train).predict(X_test)
print(classification_report(y_test, y_pred))
plt.show()
###Output
_____no_output_____
|
notebooks/Speedup-sample_time_and_space_V4.5-numba.ipynb
|
###Markdown
Optimization Strategie - STEP 4* include Numba´s Just-In-Time-Compiler into optimization - enable numba
###Code
import theano
import theano.tensor as tt
import numpy as np
import pandas as pd
import numba
print("Theano version: ",theano.__version__)
print("Numba version: ",numba.__version__)
###Output
Theano version: 1.0.4
Numba version: 0.49.0
###Markdown
A - Read/Create Input Data* output: - kw_data - day_data - time_by_day Read counties
###Code
import pickle as pkl
with open('../data_complete/counties/counties.pkl', "rb") as f:
counties = pkl.load(f)
###Output
_____no_output_____
###Markdown
Read data
###Code
disease = "covid19"
prediction_region = "germany"
def load_daily_data(disease, prediction_region, counties, seperator=","):
data = pd.read_csv("../data_complete/diseases/{}.csv".format(disease),
sep=seperator, encoding='iso-8859-1', index_col=0)
if "99999" in data.columns:
data.drop("99999", inplace=True, axis=1)
data = data.loc[:, list(
filter(lambda cid: prediction_region in counties[cid]["region"], data.columns))]
data.index = [pd.Timestamp(date) for date in data.index]
return data
indata = load_daily_data(disease, prediction_region, counties)
data = indata
data
###Output
_____no_output_____
###Markdown
Create times_by_day dictionary
###Code
import datetime
from collections import OrderedDict
rnd_tsel = np.random.Generator(np.random.PCG64(12345))
def uniform_times_by_day(days, n=10):
""" Samples n random timepoints within a day, per day. converts pd.Timestamps to datetime obj."""
res = OrderedDict()
for day in days:
time_min = datetime.datetime.combine(day, datetime.time.min)
time_max = datetime.datetime.combine(day, datetime.time.max)
res[day] = rnd_tsel.random(n) * (time_max - time_min) + time_min
return res
times_by_day=uniform_times_by_day(data.index)
#times_by_day
###Output
_____no_output_____
###Markdown
Create locations_by_county dictionary
###Code
from collections import OrderedDict
rnd_csel = np.random.Generator(np.random.PCG64(12345))
def uniform_locations_by_county(counties, n=5):
res = OrderedDict()
for (county_id, county) in counties.items():
tp = county["testpoints"]
if n == len(tp):
res[county_id] = tp
else:
idx = rnd_csel.choice(tp.shape[0], n, replace=n > len(tp))
res[county_id] = tp[idx]
return res
locations_by_county=uniform_locations_by_county(counties)
#locations_by_county
###Output
_____no_output_____
###Markdown
Define temporal_bfs and spatial_bfs
###Code
def gaussian_bf(dx, σ):
""" spatial basis function """
σ = np.float32(σ)
res = tt.zeros_like(dx)
idx = (abs(dx) < np.float32(5) * σ) # .nonzero()
return tt.set_subtensor(res[idx], tt.exp(
np.float32(-0.5 / (σ**2)) * (dx[idx])**2) / np.float32(np.sqrt(2 * np.pi * σ**2)))
def bspline_bfs(x, knots, P):
""" temporal basis function
x: t-delta distance to last knot (horizon 5)
"""
knots = knots.astype(np.float32)
idx = ((x >= knots[0]) & (x < knots[-1])) # .nonzero()
xx = x[idx]
N = {}
for p in range(P + 1):
for i in range(len(knots) - 1 - p):
if p == 0:
N[(i, p)] = tt.where((knots[i] <= xx)
* (xx < knots[i + 1]), 1.0, 0.0)
else:
N[(i, p)] = (xx - knots[i]) / (knots[i + p] - knots[i]) * N[(i, p - 1)] + \
(knots[i + p + 1] - xx) / (knots[i + p + 1] - knots[i + 1]) * N[(i + 1, p - 1)]
highest_level = []
for i in range(len(knots) - 1 - P):
res = tt.zeros_like(x)
highest_level.append(tt.set_subtensor(res[idx], N[(i, P)]))
return highest_level
#NOTE: Do we want basis functions with a longer temporal horizon? // we may want to weight them around fixed days?!
#NOTE: Split this up, so we can get multiple basis functions!
def temporal_bfs(x):
return bspline_bfs(x, np.array([0, 0, 1, 2, 3, 4, 5]) * 24 * 3600.0, 2)
def spatial_bfs(x):
return [gaussian_bf(x, σ) for σ in [6.25, 12.5, 25.0, 50.0]]
###Output
_____no_output_____
###Markdown
Define Theano function ia_bfs
###Code
def jacobian_sq(latitude, R=6365.902):
"""
jacobian_sq(latitude)
Computes the "square root" (Cholesky factor) of the Jacobian of the cartesian projection from polar coordinates (in degrees longitude, latitude) onto cartesian coordinates (in km east/west, north/south) at a given latitude (the projection's Jacobian is invariante wrt. longitude).
TODO: don't import jacobian_sq from geo_utils to remove potential conflicts
"""
return R * (np.pi / 180.0) * (abs(tt.cos(tt.deg2rad(latitude))) *
np.array([[1.0, 0.0], [0.0, 0.0]]) + np.array([[0.0, 0.0], [0.0, 1.0]]))
def build_ia_bfs(temporal_bfs, spatial_bfs, profile):
x1 = tt.fmatrix("x1")
t1 = tt.fvector("t1")
# M = tt.fmatrix("M")
x2 = tt.fmatrix("x2")
t2 = tt.fvector("t2")
lat = x1[:, 1].mean()
M = jacobian_sq(lat)**2
# (x1,t1) are the to-be-predicted points, (x2,t2) the historic cases
# spatial distance btw. each points (defined with latitude,longitude) in x1 and x2 with gramian M
# (a-b)^2 = a^2 + b^2 -2ab; with a,b=vectors
dx = tt.sqrt( (x1.dot(M) * x1).sum(axis=1).reshape((-1, 1)) # a^2
+ (x2.dot(M) * x2).sum(axis=1).reshape(( 1, -1)) # b^2
- 2 * x1.dot(M).dot(x2.T) ) # -2ab
# temporal distance btw. each times in t1 and t2
dt = t1.reshape((-1, 1)) - t2.reshape((1, -1))
ft = tt.stack(temporal_bfs(dt.reshape((-1,))), axis=0) # cast to floats?
fx = tt.stack(spatial_bfs(dx.reshape((-1,))), axis=0)
# aggregate contributions of all cases
contrib = ft.dot(fx.T).reshape((-1,)) / tt.cast(x1.shape[0], "float32")
return theano.function([t1, x1, t2, x2], contrib, allow_input_downcast=True, profile=profile)
ia_bfs = build_ia_bfs(temporal_bfs, spatial_bfs, profile=False)
profile_ia_bfs = build_ia_bfs(temporal_bfs, spatial_bfs, profile=True)
theano.printing.debugprint(profile_ia_bfs)
# test ia_bfs()
t1=[1580234892.375513, 1580224126.122202, 1580193367.920551, 1580193367.920551, 1580185641.832341, 1580194755.123367]
x1 = [ [10.435944369180099, 51.69958916804793],
[10.435944369180099, 51.69958916804793],
[10.134378974970323, 51.51153765399198],
[10.134378974970323, 51.51153765399198],
[10.435944369180099, 51.69958916804793],
[10.97023632180951, 49.35209111265112],]
t2=[1580234892.375513, 1580224428.403552, 1580182133.833636, 1580217693.876309, 1580224428.403552, 1580224428.403552,]
x2 = [ [11.38965623, 48.0657035 ],
[11.0615104 , 48.11177134],
[ 7.12902758, 51.57865701],
[ 7.12902758, 51.57865701],
[11.38965623, 48.0657035 ],
[11.0615104 , 48.11177134],]
profile_ia_bfs(t1,x1,t2,x2)
profile_ia_bfs.profile.summary()
###Output
Elemwise{TrueDiv}[(0, 0)] [id A] '' 86
|Reshape{1} [id B] '' 85
| |dot [id C] '' 84
| | |Join [id D] '' 60
| | | |TensorConstant{0} [id E]
| | | |InplaceDimShuffle{x,0} [id F] '' 56
| | | | |AdvancedBooleanIncSubtensor{inplace=False, set_instead_of_inc=True} [id G] '' 53
| | | | |Alloc [id H] '' 16
| | | | | |TensorConstant{(1,) of 0.0} [id I]
| | | | | |Elemwise{Composite{((i0 // i1) * (i2 // i3))}}[(0, 0)] [id J] '' 12
| | | | | |Shape_i{0} [id K] '' 7
| | | | | | |t1 [id L]
| | | | | |TensorConstant{1} [id M]
| | | | | |Shape_i{0} [id N] '' 5
| | | | | | |t2 [id O]
| | | | | |TensorConstant{1} [id M]
| | | | |Elemwise{Composite{((i0 * i1 * (Switch((i2 * LT(i1, i3)), (i4 * i1), i5) + Switch(i6, (i0 * (i7 - i1)), i5))) + (i8 * i9 * i10))}}[(0, 9)] [id P] '' 48
| | | | | |TensorConstant{(1,) of 1...574074e-05} [id Q]
| | | | | |AdvancedBooleanSubtensor [id R] '' 21
| | | | | | |Reshape{1} [id S] '' 15
| | | | | | | |Elemwise{sub,no_inplace} [id T] '' 11
| | | | | | | | |InplaceDimShuffle{0,x} [id U] '' 6
| | | | | | | | | |t1 [id L]
| | | | | | | | |InplaceDimShuffle{x,0} [id V] '' 4
| | | | | | | | |t2 [id O]
| | | | | | | |TensorConstant{(1,) of -1} [id W]
| | | | | | |Elemwise{Composite{AND(GE(i0, i1), LT(i0, i2))}} [id X] '' 18
| | | | | | |Reshape{1} [id S] '' 15
| | | | | | |TensorConstant{(1,) of 0.0} [id I]
| | | | | | |TensorConstant{(1,) of 432000.0} [id Y]
| | | | | |Elemwise{ge,no_inplace} [id Z] '' 33
| | | | | | |AdvancedBooleanSubtensor [id R] '' 21
| | | | | | |TensorConstant{(1,) of 0.0} [id I]
| | | | | |TensorConstant{(1,) of 0.0} [id I]
| | | | | |TensorConstant{(1,) of inf} [id BA]
| | | | | |TensorConstant{(1,) of 0} [id BB]
| | | | | |Elemwise{Composite{(i0 * LT(i1, i2))}} [id BC] '' 38
| | | | | | |Elemwise{ge,no_inplace} [id Z] '' 33
| | | | | | |AdvancedBooleanSubtensor [id R] '' 21
| | | | | | |TensorConstant{(1,) of 86400.0} [id BD]
| | | | | |TensorConstant{(1,) of 86400.0} [id BD]
| | | | | |TensorConstant{(1,) of 5.787037e-06} [id BE]
| | | | | |Elemwise{sub,no_inplace} [id BF] '' 32
| | | | | | |TensorConstant{(1,) of 172800.0} [id BG]
| | | | | | |AdvancedBooleanSubtensor [id R] '' 21
| | | | | |Elemwise{Composite{(Switch(i0, (i1 * i2), i3) + Switch(i4, (i1 * i5), i3))}} [id BH] '' 43
| | | | | |Elemwise{Composite{(i0 * LT(i1, i2))}} [id BC] '' 38
| | | | | |TensorConstant{(1,) of 1...574074e-05} [id Q]
| | | | | |AdvancedBooleanSubtensor [id R] '' 21
| | | | | |TensorConstant{(1,) of 0} [id BB]
| | | | | |Elemwise{Composite{(GE(i0, i1) * LT(i0, i2))}} [id BI] '' 31
| | | | | | |AdvancedBooleanSubtensor [id R] '' 21
| | | | | | |TensorConstant{(1,) of 86400.0} [id BD]
| | | | | | |TensorConstant{(1,) of 172800.0} [id BG]
| | | | | |Elemwise{sub,no_inplace} [id BF] '' 32
| | | | |Elemwise{Composite{AND(GE(i0, i1), LT(i0, i2))}} [id X] '' 18
| | | |InplaceDimShuffle{x,0} [id BJ] '' 58
| | | | |AdvancedBooleanIncSubtensor{inplace=False, set_instead_of_inc=True} [id BK] '' 55
| | | | |Alloc [id H] '' 16
| | | | |Elemwise{Composite{((i0 * i1 * i2) + (i0 * i3 * i4))}}[(0, 1)] [id BL] '' 50
| | | | | |TensorConstant{(1,) of 5.787037e-06} [id BE]
| | | | | |AdvancedBooleanSubtensor [id R] '' 21
| | | | | |Elemwise{Composite{(Switch(i0, (i1 * i2), i3) + Switch(i4, (i1 * i5), i3))}} [id BH] '' 43
| | | | | |Elemwise{sub,no_inplace} [id BM] '' 29
| | | | | | |TensorConstant{(1,) of 259200.0} [id BN]
| | | | | | |AdvancedBooleanSubtensor [id R] '' 21
| | | | | |Elemwise{Composite{(Switch(i0, (i1 * i2), i3) + Switch(i4, (i1 * i5), i3))}} [id BO] '' 37
| | | | | |Elemwise{Composite{(GE(i0, i1) * LT(i0, i2))}} [id BI] '' 31
| | | | | |TensorConstant{(1,) of 1...574074e-05} [id Q]
| | | | | |Elemwise{add,no_inplace} [id BP] '' 30
| | | | | | |TensorConstant{(1,) of -86400.0} [id BQ]
| | | | | | |AdvancedBooleanSubtensor [id R] '' 21
| | | | | |TensorConstant{(1,) of 0} [id BB]
| | | | | |Elemwise{Composite{(GE(i0, i1) * LT(i0, i2))}} [id BR] '' 28
| | | | | | |AdvancedBooleanSubtensor [id R] '' 21
| | | | | | |TensorConstant{(1,) of 172800.0} [id BG]
| | | | | | |TensorConstant{(1,) of 259200.0} [id BN]
| | | | | |Elemwise{sub,no_inplace} [id BM] '' 29
| | | | |Elemwise{Composite{AND(GE(i0, i1), LT(i0, i2))}} [id X] '' 18
| | | |InplaceDimShuffle{x,0} [id BS] '' 52
| | | | |AdvancedBooleanIncSubtensor{inplace=False, set_instead_of_inc=True} [id BT] '' 47
| | | | |Alloc [id H] '' 16
| | | | |Elemwise{Composite{((i0 * i1 * i2) + (i0 * i3 * i4))}}[(0, 1)] [id BU] '' 42
| | | | | |TensorConstant{(1,) of 5.787037e-06} [id BE]
| | | | | |Elemwise{add,no_inplace} [id BP] '' 30
| | | | | |Elemwise{Composite{(Switch(i0, (i1 * i2), i3) + Switch(i4, (i1 * i5), i3))}} [id BO] '' 37
| | | | | |Elemwise{sub,no_inplace} [id BV] '' 25
| | | | | | |TensorConstant{(1,) of 345600.0} [id BW]
| | | | | | |AdvancedBooleanSubtensor [id R] '' 21
| | | | | |Elemwise{Composite{(Switch(i0, (i1 * i2), i3) + Switch(i4, (i1 * i5), i3))}} [id BX] '' 36
| | | | | |Elemwise{Composite{(GE(i0, i1) * LT(i0, i2))}} [id BR] '' 28
| | | | | |TensorConstant{(1,) of 1...574074e-05} [id Q]
| | | | | |Elemwise{add,no_inplace} [id BY] '' 27
| | | | | | |TensorConstant{(1,) of -172800.0} [id BZ]
| | | | | | |AdvancedBooleanSubtensor [id R] '' 21
| | | | | |TensorConstant{(1,) of 0} [id BB]
| | | | | |Elemwise{Composite{(GE(i0, i1) * LT(i0, i2))}} [id CA] '' 26
| | | | | | |AdvancedBooleanSubtensor [id R] '' 21
| | | | | | |TensorConstant{(1,) of 259200.0} [id BN]
| | | | | | |TensorConstant{(1,) of 345600.0} [id BW]
| | | | | |Elemwise{sub,no_inplace} [id BV] '' 25
| | | | |Elemwise{Composite{AND(GE(i0, i1), LT(i0, i2))}} [id X] '' 18
| | | |InplaceDimShuffle{x,0} [id CB] '' 51
| | | |AdvancedBooleanIncSubtensor{inplace=False, set_instead_of_inc=True} [id CC] '' 46
| | | |Alloc [id H] '' 16
| | | |Elemwise{Composite{((i0 * i1 * i2) + (i0 * (i3 - i4) * (Switch(i5, (i6 * (i7 + i4)), i8) + Switch((GE(i4, i9) * LT(i4, i3)), (i6 * (i3 - i4)), i8))))}}[(0, 1)] [id CD] '' 41
| | | | |TensorConstant{(1,) of 5.787037e-06} [id BE]
| | | | |Elemwise{add,no_inplace} [id BY] '' 27
| | | | |Elemwise{Composite{(Switch(i0, (i1 * i2), i3) + Switch(i4, (i1 * i5), i3))}} [id BX] '' 36
| | | | |TensorConstant{(1,) of 432000.0} [id Y]
| | | | |AdvancedBooleanSubtensor [id R] '' 21
| | | | |Elemwise{Composite{(GE(i0, i1) * LT(i0, i2))}} [id CA] '' 26
| | | | |TensorConstant{(1,) of 1...574074e-05} [id Q]
| | | | |TensorConstant{(1,) of -259200.0} [id CE]
| | | | |TensorConstant{(1,) of 0} [id BB]
| | | | |TensorConstant{(1,) of 345600.0} [id BW]
| | | |Elemwise{Composite{AND(GE(i0, i1), LT(i0, i2))}} [id X] '' 18
| | |InplaceDimShuffle{1,0} [id CF] '' 83
| | |Join [id CG] '' 82
| | |TensorConstant{0} [id E]
| | |InplaceDimShuffle{x,0} [id CH] '' 81
| | | |AdvancedBooleanIncSubtensor{inplace=False, set_instead_of_inc=True} [id CI] '' 77
| | | |Alloc [id CJ] '' 22
| | | | |TensorConstant{(1,) of 0.0} [id CK]
| | | | |Elemwise{Composite{((i0 // i1) * (i2 // i3))}}[(0, 0)] [id CL] '' 19
| | | | |Shape_i{0} [id CM] '' 3
| | | | | |x1 [id CN]
| | | | |TensorConstant{1} [id M]
| | | | |Shape_i{0} [id CO] '' 1
| | | | | |x2 [id CP]
| | | | |TensorConstant{1} [id M]
| | | |Elemwise{Composite{(i0 * exp((i1 * sqr(i2))))}}[(0, 2)] [id CQ] '' 73
| | | | |TensorConstant{(1,) of 0...6509978214} [id CR]
| | | | |TensorConstant{(1,) of -0..9676644802} [id CS]
| | | | |AdvancedBooleanSubtensor [id CT] '' 69
| | | | |Elemwise{Sqrt}[(0, 0)] [id CU] '' 59
| | | | | |Reshape{1} [id CV] '' 57
| | | | | |Elemwise{Composite{((i0 + i1) - (i2 * i3))}}[(0, 3)] [id CW] '' 54
| | | | | | |InplaceDimShuffle{0,x} [id CX] '' 49
| | | | | | | |Sum{axis=[1], acc_dtype=float64} [id CY] '' 44
| | | | | | | |Elemwise{Mul}[(0, 0)] [id CZ] '' 39
| | | | | | | |dot [id DA] '' 23
| | | | | | | | |x1 [id CN]
| | | | | | | | |Elemwise{Composite{sqr((i0 * ((i1 * i2) + i3)))}} [id DB] '' 20
| | | | | | | | |TensorConstant{(1, 1) of ..4975929206} [id DC]
| | | | | | | | |Elemwise{Composite{Abs(cos(deg2rad((i0 / Cast{float32}(i1)))))}}[(0, 0)] [id DD] '' 17
| | | | | | | | | |InplaceDimShuffle{x,x} [id DE] '' 13
| | | | | | | | | | |Sum{acc_dtype=float64} [id DF] '' 8
| | | | | | | | | | |Subtensor{::, int64} [id DG] '' 2
| | | | | | | | | | |x1 [id CN]
| | | | | | | | | | |Constant{1} [id DH]
| | | | | | | | | |InplaceDimShuffle{x,x} [id DI] '' 9
| | | | | | | | | |Shape_i{0} [id CM] '' 3
| | | | | | | | |TensorConstant{[[1. 0.]
[0. 0.]]} [id DJ]
| | | | | | | | |TensorConstant{[[0. 0.]
[0. 1.]]} [id DK]
| | | | | | | |x1 [id CN]
| | | | | | |InplaceDimShuffle{x,0} [id DL] '' 45
| | | | | | | |Sum{axis=[1], acc_dtype=float64} [id DM] '' 40
| | | | | | | |Elemwise{Mul}[(0, 0)] [id DN] '' 35
| | | | | | | |dot [id DO] '' 24
| | | | | | | | |x2 [id CP]
| | | | | | | | |Elemwise{Composite{sqr((i0 * ((i1 * i2) + i3)))}} [id DB] '' 20
| | | | | | | |x2 [id CP]
| | | | | | |TensorConstant{(1, 1) of 2.0} [id DP]
| | | | | | |dot [id DQ] '' 34
| | | | | | |dot [id DA] '' 23
| | | | | | |InplaceDimShuffle{1,0} [id DR] 'x2.T' 0
| | | | | | |x2 [id CP]
| | | | | |TensorConstant{(1,) of -1} [id W]
| | | | |Elemwise{lt,no_inplace} [id DS] '' 65
| | | | |Elemwise{abs_,no_inplace} [id DT] '' 61
| | | | | |Elemwise{Sqrt}[(0, 0)] [id CU] '' 59
| | | | |TensorConstant{(1,) of 31.25} [id DU]
| | | |Elemwise{lt,no_inplace} [id DS] '' 65
| | |InplaceDimShuffle{x,0} [id DV] '' 80
| | | |AdvancedBooleanIncSubtensor{inplace=False, set_instead_of_inc=True} [id DW] '' 76
| | | |Alloc [id CJ] '' 22
| | | |Elemwise{Composite{(i0 * exp((i1 * sqr(i2))))}}[(0, 2)] [id DX] '' 72
| | | | |TensorConstant{(1,) of 0...8254989107} [id DY]
| | | | |TensorConstant{(1,) of -0..9191612005} [id DZ]
| | | | |AdvancedBooleanSubtensor [id EA] '' 68
| | | | |Elemwise{Sqrt}[(0, 0)] [id CU] '' 59
| | | | |Elemwise{lt,no_inplace} [id EB] '' 64
| | | | |Elemwise{abs_,no_inplace} [id DT] '' 61
| | | | |TensorConstant{(1,) of 62.5} [id EC]
| | | |Elemwise{lt,no_inplace} [id EB] '' 64
| | |InplaceDimShuffle{x,0} [id ED] '' 79
| | | |AdvancedBooleanIncSubtensor{inplace=False, set_instead_of_inc=True} [id EE] '' 75
| | | |Alloc [id CJ] '' 22
| | | |Elemwise{Composite{(i0 * exp((i1 * sqr(i2))))}}[(0, 2)] [id EF] '' 71
| | | | |TensorConstant{(1,) of 0...1274945535} [id EG]
| | | | |TensorConstant{(1,) of -0..9797903001} [id EH]
| | | | |AdvancedBooleanSubtensor [id EI] '' 67
| | | | |Elemwise{Sqrt}[(0, 0)] [id CU] '' 59
| | | | |Elemwise{lt,no_inplace} [id EJ] '' 63
| | | | |Elemwise{abs_,no_inplace} [id DT] '' 61
| | | | |TensorConstant{(1,) of 125.0} [id EK]
| | | |Elemwise{lt,no_inplace} [id EJ] '' 63
| | |InplaceDimShuffle{x,0} [id EL] '' 78
| | |AdvancedBooleanIncSubtensor{inplace=False, set_instead_of_inc=True} [id EM] '' 74
| | |Alloc [id CJ] '' 22
| | |Elemwise{Composite{(i0 * exp((i1 * sqr(i2))))}}[(0, 2)] [id EN] '' 70
| | | |TensorConstant{(1,) of 0...5637472767} [id EO]
| | | |TensorConstant{(1,) of -0..9494757503} [id EP]
| | | |AdvancedBooleanSubtensor [id EQ] '' 66
| | | |Elemwise{Sqrt}[(0, 0)] [id CU] '' 59
| | | |Elemwise{lt,no_inplace} [id ER] '' 62
| | | |Elemwise{abs_,no_inplace} [id DT] '' 61
| | | |TensorConstant{(1,) of 250.0} [id ES]
| | |Elemwise{lt,no_inplace} [id ER] '' 62
| |TensorConstant{(1,) of -1} [id W]
|Elemwise{Cast{float32}} [id ET] '' 14
|InplaceDimShuffle{x} [id EU] '' 10
|Shape_i{0} [id CM] '' 3
###Markdown
B - Do the Sampling (the old way)
###Code
# set seed to check results
rnd_time = np.random.Generator(np.random.PCG64(12345))
rnd_loc = np.random.Generator(np.random.PCG64(12345))
rnd_time_pred = np.random.Generator(np.random.PCG64(12345))
rnd_loc_pred = np.random.Generator(np.random.PCG64(12345))
# random generators:
# MT19937, PCG64, Philox, SFC64 - https://numpy.org/devdocs/reference/random/bit_generators/index.html
#%%timeit
# loop over all days of all counties
# and draw per day n-times a random time from times_by_day[day]
def sample_time_and_space(data, times_by_day, locations_by_county, rnd_t, rnd_l):
n_total = data.sum().sum()
t_all = np.empty((n_total,), dtype=object)
x_all = np.empty((n_total, 2))
i=0
for (county_id, series) in data.iteritems():
for (day, n) in series.iteritems():
#if n==0: continue
#print(i,"\n day =",day,"\n no. samples to draw = ",n)
# draw n random times
times = times_by_day[day]
#idx = rnd_time.choice(len(times), n)
idx = np.floor( (n*[len(times)]) * rnd_t.random((n,)) ).astype("int32") # replace 'rnd_time.choice' to enable compare with new optimized solution
#print(" random sample ids = ",idx)
t_all[i:i + n] = times[idx]
# draw n random locations
locs = locations_by_county[county_id]
#idx = rnd_loc.choice(locs.shape[0], n)
idx = np.floor( (n*[locs.shape[0]]) * rnd_l.random((n,)) ).astype("int32") # replace 'rnd_time.choice' to enable compare with new optimized solution
x_all[i:i + n, :] = locs[idx, :]
i += n
return t_all, x_all
t_data_0 = []
x_data_0 = []
t_pred_0 = []
x_pred_0 = []
num_tps=5
d_offs=0 # just to limit the time of test
c_offs=0 # just to limit the time of test
days = data.index[d_offs:d_offs+50]
counties = data.columns[c_offs:c_offs+50]
_to_timestamp = np.frompyfunc(datetime.datetime.timestamp, 1, 1)
num_features = len(temporal_bfs(tt.fmatrix("tmp"))) * len(spatial_bfs(tt.fmatrix("tmp")))
res_0 = np.zeros((len(days), len(counties), num_features), dtype=np.float32)
for i, day in enumerate(days):
for j, county in enumerate(counties):
idx = ((day - pd.Timedelta(days=5)) <= data.index) * (data.index < day)
t_data, x_data = sample_time_and_space(data.iloc[idx], times_by_day, locations_by_county, rnd_time, rnd_loc)
t_pred, x_pred = sample_time_and_space(pd.DataFrame(num_tps, index=[day], columns=[county]), times_by_day, locations_by_county, rnd_time_pred, rnd_loc_pred)
#print("_to_timestamp(t_pred) (types, type1, size, value): ", type(_to_timestamp(t_pred)), type(_to_timestamp(t_pred)[0]), np.shape(_to_timestamp(t_pred)), _to_timestamp(t_pred)[0])
# => _to_timestamp(t_pred) (types, type1, size, value): <class 'numpy.ndarray'> <class 'float'> (5,) 1580217693.876309
#print("x_pred (types, size, value) : ", type(x_pred), type(x_pred[0]), type(x_pred[0][0]), np.shape(x_pred), x_pred[0][0])
# => x_pred (types, size, value) : <class 'numpy.ndarray'> <class 'numpy.ndarray'> <class 'numpy.float64'> (5, 2) 10.134378974970323
res_0[i, j, :] = ia_bfs(_to_timestamp(t_pred), x_pred, _to_timestamp(t_data), x_data)
# store all to compare with old algo
t_data_0 = t_data_0 + t_data.tolist()
x_data_0 = x_data_0 + x_data.tolist()
t_pred_0 = t_pred_0 + t_pred.tolist()
x_pred_0 = x_pred_0 + x_pred.tolist()
######## output ########
#display(t_data_0[:2])
#display(x_data_0[:2])
#display(t_pred_0[:2])
#display(x_pred_0[:2])
#res_0[1:2][:][:]
###Output
_____no_output_____
###Markdown
C - Do the Sampling (the NEW way) ------ C3 - COMPACT result* requires (A) to be finished -> data, times_by_day
###Code
def sample_time_and_space__once(times_by_day, locations_by_county):
"""
Convert dictonarys to arrays for faster access in sample_time_and_space().
Random access in times_by_day and locations_by_county are very costy.
Hence they need to be converted to arrays and access must be done through indexes.
"""
# times_by_day_np[day-id] => times[n_times]
times_by_day_np = pd.DataFrame.from_dict(times_by_day,orient='index').to_numpy(dtype='datetime64') # => type=='numpy.datetime64'
t_convert_1 = np.frompyfunc(pd.Timestamp, 1, 1)
times_by_day_np = t_convert_1(times_by_day_np) # => type=='pandas._libs.tslibs.timestamps.Timestamp'
t_convert_2 = np.frompyfunc(datetime.datetime.timestamp, 1, 1)
times_by_day_np = t_convert_2(times_by_day_np) # => type=='float'
times_by_day_np = np.array(times_by_day_np, np.float64) # need to convert this to np.float64 for numba
# locations_by_county_np[county-id] => locs[m_locs[x,y]]
max_coords = 0
for item in locations_by_county.items():
max_coords = max( len(item[1]), max_coords)
locations_by_county_np = np.empty([len(locations_by_county.keys()), max_coords, 2], dtype='float64')
for i,item in enumerate(locations_by_county.items()): # counties are sorted because of OrderedDict
locations_by_county_np[i][:] = item[1][:]
return(times_by_day_np, locations_by_county_np)
#times_by_day_np, locations_by_county_np = sample_time_and_space__once(times_by_day, locations_by_county)
#print("locations_by_county_np (types, size, value) : ",
# type(locations_by_county_np),
# type(locations_by_county_np[0]),
# type(locations_by_county_np[0][0]),
# np.shape(locations_by_county_np))
def sample_time_and_space__prep(times_by_day_np, locations_by_county_np, data, idx):
"""
Recalculations for a fixed dataframe sample_time_and_space().
Calculation of helper arrays are very costy.
If the dataframe does not change, precalculated values can be reused.
"""
# subdata 'data' of 'indata' is likely to skip a few first days(rows) in 'indata',
# but as times_by_day_np represents the whole 'indata', an offsets needs to be considered when accessing 'times_by_day_np'
dayoffset = np.where(idx==True)[0][0]
n_total = data.sum().sum()
# get number of samples per county-day
smpls_per_cntyday = np.array(data.values).flatten('F')
######## t_all ########
# get list of day-ids for all county-days
dayids = np.arange(len(data.index))
day_of_cntyday = np.tile(dayids, len(data.columns))
# get list of day-ids for all samples, use numpy.array for speedup in numba
day_of_smpl = np.array([ day_of_cntyday[i] for (i,smpls) in enumerate(smpls_per_cntyday) for x in range(smpls) ])
# get available times for each sample
time_of_days = data.index.tolist() # cannot be a np.array as it needs to stay a pandas.timeformat
av_times_per_day = [len(times_by_day[d]) for d in time_of_days]
av_times_per_smpl = [ av_times_per_day[day_of_cntyday[i]] for (i,smpls) in enumerate(smpls_per_cntyday) for x in range(smpls) ]
######## x_all ########
# get list of county-ids for all county-days
cntyids = np.arange(len(data.columns))
cnty_of_cntyday = np.repeat(cntyids, len(data.index))
# get list of county-ids for all samples, use numpy.array for speedup in numba
cnty_of_smpl = np.array([ cnty_of_cntyday[i] for (i,smpl) in enumerate(smpls_per_cntyday) for x in range(smpl) ])
# get available locations for each sample
label_of_cntys = data.columns # list of countys labels
av_locs_per_cnty = [len(locations_by_county[c]) for c in label_of_cntys]
av_locs_per_smpl = [ av_locs_per_cnty[cnty_of_cntyday[i]] for (i,smpls) in enumerate(smpls_per_cntyday) for x in range(smpls) ]
return (n_total, dayoffset,
day_of_smpl, av_times_per_smpl,
cnty_of_smpl, av_locs_per_smpl)
def sample_time_and_space__pred(n_days, n_counties, d_offs, c_offs, num_tps, av_times_per_smpl, av_locs_per_smpl, rnd_time, rnd_loc):
######## t_all ########
n_total = n_days * n_counties * num_tps
rnd_timeid_per_smpl = np.floor( av_times_per_smpl * rnd_time.random( n_total ) ).astype("int32")
# collect times for each sample with its random time-id
t_all = [ times_by_day_np[d_offs+i][rnd_timeid_per_smpl[(i*n_counties+j)*num_tps+x]] for i in range(n_days) for j in range(n_counties) for x in range(num_tps) ]
######## x_all ########
# calc random location-id for each sample
rnd_locid_per_smpl = np.floor( av_locs_per_smpl * rnd_loc.random((n_total,)) ).astype("int32")
# collect locations for each sample with its random location-id
x_all = [ locations_by_county_np[c_offs+j][rnd_locid_per_smpl[(i*n_counties+j)*num_tps+x]] for i in range(n_days) for j in range(n_counties) for x in range(num_tps) ]
return t_all, x_all
#@numba.jit(nopython=True, parallel=False, cache=False)
#def sample_time_and_space_tall(n_total, n_counties, dayoffset, day_of_smpl, rnd_timeid_per_smpl_all, times_by_day_np):
# # ensure we return a numpy array for better preformance
# return np.array([ times_by_day_np[day+dayoffset][rnd_timeid_per_smpl_all[j*n_total+i]] for j in range(n_counties) for (i,day) in enumerate(day_of_smpl) ], dtype=np.float64) # [county][day][smpl]
#@numba.jit(nopython=True, parallel=False, cache=False)
#def sample_time_and_space_xall(n_total, n_counties, dayoffset, cnty_of_smpl, rnd_locid_per_smpl_all, locations_by_county_np):
# # ensure we return a numpy array for better preformance
# return [ locations_by_county_np[cnty][rnd_locid_per_smpl_all[j*n_total+i]] for j in range(n_counties) for (i,cnty) in enumerate(cnty_of_smpl)] # [county][day][smpl]
@numba.jit(nopython=True, parallel=True, cache=False)
def sample_time_and_space_tx(n_total, n_counties, dayoffset,
day_of_smpl, rnd_timeid_per_smpl_all, times_by_day_np,
cnty_of_smpl, rnd_locid_per_smpl_all, locations_by_county_np):
# https://numba.pydata.org/numba-doc/latest/user/parallel.html#explicit-parallel-loops
#t_all_np = np.array([ times_by_day_np[day+dayoffset][rnd_timeid_per_smpl_all[j*n_total+i]] for j in range(n_counties) for (i,day) in enumerate(day_of_smpl) ], dtype=np.float64) # [county][day][smpl]
t_all_np = np.empty(day_of_smpl.size * n_counties, dtype=np.float64)
for j in numba.prange(n_counties):
for (i,day) in enumerate(day_of_smpl):
t_all_np[day_of_smpl.size*j+i] = times_by_day_np[day+dayoffset][rnd_timeid_per_smpl_all[j*n_total+i]]
#x_all_np = [ locations_by_county_np[cnty][rnd_locid_per_smpl_all[j*n_total+i]] for j in range(n_counties) for (i,cnty) in enumerate(cnty_of_smpl)] # [county][day][smpl]
x_all_np = np.empty((cnty_of_smpl.size * n_counties, 2), dtype=np.float64)
for j in numba.prange(n_counties):
for (i,cnty) in enumerate(cnty_of_smpl):
x_all_np[cnty_of_smpl.size*j+i] = locations_by_county_np[cnty][rnd_locid_per_smpl_all[j*n_total+i]]
return t_all_np, x_all_np
def sample_time_and_space(times_by_day_np, n_counties, n_total, dayoffset, day_of_smpl, av_times_per_smpl, cnty_of_smpl, av_locs_per_smpl, rnd_time, rnd_loc):
"""
Calculations samples in time and space.
Calculation a hughe random number array use precalulated results to pick samples.
"""
if n_total == 0:
return np.empty((0,), dtype=np.float64), np.empty((0, 2), dtype=np.float64)
######## t_all prepare ########
# calc random time-id for each sample
n_all = n_total * n_counties
av_times_per_smpl_all = np.tile(av_times_per_smpl, n_counties)
rnd_timeid_per_smpl_all = np.floor( av_times_per_smpl_all * rnd_time.random( (n_all,) ) ).astype("int32")
######## x_all prepare ########
# calc random location-id for each sample
av_locs_per_smpl_all = np.tile(av_locs_per_smpl, n_counties)
rnd_locid_per_smpl_all = np.floor( av_locs_per_smpl_all * rnd_loc.random( (n_all,) ) ).astype("int32")
######## t_all, x_all create ########
# collect times for each sample with its random time-id
#t_all = np.empty((n_total,), dtype=object)
#t_all = [ times_by_day_np[day+dayoffset][rnd_timeid_per_smpl_all[j*n_total+i]] for j in range(n_counties) for (i,day) in enumerate(day_of_smpl) ] # [county][day][smpl]
#t_all = sample_time_and_space_tall(n_total, n_counties, dayoffset, day_of_smpl, rnd_timeid_per_smpl_all, times_by_day_np)
# collect locations for each sample with its random location-id
#x_all = np.empty((n_total, 2))
#x_all = [ locations_by_county_np[cnty][rnd_locid_per_smpl_all[j*n_total+i]] for j in range(n_counties) for (i,cnty) in enumerate(cnty_of_smpl)] # [county][day][smpl]
#x_all = sample_time_and_space_xall(n_total, n_counties, dayoffset, cnty_of_smpl, rnd_locid_per_smpl_all, locations_by_county_np)
t_all, x_all = sample_time_and_space_tx(n_total, n_counties, dayoffset,
day_of_smpl, rnd_timeid_per_smpl_all, times_by_day_np,
cnty_of_smpl, rnd_locid_per_smpl_all, locations_by_county_np)
# numba diagnostics
#sample_time_and_space_tx.parallel_diagnostics(level=4)
return t_all, x_all
#%%timeit
# set seed to check results
# Parallel Random Number Generation - https://docs.scipy.org/doc/numpy/reference/random/parallel.html
# Multithreaded Generation - https://docs.scipy.org/doc/numpy/reference/random/multithreading.html
rnd_time = np.random.Generator(np.random.PCG64(12345))
rnd_loc = np.random.Generator(np.random.PCG64(12345))
rnd_time_pred = np.random.Generator(np.random.PCG64(12345))
rnd_loc_pred = np.random.Generator(np.random.PCG64(12345))
# limit the number of threads
number_of_threads = 12
numba.set_num_threads(number_of_threads)
# Convert dictonarys to arrays for faster access in sample_time_and_space().
(times_by_day_np, locations_by_county_np,) = sample_time_and_space__once(times_by_day, locations_by_county)
t_data_1 = []
x_data_1 = []
t_pred_1 = []
x_pred_1 = []
d_offs=0 # just to limit the time of test
c_offs=0 # just to limit the time of test
days = data.index[d_offs:d_offs+50]
counties = data.columns[c_offs:c_offs+50]
num_features = len(temporal_bfs(tt.fmatrix("tmp"))) * len(spatial_bfs(tt.fmatrix("tmp")))
res_1 = np.zeros((len(days), len(counties), num_features), dtype=np.float32)
num_tps=5
n_days = len(days)
n_counties = len(counties)
# create dataframe with 'num_tps' in each cell
pred_data = pd.DataFrame(num_tps, index=days, columns=counties)
idx = np.empty([len(data.index)], dtype='bool')
idx.fill(True)
# precalculate pediction values
(n_total, dayoffset, day_of_smpl, av_times_per_smpl, cnty_of_smpl, av_locs_per_smpl,) = sample_time_and_space__prep(times_by_day_np, locations_by_county_np, pred_data, idx)
(t_pred_all, x_pred_all,) = sample_time_and_space__pred(n_days, n_counties, d_offs, c_offs, num_tps, av_times_per_smpl, av_locs_per_smpl, rnd_time_pred, rnd_loc_pred)
for i, day in enumerate(days):
# calc which sub-table will be selected
idx = ((day - pd.Timedelta(days=5)) <= data.index) * (data.index < day)
subdata = data.iloc[idx]
if subdata.size != 0:
#print(i, day)
# Recalculations for a fixed dataframe sample_time_and_space().
(n_total, dayoffset, day_of_smpl, av_times_per_smpl, cnty_of_smpl, av_locs_per_smpl,) = sample_time_and_space__prep(times_by_day_np, locations_by_county_np, subdata, idx)
# Calculate time and space samples for all counties at once
(t_data_all, x_data_all,) = sample_time_and_space(times_by_day_np, len(counties), n_total, dayoffset, day_of_smpl, av_times_per_smpl, cnty_of_smpl, av_locs_per_smpl, rnd_time, rnd_loc)
#print(np.shape(x_data_all), type(x_data_all[0][0]))
for j, county in enumerate(counties):
# calcs only for the single DataFrame.cell[day][county]
offs = (i*n_counties+j)*num_tps
t_pred = t_pred_all[offs:offs+num_tps]
x_pred = x_pred_all[offs:offs+num_tps]
# get subarray for county==j
t_data = t_data_all[j*n_total:(j+1)*n_total] # [county][smpl]
x_data = x_data_all[j*n_total:(j+1)*n_total] # [county][smpl]
# use theano.function for day==i and county==j
res_1[i, j, :] = ia_bfs(t_pred, x_pred, t_data, x_data)
#res_1[1:2][:][:]
np.array_equal(res_0, res_1)
###Output
_____no_output_____
|
AI/AshTutor.ipynb
|
###Markdown
Data Splitted into Training and Test
###Code
training_set = pd.read_csv('https://raw.githubusercontent.com/PersieB/AshTutor/master/Training_set.csv')
training_set['Label'] = training_set['Label'].apply(recode)
training_set.head()
training_set.count()
test = pd.read_csv('https://raw.githubusercontent.com/PersieB/AshTutor/master/Test_set.csv')
test.head()
test['Label'] = test['Label'].apply(recode)
test.head()
test.count()
###Output
_____no_output_____
###Markdown
Remove punctuations
###Code
import string
def remove_punctuation(review):
non_punctuated = [x for x in review if x not in string.punctuation]
non_punctuated = ''.join(non_punctuated)
return non_punctuated
###Output
_____no_output_____
###Markdown
Nltk stopwords Remove stopwords
###Code
from nltk.corpus import stopwords
def remove_stopwords(reviews):
stop_words = set(stopwords.words('english')) - set(['not', 'off'])
return [word for word in reviews.split() if word.lower() not in stop_words]
###Output
_____no_output_____
###Markdown
Tokenize reviews
###Code
def tokenization(review):
s = remove_punctuation(review)
tokens = remove_stopwords(s)
tokens = [i.lower() for i in tokens]
return tokens
###Output
_____no_output_____
###Markdown
Naive Bayes Algorithm function **train** which implements the naive Bayes algorithm. It takes in parameters D (the training document) and C (a list containing the classes) and returns the loglikelihood, logprior and V (a set of vocabulary)
###Code
def training(D, C):
N_doc = len(D) #number of documents (reviews) in D
#N_c = {} #number of documents from D in class c
#intitialising dictionary to hold logpriors
log_priors = {x:0 for x in C}
#intitialising dictionary to hold loglikelihoods
log_likelihood = {x:{} for x in C}
Vocabulary = [] #Vocabulary initialised empty
big_doc_c = {k:{} for k in C}
D['Review'] = D['Review'].apply(lambda review: tokenization(review))
for c in C:
class_reviews = training_set[training_set['Label']==c] # all reviews belonging to a class
N_c = len(class_reviews) # number of reviews belonging to a particular class
log_priors[c] = math.log(N_c/N_doc) # log prior of c
for i, j in class_reviews.iterrows():
temp = j['Review']
for word in temp:
if word not in big_doc_c[c].keys():
big_doc_c[c][word] = 0 # append(d) for d in D with class c
big_doc_c[c][word] += 1 # increase word count
Vocabulary.append(word)
count = 0
for c in C:
count = sum(big_doc_c[c].values())
Vocabulary = set(Vocabulary) # only unique words in vocabulary
for token in Vocabulary:
count_w_c_in_V = 0
if token in big_doc_c[c]:
count_w_c_in_V = big_doc_c[c][token]
log_likelihood[c][token] = math.log((count_w_c_in_V + 1)/(count + len(Vocabulary))) #loglikelihoods for each word in a partlicular class
return log_priors, log_likelihood, Vocabulary
###Output
_____no_output_____
###Markdown
Training
###Code
C = np.unique(dataset['Label'].values)
logprior, loglikelihood, V = training(training_set, C)
###Output
_____no_output_____
###Markdown
function** predict** which implements the prediction or test function for the naive Bayes algorithm.
###Code
def predict(testdoc, logprior, loglikelihood, C, V):
sum_c = {c:logprior[c] for c in C}
for c in C:
for word in tokenization(testdoc):
if word in V:
sum_c[c] += loglikelihood[c][word]
#arg_max of sum_c #key (class) with the largest value
v = list(sum_c.values())
k = list(sum_c.keys())
chosen_class = k[v.index(max(v))] #return the key with the largest values
if(chosen_class == 0):
return 0
else:
return 1
###Output
_____no_output_____
###Markdown
Testing
###Code
predictions = []
for i in test['Review'].values:
sentiment = predict(i, logprior, loglikelihood, C, V)
predictions.append(sentiment)
#print(i, sentiment)
test['Implemented Naive Bayes'] = predictions
test.tail(10)
sen = predict('very bad tutorial', logprior, loglikelihood, C, V)
print(sen)
###Output
1
|
3_Pandas/09_TimeSpeedup.ipynb
|
###Markdown
Tips and Tricks for Pandas
###Code
%load_ext Cython
import numpy as np
np.random.seed(0)
import pandas as pd
def make_df():
return pd.DataFrame(
np.random.rand(10_000, 3),
columns=["A", "B", "C"]
)
df = make_df()
print(df.head())
###Output
A B C
0 0.548814 0.715189 0.602763
1 0.544883 0.423655 0.645894
2 0.437587 0.891773 0.963663
3 0.383442 0.791725 0.528895
4 0.568045 0.925597 0.071036
###Markdown
Speed-Up ApplyUse-Case:Replace the value in a column by 0.0 if it is less than 0.5
###Code
def slow_function(df):
col = "A"
for idx, row in df.iterrows():
if row[col] < 0.5:
row[col] = 0.0
%timeit slow_function(df)
print(df.head())
def faster_function(df):
df["B"] = df["B"].apply(lambda x: 0.0 if x < 0.5 else x)
df = make_df()
%timeit faster_function(df)
print(df.head())
def even_faster_function(df):
df["C"] = np.where(df["C"] < 0.5, 0.0, df["C"])
df = make_df()
%timeit even_faster_function(df)
print(df.head())
%%cython
cimport cython
cimport numpy as np
import numpy as np
@cython.boundscheck(False)
@cython.wraparound(False)
cpdef inner(np.ndarray[double, ndim=1] array):
for i in range(array.shape[0]):
if array[i] < 0.5:
array[i] = 0.0
def cython_apply(df):
inner(df["A"].values)
df = make_df()
%timeit cython_apply(df)
print(df.head())
import numba
@numba.jit
def inner(array):
for i in range(array.shape[0]):
if array[i] < 0.5:
array[i] = 0.0
def numba_apply(df):
inner(df["A"].values)
df = make_df()
%timeit numba_apply(df)
print(df.head())
###Output
137 µs ± 30.9 µs per loop (mean ± std. dev. of 7 runs, 1 loop each)
A B C
0 0.623699 0.136427 0.893584
1 0.000000 0.579009 0.493986
2 0.620274 0.649347 0.259397
3 0.888801 0.079085 0.380168
4 0.000000 0.206583 0.707093
###Markdown
| Function | Time | Speed-Up || --- |- |- || slow_function | 1.58 * 10^6     | - || faster_function | 6.18 * 10^3 | 255x || even_faster_function   | 680     | 2,323x || cython_function | 55 | 28,727x || numba_function | 55 | 28,727x || c++ | 30 | 52,666x | Speed-Up Transform
###Code
def slow_function(df):
col = "A"
for idx, row in df.iterrows():
row[col] = row[col] + 1.0
df = make_df()
%timeit slow_function(df)
print(df.head())
def faster_function(df):
df["B"] = df["B"].transform(lambda x: x + 1.0)
df = make_df()
%timeit faster_function(df)
print(df.head())
def even_faster_function(df):
df["C"] = df["C"] + 1.0
df = make_df()
%timeit even_faster_function(df)
print(df.head())
%%cython
cimport cython
cimport numpy as np
import numpy as np
@cython.boundscheck(False)
@cython.wraparound(False)
cpdef inner(np.ndarray[double, ndim=1] array):
for i in range(array.shape[0]):
array[i] += 1.0
def cython_transform(df):
inner(df["A"].values)
df = make_df()
%timeit cython_transform(df)
print(df.head())
import numba
@numba.jit
def inner(array):
for i in range(array.shape[0]):
array[i] += 1.0
def numba_transform(df):
inner(df["A"].values)
df = make_df()
%timeit numba_transform(df)
print(df.head())
###Output
27.1 µs ± 719 ns per loop (mean ± std. dev. of 7 runs, 10000 loops each)
A B C
0 81111.205671 0.947660 0.097525
1 81111.719924 0.841147 0.863829
2 81111.439500 0.124873 0.657900
3 81111.050730 0.564794 0.142302
4 81111.403096 0.118479 0.425367
|
Cross Spectral Analysis.ipynb
|
###Markdown
Cross Spectral Analysis
###Code
import numpy as np
import matplotlib.pyplot as plt
from scipy.fft import fft, fftfreq, fftshift
t = np.linspace(0,50,1000)
t.shape[-1]
x = np.sin(t) + np.sin(2*t)+ np.sin(4*t)+ np.sin(5*t)+ np.sin(6*t)
y = np.sin(3*t)+ np.sin(2*t)+ np.sin(4*t)+ np.sin(7*t)+ np.sin(9*t)
plt.plot(x)
plt.plot(y)
###Output
_____no_output_____
###Markdown
$$F_{x}( k) \ =C_{xk} e^{i\theta _{xk}} \ e^{i\frac{2\pi }{T} kt} =\ \ \frac{1}{2}( A_{xk} -iB_{xk}) e^{i\frac{2\pi }{T} kt}$$
###Code
Fx = fftshift(fft(x))
Fy = fftshift(fft(y))
Ffreq = fftshift(fftfreq(t.shape[-1]))
plt.plot(Ffreq, Fx.real, Ffreq, Fy.real)
###Output
_____no_output_____
###Markdown
$$co-spectra\ =\ A_{xk} A_{yk} +B_{xk} B_{yk}$$
###Code
Cxy = Fx.real*Fy.real + Fx.imag*Fy.imag
plt.plot(Cxy)
###Output
_____no_output_____
###Markdown
$$quad-spectra\ =\ A_{xk} B_{yk} -A_{yk} B_{xk}$$
###Code
Qxy = Fx.real*Fy.imag - Fx.imag*Fy.real
plt.plot(Qxy)
enumerate(Cxy)
cross = [np.complex(i, j) for index, [i,j] in zip(enumerate(Cxy),enumerate(Qxy))]
plt.plot(cross)
###Output
C:\Users\starlord\anaconda3\lib\site-packages\numpy\core\_asarray.py:83: ComplexWarning: Casting complex values to real discards the imaginary part
return array(a, dtype, copy=False, order=order)
|
docs/instruments/amor/amor_reduction.ipynb
|
###Markdown
Data reduction for AmorIn this notebook, we will look at the reduction workflow for reflectometry data collected from the PSI[Amor](https://www.psi.ch/en/sinq/amor) instrument.This is a living document and there are plans to update this as necessary with changes in the data reduction methodology and code.We will begin by importing the modules that are necessary for this notebook and loading the data.The `sample.nxs` file is the experimental data file of interest,while `reference.nxs` is the reference measurement of the neutron supermirror.
###Code
import scipp as sc
from ess import amor
###Output
_____no_output_____
###Markdown
The Amor beamlineBefore we can load the data, we need to define the parameters of the beamline.We begin by defining the convention for naming angles in our set-up.We use the Fig. 5 from the paper by [Stahn & Glavic (2016)](https://doi.org/10.1016/j.nima.2016.03.007),which is reproduced below (along with its caption).The yellow area shows the incoming and reflected beam, both with the divergence $\Delta \theta$.The inclination of the sample relative to the centre of the incoming beam(here identical to the instrument horizon) is called $\omega$,and the respective angle of the reflected beam relative to the same axis is $\gamma$.In general the detector centre is located at $\gamma_{\rm D} = 2\omega$.These are instrument coordinates and should not be confused with the situation on the sample,where the take-off angle of an individual neutron trajectory is called $\theta$.The `amor` module provides a helper function that generates the default beamline parameters.This function requires the sample rotation angle ($\omega$) as an input to fully define the beamline.In the future, all these beamline parameters (including the sample rotation) will be included in the file meta data.For now, we must define this manually.
###Code
sample_rotation = sc.scalar(0.7989, unit='deg')
amor_beamline = amor.make_beamline(sample_rotation=sample_rotation)
###Output
_____no_output_____
###Markdown
Loading the dataUsing the `amor.load` function, we load the `sample.nxs` file and perform some early preprocessing:- The `tof` values are converted from nanoseconds to microseconds.- The raw data contains events coming from two pulses, and these get folded into a single `tof` range
###Code
sample = amor.load(amor.data.get_path("sample.nxs"),
beamline=amor_beamline)
sample
###Output
_____no_output_____
###Markdown
By simply plotting the data, we get a first glimpse into the data contents.
###Code
sc.plot(sample)
###Output
_____no_output_____
###Markdown
Correcting the position of the detector pixels**Note:** once new Nexus files are produced, this step should go away. The pixel positions are wrong in the `sample.nxs` file, and require an ad-hoc correction.We apply an arbitrary shift in the vertical (`y`) direction.We first move the pixels down by 0.955 degrees,so that the centre of the beam goes through the centre of the top half of the detector blades(the bottom half of the detectors was turned off).Next, we move all the pixels so that the centre of the top half of the detector pixels lies at an angle of $2 \omega$,as described in the beamline diagram.
###Code
def pixel_position_correction(data: sc.DataArray):
return data.coords['position'].fields.z * sc.tan(2.0 *
data.coords['sample_rotation'] -
(0.955 * sc.units.deg))
sample.coords['position'].fields.y += pixel_position_correction(sample)
###Output
_____no_output_____
###Markdown
We now check that the detector pixels are in the correct position by showing the instrument view
###Code
amor.instrument_view(sample)
###Output
_____no_output_____
###Markdown
Coordinate transformation graphTo compute the wavelength $\lambda$, the scattering angle $\theta$, and the $Q$ vector for our data,we construct a coordinate transformation graph.It is based on classical conversions from `tof` and pixel `position` to $\lambda$ (`wavelength`),$\theta$ (`theta`) and $Q$ (`Q`),but comprises a number of modifications.The computation of the scattering angle $\theta$ includes a correction for the Earth's gravitational field which bends the flight path of the neutrons.The angle can be found using the following expression$$\theta = \sin^{-1}\left(\frac{\left\lvert y + \frac{g m_{\rm n}}{2 h^{2}} \lambda^{2} L_{2}^{2} \right\rvert }{L_{2}}\right) - \omega$$where $m_{\rm n}$ is the neutron mass,$g$ is the acceleration due to gravity,and $h$ is Planck's constant.For a graphical representation of the above expression,we consider once again the situation with a convergent beam onto an inclined sample.The detector (in green), whose center is located at an angle $\gamma_{\rm D}$ from the horizontal plane,has a physical extent and is measuring counts at multiple scattering angles at the same time.We consider two possible paths for neutrons.The first path (cyan) is travelling horizontally from the source to the sample and subsequently,following specular reflection, hits the detector at $\gamma_{\rm D}$ from the horizontal plane.From the symmetry of Bragg's law, the scattering angle for this path is $\theta_{1} = \gamma_{\rm D} - \omega$.The second path (red) is hitting the bottom edge of the detector.Assuming that all reflections are specular,the only way the detector can record neutron events at this location is if the neutron originated from the bottom part of the convergent beam.Using the same symmetry argument as above, the scattering angle is $\theta_{2} = \gamma_{2} - \omega$. This expression differs slightly from the equation found in the computation of the $\theta$ angle in other techniques such as[SANS](https://docs.mantidproject.org/nightly/algorithms/Q1D-v2.htmlq-unit-conversion),in that the horizontal $x$ term is absent,because we assume a planar symmetry and only consider the vertical $y$ component of the displacement.The conversion graph is defined in the reflectometry module,and can be obtained via
###Code
graph = amor.conversions.specular_reflection()
sc.show_graph(graph, simplified=True)
###Output
_____no_output_____
###Markdown
Computing the wavelengthTo compute the wavelength of the neutrons,we request the `wavelength` coordinate from the `transform_coords` method by supplying our graph defined above(see [here](https://scipp.github.io/scippneutron/user-guide/coordinate-transformations.html)for more information about using `transform_coords`).We also exclude all neutrons with a wavelength lower than 2.4 &8491;.
###Code
sample_wav = sample.transform_coords(["wavelength"], graph=graph)
wavelength_edges = sc.array(dims=['wavelength'], values=[2.4, 16.0], unit='angstrom')
sample_wav = sc.bin(sample_wav, edges=[wavelength_edges])
sample_wav
sample_wav.bins.concatenate('detector_id').plot()
###Output
_____no_output_____
###Markdown
Compute the Q vectorUsing the same method, we can compute the $Q$ vector,which now depends on both detector position (id) and wavelength
###Code
sample_q = sample_wav.transform_coords(["Q"], graph=graph)
sample_q
q_edges = sc.geomspace(dim='Q', start=0.008, stop=0.08, num=201, unit='1/angstrom')
sample_q_binned = sc.bin(sample_q, edges=[q_edges])
sample_q_summed = sample_q_binned.sum('detector_id')
sc.plot(sample_q_summed["wavelength", 0], norm="log")
###Output
_____no_output_____
###Markdown
Normalize by the super-mirrorTo perform the normalization, we load the super-mirror `reference.nxs` file.
###Code
reference = amor.load(amor.data.get_path("reference.nxs"),
beamline=amor_beamline)
reference.coords['position'].fields.y += pixel_position_correction(reference)
reference
###Output
_____no_output_____
###Markdown
We convert the reference to wavelength using the same graph
###Code
reference_wav = reference.transform_coords(["wavelength"], graph=graph)
reference_wav = sc.bin(reference_wav, edges=[wavelength_edges])
reference_wav.bins.concatenate('detector_id').plot()
###Output
_____no_output_____
###Markdown
And we then convert to $Q$ as well
###Code
reference_q = reference_wav.transform_coords(["Q"], graph=graph)
reference_q_binned = sc.bin(reference_q, edges=[q_edges])
reference_q_summed = reference_q_binned.sum('detector_id')
sc.plot(reference_q_summed["wavelength", 0], norm="log")
###Output
_____no_output_____
###Markdown
Finally, we divide the sample by the reference to obtain
###Code
normalized = sample_q_summed["wavelength", 0] / reference_q_summed["wavelength", 0]
sc.plot(normalized, norm="log")
###Output
_____no_output_____
###Markdown
Make a $(\lambda, \theta)$ mapA good sanity check is to create a two-dimensional map of the counts in $\lambda$ and $\theta$ bins.To achieve this, we request two output coordinates from the `transform_coords` method.
###Code
sample_theta = sample.transform_coords(["theta", "wavelength"], graph=graph)
sample_theta
###Output
_____no_output_____
###Markdown
Then, we concatenate all the events in the `detector_id` dimension
###Code
sample_theta = sample_theta.bins.concatenate('detector_id')
sample_theta
###Output
_____no_output_____
###Markdown
Finally, we bin into the existing `theta` dimension, and into a new `wavelength` dimension,to create a 2D output
###Code
nbins = 165
theta_edges = sc.linspace(dim='theta', start=0.4, stop=1.2, num=nbins, unit='deg')
wavelength_edges = sc.linspace(dim='wavelength', start=1.0, stop=15.0, num=nbins, unit='angstrom')
binned = sc.bin(sample_theta, edges=[sc.to_unit(theta_edges, 'rad'), wavelength_edges])
binned
binned.bins.sum().plot()
###Output
_____no_output_____
|
SD-TSIA214_Machine_Learning_for_Text_Mining/lab/Lab2-Sentiment_classification_of_movie_reviews/TP Sentiment Analysis in textual movie reviews.ipynb
|
###Markdown
Set up
###Code
import os.path as op
import numpy as np
from sklearn.base import BaseEstimator, ClassifierMixin
from glob import glob
import re
import pandas as pd
import operator
import unittest
import sys
import operator
from sklearn.model_selection import *
import matplotlib.pyplot as plt
from sklearn.svm import LinearSVC
from sklearn.naive_bayes import MultinomialNB
from sklearn.feature_extraction.text import CountVectorizer
from sklearn.pipeline import Pipeline
from sklearn.linear_model import LogisticRegression
plt.style.use('ggplot')
%load_ext line_profiler
%matplotlib inline
# Load data
print("Loading dataset")
filenames_neg = sorted(glob(op.join('.', 'data', 'imdb1', 'neg', '*.txt')))
filenames_pos = sorted(glob(op.join('.', 'data', 'imdb1', 'pos', '*.txt')))
texts_neg = [open(f).read() for f in filenames_neg]
texts_pos = [open(f).read() for f in filenames_pos]
texts = texts_neg + texts_pos
y = np.ones(len(texts), dtype=np.int)
y[:len(texts_neg)] = 0.
print("%d documents" %len(texts))
###Output
Loading dataset
2000 documents
###Markdown
Implementation of the classifier Question 1Complete the count_words function that will count the number of occurrences of each distinctword in a list of string and return vocabulary (the python dictionary. The dictionary keys are thedifferent words and the values are their number of occurences).
###Code
def count_words(texts):
"""Vectorize text : return count of each word in the text snippets
Parameters
----------
texts : list of str
The texts
Returns
-------
vocabulary : dict
A dictionary that points to an index in counts for each word.
counts : ndarray, shape (n_samples, n_features)
The counts of each word in each text.
n_samples == number of documents.
n_features == number of words in vocabulary.
"""
wordList = re.subn('\W', ' ', ' '.join(texts).lower())[0].split(' ')
words = set(wordList)
vocabulary = dict(zip(words, range(len(words))))
n_features = len(words)
counts = np.zeros( (len(texts), n_features))
for l, text in enumerate(texts, 0):
wordList = re.subn('\W', ' ', text.lower())[0].split(' ')
for word in wordList:
if(word == ''): continue
counts[l][vocabulary[word]] = counts[l][vocabulary[word]] + 1
return vocabulary, counts
###Output
_____no_output_____
###Markdown
Question 2Explain how positive and negative classes have been assigned to movie reviews (see poldata.README.2.0 file)According to the poldata.README.2.0 file, movie reviews may not have an explicit note. In the case where there is a note, this one can appear to different place of the file and in different forms. In the case of our data we focus only on the most explicit notations, resulting from an ad-hoc set of rules. The classification of a file is thus established from the first note of which one is able to identify.- With a five-star system (or equivalent digital system), more than 3.5 stars lead to positive reviews, and fewer than 2 stars lead to negative reviews.- With a four-star system (or equivalent digital system), more than 3 stars lead to positive reviews, and fewer than 1.5 stars lead to negative reviews.- With a system consisting of letters, B or above leads to a positive critique, and C- or below is considered negative. Question 3Complete the NB class to implement the Naive Bayes classifier.
###Code
class NB(BaseEstimator, ClassifierMixin):
def __init__(self):
pass
def fit(self, X, y):
N = X.shape[0]
val, counter = np.unique(y, return_counts=True)
self.class_counter = dict(zip(val, counter))
nb_word = X.shape[1]
nb_class = val.shape[0]
self.condprob = dict()
self.prior = np.zeros(nb_class)
T = {c:np.sum(X[y==c, :], axis=0) for c in self.class_counter}
for c,Nc in self.class_counter.items():
self.prior[c] = Nc/N
self.condprob[c] = (T[c] +1) / ( np.sum(T[c] + 1))
return self
def predict(self, X):
X = np.array(X)
y = [self.apply(vec) for vec in X]
return np.array(y)
def apply(self, vector):
vector = np.array(vector)
score = dict()
for c in self.class_counter:
score[c] = np.log(self.prior[c])
score[c] += np.log(self.condprob[c][vector >0.0]).sum()
return max(score.items(), key=operator.itemgetter(1))[0]
def score(self, X, y):
return np.mean(self.predict(X) == y)
# Count words in text
vocabulary, X = count_words(texts)
# Try to fit, predict and score
nb = NB()
nb.fit(X[::2], y[::2])
score = nb.score(X[1::2], y[1::2])
score
###Output
_____no_output_____
###Markdown
Question 4 Evaluate the performance of your classifier in cross-validation 5-folds
###Code
# Try to fit, predict and score
nb = NB()
my_nb_res = cross_validate(nb, X, y, cv=5)
my_nb_res
###Output
D:\ProgramData\Anaconda3\lib\site-packages\sklearn\utils\deprecation.py:122: FutureWarning: You are accessing a training score ('train_score'), which will not be available by default any more in 0.21. If you need training scores, please set return_train_score=True
warnings.warn(*warn_args, **warn_kwargs)
###Markdown
Stop word
###Code
def count_words(texts, stop_word = False):
"""Vectorize text : return count of each word in the text snippets
Parameters
----------
texts : list of str
The texts
Returns
-------
vocabulary : dict
A dictionary that points to an index in counts for each word.
counts : ndarray, shape (n_samples, n_features)
The counts of each word in each text.
n_samples == number of documents.
n_features == number of words in vocabulary.
"""
wordList = re.subn('\W', ' ', ' '.join(texts).lower())[0].split(' ')
words = set(wordList)
if stop_word:
with open("./data/english.stop", 'r') as file:
stop_words = set(file.read())
words = words - stop_words
vocabulary = dict(zip(words, range(len(words))))
n_features = len(words)
counts = np.zeros( (len(texts), n_features))
for l, text in enumerate(texts, 0):
wordList = re.subn('\W', ' ', text.lower())[0].split(' ')
for word in wordList:
if(word not in words): continue
counts[l][vocabulary[word]] = counts[l][vocabulary[word]] + 1
return vocabulary, counts
# Count words in text
vocabulary, X = count_words(texts, stop_word = True)
# Try to fit, predict and score
nb = NB()
nb.fit(X[::2], y[::2])
score_no_stop_word = nb.score(X[1::2], y[1::2])
score_no_stop_word
# Try to fit, predict and score
nb = NB()
my_nb_res_stop_word = cross_validate(nb, X, y, cv=5)
my_nb_res_stop_word
v, X_stop_word = count_words(texts, stop_word = True)
v, X = count_words(texts, stop_word = False)
scores = []
no_sw_scores = []
nb = NB()
for cv in range(2,10):
scores.append(cross_val_score(nb, X, y, cv=cv).mean())
no_sw_scores.append(cross_val_score(nb, X_stop_word, y, cv=cv).mean())
# Plot all# Plot a
plt.plot(np.arange(2,10), scores, linewidth=4, alpha=0.6,
marker='o', color="mediumseagreen", label='With stop word')
plt.plot(np.arange(2,10), no_sw_scores, linewidth=4, alpha=0.6,
marker='o', color="orange", label='Withuot stop words')
plt.title("Average score of crossing validation")
plt.ylabel('Score')
plt.xlabel('Number of folds')
plt.legend()
plt.show()
print("Score with stop words : ", score)
print("Score without stop words : ", score_no_stop_word)
###Output
_____no_output_____
###Markdown
Accroding to the score, we find when we remove the stop-words in the texts, we can improve the performence of the estimator. But when we change the number of folds. The performences are not always improved when we remove the stop word. It's means that the stop words don't always have a significant influence on the results of classification. SCIKIT-LEARN USE Question 1:Compare your implementation with scikitlearn
###Code
nb_pipline = Pipeline([('count', CountVectorizer()), ('nb', MultinomialNB())])
nb_res = cross_validate(nb_pipline.set_params(
count__analyzer='word'), texts, y, cv=5)
nb_bigram_res = cross_validate(nb_pipline.set_params(
count__analyzer='word', count__ngram_range=(2, 2)), texts, y, cv=5)
nb_char_res = cross_validate(nb_pipline.set_params(
count__analyzer='char', count__ngram_range=(1, 1), count__stop_words=None), texts, y, cv=5)
nb_res_stop_word = cross_validate(nb_pipline.set_params(
count__analyzer='word', count__ngram_range=(3,6), count__stop_words='english'), texts, y, cv=5)
nb_bigram_res_stop_word = cross_validate(nb_pipline.set_params(
count__analyzer='word', count__ngram_range=(2, 2), count__stop_words='english'), texts, y, cv=5)
nb_char_res_stop_word = cross_validate(nb_pipline.set_params(
count__analyzer='char', count__ngram_range=(3, 6), count__stop_words='english'), texts, y, cv=5)
nb_score = np.mean(nb_res['test_score'])
label = ['Homemade NB', 'NB','NB/bigram', 'NB/char'
,'Homemade NB/stop word'
, 'NB/stop word' , 'NB/bigram/stop word'
, 'NB/char/stop word']
data = [my_nb_res
,nb_res
,nb_bigram_res
,nb_char_res
, my_nb_res_stop_word
,nb_res_stop_word
,nb_bigram_res_stop_word
,nb_char_res_stop_word]
fig,ax = plt.subplots(figsize=(16,9))
x = [np.mean(res['test_score']) for res in data]
rect = ax.bar(range(len(label)),x, tick_label =label)
plt.ylim(min(x) -0.01, max(x) + 0.01)
plt.title("Test Score")
def autolabel(rects):
"""
Attach a text label above each bar displaying its height
"""
for rect in rects:
height = rect.get_height()
ax.text(rect.get_x() + rect.get_width()/2., 1.01*height,
'%.3f' % height,
ha='center', va='bottom')
autolabel(rect)
plt.plot()
###Output
_____no_output_____
###Markdown
According to the results above, we can find that the the performence of homemade implementation is enough good. This the bigram not work well for our samples.With the bigram, we can improve the performence. And we also find that removing the stop word can not always improve the performences.Influcenc about When we work on substrings of characters, we get the lowest scores. Question 2Test another classification method scikitlearn (ex : LinearSVC, LogisticRegression). Setup
###Code
def test_score(estimator, texts = texts):
lable = ["Word", "Bigram", "Char"
,"Word/Stop Word", "Bigram/Stop Word", "Char/Stop Word"]
res = cross_validate(estimator, texts, y, cv=5)
bigram_res = cross_validate(estimator.set_params(
count__analyzer='word', count__ngram_range=(2, 2), count__stop_words=None), texts, y, cv=5)
char_res = cross_validate(estimator.set_params(
count__analyzer='char', count__ngram_range=(1, 1), count__stop_words=None), texts, y, cv=5)
res_stop_word = cross_validate(estimator.set_params(
count__analyzer='word', count__ngram_range=(3,6), count__stop_words='english'), texts, y, cv=5)
bigram_res_stop_word = cross_validate(estimator.set_params(
count__analyzer='word', count__ngram_range=(2, 2), count__stop_words='english'), texts, y, cv=5)
char_res_stop_word = cross_validate(estimator.set_params(
count__analyzer='char', count__ngram_range=(3, 6), count__stop_words='english'), texts, y, cv=5)
return label, [res, bigram_res,char_res, res_stop_word, bigram_res_stop_word,char_res_stop_word]
###Output
_____no_output_____
###Markdown
Linear SVC
###Code
%%time
svc_pipline = Pipeline([('count', CountVectorizer()), ('svc', LinearSVC())])
label_svc, data_svc = test_score(svc_pipline)
svc_score = np.mean(data_svc[0]['test_score'])
###Output
Wall time: 14min 31s
###Markdown
Logistic regression
###Code
%%time
lr_pipline = Pipeline([('count', CountVectorizer()),
('lr', LogisticRegression())])
label_lr, data_lr = test_score(lr_pipline)
lr_score = np.mean(data_svc[0]['test_score'])
###Output
Wall time: 11h 32min 1s
###Markdown
Comparaison
###Code
label = ["Word", "Bigram", "Char"
,"Word/Stop Word", "Bigram/Stop Word", "Char/Stop Word"]
data_nb = [nb_res
,nb_bigram_res
,nb_char_res
,nb_res_stop_word
,nb_bigram_res_stop_word
,nb_char_res_stop_word]
fig, ax = plt.subplots(figsize=(16,9))
width = 0.2
alp = 0.8
ind = np.arange(6)
print([np.mean(l['test_score']) for l in data_nb])
rects1 = ax.bar(ind, [np.mean(l['test_score']) for l in data_nb],
width = width,label='MultinomialNB', color='skyblue' ,
yerr=[np.std(l['test_score']) for l in data_nb]
)
rects2 = ax.bar(ind + width, [np.mean(l['test_score']) for l in data_lr],
width, alpha=alp, label='LinearSVC', color='gold'
,yerr=[np.std(l['test_score']) for l in data_svc])
#
rects3 = ax.bar(ind + 2 * width, [np.mean(l['test_score']) for l in data_svc],
width, alpha=alp, label='LogisticRegression', color='lightpink'
, yerr=[np.std(l['test_score']) for l in data_lr]
)
# add some text for labels, title and axes ticks
ax.set_ylabel('Scores')
ax.set_title('Mean Scores by preprocessing for MultinomialNB')
ax.set_xticks(ind + 2 * width / 2)
ax.set_xticklabels(label)
ax.set_ylim(0.4,0.9)
plt.legend(loc="lower right")
def autolabel(rects):
"""
Attach a text label above each bar displaying its height
"""
for rect in rects:
height = rect.get_height()
ax.text(rect.get_x() + rect.get_width()/2., 1.02*height,
'%.3f' % height,
ha='center', va='bottom')
autolabel(rects1)
autolabel(rects2)
autolabel(rects3)
plt.show()
###Output
[0.812, 0.8305000000000001, 0.6094999999999999, 0.6885, 0.7835, 0.8215]
###Markdown
With this figure, the performence of SVC is best when we work on word and keep the stop word. When we rmove the stop word from our texts, the performence about word and bigram decline for the three algorithms. When we segement the texts by character, the performence is so bad, but when we remove the stop word, the perfomence significantly enhance, the score even is higher than the segmentation by word. In ours sample, the bigram don't bring benefit for our clasification. Question 3 NLTKUse NLTK library in order to process a stemming. You will used the classSnowballStemmer.
###Code
from nltk import SnowballStemmer
stemmer = SnowballStemmer("english")
class StemmedCountVectorizer(CountVectorizer):
def build_analyzer(self):
analyzer = super(StemmedCountVectorizer, self).build_analyzer()
return lambda doc: ([stemmer.stem(w) for w in analyzer(doc)])
%%time
## MultinamialNB with stem
nb_stem_pipline = Pipeline([('count', StemmedCountVectorizer()),
('mnb', MultinomialNB())])
nb_stem_score = np.mean(cross_val_score(nb_stem_pipline, texts, y, cv=5))
%%time
## SVC with stem
svc_stem_pipline = Pipeline([('count', StemmedCountVectorizer()),
('svc', LinearSVC())])
svc_stem_score = np.mean(cross_val_score(svc_stem_pipline, texts, y, cv=5))
%%time
## LogisticRegression with sem
lr_stem_pipline = Pipeline([('count', StemmedCountVectorizer()),
('lr', LogisticRegression())])
lr_stem_score = np.mean(cross_val_score(lr_stem_pipline, texts, y, cv=5))
###Output
Wall time: 1min 36s
###Markdown
We will discribe the results in next part. Question 4Filter words by grammatical category (POS : Part Of Speech) and keep only nouns,verbs, adverbs and adjectives for classification.
###Code
from nltk import pos_tag
from nltk import word_tokenize
def filter_pos(texts=texts):
filtered = [list(filter(lambda x: (x[1] == 'NOUN')
or (x[1] == 'VERB')
or (x[1] == 'ADV')
or (x[1] == 'ADJ'), pos_tag(word_tokenize(txt), tagset='universal'))) for txt in texts]
return filtered
def apply_pos_tag(filtered):
postagged = []
for i, tuples in enumerate(filtered, 0):
postagged.append('')
for t in tuples:
postagged[i] += t[0] + ' '
return postagged
postText = apply_pos_tag(filter_pos())
%%time
## MultinomialNB
nb_pipline = Pipeline([('count', CountVectorizer()), ('svc', MultinomialNB())])
nb_pos_tag_score = np.mean(cross_val_score(nb_pipline, postText, y))
%%time
## SVC
svc_pipline = Pipeline([('count', CountVectorizer()),
('svc', LinearSVC())])
svc_pos_tag_score = np.mean(cross_val_score(svc_pipline, postText, y))
%%time
## LogisticRegression
lr_pipline = Pipeline([('count', CountVectorizer()),
('lr', LogisticRegression())])
lr_pos_tag_score = np.mean(cross_val_score(lr_pipline, postText, y))
nb_res_score = [nb_score, ]
label = ['MultinomialNB', 'LinearSVC', 'LogisticRegression']
ori_scores = [nb_score, svc_score, lr_score]
stem_scores = [nb_stem_score, svc_stem_score, lr_stem_score]
pos_tag_scores = [nb_pos_tag_score, svc_pos_tag_score, lr_pos_tag_score]
label = ["MultinomialNB", "LinearSVC", "LogisticRegression"]
fig, ax = plt.subplots(figsize=(16,9))
width = 0.2
alp = 0.8
ind = np.arange(3)
rects1 = ax.bar(ind, ori_scores,
width = width,label='Model', color='skyblue')
rects2 = ax.bar(ind + width, stem_scores,
width, alpha=alp, label='Model + stem', color='gold')
#
rects3 = ax.bar(ind + 2 * width, pos_tag_scores,
width, alpha=alp, label='Model + pos_tag', color='lightpink')
# add some text for labels, title and axes ticks
ax.set_ylabel('Scores')
ax.set_title('Mean Scores by preprocessing for MultinomialNB')
ax.set_xticks(ind + 2 * width / 2)
ax.set_xticklabels(label)
ax.set_ylim(0.4,0.9)
plt.legend(loc="lower right")
def autolabel(rects):
"""
Attach a text label above each bar displaying its height
"""
for rect in rects:
height = rect.get_height()
ax.text(rect.get_x() + rect.get_width()/2., 1.02*height,
'%.3f' % height,
ha='center', va='bottom')
autolabel(rects1)
autolabel(rects2)
autolabel(rects3)
plt.show()
###Output
_____no_output_____
|
Horse_or_Human_classifier_with_Validation_150_x_150_pixel_Notebook.ipynb
|
###Markdown
Copyright 2019 The TensorFlow Authors.
###Code
#@title Licensed under the Apache License, Version 2.0 (the "License");
# you may not use this file except in compliance with the License.
# You may obtain a copy of the License at
#
# https://www.apache.org/licenses/LICENSE-2.0
#
# Unless required by applicable law or agreed to in writing, software
# distributed under the License is distributed on an "AS IS" BASIS,
# WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.
# See the License for the specific language governing permissions and
# limitations under the License.
!wget --no-check-certificate \
https://storage.googleapis.com/laurencemoroney-blog.appspot.com/horse-or-human.zip \
-O /tmp/horse-or-human.zip
!wget --no-check-certificate \
https://storage.googleapis.com/laurencemoroney-blog.appspot.com/validation-horse-or-human.zip \
-O /tmp/validation-horse-or-human.zip
###Output
_____no_output_____
###Markdown
The following python code will use the OS library to use Operating System libraries, giving you access to the file system, and the zipfile library allowing you to unzip the data.
###Code
import os
import zipfile
local_zip = '/tmp/horse-or-human.zip'
zip_ref = zipfile.ZipFile(local_zip, 'r')
zip_ref.extractall('/tmp/horse-or-human')
local_zip = '/tmp/validation-horse-or-human.zip'
zip_ref = zipfile.ZipFile(local_zip, 'r')
zip_ref.extractall('/tmp/validation-horse-or-human')
zip_ref.close()
###Output
_____no_output_____
###Markdown
The contents of the .zip are extracted to the base directory `/tmp/horse-or-human`, which in turn each contain `horses` and `humans` subdirectories.In short: The training set is the data that is used to tell the neural network model that 'this is what a horse looks like', 'this is what a human looks like' etc. One thing to pay attention to in this sample: We do not explicitly label the images as horses or humans. If you remember with the handwriting example earlier, we had labelled 'this is a 1', 'this is a 7' etc. Later you'll see something called an ImageGenerator being used -- and this is coded to read images from subdirectories, and automatically label them from the name of that subdirectory. So, for example, you will have a 'training' directory containing a 'horses' directory and a 'humans' one. ImageGenerator will label the images appropriately for you, reducing a coding step. Let's define each of these directories:
###Code
# Directory with our training horse pictures
train_horse_dir = os.path.join('/tmp/horse-or-human/horses')
# Directory with our training human pictures
train_human_dir = os.path.join('/tmp/horse-or-human/humans')
# Directory with our training horse pictures
validation_horse_dir = os.path.join('/tmp/validation-horse-or-human/horses')
# Directory with our training human pictures
validation_human_dir = os.path.join('/tmp/validation-horse-or-human/humans')
train_horse_names = os.listdir(train_horse_dir)
train_human_names = os.listdir(train_human_dir)
validation_horse_hames = os.listdir(validation_horse_dir)
validation_human_names = os.listdir(validation_human_dir)
###Output
_____no_output_____
###Markdown
Building a Small Model from ScratchBut before we continue, let's start defining the model:Step 1 will be to import tensorflow.
###Code
import tensorflow as tf
###Output
_____no_output_____
###Markdown
We then add convolutional layers as in the previous example, and flatten the final result to feed into the densely connected layers. Finally we add the densely connected layers. Note that because we are facing a two-class classification problem, i.e. a *binary classification problem*, we will end our network with a [*sigmoid* activation](https://wikipedia.org/wiki/Sigmoid_function), so that the output of our network will be a single scalar between 0 and 1, encoding the probability that the current image is class 1 (as opposed to class 0).
###Code
model = tf.keras.models.Sequential([
# Note the input shape is the desired size of the image 150x150 with 3 bytes color
# This is the first convolution
tf.keras.layers.Conv2D(16, (3,3), activation='relu', input_shape=(150, 150, 3)),
tf.keras.layers.MaxPooling2D(2, 2),
# The second convolution
tf.keras.layers.Conv2D(32, (3,3), activation='relu'),
tf.keras.layers.MaxPooling2D(2,2),
# The third convolution
tf.keras.layers.Conv2D(64, (3,3), activation='relu'),
tf.keras.layers.MaxPooling2D(2,2),
# The fourth convolution
#tf.keras.layers.Conv2D(64, (3,3), activation='relu'),
#tf.keras.layers.MaxPooling2D(2,2),
# The fifth convolution
#tf.keras.layers.Conv2D(64, (3,3), activation='relu'),
#tf.keras.layers.MaxPooling2D(2,2),
# Flatten the results to feed into a DNN
tf.keras.layers.Flatten(),
# 512 neuron hidden layer
tf.keras.layers.Dense(512, activation='relu'),
# Only 1 output neuron. It will contain a value from 0-1 where 0 for 1 class ('horses') and 1 for the other ('humans')
tf.keras.layers.Dense(1, activation='sigmoid')
])
###Output
_____no_output_____
###Markdown
The model.summary() method call prints a summary of the NN
###Code
model.summary()
###Output
_____no_output_____
###Markdown
The "output shape" column shows how the size of your feature map evolves in each successive layer. The convolution layers reduce the size of the feature maps by a bit due to padding, and each pooling layer halves the dimensions. Next, we'll configure the specifications for model training. We will train our model with the `binary_crossentropy` loss, because it's a binary classification problem and our final activation is a sigmoid. (For a refresher on loss metrics, see the [Machine Learning Crash Course](https://developers.google.com/machine-learning/crash-course/descending-into-ml/video-lecture).) We will use the `rmsprop` optimizer with a learning rate of `0.001`. During training, we will want to monitor classification accuracy.**NOTE**: In this case, using the [RMSprop optimization algorithm](https://wikipedia.org/wiki/Stochastic_gradient_descentRMSProp) is preferable to [stochastic gradient descent](https://developers.google.com/machine-learning/glossary/SGD) (SGD), because RMSprop automates learning-rate tuning for us. (Other optimizers, such as [Adam](https://wikipedia.org/wiki/Stochastic_gradient_descentAdam) and [Adagrad](https://developers.google.com/machine-learning/glossary/AdaGrad), also automatically adapt the learning rate during training, and would work equally well here.)
###Code
from tensorflow.keras.optimizers import RMSprop
model.compile(loss='binary_crossentropy',
optimizer=RMSprop(lr=0.001),
metrics=['accuracy'])
###Output
_____no_output_____
###Markdown
Data PreprocessingLet's set up data generators that will read pictures in our source folders, convert them to `float32` tensors, and feed them (with their labels) to our network. We'll have one generator for the training images and one for the validation images. Our generators will yield batches of images of size 300x300 and their labels (binary).As you may already know, data that goes into neural networks should usually be normalized in some way to make it more amenable to processing by the network. (It is uncommon to feed raw pixels into a convnet.) In our case, we will preprocess our images by normalizing the pixel values to be in the `[0, 1]` range (originally all values are in the `[0, 255]` range).In Keras this can be done via the `keras.preprocessing.image.ImageDataGenerator` class using the `rescale` parameter. This `ImageDataGenerator` class allows you to instantiate generators of augmented image batches (and their labels) via `.flow(data, labels)` or `.flow_from_directory(directory)`. These generators can then be used with the Keras model methods that accept data generators as inputs: `fit`, `evaluate_generator`, and `predict_generator`.
###Code
from tensorflow.keras.preprocessing.image import ImageDataGenerator
# All images will be rescaled by 1./255
train_datagen = ImageDataGenerator(rescale=1/255)
validation_datagen = ImageDataGenerator(rescale=1/255)
# Flow training images in batches of 128 using train_datagen generator
train_generator = train_datagen.flow_from_directory(
'/tmp/horse-or-human/', # This is the source directory for training images
target_size=(150, 150), # All images will be resized to 150x150
batch_size=128,
# Since we use binary_crossentropy loss, we need binary labels
class_mode='binary')
# Flow training images in batches of 128 using train_datagen generator
validation_generator = validation_datagen.flow_from_directory(
'/tmp/validation-horse-or-human/', # This is the source directory for training images
target_size=(150, 150), # All images will be resized to 150x150
batch_size=32,
# Since we use binary_crossentropy loss, we need binary labels
class_mode='binary')
###Output
_____no_output_____
###Markdown
TrainingLet's train for 15 epochs -- this may take a few minutes to run.Do note the values per epoch.The Loss and Accuracy are a great indication of progress of training. It's making a guess as to the classification of the training data, and then measuring it against the known label, calculating the result. Accuracy is the portion of correct guesses.
###Code
history = model.fit(
train_generator,
steps_per_epoch=8,
epochs=15,
verbose=1,
validation_data = validation_generator,
validation_steps=8)
###Output
_____no_output_____
###Markdown
Running the ModelLet's now take a look at actually running a prediction using the model. This code will allow you to choose 1 or more files from your file system, it will then upload them, and run them through the model, giving an indication of whether the object is a horse or a human.
###Code
import numpy as np
from google.colab import files
from keras.preprocessing import image
uploaded = files.upload()
for fn in uploaded.keys():
# predicting images
path = '/content/' + fn
img = image.load_img(path, target_size=(150, 150))
x = image.img_to_array(img)
x = np.expand_dims(x, axis=0)
images = np.vstack([x])
classes = model.predict(images, batch_size=10)
print(classes[0])
if classes[0]>0.5:
print(fn + " is a human")
else:
print(fn + " is a horse")
###Output
_____no_output_____
###Markdown
Visualizing Intermediate RepresentationsTo get a feel for what kind of features our convnet has learned, one fun thing to do is to visualize how an input gets transformed as it goes through the convnet.Let's pick a random image from the training set, and then generate a figure where each row is the output of a layer, and each image in the row is a specific filter in that output feature map. Rerun this cell to generate intermediate representations for a variety of training images.
###Code
import matplotlib.pyplot as plt
import numpy as np
import random
from tensorflow.keras.preprocessing.image import img_to_array, load_img
# Let's define a new Model that will take an image as input, and will output
# intermediate representations for all layers in the previous model after
# the first.
successive_outputs = [layer.output for layer in model.layers[1:]]
#visualization_model = Model(img_input, successive_outputs)
visualization_model = tf.keras.models.Model(inputs = model.input, outputs = successive_outputs)
# Let's prepare a random input image from the training set.
horse_img_files = [os.path.join(train_horse_dir, f) for f in train_horse_names]
human_img_files = [os.path.join(train_human_dir, f) for f in train_human_names]
img_path = random.choice(horse_img_files + human_img_files)
img = load_img(img_path, target_size=(150, 150)) # this is a PIL image
x = img_to_array(img) # Numpy array with shape (150, 150, 3)
x = x.reshape((1,) + x.shape) # Numpy array with shape (1, 150, 150, 3)
# Rescale by 1/255
x /= 255
# Let's run our image through our network, thus obtaining all
# intermediate representations for this image.
successive_feature_maps = visualization_model.predict(x)
# These are the names of the layers, so can have them as part of our plot
layer_names = [layer.name for layer in model.layers]
# Now let's display our representations
for layer_name, feature_map in zip(layer_names, successive_feature_maps):
if len(feature_map.shape) == 4:
# Just do this for the conv / maxpool layers, not the fully-connected layers
n_features = feature_map.shape[-1] # number of features in feature map
# The feature map has shape (1, size, size, n_features)
size = feature_map.shape[1]
# We will tile our images in this matrix
display_grid = np.zeros((size, size * n_features))
for i in range(n_features):
# Postprocess the feature to make it visually palatable
x = feature_map[0, :, :, i]
x -= x.mean()
x /= x.std()
x *= 64
x += 128
x = np.clip(x, 0, 255).astype('uint8')
# We'll tile each filter into this big horizontal grid
display_grid[:, i * size : (i + 1) * size] = x
# Display the grid
scale = 20. / n_features
plt.figure(figsize=(scale * n_features, scale))
plt.title(layer_name)
plt.grid(False)
plt.imshow(display_grid, aspect='auto', cmap='viridis')
###Output
_____no_output_____
###Markdown
As you can see we go from the raw pixels of the images to increasingly abstract and compact representations. The representations downstream start highlighting what the network pays attention to, and they show fewer and fewer features being "activated"; most are set to zero. This is called "sparsity." Representation sparsity is a key feature of deep learning.These representations carry increasingly less information about the original pixels of the image, but increasingly refined information about the class of the image. You can think of a convnet (or a deep network in general) as an information distillation pipeline. Clean UpBefore running the next exercise, run the following cell to terminate the kernel and free memory resources:
###Code
# import os, signal
# os.kill(os.getpid(), signal.SIGKILL)
###Output
_____no_output_____
|
examples/tutorial/generate.ipynb
|
###Markdown
More complicated obfuscation/encryption can be done with a custome function. The following for instance concatenate the first 3 digits in clear followed by the obscured phone number
###Code
from pyspark.sql import functions as F
from datafaucet.spark.functions import obscure, unravel
def mobile_obscure(key):
return lambda c: F.concat(F.substring(c, 1, 3), F.lit('%'), obscure(key)(c))
def mobile_unravel(key):
return lambda c: unravel(key)(F.element_at(F.split(c, '%'),2))
res = df.cols.get('number').apply(mobile_obscure('mysecret'))
res.data.grid(5)
res = res.cols.get('number').apply(mobile_unravel('mysecret'))
res.data.grid(5)
###Output
_____no_output_____
|
scraping/save_Self_introduction_test.ipynb
|
###Markdown
div.stContainer 자소서 페이지div.viewTitWrap 회사이름div.adviceTotal 자소서 총평dl.qnaLists 질문내용
###Code
soup=BeautifulSoup(browser.page_source,'html.parser')
introduction_page = soup.select('div.stContainer')
introduction_page[0]
company = introduction_page[0].select('div.viewTitWrap')[0].text.strip()
group = introduction_page[0].select('div.adviceTotal')[0].text.strip()
clasificar = introduction_page[0].select('dl.qnaLists ')[0].text.strip()
# subclass = introduction_page[0].select('dd.defaultTxt')[0].text.strip()
contents = []
for scraping in introduction_page:
company = introduction_page[0].select('div.viewTitWrap')[0].text.strip()
group = introduction_page[0].select('div.adviceTotal')[0].text.strip()
clasificar = introduction_page[0].select('dl.qnaLists ')[0].text.strip()
result = [company, group, clasificar]
contents.append(result)
contents
import pandas as pd
df = pd.DataFrame(contents,columns=['company', 'group', 'clasificar'])
df
df.to_excel('./test.xls',index=False)
###Output
_____no_output_____
|
_build/html/_sources/OpTaliX_Diffraction_analysis.ipynb
|
###Markdown
OpTaliX: Diffraction analysisThese are ...
###Code
import pandas as pd
import itables
from itables import init_notebook_mode, show
import itables.options as opt
init_notebook_mode(all_interactive=True)
opt.lengthMenu = [50, 100, 200, 500]
#opt.classes = ["display", "cell-border"]
#opt.classes = ["display", "nowrap"]
opt.columnDefs = [{"className": "dt-left", "targets": "_all"}, {"width": "500px", "targets": 1}]
import os
cwd = os.getcwd()
filename = os.path.join(cwd, os.path.join('Excel', 'OpTaliX_optimization_operands.xlsx'))
df_var = pd.read_excel(filename, sheet_name = "Diffraction Analysis", header = 0, index_col = 0)
df_var = df_var.dropna() # drop nan values
df_var
###Output
_____no_output_____
|
docs/contents/tools/classes/pytraj_Trajectory/to_molsysmt_Trajectory.ipynb
|
###Markdown
To molsysmt.Trajectory
###Code
from molsysmt.tools import pytraj_Trajectory
#pytraj_Trajectory.to_molsysmt_Trajectory(item)
###Output
_____no_output_____
|
notebooks/supplemental_figure_3-4.ipynb
|
###Markdown
Imports and data setup
###Code
# %load_ext autoreload
# %autoreload 2
import os
import matplotlib.pyplot as plt
import numpy as np
import pandas as pd
import seaborn as sns
# from joblib import Parallel, delayed
from sklearn.metrics import confusion_matrix
# from tqdm import tqdm
# # Artisinal, small-batch, handmade modules
# import classification_metrics
from nb_utils import describe
from celltype_utils import SHARED_CELLTYPES, BROAD_TO_COMPARTMENT, broad_to_compartment
from path_constants import top_hit_paths, FIGURE_FOLDER, sig_outdir_base
from plot_constants import get_cmap_color
###Output
_____no_output_____
###Markdown
Assign figure folder
###Code
figure_folder = os.path.join(FIGURE_FOLDER, "kmermaid_classification_metrics")
###Output
_____no_output_____
###Markdown
Read celltype predictions file
###Code
predicted_cells = pd.read_parquet(
os.path.join(sig_outdir_base, "aggregated-predicted-cells.parquet")
)
describe(predicted_cells)
predicted_cells.database_type.value_counts()
###Output
_____no_output_____
###Markdown
Broad group Set `scoring_groupby`, ground truth, and predicted columns
###Code
scoring_groupby = ['species', 'database_type', 'search_method', 'alphabet', 'ksize']
ground_truth_celltype_col = 'groundtruth_celltype'
predicted_celltype_col = 'predicted_celltype'
###Output
_____no_output_____
###Markdown
Compute Confusion matrices
###Code
"""
By definition a confusion matrix is such that is equal to the number of
observations known to be in group and predicted to be in group
"""
top_hit_confusion_dfs = {}
for keys, df in predicted_cells.groupby(
scoring_groupby
):
ground_truth = df[ground_truth_celltype_col]
predicted = df[predicted_celltype_col]
weight = df["similarity"]
labels = sorted(list(set(ground_truth) | set(predicted)))
cm = confusion_matrix(
ground_truth,
predicted,
# labels=labels,
# sample_weight=weight
)
cm_df = pd.DataFrame(cm, index=labels, columns=labels)
cm_df.index.name = "ground_truth"
cm_df.columns.name = "predicted"
cm_df = cm_df.reindex(index=SHARED_CELLTYPES, columns=SHARED_CELLTYPES)
# cm_df = cm_df.loc[SHARED_CELLTYPES, SHARED_CELLTYPES]
top_hit_confusion_dfs[keys] = cm_df
###Output
_____no_output_____
###Markdown
Assign confusion matrix folder
###Code
broad_confusion_matrix_folder = os.path.join(
figure_folder, "confusion_matrices", "broad_group"
)
! mkdir -p $broad_confusion_matrix_folder
sns.set_context('paper')
import matplotlib as mpl
mpl.rcParams['pdf.fonttype'] = 42
for keys, confusion_df in top_hit_confusion_dfs.items():
# ksize, alphabet = keys
fig, ax = plt.subplots(figsize=(7, 6))
# Reorder by compartment
# index = broad_to_compartment.index.intersection(confusion_df.index)
# renamer = {broad: f'{broad} ({compartment[:3]})' for broad, compartment in broad_to_compartment[index].items()}
# confusion_df = confusion_df.loc[index, index]
# confusion_df = confusion_df.rename(columns=renamer, index=renamer)
# Annotate only where nonzero
annot = confusion_df[confusion_df > 0]
# Replace all NAs (previously 0s) with an empty string
annot = annot.applymap(lambda x: '' if pd.isnull(x) else str(int(x)))
# annot = True
cmap = get_cmap_color(keys)
sns.heatmap(np.log10(confusion_df + 1), annot=annot, fmt='s', cmap=cmap)
title = '--'.join(map(str, keys))
ax.set(title=title)
# ax.set(title=f"Alphabet: {alphabet}, k-mer size: {ksize}")
pdf = os.path.join(broad_confusion_matrix_folder, f'{title}.pdf')
fig.tight_layout()
fig.savefig(pdf)
# break
###Output
_____no_output_____
###Markdown
Compartment Set `scoring_groupby`, ground truth, and predicted columns
###Code
scoring_groupby = ['species', 'database_type', 'alphabet', 'ksize']
ground_truth_celltype_col = 'groundtruth_compartment'
predicted_celltype_col = 'predicted_compartment'
###Output
_____no_output_____
###Markdown
Compute Confusion matrices
###Code
"""
By definition a confusion matrix is such that is equal to the number of
observations known to be in group and predicted to be in group
"""
compartment_confusion_dfs = {}
for keys, df in predicted_cells.groupby(scoring_groupby):
ground_truth = df[ground_truth_celltype_col]
predicted = df[predicted_celltype_col]
weight = df["similarity"]
labels = sorted(list(set(ground_truth) | set(predicted)))
cm = confusion_matrix(
ground_truth,
predicted,
# labels=labels,
# sample_weight=weight
)
cm_df = pd.DataFrame(cm, index=labels, columns=labels)
cm_df.index.name = "ground_truth"
cm_df.columns.name = "predicted"
# cm_df = cm_df.reindex(index=SHARED_CELLTYPES, columns=SHARED_CELLTYPES)
# cm_df = cm_df.loc[SHARED_CELLTYPES, SHARED_CELLTYPES]
compartment_confusion_dfs[keys] = cm_df
###Output
_____no_output_____
###Markdown
@paper Separately plot confusion matrices to fine tune parameters
###Code
compartment_confusion_matrix_folder = os.path.join(
figure_folder, "confusion_matrices", "compartment_group"
)
! mkdir -p $compartment_confusion_matrix_folder
for keys, confusion_df in compartment_confusion_dfs.items():
# ksize, alphabet = keys
fig, ax = plt.subplots(figsize=(4, 3))
# Reorder by compartment
# index = broad_to_compartment.index.intersection(confusion_df.index)
# renamer = {broad: f'{broad} ({compartment[:3]})' for broad, compartment in broad_to_compartment[index].items()}
# confusion_df = confusion_df.loc[index, index]
# confusion_df = confusion_df.rename(columns=renamer, index=renamer)
# Annotate only where nonzero
annot = confusion_df[confusion_df > 0]
# Replace all NAs (previously 0s) with an empty string
annot = annot.applymap(lambda x: '' if pd.isnull(x) else str(int(x)))
# annot = True
cmap = get_cmap_color(keys)
sns.heatmap(np.log10(confusion_df + 1), annot=annot, fmt='s', cmap=cmap)
title = '--'.join(map(str, keys))
ax.set(title=title)
# ax.set(title=f"Alphabet: {alphabet}, k-mer size: {ksize}")
pdf = os.path.join(compartment_confusion_matrix_folder, f'{title}.pdf')
fig.tight_layout()
fig.savefig(pdf)
###Output
_____no_output_____
|
analysis/Average_Rating.ipynb
|
###Markdown
Prerequisite : Need to run the NLP model notebook
###Code
df = pd.read_csv("../data/googleplaystore_user_reviews.csv")
df_review = pd.read_csv('../data/googleplaystore_cleaned.csv')
df = df.dropna()
review_with_ratings = pd.merge(df,df_review,on = 'App')
review_with_ratings.head()
review_with_ratings['tokens'] = review_with_ratings['Review New'].apply(preprocess)
###Output
_____no_output_____
###Markdown
Compound Scores are added
###Code
analyzer = SentimentIntensityAnalyzer()
def sentiment_polarity(s):
global analyzer
polarity_scores = analyzer.polarity_scores(s)
compound_score = polarity_scores["compound"]
if compound_score >= 0.5:
label = "Positive"
elif compound_score > -0.05 and compound_score < 0.05:
label = "Neutral"
else:
label = "Negative"
return label, polarity_scores["neu"], polarity_scores["pos"], polarity_scores["neg"],compound_score
df = review_with_ratings
df["Sentiment"], df["Neutral Proportion"], df["Positive Proportion"], df["Negative Proportion"],df['Compound'] = zip(*df["Review"].apply(sentiment_polarity))
df.sample(3)
###Output
_____no_output_____
###Markdown
Normalize the rating to be between 1 and 5
###Code
df['Rating_pred'] = df['Compound'].apply(lambda x: (x-(-1))/(1-(-1))*(5-1)+1)
np.min(df.Rating_pred)
np.min(df.Compound)
np.max(df.Rating_pred)
np.max(df.Compound)
###Output
_____no_output_____
###Markdown
Calculate the average rating
###Code
Average_Rating = df.groupby('App')['Rating_pred'].mean()
df_Compare = pd.DataFrame(Average_Rating)
df_Compare.head()
df_Compare = pd.merge(df_Compare,df_review, on = 'App')
df_Compare.head()
df_Compare = df_Compare.drop_duplicates()
df_Compare.head()
df_Overall = pd.DataFrame(df_Compare['App'])
df_Overall['Rating Polarity'] = df_Compare['Rating_pred']
df_Overall['Rating Dataset'] = df_Compare['Rating']
df_Overall.head()
df_Overall.to_csv("../data/Compare_Rating.csv")
###Output
_____no_output_____
###Markdown
Compute the mse to compare between Overall Rating and the Rating predicted from polarity
###Code
mse = np.mean(np.multiply((df_Overall['Rating Polarity']-df_Overall['Rating Dataset']),
(df_Overall['Rating Polarity']-df_Overall['Rating Dataset'])))
mse
###Output
_____no_output_____
|
5-algorithms/1-Tree/decisionTree.ipynb
|
###Markdown
Base Model (with no hyper parameters)Cross-Validation-Score:7087.853890255971 MAE: 8194.332009158503 MSE: 1310403929.046806 Train R2: 0.9984405456569931 Test R2: 0.6715758197834594 BEST MAX DEPTH = 7 BEST min_samples_split = 0.006 BEST min_samples_leaf = 0.004 BEST max_leaf_nodes = 49 BEST min_impurity_decrease = 34.6 BEST splitter = random BEST max_features = sqrt BEST ccp_alpha = 13434 Model After Exploratory Parameter TuningCross-Validation-Score:20940.862282131988 MAE: 18995.667954555534 MSE: 1501197758.7096622 Train R2: 0.6320352403305758 Test R2: 0.62375750536267 Final Model'ccp_alpha': 13434, 'max_depth': 7,'max_features': 'auto','max_leaf_nodes': 48,'min_impurity_decrease': 0,'min_samples_leaf': 1,'min_samples_split': 0.005,'splitter': 'random' with two attrsCross-Validation-Score:6993.424569760234 MAE: 5942.955577700336 MSE: 352654781.24830395 Train R2: 0.9051753291112914 Test R2: 0.9372154037633127 with full attrsCross-Validation-Score:7544.451049013391MAE: 7839.9115567135395MSE: 921507622.8550984Train R2: 0.9134290469474539Test R2: 0.8504812921161993 Trying to find the best hyper paramteters
###Code
# Randomized Search
splitter = ['best','random']
max_depth=[None,5,6,7,8,10]
min_samples_split=[2,0.006,0.007,0.005]
min_samples_leaf = [1,0.004,0.003,0.002]
max_features = ['auto','sqrt']
max_leaf_nodes = [None,49,50,48]
ccp_alpha = [0,13434,15000,10000]
min_impurity_decrease = [0,34.6]
random_state = [41]
random_grid = {
'splitter':splitter,
'max_depth':max_depth,
'min_samples_split':min_samples_split,
'min_samples_leaf':min_samples_leaf,
'max_features':max_features,
'max_leaf_nodes':max_leaf_nodes,
'ccp_alpha':ccp_alpha,
'min_impurity_decrease':min_impurity_decrease,
'random_state':random_state
}
tree_regressor = DecisionTreeRegressor(random_state=41)
random_search = RandomizedSearchCV(
estimator=tree_regressor,
param_distributions=random_grid,
n_iter=1000,
cv=3,
random_state=42,
n_jobs=-1,
verbose=2,
scoring='neg_mean_absolute_error')
random_search.fit(X_train,y_train)
print("done")
# Grid Search
splitter = ['best']
max_depth=[None,17,18,19]
min_samples_split=[2,3,4]
min_samples_leaf = [1,2,3]
max_features = ['auto']
max_leaf_nodes = [None,2052]
ccp_alpha = [0]
min_impurity_decrease = [0]
random_state = [41]
search_grid = {
'splitter':splitter,
'max_depth':max_depth,
'min_samples_split':min_samples_split,
'min_samples_leaf':min_samples_leaf,
'max_features':max_features,
'max_leaf_nodes':max_leaf_nodes,
'ccp_alpha':ccp_alpha,
'min_impurity_decrease':min_impurity_decrease,
'random_state':random_state
}
tree_regressor = DecisionTreeRegressor()
search = GridSearchCV(
estimator=tree_regressor,
param_grid=search_grid,
cv=3,
n_jobs=-3,
verbose=2,
scoring='neg_mean_absolute_percentage_error')
search.fit(X_train,y_train)
print("done")
search.best_params_
y_pred = search.best_estimator_.predict(X_test)
MAPE(y_pred,y_test)
###Output
_____no_output_____
###Markdown
FINAL MODEL
###Code
mae = []
mape = []
mse = []
for i in range(1,20):
print('****'+str(i)+'****')
X = df.drop('Actual Cost',axis=1)
y = df['Actual Cost']
X_train,X_test,y_train,y_test = train_test_split(
X,y,test_size=0.2)
tree_regressor = search.best_estimator_
tree_regressor.fit(X_train,y_train)
y_pred = tree_regressor.predict(X_test)
mae.append(MAE(y_test,y_pred))
mse.append(MSE(y_test,y_pred))
mape.append(MAPE(y_test,y_pred))
clear_output(wait=True)
print(np.average(mae))
print(np.average(mse))
print(np.average(mape))
###Output
6020.316669812966
516692090.33566046
1.005864773554262
###Markdown
the mean mae after running for 100 times: 6676.626301399859
###Code
MAE_MSP = MAE(df['Actual Cost'],df['Total Cost'])
print("MAE MSP:")
print(MAE_MSP)
MSE_MSP = MSE(df['Actual Cost'],df['Total Cost'])
print("MSE MSP:")
print(MSE_MSP)
print("MAPE MSP: ")
print(MAPE(df['Actual Cost'],df['Total Cost']))
# Saving Tree Graph
dot_data = tree.export_graphviz(tree_regressor, out_file=None,
feature_names=['Duration','Total Cost'],class_names=['Acutal Cost'])
graph = pydotplus.graph_from_dot_data(dot_data)
graph.write_svg("tree.svg")
# Saving model
pkl_filename = "tree.pkl"
with open(pkl_filename, 'wb') as file:
pickle.dump(tree_regressor, file)
pkl_filename = "tree.pkl"
with open(pkl_filename,'rb') as file:
model = pickle.load(file)
print("MODEL IMPORTED!")
###Output
_____no_output_____
|
src/data-prep/DataAnalysis.ipynb
|
###Markdown
Explore and Organize Dataset
###Code
folderTypes = {"female": [], "male": [], "multiperson": [], "real": []}
for folder in os.listdir(inputPath):
if folder.startswith("female"):
folderTypes["female"].append(folder.split("_")[0])
if os.path.isdir(os.path.join(outputPath, folderTypes["female"][-1])):
for file in os.listdir(os.path.join(inputPath, folder)):
shutil.copy(os.path.join(inputPath, folder, file), os.path.join(outputPath, folderTypes["female"][-1], file))
elif folder.startswith("multiperson"):
folderTypes["multiperson"].append(folder.split("_")[0])
if os.path.isdir(os.path.join(outputPath, folderTypes["multiperson"][-1])):
for file in os.listdir(os.path.join(inputPath, folder)):
shutil.copy(os.path.join(inputPath, folder, file), os.path.join(outputPath, folderTypes["multiperson"][-1], file))
elif folder.startswith("male"):
folderTypes["male"].append(folder.split("_")[0])
if folder.startswith("male06") and "nolight" in folder:
outFolder = "male06_2"
elif folder.startswith("male06"):
outFolder = "male06_1"
else:
outFolder = folderTypes["male"][-1]
if os.path.isdir(os.path.join(outputPath, outFolder)):
for file in os.listdir(os.path.join(inputPath, folder)):
shutil.copy(os.path.join(inputPath, folder, file), os.path.join(outputPath, outFolder, file))
else:
if folder.startswith("real") and os.path.isdir(os.path.join(outputPath, "real")):
for file in os.listdir(os.path.join(inputPath, folder)):
shutil.copy(os.path.join(inputPath, folder, file), os.path.join(outputPath, "real", file))
for folder in labelsDist:
if len(os.listdir(os.path.join(labelsPath, folder))) != len(os.listdir(os.path.join(outputPath, folder))):
print(folder, len(os.listdir(os.path.join(labelsPath, folder))), len(os.listdir(os.path.join(outputPath, folder))))
import matplotlib.pyplot as plt
allSamples = []
_id = 0
for folder in os.listdir(outputPath):
for file in os.listdir(os.path.join(outputPath, folder)):
if file in os.listdir(os.path.join(labelsPath, folder)):
image = Image.open(os.path.join(outputPath, folder, file))
mask = Image.open(os.path.join(labelsPath, folder, file))
try:
assert image.size == mask.size
allSamples.append({"id": _id, "folder": folder, "Filename": file, "size": image.size})
_id += 1
except AssertionError:
print(folder, file, image.size, mask.size)
# f, axarr = plt.subplots(2,2)
# axarr[0,0].imshow(image)
# axarr[0,1].imshow(mask)
# plt.show()
import pandas as pd
dataSamples = pd.DataFrame(allSamples, columns=allSamples[0].keys())
dataSamples.to_csv("FinalSamples.csv", index=False)
###Output
_____no_output_____
###Markdown
Move to dataset folder
###Code
finalPath = r"../../faceSegmentation/dataset/"
for row, sample in dataSamples.iterrows():
imagePath = os.path.join(outputPath, sample["folder"], sample["Filename"])
maskPath = os.path.join(labelsPath, sample["folder"], sample["Filename"])
shutil.copy(imagePath, os.path.join(finalPath, "images", sample["folder"] + "_" + sample["Filename"]))
shutil.copy(maskPath, os.path.join(finalPath, "masks", sample["folder"] + "_" + sample["Filename"]))
###Output
_____no_output_____
###Markdown
Remove near duplicate images
###Code
from imutils import paths
import numpy as np
import argparse
import cv2
import os
def dhash(image, hashSize=8):
# convert the image to grayscale and resize the grayscale image,
# adding a single column (width) so we can compute the horizontal
# gradient
gray = cv2.cvtColor(image, cv2.COLOR_BGR2GRAY)
resized = cv2.resize(gray, (hashSize + 1, hashSize))
# compute the (relative) horizontal gradient between adjacent
# column pixels
diff = resized[:, 1:] > resized[:, :-1]
# convert the difference image to a hash and return it
return sum([2 ** i for (i, v) in enumerate(diff.flatten()) if v])
print("[INFO] computing image hashes...")
imagePaths = list(paths.list_images("../../dataset/images"))
hashes = {}
# loop over our image paths
for imagePath in imagePaths:
# load the input image and compute the hash
image = cv2.imread(imagePath)
h = dhash(image)
# grab all image paths with that hash, add the current image
# path to it, and store the list back in the hashes dictionary
p = hashes.get(h, [])
p.append(imagePath)
hashes[h] = p
remove = True
# loop over the image hashes
for (h, hashedPaths) in hashes.items():
# check to see if there is more than one image with the same hash
if len(hashedPaths) > 1:
# check to see if this is a dry run
if not remove:
# initialize a montage to store all images with the same
# hash
montage = None
# loop over all image paths with the same hash
for p in hashedPaths:
# load the input image and resize it to a fixed width
# and heightG
image = cv2.imread(p)
image = cv2.resize(image, (150, 150))
# if our montage is None, initialize it
if montage is None:
montage = image
# otherwise, horizontally stack the images
else:
montage = np.hstack([montage, image])
# show the montage for the hash
print("[INFO] hash: {}".format(h))
cv2.imshow("Montage", montage)
cv2.waitKey(0)
# otherwise, we'll be removing the duplicate images
else:
# loop over all image paths with the same hash *except*
# for the first image in the list (since we want to keep
# one, and only one, of the duplicate images)
for p in hashedPaths[1:]:
os.remove(p)
os.remove(p.replace("/images/", "/masks/"))
print(len(os.listdir(os.path.join("../../dataset/images"))))
allFilesPath = r"../../faceSegmentation/dataset/"
subSamplesPath = r"../../dataset/subsamples/"
###Output
_____no_output_____
###Markdown
Create sub sample of datset for training
###Code
oldFolders = {}
for imageFile, maskFile in zip(os.listdir(os.path.join(allFilesPath, "images")), os.listdir(os.path.join(allFilesPath, "masks"))):
if imageFile == maskFile:
try:
if oldFolders.get(imageFile.split("_")[0]):
oldFolders[imageFile.split("_")[0]] += 1
else:
oldFolders[imageFile.split("_")[0]] = 1
except:
oldFolders[imageFile.split("_")[0]] = 1
else:
print(imageFile)
oldFolders
###Output
_____no_output_____
###Markdown
Add all files from multiperson and Real images, 10 files from other types
###Code
import random
addedFiles = {k: 0 for k in oldFolders.keys()}
allImages = os.listdir(os.path.join(allFilesPath, "images"))
random.shuffle(allImages)
for image in allImages:
if image.split("_")[0] == "real" or (image.split("_")[0] == "multiperson" and addedFiles[image.split("_")[0]]<50) or addedFiles[image.split("_")[0]]<10:
addedFiles[image.split("_")[0]] += 1
shutil.copy(os.path.join(allFilesPath, "images", image), os.path.join(subSamplesPath, "images", image))
shutil.copy(os.path.join(allFilesPath, "masks", image), os.path.join(subSamplesPath, "masks", image))
addedFiles
newFolders = {}
for imageFile, maskFile in zip(os.listdir(os.path.join(subSamplesPath, "images")), os.listdir(os.path.join(subSamplesPath, "masks"))):
if imageFile == maskFile:
try:
if newFolders.get(imageFile.split("_")[0]):
newFolders[imageFile.split("_")[0]] += 1
else:
newFolders[imageFile.split("_")[0]] = 1
except:
newFolders[imageFile.split("_")[0]] = 1
else:
print(imageFile)
print("NEW DISTRIBUTION OF SAMPLES: ", len(os.listdir(os.path.join(subSamplesPath, "images"))))
print(newFolders)
###Output
NEW DISTRIBUTION OF SAMPLES: 2103
{'real': 1723, 'femalecarla2': 10, 'femalecarla': 10, 'male06': 10, 'maleelias': 10, 'multiperson': 50, 'malegaberial2': 10, 'maleandrew': 10, 'malecarlos': 10, 'female03': 10, 'femalebarbera2': 10, 'femalebarbera': 10, 'malekumar': 10, 'malecorry2': 10, 'male23': 10, 'malegaberial': 10, 'femalelaura': 10, 'maleshawn': 10, 'female10': 10, 'male09': 10, 'femalefelice': 10, 'malecorry': 10, 'malecarlos2': 10, 'femalejoyce': 10, 'femalejoyce2': 10, 'malebruce2': 10, 'femalealison1': 10, 'femaleroberta': 10, 'femalelaura2': 10, 'female23': 10, 'male01': 10, 'femalealison2': 10, 'maleandrew2': 10, 'malebruce': 10, 'maleelias2': 10}
|
notebooks/E10_PHM08-train_Elman/0.4-warm_up-navarmn.ipynb
|
###Markdown
Load Dataset
###Code
folderpath = '../../data/interim/'
data_completed = pd.read_csv(folderpath + 'data_preprocessed.csv')
data_completed.head()
###Output
_____no_output_____
###Markdown
Data preprocessing Use the pipeline and mlp
###Code
from phm08ds.data.preprocessing import OperationalCondition
data_unlabel = data_completed.drop(labels=['Health_state', 'Operational_condition'], axis=1)
tf_op_cond = OperationalCondition()
op_cond = tf_op_cond.fit_transform(data_unlabel.loc[0])
from phm08ds.features.feature_selection import RemoveSensor
tf_select_sensor = RemoveSensor(sensors=[4,5,7,9,15,16,17,18])
data_important_sensors = tf_select_sensor.fit_transform(data_unlabel).iloc[:,5:]
from sklearn.preprocessing import StandardScaler
tf_std = StandardScaler()
data_elman = tf_std.fit_transform(data_important_sensors)
data_important_sensors.head()
data_elman
labels = np.array(data_completed['Health_state'])
from sklearn.preprocessing import LabelBinarizer
tf_label_binarize = LabelBinarizer(neg_label=-1)
labels_encoded = tf_label_binarize.fit_transform(labels)
data_important_sensors.to_csv('data_op_01_cleaned.csv')
data_completed['Health_state'].to_csv('data_op_01_cleaned_labels.csv')
###Output
_____no_output_____
###Markdown
Classification steps How to use Elman network of neurolab Folllowing the example at https://pythonhosted.org/neurolab/ex_newelm.html
###Code
import neurolab as nl
min_list = []
max_list = []
for feature in range(0,data_elman.shape[1]):
min_list.append(data_elman[:,feature].min())
max_list.append(data_elman[:,feature].max())
min_max_list = list(map(list, list(zip(min_list, max_list))))
min_max_list
###Output
_____no_output_____
###Markdown
from sklearn.preprocessing import LabelBinarizertarget_tf = LabelBinarizer()labels_encoded = target_tf.fit_transform(labels_op_1)
###Code
elman_clf = nl.net.newelm(min_max_list, [50,4], [nl.trans.TanSig(), nl.trans.PureLin()])
# Set initialized functions and init
elman_clf.layers[0].initf = nl.init.InitRand([-0.01, 0.01], 'w')
elman_clf.layers[1].initf= nl.init.InitRand([-0.01, 0.01], 'w')
# elman_clf.layers[0].initf = nl.init.initnw(1)
# elman_clf.layers[1].initf= nl.init.initnw([-0.1, 0.1], 'wb')
elman_clf.init()
# Train network
error = elman_clf.train(data_elman, labels_encoded, epochs=10, goal=0.1, adapt=True, show=1)
# Simulate network
output = elman_clf.sim(data_elman)
error
plt.plot(error)
###Output
_____no_output_____
###Markdown
Test the newtwork
###Code
real_targets = labels.reshape(-1)
real_targets
predicted_targets = tf_label_binarize.inverse_transform(output)
until_to = 100
plt.figure()
plt.plot(real_targets[:until_to])
plt.plot(output[:until_to])
from sklearn.metrics import confusion_matrix
from sklearn.metrics import accuracy_score
accuracy_score(real_targets, predicted_targets)
confusion_matrix(real_targets, predicted_targets)
###Output
_____no_output_____
|
examples/.ipynb_checkpoints/intro_to_chemvae-checkpoint.ipynb
|
###Markdown
Load libraries
###Code
# tensorflow backend
from os import environ
environ['KERAS_BACKEND'] = 'tensorflow'
# vae stuff
from chemvae.vae_utils import VAEUtils
from chemvae import mol_utils as mu
# import scientific py
import numpy as np
import pandas as pd
# rdkit stuff
from rdkit.Chem import AllChem as Chem
from rdkit.Chem import PandasTools
# plotting stuff
import matplotlib.pyplot as plt
import matplotlib as mpl
from IPython.display import SVG, display
%config InlineBackend.figure_format = 'retina'
%matplotlib inline
###Output
_____no_output_____
###Markdown
Load a model
###Code
vae = VAEUtils(directory='../models/zinc_properties')
###Output
/home/beangoben/miniconda3/lib/python3.6/site-packages/keras/models.py:245: UserWarning: No training configuration found in save file: the model was *not* compiled. Compile it manually.
warnings.warn('No training configuration found in save file: '
###Markdown
Using the VAE Decode/Encode Might not be perfect (it's probabilistic), try it several times.smiles x z x_r smiles_r
###Code
smiles_1 = mu.canon_smiles('CSCC(=O)NNC(=O)c1c(C)oc(C)c1C')
X_1 = vae.smiles_to_hot(smiles_1,canonize_smiles=True)
z_1 = vae.encode(X_1)
X_r= vae.decode(z_1)
print('{:20s} : {}'.format('Input',smiles_1))
print('{:20s} : {}'.format('Reconstruction',vae.hot_to_smiles(X_r,strip=True)[0]))
print('{:20s} : {} with norm {:.3f}'.format('Z representation',z_1.shape, np.linalg.norm(z_1)))
###Output
Input : CSCC(=O)NNC(=O)c1c(C)oc(C)c1C
Reconstruction : ClCC(=O)N)C(=O)c1c(C)oc(C)c1C
Z representation : (1, 196) with norm 10.686
###Markdown
property preditor
###Code
print('Properties (qed,SAS,logP):')
y_1 = vae.predict_prop_Z(z_1)[0]
print(y_1)
###Output
Properties (qed,SAS,logP):
[ 0.77286768 2.43317604 0.95585614]
###Markdown
Decode several attemptsVAE are probabilistic
###Code
noise=5.0
print('Searching molecules randomly sampled from {:.2f} std (z-distance) from the point'.format(noise))
df = vae.z_to_smiles( z_1,decode_attempts=100,noise_norm=noise)
print('Found {:d} unique mols, out of {:d}'.format(len(set(df['smiles'])),sum(df['count'])))
print('SMILES\n',df.smiles)
display(PandasTools.FrameToGridImage(df,column='mol', legendsCol='smiles',molsPerRow=5))
df.head()
###Output
Searching molecules randomly sampled from 5.00 std (z-distance) from the point
Found 9 unique mols, out of 76
SMILES
0 CSCC(=O)NNC(=O)c1c(C)oc(C)c1C
1 COCC(=O)NNC(=O)c1c(C)oc(C)c1C
2 CSC(C=O)NNC(=O)c1c(C)oc(C)c1C
3 CSCC(=O)NCC(=O)c1c(C)oc(C)c1C
4 COCC(=O)NCC(=O)c1c(C)oc(C)c1C
5 CSC(C=O)NCC(=O)c1c(C)oc(C)c1C
6 COC(C=O)NCC(=O)c1c(C)oc(C)c1C
7 ClCC(=O)NCC(=O)c1c(C)oc(C)c1C
8 ClC(C=O)NNC(=O)c1c(C)oc(C)c1C
Name: smiles, dtype: object
###Markdown
PCA of latent spaceSample random points from the training set along with properties
###Code
Z, data, smiles = vae.ls_sampler_w_prop(size=50000,return_smiles=True)
prop_opt = 'qed'
prop_df = pd.DataFrame(data).reset_index()
prop_df['smiles']=smiles
prop_df.head()
###Output
_____no_output_____
###Markdown
Perform a PCA projection and color the points based on a property
###Code
from sklearn.decomposition import PCA
from sklearn.preprocessing import MinMaxScaler
# do pca and normalize
Z_pca = PCA(n_components=2).fit_transform(Z)
Z_pca = MinMaxScaler().fit_transform(Z_pca)
df = pd.DataFrame(np.transpose((Z_pca[:,0],Z_pca[:,1])))
df.columns = ['x','y']
df[prop_opt]=prop_df[prop_opt]
plt.scatter(x=df['x'], y=df['y'], c=df[prop_opt],
cmap= 'viridis', marker='.',
s=10,alpha=0.5, edgecolors='none')
plt.show()
###Output
_____no_output_____
###Markdown
compare with t-SNE, will take some time
###Code
from sklearn.manifold import TSNE
Z_tsne = TSNE(n_components=2).fit_transform(Z)
Z_tsne = MinMaxScaler().fit_transform(Z_tsne)
f = pd.DataFrame(np.transpose((Z_tsne[:,0],Z_tsne[:,1])))
df.columns = ['x','y']
df[prop_opt]=prop_df[prop_opt]
plt.scatter(x=df['x'], y=df['y'], c=df[prop_opt],
cmap= 'viridis', marker='.',
s=10,alpha=0.5, edgecolors='none')
plt.show()
###Output
_____no_output_____
|
fake_news_analysis.ipynb
|
###Markdown
###Code
# mount drive
from google.colab import drive
drive.mount('/drive',force_remount=True)
import pandas as pd
import numpy as np
from google.colab import files
uploaded = files.upload()
import io
df = pd.read_csv(io.BytesIO(uploaded['train.csv']))
# Dataset is now stored in a Pandas Dataframe
df.head()
df=df.dropna()
## Get the Independent & Dependent Features
X=df.drop('label',axis=1)
y=df['label']
X.shape
y.shape
import tensorflow as tf
tf.__version__
from tensorflow.keras.layers import Embedding
from tensorflow.keras.preprocessing.sequence import pad_sequences
from tensorflow.keras.models import Sequential
from tensorflow.keras.preprocessing.text import one_hot
from tensorflow.keras.layers import LSTM
from tensorflow.keras.layers import Dense
## Vocabulary size
voc_size=5000
###Output
_____no_output_____
###Markdown
Onehot Representation
###Code
messages=X.copy()
messages['title'][1]
messages.reset_index(inplace=True)
import nltk
import re
from nltk.corpus import stopwords
nltk.download('stopwords')
## Dataset Preprocessing
from nltk.stem.porter import PorterStemmer
ps = PorterStemmer()
corpus = []
for i in range(0, len(messages)):
print(i)
review = re.sub('[^a-zA-Z]', ' ', messages['title'][i])
review = review.lower()
review = review.split()
review = [ps.stem(word) for word in review if not word in stopwords.words('english')]
review = ' '.join(review)
corpus.append(review)
corpus
onehot_repr=[one_hot(words,voc_size)for words in corpus]
onehot_repr
###Output
_____no_output_____
###Markdown
Embedding Representation
###Code
sent_length=20
embedded_docs=pad_sequences(onehot_repr,padding='pre',maxlen=sent_length)
print(embedded_docs)
embedded_docs[0]
## Creating model
embedding_vector_features=40
model=Sequential()
model.add(Embedding(voc_size,embedding_vector_features,input_length=sent_length))
model.add(LSTM(100))
model.add(Dense(1,activation='sigmoid'))
model.compile(loss='binary_crossentropy',optimizer='adam',metrics=['accuracy'])
print(model.summary())
len(embedded_docs),y.shape
X_final=np.array(embedded_docs)
y_final=np.array(y)
X_final.shape,y_final.shape
from sklearn.model_selection import train_test_split
X_train, X_test, y_train, y_test = train_test_split(X_final, y_final, test_size=0.33, random_state=42)
###Output
_____no_output_____
###Markdown
Model Training
###Code
model.fit(X_train,y_train,validation_data=(X_test,y_test),epochs=10,batch_size=64)
###Output
Epoch 1/10
192/192 [==============================] - 7s 35ms/step - loss: 0.3362 - accuracy: 0.8424 - val_loss: 0.2063 - val_accuracy: 0.9157
Epoch 2/10
192/192 [==============================] - 6s 32ms/step - loss: 0.1430 - accuracy: 0.9433 - val_loss: 0.2048 - val_accuracy: 0.9208
Epoch 3/10
192/192 [==============================] - 6s 32ms/step - loss: 0.0988 - accuracy: 0.9646 - val_loss: 0.2154 - val_accuracy: 0.9130
Epoch 4/10
192/192 [==============================] - 6s 33ms/step - loss: 0.0707 - accuracy: 0.9749 - val_loss: 0.2655 - val_accuracy: 0.9193
Epoch 5/10
192/192 [==============================] - 6s 32ms/step - loss: 0.0480 - accuracy: 0.9855 - val_loss: 0.2742 - val_accuracy: 0.9165
Epoch 6/10
192/192 [==============================] - 6s 33ms/step - loss: 0.0309 - accuracy: 0.9905 - val_loss: 0.3212 - val_accuracy: 0.9143
Epoch 7/10
192/192 [==============================] - 7s 36ms/step - loss: 0.0177 - accuracy: 0.9944 - val_loss: 0.3816 - val_accuracy: 0.9178
Epoch 8/10
192/192 [==============================] - 7s 35ms/step - loss: 0.0124 - accuracy: 0.9965 - val_loss: 0.5095 - val_accuracy: 0.9171
Epoch 9/10
192/192 [==============================] - 6s 34ms/step - loss: 0.0087 - accuracy: 0.9974 - val_loss: 0.5416 - val_accuracy: 0.9148
Epoch 10/10
192/192 [==============================] - 7s 35ms/step - loss: 0.0063 - accuracy: 0.9976 - val_loss: 0.6203 - val_accuracy: 0.9145
###Markdown
Performance Metrics And Accuracy
###Code
y_pred=model.predict_classes(X_test)
from sklearn.metrics import confusion_matrix
confusion_matrix(y_test,y_pred)
from sklearn.metrics import accuracy_score
accuracy_score(y_test,y_pred)
###Output
_____no_output_____
|
optimization/Conjugate Gradient Method.ipynb
|
###Markdown
Conjugate Gradient Method
###Code
import numpy as np
import numpy.linalg as la
import scipy.optimize as sopt
import matplotlib.pyplot as pt
###Output
_____no_output_____
###Markdown
Let's make up a random linear system with an SPD $A$:
###Code
np.random.seed(25)
n = 2
Q = la.qr(np.random.randn(n, n))[0]
A = Q @ (np.diag(np.random.rand(n)) @ Q.T)
b = np.random.randn(n)
###Output
_____no_output_____
###Markdown
Here's the objective function for CG:
###Code
def phi(xvec):
x, y = xvec
return 0.5*(A[0,0]*x*x + 2*A[1,0]*x*y + A[1,1]*y*y) - x*b[0] - y*b[1]
def dphi(xvec):
x, y = xvec
return np.array([
A[0,0]*x + A[0,1]*y - b[0],
A[1,0]*x + A[1,1]*y - b[1]
])
###Output
_____no_output_____
###Markdown
Here's the function $\phi$ as a "contour plot":
###Code
xmesh, ymesh = np.mgrid[-10:10:50j,-10:10:50j]
phimesh = phi(np.array([xmesh, ymesh]))
pt.axis("equal")
pt.contour(xmesh, ymesh, phimesh, 50)
###Output
_____no_output_____
###Markdown
Running Conjugate Gradients ("CG")Initialize the method:
###Code
x0 = np.array([2, 2./5])
#x0 = np.array([2, 1])
iterates = [x0]
gradients = [dphi(x0)]
directions = [-dphi(x0)]
###Output
_____no_output_____
###Markdown
Evaluate this cell many times in-place:
###Code
x = iterates[-1]
s = directions[-1]
def f1d(alpha):
return phi(x + alpha*s)
alpha_opt = sopt.golden(f1d)
next_x = x + alpha_opt*s
g = dphi(next_x)
last_g = gradients[-1]
gradients.append(g)
beta = np.dot(g, g)/np.dot(last_g, last_g)
directions.append(-g + beta*directions[-1])
print(phi(next_x))
iterates.append(next_x)
# plot function and iterates
pt.axis("equal")
pt.contour(xmesh, ymesh, phimesh, 50)
it_array = np.array(iterates)
pt.plot(it_array.T[0], it_array.T[1], "x-")
###Output
-4.61671051783
|
tutorials/notebook/cx_site_chart_examples/fish_4.ipynb
|
###Markdown
Example: CanvasXpress fish Chart No. 4This example page demonstrates how to, using the Python package, create a chart that matches the CanvasXpress online example located at:https://www.canvasxpress.org/examples/fish-4.htmlThis example is generated using the reproducible JSON obtained from the above page and the `canvasxpress.util.generator.generate_canvasxpress_code_from_json_file()` function.Everything required for the chart to render is included in the code below. Simply run the code block.
###Code
from canvasxpress.canvas import CanvasXpress
from canvasxpress.js.collection import CXEvents
from canvasxpress.render.jupyter import CXNoteBook
cx = CanvasXpress(
render_to="fish4",
data={
"y": {
"vars": [
"C1",
"C2",
"C3",
"C4",
"C5",
"C6",
"C7"
],
"smps": [
0,
34,
69,
187,
334,
505,
530
],
"data": [
[
99,
1,
3,
1,
3,
80,
0.1
],
[
30,
0,
0,
0,
0,
0,
0
],
[
2,
0.1,
2.5,
0.9,
0.9,
76,
0.005
],
[
60,
0,
0,
0,
0,
0,
0
],
[
0,
0,
0,
0,
0.1,
60,
0.001
],
[
2,
0,
0,
0,
0,
0,
0
],
[
1,
1,
1,
10,
20,
15,
0
]
]
}
},
config={
"backgroundType": "windowSolidGradient",
"colorSpectrum": [
"#ffe4c4",
"#ffb90f",
"#cd6600"
],
"colors": [
"#888888",
"#EF0000",
"#8FFF40",
"#FF6000",
"#50FFAF",
"#FFCF00",
"#0070FF"
],
"fishAxis": [
0,
34,
69,
187,
334,
505,
530,
650,
750
],
"fishParents": [
0,
1,
1,
1,
3,
4,
0
],
"fishSeparateIndependentClones": True,
"fishShape": "spline",
"fishTimepoints": [
0,
34,
69,
187,
334,
505,
530
],
"gradientOrientation": "horizontal",
"graphType": "Fish"
},
width=613,
height=413,
events=CXEvents(),
after_render=[],
other_init_params={
"version": 35,
"events": False,
"info": False,
"afterRenderInit": False,
"noValidate": True
}
)
display = CXNoteBook(cx)
display.render(output_file="fish_4.html")
###Output
_____no_output_____
|
models/notebooks/DeepFM/01-DeepFM_baseline.ipynb
|
###Markdown
Preprocessing
###Code
for feat in sparse_features :
lbe = LabelEncoder()
df[feat] = lbe.fit_transform(df[feat])
mms = MinMaxScaler(feature_range = (0, 1))
df[dense_features] = mms.fit_transform(df[dense_features])
###Output
_____no_output_____
###Markdown
Generate Feature Columns
###Code
fixlen_feature_columns = [SparseFeat(feat, vocabulary_size = df[feat].max() +1, embedding_dim = 4) for feat in sparse_features] + [DenseFeat(feat, 1,) for feat in dense_features]
dnn_feature_columns = fixlen_feature_columns
linear_feature_columns = fixlen_feature_columns
feature_names = get_feature_names(linear_feature_columns + dnn_feature_columns)
###Output
_____no_output_____
###Markdown
Split Dataset
###Code
train, test = train_test_split(df, test_size = 0.2)
train_model_input = {name:train[name].values for name in feature_names}
test_model_input = {name:test[name].values for name in feature_names}
###Output
_____no_output_____
###Markdown
Modeling
###Code
model = DeepFM(linear_feature_columns, dnn_feature_columns, task = 'binary')
from tensorflow.keras import backend as K
def recall(y_target, y_pred):
# clip(t, clip_value_min, clip_value_max) : clip_value_min~clip_value_max 이외 가장자리를 깎아 낸다
# round : 반올림한다
y_target_yn = K.round(K.clip(y_target, 0, 1)) # 실제값을 0(Negative) 또는 1(Positive)로 설정한다
y_pred_yn = K.round(K.clip(y_pred, 0, 1)) # 예측값을 0(Negative) 또는 1(Positive)로 설정한다
# True Positive는 실제 값과 예측 값이 모두 1(Positive)인 경우이다
count_true_positive = K.sum(y_target_yn * y_pred_yn)
# (True Positive + False Negative) = 실제 값이 1(Positive) 전체
count_true_positive_false_negative = K.sum(y_target_yn)
# Recall = (True Positive) / (True Positive + False Negative)
# K.epsilon()는 'divide by zero error' 예방차원에서 작은 수를 더한다
recall = count_true_positive / (count_true_positive_false_negative + K.epsilon())
# return a single tensor value
return recall
def precision(y_target, y_pred):
# clip(t, clip_value_min, clip_value_max) : clip_value_min~clip_value_max 이외 가장자리를 깎아 낸다
# round : 반올림한다
y_pred_yn = K.round(K.clip(y_pred, 0, 1)) # 예측값을 0(Negative) 또는 1(Positive)로 설정한다
y_target_yn = K.round(K.clip(y_target, 0, 1)) # 실제값을 0(Negative) 또는 1(Positive)로 설정한다
# True Positive는 실제 값과 예측 값이 모두 1(Positive)인 경우이다
count_true_positive = K.sum(y_target_yn * y_pred_yn)
# (True Positive + False Positive) = 예측 값이 1(Positive) 전체
count_true_positive_false_positive = K.sum(y_pred_yn)
# Precision = (True Positive) / (True Positive + False Positive)
# K.epsilon()는 'divide by zero error' 예방차원에서 작은 수를 더한다
precision = count_true_positive / (count_true_positive_false_positive + K.epsilon())
# return a single tensor value
return precision
def f1score(y_target, y_pred):
_recall = recall(y_target, y_pred)
_precision = precision(y_target, y_pred)
# K.epsilon()는 'divide by zero error' 예방차원에서 작은 수를 더한다
_f1score = ( 2 * _recall * _precision) / (_recall + _precision+ K.epsilon())
# return a single tensor value
return _f1score
import tensorflow as tf
def f1(y_true, y_pred):
y_pred = K.round(y_pred)
tp = K.sum(K.cast(y_true*y_pred, 'float'), axis=0)
tn = K.sum(K.cast((1-y_true)*(1-y_pred), 'float'), axis=0)
fp = K.sum(K.cast((1-y_true)*y_pred, 'float'), axis=0)
fn = K.sum(K.cast(y_true*(1-y_pred), 'float'), axis=0)
p = tp / (tp + fp + K.epsilon())
r = tp / (tp + fn + K.epsilon())
f1 = 2*p*r / (p+r+K.epsilon())
f1 = tf.where(tf.math.is_nan(f1), tf.zeros_like(f1), f1)
return K.mean(f1)
def f1_loss(y_true, y_pred):
tp = K.sum(K.cast(y_true*y_pred, 'float'), axis=0)
tn = K.sum(K.cast((1-y_true)*(1-y_pred), 'float'), axis=0)
fp = K.sum(K.cast((1-y_true)*y_pred, 'float'), axis=0)
fn = K.sum(K.cast(y_true*(1-y_pred), 'float'), axis=0)
p = tp / (tp + fp + K.epsilon())
r = tp / (tp + fn + K.epsilon())
f1 = 2*p*r / (p+r+K.epsilon())
f1 = tf.where(tf.math.is_nan(f1), tf.zeros_like(f1), f1)
return 1 - K.mean(f1)
model.compile("adam", f1_loss, metrics=['accuracy', f1])
a = train['like_timestamp'].values
a = a.astype(np.float32)
history = model.fit(train_model_input, a,
batch_size = 256,
epochs = 5,
verbose = 1,
validation_split = 0.2,)
pred_ans = model.predict(test_model_input, batch_size = 256)
rce_like = compute_rce(pred_ans, test['like_timestamp'])
rce_like
ap_like = average_precision_score(test['like_timestamp'], pred_ans)
ap_like
save_model(model, 'DeepFM.h5')
from deepctr.utils import custom_objects
import deepctr
deepctr.__version__
###Output
_____no_output_____
|
legacy/arkady TF legacy/TF_2020_course4_Week_1_Exercise_Answer.ipynb
|
###Markdown
Now that we have the time series, let's split it so we can start forecasting
###Code
split_time = 1100
time_train = time[:split_time]
x_train = series[:split_time]
time_valid = time[split_time:]
x_valid = series[split_time:]
plt.figure(figsize=(10, 6))
plot_series(time_train, x_train)
plt.show()
plt.figure(figsize=(10, 6))
plot_series(time_valid, x_valid)
plt.show()
###Output
_____no_output_____
###Markdown
Naive Forecast
###Code
naive_forecast = series[split_time - 1:-1]
plt.figure(figsize=(10, 6))
plot_series(time_valid, x_valid)
plot_series(time_valid, naive_forecast)
###Output
_____no_output_____
###Markdown
Let's zoom in on the start of the validation period:
###Code
plt.figure(figsize=(10, 6))
plot_series(time_valid, x_valid, start=0, end=150)
plot_series(time_valid, naive_forecast, start=1, end=151)
###Output
_____no_output_____
###Markdown
You can see that the naive forecast lags 1 step behind the time series. Now let's compute the mean squared error and the mean absolute error between the forecasts and the predictions in the validation period:
###Code
print(keras.metrics.mean_squared_error(x_valid, naive_forecast).numpy())
print(keras.metrics.mean_absolute_error(x_valid, naive_forecast).numpy())
###Output
19.578304
2.6011968
###Markdown
That's our baseline, now let's try a moving average:
###Code
def moving_average_forecast(series, window_size):
"""Forecasts the mean of the last few values.
If window_size=1, then this is equivalent to naive forecast"""
forecast = []
for time in range(len(series) - window_size):
forecast.append(series[time:time + window_size].mean())
return np.array(forecast)
moving_avg = moving_average_forecast(series, 30)[split_time - 30:]
plt.figure(figsize=(10, 6))
plot_series(time_valid, x_valid)
plot_series(time_valid, moving_avg)
print(keras.metrics.mean_squared_error(x_valid, moving_avg).numpy())
print(keras.metrics.mean_absolute_error(x_valid, moving_avg).numpy())
###Output
65.786224
4.3040023
###Markdown
That's worse than naive forecast! The moving average does not anticipate trend or seasonality, so let's try to remove them by using differencing. Since the seasonality period is 365 days, we will subtract the value at time *t* – 365 from the value at time *t*.
###Code
diff_series = (series[365:] - series[:-365])
diff_time = time[365:]
plt.figure(figsize=(10, 6))
plot_series(diff_time, diff_series)
plt.show()
###Output
_____no_output_____
###Markdown
Great, the trend and seasonality seem to be gone, so now we can use the moving average:
###Code
diff_moving_avg = moving_average_forecast(diff_series, 50)[split_time - 365 - 50:]
plt.figure(figsize=(10, 6))
plot_series(time_valid, diff_series[split_time - 365:])
plot_series(time_valid, diff_moving_avg)
plt.show()
###Output
_____no_output_____
###Markdown
Now let's bring back the trend and seasonality by adding the past values from t – 365:
###Code
diff_moving_avg_plus_past = series[split_time - 365:-365] + diff_moving_avg
plt.figure(figsize=(10, 6))
plot_series(time_valid, x_valid)
plot_series(time_valid, diff_moving_avg_plus_past)
plt.show()
print(keras.metrics.mean_squared_error(x_valid, diff_moving_avg_plus_past).numpy())
print(keras.metrics.mean_absolute_error(x_valid, diff_moving_avg_plus_past).numpy())
###Output
8.498155
2.327179
###Markdown
Better than naive forecast, good. However the forecasts look a bit too random, because we're just adding past values, which were noisy. Let's use a moving averaging on past values to remove some of the noise:
###Code
diff_moving_avg_plus_smooth_past = moving_average_forecast(series[split_time - 370:-360], 10) + diff_moving_avg
plt.figure(figsize=(10, 6))
plot_series(time_valid, x_valid)
plot_series(time_valid, diff_moving_avg_plus_smooth_past)
plt.show()
print(keras.metrics.mean_squared_error(x_valid, diff_moving_avg_plus_smooth_past).numpy())
print(keras.metrics.mean_absolute_error(x_valid, diff_moving_avg_plus_smooth_past).numpy())
###Output
12.527958
2.2034433
|
Day_1_Intro/Full_Python_Tutorial_(optinal)/05_functions.ipynb
|
###Markdown
Introduction to Python V - Functions* Based on a lecture series by Rajath Kumart (https://github.com/rajathkumarmp/Python-Lectures)* Ported to Python3 and extessions added by Janis Keuper* Copyright: Creative Commons Attribution 3.0 Unported License. To view a copy of this license, visit http://creativecommons.org/licenses/by/3.0/ Functions Most of the times, In a algorithm the statements keep repeating and it will be a tedious job to execute the same statements again and again and will consume a lot of memory and is not efficient. Enter Functions. This is the basic syntax of a function def funcname(arg1, arg2,... argN): ''' Document String''' statements return Read the above syntax as, A function by name "funcname" is defined, which accepts arguements "arg1,arg2,....argN". The function is documented and it is '''Document String'''. The function after executing the statements returns a "value".
###Code
print ("Hello!")
print ("How do you do?")
###Output
_____no_output_____
###Markdown
Instead of writing the above two statements every single time it can be replaced by defining a function which would do the job in just one line. Defining a function firstfunc().
###Code
def firstfunc():
print ("Hello")
print ("How do you do?")
firstfunc()
###Output
_____no_output_____
###Markdown
**firstfunc()** every time just prints the message to a single person. We can make our function **firstfunc()** to accept arguements which will store the name and then prints respective to that accepted name. To do so, add a argument within the function as shown.
###Code
def firstfunc(username):
print ("Hey", username + '!')
print (username + ',' ,"How do you do?")
firstfunc("Hans")
###Output
_____no_output_____
###Markdown
Let us simplify this even further by defining another function **secondfunc()** which accepts the name and stores it inside a variable and then calls the **firstfunc()** from inside the function itself.
###Code
def firstfunc(username):
print ("Hey", username + '!')
print (username + ',' ,"How do you do?")
def secondfunc():
name = input("Please enter your name : ")
firstfunc(name)
secondfunc()
###Output
_____no_output_____
###Markdown
Return Statement When the function results in some value and that value has to be stored in a variable or needs to be sent back or returned for further operation to the main algorithm, return statement is used.
###Code
def times(x,y):
z = x*y
return z
###Output
_____no_output_____
###Markdown
The above defined **times( )** function accepts two arguements and return the variable z which contains the result of the product of the two arguements
###Code
c = times(4,5)
print (c)
###Output
_____no_output_____
###Markdown
The z value is stored in variable c and can be used for further operations. Instead of declaring another variable the entire statement itself can be used in the return statement as shown.
###Code
def times(x,y):
'''This multiplies the two input arguments'''
return x*y
c = times(4,5)
print (c)
###Output
_____no_output_____
###Markdown
Since the **times( )** is now defined, we can document it as shown above. This document is returned whenever **times( )** function is called under **help( )** function.
###Code
help(times)
###Output
_____no_output_____
###Markdown
Multiple variable can also be returned, But keep in mind the order.
###Code
eglist = [10,50,30,12,6,8,100]
def egfunc(eglist):
highest = max(eglist)
lowest = min(eglist)
first = eglist[0]
last = eglist[-1]
return highest,lowest,first,last
###Output
_____no_output_____
###Markdown
If the function is just called without any variable for it to be assigned to, the result is returned inside a tuple. But if the variables are mentioned then the result is assigned to the variable in a particular order which is declared in the return statement.
###Code
egfunc(eglist)
a,b,c,d = egfunc(eglist)
print (' a =',a,'\n b =',b,'\n c =',c,'\n d =',d)
###Output
_____no_output_____
###Markdown
Implicit arguments When an argument of a function is common in majority of the cases or it is "implicit" this concept is used.
###Code
def implicitadd(x,y=3):
return x+y
###Output
_____no_output_____
###Markdown
**implicitadd( )** is a function accepts two arguments but most of the times the first argument needs to be added just by 3. Hence the second argument is assigned the value 3. Here the second argument is implicit. Now if the second argument is not defined when calling the **implicitadd( )** function then it considered as 3.
###Code
implicitadd(4)
###Output
_____no_output_____
###Markdown
But if the second argument is specified then this value overrides the implicit value assigned to the argument
###Code
implicitadd(4,4)
###Output
_____no_output_____
###Markdown
Any number of arguments If the number of arguments that is to be accepted by a function is not known then a asterisk symbol is used before the argument.
###Code
def add_n(*args):
res = 0
reslist = []
for i in args:
reslist.append(i)
print (reslist)
return sum(reslist)
###Output
_____no_output_____
###Markdown
The above function accepts any number of arguments, defines a list and appends all the arguments into that list and return the sum of all the arguments.
###Code
add_n(1,2,3,4,5)
add_n(1,2,3)
def multi_reurn():
return 1,2,3,4,5
a,b,c,d,e=multi_reurn()
print (a,e)
multi_reurn()
###Output
_____no_output_____
###Markdown
Global and Local Variables Whatever variable is declared inside a function is local variable and outside the function in global variable.
###Code
eg1 = [1,2,3,4,5]
###Output
_____no_output_____
###Markdown
In the below function we are appending a element to the declared list inside the function. eg2 variable declared inside the function is a local variable.
###Code
def egfunc1():
def thirdfunc(arg1):
eg2 = arg1[:]
eg2.append(6)
print ("This is happening inside the function :", eg2 )
print ("This is happening before the function is called : ", eg1)
thirdfunc(eg1)
print ("This is happening outside the function :", eg1 )
print ("Accessing a variable declared inside the function from outside :" , eg2)
egfunc1()
###Output
_____no_output_____
###Markdown
If a **global** variable is defined as shown in the example below then that variable can be called from anywhere.
###Code
eg3 = [1,2,3,4,5]
def egfunc1():
def thirdfunc(arg1):
global eg2
eg2 = arg1[:]
eg2.append(6)
print ("This is happening inside the function :", eg2 )
print ("This is happening before the function is called : ", eg1)
thirdfunc(eg1)
print ("This is happening outside the function :", eg1)
print ("Accessing a variable declared inside the function from outside :" , eg2)
egfunc1()
###Output
_____no_output_____
|
11_MarkovAnalysis/11_13_SimulationOfAMarkovProcess.ipynb
|
###Markdown
11.13 Simulation of a Markov ProcessIn this notebook is the Python implementation of the pseudocode provided in section 11.13 (cf. figures 11.21 and 11.22). System description is provided in example 11.12 and depicted figure 11.20. IMPORT
###Code
import numpy as np
from scipy.linalg import expm
###Output
_____no_output_____
###Markdown
Subfunction *Single history*
###Code
def GetOneHistory(lambdaA, lambdaB):
# Time to failure initialization
ttf = 0
# Initial state
state = 3
# Change of notations
lambda30 = lambdaA
lambda32 = lambdaB
lambda20 = lambdaA
lambda21 = lambdaB
# Loop while any of the final states is reached
while (state!=1) and (state!=0):
# If current state is 3
if state==3:
# Draw duration until component A failure
t30 = np.random.exponential(scale=1/lambda30)
# Draw duration until component B_1 failure
t32 = np.random.exponential(scale=1/lambda32)
# If next event is component A failure
if t30<=t32:
state = 0 # Update the system state
ttf = ttf+t30 # Update the time to failure
else:
state = 2 # Update the system state
ttf = ttf+t32 # Update the time to failure
# If current state is 2
else:
# Draw duration until component A failure # (Exponential law's property)
t20 = np.random.exponential(scale=1/lambda20)
# Draw duration until component B2 failure
t21 = np.random.exponential(scale=1/lambda21)
# If next event is component A failure
if t20<=t21:
state = 0 # Update the system state
ttf = ttf+t20 # Update the time to failure
# If next event is component B_2 failure
else:
state = 1 # Update the system state
ttf = ttf+t21 # Update the time to failure
# return time to failure and final state
return (ttf, state)
###Output
_____no_output_____
###Markdown
Subfunction providing *Estimate of MTTF and failure states probabilities*
###Code
def SystemMonteCarlo(N, lambdaA, lambdaB):
# Initialize variables
mttf = 0
state0 = 0
# Loop on N histories
for i in range(0,N):
# Get outputs of a single history
ttf, state = GetOneHistory(lambdaA, lambdaB)
# Sum time to failure
mttf = mttf+ttf
if state==0:
# Sum histories ending on state 0
state0 = state0+1
# Estimate the system MTTF
mttf = mttf/N
# Estimate probability that system ends on state 0
state0 = state0/N
# Estimate probability that system ends on state 1
state1 = 1-state0
# return time to failure and probabilities estimation
return (mttf, state0, state1)
###Output
_____no_output_____
###Markdown
Computation
###Code
mttf, state0, state1 = SystemMonteCarlo(N=1000, lambdaA=1e-6, lambdaB=1e-6)
print('MTTF: {:f}'.format(mttf))
print('Ending in state 0 probability: {:f}'.format(state0))
print('Ending in state 1 probability: {:f}'.format(state1))
###Output
MTTF: 736103.721683
Ending in state 0 probability: 0.742000
Ending in state 1 probability: 0.258000
|
DiabtetesPredictions.ipynb
|
###Markdown
###Code
#import pandas
import pandas as pd
import matplotlib.pyplot as plt
import seaborn as sns
import numpy as np
from sklearn import metrics
from sklearn.neighbors import KNeighborsClassifier
# import the metrics class
from sklearn import metrics
from sklearn.model_selection import train_test_split
# import the class
from sklearn.linear_model import LogisticRegression
# load dataset
df = pd.read_csv("/content/diabetes (1).csv", sep=',', na_values=".")
df.shape
df
# target and features
y = df.Outcome
x = df.drop(columns=['Outcome'])
x.head()
y.head()
#4: Split into training and test set
x_train, x_test, y_train, y_test = train_test_split(x, y, test_size = 0.2, random_state=42)
#5: Create and fit your model using KNeighbors classification for five neighbors (sklearn)
# KNN model fit
knn = KNeighborsClassifier(n_neighbors=5)
knn.fit(x_train, y_train)
# Predict on dataset which model has not seen before
predictions = knn.predict(x_test)
#predictions
#6: Calculate the model accuracy of the model accuracy
accuracy_score = metrics.accuracy_score(y_test, predictions)
print('Accuracy Score = ', round(accuracy_score,2))
confusion_matrix = pd.crosstab(y_test, predictions,rownames=['Actual'], colnames=['Predicted'])
print (confusion_matrix)
TN = confusion_matrix.iloc[0,0]
FN = confusion_matrix.iloc[0,1]
TP = confusion_matrix.iloc[1,1]
FP = confusion_matrix.iloc[1,0]
#8: Print the TN, FN, TP, FP values
print('True Negative =',TN)
print('False Negative =',FN)
print('True Positive =',TP)
print('False Positive =',FP)
# 9: Print the model precision value
# Precision is the ratio of tp / (tp + fp)
myprecision = TP / (TP + FP)
print('Precision = ', round(myprecision,2))
#10: # Recall = the ratio tp / (tp + fn)
myrecall = TP / (TP + FN)
print('Recall = ', round(myrecall,2))
#11: Visualize the confusion matrix with a Heatmap
ax = plt.axes()
ax.set_title('Confusion Matrix with Heatmap')
sns.heatmap(confusion_matrix, annot=True)
#This is a list of rll, how often is the classifier correct?
#(TP+TN)/total = (100+50)/165 = 0.91
#Misclassificaates that are often computed from a
#confusion matrix for a binary classifier:
#Accuracy: Overation Rate: Overall, how often is it wrong?
#(FP+FN)/total = (10+5)/165 = 0.09
#equivalent to 1 minus Accuracy
#also known as "Error Rate"
#True Positive Rate: When it's actually yes, how often does it predict yes?
#TP/actual yes = 100/105 = 0.95
#also known as "Sensitivity" or "Recall"
#False Positive Rate: When it's actually no, how often does it predict yes?
#FP/actual no = 10/60 = 0.17
#True Negative Rate: When it's actually no, how often does it predict no?
#TN/actual no = 50/60 = 0.83
#equivalent to 1 minus False Positive Rate
#also known as "Specificity"
#Precision: When it predicts yes, how often is it correct?
#TP/predicted yes = 100/110 = 0.91
#Prevalence: How often does the yes condition actually occur in our sample?
#actual yes/total = 105/165 = 0.64
###Output
_____no_output_____
|
ML1/ensemble/073Ensemble_Boosting.ipynb
|
###Markdown
Ensemble methods. Boosting AdaBoostAdaBoost consists of following steps:* initialize weights to $\frac{1}{N}$, where $N$ is the number of datapoints,* loop until $\varepsilon_{t}<\frac{1}{2}$ or maximum number of iteration is reached,* train classifier on ${S,w^{(t)}}$ and get a hypothesis $h_{t}(x_{n})$ for datapoints $x_{n}$,* compute error $\varepsilon_{t}=\sum_{n=1}^{N}w_{n}^{(t)}I(y_{n}\neq h_{t}(x_{n}))$, * set $\alpha_{t}=\log(\frac{1-\varepsilon_{t}}{\varepsilon_{t}})$. * update weights $w_{n}^{(t+1)}=\frac{w_{n}^{(t)}\exp{\alpha_{t}I(y_{n}\neq h_{t}(x_{n}))}}{Z_{t}}$, where $Z_{t}$ is a normalization constant,* output $f(X)=\text{sign}(\sum_{t=1}^{T}\alpha_{t}h_{t}(x))$. Example taken from Marsland, Machine Learning: https://seat.massey.ac.nz/personal/s.r.marsland/MLBook.html.First, we need to import libraries:
###Code
import numpy as np
from sklearn.tree import DecisionTreeClassifier
###Output
_____no_output_____
###Markdown
Variables that are used by the classifier:
###Code
from sklearn.datasets import load_iris
import numpy as np
from sklearn.model_selection import train_test_split
labels = 2
dimension = 2
iris = load_iris()
data_set = iris.data
labels = iris.target
train_set, test_set, train_labels, test_labels = train_test_split(data_set, labels,
test_size=0.33, random_state=42)
###Output
_____no_output_____
###Markdown
Weights initialization:
###Code
number_of_iterations = 10
weights = np.ones((len(test_set),)) / len(test_set)
###Output
_____no_output_____
###Markdown
The data set looks as:
###Code
import matplotlib.pyplot as plt
sizes = np.ones(len(test_set))*5
plt.scatter(test_set[:, 1], test_set[:, 2], c=test_labels, s=sizes, cmap='autumn')
plt.show()
def train_model(classifier, weights):
return classifier.fit(X=test_set, y=test_labels, sample_weight=weights)
###Output
_____no_output_____
###Markdown
Accuracy vector calculation for the weights: 0 - don't change the weight, 1 - change it.
###Code
def calculate_accuracy_vector(predicted, labels):
result = []
for i in range(len(predicted)):
if predicted[i] == labels[i]:
result.append(0)
else:
result.append(1)
return result
###Output
_____no_output_____
###Markdown
Calculate the error rate $\varepsilon_{t}=\sum_{n=1}^{N}w_{n}^{(t)}I(y_{n}\neq h_{t}(x_{n}))$:
###Code
def calculate_error(weights, model):
predicted = model.predict(test_set)
return np.dot(weights,calculate_accuracy_vector(predicted, test_labels))
###Output
_____no_output_____
###Markdown
Calculate the $\alpha_{t}=\log(\frac{1-\varepsilon_{t}}{\varepsilon_{t}})$:
###Code
def set_alpha(error_rate):
return np.log((1-error_rate)/error_rate)
###Output
_____no_output_____
###Markdown
Calculate the new weights $w_{n}^{(t+1)}=\frac{w_{n}^{(t)}\exp{\alpha_{t}I(y_{n}\neq h_{t}(x_{n}))}}{Z_{t}}$:
###Code
def set_new_weights(old_weights, alpha, model):
new_weights = old_weights * np.exp(np.multiply(alpha,calculate_accuracy_vector(model.predict(test_set), test_labels)))
Zt = np.sum(new_weights)
return new_weights / Zt
###Output
_____no_output_____
###Markdown
Now, it's time to run the code and check the weights:
###Code
classifier = DecisionTreeClassifier(max_depth=1, random_state=1)
classifier.fit(X=train_set, y=train_labels)
alphas = []
classifiers = []
for iteration in range(number_of_iterations):
model = train_model(classifier, weights)
error_rate = calculate_error(weights, model)
alpha = set_alpha(error_rate)
weights = set_new_weights(weights, alpha, model)
alphas.append(alpha)
classifiers.append(model)
print(weights)
weights = (weights/np.min(weights))*2
plt.scatter(test_set[:, 1], test_set[:, 2], c=test_labels, s=weights, cmap='autumn')
plt.show()
###Output
_____no_output_____
|
data-science/scikit-learn/09/03-What-is-RBF-Kernel.ipynb
|
###Markdown
直观理解高斯核函数
###Code
import numpy as np
import matplotlib.pyplot as plt
x = np.arange(-4, 5, 1)
x
y = np.array((x >= -1) & (x <= 2), dtype='int')
y
plt.scatter(x[y==0], [0]*len(x[y==0]))
plt.scatter(x[y==1], [0]*len(x[y==1]))
plt.show()
def gaussion(x, l):
gamma = 1.0
return np.exp(-gamma * (x - l) ** 2)
l1, l2 = -1, 1
X_new = np.empty((len(x), 2))
for i, data in enumerate(x):
X_new[i, 0] = gaussion(data, l1)
X_new[i, 1] = gaussion(data, l2)
X_new
plt.scatter(X_new[y==0, 0], X_new[y==0, 1])
plt.scatter(X_new[y==1, 0], X_new[y==1, 1])
plt.show()
###Output
_____no_output_____
|
Slide Remittance Data Analysis/SLIDE 2020.ipynb
|
###Markdown
Data Analysis on SLIDE Transactions Data in 2020
###Code
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
import plotly as py
import plotly.graph_objs as go
remittance = pd.read_csv('remittance.csv', index_col= 0,parse_dates=True)
remittance
remittance['day'] = remittance.index
remittance['day'] = remittance['day'].apply(lambda x : x.weekday())
remittance
trx = remittance[['No_of_Trx', 'day']]
amt = remittance[['Total_Amt_Recd', 'day']]
###Output
_____no_output_____
###Markdown
Visualising Number of Transactions over 2020
###Code
plt.title('Line Graph for Transactions')
trx['No_of_Trx'].plot()
###Output
_____no_output_____
###Markdown
Visualising Amount (dollars) of Transactions over 2020
###Code
plt.title('Line Graph for Amount Received')
amt['Total_Amt_Recd'].plot()
###Output
_____no_output_____
###Markdown
Histogram for Transactions in 2020- It can be observed that there are between 220 to 280 transactions made in 100 days in 2020
###Code
plt.title('Histogram for Transactions')
trx['No_of_Trx'].hist()
###Output
_____no_output_____
###Markdown
Histogram for Amount (dollars) of Transactions in 2020- It can be observed that on slightly more than 70 days in 2020, the amount of transactions fell between 60k to 80k.
###Code
plt.title('Histogram for Amount Received')
amt['Total_Amt_Recd'].hist(bins=15)
grouped_trx = trx.groupby(['day']).sum()
grouped_amt = amt.groupby(['day']).sum()
###Output
_____no_output_____
###Markdown
Peak Periods- Thursday, Saturday and Sunday- Friday saw the lowest number of transactions
###Code
plt.title('Number of Transactions - Day of Week')
plt.plot(grouped_trx)
plt.show()
plt.title('Amount $ - Day of Week')
plt.plot(grouped_amt)
plt.show()
###Output
_____no_output_____
###Markdown
Time Series Forecasting for Number of Transactions
###Code
from fbprophet import Prophet
import warnings
warnings.filterwarnings("ignore")
trx['ds'] = trx.index
trx['y'] = trx['No_of_Trx']
model = Prophet(changepoint_prior_scale=0.01).fit(trx)
###Output
INFO:fbprophet:Disabling yearly seasonality. Run prophet with yearly_seasonality=True to override this.
INFO:fbprophet:Disabling daily seasonality. Run prophet with daily_seasonality=True to override this.
###Markdown
The following shows what Prophet has learnt from the input data -- as it attempts to fit a line that exhibits the trends of the transactions values.
###Code
future = model.make_future_dataframe(periods=31)
forecast = model.predict(future)
y = model.plot(forecast)
plt.show()
y.savefig('forecast.png')
###Output
_____no_output_____
###Markdown
From this model, Prophet picked up that the trend of the data is INCREASING.
###Code
forecast
###Output
_____no_output_____
###Markdown
Below graph shows the rough estimation/predictions for SLIDE transactions
###Code
# Plot Transactions Data
plt.figure(figsize=(10,6))
plt.grid(True)
plt.xlabel('Transactions')
plt.ylabel('Forecasted Transactions')
plt.plot(forecast['yhat'][-31:])
plt.title('SLIDE Transactions')
plt.show()
###Output
_____no_output_____
|
codici/old/Untitled3.ipynb
|
###Markdown
Classification mediante regression
###Code
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
import seaborn as sns
import matplotlib.pyplot as plt
import matplotlib.colors as mcolors
from matplotlib import cm
plt.style.use('fivethirtyeight')
plt.rcParams['font.family'] = 'sans-serif'
plt.rcParams['font.serif'] = 'Ubuntu'
plt.rcParams['font.monospace'] = 'Ubuntu Mono'
plt.rcParams['font.size'] = 10
plt.rcParams['axes.labelsize'] = 10
plt.rcParams['axes.labelweight'] = 'bold'
plt.rcParams['axes.titlesize'] = 10
plt.rcParams['xtick.labelsize'] = 8
plt.rcParams['ytick.labelsize'] = 8
plt.rcParams['legend.fontsize'] = 10
plt.rcParams['figure.titlesize'] = 12
plt.rcParams['image.cmap'] = 'jet'
plt.rcParams['image.interpolation'] = 'none'
plt.rcParams['figure.figsize'] = (16, 8)
plt.rcParams['lines.linewidth'] = 2
plt.rcParams['lines.markersize'] = 8
colors = ['xkcd:pale orange', 'xkcd:sea blue', 'xkcd:pale red', 'xkcd:sage green', 'xkcd:terra cotta', 'xkcd:dull purple', 'xkcd:teal', 'xkcd:goldenrod', 'xkcd:cadet blue',
'xkcd:scarlet']
cmap_big = cm.get_cmap('Spectral', 512)
cmap = mcolors.ListedColormap(cmap_big(np.linspace(0.5, 1, 128)))
def statistics(Theta,X):
y = np.asarray(classify(Theta,X))
confmat = np.zeros((2, 2))
for i in range(2):
for j in range(2):
confmat[i,j] = np.sum(np.where(y==i,1,0)*np.where(t==j,1,0))
print('Veri negativi: {0:d}'.format(int(confmat[0,0])))
print('Falsi negativi: {0:d}'.format(int(confmat[0,1])))
print('Falsi positivi: {0:d}'.format(int(confmat[1,0])))
print('Veri positivi: {0:d}'.format(int(confmat[1,1])))
print('Precision insieme 1: {0:3.3f}'.format(round(confmat[1,1]/(confmat[1,1]+confmat[1,0]),2)))
print('Recall insieme 1: {0:3.3f}'.format(round(confmat[1,1]/(confmat[1,1]+confmat[0,1]),2)))
print( 'Precision insieme 2: {0:3.3f}'.format(round(confmat[0,0]/(confmat[0,0]+confmat[0,1]),2)))
print( 'Recall insieme 2: {0:3.3f}'.format(round(confmat[0,0]/(confmat[0,0]+confmat[1,0]),2)))
print( 'Accuracy: {0:3.3f}'.format(round(np.trace(confmat)/n,2)))
return confmat
def classify(Theta, X):
# determina i valori di regressione predetti per le due classi
y = X*Theta
# restituisce la classe con valore più elevato nella regressione
return np.where(y[:,0]>y[:,1], 0, 1)
# legge i dati in dataframe pandas
data = pd.read_csv("../dataset/esami.txt", header=0, delimiter=',', names=['x1','x2','t'])
# calcola dimensione dei dati
n = len(data)
# calcola dimensionalità delle features
nfeatures = len(data.columns)-1
X = np.array(data[['x1','x2']])
# aggiunge una colonna unitaria alla matrice delle features
X=np.column_stack((np.ones(n), X))
# determina la rappresentazione 1-su-k dei valori delle classi
t = np.array(data['t']).reshape(-1,1)
t0 = np.where(t==0, 1, 0)
t1 = np.where(t==0, 0, 1)
T=np.column_stack((t0,t1))
# costruisce la rappresentazione come np.matrix delle matrici X e T
Tm=np.matrix(T)
Xm = np.matrix(X)
# calcola i coefficienti di regressione per le due classi, associati alle colonne di Theta
Theta=(Xm.T*Xm).I*Xm.T*Tm
## calcola i valori restituiti come classificazione su una griglia
delta1=max(X[:,1])-min(X[:,1])
delta2=max(X[:,2])-min(X[:,2])
min1=min(X[:,1])-delta1/10
max1=max(X[:,1])+delta1/10
min2=min(X[:,2])-delta2/10
max2=max(X[:,2])+delta2/10
u = np.linspace(min1, max1, 100)
v = np.linspace(min2, max2, 100)
z = np.zeros((len(u), len(v)))
y0 = np.zeros((len(u), len(v)))
y1 = np.zeros((len(u), len(v)))
for i in range(0, len(u)):
for j in range(0, len(v)):
r = np.matrix([1, u[i],v[j]])*Theta
y0[i,j] = np.asarray(r)[0][0]
y1[i,j] = np.asarray(r)[0][1]
z[i,j] =np.where(r[:,0]>r[:,1], 0, 1)
u, v = np.meshgrid(u, v)
c = ([colors[i+6] for i in np.nditer(t)])
fig = plt.figure(figsize=(16,16))
ax = fig.gca()
imshow_handle = plt.imshow(y0, origin='lower', extent=(min1, max1, min2, max2))
ax.scatter(np.asarray(X[:,1]), np.asarray(X[:,2]), s=30,c=c, marker='o')
plt.contour(u, v, z, [0.5], colors=[colors[9]])
plt.xlabel('Punteggio esame 1', fontsize=10)
plt.ylabel('Punteggio esame 2', fontsize=10)
plt.xticks(fontsize=10)
plt.yticks(fontsize=10)
plt.xlim(u.min(), u.max())
plt.ylim(v.min(), v.max())
plt.title('Valori regressione per C0', fontsize=12)
plt.show()
###Output
_____no_output_____
|
Section 10/SVM-2/Teclov_SVM_casestudy3.ipynb
|
###Markdown
Letter Recognition Using SVMLet's now tackle a slightly more complex problem - letter recognition. We'll first explore the dataset a bit, prepare it (scale etc.) and then experiment with linear and non-linear SVMs with various hyperparameters. Data Understanding Let's first understand the shape, attributes etc. of the dataset.
###Code
# libraries
import pandas as pd
import numpy as np
from sklearn.svm import SVC
from sklearn.model_selection import train_test_split
from sklearn import metrics
from sklearn.metrics import confusion_matrix
from sklearn.model_selection import KFold
from sklearn.model_selection import cross_val_score
from sklearn.model_selection import GridSearchCV
import matplotlib.pyplot as plt
import seaborn as sns
from sklearn.preprocessing import scale
# dataset
letters = pd.read_csv("letter-recognition.csv")
# about the dataset
# dimensions
print("Dimensions: ", letters.shape, "\n")
# data types
print(letters.info())
# head
letters.head()
# a quirky bug: the column names have a space, e.g. 'xbox ', which throws and error when indexed
print(letters.columns)
# let's 'reindex' the column names
letters.columns = ['letter', 'xbox', 'ybox', 'width', 'height', 'onpix', 'xbar',
'ybar', 'x2bar', 'y2bar', 'xybar', 'x2ybar', 'xy2bar', 'xedge',
'xedgey', 'yedge', 'yedgex']
print(letters.columns)
order = list(np.sort(letters['letter'].unique()))
print(order)
# basic plots: How do various attributes vary with the letters
plt.figure(figsize=(16, 8))
sns.barplot(x='letter', y='xbox',
data=letters,
order=order)
letter_means = letters.groupby('letter').mean()
letter_means.head()
plt.figure(figsize=(18, 10))
sns.heatmap(letter_means)
###Output
_____no_output_____
###Markdown
Data PreparationLet's conduct some data preparation steps before modeling. Firstly, let's see if it is important to **rescale** the features, since they may have varying ranges. For example, here are the average values:
###Code
# average feature values
round(letters.drop('letter', axis=1).mean(), 2)
###Output
_____no_output_____
###Markdown
In this case, the average values do not vary a lot (e.g. having a diff of an order of magnitude). Nevertheless, it is better to rescale them.
###Code
# splitting into X and y
X = letters.drop("letter", axis = 1)
y = letters['letter']
# scaling the features
X_scaled = scale(X)
# train test split
X_train, X_test, y_train, y_test = train_test_split(X_scaled, y, test_size = 0.3, random_state = 101)
###Output
_____no_output_____
###Markdown
Model BuildingLet's fist build two basic models - linear and non-linear with default hyperparameters, and compare the accuracies.
###Code
# linear model
model_linear = SVC(kernel='linear')
model_linear.fit(X_train, y_train)
# predict
y_pred = model_linear.predict(X_test)
# confusion matrix and accuracy
# accuracy
print("accuracy:", metrics.accuracy_score(y_true=y_test, y_pred=y_pred), "\n")
# cm
print(metrics.confusion_matrix(y_true=y_test, y_pred=y_pred))
###Output
accuracy: 0.8523333333333334
[[198 0 0 0 0 0 1 1 0 1 1 1 0 0 0 0 0 1
0 1 1 0 0 0 3 0]
[ 0 188 0 3 0 1 3 3 1 0 1 0 0 2 0 1 1 9
3 0 0 1 0 1 0 0]
[ 1 0 200 0 7 0 12 1 0 0 5 0 0 0 3 0 0 0
0 0 1 0 0 0 0 0]
[ 1 15 0 210 0 1 2 2 1 0 1 0 1 5 3 0 0 5
0 1 0 0 0 0 0 0]
[ 0 1 3 0 204 2 6 1 0 0 1 5 0 0 0 0 2 2
1 2 0 0 0 1 0 3]
[ 0 0 0 1 1 201 1 2 1 1 0 0 0 2 0 2 0 0
3 7 1 0 1 0 1 0]
[ 0 1 9 4 2 2 167 1 0 1 4 3 1 0 1 0 9 1
8 0 0 2 3 0 0 0]
[ 0 7 3 11 0 4 3 141 0 2 4 1 2 0 12 0 4 12
0 0 4 2 0 4 1 0]
[ 0 0 2 3 0 6 0 0 184 9 0 0 0 0 1 0 0 0
3 0 0 0 0 4 0 3]
[ 2 0 0 3 0 2 0 2 10 187 0 0 0 1 2 0 0 1
5 0 1 0 0 0 0 4]
[ 0 1 5 2 0 0 1 3 0 0 198 2 2 0 0 0 0 19
0 0 0 0 0 12 0 0]
[ 2 1 3 2 5 0 8 1 0 0 1 206 0 0 0 0 5 0
2 1 0 0 0 0 0 0]
[ 0 3 0 0 0 0 0 3 0 0 0 0 222 1 0 0 0 2
0 0 0 0 3 0 0 0]
[ 1 0 0 4 0 0 0 6 0 0 0 0 1 235 1 1 0 0
0 0 0 1 1 0 0 0]
[ 3 0 4 7 0 0 0 21 0 0 0 0 2 0 163 3 2 3
0 0 3 0 10 0 0 0]
[ 0 2 0 2 0 16 5 1 0 1 3 0 0 0 1 225 0 0
0 0 0 1 0 0 8 0]
[ 3 1 0 0 4 0 9 0 0 2 0 1 0 0 6 0 198 0
8 0 0 0 1 0 0 2]
[ 11 11 0 2 0 1 6 3 0 0 10 0 0 3 4 0 2 188
0 1 0 0 0 1 0 0]
[ 1 13 0 0 9 5 8 0 7 1 0 2 0 0 0 1 6 1
155 6 0 0 0 3 0 10]
[ 0 0 0 1 1 4 2 4 1 0 0 0 0 0 0 0 0 1
3 214 0 0 0 1 1 6]
[ 2 0 1 2 0 0 0 2 0 0 0 0 1 1 3 0 0 0
0 0 211 0 1 0 0 0]
[ 2 2 0 0 0 0 1 3 0 0 0 0 0 1 0 1 0 3
0 0 0 190 6 0 2 0]
[ 0 0 0 0 0 0 1 0 0 0 0 0 6 1 2 0 0 0
0 0 0 0 212 0 0 0]
[ 0 2 0 4 5 0 1 0 2 3 3 3 0 0 1 0 0 1
2 2 1 0 0 212 1 1]
[ 2 0 0 0 0 2 0 1 0 0 0 0 1 0 0 0 3 0
0 4 1 10 0 2 211 0]
[ 1 0 0 0 3 0 0 0 0 6 0 0 0 0 0 0 5 0
18 1 0 0 0 1 0 194]]
###Markdown
The linear model gives approx. 85% accuracy. Let's look at a sufficiently non-linear model with randomly chosen hyperparameters.
###Code
# non-linear model
# using rbf kernel, C=1, default value of gamma
# model
non_linear_model = SVC(kernel='rbf')
# fit
non_linear_model.fit(X_train, y_train)
# predict
y_pred = non_linear_model.predict(X_test)
# confusion matrix and accuracy
# accuracy
print("accuracy:", metrics.accuracy_score(y_true=y_test, y_pred=y_pred), "\n")
# cm
print(metrics.confusion_matrix(y_true=y_test, y_pred=y_pred))
###Output
accuracy: 0.9383333333333334
[[205 0 0 1 0 0 0 0 0 1 0 0 0 0 0 0 0 0
0 0 0 0 0 0 2 0]
[ 0 205 0 3 1 0 1 0 0 0 0 0 0 0 0 0 0 6
1 0 0 0 0 1 0 0]
[ 0 0 213 0 5 0 7 1 0 0 0 0 0 0 4 0 0 0
0 0 0 0 0 0 0 0]
[ 0 4 0 234 0 0 1 3 0 0 0 0 0 3 1 0 0 2
0 0 0 0 0 0 0 0]
[ 0 0 0 0 221 1 9 0 0 0 0 0 0 0 0 0 0 1
0 0 0 0 0 0 0 2]
[ 0 0 0 1 0 215 1 1 1 0 0 0 0 1 0 1 0 0
1 3 0 0 0 0 0 0]
[ 0 0 3 4 1 1 202 0 0 0 0 1 1 0 2 0 0 1
0 0 0 1 2 0 0 0]
[ 0 7 0 5 0 0 4 177 0 0 2 0 1 0 3 0 4 13
0 0 1 0 0 0 0 0]
[ 0 0 1 1 0 3 0 0 194 11 0 0 0 0 0 1 0 0
2 0 0 0 0 2 0 0]
[ 1 0 0 1 0 0 0 0 6 206 0 0 0 1 2 0 0 0
2 0 0 0 0 1 0 0]
[ 0 4 0 2 0 0 0 4 0 0 217 0 1 0 0 0 0 14
0 0 0 0 0 3 0 0]
[ 0 0 1 0 2 0 6 0 0 0 1 222 0 0 0 0 0 3
0 0 0 0 0 2 0 0]
[ 0 5 0 0 0 0 0 2 0 0 0 0 225 0 0 0 0 0
0 0 0 0 2 0 0 0]
[ 0 2 0 1 0 0 0 2 0 0 0 0 1 239 3 0 0 2
0 0 0 0 0 0 1 0]
[ 0 0 0 1 0 0 0 0 0 0 0 0 0 0 209 0 1 1
0 0 1 0 8 0 0 0]
[ 0 2 0 3 3 11 1 1 0 0 0 0 0 0 1 237 1 0
0 0 0 0 0 0 5 0]
[ 0 0 0 0 2 0 2 0 0 0 0 0 0 0 6 0 222 0
1 0 0 0 2 0 0 0]
[ 0 10 0 2 0 0 0 0 0 0 1 0 0 4 0 0 2 224
0 0 0 0 0 0 0 0]
[ 0 3 0 0 2 3 0 0 0 0 0 0 0 0 0 0 0 0
220 0 0 0 0 0 0 0]
[ 0 0 0 1 0 2 0 2 0 0 0 0 0 0 0 1 0 1
0 228 0 0 0 3 1 0]
[ 0 0 0 0 0 0 0 0 0 0 0 0 0 1 1 0 0 0
0 0 222 0 0 0 0 0]
[ 0 7 0 0 0 0 0 1 0 0 0 0 1 4 0 1 0 0
0 0 0 193 1 0 3 0]
[ 0 1 0 0 0 0 1 1 0 0 0 0 0 1 0 0 0 0
0 0 1 0 217 0 0 0]
[ 0 2 0 3 2 0 0 0 1 0 2 0 0 0 0 0 0 1
0 0 0 0 0 233 0 0]
[ 2 0 0 0 0 0 0 0 0 0 0 0 1 0 0 0 1 0
0 1 2 2 0 0 228 0]
[ 0 0 0 0 2 0 0 0 0 0 0 0 0 0 0 0 4 0
1 0 0 0 0 0 0 222]]
###Markdown
The non-linear model gives approx. 93% accuracy. Thus, going forward, let's choose hyperparameters corresponding to non-linear models. Grid Search: Hyperparameter TuningLet's now tune the model to find the optimal values of C and gamma corresponding to an RBF kernel. We'll use 5-fold cross validation.
###Code
# creating a KFold object with 5 splits
folds = KFold(n_splits = 5, shuffle = True, random_state = 101)
# specify range of hyperparameters
# Set the parameters by cross-validation
hyper_params = [ {'gamma': [1e-2, 1e-3, 1e-4],
'C': [1, 10, 100, 1000]}]
# specify model
model = SVC(kernel="rbf")
# set up GridSearchCV()
model_cv = GridSearchCV(estimator = model,
param_grid = hyper_params,
scoring= 'accuracy',
cv = folds,
verbose = 1,
return_train_score=True)
# fit the model
model_cv.fit(X_train, y_train)
# cv results
cv_results = pd.DataFrame(model_cv.cv_results_)
cv_results
# converting C to numeric type for plotting on x-axis
cv_results['param_C'] = cv_results['param_C'].astype('int')
# # plotting
plt.figure(figsize=(16,6))
# subplot 1/3
plt.subplot(131)
gamma_01 = cv_results[cv_results['param_gamma']==0.01]
plt.plot(gamma_01["param_C"], gamma_01["mean_test_score"])
plt.plot(gamma_01["param_C"], gamma_01["mean_train_score"])
plt.xlabel('C')
plt.ylabel('Accuracy')
plt.title("Gamma=0.01")
plt.ylim([0.60, 1])
plt.legend(['test accuracy', 'train accuracy'], loc='upper left')
plt.xscale('log')
# subplot 2/3
plt.subplot(132)
gamma_001 = cv_results[cv_results['param_gamma']==0.001]
plt.plot(gamma_001["param_C"], gamma_001["mean_test_score"])
plt.plot(gamma_001["param_C"], gamma_001["mean_train_score"])
plt.xlabel('C')
plt.ylabel('Accuracy')
plt.title("Gamma=0.001")
plt.ylim([0.60, 1])
plt.legend(['test accuracy', 'train accuracy'], loc='upper left')
plt.xscale('log')
# subplot 3/3
plt.subplot(133)
gamma_0001 = cv_results[cv_results['param_gamma']==0.0001]
plt.plot(gamma_0001["param_C"], gamma_0001["mean_test_score"])
plt.plot(gamma_0001["param_C"], gamma_0001["mean_train_score"])
plt.xlabel('C')
plt.ylabel('Accuracy')
plt.title("Gamma=0.0001")
plt.ylim([0.60, 1])
plt.legend(['test accuracy', 'train accuracy'], loc='upper left')
plt.xscale('log')
###Output
_____no_output_____
###Markdown
The plots above show some useful insights:- Non-linear models (high gamma) perform *much better* than the linear ones- At any value of gamma, a high value of C leads to better performance- None of the models tend to overfit (even the complex ones), since the training and test accuracies closely follow each otherThis suggests that the problem and the data is **inherently non-linear** in nature, and a complex model will outperform simple, linear models in this case. Let's now choose the best hyperparameters.
###Code
# printing the optimal accuracy score and hyperparameters
best_score = model_cv.best_score_
best_hyperparams = model_cv.best_params_
print("The best test score is {0} corresponding to hyperparameters {1}".format(best_score, best_hyperparams))
###Output
The best test score is 0.9517142857142857 corresponding to hyperparameters {'C': 1000, 'gamma': 0.01}
###Markdown
Building and Evaluating the Final ModelLet's now build and evaluate the final model, i.e. the model with highest test accuracy.
###Code
# model with optimal hyperparameters
# model
model = SVC(C=1000, gamma=0.01, kernel="rbf")
model.fit(X_train, y_train)
y_pred = model.predict(X_test)
# metrics
print("accuracy", metrics.accuracy_score(y_test, y_pred), "\n")
print(metrics.confusion_matrix(y_test, y_pred), "\n")
###Output
accuracy 0.9596666666666667
[[206 0 0 0 0 0 0 0 0 0 0 1 0 0 0 0 0 0
0 0 0 0 0 0 2 0]
[ 0 211 0 2 1 0 1 0 0 0 0 0 0 0 0 0 0 1
2 0 0 0 0 0 0 0]
[ 0 0 220 0 3 0 4 1 0 0 0 0 0 0 2 0 0 0
0 0 0 0 0 0 0 0]
[ 0 3 0 236 0 1 0 1 0 1 0 0 0 2 2 0 0 1
1 0 0 0 0 0 0 0]
[ 0 0 1 0 225 1 4 0 0 0 0 0 0 0 0 0 0 0
1 0 0 0 0 0 0 2]
[ 0 0 0 1 0 217 0 0 1 1 0 0 0 1 0 3 0 0
0 0 0 1 0 0 0 0]
[ 0 0 2 3 1 0 209 0 0 0 0 0 1 0 1 0 0 0
0 0 0 1 1 0 0 0]
[ 0 1 3 5 0 0 2 195 1 1 2 1 1 0 1 0 3 1
0 0 0 0 0 0 0 0]
[ 0 0 0 1 0 1 0 0 203 8 0 0 0 0 0 0 1 0
0 0 0 0 0 1 0 0]
[ 0 0 0 0 0 0 0 0 7 209 0 0 0 1 0 0 0 1
0 0 2 0 0 0 0 0]
[ 0 1 0 0 2 0 0 5 0 0 228 0 0 0 0 0 0 5
0 0 0 0 0 4 0 0]
[ 0 0 0 0 0 0 1 1 0 0 0 232 0 0 0 1 0 1
0 1 0 0 0 0 0 0]
[ 0 0 0 0 0 0 0 0 0 0 0 0 230 2 0 0 0 0
0 0 0 0 2 0 0 0]
[ 0 3 0 1 0 0 0 0 0 0 0 0 1 244 0 0 0 1
0 0 0 0 1 0 0 0]
[ 0 0 2 0 0 0 2 0 0 1 0 0 2 0 210 0 1 1
0 0 1 0 1 0 0 0]
[ 0 0 0 0 1 8 0 1 1 0 0 0 0 0 0 252 1 0
0 0 0 0 0 0 1 0]
[ 0 0 0 0 3 0 1 0 0 0 0 0 0 0 2 1 226 0
0 0 0 0 0 0 0 2]
[ 0 8 0 1 0 0 1 3 0 0 3 0 0 3 0 0 0 224
0 0 0 0 0 0 0 0]
[ 0 1 0 0 1 2 1 0 0 0 0 0 0 0 0 0 0 0
223 0 0 0 0 0 0 0]
[ 0 0 0 1 0 0 0 1 0 0 0 0 0 0 0 1 0 0
0 235 0 0 0 1 0 0]
[ 2 0 0 0 0 0 1 2 0 0 0 0 0 2 0 0 0 0
0 0 217 0 0 0 0 0]
[ 0 4 0 0 0 0 0 0 0 0 0 0 1 0 0 1 0 0
0 0 0 203 1 0 1 0]
[ 0 0 0 0 0 0 1 0 0 0 0 0 0 0 0 1 0 0
0 0 0 0 220 0 0 0]
[ 0 1 0 3 2 0 0 0 0 0 3 1 0 0 0 0 0 1
0 0 0 0 0 232 1 0]
[ 1 0 0 0 0 0 0 0 0 1 0 0 0 0 0 0 0 0
1 0 1 2 1 0 230 0]
[ 0 0 0 0 2 1 0 0 0 0 0 0 0 0 0 0 3 0
1 1 0 0 0 0 0 221]]
|
notebooks/split_testing.ipynb
|
###Markdown
Split TestingPractical Overview of A/B Testing
###Code
import scipy.stats as scs
import numpy as np
# preview data
# equations in LaTeX
# conclusion
###Output
_____no_output_____
###Markdown
Types of Split Testing* A/B Testing* Multivariate Testing* Multi-Armed Bandit General Procedure What can be tested? See an aggregated list below:* Headlines* Sub headlines* Paragraph Text* Testimonials* Call to Action text* Call to Action Button* Links* Images* Content near the fold* Social proof* Media mentions* Awards and badges* Traffic* App installs* Lead generation* Conversions* Video views* Catalog sales* Reach* EngagementPlease see the references Definitions* confidence interval: * a type of interval estimate, computed from the statistics of the observed data, that might contain the true value of an unknown population parameter[Wikipedia](https://en.wikipedia.org/wiki/Confidence_interval) * the interval has an associated confidence level that, loosely speaking, quantifies the level of confidence that the parameter lies in the interval * it is not a definitive range of plausible values for the sample parameter, though it may be understood as an estimate of plausible values for the population parameter * a particular confidence interval of 95% calculated from an experiment does not mean that there is a 95% probability of a sample parameter from a repeat of the experiment falling within this interval* critical value: [StatisticsHowTo](http://www.statisticshowto.com/probability-and-statistics/find-critical-values/)* effect size: * false positive: in the case for split testing, false positive results suggest that a variant will improve a metric, when actually, the metric may be unchanged or may be affected by other factors; larger sample sizes will reduce the risk of false positives * margin of error* sample size, minimum* significance level ($\alpha$): * the probability of making the wrong decision when the null hypothesis is true [StatisticsHowTo](http://www.statisticshowto.com/what-is-an-alpha-level/) * typically experiments are run with a significance level of 0.05 but ultimately the significance level will depend on the experiment* standard error of the mean: the standard deviation of the sampling mean* statistical power: * probability of finding an effect if it is real [4] * probability of rejecting the null hypothesis when the alternative hypothesis is true [Wikipedia](https://en.wikipedia.org/wiki/Power_(statistics))* statistical significance: * statistical significance occurs when the resulting p-value from an experiment is less than the level of significance, $\alpha$ * if there is statistical significance, the null hypothesis can be rejected* t-distribution or Student's t-distribution: * continuous probability distribution that arises when estimating the mean of a normally distributed population in situations where the sample size is small and population standard deviation is unknown [Wikipedia](https://en.wikipedia.org/wiki/Student%27s_t-distribution) * can be used to approximate the confidence interval of the true mean of a normal distribution* t-test, Student: [Wikipedia](https://en.wikipedia.org/wiki/Student%27s_t-test) * standard Student's t-test for two independent samples with equal sample sizes and equal variance * t-test, Welch's: [Wikipedia](https://en.wikipedia.org/wiki/Welch%27s_t-test) * Welch's t-test for two independent samples with equal sample sizes and equal variance * type I error: * false positive* type II error: * false negative * failing to reject the null hypothesis when the null hypothesis is false * probability of type II error decreases as statistical power increases* variance ($\sigma^2$): * standard deviation squared * for a binomial distribution: $np(1-p)$* z-score: a z-score is the distance measured in number of population standard deviations from a data point to the population mean [StatisticsHowTo](http://www.statisticshowto.com/probability-and-statistics/z-score/) Rule of Thumb for Estimating Minimum Sample Size [8]For a power of 80% (typical):$$ n = 16 \frac{\sigma^2}{\delta^2} $$where:* $n$ is the minimum sample size* $\sigma^2$ is the variance* $\delta$ is the minimum effect size* the constant 16 corresponds to a statistical power of 80%; use 26 for a statistical power of 95%For a binomial proportion:$$ \sigma^2 = np(1-p) $$ Power Formula$$ Z_{power} = \frac{difference}{standarderror(difference)} - Z_{\alpha/2} $$ Equation for Standard Error of the Mean$$ \sigma_{\bar{x}} = \frac{\sigma}{\sqrt{n}} $$ Equation for Minimum Sample Size Given significance level, find z-score and critical valueHow: Use [z-table](http://www.z-table.com/) or use function in the cell below For one-tailed test:1. Find the central area under the curve after subtracting the significance level from 1. 2. Find the x-value that returns the area equivalent to the central area computed in the first step. For two-tailed test (typical):1. Find the central area under the curve after subtracting half of the significance level from 1. 2. Find the x-value that returns the area equivalent to the central area computed in the first step. Common z-scores for two-tailed testsConfidence Interval | Significance Level | z-score--- | --- | ---80% | 0.20 | 1.2885% | 0.15 | 1.4490% | 0.10 | 1.6595% | 0.05 | 1.9699% | 0.01 | 2.58The z score of a raw score, x:$$ z = \frac{x - \mu}{\sigma} $$ To find the critical valueFind the sample mean and add/subtract the standard deviation multiplied by the z-score$$ cv = \bar{x} \pm z \times s_x $$ **When comparing two independent samples, the statistical power is the area of the variant's distribution to the right (if the effect is greater) of the critical value.**
###Code
def get_zscore(significance=0.05, two_tailed=True):
"""Returns the appropriate z-score given the level of significance
Arguments:
significance (float): typically 0.05 for 5% significance level but ultimately depends on the experiment
two_tailed (boolean): False if test is one-tailed
Returns:
z_score (float)
"""
norm_dist = scs.norm()
if two_tailed:
central_area = 1 - significance/2
else:
central_area = 1 - significance
return norm_dist.ppf(central_area)
get_zscore(significance=0.01, two_tailed=True)
###Output
_____no_output_____
|
Flight Ticket Price Hackathon_ EDA & Boosting.ipynb
|
###Markdown
Quick Overview of Data
###Code
train.head(2)
test.head(5)
# check that all the data was properly imported
print('shape:', train.shape, '\n')
print('dtpyes: \n' + str(train.dtypes))
# Get counts of categorical features
print('number of airlines:', len(train['Airline'].unique()))
print('number of sources:', len(train['Source'].unique()))
print('number of destinations:', len(train['Destination'].unique()))
print('number of stops:', len(train['Total_Stops'].unique()))
print('number of Additional Info:', len(train['Additional_Info'].unique()))
train['Date_of_Journey'] = pd.to_datetime(train['Date_of_Journey'],format="%d/%m/%Y")
test['Date_of_Journey'] = pd.to_datetime(test['Date_of_Journey'],format="%d/%m/%Y")
train['Dep_Time'] = pd.to_datetime(train['Dep_Time'],format="%H:%M")
test['Dep_Time'] = pd.to_datetime(test['Dep_Time'],format="%H:%M")
# Check range of dates in df
print('Date range:', train['Date_of_Journey'].max() - train['Date_of_Journey'].min())
print('Latest Date:', train['Date_of_Journey'].max())
print('Earliest Date:', train['Date_of_Journey'].min())
print(train['Date_of_Journey'].describe())
# Price analysis
print(train['Price'].describe())
train.isnull().sum()
#There is one record in train data which have missing value in route an stops. we will delete this
train.dropna(axis=0,inplace= True)
train.reset_index(inplace= True)
train.drop('index', axis=1, inplace= True)
train.head(2)
train.shape, test.shape
###Output
_____no_output_____
###Markdown
Preprocessing- Comvert the duration column to hours or minutes- Delete arrival time, Few recors looks off as it is not in sync with date of journey and departure time. We will calculate the arrival time based on the departure and duration- Explore the various categorical feature - Add some time and duration based feature
###Code
# We already have duration variable . lets delete Arrival time
train.drop('Arrival_Time', axis=1, inplace= True)
test.drop('Arrival_Time', axis=1, inplace= True)
###Output
_____no_output_____
###Markdown
Feature Engineering
###Code
train['day_d'] = train.Date_of_Journey.dt.day
train['month_d'] = train.Date_of_Journey.dt.month
train['weekday_d'] = train.Date_of_Journey.dt.weekday
train['month_start'] = train.Date_of_Journey.dt.is_month_start
train['month_end'] = train.Date_of_Journey.dt.is_month_end
test['day_d'] = test.Date_of_Journey.dt.day
test['month_d'] = test.Date_of_Journey.dt.month
test['weekday_d'] = test.Date_of_Journey.dt.weekday
test['month_start'] = test.Date_of_Journey.dt.is_month_start
test['month_end'] = test.Date_of_Journey.dt.is_month_end
train['day_offset'] = (train['Date_of_Journey'] - datetime(2019, 3, 1))
test['day_offset'] = (test['Date_of_Journey'] - datetime(2019, 3, 1))
train['day_offset'] = train['day_offset'].dt.days
test['day_offset'] = test['day_offset'].dt.days
train['day_offset'] = train['day_offset']+1
test['day_offset'] = test['day_offset']+1
for i in range(train.shape[0]):
if train.ix[i,"Duration"].find('m') < 0:
train.ix[i,"Duration_final"] = int(re.findall('\dh|\d\dh',train.ix[i,"Duration"])[0].strip('h'))*60
else:
train.ix[i,"Duration_final"] = int(re.findall('\dh|\d\dh',train.ix[i,"Duration"])[0].strip('h'))*60 + int(re.findall('\dm|\d\dm',train.ix[i,"Duration"])[0].strip('m'))
for i in range(test.shape[0]):
if test.ix[i,"Duration"].find('m') < 0:
test.ix[i,"Duration_final"] = int(re.findall('\dh|\d\dh',test.ix[i,"Duration"])[0].strip('h'))*60
else:
test.ix[i,"Duration_final"] = int(re.findall('\dh|\d\dh',test.ix[i,"Duration"])[0].strip('h'))*60 + int(re.findall('\dm|\d\dm',test.ix[i,"Duration"])[0].strip('m'))
#we already have duration in minute so we will delete this text column
train.drop(['Duration'], axis=1, inplace= True)
test.drop(['Duration'], axis=1, inplace= True)
for i in range(train.shape[0]):
train.ix[i,"Date_of_departure_ts"] = pd.datetime(train.Date_of_Journey.dt.year[i],
train.Date_of_Journey.dt.month[i],
train.Date_of_Journey.dt.day[i],
train.Dep_Time.dt.hour[i],
train.Dep_Time.dt.minute[i])
for i in range(test.shape[0]):
test.ix[i,"Date_of_departure_ts"] = pd.datetime(test.Date_of_Journey.dt.year[i],
test.Date_of_Journey.dt.month[i],
test.Date_of_Journey.dt.day[i],
test.Dep_Time.dt.hour[i],
test.Dep_Time.dt.minute[i])
train.drop(['Dep_Time'], axis=1, inplace= True)
test.drop(['Dep_Time'], axis=1, inplace= True)
train.drop(['Date_of_Journey'], axis=1, inplace= True)
test.drop(['Date_of_Journey'], axis=1, inplace= True)
# Create arrival time stamp based on departure time and duration final
for i in range(train.shape[0]):
train.ix[i,"Arrival_time_ts"] = train.Date_of_departure_ts[i] + timedelta(minutes = train.Duration_final[i])
for i in range(test.shape[0]):
test.ix[i,"Arrival_time_ts"] = test.Date_of_departure_ts[i] + timedelta(minutes = test.Duration_final[i])
train.head(1)
## creating features based on arrival time
train['day_a'] = train.Arrival_time_ts.dt.day
test['day_a'] = test.Arrival_time_ts.dt.day
train['hour_d'] = train.Date_of_departure_ts.dt.hour
test['hour_d'] = test.Date_of_departure_ts.dt.hour
train['hour_a'] = train.Arrival_time_ts.dt.hour
test['hour_a'] = test.Arrival_time_ts.dt.hour
train['is_arrival_same_day'] = train['day_d'] == train['day_a']
test['is_arrival_same_day'] = test['day_d'] == test['day_a']
train.drop(['Date_of_departure_ts','Arrival_time_ts'], axis=1, inplace= True)
test.drop(['Date_of_departure_ts','Arrival_time_ts'], axis=1, inplace= True)
train.head(1)
###Output
_____no_output_____
###Markdown
Data Cleaning
###Code
train['Total_Stops'] = train['Total_Stops'].map({'non-stop':0, '2 stops':2, '1 stop':1, '3 stops':3, '4 stops':4})
test['Total_Stops'] = test['Total_Stops'].map({'non-stop':0, '2 stops':2, '1 stop':1, '3 stops':3, '4 stops':4})
train.loc[train.Airline =='Jet Airways Business', 'Additional_Info'] = 'Business class'
test.loc[test.Airline =='Jet Airways Business', 'Additional_Info'] = 'Business class'
train.loc[train.Airline =='Jet Airways Business', 'Airline'] = 'Jet Airways'
test.loc[test.Airline =='Jet Airways Business', 'Airline'] = 'Jet Airways'
train.loc[train.Airline =='Multiple carriers Premium economy', 'Additional_Info'] = 'Premium economy'
test.loc[test.Airline =='Multiple carriers Premium economy', 'Additional_Info'] = 'Premium economy'
train.loc[train.Airline =='Vistara Premium economy', 'Additional_Info'] = 'Premium economy'
test.loc[test.Airline =='Vistara Premium economy', 'Additional_Info'] = 'Premium economy'
train.loc[train.Airline =='Multiple carriers Premium economy', 'Airline'] = 'Multiple carriers'
test.loc[test.Airline =='Multiple carriers Premium economy', 'Airline'] = 'Multiple carriers'
train.loc[train.Airline =='Vistara Premium economy', 'Airline'] = 'Vistara'
test.loc[test.Airline =='Vistara Premium economy', 'Airline'] = 'Vistara'
train.loc[train.Destination =='New Delhi', 'Destination'] = 'Delhi'
test.loc[test.Destination =='New Delhi', 'Destination'] = 'Delhi'
train['month_start'] = train['month_start'].map({False:0,True:1})
test['month_start'] = test['month_start'].map({False:0,True:1})
train['month_end'] = train['month_end'].map({False:0,True:1})
test['month_end'] = test['month_end'].map({False:0,True:1})
train['is_arrival_same_day'] = train['is_arrival_same_day'].map({False:0,True:1})
test['is_arrival_same_day'] = test['is_arrival_same_day'].map({False:0,True:1})
###Output
_____no_output_____
###Markdown
More Exploration
###Code
# Plot Histogram
sns.distplot(train['Price'] , fit=norm);
# Get the fitted parameters used by the function
(mu, sigma) = norm.fit(train['Price'])
print( '\n mu = {:.2f} and sigma = {:.2f}\n'.format(mu, sigma))
plt.legend(['Normal dist. ($\mu=$ {:.2f} and $\sigma=$ {:.2f} )'.format(mu, sigma)],
loc='best')
plt.ylabel('Frequency')
plt.title('Price distribution')
fig = plt.figure()
res = stats.probplot(train['Price'], plot=plt)
plt.show()
print("Skewness: %f" % train['Price'].skew())
print("Kurtosis: %f" % train['Price'].kurt())
# Correlation Matrix Heatmap
corrmat = train.corr()
f, ax = plt.subplots(figsize=(12, 9))
sns.heatmap(corrmat, vmax=1, square=True, annot= True);
# Overall Quality vs Sale Price
var = 'Airline'
data = pd.concat([train['Price'], train[var]], axis=1)
f, ax = plt.subplots(figsize=(12, 8))
fig = sns.boxplot(x=var, y="Price", data=data)
fig.axis(ymin=0, ymax=90000);
# Living Area vs Sale Price
sns.jointplot(x=train['Duration_final'], y=train['Price'], kind='reg')
# Living Area vs Sale Price
sns.jointplot(x=train['day_offset'], y=train['Price'], kind='reg')
train['Route_list'] = train.Route.str.split(" → ")
test['Route_list'] = test.Route.str.split(" → ")
###Output
_____no_output_____
###Markdown
adding a feature to check if flight have the stops in metro (BOM,DEL,BLR,MAA,CCU)
###Code
for i in range(train.shape[0]):
if train.Total_Stops[i] == 0:
train.ix[i,"is_stop_BOM"] = 'False'
else:
train.ix[i,"is_stop_BOM"] = str('BOM' in train.Route_list[i][1:(train.Total_Stops[i]+1)])
for i in range(test.shape[0]):
if test.Total_Stops[i] == 0:
test.ix[i,"is_stop_BOM"] = 'False'
else:
test.ix[i,"is_stop_BOM"] = str('BOM' in test.Route_list[i][1:(test.Total_Stops[i]+1)])
for i in range(train.shape[0]):
if train.Total_Stops[i] == 0:
train.ix[i,"is_stop_DEL"] = 'False'
else:
train.ix[i,"is_stop_DEL"] = str('DEL' in train.Route_list[i][1:(train.Total_Stops[i]+1)])
for i in range(test.shape[0]):
if test.Total_Stops[i] == 0:
test.ix[i,"is_stop_DEL"] = 'False'
else:
test.ix[i,"is_stop_DEL"] = str('DEL' in test.Route_list[i][1:(test.Total_Stops[i]+1)])
for i in range(train.shape[0]):
if train.Total_Stops[i] == 0:
train.ix[i,"is_stop_BLR"] = 'False'
else:
train.ix[i,"is_stop_BLR"] = str('BLR' in train.Route_list[i][1:(train.Total_Stops[i]+1)])
for i in range(test.shape[0]):
if test.Total_Stops[i] == 0:
test.ix[i,"is_stop_BLR"] = 'False'
else:
test.ix[i,"is_stop_BLR"] = str('BLR' in test.Route_list[i][1:(test.Total_Stops[i]+1)])
for i in range(train.shape[0]):
if train.Total_Stops[i] == 0:
train.ix[i,"is_stop_MAA"] = 'False'
else:
train.ix[i,"is_stop_MAA"] = str('MAA' in train.Route_list[i][1:(train.Total_Stops[i]+1)])
for i in range(test.shape[0]):
if test.Total_Stops[i] == 0:
test.ix[i,"is_stop_MAA"] = 'False'
else:
test.ix[i,"is_stop_MAA"] = str('MAA' in test.Route_list[i][1:(test.Total_Stops[i]+1)])
for i in range(train.shape[0]):
if train.Total_Stops[i] == 0:
train.ix[i,"is_stop_CCU"] = 'False'
else:
train.ix[i,"is_stop_CCU"] = str('CCU' in train.Route_list[i][1:(train.Total_Stops[i]+1)])
for i in range(test.shape[0]):
if test.Total_Stops[i] == 0:
test.ix[i,"is_stop_CCU"] = 'False'
else:
test.ix[i,"is_stop_CCU"] = str('CCU' in test.Route_list[i][1:(test.Total_Stops[i]+1)])
train.drop(['Route','Route_list'], axis=1, inplace= True)
test.drop(['Route','Route_list'], axis=1, inplace= True)
train['is_stop_CCU'] = train['is_stop_CCU'].map({'False':0,'True':1})
test['is_stop_CCU'] = test['is_stop_CCU'].map({'False':0,'True':1})
train['is_stop_BOM'] = train['is_stop_BOM'].map({'False':0,'True':1})
test['is_stop_BOM'] = test['is_stop_BOM'].map({'False':0,'True':1})
train['is_stop_MAA'] = train['is_stop_MAA'].map({'False':0,'True':1})
test['is_stop_MAA'] = test['is_stop_MAA'].map({'False':0,'True':1})
train['is_stop_DEL'] = train['is_stop_DEL'].map({'False':0,'True':1})
test['is_stop_DEL'] = test['is_stop_DEL'].map({'False':0,'True':1})
train['is_stop_BLR'] = train['is_stop_BLR'].map({'False':0,'True':1})
test['is_stop_BLR'] = test['is_stop_BLR'].map({'False':0,'True':1})
###Output
_____no_output_____
###Markdown
Statistical test to check the significance of additional info
###Code
train['Additional_Info'] = train['Additional_Info'].map({'No info':'No info',
"In-flight meal not included":"In-flight meal not included",
'No Info':'No info',
'1 Short layover':'Layover',
'1 Long layover':'Layover',
'2 Long layover':'Layover',
'Business class':'Business class',
'No check-in baggage included':'No check-in baggage included',
'Change airports':'Change airports',
'Red-eye flight':'No info'})
test['Additional_Info'] = test['Additional_Info'].map({'No info':'No info',
"In-flight meal not included":"In-flight meal not included",
'1 Long layover':'Layover',
'Business class':'Business class',
'No check-in baggage included':'No check-in baggage included',
'Change airports':'Change airports'})
# Additional_Info
anova = ols('Price ~ C(Additional_Info)', data=train).fit()
anova.summary()
train.loc[train.Airline == 'Trujet', 'Airline'] = 'IndiGo'
# train.loc[train.Price > 35000, 'Additional_Info'] = 'Business class'
train.head(2)
test.head(5)
###Output
_____no_output_____
###Markdown
Adding a feature which shows how many minutes extra a flight takes than the Usual non stop flight
###Code
max_dur = train[train.Total_Stops == 0][['Source','Destination','Duration_final']].groupby(['Source','Destination']).max().reset_index()
max_dur
index_train = np.arange(1,train.shape[0]+1,1)
train['index'] = index_train
train.head(5)
index_test = np.arange(1,test.shape[0]+1,1)
test['index'] = index_test
test.head(5)
train = train.merge(max_dur, on = ['Source','Destination'])
test = test.merge(max_dur, on = ['Source','Destination'])
train = train.sort_values(by = 'index')
train.drop('index', axis=1, inplace= True)
train.head(5)
test = test.sort_values(by = 'index')
test.drop('index', axis=1, inplace= True)
test.head(5)
train['duration_diff'] = train['Duration_final_x'] - train['Duration_final_y']
test['duration_diff'] = test['Duration_final_x'] - test['Duration_final_y']
# train.loc[train.duration_diff <0,'duration_diff'] = 0
# test.loc[train.duration_diff <0,'duration_diff'] = 0
train.drop('Duration_final_y', axis=1, inplace= True)
test.drop('Duration_final_y', axis=1, inplace= True)
# train.drop('Additional_Info', axis=1, inplace= True)
# test.drop('Additional_Info', axis=1, inplace= True)
test.head(5)
train.to_csv('train_processed.csv', index = False)
test.to_csv('test_processed.csv', index = False)
train_df = pd.get_dummies(train)
test_df = pd.get_dummies(test)
train_df.shape, test_df.shape
train_df.head(3)
###Output
_____no_output_____
###Markdown
Modelling
###Code
from sklearn.linear_model import ElasticNet, Lasso, BayesianRidge, LassoLarsIC
from sklearn.ensemble import RandomForestRegressor, GradientBoostingRegressor
from sklearn.kernel_ridge import KernelRidge
from sklearn.pipeline import make_pipeline
from sklearn.preprocessing import RobustScaler
from sklearn.base import BaseEstimator, TransformerMixin, RegressorMixin, clone
from sklearn.model_selection import KFold, cross_val_score, train_test_split, StratifiedKFold
from sklearn.metrics import mean_squared_error, r2_score
import xgboost as xgb
import lightgbm as lgb
from catboost import CatBoostRegressor
y_train = np.log(train_df.Price.values)
# y_train = train_df.Price.values
X_train = train_df.drop('Price', axis=1)
###Output
_____no_output_____
###Markdown
LightGBM
###Code
# LightGBM
folds = KFold(n_splits=10, shuffle=False, random_state=2139)
oof_preds = np.zeros(X_train.shape[0])
sub_preds = np.zeros(test_df.shape[0])
valid_score = 0
for n_fold, (trn_idx, val_idx) in enumerate(folds.split(X_train, y_train)):
print("Fold idx:{}".format(n_fold + 1))
trn_x, trn_y = X_train.iloc[trn_idx], y_train[trn_idx]
val_x, val_y = X_train.iloc[val_idx], y_train[val_idx]
train_data = lgb.Dataset(data=trn_x, label=trn_y)
valid_data = lgb.Dataset(data=val_x, label=val_y)
params = {
'objective': 'regression',
"metric" : "rmse",
'nthread': 4,
'num_leaves': 13,
'learning_rate': 0.01,
'max_depth': 7,
'subsample': 0.8,
'bagging_fraction' : 1,
'bagging_freq': 20,
'colsample_bytree': 0.6,
'scale_pos_weight':1,
'num_rounds':50000,
'early_stopping_rounds':1000,
}
# params = {"objective" : "regression",
# "metric" : "rmse",
# 'n_estimators':30000,
# 'max_depth':7,
# 'early_stopping_rounds':500,
# "num_leaves" : 13,
# "learning_rate" : 0.01,
# "bagging_fraction" : 0.9,
# "bagging_seed" : 0,
# "num_threads" : 4,
# "colsample_bytree" : 0.7
# }
lgb_model = lgb.train(params, train_data, valid_sets=[train_data, valid_data], verbose_eval=1000)
oof_preds[val_idx] = lgb_model.predict(val_x, num_iteration=lgb_model.best_iteration)
sub_pred = lgb_model.predict(test_df, num_iteration=lgb_model.best_iteration)
sub_preds += sub_pred/ folds.n_splits
r2_score(y_train,lgb_model.predict(X_train))
final_sub = np.exp(sub_preds)
#Predict from test set
# prediction = model.predict(test, num_iteration = model.best_iteration)
submission1 = pd.DataFrame({
"Price": final_sub,
})
submission1.to_excel('submission_lgb.xlsx',index=False)
submission1.head()
feature_importance = pd.DataFrame({"columns":X_train.columns, "Value":lgb_model.feature_importance()}).sort_values(by = 'Value', ascending = False)
feature_importance.head(15)
submission1.describe()
submission1.head()
###Output
_____no_output_____
###Markdown
Catboost
###Code
# Training and Validation Set
X_tr, X_val, y_tr, y_val = train_test_split(X_train, y_train, test_size=0.1, random_state=23)
# Train Model
print("Train CatBoost Decision Tree")
modelstart= time.time()
cb_model = CatBoostRegressor(iterations=100000,
learning_rate=0.01,
depth=12,
eval_metric='RMSE',
random_seed = 23,
od_type='Iter',
metric_period = 75,
od_wait=100)
X_train.columns
cat_features = []
cb_model.fit(X_tr, y_tr,eval_set=(X_val,y_val),use_best_model=True,verbose=None, verbose_eval=75)
print("Model Evaluation Stage")
print(cb_model.get_params())
print('RMSE:', np.sqrt(mean_squared_error(y_val, cb_model.predict(X_val))))
cb_sub = np.exp(cb_model.predict(test_df))
#Predict from test set
# prediction = model.predict(test, num_iteration = model.best_iteration)
submission2 = pd.DataFrame({
"Price": cb_sub,
})
submission2.to_excel('submission_cb.xlsx',index=False)
submission2.head()
submission2.describe()
###Output
_____no_output_____
|
Indians Diabetes.ipynb
|
###Markdown
diab1=diab[diab['Outcome']==1]columns=diab.columns[:8]plt.subplots(figsize=(18,15))length=len(columns)for i,j in itertools.zip_longest(columns,range(length)): plt.subplot((length/2),3,j+1) plt.subplots_adjust(wspace=0.2,hspace=0.5) diab1[i].hist(bins=20,edgecolor='black') plt.title(i)plt.show()
###Code
diab1=diab[diab['Outcome']==1]
columns=diab.columns[:8]
plt.subplots(figsize=(18,15))
length=len(columns)
for i,j in itertools.zip_longest(columns,range(length)):
plt.subplot((length/2),3,j+1)
plt.subplots_adjust(wspace=0.2,hspace=0.5)
diab1[i].hist(bins=20,edgecolor='black')
plt.title(i)
plt.show()
sns.pairplot(data=diab,hue='Outcome',diag_kind='kde')
plt.show()
from sklearn import svm
from sklearn.neighbors import KNeighborsClassifier
from sklearn.tree import DecisionTreeClassifier
from sklearn.model_selection import train_test_split
from sklearn.model_selection import KFold
from sklearn.linear_model import LogisticRegression
from sklearn import metrics
import warnings
warnings.filterwarnings('ignore')
outcome=diab['Outcome']
data=diab[diab.columns[:8]]
train,test=train_test_split(diab,test_size=0.25,random_state=0,stratify=diab['Outcome'])# stratify the outcome
train_X=train[train.columns[:8]]
test_X=test[test.columns[:8]]
train_Y=train['Outcome']
test_Y=test['Outcome']
train_X.head(2)
types=['rbf','linear']
for i in types:
model=svm.SVC(kernel=i)
model.fit(train_X,train_Y)
prediction=model.predict(test_X)
print('Accuracy for SVM kernel=',i,'is',metrics.accuracy_score(prediction,test_Y))
model = LogisticRegression()
model.fit(train_X,train_Y)
prediction=model.predict(test_X)
print('The accuracy of the Logistic Regression is',metrics.accuracy_score(prediction,test_Y))
model=DecisionTreeClassifier()
model.fit(train_X,train_Y)
prediction=model.predict(test_X)
print('The accuracy of the Decision Tree is',metrics.accuracy_score(prediction,test_Y))
a_index=list(range(1,11))
a=pd.Series()
x=[0,1,2,3,4,5,6,7,8,9,10]
for i in list(range(1,11)):
model=KNeighborsClassifier(n_neighbors=i)
model.fit(train_X,train_Y)
prediction=model.predict(test_X)
a=a.append(pd.Series(metrics.accuracy_score(prediction,test_Y)))
plt.plot(a_index, a)
plt.xticks(x)
plt.show()
print('Accuracies for different values of n are:',a.values)
abc=[]
classifiers=['Linear Svm','Radial Svm','Logistic Regression','KNN','Decision Tree']
models=[svm.SVC(kernel='linear'),svm.SVC(kernel='rbf'),LogisticRegression(),KNeighborsClassifier(n_neighbors=3),DecisionTreeClassifier()]
for i in models:
model = i
model.fit(train_X,train_Y)
prediction=model.predict(test_X)
abc.append(metrics.accuracy_score(prediction,test_Y))
models_dataframe=pd.DataFrame(abc,index=classifiers)
models_dataframe.columns=['Accuracy']
models_dataframe
sns.heatmap(diab[diab.columns[:8]].corr(),annot=True,cmap='RdYlGn')
fig=plt.gcf()
fig.set_size_inches(10,8)
plt.show()
from sklearn.ensemble import RandomForestClassifier
model= RandomForestClassifier(n_estimators=100,random_state=0)
X=diab[diab.columns[:8]]
Y=diab['Outcome']
model.fit(X,Y)
pd.Series(model.feature_importances_,index=X.columns).sort_values(ascending=False)
diab2=diab[['Glucose','BMI','Age','DiabetesPedigreeFunction','Outcome']]
from sklearn.preprocessing import StandardScaler #Standardisation
features=diab2[diab2.columns[:4]]
features_standard=StandardScaler().fit_transform(features)# Gaussian Standardisation
x=pd.DataFrame(features_standard,columns=[['Glucose','BMI','Age','DiabetesPedigreeFunction']])
x['Outcome']=diab2['Outcome']
outcome=x['Outcome']
train1,test1=train_test_split(x,test_size=0.25,random_state=0,stratify=x['Outcome'])
train_X1=train1[train1.columns[:4]]
test_X1=test1[test1.columns[:4]]
train_Y1=train1['Outcome']
test_Y1=test1['Outcome']
abc=[]
classifiers=['Linear Svm','Radial Svm','Logistic Regression','KNN','Decision Tree']
models=[svm.SVC(kernel='linear'),svm.SVC(kernel='rbf'),LogisticRegression(),KNeighborsClassifier(n_neighbors=3),DecisionTreeClassifier()]
for i in models:
model = i
model.fit(train_X1,train_Y1)
prediction=model.predict(test_X1)
abc.append(metrics.accuracy_score(prediction,test_Y1))
new_models_dataframe=pd.DataFrame(abc,index=classifiers)
new_models_dataframe.columns=['New Accuracy']
new_models_dataframe=new_models_dataframe.merge(models_dataframe,left_index=True,right_index=True,how='left')
new_models_dataframe['Increase']=new_models_dataframe['New Accuracy']-new_models_dataframe['Accuracy']
new_models_dataframe
from sklearn.model_selection import KFold #for K-fold cross validation
from sklearn.model_selection import cross_val_score #score evaluation
kfold = KFold(n_splits=10, random_state=22) # k=10, split the data into 10 equal parts
xyz=[]
accuracy=[]
classifiers=['Linear Svm','Radial Svm','Logistic Regression','KNN','Decision Tree']
models=[svm.SVC(kernel='linear'),svm.SVC(kernel='rbf'),LogisticRegression(),KNeighborsClassifier(n_neighbors=3),DecisionTreeClassifier()]
for i in models:
model = i
cv_result = cross_val_score(model,x[x.columns[:4]],x['Outcome'], cv = kfold,scoring = "accuracy")
cv_result=cv_result
xyz.append(cv_result.mean())
accuracy.append(cv_result)
new_models_dataframe2=pd.DataFrame(abc,index=classifiers)
new_models_dataframe2.columns=['CV Mean']
new_models_dataframe2
box=pd.DataFrame(accuracy,index=[classifiers])
sns.boxplot(box.T)
plt.show()
from sklearn.ensemble import VotingClassifier #for Voting Classifier
ensemble_lin_rbf=VotingClassifier(estimators=[('Linear_svm', linear_svc), ('Radial_svm', radial_svm)],
voting='soft', weights=[2,1]).fit(train_X1,train_Y1)
print('The accuracy for Linear and Radial SVM is:',ensemble_lin_rbf.score(test_X1,test_Y1))
ensemble_lin_lr=VotingClassifier(estimators=[('Linear_svm', linear_svc), ('Logistic Regression', lr)],
voting='soft', weights=[2,1]).fit(train_X1,train_Y1)
print('The accuracy for Linear SVM and Logistic Regression is:',ensemble_lin_lr.score(test_X1,test_Y1))
###Output
_____no_output_____
|
homework/day_2/monte-carlo-advanced.ipynb
|
###Markdown
Monte Carlo Simulation - AdvancedIn this homework, we will work with the Lennard Jones equation added with cutoff distance and periodic boundary conditions.$$ U(r) = 4 \epsilon \left[\left(\frac{\sigma}{r}\right)^{12} -\left(\frac{\sigma}{r}\right)^{6} \right] $$ Reduced units:$$ U^*\left(r^*_{ij} \right) = 4 \left[\left(\frac{1}{r^*_{ij}}\right)^{12} -\left(\frac{1}{r^*_{ij}}\right)^{6} \right] $$
###Code
import math, os
import matplotlib.pyplot as plt
%matplotlib notebook
def calculate_LJ(r_ij):
"""
The LJ interaction energy between two particles.
Computes the pairwise Lennard Jones interaction energy based on the separation distance in reduced unites.
Parameters
----------
r_ij : float
The distance between the particles in reduced units.
Returns
-------
pairwise_energy : float
The pairwise Lennard Jones interaction energy in reduced units.
"""
r6_term = math.pow(1/r_ij,6)
r12_term = math.pow(r6_term,2)
pairwise_energy = 4 * (r12_term - r6_term)
ax.plot(r_ij,pairwise_energy,'ob')
return pairwise_energy
def calculate_distance(coord1,coord2,box_length=None):
"""
Calculate the distance between two 3D coordinates.
Parameters
----------
coord1, coord2 : list
The atomic coordinates [x, y, z]
box_length : float, optional
The box length. This function assumes box is a cube.
Returns
-------
distance : float
The distance between the two atoms.
"""
distance = 0
vector = [0,0,0]
for i in range(3):
vector[i] = coord1[i] -coord2[i]
if box_length is None:
pass
else:
if vector[i] > box_length/2:
vector[i] -= box_length
elif vector[i] < -box_length/2:
vector[i] += box_length
dim_dist = vector[i] ** 2
distance += dim_dist
distance = math.sqrt(distance)
return distance
def calculate_total_energy(coordinates, cutoff=3, box_length=None):
"""
Calculate the total Lennard Jones energy of a system of particles.
Parameters
----------
coordinates : list
Nested list containing particle coordinates.
cutoff : float
A criteria distance for intermolecular interaction truncation
box_length : float, optional
The box length. This function assumes box is a cube.
Returns
-------
total_energy : float
The total pairwise Lennard Jones energy of the system of particles.
"""
total_energy = 0
num_atoms = len(coordinates)
for i in range(num_atoms):
for j in range(i+1,num_atoms):
# print(F'Comparing atom number {i} with atom number {j}')
dist_ij = calculate_distance(coordinates[i], coordinates[j], box_length)
if dist_ij < cutoff:
interaction_energy = calculate_LJ(dist_ij)
total_energy += interaction_energy
return total_energy
def read_xyz(filepath):
"""
Reads coordinates from an xyz file.
Parameters
----------
filepath : str
The path to the xyz file to be processed.
Returns
-------
atomic_coordinates : list
A two dimensional list containing atomic coordinates
"""
with open(filepath) as f:
box_length = float(f.readline().split()[0])
num_atoms = float(f.readline())
coordinates = f.readlines()
atomic_coordinates = []
for atom in coordinates:
split_atoms = atom.split()
float_coords = []
# We split this way to get rid of the atom label.
for coord in split_atoms[1:]:
float_coords.append(float(coord))
atomic_coordinates.append(float_coords)
return atomic_coordinates, box_length
fig = plt.figure()
ax = fig.add_subplot(111)
fig.show()
plt.ylim(-1.1,0.1)
for i in range(1, 51):
r = i * 0.1
calculate_LJ(r)
###Output
_____no_output_____
###Markdown
From this graph, it is obvious that when $r^*_{ij}$ > 3, the pairwise energy is almost 0 and the energy curve reaches a plateau. The general set of cutoff distance at 3$\sigma$ is reasonable.
###Code
assert calculate_LJ(1) == 0
assert calculate_LJ(math.pow(2,(1/6))) == -1
file_path = os.path.join('lj_sample_configurations','lj_sample_config_periodic1.txt')
coordinates, box_length = read_xyz(file_path)
calculate_total_energy(coordinates)
calculate_total_energy(coordinates,box_length=10)
assert abs(calculate_total_energy(coordinates,box_length=10) - (-4351.5)) < 0.1
###Output
_____no_output_____
|
BanglaCharRecog.ipynb
|
###Markdown
Importing the dataset using pandas where ***data*** holds the features or images of the handwritten characters and ***label*** holds the associated bangla characters in unicode corresponding to the numeric representation.*Replace the csv locations relative to your workspace*
###Code
#data = pd.read_csv('/content/drive/My Drive/Bangla letters/data.csv')
data = pd.read_csv('/Volumes/HDD/Python/python3/Workspace/Bangla NN/Bangla letters/data.csv')
label = pd.read_csv('/Volumes/HDD/Python/python3/Workspace/Bangla NN/Bangla letters/metaDataCSV.csv')
X = (data.drop(["label"], axis=1))/255.0
y = data['label']
print(label['Char Name'][label['Label'] == y[0]])
from __future__ import print_function
import pandas as pd
import cv2
import numpy as np
import keras
from keras.models import load_model
from keras.models import Sequential
from keras.layers import Dense, Dropout, Flatten
from keras.layers import Conv2D, MaxPooling2D
from keras import backend as K
from sklearn.model_selection import train_test_split
X_train, X_test, y_train, y_test = train_test_split(X, y,
stratify=y,
test_size=0.25)
img_rows = img_cols = 28
x_train = X_train.values.reshape(X_train.shape[0], img_rows, img_cols, 1)
x_test = X_test.values.reshape(X_test.shape[0], img_rows, img_cols, 1)
input_shape = (img_rows, img_cols, 1)
num_classes = len(y_test.unique())
num_classes = 60
len(y_test.unique()),len(y_train.unique())
y_train = keras.utils.to_categorical(y_train, num_classes)
y_test = keras.utils.to_categorical(y_test, num_classes)
###Output
Using TensorFlow backend.
###Markdown
Load the character recorgination model htat we trained earlier.
###Code
model = load_model("/Volumes/HDD/Python/python3/Workspace/Bangla NN/Bangla letters/banglachar.h5")
model.summary()
score = model.evaluate(x_test, y_test, verbose=0)
print('Test loss:', score[0])
print('Test accuracy:', score[1])
def predict(image):
input = cv2.resize(image, (28 , 28)).reshape((28 , 28,1)).astype('float32') / 255
prediction = model.predict_classes(np.array([input]))
character = label['Char Name'][label['Label'] == prediction[0]].values[0]
return character
###Output
_____no_output_____
###Markdown
Creates an openCV window that allows user to draw input.
###Code
canvas = np.ones((600,600), dtype="uint8") * 255
canvas[100:500,100:500] = 0
start_point = None
end_point = None
is_drawing = False
def draw_line(img,start_at,end_at):
cv2.line(img,start_at,end_at,255,15)
def on_mouse_events(event,x,y,flags,params):
global start_point
global end_point
global canvas
global is_drawing
if event == cv2.EVENT_LBUTTONDOWN:
if is_drawing:
start_point = (x,y)
elif event == cv2.EVENT_MOUSEMOVE:
if is_drawing:
end_point = (x,y)
draw_line(canvas,start_point,end_point)
start_point = end_point
elif event == cv2.EVENT_LBUTTONUP:
is_drawing = False
cv2.namedWindow("Test Canvas")
cv2.setMouseCallback("Test Canvas", on_mouse_events)
while(True):
cv2.imshow("Test Canvas", canvas)
key = cv2.waitKey(1) & 0xFF
if key == ord('q'):
break
elif key == ord('s'):
is_drawing = True
elif key == ord('c'):
canvas[100:500,100:500] = 0
elif key == ord('p'):
image = canvas[100:500,100:500]
result = predict(image)
print("PREDICTION : ",result)
cv2.destroyAllWindows()
input = cv2.resize(image,(28,28)).reshape((28,28,1)).astype(np.float32)
cv2.destroyAllWindows()
print(label['Char Name'][label['Label'] == 48].values[0])
data[data['label']== 45].shape
###Output
_____no_output_____
|
cartpole/linear_regress.ipynb
|
###Markdown
This notebook uses OpenAI baselines to train an (Inverted pendulum/Cartpole/Acrobot) to balance. We expect the policy around the equilibruim to be somewhat linear. This notebook investigates exactly how linear and trys to "extract" the linearity from the network.
###Code
from baselines.common.cmd_util import make_mujoco_env, mujoco_arg_parser
from baselines.common import tf_util as U
import tensorflow as tf
from baselines import logger
import os
import sys
from baselines.ppo1 import mlp_policy, pposgd_simple
import gym
import gym_ucsb_robolab
import policies.mlp_relu_policy as mlp_relu_policy
import numpy as np
from mpl_toolkits import mplot3d
%matplotlib inline
import matplotlib.pyplot as plt
import itertools
#Needed for saving
import errno, datetime, time, inspect
def train(env_id, num_timesteps, seed=0):
U.make_session(num_cpu=16).__enter__()
def policy_fn(name, ob_space, ac_space):
#return mlp_policy.MlpPolicy(name=name, ob_space=ob_space, ac_space=ac_space, hid_size=64, num_hid_layers=64)
return mlp_relu_policy.ReluMlpPolicy(name=name, ob_space=ob_space, ac_space=ac_space, hid_size=64, num_hid_layers=4)
env = gym.make(env_id)
pi = pposgd_simple.learn(env, policy_fn,
max_timesteps=num_timesteps,
timesteps_per_actorbatch=2048,
clip_param=0.2, entcoeff=0.0,
optim_epochs=10, optim_stepsize=3e-4, optim_batchsize=64,
gamma=0.99, lam=0.95, schedule='linear',
)
env.close()
return pi
def save_results(filename, description = None):
"""
description: saves the results of a run of the second cell (the one that calls train) in this notebook
"""
save_dir = "data/" + filename + "/"
os.makedirs(save_dir)
if description is None:
description = input("please enter a description of the run")
datetime_str = str(datetime.datetime.today())
datetime_str = datetime_str.replace(" ", "_")
runtime_str = str(datetime.timedelta(seconds = runtime))
readme = open(save_dir + "README.txt", "w+")
readme.write("datetime: " + datetime_str + "\n\n")
readme.write("enviroment: " + env_name + "\n\n")
readme.write("description: " + description + "\n\n")
readme.write("time_elapsed: " + runtime_str + "\n\n")
readme.write("num_timesteps: " + str(num_timesteps) + "\n\n")
readme.write("seed: " + str(seed) + "\n\n")
readme.close()
# TODO add code snippets that correspond to the run
# TODO somehow store the tensorboard logs here after the fact
saver = tf.train.Saver()
saver.save(tf.get_default_session(), save_dir + filename)
os.rename("./tmp_logs/", save_dir + "tensorboard")
env_name = "Pendulum-v0"
#env_name = "Acrobot-v1"
#env_name = "su_pendulum-v0"
#env_name = "InvertedPendulum-v2"
#env_name = 'InvertedPendulumPyBulletEnv-v0'
#env_name = "su_cartpole_et-v0"
#env_name = "InvertedDoublePendulum-v2"
# comment one of these lines to switch between loading weights or training them from scratch
#load_pretrained_network = True
load_pretrained_network = False
if load_pretrained_network: #load the weights
save_name = 'invertedpendulum_3layer'
pi = train(env_name, num_timesteps=1, seed=0)
# TODO eventually need to switch to .load_variables() instead of U.load_state() but this didn't work by default for me
U.load_state(os.getcwd() + '/data/'+ save_name + '/' + save_name)
else: #run the RL algorithm
num_timesteps = 2e5
seed = 0
print("training")
start_time = time.time()
logger.configure(dir = "./tmp_logs", format_strs=["tensorboard"] )
with tf.device("/cpu:0"):
pi= train(env_name, num_timesteps=num_timesteps, seed=seed)
runtime = time.time() - start_time
# Plays out a trained policy
env = make_mujoco_env(env_name,seed=0)
ob = env.reset()
ob_list = []
act_list = []
while True:
action = pi.act(stochastic=False, ob=ob)[0]
ob, _, done, _ = env.step(action)
ob_list.append(ob)
act_list.append(action)
#if reward == 1:
# print("balanced")
#env.render()
if done:
break
#ob = env.reset()
plt.figure()
plt.plot(ob_list)
plt.figure()
plt.plot(act_list)
#U.save_state("./saved/5mil_flat")
all_weights = pi.get_variables()
# I'll fix this later..
kernel0 = all_weights[13].value().eval()
bias0 = all_weights[14].value().eval()
kernel1 = all_weights[15].value().eval()
bias1 = all_weights[16].value().eval()
kernel2 = all_weights[17].value().eval()
bias2 = all_weights[18].value().eval()
name_dict = {"kernel0": kernel0, "kernel1":kernel1, "kernel2":kernel2, "bias0":bias0, "bias1":bias1, "bias2":bias2}
scipy.io.savemat("savemat_test", name_dict)
#input_iter = itertools.combinations_with_replacement(range(-10,11),4)
#input_data = np.array([np.array(x) for x in input_iter],dtype='float32')
#output_data = np.array([pi.act(0, x)[0] for x in input_data],dtype='float32')
input_iter = itertools.combinations_with_replacement(range(-10,11),2)
input_data = np.array([np.concatenate((np.zeros(2), np.array(x))) for x in input_iter],dtype='float32')
output_data = np.array([pi.act(0, x)[0] for x in input_data],dtype='float32')
from sklearn import datasets, linear_model
from sklearn.metrics import mean_squared_error, r2_score
regr = linear_model.LinearRegression()
regr.fit(input_data,output_data.T.flatten())
#regr.fit(input_data,output_data)
#regr.fit(index, output_data.T.flatten())
lin_predict = regr.predict(input_data)
#lin_predict = regr.predict(index)
print("coefs are", regr.coef_)
print("mean sqared error:", mean_squared_error(lin_predict, output_data))
plt.plot(output_data)
plt.figure()
plt.plot(lin_predict)
input_data = np.array([[x, 0, 0, 0] for x in range(-10,11)])
output_data = np.array([pi.act(0, x)[0] for x in input_data])
plt.plot(input_data[:,0], output_data)
plt.figure()
input_data = np.array([[0, x, 0, 0] for x in range(-10,11)])
output_data = np.array([pi.act(0, x)[0] for x in input_data])
plt.plot(input_data[:,1], output_data)
plt.figure()
input_data = np.array([[0, 0, x, 0] for x in range(-10,11)])
output_data = np.array([pi.act(0, x)[0] for x in input_data])
plt.plot(input_data[:,2], output_data)
plt.figure()
input_data = np.array([[0, 0, 0, x] for x in range(-10,11)])
output_data = np.array([pi.act(0, x)[0] for x in input_data])
plt.plot(input_data[:,3], output_data)
plt.figure()
input_data = np.array([[x, x, x, x] for x in range(-10,11)])
output_data = np.array([pi.act(0, x)[0] for x in input_data])
plt.plot(input_data[:,0], output_data)
x = input_data[:,2]
y = input_data[:,3]
z= output_data.flatten()
z2 = lin_predict.flatten()
ax = plt.axes(projection='3d')
ax.plot_trisurf(x, y, z, cmap='viridis', edgecolor='none');
ax2 = plt.axes(projection='3d')
ax2.plot_trisurf(x, y, z2, cmap='viridis', edgecolor='none');
###Output
_____no_output_____
|
Analysis/Rylan van der Meulen/EDA.ipynb
|
###Markdown
**My EDA will be looking at the differences between males and females.**
###Code
import pandas as pd
import seaborn as sns
import os
cwd=os.getcwd()
import sys
os.chdir("../scripts")
import project_functions
os.chdir(cwd)
df = project_functions.load_and_process(cwd)
df
df.describe(include = 'all')
df_male = df[df['sex']=='male']
df_male
#create a dataframe with just males
def average_bmi(df):
average = df["bmi"].mean()
print("Average bmi: ",average)
return
average_bmi(df_male)
df_female = df[df['sex']=='female']
df_female
average_bmi(df_female)
###Output
Average bmi: 30.37688821752266
###Markdown
**The average bmi of males and females quite similar, although males do have a slightly higher average.**
###Code
def average_charges(df):
average = df["charges"].mean()
print("Average charges: ",average)
return
average_charges(df_male)
average_charges(df_female)
###Output
Average charges: 12569.578843835347
###Markdown
**The average charges between males and females are also similar, but once again the male average is higher.**
###Code
df_male.describe(include = 'all')
df_female.describe(include = 'all')
###Output
_____no_output_____
###Markdown
**It is interesting that both sexes in this dataset are primarily from the southeast** **Additionally, it is surpising that 24% of males are smokers while only 17% of females smoke.**
###Code
def plotAvgBmi(df):
g=sns.lmplot(x='age', y='bmi',data=df,
scatter_kws={'s': 100, 'linewidth': 1.0, 'edgecolor': 'w'})
return g
plotAvgBmi(df_male)
plotAvgBmi(df_female)
###Output
_____no_output_____
###Markdown
**In these scatterplots you can see the incredible variability among people, as well as the similarity in the pattern or lack of pattern in males and females. Although, there seems to be slightly more variability in females than males**
###Code
def plotAvgCharges(df):
g=sns.lmplot(x='age', y='charges',data=df,
scatter_kws={'s': 100, 'linewidth': 1.0, 'edgecolor': 'w'})
return g
plotAvgCharges(df_male)
plotAvgCharges(df_female)
###Output
_____no_output_____
###Markdown
**Once again we see that there isnt a whole lot of difference in the pattern of charges between males and females. However, similar to bmi, the datapoints in the male plot seem to have tighter groupings.**
###Code
df.head(20)
df.tail(20)
###Output
_____no_output_____
|
Binomial_CI_Logit_Transformed.ipynb
|
###Markdown
Given the issues we saw with the confidence interval for binomial using Wald and Fisher information, let's look at the [logit or log-odds transformed version](https://onlinecourses.science.psu.edu/stat504/node/38 "Stat 504 Notes"). We begin the same way.
###Code
trials<-1000
n<-100
p<-0.1 # make this smaller to see the CI % get farther off
#rbinom(100,100,.2) draws from Bin(n,p) ten times
x<-rbinom(trials,n,p)
###Output
_____no_output_____
###Markdown
And the vector of maximum likelihood estimators is constructed in the same way.
###Code
phat = x/n
###Output
_____no_output_____
###Markdown
The difference is that now we are going to look at the variance of the maximum likelihood estimator as a random variable in a different coordinate system (a nonlinear change of coordinates). This eliminates the hard bondaries at 0 and 1. The transformation we will use is called the logit or log-odds transform.
###Code
ps = seq(from=0,to=1,by=.01)
logit_of_p = log(ps/(1-ps))
plot(ps,logit_of_p, type='l')
title("graph of logit")
###Output
_____no_output_____
###Markdown
Applying this transformation, lets define $$\hat{\phi} = \log \frac{\hat{p}}{1-\hat{p}}$$
###Code
phihat = log (phat/(1-phat)) #vector of estimates
phi = log (p/(1-p)) # true value
phi
###Output
_____no_output_____
###Markdown
And now proceed as before in confidence interval construction, obtaining a vector of Fisher informations. The wrinkle is that we need to adjust our formula for the Fisher information using a little calculus. We have $$I(\hat{\phi})=\dfrac{I(\hat{p})}{[\phi'(\hat{p})]^2}, \;\;\text{and}$$$$\phi'(\hat{p}) = \frac{1}{\hat{p}(1-\hat{p})}.$$So the Fisher information is computed from $\hat{\phi}$ as follows.
###Code
phi_prime = 1/(phat*(1-phat))
FI_of_p = n/(phat*(1-phat))
FI = FI_of_p / (phi_prime^2)
varhat = 1/FI # the vector of estimated variances for the transformed mle, in phi-coordinates
#varhat
###Output
_____no_output_____
###Markdown
Now we compute the vectors of lower and upper bounds of our confidence intervals, this time *in the* $\phi$ *coordinate:*
###Code
lower = phihat - 1.96*sqrt(varhat)
upper = phihat + 1.96*sqrt(varhat)
###Output
_____no_output_____
###Markdown
If this is a good CI, the next number should converge to .95 as trials $\rightarrow \infty$
###Code
outcome = lower< phi & phi<upper
outcome[is.na(outcome)]=FALSE # NAs happen when phat = 0, so the CI is degenerate and doesn't contain the true value
sum(outcome)/trials
meta = 10000
metatrials = rep(0,meta);
for(i in 0:meta)
{
x<-rbinom(trials,n,p); # vector of outcomes
phat = x/n; # vector of mles
phihat = log (phat/(1-phat)) # vector of estimates
phi = log (p/(1-p)) # true value in phi coordinates (scalar)
#Fisher information calculation
phi_prime = 1/(phat*(1-phat))
FI_of_p = n/(phat*(1-phat))
FI = FI_of_p / (phi_prime^2)
varhat = 1/FI;
lower = phihat - 1.96*sqrt(varhat);
upper = phihat + 1.96*sqrt(varhat);
outcome = lower< phi & phi<upper
outcome[is.na(outcome)]=FALSE # NAs happen when phat = 0, so the CI is degenerate and doesn't contain the true value
inthere = sum(outcome)/trials
metatrials[i]=inthere;
}
#metatrials
plot(density(metatrials))
###Output
_____no_output_____
|
Subsets and Splits
No community queries yet
The top public SQL queries from the community will appear here once available.