text
stringlengths 5
261k
| id
stringlengths 16
106
| metadata
dict | __index_level_0__
int64 0
266
|
|---|---|---|---|
# Copyright 2015 The TensorFlow Authors. All Rights Reserved.
#
# Licensed under the Apache License, Version 2.0 (the "License");
# you may not use this file except in compliance with the License.
# You may obtain a copy of the License at
#
# http://www.apache.org/licenses/LICENSE-2.0
#
# Unless required by applicable law or agreed to in writing, software
# distributed under the License is distributed on an "AS IS" BASIS,
# WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.
# See the License for the specific language governing permissions and
# limitations under the License.
# ==============================================================================
"""Keras backend API."""
import collections
import itertools
import json
import os
import random
import sys
import threading
import warnings
import weakref
import numpy as np
import tensorflow.compat.v2 as tf
from tf_keras import backend_config
from tf_keras.distribute import distribute_coordinator_utils as dc
from tf_keras.dtensor import dtensor_api as dtensor
from tf_keras.engine import keras_tensor
from tf_keras.utils import control_flow_util
from tf_keras.utils import object_identity
from tf_keras.utils import tf_contextlib
from tf_keras.utils import tf_inspect
from tf_keras.utils import tf_utils
# isort: off
from tensorflow.core.protobuf import config_pb2
from tensorflow.python.eager import context
from tensorflow.python.eager.context import get_config
from tensorflow.python.platform import tf_logging as logging
from tensorflow.python.util.tf_export import keras_export
from tensorflow.tools.docs import doc_controls
py_all = all
py_sum = sum
py_any = any
# INTERNAL UTILS
# The internal graph maintained by TF-Keras and used by the symbolic TF-Keras
# APIs while executing eagerly (such as the functional API for model-building).
# This is thread-local to allow building separate models in different threads
# concurrently, but comes at the cost of not being able to build one model
# across threads.
_GRAPH = threading.local()
# A graph which is used for constructing functions in eager mode.
_CURRENT_SCRATCH_GRAPH = threading.local()
# This is a thread local object that will hold the default internal TF session
# used by TF-Keras. It can be set manually via `set_session(sess)`.
class SessionLocal(threading.local):
def __init__(self):
super().__init__()
self.session = None
_SESSION = SessionLocal()
# A global dictionary mapping graph objects to an index of counters used
# for various layer/optimizer names in each graph.
# Allows to give unique autogenerated names to layers, in a graph-specific way.
PER_GRAPH_OBJECT_NAME_UIDS = weakref.WeakKeyDictionary()
# A global set tracking what object names have been seen so far.
# Optionally used as an avoid-list when generating names
OBSERVED_NAMES = set()
# _DUMMY_EAGER_GRAPH.key is used as a key in _GRAPH_LEARNING_PHASES.
# We keep a separate reference to it to make sure it does not get removed from
# _GRAPH_LEARNING_PHASES.
# _DummyEagerGraph inherits from threading.local to make its `key` attribute
# thread local. This is needed to make set_learning_phase affect only the
# current thread during eager execution (see b/123096885 for more details).
class _DummyEagerGraph(threading.local):
"""_DummyEagerGraph provides a thread local `key` attribute.
We can't use threading.local directly, i.e. without subclassing, because
gevent monkey patches threading.local and its version does not support
weak references.
"""
class _WeakReferencableClass:
"""This dummy class is needed for two reasons.
- We need something that supports weak references. Basic types like
string and ints don't.
- We need something whose hash and equality are based on object identity
to make sure they are treated as different keys to
_GRAPH_LEARNING_PHASES.
An empty Python class satisfies both of these requirements.
"""
pass
def __init__(self):
# Constructors for classes subclassing threading.local run once
# per thread accessing something in the class. Thus, each thread will
# get a different key.
super().__init__()
self.key = _DummyEagerGraph._WeakReferencableClass()
self.learning_phase_is_set = False
_DUMMY_EAGER_GRAPH = _DummyEagerGraph()
# This boolean flag can be set to True to leave variable initialization
# up to the user.
# Change its value via `manual_variable_initialization(value)`.
_MANUAL_VAR_INIT = False
# This list holds the available devices.
# It is populated when `_get_available_gpus()` is called for the first time.
# We assume our devices don't change henceforth.
_LOCAL_DEVICES = None
# The below functions are kept accessible from backend for compatibility.
epsilon = backend_config.epsilon
floatx = backend_config.floatx
image_data_format = backend_config.image_data_format
set_epsilon = backend_config.set_epsilon
set_floatx = backend_config.set_floatx
set_image_data_format = backend_config.set_image_data_format
@keras_export("keras.backend.backend")
@doc_controls.do_not_generate_docs
def backend():
"""Publicly accessible method for determining the current backend.
Only exists for API compatibility with multi-backend TF-Keras.
Returns:
The string "tensorflow".
"""
return "tensorflow"
@keras_export("keras.backend.cast_to_floatx")
@tf.__internal__.dispatch.add_dispatch_support
@doc_controls.do_not_generate_docs
def cast_to_floatx(x):
"""Cast a Numpy array to the default TF-Keras float type.
Args:
x: Numpy array or TensorFlow tensor.
Returns:
The same array (Numpy array if `x` was a Numpy array, or TensorFlow
tensor if `x` was a tensor), cast to its new type.
Example:
>>> tf.keras.backend.floatx()
'float32'
>>> arr = np.array([1.0, 2.0], dtype='float64')
>>> arr.dtype
dtype('float64')
>>> new_arr = cast_to_floatx(arr)
>>> new_arr
array([1., 2.], dtype=float32)
>>> new_arr.dtype
dtype('float32')
"""
if isinstance(x, (tf.Tensor, tf.Variable, tf.SparseTensor)):
return tf.cast(x, dtype=floatx())
return np.asarray(x, dtype=floatx())
@keras_export("keras.backend.get_uid")
def get_uid(prefix=""):
"""Associates a string prefix with an integer counter in a TensorFlow graph.
Args:
prefix: String prefix to index.
Returns:
Unique integer ID.
Example:
>>> get_uid('dense')
1
>>> get_uid('dense')
2
"""
graph = get_graph()
if graph not in PER_GRAPH_OBJECT_NAME_UIDS:
PER_GRAPH_OBJECT_NAME_UIDS[graph] = collections.defaultdict(int)
layer_name_uids = PER_GRAPH_OBJECT_NAME_UIDS[graph]
layer_name_uids[prefix] += 1
return layer_name_uids[prefix]
@keras_export("keras.backend.reset_uids")
def reset_uids():
"""Resets graph identifiers."""
PER_GRAPH_OBJECT_NAME_UIDS.clear()
OBSERVED_NAMES.clear()
@keras_export("keras.backend.clear_session")
def clear_session():
"""Resets all state generated by TF-Keras.
TF-Keras manages a global state, which it uses to implement the Functional
model-building API and to uniquify autogenerated layer names.
If you are creating many models in a loop, this global state will consume
an increasing amount of memory over time, and you may want to clear it.
Calling `clear_session()` releases the global state: this helps avoid
clutter from old models and layers, especially when memory is limited.
Example 1: calling `clear_session()` when creating models in a loop
```python
for _ in range(100):
# Without `clear_session()`, each iteration of this loop will
# slightly increase the size of the global state managed by Keras
model = tf.keras.Sequential([
tf.keras.layers.Dense(10) for _ in range(10)])
for _ in range(100):
# With `clear_session()` called at the beginning,
# TF-Keras starts with a blank state at each iteration
# and memory consumption is constant over time.
tf.keras.backend.clear_session()
model = tf.keras.Sequential([
tf.keras.layers.Dense(10) for _ in range(10)])
```
Example 2: resetting the layer name generation counter
>>> import tensorflow as tf
>>> layers = [tf.keras.layers.Dense(10) for _ in range(10)]
>>> new_layer = tf.keras.layers.Dense(10)
>>> print(new_layer.name)
dense_10
>>> tf.keras.backend.set_learning_phase(1)
>>> print(tf.keras.backend.learning_phase())
1
>>> tf.keras.backend.clear_session()
>>> new_layer = tf.keras.layers.Dense(10)
>>> print(new_layer.name)
dense
"""
global _SESSION
global _GRAPH_LEARNING_PHASES
global _GRAPH_VARIABLES
global _GRAPH_TF_OPTIMIZERS
global _GRAPH
_GRAPH.graph = None
tf.compat.v1.reset_default_graph()
reset_uids()
if _SESSION.session is not None:
_SESSION.session.close()
_SESSION.session = None
graph = get_graph()
with graph.as_default():
_DUMMY_EAGER_GRAPH.learning_phase_is_set = False
_GRAPH_LEARNING_PHASES = {}
# Create the learning phase placeholder in graph using the default
# factory
phase = _default_learning_phase()
_internal_set_learning_phase(graph, phase)
_GRAPH_VARIABLES.pop(graph, None)
_GRAPH_TF_OPTIMIZERS.pop(graph, None)
if tf.executing_eagerly():
# Clear pending nodes in eager executors, kernel caches and
# step_containers.
context.context().clear_kernel_cache()
# Inject the clear_session function to keras_deps to remove the dependency
# from TFLite to TF-Keras.
tf.__internal__.register_clear_session_function(clear_session)
@keras_export("keras.backend.manual_variable_initialization")
@doc_controls.do_not_generate_docs
def manual_variable_initialization(value):
"""Sets the manual variable initialization flag.
This boolean flag determines whether
variables should be initialized
as they are instantiated (default), or if
the user should handle the initialization
(e.g. via `tf.compat.v1.initialize_all_variables()`).
Args:
value: Python boolean.
"""
global _MANUAL_VAR_INIT
_MANUAL_VAR_INIT = value
@keras_export("keras.backend.learning_phase")
@doc_controls.do_not_generate_docs
def learning_phase():
"""Returns the learning phase flag.
The learning phase flag is a bool tensor (0 = test, 1 = train)
to be passed as input to any TF-Keras function
that uses a different behavior at train time and test time.
Returns:
Learning phase (scalar integer tensor or Python integer).
"""
graph = tf.compat.v1.get_default_graph()
if graph is getattr(_GRAPH, "graph", None):
# Don't enter an init_scope for the learning phase if eager execution
# is enabled but we're inside the TF-Keras workspace graph.
learning_phase = symbolic_learning_phase()
else:
with tf.init_scope():
# We always check & set the learning phase inside the init_scope,
# otherwise the wrong default_graph will be used to look up the
# learning phase inside of functions & defuns.
#
# This is because functions & defuns (both in graph & in eager mode)
# will always execute non-eagerly using a function-specific default
# subgraph.
if context.executing_eagerly():
if _DUMMY_EAGER_GRAPH.key not in _GRAPH_LEARNING_PHASES:
return _default_learning_phase()
else:
return _internal_get_learning_phase(_DUMMY_EAGER_GRAPH.key)
else:
learning_phase = symbolic_learning_phase()
_mark_func_graph_as_unsaveable(graph, learning_phase)
return learning_phase
def global_learning_phase_is_set():
return _DUMMY_EAGER_GRAPH.learning_phase_is_set
def _mark_func_graph_as_unsaveable(graph, learning_phase):
"""Mark graph as unsaveable due to use of symbolic keras learning phase.
Functions that capture the symbolic learning phase cannot be exported to
SavedModel. Mark the funcgraph as unsaveable, so that an error will be
raised if it is exported.
Args:
graph: Graph or FuncGraph object.
learning_phase: Learning phase placeholder or int defined in the graph.
"""
if graph.building_function and is_placeholder(learning_phase):
graph.mark_as_unsaveable(
"The keras learning phase placeholder was used inside a function. "
"Exporting placeholders is not supported when saving out a "
"SavedModel. Please call `tf.keras.backend.set_learning_phase(0)` "
"in the function to set the learning phase to a constant value."
)
def symbolic_learning_phase():
graph = get_graph()
with graph.as_default():
if graph not in _GRAPH_LEARNING_PHASES:
phase = _default_learning_phase()
_internal_set_learning_phase(graph, phase)
return _internal_get_learning_phase(graph)
def _internal_set_learning_phase(graph, value):
global _GRAPH_LEARNING_PHASES
if isinstance(value, tf.Tensor):
# The 'value' here is a tf.Tensor with attribute 'graph'.
# There is a circular reference between key 'graph' and attribute
# 'graph'. So we need use a weakref.ref to refer to the 'value' tensor
# here. Otherwise, it would lead to memory leak.
value_ref = weakref.ref(value)
_GRAPH_LEARNING_PHASES[graph] = value_ref
else:
_GRAPH_LEARNING_PHASES[graph] = value
def _internal_get_learning_phase(graph):
phase = _GRAPH_LEARNING_PHASES.get(graph, None)
if isinstance(phase, weakref.ref):
return phase()
else:
return phase
def _default_learning_phase():
if context.executing_eagerly():
return 0
else:
with name_scope(""):
return tf.compat.v1.placeholder_with_default(
False, shape=(), name="keras_learning_phase"
)
@keras_export("keras.backend.set_learning_phase")
@doc_controls.do_not_generate_docs
def set_learning_phase(value):
"""Sets the learning phase to a fixed value.
The backend learning phase affects any code that calls
`backend.learning_phase()`
In particular, all TF-Keras built-in layers use the learning phase as the
default for the `training` arg to `Layer.__call__`.
User-written layers and models can achieve the same behavior with code that
looks like:
```python
def call(self, inputs, training=None):
if training is None:
training = backend.learning_phase()
```
Args:
value: Learning phase value, either 0 or 1 (integers).
0 = test, 1 = train
Raises:
ValueError: if `value` is neither `0` nor `1`.
"""
warnings.warn(
"`tf.keras.backend.set_learning_phase` is deprecated and "
"will be removed after 2020-10-11. To update it, simply "
"pass a True/False value to the `training` argument of the "
"`__call__` method of your layer or model."
)
deprecated_internal_set_learning_phase(value)
def deprecated_internal_set_learning_phase(value):
"""A deprecated internal implementation of set_learning_phase.
This method is an internal-only version of `set_learning_phase` that
does not raise a deprecation error. It is required because
saved_model needs to keep working with user code that uses the deprecated
learning phase methods until those APIs are fully removed from the public
API.
Specifically SavedModel saving needs to make sure the learning phase is 0
during tracing even if users overwrote it to a different value.
But, we don't want to raise deprecation warnings for users when savedmodel
sets learning phase just for compatibility with code that relied on
explicitly setting the learning phase for other values.
Args:
value: Learning phase value, either 0 or 1 (integers).
0 = test, 1 = train
Raises:
ValueError: if `value` is neither `0` nor `1`.
"""
if value not in {0, 1}:
raise ValueError("Expected learning phase to be 0 or 1.")
with tf.init_scope():
if tf.executing_eagerly():
# In an eager context, the learning phase values applies to both the
# eager context and the internal TF-Keras graph.
_DUMMY_EAGER_GRAPH.learning_phase_is_set = True
_internal_set_learning_phase(_DUMMY_EAGER_GRAPH.key, value)
_internal_set_learning_phase(get_graph(), value)
@keras_export("keras.backend.learning_phase_scope")
@tf_contextlib.contextmanager
@doc_controls.do_not_generate_docs
def learning_phase_scope(value):
"""Provides a scope within which the learning phase is equal to `value`.
The learning phase gets restored to its original value upon exiting the
scope.
Args:
value: Learning phase value, either 0 or 1 (integers).
0 = test, 1 = train
Yields:
None.
Raises:
ValueError: if `value` is neither `0` nor `1`.
"""
warnings.warn(
"`tf.keras.backend.learning_phase_scope` is deprecated and "
"will be removed after 2020-10-11. To update it, simply "
"pass a True/False value to the `training` argument of the "
"`__call__` method of your layer or model.",
stacklevel=2,
)
with deprecated_internal_learning_phase_scope(value):
try:
yield
finally:
pass
@tf_contextlib.contextmanager
def deprecated_internal_learning_phase_scope(value):
"""An internal-only version of `learning_phase_scope`.
Unlike the public method, this method does not raise a deprecation warning.
This is needed because saved model saving needs to set learning phase
to maintain compatibility
with code that sets/gets the learning phase, but saved model
saving itself shouldn't raise a deprecation warning.
We can get rid of this method and its usages when the public API is
removed.
Args:
value: Learning phase value, either 0 or 1 (integers).
0 = test, 1 = train
Yields:
None.
Raises:
ValueError: if `value` is neither `0` nor `1`.
"""
global _GRAPH_LEARNING_PHASES
if value not in {0, 1}:
raise ValueError("Expected learning phase to be 0 or 1.")
with tf.init_scope():
if tf.executing_eagerly():
previous_eager_value = _internal_get_learning_phase(
_DUMMY_EAGER_GRAPH.key
)
previous_graph_value = _internal_get_learning_phase(get_graph())
learning_phase_previously_set = _DUMMY_EAGER_GRAPH.learning_phase_is_set
try:
deprecated_internal_set_learning_phase(value)
yield
finally:
# Restore learning phase to initial value.
if not learning_phase_previously_set:
_DUMMY_EAGER_GRAPH.learning_phase_is_set = False
with tf.init_scope():
if tf.executing_eagerly():
if previous_eager_value is not None:
_internal_set_learning_phase(
_DUMMY_EAGER_GRAPH.key, previous_eager_value
)
elif _DUMMY_EAGER_GRAPH.key in _GRAPH_LEARNING_PHASES:
del _GRAPH_LEARNING_PHASES[_DUMMY_EAGER_GRAPH.key]
graph = get_graph()
if previous_graph_value is not None:
_internal_set_learning_phase(graph, previous_graph_value)
elif graph in _GRAPH_LEARNING_PHASES:
del _GRAPH_LEARNING_PHASES[graph]
@tf_contextlib.contextmanager
def eager_learning_phase_scope(value):
"""Internal scope that sets the learning phase in eager / tf.function only.
Args:
value: Learning phase value, either 0 or 1 (integers).
0 = test, 1 = train
Yields:
None.
Raises:
ValueError: if `value` is neither `0` nor `1`.
"""
global _GRAPH_LEARNING_PHASES
assert value in {0, 1}
assert tf.compat.v1.executing_eagerly_outside_functions()
global_learning_phase_was_set = global_learning_phase_is_set()
if global_learning_phase_was_set:
previous_value = learning_phase()
try:
_internal_set_learning_phase(_DUMMY_EAGER_GRAPH.key, value)
yield
finally:
# Restore learning phase to initial value or unset.
if global_learning_phase_was_set:
_internal_set_learning_phase(_DUMMY_EAGER_GRAPH.key, previous_value)
else:
del _GRAPH_LEARNING_PHASES[_DUMMY_EAGER_GRAPH.key]
def _as_graph_element(obj):
"""Convert `obj` to a graph element if possible, otherwise return `None`.
Args:
obj: Object to convert.
Returns:
The result of `obj._as_graph_element()` if that method is available;
otherwise `None`.
"""
conv_fn = getattr(obj, "_as_graph_element", None)
if conv_fn and callable(conv_fn):
return conv_fn()
return None
def _assert_same_graph(original_item, item):
"""Fail if the 2 items are from different graphs.
Args:
original_item: Original item to check against.
item: Item to check.
Raises:
ValueError: if graphs do not match.
"""
original_graph = getattr(original_item, "graph", None)
graph = getattr(item, "graph", None)
if original_graph and graph and original_graph is not graph:
raise ValueError(
"%s must be from the same graph as %s (graphs are %s and %s)."
% (item, original_item, graph, original_graph)
)
def _current_graph(op_input_list, graph=None):
"""Returns the appropriate graph to use for the given inputs.
This library method provides a consistent algorithm for choosing the graph
in which an Operation should be constructed:
1. If the default graph is being used to construct a function, we
use the default graph.
2. If the "graph" is specified explicitly, we validate that all of the
inputs in "op_input_list" are compatible with that graph.
3. Otherwise, we attempt to select a graph from the first Operation-
or Tensor-valued input in "op_input_list", and validate that all other
such inputs are in the same graph.
4. If the graph was not specified and it could not be inferred from
"op_input_list", we attempt to use the default graph.
Args:
op_input_list: A list of inputs to an operation, which may include
`Tensor`, `Operation`, and other objects that may be converted to a
graph element.
graph: (Optional) The explicit graph to use.
Raises:
TypeError: If op_input_list is not a list or tuple, or if graph is not a
Graph.
ValueError: If a graph is explicitly passed and not all inputs are from
it, or if the inputs are from multiple graphs, or we could not find a
graph and there was no default graph.
Returns:
The appropriate graph to use for the given inputs.
"""
current_default_graph = tf.compat.v1.get_default_graph()
if current_default_graph.building_function:
return current_default_graph
op_input_list = tuple(op_input_list) # Handle generators correctly
if graph and not isinstance(graph, tf.Graph):
raise TypeError(f"Input graph needs to be a Graph: {graph}")
def _is_symbolic_tensor(tensor):
if hasattr(tf, "is_symbolic_tensor"):
return tf.is_symbolic_tensor(tensor)
return type(tensor) == tf.Tensor # noqa: E721
# 1. We validate that all of the inputs are from the same graph. This is
# either the supplied graph parameter, or the first one selected from one
# the graph-element-valued inputs. In the latter case, we hold onto
# that input in original_graph_element so we can provide a more
# informative error if a mismatch is found.
original_graph_element = None
for op_input in op_input_list:
if isinstance(
op_input, (tf.Operation, tf.__internal__.CompositeTensor)
) or _is_symbolic_tensor(op_input):
graph_element = op_input
else:
graph_element = _as_graph_element(op_input)
if graph_element is not None:
if not graph:
original_graph_element = graph_element
graph = getattr(graph_element, "graph", None)
elif original_graph_element is not None:
_assert_same_graph(original_graph_element, graph_element)
elif graph_element.graph is not graph:
raise ValueError(
f"{graph_element} is not from the passed-in graph."
)
# 2. If all else fails, we use the default graph, which is always there.
return graph or current_default_graph
def _get_session(op_input_list=()):
"""Returns the session object for the current thread."""
global _SESSION
default_session = tf.compat.v1.get_default_session()
if default_session is not None:
session = default_session
else:
if tf.inside_function():
raise RuntimeError(
"Cannot get session inside Tensorflow graph function."
)
# If we don't have a session, or that session does not match the current
# graph, create and cache a new session.
if getattr(
_SESSION, "session", None
) is None or _SESSION.session.graph is not _current_graph(
op_input_list
):
# If we are creating the Session inside a tf.distribute.Strategy
# scope, we ask the strategy for the right session options to use.
if tf.distribute.has_strategy():
configure_and_create_distributed_session(
tf.distribute.get_strategy()
)
else:
_SESSION.session = tf.compat.v1.Session(
config=get_default_session_config()
)
session = _SESSION.session
return session
@keras_export(v1=["keras.backend.get_session"])
def get_session(op_input_list=()):
"""Returns the TF session to be used by the backend.
If a default TensorFlow session is available, we will return it.
Else, we will return the global TF-Keras session assuming it matches
the current graph.
If no global TF-Keras session exists at this point:
we will create a new global session.
Note that you can manually set the global session
via `K.set_session(sess)`.
Args:
op_input_list: An option sequence of tensors or ops, which will be used
to determine the current graph. Otherwise the default graph will be
used.
Returns:
A TensorFlow session.
"""
session = _get_session(op_input_list)
if not _MANUAL_VAR_INIT:
with session.graph.as_default():
_initialize_variables(session)
return session
# Inject the get_session function to keras_deps to remove the dependency
# from TFLite to TF-Keras.
tf.__internal__.register_get_session_function(get_session)
# Inject the get_session function to tracking_util to avoid the backward
# dependency from TF to TF-Keras.
tf.__internal__.tracking.register_session_provider(get_session)
def get_graph():
if tf.executing_eagerly():
global _GRAPH
if not getattr(_GRAPH, "graph", None):
_GRAPH.graph = tf.__internal__.FuncGraph("keras_graph")
return _GRAPH.graph
else:
return tf.compat.v1.get_default_graph()
@tf_contextlib.contextmanager
def _scratch_graph(graph=None):
"""Retrieve a shared and temporary func graph.
The eager execution path lifts a subgraph from the keras global graph into
a scratch graph in order to create a function. DistributionStrategies, in
turn, constructs multiple functions as well as a final combined function. In
order for that logic to work correctly, all of the functions need to be
created on the same scratch FuncGraph.
Args:
graph: A graph to be used as the current scratch graph. If not set then
a scratch graph will either be retrieved or created:
Yields:
The current scratch graph.
"""
global _CURRENT_SCRATCH_GRAPH
scratch_graph = getattr(_CURRENT_SCRATCH_GRAPH, "graph", None)
# If scratch graph and `graph` are both configured, they must match.
if (
scratch_graph is not None
and graph is not None
and scratch_graph is not graph
):
raise ValueError("Multiple scratch graphs specified.")
if scratch_graph:
yield scratch_graph
return
graph = graph or tf.__internal__.FuncGraph("keras_scratch_graph")
try:
_CURRENT_SCRATCH_GRAPH.graph = graph
yield graph
finally:
_CURRENT_SCRATCH_GRAPH.graph = None
@keras_export(v1=["keras.backend.set_session"])
def set_session(session):
"""Sets the global TensorFlow session.
Args:
session: A TF Session.
"""
global _SESSION
_SESSION.session = session
def get_default_session_config():
if os.environ.get("OMP_NUM_THREADS"):
logging.warning(
"OMP_NUM_THREADS is no longer used by the default TF-Keras config. "
"To configure the number of threads, use tf.config.threading APIs."
)
config = get_config()
config.allow_soft_placement = True
return config
def get_default_graph_uid_map():
graph = tf.compat.v1.get_default_graph()
name_uid_map = PER_GRAPH_OBJECT_NAME_UIDS.get(graph, None)
if name_uid_map is None:
name_uid_map = collections.defaultdict(int)
PER_GRAPH_OBJECT_NAME_UIDS[graph] = name_uid_map
return name_uid_map
# DEVICE MANIPULATION
class _TfDeviceCaptureOp:
"""Class for capturing the TF device scope."""
def __init__(self):
self.device = None
def _set_device(self, device):
"""This method captures TF's explicit device scope setting."""
if isinstance(device, tf.DeviceSpec):
device = device.to_string()
self.device = device
def _set_device_from_string(self, device_str):
self.device = device_str
def _get_current_tf_device():
"""Return explicit device of current context, otherwise returns `None`.
Returns:
If the current device scope is explicitly set, it returns a string with
the device (`CPU` or `GPU`). If the scope is not explicitly set, it will
return `None`.
"""
graph = get_graph()
op = _TfDeviceCaptureOp()
graph._apply_device_functions(op)
if tf.__internal__.tf2.enabled():
return tf.DeviceSpec.from_string(op.device)
else:
return tf.compat.v1.DeviceSpec.from_string(op.device)
def _is_current_explicit_device(device_type):
"""Check if the current device is explicitly set to `device_type`.
Args:
device_type: A string containing `GPU` or `CPU` (case-insensitive).
Returns:
A boolean indicating if the current device scope is explicitly set on
the device type.
Raises:
ValueError: If the `device_type` string indicates an unsupported device.
"""
device_type = device_type.upper()
if device_type not in ["CPU", "GPU"]:
raise ValueError('`device_type` should be either "CPU" or "GPU".')
device = _get_current_tf_device()
return device is not None and device.device_type == device_type.upper()
def _get_available_gpus():
"""Get a list of available GPU devices (formatted as strings).
Returns:
A list of available GPU devices.
"""
if tf.compat.v1.executing_eagerly_outside_functions():
# Returns names of devices directly.
return [d.name for d in tf.config.list_logical_devices("GPU")]
global _LOCAL_DEVICES
if _LOCAL_DEVICES is None:
_LOCAL_DEVICES = get_session().list_devices()
return [x.name for x in _LOCAL_DEVICES if x.device_type == "GPU"]
def _has_nchw_support():
"""Check whether the current scope supports NCHW ops.
TensorFlow does not support NCHW on CPU. Therefore we check if we are not
explicitly put on
CPU, and have GPUs available. In this case there will be soft-placing on the
GPU device.
Returns:
bool: if the current scope device placement would support nchw
"""
explicitly_on_cpu = _is_current_explicit_device("CPU")
gpus_available = bool(_get_available_gpus())
return not explicitly_on_cpu and gpus_available
# VARIABLE MANIPULATION
def _constant_to_tensor(x, dtype):
"""Convert the input `x` to a tensor of type `dtype`.
This is slightly faster than the _to_tensor function, at the cost of
handling fewer cases.
Args:
x: An object to be converted (numpy arrays, floats, ints and lists of
them).
dtype: The destination type.
Returns:
A tensor.
"""
return tf.constant(x, dtype=dtype)
def _to_tensor(x, dtype):
"""Convert the input `x` to a tensor of type `dtype`.
Args:
x: An object to be converted (numpy array, list, tensors).
dtype: The destination type.
Returns:
A tensor.
"""
return tf.convert_to_tensor(x, dtype=dtype)
@keras_export("keras.backend.is_sparse")
@doc_controls.do_not_generate_docs
def is_sparse(tensor):
"""Returns whether a tensor is a sparse tensor.
Args:
tensor: A tensor instance.
Returns:
A boolean.
Example:
>>> a = tf.keras.backend.placeholder((2, 2), sparse=False)
>>> print(tf.keras.backend.is_sparse(a))
False
>>> b = tf.keras.backend.placeholder((2, 2), sparse=True)
>>> print(tf.keras.backend.is_sparse(b))
True
"""
spec = getattr(tensor, "_type_spec", None)
if spec is not None:
return isinstance(spec, tf.SparseTensorSpec)
return isinstance(tensor, tf.SparseTensor)
@keras_export("keras.backend.to_dense")
@tf.__internal__.dispatch.add_dispatch_support
@doc_controls.do_not_generate_docs
def to_dense(tensor):
"""Converts a sparse tensor into a dense tensor and returns it.
Args:
tensor: A tensor instance (potentially sparse).
Returns:
A dense tensor.
Examples:
>>> b = tf.keras.backend.placeholder((2, 2), sparse=True)
>>> print(tf.keras.backend.is_sparse(b))
True
>>> c = tf.keras.backend.to_dense(b)
>>> print(tf.keras.backend.is_sparse(c))
False
"""
if is_sparse(tensor):
return tf.sparse.to_dense(tensor)
else:
return tensor
@keras_export("keras.backend.name_scope", v1=[])
@doc_controls.do_not_generate_docs
def name_scope(name):
"""A context manager for use when defining a Python op.
This context manager pushes a name scope, which will make the name of all
operations added within it have a prefix.
For example, to define a new Python op called `my_op`:
def my_op(a):
with tf.name_scope("MyOp") as scope:
a = tf.convert_to_tensor(a, name="a")
# Define some computation that uses `a`.
return foo_op(..., name=scope)
When executed, the Tensor `a` will have the name `MyOp/a`.
Args:
name: The prefix to use on all names created within the name scope.
Returns:
Name scope context manager.
"""
return tf.name_scope(name)
# Export V1 version.
_v1_name_scope = tf.compat.v1.name_scope
keras_export(v1=["keras.backend.name_scope"], allow_multiple_exports=True)(
_v1_name_scope
)
@keras_export("keras.backend.variable")
@doc_controls.do_not_generate_docs
def variable(value, dtype=None, name=None, constraint=None):
"""Instantiates a variable and returns it.
Args:
value: Numpy array, initial value of the tensor.
dtype: Tensor type.
name: Optional name string for the tensor.
constraint: Optional projection function to be
applied to the variable after an optimizer update.
Returns:
A variable instance (with TF-Keras metadata included).
Examples:
>>> val = np.array([[1, 2], [3, 4]])
>>> kvar = tf.keras.backend.variable(value=val, dtype='float64',
... name='example_var')
>>> tf.keras.backend.dtype(kvar)
'float64'
>>> print(kvar)
<tf.Variable 'example_var:...' shape=(2, 2) dtype=float64, numpy=
array([[1., 2.],
[3., 4.]])>
"""
if dtype is None:
dtype = floatx()
if hasattr(value, "tocoo"):
sparse_coo = value.tocoo()
indices = np.concatenate(
(
np.expand_dims(sparse_coo.row, 1),
np.expand_dims(sparse_coo.col, 1),
),
1,
)
v = tf.SparseTensor(
indices=indices,
values=sparse_coo.data,
dense_shape=sparse_coo.shape,
)
v._keras_shape = sparse_coo.shape
return v
v = tf.Variable(
value, dtype=tf.as_dtype(dtype), name=name, constraint=constraint
)
if isinstance(value, np.ndarray):
v._keras_shape = value.shape
elif hasattr(value, "shape"):
v._keras_shape = int_shape(value)
track_variable(v)
return v
def track_tf_optimizer(tf_optimizer):
"""Tracks the given TF optimizer for initialization of its variables."""
if tf.executing_eagerly():
return
optimizers = _GRAPH_TF_OPTIMIZERS[None]
optimizers.add(tf_optimizer)
@keras_export("keras.__internal__.backend.track_variable", v1=[])
def track_variable(v):
"""Tracks the given variable for initialization."""
if tf.executing_eagerly():
return
graph = v.graph if hasattr(v, "graph") else get_graph()
_GRAPH_VARIABLES[graph].add(v)
def observe_object_name(name):
"""Observe a name and make sure it won't be used by `unique_object_name`."""
OBSERVED_NAMES.add(name)
def unique_object_name(
name,
name_uid_map=None,
avoid_names=None,
namespace="",
zero_based=False,
avoid_observed_names=False,
):
"""Makes a object name (or any string) unique within a TF-Keras session.
Args:
name: String name to make unique.
name_uid_map: An optional defaultdict(int) to use when creating unique
names. If None (default), uses a per-Graph dictionary.
avoid_names: An optional set or dict with names which should not be used.
If None (default), don't avoid any names unless `avoid_observed_names`
is True.
namespace: Gets a name which is unique within the (graph, namespace).
Layers which are not Networks use a blank namespace and so get
graph-global names.
zero_based: If True, name sequences start with no suffix (e.g. "dense",
"dense_1"). If False, naming is one-based ("dense_1", "dense_2").
avoid_observed_names: If True, avoid any names that have been observed by
`backend.observe_object_name`.
Returns:
Unique string name.
Example:
unique_object_name('dense') # dense_1
unique_object_name('dense') # dense_2
"""
if name_uid_map is None:
name_uid_map = get_default_graph_uid_map()
if avoid_names is None:
if avoid_observed_names:
avoid_names = OBSERVED_NAMES
else:
avoid_names = set()
proposed_name = None
while proposed_name is None or proposed_name in avoid_names:
name_key = (namespace, name)
if zero_based:
number = name_uid_map[name_key]
if number:
proposed_name = name + "_" + str(number)
else:
proposed_name = name
name_uid_map[name_key] += 1
else:
name_uid_map[name_key] += 1
proposed_name = name + "_" + str(name_uid_map[name_key])
return proposed_name
def _get_variables(graph=None):
"""Returns variables corresponding to the given graph for initialization."""
assert not tf.executing_eagerly()
variables = _GRAPH_VARIABLES[graph]
for opt in _GRAPH_TF_OPTIMIZERS[graph]:
variables.update(opt.optimizer.variables())
return variables
@keras_export("keras.__internal__.backend.initialize_variables", v1=[])
def _initialize_variables(session):
"""Utility to initialize uninitialized variables on the fly."""
variables = _get_variables(get_graph())
candidate_vars = []
for v in variables:
if not getattr(v, "_keras_initialized", False):
candidate_vars.append(v)
if candidate_vars:
# This step is expensive, so we only run it on variables not already
# marked as initialized.
is_initialized = session.run(
[tf.compat.v1.is_variable_initialized(v) for v in candidate_vars]
)
# TODO(kathywu): Some metric variables loaded from SavedModel are never
# actually used, and do not have an initializer.
should_be_initialized = [
(not is_initialized[n]) and v.initializer is not None
for n, v in enumerate(candidate_vars)
]
uninitialized_vars = []
for flag, v in zip(should_be_initialized, candidate_vars):
if flag:
uninitialized_vars.append(v)
v._keras_initialized = True
if uninitialized_vars:
session.run(tf.compat.v1.variables_initializer(uninitialized_vars))
@keras_export("keras.backend.constant")
@tf.__internal__.dispatch.add_dispatch_support
@doc_controls.do_not_generate_docs
def constant(value, dtype=None, shape=None, name=None):
"""Creates a constant tensor.
Args:
value: A constant value (or list)
dtype: The type of the elements of the resulting tensor.
shape: Optional dimensions of resulting tensor.
name: Optional name for the tensor.
Returns:
A Constant Tensor.
"""
if dtype is None:
dtype = floatx()
return tf.constant(value, dtype=dtype, shape=shape, name=name)
@keras_export("keras.backend.is_keras_tensor")
def is_keras_tensor(x):
"""Returns whether `x` is a TF-Keras tensor.
A "Keras tensor" is a tensor that was returned by a TF-Keras layer,
(`Layer` class) or by `Input`.
Args:
x: A candidate tensor.
Returns:
A boolean: Whether the argument is a TF-Keras tensor.
Raises:
ValueError: In case `x` is not a symbolic tensor.
Examples:
>>> np_var = np.array([1, 2])
>>> # A numpy array is not a symbolic tensor.
>>> tf.keras.backend.is_keras_tensor(np_var)
Traceback (most recent call last):
...
ValueError: Unexpectedly found an instance of type
`<class 'numpy.ndarray'>`.
Expected a symbolic tensor instance.
>>> keras_var = tf.keras.backend.variable(np_var)
>>> # A variable created with the keras backend is not a TF-Keras tensor.
>>> tf.keras.backend.is_keras_tensor(keras_var)
False
>>> keras_placeholder = tf.keras.backend.placeholder(shape=(2, 4, 5))
>>> # A placeholder is a TF-Keras tensor.
>>> tf.keras.backend.is_keras_tensor(keras_placeholder)
True
>>> keras_input = tf.keras.layers.Input([10])
>>> # An Input is a TF-Keras tensor.
>>> tf.keras.backend.is_keras_tensor(keras_input)
True
>>> keras_layer_output = tf.keras.layers.Dense(10)(keras_input)
>>> # Any TF-Keras layer output is a TF-Keras tensor.
>>> tf.keras.backend.is_keras_tensor(keras_layer_output)
True
"""
if not isinstance(
x,
(
tf.Tensor,
tf.Variable,
tf.SparseTensor,
tf.RaggedTensor,
keras_tensor.KerasTensor,
),
):
raise ValueError(
"Unexpectedly found an instance of type `"
+ str(type(x))
+ "`. Expected a symbolic tensor instance."
)
if tf.compat.v1.executing_eagerly_outside_functions():
return isinstance(x, keras_tensor.KerasTensor)
return hasattr(x, "_keras_history")
@keras_export("keras.backend.placeholder")
@doc_controls.do_not_generate_docs
def placeholder(
shape=None, ndim=None, dtype=None, sparse=False, name=None, ragged=False
):
"""Instantiates a placeholder tensor and returns it.
Args:
shape: Shape of the placeholder
(integer tuple, may include `None` entries).
ndim: Number of axes of the tensor.
At least one of {`shape`, `ndim`} must be specified.
If both are specified, `shape` is used.
dtype: Placeholder type.
sparse: Boolean, whether the placeholder should have a sparse type.
name: Optional name string for the placeholder.
ragged: Boolean, whether the placeholder should have a ragged type.
In this case, values of 'None' in the 'shape' argument represent
ragged dimensions. For more information about RaggedTensors, see
this [guide](https://www.tensorflow.org/guide/ragged_tensor).
Raises:
ValueError: If called with sparse = True and ragged = True.
Returns:
Tensor instance (with TF-Keras metadata included).
Examples:
>>> input_ph = tf.keras.backend.placeholder(shape=(2, 4, 5))
>>> input_ph
<KerasTensor: shape=(2, 4, 5) dtype=float32 (created by layer ...)>
"""
if sparse and ragged:
raise ValueError(
"Cannot set both sparse and ragged to "
"True when creating a placeholder."
)
if dtype is None:
dtype = floatx()
if not shape:
if ndim:
shape = (None,) * ndim
if tf.compat.v1.executing_eagerly_outside_functions():
if sparse:
spec = tf.SparseTensorSpec(shape=shape, dtype=dtype)
elif ragged:
ragged_rank = 0
for i in range(1, len(shape)):
# Hacky because could be tensorshape or tuple maybe?
# Or just tensorshape?
if shape[i] is None or (
hasattr(shape[i], "value") and shape[i].value is None
):
ragged_rank = i
spec = tf.RaggedTensorSpec(
shape=shape, dtype=dtype, ragged_rank=ragged_rank
)
else:
spec = tf.TensorSpec(shape=shape, dtype=dtype, name=name)
x = keras_tensor.keras_tensor_from_type_spec(spec, name=name)
else:
with get_graph().as_default():
if sparse:
x = tf.compat.v1.sparse_placeholder(
dtype, shape=shape, name=name
)
elif ragged:
ragged_rank = 0
for i in range(1, len(shape)):
if shape[i] is None:
ragged_rank = i
type_spec = tf.RaggedTensorSpec(
shape=shape, dtype=dtype, ragged_rank=ragged_rank
)
def tensor_spec_to_placeholder(tensorspec):
return tf.compat.v1.placeholder(
tensorspec.dtype, tensorspec.shape
)
x = tf.nest.map_structure(
tensor_spec_to_placeholder,
type_spec,
expand_composites=True,
)
else:
x = tf.compat.v1.placeholder(dtype, shape=shape, name=name)
if tf.executing_eagerly():
# Add keras_history connectivity information to the placeholder
# when the placeholder is built in a top-level eager context
# (intended to be used with keras.backend.function)
from tf_keras.engine import (
input_layer,
)
x = input_layer.Input(tensor=x)
x._is_backend_placeholder = True
return x
def is_placeholder(x):
"""Returns whether `x` is a placeholder.
Args:
x: A candidate placeholder.
Returns:
Boolean.
"""
try:
if tf.compat.v1.executing_eagerly_outside_functions():
return hasattr(x, "_is_backend_placeholder")
# TODO(b/246438937): Remove the special case for tf.Variable once
# tf.Variable becomes CompositeTensor and will be expanded into
# dt_resource tensors.
if tf_utils.is_extension_type(x) and not isinstance(x, tf.Variable):
flat_components = tf.nest.flatten(x, expand_composites=True)
return py_any(is_placeholder(c) for c in flat_components)
else:
return x.op.type == "Placeholder"
except AttributeError:
return False
@keras_export("keras.backend.shape")
@tf.__internal__.dispatch.add_dispatch_support
@doc_controls.do_not_generate_docs
def shape(x):
"""Returns the symbolic shape of a tensor or variable.
Args:
x: A tensor or variable.
Returns:
A symbolic shape (which is itself a tensor).
Examples:
>>> val = np.array([[1, 2], [3, 4]])
>>> kvar = tf.keras.backend.variable(value=val)
>>> tf.keras.backend.shape(kvar)
<tf.Tensor: shape=(2,), dtype=int32, numpy=array([2, 2], dtype=int32)>
>>> input = tf.keras.backend.placeholder(shape=(2, 4, 5))
>>> tf.keras.backend.shape(input)
<KerasTensor: shape=(3,) dtype=int32 inferred_value=[2, 4, 5] ...>
"""
return tf.shape(x)
@keras_export("keras.backend.int_shape")
@doc_controls.do_not_generate_docs
def int_shape(x):
"""Returns shape of tensor/variable as a tuple of int/None entries.
Args:
x: Tensor or variable.
Returns:
A tuple of integers (or None entries).
Examples:
>>> input = tf.keras.backend.placeholder(shape=(2, 4, 5))
>>> tf.keras.backend.int_shape(input)
(2, 4, 5)
>>> val = np.array([[1, 2], [3, 4]])
>>> kvar = tf.keras.backend.variable(value=val)
>>> tf.keras.backend.int_shape(kvar)
(2, 2)
"""
try:
shape = x.shape
if not isinstance(shape, tuple):
shape = tuple(shape.as_list())
return shape
except ValueError:
return None
@keras_export("keras.backend.ndim")
@doc_controls.do_not_generate_docs
def ndim(x):
"""Returns the number of axes in a tensor, as an integer.
Args:
x: Tensor or variable.
Returns:
Integer (scalar), number of axes.
Examples:
>>> input = tf.keras.backend.placeholder(shape=(2, 4, 5))
>>> val = np.array([[1, 2], [3, 4]])
>>> kvar = tf.keras.backend.variable(value=val)
>>> tf.keras.backend.ndim(input)
3
>>> tf.keras.backend.ndim(kvar)
2
"""
return x.shape.rank
@keras_export("keras.backend.dtype")
@tf.__internal__.dispatch.add_dispatch_support
@doc_controls.do_not_generate_docs
def dtype(x):
"""Returns the dtype of a TF-Keras tensor or variable, as a string.
Args:
x: Tensor or variable.
Returns:
String, dtype of `x`.
Examples:
>>> tf.keras.backend.dtype(tf.keras.backend.placeholder(shape=(2,4,5)))
'float32'
>>> tf.keras.backend.dtype(tf.keras.backend.placeholder(shape=(2,4,5),
... dtype='float32'))
'float32'
>>> tf.keras.backend.dtype(tf.keras.backend.placeholder(shape=(2,4,5),
... dtype='float64'))
'float64'
>>> kvar = tf.keras.backend.variable(np.array([[1, 2], [3, 4]]))
>>> tf.keras.backend.dtype(kvar)
'float32'
>>> kvar = tf.keras.backend.variable(np.array([[1, 2], [3, 4]]),
... dtype='float32')
>>> tf.keras.backend.dtype(kvar)
'float32'
"""
return x.dtype.base_dtype.name
@doc_controls.do_not_generate_docs
def dtype_numpy(x):
"""Returns the numpy dtype of a TF-Keras tensor or variable.
Args:
x: Tensor or variable.
Returns:
numpy.dtype, dtype of `x`.
"""
return tf.as_dtype(x.dtype).as_numpy_dtype
@keras_export("keras.backend.eval")
@doc_controls.do_not_generate_docs
def eval(x):
"""Evaluates the value of a variable.
Args:
x: A variable.
Returns:
A Numpy array.
Examples:
>>> kvar = tf.keras.backend.variable(np.array([[1, 2], [3, 4]]),
... dtype='float32')
>>> tf.keras.backend.eval(kvar)
array([[1., 2.],
[3., 4.]], dtype=float32)
"""
return get_value(to_dense(x))
@keras_export("keras.backend.zeros")
@doc_controls.do_not_generate_docs
def zeros(shape, dtype=None, name=None):
"""Instantiates an all-zeros variable and returns it.
Args:
shape: Tuple or list of integers, shape of returned TF-Keras variable
dtype: data type of returned TF-Keras variable
name: name of returned TF-Keras variable
Returns:
A variable (including TF-Keras metadata), filled with `0.0`.
Note that if `shape` was symbolic, we cannot return a variable,
and will return a dynamically-shaped tensor instead.
Example:
>>> kvar = tf.keras.backend.zeros((3,4))
>>> tf.keras.backend.eval(kvar)
array([[0., 0., 0., 0.],
[0., 0., 0., 0.],
[0., 0., 0., 0.]], dtype=float32)
>>> A = tf.constant([1,2,3])
>>> kvar2 = tf.keras.backend.zeros(A.shape) # [0., 0., 0.]
>>> tf.keras.backend.eval(kvar2)
array([0., 0., 0.], dtype=float32)
>>> kvar3 = tf.keras.backend.zeros(A.shape,dtype=tf.int32)
>>> tf.keras.backend.eval(kvar3)
array([0, 0, 0], dtype=int32)
>>> kvar4 = tf.keras.backend.zeros([2,3])
>>> tf.keras.backend.eval(kvar4)
array([[0., 0., 0.],
[0., 0., 0.]], dtype=float32)
"""
with tf.init_scope():
if dtype is None:
dtype = floatx()
tf_dtype = tf.as_dtype(dtype)
v = tf.zeros(shape=shape, dtype=tf_dtype, name=name)
if py_all(v.shape.as_list()):
return variable(v, dtype=dtype, name=name)
return v
@keras_export("keras.backend.ones")
@tf.__internal__.dispatch.add_dispatch_support
@doc_controls.do_not_generate_docs
def ones(shape, dtype=None, name=None):
"""Instantiates an all-ones variable and returns it.
Args:
shape: Tuple of integers, shape of returned TF-Keras variable.
dtype: String, data type of returned TF-Keras variable.
name: String, name of returned TF-Keras variable.
Returns:
A TF-Keras variable, filled with `1.0`.
Note that if `shape` was symbolic, we cannot return a variable,
and will return a dynamically-shaped tensor instead.
Example:
>>> kvar = tf.keras.backend.ones((3,4))
>>> tf.keras.backend.eval(kvar)
array([[1., 1., 1., 1.],
[1., 1., 1., 1.],
[1., 1., 1., 1.]], dtype=float32)
"""
with tf.init_scope():
if dtype is None:
dtype = floatx()
tf_dtype = tf.as_dtype(dtype)
v = tf.ones(shape=shape, dtype=tf_dtype, name=name)
if py_all(v.shape.as_list()):
return variable(v, dtype=dtype, name=name)
return v
@keras_export("keras.backend.eye")
@tf.__internal__.dispatch.add_dispatch_support
@doc_controls.do_not_generate_docs
def eye(size, dtype=None, name=None):
"""Instantiate an identity matrix and returns it.
Args:
size: Integer, number of rows/columns.
dtype: String, data type of returned TF-Keras variable.
name: String, name of returned TF-Keras variable.
Returns:
A TF-Keras variable, an identity matrix.
Example:
>>> kvar = tf.keras.backend.eye(3)
>>> tf.keras.backend.eval(kvar)
array([[1., 0., 0.],
[0., 1., 0.],
[0., 0., 1.]], dtype=float32)
"""
if dtype is None:
dtype = floatx()
tf_dtype = tf.as_dtype(dtype)
return variable(tf.eye(size, dtype=tf_dtype), dtype, name)
@keras_export("keras.backend.zeros_like")
@doc_controls.do_not_generate_docs
def zeros_like(x, dtype=None, name=None):
"""Instantiates an all-zeros variable of the same shape as another tensor.
Args:
x: TF-Keras variable or TF-Keras tensor.
dtype: dtype of returned TF-Keras variable.
`None` uses the dtype of `x`.
name: name for the variable to create.
Returns:
A TF-Keras variable with the shape of `x` filled with zeros.
Example:
```python
kvar = tf.keras.backend.variable(np.random.random((2,3)))
kvar_zeros = tf.keras.backend.zeros_like(kvar)
K.eval(kvar_zeros)
# array([[ 0., 0., 0.], [ 0., 0., 0.]], dtype=float32)
```
"""
return tf.zeros_like(x, dtype=dtype, name=name)
@keras_export("keras.backend.ones_like")
@tf.__internal__.dispatch.add_dispatch_support
@doc_controls.do_not_generate_docs
def ones_like(x, dtype=None, name=None):
"""Instantiates an all-ones variable of the same shape as another tensor.
Args:
x: TF-Keras variable or tensor.
dtype: String, dtype of returned TF-Keras variable.
None uses the dtype of x.
name: String, name for the variable to create.
Returns:
A TF-Keras variable with the shape of x filled with ones.
Example:
>>> kvar = tf.keras.backend.variable(np.random.random((2,3)))
>>> kvar_ones = tf.keras.backend.ones_like(kvar)
>>> tf.keras.backend.eval(kvar_ones)
array([[1., 1., 1.],
[1., 1., 1.]], dtype=float32)
"""
return tf.ones_like(x, dtype=dtype, name=name)
def identity(x, name=None):
"""Returns a tensor with the same content as the input tensor.
Args:
x: The input tensor.
name: String, name for the variable to create.
Returns:
A tensor of the same shape, type and content.
"""
return tf.identity(x, name=name)
# Global flag to enforce tf.random.Generator for RandomGenerator.
# When this is enabled, for any caller to RandomGenerator, it will use
# tf.random.Generator to generate random numbers.
# The legacy behavior is to use TF's legacy stateful RNG ops like
# tf.random.uniform.
_USE_GENERATOR_FOR_RNG = False
# The global generator to create the seed when initializing the
# tf.random.Genrator used by RandomGenerator. When tf.random.Generator becomes
# the default solution, we would like the it to be initialized in a controlable
# way, so that each client of the program could start with same seed. This is
# very important for certain use case that requires all the client to have their
# state in sync. This instance will be set when user call
# `tf.keras.utils.set_random_seed()`
_SEED_GENERATOR = threading.local()
@keras_export(
"keras.backend.experimental.is_tf_random_generator_enabled", v1=[]
)
def is_tf_random_generator_enabled():
"""Check whether `tf.random.Generator` is used for RNG in TF-Keras.
Compared to existing TF stateful random ops, `tf.random.Generator` uses
`tf.Variable` and stateless random ops to generate random numbers,
which leads to better reproducibility in distributed training.
Note enabling it might introduce some breakage to existing code,
by producing differently-seeded random number sequences
and breaking tests that rely on specific random numbers being generated.
To disable the
usage of `tf.random.Generator`, please use
`tf.keras.backend.experimental.disable_random_generator`.
We expect the `tf.random.Generator` code path to become the default, and
will remove the legacy stateful random ops such as `tf.random.uniform` in
the future (see the [TF RNG guide](
https://www.tensorflow.org/guide/random_numbers)).
This API will also be removed in a future release as well, together with
`tf.keras.backend.experimental.enable_tf_random_generator()` and
`tf.keras.backend.experimental.disable_tf_random_generator()`
Returns:
boolean: whether `tf.random.Generator` is used for random number
generation in TF-Keras.
"""
return _USE_GENERATOR_FOR_RNG
@keras_export("keras.backend.experimental.enable_tf_random_generator", v1=[])
def enable_tf_random_generator():
"""Enable the `tf.random.Generator` as the RNG for TF-Keras.
See `tf.keras.backend.experimental.is_tf_random_generator_enabled` for more
details.
"""
global _USE_GENERATOR_FOR_RNG
_USE_GENERATOR_FOR_RNG = True
@keras_export("keras.backend.experimental.disable_tf_random_generator", v1=[])
def disable_tf_random_generator():
"""Disable the `tf.random.Generator` as the RNG for TF-Keras.
See `tf.keras.backend.experimental.is_tf_random_generator_enabled` for more
details.
"""
global _USE_GENERATOR_FOR_RNG
_USE_GENERATOR_FOR_RNG = False
class RandomGenerator(tf.__internal__.tracking.AutoTrackable):
"""Random generator that selects appropriate random ops.
This class contains the logic for legacy stateful random ops, as well as the
new stateless random ops with seeds and tf.random.Generator. Any class that
relies on RNG (eg initializer, shuffle, dropout) should use this class to
handle the transition from legacy RNGs to new RNGs.
Args:
seed: Optional int seed. When `rng_type` is "stateful", the seed is used
to create `tf.random.Generator` to produce deterministic sequences.
When `rng_type` is "stateless", new seed will be created if it is not
provided by user, and it will be passed down to stateless random ops.
When `rng_type` is "legacy_stateful", the seed will be passed down to
stateful random ops.
rng_type: Type of RNG to use, one of "stateful", "stateless",
"legacy_stateful". When `None` it uses "stateful" if
`enable_tf_random_generator` has been activated, or
"legacy_stateful" otherwise.
- When using "stateless", the random ops outputs are constant (the same
inputs result in the same outputs).
- When using "stateful" or "legacy_stateful", the random ops outputs are
non-constant, but deterministic: calling the same random op multiple
times with the same inputs results in a deterministic sequence of
different outputs.
- "legacy_stateful" is backed by TF1 stateful RNG ops
(e.g. `tf.random.uniform`), while "stateful"
is backed by TF2 APIs (e.g. `tf.random.Generator.uniform`).
Defaults to `None`.
"""
RNG_STATELESS = "stateless"
RNG_STATEFUL = "stateful"
RNG_LEGACY_STATEFUL = "legacy_stateful"
def __init__(self, seed=None, rng_type=None, **kwargs):
self._seed = seed
self._set_rng_type(rng_type, **kwargs)
self._built = False
def _set_rng_type(self, rng_type, **kwargs):
# Only supported kwargs is "force_generator", which we will remove once
# we clean up all the caller.
# TODO(scottzhu): Remove the kwargs for force_generator.
if kwargs.get("force_generator", False):
rng_type = self.RNG_STATEFUL
if rng_type is None:
if is_tf_random_generator_enabled():
self._rng_type = self.RNG_STATEFUL
else:
self._rng_type = self.RNG_LEGACY_STATEFUL
else:
if rng_type not in [
self.RNG_STATEFUL,
self.RNG_LEGACY_STATEFUL,
self.RNG_STATELESS,
]:
raise ValueError(
"Invalid `rng_type` received. "
'Valid `rng_type` are ["stateless", '
'"stateful", "legacy_stateful"].'
f" Got: {rng_type}"
)
self._rng_type = rng_type
def _maybe_init(self):
"""Lazily init the RandomGenerator.
The TF API executing_eagerly_outside_functions() has some side effect,
and couldn't be used before API like tf.enable_eager_execution(). Some
of the client side code was creating the initializer at the code load
time, which triggers the creation of RandomGenerator. Lazy init this
class to walkaround this issue until it is resolved on TF side.
"""
# TODO(b/167482354): Change this back to normal init when the bug is
# fixed.
if self._built:
return
if (
self._rng_type == self.RNG_STATEFUL
and not tf.compat.v1.executing_eagerly_outside_functions()
):
# Fall back to legacy stateful since the generator need to work in
# tf2.
self._rng_type = self.RNG_LEGACY_STATEFUL
if self._rng_type == self.RNG_STATELESS:
self._seed = self._create_seed(self._seed)
self._generator = None
elif self._rng_type == self.RNG_STATEFUL:
with tf_utils.maybe_init_scope(self):
seed = self._create_seed(self._seed)
self._generator = tf.random.Generator.from_seed(
seed, alg=tf.random.Algorithm.AUTO_SELECT
)
else:
# In legacy stateful, we use stateful op, regardless whether user
# provide seed or not. Seeded stateful op will ensure generating
# same sequences.
self._generator = None
self._built = True
def make_seed_for_stateless_op(self):
"""Generate a new seed based on the init config.
Note that this will not return python ints which will be frozen in the
graph and cause stateless op to return the same value. It will only
return value when generator is used, otherwise it will return None.
Returns:
A tensor with shape [2,].
"""
self._maybe_init()
if self._rng_type == self.RNG_STATELESS:
return [self._seed, 0]
elif self._rng_type == self.RNG_STATEFUL:
return self._generator.make_seeds()[:, 0]
return None
def make_legacy_seed(self):
"""Create a new seed for the legacy stateful ops to use.
When user didn't provide any original seed, this method will return
None. Otherwise it will increment the counter and return as the new
seed.
Note that it is important to generate different seed for stateful ops in
the `tf.function`. The random ops will return same value when same seed
is provided in the `tf.function`.
Returns:
int as new seed, or None.
"""
if self._seed is not None:
result = self._seed
self._seed += 1
return result
return None
def _create_seed(self, user_specified_seed):
if user_specified_seed is not None:
return user_specified_seed
elif getattr(_SEED_GENERATOR, "generator", None):
return _SEED_GENERATOR.generator.randint(1, 1e9)
else:
return random.randint(1, int(1e9))
def random_normal(
self, shape, mean=0.0, stddev=1.0, dtype=None, nonce=None
):
"""Produce random number based on the normal distribution.
Args:
shape: The shape of the random values to generate.
mean: Floats, default to 0. Mean of the random values to generate.
stddev: Floats, default to 1. Standard deviation of the random values
to generate.
dtype: Optional dtype of the tensor. Only floating point types are
supported. If not specified, `tf.keras.backend.floatx()` is used,
which default to `float32` unless you configured it otherwise (via
`tf.keras.backend.set_floatx(float_dtype)`)
nonce: Optional integer scalar, that will be folded into the seed in
the stateless mode.
"""
self._maybe_init()
dtype = dtype or floatx()
if self._rng_type == self.RNG_STATEFUL:
return self._generator.normal(
shape=shape, mean=mean, stddev=stddev, dtype=dtype
)
elif self._rng_type == self.RNG_STATELESS:
seed = self.make_seed_for_stateless_op()
if nonce:
seed = tf.random.experimental.stateless_fold_in(seed, nonce)
return tf.random.stateless_normal(
shape=shape, mean=mean, stddev=stddev, dtype=dtype, seed=seed
)
return tf.random.normal(
shape=shape,
mean=mean,
stddev=stddev,
dtype=dtype,
seed=self.make_legacy_seed(),
)
def random_uniform(
self, shape, minval=0.0, maxval=None, dtype=None, nonce=None
):
"""Produce random number based on the uniform distribution.
Args:
shape: The shape of the random values to generate.
minval: Floats, default to 0. Lower bound of the range of
random values to generate (inclusive).
minval: Floats, default to None. Upper bound of the range of
random values to generate (exclusive).
dtype: Optional dtype of the tensor. Only floating point types are
supported. If not specified, `tf.keras.backend.floatx()` is used,
which default to `float32` unless you configured it otherwise (via
`tf.keras.backend.set_floatx(float_dtype)`)
nonce: Optional integer scalar, that will be folded into the seed in
the stateless mode.
"""
self._maybe_init()
dtype = dtype or floatx()
if self._rng_type == self.RNG_STATEFUL:
return self._generator.uniform(
shape=shape, minval=minval, maxval=maxval, dtype=dtype
)
elif self._rng_type == self.RNG_STATELESS:
seed = self.make_seed_for_stateless_op()
if nonce:
seed = tf.random.experimental.stateless_fold_in(seed, nonce)
return tf.random.stateless_uniform(
shape=shape,
minval=minval,
maxval=maxval,
dtype=dtype,
seed=seed,
)
return tf.random.uniform(
shape=shape,
minval=minval,
maxval=maxval,
dtype=dtype,
seed=self.make_legacy_seed(),
)
def truncated_normal(
self, shape, mean=0.0, stddev=1.0, dtype=None, nonce=None
):
"""Produce random number based on the truncated normal distribution.
Args:
shape: The shape of the random values to generate.
mean: Floats, default to 0. Mean of the random values to generate.
stddev: Floats, default to 1. Standard deviation of the random values
to generate.
dtype: Optional dtype of the tensor. Only floating point types are
supported. If not specified, `tf.keras.backend.floatx()` is used,
which default to `float32` unless you configured it otherwise (via
`tf.keras.backend.set_floatx(float_dtype)`)
nonce: Optional integer scalar, that will be folded into the seed in
the stateless mode.
"""
self._maybe_init()
dtype = dtype or floatx()
if self._rng_type == self.RNG_STATEFUL:
return self._generator.truncated_normal(
shape=shape, mean=mean, stddev=stddev, dtype=dtype
)
elif self._rng_type == self.RNG_STATELESS:
seed = self.make_seed_for_stateless_op()
if nonce:
seed = tf.random.experimental.stateless_fold_in(seed, nonce)
return tf.random.stateless_truncated_normal(
shape=shape, mean=mean, stddev=stddev, dtype=dtype, seed=seed
)
return tf.random.truncated_normal(
shape=shape,
mean=mean,
stddev=stddev,
dtype=dtype,
seed=self.make_legacy_seed(),
)
def dropout(self, inputs, rate, noise_shape=None):
self._maybe_init()
if self._rng_type == self.RNG_STATEFUL:
return tf.nn.experimental.general_dropout(
inputs,
rate=rate,
noise_shape=noise_shape,
uniform_sampler=self._generator.uniform,
)
elif self._rng_type == self.RNG_STATELESS:
return tf.nn.experimental.stateless_dropout(
inputs,
rate=rate,
noise_shape=noise_shape,
seed=self.make_seed_for_stateless_op(),
)
else:
return tf.nn.dropout(
inputs,
rate=rate,
noise_shape=noise_shape,
seed=self.make_legacy_seed(),
)
@keras_export("keras.backend.random_uniform_variable")
@doc_controls.do_not_generate_docs
def random_uniform_variable(shape, low, high, dtype=None, name=None, seed=None):
"""Instantiates a variable with values drawn from a uniform distribution.
Args:
shape: Tuple of integers, shape of returned TF-Keras variable.
low: Float, lower boundary of the output interval.
high: Float, upper boundary of the output interval.
dtype: String, dtype of returned TF-Keras variable.
name: String, name of returned TF-Keras variable.
seed: Integer, random seed.
Returns:
A TF-Keras variable, filled with drawn samples.
Example:
>>> kvar = tf.keras.backend.random_uniform_variable(shape=(2,3),
... low=0.0, high=1.0)
>>> kvar
<tf.Variable 'Variable:0' shape=(2, 3) dtype=float32, numpy=...,
dtype=float32)>
"""
if dtype is None:
dtype = floatx()
tf_dtype = tf.as_dtype(dtype)
if seed is None:
# ensure that randomness is conditioned by the Numpy RNG
seed = np.random.randint(10e8)
value = tf.compat.v1.random_uniform_initializer(
low, high, dtype=tf_dtype, seed=seed
)(shape)
return variable(value, dtype=dtype, name=name)
@keras_export("keras.backend.random_normal_variable")
@doc_controls.do_not_generate_docs
def random_normal_variable(
shape, mean, scale, dtype=None, name=None, seed=None
):
"""Instantiates a variable with values drawn from a normal distribution.
Args:
shape: Tuple of integers, shape of returned TF-Keras variable.
mean: Float, mean of the normal distribution.
scale: Float, standard deviation of the normal distribution.
dtype: String, dtype of returned TF-Keras variable.
name: String, name of returned TF-Keras variable.
seed: Integer, random seed.
Returns:
A TF-Keras variable, filled with drawn samples.
Example:
>>> kvar = tf.keras.backend.random_normal_variable(shape=(2,3),
... mean=0.0, scale=1.0)
>>> kvar
<tf.Variable 'Variable:0' shape=(2, 3) dtype=float32, numpy=...,
dtype=float32)>
"""
if dtype is None:
dtype = floatx()
tf_dtype = tf.as_dtype(dtype)
if seed is None:
# ensure that randomness is conditioned by the Numpy RNG
seed = np.random.randint(10e8)
value = tf.compat.v1.random_normal_initializer(
mean, scale, dtype=tf_dtype, seed=seed
)(shape)
return variable(value, dtype=dtype, name=name)
@keras_export("keras.backend.count_params")
@doc_controls.do_not_generate_docs
def count_params(x):
"""Returns the static number of elements in a variable or tensor.
Args:
x: Variable or tensor.
Returns:
Integer, the number of scalars in `x`.
Example:
>>> kvar = tf.keras.backend.zeros((2,3))
>>> tf.keras.backend.count_params(kvar)
6
>>> tf.keras.backend.eval(kvar)
array([[0., 0., 0.],
[0., 0., 0.]], dtype=float32)
"""
return np.prod(x.shape.as_list())
@keras_export("keras.backend.cast")
@tf.__internal__.dispatch.add_dispatch_support
@doc_controls.do_not_generate_docs
def cast(x, dtype):
"""Casts a tensor to a different dtype and returns it.
You can cast a TF-Keras variable but it still returns a TF-Keras tensor.
Args:
x: TF-Keras tensor (or variable).
dtype: String, either (`'float16'`, `'float32'`, or `'float64'`).
Returns:
TF-Keras tensor with dtype `dtype`.
Examples:
Cast a float32 variable to a float64 tensor
>>> input = tf.keras.backend.ones(shape=(1,3))
>>> print(input)
<tf.Variable 'Variable:0' shape=(1, 3) dtype=float32,
numpy=array([[1., 1., 1.]], dtype=float32)>
>>> cast_input = tf.keras.backend.cast(input, dtype='float64')
>>> print(cast_input)
tf.Tensor([[1. 1. 1.]], shape=(1, 3), dtype=float64)
"""
return tf.cast(x, dtype)
# UPDATES OPS
@keras_export("keras.backend.update")
@doc_controls.do_not_generate_docs
def update(x, new_x):
return tf.compat.v1.assign(x, new_x)
@keras_export("keras.backend.update_add")
@doc_controls.do_not_generate_docs
def update_add(x, increment):
"""Update the value of `x` by adding `increment`.
Args:
x: A Variable.
increment: A tensor of same shape as `x`.
Returns:
The variable `x` updated.
"""
return tf.compat.v1.assign_add(x, increment)
@keras_export("keras.backend.update_sub")
@doc_controls.do_not_generate_docs
def update_sub(x, decrement):
"""Update the value of `x` by subtracting `decrement`.
Args:
x: A Variable.
decrement: A tensor of same shape as `x`.
Returns:
The variable `x` updated.
"""
return tf.compat.v1.assign_sub(x, decrement)
@keras_export("keras.backend.moving_average_update")
@doc_controls.do_not_generate_docs
def moving_average_update(x, value, momentum):
"""Compute the exponential moving average of a value.
The moving average 'x' is updated with 'value' following:
```
x = x * momentum + value * (1 - momentum)
```
For example:
>>> x = tf.Variable(0.0)
>>> momentum=0.9
>>> moving_average_update(x, value = 2.0, momentum=momentum).numpy()
>>> x.numpy()
0.2
The result will be biased towards the initial value of the variable.
If the variable was initialized to zero, you can divide by
`1 - momentum ** num_updates` to debias it (Section 3 of
[Kingma et al., 2015](https://arxiv.org/abs/1412.6980)):
>>> num_updates = 1.0
>>> x_zdb = x/(1 - momentum**num_updates)
>>> x_zdb.numpy()
2.0
Args:
x: A Variable, the moving average.
value: A tensor with the same shape as `x`, the new value to be
averaged in.
momentum: The moving average momentum.
Returns:
The updated variable.
"""
if tf.__internal__.tf2.enabled():
momentum = tf.cast(momentum, x.dtype)
value = tf.cast(value, x.dtype)
return x.assign_sub((x - value) * (1 - momentum))
else:
return tf.__internal__.train.assign_moving_average(
x, value, momentum, zero_debias=True
)
# LINEAR ALGEBRA
@keras_export("keras.backend.dot")
@tf.__internal__.dispatch.add_dispatch_support
@doc_controls.do_not_generate_docs
def dot(x, y):
"""Multiplies 2 tensors (and/or variables) and returns a tensor.
This operation corresponds to `numpy.dot(a, b, out=None)`.
Args:
x: Tensor or variable.
y: Tensor or variable.
Returns:
A tensor, dot product of `x` and `y`.
Examples:
If inputs `x` and `y` are 2-D arrays, then it is equivalent to `tf.matmul`.
>>> x = tf.keras.backend.placeholder(shape=(2, 3))
>>> y = tf.keras.backend.placeholder(shape=(3, 4))
>>> xy = tf.keras.backend.dot(x, y)
>>> xy
<KerasTensor: shape=(2, 4) dtype=float32 ...>
>>> x = tf.keras.backend.placeholder(shape=(32, 28, 3))
>>> y = tf.keras.backend.placeholder(shape=(3, 4))
>>> xy = tf.keras.backend.dot(x, y)
>>> xy
<KerasTensor: shape=(32, 28, 4) dtype=float32 ...>
If `x` is an N-D array and `y` is an M-D array (where M>=2), it is a sum
product over the last axis of `x` and the second-to-last axis of `y`.
>>> x = tf.keras.backend.random_uniform_variable(
... shape=(2, 3), low=0., high=1.)
>>> y = tf.keras.backend.ones((4, 3, 5))
>>> xy = tf.keras.backend.dot(x, y)
>>> tf.keras.backend.int_shape(xy)
(2, 4, 5)
"""
if ndim(x) is not None and (ndim(x) > 2 or ndim(y) > 2):
x_shape = []
for i, s in zip(int_shape(x), tf.unstack(tf.shape(x))):
if i is not None:
x_shape.append(i)
else:
x_shape.append(s)
x_shape = tuple(x_shape)
y_shape = []
for i, s in zip(int_shape(y), tf.unstack(tf.shape(y))):
if i is not None:
y_shape.append(i)
else:
y_shape.append(s)
y_shape = tuple(y_shape)
y_permute_dim = list(range(ndim(y)))
y_permute_dim = [y_permute_dim.pop(-2)] + y_permute_dim
xt = tf.reshape(x, [-1, x_shape[-1]])
yt = tf.reshape(
tf.compat.v1.transpose(y, perm=y_permute_dim), [y_shape[-2], -1]
)
return tf.reshape(
tf.matmul(xt, yt), x_shape[:-1] + y_shape[:-2] + y_shape[-1:]
)
if is_sparse(x):
out = tf.sparse.sparse_dense_matmul(x, y)
else:
out = tf.matmul(x, y)
return out
@keras_export("keras.backend.batch_dot")
@tf.__internal__.dispatch.add_dispatch_support
@doc_controls.do_not_generate_docs
def batch_dot(x, y, axes=None):
"""Batchwise dot product.
`batch_dot` is used to compute dot product of `x` and `y` when
`x` and `y` are data in batch, i.e. in a shape of
`(batch_size, :)`.
`batch_dot` results in a tensor or variable with less dimensions
than the input. If the number of dimensions is reduced to 1,
we use `expand_dims` to make sure that ndim is at least 2.
Args:
x: TF-Keras tensor or variable with `ndim >= 2`.
y: TF-Keras tensor or variable with `ndim >= 2`.
axes: Tuple or list of integers with target dimensions, or single integer.
The sizes of `x.shape[axes[0]]` and `y.shape[axes[1]]` should be equal.
Returns:
A tensor with shape equal to the concatenation of `x`'s shape
(less the dimension that was summed over) and `y`'s shape
(less the batch dimension and the dimension that was summed over).
If the final rank is 1, we reshape it to `(batch_size, 1)`.
Examples:
>>> x_batch = tf.keras.backend.ones(shape=(32, 20, 1))
>>> y_batch = tf.keras.backend.ones(shape=(32, 30, 20))
>>> xy_batch_dot = tf.keras.backend.batch_dot(x_batch, y_batch, axes=(1, 2))
>>> tf.keras.backend.int_shape(xy_batch_dot)
(32, 1, 30)
Shape inference:
Let `x`'s shape be `(100, 20)` and `y`'s shape be `(100, 30, 20)`.
If `axes` is (1, 2), to find the output shape of resultant tensor,
loop through each dimension in `x`'s shape and `y`'s shape:
* `x.shape[0]` : 100 : append to output shape
* `x.shape[1]` : 20 : do not append to output shape,
dimension 1 of `x` has been summed over. (`dot_axes[0]` = 1)
* `y.shape[0]` : 100 : do not append to output shape,
always ignore first dimension of `y`
* `y.shape[1]` : 30 : append to output shape
* `y.shape[2]` : 20 : do not append to output shape,
dimension 2 of `y` has been summed over. (`dot_axes[1]` = 2)
`output_shape` = `(100, 30)`
"""
x_shape = int_shape(x)
y_shape = int_shape(y)
x_ndim = len(x_shape)
y_ndim = len(y_shape)
if x_ndim < 2 or y_ndim < 2:
raise ValueError(
"Cannot do batch_dot on inputs "
"with rank < 2. "
"Received inputs with shapes "
+ str(x_shape)
+ " and "
+ str(y_shape)
+ "."
)
x_batch_size = x_shape[0]
y_batch_size = y_shape[0]
if x_batch_size is not None and y_batch_size is not None:
if x_batch_size != y_batch_size:
raise ValueError(
"Cannot do batch_dot on inputs "
"with different batch sizes. "
"Received inputs with shapes "
+ str(x_shape)
+ " and "
+ str(y_shape)
+ "."
)
if isinstance(axes, int):
axes = [axes, axes]
if axes is None:
if y_ndim == 2:
axes = [x_ndim - 1, y_ndim - 1]
else:
axes = [x_ndim - 1, y_ndim - 2]
if py_any(isinstance(a, (list, tuple)) for a in axes):
raise ValueError(
"Multiple target dimensions are not supported. "
+ "Expected: None, int, (int, int), "
+ "Provided: "
+ str(axes)
)
# if tuple, convert to list.
axes = list(axes)
# convert negative indices.
if axes[0] < 0:
axes[0] += x_ndim
if axes[1] < 0:
axes[1] += y_ndim
# sanity checks
if 0 in axes:
raise ValueError(
"Cannot perform batch_dot over axis 0. "
"If your inputs are not batched, "
"add a dummy batch dimension to your "
"inputs using K.expand_dims(x, 0)"
)
a0, a1 = axes
d1 = x_shape[a0]
d2 = y_shape[a1]
if d1 is not None and d2 is not None and d1 != d2:
raise ValueError(
"Cannot do batch_dot on inputs with shapes "
+ str(x_shape)
+ " and "
+ str(y_shape)
+ " with axes="
+ str(axes)
+ ". x.shape[%d] != y.shape[%d] (%d != %d)."
% (axes[0], axes[1], d1, d2)
)
# backup ndims. Need them later.
orig_x_ndim = x_ndim
orig_y_ndim = y_ndim
# if rank is 2, expand to 3.
if x_ndim == 2:
x = tf.expand_dims(x, 1)
a0 += 1
x_ndim += 1
if y_ndim == 2:
y = tf.expand_dims(y, 2)
y_ndim += 1
# bring x's dimension to be reduced to last axis.
if a0 != x_ndim - 1:
pattern = list(range(x_ndim))
for i in range(a0, x_ndim - 1):
pattern[i] = pattern[i + 1]
pattern[-1] = a0
x = tf.compat.v1.transpose(x, pattern)
# bring y's dimension to be reduced to axis 1.
if a1 != 1:
pattern = list(range(y_ndim))
for i in range(a1, 1, -1):
pattern[i] = pattern[i - 1]
pattern[1] = a1
y = tf.compat.v1.transpose(y, pattern)
# normalize both inputs to rank 3.
if x_ndim > 3:
# squash middle dimensions of x.
x_shape = shape(x)
x_mid_dims = x_shape[1:-1]
x_squashed_shape = tf.stack([x_shape[0], -1, x_shape[-1]])
x = tf.reshape(x, x_squashed_shape)
x_squashed = True
else:
x_squashed = False
if y_ndim > 3:
# squash trailing dimensions of y.
y_shape = shape(y)
y_trail_dims = y_shape[2:]
y_squashed_shape = tf.stack([y_shape[0], y_shape[1], -1])
y = tf.reshape(y, y_squashed_shape)
y_squashed = True
else:
y_squashed = False
result = tf.matmul(x, y)
# if inputs were squashed, we have to reshape the matmul output.
output_shape = tf.shape(result)
do_reshape = False
if x_squashed:
output_shape = tf.concat(
[output_shape[:1], x_mid_dims, output_shape[-1:]], 0
)
do_reshape = True
if y_squashed:
output_shape = tf.concat([output_shape[:-1], y_trail_dims], 0)
do_reshape = True
if do_reshape:
result = tf.reshape(result, output_shape)
# if the inputs were originally rank 2, we remove the added 1 dim.
if orig_x_ndim == 2:
result = tf.squeeze(result, 1)
elif orig_y_ndim == 2:
result = tf.squeeze(result, -1)
return result
@keras_export("keras.backend.transpose")
@tf.__internal__.dispatch.add_dispatch_support
@doc_controls.do_not_generate_docs
def transpose(x):
"""Transposes a tensor and returns it.
Args:
x: Tensor or variable.
Returns:
A tensor.
Examples:
>>> var = tf.keras.backend.variable([[1, 2, 3], [4, 5, 6]])
>>> tf.keras.backend.eval(var)
array([[1., 2., 3.],
[4., 5., 6.]], dtype=float32)
>>> var_transposed = tf.keras.backend.transpose(var)
>>> tf.keras.backend.eval(var_transposed)
array([[1., 4.],
[2., 5.],
[3., 6.]], dtype=float32)
>>> input = tf.keras.backend.placeholder((2, 3))
>>> input
<KerasTensor: shape=(2, 3) dtype=float32 ...>
>>> input_transposed = tf.keras.backend.transpose(input)
>>> input_transposed
<KerasTensor: shape=(3, 2) dtype=float32 ...>
"""
return tf.compat.v1.transpose(x)
@keras_export("keras.backend.gather")
@tf.__internal__.dispatch.add_dispatch_support
@doc_controls.do_not_generate_docs
def gather(reference, indices):
"""Retrieves the elements of indices `indices` in the tensor `reference`.
Args:
reference: A tensor.
indices: An integer tensor of indices.
Returns:
A tensor of same type as `reference`.
Examples:
>>> var = tf.keras.backend.variable([[1, 2, 3], [4, 5, 6]])
>>> tf.keras.backend.eval(var)
array([[1., 2., 3.],
[4., 5., 6.]], dtype=float32)
>>> var_gathered = tf.keras.backend.gather(var, [0])
>>> tf.keras.backend.eval(var_gathered)
array([[1., 2., 3.]], dtype=float32)
>>> var_gathered = tf.keras.backend.gather(var, [1])
>>> tf.keras.backend.eval(var_gathered)
array([[4., 5., 6.]], dtype=float32)
>>> var_gathered = tf.keras.backend.gather(var, [0,1,0])
>>> tf.keras.backend.eval(var_gathered)
array([[1., 2., 3.],
[4., 5., 6.],
[1., 2., 3.]], dtype=float32)
"""
return tf.compat.v1.gather(reference, indices)
# ELEMENT-WISE OPERATIONS
@keras_export("keras.backend.max")
@tf.__internal__.dispatch.add_dispatch_support
@doc_controls.do_not_generate_docs
def max(x, axis=None, keepdims=False):
"""Maximum value in a tensor.
Args:
x: A tensor or variable.
axis: An integer, the axis to find maximum values.
keepdims: A boolean, whether to keep the dimensions or not.
If `keepdims` is `False`, the rank of the tensor is reduced
by 1. If `keepdims` is `True`,
the reduced dimension is retained with length 1.
Returns:
A tensor with maximum values of `x`.
"""
return tf.reduce_max(x, axis, keepdims)
@keras_export("keras.backend.min")
@tf.__internal__.dispatch.add_dispatch_support
@doc_controls.do_not_generate_docs
def min(x, axis=None, keepdims=False):
"""Minimum value in a tensor.
Args:
x: A tensor or variable.
axis: An integer, the axis to find minimum values.
keepdims: A boolean, whether to keep the dimensions or not.
If `keepdims` is `False`, the rank of the tensor is reduced
by 1. If `keepdims` is `True`,
the reduced dimension is retained with length 1.
Returns:
A tensor with minimum values of `x`.
"""
return tf.reduce_min(x, axis, keepdims)
@keras_export("keras.backend.sum")
@tf.__internal__.dispatch.add_dispatch_support
@doc_controls.do_not_generate_docs
def sum(x, axis=None, keepdims=False):
"""Sum of the values in a tensor, alongside the specified axis.
Args:
x: A tensor or variable.
axis: An integer, the axis to sum over.
keepdims: A boolean, whether to keep the dimensions or not.
If `keepdims` is `False`, the rank of the tensor is reduced
by 1. If `keepdims` is `True`,
the reduced dimension is retained with length 1.
Returns:
A tensor with sum of `x`.
"""
return tf.reduce_sum(x, axis, keepdims)
@keras_export("keras.backend.prod")
@tf.__internal__.dispatch.add_dispatch_support
@doc_controls.do_not_generate_docs
def prod(x, axis=None, keepdims=False):
"""Multiplies the values in a tensor, alongside the specified axis.
Args:
x: A tensor or variable.
axis: An integer, the axis to compute the product.
keepdims: A boolean, whether to keep the dimensions or not.
If `keepdims` is `False`, the rank of the tensor is reduced
by 1. If `keepdims` is `True`,
the reduced dimension is retained with length 1.
Returns:
A tensor with the product of elements of `x`.
"""
return tf.reduce_prod(x, axis, keepdims)
@keras_export("keras.backend.cumsum")
@tf.__internal__.dispatch.add_dispatch_support
@doc_controls.do_not_generate_docs
def cumsum(x, axis=0):
"""Cumulative sum of the values in a tensor, alongside the specified axis.
Args:
x: A tensor or variable.
axis: An integer, the axis to compute the sum.
Returns:
A tensor of the cumulative sum of values of `x` along `axis`.
"""
return tf.cumsum(x, axis=axis)
@keras_export("keras.backend.cumprod")
@tf.__internal__.dispatch.add_dispatch_support
@doc_controls.do_not_generate_docs
def cumprod(x, axis=0):
"""Cumulative product of the values in a tensor alongside `axis`.
Args:
x: A tensor or variable.
axis: An integer, the axis to compute the product.
Returns:
A tensor of the cumulative product of values of `x` along `axis`.
"""
return tf.math.cumprod(x, axis=axis)
@keras_export("keras.backend.var")
@doc_controls.do_not_generate_docs
def var(x, axis=None, keepdims=False):
"""Variance of a tensor, alongside the specified axis.
Args:
x: A tensor or variable.
axis: An integer, the axis to compute the variance.
keepdims: A boolean, whether to keep the dimensions or not.
If `keepdims` is `False`, the rank of the tensor is reduced
by 1. If `keepdims` is `True`,
the reduced dimension is retained with length 1.
Returns:
A tensor with the variance of elements of `x`.
"""
if x.dtype.base_dtype == tf.bool:
x = tf.cast(x, floatx())
return tf.math.reduce_variance(x, axis=axis, keepdims=keepdims)
@keras_export("keras.backend.std")
@tf.__internal__.dispatch.add_dispatch_support
@doc_controls.do_not_generate_docs
def std(x, axis=None, keepdims=False):
"""Standard deviation of a tensor, alongside the specified axis.
It is an alias to `tf.math.reduce_std`.
Args:
x: A tensor or variable. It should have numerical dtypes. Boolean type
inputs will be converted to float.
axis: An integer, the axis to compute the standard deviation. If `None`
(the default), reduces all dimensions. Must be in the range
`[-rank(x), rank(x))`.
keepdims: A boolean, whether to keep the dimensions or not.
If `keepdims` is `False`, the rank of the tensor is reduced
by 1. If `keepdims` is `True`, the reduced dimension is retained
with length 1.
Returns:
A tensor with the standard deviation of elements of `x` with same dtype.
Boolean type input will be converted to float.
"""
if x.dtype.base_dtype == tf.bool:
x = tf.cast(x, floatx())
return tf.math.reduce_std(x, axis=axis, keepdims=keepdims)
@keras_export("keras.backend.mean")
@tf.__internal__.dispatch.add_dispatch_support
@doc_controls.do_not_generate_docs
def mean(x, axis=None, keepdims=False):
"""Mean of a tensor, alongside the specified axis.
Args:
x: A tensor or variable.
axis: A list of integer. Axes to compute the mean.
keepdims: A boolean, whether to keep the dimensions or not.
If `keepdims` is `False`, the rank of the tensor is reduced
by 1 for each entry in `axis`. If `keepdims` is `True`,
the reduced dimensions are retained with length 1.
Returns:
A tensor with the mean of elements of `x`.
"""
if x.dtype.base_dtype == tf.bool:
x = tf.cast(x, floatx())
return tf.reduce_mean(x, axis, keepdims)
@keras_export("keras.backend.any")
@tf.__internal__.dispatch.add_dispatch_support
@doc_controls.do_not_generate_docs
def any(x, axis=None, keepdims=False):
"""Bitwise reduction (logical OR).
Args:
x: Tensor or variable.
axis: axis along which to perform the reduction.
keepdims: whether the drop or broadcast the reduction axes.
Returns:
A uint8 tensor (0s and 1s).
"""
x = tf.cast(x, tf.bool)
return tf.reduce_any(x, axis, keepdims)
@keras_export("keras.backend.all")
@tf.__internal__.dispatch.add_dispatch_support
@doc_controls.do_not_generate_docs
def all(x, axis=None, keepdims=False):
"""Bitwise reduction (logical AND).
Args:
x: Tensor or variable.
axis: axis along which to perform the reduction.
keepdims: whether the drop or broadcast the reduction axes.
Returns:
A uint8 tensor (0s and 1s).
"""
x = tf.cast(x, tf.bool)
return tf.reduce_all(x, axis, keepdims)
@keras_export("keras.backend.argmax")
@tf.__internal__.dispatch.add_dispatch_support
@doc_controls.do_not_generate_docs
def argmax(x, axis=-1):
"""Returns the index of the maximum value along an axis.
Args:
x: Tensor or variable.
axis: axis along which to perform the reduction.
Returns:
A tensor.
"""
return tf.argmax(x, axis)
@keras_export("keras.backend.argmin")
@tf.__internal__.dispatch.add_dispatch_support
@doc_controls.do_not_generate_docs
def argmin(x, axis=-1):
"""Returns the index of the minimum value along an axis.
Args:
x: Tensor or variable.
axis: axis along which to perform the reduction.
Returns:
A tensor.
"""
return tf.argmin(x, axis)
@keras_export("keras.backend.square")
@tf.__internal__.dispatch.add_dispatch_support
@doc_controls.do_not_generate_docs
def square(x):
"""Element-wise square.
Args:
x: Tensor or variable.
Returns:
A tensor.
"""
return tf.square(x)
@keras_export("keras.backend.abs")
@tf.__internal__.dispatch.add_dispatch_support
@doc_controls.do_not_generate_docs
def abs(x):
"""Element-wise absolute value.
Args:
x: Tensor or variable.
Returns:
A tensor.
"""
return tf.abs(x)
@keras_export("keras.backend.sqrt")
@tf.__internal__.dispatch.add_dispatch_support
@doc_controls.do_not_generate_docs
def sqrt(x):
"""Element-wise square root.
This function clips negative tensor values to 0 before computing the
square root.
Args:
x: Tensor or variable.
Returns:
A tensor.
"""
zero = _constant_to_tensor(0.0, x.dtype.base_dtype)
x = tf.maximum(x, zero)
return tf.sqrt(x)
@keras_export("keras.backend.exp")
@tf.__internal__.dispatch.add_dispatch_support
@doc_controls.do_not_generate_docs
def exp(x):
"""Element-wise exponential.
Args:
x: Tensor or variable.
Returns:
A tensor.
"""
return tf.exp(x)
@keras_export("keras.backend.log")
@tf.__internal__.dispatch.add_dispatch_support
@doc_controls.do_not_generate_docs
def log(x):
"""Element-wise log.
Args:
x: Tensor or variable.
Returns:
A tensor.
"""
return tf.math.log(x)
def logsumexp(x, axis=None, keepdims=False):
"""Computes log(sum(exp(elements across dimensions of a tensor))).
This function is more numerically stable than log(sum(exp(x))).
It avoids overflows caused by taking the exp of large inputs and
underflows caused by taking the log of small inputs.
Args:
x: A tensor or variable.
axis: An integer, the axis to reduce over.
keepdims: A boolean, whether to keep the dimensions or not.
If `keepdims` is `False`, the rank of the tensor is reduced
by 1. If `keepdims` is `True`, the reduced dimension is
retained with length 1.
Returns:
The reduced tensor.
"""
return tf.reduce_logsumexp(x, axis, keepdims)
@keras_export("keras.backend.round")
@tf.__internal__.dispatch.add_dispatch_support
@doc_controls.do_not_generate_docs
def round(x):
"""Element-wise rounding to the closest integer.
In case of tie, the rounding mode used is "half to even".
Args:
x: Tensor or variable.
Returns:
A tensor.
"""
return tf.round(x)
@keras_export("keras.backend.sign")
@tf.__internal__.dispatch.add_dispatch_support
@doc_controls.do_not_generate_docs
def sign(x):
"""Element-wise sign.
Args:
x: Tensor or variable.
Returns:
A tensor.
"""
return tf.sign(x)
@keras_export("keras.backend.pow")
@tf.__internal__.dispatch.add_dispatch_support
@doc_controls.do_not_generate_docs
def pow(x, a):
"""Element-wise exponentiation.
Args:
x: Tensor or variable.
a: Python integer.
Returns:
A tensor.
"""
return tf.pow(x, a)
@keras_export("keras.backend.clip")
@tf.__internal__.dispatch.add_dispatch_support
@doc_controls.do_not_generate_docs
def clip(x, min_value, max_value):
"""Element-wise value clipping.
Args:
x: Tensor or variable.
min_value: Python float, integer, or tensor.
max_value: Python float, integer, or tensor.
Returns:
A tensor.
"""
if isinstance(min_value, (int, float)) and isinstance(
max_value, (int, float)
):
if max_value < min_value:
max_value = min_value
if min_value is None:
min_value = -np.inf
if max_value is None:
max_value = np.inf
return tf.clip_by_value(x, min_value, max_value)
@keras_export("keras.backend.equal")
@tf.__internal__.dispatch.add_dispatch_support
@doc_controls.do_not_generate_docs
def equal(x, y):
"""Element-wise equality between two tensors.
Args:
x: Tensor or variable.
y: Tensor or variable.
Returns:
A bool tensor.
"""
return tf.equal(x, y)
@keras_export("keras.backend.not_equal")
@tf.__internal__.dispatch.add_dispatch_support
@doc_controls.do_not_generate_docs
def not_equal(x, y):
"""Element-wise inequality between two tensors.
Args:
x: Tensor or variable.
y: Tensor or variable.
Returns:
A bool tensor.
"""
return tf.not_equal(x, y)
@keras_export("keras.backend.greater")
@tf.__internal__.dispatch.add_dispatch_support
@doc_controls.do_not_generate_docs
def greater(x, y):
"""Element-wise truth value of (x > y).
Args:
x: Tensor or variable.
y: Tensor or variable.
Returns:
A bool tensor.
"""
return tf.greater(x, y)
@keras_export("keras.backend.greater_equal")
@tf.__internal__.dispatch.add_dispatch_support
@doc_controls.do_not_generate_docs
def greater_equal(x, y):
"""Element-wise truth value of (x >= y).
Args:
x: Tensor or variable.
y: Tensor or variable.
Returns:
A bool tensor.
"""
return tf.greater_equal(x, y)
@keras_export("keras.backend.less")
@tf.__internal__.dispatch.add_dispatch_support
@doc_controls.do_not_generate_docs
def less(x, y):
"""Element-wise truth value of (x < y).
Args:
x: Tensor or variable.
y: Tensor or variable.
Returns:
A bool tensor.
"""
return tf.less(x, y)
@keras_export("keras.backend.less_equal")
@tf.__internal__.dispatch.add_dispatch_support
@doc_controls.do_not_generate_docs
def less_equal(x, y):
"""Element-wise truth value of (x <= y).
Args:
x: Tensor or variable.
y: Tensor or variable.
Returns:
A bool tensor.
"""
return tf.less_equal(x, y)
@keras_export("keras.backend.maximum")
@tf.__internal__.dispatch.add_dispatch_support
@doc_controls.do_not_generate_docs
def maximum(x, y):
"""Element-wise maximum of two tensors.
Args:
x: Tensor or variable.
y: Tensor or variable.
Returns:
A tensor with the element wise maximum value(s) of `x` and `y`.
Examples:
>>> x = tf.Variable([[1, 2], [3, 4]])
>>> y = tf.Variable([[2, 1], [0, -1]])
>>> m = tf.keras.backend.maximum(x, y)
>>> m
<tf.Tensor: shape=(2, 2), dtype=int32, numpy=
array([[2, 2],
[3, 4]], dtype=int32)>
"""
return tf.maximum(x, y)
@keras_export("keras.backend.minimum")
@tf.__internal__.dispatch.add_dispatch_support
@doc_controls.do_not_generate_docs
def minimum(x, y):
"""Element-wise minimum of two tensors.
Args:
x: Tensor or variable.
y: Tensor or variable.
Returns:
A tensor.
"""
return tf.minimum(x, y)
@keras_export("keras.backend.sin")
@tf.__internal__.dispatch.add_dispatch_support
@doc_controls.do_not_generate_docs
def sin(x):
"""Computes sin of x element-wise.
Args:
x: Tensor or variable.
Returns:
A tensor.
"""
return tf.sin(x)
@keras_export("keras.backend.cos")
@tf.__internal__.dispatch.add_dispatch_support
@doc_controls.do_not_generate_docs
def cos(x):
"""Computes cos of x element-wise.
Args:
x: Tensor or variable.
Returns:
A tensor.
"""
return tf.cos(x)
def _regular_normalize_batch_in_training(
x, gamma, beta, reduction_axes, epsilon=1e-3
):
"""Non-fused version of `normalize_batch_in_training`.
Args:
x: Input tensor or variable.
gamma: Tensor by which to scale the input.
beta: Tensor with which to center the input.
reduction_axes: iterable of integers,
axes over which to normalize.
epsilon: Fuzz factor.
Returns:
A tuple length of 3, `(normalized_tensor, mean, variance)`.
"""
mean, var = tf.compat.v1.nn.moments(x, reduction_axes, None, None, False)
normed = tf.nn.batch_normalization(x, mean, var, beta, gamma, epsilon)
return normed, mean, var
def _broadcast_normalize_batch_in_training(
x, gamma, beta, reduction_axes, epsilon=1e-3
):
"""Non-fused, broadcast version of `normalize_batch_in_training`.
Args:
x: Input tensor or variable.
gamma: Tensor by which to scale the input.
beta: Tensor with which to center the input.
reduction_axes: iterable of integers,
axes over which to normalize.
epsilon: Fuzz factor.
Returns:
A tuple length of 3, `(normalized_tensor, mean, variance)`.
"""
mean, var = tf.compat.v1.nn.moments(x, reduction_axes, None, None, False)
target_shape = []
for axis in range(ndim(x)):
if axis in reduction_axes:
target_shape.append(1)
else:
target_shape.append(tf.shape(x)[axis])
target_shape = tf.stack(target_shape)
broadcast_mean = tf.reshape(mean, target_shape)
broadcast_var = tf.reshape(var, target_shape)
if gamma is None:
broadcast_gamma = None
else:
broadcast_gamma = tf.reshape(gamma, target_shape)
if beta is None:
broadcast_beta = None
else:
broadcast_beta = tf.reshape(beta, target_shape)
normed = tf.nn.batch_normalization(
x,
broadcast_mean,
broadcast_var,
broadcast_beta,
broadcast_gamma,
epsilon,
)
return normed, mean, var
def _fused_normalize_batch_in_training(
x, gamma, beta, reduction_axes, epsilon=1e-3
):
"""Fused version of `normalize_batch_in_training`.
Args:
x: Input tensor or variable.
gamma: Tensor by which to scale the input.
beta: Tensor with which to center the input.
reduction_axes: iterable of integers,
axes over which to normalize.
epsilon: Fuzz factor.
Returns:
A tuple length of 3, `(normalized_tensor, mean, variance)`.
"""
if list(reduction_axes) == [0, 1, 2]:
normalization_axis = 3
tf_data_format = "NHWC"
else:
normalization_axis = 1
tf_data_format = "NCHW"
if gamma is None:
gamma = tf.constant(
1.0, dtype=x.dtype, shape=[x.shape[normalization_axis]]
)
if beta is None:
beta = tf.constant(
0.0, dtype=x.dtype, shape=[x.shape[normalization_axis]]
)
return tf.compat.v1.nn.fused_batch_norm(
x, gamma, beta, epsilon=epsilon, data_format=tf_data_format
)
@keras_export("keras.backend.normalize_batch_in_training")
@doc_controls.do_not_generate_docs
def normalize_batch_in_training(x, gamma, beta, reduction_axes, epsilon=1e-3):
"""Computes mean and std for batch then apply batch_normalization on batch.
Args:
x: Input tensor or variable.
gamma: Tensor by which to scale the input.
beta: Tensor with which to center the input.
reduction_axes: iterable of integers,
axes over which to normalize.
epsilon: Fuzz factor.
Returns:
A tuple length of 3, `(normalized_tensor, mean, variance)`.
"""
if ndim(x) == 4 and list(reduction_axes) in [[0, 1, 2], [0, 2, 3]]:
if not _has_nchw_support() and list(reduction_axes) == [0, 2, 3]:
return _broadcast_normalize_batch_in_training(
x, gamma, beta, reduction_axes, epsilon=epsilon
)
return _fused_normalize_batch_in_training(
x, gamma, beta, reduction_axes, epsilon=epsilon
)
else:
if sorted(reduction_axes) == list(range(ndim(x)))[:-1]:
return _regular_normalize_batch_in_training(
x, gamma, beta, reduction_axes, epsilon=epsilon
)
else:
return _broadcast_normalize_batch_in_training(
x, gamma, beta, reduction_axes, epsilon=epsilon
)
@keras_export("keras.backend.batch_normalization")
@tf.__internal__.dispatch.add_dispatch_support
@doc_controls.do_not_generate_docs
def batch_normalization(x, mean, var, beta, gamma, axis=-1, epsilon=1e-3):
"""Applies batch normalization on x given mean, var, beta and gamma.
I.e. returns:
`output = (x - mean) / (sqrt(var) + epsilon) * gamma + beta`
Args:
x: Input tensor or variable.
mean: Mean of batch.
var: Variance of batch.
beta: Tensor with which to center the input.
gamma: Tensor by which to scale the input.
axis: Integer, the axis that should be normalized.
(typically the features axis).
epsilon: Fuzz factor.
Returns:
A tensor.
"""
if ndim(x) == 4:
# The CPU implementation of `fused_batch_norm` only supports NHWC
if axis == 1 or axis == -3:
tf_data_format = "NCHW"
elif axis == 3 or axis == -1:
tf_data_format = "NHWC"
else:
tf_data_format = None
if (
tf_data_format == "NHWC"
or tf_data_format == "NCHW"
and _has_nchw_support()
):
# The mean / var / beta / gamma tensors may be broadcasted
# so they may have extra axes of size 1, which should be squeezed.
if ndim(mean) > 1:
mean = tf.reshape(mean, [-1])
if ndim(var) > 1:
var = tf.reshape(var, [-1])
if beta is None:
beta = zeros_like(mean)
elif ndim(beta) > 1:
beta = tf.reshape(beta, [-1])
if gamma is None:
gamma = ones_like(mean)
elif ndim(gamma) > 1:
gamma = tf.reshape(gamma, [-1])
y, _, _ = tf.compat.v1.nn.fused_batch_norm(
x,
gamma,
beta,
epsilon=epsilon,
mean=mean,
variance=var,
data_format=tf_data_format,
is_training=False,
)
return y
return tf.nn.batch_normalization(x, mean, var, beta, gamma, epsilon)
# SHAPE OPERATIONS
@keras_export("keras.backend.concatenate")
@tf.__internal__.dispatch.add_dispatch_support
@doc_controls.do_not_generate_docs
def concatenate(tensors, axis=-1):
"""Concatenates a list of tensors alongside the specified axis.
Args:
tensors: list of tensors to concatenate.
axis: concatenation axis.
Returns:
A tensor.
Example:
>>> a = tf.constant([[1, 2, 3], [4, 5, 6], [7, 8, 9]])
>>> b = tf.constant([[10, 20, 30], [40, 50, 60], [70, 80, 90]])
>>> tf.keras.backend.concatenate((a, b), axis=-1)
<tf.Tensor: shape=(3, 6), dtype=int32, numpy=
array([[ 1, 2, 3, 10, 20, 30],
[ 4, 5, 6, 40, 50, 60],
[ 7, 8, 9, 70, 80, 90]], dtype=int32)>
"""
if axis < 0:
rank = ndim(tensors[0])
if rank:
axis %= rank
else:
axis = 0
if py_all(is_sparse(x) for x in tensors):
return tf.compat.v1.sparse_concat(axis, tensors)
elif py_all(isinstance(x, tf.RaggedTensor) for x in tensors):
return tf.concat(tensors, axis)
else:
return tf.concat([to_dense(x) for x in tensors], axis)
@keras_export("keras.backend.reshape")
@tf.__internal__.dispatch.add_dispatch_support
@doc_controls.do_not_generate_docs
def reshape(x, shape):
"""Reshapes a tensor to the specified shape.
Args:
x: Tensor or variable.
shape: Target shape tuple.
Returns:
A tensor.
Example:
>>> a = tf.constant([[1, 2, 3], [4, 5, 6], [7, 8, 9], [10, 11, 12]])
>>> a
<tf.Tensor: shape=(4, 3), dtype=int32, numpy=
array([[ 1, 2, 3],
[ 4, 5, 6],
[ 7, 8, 9],
[10, 11, 12]], dtype=int32)>
>>> tf.keras.backend.reshape(a, shape=(2, 6))
<tf.Tensor: shape=(2, 6), dtype=int32, numpy=
array([[ 1, 2, 3, 4, 5, 6],
[ 7, 8, 9, 10, 11, 12]], dtype=int32)>
"""
return tf.reshape(x, shape)
@keras_export("keras.backend.permute_dimensions")
@tf.__internal__.dispatch.add_dispatch_support
@doc_controls.do_not_generate_docs
def permute_dimensions(x, pattern):
"""Permutes axes in a tensor.
Args:
x: Tensor or variable.
pattern: A tuple of
dimension indices, e.g. `(0, 2, 1)`.
Returns:
A tensor.
Example:
>>> a = tf.constant([[1, 2, 3], [4, 5, 6], [7, 8, 9], [10, 11, 12]])
>>> a
<tf.Tensor: shape=(4, 3), dtype=int32, numpy=
array([[ 1, 2, 3],
[ 4, 5, 6],
[ 7, 8, 9],
[10, 11, 12]], dtype=int32)>
>>> tf.keras.backend.permute_dimensions(a, pattern=(1, 0))
<tf.Tensor: shape=(3, 4), dtype=int32, numpy=
array([[ 1, 4, 7, 10],
[ 2, 5, 8, 11],
[ 3, 6, 9, 12]], dtype=int32)>
"""
return tf.compat.v1.transpose(x, perm=pattern)
@keras_export("keras.backend.resize_images")
@tf.__internal__.dispatch.add_dispatch_support
@doc_controls.do_not_generate_docs
def resize_images(
x, height_factor, width_factor, data_format, interpolation="nearest"
):
"""Resizes the images contained in a 4D tensor.
Args:
x: Tensor or variable to resize.
height_factor: Positive integer.
width_factor: Positive integer.
data_format: One of `"channels_first"`, `"channels_last"`.
interpolation: A string, one of `"area"`, `"bicubic"`, `"bilinear"`,
`"gaussian"`, `"lanczos3"`, `"lanczos5"`, `"mitchellcubic"`,
`"nearest"`.
Returns:
A tensor.
Raises:
ValueError: in case of incorrect value for
`data_format` or `interpolation`.
"""
if data_format == "channels_first":
rows, cols = 2, 3
elif data_format == "channels_last":
rows, cols = 1, 2
else:
raise ValueError(f"Invalid `data_format` argument: {data_format}")
new_shape = x.shape[rows : cols + 1]
if new_shape.is_fully_defined():
new_shape = tf.constant(new_shape.as_list(), dtype="int32")
else:
new_shape = tf.shape(x)[rows : cols + 1]
new_shape *= tf.constant(
np.array([height_factor, width_factor], dtype="int32")
)
if data_format == "channels_first":
x = permute_dimensions(x, [0, 2, 3, 1])
interpolations = {
"area": tf.image.ResizeMethod.AREA,
"bicubic": tf.image.ResizeMethod.BICUBIC,
"bilinear": tf.image.ResizeMethod.BILINEAR,
"gaussian": tf.image.ResizeMethod.GAUSSIAN,
"lanczos3": tf.image.ResizeMethod.LANCZOS3,
"lanczos5": tf.image.ResizeMethod.LANCZOS5,
"mitchellcubic": tf.image.ResizeMethod.MITCHELLCUBIC,
"nearest": tf.image.ResizeMethod.NEAREST_NEIGHBOR,
}
interploations_list = '"' + '", "'.join(interpolations.keys()) + '"'
if interpolation in interpolations:
x = tf.image.resize(x, new_shape, method=interpolations[interpolation])
else:
raise ValueError(
"`interpolation` argument should be one of: "
f'{interploations_list}. Received: "{interpolation}".'
)
if data_format == "channels_first":
x = permute_dimensions(x, [0, 3, 1, 2])
return x
@keras_export("keras.backend.resize_volumes")
@tf.__internal__.dispatch.add_dispatch_support
@doc_controls.do_not_generate_docs
def resize_volumes(x, depth_factor, height_factor, width_factor, data_format):
"""Resizes the volume contained in a 5D tensor.
Args:
x: Tensor or variable to resize.
depth_factor: Positive integer.
height_factor: Positive integer.
width_factor: Positive integer.
data_format: One of `"channels_first"`, `"channels_last"`.
Returns:
A tensor.
Raises:
ValueError: if `data_format` is neither
`channels_last` or `channels_first`.
"""
if data_format == "channels_first":
output = repeat_elements(x, depth_factor, axis=2)
output = repeat_elements(output, height_factor, axis=3)
output = repeat_elements(output, width_factor, axis=4)
return output
elif data_format == "channels_last":
output = repeat_elements(x, depth_factor, axis=1)
output = repeat_elements(output, height_factor, axis=2)
output = repeat_elements(output, width_factor, axis=3)
return output
else:
raise ValueError("Invalid data_format: " + str(data_format))
@keras_export("keras.backend.repeat_elements")
@tf.__internal__.dispatch.add_dispatch_support
@doc_controls.do_not_generate_docs
def repeat_elements(x, rep, axis):
"""Repeats the elements of a tensor along an axis, like `np.repeat`.
If `x` has shape `(s1, s2, s3)` and `axis` is `1`, the output
will have shape `(s1, s2 * rep, s3)`.
Args:
x: Tensor or variable.
rep: Python integer, number of times to repeat.
axis: Axis along which to repeat.
Returns:
A tensor.
Example:
>>> b = tf.constant([1, 2, 3])
>>> tf.keras.backend.repeat_elements(b, rep=2, axis=0)
<tf.Tensor: shape=(6,), dtype=int32,
numpy=array([1, 1, 2, 2, 3, 3], dtype=int32)>
"""
x_shape = x.shape.as_list()
# For static axis
if x_shape[axis] is not None:
# slices along the repeat axis
splits = tf.split(value=x, num_or_size_splits=x_shape[axis], axis=axis)
# repeat each slice the given number of reps
x_rep = [s for s in splits for _ in range(rep)]
return concatenate(x_rep, axis)
# Here we use tf.tile to mimic behavior of np.repeat so that
# we can handle dynamic shapes (that include None).
# To do that, we need an auxiliary axis to repeat elements along
# it and then merge them along the desired axis.
# Repeating
auxiliary_axis = axis + 1
x_shape = tf.shape(x)
x_rep = tf.expand_dims(x, axis=auxiliary_axis)
reps = np.ones(len(x.shape) + 1)
reps[auxiliary_axis] = rep
x_rep = tf.tile(x_rep, reps)
# Merging
reps = np.delete(reps, auxiliary_axis)
reps[axis] = rep
reps = tf.constant(reps, dtype="int32")
x_shape *= reps
x_rep = tf.reshape(x_rep, x_shape)
# Fix shape representation
x_shape = x.shape.as_list()
x_rep.set_shape(x_shape)
x_rep._keras_shape = tuple(x_shape)
return x_rep
@keras_export("keras.backend.repeat")
@tf.__internal__.dispatch.add_dispatch_support
@doc_controls.do_not_generate_docs
def repeat(x, n):
"""Repeats a 2D tensor.
if `x` has shape (samples, dim) and `n` is `2`,
the output will have shape `(samples, 2, dim)`.
Args:
x: Tensor or variable.
n: Python integer, number of times to repeat.
Returns:
A tensor.
Example:
>>> b = tf.constant([[1, 2], [3, 4]])
>>> b
<tf.Tensor: shape=(2, 2), dtype=int32, numpy=
array([[1, 2],
[3, 4]], dtype=int32)>
>>> tf.keras.backend.repeat(b, n=2)
<tf.Tensor: shape=(2, 2, 2), dtype=int32, numpy=
array([[[1, 2],
[1, 2]],
[[3, 4],
[3, 4]]], dtype=int32)>
"""
assert ndim(x) == 2
x = tf.expand_dims(x, 1)
pattern = tf.stack([1, n, 1])
return tf.tile(x, pattern)
@keras_export("keras.backend.arange")
@tf.__internal__.dispatch.add_dispatch_support
@doc_controls.do_not_generate_docs
def arange(start, stop=None, step=1, dtype="int32"):
"""Creates a 1D tensor containing a sequence of integers.
The function arguments use the same convention as
Theano's arange: if only one argument is provided,
it is in fact the "stop" argument and "start" is 0.
The default type of the returned tensor is `'int32'` to
match TensorFlow's default.
Args:
start: Start value.
stop: Stop value.
step: Difference between two successive values.
dtype: Integer dtype to use.
Returns:
An integer tensor.
Example:
>>> tf.keras.backend.arange(start=0, stop=10, step=1.5)
<tf.Tensor: shape=(7,), dtype=float32,
numpy=array([0. , 1.5, 3. , 4.5, 6. , 7.5, 9. ], dtype=float32)>
"""
# Match the behavior of numpy and Theano by returning an empty sequence.
if stop is None and start < 0:
start = 0
result = tf.range(start, limit=stop, delta=step, name="arange")
if dtype != "int32":
result = cast(result, dtype)
return result
@keras_export("keras.backend.tile")
@tf.__internal__.dispatch.add_dispatch_support
@doc_controls.do_not_generate_docs
def tile(x, n):
"""Creates a tensor by tiling `x` by `n`.
Args:
x: A tensor or variable
n: A list of integer. The length must be the same as the number of
dimensions in `x`.
Returns:
A tiled tensor.
"""
if isinstance(n, int):
n = [n]
return tf.tile(x, n)
@keras_export("keras.backend.flatten")
@tf.__internal__.dispatch.add_dispatch_support
@doc_controls.do_not_generate_docs
def flatten(x):
"""Flatten a tensor.
Args:
x: A tensor or variable.
Returns:
A tensor, reshaped into 1-D
Example:
>>> b = tf.constant([[1, 2], [3, 4]])
>>> b
<tf.Tensor: shape=(2, 2), dtype=int32, numpy=
array([[1, 2],
[3, 4]], dtype=int32)>
>>> tf.keras.backend.flatten(b)
<tf.Tensor: shape=(4,), dtype=int32,
numpy=array([1, 2, 3, 4], dtype=int32)>
"""
return tf.reshape(x, [-1])
@keras_export("keras.backend.batch_flatten")
@tf.__internal__.dispatch.add_dispatch_support
@doc_controls.do_not_generate_docs
def batch_flatten(x):
"""Turn a nD tensor into a 2D tensor with same 0th dimension.
In other words, it flattens each data samples of a batch.
Args:
x: A tensor or variable.
Returns:
A tensor.
Examples:
Flattening a 3D tensor to 2D by collapsing the last dimension.
>>> x_batch = tf.keras.backend.ones(shape=(2, 3, 4, 5))
>>> x_batch_flatten = batch_flatten(x_batch)
>>> tf.keras.backend.int_shape(x_batch_flatten)
(2, 60)
"""
x = tf.reshape(x, tf.stack([-1, prod(shape(x)[1:])]))
return x
@keras_export("keras.backend.expand_dims")
@tf.__internal__.dispatch.add_dispatch_support
@doc_controls.do_not_generate_docs
def expand_dims(x, axis=-1):
"""Adds a 1-sized dimension at index "axis".
Args:
x: A tensor or variable.
axis: Position where to add a new axis.
Returns:
A tensor with expanded dimensions.
"""
return tf.expand_dims(x, axis)
@keras_export("keras.backend.squeeze")
@tf.__internal__.dispatch.add_dispatch_support
@doc_controls.do_not_generate_docs
def squeeze(x, axis):
"""Removes a 1-dimension from the tensor at index "axis".
Args:
x: A tensor or variable.
axis: Axis to drop.
Returns:
A tensor with the same data as `x` but reduced dimensions.
"""
return tf.squeeze(x, [axis])
@keras_export("keras.backend.temporal_padding")
@tf.__internal__.dispatch.add_dispatch_support
@doc_controls.do_not_generate_docs
def temporal_padding(x, padding=(1, 1)):
"""Pads the middle dimension of a 3D tensor.
Args:
x: Tensor or variable.
padding: Tuple of 2 integers, how many zeros to
add at the start and end of dim 1.
Returns:
A padded 3D tensor.
"""
assert len(padding) == 2
pattern = [[0, 0], [padding[0], padding[1]], [0, 0]]
return tf.compat.v1.pad(x, pattern)
@keras_export("keras.backend.spatial_2d_padding")
@tf.__internal__.dispatch.add_dispatch_support
@doc_controls.do_not_generate_docs
def spatial_2d_padding(x, padding=((1, 1), (1, 1)), data_format=None):
"""Pads the 2nd and 3rd dimensions of a 4D tensor.
Args:
x: Tensor or variable.
padding: Tuple of 2 tuples, padding pattern.
data_format: One of `channels_last` or `channels_first`.
Returns:
A padded 4D tensor.
Raises:
ValueError: if `data_format` is neither
`channels_last` or `channels_first`.
"""
assert len(padding) == 2
assert len(padding[0]) == 2
assert len(padding[1]) == 2
if data_format is None:
data_format = image_data_format()
if data_format not in {"channels_first", "channels_last"}:
raise ValueError("Unknown data_format: " + str(data_format))
if data_format == "channels_first":
pattern = [[0, 0], [0, 0], list(padding[0]), list(padding[1])]
else:
pattern = [[0, 0], list(padding[0]), list(padding[1]), [0, 0]]
return tf.compat.v1.pad(x, pattern)
@keras_export("keras.backend.spatial_3d_padding")
@tf.__internal__.dispatch.add_dispatch_support
@doc_controls.do_not_generate_docs
def spatial_3d_padding(x, padding=((1, 1), (1, 1), (1, 1)), data_format=None):
"""Pads 5D tensor with zeros along the depth, height, width dimensions.
Pads these dimensions with respectively
"padding[0]", "padding[1]" and "padding[2]" zeros left and right.
For 'channels_last' data_format,
the 2nd, 3rd and 4th dimension will be padded.
For 'channels_first' data_format,
the 3rd, 4th and 5th dimension will be padded.
Args:
x: Tensor or variable.
padding: Tuple of 3 tuples, padding pattern.
data_format: One of `channels_last` or `channels_first`.
Returns:
A padded 5D tensor.
Raises:
ValueError: if `data_format` is neither
`channels_last` or `channels_first`.
"""
assert len(padding) == 3
assert len(padding[0]) == 2
assert len(padding[1]) == 2
assert len(padding[2]) == 2
if data_format is None:
data_format = image_data_format()
if data_format not in {"channels_first", "channels_last"}:
raise ValueError("Unknown data_format: " + str(data_format))
if data_format == "channels_first":
pattern = [
[0, 0],
[0, 0],
[padding[0][0], padding[0][1]],
[padding[1][0], padding[1][1]],
[padding[2][0], padding[2][1]],
]
else:
pattern = [
[0, 0],
[padding[0][0], padding[0][1]],
[padding[1][0], padding[1][1]],
[padding[2][0], padding[2][1]],
[0, 0],
]
return tf.compat.v1.pad(x, pattern)
@keras_export("keras.backend.stack")
@tf.__internal__.dispatch.add_dispatch_support
@doc_controls.do_not_generate_docs
def stack(x, axis=0):
"""Stacks a list of rank `R` tensors into a rank `R+1` tensor.
Args:
x: List of tensors.
axis: Axis along which to perform stacking.
Returns:
A tensor.
Example:
>>> a = tf.constant([[1, 2],[3, 4]])
>>> b = tf.constant([[10, 20],[30, 40]])
>>> tf.keras.backend.stack((a, b))
<tf.Tensor: shape=(2, 2, 2), dtype=int32, numpy=
array([[[ 1, 2],
[ 3, 4]],
[[10, 20],
[30, 40]]], dtype=int32)>
"""
return tf.stack(x, axis=axis)
@keras_export("keras.backend.one_hot")
@tf.__internal__.dispatch.add_dispatch_support
@doc_controls.do_not_generate_docs
def one_hot(indices, num_classes):
"""Computes the one-hot representation of an integer tensor.
Args:
indices: nD integer tensor of shape
`(batch_size, dim1, dim2, ... dim(n-1))`
num_classes: Integer, number of classes to consider.
Returns:
(n + 1)D one hot representation of the input
with shape `(batch_size, dim1, dim2, ... dim(n-1), num_classes)`
Returns:
The one-hot tensor.
"""
return tf.one_hot(indices, depth=num_classes, axis=-1)
@keras_export("keras.backend.reverse")
@tf.__internal__.dispatch.add_dispatch_support
@doc_controls.do_not_generate_docs
def reverse(x, axes):
"""Reverse a tensor along the specified axes.
Args:
x: Tensor to reverse.
axes: Integer or iterable of integers.
Axes to reverse.
Returns:
A tensor.
"""
if isinstance(axes, int):
axes = [axes]
return tf.reverse(x, axes)
# VALUE MANIPULATION
_VALUE_SET_CODE_STRING = """
>>> K = tf.keras.backend # Common keras convention
>>> v = K.variable(1.)
>>> # reassign
>>> K.set_value(v, 2.)
>>> print(K.get_value(v))
2.0
>>> # increment
>>> K.set_value(v, K.get_value(v) + 1)
>>> print(K.get_value(v))
3.0
Variable semantics in TensorFlow 2 are eager execution friendly. The above
code is roughly equivalent to:
>>> v = tf.Variable(1.)
>>> v.assign(2.)
>>> print(v.numpy())
2.0
>>> v.assign_add(1.)
>>> print(v.numpy())
3.0"""[
3:
] # Prune first newline and indent to match the docstring template.
@keras_export("keras.backend.get_value")
@doc_controls.do_not_generate_docs
def get_value(x):
"""Returns the value of a variable.
`backend.get_value` is the complement of `backend.set_value`, and provides
a generic interface for reading from variables while abstracting away the
differences between TensorFlow 1.x and 2.x semantics.
{snippet}
Args:
x: input variable.
Returns:
A Numpy array.
"""
if not tf.is_tensor(x):
return x
if tf.executing_eagerly() or isinstance(x, tf.__internal__.EagerTensor):
return x.numpy()
if not getattr(x, "_in_graph_mode", True):
# This is a variable which was created in an eager context, but is being
# evaluated from a Graph.
with tf.__internal__.eager_context.eager_mode():
return x.numpy()
if tf.compat.v1.executing_eagerly_outside_functions():
# This method of evaluating works inside the TF-Keras FuncGraph.
with tf.init_scope():
return x.numpy()
with x.graph.as_default():
return x.eval(session=get_session((x,)))
@keras_export("keras.backend.batch_get_value")
@tf.__internal__.dispatch.add_dispatch_support
@doc_controls.do_not_generate_docs
def batch_get_value(tensors):
"""Returns the value of more than one tensor variable.
Args:
tensors: list of ops to run.
Returns:
A list of Numpy arrays.
Raises:
RuntimeError: If this method is called inside defun.
"""
if tf.executing_eagerly():
return [x.numpy() for x in tensors]
elif tf.inside_function():
raise RuntimeError("Cannot get value inside Tensorflow graph function.")
if tensors:
return get_session(tensors).run(tensors)
else:
return []
@keras_export("keras.backend.set_value")
@doc_controls.do_not_generate_docs
def set_value(x, value):
"""Sets the value of a variable, from a Numpy array.
`backend.set_value` is the complement of `backend.get_value`, and provides
a generic interface for assigning to variables while abstracting away the
differences between TensorFlow 1.x and 2.x semantics.
{snippet}
Args:
x: Variable to set to a new value.
value: Value to set the tensor to, as a Numpy array
(of the same shape).
"""
value = np.asarray(value, dtype=dtype_numpy(x))
if tf.compat.v1.executing_eagerly_outside_functions():
_assign_value_to_variable(x, value)
else:
with get_graph().as_default():
tf_dtype = tf.as_dtype(x.dtype.name.split("_")[0])
if hasattr(x, "_assign_placeholder"):
assign_placeholder = x._assign_placeholder
assign_op = x._assign_op
else:
# In order to support assigning weights to resizable variables
# in Keras, we make a placeholder with the correct number of
# dimensions but with None in each dimension. This way, we can
# assign weights of any size (as long as they have the correct
# dimensionality).
placeholder_shape = tf.TensorShape([None] * value.ndim)
assign_placeholder = tf.compat.v1.placeholder(
tf_dtype, shape=placeholder_shape
)
assign_op = x.assign(assign_placeholder)
x._assign_placeholder = assign_placeholder
x._assign_op = assign_op
get_session().run(assign_op, feed_dict={assign_placeholder: value})
@keras_export("keras.backend.batch_set_value")
@tf.__internal__.dispatch.add_dispatch_support
@doc_controls.do_not_generate_docs
def batch_set_value(tuples):
"""Sets the values of many tensor variables at once.
Args:
tuples: a list of tuples `(tensor, value)`.
`value` should be a Numpy array.
"""
if tf.executing_eagerly() or tf.inside_function():
for x, value in tuples:
value = np.asarray(value, dtype=dtype_numpy(x))
_assign_value_to_variable(x, value)
else:
with get_graph().as_default():
if tuples:
assign_ops = []
feed_dict = {}
for x, value in tuples:
value = np.asarray(value, dtype=dtype_numpy(x))
tf_dtype = tf.as_dtype(x.dtype.name.split("_")[0])
if hasattr(x, "_assign_placeholder"):
assign_placeholder = x._assign_placeholder
assign_op = x._assign_op
else:
# In order to support assigning weights to resizable
# variables in Keras, we make a placeholder with the
# correct number of dimensions but with None in each
# dimension. This way, we can assign weights of any size
# (as long as they have the correct dimensionality).
placeholder_shape = tf.TensorShape([None] * value.ndim)
assign_placeholder = tf.compat.v1.placeholder(
tf_dtype, shape=placeholder_shape
)
assign_op = x.assign(assign_placeholder)
x._assign_placeholder = assign_placeholder
x._assign_op = assign_op
assign_ops.append(assign_op)
feed_dict[assign_placeholder] = value
get_session().run(assign_ops, feed_dict=feed_dict)
get_value.__doc__ = get_value.__doc__.format(snippet=_VALUE_SET_CODE_STRING)
set_value.__doc__ = set_value.__doc__.format(snippet=_VALUE_SET_CODE_STRING)
def _assign_value_to_variable(variable, value):
# Helper function to assign value to variable. It handles normal tf.Variable
# as well as DTensor variable.
if isinstance(variable, dtensor.DVariable):
mesh = variable.layout.mesh
replicate_layout = dtensor.Layout.replicated(
rank=variable.shape.rank, mesh=mesh
)
# TODO(b/262894693): Avoid the broadcast of tensor to all devices.
d_value = dtensor.copy_to_mesh(value, replicate_layout)
d_value = dtensor.relayout(d_value, variable.layout)
variable.assign(d_value)
else:
# For the normal tf.Variable assign
variable.assign(value)
@keras_export("keras.backend.print_tensor")
@tf.__internal__.dispatch.add_dispatch_support
@doc_controls.do_not_generate_docs
def print_tensor(x, message="", summarize=3):
"""Prints `message` and the tensor value when evaluated.
Note that `print_tensor` returns a new tensor identical to `x`
which should be used in the following code. Otherwise the
print operation is not taken into account during evaluation.
Example:
>>> x = tf.constant([[1.0, 2.0], [3.0, 4.0]])
>>> tf.keras.backend.print_tensor(x)
<tf.Tensor: shape=(2, 2), dtype=float32, numpy=
array([[1., 2.],
[3., 4.]], dtype=float32)>
Args:
x: Tensor to print.
message: Message to print jointly with the tensor.
summarize: The first and last `summarize` elements within each dimension
are recursively printed per Tensor. If None, then the first 3 and
last 3 elements of each dimension are printed for each tensor. If
set to -1, it will print all elements of every tensor.
Returns:
The same tensor `x`, unchanged.
"""
if isinstance(x, tf.Tensor) and hasattr(x, "graph"):
with get_graph().as_default():
op = tf.print(
message, x, output_stream=sys.stdout, summarize=summarize
)
with tf.control_dependencies([op]):
return tf.identity(x)
else:
tf.print(message, x, output_stream=sys.stdout, summarize=summarize)
return x
# GRAPH MANIPULATION
class GraphExecutionFunction:
"""Runs a computation graph.
It's possible to pass arguments to `tf.Session.run()` via `session_kwargs`.
In particular additional operations via `fetches` argument and additional
tensor substitutions via `feed_dict` arguments. Note that given
substitutions are merged with substitutions from `inputs`. Even though
`feed_dict` is passed once in the constructor (called in `model.compile()`)
we can modify the values in the dictionary. Through this feed_dict we can
provide additional substitutions besides TF-Keras inputs.
Args:
inputs: Feed placeholders to the computation graph.
outputs: Output tensors to fetch.
updates: Additional update ops to be run at function call.
name: A name to help users identify what this function does.
session_kwargs: Arguments to `tf.Session.run()`:
`fetches`, `feed_dict`, `options`, `run_metadata`.
"""
def __init__(
self, inputs, outputs, updates=None, name=None, **session_kwargs
):
updates = updates or []
if not isinstance(updates, (list, tuple)):
raise TypeError(
"`updates` in a TF-Keras backend function "
"should be a list or tuple."
)
self.inputs = tf.nest.flatten(
tf_utils.convert_variables_to_tensors(inputs),
expand_composites=True,
)
self._outputs_structure = tf_utils.convert_variables_to_tensors(outputs)
self.outputs = tf.nest.flatten(
self._outputs_structure, expand_composites=True
)
# TODO(b/127668432): Consider using autograph to generate these
# dependencies in call.
# Index 0 = total loss or model output for `predict`.
with tf.control_dependencies([self.outputs[0]]):
updates_ops = []
for update in updates:
if isinstance(update, tuple):
p, new_p = update
updates_ops.append(tf.compat.v1.assign(p, new_p))
else:
# assumed already an op
updates_ops.append(update)
self.updates_op = tf.group(*updates_ops)
self.name = name
# additional tensor substitutions
self.feed_dict = session_kwargs.pop("feed_dict", None)
# additional operations
self.fetches = session_kwargs.pop("fetches", [])
if not isinstance(self.fetches, list):
self.fetches = [self.fetches]
self.run_options = session_kwargs.pop("options", None)
self.run_metadata = session_kwargs.pop("run_metadata", None)
# The main use case of `fetches` being passed to a model is the ability
# to run custom updates
# This requires us to wrap fetches in `identity` ops.
self.fetches = [tf.identity(x) for x in self.fetches]
self.session_kwargs = session_kwargs
# This mapping keeps track of the function that should receive the
# output from a fetch in `fetches`: { fetch: function(fetch_output) }
# A Callback can use this to register a function with access to the
# output values for a fetch it added.
self.fetch_callbacks = {}
if session_kwargs:
raise ValueError(
"Some keys in session_kwargs are not supported at this time: %s"
% (session_kwargs.keys(),)
)
self._callable_fn = None
self._feed_arrays = None
self._feed_symbols = None
self._symbol_vals = None
self._fetches = None
self._session = None
def _make_callable(self, feed_arrays, feed_symbols, symbol_vals, session):
"""Generates a callable that runs the graph.
Args:
feed_arrays: List of input tensors to be fed Numpy arrays at runtime.
feed_symbols: List of input tensors to be fed symbolic tensors at
runtime.
symbol_vals: List of symbolic tensors to be fed to `feed_symbols`.
session: Session to use to generate the callable.
Returns:
Function that runs the graph according to the above options.
"""
# Prepare callable options.
callable_opts = config_pb2.CallableOptions()
# Handle external-data feed.
for x in feed_arrays:
callable_opts.feed.append(x.name)
if self.feed_dict:
for key in sorted(self.feed_dict.keys()):
callable_opts.feed.append(key.name)
# Handle symbolic feed.
for x, y in zip(feed_symbols, symbol_vals):
connection = callable_opts.tensor_connection.add()
if x.dtype != y.dtype:
y = tf.cast(y, dtype=x.dtype)
from_tensor = _as_graph_element(y)
if from_tensor is None:
from_tensor = y
connection.from_tensor = from_tensor.name # Data tensor
connection.to_tensor = x.name # Placeholder
# Handle fetches.
for x in self.outputs + self.fetches:
callable_opts.fetch.append(x.name)
# Handle updates.
callable_opts.target.append(self.updates_op.name)
# Handle run_options.
if self.run_options:
callable_opts.run_options.CopyFrom(self.run_options)
# Create callable.
callable_fn = session._make_callable_from_options(callable_opts)
# Cache parameters corresponding to the generated callable, so that
# we can detect future mismatches and refresh the callable.
self._callable_fn = callable_fn
self._feed_arrays = feed_arrays
self._feed_symbols = feed_symbols
self._symbol_vals = symbol_vals
self._fetches = list(self.fetches)
self._session = session
def _call_fetch_callbacks(self, fetches_output):
for fetch, output in zip(self._fetches, fetches_output):
if fetch in self.fetch_callbacks:
self.fetch_callbacks[fetch](output)
def _eval_if_composite(self, tensor):
"""Helper method which evaluates any CompositeTensors passed to it."""
# We need to evaluate any composite tensor objects that have been
# reconstructed in 'pack_sequence_as', since otherwise they'll be output
# as actual CompositeTensor objects instead of the value(s) contained in
# the CompositeTensors. E.g., if output_structure contains a
# SparseTensor, then this ensures that we return its value as a
# SparseTensorValue rather than a SparseTensor.
if tf_utils.is_extension_type(tensor):
return self._session.run(tensor)
else:
return tensor
def __call__(self, inputs):
inputs = tf.nest.flatten(
tf_utils.convert_variables_to_tensors(inputs),
expand_composites=True,
)
session = get_session(inputs)
feed_arrays = []
array_vals = []
feed_symbols = []
symbol_vals = []
for tensor, value in zip(self.inputs, inputs):
if value is None:
continue
if tf.is_tensor(value):
# Case: feeding symbolic tensor.
feed_symbols.append(tensor)
symbol_vals.append(value)
else:
# Case: feeding Numpy array.
feed_arrays.append(tensor)
# We need to do array conversion and type casting at this level,
# since `callable_fn` only supports exact matches.
tensor_type = tf.as_dtype(tensor.dtype)
array_vals.append(
np.asarray(value, dtype=tensor_type.as_numpy_dtype)
)
if self.feed_dict:
for key in sorted(self.feed_dict.keys()):
array_vals.append(
np.asarray(
self.feed_dict[key], dtype=key.dtype.as_numpy_dtype
)
)
# Refresh callable if anything has changed.
if (
self._callable_fn is None
or feed_arrays != self._feed_arrays
or symbol_vals != self._symbol_vals
or feed_symbols != self._feed_symbols
or self.fetches != self._fetches
or session != self._session
):
self._make_callable(feed_arrays, feed_symbols, symbol_vals, session)
fetched = self._callable_fn(*array_vals, run_metadata=self.run_metadata)
self._call_fetch_callbacks(fetched[-len(self._fetches) :])
output_structure = tf.nest.pack_sequence_as(
self._outputs_structure,
fetched[: len(self.outputs)],
expand_composites=True,
)
# We need to evaluate any composite tensor objects that have been
# reconstructed in 'pack_sequence_as', since otherwise they'll be output
# as actual CompositeTensor objects instead of the value(s) contained in
# the CompositeTensors. E.g., if output_structure contains a
# SparseTensor, then this ensures that we return its value as a
# SparseTensorValue rather than a SparseTensor.
return tf.nest.map_structure(self._eval_if_composite, output_structure)
@keras_export("keras.backend.function")
@doc_controls.do_not_generate_docs
def function(inputs, outputs, updates=None, name=None, **kwargs):
"""Instantiates a TF-Keras function.
Args:
inputs: List of placeholder tensors.
outputs: List of output tensors.
updates: List of update ops.
name: String, name of function.
**kwargs: Passed to `tf.Session.run`.
Returns:
Output values as Numpy arrays.
Raises:
ValueError: if invalid kwargs are passed in or if in eager execution.
"""
if tf.compat.v1.executing_eagerly_outside_functions():
if kwargs:
raise ValueError(
"Session keyword arguments are not supported during "
"eager execution. You passed: %s" % (kwargs,)
)
if updates:
raise ValueError(
"`updates` argument is not supported during "
"eager execution. You passed: %s" % (updates,)
)
from tf_keras import models
model = models.Model(inputs=inputs, outputs=outputs)
wrap_outputs = isinstance(outputs, list) and len(outputs) == 1
def func(model_inputs):
outs = model(model_inputs)
if wrap_outputs:
outs = [outs]
return tf_utils.sync_to_numpy_or_python_type(outs)
return func
if kwargs:
for key in kwargs:
if key not in tf_inspect.getfullargspec(tf.compat.v1.Session.run)[
0
] and key not in ["inputs", "outputs", "updates", "name"]:
msg = (
'Invalid argument "%s" passed to K.function with '
"TensorFlow backend" % key
)
raise ValueError(msg)
return GraphExecutionFunction(
inputs, outputs, updates=updates, name=name, **kwargs
)
@keras_export("keras.backend.gradients")
@doc_controls.do_not_generate_docs
def gradients(loss, variables):
"""Returns the gradients of `loss` w.r.t. `variables`.
Args:
loss: Scalar tensor to minimize.
variables: List of variables.
Returns:
A gradients tensor.
"""
return tf.compat.v1.gradients(
loss, variables, colocate_gradients_with_ops=True
)
@keras_export("keras.backend.stop_gradient")
@tf.__internal__.dispatch.add_dispatch_support
@doc_controls.do_not_generate_docs
def stop_gradient(variables):
"""Returns `variables` but with zero gradient w.r.t. every other variable.
Args:
variables: Tensor or list of tensors to consider constant with respect
to any other variable.
Returns:
A single tensor or a list of tensors (depending on the passed argument)
that has no gradient with respect to any other variable.
"""
if isinstance(variables, (list, tuple)):
return map(tf.stop_gradient, variables)
return tf.stop_gradient(variables)
# CONTROL FLOW
@keras_export("keras.backend.rnn")
@tf.__internal__.dispatch.add_dispatch_support
def rnn(
step_function,
inputs,
initial_states,
go_backwards=False,
mask=None,
constants=None,
unroll=False,
input_length=None,
time_major=False,
zero_output_for_mask=False,
return_all_outputs=True,
):
"""Iterates over the time dimension of a tensor.
Args:
step_function: RNN step function.
Args;
input; Tensor with shape `(samples, ...)` (no time dimension),
representing input for the batch of samples at a certain
time step.
states; List of tensors.
Returns;
output; Tensor with shape `(samples, output_dim)`
(no time dimension).
new_states; List of tensors, same length and shapes
as 'states'. The first state in the list must be the
output tensor at the previous timestep.
inputs: Tensor of temporal data of shape `(samples, time, ...)`
(at least 3D), or nested tensors, and each of which has shape
`(samples, time, ...)`.
initial_states: Tensor with shape `(samples, state_size)`
(no time dimension), containing the initial values for the states
used in the step function. In the case that state_size is in a
nested shape, the shape of initial_states will also follow the
nested structure.
go_backwards: Boolean. If True, do the iteration over the time
dimension in reverse order and return the reversed sequence.
mask: Binary tensor with shape `(samples, time, 1)`,
with a zero for every element that is masked.
constants: List of constant values passed at each step.
unroll: Whether to unroll the RNN or to use a symbolic `while_loop`.
input_length: An integer or a 1-D Tensor, depending on whether
the time dimension is fixed-length or not. In case of variable
length input, it is used for masking in case there's no mask
specified.
time_major: Boolean. If true, the inputs and outputs will be in shape
`(timesteps, batch, ...)`, whereas in the False case, it will be
`(batch, timesteps, ...)`. Using `time_major = True` is a bit more
efficient because it avoids transposes at the beginning and end of
the RNN calculation. However, most TensorFlow data is batch-major,
so by default this function accepts input and emits output in
batch-major form.
zero_output_for_mask: Boolean. If True, the output for masked timestep
will be zeros, whereas in the False case, output from previous
timestep is returned.
return_all_outputs: Boolean. If True, return the recurrent outputs for
all timesteps in the sequence. If False, only return the output for
the last timestep (which consumes less memory).
Returns:
A tuple, `(last_output, outputs, new_states)`.
last_output: the latest output of the rnn, of shape `(samples, ...)`
outputs:
- If `return_all_outputs=True`: a tensor with shape
`(samples, time, ...)` where each entry `outputs[s, t]` is the
output of the step function at time `t` for sample `s`
- Else, a tensor equal to `last_output` with shape
`(samples, 1, ...)`
new_states: list of tensors, latest states returned by
the step function, of shape `(samples, ...)`.
Raises:
ValueError: if input dimension is less than 3.
ValueError: if `unroll` is `True` but input timestep is not a fixed
number.
ValueError: if `mask` is provided (not `None`) but states is not
provided (`len(states)` == 0).
"""
if not tf.__internal__.tf2.enabled():
return_all_outputs = True # Not supported in TF1.
def swap_batch_timestep(input_t):
# Swap the batch and timestep dim for the incoming tensor.
axes = list(range(len(input_t.shape)))
axes[0], axes[1] = 1, 0
return tf.compat.v1.transpose(input_t, axes)
if not time_major:
inputs = tf.nest.map_structure(swap_batch_timestep, inputs)
flatted_inputs = tf.nest.flatten(inputs)
time_steps = flatted_inputs[0].shape[0]
batch = flatted_inputs[0].shape[1]
time_steps_t = tf.shape(flatted_inputs[0])[0]
for input_ in flatted_inputs:
input_.shape.with_rank_at_least(3)
if mask is not None:
if mask.dtype != tf.bool:
mask = tf.cast(mask, tf.bool)
if len(mask.shape) == 2:
mask = expand_dims(mask)
if not time_major:
mask = swap_batch_timestep(mask)
if constants is None:
constants = []
# tf.where needs its condition tensor to be the same shape as its two
# result tensors, but in our case the condition (mask) tensor is
# (nsamples, 1), and inputs are (nsamples, ndimensions) or even more.
# So we need to broadcast the mask to match the shape of inputs.
# That's what the tile call does, it just repeats the mask along its
# second dimension n times.
def _expand_mask(mask_t, input_t, fixed_dim=1):
if tf.nest.is_nested(mask_t):
raise ValueError(
f"mask_t is expected to be tensor, but got {mask_t}"
)
if tf.nest.is_nested(input_t):
raise ValueError(
f"input_t is expected to be tensor, but got {input_t}"
)
rank_diff = len(input_t.shape) - len(mask_t.shape)
for _ in range(rank_diff):
mask_t = tf.expand_dims(mask_t, -1)
multiples = [1] * fixed_dim + input_t.shape.as_list()[fixed_dim:]
return tf.tile(mask_t, multiples)
if unroll:
if not time_steps:
raise ValueError("Unrolling requires a fixed number of timesteps.")
states = tuple(initial_states)
successive_states = []
successive_outputs = []
# Process the input tensors. The input tensor need to be split on the
# time_step dim, and reverse if go_backwards is True. In the case of
# nested input, the input is flattened and then transformed
# individually. The result of this will be a tuple of lists, each of
# the item in tuple is list of the tensor with shape (batch, feature)
def _process_single_input_t(input_t):
input_t = tf.unstack(input_t) # unstack for time_step dim
if go_backwards:
input_t.reverse()
return input_t
if tf.nest.is_nested(inputs):
processed_input = tf.nest.map_structure(
_process_single_input_t, inputs
)
else:
processed_input = (_process_single_input_t(inputs),)
def _get_input_tensor(time):
inp = [t_[time] for t_ in processed_input]
return tf.nest.pack_sequence_as(inputs, inp)
if mask is not None:
mask_list = tf.unstack(mask)
if go_backwards:
mask_list.reverse()
for i in range(time_steps):
inp = _get_input_tensor(i)
mask_t = mask_list[i]
output, new_states = step_function(
inp, tuple(states) + tuple(constants)
)
tiled_mask_t = _expand_mask(mask_t, output)
if not successive_outputs:
prev_output = zeros_like(output)
else:
prev_output = successive_outputs[-1]
output = tf.where(tiled_mask_t, output, prev_output)
flat_states = tf.nest.flatten(states)
flat_new_states = tf.nest.flatten(new_states)
tiled_mask_t = tuple(
_expand_mask(mask_t, s) for s in flat_states
)
flat_final_states = tuple(
tf.where(m, s, ps)
for m, s, ps in zip(
tiled_mask_t, flat_new_states, flat_states
)
)
states = tf.nest.pack_sequence_as(states, flat_final_states)
if return_all_outputs:
successive_outputs.append(output)
successive_states.append(states)
else:
successive_outputs = [output]
successive_states = [states]
last_output = successive_outputs[-1]
new_states = successive_states[-1]
outputs = tf.stack(successive_outputs)
if zero_output_for_mask:
last_output = tf.where(
_expand_mask(mask_list[-1], last_output),
last_output,
zeros_like(last_output),
)
outputs = tf.where(
_expand_mask(mask, outputs, fixed_dim=2),
outputs,
zeros_like(outputs),
)
else: # mask is None
for i in range(time_steps):
inp = _get_input_tensor(i)
output, states = step_function(
inp, tuple(states) + tuple(constants)
)
if return_all_outputs:
successive_outputs.append(output)
successive_states.append(states)
else:
successive_outputs = [output]
successive_states = [states]
last_output = successive_outputs[-1]
new_states = successive_states[-1]
outputs = tf.stack(successive_outputs)
else: # Unroll == False
states = tuple(initial_states)
# Create input tensor array, if the inputs is nested tensors, then it
# will be flattened first, and tensor array will be created one per
# flattened tensor.
input_ta = tuple(
tf.TensorArray(
dtype=inp.dtype,
size=time_steps_t,
tensor_array_name=f"input_ta_{i}",
)
for i, inp in enumerate(flatted_inputs)
)
input_ta = tuple(
ta.unstack(input_)
if not go_backwards
else ta.unstack(reverse(input_, 0))
for ta, input_ in zip(input_ta, flatted_inputs)
)
# Get the time(0) input and compute the output for that, the output will
# be used to determine the dtype of output tensor array. Don't read from
# input_ta due to TensorArray clear_after_read default to True.
input_time_zero = tf.nest.pack_sequence_as(
inputs, [inp[0] for inp in flatted_inputs]
)
# output_time_zero is used to determine the cell output shape and its
# dtype. the value is discarded.
output_time_zero, _ = step_function(
input_time_zero, tuple(initial_states) + tuple(constants)
)
output_ta_size = time_steps_t if return_all_outputs else 1
output_ta = tuple(
tf.TensorArray(
dtype=out.dtype,
size=output_ta_size,
element_shape=out.shape,
tensor_array_name=f"output_ta_{i}",
)
for i, out in enumerate(tf.nest.flatten(output_time_zero))
)
time = tf.constant(0, dtype="int32", name="time")
# We only specify the 'maximum_iterations' when building for XLA since
# that causes slowdowns on GPU in TF.
if (
not tf.executing_eagerly()
and control_flow_util.GraphOrParentsInXlaContext(
tf.compat.v1.get_default_graph()
)
):
if input_length is None:
max_iterations = time_steps_t
else:
max_iterations = tf.reduce_max(input_length)
else:
max_iterations = None
while_loop_kwargs = {
"cond": lambda time, *_: time < time_steps_t,
"maximum_iterations": max_iterations,
"parallel_iterations": 32,
"swap_memory": True,
}
if mask is not None:
if go_backwards:
mask = reverse(mask, 0)
mask_ta = tf.TensorArray(
dtype=tf.bool, size=time_steps_t, tensor_array_name="mask_ta"
)
mask_ta = mask_ta.unstack(mask)
def masking_fn(time):
return mask_ta.read(time)
def compute_masked_output(mask_t, flat_out, flat_mask):
tiled_mask_t = tuple(
_expand_mask(mask_t, o, fixed_dim=len(mask_t.shape))
for o in flat_out
)
return tuple(
tf.where(m, o, fm)
for m, o, fm in zip(tiled_mask_t, flat_out, flat_mask)
)
elif isinstance(input_length, tf.Tensor):
if go_backwards:
max_len = tf.reduce_max(input_length, axis=0)
rev_input_length = tf.subtract(max_len - 1, input_length)
def masking_fn(time):
return tf.less(rev_input_length, time)
else:
def masking_fn(time):
return tf.greater(input_length, time)
def compute_masked_output(mask_t, flat_out, flat_mask):
return tuple(
tf.compat.v1.where(mask_t, o, zo)
for (o, zo) in zip(flat_out, flat_mask)
)
else:
masking_fn = None
if masking_fn is not None:
# Mask for the T output will be base on the output of T - 1. In the
# case T = 0, a zero filled tensor will be used.
flat_zero_output = tuple(
tf.zeros_like(o) for o in tf.nest.flatten(output_time_zero)
)
def _step(time, output_ta_t, prev_output, *states):
"""RNN step function.
Args:
time: Current timestep value.
output_ta_t: TensorArray.
prev_output: tuple of outputs from time - 1.
*states: List of states.
Returns:
Tuple: `(time + 1, output_ta_t, output) + tuple(new_states)`
"""
current_input = tuple(ta.read(time) for ta in input_ta)
# maybe set shape.
current_input = tf.nest.pack_sequence_as(inputs, current_input)
mask_t = masking_fn(time)
output, new_states = step_function(
current_input, tuple(states) + tuple(constants)
)
# mask output
flat_output = tf.nest.flatten(output)
flat_mask_output = (
flat_zero_output
if zero_output_for_mask
else tf.nest.flatten(prev_output)
)
flat_new_output = compute_masked_output(
mask_t, flat_output, flat_mask_output
)
# mask states
flat_state = tf.nest.flatten(states)
flat_new_state = tf.nest.flatten(new_states)
for state, new_state in zip(flat_state, flat_new_state):
if isinstance(new_state, tf.Tensor):
new_state.set_shape(state.shape)
flat_final_state = compute_masked_output(
mask_t, flat_new_state, flat_state
)
new_states = tf.nest.pack_sequence_as(
new_states, flat_final_state
)
ta_index_to_write = time if return_all_outputs else 0
output_ta_t = tuple(
ta.write(ta_index_to_write, out)
for ta, out in zip(output_ta_t, flat_new_output)
)
return (time + 1, output_ta_t, tuple(flat_new_output)) + tuple(
new_states
)
final_outputs = tf.compat.v1.while_loop(
body=_step,
loop_vars=(time, output_ta, flat_zero_output) + states,
**while_loop_kwargs,
)
# Skip final_outputs[2] which is the output for final timestep.
new_states = final_outputs[3:]
else:
def _step(time, output_ta_t, *states):
"""RNN step function.
Args:
time: Current timestep value.
output_ta_t: TensorArray.
*states: List of states.
Returns:
Tuple: `(time + 1,output_ta_t) + tuple(new_states)`
"""
current_input = tuple(ta.read(time) for ta in input_ta)
current_input = tf.nest.pack_sequence_as(inputs, current_input)
output, new_states = step_function(
current_input, tuple(states) + tuple(constants)
)
flat_state = tf.nest.flatten(states)
flat_new_state = tf.nest.flatten(new_states)
for state, new_state in zip(flat_state, flat_new_state):
if isinstance(new_state, tf.Tensor):
new_state.set_shape(state.shape)
flat_output = tf.nest.flatten(output)
ta_index_to_write = time if return_all_outputs else 0
output_ta_t = tuple(
ta.write(ta_index_to_write, out)
for ta, out in zip(output_ta_t, flat_output)
)
new_states = tf.nest.pack_sequence_as(
initial_states, flat_new_state
)
return (time + 1, output_ta_t) + tuple(new_states)
final_outputs = tf.compat.v1.while_loop(
body=_step,
loop_vars=(time, output_ta) + states,
**while_loop_kwargs,
)
new_states = final_outputs[2:]
output_ta = final_outputs[1]
outputs = tuple(o.stack() for o in output_ta)
last_output = tuple(o[-1] for o in outputs)
outputs = tf.nest.pack_sequence_as(output_time_zero, outputs)
last_output = tf.nest.pack_sequence_as(output_time_zero, last_output)
# static shape inference
def set_shape(output_):
if isinstance(output_, tf.Tensor):
shape = output_.shape.as_list()
if return_all_outputs:
shape[0] = time_steps
else:
shape[0] = 1
shape[1] = batch
output_.set_shape(shape)
return output_
outputs = tf.nest.map_structure(set_shape, outputs)
if not time_major:
outputs = tf.nest.map_structure(swap_batch_timestep, outputs)
return last_output, outputs, new_states
@keras_export("keras.backend.switch")
@tf.__internal__.dispatch.add_dispatch_support
@doc_controls.do_not_generate_docs
def switch(condition, then_expression, else_expression):
"""Switches between two operations depending on a scalar value.
Note that both `then_expression` and `else_expression`
should be symbolic tensors of the *same shape*.
Args:
condition: tensor (`int` or `bool`).
then_expression: either a tensor, or a callable that returns a tensor.
else_expression: either a tensor, or a callable that returns a tensor.
Returns:
The selected tensor.
Raises:
ValueError: If rank of `condition` is greater than rank of expressions.
"""
if condition.dtype != tf.bool:
condition = tf.cast(condition, "bool")
cond_ndim = ndim(condition)
if not cond_ndim:
if not callable(then_expression):
def then_expression_fn():
return then_expression
else:
then_expression_fn = then_expression
if not callable(else_expression):
def else_expression_fn():
return else_expression
else:
else_expression_fn = else_expression
x = tf.compat.v1.cond(condition, then_expression_fn, else_expression_fn)
else:
# tf.where needs its condition tensor
# to be the same shape as its two
# result tensors
if callable(then_expression):
then_expression = then_expression()
if callable(else_expression):
else_expression = else_expression()
expr_ndim = ndim(then_expression)
if cond_ndim > expr_ndim:
raise ValueError(
"Rank of `condition` should be less than or"
" equal to rank of `then_expression` and "
"`else_expression`. ndim(condition)="
+ str(cond_ndim)
+ ", ndim(then_expression)="
+ str(expr_ndim)
)
if cond_ndim > 1:
ndim_diff = expr_ndim - cond_ndim
cond_shape = tf.concat(
[tf.shape(condition), [1] * ndim_diff], axis=0
)
condition = tf.reshape(condition, cond_shape)
expr_shape = tf.shape(then_expression)
shape_diff = expr_shape - cond_shape
tile_shape = tf.where(
shape_diff > 0, expr_shape, tf.ones_like(expr_shape)
)
condition = tf.tile(condition, tile_shape)
x = tf.where(condition, then_expression, else_expression)
return x
@keras_export("keras.backend.in_train_phase")
@doc_controls.do_not_generate_docs
def in_train_phase(x, alt, training=None):
"""Selects `x` in train phase, and `alt` otherwise.
Note that `alt` should have the *same shape* as `x`.
Args:
x: What to return in train phase
(tensor or callable that returns a tensor).
alt: What to return otherwise
(tensor or callable that returns a tensor).
training: Optional scalar tensor
(or Python boolean, or Python integer)
specifying the learning phase.
Returns:
Either `x` or `alt` based on the `training` flag.
the `training` flag defaults to `K.learning_phase()`.
"""
from tf_keras.engine import (
base_layer_utils,
)
if training is None:
training = base_layer_utils.call_context().training
if training is None:
training = learning_phase()
# TODO(b/138862903): Handle the case when training is tensor.
if not tf.is_tensor(training):
if training == 1 or training is True:
if callable(x):
return x()
else:
return x
elif training == 0 or training is False:
if callable(alt):
return alt()
else:
return alt
# else: assume learning phase is a placeholder tensor.
x = switch(training, x, alt)
return x
@keras_export("keras.backend.in_test_phase")
@doc_controls.do_not_generate_docs
def in_test_phase(x, alt, training=None):
"""Selects `x` in test phase, and `alt` otherwise.
Note that `alt` should have the *same shape* as `x`.
Args:
x: What to return in test phase
(tensor or callable that returns a tensor).
alt: What to return otherwise
(tensor or callable that returns a tensor).
training: Optional scalar tensor
(or Python boolean, or Python integer)
specifying the learning phase.
Returns:
Either `x` or `alt` based on `K.learning_phase`.
"""
return in_train_phase(alt, x, training=training)
# NN OPERATIONS
@keras_export("keras.backend.relu")
@tf.__internal__.dispatch.add_dispatch_support
@doc_controls.do_not_generate_docs
def relu(x, alpha=0.0, max_value=None, threshold=0.0):
"""Rectified linear unit.
With default values, it returns element-wise `max(x, 0)`.
Otherwise, it follows:
`f(x) = max_value` for `x >= max_value`,
`f(x) = x` for `threshold <= x < max_value`,
`f(x) = alpha * (x - threshold)` otherwise.
Args:
x: A tensor or variable.
alpha: A scalar, slope of negative section (default=`0.`).
max_value: float. Saturation threshold.
threshold: float. Threshold value for thresholded activation.
Returns:
A tensor.
"""
# While x can be a tensor or variable, we also see cases where
# numpy arrays, lists, tuples are passed as well.
# lists, tuples do not have 'dtype' attribute.
dtype = getattr(x, "dtype", floatx())
if alpha != 0.0:
if max_value is None and threshold == 0:
return tf.nn.leaky_relu(x, alpha=alpha)
if threshold != 0:
negative_part = tf.nn.relu(-x + threshold)
else:
negative_part = tf.nn.relu(-x)
clip_max = max_value is not None
if threshold != 0:
# computes x for x > threshold else 0
x = x * tf.cast(tf.greater(x, threshold), dtype=dtype)
elif max_value == 6:
# if no threshold, then can use nn.relu6 native TF op for performance
x = tf.nn.relu6(x)
clip_max = False
else:
x = tf.nn.relu(x)
if clip_max:
max_value = _constant_to_tensor(max_value, x.dtype.base_dtype)
zero = _constant_to_tensor(0, x.dtype.base_dtype)
x = tf.clip_by_value(x, zero, max_value)
if alpha != 0.0:
alpha = _to_tensor(alpha, x.dtype.base_dtype)
x -= alpha * negative_part
return x
@keras_export("keras.backend.elu")
@tf.__internal__.dispatch.add_dispatch_support
@doc_controls.do_not_generate_docs
def elu(x, alpha=1.0):
"""Exponential linear unit.
Args:
x: A tensor or variable to compute the activation function for.
alpha: A scalar, slope of negative section.
Returns:
A tensor.
"""
res = tf.nn.elu(x)
if alpha == 1:
return res
else:
return tf.where(x > 0, res, alpha * res)
@keras_export("keras.backend.softmax")
@tf.__internal__.dispatch.add_dispatch_support
@doc_controls.do_not_generate_docs
def softmax(x, axis=-1):
"""Softmax of a tensor.
Args:
x: A tensor or variable.
axis: The dimension softmax would be performed on.
The default is -1 which indicates the last dimension.
Returns:
A tensor.
"""
if x.shape.rank <= 1:
raise ValueError(
f"Cannot apply softmax to a tensor that is 1D. Received input: {x}"
)
if isinstance(axis, int):
output = tf.nn.softmax(x, axis=axis)
else:
# nn.softmax does not support tuple axis.
numerator = tf.exp(x - tf.reduce_max(x, axis=axis, keepdims=True))
denominator = tf.reduce_sum(numerator, axis=axis, keepdims=True)
output = numerator / denominator
# Cache the logits to use for crossentropy loss.
output._keras_logits = x
return output
@keras_export("keras.backend.softplus")
@tf.__internal__.dispatch.add_dispatch_support
@doc_controls.do_not_generate_docs
def softplus(x):
"""Softplus of a tensor.
Args:
x: A tensor or variable.
Returns:
A tensor.
"""
return tf.math.softplus(x)
@keras_export("keras.backend.softsign")
@tf.__internal__.dispatch.add_dispatch_support
@doc_controls.do_not_generate_docs
def softsign(x):
"""Softsign of a tensor.
Args:
x: A tensor or variable.
Returns:
A tensor.
"""
return tf.math.softsign(x)
def _get_logits(output, from_logits, op_type, fn_name):
output_ = output
from_logits_ = from_logits
has_keras_logits = hasattr(output, "_keras_logits")
if has_keras_logits:
output_ = output._keras_logits
from_logits_ = True
from_expected_op_type = (
not isinstance(output, (tf.__internal__.EagerTensor, tf.Variable))
and output.op.type == op_type
) and not has_keras_logits
if from_expected_op_type:
# When softmax activation function is used for output operation, we
# use logits from the softmax function directly to compute loss in order
# to prevent collapsing zero when training.
# See b/117284466
assert len(output.op.inputs) == 1
output_ = output.op.inputs[0]
from_logits_ = True
if from_logits and (has_keras_logits or from_expected_op_type):
warnings.warn(
f'"`{fn_name}` received `from_logits=True`, but '
f"the `output` argument was produced by a {op_type} "
"activation and thus does not represent logits. "
"Was this intended?",
stacklevel=2,
)
return output_, from_logits_
@keras_export("keras.backend.categorical_crossentropy")
@tf.__internal__.dispatch.add_dispatch_support
@doc_controls.do_not_generate_docs
def categorical_crossentropy(target, output, from_logits=False, axis=-1):
"""Categorical crossentropy between an output tensor and a target tensor.
Args:
target: A tensor of the same shape as `output`.
output: A tensor resulting from a softmax
(unless `from_logits` is True, in which
case `output` is expected to be the logits).
from_logits: Boolean, whether `output` is the
result of a softmax, or is a tensor of logits.
axis: Int specifying the channels axis. `axis=-1` corresponds to data
format `channels_last`, and `axis=1` corresponds to data format
`channels_first`.
Returns:
Output tensor.
Raises:
ValueError: if `axis` is neither -1 nor one of the axes of `output`.
Example:
>>> a = tf.constant([1., 0., 0., 0., 1., 0., 0., 0., 1.], shape=[3,3])
>>> print(a)
tf.Tensor(
[[1. 0. 0.]
[0. 1. 0.]
[0. 0. 1.]], shape=(3, 3), dtype=float32)
>>> b = tf.constant([.9, .05, .05, .05, .89, .06, .05, .01, .94],
... shape=[3, 3])
>>> print(b)
tf.Tensor(
[[0.9 0.05 0.05]
[0.05 0.89 0.06]
[0.05 0.01 0.94]], shape=(3, 3), dtype=float32)
>>> loss = tf.keras.backend.categorical_crossentropy(a, b)
>>> print(np.around(loss, 5))
[0.10536 0.11653 0.06188]
>>> loss = tf.keras.backend.categorical_crossentropy(a, a)
>>> print(np.around(loss, 5))
[0. 0. 0.]
"""
target = tf.convert_to_tensor(target)
output = tf.convert_to_tensor(output)
target.shape.assert_is_compatible_with(output.shape)
output, from_logits = _get_logits(
output, from_logits, "Softmax", "categorical_crossentropy"
)
if from_logits:
return tf.nn.softmax_cross_entropy_with_logits(
labels=target, logits=output, axis=axis
)
# Adjust the predictions so that the probability of
# each class for every sample adds up to 1
# This is needed to ensure that the cross entropy is
# computed correctly.
output = output / tf.reduce_sum(output, axis, True)
# Compute cross entropy from probabilities.
epsilon_ = _constant_to_tensor(epsilon(), output.dtype.base_dtype)
output = tf.clip_by_value(output, epsilon_, 1.0 - epsilon_)
return -tf.reduce_sum(target * tf.math.log(output), axis)
@keras_export("keras.backend.categorical_focal_crossentropy")
@tf.__internal__.dispatch.add_dispatch_support
@doc_controls.do_not_generate_docs
def categorical_focal_crossentropy(
target,
output,
alpha=0.25,
gamma=2.0,
from_logits=False,
axis=-1,
):
"""Computes the alpha balanced focal crossentropy loss.
According to [Lin et al., 2018](https://arxiv.org/pdf/1708.02002.pdf), it
helps to apply a focal factor to down-weight easy examples and focus more on
hard examples. The general formula for the focal loss (FL)
is as follows:
`FL(p_t) = (1 − p_t)^gamma * log(p_t)`
where `p_t` is defined as follows:
`p_t = output if y_true == 1, else 1 - output`
`(1 − p_t)^gamma` is the `modulating_factor`, where `gamma` is a focusing
parameter. When `gamma` = 0, there is no focal effect on the cross entropy.
`gamma` reduces the importance given to simple examples in a smooth manner.
The authors use alpha-balanced variant of focal loss (FL) in the paper:
`FL(p_t) = −alpha * (1 − p_t)^gamma * log(p_t)`
where `alpha` is the weight factor for the classes. If `alpha` = 1, the
loss won't be able to handle class imbalance properly as all
classes will have the same weight. This can be a constant or a list of
constants. If alpha is a list, it must have the same length as the number
of classes.
The formula above can be generalized to:
`FL(p_t) = alpha * (1 − p_t)^gamma * CrossEntropy(target, output)`
where minus comes from `CrossEntropy(target, output)` (CE).
Extending this to multi-class case is straightforward:
`FL(p_t) = alpha * (1 − p_t)^gamma * CategoricalCE(target, output)`
Args:
target: Ground truth values from the dataset.
output: Predictions of the model.
alpha: A weight balancing factor for all classes, default is `0.25` as
mentioned in the reference. It can be a list of floats or a scalar.
In the multi-class case, alpha may be set by inverse class
frequency by using `compute_class_weight` from `sklearn.utils`.
gamma: A focusing parameter, default is `2.0` as mentioned in the
reference. It helps to gradually reduce the importance given to
simple examples in a smooth manner.
from_logits: Whether `output` is expected to be a logits tensor. By
default, we consider that `output` encodes a probability
distribution.
axis: Int specifying the channels axis. `axis=-1` corresponds to data
format `channels_last`, and `axis=1` corresponds to data format
`channels_first`.
Returns:
A tensor.
"""
target = tf.convert_to_tensor(target)
output = tf.convert_to_tensor(output)
target.shape.assert_is_compatible_with(output.shape)
output, from_logits = _get_logits(
output, from_logits, "Softmax", "categorical_focal_crossentropy"
)
if from_logits:
output = tf.nn.softmax(output, axis=axis)
# Adjust the predictions so that the probability of
# each class for every sample adds up to 1
# This is needed to ensure that the cross entropy is
# computed correctly.
output = output / tf.reduce_sum(output, axis=axis, keepdims=True)
epsilon_ = _constant_to_tensor(epsilon(), output.dtype.base_dtype)
output = tf.clip_by_value(output, epsilon_, 1.0 - epsilon_)
# Calculate cross entropy
cce = -target * tf.math.log(output)
# Calculate factors
modulating_factor = tf.pow(1.0 - output, gamma)
weighting_factor = tf.multiply(modulating_factor, alpha)
# Apply weighting factor
focal_cce = tf.multiply(weighting_factor, cce)
focal_cce = tf.reduce_sum(focal_cce, axis=axis)
return focal_cce
@keras_export("keras.backend.sparse_categorical_crossentropy")
@tf.__internal__.dispatch.add_dispatch_support
@doc_controls.do_not_generate_docs
def sparse_categorical_crossentropy(
target, output, from_logits=False, axis=-1, ignore_class=None
):
"""Categorical crossentropy with integer targets.
Args:
target: An integer tensor.
output: A tensor resulting from a softmax
(unless `from_logits` is True, in which
case `output` is expected to be the logits).
from_logits: Boolean, whether `output` is the
result of a softmax, or is a tensor of logits.
axis: Int specifying the channels axis. `axis=-1` corresponds to data
format `channels_last`, and `axis=1` corresponds to data format
`channels_first`.
ignore_class: Optional integer. The ID of a class to be ignored
during loss computation. This is useful, for example, in
segmentation problems featuring a "void" class (commonly -1
or 255) in segmentation maps.
By default (`ignore_class=None`), all classes are considered.
Returns:
Output tensor.
Raises:
ValueError: if `axis` is neither -1 nor one of the axes of `output`.
"""
target = tf.convert_to_tensor(target)
output = tf.convert_to_tensor(output)
target = cast(target, "int64")
output, from_logits = _get_logits(
output, from_logits, "Softmax", "sparse_categorical_crossentropy"
)
if not from_logits:
epsilon_ = _constant_to_tensor(epsilon(), output.dtype.base_dtype)
output = tf.clip_by_value(output, epsilon_, 1 - epsilon_)
output = tf.math.log(output)
# Permute output so that the last axis contains the logits/probabilities.
if isinstance(output.shape, (tuple, list)):
output_rank = len(output.shape)
else:
output_rank = output.shape.ndims
if output_rank is not None:
axis %= output_rank
if axis != output_rank - 1:
permutation = list(
itertools.chain(
range(axis), range(axis + 1, output_rank), [axis]
)
)
output = tf.compat.v1.transpose(output, perm=permutation)
elif axis != -1:
raise ValueError(
"Cannot compute sparse categorical crossentropy with `axis={}` "
"on an output tensor with unknown rank".format(axis)
)
# Try to adjust the shape so that rank of labels = rank of logits - 1.
output_shape = tf.shape(output)
target_rank = target.shape.ndims
update_shape = (
target_rank is not None
and output_rank is not None
and target_rank != output_rank - 1
)
if update_shape:
target = flatten(target)
output = tf.reshape(output, [-1, output_shape[-1]])
if ignore_class is not None:
valid_mask = tf.not_equal(target, cast(ignore_class, target.dtype))
target = target[valid_mask]
output = output[valid_mask]
if py_any(_is_symbolic_tensor(v) for v in [target, output]):
with get_graph().as_default():
res = tf.nn.sparse_softmax_cross_entropy_with_logits(
labels=target, logits=output
)
else:
res = tf.nn.sparse_softmax_cross_entropy_with_logits(
labels=target, logits=output
)
if ignore_class is not None:
res_shape = cast(output_shape[:-1], "int64")
valid_mask = tf.reshape(valid_mask, res_shape)
res = tf.scatter_nd(tf.where(valid_mask), res, res_shape)
res._keras_mask = valid_mask
return res
if update_shape and output_rank >= 3:
# If our output includes timesteps or
# spatial dimensions we need to reshape
res = tf.reshape(res, output_shape[:-1])
return res
@keras_export("keras.backend.binary_crossentropy")
@tf.__internal__.dispatch.add_dispatch_support
@doc_controls.do_not_generate_docs
def binary_crossentropy(target, output, from_logits=False):
"""Binary crossentropy between an output tensor and a target tensor.
Args:
target: A tensor with the same shape as `output`.
output: A tensor.
from_logits: Whether `output` is expected to be a logits tensor.
By default, we consider that `output`
encodes a probability distribution.
Returns:
A tensor.
"""
target = tf.convert_to_tensor(target)
output = tf.convert_to_tensor(output)
output, from_logits = _get_logits(
output, from_logits, "Sigmoid", "binary_crossentropy"
)
if from_logits:
return tf.nn.sigmoid_cross_entropy_with_logits(
labels=target, logits=output
)
epsilon_ = _constant_to_tensor(epsilon(), output.dtype.base_dtype)
output = tf.clip_by_value(output, epsilon_, 1.0 - epsilon_)
# Compute cross entropy from probabilities.
bce = target * tf.math.log(output + epsilon())
bce += (1 - target) * tf.math.log(1 - output + epsilon())
return -bce
@keras_export("keras.backend.binary_focal_crossentropy")
@tf.__internal__.dispatch.add_dispatch_support
@doc_controls.do_not_generate_docs
def binary_focal_crossentropy(
target,
output,
apply_class_balancing=False,
alpha=0.25,
gamma=2.0,
from_logits=False,
):
"""Binary focal crossentropy between an output tensor and a target tensor.
According to [Lin et al., 2018](https://arxiv.org/pdf/1708.02002.pdf), it
helps to apply a focal factor to down-weight easy examples and focus more on
hard examples. By default, the focal tensor is computed as follows:
`focal_factor = (1 - output) ** gamma` for class 1
`focal_factor = output ** gamma` for class 0
where `gamma` is a focusing parameter. When `gamma` = 0, there is no focal
effect on the binary crossentropy.
If `apply_class_balancing == True`, this function also takes into account a
weight balancing factor for the binary classes 0 and 1 as follows:
`weight = alpha` for class 1 (`target == 1`)
`weight = 1 - alpha` for class 0
where `alpha` is a float in the range of `[0, 1]`.
Args:
target: A tensor with the same shape as `output`.
output: A tensor.
apply_class_balancing: A bool, whether to apply weight balancing on the
binary classes 0 and 1.
alpha: A weight balancing factor for class 1, default is `0.25` as
mentioned in the reference. The weight for class 0 is `1.0 - alpha`.
gamma: A focusing parameter, default is `2.0` as mentioned in the
reference.
from_logits: Whether `output` is expected to be a logits tensor. By
default, we consider that `output` encodes a probability
distribution.
Returns:
A tensor.
"""
sigmoidal = sigmoid(output) if from_logits else output
p_t = target * sigmoidal + (1 - target) * (1 - sigmoidal)
# Calculate focal factor
focal_factor = tf.pow(1.0 - p_t, gamma)
# Binary crossentropy
bce = binary_crossentropy(
target=target,
output=output,
from_logits=from_logits,
)
focal_bce = focal_factor * bce
if apply_class_balancing:
weight = target * alpha + (1 - target) * (1 - alpha)
focal_bce = weight * focal_bce
return focal_bce
@keras_export("keras.backend.sigmoid")
@tf.__internal__.dispatch.add_dispatch_support
@doc_controls.do_not_generate_docs
def sigmoid(x):
"""Element-wise sigmoid.
Args:
x: A tensor or variable.
Returns:
A tensor.
"""
output = tf.sigmoid(x)
# Cache the logits to use for crossentropy loss.
output._keras_logits = x
return output
@keras_export("keras.backend.hard_sigmoid")
@tf.__internal__.dispatch.add_dispatch_support
@doc_controls.do_not_generate_docs
def hard_sigmoid(x):
"""Segment-wise linear approximation of sigmoid.
Faster than sigmoid.
Returns `0.` if `x < -2.5`, `1.` if `x > 2.5`.
In `-2.5 <= x <= 2.5`, returns `0.2 * x + 0.5`.
Args:
x: A tensor or variable.
Returns:
A tensor.
"""
point_two = _constant_to_tensor(0.2, x.dtype.base_dtype)
point_five = _constant_to_tensor(0.5, x.dtype.base_dtype)
x = tf.multiply(x, point_two)
x = tf.add(x, point_five)
x = tf.clip_by_value(x, 0.0, 1.0)
return x
@keras_export("keras.backend.tanh")
@tf.__internal__.dispatch.add_dispatch_support
@doc_controls.do_not_generate_docs
def tanh(x):
"""Element-wise tanh.
Args:
x: A tensor or variable.
Returns:
A tensor.
"""
return tf.tanh(x)
@keras_export("keras.backend.dropout")
@tf.__internal__.dispatch.add_dispatch_support
@doc_controls.do_not_generate_docs
def dropout(x, level, noise_shape=None, seed=None):
"""Sets entries in `x` to zero at random, while scaling the entire tensor.
Args:
x: tensor
level: fraction of the entries in the tensor
that will be set to 0.
noise_shape: shape for randomly generated keep/drop flags,
must be broadcastable to the shape of `x`
seed: random seed to ensure determinism.
Returns:
A tensor.
"""
if seed is None:
seed = np.random.randint(10e6)
return tf.nn.dropout(x, rate=level, noise_shape=noise_shape, seed=seed)
@keras_export("keras.backend.l2_normalize")
@tf.__internal__.dispatch.add_dispatch_support
@doc_controls.do_not_generate_docs
def l2_normalize(x, axis=None):
"""Normalizes a tensor wrt the L2 norm alongside the specified axis.
Args:
x: Tensor or variable.
axis: axis along which to perform normalization.
Returns:
A tensor.
"""
return tf.linalg.l2_normalize(x, axis=axis)
@keras_export("keras.backend.in_top_k")
@tf.__internal__.dispatch.add_dispatch_support
@doc_controls.do_not_generate_docs
def in_top_k(predictions, targets, k):
"""Returns whether the `targets` are in the top `k` `predictions`.
Args:
predictions: A tensor of shape `(batch_size, classes)` and type
`float32`.
targets: A 1D tensor of length `batch_size` and type `int32` or `int64`.
k: An `int`, number of top elements to consider.
Returns:
A 1D tensor of length `batch_size` and type `bool`.
`output[i]` is `True` if `predictions[i, targets[i]]` is within top-`k`
values of `predictions[i]`.
"""
return tf.compat.v1.math.in_top_k(predictions, targets, k)
# CONVOLUTIONS
def _preprocess_conv1d_input(x, data_format):
"""Transpose and cast the input before the conv1d.
Args:
x: input tensor.
data_format: string, `"channels_last"` or `"channels_first"`.
Returns:
A tensor.
"""
tf_data_format = "NWC" # to pass TF Conv2dNative operations
if data_format == "channels_first":
if not _has_nchw_support():
x = tf.compat.v1.transpose(x, (0, 2, 1)) # NCW -> NWC
else:
tf_data_format = "NCW"
return x, tf_data_format
def _preprocess_conv2d_input(x, data_format, force_transpose=False):
"""Transpose and cast the input before the conv2d.
Args:
x: input tensor.
data_format: string, `"channels_last"` or `"channels_first"`.
force_transpose: Boolean. If True, the input will always be transposed
from NCHW to NHWC if `data_format` is `"channels_first"`.
If False, the transposition only occurs on CPU (GPU ops are
assumed to support NCHW).
Returns:
A tensor.
"""
tf_data_format = "NHWC"
if data_format == "channels_first":
if not _has_nchw_support() or force_transpose:
x = tf.compat.v1.transpose(x, (0, 2, 3, 1)) # NCHW -> NHWC
else:
tf_data_format = "NCHW"
return x, tf_data_format
def _preprocess_conv3d_input(x, data_format):
"""Transpose and cast the input before the conv3d.
Args:
x: input tensor.
data_format: string, `"channels_last"` or `"channels_first"`.
Returns:
A tensor.
"""
tf_data_format = "NDHWC"
if data_format == "channels_first":
if not _has_nchw_support():
x = tf.compat.v1.transpose(x, (0, 2, 3, 4, 1))
else:
tf_data_format = "NCDHW"
return x, tf_data_format
def _preprocess_padding(padding):
"""Convert keras' padding to TensorFlow's padding.
Args:
padding: string, one of 'same' , 'valid'
Returns:
a string, one of 'SAME', 'VALID'.
Raises:
ValueError: if invalid `padding'`
"""
if padding == "same":
padding = "SAME"
elif padding == "valid":
padding = "VALID"
else:
raise ValueError("Invalid padding: " + str(padding))
return padding
@keras_export("keras.backend.conv1d")
@tf.__internal__.dispatch.add_dispatch_support
@doc_controls.do_not_generate_docs
def conv1d(
x, kernel, strides=1, padding="valid", data_format=None, dilation_rate=1
):
"""1D convolution.
Args:
x: Tensor or variable.
kernel: kernel tensor.
strides: stride integer.
padding: string, `"same"`, `"causal"` or `"valid"`.
data_format: string, one of "channels_last", "channels_first".
dilation_rate: integer dilate rate.
Returns:
A tensor, result of 1D convolution.
Raises:
ValueError: if `data_format` is neither `channels_last` or
`channels_first`.
"""
if data_format is None:
data_format = image_data_format()
if data_format not in {"channels_first", "channels_last"}:
raise ValueError("Unknown data_format: " + str(data_format))
kernel_shape = kernel.shape.as_list()
if padding == "causal":
# causal (dilated) convolution:
left_pad = dilation_rate * (kernel_shape[0] - 1)
x = temporal_padding(x, (left_pad, 0))
padding = "valid"
padding = _preprocess_padding(padding)
x, tf_data_format = _preprocess_conv1d_input(x, data_format)
x = tf.compat.v1.nn.convolution(
input=x,
filter=kernel,
dilation_rate=dilation_rate,
strides=strides,
padding=padding,
data_format=tf_data_format,
)
if data_format == "channels_first" and tf_data_format == "NWC":
x = tf.compat.v1.transpose(x, (0, 2, 1)) # NWC -> NCW
return x
@keras_export("keras.backend.conv2d")
@tf.__internal__.dispatch.add_dispatch_support
@doc_controls.do_not_generate_docs
def conv2d(
x,
kernel,
strides=(1, 1),
padding="valid",
data_format=None,
dilation_rate=(1, 1),
):
"""2D convolution.
Args:
x: Tensor or variable.
kernel: kernel tensor.
strides: strides tuple.
padding: string, `"same"` or `"valid"`.
data_format: `"channels_last"` or `"channels_first"`.
dilation_rate: tuple of 2 integers.
Returns:
A tensor, result of 2D convolution.
Raises:
ValueError: if `data_format` is neither `channels_last` or
`channels_first`.
"""
if data_format is None:
data_format = image_data_format()
if data_format not in {"channels_first", "channels_last"}:
raise ValueError("Unknown data_format: " + str(data_format))
x, tf_data_format = _preprocess_conv2d_input(x, data_format)
padding = _preprocess_padding(padding)
x = tf.compat.v1.nn.convolution(
input=x,
filter=kernel,
dilation_rate=dilation_rate,
strides=strides,
padding=padding,
data_format=tf_data_format,
)
if data_format == "channels_first" and tf_data_format == "NHWC":
x = tf.compat.v1.transpose(x, (0, 3, 1, 2)) # NHWC -> NCHW
return x
@keras_export("keras.backend.conv2d_transpose")
@tf.__internal__.dispatch.add_dispatch_support
@doc_controls.do_not_generate_docs
def conv2d_transpose(
x,
kernel,
output_shape,
strides=(1, 1),
padding="valid",
data_format=None,
dilation_rate=(1, 1),
):
"""2D deconvolution (i.e.
transposed convolution).
Args:
x: Tensor or variable.
kernel: kernel tensor.
output_shape: 1D int tensor for the output shape.
strides: strides tuple.
padding: string, `"same"` or `"valid"`.
data_format: string, `"channels_last"` or `"channels_first"`.
dilation_rate: Tuple of 2 integers.
Returns:
A tensor, result of transposed 2D convolution.
Raises:
ValueError: if `data_format` is neither `channels_last` or
`channels_first`.
"""
if data_format is None:
data_format = image_data_format()
if data_format not in {"channels_first", "channels_last"}:
raise ValueError("Unknown data_format: " + str(data_format))
# `atrous_conv2d_transpose` only supports NHWC format, even on GPU.
if data_format == "channels_first" and dilation_rate != (1, 1):
force_transpose = True
else:
force_transpose = False
x, tf_data_format = _preprocess_conv2d_input(
x, data_format, force_transpose
)
if data_format == "channels_first" and tf_data_format == "NHWC":
output_shape = (
output_shape[0],
output_shape[2],
output_shape[3],
output_shape[1],
)
if output_shape[0] is None:
output_shape = (shape(x)[0],) + tuple(output_shape[1:])
if isinstance(output_shape, (tuple, list)):
output_shape = tf.stack(list(output_shape))
padding = _preprocess_padding(padding)
if tf_data_format == "NHWC":
strides = (1,) + strides + (1,)
else:
strides = (1, 1) + strides
if dilation_rate == (1, 1):
x = tf.compat.v1.nn.conv2d_transpose(
x,
kernel,
output_shape,
strides,
padding=padding,
data_format=tf_data_format,
)
else:
if dilation_rate[0] != dilation_rate[1]:
raise ValueError(
"Expected the 2 dimensions of the `dilation_rate` argument "
"to be equal to each other. "
f"Received: dilation_rate={dilation_rate}"
)
x = tf.nn.atrous_conv2d_transpose(
x, kernel, output_shape, rate=dilation_rate[0], padding=padding
)
if data_format == "channels_first" and tf_data_format == "NHWC":
x = tf.compat.v1.transpose(x, (0, 3, 1, 2)) # NHWC -> NCHW
return x
def separable_conv1d(
x,
depthwise_kernel,
pointwise_kernel,
strides=1,
padding="valid",
data_format=None,
dilation_rate=1,
):
"""1D convolution with separable filters.
Args:
x: input tensor
depthwise_kernel: convolution kernel for the depthwise convolution.
pointwise_kernel: kernel for the 1x1 convolution.
strides: stride integer.
padding: string, `"same"` or `"valid"`.
data_format: string, `"channels_last"` or `"channels_first"`.
dilation_rate: integer dilation rate.
Returns:
Output tensor.
Raises:
ValueError: if `data_format` is neither `channels_last` or
`channels_first`.
"""
if data_format is None:
data_format = image_data_format()
if data_format not in {"channels_first", "channels_last"}:
raise ValueError("Unknown data_format: " + str(data_format))
if isinstance(strides, int):
strides = (strides,)
if isinstance(dilation_rate, int):
dilation_rate = (dilation_rate,)
x, tf_data_format = _preprocess_conv1d_input(x, data_format)
padding = _preprocess_padding(padding)
if not isinstance(strides, tuple):
strides = tuple(strides)
if tf_data_format == "NWC":
spatial_start_dim = 1
strides = (1,) + strides * 2 + (1,)
else:
spatial_start_dim = 2
strides = (1, 1) + strides * 2
x = tf.expand_dims(x, spatial_start_dim)
depthwise_kernel = tf.expand_dims(depthwise_kernel, 0)
pointwise_kernel = tf.expand_dims(pointwise_kernel, 0)
dilation_rate = (1,) + dilation_rate
x = tf.nn.separable_conv2d(
x,
depthwise_kernel,
pointwise_kernel,
strides=strides,
padding=padding,
dilations=dilation_rate,
data_format=tf_data_format,
)
x = tf.squeeze(x, [spatial_start_dim])
if data_format == "channels_first" and tf_data_format == "NWC":
x = tf.compat.v1.transpose(x, (0, 2, 1)) # NWC -> NCW
return x
@keras_export("keras.backend.separable_conv2d")
@tf.__internal__.dispatch.add_dispatch_support
@doc_controls.do_not_generate_docs
def separable_conv2d(
x,
depthwise_kernel,
pointwise_kernel,
strides=(1, 1),
padding="valid",
data_format=None,
dilation_rate=(1, 1),
):
"""2D convolution with separable filters.
Args:
x: input tensor
depthwise_kernel: convolution kernel for the depthwise convolution.
pointwise_kernel: kernel for the 1x1 convolution.
strides: strides tuple (length 2).
padding: string, `"same"` or `"valid"`.
data_format: string, `"channels_last"` or `"channels_first"`.
dilation_rate: tuple of integers,
dilation rates for the separable convolution.
Returns:
Output tensor.
Raises:
ValueError: if `data_format` is neither `channels_last` or
`channels_first`.
ValueError: if `strides` is not a tuple of 2 integers.
"""
if data_format is None:
data_format = image_data_format()
if data_format not in {"channels_first", "channels_last"}:
raise ValueError("Unknown data_format: " + str(data_format))
if len(strides) != 2:
raise ValueError("`strides` must be a tuple of 2 integers.")
x, tf_data_format = _preprocess_conv2d_input(x, data_format)
padding = _preprocess_padding(padding)
if not isinstance(strides, tuple):
strides = tuple(strides)
if tf_data_format == "NHWC":
strides = (1,) + strides + (1,)
else:
strides = (1, 1) + strides
x = tf.nn.separable_conv2d(
x,
depthwise_kernel,
pointwise_kernel,
strides=strides,
padding=padding,
dilations=dilation_rate,
data_format=tf_data_format,
)
if data_format == "channels_first" and tf_data_format == "NHWC":
x = tf.compat.v1.transpose(x, (0, 3, 1, 2)) # NHWC -> NCHW
return x
@keras_export("keras.backend.depthwise_conv2d")
@tf.__internal__.dispatch.add_dispatch_support
@doc_controls.do_not_generate_docs
def depthwise_conv2d(
x,
depthwise_kernel,
strides=(1, 1),
padding="valid",
data_format=None,
dilation_rate=(1, 1),
):
"""2D convolution with separable filters.
Args:
x: input tensor
depthwise_kernel: convolution kernel for the depthwise convolution.
strides: strides tuple (length 2).
padding: string, `"same"` or `"valid"`.
data_format: string, `"channels_last"` or `"channels_first"`.
dilation_rate: tuple of integers,
dilation rates for the separable convolution.
Returns:
Output tensor.
Raises:
ValueError: if `data_format` is neither `channels_last` or
`channels_first`.
"""
if data_format is None:
data_format = image_data_format()
if data_format not in {"channels_first", "channels_last"}:
raise ValueError("Unknown data_format: " + str(data_format))
x, tf_data_format = _preprocess_conv2d_input(x, data_format)
padding = _preprocess_padding(padding)
if tf_data_format == "NHWC":
strides = (1,) + strides + (1,)
else:
strides = (1, 1) + strides
x = tf.nn.depthwise_conv2d(
x,
depthwise_kernel,
strides=strides,
padding=padding,
dilations=dilation_rate,
data_format=tf_data_format,
)
if data_format == "channels_first" and tf_data_format == "NHWC":
x = tf.compat.v1.transpose(x, (0, 3, 1, 2)) # NHWC -> NCHW
return x
@keras_export("keras.backend.conv3d")
@tf.__internal__.dispatch.add_dispatch_support
@doc_controls.do_not_generate_docs
def conv3d(
x,
kernel,
strides=(1, 1, 1),
padding="valid",
data_format=None,
dilation_rate=(1, 1, 1),
):
"""3D convolution.
Args:
x: Tensor or variable.
kernel: kernel tensor.
strides: strides tuple.
padding: string, `"same"` or `"valid"`.
data_format: string, `"channels_last"` or `"channels_first"`.
dilation_rate: tuple of 3 integers.
Returns:
A tensor, result of 3D convolution.
Raises:
ValueError: if `data_format` is neither `channels_last` or
`channels_first`.
"""
if data_format is None:
data_format = image_data_format()
if data_format not in {"channels_first", "channels_last"}:
raise ValueError("Unknown data_format: " + str(data_format))
x, tf_data_format = _preprocess_conv3d_input(x, data_format)
padding = _preprocess_padding(padding)
x = tf.compat.v1.nn.convolution(
input=x,
filter=kernel,
dilation_rate=dilation_rate,
strides=strides,
padding=padding,
data_format=tf_data_format,
)
if data_format == "channels_first" and tf_data_format == "NDHWC":
x = tf.compat.v1.transpose(x, (0, 4, 1, 2, 3))
return x
def conv3d_transpose(
x,
kernel,
output_shape,
strides=(1, 1, 1),
padding="valid",
data_format=None,
):
"""3D deconvolution (i.e.
transposed convolution).
Args:
x: input tensor.
kernel: kernel tensor.
output_shape: 1D int tensor for the output shape.
strides: strides tuple.
padding: string, "same" or "valid".
data_format: string, `"channels_last"` or `"channels_first"`.
Returns:
A tensor, result of transposed 3D convolution.
Raises:
ValueError: if `data_format` is neither `channels_last` or
`channels_first`.
"""
if data_format is None:
data_format = image_data_format()
if data_format not in {"channels_first", "channels_last"}:
raise ValueError("Unknown data_format: " + str(data_format))
if isinstance(output_shape, (tuple, list)):
output_shape = tf.stack(output_shape)
x, tf_data_format = _preprocess_conv3d_input(x, data_format)
if data_format == "channels_first" and tf_data_format == "NDHWC":
output_shape = (
output_shape[0],
output_shape[2],
output_shape[3],
output_shape[4],
output_shape[1],
)
if output_shape[0] is None:
output_shape = (tf.shape(x)[0],) + tuple(output_shape[1:])
output_shape = tf.stack(list(output_shape))
padding = _preprocess_padding(padding)
if tf_data_format == "NDHWC":
strides = (1,) + strides + (1,)
else:
strides = (1, 1) + strides
x = tf.compat.v1.nn.conv3d_transpose(
x,
kernel,
output_shape,
strides,
padding=padding,
data_format=tf_data_format,
)
if data_format == "channels_first" and tf_data_format == "NDHWC":
x = tf.compat.v1.transpose(x, (0, 4, 1, 2, 3))
return x
@keras_export("keras.backend.pool2d")
@tf.__internal__.dispatch.add_dispatch_support
@doc_controls.do_not_generate_docs
def pool2d(
x,
pool_size,
strides=(1, 1),
padding="valid",
data_format=None,
pool_mode="max",
):
"""2D Pooling.
Args:
x: Tensor or variable.
pool_size: tuple of 2 integers.
strides: tuple of 2 integers.
padding: string, `"same"` or `"valid"`.
data_format: string, `"channels_last"` or `"channels_first"`.
pool_mode: string, `"max"` or `"avg"`.
Returns:
A tensor, result of 2D pooling.
Raises:
ValueError: if `data_format` is neither `"channels_last"` or
`"channels_first"`.
ValueError: if `pool_size` is not a tuple of 2 integers.
ValueError: if `strides` is not a tuple of 2 integers.
ValueError: if `pool_mode` is neither `"max"` or `"avg"`.
"""
if data_format is None:
data_format = image_data_format()
if data_format not in {"channels_first", "channels_last"}:
raise ValueError("Unknown data_format: " + str(data_format))
if len(pool_size) != 2:
raise ValueError("`pool_size` must be a tuple of 2 integers.")
if len(strides) != 2:
raise ValueError("`strides` must be a tuple of 2 integers.")
x, tf_data_format = _preprocess_conv2d_input(x, data_format)
padding = _preprocess_padding(padding)
if tf_data_format == "NHWC":
strides = (1,) + strides + (1,)
pool_size = (1,) + pool_size + (1,)
else:
strides = (1, 1) + strides
pool_size = (1, 1) + pool_size
if pool_mode == "max":
x = tf.compat.v1.nn.max_pool(
x, pool_size, strides, padding=padding, data_format=tf_data_format
)
elif pool_mode == "avg":
x = tf.compat.v1.nn.avg_pool(
x, pool_size, strides, padding=padding, data_format=tf_data_format
)
else:
raise ValueError("Invalid pooling mode: " + str(pool_mode))
if data_format == "channels_first" and tf_data_format == "NHWC":
x = tf.compat.v1.transpose(x, (0, 3, 1, 2)) # NHWC -> NCHW
return x
@keras_export("keras.backend.pool3d")
@tf.__internal__.dispatch.add_dispatch_support
@doc_controls.do_not_generate_docs
def pool3d(
x,
pool_size,
strides=(1, 1, 1),
padding="valid",
data_format=None,
pool_mode="max",
):
"""3D Pooling.
Args:
x: Tensor or variable.
pool_size: tuple of 3 integers.
strides: tuple of 3 integers.
padding: string, `"same"` or `"valid"`.
data_format: string, `"channels_last"` or `"channels_first"`.
pool_mode: string, `"max"` or `"avg"`.
Returns:
A tensor, result of 3D pooling.
Raises:
ValueError: if `data_format` is neither `"channels_last"` or
`"channels_first"`.
ValueError: if `pool_mode` is neither `"max"` or `"avg"`.
"""
if data_format is None:
data_format = image_data_format()
if data_format not in {"channels_first", "channels_last"}:
raise ValueError("Unknown data_format: " + str(data_format))
x, tf_data_format = _preprocess_conv3d_input(x, data_format)
padding = _preprocess_padding(padding)
if tf_data_format == "NDHWC":
strides = (1,) + strides + (1,)
pool_size = (1,) + pool_size + (1,)
else:
strides = (1, 1) + strides
pool_size = (1, 1) + pool_size
if pool_mode == "max":
x = tf.nn.max_pool3d(
x, pool_size, strides, padding=padding, data_format=tf_data_format
)
elif pool_mode == "avg":
x = tf.nn.avg_pool3d(
x, pool_size, strides, padding=padding, data_format=tf_data_format
)
else:
raise ValueError("Invalid pooling mode: " + str(pool_mode))
if data_format == "channels_first" and tf_data_format == "NDHWC":
x = tf.compat.v1.transpose(x, (0, 4, 1, 2, 3))
return x
def local_conv(
inputs, kernel, kernel_size, strides, output_shape, data_format=None
):
"""Apply N-D convolution with un-shared weights.
Args:
inputs: (N+2)-D tensor with shape
(batch_size, channels_in, d_in1, ..., d_inN)
if data_format='channels_first', or
(batch_size, d_in1, ..., d_inN, channels_in)
if data_format='channels_last'.
kernel: the unshared weight for N-D convolution,
with shape (output_items, feature_dim, channels_out), where
feature_dim = np.prod(kernel_size) * channels_in,
output_items = np.prod(output_shape).
kernel_size: a tuple of N integers, specifying the
spatial dimensions of the N-D convolution window.
strides: a tuple of N integers, specifying the strides
of the convolution along the spatial dimensions.
output_shape: a tuple of (d_out1, ..., d_outN) specifying the spatial
dimensionality of the output.
data_format: string, "channels_first" or "channels_last".
Returns:
An (N+2)-D tensor with shape:
(batch_size, channels_out) + output_shape
if data_format='channels_first', or:
(batch_size,) + output_shape + (channels_out,)
if data_format='channels_last'.
Raises:
ValueError: if `data_format` is neither
`channels_last` nor `channels_first`.
"""
if data_format is None:
data_format = image_data_format()
if data_format not in {"channels_first", "channels_last"}:
raise ValueError("Unknown data_format: " + str(data_format))
kernel_shape = int_shape(kernel)
feature_dim = kernel_shape[1]
channels_out = kernel_shape[-1]
ndims = len(output_shape)
spatial_dimensions = list(range(ndims))
xs = []
output_axes_ticks = [range(axis_max) for axis_max in output_shape]
for position in itertools.product(*output_axes_ticks):
slices = [slice(None)]
if data_format == "channels_first":
slices.append(slice(None))
slices.extend(
slice(
position[d] * strides[d],
position[d] * strides[d] + kernel_size[d],
)
for d in spatial_dimensions
)
if data_format == "channels_last":
slices.append(slice(None))
xs.append(reshape(inputs[slices], (1, -1, feature_dim)))
x_aggregate = concatenate(xs, axis=0)
output = batch_dot(x_aggregate, kernel)
output = reshape(output, output_shape + (-1, channels_out))
if data_format == "channels_first":
permutation = [ndims, ndims + 1] + spatial_dimensions
else:
permutation = [ndims] + spatial_dimensions + [ndims + 1]
return permute_dimensions(output, permutation)
@keras_export("keras.backend.local_conv1d")
@tf.__internal__.dispatch.add_dispatch_support
@doc_controls.do_not_generate_docs
def local_conv1d(inputs, kernel, kernel_size, strides, data_format=None):
"""Apply 1D conv with un-shared weights.
Args:
inputs: 3D tensor with shape:
(batch_size, steps, input_dim)
if data_format is "channels_last" or
(batch_size, input_dim, steps)
if data_format is "channels_first".
kernel: the unshared weight for convolution,
with shape (output_length, feature_dim, filters).
kernel_size: a tuple of a single integer,
specifying the length of the 1D convolution window.
strides: a tuple of a single integer,
specifying the stride length of the convolution.
data_format: the data format, channels_first or channels_last.
Returns:
A 3d tensor with shape:
(batch_size, output_length, filters)
if data_format='channels_first'
or 3D tensor with shape:
(batch_size, filters, output_length)
if data_format='channels_last'.
"""
output_shape = (kernel.shape[0],)
return local_conv(
inputs, kernel, kernel_size, strides, output_shape, data_format
)
@keras_export("keras.backend.local_conv2d")
@tf.__internal__.dispatch.add_dispatch_support
@doc_controls.do_not_generate_docs
def local_conv2d(
inputs, kernel, kernel_size, strides, output_shape, data_format=None
):
"""Apply 2D conv with un-shared weights.
Args:
inputs: 4D tensor with shape:
(batch_size, filters, new_rows, new_cols)
if data_format='channels_first'
or 4D tensor with shape:
(batch_size, new_rows, new_cols, filters)
if data_format='channels_last'.
kernel: the unshared weight for convolution,
with shape (output_items, feature_dim, filters).
kernel_size: a tuple of 2 integers, specifying the
width and height of the 2D convolution window.
strides: a tuple of 2 integers, specifying the strides
of the convolution along the width and height.
output_shape: a tuple with (output_row, output_col).
data_format: the data format, channels_first or channels_last.
Returns:
A 4D tensor with shape:
(batch_size, filters, new_rows, new_cols)
if data_format='channels_first'
or 4D tensor with shape:
(batch_size, new_rows, new_cols, filters)
if data_format='channels_last'.
"""
return local_conv(
inputs, kernel, kernel_size, strides, output_shape, data_format
)
@keras_export("keras.backend.bias_add")
@tf.__internal__.dispatch.add_dispatch_support
@doc_controls.do_not_generate_docs
def bias_add(x, bias, data_format=None):
"""Adds a bias vector to a tensor.
Args:
x: Tensor or variable.
bias: Bias tensor to add.
data_format: string, `"channels_last"` or `"channels_first"`.
Returns:
Output tensor.
Raises:
ValueError: In one of the two cases below:
1. invalid `data_format` argument.
2. invalid bias shape.
the bias should be either a vector or
a tensor with ndim(x) - 1 dimension
"""
if data_format is None:
data_format = image_data_format()
if data_format not in {"channels_first", "channels_last"}:
raise ValueError("Unknown data_format: " + str(data_format))
bias_shape = int_shape(bias)
if len(bias_shape) != 1 and len(bias_shape) != ndim(x) - 1:
raise ValueError(
"Unexpected bias dimensions %d, expect to be 1 or %d dimensions"
% (len(bias_shape), ndim(x) - 1)
)
if len(bias_shape) == 1:
if data_format == "channels_first":
return tf.nn.bias_add(x, bias, data_format="NCHW")
return tf.nn.bias_add(x, bias, data_format="NHWC")
if ndim(x) in (3, 4, 5):
if data_format == "channels_first":
bias_reshape_axis = (1, bias_shape[-1]) + bias_shape[:-1]
return x + reshape(bias, bias_reshape_axis)
return x + reshape(bias, (1,) + bias_shape)
return tf.nn.bias_add(x, bias)
# RANDOMNESS
@keras_export("keras.backend.random_normal")
@tf.__internal__.dispatch.add_dispatch_support
@doc_controls.do_not_generate_docs
def random_normal(shape, mean=0.0, stddev=1.0, dtype=None, seed=None):
"""Returns a tensor with normal distribution of values.
It is an alias to `tf.random.normal`.
Args:
shape: A tuple of integers, the shape of tensor to create.
mean: A float, the mean value of the normal distribution to draw
samples. Defaults to `0.0`.
stddev: A float, the standard deviation of the normal distribution
to draw samples. Defaults to `1.0`.
dtype: `tf.dtypes.DType`, dtype of returned tensor. None uses Keras
backend dtype which is float32. Defaults to `None`.
seed: Integer, random seed. Will use a random numpy integer when not
specified.
Returns:
A tensor with normal distribution of values.
Example:
>>> random_normal_tensor = tf.keras.backend.random_normal(shape=(2,3),
... mean=0.0, stddev=1.0)
>>> random_normal_tensor
<tf.Tensor: shape=(2, 3), dtype=float32, numpy=...,
dtype=float32)>
"""
if dtype is None:
dtype = floatx()
if seed is None:
seed = np.random.randint(10e6)
return tf.random.normal(
shape, mean=mean, stddev=stddev, dtype=dtype, seed=seed
)
@keras_export("keras.backend.random_uniform")
@tf.__internal__.dispatch.add_dispatch_support
@doc_controls.do_not_generate_docs
def random_uniform(shape, minval=0.0, maxval=1.0, dtype=None, seed=None):
"""Returns a tensor with uniform distribution of values.
Args:
shape: A tuple of integers, the shape of tensor to create.
minval: A float, lower boundary of the uniform distribution
to draw samples.
maxval: A float, upper boundary of the uniform distribution
to draw samples.
dtype: String, dtype of returned tensor.
seed: Integer, random seed.
Returns:
A tensor.
Example:
>>> random_uniform_tensor = tf.keras.backend.random_uniform(shape=(2,3),
... minval=0.0, maxval=1.0)
>>> random_uniform_tensor
<tf.Tensor: shape=(2, 3), dtype=float32, numpy=...,
dtype=float32)>
"""
if dtype is None:
dtype = floatx()
if seed is None:
seed = np.random.randint(10e6)
return tf.random.uniform(
shape, minval=minval, maxval=maxval, dtype=dtype, seed=seed
)
@keras_export("keras.backend.random_binomial")
@tf.__internal__.dispatch.add_dispatch_support
@doc_controls.do_not_generate_docs
def random_binomial(shape, p=0.0, dtype=None, seed=None):
"""Returns a tensor with random binomial distribution of values.
DEPRECATED, use `tf.keras.backend.random_bernoulli` instead.
The binomial distribution with parameters `n` and `p` is the probability
distribution of the number of successful Bernoulli process. Only supports
`n` = 1 for now.
Args:
shape: A tuple of integers, the shape of tensor to create.
p: A float, `0. <= p <= 1`, probability of binomial distribution.
dtype: String, dtype of returned tensor.
seed: Integer, random seed.
Returns:
A tensor.
Example:
>>> random_binomial_tensor = tf.keras.backend.random_binomial(shape=(2,3),
... p=0.5)
>>> random_binomial_tensor
<tf.Tensor: shape=(2, 3), dtype=float32, numpy=...,
dtype=float32)>
"""
warnings.warn(
"`tf.keras.backend.random_binomial` is deprecated, "
"and will be removed in a future version."
"Please use `tf.keras.backend.random_bernoulli` instead.",
stacklevel=2,
)
return random_bernoulli(shape, p, dtype, seed)
@keras_export("keras.backend.random_bernoulli")
@tf.__internal__.dispatch.add_dispatch_support
@doc_controls.do_not_generate_docs
def random_bernoulli(shape, p=0.0, dtype=None, seed=None):
"""Returns a tensor with random bernoulli distribution of values.
Args:
shape: A tuple of integers, the shape of tensor to create.
p: A float, `0. <= p <= 1`, probability of bernoulli distribution.
dtype: String, dtype of returned tensor.
seed: Integer, random seed.
Returns:
A tensor.
"""
if dtype is None:
dtype = floatx()
if seed is None:
seed = np.random.randint(10e6)
return tf.where(
tf.random.uniform(shape, dtype=dtype, seed=seed) <= p,
tf.ones(shape, dtype=dtype),
tf.zeros(shape, dtype=dtype),
)
@keras_export("keras.backend.truncated_normal")
@tf.__internal__.dispatch.add_dispatch_support
@doc_controls.do_not_generate_docs
def truncated_normal(shape, mean=0.0, stddev=1.0, dtype=None, seed=None):
"""Returns a tensor with truncated random normal distribution of values.
The generated values follow a normal distribution
with specified mean and standard deviation,
except that values whose magnitude is more than
two standard deviations from the mean are dropped and re-picked.
Args:
shape: A tuple of integers, the shape of tensor to create.
mean: Mean of the values.
stddev: Standard deviation of the values.
dtype: String, dtype of returned tensor.
seed: Integer, random seed.
Returns:
A tensor.
"""
if dtype is None:
dtype = floatx()
if seed is None:
seed = np.random.randint(10e6)
return tf.random.truncated_normal(
shape, mean, stddev, dtype=dtype, seed=seed
)
# CTC
# TensorFlow has a native implementation, but it uses sparse tensors
# and therefore requires a wrapper for TF-Keras. The functions below convert
# dense to sparse tensors and also wraps up the beam search code that is
# in TensorFlow's CTC implementation
@keras_export("keras.backend.ctc_label_dense_to_sparse")
@tf.__internal__.dispatch.add_dispatch_support
@doc_controls.do_not_generate_docs
def ctc_label_dense_to_sparse(labels, label_lengths):
"""Converts CTC labels from dense to sparse.
Args:
labels: dense CTC labels.
label_lengths: length of the labels.
Returns:
A sparse tensor representation of the labels.
"""
label_shape = tf.shape(labels)
num_batches_tns = tf.stack([label_shape[0]])
max_num_labels_tns = tf.stack([label_shape[1]])
def range_less_than(old_input, current_input):
return tf.expand_dims(tf.range(tf.shape(old_input)[1]), 0) < tf.fill(
max_num_labels_tns, current_input
)
init = tf.cast(tf.fill([1, label_shape[1]], 0), tf.bool)
dense_mask = tf.compat.v1.scan(
range_less_than, label_lengths, initializer=init, parallel_iterations=1
)
dense_mask = dense_mask[:, 0, :]
label_array = tf.reshape(
tf.tile(tf.range(0, label_shape[1]), num_batches_tns), label_shape
)
label_ind = tf.compat.v1.boolean_mask(label_array, dense_mask)
batch_array = tf.compat.v1.transpose(
tf.reshape(
tf.tile(tf.range(0, label_shape[0]), max_num_labels_tns),
reverse(label_shape, 0),
)
)
batch_ind = tf.compat.v1.boolean_mask(batch_array, dense_mask)
indices = tf.compat.v1.transpose(
tf.reshape(concatenate([batch_ind, label_ind], axis=0), [2, -1])
)
vals_sparse = tf.compat.v1.gather_nd(labels, indices)
return tf.SparseTensor(
tf.cast(indices, tf.int64), vals_sparse, tf.cast(label_shape, tf.int64)
)
@keras_export("keras.backend.ctc_batch_cost")
@tf.__internal__.dispatch.add_dispatch_support
@doc_controls.do_not_generate_docs
def ctc_batch_cost(y_true, y_pred, input_length, label_length):
"""Runs CTC loss algorithm on each batch element.
Args:
y_true: tensor `(samples, max_string_length)`
containing the truth labels.
y_pred: tensor `(samples, time_steps, num_categories)`
containing the prediction, or output of the softmax.
input_length: tensor `(samples, 1)` containing the sequence length for
each batch item in `y_pred`.
label_length: tensor `(samples, 1)` containing the sequence length for
each batch item in `y_true`.
Returns:
Tensor with shape (samples,1) containing the
CTC loss of each element.
"""
label_length = tf.cast(tf.squeeze(label_length, axis=-1), tf.int32)
input_length = tf.cast(tf.squeeze(input_length, axis=-1), tf.int32)
sparse_labels = tf.cast(
ctc_label_dense_to_sparse(y_true, label_length), tf.int32
)
y_pred = tf.math.log(
tf.compat.v1.transpose(y_pred, perm=[1, 0, 2]) + epsilon()
)
return tf.expand_dims(
tf.compat.v1.nn.ctc_loss(
inputs=y_pred, labels=sparse_labels, sequence_length=input_length
),
1,
)
@keras_export("keras.backend.ctc_decode")
@tf.__internal__.dispatch.add_dispatch_support
@doc_controls.do_not_generate_docs
def ctc_decode(y_pred, input_length, greedy=True, beam_width=100, top_paths=1):
"""Decodes the output of a softmax.
Can use either greedy search (also known as best path)
or a constrained dictionary search.
Args:
y_pred: tensor `(samples, time_steps, num_categories)`
containing the prediction, or output of the softmax.
input_length: tensor `(samples, )` containing the sequence length for
each batch item in `y_pred`.
greedy: perform much faster best-path search if `true`.
This does not use a dictionary.
beam_width: if `greedy` is `false`: a beam search decoder will be used
with a beam of this width.
top_paths: if `greedy` is `false`,
how many of the most probable paths will be returned.
Returns:
Tuple:
List: if `greedy` is `true`, returns a list of one element that
contains the decoded sequence.
If `false`, returns the `top_paths` most probable
decoded sequences.
Each decoded sequence has shape (samples, time_steps).
Important: blank labels are returned as `-1`.
Tensor `(top_paths, )` that contains
the log probability of each decoded sequence.
"""
input_shape = shape(y_pred)
num_samples, num_steps = input_shape[0], input_shape[1]
y_pred = tf.math.log(
tf.compat.v1.transpose(y_pred, perm=[1, 0, 2]) + epsilon()
)
input_length = tf.cast(input_length, tf.int32)
if greedy:
(decoded, log_prob) = tf.nn.ctc_greedy_decoder(
inputs=y_pred, sequence_length=input_length
)
else:
(decoded, log_prob) = tf.compat.v1.nn.ctc_beam_search_decoder(
inputs=y_pred,
sequence_length=input_length,
beam_width=beam_width,
top_paths=top_paths,
)
decoded_dense = []
for st in decoded:
st = tf.SparseTensor(st.indices, st.values, (num_samples, num_steps))
decoded_dense.append(tf.sparse.to_dense(sp_input=st, default_value=-1))
return (decoded_dense, log_prob)
# HIGH ORDER FUNCTIONS
@keras_export("keras.backend.map_fn")
@doc_controls.do_not_generate_docs
def map_fn(fn, elems, name=None, dtype=None):
"""Map the function fn over the elements elems and return the outputs.
Args:
fn: Callable that will be called upon each element in elems
elems: tensor
name: A string name for the map node in the graph
dtype: Output data type.
Returns:
Tensor with dtype `dtype`.
"""
return tf.compat.v1.map_fn(fn, elems, name=name, dtype=dtype)
@keras_export("keras.backend.foldl")
@doc_controls.do_not_generate_docs
def foldl(fn, elems, initializer=None, name=None):
"""Reduce elems using fn to combine them from left to right.
Args:
fn: Callable that will be called upon each element in elems and an
accumulator, for instance `lambda acc, x: acc + x`
elems: tensor
initializer: The first value used (`elems[0]` in case of None)
name: A string name for the foldl node in the graph
Returns:
Tensor with same type and shape as `initializer`.
"""
return tf.compat.v1.foldl(fn, elems, initializer=initializer, name=name)
@keras_export("keras.backend.foldr")
@doc_controls.do_not_generate_docs
def foldr(fn, elems, initializer=None, name=None):
"""Reduce elems using fn to combine them from right to left.
Args:
fn: Callable that will be called upon each element in elems and an
accumulator, for instance `lambda acc, x: acc + x`
elems: tensor
initializer: The first value used (`elems[-1]` in case of None)
name: A string name for the foldr node in the graph
Returns:
Same type and shape as initializer
"""
return tf.compat.v1.foldr(fn, elems, initializer=initializer, name=name)
# Load TF-Keras default configuration from config file if present.
# Set TF-Keras base dir path given KERAS_HOME env variable, if applicable.
# Otherwise either ~/.keras or /tmp.
if "KERAS_HOME" in os.environ:
_keras_dir = os.environ.get("KERAS_HOME")
else:
_keras_base_dir = os.path.expanduser("~")
_keras_dir = os.path.join(_keras_base_dir, ".keras")
_config_path = os.path.expanduser(os.path.join(_keras_dir, "keras.json"))
if os.path.exists(_config_path):
try:
with open(_config_path) as fh:
_config = json.load(fh)
except ValueError:
_config = {}
_floatx = _config.get("floatx", floatx())
assert _floatx in {"float16", "float32", "float64"}
_epsilon = _config.get("epsilon", epsilon())
assert isinstance(_epsilon, float)
_image_data_format = _config.get("image_data_format", image_data_format())
assert _image_data_format in {"channels_last", "channels_first"}
set_floatx(_floatx)
set_epsilon(_epsilon)
set_image_data_format(_image_data_format)
# Save config file.
if not os.path.exists(_keras_dir):
try:
os.makedirs(_keras_dir)
except OSError:
# Except permission denied and potential race conditions
# in multi-threaded environments.
pass
if not os.path.exists(_config_path):
_config = {
"floatx": floatx(),
"epsilon": epsilon(),
"backend": "tensorflow",
"image_data_format": image_data_format(),
}
try:
with open(_config_path, "w") as f:
f.write(json.dumps(_config, indent=4))
except IOError:
# Except permission denied.
pass
def configure_and_create_distributed_session(distribution_strategy):
"""Configure session config and create a session with it."""
def _create_session(distribution_strategy):
"""Create the Distributed Strategy session."""
session_config = get_default_session_config()
# If a session already exists, merge in its config; in the case there is
# a conflict, take values of the existing config.
global _SESSION
if getattr(_SESSION, "session", None) and _SESSION.session._config:
session_config.MergeFrom(_SESSION.session._config)
if is_tpu_strategy(distribution_strategy):
# TODO(priyag, yuefengz): Remove this workaround when Distribute
# Coordinator is integrated with keras and we can create a session
# from there.
distribution_strategy.configure(session_config)
master = (
distribution_strategy.extended._tpu_cluster_resolver.master()
)
session = tf.compat.v1.Session(config=session_config, target=master)
else:
worker_context = dc.get_current_worker_context()
if worker_context:
dc_session_config = worker_context.session_config
# Merge the default session config to the one from distribute
# coordinator, which is fine for now since they don't have
# conflicting configurations.
dc_session_config.MergeFrom(session_config)
session = tf.compat.v1.Session(
config=dc_session_config,
target=worker_context.master_target,
)
else:
distribution_strategy.configure(session_config)
session = tf.compat.v1.Session(config=session_config)
set_session(session)
if distribution_strategy.extended._in_multi_worker_mode():
dc.run_distribute_coordinator(_create_session, distribution_strategy)
else:
_create_session(distribution_strategy)
def _is_tpu_strategy_class(clz):
is_tpu_strat = lambda k: k.__name__.startswith("TPUStrategy")
if is_tpu_strat(clz):
return True
return py_any(map(_is_tpu_strategy_class, clz.__bases__))
def is_tpu_strategy(strategy):
"""Returns whether input is a TPUStrategy instance or subclass instance."""
return _is_tpu_strategy_class(strategy.__class__)
def _is_symbolic_tensor(x):
return tf.is_tensor(x) and not isinstance(x, tf.__internal__.EagerTensor)
def convert_inputs_if_ragged(inputs):
"""Converts any ragged tensors to dense."""
def _convert_ragged_input(inputs):
if isinstance(inputs, tf.RaggedTensor):
return inputs.to_tensor()
return inputs
flat_inputs = tf.nest.flatten(inputs)
contains_ragged = py_any(
isinstance(i, tf.RaggedTensor) for i in flat_inputs
)
if not contains_ragged:
return inputs, None
inputs = tf.nest.map_structure(_convert_ragged_input, inputs)
# Multiple mask are not yet supported, so one mask is used on all inputs.
# We approach this similarly when using row lengths to ignore steps.
nested_row_lengths = tf.cast(
flat_inputs[0].nested_row_lengths()[0], "int32"
)
return inputs, nested_row_lengths
def maybe_convert_to_ragged(
is_ragged_input, output, nested_row_lengths, go_backwards=False
):
"""Converts any ragged input back to its initial structure."""
if not is_ragged_input:
return output
if go_backwards:
# Reverse based on the timestep dim, so that nested_row_lengths will
# mask from the correct direction. Return the reverse ragged tensor.
output = reverse(output, [1])
ragged = tf.RaggedTensor.from_tensor(output, nested_row_lengths)
return reverse(ragged, [1])
else:
return tf.RaggedTensor.from_tensor(output, nested_row_lengths)
class ContextValueCache(weakref.WeakKeyDictionary):
"""Container that caches (possibly tensor) values based on the context.
This class is similar to defaultdict, where values may be produced by the
default factory specified during initialization. This class also has a
default value for the key (when key is `None`) -- the key is set to the
current graph or eager context. The default factories for key and value are
only used in `__getitem__` and `setdefault`. The `.get()` behavior remains
the same.
This object will return the value of the current graph or closest parent
graph if the current graph is a function. This is to reflect the fact that
if a tensor is created in eager/graph, child functions may capture that
tensor.
The default factory method may accept keyword arguments (unlike defaultdict,
which only accepts callables with 0 arguments). To pass keyword arguments to
`default_factory`, use the `setdefault` method instead of `__getitem__`.
An example of how this class can be used in different contexts:
```
cache = ContextValueCache(int)
# Eager mode
cache[None] += 2
cache[None] += 4
assert cache[None] == 6
# Graph mode
with tf.Graph().as_default() as g:
cache[None] += 5
cache[g] += 3
assert cache[g] == 8
```
Example of a default factory with arguments:
```
cache = ContextValueCache(lambda x: x + 1)
g = tf.get_default_graph()
# Example with keyword argument.
value = cache.setdefault(key=g, kwargs={'x': 3})
assert cache[g] == 4
```
"""
def __init__(self, default_factory):
self.default_factory = default_factory
weakref.WeakKeyDictionary.__init__(self)
def _key(self):
if tf.executing_eagerly():
return _DUMMY_EAGER_GRAPH.key
else:
return tf.compat.v1.get_default_graph()
def _get_parent_graph(self, graph):
"""Returns the parent graph or dummy eager object."""
# TODO(b/149317164): Currently FuncGraphs use ops.get_default_graph() as
# the outer graph. This results in outer_graph always being a Graph,
# even in eager mode (get_default_graph will create a new Graph if there
# isn't a default graph). Because of this bug, we have to specially set
# the key when eager execution is enabled.
parent_graph = graph.outer_graph
if (
not isinstance(parent_graph, tf.__internal__.FuncGraph)
and tf.compat.v1.executing_eagerly_outside_functions()
):
return _DUMMY_EAGER_GRAPH.key
return parent_graph
def _get_recursive(self, key):
"""Gets the value at key or the closest parent graph."""
value = self.get(key)
if value is not None:
return value
# Since FuncGraphs are able to capture tensors and variables from their
# parent graphs, recursively search to see if there is a value stored
# for one of the parent graphs.
if isinstance(key, tf.__internal__.FuncGraph):
return self._get_recursive(self._get_parent_graph(key))
return None
def __getitem__(self, key):
"""Gets the value at key (or current context), or sets default value.
Args:
key: May be `None` or `Graph`object. When `None`, the key is set to
the current context.
Returns:
Either the cached or default value.
"""
if key is None:
key = self._key()
value = self._get_recursive(key)
if value is None:
value = self[key] = self.default_factory()
return value
def setdefault(self, key=None, default=None, kwargs=None):
"""Sets the default value if key is not in dict, and returns the
value."""
if key is None:
key = self._key()
kwargs = kwargs or {}
if default is None and key not in self:
default = self.default_factory(**kwargs)
return weakref.WeakKeyDictionary.setdefault(self, key, default)
# This dictionary holds a mapping {graph: learning_phase}. In eager mode, a
# dummy object is used.
# A learning phase is a bool tensor used to run TF-Keras models in
# either train mode (learning_phase == 1) or test mode (learning_phase == 0).
_GRAPH_LEARNING_PHASES = ContextValueCache(
object_identity.ObjectIdentityWeakSet
)
# This dictionary holds a mapping between a graph and variables to initialize
# in the graph.
_GRAPH_VARIABLES = ContextValueCache(object_identity.ObjectIdentityWeakSet)
# This dictionary holds a mapping between a graph and TF optimizers created in
# the graph.
_GRAPH_TF_OPTIMIZERS = ContextValueCache(object_identity.ObjectIdentityWeakSet)
|
tf-keras/tf_keras/backend.py/0
|
{
"file_path": "tf-keras/tf_keras/backend.py",
"repo_id": "tf-keras",
"token_count": 108654
}
| 183 |
# Copyright 2020 The TensorFlow Authors. All Rights Reserved.
#
# Licensed under the Apache License, Version 2.0 (the "License");
# you may not use this file except in compliance with the License.
# You may obtain a copy of the License at
#
# http://www.apache.org/licenses/LICENSE-2.0
#
# Unless required by applicable law or agreed to in writing, software
# distributed under the License is distributed on an "AS IS" BASIS,
# WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.
# See the License for the specific language governing permissions and
# limitations under the License.
# ==============================================================================
"""Benchmarks on CNN on cifar10 dataset."""
from __future__ import absolute_import
from __future__ import division
from __future__ import print_function
import tensorflow.compat.v2 as tf
import tf_keras as keras
from tf_keras.benchmarks import benchmark_util
class Cifar10CNNBenchmark(tf.test.Benchmark):
"""Benchmarks for CNN using `tf.test.Benchmark`."""
def __init__(self):
super().__init__()
self.num_classes = 10
(self.x_train, self.y_train), _ = keras.datasets.cifar10.load_data()
self.x_train = self.x_train.astype("float32") / 255
self.y_train = keras.utils.to_categorical(
self.y_train, self.num_classes
)
self.epochs = 5
def _build_model(self):
"""Model from
https://github.com/keras-team/tf-keras/blob/master/examples/cifar10_cnn.py.
"""
model = keras.Sequential()
model.add(
keras.layers.Conv2D(
32, (3, 3), padding="same", input_shape=self.x_train.shape[1:]
)
)
model.add(keras.layers.Activation("relu"))
model.add(keras.layers.Conv2D(32, (3, 3)))
model.add(keras.layers.Activation("relu"))
model.add(keras.layers.MaxPooling2D(pool_size=(2, 2)))
model.add(keras.layers.Dropout(0.25))
model.add(keras.layers.Conv2D(64, (3, 3), padding="same"))
model.add(keras.layers.Activation("relu"))
model.add(keras.layers.Conv2D(64, (3, 3)))
model.add(keras.layers.Activation("relu"))
model.add(keras.layers.MaxPooling2D(pool_size=(2, 2)))
model.add(keras.layers.Dropout(0.25))
model.add(keras.layers.Flatten())
model.add(keras.layers.Dense(512))
model.add(keras.layers.Activation("relu"))
model.add(keras.layers.Dropout(0.5))
model.add(keras.layers.Dense(self.num_classes))
model.add(keras.layers.Activation("softmax"))
return model
# In each benchmark test, the required arguments for the
# method `measure_performance` include:
# x: Input data, it could be Numpy or loaded from tfds.
# y: Target data. If `x` is a dataset or generator instance,
# `y` should not be specified.
# loss: Loss function for model.
# optimizer: Optimizer for model.
# Check more details in `measure_performance()` method of
# benchmark_util.
def benchmark_cnn_cifar10_bs_256(self):
"""Measure performance with batch_size=256."""
batch_size = 256
metrics, wall_time, extras = benchmark_util.measure_performance(
self._build_model,
x=self.x_train,
y=self.y_train,
batch_size=batch_size,
epochs=self.epochs,
optimizer=keras.optimizers.RMSprop(
learning_rate=0.0001, decay=1e-6
),
loss="categorical_crossentropy",
metrics=["accuracy"],
)
metadata = benchmark_util.get_keras_examples_metadata("cnn", batch_size)
extras.update(metadata)
self.report_benchmark(
wall_time=wall_time, metrics=metrics, extras=extras
)
def benchmark_cnn_cifar10_bs_512(self):
"""Measure performance with batch_size=512."""
batch_size = 512
metrics, wall_time, extras = benchmark_util.measure_performance(
self._build_model,
x=self.x_train,
y=self.y_train,
batch_size=batch_size,
epochs=self.epochs,
optimizer=keras.optimizers.RMSprop(
learning_rate=0.0001, decay=1e-6
),
loss="categorical_crossentropy",
metrics=["accuracy"],
)
metadata = benchmark_util.get_keras_examples_metadata("cnn", batch_size)
extras.update(metadata)
self.report_benchmark(
wall_time=wall_time, metrics=metrics, extras=extras
)
def benchmark_cnn_cifar10_bs_1024(self):
"""Measure performance with batch_size=1024."""
batch_size = 1024
metrics, wall_time, extras = benchmark_util.measure_performance(
self._build_model,
x=self.x_train,
y=self.y_train,
batch_size=batch_size,
epochs=self.epochs,
optimizer=keras.optimizers.RMSprop(
learning_rate=0.0001, decay=1e-6
),
loss="categorical_crossentropy",
metrics=["accuracy"],
)
metadata = benchmark_util.get_keras_examples_metadata("cnn", batch_size)
extras.update(metadata)
self.report_benchmark(
wall_time=wall_time, metrics=metrics, extras=extras
)
def benchmark_cnn_cifar10_bs_1024_gpu_2(self):
"""Measure performance with batch_size=1024, gpu=2 and
distribution_strategy=`mirrored`.
"""
batch_size = 1024
metrics, wall_time, extras = benchmark_util.measure_performance(
self._build_model,
x=self.x_train,
y=self.y_train,
batch_size=batch_size,
num_gpus=2,
distribution_strategy="mirrored",
epochs=self.epochs,
optimizer=keras.optimizers.RMSprop(
learning_rate=0.0001, decay=1e-6
),
loss="categorical_crossentropy",
metrics=["accuracy"],
)
metadata = benchmark_util.get_keras_examples_metadata("cnn", batch_size)
extras.update(metadata)
self.report_benchmark(
wall_time=wall_time, metrics=metrics, extras=extras
)
if __name__ == "__main__":
tf.test.main()
|
tf-keras/tf_keras/benchmarks/keras_examples_benchmarks/cifar10_cnn_benchmark_test.py/0
|
{
"file_path": "tf-keras/tf_keras/benchmarks/keras_examples_benchmarks/cifar10_cnn_benchmark_test.py",
"repo_id": "tf-keras",
"token_count": 2916
}
| 184 |
# Copyright 2020 The TensorFlow Authors. All Rights Reserved.
#
# Licensed under the Apache License, Version 2.0 (the "License");
# you may not use this file except in compliance with the License.
# You may obtain a copy of the License at
#
# http://www.apache.org/licenses/LICENSE-2.0
#
# Unless required by applicable law or agreed to in writing, software
# distributed under the License is distributed on an "AS IS" BASIS,
# WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.
# See the License for the specific language governing permissions and
# limitations under the License.
# ==============================================================================
"""Benchmarks for saved model on DenseNet201."""
from __future__ import absolute_import
from __future__ import division
from __future__ import print_function
import tensorflow.compat.v2 as tf
import tf_keras as keras
from tf_keras.benchmarks.saved_model_benchmarks import (
saved_model_benchmark_util,
)
class BenchmarkSaveApplications(tf.test.Benchmark):
def benchmark_save_and_load_densenet_201(self):
app = keras.applications.DenseNet201
(
save_result,
load_result,
) = saved_model_benchmark_util.save_and_load_benchmark(app)
self.report_benchmark(
iters=save_result["iters"],
wall_time=save_result["wall_time"],
name=save_result["name"],
)
self.report_benchmark(
iters=load_result["iters"],
wall_time=load_result["wall_time"],
name=load_result["name"],
)
if __name__ == "__main__":
tf.test.main()
|
tf-keras/tf_keras/benchmarks/saved_model_benchmarks/densenet_benchmark_test.py/0
|
{
"file_path": "tf-keras/tf_keras/benchmarks/saved_model_benchmarks/densenet_benchmark_test.py",
"repo_id": "tf-keras",
"token_count": 589
}
| 185 |
"""Small NumPy datasets for debugging/testing."""
|
tf-keras/tf_keras/datasets/__init__.py/0
|
{
"file_path": "tf-keras/tf_keras/datasets/__init__.py",
"repo_id": "tf-keras",
"token_count": 12
}
| 186 |
# Copyright 2019 The TensorFlow Authors. All Rights Reserved.
#
# Licensed under the Apache License, Version 2.0 (the "License");
# you may not use this file except in compliance with the License.
# You may obtain a copy of the License at
#
# http://www.apache.org/licenses/LICENSE-2.0
#
# Unless required by applicable law or agreed to in writing, software
# distributed under the License is distributed on an "AS IS" BASIS,
# WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.
# See the License for the specific language governing permissions and
# limitations under the License.
# ==============================================================================
"""Tests for custom training loops."""
import os
import numpy as np
import tensorflow.compat.v2 as tf
from absl.testing import parameterized
import tf_keras as keras
from tf_keras.distribute import strategy_combinations
from tf_keras.layers import core
from tf_keras.optimizers.legacy import gradient_descent
class CustomModel(tf.Module):
def __init__(self, name=None):
super().__init__(name=name)
with self.name_scope:
self._layers = [
keras.layers.Dense(4, name="dense"),
]
@tf.Module.with_name_scope
def __call__(self, x):
for layer in self._layers:
x = layer(x)
return x
@tf.__internal__.distribute.combinations.generate(
tf.__internal__.test.combinations.combine(
distribution=(
strategy_combinations.all_strategies
+ strategy_combinations.multiworker_strategies
),
mode=["eager"],
)
)
class KerasModelsTest(tf.test.TestCase, parameterized.TestCase):
def test_single_keras_layer_run(self, distribution):
dataset = _get_dataset()
input_iterator = iter(
distribution.experimental_distribute_dataset(dataset)
)
with distribution.scope():
model = keras.layers.Dense(4, name="dense")
@tf.function
def train_step(iterator):
def step_fn(inputs):
images, targets = inputs
with tf.GradientTape() as tape:
outputs = model(images)
loss = keras.losses.mean_squared_error(targets, outputs)
grads = tape.gradient(loss, model.variables)
return grads
outputs = distribution.run(step_fn, args=(next(iterator),))
return tf.nest.map_structure(
distribution.experimental_local_results, outputs
)
train_step(input_iterator)
def test_keras_model_optimizer_run(self, distribution):
dataset = _get_dataset()
input_iterator = iter(
distribution.experimental_distribute_dataset(dataset)
)
with distribution.scope():
model = _get_model()
optimizer = keras.optimizers.legacy.rmsprop.RMSprop()
@tf.function
def train_step(replicated_inputs):
def step_fn(inputs):
images, targets = inputs
with tf.GradientTape() as tape:
outputs = model(images)
loss = keras.losses.mean_squared_error(targets, outputs)
grads = tape.gradient(loss, model.variables)
optimizer.apply_gradients(zip(grads, model.variables))
return loss
outputs = distribution.run(step_fn, args=(replicated_inputs,))
return tf.nest.map_structure(
distribution.experimental_local_results, outputs
)
for x in input_iterator:
train_step(x)
def test_keras_subclass_model_optimizer_run(self, distribution):
def get_subclass_model():
class KerasSubclassModel(keras.Model):
def __init__(self):
super().__init__()
self.l = keras.layers.Dense(4, name="dense")
def call(self, x):
return self.l(x)
return KerasSubclassModel()
dataset = _get_dataset()
input_iterator = iter(
distribution.experimental_distribute_dataset(dataset)
)
with distribution.scope():
model = get_subclass_model()
optimizer = keras.optimizers.legacy.rmsprop.RMSprop()
@tf.function
def train_step(iterator):
def step_fn(inputs):
images, targets = inputs
with tf.GradientTape() as tape:
outputs = model(images)
loss = keras.losses.mean_squared_error(targets, outputs)
grads = tape.gradient(loss, model.variables)
optimizer.apply_gradients(zip(grads, model.variables))
return loss
outputs = distribution.run(step_fn, args=(next(iterator),))
return tf.nest.map_structure(
distribution.experimental_local_results, outputs
)
train_step(input_iterator)
def test_keras_model_optimizer_run_loop(self, distribution):
dataset = _get_dataset()
input_iterator = iter(
distribution.experimental_distribute_dataset(dataset)
)
with distribution.scope():
model = _get_model()
optimizer = keras.optimizers.legacy.rmsprop.RMSprop()
@tf.function
def train_step(iterator):
def step_fn(inputs):
images, targets = inputs
with tf.GradientTape() as tape:
outputs = model(images)
loss = keras.losses.mean_squared_error(targets, outputs)
grads = tape.gradient(loss, model.variables)
optimizer.apply_gradients(zip(grads, model.variables))
return loss
for _ in tf.range(4):
distribution.run(step_fn, args=(next(iterator),))
train_step(input_iterator)
def test_batch_norm_with_dynamic_batch(self, distribution):
inputs = np.zeros((10, 3, 3, 3), dtype=np.float32)
targets = np.zeros((10, 4), dtype=np.float32)
dataset = tf.data.Dataset.from_tensor_slices((inputs, targets))
dataset = dataset.repeat()
dataset = dataset.batch(10)
input_iterator = iter(
distribution.experimental_distribute_dataset(dataset)
)
with distribution.scope():
x = keras.layers.Input(shape=(3, 3, 3), name="input")
y = keras.layers.BatchNormalization(fused=True, name="bn")(x)
y = keras.layers.Flatten()(y)
y = keras.layers.Dense(4, name="dense")(y)
model = keras.Model(x, y)
optimizer = keras.optimizers.legacy.rmsprop.RMSprop()
@tf.function
def train_step(iterator):
def step_fn(inputs):
images, targets = inputs
with tf.GradientTape() as tape:
outputs = model(images, training=True)
loss = keras.losses.mean_squared_error(targets, outputs)
grads = tape.gradient(loss, model.variables)
optimizer.apply_gradients(zip(grads, model.variables))
return loss
distribution.run(step_fn, args=(next(iterator),))
train_step(input_iterator)
def test_lstm(self, distribution):
batch_size = 32
def create_lstm_model():
model = keras.models.Sequential()
# We only have LSTM variables so we can detect no gradient issues
# more easily.
model.add(
keras.layers.LSTM(
1, return_sequences=False, input_shape=(10, 1)
)
)
return model
def create_lstm_data():
seq_length = 10
x_train = np.random.rand(batch_size, seq_length, 1).astype(
"float32"
)
y_train = np.random.rand(batch_size, 1).astype("float32")
return x_train, y_train
x, y = create_lstm_data()
dataset = tf.data.Dataset.from_tensor_slices((x, y))
dataset = dataset.batch(batch_size)
input_iterator = iter(
distribution.experimental_distribute_dataset(dataset)
)
with distribution.scope():
model = create_lstm_model()
optimizer = keras.optimizers.legacy.gradient_descent.SGD()
@tf.function
def train_step(input_iterator):
def step_fn(inputs):
inps, targ = inputs
with tf.GradientTape() as tape:
output = model(inps)
loss = tf.reduce_mean(
keras.losses.binary_crossentropy(
y_true=targ, y_pred=output, from_logits=False
)
)
grads = tape.gradient(loss, model.variables)
optimizer.apply_gradients(zip(grads, model.variables))
return loss
outputs = distribution.run(step_fn, args=(next(input_iterator),))
return distribution.experimental_local_results(outputs)
train_step(input_iterator)
def test_nested_tf_functions(self, distribution):
# The test builds two computations with keras layers, one with nested
# tf.function, and the other without nested tf.function. We run these
# computations independently on the model with same weights, and make
# sure the variables are still the same after one training step.
inputs = np.random.random((10, 3)).astype(np.float32)
targets = np.ones((10, 4), dtype=np.float32)
dataset = tf.data.Dataset.from_tensor_slices((inputs, targets)).repeat()
dataset = dataset.batch(10)
input_iterator = iter(
distribution.experimental_distribute_dataset(dataset)
)
def get_model():
x = keras.layers.Input(shape=(3,), name="input")
y = keras.layers.Dense(4, name="dense")(x)
model = keras.Model(x, y)
return model
with distribution.scope():
model = get_model()
optimizer = keras.optimizers.legacy.gradient_descent.SGD(
0.1, momentum=0.01
)
weights_file = os.path.join(self.get_temp_dir(), ".h5")
model.save_weights(weights_file)
model2 = get_model()
model2.load_weights(weights_file)
# Make sure model and model2 variables are in sync when initialized.
for model_v, model2_v in zip(model.variables, model2.variables):
self.assertAllClose(model_v.numpy(), model2_v.numpy())
def compute_loss(images, targets):
outputs = model(images)
return keras.losses.mean_squared_error(targets, outputs)
@tf.function
def train_step_without_nested_tf_function(inputs):
def step_fn(inputs):
images, targets = inputs
with tf.GradientTape() as tape:
loss = compute_loss(images, targets)
grads = tape.gradient(loss, model.variables)
optimizer.apply_gradients(zip(grads, model.variables))
distribution.run(step_fn, args=(inputs,))
@tf.function
def compute_loss2(images, targets):
outputs = model2(images)
return keras.losses.mean_squared_error(targets, outputs)
@tf.function
def train_step_with_nested_tf_function(inputs):
def step_fn(inputs):
images, targets = inputs
with tf.GradientTape() as tape:
loss = compute_loss2(images, targets)
grads = tape.gradient(loss, model2.variables)
optimizer.apply_gradients(zip(grads, model2.variables))
distribution.run(step_fn, args=(inputs,))
inputs = next(input_iterator)
train_step_without_nested_tf_function(inputs)
train_step_with_nested_tf_function(inputs)
# Make sure model and model2 variables are still in sync.
for model_v, model2_v in zip(model.variables, model2.variables):
self.assertAllClose(model_v.numpy(), model2_v.numpy())
def test_nested_tf_functions_with_control_flow(self, distribution):
inputs = np.random.random((10, 3)).astype(np.float32)
targets = np.ones((10, 4), dtype=np.float32)
dataset = tf.data.Dataset.from_tensor_slices((inputs, targets)).repeat()
dataset = dataset.batch(10)
input_iterator = iter(
distribution.experimental_distribute_dataset(dataset)
)
def get_model():
x = keras.layers.Input(shape=(3,), name="input")
y = keras.layers.Dense(4, name="dense")(x)
model = keras.Model(x, y)
return model
with distribution.scope():
model = get_model()
optimizer = keras.optimizers.legacy.gradient_descent.SGD(
0.1, momentum=0.01
)
@tf.function
def train_step(iterator):
def step_fn(inputs):
images, targets = inputs
with tf.GradientTape() as tape:
outputs = model(images)
loss = keras.losses.mean_squared_error(targets, outputs)
grads = tape.gradient(loss, model.variables)
optimizer.apply_gradients(zip(grads, model.variables))
distribution.run(step_fn, args=(next(iterator),))
@tf.function
def train_steps(iterator):
for _ in tf.range(10):
train_step(iterator)
train_steps(input_iterator)
def test_nested_tf_functions_with_tf_function_passing_to_strategy_run(
self, distribution
):
self.skipTest("b/190608193")
inputs = np.random.random((10, 3)).astype(np.float32)
targets = np.ones((10, 4), dtype=np.float32)
dataset = tf.data.Dataset.from_tensor_slices((inputs, targets)).repeat()
dataset = dataset.batch(10)
input_iterator = iter(
distribution.experimental_distribute_dataset(dataset)
)
def get_model():
x = keras.layers.Input(shape=(3,), name="input")
y = keras.layers.Dense(4, name="dense")(x)
model = keras.Model(x, y)
return model
with distribution.scope():
model = get_model()
optimizer = keras.optimizers.legacy.gradient_descent.SGD(
0.1, momentum=0.01
)
@tf.function
def compute_loss(images, targets):
outputs = model(images)
return keras.losses.mean_squared_error(targets, outputs)
@tf.function
def step_fn(inputs):
images, targets = inputs
with tf.GradientTape() as tape:
loss = compute_loss(images, targets)
grads = tape.gradient(loss, model.variables)
optimizer.apply_gradients(zip(grads, model.variables))
inputs = next(input_iterator)
distribution.run(step_fn, args=(inputs,))
def test_customized_tf_module_run(self, distribution):
dataset = _get_dataset()
input_iterator = iter(
distribution.experimental_distribute_dataset(dataset)
)
with distribution.scope():
model = CustomModel()
@tf.function
def train_step(iterator):
def step_fn(inputs):
images, targets = inputs
with tf.GradientTape() as tape:
outputs = model(images)
loss = keras.losses.mean_squared_error(targets, outputs)
grads = tape.gradient(loss, model.variables)
return grads
outputs = distribution.run(step_fn, args=(next(iterator),))
return tf.nest.map_structure(
distribution.experimental_local_results, outputs
)
train_step(input_iterator)
def test_reduce_loss(self, distribution):
inputs = np.zeros((10, 4), dtype=np.float32)
targets = np.zeros((10, 1), dtype=np.float32)
dataset = tf.data.Dataset.from_tensor_slices((inputs, targets))
dataset = dataset.batch(10)
input_iterator = iter(
distribution.experimental_distribute_dataset(dataset)
)
with distribution.scope():
x = keras.layers.Input(shape=(4), name="input")
y = keras.layers.Dense(3, name="dense")(x)
model = keras.Model(x, y)
@tf.function
def train_step(iterator):
def step_fn(inputs):
images, targets = inputs
outputs = model(images)
loss = keras.losses.sparse_categorical_crossentropy(
targets, outputs
)
return loss
return distribution.run(step_fn, args=(next(iterator),))
loss = train_step(input_iterator)
loss = distribution.reduce(tf.distribute.ReduceOp.MEAN, loss, axis=0)
def test_variable_run_argument(self, distribution):
# Test that variables passed to run() remain variables. Previous
# behavior in TPUStrategy was to cast to Tensor.
with distribution.scope():
optimizer = gradient_descent.SGD(0.1)
net = core.Dense(1, trainable=True)
dataset = tf.data.Dataset.from_tensors([[1.0]])
dataset = dataset.repeat()
dataset = dataset.batch(2, drop_remainder=True)
def replica_step(trainable_variables, features):
with tf.GradientTape() as tape:
net_out = net(features[0], training=True)
loss = (net_out - 1.0) * (net_out - 1.0)
gradients = tape.gradient(loss, trainable_variables)
optimizer.apply_gradients(zip(gradients, trainable_variables))
return loss
@tf.function
def step(features):
per_replica_losses = distribution.run(
replica_step,
(net.trainable_variables, features),
)
loss = distribution.reduce(
tf.distribute.ReduceOp.SUM, per_replica_losses, axis=None
)
return loss
step(next(iter(dataset)))
class KerasModelsXLATest(tf.test.TestCase, parameterized.TestCase):
@tf.__internal__.distribute.combinations.generate(
tf.__internal__.test.combinations.combine(
distribution=strategy_combinations.tpu_strategies, mode=["eager"]
)
)
def test_tf_function_jit_compile(self, distribution):
dataset = _get_dataset()
input_iterator = iter(
distribution.experimental_distribute_dataset(dataset)
)
class CustomDense(keras.layers.Layer):
def __init__(self, num_outputs):
super().__init__()
self.num_outputs = num_outputs
def build(self, input_shape):
self.kernel = self.add_weight(
"kernel", shape=[int(input_shape[-1]), self.num_outputs]
)
@tf.function(jit_compile=True)
def call(self, inputs):
return tf.matmul(inputs, self.kernel)
with distribution.scope():
x = keras.layers.Input(shape=(3,))
y = CustomDense(4)(x)
model = keras.Model(x, y)
@tf.function
def train_step(iterator):
def step_fn(inputs):
images, targets = inputs
with tf.GradientTape() as tape:
outputs = model(images)
loss = keras.losses.mean_squared_error(targets, outputs)
grads = tape.gradient(loss, model.variables)
return grads
outputs = distribution.run(step_fn, args=(next(iterator),))
return tf.nest.map_structure(
distribution.experimental_local_results, outputs
)
train_step(input_iterator)
def _get_dataset():
inputs = np.zeros((31, 3), dtype=np.float32)
targets = np.zeros((31, 4), dtype=np.float32)
dataset = tf.data.Dataset.from_tensor_slices((inputs, targets))
dataset = dataset.batch(10)
return dataset
def _get_model():
x = keras.layers.Input(shape=(3,), name="input")
y = keras.layers.Dense(4, name="dense")(x)
model = keras.Model(x, y)
return model
if __name__ == "__main__":
tf.__internal__.distribute.multi_process_runner.test_main()
|
tf-keras/tf_keras/distribute/custom_training_loop_models_test.py/0
|
{
"file_path": "tf-keras/tf_keras/distribute/custom_training_loop_models_test.py",
"repo_id": "tf-keras",
"token_count": 9930
}
| 187 |
# Copyright 2018 The TensorFlow Authors. All Rights Reserved.
#
# Licensed under the Apache License, Version 2.0 (the "License");
# you may not use this file except in compliance with the License.
# You may obtain a copy of the License at
#
# http://www.apache.org/licenses/LICENSE-2.0
#
# Unless required by applicable law or agreed to in writing, software
# distributed under the License is distributed on an "AS IS" BASIS,
# WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.
# See the License for the specific language governing permissions and
# limitations under the License.
# ==============================================================================
"""Tests for TF-Keras metrics."""
import tensorflow.compat.v2 as tf
from absl.testing import parameterized
from tf_keras import metrics
from tf_keras.engine import base_layer
combinations = tf.__internal__.distribute.combinations
def _labeled_dataset_fn():
# First four batches of x: labels, predictions -> (labels == predictions)
# 0: 0, 0 -> True; 1: 1, 1 -> True; 2: 2, 2 -> True; 3: 3, 0 -> False
# 4: 4, 1 -> False; 5: 0, 2 -> False; 6: 1, 0 -> False; 7: 2, 1 -> False
# 8: 3, 2 -> False; 9: 4, 0 -> False; 10: 0, 1 -> False; 11: 1, 2 -> False
# 12: 2, 0 -> False; 13: 3, 1 -> False; 14: 4, 2 -> False; 15: 0, 0 -> True
return (
tf.data.Dataset.range(1000)
.map(lambda x: {"labels": x % 5, "predictions": x % 3})
.batch(4, drop_remainder=True)
)
def _boolean_dataset_fn():
# First four batches of labels, predictions: {TP, FP, TN, FN}
# with a threshold of 0.5:
# T, T -> TP; F, T -> FP; T, F -> FN
# F, F -> TN; T, T -> TP; F, T -> FP
# T, F -> FN; F, F -> TN; T, T -> TP
# F, T -> FP; T, F -> FN; F, F -> TN
return (
tf.data.Dataset.from_tensor_slices(
{
"labels": [True, False, True, False],
"predictions": [True, True, False, False],
}
)
.repeat()
.batch(3, drop_remainder=True)
)
def _threshold_dataset_fn():
# First four batches of labels, predictions: {TP, FP, TN, FN}
# with a threshold of 0.5:
# True, 1.0 -> TP; False, .75 -> FP; True, .25 -> FN
# False, 0.0 -> TN; True, 1.0 -> TP; False, .75 -> FP
# True, .25 -> FN; False, 0.0 -> TN; True, 1.0 -> TP
# False, .75 -> FP; True, .25 -> FN; False, 0.0 -> TN
return (
tf.data.Dataset.from_tensor_slices(
{
"labels": [True, False, True, False],
"predictions": [1.0, 0.75, 0.25, 0.0],
}
)
.repeat()
.batch(3, drop_remainder=True)
)
def _regression_dataset_fn():
return tf.data.Dataset.from_tensor_slices(
{"labels": [1.0, 0.5, 1.0, 0.0], "predictions": [1.0, 0.75, 0.25, 0.0]}
).repeat()
def all_combinations():
return tf.__internal__.test.combinations.combine(
distribution=[
combinations.default_strategy,
combinations.one_device_strategy,
combinations.mirrored_strategy_with_gpu_and_cpu,
combinations.mirrored_strategy_with_two_gpus,
],
mode=["graph", "eager"],
)
def tpu_combinations():
return tf.__internal__.test.combinations.combine(
distribution=[
combinations.tpu_strategy,
],
mode=["graph"],
)
class KerasMetricsTest(tf.test.TestCase, parameterized.TestCase):
def _test_metric(
self, distribution, dataset_fn, metric_init_fn, expected_fn
):
with tf.Graph().as_default(), distribution.scope():
metric = metric_init_fn()
iterator = distribution.make_input_fn_iterator(
lambda _: dataset_fn()
)
updates = distribution.experimental_local_results(
distribution.run(metric, args=(iterator.get_next(),))
)
batches_per_update = distribution.num_replicas_in_sync
self.evaluate(iterator.initializer)
self.evaluate([v.initializer for v in metric.variables])
batches_consumed = 0
for i in range(4):
batches_consumed += batches_per_update
self.evaluate(updates)
self.assertAllClose(
expected_fn(batches_consumed),
self.evaluate(metric.result()),
0.001,
msg="After update #" + str(i + 1),
)
if batches_consumed >= 4: # Consume 4 input batches in total.
break
@combinations.generate(all_combinations() + tpu_combinations())
def testMean(self, distribution):
def _dataset_fn():
return (
tf.data.Dataset.range(1000)
.map(tf.compat.v1.to_float)
.batch(4, drop_remainder=True)
)
def _expected_fn(num_batches):
# Mean(0..3) = 1.5, Mean(0..7) = 3.5, Mean(0..11) = 5.5, etc.
return num_batches * 2 - 0.5
self._test_metric(distribution, _dataset_fn, metrics.Mean, _expected_fn)
@combinations.generate(
tf.__internal__.test.combinations.combine(
distribution=[
combinations.mirrored_strategy_with_one_cpu,
combinations.mirrored_strategy_with_gpu_and_cpu,
combinations.mirrored_strategy_with_two_gpus,
combinations.tpu_strategy_packed_var,
combinations.parameter_server_strategy_1worker_2ps_cpu,
combinations.parameter_server_strategy_1worker_2ps_1gpu,
],
mode=["eager"],
jit_compile=[False],
)
+ tf.__internal__.test.combinations.combine(
distribution=[combinations.mirrored_strategy_with_two_gpus],
mode=["eager"],
jit_compile=[True],
)
)
def testAddMetric(self, distribution, jit_compile):
if not tf.__internal__.tf2.enabled():
self.skipTest(
"Skip test since tf2 is not enabled. Pass "
" --test_env=TF2_BEHAVIOR=1 to enable tf2 behavior."
)
class MetricLayer(base_layer.Layer):
def __init__(self):
super().__init__(name="metric_layer")
self.sum = metrics.Sum(name="sum")
# Using aggregation for jit_compile results in failure. Thus
# only set aggregation for PS Strategy for multi-gpu tests.
if isinstance(
distribution,
tf.distribute.experimental.ParameterServerStrategy,
):
self.sum_var = tf.Variable(
1.0,
aggregation=tf.VariableAggregation.ONLY_FIRST_REPLICA,
)
else:
self.sum_var = tf.Variable(1.0)
def call(self, inputs):
self.add_metric(self.sum(inputs))
self.add_metric(
tf.reduce_mean(inputs), name="mean", aggregation="mean"
)
self.sum_var.assign(self.sum.result())
return inputs
with distribution.scope():
layer = MetricLayer()
def func():
return layer(tf.ones(()))
if jit_compile:
func = tf.function(jit_compile=True)(func)
@tf.function
def run():
return distribution.run(func)
if distribution._should_use_with_coordinator:
coord = tf.distribute.experimental.coordinator.ClusterCoordinator(
distribution
)
coord.schedule(run)
coord.join()
else:
run()
self.assertEqual(
layer.metrics[0].result().numpy(),
1.0 * distribution.num_replicas_in_sync,
)
self.assertEqual(layer.metrics[1].result().numpy(), 1.0)
self.assertEqual(
layer.sum_var.read_value().numpy(),
1.0 * distribution.num_replicas_in_sync,
)
@combinations.generate(all_combinations())
def test_precision(self, distribution):
# True positive is 2, false positive 1, precision is 2/3 = 0.6666667
label_prediction = ([0, 1, 1, 1], [1, 0, 1, 1])
with distribution.scope():
precision = metrics.Precision()
self.evaluate([v.initializer for v in precision.variables])
updates = distribution.run(precision, args=label_prediction)
self.evaluate(updates)
self.assertAllClose(precision.result(), 0.6666667)
@combinations.generate(all_combinations())
def test_recall(self, distribution):
# True positive is 2, false negative 1, precision is 2/3 = 0.6666667
label_prediction = ([0, 1, 1, 1], [1, 0, 1, 1])
with distribution.scope():
recall = metrics.Recall()
self.evaluate([v.initializer for v in recall.variables])
updates = distribution.run(recall, args=label_prediction)
self.evaluate(updates)
self.assertAllClose(recall.result(), 0.6666667)
@combinations.generate(all_combinations())
def test_SensitivityAtSpecificity(self, distribution):
label_prediction = ([0, 0, 0, 1, 1], [0, 0.3, 0.8, 0.3, 0.8])
with distribution.scope():
metric = metrics.SensitivityAtSpecificity(0.5)
self.evaluate([v.initializer for v in metric.variables])
updates = distribution.run(metric, args=label_prediction)
self.evaluate(updates)
self.assertAllClose(metric.result(), 0.5)
@combinations.generate(all_combinations())
def test_SpecificityAtSensitivity(self, distribution):
label_prediction = ([0, 0, 0, 1, 1], [0, 0.3, 0.8, 0.3, 0.8])
with distribution.scope():
metric = metrics.SpecificityAtSensitivity(0.5)
self.evaluate([v.initializer for v in metric.variables])
updates = distribution.run(metric, args=label_prediction)
self.evaluate(updates)
self.assertAllClose(metric.result(), 0.66666667)
@combinations.generate(all_combinations())
def test_PrecisionAtRecall(self, distribution):
label_prediction = ([0, 0, 0, 1, 1], [0, 0.3, 0.8, 0.3, 0.8])
with distribution.scope():
metric = metrics.PrecisionAtRecall(0.5)
self.evaluate([v.initializer for v in metric.variables])
updates = distribution.run(metric, args=label_prediction)
self.evaluate(updates)
self.assertAllClose(metric.result(), 0.5)
@combinations.generate(all_combinations())
def test_RecallAtPrecision(self, distribution):
label_prediction = ([0, 0, 1, 1], [0, 0.5, 0.3, 0.9])
with distribution.scope():
metric = metrics.RecallAtPrecision(0.8)
self.evaluate([v.initializer for v in metric.variables])
updates = distribution.run(metric, args=label_prediction)
self.evaluate(updates)
self.assertAllClose(metric.result(), 0.5)
@combinations.generate(all_combinations())
def test_auc(self, distribution):
label_prediction = ([0, 0, 1, 1], [0, 0.5, 0.3, 0.9])
with distribution.scope():
metric = metrics.AUC(num_thresholds=3)
self.evaluate([v.initializer for v in metric.variables])
updates = distribution.run(metric, args=label_prediction)
self.evaluate(updates)
self.assertAllClose(metric.result(), 0.75)
if __name__ == "__main__":
tf.__internal__.distribute.multi_process_runner.test_main()
|
tf-keras/tf_keras/distribute/keras_metrics_test.py/0
|
{
"file_path": "tf-keras/tf_keras/distribute/keras_metrics_test.py",
"repo_id": "tf-keras",
"token_count": 5529
}
| 188 |
# Copyright 2019 The TensorFlow Authors. All Rights Reserved.
#
# Licensed under the Apache License, Version 2.0 (the "License");
# you may not use this file except in compliance with the License.
# You may obtain a copy of the License at
#
# http://www.apache.org/licenses/LICENSE-2.0
#
# Unless required by applicable law or agreed to in writing, software
# distributed under the License is distributed on an "AS IS" BASIS,
# WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.
# See the License for the specific language governing permissions and
# limitations under the License.
# ==============================================================================
"""Utilities for testing multi-worker distribution strategies with TF-Keras."""
import threading
import unittest
import tensorflow.compat.v2 as tf
import tf_keras as keras
from tf_keras.optimizers.legacy import gradient_descent
# isort: off
from tensorflow.python.distribute.cluster_resolver import (
SimpleClusterResolver,
)
from tensorflow.python.platform import tf_logging as logging
from tensorflow.python.training.server_lib import (
ClusterSpec,
)
_portpicker_import_error = None
try:
import portpicker
except (
ImportError,
ModuleNotFoundError,
) as _error:
_portpicker_import_error = _error
portpicker = None
ASSIGNED_PORTS = set()
lock = threading.Lock()
def mnist_synthetic_dataset(
batch_size, steps_per_epoch, target_values="constant"
):
"""Generate synthetic MNIST dataset for testing."""
# train dataset
x_train = tf.ones(
[batch_size * steps_per_epoch, 28, 28, 1], dtype=tf.float32
)
if target_values == "constant":
y_train = tf.ones([batch_size * steps_per_epoch, 1], dtype=tf.int32)
elif target_values == "increasing":
y_train = tf.reshape(
tf.range(batch_size * steps_per_epoch, dtype=tf.int32), (-1, 1)
)
else:
raise ValueError(
'Unknown value for `target_values` "'
+ str(target_values)
+ '". Valid options are "constant" and "increasing".'
)
train_ds = tf.data.Dataset.from_tensor_slices((x_train, y_train))
train_ds = train_ds.repeat()
# train_ds = train_ds.shuffle(100)
train_ds = train_ds.batch(batch_size, drop_remainder=True)
# eval dataset
x_test = tf.random.uniform([10000, 28, 28, 1], dtype=tf.float32)
y_test = tf.random.uniform([10000, 1], minval=0, maxval=9, dtype=tf.int32)
eval_ds = tf.data.Dataset.from_tensor_slices((x_test, y_test))
eval_ds = eval_ds.batch(batch_size, drop_remainder=True)
return train_ds, eval_ds
def get_mnist_model(input_shape):
"""Define a deterministically-initialized CNN model for MNIST testing."""
inputs = keras.Input(shape=input_shape)
x = keras.layers.Conv2D(
32,
kernel_size=(3, 3),
activation="relu",
kernel_initializer=keras.initializers.TruncatedNormal(seed=99),
)(inputs)
x = keras.layers.BatchNormalization()(x)
x = keras.layers.Flatten()(x) + keras.layers.Flatten()(x)
x = keras.layers.Dense(
10,
activation="softmax",
kernel_initializer=keras.initializers.TruncatedNormal(seed=99),
)(x)
model = keras.Model(inputs=inputs, outputs=x)
# TODO(yuefengz): optimizer with slot variables doesn't work because of
# optimizer's bug.
# TODO(yuefengz): we should not allow non-v2 optimizer.
model.compile(
loss=keras.losses.sparse_categorical_crossentropy,
optimizer=gradient_descent.SGD(learning_rate=0.001),
metrics=["accuracy"],
)
return model
def make_parameter_server_cluster(num_workers, num_ps):
cluster_def = create_in_process_cluster(
num_workers=num_workers, num_ps=num_ps, rpc_layer="grpc"
)
return SimpleClusterResolver(ClusterSpec(cluster_def), rpc_layer="grpc")
def pick_unused_port():
"""Returns an unused and unassigned local port."""
if _portpicker_import_error:
raise _portpicker_import_error
global ASSIGNED_PORTS
with lock:
while True:
try:
port = portpicker.pick_unused_port()
except portpicker.NoFreePortFoundError:
raise unittest.SkipTest(
"Flakes in portpicker library do not represent "
"TensorFlow errors."
)
if port > 10000 and port not in ASSIGNED_PORTS:
ASSIGNED_PORTS.add(port)
logging.info("Using local port %r", port)
return port
def _create_cluster(
num_workers,
num_ps,
has_chief=False,
has_eval=False,
protocol="grpc",
worker_config=None,
ps_config=None,
eval_config=None,
worker_name="worker",
ps_name="ps",
chief_name="chief",
):
"""Creates and starts local servers and returns the cluster_spec dict."""
if _portpicker_import_error:
raise _portpicker_import_error
worker_ports = [pick_unused_port() for _ in range(num_workers)]
ps_ports = [pick_unused_port() for _ in range(num_ps)]
cluster_dict = {}
if num_workers > 0:
cluster_dict[worker_name] = [
f"localhost:{port}" for port in worker_ports
]
if num_ps > 0:
cluster_dict[ps_name] = [f"localhost:{port}" for port in ps_ports]
if has_eval:
cluster_dict["evaluator"] = [f"localhost:{pick_unused_port()}"]
if has_chief:
cluster_dict[chief_name] = [f"localhost:{pick_unused_port()}"]
cs = tf.train.ClusterSpec(cluster_dict)
for i in range(num_workers):
tf.distribute.Server(
cs,
job_name=worker_name,
protocol=protocol,
task_index=i,
config=worker_config,
start=True,
)
for i in range(num_ps):
tf.distribute.Server(
cs,
job_name=ps_name,
protocol=protocol,
task_index=i,
config=ps_config,
start=True,
)
if has_chief:
tf.distribute.Server(
cs,
job_name=chief_name,
protocol=protocol,
task_index=0,
config=worker_config,
start=True,
)
if has_eval:
tf.distribute.Server(
cs,
job_name="evaluator",
protocol=protocol,
task_index=0,
config=eval_config,
start=True,
)
return cluster_dict
def create_in_process_cluster(
num_workers, num_ps, has_chief=False, has_eval=False, rpc_layer="grpc"
):
"""Create an in-process cluster that consists of only standard server."""
# Leave some memory for cuda runtime.
gpu_mem_frac = 0.7 / (num_workers + int(has_chief) + int(has_eval))
worker_config = tf.compat.v1.ConfigProto()
worker_config.gpu_options.per_process_gpu_memory_fraction = gpu_mem_frac
# The cluster may hang if workers don't have enough inter_op threads. See
# b/172296720 for more details.
if worker_config.inter_op_parallelism_threads < num_workers + 1:
worker_config.inter_op_parallelism_threads = num_workers + 1
# Enable collective ops which has no impact on non-collective ops.
if has_chief:
worker_config.experimental.collective_group_leader = (
"/job:chief/replica:0/task:0"
)
else:
worker_config.experimental.collective_group_leader = (
"/job:worker/replica:0/task:0"
)
ps_config = tf.compat.v1.ConfigProto()
ps_config.device_count["GPU"] = 0
eval_config = tf.compat.v1.ConfigProto()
eval_config.experimental.collective_group_leader = ""
# Create in-process servers. Once an in-process tensorflow server is
# created, there is no way to terminate it. So we create one cluster per
# test process. We could've started the server in another process, we could
# then kill that process to terminate the server. The reasons why we don"t
# want multiple processes are
# 1) it is more difficult to manage these processes;
# 2) there is something global in CUDA such that if we initialize CUDA in
# the parent process, the child process cannot initialize it again and thus
# cannot use GPUs (https://stackoverflow.com/questions/22950047).
cluster = None
try:
cluster = _create_cluster(
num_workers,
num_ps=num_ps,
has_chief=has_chief,
has_eval=has_eval,
worker_config=worker_config,
ps_config=ps_config,
eval_config=eval_config,
protocol=rpc_layer,
)
except tf.errors.UnknownError as e:
if "Could not start gRPC server" in e.message:
raise unittest.SkipTest("Cannot start std servers.")
else:
raise
return cluster
|
tf-keras/tf_keras/distribute/multi_worker_testing_utils.py/0
|
{
"file_path": "tf-keras/tf_keras/distribute/multi_worker_testing_utils.py",
"repo_id": "tf-keras",
"token_count": 3786
}
| 189 |
# Copyright 2022 The TensorFlow Authors. All Rights Reserved.
#
# Licensed under the Apache License, Version 2.0 (the "License");
# you may not use this file except in compliance with the License.
# You may obtain a copy of the License at
#
# http://www.apache.org/licenses/LICENSE-2.0
#
# Unless required by applicable law or agreed to in writing, software
# distributed under the License is distributed on an "AS IS" BASIS,
# WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.
# See the License for the specific language governing permissions and
# limitations under the License.
# ==============================================================================
"""Tests for initializers."""
import numpy as np
import tensorflow.compat.v2 as tf
from absl.testing import parameterized
from tf_keras import backend
from tf_keras import initializers
from tf_keras.dtensor import dtensor_api as dtensor
from tf_keras.dtensor import test_util
from tf_keras.utils import tf_utils
class InitializersTest(test_util.DTensorBaseTest):
def setUp(self):
super().setUp()
global_ids = test_util.create_device_ids_array((2, 2))
local_device_ids = np.ravel(global_ids).tolist()
mesh_dict = {
"CPU": dtensor.Mesh(
["X", "Y"],
global_ids,
local_device_ids,
test_util.create_device_list((2, 2), "CPU"),
)
}
self.mesh = self.configTestMesh(mesh_dict)
@parameterized.named_parameters(
("Zeros", initializers.Zeros, {}),
("Ones", initializers.Ones, {}),
("Constant", initializers.Constant, {"value": 3.0}),
# TODO(b/222160686): Add Identity after after we have SPMD support for
# tf.MatrixDiagV3
# ('Identity', initializers.Identity, {}),
)
def test_static_value_initializer(self, initializer_cls, init_args):
layout = dtensor.Layout(
[dtensor.UNSHARDED, dtensor.UNSHARDED], self.mesh
)
shape = (4, 4)
initializer = initializer_cls(**init_args)
value = initializer(shape=shape, layout=layout)
normal_tensor_value = initializer(shape=shape)
self.assertEqual(value.shape, shape)
fetched_layout = dtensor.fetch_layout(value)
self.assertEqual(layout, fetched_layout)
self.assertAllClose(value, normal_tensor_value)
@parameterized.named_parameters(
("RandomUniform", initializers.RandomUniform, {}),
("RandomUniform_seeded", initializers.RandomUniform, {"seed": 1}),
("RandomNormal", initializers.RandomNormal, {}),
("RandomNormal_seeded", initializers.RandomNormal, {"seed": 1}),
("TruncatedNormal", initializers.TruncatedNormal, {}),
("TruncatedNormal_seeded", initializers.TruncatedNormal, {"seed": 1}),
("Orthogonal", initializers.Orthogonal, {}),
("Orthogonal_seeded", initializers.Orthogonal, {"seed": 1}),
("VarianceScaling", initializers.VarianceScaling, {}),
("VarianceScaling_seeded", initializers.VarianceScaling, {"seed": 1}),
("GlorotUniform", initializers.GlorotUniform, {}),
("GlorotUniform_seeded", initializers.GlorotUniform, {"seed": 1}),
("GlorotNormal", initializers.GlorotNormal, {}),
("GlorotNormal_seeded", initializers.GlorotNormal, {"seed": 1}),
("LecunNormal", initializers.LecunNormal, {}),
("LecunNormal_seeded", initializers.LecunNormal, {"seed": 1}),
("LecunUniform", initializers.LecunUniform, {}),
("LecunUniform_seeded", initializers.LecunUniform, {"seed": 1}),
("HeNormal", initializers.HeNormal, {}),
("HeNormal_seeded", initializers.HeNormal, {"seed": 1}),
("HeUniform", initializers.HeUniform, {}),
("HeUniform_seeded", initializers.HeUniform, {"seed": 1}),
)
def test_random_value_initializer(self, initializer_cls, init_args):
layout = dtensor.Layout(
[dtensor.UNSHARDED, dtensor.UNSHARDED], self.mesh
)
shape = (4, 4)
initializer = initializer_cls(**init_args)
# Make sure to raise error when keras global seed is not set.
with self.assertRaisesRegex(ValueError, "set the global seed"):
initializer(shape=shape, layout=layout)
try:
tf_utils.set_random_seed(1337)
value = initializer(shape=shape, layout=layout)
self.assertEqual(value.shape, shape)
fetched_layout = dtensor.fetch_layout(value)
self.assertEqual(layout, fetched_layout)
# Make sure when same seed is set again, the new initializer should
# generate same result
tf_utils.set_random_seed(1337)
initializer = initializer_cls(**init_args)
new_value = initializer(shape=shape, layout=layout)
self.assertAllClose(value, new_value)
finally:
# Unset the keras global generator so that it doesn't affect other
# tests that need to verify the existence of global generator.
backend._SEED_GENERATOR.generator = None
@parameterized.named_parameters(
("zeros", "zeros", initializers.Zeros),
("Zeros", "Zeros", initializers.Zeros),
("ones", "ones", initializers.Ones),
("Ones", "Ones", initializers.Ones),
("constant", "constant", initializers.Constant),
("Constant", "Constant", initializers.Constant),
("random_uniform", "random_uniform", initializers.RandomUniform),
("RandomUniform", "RandomUniform", initializers.RandomUniform),
("random_normal", "random_normal", initializers.RandomNormal),
("RandomNormal", "RandomNormal", initializers.RandomNormal),
("truncated_normal", "truncated_normal", initializers.TruncatedNormal),
("TruncatedNormal", "TruncatedNormal", initializers.TruncatedNormal),
("Identity", "Identity", initializers.Identity),
("identity", "identity", initializers.Identity),
("Orthogonal", "Orthogonal", initializers.Orthogonal),
("orthogonal", "orthogonal", initializers.Orthogonal),
("variance_scaling", "variance_scaling", initializers.VarianceScaling),
("VarianceScaling", "VarianceScaling", initializers.VarianceScaling),
("glorot_uniform", "glorot_uniform", initializers.GlorotUniform),
("GlorotUniform", "GlorotUniform", initializers.GlorotUniform),
("glorot_normal", "glorot_normal", initializers.GlorotNormal),
("GlorotNormal", "GlorotNormal", initializers.GlorotNormal),
("lecun_normal", "lecun_normal", initializers.LecunNormal),
("LecunNormal", "LecunNormal", initializers.LecunNormal),
("lecun_uniform", "lecun_uniform", initializers.LecunUniform),
("LecunUniform", "LecunUniform", initializers.LecunUniform),
("he_normal", "he_normal", initializers.HeNormal),
("HeNormal", "HeNormal", initializers.HeNormal),
("he_uniform", "he_uniform", initializers.HeUniform),
("HeUniform", "HeUniform", initializers.HeUniform),
)
def test_serialization_deserialization(self, cls_name, expected_cls):
initializer = initializers.get(cls_name)
self.assertIsInstance(initializer, expected_cls)
config = initializers.serialize(initializer)
recreated = initializers.deserialize(config)
self.assertIsInstance(recreated, expected_cls)
self.assertEqual(config, initializers.serialize(recreated))
if __name__ == "__main__":
tf.test.main()
|
tf-keras/tf_keras/dtensor/initializers_test.py/0
|
{
"file_path": "tf-keras/tf_keras/dtensor/initializers_test.py",
"repo_id": "tf-keras",
"token_count": 3086
}
| 190 |
# Copyright 2015 The TensorFlow Authors. All Rights Reserved.
#
# Licensed under the Apache License, Version 2.0 (the "License");
# you may not use this file except in compliance with the License.
# You may obtain a copy of the License at
#
# http://www.apache.org/licenses/LICENSE-2.0
#
# Unless required by applicable law or agreed to in writing, software
# distributed under the License is distributed on an "AS IS" BASIS,
# WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.
# See the License for the specific language governing permissions and
# limitations under the License.
# ==============================================================================
"""Contains the base Layer class, from which all layers inherit."""
import collections
import contextlib
import functools
import itertools
import textwrap
import threading
import warnings
import weakref
import numpy as np
import tensorflow.compat.v2 as tf
from tf_keras import backend
from tf_keras import constraints
from tf_keras import initializers
from tf_keras import regularizers
from tf_keras.dtensor import lazy_variable
from tf_keras.engine import base_layer_utils
from tf_keras.engine import input_spec
from tf_keras.engine import keras_tensor
from tf_keras.engine import node as node_module
from tf_keras.mixed_precision import autocast_variable
from tf_keras.mixed_precision import policy
from tf_keras.saving import serialization_lib
from tf_keras.saving.legacy.saved_model import layer_serialization
from tf_keras.utils import generic_utils
from tf_keras.utils import layer_utils
from tf_keras.utils import object_identity
from tf_keras.utils import tf_inspect
from tf_keras.utils import tf_utils
from tf_keras.utils import traceback_utils
from tf_keras.utils import version_utils
# A module that only depends on `keras.layers` import these from here.
from tf_keras.utils.generic_utils import to_snake_case # noqa: F401
from tf_keras.utils.tf_utils import is_tensor_or_tensor_list # noqa: F401
# isort: off
from google.protobuf import json_format
from tensorflow.python.platform import tf_logging
from tensorflow.python.util.tf_export import (
get_canonical_name_for_symbol,
)
from tensorflow.python.util.tf_export import keras_export
from tensorflow.tools.docs import doc_controls
metrics_mod = generic_utils.LazyLoader(
"metrics_mod", globals(), "tf_keras.metrics"
)
# Prefix that is added to the TF op layer names.
_TF_OP_LAYER_NAME_PREFIX = "tf_op_layer_"
# TODO(mdan): Should we have a single generic type for types that can be passed
# to tf.cast?
_AUTOCAST_TYPES = (tf.Tensor, tf.SparseTensor, tf.RaggedTensor)
_is_name_scope_on_model_declaration_enabled = False
_name_scope_unnester_stack = threading.local()
@contextlib.contextmanager
def _name_scope_unnester(full_name_scope):
"""Helper to get relative name scope from fully-speced nested name scopes.
Args:
full_name_scope: full(absolute) name scope path.
Yields:
Relative name scope path from the parent `_name_scope_unnester` context
manager.
Example:
```
with _name_scope_unnester('a') as name1: # name1 == 'a'
with _name_scope_unnester('a/b') as name2: # name2 == 'b'
with _name_scope_unnester('a/b/c') as name3: # name3 == 'c'
pass
```
"""
if not getattr(_name_scope_unnester_stack, "value", None):
_name_scope_unnester_stack.value = [""]
_name_scope_unnester_stack.value.append(full_name_scope)
try:
full_name_scope = _name_scope_unnester_stack.value[-1]
outer_name_scope = _name_scope_unnester_stack.value[-2]
relative_name_scope = full_name_scope.lstrip(outer_name_scope)
relative_name_scope = relative_name_scope.lstrip("/")
yield relative_name_scope
finally:
_name_scope_unnester_stack.value.pop()
@keras_export("keras.layers.Layer")
class Layer(tf.Module, version_utils.LayerVersionSelector):
"""This is the class from which all layers inherit.
A layer is a callable object that takes as input one or more tensors and
that outputs one or more tensors. It involves *computation*, defined
in the `call()` method, and a *state* (weight variables). State can be
created in various places, at the convenience of the subclass implementer:
* in `__init__()`;
* in the optional `build()` method, which is invoked by the first
`__call__()` to the layer, and supplies the shape(s) of the input(s),
which may not have been known at initialization time;
* in the first invocation of `call()`, with some caveats discussed
below.
Layers are recursively composable: If you assign a Layer instance as an
attribute of another Layer, the outer layer will start tracking the weights
created by the inner layer. Nested layers should be instantiated in the
`__init__()` method.
Users will just instantiate a layer and then treat it as a callable.
Args:
trainable: Boolean, whether the layer's variables should be trainable.
name: String name of the layer.
dtype: The dtype of the layer's computations and weights. Can also be a
`tf.keras.mixed_precision.Policy`, which allows the computation and
weight dtype to differ. Default of `None` means to use
`tf.keras.mixed_precision.global_policy()`, which is a float32 policy
unless set to different value.
dynamic: Set this to `True` if your layer should only be run eagerly, and
should not be used to generate a static computation graph.
This would be the case for a Tree-RNN or a recursive network,
for example, or generally for any layer that manipulates tensors
using Python control flow. If `False`, we assume that the layer can
safely be used to generate a static computation graph.
Attributes:
name: The name of the layer (string).
dtype: The dtype of the layer's weights.
variable_dtype: Alias of `dtype`.
compute_dtype: The dtype of the layer's computations. Layers automatically
cast inputs to this dtype which causes the computations and output to
also be in this dtype. When mixed precision is used with a
`tf.keras.mixed_precision.Policy`, this will be different than
`variable_dtype`.
dtype_policy: The layer's dtype policy. See the
`tf.keras.mixed_precision.Policy` documentation for details.
trainable_weights: List of variables to be included in backprop.
non_trainable_weights: List of variables that should not be
included in backprop.
weights: The concatenation of the lists trainable_weights and
non_trainable_weights (in this order).
trainable: Whether the layer should be trained (boolean), i.e. whether
its potentially-trainable weights should be returned as part of
`layer.trainable_weights`.
input_spec: Optional (list of) `InputSpec` object(s) specifying the
constraints on inputs that can be accepted by the layer.
We recommend that descendants of `Layer` implement the following methods:
* `__init__()`: Defines custom layer attributes, and creates layer weights
that do not depend on input shapes, using `add_weight()`, or other state.
* `build(self, input_shape)`: This method can be used to create weights that
depend on the shape(s) of the input(s), using `add_weight()`, or other
state. `__call__()` will automatically build the layer (if it has not been
built yet) by calling `build()`.
* `call(self, inputs, *args, **kwargs)`: Called in `__call__` after making
sure `build()` has been called. `call()` performs the logic of applying
the layer to the `inputs`. The first invocation may additionally create
state that could not be conveniently created in `build()`; see its
docstring for details.
Two reserved keyword arguments you can optionally use in `call()` are:
- `training` (boolean, whether the call is in inference mode or training
mode). See more details in [the layer/model subclassing guide](
https://www.tensorflow.org/guide/keras/custom_layers_and_models#privileged_training_argument_in_the_call_method)
- `mask` (boolean tensor encoding masked timesteps in the input, used
in RNN layers). See more details in
[the layer/model subclassing guide](
https://www.tensorflow.org/guide/keras/custom_layers_and_models#privileged_mask_argument_in_the_call_method)
A typical signature for this method is `call(self, inputs)`, and user
could optionally add `training` and `mask` if the layer need them. `*args`
and `**kwargs` is only useful for future extension when more input
parameters are planned to be added.
* `get_config(self)`: Returns a dictionary containing the configuration used
to initialize this layer. If the keys differ from the arguments
in `__init__`, then override `from_config(self)` as well.
This method is used when saving
the layer or a model that contains this layer.
Examples:
Here's a basic example: a layer with two variables, `w` and `b`,
that returns `y = w . x + b`.
It shows how to implement `build()` and `call()`.
Variables set as attributes of a layer are tracked as weights
of the layers (in `layer.weights`).
```python
class SimpleDense(Layer):
def __init__(self, units=32):
super(SimpleDense, self).__init__()
self.units = units
def build(self, input_shape): # Create the state of the layer (weights)
w_init = tf.random_normal_initializer()
self.w = tf.Variable(
initial_value=w_init(shape=(input_shape[-1], self.units),
dtype='float32'),
trainable=True)
b_init = tf.zeros_initializer()
self.b = tf.Variable(
initial_value=b_init(shape=(self.units,), dtype='float32'),
trainable=True)
def call(self, inputs): # Defines the computation from inputs to outputs
return tf.matmul(inputs, self.w) + self.b
# Instantiates the layer.
linear_layer = SimpleDense(4)
# This will also call `build(input_shape)` and create the weights.
y = linear_layer(tf.ones((2, 2)))
assert len(linear_layer.weights) == 2
# These weights are trainable, so they're listed in `trainable_weights`:
assert len(linear_layer.trainable_weights) == 2
```
Note that the method `add_weight()` offers a shortcut to create weights:
```python
class SimpleDense(Layer):
def __init__(self, units=32):
super(SimpleDense, self).__init__()
self.units = units
def build(self, input_shape):
self.w = self.add_weight(shape=(input_shape[-1], self.units),
initializer='random_normal',
trainable=True)
self.b = self.add_weight(shape=(self.units,),
initializer='random_normal',
trainable=True)
def call(self, inputs):
return tf.matmul(inputs, self.w) + self.b
```
Besides trainable weights, updated via backpropagation during training,
layers can also have non-trainable weights. These weights are meant to
be updated manually during `call()`. Here's a example layer that computes
the running sum of its inputs:
```python
class ComputeSum(Layer):
def __init__(self, input_dim):
super(ComputeSum, self).__init__()
# Create a non-trainable weight.
self.total = tf.Variable(initial_value=tf.zeros((input_dim,)),
trainable=False)
def call(self, inputs):
self.total.assign_add(tf.reduce_sum(inputs, axis=0))
return self.total
my_sum = ComputeSum(2)
x = tf.ones((2, 2))
y = my_sum(x)
print(y.numpy()) # [2. 2.]
y = my_sum(x)
print(y.numpy()) # [4. 4.]
assert my_sum.weights == [my_sum.total]
assert my_sum.non_trainable_weights == [my_sum.total]
assert my_sum.trainable_weights == []
```
For more information about creating layers, see the guide
[Making new Layers and Models via subclassing](
https://www.tensorflow.org/guide/keras/custom_layers_and_models)
"""
@tf.__internal__.tracking.no_automatic_dependency_tracking
def __init__(
self, trainable=True, name=None, dtype=None, dynamic=False, **kwargs
):
self._instrument_layer_creation()
# These properties should be set by the user via keyword arguments.
# note that 'dtype', 'input_shape' and 'batch_input_shape'
# are only applicable to input layers: do not pass these keywords
# to non-input layers.
allowed_kwargs = {
"input_dim",
"input_shape",
"batch_input_shape",
"batch_size",
"weights",
"activity_regularizer",
"autocast",
"implementation",
}
# Validate optional keyword arguments.
generic_utils.validate_kwargs(kwargs, allowed_kwargs)
# Mutable properties
# Indicates whether the layer's weights are updated during training
# and whether the layer's updates are run during training.
if not (
isinstance(trainable, bool)
or (
isinstance(trainable, (tf.Tensor, tf.Variable))
and trainable.dtype is tf.bool
)
):
raise TypeError(
"Expected `trainable` argument to be a boolean, "
f"but got: {trainable}"
)
self._trainable = trainable
# A stateful layer is a layer whose updates are run during inference
# too, for instance stateful RNNs.
self._stateful = False
# Indicates whether `build` needs to be called upon layer call, to
# create the layer's weights. (Note that the first call() may also
# create weights, independent of build().)
self.built = False
# Provides information about which inputs are compatible with the layer.
self._input_spec = None
# SavedModel-related attributes.
# Record the build input shape for loading purposes.
# TODO(kathywu): Move this to Layer._set_save_spec once cl/290121460 is
# submitted.
self._build_input_shape = None
self._saved_model_inputs_spec = None
self._saved_model_arg_spec = None
# `Layer.compute_mask` will be called at the end of `Layer.__call__` if
# `Layer.compute_mask` is overridden, or if the `Layer` subclass sets
# `self.supports_masking=True`.
self._supports_masking = not generic_utils.is_default(self.compute_mask)
self._init_set_name(name)
self._activity_regularizer = regularizers.get(
kwargs.pop("activity_regularizer", None)
)
self._maybe_create_attribute("_trainable_weights", [])
self._maybe_create_attribute("_non_trainable_weights", [])
self._updates = []
# Object to store all thread local layer properties.
self._thread_local = threading.local()
# A list of zero-argument lambdas which return Tensors, used for
# variable regularizers.
self._callable_losses = []
# A list of symbolic Tensors containing activity regularizers and losses
# manually added through `add_loss` in graph-building mode.
self._losses = []
# A list of metric instances corresponding to the symbolic metric
# tensors added using the `add_metric` API.
self._metrics = []
# Ensures the same metric is not added multiple times in
# `MirroredStrategy`.
self._metrics_lock = threading.Lock()
# Note that models also have a dtype policy, as they are layers. For
# functional models, the policy is only used in Model.compile, which
# wraps the optimizer with a LossScaleOptimizer if the policy name is
# "mixed_float16". Subclassed models additionally use the policy's
# compute and variable dtypes, as like any ordinary layer.
self._set_dtype_policy(dtype)
# Boolean indicating whether the layer automatically casts its inputs to
# the layer's compute_dtype.
self._autocast = kwargs.get(
"autocast", base_layer_utils.v2_dtype_behavior_enabled()
)
# Tracks `TrackableDataStructure`s, `Module`s, and `Layer`s.
# Ordered by when the object was assigned as an attr.
# Entries are unique.
self._maybe_create_attribute("_self_tracked_trackables", [])
# These lists will be filled via successive calls
# to self._add_inbound_node().
# Used in symbolic mode only, only in conjunction with graph-networks
self._inbound_nodes_value = []
self._outbound_nodes_value = []
self._init_call_fn_args()
# Whether the `call` method can be used to build a TF graph without
# issues. This attribute has no effect if the model is created using
# the Functional API. Instead, `model.dynamic` is determined based on
# the internal layers.
if not isinstance(dynamic, bool):
raise TypeError(
"Expected `dynamic` argument to be a boolean, "
f"but got: {dynamic}"
)
self._dynamic = dynamic
# Manage input shape information if passed.
if "input_dim" in kwargs and "input_shape" not in kwargs:
# Backwards compatibility: alias 'input_dim' to 'input_shape'.
kwargs["input_shape"] = (kwargs["input_dim"],)
if "input_shape" in kwargs or "batch_input_shape" in kwargs:
# In this case we will later create an input layer
# to insert before the current layer
if "batch_input_shape" in kwargs:
batch_input_shape = tuple(kwargs["batch_input_shape"])
elif "input_shape" in kwargs:
if "batch_size" in kwargs:
batch_size = kwargs["batch_size"]
else:
batch_size = None
batch_input_shape = (batch_size,) + tuple(kwargs["input_shape"])
self._batch_input_shape = batch_input_shape
# Manage initial weight values if passed.
self._initial_weights = kwargs.get("weights", None)
# Whether the layer will track any layers that is set as attribute on
# itself as sub-layers, the weights from the sub-layers will be included
# in the parent layer's variables() as well. Defaults to `True`, which
# means auto tracking is turned on. Certain subclass might want to turn
# it off, like Sequential model.
self._auto_track_sub_layers = True
# For backwards compat reasons, most built-in layers do not guarantee
# That they will 100% preserve the structure of input args when saving
# / loading configs. E.g. they may un-nest an arg that is
# a list with one element.
self._preserve_input_structure_in_config = False
# Save outer name scope at layer declaration so that it is preserved at
# the actual layer construction.
self._name_scope_on_declaration = tf.get_current_name_scope()
# Save the temp regularization losses created in the DTensor use case.
# When DTensor is enable, we will first create LazyInitVariable and then
# DVariable with proper layout afterward. For the weights regularization
# loss, we have to create against the DVariable as well.
self._captured_weight_regularizer = []
@tf.__internal__.tracking.no_automatic_dependency_tracking
@generic_utils.default
def build(self, input_shape):
"""Creates the variables of the layer (for subclass implementers).
This is a method that implementers of subclasses of `Layer` or `Model`
can override if they need a state-creation step in-between
layer instantiation and layer call. It is invoked automatically before
the first execution of `call()`.
This is typically used to create the weights of `Layer` subclasses
(at the discretion of the subclass implementer).
Args:
input_shape: Instance of `TensorShape`, or list of instances of
`TensorShape` if the layer expects a list of inputs
(one instance per input).
"""
self._build_input_shape = input_shape
self.built = True
@doc_controls.for_subclass_implementers
def call(self, inputs, *args, **kwargs):
"""This is where the layer's logic lives.
The `call()` method may not create state (except in its first
invocation, wrapping the creation of variables or other resources in
`tf.init_scope()`). It is recommended to create state, including
`tf.Variable` instances and nested `Layer` instances,
in `__init__()`, or in the `build()` method that is
called automatically before `call()` executes for the first time.
Args:
inputs: Input tensor, or dict/list/tuple of input tensors.
The first positional `inputs` argument is subject to special rules:
- `inputs` must be explicitly passed. A layer cannot have zero
arguments, and `inputs` cannot be provided via the default value
of a keyword argument.
- NumPy array or Python scalar values in `inputs` get cast as
tensors.
- TF-Keras mask metadata is only collected from `inputs`.
- Layers are built (`build(input_shape)` method)
using shape info from `inputs` only.
- `input_spec` compatibility is only checked against `inputs`.
- Mixed precision input casting is only applied to `inputs`.
If a layer has tensor arguments in `*args` or `**kwargs`, their
casting behavior in mixed precision should be handled manually.
- The SavedModel input specification is generated using `inputs`
only.
- Integration with various ecosystem packages like TFMOT, TFLite,
TF.js, etc is only supported for `inputs` and not for tensors in
positional and keyword arguments.
*args: Additional positional arguments. May contain tensors, although
this is not recommended, for the reasons above.
**kwargs: Additional keyword arguments. May contain tensors, although
this is not recommended, for the reasons above.
The following optional keyword arguments are reserved:
- `training`: Boolean scalar tensor of Python boolean indicating
whether the `call` is meant for training or inference.
- `mask`: Boolean input mask. If the layer's `call()` method takes a
`mask` argument, its default value will be set to the mask
generated for `inputs` by the previous layer (if `input` did come
from a layer that generated a corresponding mask, i.e. if it came
from a TF-Keras layer with masking support).
Returns:
A tensor or list/tuple of tensors.
"""
return inputs
@doc_controls.for_subclass_implementers
def add_weight(
self,
name=None,
shape=None,
dtype=None,
initializer=None,
regularizer=None,
trainable=None,
constraint=None,
use_resource=None,
synchronization=tf.VariableSynchronization.AUTO,
aggregation=tf.VariableAggregation.NONE,
**kwargs,
):
"""Adds a new variable to the layer.
Args:
name: Variable name.
shape: Variable shape. Defaults to scalar if unspecified.
dtype: The type of the variable. Defaults to `self.dtype`.
initializer: Initializer instance (callable).
regularizer: Regularizer instance (callable).
trainable: Boolean, whether the variable should be part of the layer's
"trainable_variables" (e.g. variables, biases)
or "non_trainable_variables" (e.g. BatchNorm mean and variance).
Note that `trainable` cannot be `True` if `synchronization`
is set to `ON_READ`.
constraint: Constraint instance (callable).
use_resource: Whether to use a `ResourceVariable` or not.
See [this guide](
https://www.tensorflow.org/guide/migrate/tf1_vs_tf2#resourcevariables_instead_of_referencevariables)
for more information.
synchronization: Indicates when a distributed a variable will be
aggregated. Accepted values are constants defined in the class
`tf.VariableSynchronization`. By default the synchronization is set
to `AUTO` and the current `DistributionStrategy` chooses when to
synchronize. If `synchronization` is set to `ON_READ`, `trainable`
must not be set to `True`.
aggregation: Indicates how a distributed variable will be aggregated.
Accepted values are constants defined in the class
`tf.VariableAggregation`.
**kwargs: Additional keyword arguments. Accepted values are `getter`,
`collections`, `experimental_autocast` and `caching_device`.
Returns:
The variable created.
Raises:
ValueError: When giving unsupported dtype and no initializer or when
trainable has been set to True with synchronization set as
`ON_READ`.
"""
if shape is None:
shape = ()
kwargs.pop("partitioner", None) # Ignored.
# Validate optional keyword arguments.
for kwarg in kwargs:
if kwarg not in [
"collections",
"experimental_autocast",
"caching_device",
"getter",
"layout",
"experimental_enable_variable_lifting",
]:
raise TypeError("Unknown keyword argument:", kwarg)
collections_arg = kwargs.pop("collections", None)
# 'experimental_autocast' can be set to False by the caller to indicate
# an AutoCastVariable should never be created.
autocast = kwargs.pop("experimental_autocast", True)
# See the docstring for tf.Variable about the details for
# caching_device.
caching_device = kwargs.pop("caching_device", None)
layout = kwargs.pop("layout", None)
# Specially handling of auto layout fetch, based on the variable name
# and attribute name. For built-in keras layers, usually the variable
# name, eg 'kernel', will match with a 'kernel_layout' attribute name on
# the instance. We will try to do this auto fetch if layout is not
# explicitly specified. This is mainly a quick workaround for not
# applying too many interface change to built-in layers, until DTensor
# is a public API. Also see dtensor.utils.allow_initializer_layout for
# more details.
# TODO(scottzhu): Remove this once dtensor is public to end user.
if not layout and name:
layout = getattr(self, name + "_layout", None)
if dtype is None:
dtype = self.dtype or backend.floatx()
dtype = tf.as_dtype(dtype)
if self._dtype_policy.variable_dtype is None:
# The policy is "_infer", so we infer the policy from the variable
# dtype.
self._set_dtype_policy(policy.Policy(dtype.base_dtype.name))
initializer = initializers.get(initializer)
regularizer = regularizers.get(regularizer)
constraint = constraints.get(constraint)
if synchronization == tf.VariableSynchronization.ON_READ:
if trainable:
raise ValueError(
"Synchronization value can be set to "
"VariableSynchronization.ON_READ only for non-trainable "
"variables. You have specified trainable=True and "
"synchronization=VariableSynchronization.ON_READ."
)
else:
# Set trainable to be false when variable is to be synced on
# read.
trainable = False
elif trainable is None:
trainable = True
# Initialize variable when no initializer provided
if initializer is None:
# If dtype is DT_FLOAT, provide a uniform unit scaling initializer
if dtype.is_floating:
initializer = initializers.get("glorot_uniform")
# If dtype is DT_INT/DT_UINT, provide a default value `zero`
# If dtype is DT_BOOL, provide a default value `FALSE`
elif dtype.is_integer or dtype.is_unsigned or dtype.is_bool:
initializer = initializers.get("zeros")
# NOTES:Do we need to support for handling DT_STRING and DT_COMPLEX
# here?
elif "getter" not in kwargs:
# When `getter` is specified, it's possibly fine for
# `initializer` to be None since it's up to the custom `getter`
# to raise error in case it indeed needs `initializer`.
raise ValueError(
f"An initializer for variable {name} of type "
f"{dtype.base_dtype} is required for layer "
f"{self.name}. Received: {initializer}."
)
getter = kwargs.pop("getter", base_layer_utils.make_variable)
if (
autocast
and self._dtype_policy.compute_dtype
!= self._dtype_policy.variable_dtype
and dtype.is_floating
):
old_getter = getter
# Wrap variable constructor to return an AutoCastVariable.
def getter(*args, **kwargs):
variable = old_getter(*args, **kwargs)
return autocast_variable.create_autocast_variable(variable)
# Also the caching_device does not work with the mixed precision
# API, disable it if it is specified.
# TODO(b/142020079): Re-enable it once the bug is fixed.
if caching_device is not None:
tf_logging.warning(
"`caching_device` does not work with mixed precision API. "
"Ignoring user specified `caching_device`."
)
caching_device = None
if layout:
getter = functools.partial(getter, layout=layout)
variable = self._add_variable_with_custom_getter(
name=name,
shape=shape,
# TODO(allenl): a `make_variable` equivalent should be added as a
# `Trackable` method.
getter=getter,
# Manage errors in Layer rather than Trackable.
overwrite=True,
initializer=initializer,
dtype=dtype,
constraint=constraint,
trainable=trainable,
use_resource=use_resource,
collections=collections_arg,
synchronization=synchronization,
aggregation=aggregation,
caching_device=caching_device,
)
if regularizer is not None:
# TODO(fchollet): in the future, this should be handled at the
# level of variable creation, and weight regularization losses
# should be variable attributes.
name_in_scope = variable.name[: variable.name.find(":")]
self._handle_weight_regularization(
name_in_scope, variable, regularizer
)
if base_layer_utils.is_split_variable(variable):
for v in variable:
backend.track_variable(v)
if trainable:
self._trainable_weights.append(v)
else:
self._non_trainable_weights.append(v)
else:
backend.track_variable(variable)
if trainable:
self._trainable_weights.append(variable)
else:
self._non_trainable_weights.append(variable)
return variable
def __new__(cls, *args, **kwargs):
# Generate a config to be returned by default by `get_config()`.
arg_names = tf_inspect.getfullargspec(cls.__init__).args
kwargs.update(dict(zip(arg_names[1 : len(args) + 1], args)))
instance = super(Layer, cls).__new__(cls, *args, **kwargs)
# For safety, we only rely on auto-configs for a small set of
# serializable types.
supported_types = (str, int, float, bool, type(None))
try:
flat_arg_values = tf.nest.flatten(kwargs)
auto_get_config = True
for value in flat_arg_values:
if not isinstance(value, supported_types):
auto_get_config = False
break
except TypeError:
auto_get_config = False
try:
instance._auto_get_config = auto_get_config
if auto_get_config:
instance._auto_config = serialization_lib.Config(**kwargs)
except RecursionError:
# Setting an instance attribute in __new__ has the potential
# to trigger an infinite recursion if a subclass overrides
# setattr in an unsafe way.
pass
return instance
@generic_utils.default
def get_config(self):
"""Returns the config of the layer.
A layer config is a Python dictionary (serializable)
containing the configuration of a layer.
The same layer can be reinstantiated later
(without its trained weights) from this configuration.
The config of a layer does not include connectivity
information, nor the layer class name. These are handled
by `Network` (one layer of abstraction above).
Note that `get_config()` does not guarantee to return a fresh copy of
dict every time it is called. The callers should make a copy of the
returned dict if they want to modify it.
Returns:
Python dictionary.
"""
config = {
"name": self.name,
"trainable": self.trainable,
}
config["dtype"] = policy.serialize(self._dtype_policy)
if hasattr(self, "_batch_input_shape"):
config["batch_input_shape"] = self._batch_input_shape
if not generic_utils.is_default(self.get_config):
# In this case the subclass implements get_config()
return config
# In this case the subclass doesn't implement get_config():
# Let's see if we can autogenerate it.
if getattr(self, "_auto_get_config", False):
xtra_args = set(config.keys())
config.update(self._auto_config.config)
# Remove args non explicitly supported
argspec = tf_inspect.getfullargspec(self.__init__)
if argspec.varkw != "kwargs":
for key in xtra_args - xtra_args.intersection(argspec.args[1:]):
config.pop(key, None)
return config
else:
raise NotImplementedError(
textwrap.dedent(
f"""
Layer {self.__class__.__name__} was created by passing
non-serializable argument values in `__init__()`,
and therefore the layer must override `get_config()` in
order to be serializable. Please implement `get_config()`.
Example:
class CustomLayer(keras.layers.Layer):
def __init__(self, arg1, arg2, **kwargs):
super().__init__(**kwargs)
self.arg1 = arg1
self.arg2 = arg2
def get_config(self):
config = super().get_config()
config.update({{
"arg1": self.arg1,
"arg2": self.arg2,
}})
return config"""
)
)
@classmethod
def from_config(cls, config):
"""Creates a layer from its config.
This method is the reverse of `get_config`,
capable of instantiating the same layer from the config
dictionary. It does not handle layer connectivity
(handled by Network), nor weights (handled by `set_weights`).
Args:
config: A Python dictionary, typically the
output of get_config.
Returns:
A layer instance.
"""
try:
return cls(**config)
except Exception as e:
raise TypeError(
f"Error when deserializing class '{cls.__name__}' using "
f"config={config}.\n\nException encountered: {e}"
)
def compute_output_shape(self, input_shape):
"""Computes the output shape of the layer.
This method will cause the layer's state to be built, if that has not
happened before. This requires that the layer will later be used with
inputs that match the input shape provided here.
Args:
input_shape: Shape tuple (tuple of integers) or `tf.TensorShape`,
or structure of shape tuples / `tf.TensorShape` instances
(one per output tensor of the layer).
Shape tuples can include None for free dimensions,
instead of an integer.
Returns:
A `tf.TensorShape` instance
or structure of `tf.TensorShape` instances.
"""
if tf.executing_eagerly():
# In this case we build the model first in order to do shape
# inference. This is acceptable because the framework only calls
# `compute_output_shape` on shape values that the layer would later
# be built for. It would however cause issues in case a user
# attempts to use `compute_output_shape` manually with shapes that
# are incompatible with the shape the Layer will be called on (these
# users will have to implement `compute_output_shape` themselves).
self._maybe_build(input_shape)
graph_name = str(self.name) + "_scratch_graph"
with tf.__internal__.FuncGraph(graph_name).as_default():
input_shape = tf_utils.convert_shapes(
input_shape, to_tuples=False
)
def _make_placeholder_like(shape):
ph = backend.placeholder(shape=shape, dtype=self.dtype)
ph._keras_mask = None
return ph
inputs = tf.nest.map_structure(
_make_placeholder_like, input_shape
)
try:
outputs = self(inputs, training=False)
except TypeError as e:
raise NotImplementedError(
"We could not automatically infer the static shape of "
"the layer's output. Please implement the "
"`compute_output_shape` method on your layer (%s)."
% self.__class__.__name__
) from e
return tf.nest.map_structure(lambda t: t.shape, outputs)
raise NotImplementedError(
"Please run in eager mode or implement the `compute_output_shape` "
"method on your layer (%s)." % self.__class__.__name__
)
@doc_controls.for_subclass_implementers
def compute_output_signature(self, input_signature):
"""Compute the output tensor signature of the layer based on the inputs.
Unlike a TensorShape object, a TensorSpec object contains both shape
and dtype information for a tensor. This method allows layers to provide
output dtype information if it is different from the input dtype.
For any layer that doesn't implement this function,
the framework will fall back to use `compute_output_shape`, and will
assume that the output dtype matches the input dtype.
Args:
input_signature: Single TensorSpec or nested structure of TensorSpec
objects, describing a candidate input for the layer.
Returns:
Single TensorSpec or nested structure of TensorSpec objects,
describing how the layer would transform the provided input.
Raises:
TypeError: If input_signature contains a non-TensorSpec object.
"""
def check_type_return_shape(s):
if not isinstance(s, tf.TensorSpec):
raise TypeError(
"Only TensorSpec signature types are supported. "
f"Received: {s}."
)
return s.shape
input_shape = tf.nest.map_structure(
check_type_return_shape, input_signature
)
output_shape = self.compute_output_shape(input_shape)
try:
dtype = self.output.dtype
except AttributeError:
dtype = self._compute_dtype
if dtype is None:
input_dtypes = [s.dtype for s in tf.nest.flatten(input_signature)]
# Default behavior when self.dtype is None, is to use the first
# input's dtype.
dtype = input_dtypes[0]
return tf.nest.map_structure(
lambda s: tf.TensorSpec(dtype=dtype, shape=s), output_shape
)
@generic_utils.default
def compute_mask(self, inputs, mask=None):
"""Computes an output mask tensor.
Args:
inputs: Tensor or list of tensors.
mask: Tensor or list of tensors.
Returns:
None or a tensor (or list of tensors,
one per output tensor of the layer).
"""
if not self._supports_masking:
if any(m is not None for m in tf.nest.flatten(mask)):
raise TypeError(
"Layer " + self.name + " does not support masking, "
"but was passed an input_mask: " + str(mask)
)
# masking not explicitly supported: return None as mask.
return None
# if masking is explicitly supported, by default
# carry over the input mask
return mask
@traceback_utils.filter_traceback
def __call__(self, *args, **kwargs):
"""Wraps `call`, applying pre- and post-processing steps.
Args:
*args: Positional arguments to be passed to `self.call`.
**kwargs: Keyword arguments to be passed to `self.call`.
Returns:
Output tensor(s).
Note:
- The following optional keyword arguments are reserved for specific
uses:
* `training`: Boolean scalar tensor of Python boolean indicating
whether the `call` is meant for training or inference.
* `mask`: Boolean input mask.
- If the layer's `call` method takes a `mask` argument (as some Keras
layers do), its default value will be set to the mask generated
for `inputs` by the previous layer (if `input` did come from
a layer that generated a corresponding mask, i.e. if it came from
a TF-Keras layer with masking support.
- If the layer is not built, the method will call `build`.
Raises:
ValueError: if the layer's `call` method returns None (an invalid
value).
RuntimeError: if `super().__init__()` was not called in the
constructor.
"""
if not hasattr(self, "_thread_local"):
raise RuntimeError(
"You must call `super().__init__()` in the layer constructor."
)
# `inputs` (the first arg in the method spec) is special cased in
# layer call due to historical reasons.
# This special casing currently takes the form of:
# - 'inputs' must be explicitly passed. A layer cannot have zero
# arguments, and inputs cannot have been provided via the default
# value of a kwarg.
# - numpy/scalar values in `inputs` get converted to tensors
# - implicit masks / mask metadata are only collected from 'inputs`
# - Layers are built using shape info from 'inputs' only
# - input_spec compatibility is only checked against `inputs`
# - mixed precision casting (autocast) is only applied to `inputs`,
# not to any other argument.
inputs, args, kwargs = self._call_spec.split_out_first_arg(args, kwargs)
input_list = tf.nest.flatten(inputs)
# Functional Model construction mode is invoked when `Layer`s are called
# on symbolic `KerasTensor`s, i.e.:
# >> inputs = tf.keras.Input(10)
# >> outputs = MyLayer()(inputs) # Functional construction mode.
# >> model = tf.keras.Model(inputs, outputs)
if _in_functional_construction_mode(
self, inputs, args, kwargs, input_list
):
return self._functional_construction_call(
inputs, args, kwargs, input_list
)
# Maintains info about the `Layer.call` stack.
call_context = base_layer_utils.call_context()
# Accept NumPy and scalar inputs by converting to Tensors.
if any(
isinstance(x, (tf.Tensor, np.ndarray, float, int))
for x in input_list
):
inputs = tf.nest.map_structure(
_convert_numpy_or_python_types, inputs
)
input_list = tf.nest.flatten(inputs)
# Handle `mask` propagation from previous layer to current layer. Masks
# can be propagated explicitly via the `mask` argument, or implicitly
# via setting the `_keras_mask` attribute on the inputs to a Layer.
# Masks passed explicitly take priority.
input_masks, mask_is_implicit = self._get_input_masks(
inputs, input_list, args, kwargs
)
if self._expects_mask_arg and mask_is_implicit:
kwargs["mask"] = input_masks
# Training mode for `Layer.call` is set via (in order of priority):
# (1) The `training` argument passed to this `Layer.call`, if it is not
# None
# (2) The training mode of an outer `Layer.call`.
# (3) The default mode set by `tf.keras.backend.set_learning_phase` (if
# set)
# (4) Any non-None default value for `training` specified in the call
# signature
# (5) False (treating the layer as if it's in inference)
args, kwargs, training_mode = self._set_training_mode(
args, kwargs, call_context
)
# Losses are cleared for all sublayers on the outermost `Layer.call`.
# Losses are not cleared on inner `Layer.call`s, because sublayers can
# be called multiple times.
if not call_context.in_call:
self._clear_losses()
eager = tf.executing_eagerly()
with call_context.enter(
layer=self,
inputs=inputs,
build_graph=not eager,
training=training_mode,
):
input_spec.assert_input_compatibility(
self.input_spec, inputs, self.name
)
if eager:
call_fn = self.call
name_scope = self._name
else:
name_scope = self._get_unnested_name_scope()
call_fn = self._autographed_call()
call_fn = traceback_utils.inject_argument_info_in_traceback(
call_fn,
object_name=(
f"layer '{self.name}' (type {self.__class__.__name__})"
),
)
with contextlib.ExitStack() as namescope_stack:
if _is_name_scope_on_model_declaration_enabled:
namescope_stack.enter_context(
_name_scope_unnester(self._name_scope_on_declaration)
)
namescope_stack.enter_context(tf.name_scope(name_scope))
if not self.built:
self._maybe_build(inputs)
if self._autocast:
inputs = self._maybe_cast_inputs(inputs, input_list)
with autocast_variable.enable_auto_cast_variables(
self._compute_dtype_object
):
outputs = call_fn(inputs, *args, **kwargs)
if self._activity_regularizer:
self._handle_activity_regularization(inputs, outputs)
if self._supports_masking:
self._set_mask_metadata(
inputs, outputs, input_masks, not eager
)
if self._saved_model_inputs_spec is None:
self._set_save_spec(inputs, args, kwargs)
return outputs
def _get_unnested_name_scope(self):
if _is_name_scope_on_model_declaration_enabled:
with _name_scope_unnester(
self._name_scope_on_declaration
) as relative_name_scope_on_declaration:
# To avoid `tf.name_scope` autoincrement, use absolute path.
relative_name_scope = filter(
None,
[
tf.get_current_name_scope(),
relative_name_scope_on_declaration,
],
)
current_name_scope = "/".join(relative_name_scope) + "/"
if current_name_scope == "/":
current_name_scope = self._name_scope_on_declaration
with tf.name_scope(current_name_scope):
name_scope = self._name_scope() # Avoid autoincrementing.
else:
name_scope = self._name_scope()
return name_scope
@property
def dtype(self):
"""The dtype of the layer weights.
This is equivalent to `Layer.dtype_policy.variable_dtype`. Unless
mixed precision is used, this is the same as `Layer.compute_dtype`, the
dtype of the layer's computations.
"""
return self._dtype_policy.variable_dtype
@property
def name(self):
"""Name of the layer (string), set in the constructor."""
return self._name
@property
def supports_masking(self):
"""Whether this layer supports computing a mask using `compute_mask`."""
return self._supports_masking
@supports_masking.setter
def supports_masking(self, value):
self._supports_masking = value
@property
def dynamic(self):
"""Whether the layer is dynamic (eager-only); set in the constructor."""
return any(layer._dynamic for layer in self._flatten_layers())
@property
@doc_controls.do_not_doc_inheritable
def stateful(self):
return any(layer._stateful for layer in self._flatten_layers())
@stateful.setter
def stateful(self, value):
self._stateful = value
@property
def trainable(self):
return self._trainable
@trainable.setter
def trainable(self, value):
"""Sets trainable attribute for the layer and its sublayers.
When this value is changed during training (e.g. with a
`tf.keras.callbacks.Callback`) you need to call the parent
`tf.keras.Model.make_train_function` with `force=True` in order to
recompile the training graph.
Args:
value: Boolean with the desired state for the layer's trainable
attribute.
"""
for layer in self._flatten_layers():
layer._trainable = value
@property
def activity_regularizer(self):
"""Optional regularizer function for the output of this layer."""
return self._activity_regularizer
@activity_regularizer.setter
def activity_regularizer(self, regularizer):
"""Optional regularizer function for the output of this layer."""
self._activity_regularizer = regularizer
@property
def input_spec(self):
"""`InputSpec` instance(s) describing the input format for this layer.
When you create a layer subclass, you can set `self.input_spec` to
enable the layer to run input compatibility checks when it is called.
Consider a `Conv2D` layer: it can only be called on a single input
tensor of rank 4. As such, you can set, in `__init__()`:
```python
self.input_spec = tf.keras.layers.InputSpec(ndim=4)
```
Now, if you try to call the layer on an input that isn't rank 4
(for instance, an input of shape `(2,)`, it will raise a
nicely-formatted error:
```
ValueError: Input 0 of layer conv2d is incompatible with the layer:
expected ndim=4, found ndim=1. Full shape received: [2]
```
Input checks that can be specified via `input_spec` include:
- Structure (e.g. a single input, a list of 2 inputs, etc)
- Shape
- Rank (ndim)
- Dtype
For more information, see `tf.keras.layers.InputSpec`.
Returns:
A `tf.keras.layers.InputSpec` instance, or nested structure thereof.
"""
return self._input_spec
@input_spec.setter
# Must be decorated to prevent tracking, since the input_spec can be nested
# InputSpec objects.
@tf.__internal__.tracking.no_automatic_dependency_tracking
def input_spec(self, value):
for v in tf.nest.flatten(value):
if v is not None and not isinstance(v, input_spec.InputSpec):
raise TypeError(
"Layer input_spec must be an instance of InputSpec. "
"Got: {}".format(v)
)
self._input_spec = value
@property
def trainable_weights(self):
"""List of all trainable weights tracked by this layer.
Trainable weights are updated via gradient descent during training.
Returns:
A list of trainable variables.
"""
self._update_trackables()
if self.trainable:
children_weights = self._gather_children_attribute(
"trainable_variables"
)
return self._dedup_weights(
self._trainable_weights + children_weights
)
else:
return []
@property
def non_trainable_weights(self):
"""List of all non-trainable weights tracked by this layer.
Non-trainable weights are *not* updated during training. They are
expected to be updated manually in `call()`.
Returns:
A list of non-trainable variables.
"""
self._update_trackables()
if self.trainable:
children_weights = self._gather_children_attribute(
"non_trainable_variables"
)
non_trainable_weights = (
self._non_trainable_weights + children_weights
)
else:
children_weights = self._gather_children_attribute("variables")
non_trainable_weights = (
self._trainable_weights
+ self._non_trainable_weights
+ children_weights
)
return self._dedup_weights(non_trainable_weights)
@property
def weights(self):
"""Returns the list of all layer variables/weights.
Returns:
A list of variables.
"""
return self.trainable_weights + self.non_trainable_weights
@property
@doc_controls.do_not_generate_docs
def updates(self):
warnings.warn(
"`layer.updates` will be removed in a future version. "
"This property should not be used in TensorFlow 2.0, "
"as `updates` are applied automatically.",
stacklevel=2,
)
return []
@property
def losses(self):
"""List of losses added using the `add_loss()` API.
Variable regularization tensors are created when this property is
accessed, so it is eager safe: accessing `losses` under a
`tf.GradientTape` will propagate gradients back to the corresponding
variables.
Examples:
>>> class MyLayer(tf.keras.layers.Layer):
... def call(self, inputs):
... self.add_loss(tf.abs(tf.reduce_mean(inputs)))
... return inputs
>>> l = MyLayer()
>>> l(np.ones((10, 1)))
>>> l.losses
[1.0]
>>> inputs = tf.keras.Input(shape=(10,))
>>> x = tf.keras.layers.Dense(10)(inputs)
>>> outputs = tf.keras.layers.Dense(1)(x)
>>> model = tf.keras.Model(inputs, outputs)
>>> # Activity regularization.
>>> len(model.losses)
0
>>> model.add_loss(tf.abs(tf.reduce_mean(x)))
>>> len(model.losses)
1
>>> inputs = tf.keras.Input(shape=(10,))
>>> d = tf.keras.layers.Dense(10, kernel_initializer='ones')
>>> x = d(inputs)
>>> outputs = tf.keras.layers.Dense(1)(x)
>>> model = tf.keras.Model(inputs, outputs)
>>> # Weight regularization.
>>> model.add_loss(lambda: tf.reduce_mean(d.kernel))
>>> model.losses
[<tf.Tensor: shape=(), dtype=float32, numpy=1.0>]
Returns:
A list of tensors.
"""
collected_losses = []
for layer in self._flatten_layers():
# If any eager losses are present, we assume the model to be part of
# an eager training loop (either a custom one or the one used when
# `run_eagerly=True`) and so we always return just the eager losses.
if layer._eager_losses:
# Filter placeholder losses that may have been added by revived
# layers. (see base_layer_utils for details).
if (
layer._eager_losses[0]
is not base_layer_utils.REVIVED_LOSS_PLACEHOLDER
):
collected_losses.extend(layer._eager_losses)
else:
collected_losses.extend(layer._losses)
for regularizer in layer._callable_losses:
loss_tensor = regularizer()
if loss_tensor is not None:
collected_losses.append(loss_tensor)
return collected_losses
def add_loss(self, losses, **kwargs):
"""Add loss tensor(s), potentially dependent on layer inputs.
Some losses (for instance, activity regularization losses) may be
dependent on the inputs passed when calling a layer. Hence, when reusing
the same layer on different inputs `a` and `b`, some entries in
`layer.losses` may be dependent on `a` and some on `b`. This method
automatically keeps track of dependencies.
This method can be used inside a subclassed layer or model's `call`
function, in which case `losses` should be a Tensor or list of Tensors.
Example:
```python
class MyLayer(tf.keras.layers.Layer):
def call(self, inputs):
self.add_loss(tf.abs(tf.reduce_mean(inputs)))
return inputs
```
The same code works in distributed training: the input to `add_loss()`
is treated like a regularization loss and averaged across replicas
by the training loop (both built-in `Model.fit()` and compliant custom
training loops).
The `add_loss` method can also be called directly on a Functional Model
during construction. In this case, any loss Tensors passed to this Model
must be symbolic and be able to be traced back to the model's `Input`s.
These losses become part of the model's topology and are tracked in
`get_config`.
Example:
```python
inputs = tf.keras.Input(shape=(10,))
x = tf.keras.layers.Dense(10)(inputs)
outputs = tf.keras.layers.Dense(1)(x)
model = tf.keras.Model(inputs, outputs)
# Activity regularization.
model.add_loss(tf.abs(tf.reduce_mean(x)))
```
If this is not the case for your loss (if, for example, your loss
references a `Variable` of one of the model's layers), you can wrap your
loss in a zero-argument lambda. These losses are not tracked as part of
the model's topology since they can't be serialized.
Example:
```python
inputs = tf.keras.Input(shape=(10,))
d = tf.keras.layers.Dense(10)
x = d(inputs)
outputs = tf.keras.layers.Dense(1)(x)
model = tf.keras.Model(inputs, outputs)
# Weight regularization.
model.add_loss(lambda: tf.reduce_mean(d.kernel))
```
Args:
losses: Loss tensor, or list/tuple of tensors. Rather than tensors,
losses may also be zero-argument callables which create a loss
tensor.
**kwargs: Used for backwards compatibility only.
"""
kwargs.pop("inputs", None)
if kwargs:
raise TypeError(f"Unknown keyword arguments: {kwargs.keys()}")
def _tag_callable(loss):
"""Tags callable loss tensor as `_unconditional_loss`."""
if callable(loss):
# We run the loss without autocasting, as regularizers are often
# numerically unstable in float16.
with autocast_variable.enable_auto_cast_variables(None):
loss = loss()
if loss is None:
# Will be filtered out when computing the .losses property
return None
if not tf.is_tensor(loss):
loss = tf.convert_to_tensor(loss, dtype=backend.floatx())
loss._unconditional_loss = True
return loss
losses = tf.nest.flatten(losses)
callable_losses = []
eager_losses = []
symbolic_losses = []
for loss in losses:
if callable(loss):
callable_losses.append(functools.partial(_tag_callable, loss))
continue
if loss is None:
continue
if not tf.is_tensor(loss) and not isinstance(
loss, keras_tensor.KerasTensor
):
loss = tf.convert_to_tensor(loss, dtype=backend.floatx())
# TF Functions should take the eager path.
if (
tf_utils.is_symbolic_tensor(loss)
or isinstance(loss, keras_tensor.KerasTensor)
) and not base_layer_utils.is_in_tf_function():
symbolic_losses.append(loss)
elif tf.is_tensor(loss):
eager_losses.append(loss)
self._callable_losses.extend(callable_losses)
in_call_context = base_layer_utils.call_context().in_call
if eager_losses and not in_call_context:
raise ValueError(
"Expected a symbolic Tensors or a callable for the loss value. "
"Please wrap your loss computation in a zero argument `lambda`."
)
self._eager_losses.extend(eager_losses)
for symbolic_loss in symbolic_losses:
if getattr(self, "_is_graph_network", False):
self._graph_network_add_loss(symbolic_loss)
else:
# Possible a loss was added in a Layer's `build`.
self._losses.append(symbolic_loss)
@property
def metrics(self):
"""List of metrics attached to the layer.
Returns:
A list of `Metric` objects.
"""
collected_metrics = []
for layer in self._flatten_layers():
if not hasattr(layer, "_metrics_lock"):
continue
with layer._metrics_lock:
collected_metrics.extend(layer._metrics)
return collected_metrics
@doc_controls.do_not_generate_docs
def add_metric(self, value, name=None, **kwargs):
"""Adds metric tensor to the layer.
This method can be used inside the `call()` method of a subclassed layer
or model.
```python
class MyMetricLayer(tf.keras.layers.Layer):
def __init__(self):
super(MyMetricLayer, self).__init__(name='my_metric_layer')
self.mean = tf.keras.metrics.Mean(name='metric_1')
def call(self, inputs):
self.add_metric(self.mean(inputs))
self.add_metric(tf.reduce_sum(inputs), name='metric_2')
return inputs
```
This method can also be called directly on a Functional Model during
construction. In this case, any tensor passed to this Model must
be symbolic and be able to be traced back to the model's `Input`s. These
metrics become part of the model's topology and are tracked when you
save the model via `save()`.
```python
inputs = tf.keras.Input(shape=(10,))
x = tf.keras.layers.Dense(10)(inputs)
outputs = tf.keras.layers.Dense(1)(x)
model = tf.keras.Model(inputs, outputs)
model.add_metric(math_ops.reduce_sum(x), name='metric_1')
```
Note: Calling `add_metric()` with the result of a metric object on a
Functional Model, as shown in the example below, is not supported. This
is because we cannot trace the metric result tensor back to the model's
inputs.
```python
inputs = tf.keras.Input(shape=(10,))
x = tf.keras.layers.Dense(10)(inputs)
outputs = tf.keras.layers.Dense(1)(x)
model = tf.keras.Model(inputs, outputs)
model.add_metric(tf.keras.metrics.Mean()(x), name='metric_1')
```
Args:
value: Metric tensor.
name: String metric name.
**kwargs: Additional keyword arguments for backward compatibility.
Accepted values:
`aggregation` - When the `value` tensor provided is not the result
of calling a `keras.Metric` instance, it will be aggregated by
default using a `keras.Metric.Mean`.
"""
kwargs_keys = list(kwargs.keys())
if len(kwargs_keys) > 1 or (
len(kwargs_keys) == 1 and kwargs_keys[0] != "aggregation"
):
raise TypeError(
f"Unknown keyword arguments: {kwargs.keys()}. "
"Expected `aggregation`."
)
from_metric_obj = hasattr(value, "_metric_obj")
is_symbolic = isinstance(value, keras_tensor.KerasTensor)
in_call_context = base_layer_utils.call_context().in_call
if name is None and not from_metric_obj:
# Eg. `self.add_metric(math_ops.reduce_sum(x))` In eager mode, we
# use metric name to lookup a metric. Without a name, a new Mean
# metric wrapper will be created on every model/layer call. So, we
# raise an error when no name is provided. We will do the same for
# symbolic mode for consistency although a name will be generated if
# no name is provided.
# We will not raise this error in the foll use case for the sake of
# consistency as name in provided in the metric constructor.
# mean = metrics.Mean(name='my_metric')
# model.add_metric(mean(outputs))
raise ValueError(
"Please provide a name for your metric like "
"`self.add_metric(tf.reduce_sum(inputs), "
"name='mean_activation')`"
)
elif from_metric_obj:
name = value._metric_obj.name
if not in_call_context and not is_symbolic:
raise ValueError(
"Expected a symbolic Tensor for the metric value, received: "
+ str(value)
)
# If a metric was added in a Layer's `call` or `build`.
if in_call_context or not getattr(self, "_is_graph_network", False):
# TF Function path should take the eager path.
# If the given metric is available in `metrics` list we just update
# state on it, otherwise we create a new metric instance and
# add it to the `metrics` list.
metric_obj = getattr(value, "_metric_obj", None)
# Tensors that come from a Metric object already updated the Metric
# state.
should_update_state = not metric_obj
name = metric_obj.name if metric_obj else name
with self._metrics_lock:
match = self._get_existing_metric(name)
if match:
metric_obj = match
elif metric_obj:
self._metrics.append(metric_obj)
else:
# Build the metric object with the value's dtype if it
# defines one
metric_obj = metrics_mod.Mean(
name=name, dtype=getattr(value, "dtype", None)
)
self._metrics.append(metric_obj)
if should_update_state:
metric_obj(value)
else:
if from_metric_obj:
raise ValueError(
"Using the result of calling a `Metric` object "
"when calling `add_metric` on a Functional "
"Model is not supported. Please pass the "
"Tensor to monitor directly."
)
# Insert layers into the TF-Keras Graph Network.
aggregation = None if from_metric_obj else "mean"
self._graph_network_add_metric(value, aggregation, name)
@doc_controls.do_not_doc_inheritable
def add_update(self, updates):
"""Add update op(s), potentially dependent on layer inputs.
Weight updates (for instance, the updates of the moving mean and
variance in a BatchNormalization layer) may be dependent on the inputs
passed when calling a layer. Hence, when reusing the same layer on
different inputs `a` and `b`, some entries in `layer.updates` may be
dependent on `a` and some on `b`. This method automatically keeps track
of dependencies.
This call is ignored when eager execution is enabled (in that case,
variable updates are run on the fly and thus do not need to be tracked
for later execution).
Args:
updates: Update op, or list/tuple of update ops, or zero-arg callable
that returns an update op. A zero-arg callable should be passed in
order to disable running the updates by setting `trainable=False`
on this Layer, when executing in Eager mode.
"""
call_context = base_layer_utils.call_context()
# No need to run updates during Functional API construction.
if call_context.in_keras_graph:
return
# Callable updates are disabled by setting `trainable=False`.
if not call_context.frozen:
for update in tf.nest.flatten(updates):
if callable(update):
update()
def set_weights(self, weights):
"""Sets the weights of the layer, from NumPy arrays.
The weights of a layer represent the state of the layer. This function
sets the weight values from numpy arrays. The weight values should be
passed in the order they are created by the layer. Note that the layer's
weights must be instantiated before calling this function, by calling
the layer.
For example, a `Dense` layer returns a list of two values: the kernel
matrix and the bias vector. These can be used to set the weights of
another `Dense` layer:
>>> layer_a = tf.keras.layers.Dense(1,
... kernel_initializer=tf.constant_initializer(1.))
>>> a_out = layer_a(tf.convert_to_tensor([[1., 2., 3.]]))
>>> layer_a.get_weights()
[array([[1.],
[1.],
[1.]], dtype=float32), array([0.], dtype=float32)]
>>> layer_b = tf.keras.layers.Dense(1,
... kernel_initializer=tf.constant_initializer(2.))
>>> b_out = layer_b(tf.convert_to_tensor([[10., 20., 30.]]))
>>> layer_b.get_weights()
[array([[2.],
[2.],
[2.]], dtype=float32), array([0.], dtype=float32)]
>>> layer_b.set_weights(layer_a.get_weights())
>>> layer_b.get_weights()
[array([[1.],
[1.],
[1.]], dtype=float32), array([0.], dtype=float32)]
Args:
weights: a list of NumPy arrays. The number
of arrays and their shape must match
number of the dimensions of the weights
of the layer (i.e. it should match the
output of `get_weights`).
Raises:
ValueError: If the provided weights list does not match the
layer's specifications.
"""
params = self.weights
expected_num_weights = 0
for param in params:
if isinstance(param, base_layer_utils.TrackableWeightHandler):
expected_num_weights += param.num_tensors
else:
expected_num_weights += 1
if expected_num_weights != len(weights):
raise ValueError(
'You called `set_weights(weights)` on layer "%s" '
"with a weight list of length %s, but the layer was "
"expecting %s weights. Provided weights: %s..."
% (
self.name,
len(weights),
expected_num_weights,
str(weights)[:50],
)
)
weight_index = 0
weight_value_tuples = []
for param in params:
if isinstance(param, base_layer_utils.TrackableWeightHandler):
num_tensors = param.num_tensors
tensors = weights[weight_index : weight_index + num_tensors]
param.set_weights(tensors)
weight_index += num_tensors
else:
weight = weights[weight_index]
weight_shape = weight.shape if hasattr(weight, "shape") else ()
ref_shape = param.shape
if not ref_shape.is_compatible_with(weight_shape):
raise ValueError(
f"Layer {self.name} weight shape {ref_shape} "
"is not compatible with provided weight "
f"shape {weight_shape}."
)
weight_value_tuples.append((param, weight))
weight_index += 1
backend.batch_set_value(weight_value_tuples)
# Perform any layer defined finalization of the layer state.
for layer in self._flatten_layers():
layer.finalize_state()
def get_weights(self):
"""Returns the current weights of the layer, as NumPy arrays.
The weights of a layer represent the state of the layer. This function
returns both trainable and non-trainable weight values associated with
this layer as a list of NumPy arrays, which can in turn be used to load
state into similarly parameterized layers.
For example, a `Dense` layer returns a list of two values: the kernel
matrix and the bias vector. These can be used to set the weights of
another `Dense` layer:
>>> layer_a = tf.keras.layers.Dense(1,
... kernel_initializer=tf.constant_initializer(1.))
>>> a_out = layer_a(tf.convert_to_tensor([[1., 2., 3.]]))
>>> layer_a.get_weights()
[array([[1.],
[1.],
[1.]], dtype=float32), array([0.], dtype=float32)]
>>> layer_b = tf.keras.layers.Dense(1,
... kernel_initializer=tf.constant_initializer(2.))
>>> b_out = layer_b(tf.convert_to_tensor([[10., 20., 30.]]))
>>> layer_b.get_weights()
[array([[2.],
[2.],
[2.]], dtype=float32), array([0.], dtype=float32)]
>>> layer_b.set_weights(layer_a.get_weights())
>>> layer_b.get_weights()
[array([[1.],
[1.],
[1.]], dtype=float32), array([0.], dtype=float32)]
Returns:
Weights values as a list of NumPy arrays.
"""
weights = self.weights
output_weights = []
for weight in weights:
if isinstance(weight, base_layer_utils.TrackableWeightHandler):
output_weights.extend(weight.get_tensors())
else:
output_weights.append(weight)
return backend.batch_get_value(output_weights)
@doc_controls.do_not_generate_docs
def finalize_state(self):
"""Finalizes the layers state after updating layer weights.
This function can be subclassed in a layer and will be called after
updating a layer weights. It can be overridden to finalize any
additional layer state after a weight update.
This function will be called after weights of a layer have been restored
from a loaded model.
"""
pass
@doc_controls.do_not_doc_inheritable
def get_input_mask_at(self, node_index):
"""Retrieves the input mask tensor(s) of a layer at a given node.
Args:
node_index: Integer, index of the node
from which to retrieve the attribute.
E.g. `node_index=0` will correspond to the
first time the layer was called.
Returns:
A mask tensor
(or list of tensors if the layer has multiple inputs).
"""
inputs = self.get_input_at(node_index)
if isinstance(inputs, list):
return [getattr(x, "_keras_mask", None) for x in inputs]
else:
return getattr(inputs, "_keras_mask", None)
@doc_controls.do_not_doc_inheritable
def get_output_mask_at(self, node_index):
"""Retrieves the output mask tensor(s) of a layer at a given node.
Args:
node_index: Integer, index of the node
from which to retrieve the attribute.
E.g. `node_index=0` will correspond to the
first time the layer was called.
Returns:
A mask tensor
(or list of tensors if the layer has multiple outputs).
"""
output = self.get_output_at(node_index)
if isinstance(output, list):
return [getattr(x, "_keras_mask", None) for x in output]
else:
return getattr(output, "_keras_mask", None)
@property
@doc_controls.do_not_doc_inheritable
def input_mask(self):
"""Retrieves the input mask tensor(s) of a layer.
Only applicable if the layer has exactly one inbound node,
i.e. if it is connected to one incoming layer.
Returns:
Input mask tensor (potentially None) or list of input
mask tensors.
Raises:
AttributeError: if the layer is connected to
more than one incoming layers.
"""
inputs = self.input
if isinstance(inputs, list):
return [getattr(x, "_keras_mask", None) for x in inputs]
else:
return getattr(inputs, "_keras_mask", None)
@property
@doc_controls.do_not_doc_inheritable
def output_mask(self):
"""Retrieves the output mask tensor(s) of a layer.
Only applicable if the layer has exactly one inbound node,
i.e. if it is connected to one incoming layer.
Returns:
Output mask tensor (potentially None) or list of output
mask tensors.
Raises:
AttributeError: if the layer is connected to
more than one incoming layers.
"""
output = self.output
if isinstance(output, list):
return [getattr(x, "_keras_mask", None) for x in output]
else:
return getattr(output, "_keras_mask", None)
@doc_controls.do_not_doc_inheritable
def get_input_shape_at(self, node_index):
"""Retrieves the input shape(s) of a layer at a given node.
Args:
node_index: Integer, index of the node
from which to retrieve the attribute.
E.g. `node_index=0` will correspond to the
first time the layer was called.
Returns:
A shape tuple
(or list of shape tuples if the layer has multiple inputs).
Raises:
RuntimeError: If called in Eager mode.
"""
return self._get_node_attribute_at_index(
node_index, "input_shapes", "input shape"
)
@doc_controls.do_not_doc_inheritable
def get_output_shape_at(self, node_index):
"""Retrieves the output shape(s) of a layer at a given node.
Args:
node_index: Integer, index of the node
from which to retrieve the attribute.
E.g. `node_index=0` will correspond to the
first time the layer was called.
Returns:
A shape tuple
(or list of shape tuples if the layer has multiple outputs).
Raises:
RuntimeError: If called in Eager mode.
"""
return self._get_node_attribute_at_index(
node_index, "output_shapes", "output shape"
)
@doc_controls.do_not_doc_inheritable
def get_input_at(self, node_index):
"""Retrieves the input tensor(s) of a layer at a given node.
Args:
node_index: Integer, index of the node
from which to retrieve the attribute.
E.g. `node_index=0` will correspond to the
first input node of the layer.
Returns:
A tensor (or list of tensors if the layer has multiple inputs).
Raises:
RuntimeError: If called in Eager mode.
"""
return self._get_node_attribute_at_index(
node_index, "input_tensors", "input"
)
@doc_controls.do_not_doc_inheritable
def get_output_at(self, node_index):
"""Retrieves the output tensor(s) of a layer at a given node.
Args:
node_index: Integer, index of the node
from which to retrieve the attribute.
E.g. `node_index=0` will correspond to the
first output node of the layer.
Returns:
A tensor (or list of tensors if the layer has multiple outputs).
Raises:
RuntimeError: If called in Eager mode.
"""
return self._get_node_attribute_at_index(
node_index, "output_tensors", "output"
)
@property
def input(self):
"""Retrieves the input tensor(s) of a layer.
Only applicable if the layer has exactly one input,
i.e. if it is connected to one incoming layer.
Returns:
Input tensor or list of input tensors.
Raises:
RuntimeError: If called in Eager mode.
AttributeError: If no inbound nodes are found.
"""
if not self._inbound_nodes:
raise AttributeError(
"Layer " + self.name + " is not connected, no input to return."
)
return self._get_node_attribute_at_index(0, "input_tensors", "input")
@property
def output(self):
"""Retrieves the output tensor(s) of a layer.
Only applicable if the layer has exactly one output,
i.e. if it is connected to one incoming layer.
Returns:
Output tensor or list of output tensors.
Raises:
AttributeError: if the layer is connected to more than one incoming
layers.
RuntimeError: if called in Eager mode.
"""
if not self._inbound_nodes:
raise AttributeError(
"Layer " + self.name + " has no inbound nodes."
)
return self._get_node_attribute_at_index(0, "output_tensors", "output")
@property
@doc_controls.do_not_doc_inheritable
def input_shape(self):
"""Retrieves the input shape(s) of a layer.
Only applicable if the layer has exactly one input,
i.e. if it is connected to one incoming layer, or if all inputs
have the same shape.
Returns:
Input shape, as an integer shape tuple
(or list of shape tuples, one tuple per input tensor).
Raises:
AttributeError: if the layer has no defined input_shape.
RuntimeError: if called in Eager mode.
"""
if not self._inbound_nodes:
raise AttributeError(
f'The layer "{self.name}" has never been called '
"and thus has no defined input shape. Note that the "
"`input_shape` property is only available for "
"Functional and Sequential models."
)
all_input_shapes = set(
[str(node.input_shapes) for node in self._inbound_nodes]
)
if len(all_input_shapes) == 1:
return self._inbound_nodes[0].input_shapes
else:
raise AttributeError(
'The layer "'
+ str(self.name)
+ '" has multiple inbound nodes, '
"with different input shapes. Hence "
'the notion of "input shape" is '
"ill-defined for the layer. "
"Use `get_input_shape_at(node_index)` "
"instead."
)
def count_params(self):
"""Count the total number of scalars composing the weights.
Returns:
An integer count.
Raises:
ValueError: if the layer isn't yet built
(in which case its weights aren't yet defined).
"""
if not self.built:
if getattr(self, "_is_graph_network", False):
with tf_utils.maybe_init_scope(self):
self._maybe_build(self.inputs)
else:
raise ValueError(
"You tried to call `count_params` "
f"on layer {self.name}"
", but the layer isn't built. "
"You can build it manually via: "
f"`{self.name}.build(batch_input_shape)`."
)
return layer_utils.count_params(self.weights)
@property
@doc_controls.do_not_doc_inheritable
def output_shape(self):
"""Retrieves the output shape(s) of a layer.
Only applicable if the layer has one output,
or if all outputs have the same shape.
Returns:
Output shape, as an integer shape tuple
(or list of shape tuples, one tuple per output tensor).
Raises:
AttributeError: if the layer has no defined output shape.
RuntimeError: if called in Eager mode.
"""
if not self._inbound_nodes:
raise AttributeError(
f'The layer "{self.name}" has never been called '
"and thus has no defined output shape."
)
all_output_shapes = set(
[str(node.output_shapes) for node in self._inbound_nodes]
)
if len(all_output_shapes) == 1:
return self._inbound_nodes[0].output_shapes
else:
raise AttributeError(
'The layer "%s"'
" has multiple inbound nodes, "
"with different output shapes. Hence "
'the notion of "output shape" is '
"ill-defined for the layer. "
"Use `get_output_shape_at(node_index)` "
"instead." % self.name
)
@property
def dtype_policy(self):
"""The dtype policy associated with this layer.
This is an instance of a `tf.keras.mixed_precision.Policy`.
"""
return self._dtype_policy
@property
def compute_dtype(self):
"""The dtype of the layer's computations.
This is equivalent to `Layer.dtype_policy.compute_dtype`. Unless
mixed precision is used, this is the same as `Layer.dtype`, the dtype of
the weights.
Layers automatically cast their inputs to the compute dtype, which
causes computations and the output to be in the compute dtype as well.
This is done by the base Layer class in `Layer.__call__`, so you do not
have to insert these casts if implementing your own layer.
Layers often perform certain internal computations in higher precision
when `compute_dtype` is float16 or bfloat16 for numeric stability. The
output will still typically be float16 or bfloat16 in such cases.
Returns:
The layer's compute dtype.
"""
return self._dtype_policy.compute_dtype
@property
def variable_dtype(self):
"""Alias of `Layer.dtype`, the dtype of the weights."""
return self.dtype
@property
@doc_controls.do_not_doc_inheritable
def inbound_nodes(self):
"""Return Functional API nodes upstream of this layer."""
return self._inbound_nodes
@property
@doc_controls.do_not_doc_inheritable
def outbound_nodes(self):
"""Return Functional API nodes downstream of this layer."""
return self._outbound_nodes
############################################################################
# Methods & attributes below are public aliases of other methods. #
############################################################################
@property
@doc_controls.do_not_generate_docs
def variables(self):
"""Returns the list of all layer variables/weights.
Alias of `self.weights`.
Note: This will not track the weights of nested `tf.Modules` that are
not themselves TF-Keras layers.
Returns:
A list of variables.
"""
return self.weights
@property
@doc_controls.do_not_generate_docs
def trainable_variables(self):
return self.trainable_weights
@property
@doc_controls.do_not_generate_docs
def non_trainable_variables(self):
return self.non_trainable_weights
@doc_controls.do_not_doc_inheritable
def add_variable(self, *args, **kwargs):
"""Deprecated, do NOT use! Alias for `add_weight`."""
warnings.warn(
"`layer.add_variable` is deprecated and "
"will be removed in a future version. "
"Please use the `layer.add_weight()` method instead.",
stacklevel=2,
)
return self.add_weight(*args, **kwargs)
def get_build_config(self):
"""Returns a dictionary with the layer's input shape.
This method returns a config dict that can be used by
`build_from_config(config)` to create all states (e.g. Variables and
Lookup tables) needed by the layer.
By default, the config only contains the input shape that the layer
was built with. If you're writing a custom layer that creates state in
an unusual way, you should override this method to make sure this state
is already created when TF-Keras attempts to load its value upon model
loading.
Returns:
A dict containing the input shape associated with the layer.
"""
if self._build_input_shape is not None:
def convert_tensorshapes(x):
if isinstance(x, tf.TensorShape) and x._dims:
return tuple(x.as_list())
return x
return {
"input_shape": tf.nest.map_structure(
convert_tensorshapes, self._build_input_shape
)
}
def build_from_config(self, config):
"""Builds the layer's states with the supplied config dict.
By default, this method calls the `build(config["input_shape"])` method,
which creates weights based on the layer's input shape in the supplied
config. If your config contains other information needed to load the
layer's state, you should override this method.
Args:
config: Dict containing the input shape associated with this layer.
"""
input_shape = config["input_shape"]
if input_shape is not None:
self.build(input_shape)
############################################################################
# Methods & attributes below are all private and only used by the framework.
############################################################################
# See tf.Module for the usage of this property.
# The key for _obj_reference_counts_dict is a Trackable, which could be a
# variable or layer etc. tf.Module._flatten will fail to flatten the key
# since it is trying to convert Trackable to a string. This attribute can be
# ignored even after the fix of nest lib, since the trackable object should
# already been available as individual attributes.
# _obj_reference_counts_dict just contains a copy of them.
_TF_MODULE_IGNORED_PROPERTIES = frozenset(
itertools.chain(
("_obj_reference_counts_dict",),
tf.Module._TF_MODULE_IGNORED_PROPERTIES,
)
)
# When loading from a SavedModel, Layers typically can be revived into a
# generic Layer wrapper. Sometimes, however, layers may implement methods
# that go beyond this wrapper, as in the case of PreprocessingLayers'
# `adapt` method. When this is the case, layer implementers can override
# must_restore_from_config to return True; layers with this property must
# be restored into their actual objects (and will fail if the object is
# not available to the restoration code).
_must_restore_from_config = False
def _get_cell_name(self):
canonical_name = get_canonical_name_for_symbol(
self.__class__, api_name="keras", add_prefix_to_v1_names=True
)
if canonical_name is not None:
return f"tf.{canonical_name}"
return self.__class__.__module__ + "." + self.__class__.__name__
def _instrument_layer_creation(self):
self._instrumented_keras_api = False
self._instrumented_keras_layer_class = False
self._instrumented_keras_model_class = False
if not getattr(self, "_disable_keras_instrumentation", False):
self._instrumented_keras_api = True
if getattr(self, "_is_model_for_instrumentation", False):
self._instrumented_keras_model_class = True
else:
self._instrumented_keras_layer_class = True
@doc_controls.for_subclass_implementers
def _add_trackable(self, trackable_object, trainable):
"""Adds a Trackable object to this layer's state.
Args:
trackable_object: The tf.tracking.Trackable object to add.
trainable: Boolean, whether the variable should be part of the layer's
"trainable_variables" (e.g. variables, biases) or
"non_trainable_variables" (e.g. BatchNorm mean and variance).
Returns:
The TrackableWeightHandler used to track this object.
"""
if isinstance(
trackable_object, base_layer_utils.TrackableWeightHandler
):
handler = trackable_object
else:
handler = base_layer_utils.TrackableWeightHandler(trackable_object)
if trainable:
self._trainable_weights.append(handler)
else:
self._non_trainable_weights.append(handler)
return handler
def _clear_losses(self):
"""Used every step in eager to reset losses."""
# Set to thread local directly to avoid Layer.__setattr__ overhead.
if not getattr(
self, "_self_tracked_trackables", None
): # Fast path for single Layer.
self._thread_local._eager_losses = []
else:
for layer in self._flatten_layers():
layer._thread_local._eager_losses = []
def _keras_tensor_symbolic_call(self, inputs, input_masks, args, kwargs):
if self.dynamic:
# We will use static shape inference to return symbolic tensors
# matching the specifications of the layer outputs.
# Since `self.dynamic` is True, we will never attempt to
# run the underlying TF graph (which is disconnected).
# TODO(fchollet): consider py_func as an alternative, which
# would enable us to run the underlying graph if needed.
input_signature = tf.nest.map_structure(
lambda x: tf.TensorSpec(shape=x.shape, dtype=x.dtype), inputs
)
output_signature = self.compute_output_signature(input_signature)
return tf.nest.map_structure(
keras_tensor.KerasTensor, output_signature
)
else:
return self._infer_output_signature(
inputs, args, kwargs, input_masks
)
def _should_use_autograph(self):
if base_layer_utils.from_saved_model(self):
return False
if base_layer_utils.is_subclassed(self):
return True
return False
def _infer_output_signature(self, inputs, args, kwargs, input_masks):
"""Call the layer on input KerasTensors, returns output KerasTensors."""
keras_tensor_inputs = inputs
call_fn = self.call
# Wrapping `call` function in autograph to allow for dynamic control
# flow and control dependencies in call. We are limiting this to
# subclassed layers as autograph is strictly needed only for
# subclassed layers and models.
# tf_convert will respect the value of autograph setting in the
# enclosing tf.function, if any.
if self._should_use_autograph():
call_fn = tf.__internal__.autograph.tf_convert(
self.call, tf.__internal__.autograph.control_status_ctx()
)
call_fn = traceback_utils.inject_argument_info_in_traceback(
call_fn,
object_name=f'layer "{self.name}" (type {self.__class__.__name__})',
)
# We enter a scratch graph and build placeholder inputs inside of it
# that match the input args.
# We then call the layer inside of the scratch graph to identify the
# output signatures, then we build KerasTensors corresponding to those
# outputs.
scratch_graph = tf.__internal__.FuncGraph(
str(self.name) + "_scratch_graph"
)
with scratch_graph.as_default():
inputs = tf.nest.map_structure(
keras_tensor.keras_tensor_to_placeholder, inputs
)
args = tf.nest.map_structure(
keras_tensor.keras_tensor_to_placeholder, args
)
kwargs = tf.nest.map_structure(
keras_tensor.keras_tensor_to_placeholder, kwargs
)
input_masks = tf.nest.map_structure(
keras_tensor.keras_tensor_to_placeholder, input_masks
)
with backend.name_scope(self._name_scope()):
with autocast_variable.enable_auto_cast_variables(
self._compute_dtype_object
):
# Build layer if applicable (if the `build` method has been
# overridden).
# TODO(kaftan): do we maybe_build here, or have we already
# done it?
self._maybe_build(inputs)
inputs = self._maybe_cast_inputs(inputs)
outputs = call_fn(inputs, *args, **kwargs)
self._handle_activity_regularization(inputs, outputs)
self._set_mask_metadata(
inputs, outputs, input_masks, build_graph=False
)
outputs = tf.nest.map_structure(
keras_tensor.keras_tensor_from_tensor, outputs
)
self._set_save_spec(keras_tensor_inputs, args, kwargs)
if hasattr(self, "_set_inputs") and not self.inputs:
# TODO(kaftan): figure out if we need to do this at all
# Subclassed network: explicitly set metadata normally set by
# a call to self._set_inputs().
self._set_inputs(inputs, outputs)
del scratch_graph
return outputs
def _functional_construction_call(self, inputs, args, kwargs, input_list):
call_context = base_layer_utils.call_context()
# Accept NumPy and scalar inputs by converting to Tensors.
if any(
isinstance(x, (tf.Tensor, np.ndarray, float, int))
for x in input_list
):
def _convert_non_tensor(x):
# Don't call `ops.convert_to_tensor` on all `inputs` because
# `SparseTensors` can't be converted to `Tensor`.
if isinstance(x, (tf.Tensor, np.ndarray, float, int)):
return tf.convert_to_tensor(x)
return x
inputs = tf.nest.map_structure(_convert_non_tensor, inputs)
input_list = tf.nest.flatten(inputs)
# Handle `mask` propagation from previous layer to current layer. Masks
# can be propagated explicitly via the `mask` argument, or implicitly
# via setting the `_keras_mask` attribute on the inputs to a Layer.
# Masks passed explicitly take priority.
mask_arg_passed_by_framework = False
input_masks, mask_is_implicit = self._get_input_masks(
inputs, input_list, args, kwargs
)
if self._expects_mask_arg and mask_is_implicit:
kwargs["mask"] = input_masks
mask_arg_passed_by_framework = True
# If `training` argument is None or not explicitly passed,
# propagate `training` value from this layer's calling layer.
training_value = None
training_arg_passed_by_framework = False
# Priority 1: `training` was explicitly passed a non-None value.
if self._call_spec.arg_was_passed("training", args, kwargs):
training_value = self._call_spec.get_arg_value(
"training", args, kwargs
)
if not self._expects_training_arg:
kwargs.pop("training")
if training_value is None:
# Priority 2: `training` was passed to a parent layer.
if call_context.training is not None:
training_value = call_context.training
# Priority 3: `learning_phase()` has been set.
elif backend.global_learning_phase_is_set():
training_value = backend.learning_phase()
# Force the training_value to be bool type which matches to the
# contract for layer/model call args.
if tf.is_tensor(training_value):
training_value = tf.cast(training_value, tf.bool)
else:
training_value = bool(training_value)
# Priority 4: trace layer with the default training argument
# specified in the `call` signature (or in inference mode if the
# `call` signature specifies no non-None default).
else:
training_value = self._call_spec.default_training_arg
# In cases (2), (3), (4) the training argument is passed
# automatically by the framework, and will not be hard-coded into
# the model.
if self._expects_training_arg:
args, kwargs = self._call_spec.set_arg_value(
"training", training_value, args, kwargs
)
training_arg_passed_by_framework = True
with call_context.enter(
layer=self, inputs=inputs, build_graph=True, training=training_value
):
# Check input assumptions set after layer building, e.g. input
# shape.
try:
outputs = self._keras_tensor_symbolic_call(
inputs, input_masks, args, kwargs
)
except TypeError as e:
if "DictWrapper" in str(e):
raise TypeError(
f"{self} could not be deserialized properly. Please"
" ensure that components that are Python object"
" instances (layers, models, etc.) returned by"
" `get_config()` are explicitly deserialized in the"
" model's `from_config()` method."
) from e
else:
raise e
if outputs is None:
raise ValueError(
"A layer's `call` method should return a "
"Tensor or a list of Tensors, not None "
"(layer: " + self.name + ")."
)
if training_arg_passed_by_framework:
args, kwargs = self._call_spec.set_arg_value(
"training", None, args, kwargs, pop_kwarg_if_none=True
)
if mask_arg_passed_by_framework:
kwargs.pop("mask")
# Node connectivity does not special-case the first argument.
outputs = self._set_connectivity_metadata(
(inputs,) + args, kwargs, outputs
)
return outputs
def _set_training_mode(self, args, kwargs, call_context):
training_mode = None
if self._expects_training_arg:
# (1) `training` was passed to this `Layer.call`.
if self._call_spec.arg_was_passed("training", args, kwargs):
training_mode = self._call_spec.get_arg_value(
"training", args, kwargs
)
# If no `training` arg was passed, or `None` was explicitly passed,
# the framework will make a decision about the training mode is.
if training_mode is None:
call_ctx_training = call_context.training
# (2) `training` mode is inferred from an outer `Layer.call`.
if call_ctx_training is not None:
training_mode = call_ctx_training
# (3) User set `tf.keras.backend.set_learning_phase`.
elif backend.global_learning_phase_is_set():
training_mode = backend.learning_phase()
# Ensure value is a `bool` or `tf.bool`.
if isinstance(training_mode, bool):
pass
elif tf.is_tensor(training_mode):
training_mode = tf.cast(training_mode, tf.bool)
else:
training_mode = bool(training_mode)
# (4) We default to using `call`'s default value for `training`,
# or treating the layer as if it is in inference if no non-None
# default is specified in the `call` signature.
else:
training_mode = self._call_spec.default_training_arg
# For case (2), (3), (4) `training` arg is passed by framework.
args, kwargs = self._call_spec.set_arg_value(
"training", training_mode, args, kwargs
)
else:
if "training" in kwargs:
# `training` was passed to this `Layer` but is not needed for
# `Layer.call`. It will set the default mode for inner
# `Layer.call`s.
training_mode = kwargs.pop("training")
else:
# Grab the current `training` mode from any outer `Layer.call`.
training_mode = call_context.training
return args, kwargs, training_mode
def _autographed_call(self):
# Wrapping `call` function in autograph to allow for dynamic control
# flow and control dependencies in call. We are limiting this to
# subclassed layers as autograph is strictly needed only for
# subclassed layers and models.
# tf_convert will respect the value of autograph setting in the
# enclosing tf.function, if any.
if self._should_use_autograph():
return tf.__internal__.autograph.tf_convert(
self.call, tf.__internal__.autograph.control_status_ctx()
)
return self.call
@property
def _inbound_nodes(self):
return self._inbound_nodes_value
@_inbound_nodes.setter
@tf.__internal__.tracking.no_automatic_dependency_tracking
def _inbound_nodes(self, value):
self._inbound_nodes_value = value
@property
def _outbound_nodes(self):
return self._outbound_nodes_value
@_outbound_nodes.setter
@tf.__internal__.tracking.no_automatic_dependency_tracking
def _outbound_nodes(self, value):
self._outbound_nodes_value = value
def _set_dtype_policy(self, dtype):
"""Sets self._dtype_policy."""
self._dtype_policy = policy.get_policy(dtype)
# Performance optimization: cache the compute dtype as a Dtype object or
# None, so that str to Dtype conversion doesn't happen in
# Layer.__call__.
# TODO(b/157486353): Investigate returning DTypes in Policy.
if self._dtype_policy.compute_dtype:
self._compute_dtype_object = tf.as_dtype(
self._dtype_policy.compute_dtype
)
else:
self._compute_dtype_object = None
@property
def _compute_dtype(self):
"""Deprecated alias of `compute_dtype`."""
return self._dtype_policy.compute_dtype
def _maybe_cast_inputs(self, inputs, input_list=None):
"""Maybe casts the inputs to the compute dtype.
If self._compute_dtype is floating-point, and self_autocast is True,
floating-point inputs are casted to self._compute_dtype.
Args:
inputs: Input tensor, or structure of input tensors.
input_list: Flat list of input tensors.
Returns:
`inputs`, but tensors may have been casted to self._compute_dtype
"""
if not input_list:
input_list = tf.nest.flatten(inputs)
compute_dtype_object = self._compute_dtype_object
should_autocast = (
self._autocast
and compute_dtype_object
and compute_dtype_object.is_floating
)
if should_autocast and any(
map(self._should_cast_single_input, input_list)
):
# Only perform expensive `nest` operation when needed.
return tf.nest.map_structure(self._cast_single_input, inputs)
else:
return inputs
def _should_cast_single_input(self, x):
if isinstance(x, _AUTOCAST_TYPES):
return (
self._compute_dtype_object
and x.dtype != self._compute_dtype_object
and x.dtype.is_floating
)
return False
def _cast_single_input(self, x):
"""Cast a single Tensor or TensorSpec to the compute dtype."""
if self._should_cast_single_input(x):
return tf.cast(x, self._compute_dtype_object)
else:
return x
# _dtype used to be an attribute set in the constructor. We still expose it
# because some clients still use it.
# TODO(reedwm): Deprecate, then remove the _dtype property.
@property
def _dtype(self):
# This is equivalent to returning self.dtype . We do not return
# self.dtype as it would cause infinite recursion in a few subclasses,
# which override "dtype" to return self._dtype.
return self._dtype_policy.variable_dtype
@_dtype.setter
def _dtype(self, value):
value = tf.as_dtype(value).name
self._set_dtype_policy(policy.Policy(value))
def _name_scope(self):
if not tf.__internal__.tf2.enabled():
return self.name
name_scope = self.name
current_name_scope = tf.__internal__.get_name_scope()
if current_name_scope:
name_scope = current_name_scope + "/" + name_scope
if name_scope:
# Note that the trailing `/` prevents autogenerated
# numerical suffixes to get appended. It will also fully reset
# nested name scope (i.e. the outer name scope has no effect).
name_scope += "/"
return name_scope
def _init_set_name(self, name, zero_based=True):
if name is None:
self._name = backend.unique_object_name(
generic_utils.to_snake_case(self.__class__.__name__),
zero_based=zero_based,
)
elif isinstance(name, str):
backend.observe_object_name(name)
self._name = name
else:
raise TypeError(
f"Expected `name` argument to be a string, but got: {name}"
)
def _get_existing_metric(self, name=None):
match = [m for m in self._metrics if m.name == name]
if not match:
return
if len(match) > 1:
raise ValueError(
"Please provide different names for the metrics you have "
'added. We found {} metrics with the name: "{}"'.format(
len(match), name
)
)
return match[0]
def _handle_weight_regularization(self, name, variable, regularizer):
"""Create lambdas which compute regularization losses."""
def _loss_for_variable(v):
"""Creates a regularization loss `Tensor` for variable `v`."""
with backend.name_scope(name + "/Regularizer"):
regularization = regularizer(v)
return regularization
if base_layer_utils.is_split_variable(variable):
for v in variable:
self.add_loss(functools.partial(_loss_for_variable, v))
elif isinstance(variable, lazy_variable.LazyInitVariable):
self._captured_weight_regularizer.append(
(name, variable, regularizer)
)
else:
self.add_loss(functools.partial(_loss_for_variable, variable))
def _handle_activity_regularization(self, inputs, outputs):
# Apply activity regularization.
# Note that it should be applied every time the layer creates a new
# output, since it is output-specific.
if self._activity_regularizer:
output_list = tf.nest.flatten(outputs)
with backend.name_scope("ActivityRegularizer"):
for output in output_list:
activity_loss = tf.convert_to_tensor(
self._activity_regularizer(output)
)
batch_size = tf.cast(
tf.shape(output)[0], activity_loss.dtype
)
# Make activity regularization strength batch-agnostic.
mean_activity_loss = tf.math.divide_no_nan(
activity_loss, batch_size
)
self.add_loss(mean_activity_loss)
def _set_mask_metadata(self, inputs, outputs, previous_mask, build_graph):
# Many `Layer`s don't need to call `compute_mask`.
# This method is optimized to do as little work as needed for the common
# case.
if not self._supports_masking:
return
flat_outputs = tf.nest.flatten(outputs)
mask_already_computed = getattr(
self, "_compute_output_and_mask_jointly", False
) or all(
getattr(x, "_keras_mask", None) is not None for x in flat_outputs
)
if mask_already_computed:
if build_graph:
self._set_mask_keras_history_checked(flat_outputs)
return
output_masks = self.compute_mask(inputs, previous_mask)
if output_masks is None:
return
flat_masks = tf.nest.flatten(output_masks)
for tensor, mask in zip(flat_outputs, flat_masks):
try:
tensor._keras_mask = mask
except AttributeError:
# C Type such as np.ndarray.
pass
if build_graph:
self._set_mask_keras_history_checked(flat_outputs)
def _set_mask_keras_history_checked(self, flat_outputs):
for output in flat_outputs:
if getattr(output, "_keras_mask", None) is not None:
# Do not track masks for `TensorFlowOpLayer` construction.
output._keras_mask._keras_history_checked = True
def _get_input_masks(self, inputs, input_list, args, kwargs):
if not self._supports_masking and not self._expects_mask_arg:
# Input masks only need to be retrieved if they are needed for
# `call` or `compute_mask`.
input_masks = None
implicit_mask = False
elif self._call_spec.arg_was_passed("mask", args, kwargs):
input_masks = self._call_spec.get_arg_value("mask", args, kwargs)
implicit_mask = False
else:
input_masks = [getattr(t, "_keras_mask", None) for t in input_list]
if all(mask is None for mask in input_masks):
input_masks = None
implicit_mask = False
else:
# Only do expensive `nest` op when masking is actually being
# used.
input_masks = tf.nest.pack_sequence_as(inputs, input_masks)
implicit_mask = True
return input_masks, implicit_mask
def _set_connectivity_metadata(self, args, kwargs, outputs):
# If the layer returns tensors from its inputs unmodified,
# we copy them to avoid loss of KerasHistory metadata.
flat_outputs = tf.nest.flatten(outputs)
flat_inputs = tf.nest.flatten((args, kwargs))
input_ids_set = {id(i) for i in flat_inputs}
outputs_copy = []
for x in flat_outputs:
if id(x) in input_ids_set:
with backend.name_scope(self.name):
x = tf.identity(x)
outputs_copy.append(x)
outputs = tf.nest.pack_sequence_as(outputs, outputs_copy)
# Create node, Node wires itself to inbound and outbound layers. The
# Node constructor actually updates this layer's self._inbound_nodes,
# sets _keras_history on the outputs, and adds itself to the
# `_outbound_nodes` of the layers that produced the inputs to this layer
# call.
node_module.Node(
self, call_args=args, call_kwargs=kwargs, outputs=outputs
)
return outputs
def _get_node_attribute_at_index(self, node_index, attr, attr_name):
"""Private utility to retrieves an attribute (e.g. inputs) from a node.
This is used to implement the methods:
- get_input_shape_at
- get_output_shape_at
- get_input_at
etc...
Args:
node_index: Integer index of the node from which
to retrieve the attribute.
attr: Exact node attribute name.
attr_name: Human-readable attribute name, for error messages.
Returns:
The layer's attribute `attr` at the node of index `node_index`.
Raises:
RuntimeError: If the layer has no inbound nodes, or if called in
Eager mode.
ValueError: If the index provided does not match any node.
"""
if not self._inbound_nodes:
raise RuntimeError(
f"The layer {self.name} has never been called "
f"and thus has no defined {attr_name}."
)
if not len(self._inbound_nodes) > node_index:
raise ValueError(
f"Asked to get {attr_name} at node "
f"{node_index}, but the layer has only "
f"{len(self._inbound_nodes)} inbound nodes."
)
values = getattr(self._inbound_nodes[node_index], attr)
if isinstance(values, list) and len(values) == 1:
return values[0]
else:
return values
def _maybe_build(self, inputs):
# Check input assumptions set before layer building, e.g. input rank.
if not self.built:
input_spec.assert_input_compatibility(
self.input_spec, inputs, self.name
)
input_list = tf.nest.flatten(inputs)
if input_list and self._dtype_policy.compute_dtype is None:
try:
dtype = input_list[0].dtype.base_dtype.name
except AttributeError:
pass
else:
self._set_dtype_policy(policy.Policy(dtype))
input_shapes = None
# Converts Tensors / CompositeTensors to TensorShapes.
if any(hasattr(x, "shape") for x in input_list):
input_shapes = tf_utils.get_shapes(inputs)
else:
# Converts input shape to TensorShapes.
try:
input_shapes = tf_utils.convert_shapes(
inputs, to_tuples=False
)
except ValueError:
pass
# Only call `build` if the user has manually overridden the build
# method.
if not hasattr(self.build, "_is_default"):
# Any setup work performed only once should happen in an
# `init_scope` to avoid creating symbolic Tensors that will
# later pollute any eager operations.
with tf_utils.maybe_init_scope(self):
self.build(input_shapes)
# We must set also ensure that the layer is marked as built, and the
# build shape is stored since user defined build functions may not
# be calling `super.build()`
Layer.build(self, input_shapes)
# Optionally load weight values specified at layer instantiation.
if self._initial_weights is not None:
with tf.init_scope():
# Using `init_scope` since we want variable assignment in
# `set_weights` to be treated like variable initialization.
self.set_weights(self._initial_weights)
self._initial_weights = None
def _get_trainable_state(self):
"""Get the `trainable` state of each sublayer.
Returns:
A dict mapping all sublayers to their `trainable` value.
"""
trainable_state = weakref.WeakKeyDictionary()
for layer in self._flatten_layers():
trainable_state[layer] = layer.trainable
return trainable_state
def _set_trainable_state(self, trainable_state):
"""Set `trainable` state for each sublayer."""
for layer in self._flatten_layers():
if layer in trainable_state:
layer.trainable = trainable_state[layer]
@property
def _obj_reference_counts(self):
"""A dict counting the number of attributes referencing an object."""
self._maybe_create_attribute(
"_obj_reference_counts_dict",
object_identity.ObjectIdentityDictionary(),
)
return self._obj_reference_counts_dict
@tf.__internal__.tracking.no_automatic_dependency_tracking
def _maybe_create_attribute(self, name, default_value):
"""Create attribute (with the default value) if it hasn't been created.
This is useful for fields that is used for tracking purpose,
_trainable_weights, or _layers. Note that user could create a layer
subclass and assign an internal field before invoking the
Layer.__init__(), the __setattr__() need to create the tracking fields
and __init__() need to not override them.
Args:
name: String, the name of the attribute.
default_value: Object, the default value of the attribute.
"""
if not hasattr(self, name):
self.__setattr__(name, default_value)
def __delattr__(self, name):
# For any super.__delattr__() call, we will directly use the
# implementation in Trackable and skip the behavior in AutoTrackable.
# The Layer was originally use Trackable as base class, the change of
# using Module as base class forced us to have AutoTrackable in the
# class hierarchy.
#
# TODO(b/180760306) Keeping the status quo of skipping _delattr__ and
# __setattr__ in AutoTrackable may be unsustainable.
existing_value = getattr(self, name, None)
# If this value is replacing an existing object assigned to an
# attribute, we should clean it out to avoid leaking memory. First we
# check if there are other attributes referencing it.
reference_counts = self._obj_reference_counts
if existing_value not in reference_counts:
super(tf.__internal__.tracking.AutoTrackable, self).__delattr__(
name
)
return
reference_count = reference_counts[existing_value]
if reference_count > 1:
# There are other remaining references. We can't remove this object
# from _layers etc.
reference_counts[existing_value] = reference_count - 1
super(tf.__internal__.tracking.AutoTrackable, self).__delattr__(
name
)
return
else:
# This is the last remaining reference.
del reference_counts[existing_value]
super(tf.__internal__.tracking.AutoTrackable, self).__delattr__(name)
if isinstance(existing_value, Layer) or base_layer_utils.has_weights(
existing_value
):
super(tf.__internal__.tracking.AutoTrackable, self).__setattr__(
"_self_tracked_trackables",
[
l
for l in self._self_tracked_trackables
if l is not existing_value
],
)
if isinstance(existing_value, tf.Variable):
super(tf.__internal__.tracking.AutoTrackable, self).__setattr__(
"_trainable_weights",
[w for w in self._trainable_weights if w is not existing_value],
)
super(tf.__internal__.tracking.AutoTrackable, self).__setattr__(
"_non_trainable_weights",
[
w
for w in self._non_trainable_weights
if w is not existing_value
],
)
def __setattr__(self, name, value):
if (
name == "_self_setattr_tracking"
or not getattr(self, "_self_setattr_tracking", True)
# Exclude @property.setters from tracking
or hasattr(self.__class__, name)
):
try:
super(tf.__internal__.tracking.AutoTrackable, self).__setattr__(
name, value
)
except AttributeError:
raise AttributeError(
(
'Can\'t set the attribute "{}", likely because it '
"conflicts with an existing read-only @property of the "
"object. Please choose a different name."
).format(name)
)
return
# Wraps data structures in `Trackable`, unwraps `NoDependency` objects.
value = tf.__internal__.tracking.sticky_attribute_assignment(
trackable=self, value=value, name=name
)
reference_counts = self._obj_reference_counts
reference_counts[value] = reference_counts.get(value, 0) + 1
# When replacing an existing tf.Variable with a new one, we want to
# check its existing position in the
# self._trainable/non_trainable_variable, so that we can put it back to
# the original position.
if isinstance(value, tf.Variable) and isinstance(
getattr(self, name, None), tf.Variable
):
existing_variable = getattr(self, name)
def _get_variable_from_list(var_list, var):
# helper function to get the tf.variable from the list
# the default list.index() use == for comparison, which will
# cause issue for eager tensor.
for i in range(len(var_list)):
if var_list[i] is var:
return i
return None
if existing_variable.trainable:
self._maybe_create_attribute("_trainable_weights", [])
position = _get_variable_from_list(
self._trainable_weights, existing_variable
)
else:
self._maybe_create_attribute("_non_trainable_variable", [])
position = _get_variable_from_list(
self._non_trainable_variable, existing_variable
)
else:
position = None
# Clean out the old attribute, which clears _layers and
# _trainable_weights if necessary.
try:
self.__delattr__(name)
except AttributeError:
pass
# Keep track of metric instance created in subclassed layer.
for val in tf.nest.flatten(value):
if isinstance(val, metrics_mod.Metric) and hasattr(
self, "_metrics"
):
self._metrics.append(val)
# Append value to self._self_tracked_trackables if relevant
if getattr(self, "_auto_track_sub_layers", True) and (
isinstance(value, tf.Module) or base_layer_utils.has_weights(value)
):
self._maybe_create_attribute("_self_tracked_trackables", [])
# We need to check object identity to avoid de-duplicating empty
# container types which compare equal.
if not any(
(layer is value for layer in self._self_tracked_trackables)
):
self._self_tracked_trackables.append(value)
if hasattr(value, "_use_resource_variables"):
# Legacy layers (V1 tf.layers) must always use
# resource variables.
value._use_resource_variables = True
# Append value to list of trainable / non-trainable weights if relevant
# TODO(b/125122625): This won't pick up on any variables added to a
# list/dict after creation.
self._track_variables(value, position=position)
# TODO(b/180760306) Skip the auto trackable from tf.Module to keep
# status quo. See the comment at __delattr__.
super(tf.__internal__.tracking.AutoTrackable, self).__setattr__(
name, value
)
def _update_trackables(self):
"""Track variables added to lists/dicts after creation"""
for trackable_obj in self._self_tracked_trackables:
if isinstance(
trackable_obj, tf.__internal__.tracking.TrackableDataStructure
):
self._track_variables(trackable_obj)
def _track_variables(self, value, position=None):
"""Tracks `Variable`s including `Variable`s in `CompositeTensor`s."""
for val in tf.nest.flatten(value):
if isinstance(val, tf.Variable):
self._track_variable(val, position=position)
elif tf_utils.is_extension_type(val):
# Manually expand extension types to track resource variables.
nested_vals = tf_utils.type_spec_from_value(val)._to_components(
val
)
self._track_variables(nested_vals, position=position)
def _track_variable(self, val, position=None):
"""Tracks the given `tf.Variable`."""
# Users may add extra weights/variables simply by assigning them to
# attributes (invalid for graph networks)
self._maybe_create_attribute("_trainable_weights", [])
self._maybe_create_attribute("_non_trainable_weights", [])
if val.trainable:
if any(val is w for w in self._trainable_weights):
return
if position is None:
self._trainable_weights.append(val)
else:
self._trainable_weights.insert(position, val)
else:
if any(val is w for w in self._non_trainable_weights):
return
if position is None:
self._non_trainable_weights.append(val)
else:
self._non_trainable_weights.insert(position, val)
backend.track_variable(val)
def _gather_children_attribute(self, attribute):
assert attribute in {
"variables",
"trainable_variables",
"non_trainable_variables",
}
if hasattr(self, "_self_tracked_trackables"):
nested_layers = self._flatten_modules(
include_self=False, recursive=False
)
return list(
itertools.chain.from_iterable(
getattr(layer, attribute) for layer in nested_layers
)
)
return []
def _flatten_layers(self, recursive=True, include_self=True):
for m in self._flatten_modules(
recursive=recursive, include_self=include_self
):
if isinstance(m, Layer):
yield m
def _flatten_modules(self, recursive=True, include_self=True):
"""Flattens `tf.Module` instances (excluding `Metrics`).
Args:
recursive: Whether to recursively flatten through submodules.
include_self: Whether to include this `Layer` instance.
Yields:
`tf.Module` instance tracked by this `Layer`.
"""
if include_self:
yield self
# Only instantiate set and deque if needed.
trackables = getattr(self, "_self_tracked_trackables", None)
if trackables:
seen_object_ids = set()
deque = collections.deque(trackables)
while deque:
trackable_obj = deque.popleft()
trackable_id = id(trackable_obj)
if trackable_id in seen_object_ids:
continue
seen_object_ids.add(trackable_id)
# Metrics are not considered part of the Layer's topology.
if isinstance(trackable_obj, tf.Module) and not isinstance(
trackable_obj, metrics_mod.Metric
):
yield trackable_obj
# Introspect recursively through sublayers.
if recursive:
subtrackables = getattr(
trackable_obj, "_self_tracked_trackables", None
)
if subtrackables:
deque.extendleft(reversed(subtrackables))
elif isinstance(
trackable_obj,
tf.__internal__.tracking.TrackableDataStructure,
):
# Data structures are introspected even with
# `recursive=False`.
tracked_values = trackable_obj._values
if tracked_values:
deque.extendleft(reversed(tracked_values))
# This is a hack so that the is_layer (within
# training/trackable/layer_utils.py) check doesn't get the weights attr.
# TODO(b/110718070): Remove when fixed.
def _is_layer(self):
return True
def _init_call_fn_args(self, expects_training_arg=None):
self._call_spec = layer_utils.CallFunctionSpec(
tf_inspect.getfullargspec(self.call)
)
if expects_training_arg is not None:
self._call_spec.expects_training_arg = expects_training_arg
@property
def _expects_training_arg(self):
"""Whether the call function uses 'training' as a parameter."""
return self._call_spec.expects_training_arg
@property
def _expects_mask_arg(self):
return self._call_spec.expects_mask_arg
@property
def _eager_losses(self):
# A list of loss values containing activity regularizers and losses
# manually added through `add_loss` during eager execution. It is
# cleared after every batch. Because we plan on eventually allowing a
# same model instance to be trained in eager mode or graph mode
# alternatively, we need to keep track of eager losses and symbolic
# losses via separate attributes.
if not hasattr(self._thread_local, "_eager_losses"):
self._thread_local._eager_losses = []
return self._thread_local._eager_losses
@_eager_losses.setter
def _eager_losses(self, losses):
self._thread_local._eager_losses = losses
def _dedup_weights(self, weights):
"""Dedupe weights while maintaining order as much as possible."""
output, seen_ids = [], set()
for w in weights:
if id(w) not in seen_ids:
output.append(w)
# Track the Variable's identity to avoid __eq__ issues.
seen_ids.add(id(w))
return output
# SavedModel properties. Please see keras/saving/saved_model for details.
@tf.__internal__.tracking.no_automatic_dependency_tracking
def _set_save_spec(self, inputs, args=None, kwargs=None):
"""Defines the save spec so that serialization can trace layer calls.
The TensorSpecs of the call function `inputs`, `args`, and `kwargs` are
saved into a tuple of `([inputs] + args, kwargs)`.
Args:
inputs: possibly nested inputs passed into the call function.
args: a list of positional arguments passed into call.
kwargs: a dictionary of keyword arguments passed into call.
"""
if self._saved_model_inputs_spec is not None:
return # Already set.
inputs_spec = tf.nest.map_structure(tf_utils.get_tensor_spec, inputs)
args_spec = tf.nest.map_structure(tf_utils.get_tensor_spec, args or [])
kwargs_spec = {}
# Filter out non-tensor arguments from kwargs.
for key, kwarg in kwargs.items():
flat_kwarg = tf.nest.flatten(kwarg)
flat_specs = [tf_utils.get_tensor_spec(x) for x in flat_kwarg]
if any(s is None for s in flat_specs):
continue
kwargs_spec[key] = tf.nest.pack_sequence_as(kwarg, flat_specs)
self._saved_model_inputs_spec = inputs_spec
self._saved_model_arg_spec = (
[inputs_spec] + list(args_spec),
kwargs_spec,
)
def _get_save_spec(self, dynamic_batch=True, inputs_only=True):
if self._saved_model_inputs_spec is None:
return None
spec = tf.nest.map_structure(
lambda t: tf_utils.get_tensor_spec(t, dynamic_batch=dynamic_batch),
self._saved_model_arg_spec,
)
return spec[0][0] if inputs_only else spec
@property
def _trackable_saved_model_saver(self):
return layer_serialization.LayerSavedModelSaver(self)
@property
def _object_identifier(self):
return self._trackable_saved_model_saver.object_identifier
@property
def _tracking_metadata(self):
"""Info about this layer to be saved into the SavedModel."""
return self._trackable_saved_model_saver.tracking_metadata
def _trackable_children(self, save_type="checkpoint", **kwargs):
if save_type == "savedmodel":
cache = kwargs["cache"]
# TODO(b/213628533): This must be called before super() to ensure
# that any input shape changes are applied before getting the config
# of the model.
children = self._trackable_saved_model_saver.trackable_children(
cache
)
else:
children = {}
children.update(super()._trackable_children(save_type, **kwargs))
return children
@property
def _use_input_spec_as_call_signature(self):
# Whether input spec can be used as the call signature when tracing the
# Layer for SavedModel. By default, this is set to `True` for layers
# exported from the TF-Keras library, because the layers more rigidly
# define the `input_specs` property (many custom layers only set the
# `ndims`)
return (
get_canonical_name_for_symbol(type(self), api_name="keras")
is not None
)
def __getstate__(self):
# Override to support `copy.deepcopy` and pickling.
# Thread-local objects cannot be copied in Python 3, so pop these.
# Thread-local objects are used to cache losses in MirroredStrategy, and
# so shouldn't be copied.
state = self.__dict__.copy()
state.pop("_thread_local", None)
state.pop("_metrics_lock", None)
return state
def __setstate__(self, state):
state["_thread_local"] = threading.local()
state["_metrics_lock"] = threading.Lock()
# Bypass Trackable logic as `__dict__` already contains this info.
object.__setattr__(self, "__dict__", state)
def save_own_variables(self, store):
"""Saves the state of the layer.
You can override this method to take full control of how the state of
the layer is saved upon calling `model.save()`.
Args:
store: Dict where the state of the model will be saved.
"""
all_vars = self._trainable_weights + self._non_trainable_weights
for i, v in enumerate(all_vars):
store[f"{i}"] = v.numpy()
def load_own_variables(self, store):
"""Loads the state of the layer.
You can override this method to take full control of how the state of
the layer is loaded upon calling `keras.models.load_model()`.
Args:
store: Dict from which the state of the model will be loaded.
"""
self._update_trackables()
all_vars = self._trainable_weights + self._non_trainable_weights
if len(store.keys()) != len(all_vars):
raise ValueError(
f"Layer '{self.name}' expected {len(all_vars)} variables, "
"but received "
f"{len(store.keys())} variables during loading. "
f"Expected: {[v.name for v in all_vars]}"
)
for i, v in enumerate(all_vars):
# TODO(rchao): check shapes and raise errors.
v.assign(store[f"{i}"])
class TensorFlowOpLayer(Layer):
"""Wraps a TensorFlow Operation in a Layer.
This class is used internally by the Functional API. When a user
uses a raw TensorFlow Operation on symbolic tensors originating
from an `Input` Layer, the resultant operation will be wrapped
with this Layer object in order to make the operation compatible
with the TF-Keras API.
This Layer will create a new, identical operation (except for inputs
and outputs) every time it is called. If `run_eagerly` is `True`,
the op creation and calculation will happen inside an Eager function.
Instances of this Layer are created when `autolambda` is called, which
is whenever a Layer's `__call__` encounters symbolic inputs that do
not have TF-Keras metadata, or when a Network's `__init__` encounters
outputs that do not have TF-Keras metadata.
Attributes:
node_def: String, the serialized NodeDef of the Op this layer will wrap.
name: String, the name of the Layer.
constants: Dict of NumPy arrays, the values of any Tensors needed for this
Operation that do not originate from a TF-Keras `Input` Layer. Since all
placeholders must come from TF-Keras `Input` Layers, these Tensors must
be treated as constant in the Functional API.
trainable: Bool, whether this Layer is trainable. Currently Variables are
not supported, and so this parameter has no effect.
dtype: The default dtype of this Layer. Inherited from `Layer` and has no
effect on this class, however is used in `get_config`.
"""
@tf.__internal__.tracking.no_automatic_dependency_tracking
def __init__(
self, node_def, name, constants=None, trainable=True, dtype=None
):
# Pass autocast=False, as if inputs are cast, input types might not
# match Operation type.
super(TensorFlowOpLayer, self).__init__(
name=_TF_OP_LAYER_NAME_PREFIX + name,
trainable=trainable,
dtype=dtype,
autocast=False,
)
if isinstance(node_def, dict):
self.node_def = json_format.ParseDict(
node_def, tf.compat.v1.NodeDef()
)
else:
if not isinstance(node_def, bytes):
node_def = node_def.encode("utf-8")
self.node_def = tf.compat.v1.NodeDef.FromString(node_def)
# JSON serialization stringifies keys which are integer input indices.
self.constants = (
{int(index): constant for index, constant in constants.items()}
if constants is not None
else {}
)
# Layer uses original op unless it is called on new inputs.
# This means `built` is not set in `__call__`.
self.built = True
# Do not individually trace TensorflowOpLayers in the SavedModel.
self._must_restore_from_config = True
def call(self, inputs):
if tf.executing_eagerly():
return self._defun_call(inputs)
return self._make_op(inputs)
def _make_node_def(self, graph):
node_def = tf.compat.v1.NodeDef()
node_def.CopyFrom(self.node_def)
# Used in TPUReplicateContext to indicate whether this node has been
# cloned and to not add TPU attributes.
node_def.attr["_cloned"].b = True
node_def.name = graph.unique_name(node_def.name)
return node_def
def _make_op(self, inputs):
inputs = tf.nest.flatten(inputs)
graph = inputs[0].graph
node_def = self._make_node_def(graph)
with graph.as_default():
for index, constant in self.constants.items():
# Recreate constant in graph to add distribution context.
value = tf.get_static_value(constant)
if value is not None:
if isinstance(value, dict):
value = serialization_lib.deserialize_keras_object(
value
)
constant = tf.constant(value, name=node_def.input[index])
inputs.insert(index, constant)
# TODO(b/183990973): We should drop or consolidate these private api
# calls for adding an op to the graph and recording its gradient.
c_op = tf.__internal__.create_c_op(
graph, node_def, inputs, control_inputs=[]
)
op = graph._create_op_from_tf_operation(c_op)
op._control_flow_post_processing()
# Record the gradient because custom-made ops don't go through the
# code-gen'd eager call path
op_type = tf.compat.as_str(op.op_def.name)
attr_names = [
tf.compat.as_str(attr.name) for attr in op.op_def.attr
]
attrs = []
for attr_name in attr_names:
attrs.append(attr_name)
attrs.append(op.get_attr(attr_name))
attrs = tuple(attrs)
tf.__internal__.record_gradient(
op_type, op.inputs, attrs, op.outputs
)
if len(op.outputs) == 1:
return op.outputs[0]
return op.outputs
@tf.function
def _defun_call(self, inputs):
"""Wraps op creation method in an Eager function for `run_eagerly`."""
return self._make_op(inputs)
def get_config(self):
config = super(TensorFlowOpLayer, self).get_config()
config.update(
{
# `__init__` prefixes the name. Revert to the constructor
# argument.
"name": config["name"][len(_TF_OP_LAYER_NAME_PREFIX) :],
"node_def": json_format.MessageToDict(self.node_def),
"constants": {
i: backend.get_value(c) for i, c in self.constants.items()
},
}
)
return config
class AddLoss(Layer):
"""Adds its inputs as a loss.
Attributes:
unconditional: Whether or not the loss should be conditioned on the
inputs.
"""
def __init__(self, unconditional, **kwargs):
# Pass autocast=False, as there is no reason to cast loss to a different
# dtype.
kwargs["autocast"] = False
super(AddLoss, self).__init__(**kwargs)
self.unconditional = unconditional
def call(self, inputs):
self.add_loss(inputs, inputs=(not self.unconditional))
return inputs
def get_config(self):
config = super(AddLoss, self).get_config()
config.update({"unconditional": self.unconditional})
return config
class AddMetric(Layer):
"""Adds its inputs as a metric.
Attributes:
aggregation: 'mean' or None. How the inputs should be aggregated.
metric_name: The name to use for this metric.
"""
def __init__(self, aggregation=None, metric_name=None, **kwargs):
super(AddMetric, self).__init__(**kwargs)
self.aggregation = aggregation
self.metric_name = metric_name
def call(self, inputs):
self.add_metric(
inputs, aggregation=self.aggregation, name=self.metric_name
)
return inputs
def get_config(self):
config = super(AddMetric, self).get_config()
config.update(
{"aggregation": self.aggregation, "metric_name": self.metric_name}
)
return config
def _in_functional_construction_mode(layer, inputs, args, kwargs, input_list):
"""Check the arguments to see if we are constructing a functional model."""
# We are constructing a functional model if any of the inputs
# are KerasTensors
return any(
isinstance(tensor, keras_tensor.KerasTensor)
for tensor in tf.nest.flatten([inputs, args, kwargs])
)
def _convert_numpy_or_python_types(x):
if isinstance(x, (tf.Tensor, np.ndarray, float, int)):
return tf.convert_to_tensor(x)
return x
@keras_export("keras.__internal__.apply_name_scope_on_model_declaration", v1=[])
def _apply_name_scope_on_model_declaration(enable):
"""Apply `with tf.name_scope(...)` on model declaration.
```python
tf.keras.__internal__.apply_name_scope_on_model_declaration(True)
inputs = input_layer.Input((3,))
with tf.name_scope('MyScope'):
outputs = layers.Dense(10, name='MyDense')(inputs)
model = tf.keras.Model(inputs, outputs)
# with `tf.keras.__internal__.apply_name_scope_on_model_declaration(True)`,
# The name of the dense layer is "model/MyScope/MyDense/*", and without,
# "model/MyDense/*"
```
Args:
enable: Enables if `True`, disables if `False`.
"""
if not isinstance(enable, bool):
raise TypeError(
f"`enable` argument must be `True` or `False`, got {enable}"
)
global _is_name_scope_on_model_declaration_enabled
_is_name_scope_on_model_declaration_enabled = enable
@keras_export("keras.__internal__.layers.BaseRandomLayer")
class BaseRandomLayer(Layer):
"""A layer handle the random number creation and savemodel behavior."""
@tf.__internal__.tracking.no_automatic_dependency_tracking
def __init__(
self, seed=None, force_generator=False, rng_type=None, **kwargs
):
"""Initialize the BaseRandomLayer.
Note that the constructor is annotated with
@no_automatic_dependency_tracking. This is to skip the auto
tracking of self._random_generator instance, which is an AutoTrackable.
The backend.RandomGenerator could contain a tf.random.Generator instance
which will have tf.Variable as the internal state. We want to avoid
saving that state into model.weights and checkpoints for backward
compatibility reason. In the meantime, we still need to make them
visible to SavedModel when it is tracing the tf.function for the
`call()`.
See _list_extra_dependencies_for_serialization below for more details.
Args:
seed: optional integer, used to create RandomGenerator.
force_generator: boolean, default to False, whether to force the
RandomGenerator to use the code branch of tf.random.Generator.
rng_type: string, the rng type that will be passed to backend
RandomGenerator. `None` will allow RandomGenerator to choose
types by itself. Valid values are "stateful", "stateless",
"legacy_stateful". Defaults to `None`.
**kwargs: other keyword arguments that will be passed to the parent
*class
"""
super().__init__(**kwargs)
self._random_generator = backend.RandomGenerator(
seed, force_generator=force_generator, rng_type=rng_type
)
def build(self, input_shape):
super().build(input_shape)
self._random_generator._maybe_init()
def _trackable_children(self, save_type="checkpoint", **kwargs):
if save_type == "savedmodel":
cache = kwargs["cache"]
# TODO(b/213628533): This must be called before super() to ensure
# that any input shape changes are applied before getting the config
# of the model.
children = self._trackable_saved_model_saver.trackable_children(
cache
)
# This method exposes the self._random_generator to SavedModel only
# (not layer.weights and checkpoint).
children["_random_generator"] = self._random_generator
else:
children = {}
children.update(super()._trackable_children(save_type, **kwargs))
return children
def _lookup_dependency(self, name, cached_dependencies=None):
# When loading from a TF-Keras SavedModel load, make sure that the
# loader can find the random generator, otherwise the loader will assume
# that it does not exist, and will try to create a new generator.
if name == "_random_generator":
return self._random_generator
elif cached_dependencies is not None:
return cached_dependencies.get(name)
else:
return super()._lookup_dependency(name)
|
tf-keras/tf_keras/engine/base_layer.py/0
|
{
"file_path": "tf-keras/tf_keras/engine/base_layer.py",
"repo_id": "tf-keras",
"token_count": 68681
}
| 191 |
# Copyright 2016 The TensorFlow Authors. All Rights Reserved.
#
# Licensed under the Apache License, Version 2.0 (the "License");
# you may not use this file except in compliance with the License.
# You may obtain a copy of the License at
#
# http://www.apache.org/licenses/LICENSE-2.0
#
# Unless required by applicable law or agreed to in writing, software
# distributed under the License is distributed on an "AS IS" BASIS,
# WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.
# See the License for the specific language governing permissions and
# limitations under the License.
# ,============================================================================
"""Tests for layer graphs construction & handling."""
import warnings
import numpy as np
import tensorflow.compat.v2 as tf
from tf_keras import backend
from tf_keras import layers
from tf_keras import losses
from tf_keras import models
from tf_keras.engine import base_layer
from tf_keras.engine import functional
from tf_keras.engine import input_layer as input_layer_lib
from tf_keras.engine import sequential
from tf_keras.engine import training as training_lib
from tf_keras.saving import object_registration
from tf_keras.saving.legacy import save
from tf_keras.testing_infra import test_combinations
from tf_keras.testing_infra import test_utils
from tf_keras.utils import layer_utils
from tf_keras.utils import tf_utils
# isort: off
from tensorflow.python.checkpoint.checkpoint import (
Checkpoint,
)
from tensorflow.python.framework import extension_type
class NetworkConstructionTest(test_combinations.TestCase):
def test_default_model_name(self):
inputs = input_layer_lib.Input(shape=(1,))
outputs = layers.Dense(1, activation="relu")(inputs)
model = training_lib.Model(inputs=inputs, outputs=outputs)
self.assertEqual(model.name, "model")
model_2 = training_lib.Model(inputs=inputs, outputs=outputs)
self.assertEqual(model_2.name, "model_1")
model_3 = training_lib.Model(inputs=inputs, outputs=outputs)
self.assertEqual(model_3.name, "model_2")
def test_get_updates(self):
class MyLayer(layers.Layer):
def build(self, input_shape):
self.a = self.add_weight(
"a", (1, 1), "float32", trainable=False
)
self.b = self.add_weight(
"b", (1, 1), "float32", trainable=False
)
self.add_update(
tf.compat.v1.assign_add(
self.a, [[1.0]], name="unconditional_update"
)
)
self.built = True
def call(self, inputs):
self.add_update(
tf.compat.v1.assign_add(
self.b, inputs, name="conditional_update"
)
)
return inputs + 1
with tf.Graph().as_default():
x1 = input_layer_lib.Input(shape=(1,))
layer = MyLayer()
_ = layer(x1)
self.assertEqual(len(layer.updates), 2)
x2 = input_layer_lib.Input(shape=(1,))
y2 = layer(x2)
self.assertEqual(len(layer.updates), 3)
network = functional.Functional(x2, y2)
self.assertEqual(len(network.updates), 3)
x3 = input_layer_lib.Input(shape=(1,))
_ = layer(x3)
self.assertEqual(len(network.updates), 4)
x4 = input_layer_lib.Input(shape=(1,))
_ = network(x4)
self.assertEqual(len(network.updates), 5)
network.add_update(tf.compat.v1.assign_add(layer.a, [[1]]))
self.assertEqual(len(network.updates), 6)
network.add_update(tf.compat.v1.assign_add(layer.b, x4))
self.assertEqual(len(network.updates), 7)
@test_combinations.generate(test_combinations.combine(mode=["graph"]))
def test_get_updates_bn(self):
x1 = input_layer_lib.Input(shape=(1,))
layer = layers.BatchNormalization()
_ = layer(x1)
self.assertEqual(len(layer.updates), 2)
def test_get_layer(self):
# create a simple network
x = input_layer_lib.Input(shape=(32,))
dense_a = layers.Dense(4, name="dense_a")
dense_b = layers.Dense(2, name="dense_b")
y = dense_b(dense_a(x))
network = functional.Functional(x, y, name="dense_network")
# test various get_layer by index
self.assertEqual(network.get_layer(index=1), dense_a)
# test invalid get_layer by index
with self.assertRaisesRegex(
ValueError,
"Was asked to retrieve layer at index "
+ str(3)
+ " but model only has "
+ str(len(network.layers))
+ " layers.",
):
network.get_layer(index=3)
# test that only one between name and index is requested
with self.assertRaisesRegex(
ValueError, "Provide only a layer name or a layer index"
):
network.get_layer(index=1, name="dense_b")
# test that a name or an index must be provided
with self.assertRaisesRegex(
ValueError, "Provide either a layer name or layer index."
):
network.get_layer()
# test various get_layer by name
self.assertEqual(network.get_layer(name="dense_a"), dense_a)
# test invalid get_layer by name
with self.assertRaisesRegex(ValueError, "No such layer: dense_c."):
network.get_layer(name="dense_c")
@test_combinations.generate(
test_combinations.combine(mode=["graph", "eager"])
)
def testTopologicalAttributes(self):
# test layer attributes / methods related to cross-layer connectivity.
a = input_layer_lib.Input(shape=(32,), name="input_a")
b = input_layer_lib.Input(shape=(32,), name="input_b")
# test input, output, input_shape, output_shape
test_layer = layers.Dense(16, name="test_layer")
a_test = test_layer(a)
self.assertIs(test_layer.input, a)
self.assertIs(test_layer.output, a_test)
self.assertEqual(test_layer.input_shape, (None, 32))
self.assertEqual(test_layer.output_shape, (None, 16))
# test `get_*_at` methods
dense = layers.Dense(16, name="dense_1")
a_2 = dense(a)
b_2 = dense(b)
self.assertIs(dense.get_input_at(0), a)
self.assertIs(dense.get_input_at(1), b)
self.assertIs(dense.get_output_at(0), a_2)
self.assertIs(dense.get_output_at(1), b_2)
self.assertEqual(dense.get_input_shape_at(0), (None, 32))
self.assertEqual(dense.get_input_shape_at(1), (None, 32))
self.assertEqual(dense.get_output_shape_at(0), (None, 16))
self.assertEqual(dense.get_output_shape_at(1), (None, 16))
# Test invalid value for attribute retrieval.
with self.assertRaises(ValueError):
dense.get_input_at(2)
with self.assertRaises(AttributeError):
new_dense = layers.Dense(16)
_ = new_dense.input
with self.assertRaises(AttributeError):
new_dense = layers.Dense(16)
_ = new_dense.output
with self.assertRaises(AttributeError):
new_dense = layers.Dense(16)
_ = new_dense.output_shape
with self.assertRaises(AttributeError):
new_dense = layers.Dense(16)
_ = new_dense.input_shape
with self.assertRaises(AttributeError):
new_dense = layers.Dense(16)
a = input_layer_lib.Input(shape=(3, 32))
a = input_layer_lib.Input(shape=(5, 32))
a_2 = dense(a)
b_2 = dense(b)
_ = new_dense.input_shape
with self.assertRaises(AttributeError):
new_dense = layers.Dense(16)
a = input_layer_lib.Input(shape=(3, 32))
a = input_layer_lib.Input(shape=(5, 32))
a_2 = dense(a)
b_2 = dense(b)
_ = new_dense.output_shape
def _assertAllIs(self, a, b):
self.assertTrue(all(x is y for x, y in zip(a, b)))
@test_combinations.generate(
test_combinations.combine(mode=["graph", "eager"])
)
def testTopologicalAttributesMultiOutputLayer(self):
class PowersLayer(layers.Layer):
def call(self, inputs):
return [inputs**2, inputs**3]
x = input_layer_lib.Input(shape=(32,))
test_layer = PowersLayer()
p1, p2 = test_layer(x)
self.assertIs(test_layer.input, x)
self._assertAllIs(test_layer.output, [p1, p2])
self.assertEqual(test_layer.input_shape, (None, 32))
self.assertEqual(test_layer.output_shape, [(None, 32), (None, 32)])
@test_combinations.generate(
test_combinations.combine(mode=["graph", "eager"])
)
def testTopologicalAttributesMultiInputLayer(self):
class AddLayer(layers.Layer):
def call(self, inputs):
assert len(inputs) == 2
return inputs[0] + inputs[1]
a = input_layer_lib.Input(shape=(32,))
b = input_layer_lib.Input(shape=(32,))
test_layer = AddLayer()
y = test_layer([a, b])
self._assertAllIs(test_layer.input, [a, b])
self.assertIs(test_layer.output, y)
self.assertEqual(test_layer.input_shape, [(None, 32), (None, 32)])
self.assertEqual(test_layer.output_shape, (None, 32))
def testBasicNetwork(self):
with tf.Graph().as_default():
# minimum viable network
x = input_layer_lib.Input(shape=(32,))
dense = layers.Dense(2)
y = dense(x)
network = functional.Functional(x, y, name="dense_network")
# test basic attributes
self.assertEqual(network.name, "dense_network")
self.assertEqual(len(network.layers), 2) # InputLayer + Dense
self.assertEqual(network.layers[1], dense)
self._assertAllIs(network.weights, dense.weights)
self._assertAllIs(
network.trainable_weights, dense.trainable_weights
)
self._assertAllIs(
network.non_trainable_weights, dense.non_trainable_weights
)
# test callability on Input
x_2 = input_layer_lib.Input(shape=(32,))
y_2 = network(x_2)
self.assertEqual(y_2.shape.as_list(), [None, 2])
# test callability on regular tensor
x_2 = tf.compat.v1.placeholder(dtype="float32", shape=(None, 32))
y_2 = network(x_2)
self.assertEqual(y_2.shape.as_list(), [None, 2])
# test network `trainable` attribute
network.trainable = False
self._assertAllIs(network.weights, dense.weights)
self.assertEqual(network.trainable_weights, [])
self._assertAllIs(
network.non_trainable_weights,
dense.trainable_weights + dense.non_trainable_weights,
)
@test_combinations.generate(
test_combinations.combine(mode=["graph", "eager"])
)
def test_trainable_weights(self):
a = layers.Input(shape=(2,))
b = layers.Dense(1)(a)
model = training_lib.Model(a, b)
weights = model.weights
self._assertAllIs(model.trainable_weights, weights)
self.assertListEqual(model.non_trainable_weights, [])
model.trainable = False
self.assertListEqual(model.trainable_weights, [])
self._assertAllIs(model.non_trainable_weights, weights)
model.trainable = True
self._assertAllIs(model.trainable_weights, weights)
self.assertListEqual(model.non_trainable_weights, [])
model.layers[1].trainable = False
self.assertListEqual(model.trainable_weights, [])
self._assertAllIs(model.non_trainable_weights, weights)
# sequential model
model = sequential.Sequential()
model.add(layers.Dense(1, input_dim=2))
weights = model.weights
self._assertAllIs(model.trainable_weights, weights)
self.assertListEqual(model.non_trainable_weights, [])
model.trainable = False
self.assertListEqual(model.trainable_weights, [])
self._assertAllIs(model.non_trainable_weights, weights)
model.trainable = True
self._assertAllIs(model.trainable_weights, weights)
self.assertListEqual(model.non_trainable_weights, [])
model.layers[0].trainable = False
self.assertListEqual(model.trainable_weights, [])
self._assertAllIs(model.non_trainable_weights, weights)
def test_layer_call_arguments(self):
with tf.Graph().as_default():
# Test the ability to pass and serialize arguments to `call`.
inp = layers.Input(shape=(2,))
x = layers.Dense(3)(inp)
x = layers.Dropout(0.5)(x, training=True)
model = training_lib.Model(inp, x)
# Would be `dropout/cond/Merge` by default
self.assertIn("dropout", model.output.op.name)
# Test that argument is kept when applying the model
inp2 = layers.Input(shape=(2,))
out2 = model(inp2)
self.assertIn("dropout", out2.op.name)
# Test that argument is kept after loading a model
config = model.get_config()
model = training_lib.Model.from_config(config)
self.assertIn("dropout", model.output.op.name)
def test_node_construction(self):
# test basics
a = layers.Input(shape=(32,), name="input_a")
b = layers.Input(shape=(32,), name="input_b")
with self.assertRaises(ValueError):
_ = layers.Input(shape=(32,), batch_shape=(10, 32))
with self.assertRaises(ValueError):
_ = layers.Input(shape=(32,), unknown_kwarg=None)
self.assertListEqual(a.shape.as_list(), [None, 32])
a_layer, a_node_index, a_tensor_index = a._keras_history
b_layer, _, _ = b._keras_history
self.assertEqual(len(a_layer._inbound_nodes), 1)
self.assertEqual(a_tensor_index, 0)
node = a_layer._inbound_nodes[a_node_index]
self.assertEqual(node.outbound_layer, a_layer)
self.assertListEqual(node.inbound_layers, [])
self.assertListEqual(node.input_tensors, [a])
self.assertListEqual(node.input_shapes, [(None, 32)])
self.assertListEqual(node.output_tensors, [a])
self.assertListEqual(node.output_shapes, [(None, 32)])
dense = layers.Dense(16, name="dense_1")
a_2 = dense(a)
b_2 = dense(b)
self.assertEqual(len(dense._inbound_nodes), 2)
self.assertEqual(len(dense._outbound_nodes), 0)
self.assertEqual(dense._inbound_nodes[0].inbound_layers, a_layer)
self.assertEqual(dense._inbound_nodes[0].outbound_layer, dense)
self.assertEqual(dense._inbound_nodes[1].inbound_layers, b_layer)
self.assertEqual(dense._inbound_nodes[1].outbound_layer, dense)
self.assertIs(dense._inbound_nodes[0].input_tensors, a)
self.assertIs(dense._inbound_nodes[1].input_tensors, b)
# test layer properties
test_layer = layers.Dense(16, name="test_layer")
a_test = test_layer(a)
self.assertListEqual(test_layer.kernel.shape.as_list(), [32, 16])
self.assertIs(test_layer.input, a)
self.assertIs(test_layer.output, a_test)
self.assertEqual(test_layer.input_shape, (None, 32))
self.assertEqual(test_layer.output_shape, (None, 16))
self.assertIs(dense.get_input_at(0), a)
self.assertIs(dense.get_input_at(1), b)
self.assertIs(dense.get_output_at(0), a_2)
self.assertIs(dense.get_output_at(1), b_2)
self.assertEqual(dense.get_input_shape_at(0), (None, 32))
self.assertEqual(dense.get_input_shape_at(1), (None, 32))
self.assertEqual(dense.get_output_shape_at(0), (None, 16))
self.assertEqual(dense.get_output_shape_at(1), (None, 16))
self.assertEqual(dense.get_input_mask_at(0), None)
self.assertEqual(dense.get_input_mask_at(1), None)
self.assertEqual(dense.get_output_mask_at(0), None)
self.assertEqual(dense.get_output_mask_at(1), None)
@test_combinations.generate(
test_combinations.combine(mode=["graph", "eager"])
)
def test_multi_input_layer(self):
with self.cached_session():
# test multi-input layer
a = layers.Input(shape=(32,), name="input_a")
b = layers.Input(shape=(32,), name="input_b")
dense = layers.Dense(16, name="dense_1")
a_2 = dense(a)
b_2 = dense(b)
merged = layers.concatenate([a_2, b_2], name="merge")
self.assertListEqual(merged.shape.as_list(), [None, 16 * 2])
(
merge_layer,
merge_node_index,
merge_tensor_index,
) = merged._keras_history
self.assertEqual(merge_node_index, 0)
self.assertEqual(merge_tensor_index, 0)
self.assertEqual(len(merge_layer._inbound_nodes), 1)
self.assertEqual(len(merge_layer._outbound_nodes), 0)
self.assertEqual(
len(merge_layer._inbound_nodes[0].input_tensors), 2
)
self.assertEqual(
len(merge_layer._inbound_nodes[0].inbound_layers), 2
)
c = layers.Dense(64, name="dense_2")(merged)
d = layers.Dense(5, name="dense_3")(c)
model = training_lib.Model(
inputs=[a, b], outputs=[c, d], name="model"
)
self.assertEqual(len(model.layers), 6)
output_shapes = model.compute_output_shape([(None, 32), (None, 32)])
self.assertListEqual(output_shapes[0].as_list(), [None, 64])
self.assertListEqual(output_shapes[1].as_list(), [None, 5])
self.assertListEqual(
model.compute_mask([a, b], [None, None]), [None, None]
)
# we don't check names of first 2 layers (inputs) because
# ordering of same-level layers is not fixed
self.assertListEqual(
[l.name for l in model.layers][2:],
["dense_1", "merge", "dense_2", "dense_3"],
)
self.assertListEqual(
[l.name for l in model._input_layers], ["input_a", "input_b"]
)
self.assertListEqual(
[l.name for l in model._output_layers], ["dense_2", "dense_3"]
)
# actually run model
fn = backend.function(model.inputs, model.outputs)
input_a_np = np.random.random((10, 32))
input_b_np = np.random.random((10, 32))
fn_outputs = fn([input_a_np, input_b_np])
self.assertListEqual(
[x.shape for x in fn_outputs], [(10, 64), (10, 5)]
)
# test get_source_inputs
self._assertAllIs(layer_utils.get_source_inputs(c), [a, b])
# serialization / deserialization
json_config = model.to_json()
recreated_model = models.model_from_json(json_config)
recreated_model.compile("rmsprop", "mse")
self.assertListEqual(
[l.name for l in recreated_model.layers][2:],
["dense_1", "merge", "dense_2", "dense_3"],
)
self.assertListEqual(
[l.name for l in recreated_model._input_layers],
["input_a", "input_b"],
)
self.assertListEqual(
[l.name for l in recreated_model._output_layers],
["dense_2", "dense_3"],
)
fn = backend.function(
recreated_model.inputs, recreated_model.outputs
)
input_a_np = np.random.random((10, 32))
input_b_np = np.random.random((10, 32))
fn_outputs = fn([input_a_np, input_b_np])
self.assertListEqual(
[x.shape for x in fn_outputs], [(10, 64), (10, 5)]
)
def test_multi_output_layer_output_names(self):
inp = layers.Input(name="inp", shape=(None,), dtype=tf.float32)
class _MultiOutput(layers.Layer):
def call(self, x):
return x + 1.0, x + 2.0
out = _MultiOutput(name="out")(inp)
model = training_lib.Model(inp, out)
self.assertEqual(["out", "out_1"], model.output_names)
self.assertAllClose([2.0, 3.0], model(1.0))
def test_recursion(self):
with tf.Graph().as_default(), self.cached_session():
a = layers.Input(shape=(32,), name="input_a")
b = layers.Input(shape=(32,), name="input_b")
dense = layers.Dense(16, name="dense_1")
a_2 = dense(a)
b_2 = dense(b)
merged = layers.concatenate([a_2, b_2], name="merge")
c = layers.Dense(64, name="dense_2")(merged)
d = layers.Dense(5, name="dense_3")(c)
model = training_lib.Model(
inputs=[a, b], outputs=[c, d], name="model"
)
e = layers.Input(shape=(32,), name="input_e")
f = layers.Input(shape=(32,), name="input_f")
self.assertEqual(len(model.inputs), 2)
g, h = model([e, f])
self.assertEqual(len(model.inputs), 2)
self.assertEqual(g.name, "model/dense_2/BiasAdd:0")
self.assertListEqual(g.shape.as_list(), c.shape.as_list())
self.assertListEqual(h.shape.as_list(), d.shape.as_list())
# test separate manipulation of different layer outputs
i = layers.Dense(7, name="dense_4")(h)
final_model = training_lib.Model(
inputs=[e, f], outputs=[i, g], name="final"
)
self.assertEqual(len(final_model.inputs), 2)
self.assertEqual(len(final_model.outputs), 2)
self.assertEqual(len(final_model.layers), 4)
# we don't check names of first 2 layers (inputs) because
# ordering of same-level layers is not fixed
self.assertListEqual(
[layer.name for layer in final_model.layers][2:],
["model", "dense_4"],
)
self.assertListEqual(
model.compute_mask([e, f], [None, None]), [None, None]
)
self.assertListEqual(
final_model.compute_output_shape([(10, 32), (10, 32)]),
[(10, 7), (10, 64)],
)
# run recursive model
fn = backend.function(final_model.inputs, final_model.outputs)
input_a_np = np.random.random((10, 32))
input_b_np = np.random.random((10, 32))
fn_outputs = fn([input_a_np, input_b_np])
self.assertListEqual(
[x.shape for x in fn_outputs], [(10, 7), (10, 64)]
)
# test serialization
model_config = final_model.get_config()
recreated_model = models.Model.from_config(model_config)
fn = backend.function(
recreated_model.inputs, recreated_model.outputs
)
input_a_np = np.random.random((10, 32))
input_b_np = np.random.random((10, 32))
fn_outputs = fn([input_a_np, input_b_np])
self.assertListEqual(
[x.shape for x in fn_outputs], [(10, 7), (10, 64)]
)
@test_combinations.generate(
test_combinations.combine(mode=["graph", "eager"])
)
def test_multi_input_multi_output_recursion(self):
with self.cached_session():
# test multi-input multi-output
a = layers.Input(shape=(32,), name="input_a")
b = layers.Input(shape=(32,), name="input_b")
dense = layers.Dense(16, name="dense_1")
a_2 = dense(a)
b_2 = dense(b)
merged = layers.concatenate([a_2, b_2], name="merge")
c = layers.Dense(64, name="dense_2")(merged)
d = layers.Dense(5, name="dense_3")(c)
model = training_lib.Model(
inputs=[a, b], outputs=[c, d], name="model"
)
j = layers.Input(shape=(32,), name="input_j")
k = layers.Input(shape=(32,), name="input_k")
_, n = model([j, k])
o = layers.Input(shape=(32,), name="input_o")
p = layers.Input(shape=(32,), name="input_p")
q, _ = model([o, p])
self.assertListEqual(n.shape.as_list(), [None, 5])
self.assertListEqual(q.shape.as_list(), [None, 64])
s = layers.concatenate([n, q], name="merge_nq")
self.assertListEqual(s.shape.as_list(), [None, 64 + 5])
# test with single output as 1-elem list
multi_io_model = training_lib.Model([j, k, o, p], [s])
fn = backend.function(multi_io_model.inputs, multi_io_model.outputs)
fn_outputs = fn(
[
np.random.random((10, 32)),
np.random.random((10, 32)),
np.random.random((10, 32)),
np.random.random((10, 32)),
]
)
self.assertListEqual([x.shape for x in fn_outputs], [(10, 69)])
# test with single output as tensor
multi_io_model = training_lib.Model([j, k, o, p], s)
fn = backend.function(multi_io_model.inputs, multi_io_model.outputs)
fn_outputs = fn(
[
np.random.random((10, 32)),
np.random.random((10, 32)),
np.random.random((10, 32)),
np.random.random((10, 32)),
]
)
# note that the output of the function will still be a 1-elem list
self.assertListEqual([x.shape for x in fn_outputs], [(10, 69)])
# test serialization
model_config = multi_io_model.get_config()
recreated_model = models.Model.from_config(model_config)
fn = backend.function(
recreated_model.inputs, recreated_model.outputs
)
fn_outputs = fn(
[
np.random.random((10, 32)),
np.random.random((10, 32)),
np.random.random((10, 32)),
np.random.random((10, 32)),
]
)
# note that the output of the function will still be a 1-elem list
self.assertListEqual([x.shape for x in fn_outputs], [(10, 69)])
config = model.get_config()
models.Model.from_config(config)
model.summary()
json_str = model.to_json()
models.model_from_json(json_str)
@test_combinations.generate(
test_combinations.combine(mode=["graph", "eager"])
)
def test_invalid_graphs(self):
a = layers.Input(shape=(32,), name="input_a")
b = layers.Input(shape=(32,), name="input_b")
dense = layers.Dense(16, name="dense_1")
a_2 = dense(a)
b_2 = dense(b)
merged = layers.concatenate([a_2, b_2], name="merge")
c = layers.Dense(64, name="dense_2")(merged)
d = layers.Dense(5, name="dense_3")(c)
model = training_lib.Model(inputs=[a, b], outputs=[c, d], name="model")
# disconnected graph
j = layers.Input(shape=(32,), name="input_j")
k = layers.Input(shape=(32,), name="input_k")
m, n = model([j, k])
with self.assertRaises(Exception):
training_lib.Model([j], [m, n])
# redundant outputs
j = layers.Input(shape=(32,), name="input_j")
k = layers.Input(shape=(32,), name="input_k")
m, n = model([j, k])
training_lib.Model([j, k], [m, n, n])
# redundant inputs
j = layers.Input(shape=(32,), name="input_j")
k = layers.Input(shape=(32,), name="input_k")
m, n = model([j, k])
with self.assertRaises(Exception):
training_lib.Model([j, k, j], [m, n])
# i have not idea what I'm doing: garbage as inputs/outputs
j = layers.Input(shape=(32,), name="input_j")
k = layers.Input(shape=(32,), name="input_k")
m, n = model([j, k])
with self.assertRaises(Exception):
training_lib.Model([j, k], [m, n, 0])
def test_raw_tf_compatibility(self):
with tf.Graph().as_default():
# test calling layers/models on TF tensors
a = layers.Input(shape=(32,), name="input_a")
b = layers.Input(shape=(32,), name="input_b")
dense = layers.Dense(16, name="dense_1")
a_2 = dense(a)
b_2 = dense(b)
merged = layers.concatenate([a_2, b_2], name="merge")
c = layers.Dense(64, name="dense_2")(merged)
d = layers.Dense(5, name="dense_3")(c)
model = training_lib.Model(
inputs=[a, b], outputs=[c, d], name="model"
)
j = layers.Input(shape=(32,), name="input_j")
k = layers.Input(shape=(32,), name="input_k")
self.assertEqual(len(model.inputs), 2)
m, n = model([j, k])
self.assertEqual(len(model.inputs), 2)
tf_model = training_lib.Model([j, k], [m, n])
j_tf = tf.compat.v1.placeholder(dtype=tf.float32, shape=(None, 32))
k_tf = tf.compat.v1.placeholder(dtype=tf.float32, shape=(None, 32))
m_tf, n_tf = tf_model([j_tf, k_tf])
self.assertListEqual(m_tf.shape.as_list(), [None, 64])
self.assertListEqual(n_tf.shape.as_list(), [None, 5])
# test merge
layers.concatenate([j_tf, k_tf], axis=1)
layers.add([j_tf, k_tf])
# test tensor input
x = tf.compat.v1.placeholder(shape=(None, 2), dtype=tf.float32)
layers.InputLayer(input_tensor=x)
x = layers.Input(tensor=x)
layers.Dense(2)(x)
@test_combinations.generate(
test_combinations.combine(mode=["graph", "eager"])
)
def test_basic_masking(self):
a = layers.Input(shape=(10, 32), name="input_a")
b = layers.Masking()(a)
model = training_lib.Model(a, b)
self.assertEqual(model.output_mask.shape.as_list(), [None, 10])
def testMaskingSingleInput(self):
class MaskedLayer(layers.Layer):
def call(self, inputs, mask=None):
if mask is not None:
return inputs * mask
return inputs
def compute_mask(self, inputs, mask=None):
return tf.ones_like(inputs)
if tf.executing_eagerly():
a = tf.constant([2] * 32)
mask = tf.constant([0, 1] * 16)
a._keras_mask = mask
b = MaskedLayer()(a)
self.assertTrue(hasattr(b, "_keras_mask"))
self.assertAllEqual(
self.evaluate(tf.ones_like(mask)),
self.evaluate(getattr(b, "_keras_mask")),
)
self.assertAllEqual(self.evaluate(a * mask), self.evaluate(b))
else:
x = input_layer_lib.Input(shape=(32,))
y = MaskedLayer()(x)
network = functional.Functional(x, y)
# test callability on Input
x_2 = input_layer_lib.Input(shape=(32,))
y_2 = network(x_2)
self.assertEqual(y_2.shape.as_list(), [None, 32])
# test callability on regular tensor
x_2 = tf.compat.v1.placeholder(dtype="float32", shape=(None, 32))
y_2 = network(x_2)
self.assertEqual(y_2.shape.as_list(), [None, 32])
def test_activity_regularization_with_model_composition(self):
def reg(x):
return tf.reduce_sum(x)
net_a_input = input_layer_lib.Input((2,))
net_a = net_a_input
net_a = layers.Dense(
2,
kernel_initializer="ones",
use_bias=False,
activity_regularizer=reg,
)(net_a)
model_a = training_lib.Model([net_a_input], [net_a])
net_b_input = input_layer_lib.Input((2,))
net_b = model_a(net_b_input)
model_b = training_lib.Model([net_b_input], [net_b])
model_b.compile(optimizer="sgd", loss=None)
x = np.ones((1, 2))
loss = model_b.evaluate(x)
self.assertEqual(loss, 4.0)
@test_combinations.generate(test_combinations.keras_mode_combinations())
def test_layer_sharing_at_heterogenous_depth(self):
x_val = np.random.random((10, 5))
x = input_layer_lib.Input(shape=(5,))
a = layers.Dense(5, name="A")
b = layers.Dense(5, name="B")
output = a(b(a(b(x))))
m = training_lib.Model(x, output)
m.run_eagerly = test_utils.should_run_eagerly()
output_val = m.predict(x_val)
config = m.get_config()
weights = m.get_weights()
m2 = models.Model.from_config(config)
m2.set_weights(weights)
output_val_2 = m2.predict(x_val)
self.assertAllClose(output_val, output_val_2, atol=1e-6)
@test_combinations.generate(test_combinations.keras_mode_combinations())
def test_layer_sharing_at_heterogenous_depth_with_concat(self):
input_shape = (16, 9, 3)
input_layer = input_layer_lib.Input(shape=input_shape)
a = layers.Dense(3, name="dense_A")
b = layers.Dense(3, name="dense_B")
c = layers.Dense(3, name="dense_C")
x1 = b(a(input_layer))
x2 = a(c(input_layer))
output = layers.concatenate([x1, x2])
m = training_lib.Model(inputs=input_layer, outputs=output)
m.run_eagerly = test_utils.should_run_eagerly()
x_val = np.random.random((10, 16, 9, 3))
output_val = m.predict(x_val)
config = m.get_config()
weights = m.get_weights()
m2 = models.Model.from_config(config)
m2.set_weights(weights)
output_val_2 = m2.predict(x_val)
self.assertAllClose(output_val, output_val_2, atol=1e-6)
def test_layer_sharing_maintains_node_order(self):
# See https://github.com/keras-team/tf-keras/issues/14838.
inp = input_layer_lib.Input(shape=[5], name="main_input")
shared_layer = layers.Layer(name="shared")
ones_result = shared_layer(tf.ones_like(inp))
zeros_result = shared_layer(tf.zeros_like(inp))
zeros_result = layers.Layer(name="blank")(zeros_result)
m = training_lib.Model(
inputs=[inp], outputs=[zeros_result, ones_result]
)
m2 = models.Model.from_config(m.get_config())
self.assertAllClose(
m2.predict_on_batch(tf.zeros([1, 5])),
m.predict_on_batch(tf.zeros([1, 5])),
)
@test_combinations.generate(test_combinations.keras_mode_combinations())
def test_explicit_training_argument(self):
a = layers.Input(shape=(2,))
b = layers.Dropout(0.5)(a)
base_model = training_lib.Model(a, b)
a = layers.Input(shape=(2,))
b = base_model(a, training=False)
model = training_lib.Model(a, b)
x = np.ones((100, 2))
y = np.ones((100, 2))
model.compile(
optimizer="sgd",
loss="mse",
run_eagerly=test_utils.should_run_eagerly(),
)
loss = model.train_on_batch(x, y)
self.assertEqual(
loss, 0
) # In inference mode, output is equal to input.
a = layers.Input(shape=(2,))
b = base_model(a, training=True)
model = training_lib.Model(a, b)
preds = model.predict(x)
self.assertEqual(np.min(preds), 0.0) # At least one unit was dropped.
@test_combinations.generate(test_combinations.keras_mode_combinations())
def test_mask_derived_from_keras_layer(self):
inputs = input_layer_lib.Input((5, 10))
mask = input_layer_lib.Input((5,))
outputs = layers.RNN(layers.LSTMCell(100))(inputs, mask=mask)
model = training_lib.Model([inputs, mask], outputs)
model.compile("sgd", "mse", run_eagerly=test_utils.should_run_eagerly())
history = model.fit(
x=[np.ones((10, 5, 10)), np.zeros((10, 5))],
y=np.zeros((10, 100)),
batch_size=2,
)
# All data is masked, returned values are 0's.
self.assertEqual(history.history["loss"][0], 0.0)
history = model.fit(
x=[np.ones((10, 5, 10)), np.ones((10, 5))],
y=np.zeros((10, 100)),
batch_size=2,
)
# Data is not masked, returned values are random.
self.assertGreater(history.history["loss"][0], 0.0)
model = training_lib.Model.from_config(model.get_config())
model.compile("sgd", "mse", run_eagerly=test_utils.should_run_eagerly())
history = model.fit(
x=[np.ones((10, 5, 10)), np.zeros((10, 5))],
y=np.zeros((10, 100)),
batch_size=2,
)
# All data is masked, returned values are 0's.
self.assertEqual(history.history["loss"][0], 0.0)
history = model.fit(
x=[np.ones((10, 5, 10)), np.ones((10, 5))],
y=np.zeros((10, 100)),
batch_size=2,
)
# Data is not masked, returned values are random.
self.assertGreater(history.history["loss"][0], 0.0)
@test_combinations.generate(test_combinations.keras_mode_combinations())
def test_call_arg_derived_from_keras_layer(self):
class MyAdd(layers.Layer):
def call(self, x1, x2):
return x1 + x2
input1 = input_layer_lib.Input(10)
input2 = input_layer_lib.Input(10)
outputs = MyAdd()(input1, input2)
model = training_lib.Model([input1, input2], outputs)
model.compile("sgd", "mse", run_eagerly=test_utils.should_run_eagerly())
history = model.fit(
x=[3 * np.ones((10, 10)), 7 * np.ones((10, 10))],
y=10 * np.ones((10, 10)),
batch_size=2,
)
# Check that second input was correctly added to first.
self.assertEqual(history.history["loss"][0], 0.0)
# Check serialization.
model = training_lib.Model.from_config(
model.get_config(), custom_objects={"MyAdd": MyAdd}
)
model.compile("sgd", "mse", run_eagerly=test_utils.should_run_eagerly())
history = model.fit(
x=[3 * np.ones((10, 10)), 7 * np.ones((10, 10))],
y=10 * np.ones((10, 10)),
batch_size=2,
)
# Check that second input was correctly added to first.
self.assertEqual(history.history["loss"][0], 0.0)
@test_combinations.generate(
test_combinations.keras_mode_combinations(mode="eager"),
)
def test_only_some_in_first_arg_derived_from_keras_layer_keras_tensors(
self,
):
# This functionality is unsupported in v1 graphs
class MyAddAll(layers.Layer):
def call(self, inputs):
x = inputs[0]
for inp in inputs[1:]:
if inp is not None:
x = x + inp
return x
input1 = input_layer_lib.Input(10)
input2 = input_layer_lib.Input(10)
layer = MyAddAll()
outputs = layer([0.0, input1, None, input2, None])
model = training_lib.Model([input1, input2], outputs)
self.assertIn(layer, model.layers)
model.compile("sgd", "mse", run_eagerly=test_utils.should_run_eagerly())
history = model.fit(
x=[3 * np.ones((10, 10)), 7 * np.ones((10, 10))],
y=10 * np.ones((10, 10)),
batch_size=2,
)
# Check that second input was correctly added to first.
self.assertEqual(history.history["loss"][0], 0.0)
# Check serialization.
model = training_lib.Model.from_config(
model.get_config(), custom_objects={"MyAddAll": MyAddAll}
)
model.compile("sgd", "mse", run_eagerly=test_utils.should_run_eagerly())
history = model.fit(
x=[3 * np.ones((10, 10)), 7 * np.ones((10, 10))],
y=10 * np.ones((10, 10)),
batch_size=2,
)
# Check that second input was correctly added to first.
self.assertEqual(history.history["loss"][0], 0.0)
@test_combinations.generate(
test_combinations.times(
test_combinations.keras_mode_combinations(),
test_combinations.combine(share_already_used_layer=[True, False]),
)
)
def test_call_kwarg_derived_from_keras_layer(
self, share_already_used_layer
):
class MaybeAdd(layers.Layer):
def call(self, x1, x2=None):
if x2 is not None:
return x1 + x2
return x1
class IdentityLayer(layers.Layer):
def call(self, x):
return x
input1 = input_layer_lib.Input(10)
input2 = input_layer_lib.Input(10)
identity_layer = IdentityLayer()
if share_already_used_layer:
# We have had model serialization/deserialization break in the past:
# when a layer was previously used to construct other functional
# models and had a non-empty list of inbound nodes before being used
# to define the model being serialized/deserialized. (The
# serialization/deserialization was not correctly adjusting the
# node_index serialization/deserialization). So, we explicitly test
# this case.
training_lib.Model([input1], identity_layer(input1))
outputs = MaybeAdd()(input1, x2=identity_layer(input2))
model = training_lib.Model([input1, input2], outputs)
model.compile("sgd", "mse", run_eagerly=test_utils.should_run_eagerly())
history = model.fit(
x=[3 * np.ones((10, 10)), 7 * np.ones((10, 10))],
y=10 * np.ones((10, 10)),
batch_size=2,
)
# Check that second input was correctly added to first.
self.assertEqual(history.history["loss"][0], 0.0)
model = training_lib.Model.from_config(
model.get_config(),
custom_objects={
"MaybeAdd": MaybeAdd,
"IdentityLayer": IdentityLayer,
},
)
model.compile("sgd", "mse", run_eagerly=test_utils.should_run_eagerly())
history = model.fit(
x=[3 * np.ones((10, 10)), 7 * np.ones((10, 10))],
y=10 * np.ones((10, 10)),
batch_size=2,
)
# Check that second input was correctly added to first.
self.assertEqual(history.history["loss"][0], 0.0)
@test_combinations.generate(test_combinations.keras_mode_combinations())
def test_call_kwarg_dtype_serialization(self):
class Double(layers.Layer):
def call(self, x1, dtype=None):
return tf.cast(x1 + x1, dtype=dtype)
input1 = input_layer_lib.Input(10)
outputs = Double()(input1, dtype=tf.float16)
model = training_lib.Model([input1], outputs)
model.compile("sgd", "mse", run_eagerly=test_utils.should_run_eagerly())
history = model.fit(
x=[3 * np.ones((10, 10))], y=6 * np.ones((10, 10)), batch_size=2
)
# Check that input was correctly doubled.
self.assertEqual(history.history["loss"][0], 0.0)
# Check the output dtype
self.assertEqual(model(tf.ones((3, 10))).dtype, tf.float16)
model = training_lib.Model.from_config(
model.get_config(), custom_objects={"Double": Double}
)
model.compile("sgd", "mse", run_eagerly=test_utils.should_run_eagerly())
history = model.fit(
x=[3 * np.ones((10, 10))], y=6 * np.ones((10, 10)), batch_size=2
)
# Check that input was correctly doubled.
self.assertEqual(history.history["loss"][0], 0.0)
# Check the output dtype
self.assertEqual(model(tf.ones((3, 10))).dtype, tf.float16)
@test_combinations.generate(test_combinations.keras_mode_combinations())
def test_call_kwarg_nonserializable(self):
class Double(layers.Layer):
def call(self, x1, kwarg=None):
return x1 + x1
class NonSerializable:
def __init__(self, foo=None):
self.foo = foo
input1 = input_layer_lib.Input(10)
outputs = Double()(input1, kwarg=NonSerializable())
model = training_lib.Model([input1], outputs)
model.compile("sgd", "mse", run_eagerly=test_utils.should_run_eagerly())
history = model.fit(
x=[3 * np.ones((10, 10))], y=6 * np.ones((10, 10)), batch_size=2
)
# Check that input was correctly doubled.
self.assertEqual(history.history["loss"][0], 0.0)
with self.assertRaisesRegex(
TypeError,
"Layer double was passed non-JSON-serializable arguments.",
):
model.get_config()
@test_combinations.generate(
test_combinations.times(
test_combinations.keras_mode_combinations(),
test_combinations.combine(share_already_used_layer=[True, False]),
)
)
def test_call_kwarg_derived_from_keras_layer_and_first_arg_is_constant(
self, share_already_used_layer
):
class IdentityLayer(layers.Layer):
def call(self, x):
return x
class MaybeAdd(layers.Layer):
def call(self, x1, x2=None):
if x2 is not None:
return x1 + x2
return x1
input2 = input_layer_lib.Input(10)
identity_layer = IdentityLayer()
if share_already_used_layer:
# We have had model serialization/deserialization break in the past:
# when a layer was previously used to construct other functional
# models and had a non-empty list of inbound nodes before being used
# to define the model being serialized/deserialized. (The
# serialization/deserialization was not correctly adjusting the
# node_index serialization/deserialization). So, we explicitly test
# this case.
training_lib.Model([input2], identity_layer(input2))
outputs = MaybeAdd()(3.0, x2=identity_layer(input2))
model = training_lib.Model([input2], outputs)
model.compile("sgd", "mse", run_eagerly=test_utils.should_run_eagerly())
history = model.fit(
x=7 * np.ones((10, 10)), y=10 * np.ones((10, 10)), batch_size=2
)
# Check that second input was correctly added to first.
self.assertEqual(history.history["loss"][0], 0.0)
model = training_lib.Model.from_config(
model.get_config(),
custom_objects={
"MaybeAdd": MaybeAdd,
"IdentityLayer": IdentityLayer,
},
)
model.compile("sgd", "mse", run_eagerly=test_utils.should_run_eagerly())
history = model.fit(
x=7 * np.ones((10, 10)), y=10 * np.ones((10, 10)), batch_size=2
)
# Check that second input was correctly added to first.
self.assertEqual(history.history["loss"][0], 0.0)
@test_combinations.generate(test_combinations.keras_mode_combinations())
def test_dont_cast_composite_unless_necessary(self):
if not tf.executing_eagerly():
# Creating TF-Keras inputs from a type_spec only supported in eager.
return
# TODO(edloper): Change this to tf.experimental.ExtensionTyep once
# it's been released.
class MyType(extension_type.ExtensionType):
# TODO(edloper) Remove _shape and _dtype once TF-Keras has been
# switched to use .shape and .dtype instead.
value: tf.Tensor
_shape = property(lambda self: self.value.shape)
shape = property(lambda self: self.value.shape)
_dtype = property(lambda self: self.value.dtype)
dtype = property(lambda self: self.value.dtype)
class Spec:
_shape = property(lambda self: self.value.shape)
shape = property(lambda self: self.value.shape)
_dtype = property(lambda self: self.value.dtype)
dtype = property(lambda self: self.value.dtype)
my_spec = MyType.Spec(tf.TensorSpec([5], tf.float32))
input1 = input_layer_lib.Input(type_spec=my_spec)
model = training_lib.Model([input1], input1)
model.compile(run_eagerly=test_utils.should_run_eagerly())
model(MyType([1.0, 2.0, 3.0, 4.0, 5.0])) # Does not require cast.
with self.assertRaises((ValueError, TypeError)):
model(MyType([1, 2, 3, 4, 5]))
@test_combinations.generate(test_combinations.keras_mode_combinations())
def test_composite_call_kwarg_derived_from_keras_layer(self):
# Create a test layer that accepts composite tensor inputs.
class MaybeAdd(layers.Layer):
def call(self, x1, x2=None):
# We need to convert this to a tensor for loss calculations -
# losses don't play nicely with ragged tensors yet.
if x2 is not None:
return (x1 + x2).to_tensor(default_value=0)
return x1.to_tensor(default_value=0)
input1 = input_layer_lib.Input((None,), ragged=True)
input2 = input_layer_lib.Input((None,), ragged=True)
outputs = MaybeAdd()(input1, x2=input2)
model = training_lib.Model([input1, input2], outputs)
model.compile("sgd", "mse", run_eagerly=test_utils.should_run_eagerly())
input_data = [
tf.ragged.constant([[3.0, 3.0], [3.0, 3.0], [3.0]]),
tf.ragged.constant([[7.0, 7.0], [7.0, 7.0], [7.0]]),
]
expected_data = np.array([[10.0, 10.0], [10.0, 10.0], [10.0, 0.0]])
history = model.fit(x=input_data, y=expected_data)
# Check that second input was correctly added to first.
self.assertEqual(history.history["loss"][0], 0.0)
model = training_lib.Model.from_config(
model.get_config(), custom_objects={"MaybeAdd": MaybeAdd}
)
model.compile("sgd", "mse", run_eagerly=test_utils.should_run_eagerly())
history = model.fit(x=input_data, y=expected_data)
# Check that second input was correctly added to first.
self.assertEqual(history.history["loss"][0], 0.0)
@test_combinations.generate(
test_combinations.keras_mode_combinations(mode="eager")
)
def test_call_some_not_all_nested_in_first_arg_derived_from_keras_layer(
self,
):
# This functionality is unsupported in v1 graphs
class AddAll(layers.Layer):
def call(self, x1_x2, x3):
x1, x2 = x1_x2
out = x1 + x2
if x3 is not None:
for t in x3.values():
out += t
return out
input1 = input_layer_lib.Input(10)
input2 = input_layer_lib.Input(10)
input3 = input_layer_lib.Input(10)
layer = AddAll()
outputs = layer(
[input1, 4 * tf.ones((1, 10))],
x3={"a": input2, "b": input3, "c": 5 * tf.ones((1, 10))},
)
model = training_lib.Model([input1, input2, input3], outputs)
self.assertIn(layer, model.layers)
model.compile("sgd", "mse", run_eagerly=test_utils.should_run_eagerly())
history = model.fit(
x=[np.ones((10, 10)), 2 * np.ones((10, 10)), 3 * np.ones((10, 10))],
y=15 * np.ones((10, 10)),
batch_size=2,
)
# Check that all inputs were correctly added.
self.assertEqual(history.history["loss"][0], 0.0)
model = training_lib.Model.from_config(
model.get_config(), custom_objects={"AddAll": AddAll}
)
model.compile("sgd", "mse", run_eagerly=test_utils.should_run_eagerly())
history = model.fit(
x=[np.ones((10, 10)), 2 * np.ones((10, 10)), 3 * np.ones((10, 10))],
y=15 * np.ones((10, 10)),
batch_size=2,
)
# Check that all inputs were correctly added.
self.assertEqual(history.history["loss"][0], 0.0)
@test_combinations.generate(test_combinations.keras_mode_combinations())
def test_call_nested_arg_derived_from_keras_layer(self):
class AddAll(layers.Layer):
def call(self, x1, x2, x3=None):
out = x1 + x2
if x3 is not None:
for t in x3.values():
out += t
return out
input1 = input_layer_lib.Input(10)
input2 = input_layer_lib.Input(10)
input3 = input_layer_lib.Input(10)
outputs = AddAll()(
input1,
4 * tf.ones((1, 10)),
x3={"a": input2, "b": input3, "c": 5 * tf.ones((1, 10))},
)
model = training_lib.Model([input1, input2, input3], outputs)
model.compile("sgd", "mse", run_eagerly=test_utils.should_run_eagerly())
history = model.fit(
x=[np.ones((10, 10)), 2 * np.ones((10, 10)), 3 * np.ones((10, 10))],
y=15 * np.ones((10, 10)),
batch_size=2,
)
# Check that all inputs were correctly added.
self.assertEqual(history.history["loss"][0], 0.0)
model = training_lib.Model.from_config(
model.get_config(), custom_objects={"AddAll": AddAll}
)
model.compile("sgd", "mse", run_eagerly=test_utils.should_run_eagerly())
history = model.fit(
x=[np.ones((10, 10)), 2 * np.ones((10, 10)), 3 * np.ones((10, 10))],
y=15 * np.ones((10, 10)),
batch_size=2,
)
# Check that all inputs were correctly added.
self.assertEqual(history.history["loss"][0], 0.0)
@test_combinations.generate(test_combinations.keras_mode_combinations())
def test_multi_output_model_with_none_masking(self):
def func(x):
return [x * 0.2, x * 0.3]
def output_shape(input_shape):
return [input_shape, input_shape]
i = layers.Input(shape=(3, 2, 1))
o = layers.Lambda(function=func, output_shape=output_shape)(i)
self.assertEqual(backend.int_shape(o[0]), (None, 3, 2, 1))
self.assertEqual(backend.int_shape(o[1]), (None, 3, 2, 1))
o = layers.add(o)
model = training_lib.Model(i, o)
model.run_eagerly = test_utils.should_run_eagerly()
i2 = layers.Input(shape=(3, 2, 1))
o2 = model(i2)
model2 = training_lib.Model(i2, o2)
model2.run_eagerly = test_utils.should_run_eagerly()
x = np.random.random((4, 3, 2, 1))
out = model2.predict(x)
assert out.shape == (4, 3, 2, 1)
self.assertAllClose(out, x * 0.2 + x * 0.3, atol=1e-4)
@test_combinations.generate(test_combinations.keras_mode_combinations())
def test_constant_initializer_with_numpy(self):
initializer = tf.compat.v1.constant_initializer(np.ones((3, 2)))
model = sequential.Sequential()
model.add(
layers.Dense(2, input_shape=(3,), kernel_initializer=initializer)
)
model.add(layers.Dense(3))
model.compile(
loss="mse",
optimizer="sgd",
metrics=["acc"],
run_eagerly=test_utils.should_run_eagerly(),
)
json_str = model.to_json()
models.model_from_json(json_str)
def test_subclassed_error_if_init_not_called(self):
class MyNetwork(training_lib.Model):
def __init__(self):
self._foo = [layers.Dense(10), layers.Dense(10)]
with self.assertRaisesRegex(RuntimeError, "forgot to call"):
MyNetwork()
@test_combinations.generate(
test_combinations.combine(mode=["graph", "eager"])
)
def test_int_input_shape(self):
inputs = input_layer_lib.Input(10)
self.assertEqual([None, 10], inputs.shape.as_list())
inputs_with_batch = input_layer_lib.Input(batch_size=20, shape=5)
self.assertEqual([20, 5], inputs_with_batch.shape.as_list())
@test_combinations.generate(
test_combinations.combine(mode=["graph", "eager"])
)
def test_model_initialization(self):
# Functional model
inputs = input_layer_lib.Input(shape=(32,))
outputs = layers.Dense(4)(inputs)
with self.assertRaisesRegex(
TypeError, "Keyword argument not understood"
):
model = training_lib.Model(
inputs, outputs, name="m", trainable=False, dtype="int64"
)
with self.assertRaisesRegex(
TypeError, "Keyword argument not understood"
):
model = training_lib.Model(
inputs, outputs, name="m", trainable=False, dynamic=False
)
model = training_lib.Model(inputs, outputs, name="m", trainable=False)
self.assertEqual("m", model.name)
self.assertFalse(model.trainable)
self.assertFalse(model.dynamic)
class SubclassModel(training_lib.Model):
pass
# Subclassed model
model = SubclassModel(
name="subclassed", trainable=True, dtype="int64", dynamic=True
)
self.assertEqual("subclassed", model.name)
self.assertTrue(model.dynamic)
self.assertTrue(model.trainable)
w = model.add_weight(
"w", [], initializer=tf.compat.v1.constant_initializer(1)
)
self.assertEqual(tf.int64, w.dtype)
def test_disconnected_inputs(self):
input_tensor1 = input_layer_lib.Input(shape=[200], name="a")
input_tensor2 = input_layer_lib.Input(shape=[10], name="b")
output_tensor1 = layers.Dense(units=10)(input_tensor1)
net = functional.Functional(
inputs=[input_tensor1, input_tensor2], outputs=[output_tensor1]
)
net2 = functional.Functional.from_config(net.get_config())
self.assertLen(net2.inputs, 2)
self.assertEqual("a", net2.layers[0].name)
self.assertEqual("b", net2.layers[1].name)
@test_combinations.generate(
test_combinations.keras_model_type_combinations()
)
def test_dependency_tracking(self):
model = test_utils.get_small_mlp(1, 4, input_dim=3)
model.trackable = Checkpoint()
self.assertIn("trackable", model._unconditional_dependency_names)
self.assertEqual(model.trackable, model._lookup_dependency("trackable"))
@test_combinations.generate(
test_combinations.combine(mode=["graph", "eager"])
)
def test_model_construction_in_tf_function(self):
d = {"model": None}
@tf.function
def fn(x):
if d["model"] is None:
# Check that Functional can be built in a `tf.function`.
inputs = input_layer_lib.Input(10)
outputs = layers.Dense(1)(inputs)
model = functional.Functional(inputs, outputs)
d["model"] = model
else:
model = d["model"]
return model(x)
x = tf.ones((10, 10))
y = fn(x)
self.assertEqual(y.shape.as_list(), [10, 1])
def test_save_spec(self):
"""Tests that functional model generates the correct save spec."""
class MultiInputModel(training_lib.Model):
def call(self, x, y):
return x
inp = input_layer_lib.Input(shape=(1,))
inp2 = input_layer_lib.Input(shape=(1,), batch_size=5, dtype=tf.int32)
out = MultiInputModel()(inp, inp2)
m = training_lib.Model(inputs={"x": inp, "y": inp2}, outputs=out)
input_spec = m.save_spec(dynamic_batch=False)[0][0]
self.assertIn("x", input_spec)
self.assertIn("y", input_spec)
self.assertAllEqual([None, 1], input_spec["x"].shape.as_list())
self.assertAllEqual(tf.float32, input_spec["x"].dtype)
self.assertAllEqual([5, 1], input_spec["y"].shape.as_list())
self.assertAllEqual(tf.int32, input_spec["y"].dtype)
def test_layer_ordering_checkpoint_compatibility(self):
class MLPKeras(layers.Layer):
def __init__(self, name: str) -> None:
super(MLPKeras, self).__init__(name=name)
self.layer_1 = layers.Dense(
10, activation="relu", name=f"{name}_dense_1"
)
self.layer_2 = layers.Dense(
10, activation="relu", name=f"{name}_dense_2"
)
def call(self, inputs: tf.Tensor) -> tf.Tensor:
return self.layer_2(self.layer_1(inputs))
mlp_keras_1 = MLPKeras("mlp_1")
mlp_keras_2 = MLPKeras("mlp_2")
inputs = input_layer_lib.Input((5,))
# Make model which is the sum of two MLPs.
outputs_1 = mlp_keras_1(inputs) + mlp_keras_2(inputs)
functional_model_1 = functional.Functional(
inputs=inputs, outputs=outputs_1
)
ckpt_1 = Checkpoint(model=functional_model_1)
filepath = tf.io.gfile.join(self.get_temp_dir(), "model_1_ckpt")
ckpt_path = ckpt_1.save(filepath)
# Swap order of MLPs.
outputs_2 = mlp_keras_2(inputs) + mlp_keras_1(inputs)
functional_model_2 = functional.Functional(
inputs=inputs, outputs=outputs_2
)
Checkpoint(model=functional_model_2).restore(
ckpt_path
).assert_consumed()
class DeferredModeTest(test_combinations.TestCase):
@test_combinations.generate(
test_combinations.combine(mode=["graph", "eager"])
)
def testSimpleNetworkBuilding(self):
inputs = input_layer_lib.Input(shape=(32,))
if tf.executing_eagerly():
self.assertEqual(inputs.dtype.name, "float32")
self.assertEqual(inputs.shape.as_list(), [None, 32])
x = layers.Dense(2)(inputs)
if tf.executing_eagerly():
self.assertEqual(x.dtype.name, "float32")
self.assertEqual(x.shape.as_list(), [None, 2])
outputs = layers.Dense(4)(x)
network = functional.Functional(inputs, outputs)
self.assertIsInstance(network, functional.Functional)
if tf.executing_eagerly():
# It should be possible to call such a network on EagerTensors.
inputs = tf.constant(np.random.random((10, 32)).astype("float32"))
outputs = network(inputs)
self.assertEqual(outputs.shape.as_list(), [10, 4])
@test_combinations.generate(
test_combinations.combine(mode=["graph", "eager"])
)
def testMultiIONetworkBuilding(self):
input_a = input_layer_lib.Input(shape=(32,))
input_b = input_layer_lib.Input(shape=(16,))
a = layers.Dense(16)(input_a)
class AddLayer(layers.Layer):
def call(self, inputs):
return inputs[0] + inputs[1]
c = AddLayer()([a, input_b])
c = layers.Dense(2)(c)
network = functional.Functional([input_a, input_b], [a, c])
if tf.executing_eagerly():
a_val = tf.constant(np.random.random((10, 32)).astype("float32"))
b_val = tf.constant(np.random.random((10, 16)).astype("float32"))
outputs = network([a_val, b_val])
self.assertEqual(len(outputs), 2)
self.assertEqual(outputs[0].shape.as_list(), [10, 16])
self.assertEqual(outputs[1].shape.as_list(), [10, 2])
class DefaultShapeInferenceBehaviorTest(test_combinations.TestCase):
def _testShapeInference(self, model, input_shape, expected_output_shape):
input_value = np.random.random(input_shape)
output_value = model.predict(input_value)
self.assertEqual(output_value.shape, expected_output_shape)
@test_combinations.generate(
test_combinations.combine(mode=["graph", "eager"])
)
def testSingleInputCase(self):
class LayerWithOneInput(layers.Layer):
def build(self, input_shape):
self.w = tf.ones(shape=(3, 4))
def call(self, inputs):
return backend.dot(inputs, self.w)
inputs = input_layer_lib.Input(shape=(3,))
layer = LayerWithOneInput()
if tf.executing_eagerly():
self.assertEqual(
layer.compute_output_shape((None, 3)).as_list(), [None, 4]
)
# As a side-effect, compute_output_shape builds the layer.
self.assertTrue(layer.built)
# We can still query the layer's compute_output_shape with
# compatible input shapes.
self.assertEqual(
layer.compute_output_shape((6, 3)).as_list(), [6, 4]
)
outputs = layer(inputs)
model = training_lib.Model(inputs, outputs)
self._testShapeInference(model, (2, 3), (2, 4))
@test_combinations.generate(
test_combinations.combine(mode=["graph", "eager"])
)
def testMultiInputOutputCase(self):
class MultiInputOutputLayer(layers.Layer):
def build(self, input_shape):
self.w = tf.ones(shape=(3, 4))
def call(self, inputs):
a = backend.dot(inputs[0], self.w)
b = a + inputs[1]
return [a, b]
input_a = input_layer_lib.Input(shape=(3,))
input_b = input_layer_lib.Input(shape=(4,))
output_a, output_b = MultiInputOutputLayer()([input_a, input_b])
model = training_lib.Model([input_a, input_b], [output_a, output_b])
output_a_val, output_b_val = model.predict(
[np.random.random((2, 3)), np.random.random((2, 4))]
)
self.assertEqual(output_a_val.shape, (2, 4))
self.assertEqual(output_b_val.shape, (2, 4))
@test_combinations.generate(
test_combinations.combine(mode=["graph", "eager"])
)
def testTrainingArgument(self):
class LayerWithTrainingArg(layers.Layer):
def build(self, input_shape):
self.w = tf.ones(shape=(3, 4))
def call(self, inputs, training):
return backend.dot(inputs, self.w)
inputs = input_layer_lib.Input(shape=(3,))
outputs = LayerWithTrainingArg()(inputs, training=False)
model = training_lib.Model(inputs, outputs)
self._testShapeInference(model, (2, 3), (2, 4))
@test_combinations.generate(
test_combinations.combine(mode=["graph", "eager"])
)
def testNoneInShape(self):
class Model(training_lib.Model):
def __init__(self):
super().__init__()
self.conv1 = layers.Conv2D(8, 3)
self.pool = layers.GlobalAveragePooling2D()
self.fc = layers.Dense(3)
def call(self, x):
x = self.conv1(x)
x = self.pool(x)
x = self.fc(x)
return x
model = Model()
model.build(tf.TensorShape((None, None, None, 1)))
self.assertTrue(model.built, "Model should be built")
self.assertTrue(
model.weights,
"Model should have its weights created as it has been built",
)
sample_input = tf.ones((1, 10, 10, 1))
output = model(sample_input)
self.assertEqual(output.shape, (1, 3))
@test_combinations.generate(
test_combinations.combine(mode=["graph", "eager"])
)
def testNoneInShapeWithCompoundModel(self):
class BasicBlock(training_lib.Model):
def __init__(self):
super().__init__()
self.conv1 = layers.Conv2D(8, 3)
self.pool = layers.GlobalAveragePooling2D()
self.dense = layers.Dense(3)
def call(self, x):
x = self.conv1(x)
x = self.pool(x)
x = self.dense(x)
return x
class CompoundModel(training_lib.Model):
def __init__(self):
super().__init__()
self.block = BasicBlock()
def call(self, x):
x = self.block(x)
return x
model = CompoundModel()
model.build(tf.TensorShape((None, None, None, 1)))
self.assertTrue(model.built, "Model should be built")
self.assertTrue(
model.weights,
"Model should have its weights created as it has been built",
)
sample_input = tf.ones((1, 10, 10, 1))
output = model(sample_input)
self.assertEqual(output.shape, (1, 3))
@test_combinations.generate(
test_combinations.combine(mode=["graph", "eager"])
)
def testNoneInShapeWithFunctionalAPI(self):
class BasicBlock(training_lib.Model):
# Inheriting from layers.Layer since we are calling this layer
# inside a model created using functional API.
def __init__(self):
super().__init__()
self.conv1 = layers.Conv2D(8, 3)
def call(self, x):
x = self.conv1(x)
return x
input_layer = layers.Input(shape=(None, None, 1))
x = BasicBlock()(input_layer)
x = layers.GlobalAveragePooling2D()(x)
output_layer = layers.Dense(3)(x)
model = training_lib.Model(inputs=input_layer, outputs=output_layer)
model.build(tf.TensorShape((None, None, None, 1)))
self.assertTrue(model.built, "Model should be built")
self.assertTrue(
model.weights,
"Model should have its weights created as it has been built",
)
sample_input = tf.ones((1, 10, 10, 1))
output = model(sample_input)
self.assertEqual(output.shape, (1, 3))
@test_combinations.generate(test_combinations.keras_mode_combinations())
def test_sequential_as_downstream_of_masking_layer(self):
inputs = layers.Input(shape=(3, 4))
x = layers.Masking(mask_value=0.0, input_shape=(3, 4))(inputs)
s = sequential.Sequential()
s.add(layers.Dense(5, input_shape=(4,)))
x = layers.TimeDistributed(s)(x)
model = training_lib.Model(inputs=inputs, outputs=x)
model.compile(
optimizer="rmsprop",
loss="mse",
run_eagerly=test_utils.should_run_eagerly(),
)
model_input = np.random.randint(low=1, high=5, size=(10, 3, 4)).astype(
"float32"
)
for i in range(4):
model_input[i, i:, :] = 0.0
model.fit(
model_input, np.random.random((10, 3, 5)), epochs=1, batch_size=6
)
if not tf.executing_eagerly():
# Note: this doesn't work in eager due to DeferredTensor/ops
# compatibility issue.
mask_outputs = [model.layers[1].compute_mask(model.layers[1].input)]
mask_outputs += [
model.layers[2].compute_mask(
model.layers[2].input, mask_outputs[-1]
)
]
func = backend.function([model.input], mask_outputs)
mask_outputs_val = func([model_input])
self.assertAllClose(
mask_outputs_val[0], np.any(model_input, axis=-1)
)
self.assertAllClose(
mask_outputs_val[1], np.any(model_input, axis=-1)
)
@test_combinations.generate(
test_combinations.combine(mode=["graph", "eager"])
)
def test_external_keras_serialization_compat_input_layers(self):
inputs = input_layer_lib.Input(shape=(10,))
outputs = layers.Dense(1)(inputs)
model = training_lib.Model(inputs, outputs)
config = model.get_config()
# Checks that single inputs and outputs are still saved as 1-element
# lists. Saving as 1-element lists or not is equivalent in TF Keras,
# but only the 1-element list format is supported in TF.js and
# keras-team/Keras.
self.assertLen(config["input_layers"], 1)
self.assertLen(config["output_layers"], 1)
@test_combinations.generate(
test_combinations.combine(mode=["graph", "eager"])
)
@test_utils.run_v2_only
def test_save_load_with_single_elem_list_inputs_saved_model(self):
class MyLayer(layers.Layer):
def __init__(self):
super().__init__()
self._preserve_input_structure_in_config = True
def call(self, inputs):
return inputs[0]
inputs = input_layer_lib.Input(shape=(3,))
layer = MyLayer()
outputs = layer([inputs])
model = training_lib.Model(inputs=inputs, outputs=outputs)
model.save("/tmp/km2")
save.load_model("/tmp/km2")
@test_utils.run_v2_only
def test_save_load_with_single_elem_list_inputs_keras_v3(self):
@object_registration.register_keras_serializable()
class MyLayer(layers.Layer):
def __init__(self):
super().__init__()
self._preserve_input_structure_in_config = True
def call(self, inputs):
return inputs[0]
inputs = input_layer_lib.Input(shape=(3,))
layer = MyLayer()
outputs = layer([inputs])
model = training_lib.Model(inputs=inputs, outputs=outputs)
model.save("/tmp/model.keras")
models.load_model("/tmp/model.keras")
@test_combinations.generate(
test_combinations.combine(mode=["graph", "eager"])
)
def test_external_keras_serialization_compat_inbound_nodes(self):
# Check single Tensor input.
inputs = input_layer_lib.Input(shape=(10,), name="in")
outputs = layers.Dense(1)(inputs)
model = training_lib.Model(inputs, outputs)
config = model.get_config()
self.assertEqual(
config["layers"][1]["inbound_nodes"], [[["in", 0, 0, {}]]]
)
# Check multiple Tensor input.
inputs1 = input_layer_lib.Input(shape=(10,), name="in1")
inputs2 = input_layer_lib.Input(shape=(10,), name="in2")
outputs = layers.Add()([inputs1, inputs2])
model = training_lib.Model([inputs1, inputs2], outputs)
config = model.get_config()
self.assertEqual(
config["layers"][2]["inbound_nodes"],
[[["in1", 0, 0, {}], ["in2", 0, 0, {}]]],
)
@test_combinations.generate(test_combinations.combine(mode=["eager"]))
def test_dict_inputs_tensors(self):
# Note that this test is running with v2 eager only, since the v1
# will behave differently wrt to dict input for training.
inputs = {
"sentence2": input_layer_lib.Input(
shape=(), name="a", dtype=tf.string
),
"sentence1": input_layer_lib.Input(
shape=(), name="b", dtype=tf.string
),
}
strlen = layers.Lambda(tf.strings.length)
diff = layers.Subtract()(
[strlen(inputs["sentence1"]), strlen(inputs["sentence2"])]
)
diff = tf.cast(diff, tf.float32)
model = training_lib.Model(inputs, diff)
extra_keys = {
"sentence1": tf.constant(["brown fox", "lazy dog"]),
"sentence2": tf.constant(["owl", "cheeky cat"]),
"label": tf.constant([0, 1]),
}
with warnings.catch_warnings(record=True) as w:
warnings.simplefilter("always")
model(extra_keys)
self.assertIn("ignored by the model", str(w[-1].message))
model.compile("sgd", "mse")
with warnings.catch_warnings(record=True) as w:
warnings.simplefilter("always")
model.fit(extra_keys, y=tf.constant([0, 1]), steps_per_epoch=1)
self.assertIn("ignored by the model", str(w[-1].message))
with warnings.catch_warnings(record=True) as w:
warnings.simplefilter("always")
model.evaluate(extra_keys, tf.constant([0, 1]))
self.assertIn("ignored by the model", str(w[-1].message))
# Make sure the model inputs are sorted with the dict keys.
self.assertEqual(model.inputs[0]._keras_history.layer.name, "b")
self.assertEqual(model.inputs[1]._keras_history.layer.name, "a")
class GraphUtilsTest(tf.test.TestCase):
def testGetReachableFromInputs(self):
with tf.Graph().as_default(), self.cached_session():
pl_1 = tf.compat.v1.placeholder(shape=None, dtype="float32")
pl_2 = tf.compat.v1.placeholder(shape=None, dtype="float32")
pl_3 = tf.compat.v1.placeholder(shape=None, dtype="float32")
x_1 = pl_1 + pl_2
x_2 = pl_2 * 2
x_3 = pl_3 + 1
x_4 = x_1 + x_2
x_5 = x_3 * pl_1
self.assertEqual(
tf_utils.get_reachable_from_inputs([pl_1]),
{pl_1, x_1, x_4, x_5, x_1.op, x_4.op, x_5.op},
)
self.assertEqual(
tf_utils.get_reachable_from_inputs([pl_1, pl_2]),
{
pl_1,
pl_2,
x_1,
x_2,
x_4,
x_5,
x_1.op,
x_2.op,
x_4.op,
x_5.op,
},
)
self.assertEqual(
tf_utils.get_reachable_from_inputs([pl_3]),
{pl_3, x_3, x_5, x_3.op, x_5.op},
)
self.assertEqual(
tf_utils.get_reachable_from_inputs([x_3]), {x_3, x_5, x_5.op}
)
class NestedNetworkTest(test_combinations.TestCase):
@test_combinations.generate(
test_combinations.combine(mode=["graph", "eager"])
)
def test_nested_inputs_network(self):
inputs = {
"x1": input_layer_lib.Input(shape=(1,)),
"x2": input_layer_lib.Input(shape=(1,)),
}
outputs = layers.Add()([inputs["x1"], inputs["x2"]])
network = functional.Functional(inputs, outputs)
network = functional.Functional.from_config(network.get_config())
result_tensor = network(
{"x1": tf.ones((1, 1), "float32"), "x2": tf.ones((1, 1), "float32")}
)
result = self.evaluate(result_tensor)
self.assertAllEqual(result, [[2.0]])
# TODO(b/122726584): Investigate why concrete batch is flaky in some
# builds.
output_shape = network.compute_output_shape(
{"x1": (None, 1), "x2": (None, 1)}
)
self.assertListEqual(output_shape.as_list(), [None, 1])
@test_combinations.generate(
test_combinations.combine(mode=["graph", "eager"])
)
def test_nested_outputs_network(self):
inputs = input_layer_lib.Input(shape=(1,))
outputs = {
"x+x": layers.Add()([inputs, inputs]),
"x*x": layers.Multiply()([inputs, inputs]),
}
network = functional.Functional(inputs, outputs)
network = functional.Functional.from_config(network.get_config())
result_tensor = network(tf.ones((1, 1), "float32"))
result = self.evaluate(result_tensor)
self.assertAllEqual(result["x+x"], [[2.0]])
self.assertAllEqual(result["x*x"], [[1.0]])
output_shape = network.compute_output_shape((None, 1))
self.assertListEqual(output_shape["x+x"].as_list(), [None, 1])
self.assertListEqual(output_shape["x*x"].as_list(), [None, 1])
@test_combinations.generate(
test_combinations.combine(mode=["graph", "eager"])
)
def test_nested_network_inside_network(self):
inner_inputs = {
"x1": input_layer_lib.Input(shape=(1,)),
"x2": input_layer_lib.Input(shape=(1,)),
}
inner_outputs = {
"x1+x2": layers.Add()([inner_inputs["x1"], inner_inputs["x2"]]),
"x1*x2": layers.Multiply()(
[inner_inputs["x1"], inner_inputs["x2"]]
),
}
inner_network = functional.Functional(inner_inputs, inner_outputs)
inputs = [
input_layer_lib.Input(shape=(1,)),
input_layer_lib.Input(shape=(1,)),
]
middle = inner_network({"x1": inputs[0], "x2": inputs[1]})
outputs = layers.Add()([middle["x1+x2"], middle["x1*x2"]])
network = functional.Functional(inputs, outputs)
network = functional.Functional.from_config(network.get_config())
# Computes: `(x1+x2) + (x1*x2)`
result_tensor = network(
[tf.ones((1, 1), "float32"), tf.ones((1, 1), "float32")]
)
result = self.evaluate(result_tensor)
self.assertAllEqual(result, [[3.0]])
output_shape = network.compute_output_shape([(None, 1), (None, 1)])
self.assertListEqual(output_shape.as_list(), [None, 1])
@test_combinations.generate(test_combinations.combine(mode=["graph"]))
def test_updates_with_direct_call(self):
inputs = input_layer_lib.Input(shape=(10,))
x = layers.BatchNormalization()(inputs)
x = layers.Dense(10)(x)
model = training_lib.Model(inputs, x)
ph = backend.placeholder(shape=(10, 10))
model(ph)
self.assertLen(model.updates, 4)
@test_combinations.generate(
test_combinations.combine(mode=["graph", "eager"])
)
def test_dict_mapping_input(self):
class ReturnFirst(layers.Layer):
def call(self, inputs):
b, _ = inputs
return b
# Checks that inputs are put in same order as the
# Model was constructed with.
b = input_layer_lib.Input(shape=(10,), name="b")
a = input_layer_lib.Input(shape=(10,), name="a")
outputs = ReturnFirst()([b, a])
b_val = tf.ones((10, 10))
a_val = tf.zeros((10, 10))
model = training_lib.Model([b, a], outputs)
res = model({"a": a_val, "b": b_val})
self.assertAllClose(self.evaluate(res), self.evaluate(b_val))
reversed_model = training_lib.Model([a, b], outputs)
res = reversed_model({"a": a_val, "b": b_val})
self.assertAllClose(self.evaluate(res), self.evaluate(b_val))
@test_combinations.generate(
test_combinations.combine(mode=["graph", "eager"])
)
def test_dict_mapping_single_input(self):
b = input_layer_lib.Input(shape=(1,), name="b")
outputs = b * 2
model = training_lib.Model(b, outputs)
b_val = tf.ones((1, 1))
extra_val = tf.ones((1, 10))
inputs = {"a": extra_val, "b": b_val}
res = model(inputs)
# Check that 'b' was used and 'a' was ignored.
self.assertEqual(res.shape.as_list(), [1, 1])
@test_combinations.generate(
test_combinations.combine(mode=["graph", "eager"])
)
def test_nested_dict_mapping(self):
a = input_layer_lib.Input(shape=(1,), dtype="int32", name="a")
b = input_layer_lib.Input(shape=(1,), dtype="int32", name="b")
c = input_layer_lib.Input(shape=(1,), dtype="int32", name="c")
d = input_layer_lib.Input(shape=(1,), dtype="int32", name="d")
inputs = {"a": (a, b), "c": (c, d)}
outputs = 1000 * a + 100 * b + 10 * c + d
model = training_lib.Model(inputs, outputs)
a_val = tf.ones((1, 1), dtype="int32")
b_val = 2 * tf.ones((1, 1), dtype="int32")
c_val = 3 * tf.ones((1, 1), dtype="int32")
d_val = 4 * tf.ones((1, 1), dtype="int32")
inputs_val = {"a": (a_val, b_val), "c": (c_val, d_val)}
res = model(inputs_val)
# Check that inputs were flattened in the correct order.
self.assertFalse(model._enable_dict_to_input_mapping)
self.assertEqual(self.evaluate(res), [1234])
@test_combinations.generate(test_combinations.keras_mode_combinations())
class AddLossTest(test_combinations.TestCase):
def test_add_loss_outside_call_only_loss(self):
inputs = input_layer_lib.Input((10,))
mid = layers.Dense(10)(inputs)
outputs = layers.Dense(1)(mid)
model = training_lib.Model(inputs, outputs)
model.add_loss(tf.reduce_mean(outputs))
self.assertLen(model.losses, 1)
initial_weights = model.get_weights()
x = np.ones((10, 10))
model.compile("sgd", run_eagerly=test_utils.should_run_eagerly())
model.fit(x, batch_size=2, epochs=1)
model2 = model.from_config(model.get_config())
model2.compile("sgd", run_eagerly=test_utils.should_run_eagerly())
model2.set_weights(initial_weights)
model2.fit(x, batch_size=2, epochs=1)
# The TFOpLayer and the AddLoss layer are serialized.
self.assertLen(model2.layers, 5)
self.assertAllClose(model.get_weights(), model2.get_weights())
def test_add_loss_outside_call_multiple_losses(self):
inputs = input_layer_lib.Input((10,))
x1 = layers.Dense(10)(inputs)
x2 = layers.Dense(10)(x1)
outputs = layers.Dense(1)(x2)
model = training_lib.Model(inputs, outputs)
model.add_loss(tf.reduce_sum(x1 * x2))
model.add_loss(tf.reduce_mean(outputs))
self.assertLen(model.losses, 2)
initial_weights = model.get_weights()
x, y = np.ones((10, 10)), np.ones((10, 1))
model.compile("sgd", "mse", run_eagerly=test_utils.should_run_eagerly())
model.fit(x, y, batch_size=2, epochs=1)
model2 = model.from_config(model.get_config())
model2.compile(
"sgd", "mse", run_eagerly=test_utils.should_run_eagerly()
)
model2.set_weights(initial_weights)
model2.fit(x, y, batch_size=2, epochs=1)
self.assertAllClose(model.get_weights(), model2.get_weights())
def test_add_loss_crossentropy_backtracking(self):
inputs = input_layer_lib.Input((2,))
labels = input_layer_lib.Input((1,))
outputs = layers.Dense(1, activation="sigmoid")(inputs)
model = functional.Functional([inputs, labels], outputs)
model.add_loss(losses.binary_crossentropy(labels, outputs))
model.compile("adam")
x = np.random.random((2, 2))
y = np.random.random((2, 1))
model.fit([x, y])
inputs = input_layer_lib.Input((2,))
labels = input_layer_lib.Input((2,))
outputs = layers.Dense(2, activation="softmax")(inputs)
model = functional.Functional([inputs, labels], outputs)
model.add_loss(losses.categorical_crossentropy(labels, outputs))
model.compile("adam")
x = np.random.random((2, 2))
y = np.random.random((2, 2))
model.fit([x, y])
inputs = input_layer_lib.Input((2,))
labels = input_layer_lib.Input((1,), dtype="int32")
outputs = layers.Dense(2, activation="softmax")(inputs)
model = functional.Functional([inputs, labels], outputs)
model.add_loss(losses.sparse_categorical_crossentropy(labels, outputs))
model.compile("adam")
x = np.random.random((2, 2))
y = np.random.randint(0, 2, size=(2, 1))
model.fit([x, y])
@test_combinations.generate(test_combinations.keras_mode_combinations())
class WeightAccessTest(test_combinations.TestCase):
def test_functional_model(self):
inputs = input_layer_lib.Input((10,))
x1 = layers.Dense(10)(inputs)
x2 = layers.Dense(10)(x1)
outputs = layers.Dense(1)(x2)
model = training_lib.Model(inputs, outputs)
self.assertEqual(len(model.weights), 6)
def test_sequential_model_with_input_shape(self):
x1 = layers.Dense(10, input_shape=(10,))
x2 = layers.Dense(10)
x3 = layers.Dense(1)
model = sequential.Sequential([x1, x2, x3])
self.assertEqual(len(model.weights), 6)
def test_sequential_model_without_input_shape(self):
x1 = layers.Dense(10)
x2 = layers.Dense(10)
x3 = layers.Dense(1)
model = sequential.Sequential([x1, x2, x3])
with self.assertRaisesRegex(
ValueError, "Weights for model .* have not yet been created"
):
_ = model.weights
def test_subclass_model_with_build_method(self):
class SubclassModel(models.Model):
def build(self, input_shape):
self.w = self.add_weight(
shape=input_shape[-1], initializer="ones"
)
def call(self, inputs):
return inputs * self.w
model = SubclassModel()
with self.assertRaisesRegex(
ValueError, "Weights for model .* have not yet been created"
):
_ = model.weights
model(input_layer_lib.Input((10,)))
self.assertEqual(len(model.weights), 1)
def test_subclass_model_without_build_method(self):
class SubclassModel(models.Model):
def __init__(self):
super().__init__()
self.w = self.add_weight(shape=(), initializer="ones")
def call(self, inputs):
return inputs * self.w
model = SubclassModel()
self.assertEqual(len(model.weights), 1)
@test_combinations.generate(test_combinations.combine(mode=["graph", "eager"]))
class DTypeTest(test_combinations.TestCase):
@test_utils.enable_v2_dtype_behavior
def test_graph_network_dtype(self):
inputs = input_layer_lib.Input((10,))
outputs = layers.Dense(10)(inputs)
network = functional.Functional(inputs, outputs)
self.assertEqual(network.dtype, "float32")
@test_utils.enable_v2_dtype_behavior
def test_subclassed_network_dtype(self):
class IdentityNetwork(training_lib.Model):
def call(self, inputs):
return inputs
network = IdentityNetwork()
self.assertEqual(network.dtype, "float32")
self.assertEqual(network(tf.constant(1, "float64")).dtype, "float32")
network = IdentityNetwork(dtype="float16")
self.assertEqual(network.dtype, "float16")
self.assertEqual(network(tf.constant(1, "float64")).dtype, "float16")
network = IdentityNetwork(autocast=False)
self.assertEqual(network.dtype, "float32")
self.assertEqual(network(tf.constant(1, "float64")).dtype, "float64")
class AttrTrackingLayer(base_layer.Layer):
"""Count how many times `dynamic` and `stateful` are called.
These counts are used to test that the attribute cache behaves as expected.
"""
def __init__(self, *args, **kwargs):
self.stateful_count = 0
self.dynamic_count = 0
super().__init__(*args, **kwargs)
@base_layer.Layer.stateful.getter
def stateful(self):
self.stateful_count += 1
return super().stateful
@property
def dynamic(self):
self.dynamic_count += 1
return super().dynamic
@test_combinations.generate(test_combinations.combine(mode=["graph", "eager"]))
class CacheCorrectnessTest(test_combinations.TestCase):
def layer_and_network_test(self):
# Top level layer
network = functional.Functional()
layer_0 = AttrTrackingLayer()
sub_network = functional.Functional()
layer_1 = AttrTrackingLayer(dynamic=True)
layer_2 = AttrTrackingLayer()
sub_network.sub_layers = [layer_1, layer_2]
network.sub_layer = layer_0
for _ in range(2):
self.assertEqual(network.dynamic, False)
self.assertEqual(network.stateful, False)
# The second pass should be a cache hit.
self.assertEqual(layer_0.dynamic_count, 1)
self.assertEqual(layer_0.stateful_count, 1)
# Mutations of the sub-layer should force recalculation of the network's
# stateful attribute. (mutations bubble up.)
layer_0.stateful = True
self.assertEqual(network.stateful, True)
self.assertEqual(layer_0.stateful_count, 2)
layer_0.stateful = False
self.assertEqual(network.stateful, False)
self.assertEqual(layer_0.stateful_count, 3)
# But changing stateful should not affect dynamic.
self.assertEqual(network.dynamic, False)
self.assertEqual(layer_0.dynamic_count, 1)
network.sub_network = sub_network
# Adding to the topology should invalidate the cache and reflect in the
# top level network.
self.assertEqual(network.dynamic, True)
self.assertEqual(layer_0.dynamic_count, 2)
self.assertEqual(layer_1.dynamic_count, 1)
# Still dynamic, but we need to recompute.
sub_network.sub_layers.pop()
self.assertEqual(network.dynamic, True)
self.assertEqual(layer_0.dynamic_count, 3)
self.assertEqual(layer_1.dynamic_count, 2)
# Now that we've removed the dynamic layer deep in the layer hierarchy,
# we need to make sure that that bubbles up through all the levels.
sub_network.sub_layers.pop()
self.assertEqual(network.dynamic, False)
self.assertEqual(layer_0.dynamic_count, 4)
self.assertEqual(layer_1.dynamic_count, 2)
# Now check with a tracked dict.
sub_network.sub_layers = {
"layer_1": layer_1,
"layer_2": layer_2,
}
self.assertEqual(network.dynamic, True)
self.assertEqual(layer_0.dynamic_count, 5)
self.assertEqual(layer_1.dynamic_count, 3)
# In-place assignment should still invalidate the cache.
sub_network.sub_layers["layer_1"] = layer_1
self.assertEqual(network.dynamic, True)
self.assertEqual(layer_0.dynamic_count, 6)
self.assertEqual(layer_1.dynamic_count, 4)
sub_network.sub_layers["layer_1"] = None
for _ in range(2):
self.assertEqual(network.dynamic, False)
self.assertEqual(layer_0.dynamic_count, 7)
self.assertEqual(layer_1.dynamic_count, 4)
layer_3 = AttrTrackingLayer()
layer_3.stateful = True
sub_network.sub_layers = None
self.assertEqual(network.dynamic, False)
self.assertEqual(network.stateful, False)
# Test duplicate layers.
sub_network.sub_layers = [layer_1, layer_1, layer_1, layer_3]
self.assertEqual(network.dynamic, True)
self.assertEqual(network.stateful, True)
for _ in range(3):
sub_network.sub_layers.pop()
self.assertEqual(network.dynamic, True)
self.assertEqual(network.stateful, False)
sub_network.sub_layers.pop()
self.assertEqual(network.dynamic, False)
self.assertEqual(network.stateful, False)
def test_compute_output_shape_cache(self):
# See https://github.com/tensorflow/tensorflow/issues/32029.
x = input_layer_lib.Input(shape=(None, 32))
dense = layers.Dense(2)
y = dense(x)
network = functional.Functional(x, y, name="dense_network")
for i in range(999, 1024):
self.assertEqual(
network.compute_output_shape((1, i, 32)), (1, i, 2)
)
def test_2d_inputs_squeezed_to_1d(self):
input_1d = input_layer_lib.Input(shape=())
outputs = input_1d * 2.0
net = functional.Functional(input_1d, outputs)
x = np.ones((10, 1))
y = net(x)
self.assertEqual(y.shape.rank, 1)
def test_1d_inputs_expanded_to_2d(self):
input_1d = input_layer_lib.Input(shape=(1,))
outputs = input_1d * 2.0
net = functional.Functional(input_1d, outputs)
x = np.ones((10,))
y = net(x)
self.assertEqual(y.shape.rank, 2)
def test_training_passed_during_construction(self):
def _call(inputs, training):
if training is None:
return inputs * -1.0
elif training:
return inputs
else:
return inputs * 0.0
class MyLayer(base_layer.Layer):
def call(self, inputs, training=True):
return _call(inputs, training)
my_layer = MyLayer()
x = np.ones((1, 10))
# Hard-coded `true` value passed during construction is respected.
inputs = input_layer_lib.Input(10)
outputs = my_layer(inputs, training=True)
network = functional.Functional(inputs, outputs)
self.assertAllEqual(network(x, training=True), _call(x, True))
self.assertAllEqual(network(x, training=False), _call(x, True))
self.assertAllEqual(network(x), _call(x, True))
# Hard-coded `false` value passed during construction is respected.
inputs = input_layer_lib.Input(10)
outputs = my_layer(inputs, training=False)
network = functional.Functional(inputs, outputs)
self.assertAllEqual(network(x, training=True), _call(x, False))
self.assertAllEqual(network(x, training=False), _call(x, False))
self.assertAllEqual(network(x), _call(x, False))
if tf.executing_eagerly():
# In v2, construction still works when no `training` is specified
# When no value passed during construction, it uses the local
# default.
inputs = input_layer_lib.Input(10)
outputs = my_layer(inputs)
network = functional.Functional(inputs, outputs)
self.assertAllEqual(network(x, training=True), _call(x, True))
self.assertAllEqual(network(x, training=False), _call(x, False))
self.assertAllEqual(network(x), _call(x, True)) # Use local default
# `None` value passed positionally during construction is ignored at
# runtime
inputs = input_layer_lib.Input(10)
outputs = my_layer(inputs, None)
network = functional.Functional(inputs, outputs)
self.assertAllEqual(network(x, training=True), _call(x, True))
self.assertAllEqual(network(x, training=False), _call(x, False))
if tf.executing_eagerly():
self.assertAllEqual(network(x), _call(x, True)) # Use local default
else:
# in v1 training would have defaulted to using the `None` inside the
# layer if training is not passed at runtime
self.assertAllEqual(network(x), _call(x, None))
# `None` value passed as kwarg during construction is ignored at
# runtime.
inputs = input_layer_lib.Input(10)
outputs = my_layer(inputs, training=None)
network = functional.Functional(inputs, outputs)
self.assertAllEqual(network(x, training=True), _call(x, True))
self.assertAllEqual(network(x, training=False), _call(x, False))
if tf.executing_eagerly():
self.assertAllEqual(network(x), _call(x, True)) # Use local default
else:
# in v1 training would have defaulted to using the `None` inside the
# layer if training is not passed at runtime
self.assertAllEqual(network(x), _call(x, None))
class InputsOutputsErrorTest(test_combinations.TestCase):
@test_utils.enable_v2_dtype_behavior
def test_input_error(self):
inputs = input_layer_lib.Input((10,))
outputs = layers.Dense(10)(inputs)
with self.assertRaisesRegex(
TypeError, "('Keyword argument not understood:', 'input')"
):
models.Model(input=inputs, outputs=outputs)
@test_utils.enable_v2_dtype_behavior
def test_output_error(self):
inputs = input_layer_lib.Input((10,))
outputs = layers.Dense(10)(inputs)
with self.assertRaisesRegex(
TypeError, "('Keyword argument not understood:', 'output')"
):
models.Model(inputs=inputs, output=outputs)
def test_input_spec(self):
if not tf.executing_eagerly():
return
inputs = input_layer_lib.Input((10,))
outputs = layers.Dense(10)(inputs)
model = models.Model(inputs, outputs)
with self.assertRaisesRegex(ValueError, r".*expected shape=.*"):
model(np.zeros((3, 11)))
def test_input_spec_list_of_inputs(self):
if not tf.executing_eagerly():
return
input_1 = input_layer_lib.Input((10,), name="1")
input_2 = input_layer_lib.Input((5,), name="2")
x = layers.Concatenate()([input_1, input_2])
outputs = layers.Dense(10)(x)
model = models.Model([input_1, input_2], outputs)
with self.assertRaisesRegex(ValueError, r".*expects 2 input.*"):
model(np.zeros((3, 10)))
with self.assertRaisesRegex(ValueError, r".*expects 2 input.*"):
model([np.zeros((3, 10)), np.zeros((3, 5)), np.zeros((3, 10))])
with self.assertRaisesRegex(ValueError, r".*expected shape=.*"):
model([np.zeros((3, 10)), np.zeros((3, 6))])
# Test passing data via dict keyed by input name
with self.assertRaisesRegex(ValueError, r"Missing data for input.*"):
model({"1": np.zeros((3, 10))})
with self.assertRaisesRegex(ValueError, r".*expected shape=.*"):
model({"1": np.zeros((3, 10)), "2": np.zeros((3, 6))})
def test_input_spec_dict(self):
if not tf.executing_eagerly():
return
input_1 = input_layer_lib.Input((10,))
input_2 = input_layer_lib.Input((5,))
x = layers.Concatenate()([input_1, input_2])
outputs = layers.Dense(10)(x)
model = models.Model({"1": input_1, "2": input_2}, outputs)
with self.assertRaisesRegex(ValueError, r"Missing data for input.*"):
model({"1": np.zeros((3, 10))})
with self.assertRaisesRegex(ValueError, r".*expected shape=.*"):
model({"1": np.zeros((3, 10)), "2": np.zeros((3, 6))})
class FunctionalSubclassModel(training_lib.Model):
def __init__(self, *args, **kwargs):
self.foo = {"foo": "bar"} # Make sure users can assign dict attributes
my_input = input_layer_lib.Input(shape=(16,))
dense = layers.Dense(32, activation="relu")
output = dense(my_input)
outputs = {"output": output}
super().__init__(inputs=[my_input], outputs=outputs, *args, **kwargs)
class MixinClass:
def __init__(self, foo, **kwargs):
self._foo = foo
super().__init__(**kwargs)
def get_foo(self):
return self._foo
class SubclassedModel(training_lib.Model):
def __init__(self, bar, **kwargs):
self._bar = bar
super().__init__(**kwargs)
def get_bar(self):
return self._bar
class MultipleInheritanceModelTest(test_combinations.TestCase):
def testFunctionalSubclass(self):
m = FunctionalSubclassModel()
# Some smoke test for the weights and output shape of the model
self.assertLen(m.weights, 2)
self.assertEqual(m.outputs[0].shape.as_list(), [None, 32])
def testFunctionalSubclassPreMixin(self):
class MixedFunctionalSubclassModel(MixinClass, FunctionalSubclassModel):
pass
m = MixedFunctionalSubclassModel(foo="123")
self.assertTrue(m._is_graph_network)
self.assertLen(m.weights, 2)
self.assertEqual(m.outputs[0].shape.as_list(), [None, 32])
self.assertEqual(m.get_foo(), "123")
def testFunctionalSubclassPostMixin(self):
# Make sure the the mixin class is also init correct when the order
# changed.
class MixedFunctionalSubclassModel(FunctionalSubclassModel, MixinClass):
pass
m = MixedFunctionalSubclassModel(foo="123")
self.assertTrue(m._is_graph_network)
self.assertLen(m.weights, 2)
self.assertEqual(m.outputs[0].shape.as_list(), [None, 32])
self.assertEqual(m.get_foo(), "123")
def testSubclassModelPreMixin(self):
class MixedSubclassModel(MixinClass, SubclassedModel):
pass
m = MixedSubclassModel(foo="123", bar="456")
self.assertFalse(m._is_graph_network)
self.assertEqual(m.get_foo(), "123")
self.assertEqual(m.get_bar(), "456")
if __name__ == "__main__":
tf.test.main()
|
tf-keras/tf_keras/engine/functional_test.py/0
|
{
"file_path": "tf-keras/tf_keras/engine/functional_test.py",
"repo_id": "tf-keras",
"token_count": 50491
}
| 192 |
# Copyright 2015 The TensorFlow Authors. All Rights Reserved.
#
# Licensed under the Apache License, Version 2.0 (the "License");
# you may not use this file except in compliance with the License.
# You may obtain a copy of the License at
#
# http://www.apache.org/licenses/LICENSE-2.0
#
# Unless required by applicable law or agreed to in writing, software
# distributed under the License is distributed on an "AS IS" BASIS,
# WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.
# See the License for the specific language governing permissions and
# limitations under the License.
# ==============================================================================
"""Training-related part of the TF-Keras engine."""
import copy
import itertools
import json
import warnings
import weakref
import numpy as np
import tensorflow.compat.v2 as tf
from tensorflow.python.distribute import distribute_utils
from tensorflow.python.distribute import input_ops
from tensorflow.python.eager import context
from tensorflow.python.platform import tf_logging as logging
from tensorflow.python.util.tf_export import keras_export
from tensorflow.tools.docs import doc_controls
from tf_keras import backend
from tf_keras import callbacks as callbacks_module
from tf_keras import optimizers
from tf_keras.dtensor import dtensor_api
from tf_keras.dtensor import layout_map as layout_map_lib
from tf_keras.engine import base_layer
from tf_keras.engine import base_layer_utils
from tf_keras.engine import compile_utils
from tf_keras.engine import data_adapter
from tf_keras.engine import input_layer as input_layer_module
from tf_keras.engine import training_utils
from tf_keras.metrics import base_metric
from tf_keras.mixed_precision import loss_scale_optimizer as lso
from tf_keras.optimizers import optimizer
from tf_keras.optimizers import optimizer_v1
from tf_keras.saving import pickle_utils
from tf_keras.saving import saving_api
from tf_keras.saving import saving_lib
from tf_keras.saving import serialization_lib
from tf_keras.saving.legacy import serialization
from tf_keras.saving.legacy.saved_model import json_utils
from tf_keras.saving.legacy.saved_model import model_serialization
from tf_keras.utils import generic_utils
from tf_keras.utils import io_utils
from tf_keras.utils import layer_utils
from tf_keras.utils import steps_per_execution_tuning
from tf_keras.utils import tf_inspect
from tf_keras.utils import tf_utils
from tf_keras.utils import traceback_utils
from tf_keras.utils import version_utils
from tf_keras.utils.mode_keys import ModeKeys
try:
import h5py
except ImportError:
h5py = None
@keras_export("keras.Model", "keras.models.Model")
class Model(base_layer.Layer, version_utils.ModelVersionSelector):
"""A model grouping layers into an object with training/inference features.
Args:
inputs: The input(s) of the model: a `keras.Input` object or a
combination of `keras.Input` objects in a dict, list or tuple.
outputs: The output(s) of the model: a tensor that originated from
`keras.Input` objects or a combination of such tensors in a dict,
list or tuple. See Functional API example below.
name: String, the name of the model.
There are two ways to instantiate a `Model`:
1 - With the "Functional API", where you start from `Input`,
you chain layer calls to specify the model's forward pass,
and finally you create your model from inputs and outputs:
```python
import tensorflow as tf
inputs = tf.keras.Input(shape=(3,))
x = tf.keras.layers.Dense(4, activation=tf.nn.relu)(inputs)
outputs = tf.keras.layers.Dense(5, activation=tf.nn.softmax)(x)
model = tf.keras.Model(inputs=inputs, outputs=outputs)
```
Note: Only dicts, lists, and tuples of input tensors are supported. Nested
inputs are not supported (e.g. lists of list or dicts of dict).
A new Functional API model can also be created by using the
intermediate tensors. This enables you to quickly extract sub-components
of the model.
Example:
```python
inputs = keras.Input(shape=(None, None, 3))
processed = keras.layers.RandomCrop(width=32, height=32)(inputs)
conv = keras.layers.Conv2D(filters=2, kernel_size=3)(processed)
pooling = keras.layers.GlobalAveragePooling2D()(conv)
feature = keras.layers.Dense(10)(pooling)
full_model = keras.Model(inputs, feature)
backbone = keras.Model(processed, conv)
activations = keras.Model(conv, feature)
```
Note that the `backbone` and `activations` models are not
created with `keras.Input` objects, but with the tensors that are originated
from `keras.Input` objects. Under the hood, the layers and weights will
be shared across these models, so that user can train the `full_model`, and
use `backbone` or `activations` to do feature extraction.
The inputs and outputs of the model can be nested structures of tensors as
well, and the created models are standard Functional API models that support
all the existing APIs.
2 - By subclassing the `Model` class: in that case, you should define your
layers in `__init__()` and you should implement the model's forward pass
in `call()`.
```python
import tensorflow as tf
class MyModel(tf.keras.Model):
def __init__(self):
super().__init__()
self.dense1 = tf.keras.layers.Dense(4, activation=tf.nn.relu)
self.dense2 = tf.keras.layers.Dense(5, activation=tf.nn.softmax)
def call(self, inputs):
x = self.dense1(inputs)
return self.dense2(x)
model = MyModel()
```
If you subclass `Model`, you can optionally have
a `training` argument (boolean) in `call()`, which you can use to specify
a different behavior in training and inference:
```python
import tensorflow as tf
class MyModel(tf.keras.Model):
def __init__(self):
super().__init__()
self.dense1 = tf.keras.layers.Dense(4, activation=tf.nn.relu)
self.dense2 = tf.keras.layers.Dense(5, activation=tf.nn.softmax)
self.dropout = tf.keras.layers.Dropout(0.5)
def call(self, inputs, training=False):
x = self.dense1(inputs)
if training:
x = self.dropout(x, training=training)
return self.dense2(x)
model = MyModel()
```
Once the model is created, you can config the model with losses and metrics
with `model.compile()`, train the model with `model.fit()`, or use the model
to do prediction with `model.predict()`.
"""
_TF_MODULE_IGNORED_PROPERTIES = frozenset(
itertools.chain(
(
"_train_counter",
"_test_counter",
"_predict_counter",
"_steps_per_execution",
"_compiled_trainable_state",
),
base_layer.Layer._TF_MODULE_IGNORED_PROPERTIES,
)
)
_SCALAR_UPRANKING_ON = False
def __new__(cls, *args, **kwargs):
# Signature detection
if is_functional_model_init_params(args, kwargs) and cls == Model:
# Functional model
from tf_keras.engine import functional
return functional.Functional(skip_init=True, *args, **kwargs)
else:
return super(Model, cls).__new__(cls, *args, **kwargs)
@tf.__internal__.tracking.no_automatic_dependency_tracking
@traceback_utils.filter_traceback
def __init__(self, *args, **kwargs):
self._is_model_for_instrumentation = True
# Special case for Subclassed Functional Model, which we couldn't detect
# when __new__ is called. We only realize it is a functional model when
# it calls super.__init__ with input and output tensor.
from tf_keras.engine import functional
if is_functional_model_init_params(args, kwargs) and not isinstance(
self, functional.Functional
):
# Filter the kwargs for multiple inheritance.
supported_kwargs = [
"inputs",
"outputs",
"name",
"trainable",
"skip_init",
]
model_kwargs = {
k: kwargs[k] for k in kwargs if k in supported_kwargs
}
other_kwargs = {
k: kwargs[k] for k in kwargs if k not in supported_kwargs
}
inject_functional_model_class(self.__class__)
functional.Functional.__init__(self, *args, **model_kwargs)
# In case there is any multiple inheritance here, we need to call
# the __init__ for any class that appears after the Functional
# class.
clz_to_init = []
found_functional_class = False
for clz in self.__class__.__bases__:
if issubclass(clz, functional.Functional):
found_functional_class = True
continue
if found_functional_class:
clz_to_init.append(clz)
if clz_to_init:
for clz in clz_to_init:
clz.__init__(self, *args, **other_kwargs)
elif other_kwargs:
# In case there are unused kwargs, we should raise an error to
# user, in case they have a typo in the param name.
raise TypeError(
"The following keyword arguments passed to `Model` aren't "
"supported: {}.".format(other_kwargs)
)
return
# The following are implemented as property functions:
# self.trainable_weights
# self.non_trainable_weights
# `inputs` / `outputs` will only appear in kwargs if either are
# misspelled.
generic_utils.validate_kwargs(
kwargs,
{
"trainable",
"dtype",
"dynamic",
"name",
"autocast",
"inputs",
"outputs",
},
)
super().__init__(**kwargs)
# By default, Model is a subclass model, which is not in graph network.
self._is_graph_network = False
self.inputs = None
self.outputs = None
self.input_names = None
self.output_names = None
# stop_training is used by callback to stop training when error happens
self.stop_training = False
self.history = None
# These objects are used in the default `Model.compile`. They are not
# guaranteed to be set after `Model.compile` is called, as users can
# override compile with custom logic.
self.compiled_loss = None
self.compiled_metrics = None
# This is True for Sequential networks and Functional networks.
self._compute_output_and_mask_jointly = False
# Don't reset compilation if already done. This may occur if calling
# `__init__` (or `_init_graph_network`) on an already-compiled model
# such as a Sequential model. Sequential models may need to rebuild
# themselves after compilation.
self._maybe_create_attribute("_is_compiled", False)
self._maybe_create_attribute("optimizer", None)
# Model must be created under scope of DistStrat it will be trained
# with.
if tf.distribute.has_strategy():
self._distribution_strategy = tf.distribute.get_strategy()
else:
self._distribution_strategy = None
self._distribute_reduction_method = None
self._cluster_coordinator = None
# Defaults to value of `tf.config.experimental_functions_run_eagerly`.
self._run_eagerly = None
# Initialize cache attrs.
self._reset_compile_cache()
# Fault-tolerance handler. Set in `ModelCheckpoint`.
self._training_state = None
self._saved_model_inputs_spec = None
self._saved_model_arg_spec = None
self._checkpoint = tf.train.Checkpoint(root=weakref.ref(self))
self._steps_per_execution = None
self._steps_per_execution_tuner = None
self._autotune_steps_per_execution = False
self._layout_map = layout_map_lib.get_current_layout_map()
self._init_batch_counters()
self._base_model_initialized = True
# `jit_compile` starts off with None as default and gets overwritten by
# the value specified in `Model.compile`, and this is effective for
# `fit`, `evaluate`, and `predict`.
self._jit_compile = None
def _create_counter_variable(self, init_value):
"""Helper function for counter variable creation.
For the DTensor use case with layout map, since the variable are not
tracked by model, they can't be visited by the layout map, and need to
be properly initialized as DVariable.
"""
# This function should be removed after we move to the strategy based
# implementation for DTensor.
if self._layout_map is None:
agg = tf.VariableAggregation.ONLY_FIRST_REPLICA
return tf.Variable(init_value, dtype="int64", aggregation=agg)
else:
layout = dtensor_api.Layout.replicated(
mesh=self._layout_map.get_default_mesh(), rank=0
)
return dtensor_api.DVariable(
init_value, dtype="int64", layout=layout
)
@tf.__internal__.tracking.no_automatic_dependency_tracking
def _init_batch_counters(self):
# Untracked Variables, used to keep track of mini-batches seen in `fit`,
# `evaluate`, and `predict`.
if not tf.inside_function():
# Creating variables inside tf.function is not allowed, hence
# these would otherwise prevent users from creating TF-Keras layers
# inside tf.function.
# These variables are not connected to outputs so they have no
# effect on graph generation anyway.
self._train_counter = self._create_counter_variable(0)
self._test_counter = self._create_counter_variable(0)
self._predict_counter = self._create_counter_variable(0)
def __setattr__(self, name, value):
if not getattr(self, "_self_setattr_tracking", True):
super().__setattr__(name, value)
return
if all(
isinstance(v, (base_layer.Layer, tf.Variable))
or base_layer_utils.has_weights(v)
for v in tf.nest.flatten(value)
):
try:
self._base_model_initialized
except AttributeError:
raise RuntimeError(
"It looks like you are subclassing `Model` and you "
"forgot to call `super().__init__()`."
" Always start with this line."
)
super().__setattr__(name, value)
def __reduce__(self):
if self.built:
return (
pickle_utils.deserialize_model_from_bytecode,
(pickle_utils.serialize_model_as_bytecode(self),),
)
else:
# SavedModel (and hence serialize_model_as_bytecode) only support
# built models, but if the model is not built,
# it may be possible to serialize as a plain Python object,
# as long as the constituent parts (layers, optimizers, losses,
# etc.) can be serialized as plain Python objects. Thus we call up
# the superclass hierarchy to get an implementation of __reduce__
# that can pickle this Model as a plain Python object.
return super().__reduce__()
def __deepcopy__(self, memo):
if self.built:
new = pickle_utils.deserialize_model_from_bytecode(
pickle_utils.serialize_model_as_bytecode(self)
)
memo[id(self)] = new
else:
# See comment in __reduce__ for explanation
deserializer, serialized, *rest = super().__reduce__()
new = deserializer(*serialized)
memo[id(self)] = new
if rest:
state = copy.deepcopy(rest[0], memo=memo)
new.__setstate__(state)
return new
def __copy__(self):
return self.__deepcopy__({})
@generic_utils.default
def build(self, input_shape):
"""Builds the model based on input shapes received.
This is to be used for subclassed models, which do not know at
instantiation time what their inputs look like.
This method only exists for users who want to call `model.build()` in a
standalone way (as a substitute for calling the model on real data to
build it). It will never be called by the framework (and thus it will
never throw unexpected errors in an unrelated workflow).
Args:
input_shape: Single tuple, `TensorShape` instance, or list/dict of
shapes, where shapes are tuples, integers, or `TensorShape`
instances.
Raises:
ValueError:
1. In case of invalid user-provided data (not of type tuple,
list, `TensorShape`, or dict).
2. If the model requires call arguments that are agnostic
to the input shapes (positional or keyword arg in call
signature).
3. If not all layers were properly built.
4. If float type inputs are not supported within the layers.
In each of these cases, the user should build their model by calling
it on real tensor data.
"""
if self._is_graph_network:
super().build(input_shape)
return
if input_shape is None:
raise ValueError(
"Input shape must be defined when calling `build()` on "
"a `Model` subclass."
)
valid_types = (tuple, list, tf.TensorShape, dict)
if not isinstance(input_shape, valid_types):
raise ValueError(
"Specified input shape is not one of the valid types. "
"Please specify a batch input shape of type tuple or "
"list of input shapes. User provided "
"input type: {}.".format(type(input_shape))
)
if input_shape and not self.inputs:
# We create placeholders for the `None`s in the shape and build the
# model in a Graph. Since tf.Variable is compatible with both eager
# execution and graph building, the variables created after building
# the model in a Graph are still valid when executing eagerly.
if tf.executing_eagerly():
graph = tf.__internal__.FuncGraph("build_graph")
else:
graph = backend.get_graph()
with graph.as_default():
if isinstance(input_shape, list) and all(
d is None or isinstance(d, int) for d in input_shape
):
input_shape = tuple(input_shape)
if isinstance(input_shape, list):
x = [
base_layer_utils.generate_placeholders_from_shape(shape)
for shape in input_shape
]
elif isinstance(input_shape, dict):
x = {
k: base_layer_utils.generate_placeholders_from_shape(
shape
)
for k, shape in input_shape.items()
}
else:
x = base_layer_utils.generate_placeholders_from_shape(
input_shape
)
kwargs = {}
call_signature = self._call_spec.full_argspec
call_args = call_signature.args
# Exclude `self`, `inputs`, and any argument with a default
# value.
if len(call_args) > 2:
if call_signature.defaults:
call_args = call_args[2 : -len(call_signature.defaults)]
else:
call_args = call_args[2:]
for arg in call_args:
if arg == "training":
# Case where `training` is a positional arg with no
# default.
kwargs["training"] = False
else:
# Has invalid call signature with unknown positional
# arguments.
raise ValueError(
"Currently, you cannot build your model if it "
"has positional or keyword arguments that are "
"not inputs to the model, but are required for "
"its `call()` method. Instead, in order to "
"instantiate and build your model, `call()` "
"your model on real tensor data with all "
"expected call arguments. The argument "
"for `call()` can be a single list/tuple that "
"contains multiple inputs."
)
elif len(call_args) < 2:
# Signature without `inputs`.
raise ValueError(
"You can only call `build()` on a model if its "
"`call()` method accepts an `inputs` argument."
)
try:
self.call(x, **kwargs)
except (tf.errors.InvalidArgumentError, TypeError) as e:
raise ValueError(
"You cannot build your model by calling `build` "
"if your layers do not support float type inputs. "
"Instead, in order to instantiate and build your "
"model, call your model on real tensor data (of "
"the correct dtype).\n\nThe actual error from "
f"`call` is: {e}."
)
super().build(input_shape)
@traceback_utils.filter_traceback
def __call__(self, *args, **kwargs):
if self._layout_map is not None and not self.built:
# Note that this method is only overridden for DTensor and layout
# injection purpose.
# Capture the inputs and create graph input as replacement for model
# to initialize its weights first.
copied_args = copy.copy(args)
copied_kwargs = copy.copy(kwargs)
(
inputs,
copied_args,
copied_kwargs,
) = self._call_spec.split_out_first_arg(copied_args, copied_kwargs)
def _convert_to_graph_inputs(x):
if isinstance(x, (tf.Tensor, np.ndarray, float, int)):
x = tf.convert_to_tensor(x)
return input_layer_module.Input(x.shape)
# TODO(scottzhu): maybe better handle mask and training flag.
inputs = tf.nest.map_structure(_convert_to_graph_inputs, inputs)
copied_args = tf.nest.map_structure(
_convert_to_graph_inputs, copied_args
)
copied_kwargs = tf.nest.map_structure(
_convert_to_graph_inputs, copied_kwargs
)
with layout_map_lib.layout_map_scope(self._layout_map):
# We ignore the result here.
super().__call__(inputs, *copied_args, **copied_kwargs)
layout_map_lib._map_subclass_model_variable(self, self._layout_map)
return super().__call__(*args, **kwargs)
@doc_controls.doc_in_current_and_subclasses
def call(self, inputs, training=None, mask=None):
"""Calls the model on new inputs and returns the outputs as tensors.
In this case `call()` just reapplies
all ops in the graph to the new inputs
(e.g. build a new computational graph from the provided inputs).
Note: This method should not be called directly. It is only meant to be
overridden when subclassing `tf.keras.Model`.
To call a model on an input, always use the `__call__()` method,
i.e. `model(inputs)`, which relies on the underlying `call()` method.
Args:
inputs: Input tensor, or dict/list/tuple of input tensors.
training: Boolean or boolean scalar tensor, indicating whether to
run the `Network` in training mode or inference mode.
mask: A mask or list of masks. A mask can be either a boolean tensor
or None (no mask). For more details, check the guide
[here](https://www.tensorflow.org/guide/keras/masking_and_padding).
Returns:
A tensor if there is a single output, or
a list of tensors if there are more than one outputs.
"""
raise NotImplementedError(
"Unimplemented `tf.keras.Model.call()`: if you "
"intend to create a `Model` with the Functional "
"API, please provide `inputs` and `outputs` "
"arguments. Otherwise, subclass `Model` with an "
"overridden `call()` method."
)
@traceback_utils.filter_traceback
def compile(
self,
optimizer="rmsprop",
loss=None,
metrics=None,
loss_weights=None,
weighted_metrics=None,
run_eagerly=None,
steps_per_execution=None,
jit_compile=None,
pss_evaluation_shards=0,
**kwargs,
):
"""Configures the model for training.
Example:
```python
model.compile(optimizer=tf.keras.optimizers.Adam(learning_rate=1e-3),
loss=tf.keras.losses.BinaryCrossentropy(),
metrics=[tf.keras.metrics.BinaryAccuracy(),
tf.keras.metrics.FalseNegatives()])
```
Args:
optimizer: String (name of optimizer) or optimizer instance. See
`tf.keras.optimizers`.
loss: Loss function. May be a string (name of loss function), or
a `tf.keras.losses.Loss` instance. See `tf.keras.losses`. A loss
function is any callable with the signature `loss = fn(y_true,
y_pred)`, where `y_true` are the ground truth values, and
`y_pred` are the model's predictions.
`y_true` should have shape
`(batch_size, d0, .. dN)` (except in the case of
sparse loss functions such as
sparse categorical crossentropy which expects integer arrays of
shape `(batch_size, d0, .. dN-1)`).
`y_pred` should have shape `(batch_size, d0, .. dN)`.
The loss function should return a float tensor.
If a custom `Loss` instance is
used and reduction is set to `None`, return value has shape
`(batch_size, d0, .. dN-1)` i.e. per-sample or per-timestep loss
values; otherwise, it is a scalar. If the model has multiple
outputs, you can use a different loss on each output by passing a
dictionary or a list of losses. The loss value that will be
minimized by the model will then be the sum of all individual
losses, unless `loss_weights` is specified.
metrics: List of metrics to be evaluated by the model during
training and testing. Each of this can be a string (name of a
built-in function), function or a `tf.keras.metrics.Metric`
instance. See `tf.keras.metrics`. Typically you will use
`metrics=['accuracy']`.
A function is any callable with the signature `result = fn(y_true,
y_pred)`. To specify different metrics for different outputs of a
multi-output model, you could also pass a dictionary, such as
`metrics={'output_a':'accuracy', 'output_b':['accuracy', 'mse']}`.
You can also pass a list to specify a metric or a list of metrics
for each output, such as
`metrics=[['accuracy'], ['accuracy', 'mse']]`
or `metrics=['accuracy', ['accuracy', 'mse']]`. When you pass the
strings 'accuracy' or 'acc', we convert this to one of
`tf.keras.metrics.BinaryAccuracy`,
`tf.keras.metrics.CategoricalAccuracy`,
`tf.keras.metrics.SparseCategoricalAccuracy` based on the shapes
of the targets and of the model output. We do a similar
conversion for the strings 'crossentropy' and 'ce' as well.
The metrics passed here are evaluated without sample weighting; if
you would like sample weighting to apply, you can specify your
metrics via the `weighted_metrics` argument instead.
loss_weights: Optional list or dictionary specifying scalar
coefficients (Python floats) to weight the loss contributions of
different model outputs. The loss value that will be minimized by
the model will then be the *weighted sum* of all individual
losses, weighted by the `loss_weights` coefficients. If a list,
it is expected to have a 1:1 mapping to the model's outputs. If a
dict, it is expected to map output names (strings) to scalar
coefficients.
weighted_metrics: List of metrics to be evaluated and weighted by
`sample_weight` or `class_weight` during training and testing.
run_eagerly: Bool. If `True`, this `Model`'s logic will not be
wrapped in a `tf.function`. Recommended to leave this as `None`
unless your `Model` cannot be run inside a `tf.function`.
`run_eagerly=True` is not supported when using
`tf.distribute.experimental.ParameterServerStrategy`. Defaults to
`False`.
steps_per_execution: Int or `'auto'`. The number of batches to
run during each `tf.function` call. If set to "auto", keras will
automatically tune `steps_per_execution` during runtime. Running
multiple batches inside a single `tf.function` call can greatly
improve performance on TPUs, when used with distributed strategies
such as `ParameterServerStrategy`, or with small models with a
large Python overhead. At most, one full epoch will be run each
execution. If a number larger than the size of the epoch is
passed, the execution will be truncated to the size of the epoch.
Note that if `steps_per_execution` is set to `N`,
`Callback.on_batch_begin` and `Callback.on_batch_end` methods will
only be called every `N` batches (i.e. before/after each
`tf.function` execution). Defaults to `1`.
jit_compile: If `True`, compile the model training step with XLA.
[XLA](https://www.tensorflow.org/xla) is an optimizing compiler
for machine learning.
`jit_compile` is not enabled for by default.
Note that `jit_compile=True`
may not necessarily work for all models.
For more information on supported operations please refer to the
[XLA documentation](https://www.tensorflow.org/xla).
Also refer to
[known XLA issues](https://www.tensorflow.org/xla/known_issues)
for more details.
pss_evaluation_shards: Integer or 'auto'. Used for
`tf.distribute.ParameterServerStrategy` training only. This arg
sets the number of shards to split the dataset into, to enable an
exact visitation guarantee for evaluation, meaning the model will
be applied to each dataset element exactly once, even if workers
fail. The dataset must be sharded to ensure separate workers do
not process the same data. The number of shards should be at least
the number of workers for good performance. A value of 'auto'
turns on exact evaluation and uses a heuristic for the number of
shards based on the number of workers. 0, meaning no
visitation guarantee is provided. NOTE: Custom implementations of
`Model.test_step` will be ignored when doing exact evaluation.
Defaults to `0`.
**kwargs: Arguments supported for backwards compatibility only.
"""
if jit_compile and not tf_utils.can_jit_compile(warn=True):
jit_compile = False
self._compile_config = serialization_lib.Config(
optimizer=optimizer,
loss=loss,
metrics=metrics,
loss_weights=loss_weights,
weighted_metrics=weighted_metrics,
run_eagerly=run_eagerly,
steps_per_execution=steps_per_execution,
jit_compile=jit_compile,
)
with self.distribute_strategy.scope():
if "experimental_steps_per_execution" in kwargs:
logging.warning(
"The argument `steps_per_execution` is no longer "
"experimental. Pass `steps_per_execution` instead of "
"`experimental_steps_per_execution`."
)
if not steps_per_execution:
steps_per_execution = kwargs.pop(
"experimental_steps_per_execution"
)
# When compiling from an already-serialized model, we do not want to
# reapply some processing steps (e.g. metric renaming for
# multi-output models, which have prefixes added for each
# corresponding output name).
from_serialized = kwargs.pop("from_serialized", False)
self._validate_compile(optimizer, metrics, **kwargs)
self._run_eagerly = run_eagerly
self.optimizer = self._get_optimizer(optimizer)
mesh = None
if self._layout_map is not None:
mesh = self._layout_map.get_default_mesh()
if isinstance(loss, compile_utils.LossesContainer):
self.compiled_loss = loss
else:
self.compiled_loss = compile_utils.LossesContainer(
loss,
loss_weights,
output_names=self.output_names,
mesh=mesh,
)
self.compiled_metrics = compile_utils.MetricsContainer(
metrics,
weighted_metrics,
output_names=self.output_names,
from_serialized=from_serialized,
mesh=mesh,
)
if steps_per_execution == "auto":
if self._steps_per_execution is None:
self._configure_steps_per_execution(1)
self._steps_per_execution_tuner = (
steps_per_execution_tuning.StepsPerExecutionTuner(
self.optimizer, self._steps_per_execution
)
)
self._autotune_steps_per_execution = True
else:
self._configure_steps_per_execution(steps_per_execution or 1)
self._pss_evaluation_shards = self._infer_exact_eval_shards(
pss_evaluation_shards
)
# Initializes attrs that are reset each time `compile` is called.
self._reset_compile_cache()
self._is_compiled = True
self.loss = loss or {}
if (self._run_eagerly or self.dynamic) and jit_compile:
raise ValueError(
"You cannot enable `run_eagerly` and `jit_compile` "
"at the same time."
)
else:
self._jit_compile = jit_compile
def _get_optimizer(self, optimizer):
"""Wraps `optimizer` in `LossScaleOptimizer` if necessary."""
def _get_single_optimizer(opt):
opt = optimizers.get(opt)
if self.dtype_policy.name == "mixed_float16" and not isinstance(
opt, lso.BaseLossScaleOptimizer
):
# Loss scaling is necessary with mixed_float16 for models to
# converge to the same accuracy as with float32.
opt = lso.BaseLossScaleOptimizer(opt)
return opt
return tf.nest.map_structure(_get_single_optimizer, optimizer)
@tf.__internal__.tracking.no_automatic_dependency_tracking
def _reset_compile_cache(self):
self.train_function = None
self.test_function = None
self.predict_function = None
# Used to cache the `tf.function`'ed `train_function` to be logged in
# TensorBoard, since the original `train_function` is not necessarily
# a `tf.function` (e.g., with ParameterServerStrategy, the
# `train_function` is a scheduling of the actual training function to a
# remote worker).
self.train_tf_function = None
# Used to cache `trainable` attr of `Layer`s for `fit`.
self._compiled_trainable_state = self._get_trainable_state()
@tf.__internal__.tracking.no_automatic_dependency_tracking
def _configure_steps_per_execution(self, steps_per_execution):
self._steps_per_execution = self._create_counter_variable(
steps_per_execution
)
@property
def _should_compute_mask(self):
return False
@property
def metrics(self):
"""Return metrics added using `compile()` or `add_metric()`.
Note: Metrics passed to `compile()` are available only after a
`keras.Model` has been trained/evaluated on actual data.
Examples:
>>> inputs = tf.keras.layers.Input(shape=(3,))
>>> outputs = tf.keras.layers.Dense(2)(inputs)
>>> model = tf.keras.models.Model(inputs=inputs, outputs=outputs)
>>> model.compile(optimizer="Adam", loss="mse", metrics=["mae"])
>>> [m.name for m in model.metrics]
[]
>>> x = np.random.random((2, 3))
>>> y = np.random.randint(0, 2, (2, 2))
>>> model.fit(x, y)
>>> [m.name for m in model.metrics]
['loss', 'mae']
>>> inputs = tf.keras.layers.Input(shape=(3,))
>>> d = tf.keras.layers.Dense(2, name='out')
>>> output_1 = d(inputs)
>>> output_2 = d(inputs)
>>> model = tf.keras.models.Model(
... inputs=inputs, outputs=[output_1, output_2])
>>> model.add_metric(
... tf.reduce_sum(output_2), name='mean', aggregation='mean')
>>> model.compile(optimizer="Adam", loss="mse", metrics=["mae", "acc"])
>>> model.fit(x, (y, y))
>>> [m.name for m in model.metrics]
['loss', 'out_loss', 'out_1_loss', 'out_mae', 'out_acc', 'out_1_mae',
'out_1_acc', 'mean']
"""
metrics = []
if self._is_compiled:
if self.compiled_loss is not None:
metrics += self.compiled_loss.metrics
if self.compiled_metrics is not None:
metrics += self.compiled_metrics.metrics
for l in self._flatten_layers():
metrics.extend(l._metrics)
return metrics
@property
def metrics_names(self):
"""Returns the model's display labels for all outputs.
Note: `metrics_names` are available only after a `keras.Model` has been
trained/evaluated on actual data.
Examples:
>>> inputs = tf.keras.layers.Input(shape=(3,))
>>> outputs = tf.keras.layers.Dense(2)(inputs)
>>> model = tf.keras.models.Model(inputs=inputs, outputs=outputs)
>>> model.compile(optimizer="Adam", loss="mse", metrics=["mae"])
>>> model.metrics_names
[]
>>> x = np.random.random((2, 3))
>>> y = np.random.randint(0, 2, (2, 2))
>>> model.fit(x, y)
>>> model.metrics_names
['loss', 'mae']
>>> inputs = tf.keras.layers.Input(shape=(3,))
>>> d = tf.keras.layers.Dense(2, name='out')
>>> output_1 = d(inputs)
>>> output_2 = d(inputs)
>>> model = tf.keras.models.Model(
... inputs=inputs, outputs=[output_1, output_2])
>>> model.compile(optimizer="Adam", loss="mse", metrics=["mae", "acc"])
>>> model.fit(x, (y, y))
>>> model.metrics_names
['loss', 'out_loss', 'out_1_loss', 'out_mae', 'out_acc', 'out_1_mae',
'out_1_acc']
"""
# This property includes all output names including `loss` and
# per-output losses for backward compatibility.
return [m.name for m in self.metrics]
@property
def distribute_strategy(self):
"""The `tf.distribute.Strategy` this model was created under."""
return self._distribution_strategy or tf.distribute.get_strategy()
@property
def run_eagerly(self):
"""Settable attribute indicating whether the model should run eagerly.
Running eagerly means that your model will be run step by step,
like Python code. Your model might run slower, but it should become
easier for you to debug it by stepping into individual layer calls.
By default, we will attempt to compile your model to a static graph to
deliver the best execution performance.
Returns:
Boolean, whether the model should run eagerly.
"""
if self.dynamic and self._run_eagerly == False:
# TODO(fchollet): consider using py_func to enable this.
raise ValueError(
"Your model contains layers that can only be "
"successfully run in eager execution (layers "
"constructed with `dynamic=True`). "
"You cannot set `run_eagerly=False`."
)
if self._cluster_coordinator and self._run_eagerly:
raise ValueError(
"When using `Model` with `ParameterServerStrategy`, "
"`run_eagerly` is not supported."
)
# Run eagerly logic, by priority:
# (1) Dynamic models must be run eagerly.
# (2) Explicitly setting run_eagerly causes a Model to be run eagerly.
# (3) Not explicitly setting run_eagerly defaults to TF's global
# setting.
return (
self.dynamic
or self._run_eagerly
or (tf.config.functions_run_eagerly() and self._run_eagerly is None)
)
@run_eagerly.setter
def run_eagerly(self, value):
self._run_eagerly = value
@property
def autotune_steps_per_execution(self):
"""Settable property to enable tuning for steps_per_execution"""
return self._autotune_steps_per_execution
@autotune_steps_per_execution.setter
def autotune_steps_per_execution(self, value):
self._autotune_steps_per_execution = value
if value and self._steps_per_execution_tuner is None:
if self._steps_per_execution is None:
self._configure_steps_per_execution(1)
self._steps_per_execution_tuner = (
steps_per_execution_tuning.StepsPerExecutionTuner(
self.optimizer, self._steps_per_execution
)
)
@property
def steps_per_execution(self):
"""Settable `steps_per_execution variable. Requires a compiled model."""
return self._steps_per_execution
@steps_per_execution.setter
def steps_per_execution(self, value):
if self._steps_per_execution is None:
self._configure_steps_per_execution(value)
else:
self._steps_per_execution.assign(value)
@property
def jit_compile(self):
"""Specify whether to compile the model with XLA.
[XLA](https://www.tensorflow.org/xla) is an optimizing compiler
for machine learning. `jit_compile` is not enabled by default.
Note that `jit_compile=True` may not necessarily work for all models.
For more information on supported operations please refer to the
[XLA documentation](https://www.tensorflow.org/xla). Also refer to
[known XLA issues](https://www.tensorflow.org/xla/known_issues)
for more details.
"""
return self._jit_compile
@jit_compile.setter
def jit_compile(self, value):
# Function remains cached with previous jit_compile settings
if self._jit_compile == value:
# Avoid resetting compiler cache if possible if the value is the
# same
return
# Check if TensorFlow is compiled with XLA before setting the value
if value and not tf_utils.can_jit_compile(warn=True):
self._jit_compile = False
return
self._jit_compile = value
# Setting `jit_compile` should invalidate previously cached functions.
self._reset_compile_cache()
@property
def distribute_reduction_method(self):
"""The method employed to reduce per-replica values during training.
Unless specified, the value "auto" will be assumed, indicating that
the reduction strategy should be chosen based on the current
running environment.
See `reduce_per_replica` function for more details.
"""
return self._distribute_reduction_method or "auto"
@distribute_reduction_method.setter
def distribute_reduction_method(self, value):
self._distribute_reduction_method = value
def _validate_target_and_loss(self, y, loss):
"""Raises error if target or loss is not found.
This method verifies that the target and loss are properly populated
when applicable, or raises errors.
Args:
y: the target for training.
loss: the total loss tensor including loss added via `compile` and
`add_loss`.
"""
# `self.loss` references the loss added via `compile` call. If users
# have provided such, the target must be provided; otherwise it's a user
# error. Note that `self.loss` does not include losses added via
# `add_loss`, and it is a valid use when such loss from `add_loss`
# exists and target does not.
if self.loss and y is None:
raise ValueError(
"Target data is missing. Your model was compiled with "
f"loss={self.loss}, "
"and therefore expects target data to be provided in `fit()`."
)
# For training, there must be compiled loss or regularization loss to
# exist in order to apply the gradients. If one is not found, it means
# no loss was supplied via `compile` or `add_loss`.
elif loss is None:
raise ValueError(
"No loss found. You may have forgotten to provide a `loss` "
"argument in the `compile()` method."
)
def train_step(self, data):
"""The logic for one training step.
This method can be overridden to support custom training logic.
For concrete examples of how to override this method see
[Customizing what happens in fit](
https://www.tensorflow.org/guide/keras/customizing_what_happens_in_fit).
This method is called by `Model.make_train_function`.
This method should contain the mathematical logic for one step of
training. This typically includes the forward pass, loss calculation,
backpropagation, and metric updates.
Configuration details for *how* this logic is run (e.g. `tf.function`
and `tf.distribute.Strategy` settings), should be left to
`Model.make_train_function`, which can also be overridden.
Args:
data: A nested structure of `Tensor`s.
Returns:
A `dict` containing values that will be passed to
`tf.keras.callbacks.CallbackList.on_train_batch_end`. Typically, the
values of the `Model`'s metrics are returned. Example:
`{'loss': 0.2, 'accuracy': 0.7}`.
"""
x, y, sample_weight = data_adapter.unpack_x_y_sample_weight(data)
# Run forward pass.
with tf.GradientTape() as tape:
y_pred = self(x, training=True)
loss = self.compute_loss(x, y, y_pred, sample_weight)
self._validate_target_and_loss(y, loss)
# Run backwards pass.
self.optimizer.minimize(loss, self.trainable_variables, tape=tape)
return self.compute_metrics(x, y, y_pred, sample_weight)
def compute_loss(self, x=None, y=None, y_pred=None, sample_weight=None):
"""Compute the total loss, validate it, and return it.
Subclasses can optionally override this method to provide custom loss
computation logic.
Example:
```python
class MyModel(tf.keras.Model):
def __init__(self, *args, **kwargs):
super(MyModel, self).__init__(*args, **kwargs)
self.loss_tracker = tf.keras.metrics.Mean(name='loss')
def compute_loss(self, x, y, y_pred, sample_weight):
loss = tf.reduce_mean(tf.math.squared_difference(y_pred, y))
loss += tf.add_n(self.losses)
self.loss_tracker.update_state(loss)
return loss
def reset_metrics(self):
self.loss_tracker.reset_states()
@property
def metrics(self):
return [self.loss_tracker]
tensors = tf.random.uniform((10, 10)), tf.random.uniform((10,))
dataset = tf.data.Dataset.from_tensor_slices(tensors).repeat().batch(1)
inputs = tf.keras.layers.Input(shape=(10,), name='my_input')
outputs = tf.keras.layers.Dense(10)(inputs)
model = MyModel(inputs, outputs)
model.add_loss(tf.reduce_sum(outputs))
optimizer = tf.keras.optimizers.SGD()
model.compile(optimizer, loss='mse', steps_per_execution=10)
model.fit(dataset, epochs=2, steps_per_epoch=10)
print('My custom loss: ', model.loss_tracker.result().numpy())
```
Args:
x: Input data.
y: Target data.
y_pred: Predictions returned by the model (output of `model(x)`)
sample_weight: Sample weights for weighting the loss function.
Returns:
The total loss as a `tf.Tensor`, or `None` if no loss results (which
is the case when called by `Model.test_step`).
"""
del x # The default implementation does not use `x`.
return self.compiled_loss(
y, y_pred, sample_weight, regularization_losses=self.losses
)
def compute_metrics(self, x, y, y_pred, sample_weight):
"""Update metric states and collect all metrics to be returned.
Subclasses can optionally override this method to provide custom metric
updating and collection logic.
Example:
```python
class MyModel(tf.keras.Sequential):
def compute_metrics(self, x, y, y_pred, sample_weight):
# This super call updates `self.compiled_metrics` and returns
# results for all metrics listed in `self.metrics`.
metric_results = super(MyModel, self).compute_metrics(
x, y, y_pred, sample_weight)
# Note that `self.custom_metric` is not listed in `self.metrics`.
self.custom_metric.update_state(x, y, y_pred, sample_weight)
metric_results['custom_metric_name'] = self.custom_metric.result()
return metric_results
```
Args:
x: Input data.
y: Target data.
y_pred: Predictions returned by the model (output of `model.call(x)`)
sample_weight: Sample weights for weighting the loss function.
Returns:
A `dict` containing values that will be passed to
`tf.keras.callbacks.CallbackList.on_train_batch_end()`. Typically, the
values of the metrics listed in `self.metrics` are returned. Example:
`{'loss': 0.2, 'accuracy': 0.7}`.
"""
del x # The default implementation does not use `x`.
self.compiled_metrics.update_state(y, y_pred, sample_weight)
return self.get_metrics_result()
def get_metrics_result(self):
"""Returns the model's metrics values as a dict.
If any of the metric result is a dict (containing multiple metrics),
each of them gets added to the top level returned dict of this method.
Returns:
A `dict` containing values of the metrics listed in `self.metrics`.
Example:
`{'loss': 0.2, 'accuracy': 0.7}`.
"""
# Collect metrics to return
return_metrics = {}
for metric in self.metrics:
result = metric.result()
if isinstance(result, dict):
return_metrics.update(result)
else:
return_metrics[metric.name] = result
return return_metrics
def _validate_and_get_metrics_result(self, logs):
"""Returns model metrics as a dict if the keys match with input logs.
When the training / evalution is performed with asynchronous steps, such
as the case with `tf.distribute.ParameterServerStrategy`, the last
scheduled `train / test_step` may not give the latest metrics because it
is not guaranteed to be executed the last. This method gets metrics from
the model directly instead of relying on the return from last step
function.
It logs a warning if the metric results could not be overridden when
used with `tf.distribute.ParameterServerStrategy`.
When the user has custom train / test step functions, the metrics
returned may be different from `Model.metrics`. In those instances,
this function will be no-op and return the logs.
Args:
logs: A `dict` of metrics returned by train / test step function.
Returns:
A `dict` containing values of the metrics listed in `self.metrics`
when logs and model metrics keys match. Otherwise it returns input
`logs`.
"""
PSS_WARN_MSG = "Could not get Model metric results. \
Using the results of last step function could lead to incorrect \
results when used with ParameterServerStrategy"
try:
metric_logs = self.get_metrics_result()
except TypeError:
if self._cluster_coordinator:
logging.warning(PSS_WARN_MSG)
else:
# Verify that train / test step logs passed and metric logs have
# matching keys. Could be different when using custom step functions
if isinstance(logs, dict) and set(logs.keys()) == set(
metric_logs.keys()
):
logs = tf_utils.sync_to_numpy_or_python_type(metric_logs)
elif self._cluster_coordinator:
logging.warning(PSS_WARN_MSG)
return logs
def _aggregate_exact_metrics(self, logs):
# When doing exact evaluation, `logs` is a list of each data shard's
# metric variables, which will be used to update the metrics.
for shard_result in logs:
for metric in self.metrics:
if metric.name not in shard_result.keys():
logging.log_first_n(
logging.WARN,
f"No matching result found for metric {metric.name}. "
"This metric's computed result may be incorrect.",
3,
)
continue
metric_result = shard_result[metric.name]
if len(metric_result) != len(metric.weights):
raise ValueError(
f"Expected {len(metric.weights)} variables in result "
f"for metric {metric.name}, but found "
f"{len(metric_result)}."
)
for weight, val in zip(metric.weights, metric_result):
weight.assign_add(val)
return self.get_metrics_result()
def make_train_function(self, force=False):
"""Creates a function that executes one step of training.
This method can be overridden to support custom training logic.
This method is called by `Model.fit` and `Model.train_on_batch`.
Typically, this method directly controls `tf.function` and
`tf.distribute.Strategy` settings, and delegates the actual training
logic to `Model.train_step`.
This function is cached the first time `Model.fit` or
`Model.train_on_batch` is called. The cache is cleared whenever
`Model.compile` is called. You can skip the cache and generate again the
function with `force=True`.
Args:
force: Whether to regenerate the train function and skip the cached
function if available.
Returns:
Function. The function created by this method should accept a
`tf.data.Iterator`, and return a `dict` containing values that will
be passed to `tf.keras.Callbacks.on_train_batch_end`, such as
`{'loss': 0.2, 'accuracy': 0.7}`.
"""
if self.train_function is not None and not force:
return self.train_function
def step_function(model, iterator):
"""Runs a single training step."""
def run_step(data):
outputs = model.train_step(data)
# Ensure counter is updated only if `train_step` succeeds.
with tf.control_dependencies(_minimum_control_deps(outputs)):
model._train_counter.assign_add(1)
return outputs
if self.jit_compile:
run_step = tf.function(
run_step, jit_compile=True, reduce_retracing=True
)
data = next(iterator)
outputs = model.distribute_strategy.run(run_step, args=(data,))
outputs = reduce_per_replica(
outputs,
self.distribute_strategy,
reduction=self.distribute_reduction_method,
)
return outputs
# Special case if steps_per_execution is one.
if (
self._steps_per_execution is None
or self._steps_per_execution.numpy().item() == 1
and not self.autotune_steps_per_execution
):
def train_function(iterator):
"""Runs a training execution with a single step."""
return step_function(self, iterator)
if not self.run_eagerly:
train_function = tf.function(
train_function, reduce_retracing=True
)
self.train_tf_function = train_function
if self._cluster_coordinator:
self.train_function = (
lambda it: self._cluster_coordinator.schedule(
train_function, args=(it,)
)
)
else:
self.train_function = train_function
# If we're using a coordinator, use the value of
# self._steps_per_execution at the time the function is
# called/scheduled, and not when it is actually executed.
elif self._cluster_coordinator:
def train_function(iterator, steps_per_execution):
"""Runs a training execution with multiple steps."""
for _ in tf.range(steps_per_execution):
outputs = step_function(self, iterator)
return outputs
if not self.run_eagerly:
train_function = tf.function(
train_function, reduce_retracing=True
)
self.train_tf_function = train_function
self.train_function = lambda it: self._cluster_coordinator.schedule(
train_function, args=(it, self._steps_per_execution.value())
)
else:
def train_function(iterator):
"""Runs a training execution with multiple steps."""
for _ in tf.range(self._steps_per_execution):
outputs = step_function(self, iterator)
return outputs
if not self.run_eagerly:
train_function = tf.function(
train_function, reduce_retracing=True
)
self.train_tf_function = train_function
self.train_function = train_function
return self.train_function
@traceback_utils.filter_traceback
def fit(
self,
x=None,
y=None,
batch_size=None,
epochs=1,
verbose="auto",
callbacks=None,
validation_split=0.0,
validation_data=None,
shuffle=True,
class_weight=None,
sample_weight=None,
initial_epoch=0,
steps_per_epoch=None,
validation_steps=None,
validation_batch_size=None,
validation_freq=1,
max_queue_size=10,
workers=1,
use_multiprocessing=False,
):
"""Trains the model for a fixed number of epochs (dataset iterations).
Args:
x: Input data. It could be:
- A Numpy array (or array-like), or a list of arrays
(in case the model has multiple inputs).
- A TensorFlow tensor, or a list of tensors
(in case the model has multiple inputs).
- A dict mapping input names to the corresponding array/tensors,
if the model has named inputs.
- A `tf.data` dataset. Should return a tuple
of either `(inputs, targets)` or
`(inputs, targets, sample_weights)`.
- A generator or `keras.utils.Sequence` returning `(inputs,
targets)` or `(inputs, targets, sample_weights)`.
- A `tf.keras.utils.experimental.DatasetCreator`, which wraps a
callable that takes a single argument of type
`tf.distribute.InputContext`, and returns a `tf.data.Dataset`.
`DatasetCreator` should be used when users prefer to specify the
per-replica batching and sharding logic for the `Dataset`.
See `tf.keras.utils.experimental.DatasetCreator` doc for more
information.
A more detailed description of unpacking behavior for iterator
types (Dataset, generator, Sequence) is given below. If these
include `sample_weights` as a third component, note that sample
weighting applies to the `weighted_metrics` argument but not the
`metrics` argument in `compile()`. If using
`tf.distribute.experimental.ParameterServerStrategy`, only
`DatasetCreator` type is supported for `x`.
y: Target data. Like the input data `x`,
it could be either Numpy array(s) or TensorFlow tensor(s).
It should be consistent with `x` (you cannot have Numpy inputs and
tensor targets, or inversely). If `x` is a dataset, generator,
or `keras.utils.Sequence` instance, `y` should
not be specified (since targets will be obtained from `x`).
batch_size: Integer or `None`.
Number of samples per gradient update.
If unspecified, `batch_size` will default to 32.
Do not specify the `batch_size` if your data is in the
form of datasets, generators, or `keras.utils.Sequence`
instances (since they generate batches).
epochs: Integer. Number of epochs to train the model.
An epoch is an iteration over the entire `x` and `y`
data provided
(unless the `steps_per_epoch` flag is set to
something other than None).
Note that in conjunction with `initial_epoch`,
`epochs` is to be understood as "final epoch".
The model is not trained for a number of iterations
given by `epochs`, but merely until the epoch
of index `epochs` is reached.
verbose: 'auto', 0, 1, or 2. Verbosity mode.
0 = silent, 1 = progress bar, 2 = one line per epoch.
'auto' becomes 1 for most cases, but 2 when used with
`ParameterServerStrategy`. Note that the progress bar is not
particularly useful when logged to a file, so verbose=2 is
recommended when not running interactively (eg, in a production
environment). Defaults to 'auto'.
callbacks: List of `keras.callbacks.Callback` instances.
List of callbacks to apply during training.
See `tf.keras.callbacks`. Note
`tf.keras.callbacks.ProgbarLogger` and
`tf.keras.callbacks.History` callbacks are created automatically
and need not be passed into `model.fit`.
`tf.keras.callbacks.ProgbarLogger` is created or not based on
`verbose` argument to `model.fit`.
Callbacks with batch-level calls are currently unsupported with
`tf.distribute.experimental.ParameterServerStrategy`, and users
are advised to implement epoch-level calls instead with an
appropriate `steps_per_epoch` value.
validation_split: Float between 0 and 1.
Fraction of the training data to be used as validation data.
The model will set apart this fraction of the training data,
will not train on it, and will evaluate
the loss and any model metrics
on this data at the end of each epoch.
The validation data is selected from the last samples
in the `x` and `y` data provided, before shuffling. This
argument is not supported when `x` is a dataset, generator or
`keras.utils.Sequence` instance.
If both `validation_data` and `validation_split` are provided,
`validation_data` will override `validation_split`.
`validation_split` is not yet supported with
`tf.distribute.experimental.ParameterServerStrategy`.
validation_data: Data on which to evaluate
the loss and any model metrics at the end of each epoch.
The model will not be trained on this data. Thus, note the fact
that the validation loss of data provided using
`validation_split` or `validation_data` is not affected by
regularization layers like noise and dropout.
`validation_data` will override `validation_split`.
`validation_data` could be:
- A tuple `(x_val, y_val)` of Numpy arrays or tensors.
- A tuple `(x_val, y_val, val_sample_weights)` of NumPy
arrays.
- A `tf.data.Dataset`.
- A Python generator or `keras.utils.Sequence` returning
`(inputs, targets)` or `(inputs, targets, sample_weights)`.
`validation_data` is not yet supported with
`tf.distribute.experimental.ParameterServerStrategy`.
shuffle: Boolean (whether to shuffle the training data
before each epoch) or str (for 'batch'). This argument is
ignored when `x` is a generator or an object of tf.data.Dataset.
'batch' is a special option for dealing
with the limitations of HDF5 data; it shuffles in batch-sized
chunks. Has no effect when `steps_per_epoch` is not `None`.
class_weight: Optional dictionary mapping class indices (integers)
to a weight (float) value, used for weighting the loss function
(during training only).
This can be useful to tell the model to
"pay more attention" to samples from
an under-represented class. When `class_weight` is specified
and targets have a rank of 2 or greater, either `y` must be
one-hot encoded, or an explicit final dimension of `1` must
be included for sparse class labels.
sample_weight: Optional Numpy array of weights for
the training samples, used for weighting the loss function
(during training only). You can either pass a flat (1D)
Numpy array with the same length as the input samples
(1:1 mapping between weights and samples),
or in the case of temporal data,
you can pass a 2D array with shape
`(samples, sequence_length)`,
to apply a different weight to every timestep of every sample.
This argument is not supported when `x` is a dataset, generator,
or `keras.utils.Sequence` instance, instead provide the
sample_weights as the third element of `x`.
Note that sample weighting does not apply to metrics specified
via the `metrics` argument in `compile()`. To apply sample
weighting to your metrics, you can specify them via the
`weighted_metrics` in `compile()` instead.
initial_epoch: Integer.
Epoch at which to start training
(useful for resuming a previous training run).
steps_per_epoch: Integer or `None`.
Total number of steps (batches of samples)
before declaring one epoch finished and starting the
next epoch. When training with input tensors such as
TensorFlow data tensors, the default `None` is equal to
the number of samples in your dataset divided by
the batch size, or 1 if that cannot be determined. If x is a
`tf.data` dataset, and 'steps_per_epoch'
is None, the epoch will run until the input dataset is
exhausted. When passing an infinitely repeating dataset, you
must specify the `steps_per_epoch` argument. If
`steps_per_epoch=-1` the training will run indefinitely with an
infinitely repeating dataset. This argument is not supported
with array inputs.
When using `tf.distribute.experimental.ParameterServerStrategy`:
* `steps_per_epoch=None` is not supported.
validation_steps: Only relevant if `validation_data` is provided and
is a `tf.data` dataset. Total number of steps (batches of
samples) to draw before stopping when performing validation
at the end of every epoch. If 'validation_steps' is None,
validation will run until the `validation_data` dataset is
exhausted. In the case of an infinitely repeated dataset, it
will run into an infinite loop. If 'validation_steps' is
specified and only part of the dataset will be consumed, the
evaluation will start from the beginning of the dataset at each
epoch. This ensures that the same validation samples are used
every time.
validation_batch_size: Integer or `None`.
Number of samples per validation batch.
If unspecified, will default to `batch_size`.
Do not specify the `validation_batch_size` if your data is in
the form of datasets, generators, or `keras.utils.Sequence`
instances (since they generate batches).
validation_freq: Only relevant if validation data is provided.
Integer or `collections.abc.Container` instance (e.g. list, tuple,
etc.). If an integer, specifies how many training epochs to run
before a new validation run is performed, e.g. `validation_freq=2`
runs validation every 2 epochs. If a Container, specifies the
epochs on which to run validation, e.g.
`validation_freq=[1, 2, 10]` runs validation at the end of the
1st, 2nd, and 10th epochs.
max_queue_size: Integer. Used for generator or
`keras.utils.Sequence` input only. Maximum size for the generator
queue. If unspecified, `max_queue_size` will default to 10.
workers: Integer. Used for generator or `keras.utils.Sequence` input
only. Maximum number of processes to spin up
when using process-based threading. If unspecified, `workers`
will default to 1.
use_multiprocessing: Boolean. Used for generator or
`keras.utils.Sequence` input only. If `True`, use process-based
threading. If unspecified, `use_multiprocessing` will default to
`False`. Note that because this implementation relies on
multiprocessing, you should not pass non-pickleable arguments to
the generator as they can't be passed easily to children
processes.
Unpacking behavior for iterator-like inputs:
A common pattern is to pass a tf.data.Dataset, generator, or
tf.keras.utils.Sequence to the `x` argument of fit, which will in fact
yield not only features (x) but optionally targets (y) and sample
weights. TF-Keras requires that the output of such iterator-likes be
unambiguous. The iterator should return a tuple of length 1, 2, or 3,
where the optional second and third elements will be used for y and
sample_weight respectively. Any other type provided will be wrapped in
a length one tuple, effectively treating everything as 'x'. When
yielding dicts, they should still adhere to the top-level tuple
structure.
e.g. `({"x0": x0, "x1": x1}, y)`. TF-Keras will not attempt to
separate features, targets, and weights from the keys of a single
dict.
A notable unsupported data type is the namedtuple. The reason is
that it behaves like both an ordered datatype (tuple) and a mapping
datatype (dict). So given a namedtuple of the form:
`namedtuple("example_tuple", ["y", "x"])`
it is ambiguous whether to reverse the order of the elements when
interpreting the value. Even worse is a tuple of the form:
`namedtuple("other_tuple", ["x", "y", "z"])`
where it is unclear if the tuple was intended to be unpacked into x,
y, and sample_weight or passed through as a single element to `x`. As
a result the data processing code will simply raise a ValueError if it
encounters a namedtuple. (Along with instructions to remedy the
issue.)
Returns:
A `History` object. Its `History.history` attribute is
a record of training loss values and metrics values
at successive epochs, as well as validation loss values
and validation metrics values (if applicable).
Raises:
RuntimeError: 1. If the model was never compiled or,
2. If `model.fit` is wrapped in `tf.function`.
ValueError: In case of mismatch between the provided input data
and what the model expects or when the input data is empty.
"""
# Legacy graph support is contained in `training_v1.Model`.
version_utils.disallow_legacy_graph("Model", "fit")
self._assert_compile_was_called()
self._check_call_args("fit")
_disallow_inside_tf_function("fit")
verbose = _get_verbosity(verbose, self.distribute_strategy)
if validation_split and validation_data is None:
# Create the validation data using the training data. Only supported
# for `Tensor` and `NumPy` input.
(
x,
y,
sample_weight,
), validation_data = data_adapter.train_validation_split(
(x, y, sample_weight), validation_split=validation_split
)
if validation_data:
(
val_x,
val_y,
val_sample_weight,
) = data_adapter.unpack_x_y_sample_weight(validation_data)
if self.distribute_strategy._should_use_with_coordinator:
self._cluster_coordinator = (
tf.distribute.experimental.coordinator.ClusterCoordinator(
self.distribute_strategy
)
)
with self.distribute_strategy.scope(), training_utils.RespectCompiledTrainableState( # noqa: E501
self
):
# Creates a `tf.data.Dataset` and handles batch and epoch iteration.
data_handler = data_adapter.get_data_handler(
x=x,
y=y,
sample_weight=sample_weight,
batch_size=batch_size,
steps_per_epoch=steps_per_epoch,
initial_epoch=initial_epoch,
epochs=epochs,
shuffle=shuffle,
class_weight=class_weight,
max_queue_size=max_queue_size,
workers=workers,
use_multiprocessing=use_multiprocessing,
model=self,
steps_per_execution=self._steps_per_execution,
)
# Container that configures and calls `tf.keras.Callback`s.
if not isinstance(callbacks, callbacks_module.CallbackList):
callbacks = callbacks_module.CallbackList(
callbacks,
add_history=True,
add_progbar=verbose != 0,
model=self,
verbose=verbose,
epochs=epochs,
steps=data_handler.inferred_steps,
)
self.stop_training = False
self.train_function = self.make_train_function()
self._train_counter.assign(0)
callbacks.on_train_begin()
training_logs = None
if self.autotune_steps_per_execution:
self._steps_per_execution_tuner.start()
# Handle fault-tolerance for multi-worker.
# TODO(omalleyt): Fix the ordering issues that mean this has to
# happen after `callbacks.on_train_begin`.
steps_per_epoch_inferred = (
steps_per_epoch or data_handler.inferred_steps
)
(
data_handler._initial_epoch,
data_handler._initial_step,
) = self._maybe_load_initial_counters_from_ckpt(
steps_per_epoch_inferred, initial_epoch
)
logs = None
for epoch, iterator in data_handler.enumerate_epochs():
self.reset_metrics()
callbacks.on_epoch_begin(epoch)
with data_handler.catch_stop_iteration():
for step in data_handler.steps():
with tf.profiler.experimental.Trace(
"train",
epoch_num=epoch,
step_num=step,
batch_size=batch_size,
_r=1,
):
callbacks.on_train_batch_begin(step)
tmp_logs = self.train_function(iterator)
if data_handler.should_sync:
context.async_wait()
# No error, now safe to assign to logs.
logs = tmp_logs
end_step = step + data_handler.step_increment
callbacks.on_train_batch_end(end_step, logs)
if self.stop_training:
break
logs = tf_utils.sync_to_numpy_or_python_type(logs)
if logs is None:
raise ValueError(
"Unexpected result of `train_function` "
"(Empty logs). This could be due to issues in input "
"pipeline that resulted in an empty dataset. "
"Otherwise, please use "
"`Model.compile(..., run_eagerly=True)`, or "
"`tf.config.run_functions_eagerly(True)` for more "
"information of where went wrong, or file a "
"issue/bug to `tf.keras`."
)
# Override with model metrics instead of last step logs
logs = self._validate_and_get_metrics_result(logs)
epoch_logs = copy.copy(logs)
# Run validation.
if validation_data and self._should_eval(
epoch, validation_freq
):
if self._pss_evaluation_shards:
self._disallow_exact_eval_with_add_metrics()
# Create data_handler for evaluation and cache it.
if getattr(self, "_eval_data_handler", None) is None:
self._eval_data_handler = data_adapter.get_data_handler(
x=val_x,
y=val_y,
sample_weight=val_sample_weight,
batch_size=validation_batch_size or batch_size,
steps_per_epoch=validation_steps,
initial_epoch=0,
epochs=1,
max_queue_size=max_queue_size,
workers=workers,
use_multiprocessing=use_multiprocessing,
model=self,
steps_per_execution=self._steps_per_execution,
pss_evaluation_shards=self._pss_evaluation_shards,
)
val_logs = self.evaluate(
x=val_x,
y=val_y,
sample_weight=val_sample_weight,
batch_size=validation_batch_size or batch_size,
steps=validation_steps,
callbacks=callbacks,
max_queue_size=max_queue_size,
workers=workers,
use_multiprocessing=use_multiprocessing,
return_dict=True,
_use_cached_eval_dataset=True,
)
val_logs = {
"val_" + name: val for name, val in val_logs.items()
}
epoch_logs.update(val_logs)
callbacks.on_epoch_end(epoch, epoch_logs)
training_logs = epoch_logs
if self.stop_training:
break
if isinstance(self.optimizer, optimizer.Optimizer) and epochs > 0:
self.optimizer.finalize_variable_values(
self.trainable_variables
)
# If eval data_handler exists, delete it after all epochs are done.
if getattr(self, "_eval_data_handler", None) is not None:
del self._eval_data_handler
if self.autotune_steps_per_execution:
self._steps_per_execution_tuner.stop()
callbacks.on_train_end(logs=training_logs)
return self.history
def test_step(self, data):
"""The logic for one evaluation step.
This method can be overridden to support custom evaluation logic.
This method is called by `Model.make_test_function`.
This function should contain the mathematical logic for one step of
evaluation.
This typically includes the forward pass, loss calculation, and metrics
updates.
Configuration details for *how* this logic is run (e.g. `tf.function`
and `tf.distribute.Strategy` settings), should be left to
`Model.make_test_function`, which can also be overridden.
Args:
data: A nested structure of `Tensor`s.
Returns:
A `dict` containing values that will be passed to
`tf.keras.callbacks.CallbackList.on_train_batch_end`. Typically, the
values of the `Model`'s metrics are returned.
"""
x, y, sample_weight = data_adapter.unpack_x_y_sample_weight(data)
y_pred = self(x, training=False)
# Updates stateful loss metrics.
self.compute_loss(x, y, y_pred, sample_weight)
return self.compute_metrics(x, y, y_pred, sample_weight)
def _make_test_function_exact(self):
if getattr(self, "_shard_test_function", None):
return self._shard_test_function
def step_function(batch):
def run_step(data):
# TODO(b/272050910): Use sample_weight for weighted metrics.
x, y, sample_weight = data_adapter.unpack_x_y_sample_weight(
data
)
y_pred = self(x, training=False)
return x, y, y_pred, sample_weight
if self._jit_compile:
run_step = tf.function(
run_step, jit_compile=True, reduce_retracing=True
)
outputs = self.distribute_strategy.run(run_step, args=(batch,))
outputs = reduce_per_replica(
outputs,
self.distribute_strategy,
reduction=self.distribute_reduction_method,
)
return outputs
def shard_test_function(dataset, total_shards, shard_idx):
# Copy loss and metric variables to the worker and work with them
# locally. This ensures each shard function is atomic: if a worker
# is preempted, the intermediate progress is discarded and that
# shard is retried. This in turn guarantees exactly-once visitation.
local_unweighted_metrics, local_weighted_metrics = [], []
with tf_utils.with_metric_local_vars_scope():
# TODO(jmullenbach): implement and use a clone for
# `MetricsContainer` and use its `update_state` method directly.
for metric in self.compiled_metrics.unweighted_metrics:
if metric is not None:
local_unweighted_metrics.append(
base_metric.clone_metric(metric)
)
for metric in self.compiled_metrics.weighted_metrics:
if metric is not None:
local_weighted_metrics.append(
base_metric.clone_metric(metric)
)
local_loss = compile_utils.LossesContainer.from_config(
self.compiled_loss.get_config()
)
dataset = input_ops.auto_shard_dataset(
dataset, total_shards, shard_idx
)
iterator = iter(dataset)
with distribute_utils.cache_variable_reads():
for batch in iterator:
x, y, y_pred, sample_weight = step_function(batch)
for weighted_metric in local_weighted_metrics:
weighted_metric.update_state(y, y_pred, sample_weight)
for unweighted_metric in local_unweighted_metrics:
unweighted_metric.update_state(y, y_pred)
local_loss(y, y_pred, sample_weight)
local_metrics = (
local_unweighted_metrics
+ local_weighted_metrics
+ local_loss.metrics
)
outputs = {metric.name: metric.weights for metric in local_metrics}
with tf.control_dependencies(_minimum_control_deps(outputs)):
self._test_counter.assign_add(1)
return outputs
if not self.run_eagerly:
shard_test_function = tf.function(
shard_test_function, reduce_retracing=True
)
self._shard_test_function = (
lambda *args: self._cluster_coordinator.schedule(
shard_test_function,
args=args,
)
)
return self._shard_test_function
def make_test_function(self, force=False):
"""Creates a function that executes one step of evaluation.
This method can be overridden to support custom evaluation logic.
This method is called by `Model.evaluate` and `Model.test_on_batch`.
Typically, this method directly controls `tf.function` and
`tf.distribute.Strategy` settings, and delegates the actual evaluation
logic to `Model.test_step`.
This function is cached the first time `Model.evaluate` or
`Model.test_on_batch` is called. The cache is cleared whenever
`Model.compile` is called. You can skip the cache and generate again the
function with `force=True`.
Args:
force: Whether to regenerate the test function and skip the cached
function if available.
Returns:
Function. The function created by this method should accept a
`tf.data.Iterator`, and return a `dict` containing values that will
be passed to `tf.keras.Callbacks.on_test_batch_end`.
"""
if self.test_function is not None and not force:
return self.test_function
def step_function(model, iterator):
"""Runs a single evaluation step."""
def run_step(data):
outputs = model.test_step(data)
# Ensure counter is updated only if `test_step` succeeds.
with tf.control_dependencies(_minimum_control_deps(outputs)):
model._test_counter.assign_add(1)
return outputs
if self.jit_compile:
run_step = tf.function(
run_step, jit_compile=True, reduce_retracing=True
)
data = next(iterator)
outputs = model.distribute_strategy.run(run_step, args=(data,))
outputs = reduce_per_replica(
outputs,
self.distribute_strategy,
reduction=self.distribute_reduction_method,
)
return outputs
# Special case if steps_per_execution is one.
if (
self._steps_per_execution is None
or self._steps_per_execution.numpy().item() == 1
and not self.autotune_steps_per_execution
):
def test_function(iterator):
"""Runs a test execution with a single step."""
return step_function(self, iterator)
if not self.run_eagerly:
test_function = tf.function(
test_function, reduce_retracing=True
)
if self._cluster_coordinator:
self.test_function = (
lambda it: self._cluster_coordinator.schedule(
test_function, args=(it,)
)
)
else:
self.test_function = test_function
# If we're using a coordinator, use the value of
# self._steps_per_execution at the time the function is
# called/scheduled, and not when it is actually executed.
elif self._cluster_coordinator:
def test_function(iterator, steps_per_execution):
"""Runs a test execution with multiple steps."""
for _ in tf.range(steps_per_execution):
outputs = step_function(self, iterator)
return outputs
if not self.run_eagerly:
test_function = tf.function(
test_function, reduce_retracing=True
)
self.test_function = lambda it: self._cluster_coordinator.schedule(
test_function, args=(it, self._steps_per_execution.value())
)
else:
def test_function(iterator):
"""Runs a test execution with multiple steps."""
for _ in tf.range(self._steps_per_execution):
outputs = step_function(self, iterator)
return outputs
if not self.run_eagerly:
test_function = tf.function(
test_function, reduce_retracing=True
)
self.test_function = test_function
return self.test_function
@traceback_utils.filter_traceback
def evaluate(
self,
x=None,
y=None,
batch_size=None,
verbose="auto",
sample_weight=None,
steps=None,
callbacks=None,
max_queue_size=10,
workers=1,
use_multiprocessing=False,
return_dict=False,
**kwargs,
):
"""Returns the loss value & metrics values for the model in test mode.
Computation is done in batches (see the `batch_size` arg.)
Args:
x: Input data. It could be:
- A Numpy array (or array-like), or a list of arrays
(in case the model has multiple inputs).
- A TensorFlow tensor, or a list of tensors
(in case the model has multiple inputs).
- A dict mapping input names to the corresponding array/tensors,
if the model has named inputs.
- A `tf.data` dataset. Should return a tuple
of either `(inputs, targets)` or
`(inputs, targets, sample_weights)`.
- A generator or `keras.utils.Sequence` returning `(inputs,
targets)` or `(inputs, targets, sample_weights)`.
A more detailed description of unpacking behavior for iterator
types (Dataset, generator, Sequence) is given in the `Unpacking
behavior for iterator-like inputs` section of `Model.fit`.
y: Target data. Like the input data `x`, it could be either Numpy
array(s) or TensorFlow tensor(s). It should be consistent with `x`
(you cannot have Numpy inputs and tensor targets, or inversely).
If `x` is a dataset, generator or `keras.utils.Sequence` instance,
`y` should not be specified (since targets will be obtained from
the iterator/dataset).
batch_size: Integer or `None`. Number of samples per batch of
computation. If unspecified, `batch_size` will default to 32. Do
not specify the `batch_size` if your data is in the form of a
dataset, generators, or `keras.utils.Sequence` instances (since
they generate batches).
verbose: `"auto"`, 0, 1, or 2. Verbosity mode.
0 = silent, 1 = progress bar, 2 = single line.
`"auto"` becomes 1 for most cases, and to 2 when used with
`ParameterServerStrategy`. Note that the progress bar is not
particularly useful when logged to a file, so `verbose=2` is
recommended when not running interactively (e.g. in a production
environment). Defaults to 'auto'.
sample_weight: Optional Numpy array of weights for the test samples,
used for weighting the loss function. You can either pass a flat
(1D) Numpy array with the same length as the input samples
(1:1 mapping between weights and samples), or in the case of
temporal data, you can pass a 2D array with shape `(samples,
sequence_length)`, to apply a different weight to every
timestep of every sample. This argument is not supported when
`x` is a dataset, instead pass sample weights as the third
element of `x`.
steps: Integer or `None`. Total number of steps (batches of samples)
before declaring the evaluation round finished. Ignored with the
default value of `None`. If x is a `tf.data` dataset and `steps`
is None, 'evaluate' will run until the dataset is exhausted. This
argument is not supported with array inputs.
callbacks: List of `keras.callbacks.Callback` instances. List of
callbacks to apply during evaluation. See
[callbacks](https://www.tensorflow.org/api_docs/python/tf/tf_keras/callbacks).
max_queue_size: Integer. Used for generator or
`keras.utils.Sequence` input only. Maximum size for the generator
queue. If unspecified, `max_queue_size` will default to 10.
workers: Integer. Used for generator or `keras.utils.Sequence` input
only. Maximum number of processes to spin up when using
process-based threading. If unspecified, `workers` will default to
1.
use_multiprocessing: Boolean. Used for generator or
`keras.utils.Sequence` input only. If `True`, use process-based
threading. If unspecified, `use_multiprocessing` will default to
`False`. Note that because this implementation relies on
multiprocessing, you should not pass non-pickleable arguments to
the generator as they can't be passed easily to children
processes.
return_dict: If `True`, loss and metric results are returned as a
dict, with each key being the name of the metric. If `False`, they
are returned as a list.
**kwargs: Unused at this time.
See the discussion of `Unpacking behavior for iterator-like inputs` for
`Model.fit`.
Returns:
Scalar test loss (if the model has a single output and no metrics)
or list of scalars (if the model has multiple outputs
and/or metrics). The attribute `model.metrics_names` will give you
the display labels for the scalar outputs.
Raises:
RuntimeError: If `model.evaluate` is wrapped in a `tf.function`.
"""
version_utils.disallow_legacy_graph("Model", "evaluate")
self._assert_compile_was_called()
self._check_call_args("evaluate")
self._check_sample_weight_warning(x, sample_weight)
_disallow_inside_tf_function("evaluate")
use_cached_eval_dataset = kwargs.pop("_use_cached_eval_dataset", False)
if kwargs:
raise TypeError(f"Invalid keyword arguments: {list(kwargs.keys())}")
if self.distribute_strategy._should_use_with_coordinator:
self._cluster_coordinator = (
tf.distribute.experimental.coordinator.ClusterCoordinator(
self.distribute_strategy
)
)
verbose = _get_verbosity(verbose, self.distribute_strategy)
if self._pss_evaluation_shards:
self._disallow_exact_eval_with_add_metrics()
with self.distribute_strategy.scope():
# Use cached evaluation data only when it's called in `Model.fit`
if (
use_cached_eval_dataset
and getattr(self, "_eval_data_handler", None) is not None
):
data_handler = self._eval_data_handler
else:
# Creates a `tf.data.Dataset` and handles batch and epoch
# iteration.
data_handler = data_adapter.get_data_handler(
x=x,
y=y,
sample_weight=sample_weight,
batch_size=batch_size,
steps_per_epoch=steps,
initial_epoch=0,
epochs=1,
max_queue_size=max_queue_size,
workers=workers,
use_multiprocessing=use_multiprocessing,
model=self,
steps_per_execution=self._steps_per_execution,
pss_evaluation_shards=self._pss_evaluation_shards,
)
# Container that configures and calls `tf.keras.Callback`s.
if not isinstance(callbacks, callbacks_module.CallbackList):
callbacks = callbacks_module.CallbackList(
callbacks,
add_history=True,
add_progbar=verbose != 0,
model=self,
verbose=verbose,
epochs=1,
steps=data_handler.inferred_steps,
)
# Initialize to prevent errors if 0 epochs are evaluated.
logs = {}
test_function_runner = self._get_test_function_runner(callbacks)
self._test_counter.assign(0)
callbacks.on_test_begin()
if self.autotune_steps_per_execution:
self._steps_per_execution_tuner.start()
for (
_,
dataset_or_iterator,
) in data_handler.enumerate_epochs(): # Single epoch.
self.reset_metrics()
with data_handler.catch_stop_iteration():
for step in data_handler.steps():
with tf.profiler.experimental.Trace(
"test", step_num=step, _r=1
):
callbacks.on_test_batch_begin(step)
logs = test_function_runner.run_step(
dataset_or_iterator,
data_handler,
step,
self._pss_evaluation_shards,
)
logs = tf_utils.sync_to_numpy_or_python_type(logs)
# Override with model metrics instead of last step logs
if self._pss_evaluation_shards:
logs = self._aggregate_exact_metrics(logs)
else:
logs = self._validate_and_get_metrics_result(logs)
if self.autotune_steps_per_execution:
self._steps_per_execution_tuner.stop()
callbacks.on_test_end(logs=logs)
if return_dict:
return logs
else:
return flatten_metrics_in_order(logs, self.metrics_names)
def _disallow_exact_eval_with_add_metrics(self):
metrics_from_add_metric = [
metric
for layer in self._flatten_layers()
for metric in layer._metrics
]
compiled_metrics = self.compiled_metrics.metrics
if any(
[
metric not in compiled_metrics
for metric in metrics_from_add_metric
]
):
raise ValueError(
"Detected that a metric was added to this model "
"via `Model.add_metric`. This is not currently "
"supported when using exact evaluation with "
"`tf.distribute.ParameterServerStrategy`."
)
def _infer_exact_eval_shards(self, pss_evaluation_shards):
if not self.distribute_strategy._should_use_with_coordinator:
return 0
if pss_evaluation_shards == "auto":
# TODO(b/264265138) evaluate and improve this heuristic
return self.distribute_strategy._num_workers * 5
return pss_evaluation_shards
def _get_test_function_runner(self, callbacks):
if (
self._pss_evaluation_shards
and self.distribute_strategy._should_use_with_coordinator
):
self.test_function = self._make_test_function_exact()
test_function_runner = _ExactTestFunction(
self.test_function, callbacks
)
else:
self.test_function = self.make_test_function()
test_function_runner = _TestFunction(self.test_function, callbacks)
return test_function_runner
def predict_step(self, data):
"""The logic for one inference step.
This method can be overridden to support custom inference logic.
This method is called by `Model.make_predict_function`.
This method should contain the mathematical logic for one step of
inference. This typically includes the forward pass.
Configuration details for *how* this logic is run (e.g. `tf.function`
and `tf.distribute.Strategy` settings), should be left to
`Model.make_predict_function`, which can also be overridden.
Args:
data: A nested structure of `Tensor`s.
Returns:
The result of one inference step, typically the output of calling the
`Model` on data.
"""
x, _, _ = data_adapter.unpack_x_y_sample_weight(data)
return self(x, training=False)
def make_predict_function(self, force=False):
"""Creates a function that executes one step of inference.
This method can be overridden to support custom inference logic.
This method is called by `Model.predict` and `Model.predict_on_batch`.
Typically, this method directly controls `tf.function` and
`tf.distribute.Strategy` settings, and delegates the actual evaluation
logic to `Model.predict_step`.
This function is cached the first time `Model.predict` or
`Model.predict_on_batch` is called. The cache is cleared whenever
`Model.compile` is called. You can skip the cache and generate again the
function with `force=True`.
Args:
force: Whether to regenerate the predict function and skip the cached
function if available.
Returns:
Function. The function created by this method should accept a
`tf.data.Iterator`, and return the outputs of the `Model`.
"""
if self.predict_function is not None and not force:
return self.predict_function
def step_function(model, iterator):
"""Runs a single evaluation step."""
def run_step(data):
outputs = model.predict_step(data)
# Ensure counter is updated only if `test_step` succeeds.
with tf.control_dependencies(_minimum_control_deps(outputs)):
model._predict_counter.assign_add(1)
return outputs
if self.jit_compile:
run_step = tf.function(
run_step, jit_compile=True, reduce_retracing=True
)
data = next(iterator)
outputs = model.distribute_strategy.run(run_step, args=(data,))
outputs = reduce_per_replica(
outputs, self.distribute_strategy, reduction="concat"
)
return outputs
# Special case if steps_per_execution is one.
if (
self._steps_per_execution is None
or self._steps_per_execution.numpy().item() == 1
and not self.autotune_steps_per_execution
):
def predict_function(iterator):
"""Runs an evaluation execution with a single step."""
return step_function(self, iterator)
else:
def predict_function(iterator):
"""Runs an evaluation execution with multiple steps."""
outputs = step_function(self, iterator)
for _ in tf.range(self._steps_per_execution - 1):
tf.autograph.experimental.set_loop_options(
shape_invariants=[
(
outputs,
tf.nest.map_structure(
lambda t: tf_utils.get_tensor_spec(
t, dynamic_batch=True
).shape,
outputs,
),
)
]
)
step_outputs = step_function(self, iterator)
outputs = tf.nest.map_structure(
lambda t1, t2: concat([t1, t2]), outputs, step_outputs
)
return outputs
if not self.run_eagerly:
predict_function = tf.function(
predict_function, reduce_retracing=True
)
self.predict_function = predict_function
return self.predict_function
@traceback_utils.filter_traceback
def predict(
self,
x,
batch_size=None,
verbose="auto",
steps=None,
callbacks=None,
max_queue_size=10,
workers=1,
use_multiprocessing=False,
):
"""Generates output predictions for the input samples.
Computation is done in batches. This method is designed for batch
processing of large numbers of inputs. It is not intended for use inside
of loops that iterate over your data and process small numbers of inputs
at a time.
For small numbers of inputs that fit in one batch,
directly use `__call__()` for faster execution, e.g.,
`model(x)`, or `model(x, training=False)` if you have layers such as
`tf.keras.layers.BatchNormalization` that behave differently during
inference. You may pair the individual model call with a `tf.function`
for additional performance inside your inner loop.
If you need access to numpy array values instead of tensors after your
model call, you can use `tensor.numpy()` to get the numpy array value of
an eager tensor.
Also, note the fact that test loss is not affected by
regularization layers like noise and dropout.
Note: See [this FAQ entry](
https://keras.io/getting_started/faq/#whats-the-difference-between-model-methods-predict-and-call)
for more details about the difference between `Model` methods
`predict()` and `__call__()`.
Args:
x: Input samples. It could be:
- A Numpy array (or array-like), or a list of arrays
(in case the model has multiple inputs).
- A TensorFlow tensor, or a list of tensors
(in case the model has multiple inputs).
- A `tf.data` dataset.
- A generator or `keras.utils.Sequence` instance.
A more detailed description of unpacking behavior for iterator
types (Dataset, generator, Sequence) is given in the `Unpacking
behavior for iterator-like inputs` section of `Model.fit`.
batch_size: Integer or `None`.
Number of samples per batch.
If unspecified, `batch_size` will default to 32.
Do not specify the `batch_size` if your data is in the
form of dataset, generators, or `keras.utils.Sequence` instances
(since they generate batches).
verbose: `"auto"`, 0, 1, or 2. Verbosity mode.
0 = silent, 1 = progress bar, 2 = single line.
`"auto"` becomes 1 for most cases, and to 2 when used with
`ParameterServerStrategy`. Note that the progress bar is not
particularly useful when logged to a file, so `verbose=2` is
recommended when not running interactively (e.g. in a production
environment). Defaults to 'auto'.
steps: Total number of steps (batches of samples)
before declaring the prediction round finished.
Ignored with the default value of `None`. If x is a `tf.data`
dataset and `steps` is None, `predict()` will
run until the input dataset is exhausted.
callbacks: List of `keras.callbacks.Callback` instances.
List of callbacks to apply during prediction.
See [callbacks](
https://www.tensorflow.org/api_docs/python/tf/tf_keras/callbacks).
max_queue_size: Integer. Used for generator or
`keras.utils.Sequence` input only. Maximum size for the
generator queue. If unspecified, `max_queue_size` will default
to 10.
workers: Integer. Used for generator or `keras.utils.Sequence` input
only. Maximum number of processes to spin up when using
process-based threading. If unspecified, `workers` will default
to 1.
use_multiprocessing: Boolean. Used for generator or
`keras.utils.Sequence` input only. If `True`, use process-based
threading. If unspecified, `use_multiprocessing` will default to
`False`. Note that because this implementation relies on
multiprocessing, you should not pass non-pickleable arguments to
the generator as they can't be passed easily to children
processes.
See the discussion of `Unpacking behavior for iterator-like inputs` for
`Model.fit`. Note that Model.predict uses the same interpretation rules
as `Model.fit` and `Model.evaluate`, so inputs must be unambiguous for
all three methods.
Returns:
Numpy array(s) of predictions.
Raises:
RuntimeError: If `model.predict` is wrapped in a `tf.function`.
ValueError: In case of mismatch between the provided
input data and the model's expectations,
or in case a stateful model receives a number of samples
that is not a multiple of the batch size.
"""
version_utils.disallow_legacy_graph("Model", "predict")
self._check_call_args("predict")
_disallow_inside_tf_function("predict")
# TODO(yashkatariya): Cache model on the coordinator for faster
# prediction. If running under PSS, then swap it with OneDeviceStrategy
# so that execution will run on the coordinator.
original_pss_strategy = None
if self.distribute_strategy._should_use_with_coordinator:
original_pss_strategy = self.distribute_strategy
self._distribution_strategy = None
# Cluster coordinator is set by `.fit()` and `.evaluate()` which is not
# needed in `.predict()` because all the predictions happen on the
# coordinator/locally.
if self._cluster_coordinator:
self._cluster_coordinator = None
verbose = _get_verbosity(verbose, self.distribute_strategy)
outputs = None
with self.distribute_strategy.scope():
# Creates a `tf.data.Dataset` and handles batch and epoch iteration.
dataset_types = (tf.compat.v1.data.Dataset, tf.data.Dataset)
if (
self._in_multi_worker_mode()
or _is_tpu_multi_host(self.distribute_strategy)
) and isinstance(x, dataset_types):
try:
options = tf.data.Options()
data_option = tf.data.experimental.AutoShardPolicy.DATA
options.experimental_distribute.auto_shard_policy = (
data_option
)
x = x.with_options(options)
except ValueError:
warnings.warn(
"Using Model.predict with MultiWorkerMirroredStrategy "
"or TPUStrategy and AutoShardPolicy.FILE might lead to "
"out-of-order result. Consider setting it to "
"AutoShardPolicy.DATA.",
stacklevel=2,
)
data_handler = data_adapter.get_data_handler(
x=x,
batch_size=batch_size,
steps_per_epoch=steps,
initial_epoch=0,
epochs=1,
max_queue_size=max_queue_size,
workers=workers,
use_multiprocessing=use_multiprocessing,
model=self,
steps_per_execution=self._steps_per_execution,
)
# Container that configures and calls `tf.keras.Callback`s.
if not isinstance(callbacks, callbacks_module.CallbackList):
callbacks = callbacks_module.CallbackList(
callbacks,
add_history=True,
add_progbar=verbose != 0,
model=self,
verbose=verbose,
epochs=1,
steps=data_handler.inferred_steps,
)
self.predict_function = self.make_predict_function()
self._predict_counter.assign(0)
callbacks.on_predict_begin()
if self.autotune_steps_per_execution:
self._steps_per_execution_tuner.start()
batch_outputs = None
for _, iterator in data_handler.enumerate_epochs(): # Single epoch.
with data_handler.catch_stop_iteration():
for step in data_handler.steps():
callbacks.on_predict_batch_begin(step)
tmp_batch_outputs = self.predict_function(iterator)
if data_handler.should_sync:
context.async_wait()
batch_outputs = (
tmp_batch_outputs # No error, now safe to assign.
)
if outputs is None:
outputs = tf.nest.map_structure(
lambda batch_output: [batch_output],
batch_outputs,
)
else:
tf.__internal__.nest.map_structure_up_to(
batch_outputs,
lambda output, batch_output: output.append(
batch_output
),
outputs,
batch_outputs,
)
end_step = step + data_handler.step_increment
callbacks.on_predict_batch_end(
end_step, {"outputs": batch_outputs}
)
if batch_outputs is None:
raise ValueError(
"Unexpected result of `predict_function` "
"(Empty batch_outputs). Please use "
"`Model.compile(..., run_eagerly=True)`, or "
"`tf.config.run_functions_eagerly(True)` for more "
"information of where went wrong, or file a "
"issue/bug to `tf.keras`."
)
if self.autotune_steps_per_execution:
self._steps_per_execution_tuner.stop()
callbacks.on_predict_end()
all_outputs = tf.__internal__.nest.map_structure_up_to(
batch_outputs, potentially_ragged_concat, outputs
)
# If originally PSS strategy was used, then replace it back since
# predict is running under `OneDeviceStrategy` after the swap and once
# its done we need to replace it back to PSS again.
if original_pss_strategy is not None:
self._distribution_strategy = original_pss_strategy
return tf_utils.sync_to_numpy_or_python_type(all_outputs)
def reset_metrics(self):
"""Resets the state of all the metrics in the model.
Examples:
>>> inputs = tf.keras.layers.Input(shape=(3,))
>>> outputs = tf.keras.layers.Dense(2)(inputs)
>>> model = tf.keras.models.Model(inputs=inputs, outputs=outputs)
>>> model.compile(optimizer="Adam", loss="mse", metrics=["mae"])
>>> x = np.random.random((2, 3))
>>> y = np.random.randint(0, 2, (2, 2))
>>> _ = model.fit(x, y, verbose=0)
>>> assert all(float(m.result()) for m in model.metrics)
>>> model.reset_metrics()
>>> assert all(float(m.result()) == 0 for m in model.metrics)
"""
for m in self.metrics:
m.reset_state()
def train_on_batch(
self,
x,
y=None,
sample_weight=None,
class_weight=None,
reset_metrics=True,
return_dict=False,
):
"""Runs a single gradient update on a single batch of data.
Args:
x: Input data. It could be:
- A Numpy array (or array-like), or a list of arrays
(in case the model has multiple inputs).
- A TensorFlow tensor, or a list of tensors
(in case the model has multiple inputs).
- A dict mapping input names to the corresponding array/tensors,
if the model has named inputs.
y: Target data. Like the input data `x`, it could be either Numpy
array(s) or TensorFlow tensor(s).
sample_weight: Optional array of the same length as x, containing
weights to apply to the model's loss for each sample. In the case
of temporal data, you can pass a 2D array with shape (samples,
sequence_length), to apply a different weight to every timestep of
every sample.
class_weight: Optional dictionary mapping class indices (integers)
to a weight (float) to apply to the model's loss for the samples
from this class during training. This can be useful to tell the
model to "pay more attention" to samples from an under-represented
class. When `class_weight` is specified and targets have a rank of
2 or greater, either `y` must be one-hot encoded, or an explicit
final dimension of `1` must be included for sparse class labels.
reset_metrics: If `True`, the metrics returned will be only for this
batch. If `False`, the metrics will be statefully accumulated
across batches.
return_dict: If `True`, loss and metric results are returned as a
dict, with each key being the name of the metric. If `False`, they
are returned as a list.
Returns:
Scalar training loss
(if the model has a single output and no metrics)
or list of scalars (if the model has multiple outputs
and/or metrics). The attribute `model.metrics_names` will give you
the display labels for the scalar outputs.
Raises:
RuntimeError: If `model.train_on_batch` is wrapped in a `tf.function`.
"""
self._assert_compile_was_called()
self._check_call_args("train_on_batch")
_disallow_inside_tf_function("train_on_batch")
if reset_metrics:
self.reset_metrics()
with self.distribute_strategy.scope(), training_utils.RespectCompiledTrainableState( # noqa: E501
self
):
iterator = data_adapter.single_batch_iterator(
self.distribute_strategy, x, y, sample_weight, class_weight
)
self.train_function = self.make_train_function()
logs = self.train_function(iterator)
logs = tf_utils.sync_to_numpy_or_python_type(logs)
if return_dict:
return logs
else:
return flatten_metrics_in_order(logs, self.metrics_names)
def test_on_batch(
self,
x,
y=None,
sample_weight=None,
reset_metrics=True,
return_dict=False,
):
"""Test the model on a single batch of samples.
Args:
x: Input data. It could be:
- A Numpy array (or array-like), or a list of arrays (in case the
model has multiple inputs).
- A TensorFlow tensor, or a list of tensors (in case the model has
multiple inputs).
- A dict mapping input names to the corresponding array/tensors,
if the model has named inputs.
y: Target data. Like the input data `x`, it could be either Numpy
array(s) or TensorFlow tensor(s). It should be consistent with `x`
(you cannot have Numpy inputs and tensor targets, or inversely).
sample_weight: Optional array of the same length as x, containing
weights to apply to the model's loss for each sample. In the case
of temporal data, you can pass a 2D array with shape (samples,
sequence_length), to apply a different weight to every timestep of
every sample.
reset_metrics: If `True`, the metrics returned will be only for this
batch. If `False`, the metrics will be statefully accumulated
across batches.
return_dict: If `True`, loss and metric results are returned as a
dict, with each key being the name of the metric. If `False`, they
are returned as a list.
Returns:
Scalar test loss (if the model has a single output and no metrics)
or list of scalars (if the model has multiple outputs
and/or metrics). The attribute `model.metrics_names` will give you
the display labels for the scalar outputs.
Raises:
RuntimeError: If `model.test_on_batch` is wrapped in a
`tf.function`.
"""
self._assert_compile_was_called()
self._check_call_args("test_on_batch")
_disallow_inside_tf_function("test_on_batch")
if reset_metrics:
self.reset_metrics()
with self.distribute_strategy.scope():
iterator = data_adapter.single_batch_iterator(
self.distribute_strategy, x, y, sample_weight
)
self.test_function = self.make_test_function()
logs = self.test_function(iterator)
logs = tf_utils.sync_to_numpy_or_python_type(logs)
if return_dict:
return logs
else:
return flatten_metrics_in_order(logs, self.metrics_names)
def predict_on_batch(self, x):
"""Returns predictions for a single batch of samples.
Args:
x: Input data. It could be:
- A Numpy array (or array-like), or a list of arrays (in case the
model has multiple inputs).
- A TensorFlow tensor, or a list of tensors (in case the model has
multiple inputs).
Returns:
Numpy array(s) of predictions.
Raises:
RuntimeError: If `model.predict_on_batch` is wrapped in a
`tf.function`.
"""
self._check_call_args("predict_on_batch")
_disallow_inside_tf_function("predict_on_batch")
with self.distribute_strategy.scope():
iterator = data_adapter.single_batch_iterator(
self.distribute_strategy, x
)
self.predict_function = self.make_predict_function()
outputs = self.predict_function(iterator)
return tf_utils.sync_to_numpy_or_python_type(outputs)
@doc_controls.do_not_generate_docs
def fit_generator(
self,
generator,
steps_per_epoch=None,
epochs=1,
verbose=1,
callbacks=None,
validation_data=None,
validation_steps=None,
validation_freq=1,
class_weight=None,
max_queue_size=10,
workers=1,
use_multiprocessing=False,
shuffle=True,
initial_epoch=0,
):
"""Fits the model on data yielded batch-by-batch by a Python generator.
DEPRECATED:
`Model.fit` now supports generators, so there is no longer any need to
use this endpoint.
"""
warnings.warn(
"`Model.fit_generator` is deprecated and "
"will be removed in a future version. "
"Please use `Model.fit`, which supports generators.",
stacklevel=2,
)
return self.fit(
generator,
steps_per_epoch=steps_per_epoch,
epochs=epochs,
verbose=verbose,
callbacks=callbacks,
validation_data=validation_data,
validation_steps=validation_steps,
validation_freq=validation_freq,
class_weight=class_weight,
max_queue_size=max_queue_size,
workers=workers,
use_multiprocessing=use_multiprocessing,
shuffle=shuffle,
initial_epoch=initial_epoch,
)
@doc_controls.do_not_generate_docs
def evaluate_generator(
self,
generator,
steps=None,
callbacks=None,
max_queue_size=10,
workers=1,
use_multiprocessing=False,
verbose=0,
):
"""Evaluates the model on a data generator.
DEPRECATED:
`Model.evaluate` now supports generators, so there is no longer any
need to use this endpoint.
"""
warnings.warn(
"`Model.evaluate_generator` is deprecated and "
"will be removed in a future version. "
"Please use `Model.evaluate`, which supports generators.",
stacklevel=2,
)
self._check_call_args("evaluate_generator")
return self.evaluate(
generator,
steps=steps,
max_queue_size=max_queue_size,
workers=workers,
use_multiprocessing=use_multiprocessing,
verbose=verbose,
callbacks=callbacks,
)
@doc_controls.do_not_generate_docs
def predict_generator(
self,
generator,
steps=None,
callbacks=None,
max_queue_size=10,
workers=1,
use_multiprocessing=False,
verbose=0,
):
"""Generates predictions for the input samples from a data generator.
DEPRECATED:
`Model.predict` now supports generators, so there is no longer any
need to use this endpoint.
"""
warnings.warn(
"`Model.predict_generator` is deprecated and "
"will be removed in a future version. "
"Please use `Model.predict`, which supports generators.",
stacklevel=2,
)
return self.predict(
generator,
steps=steps,
max_queue_size=max_queue_size,
workers=workers,
use_multiprocessing=use_multiprocessing,
verbose=verbose,
callbacks=callbacks,
)
######################################################################
# Functions below are not training related. They are for model weights
# tracking, save/load, serialization, etc.
######################################################################
@property
def trainable_weights(self):
self._assert_weights_created()
if not self._trainable:
return []
trainable_variables = []
for trackable_obj in self._self_tracked_trackables:
trainable_variables += trackable_obj.trainable_variables
trainable_variables += self._trainable_weights
return self._dedup_weights(trainable_variables)
@property
def non_trainable_weights(self):
self._assert_weights_created()
non_trainable_variables = []
for trackable_obj in self._self_tracked_trackables:
non_trainable_variables += trackable_obj.non_trainable_variables
if not self._trainable:
# Return order is all trainable vars, then all non-trainable vars.
trainable_variables = []
for trackable_obj in self._self_tracked_trackables:
trainable_variables += trackable_obj.trainable_variables
non_trainable_variables = (
trainable_variables
+ self._trainable_weights
+ non_trainable_variables
+ self._non_trainable_weights
)
else:
non_trainable_variables = (
non_trainable_variables + self._non_trainable_weights
)
return self._dedup_weights(non_trainable_variables)
def get_weights(self):
"""Retrieves the weights of the model.
Returns:
A flat list of Numpy arrays.
"""
with self.distribute_strategy.scope():
return super().get_weights()
@traceback_utils.filter_traceback
def save(self, filepath, overwrite=True, save_format=None, **kwargs):
"""Saves a model as a TensorFlow SavedModel or HDF5 file.
See the [Serialization and Saving guide](
https://keras.io/guides/serialization_and_saving/) for details.
Args:
model: TF-Keras model instance to be saved.
filepath: `str` or `pathlib.Path` object. Path where to save the
model.
overwrite: Whether we should overwrite any existing model at the
target location, or instead ask the user via an interactive
prompt.
save_format: Either `"keras"`, `"tf"`, `"h5"`,
indicating whether to save the model
in the native TF-Keras format (`.keras`),
in the TensorFlow SavedModel format
(referred to as "SavedModel" below),
or in the legacy HDF5 format (`.h5`).
Defaults to `"tf"` in TF 2.X, and `"h5"` in TF 1.X.
SavedModel format arguments:
include_optimizer: Only applied to SavedModel and legacy HDF5
formats. If False, do not save the optimizer state.
Defaults to `True`.
signatures: Only applies to SavedModel format. Signatures to save
with the SavedModel. See the `signatures` argument in
`tf.saved_model.save` for details.
options: Only applies to SavedModel format.
`tf.saved_model.SaveOptions` object that specifies SavedModel
saving options.
save_traces: Only applies to SavedModel format. When enabled, the
SavedModel will store the function traces for each layer. This
can be disabled, so that only the configs of each layer are
stored. Defaults to `True`.
Disabling this will decrease serialization time
and reduce file size, but it requires that all custom
layers/models implement a `get_config()` method.
Example:
```python
model = tf.keras.Sequential([
tf.keras.layers.Dense(5, input_shape=(3,)),
tf.keras.layers.Softmax()])
model.save("model.keras")
loaded_model = tf.keras.models.load_model("model.keras")
x = tf.random.uniform((10, 3))
assert np.allclose(model.predict(x), loaded_model.predict(x))
```
Note that `model.save()` is an alias for `tf.keras.models.save_model()`.
"""
saving_api.save_model(
self,
filepath=filepath,
overwrite=overwrite,
save_format=save_format,
**kwargs,
)
@traceback_utils.filter_traceback
def save_weights(
self, filepath, overwrite=True, save_format=None, options=None
):
"""Saves all layer weights.
Either saves in HDF5 or in TensorFlow format based on the `save_format`
argument.
When saving in HDF5 format, the weight file has:
- `layer_names` (attribute), a list of strings
(ordered names of model layers).
- For every layer, a `group` named `layer.name`
- For every such layer group, a group attribute `weight_names`,
a list of strings
(ordered names of weights tensor of the layer).
- For every weight in the layer, a dataset
storing the weight value, named after the weight tensor.
When saving in TensorFlow format, all objects referenced by the network
are saved in the same format as `tf.train.Checkpoint`, including any
`Layer` instances or `Optimizer` instances assigned to object
attributes. For networks constructed from inputs and outputs using
`tf.keras.Model(inputs, outputs)`, `Layer` instances used by the network
are tracked/saved automatically. For user-defined classes which inherit
from `tf.keras.Model`, `Layer` instances must be assigned to object
attributes, typically in the constructor. See the documentation of
`tf.train.Checkpoint` and `tf.keras.Model` for details.
While the formats are the same, do not mix `save_weights` and
`tf.train.Checkpoint`. Checkpoints saved by `Model.save_weights` should
be loaded using `Model.load_weights`. Checkpoints saved using
`tf.train.Checkpoint.save` should be restored using the corresponding
`tf.train.Checkpoint.restore`. Prefer `tf.train.Checkpoint` over
`save_weights` for training checkpoints.
The TensorFlow format matches objects and variables by starting at a
root object, `self` for `save_weights`, and greedily matching attribute
names. For `Model.save` this is the `Model`, and for `Checkpoint.save`
this is the `Checkpoint` even if the `Checkpoint` has a model attached.
This means saving a `tf.keras.Model` using `save_weights` and loading
into a `tf.train.Checkpoint` with a `Model` attached (or vice versa)
will not match the `Model`'s variables. See the
[guide to training checkpoints](
https://www.tensorflow.org/guide/checkpoint) for details on
the TensorFlow format.
Args:
filepath: String or PathLike, path to the file to save the weights
to. When saving in TensorFlow format, this is the prefix used
for checkpoint files (multiple files are generated). Note that
the '.h5' suffix causes weights to be saved in HDF5 format.
overwrite: Whether to silently overwrite any existing file at the
target location, or provide the user with a manual prompt.
save_format: Either 'tf' or 'h5'. A `filepath` ending in '.h5' or
'.keras' will default to HDF5 if `save_format` is `None`.
Otherwise, `None` becomes 'tf'. Defaults to `None`.
options: Optional `tf.train.CheckpointOptions` object that specifies
options for saving weights.
Raises:
ImportError: If `h5py` is not available when attempting to save in
HDF5 format.
"""
saving_api.save_weights(
self,
filepath=filepath,
overwrite=overwrite,
save_format=save_format,
options=options,
)
@traceback_utils.filter_traceback
def load_weights(
self, filepath, skip_mismatch=False, by_name=False, options=None
):
"""Loads all layer weights from a saved files.
The saved file could be a SavedModel file, a `.keras` file (v3 saving
format), or a file created via `model.save_weights()`.
By default, weights are loaded based on the network's
topology. This means the architecture should be the same as when the
weights were saved. Note that layers that don't have weights are not
taken into account in the topological ordering, so adding or removing
layers is fine as long as they don't have weights.
**Partial weight loading**
If you have modified your model, for instance by adding a new layer
(with weights) or by changing the shape of the weights of a layer,
you can choose to ignore errors and continue loading
by setting `skip_mismatch=True`. In this case any layer with
mismatching weights will be skipped. A warning will be displayed
for each skipped layer.
**Weight loading by name**
If your weights are saved as a `.h5` file created
via `model.save_weights()`, you can use the argument `by_name=True`.
In this case, weights are loaded into layers only if they share
the same name. This is useful for fine-tuning or transfer-learning
models where some of the layers have changed.
Note that only topological loading (`by_name=False`) is supported when
loading weights from the `.keras` v3 format or from the TensorFlow
SavedModel format.
Args:
filepath: String, path to the weights file to load. For weight files
in TensorFlow format, this is the file prefix (the same as was
passed to `save_weights()`). This can also be a path to a
SavedModel or a `.keras` file (v3 saving format) saved
via `model.save()`.
skip_mismatch: Boolean, whether to skip loading of layers where
there is a mismatch in the number of weights, or a mismatch in
the shape of the weights.
by_name: Boolean, whether to load weights by name or by topological
order. Only topological loading is supported for weight files in
the `.keras` v3 format or in the TensorFlow SavedModel format.
options: Optional `tf.train.CheckpointOptions` object that specifies
options for loading weights (only valid for a SavedModel file).
"""
return saving_api.load_weights(
self,
filepath=filepath,
by_name=by_name,
skip_mismatch=skip_mismatch,
options=options,
)
def _updated_config(self):
"""Util shared between different serialization methods.
Returns:
Model config with TF-Keras version information added.
"""
from tf_keras import __version__ as keras_version
config = self.get_config()
model_config = {
"class_name": self.__class__.__name__,
"config": config,
"keras_version": keras_version,
"backend": backend.backend(),
}
return model_config
@generic_utils.default
def get_config(self):
"""Returns the config of the `Model`.
Config is a Python dictionary (serializable) containing the
configuration of an object, which in this case is a `Model`. This allows
the `Model` to be be reinstantiated later (without its trained weights)
from this configuration.
Note that `get_config()` does not guarantee to return a fresh copy of
dict every time it is called. The callers should make a copy of the
returned dict if they want to modify it.
Developers of subclassed `Model` are advised to override this method,
and continue to update the dict from `super(MyModel, self).get_config()`
to provide the proper configuration of this `Model`. The default config
will return config dict for init parameters if they are basic types.
Raises `NotImplementedError` when in cases where a custom
`get_config()` implementation is required for the subclassed model.
Returns:
Python dictionary containing the configuration of this `Model`.
"""
# If sublcass doesn't implement `get_config()` parse from init args
# otherwise default to empty dict
if generic_utils.is_default(self.get_config):
try:
config = base_layer.Layer.get_config(self)
except NotImplementedError:
config = {}
logging.warning(
"Model's `__init__()` arguments contain non-serializable "
"objects. Please implement a `get_config()` method in the "
"subclassed Model for proper saving and loading. "
"Defaulting to empty config."
)
else:
config = {}
return config
@classmethod
def from_config(cls, config, custom_objects=None):
# `from_config` assumes `cls` is either `Functional` or a child class of
# `Functional`. In the case that `cls` is meant to behave like a child
# class of `Functional` but only inherits from the `Model` class, we
# have to call `cls(...)` instead of `Functional.from_config`.
from tf_keras.engine import functional
with serialization.SharedObjectLoadingScope():
functional_config_keys = [
"name",
"layers",
"input_layers",
"output_layers",
]
is_functional_config = all(
key in config for key in functional_config_keys
)
argspec = tf_inspect.getfullargspec(cls.__init__)
functional_init_args = tf_inspect.getfullargspec(
functional.Functional.__init__
).args[1:]
revivable_as_functional = (
cls in {functional.Functional, Model}
or argspec.args[1:] == functional_init_args
or (argspec.varargs == "args" and argspec.varkw == "kwargs")
)
if is_functional_config and revivable_as_functional:
# Revive Functional model
# (but not Functional subclasses with a custom __init__)
inputs, outputs, layers = functional.reconstruct_from_config(
config, custom_objects
)
model = cls(
inputs=inputs, outputs=outputs, name=config.get("name")
)
functional.connect_ancillary_layers(model, layers)
else:
# Either the model has a custom __init__, or the config
# does not contain all the information necessary to
# revive a Functional model. This happens when the user creates
# subclassed models where `get_config()` is returning
# insufficient information to be considered a Functional model.
# In this case, we fall back to provide all config into the
# constructor of the class.
try:
model = cls(**config)
except TypeError as e:
raise TypeError(
"Unable to revive model from config. When overriding "
"the `get_config()` method, make sure that the "
"returned config contains all items used as arguments "
f"in the constructor to {cls}, "
"which is the default behavior. "
"You can override this default behavior by defining a "
"`from_config(cls, config)` class method to specify "
"how to create an "
f"instance of {cls.__name__} from its config.\n\n"
f"Received config={config}\n\n"
f"Error encountered during deserialization: {e}"
)
return model
def to_json(self, **kwargs):
"""Returns a JSON string containing the network configuration.
To load a network from a JSON save file, use
`keras.models.model_from_json(json_string, custom_objects={})`.
Args:
**kwargs: Additional keyword arguments to be passed to
*`json.dumps()`.
Returns:
A JSON string.
"""
model_config = self._updated_config()
return json.dumps(
model_config, default=json_utils.get_json_type, **kwargs
)
def to_yaml(self, **kwargs):
"""Returns a yaml string containing the network configuration.
Note: Since TF 2.6, this method is no longer supported and will raise a
RuntimeError.
To load a network from a yaml save file, use
`keras.models.model_from_yaml(yaml_string, custom_objects={})`.
`custom_objects` should be a dictionary mapping
the names of custom losses / layers / etc to the corresponding
functions / classes.
Args:
**kwargs: Additional keyword arguments
to be passed to `yaml.dump()`.
Returns:
A YAML string.
Raises:
RuntimeError: announces that the method poses a security risk
"""
raise RuntimeError(
"Method `model.to_yaml()` has been removed due to security risk of "
"arbitrary code execution. Please use `model.to_json()` instead."
)
def reset_states(self):
for layer in self.layers:
if hasattr(layer, "reset_states") and getattr(
layer, "stateful", False
):
layer.reset_states()
@property
@doc_controls.do_not_generate_docs
def state_updates(self):
"""Deprecated, do NOT use!
Returns the `updates` from all layers that are stateful.
This is useful for separating training updates and
state updates, e.g. when we need to update a layer's internal state
during prediction.
Returns:
A list of update ops.
"""
warnings.warn(
"`Model.state_updates` will be removed in a future version. "
"This property should not be used in TensorFlow 2.0, "
"as `updates` are applied automatically.",
stacklevel=2,
)
state_updates = []
for layer in self.layers:
if getattr(layer, "stateful", False):
if hasattr(layer, "updates"):
state_updates += layer.updates
return state_updates
@property
def weights(self):
"""Returns the list of all layer variables/weights.
Note: This will not track the weights of nested `tf.Modules` that are
not themselves TF-Keras layers.
Returns:
A list of variables.
"""
return self._dedup_weights(self._undeduplicated_weights)
@property
def _undeduplicated_weights(self):
"""Returns the undeduplicated list of all layer variables/weights."""
self._assert_weights_created()
weights = []
for layer in self._self_tracked_trackables:
weights += layer.variables
weights += self._trainable_weights + self._non_trainable_weights
return weights
def summary(
self,
line_length=None,
positions=None,
print_fn=None,
expand_nested=False,
show_trainable=False,
layer_range=None,
):
"""Prints a string summary of the network.
Args:
line_length: Total length of printed lines
(e.g. set this to adapt the display to different
terminal window sizes).
positions: Relative or absolute positions of log elements
in each line. If not provided, becomes
`[0.3, 0.6, 0.70, 1.]`. Defaults to `None`.
print_fn: Print function to use. By default, prints to `stdout`.
If `stdout` doesn't work in your environment, change to `print`.
It will be called on each line of the summary.
You can set it to a custom function
in order to capture the string summary.
expand_nested: Whether to expand the nested models.
Defaults to `False`.
show_trainable: Whether to show if a layer is trainable.
Defaults to `False`.
layer_range: a list or tuple of 2 strings,
which is the starting layer name and ending layer name
(both inclusive) indicating the range of layers to be printed
in summary. It also accepts regex patterns instead of exact
name. In such case, start predicate will be the first element
it matches to `layer_range[0]` and the end predicate will be
the last element it matches to `layer_range[1]`.
By default `None` which considers all layers of model.
Raises:
ValueError: if `summary()` is called before the model is built.
"""
if not self.built:
raise ValueError(
"This model has not yet been built. "
"Build the model first by calling `build()` or by calling "
"the model on a batch of data."
)
layer_utils.print_summary(
self,
line_length=line_length,
positions=positions,
print_fn=print_fn,
expand_nested=expand_nested,
show_trainable=show_trainable,
layer_range=layer_range,
)
@property
def layers(self):
return list(self._flatten_layers(include_self=False, recursive=False))
@layers.setter
def layers(self, _):
raise AttributeError(
"`Model.layers` attribute is reserved and should not be used. "
"Please use another name."
)
def get_layer(self, name=None, index=None):
"""Retrieves a layer based on either its name (unique) or index.
If `name` and `index` are both provided, `index` will take precedence.
Indices are based on order of horizontal graph traversal (bottom-up).
Args:
name: String, name of layer.
index: Integer, index of layer.
Returns:
A layer instance.
"""
# TODO(fchollet): We could build a dictionary based on layer names
# since they are constant, but we have not done that yet.
if index is not None and name is not None:
raise ValueError(
"Provide only a layer name or a layer index. Received: "
f"index={index}, name={name}."
)
if index is not None:
if len(self.layers) <= index:
raise ValueError(
f"Was asked to retrieve layer at index {index}"
f" but model only has {len(self.layers)}"
" layers."
)
else:
return self.layers[index]
if name is not None:
for layer in self.layers:
if layer.name == name:
return layer
raise ValueError(
f"No such layer: {name}. Existing layers are: "
f"{list(layer.name for layer in self.layers)}."
)
raise ValueError(
"Provide either a layer name or layer index at `get_layer`."
)
def get_weight_paths(self):
"""Retrieve all the variables and their paths for the model.
The variable path (string) is a stable key to identify a `tf.Variable`
instance owned by the model. It can be used to specify variable-specific
configurations (e.g. DTensor, quantization) from a global view.
This method returns a dict with weight object paths as keys
and the corresponding `tf.Variable` instances as values.
Note that if the model is a subclassed model and the weights haven't
been initialized, an empty dict will be returned.
Returns:
A dict where keys are variable paths and values are `tf.Variable`
instances.
Example:
```python
class SubclassModel(tf.keras.Model):
def __init__(self, name=None):
super().__init__(name=name)
self.d1 = tf.keras.layers.Dense(10)
self.d2 = tf.keras.layers.Dense(20)
def call(self, inputs):
x = self.d1(inputs)
return self.d2(x)
model = SubclassModel()
model(tf.zeros((10, 10)))
weight_paths = model.get_weight_paths()
# weight_paths:
# {
# 'd1.kernel': model.d1.kernel,
# 'd1.bias': model.d1.bias,
# 'd2.kernel': model.d2.kernel,
# 'd2.bias': model.d2.bias,
# }
# Functional model
inputs = tf.keras.Input((10,), batch_size=10)
x = tf.keras.layers.Dense(20, name='d1')(inputs)
output = tf.keras.layers.Dense(30, name='d2')(x)
model = tf.keras.Model(inputs, output)
d1 = model.layers[1]
d2 = model.layers[2]
weight_paths = model.get_weight_paths()
# weight_paths:
# {
# 'd1.kernel': d1.kernel,
# 'd1.bias': d1.bias,
# 'd2.kernel': d2.kernel,
# 'd2.bias': d2.bias,
# }
```
"""
result = {}
(
descendants,
object_paths_dict,
) = tf.__internal__.tracking.ObjectGraphView(
self
).breadth_first_traversal()
for descendant in descendants:
if isinstance(descendant, tf.Variable):
trackable_references = object_paths_dict[descendant]
object_path = ".".join([t.name for t in trackable_references])
result[object_path] = descendant
return result
def get_compile_config(self):
"""Returns a serialized config with information for compiling the model.
This method returns a config dictionary containing all the information
(optimizer, loss, metrics, etc.) with which the model was compiled.
Returns:
A dict containing information for compiling the model.
"""
if self._is_compiled and hasattr(self, "_compile_config"):
return self._compile_config.serialize()
def compile_from_config(self, config):
"""Compiles the model with the information given in config.
This method uses the information in the config (optimizer, loss,
metrics, etc.) to compile the model.
Args:
config: Dict containing information for compiling the model.
"""
has_overridden_compile = self.__class__.compile != Model.compile
if has_overridden_compile:
logging.warning(
"`compile()` was not called as part of model loading "
"because the model's `compile()` method is custom. "
"All subclassed Models that have `compile()` "
"overridden should also override "
"`get_compile_config()` and `compile_from_config(config)`. "
"Alternatively, you can "
"call `compile()` manually after loading."
)
return
config = saving_lib.deserialize_keras_object(config)
self.compile(**config)
if (
hasattr(self, "optimizer")
# Exempt legacy optimizers.
and isinstance(self.optimizer, optimizer.Optimizer)
and self.built
):
# Create optimizer variables.
self.optimizer.build(self.trainable_variables)
def export(self, filepath):
"""Create a SavedModel artifact for inference (e.g. via TF-Serving).
This method lets you export a model to a lightweight SavedModel artifact
that contains the model's forward pass only (its `call()` method)
and can be served via e.g. TF-Serving. The forward pass is registered
under the name `serve()` (see example below).
The original code of the model (including any custom layers you may
have used) is *no longer* necessary to reload the artifact -- it is
entirely standalone.
Args:
filepath: `str` or `pathlib.Path` object. Path where to save
the artifact.
Example:
```python
# Create the artifact
model.export("path/to/location")
# Later, in a different process / environment...
reloaded_artifact = tf.saved_model.load("path/to/location")
predictions = reloaded_artifact.serve(input_data)
```
If you would like to customize your serving endpoints, you can
use the lower-level `keras.export.ExportArchive` class. The `export()`
method relies on `ExportArchive` internally.
"""
from tf_keras.export import export_lib
export_lib.export_model(self, filepath)
@tf.__internal__.tracking.no_automatic_dependency_tracking
def _set_save_spec(self, inputs, args=None, kwargs=None):
"""Defines the save spec so that serialization can trace `call()`.
The TensorSpecs of the call function `inputs`, `args`, and `kwargs` are
saved into a tuple of `([inputs] + args, kwargs)`. The input
`TensorSpec` names are updated to match the built `input_names`.
The specs can be retrieved with the `save_spec` property.
Args:
inputs: possibly nested inputs passed into the call function.
args: a list of positional arguments passed into call.
kwargs: a dictionary of keyword arguments passed into call.
"""
if self._saved_model_inputs_spec is not None:
return # Already set.
args = args or []
kwargs = kwargs or {}
input_names = self.input_names
if not input_names:
input_names = compile_utils.create_pseudo_input_names(inputs)
flat_inputs = tf.nest.flatten(inputs)
inputs_spec = []
for name, tensor in zip(input_names, flat_inputs):
inputs_spec.append(
tf_utils.get_tensor_spec(tensor, dynamic_batch=False, name=name)
)
inputs_spec = tf.nest.pack_sequence_as(inputs, inputs_spec)
super()._set_save_spec(inputs_spec, args, kwargs)
# Store the input shapes
if (
self.__class__.__name__ == "Sequential"
and self._build_input_shape is None
):
self._build_input_shape = tf.nest.map_structure(
lambda x: None if x is None else x.shape, inputs_spec
)
def save_spec(self, dynamic_batch=True):
"""Returns the `tf.TensorSpec` of call args as a tuple `(args, kwargs)`.
This value is automatically defined after calling the model for the
first time. Afterwards, you can use it when exporting the model for
serving:
```python
model = tf.keras.Model(...)
@tf.function
def serve(*args, **kwargs):
outputs = model(*args, **kwargs)
# Apply postprocessing steps, or add additional outputs.
...
return outputs
# arg_specs is `[tf.TensorSpec(...), ...]`. kwarg_specs, in this
# example, is an empty dict since functional models do not use keyword
# arguments.
arg_specs, kwarg_specs = model.save_spec()
model.save(path, signatures={
'serving_default': serve.get_concrete_function(*arg_specs,
**kwarg_specs)
})
```
Args:
dynamic_batch: Whether to set the batch sizes of all the returned
`tf.TensorSpec` to `None`. (Note that when defining functional or
Sequential models with `tf.keras.Input([...], batch_size=X)`, the
batch size will always be preserved). Defaults to `True`.
Returns:
If the model inputs are defined, returns a tuple `(args, kwargs)`. All
elements in `args` and `kwargs` are `tf.TensorSpec`.
If the model inputs are not defined, returns `None`.
The model inputs are automatically set when calling the model,
`model.fit`, `model.evaluate` or `model.predict`.
"""
return self._get_save_spec(dynamic_batch, inputs_only=False)
def _assert_weights_created(self):
"""Asserts that all the weights for the model have been created.
For a non-dynamic model, the weights must already be created after the
layer has been called. For a dynamic model, the exact list of weights
can never be known for certain since it may change at any time during
execution.
We run this check right before accessing weights or getting the Numpy
value for the current weights. Otherwise, if the layer has never been
called, the user would just get an empty list, which is misleading.
Raises:
ValueError: if the weights of the network have not yet been created.
"""
if self.dynamic:
return
if (
"build" in self.__class__.__dict__
and self.__class__ != Model
and not self.built
):
# For any model that has customized build() method but hasn't been
# invoked yet, this will cover both sequential and subclass model.
# Also make sure to exclude Model class itself which has build()
# defined.
raise ValueError(
f"Weights for model '{self.name}' have not yet been "
"created. "
"Weights are created when the model is first called on "
"inputs or `build()` is called with an `input_shape`."
)
def _check_call_args(self, method_name):
"""Check that `call()` has only one positional arg."""
# Always allow first arg, regardless of arg name.
fullargspec = self._call_spec.full_argspec
if fullargspec.defaults:
positional_args = fullargspec.args[: -len(fullargspec.defaults)]
else:
positional_args = fullargspec.args
if "training" in positional_args:
positional_args.remove("training")
# self and first arg can be positional.
if len(positional_args) > 2:
extra_args = positional_args[2:]
raise ValueError(
f"Models passed to `{method_name}` can only have `training` "
"and the first argument in `call()` as positional arguments, "
f"found: {extra_args}."
)
def _validate_compile(self, optimizer, metrics, **kwargs):
"""Performs validation checks for the default `compile()`."""
if any(
isinstance(opt, optimizer_v1.Optimizer)
for opt in tf.nest.flatten(optimizer)
):
raise ValueError(
f"`tf.compat.v1.keras` Optimizer ({optimizer}) is "
"not supported when eager execution is enabled. Use a "
"`tf.keras` Optimizer instead, or disable eager "
"execution."
)
kwargs.pop("cloning", None) # Legacy DistStrat argument, never used.
kwargs.pop("experimental_run_tf_function", None) # Always `True`.
distribute_arg = kwargs.pop("distribute", None)
if distribute_arg is not None:
raise ValueError(
"`distribute` argument in compile is not available in TF 2.0. "
"Please create the model under the `strategy.scope()`. "
f"Received: {distribute_arg}."
)
target_tensor_arg = kwargs.pop("target_tensors", None)
if target_tensor_arg is not None:
raise ValueError(
"`target_tensors` argument is not supported when executing "
f"eagerly. Received: {target_tensor_arg}."
)
invalid_kwargs = set(kwargs) - {"sample_weight_mode"}
if invalid_kwargs:
raise TypeError(
"Invalid keyword argument(s) in `compile()`: "
f"{(invalid_kwargs,)}. Valid keyword arguments include "
'"cloning", "experimental_run_tf_function", "distribute",'
' "target_tensors", or "sample_weight_mode".'
)
# Model must be created and compiled with the same DistStrat.
if self.built and tf.distribute.has_strategy():
strategy = tf.distribute.get_strategy()
for v in self.variables:
if not strategy.extended.variable_created_in_scope(v):
raise ValueError(
f"Variable ({v}) was not created in the distribution "
f"strategy scope of ({strategy}). It is most likely "
"because some layers, model, or optimizer was being "
"created outside the distribution strategy scope. Try "
"to make sure your code looks similar "
"to the following.\nwith strategy.scope():\n"
" model=_create_model()\n"
" model.compile(...)"
)
# Model metrics must be created in the same distribution strategy scope
# as the model.
strategy = self.distribute_strategy
for metric in tf.nest.flatten(metrics):
for v in getattr(metric, "variables", []):
if not strategy.extended.variable_created_in_scope(v):
raise ValueError(
f"Metric ({metric}) passed to `model.compile` was "
"created inside a different distribution strategy "
"scope than the model. All metrics must be created "
"in the same distribution strategy "
f"scope as the model (in this case {strategy}). "
"If you pass in a string identifier for a metric to "
"compile, the metric will automatically be created "
"in the correct distribution strategy scope."
)
# Model metrics must be created in the same distribution strategy scope
# as the model.
for opt in tf.nest.flatten(optimizer):
for v in getattr(opt, "_weights", []):
if not strategy.extended.variable_created_in_scope(v):
raise ValueError(
f"Optimizer ({optimizer}) passed to `model.compile` "
"was created inside a different distribution strategy "
"scope than the model. All optimizers must be created "
"in the same distribution strategy scope as the model "
f"(in this case {strategy}). If you pass in a string "
"identifier for an optimizer to compile, the optimizer "
"will automatically be created in the correct "
"distribution strategy scope."
)
def _maybe_load_initial_counters_from_ckpt(
self, steps_per_epoch, initial_epoch
):
"""Maybe load initial epoch from ckpt, considering worker recovery.
Refer to tensorflow/python/tf_keras/distribute/worker_training_state.py
for more information.
Args:
steps_per_epoch: The number of step per epoch.
initial_epoch: The original initial_epoch user passes in `fit()`.
mode: The mode for running `model.fit()`.
Returns:
If the training is recovering from previous failure under multi-worker
training setting, return the (epoch, step) the training is supposed to
continue at. Otherwise, return the `initial_epoch, initial_step` the
user passes in.
"""
initial_step = 0
if self._training_state is not None:
return self._training_state.maybe_load_initial_counters_from_ckpt(
steps_per_epoch, initial_epoch, mode=ModeKeys.TRAIN
)
return (initial_epoch, initial_step)
def _assert_compile_was_called(self):
# Checks whether `compile` has been called. If it has been called,
# then the optimizer is set. This is different from whether the
# model is compiled
# (i.e. whether the model is built and its inputs/outputs are set).
if not self._is_compiled:
raise RuntimeError(
"You must compile your model before "
"training/testing. "
"Use `model.compile(optimizer, loss)`."
)
def _check_sample_weight_warning(self, x, sample_weight):
# Datasets can include sample weight, by returning a tuple with the
# structure of `(x, y, sample_weight)`.
sample_weight_present = sample_weight is not None or (
isinstance(x, tf.data.Dataset)
and isinstance(x.element_spec, tuple)
and len(x.element_spec) == 3
)
if (
sample_weight_present
and self.compiled_metrics._user_weighted_metrics is None
):
logging.warning(
"`evaluate()` received a value for `sample_weight`, but "
"`weighted_metrics` were not provided. Did you mean to pass "
"metrics to `weighted_metrics` in `compile()`? If this is "
"intentional you can pass `weighted_metrics=[]` to `compile()` "
"in order to silence this warning."
)
def _set_inputs(self, inputs, outputs=None, training=None):
"""This method is for compat with Modelv1. Only inputs are needed
here."""
self._set_save_spec(inputs)
@property
def _trackable_saved_model_saver(self):
return model_serialization.ModelSavedModelSaver(self)
def _trackable_children(self, save_type="checkpoint", **kwargs):
if save_type == "savedmodel":
# SavedModel needs to ignore the execution functions.
train_function = self.train_function
test_function = self.test_function
predict_function = self.predict_function
train_tf_function = self.train_tf_function
self.train_function = None
self.test_function = None
self.predict_function = None
self.train_tf_function = None
children = super()._trackable_children(save_type, **kwargs)
if save_type == "savedmodel":
self.train_function = train_function
self.test_function = test_function
self.predict_function = predict_function
self.train_tf_function = train_tf_function
return children
def _should_eval(self, epoch, validation_freq):
epoch = epoch + 1 # one-index the user-facing epoch.
if isinstance(validation_freq, int):
return epoch % validation_freq == 0
elif isinstance(validation_freq, list):
return epoch in validation_freq
else:
raise ValueError(
"Expected `validation_freq` to be a list or int. "
f"Received: validation_freq={validation_freq} of the "
f"type {type(validation_freq)}."
)
######################################################################
# Functions below exist only as v1 / v2 compatibility shims.
######################################################################
def _get_compile_args(self, user_metrics=True):
"""Used for saving or cloning a Model.
Args:
user_metrics: Whether to return user-supplied metrics or `Metric`
objects. If True, returns the user-supplied metrics.
Defaults to `True`.
Returns:
Dictionary of arguments that were used when compiling the model.
"""
self._assert_compile_was_called()
saved_metrics = self.compiled_metrics._user_metrics
saved_weighted_metrics = self.compiled_metrics._user_weighted_metrics
if not user_metrics:
if saved_metrics is not None:
saved_metrics = self.compiled_metrics._metrics
if saved_weighted_metrics is not None:
saved_weighted_metrics = self.compiled_metrics._weighted_metrics
compile_args = {
"optimizer": self.optimizer,
"loss": self.compiled_loss._user_losses,
"metrics": saved_metrics,
"weighted_metrics": saved_weighted_metrics,
"loss_weights": self.compiled_loss._user_loss_weights,
}
return compile_args
def _get_callback_model(self):
return self
def _in_multi_worker_mode(self):
return self.distribute_strategy.extended._in_multi_worker_mode()
@property
def _compile_was_called(self):
return self._is_compiled
class _TestFunction:
def __init__(self, function, callbacks):
self._function = function
self._callbacks = callbacks
def run_step(self, dataset_or_iterator, data_handler, step, unused_shards):
tmp_logs = self._function(dataset_or_iterator)
if data_handler.should_sync:
context.async_wait()
logs = tmp_logs
end_step = step + data_handler.step_increment
self._callbacks.on_test_batch_end(end_step, logs)
return logs
class _ExactTestFunction(_TestFunction):
def __init__(self, function, callbacks):
super().__init__(function, callbacks)
self._logs = []
def run_step(self, dataset_or_iterator, data_handler, step, shards):
tmp_logs = self._function(
dataset_or_iterator,
tf.constant(shards, dtype=tf.int64),
tf.constant(step, dtype=tf.int64),
)
if data_handler.should_sync:
context.async_wait()
self._logs.append(tmp_logs)
return self._logs
def reduce_per_replica(values, strategy, reduction):
"""Attempt to reduce the structure `values` to single values.
Given `values` (a `tf.Tensor` or a `PerReplica` structure),
which represents the values across all the replicas, `reduce_per_replica`
attempts to "reduce" those values and returns the corresponding structure
that represents only single values.
Currently, `reduce_per_replica` is only used for reducing the metric results
from `tf.distribute.Strategy.run()`. Depending on the underlying
`Strategy` implementation, `values` may be a `PerReplica` object,
which can be thought of as a collection of values across the replicas,
or a `tf.Tensor`, if the strategy has already conducted the reduction
for the downstream library.
There are five possible outcomes of reduction:
1) if the `values` is a structure of simple `tf.Tensor`s, meaning that
reduction is not actually needed, `reduce_per_replica` returns the
structure as-is.
2) else, if `reduction="auto"`, then the best reduction strategy is
chosen based on the current environment. This should only be used
for training cases (`fit()`).
3) else, if `reduction="first"`, then `reduce_per_replica`
returns the values of the first replica. This is used in the case of
training and evaluation, where `values` is expected to hold the same
value across the replicas as a result of `Strategy`'s synchronization
across the replicas.
`reduce_per_replica` does not synchronize the values.
4) else, if `reduction="sum"`, then `reduce_per_replica` returns the sum
of values for all replicas. This may be used in the custom training loop
case, where each replica contain different values which are not
synchronized.
5) else, if `reduction="concat"`, then `reduce_per_replica`
returns the concatenation of the values across the replicas, along the
axis of dimension 0. This is used in the inference case (`predict()`).
Args:
values: Structure of `PerReplica` objects or `tf.Tensor`s. `tf.Tensor`s
are returned as-is.
strategy: `tf.distribute.Strategy` object.
reduction: One of `"auto"`, `"first"`, `"concat"`, or `"sum"`.
`"auto"` will select `"first"` when used under a TPUStrategy, or
`"sum"` otherwise.
Returns:
Structure of `Tensor`s, representing the result of reduction.
Raises:
ValueError: if the reduction method is not supported.
"""
if reduction == "auto":
reduction = "first" if backend.is_tpu_strategy(strategy) else "sum"
def _reduce(v):
"""Reduce a single `PerReplica` object."""
if _collective_all_reduce_multi_worker(strategy):
if reduction == "concat":
return _multi_worker_concat(v, strategy)
elif reduction == "sum":
return strategy.reduce("SUM", v, axis=None)
if _is_dtensor_per_replica_instance(v):
return _reduce_dtensor_per_replica(v, strategy, reduction)
elif not _is_per_replica_instance(v):
return v
elif reduction == "first":
return strategy.experimental_local_results(v)[0]
elif reduction == "concat":
if _is_tpu_multi_host(strategy):
return _tpu_multi_host_concat(v, strategy)
else:
return concat(strategy.experimental_local_results(v))
elif reduction == "sum":
return tf.reduce_sum(strategy.experimental_local_results(v))
else:
raise ValueError(
'`reduction` must be "first", "concat", "sum", or "auto". '
f"Received: reduction={reduction}."
)
return tf.nest.map_structure(_reduce, values)
def concat(tensors, axis=0):
"""Concats `tensor`s along `axis`."""
if isinstance(tensors[0], tf.SparseTensor):
return tf.sparse.concat(axis=axis, sp_inputs=tensors)
elif _is_scalar(tensors[0]):
return tf.stack(tensors, axis=axis)
else:
return tf.concat(tensors, axis=axis)
def potentially_ragged_concat(tensors):
"""Concats `Tensor`s along their first dimension.
Args:
tensors: List of `Tensor`s.
Returns:
Concatenation of the inputs along the first dimension -- of type `Tensor`
if all input shapes are compatible, or `RaggedTensor` if not.
"""
if len(tensors) == 1:
return tensors[0]
if isinstance(tensors[0], tf.SparseTensor):
return tf.sparse.concat(axis=0, sp_inputs=tensors)
elif isinstance(tensors[0], tf.RaggedTensor):
return tf.concat(tensors, axis=0)
elif not tf.__internal__.tf2.enabled():
return tf.concat(tensors, axis=0)
non_batch_shapes = tf.stack([tf.shape(tensor)[1:] for tensor in tensors])
constant_dims = tf.math.reduce_all(
non_batch_shapes == non_batch_shapes[:1], axis=0
)
if tf.math.reduce_all(constant_dims).numpy().item():
# All non-batch dims are constant
if _is_scalar(tensors[0]):
return tf.stack(tensors, axis=0)
else:
return tf.concat(tensors, axis=0)
# First, identify constant inner dimensions by finding the
# rightmost dimension that is not constant
constant_inner_dimensions = (
constant_dims.numpy().tolist()[::-1].index(False)
)
# If there are constant inner dimensions, define a constant inner shape
if constant_inner_dimensions == 0:
constant_inner_shape = None
else:
constant_inner_shape = tensors[0].shape[-constant_inner_dimensions:]
return tf.ragged.constant(
[tensor.numpy() for tensor in tensors], inner_shape=constant_inner_shape
).merge_dims(0, 1)
def _reduce_dtensor_per_replica(value, strategy, reduction):
# Note that this function could happen in graph, so we can't just access
# the per-replica.values(), which will trigger unpack in graph and result
# into error.
# For now we will perform ops on dtensor instance directly on a global
# context.
dtensor = value._dtensor
if reduction == "first":
num_replica = strategy.num_replicas_in_sync
return tf.split(dtensor, num_replica, axis=0)[0]
elif reduction == "concat":
# Since dtensor is already in global context, the concat is a no-op
return dtensor
elif reduction == "sum":
return tf.reduce_sum(dtensor)
else:
raise ValueError(
'`reduction` must be one of "first", "concat", "sum", or "auto". '
f"Received: reduction={reduction}."
)
def _get_verbosity(verbose, distribute_strategy):
"""Find the right verbosity value for 'auto'."""
if verbose == 1 and distribute_strategy._should_use_with_coordinator:
raise ValueError(
"`verbose=1` is not allowed with `ParameterServerStrategy` for "
f"performance reasons. Received: verbose={verbose}"
)
if verbose == "auto":
if (
distribute_strategy._should_use_with_coordinator
or not io_utils.is_interactive_logging_enabled()
):
# Defaults to epoch-level logging for PSStrategy or using absl
# logging.
return 2
else:
return 1 # Defaults to batch-level logging otherwise.
return verbose
def _is_tpu_multi_host(strategy):
return backend.is_tpu_strategy(strategy) and strategy.extended.num_hosts > 1
def _tpu_multi_host_concat(v, strategy):
"""Correctly order TPU PerReplica objects."""
replicas = strategy.experimental_local_results(v)
# When distributed datasets are created from Tensors / NumPy,
# TPUStrategy.experimental_distribute_dataset shards data in
# (Replica, Host) order, and TPUStrategy.experimental_local_results returns
# it in (Host, Replica) order.
# TODO(b/150317897): Figure out long-term plan here.
num_replicas_per_host = strategy.extended.num_replicas_per_host
ordered_replicas = []
for replica_id in range(num_replicas_per_host):
ordered_replicas += replicas[replica_id::num_replicas_per_host]
return concat(ordered_replicas)
def _collective_all_reduce_multi_worker(strategy):
return (
isinstance(strategy, tf.distribute.MultiWorkerMirroredStrategy)
) and strategy.extended._in_multi_worker_mode()
# TODO(wxinyi): merge this with _tpu_multi_host_concat once we have all_gather
# for all strategies
def _multi_worker_concat(v, strategy):
"""Order PerReplica objects for CollectiveAllReduceStrategy and concat."""
replicas = strategy.gather(v, axis=0)
# v might not have the same shape on different replicas
if _is_per_replica_instance(v):
shapes = tf.concat(
[
tf.expand_dims(tf.shape(single_value)[0], axis=0)
for single_value in v.values
],
axis=0,
)
all_shapes = strategy.gather(shapes, axis=0)
else:
# v is a tensor. This may happen when, say, we have 2x1 multi-worker.
all_shapes = strategy.gather(
tf.expand_dims(tf.shape(v)[0], axis=0), axis=0
)
replicas = tf.split(
replicas,
num_or_size_splits=all_shapes,
num=strategy.num_replicas_in_sync,
)
ordered_replicas = []
num_replicas_per_worker = len(strategy.extended.worker_devices)
for replica_id in range(num_replicas_per_worker):
ordered_replicas += replicas[replica_id::num_replicas_per_worker]
return concat(ordered_replicas)
def _is_scalar(x):
return isinstance(x, (tf.Tensor, tf.Variable)) and x.shape.rank == 0
def _minimum_control_deps(outputs):
"""Returns the minimum control dependencies to ensure step succeeded."""
if tf.executing_eagerly():
return [] # Control dependencies not needed.
outputs = tf.nest.flatten(outputs, expand_composites=True)
for out in outputs:
# Variables can't be control dependencies.
if not isinstance(out, tf.Variable):
return [out] # Return first Tensor or Op from outputs.
return [] # No viable Tensor or Op to use for control deps.
def _disallow_inside_tf_function(method_name):
if tf.inside_function():
error_msg = (
"Detected a call to `Model.{method_name}` inside a `tf.function`. "
"`Model.{method_name} is a high-level endpoint that manages its "
"own `tf.function`. Please move the call to `Model.{method_name}` "
"outside of all enclosing `tf.function`s. Note that you can call a "
"`Model` directly on `Tensor`s inside a `tf.function` like: "
"`model(x)`."
).format(method_name=method_name)
raise RuntimeError(error_msg)
def flatten_metrics_in_order(logs, metrics_names):
"""Turns the `logs` dict into a list as per key order of `metrics_names`."""
results = []
for name in metrics_names:
if name in logs:
results.append(logs[name])
for key in sorted(logs.keys()):
if key not in metrics_names:
results.append(logs[key])
if len(results) == 1:
return results[0]
return results
def _is_per_replica_instance(obj):
return isinstance(obj, tf.distribute.DistributedValues) and isinstance(
obj, tf.__internal__.CompositeTensor
)
def _is_dtensor_per_replica_instance(obj):
# This is a temp check for DTensorDistributedValue, which is not public API
# yet.
# TODO(scottzhu): Move to more stable API when dtensor based strategy is
# ready.
return isinstance(obj, tf.distribute.DistributedValues) and hasattr(
obj, "_dtensor"
)
def disable_multi_worker(method):
"""Decorator that disallows multi-worker use of `method`."""
def _method_wrapper(self, *args, **kwargs):
if self._in_multi_worker_mode():
raise ValueError(
f"{method.__name__} is not supported in multi-worker "
"mode. Please use a non-multi-worker "
"`tf.distribute.Strategy` such as "
"`tf.distribute.MirroredStrategy`."
)
return method(self, *args, **kwargs)
return tf.__internal__.decorator.make_decorator(
target=method, decorator_func=_method_wrapper
)
def inject_functional_model_class(cls):
"""Inject `Functional` into the hierarchy of this class if needed."""
from tf_keras.engine import functional
from tf_keras.engine import training_v1
if cls == Model or cls == training_v1.Model:
return functional.Functional
# In case there is any multiple inheritance, we stop injecting the
# class if keras model is not in its class hierarchy.
if cls == object:
return object
cls.__bases__ = tuple(
inject_functional_model_class(base) for base in cls.__bases__
)
# Trigger any `__new__` class swapping that needed to happen on `Functional`
# but did not because functional was not in the class hierarchy.
cls.__new__(cls)
return cls
def is_functional_model_init_params(args, kwargs):
# Both inputs and outputs in args
if len(args) == 2:
return True
# Both inputs in args, outputs in kwargs
if len(args) == 1 and "outputs" in kwargs:
return True
# Both in kwargs
if "inputs" in kwargs and "outputs" in kwargs:
return True
return False
|
tf-keras/tf_keras/engine/training.py/0
|
{
"file_path": "tf-keras/tf_keras/engine/training.py",
"repo_id": "tf-keras",
"token_count": 87211
}
| 193 |
# Description:
# Contains TF-Keras models to Estimator converter
# Placeholder: load unaliased py_library
package(
# copybara:uncomment default_applicable_licenses = ["//tf_keras:license"],
default_visibility = [
"//tf_keras:friends",
],
licenses = ["notice"],
)
py_library(
name = "estimator",
srcs = [
"__init__.py",
],
srcs_version = "PY3",
deps = [
"//:expect_tensorflow_installed",
],
)
|
tf-keras/tf_keras/estimator/BUILD/0
|
{
"file_path": "tf-keras/tf_keras/estimator/BUILD",
"repo_id": "tf-keras",
"token_count": 202
}
| 194 |
# Description:
# Contains the TF-Keras initializer API (internal TensorFlow version).
# Placeholder: load unaliased py_library
load("@org_keras//tf_keras:tf_keras.bzl", "tf_py_test")
package(
# copybara:uncomment default_applicable_licenses = ["//tf_keras:license"],
default_visibility = [
"//tf_keras:friends",
],
licenses = ["notice"],
)
py_library(
name = "initializers",
srcs = [
"__init__.py",
"initializers.py",
"initializers_v1.py",
],
srcs_version = "PY3",
deps = [
"//:expect_tensorflow_installed",
"//tf_keras:backend",
"//tf_keras/dtensor:utils",
"//tf_keras/saving:serialization_lib",
"//tf_keras/utils:generic_utils",
"//tf_keras/utils:tf_inspect",
],
)
tf_py_test(
name = "initializers_test",
size = "small",
srcs = ["initializers_test.py"],
python_version = "PY3",
deps = [
":initializers",
"//:expect_numpy_installed",
"//:expect_tensorflow_installed",
"//tf_keras:backend",
"//tf_keras/engine",
"//tf_keras/models",
"//tf_keras/testing_infra:test_combinations",
],
)
|
tf-keras/tf_keras/initializers/BUILD/0
|
{
"file_path": "tf-keras/tf_keras/initializers/BUILD",
"repo_id": "tf-keras",
"token_count": 570
}
| 195 |
# Copyright 2020 The TensorFlow Authors. All Rights Reserved.
#
# Licensed under the Apache License, Version 2.0 (the "License");
# you may not use this file except in compliance with the License.
# You may obtain a copy of the License at
#
# http://www.apache.org/licenses/LICENSE-2.0
#
# Unless required by applicable law or agreed to in writing, software
# distributed under the License is distributed on an "AS IS" BASIS,
# WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.
# See the License for the specific language governing permissions and
# limitations under the License.
# ==============================================================================
import gc
import tensorflow.compat.v2 as tf
# isort: off
from tensorflow.python.framework import (
test_util as tf_test_utils,
)
from tensorflow.python.platform import test as test_lib
layers = tf.keras.layers
optimizers = tf.keras.optimizers
def _get_big_cnn_model(
img_dim, n_channels, num_partitions, blocks_per_partition
):
"""Creates a test model whose activations are significantly larger than
model size."""
model = tf.keras.Sequential()
model.add(layers.Input(shape=(img_dim, img_dim, n_channels)))
for _ in range(num_partitions):
for _ in range(blocks_per_partition):
model.add(
layers.Conv2D(10, 5, padding="same", activation=tf.nn.relu)
)
model.add(layers.MaxPooling2D((1, 1), padding="same"))
model.add(
layers.Conv2D(40, 5, padding="same", activation=tf.nn.relu)
)
model.add(layers.MaxPooling2D((1, 1), padding="same"))
model.add(
layers.Conv2D(20, 5, padding="same", activation=tf.nn.relu)
)
model.add(layers.MaxPooling2D((1, 1), padding="same"))
model.add(layers.Flatten())
model.add(layers.Dense(32, activation=tf.nn.relu))
model.add(layers.Dense(10))
return model
def _get_split_cnn_model(
img_dim, n_channels, num_partitions, blocks_per_partition
):
"""Creates a test model that is split into `num_partitions` smaller
models."""
models = [tf.keras.Sequential() for _ in range(num_partitions)]
models[0].add(layers.Input(shape=(img_dim, img_dim, n_channels)))
for i in range(num_partitions):
model = models[i]
if i > 0:
last_shape = models[i - 1].layers[-1].output_shape
model.add(layers.Input(shape=last_shape[1:]))
for _ in range(blocks_per_partition):
model.add(
layers.Conv2D(10, 5, padding="same", activation=tf.nn.relu)
)
model.add(layers.MaxPooling2D((1, 1), padding="same"))
model.add(
layers.Conv2D(40, 5, padding="same", activation=tf.nn.relu)
)
model.add(layers.MaxPooling2D((1, 1), padding="same"))
model.add(
layers.Conv2D(20, 5, padding="same", activation=tf.nn.relu)
)
model.add(layers.MaxPooling2D((1, 1), padding="same"))
models[-1].add(layers.Flatten())
models[-1].add(layers.Dense(32, activation=tf.nn.relu))
models[-1].add(layers.Dense(10))
return models
def _compute_loss(logits, labels):
return tf.reduce_mean(
tf.nn.sparse_softmax_cross_entropy_with_logits(
logits=logits, labels=labels
)
)
def _limit_gpu_memory():
"""Helper function to limit GPU memory for testing."""
gpus = tf.config.experimental.list_physical_devices("GPU")
if gpus:
tf.config.experimental.set_virtual_device_configuration(
gpus[0],
[
tf.config.experimental.VirtualDeviceConfiguration(
memory_limit=2048
)
],
)
return True
return False
def _get_dummy_data(img_dim, n_channels, batch_size):
inputs = tf.ones([batch_size, img_dim, img_dim, n_channels])
labels = tf.ones([batch_size], dtype=tf.int64)
return inputs, labels
def _train_no_recompute(n_steps):
"""Trains a single large model without gradient checkpointing."""
img_dim, n_channels, batch_size = 256, 1, 4
x, y = _get_dummy_data(img_dim, n_channels, batch_size)
model = _get_big_cnn_model(
img_dim, n_channels, num_partitions=3, blocks_per_partition=2
)
optimizer = optimizers.SGD()
losses = []
tr_vars = model.trainable_variables
for _ in range(n_steps):
with tf.GradientTape() as tape:
logits = model(x)
loss = _compute_loss(logits, y)
losses.append(loss)
grads = tape.gradient(loss, tr_vars) # tr_vars
optimizer.apply_gradients(zip(grads, tr_vars))
del grads
return losses
def _train_with_recompute(n_steps):
"""Trains a single large model with gradient checkpointing using
tf.recompute_grad."""
img_dim, n_channels, batch_size = 256, 1, 4
x, y = _get_dummy_data(img_dim, n_channels, batch_size)
# This model is the same model as _get_big_cnn_model but split into 3 parts.
models = _get_split_cnn_model(
img_dim, n_channels, num_partitions=3, blocks_per_partition=2
)
model1, model2, model3 = models
# Apply gradient checkpointing to the submodels using tf.recompute_grad.
model1_re = tf.recompute_grad(model1)
model2_re = tf.recompute_grad(model2)
model3_re = tf.recompute_grad(model3)
optimizer = optimizers.SGD()
tr_vars = (
model1.trainable_variables
+ model2.trainable_variables
+ model3.trainable_variables
)
losses = []
for _ in range(n_steps):
with tf.GradientTape() as tape:
logits1 = model1_re(x)
logits2 = model2_re(logits1)
logits3 = model3_re(logits2)
loss = _compute_loss(logits3, y)
losses.append(loss)
grads = tape.gradient(loss, tr_vars) # tr_vars
optimizer.apply_gradients(zip(grads, tr_vars))
del grads
return losses
@tf_test_utils.with_eager_op_as_function
class GradientCheckpointTest(tf.test.TestCase):
def test_raises_oom_exception(self):
self.skipTest("b/232015009: flaky test")
if not _limit_gpu_memory():
self.skipTest("No virtual GPUs found")
with self.assertRaises(Exception) as context:
_train_no_recompute(1)
self.assertIsInstance(
context.exception, tf.errors.ResourceExhaustedError
)
@tf_test_utils.disable_xla(
"xla does not support searching for memory-limited solvers."
)
def test_does_not_raise_oom_exception(self):
if not _limit_gpu_memory():
self.skipTest("No virtual GPUs found")
if test_lib.is_built_with_rocm():
self.skipTest(
"ROCm MIOpen does not support searching for memory-limited"
"solvers yet so skip the subtest which would result in OOM."
)
n_step = 2
losses = _train_with_recompute(n_step)
self.assertLen(losses, n_step)
def tearDown(self):
super().tearDown()
# Make sure all the models created in keras has been deleted and cleared
# from the global keras grpah, also do a force GC to recycle the GPU
# memory.
tf.keras.backend.clear_session()
gc.collect()
if __name__ == "__main__":
tf.test.main()
|
tf-keras/tf_keras/integration_test/gradient_checkpoint_test.py/0
|
{
"file_path": "tf-keras/tf_keras/integration_test/gradient_checkpoint_test.py",
"repo_id": "tf-keras",
"token_count": 3314
}
| 196 |
"""Text classification model.
Adapted from https://keras.io/examples/nlp/text_classification_from_scratch/
"""
import re
import string
import tensorflow as tf
from tensorflow import keras
from tf_keras.integration_test.models.input_spec import InputSpec
MAX_FEATURES = 1000
EMBEDDING_DIM = 64
SEQUENCE_LENGTH = 32
def get_data_spec(batch_size):
return (
InputSpec((batch_size,), dtype="string"),
InputSpec((batch_size, 1), dtype="int32", range=[0, 2]),
)
def custom_standardization(input_data):
lowercase = tf.strings.lower(input_data)
stripped_html = tf.strings.regex_replace(lowercase, "<br />", " ")
return tf.strings.regex_replace(
stripped_html, f"[{re.escape(string.punctuation)}]", ""
)
def get_input_preprocessor():
input_vectorizer = keras.layers.TextVectorization(
standardize=custom_standardization,
max_tokens=MAX_FEATURES,
output_mode="int",
output_sequence_length=SEQUENCE_LENGTH,
)
text_ds = tf.data.Dataset.from_tensor_slices(
[
"Lorem ipsum dolor sit amet",
"consectetur adipiscing elit",
"sed do eiusmod tempor incididunt ut",
"labore et dolore magna aliqua.",
"Ut enim ad minim veniam",
"quis nostrud exercitation ullamco",
"laboris nisi ut aliquip ex ea commodo consequat.",
]
)
input_vectorizer.adapt(text_ds)
return input_vectorizer
def get_model(
build=False, compile=False, jit_compile=False, include_preprocessing=True
):
if include_preprocessing:
inputs = keras.Input(shape=(), dtype="string")
x = get_input_preprocessor()(inputs)
else:
inputs = keras.Input(shape=(None,), dtype="int64")
x = inputs
x = keras.layers.Embedding(MAX_FEATURES, EMBEDDING_DIM)(x)
x = keras.layers.Dropout(0.5)(x)
x = keras.layers.Conv1D(
128, 7, padding="valid", activation="relu", strides=3
)(x)
x = keras.layers.Conv1D(
128, 7, padding="valid", activation="relu", strides=3
)(x)
x = keras.layers.GlobalMaxPooling1D()(x)
x = keras.layers.Dense(128, activation="relu")(x)
x = keras.layers.Dropout(0.5)(x)
predictions = keras.layers.Dense(
1, activation="sigmoid", name="predictions"
)(x)
model = keras.Model(inputs, predictions)
if compile:
model.compile(
loss="binary_crossentropy",
optimizer="adam",
metrics=["accuracy"],
jit_compile=jit_compile,
)
return model
def get_custom_objects():
return {"custom_standardization": custom_standardization}
|
tf-keras/tf_keras/integration_test/models/text_classification.py/0
|
{
"file_path": "tf-keras/tf_keras/integration_test/models/text_classification.py",
"repo_id": "tf-keras",
"token_count": 1183
}
| 197 |
"""Test Model.fit across a diverse range of models."""
import os
import tensorflow.compat.v2 as tf
from absl.testing import parameterized
from tf_keras.integration_test.models import bert
from tf_keras.integration_test.models import dcgan
from tf_keras.integration_test.models import edge_case_model
from tf_keras.integration_test.models import input_spec
from tf_keras.integration_test.models import low_level_model
from tf_keras.integration_test.models import mini_unet
from tf_keras.integration_test.models import mini_xception
from tf_keras.integration_test.models import retinanet
from tf_keras.integration_test.models import structured_data_classification
from tf_keras.integration_test.models import text_classification
from tf_keras.integration_test.models import timeseries_forecasting
from tf_keras.integration_test.models import vae
from tf_keras.testing_infra import test_combinations
from tf_keras.testing_infra import test_utils
def get_dataset(data_specs, batch_size):
values = tf.nest.map_structure(input_spec.spec_to_value, data_specs)
dataset = (
tf.data.Dataset.from_tensor_slices(values)
.prefetch(batch_size * 2)
.batch(batch_size)
)
return dataset
@test_utils.run_v2_only
class SavingV3Test(test_combinations.TestCase):
@parameterized.named_parameters(
("bert", bert),
("edge_case_model", edge_case_model),
# ("efficientnet_v2", efficientnet_v2), # Too expensive to run on CI
("low_level_model", low_level_model),
("mini_unet", mini_unet),
("mini_xception", mini_xception),
("retinanet", retinanet),
("structured_data_classification", structured_data_classification),
("text_classification", text_classification),
("timeseries_forecasting", timeseries_forecasting),
)
def test_saving_v3(self, module):
batch_size = 2
data_specs = module.get_data_spec(batch_size * 2)
dataset = get_dataset(data_specs, batch_size)
for batch in dataset.take(1):
pass
if isinstance(batch, tuple):
batch = batch[0]
model = module.get_model(
build=True,
compile=True,
jit_compile=False,
include_preprocessing=True,
)
model.fit(dataset, epochs=1, steps_per_epoch=1)
temp_filepath = os.path.join(
self.get_temp_dir(), f"{module.__name__}.keras"
)
model.save(temp_filepath, save_format="keras_v3")
with tf.keras.utils.custom_object_scope(module.get_custom_objects()):
new_model = tf.keras.models.load_model(temp_filepath)
# Test model weights
self.assertIs(new_model.__class__, model.__class__)
self.assertEqual(len(model.get_weights()), len(new_model.get_weights()))
for w1, w2 in zip(model.get_weights(), new_model.get_weights()):
if w1.dtype == "object":
self.assertEqual(str(w1), str(w2))
else:
self.assertAllClose(w1, w2, atol=1e-6)
# Test forward pass
self.assertAllClose(new_model(batch), model(batch), atol=1e-6)
# Test optimizer state
if hasattr(model, "optimizer"):
self.assertEqual(
len(model.optimizer.variables()),
len(new_model.optimizer.variables()),
)
for v1, v2 in zip(
model.optimizer.variables(), new_model.optimizer.variables()
):
self.assertAllClose(v1.numpy(), v2.numpy(), atol=1e-6)
# Test training still works
new_model.fit(dataset, epochs=1, steps_per_epoch=1)
@parameterized.named_parameters(("dcgan", dcgan), ("vae", vae))
def test_saving_v3_no_call(self, module):
batch_size = 2
data_specs = module.get_data_spec(batch_size * 2)
dataset = get_dataset(data_specs, batch_size)
model = module.get_model(
build=True,
compile=True,
jit_compile=False,
include_preprocessing=True,
)
temp_filepath = os.path.join(
self.get_temp_dir(), f"{module.__name__}.keras"
)
model.save(temp_filepath, save_format="keras_v3")
with tf.keras.utils.custom_object_scope(module.get_custom_objects()):
new_model = tf.keras.models.load_model(temp_filepath)
# Test model weights
self.assertIs(new_model.__class__, model.__class__)
self.assertEqual(len(model.get_weights()), len(new_model.get_weights()))
for w1, w2 in zip(model.get_weights(), new_model.get_weights()):
if w1.dtype == "object":
self.assertEqual(str(w1), str(w2))
else:
self.assertAllClose(w1, w2, atol=1e-6)
# Test training still works
new_model.fit(dataset, epochs=1, steps_per_epoch=1)
if __name__ == "__main__":
tf.test.main()
|
tf-keras/tf_keras/integration_test/saving_v3_test.py/0
|
{
"file_path": "tf-keras/tf_keras/integration_test/saving_v3_test.py",
"repo_id": "tf-keras",
"token_count": 2251
}
| 198 |
# Copyright 2015 The TensorFlow Authors. All Rights Reserved.
#
# Licensed under the Apache License, Version 2.0 (the "License");
# you may not use this file except in compliance with the License.
# You may obtain a copy of the License at
#
# http://www.apache.org/licenses/LICENSE-2.0
#
# Unless required by applicable law or agreed to in writing, software
# distributed under the License is distributed on an "AS IS" BASIS,
# WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.
# See the License for the specific language governing permissions and
# limitations under the License.
# ==============================================================================
"""Leaky version of a Rectified Linear Unit activation layer."""
from tf_keras import backend
from tf_keras.engine.base_layer import Layer
from tf_keras.utils import tf_utils
# isort: off
from tensorflow.python.util.tf_export import keras_export
@keras_export("keras.layers.LeakyReLU")
class LeakyReLU(Layer):
"""Leaky version of a Rectified Linear Unit.
It allows a small gradient when the unit is not active:
```
f(x) = alpha * x if x < 0
f(x) = x if x >= 0
```
Usage:
>>> layer = tf.keras.layers.LeakyReLU()
>>> output = layer([-3.0, -1.0, 0.0, 2.0])
>>> list(output.numpy())
[-0.9, -0.3, 0.0, 2.0]
>>> layer = tf.keras.layers.LeakyReLU(alpha=0.1)
>>> output = layer([-3.0, -1.0, 0.0, 2.0])
>>> list(output.numpy())
[-0.3, -0.1, 0.0, 2.0]
Input shape:
Arbitrary. Use the keyword argument `input_shape`
(tuple of integers, does not include the batch axis)
when using this layer as the first layer in a model.
Output shape:
Same shape as the input.
Args:
alpha: Float >= `0.`. Negative slope coefficient. Defaults to `0.3`.
"""
def __init__(self, alpha=0.3, **kwargs):
super().__init__(**kwargs)
if alpha is None:
raise ValueError(
"The alpha value of a Leaky ReLU layer cannot be None, "
f"Expecting a float. Received: {alpha}"
)
self.supports_masking = True
self.alpha = backend.cast_to_floatx(alpha)
def call(self, inputs):
return backend.relu(inputs, alpha=self.alpha)
def get_config(self):
config = {"alpha": float(self.alpha)}
base_config = super().get_config()
return dict(list(base_config.items()) + list(config.items()))
@tf_utils.shape_type_conversion
def compute_output_shape(self, input_shape):
return input_shape
|
tf-keras/tf_keras/layers/activation/leaky_relu.py/0
|
{
"file_path": "tf-keras/tf_keras/layers/activation/leaky_relu.py",
"repo_id": "tf-keras",
"token_count": 982
}
| 199 |
# Copyright 2019 The TensorFlow Authors. All Rights Reserved.
#
# Licensed under the Apache License, Version 2.0 (the "License");
# you may not use this file except in compliance with the License.
# You may obtain a copy of the License at
#
# http://www.apache.org/licenses/LICENSE-2.0
#
# Unless required by applicable law or agreed to in writing, software
# distributed under the License is distributed on an "AS IS" BASIS,
# WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.
# See the License for the specific language governing permissions and
# limitations under the License.
# ==============================================================================
"""Base class for attention layers that can be used in sequence DNN/CNN models.
This file follows the terminology of https://arxiv.org/abs/1706.03762 Figure 2.
Attention is formed by three tensors: Query, Key and Value.
"""
import tensorflow.compat.v2 as tf
from absl import logging
from tf_keras import backend
from tf_keras.engine import base_layer
from tf_keras.utils import control_flow_util
# isort: off
from tensorflow.python.util.tf_export import keras_export
@keras_export("keras.__internal__.layers.BaseDenseAttention", v1=[])
class BaseDenseAttention(base_layer.BaseRandomLayer):
"""Base Attention class for Dense networks.
This class is suitable for Dense or CNN networks, and not for RNN networks.
Implementations of attention mechanisms should inherit from this class, and
reuse the `apply_attention_scores()` method.
Args:
dropout: Float between 0 and 1. Fraction of the units to drop for the
attention scores.
Call arguments:
inputs: List of the following tensors:
* query: Query `Tensor` of shape `[batch_size, Tq, dim]`.
* value: Value `Tensor` of shape `[batch_size, Tv, dim]`.
* key: Optional key `Tensor` of shape `[batch_size, Tv, dim]`. If
not given, will use `value` for both `key` and `value`, which is
the most common case.
mask: List of the following tensors:
* query_mask: A boolean mask `Tensor` of shape `[batch_size, Tq]`.
If given, the output will be zero at the positions where
`mask==False`.
* value_mask: A boolean mask `Tensor` of shape `[batch_size, Tv]`.
If given, will apply the mask such that values at positions
where `mask==False` do not contribute to the result.
training: Python boolean indicating whether the layer should behave in
training mode (adding dropout) or in inference mode (no dropout).
return_attention_scores: bool, if `True`, returns the attention scores
(after masking and softmax) as an additional output argument.
Output:
Attention outputs of shape `[batch_size, Tq, dim]`.
[Optional] Attention scores after masking and softmax with shape
`[batch_size, Tq, Tv]`.
"""
def __init__(self, dropout=0.0, **kwargs):
# Deprecated field `causal` determines whether to using causal masking.
# Use `use_causal_mask` in call() method instead.
if "causal" in kwargs:
logging.warning(
"`causal` argument is deprecated. Please use `use_causal_mask` "
"in call() method to specify causal masking."
)
self.causal = kwargs.pop("causal", False)
super().__init__(**kwargs)
self.dropout = dropout
self.supports_masking = True
def build(self, input_shape):
# Skip RNG initialization if dropout rate is 0. This will let the layer
# be purely stateless, with no reference to any variable.
if self.dropout > 0:
super().build(input_shape)
self.built = True
def _calculate_scores(self, query, key):
"""Calculates attention scores.
Args:
query: Query tensor of shape `[batch_size, Tq, dim]`.
key: Key tensor of shape `[batch_size, Tv, dim]`.
Returns:
Tensor of shape `[batch_size, Tq, Tv]`.
"""
return NotImplementedError
def _apply_scores(self, scores, value, scores_mask=None, training=None):
"""Applies attention scores to the given value tensor.
To use this method in your attention layer, follow the steps:
* Use `query` tensor of shape `[batch_size, Tq]` and `key` tensor of
shape `[batch_size, Tv]` to calculate the attention `scores`.
* Pass `scores` and `value` tensors to this method. The method applies
`scores_mask`, calculates
`attention_distribution = softmax(scores)`, then returns
`matmul(attention_distribution, value).
* Apply `query_mask` and return the result.
Args:
scores: Scores float tensor of shape `[batch_size, Tq, Tv]`.
value: Value tensor of shape `[batch_size, Tv, dim]`.
scores_mask: A boolean mask `Tensor` of shape `[batch_size, 1, Tv]`
or `[batch_size, Tq, Tv]`. If given, scores at positions where
`scores_mask==False` do not contribute to the result. It must
contain at least one `True` value in each line along the last
dimension.
training: Python boolean indicating whether the layer should behave
in training mode (adding dropout) or in inference mode
(no dropout).
Returns:
Tensor of shape `[batch_size, Tq, dim]`.
Attention scores after masking and softmax with shape
`[batch_size, Tq, Tv]`.
"""
if scores_mask is not None:
padding_mask = tf.logical_not(scores_mask)
# Bias so padding positions do not contribute to attention
# distribution. Note 65504. is the max float16 value.
if scores.dtype is tf.float16:
scores -= 65504.0 * tf.cast(padding_mask, dtype=scores.dtype)
else:
scores -= 1.0e9 * tf.cast(padding_mask, dtype=scores.dtype)
if training is None:
training = backend.learning_phase()
weights = tf.nn.softmax(scores)
if self.dropout > 0:
def dropped_weights():
return self._random_generator.dropout(
weights, rate=self.dropout
)
weights = control_flow_util.smart_cond(
training, dropped_weights, lambda: tf.identity(weights)
)
return tf.matmul(weights, value), weights
# TODO(b/125916026): Consider exposing a __call__ method with named args.
def call(
self,
inputs,
mask=None,
training=None,
return_attention_scores=False,
use_causal_mask=False,
):
self._validate_call_args(inputs=inputs, mask=mask)
q = inputs[0]
v = inputs[1]
k = inputs[2] if len(inputs) > 2 else v
q_mask = mask[0] if mask else None
v_mask = mask[1] if mask else None
scores = self._calculate_scores(query=q, key=k)
if v_mask is not None:
# Mask of shape [batch_size, 1, Tv].
v_mask = tf.expand_dims(v_mask, axis=-2)
if self.causal or use_causal_mask:
# Creates a lower triangular mask, so position i cannot attend to
# positions j>i. This prevents the flow of information from the
# future into the past.
scores_shape = tf.shape(scores)
# causal_mask_shape = [1, Tq, Tv].
causal_mask_shape = tf.concat(
[tf.ones_like(scores_shape[:-2]), scores_shape[-2:]], axis=0
)
causal_mask = _lower_triangular_mask(causal_mask_shape)
else:
causal_mask = None
scores_mask = _merge_masks(v_mask, causal_mask)
result, attention_scores = self._apply_scores(
scores=scores, value=v, scores_mask=scores_mask, training=training
)
if q_mask is not None:
# Mask of shape [batch_size, Tq, 1].
q_mask = tf.expand_dims(q_mask, axis=-1)
result *= tf.cast(q_mask, dtype=result.dtype)
if return_attention_scores:
return result, attention_scores
return result
def compute_mask(self, inputs, mask=None):
self._validate_call_args(inputs=inputs, mask=mask)
if mask:
q_mask = mask[0]
if q_mask is None:
return None
return tf.convert_to_tensor(q_mask)
return None
def compute_output_shape(self, input_shape):
# return_attention_scores argument of BaseDenseAttention.call method
# is ignored. Output shape of attention_scores cannot be returned.
return tf.TensorShape(input_shape[0])
def _validate_call_args(self, inputs, mask):
"""Validates arguments of the call method."""
class_name = self.__class__.__name__
if not isinstance(inputs, list):
raise ValueError(
f"{class_name} layer must be called on a list of inputs, "
"namely [query, value] or [query, value, key]. "
f"Received: {inputs}."
)
if len(inputs) < 2 or len(inputs) > 3:
raise ValueError(
f"{class_name} layer accepts inputs list of length 2 or 3, "
"namely [query, value] or [query, value, key]. "
f"Received length: {len(inputs)}."
)
if mask:
if not isinstance(mask, list):
raise ValueError(
f"{class_name} layer mask must be a list, "
f"namely [query_mask, value_mask]. Received: {mask}."
)
if len(mask) < 2 or len(mask) > len(inputs):
raise ValueError(
f"{class_name} layer mask must be a list of length 2, "
"namely [query_mask, value_mask]. "
f"Received length: {len(mask)}."
)
def get_config(self):
config = {
"dropout": self.dropout,
}
base_config = super().get_config()
return dict(list(base_config.items()) + list(config.items()))
def _lower_triangular_mask(shape):
"""Creates a lower-triangular boolean mask over the last 2 dimensions."""
row_index = tf.cumsum(tf.ones(shape=shape, dtype=tf.int32), axis=-2)
col_index = tf.cumsum(tf.ones(shape=shape, dtype=tf.int32), axis=-1)
return tf.greater_equal(row_index, col_index)
def _merge_masks(x, y):
if x is None:
return y
if y is None:
return x
return tf.logical_and(x, y)
|
tf-keras/tf_keras/layers/attention/base_dense_attention.py/0
|
{
"file_path": "tf-keras/tf_keras/layers/attention/base_dense_attention.py",
"repo_id": "tf-keras",
"token_count": 4707
}
| 200 |
# Copyright 2018 The TensorFlow Authors. All Rights Reserved.
#
# Licensed under the Apache License, Version 2.0 (the "License");
# you may not use this file except in compliance with the License.
# You may obtain a copy of the License at
#
# http://www.apache.org/licenses/LICENSE-2.0
#
# Unless required by applicable law or agreed to in writing, software
# distributed under the License is distributed on an "AS IS" BASIS,
# WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.
# See the License for the specific language governing permissions and
# limitations under the License.
# ==============================================================================
"""Tests for convolutional transpose layers."""
import numpy as np
import tensorflow.compat.v2 as tf
from absl.testing import parameterized
import tf_keras as keras
from tf_keras.testing_infra import test_combinations
from tf_keras.testing_infra import test_utils
@test_combinations.run_all_keras_modes
class Conv1DTransposeTest(test_combinations.TestCase):
def _run_test(self, kwargs, expected_output_shape):
num_samples = 2
stack_size = 3
num_col = 6
with test_utils.use_gpu():
test_utils.layer_test(
keras.layers.Conv1DTranspose,
kwargs=kwargs,
input_shape=(num_samples, num_col, stack_size),
expected_output_shape=expected_output_shape,
)
@parameterized.named_parameters(
("padding_valid", {"padding": "valid"}, (None, 8, 2)),
("padding_same", {"padding": "same"}, (None, 6, 2)),
("strides", {"strides": 2}, (None, 13, 2)),
# Only runs on GPU with CUDA, dilation_rate>1 is not supported on CPU.
("dilation_rate", {"dilation_rate": 2}, (None, 10, 2)),
# Only runs on GPU with CUDA, channels_first is not supported on CPU.
# TODO(b/62340061): Support channels_first on CPU.
("data_format", {"data_format": "channels_first"}),
)
def test_conv1d_transpose(self, kwargs, expected_output_shape=None):
kwargs["filters"] = 2
kwargs["kernel_size"] = 3
if (
"data_format" not in kwargs and "dilation_rate" not in kwargs
) or tf.test.is_gpu_available(cuda_only=True):
self._run_test(kwargs, expected_output_shape)
def test_conv1d_transpose_invalid_strides_and_dilation_rate(self):
kwargs = {"strides": 2, "dilation_rate": 2}
with self.assertRaisesRegex(
ValueError, r"""`strides > 1` not supported in conjunction"""
):
keras.layers.Conv1DTranspose(filters=1, kernel_size=2, **kwargs)
@test_combinations.run_all_keras_modes
class Conv2DTransposeTest(test_combinations.TestCase):
def _run_test(self, kwargs):
num_samples = 2
stack_size = 3
num_row = 7
num_col = 6
with self.cached_session():
test_utils.layer_test(
keras.layers.Conv2DTranspose,
kwargs=kwargs,
input_shape=(num_samples, num_row, num_col, stack_size),
)
@parameterized.named_parameters(
("padding_valid", {"padding": "valid"}),
("padding_same", {"padding": "same"}),
("strides", {"strides": (2, 2)}),
# Only runs on GPU with CUDA, channels_first is not supported on CPU.
# TODO(b/62340061): Support channels_first on CPU.
("data_format", {"data_format": "channels_first"}),
(
"strides_output_padding",
{"strides": (2, 2), "output_padding": (1, 1)},
),
)
def test_conv2d_transpose(self, kwargs):
kwargs["filters"] = 2
kwargs["kernel_size"] = (3, 3)
if "data_format" not in kwargs or tf.test.is_gpu_available(
cuda_only=True
):
self._run_test(kwargs)
def test_conv2d_transpose_regularizers(self):
kwargs = {
"filters": 3,
"kernel_size": 3,
"padding": "valid",
"kernel_regularizer": "l2",
"bias_regularizer": "l2",
"activity_regularizer": "l2",
"strides": 1,
}
with self.cached_session():
layer = keras.layers.Conv2DTranspose(**kwargs)
layer.build((None, 5, 5, 2))
self.assertEqual(len(layer.losses), 2)
layer(keras.backend.variable(np.ones((1, 5, 5, 2))))
self.assertEqual(len(layer.losses), 3)
def test_conv2d_transpose_constraints(self):
k_constraint = lambda x: x
b_constraint = lambda x: x
kwargs = {
"filters": 3,
"kernel_size": 3,
"padding": "valid",
"kernel_constraint": k_constraint,
"bias_constraint": b_constraint,
"strides": 1,
}
with self.cached_session():
layer = keras.layers.Conv2DTranspose(**kwargs)
layer.build((None, 5, 5, 2))
self.assertEqual(layer.kernel.constraint, k_constraint)
self.assertEqual(layer.bias.constraint, b_constraint)
def test_conv2d_transpose_dilation(self):
test_utils.layer_test(
keras.layers.Conv2DTranspose,
kwargs={
"filters": 2,
"kernel_size": 3,
"padding": "same",
"data_format": "channels_last",
"dilation_rate": (2, 2),
},
input_shape=(2, 5, 6, 3),
)
input_data = np.arange(48).reshape((1, 4, 4, 3)).astype(np.float32)
expected_output = np.float32(
[
[192, 228, 192, 228],
[336, 372, 336, 372],
[192, 228, 192, 228],
[336, 372, 336, 372],
]
).reshape((1, 4, 4, 1))
test_utils.layer_test(
keras.layers.Conv2DTranspose,
input_data=input_data,
kwargs={
"filters": 1,
"kernel_size": 3,
"padding": "same",
"data_format": "channels_last",
"dilation_rate": (2, 2),
"kernel_initializer": "ones",
},
expected_output=expected_output,
)
def test_conv2d_transpose_invalid_strides_and_dilation_rate(self):
kwargs = {"strides": [2, 1], "dilation_rate": [2, 1]}
with self.assertRaisesRegex(
ValueError, r"""`strides > 1` not supported in conjunction"""
):
keras.layers.Conv2DTranspose(filters=1, kernel_size=2, **kwargs)
@test_combinations.run_all_keras_modes
class Conv3DTransposeTest(test_combinations.TestCase):
def _run_test(self, kwargs, expected_output_shape):
num_samples = 2
stack_size = 3
num_row = 7
num_col = 6
depth = 5
with test_utils.use_gpu():
test_utils.layer_test(
keras.layers.Conv3DTranspose,
kwargs=kwargs,
input_shape=(num_samples, depth, num_row, num_col, stack_size),
expected_output_shape=expected_output_shape,
)
@parameterized.named_parameters(
("padding_valid", {"padding": "valid"}, (None, 7, 9, 8, 2)),
("padding_same", {"padding": "same"}, (None, 5, 7, 6, 2)),
("strides", {"strides": (2, 2, 2)}, (None, 11, 15, 13, 2)),
("dilation_rate", {"dilation_rate": (2, 2, 2)}, (None, 7, 9, 8, 2)),
# Only runs on GPU with CUDA, channels_first is not supported on CPU.
# TODO(b/62340061): Support channels_first on CPU.
("data_format", {"data_format": "channels_first"}),
(
"strides_output_padding",
{"strides": (2, 2, 2), "output_padding": (1, 1, 1)},
(None, 12, 16, 14, 2),
),
)
def test_conv3d_transpose(self, kwargs, expected_output_shape=None):
kwargs["filters"] = 2
kwargs["kernel_size"] = (3, 3, 3)
if "data_format" not in kwargs or tf.test.is_gpu_available(
cuda_only=True
):
self._run_test(kwargs, expected_output_shape)
def test_conv3d_transpose_regularizers(self):
kwargs = {
"filters": 3,
"kernel_size": 3,
"padding": "valid",
"kernel_regularizer": "l2",
"bias_regularizer": "l2",
"activity_regularizer": "l2",
"strides": 1,
}
with self.cached_session():
layer = keras.layers.Conv3DTranspose(**kwargs)
layer.build((None, 5, 5, 5, 2))
self.assertEqual(len(layer.losses), 2)
layer(keras.backend.variable(np.ones((1, 5, 5, 5, 2))))
self.assertEqual(len(layer.losses), 3)
def test_conv3d_transpose_constraints(self):
k_constraint = lambda x: x
b_constraint = lambda x: x
kwargs = {
"filters": 3,
"kernel_size": 3,
"padding": "valid",
"kernel_constraint": k_constraint,
"bias_constraint": b_constraint,
"strides": 1,
}
with self.cached_session():
layer = keras.layers.Conv3DTranspose(**kwargs)
layer.build((None, 5, 5, 5, 2))
self.assertEqual(layer.kernel.constraint, k_constraint)
self.assertEqual(layer.bias.constraint, b_constraint)
def test_conv3d_transpose_dynamic_shape(self):
input_data = np.random.random((1, 3, 3, 3, 3)).astype(np.float32)
with self.cached_session():
# Won't raise error here.
test_utils.layer_test(
keras.layers.Conv3DTranspose,
kwargs={
"data_format": "channels_last",
"filters": 3,
"kernel_size": 3,
},
input_shape=(None, None, None, None, 3),
input_data=input_data,
)
if tf.test.is_gpu_available(cuda_only=True):
test_utils.layer_test(
keras.layers.Conv3DTranspose,
kwargs={
"data_format": "channels_first",
"filters": 3,
"kernel_size": 3,
},
input_shape=(None, 3, None, None, None),
input_data=input_data,
)
def test_conv3d_transpose_invalid_strides_and_dilation_rate(self):
kwargs = {"strides": [2, 2, 1], "dilation_rate": [2, 2, 1]}
with self.assertRaisesRegex(
ValueError, r"""`strides > 1` not supported in conjunction"""
):
keras.layers.Conv3DTranspose(filters=1, kernel_size=2, **kwargs)
if __name__ == "__main__":
tf.test.main()
|
tf-keras/tf_keras/layers/convolutional/conv_transpose_test.py/0
|
{
"file_path": "tf-keras/tf_keras/layers/convolutional/conv_transpose_test.py",
"repo_id": "tf-keras",
"token_count": 5459
}
| 201 |
# Copyright 2022 The TensorFlow Authors. All Rights Reserved.
#
# Licensed under the Apache License, Version 2.0 (the "License");
# you may not use this file except in compliance with the License.
# You may obtain a copy of the License at
#
# http://www.apache.org/licenses/LICENSE-2.0
#
# Unless required by applicable law or agreed to in writing, software
# distributed under the License is distributed on an "AS IS" BASIS,
# WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.
# See the License for the specific language governing permissions and
# limitations under the License.
# ==============================================================================
"""Contains the Identity layer."""
import tensorflow.compat.v2 as tf
from tf_keras.engine.base_layer import Layer
# isort: off
from tensorflow.python.util.tf_export import keras_export
@keras_export("keras.layers.Identity")
class Identity(Layer):
"""Identity layer.
This layer should be used as a placeholder when no operation is to be
performed. The layer is argument insensitive, and returns its `inputs`
argument as output.
Args:
name: Optional name for the layer instance.
"""
def call(self, inputs):
return tf.nest.map_structure(tf.identity, inputs)
|
tf-keras/tf_keras/layers/core/identity.py/0
|
{
"file_path": "tf-keras/tf_keras/layers/core/identity.py",
"repo_id": "tf-keras",
"token_count": 367
}
| 202 |
# Copyright 2015 The TensorFlow Authors. All Rights Reserved.
#
# Licensed under the Apache License, Version 2.0 (the "License");
# you may not use this file except in compliance with the License.
# You may obtain a copy of the License at
#
# http://www.apache.org/licenses/LICENSE-2.0
#
# Unless required by applicable law or agreed to in writing, software
# distributed under the License is distributed on an "AS IS" BASIS,
# WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.
# See the License for the specific language governing permissions and
# limitations under the License.
# ==============================================================================
"""Layer that subtracts two inputs."""
from tf_keras.layers.merging.base_merge import _Merge
from tf_keras.utils import tf_utils
# isort: off
from tensorflow.python.util.tf_export import keras_export
@keras_export("keras.layers.Subtract")
class Subtract(_Merge):
"""Layer that subtracts two inputs.
It takes as input a list of tensors of size 2, both of the same shape, and
returns a single tensor, (inputs[0] - inputs[1]), also of the same shape.
Examples:
```python
import tf_keras as keras
input1 = keras.layers.Input(shape=(16,))
x1 = keras.layers.Dense(8, activation='relu')(input1)
input2 = keras.layers.Input(shape=(32,))
x2 = keras.layers.Dense(8, activation='relu')(input2)
# Equivalent to subtracted = keras.layers.subtract([x1, x2])
subtracted = keras.layers.Subtract()([x1, x2])
out = keras.layers.Dense(4)(subtracted)
model = keras.models.Model(inputs=[input1, input2], outputs=out)
```
"""
@tf_utils.shape_type_conversion
def build(self, input_shape):
super().build(input_shape)
if len(input_shape) != 2:
raise ValueError(
"A `Subtract` layer should be called on exactly 2 inputs. "
f"Received: input_shape={input_shape}"
)
def _merge_function(self, inputs):
if len(inputs) != 2:
raise ValueError(
"A `Subtract` layer should be called on exactly 2 inputs. "
f"Received: inputs={inputs}"
)
return inputs[0] - inputs[1]
@keras_export("keras.layers.subtract")
def subtract(inputs, **kwargs):
"""Functional interface to the `Subtract` layer.
Args:
inputs: A list of input tensors (exactly 2).
**kwargs: Standard layer keyword arguments.
Returns:
A tensor, the difference of the inputs.
Examples:
```python
import tf_keras as keras
input1 = keras.layers.Input(shape=(16,))
x1 = keras.layers.Dense(8, activation='relu')(input1)
input2 = keras.layers.Input(shape=(32,))
x2 = keras.layers.Dense(8, activation='relu')(input2)
subtracted = keras.layers.subtract([x1, x2])
out = keras.layers.Dense(4)(subtracted)
model = keras.models.Model(inputs=[input1, input2], outputs=out)
```
"""
return Subtract(**kwargs)(inputs)
|
tf-keras/tf_keras/layers/merging/subtract.py/0
|
{
"file_path": "tf-keras/tf_keras/layers/merging/subtract.py",
"repo_id": "tf-keras",
"token_count": 1238
}
| 203 |
# Description:
# Contains the TF-Keras pooling layers.
# Placeholder: load unaliased py_library
load("@org_keras//tf_keras:tf_keras.bzl", "tf_py_test")
package(
# copybara:uncomment default_applicable_licenses = ["//tf_keras:license"],
default_visibility = [
"//tf_keras:friends",
"//third_party/py/tensorflow_gnn:__subpackages__",
"//third_party/tensorflow/python/distribute:__pkg__",
"//third_party/tensorflow/python/feature_column:__pkg__",
"//third_party/tensorflow/python/trackable:__pkg__",
"//third_party/tensorflow/tools/pip_package:__pkg__",
"//third_party/tensorflow_models/official/projects/residual_mobilenet/modeling/backbones:__pkg__",
],
licenses = ["notice"],
)
py_library(
name = "pooling",
srcs = ["__init__.py"],
srcs_version = "PY3",
deps = [
":average_pooling1d",
":average_pooling2d",
":average_pooling3d",
":global_average_pooling1d",
":global_average_pooling2d",
":global_average_pooling3d",
":global_max_pooling1d",
":global_max_pooling2d",
":global_max_pooling3d",
":max_pooling1d",
":max_pooling2d",
":max_pooling3d",
],
)
py_library(
name = "base_pooling1d",
srcs = ["base_pooling1d.py"],
srcs_version = "PY3",
deps = [
"//:expect_tensorflow_installed",
"//tf_keras:backend",
"//tf_keras/engine:base_layer",
"//tf_keras/engine:input_spec",
"//tf_keras/utils:engine_utils",
],
)
py_library(
name = "base_pooling2d",
srcs = ["base_pooling2d.py"],
srcs_version = "PY3",
deps = [
"//:expect_tensorflow_installed",
"//tf_keras:backend",
"//tf_keras/engine:base_layer",
"//tf_keras/engine:input_spec",
"//tf_keras/utils:engine_utils",
],
)
py_library(
name = "base_pooling3d",
srcs = ["base_pooling3d.py"],
srcs_version = "PY3",
deps = [
"//:expect_tensorflow_installed",
"//tf_keras:backend",
"//tf_keras/engine:base_layer",
"//tf_keras/engine:input_spec",
"//tf_keras/utils:engine_utils",
],
)
py_library(
name = "base_global_pooling1d",
srcs = ["base_global_pooling1d.py"],
srcs_version = "PY3",
deps = [
"//:expect_tensorflow_installed",
"//tf_keras/engine:base_layer",
"//tf_keras/engine:input_spec",
"//tf_keras/utils:engine_utils",
],
)
py_library(
name = "base_global_pooling2d",
srcs = ["base_global_pooling2d.py"],
srcs_version = "PY3",
deps = [
"//:expect_tensorflow_installed",
"//tf_keras/engine:base_layer",
"//tf_keras/engine:input_spec",
"//tf_keras/utils:engine_utils",
],
)
py_library(
name = "base_global_pooling3d",
srcs = ["base_global_pooling3d.py"],
srcs_version = "PY3",
deps = [
"//:expect_tensorflow_installed",
"//tf_keras/engine:base_layer",
"//tf_keras/engine:input_spec",
"//tf_keras/utils:engine_utils",
],
)
py_library(
name = "max_pooling1d",
srcs = ["max_pooling1d.py"],
srcs_version = "PY3",
deps = [
":base_pooling1d",
"//tf_keras:backend",
],
)
py_library(
name = "max_pooling2d",
srcs = ["max_pooling2d.py"],
srcs_version = "PY3",
deps = [
":base_pooling2d",
"//:expect_tensorflow_installed",
],
)
py_library(
name = "max_pooling3d",
srcs = ["max_pooling3d.py"],
srcs_version = "PY3",
deps = [
":base_pooling3d",
"//:expect_tensorflow_installed",
],
)
py_library(
name = "average_pooling1d",
srcs = ["average_pooling1d.py"],
srcs_version = "PY3",
deps = [
":base_pooling1d",
"//tf_keras:backend",
],
)
py_library(
name = "average_pooling2d",
srcs = ["average_pooling2d.py"],
srcs_version = "PY3",
deps = [
":base_pooling2d",
"//:expect_tensorflow_installed",
],
)
py_library(
name = "average_pooling3d",
srcs = ["average_pooling3d.py"],
srcs_version = "PY3",
deps = [
":base_pooling3d",
"//:expect_tensorflow_installed",
],
)
py_library(
name = "global_max_pooling1d",
srcs = ["global_max_pooling1d.py"],
srcs_version = "PY3",
deps = [
":base_global_pooling1d",
"//tf_keras:backend",
],
)
py_library(
name = "global_max_pooling2d",
srcs = ["global_max_pooling2d.py"],
srcs_version = "PY3",
deps = [
":base_global_pooling2d",
"//tf_keras:backend",
],
)
py_library(
name = "global_max_pooling3d",
srcs = ["global_max_pooling3d.py"],
srcs_version = "PY3",
deps = [
":base_global_pooling3d",
"//tf_keras:backend",
],
)
py_library(
name = "global_average_pooling1d",
srcs = ["global_average_pooling1d.py"],
srcs_version = "PY3",
deps = [
":base_global_pooling1d",
"//:expect_tensorflow_installed",
"//tf_keras:backend",
],
)
py_library(
name = "global_average_pooling2d",
srcs = ["global_average_pooling2d.py"],
srcs_version = "PY3",
deps = [
":base_global_pooling2d",
"//tf_keras:backend",
],
)
py_library(
name = "global_average_pooling3d",
srcs = ["global_average_pooling3d.py"],
srcs_version = "PY3",
deps = [
":base_global_pooling3d",
"//tf_keras:backend",
],
)
tf_py_test(
name = "average_pooling_test",
size = "medium",
srcs = ["average_pooling_test.py"],
python_version = "PY3",
shard_count = 8,
tags = [
"notsan", # TODO(b/183962355)
],
deps = [
"//:expect_absl_installed", # absl/testing:parameterized
"//:expect_tensorflow_installed",
"//tf_keras",
"//tf_keras/testing_infra:test_combinations",
"//tf_keras/testing_infra:test_utils",
],
)
tf_py_test(
name = "max_pooling_test",
size = "medium",
srcs = ["max_pooling_test.py"],
python_version = "PY3",
shard_count = 8,
tags = [
"notsan", # TODO(b/183962355)
],
deps = [
"//:expect_absl_installed", # absl/testing:parameterized
"//:expect_tensorflow_installed",
"//tf_keras",
"//tf_keras/testing_infra:test_combinations",
"//tf_keras/testing_infra:test_utils",
],
)
tf_py_test(
name = "global_average_pooling_test",
size = "medium",
srcs = ["global_average_pooling_test.py"],
python_version = "PY3",
shard_count = 8,
tags = [
"notsan", # TODO(b/183962355)
],
deps = [
"//:expect_absl_installed", # absl/testing:parameterized
"//:expect_tensorflow_installed",
"//tf_keras",
"//tf_keras/testing_infra:test_combinations",
"//tf_keras/testing_infra:test_utils",
],
)
tf_py_test(
name = "global_max_pooling_test",
size = "medium",
srcs = ["global_max_pooling_test.py"],
python_version = "PY3",
shard_count = 8,
tags = [
"notsan", # TODO(b/183962355)
],
deps = [
"//:expect_absl_installed", # absl/testing:parameterized
"//:expect_tensorflow_installed",
"//tf_keras",
"//tf_keras/testing_infra:test_combinations",
"//tf_keras/testing_infra:test_utils",
],
)
|
tf-keras/tf_keras/layers/pooling/BUILD/0
|
{
"file_path": "tf-keras/tf_keras/layers/pooling/BUILD",
"repo_id": "tf-keras",
"token_count": 3780
}
| 204 |
# Copyright 2015 The TensorFlow Authors. All Rights Reserved.
#
# Licensed under the Apache License, Version 2.0 (the "License");
# you may not use this file except in compliance with the License.
# You may obtain a copy of the License at
#
# http://www.apache.org/licenses/LICENSE-2.0
#
# Unless required by applicable law or agreed to in writing, software
# distributed under the License is distributed on an "AS IS" BASIS,
# WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.
# See the License for the specific language governing permissions and
# limitations under the License.
# ==============================================================================
"""Global max pooling 1D layer."""
from tf_keras import backend
from tf_keras.layers.pooling.base_global_pooling1d import GlobalPooling1D
# isort: off
from tensorflow.python.util.tf_export import keras_export
@keras_export("keras.layers.GlobalMaxPooling1D", "keras.layers.GlobalMaxPool1D")
class GlobalMaxPooling1D(GlobalPooling1D):
"""Global max pooling operation for 1D temporal data.
Downsamples the input representation by taking the maximum value over
the time dimension.
For example:
>>> x = tf.constant([[1., 2., 3.], [4., 5., 6.], [7., 8., 9.]])
>>> x = tf.reshape(x, [3, 3, 1])
>>> x
<tf.Tensor: shape=(3, 3, 1), dtype=float32, numpy=
array([[[1.], [2.], [3.]],
[[4.], [5.], [6.]],
[[7.], [8.], [9.]]], dtype=float32)>
>>> max_pool_1d = tf.keras.layers.GlobalMaxPooling1D()
>>> max_pool_1d(x)
<tf.Tensor: shape=(3, 1), dtype=float32, numpy=
array([[3.],
[6.],
[9.], dtype=float32)>
Args:
data_format: A string,
one of `channels_last` (default) or `channels_first`.
The ordering of the dimensions in the inputs.
`channels_last` corresponds to inputs with shape
`(batch, steps, features)` while `channels_first`
corresponds to inputs with shape
`(batch, features, steps)`.
keepdims: A boolean, whether to keep the temporal dimension or not.
If `keepdims` is `False` (default), the rank of the tensor is reduced
for spatial dimensions.
If `keepdims` is `True`, the temporal dimension are retained with
length 1.
The behavior is the same as for `tf.reduce_max` or `np.max`.
Input shape:
- If `data_format='channels_last'`:
3D tensor with shape:
`(batch_size, steps, features)`
- If `data_format='channels_first'`:
3D tensor with shape:
`(batch_size, features, steps)`
Output shape:
- If `keepdims`=False:
2D tensor with shape `(batch_size, features)`.
- If `keepdims`=True:
- If `data_format='channels_last'`:
3D tensor with shape `(batch_size, 1, features)`
- If `data_format='channels_first'`:
3D tensor with shape `(batch_size, features, 1)`
"""
def call(self, inputs):
steps_axis = 1 if self.data_format == "channels_last" else 2
return backend.max(inputs, axis=steps_axis, keepdims=self.keepdims)
# Alias
GlobalMaxPool1D = GlobalMaxPooling1D
|
tf-keras/tf_keras/layers/pooling/global_max_pooling1d.py/0
|
{
"file_path": "tf-keras/tf_keras/layers/pooling/global_max_pooling1d.py",
"repo_id": "tf-keras",
"token_count": 1236
}
| 205 |
# Copyright 2020 The TensorFlow Authors. All Rights Reserved.
#
# Licensed under the Apache License, Version 2.0 (the "License");
# you may not use this file except in compliance with the License.
# You may obtain a copy of the License at
#
# http://www.apache.org/licenses/LICENSE-2.0
#
# Unless required by applicable law or agreed to in writing, software
# distributed under the License is distributed on an "AS IS" BASIS,
# WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.
# See the License for the specific language governing permissions and
# limitations under the License.
# ==============================================================================
"""Benchmark for KPL implementation of vocabulary columns from files with dense
inputs."""
import os
import tensorflow.compat.v2 as tf
import tf_keras as keras
from tf_keras.layers.preprocessing import string_lookup
from tf_keras.layers.preprocessing.benchmarks import (
feature_column_benchmark as fc_bm,
)
# isort: off
from tensorflow.python.eager.def_function import (
function as tf_function,
)
NUM_REPEATS = 10
BATCH_SIZES = [32, 256]
class BenchmarkLayer(tf.test.TestCase, fc_bm.LayerBenchmark):
"""Benchmark the layer forward pass."""
def _write_to_temp_file(self, file_name, vocab_list):
vocab_path = os.path.join(self.get_temp_dir(), file_name + ".txt")
with tf.io.gfile.GFile(vocab_path, "w") as writer:
for vocab in vocab_list:
writer.write(vocab + "\n")
writer.flush()
writer.close()
return vocab_path
def embedding_varlen(self, batch_size, max_length):
"""Benchmark a variable-length embedding."""
# Data and constants.
vocab = fc_bm.create_vocabulary(32768)
path = self._write_to_temp_file("tmp", vocab)
data = fc_bm.create_string_data(
max_length, batch_size * NUM_REPEATS, vocab, pct_oov=0.15
)
# TF-Keras implementation
model = keras.Sequential()
model.add(
keras.Input(shape=(max_length,), name="data", dtype=tf.string)
)
model.add(string_lookup.StringLookup(vocabulary=path, mask_token=None))
# FC implementation
fc = tf.feature_column.categorical_column_with_vocabulary_list(
key="data", vocabulary_list=vocab, num_oov_buckets=1
)
# Wrap the FC implementation in a tf.function for a fair comparison
@tf_function()
def fc_fn(tensors):
fc.transform_feature(
tf.__internal__.feature_column.FeatureTransformationCache(
tensors
),
None,
)
# Benchmark runs
keras_data = {
"data": data.to_tensor(
default_value="", shape=(batch_size, max_length)
)
}
k_avg_time = fc_bm.run_keras(keras_data, model, batch_size, NUM_REPEATS)
fc_data = {
"data": data.to_tensor(
default_value="", shape=(batch_size, max_length)
)
}
fc_avg_time = fc_bm.run_fc(fc_data, fc_fn, batch_size, NUM_REPEATS)
return k_avg_time, fc_avg_time
def benchmark_layer(self):
for batch in BATCH_SIZES:
name = f"vocab_list|dense|batch_{batch}"
k_time, f_time = self.embedding_varlen(
batch_size=batch, max_length=256
)
self.report(name, k_time, f_time, NUM_REPEATS)
if __name__ == "__main__":
tf.test.main()
|
tf-keras/tf_keras/layers/preprocessing/benchmarks/category_vocab_file_dense_benchmark.py/0
|
{
"file_path": "tf-keras/tf_keras/layers/preprocessing/benchmarks/category_vocab_file_dense_benchmark.py",
"repo_id": "tf-keras",
"token_count": 1559
}
| 206 |
# Copyright 2020 The TensorFlow Authors. All Rights Reserved.
#
# Licensed under the Apache License, Version 2.0 (the "License");
# you may not use this file except in compliance with the License.
# You may obtain a copy of the License at
#
# http://www.apache.org/licenses/LICENSE-2.0
#
# Unless required by applicable law or agreed to in writing, software
# distributed under the License is distributed on an "AS IS" BASIS,
# WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.
# See the License for the specific language governing permissions and
# limitations under the License.
# ==============================================================================
"""Benchmark for KPL implementation of weighted embedding column with
varying-length inputs."""
import tensorflow.compat.v2 as tf
import tf_keras as keras
from tf_keras.layers.preprocessing.benchmarks import (
feature_column_benchmark as fc_bm,
)
# isort: off
from tensorflow.python.eager.def_function import (
function as tf_function,
)
NUM_REPEATS = 10
BATCH_SIZES = [32, 256]
### KPL AND FC IMPLEMENTATION BENCHMARKS ###
def embedding_varlen(batch_size, max_length):
"""Benchmark a variable-length embedding."""
# Data and constants.
embedding_size = 32768
data = fc_bm.create_data(
max_length, batch_size * NUM_REPEATS, embedding_size - 1, dtype=int
)
weight = tf.ones_like(data, dtype=tf.float32)
# TF-Keras implementation
data_input = keras.Input(
shape=(None,), ragged=True, name="data", dtype=tf.int64
)
weight_input = keras.Input(
shape=(None,), ragged=True, name="weight", dtype=tf.float32
)
embedded_data = keras.layers.Embedding(embedding_size, 256)(data_input)
weighted_embedding = tf.multiply(
embedded_data, tf.expand_dims(weight_input, -1)
)
reduced_embedding = tf.reduce_sum(weighted_embedding, axis=1)
model = keras.Model([data_input, weight_input], reduced_embedding)
# FC implementation
fc = tf.feature_column.embedding_column(
tf.feature_column.weighted_categorical_column(
tf.feature_column.categorical_column_with_identity(
"data", num_buckets=embedding_size - 1
),
weight_feature_key="weight",
),
dimension=256,
)
# Wrap the FC implementation in a tf.function for a fair comparison
@tf_function()
def fc_fn(tensors):
fc.transform_feature(
tf.__internal__.feature_column.FeatureTransformationCache(tensors),
None,
)
# Benchmark runs
keras_data = {"data": data, "weight": weight}
k_avg_time = fc_bm.run_keras(keras_data, model, batch_size, NUM_REPEATS)
fc_data = {"data": data.to_sparse(), "weight": weight.to_sparse()}
fc_avg_time = fc_bm.run_fc(fc_data, fc_fn, batch_size, NUM_REPEATS)
return k_avg_time, fc_avg_time
class BenchmarkLayer(fc_bm.LayerBenchmark):
"""Benchmark the layer forward pass."""
def benchmark_layer(self):
for batch in BATCH_SIZES:
name = f"weighted_embedding|varlen|batch_{batch}"
k_time, f_time = embedding_varlen(batch_size=batch, max_length=256)
self.report(name, k_time, f_time, NUM_REPEATS)
if __name__ == "__main__":
tf.test.main()
|
tf-keras/tf_keras/layers/preprocessing/benchmarks/weighted_embedding_varlen_benchmark.py/0
|
{
"file_path": "tf-keras/tf_keras/layers/preprocessing/benchmarks/weighted_embedding_varlen_benchmark.py",
"repo_id": "tf-keras",
"token_count": 1285
}
| 207 |
# Copyright 2019 The TensorFlow Authors. All Rights Reserved.
#
# Licensed under the Apache License, Version 2.0 (the "License");
# you may not use this file except in compliance with the License.
# You may obtain a copy of the License at
#
# http://www.apache.org/licenses/LICENSE-2.0
#
# Unless required by applicable law or agreed to in writing, software
# distributed under the License is distributed on an "AS IS" BASIS,
# WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.
# See the License for the specific language governing permissions and
# limitations under the License.
# ==============================================================================
"""Distribution tests for keras.layers.preprocessing.index_lookup."""
import os
import numpy as np
import tensorflow.compat.v2 as tf
import tf_keras as keras
from tf_keras import backend
from tf_keras.distribute import strategy_combinations
from tf_keras.layers.preprocessing import index_lookup
from tf_keras.layers.preprocessing import preprocessing_test_utils
from tf_keras.testing_infra import test_combinations
from tf_keras.testing_infra import test_utils
# isort: off
from tensorflow.python.framework import (
test_util as tf_test_utils,
)
@test_utils.run_v2_only
@tf.__internal__.distribute.combinations.generate(
tf.__internal__.test.combinations.combine(
strategy=strategy_combinations.all_strategies
+ strategy_combinations.multi_worker_mirrored_strategies
+ strategy_combinations.parameter_server_strategies_single_worker
+ strategy_combinations.parameter_server_strategies_multi_worker,
mode=["eager"],
)
)
class IndexLookupDistributionTest(
test_combinations.TestCase, preprocessing_test_utils.PreprocessingLayerTest
):
def _write_to_temp_file(self, file_name, vocab_list):
vocab_path = os.path.join(self.get_temp_dir(), file_name + ".txt")
with tf.io.gfile.GFile(vocab_path, "w") as writer:
for vocab in vocab_list:
writer.write(vocab + "\n")
writer.flush()
writer.close()
return vocab_path
def test_strategy(self, strategy):
if (
backend.is_tpu_strategy(strategy)
and not tf_test_utils.is_mlir_bridge_enabled()
):
self.skipTest("TPU tests require MLIR bridge")
vocab_data = [
[
"earth",
"earth",
"earth",
"earth",
"wind",
"wind",
"wind",
"and",
"and",
"fire",
]
]
vocab_dataset = tf.data.Dataset.from_tensors(vocab_data)
input_array = np.array(
[
["earth", "wind", "and", "fire"],
["fire", "and", "earth", "michigan"],
]
)
input_dataset = tf.data.Dataset.from_tensor_slices(input_array).batch(
2, drop_remainder=True
)
expected_output = [[2, 3, 4, 5], [5, 4, 2, 1]]
tf.config.set_soft_device_placement(True)
with strategy.scope():
input_data = keras.Input(shape=(None,), dtype=tf.string)
layer = index_lookup.IndexLookup(
max_tokens=None,
num_oov_indices=1,
mask_token="",
oov_token="[OOV]",
vocabulary_dtype=tf.string,
)
layer.adapt(vocab_dataset)
int_data = layer(input_data)
model = keras.Model(inputs=input_data, outputs=int_data)
model.compile(loss="mse")
output_dataset = model.predict(input_dataset)
self.assertAllEqual(expected_output, output_dataset)
def test_strategy_with_file(self, strategy):
if (
backend.is_tpu_strategy(strategy)
and not tf_test_utils.is_mlir_bridge_enabled()
):
self.skipTest("TPU tests require MLIR bridge")
vocab_data = ["earth", "wind", "and", "fire"]
vocab_file = self._write_to_temp_file("temp", vocab_data)
input_array = np.array(
[
["earth", "wind", "and", "fire"],
["fire", "and", "earth", "michigan"],
]
)
input_dataset = tf.data.Dataset.from_tensor_slices(input_array).batch(
2, drop_remainder=True
)
expected_output = [[2, 3, 4, 5], [5, 4, 2, 1]]
tf.config.set_soft_device_placement(True)
with strategy.scope():
input_data = keras.Input(shape=(None,), dtype=tf.string)
layer = index_lookup.IndexLookup(
max_tokens=None,
num_oov_indices=1,
mask_token="",
oov_token="[OOV]",
vocabulary_dtype=tf.string,
vocabulary=vocab_file,
)
int_data = layer(input_data)
model = keras.Model(inputs=input_data, outputs=int_data)
model.compile(loss="mse")
output_dataset = model.predict(input_dataset)
self.assertAllEqual(expected_output, output_dataset)
def test_tpu_with_multiple_oov(self, strategy):
# TODO(b/180614455): remove this check when MLIR bridge is always
# enabled.
if backend.is_tpu_strategy(strategy):
self.skipTest("This test needs MLIR bridge on TPU.")
vocab_data = [
[
"earth",
"earth",
"earth",
"earth",
"wind",
"wind",
"wind",
"and",
"and",
"fire",
]
]
vocab_dataset = tf.data.Dataset.from_tensors(vocab_data)
input_array = np.array(
[
["earth", "wind", "and", "fire"],
["fire", "and", "earth", "michigan"],
]
)
input_dataset = tf.data.Dataset.from_tensor_slices(input_array).batch(
2, drop_remainder=True
)
expected_output = [[3, 4, 5, 6], [6, 5, 3, 1]]
tf.config.set_soft_device_placement(True)
with strategy.scope():
input_data = keras.Input(shape=(None,), dtype=tf.string)
layer = index_lookup.IndexLookup(
max_tokens=None,
num_oov_indices=2,
mask_token="",
oov_token="[OOV]",
vocabulary_dtype=tf.string,
)
layer.adapt(vocab_dataset)
int_data = layer(input_data)
model = keras.Model(inputs=input_data, outputs=int_data)
output_dataset = model.predict(input_dataset)
self.assertAllEqual(expected_output, output_dataset)
if __name__ == "__main__":
tf.__internal__.distribute.multi_process_runner.test_main()
|
tf-keras/tf_keras/layers/preprocessing/index_lookup_distribution_test.py/0
|
{
"file_path": "tf-keras/tf_keras/layers/preprocessing/index_lookup_distribution_test.py",
"repo_id": "tf-keras",
"token_count": 3410
}
| 208 |
# Copyright 2015 The TensorFlow Authors. All Rights Reserved.
#
# Licensed under the Apache License, Version 2.0 (the "License");
# you may not use this file except in compliance with the License.
# You may obtain a copy of the License at
#
# http://www.apache.org/licenses/LICENSE-2.0
#
# Unless required by applicable law or agreed to in writing, software
# distributed under the License is distributed on an "AS IS" BASIS,
# WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.
# See the License for the specific language governing permissions and
# limitations under the License.
# ==============================================================================
"""Contains the SpatialDropout3D layer."""
import tensorflow.compat.v2 as tf
from tf_keras import backend
from tf_keras.engine.input_spec import InputSpec
from tf_keras.layers.regularization.dropout import Dropout
# isort: off
from tensorflow.python.util.tf_export import keras_export
@keras_export("keras.layers.SpatialDropout3D")
class SpatialDropout3D(Dropout):
"""Spatial 3D version of Dropout.
This version performs the same function as Dropout, however, it drops
entire 3D feature maps instead of individual elements. If adjacent voxels
within feature maps are strongly correlated (as is normally the case in
early convolution layers) then regular dropout will not regularize the
activations and will otherwise just result in an effective learning rate
decrease. In this case, SpatialDropout3D will help promote independence
between feature maps and should be used instead.
Args:
rate: Float between 0 and 1. Fraction of the input units to drop.
data_format: 'channels_first' or 'channels_last'. In 'channels_first'
mode, the channels dimension (the depth) is at index 1, in
'channels_last' mode is it at index 4. When unspecified, uses
`image_data_format` value found in your TF-Keras config file at
`~/.keras/keras.json` (if exists) else 'channels_last'.
Defaults to 'channels_last'.
Call arguments:
inputs: A 5D tensor.
training: Python boolean indicating whether the layer should behave in
training mode (adding dropout) or in inference mode (doing nothing).
Input shape:
5D tensor with shape: `(samples, channels, dim1, dim2, dim3)` if
data_format='channels_first'
or 5D tensor with shape: `(samples, dim1, dim2, dim3, channels)` if
data_format='channels_last'.
Output shape: Same as input.
References: - [Efficient Object Localization Using Convolutional
Networks](https://arxiv.org/abs/1411.4280)
"""
def __init__(self, rate, data_format=None, **kwargs):
super().__init__(rate, **kwargs)
if data_format is None:
data_format = backend.image_data_format()
if data_format not in {"channels_last", "channels_first"}:
raise ValueError(
'`data_format` must be "channels_last" or "channels_first". '
f"Received: data_format={data_format}."
)
self.data_format = data_format
self.input_spec = InputSpec(ndim=5)
def _get_noise_shape(self, inputs):
input_shape = tf.shape(inputs)
if self.data_format == "channels_first":
return (input_shape[0], input_shape[1], 1, 1, 1)
elif self.data_format == "channels_last":
return (input_shape[0], 1, 1, 1, input_shape[4])
|
tf-keras/tf_keras/layers/regularization/spatial_dropout3d.py/0
|
{
"file_path": "tf-keras/tf_keras/layers/regularization/spatial_dropout3d.py",
"repo_id": "tf-keras",
"token_count": 1218
}
| 209 |
# Copyright 2015 The TensorFlow Authors. All Rights Reserved.
#
# Licensed under the Apache License, Version 2.0 (the "License");
# you may not use this file except in compliance with the License.
# You may obtain a copy of the License at
#
# http://www.apache.org/licenses/LICENSE-2.0
#
# Unless required by applicable law or agreed to in writing, software
# distributed under the License is distributed on an "AS IS" BASIS,
# WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.
# See the License for the specific language governing permissions and
# limitations under the License.
# ==============================================================================
"""Keras upsampling layer for 1D inputs."""
import tensorflow.compat.v2 as tf
from tf_keras import backend
from tf_keras.engine.base_layer import Layer
from tf_keras.engine.input_spec import InputSpec
# isort: off
from tensorflow.python.util.tf_export import keras_export
@keras_export("keras.layers.UpSampling1D")
class UpSampling1D(Layer):
"""Upsampling layer for 1D inputs.
Repeats each temporal step `size` times along the time axis.
Examples:
>>> input_shape = (2, 2, 3)
>>> x = np.arange(np.prod(input_shape)).reshape(input_shape)
>>> print(x)
[[[ 0 1 2]
[ 3 4 5]]
[[ 6 7 8]
[ 9 10 11]]]
>>> y = tf.keras.layers.UpSampling1D(size=2)(x)
>>> print(y)
tf.Tensor(
[[[ 0 1 2]
[ 0 1 2]
[ 3 4 5]
[ 3 4 5]]
[[ 6 7 8]
[ 6 7 8]
[ 9 10 11]
[ 9 10 11]]], shape=(2, 4, 3), dtype=int64)
Args:
size: Integer. Upsampling factor.
Input shape:
3D tensor with shape: `(batch_size, steps, features)`.
Output shape:
3D tensor with shape: `(batch_size, upsampled_steps, features)`.
"""
def __init__(self, size=2, **kwargs):
super().__init__(**kwargs)
self.size = int(size)
self.input_spec = InputSpec(ndim=3)
def compute_output_shape(self, input_shape):
input_shape = tf.TensorShape(input_shape).as_list()
size = (
self.size * input_shape[1] if input_shape[1] is not None else None
)
return tf.TensorShape([input_shape[0], size, input_shape[2]])
def call(self, inputs):
output = backend.repeat_elements(inputs, self.size, axis=1)
return output
def get_config(self):
config = {"size": self.size}
base_config = super().get_config()
return dict(list(base_config.items()) + list(config.items()))
|
tf-keras/tf_keras/layers/reshaping/up_sampling1d.py/0
|
{
"file_path": "tf-keras/tf_keras/layers/reshaping/up_sampling1d.py",
"repo_id": "tf-keras",
"token_count": 1015
}
| 210 |
# Copyright 2015 The TensorFlow Authors. All Rights Reserved.
#
# Licensed under the Apache License, Version 2.0 (the "License");
# you may not use this file except in compliance with the License.
# You may obtain a copy of the License at
#
# http://www.apache.org/licenses/LICENSE-2.0
#
# Unless required by applicable law or agreed to in writing, software
# distributed under the License is distributed on an "AS IS" BASIS,
# WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.
# See the License for the specific language governing permissions and
# limitations under the License.
# ==============================================================================
"""Base class for wrapper layers.
Wrappers are layers that augment the functionality of another layer.
"""
import copy
from tf_keras.engine.base_layer import Layer
from tf_keras.saving import serialization_lib
from tf_keras.saving.legacy import serialization as legacy_serialization
# isort: off
from tensorflow.python.util.tf_export import keras_export
@keras_export("keras.layers.Wrapper")
class Wrapper(Layer):
"""Abstract wrapper base class.
Wrappers take another layer and augment it in various ways.
Do not use this class as a layer, it is only an abstract base class.
Two usable wrappers are the `TimeDistributed` and `Bidirectional` wrappers.
Args:
layer: The layer to be wrapped.
"""
def __init__(self, layer, **kwargs):
try:
assert isinstance(layer, Layer)
except Exception:
raise ValueError(
f"Layer {layer} supplied to wrapper is"
" not a supported layer type. Please"
" ensure wrapped layer is a valid TF-Keras layer."
)
self.layer = layer
super().__init__(**kwargs)
def build(self, input_shape=None):
if not self.layer.built:
self.layer.build(input_shape)
self.layer.built = True
self.built = True
@property
def activity_regularizer(self):
if hasattr(self.layer, "activity_regularizer"):
return self.layer.activity_regularizer
else:
return None
def get_config(self):
try:
config = {
"layer": serialization_lib.serialize_keras_object(self.layer)
}
except TypeError: # Case of incompatible custom wrappers
config = {
"layer": legacy_serialization.serialize_keras_object(self.layer)
}
base_config = super().get_config()
return dict(list(base_config.items()) + list(config.items()))
@classmethod
def from_config(cls, config, custom_objects=None):
from tf_keras.layers import deserialize as deserialize_layer
# Avoid mutating the input dict
config = copy.deepcopy(config)
use_legacy_format = "module" not in config
layer = deserialize_layer(
config.pop("layer"),
custom_objects=custom_objects,
use_legacy_format=use_legacy_format,
)
return cls(layer, **config)
|
tf-keras/tf_keras/layers/rnn/base_wrapper.py/0
|
{
"file_path": "tf-keras/tf_keras/layers/rnn/base_wrapper.py",
"repo_id": "tf-keras",
"token_count": 1180
}
| 211 |
# Copyright 2019 The TensorFlow Authors. All Rights Reserved.
#
# Licensed under the Apache License, Version 2.0 (the "License");
# you may not use this file except in compliance with the License.
# You may obtain a copy of the License at
#
# http://www.apache.org/licenses/LICENSE-2.0
#
# Unless required by applicable law or agreed to in writing, software
# distributed under the License is distributed on an "AS IS" BASIS,
# WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.
# See the License for the specific language governing permissions and
# limitations under the License.
# ==============================================================================
"""Utilities used by both the GRU and LSTM classes."""
import uuid
import tensorflow.compat.v2 as tf
# isort: off
from tensorflow.python.eager.context import get_device_name
# The following string constants are used by Defun approach for unified backend
# of LSTM and GRU.
_FUNCTION_API_NAME_ATTRIBUTE = "api_implements"
_FUNCTION_DEVICE_ATTRIBUTE = "api_preferred_device"
CPU_DEVICE_NAME = "CPU"
GPU_DEVICE_NAME = "GPU"
# The following number constants are used to represent the runtime of the defun
# backend function. Since the CPU/GPU implementation are mathematically same, we
# need some signal for the function to indicate which function is executed. This
# is for testing purpose to verify the correctness of swapping backend function.
RUNTIME_UNKNOWN = 0
RUNTIME_CPU = 1
RUNTIME_GPU = 2
CUDNN_AVAILABLE_MSG = "Layer %s will use cuDNN kernels when running on GPU."
CUDNN_NOT_AVAILABLE_MSG = (
"Layer %s will not use cuDNN kernels since it "
"doesn't meet the criteria. It will "
"use a generic GPU kernel as fallback when running "
"on GPU."
)
def use_new_gru_lstm_impl():
return False
# TODO(b/169707691): The wrapper can be removed if TFLite doesn't need to rely
# on supportive attributes from LSTM/GRU.
class DefunWrapper:
"""A wrapper with no deep copy of the Defun in LSTM/GRU layer."""
def __init__(self, time_major, go_backwards, layer_name):
self.time_major = time_major
self.go_backwards = go_backwards
self.layer_name = layer_name
if self.layer_name not in ["lstm", "gru"]:
raise ValueError(
"Defun wrapper only applies to LSTM and GRU layer, "
"but given {}".format(self.layer_name)
)
# The first two attributes are added to support TFLite use case.
supportive_attributes = {
"time_major": self.time_major,
"go_backwards": self.go_backwards,
_FUNCTION_API_NAME_ATTRIBUTE: self.layer_name
+ "_"
+ str(uuid.uuid4()),
}
if self.layer_name == "lstm":
from tf_keras.layers.rnn import (
lstm,
)
layer_func = lstm.lstm_with_backend_selection
else:
from tf_keras.layers.rnn import (
gru,
)
layer_func = gru.gru_with_backend_selection
self.defun_layer = tf.function(
layer_func,
autograph=False,
experimental_attributes=supportive_attributes,
)
def __deepcopy__(self, memo):
new_wrapper = type(self)(
self.time_major, self.go_backwards, self.layer_name
)
memo[id(self)] = new_wrapper
return new_wrapper
def canonical_to_params(weights, biases, shape, transpose_weights=False):
"""Utility function convert variable to cuDNN compatible parameter.
Note that TF-Keras weights for kernels are different from the cuDNN format.
Eg.:
```
TF-Keras cuDNN
[[0, 1, 2], <---> [[0, 2, 4],
[3, 4, 5]] [1, 3, 5]]
```
If the input weights need to be in a unified format, then set
`transpose_weights=True` to convert the weights.
Args:
weights: list of weights for the individual kernels and recurrent kernels.
biases: list of biases for individual gate.
shape: the shape for the converted variables that will be feed to cuDNN.
transpose_weights: boolean, whether to transpose the weights.
Returns:
The converted weights that can be feed to cuDNN ops as param.
"""
def convert(w):
return tf.transpose(w) if transpose_weights else w
weights = [tf.reshape(convert(x), shape) for x in weights]
biases = [tf.reshape(x, shape) for x in biases]
return tf.concat(weights + biases, axis=0)
def is_sequence_right_padded(mask):
"""Check the mask tensor and see if it right padded.
For cuDNN kernel, it uses the sequence length param to skip the tailing
timestep. If the data is left padded, or not a strict right padding (has
masked value in the middle of the sequence), then cuDNN kernel won't be work
properly in those cases.
Left padded data: [[False, False, True, True, True]].
Right padded data: [[True, True, True, False, False]].
Mixture of mask/unmasked data: [[True, False, True, False, False]].
Note that for the mixed data example above, the actually data RNN should see
are those 2 Trues (index 0 and 2), the index 1 False should be ignored and
not pollute the internal states.
Args:
mask: the Boolean tensor with shape [batch, timestep]
Returns:
boolean scalar tensor, whether the mask is strictly right padded.
"""
max_seq_length = tf.shape(mask)[1]
count_of_true = tf.reduce_sum(tf.cast(mask, tf.int32), axis=1)
right_padded_mask = tf.sequence_mask(count_of_true, maxlen=max_seq_length)
return tf.reduce_all(tf.equal(mask, right_padded_mask))
def has_fully_masked_sequence(mask):
# See https://github.com/tensorflow/tensorflow/issues/33148 for more
# details. Cudnn kernel will error out if the input sequence contains any
# fully masked data. We walk around this issue by rerouting the computation
# to standard kernel, until the issue on cudnn side has been fixed. For a
# fully masked sequence, it will contain all Falses. To make it easy to
# check, we inverse the boolean, check if any of the sequence has all True.
return tf.reduce_any(tf.reduce_all(tf.logical_not(mask), axis=1))
def is_cudnn_supported_inputs(mask, time_major, sequence_lengths):
if tf.sysconfig.get_build_info()["is_rocm_build"]:
if (not time_major) and (sequence_lengths is not None):
return False
if mask is not None:
return tf.reduce_all(mask)
elif sequence_lengths is not None:
return tf.math.equal(
tf.reduce_min(sequence_lengths), tf.reduce_max(sequence_lengths)
)
else:
return True
if mask is None:
return True
if time_major:
mask = tf.transpose(mask)
return tf.logical_and(
is_sequence_right_padded(mask),
tf.logical_not(has_fully_masked_sequence(mask)),
)
def calculate_sequence_by_mask(mask, time_major):
"""Calculate the sequence length tensor (1-D) based on the masking tensor.
The masking tensor is a 2D boolean tensor with shape [batch, timestep]. For
any timestep that should be masked, the corresponding field will be False.
Consider the following example:
a = [[True, True, False, False],
[True, True, True, False]]
It is a (2, 4) tensor, and the corresponding sequence length result should
be 1D tensor with value [2, 3]. Note that the masking tensor must be right
padded that could be checked by, e.g., `is_sequence_right_padded()`.
Args:
mask: Boolean tensor with shape [batch, timestep] or [timestep, batch] if
time_major=True.
time_major: Boolean, which indicates whether the mask is time major or
batch major.
Returns:
sequence_length: 1D int32 tensor.
"""
timestep_index = 0 if time_major else 1
return tf.reduce_sum(tf.cast(mask, tf.int32), axis=timestep_index)
def generate_defun_backend(
unique_api_name, preferred_device, func, supportive_attributes
):
function_attributes = {
_FUNCTION_API_NAME_ATTRIBUTE: unique_api_name,
_FUNCTION_DEVICE_ATTRIBUTE: preferred_device,
}
function_attributes.update(supportive_attributes)
return tf.function(
func, autograph=False, experimental_attributes=function_attributes
)
def get_context_device_type():
"""Parse the current context and return the device type, eg CPU/GPU."""
current_device = get_device_name()
if current_device is None:
return None
return tf.compat.v1.DeviceSpec.from_string(current_device).device_type
def runtime(runtime_name):
with tf.device("/cpu:0"):
return tf.constant(runtime_name, dtype=tf.float32, name="runtime")
def read_variable_value(v):
"""Read the value of a variable if it is variable."""
if isinstance(v, tf.Variable):
return v.read_value()
return v
def function_register(func, *args, **kwargs):
"""Register a specialization of a `Function` into the graph.
This won't actually call the function with the inputs, and only put the
function definition into graph. Register function with different input param
will result into multiple version of functions registered in graph.
Args:
func: the `Function` instance that generated by a @defun
*args: input arguments for the Python function.
**kwargs: input keyword arguments for the Python function.
Returns:
a `ConcreteFunction` object specialized to inputs and execution context.
Raises:
ValueError: When the input function is not a defun wrapped python
function.
"""
concrete_func = func.get_concrete_function(*args, **kwargs)
concrete_func.add_to_graph()
concrete_func.add_gradient_functions_to_graph()
return concrete_func
|
tf-keras/tf_keras/layers/rnn/gru_lstm_utils.py/0
|
{
"file_path": "tf-keras/tf_keras/layers/rnn/gru_lstm_utils.py",
"repo_id": "tf-keras",
"token_count": 3677
}
| 212 |
# Copyright 2016 The TensorFlow Authors. All Rights Reserved.
#
# Licensed under the Apache License, Version 2.0 (the "License");
# you may not use this file except in compliance with the License.
# You may obtain a copy of the License at
#
# http://www.apache.org/licenses/LICENSE-2.0
#
# Unless required by applicable law or agreed to in writing, software
# distributed under the License is distributed on an "AS IS" BASIS,
# WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.
# See the License for the specific language governing permissions and
# limitations under the License.
# ==============================================================================
"""Tests for TimeDistributed wrapper."""
import numpy as np
import tensorflow.compat.v2 as tf
from absl.testing import parameterized
import tf_keras as keras
from tf_keras.testing_infra import test_combinations
from tf_keras.testing_infra import test_utils
# isort: off
from tensorflow.python.checkpoint import (
checkpoint as trackable_util,
)
class TimeDistributedTest(test_combinations.TestCase):
@test_combinations.generate(
test_combinations.combine(mode=["graph", "eager"])
)
def test_timedistributed_dense(self):
model = keras.models.Sequential()
model.add(
keras.layers.TimeDistributed(
keras.layers.Dense(2), input_shape=(3, 4)
)
)
model.compile(optimizer="rmsprop", loss="mse")
model.fit(
np.random.random((10, 3, 4)),
np.random.random((10, 3, 2)),
epochs=1,
batch_size=10,
)
# test config
model.get_config()
# check whether the model variables are present in the
# trackable list of objects
checkpointed_object_ids = {
id(o) for o in trackable_util.list_objects(model)
}
for v in model.variables:
self.assertIn(id(v), checkpointed_object_ids)
def test_timedistributed_static_batch_size(self):
model = keras.models.Sequential()
model.add(
keras.layers.TimeDistributed(
keras.layers.Dense(2), input_shape=(3, 4), batch_size=10
)
)
model.compile(optimizer="rmsprop", loss="mse")
model.fit(
np.random.random((10, 3, 4)),
np.random.random((10, 3, 2)),
epochs=1,
batch_size=10,
)
def test_timedistributed_invalid_init(self):
x = tf.constant(np.zeros((1, 1)).astype("float32"))
with self.assertRaisesRegex(
ValueError,
"Please initialize `TimeDistributed` layer with a "
"`tf.keras.layers.Layer` instance.",
):
keras.layers.TimeDistributed(x)
def test_timedistributed_conv2d(self):
with self.cached_session():
model = keras.models.Sequential()
model.add(
keras.layers.TimeDistributed(
keras.layers.Conv2D(5, (2, 2), padding="same"),
input_shape=(2, 4, 4, 3),
)
)
model.add(keras.layers.Activation("relu"))
model.compile(optimizer="rmsprop", loss="mse")
model.train_on_batch(
np.random.random((1, 2, 4, 4, 3)),
np.random.random((1, 2, 4, 4, 5)),
)
model = keras.models.model_from_json(model.to_json())
model.summary()
def test_timedistributed_stacked(self):
with self.cached_session():
model = keras.models.Sequential()
model.add(
keras.layers.TimeDistributed(
keras.layers.Dense(2), input_shape=(3, 4)
)
)
model.add(keras.layers.TimeDistributed(keras.layers.Dense(3)))
model.add(keras.layers.Activation("relu"))
model.compile(optimizer="rmsprop", loss="mse")
model.fit(
np.random.random((10, 3, 4)),
np.random.random((10, 3, 3)),
epochs=1,
batch_size=10,
)
def test_regularizers(self):
with self.cached_session():
model = keras.models.Sequential()
model.add(
keras.layers.TimeDistributed(
keras.layers.Dense(
2, kernel_regularizer="l1", activity_regularizer="l1"
),
input_shape=(3, 4),
)
)
model.add(keras.layers.Activation("relu"))
model.compile(optimizer="rmsprop", loss="mse")
self.assertEqual(len(model.losses), 2)
def test_TimeDistributed_learning_phase(self):
with self.cached_session():
keras.utils.set_random_seed(0)
x = keras.layers.Input(shape=(3, 2))
y = keras.layers.TimeDistributed(keras.layers.Dropout(0.999))(
x, training=True
)
model = keras.models.Model(x, y)
y = model.predict(np.random.random((10, 3, 2)))
self.assertAllClose(np.mean(y), 0.0, atol=1e-1, rtol=1e-1)
def test_TimeDistributed_batchnorm(self):
with self.cached_session():
# test that wrapped BN updates still work.
model = keras.models.Sequential()
model.add(
keras.layers.TimeDistributed(
keras.layers.BatchNormalization(center=True, scale=True),
name="bn",
input_shape=(10, 2),
)
)
model.compile(optimizer="rmsprop", loss="mse")
# Assert that mean and variance are 0 and 1.
td = model.layers[0]
self.assertAllClose(td.get_weights()[2], np.array([0, 0]))
assert np.array_equal(td.get_weights()[3], np.array([1, 1]))
# Train
model.train_on_batch(
np.random.normal(loc=2, scale=2, size=(1, 10, 2)),
np.broadcast_to(np.array([0, 1]), (1, 10, 2)),
)
# Assert that mean and variance changed.
assert not np.array_equal(td.get_weights()[2], np.array([0, 0]))
assert not np.array_equal(td.get_weights()[3], np.array([1, 1]))
def test_TimeDistributed_trainable(self):
# test layers that need learning_phase to be set
x = keras.layers.Input(shape=(3, 2))
layer = keras.layers.TimeDistributed(keras.layers.BatchNormalization())
_ = layer(x)
self.assertEqual(len(layer.trainable_weights), 2)
layer.trainable = False
assert not layer.trainable_weights
layer.trainable = True
assert len(layer.trainable_weights) == 2
def test_TimeDistributed_with_masked_embedding_and_unspecified_shape(self):
with self.cached_session():
# test with unspecified shape and Embeddings with mask_zero
model = keras.models.Sequential()
model.add(
keras.layers.TimeDistributed(
keras.layers.Embedding(5, 6, mask_zero=True),
input_shape=(None, None),
)
) # N by t_1 by t_2 by 6
model.add(
keras.layers.TimeDistributed(
keras.layers.SimpleRNN(7, return_sequences=True)
)
)
model.add(
keras.layers.TimeDistributed(
keras.layers.SimpleRNN(8, return_sequences=False)
)
)
model.add(keras.layers.SimpleRNN(1, return_sequences=False))
model.compile(optimizer="rmsprop", loss="mse")
model_input = np.random.randint(
low=1, high=5, size=(10, 3, 4), dtype="int32"
)
for i in range(4):
model_input[i, i:, i:] = 0
model.fit(
model_input, np.random.random((10, 1)), epochs=1, batch_size=10
)
mask_outputs = [model.layers[0].compute_mask(model.input)]
for layer in model.layers[1:]:
mask_outputs.append(
layer.compute_mask(layer.input, mask_outputs[-1])
)
func = keras.backend.function([model.input], mask_outputs[:-1])
mask_outputs_val = func([model_input])
ref_mask_val_0 = model_input > 0 # embedding layer
ref_mask_val_1 = ref_mask_val_0 # first RNN layer
ref_mask_val_2 = np.any(ref_mask_val_1, axis=-1) # second RNN layer
ref_mask_val = [ref_mask_val_0, ref_mask_val_1, ref_mask_val_2]
for i in range(3):
self.assertAllEqual(mask_outputs_val[i], ref_mask_val[i])
self.assertIs(mask_outputs[-1], None) # final layer
@test_combinations.generate(
test_combinations.combine(mode=["graph", "eager"])
)
def test_TimeDistributed_with_masking_layer(self):
# test with Masking layer
model = keras.models.Sequential()
model.add(
keras.layers.TimeDistributed(
keras.layers.Masking(
mask_value=0.0,
),
input_shape=(None, 4),
)
)
model.add(keras.layers.TimeDistributed(keras.layers.Dense(5)))
model.compile(optimizer="rmsprop", loss="mse")
model_input = np.random.randint(low=1, high=5, size=(10, 3, 4))
for i in range(4):
model_input[i, i:, :] = 0.0
model.compile(optimizer="rmsprop", loss="mse")
model.fit(
model_input, np.random.random((10, 3, 5)), epochs=1, batch_size=6
)
mask_outputs = [model.layers[0].compute_mask(model.input)]
mask_outputs += [
model.layers[1].compute_mask(
model.layers[1].input, mask_outputs[-1]
)
]
func = keras.backend.function([model.input], mask_outputs)
mask_outputs_val = func([model_input])
self.assertEqual((mask_outputs_val[0]).all(), model_input.all())
self.assertEqual((mask_outputs_val[1]).all(), model_input.all())
def test_TimeDistributed_with_different_time_shapes(self):
time_dist = keras.layers.TimeDistributed(keras.layers.Dense(5))
ph_1 = keras.backend.placeholder(shape=(None, 10, 13))
out_1 = time_dist(ph_1)
self.assertEqual(out_1.shape.as_list(), [None, 10, 5])
ph_2 = keras.backend.placeholder(shape=(None, 1, 13))
out_2 = time_dist(ph_2)
self.assertEqual(out_2.shape.as_list(), [None, 1, 5])
ph_3 = keras.backend.placeholder(shape=(None, 1, 18))
with self.assertRaisesRegex(ValueError, "is incompatible with"):
time_dist(ph_3)
def test_TimeDistributed_with_invalid_dimensions(self):
time_dist = keras.layers.TimeDistributed(keras.layers.Dense(5))
ph = keras.backend.placeholder(shape=(None, 10))
with self.assertRaisesRegex(
ValueError,
"`TimeDistributed` Layer should be passed an `input_shape `",
):
time_dist(ph)
@test_combinations.generate(
test_combinations.combine(mode=["graph", "eager"])
)
def test_TimeDistributed_reshape(self):
class NoReshapeLayer(keras.layers.Layer):
def call(self, inputs):
return inputs
# Built-in layers that aren't stateful use the reshape implementation.
td1 = keras.layers.TimeDistributed(keras.layers.Dense(5))
self.assertTrue(td1._always_use_reshape)
# Built-in layers that are stateful don't use the reshape
# implementation.
td2 = keras.layers.TimeDistributed(
keras.layers.RNN(keras.layers.SimpleRNNCell(10), stateful=True)
)
self.assertFalse(td2._always_use_reshape)
# Custom layers are not allowlisted for the fast reshape implementation.
td3 = keras.layers.TimeDistributed(NoReshapeLayer())
self.assertFalse(td3._always_use_reshape)
@test_combinations.run_all_keras_modes
@parameterized.named_parameters(
("fully_defined", [3, 2, 4], [3, 2, 8]),
("dynamic_batch_size", [None, 2, 4], [None, 2, 8]),
("two_dynamic_dims", [None, None, 4], [None, None, 8]),
("rank_only", [None, None, None], [None, None, None]),
)
def test_TimeDistributed_output_shape_return_types(
self, input_shape, expected_output_shape
):
class TestLayer(keras.layers.Layer):
def call(self, inputs):
return tf.concat([inputs, inputs], axis=-1)
def compute_output_shape(self, input_shape):
output_shape = tf.TensorShape(input_shape).as_list()
if output_shape[-1] is not None:
output_shape[-1] = output_shape[-1] * 2
output_shape = tf.TensorShape(output_shape)
return output_shape
class TestListLayer(TestLayer):
def compute_output_shape(self, input_shape):
shape = super().compute_output_shape(input_shape)
return shape.as_list()
class TestTupleLayer(TestLayer):
def compute_output_shape(self, input_shape):
shape = super().compute_output_shape(input_shape)
return tuple(shape.as_list())
# Layers can specify output shape as list/tuple/TensorShape
test_layers = [TestLayer, TestListLayer, TestTupleLayer]
for layer in test_layers:
input_layer = keras.layers.TimeDistributed(layer())
inputs = keras.backend.placeholder(shape=input_shape)
output = input_layer(inputs)
self.assertEqual(output.shape.as_list(), expected_output_shape)
self.assertEqual(
input_layer.compute_output_shape(input_shape).as_list(),
expected_output_shape,
)
@test_combinations.run_all_keras_modes(always_skip_v1=True)
# TODO(scottzhu): check why v1 session failed.
def test_TimeDistributed_with_mask_first_implementation(self):
np.random.seed(100)
rnn_layer = keras.layers.LSTM(4, return_sequences=True, stateful=True)
data = np.array(
[
[[[1.0], [1.0]], [[0.0], [1.0]]],
[[[1.0], [0.0]], [[1.0], [1.0]]],
[[[1.0], [0.0]], [[1.0], [1.0]]],
]
)
x = keras.layers.Input(shape=(2, 2, 1), batch_size=3)
x_masking = keras.layers.Masking()(x)
y = keras.layers.TimeDistributed(rnn_layer)(x_masking)
model_1 = keras.models.Model(x, y)
model_1.compile(
"rmsprop", "mse", run_eagerly=test_utils.should_run_eagerly()
)
output_with_mask = model_1.predict(data, steps=1)
y = keras.layers.TimeDistributed(rnn_layer)(x)
model_2 = keras.models.Model(x, y)
model_2.compile(
"rmsprop", "mse", run_eagerly=test_utils.should_run_eagerly()
)
output = model_2.predict(data, steps=1)
self.assertNotAllClose(output_with_mask, output, atol=1e-7)
@test_combinations.run_all_keras_modes
@parameterized.named_parameters(
*test_utils.generate_combinations_with_testcase_name(
layer=[keras.layers.LSTM, keras.layers.Dense]
)
)
def test_TimeDistributed_with_ragged_input(self, layer):
if tf.executing_eagerly():
self.skipTest("b/143103634")
np.random.seed(100)
layer = layer(4)
ragged_data = tf.ragged.constant(
[
[[[1.0], [1.0]], [[2.0], [2.0]]],
[[[4.0], [4.0]], [[5.0], [5.0]], [[6.0], [6.0]]],
[[[7.0], [7.0]], [[8.0], [8.0]], [[9.0], [9.0]]],
],
ragged_rank=1,
)
x_ragged = keras.Input(shape=(None, 2, 1), dtype="float32", ragged=True)
y_ragged = keras.layers.TimeDistributed(layer)(x_ragged)
model_1 = keras.models.Model(x_ragged, y_ragged)
model_1._run_eagerly = test_utils.should_run_eagerly()
output_ragged = model_1.predict(ragged_data, steps=1)
x_dense = keras.Input(shape=(None, 2, 1), dtype="float32")
masking = keras.layers.Masking()(x_dense)
y_dense = keras.layers.TimeDistributed(layer)(masking)
model_2 = keras.models.Model(x_dense, y_dense)
dense_data = ragged_data.to_tensor()
model_2._run_eagerly = test_utils.should_run_eagerly()
output_dense = model_2.predict(dense_data, steps=1)
output_ragged = convert_ragged_tensor_value(output_ragged)
self.assertAllEqual(output_ragged.to_tensor(), output_dense)
@test_combinations.run_all_keras_modes
def test_TimeDistributed_with_ragged_input_with_batch_size(self):
np.random.seed(100)
layer = keras.layers.Dense(16)
ragged_data = tf.ragged.constant(
[
[[[1.0], [1.0]], [[2.0], [2.0]]],
[[[4.0], [4.0]], [[5.0], [5.0]], [[6.0], [6.0]]],
[[[7.0], [7.0]], [[8.0], [8.0]], [[9.0], [9.0]]],
],
ragged_rank=1,
)
# Use the first implementation by specifying batch_size
x_ragged = keras.Input(
shape=(None, 2, 1), batch_size=3, dtype="float32", ragged=True
)
y_ragged = keras.layers.TimeDistributed(layer)(x_ragged)
model_1 = keras.models.Model(x_ragged, y_ragged)
output_ragged = model_1.predict(ragged_data, steps=1)
x_dense = keras.Input(shape=(None, 2, 1), batch_size=3, dtype="float32")
masking = keras.layers.Masking()(x_dense)
y_dense = keras.layers.TimeDistributed(layer)(masking)
model_2 = keras.models.Model(x_dense, y_dense)
dense_data = ragged_data.to_tensor()
output_dense = model_2.predict(dense_data, steps=1)
output_ragged = convert_ragged_tensor_value(output_ragged)
self.assertAllEqual(output_ragged.to_tensor(), output_dense)
def test_TimeDistributed_set_static_shape(self):
layer = keras.layers.TimeDistributed(keras.layers.Conv2D(16, (3, 3)))
inputs = keras.Input(batch_shape=(1, None, 32, 32, 1))
outputs = layer(inputs)
# Make sure the batch dim is not lost after array_ops.reshape.
self.assertListEqual(outputs.shape.as_list(), [1, None, 30, 30, 16])
@test_combinations.run_all_keras_modes
def test_TimeDistributed_with_mimo(self):
dense_1 = keras.layers.Dense(8)
dense_2 = keras.layers.Dense(16)
class TestLayer(keras.layers.Layer):
def __init__(self):
super().__init__()
self.dense_1 = dense_1
self.dense_2 = dense_2
def call(self, inputs):
return self.dense_1(inputs[0]), self.dense_2(inputs[1])
def compute_output_shape(self, input_shape):
output_shape_1 = self.dense_1.compute_output_shape(
input_shape[0]
)
output_shape_2 = self.dense_2.compute_output_shape(
input_shape[1]
)
return output_shape_1, output_shape_2
np.random.seed(100)
layer = TestLayer()
data_1 = tf.constant(
[
[[[1.0], [1.0]], [[2.0], [2.0]]],
[[[4.0], [4.0]], [[5.0], [5.0]]],
[[[7.0], [7.0]], [[8.0], [8.0]]],
]
)
data_2 = tf.constant(
[
[[[1.0], [1.0]], [[2.0], [2.0]]],
[[[4.0], [4.0]], [[5.0], [5.0]]],
[[[7.0], [7.0]], [[8.0], [8.0]]],
]
)
x1 = keras.Input(shape=(None, 2, 1), dtype="float32")
x2 = keras.Input(shape=(None, 2, 1), dtype="float32")
y1, y2 = keras.layers.TimeDistributed(layer)([x1, x2])
model_1 = keras.models.Model([x1, x2], [y1, y2])
model_1.compile(
optimizer="rmsprop",
loss="mse",
run_eagerly=test_utils.should_run_eagerly(),
)
output_1 = model_1.predict((data_1, data_2), steps=1)
y1 = dense_1(x1)
y2 = dense_2(x2)
model_2 = keras.models.Model([x1, x2], [y1, y2])
output_2 = model_2.predict((data_1, data_2), steps=1)
self.assertAllClose(output_1, output_2)
model_1.fit(
x=[
np.random.random((10, 2, 2, 1)),
np.random.random((10, 2, 2, 1)),
],
y=[
np.random.random((10, 2, 2, 8)),
np.random.random((10, 2, 2, 16)),
],
epochs=1,
batch_size=3,
)
def test_TimeDistributed_Attention(self):
query_input = keras.layers.Input(shape=(None, 1, 10), dtype="float32")
value_input = keras.layers.Input(shape=(None, 4, 10), dtype="float32")
# Query-value attention of shape [batch_size, Tq, filters].
query_value_attention_seq = keras.layers.TimeDistributed(
keras.layers.Attention()
)([query_input, value_input])
model = keras.models.Model(
[query_input, value_input], query_value_attention_seq
)
model.compile(optimizer="rmsprop", loss="mse")
model.fit(
[
np.random.random((10, 8, 1, 10)),
np.random.random((10, 8, 4, 10)),
],
np.random.random((10, 8, 1, 10)),
epochs=1,
batch_size=10,
)
# test config and serialization/deserialization
model.get_config()
model = keras.models.model_from_json(model.to_json())
model.summary()
def convert_ragged_tensor_value(inputs):
if isinstance(inputs, tf.compat.v1.ragged.RaggedTensorValue):
flat_values = tf.convert_to_tensor(
value=inputs.flat_values, name="flat_values"
)
return tf.RaggedTensor.from_nested_row_splits(
flat_values, inputs.nested_row_splits, validate=False
)
return inputs
if __name__ == "__main__":
tf.test.main()
|
tf-keras/tf_keras/layers/rnn/time_distributed_test.py/0
|
{
"file_path": "tf-keras/tf_keras/layers/rnn/time_distributed_test.py",
"repo_id": "tf-keras",
"token_count": 11405
}
| 213 |
# Copyright 2015 The TensorFlow Authors. All Rights Reserved.
#
# Licensed under the Apache License, Version 2.0 (the "License");
# you may not use this file except in compliance with the License.
# You may obtain a copy of the License at
#
# http://www.apache.org/licenses/LICENSE-2.0
#
# Unless required by applicable law or agreed to in writing, software
# distributed under the License is distributed on an "AS IS" BASIS,
# WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.
# See the License for the specific language governing permissions and
# limitations under the License.
# ==============================================================================
"""Tests for tf.layers.normalization."""
from __future__ import absolute_import
from __future__ import division
from __future__ import print_function
import os
import numpy as np
import tensorflow.compat.v2 as tf
from tf_keras.legacy_tf_layers import convolutional as conv_layers
from tf_keras.legacy_tf_layers import normalization as normalization_layers
# isort: off
from tensorflow.core.protobuf import saver_pb2
from tensorflow.python.framework import (
test_util as tf_test_utils,
)
@tf_test_utils.run_v1_only("b/120545219")
class BNTest(tf.test.TestCase):
def _simple_model(self, image, fused, freeze_mode):
output_channels, kernel_size = 2, 3
conv = conv_layers.conv2d(
image,
output_channels,
kernel_size,
use_bias=False,
kernel_initializer=tf.compat.v1.ones_initializer(),
)
bn_layer = normalization_layers.BatchNormalization(fused=fused)
bn_layer._bessels_correction_test_only = False
training = not freeze_mode
bn = bn_layer(conv, training=training)
loss = tf.reduce_sum(tf.abs(bn))
optimizer = tf.compat.v1.train.GradientDescentOptimizer(0.01)
if not freeze_mode:
update_ops = tf.compat.v1.get_collection(
tf.compat.v1.GraphKeys.UPDATE_OPS
)
with tf.control_dependencies(update_ops):
train_op = optimizer.minimize(loss)
else:
train_op = optimizer.minimize(loss)
saver = tf.compat.v1.train.Saver(write_version=saver_pb2.SaverDef.V2)
return loss, train_op, saver
def _train(
self,
checkpoint_path,
shape,
use_gpu,
is_fused,
restore=False,
freeze_mode=False,
dtype=tf.float32,
):
tf.compat.v1.reset_default_graph()
graph = tf.compat.v1.get_default_graph()
with self.session(graph=graph, use_gpu=use_gpu) as sess:
image = tf.compat.v1.placeholder(dtype=dtype, shape=shape)
loss, train_op, saver = self._simple_model(
image, is_fused, freeze_mode
)
if restore:
saver.restore(sess, checkpoint_path)
else:
self.evaluate(tf.compat.v1.global_variables_initializer())
np.random.seed(0)
for _ in range(2):
image_val = np.random.rand(*shape).astype(dtype.as_numpy_dtype)
sess.run([loss, train_op], feed_dict={image: image_val})
if restore:
all_vars = tf.compat.v1.get_collection(
tf.compat.v1.GraphKeys.GLOBAL_VARIABLES
)
all_vars_values = [var.eval() for var in all_vars]
return all_vars_values
else:
saver.save(sess, checkpoint_path)
def _infer(self, checkpoint_path, image_val, shape, use_gpu, is_fused):
dtype = image_val.dtype
tf.compat.v1.reset_default_graph()
graph = tf.compat.v1.get_default_graph()
with self.session(graph=graph, use_gpu=use_gpu) as sess:
image = tf.compat.v1.placeholder(dtype=dtype, shape=shape)
loss, _, saver = self._simple_model(image, is_fused, True)
saver.restore(sess, checkpoint_path)
loss_val = sess.run(loss, feed_dict={image: image_val})
return loss_val
def _trainEvalSequence(
self, dtype, train1_use_gpu, train2_use_gpu, infer_use_gpu
):
batch, height, width, input_channels = 2, 4, 5, 3
shape = [batch, height, width, input_channels]
# Not all characters in a dtype string representation are allowed in
# filenames in all operating systems. This map will sanitize these.
dtype_to_valid_fn = {
tf.float16: "float16",
tf.float32: "float32",
}
checkpoint = os.path.join(
self.get_temp_dir(),
"cp_%s_%s_%s_%s"
% (
dtype_to_valid_fn[dtype],
train1_use_gpu,
train2_use_gpu,
infer_use_gpu,
),
)
self._train(
checkpoint,
shape,
use_gpu=train1_use_gpu,
is_fused=True,
restore=False,
freeze_mode=False,
dtype=dtype,
)
train_vars = self._train(
checkpoint,
shape,
use_gpu=train2_use_gpu,
is_fused=True,
restore=True,
freeze_mode=False,
dtype=dtype,
)
np.random.seed(0)
image_val = np.random.rand(batch, height, width, input_channels).astype(
dtype.as_numpy_dtype
)
loss_val = self._infer(
checkpoint, image_val, shape, use_gpu=infer_use_gpu, is_fused=True
)
return train_vars, loss_val
def testHalfPrecision(self):
ref_vars, ref_loss = self._trainEvalSequence(
dtype=tf.float32,
train1_use_gpu=True,
train2_use_gpu=True,
infer_use_gpu=True,
)
self.assertEqual(len(ref_vars), 5)
for train1_use_gpu in [True, False]:
for train2_use_gpu in [True, False]:
for infer_use_gpu in [True, False]:
test_vars, test_loss = self._trainEvalSequence(
tf.float16,
train1_use_gpu,
train2_use_gpu,
infer_use_gpu,
)
self.assertEqual(len(test_vars), 5)
for test_var, ref_var in zip(test_vars, ref_vars):
self.assertAllClose(
test_var, ref_var, rtol=1.0e-3, atol=1.0e-3
)
self.assertAllClose(
test_loss, ref_loss, rtol=1.0e-3, atol=1.0e-3
)
def _testCheckpoint(
self,
is_fused_checkpoint_a,
is_fused_checkpoint_b,
use_gpu_checkpoint_a,
use_gpu_checkpoint_b,
use_gpu_test_a,
use_gpu_test_b,
freeze_mode,
):
batch, height, width, input_channels = 2, 4, 5, 3
shape = [batch, height, width, input_channels]
base_path = "%s_%s_%s_%s_%s_%s" % (
is_fused_checkpoint_a,
is_fused_checkpoint_b,
use_gpu_checkpoint_a,
use_gpu_checkpoint_b,
use_gpu_test_a,
use_gpu_test_b,
)
checkpoint_path_a = os.path.join(
self.get_temp_dir(), f"checkpoint_a_{base_path}"
)
self._train(
checkpoint_path_a,
shape,
use_gpu_checkpoint_a,
is_fused_checkpoint_a,
restore=False,
freeze_mode=freeze_mode,
)
checkpoint_path_b = os.path.join(
self.get_temp_dir(), f"checkpoint_b_{base_path}"
)
self._train(
checkpoint_path_b,
shape,
use_gpu_checkpoint_b,
is_fused_checkpoint_b,
restore=False,
freeze_mode=freeze_mode,
)
vars_fused = self._train(
checkpoint_path_a,
shape,
use_gpu_test_a,
True,
restore=True,
freeze_mode=freeze_mode,
)
vars_nonfused = self._train(
checkpoint_path_b,
shape,
use_gpu_test_b,
False,
restore=True,
freeze_mode=freeze_mode,
)
self.assertEqual(len(vars_fused), 5)
self.assertEqual(len(vars_nonfused), 5)
for var_fused, var_nonfused in zip(vars_fused, vars_nonfused):
self.assertAllClose(var_fused, var_nonfused, atol=1e-5)
image_val = np.random.rand(batch, height, width, input_channels).astype(
np.float32
)
loss_fused_val = self._infer(
checkpoint_path_a, image_val, shape, use_gpu_test_a, True
)
loss_nonfused_val = self._infer(
checkpoint_path_b, image_val, shape, use_gpu_test_b, False
)
self.assertAllClose(
loss_fused_val, loss_nonfused_val, atol=1e-6, rtol=3e-4
)
def _testCheckpointCrossDevice(
self, ckpt_a_fused, ckpt_a_use_gpu, ckpt_b_fused, ckpt_b_use_gpu
):
for use_gpu_test_a in [True, False]:
for use_gpu_test_b in [True, False]:
for freeze_mode in [True, False]:
self._testCheckpoint(
ckpt_a_fused,
ckpt_a_use_gpu,
ckpt_b_fused,
ckpt_b_use_gpu,
use_gpu_test_a,
use_gpu_test_b,
freeze_mode,
)
def testCheckpointFusedCPUAndFusedGPU(self):
self._testCheckpointCrossDevice(True, False, True, True)
def testCheckpointFusedCPUAndFusedCPU(self):
self._testCheckpointCrossDevice(True, False, True, False)
def testCheckpointFusedGPUAndFusedGPU(self):
self._testCheckpointCrossDevice(True, True, True, True)
def testCheckpointNonFusedCPUAndNonFusedGPU(self):
self._testCheckpointCrossDevice(False, False, False, True)
def testCheckpointNonFusedCPUAndNonFusedCPU(self):
self._testCheckpointCrossDevice(False, False, False, False)
def testCheckpointNonFusedGPUAndNonFusedGPU(self):
self._testCheckpointCrossDevice(False, True, False, True)
def testCheckpointNonFusedGPUAndFusedGPU(self):
self._testCheckpointCrossDevice(False, True, True, True)
def testCheckpointNonFusedGPUAndFusedCPU(self):
self._testCheckpointCrossDevice(False, True, True, False)
def testCheckpointNonFusedCPUAndFusedCPU(self):
self._testCheckpointCrossDevice(False, False, True, False)
def testCreateBN(self):
# Call layer.
bn = normalization_layers.BatchNormalization(axis=1)
inputs = tf.random.uniform((5, 4, 3), seed=1)
training = tf.compat.v1.placeholder(dtype="bool")
outputs = bn(inputs, training=training)
# Verify shape.
self.assertListEqual(outputs.get_shape().as_list(), [5, 4, 3])
# Verify layer attributes.
self.assertEqual(len(bn.updates), 2)
self.assertEqual(len(bn.variables), 4)
self.assertEqual(len(bn.trainable_variables), 2)
self.assertEqual(len(bn.non_trainable_variables), 2)
# Test that updates were created and added to UPDATE_OPS.
self.assertEqual(len(bn.updates), 2)
self.assertListEqual(
tf.compat.v1.get_collection(tf.compat.v1.GraphKeys.UPDATE_OPS),
bn.updates,
)
# Test that weights were created and added to TRAINABLE_VARIABLES.
self.assertListEqual(
tf.compat.v1.get_collection(
tf.compat.v1.GraphKeys.TRAINABLE_VARIABLES
),
bn.trainable_variables,
)
def testCreateFusedBNFloat16(self):
# Call layer.
bn = normalization_layers.BatchNormalization(axis=1, fused=True)
inputs = tf.random.uniform((5, 4, 3, 3), seed=1, dtype=tf.float16)
training = tf.compat.v1.placeholder(dtype="bool")
outputs = bn(inputs, training=training)
# Verify shape.
self.assertListEqual(outputs.get_shape().as_list(), [5, 4, 3, 3])
# Verify layer attributes.
self.assertEqual(len(bn.updates), 2)
self.assertEqual(len(bn.variables), 4)
self.assertEqual(len(bn.trainable_variables), 2)
self.assertEqual(len(bn.non_trainable_variables), 2)
for var in bn.variables:
self.assertTrue(var.dtype._is_ref_dtype)
# Test that updates were created and added to UPDATE_OPS.
self.assertEqual(len(bn.updates), 2)
self.assertListEqual(
tf.compat.v1.get_collection(tf.compat.v1.GraphKeys.UPDATE_OPS),
bn.updates,
)
# Test that weights were created and added to TRAINABLE_VARIABLES.
self.assertListEqual(
tf.compat.v1.get_collection(
tf.compat.v1.GraphKeys.TRAINABLE_VARIABLES
),
bn.trainable_variables,
)
def test3DInputAxis1(self):
epsilon = 1e-3
bn = normalization_layers.BatchNormalization(
axis=1, epsilon=epsilon, momentum=0.9
)
inputs = tf.Variable(
np.random.random((5, 4, 3)) + 100, dtype=tf.float32
)
training = tf.compat.v1.placeholder(dtype="bool")
outputs = bn(inputs, training=training)
with self.cached_session() as sess:
# Test training with placeholder learning phase.
self.evaluate(tf.compat.v1.global_variables_initializer())
np_gamma, np_beta = self.evaluate([bn.gamma, bn.beta])
np_gamma = np.reshape(np_gamma, (1, 4, 1))
np_beta = np.reshape(np_beta, (1, 4, 1))
for _ in range(100):
np_output, _, _ = sess.run(
[outputs] + bn.updates, feed_dict={training: True}
)
# Verify that the axis is normalized during training.
normed_np_output = ((np_output - epsilon) * np_gamma) + np_beta
self.assertAlmostEqual(np.mean(normed_np_output), 0.0, places=1)
self.assertAlmostEqual(np.std(normed_np_output), 1.0, places=1)
# Verify that the statistics are updated during training.
moving_mean, moving_var = self.evaluate(
[bn.moving_mean, bn.moving_variance]
)
np_inputs = self.evaluate(inputs)
mean = np.mean(np_inputs, axis=(0, 2))
std = np.std(np_inputs, axis=(0, 2))
variance = np.square(std)
self.assertAllClose(mean, moving_mean, atol=1e-2)
self.assertAllClose(variance, moving_var, atol=1e-2)
# Test inference with placeholder learning phase.
np_output = sess.run(outputs, feed_dict={training: False})
# Verify that the axis is normalized during inference.
normed_np_output = ((np_output - epsilon) * np_gamma) + np_beta
self.assertAlmostEqual(np.mean(normed_np_output), 0.0, places=1)
self.assertAlmostEqual(np.std(normed_np_output), 1.0, places=1)
def test3DInputAxis2(self):
epsilon = 1e-3
bn = normalization_layers.BatchNormalization(
axis=2, epsilon=epsilon, momentum=0.9
)
inputs = tf.Variable(
np.random.random((5, 4, 3)) + 100, dtype=tf.float32
)
training = tf.compat.v1.placeholder(dtype="bool")
outputs = bn(inputs, training=training)
with self.cached_session() as sess:
# Test training with placeholder learning phase.
self.evaluate(tf.compat.v1.global_variables_initializer())
np_gamma, np_beta = self.evaluate([bn.gamma, bn.beta])
np_gamma = np.reshape(np_gamma, (1, 1, 3))
np_beta = np.reshape(np_beta, (1, 1, 3))
for _ in range(100):
np_output, _, _ = sess.run(
[outputs] + bn.updates, feed_dict={training: True}
)
# Verify that the axis is normalized during training.
normed_np_output = ((np_output - epsilon) * np_gamma) + np_beta
self.assertAlmostEqual(np.mean(normed_np_output), 0.0, places=1)
self.assertAlmostEqual(np.std(normed_np_output), 1.0, places=1)
# Verify that the statistics are updated during training.
moving_mean, moving_var = self.evaluate(
[bn.moving_mean, bn.moving_variance]
)
np_inputs = self.evaluate(inputs)
mean = np.mean(np_inputs, axis=(0, 1))
std = np.std(np_inputs, axis=(0, 1))
variance = np.square(std)
self.assertAllClose(mean, moving_mean, atol=1e-2)
self.assertAllClose(variance, moving_var, atol=1e-2)
# Test inference with placeholder learning phase.
np_output = sess.run(outputs, feed_dict={training: False})
# Verify that the axis is normalized during inference.
normed_np_output = ((np_output - epsilon) * np_gamma) + np_beta
self.assertAlmostEqual(np.mean(normed_np_output), 0.0, places=1)
self.assertAlmostEqual(np.std(normed_np_output), 1.0, places=1)
def test4DInputAxis1(self):
if tf.test.is_gpu_available(cuda_only=True):
epsilon = 1e-3
bn = normalization_layers.BatchNormalization(
axis=1, epsilon=epsilon, momentum=0.9
)
inputs = tf.Variable(
np.random.random((5, 4, 3, 6)) + 100, dtype=tf.float32
)
training = tf.compat.v1.placeholder(dtype="bool")
outputs = bn(inputs, training=training)
with self.session() as sess:
# Test training with placeholder learning phase.
self.evaluate(tf.compat.v1.global_variables_initializer())
np_gamma, np_beta = self.evaluate([bn.gamma, bn.beta])
np_gamma = np.reshape(np_gamma, (1, 4, 1, 1))
np_beta = np.reshape(np_beta, (1, 4, 1, 1))
for _ in range(100):
np_output, _, _ = sess.run(
[outputs] + bn.updates, feed_dict={training: True}
)
# Verify that the axis is normalized during training.
normed_np_output = (
(np_output - epsilon) * np_gamma
) + np_beta
self.assertAlmostEqual(
np.mean(normed_np_output), 0.0, places=1
)
self.assertAlmostEqual(
np.std(normed_np_output), 1.0, places=1
)
# Verify that the statistics are updated during training.
moving_mean, moving_var = self.evaluate(
[bn.moving_mean, bn.moving_variance]
)
np_inputs = self.evaluate(inputs)
mean = np.mean(np_inputs, axis=(0, 2, 3))
std = np.std(np_inputs, axis=(0, 2, 3))
variance = np.square(std)
self.assertAllClose(mean, moving_mean, atol=1e-2)
self.assertAllClose(variance, moving_var, atol=1e-2)
# Test inference with placeholder learning phase.
np_output = sess.run(outputs, feed_dict={training: False})
# Verify that the axis is normalized during inference.
normed_np_output = ((np_output - epsilon) * np_gamma) + np_beta
self.assertAlmostEqual(np.mean(normed_np_output), 0.0, places=1)
self.assertAlmostEqual(np.std(normed_np_output), 1.0, places=1)
def test4DInputAxis2(self):
epsilon = 1e-3
bn = normalization_layers.BatchNormalization(
axis=2, epsilon=epsilon, momentum=0.9
)
inputs = tf.Variable(
np.random.random((5, 4, 3, 6)) + 100, dtype=tf.float32
)
training = tf.compat.v1.placeholder(dtype="bool")
outputs = bn(inputs, training=training)
with self.cached_session() as sess:
# Test training with placeholder learning phase.
self.evaluate(tf.compat.v1.global_variables_initializer())
np_gamma, np_beta = self.evaluate([bn.gamma, bn.beta])
np_gamma = np.reshape(np_gamma, (1, 1, 3, 1))
np_beta = np.reshape(np_beta, (1, 1, 3, 1))
for _ in range(100):
np_output, _, _ = sess.run(
[outputs] + bn.updates, feed_dict={training: True}
)
# Verify that the axis is normalized during training.
normed_np_output = ((np_output - epsilon) * np_gamma) + np_beta
self.assertAlmostEqual(np.mean(normed_np_output), 0.0, places=1)
self.assertAlmostEqual(np.std(normed_np_output), 1.0, places=1)
# Verify that the statistics are updated during training.
moving_mean, moving_var = self.evaluate(
[bn.moving_mean, bn.moving_variance]
)
np_inputs = self.evaluate(inputs)
mean = np.mean(np_inputs, axis=(0, 1, 3))
std = np.std(np_inputs, axis=(0, 1, 3))
variance = np.square(std)
self.assertAllClose(mean, moving_mean, atol=1e-2)
self.assertAllClose(variance, moving_var, atol=1e-2)
# Test inference with placeholder learning phase.
np_output = sess.run(outputs, feed_dict={training: False})
# Verify that the axis is normalized during inference.
normed_np_output = ((np_output - epsilon) * np_gamma) + np_beta
self.assertAlmostEqual(np.mean(normed_np_output), 0.0, places=1)
self.assertAlmostEqual(np.std(normed_np_output), 1.0, places=1)
def test4DInputAxis3(self):
epsilon = 1e-3
bn = normalization_layers.BatchNormalization(
axis=3, epsilon=epsilon, momentum=0.9
)
inputs = tf.Variable(
np.random.random((5, 4, 3, 6)) + 100, dtype=tf.float32
)
training = tf.compat.v1.placeholder(dtype="bool")
outputs = bn(inputs, training=training)
with self.cached_session() as sess:
# Test training with placeholder learning phase.
self.evaluate(tf.compat.v1.global_variables_initializer())
np_gamma, np_beta = self.evaluate([bn.gamma, bn.beta])
np_gamma = np.reshape(np_gamma, (1, 1, 1, 6))
np_beta = np.reshape(np_beta, (1, 1, 1, 6))
for _ in range(100):
np_output, _, _ = sess.run(
[outputs] + bn.updates, feed_dict={training: True}
)
# Verify that the axis is normalized during training.
normed_np_output = ((np_output - epsilon) * np_gamma) + np_beta
self.assertAlmostEqual(np.mean(normed_np_output), 0.0, places=1)
self.assertAlmostEqual(np.std(normed_np_output), 1.0, places=1)
# Verify that the statistics are updated during training.
moving_mean, moving_var = self.evaluate(
[bn.moving_mean, bn.moving_variance]
)
np_inputs = self.evaluate(inputs)
mean = np.mean(np_inputs, axis=(0, 1, 2))
std = np.std(np_inputs, axis=(0, 1, 2))
variance = np.square(std)
self.assertAllClose(mean, moving_mean, atol=1e-2)
self.assertAllClose(variance, moving_var, atol=1e-2)
# Test inference with placeholder learning phase.
np_output = sess.run(outputs, feed_dict={training: False})
# Verify that the axis is normalized during inference.
normed_np_output = ((np_output - epsilon) * np_gamma) + np_beta
self.assertAlmostEqual(np.mean(normed_np_output), 0.0, places=1)
self.assertAlmostEqual(np.std(normed_np_output), 1.0, places=1)
def test4DInputAxis3Fused(self):
epsilon = 1e-3
bn = normalization_layers.BatchNormalization(
axis=3, epsilon=epsilon, momentum=0.9, fused=True
)
inputs = tf.Variable(
np.random.random((5, 4, 3, 6)) + 100, dtype=tf.float32
)
training = tf.compat.v1.placeholder(dtype="bool")
outputs = bn(inputs, training=training)
with self.cached_session() as sess:
# Test training with placeholder learning phase.
self.evaluate(tf.compat.v1.global_variables_initializer())
np_gamma, np_beta = self.evaluate([bn.gamma, bn.beta])
np_gamma = np.reshape(np_gamma, (1, 1, 1, 6))
np_beta = np.reshape(np_beta, (1, 1, 1, 6))
for _ in range(100):
np_output, _, _ = sess.run(
[outputs] + bn.updates, feed_dict={training: True}
)
# Verify that the axis is normalized during training.
normed_np_output = ((np_output - epsilon) * np_gamma) + np_beta
self.assertAlmostEqual(np.mean(normed_np_output), 0.0, places=1)
self.assertAlmostEqual(np.std(normed_np_output), 1.0, places=1)
# Verify that the statistics are updated during training.
moving_mean, moving_var = self.evaluate(
[bn.moving_mean, bn.moving_variance]
)
np_inputs = self.evaluate(inputs)
mean = np.mean(np_inputs, axis=(0, 1, 2))
std = np.std(np_inputs, axis=(0, 1, 2))
variance = np.square(std)
self.assertAllClose(mean, moving_mean, atol=1e-2)
self.assertAllClose(variance, moving_var, atol=1e-2)
# Test inference with placeholder learning phase.
np_output = sess.run(outputs, feed_dict={training: False})
# Verify that the axis is normalized during inference.
normed_np_output = ((np_output - epsilon) * np_gamma) + np_beta
self.assertAlmostEqual(np.mean(normed_np_output), 0.0, places=1)
self.assertAlmostEqual(np.std(normed_np_output), 1.0, places=1)
def test4DInputAxis1Fused(self):
if tf.test.is_gpu_available(cuda_only=True):
epsilon = 1e-3
bn = normalization_layers.BatchNormalization(
axis=1, epsilon=epsilon, momentum=0.9, fused=True
)
inputs = tf.Variable(
np.random.random((5, 4, 3, 6)) + 100, dtype=tf.float32
)
training = tf.compat.v1.placeholder(dtype="bool")
outputs = bn(inputs, training=training)
with self.cached_session() as sess:
# Test training with placeholder learning phase.
self.evaluate(tf.compat.v1.global_variables_initializer())
np_gamma, np_beta = self.evaluate([bn.gamma, bn.beta])
np_gamma = np.reshape(np_gamma, (1, 4, 1, 1))
np_beta = np.reshape(np_beta, (1, 4, 1, 1))
for _ in range(100):
np_output, _, _ = sess.run(
[outputs] + bn.updates, feed_dict={training: True}
)
# Verify that the axis is normalized during training.
normed_np_output = (
(np_output - epsilon) * np_gamma
) + np_beta
self.assertAlmostEqual(
np.mean(normed_np_output), 0.0, places=1
)
self.assertAlmostEqual(
np.std(normed_np_output), 1.0, places=1
)
# Verify that the statistics are updated during training.
moving_mean, moving_var = self.evaluate(
[bn.moving_mean, bn.moving_variance]
)
np_inputs = self.evaluate(inputs)
mean = np.mean(np_inputs, axis=(0, 2, 3))
std = np.std(np_inputs, axis=(0, 2, 3))
variance = np.square(std)
self.assertAllClose(mean, moving_mean, atol=1e-2)
self.assertAllClose(variance, moving_var, atol=1e-2)
# Test inference with placeholder learning phase.
np_output = sess.run(outputs, feed_dict={training: False})
# Verify that the axis is normalized during inference.
normed_np_output = ((np_output - epsilon) * np_gamma) + np_beta
self.assertAlmostEqual(np.mean(normed_np_output), 0.0, places=1)
self.assertAlmostEqual(np.std(normed_np_output), 1.0, places=1)
def testNegativeAxis(self):
epsilon = 1e-3
bn = normalization_layers.BatchNormalization(
axis=-1, epsilon=epsilon, momentum=0.9
)
inputs = tf.Variable(
np.random.random((5, 4, 3, 6)) + 100, dtype=tf.float32
)
training = tf.compat.v1.placeholder(dtype="bool")
outputs = bn(inputs, training=training)
with self.cached_session() as sess:
# Test training with placeholder learning phase.
self.evaluate(tf.compat.v1.global_variables_initializer())
np_gamma, np_beta = self.evaluate([bn.gamma, bn.beta])
np_gamma = np.reshape(np_gamma, (1, 1, 1, 6))
np_beta = np.reshape(np_beta, (1, 1, 1, 6))
for _ in range(100):
np_output, _, _ = sess.run(
[outputs] + bn.updates, feed_dict={training: True}
)
# Verify that the axis is normalized during training.
normed_np_output = ((np_output - epsilon) * np_gamma) + np_beta
self.assertAlmostEqual(np.mean(normed_np_output), 0.0, places=1)
self.assertAlmostEqual(np.std(normed_np_output), 1.0, places=1)
# Verify that the statistics are updated during training.
moving_mean, moving_var = self.evaluate(
[bn.moving_mean, bn.moving_variance]
)
np_inputs = self.evaluate(inputs)
mean = np.mean(np_inputs, axis=(0, 1, 2))
std = np.std(np_inputs, axis=(0, 1, 2))
variance = np.square(std)
self.assertAllClose(mean, moving_mean, atol=1e-2)
self.assertAllClose(variance, moving_var, atol=1e-2)
# Test inference with placeholder learning phase.
np_output = sess.run(outputs, feed_dict={training: False})
# Verify that the axis is normalized during inference.
normed_np_output = ((np_output - epsilon) * np_gamma) + np_beta
self.assertAlmostEqual(np.mean(normed_np_output), 0.0, places=1)
self.assertAlmostEqual(np.std(normed_np_output), 1.0, places=1)
def testBooleanLearningPhase(self):
epsilon = 1e-3
bn = normalization_layers.BatchNormalization(
axis=-1, epsilon=epsilon, momentum=0.9
)
inputs = tf.Variable(
np.random.random((5, 4, 3, 6)) + 100, dtype=tf.float32
)
outputs_training = bn(inputs, training=True)
outputs_infer = bn(inputs, training=False)
with self.cached_session() as sess:
# Test training with placeholder learning phase.
self.evaluate(tf.compat.v1.global_variables_initializer())
np_gamma, np_beta = self.evaluate([bn.gamma, bn.beta])
np_gamma = np.reshape(np_gamma, (1, 1, 1, 6))
np_beta = np.reshape(np_beta, (1, 1, 1, 6))
for _ in range(100):
np_output, _, _ = sess.run([outputs_training] + bn.updates)
# Verify that the axis is normalized during training.
normed_np_output = ((np_output - epsilon) * np_gamma) + np_beta
self.assertAlmostEqual(np.mean(normed_np_output), 0.0, places=2)
self.assertAlmostEqual(np.std(normed_np_output), 1.0, places=1)
# Verify that the statistics are updated during training.
moving_mean, moving_var = self.evaluate(
[bn.moving_mean, bn.moving_variance]
)
np_inputs = self.evaluate(inputs)
mean = np.mean(np_inputs, axis=(0, 1, 2))
std = np.std(np_inputs, axis=(0, 1, 2))
variance = np.square(std)
self.assertAllClose(mean, moving_mean, atol=1e-2)
self.assertAllClose(variance, moving_var, atol=1e-2)
# Test inference with placeholder learning phase.
np_output = self.evaluate(outputs_infer)
# Verify that the axis is normalized during inference.
normed_np_output = ((np_output - epsilon) * np_gamma) + np_beta
self.assertAlmostEqual(np.mean(normed_np_output), 0.0, places=1)
self.assertAlmostEqual(np.std(normed_np_output), 1.0, places=1)
def testFunctionalNoReuse(self):
inputs = tf.Variable(np.random.random((5, 4, 3, 6)), dtype=tf.float32)
epsilon = 1e-3
training = tf.compat.v1.placeholder(dtype="bool")
outputs = normalization_layers.batch_norm(
inputs,
axis=-1,
momentum=0.9,
epsilon=epsilon,
training=training,
name="bn",
)
updates = tf.compat.v1.get_collection(tf.compat.v1.GraphKeys.UPDATE_OPS)
all_vars = {v.name: v for v in tf.compat.v1.global_variables()}
moving_mean = all_vars["bn/moving_mean:0"]
moving_variance = all_vars["bn/moving_variance:0"]
beta = all_vars["bn/beta:0"]
gamma = all_vars["bn/gamma:0"]
with self.cached_session() as sess:
# Test training with placeholder learning phase.
self.evaluate(tf.compat.v1.global_variables_initializer())
np_gamma, np_beta = self.evaluate([gamma, beta])
np_gamma = np.reshape(np_gamma, (1, 1, 1, 6))
np_beta = np.reshape(np_beta, (1, 1, 1, 6))
for _ in range(100):
np_output, _, _ = sess.run(
[outputs] + updates, feed_dict={training: True}
)
# Verify that the axis is normalized during training.
normed_np_output = ((np_output - epsilon) * np_gamma) + np_beta
self.assertAlmostEqual(np.mean(normed_np_output), 0.0, places=1)
self.assertAlmostEqual(np.std(normed_np_output), 1.0, places=1)
# Verify that the statistics are updated during training.
np_moving_mean, np_moving_var = self.evaluate(
[moving_mean, moving_variance]
)
np_inputs = self.evaluate(inputs)
np_mean = np.mean(np_inputs, axis=(0, 1, 2))
np_std = np.std(np_inputs, axis=(0, 1, 2))
np_variance = np.square(np_std)
self.assertAllClose(np_mean, np_moving_mean, atol=1e-2)
self.assertAllClose(np_variance, np_moving_var, atol=1e-2)
# Test inference with placeholder learning phase.
np_output = sess.run(outputs, feed_dict={training: False})
# Verify that the axis is normalized during inference.
normed_np_output = ((np_output - epsilon) * np_gamma) + np_beta
self.assertAlmostEqual(np.mean(normed_np_output), 0.0, places=1)
self.assertAlmostEqual(np.std(normed_np_output), 1.0, places=1)
def testFunctionalReuse(self):
inputs1 = tf.Variable(np.random.random((5, 4, 3, 6)), dtype=tf.float32)
inputs2 = tf.Variable(np.random.random((5, 4, 3, 6)), dtype=tf.float32)
epsilon = 1e-3
training = tf.compat.v1.placeholder(dtype="bool")
_ = normalization_layers.batch_norm(
inputs1,
axis=-1,
momentum=0.9,
epsilon=epsilon,
training=training,
name="bn",
)
outputs2 = normalization_layers.batch_norm(
inputs2,
axis=-1,
momentum=0.9,
epsilon=epsilon,
training=training,
name="bn",
reuse=True,
)
# Last 2 update ops
updates = tf.compat.v1.get_collection(
tf.compat.v1.GraphKeys.UPDATE_OPS
)[-2:]
all_vars = {v.name: v for v in tf.compat.v1.global_variables()}
moving_mean = all_vars["bn/moving_mean:0"]
moving_variance = all_vars["bn/moving_variance:0"]
beta = all_vars["bn/beta:0"]
gamma = all_vars["bn/gamma:0"]
with self.cached_session() as sess:
# Test training with placeholder learning phase.
self.evaluate(tf.compat.v1.global_variables_initializer())
for _ in range(100):
np_output, _, _ = sess.run(
[outputs2] + updates, feed_dict={training: True}
)
# Verify that the statistics are updated during training.
np_moving_mean, np_moving_var = self.evaluate(
[moving_mean, moving_variance]
)
np_inputs = self.evaluate(inputs2)
np_mean = np.mean(np_inputs, axis=(0, 1, 2))
np_std = np.std(np_inputs, axis=(0, 1, 2))
np_variance = np.square(np_std)
self.assertAllClose(np_mean, np_moving_mean, atol=1e-2)
self.assertAllClose(np_variance, np_moving_var, atol=1e-2)
# Verify that the axis is normalized during training.
np_gamma, np_beta = self.evaluate([gamma, beta])
np_gamma = np.reshape(np_gamma, (1, 1, 1, 6))
np_beta = np.reshape(np_beta, (1, 1, 1, 6))
normed_np_output = ((np_output - epsilon) * np_gamma) + np_beta
self.assertAlmostEqual(np.mean(normed_np_output), 0.0, places=2)
self.assertAlmostEqual(np.std(normed_np_output), 1.0, places=1)
# Test inference with placeholder learning phase.
np_output = sess.run(outputs2, feed_dict={training: False})
# Verify that the axis is normalized during inference.
normed_np_output = ((np_output - epsilon) * np_gamma) + np_beta
self.assertAlmostEqual(np.mean(normed_np_output), 0.0, places=2)
self.assertAlmostEqual(np.std(normed_np_output), 1.0, places=1)
def testFunctionalReuseFromScope(self):
inputs = tf.Variable(np.random.random((5, 4, 3, 6)), dtype=tf.float32)
epsilon = 1e-3
training = tf.compat.v1.placeholder(dtype="bool")
with tf.compat.v1.variable_scope("scope"):
_ = normalization_layers.batch_norm(
inputs,
axis=-1,
momentum=0.9,
epsilon=epsilon,
training=training,
)
self.assertEqual(len(tf.compat.v1.global_variables()), 5)
with tf.compat.v1.variable_scope("scope", reuse=True):
_ = normalization_layers.batch_norm(
inputs,
axis=-1,
momentum=0.9,
epsilon=epsilon,
training=training,
)
self.assertEqual(len(tf.compat.v1.global_variables()), 5)
def testNoCenter(self):
bn = normalization_layers.BatchNormalization(axis=1, center=False)
inputs = tf.random.uniform((5, 4, 3), seed=1)
training = tf.compat.v1.placeholder(dtype="bool")
outputs = bn(inputs, training=training)
# Verify shape.
self.assertListEqual(outputs.get_shape().as_list(), [5, 4, 3])
# Verify layer attributes.
self.assertEqual(len(bn.updates), 2)
self.assertEqual(len(bn.variables), 3)
self.assertEqual(len(bn.trainable_variables), 1)
self.assertEqual(len(bn.non_trainable_variables), 2)
def testNoScale(self):
bn = normalization_layers.BatchNormalization(axis=1, scale=False)
inputs = tf.random.uniform((5, 4, 3), seed=1)
training = tf.compat.v1.placeholder(dtype="bool")
outputs = bn(inputs, training=training)
# Verify shape.
self.assertListEqual(outputs.get_shape().as_list(), [5, 4, 3])
# Verify layer attributes.
self.assertEqual(len(bn.updates), 2)
self.assertEqual(len(bn.variables), 3)
self.assertEqual(len(bn.trainable_variables), 1)
self.assertEqual(len(bn.non_trainable_variables), 2)
def testRegularizers(self):
reg = lambda x: 0.1 * tf.reduce_sum(x)
bn = normalization_layers.BatchNormalization(
axis=1, beta_regularizer=reg
)
inputs = tf.random.uniform((5, 4, 3), seed=1)
training = tf.compat.v1.placeholder(dtype="bool")
_ = bn(inputs, training=training)
self.assertEqual(len(bn.losses), 1)
bn = normalization_layers.BatchNormalization(
axis=1, gamma_regularizer=reg
)
inputs = tf.random.uniform((5, 4, 3), seed=1)
training = tf.compat.v1.placeholder(dtype="bool")
_ = bn(inputs, training=training)
self.assertEqual(len(bn.losses), 1)
def testConstraints(self):
g_constraint = lambda x: x / tf.reduce_sum(x)
b_constraint = lambda x: x / tf.reduce_max(x)
bn = normalization_layers.BatchNormalization(
axis=1, gamma_constraint=g_constraint, beta_constraint=b_constraint
)
inputs = tf.random.uniform((5, 4, 3), seed=1)
bn(inputs)
self.assertEqual(bn.gamma_constraint, g_constraint)
self.assertEqual(bn.beta_constraint, b_constraint)
def testRenorm(self):
shape = (4, 3)
xt = tf.compat.v1.placeholder(tf.float32, shape)
momentum = 0.99
renorm_momentum = 0.8
rmax = 1.1
rmin = 0.9
dmax = 0.1
gamma = 2.0
beta = 3.0
epsilon = 0.001
bn = normalization_layers.BatchNormalization(
axis=1,
gamma_initializer=tf.compat.v1.constant_initializer(gamma),
beta_initializer=tf.compat.v1.constant_initializer(beta),
epsilon=epsilon,
momentum=momentum,
renorm=True,
renorm_clipping={"rmax": rmax, "rmin": rmin, "dmax": dmax},
renorm_momentum=renorm_momentum,
)
training = tf.compat.v1.placeholder(tf.bool)
yt = bn(xt, training=training)
moving_mean = 0.0
moving_stddev = 1.0
renorm_mean = 0.0
renorm_stddev = 1.0
with self.session() as sess:
self.evaluate(tf.compat.v1.global_variables_initializer())
for _ in range(5):
x = np.random.random(shape)
mean = x.mean(0)
variance = x.var(0)
stddev = np.sqrt(variance + epsilon)
r = (stddev / renorm_stddev).clip(rmin, rmax)
d = ((mean - renorm_mean) / renorm_stddev).clip(-dmax, dmax)
y_train = ((x - mean) / stddev * r + d) * gamma + beta
renorm_mean += (mean - renorm_mean) * (1.0 - renorm_momentum)
renorm_stddev += (stddev - renorm_stddev) * (
1.0 - renorm_momentum
)
moving_mean += (mean - moving_mean) * (1.0 - momentum)
moving_stddev += (stddev - moving_stddev) * (1.0 - momentum)
y_test = (
(x - moving_mean)
/ (moving_stddev * moving_stddev) ** 0.5
* gamma
) + beta
yt_val_train, _, _ = sess.run(
[yt] + bn.updates, feed_dict={xt: x, training: True}
)
yt_val_test, _, _ = sess.run(
[yt] + bn.updates, feed_dict={xt: x, training: False}
)
self.assertAllClose(y_train, yt_val_train, atol=1e-5)
self.assertAllClose(y_test, yt_val_test, atol=1e-5)
def testRenormNoClippingSameMomentumGivesSameTestTrain(self):
shape = (4, 3)
xt = tf.compat.v1.placeholder(tf.float32, shape)
momentum = 0.9
renorm_momentum = 0.9
gamma = 2.0
beta = 3.0
epsilon = 0.001
bn = normalization_layers.BatchNormalization(
axis=1,
gamma_initializer=tf.compat.v1.constant_initializer(gamma),
beta_initializer=tf.compat.v1.constant_initializer(beta),
epsilon=epsilon,
momentum=momentum,
renorm=True,
renorm_clipping=None,
renorm_momentum=momentum,
)
training = tf.compat.v1.placeholder(tf.bool)
yt = bn(xt, training=training)
moving_mean = 0.0
moving_stddev = 1.0
renorm_mean = 0.0
renorm_stddev = 1.0
with self.session() as sess:
self.evaluate(tf.compat.v1.global_variables_initializer())
for step in range(6):
x = np.random.random(shape)
mean = x.mean(0)
variance = x.var(0)
stddev = np.sqrt(variance + epsilon)
r = stddev / renorm_stddev
d = (mean - renorm_mean) / renorm_stddev
y_test = (
(x - moving_mean)
/ (moving_stddev * moving_stddev) ** 0.5
* gamma
) + beta
y_train = ((x - mean) / stddev * r + d) * gamma + beta
renorm_mean += (mean - renorm_mean) * (1.0 - renorm_momentum)
renorm_stddev += (stddev - renorm_stddev) * (
1.0 - renorm_momentum
)
moving_mean += (mean - moving_mean) * (1.0 - momentum)
moving_stddev += (stddev - moving_stddev) * (1.0 - momentum)
# Compute test values first, before the train mode updates the
# moving averages.
yt_val_test, _, _ = sess.run(
[yt] + bn.updates, feed_dict={xt: x, training: False}
)
yt_val_train, _, _ = sess.run(
[yt] + bn.updates, feed_dict={xt: x, training: True}
)
# Due to initialization inconsistencies, values may not be
# identical on the first iteration (but shouldn't be different
# by much more than epsilon). After the first iteration they
# should be identical.
atol = epsilon * 1.5 if step == 0 else 1e-5
self.assertAllClose(y_train, yt_val_train, atol=atol)
self.assertAllClose(y_test, yt_val_test, atol=atol)
self.assertAllClose(yt_val_train, yt_val_test, atol=atol)
def testAdjustment(self):
shape = (4, 3)
xt = tf.compat.v1.placeholder(tf.float32, shape)
momentum = 0.99
gamma = 2.0
beta = 3.0
epsilon = 0.001
adjust_scale = tf.random.uniform(shape[-1:], 0.5, 1.5)
adjust_bias = tf.random.uniform(shape[-1:], -0.2, 0.2)
bn = normalization_layers.BatchNormalization(
axis=1,
gamma_initializer=tf.compat.v1.constant_initializer(gamma),
beta_initializer=tf.compat.v1.constant_initializer(beta),
epsilon=epsilon,
momentum=momentum,
adjustment=lambda _: (adjust_scale, adjust_bias),
)
training = tf.compat.v1.placeholder(tf.bool)
yt = bn(xt, training=training)
moving_mean = 0.0
moving_variance = 1.0
with self.session() as sess:
self.evaluate(tf.compat.v1.global_variables_initializer())
for _ in range(5):
x = np.random.random(shape)
yt_val_train, adj_scale_val, adj_bias_val = sess.run(
[yt, adjust_scale, adjust_bias] + bn.updates,
feed_dict={xt: x, training: True},
)[:3]
yt_val_test = sess.run(
[yt] + bn.updates, feed_dict={xt: x, training: False}
)[0]
mean = x.mean(0)
variance = x.var(0)
y_train = (
((x - mean) / (variance + epsilon) ** 0.5) * adj_scale_val
+ adj_bias_val
) * gamma + beta
moving_mean += (mean - moving_mean) * (1.0 - momentum)
moving_variance += (variance - moving_variance) * (
1.0 - momentum
)
y_test = (
(x - moving_mean)
/ (moving_variance + epsilon) ** 0.5
* gamma
) + beta
self.assertAllClose(y_train, yt_val_train, atol=1e-5)
self.assertAllClose(y_test, yt_val_test, atol=1e-5)
def testRenormWithAdjustment(self):
shape = (4, 3)
xt = tf.compat.v1.placeholder(tf.float32, shape)
momentum = 0.99
renorm_momentum = 0.8
rmax = 1.1
rmin = 0.9
dmax = 0.1
gamma = 2.0
beta = 3.0
epsilon = 0.001
adjust_scale = tf.random.uniform(shape[-1:], 0.5, 1.5)
adjust_bias = tf.random.uniform(shape[-1:], -0.2, 0.2)
bn = normalization_layers.BatchNormalization(
axis=1,
gamma_initializer=tf.compat.v1.constant_initializer(gamma),
beta_initializer=tf.compat.v1.constant_initializer(beta),
epsilon=epsilon,
momentum=momentum,
renorm=True,
renorm_clipping={"rmax": rmax, "rmin": rmin, "dmax": dmax},
renorm_momentum=renorm_momentum,
adjustment=lambda _: (adjust_scale, adjust_bias),
)
training = tf.compat.v1.placeholder(tf.bool)
yt = bn(xt, training=training)
moving_mean = 0.0
moving_stddev = 1.0
renorm_mean = 0.0
renorm_stddev = 1.0
with self.session() as sess:
self.evaluate(tf.compat.v1.global_variables_initializer())
for _ in range(5):
x = np.random.random(shape)
yt_val_train, adj_scale_val, adj_bias_val = sess.run(
[yt, adjust_scale, adjust_bias] + bn.updates,
feed_dict={xt: x, training: True},
)[:3]
yt_val_test = sess.run(
[yt] + bn.updates, feed_dict={xt: x, training: False}
)[0]
mean = x.mean(0)
variance = x.var(0)
stddev = np.sqrt(variance + epsilon)
r = (stddev / renorm_stddev).clip(rmin, rmax)
d = ((mean - renorm_mean) / renorm_stddev).clip(-dmax, dmax)
y_train = (
((x - mean) / stddev * r + d) * adj_scale_val + adj_bias_val
) * gamma + beta
renorm_mean += (mean - renorm_mean) * (1.0 - renorm_momentum)
renorm_stddev += (stddev - renorm_stddev) * (
1.0 - renorm_momentum
)
moving_mean += (mean - moving_mean) * (1.0 - momentum)
moving_stddev += (stddev - moving_stddev) * (1.0 - momentum)
y_test = (
(x - moving_mean)
/ (moving_stddev * moving_stddev) ** 0.5
* gamma
) + beta
self.assertAllClose(y_train, yt_val_train, atol=1e-5)
self.assertAllClose(y_test, yt_val_test, atol=1e-5)
def testGhostBNNegativeVirtualBatch(self):
shape = [6, 5, 4, 3]
inp = tf.random.uniform(shape, seed=1)
with self.assertRaises(ValueError):
normalization_layers.batch_normalization(inp, virtual_batch_size=-1)
def testGhostBNVirtualBatchFull(self):
shape = [6, 5, 4, 3]
inp = tf.random.uniform(shape, seed=1)
out1 = normalization_layers.batch_normalization(inp)
out2 = normalization_layers.batch_normalization(
inp, virtual_batch_size=6
)
self.assertListEqual(out1.shape.as_list(), out2.shape.as_list())
with self.session() as sess:
self.evaluate(tf.compat.v1.global_variables_initializer())
x = np.random.random(shape)
y1, y2 = sess.run([out1, out2], feed_dict={inp: x})
self.assertAllClose(y1, y2, atol=1e-5)
def testGhostBNInputOutputShapesMatch(self):
shape = [6, 4, 3]
inp = tf.random.uniform(shape, seed=1)
out = normalization_layers.batch_normalization(
inp, virtual_batch_size=3
)
self.assertListEqual(out.shape.as_list(), shape)
def testGhostBNUnknownBatchSize(self):
np_shape = [10, 5, 4]
tf_shape = [None, 5, 4]
inp = tf.compat.v1.placeholder(tf.float32, tf_shape)
out = normalization_layers.batch_normalization(
inp, virtual_batch_size=2
)
with self.session() as sess:
self.evaluate(tf.compat.v1.global_variables_initializer())
x = np.random.random(np_shape)
y = sess.run(out, feed_dict={inp: x})
self.assertListEqual(list(y.shape), np_shape)
def testGhostBN2Dims(self):
shape = [6, 2]
virtual_batch_size = 3
beta = 2.0
gamma = 3.0
momentum = 0.8
epsilon = 1e-3
moving_means = np.zeros([2, 2], dtype=np.float32)
moving_vars = np.ones([2, 2], dtype=np.float32)
inp = tf.compat.v1.placeholder(tf.float32, shape)
is_training = tf.compat.v1.placeholder(tf.bool)
bn = normalization_layers.BatchNormalization(
momentum=momentum,
epsilon=epsilon,
beta_initializer=tf.compat.v1.constant_initializer(beta),
gamma_initializer=tf.compat.v1.constant_initializer(gamma),
virtual_batch_size=virtual_batch_size,
)
out = bn(inp, training=is_training)
ghost_shape = [
virtual_batch_size,
shape[0] // virtual_batch_size,
shape[1],
]
with self.session() as sess:
self.evaluate(tf.compat.v1.global_variables_initializer())
for _ in range(5):
x = np.random.random(shape)
sub_batched = np.reshape(x, ghost_shape)
means = np.mean(sub_batched, axis=0, keepdims=True)
variances = np.var(sub_batched, axis=0, keepdims=True)
avg_means = np.mean(means, axis=1, keepdims=True)
avg_variances = np.mean(variances, axis=1, keepdims=True)
moving_means = moving_means * momentum + avg_means * (
1.0 - momentum
)
moving_vars = moving_vars * momentum + avg_variances * (
1.0 - momentum
)
y_train = (
(sub_batched - means) / (variances + epsilon) ** 0.5 * gamma
) + beta
y_test = (
(sub_batched - moving_means)
/ (moving_vars + epsilon) ** 0.5
* gamma
) + beta
y_train = np.reshape(y_train, shape)
y_test = np.reshape(y_test, shape)
y_val_train, _, _ = sess.run(
[out] + bn.updates, feed_dict={inp: x, is_training: True}
)
y_val_test = sess.run(
out, feed_dict={inp: x, is_training: False}
)
self.assertAllClose(y_train, y_val_train, atol=1e-5)
self.assertAllClose(y_test, y_val_test, atol=1e-5)
def testGhostBN4DimsAxis3(self):
shape = [6, 10, 10, 3]
virtual_batch_size = 2
beta = 2.0
gamma = 3.0
momentum = 0.8
epsilon = 1e-3
moving_means = np.zeros([1, 1, 1, 1, 3], dtype=np.float32)
moving_vars = np.ones([1, 1, 1, 1, 3], dtype=np.float32)
inp = tf.compat.v1.placeholder(tf.float32, shape)
is_training = tf.compat.v1.placeholder(tf.bool)
bn = normalization_layers.BatchNormalization(
axis=3,
momentum=momentum,
epsilon=epsilon,
beta_initializer=tf.compat.v1.constant_initializer(beta),
gamma_initializer=tf.compat.v1.constant_initializer(gamma),
virtual_batch_size=virtual_batch_size,
)
out = bn(inp, training=is_training)
ghost_shape = [
virtual_batch_size,
shape[0] // virtual_batch_size,
] + shape[1:]
with self.session() as sess:
self.evaluate(tf.compat.v1.global_variables_initializer())
for _ in range(5):
x = np.random.random(shape)
sub_batched = np.reshape(x, ghost_shape)
means = np.mean(sub_batched, axis=(0, 2, 3), keepdims=True)
variances = np.var(sub_batched, axis=(0, 2, 3), keepdims=True)
avg_means = np.mean(means, axis=1, keepdims=True)
avg_variances = np.mean(variances, axis=1, keepdims=True)
moving_means = moving_means * momentum + avg_means * (
1.0 - momentum
)
moving_vars = moving_vars * momentum + avg_variances * (
1.0 - momentum
)
y_train = (
(sub_batched - means) / (variances + epsilon) ** 0.5 * gamma
) + beta
y_test = (
(sub_batched - moving_means)
/ (moving_vars + epsilon) ** 0.5
* gamma
) + beta
y_train = np.reshape(y_train, shape)
y_test = np.reshape(y_test, shape)
y_val_train, _, _ = sess.run(
[out] + bn.updates, feed_dict={inp: x, is_training: True}
)
y_val_test = sess.run(
out, feed_dict={inp: x, is_training: False}
)
self.assertAllClose(y_train, y_val_train, atol=1e-2)
self.assertAllClose(y_test, y_val_test, atol=1e-2)
def testGhostBN4DimsAxis1(self):
shape = [6, 3, 10, 10]
virtual_batch_size = 2
beta = 2.0
gamma = 3.0
momentum = 0.8
epsilon = 1e-3
moving_means = np.zeros([1, 1, 3, 1, 1], dtype=np.float32)
moving_vars = np.ones([1, 1, 3, 1, 1], dtype=np.float32)
inp = tf.compat.v1.placeholder(tf.float32, shape)
is_training = tf.compat.v1.placeholder(tf.bool)
bn = normalization_layers.BatchNormalization(
axis=1,
momentum=momentum,
epsilon=epsilon,
beta_initializer=tf.compat.v1.constant_initializer(beta),
gamma_initializer=tf.compat.v1.constant_initializer(gamma),
virtual_batch_size=virtual_batch_size,
fused=False,
) # NCHW is unsupported by CPU fused batch norm
out = bn(inp, training=is_training)
ghost_shape = [
virtual_batch_size,
shape[0] // virtual_batch_size,
] + shape[1:]
with self.session() as sess:
self.evaluate(tf.compat.v1.global_variables_initializer())
for _ in range(5):
x = np.random.random(shape)
sub_batched = np.reshape(x, ghost_shape)
means = np.mean(sub_batched, axis=(0, 3, 4), keepdims=True)
variances = np.var(sub_batched, axis=(0, 3, 4), keepdims=True)
avg_means = np.mean(means, axis=1, keepdims=True)
avg_variances = np.mean(variances, axis=1, keepdims=True)
moving_means = moving_means * momentum + avg_means * (
1.0 - momentum
)
moving_vars = moving_vars * momentum + avg_variances * (
1.0 - momentum
)
y_train = (
(sub_batched - means) / (variances + epsilon) ** 0.5 * gamma
) + beta
y_test = (
(sub_batched - moving_means)
/ (moving_vars + epsilon) ** 0.5
* gamma
) + beta
y_train = np.reshape(y_train, shape)
y_test = np.reshape(y_test, shape)
y_val_train, _, _ = sess.run(
[out] + bn.updates, feed_dict={inp: x, is_training: True}
)
y_val_test = sess.run(
out, feed_dict={inp: x, is_training: False}
)
self.assertAllClose(y_train, y_val_train, atol=1e-2)
self.assertAllClose(y_test, y_val_test, atol=1e-2)
def testMultiAxisInvalid(self):
shape = [6, 5, 4, 3]
inp = tf.random.uniform(shape, seed=1)
with self.assertRaises(ValueError):
normalization_layers.batch_normalization(
inp, axis=[1, 4]
) # out of bounds
with self.assertRaises(ValueError):
normalization_layers.batch_normalization(
inp, axis=[-5, 1]
) # out of bounds
with self.assertRaises(ValueError):
normalization_layers.batch_normalization(
inp, axis=[1, 2, 1]
) # duplicate
def test3DInputMultiAxis12(self):
epsilon = 1e-3
bn = normalization_layers.BatchNormalization(
axis=[1, 2], epsilon=epsilon, momentum=0.9
)
inputs = tf.Variable(
np.random.random((5, 4, 3)) + 100, dtype=tf.float32
)
training = tf.compat.v1.placeholder(dtype="bool")
outputs = bn(inputs, training=training)
with self.cached_session() as sess:
# Test training with placeholder learning phase.
self.evaluate(tf.compat.v1.global_variables_initializer())
np_gamma, np_beta = self.evaluate([bn.gamma, bn.beta])
for _ in range(100):
np_output, _, _ = sess.run(
[outputs] + bn.updates, feed_dict={training: True}
)
# Verify that the axis is normalized during training.
normed_np_output = ((np_output - epsilon) * np_gamma) + np_beta
self.assertAlmostEqual(np.mean(normed_np_output), 0.0, places=1)
self.assertAlmostEqual(np.std(normed_np_output), 1.0, places=1)
# Verify that the statistics are updated during training.
moving_mean, moving_var = self.evaluate(
[bn.moving_mean, bn.moving_variance]
)
np_inputs = self.evaluate(inputs)
mean = np.mean(np_inputs, axis=0, keepdims=True)
std = np.std(np_inputs, axis=0, keepdims=True)
variance = np.square(std)
self.assertAllClose(mean, moving_mean, atol=1e-2)
self.assertAllClose(variance, moving_var, atol=1e-2)
# Test inference with placeholder learning phase.
np_output = sess.run(outputs, feed_dict={training: False})
# Verify that the axis is normalized during inference.
normed_np_output = ((np_output - epsilon) * np_gamma) + np_beta
self.assertAlmostEqual(np.mean(normed_np_output), 0.0, places=1)
self.assertAlmostEqual(np.std(normed_np_output), 1.0, places=1)
def test5DInputMultiAxis123(self):
epsilon = 1e-3
bn = normalization_layers.BatchNormalization(
axis=[1, 2, 3], epsilon=epsilon, momentum=0.9
)
inputs = tf.Variable(
np.random.random((5, 3, 4, 4, 3)) + 100, dtype=tf.float32
)
training = tf.compat.v1.placeholder(dtype="bool")
outputs = bn(inputs, training=training)
with self.cached_session() as sess:
# Test training with placeholder learning phase.
self.evaluate(tf.compat.v1.global_variables_initializer())
np_gamma, np_beta = self.evaluate([bn.gamma, bn.beta])
for _ in range(100):
np_output, _, _ = sess.run(
[outputs] + bn.updates, feed_dict={training: True}
)
# Verify that the axis is normalized during training.
normed_np_output = ((np_output - epsilon) * np_gamma) + np_beta
self.assertAlmostEqual(np.mean(normed_np_output), 0.0, places=1)
self.assertAlmostEqual(np.std(normed_np_output), 1.0, places=1)
# Verify that the statistics are updated during training.
moving_mean, moving_var = self.evaluate(
[bn.moving_mean, bn.moving_variance]
)
np_inputs = self.evaluate(inputs)
mean = np.mean(np_inputs, axis=(0, 4), keepdims=True)
std = np.std(np_inputs, axis=(0, 4), keepdims=True)
variance = np.square(std)
self.assertAllClose(mean, moving_mean, atol=1e-2)
self.assertAllClose(variance, moving_var, atol=1e-2)
# Test inference with placeholder learning phase.
np_output = sess.run(outputs, feed_dict={training: False})
# Verify that the axis is normalized during inference.
normed_np_output = ((np_output - epsilon) * np_gamma) + np_beta
self.assertAlmostEqual(np.mean(normed_np_output), 0.0, places=1)
self.assertAlmostEqual(np.std(normed_np_output), 1.0, places=1)
def testGhostBN5DimsMultiAxis14(self):
shape = [6, 3, 10, 10, 4]
virtual_batch_size = 3
beta = 2.0
gamma = 3.0
momentum = 0.8
epsilon = 1e-3
moving_means = np.zeros([1, 1, 3, 1, 1, 4], dtype=np.float32)
moving_vars = np.ones([1, 1, 3, 1, 1, 4], dtype=np.float32)
inp = tf.compat.v1.placeholder(tf.float32, shape)
is_training = tf.compat.v1.placeholder(tf.bool)
bn = normalization_layers.BatchNormalization(
axis=[1, 4],
momentum=momentum,
epsilon=epsilon,
beta_initializer=tf.compat.v1.constant_initializer(beta),
gamma_initializer=tf.compat.v1.constant_initializer(gamma),
virtual_batch_size=virtual_batch_size,
fused=False,
)
out = bn(inp, training=is_training)
ghost_shape = [
virtual_batch_size,
shape[0] // virtual_batch_size,
] + shape[1:]
with self.session() as sess:
self.evaluate(tf.compat.v1.global_variables_initializer())
for _ in range(5):
x = np.random.random(shape)
sub_batched = np.reshape(x, ghost_shape)
means = np.mean(sub_batched, axis=(0, 3, 4), keepdims=True)
variances = np.var(sub_batched, axis=(0, 3, 4), keepdims=True)
avg_means = np.mean(means, axis=1, keepdims=True)
avg_variances = np.mean(variances, axis=1, keepdims=True)
moving_means = moving_means * momentum + avg_means * (
1.0 - momentum
)
moving_vars = moving_vars * momentum + avg_variances * (
1.0 - momentum
)
y_train = (
(sub_batched - means) / (variances + epsilon) ** 0.5 * gamma
) + beta
y_test = (
(sub_batched - moving_means)
/ (moving_vars + epsilon) ** 0.5
* gamma
) + beta
y_train = np.reshape(y_train, shape)
y_test = np.reshape(y_test, shape)
y_val_train, _, _ = sess.run(
[out] + bn.updates, feed_dict={inp: x, is_training: True}
)
y_val_test = sess.run(
out, feed_dict={inp: x, is_training: False}
)
self.assertAllClose(y_train, y_val_train, atol=1e-2)
self.assertAllClose(y_test, y_val_test, atol=1e-2)
if __name__ == "__main__":
tf.test.main()
|
tf-keras/tf_keras/legacy_tf_layers/normalization_test.py/0
|
{
"file_path": "tf-keras/tf_keras/legacy_tf_layers/normalization_test.py",
"repo_id": "tf-keras",
"token_count": 36245
}
| 214 |
# Copyright 2019 The TensorFlow Authors. All Rights Reserved.
#
# Licensed under the Apache License, Version 2.0 (the "License");
# you may not use this file except in compliance with the License.
# You may obtain a copy of the License at
#
# http://www.apache.org/licenses/LICENSE-2.0
#
# Unless required by applicable law or agreed to in writing, software
# distributed under the License is distributed on an "AS IS" BASIS,
# WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.
# See the License for the specific language governing permissions and
# limitations under the License.
# ==============================================================================
"""Tests for AutoCastVariable."""
import os
import threading
import numpy as np
import tensorflow.compat.v2 as tf
from absl.testing import parameterized
from tensorflow.python.eager import test
from tf_keras.layers import Dense
from tf_keras.mixed_precision import autocast_variable
from tf_keras.optimizers.legacy import adadelta
from tf_keras.optimizers.legacy import adagrad
from tf_keras.optimizers.legacy import adam
from tf_keras.optimizers.legacy import adamax
from tf_keras.optimizers.legacy import ftrl
from tf_keras.optimizers.legacy import gradient_descent as gradient_descent_v2
from tf_keras.optimizers.legacy import nadam
from tf_keras.optimizers.legacy import rmsprop
maybe_distribute = tf.__internal__.test.combinations.combine(
distribution=[
tf.__internal__.distribute.combinations.default_strategy,
tf.__internal__.distribute.combinations.mirrored_strategy_with_two_cpus, # noqa: E501
]
)
def get_var(val, dtype, name=None):
return tf.Variable(val, dtype=dtype, name=name)
def checkpoint_test_helper(test_case, distribution, enable_async_ckpt):
with test_case.test_session():
with distribution.scope():
x = get_var(1.0, tf.float32)
x = autocast_variable.create_autocast_variable(x)
test_case.evaluate(x.initializer)
test_case.evaluate(x.assign(123.0))
checkpoint = tf.train.Checkpoint(x=x)
ckpt_options = tf.train.CheckpointOptions(
experimental_enable_async_checkpoint=enable_async_ckpt
)
prefix = os.path.join(test_case.get_temp_dir(), "ckpt")
save_path = checkpoint.save(prefix, options=ckpt_options)
test_case.evaluate(x.assign(234.0))
test_case.assertNotAllClose(123.0, x.read_value())
checkpoint.restore(save_path).assert_consumed().run_restore_ops()
test_case.assertEqual(test_case.evaluate(x), 123.0)
# Another round of saving/restoring to ensure that the logic of
# _copy_trackable_to_cpu works when a copy is already created in
# object_map.
test_case.evaluate(x.assign(345.0))
save_path = checkpoint.save(prefix, options=ckpt_options)
test_case.evaluate(x.assign(456.0))
checkpoint.restore(save_path).assert_consumed().run_restore_ops()
test_case.assertEqual(test_case.evaluate(x), 345.0)
@tf.__internal__.distribute.combinations.generate(
tf.__internal__.test.combinations.combine(mode=["graph", "eager"])
)
class AutoCastVariableTest(test.TestCase, parameterized.TestCase):
@tf.__internal__.distribute.combinations.generate(maybe_distribute)
def test_read(self, distribution):
with distribution.scope():
x = get_var(1.0, tf.float32)
x = autocast_variable.create_autocast_variable(x)
self.evaluate(x.initializer)
# outside of auto cast scope.
self.assertEqual(x.dtype, tf.float32)
self.assertEqual(x.value().dtype, tf.float32)
self.assertEqual(x.read_value().dtype, tf.float32)
self.assertEqual(tf.identity(x).dtype, tf.float32)
# within auto cast scope of different dtype
with autocast_variable.enable_auto_cast_variables(tf.float16):
self.assertEqual(x.dtype, tf.float32)
self.assertEqual(x.value().dtype, tf.float16)
self.assertEqual(x.read_value().dtype, tf.float16)
self.assertEqual(tf.identity(x).dtype, tf.float16)
# within auto cast scope of same dtype
with autocast_variable.enable_auto_cast_variables(tf.float32):
self.assertEqual(x.dtype, tf.float32)
self.assertEqual(x.value().dtype, tf.float32)
self.assertEqual(x.read_value().dtype, tf.float32)
self.assertEqual(tf.identity(x).dtype, tf.float32)
def test_sparse_reads(self):
x = get_var([1.0, 2], tf.float32)
# DistributedVariables do not support sparse_read or gather_nd, so we
# pass distribute=False
x = autocast_variable.create_autocast_variable(x)
self.evaluate(x.initializer)
self.assertEqual(x.sparse_read([0]).dtype, tf.float32)
self.assertEqual(x.gather_nd([0]).dtype, tf.float32)
with autocast_variable.enable_auto_cast_variables(tf.float16):
self.assertEqual(x.sparse_read([0]).dtype, tf.float16)
self.assertEqual(x.gather_nd([0]).dtype, tf.float16)
def test_tf_function_with_variable_and_autocast_variable(self):
ones = tf.ones((2, 2))
layer1 = Dense(2, dtype="float32")
layer2 = Dense(2, dtype="mixed_float16")
layer1(ones)
layer2(ones)
@tf.function
def f(x):
return x + 1
self.assertEqual(f(layer1.kernel).dtype, tf.dtypes.float32)
self.assertEqual(f(layer2.kernel).dtype, tf.dtypes.float32)
@tf.__internal__.distribute.combinations.generate(maybe_distribute)
def test_read_nested_scopes(self, distribution):
with distribution.scope():
x = get_var(1.0, tf.float32)
x = autocast_variable.create_autocast_variable(x)
self.evaluate(x.initializer)
with autocast_variable.enable_auto_cast_variables(tf.float16):
self.assertEqual(x.read_value().dtype, tf.float16)
with autocast_variable.enable_auto_cast_variables(tf.float32):
self.assertEqual(x.read_value().dtype, tf.float32)
self.assertEqual(x.read_value().dtype, tf.float16)
@tf.__internal__.distribute.combinations.generate(maybe_distribute)
def test_dtype_is_not_string(self, distribution):
with distribution.scope():
x = get_var(1.0, tf.float32)
x = autocast_variable.create_autocast_variable(x)
self.assertEqual(x.dtype, tf.float32)
self.assertIsInstance(x.dtype, tf.DType)
self.assertEqual(x.true_dtype, tf.float32)
self.assertIsInstance(x.true_dtype, tf.DType)
dtype = tf.float16
with autocast_variable.enable_auto_cast_variables(dtype):
self.assertEqual(x.dtype, tf.float32)
self.assertIsInstance(x.dtype, tf.DType)
self.assertEqual(x.true_dtype, tf.float32)
self.assertIsInstance(x.true_dtype, tf.DType)
@tf.__internal__.distribute.combinations.generate(maybe_distribute)
def test_method_delegations(self, distribution):
# Test AutoCastVariable correctly delegates Variable methods to the
# underlying variable.
with self.test_session(), distribution.scope():
for read_dtype in (tf.float32, tf.float16):
if tf.distribute.has_strategy() and not tf.executing_eagerly():
# MirroredVariable.assign will (incorrectly) return a
# Mirrored value instead of a MirroredVariable in graph
# mode. So we cannot properly wrap it in an
# AutoCastVariable.
evaluate = self.evaluate
else:
def evaluate(var):
self.assertIsInstance(
var, autocast_variable.AutoCastVariable
)
self.assertEqual(tf.identity(var).dtype, read_dtype)
return self.evaluate(var)
x = get_var(7.0, tf.float32)
x = autocast_variable.create_autocast_variable(x)
with autocast_variable.enable_auto_cast_variables(read_dtype):
self.evaluate(x.initializer)
self.assertEqual(self.evaluate(x.value()), 7)
self.assertEqual(self.evaluate(x.read_value()), 7)
self.assertTrue(x.trainable)
self.assertEqual(
x.synchronization, x._variable.synchronization
)
self.assertEqual(x.aggregation, x._variable.aggregation)
self.assertEqual(self.evaluate(x.read_value()), 7)
if not tf.executing_eagerly():
if not tf.distribute.has_strategy():
# These functions are not supported for
# DistributedVariables
x.load(9)
self.assertEqual(x.eval(), 9)
self.assertEqual(self.evaluate(x.initial_value), 7)
self.assertEqual(x.op, x._variable.op)
self.assertEqual(x.graph, x._variable.graph)
if not tf.distribute.has_strategy():
# These attributes are not supported for
# DistributedVariables
self.assertIsNone(x.constraint)
self.assertEqual(x.initializer, x._variable.initializer)
self.assertEqual(evaluate(x.assign(8)), 8)
self.assertEqual(evaluate(x.assign_add(2)), 10)
self.assertEqual(evaluate(x.assign_sub(3)), 7)
self.assertEqual(x.name, x._variable.name)
self.assertEqual(x.device, x._variable.device)
self.assertEqual(x.shape, ())
self.assertEqual(x.get_shape(), ())
if not tf.distribute.has_strategy():
# Test scatter_* methods. These are not supported for
# DistributedVariables
x = get_var([7, 8], tf.float32)
x = autocast_variable.create_autocast_variable(x)
with autocast_variable.enable_auto_cast_variables(
read_dtype
):
self.evaluate(x.initializer)
self.assertAllEqual(self.evaluate(x.value()), [7, 8])
def slices(val, index):
return tf.IndexedSlices(
values=tf.constant(val, dtype=tf.float32),
indices=tf.constant(index, dtype=tf.int32),
dense_shape=tf.constant([2], dtype=tf.int32),
)
self.assertAllEqual(
evaluate(x.scatter_sub(slices(1.0, 0))), [6, 8]
)
self.assertAllEqual(
evaluate(x.scatter_add(slices(1.0, 0))), [7, 8]
)
self.assertAllEqual(
evaluate(x.scatter_max(slices(9.0, 1))), [7, 9]
)
self.assertAllEqual(
evaluate(x.scatter_min(slices(8.0, 1))), [7, 8]
)
self.assertAllEqual(
evaluate(x.scatter_mul(slices(2.0, 1))), [7, 16]
)
self.assertAllEqual(
evaluate(x.scatter_div(slices(2.0, 1))), [7, 8]
)
self.assertAllEqual(
evaluate(x.scatter_update(slices(4.0, 1))), [7, 4]
)
self.assertAllEqual(
evaluate(x.scatter_nd_sub([[0], [1]], [1.0, 2.0])),
[6, 2],
)
self.assertAllEqual(
evaluate(x.scatter_nd_add([[0], [1]], [1.0, 2.0])),
[7, 4],
)
self.assertAllEqual(
evaluate(
x.scatter_nd_update([[0], [1]], [1.0, 2.0])
),
[1, 2],
)
@tf.__internal__.distribute.combinations.generate(maybe_distribute)
def test_operator_overloads(self, distribution):
with distribution.scope():
for read_dtype in (tf.float32, tf.float16):
x = get_var(7.0, tf.float32)
x = autocast_variable.create_autocast_variable(x)
with autocast_variable.enable_auto_cast_variables(read_dtype):
self.evaluate(x.initializer)
self.assertAlmostEqual(8, self.evaluate(x + 1))
self.assertAlmostEqual(10, self.evaluate(3 + x))
self.assertAlmostEqual(14, self.evaluate(x + x))
self.assertAlmostEqual(5, self.evaluate(x - 2))
self.assertAlmostEqual(6, self.evaluate(13 - x))
self.assertAlmostEqual(0, self.evaluate(x - x))
self.assertAlmostEqual(14, self.evaluate(x * 2))
self.assertAlmostEqual(21, self.evaluate(3 * x))
self.assertAlmostEqual(49, self.evaluate(x * x))
self.assertAlmostEqual(3.5, self.evaluate(x / 2))
self.assertAlmostEqual(1.5, self.evaluate(10.5 / x))
self.assertAlmostEqual(3, self.evaluate(x // 2))
self.assertAlmostEqual(2, self.evaluate(15 // x))
if read_dtype == tf.float32:
# The "mod" operator does not support float16
self.assertAlmostEqual(1, self.evaluate(x % 2))
self.assertAlmostEqual(2, self.evaluate(16 % x))
self.assertTrue(self.evaluate(x < 12))
self.assertTrue(self.evaluate(x <= 12))
self.assertFalse(self.evaluate(x > 12))
self.assertFalse(self.evaluate(x >= 12))
self.assertFalse(self.evaluate(12 < x))
self.assertFalse(self.evaluate(12 <= x))
self.assertTrue(self.evaluate(12 > x))
self.assertTrue(self.evaluate(12 >= x))
self.assertAlmostEqual(
343, self.evaluate(pow(x, 3)), places=4
)
self.assertAlmostEqual(
128, self.evaluate(pow(2, x)), places=4
)
self.assertAlmostEqual(-7, self.evaluate(-x))
self.assertAlmostEqual(7, self.evaluate(abs(x)))
x = get_var([7, 8, 9], tf.float32)
x = autocast_variable.create_autocast_variable(x)
self.evaluate(x.initializer)
self.assertEqual(self.evaluate(x[1]), 8)
if tf.__internal__.tf2.enabled() and tf.executing_eagerly():
self.assertAllEqual(
x == [7.0, 8.0, 10.0], [True, True, False]
)
self.assertAllEqual(
x != [7.0, 8.0, 10.0], [False, False, True]
)
@tf.__internal__.distribute.combinations.generate(maybe_distribute)
def test_assign(self, distribution):
with distribution.scope():
x = get_var(0.0, tf.float32)
x = autocast_variable.create_autocast_variable(x)
self.evaluate(x.initializer)
# outside of auto cast scope.
v1 = tf.constant(3.0, dtype=tf.float32)
v2 = tf.constant(3.0, dtype=tf.float16)
def run_and_check():
# Assign float32 values
self.assertAllClose(3.0, self.evaluate(x.assign(v1)))
self.assertAllClose(3.0 * 2, self.evaluate(x.assign_add(v1)))
self.assertAllClose(3.0, self.evaluate(x.assign_sub(v1)))
# Attempt to assign float16 values
with self.assertRaisesRegex(
ValueError,
"conversion requested dtype float32 for Tensor with dtype "
"float16",
):
self.evaluate(x.assign(v2))
with self.assertRaisesRegex(
ValueError,
"conversion requested dtype float32 for Tensor with dtype "
"float16",
):
self.evaluate(x.assign_add(v2))
with self.assertRaisesRegex(
ValueError,
"conversion requested dtype float32 for Tensor with dtype "
"float16",
):
self.evaluate(x.assign_sub(v2))
# Assign Python floats
self.assertAllClose(0.0, self.evaluate(x.assign(0.0)))
self.assertAllClose(3.0, self.evaluate(x.assign(3.0)))
self.assertAllClose(3.0 * 2, self.evaluate(x.assign_add(3.0)))
self.assertAllClose(3.0, self.evaluate(x.assign_sub(3.0)))
# Assign multiple times
# This currently doesn't work in graph mode if a strategy is
# used
if not tf.distribute.has_strategy() or tf.executing_eagerly():
assign = x.assign(1.0)
self.assertAllClose(1.0, self.evaluate(assign))
self.assertAllClose(0.0, self.evaluate(assign.assign(0.0)))
assign_add = x.assign_add(3.0)
self.assertAllClose(3.0, self.evaluate(assign_add))
self.assertAllClose(
3.0 * 3,
self.evaluate(x.assign_add(3.0).assign_add(3.0)),
)
self.assertAllClose(3.0 * 3, x)
assign_sub = x.assign_sub(3.0)
self.assertAllClose(3.0 * 2, self.evaluate(assign_sub))
self.assertAllClose(
0.0, self.evaluate(x.assign_sub(3.0).assign_sub(3.0))
)
# Assign with read_value=False
self.assertIsNone(
self.evaluate(x.assign(1.0, read_value=False))
)
self.assertAllClose(1.0, self.evaluate(x))
self.assertIsNone(
self.evaluate(x.assign_add(2.0, read_value=False))
)
self.assertAllClose(3.0, self.evaluate(x))
self.assertIsNone(
self.evaluate(x.assign_sub(3.0, read_value=False))
)
self.assertAllClose(0.0, self.evaluate(x))
# Use the tf.assign functions instead of the var.assign methods.
self.assertAllClose(
0.0, self.evaluate(tf.compat.v1.assign(x, 0.0))
)
self.assertAllClose(
3.0, self.evaluate(tf.compat.v1.assign(x, 3.0))
)
self.assertAllClose(
3.0 * 2, self.evaluate(tf.compat.v1.assign_add(x, 3.0))
)
self.assertAllClose(
3.0, self.evaluate(tf.compat.v1.assign_sub(x, 3.0))
)
run_and_check()
# reset x
self.evaluate(x.assign(0.0))
# within auto cast scope.
with autocast_variable.enable_auto_cast_variables(tf.float16):
# assign still expect float32 value even if in float16 scope
run_and_check()
@tf.__internal__.distribute.combinations.generate(maybe_distribute)
def test_assign_tf_function(self, distribution):
if not tf.executing_eagerly():
self.skipTest("Test is not compatible with graph mode")
with distribution.scope():
x = get_var(0.0, tf.float32)
x = autocast_variable.create_autocast_variable(x)
@tf.function
def run_assign():
return (
x.assign(1.0)
.assign_add(3.0)
.assign_add(3.0)
.assign_sub(2.0)
)
with autocast_variable.enable_auto_cast_variables(tf.float16):
self.assertAllClose(5.0, self.evaluate(run_assign()))
@tf.__internal__.distribute.combinations.generate(maybe_distribute)
def test_op_attribute(self, distribution):
with distribution.scope():
x = get_var(0.0, tf.float32)
x = autocast_variable.create_autocast_variable(x)
# Variable.op raises an AttributeError in Eager mode and is an op in
# graph mode. Variable.assign(...).op is None in Eager mode and an
# op in Graph mode or a tf.function. We test this is also true of
# AutoCastVariable.
if tf.executing_eagerly():
with self.assertRaises(AttributeError):
x.op
self.assertIsNone(x.assign(1.0).op)
self.assertIsNone(x.assign_add(1.0).op)
self.assertIsNone(x.assign_sub(1.0).op)
else:
self.assertIsNotNone(x.op)
self.assertIsNotNone(x.assign(1.0).op)
self.assertIsNotNone(x.assign_add(1.0).op)
self.assertIsNotNone(x.assign_sub(1.0).op)
@tf.function
def func():
self.assertIsNotNone(x.assign(1.0).op)
self.assertIsNotNone(x.assign_add(1.0).op)
self.assertIsNotNone(x.assign_sub(1.0).op)
func()
@tf.__internal__.distribute.combinations.generate(maybe_distribute)
def test_tf_function_control_dependencies(self, distribution):
if not tf.executing_eagerly():
self.skipTest("Test is not compatible with graph mode")
with distribution.scope():
x = get_var(0.0, tf.float32)
x = autocast_variable.create_autocast_variable(x)
@tf.function
def func():
update = x.assign_add(1.0)
with tf.control_dependencies([update]):
x.assign_add(1.0)
func()
self.assertAllClose(2.0, self.evaluate(x))
@tf.__internal__.distribute.combinations.generate(maybe_distribute)
def test_assign_stays_in_true_dtype(self, distribution):
with distribution.scope():
x = get_var(1.0, tf.float32)
x = autocast_variable.create_autocast_variable(x)
self.evaluate(x.initializer)
# small_val is a value such that 1.0 + small_val == 1.0 in fp16, but
# not in fp32
small_val = np.finfo("float16").eps / 2
small_tensor = tf.constant(small_val, dtype=tf.float32)
with autocast_variable.enable_auto_cast_variables(tf.float16):
# Variable should be increased, despite it appearing to be the
# same float16 value.
self.evaluate(x.assign(1.0 + small_tensor))
self.assertEqual(1.0, self.evaluate(x.value()))
self.assertEqual(1.0 + small_val, self.evaluate(x))
self.evaluate(x.assign(1.0))
with autocast_variable.enable_auto_cast_variables(tf.float16):
self.evaluate(x.assign_add(small_tensor))
self.assertEqual(1.0, self.evaluate(x.value()))
self.assertEqual(1.0 + small_val, self.evaluate(x))
def test_thread_local_autocast_dtype(self):
x = get_var(1.0, tf.float32)
x = autocast_variable.create_autocast_variable(x)
self.evaluate(x.initializer)
with autocast_variable.enable_auto_cast_variables(tf.float16):
self.assertEqual(tf.identity(x).dtype, tf.float16)
# New threads should not see the modified value of the autocast
# dtype.
var_dtype = None
def f():
nonlocal var_dtype
var_dtype = x._cast_dtype
thread = threading.Thread(target=f)
thread.start()
thread.join()
self.assertEqual(var_dtype, tf.float32)
@tf.__internal__.distribute.combinations.generate(maybe_distribute)
def test_checkpoint(self, distribution):
checkpoint_test_helper(self, distribution, enable_async_ckpt=False)
@tf.__internal__.distribute.combinations.generate(maybe_distribute)
def test_asyncCheckpoint(self, distribution):
checkpoint_test_helper(self, distribution, enable_async_ckpt=True)
@tf.__internal__.distribute.combinations.generate(maybe_distribute)
def test_invalid_wrapped_variable(self, distribution):
with distribution.scope():
# Wrap a non-variable
with self.assertRaisesRegex(ValueError, "variable must be of type"):
x = tf.constant([1.0], dtype=tf.float32)
autocast_variable.create_autocast_variable(x)
# Wrap a non-floating point variable
with self.assertRaisesRegex(
ValueError, "variable must be a floating point"
):
x = get_var(1, tf.int32)
autocast_variable.create_autocast_variable(x)
def test_repr(self):
# We do not test with DistributionStrategy because we do not want to
# rely on the exact __repr__ output of a DistributedVariable.
x = get_var(1.0, tf.float32, name="x")
x = autocast_variable.create_autocast_variable(x)
if tf.executing_eagerly():
self.assertStartsWith(
repr(x),
"<AutoCastVariable 'x:0' shape=() dtype=float32 "
"dtype_to_cast_to=float32, numpy=",
)
with autocast_variable.enable_auto_cast_variables(tf.float16):
self.assertStartsWith(
repr(x),
"<AutoCastVariable 'x:0' shape=() dtype=float32 "
"dtype_to_cast_to=float16, numpy=",
)
else:
self.assertEqual(
repr(x),
"<AutoCastVariable 'x:0' shape=() dtype=float32 "
"dtype_to_cast_to=float32>",
)
with autocast_variable.enable_auto_cast_variables(tf.float16):
self.assertEqual(
repr(x),
"<AutoCastVariable 'x:0' shape=() dtype=float32 "
"dtype_to_cast_to=float16>",
)
@tf.__internal__.distribute.combinations.generate(
tf.__internal__.test.combinations.combine(
distribution=[
tf.__internal__.distribute.combinations.mirrored_strategy_with_two_cpus, # noqa: E501
]
)
)
def test_repr_distributed(self, distribution):
with distribution.scope():
x = get_var(1.0, tf.float32)
x = autocast_variable.create_autocast_variable(x)
use_policy = getattr(
distribution.extended, "_use_var_policy", False
)
if use_policy:
self.assertRegex(
repr(x).replace("\n", " "),
"<AutoCastDistributedVariable dtype=float32 "
"dtype_to_cast_to=float32 "
"inner_variable=DistributedVariable.*>",
)
else:
self.assertRegex(
repr(x).replace("\n", " "),
"<AutoCastDistributedVariable dtype=float32 "
"dtype_to_cast_to=float32 "
"inner_variable=MirroredVariable.*>",
)
@tf.__internal__.distribute.combinations.generate(
tf.__internal__.test.combinations.combine(
optimizer_class=[
adadelta.Adadelta,
adagrad.Adagrad,
adam.Adam,
adamax.Adamax,
ftrl.Ftrl,
gradient_descent_v2.SGD,
nadam.Nadam,
rmsprop.RMSprop,
tf.compat.v1.train.GradientDescentOptimizer,
],
use_tf_function=[False, True],
)
)
def test_optimizer(self, optimizer_class, use_tf_function):
if use_tf_function and not tf.executing_eagerly():
self.skipTest("Test does not support graph mode with tf.function")
x = get_var(1.0, tf.float32)
x = autocast_variable.create_autocast_variable(x)
y = get_var(1.0, tf.float32)
opt = optimizer_class(learning_rate=1.0)
def f():
# Minimize both the AutoCastVariable and the normal tf.Variable.
# Both variables should be updated to the same value.
op = opt.minimize(lambda: x + y, var_list=[x, y])
return (
None
if tf.compat.v1.executing_eagerly_outside_functions()
else op
)
if use_tf_function:
f = tf.function(f)
if tf.executing_eagerly():
f()
else:
op = f()
self.evaluate(tf.compat.v1.global_variables_initializer())
self.evaluate(op)
# Assert the AutoCastVariable has changed from its initial value
self.assertNotEqual(self.evaluate(x), 1.0)
# Assert AutoCastVariable is updated correctly by comparing it to the
# normal variable
self.assertAlmostEqual(self.evaluate(x), self.evaluate(y))
if optimizer_class in (
gradient_descent_v2.SGD,
tf.compat.v1.train.GradientDescentOptimizer,
):
# With SGD, the variables decreases by exactly 1
self.assertEqual(self.evaluate(x), 0)
if __name__ == "__main__":
test.main()
|
tf-keras/tf_keras/mixed_precision/autocast_variable_test.py/0
|
{
"file_path": "tf-keras/tf_keras/mixed_precision/autocast_variable_test.py",
"repo_id": "tf-keras",
"token_count": 16159
}
| 215 |
# Copyright 2021 The TensorFlow Authors. All Rights Reserved.
#
# Licensed under the Apache License, Version 2.0 (the "License");
# you may not use this file except in compliance with the License.
# You may obtain a copy of the License at
#
# http://www.apache.org/licenses/LICENSE-2.0
#
# Unless required by applicable law or agreed to in writing, software
# distributed under the License is distributed on an "AS IS" BASIS,
# WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.
# See the License for the specific language governing permissions and
# limitations under the License.
# ==============================================================================
"""Adagrad optimizer implementation."""
import tensorflow.compat.v2 as tf
from tf_keras import initializers
from tf_keras.optimizers import optimizer
from tf_keras.saving.object_registration import register_keras_serializable
# isort: off
from tensorflow.python.util.tf_export import keras_export
@register_keras_serializable()
@keras_export(
"keras.optimizers.Adagrad",
"keras.optimizers.experimental.Adagrad",
"keras.dtensor.experimental.optimizers.Adagrad",
v1=[],
)
class Adagrad(optimizer.Optimizer):
r"""Optimizer that implements the Adagrad algorithm.
Adagrad is an optimizer with parameter-specific learning rates,
which are adapted relative to how frequently a parameter gets
updated during training. The more updates a parameter receives,
the smaller the updates.
Args:
learning_rate: Initial value for the learning rate:
either a floating point value,
or a `tf.keras.optimizers.schedules.LearningRateSchedule` instance.
Defaults to 0.001. Note that `Adagrad` tends to benefit from higher
initial learning rate values compared to other optimizers. To match
the exact form in the original paper, use 1.0.
initial_accumulator_value: Floating point value.
Starting value for the accumulators (per-parameter momentum values).
Must be non-negative.
epsilon: Small floating point value used to maintain numerical
stability.
{{base_optimizer_keyword_args}}
Reference:
- [Duchi et al., 2011](
http://www.jmlr.org/papers/volume12/duchi11a/duchi11a.pdf).
"""
def __init__(
self,
learning_rate=0.001,
initial_accumulator_value=0.1,
epsilon=1e-7,
weight_decay=None,
clipnorm=None,
clipvalue=None,
global_clipnorm=None,
use_ema=False,
ema_momentum=0.99,
ema_overwrite_frequency=None,
jit_compile=True,
name="Adagrad",
**kwargs
):
super().__init__(
weight_decay=weight_decay,
clipnorm=clipnorm,
clipvalue=clipvalue,
global_clipnorm=global_clipnorm,
use_ema=use_ema,
ema_momentum=ema_momentum,
ema_overwrite_frequency=ema_overwrite_frequency,
jit_compile=jit_compile,
name=name,
**kwargs
)
self._learning_rate = self._build_learning_rate(learning_rate)
self.initial_accumulator_value = initial_accumulator_value
self.epsilon = epsilon
def build(self, var_list):
super().build(var_list)
if hasattr(self, "_built") and self._built:
return
self._built = True
self._accumulators = []
initializer = initializers.Constant(self.initial_accumulator_value)
for var in var_list:
self._accumulators.append(
self.add_variable_from_reference(
var,
"accumulator",
initial_value=initializer(shape=var.shape, dtype=var.dtype),
)
)
def update_step(self, grad, variable):
"""Update step given gradient and the associated model variable."""
lr = tf.cast(self.learning_rate, variable.dtype)
var_key = self._var_key(variable)
accumulator = self._accumulators[self._index_dict[var_key]]
if isinstance(grad, tf.IndexedSlices):
# Sparse gradients.
accumulator.scatter_add(
tf.IndexedSlices(grad.values * grad.values, grad.indices)
)
sparse_accumulator = tf.gather(accumulator, indices=grad.indices)
sparse_denominator = tf.sqrt(sparse_accumulator + self.epsilon)
variable.scatter_add(
tf.IndexedSlices(
-lr * grad.values / sparse_denominator, grad.indices
)
)
else:
# Dense gradients.
accumulator.assign_add(grad * grad)
variable.assign_sub(lr * grad / tf.sqrt(accumulator + self.epsilon))
def get_config(self):
config = super().get_config()
config.update(
{
"learning_rate": self._serialize_hyperparameter(
self._learning_rate
),
"initial_accumulator_value": self.initial_accumulator_value,
"epsilon": self.epsilon,
}
)
return config
Adagrad.__doc__ = Adagrad.__doc__.replace(
"{{base_optimizer_keyword_args}}", optimizer.base_optimizer_keyword_args
)
|
tf-keras/tf_keras/optimizers/adagrad.py/0
|
{
"file_path": "tf-keras/tf_keras/optimizers/adagrad.py",
"repo_id": "tf-keras",
"token_count": 2340
}
| 216 |
# Copyright 2015 The TensorFlow Authors. All Rights Reserved.
#
# Licensed under the Apache License, Version 2.0 (the "License");
# you may not use this file except in compliance with the License.
# You may obtain a copy of the License at
#
# http://www.apache.org/licenses/LICENSE-2.0
#
# Unless required by applicable law or agreed to in writing, software
# distributed under the License is distributed on an "AS IS" BASIS,
# WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.
# See the License for the specific language governing permissions and
# limitations under the License.
# ==============================================================================
"""Functional tests for Ftrl operations."""
import numpy as np
import tensorflow.compat.v2 as tf
from tf_keras.optimizers.legacy import ftrl
class FtrlOptimizerTest(tf.test.TestCase):
def doTestFtrlwithoutRegularization(self, use_resource=False):
# TODO(tanzheny, omalleyt): Fix test in eager mode.
for dtype in [tf.float32]:
with tf.Graph().as_default(), self.cached_session():
if use_resource:
var0 = tf.Variable([0.0, 0.0], dtype=dtype)
var1 = tf.Variable([0.0, 0.0], dtype=dtype)
else:
var0 = tf.Variable([0.0, 0.0], dtype=dtype)
var1 = tf.Variable([0.0, 0.0], dtype=dtype)
grads0 = tf.constant([0.1, 0.2], dtype=dtype)
grads1 = tf.constant([0.01, 0.02], dtype=dtype)
opt = ftrl.Ftrl(
3.0,
initial_accumulator_value=0.1,
l1_regularization_strength=0.0,
l2_regularization_strength=0.0,
)
update = opt.apply_gradients(
zip([grads0, grads1], [var0, var1])
)
self.evaluate(tf.compat.v1.global_variables_initializer())
v0_val, v1_val = self.evaluate([var0, var1])
self.assertAllClose([0.0, 0.0], v0_val)
self.assertAllClose([0.0, 0.0], v1_val)
# Run 3 steps FTRL
for _ in range(3):
update.run()
v0_val, v1_val = self.evaluate([var0, var1])
self.assertAllCloseAccordingToType(
np.array([-2.60260963, -4.29698515]), v0_val
)
self.assertAllCloseAccordingToType(
np.array([-0.28432083, -0.56694895]), v1_val
)
def testFtrlWithoutRegularization(self):
self.doTestFtrlwithoutRegularization(use_resource=False)
def testResourceFtrlWithoutRegularization(self):
self.doTestFtrlwithoutRegularization(use_resource=True)
def testFtrlwithoutRegularization2(self):
# TODO(tanzheny, omalleyt): Fix test in eager mode.
for dtype in [tf.half, tf.float32]:
with tf.Graph().as_default(), self.cached_session():
var0 = tf.Variable([1.0, 2.0], dtype=dtype)
var1 = tf.Variable([4.0, 3.0], dtype=dtype)
grads0 = tf.constant([0.1, 0.2], dtype=dtype)
grads1 = tf.constant([0.01, 0.02], dtype=dtype)
opt = ftrl.Ftrl(
3.0,
initial_accumulator_value=0.1,
l1_regularization_strength=0.0,
l2_regularization_strength=0.0,
)
update = opt.apply_gradients(
zip([grads0, grads1], [var0, var1])
)
self.evaluate(tf.compat.v1.global_variables_initializer())
v0_val, v1_val = self.evaluate([var0, var1])
self.assertAllCloseAccordingToType([1.0, 2.0], v0_val)
self.assertAllCloseAccordingToType([4.0, 3.0], v1_val)
# Run 3 steps FTRL
for _ in range(3):
update.run()
v0_val, v1_val = self.evaluate([var0, var1])
self.assertAllCloseAccordingToType(
np.array([-2.55607247, -3.98729396]), v0_val
)
self.assertAllCloseAccordingToType(
np.array([-0.28232238, -0.56096673]), v1_val
)
def testMinimizeSparseResourceVariable(self):
# TODO(tanzheny, omalleyt): Fix test in eager mode.
for dtype in [tf.half, tf.float32, tf.float64]:
with tf.Graph().as_default(), self.cached_session():
var0 = tf.Variable([[1.0, 2.0]], dtype=dtype)
x = tf.constant([[4.0], [5.0]], dtype=dtype)
def loss():
pred = tf.matmul(
tf.compat.v1.nn.embedding_lookup([var0], [0]), x
)
return pred * pred
sgd_op = ftrl.Ftrl(1.0).minimize(loss, var_list=[var0])
self.evaluate(tf.compat.v1.global_variables_initializer())
# Fetch params to validate initial values
self.assertAllCloseAccordingToType(
[[1.0, 2.0]], self.evaluate(var0)
)
# Run 1 step of sgd
sgd_op.run()
# Validate updated params
self.assertAllCloseAccordingToType(
[[0, 1]], self.evaluate(var0), atol=0.01
)
def testFtrlWithL1(self):
# TODO(tanzheny, omalleyt): Fix test in eager mode.
for dtype in [tf.half, tf.float32]:
with tf.Graph().as_default(), self.cached_session():
var0 = tf.Variable([1.0, 2.0], dtype=dtype)
var1 = tf.Variable([4.0, 3.0], dtype=dtype)
grads0 = tf.constant([0.1, 0.2], dtype=dtype)
grads1 = tf.constant([0.01, 0.02], dtype=dtype)
opt = ftrl.Ftrl(
3.0,
initial_accumulator_value=0.1,
l1_regularization_strength=0.001,
l2_regularization_strength=0.0,
)
update = opt.apply_gradients(
zip([grads0, grads1], [var0, var1])
)
self.evaluate(tf.compat.v1.global_variables_initializer())
v0_val, v1_val = self.evaluate([var0, var1])
self.assertAllCloseAccordingToType([1.0, 2.0], v0_val)
self.assertAllCloseAccordingToType([4.0, 3.0], v1_val)
# Run 10 steps FTRL
for _ in range(10):
update.run()
v0_val, v1_val = self.evaluate([var0, var1])
self.assertAllCloseAccordingToType(
np.array([-7.66718769, -10.91273689]), v0_val
)
self.assertAllCloseAccordingToType(
np.array([-0.93460727, -1.86147261]), v1_val
)
def testFtrlWithBeta(self):
# TODO(tanzheny, omalleyt): Fix test in eager mode.
for dtype in [tf.half, tf.float32]:
with tf.Graph().as_default(), self.cached_session():
var0 = tf.Variable([1.0, 2.0], dtype=dtype)
var1 = tf.Variable([4.0, 3.0], dtype=dtype)
grads0 = tf.constant([0.1, 0.2], dtype=dtype)
grads1 = tf.constant([0.01, 0.02], dtype=dtype)
opt = ftrl.Ftrl(3.0, initial_accumulator_value=0.1, beta=0.1)
update = opt.apply_gradients(
zip([grads0, grads1], [var0, var1])
)
self.evaluate(tf.compat.v1.global_variables_initializer())
v0_val, v1_val = self.evaluate([var0, var1])
self.assertAllCloseAccordingToType([1.0, 2.0], v0_val)
self.assertAllCloseAccordingToType([4.0, 3.0], v1_val)
# Run 10 steps FTRL
for _ in range(10):
update.run()
v0_val, v1_val = self.evaluate([var0, var1])
self.assertAllCloseAccordingToType(
np.array([-6.096838, -9.162214]), v0_val
)
self.assertAllCloseAccordingToType(
np.array([-0.717741, -1.425132]), v1_val
)
def testFtrlWithL2_Beta(self):
# TODO(tanzheny, omalleyt): Fix test in eager mode.
for dtype in [tf.half, tf.float32]:
with tf.Graph().as_default(), self.cached_session():
var0 = tf.Variable([1.0, 2.0], dtype=dtype)
var1 = tf.Variable([4.0, 3.0], dtype=dtype)
grads0 = tf.constant([0.1, 0.2], dtype=dtype)
grads1 = tf.constant([0.01, 0.02], dtype=dtype)
opt = ftrl.Ftrl(
3.0,
initial_accumulator_value=0.1,
l1_regularization_strength=0.0,
l2_regularization_strength=0.1,
beta=0.1,
)
update = opt.apply_gradients(
zip([grads0, grads1], [var0, var1])
)
self.evaluate(tf.compat.v1.global_variables_initializer())
v0_val, v1_val = self.evaluate([var0, var1])
self.assertAllCloseAccordingToType([1.0, 2.0], v0_val)
self.assertAllCloseAccordingToType([4.0, 3.0], v1_val)
# Run 10 steps FTRL
for _ in range(10):
update.run()
v0_val, v1_val = self.evaluate([var0, var1])
self.assertAllCloseAccordingToType(
np.array([-2.735487, -4.704625]), v0_val
)
self.assertAllCloseAccordingToType(
np.array([-0.294335, -0.586556]), v1_val
)
def testFtrlWithL1_L2(self):
# TODO(tanzheny, omalleyt): Fix test in eager mode.
for dtype in [tf.half, tf.float32]:
with tf.Graph().as_default(), self.cached_session():
var0 = tf.Variable([1.0, 2.0], dtype=dtype)
var1 = tf.Variable([4.0, 3.0], dtype=dtype)
grads0 = tf.constant([0.1, 0.2], dtype=dtype)
grads1 = tf.constant([0.01, 0.02], dtype=dtype)
opt = ftrl.Ftrl(
3.0,
initial_accumulator_value=0.1,
l1_regularization_strength=0.001,
l2_regularization_strength=2.0,
)
update = opt.apply_gradients(
zip([grads0, grads1], [var0, var1])
)
self.evaluate(tf.compat.v1.global_variables_initializer())
v0_val, v1_val = self.evaluate([var0, var1])
self.assertAllCloseAccordingToType([1.0, 2.0], v0_val)
self.assertAllCloseAccordingToType([4.0, 3.0], v1_val)
# Run 10 steps FTRL
for _ in range(10):
update.run()
v0_val, v1_val = self.evaluate([var0, var1])
self.assertAllCloseAccordingToType(
np.array([-0.24059935, -0.46829352]), v0_val
)
self.assertAllCloseAccordingToType(
np.array([-0.02406147, -0.04830509]), v1_val
)
def testFtrlWithL1_L2_L2Shrinkage(self):
"""Test the new FTRL op with support for l2 shrinkage.
The addition of this parameter which places a constant pressure on
weights towards the origin causes the gradient descent trajectory to
differ. The weights will tend to have smaller magnitudes with this
parameter set.
"""
# TODO(tanzheny, omalleyt): Fix test in eager mode.
for dtype in [tf.half, tf.float32]:
with tf.Graph().as_default(), self.cached_session():
var0 = tf.Variable([1.0, 2.0], dtype=dtype)
var1 = tf.Variable([4.0, 3.0], dtype=dtype)
grads0 = tf.constant([0.1, 0.2], dtype=dtype)
grads1 = tf.constant([0.01, 0.02], dtype=dtype)
opt = ftrl.Ftrl(
3.0,
initial_accumulator_value=0.1,
l1_regularization_strength=0.001,
l2_regularization_strength=2.0,
l2_shrinkage_regularization_strength=0.1,
)
update = opt.apply_gradients(
zip([grads0, grads1], [var0, var1])
)
self.evaluate(tf.compat.v1.global_variables_initializer())
v0_val, v1_val = self.evaluate([var0, var1])
self.assertAllCloseAccordingToType([1.0, 2.0], v0_val)
self.assertAllCloseAccordingToType([4.0, 3.0], v1_val)
# Run 10 steps FTRL
for _ in range(10):
update.run()
v0_val, v1_val = self.evaluate([var0, var1])
self.assertAllCloseAccordingToType(
np.array([-0.22578995, -0.44345796]), v0_val
)
self.assertAllCloseAccordingToType(
np.array([-0.14378493, -0.13229476]), v1_val
)
def testFtrlWithL1_L2_L2ShrinkageSparse(self):
"""Tests the new FTRL op with support for l2 shrinkage on sparse
grads."""
# TODO(tanzheny, omalleyt): Fix test in eager mode.
for dtype in [tf.half, tf.float32]:
with tf.Graph().as_default(), self.cached_session():
var0 = tf.Variable([[1.0], [2.0]], dtype=dtype)
var1 = tf.Variable([[4.0], [3.0]], dtype=dtype)
grads0 = tf.IndexedSlices(
tf.constant([0.1], shape=[1, 1], dtype=dtype),
tf.constant([0]),
tf.constant([2, 1]),
)
grads1 = tf.IndexedSlices(
tf.constant([0.02], shape=[1, 1], dtype=dtype),
tf.constant([1]),
tf.constant([2, 1]),
)
opt = ftrl.Ftrl(
3.0,
initial_accumulator_value=0.1,
l1_regularization_strength=0.001,
l2_regularization_strength=2.0,
l2_shrinkage_regularization_strength=0.1,
)
update = opt.apply_gradients(
zip([grads0, grads1], [var0, var1])
)
self.evaluate(tf.compat.v1.global_variables_initializer())
v0_val, v1_val = self.evaluate([var0, var1])
self.assertAllCloseAccordingToType([[1.0], [2.0]], v0_val)
self.assertAllCloseAccordingToType([[4.0], [3.0]], v1_val)
# Run 10 steps FTRL
for _ in range(10):
update.run()
v0_val, v1_val = self.evaluate([var0, var1])
self.assertAllCloseAccordingToType(
[[-0.22578995], [2.0]], v0_val
)
self.assertAllCloseAccordingToType(
[[4.0], [-0.13229476]], v1_val
)
def testFtrlWithL2ShrinkageDoesNotChangeLrSchedule(self):
"""Verifies that l2 shrinkage in FTRL does not change lr schedule."""
# TODO(tanzheny, omalleyt): Fix test in eager mode.
for dtype in [tf.half, tf.float32]:
with tf.Graph().as_default(), self.cached_session() as sess:
var0 = tf.Variable([1.0, 2.0], dtype=dtype)
var1 = tf.Variable([1.0, 2.0], dtype=dtype)
grads0 = tf.constant([0.1, 0.2], dtype=dtype)
grads1 = tf.constant([0.1, 0.2], dtype=dtype)
opt0 = ftrl.Ftrl(
3.0,
initial_accumulator_value=0.1,
l1_regularization_strength=0.001,
l2_regularization_strength=2.0,
l2_shrinkage_regularization_strength=0.1,
)
opt1 = ftrl.Ftrl(
3.0,
initial_accumulator_value=0.1,
l1_regularization_strength=0.001,
l2_regularization_strength=2.0,
)
update0 = opt0.apply_gradients([(grads0, var0)])
update1 = opt1.apply_gradients([(grads1, var1)])
self.evaluate(tf.compat.v1.global_variables_initializer())
v0_val, v1_val = self.evaluate([var0, var1])
self.assertAllCloseAccordingToType([1.0, 2.0], v0_val)
self.assertAllCloseAccordingToType([1.0, 2.0], v1_val)
# Run 10 steps FTRL
for _ in range(10):
update0.run()
update1.run()
v0_val, v1_val = self.evaluate([var0, var1])
# var0 is experiencing L2 shrinkage so it should be smaller than
# var1 in magnitude.
self.assertTrue((v0_val**2 < v1_val**2).all())
accum0 = sess.run(opt0.get_slot(var0, "accumulator"))
accum1 = sess.run(opt1.get_slot(var1, "accumulator"))
# L2 shrinkage should not change how we update grad accumulator.
self.assertAllCloseAccordingToType(accum0, accum1)
def applyOptimizer(self, opt, dtype, steps=5, is_sparse=False):
if is_sparse:
var0 = tf.Variable([[0.0], [0.0]], dtype=dtype)
var1 = tf.Variable([[0.0], [0.0]], dtype=dtype)
grads0 = tf.IndexedSlices(
tf.constant([0.1], shape=[1, 1], dtype=dtype),
tf.constant([0]),
tf.constant([2, 1]),
)
grads1 = tf.IndexedSlices(
tf.constant([0.02], shape=[1, 1], dtype=dtype),
tf.constant([1]),
tf.constant([2, 1]),
)
else:
var0 = tf.Variable([0.0, 0.0], dtype=dtype)
var1 = tf.Variable([0.0, 0.0], dtype=dtype)
grads0 = tf.constant([0.1, 0.2], dtype=dtype)
grads1 = tf.constant([0.01, 0.02], dtype=dtype)
update = opt.apply_gradients(zip([grads0, grads1], [var0, var1]))
self.evaluate(tf.compat.v1.global_variables_initializer())
v0_val, v1_val = self.evaluate([var0, var1])
if is_sparse:
self.assertAllCloseAccordingToType([[0.0], [0.0]], v0_val)
self.assertAllCloseAccordingToType([[0.0], [0.0]], v1_val)
else:
self.assertAllCloseAccordingToType([0.0, 0.0], v0_val)
self.assertAllCloseAccordingToType([0.0, 0.0], v1_val)
# Run Ftrl for a few steps
for _ in range(steps):
update.run()
v0_val, v1_val = self.evaluate([var0, var1])
return v0_val, v1_val
# When variables are initialized with Zero, FTRL-Proximal has two
# properties:
# 1. Without L1&L2 but with fixed learning rate, FTRL-Proximal is identical
# with GradientDescent.
# 2. Without L1&L2 but with adaptive learning rate, FTRL-Proximal is
# identical with Adagrad.
# So, basing on these two properties, we test if our implementation of
# FTRL-Proximal performs same updates as Adagrad or GradientDescent.
def testEquivAdagradwithoutRegularization(self):
# TODO(tanzheny, omalleyt): Fix test in eager mode.
for dtype in [tf.half, tf.float32]:
with tf.Graph().as_default(), self.cached_session():
val0, val1 = self.applyOptimizer(
ftrl.Ftrl(
3.0,
# Adagrad learning rate
learning_rate_power=-0.5,
initial_accumulator_value=0.1,
l1_regularization_strength=0.0,
l2_regularization_strength=0.0,
),
dtype,
)
with tf.Graph().as_default(), self.cached_session():
val2, val3 = self.applyOptimizer(
tf.compat.v1.train.AdagradOptimizer(
3.0, initial_accumulator_value=0.1
),
dtype,
)
self.assertAllCloseAccordingToType(val0, val2)
self.assertAllCloseAccordingToType(val1, val3)
def testEquivSparseAdagradwithoutRegularization(self):
# TODO(tanzheny, omalleyt): Fix test in eager mode.
for dtype in [tf.half, tf.float32]:
with tf.Graph().as_default(), self.cached_session():
val0, val1 = self.applyOptimizer(
ftrl.Ftrl(
3.0,
# Adagrad learning rate
learning_rate_power=-0.5,
initial_accumulator_value=0.1,
l1_regularization_strength=0.0,
l2_regularization_strength=0.0,
),
dtype,
is_sparse=True,
)
with tf.Graph().as_default(), self.cached_session():
val2, val3 = self.applyOptimizer(
tf.compat.v1.train.AdagradOptimizer(
3.0, initial_accumulator_value=0.1
),
dtype,
is_sparse=True,
)
self.assertAllCloseAccordingToType(val0, val2)
self.assertAllCloseAccordingToType(val1, val3)
def testEquivSparseGradientDescentwithoutRegularization(self):
# TODO(tanzheny, omalleyt): Fix test in eager mode.
for dtype in [tf.half, tf.float32]:
with tf.Graph().as_default(), self.cached_session():
val0, val1 = self.applyOptimizer(
ftrl.Ftrl(
3.0,
# Fixed learning rate
learning_rate_power=-0.0,
initial_accumulator_value=0.1,
l1_regularization_strength=0.0,
l2_regularization_strength=0.0,
),
dtype,
is_sparse=True,
)
with tf.Graph().as_default(), self.cached_session():
val2, val3 = self.applyOptimizer(
tf.compat.v1.train.GradientDescentOptimizer(3.0),
dtype,
is_sparse=True,
)
self.assertAllCloseAccordingToType(val0, val2)
self.assertAllCloseAccordingToType(val1, val3)
def testEquivGradientDescentwithoutRegularization(self):
# TODO(tanzheny, omalleyt): Fix test in eager mode.
for dtype in [tf.half, tf.float32]:
with tf.Graph().as_default(), self.cached_session():
val0, val1 = self.applyOptimizer(
ftrl.Ftrl(
3.0,
# Fixed learning rate
learning_rate_power=-0.0,
initial_accumulator_value=0.1,
l1_regularization_strength=0.0,
l2_regularization_strength=0.0,
),
dtype,
)
with tf.Graph().as_default(), self.cached_session():
val2, val3 = self.applyOptimizer(
tf.compat.v1.train.GradientDescentOptimizer(3.0), dtype
)
self.assertAllCloseAccordingToType(val0, val2)
self.assertAllCloseAccordingToType(val1, val3)
if __name__ == "__main__":
tf.test.main()
|
tf-keras/tf_keras/optimizers/legacy/ftrl_test.py/0
|
{
"file_path": "tf-keras/tf_keras/optimizers/legacy/ftrl_test.py",
"repo_id": "tf-keras",
"token_count": 13544
}
| 217 |
"""Tests for the reworked optimizer.
More context in go/new-keras-optimizer
"""
import os
from unittest import mock
import numpy as np
import tensorflow.compat.v2 as tf
from absl.testing import parameterized
import tf_keras as keras
from tf_keras.optimizers import adadelta as adadelta_new
from tf_keras.optimizers import adafactor as adafactor_new
from tf_keras.optimizers import adagrad as adagrad_new
from tf_keras.optimizers import adam as adam_new
from tf_keras.optimizers import adamax as adamax_new
from tf_keras.optimizers import adamw as adamw_new
from tf_keras.optimizers import ftrl as ftrl_new
from tf_keras.optimizers import lion as lion_new
from tf_keras.optimizers import nadam as nadam_new
from tf_keras.optimizers import rmsprop as rmsprop_new
from tf_keras.optimizers import sgd as sgd_new
from tf_keras.optimizers.legacy import adadelta as adadelta_old
from tf_keras.optimizers.legacy import adagrad as adagrad_old
from tf_keras.optimizers.legacy import adam as adam_old
from tf_keras.optimizers.legacy import ftrl as ftrl_old
from tf_keras.optimizers.legacy import gradient_descent as sgd_old
from tf_keras.optimizers.legacy import rmsprop as rmsprop_old
from tf_keras.optimizers.schedules import learning_rate_schedule
from tf_keras.testing_infra import test_utils
from tf_keras.utils import losses_utils
ds_combinations = tf.__internal__.distribute.combinations
STRATEGIES = [
# TODO(b/202992598): Add PSS strategy once the XLA issues is resolved.
ds_combinations.one_device_strategy,
ds_combinations.mirrored_strategy_with_two_cpus,
ds_combinations.mirrored_strategy_with_two_gpus,
ds_combinations.tpu_strategy,
ds_combinations.cloud_tpu_strategy,
ds_combinations.multi_worker_mirrored_2x1_cpu,
ds_combinations.multi_worker_mirrored_2x2_gpu,
ds_combinations.central_storage_strategy_with_two_gpus,
]
adadelta_new_fn = tf.__internal__.test.combinations.NamedObject(
"experimentaladadelta",
lambda: adadelta_new.Adadelta(
0.002, use_ema=True, ema_overwrite_frequency=None
),
)
adagrad_new_fn = tf.__internal__.test.combinations.NamedObject(
"experimentaladagrad", lambda: adagrad_new.Adagrad(0.002)
)
adafactor_new_fn = tf.__internal__.test.combinations.NamedObject(
"adafactor", lambda: adafactor_new.Adafactor(0.002)
)
adam_new_fn = tf.__internal__.test.combinations.NamedObject(
"experimentaladam", lambda: adam_new.Adam(0.002)
)
adamax_new_fn = tf.__internal__.test.combinations.NamedObject(
"experimentaladamax", lambda: adamax_new.Adamax(0.002)
)
adamw_new_fn = tf.__internal__.test.combinations.NamedObject(
"experimentaladamw", lambda: adamw_new.AdamW(0.002, weight_decay=0.004)
)
ftrl_new_fn = tf.__internal__.test.combinations.NamedObject(
"experimentalftrl", lambda: ftrl_new.Ftrl(0.002)
)
lion_new_fn = tf.__internal__.test.combinations.NamedObject(
"lion", lambda: lion_new.Lion(0.002)
)
nadam_new_fn = tf.__internal__.test.combinations.NamedObject(
"experimentnadam", lambda: nadam_new.Nadam(0.002)
)
rmsprop_new_fn = tf.__internal__.test.combinations.NamedObject(
"experimentalrmsprop", lambda: rmsprop_new.RMSprop(0.002)
)
sgd_new_fn = tf.__internal__.test.combinations.NamedObject(
"experimentalsgdaverage",
lambda: sgd_new.SGD(
0.002, weight_decay=0.004, use_ema=True, ema_overwrite_frequency=1
),
)
OPTIMIZER_FN = [
adadelta_new_fn,
adagrad_new_fn,
adafactor_new_fn,
adam_new_fn,
adamax_new_fn,
adamw_new_fn,
ftrl_new_fn,
lion_new_fn,
nadam_new_fn,
rmsprop_new_fn,
sgd_new_fn,
]
class OptimizerFuntionalityTest(tf.test.TestCase, parameterized.TestCase):
"""Test the functionality of optimizer."""
def testAddVariableFromReference(self):
optimizer = adam_new.Adam()
variable = optimizer.add_variable_from_reference(
tf.Variable(1.0, name="tmp"), "test"
)
self.assertEqual(variable._shared_name, "test/tmp")
self.assertEqual(self.evaluate(variable), 0)
def testAddVarialeWithCustomShape(self):
optimizer = adam_new.Adam()
variable = optimizer.add_variable_from_reference(
tf.Variable([1.0, 2.0], name="tmp"), "test", shape=[]
)
self.assertEqual(variable, tf.Variable(0.0))
def testBuildIndexDict(self):
optimizer = adam_new.Adam()
var_list = [tf.Variable(0, name=f"var{i}") for i in range(10)]
optimizer._build_index_dict(var_list)
self.assertEqual(
optimizer._index_dict[optimizer._var_key(var_list[7])], 7
)
def testComputeGradients(self):
optimizer = adam_new.Adam()
x = tf.Variable([1.0, 2.0], dtype=tf.float32)
loss_fn = lambda: x
# Test Tensor-type var_list.
var_list = [x]
grads_and_vars = optimizer.compute_gradients(loss_fn, var_list)
grads, _ = zip(*grads_and_vars)
self.assertAllEqual(grads[0], tf.constant([1.0, 1.0]))
# Test callable-type var_list, and create variable in loss fn.
x = []
def loss_fn():
variable = tf.Variable([1.0, 2.0], dtype=tf.float32)
x.append(variable)
return variable
var_list = lambda: x
grads_and_vars = optimizer.compute_gradients(loss_fn, var_list)
grads, _ = zip(*grads_and_vars)
self.assertAllEqual(grads[0], tf.constant([1.0, 1.0]))
def testClipNorm(self):
optimizer = adam_new.Adam(clipnorm=1)
grad = [tf.convert_to_tensor([100.0, 100.0])]
clipped_grad = optimizer._clip_gradients(grad)
self.assertAllClose(clipped_grad[0], [2**0.5 / 2, 2**0.5 / 2])
def testClipValue(self):
optimizer = adam_new.Adam(clipvalue=1)
grad = [tf.convert_to_tensor([100.0, 100.0])]
clipped_grad = optimizer._clip_gradients(grad)
self.assertAllEqual(clipped_grad[0], [1.0, 1.0])
def testWeightDecay(self):
grads, var1, var2, var3 = (
tf.zeros(()),
tf.Variable(2.0),
tf.Variable(2.0, name="exclude"),
tf.Variable(2.0),
)
optimizer_1 = adamw_new.AdamW(learning_rate=1, weight_decay=0.004)
optimizer_1.apply_gradients(zip([grads], [var1]))
optimizer_2 = adamw_new.AdamW(learning_rate=1, weight_decay=0.004)
optimizer_2.exclude_from_weight_decay(var_names=["exclude"])
optimizer_2.apply_gradients(zip([grads, grads], [var1, var2]))
optimizer_3 = adamw_new.AdamW(learning_rate=1, weight_decay=0.004)
optimizer_3.exclude_from_weight_decay(var_list=[var3])
optimizer_3.apply_gradients(zip([grads, grads], [var1, var3]))
self.assertEqual(var1, 1.9760959)
self.assertEqual(var2, 2.0)
self.assertEqual(var3, 2.0)
grads, var1, var2, var3 = (
tf.zeros(()),
tf.Variable(2.0),
tf.Variable(2.0, name="exclude"),
tf.Variable(2.0),
)
optimizer_1 = sgd_new.SGD(learning_rate=1, weight_decay=0.004)
optimizer_1.apply_gradients(zip([grads], [var1]))
optimizer_2 = sgd_new.SGD(learning_rate=1, weight_decay=0.004)
optimizer_2.exclude_from_weight_decay(var_names=["exclude"])
optimizer_2.apply_gradients(zip([grads, grads], [var1, var2]))
optimizer_3 = sgd_new.SGD(learning_rate=1, weight_decay=0.004)
optimizer_3.exclude_from_weight_decay(var_list=[var3])
optimizer_3.apply_gradients(zip([grads, grads], [var1, var3]))
self.assertEqual(var1, 1.9760959)
self.assertEqual(var2, 2.0)
self.assertEqual(var3, 2.0)
def testClipGlobalNorm(self):
optimizer = adam_new.Adam(global_clipnorm=1)
grad = [
tf.cast([100.0, 100.0], dtype=tf.float32),
tf.cast([100.0, 100.0], dtype=tf.float32),
]
clipped_grad = optimizer._clip_gradients(grad)
self.assertAllClose(clipped_grad[0], [0.5, 0.5])
def testPassingLegacyArgsRaiseError(self):
with self.assertRaisesRegex(ValueError, "decay is deprecated*"):
_ = adam_new.Adam(clipnorm=1, decay=0.5)
def testPassingLegacyClipnorm(self):
optimizer = adam_new.Adam(clipnorm=1)
self.assertEqual(optimizer.clipnorm, 1)
def testReturnAllOptimizerVariables(self):
x = tf.Variable([[1.0, 2.0], [3.0, 4.0]], dtype=tf.float32)
optimizer = adam_new.Adam()
grads = tf.convert_to_tensor([[1.0, 2.0], [3.0, 4.0]])
optimizer.apply_gradients(zip([grads], [x]))
optimizer_variables = optimizer.variables
all_names = [var._shared_name for var in optimizer_variables]
self.assertLen(optimizer_variables, 3)
self.assertCountEqual(
all_names,
[
"iteration",
"Adam/m/Variable",
"Adam/v/Variable",
],
)
def testSetWeights(self):
x = tf.Variable([[1.0, 2.0], [3.0, 4.0]], dtype=tf.float32)
optimizer_1 = adam_new.Adam()
grads = tf.convert_to_tensor([[1.0, 2.0], [3.0, 4.0]])
optimizer_1.apply_gradients(zip([grads], [x]))
optimizer_2 = adam_new.Adam()
with self.assertRaisesRegex(ValueError, "You are calling*"):
optimizer_2.set_weights(optimizer_1.variables)
optimizer_2.build([x])
optimizer_2.set_weights(optimizer_1.variables)
self.assertAllClose(optimizer_1.variables, optimizer_2.variables)
def testSetLearningRate(self):
optimizer = adam_new.Adam(learning_rate=1.0)
self.assertIsInstance(optimizer._learning_rate, tf.Variable)
self.assertEqual(self.evaluate(optimizer.learning_rate), 1.0)
optimizer.learning_rate = 2.0
self.assertEqual(self.evaluate(optimizer.learning_rate), 2.0)
# Test the legacy setter.
optimizer.lr = 3.0
self.assertEqual(self.evaluate(optimizer.learning_rate), 3.0)
lr_schedule = learning_rate_schedule.ExponentialDecay(
initial_learning_rate=1e-2, decay_steps=10000, decay_rate=0.9
)
optimizer = adam_new.Adam(learning_rate=lr_schedule)
self.assertIsInstance(
optimizer._learning_rate, learning_rate_schedule.ExponentialDecay
)
self.assertEqual(optimizer.learning_rate, 0.01)
# Test the legacy property.
self.assertEqual(optimizer.lr, 0.01)
x = tf.Variable([1.0, 2.0], dtype=tf.float32)
grads = tf.convert_to_tensor([1.0, 2.0])
for _ in range(2):
optimizer.apply_gradients(zip([grads], [x]))
self.assertTrue(
optimizer.learning_rate < 0.01 and optimizer.learning_rate > 0.00999
)
# Check it does not throw error to set `learning_rate` by a
# LearningRateScheduler instance.
optimizer.learning_rate = learning_rate_schedule.ExponentialDecay(
initial_learning_rate=1e-2, decay_steps=10000, decay_rate=0.9
)
with self.assertRaisesRegex(
TypeError, "This optimizer was created with*"
):
optimizer.learning_rate = 2.0
def testSetIterations(self):
optimizer = adam_new.Adam(jit_compile=False)
optimizer.iterations = tf.Variable(2, dtype=tf.int32)
self.assertEqual(optimizer.iterations, 2)
var_list = [tf.Variable(2.0), tf.Variable(2.0)]
grads = tf.convert_to_tensor([1.0, 1.0])
iterations = optimizer.apply_gradients(zip(grads, var_list))
self.assertEqual(iterations, 3)
self.assertEqual(optimizer.iterations, 3)
with self.assertRaisesRegex(RuntimeError, "Cannot set*"):
optimizer.iterations = 2
def testVariableConstraints(self):
optimizer = adam_new.Adam()
inputs = keras.layers.Input(shape=[1])
outputs = keras.layers.Dense(1, kernel_constraint="NonNeg")(inputs)
model = keras.models.Model(inputs=inputs, outputs=outputs)
model.trainable_variables[0] = -999999 # Set as a negative number.
grads = [tf.zeros(1, 1), tf.zeros(1)]
optimizer.apply_gradients(zip(grads, model.trainable_variables))
self.assertEqual(model.trainable_variables[0], 0.0)
def testNoGradients(self):
optimizer = adam_new.Adam(jit_compile=False)
optimizer.apply_gradients(zip([], []))
def testApplyGradientsNameArg(self):
optimizer = adam_new.Adam(jit_compile=False)
var_list = [tf.Variable(2.0), tf.Variable(2.0)]
grads = tf.convert_to_tensor([1.0, 1.0])
optimizer.apply_gradients(zip(grads, var_list), name="dummy")
self.assertIn("dummy", optimizer._velocities[0].name)
def testPassingMissingWDError(self):
with self.assertRaises(ValueError):
_ = adamw_new.AdamW(0.01, weight_decay=None)
with self.assertRaisesRegex(ValueError, "Missing value of"):
_ = adamw_new.AdamW(0.01, weight_decay=None)
def testMovingAverageOptimizer(self):
optimizer = sgd_new.SGD(
learning_rate=1,
use_ema=True,
ema_momentum=0.5,
ema_overwrite_frequency=3,
)
# `var2` does not produce gradients.
var1, var2, var3 = tf.Variable(2.0), tf.Variable(2.0), tf.Variable(2.0)
with tf.GradientTape() as tape:
loss = var1 + var3
grads = tape.gradient(loss, [var1, var2, var3])
# First iteration: [var1, var2, var3] = [1.0, 2.0, 1.0]
optimizer.apply_gradients(zip(grads, [var1, var2, var3]))
self.assertAllEqual(
[var1.numpy(), var2.numpy(), var3.numpy()],
[1.0, 2.0, 1.0],
)
# Second iteration: [var1, var2, var3] = [0.0, 2.0, 0.0]
optimizer.apply_gradients(zip(grads, [var1, var2, var3]))
self.assertAllEqual(
[var1.numpy(), var2.numpy(), var3.numpy()],
[0.0, 2.0, 0.0],
)
# Third iteration, without EMA, we should see [var1, var2, var3] =
# [-1.0, 2.0 -1.0], but overwriting results in [var1, var2] =
# [-0.125, 2.0, -0.125].
optimizer.apply_gradients(zip(grads, [var1, var2, var3]))
self.assertAllEqual(
[var1.numpy(), var2.numpy(), var3.numpy()],
[-0.125, 2.0, -0.125],
)
def testGetAndFromConfig(self):
class CustomLRSchedule(learning_rate_schedule.LearningRateSchedule):
def __init__(self, initial_learning_rate):
self.initial_learning_rate = initial_learning_rate
def __call__(self, step):
step = tf.cast(step, tf.float32)
return self.initial_learning_rate / (step + 1)
def get_config(self):
return {"initial_learning_rate": self.initial_learning_rate}
learning_rate = CustomLRSchedule(0.05)
optimizer = adam_new.Adam(
learning_rate=learning_rate,
beta_1=0.7,
beta_2=0.77,
amsgrad=True,
epsilon=0.001,
clipnorm=0.5,
use_ema=True,
ema_momentum=0.5,
ema_overwrite_frequency=50,
name="custom_adam",
)
config = optimizer.get_config()
expected_config = {
"name": "custom_adam",
"beta_1": 0.7,
"beta_2": 0.77,
"epsilon": 0.001,
"amsgrad": True,
"clipnorm": 0.5,
"global_clipnorm": None,
"clipvalue": None,
"use_ema": True,
"ema_momentum": 0.5,
"ema_overwrite_frequency": 50,
"is_legacy_optimizer": False,
}
expected_learning_rate = {
"class_name": "CustomLRSchedule",
"config": {"initial_learning_rate": 0.05},
"module": None,
"registered_name": "CustomLRSchedule",
}
self.assertDictContainsSubset(expected_config, config)
self.assertDictEqual(expected_learning_rate, config["learning_rate"])
restored_optimizer = adam_new.Adam.from_config(
config, custom_objects={"CustomLRSchedule": CustomLRSchedule}
)
self.assertDictEqual(
restored_optimizer.get_config(), optimizer.get_config()
)
def testCheckpointOptimizer(self):
x = tf.Variable([[1.0, 2.0], [3.0, 4.0]], dtype=tf.float32)
lr_schedule = learning_rate_schedule.ExponentialDecay(
initial_learning_rate=1e-2, decay_steps=10000, decay_rate=0.9
)
optimizer_1 = adam_new.Adam(
learning_rate=lr_schedule, beta_1=0.8, beta_2=0.888
)
grads = tf.convert_to_tensor([[1.0, 2.0], [3.0, 4.0]])
for _ in range(1):
optimizer_1.apply_gradients(zip([grads], [x]))
# Then save the variable and optimizer to a checkpoint.
checkpoint_1 = tf.train.Checkpoint(var=x, optimizer=optimizer_1)
checkpoint_path = checkpoint_1.save(self.get_temp_dir())
# Create a new optimizer and call restore on it (and x)
x2 = tf.Variable([[0.0, 0.0], [0.0, 0.0]], dtype=x.dtype)
optimizer_2 = adam_new.Adam(
learning_rate=lr_schedule, beta_1=0.8, beta_2=0.888
)
checkpoint_2 = tf.train.Checkpoint(var=x2, optimizer=optimizer_2)
checkpoint_2.restore(checkpoint_path)
for _ in range(2):
optimizer_1.apply_gradients(zip([grads], [x]))
optimizer_2.apply_gradients(zip([grads], [x]))
self.assertTrue(
(
self.evaluate(optimizer_1._momentums._storage[0])
== self.evaluate(optimizer_2._momentums._storage[0])
).all()
)
self.assertEqual(
self.evaluate(optimizer_1._iterations),
self.evaluate(optimizer_2._iterations),
)
def testCheckpointOptimizerWithModel(self):
inputs = keras.layers.Input(shape=(1,))
outputs = keras.layers.Dense(1)(inputs)
model = keras.Model(inputs=inputs, outputs=outputs)
optimizer = adamax_new_fn()
x = tf.expand_dims(tf.convert_to_tensor([1, 1, 1, 0, 0, 0]), axis=1)
y = tf.expand_dims(tf.convert_to_tensor([1, 1, 1, 0, 0, 0]), axis=1)
model.compile(loss="mse", optimizer=optimizer)
path = os.path.join(self.get_temp_dir(), "ckpt")
checkpoint_callback = keras.callbacks.ModelCheckpoint(path)
model.fit(x, y, callbacks=[checkpoint_callback])
new_model = keras.Model(inputs=inputs, outputs=outputs)
new_optimizer = adamax_new_fn()
new_model.compile(loss="mse", optimizer=new_optimizer)
new_model.load_weights(path)
self.assertEqual(
new_model.optimizer.iterations.numpy(),
model.optimizer.iterations.numpy(),
)
def testRestoreOldOptimizerCheckpoint(self):
inputs = keras.layers.Input(shape=(1,))
outputs = keras.layers.Dense(1)(inputs)
model = keras.Model(inputs=inputs, outputs=outputs)
optimizer = adam_old.Adam()
x = tf.expand_dims(tf.convert_to_tensor([1, 1, 1, 0, 0, 0]), axis=1)
y = tf.expand_dims(tf.convert_to_tensor([1, 1, 1, 0, 0, 0]), axis=1)
model.compile(loss="mse", optimizer=optimizer)
path = os.path.join(self.get_temp_dir(), "ckpt")
checkpoint_callback = keras.callbacks.ModelCheckpoint(path)
model.fit(x, y, callbacks=[checkpoint_callback])
new_model = keras.Model(inputs=inputs, outputs=outputs)
new_optimizer = adam_new.Adam()
new_model.compile(loss="mse", optimizer=new_optimizer)
with self.assertRaisesRegex(
ValueError, "You are trying to restore a checkpoint.*Adam.*"
):
new_model.load_weights(path)
@parameterized.product(optimizer_fn=OPTIMIZER_FN)
def testSaveAndLoadOptimizerWithModel(self, optimizer_fn):
inputs = keras.layers.Input(shape=(1,))
outputs = keras.layers.Dense(1)(inputs)
model = keras.Model(inputs=inputs, outputs=outputs)
optimizer = optimizer_fn()
optimizer.clipnorm = 0.1
x = tf.expand_dims(tf.convert_to_tensor([1, 1, 1, 0, 0, 0]), axis=1)
y = tf.expand_dims(tf.convert_to_tensor([1, 1, 1, 0, 0, 0]), axis=1)
model.compile(loss="mse", optimizer=optimizer)
model.fit(x, y)
# Save in h5 format.
path = os.path.join(self.get_temp_dir(), "model.h5")
model.save(path)
loaded_model = keras.models.load_model(path)
loaded_model.load_weights(path)
loaded_optimizer = loaded_model.optimizer
self.assertEqual(type(optimizer), type(loaded_optimizer))
self.assertEqual(loaded_optimizer.learning_rate, 0.002)
self.assertEqual(loaded_optimizer.clipnorm, 0.1)
self.assertAllClose(optimizer.variables, loaded_optimizer.variables)
# Save in TF-Keras SavedModel format.
model.fit(x, y)
path = os.path.join(self.get_temp_dir(), "model")
model.save(path)
loaded_model = keras.models.load_model(path)
loaded_model.load_weights(path)
loaded_optimizer = loaded_model.optimizer
self.assertEqual(type(optimizer), type(loaded_optimizer))
self.assertEqual(loaded_optimizer.learning_rate, 0.002)
self.assertEqual(loaded_optimizer.clipnorm, 0.1)
loaded_optimizer.build(loaded_model.trainable_variables)
self.assertAllClose(optimizer.variables, loaded_optimizer.variables)
# Save in `.keras` format.
path = os.path.join(self.get_temp_dir(), "model.keras")
model.save(path)
loaded_model = keras.models.load_model(path)
loaded_model.load_weights(path)
loaded_optimizer = loaded_model.optimizer
self.assertEqual(type(optimizer), type(loaded_optimizer))
self.assertEqual(loaded_optimizer.learning_rate, 0.002)
self.assertEqual(loaded_optimizer.clipnorm, 0.1)
self.assertAllClose(optimizer.variables, loaded_optimizer.variables)
@parameterized.product(optimizer_fn=OPTIMIZER_FN)
def testSparseGradientsWorkAsExpected(self, optimizer_fn):
optimizer_1 = optimizer_fn()
optimizer_2 = optimizer_fn()
x1 = tf.Variable(np.ones([5]), dtype=tf.float64)
x2 = tf.Variable(np.ones([5]), dtype=tf.float64)
grads = tf.convert_to_tensor([0, 1.0, 1.5, 0, 0], dtype=tf.float64)
sparse_grads = tf.IndexedSlices(
tf.convert_to_tensor([1.0, 1.5], dtype=tf.float64),
tf.convert_to_tensor([1, 2]),
dense_shape=tf.convert_to_tensor([len(grads)]),
)
for _ in range(5):
optimizer_1.apply_gradients(zip([grads], [x1]))
optimizer_2.apply_gradients(zip([sparse_grads], [x2]))
self.assertAllClose(x1, x2)
@test_utils.run_v2_only
def test_convert_to_legacy_optimizer(self):
if not tf.executing_eagerly():
# The conversion could only happen in eager mode.
return
optimizer_list = [
"adadelta",
"adagrad",
"adam",
"adamax",
"nadam",
"rmsprop",
"sgd",
"ftrl",
]
# Test conversion does not throw errors.
for name in optimizer_list:
experimental_optimizer = keras.optimizers.get(
name, use_legacy_optimizer=False
)
reference_legacy_optimizer = keras.optimizers.get(
name, use_legacy_optimizer=True
)
converted_legacy_optimizer = (
keras.optimizers.convert_to_legacy_optimizer(
experimental_optimizer
)
)
self.assertEqual(
type(reference_legacy_optimizer),
type(converted_legacy_optimizer),
)
self.assertDictEqual(
reference_legacy_optimizer.get_config(),
converted_legacy_optimizer.get_config(),
)
lr_schedule = learning_rate_schedule.ExponentialDecay(
initial_learning_rate=1e-2, decay_steps=10000, decay_rate=0.9
)
optimizer = adam_new.Adam(learning_rate=lr_schedule)
legacy_optimizer = keras.optimizers.convert_to_legacy_optimizer(
optimizer
)
self.assertDictEqual(
optimizer.get_config()["learning_rate"],
legacy_optimizer.get_config()["learning_rate"],
)
class CustomLRSchedule(learning_rate_schedule.LearningRateSchedule):
def __init__(self, initial_learning_rate):
self.initial_learning_rate = initial_learning_rate
def __call__(self, step):
step = tf.cast(step, tf.float32)
return self.initial_learning_rate / (step + 1)
def get_config(self):
return {"initial_learning_rate": self.initial_learning_rate}
lr_schedule = CustomLRSchedule(0.001)
optimizer = adam_new.Adam(learning_rate=lr_schedule)
legacy_optimizer = keras.optimizers.convert_to_legacy_optimizer(
optimizer
)
self.assertDictEqual(
optimizer.get_config()["learning_rate"],
legacy_optimizer.get_config()["learning_rate"],
)
@test_utils.run_v2_only
def test_arm_mac_get_legacy_optimizer(self):
with mock.patch(
"platform.system",
mock.MagicMock(return_value="Darwin"),
):
with mock.patch(
"platform.processor",
mock.MagicMock(return_value="arm"),
):
optimizer = keras.optimizers.get("adam")
self.assertIsInstance(optimizer, adam_old.Adam)
class OptimizerRegressionTest(tf.test.TestCase, parameterized.TestCase):
"""Test optimizer outputs the same numerical results as optimizer_v2."""
def _compare_numerical(self, old_optimizer, new_optimizer):
x1 = tf.Variable(np.ones([10]), dtype=tf.float64)
x2 = tf.Variable(np.ones([10]), dtype=tf.float64)
grads = tf.convert_to_tensor(np.arange(0.1, 1.1, 0.1))
first_grads = tf.constant([0.01] * 10, dtype=tf.float64)
sparse_grads = tf.IndexedSlices(
tf.convert_to_tensor([0, 0.2, 0.4, 0.8, 0.8], dtype=tf.float64),
tf.convert_to_tensor([0, 2, 4, 6, 6]),
dense_shape=tf.convert_to_tensor([len(grads)]),
)
old_optimizer.apply_gradients(zip([first_grads], [x1]))
new_optimizer.apply_gradients(zip([first_grads], [x2]))
for _ in range(5):
self.assertAllClose(x1, x2, rtol=5e-4, atol=5e-4)
old_optimizer.apply_gradients(zip([grads], [x1]))
new_optimizer.apply_gradients(zip([grads], [x2]))
for _ in range(5):
self.assertAllClose(x1, x2, rtol=5e-4, atol=5e-4)
old_optimizer.apply_gradients(zip([sparse_grads], [x1]))
new_optimizer.apply_gradients(zip([sparse_grads], [x2]))
def testAdam(self):
self._compare_numerical(
adam_old.Adam(amsgrad=True), adam_new.Adam(amsgrad=True)
)
def testAdadelta(self):
self._compare_numerical(
adadelta_old.Adadelta(), adadelta_new.Adadelta()
)
def testAdagrad(self):
self._compare_numerical(adagrad_old.Adagrad(), adagrad_new.Adagrad())
def testFtrl(self):
self._compare_numerical(ftrl_old.Ftrl(), ftrl_new.Ftrl())
def testRMSprop(self):
self._compare_numerical(
rmsprop_old.RMSprop(centered=True),
rmsprop_new.RMSprop(centered=True),
)
@parameterized.product(nesterov=[True, False])
def testSgd(self, nesterov):
self._compare_numerical(
sgd_old.SGD(nesterov=nesterov), sgd_new.SGD(nesterov=nesterov)
)
def testWeightDecay(self):
self._compare_numerical(
adam_new.Adam(learning_rate=1, weight_decay=0.5, epsilon=0),
adamw_new.AdamW(learning_rate=1, weight_decay=0.5, epsilon=0),
)
class DistributedTrainingTest(tf.test.TestCase, parameterized.TestCase):
@ds_combinations.generate(
tf.__internal__.test.combinations.combine(
strategy=STRATEGIES, optimizer_fn=OPTIMIZER_FN
)
)
def testGetGradientsInModel(self, strategy, optimizer_fn):
with strategy.scope():
model = keras.Sequential(
[keras.layers.Input(shape=(1,)), keras.layers.Dense(1)]
)
optimizer = optimizer_fn()
x = tf.expand_dims(tf.convert_to_tensor([1, 1, 1, 0, 0, 0]), axis=1)
y = tf.expand_dims(tf.convert_to_tensor([1, 1, 1, 0, 0, 0]), axis=1)
model.compile(loss="mse", optimizer=optimizer)
model.fit(x, y, epochs=1, steps_per_epoch=5)
if optimizer.name == "Adam":
# Assert the momentum variable is not 0.
self.assertNotEqual(
self.evaluate(optimizer._momentums._storage[0]), 0
)
elif optimizer.name == "Adadelta":
# Assert the accumulated variable is not 0.
self.assertNotEqual(
self.evaluate(optimizer._accumulated_grads._storage[0]), 0
)
elif optimizer.name == "Adagrad":
# Assert the accumulated variable is not 0.
self.assertNotEqual(
self.evaluate(optimizer._accumulators._storage[0]), 0
)
@ds_combinations.generate(
tf.__internal__.test.combinations.combine(
strategy=STRATEGIES, optimizer_fn=OPTIMIZER_FN
)
)
def testGetGradientsInCustomTrainingLoop(self, strategy, optimizer_fn):
with strategy.scope():
model = keras.Sequential(
[keras.layers.Input(shape=(1,)), keras.layers.Dense(1)]
)
optimizer = optimizer_fn()
def per_worker_dataset_fn():
def dataset_fn(_):
x, y = [1, 1, 1, 0, 0, 0], [1, 1, 1, 0, 0, 0]
ds = tf.data.Dataset.from_tensor_slices((x, y))
ds = ds.repeat().batch(6)
return ds
return strategy.distribute_datasets_from_function(dataset_fn)
ds = per_worker_dataset_fn()
@tf.function
def train_step(ds):
def replica_fn(data):
features, labels = data
with tf.GradientTape() as tape:
output = model(tf.expand_dims(features, axis=1))
loss = keras.losses.MeanSquaredError(
reduction=losses_utils.ReductionV2.NONE
)(labels, output)
grads = tape.gradient(loss, model.trainable_variables)
optimizer.apply_gradients(
zip(grads, model.trainable_variables)
)
strategy.run(replica_fn, args=(next(iter(ds)),))
for _ in range(3):
train_step(ds)
self.assertEqual(self.evaluate(optimizer.iterations), 3)
@ds_combinations.generate(
tf.__internal__.test.combinations.combine(
strategy=[
ds_combinations.mirrored_strategy_with_two_gpus,
ds_combinations.tpu_strategy,
ds_combinations.multi_worker_mirrored_2x2_gpu,
ds_combinations.central_storage_strategy_with_two_gpus,
]
)
)
def testJitCompile(self, strategy):
# Test the optimizer yields same numerical results when jit_compile is
# on and off.
with strategy.scope():
optimizer_1 = adam_new.Adam(
jit_compile=False, use_ema=True, ema_overwrite_frequency=1
)
optimizer_2 = adam_new.Adam(
jit_compile=True, use_ema=True, ema_overwrite_frequency=1
)
model_1 = keras.Sequential(
[
keras.layers.Input(shape=(2,)),
keras.layers.Dense(5),
keras.layers.Dense(1),
]
)
model_2 = keras.models.clone_model(model_1)
model_2.set_weights(model_1.get_weights())
def per_worker_dataset_fn():
def dataset_fn(_):
x = np.random.rand(6, 2)
y = [1, 1, 1, 0, 0, 0]
ds = tf.data.Dataset.from_tensor_slices((x, y))
ds = ds.repeat().batch(6)
return ds
return strategy.distribute_datasets_from_function(dataset_fn)
ds = per_worker_dataset_fn()
@tf.function
def train_step(ds):
def replica_fn(data):
features, labels = data
with tf.GradientTape() as tape:
output_1 = model_1(features)
loss_1 = keras.losses.MeanSquaredError(
reduction=losses_utils.ReductionV2.NONE
)(labels, output_1)
grads_1 = tape.gradient(loss_1, model_1.trainable_variables)
optimizer_1.apply_gradients(
zip(grads_1, model_1.trainable_variables),
skip_gradients_aggregation=False,
)
with tf.GradientTape() as tape:
output_2 = model_2(features)
loss_2 = keras.losses.MeanSquaredError(
reduction=losses_utils.ReductionV2.NONE
)(labels, output_2)
grads_2 = tape.gradient(loss_2, model_2.trainable_variables)
optimizer_2.apply_gradients(
zip(grads_2, model_2.trainable_variables),
experimental_aggregate_gradients=True,
)
strategy.run(replica_fn, args=(next(iter(ds)),))
for _ in range(3):
train_step(ds)
self.assertAllClose(
model_1.trainable_variables[0][0],
model_2.trainable_variables[0][0],
)
if __name__ == "__main__":
tf.__internal__.distribute.multi_process_runner.test_main()
|
tf-keras/tf_keras/optimizers/optimizer_test.py/0
|
{
"file_path": "tf-keras/tf_keras/optimizers/optimizer_test.py",
"repo_id": "tf-keras",
"token_count": 17164
}
| 218 |
# Description:
# Contains the TF-Keras preprocessing layers (internal TensorFlow version).
# Placeholder: load unaliased py_library
load("@org_keras//tf_keras:tf_keras.bzl", "tf_py_test")
package(
# copybara:uncomment default_applicable_licenses = ["//tf_keras:license"],
default_visibility = [
"//tf_keras:friends",
],
licenses = ["notice"],
)
py_library(
name = "preprocessing",
srcs = [
"__init__.py",
],
srcs_version = "PY3",
deps = [
":image",
":sequence",
":text",
"//tf_keras/utils",
],
)
py_library(
name = "image",
srcs = [
"image.py",
],
srcs_version = "PY3",
deps = [
"//:expect_numpy_installed",
"//:expect_pillow_installed",
"//:expect_scipy_installed",
"//:expect_tensorflow_installed",
"//tf_keras:backend",
"//tf_keras/utils:data_utils",
"//tf_keras/utils:image_utils",
],
)
py_library(
name = "sequence",
srcs = [
"sequence.py",
],
srcs_version = "PY3",
deps = [
"//:expect_tensorflow_installed",
"//tf_keras/utils:data_utils",
],
)
py_library(
name = "text",
srcs = [
"text.py",
],
srcs_version = "PY3",
deps = ["//:expect_tensorflow_installed"],
)
tf_py_test(
name = "image_test",
size = "medium",
srcs = ["image_test.py"],
python_version = "PY3",
tags = [
"no_oss", # TODO(scottzhu): Fix for multiple export issue.
],
deps = [
":image",
"//:expect_numpy_installed",
"//:expect_pandas_installed",
"//:expect_tensorflow_installed",
"//tf_keras",
"//tf_keras/testing_infra:test_combinations",
],
)
tf_py_test(
name = "sequence_test",
size = "small",
srcs = ["sequence_test.py"],
python_version = "PY3",
deps = [
":sequence",
"//:expect_numpy_installed",
"//:expect_tensorflow_installed",
"//tf_keras/testing_infra:test_combinations",
],
)
tf_py_test(
name = "text_test",
size = "small",
srcs = ["text_test.py"],
python_version = "PY3",
deps = [
":text",
"//:expect_numpy_installed",
"//:expect_tensorflow_installed",
"//tf_keras/testing_infra:test_combinations",
],
)
|
tf-keras/tf_keras/preprocessing/BUILD/0
|
{
"file_path": "tf-keras/tf_keras/preprocessing/BUILD",
"repo_id": "tf-keras",
"token_count": 1177
}
| 219 |
# Copyright 2020 The TensorFlow Authors. All Rights Reserved.
#
# Licensed under the Apache License, Version 2.0 (the "License");
# you may not use this file except in compliance with the License.
# You may obtain a copy of the License at
#
# http://www.apache.org/licenses/LICENSE-2.0
#
# Unless required by applicable law or agreed to in writing, software
# distributed under the License is distributed on an "AS IS" BASIS,
# WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.
# See the License for the specific language governing permissions and
# limitations under the License.
# ==============================================================================
"""Tests the JSON encoder and decoder."""
import enum
import tensorflow.compat.v2 as tf
from tf_keras.saving.legacy.saved_model import json_utils
from tf_keras.testing_infra import test_combinations
from tf_keras.testing_infra import test_utils
class JsonUtilsTest(test_combinations.TestCase):
def test_encode_decode_tensor_shape(self):
metadata = {
"key1": tf.TensorShape(None),
"key2": [tf.TensorShape([None]), tf.TensorShape([3, None, 5])],
}
string = json_utils.Encoder().encode(metadata)
loaded = json_utils.decode(string)
self.assertEqual(set(loaded.keys()), {"key1", "key2"})
self.assertAllEqual(loaded["key1"].rank, None)
self.assertAllEqual(loaded["key2"][0].as_list(), [None])
self.assertAllEqual(loaded["key2"][1].as_list(), [3, None, 5])
def test_encode_decode_tuple(self):
metadata = {"key1": (3, 5), "key2": [(1, (3, 4)), (1,)]}
string = json_utils.Encoder().encode(metadata)
loaded = json_utils.decode(string)
self.assertEqual(set(loaded.keys()), {"key1", "key2"})
self.assertAllEqual(loaded["key1"], (3, 5))
self.assertAllEqual(loaded["key2"], [(1, (3, 4)), (1,)])
def test_encode_decode_type_spec(self):
spec = tf.TensorSpec((1, 5), tf.float32)
string = json_utils.Encoder().encode(spec)
loaded = json_utils.decode(string)
self.assertEqual(spec, loaded)
invalid_type_spec = {
"class_name": "TypeSpec",
"type_spec": "Invalid Type",
"serialized": None,
}
string = json_utils.Encoder().encode(invalid_type_spec)
with self.assertRaisesRegexp(
ValueError, "No TypeSpec has been registered"
):
loaded = json_utils.decode(string)
def test_encode_decode_enum(self):
class Enum(enum.Enum):
CLASS_A = "a"
CLASS_B = "b"
config = {"key": Enum.CLASS_A, "key2": Enum.CLASS_B}
string = json_utils.Encoder().encode(config)
loaded = json_utils.decode(string)
self.assertAllEqual({"key": "a", "key2": "b"}, loaded)
@test_utils.run_v2_only
def test_encode_decode_ragged_tensor(self):
x = tf.ragged.constant([[1.0, 2.0], [3.0]])
string = json_utils.Encoder().encode(x)
loaded = json_utils.decode(string)
self.assertAllEqual(loaded, x)
@test_utils.run_v2_only
def test_encode_decode_extension_type_tensor(self):
class MaskedTensor(tf.experimental.ExtensionType):
__name__ = "MaskedTensor"
values: tf.Tensor
mask: tf.Tensor
x = MaskedTensor(
values=[[1, 2, 3], [4, 5, 6]],
mask=[[True, True, False], [True, False, True]],
)
string = json_utils.Encoder().encode(x)
loaded = json_utils.decode(string)
self.assertAllEqual(loaded, x)
def test_encode_decode_bytes(self):
b_string = b"abc"
json_string = json_utils.Encoder().encode(b_string)
loaded = json_utils.decode(json_string)
self.assertAllEqual(b_string, loaded)
if __name__ == "__main__":
tf.test.main()
|
tf-keras/tf_keras/saving/legacy/saved_model/json_utils_test.py/0
|
{
"file_path": "tf-keras/tf_keras/saving/legacy/saved_model/json_utils_test.py",
"repo_id": "tf-keras",
"token_count": 1680
}
| 220 |
# Copyright 2015 The TensorFlow Authors. All Rights Reserved.
#
# Licensed under the Apache License, Version 2.0 (the "License");
# you may not use this file except in compliance with the License.
# You may obtain a copy of the License at
#
# http://www.apache.org/licenses/LICENSE-2.0
#
# Unless required by applicable law or agreed to in writing, software
# distributed under the License is distributed on an "AS IS" BASIS,
# WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.
# See the License for the specific language governing permissions and
# limitations under the License.
# ==============================================================================
"""Python utilities required by TF-Keras."""
import inspect
import threading
# isort: off
from tensorflow.python.util.tf_export import keras_export
_GLOBAL_CUSTOM_OBJECTS = {}
_GLOBAL_CUSTOM_NAMES = {}
# Thread-local custom objects set by custom_object_scope.
_THREAD_LOCAL_CUSTOM_OBJECTS = threading.local()
@keras_export(
"keras.saving.custom_object_scope",
"keras.utils.custom_object_scope",
"keras.utils.CustomObjectScope",
)
class CustomObjectScope:
"""Exposes custom classes/functions to TF-Keras deserialization internals.
Under a scope `with custom_object_scope(objects_dict)`, TF-Keras methods
such as `tf.keras.models.load_model` or `tf.keras.models.model_from_config`
will be able to deserialize any custom object referenced by a
saved config (e.g. a custom layer or metric).
Example:
Consider a custom regularizer `my_regularizer`:
```python
layer = Dense(3, kernel_regularizer=my_regularizer)
# Config contains a reference to `my_regularizer`
config = layer.get_config()
...
# Later:
with custom_object_scope({'my_regularizer': my_regularizer}):
layer = Dense.from_config(config)
```
Args:
*args: Dictionary or dictionaries of `{name: object}` pairs.
"""
def __init__(self, *args):
self.custom_objects = args
self.backup = None
def __enter__(self):
self.backup = _THREAD_LOCAL_CUSTOM_OBJECTS.__dict__.copy()
for objects in self.custom_objects:
_THREAD_LOCAL_CUSTOM_OBJECTS.__dict__.update(objects)
return self
def __exit__(self, *args, **kwargs):
_THREAD_LOCAL_CUSTOM_OBJECTS.__dict__.clear()
_THREAD_LOCAL_CUSTOM_OBJECTS.__dict__.update(self.backup)
@keras_export(
"keras.saving.get_custom_objects", "keras.utils.get_custom_objects"
)
def get_custom_objects():
"""Retrieves a live reference to the global dictionary of custom objects.
Custom objects set using using `custom_object_scope` are not added to the
global dictionary of custom objects, and will not appear in the returned
dictionary.
Example:
```python
get_custom_objects().clear()
get_custom_objects()['MyObject'] = MyObject
```
Returns:
Global dictionary mapping registered class names to classes.
"""
return _GLOBAL_CUSTOM_OBJECTS
@keras_export(
"keras.saving.register_keras_serializable",
"keras.utils.register_keras_serializable",
)
def register_keras_serializable(package="Custom", name=None):
"""Registers an object with the TF-Keras serialization framework.
This decorator injects the decorated class or function into the TF-Keras
custom object dictionary, so that it can be serialized and deserialized
without needing an entry in the user-provided custom object dict. It also
injects a function that TF-Keras will call to get the object's serializable
string key.
Note that to be serialized and deserialized, classes must implement the
`get_config()` method. Functions do not have this requirement.
The object will be registered under the key 'package>name' where `name`,
defaults to the object name if not passed.
Example:
```python
# Note that `'my_package'` is used as the `package` argument here, and since
# the `name` argument is not provided, `'MyDense'` is used as the `name`.
@keras.saving.register_keras_serializable('my_package')
class MyDense(keras.layers.Dense):
pass
assert keras.saving.get_registered_object('my_package>MyDense') == MyDense
assert keras.saving.get_registered_name(MyDense) == 'my_package>MyDense'
```
Args:
package: The package that this class belongs to. This is used for the
`key` (which is `"package>name"`) to idenfify the class. Note that this
is the first argument passed into the decorator.
name: The name to serialize this class under in this package. If not
provided or `None`, the class' name will be used (note that this is the
case when the decorator is used with only one argument, which becomes
the `package`).
Returns:
A decorator that registers the decorated class with the passed names.
"""
def decorator(arg):
"""Registers a class with the TF-Keras serialization framework."""
class_name = name if name is not None else arg.__name__
registered_name = package + ">" + class_name
if inspect.isclass(arg) and not hasattr(arg, "get_config"):
raise ValueError(
"Cannot register a class that does not have a "
"get_config() method."
)
_GLOBAL_CUSTOM_OBJECTS[registered_name] = arg
_GLOBAL_CUSTOM_NAMES[arg] = registered_name
return arg
return decorator
@keras_export(
"keras.saving.get_registered_name", "keras.utils.get_registered_name"
)
def get_registered_name(obj):
"""Returns the name registered to an object within the TF-Keras framework.
This function is part of the TF-Keras serialization and deserialization
framework. It maps objects to the string names associated with those objects
for serialization/deserialization.
Args:
obj: The object to look up.
Returns:
The name associated with the object, or the default Python name if the
object is not registered.
"""
if obj in _GLOBAL_CUSTOM_NAMES:
return _GLOBAL_CUSTOM_NAMES[obj]
else:
return obj.__name__
@keras_export(
"keras.saving.get_registered_object", "keras.utils.get_registered_object"
)
def get_registered_object(name, custom_objects=None, module_objects=None):
"""Returns the class associated with `name` if it is registered with
TF-Keras.
This function is part of the TF-Keras serialization and deserialization
framework. It maps strings to the objects associated with them for
serialization/deserialization.
Example:
```python
def from_config(cls, config, custom_objects=None):
if 'my_custom_object_name' in config:
config['hidden_cls'] = tf.keras.saving.get_registered_object(
config['my_custom_object_name'], custom_objects=custom_objects)
```
Args:
name: The name to look up.
custom_objects: A dictionary of custom objects to look the name up in.
Generally, custom_objects is provided by the user.
module_objects: A dictionary of custom objects to look the name up in.
Generally, module_objects is provided by midlevel library implementers.
Returns:
An instantiable class associated with `name`, or `None` if no such class
exists.
"""
if name in _THREAD_LOCAL_CUSTOM_OBJECTS.__dict__:
return _THREAD_LOCAL_CUSTOM_OBJECTS.__dict__[name]
elif name in _GLOBAL_CUSTOM_OBJECTS:
return _GLOBAL_CUSTOM_OBJECTS[name]
elif custom_objects and name in custom_objects:
return custom_objects[name]
elif module_objects and name in module_objects:
return module_objects[name]
return None
# Aliases
custom_object_scope = CustomObjectScope
|
tf-keras/tf_keras/saving/object_registration.py/0
|
{
"file_path": "tf-keras/tf_keras/saving/object_registration.py",
"repo_id": "tf-keras",
"token_count": 2776
}
| 221 |
# Description:
# Contains TF-Keras test utils and integration tests.
# Placeholder: load unaliased py_library
# Placeholder: load unaliased py_test
# buildifier: disable=same-origin-load
load("@org_keras//tf_keras:tf_keras.bzl", "cuda_py_test")
# buildifier: disable=same-origin-load
load("@org_keras//tf_keras:tf_keras.bzl", "tf_py_test")
load("@org_keras//tf_keras:tf_keras.bzl", "tpu_py_test")
package(
# copybara:uncomment default_applicable_licenses = ["//tf_keras:license"],
default_visibility = [
"//tf_keras:friends",
"//third_party/tensorflow/tools/pip_package:__pkg__",
],
licenses = ["notice"],
)
tf_py_test(
name = "get_config_test",
srcs = ["get_config_test.py"],
python_version = "PY3",
shard_count = 4,
tags = [
"no_pip",
],
deps = [
":get_config_samples",
"//:expect_absl_installed", # absl/testing:parameterized
"//:expect_tensorflow_installed",
"//tf_keras",
"//tf_keras/api:tf_keras_api",
"//tf_keras/testing_infra:test_combinations",
],
)
tf_py_test(
name = "add_loss_correctness_test",
srcs = ["add_loss_correctness_test.py"],
python_version = "PY3",
shard_count = 4,
deps = [
"//:expect_absl_installed", # absl/testing:parameterized
"//:expect_numpy_installed",
"//:expect_tensorflow_installed",
"//tf_keras",
"//tf_keras/api:tf_keras_api",
"//tf_keras/testing_infra:test_combinations",
],
)
tpu_py_test(
name = "automatic_outside_compilation_test",
srcs = [
"automatic_outside_compilation_test.py",
],
disable_experimental = True,
disable_mlir_bridge = False,
python_version = "PY3",
tags = ["no_oss"],
deps = [
"//:expect_tensorboard_installed",
"//:expect_tensorflow_installed",
"//tf_keras",
"//tf_keras/api:tf_keras_api",
"//tf_keras/distribute:distribute_strategy_test_lib",
],
)
tf_py_test(
name = "convert_to_constants_test",
srcs = ["convert_to_constants_test.py"],
python_version = "PY3",
shard_count = 4,
deps = [
"//:expect_tensorflow_installed",
"//tf_keras",
"//tf_keras/api:tf_keras_api",
"//tf_keras/testing_infra:test_utils",
],
)
tf_py_test(
name = "custom_training_loop_test",
srcs = ["custom_training_loop_test.py"],
python_version = "PY3",
shard_count = 4,
tags = ["notsan"],
deps = [
"//:expect_absl_installed", # absl/testing:parameterized
"//:expect_numpy_installed",
"//:expect_tensorflow_installed",
"//tf_keras",
"//tf_keras/api:tf_keras_api",
"//tf_keras/testing_infra:test_combinations",
],
)
tf_py_test(
name = "graph_util_test",
srcs = ["graph_util_test.py"],
python_version = "PY3",
deps = [
"//:expect_tensorflow_installed",
"//tf_keras",
"//tf_keras/api:tf_keras_api",
],
)
tf_py_test(
name = "integration_test",
size = "medium",
srcs = ["integration_test.py"],
python_version = "PY3",
shard_count = 16,
tags = [
"notsan",
],
deps = [
"//:expect_absl_installed", # absl/testing:parameterized
"//:expect_numpy_installed",
"//:expect_tensorflow_installed",
"//tf_keras",
"//tf_keras/api:tf_keras_api",
"//tf_keras/layers/rnn:legacy_cells",
"//tf_keras/legacy_tf_layers:layers_base",
"//tf_keras/testing_infra:test_combinations",
],
)
py_library(
name = "model_architectures",
srcs = [
"model_architectures.py",
],
srcs_version = "PY3",
deps = [
"//tf_keras",
],
)
tf_py_test(
name = "model_architectures_test",
srcs = ["model_architectures_test.py"],
python_version = "PY3",
shard_count = 16,
deps = [
":model_architectures",
"//:expect_absl_installed", # absl/testing:parameterized
"//:expect_numpy_installed",
"//tf_keras",
"//tf_keras/api:tf_keras_api",
"//tf_keras/testing_infra:test_combinations",
],
)
py_library(
name = "model_subclassing_test_util",
srcs = ["model_subclassing_test_util.py"],
srcs_version = "PY3",
deps = [
"//tf_keras",
"//tf_keras/testing_infra:test_utils",
],
)
tf_py_test(
name = "model_subclassing_test",
srcs = ["model_subclassing_test.py"],
python_version = "PY3",
shard_count = 4,
tags = [
"no_windows",
"notsan",
],
deps = [
":model_subclassing_test_util",
"//:expect_absl_installed", # absl/testing:parameterized
"//:expect_numpy_installed",
"//:expect_tensorflow_installed",
"//tf_keras",
"//tf_keras/api:tf_keras_api",
"//tf_keras/testing_infra:test_combinations",
],
)
tf_py_test(
name = "model_subclassing_compiled_test",
srcs = ["model_subclassing_compiled_test.py"],
python_version = "PY3",
shard_count = 4,
tags = [
"no_windows",
"notsan",
],
deps = [
":model_subclassing_test_util",
"//:expect_absl_installed", # absl/testing:parameterized
"//:expect_numpy_installed",
"//:expect_tensorflow_installed",
"//tf_keras",
"//tf_keras/api:tf_keras_api",
"//tf_keras/testing_infra:test_combinations",
],
)
cuda_py_test(
name = "memory_test",
size = "medium",
srcs = ["memory_test.py"],
tags = [
"no_oss",
"optonly", # The test is too slow in non-opt mode
],
# TODO(b/140065350): Re-enable
xla_enable_strict_auto_jit = False,
deps = [
"//:expect_tensorflow_installed",
"//tf_keras",
"//tf_keras/api:tf_keras_api",
"//third_party/tensorflow/python/eager/memory_tests:memory_test_util",
],
)
tf_py_test(
name = "memory_checker_test",
size = "medium",
srcs = ["memory_checker_test.py"],
python_version = "PY3",
shard_count = 8,
tags = [
"no_oss",
"no_pip",
"no_windows",
"noasan", # TODO(b/149948895): Re-enable.
"nomsan", # TODO(b/149948895): Re-enable.
"notsan", # TODO(b/149948895): Re-enable.
],
deps = [
"//:expect_tensorflow_installed",
"//tf_keras/api:tf_keras_api",
],
)
tf_py_test(
name = "saved_model_test",
size = "small",
srcs = ["saved_model_test.py"],
tags = [
"no_oss", # TODO(b/170766453)
"notap", # TODO(b/170766453)
],
deps = [
"//:expect_tensorflow_installed",
"//tf_keras",
"//tf_keras/api:tf_keras_api",
"//tf_keras/layers/core",
"//tf_keras/metrics",
"//tf_keras/optimizers/legacy:optimizers",
],
)
cuda_py_test(
name = "saver_test",
size = "medium",
srcs = ["saver_test.py"],
python_version = "PY3",
deps = [
"//:expect_tensorflow_installed",
"//tf_keras/api:tf_keras_api",
"//tf_keras/engine",
"//tf_keras/layers/core",
],
)
tf_py_test(
name = "serialization_util_test",
size = "small",
srcs = ["serialization_util_test.py"],
python_version = "PY3",
deps = [
"//:expect_tensorflow_installed",
"//tf_keras",
"//tf_keras/api:tf_keras_api",
"//tf_keras/engine",
"//tf_keras/layers/core",
"//tf_keras/testing_infra:test_combinations",
],
)
tf_py_test(
name = "temporal_sample_weights_correctness_test",
srcs = ["temporal_sample_weights_correctness_test.py"],
python_version = "PY3",
shard_count = 20,
deps = [
"//:expect_numpy_installed",
"//:expect_tensorflow_installed",
"//tf_keras",
"//tf_keras/api:tf_keras_api",
"//tf_keras/testing_infra:test_combinations",
],
)
tf_py_test(
name = "tracking_test",
srcs = ["tracking_test.py"],
python_version = "PY3",
tags = [
"no_windows",
"nomac",
],
deps = [
"//:expect_absl_installed", # absl/testing:parameterized
"//:expect_tensorflow_installed",
"//tf_keras",
"//tf_keras/api:tf_keras_api",
"//tf_keras/engine",
"//tf_keras/layers/core",
"//tf_keras/layers/normalization",
"//tf_keras/testing_infra:test_combinations",
],
)
tf_py_test(
name = "tracking_util_test",
srcs = ["tracking_util_test.py"],
python_version = "PY3",
tags = ["notsan"], # b/74395663
deps = [
"//:expect_absl_installed", # absl/testing:parameterized
"//:expect_tensorflow_installed",
"//tf_keras",
"//tf_keras/api:tf_keras_api",
"//tf_keras/engine",
"//tf_keras/layers/core",
"//tf_keras/optimizers/legacy:optimizers",
"//tf_keras/testing_infra:test_combinations",
],
)
tf_py_test(
name = "tracking_util_with_v1_optimizers_test",
srcs = ["tracking_util_with_v1_optimizers_test.py"],
tags = [
"no_windows", # TODO(b/184424727): Re-enable this.
"notsan", # b/74395663
],
deps = [
"//:expect_tensorflow_installed",
"//tf_keras",
"//tf_keras/api:tf_keras_api",
"//tf_keras/engine",
"//tf_keras/layers/core",
"//tf_keras/testing_infra:test_combinations",
],
)
py_library(
name = "get_config_samples",
srcs = ["get_config_samples.py"],
srcs_version = "PY3",
deps = [],
)
py_test(
name = "keras_doctest",
srcs = ["keras_doctest.py"],
python_version = "PY3",
tags = [
"no_pip",
"noasan",
"nomsan",
"notsan",
],
deps = [
"//:expect_tensorflow_installed",
"//tf_keras/testing_infra:keras_doctest_lib",
],
)
|
tf-keras/tf_keras/tests/BUILD/0
|
{
"file_path": "tf-keras/tf_keras/tests/BUILD",
"repo_id": "tf-keras",
"token_count": 5008
}
| 222 |
# Copyright 2018 The TensorFlow Authors. All Rights Reserved.
#
# Licensed under the Apache License, Version 2.0 (the "License");
# you may not use this file except in compliance with the License.
# You may obtain a copy of the License at
#
# http://www.apache.org/licenses/LICENSE-2.0
#
# Unless required by applicable law or agreed to in writing, software
# distributed under the License is distributed on an "AS IS" BASIS,
# WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.
# See the License for the specific language governing permissions and
# limitations under the License.
# ==============================================================================
"""Tests for Model subclassing."""
import copy
import os
import numpy as np
import tensorflow.compat.v2 as tf
from absl.testing import parameterized
import tf_keras as keras
from tf_keras.testing_infra import test_combinations
from tf_keras.testing_infra import test_utils
from tf_keras.tests import model_subclassing_test_util as model_util
# isort: off
from tensorflow.python.framework import (
test_util as tf_test_utils,
)
from tensorflow.python.trackable import data_structures
try:
import h5py
except ImportError:
h5py = None
@test_combinations.run_all_keras_modes
class ModelSubclassingTest(test_combinations.TestCase):
def test_custom_build(self):
class DummyModel(keras.Model):
def __init__(self):
super().__init__()
self.dense1 = keras.layers.Dense(32, activation="relu")
self.uses_custom_build = False
def call(self, inputs):
return self.dense1(inputs)
def build(self, input_shape):
self.uses_custom_build = True
test_model = DummyModel()
dummy_data = tf.ones((32, 50))
test_model(dummy_data)
self.assertTrue(
test_model.uses_custom_build,
"Model should use user defined build when called.",
)
def test_attribute_conflict_error(self):
class ModelWithProperty(keras.Model):
@property
def read_only(self):
return 1.0
m = ModelWithProperty()
with self.assertRaisesRegex(AttributeError, "read_only"):
m.read_only = 2.0
def test_custom_build_with_fit(self):
class DummyModel(keras.Model):
def __init__(self):
super().__init__()
self.layer1 = keras.layers.Dense(10, activation="relu")
def build(self, input_shape):
self.layer2 = keras.layers.Dense(1, activation="relu")
def call(self, inputs):
return self.layer2(self.layer1(inputs))
model = DummyModel()
model.compile("sgd", "mse", run_eagerly=test_utils.should_run_eagerly())
model.fit(np.ones((10, 10)), np.ones((10, 1)), batch_size=2, epochs=2)
self.assertLen(model.layers, 2)
self.assertLen(model.trainable_variables, 4)
def test_dataset_dict_with_fit(self):
class MyModel(keras.Model):
def __init__(self):
super().__init__()
self.dense1 = keras.layers.Dense(1)
self.dense2 = keras.layers.Dense(1)
self.add = keras.layers.Add()
def call(self, x):
return self.add([self.dense1(x["a"]), self.dense2(x["b"])])
model = MyModel()
model.compile("sgd", "mse", run_eagerly=test_utils.should_run_eagerly())
data = tf.data.Dataset.from_tensor_slices(
({"a": np.ones((32, 10)), "b": np.ones((32, 20))}, np.ones((32, 1)))
).batch(2)
model.fit(data, epochs=2)
def test_invalid_input_shape_build(self):
num_classes = 2
input_dim = 50
model = test_utils.SmallSubclassMLP(
num_hidden=32, num_classes=num_classes, use_dp=True, use_bn=True
)
self.assertFalse(model.built, "Model should not have been built")
self.assertFalse(
model.weights,
"Model should have no weights since it has not been built.",
)
with self.assertRaisesRegex(
ValueError, "input shape is not one of the valid types"
):
model.build(input_shape=tf.compat.v1.Dimension(input_dim))
def test_embed_dtype_with_subclass_build(self):
class Embedding(keras.layers.Layer):
"""An Embedding layer."""
def __init__(self, vocab_size, embedding_dim, **kwargs):
super().__init__(**kwargs)
self.vocab_size = vocab_size
self.embedding_dim = embedding_dim
def build(self, _):
self.embedding = self.add_weight(
"embedding_kernel",
shape=[self.vocab_size, self.embedding_dim],
dtype=np.float32,
initializer=tf.compat.v1.random_uniform_initializer(
-0.1, 0.1
),
trainable=True,
)
def call(self, x):
return tf.compat.v1.nn.embedding_lookup(self.embedding, x)
class EmbedModel(keras.Model):
def __init__(self, vocab_size, embed_size):
super().__init__()
self.embed1 = Embedding(vocab_size, embed_size)
def call(self, inputs):
return self.embed1(inputs)
model = EmbedModel(100, 20)
self.assertFalse(model.built, "Model should not have been built")
self.assertFalse(
model.weights,
"Model should have no weights since it has not been built.",
)
with self.assertRaisesRegex(
ValueError, "if your layers do not support float type inputs"
):
model.build(input_shape=(35, 20))
def test_single_time_step_rnn_build(self):
dim = 4
timesteps = 1
batch_input_shape = (None, timesteps, dim)
units = 3
class SimpleRNNModel(keras.Model):
def __init__(self):
super().__init__()
self.lstm = keras.layers.LSTM(units)
def call(self, inputs):
return self.lstm(inputs)
model = SimpleRNNModel()
self.assertFalse(model.built, "Model should not have been built")
self.assertFalse(
model.weights,
"Model should have no weights since it has not been built.",
)
model.build(batch_input_shape)
self.assertTrue(
model.weights,
"Model should have weights now that it has been properly built.",
)
self.assertTrue(
model.built, "Model should be built after calling `build`."
)
model(tf.ones((32, timesteps, dim)))
def test_single_io_subclass_build(self):
num_classes = 2
input_dim = 50
batch_size = None
model = test_utils.SmallSubclassMLP(
num_hidden=32, num_classes=num_classes, use_dp=True, use_bn=True
)
self.assertFalse(model.built, "Model should not have been built")
self.assertFalse(
model.weights,
"Model should have no weights since it has not been built.",
)
model.build(input_shape=(batch_size, input_dim))
self.assertTrue(
model.weights,
"Model should have weights now that it has been properly built.",
)
self.assertTrue(
model.built, "Model should be built after calling `build`."
)
model(tf.ones((32, input_dim)))
def test_single_io_dimension_subclass_build(self):
num_classes = 2
input_dim = tf.compat.v1.Dimension(50)
batch_size = tf.compat.v1.Dimension(None)
model = test_utils.SmallSubclassMLP(
num_hidden=32, num_classes=num_classes, use_dp=True, use_bn=True
)
self.assertFalse(model.built, "Model should not have been built")
self.assertFalse(
model.weights,
"Model should have no weights since it has not been built.",
)
model.build(input_shape=(batch_size, input_dim))
self.assertTrue(
model.weights,
"Model should have weights now that it has been properly built.",
)
self.assertTrue(
model.built, "Model should be built after calling `build`."
)
model(tf.ones((32, input_dim)))
def test_multidim_io_subclass_build(self):
num_classes = 10
# Input size, e.g. image
batch_size = 32
input_shape = (32, 32, 3)
model = model_util.SimpleConvTestModel(num_classes)
self.assertFalse(model.built, "Model should not have been built")
self.assertFalse(
model.weights,
"Model should have no weights since it has not been built.",
)
batch_input_shape = (batch_size,) + input_shape
model.build(input_shape=batch_input_shape)
self.assertTrue(
model.weights,
"Model should have weights now that it has been properly built.",
)
self.assertTrue(
model.built, "Model should be built after calling `build`."
)
model(tf.ones(batch_input_shape))
def test_tensorshape_io_subclass_build(self):
num_classes = 10
# Input size, e.g. image
batch_size = None
input_shape = (32, 32, 3)
model = model_util.SimpleConvTestModel(num_classes)
self.assertFalse(model.built, "Model should not have been built")
self.assertFalse(
model.weights,
"Model should have no weights since it has not been built.",
)
model.build(input_shape=tf.TensorShape((batch_size,) + input_shape))
self.assertTrue(
model.weights,
"Model should have weights now that it has been properly built.",
)
self.assertTrue(
model.built, "Model should be built after calling `build`."
)
model(tf.ones((32,) + input_shape))
def test_subclass_save_model(self):
num_classes = 10
# Input size, e.g. image
batch_size = None
input_shape = (32, 32, 3)
model = model_util.SimpleConvTestModel(num_classes)
self.assertFalse(model.built, "Model should not have been built")
self.assertFalse(
model.weights,
"Model should have no weights since it has not been built.",
)
model.build(input_shape=tf.TensorShape((batch_size,) + input_shape))
self.assertTrue(
model.weights,
"Model should have weights now that it has been properly built.",
)
self.assertTrue(
model.built, "Model should be built after calling `build`."
)
weights = model.get_weights()
tf_format_name = os.path.join(self.get_temp_dir(), "ckpt")
model.save_weights(tf_format_name)
if h5py is not None:
hdf5_format_name = os.path.join(self.get_temp_dir(), "weights.h5")
model.save_weights(hdf5_format_name)
model = model_util.SimpleConvTestModel(num_classes)
model.build(input_shape=tf.TensorShape((batch_size,) + input_shape))
if h5py is not None:
model.load_weights(hdf5_format_name)
self.assertAllClose(weights, model.get_weights())
model.load_weights(tf_format_name)
self.assertAllClose(weights, model.get_weights())
def test_multi_io_subclass_build(self):
batch_size = None
num_samples = 1000
input_dim = 50
model = model_util.get_multi_io_subclass_model()
self.assertFalse(model.built, "Model should not have been built")
self.assertFalse(
model.weights,
"Model should have no weights since it has not been built.",
)
batch_input_shape = tf.TensorShape((batch_size, input_dim))
model.build(input_shape=[batch_input_shape, batch_input_shape])
self.assertTrue(
model.weights,
"Model should have weights now that it has been properly built.",
)
self.assertTrue(
model.built, "Model should be built after calling `build`."
)
x1 = tf.ones((num_samples, input_dim))
x2 = tf.ones((num_samples, input_dim))
model([x1, x2])
def test_summary(self):
class ToString:
def __init__(self):
self.contents = ""
def __call__(self, msg):
self.contents += msg + "\n"
# Single-io
model = test_utils.SmallSubclassMLP(
num_hidden=32, num_classes=4, use_bn=True, use_dp=True
)
model(np.ones((3, 4))) # need to build model first
print_fn = ToString()
model.summary(print_fn=print_fn)
self.assertIn("Trainable params: 356", print_fn.contents)
# Multi-io
model = model_util.get_multi_io_subclass_model(
num_classes=(5, 6), use_bn=True, use_dp=True
)
model([np.ones((3, 4)), np.ones((3, 4))]) # need to build model first
print_fn = ToString()
model.summary(print_fn=print_fn)
self.assertIn("Trainable params: 587", print_fn.contents)
# Single-io with unused layer
model = test_utils.SmallSubclassMLP(
num_hidden=32, num_classes=4, use_bn=True, use_dp=True
)
model.unused_layer = keras.layers.Dense(10)
model(np.ones((3, 4))) # need to build model first
print_fn = ToString()
model.summary(print_fn=print_fn)
self.assertIn("Trainable params: 356", print_fn.contents)
self.assertIn("0 (unused)", print_fn.contents)
def test_no_dependency(self):
class Foo(keras.Model):
def __init__(self):
super().__init__()
self.isdep = keras.layers.Dense(1)
self.notdep = data_structures.NoDependency(
keras.layers.Dense(2)
)
self.notdep_var = data_structures.NoDependency(
tf.Variable(1.0, name="notdep_var")
)
m = Foo()
self.assertEqual([m.isdep, m.notdep], m.layers)
self.assertEqual(1, len(m._trackable_children()))
self.assertIs(m.isdep, m._trackable_children()["isdep"])
self.assertEqual("notdep_var:0", m.notdep_var.name)
def test_extra_variable(self):
class ExtraVar(keras.Model):
def __init__(self):
super().__init__()
self.dense = keras.layers.Dense(1)
self.var = tf.Variable(1.0)
self.not_trainable_var = tf.Variable(2.0, trainable=False)
def call(self, inputs):
return self.dense(inputs + self.var)
m = ExtraVar()
self.assertTrue(m.trainable)
self.assertEqual([m.dense], m.layers)
self.assertEqual([m.var, m.not_trainable_var], m.variables)
self.assertEqual([m.var], m.trainable_variables)
self.assertEqual([m.not_trainable_var], m.non_trainable_variables)
self.assertLen(m.get_weights(), 2)
m.trainable = False
self.assertEqual([m.var, m.not_trainable_var], m.variables)
self.assertEqual([], m.trainable_variables)
self.assertEqual(
[m.var, m.not_trainable_var], m.non_trainable_variables
)
self.assertLen(m.get_weights(), 2)
m.trainable = True
m(tf.ones([1, 1]))
self.assertEqual([m.dense.kernel, m.dense.bias], m.dense.variables)
self.assertEqual([m.dense.kernel, m.dense.bias], m.dense.weights)
self.assertLen(m.get_weights(), 4)
self.assertEqual(
[m.dense.kernel, m.dense.bias, m.var, m.not_trainable_var],
m.variables,
)
self.assertEqual(
[m.dense.kernel, m.dense.bias, m.var], m.trainable_variables
)
self.assertEqual([m.not_trainable_var], m.non_trainable_variables)
m.dense.trainable = False
self.assertEqual(
[m.dense.kernel, m.dense.bias, m.var, m.not_trainable_var],
m.variables,
)
self.assertEqual([m.var], m.trainable_variables)
self.assertEqual(
[m.dense.kernel, m.dense.bias, m.not_trainable_var],
m.non_trainable_variables,
)
self.assertLen(m.get_weights(), 4)
def test_add_weight_in_model(self):
class MyModel(keras.Model):
def __init__(self):
super().__init__()
self.b = self.add_weight("bias", (10,))
self.c = self.add_weight("bias2", (10,), trainable=False)
def call(self, inputs):
return inputs + self.b + self.c
x = tf.convert_to_tensor(np.ones((10, 10), "float32"))
model = MyModel()
model(x)
self.assertEqual(1, len(model.trainable_weights))
self.assertEqual(1, len(model.non_trainable_weights))
self.assertEqual(2, len(model.weights))
class MyModelCustomBuild(keras.Model):
def build(self, input_shape):
self.b = self.add_weight("bias", (10,))
self.c = self.add_weight("bias2", (10,), trainable=False)
def call(self, inputs):
return inputs + self.b + self.c
x = tf.convert_to_tensor(np.ones((10, 10), "float32"))
model = MyModelCustomBuild()
model(x)
self.assertEqual(1, len(model.trainable_weights))
self.assertEqual(1, len(model.non_trainable_weights))
self.assertEqual(2, len(model.weights))
def test_add_update_in_model(self):
class MyModel(keras.Model):
def __init__(self):
super().__init__()
self.b = self.add_weight("bias", (10,))
self.c = self.add_weight("bias2", (10,))
def call(self, inputs):
# Unconditional
self.add_update(self.b.assign(self.b * 2))
# Conditional
self.add_update(self.c.assign(inputs[1, :]))
return inputs + self.b + self.c
x = tf.convert_to_tensor(np.ones((10, 10), "float32"))
model = MyModel()
model(x)
if tf.executing_eagerly():
self.assertLen(model.updates, 0)
else:
self.assertLen(model.updates, 2)
class GraphSpecificModelSubclassingTests(tf.test.TestCase):
def test_single_io_workflow_with_tensors(self):
num_classes = 2
num_samples = 10
input_dim = 50
with tf.Graph().as_default(), self.cached_session():
model = test_utils.SmallSubclassMLP(
num_hidden=32, num_classes=num_classes, use_dp=True, use_bn=True
)
model.compile(loss="mse", optimizer="rmsprop")
x = tf.ones((num_samples, input_dim))
y = tf.zeros((num_samples, num_classes))
model.fit(x, y, epochs=2, steps_per_epoch=10, verbose=0)
_ = model.evaluate(steps=10, verbose=0)
def test_multi_io_workflow_with_tensors(self):
num_classes = (2, 3)
num_samples = 10
input_dim = 50
with tf.Graph().as_default(), self.cached_session():
model = model_util.get_multi_io_subclass_model(
num_classes=num_classes, use_dp=True, use_bn=True
)
model.compile(loss="mse", optimizer="rmsprop")
x1 = tf.ones((num_samples, input_dim))
x2 = tf.ones((num_samples, input_dim))
y1 = tf.zeros((num_samples, num_classes[0]))
y2 = tf.zeros((num_samples, num_classes[1]))
model.fit(
[x1, x2], [y1, y2], epochs=2, steps_per_epoch=10, verbose=0
)
_ = model.evaluate(steps=10, verbose=0)
def test_updates_and_losses_for_nested_models_in_subclassed_model(self):
# Case 1: deferred-build sequential nested in subclass.
class TestModel1(keras.Model):
def __init__(self):
super().__init__()
self.fc = keras.layers.Dense(
10, input_shape=(784,), activity_regularizer="l1"
)
self.bn = keras.Sequential(
[keras.layers.BatchNormalization(axis=1)]
)
def call(self, x):
return self.bn(self.fc(x))
with tf.compat.v1.get_default_graph().as_default(), self.cached_session(): # noqa: E501
model = TestModel1()
x = tf.ones(shape=[100, 784], dtype="float32")
model(x)
self.assertLen(model.updates, 2)
self.assertLen(model.losses, 1)
# Case 2: placeholder-sequential nested in subclass.
class TestModel2(keras.Model):
def __init__(self):
super().__init__()
self.fc = keras.layers.Dense(
10, input_shape=(784,), activity_regularizer="l1"
)
self.bn = keras.Sequential(
[keras.layers.BatchNormalization(axis=1, input_shape=(10,))]
)
def call(self, x):
return self.bn(self.fc(x))
with tf.compat.v1.get_default_graph().as_default(), self.cached_session(): # noqa: E501
model = TestModel2()
x = tf.ones(shape=[100, 784], dtype="float32")
model(x)
self.assertEqual(len(model.get_updates_for(x)), 2)
self.assertEqual(len(model.get_losses_for(x)), 1)
# Case 3: functional-API model nested in subclass.
with tf.compat.v1.get_default_graph().as_default():
inputs = keras.Input((10,))
outputs = keras.layers.BatchNormalization(axis=1)(inputs)
bn = keras.Model(inputs, outputs)
class TestModel3(keras.Model):
def __init__(self):
super().__init__()
self.fc = keras.layers.Dense(
10, input_shape=(784,), activity_regularizer="l1"
)
self.bn = bn
def call(self, x):
return self.bn(self.fc(x))
with self.cached_session():
model = TestModel3()
x = tf.ones(shape=[100, 784], dtype="float32")
model(x)
self.assertEqual(len(model.get_updates_for(x)), 2)
self.assertEqual(len(model.get_losses_for(x)), 1)
def test_multi_io_workflow_with_numpy_arrays_and_custom_placeholders(self):
num_classes = (2, 3)
num_samples = 1000
input_dim = 50
with tf.Graph().as_default(), self.cached_session():
model = model_util.get_multi_io_subclass_model(
num_classes=num_classes, use_dp=True, use_bn=True
)
model.compile(loss="mse", optimizer="rmsprop")
x1 = np.ones((num_samples, input_dim))
x2 = np.ones((num_samples, input_dim))
y1 = np.zeros((num_samples, num_classes[0]))
y2 = np.zeros((num_samples, num_classes[1]))
x2_placeholder = tf.compat.v1.placeholder(
dtype="float32", shape=(None, input_dim)
)
model._set_inputs([x1, x2_placeholder])
model.fit([x1, x2], [y1, y2], epochs=2, batch_size=32, verbose=0)
_ = model.evaluate([x1, x2], [y1, y2], verbose=0)
@test_combinations.generate(test_combinations.combine(mode=["graph", "eager"]))
class CustomCallSignatureTests(tf.test.TestCase, parameterized.TestCase):
def test_no_inputs_in_signature(self):
model = model_util.CustomCallModel()
first = tf.ones([2, 3])
second = tf.ones([2, 5])
output = model(first, second)
self.evaluate([v.initializer for v in model.variables])
expected_output = self.evaluate(
model.dense1(first) + model.dense2(second)
)
self.assertAllClose(expected_output, self.evaluate(output))
output = model(first, second, fiddle_with_output="yes")
self.assertAllClose(10.0 * expected_output, self.evaluate(output))
output = model(first, second=second, training=False)
self.assertAllClose(expected_output, self.evaluate(output))
def test_training_args_call_build(self):
input_dim = 2
model = model_util.TrainingNoDefaultModel()
self.assertFalse(model.built, "Model should not have been built")
self.assertFalse(
model.weights,
"Model should have no weights since it has not been built.",
)
model.build((None, input_dim))
self.assertTrue(
model.weights,
"Model should have weights now that it has been properly built.",
)
self.assertTrue(
model.built, "Model should be built after calling `build`."
)
def test_training_and_mask_args_call_build(self):
input_dim = 2
model = model_util.TrainingMaskingModel()
self.assertFalse(model.built, "Model should not have been built")
self.assertFalse(
model.weights,
"Model should have no weights since it has not been built.",
)
model.build((None, input_dim))
self.assertTrue(
model.weights,
"Model should have weights now that it has been properly built.",
)
self.assertTrue(
model.built, "Model should be built after calling `build`."
)
def test_custom_call_kwargs_and_build(self):
first_input_shape = (2, 3)
second_input_shape = (2, 5)
model = model_util.CustomCallModel()
self.assertFalse(model.built, "Model should not have been built")
self.assertFalse(
model.weights,
"Model should have no weights since it has not been built.",
)
with self.assertRaisesRegex(
ValueError, "cannot build your model if it has positional"
):
model.build(input_shape=[first_input_shape, second_input_shape])
def test_kwargs_in_signature(self):
class HasKwargs(keras.Model):
def call(self, x, y=3, **kwargs):
return x
model = HasKwargs()
arg = tf.ones([1])
model(arg, a=3)
if not tf.executing_eagerly():
self.assertLen(model.inputs, 1)
@tf_test_utils.assert_no_new_tensors
@tf_test_utils.assert_no_garbage_created
def test_training_no_default(self):
if not tf.executing_eagerly():
return
model = model_util.TrainingNoDefaultModel()
arg = tf.ones([1, 1])
model(arg, True)
def test_positional_arg_in_call(self):
class ModelWithPositionalArgs(keras.Model):
def call(self, x, x2, x3=None):
return x + x2
x = np.ones((10, 1))
y = np.ones((10, 1))
m = ModelWithPositionalArgs()
m.compile("sgd", "mse")
with self.assertRaisesRegex(ValueError, r"Models passed to `fit`"):
m.fit(x, y, batch_size=2)
with self.assertRaisesRegex(ValueError, r"Models passed to `evaluate`"):
m.evaluate(x, y, batch_size=2)
with self.assertRaisesRegex(ValueError, r"Models passed to `predict`"):
m.predict(x, batch_size=2)
with self.assertRaisesRegex(
ValueError, r"Models passed to `train_on_batch`"
):
m.train_on_batch(x, y)
with self.assertRaisesRegex(
ValueError, r"Models passed to `test_on_batch`"
):
m.test_on_batch(x, y)
with self.assertRaisesRegex(
ValueError, r"Models passed to `predict_on_batch`"
):
m.predict_on_batch(x)
def test_deepcopy(self):
if not tf.executing_eagerly():
self.skipTest("Run in eager mode only.")
class MyModel(keras.Model):
def __init__(self):
super().__init__()
self.my_variable = tf.Variable(0.0, trainable=False)
self.layer = keras.layers.Dense(4)
def call(self, obs):
return self.layer(obs)
model = MyModel()
model.my_variable.assign_add(1.0)
new_model = copy.deepcopy(model)
self.assertEqual(model.my_variable.numpy(), 1.0)
self.assertEqual(new_model.my_variable.numpy(), 1.0)
model.my_variable.assign_add(1.0)
self.assertEqual(model.my_variable.numpy(), 2.0)
self.assertEqual(new_model.my_variable.numpy(), 1.0)
# Check that Trackable logic still works.
self.assertLen(new_model.variables, 1)
self.assertLen(new_model.layers, 1)
def test_batch_counters_not_in_variables(self):
class MyModel(keras.Model):
def __init__(self):
super().__init__()
self.layer = keras.layers.Dense(4)
def call(self, obs):
return self.layer(obs)
model = MyModel()
model(np.ones((10, 10)))
self.assertLen(model.variables, 2)
if __name__ == "__main__":
tf.test.main()
|
tf-keras/tf_keras/tests/model_subclassing_test.py/0
|
{
"file_path": "tf-keras/tf_keras/tests/model_subclassing_test.py",
"repo_id": "tf-keras",
"token_count": 14345
}
| 223 |
# Copyright 2019 The TensorFlow Authors. All Rights Reserved.
#
# Licensed under the Apache License, Version 2.0 (the "License");
# you may not use this file except in compliance with the License.
# You may obtain a copy of the License at
#
# http://www.apache.org/licenses/LICENSE-2.0
#
# Unless required by applicable law or agreed to in writing, software
# distributed under the License is distributed on an "AS IS" BASIS,
# WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.
# See the License for the specific language governing permissions and
# limitations under the License.
# ==============================================================================
"""Utils to help build and verify pip package for TF-Keras."""
from __future__ import absolute_import
from __future__ import division
from __future__ import print_function
import argparse
import fnmatch
import os
PIP_EXCLUDED_FILES = frozenset(
[
"tf_keras/api/extractor_wrapper.py",
"tf_keras/api/generator_wrapper.py",
"tf_keras/applications/efficientnet_weight_update_util.py",
"tf_keras/distribute/tpu_strategy_test_utils.py",
"tf_keras/saving/legacy/saved_model/create_test_saved_model.py",
"tf_keras/tools/pip_package/setup.py",
"tf_keras/tools/pip_package/create_pip_helper.py",
]
)
PIP_EXCLUDED_DIRS = frozenset(
[
"tf_keras/benchmarks",
"tf_keras/tests",
]
)
# Directories that should not have __init__.py files generated within them.
EXCLUDED_INIT_FILE_DIRECTORIES = frozenset(
[
"tf_keras/benchmarks",
"tf_keras/tools",
]
)
class PipPackagingError(Exception):
pass
def create_init_files(pip_root):
"""Create __init__.py in pip directory tree.
These files are auto-generated by Bazel when doing typical build/test, but
do not get auto-generated by the pip build process. Currently, the entire
directory tree is just python files, so its fine to just create all of the
init files.
Args:
pip_root: Root directory of code being packaged into pip.
"""
for path, subdirs, _ in os.walk(pip_root):
for subdir in subdirs:
init_file_path = os.path.join(path, subdir, "__init__.py")
if any(
excluded_path in init_file_path
for excluded_path in EXCLUDED_INIT_FILE_DIRECTORIES
):
continue
if not os.path.exists(init_file_path):
# Create empty file
open(init_file_path, "w").close()
def verify_python_files_in_pip(pip_root, bazel_root):
"""Verifies all expected files are packaged into Pip.
Args:
pip_root: Root directory of code being packaged into pip.
bazel_root: Root directory of TF-Keras Bazel workspace.
Raises:
PipPackagingError: Missing file in pip.
"""
for path, _, files in os.walk(bazel_root):
if any(d for d in PIP_EXCLUDED_DIRS if d in path):
# Skip any directories that are exclude from PIP, eg tests.
continue
python_files = set(fnmatch.filter(files, "*.py"))
python_test_files = set(fnmatch.filter(files, "*test.py"))
python_benchmark_files = set(fnmatch.filter(files, "*benchmark.py"))
# We only care about python files in the pip package, see
# create_init_files.
files = python_files - python_test_files - python_benchmark_files
for f in files:
pip_path = os.path.join(
pip_root, os.path.relpath(path, bazel_root), f
)
file_name = os.path.join(path, f)
path_exists = os.path.exists(pip_path)
file_excluded = file_name.lstrip("./") in PIP_EXCLUDED_FILES
if not path_exists and not file_excluded:
raise PipPackagingError(
"Pip package missing the file %s. If this is expected, "
"add it to PIP_EXCLUDED_FILES in "
"create_pip_helper.py. Otherwise, "
"make sure it is a build dependency of the pip package"
% file_name
)
if path_exists and file_excluded:
raise PipPackagingError(
f"File in PIP_EXCLUDED_FILES included in pip. {file_name}"
)
def main():
parser = argparse.ArgumentParser()
parser.add_argument(
"--bazel-root",
type=str,
required=True,
help="Root directory of TF-Keras Bazel workspace.",
)
parser.add_argument(
"--pip-root",
type=str,
required=True,
help="Root directory of code being packaged into pip.",
)
args = parser.parse_args()
create_init_files(args.pip_root)
verify_python_files_in_pip(args.pip_root, args.bazel_root)
if __name__ == "__main__":
main()
|
tf-keras/tf_keras/tools/pip_package/create_pip_helper.py/0
|
{
"file_path": "tf-keras/tf_keras/tools/pip_package/create_pip_helper.py",
"repo_id": "tf-keras",
"token_count": 2082
}
| 224 |
"""Tests for Dataset Utils"""
import os
import shutil
import numpy as np
import tensorflow.compat.v2 as tf
from tf_keras.testing_infra import test_utils
from tf_keras.utils import dataset_utils
@test_utils.run_v2_only
class SplitDatasetTest(tf.test.TestCase):
def test_numpy_array(self):
dataset = np.ones(shape=(200, 32))
res = dataset_utils.split_dataset(
dataset, left_size=0.8, right_size=0.2
)
self.assertLen(res, 2)
left_split, right_split = res
self.assertIsInstance(left_split, tf.data.Dataset)
self.assertIsInstance(right_split, tf.data.Dataset)
self.assertLen(left_split, 160)
self.assertLen(right_split, 40)
self.assertAllEqual(dataset[:160], list(left_split))
self.assertAllEqual(dataset[-40:], list(right_split))
def test_list_of_numpy_arrays(self):
# test with list of np arrays with same shapes
dataset = [np.ones(shape=(200, 32)), np.zeros(shape=(200, 32))]
res = dataset_utils.split_dataset(dataset, left_size=4)
self.assertLen(res, 2)
left_split, right_split = res
self.assertIsInstance(left_split, tf.data.Dataset)
self.assertIsInstance(right_split, tf.data.Dataset)
self.assertEqual(np.array(list(left_split)).shape, (4, 2, 32))
self.assertEqual(np.array(list(right_split)).shape, (196, 2, 32))
# test with different shapes
dataset = [np.ones(shape=(5, 3)), np.ones(shape=(5,))]
left_split, right_split = dataset_utils.split_dataset(
dataset, left_size=0.3
)
self.assertEqual(np.array(list(left_split), dtype=object).shape, (2, 2))
self.assertEqual(
np.array(list(right_split), dtype=object).shape, (3, 2)
)
self.assertEqual(
np.array(list(left_split)[0], dtype=object).shape, (2,)
)
self.assertEqual(np.array(list(left_split)[0][0]).shape, (3,))
self.assertEqual(np.array(list(left_split)[0][1]).shape, ())
self.assertEqual(
np.array(list(right_split)[0], dtype=object).shape, (2,)
)
self.assertEqual(np.array(list(right_split)[0][0]).shape, (3,))
self.assertEqual(np.array(list(right_split)[0][1]).shape, ())
def test_dataset_with_invalid_shape(self):
with self.assertRaisesRegex(
ValueError,
"Received a list of NumPy arrays with different lengths",
):
dataset = [np.ones(shape=(200, 32)), np.zeros(shape=(100, 32))]
dataset_utils.split_dataset(dataset, left_size=4)
with self.assertRaisesRegex(
ValueError,
"Received a tuple of NumPy arrays with different lengths",
):
dataset = (np.ones(shape=(200, 32)), np.zeros(shape=(201, 32)))
dataset_utils.split_dataset(dataset, left_size=4)
def test_tuple_of_numpy_arrays(self):
dataset = (np.random.rand(4, 3), np.random.rand(4, 3))
left_split, right_split = dataset_utils.split_dataset(
dataset, left_size=2
)
self.assertIsInstance(left_split, tf.data.Dataset)
self.assertIsInstance(right_split, tf.data.Dataset)
self.assertEqual(len(left_split), 2)
self.assertEqual(len(right_split), 2)
self.assertEqual(np.array(list(left_split)[0]).shape, (2, 3))
self.assertEqual(np.array(list(left_split)[1]).shape, (2, 3))
# test with fractional size
dataset = (np.random.rand(5, 32, 32), np.random.rand(5, 32, 32))
left_split, right_split = dataset_utils.split_dataset(
dataset, right_size=0.4
)
self.assertIsInstance(left_split, tf.data.Dataset)
self.assertIsInstance(right_split, tf.data.Dataset)
self.assertEqual(np.array(list(left_split)).shape, (3, 2, 32, 32))
self.assertEqual(np.array(list(right_split)).shape, (2, 2, 32, 32))
self.assertEqual(np.array(list(left_split))[0].shape, (2, 32, 32))
self.assertEqual(np.array(list(left_split))[1].shape, (2, 32, 32))
self.assertEqual(np.array(list(right_split))[0].shape, (2, 32, 32))
self.assertEqual(np.array(list(right_split))[1].shape, (2, 32, 32))
# test with tuple of np arrays with different shapes
dataset = (
np.random.rand(5, 32, 32),
np.random.rand(
5,
),
)
left_split, right_split = dataset_utils.split_dataset(
dataset, left_size=2, right_size=3
)
self.assertIsInstance(left_split, tf.data.Dataset)
self.assertIsInstance(right_split, tf.data.Dataset)
self.assertEqual(np.array(list(left_split), dtype=object).shape, (2, 2))
self.assertEqual(
np.array(list(right_split), dtype=object).shape, (3, 2)
)
self.assertEqual(
np.array(list(left_split)[0], dtype=object).shape, (2,)
)
self.assertEqual(np.array(list(left_split)[0][0]).shape, (32, 32))
self.assertEqual(np.array(list(left_split)[0][1]).shape, ())
self.assertEqual(
np.array(list(right_split)[0], dtype=object).shape, (2,)
)
self.assertEqual(np.array(list(right_split)[0][0]).shape, (32, 32))
self.assertEqual(np.array(list(right_split)[0][1]).shape, ())
def test_batched_tf_dataset_of_vectors(self):
vectors = np.ones(shape=(100, 32, 32, 1))
dataset = tf.data.Dataset.from_tensor_slices(vectors)
dataset = dataset.batch(10)
left_split, right_split = dataset_utils.split_dataset(
dataset, left_size=2
)
# Ensure that the splits are batched
self.assertEqual(len(list(right_split)), 10)
left_split, right_split = left_split.unbatch(), right_split.unbatch()
self.assertAllEqual(np.array(list(left_split)).shape, (2, 32, 32, 1))
self.assertAllEqual(np.array(list(right_split)).shape, (98, 32, 32, 1))
dataset = dataset.unbatch()
self.assertAllEqual(list(dataset), list(left_split) + list(right_split))
def test_batched_tf_dataset_of_tuple_of_vectors(self):
tuple_of_vectors = (
np.random.rand(10, 32, 32),
np.random.rand(10, 32, 32),
)
dataset = tf.data.Dataset.from_tensor_slices(tuple_of_vectors)
dataset = dataset.batch(2)
left_split, right_split = dataset_utils.split_dataset(
dataset, left_size=4
)
# Ensure that the splits are batched
self.assertEqual(np.array(list(right_split)).shape, (3, 2, 2, 32, 32))
self.assertEqual(np.array(list(left_split)).shape, (2, 2, 2, 32, 32))
left_split, right_split = left_split.unbatch(), right_split.unbatch()
self.assertAllEqual(np.array(list(left_split)).shape, (4, 2, 32, 32))
self.assertAllEqual(np.array(list(right_split)).shape, (6, 2, 32, 32))
dataset = dataset.unbatch()
self.assertAllEqual(list(dataset), list(left_split) + list(right_split))
def test_batched_tf_dataset_of_dict_of_vectors(self):
dict_samples = {"X": np.random.rand(10, 3), "Y": np.random.rand(10, 3)}
dataset = tf.data.Dataset.from_tensor_slices(dict_samples)
dataset = dataset.batch(2)
left_split, right_split = dataset_utils.split_dataset(
dataset, left_size=2
)
self.assertAllEqual(np.array(list(left_split)).shape, (1,))
self.assertAllEqual(np.array(list(right_split)).shape, (4,))
left_split, right_split = left_split.unbatch(), right_split.unbatch()
self.assertEqual(len(list(left_split)), 2)
self.assertEqual(len(list(right_split)), 8)
for i in range(10):
if i < 2:
self.assertEqual(
list(left_split)[i], list(dataset.unbatch())[i]
)
else:
self.assertEqual(
list(right_split)[i - 2], list(dataset.unbatch())[i]
)
# test with dict of np arrays with different shapes
dict_samples = {
"images": np.random.rand(10, 16, 16, 3),
"labels": np.random.rand(
10,
),
}
dataset = tf.data.Dataset.from_tensor_slices(dict_samples)
dataset = dataset.batch(1)
left_split, right_split = dataset_utils.split_dataset(
dataset, right_size=0.3
)
self.assertAllEqual(np.array(list(left_split)).shape, (7,))
self.assertAllEqual(np.array(list(right_split)).shape, (3,))
dataset = dataset.unbatch()
left_split, right_split = left_split.unbatch(), right_split.unbatch()
self.assertEqual(len(list(left_split)), 7)
self.assertEqual(len(list(right_split)), 3)
for i in range(10):
if i < 7:
self.assertEqual(list(left_split)[i], list(dataset)[i])
else:
self.assertEqual(list(right_split)[i - 7], list(dataset)[i])
def test_unbatched_tf_dataset_of_vectors(self):
vectors = np.ones(shape=(100, 16, 16, 3))
dataset = tf.data.Dataset.from_tensor_slices(vectors)
left_split, right_split = dataset_utils.split_dataset(
dataset, left_size=0.25
)
self.assertAllEqual(np.array(list(left_split)).shape, (25, 16, 16, 3))
self.assertAllEqual(np.array(list(right_split)).shape, (75, 16, 16, 3))
self.assertAllEqual(list(dataset), list(left_split) + list(right_split))
dataset = [np.random.rand(10, 3, 3) for _ in range(5)]
dataset = tf.data.Dataset.from_tensor_slices(dataset)
left_split, right_split = dataset_utils.split_dataset(
dataset, left_size=2
)
self.assertAllEqual(list(dataset), list(left_split) + list(right_split))
def test_unbatched_tf_dataset_of_tuple_of_vectors(self):
# test with tuple of np arrays with same shape
X, Y = (np.random.rand(10, 32, 32, 1), np.random.rand(10, 32, 32, 1))
dataset = tf.data.Dataset.from_tensor_slices((X, Y))
left_split, right_split = dataset_utils.split_dataset(
dataset, left_size=5
)
self.assertEqual(len(list(left_split)), 5)
self.assertEqual(len(list(right_split)), 5)
self.assertAllEqual(list(dataset), list(left_split) + list(right_split))
# test with tuple of np arrays with different shapes
X, Y = (
np.random.rand(5, 3, 3),
np.random.rand(
5,
),
)
dataset = tf.data.Dataset.from_tensor_slices((X, Y))
left_split, right_split = dataset_utils.split_dataset(
dataset, left_size=0.5
)
self.assertEqual(len(list(left_split)), 2)
self.assertEqual(len(list(right_split)), 3)
self.assertEqual(np.array(list(left_split)[0][0]).shape, (3, 3))
self.assertEqual(np.array(list(left_split)[0][1]).shape, ())
def test_unbatched_tf_dataset_of_dict_of_vectors(self):
# test with dict of np arrays of same shape
dict_samples = {"X": np.random.rand(10, 2), "Y": np.random.rand(10, 2)}
dataset = tf.data.Dataset.from_tensor_slices(dict_samples)
left_split, right_split = dataset_utils.split_dataset(
dataset, left_size=2
)
self.assertEqual(len(list(left_split)), 2)
self.assertEqual(len(list(right_split)), 8)
for i in range(10):
if i < 2:
self.assertEqual(list(left_split)[i], list(dataset)[i])
else:
self.assertEqual(list(right_split)[i - 2], list(dataset)[i])
# test with dict of np arrays with different shapes
dict_samples = {
"images": np.random.rand(10, 16, 16, 3),
"labels": np.random.rand(
10,
),
}
dataset = tf.data.Dataset.from_tensor_slices(dict_samples)
left_split, right_split = dataset_utils.split_dataset(
dataset, left_size=0.3
)
self.assertEqual(len(list(left_split)), 3)
self.assertEqual(len(list(right_split)), 7)
for i in range(10):
if i < 3:
self.assertEqual(list(left_split)[i], list(dataset)[i])
else:
self.assertEqual(list(right_split)[i - 3], list(dataset)[i])
# test with dict of text arrays
txt_feature = ["abb", "bb", "cc", "d", "e", "f", "g", "h", "i", "j"]
dict_samples = {
"txt_feature": txt_feature,
"label": [1, 2, 3, 4, 5, 6, 7, 8, 9, 10],
}
dataset = tf.data.Dataset.from_tensor_slices(dict_samples)
left_split, right_split = dataset_utils.split_dataset(
dataset, left_size=0.45, right_size=0.55
)
self.assertEqual(len(list(left_split)), 4)
self.assertEqual(len(list(right_split)), 6)
for i in range(10):
if i < 4:
self.assertEqual(list(left_split)[i], list(dataset)[i])
else:
self.assertEqual(list(right_split)[i - 4], list(dataset)[i])
def test_list_dataset(self):
dataset = [np.ones(shape=(10, 10, 10)) for _ in range(10)]
left_split, right_split = dataset_utils.split_dataset(
dataset, left_size=5, right_size=5
)
self.assertEqual(len(left_split), len(right_split))
self.assertIsInstance(left_split, tf.data.Dataset)
self.assertIsInstance(left_split, tf.data.Dataset)
dataset = [np.ones(shape=(10, 10, 10)) for _ in range(10)]
left_split, right_split = dataset_utils.split_dataset(
dataset, left_size=0.6, right_size=0.4
)
self.assertEqual(len(left_split), 6)
self.assertEqual(len(right_split), 4)
def test_invalid_dataset(self):
with self.assertRaisesRegex(
TypeError,
"The `dataset` argument must be either a `tf.data.Dataset` "
"object or a list/tuple of arrays.",
):
dataset_utils.split_dataset(dataset=None, left_size=5)
with self.assertRaisesRegex(
TypeError,
"The `dataset` argument must be either a `tf.data.Dataset` "
"object or a list/tuple of arrays.",
):
dataset_utils.split_dataset(dataset=1, left_size=5)
with self.assertRaisesRegex(
TypeError,
"The `dataset` argument must be either a `tf.data.Dataset` "
"object or a list/tuple of arrays.",
):
dataset_utils.split_dataset(dataset=float(1.2), left_size=5)
with self.assertRaisesRegex(
TypeError,
"The `dataset` argument must be either a `tf.data.Dataset` "
"object or a list/tuple of arrays.",
):
dataset_utils.split_dataset(dataset=dict({}), left_size=5)
with self.assertRaisesRegex(
TypeError,
"The `dataset` argument must be either a `tf.data.Dataset` "
"object or a list/tuple of arrays.",
):
dataset_utils.split_dataset(dataset=float("INF"), left_size=5)
def test_valid_left_and_right_sizes(self):
dataset = np.array([1, 2, 3])
splitted_dataset = dataset_utils.split_dataset(dataset, 1, 2)
self.assertLen(splitted_dataset, 2)
left_split, right_split = splitted_dataset
self.assertEqual(len(left_split), 1)
self.assertEqual(len(right_split), 2)
self.assertEqual(list(left_split), [1])
self.assertEqual(list(right_split), [2, 3])
dataset = np.ones(shape=(200, 32))
res = dataset_utils.split_dataset(dataset, left_size=150, right_size=50)
self.assertLen(res, 2)
self.assertIsInstance(res[0], tf.data.Dataset)
self.assertIsInstance(res[1], tf.data.Dataset)
self.assertLen(res[0], 150)
self.assertLen(res[1], 50)
dataset = np.ones(shape=(200, 32))
res = dataset_utils.split_dataset(dataset, left_size=120)
self.assertLen(res, 2)
self.assertIsInstance(res[0], tf.data.Dataset)
self.assertIsInstance(res[1], tf.data.Dataset)
self.assertLen(res[0], 120)
self.assertLen(res[1], 80)
dataset = np.ones(shape=(10000, 16))
res = dataset_utils.split_dataset(dataset, right_size=20)
self.assertLen(res, 2)
self.assertIsInstance(res[0], tf.data.Dataset)
self.assertIsInstance(res[1], tf.data.Dataset)
self.assertLen(res[0], 9980)
self.assertLen(res[1], 20)
dataset = np.array([1, 2, 3, 4, 5, 6, 7, 8, 9, 10])
splitted_dataset = dataset_utils.split_dataset(
dataset, left_size=0.1, right_size=0.9
)
self.assertLen(splitted_dataset, 2)
left_split, right_split = splitted_dataset
self.assertEqual(len(left_split), 1)
self.assertEqual(len(right_split), 9)
self.assertEqual(list(left_split), [1])
self.assertEqual(list(right_split), [2, 3, 4, 5, 6, 7, 8, 9, 10])
dataset = np.array([1, 2, 3, 4, 5, 6, 7, 8, 9, 10])
splitted_dataset = dataset_utils.split_dataset(
dataset, left_size=2, right_size=5
)
self.assertLen(splitted_dataset, 2)
left_split, right_split = splitted_dataset
self.assertEqual(len(left_split), 2)
self.assertEqual(len(right_split), 5)
self.assertEqual(list(left_split), [1, 2])
self.assertEqual(list(right_split), [6, 7, 8, 9, 10])
def test_float_left_and_right_sizes(self):
X = np.array([[0.1, 0.2, 0.3], [0.4, 0.5, 0.6], [0.7, 0.8, 0.9]])
dataset = tf.data.Dataset.from_tensor_slices(X)
left_split, right_split = dataset_utils.split_dataset(
dataset, left_size=0.8, right_size=0.2
)
self.assertEqual(len(left_split), 2)
self.assertEqual(len(right_split), 1)
def test_invalid_float_left_and_right_sizes(self):
expected_regex = (
r"^(.*?(\bleft_size\b).*?(\bshould be\b)"
r".*?(\bwithin the range\b).*?(\b0\b).*?(\b1\b))"
)
with self.assertRaisesRegexp(ValueError, expected_regex):
dataset = [
np.ones(shape=(200, 32, 32)),
np.zeros(shape=(200, 32, 32)),
]
dataset_utils.split_dataset(dataset, left_size=1.5, right_size=0.2)
expected_regex = (
r"^(.*?(\bright_size\b).*?(\bshould be\b)"
r".*?(\bwithin the range\b).*?(\b0\b).*?(\b1\b))"
)
with self.assertRaisesRegex(ValueError, expected_regex):
dataset = [np.ones(shape=(200, 32)), np.zeros(shape=(200, 32))]
dataset_utils.split_dataset(dataset, left_size=0.8, right_size=-0.8)
def test_None_and_zero_left_and_right_size(self):
expected_regex = (
r"^.*?(\bleft_size\b).*?(\bright_size\b).*?(\bmust "
r"be specified\b).*?(\bReceived: left_size=None and"
r" right_size=None\b)"
)
with self.assertRaisesRegex(ValueError, expected_regex):
dataset_utils.split_dataset(
dataset=np.array([1, 2, 3]), left_size=None
)
with self.assertRaisesRegex(ValueError, expected_regex):
dataset_utils.split_dataset(
np.array([1, 2, 3]), left_size=None, right_size=None
)
expected_regex = (
r"^.*?(\bleft_size\b).*?(\bshould be\b)"
r".*?(\bpositive\b).*?(\bsmaller than 3\b)"
)
with self.assertRaisesRegex(ValueError, expected_regex):
dataset_utils.split_dataset(np.array([1, 2, 3]), left_size=3)
expected_regex = (
"Both `left_size` and `right_size` are zero. "
"At least one of the split sizes must be non-zero."
)
with self.assertRaisesRegex(ValueError, expected_regex):
dataset_utils.split_dataset(
np.array([1, 2, 3]), left_size=0, right_size=0
)
def test_invalid_left_and_right_size_types(self):
expected_regex = (
r"^.*?(\bInvalid `left_size` and `right_size` Types"
r"\b).*?(\bExpected: integer or float or None\b)"
)
with self.assertRaisesRegex(TypeError, expected_regex):
dataset_utils.split_dataset(
np.array([1, 2, 3]), left_size="1", right_size="1"
)
expected_regex = r"^.*?(\bInvalid `right_size` Type\b)"
with self.assertRaisesRegex(TypeError, expected_regex):
dataset_utils.split_dataset(
np.array([1, 2, 3]), left_size=0, right_size="1"
)
expected_regex = r"^.*?(\bInvalid `left_size` Type\b)"
with self.assertRaisesRegex(TypeError, expected_regex):
dataset_utils.split_dataset(
np.array([1, 2, 3]), left_size="100", right_size=None
)
expected_regex = r"^.*?(\bInvalid `right_size` Type\b)"
with self.assertRaisesRegex(TypeError, expected_regex):
dataset_utils.split_dataset(np.array([1, 2, 3]), right_size="1")
expected_regex = r"^.*?(\bInvalid `right_size` Type\b)"
with self.assertRaisesRegex(TypeError, expected_regex):
dataset_utils.split_dataset(
np.array([1, 2, 3]), left_size=0.5, right_size="1"
)
def test_end_to_end(self):
x_train = np.random.random((10000, 28, 28))
y_train = np.random.randint(0, 10, size=(10000,))
left_split, right_split = dataset_utils.split_dataset(
(x_train, y_train), left_size=0.8
)
self.assertIsInstance(left_split, tf.data.Dataset)
self.assertIsInstance(right_split, tf.data.Dataset)
self.assertEqual(len(left_split), 8000)
self.assertEqual(len(right_split), 2000)
@test_utils.run_v2_only
class IndexDirectoryStructureTest(tf.test.TestCase):
def test_explicit_labels_and_unnested_files(self):
# Get a unique temp directory
temp_dir = os.path.join(
self.get_temp_dir(), str(np.random.randint(1e6))
)
os.mkdir(temp_dir)
self.addCleanup(shutil.rmtree, temp_dir)
# Number of temp files, each of which
# will have its own explicit label
num_files = 10
explicit_labels = np.random.randint(0, 10, size=num_files).tolist()
# Save empty text files to root of temp directory
# (content is not important, only location)
for i in range(len(explicit_labels)):
with open(os.path.join(temp_dir, f"file{i}.txt"), "w"):
pass
file_paths, labels, class_names = dataset_utils.index_directory(
temp_dir, labels=explicit_labels, formats=".txt"
)
# Files are found at the root of the temp directory, when
# `labels` are passed explicitly to `index_directory` and
# the number of returned and passed labels match
self.assertLen(file_paths, num_files)
self.assertLen(labels, num_files)
# Class names are returned as a sorted list
expected_class_names = sorted(set(explicit_labels))
self.assertEqual(expected_class_names, class_names)
if __name__ == "__main__":
tf.test.main()
|
tf-keras/tf_keras/utils/dataset_utils_test.py/0
|
{
"file_path": "tf-keras/tf_keras/utils/dataset_utils_test.py",
"repo_id": "tf-keras",
"token_count": 11494
}
| 225 |
# Copyright 2020 The TensorFlow Authors. All Rights Reserved.
#
# Licensed under the Apache License, Version 2.0 (the "License");
# you may not use this file except in compliance with the License.
# You may obtain a copy of the License at
#
# http://www.apache.org/licenses/LICENSE-2.0
#
# Unless required by applicable law or agreed to in writing, software
# distributed under the License is distributed on an "AS IS" BASIS,
# WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.
# See the License for the specific language governing permissions and
# limitations under the License.
# ==============================================================================
"""Tests for layer_utils."""
import collections
import contextlib
import io
import multiprocessing.dummy
import os
import pickle
import shutil
import sys
import tempfile
import time
import timeit
import numpy as np
import tensorflow.compat.v2 as tf
from absl.testing import parameterized
import tf_keras as keras
from tf_keras import backend
from tf_keras import layers
from tf_keras.dtensor import dtensor_api as dtensor
from tf_keras.dtensor import layout_map as layout_map_lib
from tf_keras.dtensor import test_util
from tf_keras.testing_infra import test_utils
from tf_keras.utils import io_utils
from tf_keras.utils import layer_utils
from tf_keras.utils import tf_utils
_PICKLEABLE_CALL_COUNT = collections.Counter()
class MyPickleableObject(tf.__internal__.tracking.AutoTrackable):
"""Needed for InterfaceTests.test_property_cache_serialization.
This class must be at the top level. This is a constraint of pickle,
unrelated to `cached_per_instance`.
"""
@property
@layer_utils.cached_per_instance
def my_id(self):
_PICKLEABLE_CALL_COUNT[self] += 1
return id(self)
class LayerUtilsTest(tf.test.TestCase, parameterized.TestCase):
def setUp(self):
super().setUp()
# Reset the UID so that all the layer/model ID will always start with 1.
# This will help remove the undetermined IDs from the model.summary()
backend.reset_uids()
def test_print_summary(self):
model = keras.Sequential()
model.add(
keras.layers.Conv2D(
filters=2,
kernel_size=(2, 3),
input_shape=(3, 5, 5),
name="conv",
)
)
model.add(keras.layers.Flatten(name="flat"))
model.add(keras.layers.Dense(5, name="dense"))
file_name = "model_1.txt"
temp_dir = self.get_temp_dir()
self.addCleanup(shutil.rmtree, temp_dir, ignore_errors=True)
fpath = os.path.join(temp_dir, file_name)
writer = open(fpath, "w")
def print_to_file(text):
print(text, file=writer)
try:
layer_utils.print_summary(model, print_fn=print_to_file)
self.assertTrue(tf.io.gfile.exists(fpath))
writer.close()
with open(fpath, "r") as reader:
lines = reader.readlines()
self.assertEqual(len(lines), 15)
except ImportError:
pass
def test_print_summary_without_print_fn(self):
model = keras.Sequential(
[keras.layers.Dense(5, input_shape=(10,), name="dense")]
)
io_utils.enable_interactive_logging()
with self.captureWritesToStream(sys.stdout) as printed:
layer_utils.print_summary(model)
self.assertIn("dense (Dense)", printed.contents())
def test_print_summary_format_long_names(self):
shape = (8, 8, 3)
model = keras.Sequential(
[
keras.Input(shape),
keras.layers.Conv2D(4, 3, name="Really-Long-name-test"),
keras.layers.Conv2D(4, 3, name="Another-long-name-test"),
keras.layers.Flatten(),
keras.layers.Dense(2, name="long-name-test-output"),
]
)
file_name = "sequential.txt"
temp_dir = self.get_temp_dir()
self.addCleanup(shutil.rmtree, temp_dir, ignore_errors=True)
fpath = os.path.join(temp_dir, file_name)
writer = open(fpath, "w")
def print_to_file(text):
print(text, file=writer)
layer_utils.print_summary(model, print_fn=print_to_file)
self.assertTrue(tf.io.gfile.exists(fpath))
writer.close()
reader = open(fpath, "r")
lines = reader.readlines()
reader.close()
check_str = (
'Model: "sequential"\n'
"_________________________________________________________________\n" # noqa: E501
" Layer (type) Output Shape Param # \n" # noqa: E501
"=================================================================\n" # noqa: E501
" Really-Long-name-test (Con (None, 6, 6, 4) 112 \n" # noqa: E501
" v2D) \n" # noqa: E501
" \n" # noqa: E501
" Another-long-name-test (Co (None, 4, 4, 4) 148 \n" # noqa: E501
" nv2D) \n" # noqa: E501
" \n" # noqa: E501
" flatten (Flatten) (None, 64) 0 \n" # noqa: E501
" \n" # noqa: E501
" long-name-test-output (Den (None, 2) 130 \n" # noqa: E501
" se) \n" # noqa: E501
" \n" # noqa: E501
"=================================================================\n" # noqa: E501
"Total params: 390 (1.52 KB)\n"
"Trainable params: 390 (1.52 KB)\n"
"Non-trainable params: 0 (0.00 Byte)\n"
"_________________________________________________________________\n" # noqa: E501
)
fin_str = "".join(lines)
self.assertIn(fin_str, check_str)
self.assertEqual(len(lines), 20)
def test_print_summary_expand_nested(self):
shape = (None, None, 3)
def make_model():
x = inputs = keras.Input(shape)
x = keras.layers.Conv2D(3, 1)(x)
x = keras.layers.BatchNormalization()(x)
return keras.Model(inputs, x)
x = inner_inputs = keras.Input(shape)
x = make_model()(x)
inner_model = keras.Model(inner_inputs, x)
inputs = keras.Input(shape)
model = keras.Model(inputs, inner_model(inputs))
file_name = "model_2.txt"
temp_dir = self.get_temp_dir()
self.addCleanup(shutil.rmtree, temp_dir, ignore_errors=True)
fpath = os.path.join(temp_dir, file_name)
writer = open(fpath, "w")
def print_to_file(text):
print(text, file=writer)
try:
layer_utils.print_summary(
model, print_fn=print_to_file, expand_nested=True
)
self.assertTrue(tf.io.gfile.exists(fpath))
writer.close()
reader = open(fpath, "r")
lines = reader.readlines()
reader.close()
check_str = (
'Model: "model_2"\n'
"_________________________________________________________________\n" # noqa: E501
" Layer (type) Output Shape Param # \n" # noqa: E501
"=================================================================\n" # noqa: E501
" input_3 (InputLayer) [(None, None, None, 3)] 0 \n" # noqa: E501
" \n" # noqa: E501
" model_1 (Functional) (None, None, None, 3) 24 \n" # noqa: E501
"|¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯|\n" # noqa: E501
"| input_1 (InputLayer) [(None, None, None, 3)] 0 |\n" # noqa: E501
"| |\n" # noqa: E501
"| model (Functional) (None, None, None, 3) 24 |\n" # noqa: E501
"||¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯||\n" # noqa: E501
"|| input_2 (InputLayer) [(None, None, None, 3)] 0 ||\n" # noqa: E501
"|| ||\n" # noqa: E501
"|| conv2d (Conv2D) (None, None, None, 3) 12 ||\n" # noqa: E501
"|| ||\n" # noqa: E501
"|| batch_normalization (Bat (None, None, None, 3) 12 ||\n" # noqa: E501
"|| chNormalization) ||\n" # noqa: E501
"|¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯|\n" # noqa: E501
"¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯\n" # noqa: E501
"=================================================================\n" # noqa: E501
"Total params: 24 (96.00 Byte)\n"
"Trainable params: 18 (72.00 Byte)\n"
"Non-trainable params: 6 (24.00 Byte)\n"
"_________________________________________________________________\n" # noqa: E501
)
fin_str = "".join(lines)
self.assertIn(fin_str, check_str)
self.assertEqual(len(lines), 25)
except ImportError:
pass
def test_summary_subclass_model_expand_nested(self):
class Sequential(keras.Model):
def __init__(self, *args):
super().__init__()
self.module_list = list(args) if args else []
def call(self, x):
for module in self.module_list:
x = module(x)
return x
class Block(keras.Model):
def __init__(self):
super().__init__()
self.module = Sequential(
keras.layers.Dense(10),
keras.layers.Dense(10),
)
def call(self, input_tensor):
x = self.module(input_tensor)
return x
class Base(keras.Model):
def __init__(self):
super().__init__()
self.module = Sequential(Block(), Block())
def call(self, input_tensor):
x = self.module(input_tensor)
y = self.module(x)
return x, y
class Network(keras.Model):
def __init__(self):
super().__init__()
self.child = Base()
def call(self, inputs):
return self.child(inputs)
net = Network()
inputs = keras.Input(shape=(10,))
outputs = net(inputs)
model = keras.models.Model(inputs=inputs, outputs=outputs)
file_name = "model_3.txt"
temp_dir = self.get_temp_dir()
self.addCleanup(shutil.rmtree, temp_dir, ignore_errors=True)
fpath = os.path.join(temp_dir, file_name)
writer = open(fpath, "w")
def print_to_file(text):
print(text, file=writer)
try:
layer_utils.print_summary(
model,
line_length=120,
print_fn=print_to_file,
expand_nested=True,
)
self.assertTrue(tf.io.gfile.exists(fpath))
writer.close()
with open(fpath, "r") as reader:
lines = reader.readlines()
# The output content are slightly different for the input shapes
# between v1 and v2.
if tf.__internal__.tf2.enabled():
self.assertEqual(len(lines), 39)
else:
self.assertEqual(len(lines), 40)
except ImportError:
pass
def test_print_summary_show_trainable(self):
model = keras.Sequential(name="trainable")
untrained = keras.layers.Conv2D(
filters=2, kernel_size=(2, 3), input_shape=(3, 5, 5), name="conv"
)
model.add(untrained)
model.add(keras.layers.Flatten(name="flat"))
model.add(keras.layers.Dense(5, name="dense"))
untrained.trainable = False
file_name = "model_4.txt"
temp_dir = self.get_temp_dir()
self.addCleanup(shutil.rmtree, temp_dir, ignore_errors=True)
fpath = os.path.join(temp_dir, file_name)
writer = open(fpath, "w")
def print_to_file(text):
print(text, file=writer)
try:
layer_utils.print_summary(
model, print_fn=print_to_file, show_trainable=True
)
self.assertTrue(tf.io.gfile.exists(fpath))
writer.close()
with open(fpath, "r") as reader:
lines = reader.readlines()
check_str = (
'Model: "trainable"\n'
"____________________________________________________________________________\n" # noqa: E501
" Layer (type) Output Shape Param # Trainable \n" # noqa: E501
"============================================================================\n" # noqa: E501
" conv (Conv2D) (None, 2, 3, 2) 62 N \n" # noqa: E501
" \n" # noqa: E501
" flat (Flatten) (None, 12) 0 Y \n" # noqa: E501
" \n" # noqa: E501
" dense (Dense) (None, 5) 65 Y \n" # noqa: E501
" \n" # noqa: E501
"============================================================================\n" # noqa: E501
"Total params: 127 (508.00 Byte)\n"
"Trainable params: 65 (260.00 Byte)\n"
"Non-trainable params: 62 (248.00 Byte)\n"
"____________________________________________________________________________\n" # noqa: E501
"____________________________________________________________________________\n" # noqa: E501
)
fin_str = "".join(lines)
self.assertIn(fin_str, check_str)
self.assertEqual(len(lines), 15)
except ImportError:
pass
def test_print_summary_expand_nested_show_trainable(self):
shape = (None, None, 3)
def make_model():
x = inputs = keras.Input(shape, name="input2")
untrainable = keras.layers.Conv2D(3, 1)
untrainable.trainable = False
x = untrainable(x)
x = keras.layers.BatchNormalization()(x)
return keras.Model(inputs, x)
x = inner_inputs = keras.Input(shape, name="input1")
x = make_model()(x)
inner_model = keras.Model(inner_inputs, x)
inputs = keras.Input(shape, name="input3")
model = keras.Model(inputs, inner_model(inputs))
file_name = "model_6.txt"
temp_dir = self.get_temp_dir()
self.addCleanup(shutil.rmtree, temp_dir, ignore_errors=True)
fpath = os.path.join(temp_dir, file_name)
writer = open(fpath, "w")
def print_to_file(text):
print(text, file=writer)
try:
layer_utils.print_summary(
model,
print_fn=print_to_file,
expand_nested=True,
show_trainable=True,
)
self.assertTrue(tf.io.gfile.exists(fpath))
writer.close()
with open(fpath, "r") as reader:
lines = reader.readlines()
check_str = (
'Model: "model_2"\n'
"____________________________________________________________________________\n" # noqa: E501
" Layer (type) Output Shape Param # Trainable \n" # noqa: E501
"============================================================================\n" # noqa: E501
" input3 (InputLayer) [(None, None, None, 3)] 0 Y \n" # noqa: E501
" \n" # noqa: E501
" model_1 (Functional) (None, None, None, 3) 24 Y \n" # noqa: E501
"|¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯|\n" # noqa: E501
"| input1 (InputLayer) [(None, None, None, 3)] 0 Y |\n" # noqa: E501
"| |\n" # noqa: E501
"| model (Functional) (None, None, None, 3) 24 Y |\n" # noqa: E501
"||¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯||\n" # noqa: E501
"|| input2 (InputLayer) [(None, None, None, 3)] 0 Y ||\n" # noqa: E501
"|| ||\n" # noqa: E501
"|| conv2d (Conv2D) (None, None, None, 3) 12 N ||\n" # noqa: E501
"|| ||\n" # noqa: E501
"|| batch_normalization (Bat (None, None, None, 3) 12 Y ||\n" # noqa: E501
"|| chNormalization) ||\n" # noqa: E501
"|¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯|\n" # noqa: E501
"¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯\n" # noqa: E501
"============================================================================\n" # noqa: E501
"Total params: 24 (96.00 Byte)\n"
"Trainable params: 6 (24.00 Byte)\n"
"Non-trainable params: 18 (72.00 Byte)\n"
"____________________________________________________________________________\n" # noqa: E501
)
fin_str = "".join(lines)
self.assertIn(fin_str, check_str)
self.assertEqual(len(lines), 25)
except ImportError:
pass
def test_print_summary_layer_range(self):
model = keras.Sequential()
model.add(
keras.layers.Conv2D(
filters=2,
kernel_size=(2, 3),
input_shape=(3, 5, 5),
name="conv",
)
)
model.add(keras.layers.Flatten(name="flat"))
model.add(keras.layers.Dense(5, name="dense"))
file_name = "model_7.txt"
temp_dir = self.get_temp_dir()
self.addCleanup(shutil.rmtree, temp_dir, ignore_errors=True)
fpath = os.path.join(temp_dir, file_name)
writer = open(fpath, "w")
def print_to_file(text):
print(text, file=writer)
try:
layer_utils.print_summary(
model, print_fn=print_to_file, layer_range=["conv", "flat"]
)
self.assertTrue(tf.io.gfile.exists(fpath))
writer.close()
with open(fpath, "r") as reader:
lines = reader.readlines()
# The expected lenght with no layer filter is 15
# we filtered out 2 lines by excluding the layer 'dense'
self.assertEqual(len(lines), 15 - 2)
except ImportError:
pass
def test_print_summary_layer_range_with_expand_nested(self):
shape = (None, None, 3)
def make_model():
x = inputs = keras.Input(shape, name="input_2")
x = keras.layers.Conv2D(3, 1)(x)
x = keras.layers.BatchNormalization()(x)
return keras.Model(inputs, x, name="2nd_inner")
x = inner_inputs = keras.Input(shape, name="input_1")
x = make_model()(x)
inner_model = keras.Model(inner_inputs, x, name="1st_inner")
inputs = keras.Input(shape, name="input_3")
model = keras.Model(inputs, inner_model(inputs))
file_name = "model_8.txt"
temp_dir = self.get_temp_dir()
self.addCleanup(shutil.rmtree, temp_dir, ignore_errors=True)
fpath = os.path.join(temp_dir, file_name)
writer = open(fpath, "w")
def print_to_file(text):
print(text, file=writer)
try:
layer_utils.print_summary(
model,
print_fn=print_to_file,
expand_nested=True,
layer_range=["1st_inner", "1st_inner"],
)
layer_utils.print_summary(
model,
expand_nested=True,
layer_range=["1st_inner", "1st_inner"],
)
self.assertTrue(tf.io.gfile.exists(fpath))
writer.close()
with open(fpath, "r") as reader:
lines = reader.readlines()
check_str = (
'Model: "model"\n'
"_________________________________________________________________\n" # noqa: E501
" Layer (type) Output Shape Param # \n" # noqa: E501
"=================================================================\n" # noqa: E501
" 1st_inner (Functional) (None, None, None, 3) 24 \n" # noqa: E501
"|¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯|\n" # noqa: E501
"| input_1 (InputLayer) [(None, None, None, 3)] 0 |\n" # noqa: E501
"| |\n" # noqa: E501
"| 2nd_inner (Functional) (None, None, None, 3) 24 |\n" # noqa: E501
"||¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯||\n" # noqa: E501
"|| input_2 (InputLayer) [(None, None, None, 3)] 0 ||\n" # noqa: E501
"|| ||\n" # noqa: E501
"|| conv2d (Conv2D) (None, None, None, 3) 12 ||\n" # noqa: E501
"|| ||\n" # noqa: E501
"|| batch_normalization (Bat (None, None, None, 3) 12 ||\n" # noqa: E501
"|| chNormalization) ||\n" # noqa: E501
"|¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯|\n" # noqa: E501
"¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯\n" # noqa: E501
"=================================================================\n" # noqa: E501
"Total params: 24 (96.00 Byte)\n"
"Trainable params: 18 (72.00 Byte)\n"
"Non-trainable params: 6 (24.00 Byte)\n"
"_________________________________________________________________\n" # noqa: E501
)
check_lines = check_str.split("\n")[
:-1
] # Removing final empty string which is not a line
fin_str = "".join(lines)
self.assertIn(fin_str, check_str)
self.assertEqual(len(lines), len(check_lines))
except ImportError:
pass
def test_weight_memory_size(self):
v1 = tf.Variable(tf.zeros(shape=(1, 2), dtype=tf.float32))
v2 = tf.Variable(tf.zeros(shape=(2, 3), dtype=tf.float64))
v3 = tf.Variable(tf.zeros(shape=(4, 5), dtype=tf.int16))
v4 = tf.Variable(tf.zeros(shape=(6,), dtype=tf.uint8))
weights = [v1, v1, v2, v3, v4]
weight_memory_size = layer_utils.weight_memory_size(weights)
expected_memory_size = 1 * 2 * 4 + 2 * 3 * 8 + 4 * 5 * 2 + 6 * 1
self.assertEqual(weight_memory_size, expected_memory_size)
@parameterized.parameters(
(0, "0.00 Byte"),
(1000, "1000.00 Byte"),
(1024, "1.00 KB"),
(1024 * 2 - 1, "2.00 KB"),
(1024 * 2 + 1, "2.00 KB"),
(1024**2 + 1, "1.00 MB"),
(1024**3 - 1, "1024.00 MB"),
(1024**3, "1.00 GB"),
(1024**4, "1.00 TB"),
(1024**5, "1.00 PB"),
(1024**5 * 1.41415, "1.41 PB"),
)
def test_readable_weight_memory_size(self, size, expected_result):
result = layer_utils.readable_memory_size(size)
self.assertEqual(result, expected_result)
def test_property_cache(self):
test_counter = collections.Counter()
class MyObject(tf.__internal__.tracking.AutoTrackable):
def __init__(self):
super().__init__()
self._frozen = True
def __setattr__(self, key, value):
"""Enforce that cache does not set attribute on MyObject."""
if getattr(self, "_frozen", False):
raise ValueError("Cannot mutate when frozen.")
return super().__setattr__(key, value)
@property
@layer_utils.cached_per_instance
def test_property(self):
test_counter[id(self)] += 1
return id(self)
first_object = MyObject()
second_object = MyObject()
# Make sure the objects return the correct values
self.assertEqual(first_object.test_property, id(first_object))
self.assertEqual(second_object.test_property, id(second_object))
# Make sure the cache does not share across objects
self.assertNotEqual(
first_object.test_property, second_object.test_property
)
# Check again (Now the values should be cached.)
self.assertEqual(first_object.test_property, id(first_object))
self.assertEqual(second_object.test_property, id(second_object))
# Count the function calls to make sure the cache is actually being
# used.
self.assertAllEqual(tuple(test_counter.values()), (1, 1))
def test_property_cache_threaded(self):
call_count = collections.Counter()
class MyObject(tf.__internal__.tracking.AutoTrackable):
@property
@layer_utils.cached_per_instance
def test_property(self):
# Random sleeps to ensure that the execution thread changes
# mid-computation.
call_count["test_property"] += 1
time.sleep(np.random.random() + 1.0)
# Use a RandomState which is seeded off the instance's id (the
# mod is because numpy limits the range of seeds) to ensure that
# an instance returns the same value in different threads, but
# different instances return different values.
return int(
np.random.RandomState(id(self) % (2**31)).randint(2**16)
)
def get_test_property(self, _):
"""Function provided to .map for threading test."""
return self.test_property
# Test that multiple threads return the same value. This requires that
# the underlying function is repeatable, as cached_property makes no
# attempt to prioritize the first call.
test_obj = MyObject()
with contextlib.closing(multiprocessing.dummy.Pool(32)) as pool:
# Intentionally make a large pool (even when there are only a small
# number of cpus) to ensure that the runtime switches threads.
results = pool.map(test_obj.get_test_property, range(64))
self.assertEqual(len(set(results)), 1)
# Make sure we actually are testing threaded behavior.
self.assertGreater(call_count["test_property"], 1)
# Make sure new threads still cache hit.
with contextlib.closing(multiprocessing.dummy.Pool(2)) as pool:
start_time = (
timeit.default_timer()
) # Don't time pool instantiation.
results = pool.map(test_obj.get_test_property, range(4))
total_time = timeit.default_timer() - start_time
# Note(taylorrobie): The reason that it is safe to time a unit test is
# that a cache hit will be << 1 second, and a cache miss is guaranteed
# to be >= 1 second. Empirically confirmed by 100,000 runs with no
# flakes.
self.assertLess(total_time, 0.95)
def test_property_cache_serialization(self):
# Reset call count. .keys() must be wrapped in a list, because otherwise
# we would mutate the iterator while iterating.
for k in list(_PICKLEABLE_CALL_COUNT.keys()):
_PICKLEABLE_CALL_COUNT.pop(k)
first_instance = MyPickleableObject()
self.assertEqual(id(first_instance), first_instance.my_id)
# Test that we can pickle and un-pickle
second_instance = pickle.loads(pickle.dumps(first_instance))
self.assertEqual(id(second_instance), second_instance.my_id)
self.assertNotEqual(first_instance.my_id, second_instance.my_id)
# Make sure de-serialized object uses the cache.
self.assertEqual(_PICKLEABLE_CALL_COUNT[second_instance], 1)
# Make sure the decorator cache is not being serialized with the object.
expected_size = len(pickle.dumps(second_instance))
for _ in range(5):
# Add some more entries to the cache.
_ = MyPickleableObject().my_id
self.assertEqual(len(_PICKLEABLE_CALL_COUNT), 7)
size_check_instance = MyPickleableObject()
_ = size_check_instance.my_id
self.assertEqual(expected_size, len(pickle.dumps(size_check_instance)))
def test_warmstart_embedding_matrix_with_list(self):
vocab_base = ["unk", "a", "b", "c"]
vocab_new = ["unk", "unk", "a", "b", "c", "d", "e"]
vectorized_vocab_base = np.random.rand(len(vocab_base), 3)
vectorized_vocab_new = np.random.rand(len(vocab_new), 3)
warmstarted_embedding_matrix = layer_utils.warmstart_embedding_matrix(
base_vocabulary=vocab_base,
new_vocabulary=vocab_new,
base_embeddings=vectorized_vocab_base,
new_embeddings_initializer=keras.initializers.Constant(
vectorized_vocab_new
),
)
self.assertAllEqual(
warmstarted_embedding_matrix[2],
vectorized_vocab_base[1],
)
def test_warmstart_embedding_matrix_with_nparray(self):
vocab_base = np.array(["unk", "a", "b", "c"])
vocab_new = np.array(["unk", "unk", "a", "b", "c", "d", "e"])
vectorized_vocab_base = np.random.rand(len(vocab_base), 3)
vectorized_vocab_new = np.random.rand(len(vocab_new), 3)
warmstarted_embedding_matrix = layer_utils.warmstart_embedding_matrix(
base_vocabulary=vocab_base,
new_vocabulary=vocab_new,
base_embeddings=vectorized_vocab_base,
new_embeddings_initializer=keras.initializers.Constant(
vectorized_vocab_new
),
)
self.assertAllEqual(
warmstarted_embedding_matrix[2],
vectorized_vocab_base[1],
)
@test_utils.run_v2_only
def test_warmstart_embedding_matrix_with_tensor(self):
vocab_base = tf.convert_to_tensor(["unk", "a", "b", "c"])
vocab_new = tf.convert_to_tensor(
["unk", "unk", "a", "b", "c", "d", "e"]
)
vectorized_vocab_base = np.random.rand(vocab_base.shape[0], 3)
vectorized_vocab_new = np.random.rand(vocab_new.shape[0], 3)
warmstarted_embedding_matrix = layer_utils.warmstart_embedding_matrix(
base_vocabulary=vocab_base,
new_vocabulary=vocab_new,
base_embeddings=vectorized_vocab_base,
new_embeddings_initializer=keras.initializers.Constant(
vectorized_vocab_new
),
)
self.assertAllEqual(
warmstarted_embedding_matrix[2],
vectorized_vocab_base[1],
)
def test_warmstart_embedding_matrix_with_file_name(self):
def _write_list_to_file(filename, content_list):
with tf.io.gfile.GFile(filename, "w") as output_file:
for line in content_list:
output_file.write(line + "\n")
vocab_base = ["UNK", "a", "b", "c"]
vocab_base_file = tempfile.mktemp(".tsv")
_write_list_to_file(vocab_base_file, vocab_base)
vocab_new = ["UNK", "UNK", "a", "b", "c", "d", "e"]
vocab_new_file = tempfile.mktemp(".tsv")
vectorized_vocab_base = np.random.rand(len(vocab_base), 3)
vectorized_vocab_new = np.random.rand(len(vocab_new), 3)
_write_list_to_file(vocab_new_file, vocab_new)
warmstarted_embedding_matrix = layer_utils.warmstart_embedding_matrix(
base_vocabulary=vocab_base_file,
new_vocabulary=vocab_new_file,
base_embeddings=vectorized_vocab_base,
new_embeddings_initializer=keras.initializers.Constant(
vectorized_vocab_new
),
)
self.assertAllEqual(
warmstarted_embedding_matrix[3],
vectorized_vocab_base[2],
)
def test_warmstart_default_initialization(self):
def _write_list_to_file(filename, content_list):
with tf.io.gfile.GFile(filename, "w") as output_file:
for line in content_list:
output_file.write(line + "\n")
vocab_base = ["UNK", "a", "b", "c"]
vocab_base_file = tempfile.mktemp(".tsv")
_write_list_to_file(vocab_base_file, vocab_base)
vocab_new = ["UNK", "UNK", "a", "b", "c", "d", "e"]
vocab_new_file = tempfile.mktemp(".tsv")
vectorized_vocab_base = np.random.rand(len(vocab_base), 3)
_write_list_to_file(vocab_new_file, vocab_new)
warmstarted_embedding_matrix = layer_utils.warmstart_embedding_matrix(
base_vocabulary=vocab_base_file,
new_vocabulary=vocab_new_file,
base_embeddings=vectorized_vocab_base,
)
self.assertAllEqual(
warmstarted_embedding_matrix[3],
vectorized_vocab_base[2],
)
def test_warmstart_default_value(self):
vocab_base = np.array(["unk", "a", "b", "c"])
vocab_new = np.array(["unk", "unk", "a", "b", "c", "d", "e"])
vectorized_vocab_base = np.random.rand(len(vocab_base), 3)
warmstarted_embedding_matrix = layer_utils.warmstart_embedding_matrix(
base_vocabulary=vocab_base,
new_vocabulary=vocab_new,
base_embeddings=vectorized_vocab_base,
)
self.assertAllEqual(
warmstarted_embedding_matrix[2],
vectorized_vocab_base[1],
)
def test_warmstart_with_randomuniform_initializer(self):
vocab_base = np.array(["unk", "a", "b", "c"])
vocab_new = np.array(["unk", "unk", "a", "b", "c", "d", "e"])
vectorized_vocab_base = np.random.rand(len(vocab_base), 3)
warmstarted_embedding_matrix = layer_utils.warmstart_embedding_matrix(
base_vocabulary=vocab_base,
new_vocabulary=vocab_new,
base_embeddings=vectorized_vocab_base,
new_embeddings_initializer="RandomUniform",
)
self.assertAllEqual(
warmstarted_embedding_matrix[2],
vectorized_vocab_base[1],
)
def test_warmstart_with_nothing_in_common(self):
vocab_base = np.array(["unk", "a", "b", "c"])
vocab_new = np.array(["d", "e", "f", "g", "h"])
vectorized_vocab_base = np.random.rand(len(vocab_base), 3)
vectorized_vocab_new = np.random.rand(len(vocab_new), 3)
warmstarted_embedding_matrix = layer_utils.warmstart_embedding_matrix(
base_vocabulary=vocab_base,
new_vocabulary=vocab_new,
base_embeddings=vectorized_vocab_base,
new_embeddings_initializer=keras.initializers.Constant(
vectorized_vocab_new
),
)
self.assertAllEqual(
warmstarted_embedding_matrix,
vectorized_vocab_new,
)
def test_warmstart_with_new_vocab_smaller(self):
vocab_base = np.array(["unk", "a", "b", "c"])
vocab_new = np.array(["d", "e", "f", "a"])
vectorized_vocab_base = np.random.rand(len(vocab_base), 3)
warmstarted_embedding_matrix = layer_utils.warmstart_embedding_matrix(
base_vocabulary=vocab_base,
new_vocabulary=vocab_new,
base_embeddings=vectorized_vocab_base,
new_embeddings_initializer="uniform",
)
self.assertAllEqual(
warmstarted_embedding_matrix[3],
vectorized_vocab_base[1],
)
@test_utils.run_v2_only
class DTensorVariableSummaryTest(test_util.DTensorBaseTest):
def setUp(self):
super().setUp()
backend.reset_uids()
backend.enable_tf_random_generator()
tf_utils.set_random_seed(1337)
global_ids = test_util.create_device_ids_array((2, 2))
local_device_ids = np.ravel(global_ids).tolist()
mesh_dict = {
"CPU": dtensor.Mesh(
["batch", "model"],
global_ids,
local_device_ids,
test_util.create_device_list((2, 2), "CPU"),
)
}
self.mesh = self.configTestMesh(mesh_dict)
self.replicated_2d = dtensor.Layout.replicated(self.mesh, rank=2)
self.replicated_1d = dtensor.Layout.replicated(self.mesh, rank=1)
self.sharded_2d = dtensor.Layout(["model", "batch"], self.mesh)
self.sharded_1d = dtensor.Layout(["model"], self.mesh)
def test_model_summary(self):
layout_map = layout_map_lib.LayoutMap(mesh=self.mesh)
layout_map["d1.kernel"] = self.replicated_2d
layout_map["d1.bias"] = self.replicated_1d
layout_map["d2.kernel"] = self.sharded_2d
layout_map["d2.bias"] = self.sharded_1d
with layout_map.scope():
inputs = layers.Input((10,), batch_size=10)
x = layers.Dense(20, name="d1")(inputs)
x = layers.Dropout(0.1)(x)
output = layers.Dense(30, name="d2")(x)
model = keras.Model(inputs, output)
# For dtype = float32, following value are expected from memory stats
expected_result = {}
replicated_var_count = 10 * 20 + 20 # For d1 kernel and bias
model_batch_shard_var_count = 30 * 20 # For d2 kernel
model_shard_var_count = 30 # For d2 bias
expected_result[()] = (replicated_var_count, replicated_var_count * 4)
expected_result[("batch", "model")] = (
model_batch_shard_var_count,
model_batch_shard_var_count * 4,
)
expected_result[("model",)] = (
model_shard_var_count,
model_shard_var_count * 4,
)
expected_total_weight_count = (
replicated_var_count
+ model_batch_shard_var_count
+ model_shard_var_count
)
expected_total_memory_size = expected_total_weight_count * 4
(
total_weight_count,
total_memory_size,
per_sharing_spec_result,
) = layer_utils.dtensor_variable_summary(model.weights)
self.assertEqual(total_weight_count, expected_total_weight_count)
self.assertEqual(total_memory_size, expected_total_memory_size)
self.assertDictEqual(per_sharing_spec_result, expected_result)
output_buffer = io.StringIO()
def print_to_buffer(content):
output_buffer.write(content)
model.summary(print_fn=print_to_buffer)
self.assertRegex(
output_buffer.getvalue(),
f"{replicated_var_count} / {expected_total_weight_count} params "
".* are fully replicated",
)
self.assertRegex(
output_buffer.getvalue(),
f"{model_batch_shard_var_count} / {expected_total_weight_count} "
r"params .* are sharded based on spec .*batch.*model"
r".* across 4 devices",
)
self.assertRegex(
output_buffer.getvalue(),
f"{model_shard_var_count} / {expected_total_weight_count} "
r"params .* are sharded based on spec .*model"
r".* across 2 devices",
)
self.assertIn(
"Overall per device memory usage: 1.50 KB", output_buffer.getvalue()
)
self.assertIn("Overall sharding factor: 2.21", output_buffer.getvalue())
if __name__ == "__main__":
tf.test.main()
|
tf-keras/tf_keras/utils/layer_utils_test.py/0
|
{
"file_path": "tf-keras/tf_keras/utils/layer_utils_test.py",
"repo_id": "tf-keras",
"token_count": 21338
}
| 226 |
# Copyright 2017 The TensorFlow Authors. All Rights Reserved.
#
# Licensed under the Apache License, Version 2.0 (the "License");
# you may not use this file except in compliance with the License.
# You may obtain a copy of the License at
#
# http://www.apache.org/licenses/LICENSE-2.0
#
# Unless required by applicable law or agreed to in writing, software
# distributed under the License is distributed on an "AS IS" BASIS,
# WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.
# See the License for the specific language governing permissions and
# limitations under the License.
# ==============================================================================
"""TFDecorator-aware replacements for the contextlib module."""
import contextlib as _contextlib
import tensorflow.compat.v2 as tf
def contextmanager(target):
"""A tf_decorator-aware wrapper for `contextlib.contextmanager`.
Usage is identical to `contextlib.contextmanager`.
Args:
target: A callable to be wrapped in a contextmanager.
Returns:
A callable that can be used inside of a `with` statement.
"""
context_manager = _contextlib.contextmanager(target)
return tf.__internal__.decorator.make_decorator(
target, context_manager, "contextmanager"
)
|
tf-keras/tf_keras/utils/tf_contextlib.py/0
|
{
"file_path": "tf-keras/tf_keras/utils/tf_contextlib.py",
"repo_id": "tf-keras",
"token_count": 364
}
| 227 |
# Copyright 2020 The AutoKeras Authors.
#
# Licensed under the Apache License, Version 2.0 (the "License");
# you may not use this file except in compliance with the License.
# You may obtain a copy of the License at
#
# http://www.apache.org/licenses/LICENSE-2.0
#
# Unless required by applicable law or agreed to in writing, software
# distributed under the License is distributed on an "AS IS" BASIS,
# WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.
# See the License for the specific language governing permissions and
# limitations under the License.
import tensorflow as tf
from tensorflow import keras
from autokeras.blocks.basic import BertBlock
from autokeras.blocks.basic import ConvBlock
from autokeras.blocks.basic import DenseBlock
from autokeras.blocks.basic import EfficientNetBlock
from autokeras.blocks.basic import Embedding
from autokeras.blocks.basic import ResNetBlock
from autokeras.blocks.basic import RNNBlock
from autokeras.blocks.basic import Transformer
from autokeras.blocks.basic import XceptionBlock
from autokeras.blocks.heads import ClassificationHead
from autokeras.blocks.heads import RegressionHead
from autokeras.blocks.heads import SegmentationHead
from autokeras.blocks.preprocessing import CategoricalToNumerical
from autokeras.blocks.preprocessing import ImageAugmentation
from autokeras.blocks.preprocessing import Normalization
from autokeras.blocks.preprocessing import TextToIntSequence
from autokeras.blocks.preprocessing import TextToNgramVector
from autokeras.blocks.reduction import Flatten
from autokeras.blocks.reduction import Merge
from autokeras.blocks.reduction import SpatialReduction
from autokeras.blocks.reduction import TemporalReduction
from autokeras.blocks.wrapper import GeneralBlock
from autokeras.blocks.wrapper import ImageBlock
from autokeras.blocks.wrapper import StructuredDataBlock
from autokeras.blocks.wrapper import TextBlock
from autokeras.blocks.wrapper import TimeseriesBlock
from autokeras.utils import utils
def serialize(obj):
return utils.serialize_keras_object(obj)
def deserialize(config, custom_objects=None):
return utils.deserialize_keras_object(
config,
module_objects=globals(),
custom_objects=custom_objects,
printable_module_name="hypermodels",
)
|
autokeras/autokeras/blocks/__init__.py/0
|
{
"file_path": "autokeras/autokeras/blocks/__init__.py",
"repo_id": "autokeras",
"token_count": 652
}
| 0 |
# Copyright 2020 The AutoKeras Authors.
#
# Licensed under the Apache License, Version 2.0 (the "License");
# you may not use this file except in compliance with the License.
# You may obtain a copy of the License at
#
# http://www.apache.org/licenses/LICENSE-2.0
#
# Unless required by applicable law or agreed to in writing, software
# distributed under the License is distributed on an "AS IS" BASIS,
# WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.
# See the License for the specific language governing permissions and
# limitations under the License.
from tensorflow import nest
from autokeras.engine import named_hypermodel
from autokeras.engine import node as node_module
class Block(named_hypermodel.NamedHyperModel):
"""The base class for different Block.
The Block can be connected together to build the search space for an
AutoModel. Notably, many args in the __init__ function are defaults to be a
tunable variable when not specified by the user.
"""
def __init__(self, **kwargs):
super().__init__(**kwargs)
self.inputs = None
self.outputs = None
self._num_output_node = 1
def _build_wrapper(self, hp, *args, **kwargs):
with hp.name_scope(self.name):
return super()._build_wrapper(hp, *args, **kwargs)
def __call__(self, inputs):
"""Functional API.
# Arguments
inputs: A list of input node(s) or a single input node for the
block.
# Returns
list: A list of output node(s) of the Block.
"""
self.inputs = nest.flatten(inputs)
for input_node in self.inputs:
if not isinstance(input_node, node_module.Node):
raise TypeError(
"Expect the inputs to block {name} to be "
"a Node, but got {type}.".format(
name=self.name, type=type(input_node)
)
)
input_node.add_out_block(self)
self.outputs = []
for _ in range(self._num_output_node):
output_node = node_module.Node()
output_node.add_in_block(self)
self.outputs.append(output_node)
return self.outputs
def build(self, hp, inputs=None):
"""Build the Block into a real Keras Model.
The subclasses should override this function and return the output node.
# Arguments
hp: HyperParameters. The hyperparameters for building the model.
inputs: A list of input node(s).
"""
raise NotImplementedError
|
autokeras/autokeras/engine/block.py/0
|
{
"file_path": "autokeras/autokeras/engine/block.py",
"repo_id": "autokeras",
"token_count": 1054
}
| 1 |
# Copyright 2020 The AutoKeras Authors.
#
# Licensed under the Apache License, Version 2.0 (the "License");
# you may not use this file except in compliance with the License.
# You may obtain a copy of the License at
#
# http://www.apache.org/licenses/LICENSE-2.0
#
# Unless required by applicable law or agreed to in writing, software
# distributed under the License is distributed on an "AS IS" BASIS,
# WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.
# See the License for the specific language governing permissions and
# limitations under the License.
import numpy as np
from autokeras.engine import preprocessor
class PostProcessor(preprocessor.TargetPreprocessor):
def transform(self, dataset):
return dataset
class SigmoidPostprocessor(PostProcessor):
"""Postprocessor for sigmoid outputs."""
def postprocess(self, data):
"""Transform probabilities to zeros and ones.
# Arguments
data: numpy.ndarray. The output probabilities of the classification
head.
# Returns
numpy.ndarray. The zeros and ones predictions.
"""
data[data < 0.5] = 0
data[data > 0.5] = 1
return data
class SoftmaxPostprocessor(PostProcessor):
"""Postprocessor for softmax outputs."""
def postprocess(self, data):
"""Transform probabilities to zeros and ones.
# Arguments
data: numpy.ndarray. The output probabilities of the classification
head.
# Returns
numpy.ndarray. The zeros and ones predictions.
"""
idx = np.argmax(data, axis=-1)
data = np.zeros(data.shape)
data[np.arange(data.shape[0]), idx] = 1
return data
|
autokeras/autokeras/preprocessors/postprocessors.py/0
|
{
"file_path": "autokeras/autokeras/preprocessors/postprocessors.py",
"repo_id": "autokeras",
"token_count": 637
}
| 2 |
# Copyright 2020 The AutoKeras Authors.
#
# Licensed under the Apache License, Version 2.0 (the "License");
# you may not use this file except in compliance with the License.
# You may obtain a copy of the License at
#
# http://www.apache.org/licenses/LICENSE-2.0
#
# Unless required by applicable law or agreed to in writing, software
# distributed under the License is distributed on an "AS IS" BASIS,
# WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.
# See the License for the specific language governing permissions and
# limitations under the License.
import pathlib
from typing import Dict
from typing import List
from typing import Optional
from typing import Type
from typing import Union
import pandas as pd
import tensorflow as tf
from tensorflow import nest
from autokeras import auto_model
from autokeras import blocks
from autokeras import nodes as input_module
from autokeras.engine import tuner
from autokeras.tuners import task_specific
from autokeras.utils import types
class BaseStructuredDataPipeline(auto_model.AutoModel):
def __init__(self, inputs, outputs, **kwargs):
self.check(inputs.column_names, inputs.column_types)
super().__init__(inputs=inputs, outputs=outputs, **kwargs)
self._target_col_name = None
@staticmethod
def _read_from_csv(x, y):
df = pd.read_csv(x)
target = df.pop(y).to_numpy()
return df, target
def check(self, column_names, column_types):
if column_types:
for column_type in column_types.values():
if column_type not in ["categorical", "numerical"]:
raise ValueError(
'column_types should be either "categorical" '
'or "numerical", but got {name}'.format(
name=column_type
)
)
def check_in_fit(self, x):
input_node = nest.flatten(self.inputs)[0]
# Extract column_names from pd.DataFrame.
if isinstance(x, pd.DataFrame) and input_node.column_names is None:
input_node.column_names = list(x.columns)
if input_node.column_names and input_node.column_types:
for column_name in input_node.column_types:
if column_name not in input_node.column_names:
raise ValueError(
"column_names and column_types are "
"mismatched. Cannot find column name "
"{name} in the data.".format(name=column_name)
)
def read_for_predict(self, x):
if isinstance(x, str):
x = pd.read_csv(x)
if self._target_col_name in x:
x.pop(self._target_col_name)
return x
def fit(
self,
x=None,
y=None,
epochs=None,
callbacks=None,
validation_split=0.2,
validation_data=None,
**kwargs
):
"""Search for the best model and hyperparameters for the AutoModel.
# Arguments
x: String, numpy.ndarray, pandas.DataFrame or tensorflow.Dataset.
Training data x. If the data is from a csv file, it should be a
string specifying the path of the csv file of the training data.
y: String, numpy.ndarray, or tensorflow.Dataset. Training data y.
If the data is from a csv file, it should be a string, which is
the name of the target column. Otherwise, it can be
single-column or multi-column. The values should all be
numerical.
epochs: Int. The number of epochs to train each model during the
search. If unspecified, we would use epochs equal to 1000 and
early stopping with patience equal to 10.
callbacks: List of Keras callbacks to apply during training and
validation.
validation_split: Float between 0 and 1. Defaults to 0.2. Fraction
of the training data to be used as validation data. The model
will set apart this fraction of the training data, will not
train on it, and will evaluate the loss and any model metrics on
this data at the end of each epoch. The validation data is
selected from the last samples in the `x` and `y` data provided,
before shuffling. This argument is not supported when `x` is a
dataset. The best model found would be fit on the entire
dataset including the validation data.
validation_data: Data on which to evaluate the loss and any model
metrics at the end of each epoch. The model will not be trained
on this data. `validation_data` will override
`validation_split`. The type of the validation data should be
the same as the training data. The best model found would be
fit on the training dataset without the validation data.
**kwargs: Any arguments supported by
[keras.Model.fit](https://www.tensorflow.org/api_docs/python/tf/keras/Model#fit).
# Returns
history: A Keras History object corresponding to the best model.
Its History.history attribute is a record of training
loss values and metrics values at successive epochs, as well as
validation loss values and validation metrics values (if
applicable).
"""
# x is file path of training data
if isinstance(x, str):
self._target_col_name = y
x, y = self._read_from_csv(x, y)
if validation_data and not isinstance(validation_data, tf.data.Dataset):
x_val, y_val = validation_data
if isinstance(x_val, str):
validation_data = self._read_from_csv(x_val, y_val)
self.check_in_fit(x)
history = super().fit(
x=x,
y=y,
epochs=epochs,
callbacks=callbacks,
validation_split=validation_split,
validation_data=validation_data,
**kwargs
)
return history
def predict(self, x, **kwargs):
"""Predict the output for a given testing data.
# Arguments
x: String, numpy.ndarray, pandas.DataFrame or tensorflow.Dataset.
Testing data x. If the data is from a csv file, it should be a
string specifying the path of the csv file of the testing data.
**kwargs: Any arguments supported by keras.Model.predict.
# Returns
A list of numpy.ndarray objects or a single numpy.ndarray.
The predicted results.
"""
x = self.read_for_predict(x)
return super().predict(x=x, **kwargs)
def evaluate(self, x, y=None, **kwargs):
"""Evaluate the best model for the given data.
# Arguments
x: String, numpy.ndarray, pandas.DataFrame or tensorflow.Dataset.
Testing data x. If the data is from a csv file, it should be a
string specifying the path of the csv file of the testing data.
y: String, numpy.ndarray, or tensorflow.Dataset. Testing data y.
If the data is from a csv file, it should be a string
corresponding to the label column.
**kwargs: Any arguments supported by keras.Model.evaluate.
# Returns
Scalar test loss (if the model has a single output and no metrics)
or list of scalars (if the model has multiple outputs and/or
metrics). The attribute model.metrics_names will give you the
display labels for the scalar outputs.
"""
if isinstance(x, str):
x, y = self._read_from_csv(x, y)
return super().evaluate(x=x, y=y, **kwargs)
class SupervisedStructuredDataPipeline(BaseStructuredDataPipeline):
def __init__(self, outputs, column_names, column_types, **kwargs):
inputs = input_module.StructuredDataInput(
column_names=column_names, column_types=column_types
)
super().__init__(inputs=inputs, outputs=outputs, **kwargs)
class StructuredDataClassifier(SupervisedStructuredDataPipeline):
"""AutoKeras structured data classification class.
# Arguments
column_names: A list of strings specifying the names of the columns. The
length of the list should be equal to the number of columns of the
data excluding the target column. Defaults to None. If None, it will
obtained from the header of the csv file or the pandas.DataFrame.
column_types: Dict. The keys are the column names. The values should
either be 'numerical' or 'categorical', indicating the type of that
column. Defaults to None. If not None, the column_names need to be
specified. If None, it will be inferred from the data.
num_classes: Int. Defaults to None. If None, it will be inferred from
the data.
multi_label: Boolean. Defaults to False.
loss: A Keras loss function. Defaults to use 'binary_crossentropy' or
'categorical_crossentropy' based on the number of classes.
metrics: A list of Keras metrics. Defaults to use 'accuracy'.
project_name: String. The name of the AutoModel. Defaults to
'structured_data_classifier'.
max_trials: Int. The maximum number of different Keras Models to try.
The search may finish before reaching the max_trials. Defaults to
100.
directory: String. The path to a directory for storing the search
outputs. Defaults to None, which would create a folder with the
name of the AutoModel in the current directory.
objective: String. Name of model metric to minimize
or maximize. Defaults to 'val_accuracy'.
tuner: String or subclass of AutoTuner. If string, it should be one of
'greedy', 'bayesian', 'hyperband' or 'random'. It can also be a
subclass of AutoTuner. If left unspecified, it uses a task specific
tuner, which first evaluates the most commonly used models for the
task before exploring other models.
overwrite: Boolean. Defaults to `False`. If `False`, reloads an existing
project of the same name if one is found. Otherwise, overwrites the
project.
seed: Int. Random seed.
max_model_size: Int. Maximum number of scalars in the parameters of a
model. Models larger than this are rejected.
**kwargs: Any arguments supported by AutoModel.
"""
def __init__(
self,
column_names: Optional[List[str]] = None,
column_types: Optional[Dict] = None,
num_classes: Optional[int] = None,
multi_label: bool = False,
loss: Optional[types.LossType] = None,
metrics: Optional[types.MetricsType] = None,
project_name: str = "structured_data_classifier",
max_trials: int = 100,
directory: Optional[Union[str, pathlib.Path]] = None,
objective: str = "val_accuracy",
tuner: Union[str, Type[tuner.AutoTuner]] = None,
overwrite: bool = False,
seed: Optional[int] = None,
max_model_size: Optional[int] = None,
**kwargs
):
if tuner is None:
tuner = task_specific.StructuredDataClassifierTuner
super().__init__(
outputs=blocks.ClassificationHead(
num_classes=num_classes,
multi_label=multi_label,
loss=loss,
metrics=metrics,
),
column_names=column_names,
column_types=column_types,
max_trials=max_trials,
directory=directory,
project_name=project_name,
objective=objective,
tuner=tuner,
overwrite=overwrite,
seed=seed,
max_model_size=max_model_size,
**kwargs
)
def fit(
self,
x=None,
y=None,
epochs=None,
callbacks=None,
validation_split=0.2,
validation_data=None,
**kwargs
):
"""Search for the best model and hyperparameters for the AutoModel.
# Arguments
x: String, numpy.ndarray, pandas.DataFrame or tensorflow.Dataset.
Training data x. If the data is from a csv file, it should be a
string specifying the path of the csv file of the training data.
y: String, numpy.ndarray, or tensorflow.Dataset. Training data y.
If the data is from a csv file, it should be a string, which is
the name of the target column. Otherwise, It can be raw labels,
one-hot encoded if more than two classes, or binary encoded for
binary classification.
epochs: Int. The number of epochs to train each model during the
search. If unspecified, we would use epochs equal to 1000 and
early stopping with patience equal to 10.
callbacks: List of Keras callbacks to apply during training and
validation.
validation_split: Float between 0 and 1. Defaults to 0.2. Fraction
of the training data to be used as validation data. The model
will set apart this fraction of the training data, will not
train on it, and will evaluate the loss and any model metrics on
this data at the end of each epoch. The validation data is
selected from the last samples in the `x` and `y` data provided,
before shuffling. This argument is not supported when `x` is a
dataset.
validation_data: Data on which to evaluate the loss and any model
metrics at the end of each epoch. The model will not be trained
on this data. `validation_data` will override
`validation_split`. The type of the validation data should be
the same as the training data.
**kwargs: Any arguments supported by
[keras.Model.fit](https://www.tensorflow.org/api_docs/python/tf/keras/Model#fit).
# Returns
history: A Keras History object corresponding to the best model.
Its History.history attribute is a record of training
loss values and metrics values at successive epochs, as well as
validation loss values and validation metrics values (if
applicable).
"""
history = super().fit(
x=x,
y=y,
epochs=epochs,
callbacks=callbacks,
validation_split=validation_split,
validation_data=validation_data,
**kwargs
)
return history
class StructuredDataRegressor(SupervisedStructuredDataPipeline):
"""AutoKeras structured data regression class.
# Arguments
column_names: A list of strings specifying the names of the columns. The
length of the list should be equal to the number of columns of the
data excluding the target column. Defaults to None. If None, it will
obtained from the header of the csv file or the pandas.DataFrame.
column_types: Dict. The keys are the column names. The values should
either be 'numerical' or 'categorical', indicating the type of that
column. Defaults to None. If not None, the column_names need to be
specified. If None, it will be inferred from the data.
output_dim: Int. The number of output dimensions. Defaults to None.
If None, it will be inferred from the data.
loss: A Keras loss function. Defaults to use 'mean_squared_error'.
metrics: A list of Keras metrics. Defaults to use 'mean_squared_error'.
project_name: String. The name of the AutoModel. Defaults to
'structured_data_regressor'.
max_trials: Int. The maximum number of different Keras Models to try.
The search may finish before reaching the max_trials. Defaults to
100.
directory: String. The path to a directory for storing the search
outputs. Defaults to None, which would create a folder with the
name of the AutoModel in the current directory.
objective: String. Name of model metric to minimize
or maximize, e.g. 'val_accuracy'. Defaults to 'val_loss'.
tuner: String or subclass of AutoTuner. If string, it should be one of
'greedy', 'bayesian', 'hyperband' or 'random'. It can also be a
subclass of AutoTuner. If left unspecified, it uses a task specific
tuner, which first evaluates the most commonly used models for the
task before exploring other models.
overwrite: Boolean. Defaults to `False`. If `False`, reloads an existing
project of the same name if one is found. Otherwise, overwrites the
project.
seed: Int. Random seed.
max_model_size: Int. Maximum number of scalars in the parameters of a
model. Models larger than this are rejected.
**kwargs: Any arguments supported by AutoModel.
"""
def __init__(
self,
column_names: Optional[List[str]] = None,
column_types: Optional[Dict[str, str]] = None,
output_dim: Optional[int] = None,
loss: types.LossType = "mean_squared_error",
metrics: Optional[types.MetricsType] = None,
project_name: str = "structured_data_regressor",
max_trials: int = 100,
directory: Union[str, pathlib.Path, None] = None,
objective: str = "val_loss",
tuner: Union[str, Type[tuner.AutoTuner]] = None,
overwrite: bool = False,
seed: Optional[int] = None,
max_model_size: Optional[int] = None,
**kwargs
):
if tuner is None:
tuner = task_specific.StructuredDataRegressorTuner
super().__init__(
outputs=blocks.RegressionHead(
output_dim=output_dim, loss=loss, metrics=metrics
),
column_names=column_names,
column_types=column_types,
max_trials=max_trials,
directory=directory,
project_name=project_name,
objective=objective,
tuner=tuner,
overwrite=overwrite,
seed=seed,
max_model_size=max_model_size,
**kwargs
)
|
autokeras/autokeras/tasks/structured_data.py/0
|
{
"file_path": "autokeras/autokeras/tasks/structured_data.py",
"repo_id": "autokeras",
"token_count": 7954
}
| 3 |
# Copyright 2020 The AutoKeras Authors.
#
# Licensed under the Apache License, Version 2.0 (the "License");
# you may not use this file except in compliance with the License.
# You may obtain a copy of the License at
#
# http://www.apache.org/licenses/LICENSE-2.0
#
# Unless required by applicable law or agreed to in writing, software
# distributed under the License is distributed on an "AS IS" BASIS,
# WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.
# See the License for the specific language governing permissions and
# limitations under the License.
import copy
import keras_tuner
import autokeras as ak
from autokeras.tuners import task_specific
def test_img_clf_init_hp0_equals_hp_of_a_model(tmp_path):
clf = ak.ImageClassifier(directory=tmp_path)
clf.inputs[0].shape = (32, 32, 3)
clf.outputs[0].in_blocks[0].shape = (10,)
init_hp = task_specific.IMAGE_CLASSIFIER[0]
hp = keras_tuner.HyperParameters()
hp.values = copy.copy(init_hp)
clf.tuner.hypermodel.build(hp)
assert set(init_hp.keys()) == set(hp._hps.keys())
def test_img_clf_init_hp1_equals_hp_of_a_model(tmp_path):
clf = ak.ImageClassifier(directory=tmp_path)
clf.inputs[0].shape = (32, 32, 3)
clf.outputs[0].in_blocks[0].shape = (10,)
init_hp = task_specific.IMAGE_CLASSIFIER[1]
hp = keras_tuner.HyperParameters()
hp.values = copy.copy(init_hp)
clf.tuner.hypermodel.build(hp)
assert set(init_hp.keys()) == set(hp._hps.keys())
def test_img_clf_init_hp2_equals_hp_of_a_model(tmp_path):
clf = ak.ImageClassifier(directory=tmp_path)
clf.inputs[0].shape = (32, 32, 3)
clf.outputs[0].in_blocks[0].shape = (10,)
init_hp = task_specific.IMAGE_CLASSIFIER[2]
hp = keras_tuner.HyperParameters()
hp.values = copy.copy(init_hp)
clf.tuner.hypermodel.build(hp)
assert set(init_hp.keys()) == set(hp._hps.keys())
def test_txt_clf_init_hp2_equals_hp_of_a_model(tmp_path):
clf = ak.TextClassifier(directory=tmp_path)
clf.inputs[0].shape = (1,)
clf.inputs[0].batch_size = 6
clf.inputs[0].num_samples = 1000
clf.outputs[0].in_blocks[0].shape = (10,)
clf.tuner.hypermodel.epochs = 1000
clf.tuner.hypermodel.num_samples = 20000
init_hp = task_specific.TEXT_CLASSIFIER[2]
hp = keras_tuner.HyperParameters()
hp.values = copy.copy(init_hp)
clf.tuner.hypermodel.build(hp)
assert set(init_hp.keys()) == set(hp._hps.keys())
def test_txt_clf_init_hp1_equals_hp_of_a_model(tmp_path):
clf = ak.TextClassifier(directory=tmp_path)
clf.inputs[0].shape = (1,)
clf.outputs[0].in_blocks[0].shape = (10,)
init_hp = task_specific.TEXT_CLASSIFIER[1]
hp = keras_tuner.HyperParameters()
hp.values = copy.copy(init_hp)
clf.tuner.hypermodel.build(hp)
assert set(init_hp.keys()) == set(hp._hps.keys())
def test_txt_clf_init_hp0_equals_hp_of_a_model(tmp_path):
clf = ak.TextClassifier(directory=tmp_path)
clf.inputs[0].shape = (1,)
clf.outputs[0].in_blocks[0].shape = (10,)
init_hp = task_specific.TEXT_CLASSIFIER[0]
hp = keras_tuner.HyperParameters()
hp.values = copy.copy(init_hp)
clf.tuner.hypermodel.build(hp)
assert set(init_hp.keys()) == set(hp._hps.keys())
def test_sd_clf_init_hp0_equals_hp_of_a_model(tmp_path):
clf = ak.StructuredDataClassifier(
directory=tmp_path,
column_names=["a", "b"],
column_types={"a": "numerical", "b": "numerical"},
)
clf.inputs[0].shape = (2,)
clf.outputs[0].in_blocks[0].shape = (10,)
init_hp = task_specific.STRUCTURED_DATA_CLASSIFIER[0]
hp = keras_tuner.HyperParameters()
hp.values = copy.copy(init_hp)
clf.tuner.hypermodel.build(hp)
assert set(init_hp.keys()) == set(hp._hps.keys())
def test_sd_reg_init_hp0_equals_hp_of_a_model(tmp_path):
clf = ak.StructuredDataRegressor(
directory=tmp_path,
column_names=["a", "b"],
column_types={"a": "numerical", "b": "numerical"},
)
clf.inputs[0].shape = (2,)
clf.outputs[0].in_blocks[0].shape = (10,)
init_hp = task_specific.STRUCTURED_DATA_REGRESSOR[0]
hp = keras_tuner.HyperParameters()
hp.values = copy.copy(init_hp)
clf.tuner.hypermodel.build(hp)
assert set(init_hp.keys()) == set(hp._hps.keys())
|
autokeras/autokeras/tuners/task_specific_test.py/0
|
{
"file_path": "autokeras/autokeras/tuners/task_specific_test.py",
"repo_id": "autokeras",
"token_count": 1843
}
| 4 |
# Copyright 2020 The AutoKeras Authors.
#
# Licensed under the Apache License, Version 2.0 (the "License");
# you may not use this file except in compliance with the License.
# You may obtain a copy of the License at
#
# http://www.apache.org/licenses/LICENSE-2.0
#
# Unless required by applicable law or agreed to in writing, software
# distributed under the License is distributed on an "AS IS" BASIS,
# WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.
# See the License for the specific language governing permissions and
# limitations under the License.
import numpy as np
import pandas as pd
import sklearn
import tensorflow as tf
import autokeras as ak
from benchmark.experiments import experiment
class StructuredDataClassifierExperiment(experiment.Experiment):
def get_auto_model(self):
return ak.StructuredDataClassifier(
max_trials=10, directory=self.tmp_dir, overwrite=True
)
class Titanic(StructuredDataClassifierExperiment):
def __init__(self):
super().__init__(name="Titanic")
@staticmethod
def load_data():
TRAIN_DATA_URL = (
"https://storage.googleapis.com/tf-datasets/titanic/train.csv"
)
TEST_DATA_URL = (
"https://storage.googleapis.com/tf-datasets/titanic/eval.csv"
)
x_train = tf.keras.utils.get_file("titanic_train.csv", TRAIN_DATA_URL)
x_test = tf.keras.utils.get_file("titanic_eval.csv", TEST_DATA_URL)
return (x_train, "survived"), (x_test, "survived")
class Iris(StructuredDataClassifierExperiment):
def __init__(self):
super().__init__(name="Iris")
@staticmethod
def load_data():
# Prepare the dataset.
TRAIN_DATA_URL = (
"https://storage.googleapis.com/"
"download.tensorflow.org/data/iris_training.csv"
)
x_train = tf.keras.utils.get_file("iris_train.csv", TRAIN_DATA_URL)
TEST_DATA_URL = (
"https://storage.googleapis.com/"
"download.tensorflow.org/data/iris_test.csv"
)
x_test = tf.keras.utils.get_file("iris_test.csv", TEST_DATA_URL)
return (x_train, "virginica"), (x_test, "virginica")
class Wine(StructuredDataClassifierExperiment):
def __init__(self):
super().__init__(name="Wine")
@staticmethod
def load_data():
DATASET_URL = (
"https://archive.ics.uci.edu/ml/"
"machine-learning-databases/wine/wine.data"
)
# save data
dataset = tf.keras.utils.get_file("wine.csv", DATASET_URL)
data = pd.read_csv(dataset, header=None).sample(frac=1, random_state=5)
split_length = int(data.shape[0] * 0.8) # 141
return (data.iloc[:split_length, 1:], data.iloc[:split_length, 0]), (
data.iloc[split_length:, 1:],
data.iloc[split_length:, 0],
)
class StructuredDataRegressorExperiment(experiment.Experiment):
def get_auto_model(self):
return ak.StructuredDataRegressor(
max_trials=10, directory=self.tmp_dir, overwrite=True
)
class CaliforniaHousing(StructuredDataRegressorExperiment):
@staticmethod
def load_data():
house_dataset = sklearn.datasets.fetch_california_housing()
(
x_train,
x_test,
y_train,
y_test,
) = sklearn.model_selection.train_test_split(
house_dataset.data,
np.array(house_dataset.target),
test_size=0.2,
random_state=42,
)
return (x_train, y_train), (x_test, y_test)
|
autokeras/benchmark/experiments/structured_data.py/0
|
{
"file_path": "autokeras/benchmark/experiments/structured_data.py",
"repo_id": "autokeras",
"token_count": 1586
}
| 5 |
<jupyter_start><jupyter_code>!pip install autokeras
import tensorflow as tf
from tensorflow.keras.datasets import mnist
import autokeras as ak<jupyter_output><empty_output><jupyter_text>To make this tutorial easy to follow, we just treat MNIST dataset as aregression dataset. It means we will treat prediction targets of MNIST dataset,which are integers ranging from 0 to 9 as numerical values, so that they can bedirectly used as the regression targets. A Simple ExampleThe first step is to prepare your data. Here we use the MNIST dataset as anexample<jupyter_code>(x_train, y_train), (x_test, y_test) = mnist.load_data()
x_train = x_train[:100]
y_train = y_train[:100]
print(x_train.shape) # (60000, 28, 28)
print(y_train.shape) # (60000,)
print(y_train[:3]) # array([7, 2, 1], dtype=uint8)<jupyter_output><empty_output><jupyter_text>The second step is to run the ImageRegressor. It is recommended have moretrials for more complicated datasets. This is just a quick demo of MNIST, sowe set max_trials to 1. For the same reason, we set epochs to 2. You can alsoleave the epochs unspecified for an adaptive number of epochs.<jupyter_code># Initialize the image regressor.
reg = ak.ImageRegressor(overwrite=True, max_trials=1)
# Feed the image regressor with training data.
reg.fit(x_train, y_train, epochs=2)
# Predict with the best model.
predicted_y = reg.predict(x_test)
print(predicted_y)
# Evaluate the best model with testing data.
print(reg.evaluate(x_test, y_test))<jupyter_output><empty_output><jupyter_text>Validation DataBy default, AutoKeras use the last 20% of training data as validation data. Asshown in the example below, you can use validation_split to specify thepercentage.<jupyter_code>reg.fit(
x_train,
y_train,
# Split the training data and use the last 15% as validation data.
validation_split=0.15,
epochs=2,
)<jupyter_output><empty_output><jupyter_text>You can also use your own validation set instead of splitting it from thetraining data with validation_data.<jupyter_code>split = 50000
x_val = x_train[split:]
y_val = y_train[split:]
x_train = x_train[:split]
y_train = y_train[:split]
reg.fit(
x_train,
y_train,
# Use your own validation set.
validation_data=(x_val, y_val),
epochs=2,
)<jupyter_output><empty_output><jupyter_text>Customized Search SpaceFor advanced users, you may customize your search space by using AutoModelinstead of ImageRegressor. You can configure the ImageBlock for some high-levelconfigurations, e.g., block_type for the type of neural network to search,normalize for whether to do data normalization, augment for whether to do dataaugmentation. You can also do not specify these arguments, which would leavethe different choices to be tuned automatically. See the following example fordetail.<jupyter_code>input_node = ak.ImageInput()
output_node = ak.ImageBlock(
# Only search ResNet architectures.
block_type="resnet",
# Normalize the dataset.
normalize=False,
# Do not do data augmentation.
augment=False,
)(input_node)
output_node = ak.RegressionHead()(output_node)
reg = ak.AutoModel(
inputs=input_node, outputs=output_node, overwrite=True, max_trials=1
)
reg.fit(x_train, y_train, epochs=2)<jupyter_output><empty_output><jupyter_text>The usage of AutoModel is similar to the functional API of Keras. Basically,you are building a graph, whose edges are blocks and the nodes are intermediateoutputs of blocks. To add an edge from input_node to output_node withoutput_node = ak.[some_block]([block_args])(input_node).You can even also use more fine grained blocks to customize the search spaceeven further. See the following example.<jupyter_code>input_node = ak.ImageInput()
output_node = ak.Normalization()(input_node)
output_node = ak.ImageAugmentation(horizontal_flip=False)(output_node)
output_node = ak.ResNetBlock(version="v2")(output_node)
output_node = ak.RegressionHead()(output_node)
reg = ak.AutoModel(
inputs=input_node, outputs=output_node, overwrite=True, max_trials=1
)
reg.fit(x_train, y_train, epochs=2)<jupyter_output><empty_output><jupyter_text>Data FormatThe AutoKeras ImageRegressor is quite flexible for the data format.For the image, it accepts data formats both with and without the channeldimension. The images in the MNIST dataset do not have the channel dimension.Each image is a matrix with shape (28, 28). AutoKeras also accepts images ofthree dimensions with the channel dimension at last, e.g., (32, 32, 3), (28,28, 1).For the regression targets, it should be a vector of numerical values.AutoKeras accepts numpy.ndarray.We also support using tf.data.Dataset format for the training data. In thiscase, the images would have to be 3-dimentional.<jupyter_code>(x_train, y_train), (x_test, y_test) = mnist.load_data()
# Reshape the images to have the channel dimension.
x_train = x_train.reshape(x_train.shape + (1,))
x_test = x_test.reshape(x_test.shape + (1,))
y_train = y_train.reshape(y_train.shape + (1,))
y_test = y_test.reshape(y_test.shape + (1,))
print(x_train.shape) # (60000, 28, 28, 1)
print(y_train.shape) # (60000, 10)
train_set = tf.data.Dataset.from_tensor_slices(((x_train,), (y_train,)))
test_set = tf.data.Dataset.from_tensor_slices(((x_test,), (y_test,)))
reg = ak.ImageRegressor(overwrite=True, max_trials=1)
# Feed the tensorflow Dataset to the regressor.
reg.fit(train_set, epochs=2)
# Predict with the best model.
predicted_y = reg.predict(test_set)
# Evaluate the best model with testing data.
print(reg.evaluate(test_set))<jupyter_output><empty_output>
|
autokeras/docs/ipynb/image_regression.ipynb/0
|
{
"file_path": "autokeras/docs/ipynb/image_regression.ipynb",
"repo_id": "autokeras",
"token_count": 1838
}
| 6 |
"""shell
pip install autokeras
"""
import numpy as np
import tensorflow as tf
from tensorflow.keras.datasets import mnist
import autokeras as ak
"""
In this tutorial, we show how to customize your search space with
[AutoModel](/auto_model/#automodel-class) and how to implement your own block
as search space. This API is mainly for advanced users who already know what
their model should look like.
## Customized Search Space
First, let us see how we can build the following neural network using the
building blocks in AutoKeras.
<div class="mermaid">
graph LR
id1(ImageInput) --> id2(Normalization)
id2 --> id3(Image Augmentation)
id3 --> id4(Convolutional)
id3 --> id5(ResNet V2)
id4 --> id6(Merge)
id5 --> id6
id6 --> id7(Classification Head)
</div>
We can make use of the [AutoModel](/auto_model/#automodel-class) API in
AutoKeras to implemented as follows.
The usage is the same as the [Keras functional
API](https://www.tensorflow.org/guide/keras/functional).
Since this is just a demo, we use small amount of `max_trials` and `epochs`.
"""
input_node = ak.ImageInput()
output_node = ak.Normalization()(input_node)
output_node1 = ak.ConvBlock()(output_node)
output_node2 = ak.ResNetBlock(version="v2")(output_node)
output_node = ak.Merge()([output_node1, output_node2])
output_node = ak.ClassificationHead()(output_node)
auto_model = ak.AutoModel(
inputs=input_node, outputs=output_node, overwrite=True, max_trials=1
)
"""
Whild building the model, the blocks used need to follow this topology:
`Preprocessor` -> `Block` -> `Head`. `Normalization` and `ImageAugmentation`
are `Preprocessor`s.
`ClassificationHead` is `Head`. The rest are `Block`s.
In the code above, we use `ak.ResNetBlock(version='v2')` to specify the version
of ResNet to use. There are many other arguments to specify for each building
block. For most of the arguments, if not specified, they would be tuned
automatically. Please refer to the documentation links at the bottom of the
page for more details.
Then, we prepare some data to run the model.
"""
(x_train, y_train), (x_test, y_test) = mnist.load_data()
print(x_train.shape) # (60000, 28, 28)
print(y_train.shape) # (60000,)
print(y_train[:3]) # array([7, 2, 1], dtype=uint8)
# Feed the AutoModel with training data.
auto_model.fit(x_train[:100], y_train[:100], epochs=1)
# Predict with the best model.
predicted_y = auto_model.predict(x_test)
# Evaluate the best model with testing data.
print(auto_model.evaluate(x_test, y_test))
"""
For multiple input nodes and multiple heads search space, you can refer to
[this section](/tutorial/multi/#customized-search-space).
## Validation Data
If you would like to provide your own validation data or change the ratio of
the validation data, please refer to the Validation Data section of the
tutorials of [Image
Classification](/tutorial/image_classification/#validation-data), [Text
Classification](/tutorial/text_classification/#validation-data), [Structured
Data
Classification](/tutorial/structured_data_classification/#validation-data),
[Multi-task and Multiple Validation](/tutorial/multi/#validation-data).
## Data Format
You can refer to the documentation of
[ImageInput](/node/#imageinput-class),
[StructuredDataInput](/node/#structureddatainput-class),
[TextInput](/node/#textinput-class),
[RegressionHead](/block/#regressionhead-class),
[ClassificationHead](/block/#classificationhead-class),
for the format of different types of data.
You can also refer to the Data Format section of the tutorials of
[Image Classification](/tutorial/image_classification/#data-format),
[Text Classification](/tutorial/text_classification/#data-format),
[Structured Data
Classification](/tutorial/structured_data_classification/#data-format).
## Implement New Block
You can extend the [Block](/base/#block-class)
class to implement your own building blocks and use it with
[AutoModel](/auto_model/#automodel-class).
The first step is to learn how to write a build function for
[KerasTuner](https://keras-team.github.io/keras-tuner/#usage-the-basics). You
need to override the [build function](/base/#build-method) of the block. The
following example shows how to implement a single Dense layer block whose
number of neurons is tunable.
"""
class SingleDenseLayerBlock(ak.Block):
def build(self, hp, inputs=None):
# Get the input_node from inputs.
input_node = tf.nest.flatten(inputs)[0]
layer = tf.keras.layers.Dense(
hp.Int("num_units", min_value=32, max_value=512, step=32)
)
output_node = layer(input_node)
return output_node
"""
You can connect it with other blocks and build it into an
[AutoModel](/auto_model/#automodel-class).
"""
# Build the AutoModel
input_node = ak.Input()
output_node = SingleDenseLayerBlock()(input_node)
output_node = ak.RegressionHead()(output_node)
auto_model = ak.AutoModel(input_node, output_node, overwrite=True, max_trials=1)
# Prepare Data
num_instances = 100
x_train = np.random.rand(num_instances, 20).astype(np.float32)
y_train = np.random.rand(num_instances, 1).astype(np.float32)
x_test = np.random.rand(num_instances, 20).astype(np.float32)
y_test = np.random.rand(num_instances, 1).astype(np.float32)
# Train the model
auto_model.fit(x_train, y_train, epochs=1)
print(auto_model.evaluate(x_test, y_test))
"""
## Reference
[AutoModel](/auto_model/#automodel-class)
**Nodes**:
[ImageInput](/node/#imageinput-class),
[Input](/node/#input-class),
[StructuredDataInput](/node/#structureddatainput-class),
[TextInput](/node/#textinput-class).
**Preprocessors**:
[FeatureEngineering](/block/#featureengineering-class),
[ImageAugmentation](/block/#imageaugmentation-class),
[LightGBM](/block/#lightgbm-class),
[Normalization](/block/#normalization-class),
[TextToIntSequence](/block/#texttointsequence-class),
[TextToNgramVector](/block/#texttongramvector-class).
**Blocks**:
[ConvBlock](/block/#convblock-class),
[DenseBlock](/block/#denseblock-class),
[Embedding](/block/#embedding-class),
[Merge](/block/#merge-class),
[ResNetBlock](/block/#resnetblock-class),
[RNNBlock](/block/#rnnblock-class),
[SpatialReduction](/block/#spatialreduction-class),
[TemporalReduction](/block/#temporalreduction-class),
[XceptionBlock](/block/#xceptionblock-class),
[ImageBlock](/block/#imageblock-class),
[StructuredDataBlock](/block/#structureddatablock-class),
[TextBlock](/block/#textblock-class).
"""
|
autokeras/docs/py/customized.py/0
|
{
"file_path": "autokeras/docs/py/customized.py",
"repo_id": "autokeras",
"token_count": 2139
}
| 7 |
# TensorFlow Cloud
[TensorFlow Cloud](https://github.com/tensorflow/cloud) allows you to run your
TensorFlow program leveraging the computing power on Google Cloud easily.
Please follow the [instructions](https://github.com/tensorflow/cloud) to setup your account.
AutoKeras has successfully integrated with this service.
Now you can run your program on Google Cloud only by inserting a few more lines of code.
Please see the example below.
```python
import argparse
import os
import autokeras as ak
import tensorflow_cloud as tfc
from tensorflow.keras.datasets import mnist
parser = argparse.ArgumentParser(description="Model save path arguments.")
parser.add_argument("--path", required=True, type=str, help="Keras model save path")
args = parser.parse_args()
tfc.run(
chief_config=tfc.COMMON_MACHINE_CONFIGS["V100_1X"],
docker_base_image="haifengjin/autokeras:1.0.3",
)
# Prepare the dataset.
(x_train, y_train), (x_test, y_test) = mnist.load_data()
print(x_train.shape) # (60000, 28, 28)
print(y_train.shape) # (60000,)
print(y_train[:3]) # array([7, 2, 1], dtype=uint8)
# Initialize the ImageClassifier.
clf = ak.ImageClassifier(max_trials=2)
# Search for the best model.
clf.fit(x_train, y_train, epochs=10)
# Evaluate on the testing data.
print("Accuracy: {accuracy}".format(accuracy=clf.evaluate(x_test, y_test)[1]))
clf.export_model().save(os.path.join(args.path, "model.h5"))
```
You can find the code above [here](https://github.com/tensorflow/cloud/blob/master/tensorflow_cloud/python/tests/integration/call_run_within_script_with_autokeras.py)
|
autokeras/docs/templates/extensions/tf_cloud.md/0
|
{
"file_path": "autokeras/docs/templates/extensions/tf_cloud.md",
"repo_id": "autokeras",
"token_count": 550
}
| 8 |
"""
Search for a good model for the
[IMDB](
https://keras.io/datasets/#imdb-movie-reviews-sentiment-classification) dataset.
"""
import numpy as np
import tensorflow as tf
import autokeras as ak
def imdb_raw():
max_features = 20000
index_offset = 3 # word index offset
(x_train, y_train), (x_test, y_test) = tf.keras.datasets.imdb.load_data(
num_words=max_features, index_from=index_offset
)
x_train = x_train
y_train = y_train.reshape(-1, 1)
x_test = x_test
y_test = y_test.reshape(-1, 1)
word_to_id = tf.keras.datasets.imdb.get_word_index()
word_to_id = {k: (v + index_offset) for k, v in word_to_id.items()}
word_to_id["<PAD>"] = 0
word_to_id["<START>"] = 1
word_to_id["<UNK>"] = 2
id_to_word = {value: key for key, value in word_to_id.items()}
x_train = list(
map(lambda sentence: " ".join(id_to_word[i] for i in sentence), x_train)
)
x_test = list(
map(lambda sentence: " ".join(id_to_word[i] for i in sentence), x_test)
)
x_train = np.array(x_train, dtype=np.str)
x_test = np.array(x_test, dtype=np.str)
return (x_train, y_train), (x_test, y_test)
# Prepare the data.
(x_train, y_train), (x_test, y_test) = imdb_raw()
print(x_train.shape) # (25000,)
print(y_train.shape) # (25000, 1)
print(x_train[0][:50]) # <START> this film was just brilliant casting <UNK>
# Initialize the TextClassifier
clf = ak.TextClassifier(max_trials=3)
# Search for the best model.
clf.fit(x_train, y_train, epochs=2, batch_size=8)
# Evaluate on the testing data.
print("Accuracy: {accuracy}".format(accuracy=clf.evaluate(x_test, y_test)))
|
autokeras/examples/imdb.py/0
|
{
"file_path": "autokeras/examples/imdb.py",
"repo_id": "autokeras",
"token_count": 722
}
| 9 |
isort .
black .
for i in $(find autokeras benchmark -name '*.py')
do
if ! grep -q Copyright $i
then
echo $i
cat shell/copyright.txt $i >$i.new && mv $i.new $i
fi
done
flake8 .
|
autokeras/shell/format.sh/0
|
{
"file_path": "autokeras/shell/format.sh",
"repo_id": "autokeras",
"token_count": 83
}
| 10 |
# Keras governance structure

---
## Design review process
Design-related communications are expected to happen primarily asynchronously via:
- The Pull Requests used for API proposals.
- [The Keras mailing list](https://groups.google.com/forum/#!forum/keras-users).
The process for writing and submitting design proposals is same as the [TensorFlow RFC process](https://github.com/tensorflow/community/blob/master/governance/TF-RFCs.md).
- Start from [this template](https://github.com/keras-team/governance/blob/master/rfcs/yyyymmdd-rfc-template.md).
- Fill in the content. Note that you will need to insert code examples.
- Provide enough context information for anyone to undertsand what's going on.
- Provide a solid argument as for why the feature is neeed.
- Include a code example of the **end-to-end workflow** you have in mind.
- Open a Pull Request in the [Keras API proposals folder in this repository](https://github.com/keras-team/governance/tree/master/rfcs).
- Send the Pull Request link to `[email protected]` with a subject that starts with `[API DESIGN REVIEW]` (all caps) so that we notice it.
- Wait for comments, and answer them as they come. Edit the proposal as necessary.
- The proposal will finally be approved or rejected. Once approved, you can send out Pull Requests to implement the API changes or ask others to write Pull Requests (targeting `keras-team/keras`).
Note that:
- Anyone is free to send out API proposals.
- Anyone is free to comment on API proposals or ask questions.
---
## Leadership
### BDFL
Role: final call in decisions related to the Keras API.
- Francois Chollet ([email protected])
---
## Our mission
The purpose of our work is to democratize access to machine learning through dependable standards and usable, productive APIs.
We seek to empower as many people as possible, from a wide diversity of backgrounds, to take ownership of ML technology and to use it to build their own solutions to their own problems.
Existing machine learning technology has the potential to solve a huge amount of problems in the world today, across every industry, and to help a tremendous amount of people. The potential is sky-high. We've barely even started. So how do we fully realize this potential?
We believe that we will only fully realize the potential of machine learning if it becomes a tool in everyone's hands -- not just a technology developed behind closed doors by an "AI industry", that you could only deploy by waiting for a turnkey cloud API to become available commercially, or by contracting an expensive consulting firm. We can't wait for experts to solve every problem -- experts at large tech companies don't even have visibility into a tiny fraction of the problems that can be solved. End users should solve their own problems. And our mission is to empower them to do just that.
Our mission is to make these capabilities available to anyone with basic computer literacy, for free. This is how we maximize the realized potential of these technologies, and how we maximize our positive impact on the world.
---
## Code of conduct
In the interest of fostering an open and welcoming environment,
we as contributors and maintainers pledge to making participation in our project
and our community a harassment-free experience for everyone.
All activity will abide by the [Code of Conduct](https://github.com/tensorflow/tensorflow/blob/master/CODE_OF_CONDUCT.md).
---
## Our values and our strengths
We will be able to reach our milestones because we yield superpowers of a kind that is quite uncommon among developers of ML tools:
- We have empathy for our users.
- We value and practice good design.
- We embrace openness and we are dedicated to foster our developer community.
- We value and practice good communication.
- We have a unique brand that users love.
**1) We have empathy for our users.** We know that every decision we make should be made with the user in mind, whether a design decision or a strategy decision. "What will be the total impact of our choices on the people who rely on our software?" is the question behind everything we do.
Having empathy for our users means:
- Being users ourselves -- actively using our product in similar scenarios as what our users face.
- Understanding our users by being closely in touch with them and having clear visibility into the developer experience: actively seeking community input in everything we do, listening to feedback, talking with users at external talks and developer events.
- Putting ourselves in our users' shoes: always going above and beyond to be helpful to our users and to improve the user experience of our products.
**2) We value good design.** We are fully aware that a delightful UX is what has made us successful so far and what will keep making us successful in the future. We know that things should be as simple as possible (but no simpler). We prefer elegance and minimalism over technical prowess. We follow formal principles for good design.
**3) We embrace openness and we are dedicated to foster our developer community.** We know that long-term community building is critical to the success of our project, and we know that developer communities don't get built behind closed doors. We understand the necessity of doing our work in the open and the importance of involving open-source contributors at all stages of the development process.
**4) We value and practice good communication.** We understand that great documentation, great code examples, and transparency of governance are essential to Keras adoption and contribute meaningfully to the Keras UX. We value external communication, documentation, and developers relations as much as we value technical contributions.
**5) We value what makes us different and unique, and we value our brand that users love, Keras.** The Keras brand is an essential tool in reaching our goals: it stands for user-friendliness, accessibility, and good design. We are proud of our banner and we will carry it forward.
|
governance/README.md/0
|
{
"file_path": "governance/README.md",
"repo_id": "governance",
"token_count": 1426
}
| 11 |
"""DenseNet models for Keras.
# Reference paper
- [Densely Connected Convolutional Networks]
(https://arxiv.org/abs/1608.06993) (CVPR 2017 Best Paper Award)
# Reference implementation
- [Torch DenseNets]
(https://github.com/liuzhuang13/DenseNet/blob/master/models/densenet.lua)
- [TensorNets]
(https://github.com/taehoonlee/tensornets/blob/master/tensornets/densenets.py)
"""
from __future__ import absolute_import
from __future__ import division
from __future__ import print_function
import os
from . import get_submodules_from_kwargs
from . import imagenet_utils
from .imagenet_utils import decode_predictions
from .imagenet_utils import _obtain_input_shape
BASE_WEIGTHS_PATH = (
'https://github.com/keras-team/keras-applications/'
'releases/download/densenet/')
DENSENET121_WEIGHT_PATH = (
BASE_WEIGTHS_PATH +
'densenet121_weights_tf_dim_ordering_tf_kernels.h5')
DENSENET121_WEIGHT_PATH_NO_TOP = (
BASE_WEIGTHS_PATH +
'densenet121_weights_tf_dim_ordering_tf_kernels_notop.h5')
DENSENET169_WEIGHT_PATH = (
BASE_WEIGTHS_PATH +
'densenet169_weights_tf_dim_ordering_tf_kernels.h5')
DENSENET169_WEIGHT_PATH_NO_TOP = (
BASE_WEIGTHS_PATH +
'densenet169_weights_tf_dim_ordering_tf_kernels_notop.h5')
DENSENET201_WEIGHT_PATH = (
BASE_WEIGTHS_PATH +
'densenet201_weights_tf_dim_ordering_tf_kernels.h5')
DENSENET201_WEIGHT_PATH_NO_TOP = (
BASE_WEIGTHS_PATH +
'densenet201_weights_tf_dim_ordering_tf_kernels_notop.h5')
backend = None
layers = None
models = None
keras_utils = None
def dense_block(x, blocks, name):
"""A dense block.
# Arguments
x: input tensor.
blocks: integer, the number of building blocks.
name: string, block label.
# Returns
output tensor for the block.
"""
for i in range(blocks):
x = conv_block(x, 32, name=name + '_block' + str(i + 1))
return x
def transition_block(x, reduction, name):
"""A transition block.
# Arguments
x: input tensor.
reduction: float, compression rate at transition layers.
name: string, block label.
# Returns
output tensor for the block.
"""
bn_axis = 3 if backend.image_data_format() == 'channels_last' else 1
x = layers.BatchNormalization(axis=bn_axis, epsilon=1.001e-5,
name=name + '_bn')(x)
x = layers.Activation('relu', name=name + '_relu')(x)
x = layers.Conv2D(int(backend.int_shape(x)[bn_axis] * reduction), 1,
use_bias=False,
name=name + '_conv')(x)
x = layers.AveragePooling2D(2, strides=2, name=name + '_pool')(x)
return x
def conv_block(x, growth_rate, name):
"""A building block for a dense block.
# Arguments
x: input tensor.
growth_rate: float, growth rate at dense layers.
name: string, block label.
# Returns
Output tensor for the block.
"""
bn_axis = 3 if backend.image_data_format() == 'channels_last' else 1
x1 = layers.BatchNormalization(axis=bn_axis,
epsilon=1.001e-5,
name=name + '_0_bn')(x)
x1 = layers.Activation('relu', name=name + '_0_relu')(x1)
x1 = layers.Conv2D(4 * growth_rate, 1,
use_bias=False,
name=name + '_1_conv')(x1)
x1 = layers.BatchNormalization(axis=bn_axis, epsilon=1.001e-5,
name=name + '_1_bn')(x1)
x1 = layers.Activation('relu', name=name + '_1_relu')(x1)
x1 = layers.Conv2D(growth_rate, 3,
padding='same',
use_bias=False,
name=name + '_2_conv')(x1)
x = layers.Concatenate(axis=bn_axis, name=name + '_concat')([x, x1])
return x
def DenseNet(blocks,
include_top=True,
weights='imagenet',
input_tensor=None,
input_shape=None,
pooling=None,
classes=1000,
**kwargs):
"""Instantiates the DenseNet architecture.
Optionally loads weights pre-trained on ImageNet.
Note that the data format convention used by the model is
the one specified in your Keras config at `~/.keras/keras.json`.
# Arguments
blocks: numbers of building blocks for the four dense layers.
include_top: whether to include the fully-connected
layer at the top of the network.
weights: one of `None` (random initialization),
'imagenet' (pre-training on ImageNet),
or the path to the weights file to be loaded.
input_tensor: optional Keras tensor
(i.e. output of `layers.Input()`)
to use as image input for the model.
input_shape: optional shape tuple, only to be specified
if `include_top` is False (otherwise the input shape
has to be `(224, 224, 3)` (with `'channels_last'` data format)
or `(3, 224, 224)` (with `'channels_first'` data format).
It should have exactly 3 inputs channels,
and width and height should be no smaller than 32.
E.g. `(200, 200, 3)` would be one valid value.
pooling: optional pooling mode for feature extraction
when `include_top` is `False`.
- `None` means that the output of the model will be
the 4D tensor output of the
last convolutional block.
- `avg` means that global average pooling
will be applied to the output of the
last convolutional block, and thus
the output of the model will be a 2D tensor.
- `max` means that global max pooling will
be applied.
classes: optional number of classes to classify images
into, only to be specified if `include_top` is True, and
if no `weights` argument is specified.
# Returns
A Keras model instance.
# Raises
ValueError: in case of invalid argument for `weights`,
or invalid input shape.
"""
global backend, layers, models, keras_utils
backend, layers, models, keras_utils = get_submodules_from_kwargs(kwargs)
if not (weights in {'imagenet', None} or os.path.exists(weights)):
raise ValueError('The `weights` argument should be either '
'`None` (random initialization), `imagenet` '
'(pre-training on ImageNet), '
'or the path to the weights file to be loaded.')
if weights == 'imagenet' and include_top and classes != 1000:
raise ValueError('If using `weights` as `"imagenet"` with `include_top`'
' as true, `classes` should be 1000')
# Determine proper input shape
input_shape = _obtain_input_shape(input_shape,
default_size=224,
min_size=32,
data_format=backend.image_data_format(),
require_flatten=include_top,
weights=weights)
if input_tensor is None:
img_input = layers.Input(shape=input_shape)
else:
if not backend.is_keras_tensor(input_tensor):
img_input = layers.Input(tensor=input_tensor, shape=input_shape)
else:
img_input = input_tensor
bn_axis = 3 if backend.image_data_format() == 'channels_last' else 1
x = layers.ZeroPadding2D(padding=((3, 3), (3, 3)))(img_input)
x = layers.Conv2D(64, 7, strides=2, use_bias=False, name='conv1/conv')(x)
x = layers.BatchNormalization(
axis=bn_axis, epsilon=1.001e-5, name='conv1/bn')(x)
x = layers.Activation('relu', name='conv1/relu')(x)
x = layers.ZeroPadding2D(padding=((1, 1), (1, 1)))(x)
x = layers.MaxPooling2D(3, strides=2, name='pool1')(x)
x = dense_block(x, blocks[0], name='conv2')
x = transition_block(x, 0.5, name='pool2')
x = dense_block(x, blocks[1], name='conv3')
x = transition_block(x, 0.5, name='pool3')
x = dense_block(x, blocks[2], name='conv4')
x = transition_block(x, 0.5, name='pool4')
x = dense_block(x, blocks[3], name='conv5')
x = layers.BatchNormalization(
axis=bn_axis, epsilon=1.001e-5, name='bn')(x)
x = layers.Activation('relu', name='relu')(x)
if include_top:
x = layers.GlobalAveragePooling2D(name='avg_pool')(x)
x = layers.Dense(classes, activation='softmax', name='fc1000')(x)
else:
if pooling == 'avg':
x = layers.GlobalAveragePooling2D(name='avg_pool')(x)
elif pooling == 'max':
x = layers.GlobalMaxPooling2D(name='max_pool')(x)
# Ensure that the model takes into account
# any potential predecessors of `input_tensor`.
if input_tensor is not None:
inputs = keras_utils.get_source_inputs(input_tensor)
else:
inputs = img_input
# Create model.
if blocks == [6, 12, 24, 16]:
model = models.Model(inputs, x, name='densenet121')
elif blocks == [6, 12, 32, 32]:
model = models.Model(inputs, x, name='densenet169')
elif blocks == [6, 12, 48, 32]:
model = models.Model(inputs, x, name='densenet201')
else:
model = models.Model(inputs, x, name='densenet')
# Load weights.
if weights == 'imagenet':
if include_top:
if blocks == [6, 12, 24, 16]:
weights_path = keras_utils.get_file(
'densenet121_weights_tf_dim_ordering_tf_kernels.h5',
DENSENET121_WEIGHT_PATH,
cache_subdir='models',
file_hash='9d60b8095a5708f2dcce2bca79d332c7')
elif blocks == [6, 12, 32, 32]:
weights_path = keras_utils.get_file(
'densenet169_weights_tf_dim_ordering_tf_kernels.h5',
DENSENET169_WEIGHT_PATH,
cache_subdir='models',
file_hash='d699b8f76981ab1b30698df4c175e90b')
elif blocks == [6, 12, 48, 32]:
weights_path = keras_utils.get_file(
'densenet201_weights_tf_dim_ordering_tf_kernels.h5',
DENSENET201_WEIGHT_PATH,
cache_subdir='models',
file_hash='1ceb130c1ea1b78c3bf6114dbdfd8807')
else:
if blocks == [6, 12, 24, 16]:
weights_path = keras_utils.get_file(
'densenet121_weights_tf_dim_ordering_tf_kernels_notop.h5',
DENSENET121_WEIGHT_PATH_NO_TOP,
cache_subdir='models',
file_hash='30ee3e1110167f948a6b9946edeeb738')
elif blocks == [6, 12, 32, 32]:
weights_path = keras_utils.get_file(
'densenet169_weights_tf_dim_ordering_tf_kernels_notop.h5',
DENSENET169_WEIGHT_PATH_NO_TOP,
cache_subdir='models',
file_hash='b8c4d4c20dd625c148057b9ff1c1176b')
elif blocks == [6, 12, 48, 32]:
weights_path = keras_utils.get_file(
'densenet201_weights_tf_dim_ordering_tf_kernels_notop.h5',
DENSENET201_WEIGHT_PATH_NO_TOP,
cache_subdir='models',
file_hash='c13680b51ded0fb44dff2d8f86ac8bb1')
model.load_weights(weights_path)
elif weights is not None:
model.load_weights(weights)
return model
def DenseNet121(include_top=True,
weights='imagenet',
input_tensor=None,
input_shape=None,
pooling=None,
classes=1000,
**kwargs):
return DenseNet([6, 12, 24, 16],
include_top, weights,
input_tensor, input_shape,
pooling, classes,
**kwargs)
def DenseNet169(include_top=True,
weights='imagenet',
input_tensor=None,
input_shape=None,
pooling=None,
classes=1000,
**kwargs):
return DenseNet([6, 12, 32, 32],
include_top, weights,
input_tensor, input_shape,
pooling, classes,
**kwargs)
def DenseNet201(include_top=True,
weights='imagenet',
input_tensor=None,
input_shape=None,
pooling=None,
classes=1000,
**kwargs):
return DenseNet([6, 12, 48, 32],
include_top, weights,
input_tensor, input_shape,
pooling, classes,
**kwargs)
def preprocess_input(x, data_format=None, **kwargs):
"""Preprocesses a numpy array encoding a batch of images.
# Arguments
x: a 3D or 4D numpy array consists of RGB values within [0, 255].
data_format: data format of the image tensor.
# Returns
Preprocessed array.
"""
return imagenet_utils.preprocess_input(x, data_format,
mode='torch', **kwargs)
setattr(DenseNet121, '__doc__', DenseNet.__doc__)
setattr(DenseNet169, '__doc__', DenseNet.__doc__)
setattr(DenseNet201, '__doc__', DenseNet.__doc__)
|
keras-applications/keras_applications/densenet.py/0
|
{
"file_path": "keras-applications/keras_applications/densenet.py",
"repo_id": "keras-applications",
"token_count": 6680
}
| 12 |
"""Xception V1 model for Keras.
On ImageNet, this model gets to a top-1 validation accuracy of 0.790
and a top-5 validation accuracy of 0.945.
Do note that the input image format for this model is different than for
the VGG16 and ResNet models (299x299 instead of 224x224),
and that the input preprocessing function
is also different (same as Inception V3).
# Reference
- [Xception: Deep Learning with Depthwise Separable Convolutions](
https://arxiv.org/abs/1610.02357) (CVPR 2017)
"""
from __future__ import absolute_import
from __future__ import division
from __future__ import print_function
import os
import warnings
from . import get_submodules_from_kwargs
from . import imagenet_utils
from .imagenet_utils import decode_predictions
from .imagenet_utils import _obtain_input_shape
TF_WEIGHTS_PATH = (
'https://github.com/fchollet/deep-learning-models/'
'releases/download/v0.4/'
'xception_weights_tf_dim_ordering_tf_kernels.h5')
TF_WEIGHTS_PATH_NO_TOP = (
'https://github.com/fchollet/deep-learning-models/'
'releases/download/v0.4/'
'xception_weights_tf_dim_ordering_tf_kernels_notop.h5')
def Xception(include_top=True,
weights='imagenet',
input_tensor=None,
input_shape=None,
pooling=None,
classes=1000,
**kwargs):
"""Instantiates the Xception architecture.
Optionally loads weights pre-trained on ImageNet.
Note that the data format convention used by the model is
the one specified in your Keras config at `~/.keras/keras.json`.
Note that the default input image size for this model is 299x299.
# Arguments
include_top: whether to include the fully-connected
layer at the top of the network.
weights: one of `None` (random initialization),
'imagenet' (pre-training on ImageNet),
or the path to the weights file to be loaded.
input_tensor: optional Keras tensor
(i.e. output of `layers.Input()`)
to use as image input for the model.
input_shape: optional shape tuple, only to be specified
if `include_top` is False (otherwise the input shape
has to be `(299, 299, 3)`.
It should have exactly 3 inputs channels,
and width and height should be no smaller than 71.
E.g. `(150, 150, 3)` would be one valid value.
pooling: Optional pooling mode for feature extraction
when `include_top` is `False`.
- `None` means that the output of the model will be
the 4D tensor output of the
last convolutional block.
- `avg` means that global average pooling
will be applied to the output of the
last convolutional block, and thus
the output of the model will be a 2D tensor.
- `max` means that global max pooling will
be applied.
classes: optional number of classes to classify images
into, only to be specified if `include_top` is True,
and if no `weights` argument is specified.
# Returns
A Keras model instance.
# Raises
ValueError: in case of invalid argument for `weights`,
or invalid input shape.
RuntimeError: If attempting to run this model with a
backend that does not support separable convolutions.
"""
backend, layers, models, keras_utils = get_submodules_from_kwargs(kwargs)
if not (weights in {'imagenet', None} or os.path.exists(weights)):
raise ValueError('The `weights` argument should be either '
'`None` (random initialization), `imagenet` '
'(pre-training on ImageNet), '
'or the path to the weights file to be loaded.')
if weights == 'imagenet' and include_top and classes != 1000:
raise ValueError('If using `weights` as `"imagenet"` with `include_top`'
' as true, `classes` should be 1000')
# Determine proper input shape
input_shape = _obtain_input_shape(input_shape,
default_size=299,
min_size=71,
data_format=backend.image_data_format(),
require_flatten=include_top,
weights=weights)
if input_tensor is None:
img_input = layers.Input(shape=input_shape)
else:
if not backend.is_keras_tensor(input_tensor):
img_input = layers.Input(tensor=input_tensor, shape=input_shape)
else:
img_input = input_tensor
channel_axis = 1 if backend.image_data_format() == 'channels_first' else -1
x = layers.Conv2D(32, (3, 3),
strides=(2, 2),
use_bias=False,
name='block1_conv1')(img_input)
x = layers.BatchNormalization(axis=channel_axis, name='block1_conv1_bn')(x)
x = layers.Activation('relu', name='block1_conv1_act')(x)
x = layers.Conv2D(64, (3, 3), use_bias=False, name='block1_conv2')(x)
x = layers.BatchNormalization(axis=channel_axis, name='block1_conv2_bn')(x)
x = layers.Activation('relu', name='block1_conv2_act')(x)
residual = layers.Conv2D(128, (1, 1),
strides=(2, 2),
padding='same',
use_bias=False)(x)
residual = layers.BatchNormalization(axis=channel_axis)(residual)
x = layers.SeparableConv2D(128, (3, 3),
padding='same',
use_bias=False,
name='block2_sepconv1')(x)
x = layers.BatchNormalization(axis=channel_axis, name='block2_sepconv1_bn')(x)
x = layers.Activation('relu', name='block2_sepconv2_act')(x)
x = layers.SeparableConv2D(128, (3, 3),
padding='same',
use_bias=False,
name='block2_sepconv2')(x)
x = layers.BatchNormalization(axis=channel_axis, name='block2_sepconv2_bn')(x)
x = layers.MaxPooling2D((3, 3),
strides=(2, 2),
padding='same',
name='block2_pool')(x)
x = layers.add([x, residual])
residual = layers.Conv2D(256, (1, 1), strides=(2, 2),
padding='same', use_bias=False)(x)
residual = layers.BatchNormalization(axis=channel_axis)(residual)
x = layers.Activation('relu', name='block3_sepconv1_act')(x)
x = layers.SeparableConv2D(256, (3, 3),
padding='same',
use_bias=False,
name='block3_sepconv1')(x)
x = layers.BatchNormalization(axis=channel_axis, name='block3_sepconv1_bn')(x)
x = layers.Activation('relu', name='block3_sepconv2_act')(x)
x = layers.SeparableConv2D(256, (3, 3),
padding='same',
use_bias=False,
name='block3_sepconv2')(x)
x = layers.BatchNormalization(axis=channel_axis, name='block3_sepconv2_bn')(x)
x = layers.MaxPooling2D((3, 3), strides=(2, 2),
padding='same',
name='block3_pool')(x)
x = layers.add([x, residual])
residual = layers.Conv2D(728, (1, 1),
strides=(2, 2),
padding='same',
use_bias=False)(x)
residual = layers.BatchNormalization(axis=channel_axis)(residual)
x = layers.Activation('relu', name='block4_sepconv1_act')(x)
x = layers.SeparableConv2D(728, (3, 3),
padding='same',
use_bias=False,
name='block4_sepconv1')(x)
x = layers.BatchNormalization(axis=channel_axis, name='block4_sepconv1_bn')(x)
x = layers.Activation('relu', name='block4_sepconv2_act')(x)
x = layers.SeparableConv2D(728, (3, 3),
padding='same',
use_bias=False,
name='block4_sepconv2')(x)
x = layers.BatchNormalization(axis=channel_axis, name='block4_sepconv2_bn')(x)
x = layers.MaxPooling2D((3, 3), strides=(2, 2),
padding='same',
name='block4_pool')(x)
x = layers.add([x, residual])
for i in range(8):
residual = x
prefix = 'block' + str(i + 5)
x = layers.Activation('relu', name=prefix + '_sepconv1_act')(x)
x = layers.SeparableConv2D(728, (3, 3),
padding='same',
use_bias=False,
name=prefix + '_sepconv1')(x)
x = layers.BatchNormalization(axis=channel_axis,
name=prefix + '_sepconv1_bn')(x)
x = layers.Activation('relu', name=prefix + '_sepconv2_act')(x)
x = layers.SeparableConv2D(728, (3, 3),
padding='same',
use_bias=False,
name=prefix + '_sepconv2')(x)
x = layers.BatchNormalization(axis=channel_axis,
name=prefix + '_sepconv2_bn')(x)
x = layers.Activation('relu', name=prefix + '_sepconv3_act')(x)
x = layers.SeparableConv2D(728, (3, 3),
padding='same',
use_bias=False,
name=prefix + '_sepconv3')(x)
x = layers.BatchNormalization(axis=channel_axis,
name=prefix + '_sepconv3_bn')(x)
x = layers.add([x, residual])
residual = layers.Conv2D(1024, (1, 1), strides=(2, 2),
padding='same', use_bias=False)(x)
residual = layers.BatchNormalization(axis=channel_axis)(residual)
x = layers.Activation('relu', name='block13_sepconv1_act')(x)
x = layers.SeparableConv2D(728, (3, 3),
padding='same',
use_bias=False,
name='block13_sepconv1')(x)
x = layers.BatchNormalization(axis=channel_axis, name='block13_sepconv1_bn')(x)
x = layers.Activation('relu', name='block13_sepconv2_act')(x)
x = layers.SeparableConv2D(1024, (3, 3),
padding='same',
use_bias=False,
name='block13_sepconv2')(x)
x = layers.BatchNormalization(axis=channel_axis, name='block13_sepconv2_bn')(x)
x = layers.MaxPooling2D((3, 3),
strides=(2, 2),
padding='same',
name='block13_pool')(x)
x = layers.add([x, residual])
x = layers.SeparableConv2D(1536, (3, 3),
padding='same',
use_bias=False,
name='block14_sepconv1')(x)
x = layers.BatchNormalization(axis=channel_axis, name='block14_sepconv1_bn')(x)
x = layers.Activation('relu', name='block14_sepconv1_act')(x)
x = layers.SeparableConv2D(2048, (3, 3),
padding='same',
use_bias=False,
name='block14_sepconv2')(x)
x = layers.BatchNormalization(axis=channel_axis, name='block14_sepconv2_bn')(x)
x = layers.Activation('relu', name='block14_sepconv2_act')(x)
if include_top:
x = layers.GlobalAveragePooling2D(name='avg_pool')(x)
x = layers.Dense(classes, activation='softmax', name='predictions')(x)
else:
if pooling == 'avg':
x = layers.GlobalAveragePooling2D()(x)
elif pooling == 'max':
x = layers.GlobalMaxPooling2D()(x)
# Ensure that the model takes into account
# any potential predecessors of `input_tensor`.
if input_tensor is not None:
inputs = keras_utils.get_source_inputs(input_tensor)
else:
inputs = img_input
# Create model.
model = models.Model(inputs, x, name='xception')
# Load weights.
if weights == 'imagenet':
if include_top:
weights_path = keras_utils.get_file(
'xception_weights_tf_dim_ordering_tf_kernels.h5',
TF_WEIGHTS_PATH,
cache_subdir='models',
file_hash='0a58e3b7378bc2990ea3b43d5981f1f6')
else:
weights_path = keras_utils.get_file(
'xception_weights_tf_dim_ordering_tf_kernels_notop.h5',
TF_WEIGHTS_PATH_NO_TOP,
cache_subdir='models',
file_hash='b0042744bf5b25fce3cb969f33bebb97')
model.load_weights(weights_path)
if backend.backend() == 'theano':
keras_utils.convert_all_kernels_in_model(model)
elif weights is not None:
model.load_weights(weights)
return model
def preprocess_input(x, **kwargs):
"""Preprocesses a numpy array encoding a batch of images.
# Arguments
x: a 4D numpy array consists of RGB values within [0, 255].
# Returns
Preprocessed array.
"""
return imagenet_utils.preprocess_input(x, mode='tf', **kwargs)
|
keras-applications/keras_applications/xception.py/0
|
{
"file_path": "keras-applications/keras_applications/xception.py",
"repo_id": "keras-applications",
"token_count": 6980
}
| 13 |
/*
* Wrap inline code samples otherwise they shoot of the side and
* can't be read at all.
*
* https://github.com/mkdocs/mkdocs/issues/313
* https://github.com/mkdocs/mkdocs/issues/233
* https://github.com/mkdocs/mkdocs/issues/834
*/
.rst-content code {
white-space: pre-wrap;
word-wrap: break-word;
padding: 2px 5px;
}
/**
* Make code blocks display as blocks and give them the appropriate
* font size and padding.
*
* https://github.com/mkdocs/mkdocs/issues/855
* https://github.com/mkdocs/mkdocs/issues/834
* https://github.com/mkdocs/mkdocs/issues/233
*/
.rst-content pre code {
white-space: pre;
word-wrap: normal;
display: block;
padding: 12px;
font-size: 12px;
}
/*
* Fix link colors when the link text is inline code.
*
* https://github.com/mkdocs/mkdocs/issues/718
*/
a code {
color: #2980B9;
}
a:hover code {
color: #3091d1;
}
a:visited code {
color: #9B59B6;
}
/*
* The CSS classes from highlight.js seem to clash with the
* ReadTheDocs theme causing some code to be incorrectly made
* bold and italic.
*
* https://github.com/mkdocs/mkdocs/issues/411
*/
pre .cs, pre .c {
font-weight: inherit;
font-style: inherit;
}
/*
* Fix some issues with the theme and non-highlighted code
* samples. Without and highlighting styles attached the
* formatting is broken.
*
* https://github.com/mkdocs/mkdocs/issues/319
*/
.rst-content .no-highlight {
display: block;
padding: 0.5em;
color: #333;
}
/*
* Additions specific to the search functionality provided by MkDocs
*/
.search-results {
margin-top: 23px;
}
.search-results article {
border-top: 1px solid #E1E4E5;
padding-top: 24px;
}
.search-results article:first-child {
border-top: none;
}
form .search-query {
width: 100%;
border-radius: 50px;
padding: 6px 12px; /* csslint allow: box-model */
border-color: #D1D4D5;
}
/*
* Improve inline code blocks within admonitions.
*
* https://github.com/mkdocs/mkdocs/issues/656
*/
.rst-content .admonition code {
color: #404040;
border: 1px solid #c7c9cb;
border: 1px solid rgba(0, 0, 0, 0.2);
background: #f8fbfd;
background: rgba(255, 255, 255, 0.7);
}
/*
* Account for wide tables which go off the side.
* Override borders to avoid wierdness on narrow tables.
*
* https://github.com/mkdocs/mkdocs/issues/834
* https://github.com/mkdocs/mkdocs/pull/1034
*/
.rst-content .section .docutils {
width: 100%;
overflow: auto;
display: block;
border: none;
}
td, th {
border: 1px solid #e1e4e5 !important; /* csslint allow: important */
border-collapse: collapse;
}
/*Keras extras*/
.keras-logo {
max-height: 55px;
width: 100%;
background: #d00000;
font-size: 140%;
color: white;
font-family: "Source Sans Pro", "ff-tisa-web-pro", "Georgia", Arial, sans-serif;
}
.keras-logo-img {
max-width: 45px;
margin: 10px;
}
h1, h2, h3, h4, h5, h6, legend {
font-family: "Source Sans Pro", "ff-tisa-web-pro", "Georgia", Arial, sans-serif;,
}
|
keras-contrib/contrib_docs/theme/css/theme_extra.css/0
|
{
"file_path": "keras-contrib/contrib_docs/theme/css/theme_extra.css",
"repo_id": "keras-contrib",
"token_count": 1171
}
| 14 |
"""
Adapted from keras example cifar10_cnn.py and github.com/raghakot/keras-resnet
Train ResNet-18 on the CIFAR10 small images dataset.
GPU run command with Theano backend (with TensorFlow, the GPU is automatically used):
THEANO_FLAGS=mode=FAST_RUN,device=gpu,floatX=float32 python cifar10.py
"""
from __future__ import print_function
from keras.datasets import cifar10
from keras.preprocessing.image import ImageDataGenerator
from keras.utils import np_utils
from keras.callbacks import ModelCheckpoint
from keras.callbacks import ReduceLROnPlateau
from keras.callbacks import CSVLogger
from keras.callbacks import EarlyStopping
from keras_contrib.applications.resnet import ResNet18
import numpy as np
weights_file = 'ResNet18v2-CIFAR-10.h5'
lr_reducer = ReduceLROnPlateau(factor=np.sqrt(0.1), cooldown=0,
patience=5, min_lr=0.5e-6)
early_stopper = EarlyStopping(min_delta=0.001, patience=10)
csv_logger = CSVLogger('ResNet18v2-CIFAR-10.csv')
model_checkpoint = ModelCheckpoint(weights_file, monitor='val_acc', save_best_only=True,
save_weights_only=True, mode='auto')
batch_size = 32
nb_classes = 10
nb_epoch = 200
data_augmentation = True
# input image dimensions
img_rows, img_cols = 32, 32
# The CIFAR10 images are RGB.
img_channels = 3
# The data, shuffled and split between train and test sets:
(X_train, y_train), (X_test, y_test) = cifar10.load_data()
# Convert class vectors to binary class matrices.
Y_train = np_utils.to_categorical(y_train, nb_classes)
Y_test = np_utils.to_categorical(y_test, nb_classes)
X_train = X_train.astype('float32')
X_test = X_test.astype('float32')
# subtract mean and normalize
mean_image = np.mean(X_train, axis=0)
X_train -= mean_image
X_test -= mean_image
X_train /= 128.
X_test /= 128.
model = ResNet18((img_rows, img_cols, img_channels), nb_classes)
model.compile(loss='categorical_crossentropy',
optimizer='adam',
metrics=['accuracy'])
if not data_augmentation:
print('Not using data augmentation.')
model.fit(X_train, Y_train,
batch_size=batch_size,
nb_epoch=nb_epoch,
validation_data=(X_test, Y_test),
shuffle=True,
callbacks=[lr_reducer, early_stopper, csv_logger, model_checkpoint])
else:
print('Using real-time data augmentation.')
# This will do preprocessing and realtime data augmentation:
datagen = ImageDataGenerator(
featurewise_center=False, # set input mean to 0 over the dataset
samplewise_center=False, # set each sample mean to 0
featurewise_std_normalization=False, # divide inputs by std of the dataset
samplewise_std_normalization=False, # divide each input by its std
zca_whitening=False, # apply ZCA whitening
rotation_range=0, # randomly rotate images in the range (degrees, 0 to 180)
width_shift_range=0.1, # randomly shift images horizontally
height_shift_range=0.1, # randomly shift images vertically
horizontal_flip=True, # randomly flip images
vertical_flip=False) # randomly flip images
# Compute quantities required for featurewise normalization
# (std, mean, and principal components if ZCA whitening is applied).
datagen.fit(X_train)
callbacks = [lr_reducer, early_stopper, csv_logger, model_checkpoint]
# Fit the model on the batches generated by datagen.flow().
model.fit_generator(datagen.flow(X_train, Y_train, batch_size=batch_size),
steps_per_epoch=X_train.shape[0] // batch_size,
validation_data=(X_test, Y_test),
epochs=nb_epoch, verbose=2,
callbacks=callbacks)
scores = model.evaluate(X_test, Y_test, batch_size=batch_size)
print('Test loss : ', scores[0])
print('Test accuracy : ', scores[1])
|
keras-contrib/examples/cifar10_resnet.py/0
|
{
"file_path": "keras-contrib/examples/cifar10_resnet.py",
"repo_id": "keras-contrib",
"token_count": 1567
}
| 15 |
import numpy as np
from keras import backend as K
def extract_image_patches(X, ksizes, strides,
padding='valid',
data_format='channels_first'):
raise NotImplementedError
def depth_to_space(input, scale, data_format=None):
raise NotImplementedError
def moments(x, axes, shift=None, keep_dims=False):
mean_batch = np.mean(x, axis=tuple(axes), keepdims=keep_dims)
var_batch = np.var(x, axis=tuple(axes), keepdims=keep_dims)
return mean_batch, var_batch
|
keras-contrib/keras_contrib/backend/numpy_backend.py/0
|
{
"file_path": "keras-contrib/keras_contrib/backend/numpy_backend.py",
"repo_id": "keras-contrib",
"token_count": 231
}
| 16 |
from __future__ import absolute_import
from .advanced_activations.pelu import PELU
from .advanced_activations.srelu import SReLU
from .advanced_activations.swish import Swish
from .advanced_activations.sinerelu import SineReLU
from .convolutional.cosineconvolution2d import CosineConv2D
from .convolutional.cosineconvolution2d import CosineConvolution2D
from .convolutional.subpixelupscaling import SubPixelUpscaling
from .core import CosineDense
from .crf import CRF
from .capsule import Capsule
from .normalization.instancenormalization import InstanceNormalization
from .normalization.groupnormalization import GroupNormalization
|
keras-contrib/keras_contrib/layers/__init__.py/0
|
{
"file_path": "keras-contrib/keras_contrib/layers/__init__.py",
"repo_id": "keras-contrib",
"token_count": 191
}
| 17 |
from keras import backend as K
from keras.losses import categorical_crossentropy
from keras.losses import sparse_categorical_crossentropy
def crf_nll(y_true, y_pred):
"""The negative log-likelihood for linear chain Conditional Random Field (CRF).
This loss function is only used when the `layers.CRF` layer
is trained in the "join" mode.
# Arguments
y_true: tensor with true targets.
y_pred: tensor with predicted targets.
# Returns
A scalar representing corresponding to the negative log-likelihood.
# Raises
TypeError: If CRF is not the last layer.
# About GitHub
If you open an issue or a pull request about CRF, please
add `cc @lzfelix` to notify Luiz Felix.
"""
crf, idx = y_pred._keras_history[:2]
if crf._outbound_nodes:
raise TypeError('When learn_model="join", CRF must be the last layer.')
if crf.sparse_target:
y_true = K.one_hot(K.cast(y_true[:, :, 0], 'int32'), crf.units)
X = crf._inbound_nodes[idx].input_tensors[0]
mask = crf._inbound_nodes[idx].input_masks[0]
nloglik = crf.get_negative_log_likelihood(y_true, X, mask)
return nloglik
def crf_loss(y_true, y_pred):
"""General CRF loss function depending on the learning mode.
# Arguments
y_true: tensor with true targets.
y_pred: tensor with predicted targets.
# Returns
If the CRF layer is being trained in the join mode, returns the negative
log-likelihood. Otherwise returns the categorical crossentropy implemented
by the underlying Keras backend.
# About GitHub
If you open an issue or a pull request about CRF, please
add `cc @lzfelix` to notify Luiz Felix.
"""
crf, idx = y_pred._keras_history[:2]
if crf.learn_mode == 'join':
return crf_nll(y_true, y_pred)
else:
if crf.sparse_target:
return sparse_categorical_crossentropy(y_true, y_pred)
else:
return categorical_crossentropy(y_true, y_pred)
|
keras-contrib/keras_contrib/losses/crf_losses.py/0
|
{
"file_path": "keras-contrib/keras_contrib/losses/crf_losses.py",
"repo_id": "keras-contrib",
"token_count": 822
}
| 18 |
import numpy as np
from keras.datasets import mnist
from keras.layers import Activation
from keras.layers import Dense
from keras.models import Sequential
from keras.utils import np_utils
np.random.seed(1337)
nb_classes = 10
batch_size = 128
nb_epoch = 5
weighted_class = 9
standard_weight = 1
high_weight = 5
max_train_samples = 5000
max_test_samples = 1000
def get_data():
# the data, shuffled and split between tran and test sets
(X_train, y_train), (X_test, y_test) = mnist.load_data()
X_train = X_train.reshape(60000, 784)[:max_train_samples]
X_test = X_test.reshape(10000, 784)[:max_test_samples]
X_train = X_train.astype('float32') / 255
X_test = X_test.astype('float32') / 255
# convert class vectors to binary class matrices
y_train = y_train[:max_train_samples]
y_test = y_test[:max_test_samples]
Y_train = np_utils.to_categorical(y_train, nb_classes)
Y_test = np_utils.to_categorical(y_test, nb_classes)
test_ids = np.where(y_test == np.array(weighted_class))[0]
return (X_train, Y_train), (X_test, Y_test), test_ids
def validate_regularizer(weight_reg=None, activity_reg=None):
model = Sequential()
model.add(Dense(50, input_shape=(784,)))
model.add(Activation('relu'))
model.add(Dense(10, W_regularizer=weight_reg,
activity_regularizer=activity_reg))
model.add(Activation('softmax'))
return model
|
keras-contrib/keras_contrib/tests/regularizers.py/0
|
{
"file_path": "keras-contrib/keras_contrib/tests/regularizers.py",
"repo_id": "keras-contrib",
"token_count": 575
}
| 19 |
from __future__ import print_function
import pytest
import time
import random
from keras_contrib import datasets
if __name__ == '__main__':
pytest.main([__file__])
|
keras-contrib/tests/keras_contrib/datasets/test_datasets.py/0
|
{
"file_path": "keras-contrib/tests/keras_contrib/datasets/test_datasets.py",
"repo_id": "keras-contrib",
"token_count": 53
}
| 20 |
from __future__ import print_function
import pytest
from keras_contrib.utils.test_utils import is_tf_keras
from keras_contrib.tests import optimizers
from keras_contrib.optimizers import ftml
@pytest.mark.xfail(is_tf_keras,
reason='TODO fix this.',
strict=True)
def test_ftml():
optimizers._test_optimizer(ftml())
optimizers._test_optimizer(ftml(lr=0.003, beta_1=0.8,
beta_2=0.9, epsilon=1e-5,
decay=1e-3))
|
keras-contrib/tests/keras_contrib/optimizers/ftml_test.py/0
|
{
"file_path": "keras-contrib/tests/keras_contrib/optimizers/ftml_test.py",
"repo_id": "keras-contrib",
"token_count": 274
}
| 21 |
"""Benchmark conv layers.
To run benchmarks, see the following command for an example, please change the
flag to your custom value:
```
python3 -m benchmarks.layer_benchmark.conv_benchmark \
--benchmark_name=benchmark_conv2D \
--num_samples=2046 \
--batch_size=256 \
--jit_compile=True
```
"""
from absl import app
from absl import flags
from benchmarks.layer_benchmark.base_benchmark import LayerBenchmark
FLAGS = flags.FLAGS
def benchmark_conv1D(
num_samples,
batch_size,
jit_compile=True,
):
layer_name = "Conv1D"
init_args = {
"filters": 64,
"kernel_size": 2,
}
benchmark = LayerBenchmark(
layer_name,
init_args,
input_shape=[1024, 256],
jit_compile=jit_compile,
)
benchmark.benchmark_predict(
num_samples=num_samples,
batch_size=batch_size,
)
benchmark.benchmark_train(
num_samples=num_samples,
batch_size=batch_size,
)
def benchmark_conv2D(
num_samples,
batch_size,
jit_compile=True,
):
layer_name = "Conv2D"
init_args = {
"filters": 16,
"kernel_size": 2,
}
benchmark = LayerBenchmark(
layer_name,
init_args,
input_shape=[128, 128, 4],
jit_compile=jit_compile,
)
benchmark.benchmark_predict(
num_samples=num_samples,
batch_size=batch_size,
)
benchmark.benchmark_train(
num_samples=num_samples,
batch_size=batch_size,
)
def benchmark_conv3D(
num_samples,
batch_size,
jit_compile=True,
):
layer_name = "Conv3D"
init_args = {
"filters": 16,
"kernel_size": 2,
}
benchmark = LayerBenchmark(
layer_name,
init_args,
input_shape=[32, 32, 32, 4],
jit_compile=jit_compile,
)
benchmark.benchmark_predict(
num_samples=num_samples,
batch_size=batch_size,
)
benchmark.benchmark_train(
num_samples=num_samples,
batch_size=batch_size,
)
def benchmark_depthwise_conv1D(
num_samples,
batch_size,
jit_compile=True,
):
layer_name = "DepthwiseConv1D"
init_args = {
"kernel_size": 16,
"depth_multiplier": 2,
}
benchmark = LayerBenchmark(
layer_name,
init_args,
input_shape=[256, 64],
jit_compile=jit_compile,
)
benchmark.benchmark_predict(
num_samples=num_samples,
batch_size=batch_size,
)
benchmark.benchmark_train(
num_samples=num_samples,
batch_size=batch_size,
)
def benchmark_depthwise_conv2D(
num_samples,
batch_size,
jit_compile=True,
):
layer_name = "DepthwiseConv2D"
init_args = {
"kernel_size": 16,
"depth_multiplier": 2,
}
benchmark = LayerBenchmark(
layer_name,
init_args,
input_shape=[128, 128, 4],
jit_compile=jit_compile,
)
benchmark.benchmark_predict(
num_samples=num_samples,
batch_size=batch_size,
)
benchmark.benchmark_train(
num_samples=num_samples,
batch_size=batch_size,
)
def benchmark_separable_conv1D(
num_samples,
batch_size,
jit_compile=True,
):
layer_name = "SeparableConv1D"
init_args = {
"kernel_size": 16,
"depth_multiplier": 2,
"filters": 3,
}
benchmark = LayerBenchmark(
layer_name,
init_args,
input_shape=[256, 64],
jit_compile=jit_compile,
)
benchmark.benchmark_predict(
num_samples=num_samples,
batch_size=batch_size,
)
benchmark.benchmark_train(
num_samples=num_samples,
batch_size=batch_size,
)
def benchmark_separable_conv2D(
num_samples,
batch_size,
jit_compile=True,
):
layer_name = "SeparableConv2D"
init_args = {
"kernel_size": 16,
"depth_multiplier": 2,
"filters": 3,
}
benchmark = LayerBenchmark(
layer_name,
init_args,
input_shape=[128, 128, 4],
jit_compile=jit_compile,
)
benchmark.benchmark_predict(
num_samples=num_samples,
batch_size=batch_size,
)
benchmark.benchmark_train(
num_samples=num_samples,
batch_size=batch_size,
)
def benchmark_conv1D_transpose(
num_samples,
batch_size,
jit_compile=True,
):
layer_name = "Conv1DTranspose"
init_args = {
"filters": 32,
"kernel_size": 4,
}
benchmark = LayerBenchmark(
layer_name,
init_args,
input_shape=[256, 256],
jit_compile=jit_compile,
)
benchmark.benchmark_predict(
num_samples=num_samples,
batch_size=batch_size,
)
benchmark.benchmark_train(
num_samples=num_samples,
batch_size=batch_size,
)
def benchmark_conv2D_transpose(
num_samples,
batch_size,
jit_compile=True,
):
layer_name = "Conv2DTranspose"
init_args = {
"filters": 16,
"kernel_size": 2,
}
benchmark = LayerBenchmark(
layer_name,
init_args,
input_shape=[128, 128, 4],
jit_compile=jit_compile,
)
benchmark.benchmark_predict(
num_samples=num_samples,
batch_size=batch_size,
)
benchmark.benchmark_train(
num_samples=num_samples,
batch_size=batch_size,
)
def benchmark_conv3D_transpose(
num_samples,
batch_size,
jit_compile=True,
):
layer_name = "Conv3DTranspose"
init_args = {
"filters": 16,
"kernel_size": 2,
}
benchmark = LayerBenchmark(
layer_name,
init_args,
input_shape=[32, 32, 32, 4],
jit_compile=jit_compile,
)
benchmark.benchmark_predict(
num_samples=num_samples,
batch_size=batch_size,
)
benchmark.benchmark_train(
num_samples=num_samples,
batch_size=batch_size,
)
BENCHMARK_NAMES = {
"benchmark_conv1D": benchmark_conv1D,
"benchmark_conv2D": benchmark_conv2D,
"benchmark_conv3D": benchmark_conv3D,
"benchmark_depthwise_conv1D": benchmark_depthwise_conv1D,
"benchmark_depthwise_conv2D": benchmark_depthwise_conv2D,
"benchmark_separable_conv1D": benchmark_separable_conv1D,
"benchmark_separable_conv2D": benchmark_separable_conv2D,
"benchmark_conv1D_transpose": benchmark_conv1D_transpose,
"benchmark_conv2D_transpose": benchmark_conv2D_transpose,
"benchmark_conv3D_transpose": benchmark_conv3D_transpose,
}
def main(_):
benchmark_name = FLAGS.benchmark_name
num_samples = FLAGS.num_samples
batch_size = FLAGS.batch_size
jit_compile = FLAGS.jit_compile
if benchmark_name is None:
for name, benchmark_fn in BENCHMARK_NAMES:
benchmark_fn(num_samples, batch_size, jit_compile)
return
if benchmark_name not in BENCHMARK_NAMES:
raise ValueError(
f"Invalid benchmark name: {benchmark_name}, `benchmark_name` must "
f"be one of {BENCHMARK_NAMES.keys()}"
)
benchmark_fn = BENCHMARK_NAMES[benchmark_name]
benchmark_fn(num_samples, batch_size, jit_compile)
if __name__ == "__main__":
app.run(main)
|
keras-core/benchmarks/layer_benchmark/conv_benchmark.py/0
|
{
"file_path": "keras-core/benchmarks/layer_benchmark/conv_benchmark.py",
"repo_id": "keras-core",
"token_count": 3507
}
| 22 |
"""Benchmark Keras performance with torch custom training loop.
In this file we use a model with 3 dense layers. Training loop is written in the
vanilla torch way, and we compare the performance between building model with
Keras and torch.
"""
import numpy as np
import torch
import torch.nn as nn
import torch.optim as optim
import keras_core
from benchmarks.torch_ctl_benchmark.benchmark_utils import train_loop
from keras_core import layers
num_classes = 2
input_shape = (8192,)
batch_size = 4096
num_batches = 20
num_epochs = 1
x_train = np.random.normal(
size=(num_batches * batch_size, *input_shape)
).astype(np.float32)
y_train = np.random.randint(0, num_classes, size=(num_batches * batch_size,))
# Create a TensorDataset
dataset = torch.utils.data.TensorDataset(
torch.from_numpy(x_train), torch.from_numpy(y_train)
)
# Create a DataLoader
train_loader = torch.utils.data.DataLoader(
dataset, batch_size=batch_size, shuffle=False
)
class TorchModel(torch.nn.Module):
def __init__(self):
super().__init__()
self.dense1 = torch.nn.Linear(8192, 64)
self.activation1 = torch.nn.ReLU()
self.dense2 = torch.nn.Linear(64, 8)
self.activation2 = torch.nn.ReLU()
self.dense3 = torch.nn.Linear(8, num_classes)
self.softmax = torch.nn.Softmax(dim=1)
def forward(self, x):
x = self.dense1(x)
x = self.activation1(x)
x = self.dense2(x)
x = self.activation2(x)
x = self.dense3(x)
x = self.softmax(x)
return x
def run_keras_core_custom_training_loop():
keras_model = keras_core.Sequential(
[
layers.Input(shape=input_shape),
layers.Dense(64, activation="relu"),
layers.Dense(8, activation="relu"),
layers.Dense(num_classes),
layers.Softmax(),
]
)
optimizer = optim.Adam(keras_model.parameters(), lr=0.001)
loss_fn = nn.CrossEntropyLoss()
train_loop(
keras_model,
train_loader,
num_epochs=num_epochs,
optimizer=optimizer,
loss_fn=loss_fn,
framework="keras_core",
)
def run_torch_custom_training_loop():
torch_model = TorchModel()
optimizer = optim.Adam(torch_model.parameters(), lr=0.001)
loss_fn = nn.CrossEntropyLoss()
train_loop(
torch_model,
train_loader,
num_epochs=num_epochs,
optimizer=optimizer,
loss_fn=loss_fn,
framework="torch",
)
if __name__ == "__main__":
run_keras_core_custom_training_loop()
run_torch_custom_training_loop()
|
keras-core/benchmarks/torch_ctl_benchmark/dense_model_benchmark.py/0
|
{
"file_path": "keras-core/benchmarks/torch_ctl_benchmark/dense_model_benchmark.py",
"repo_id": "keras-core",
"token_count": 1147
}
| 23 |
"""
Title: MultipleChoice Task with Transfer Learning
Author: Md Awsafur Rahman
Date created: 2023/09/14
Last modified: 2023/09/14
Description: Use pre-trained nlp models for multiplechoice task.
Accelerator: GPU
"""
"""
## Introduction
In this example, we will demonstrate how to perform the **MultipleChoice** task by
finetuning pre-trained DebertaV3 model. In this task, several candidate answers are
provided along with a context and the model is trained to select the correct answer
unlike question answering. We will use SWAG dataset to demonstrate this example.
"""
"""
## Setup
"""
"""shell
pip install -q keras-core --upgrade
pip install -q keras-nlp --upgrade
"""
import os
os.environ["KERAS_BACKEND"] = "jax" # or "tensorflow" or "torch"
import keras_nlp
import keras_core as keras
import tensorflow as tf
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
"""
## Dataset
In this example we'll use **SWAG** dataset for multiplechoice task.
"""
"""shell
wget "https://github.com/rowanz/swagaf/archive/refs/heads/master.zip" -O swag.zip
unzip -q /content/swag.zip
"""
"""shell
ls /content/swagaf-master/data
"""
"""
## Configuration
"""
class CFG:
preset = "deberta_v3_extra_small_en" # Name of pretrained models
sequence_length = 200 # Input sequence length
seed = 42 # Random seed
epochs = 5 # Training epochs
batch_size = 8 # Batch size
augment = True # Augmentation (Shuffle Options)
"""
## Reproducibility
Sets value for random seed to produce similar result in each run.
"""
keras.utils.set_random_seed(CFG.seed)
"""
## Meta Data
* **train.csv** - will be used for training.
* `sent1` and `sent2`: these fields show how a sentence starts, and if you put the two
together, you get the `startphrase` field.
* `ending_<i>`: suggests a possible ending for how a sentence can end, but only one of
them is correct.
* `label`: identifies the correct sentence ending.
* **val.csv** - similar to `train.csv` but will be used for validation.
"""
# Train data
train_df = pd.read_csv(
"/content/swagaf-master/data/train.csv", index_col=0
) # Read CSV file into a DataFrame
train_df = train_df.sample(frac=0.02)
print("# Train Data: {:,}".format(len(train_df)))
# Valid data
valid_df = pd.read_csv(
"/content/swagaf-master/data/val.csv", index_col=0
) # Read CSV file into a DataFrame
valid_df = valid_df.sample(frac=0.02)
print("# Valid Data: {:,}".format(len(valid_df)))
"""
## Contextualize Options
Our approach entails furnishing the model with question and answer pairs, as opposed to
employing a single question for all five options. In practice, this signifies that for
the five options, we will supply the model with the same set of five questions combined
with each respective answer choice (e.g., `(Q + A)`, `(Q + B)`, and so on). This analogy
draws parallels to the practice of revisiting a question multiple times during an exam to
promote a deeper understanding of the problem at hand.
> Notably, in the context of SWAG dataset, question is the start of a sentence and
options are possible ending of that sentence.
"""
# Define a function to create options based on the prompt and choices
def make_options(row):
row["options"] = [
f"{row.startphrase}\n{row.ending0}", # Option 0
f"{row.startphrase}\n{row.ending1}", # Option 1
f"{row.startphrase}\n{row.ending2}", # Option 2
f"{row.startphrase}\n{row.ending3}",
] # Option 3
return row
"""
Apply the `make_options` function to each row of the dataframe
"""
train_df = train_df.apply(make_options, axis=1)
valid_df = valid_df.apply(make_options, axis=1)
"""
## Preprocessing
**What it does:** The preprocessor takes input strings and transforms them into a
dictionary (`token_ids`, `padding_mask`) containing preprocessed tensors. This process
starts with tokenization, where input strings are converted into sequences of token IDs.
**Why it's important:** Initially, raw text data is complex and challenging for modeling
due to its high dimensionality. By converting text into a compact set of tokens, such as
transforming `"The quick brown fox"` into `["the", "qu", "##ick", "br", "##own", "fox"]`,
we simplify the data. Many models rely on special tokens and additional tensors to
understand input. These tokens help divide input and identify padding, among other tasks.
Making all sequences the same length through padding boosts computational efficiency,
making subsequent steps smoother.
Explore the following pages to access the available preprocessing and tokenizer layers in
**KerasNLP**:
- [Preprocessing](https://keras.io/api/keras_nlp/preprocessing_layers/)
- [Tokenizers](https://keras.io/api/keras_nlp/tokenizers/)
"""
preprocessor = keras_nlp.models.DebertaV3Preprocessor.from_preset(
preset=CFG.preset, # Name of the model
sequence_length=CFG.sequence_length, # Max sequence length, will be padded if shorter
)
"""
Now, let's examine what the output shape of the preprocessing layer looks like. The
output shape of the layer can be represented as $(num\_choices, sequence\_length)$.
"""
outs = preprocessor(
train_df.options.iloc[0]
) # Process options for the first row
# Display the shape of each processed output
for k, v in outs.items():
print(k, ":", v.shape)
"""
We'll use the `preprocessing_fn` function to transform each text option using the
`dataset.map(preprocessing_fn)` method.
"""
def preprocess_fn(text, label=None):
text = preprocessor(text) # Preprocess text
return (
(text, label) if label is not None else text
) # Return processed text and label if available
"""
## Augmentation
In this notebook, we'll experiment with an interesting augmentation technique,
`option_shuffle`. Since we're providing the model with one option at a time, we can
introduce a shuffle to the order of options. For instance, options `[A, C, E, D, B]`
would be rearranged as `[D, B, A, E, C]`. This practice will help the model focus on the
content of the options themselves, rather than being influenced by their positions.
**Note:** Even though `option_shuffle` function is written in pure
tensorflow, it can be used with any backend (e.g. JAX, PyTorch) as it is only used
in `tf.data.Dataset` pipeline which is compatible with Keras Core routines.
"""
def option_shuffle(options, labels, prob=0.50, seed=None):
if tf.random.uniform([]) > prob: # Shuffle probability check
return options, labels
# Shuffle indices of options and labels in the same order
indices = tf.random.shuffle(tf.range(tf.shape(options)[0]), seed=seed)
# Shuffle options and labels
options = tf.gather(options, indices)
labels = tf.gather(labels, indices)
return options, labels
"""
In the following function, we'll merge all augmentation functions to apply to the text.
These augmentations will be applied to the data using the `dataset.map(augment_fn)`
approach.
"""
def augment_fn(text, label=None):
text, label = option_shuffle(text, label, prob=0.5 # Shuffle the options
return (text, label) if label is not None else text
"""
## DataLoader
The code below sets up a robust data flow pipeline using `tf.data.Dataset` for data
processing. Notable aspects of `tf.data` include its ability to simplify pipeline
construction and represent components in sequences.
To learn more about `tf.data`, refer to this
[documentation](https://www.tensorflow.org/guide/data).
"""
def build_dataset(
texts,
labels=None,
batch_size=32,
cache=False,
augment=False,
repeat=False,
shuffle=1024,
):
AUTO = tf.data.AUTOTUNE # AUTOTUNE option
slices = (
(texts,)
if labels is None
else (texts, keras.utils.to_categorical(labels, num_classes=4))
) # Create slices
ds = tf.data.Dataset.from_tensor_slices(
slices
) # Create dataset from slices
ds = ds.cache() if cache else ds # Cache dataset if enabled
if augment: # Apply augmentation if enabled
ds = ds.map(augment_fn, num_parallel_calls=AUTO)
ds = ds.map(
preprocess_fn, num_parallel_calls=AUTO
) # Map preprocessing function
ds = ds.repeat() if repeat else ds # Repeat dataset if enabled
opt = tf.data.Options() # Create dataset options
if shuffle:
ds = ds.shuffle(shuffle, seed=CFG.seed) # Shuffle dataset if enabled
opt.experimental_deterministic = False
ds = ds.with_options(opt) # Set dataset options
ds = ds.batch(batch_size, drop_remainder=True) # Batch dataset
ds = ds.prefetch(AUTO) # Prefetch next batch
return ds # Return the built dataset
"""
Now let's create train and valid dataloader using above funciton.
"""
# Build train dataloader
train_texts = train_df.options.tolist() # Extract training texts
train_labels = train_df.label.tolist() # Extract training labels
train_ds = build_dataset(
train_texts,
train_labels,
batch_size=CFG.batch_size,
cache=True,
shuffle=True,
repeat=True,
augment=CFG.augment,
)
# Build valid dataloader
valid_texts = valid_df.options.tolist() # Extract validation texts
valid_labels = valid_df.label.tolist() # Extract validation labels
valid_ds = build_dataset(
valid_texts,
valid_labels,
batch_size=CFG.batch_size,
cache=True,
shuffle=False,
repeat=False,
augment=False,
)
"""
## LR Schedule
Implementing a learning rate scheduler is crucial for transfer learning. The learning
rate initiates at `lr_start` and gradually tapers down to `lr_min` using **cosine**
curve.
**Importance:** A well-structured learning rate schedule is essential for efficient model
training, ensuring optimal convergence and avoiding issues such as overshooting or
stagnation.
"""
import math
def get_lr_callback(batch_size=8, mode="cos", epochs=10, plot=False):
lr_start, lr_max, lr_min = 1.0e-6, 0.6e-6 * batch_size, 1e-6
lr_ramp_ep, lr_sus_ep = 2, 0
def lrfn(epoch): # Learning rate update function
if epoch < lr_ramp_ep:
lr = (lr_max - lr_start) / lr_ramp_ep * epoch + lr_start
elif epoch < lr_ramp_ep + lr_sus_ep:
lr = lr_max
else:
decay_total_epochs, decay_epoch_index = (
epochs - lr_ramp_ep - lr_sus_ep + 3,
epoch - lr_ramp_ep - lr_sus_ep,
)
phase = math.pi * decay_epoch_index / decay_total_epochs
lr = (lr_max - lr_min) * 0.5 * (1 + math.cos(phase)) + lr_min
return lr
if plot: # Plot lr curve if plot is True
plt.figure(figsize=(10, 5))
plt.plot(
np.arange(epochs),
[lrfn(epoch) for epoch in np.arange(epochs)],
marker="o",
)
plt.xlabel("epoch")
plt.ylabel("lr")
plt.title("LR Scheduler")
plt.show()
return keras.callbacks.LearningRateScheduler(
lrfn, verbose=False
) # Create lr callback
_ = get_lr_callback(CFG.batch_size, plot=True)
"""
## Callbacks
The function below will gather all the training callbacks, such as `lr_scheduler`,
`model_checkpoint`.
"""
def get_callbacks():
callbacks = []
lr_cb = get_lr_callback(CFG.batch_size) # Get lr callback
ckpt_cb = keras.callbacks.ModelCheckpoint(
f"best.keras",
monitor="val_accuracy",
save_best_only=True,
save_weights_only=False,
mode="max",
) # Get Model checkpoint callback
callbacks.extend([lr_cb, ckpt_cb]) # Add lr and checkpoint callbacks
return callbacks # Return the list of callbacks
callbacks = get_callbacks()
"""
## MultipleChoice Model
"""
"""
### Pre-trained Models
The `KerasNLP` library provides comprehensive, ready-to-use implementations of popular
NLP model architectures. It features a variety of pre-trained models including `Bert`,
`Roberta`, `DebertaV3`, and more. In this notebook, we'll showcase the usage of
`DistillBert`. However, feel free to explore all available models in the [KerasNLP
documentation](https://keras.io/api/keras_nlp/models/). Also for a deeper understanding
of `KerasNLP`, refer to the informative [getting started
guide](https://keras.io/guides/keras_nlp/getting_started/).
Our approach involves using `keras_nlp.models.XXClassifier` to process each question and
option pari (e.g. (Q+A), (Q+B), etc.), generating logits. These logits are then combined
and passed through a softmax function to produce the final output.
"""
"""
### Classifier for Multiple-Choice Tasks
When dealing with multiple-choice questions, instead of giving the model the question and
all options together `(Q + A + B + C ...)`, we provide the model with one option at a
time along with the question. For instance, `(Q + A)`, `(Q + B)`, and so on. Once we have
the prediction scores (logits) for all options, we combine them using the `Softmax`
function to get the ultimate result. If we had given all options at once to the model,
the text's length would increase, making it harder for the model to handle. The picture
below illustrates this idea:
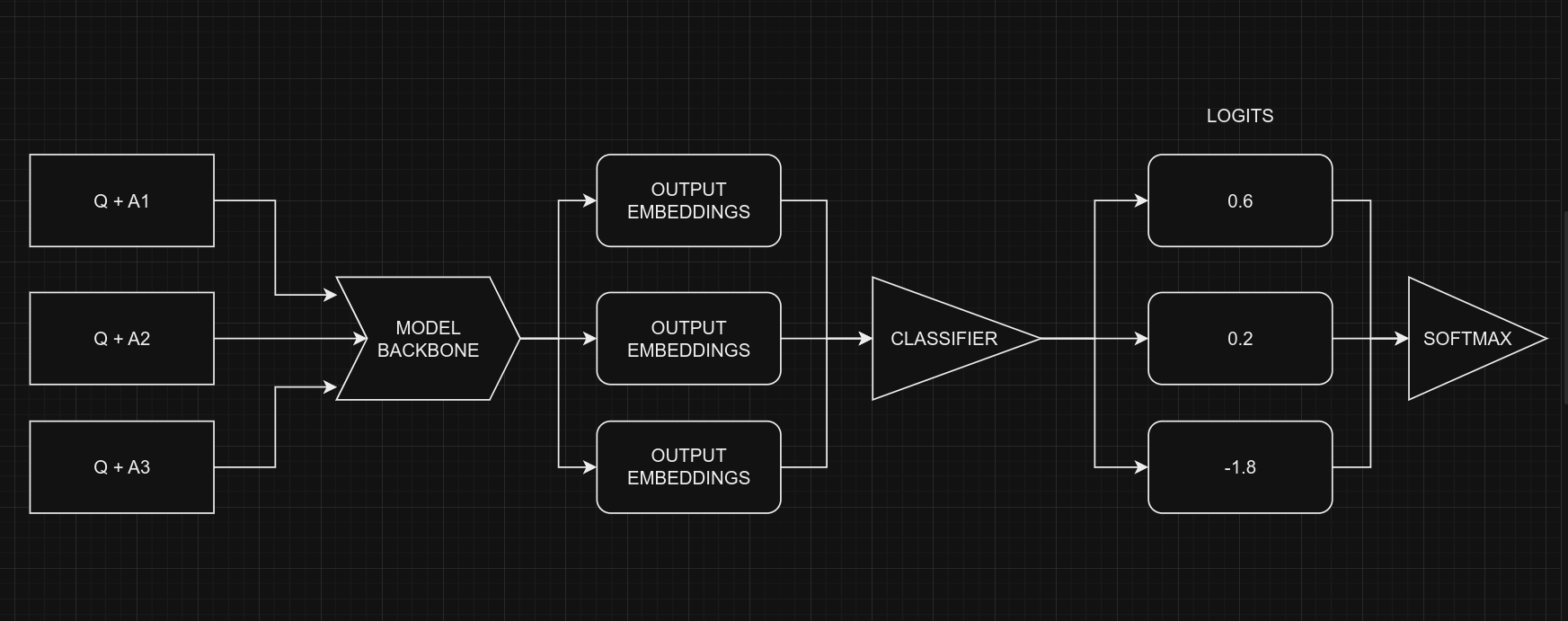
<div align="center"><b> Picture Credict: </b> <a
href="https://twitter.com/johnowhitaker"> @johnowhitaker </a> </div></div><br>
From a coding perspective, remember that we use the same model for all five options, with
shared weights. Despite the figure suggesting five separate models, they are, in fact,
one model with shared weights. Another point to consider is the the input shapes of
Classifier and MultipleChoice.
* Input shape for **Multiple Choice**: $(batch\_size, num\_choices, seq\_length)$
* Input shape for **Classifier**: $(batch\_size, seq\_length)$
Certainly, it's clear that we can't directly give the data for the multiple-choice task
to the model because the input shapes don't match. To handle this, we'll use **slicing**.
This means we'll separate the features of each option, like $feature_{(Q + A)}$ and
$feature_{(Q + B)}$, and give them one by one to the NLP classifier. After we get the
prediction scores $logits_{(Q + A)}$ and $logits_{(Q + B)}$ for all the options, we'll
use the Softmax function, like $\operatorname{Softmax}([logits_{(Q + A)}, logits_{(Q +
B)}])$, to combine them. This final step helps us make the ultimate decision or choice.
> Note that in the classifier, we set `num_classes=1` instead of `5`. This is because the
classifier produces a single output for each option. When dealing with five options,
these individual outputs are joined together and then processed through a softmax
function to generate the final result, which has a dimension of `5`.
"""
# Selects one option from five
class SelectOption(keras.layers.Layer):
def __init__(self, index, **kwargs):
super().__init__(**kwargs)
self.index = index
def call(self, inputs):
# Selects a specific slice from the inputs tensor
return inputs[:, self.index, :]
def get_config(self):
# For serialize the model
base_config = super().get_config()
config = {
"index": self.index,
}
return {**base_config, **config}
def build_model():
# Define input layers
inputs = {
"token_ids": keras.Input(
shape=(4, None), dtype=t"int32", name="token_ids"
),
"padding_mask": keras.Input(
shape=(4, None), dtype="int32", name="padding_mask"
),
}
# Create a DebertaV3Classifier model
classifier = keras_nlp.models.DebertaV3Classifier.from_preset(
CFG.preset,
preprocessor=None,
num_classes=1, # one output per one option, for five options total 5 outputs
)
logits = []
# Loop through each option (Q+A), (Q+B) etc and compute associted logits
for option_idx in range(4):
option = {
k: SelectOption(option_idx, name=f"{k}_{option_idx}")(v)
for k, v in inputs.items()
}
logit = classifier(option)
logits.append(logit)
# Compute final output
logits = keras.layers.Concatenate(axis=-1)(logits)
outputs = keras.layers.Softmax(axis=-1)(logits)
model = keras.Model(inputs, outputs)
# Compile the model with optimizer, loss, and metrics
model.compile(
optimizer=keras.optimizers.AdamW(5e-6),
loss=keras.losses.CategoricalCrossentropy(label_smoothing=0.02),
metrics=[
keras.metrics.CategoricalAccuracy(name="accuracy"),
],
jit_compile=True,
)
return model
# Build the Build
model = build_model()
"""
Let's checkout the model summary to have a better insight on the model.
"""
model.summary()
"""
Finally, let's check the model structure visually if everything is in place.
"""
keras.utils.plot_model(model, show_shapes=True)
"""
## Training
"""
# Start training the model
history = model.fit(
train_ds,
epochs=CFG.epochs,
validation_data=valid_ds,
callbacks=callbacks,
steps_per_epoch=int(len(train_df) / CFG.batch_size),
verbose=1,
)
"""
## Inference
"""
# Make predictions using the trained model on last validation data
predictions = model.predict(
valid_ds,
batch_size=CFG.batch_size, # max batch size = valid size
verbose=1,
)
# Format predictions and true answers
pred_answers = np.arange(4)[np.argsort(-predictions)][:, 0]
true_answers = valid_df.label.values
# Check 5 Predictions
print("# Predictions\n")
for i in range(0, 50, 10):
row = valid_df.iloc[i]
question = row.startphrase
pred_answer = f"ending{pred_answers[i]}"
true_answer = f"ending{true_answers[i]}"
print(f"❓ Sentence {i+1}:\n{question}\n")
print(f"✅ True Ending: {true_answer}\n >> {row[true_answer]}\n")
print(f"🤖 Predicted Ending: {pred_answer}\n >> {row[pred_answer]}\n")
print("-" * 90, "\n")
"""
## Reference
* [Multiple Choice with
HF](https://twitter.com/johnowhitaker/status/1689790373454041089?s=20)
* [Keras NLP](https://keras.io/api/keras_nlp/)
* [BirdCLEF23: Pretraining is All you Need
[Train]](https://www.kaggle.com/code/awsaf49/birdclef23-pretraining-is-all-you-need-train)
[Train]](https://www.kaggle.com/code/awsaf49/birdclef23-pretraining-is-all-you-need-train)
* [Triple Stratified KFold with
TFRecords](https://www.kaggle.com/code/cdeotte/triple-stratified-kfold-with-tfrecords)
"""
|
keras-core/examples/keras_io/nlp/multiple_choice_task_with_transfer_learning.py/0
|
{
"file_path": "keras-core/examples/keras_io/nlp/multiple_choice_task_with_transfer_learning.py",
"repo_id": "keras-core",
"token_count": 6555
}
| 24 |
"""
Title: MixUp augmentation for image classification
Author: [Sayak Paul](https://twitter.com/RisingSayak)
Date created: 2021/03/06
Last modified: 2023/07/24
Description: Data augmentation using the mixup technique for image classification.
Accelerator: GPU
"""
"""
## Introduction
"""
"""
_mixup_ is a *domain-agnostic* data augmentation technique proposed in [mixup: Beyond Empirical Risk Minimization](https://arxiv.org/abs/1710.09412)
by Zhang et al. It's implemented with the following formulas:

(Note that the lambda values are values with the [0, 1] range and are sampled from the
[Beta distribution](https://en.wikipedia.org/wiki/Beta_distribution).)
The technique is quite systematically named. We are literally mixing up the features and
their corresponding labels. Implementation-wise it's simple. Neural networks are prone
to [memorizing corrupt labels](https://arxiv.org/abs/1611.03530). mixup relaxes this by
combining different features with one another (same happens for the labels too) so that
a network does not get overconfident about the relationship between the features and
their labels.
mixup is specifically useful when we are not sure about selecting a set of augmentation
transforms for a given dataset, medical imaging datasets, for example. mixup can be
extended to a variety of data modalities such as computer vision, naturallanguage
processing, speech, and so on.
"""
"""
## Setup
"""
import numpy as np
import keras_core as keras
import matplotlib.pyplot as plt
from keras_core import layers
# TF imports related to tf.data preprocessing
from tensorflow import data as tf_data
from tensorflow import image as tf_image
from tensorflow.random import gamma as tf_random_gamma
"""
## Prepare the dataset
In this example, we will be using the [FashionMNIST](https://github.com/zalandoresearch/fashion-mnist) dataset. But this same recipe can
be used for other classification datasets as well.
"""
(x_train, y_train), (x_test, y_test) = keras.datasets.fashion_mnist.load_data()
x_train = x_train.astype("float32") / 255.0
x_train = np.reshape(x_train, (-1, 28, 28, 1))
y_train = keras.ops.one_hot(y_train, 10)
x_test = x_test.astype("float32") / 255.0
x_test = np.reshape(x_test, (-1, 28, 28, 1))
y_test = keras.ops.one_hot(y_test, 10)
"""
## Define hyperparameters
"""
AUTO = tf_data.AUTOTUNE
BATCH_SIZE = 64
EPOCHS = 10
"""
## Convert the data into TensorFlow `Dataset` objects
"""
# Put aside a few samples to create our validation set
val_samples = 2000
x_val, y_val = x_train[:val_samples], y_train[:val_samples]
new_x_train, new_y_train = x_train[val_samples:], y_train[val_samples:]
train_ds_one = (
tf_data.Dataset.from_tensor_slices((new_x_train, new_y_train))
.shuffle(BATCH_SIZE * 100)
.batch(BATCH_SIZE)
)
train_ds_two = (
tf_data.Dataset.from_tensor_slices((new_x_train, new_y_train))
.shuffle(BATCH_SIZE * 100)
.batch(BATCH_SIZE)
)
# Because we will be mixing up the images and their corresponding labels, we will be
# combining two shuffled datasets from the same training data.
train_ds = tf_data.Dataset.zip((train_ds_one, train_ds_two))
val_ds = tf_data.Dataset.from_tensor_slices((x_val, y_val)).batch(BATCH_SIZE)
test_ds = tf_data.Dataset.from_tensor_slices((x_test, y_test)).batch(BATCH_SIZE)
"""
## Define the mixup technique function
To perform the mixup routine, we create new virtual datasets using the training data from
the same dataset, and apply a lambda value within the [0, 1] range sampled from a [Beta distribution](https://en.wikipedia.org/wiki/Beta_distribution)
— such that, for example, `new_x = lambda * x1 + (1 - lambda) * x2` (where
`x1` and `x2` are images) and the same equation is applied to the labels as well.
"""
def sample_beta_distribution(size, concentration_0=0.2, concentration_1=0.2):
gamma_1_sample = tf_random_gamma(shape=[size], alpha=concentration_1)
gamma_2_sample = tf_random_gamma(shape=[size], alpha=concentration_0)
return gamma_1_sample / (gamma_1_sample + gamma_2_sample)
def mix_up(ds_one, ds_two, alpha=0.2):
# Unpack two datasets
images_one, labels_one = ds_one
images_two, labels_two = ds_two
batch_size = keras.backend.shape(images_one)[0]
# Sample lambda and reshape it to do the mixup
l = sample_beta_distribution(batch_size, alpha, alpha)
x_l = keras.ops.reshape(l, (batch_size, 1, 1, 1))
y_l = keras.ops.reshape(l, (batch_size, 1))
# Perform mixup on both images and labels by combining a pair of images/labels
# (one from each dataset) into one image/label
images = images_one * x_l + images_two * (1 - x_l)
labels = labels_one * y_l + labels_two * (1 - y_l)
return (images, labels)
"""
**Note** that here , we are combining two images to create a single one. Theoretically,
we can combine as many we want but that comes at an increased computation cost. In
certain cases, it may not help improve the performance as well.
"""
"""
## Visualize the new augmented dataset
"""
# First create the new dataset using our `mix_up` utility
train_ds_mu = train_ds.map(
lambda ds_one, ds_two: mix_up(ds_one, ds_two, alpha=0.2),
num_parallel_calls=AUTO,
)
# Let's preview 9 samples from the dataset
sample_images, sample_labels = next(iter(train_ds_mu))
plt.figure(figsize=(10, 10))
for i, (image, label) in enumerate(zip(sample_images[:9], sample_labels[:9])):
ax = plt.subplot(3, 3, i + 1)
plt.imshow(image.numpy().squeeze())
print(label.numpy().tolist())
plt.axis("off")
"""
## Model building
"""
def get_training_model():
model = keras.Sequential(
[
layers.Conv2D(
16, (5, 5), activation="relu", input_shape=(28, 28, 1)
),
layers.MaxPooling2D(pool_size=(2, 2)),
layers.Conv2D(32, (5, 5), activation="relu"),
layers.MaxPooling2D(pool_size=(2, 2)),
layers.Dropout(0.2),
layers.GlobalAveragePooling2D(),
layers.Dense(128, activation="relu"),
layers.Dense(10, activation="softmax"),
]
)
return model
"""
For the sake of reproducibility, we serialize the initial random weights of our shallow
network.
"""
initial_model = get_training_model()
initial_model.save_weights("initial_weights.weights.h5")
"""
## 1. Train the model with the mixed up dataset
"""
model = get_training_model()
model.load_weights("initial_weights.weights.h5")
model.compile(
loss="categorical_crossentropy", optimizer="adam", metrics=["accuracy"]
)
model.fit(train_ds_mu, validation_data=val_ds, epochs=EPOCHS)
_, test_acc = model.evaluate(test_ds)
print("Test accuracy: {:.2f}%".format(test_acc * 100))
"""
## 2. Train the model *without* the mixed up dataset
"""
model = get_training_model()
model.load_weights("initial_weights.weights.h5")
model.compile(
loss="categorical_crossentropy", optimizer="adam", metrics=["accuracy"]
)
# Notice that we are NOT using the mixed up dataset here
model.fit(train_ds_one, validation_data=val_ds, epochs=EPOCHS)
_, test_acc = model.evaluate(test_ds)
print("Test accuracy: {:.2f}%".format(test_acc * 100))
"""
Readers are encouraged to try out mixup on different datasets from different domains and
experiment with the lambda parameter. You are strongly advised to check out the
[original paper](https://arxiv.org/abs/1710.09412) as well - the authors present several ablation studies on mixup
showing how it can improve generalization, as well as show their results of combining
more than two images to create a single one.
"""
"""
## Notes
* With mixup, you can create synthetic examples — especially when you lack a large
dataset - without incurring high computational costs.
* [Label smoothing](https://www.pyimagesearch.com/2019/12/30/label-smoothing-with-keras-tensorflow-and-deep-learning/) and mixup usually do not work well together because label smoothing
already modifies the hard labels by some factor.
* mixup does not work well when you are using [Supervised Contrastive
Learning](https://arxiv.org/abs/2004.11362) (SCL) since SCL expects the true labels
during its pre-training phase.
* A few other benefits of mixup include (as described in the [paper](https://arxiv.org/abs/1710.09412)) robustness to
adversarial examples and stabilized GAN (Generative Adversarial Networks) training.
* There are a number of data augmentation techniques that extend mixup such as
[CutMix](https://arxiv.org/abs/1905.04899) and [AugMix](https://arxiv.org/abs/1912.02781).
"""
|
keras-core/examples/keras_io/tensorflow/vision/mixup.py/0
|
{
"file_path": "keras-core/examples/keras_io/tensorflow/vision/mixup.py",
"repo_id": "keras-core",
"token_count": 2954
}
| 25 |
import numpy as np
import tensorflow as tf
from keras_core import layers
from keras_core import losses
from keras_core import metrics
from keras_core import models
from keras_core import optimizers
from keras_core.utils import rng_utils
def test_model_fit():
cpus = tf.config.list_physical_devices("CPU")
tf.config.set_logical_device_configuration(
cpus[0],
[
tf.config.LogicalDeviceConfiguration(),
tf.config.LogicalDeviceConfiguration(),
],
)
rng_utils.set_random_seed(1337)
strategy = tf.distribute.MirroredStrategy(["CPU:0", "CPU:1"])
with strategy.scope():
inputs = layers.Input((100,), batch_size=32)
x = layers.Dense(256, activation="relu")(inputs)
x = layers.Dense(256, activation="relu")(x)
x = layers.Dense(256, activation="relu")(x)
x = layers.BatchNormalization()(x)
outputs = layers.Dense(16)(x)
model = models.Model(inputs, outputs)
model.summary()
x = np.random.random((50000, 100))
y = np.random.random((50000, 16))
batch_size = 32
epochs = 5
with strategy.scope():
model.compile(
optimizer=optimizers.SGD(learning_rate=0.001, momentum=0.01),
loss=losses.MeanSquaredError(),
metrics=[metrics.MeanSquaredError()],
# TODO(scottzhu): Find out where is the variable
# that is not created eagerly and break the usage of XLA.
jit_compile=False,
)
history = model.fit(
x, y, batch_size=batch_size, epochs=epochs, validation_split=0.2
)
print("History:")
print(history.history)
if __name__ == "__main__":
test_model_fit()
|
keras-core/integration_tests/distribute_training_test.py/0
|
{
"file_path": "keras-core/integration_tests/distribute_training_test.py",
"repo_id": "keras-core",
"token_count": 751
}
| 26 |
import json
import warnings
import numpy as np
from keras_core import activations
from keras_core import backend
from keras_core import ops
from keras_core.api_export import keras_core_export
from keras_core.utils import file_utils
CLASS_INDEX = None
CLASS_INDEX_PATH = (
"https://storage.googleapis.com/download.tensorflow.org/"
"data/imagenet_class_index.json"
)
PREPROCESS_INPUT_DOC = """
Preprocesses a tensor or Numpy array encoding a batch of images.
Usage example with `applications.MobileNet`:
```python
i = keras_core.layers.Input([None, None, 3], dtype="uint8")
x = ops.cast(i, "float32")
x = keras_core.applications.mobilenet.preprocess_input(x)
core = keras_core.applications.MobileNet()
x = core(x)
model = keras_core.Model(inputs=[i], outputs=[x])
result = model(image)
```
Args:
x: A floating point `numpy.array` or a backend-native tensor,
3D or 4D with 3 color
channels, with values in the range [0, 255].
The preprocessed data are written over the input data
if the data types are compatible. To avoid this
behaviour, `numpy.copy(x)` can be used.
data_format: Optional data format of the image tensor/array. None, means
the global setting `keras_core.backend.image_data_format()` is used
(unless you changed it, it uses "channels_last").{mode}
Defaults to `None`.
Returns:
Preprocessed array with type `float32`.
{ret}
Raises:
{error}
"""
PREPROCESS_INPUT_MODE_DOC = """
mode: One of "caffe", "tf" or "torch".
- caffe: will convert the images from RGB to BGR,
then will zero-center each color channel with
respect to the ImageNet dataset,
without scaling.
- tf: will scale pixels between -1 and 1,
sample-wise.
- torch: will scale pixels between 0 and 1 and then
will normalize each channel with respect to the
ImageNet dataset.
Defaults to `"caffe"`.
"""
PREPROCESS_INPUT_DEFAULT_ERROR_DOC = """
ValueError: In case of unknown `mode` or `data_format` argument."""
PREPROCESS_INPUT_ERROR_DOC = """
ValueError: In case of unknown `data_format` argument."""
PREPROCESS_INPUT_RET_DOC_TF = """
The inputs pixel values are scaled between -1 and 1, sample-wise."""
PREPROCESS_INPUT_RET_DOC_TORCH = """
The input pixels values are scaled between 0 and 1 and each channel is
normalized with respect to the ImageNet dataset."""
PREPROCESS_INPUT_RET_DOC_CAFFE = """
The images are converted from RGB to BGR, then each color channel is
zero-centered with respect to the ImageNet dataset, without scaling."""
@keras_core_export("keras_core.applications.imagenet_utils.preprocess_input")
def preprocess_input(x, data_format=None, mode="caffe"):
"""Preprocesses a tensor or Numpy array encoding a batch of images."""
if mode not in {"caffe", "tf", "torch"}:
raise ValueError(
"Expected mode to be one of `caffe`, `tf` or `torch`. "
f"Received: mode={mode}"
)
if data_format is None:
data_format = backend.image_data_format()
elif data_format not in {"channels_first", "channels_last"}:
raise ValueError(
"Expected data_format to be one of `channels_first` or "
f"`channels_last`. Received: data_format={data_format}"
)
if isinstance(x, np.ndarray):
return _preprocess_numpy_input(x, data_format=data_format, mode=mode)
else:
return _preprocess_tensor_input(x, data_format=data_format, mode=mode)
preprocess_input.__doc__ = PREPROCESS_INPUT_DOC.format(
mode=PREPROCESS_INPUT_MODE_DOC,
ret="",
error=PREPROCESS_INPUT_DEFAULT_ERROR_DOC,
)
@keras_core_export("keras_core.applications.imagenet_utils.decode_predictions")
def decode_predictions(preds, top=5):
"""Decodes the prediction of an ImageNet model.
Args:
preds: NumPy array encoding a batch of predictions.
top: Integer, how many top-guesses to return. Defaults to `5`.
Returns:
A list of lists of top class prediction tuples
`(class_name, class_description, score)`.
One list of tuples per sample in batch input.
Raises:
ValueError: In case of invalid shape of the `pred` array
(must be 2D).
"""
global CLASS_INDEX
if len(preds.shape) != 2 or preds.shape[1] != 1000:
raise ValueError(
"`decode_predictions` expects "
"a batch of predictions "
"(i.e. a 2D array of shape (samples, 1000)). "
f"Received array with shape: {preds.shape}"
)
if CLASS_INDEX is None:
fpath = file_utils.get_file(
"imagenet_class_index.json",
CLASS_INDEX_PATH,
cache_subdir="models",
file_hash="c2c37ea517e94d9795004a39431a14cb",
)
with open(fpath) as f:
CLASS_INDEX = json.load(f)
results = []
preds = ops.convert_to_numpy(preds)
for pred in preds:
top_indices = pred.argsort()[-top:][::-1]
result = [tuple(CLASS_INDEX[str(i)]) + (pred[i],) for i in top_indices]
result.sort(key=lambda x: x[2], reverse=True)
results.append(result)
return results
def _preprocess_numpy_input(x, data_format, mode):
"""Preprocesses a NumPy array encoding a batch of images.
Args:
x: Input array, 3D or 4D.
data_format: Data format of the image array.
mode: One of "caffe", "tf" or "torch".
- caffe: will convert the images from RGB to BGR,
then will zero-center each color channel with
respect to the ImageNet dataset,
without scaling.
- tf: will scale pixels between -1 and 1,
sample-wise.
- torch: will scale pixels between 0 and 1 and then
will normalize each channel with respect to the
ImageNet dataset.
Returns:
Preprocessed Numpy array.
"""
if not issubclass(x.dtype.type, np.floating):
x = x.astype(backend.floatx(), copy=False)
if mode == "tf":
x /= 127.5
x -= 1.0
return x
elif mode == "torch":
x /= 255.0
mean = [0.485, 0.456, 0.406]
std = [0.229, 0.224, 0.225]
else:
if data_format == "channels_first":
# 'RGB'->'BGR'
if len(x.shape) == 3:
x = x[::-1, ...]
else:
x = x[:, ::-1, ...]
else:
# 'RGB'->'BGR'
x = x[..., ::-1]
mean = [103.939, 116.779, 123.68]
std = None
# Zero-center by mean pixel
if data_format == "channels_first":
if len(x.shape) == 3:
x[0, :, :] -= mean[0]
x[1, :, :] -= mean[1]
x[2, :, :] -= mean[2]
if std is not None:
x[0, :, :] /= std[0]
x[1, :, :] /= std[1]
x[2, :, :] /= std[2]
else:
x[:, 0, :, :] -= mean[0]
x[:, 1, :, :] -= mean[1]
x[:, 2, :, :] -= mean[2]
if std is not None:
x[:, 0, :, :] /= std[0]
x[:, 1, :, :] /= std[1]
x[:, 2, :, :] /= std[2]
else:
x[..., 0] -= mean[0]
x[..., 1] -= mean[1]
x[..., 2] -= mean[2]
if std is not None:
x[..., 0] /= std[0]
x[..., 1] /= std[1]
x[..., 2] /= std[2]
return x
def _preprocess_tensor_input(x, data_format, mode):
"""Preprocesses a tensor encoding a batch of images.
Args:
x: Input tensor, 3D or 4D.
data_format: Data format of the image tensor.
mode: One of "caffe", "tf" or "torch".
- caffe: will convert the images from RGB to BGR,
then will zero-center each color channel with
respect to the ImageNet dataset,
without scaling.
- tf: will scale pixels between -1 and 1,
sample-wise.
- torch: will scale pixels between 0 and 1 and then
will normalize each channel with respect to the
ImageNet dataset.
Returns:
Preprocessed tensor.
"""
ndim = len(x.shape)
if mode == "tf":
x /= 127.5
x -= 1.0
return x
elif mode == "torch":
x /= 255.0
mean = [0.485, 0.456, 0.406]
std = [0.229, 0.224, 0.225]
else:
if data_format == "channels_first":
# 'RGB'->'BGR'
if len(x.shape) == 3:
x = ops.stack([x[i, ...] for i in (2, 1, 0)], axis=0)
else:
x = ops.stack([x[:, i, :] for i in (2, 1, 0)], axis=1)
else:
# 'RGB'->'BGR'
x = ops.stack([x[..., i] for i in (2, 1, 0)], axis=-1)
mean = [103.939, 116.779, 123.68]
std = None
mean_tensor = ops.convert_to_tensor(-np.array(mean), dtype=x.dtype)
# Zero-center by mean pixel
if data_format == "channels_first":
mean_tensor = ops.reshape(mean_tensor, (1, 3) + (1,) * (ndim - 2))
else:
mean_tensor = ops.reshape(mean_tensor, (1,) * (ndim - 1) + (3,))
x += mean_tensor
if std is not None:
std_tensor = ops.convert_to_tensor(np.array(std), dtype=x.dtype)
if data_format == "channels_first":
std_tensor = ops.reshape(std_tensor, (-1, 1, 1))
x /= std_tensor
return x
def obtain_input_shape(
input_shape,
default_size,
min_size,
data_format,
require_flatten,
weights=None,
):
"""Internal utility to compute/validate a model's input shape.
Args:
input_shape: Either None (will return the default network input shape),
or a user-provided shape to be validated.
default_size: Default input width/height for the model.
min_size: Minimum input width/height accepted by the model.
data_format: Image data format to use.
require_flatten: Whether the model is expected to
be linked to a classifier via a Flatten layer.
weights: One of `None` (random initialization)
or 'imagenet' (pre-training on ImageNet).
If weights='imagenet' input channels must be equal to 3.
Returns:
An integer shape tuple (may include None entries).
Raises:
ValueError: In case of invalid argument values.
"""
if weights != "imagenet" and input_shape and len(input_shape) == 3:
if data_format == "channels_first":
if input_shape[0] not in {1, 3}:
warnings.warn(
"This model usually expects 1 or 3 input channels. "
"However, it was passed an input_shape "
f"with {input_shape[0]} input channels.",
stacklevel=2,
)
default_shape = (input_shape[0], default_size, default_size)
else:
if input_shape[-1] not in {1, 3}:
warnings.warn(
"This model usually expects 1 or 3 input channels. "
"However, it was passed an input_shape "
f"with {input_shape[-1]} input channels.",
stacklevel=2,
)
default_shape = (default_size, default_size, input_shape[-1])
else:
if data_format == "channels_first":
default_shape = (3, default_size, default_size)
else:
default_shape = (default_size, default_size, 3)
if weights == "imagenet" and require_flatten:
if input_shape is not None:
if input_shape != default_shape:
raise ValueError(
"When setting `include_top=True` "
"and loading `imagenet` weights, "
f"`input_shape` should be {default_shape}. "
f"Received: input_shape={input_shape}"
)
return default_shape
if input_shape:
if data_format == "channels_first":
if input_shape is not None:
if len(input_shape) != 3:
raise ValueError(
"`input_shape` must be a tuple of three integers."
)
if input_shape[0] != 3 and weights == "imagenet":
raise ValueError(
"The input must have 3 channels; Received "
f"`input_shape={input_shape}`"
)
if (
input_shape[1] is not None and input_shape[1] < min_size
) or (input_shape[2] is not None and input_shape[2] < min_size):
raise ValueError(
f"Input size must be at least {min_size}"
f"x{min_size}; Received: "
f"input_shape={input_shape}"
)
else:
if input_shape is not None:
if len(input_shape) != 3:
raise ValueError(
"`input_shape` must be a tuple of three integers."
)
if input_shape[-1] != 3 and weights == "imagenet":
raise ValueError(
"The input must have 3 channels; Received "
f"`input_shape={input_shape}`"
)
if (
input_shape[0] is not None and input_shape[0] < min_size
) or (input_shape[1] is not None and input_shape[1] < min_size):
raise ValueError(
"Input size must be at least "
f"{min_size}x{min_size}; Received: "
f"input_shape={input_shape}"
)
else:
if require_flatten:
input_shape = default_shape
else:
if data_format == "channels_first":
input_shape = (3, None, None)
else:
input_shape = (None, None, 3)
if require_flatten:
if None in input_shape:
raise ValueError(
"If `include_top` is True, "
"you should specify a static `input_shape`. "
f"Received: input_shape={input_shape}"
)
return input_shape
def correct_pad(inputs, kernel_size):
"""Returns a tuple for zero-padding for 2D convolution with downsampling.
Args:
inputs: Input tensor.
kernel_size: An integer or tuple/list of 2 integers.
Returns:
A tuple.
"""
img_dim = 2 if backend.image_data_format() == "channels_first" else 1
input_size = inputs.shape[img_dim : (img_dim + 2)]
if isinstance(kernel_size, int):
kernel_size = (kernel_size, kernel_size)
if input_size[0] is None:
adjust = (1, 1)
else:
adjust = (1 - input_size[0] % 2, 1 - input_size[1] % 2)
correct = (kernel_size[0] // 2, kernel_size[1] // 2)
return (
(correct[0] - adjust[0], correct[0]),
(correct[1] - adjust[1], correct[1]),
)
def validate_activation(classifier_activation, weights):
"""validates that the classifer_activation is compatible with the weights.
Args:
classifier_activation: str or callable activation function
weights: The pretrained weights to load.
Raises:
ValueError: if an activation other than `None` or `softmax` are used with
pretrained weights.
"""
if weights is None:
return
classifier_activation = activations.get(classifier_activation)
if classifier_activation not in {
activations.get("softmax"),
activations.get(None),
}:
raise ValueError(
"Only `None` and `softmax` activations are allowed "
"for the `classifier_activation` argument when using "
"pretrained weights, with `include_top=True`; Received: "
f"classifier_activation={classifier_activation}"
)
|
keras-core/keras_core/applications/imagenet_utils.py/0
|
{
"file_path": "keras-core/keras_core/applications/imagenet_utils.py",
"repo_id": "keras-core",
"token_count": 7599
}
| 27 |
from keras_core.backend.common.backend_utils import (
_convert_conv_tranpose_padding_args_from_keras_to_jax,
)
from keras_core.backend.common.backend_utils import (
_convert_conv_tranpose_padding_args_from_keras_to_torch,
)
from keras_core.backend.common.backend_utils import (
_get_output_shape_given_tf_padding,
)
from keras_core.backend.common.backend_utils import (
compute_conv_transpose_padding_args_for_jax,
)
from keras_core.backend.common.backend_utils import (
compute_conv_transpose_padding_args_for_torch,
)
from keras_core.testing import test_case
class ConvertConvTransposePaddingArgsJAXTest(test_case.TestCase):
def test_valid_padding_without_output_padding(self):
"""Test conversion with 'valid' padding and no output padding"""
(
left_pad,
right_pad,
) = _convert_conv_tranpose_padding_args_from_keras_to_jax(
kernel_size=3,
stride=2,
dilation_rate=1,
padding="valid",
output_padding=None,
)
self.assertEqual(left_pad, 2)
self.assertEqual(right_pad, 2)
def test_same_padding_without_output_padding(self):
"""Test conversion with 'same' padding and no output padding."""
(
left_pad,
right_pad,
) = _convert_conv_tranpose_padding_args_from_keras_to_jax(
kernel_size=3,
stride=2,
dilation_rate=1,
padding="same",
output_padding=None,
)
self.assertEqual(left_pad, 2)
self.assertEqual(right_pad, 1)
class ConvertConvTransposePaddingArgsTorchTest(test_case.TestCase):
def test_valid_padding_without_output_padding(self):
"""Test conversion with 'valid' padding and no output padding"""
(
torch_padding,
torch_output_padding,
) = _convert_conv_tranpose_padding_args_from_keras_to_torch(
kernel_size=3,
stride=2,
dilation_rate=1,
padding="valid",
output_padding=None,
)
self.assertEqual(torch_padding, 0)
self.assertEqual(torch_output_padding, 0)
def test_same_padding_without_output_padding(self):
"""Test conversion with 'same' padding and no output padding"""
(
torch_padding,
torch_output_padding,
) = _convert_conv_tranpose_padding_args_from_keras_to_torch(
kernel_size=3,
stride=2,
dilation_rate=1,
padding="same",
output_padding=None,
)
self.assertEqual(torch_padding, 1)
self.assertEqual(torch_output_padding, 1)
class ComputeConvTransposePaddingArgsForJAXTest(test_case.TestCase):
def test_valid_padding_without_output_padding(self):
"""Test computation with 'valid' padding and no output padding"""
jax_padding = compute_conv_transpose_padding_args_for_jax(
input_shape=(1, 5, 5, 3),
kernel_shape=(3, 3, 3, 3),
strides=2,
padding="valid",
output_padding=None,
dilation_rate=1,
)
self.assertEqual(jax_padding, [(2, 2), (2, 2)])
def test_same_padding_without_output_padding(self):
"""Test computation with 'same' padding and no output padding"""
jax_padding = compute_conv_transpose_padding_args_for_jax(
input_shape=(1, 5, 5, 3),
kernel_shape=(3, 3, 3, 3),
strides=2,
padding="same",
output_padding=None,
dilation_rate=1,
)
self.assertEqual(jax_padding, [(2, 1), (2, 1)])
class ComputeConvTransposePaddingArgsForTorchTest(test_case.TestCase):
def test_valid_padding_without_output_padding(self):
"""Test computation with 'valid' padding and no output padding"""
(
torch_paddings,
torch_output_paddings,
) = compute_conv_transpose_padding_args_for_torch(
input_shape=(1, 5, 5, 3),
kernel_shape=(3, 3, 3, 3),
strides=2,
padding="valid",
output_padding=None,
dilation_rate=1,
)
self.assertEqual(torch_paddings, [0, 0])
self.assertEqual(torch_output_paddings, [0, 0])
def test_same_padding_without_output_padding(self):
"""Test computation with 'same' padding and no output padding"""
(
torch_paddings,
torch_output_paddings,
) = compute_conv_transpose_padding_args_for_torch(
input_shape=(1, 5, 5, 3),
kernel_shape=(3, 3, 3, 3),
strides=2,
padding="same",
output_padding=None,
dilation_rate=1,
)
self.assertEqual(torch_paddings, [1, 1])
self.assertEqual(torch_output_paddings, [1, 1])
def test_valid_padding_with_none_output_padding(self):
"""Test conversion with 'valid' padding and no output padding"""
(
torch_padding,
torch_output_padding,
) = _convert_conv_tranpose_padding_args_from_keras_to_torch(
kernel_size=3,
stride=2,
dilation_rate=1,
padding="valid",
output_padding=None,
)
self.assertEqual(torch_padding, 0)
self.assertEqual(torch_output_padding, 0)
def test_valid_padding_with_output_padding(self):
"""Test conversion with 'valid' padding and output padding for Torch."""
(
torch_padding,
torch_output_padding,
) = _convert_conv_tranpose_padding_args_from_keras_to_torch(
kernel_size=3,
stride=2,
dilation_rate=1,
padding="valid",
output_padding=1,
)
self.assertEqual(torch_padding, 0)
self.assertEqual(torch_output_padding, 1)
class GetOutputShapeGivenTFPaddingTest(test_case.TestCase):
def test_valid_padding_without_output_padding(self):
"""Test computation with 'valid' padding and no output padding."""
output_shape = _get_output_shape_given_tf_padding(
input_size=5,
kernel_size=3,
strides=2,
padding="valid",
output_padding=None,
dilation_rate=1,
)
self.assertEqual(output_shape, 11)
def test_same_padding_without_output_padding(self):
"""Test computation with 'same' padding and no output padding."""
output_shape = _get_output_shape_given_tf_padding(
input_size=5,
kernel_size=3,
strides=2,
padding="same",
output_padding=None,
dilation_rate=1,
)
self.assertEqual(output_shape, 10)
def test_valid_padding_with_output_padding(self):
"""Test computation with 'valid' padding and output padding."""
output_shape = _get_output_shape_given_tf_padding(
input_size=5,
kernel_size=3,
strides=2,
padding="valid",
output_padding=1,
dilation_rate=1,
)
self.assertEqual(output_shape, 12)
def test_warning_for_inconsistencies(self):
"""Test that a warning is raised for potential inconsistencies"""
with self.assertWarns(Warning):
_convert_conv_tranpose_padding_args_from_keras_to_torch(
kernel_size=3,
stride=2,
dilation_rate=1,
padding="same",
output_padding=1,
)
def test_same_padding_without_output_padding_for_torch_(self):
"""Test conversion with 'same' padding and no output padding."""
(
torch_padding,
torch_output_padding,
) = _convert_conv_tranpose_padding_args_from_keras_to_torch(
kernel_size=3,
stride=2,
dilation_rate=1,
padding="same",
output_padding=None,
)
self.assertEqual(torch_padding, max(-((3 % 2 - 3) // 2), 0))
self.assertEqual(torch_output_padding, 1)
|
keras-core/keras_core/backend/common/backend_utils_test.py/0
|
{
"file_path": "keras-core/keras_core/backend/common/backend_utils_test.py",
"repo_id": "keras-core",
"token_count": 3992
}
| 28 |
"""!!!DO NOT USE!!!
Distribution related class for JAX backend.
This is just a prototype and we might want to unify it
with other backends in the future.
"""
import jax
def list_devices(device_type=None):
"""Return all the available devices based on the device type.
Note that this should return the global devices in a distributed setting.
Args:
device_type: string of `"cpu"`, `"gpu"` or `"tpu"`. Defaults to `"gpu"`
or `"tpu"` if available when device_type is not provided. Otherwise
will return the `"cpu"` devices.
Return:
List of devices that are available for distribute computation.
"""
device_type = device_type.lower() if device_type else None
return jax.devices(backend=device_type)
def to_jax_mesh(device_mesh):
"""Convert the DeviceMesh to JAX backend specific Mesh.
Args:
device_mesh: DeviceMesh instance to convert.
Returns:
A `jax.sharding.Mesh` instance.
"""
return jax.sharding.Mesh(device_mesh.devices, device_mesh.axis_names)
def to_jax_layout(tensor_layout):
"""Convert the TensorLayout to JAX backend specific Sharding.
Args:
tensor_layout: TensorLayout instance to convert.
Returns:
A `jax.sharding.NamedSharding` instance.
"""
if tensor_layout.device_mesh is None:
raise ValueError(
"Cannot create sharding when device mesh is not set "
"for TensorLayout."
)
partition_spec = jax.sharding.PartitionSpec(*tensor_layout.axes)
jax_mesh = to_jax_mesh(tensor_layout.device_mesh)
return jax.sharding.NamedSharding(jax_mesh, partition_spec)
def distribute_value(value, tensor_layout):
"""Distribute the value based on the layout.
Args:
value: `jax.Array` that need to be distributed.
tensor_layout: `TensorLayout` for the distribution information, or a
`jax.sharding.Sharding` instance.
Returns:
Distributed value.
"""
if not isinstance(tensor_layout, jax.sharding.Sharding):
tensor_layout = to_jax_layout(tensor_layout)
return jax.device_put(value, tensor_layout)
|
keras-core/keras_core/backend/jax/distribution_lib.py/0
|
{
"file_path": "keras-core/keras_core/backend/jax/distribution_lib.py",
"repo_id": "keras-core",
"token_count": 829
}
| 29 |
"""Tests for SavedModel functionality under tf implementation."""
import os
import numpy as np
import pytest
import tensorflow as tf
from keras_core import backend
from keras_core import layers
from keras_core import metrics
from keras_core import models
from keras_core import testing
from keras_core.saving import object_registration
@object_registration.register_keras_serializable(package="my_package")
class CustomModelX(models.Model):
def __init__(self, *args, **kwargs):
super().__init__(*args, **kwargs)
self.dense1 = layers.Dense(1)
self.dense2 = layers.Dense(1)
def call(self, inputs):
out = self.dense1(inputs)
return self.dense2(out)
def one(self):
return 1
@pytest.mark.skipif(
backend.backend() != "tensorflow",
reason="The SavedModel test can only run with TF backend.",
)
class SavedModelTest(testing.TestCase):
def test_sequential(self):
model = models.Sequential([layers.Dense(1)])
model.compile(loss="mse", optimizer="adam")
X_train = np.random.rand(100, 3)
y_train = np.random.rand(100, 1)
model.fit(X_train, y_train)
path = os.path.join(self.get_temp_dir(), "my_keras_core_model")
tf.saved_model.save(model, path)
restored_model = tf.saved_model.load(path)
self.assertAllClose(
model(X_train),
restored_model.signatures["serving_default"](
tf.convert_to_tensor(X_train, dtype=tf.float32)
)["output_0"],
rtol=1e-4,
atol=1e-4,
)
def test_functional(self):
inputs = layers.Input(shape=(3,))
x = layers.Dense(1, name="first_dense")(inputs)
outputs = layers.Dense(1, name="second_dense")(x)
model = models.Model(inputs, outputs)
model.compile(
optimizer="adam",
loss="mse",
)
X_train = np.random.rand(100, 3)
y_train = np.random.rand(100, 1)
model.fit(X_train, y_train)
path = os.path.join(self.get_temp_dir(), "my_keras_core_model")
tf.saved_model.save(model, path)
restored_model = tf.saved_model.load(path)
self.assertAllClose(
model(X_train),
restored_model.signatures["serving_default"](
tf.convert_to_tensor(X_train, dtype=tf.float32)
)["output_0"],
rtol=1e-4,
atol=1e-4,
)
def test_subclassed(self):
model = CustomModelX()
model.compile(
optimizer="adam",
loss="mse",
metrics=[metrics.Hinge(), "mse"],
)
X_train = np.random.rand(100, 3)
y_train = np.random.rand(100, 1)
model.fit(X_train, y_train)
path = os.path.join(self.get_temp_dir(), "my_keras_core_model")
tf.saved_model.save(model, path)
restored_model = tf.saved_model.load(path)
self.assertAllClose(
model(X_train),
restored_model.signatures["serving_default"](
tf.convert_to_tensor(X_train, dtype=tf.float32)
)["output_0"],
rtol=1e-4,
atol=1e-4,
)
def test_custom_model_and_layer(self):
@object_registration.register_keras_serializable(package="my_package")
class CustomLayer(layers.Layer):
def __call__(self, inputs):
return inputs
@object_registration.register_keras_serializable(package="my_package")
class Model(models.Model):
def __init__(self):
super().__init__()
self.layer = CustomLayer()
@tf.function(input_signature=[tf.TensorSpec([None, 1])])
def call(self, inputs):
return self.layer(inputs)
model = Model()
inp = np.array([[1.0]])
result = model(inp)
path = os.path.join(self.get_temp_dir(), "my_keras_core_model")
tf.saved_model.save(model, path)
restored_model = tf.saved_model.load(path)
self.assertAllClose(
result,
restored_model.call(inp),
rtol=1e-4,
atol=1e-4,
)
def test_multi_input_model(self):
input_1 = layers.Input(shape=(3,))
input_2 = layers.Input(shape=(5,))
model = models.Model([input_1, input_2], [input_1, input_2])
path = os.path.join(self.get_temp_dir(), "my_keras_core_model")
tf.saved_model.save(model, path)
restored_model = tf.saved_model.load(path)
input_arr_1 = np.random.random((1, 3)).astype("float32")
input_arr_2 = np.random.random((1, 5)).astype("float32")
outputs = restored_model.signatures["serving_default"](
inputs=tf.convert_to_tensor(input_arr_1, dtype=tf.float32),
inputs_1=tf.convert_to_tensor(input_arr_2, dtype=tf.float32),
)
self.assertAllClose(
input_arr_1, outputs["output_0"], rtol=1e-4, atol=1e-4
)
self.assertAllClose(
input_arr_2, outputs["output_1"], rtol=1e-4, atol=1e-4
)
def test_multi_input_custom_model_and_layer(self):
@object_registration.register_keras_serializable(package="my_package")
class CustomLayer(layers.Layer):
def __call__(self, *input_list):
self.add_loss(input_list[-2] * 2)
return sum(input_list)
@object_registration.register_keras_serializable(package="my_package")
class CustomModel(models.Model):
def build(self, input_shape):
super().build(input_shape)
self.layer = CustomLayer()
@tf.function
def call(self, *inputs):
inputs = list(inputs)
return self.layer(*inputs)
model = CustomModel()
inp = [
tf.constant(i, shape=[1, 1], dtype=tf.float32) for i in range(1, 4)
]
expected = model(*inp)
path = os.path.join(self.get_temp_dir(), "my_keras_core_model")
tf.saved_model.save(model, path)
restored_model = tf.saved_model.load(path)
output = restored_model.call(*inp)
self.assertAllClose(expected, output, rtol=1e-4, atol=1e-4)
|
keras-core/keras_core/backend/tensorflow/saved_model_test.py/0
|
{
"file_path": "keras-core/keras_core/backend/tensorflow/saved_model_test.py",
"repo_id": "keras-core",
"token_count": 3101
}
| 30 |
from keras_core import optimizers
from keras_core.backend.torch.optimizers import torch_adam
class AdamW(torch_adam.Adam, optimizers.AdamW):
pass
|
keras-core/keras_core/backend/torch/optimizers/torch_adamw.py/0
|
{
"file_path": "keras-core/keras_core/backend/torch/optimizers/torch_adamw.py",
"repo_id": "keras-core",
"token_count": 54
}
| 31 |
import logging
import os
import sys
import time
import warnings
import tree
from keras_core import backend
from keras_core import ops
from keras_core.api_export import keras_core_export
from keras_core.callbacks.callback import Callback
from keras_core.layers import Embedding
from keras_core.optimizers import Optimizer
from keras_core.utils import file_utils
@keras_core_export("keras_core.callbacks.TensorBoard")
class TensorBoard(Callback):
"""Enable visualizations for TensorBoard.
TensorBoard is a visualization tool provided with TensorFlow. A TensorFlow
installation is required to use this callback.
This callback logs events for TensorBoard, including:
* Metrics summary plots
* Training graph visualization
* Weight histograms
* Sampled profiling
When used in `model.evaluate()` or regular validation
in addition to epoch summaries, there will be a summary that records
evaluation metrics vs `model.optimizer.iterations` written. The metric names
will be prepended with `evaluation`, with `model.optimizer.iterations` being
the step in the visualized TensorBoard.
If you have installed TensorFlow with pip, you should be able
to launch TensorBoard from the command line:
```
tensorboard --logdir=path_to_your_logs
```
You can find more information about TensorBoard
[here](https://www.tensorflow.org/get_started/summaries_and_tensorboard).
Args:
log_dir: the path of the directory where to save the log files to be
parsed by TensorBoard. e.g.,
`log_dir = os.path.join(working_dir, 'logs')`.
This directory should not be reused by any other callbacks.
histogram_freq: frequency (in epochs) at which to compute
weight histograms for the layers of the model. If set to 0,
histograms won't be computed. Validation data (or split) must be
specified for histogram visualizations.
write_graph: (Not supported at this time)
Whether to visualize the graph in TensorBoard.
Note that the log file can become quite large
when `write_graph` is set to `True`.
write_images: whether to write model weights to visualize as image in
TensorBoard.
write_steps_per_second: whether to log the training steps per second
into TensorBoard. This supports both epoch and batch frequency
logging.
update_freq: `"batch"` or `"epoch"` or integer. When using `"epoch"`,
writes the losses and metrics to TensorBoard after every epoch.
If using an integer, let's say `1000`, all metrics and losses
(including custom ones added by `Model.compile`) will be logged to
TensorBoard every 1000 batches. `"batch"` is a synonym for 1,
meaning that they will be written every batch.
Note however that writing too frequently to TensorBoard can slow
down your training, especially when used with distribution
strategies as it will incur additional synchronization overhead.
Batch-level summary writing is also available via `train_step`
override. Please see
[TensorBoard Scalars tutorial](
https://www.tensorflow.org/tensorboard/scalars_and_keras#batch-level_logging) # noqa: E501
for more details.
profile_batch: (Not supported at this time)
Profile the batch(es) to sample compute characteristics.
profile_batch must be a non-negative integer or a tuple of integers.
A pair of positive integers signify a range of batches to profile.
By default, profiling is disabled.
embeddings_freq: frequency (in epochs) at which embedding layers will be
visualized. If set to 0, embeddings won't be visualized.
embeddings_metadata: Dictionary which maps embedding layer names to the
filename of a file in which to save metadata for the embedding layer.
In case the same metadata file is to be
used for all embedding layers, a single filename can be passed.
Examples:
Basic usage:
```python
tensorboard_callback = keras_core.callbacks.TensorBoard(log_dir="./logs")
model.fit(x_train, y_train, epochs=2, callbacks=[tensorboard_callback])
# Then run the tensorboard command to view the visualizations.
```
Custom batch-level summaries in a subclassed Model:
```python
class MyModel(keras_core.Model):
def build(self, _):
self.dense = keras_core.layers.Dense(10)
def call(self, x):
outputs = self.dense(x)
tf.summary.histogram('outputs', outputs)
return outputs
model = MyModel()
model.compile('sgd', 'mse')
# Make sure to set `update_freq=N` to log a batch-level summary every N
# batches. In addition to any `tf.summary` contained in `model.call()`,
# metrics added in `Model.compile` will be logged every N batches.
tb_callback = keras_core.callbacks.TensorBoard('./logs', update_freq=1)
model.fit(x_train, y_train, callbacks=[tb_callback])
```
Custom batch-level summaries in a Functional API Model:
```python
def my_summary(x):
tf.summary.histogram('x', x)
return x
inputs = keras_core.Input(10)
x = keras_core.layers.Dense(10)(inputs)
outputs = keras_core.layers.Lambda(my_summary)(x)
model = keras_core.Model(inputs, outputs)
model.compile('sgd', 'mse')
# Make sure to set `update_freq=N` to log a batch-level summary every N
# batches. In addition to any `tf.summary` contained in `Model.call`,
# metrics added in `Model.compile` will be logged every N batches.
tb_callback = keras_core.callbacks.TensorBoard('./logs', update_freq=1)
model.fit(x_train, y_train, callbacks=[tb_callback])
```
Profiling:
```python
# Profile a single batch, e.g. the 5th batch.
tensorboard_callback = keras_core.callbacks.TensorBoard(
log_dir='./logs', profile_batch=5)
model.fit(x_train, y_train, epochs=2, callbacks=[tensorboard_callback])
# Profile a range of batches, e.g. from 10 to 20.
tensorboard_callback = keras_core.callbacks.TensorBoard(
log_dir='./logs', profile_batch=(10,20))
model.fit(x_train, y_train, epochs=2, callbacks=[tensorboard_callback])
```
"""
def __init__(
self,
log_dir="logs",
histogram_freq=0,
write_graph=True,
write_images=False,
write_steps_per_second=False,
update_freq="epoch",
profile_batch=0,
embeddings_freq=0,
embeddings_metadata=None,
):
super().__init__()
self.log_dir = str(log_dir)
self.histogram_freq = histogram_freq
self.write_graph = write_graph
self.write_images = write_images
self.write_steps_per_second = write_steps_per_second
self.update_freq = 1 if update_freq == "batch" else update_freq
self.embeddings_freq = embeddings_freq
self.embeddings_metadata = embeddings_metadata
if profile_batch and backend.backend() != "tensorflow":
# TODO: profiling not available in JAX/torch
raise ValueError(
"Profiling is not yet available with the "
f"{backend.backend()} backend. Please open a PR "
"if you'd like to add this feature. Received: "
f"profile_batch={profile_batch} (must be 0)"
)
self._init_profile_batch(profile_batch)
self._global_train_batch = 0
self._previous_epoch_iterations = 0
self._train_accumulated_time = 0
self._batch_start_time = 0
self._summary_module = None
# Lazily initialized in order to avoid creating event files when
# not needed.
self._writers = {}
# Used to restore any existing `SummaryWriter` after training ends.
self._prev_summary_state = []
def set_model(self, model):
"""Sets Keras model and writes graph if specified."""
self._model = model
self._log_write_dir = self.log_dir
self._train_dir = os.path.join(self._log_write_dir, "train")
self._train_step = 0
self._val_dir = os.path.join(self._log_write_dir, "validation")
self._val_step = 0
self._writers = {} # Resets writers.
self._should_write_train_graph = False
if self.write_graph:
self._write_keras_model_summary()
self._should_write_train_graph = True
if self.embeddings_freq:
self._configure_embeddings()
@property
def summary(self):
if self._summary_module is None:
import tensorflow.summary as summary
self._summary_module = summary
return self._summary_module
@property
def _train_writer(self):
if "train" not in self._writers:
self._writers["train"] = self.summary.create_file_writer(
self._train_dir
)
return self._writers["train"]
@property
def _val_writer(self):
if "val" not in self._writers:
self._writers["val"] = self.summary.create_file_writer(
self._val_dir
)
return self._writers["val"]
def _write_keras_model_train_graph(self):
"""Writes Keras model train_function graph to TensorBoard."""
with self._train_writer.as_default():
with self.summary.record_if(True):
train_fn = self.model.train_function
# If the train_function is a `tf.function`, we can write out a
# graph
if hasattr(train_fn, "function_spec"):
# TODO(b/243822285): Use _variable_creation_fn directly.
if hasattr(train_fn, "_concrete_stateful_fn"):
self.summary.graph(train_fn._concrete_stateful_fn.graph)
else:
self.summary.graph(
train_fn._concrete_variable_creation_fn.graph
)
def _write_keras_model_summary(self):
"""Writes Keras graph network summary to TensorBoard."""
with self._train_writer.as_default():
with self.summary.record_if(True):
if (
self.model.__class__.__name__ == "Functional"
or self.model.__class__.__name__ == "Sequential"
):
keras_model_summary("keras", self.model, step=0)
def _configure_embeddings(self):
"""Configure the Projector for embeddings."""
from google.protobuf import text_format
from tensorboard.plugins import projector
config = projector.ProjectorConfig()
for layer in self.model.layers:
if isinstance(layer, Embedding):
embedding = config.embeddings.add()
# Embeddings are always the first layer, so this naming should
# be consistent in any keras models checkpoints.
name = (
"layer_with_weights-0/embeddings/.ATTRIBUTES/VARIABLE_VALUE"
)
embedding.tensor_name = name
if self.embeddings_metadata is not None:
if isinstance(self.embeddings_metadata, str):
embedding.metadata_path = self.embeddings_metadata
else:
if layer.name in self.embeddings_metadata.keys():
embedding.metadata_path = (
self.embeddings_metadata.pop(layer.name)
)
if self.embeddings_metadata and not isinstance(
self.embeddings_metadata, str
):
raise ValueError(
"Unrecognized `Embedding` layer names passed to "
"`keras_core.callbacks.TensorBoard` `embeddings_metadata` "
f"argument: {self.embeddings_metadata.keys()}"
)
config_pbtxt = text_format.MessageToString(config)
path = os.path.join(self._log_write_dir, "projector_config.pbtxt")
with file_utils.File(path, "w") as f:
f.write(config_pbtxt)
def _push_writer(self, writer, step):
"""Sets the default writer for custom batch-level summaries."""
if self.update_freq == "epoch":
return
def should_record():
return step % self.update_freq == 0
summary_context = (
writer.as_default(step),
self.summary.record_if(should_record),
)
self._prev_summary_state.append(summary_context)
summary_context[0].__enter__()
summary_context[1].__enter__()
def _pop_writer(self):
"""Pops the current writer."""
if self.update_freq == "epoch":
return
# See _push_writer for the content of the previous_context, which is
# pair of context.
previous_context = self._prev_summary_state.pop()
previous_context[1].__exit__(*sys.exc_info())
previous_context[0].__exit__(*sys.exc_info())
def _close_writers(self):
for writer in self._writers.values():
writer.close()
def _init_profile_batch(self, profile_batch):
"""Validate profile_batch value and set the range of batches to profile.
Sets values of _start_batch and _stop_batch attributes,
specifying the start and stop batch to profile.
Setting `profile_batch=0` disables profiling.
Args:
profile_batch: The range of batches to profile. Should be a
non-negative integer or a comma separated string of pair of positive
integers. A pair of positive integers signify a range of batches to
profile.
Raises:
ValueError: If profile_batch is not an integer or a comma separated
pair of positive integers.
"""
profile_batch_error_message = (
"profile_batch must be a non-negative integer or "
"2-tuple of positive "
"integers. A pair of positive integers "
"signifies a range of batches "
f"to profile. Found: {profile_batch}"
)
# Support legacy way of specifying "start,stop" or "start" as str.
if isinstance(profile_batch, str):
profile_batch = str(profile_batch).split(",")
profile_batch = tree.map_structure(int, profile_batch)
if isinstance(profile_batch, int):
self._start_batch = profile_batch
self._stop_batch = profile_batch
elif (
isinstance(profile_batch, (tuple, list)) and len(profile_batch) == 2
):
self._start_batch, self._stop_batch = profile_batch
else:
raise ValueError(profile_batch_error_message)
if self._start_batch < 0 or self._stop_batch < self._start_batch:
raise ValueError(profile_batch_error_message)
# True when the profiler was successfully started by this callback.
# We track the status here to make sure callbacks do not interfere with
# each other. The callback will only stop the profiler it started.
self._profiler_started = False
if self._start_batch > 0:
# Warm up and improve the profiling accuracy.
self._start_profiler(logdir="")
self._stop_profiler(save=False)
# True when a trace is running.
self._is_tracing = False
# Setting `profile_batch=0` disables profiling.
self._should_trace = not (
self._start_batch == 0 and self._stop_batch == 0
)
def on_train_begin(self, logs=None):
self._global_train_batch = 0
self._previous_epoch_iterations = 0
self._push_writer(self._train_writer, self._train_step)
def on_train_end(self, logs=None):
self._pop_writer()
if self._is_tracing:
self._stop_trace()
self._close_writers()
def on_test_begin(self, logs=None):
self._push_writer(self._val_writer, self._val_step)
def on_test_end(self, logs=None):
if self.model.optimizer and hasattr(self.model.optimizer, "iterations"):
with self.summary.record_if(True), self._val_writer.as_default():
for name, value in logs.items():
self.summary.scalar(
"evaluation_" + name + "_vs_iterations",
value,
step=self.model.optimizer.iterations,
)
self._pop_writer()
def _implements_train_batch_hooks(self):
# Only call batch hooks when tracing or write_steps_per_second are
# enabled
return self._should_trace or self.write_steps_per_second
def on_train_batch_begin(self, batch, logs=None):
self._global_train_batch += 1
if self.write_steps_per_second:
self._batch_start_time = time.time()
if not self._should_trace:
return
if self._global_train_batch == self._start_batch:
self._start_trace()
def on_train_batch_end(self, batch, logs=None):
if self._should_write_train_graph:
self._write_keras_model_train_graph()
self._should_write_train_graph = False
if self.write_steps_per_second:
batch_run_time = time.time() - self._batch_start_time
self.summary.scalar(
"batch_steps_per_second",
1.0 / batch_run_time,
step=self._train_step,
)
# `logs` isn't necessarily always a dict
if isinstance(logs, dict):
for name, value in logs.items():
self.summary.scalar(
"batch_" + name, value, step=self._train_step
)
if not self._should_trace:
return
if self._is_tracing and self._global_train_batch >= self._stop_batch:
self._stop_trace()
def on_epoch_begin(self, epoch, logs=None):
# Keeps track of epoch for profiling.
if self.write_steps_per_second:
self._previous_epoch_iterations = self.model.optimizer.iterations
self._epoch_start_time = time.time()
def on_epoch_end(self, epoch, logs=None):
"""Runs metrics and histogram summaries at epoch end."""
self._log_epoch_metrics(epoch, logs)
if self.histogram_freq and epoch % self.histogram_freq == 0:
self._log_weights(epoch)
if self.embeddings_freq and epoch % self.embeddings_freq == 0:
self._log_embeddings(epoch)
def _start_trace(self):
self.summary.trace_on(graph=True, profiler=False)
self._start_profiler(logdir=self.log_dir)
self._is_tracing = True
def _stop_trace(self, batch=None):
"""Logs the trace graph to TensorBoard."""
if batch is None:
batch = self._stop_batch
with self._train_writer.as_default():
with self.summary.record_if(True):
# TODO(b/126388999): Remove step info in the summary name.
self.summary.trace_export(name="batch_%d" % batch, step=batch)
self._stop_profiler()
self._is_tracing = False
def _collect_learning_rate(self, logs):
if isinstance(self.model.optimizer, Optimizer):
logs["learning_rate"] = float(
ops.convert_to_numpy(self.model.optimizer.learning_rate)
)
return logs
def _compute_steps_per_second(self):
current_iteration = self.model.optimizer.iterations
time_since_epoch_begin = time.time() - self._epoch_start_time
current_iteration = ops.convert_to_tensor(current_iteration, "float32")
self._previous_epoch_iterations = ops.convert_to_tensor(
self._previous_epoch_iterations, "float32"
)
time_since_epoch_begin = ops.convert_to_tensor(
time_since_epoch_begin, "float32"
)
steps_per_second = (
current_iteration - self._previous_epoch_iterations
) / time_since_epoch_begin
return float(steps_per_second)
def _log_epoch_metrics(self, epoch, logs):
"""Writes epoch metrics out as scalar summaries.
Args:
epoch: Int. The global step to use for TensorBoard.
logs: Dict. Keys are scalar summary names, values are scalars.
"""
if not logs:
return
train_logs = {k: v for k, v in logs.items() if not k.startswith("val_")}
val_logs = {k: v for k, v in logs.items() if k.startswith("val_")}
train_logs = self._collect_learning_rate(train_logs)
if self.write_steps_per_second:
train_logs["steps_per_second"] = self._compute_steps_per_second()
with self.summary.record_if(True):
if train_logs:
with self._train_writer.as_default():
for name, value in train_logs.items():
self.summary.scalar("epoch_" + name, value, step=epoch)
if val_logs:
with self._val_writer.as_default():
for name, value in val_logs.items():
name = name[4:] # Remove 'val_' prefix.
self.summary.scalar("epoch_" + name, value, step=epoch)
def _log_weights(self, epoch):
"""Logs the weights of the Model to TensorBoard."""
with self._train_writer.as_default():
with self.summary.record_if(True):
for layer in self.model.layers:
for weight in layer.weights:
weight_name = weight.name.replace(":", "_")
# Add a suffix to prevent summary tag name collision.
histogram_weight_name = weight_name + "/histogram"
self.summary.histogram(
histogram_weight_name, weight, step=epoch
)
if self.write_images:
# Add a suffix to prevent summary tag name
# collision.
image_weight_name = weight_name + "/image"
self._log_weight_as_image(
weight, image_weight_name, epoch
)
self._train_writer.flush()
def _log_weight_as_image(self, weight, weight_name, epoch):
"""Logs a weight as a TensorBoard image."""
w_img = ops.squeeze(weight)
shape = w_img.shape
if len(shape) == 1: # Bias case
w_img = ops.reshape(w_img, [1, shape[0], 1, 1])
elif len(shape) == 2: # Dense layer kernel case
if shape[0] > shape[1]:
w_img = ops.transpose(w_img)
shape = w_img.shape
w_img = ops.reshape(w_img, [1, shape[0], shape[1], 1])
elif len(shape) == 3: # ConvNet case
if backend.image_data_format() == "channels_last":
# Switch to channels_first to display every kernel as a separate
# image.
w_img = ops.transpose(w_img, [2, 0, 1])
shape = w_img.shape
w_img = ops.reshape(w_img, [shape[0], shape[1], shape[2], 1])
w_img = backend.convert_to_numpy(w_img)
shape = w_img.shape
# Not possible to handle 3D convnets etc.
if len(shape) == 4 and shape[-1] in [1, 3, 4]:
self.summary.image(weight_name, w_img, step=epoch)
def _log_embeddings(self, epoch):
embeddings_ckpt = os.path.join(
self._log_write_dir,
"train",
f"keras_embedding.ckpt-{epoch}.weights.h5",
)
self.model.save_weights(embeddings_ckpt)
def _start_profiler(self, logdir):
"""Starts the profiler if currently inactive.
Args:
logdir: Directory where profiler results will be saved.
"""
if self._profiler_started:
return
try:
backend.tensorboard.start_trace(logdir)
self._profiler_started = True
except Exception as e:
# Profiler errors should not be fatal.
logging.error("Failed to start profiler: %s", e)
def _stop_profiler(self, save=True):
"""Stops the profiler if currently active.
Args:
save: Whether to save the profiler results to TensorBoard.
"""
if not self._profiler_started:
return
try:
backend.tensorboard.stop_trace(save=save)
except Exception as e:
# Profiler errors should not be fatal.
logging.error("Failed to stop profiler: %s", e)
finally:
self._profiler_started = False
def keras_model_summary(name, data, step=None):
"""Writes a Keras model as JSON to as a Summary.
Writing the Keras model configuration allows the TensorBoard graph plugin to
render a conceptual graph, as opposed to graph of ops. In case the model
fails to serialize as JSON, it ignores and returns False.
Args:
name: A name for this summary. The summary tag used for TensorBoard will
be this name prefixed by any active name scopes.
data: A Keras Model to write.
step: Explicit `int64`-castable monotonic step value for this summary.
If omitted, this defaults to `tf.summary.experimental.get_step()`,
which must not be `None`.
Returns:
True on success, or False if no summary was written because no default
summary writer was available.
Raises:
ValueError: if a default writer exists, but no step was provided and
`tf.summary.experimental.get_step()` is `None`.
"""
import tensorflow.summary as summary
from tensorflow.compat.v1 import SummaryMetadata
summary_metadata = SummaryMetadata()
# Hard coding a plugin name. Please refer to go/tb-plugin-name-hardcode for
# the rationale.
summary_metadata.plugin_data.plugin_name = "graph_keras_model"
# version number = 1
summary_metadata.plugin_data.content = b"1"
try:
json_string = data.to_json()
except Exception as exc:
# An exception should not break a model code.
warnings.warn(f"Model failed to serialize as JSON. Ignoring... {exc}")
return False
with summary.experimental.summary_scope(
name, "graph_keras_model", [data, step]
) as (tag, _):
return summary.write(
tag=tag, tensor=json_string, step=step, metadata=summary_metadata
)
|
keras-core/keras_core/callbacks/tensorboard.py/0
|
{
"file_path": "keras-core/keras_core/callbacks/tensorboard.py",
"repo_id": "keras-core",
"token_count": 11924
}
| 32 |
"""Reuters topic classification dataset."""
import json
import numpy as np
from keras_core.api_export import keras_core_export
from keras_core.utils.file_utils import get_file
from keras_core.utils.python_utils import remove_long_seq
@keras_core_export("keras_core.datasets.reuters.load_data")
def load_data(
path="reuters.npz",
num_words=None,
skip_top=0,
maxlen=None,
test_split=0.2,
seed=113,
start_char=1,
oov_char=2,
index_from=3,
):
"""Loads the Reuters newswire classification dataset.
This is a dataset of 11,228 newswires from Reuters, labeled over 46 topics.
This was originally generated by parsing and preprocessing the classic
Reuters-21578 dataset, but the preprocessing code is no longer packaged
with Keras. See this
[GitHub discussion](https://github.com/keras-team/keras/issues/12072)
for more info.
Each newswire is encoded as a list of word indexes (integers).
For convenience, words are indexed by overall frequency in the dataset,
so that for instance the integer "3" encodes the 3rd most frequent word in
the data. This allows for quick filtering operations such as:
"only consider the top 10,000 most
common words, but eliminate the top 20 most common words".
As a convention, "0" does not stand for a specific word, but instead is used
to encode any unknown word.
Args:
path: where to cache the data (relative to `~/.keras/dataset`).
num_words: integer or None. Words are
ranked by how often they occur (in the training set) and only
the `num_words` most frequent words are kept. Any less frequent word
will appear as `oov_char` value in the sequence data. If None,
all words are kept. Defaults to `None`.
skip_top: skip the top N most frequently occurring words
(which may not be informative). These words will appear as
`oov_char` value in the dataset. 0 means no words are
skipped. Defaults to `0`.
maxlen: int or None. Maximum sequence length.
Any longer sequence will be truncated. None means no truncation.
Defaults to `None`.
test_split: Float between `0.` and `1.`. Fraction of the dataset to be
used as test data. `0.2` means that 20% of the dataset is used as
test data. Defaults to `0.2`.
seed: int. Seed for reproducible data shuffling.
start_char: int. The start of a sequence will be marked with this
character. 0 is usually the padding character. Defaults to `1`.
oov_char: int. The out-of-vocabulary character.
Words that were cut out because of the `num_words` or
`skip_top` limits will be replaced with this character.
index_from: int. Index actual words with this index and higher.
Returns:
Tuple of Numpy arrays: `(x_train, y_train), (x_test, y_test)`.
**`x_train`, `x_test`**: lists of sequences, which are lists of indexes
(integers). If the num_words argument was specific, the maximum
possible index value is `num_words - 1`. If the `maxlen` argument was
specified, the largest possible sequence length is `maxlen`.
**`y_train`, `y_test`**: lists of integer labels (1 or 0).
**Note**: The 'out of vocabulary' character is only used for
words that were present in the training set but are not included
because they're not making the `num_words` cut here.
Words that were not seen in the training set but are in the test set
have simply been skipped.
"""
origin_folder = (
"https://storage.googleapis.com/tensorflow/tf-keras-datasets/"
)
path = get_file(
fname=path,
origin=origin_folder + "reuters.npz",
file_hash=( # noqa: E501
"d6586e694ee56d7a4e65172e12b3e987c03096cb01eab99753921ef915959916"
),
)
with np.load(path, allow_pickle=True) as f:
xs, labels = f["x"], f["y"]
rng = np.random.RandomState(seed)
indices = np.arange(len(xs))
rng.shuffle(indices)
xs = xs[indices]
labels = labels[indices]
if start_char is not None:
xs = [[start_char] + [w + index_from for w in x] for x in xs]
elif index_from:
xs = [[w + index_from for w in x] for x in xs]
if maxlen:
xs, labels = remove_long_seq(maxlen, xs, labels)
if not num_words:
num_words = max(max(x) for x in xs)
# by convention, use 2 as OOV word
# reserve 'index_from' (=3 by default) characters:
# 0 (padding), 1 (start), 2 (OOV)
if oov_char is not None:
xs = [
[w if skip_top <= w < num_words else oov_char for w in x]
for x in xs
]
else:
xs = [[w for w in x if skip_top <= w < num_words] for x in xs]
idx = int(len(xs) * (1 - test_split))
x_train, y_train = np.array(xs[:idx], dtype="object"), np.array(
labels[:idx]
)
x_test, y_test = np.array(xs[idx:], dtype="object"), np.array(labels[idx:])
return (x_train, y_train), (x_test, y_test)
@keras_core_export("keras_core.datasets.reuters.get_word_index")
def get_word_index(path="reuters_word_index.json"):
"""Retrieves a dict mapping words to their index in the Reuters dataset.
Actual word indices starts from 3, with 3 indices reserved for:
0 (padding), 1 (start), 2 (oov).
E.g. word index of 'the' is 1, but the in the actual training data, the
index of 'the' will be 1 + 3 = 4. Vice versa, to translate word indices in
training data back to words using this mapping, indices need to substract 3.
Args:
path: where to cache the data (relative to `~/.keras/dataset`).
Returns:
The word index dictionary. Keys are word strings, values are their
index.
"""
origin_folder = (
"https://storage.googleapis.com/tensorflow/tf-keras-datasets/"
)
path = get_file(
path,
origin=origin_folder + "reuters_word_index.json",
file_hash="4d44cc38712099c9e383dc6e5f11a921",
)
with open(path) as f:
return json.load(f)
@keras_core_export("keras_core.datasets.reuters.get_label_names")
def get_label_names():
"""Returns labels as a list of strings with indices matching training data.
Reference:
- [Reuters Dataset](https://martin-thoma.com/nlp-reuters/)
"""
return (
"cocoa",
"grain",
"veg-oil",
"earn",
"acq",
"wheat",
"copper",
"housing",
"money-supply",
"coffee",
"sugar",
"trade",
"reserves",
"ship",
"cotton",
"carcass",
"crude",
"nat-gas",
"cpi",
"money-fx",
"interest",
"gnp",
"meal-feed",
"alum",
"oilseed",
"gold",
"tin",
"strategic-metal",
"livestock",
"retail",
"ipi",
"iron-steel",
"rubber",
"heat",
"jobs",
"lei",
"bop",
"zinc",
"orange",
"pet-chem",
"dlr",
"gas",
"silver",
"wpi",
"hog",
"lead",
)
|
keras-core/keras_core/datasets/reuters.py/0
|
{
"file_path": "keras-core/keras_core/datasets/reuters.py",
"repo_id": "keras-core",
"token_count": 3069
}
| 33 |
import pytest
from keras_core import activations
from keras_core import layers
from keras_core import testing
class ActivationTest(testing.TestCase):
@pytest.mark.requires_trainable_backend
def test_activation_basics(self):
self.run_layer_test(
layers.Activation,
init_kwargs={
"activation": "relu",
},
input_shape=(2, 3),
expected_output_shape=(2, 3),
expected_num_trainable_weights=0,
expected_num_non_trainable_weights=0,
expected_num_seed_generators=0,
expected_num_losses=0,
supports_masking=True,
)
self.run_layer_test(
layers.Activation,
init_kwargs={
"activation": activations.gelu,
},
input_shape=(2, 2),
expected_output_shape=(2, 2),
expected_num_trainable_weights=0,
expected_num_non_trainable_weights=0,
expected_num_seed_generators=0,
expected_num_losses=0,
supports_masking=True,
)
|
keras-core/keras_core/layers/activations/activation_test.py/0
|
{
"file_path": "keras-core/keras_core/layers/activations/activation_test.py",
"repo_id": "keras-core",
"token_count": 571
}
| 34 |
import collections
import math
import string
import numpy as np
from keras_core import constraints
from keras_core import initializers
from keras_core import ops
from keras_core import regularizers
from keras_core.api_export import keras_core_export
from keras_core.layers.activations.softmax import Softmax
from keras_core.layers.core.einsum_dense import EinsumDense
from keras_core.layers.layer import Layer
from keras_core.layers.regularization.dropout import Dropout
@keras_core_export("keras_core.layers.MultiHeadAttention")
class MultiHeadAttention(Layer):
"""MultiHeadAttention layer.
This is an implementation of multi-headed attention as described in the
paper "Attention is all you Need"
[Vaswani et al., 2017](https://arxiv.org/abs/1706.03762).
If `query`, `key,` `value` are the same, then
this is self-attention. Each timestep in `query` attends to the
corresponding sequence in `key`, and returns a fixed-width vector.
This layer first projects `query`, `key` and `value`. These are
(effectively) a list of tensors of length `num_attention_heads`, where the
corresponding shapes are `(batch_size, <query dimensions>, key_dim)`,
`(batch_size, <key/value dimensions>, key_dim)`,
`(batch_size, <key/value dimensions>, value_dim)`.
Then, the query and key tensors are dot-producted and scaled. These are
softmaxed to obtain attention probabilities. The value tensors are then
interpolated by these probabilities, then concatenated back to a single
tensor.
Finally, the result tensor with the last dimension as `value_dim` can take
a linear projection and return.
Args:
num_heads: Number of attention heads.
key_dim: Size of each attention head for query and key.
value_dim: Size of each attention head for value.
dropout: Dropout probability.
use_bias: Boolean, whether the dense layers use bias vectors/matrices.
output_shape: The expected shape of an output tensor, besides the batch
and sequence dims. If not specified, projects back to the query
feature dim (the query input's last dimension).
attention_axes: axes over which the attention is applied. `None` means
attention over all axes, but batch, heads, and features.
kernel_initializer: Initializer for dense layer kernels.
bias_initializer: Initializer for dense layer biases.
kernel_regularizer: Regularizer for dense layer kernels.
bias_regularizer: Regularizer for dense layer biases.
activity_regularizer: Regularizer for dense layer activity.
kernel_constraint: Constraint for dense layer kernels.
bias_constraint: Constraint for dense layer kernels.
Call arguments:
query: Query tensor of shape `(B, T, dim)`, where `B` is the batch size,
`T` is the target sequence length, and dim is the feature dimension.
value: Value tensor of shape `(B, S, dim)`, where `B` is the batch size,
`S` is the source sequence length, and dim is the feature dimension.
key: Optional key tensor of shape `(B, S, dim)`. If not given, will
use `value` for both `key` and `value`, which is the most common
case.
attention_mask: a boolean mask of shape `(B, T, S)`, that prevents
attention to certain positions. The boolean mask specifies which
query elements can attend to which key elements, 1 indicates
attention and 0 indicates no attention. Broadcasting can happen for
the missing batch dimensions and the head dimension.
return_attention_scores: A boolean to indicate whether the output should
be `(attention_output, attention_scores)` if `True`, or
`attention_output` if `False`. Defaults to `False`.
training: Python boolean indicating whether the layer should behave in
training mode (adding dropout) or in inference mode (no dropout).
Will go with either using the training mode of the parent
layer/model, or `False` (inference) if there is no parent layer.
use_causal_mask: A boolean to indicate whether to apply a causal mask to
prevent tokens from attending to future tokens (e.g., used in a
decoder Transformer).
Returns:
attention_output: The result of the computation, of shape `(B, T, E)`,
where `T` is for target sequence shapes and `E` is the query input
last dimension if `output_shape` is `None`. Otherwise, the
multi-head outputs are projected to the shape specified by
`output_shape`.
attention_scores: (Optional) multi-head attention coefficients over
attention axes.
"""
def __init__(
self,
num_heads,
key_dim,
value_dim=None,
dropout=0.0,
use_bias=True,
output_shape=None,
attention_axes=None,
kernel_initializer="glorot_uniform",
bias_initializer="zeros",
kernel_regularizer=None,
bias_regularizer=None,
activity_regularizer=None,
kernel_constraint=None,
bias_constraint=None,
**kwargs,
):
super().__init__(**kwargs)
self.supports_masking = True
self._num_heads = num_heads
self._key_dim = key_dim
# Cache 1.0 / math.sqrt(self._key_dim).
self._inverse_sqrt_key_dim = None
self._value_dim = value_dim if value_dim else key_dim
self._dropout = dropout
self._use_bias = use_bias
self._output_shape = output_shape
self._kernel_initializer = initializers.get(kernel_initializer)
self._bias_initializer = initializers.get(bias_initializer)
self._kernel_regularizer = regularizers.get(kernel_regularizer)
self._bias_regularizer = regularizers.get(bias_regularizer)
self._activity_regularizer = regularizers.get(activity_regularizer)
self._kernel_constraint = constraints.get(kernel_constraint)
self._bias_constraint = constraints.get(bias_constraint)
if isinstance(attention_axes, int):
attention_axes = (attention_axes,)
elif attention_axes and not isinstance(attention_axes, (list, tuple)):
raise ValueError(
"`attention_axes` must be an int, list, or tuple."
f"Received: attention_axes={attention_axes}"
)
self._attention_axes = attention_axes
def get_config(self):
base_config = super().get_config()
config = {
"num_heads": self._num_heads,
"key_dim": self._key_dim,
"value_dim": self._value_dim,
"dropout": self._dropout,
"use_bias": self._use_bias,
"output_shape": self._output_shape,
"attention_axes": self._attention_axes,
"kernel_initializer": initializers.serialize(
self._kernel_initializer
),
"bias_initializer": initializers.serialize(self._bias_initializer),
"kernel_regularizer": regularizers.serialize(
self._kernel_regularizer
),
"bias_regularizer": regularizers.serialize(self._bias_regularizer),
"activity_regularizer": regularizers.serialize(
self._activity_regularizer
),
"kernel_constraint": constraints.serialize(self._kernel_constraint),
"bias_constraint": constraints.serialize(self._bias_constraint),
}
return {**base_config, **config}
def build(
self,
query_shape,
value_shape,
key_shape=None,
):
"""Builds layers and variables.
Args:
query_shape: Shape of the `query` tensor.
value_shape: Shape of the `value` tensor.
key: Optional shape of the `key` tensor.
"""
key_shape = value_shape if key_shape is None else key_shape
query_rank = len(query_shape)
value_rank = len(value_shape)
key_rank = len(key_shape)
einsum_equation, bias_axes, output_rank = _build_proj_equation(
query_rank - 1, bound_dims=1, output_dims=2
)
self._query_dense = EinsumDense(
einsum_equation,
output_shape=_get_output_shape(
output_rank - 1, [self._num_heads, self._key_dim]
),
bias_axes=bias_axes if self._use_bias else None,
name="query",
**self._get_common_kwargs_for_sublayer(),
)
self._query_dense.build(query_shape)
einsum_equation, bias_axes, output_rank = _build_proj_equation(
key_rank - 1, bound_dims=1, output_dims=2
)
self._key_dense = EinsumDense(
einsum_equation,
output_shape=_get_output_shape(
output_rank - 1, [self._num_heads, self._key_dim]
),
bias_axes=bias_axes if self._use_bias else None,
name="key",
**self._get_common_kwargs_for_sublayer(),
)
self._key_dense.build(key_shape)
einsum_equation, bias_axes, output_rank = _build_proj_equation(
value_rank - 1, bound_dims=1, output_dims=2
)
self._value_dense = EinsumDense(
einsum_equation,
output_shape=_get_output_shape(
output_rank - 1, [self._num_heads, self._value_dim]
),
bias_axes=bias_axes if self._use_bias else None,
name="value",
**self._get_common_kwargs_for_sublayer(),
)
self._value_dense.build(value_shape)
# Builds the attention computations for multi-head dot product
# attention. These computations could be wrapped into the keras
# attention layer once it supports multi-head einsum computations.
self._build_attention(output_rank)
self._output_dense = self._make_output_dense(
query_shape,
self._get_common_kwargs_for_sublayer(),
"attention_output",
)
output_dense_input_shape = list(
self._query_dense.compute_output_shape(query_shape)
)
output_dense_input_shape[-1] = self._value_dim
self._output_dense.build(tuple(output_dense_input_shape))
self.built = True
def _get_common_kwargs_for_sublayer(self):
common_kwargs = dict(
kernel_regularizer=self._kernel_regularizer,
bias_regularizer=self._bias_regularizer,
activity_regularizer=self._activity_regularizer,
kernel_constraint=self._kernel_constraint,
bias_constraint=self._bias_constraint,
dtype=self.dtype_policy,
)
# Create new clone of kernel/bias initializer, so that we don't reuse
# the initializer instance, which could lead to same init value since
# initializer is stateless.
kernel_initializer = self._kernel_initializer.__class__.from_config(
self._kernel_initializer.get_config()
)
bias_initializer = self._bias_initializer.__class__.from_config(
self._bias_initializer.get_config()
)
common_kwargs["kernel_initializer"] = kernel_initializer
common_kwargs["bias_initializer"] = bias_initializer
return common_kwargs
def _make_output_dense(self, query_shape, common_kwargs, name=None):
"""Builds the output projection matrix.
Args:
free_dims: Number of free dimensions for einsum equation building.
common_kwargs: Common keyword arguments for einsum layer.
name: Name for the projection layer.
Returns:
Projection layer.
"""
query_rank = len(query_shape)
if self._output_shape:
if not isinstance(self._output_shape, collections.abc.Sized):
output_shape = [self._output_shape]
else:
output_shape = self._output_shape
else:
output_shape = [query_shape[-1]]
einsum_equation, bias_axes, output_rank = _build_proj_equation(
query_rank - 1, bound_dims=2, output_dims=len(output_shape)
)
return EinsumDense(
einsum_equation,
output_shape=_get_output_shape(output_rank - 1, output_shape),
bias_axes=bias_axes if self._use_bias else None,
name=name,
**common_kwargs,
)
def _build_attention(self, rank):
"""Builds multi-head dot-product attention computations.
This function builds attributes necessary for `_compute_attention` to
customize attention computation to replace the default dot-product
attention.
Args:
rank: the rank of query, key, value tensors.
"""
if self._attention_axes is None:
self._attention_axes = tuple(range(1, rank - 2))
else:
self._attention_axes = tuple(self._attention_axes)
(
self._dot_product_equation,
self._combine_equation,
attn_scores_rank,
) = _build_attention_equation(rank, attn_axes=self._attention_axes)
norm_axes = tuple(
range(
attn_scores_rank - len(self._attention_axes), attn_scores_rank
)
)
self._softmax = Softmax(axis=norm_axes, dtype=self.dtype_policy)
self._dropout_layer = Dropout(
rate=self._dropout, dtype=self.dtype_policy
)
self._inverse_sqrt_key_dim = 1.0 / math.sqrt(float(self._key_dim))
def _masked_softmax(self, attention_scores, attention_mask=None):
# Normalize the attention scores to probabilities.
# attention_scores = [B, N, T, S]
if attention_mask is not None:
# The expand dim happens starting from the `num_heads` dimension,
# (<batch_dims>, num_heads, <query_attention_dims,
# key_attention_dims>)
mask_expansion_axis = -len(self._attention_axes) * 2 - 1
for _ in range(
len(attention_scores.shape) - len(attention_mask.shape)
):
attention_mask = ops.expand_dims(
attention_mask, axis=mask_expansion_axis
)
return self._softmax(attention_scores, mask=attention_mask)
def _compute_attention(
self, query, key, value, attention_mask=None, training=None
):
"""Applies Dot-product attention with query, key, value tensors.
This function defines the computation inside `call` with projected
multi-head Q, K, V inputs. Users can override this function for
customized attention implementation.
Args:
query: Projected query tensor of shape `(B, T, N, key_dim)`.
key: Projected key tensor of shape `(B, S, N, key_dim)`.
value: Projected value tensor of shape `(B, S, N, value_dim)`.
attention_mask: a boolean mask of shape `(B, T, S)`, that prevents
attention to certain positions. It is generally not needed if
the `query` and `value` (and/or `key`) are masked.
training: Python boolean indicating whether the layer should behave
in training mode (adding dropout) or in inference mode (doing
nothing).
Returns:
attention_output: Multi-headed outputs of attention computation.
attention_scores: Multi-headed attention weights.
"""
# Note: Applying scalar multiply at the smaller end of einsum improves
# XLA performance, but may introduce slight numeric differences in
# the Transformer attention head.
query = ops.multiply(
query, ops.cast(self._inverse_sqrt_key_dim, query.dtype)
)
# Take the dot product between "query" and "key" to get the raw
# attention scores.
attention_scores = ops.einsum(self._dot_product_equation, key, query)
attention_scores = self._masked_softmax(
attention_scores, attention_mask
)
# This is actually dropping out entire tokens to attend to, which might
# seem a bit unusual, but is taken from the original Transformer paper.
attention_scores_dropout = self._dropout_layer(
attention_scores, training=training
)
# `context_layer` = [B, T, N, H]
attention_output = ops.einsum(
self._combine_equation, attention_scores_dropout, value
)
return attention_output, attention_scores
def call(
self,
query,
value,
key=None,
query_mask=None,
value_mask=None,
key_mask=None,
attention_mask=None,
return_attention_scores=False,
training=None,
use_causal_mask=False,
):
if key is None:
key = value
attention_mask = self._compute_attention_mask(
query,
value,
query_mask=query_mask,
value_mask=value_mask,
key_mask=key_mask,
attention_mask=attention_mask,
use_causal_mask=use_causal_mask,
)
# N = `num_attention_heads`
# H = `size_per_head`
# `query` = [B, T, N ,H]
query = self._query_dense(query)
# `key` = [B, S, N, H]
key = self._key_dense(key)
# `value` = [B, S, N, H]
value = self._value_dense(value)
attention_output, attention_scores = self._compute_attention(
query, key, value, attention_mask, training
)
attention_output = self._output_dense(attention_output)
if return_attention_scores:
return attention_output, attention_scores
return attention_output
def _compute_attention_mask(
self,
query,
value,
query_mask=None,
value_mask=None,
key_mask=None,
attention_mask=None,
use_causal_mask=False,
):
"""Computes the attention mask, using the Keras masks of the inputs.
* The `query`'s mask is reshaped from [B, T] to [B, T, 1].
* The `value`'s mask is reshaped from [B, S] to [B, 1, S].
* The `key`'s mask is reshaped from [B, S] to [B, 1, S]. The `key`'s
mask is ignored if `key` is `None` or if `key is value`.
* If `use_causal_mask=True`, then the causal mask is computed. Its shape
is [1, T, S].
All defined masks are merged using a logical AND operation (`&`).
In general, if the `query` and `value` are masked, then there is no need
to define the `attention_mask`.
Args:
query: Projected query tensor of shape `(B, T, N, key_dim)`.
key: Projected key tensor of shape `(B, T, N, key_dim)`.
value: Projected value tensor of shape `(B, T, N, value_dim)`.
attention_mask: a boolean mask of shape `(B, T, S)`, that prevents
attention to certain positions.
use_causal_mask: A boolean to indicate whether to apply a causal
mask to prevent tokens from attending to future tokens (e.g.,
used in a decoder Transformer).
Returns:
attention_mask: a boolean mask of shape `(B, T, S)`, that prevents
attention to certain positions, based on the Keras masks of the
`query`, `key`, `value`, and `attention_mask` tensors, and the
causal mask if `use_causal_mask=True`.
"""
auto_mask = None
if query_mask is not None:
query_mask = ops.cast(query_mask, "bool") # defensive casting
# B = batch size, T = max query length
auto_mask = ops.expand_dims(query_mask, -1) # shape is [B, T, 1]
if value_mask is not None:
value_mask = ops.cast(value_mask, "bool") # defensive casting
# B = batch size, S == max value length
mask = ops.expand_dims(value_mask, -2) # shape is [B, 1, S]
auto_mask = mask if auto_mask is None else auto_mask & mask
if key_mask is not None:
key_mask = ops.cast(key_mask, "bool") # defensive casting
# B == batch size, S == max key length == max value length
mask = ops.expand_dims(key_mask, -2) # shape is [B, 1, S]
auto_mask = mask if auto_mask is None else auto_mask & mask
if use_causal_mask:
# the shape of the causal mask is [1, T, S]
mask = self._compute_causal_mask(query, value)
auto_mask = mask if auto_mask is None else auto_mask & mask
if auto_mask is not None:
# merge attention_mask & automatic mask, to shape [B, T, S]
attention_mask = (
auto_mask
if attention_mask is None
else ops.cast(attention_mask, bool) & auto_mask
)
return attention_mask
def _compute_causal_mask(self, query, value=None):
"""Computes a causal mask (e.g., for masked self-attention layers).
For example, if query and value both contain sequences of length 4,
this function returns a boolean tensor equal to:
```
[[[True, False, False, False],
[True, True, False, False],
[True, True, True, False],
[True, True, True, True]]]
```
Args:
query: query tensor of shape `(B, T, ...)`.
value: value tensor of shape `(B, S, ...)` (optional, defaults to
query).
Returns:
mask: a boolean tensor of shape `(1, T, S)` containing a lower
triangular matrix of shape `(T, S)`.
"""
q_seq_length = ops.shape(query)[1]
v_seq_length = q_seq_length if value is None else ops.shape(value)[1]
ones_mask = ops.ones((1, q_seq_length, v_seq_length), dtype="int32")
row_index = ops.cumsum(ones_mask, axis=-2)
col_index = ops.cumsum(ones_mask, axis=-1)
return ops.greater_equal(row_index, col_index)
def compute_output_shape(
self,
query_shape,
value_shape,
key_shape=None,
):
if key_shape is None:
key_shape = value_shape
if query_shape[-1] != value_shape[-1]:
raise ValueError(
"The last dimension of `query_shape` and `value_shape` "
f"must be equal, but are {query_shape[-1]}, {value_shape[-1]}. "
"Received: query_shape={query_shape}, value_shape={value_shape}"
)
if value_shape[1:-1] != key_shape[1:-1]:
raise ValueError(
"All dimensions of `value` and `key`, except the last one, "
f"must be equal. Received: value_shape={value_shape} and "
f"key_shape={key_shape}"
)
if self._output_shape:
return query_shape[:-1] + self._output_shape
return query_shape
def _index_to_einsum_variable(i):
"""Coverts an index to a einsum variable name.
We simply map indices to lowercase characters, e.g. 0 -> 'a', 1 -> 'b'.
"""
return string.ascii_lowercase[i]
def _build_attention_equation(rank, attn_axes):
"""Builds einsum equations for the attention computation.
Query, key, value inputs after projection are expected to have the shape as:
`(bs, <non-attention dims>, <attention dims>, num_heads, channels)`.
`bs` and `<non-attention dims>` are treated as `<batch dims>`.
The attention operations can be generalized:
1. Query-key dot product:
(<batch dims>, <query attention dims>, num_heads, channels),
(<batch dims>, <key attention dims>, num_heads, channels) ->
(<batch dims>, num_heads, <query attention dims>, <key attention dims>)
2. Combination:
(<batch dims>, num_heads, <query attention dims>, <key attention dims>),
(<batch dims>, <value attention dims>, num_heads, channels) -> (<batch
dims>, <query attention dims>, num_heads, channels)
Args:
rank: Rank of query, key, value tensors.
attn_axes: List/tuple of axes, `[-1, rank)`,
that attention will be applied to.
Returns:
Einsum equations.
"""
target_notation = ""
for i in range(rank):
target_notation += _index_to_einsum_variable(i)
# `batch_dims` includes the head dim.
batch_dims = tuple(np.delete(range(rank), attn_axes + (rank - 1,)))
letter_offset = rank
source_notation = ""
for i in range(rank):
if i in batch_dims or i == rank - 1:
source_notation += target_notation[i]
else:
source_notation += _index_to_einsum_variable(letter_offset)
letter_offset += 1
product_notation = "".join(
[target_notation[i] for i in batch_dims]
+ [target_notation[i] for i in attn_axes]
+ [source_notation[i] for i in attn_axes]
)
dot_product_equation = "%s,%s->%s" % (
source_notation,
target_notation,
product_notation,
)
attn_scores_rank = len(product_notation)
combine_equation = "%s,%s->%s" % (
product_notation,
source_notation,
target_notation,
)
return dot_product_equation, combine_equation, attn_scores_rank
def _build_proj_equation(free_dims, bound_dims, output_dims):
"""Builds an einsum equation for projections inside multi-head attention."""
input_str = ""
kernel_str = ""
output_str = ""
bias_axes = ""
letter_offset = 0
for i in range(free_dims):
char = _index_to_einsum_variable(i + letter_offset)
input_str += char
output_str += char
letter_offset += free_dims
for i in range(bound_dims):
char = _index_to_einsum_variable(i + letter_offset)
input_str += char
kernel_str += char
letter_offset += bound_dims
for i in range(output_dims):
char = _index_to_einsum_variable(i + letter_offset)
kernel_str += char
output_str += char
bias_axes += char
equation = f"{input_str},{kernel_str}->{output_str}"
return equation, bias_axes, len(output_str)
def _get_output_shape(output_rank, known_last_dims):
return [None] * (output_rank - len(known_last_dims)) + list(known_last_dims)
|
keras-core/keras_core/layers/attention/multi_head_attention.py/0
|
{
"file_path": "keras-core/keras_core/layers/attention/multi_head_attention.py",
"repo_id": "keras-core",
"token_count": 11675
}
| 35 |
import inspect
import types
import tree
from keras_core import backend
from keras_core.api_export import keras_core_export
from keras_core.layers.layer import Layer
from keras_core.saving import serialization_lib
from keras_core.utils import python_utils
from keras_core.utils import shape_utils
@keras_core_export("keras_core.layers.Lambda")
class Lambda(Layer):
"""Wraps arbitrary expressions as a `Layer` object.
The `Lambda` layer exists so that arbitrary expressions can be used
as a `Layer` when constructing Sequential
and Functional API models. `Lambda` layers are best suited for simple
operations or quick experimentation. For more advanced use cases,
prefer writing new subclasses of `Layer`.
WARNING: `Lambda` layers have (de)serialization limitations!
The main reason to subclass `Layer` instead of using a
`Lambda` layer is saving and inspecting a model. `Lambda` layers
are saved by serializing the Python bytecode, which is fundamentally
non-portable and potentially unsafe.
They should only be loaded in the same environment where
they were saved. Subclassed layers can be saved in a more portable way
by overriding their `get_config()` method. Models that rely on
subclassed Layers are also often easier to visualize and reason about.
Example:
```python
# add a x -> x^2 layer
model.add(Lambda(lambda x: x ** 2))
```
Args:
function: The function to be evaluated. Takes input tensor as first
argument.
output_shape: Expected output shape from function. This argument
can usually be inferred if not explicitly provided.
Can be a tuple or function. If a tuple, it only specifies
the first dimension onward; sample dimension is assumed
either the same as the input:
`output_shape = (input_shape[0], ) + output_shape` or,
the input is `None` and the sample dimension is also `None`:
`output_shape = (None, ) + output_shape`.
If a function, it specifies the
entire shape as a function of the input shape:
`output_shape = f(input_shape)`.
mask: Either None (indicating no masking) or a callable with the same
signature as the `compute_mask` layer method, or a tensor
that will be returned as output mask regardless
of what the input is.
arguments: Optional dictionary of keyword arguments to be passed to the
function.
"""
def __init__(
self, function, output_shape=None, mask=None, arguments=None, **kwargs
):
super().__init__(**kwargs)
self.arguments = arguments or {}
self.function = function
if mask is not None:
self.supports_masking = True
else:
self.supports_masking = False
self.mask = mask
self._output_shape = output_shape
# Warning on every invocation will be quite irksome in Eager mode.
self._already_warned = False
function_args = inspect.getfullargspec(function).args
self._fn_expects_training_arg = "training" in function_args
self._fn_expects_mask_arg = "mask" in function_args
def compute_output_shape(self, input_shape):
if self._output_shape is None:
# Leverage backend shape inference
try:
inputs = shape_utils.map_shape_structure(
lambda x: backend.KerasTensor(x, dtype=self.compute_dtype),
input_shape,
)
output_spec = backend.compute_output_spec(self.call, inputs)
return tree.map_structure(lambda x: x.shape, output_spec)
except:
raise NotImplementedError(
"We could not automatically infer the shape of "
"the Lambda's output. Please specify the `output_shape` "
"argument for this Lambda layer."
)
if callable(self._output_shape):
return self._output_shape(input_shape)
# Output shapes are passed directly and don't include batch dimension.
batch_size = tree.flatten(input_shape)[0]
def _add_batch(shape):
return (batch_size,) + shape
return shape_utils.map_shape_structure(_add_batch, self._output_shape)
def call(self, inputs, mask=None, training=None):
# We must copy for thread safety,
# but it only needs to be a shallow copy.
kwargs = {k: v for k, v in self.arguments.items()}
if self._fn_expects_mask_arg:
kwargs["mask"] = mask
if self._fn_expects_training_arg:
kwargs["training"] = training
return self.function(inputs, **kwargs)
def compute_mask(self, inputs, mask=None):
if callable(self.mask):
return self.mask(inputs, mask)
return self.mask
def get_config(self):
config = {
"function": self._serialize_function_to_config(self.function),
}
if self._output_shape is not None:
if callable(self._output_shape):
output_shape = self._serialize_function_to_config(
self._output_shape
)
else:
output_shape = self._output_shape
config["output_shape"] = output_shape
if self.mask is not None:
if callable(self.mask):
mask = self._serialize_function_to_config(self.mask)
else:
mask = serialization_lib.serialize_keras_object(self.mask)
config["mask"] = mask
config["arguments"] = serialization_lib.serialize_keras_object(
self.arguments
)
base_config = super().get_config()
return {**base_config, **config}
def _serialize_function_to_config(self, fn):
if isinstance(fn, types.LambdaType) and fn.__name__ == "<lambda>":
code, defaults, closure = python_utils.func_dump(fn)
return {
"class_name": "__lambda__",
"config": {
"code": code,
"defaults": defaults,
"closure": closure,
},
}
elif callable(fn):
return serialization_lib.serialize_keras_object(fn)
raise ValueError(
"Invalid input type for serialization. "
f"Received: {fn} of type {type(fn)}."
)
@staticmethod
def _raise_for_lambda_deserialization(arg_name, safe_mode):
if safe_mode:
raise ValueError(
"The `{arg_name}` of this `Lambda` layer is a Python lambda. "
"Deserializing it is unsafe. If you trust the source of the "
"config artifact, you can override this error "
"by passing `safe_mode=False` "
"to `from_config()`, or calling "
"`keras_core.config.enable_unsafe_deserialization()."
)
@classmethod
def from_config(cls, config, custom_objects=None, safe_mode=None):
safe_mode = safe_mode or serialization_lib.in_safe_mode()
fn_config = config["function"]
if (
isinstance(fn_config, dict)
and "class_name" in fn_config
and fn_config["class_name"] == "__lambda__"
):
cls._raise_for_lambda_deserialization("function", safe_mode)
inner_config = fn_config["config"]
fn = python_utils.func_load(
inner_config["code"],
defaults=inner_config["defaults"],
closure=inner_config["closure"],
)
config["function"] = fn
else:
config["function"] = serialization_lib.deserialize_keras_object(
fn_config, custom_objects=custom_objects
)
if "output_shape" in config:
fn_config = config["output_shape"]
if (
isinstance(fn_config, dict)
and "class_name" in fn_config
and fn_config["class_name"] == "__lambda__"
):
cls._raise_for_lambda_deserialization("function", safe_mode)
inner_config = fn_config["config"]
fn = python_utils.func_load(
inner_config["code"],
defaults=inner_config["defaults"],
closure=inner_config["closure"],
)
config["output_shape"] = fn
else:
config[
"output_shape"
] = serialization_lib.deserialize_keras_object(
fn_config, custom_objects=custom_objects
)
if "arguments" in config:
config["arguments"] = serialization_lib.deserialize_keras_object(
config["arguments"], custom_objects=custom_objects
)
return cls(**config)
|
keras-core/keras_core/layers/core/lambda_layer.py/0
|
{
"file_path": "keras-core/keras_core/layers/core/lambda_layer.py",
"repo_id": "keras-core",
"token_count": 4053
}
| 36 |
import numpy as np
import pytest
from absl.testing import parameterized
from keras_core import backend
from keras_core import layers
from keras_core import models
from keras_core import testing
def np_dot(a, b, axes):
if isinstance(axes, int):
axes = (axes, axes)
axes = [axis if axis < 0 else axis - 1 for axis in axes]
res = np.stack([np.tensordot(a[i], b[i], axes) for i in range(a.shape[0])])
if len(res.shape) == 1:
res = np.expand_dims(res, axis=1)
return res
TEST_PARAMETERS = [
{
"testcase_name": "add",
"layer_class": layers.Add,
"np_op": np.add,
},
{
"testcase_name": "substract",
"layer_class": layers.Subtract,
"np_op": np.subtract,
},
{
"testcase_name": "minimum",
"layer_class": layers.Minimum,
"np_op": np.minimum,
},
{
"testcase_name": "maximum",
"layer_class": layers.Maximum,
"np_op": np.maximum,
},
{
"testcase_name": "multiply",
"layer_class": layers.Multiply,
"np_op": np.multiply,
},
{
"testcase_name": "average",
"layer_class": layers.Average,
"np_op": lambda a, b: np.multiply(np.add(a, b), 0.5),
},
{
"testcase_name": "concat",
"layer_class": layers.Concatenate,
"np_op": lambda a, b, **kwargs: np.concatenate((a, b), **kwargs),
"init_kwargs": {"axis": -1},
"expected_output_shape": (2, 4, 10),
},
{
"testcase_name": "dot_2d",
"layer_class": layers.Dot,
"np_op": np_dot,
"init_kwargs": {"axes": -1},
"input_shape": (2, 4),
"expected_output_shape": (2, 1),
"skip_mask_test": True,
},
{
"testcase_name": "dot_3d",
"layer_class": layers.Dot,
"np_op": np_dot,
"init_kwargs": {"axes": -1},
"expected_output_shape": (2, 4, 4),
"skip_mask_test": True,
},
]
@pytest.mark.requires_trainable_backend
class MergingLayersTest(testing.TestCase, parameterized.TestCase):
@parameterized.named_parameters(TEST_PARAMETERS)
def test_basic(
self,
layer_class,
init_kwargs={},
input_shape=(2, 4, 5),
expected_output_shape=(2, 4, 5),
**kwargs
):
self.run_layer_test(
layer_class,
init_kwargs=init_kwargs,
input_shape=[input_shape, input_shape],
expected_output_shape=expected_output_shape,
expected_num_trainable_weights=0,
expected_num_non_trainable_weights=0,
expected_num_seed_generators=0,
expected_num_losses=0,
supports_masking=True,
)
@parameterized.named_parameters(TEST_PARAMETERS)
def test_correctness_static(
self,
layer_class,
np_op,
init_kwargs={},
input_shape=(2, 4, 5),
expected_output_shape=(2, 4, 5),
skip_mask_test=False,
):
batch_size = input_shape[0]
shape = input_shape[1:]
x1 = np.random.rand(*input_shape)
x2 = np.random.rand(*input_shape)
x3 = np_op(x1, x2, **init_kwargs)
input_1 = layers.Input(shape=shape, batch_size=batch_size)
input_2 = layers.Input(shape=shape, batch_size=batch_size)
layer = layer_class(**init_kwargs)
out = layer([input_1, input_2])
model = models.Model([input_1, input_2], out)
res = model([x1, x2])
self.assertEqual(res.shape, expected_output_shape)
self.assertAllClose(res, x3, atol=1e-4)
self.assertIsNone(layer.compute_mask([input_1, input_2], [None, None]))
if not skip_mask_test:
self.assertTrue(
np.all(
backend.convert_to_numpy(
layer.compute_mask(
[input_1, input_2],
[backend.Variable(x1), backend.Variable(x2)],
)
)
)
)
@parameterized.named_parameters(TEST_PARAMETERS)
def test_correctness_dynamic(
self,
layer_class,
np_op,
init_kwargs={},
input_shape=(2, 4, 5),
expected_output_shape=(2, 4, 5),
skip_mask_test=False,
):
shape = input_shape[1:]
x1 = np.random.rand(*input_shape)
x2 = np.random.rand(*input_shape)
x3 = np_op(x1, x2, **init_kwargs)
input_1 = layers.Input(shape=shape)
input_2 = layers.Input(shape=shape)
layer = layer_class(**init_kwargs)
out = layer([input_1, input_2])
model = models.Model([input_1, input_2], out)
res = model([x1, x2])
self.assertEqual(res.shape, expected_output_shape)
self.assertAllClose(res, x3, atol=1e-4)
self.assertIsNone(layer.compute_mask([input_1, input_2], [None, None]))
if not skip_mask_test:
self.assertTrue(
np.all(
backend.convert_to_numpy(
layer.compute_mask(
[input_1, input_2],
[backend.Variable(x1), backend.Variable(x2)],
)
)
)
)
@parameterized.named_parameters(TEST_PARAMETERS)
def test_errors(
self,
layer_class,
init_kwargs={},
input_shape=(2, 4, 5),
skip_mask_test=False,
**kwargs
):
if skip_mask_test:
pytest.skip("Masking not supported")
batch_size = input_shape[0]
shape = input_shape[1:]
x1 = np.random.rand(*input_shape)
x1 = np.random.rand(batch_size, *shape)
input_1 = layers.Input(shape=shape, batch_size=batch_size)
input_2 = layers.Input(shape=shape, batch_size=batch_size)
layer = layer_class(**init_kwargs)
with self.assertRaisesRegex(ValueError, "`mask` should be a list."):
layer.compute_mask([input_1, input_2], x1)
with self.assertRaisesRegex(ValueError, "`inputs` should be a list."):
layer.compute_mask(input_1, [None, None])
with self.assertRaisesRegex(
ValueError, " should have the same length."
):
layer.compute_mask([input_1, input_2], [None])
def test_subtract_layer_inputs_length_errors(self):
shape = (4, 5)
input_1 = layers.Input(shape=shape)
input_2 = layers.Input(shape=shape)
input_3 = layers.Input(shape=shape)
with self.assertRaisesRegex(
ValueError, "layer should be called on exactly 2 inputs"
):
layers.Subtract()([input_1, input_2, input_3])
with self.assertRaisesRegex(
ValueError, "layer should be called on exactly 2 inputs"
):
layers.Subtract()([input_1])
@parameterized.named_parameters(TEST_PARAMETERS)
@pytest.mark.skipif(
not backend.SUPPORTS_SPARSE_TENSORS,
reason="Backend does not support sparse tensors.",
)
def test_sparse(
self,
layer_class,
np_op,
init_kwargs={},
input_shape=(2, 4, 5),
expected_output_shape=(2, 4, 5),
**kwargs
):
import tensorflow as tf
self.run_layer_test(
layer_class,
init_kwargs=init_kwargs,
input_shape=[input_shape, input_shape],
input_sparse=True,
expected_output_shape=expected_output_shape,
expected_output_sparse=True,
expected_num_trainable_weights=0,
expected_num_non_trainable_weights=0,
expected_num_seed_generators=0,
expected_num_losses=0,
supports_masking=True,
run_training_check=False,
run_mixed_precision_check=False,
)
layer = layer_class(**init_kwargs)
# Merging a sparse tensor with a dense tensor, or a dense tensor with a
# sparse tensor produces a dense tensor
x1 = tf.SparseTensor(
indices=[[0, 0], [1, 2]], values=[1.0, 2.0], dense_shape=(2, 3)
)
x1_np = tf.sparse.to_dense(x1).numpy()
x2 = np.random.rand(2, 3)
self.assertAllClose(layer([x1, x2]), np_op(x1_np, x2, **init_kwargs))
self.assertAllClose(layer([x2, x1]), np_op(x2, x1_np, **init_kwargs))
# Merging a sparse tensor with a sparse tensor produces a sparse tensor
x3 = tf.SparseTensor(
indices=[[0, 0], [1, 1]], values=[4.0, 5.0], dense_shape=(2, 3)
)
x3_np = tf.sparse.to_dense(x3).numpy()
self.assertIsInstance(layer([x1, x3]), tf.SparseTensor)
self.assertAllClose(layer([x1, x3]), np_op(x1_np, x3_np, **init_kwargs))
|
keras-core/keras_core/layers/merging/merging_test.py/0
|
{
"file_path": "keras-core/keras_core/layers/merging/merging_test.py",
"repo_id": "keras-core",
"token_count": 4557
}
| 37 |
import os
import numpy as np
import pytest
from absl.testing import parameterized
from tensorflow import data as tf_data
from keras_core import backend
from keras_core import layers
from keras_core import models
from keras_core import testing
from keras_core.saving import saving_api
@pytest.mark.skipif(
backend.backend() == "numpy", reason="Failing for numpy backend."
)
class IndexLookupLayerTest(testing.TestCase, parameterized.TestCase):
def test_basics_string_vocab(self):
# Case: adapt + list inputs
adapt_data = ["one", "one", "one", "two", "two", "three"]
input_data = ["one", "two", "four"]
kwargs = {
"max_tokens": 7,
"num_oov_indices": 1,
"mask_token": "",
"oov_token": "[OOV]",
"vocabulary_dtype": "string",
}
layer = layers.IndexLookup(**kwargs)
layer.adapt(adapt_data)
self.assertEqual(
layer.get_vocabulary(), ["", "[OOV]", "one", "two", "three"]
)
self.assertEqual(
layer.get_vocabulary(include_special_tokens=False),
["one", "two", "three"],
)
output = layer(input_data)
self.assertEqual(list(output), [2, 3, 1])
if backend.backend() != "torch":
self.run_class_serialization_test(layer)
# Case: numpy array input
output = layer(np.array(input_data))
self.assertEqual(list(output), [2, 3, 1])
# Case: fixed vocab + list inputs
vocabulary = ["one", "two", "three"]
layer = layers.IndexLookup(vocabulary=vocabulary, **kwargs)
self.assertEqual(
layer.get_vocabulary(), ["", "[OOV]", "one", "two", "three"]
)
self.assertEqual(
layer.get_vocabulary(include_special_tokens=False),
["one", "two", "three"],
)
output = layer(input_data)
self.assertEqual(list(output), [2, 3, 1])
if backend.backend() != "torch":
self.run_class_serialization_test(layer)
# Case: fixed vocab with special tokens + list inputs
vocabulary_with_special_tokens = ["", "[OOV]", "one", "two", "three"]
layer = layers.IndexLookup(
vocabulary=vocabulary_with_special_tokens, **kwargs
)
self.assertEqual(
layer.get_vocabulary(), ["", "[OOV]", "one", "two", "three"]
)
self.assertEqual(
layer.get_vocabulary(include_special_tokens=False),
["one", "two", "three"],
)
output = layer(input_data)
self.assertEqual(list(output), [2, 3, 1])
if backend.backend() != "torch":
self.run_class_serialization_test(layer)
# Case: set vocabulary
layer = layers.IndexLookup(**kwargs)
layer.set_vocabulary(vocabulary)
self.assertEqual(
layer.get_vocabulary(), ["", "[OOV]", "one", "two", "three"]
)
self.assertEqual(
layer.get_vocabulary(include_special_tokens=False),
["one", "two", "three"],
)
output = layer(input_data)
self.assertEqual(list(output), [2, 3, 1])
if backend.backend() != "torch":
self.run_class_serialization_test(layer)
# Case: set vocabulary (with special tokens)
layer = layers.IndexLookup(**kwargs)
layer.set_vocabulary(vocabulary_with_special_tokens)
self.assertEqual(
layer.get_vocabulary(), ["", "[OOV]", "one", "two", "three"]
)
self.assertEqual(
layer.get_vocabulary(include_special_tokens=False),
["one", "two", "three"],
)
output = layer(input_data)
self.assertEqual(list(output), [2, 3, 1])
if backend.backend() != "torch":
self.run_class_serialization_test(layer)
def test_basics_integer_vocab(self):
# Case: adapt + list inputs
adapt_data = [1, 1, 1, 2, 2, 3]
input_data = [1, 2, 4]
kwargs = {
"max_tokens": 7,
"num_oov_indices": 1,
"mask_token": 0,
"oov_token": -1,
"vocabulary_dtype": "int64",
}
layer = layers.IndexLookup(**kwargs)
layer.adapt(adapt_data)
self.assertEqual(layer.get_vocabulary(), [0, -1, 1, 2, 3])
self.assertEqual(
layer.get_vocabulary(include_special_tokens=False),
[1, 2, 3],
)
output = layer(input_data)
self.assertEqual(list(output), [2, 3, 1])
if backend.backend() != "torch":
self.run_class_serialization_test(layer)
# Case: numpy array input
output = layer(np.array(input_data))
self.assertEqual(list(output), [2, 3, 1])
# Case: fixed vocab + list inputs
vocabulary = [1, 2, 3]
layer = layers.IndexLookup(vocabulary=vocabulary, **kwargs)
self.assertEqual(layer.get_vocabulary(), [0, -1, 1, 2, 3])
self.assertEqual(
layer.get_vocabulary(include_special_tokens=False),
[1, 2, 3],
)
output = layer(input_data)
self.assertEqual(list(output), [2, 3, 1])
if backend.backend() != "torch":
self.run_class_serialization_test(layer)
# Case: fixed vocab with special tokens + list inputs
vocabulary_with_special_tokens = [0, -1, 1, 2, 3]
layer = layers.IndexLookup(
vocabulary=vocabulary_with_special_tokens, **kwargs
)
self.assertEqual(layer.get_vocabulary(), [0, -1, 1, 2, 3])
self.assertEqual(
layer.get_vocabulary(include_special_tokens=False),
[1, 2, 3],
)
output = layer(input_data)
self.assertEqual(list(output), [2, 3, 1])
if backend.backend() != "torch":
self.run_class_serialization_test(layer)
# Case: set vocabulary
layer = layers.IndexLookup(**kwargs)
layer.set_vocabulary(vocabulary)
self.assertEqual(layer.get_vocabulary(), [0, -1, 1, 2, 3])
self.assertEqual(
layer.get_vocabulary(include_special_tokens=False),
[1, 2, 3],
)
output = layer(input_data)
self.assertEqual(list(output), [2, 3, 1])
if backend.backend() != "torch":
self.run_class_serialization_test(layer)
# Case: set vocabulary (with special tokens)
layer = layers.IndexLookup(**kwargs)
layer.set_vocabulary(vocabulary_with_special_tokens)
self.assertEqual(layer.get_vocabulary(), [0, -1, 1, 2, 3])
self.assertEqual(
layer.get_vocabulary(include_special_tokens=False),
[1, 2, 3],
)
output = layer(input_data)
self.assertEqual(list(output), [2, 3, 1])
if backend.backend() != "torch":
self.run_class_serialization_test(layer)
def test_max_tokens_adapt(self):
adapt_data = [1, 1, 1, 2, 2, 3]
input_data = [1, 2, 3, 4]
kwargs = {
"max_tokens": 4,
"num_oov_indices": 1,
"mask_token": 0,
"oov_token": -1,
"vocabulary_dtype": "int64",
}
layer = layers.IndexLookup(**kwargs)
layer.adapt(adapt_data)
self.assertEqual(layer.get_vocabulary(), [0, -1, 1, 2])
self.assertEqual(
layer.get_vocabulary(include_special_tokens=False),
[1, 2],
)
output = layer(input_data)
self.assertEqual(list(output), [2, 3, 1, 1])
if backend.backend() != "torch":
self.run_class_serialization_test(layer)
def test_pad_to_max_tokens(self):
vocabulary = [1, 2]
input_data = [1, 2]
kwargs = {
"max_tokens": 5,
"num_oov_indices": 1,
"mask_token": 0,
"oov_token": -1,
"vocabulary_dtype": "int64",
"vocabulary": vocabulary,
"pad_to_max_tokens": True,
"output_mode": "multi_hot",
}
layer = layers.IndexLookup(**kwargs)
output = layer(input_data)
self.assertAllClose(output, [0, 1, 1, 0, 0])
if backend.backend() != "torch":
self.run_class_serialization_test(layer)
def test_output_modes(self):
vocabulary = ["one", "two", "three"]
single_sample_input_data = ["one", "two", "four"]
batch_input_data = [["one", "two", "four", "two"]]
kwargs = {
"max_tokens": 7,
"num_oov_indices": 1,
"mask_token": "",
"oov_token": "[OOV]",
"vocabulary_dtype": "string",
"vocabulary": vocabulary,
}
# int
kwargs["output_mode"] = "int"
layer = layers.IndexLookup(**kwargs)
output = layer(single_sample_input_data)
self.assertAllClose(output, [2, 3, 1])
output = layer(batch_input_data)
self.assertAllClose(output, [[2, 3, 1, 3]])
# multi-hot
kwargs["output_mode"] = "multi_hot"
layer = layers.IndexLookup(**kwargs)
output = layer(single_sample_input_data)
self.assertAllClose(output, [1, 1, 1, 0])
output = layer(batch_input_data)
self.assertAllClose(output, [[1, 1, 1, 0]])
# one-hot
kwargs["output_mode"] = "one_hot"
layer = layers.IndexLookup(**kwargs)
output = layer(single_sample_input_data)
self.assertAllClose(output, [[0, 1, 0, 0], [0, 0, 1, 0], [1, 0, 0, 0]])
# count
kwargs["output_mode"] = "count"
layer = layers.IndexLookup(**kwargs)
output = layer(single_sample_input_data)
self.assertAllClose(output, [1, 1, 1, 0])
output = layer(batch_input_data)
self.assertAllClose(output, [[1, 1, 2, 0]])
# tf-idf
kwargs["output_mode"] = "tf_idf"
kwargs["idf_weights"] = np.array([0.1, 0.2, 0.3])
layer = layers.IndexLookup(**kwargs)
output = layer(single_sample_input_data)
self.assertAllClose(output, [0.2, 0.1, 0.2, 0.0])
output = layer(batch_input_data)
self.assertAllClose(output, [[0.2, 0.1, 0.4, 0.0]])
def test_sparse_outputs(self):
# TODO
pass
def test_adapt_tf_idf(self):
# Case: unbatched data
adapt_data = ["one", "one", "one", "two", "two", "three"]
input_data = ["one", "two", "four"]
kwargs = {
"max_tokens": 7,
"num_oov_indices": 1,
"mask_token": "",
"oov_token": "[OOV]",
"vocabulary_dtype": "string",
"output_mode": "tf_idf",
}
layer = layers.IndexLookup(**kwargs)
layer.adapt(adapt_data)
output = layer(input_data)
# Document counts for one, two, three = [3, 2, 1]
idf_weights = np.log(1 + len(adapt_data) / (1 + np.array([3, 2, 1])))
self.assertAllClose(layer.idf_weights[1:], idf_weights)
self.assertAllClose(output, [1.1337324, 0.91629076, 1.0986123, 0.0])
# Case: batched data
adapt_data = [["one", "one"], ["one", "two"], ["two", "three"]]
input_data = [["one", "two"], ["two", "four"]]
kwargs = {
"max_tokens": 7,
"num_oov_indices": 1,
"mask_token": "",
"oov_token": "[OOV]",
"vocabulary_dtype": "string",
"output_mode": "tf_idf",
}
layer = layers.IndexLookup(**kwargs)
layer.adapt(adapt_data)
# Document counts for one, two, three = [2, 2, 1]
idf_weights = np.log(1 + len(adapt_data) / (1 + np.array([2, 2, 1])))
self.assertAllClose(layer.idf_weights[1:], idf_weights)
output = layer(input_data)
self.assertAllClose(
output,
[
[0.0, 0.6931472, 0.6931472, 0.0],
[0.76752836, 0.0, 0.6931472, 0.0],
],
)
def test_invert(self):
vocabulary = ["one", "two", "three"]
single_sample_input_data = [2, 3, 1]
batch_input_data = [[2, 3, 1, 3]]
kwargs = {
"max_tokens": 7,
"num_oov_indices": 1,
"mask_token": "",
"oov_token": "[OOV]",
"vocabulary_dtype": "string",
"vocabulary": vocabulary,
"invert": True,
"output_mode": "int",
}
layer = layers.IndexLookup(**kwargs)
output = layer(single_sample_input_data)
self.assertEqual(
[w.decode("utf-8") for w in output.numpy()], ["one", "two", "[OOV]"]
)
output = layer(batch_input_data)
self.assertEqual(
[w.decode("utf-8") for w in output.numpy()[0]],
["one", "two", "[OOV]", "two"],
)
@pytest.mark.skipif(
backend.backend() != "tensorflow", reason="Requires string input dtype"
)
def test_saving(self):
# Test with adapt()
vocabulary = ["one", "two", "three"]
adapt_data = ["one", "one", "one", "two", "two", "three"]
batch_input_data = np.array([["one", "two", "four"]])
kwargs = {
"max_tokens": 7,
"num_oov_indices": 1,
"mask_token": "",
"oov_token": "[OOV]",
"vocabulary_dtype": "string",
"output_mode": "int",
}
layer = layers.IndexLookup(**kwargs)
layer.adapt(adapt_data)
model = models.Sequential(
[
layers.Input(shape=(None,), dtype="string"),
layer,
]
)
output_1 = model(batch_input_data)
path = os.path.join(self.get_temp_dir(), "model.keras")
model.save(path)
model = saving_api.load_model(path)
output_2 = model(batch_input_data)
self.assertAllClose(output_1, output_2)
# Test when vocabulary is provided
kwargs["vocabulary"] = vocabulary
layer = layers.IndexLookup(**kwargs)
model = models.Sequential(
[
layers.Input(shape=(None,), dtype="string"),
layer,
]
)
output_1 = model(batch_input_data)
path = os.path.join(self.get_temp_dir(), "model.keras")
model.save(path)
model = saving_api.load_model(path)
output_2 = model(batch_input_data)
self.assertAllClose(output_1, output_2)
def test_adapt_with_tf_data(self):
# Case: adapt + list inputs
adapt_data = tf_data.Dataset.from_tensor_slices(
["one", "one", "one", "two", "two", "three"]
).batch(2)
input_data = ["one", "two", "four"]
kwargs = {
"max_tokens": 7,
"num_oov_indices": 1,
"mask_token": "",
"oov_token": "[OOV]",
"vocabulary_dtype": "string",
}
layer = layers.IndexLookup(**kwargs)
layer.adapt(adapt_data)
self.assertEqual(
layer.get_vocabulary(), ["", "[OOV]", "one", "two", "three"]
)
self.assertEqual(
layer.get_vocabulary(include_special_tokens=False),
["one", "two", "three"],
)
output = layer(input_data)
self.assertEqual(list(output), [2, 3, 1])
if backend.backend() != "torch":
self.run_class_serialization_test(layer)
|
keras-core/keras_core/layers/preprocessing/index_lookup_test.py/0
|
{
"file_path": "keras-core/keras_core/layers/preprocessing/index_lookup_test.py",
"repo_id": "keras-core",
"token_count": 7836
}
| 38 |
import numpy as np
from absl.testing import parameterized
from tensorflow import data as tf_data
from keras_core import backend
from keras_core import layers
from keras_core import testing
class RandomTranslationTest(testing.TestCase, parameterized.TestCase):
@parameterized.named_parameters(
("random_translate_4_by_6", 0.4, 0.6),
("random_translate_3_by_2", 0.3, 0.2),
("random_translate_tuple_factor", (-0.5, 0.4), (0.2, 0.3)),
)
def test_random_translation(self, height_factor, width_factor):
self.run_layer_test(
layers.RandomTranslation,
init_kwargs={
"height_factor": height_factor,
"width_factor": width_factor,
},
input_shape=(2, 3, 4),
expected_output_shape=(2, 3, 4),
supports_masking=False,
run_training_check=False,
)
@parameterized.named_parameters(
("bad_len", [0.1, 0.2, 0.3], 0.0),
("bad_type", {"dummy": 0.3}, 0.0),
("exceed_range_single", -1.1, 0.0),
("exceed_range_tuple", (-1.1, 0.0), 0.0),
)
def test_random_translation_with_bad_factor(
self, height_factor, width_factor
):
with self.assertRaises(ValueError):
self.run_layer_test(
layers.RandomTranslation,
init_kwargs={
"height_factor": height_factor,
"width_factor": width_factor,
},
input_shape=(2, 3, 4),
expected_output_shape=(2, 3, 4),
supports_masking=False,
run_training_check=False,
)
def test_random_translation_with_inference_mode(self):
input_data = np.random.random((1, 4, 4, 3))
expected_output = input_data
layer = layers.RandomTranslation(0.2, 0.1)
output = layer(input_data, training=False)
self.assertAllClose(output, expected_output)
@parameterized.parameters(["channels_first", "channels_last"])
def test_random_translation_up_numeric_reflect(self, data_format):
input_image = np.arange(0, 25)
expected_output = np.asarray(
[
[5, 6, 7, 8, 9],
[10, 11, 12, 13, 14],
[15, 16, 17, 18, 19],
[20, 21, 22, 23, 24],
[20, 21, 22, 23, 24],
]
)
if data_format == "channels_last":
input_image = np.reshape(input_image, (1, 5, 5, 1))
expected_output = backend.convert_to_tensor(
np.reshape(expected_output, (1, 5, 5, 1))
)
else:
input_image = np.reshape(input_image, (1, 1, 5, 5))
expected_output = backend.convert_to_tensor(
np.reshape(expected_output, (1, 1, 5, 5))
)
self.run_layer_test(
layers.RandomTranslation,
init_kwargs={
"height_factor": (-0.2, -0.2),
"width_factor": 0.0,
"data_format": data_format,
},
input_shape=None,
input_data=input_image,
expected_output=expected_output,
supports_masking=False,
run_training_check=False,
)
@parameterized.parameters(["channels_first", "channels_last"])
def test_random_translation_up_numeric_constant(self, data_format):
input_image = np.arange(0, 25).astype("float32")
# Shifting by -.2 * 5 = 1 pixel.
expected_output = np.asarray(
[
[5, 6, 7, 8, 9],
[10, 11, 12, 13, 14],
[15, 16, 17, 18, 19],
[20, 21, 22, 23, 24],
[0, 0, 0, 0, 0],
]
)
if data_format == "channels_last":
input_image = np.reshape(input_image, (1, 5, 5, 1))
expected_output = backend.convert_to_tensor(
np.reshape(expected_output, (1, 5, 5, 1)), dtype="float32"
)
else:
input_image = np.reshape(input_image, (1, 1, 5, 5))
expected_output = backend.convert_to_tensor(
np.reshape(expected_output, (1, 1, 5, 5)), dtype="float32"
)
self.run_layer_test(
layers.RandomTranslation,
init_kwargs={
"height_factor": (-0.2, -0.2),
"width_factor": 0.0,
"fill_mode": "constant",
"data_format": data_format,
},
input_shape=None,
input_data=input_image,
expected_output=expected_output,
supports_masking=False,
run_training_check=False,
)
@parameterized.parameters(["channels_first", "channels_last"])
def test_random_translation_down_numeric_reflect(self, data_format):
input_image = np.arange(0, 25)
# Shifting by .2 * 5 = 1 pixel.
expected_output = np.asarray(
[
[0, 1, 2, 3, 4],
[0, 1, 2, 3, 4],
[5, 6, 7, 8, 9],
[10, 11, 12, 13, 14],
[15, 16, 17, 18, 19],
]
)
if data_format == "channels_last":
input_image = np.reshape(input_image, (1, 5, 5, 1))
expected_output = backend.convert_to_tensor(
np.reshape(expected_output, (1, 5, 5, 1))
)
else:
input_image = np.reshape(input_image, (1, 1, 5, 5))
expected_output = backend.convert_to_tensor(
np.reshape(expected_output, (1, 1, 5, 5))
)
self.run_layer_test(
layers.RandomTranslation,
init_kwargs={
"height_factor": (0.2, 0.2),
"width_factor": 0.0,
"data_format": data_format,
},
input_shape=None,
input_data=input_image,
expected_output=expected_output,
supports_masking=False,
run_training_check=False,
)
@parameterized.parameters(["channels_first", "channels_last"])
def test_random_translation_asymmetric_size_numeric_reflect(
self, data_format
):
input_image = np.arange(0, 16)
# Shifting by .2 * 5 = 1 pixel.
expected_output = np.asarray(
[
[6, 7],
[4, 5],
[2, 3],
[0, 1],
[0, 1],
[2, 3],
[4, 5],
[6, 7],
]
)
if data_format == "channels_last":
input_image = np.reshape(input_image, (1, 8, 2, 1))
expected_output = backend.convert_to_tensor(
np.reshape(expected_output, (1, 8, 2, 1))
)
else:
input_image = np.reshape(input_image, (1, 1, 8, 2))
expected_output = backend.convert_to_tensor(
np.reshape(expected_output, (1, 1, 8, 2))
)
self.run_layer_test(
layers.RandomTranslation,
init_kwargs={
"height_factor": (0.5, 0.5),
"width_factor": 0.0,
"data_format": data_format,
},
input_shape=None,
input_data=input_image,
expected_output=expected_output,
supports_masking=False,
run_training_check=False,
)
@parameterized.parameters(["channels_first", "channels_last"])
def test_random_translation_down_numeric_constant(self, data_format):
input_image = np.arange(0, 25)
# Shifting by .2 * 5 = 1 pixel.
expected_output = np.asarray(
[
[0, 0, 0, 0, 0],
[0, 1, 2, 3, 4],
[5, 6, 7, 8, 9],
[10, 11, 12, 13, 14],
[15, 16, 17, 18, 19],
]
)
if data_format == "channels_last":
input_image = np.reshape(input_image, (1, 5, 5, 1))
expected_output = backend.convert_to_tensor(
np.reshape(expected_output, (1, 5, 5, 1))
)
else:
input_image = np.reshape(input_image, (1, 1, 5, 5))
expected_output = backend.convert_to_tensor(
np.reshape(expected_output, (1, 1, 5, 5))
)
self.run_layer_test(
layers.RandomTranslation,
init_kwargs={
"height_factor": (0.2, 0.2),
"width_factor": 0.0,
"fill_mode": "constant",
"fill_value": 0.0,
"data_format": data_format,
},
input_shape=None,
input_data=input_image,
expected_output=expected_output,
supports_masking=False,
run_training_check=False,
)
@parameterized.parameters(["channels_first", "channels_last"])
def test_random_translation_left_numeric_reflect(self, data_format):
input_image = np.arange(0, 25)
# Shifting by .2 * 5 = 1 pixel.
expected_output = np.asarray(
[
[1, 2, 3, 4, 4],
[6, 7, 8, 9, 9],
[11, 12, 13, 14, 14],
[16, 17, 18, 19, 19],
[21, 22, 23, 24, 24],
]
)
if data_format == "channels_last":
input_image = np.reshape(input_image, (1, 5, 5, 1))
expected_output = backend.convert_to_tensor(
np.reshape(expected_output, (1, 5, 5, 1))
)
else:
input_image = np.reshape(input_image, (1, 1, 5, 5))
expected_output = backend.convert_to_tensor(
np.reshape(expected_output, (1, 1, 5, 5))
)
self.run_layer_test(
layers.RandomTranslation,
init_kwargs={
"height_factor": 0.0,
"width_factor": (-0.2, -0.2),
"data_format": data_format,
},
input_shape=None,
input_data=input_image,
expected_output=expected_output,
supports_masking=False,
run_training_check=False,
)
@parameterized.parameters(["channels_first", "channels_last"])
def test_random_translation_left_numeric_constant(self, data_format):
input_image = np.arange(0, 25)
# Shifting by .2 * 5 = 1 pixel.
expected_output = np.asarray(
[
[1, 2, 3, 4, 0],
[6, 7, 8, 9, 0],
[11, 12, 13, 14, 0],
[16, 17, 18, 19, 0],
[21, 22, 23, 24, 0],
]
)
if data_format == "channels_last":
input_image = np.reshape(input_image, (1, 5, 5, 1))
expected_output = backend.convert_to_tensor(
np.reshape(expected_output, (1, 5, 5, 1))
)
else:
input_image = np.reshape(input_image, (1, 1, 5, 5))
expected_output = backend.convert_to_tensor(
np.reshape(expected_output, (1, 1, 5, 5))
)
self.run_layer_test(
layers.RandomTranslation,
init_kwargs={
"height_factor": 0.0,
"width_factor": (-0.2, -0.2),
"fill_mode": "constant",
"fill_value": 0.0,
"data_format": data_format,
},
input_shape=None,
input_data=input_image,
expected_output=expected_output,
supports_masking=False,
run_training_check=False,
)
def test_tf_data_compatibility(self):
layer = layers.RandomTranslation(0.2, 0.1)
input_data = np.random.random((1, 4, 4, 3))
ds = tf_data.Dataset.from_tensor_slices(input_data).batch(1).map(layer)
for output in ds.take(1):
output.numpy()
|
keras-core/keras_core/layers/preprocessing/random_translation_test.py/0
|
{
"file_path": "keras-core/keras_core/layers/preprocessing/random_translation_test.py",
"repo_id": "keras-core",
"token_count": 6673
}
| 39 |
import numpy as np
import pytest
from keras_core import backend
from keras_core import layers
from keras_core import testing
class DropoutTest(testing.TestCase):
@pytest.mark.requires_trainable_backend
def test_dropout_basics(self):
self.run_layer_test(
layers.Dropout,
init_kwargs={
"rate": 0.2,
},
input_shape=(2, 3),
expected_output_shape=(2, 3),
expected_num_trainable_weights=0,
expected_num_non_trainable_weights=0,
expected_num_seed_generators=1,
expected_num_losses=0,
supports_masking=True,
)
def test_dropout_rescaling(self):
inputs = np.ones((20, 500))
layer = layers.Dropout(0.5, seed=1337)
outputs = layer(inputs, training=True)
outputs = backend.convert_to_numpy(outputs)
self.assertAllClose(np.mean(outputs), 1.0, atol=0.02)
self.assertAllClose(np.max(outputs), 2.0)
def test_dropout_partial_noise_shape_dynamic(self):
inputs = np.ones((20, 5, 10))
layer = layers.Dropout(0.5, noise_shape=(None, 1, None))
outputs = layer(inputs, training=True)
self.assertAllClose(outputs[:, 0, :], outputs[:, 1, :])
def test_dropout_partial_noise_shape_static(self):
inputs = np.ones((20, 5, 10))
layer = layers.Dropout(0.5, noise_shape=(20, 1, 10))
outputs = layer(inputs, training=True)
self.assertAllClose(outputs[:, 0, :], outputs[:, 1, :])
|
keras-core/keras_core/layers/regularization/dropout_test.py/0
|
{
"file_path": "keras-core/keras_core/layers/regularization/dropout_test.py",
"repo_id": "keras-core",
"token_count": 721
}
| 40 |
from keras_core.api_export import keras_core_export
from keras_core.metrics.accuracy_metrics import Accuracy
from keras_core.metrics.accuracy_metrics import BinaryAccuracy
from keras_core.metrics.accuracy_metrics import CategoricalAccuracy
from keras_core.metrics.accuracy_metrics import SparseCategoricalAccuracy
from keras_core.metrics.accuracy_metrics import SparseTopKCategoricalAccuracy
from keras_core.metrics.accuracy_metrics import TopKCategoricalAccuracy
from keras_core.metrics.confusion_metrics import AUC
from keras_core.metrics.confusion_metrics import FalseNegatives
from keras_core.metrics.confusion_metrics import FalsePositives
from keras_core.metrics.confusion_metrics import Precision
from keras_core.metrics.confusion_metrics import PrecisionAtRecall
from keras_core.metrics.confusion_metrics import Recall
from keras_core.metrics.confusion_metrics import RecallAtPrecision
from keras_core.metrics.confusion_metrics import SensitivityAtSpecificity
from keras_core.metrics.confusion_metrics import SpecificityAtSensitivity
from keras_core.metrics.confusion_metrics import TrueNegatives
from keras_core.metrics.confusion_metrics import TruePositives
from keras_core.metrics.f_score_metrics import F1Score
from keras_core.metrics.f_score_metrics import FBetaScore
from keras_core.metrics.hinge_metrics import CategoricalHinge
from keras_core.metrics.hinge_metrics import Hinge
from keras_core.metrics.hinge_metrics import SquaredHinge
from keras_core.metrics.iou_metrics import BinaryIoU
from keras_core.metrics.iou_metrics import IoU
from keras_core.metrics.iou_metrics import MeanIoU
from keras_core.metrics.iou_metrics import OneHotIoU
from keras_core.metrics.iou_metrics import OneHotMeanIoU
from keras_core.metrics.metric import Metric
from keras_core.metrics.probabilistic_metrics import BinaryCrossentropy
from keras_core.metrics.probabilistic_metrics import CategoricalCrossentropy
from keras_core.metrics.probabilistic_metrics import KLDivergence
from keras_core.metrics.probabilistic_metrics import Poisson
from keras_core.metrics.probabilistic_metrics import (
SparseCategoricalCrossentropy,
)
from keras_core.metrics.reduction_metrics import Mean
from keras_core.metrics.reduction_metrics import MeanMetricWrapper
from keras_core.metrics.reduction_metrics import Sum
from keras_core.metrics.regression_metrics import CosineSimilarity
from keras_core.metrics.regression_metrics import LogCoshError
from keras_core.metrics.regression_metrics import MeanAbsoluteError
from keras_core.metrics.regression_metrics import MeanAbsolutePercentageError
from keras_core.metrics.regression_metrics import MeanSquaredError
from keras_core.metrics.regression_metrics import MeanSquaredLogarithmicError
from keras_core.metrics.regression_metrics import R2Score
from keras_core.metrics.regression_metrics import RootMeanSquaredError
from keras_core.saving import serialization_lib
from keras_core.utils.naming import to_snake_case
ALL_OBJECTS = {
# Base
Metric,
Mean,
Sum,
MeanMetricWrapper,
# Regression
MeanSquaredError,
RootMeanSquaredError,
MeanAbsoluteError,
MeanAbsolutePercentageError,
MeanSquaredLogarithmicError,
CosineSimilarity,
LogCoshError,
R2Score,
# Classification
AUC,
FalseNegatives,
FalsePositives,
Precision,
PrecisionAtRecall,
Recall,
RecallAtPrecision,
SensitivityAtSpecificity,
SpecificityAtSensitivity,
TrueNegatives,
TruePositives,
# Hinge
Hinge,
SquaredHinge,
CategoricalHinge,
# Probabilistic
KLDivergence,
Poisson,
BinaryCrossentropy,
CategoricalCrossentropy,
SparseCategoricalCrossentropy,
# Accuracy
Accuracy,
BinaryAccuracy,
CategoricalAccuracy,
SparseCategoricalAccuracy,
TopKCategoricalAccuracy,
SparseTopKCategoricalAccuracy,
# F-Score
F1Score,
FBetaScore,
# IoU
IoU,
BinaryIoU,
MeanIoU,
OneHotIoU,
OneHotMeanIoU,
}
ALL_OBJECTS_DICT = {cls.__name__: cls for cls in ALL_OBJECTS}
ALL_OBJECTS_DICT.update(
{to_snake_case(cls.__name__): cls for cls in ALL_OBJECTS}
)
# TODO: Align with `tf.keras` and set the name attribute of metrics
# with the key name. Currently it uses default name of class definitions.
ALL_OBJECTS_DICT.update(
{
"bce": BinaryCrossentropy,
"BCE": BinaryCrossentropy,
"mse": MeanSquaredError,
"MSE": MeanSquaredError,
"mae": MeanAbsoluteError,
"MAE": MeanAbsoluteError,
"mape": MeanAbsolutePercentageError,
"MAPE": MeanAbsolutePercentageError,
"msle": MeanSquaredLogarithmicError,
"MSLE": MeanSquaredLogarithmicError,
}
)
@keras_core_export("keras_core.metrics.serialize")
def serialize(metric):
"""Serializes metric function or `Metric` instance.
Args:
metric: A Keras `Metric` instance or a metric function.
Returns:
Metric configuration dictionary.
"""
return serialization_lib.serialize_keras_object(metric)
@keras_core_export("keras_core.metrics.deserialize")
def deserialize(config, custom_objects=None):
"""Deserializes a serialized metric class/function instance.
Args:
config: Metric configuration.
custom_objects: Optional dictionary mapping names (strings)
to custom objects (classes and functions) to be
considered during deserialization.
Returns:
A Keras `Metric` instance or a metric function.
"""
return serialization_lib.deserialize_keras_object(
config,
module_objects=ALL_OBJECTS_DICT,
custom_objects=custom_objects,
)
@keras_core_export("keras_core.metrics.get")
def get(identifier):
"""Retrieves a Keras metric as a `function`/`Metric` class instance.
The `identifier` may be the string name of a metric function or class.
>>> metric = metrics.get("categorical_crossentropy")
>>> type(metric)
<class 'function'>
>>> metric = metrics.get("CategoricalCrossentropy")
>>> type(metric)
<class '...metrics.CategoricalCrossentropy'>
You can also specify `config` of the metric to this function by passing dict
containing `class_name` and `config` as an identifier. Also note that the
`class_name` must map to a `Metric` class
>>> identifier = {"class_name": "CategoricalCrossentropy",
... "config": {"from_logits": True}}
>>> metric = metrics.get(identifier)
>>> type(metric)
<class '...metrics.CategoricalCrossentropy'>
Args:
identifier: A metric identifier. One of None or string name of a metric
function/class or metric configuration dictionary or a metric
function or a metric class instance
Returns:
A Keras metric as a `function`/ `Metric` class instance.
"""
if identifier is None:
return None
if isinstance(identifier, dict):
obj = deserialize(identifier)
elif isinstance(identifier, str):
obj = deserialize(identifier)
else:
obj = identifier
if callable(obj):
return obj
else:
raise ValueError(f"Could not interpret metric identifier: {identifier}")
|
keras-core/keras_core/metrics/__init__.py/0
|
{
"file_path": "keras-core/keras_core/metrics/__init__.py",
"repo_id": "keras-core",
"token_count": 2653
}
| 41 |
import numpy as np
from keras_core import testing
from keras_core.saving import saving_lib_test
class VariableMappingTest(testing.TestCase):
def test_basics(self):
model = saving_lib_test._get_basic_functional_model()
model.optimizer.build(model.trainable_variables)
variable_map = model._get_variable_map()
self.assertIn("first_dense/kernel", variable_map)
self.assertIn("second_dense/bias", variable_map)
self.assertIn("adam/learning_rate", variable_map)
model = saving_lib_test._get_basic_sequential_model()
model.build((None, 1))
model.optimizer.build(model.trainable_variables)
variable_map = model._get_variable_map()
self.assertIn("sequential/dense_1/bias", variable_map)
self.assertIn("adam/learning_rate", variable_map)
model = saving_lib_test._get_subclassed_model()
model(np.ones((1, 1)))
model.optimizer.build(model.trainable_variables)
variable_map = model._get_variable_map()
self.assertIn("custom_model_x/my_dense_1/dense/kernel", variable_map)
self.assertIn("custom_model_x/my_dense_1/my_dict_weight", variable_map)
self.assertIn(
"custom_model_x/my_dense_1/my_additional_weight", variable_map
)
self.assertIn("adam/learning_rate", variable_map)
|
keras-core/keras_core/models/variable_mapping_test.py/0
|
{
"file_path": "keras-core/keras_core/models/variable_mapping_test.py",
"repo_id": "keras-core",
"token_count": 568
}
| 42 |
from keras_core.optimizers.schedules.learning_rate_schedule import CosineDecay
from keras_core.optimizers.schedules.learning_rate_schedule import (
CosineDecayRestarts,
)
from keras_core.optimizers.schedules.learning_rate_schedule import (
ExponentialDecay,
)
from keras_core.optimizers.schedules.learning_rate_schedule import (
InverseTimeDecay,
)
from keras_core.optimizers.schedules.learning_rate_schedule import (
PiecewiseConstantDecay,
)
from keras_core.optimizers.schedules.learning_rate_schedule import (
PolynomialDecay,
)
|
keras-core/keras_core/optimizers/schedules/__init__.py/0
|
{
"file_path": "keras-core/keras_core/optimizers/schedules/__init__.py",
"repo_id": "keras-core",
"token_count": 196
}
| 43 |
import os
import unittest.mock as mock
import numpy as np
from absl import logging
from keras_core import layers
from keras_core.models import Sequential
from keras_core.saving import saving_api
from keras_core.testing import test_case
class SaveModelTests(test_case.TestCase):
def get_model(self):
return Sequential(
[
layers.Dense(5, input_shape=(3,)),
layers.Softmax(),
]
)
def test_basic_saving(self):
"""Test basic model saving and loading."""
model = self.get_model()
filepath = os.path.join(self.get_temp_dir(), "test_model.keras")
saving_api.save_model(model, filepath)
loaded_model = saving_api.load_model(filepath)
x = np.random.uniform(size=(10, 3))
self.assertTrue(np.allclose(model.predict(x), loaded_model.predict(x)))
def test_invalid_save_format(self):
"""Test deprecated save_format argument."""
model = self.get_model()
with self.assertRaisesRegex(
ValueError, "The `save_format` argument is deprecated"
):
saving_api.save_model(model, "model.txt", save_format=True)
def test_unsupported_arguments(self):
"""Test unsupported argument during model save."""
model = self.get_model()
filepath = os.path.join(self.get_temp_dir(), "test_model.keras")
with self.assertRaisesRegex(
ValueError, r"The following argument\(s\) are not supported"
):
saving_api.save_model(model, filepath, random_arg=True)
def test_save_h5_format(self):
"""Test saving model in h5 format."""
model = self.get_model()
filepath_h5 = os.path.join(self.get_temp_dir(), "test_model.h5")
saving_api.save_model(model, filepath_h5)
self.assertTrue(os.path.exists(filepath_h5))
os.remove(filepath_h5)
def test_save_unsupported_extension(self):
"""Test saving model with unsupported extension."""
model = self.get_model()
with self.assertRaisesRegex(
ValueError, "Invalid filepath extension for saving"
):
saving_api.save_model(model, "model.png")
class LoadModelTests(test_case.TestCase):
def get_model(self):
return Sequential(
[
layers.Dense(5, input_shape=(3,)),
layers.Softmax(),
]
)
def test_basic_load(self):
"""Test basic model loading."""
model = self.get_model()
filepath = os.path.join(self.get_temp_dir(), "test_model.keras")
saving_api.save_model(model, filepath)
loaded_model = saving_api.load_model(filepath)
x = np.random.uniform(size=(10, 3))
self.assertTrue(np.allclose(model.predict(x), loaded_model.predict(x)))
def test_load_unsupported_format(self):
"""Test loading model with unsupported format."""
with self.assertRaisesRegex(ValueError, "File format not supported"):
saving_api.load_model("model.pkl")
def test_load_keras_not_zip(self):
"""Test loading keras file that's not a zip."""
with self.assertRaisesRegex(ValueError, "File not found"):
saving_api.load_model("not_a_zip.keras")
def test_load_h5_format(self):
"""Test loading model in h5 format."""
model = self.get_model()
filepath_h5 = os.path.join(self.get_temp_dir(), "test_model.h5")
saving_api.save_model(model, filepath_h5)
loaded_model = saving_api.load_model(filepath_h5)
x = np.random.uniform(size=(10, 3))
self.assertTrue(np.allclose(model.predict(x), loaded_model.predict(x)))
os.remove(filepath_h5)
def test_load_model_with_custom_objects(self):
"""Test loading model with custom objects."""
class CustomLayer(layers.Layer):
def call(self, inputs):
return inputs
model = Sequential([CustomLayer(input_shape=(3,))])
filepath = os.path.join(self.get_temp_dir(), "custom_model.keras")
model.save(filepath)
loaded_model = saving_api.load_model(
filepath, custom_objects={"CustomLayer": CustomLayer}
)
self.assertIsInstance(loaded_model.layers[0], CustomLayer)
os.remove(filepath)
class LoadWeightsTests(test_case.TestCase):
def get_model(self):
return Sequential(
[
layers.Dense(5, input_shape=(3,)),
layers.Softmax(),
]
)
def test_load_keras_weights(self):
"""Test loading keras weights."""
model = self.get_model()
filepath = os.path.join(self.get_temp_dir(), "test_weights.weights.h5")
model.save_weights(filepath)
original_weights = model.get_weights()
model.load_weights(filepath)
loaded_weights = model.get_weights()
for orig, loaded in zip(original_weights, loaded_weights):
self.assertTrue(np.array_equal(orig, loaded))
def test_load_h5_weights_by_name(self):
"""Test loading h5 weights by name."""
model = self.get_model()
filepath = os.path.join(self.get_temp_dir(), "test_weights.weights.h5")
model.save_weights(filepath)
with self.assertRaisesRegex(ValueError, "Invalid keyword arguments"):
model.load_weights(filepath, by_name=True)
def test_load_weights_invalid_extension(self):
"""Test loading weights with unsupported extension."""
model = self.get_model()
with self.assertRaisesRegex(ValueError, "File format not supported"):
model.load_weights("invalid_extension.pkl")
class SaveModelTestsWarning(test_case.TestCase):
def get_model(self):
return Sequential(
[
layers.Dense(5, input_shape=(3,)),
layers.Softmax(),
]
)
def test_h5_deprecation_warning(self):
"""Test deprecation warning for h5 format."""
model = self.get_model()
filepath = os.path.join(self.get_temp_dir(), "test_model.h5")
with mock.patch.object(logging, "warning") as mock_warn:
saving_api.save_model(model, filepath)
mock_warn.assert_called_once_with(
"You are saving your model as an HDF5 file via `model.save()`. "
"This file format is considered legacy. "
"We recommend using instead the native Keras format, "
"e.g. `model.save('my_model.keras')`."
)
|
keras-core/keras_core/saving/saving_api_test.py/0
|
{
"file_path": "keras-core/keras_core/saving/saving_api_test.py",
"repo_id": "keras-core",
"token_count": 2921
}
| 44 |
import itertools
import tree
from keras_core.trainers.data_adapters.data_adapter import DataAdapter
class GeneratorDataAdapter(DataAdapter):
"""Adapter for Python generators."""
def __init__(self, generator):
data, generator = peek_and_restore(generator)
self.generator = generator
self._output_signature = None
if not isinstance(data, tuple):
raise ValueError(
"When passing a Python generator to a Keras model, "
"the generator must return a tuple, either "
"(input,) or (inputs, targets) or "
"(inputs, targets, sample_weights). "
f"Received: {data}"
)
def _set_tf_output_signature(self):
from keras_core.utils.module_utils import tensorflow as tf
data, generator = peek_and_restore(self.generator)
self.generator = generator
def get_tensor_spec(x):
shape = x.shape
if len(shape) < 1:
raise ValueError(
"When passing a Python generator to a Keras model, "
"the arrays returned by the generator "
"must be at least rank 1. Received: "
f"{x} of rank {len(x.shape)}"
)
shape = list(shape)
shape[0] = None # The batch size is not guaranteed to be static.
return tf.TensorSpec(shape=shape, dtype=x.dtype.name)
self._output_signature = tree.map_structure(get_tensor_spec, data)
def get_numpy_iterator(self):
for batch in self.generator:
yield batch
def get_tf_dataset(self):
from keras_core.utils.module_utils import tensorflow as tf
if self._output_signature is None:
self._set_tf_output_signature()
ds = tf.data.Dataset.from_generator(
self.get_numpy_iterator,
output_signature=self._output_signature,
)
ds = ds.prefetch(tf.data.AUTOTUNE)
return ds
@property
def num_batches(self):
return None
@property
def batch_size(self):
return None
def peek_and_restore(generator):
element = next(generator)
return element, itertools.chain([element], generator)
|
keras-core/keras_core/trainers/data_adapters/generator_data_adapter.py/0
|
{
"file_path": "keras-core/keras_core/trainers/data_adapters/generator_data_adapter.py",
"repo_id": "keras-core",
"token_count": 1045
}
| 45 |
import sys
from keras_core import backend as backend_module
from keras_core.backend.common import global_state
def in_tf_graph():
if global_state.get_global_attribute("in_tf_graph_scope", False):
return True
if "tensorflow" in sys.modules:
from keras_core.utils.module_utils import tensorflow as tf
return not tf.executing_eagerly()
return False
class TFGraphScope:
def __init__(self):
self._original_value = global_state.get_global_attribute(
"in_tf_graph_scope", False
)
def __enter__(self):
global_state.set_global_attribute("in_tf_graph_scope", True)
def __exit__(self, *args, **kwargs):
global_state.set_global_attribute(
"in_tf_graph_scope", self._original_value
)
class DynamicBackend:
"""A class that can be used to switch from one backend to another.
Usage:
```python
backend = DynamicBackend("tensorflow")
y = backend.square(tf.constant(...))
backend.set_backend("jax")
y = backend.square(jax.numpy.array(...))
```
Args:
backend: Initial backend to use (string).
"""
def __init__(self, backend=None):
self._backend = backend or backend_module.backend()
def set_backend(self, backend):
self._backend = backend
def reset(self):
self._backend = backend_module.backend()
def __getattr__(self, name):
if self._backend == "tensorflow":
from keras_core.backend import tensorflow as tf_backend
return getattr(tf_backend, name)
if self._backend == "jax":
from keras_core.backend import jax as jax_backend
return getattr(jax_backend, name)
if self._backend == "torch":
from keras_core.backend import torch as torch_backend
return getattr(torch_backend, name)
if self._backend == "numpy":
# TODO (ariG23498):
# The import `from keras_core.backend import numpy as numpy_backend`
# is not working. This is a temporary fix.
# The import is redirected to `keras_core.backend.numpy.numpy.py`
from keras_core import backend as numpy_backend
return getattr(numpy_backend, name)
|
keras-core/keras_core/utils/backend_utils.py/0
|
{
"file_path": "keras-core/keras_core/utils/backend_utils.py",
"repo_id": "keras-core",
"token_count": 961
}
| 46 |
# Copyright 2022 The KerasCV Authors
#
# Licensed under the Apache License, Version 2.0 (the "License");
# you may not use this file except in compliance with the License.
# You may obtain a copy of the License at
#
# https://www.apache.org/licenses/LICENSE-2.0
#
# Unless required by applicable law or agreed to in writing, software
# distributed under the License is distributed on an "AS IS" BASIS,
# WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.
# See the License for the specific language governing permissions and
# limitations under the License.
"""
Title: Train a Semantic Segmentation Model on Pascal VOC 2012 using KerasCV
Author: [tanzhenyu](https://github.com/tanzhenyu)
Date created: 2022/10/25
Last modified: 2022/10/25
Description: Use KerasCV to train a DeepLabV3 on Pascal VOC 2012.
"""
import sys
import tensorflow as tf
from absl import flags
from absl import logging
from tensorflow import keras
from keras_cv import models
from keras_cv.datasets.pascal_voc.segmentation import load
flags.DEFINE_string(
"weights_path",
"weights_{epoch:02d}.h5",
"Directory which will be used to store weight checkpoints.",
)
flags.DEFINE_boolean(
"mixed_precision",
True,
"whether to use FP16 mixed precision for training.",
)
flags.DEFINE_string(
"tensorboard_path",
"logs",
"Directory which will be used to store tensorboard logs.",
)
flags.DEFINE_integer(
"epochs",
100,
"Number of epochs to run for.",
)
flags.DEFINE_string(
"model_name",
None,
"The model name to be trained",
)
flags.DEFINE_string(
"model_kwargs",
"{}",
"Keyword argument dictionary to pass to the constructor of the model being"
" trained",
)
FLAGS = flags.FLAGS
FLAGS(sys.argv)
if FLAGS.mixed_precision:
logging.info("mixed precision training enabled")
keras.mixed_precision.set_global_policy("mixed_float16")
# Try to detect an available TPU. If none is present, defaults to
# MirroredStrategy
try:
tpu = tf.distribute.cluster_resolver.TPUClusterResolver.connect()
strategy = tf.distribute.TPUStrategy(tpu)
except ValueError:
# MirroredStrategy is best for a single machine with one or multiple GPUs
strategy = tf.distribute.MirroredStrategy()
print("Number of accelerators: ", strategy.num_replicas_in_sync)
# parameters from FasterRCNN [paper](https://arxiv.org/pdf/1506.01497.pdf)
local_batch = 4
global_batch = local_batch * strategy.num_replicas_in_sync
base_lr = 0.007 * global_batch / 16
# TODO(tanzhenyu): add a diff dataset.
# all_ds = load(split="sbd_train", data_dir=None)
# all_ds = all_ds.concatenate(load(split="sbd_eval", data_dir=None))
# train_ds = all_ds.take(10000)
# eval_ds = all_ds.skip(10000).concatenate(load(split="diff", data_dir=None))
train_ds = load(split="sbd_train", data_dir=None)
eval_ds = load(split="sbd_eval", data_dir=None)
resize_layer = keras.layers.Resizing(512, 512, interpolation="nearest")
image_size = [512, 512, 3]
# TODO(tanzhenyu): move to KPL.
def flip_fn(image, cls_seg):
if tf.random.uniform([], minval=0, maxval=1, dtype=tf.float32) > 0.5:
image = tf.image.flip_left_right(image)
cls_seg = tf.reverse(cls_seg, axis=[1])
return image, cls_seg
def proc_train_fn(examples):
image = examples.pop("image")
image = tf.cast(image, tf.float32)
image = resize_layer(image)
cls_seg = examples.pop("class_segmentation")
cls_seg = tf.cast(cls_seg, tf.float32)
cls_seg = resize_layer(cls_seg)
image, cls_seg = flip_fn(image, cls_seg)
cls_seg = tf.cast(cls_seg, tf.uint8)
sample_weight = tf.equal(cls_seg, 255)
zeros = tf.zeros_like(cls_seg)
cls_seg = tf.where(sample_weight, zeros, cls_seg)
return image, cls_seg
def proc_eval_fn(examples):
image = examples.pop("image")
image = tf.cast(image, tf.float32)
image = resize_layer(image)
cls_seg = examples.pop("class_segmentation")
cls_seg = tf.cast(cls_seg, tf.float32)
cls_seg = resize_layer(cls_seg)
cls_seg = tf.cast(cls_seg, tf.uint8)
sample_weight = tf.equal(cls_seg, 255)
zeros = tf.zeros_like(cls_seg)
cls_seg = tf.where(sample_weight, zeros, cls_seg)
return image, cls_seg
train_ds = train_ds.map(proc_train_fn, num_parallel_calls=tf.data.AUTOTUNE)
train_ds = train_ds.batch(global_batch, drop_remainder=True)
eval_ds = eval_ds.map(proc_eval_fn, num_parallel_calls=tf.data.AUTOTUNE)
eval_ds = eval_ds.batch(global_batch, drop_remainder=True)
train_ds = train_ds.shuffle(8)
train_ds = train_ds.prefetch(2)
with strategy.scope():
lr_decay = keras.optimizers.schedules.PiecewiseConstantDecay(
boundaries=[30000 * 16 / global_batch],
values=[base_lr, 0.1 * base_lr],
)
backbone = models.ResNet50V2(
include_rescaling=True,
# This argument gives a 2% mIoU increase
stackwise_dilations=[1, 1, 1, 2],
input_shape=(512, 512, 3),
include_top=False,
weights="imagenet",
)
model = models.__dict__[FLAGS.model_name]
model = model(num_classes=21, backbone=backbone, **eval(FLAGS.model_kwargs))
optimizer = keras.optimizers.SGD(
learning_rate=lr_decay, momentum=0.9, clipnorm=10.0
)
# ignore 255 as the class for semantic boundary.
loss_fn = keras.losses.SparseCategoricalCrossentropy(ignore_class=255)
metrics = [
keras.metrics.SparseCategoricalCrossentropy(ignore_class=255),
keras.metrics.MeanIoU(num_classes=21, sparse_y_pred=False),
keras.metrics.SparseCategoricalAccuracy(),
]
callbacks = [
keras.callbacks.ModelCheckpoint(
filepath=FLAGS.weights_path,
monitor="val_mean_io_u",
save_best_only=True,
save_weights_only=True,
),
keras.callbacks.TensorBoard(
log_dir=FLAGS.tensorboard_path, write_steps_per_second=True
),
]
model.compile(optimizer=optimizer, loss=loss_fn, metrics=metrics)
model.fit(
train_ds, epochs=FLAGS.epochs, validation_data=eval_ds, callbacks=callbacks
)
|
keras-cv/examples/training/semantic_segmentation/pascal_voc/basic_training.py/0
|
{
"file_path": "keras-cv/examples/training/semantic_segmentation/pascal_voc/basic_training.py",
"repo_id": "keras-cv",
"token_count": 2420
}
| 47 |
# Copyright 2023 The KerasCV Authors
#
# Licensed under the Apache License, Version 2.0 (the "License");
# you may not use this file except in compliance with the License.
# You may obtain a copy of the License at
#
# https://www.apache.org/licenses/LICENSE-2.0
#
# Unless required by applicable law or agreed to in writing, software
# distributed under the License is distributed on an "AS IS" BASIS,
# WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.
# See the License for the specific language governing permissions and
# limitations under the License.
"""Converter functions for working with bounding box formats."""
import tensorflow as tf
from keras_cv.api_export import keras_cv_export
from keras_cv.backend import keras
from keras_cv.backend import ops
from keras_cv.backend.scope import tf_data
# Internal exception to propagate the fact images was not passed to a converter
# that needs it.
class RequiresImagesException(Exception):
pass
ALL_AXES = 4
def _encode_box_to_deltas(
anchors,
boxes,
anchor_format: str,
box_format: str,
variance=None,
image_shape=None,
):
"""Converts bounding_boxes from `center_yxhw` to delta format."""
if variance is not None:
variance = ops.convert_to_tensor(variance, "float32")
var_len = variance.shape[-1]
if var_len != 4:
raise ValueError(f"`variance` must be length 4, got {variance}")
encoded_anchors = convert_format(
anchors,
source=anchor_format,
target="center_yxhw",
image_shape=image_shape,
)
boxes = convert_format(
boxes, source=box_format, target="center_yxhw", image_shape=image_shape
)
anchor_dimensions = ops.maximum(
encoded_anchors[..., 2:], keras.backend.epsilon()
)
box_dimensions = ops.maximum(boxes[..., 2:], keras.backend.epsilon())
# anchors be unbatched, boxes can either be batched or unbatched.
boxes_delta = ops.concatenate(
[
(boxes[..., :2] - encoded_anchors[..., :2]) / anchor_dimensions,
ops.log(box_dimensions / anchor_dimensions),
],
axis=-1,
)
if variance is not None:
boxes_delta /= variance
return boxes_delta
def _decode_deltas_to_boxes(
anchors,
boxes_delta,
anchor_format: str,
box_format: str,
variance=None,
image_shape=None,
):
"""Converts bounding_boxes from delta format to `center_yxhw`."""
if variance is not None:
variance = ops.convert_to_tensor(variance, "float32")
var_len = variance.shape[-1]
if var_len != 4:
raise ValueError(f"`variance` must be length 4, got {variance}")
def decode_single_level(anchor, box_delta):
encoded_anchor = convert_format(
anchor,
source=anchor_format,
target="center_yxhw",
image_shape=image_shape,
)
if variance is not None:
box_delta = box_delta * variance
# anchors be unbatched, boxes can either be batched or unbatched.
box = ops.concatenate(
[
box_delta[..., :2] * encoded_anchor[..., 2:]
+ encoded_anchor[..., :2],
ops.exp(box_delta[..., 2:]) * encoded_anchor[..., 2:],
],
axis=-1,
)
box = convert_format(
box,
source="center_yxhw",
target=box_format,
image_shape=image_shape,
)
return box
if isinstance(anchors, dict) and isinstance(boxes_delta, dict):
boxes = {}
for lvl, anchor in anchors.items():
boxes[lvl] = decode_single_level(anchor, boxes_delta[lvl])
return boxes
else:
return decode_single_level(anchors, boxes_delta)
def _center_yxhw_to_xyxy(boxes, images=None, image_shape=None):
y, x, height, width = ops.split(boxes, ALL_AXES, axis=-1)
return ops.concatenate(
[x - width / 2.0, y - height / 2.0, x + width / 2.0, y + height / 2.0],
axis=-1,
)
def _center_xywh_to_xyxy(boxes, images=None, image_shape=None):
x, y, width, height = ops.split(boxes, ALL_AXES, axis=-1)
return ops.concatenate(
[x - width / 2.0, y - height / 2.0, x + width / 2.0, y + height / 2.0],
axis=-1,
)
def _xywh_to_xyxy(boxes, images=None, image_shape=None):
x, y, width, height = ops.split(boxes, ALL_AXES, axis=-1)
return ops.concatenate([x, y, x + width, y + height], axis=-1)
def _xyxy_to_center_yxhw(boxes, images=None, image_shape=None):
left, top, right, bottom = ops.split(boxes, ALL_AXES, axis=-1)
return ops.concatenate(
[
(top + bottom) / 2.0,
(left + right) / 2.0,
bottom - top,
right - left,
],
axis=-1,
)
def _rel_xywh_to_xyxy(boxes, images=None, image_shape=None):
image_height, image_width = _image_shape(images, image_shape, boxes)
x, y, width, height = ops.split(boxes, ALL_AXES, axis=-1)
return ops.concatenate(
[
image_width * x,
image_height * y,
image_width * (x + width),
image_height * (y + height),
],
axis=-1,
)
def _xyxy_no_op(boxes, images=None, image_shape=None):
return boxes
def _xyxy_to_xywh(boxes, images=None, image_shape=None):
left, top, right, bottom = ops.split(boxes, ALL_AXES, axis=-1)
return ops.concatenate(
[left, top, right - left, bottom - top],
axis=-1,
)
def _xyxy_to_rel_xywh(boxes, images=None, image_shape=None):
image_height, image_width = _image_shape(images, image_shape, boxes)
left, top, right, bottom = ops.split(boxes, ALL_AXES, axis=-1)
left, right = (
left / image_width,
right / image_width,
)
top, bottom = top / image_height, bottom / image_height
return ops.concatenate(
[left, top, right - left, bottom - top],
axis=-1,
)
def _xyxy_to_center_xywh(boxes, images=None, image_shape=None):
left, top, right, bottom = ops.split(boxes, ALL_AXES, axis=-1)
return ops.concatenate(
[
(left + right) / 2.0,
(top + bottom) / 2.0,
right - left,
bottom - top,
],
axis=-1,
)
def _rel_xyxy_to_xyxy(boxes, images=None, image_shape=None):
image_height, image_width = _image_shape(images, image_shape, boxes)
left, top, right, bottom = ops.split(
boxes,
ALL_AXES,
axis=-1,
)
left, right = left * image_width, right * image_width
top, bottom = top * image_height, bottom * image_height
return ops.concatenate(
[left, top, right, bottom],
axis=-1,
)
def _xyxy_to_rel_xyxy(boxes, images=None, image_shape=None):
image_height, image_width = _image_shape(images, image_shape, boxes)
left, top, right, bottom = ops.split(
boxes,
ALL_AXES,
axis=-1,
)
left, right = left / image_width, right / image_width
top, bottom = top / image_height, bottom / image_height
return ops.concatenate(
[left, top, right, bottom],
axis=-1,
)
def _yxyx_to_xyxy(boxes, images=None, image_shape=None):
y1, x1, y2, x2 = ops.split(boxes, ALL_AXES, axis=-1)
return ops.concatenate([x1, y1, x2, y2], axis=-1)
def _rel_yxyx_to_xyxy(boxes, images=None, image_shape=None):
image_height, image_width = _image_shape(images, image_shape, boxes)
top, left, bottom, right = ops.split(
boxes,
ALL_AXES,
axis=-1,
)
left, right = left * image_width, right * image_width
top, bottom = top * image_height, bottom * image_height
return ops.concatenate(
[left, top, right, bottom],
axis=-1,
)
def _xyxy_to_yxyx(boxes, images=None, image_shape=None):
x1, y1, x2, y2 = ops.split(boxes, ALL_AXES, axis=-1)
return ops.concatenate([y1, x1, y2, x2], axis=-1)
def _xyxy_to_rel_yxyx(boxes, images=None, image_shape=None):
image_height, image_width = _image_shape(images, image_shape, boxes)
left, top, right, bottom = ops.split(boxes, ALL_AXES, axis=-1)
left, right = left / image_width, right / image_width
top, bottom = top / image_height, bottom / image_height
return ops.concatenate(
[top, left, bottom, right],
axis=-1,
)
TO_XYXY_CONVERTERS = {
"xywh": _xywh_to_xyxy,
"center_xywh": _center_xywh_to_xyxy,
"center_yxhw": _center_yxhw_to_xyxy,
"rel_xywh": _rel_xywh_to_xyxy,
"xyxy": _xyxy_no_op,
"rel_xyxy": _rel_xyxy_to_xyxy,
"yxyx": _yxyx_to_xyxy,
"rel_yxyx": _rel_yxyx_to_xyxy,
}
FROM_XYXY_CONVERTERS = {
"xywh": _xyxy_to_xywh,
"center_xywh": _xyxy_to_center_xywh,
"center_yxhw": _xyxy_to_center_yxhw,
"rel_xywh": _xyxy_to_rel_xywh,
"xyxy": _xyxy_no_op,
"rel_xyxy": _xyxy_to_rel_xyxy,
"yxyx": _xyxy_to_yxyx,
"rel_yxyx": _xyxy_to_rel_yxyx,
}
@keras_cv_export("keras_cv.bounding_box.convert_format")
@tf_data
def convert_format(
boxes, source, target, images=None, image_shape=None, dtype="float32"
):
f"""Converts bounding_boxes from one format to another.
Supported formats are:
- `"xyxy"`, also known as `corners` format. In this format the first four
axes represent `[left, top, right, bottom]` in that order.
- `"rel_xyxy"`. In this format, the axes are the same as `"xyxy"` but the x
coordinates are normalized using the image width, and the y axes the
image height. All values in `rel_xyxy` are in the range `(0, 1)`.
- `"xywh"`. In this format the first four axes represent
`[left, top, width, height]`.
- `"rel_xywh". In this format the first four axes represent
[left, top, width, height], just like `"xywh"`. Unlike `"xywh"`, the
values are in the range (0, 1) instead of absolute pixel values.
- `"center_xyWH"`. In this format the first two coordinates represent the x
and y coordinates of the center of the bounding box, while the last two
represent the width and height of the bounding box.
- `"center_yxHW"`. In this format the first two coordinates represent the y
and x coordinates of the center of the bounding box, while the last two
represent the height and width of the bounding box.
- `"yxyx"`. In this format the first four axes represent
[top, left, bottom, right] in that order.
- `"rel_yxyx"`. In this format, the axes are the same as `"yxyx"` but the x
coordinates are normalized using the image width, and the y axes the
image height. All values in `rel_yxyx` are in the range (0, 1).
Formats are case insensitive. It is recommended that you capitalize width
and height to maximize the visual difference between `"xyWH"` and `"xyxy"`.
Relative formats, abbreviated `rel`, make use of the shapes of the `images`
passed. In these formats, the coordinates, widths, and heights are all
specified as percentages of the host image. `images` may be a ragged
Tensor. Note that using a ragged Tensor for images may cause a substantial
performance loss, as each image will need to be processed separately due to
the mismatching image shapes.
Usage:
```python
boxes = load_coco_dataset()
boxes_in_xywh = keras_cv.bounding_box.convert_format(
boxes,
source='xyxy',
target='xyWH'
)
```
Args:
boxes: tensor representing bounding boxes in the format specified in
the `source` parameter. `boxes` can optionally have extra
dimensions stacked on the final axis to store metadata. boxes
should be a 3D tensor, with the shape `[batch_size, num_boxes, 4]`.
Alternatively, boxes can be a dictionary with key 'boxes' containing
a tensor matching the aforementioned spec.
source:One of {" ".join([f'"{f}"' for f in TO_XYXY_CONVERTERS.keys()])}.
Used to specify the original format of the `boxes` parameter.
target:One of {" ".join([f'"{f}"' for f in TO_XYXY_CONVERTERS.keys()])}.
Used to specify the destination format of the `boxes` parameter.
images: (Optional) a batch of images aligned with `boxes` on the first
axis. Should be at least 3 dimensions, with the first 3 dimensions
representing: `[batch_size, height, width]`. Used in some
converters to compute relative pixel values of the bounding box
dimensions. Required when transforming from a rel format to a
non-rel format.
dtype: the data type to use when transforming the boxes, defaults to
`"float32"`.
"""
if isinstance(boxes, dict):
converted_boxes = boxes.copy()
converted_boxes["boxes"] = convert_format(
boxes["boxes"],
source=source,
target=target,
images=images,
image_shape=image_shape,
dtype=dtype,
)
return converted_boxes
if boxes.shape[-1] is not None and boxes.shape[-1] != 4:
raise ValueError(
"Expected `boxes` to be a Tensor with a final dimension of "
f"`4`. Instead, got `boxes.shape={boxes.shape}`."
)
if images is not None and image_shape is not None:
raise ValueError(
"convert_format() expects either `images` or `image_shape`, but "
f"not both. Received images={images} image_shape={image_shape}"
)
_validate_image_shape(image_shape)
source = source.lower()
target = target.lower()
if source not in TO_XYXY_CONVERTERS:
raise ValueError(
"`convert_format()` received an unsupported format for the "
"argument `source`. `source` should be one of "
f"{TO_XYXY_CONVERTERS.keys()}. Got source={source}"
)
if target not in FROM_XYXY_CONVERTERS:
raise ValueError(
"`convert_format()` received an unsupported format for the "
"argument `target`. `target` should be one of "
f"{FROM_XYXY_CONVERTERS.keys()}. Got target={target}"
)
boxes = ops.cast(boxes, dtype)
if source == target:
return boxes
# rel->rel conversions should not require images
if source.startswith("rel") and target.startswith("rel"):
source = source.replace("rel_", "", 1)
target = target.replace("rel_", "", 1)
boxes, images, squeeze = _format_inputs(boxes, images)
to_xyxy_fn = TO_XYXY_CONVERTERS[source]
from_xyxy_fn = FROM_XYXY_CONVERTERS[target]
try:
in_xyxy = to_xyxy_fn(boxes, images=images, image_shape=image_shape)
result = from_xyxy_fn(in_xyxy, images=images, image_shape=image_shape)
except RequiresImagesException:
raise ValueError(
"convert_format() must receive `images` or `image_shape` when "
"transforming between relative and absolute formats."
f"convert_format() received source=`{format}`, target=`{format}, "
f"but images={images} and image_shape={image_shape}."
)
return _format_outputs(result, squeeze)
def _format_inputs(boxes, images):
boxes_rank = len(boxes.shape)
if boxes_rank > 3:
raise ValueError(
"Expected len(boxes.shape)=2, or len(boxes.shape)=3, got "
f"len(boxes.shape)={boxes_rank}"
)
boxes_includes_batch = boxes_rank == 3
# Determine if images needs an expand_dims() call
if images is not None:
images_rank = len(images.shape)
if images_rank > 4:
raise ValueError(
"Expected len(images.shape)=2, or len(images.shape)=3, got "
f"len(images.shape)={images_rank}"
)
images_include_batch = images_rank == 4
if boxes_includes_batch != images_include_batch:
raise ValueError(
"convert_format() expects both boxes and images to be batched, "
"or both boxes and images to be unbatched. Received "
f"len(boxes.shape)={boxes_rank}, "
f"len(images.shape)={images_rank}. Expected either "
"len(boxes.shape)=2 AND len(images.shape)=3, or "
"len(boxes.shape)=3 AND len(images.shape)=4."
)
if not images_include_batch:
images = ops.expand_dims(images, axis=0)
if not boxes_includes_batch:
return ops.expand_dims(boxes, axis=0), images, True
return boxes, images, False
def _validate_image_shape(image_shape):
# Escape early if image_shape is None and skip validation.
if image_shape is None:
return
# tuple/list
if isinstance(image_shape, (tuple, list)):
if len(image_shape) != 3:
raise ValueError(
"image_shape should be of length 3, but got "
f"image_shape={image_shape}"
)
return
# tensor
if ops.is_tensor(image_shape):
if len(image_shape.shape) > 1:
raise ValueError(
"image_shape.shape should be (3), but got "
f"image_shape.shape={image_shape.shape}"
)
if image_shape.shape[0] != 3:
raise ValueError(
"image_shape.shape should be (3), but got "
f"image_shape.shape={image_shape.shape}"
)
return
# Warn about failure cases
raise ValueError(
"Expected image_shape to be either a tuple, list, Tensor. "
f"Received image_shape={image_shape}"
)
def _format_outputs(boxes, squeeze):
if squeeze:
return ops.squeeze(boxes, axis=0)
return boxes
def _image_shape(images, image_shape, boxes):
if images is None and image_shape is None:
raise RequiresImagesException()
if image_shape is None:
if not isinstance(images, tf.RaggedTensor):
image_shape = ops.shape(images)
height, width = image_shape[1], image_shape[2]
else:
height = ops.reshape(images.row_lengths(), (-1, 1))
width = ops.reshape(ops.max(images.row_lengths(axis=2), 1), (-1, 1))
height = ops.expand_dims(height, axis=-1)
width = ops.expand_dims(width, axis=-1)
else:
height, width = image_shape[0], image_shape[1]
return ops.cast(height, boxes.dtype), ops.cast(width, boxes.dtype)
|
keras-cv/keras_cv/bounding_box/converters.py/0
|
{
"file_path": "keras-cv/keras_cv/bounding_box/converters.py",
"repo_id": "keras-cv",
"token_count": 7974
}
| 48 |
# Copyright 2022 The KerasCV Authors
#
# Licensed under the Apache License, Version 2.0 (the "License");
# you may not use this file except in compliance with the License.
# You may obtain a copy of the License at
#
# https://www.apache.org/licenses/LICENSE-2.0
#
# Unless required by applicable law or agreed to in writing, software
# distributed under the License is distributed on an "AS IS" BASIS,
# WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.
# See the License for the specific language governing permissions and
# limitations under the License.
import numpy as np
from keras_cv import bounding_box
from keras_cv.tests.test_case import TestCase
class ValidateTest(TestCase):
def test_raises_nondict(self):
with self.assertRaisesRegex(
ValueError, "Expected `bounding_boxes` to be a dictionary, got "
):
bounding_box.validate_format(np.ones((4, 3, 6)))
def test_mismatch_dimensions(self):
with self.assertRaisesRegex(
ValueError,
"Expected `boxes` and `classes` to have matching dimensions",
):
bounding_box.validate_format(
{"boxes": np.ones((4, 3, 6)), "classes": np.ones((4, 6))}
)
def test_bad_keys(self):
with self.assertRaisesRegex(ValueError, "containing keys"):
bounding_box.validate_format(
{
"box": [
1,
2,
3,
],
"class": [1234],
}
)
|
keras-cv/keras_cv/bounding_box/validate_format_test.py/0
|
{
"file_path": "keras-cv/keras_cv/bounding_box/validate_format_test.py",
"repo_id": "keras-cv",
"token_count": 716
}
| 49 |
# Copyright 2022 The KerasCV Authors
#
# Licensed under the Apache License, Version 2.0 (the "License");
# you may not use this file except in compliance with the License.
# You may obtain a copy of the License at
#
# https://www.apache.org/licenses/LICENSE-2.0
#
# Unless required by applicable law or agreed to in writing, software
# distributed under the License is distributed on an "AS IS" BASIS,
# WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.
# See the License for the specific language governing permissions and
# limitations under the License.
"""
formats.py contains axis information for each supported format.
"""
from keras_cv.api_export import keras_cv_export
@keras_cv_export("keras_cv.keypoint.XY")
class XY:
"""XY contains axis indices for the XY format.
All values in the XY format should be absolute pixel values.
The XY format consists of the following required indices:
- X: the width position
- Y: the height position
and the following optional indices, used in some KerasCV components:
- CLASS: class of the keypoints
- CONFIDENCE: confidence of the keypoints
"""
X = 0
Y = 1
CLASS = 2
CONFIDENCE = 3
@keras_cv_export("keras_cv.keypoint.REL_XY")
class REL_XY:
"""REL_XY contains axis indices for the REL_XY format.
REL_XY is like XY, but each value is relative to the width and height of the
origin image. Values are percentages of the origin images' width and height
respectively.
The REL_XY format consists of the following required indices:
- X: the width position
- Y: the height position
and the following optional indices, used in some KerasCV components:
- CLASS: class of the keypoints
- CONFIDENCE: confidence of the keypoints
"""
X = 0
Y = 1
CLASS = 2
CONFIDENCE = 3
|
keras-cv/keras_cv/keypoint/formats.py/0
|
{
"file_path": "keras-cv/keras_cv/keypoint/formats.py",
"repo_id": "keras-cv",
"token_count": 566
}
| 50 |
# Copyright 2022 The KerasCV Authors
#
# Licensed under the Apache License, Version 2.0 (the "License");
# you may not use this file except in compliance with the License.
# You may obtain a copy of the License at
#
# https://www.apache.org/licenses/LICENSE-2.0
#
# Unless required by applicable law or agreed to in writing, software
# distributed under the License is distributed on an "AS IS" BASIS,
# WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.
# See the License for the specific language governing permissions and
# limitations under the License.
from typing import List
from keras_cv.api_export import keras_cv_export
from keras_cv.backend import keras
from keras_cv.backend import ops
@keras_cv_export("keras_cv.layers.BoxMatcher")
class BoxMatcher(keras.layers.Layer):
"""Box matching logic based on argmax of highest value (e.g., IOU).
This class computes matches from a similarity matrix. Each row will be
matched to at least one column, the matched result can either be positive
/ negative, or simply ignored depending on the setting.
The settings include `thresholds` and `match_values`, for example if:
1) thresholds=[negative_threshold, positive_threshold], and
match_values=[negative_value=0, ignore_value=-1, positive_value=1]: the
rows will be assigned to positive_value if its argmax result >=
positive_threshold; the rows will be assigned to negative_value if its
argmax result < negative_threshold, and the rows will be assigned to
ignore_value if its argmax result is between [negative_threshold,
positive_threshold).
2) thresholds=[negative_threshold, positive_threshold], and
match_values=[ignore_value=-1, negative_value=0, positive_value=1]: the
rows will be assigned to positive_value if its argmax result >=
positive_threshold; the rows will be assigned to ignore_value if its
argmax result < negative_threshold, and the rows will be assigned to
negative_value if its argmax result is between [negative_threshold,
positive_threshold). This is different from case 1) by swapping first two
values.
3) thresholds=[positive_threshold], and
match_values=[negative_values, positive_value]: the rows will be assigned
to positive value if its argmax result >= positive_threshold; the rows
will be assigned to negative_value if its argmax result <
negative_threshold.
Args:
thresholds: A sorted list of floats to classify the matches into
different results (e.g. positive or negative or ignored match). The
list will be prepended with -Inf and and appended with +Inf.
match_values: A list of integers representing matched results (e.g.
positive or negative or ignored match). len(`match_values`) must
equal to len(`thresholds`) + 1.
force_match_for_each_col: each row will be argmax matched to at
least one column. This means some columns will be matched to
multiple rows while some columns will not be matched to any rows.
Filtering by `thresholds` will make less columns match to positive
result. Setting this to True guarantees that each column will be
matched to positive result to at least one row.
Raises:
ValueError: if `thresholds` not sorted or
len(`match_values`) != len(`thresholds`) + 1
Usage:
```python
box_matcher = keras_cv.layers.BoxMatcher([0.3, 0.7], [-1, 0, 1])
iou_metric = keras_cv.bounding_box.compute_iou(anchors, boxes)
matched_columns, matched_match_values = box_matcher(iou_metric)
cls_mask = ops.less_equal(matched_match_values, 0)
```
TODO(tanzhenyu): document when to use which mode.
"""
def __init__(
self,
thresholds: List[float],
match_values: List[int],
force_match_for_each_col: bool = False,
**kwargs,
):
super().__init__(**kwargs)
if sorted(thresholds) != thresholds:
raise ValueError(f"`threshold` must be sorted, got {thresholds}")
self.match_values = match_values
if len(match_values) != len(thresholds) + 1:
raise ValueError(
f"len(`match_values`) must be len(`thresholds`) + 1, got "
f"match_values {match_values}, thresholds {thresholds}"
)
thresholds.insert(0, -float("inf"))
thresholds.append(float("inf"))
self.thresholds = thresholds
self.force_match_for_each_col = force_match_for_each_col
self.built = True
def call(self, similarity_matrix):
"""Matches each row to a column based on argmax
TODO(tanzhenyu): consider swapping rows and cols.
Args:
similarity_matrix: A float Tensor of shape [num_rows, num_cols] or
[batch_size, num_rows, num_cols] representing any similarity metric.
Returns:
matched_columns: An integer tensor of shape [num_rows] or [batch_size,
num_rows] storing the index of the matched colum for each row.
matched_values: An integer tensor of shape [num_rows] or [batch_size,
num_rows] storing the match result (positive match, negative match,
ignored match).
"""
squeeze_result = False
if len(similarity_matrix.shape) == 2:
squeeze_result = True
similarity_matrix = ops.expand_dims(similarity_matrix, axis=0)
static_shape = list(similarity_matrix.shape)
num_rows = static_shape[1] or ops.shape(similarity_matrix)[1]
batch_size = static_shape[0] or ops.shape(similarity_matrix)[0]
def _match_when_cols_are_empty():
"""Performs matching when the rows of similarity matrix are empty.
When the rows are empty, all detections are false positives. So we
return a tensor of -1's to indicate that the rows do not match to
any columns.
Returns:
matched_columns: An integer tensor of shape [batch_size,
num_rows] storing the index of the matched column for each
row.
matched_values: An integer tensor of shape [batch_size,
num_rows] storing the match type indicator (e.g. positive or
negative or ignored match).
"""
with ops.name_scope("empty_boxes"):
matched_columns = ops.zeros(
[batch_size, num_rows], dtype="int32"
)
matched_values = -ops.ones(
[batch_size, num_rows], dtype="int32"
)
return matched_columns, matched_values
def _match_when_cols_are_non_empty():
"""Performs matching when the rows of similarity matrix are
non-empty.
Returns:
matched_columns: An integer tensor of shape [batch_size,
num_rows] storing the index of the matched column for each
row.
matched_values: An integer tensor of shape [batch_size,
num_rows] storing the match type indicator (e.g. positive or
negative or ignored match).
"""
with ops.name_scope("non_empty_boxes"):
# Jax traces this function even when running eagerly and the
# columns are non-empty. Therefore, we need to handle the case
# where the similarity matrix is empty. We do this by padding
# some -1s to the end. -1s are guaranteed to not affect argmax
# matching because all values in a similarity matrix are [0,1]
# and the indexing won't change because these are added at the
# end.
padded_similarity_matrix = ops.concatenate(
[similarity_matrix, -ops.ones((batch_size, num_rows, 1))],
axis=-1,
)
matched_columns = ops.argmax(
padded_similarity_matrix,
axis=-1,
)
# Get logical indices of ignored and unmatched columns as int32
matched_vals = ops.max(padded_similarity_matrix, axis=-1)
matched_values = ops.zeros([batch_size, num_rows], "int32")
match_dtype = matched_vals.dtype
for ind, low, high in zip(
self.match_values, self.thresholds[:-1], self.thresholds[1:]
):
low_threshold = ops.cast(low, match_dtype)
high_threshold = ops.cast(high, match_dtype)
mask = ops.logical_and(
ops.greater_equal(matched_vals, low_threshold),
ops.less(matched_vals, high_threshold),
)
matched_values = self._set_values_using_indicator(
matched_values, mask, ind
)
if self.force_match_for_each_col:
# [batch_size, num_cols], for each column (groundtruth_box),
# find the best matching row (anchor).
matching_rows = ops.argmax(
padded_similarity_matrix,
axis=1,
)
# [batch_size, num_cols, num_rows], a transposed 0-1 mapping
# matrix M, where M[j, i] = 1 means column j is matched to
# row i.
column_to_row_match_mapping = ops.one_hot(
matching_rows, num_rows
)
# [batch_size, num_rows], for each row (anchor), find the
# matched column (groundtruth_box).
force_matched_columns = ops.argmax(
column_to_row_match_mapping,
axis=1,
)
# [batch_size, num_rows]
force_matched_column_mask = ops.cast(
ops.max(column_to_row_match_mapping, axis=1),
"bool",
)
# [batch_size, num_rows]
matched_columns = ops.where(
force_matched_column_mask,
force_matched_columns,
matched_columns,
)
matched_values = ops.where(
force_matched_column_mask,
self.match_values[-1]
* ops.ones([batch_size, num_rows], dtype="int32"),
matched_values,
)
return ops.cast(matched_columns, "int32"), matched_values
num_boxes = (
similarity_matrix.shape[-1] or ops.shape(similarity_matrix)[-1]
)
matched_columns, matched_values = ops.cond(
pred=ops.greater(num_boxes, 0),
true_fn=_match_when_cols_are_non_empty,
false_fn=_match_when_cols_are_empty,
)
if squeeze_result:
matched_columns = ops.squeeze(matched_columns, axis=0)
matched_values = ops.squeeze(matched_values, axis=0)
return matched_columns, matched_values
def _set_values_using_indicator(self, x, indicator, val):
"""Set the indicated fields of x to val.
Args:
x: tensor.
indicator: boolean with same shape as x.
val: scalar with value to set.
Returns:
modified tensor.
"""
indicator = ops.cast(indicator, x.dtype)
return ops.add(ops.multiply(x, 1 - indicator), val * indicator)
def get_config(self):
config = {
"thresholds": self.thresholds[1:-1],
"match_values": self.match_values,
"force_match_for_each_col": self.force_match_for_each_col,
}
return config
|
keras-cv/keras_cv/layers/object_detection/box_matcher.py/0
|
{
"file_path": "keras-cv/keras_cv/layers/object_detection/box_matcher.py",
"repo_id": "keras-cv",
"token_count": 5538
}
| 51 |
# Copyright 2022 The KerasCV Authors
#
# Licensed under the Apache License, Version 2.0 (the "License");
# you may not use this file except in compliance with the License.
# You may obtain a copy of the License at
#
# https://www.apache.org/licenses/LICENSE-2.0
#
# Unless required by applicable law or agreed to in writing, software
# distributed under the License is distributed on an "AS IS" BASIS,
# WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.
# See the License for the specific language governing permissions and
# limitations under the License.
import pytest
import tensorflow as tf
from keras_cv.layers.object_detection.sampling import balanced_sample
from keras_cv.tests.test_case import TestCase
@pytest.mark.tf_keras_only
class BalancedSamplingTest(TestCase):
def test_balanced_sampling(self):
positive_matches = tf.constant(
[
True,
False,
False,
False,
False,
False,
False,
False,
False,
False,
]
)
negative_matches = tf.constant(
[False, True, True, True, True, True, True, True, True, True]
)
num_samples = 5
positive_fraction = 0.2
res = balanced_sample(
positive_matches, negative_matches, num_samples, positive_fraction
)
# The 1st element must be selected, given it's the only one.
self.assertAllClose(res[0], 1)
def test_balanced_batched_sampling(self):
positive_matches = tf.constant(
[
[
True,
False,
False,
False,
False,
False,
False,
False,
False,
False,
],
[
False,
False,
False,
False,
False,
False,
True,
False,
False,
False,
],
]
)
negative_matches = tf.constant(
[
[False, True, True, True, True, True, True, True, True, True],
[True, True, True, True, True, True, False, True, True, True],
]
)
num_samples = 5
positive_fraction = 0.2
res = balanced_sample(
positive_matches, negative_matches, num_samples, positive_fraction
)
# the 1st element from the 1st batch must be selected, given it's the
# only one
self.assertAllClose(res[0][0], 1)
# the 7th element from the 2nd batch must be selected, given it's the
# only one
self.assertAllClose(res[1][6], 1)
def test_balanced_sampling_over_positive_fraction(self):
positive_matches = tf.constant(
[
True,
False,
False,
False,
False,
False,
False,
False,
False,
False,
]
)
negative_matches = tf.constant(
[False, True, True, True, True, True, True, True, True, True]
)
num_samples = 5
positive_fraction = 0.4
res = balanced_sample(
positive_matches, negative_matches, num_samples, positive_fraction
)
# only 1 positive sample exists, thus it is chosen
self.assertAllClose(res[0], 1)
def test_balanced_sampling_under_positive_fraction(self):
positive_matches = tf.constant(
[
True,
False,
False,
False,
False,
False,
False,
False,
False,
False,
]
)
negative_matches = tf.constant(
[False, True, True, True, True, True, True, True, True, True]
)
num_samples = 5
positive_fraction = 0.1
res = balanced_sample(
positive_matches, negative_matches, num_samples, positive_fraction
)
# no positive is chosen
self.assertAllClose(res[0], 0)
self.assertAllClose(tf.reduce_sum(res), 5)
def test_balanced_sampling_over_num_samples(self):
positive_matches = tf.constant(
[
True,
False,
False,
False,
False,
False,
False,
False,
False,
False,
]
)
negative_matches = tf.constant(
[False, True, True, True, True, True, True, True, True, True]
)
# users want to get 20 samples, but only 10 are available
num_samples = 20
positive_fraction = 0.1
with self.assertRaisesRegex(ValueError, "has less element"):
_ = balanced_sample(
positive_matches,
negative_matches,
num_samples,
positive_fraction,
)
def test_balanced_sampling_no_positive(self):
positive_matches = tf.constant(
[
False,
False,
False,
False,
False,
False,
False,
False,
False,
False,
]
)
# the rest are neither positive nor negative, but ignored matches
negative_matches = tf.constant(
[False, False, True, False, False, True, False, False, True, False]
)
num_samples = 5
positive_fraction = 0.5
res = balanced_sample(
positive_matches, negative_matches, num_samples, positive_fraction
)
# given only 3 negative and 0 positive, select all of them
self.assertAllClose(res, [0, 0, 1, 0, 0, 1, 0, 0, 1, 0])
def test_balanced_sampling_no_negative(self):
positive_matches = tf.constant(
[True, True, False, False, False, False, False, False, False, False]
)
# 2-9 indices are neither positive nor negative, they're ignored matches
negative_matches = tf.constant([False] * 10)
num_samples = 5
positive_fraction = 0.5
res = balanced_sample(
positive_matches, negative_matches, num_samples, positive_fraction
)
# given only 2 positive and 0 negative, select all of them.
self.assertAllClose(res, [1, 1, 0, 0, 0, 0, 0, 0, 0, 0])
def test_balanced_sampling_many_samples(self):
positive_matches = tf.random.uniform(
[2, 1000], minval=0, maxval=1, dtype=tf.float32
)
positive_matches = positive_matches > 0.98
negative_matches = tf.logical_not(positive_matches)
num_samples = 256
positive_fraction = 0.25
_ = balanced_sample(
positive_matches, negative_matches, num_samples, positive_fraction
)
|
keras-cv/keras_cv/layers/object_detection/sampling_test.py/0
|
{
"file_path": "keras-cv/keras_cv/layers/object_detection/sampling_test.py",
"repo_id": "keras-cv",
"token_count": 3917
}
| 52 |
# Copyright 2023 The KerasCV Authors
#
# Licensed under the Apache License, Version 2.0 (the "License");
# you may not use this file except in compliance with the License.
# You may obtain a copy of the License at
#
# https://www.apache.org/licenses/LICENSE-2.0
#
# Unless required by applicable law or agreed to in writing, software
# distributed under the License is distributed on an "AS IS" BASIS,
# WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.
# See the License for the specific language governing permissions and
# limitations under the License.
import keras
import tensorflow as tf
import tree
if hasattr(keras, "src"):
keras_backend = keras.src.backend
else:
keras_backend = keras.backend
from keras_cv import bounding_box
from keras_cv.api_export import keras_cv_export
from keras_cv.backend import config
from keras_cv.backend import keras
from keras_cv.backend import ops
from keras_cv.backend import scope
from keras_cv.utils import preprocessing
# In order to support both unbatched and batched inputs, the horizontal
# and vertical axis is reverse indexed
H_AXIS = -3
W_AXIS = -2
IMAGES = "images"
LABELS = "labels"
TARGETS = "targets"
BOUNDING_BOXES = "bounding_boxes"
KEYPOINTS = "keypoints"
SEGMENTATION_MASKS = "segmentation_masks"
IS_DICT = "is_dict"
USE_TARGETS = "use_targets"
@keras_cv_export("keras_cv.layers.BaseImageAugmentationLayer")
class BaseImageAugmentationLayer(keras.layers.Layer):
"""Abstract base layer for image augmentation.
This layer contains base functionalities for preprocessing layers which
augment image related data, e.g. image and in the future, label and bounding
boxes. The subclasses could avoid making certain mistakes and reduce code
duplications.
This layer requires you to implement one method: `augment_image()`, which
augments one single image during the training. There are a few additional
methods that you can implement for added functionality on the layer:
`augment_label()`, which handles label augmentation if the layer supports
that.
`augment_bounding_boxes()`, which handles the bounding box augmentation, if
the layer supports that.
`get_random_transformation()`, which should produce a random transformation
setting. The transformation object, which could be of any type, will be
passed to `augment_image`, `augment_label` and `augment_bounding_boxes`, to
coordinate the randomness behaviour, e.g., in the RandomFlip layer, the
image and bounding_boxes should be changed in the same way.
The `call()` method supports two formats of inputs:
1. A single image tensor with shape (height, width, channels) or
(batch_size, height, width, channels)
2. A dict of tensors with any of the following keys (note that `"images"`
must be present):
* `"images"` - Image Tensor with shape (height, width, channels) or
(batch_size, height, width, channels)
* `"labels"` - One-hot encoded classification labels Tensor with shape
(num_classes) or (batch_size, num_classes)
* `"bounding_boxes"` - A dictionary with keys:
* `"boxes"` - Tensor with shape (num_boxes, 4) or (batch_size,
num_boxes, 4)
* `"classes"` - Tensor of class labels for boxes with shape (num_boxes,
num_classes) or (batch_size, num_boxes, num_classes).
Any other keys included in this dictionary will be ignored and unmodified
by an augmentation layer.
The output of the `call()` will be the same structure as the inputs.
The `call()` will unpack the inputs, forward to the correct function, and
pack the output back to the same structure as the inputs.
By default, the `call()` method leverages the `tf.vectorized_map()`
function. Auto-vectorization can be disabled by setting
`self.auto_vectorize = False` in your `__init__()` method. When disabled,
`call()` instead relies on `tf.map_fn()`. For example:
```python
class SubclassLayer(keras_cv.BaseImageAugmentationLayer):
def __init__(self):
super().__init__()
self.auto_vectorize = False
```
Example:
```python
class RandomContrast(keras_cv.BaseImageAugmentationLayer):
def __init__(self, factor=(0.5, 1.5), **kwargs):
super().__init__(**kwargs)
self._factor = factor
def augment_image(self, image, transformation):
random_factor = tf.random.uniform([], self._factor[0], self._factor[1])
mean = tf.math.reduced_mean(inputs, axis=-1, keep_dim=True)
return (inputs - mean) * random_factor + mean
```
Note that since the randomness is also a common functionality, this layer
also includes a keras_backend.RandomGenerator, which can be used to
produce the random numbers. The random number generator is stored in the
`self._random_generator` attribute.
"""
def __init__(self, seed=None, **kwargs):
if seed is not None:
self._random_generator = tf.random.Generator.from_seed(seed=seed)
else:
self._random_generator = tf.random.get_global_generator()
super().__init__(**kwargs)
self._allow_non_tensor_positional_args = True
self.built = True
self._convert_input_args = False
@property
def force_output_ragged_images(self):
"""Control whether to force outputting of ragged images."""
return getattr(self, "_force_output_ragged_images", False)
@force_output_ragged_images.setter
def force_output_ragged_images(self, force_output_ragged_images):
self._force_output_ragged_images = force_output_ragged_images
@property
def force_output_dense_images(self):
"""Control whether to force outputting of dense images."""
return getattr(self, "_force_output_dense_images", False)
@force_output_dense_images.setter
def force_output_dense_images(self, force_output_dense_images):
self._force_output_dense_images = force_output_dense_images
@property
def auto_vectorize(self):
"""Control whether automatic vectorization occurs.
By default, the `call()` method leverages the `tf.vectorized_map()`
function. Auto-vectorization can be disabled by setting
`self.auto_vectorize = False` in your `__init__()` method. When
disabled, `call()` instead relies on `tf.map_fn()`. For example:
```python
class SubclassLayer(BaseImageAugmentationLayer):
def __init__(self):
super().__init__()
self.auto_vectorize = False
```
"""
return getattr(self, "_auto_vectorize", True)
@auto_vectorize.setter
def auto_vectorize(self, auto_vectorize):
self._auto_vectorize = auto_vectorize
def compute_image_signature(self, images):
"""Computes the output image signature for the `augment_image()`
function.
Must be overridden to return tensors with different shapes than the
input images. By default, returns either a `tf.RaggedTensorSpec`
matching the input image spec, or a `tf.TensorSpec` matching the input
image spec.
"""
if self.force_output_dense_images:
return tf.TensorSpec(images.shape[1:], self.compute_dtype)
if self.force_output_ragged_images or isinstance(
images, tf.RaggedTensor
):
ragged_spec = tf.RaggedTensorSpec(
shape=images.shape[1:],
ragged_rank=1,
dtype=self.compute_dtype,
)
return ragged_spec
return tf.TensorSpec(images.shape[1:], self.compute_dtype)
# TODO(lukewood): promote to user facing API if needed
def _compute_bounding_box_signature(self, bounding_boxes):
return {
"boxes": tf.RaggedTensorSpec(
shape=[None, 4],
ragged_rank=1,
dtype=self.compute_dtype,
),
"classes": tf.RaggedTensorSpec(
shape=[None], dtype=self.compute_dtype
),
}
# TODO(lukewood): promote to user facing API if needed
def _compute_keypoints_signature(self, keypoints):
if isinstance(keypoints, tf.RaggedTensor):
ragged_spec = tf.RaggedTensorSpec(
shape=keypoints.shape[1:],
ragged_rank=1,
dtype=self.compute_dtype,
)
return ragged_spec
return tf.TensorSpec(
shape=keypoints.shape[1:],
dtype=self.compute_dtype,
)
# TODO(lukewood): promote to user facing API if needed
def _compute_target_signature(self, targets):
return tf.TensorSpec(targets.shape[1:], self.compute_dtype)
def _compute_output_signature(self, inputs):
fn_output_signature = {
IMAGES: self.compute_image_signature(inputs[IMAGES])
}
bounding_boxes = inputs.get(BOUNDING_BOXES, None)
if bounding_boxes is not None:
fn_output_signature[BOUNDING_BOXES] = (
self._compute_bounding_box_signature(bounding_boxes)
)
segmentation_masks = inputs.get(SEGMENTATION_MASKS, None)
if segmentation_masks is not None:
fn_output_signature[SEGMENTATION_MASKS] = (
self.compute_image_signature(segmentation_masks)
)
keypoints = inputs.get(KEYPOINTS, None)
if keypoints is not None:
fn_output_signature[KEYPOINTS] = self._compute_keypoints_signature(
keypoints
)
labels = inputs.get(LABELS, None)
if labels is not None:
fn_output_signature[LABELS] = self._compute_target_signature(labels)
return fn_output_signature
@staticmethod
def _any_ragged(inputs):
if isinstance(inputs[IMAGES], tf.RaggedTensor):
return True
if BOUNDING_BOXES in inputs:
return True
if KEYPOINTS in inputs:
return True
return False
def _map_fn(self, func, inputs):
"""Returns either tf.map_fn or tf.vectorized_map based on the provided
inputs.
Args:
inputs: dictionary of inputs provided to map_fn.
"""
if self._any_ragged(inputs) or self.force_output_ragged_images:
return tf.map_fn(
func,
inputs,
fn_output_signature=self._compute_output_signature(inputs),
)
if self.auto_vectorize:
return tf.vectorized_map(func, inputs)
return tf.map_fn(func, inputs)
def augment_image(self, image, transformation, **kwargs):
"""Augment a single image during training.
Args:
image: 3D image input tensor to the layer. Forwarded from
`layer.call()`.
transformation: The transformation object produced by
`get_random_transformation`. Used to coordinate the randomness
between image, label, bounding box, keypoints, and segmentation
mask.
Returns:
output 3D tensor, which will be forward to `layer.call()`.
"""
raise NotImplementedError()
def augment_label(self, label, transformation, **kwargs):
"""Augment a single label during training.
Args:
label: 1D label to the layer. Forwarded from `layer.call()`.
transformation: The transformation object produced by
`get_random_transformation`. Used to coordinate the randomness
between image, label, bounding box, keypoints, and segmentation
mask.
Returns:
output 1D tensor, which will be forward to `layer.call()`.
"""
raise NotImplementedError()
def augment_target(self, target, transformation, **kwargs):
"""Augment a single target during training.
Args:
target: 1D label to the layer. Forwarded from `layer.call()`.
transformation: The transformation object produced by
`get_random_transformation`. Used to coordinate the randomness
between image, label, bounding box, keypoints, and segmentation
mask.
Returns:
output 1D tensor, which will be forward to `layer.call()`.
"""
return self.augment_label(target, transformation)
def augment_bounding_boxes(self, bounding_boxes, transformation, **kwargs):
"""Augment bounding boxes for one image during training.
Args:
bounding_boxes: 2D bounding boxes to the layer. Forwarded from
`call()`.
transformation: The transformation object produced by
`get_random_transformation`. Used to coordinate the randomness
between image, label, bounding box, keypoints, and segmentation
mask.
Returns:
output 2D tensor, which will be forward to `layer.call()`.
"""
raise NotImplementedError()
def augment_keypoints(self, keypoints, transformation, **kwargs):
"""Augment keypoints for one image during training.
Args:
keypoints: 2D keypoints input tensor to the layer. Forwarded from
`layer.call()`.
transformation: The transformation object produced by
`get_random_transformation`. Used to coordinate the randomness
between image, label, bounding box, keypoints, and segmentation
mask.
Returns:
output 2D tensor, which will be forward to `layer.call()`.
"""
raise NotImplementedError()
def augment_segmentation_mask(
self, segmentation_mask, transformation, **kwargs
):
"""Augment a single image's segmentation mask during training.
Args:
segmentation_mask: 3D segmentation mask input tensor to the layer.
This should generally have the shape [H, W, 1], or in some cases
[H, W, C] for multilabeled data. Forwarded from `layer.call()`.
transformation: The transformation object produced by
`get_random_transformation`. Used to coordinate the randomness
between image, label, bounding box, keypoints, and segmentation
mask.
Returns:
output 3D tensor containing the augmented segmentation mask, which
will be forward to `layer.call()`.
"""
raise NotImplementedError()
def get_random_transformation(
self,
image=None,
label=None,
bounding_boxes=None,
keypoints=None,
segmentation_mask=None,
):
"""Produce random transformation config for one single input.
This is used to produce same randomness between
image/label/bounding_box.
Args:
image: 3D image tensor from inputs.
label: optional 1D label tensor from inputs.
bounding_boxes: optional 2D bounding boxes tensor from inputs.
segmentation_mask: optional 3D segmentation mask tensor from inputs.
Returns:
Any type of object, which will be forwarded to `augment_image`,
`augment_label` and `augment_bounding_box` as the `transformation`
parameter.
"""
return None
def call(self, inputs):
# try to convert a given backend native tensor to TensorFlow tensor
# before passing it over to TFDataScope
is_tf_backend = config.backend() == "tensorflow"
is_in_tf_graph = not tf.executing_eagerly()
contains_ragged = lambda y: any(
tree.map_structure(
lambda x: isinstance(x, (tf.RaggedTensor, tf.SparseTensor)),
tree.flatten(y),
)
)
inputs_contain_ragged = contains_ragged(inputs)
if not is_tf_backend and not inputs_contain_ragged:
inputs = tree.map_structure(
lambda x: tf.convert_to_tensor(x), inputs
)
with scope.TFDataScope():
inputs = self._ensure_inputs_are_compute_dtype(inputs)
inputs, metadata = self._format_inputs(inputs)
images = inputs[IMAGES]
if images.shape.rank == 3:
outputs = self._format_output(self._augment(inputs), metadata)
elif images.shape.rank == 4:
outputs = self._format_output(
self._batch_augment(inputs), metadata
)
else:
raise ValueError(
"Image augmentation layers are expecting inputs to be "
"rank 3 (HWC) or 4D (NHWC) tensors. Got shape: "
f"{images.shape}"
)
# convert the outputs to backend native tensors if none of them
# contain RaggedTensors. Note that if the user passed in Raggeds
# but the outputs are dense, we still don't want to convert to
# backend native tensors. This is to avoid breaking TF data
# pipelines that can't easily be ported to become backend
# agnostic.
# Skip this step for TF backend or if in `tf.graph` like `tf.data`.
if not is_tf_backend and not is_in_tf_graph:
if not inputs_contain_ragged and not contains_ragged(outputs):
outputs = tree.map_structure(
# some layers return None, handle that case when
# converting to tensors
lambda x: ops.convert_to_tensor(x) if x is not None else x,
outputs,
)
return outputs
def _augment(self, inputs):
raw_image = inputs.get(IMAGES, None)
image = raw_image
label = inputs.get(LABELS, None)
bounding_boxes = inputs.get(BOUNDING_BOXES, None)
keypoints = inputs.get(KEYPOINTS, None)
segmentation_mask = inputs.get(SEGMENTATION_MASKS, None)
image_ragged = isinstance(image, tf.RaggedTensor)
# At this point, the tensor is not actually ragged as we have mapped
# over the batch axis. This call is required to make `tf.shape()` behave
# as users subclassing the layer expect.
if image_ragged:
image = image.to_tensor()
transformation = self.get_random_transformation(
image=image,
label=label,
bounding_boxes=bounding_boxes,
keypoints=keypoints,
segmentation_mask=segmentation_mask,
)
image = self.augment_image(
image,
transformation=transformation,
bounding_boxes=bounding_boxes,
label=label,
)
if (
image_ragged and not self.force_output_dense_images
) or self.force_output_ragged_images:
image = tf.RaggedTensor.from_tensor(image)
result = {IMAGES: image}
if label is not None:
label = self.augment_target(
label,
transformation=transformation,
bounding_boxes=bounding_boxes,
image=image,
)
result[LABELS] = label
if bounding_boxes is not None:
bounding_boxes = bounding_box.to_dense(bounding_boxes)
bounding_boxes = self.augment_bounding_boxes(
bounding_boxes,
transformation=transformation,
label=label,
image=raw_image,
)
bounding_boxes = bounding_box.to_ragged(
bounding_boxes, dtype=self.compute_dtype
)
result[BOUNDING_BOXES] = bounding_boxes
if keypoints is not None:
keypoints = self.augment_keypoints(
keypoints,
transformation=transformation,
label=label,
bounding_boxes=bounding_boxes,
image=image,
)
result[KEYPOINTS] = keypoints
if segmentation_mask is not None:
segmentation_mask = self.augment_segmentation_mask(
segmentation_mask,
transformation=transformation,
)
result[SEGMENTATION_MASKS] = segmentation_mask
# preserve any additional inputs unmodified by this layer.
for key in inputs.keys() - result.keys():
result[key] = inputs[key]
return result
def _batch_augment(self, inputs):
return self._map_fn(self._augment, inputs)
def _format_inputs(self, inputs):
metadata = {IS_DICT: True, USE_TARGETS: False}
if tf.is_tensor(inputs):
# single image input tensor
metadata[IS_DICT] = False
inputs = {IMAGES: inputs}
return inputs, metadata
if not isinstance(inputs, dict):
raise ValueError(
"Expect the inputs to be image tensor or dict. Got "
f"inputs={inputs} of type {type(inputs)}"
)
if BOUNDING_BOXES in inputs:
inputs[BOUNDING_BOXES] = self._format_bounding_boxes(
inputs[BOUNDING_BOXES]
)
if isinstance(inputs, dict) and TARGETS in inputs:
# TODO(scottzhu): Check if it only contains the valid keys
inputs[LABELS] = inputs[TARGETS]
del inputs[TARGETS]
metadata[USE_TARGETS] = True
return inputs, metadata
return inputs, metadata
def _format_bounding_boxes(self, bounding_boxes):
# We can't catch the case where this is None, sometimes RaggedTensor
# drops this dimension
if "classes" not in bounding_boxes:
raise ValueError(
"Bounding boxes are missing class_id. If you would like to pad "
"the bounding boxes with class_id, use: "
"`bounding_boxes['classes'] = "
"tf.ones_like(bounding_boxes['boxes'])`."
)
return bounding_boxes
def _format_output(self, output, metadata):
if not metadata[IS_DICT]:
return output[IMAGES]
elif metadata[USE_TARGETS]:
output[TARGETS] = output[LABELS]
del output[LABELS]
return output
def _ensure_inputs_are_compute_dtype(self, inputs):
if not isinstance(inputs, dict):
return preprocessing.ensure_tensor(
inputs,
self.compute_dtype,
)
# Copy the input dict before we mutate it.
inputs = dict(inputs)
inputs[IMAGES] = preprocessing.ensure_tensor(
inputs[IMAGES],
self.compute_dtype,
)
if BOUNDING_BOXES in inputs:
inputs[BOUNDING_BOXES] = bounding_box.ensure_tensor(
inputs[BOUNDING_BOXES], dtype=self.compute_dtype
)
return inputs
|
keras-cv/keras_cv/layers/preprocessing/base_image_augmentation_layer.py/0
|
{
"file_path": "keras-cv/keras_cv/layers/preprocessing/base_image_augmentation_layer.py",
"repo_id": "keras-cv",
"token_count": 9825
}
| 53 |
# Copyright 2023 The KerasCV Authors
#
# Licensed under the Apache License, Version 2.0 (the "License");
# you may not use this file except in compliance with the License.
# You may obtain a copy of the License at
#
# https://www.apache.org/licenses/LICENSE-2.0
#
# Unless required by applicable law or agreed to in writing, software
# distributed under the License is distributed on an "AS IS" BASIS,
# WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.
# See the License for the specific language governing permissions and
# limitations under the License.
import tensorflow as tf
from keras_cv import bounding_box
from keras_cv.api_export import keras_cv_export
from keras_cv.layers.preprocessing.base_image_augmentation_layer import (
BaseImageAugmentationLayer,
)
@keras_cv_export("keras_cv.layers.MixUp")
class MixUp(BaseImageAugmentationLayer):
"""MixUp implements the MixUp data augmentation technique.
Args:
alpha: Float between 0 and 1. Inverse scale parameter for the gamma
distribution. This controls the shape of the distribution from which
the smoothing values are sampled. Defaults to 0.2, which is a
recommended value when training an imagenet1k classification model.
seed: Integer. Used to create a random seed.
References:
- [MixUp paper](https://arxiv.org/abs/1710.09412).
- [MixUp for Object Detection paper](https://arxiv.org/pdf/1902.04103).
Sample usage:
```python
(images, labels), _ = keras.datasets.cifar10.load_data()
images, labels = images[:10], labels[:10]
# Labels must be floating-point and one-hot encoded
labels = tf.cast(tf.one_hot(labels, 10), tf.float32)
mixup = keras_cv.layers.preprocessing.MixUp(10)
augmented_images, updated_labels = mixup(
{'images': images, 'labels': labels}
)
# output == {'images': updated_images, 'labels': updated_labels}
```
"""
def __init__(self, alpha=0.2, seed=None, **kwargs):
super().__init__(seed=seed, **kwargs)
self.alpha = alpha
self.seed = seed
def _sample_from_beta(self, alpha, beta, shape):
sample_alpha = tf.random.gamma(
shape,
alpha=alpha,
)
sample_beta = tf.random.gamma(
shape,
alpha=beta,
)
return sample_alpha / (sample_alpha + sample_beta)
def _batch_augment(self, inputs):
self._validate_inputs(inputs)
images = inputs.get("images", None)
labels = inputs.get("labels", None)
bounding_boxes = inputs.get("bounding_boxes", None)
segmentation_masks = inputs.get("segmentation_masks", None)
images, lambda_sample, permutation_order = self._mixup(images)
if labels is not None:
labels = self._update_labels(
tf.cast(labels, dtype=self.compute_dtype),
lambda_sample,
permutation_order,
)
inputs["labels"] = labels
if bounding_boxes is not None:
bounding_boxes = self._update_bounding_boxes(
bounding_boxes, permutation_order
)
inputs["bounding_boxes"] = bounding_boxes
inputs["images"] = images
if segmentation_masks is not None:
segmentation_masks = self._update_segmentation_masks(
segmentation_masks, lambda_sample, permutation_order
)
inputs["segmentation_masks"] = segmentation_masks
return inputs
def _augment(self, inputs):
raise ValueError(
"MixUp received a single image to `call`. The layer relies on "
"combining multiple examples, and as such will not behave as "
"expected. Please call the layer with 2 or more samples."
)
def _mixup(self, images):
batch_size = tf.shape(images)[0]
permutation_order = tf.random.shuffle(
tf.range(0, batch_size), seed=self.seed
)
lambda_sample = self._sample_from_beta(
self.alpha, self.alpha, (batch_size,)
)
lambda_sample = tf.cast(
tf.reshape(lambda_sample, [-1, 1, 1, 1]), dtype=self.compute_dtype
)
mixup_images = tf.cast(
tf.gather(images, permutation_order), dtype=self.compute_dtype
)
images = lambda_sample * images + (1.0 - lambda_sample) * mixup_images
return images, tf.squeeze(lambda_sample), permutation_order
def _update_labels(self, labels, lambda_sample, permutation_order):
labels_for_mixup = tf.gather(labels, permutation_order)
lambda_sample = tf.reshape(lambda_sample, [-1, 1])
labels = (
lambda_sample * labels + (1.0 - lambda_sample) * labels_for_mixup
)
return labels
def _update_bounding_boxes(self, bounding_boxes, permutation_order):
boxes, classes = bounding_boxes["boxes"], bounding_boxes["classes"]
boxes_for_mixup = tf.gather(boxes, permutation_order)
classes_for_mixup = tf.gather(classes, permutation_order)
boxes = tf.concat([boxes, boxes_for_mixup], axis=1)
classes = tf.concat([classes, classes_for_mixup], axis=1)
return {"boxes": boxes, "classes": classes}
def _update_segmentation_masks(
self, segmentation_masks, lambda_sample, permutation_order
):
lambda_sample = tf.reshape(lambda_sample, [-1, 1, 1, 1])
segmentation_masks_for_mixup = tf.gather(
segmentation_masks, permutation_order
)
segmentation_masks = (
lambda_sample * segmentation_masks
+ (1.0 - lambda_sample) * segmentation_masks_for_mixup
)
return segmentation_masks
def _validate_inputs(self, inputs):
images = inputs.get("images", None)
labels = inputs.get("labels", None)
bounding_boxes = inputs.get("bounding_boxes", None)
segmentation_masks = inputs.get("segmentation_masks", None)
if images is None or (
labels is None
and bounding_boxes is None
and segmentation_masks is None
):
raise ValueError(
"MixUp expects inputs in a dictionary with format "
'{"images": images, "labels": labels}. or'
'{"images": images, "bounding_boxes": bounding_boxes}. or'
'{"images": images, "segmentation_masks": segmentation_masks}. '
f"Got: inputs = {inputs}."
)
if labels is not None and not labels.dtype.is_floating:
raise ValueError(
f"MixUp received labels with type {labels.dtype}. "
"Labels must be of type float."
)
if bounding_boxes is not None:
_ = bounding_box.validate_format(bounding_boxes)
if segmentation_masks is not None:
if len(segmentation_masks.shape) != 4:
raise ValueError(
"MixUp expects shape of segmentation_masks as "
"[batch, h, w, num_classes]. "
f"Got: shape = {segmentation_masks.shape}. "
)
def get_config(self):
config = {
"alpha": self.alpha,
"seed": self.seed,
}
base_config = super().get_config()
return dict(list(base_config.items()) + list(config.items()))
|
keras-cv/keras_cv/layers/preprocessing/mix_up.py/0
|
{
"file_path": "keras-cv/keras_cv/layers/preprocessing/mix_up.py",
"repo_id": "keras-cv",
"token_count": 3236
}
| 54 |
Subsets and Splits
No community queries yet
The top public SQL queries from the community will appear here once available.