text
stringlengths 226
34.5k
|
|---|
Possible incongruence in Sympy when assuming positive numbers
Question: Using Sympy package version 1.0 on Python 2.7.11 I found what (to me) is an
incongruence. This is the code I'm using:
import sympy as sy
from sympy.stats import Normal, density
from sympy.assumptions import assuming, Q, ask
sy.init_printing()
a = sy.symbols('a', real=True)
with assuming(Q.positive(a)):
print ask(Q.positive(a))
N = Normal('N', 0, a)
What I got is
True
from the first print as expected but an exception when creating the Normal
object
ValueError: Standard deviation must be positive
Can anyone, please, explain if it is intended to be like this and why? Thanks!
PS: I'm aware that I could declare the symbols to be positive
Answer: The problem is simple: there are two assumptions systems in SymPy, called the
old-style and new-style assumptions. They don't interact quite well, yet.
The old-style assumptions define predicates on symbols, e.g.
x = Symbol("x", positive=True)
deduction is then performed on generic expressions with methods such as
`.is_positive`
>>> x.is_positive
True
The latest version of SymPy has linked the old-style assumptions to the new-
style ones, so you can now query
>>> ask(Q.positive(x))
True
Older versions of SymPy would return `None`, as the two assumptions systems
were not linked at all.
The problem is that this relation is **not yet** reciprocal: the old-style
assumptions system is not aware of assumptions defined with the new-style
assumptions system. You can verify it yourself:
>>> with assuming(Q.positive(y)):
... print y.is_positive
None
The random variable _Normal_ requires the standard deviation parameter to be
positive, verification is done with the old-style assumptions. Therefore your
case fails.
Note that the positivity condition on the standard deviation is likely to get
relaxed to a non-negativity condition in the next SymPy version (that is,
allow the positivity-indefinite case to be accepted).
|
Frequency Response Scipy.signal
Question: I'm learning digital signal processing to implement filters and am using
python to easily implement a test ideas. So I just started using the
scipy.signal library to find the impulse response and frequency response of
different filters.
Currently I am working through the book "Digital Signals, Processors and Noise
by Paul A. Lynn (1992)" (and finding it an amazing resource for learning this
stuff). In this book they have a filter with the transfer functions shown
below:
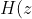&space;=&space;%5Cfrac%7Bz%5E5&space;-&space;z%5E4&space;+&space;z%5E3&space;-&space;z%5E2%7D%7Bz%5E5&space;+&space;0.54048z%5E4&space;-&space;0.62519z%5E3&space;-&space;0.66354z%5E2&space;+&space;0.60317z&space;+0.69341%7D)
I divided the numerator and denominator by
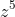 in order to get the following
equation:
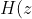&space;=&space;%5Cdfrac%7B1&space;-&space;z%5E%7B-1%7D&space;+&space;z%5E%7B-2%7D&space;-&space;z%5E%7B-3%7D%7D%7B1&space;+&space;0.54048z%5E%7B-1%7D&space;-&space;0.62519z%5E%7B-2%7D&space;-&space;0.66354z%5E%7B-3%7D&space;+&space;0.60317z%5E%7B-4%7D&space;+0.69341z%5E%7B-5%7D%7D)
I then implemented this with Scipy using:
NumeratorZcoefs = [1, -1, 1, -1]
DenominatorZcoefs = [1, 0.54048, -0.62519, -0.66354, 0.60317, 0.69341]
FreqResponse = scipy.signal.freqz(NumeratorZcoefs, DenominatorZcoefs)
fig = plt.figure(figsize = [8, 6])
ax = fig.add_subplot(111)
ax.plot(FreqResponse[0], abs(np.array(FreqResponse[1])))
ax.set_xlim(0, 2*np.pi)
ax.set_xlabel("$\Omega$")
and produce the plot shown below:
[](http://i.stack.imgur.com/OLLSY.png)
However in the book the frequency response is shown to be the following:
[](http://i.stack.imgur.com/cdiy7.jpg)
They are the same shape but the ratio of the peaks at ~2.3 and 0.5 are very
different for the 2 plots, could someone suggest why this is?
Edit:
To add to this, I've just implemented a function to calculate the frequency
response by hand (by calculating the distance from the poles and zeros of the
function) and I get a similar ratio to the plot generated by scipy.signal,
however the numbers are not the same, does anyone know why this might by?
Implementation is as follows:
def H(omega):
z1 = np.array([0,0]) # zero at 0, 0
z2 = np.array([0,0]) # Another zero at 0, 0
z3 = np.array([0, 1]) # zero at i
z4 = np.array([0, -1]) # zero at -i
z5 = np.array([1, 0]) # zero at 1
z = np.array([z1, z2, z3, z4, z5])
p1 = np.array([-0.8, 0])
p = cmath.rect(0.98, np.pi/4)
p2 = np.array([p.real, p.imag])
p = cmath.rect(0.98, -np.pi/4)
p3 = np.array([p.real, p.imag])
p = cmath.rect(0.95, 5*np.pi/6)
p4 = np.array([p.real, p.imag])
p = cmath.rect(0.95, -5*np.pi/6)
p5 = np.array([p.real, p.imag])
p = np.array([p1, p2, p3, p4, p5])
a = cmath.rect(1,omega)
a_2dvector = np.array([a.real, a.imag])
dz = z-a_2dvector
dp = p-a_2dvector
dzmag = []
for dis in dz:
dzmag.append(np.sqrt(dis.dot(dis)))
dpmag = []
for dis in dp:
dpmag.append(np.sqrt(dis.dot(dis)))
return(np.product(dzmag)/np.product(dpmag))
I then plot the frequency response like so:
omegalist = np.linspace(0,2*np.pi,5000)
Hlist = []
for omega in omegalist:
Hlist.append(H(omega))
fig = plt.figure()
ax = fig.add_subplot(111)
ax.plot(omegalist, Hlist)
ax.set_xlabel("$\Omega$")
ax.set_ylabel("$|H(\Omega)|$")
and get the following plot:
[](http://i.stack.imgur.com/nx1hZ.png)
Answer: The SciPy generated frequency response is correct. In any case, I wouldn't
trust the book's figure which appears to have been drawn by hand.
If you want to find the frequency response "manually", this can be simply done
by defining a function returning the original Z-transform and evaluating it on
the unit circle as follows
def H(z):
num = z**5 - z**4 + z**3 - z**2
denom = z**5 + 0.54048*z**4 - 0.62519*z**3 - 0.66354*z**2 + 0.60317*z + 0.69341
return num/denom
import numpy as np
import matplotlib.pyplot as plt
w_range = np.linspace(0, 2*np.pi, 1000)
plt.plot(w_range, np.abs(H(np.exp(1j*w_range))))
The result is exactly the same as SciPy.
|
Enabling `xrange` in Python3 for portability
Question: I wrote a script which I wanted to enable for both Python 2 and Python 3.
After importing `division` and `print_function` from `__future__`, my only
concern was that my `range` returns a whole array in Python 2, wasting time
and memory.
I added the following 3 lines at the beginning of the script, as a workaround:
if sys.version_info[0] == 3:
def xrange(i):
return range(i)
Then, I only used `xrange` in my code.
Is there some more elegant way to do it rather than my workaround?
Answer: You can simplify it a bit:
if sys.version_info[0] == 3:
xrange = range
I would do it the other way around:
if sys.version_info[0] == 2:
range = xrange
If you ever want to drop Python 2.x support, you can just remove those two
lines without going through all your code.
However, I strongly suggest considering the [`six`
library](https://pythonhosted.org/six/). It is the de-facto standard for
enabling Python 2 and 3 compatibility.
from six.moves import range
|
What is the RegEx pattern for 24-06-2015 10:15:45: Aditya Krishnakant:?
Question: What is the RegEx pattern for `24-06-2015 10:15:45: Aditya Krishnakant:`
If you look at the whatsapp chat transcript, it looks like a mess. The purpose
of this code is to print messages sent by a person in a new line (for better
readability). This is my code
import re
f = open("wa_chat.txt", "r")
match = re.findall(r'(\d{2})\:(\d{2})\:(\d{4})\s(\d{2})\:(\d{2})\:(\d{2})\:\s(\w)\s(\w)\:', f)
for content in match:
print(f.readlines(), '\n')
f.close()
I am getting the following error message:
Traceback (most recent call last):
File "whatsapp.py", line 4, in <module>
match = re.findall(r'(\d{2})\:(\d{2})\:(\d{4})\s(\d{2})\:(\d{2})\:(\d{2})\:\s(\w)\s(\w)\:', f)
File "/usr/lib/python2.7/re.py", line 177, in findall
return_compile(pattern, flags).findall(string)
TypeError: expected string or buffer
Where am I going wrong?
Answer: For some reason you're putting `\:` where `-` should be. Also, instead of `\s`
you can be more specific and just use a space. You can be more specific with
those kinds of things because you know exactly what the format is. Your other
big problem is that you're only using `\w`, which only matches one
alphanumeric character, when you should use `\w+`, matching the whole word.
Lastly, your actual error is coming from the fact that you're passing in a
file object instead of the string containing its contents, i.e. `f.read()`.
Here's some code that should work:
import re
f = open("wa_chat.txt", 'r')
match = re.findall(r'(\d{2})-(\d{2})-(\d{4}) (\d{2}):(\d{2}):(\d{2}): (\w+) (\w+):', f.read())
print match #or do whatever you want with it
Note that `match` will be a list of tuples since you wanted to use grouping.
|
Drawing lines between clicks on image with Matplotlib
Question: I am trying to allow for a user to right click on an image once, and then
again, and then the program will draw a line from the first click to the
second.
However, what I have now, inserts lines into my image seemingly randomly. They
pop up and they are nowhere near my clicks, and are of random lengths and
angles.
I'm sort of a beginner to python and definitely to `matplotlib`, so any help
would be appreciated. Below is my code, with the relevant area marked with a
line of #s:
from pymouse import PyMouse
import matplotlib.pyplot as plt
import matplotlib.lines as lines
import numpy
im1 = plt.imread('xexample1.PNG')
im2 = plt.imread('xexample2.PNG')
im3 = plt.imread('xexample3.PNG')
data_images = [im1,im2,im3]
index = 0
ax = plt.gca()
fig = plt.gcf()
plt.imshow(data_images[index])
linepoints = numpy.array([])
print linepoints
#on click event- print x,y coords
def onclick(event):
# if event.xdata != None and event.ydata != None:
plot = numpy.asarray(data_images[index])
if event.button == 1:
print("IMAGE: %d" %index, event.xdata, event.ydata,(plot[event.xdata][event.ydata])*255)
######################################################################
if event.button == 3:
global linepoints
x = event.xdata
y = event.ydata
tup1 = [(x, y)]
linepoints = numpy.append(linepoints, x)
linepoints = numpy.append(linepoints, y)
if numpy.size(linepoints) == 4:
# print "full"
#l1 = lines.Line2D([linepoints[0], linepoints[1]], [linepoints[2],linepoints[3]], transform=fig.transFigure, figure=plt)
#fig.canvas.draw()
plt.plot((linepoints[0], linepoints[1]), (linepoints[2], linepoints[3]), '-')
print linepoints
linepoints = numpy.array([])
print linepoints
# plt.show()
######################################################################
def toggle_images(event):
global index
if event.key == 'x':
index += 1
if index < len(data_images) and index >= 0:
plt.imshow(data_images[index])
plt.draw()
else:
#plt.close()
print 'out of range'
index -= 1
if event.key == 'z':
index -= 1
if index < len(data_images) and index >= 0:
plt.imshow(data_images[index])
plt.draw()
else:
#plt.close()
print 'out of range'
index += 1
plt.imshow(data_images[index])
plt.connect('key_press_event',toggle_images)
cid = fig.canvas.mpl_connect('button_press_event', onclick)
plt.show()
Answer: I created a dressed-down version attached below, but in the end there was only
a very minor mistake in your code:
plt.plot((linepoints[0], linepoints[1]), (linepoints[2], linepoints[3]), '-')
needed to be:
plt.plot((linepoints[0], linepoints[2]), (linepoints[1], linepoints[3]), '-')
I.e.; your 1st (index `0`) and 3rd (index `2`) values are the `x` values and
2nd (index `1`) and fourth (index `3`) are the `y` values, now you were
actually plotting `(x0,y0),(x1,y1)` instead of `(x0,x1),(y0,y1)`
My minimal example:
import matplotlib.pyplot as plt
import numpy
plt.close('all')
fake_img = numpy.random.random((10,10))
plt.imshow(fake_img, interpolation='none')
ax = plt.gca()
fig = plt.gcf()
linepoints = numpy.array([])
def onclick(event):
if event.button == 3:
global linepoints
x = event.xdata
y = event.ydata
linepoints = numpy.append(linepoints, x)
linepoints = numpy.append(linepoints, y)
if numpy.size(linepoints) == 4:
plt.plot((linepoints[0], linepoints[2]), (linepoints[1], linepoints[3]), '-')
linepoints = numpy.array([])
plt.show()
cid = fig.canvas.mpl_connect('button_press_event', onclick)
plt.show()
|
Edit a .txt file, then convert to valid xml with python
Question: I have lots of text files that I need to convert to .xml in order to be able
to work with more efficiently (I am supposed to be doing a couple of language
models to analyze English dialects)
the files go like this:
<I> <IFL-IDN W2C-001 #1:1> <#> <h> <bold> Some Statement that I can edit </bold> <bold> followed by another </bold> </h>
<IFL-IDN W2C-001 #2:1> <p> <#> more and more text that is not very relevant . </p></I>
There are about 500 words per file, what I want to do is to identify the tags,
and close the unclosed ones like <#> and at the end of the sentence.
then I'd like to convert the whole .txt files to valid xml files with before
and after every word. I could have separated that with .split() but the
problem is those kind of tags have spaces in them.
The best code I could come up with is to splilines(), then .split() on a
sentence, then try to Identify the
here is the code for that
Korpus = open("w2c-001.txt").read().splitlines()
for i in Korpus:
Sentence = i.split()
for j in range(0,len(Sentence)-2):
if((Sentence[j][0]=='<' and Sentence[j][len(Sentence[j])-1]!='>') or( Sentence[j][0]!='<' and Sentence[j][len(Sentence[j])-1]=='>')):
Sentence[j] = Sentence[j] + " " + Sentence[j+1] +" " + Sentence[j+2]
Sentence.remove(Sentence[j+1])
Sentence.remove(Sentence[j+2])
#print(Sentence[j])
print(Sentence[j])
My intial thought was If I can write something even to save a valid xml in a
.txt file, converting that file to a .xml shouldn't be a big porblem. I can't
find a python library that can do this, eltree library can create xml, but I
found nothing to identify it and convert it.
Thank you in advance, any help would be very appreciated.
Answer: First, you don't have to load the file and split lines, you can iterate over
the lines. An xml parser can be applied to each line separatly.
Korpus = open("w2c-001.txt")
for line in Korpus:
...
If you want to parse it yourself, use regular expressions to find the tags
import re
re.findall(r'<[a-z]*>','<h> <bold> Some Statement that I can edit </bold> <bold> followed by another </bold> </h>')
XML is not a file format, it is a langage, just write plain text to .xml file
and you're done.
|
How can I get a random unicode string
Question: I am testing a REST based service and one of the inputs is a text string. So I
am sending it random unicode strings from my python code. So far the unicode
strings that I sent were in the ascii range, so everything worked.
Now I am attempting to send characters beyond the ascii range and I am getting
an encoding error. Here is my code. I have been through this
[link](http://nedbatchelder.com/text/unipain.html) and still unable to wrap my
head around it.
# coding=utf-8
import os, random, string
import json
junk_len = 512
junk = (("%%0%dX" % junk_len) % random.getrandbits(junk_len * 8))
for i in xrange(1,5):
if(len(junk) % 8 == 0):
print u'decoding to hex'
message = junk.decode("hex")
print 'Hex chars %s' %message
print u' '.join(message.encode("utf-8").strip())
The first line prints without any issues, but I can't send it to the REST
service without encoding it. Hence the second line where I am attempting to
encode it to utf-8. This is the line of code that fails with the following
message.
> UnicodeDecodeError: 'ascii' codec can't decode byte 0x81 in position 7:
> ordinal not in range(128)
Answer: UTF-8 only allows certain bit patterns. You appear to be using UTF-8 in your
code, so you will need to conform to the allowed UTF-8 patterns.
1 byte: 0b0xxxxxxx
2 byte: 0b110xxxxx 0b10xxxxxx
3 byte: 0b1110xxxx 0b10xxxxxx 0b10xxxxxx
4 byte: 0b11110xxx 0b10xxxxxx 0b10xxxxxx 0b10xxxxxx
In the multi-byte patterns, the first byte indicates the number of bytes in
the whole pattern with leading 1s followed by 0 and data bits `x`. The non-
leading bytes all follow the same pattern: 0b10xxxxxx with two leading
indicator bits `10` and six data bits `xxxxxx`.
In general, randomly generated bytes will not follow these patterns. You can
only generate the data bits `x` randomly.
|
AttributeError: 'file' object has no attribute 'encode'
Question:
Traceback (most recent call last):
File "E:\blahblahblah\emailsend.py", line 26, in <module>
msg.attach(MIMEText(file))
File "E:\blahblahblah\Python 2.7.11\lib\email\mime\text.py", line 30, in __init__
self.set_payload(_text, _charset)
File "E:\blahblahblah\Python 2.7.11\lib\email\message.py", line 226, in set_payload
self.set_charset(charset)
File "E:\blahblahblah\Python 2.7.11\lib\email\message.py", line 268, in set_charset
cte(self)
File "E:\blahblahblah\Python 2.7.11\lib\email\encoders.py", line 73, in encode_7or8bit
orig.encode('ascii')
AttributeError: 'file' object has no attribute 'encode'https://stackoverflow.com/questions/ask#
I've been looking this up a lot but I haven't found an answer.
The only important parts of the code is this:
file = open('newfile.txt')
msg.attach(MIMEText(file))
There are other parts but I've debugged it and I get the error at the
'msg.attach(MIMEText(file))' line.
Any help?
Answer: MIMEText takes the _content_ of the file, not the file object.
msg.attach(MIMEText(open("newfile.txt").read()))
|
Python update last line of file (stdout)
Question: In Python program, I am redirecting `stdout` using below:
sys.stdout = open("log_file.txt", "a",0)
On certain condition I want rewrite the last line of the file.
I have tried below:
if (status=='SAME'):
print '\r'+'Above status doesnot change and last checked @'+str(datetime.datetime.fromtimestamp(time.time())),
This seems to work when I look at file using tail command.
tail -f log_file.txt
However when the I look at the original content of the file, it is not
overwriting the last line but it is appending.
Please suggest me any other approaches keeping `sys.stdout =
open("log_file.txt", "a",0)` as is.
My code is Producing output as :
0007505-160614083053377-oozie-oozi-W-> Started : 2016-06-14 16:15:32
0007505-160614083053377-oozie-oozi-W@HiveScript RUNNING job_1465907358342_1346 RUNNING -
Above status doesnot change and last checked @2016-06-14 16:15:43.096288
Above status doesnot change and last checked @2016-06-14 16:15:53.344065
Above status doesnot change and last checked @2016-06-14 16:16:03.672789
0007505-160614083053377-oozie-oozi-W@end OK - OK -
I want it to be
0007505-160614083053377-oozie-oozi-W-> Started : 2016-06-14 16:15:32
0007505-160614083053377-oozie-oozi-W@HiveScript RUNNING job_1465907358342_1346 RUNNING -
Above status doesnot change and last checked @2016-06-14 16:16:03.672789
0007505-160614083053377-oozie-oozi-W@end OK - OK -
Answer: Without actually running a test case, I'd try:
* `open` the file in `append` mode
* read lines, noting the file position at the start of each line
* after reading the last line, use `seek` to move back to the start of that line
* `write` the new text (make sure it is at least as long as the original).
* `close` the file
===================
from __future__ import print_function # for 2.7
f = open('log.txt', mode='r+') # for preexisting file
for i in range(10):
ref = f.tell()
print('%s line %s'%(i,ref), file=f)
if (i % 3)==0:
f.seek(ref)
print('%s oops %s'%(i,ref), file=f)
ref = f.tell()
print('ending at %3d'%100, file=f)
f.seek(ref)
print('ending at %3d'%f.tell(), file=f)
f.close()
produces:
2200:~/mypy$ cat log.txt
0 oops 0
1 line 9
2 line 18
3 oops 28
4 line 38
5 line 48
6 oops 58
7 line 68
8 line 78
9 oops 88
ending at 98
In 2.7 this form also works:
sys.stdout = f
for i in range(10):
ref = f.tell() # or ref = sys.stdout.tell()
print '%s line %s'%(i,ref)
if (i % 3)==0:
f.seek(ref)
print '%s oops %s'%(i,ref)
|
Complex parsing of a string in Python
Question: I want to parse a string with a format like this:
[{text1}]{quantity}[{text2}]
This rule means that at the beginning there is some text that can optionally
be there or not, followed by a {quantity} whose syntax I describe just below,
followed by more optional text.
The {quantity} can take a variety of forms, with {n} being any positive
integer
{n}
{n}PCS
{n}PC
{n}PCS.
{n}PC.
Lot of {n}
Also, it should accept this additional rule:
{n} {text2}
In this rule, {n} is followed by a space then {text2}
In the cases where PC or PCS appears
* it may or may not be followed by a dot
* case insensitive
* a space can optionally appear between {n} and PCS
* The following are all stripped: PC or PCS, the optional dot, and the optional space
The desired output is normalized to two variables:
* {n} as an integer
* [{text1}] [{text2}], that is, first {text1} (if present), then a space, then {text2} (if present), concatenated to one string. A space to separate the text pieces is only used if there are two of them.
If the {quantity} includes anything besides a positive integer, {n} consists
only of the the integer, and the rest of {quantity} (e.g. " PCS.") is stripped
from both {n} and the resultant text string.
In the text parts, more integers could appear. Any other than the {quantity}
found should be regarded as just part of the text, not interpreted as another
quantity.
I am a former C/C++ programmer. If I had to solve this with those languages, I
would probably use rules in lex and yacc, or else I would have to write a lot
of nasty code to hand-parse it.
I would like to learn a clean approach for coding this efficiently in Python,
probably using rules in some form to easily support more cases. I think I
could use lex and yacc with Python, but I wonder if there is an easier way.
I'm a Python newbie; I don't even know where to start with this.
I am not asking anyone to write code for a complete solution, rather, I need
an approach or two, and perhaps some sample code showing part of how to do it.
Answer: Pyparsing let's you build up a parser by stitching together smaller parsers
using '+' and '|' operators (among others). You can also attach names to the
individual elements in the parser, to make it easier to get at the values
afterward.
from pyparsing import (pyparsing_common, CaselessKeyword, Optional, ungroup, restOfLine,
oneOf, SkipTo)
int_qty = pyparsing_common.integer
# compose an expression for the quantity, in its various forms
"""
{n}
{n}PCS
{n}PC
{n}PCS.
{n}PC.
Lot of {n}
"""
LOT = CaselessKeyword("lot")
OF = CaselessKeyword("of")
pieces = oneOf("PC PCS PC. PCS.", caseless=True)
qty_expr = Optional(LOT + OF).suppress() + int_qty("qty") + Optional(pieces).suppress()
# compose expression for entire line
line_expr = SkipTo(qty_expr)("text1") + qty_expr + restOfLine("text2")
tests = """
Send me 1000 widgets pronto!
Deliver a Lot of 50 barrels of maple syrup by Monday, June 10.
My shipment was short by 25 pcs.
"""
line_expr.runTests(tests)
Prints:
Send me 1000 widgets pronto!
['Send me', 1000, ' widgets pronto!']
- qty: 1000
- text1: ['Send me']
- text2: widgets pronto!
Deliver a Lot of 50 barrels of maple syrup by Monday, June 10.
['Deliver a ', 50, ' barrels of maple syrup by Monday, June 10.']
- qty: 50
- text1: ['Deliver a ']
- text2: barrels of maple syrup by Monday, June 10.
My shipment was short by 25 pcs.
['My shipment was short by', 25, '']
- qty: 25
- text1: ['My shipment was short by']
- text2:
EDIT: Pyparsing supports two forms of alternatives for matching: MatchFirst,
which stops on the first matched alternative (which is defined using the '|'
operator), and Or, which evaluates all alternatives and selects the longest
match (defined using '^' operator). So if you need a priority of the quantity
expression, then you define it explicitly:
qty_pcs_expr = int_qty("qty") + White().suppress() + pieces.suppress()
qty_expr = Optional(LOT + OF).suppress() + int_qty("qty") + FollowedBy(White())
# compose expression for entire line
line_expr = (SkipTo(qty_pcs_expr)("text1") + qty_pcs_expr + restOfLine("text2") |
SkipTo(qty_expr)("text1") + qty_expr + restOfLine("text2"))
Here are the new tests:
tests = """
Send me 1000 widgets pronto!
Deliver a Lot of 50 barrels of maple syrup by Monday, June 10.
My shipment was short by 25 pcs.
2. I expect 22 pcs delivered in the morning
On May 15 please deliver 1000 PCS.
"""
Giving:
2. I expect 22 pcs delivered in the morning
['2. I expect ', 22, ' delivered in the morning']
- qty: 22
- text1: ['2. I expect ']
- text2: delivered in the morning
On May 15 please deliver 1000 PCS.
['On May 15 please deliver ', 1000, '']
- qty: 1000
- text1: ['On May 15 please deliver ']
- text2:
|
Cross-validation on XGBClassifier for multiclass classification in python
Question: I'm trying to perform cross-validation on a XGBClassifier for a multi-class
classification problem using the following code adapted from
<http://www.analyticsvidhya.com/blog/2016/03/complete-guide-parameter-tuning-
xgboost-with-codes-python/>
import numpy as np
import pandas as pd
import xgboost as xgb
from xgboost.sklearn import XGBClassifier
from sklearn.preprocessing import LabelEncoder
from sklearn import cross_validation, metrics
from sklearn.grid_search import GridSearchCV
def modelFit(alg, X, y, useTrainCV=True, cvFolds=5, early_stopping_rounds=50):
if useTrainCV:
xgbParams = alg.get_xgb_params()
xgTrain = xgb.DMatrix(X, label=y)
cvresult = xgb.cv(xgbParams,
xgTrain,
num_boost_round=alg.get_params()['n_estimators'],
nfold=cvFolds,
stratified=True,
metrics={'mlogloss'},
early_stopping_rounds=early_stopping_rounds,
seed=0,
callbacks=[xgb.callback.print_evaluation(show_stdv=False), xgb.callback.early_stop(3)])
print cvresult
alg.set_params(n_estimators=cvresult.shape[0])
# Fit the algorithm
alg.fit(X, y, eval_metric='mlogloss')
# Predict
dtrainPredictions = alg.predict(X)
dtrainPredProb = alg.predict_proba(X)
# Print model report:
print "\nModel Report"
print "Classification report: \n"
print(classification_report(y_val, y_val_pred))
print "Accuracy : %.4g" % metrics.accuracy_score(y, dtrainPredictions)
print "Log Loss Score (Train): %f" % metrics.log_loss(y, dtrainPredProb)
feat_imp = pd.Series(alg.booster().get_fscore()).sort_values(ascending=False)
feat_imp.plot(kind='bar', title='Feature Importances')
plt.ylabel('Feature Importance Score')
# 1) Read training set
print('>> Read training set')
train = pd.read_csv(trainFile)
# 2) Extract target attribute and convert to numeric
print('>> Preprocessing')
y_train = train['OutcomeType'].values
le_y = LabelEncoder()
y_train = le_y.fit_transform(y_train)
train.drop('OutcomeType', axis=1, inplace=True)
# 4) Extract features and target from training set
X_train = train.values
# 5) First classifier
xgb = XGBClassifier(learning_rate =0.1,
n_estimators=1000,
max_depth=5,
min_child_weight=1,
gamma=0,
subsample=0.8,
colsample_bytree=0.8,
scale_pos_weight=1,
objective='multi:softprob',
seed=27)
modelFit(xgb, X_train, y_train)
where `y_train` contains labels from 0 to 4. However, when I run this code I
get the following error from the `xgb.cv` function `xgboost.core.XGBoostError:
value 0for Parameter num_class should be greater equal to 1`. On the XGBoost
doc I read that in the multiclass case xgb extrapolates the number of classes
from the labels in the target vector, so I don't understand what is going on.
Answer: You have to add the parameter ‘num_class’ to the xgb_param dictionary. This is
also mentioned in the parameters description and in a comment from the link
you provided above.
|
Celery: Received unregistered task of type <AsyncResult: [hash]>
Question: In all of the similar problems I've seen on stackOverflow:
* [Celery Received unregistered task of type (run example)](http://stackoverflow.com/questions/9769496/celery-received-unregistered-task-of-type-run-example)
* [getting error Received unregistered task of type 'mytasks.add'](http://stackoverflow.com/questions/12108639/getting-error-received-unregistered-task-of-type-mytasks-add)
* [Received unregistered task for celery](http://stackoverflow.com/questions/25385601/received-unregistered-task-for-celery)
* <http://serverfault.com/questions/416888/celery-daemon-receives-unregistered-tasks>
* <https://github.com/duointeractive/sea-cucumber/issues/15>
the error states the name of the task that's unregistered. I'm having a
different issue. The name of the task isn't being displayed, but rather
`Received unregistered task of type <AsyncResult: e8018fcb-cd15-4dca-
ae6d-6eb906055f13>`, resulting in `KeyError: <AsyncResult: e8018fcb-cd15-4dca-
ae6d-6eb906055f13>`.
Here's my traceback:
KeyError: <AsyncResult: 4aca05f8-14c6-4a25-988a-ff605a27871d>
[2016-06-15 14:11:46,016: ERROR/MainProcess] Received unregistered task of type <AsyncResult: e8018fcb-cd15-4dca-ae6d-6eb906055f13>.
The message has been ignored and discarded.
Did you remember to import the module containing this task?
Or maybe you are using relative imports?
The full contents of the message body was:
{'utc': True, 'chord': None, 'args': [], 'retries': 0, 'expires': None, 'task': <AsyncResult: e8018fcb-cd15-4dca-ae6d-6eb906055f13>, 'callbacks': None, 'errbacks': None, 'timelimit': (None, None), 'taskset': 'a6e8d1c0-c75b-471e-b21f-af8492592aeb', 'kwargs': {}, 'eta': None, 'id': '0dffed5f-3090-417c-a9ec-c99e11bc9579'} (568b)
Traceback (most recent call last):
File "/Users/me/Developer/virtualenvironments/project_name/lib/python2.7/site-packages/celery/worker/consumer.py", line 456, in on_task_received
strategies[name](message, body,
KeyError: <AsyncResult: e8018fcb-cd15-4dca-ae6d-6eb906055f13>
My celery app is including the file where my only 3 tasks are:
app/celery_app.py:
celery_app = Celery('app',
broker='amqp://ip', # RabbitMQ
backend='redis://ip', #Redis
include=['app.tasks.assets'])
celery_app.conf.update(
CELERY_DEFAULT_QUEUE = 'local_testing',
CELERY_TASK_RESULT_EXPIRES=86400, # 24 hours
CELERY_ROUTES={
'app.tasks.assets.list_assets': {'queue': 'gatherAPI'},
'app.tasks.assets.massage_assets': {'queue':'computation'},
'app.tasks.assets.save_assets': {'queue':'database_writes'},
}
)
app/tasks/assets.py:
from __future__ import absolute_import
from celery import current_app
@current_app.task(name='app.tasks.assets.list_assets')
def list_assets(*args, **kwargs):
print "list assets"
@current_app.task(name='app.tasks.assets.massage_assets')
def massage_assets(assets):
print "massaging assets"
@current_app.task(name='app.tasks.assets.save_assets', ignore_result=True)
def save_assets(assets):
print "saving assets..."
These errors occur _only_ in the queues "celery" (which I'm not using) and
"local_testing".
**The appropriate queues for all of these tasks print out and work as
intended** , but somehow, the queues named "celery" and "local_testing" are
filling up (same queue size) and spitting out nothing but this traceback over
and over again.
Here's how I'm calling the tasks...
app/processes/processes.py:
from celery import group
class Process(object):
def run_process(self, resource_generator, chain_signature):
tasks = []
for resources in resource_generator:
tasks.append(chain_signature(resources))
group(tasks)()
app/processes/assets.py:
from __future__ import absolute_import
from app.processes.processes import Process
from app.indexes.asset import AssetIndex
from app.tasks.assets import *
class AssetProcess(Process):
def run(self):
Process.run_process(self,
resource_generator=AssetIndex.asset_generator(),
chain_signature=(
list_assets.s() |
massage_assets.s() |
save_assets.s()))
Again, the default queue is set to "local_testing", so I'm not sure how
anything's being piped to the "celery" queue. The traceback I'm getting is
also fairly unhelpful.
I'm launching the celery worker (with the "celery" queue, or with the
local_testing queue (-Q local_testing)) from the directory above app/, like
so:
`celery -A app.celery_app worker -l info -n worker3.%h`
Any help is greatly appreciated.
Cheers!
Answer: I've determined the problem, and it's from using group.
By passing the chain signature an argument, it's automatically applied
asynchronously. By using group, I'm grouping the asyncResult object, which
doesn't make any sense. I've altered the execution thusly:
def run_process(self, resource_generator, chain_signature):
for resources in resource_generator:
chain_signature(resources)
This effectively does what I wanted anyway.
Cheers
|
moto with boto3 - Cannot build mock ELB
Question: I'm new to moto and aws so I'm trying to wrtie some simple tests cases for a
simple ELB checker in aws.
I have read the moto page here:
<https://github.com/spulec/moto>
and I am following the boto3 guide here:
<https://boto3.readthedocs.io/en/latest/reference/services/elb.html>
But I dont think I am understanding how to use moto with boto3. Below is my
code and error, any help would be greatly appreciated.
# # -*- coding: utf-8 -*-
from .context import elb_tools
from nose.tools import set_trace;
import unittest
from moto.elb import mock_elb
import boto3
class TestElbTools(unittest.TestCase):
"""Basic test cases."""
def setUp(self):
pass
@mock_elb
def test_check_elb_has_attached_instances(self):
empty_elb = mock_elb
mock_elb.describe_load_balancers()
if __name__ == '__main__':
unittest.main()
output:
D:\dev\git_repos\platform-health>nosetests
E
======================================================================
ERROR: test_check_elb_has_attached_instances (test_orphan_elb_finder.test_elb_tools.TestElbTools)
----------------------------------------------------------------------
Traceback (most recent call last):
File "d:\apps\python\python27\lib\site-packages\moto\core\models.py", line 71, in wrapper
result = func(*args, **kwargs)
File "D:\dev\git_repos\platform-health\tests\unit_tests\test_orphan_elb_finder\test_elb_tools.py", line 22, in test_check_elb_has_attached_instances
mock_elb.describe_load_balancers()
AttributeError: 'function' object has no attribute 'describe_load_balancers'
----------------------------------------------------------------------
Ran 1 test in 0.561s
FAILED (errors=1)
Answer: Ok so I got some help from a friend. The below should do the trick
# # -*- coding: utf-8 -*-
from .context import elb_tools
from nose.tools import set_trace;
import unittest
from moto.elb import mock_elb
import boto3
class TestElbTools(unittest.TestCase):
"""Basic test cases."""
def setUp(self):
#pass
self.region = 'ap-southeast-2'
@mock_elb
def test_check_elb_has_attached_instances(self):
elb_client = boto3.client('elb', region_name=self.region)
set_trace()
elb_client.describe_load_balancers()
if __name__ == '__main__':
unittest.main()
|
Efficiently convert a string of comma separated values to bytes
Question: My `python3` program is receiving data from elsewhere as a string in the
following format (the `...` means more data that I care to type out):
data = "0,12,145,234;1,0,0,128;2,255,255,255;...;909,100,100,100;"
I want to convert this to packed binary data where I disregard the `,` and `;`
characters. Currently, I am doing the following:
splitData = data.split(';')[:-1] # ignore the last ';'
buff = []
for item in splitData:
addr, R, G, B = item.split(',')
addr = int(addr) # two bytes
R = int(R) # one byte
G = int(G) # one byte
B = int(B) # one byte
packed = struct.pack('HBBB', addr, R, G, B)
buff.append(packed)
dataBytes = b''.join(buff)
For my example data above, this process gives me the following:
dataBytes = b'\x00\x00\x0c\x91\xea\x01\x00\x00\x00\x80...\x8d\x03ddd'
which is what I want (and is about one third the size of the original string).
However, this process is taking about `0.002` seconds. I need to do this
process 33 times per frame, which leads to about `0.05` seconds to compute,
amounting to about 20 frames per second. I would like to speed this up, if
possible.
Is there a way to convert from the string data to byte data which is faster
than the method above?
Answer: Using itertools, doing a replace then splitting, mapping to int and finally
zipping in fours is about 25 percent faster:
In [82]: data = "0,12,145,234;1,0,0,128;2,255,255,255;909,100,100,100;" * 1000
In [83]: from itertools import imap, izip
[84]: %%timeit
splitData = data.split(';')[:-1] # ignore the last ';'
buff = []
for item in splitData:
addr, R, G, B = item.split(',')
addr = int(addr) # two bytes
R = int(R) # one byte
G = int(G) # one byte
B = int(B) # one byte
packed = struct.pack('HBBB', addr, R, G, B)
buff.append(packed)
dataBytes = b''.join(buff)
....:
100 loops, best of 3: 8.61 ms per loop
In [85]: %%timeit
mapped = imap(int, data[:-1].replace(";", ",").split(","))
b"".join([struct.pack('HBBB', *sub) for sub in izip(mapped, mapped, mapped, mapped)])
....:
100 loops, best of 3: 6.27 ms per loop
Using python3, just use map and zip:
In [4]: %%timeit
mapped = map(int, data[:-1].replace(";", ",").split(","))
b"".join([struct.pack('HBBB', *sub) for sub in zip(mapped, mapped, mapped, mapped)])
...:
100 loops, best of 3: 3.61 ms per loop
In [5]: %%timeit
splitData = data.split(';')[:-1] # ignore the last ';'
buff = [] for item in splitData:
addr, R, G, B = item.split(',')
addr = int(addr) # two bytes
R = int(R) # one byte
G = int(G) # one byte
B = int(B) # one byte
packed = struct.pack('HBBB', addr, R, G, B)
buff.append(packed)
dataBytes = b''.join(buff)
...:
100 loops, best of 3: 4.89 ms per loop
|
Including external shared intel's mkl library in c extension for python
Question: I wrote a python c extension that uses routine from Intel's math kernel
library (mkl). This is the first time that I write a c extension. I just
learned that today.
The c extension compiled. But when I import it in python, it says undefined
symbol, and can't find a function that is defined in the mkl.
How to include any external c library in a python c extension?
Thank you for your help.
mkl_helper.c:
#include "Python.h"
#include "numpy/arrayobject.h"
#include "mkl.h"
static PyObject* test4 (PyObject *self, PyObject *args)
{
// test4 (m, n,
// a, ja, ia,
// c, jc, ic)
PyArrayObject *shape_array;
PyArrayObject *a_array; // csr_matrix.data
PyArrayObject *ja_array; // csr_matrix.indices
PyArrayObject *ia_array; // csr_matrix.indptr
PyArrayObject *c_array;
PyArrayObject *jc_array;
PyArrayObject *ic_array;
if (!PyArg_ParseTuple(args, "O!O!O!O!O!O!O!",
&PyArray_Type, &shape_array,
&PyArray_Type, &a_array,
&PyArray_Type, &ja_array,
&PyArray_Type, &ia_array,
&PyArray_Type, &c_array,
&PyArray_Type, &jc_array,
&PyArray_Type, &ic_array))
{
return NULL;
}
long * ptr_int = shape_array->data;
int m = ptr_int[0];
int n = ptr_int[1];
int k = n;
float * a_data_ptr = a_array->data;
float * ja_data_ptr = ja_array->data;
float * ia_data_ptr = ia_array->data;
float * c_data_ptr = c_array->data;
float * jc_data_ptr = jc_array->data;
float * ic_data_ptr = ic_array->data;
char trans = 'T';
int sort = 0;
int nzmax = n*n;
int info = -3;
int request = 0;
mkl_scsrmultcsr(&trans, &request, &sort,
&m, &n, &k,
a_data_ptr, ja_data_ptr, ia_data_ptr,
a_data_ptr, ja_data_ptr, ia_data_ptr,
c_data_ptr, jc_data_ptr, ic_data_ptr,
&nzmax, &info);
return PyInt_FromLong(info);
}
static struct PyMethodDef methods[] = {
{"test4", test4, METH_VARARGS, "test2(arr1)\n take a numpy array and return its shape as a tuple"},
{NULL, NULL, 0, NULL}
};
PyMODINIT_FUNC
initmkl_helper (void)
{
(void)Py_InitModule("mkl_helper", methods);
import_array();
}
setup.py:
from distutils.core import setup, Extension
import numpy as np
ext_modules = [ Extension('mkl_helper', sources = ['mkl_helper.c']) ]
setup(
name = 'mkl_helper',
version = '1.0',
include_dirs = [np.get_include()], #Add Include path of numpy
ext_modules = ext_modules
)
test.py:
import mkl_helper
result of running test.py:
Traceback (most recent call last):
File "<string>", line 1, in <module>
ImportError: /home/rxu/local/lib/python2.7/site-packages/mkl_helper.so: undefined symbol: mkl_scsrmultcsr
Update 6/16/2016:
this seems to be useful:
1.12. Providing a C API for an Extension Module in
<https://docs.python.org/2/extending/extending.html> says even including one c
extension in another c extension can have problems if linked as share library.
so, i guess I have to link the mkl as a static library? or add inlcude mkl.h
to the python.h?
But then, in python (without c), I can use
ctypes.cdll.LoadLibrary("./mkl_rt.so") to load the mkl's shared library and
then use c function from the shared library without problem (as in
[here](http://stackoverflow.com/questions/37536106/directly-use-intel-mkl-
library-on-scipy-sparse-matrix-to-calculate-a-dot-a-t-wit)). Yet Python/C api
cannot do the same thing in c?
For linking external c library staticly, the setup.py might need: extra
objects in class distutils.core.Extensions at
<https://docs.python.org/2/distutils/apiref.html?highlight=include#distutils.ccompiler.CCompiler.add_include_dir>
Related question about cython with no answer: [Combining Cython with
MKL](http://stackoverflow.com/questions/33596817/combining-cython-with-mkl)
This one seems more helpful: [Python, ImportError: undefined symbol:
g_utf8_skip](http://stackoverflow.com/questions/27149849/python-importerror-
undefined-symbol-g-utf8-skip)
This one use dlopen which is deprecated: [Undefined Symbol in C++ When Loading
a Python Shared Library](http://stackoverflow.com/questions/8302810/undefined-
symbol-in-c-when-loading-a-python-shared-library?rq=1)
Answer: oopcode's answer in [Python, ImportError: undefined symbol:
g_utf8_skip](http://stackoverflow.com/questions/27149849/python-importerror-
undefined-symbol-g-utf8-skip) works. The situation improved with the
following.
importing the c extension into python has no error. Calling the c extension
from python give the following error: Intel MKL FATAL ERROR: Cannot load
libmkl_mc.so or libmkl_def.so.
I remember when I manually compiled numpy with mkl, the site.cfg file asked
for library path and include path for the intel's mkl. Guess I need to add the
library path to the extra_link_args as well... But that didn't work.
Someone with anaconda has that error too as in
[here](https://github.com/SiLab-Bonn/pyBAR/issues/54). Similar case at intel's
forum [here](https://software.intel.com/en-us/forums/intel-math-kernel-
library/topic/296094).
This stackoverflow question says extra_compile_args is also needed: [How to
pass flag to gcc in Python setup.py
script](http://stackoverflow.com/questions/1676384/how-to-pass-flag-to-gcc-in-
python-setup-py-script)
setup.py
from distutils.core import setup, Extension
import numpy as np
extra_link_args=["-I", "(intel's dir)/intel/compilers_and_libraries_2016.3.210/linux/mkl/include", "-L", "(intel's dir)/intel/mkl/lib/intel64/libmkl_mc.so", "-mkl"]
ext_modules = [ Extension('mkl_helper', sources = ['mkl_helper.c'], extra_link_args=extra_link_args) ]
setup(
name = 'mkl_helper',
version = '1.0',
include_dirs = [np.get_include()], #Add Include path of numpy
ext_modules = ext_modules
)
Update: I finally got it working as in [here](https://software.intel.com/en-
us/forums/intel-math-kernel-library/topic/640387) But mkl stillll just use
only one of the 12 cpu.
|
openpyxl no attribution error
Question: ## Python 3.5 openpyxl 2.4
Hi everyone, I got a simple but confusing problem here. FYI the API doc
relating to worksheet is
<http://openpyxl.readthedocs.io/en/default/api/openpyxl.worksheet.worksheet.html>
Here is some simple code for testing.
# -*- coding: utf-8 -*-
from openpyxl import load_workbook
wb2 = load_workbook('example.xlsx')
print (wb2.get_sheet_names())
ws = wb2.get_sheet_by_name('Sheet1')
print (type(ws))
print (ws.calculate_dimension())
list = []
for i in ws.rows:
print ('\n')
for cell in i:
list.append(cell.value)
print(str(cell.value).encode('utf-8'))
print (type(ws))
ws.get_highest_row()
here's what turned out eventually
<class 'openpyxl.worksheet.worksheet.Worksheet'>
Traceback (most recent call last):
File "script.py", line 17, in <module>
ws.get_highest_row()
AttributeError: 'Worksheet' object has no attribute 'get_highest_row'
I run into the problem where it says that `get_highest_row` is not an
attribute. This seems correct since this function is under class
`worksheet.worksheet` (from API doc), and `ws` is
`worksheet.worksheet.Worksheet` (I've no idea what that is) may inherits some
functions so it can still call `dimension()`, but can someone tell me how to
fix this? I want to check through one specific row or column and do some
sorting with varying length of cols and rows. Any help is appreciated!
Answer: I tried it with openpyxl 2.3.5 and got the following
> /usr/local/lib/python3.5/site-packages/openpyxl/worksheet/worksheet.py:350:
> **UserWarning: Call to deprecated function or class get_highest_row (Use the
> max_row property).** def get_highest_row(self):
So as you are using 2.4 they probably removed it from there as it was
deprecated already in 2.3.5.
EDIT: In the
[documentation](http://openpyxl.readthedocs.io/en/2.4/api/openpyxl.worksheet.worksheet.html)
for 2.4 this method is not mentioned any longer
|
generating UUID creation token for EFS
Question: Is this sufficient for generating a UUID for using as a creation token for AWS
CLI EFS ~ Elastic File System? Assuming you've been granted appropriate
permissions.
Python 2.7.9 (default, Dec 10 2014, 12:24:55) [MSC v.1500 32 bit (Intel)] on win32
Type "help", "copyright", "credits" or "license" for more information.
>>> import uuid
>>> uuid.uuid4()
UUID('c3505347-ec30-4f74-9597-e4180de6c56f')
>>>
Answer: That should be fine. If the file system made with that token already exists, a
`FileSystemAlreadyExists` error will be returned. Ideally you should keep
track of tokens you've sent over the wire at least until the EFS has been
successfully created.
|
Sum over parametric function
Question: I am dealing with a parametric function like this:
[](http://i.stack.imgur.com/RA0Dy.png)
and Ideally I would like to sum over the repeated x axis, as shown in the
example. It is to say, for x~4.75, I see that the function can be 0.04, 0.06
or 0.16, and I would like to add a point to the sum of 0.06+0.04+0.16 = 0.26.
I need to do that for every point, so that I can construct a function which is
a kind of 'projection' of the parametric one. Anyone has an idea on how can I
do that in Python?
Answer: Look at the example:
import numpy as np
import matplotlib.pyplot as plt
# set x, y
x = np.arange(-3.,3.,.1)
N = x.size
x[10:13] = x[10]
y = x ** 3 + np.random.rand(N)
# plot curve
fig, ax = plt.subplots()
plt.plot(x,y,'b-')
curve = ax.lines[0]
# get data of plotted curve
xvalues = curve.get_xdata()
yvalues = curve.get_ydata()
# get y for given x
indexes = np. where(xvalues == x[10])
# test print
print xvalues[indexes]
print yvalues[indexes]
print "Sum of y(x) = ",np.sum(yvalues[indexes]) , " where x = ", x[10]
# define markers
xm = []
ym = []
for x1 in x:
indexes = np.where(xvalues == x1)
print x1, yvalues[indexes]
if len(yvalues[indexes]) > 1:
xm += [xvalues[indexes],]
ym += [np.sum(yvalues[indexes]),]
plt.plot(xm, ym, linestyle = 'None', marker='o', color='g')
plt.show()
Test output:
x: [-2. -2. -2.]
y: [-7.0936372 -7.42647923 -7.56571131]
Sum of y(x) = -22.0858277351 where x = -2.0
[](http://i.stack.imgur.com/9CKRR.png)
|
Find Customer from shopify with the particular range of date using shopify python api
Question: How to import the customers from shopify with the particular range of date i.e
importing the customers with the date filter ?
Please refer the code below for importing customers without filter:
shop_url = "https://%s:%s@%s.myshopify.com/admin/" % (self.api_key_shopify,
self.password_shopify, self.name)
shopify.ShopifyResource.set_site(shop_url)
customer_list = shopify.Customer.find()
Answer: [As you can see from the
documentation](https://help.shopify.com/api/reference/customer#index), the
parameters you need to pass with the request in order to obtain customers
created within a specific range are `created_at_min` and `created_at_max`.
Unless you want to type the timestamps manually, we're going to need the
[`datetime`](https://docs.python.org/2/library/datetime.html#datetime.datetime)
object from the [`datetime`](https://docs.python.org/2/library/datetime.html)
module.
from datetime import datetime
time_format = "%Y-%m-%dT%H:%M:%S+00:00"
min_date = datetime(year=2016, month=5, day=1).strftime(time_format)
max_date = datetime(year=2016, month=6, day=1).strftime(time_format)
The formatting string we're using with the `strftime` method will give us our
time in the format required by the Shopify documentation, with UTC timezone
hard-coded in. If you want to use a different timezone, you can either hard-
code in a different time offset, or use the [pytz
module](https://docs.python.org/2/library/datetime.html#datetime.datetime).
Now, for actually calling the API. When using Shopify resources' `find`
methods, you pass property name/value pairs as keyword arguments, like so:
customer_list = shopify.Customer.find(
created_at_min = date1,
created_at_max = date2
)
And voila, this should return you a list of `Customer` resources, or an empty
list if there are no matches.
|
Python OS symlink and .json.gz files
Question: I am creating a symlink with Python. First, I check if the file exists and
then create the symlink.
import os
project = 'test'
if os.path.isfile("../../project/json/nodes1.json.gz"):
os.symlink("../../project/json/nodes1.json.gz","../simulations/"+project+"/nodes1.json.gz")
However, when I ran a script that required this symlink, it failed. On
inspection with finder I get `The operation can’t be completed because the
original item for “nodes1.json.gz” can’t be found.`
I am sure I have the right paths and it is correctly setting the symlink. Is
there an issue with using `.gz` files?
Answer: Unless you specify the `dir_fd` argument to the
[`os.symlink()`](https://docs.python.org/3/library/os.html#os.symlink)
function, you need to specify the absolute path for both `src` and `dst`
argument.
|
How to extract word frequency from document-term matrix?
Question: I am doing LDA analysis with Python. And I used the following code to create a
document-term matrix
corpus = [dictionary.doc2bow(text) for text in texts].
Is there any easy ways to count the word frequency over the whole corpus.
Since I do have the dictionary which is a term-id list, I think I can match
the word frequency with term-id.
Answer: You can use `nltk` in order to count word frequency in string `texts`
from nltk import FreqDist
import nltk
texts = 'hi there hello there'
words = nltk.tokenize.word_tokenize(texts)
fdist = FreqDist(words)
`fdist` will give you word frequency of given string `texts`.
However, you have a list of text. One way to count frequency is to use
`CountVectorizer` from `scikit-learn` for list of strings.
import numpy as np
from sklearn.feature_extraction.text import CountVectorizer
texts = ['hi there', 'hello there', 'hello here you are']
vectorizer = CountVectorizer()
X = vectorizer.fit_transform(texts)
freq = np.ravel(X.sum(axis=0)) # sum each columns to get total counts for each word
this `freq` will correspond to value in dictionary `vectorizer.vocabulary_`
import operator
# get vocabulary keys, sorted by value
vocab = [v[0] for v in sorted(vectorizer.vocabulary_.items(), key=operator.itemgetter(1))]
fdist = dict(zip(vocab, freq)) # return same format as nltk
|
Suggest a better counting process using Python | Pandas?
Question: I'm using Python and Pandas to pull metrics (inbound calls, abandoned calls,
etc.) from our call switch (csv format). The code works well but how I'm
calculating the metrics feels kludige and I'm hoping someone can suggest a
better way.
Data looks like:
Date/Time VPSNumber Duration CallerID ConnectingNum Extension Direction Type
2016-05-31 12:52:35 1-555-555-5555 1:00 1-555-555-0000 0 3 - Support » In Inbound leg of forwarded call
2016-05-31 12:53:19 1-555-555-5555 0:18 Unknown 1-555-555-5555 3 - Support « Out Forwarded call connected
2016-05-31 11:13:13 1-555-555-5555 1:18 1-555-555-1234 0 3 - Support » In Inbound leg of forwarded call
Code looks like:
import pandas as pd
allData = r'phoneSwitch.csv'
phone_df = pd.read_csv((allData),parse_dates=['Date/Time'],index_col='Date/Time')
phone_df.columns = ["VPSNumber","Duration","CallerID","ConnectingNum","Extension","Direction","Type"]
phone_df.fillna(0,inplace=True)
# USE 2016, WHOLE MONTH DATA ONLY
phone_2016_df = (phone_df.loc['2016-01-01':'2016-05-31'])
allInboundCalls = phone_2016_df[(phone_2016_df.Extension == "3") & (phone_2016_df.Direction == "» In") | \
(phone_2016_df.Extension == "717") & (phone_2016_df.Direction == "» In") | \
(phone_2016_df.Extension == "726") & (phone_2016_df.Direction == "» In")].count()["Extension"]
Again, rolling up the data works fine this way - it's more or less a big
countifs statement - but it feels a little bush league and I was hoping there
are any suggestions for improvment and or making the code simpler.
Thanks!
Answer: First filter for inbound calls using a boolean mask `df[df.Direction == "»
In"]`. You can then is `loc` with `isin(...)` to located all those inbound
calls with an extension in your desired group (e.g. 3, 717 and 726).
df = (phone_df.loc['2016-01-01':'2016-05-31'])
allInboundCalls = \
df[df.Direction == "» In"].loc[df.Extension.isin(["3", "717", "726"])].count()["Extension"]
|
Scapy Installation fail on Windows 7 64 bit Python 3.5
Question: I have been going crazy with this installation but nothing seems to work.
Python 3.5 is installed under : "C:\Program Files (x86)\Python35-32", so there
is a space in between.
I installed scapy in 2 ways:
1. pip3 install scapy-python3
2. From the website <http://www.secdev.org/projects/scapy>, I downloaded the link for Scapy-2.3.2 and then compiling it as python setup.py install (This was most likely for Python 2 because first, it failed on fname(chmod,0755) which I corrected to fname(chmod,0o755) but it expected some more arguments)
With each I tried running scapy terminal but to no-avail. It returns the
following error:
python: can't open file 'C:\Program': [Errno 2] No such file or directory
Tried running the following code (want to ping google):
from scapy.all import sr1,IP,ICMP
p=sr1(IP(dst="8.8.8.8")/ICMP())
if p:
p.show()
But get the following error:
WARNING: Windows support for scapy3k is currently in testing.
Sniffing/sending/receiving packets should be working with WinPcap driver and
Powershell. Create issues at https://github.com/phaethon/scapy
Traceback (most recent call last):
File "C:\Users\rads x\Desktop\FIX\connectivity.py", line 13, in
<module>
from scapy.all import sr1,IP,ICMP
File "C:\Program Files (x86)\Python35-32\lib\site-packages\scapy\all.py",
line 16, in <module>
from .arch import *
File "C:\Program Files (x86)\Python35-32\lib\site-
packages\scapy\arch\__init__.py", line 88, in <module>
from .windows import *
File "C:\Program Files (x86)\Python35-32\lib\site-
packages\scapy\arch\windows\__init__.py", line 23, in <module>
from scapy.arch import pcapdnet
File "C:\Program Files (x86)\Python35-32\lib\site-
packages\scapy\arch\pcapdnet.py", line 32, in <module>
from .winpcapy import *
File "C:\Program Files (x86)\Python35-32\lib\site-\
packages\scapy\arch\winpcapy.py", line 26, in <module>
_lib=CDLL('wpcap.dll')
File "C:\Program Files (x86)\Python35-32\lib\ctypes\__init__.py", line 347,
in __init__
self._handle = _dlopen(self._name, mode)
OSError: [WinError 126] The specified module could not be found
Answer: According to the docs scrapy is not supported on Windows with Python 3.x due
to missing dependency (Twisted)
Source: <http://doc.scrapy.org/en/latest/intro/install.html>
|
Python is unable to import library compiled with boost_python
Question: I have the following sample program:
// src/main.cpp
#include <boost/python.hpp>
char const* func()
{
return "String";
}
BOOST_PYTHON_MODULE(bridge)
{
boost::python::def("func", func);
}
When built using the following CMakeLists.txt, no compiler errors are given:
project(bridge)
cmake_minimum_required(VERSION 3.5)
set(PROJECT_SOURCE_DIR ${CMAKE_SOURCE_DIR}/src)
set(CMAKE_BINARY_DIR ${CMAKE_SOURCE_DIR}/bin)
set(EXECUTABLE_OUTPUT_PATH ${CMAKE_BINARY_DIR})
set(LIBRARY_OUTPUT_PATH ${CMAKE_BINARY_DIR})
set(SOURCE_FILES
${PROJECT_SOURCE_DIR}/main.cpp
)
# Include Python
#set(Python_ADDITIONAL_VERSIONS 3.5)
find_package(PythonLibs)
if (${PYTHONLIBS_FOUND})
include_directories(${PYTHON_INCLUDE_DIRS})
link_directories(${PYTHON_LIBRARIES})
endif()
# Include Boost
find_package(Boost 1.61.0 COMPONENTS python REQUIRED)
if (${Boost_FOUND})
include_directories(${Boost_INCLUDE_DIRS})
link_directories(${Boost_LIBRARY_DIR})
endif()
# Enable C++ 11
add_compile_options(-std=c++11)
add_compile_options("-lboost_python")
add_library(bridge SHARED ${SOURCE_FILES})
target_link_libraries(bridge ${PYTHON_LIBRARIES})
target_link_libraries(bridge ${Boost_LIBRARIES})
However, importing the shared library (libbridge.so) gives the following
error:
/bin$ python
Python 2.7.11+ (default, Apr 17 2016, 14:00:29)
[GCC 5.3.1 20160413] on linux2
Type "help", "copyright", "credits" or "license" for more information.
>>> import libbridge
Traceback (most recent call last):
File "<stdin>", line 1, in <module>
ImportError: ./libbridge.so: undefined symbol: _ZN5boost6python6detail11init_moduleER11PyModuleDefPFvvE
I have compiled boost and boost_python without any problem, and other boost
libraries are fully functional. What is wrong here?
_Edit_ : In another post a solution was given by making the filename the same
as the argument fed into `BOOST_PYTHON_MODULE`. After these modifications, the
following error is now given by `import libbridge`:
ImportError: ./libbridge.so: invalid ELF header
Exporting the environment variable `$LD_LIBRARY_PATH=$BOOST_ROOT/stage/lib`
does not seem to create a difference.
Answer: I have found a solution. The problem is due to Python version mismatch inside
Boost. I decided to compile everything in Python 3 and it solves the problem.
I proceeded as follows:
1. I uncommented the following line to the auto-generated `user-config.jam` located in `$BOOST_ROOT/tools/build/example/`
using python : 3.5 : /usr/bin/python3 : /usr/include/python3.5 : /usr/lib;
2. Boost.Python was built from scratch using the commands (executed in sudo to gain permission to `/usr/local`)
$BOOST_ROOT : ./b2 --with-python --clean $BOOST_ROOT : ./b2 --with-python
--install
I verified that the libraries are indeed Python 3 using
$BOOST_ROOT : nm -D stage/lib/libboost_python-3.so | grep PyClass_Type
No output should be given. If the library was compiled with Python 2, then `U
PyClass_Type` would show up.
3. The `CMakeLists.txt` file in the sample project was slightly modified:
set(Python_ADDITIONAL_VERSIONS 3.5) // uncommented find_package(Boost 1.61.0
COMPONENTS python3 REQUIRED) // python3 instead of python
add_compile_options("-lboost_python") // removed
4. Now `python3` (not `python`) should be able to link against the compiled `libbridge.so` library.
|
TypeError: a bytes-like object is required, not 'str' for Python Code
Question: I get this when I try to run a code called NUT_check.py and this comes from
line 69. I'm very unsure of what is happening as I didn't make this code
myself, nor have I really used Python extensively. Line 69 is ending with a
line of hyphens to mark it
import sys
import struct
from struct import pack
import os
from util import *
import string
import math
nut = open(sys.argv[1], "rb+")
NTWU = readu32be(nut)
Version = readu16be(nut)
fileTotal = readu16be(nut)
nut.seek(0x10)
paddingFix = 0
for i in range(fileTotal):
if i > 0:
paddingFix = paddingFix + headerSize
fullSize = readu32be(nut)
nut.seek(4,1)
size = readu32be(nut)
headerSize = readu16be(nut)
nut.seek(2,1)
mipsFlag = readu16be(nut)
gfxFormat = readu16be(nut)
if NTWU == 0x4E545755:
width = readu16be(nut)
height = readu16be(nut)
if NTWU == 0x4E545033:
width2 = readByte(nut)
width1 = readByte(nut)
height2 = readByte(nut)
height1 = readByte(nut)
numOfMips = readu32be(nut)
nut.seek(4,1)
offset1 = (readu32be(nut) + 16)
offset2 = (readu32be(nut) + 16)
offset3 = (readu32be(nut) + 16)
nut.seek(4,1)
if headerSize == 0x60:
size1 = readu32be(nut)
nut.seek(12,1)
if headerSize == 0x70:
size1 = readu32be(nut)
nut.seek(0x1C,1)
if headerSize == 0x80:
size1 = readu32be(nut)
nut.seek(0x2C,1)
if headerSize == 0x90:
size1 = readu32be(nut)
nut.seek(0x3C,1)
eXt = readu32be(nut)
nut.seek(12,1)
GIDX = readu32be(nut)
nut.seek(6,1)
skinNum = readByte(nut)
fileNum = readByte(nut)
nut.seek(4,1)
print("Slot Number %i Texture id %s,%i has %i mipmaps. Format is %i" % (skinNum/4,hex(fileNum),fileNum,mipsFlag,gfxFormat))
if i == 0:
offsetHeader = offset3
if i > 0:
offset1 += paddingFix
offsetHeader += 0x80
backNTime = nut.tell()
nut.seek(offsetHeader)
fileStr = ("%d" % fileNum)
outfile = open("Convert" + "/" + fileStr + ".gtx", "wb")
outfile.write("\x47\x66\x78\x32\x00\x00\x00\x20\x00\x00\x00\x07\x00\x00\x00\x01\x00\x00\x00\x02\x00\x00\x00\x00\x00\x00\x00\x00\x00\x00\x00\x00\x42\x4C\x4B\x7B\x00\x00\x00\x20\x00\x00\x00\x01\x00\x00\x00\x00\x00\x00\x00\x0B\x00\x00\x00\x9C\x00\x00\x00\x00\x00\x00\x00\x00") ----------------------------------------------
outfile.write(nut.read(0x80))
outfile.write("\x00\x00\x00\x01\x00\x01\x02\x03\x1F\xF8\x7F\x21\xC4\x00\x03\xFF\x06\x88\x80\x00\x00\x00\x00\x0A\x80\x00\x00\x10\x42\x4C\x4B\x7B\x00\x00\x00\x20\x00\x00\x00\x01\x00\x00\x00\x00\x00\x00\x00\x0C\x00\x08\x00\x00\x00\x00\x00\x00\x00\x00\x00\x00")
outfile.seek(0x50)
outfile.write(struct.pack(">I",1))
outfile.seek(0xf0)
outfile.write(struct.pack(">I",size))
outfile.seek(8,1)
nut.seek(offset1)
outfile.write(nut.read(size))
outfile.write("\x42\x4C\x4B\x7B\x00\x00\x00\x20\x00\x00\x00\x01\x00\x00\x00\x00\x00\x00\x00\x01\x00\x00\x00\x00\x00\x00\x00\x00\x00\x00\x00\x00")
outfile.close()
nut.seek(backNTime)
nut.close()
Answer: I suspect you're using python3 instead of python2. If you continue to use
python3, trying change the binary-ish string constants to be prefixed with
`b`:
>>> outfile = open("/tmp/foo", "wb")
>>> outfile.write("\x47\x66\x78\x32\x00\x00\x00\x20\x00\x00\x00\x07\x00\x00\x00\x01\x00\x00\x00\x02\x00\x00\x00\x00\x00\x00\x00\x00\x00\x00\x00\x00\x42\x4C\x4B\x7B\x00\x00\x00\x20\x00\x00\x00\x01\x00\x00\x00\x00\x00\x00\x00\x0B\x00\x00\x00\x9C\x00\x00\x00\x00\x00\x00\x00\x00")
Traceback (most recent call last):
File "<stdin>", line 1, in <module>
TypeError: 'str' does not support the buffer interface
>>> outfile.write(b"\x47\x66\x78\x32\x00\x00\x00\x20\x00\x00\x00\x07\x00\x00\x00\x01\x00\x00\x00\x02\x00\x00\x00\x00\x00\x00\x00\x00\x00\x00\x00\x00\x42\x4C\x4B\x7B\x00\x00\x00\x20\x00\x00\x00\x01\x00\x00\x00\x00\x00\x00\x00\x0B\x00\x00\x00\x9C\x00\x00\x00\x00\x00\x00\x00\x00")
64
>>>
|
Can't load all friend Facebook API python
Question: I'm new to Facebook API. I try the below code but it shows only 27 friends in
my list friend(I have about 200). Why? Thanks for your help.
import facebook
token = 'my_token'
graph = facebook.GraphAPI(token)
friends = graph.get_connections(id='me', connection_name='friends')
numberOfFriends = len(friends['data'])
print numberOfFriends
for fr in friends['data']:
print fr['name'] +" "+ fr['id']
Answer: Only friends who authorized your app will show up. If you did not select your
own app in the explorer, it will be the friends who authorized the explorer
app with the user_friends permission.
|
How to convert Decimal() to a number in Python?
Question: I have a JSON output that is `Decimal('142.68500203')`.
I can use str() to get a string which is `"142.68500203"`. But I want to get a
number output which is `142.68500203`.
How to make the conversion in Python?
Answer: It depends what you mean by "number", it could be argued that `Decimal` is a
number class. If you mean into an object of `float` class then:
from decimal import Decimal
x = Decimal('142.68500203')
y = float(x)
print x, y
Gives:
142.68500203 142.68500203
But beware! There are good reasons for using `Decimal`, read [the Decimal
documentation](https://docs.python.org/2/library/decimal.html) before deciding
you want `float`.
|
Python - I dont know how to x works in this code
Question:
def test(x):
def add(y):
return x+y
def mul(y):
return x*y
return "test"
will var `x` go to GC?
Answer: Python uses reference counting to manage _objects_ , not variables. If you
want to know when the object referenced by `x` is cleared from memory, you
need to keep track of how many references there are to it. `x` is just one
such reference.
But, if there are _no other references_ to the same object (`x` is the only
reference to it), then yes, that object will be deleted from memory, because
both `add` and `mul` are not returned by `test`.
`x` is used as a closure by those two nested functions, and thus the object
referenced by `x` would gain two additional references (via the function
object closures). But since the function objects are deleted when the function
ends (as they are just locals within `test`), so will their closures, and so
the end result is that there are no more references to the object referenced
by `x` at the end of the `test()` call as there were at the start of the call.
It doesn't matter here that closure cells are somewhat special in that they
track a variable, not an object directly. Closures are just objects too to
Python (albeit with special support in the interpreter to help track the
original reference), so they are subject to reference counting too.
You can use [`sys.getrefcount()`
function](https://docs.python.org/3/library/sys.html#sys.getrefcount) to see
how many references there are to an object:
>>> import sys
>>> def test(x):
... def add(y):
... return x+y
... def mul(y):
... return x*y
... return "test"
...
>>> value = 'foo bar'
>>> sys.getrefcount(value)
2
>>> test(value)
'test'
>>> sys.getrefcount(value)
2
The refcount did not change, so `test` never resulted in additional references
after the function completed. This changes if you actually returned the nested
functions:
>>> def test2(x):
... def add(y):
... return x+y
... def mul(y):
... return x*y
... return {'+': add, '*': add}
...
>>> test_result = test2(value)
>>> test_result
{'+': <function add at 0x104e85b90>, '*': <function add at 0x104e85b90>}
>>> sys.getrefcount(value)
3
>>> del test_result['+']
>>> sys.getrefcount(value)
3
>>> del test_result['*']
>>> sys.getrefcount(value)
2
The two functions both reference the same closure cell, which in turn
references the value of `x`, so while those two function objects still exist
the reference count goes up by one. Deleting both functions cleared the
closure cell (the refcount for that object went down to 0), and the reference
count dropped again.
As soon as the refcount for the `'foo bar'` string goes to 0, it'll too be
deleted. In the above examples it is always 2, because both the name `value`
and the `sys.getrefcount()` function reference it whenever I try to access the
reference count.
|
Python script to check server status
Question: I want to check some service status (like MySQL, Apache2) using python. I
wrote a simple script.
#!/usr/bin/python
from subprocess import call
command = raw_input('Please enter service name : ')
call(["/etc/init.d/"+command, "status"])
This script will request the user to enter the service and display its results
on terminal as follows.
● apache2.service - LSB: Apache2 web server
Loaded: loaded (/etc/init.d/apache2)
Drop-In: /lib/systemd/system/apache2.service.d
└─apache2-systemd.conf
Active: active (running) since සි 2016-06-17 09:16:10 IST; 5h 43min ago
Docs: man:systemd-sysv-generator(8)
Process: 2313 ExecReload=/etc/init.d/apache2 reload (code=exited, status=0/SUCCESS)
Process: 1560 ExecStart=/etc/init.d/apache2 start (code=exited, status=0/SUCCESS)
CGroup: /system.slice/apache2.service
├─1941 /usr/sbin/apache2 -k start
├─2332 /usr/sbin/apache2 -k start
├─2333 /usr/sbin/apache2 -k start
├─2334 /usr/sbin/apache2 -k start
├─2335 /usr/sbin/apache2 -k start
└─2336 /usr/sbin/apache2 -k start
I just want to take this line for each service `Active: active (running)` and
check if that is running and if not I want to ask do you want to start it.
Can some one help me to do this? Thanks in advance
Answer: I think what you want is to capture the output. Something along the lines of:
status = subprocess.check_output("YOUR COMMAND", shell=True)
if ("Active: active (running)" in status):
...
|
Python dynamic import
Question: I have a list of tuples like `[(module_name, module_abs_path)(mod2, path2),
...]`
The modules are located in the 'modules' subdir where my script lives. I am
trying to write a script which reads a conf file and makes some variables from
the conf file available to the modules from the modules dir. My intention is
to load and run all the modules from this script, so they get access to these
variables.
Things I have tried so far but failed:
1. Tried using `__import__()` but it says running by file name is not allowed
2. Tried `importlib.import_module()` but gives the same error.
How should I go about doing this?
Answer: Have you tried to fix up the path before importing?
from __future__ import print_function
import importlib
import sys
def import_modules(modules):
modules_dict = dict()
for module_name, module_path in modules:
sys.path.append(module_path) # Fix the path
modules_dict[module_name] = importlib.import_module(module_name)
sys.path.pop() # Undo the path.append
return modules_dict
if __name__ == '__main__':
modules_info = [
('module1', '/abs/path/to/module1'),
]
modules_dict = import_modules(modules_info)
# At this point, we can access the module as
# modules_dict['module1']
# or...
globals().update(modules_dict)
# ... simply as module1
|
How would you simplify this program? Python
Question: I wrote this program, which purpose is to visit the 18th link on the list of
links and then on the new page visit the 18th link again.
This program works as intended, but it's a little repetitive and inelegant.
I was wondering if you have any ideas on how to make it simpler, without using
any functions. If I wanted to repeat the process 10 or 100 times, this would
become very long.
Thanks for any suggestions!
# Note - this code must run in Python 2.x and you must download
# http://www.pythonlearn.com/code/BeautifulSoup.py
# Into the same folder as this program
import urllib
from BeautifulSoup import *
url = raw_input('Enter - ')
if len(url) < 1 :
url='http://python-data.dr-chuck.net/known_by_Oluwanifemi.html'
html = urllib.urlopen(url).read()
soup = BeautifulSoup(html)
# Retrieve all of the anchor tags
tags = soup('a')
urllist = list()
count = 0
loopcount = 0
for tag in tags:
count = count + 1
tg = tag.get('href', None)
if count == 18:
print count, tg
urllist.append(tg)
url2 = (urllist[0])
html2 = urllib.urlopen(url2).read()
soup2 = BeautifulSoup(html2)
tags2 = soup2('a')
count2 = 0
for tag2 in tags2:
count2 = count2 + 1
tg2 = tag2.get('href', None)
if count2 == 18:
print count2, tg2
urllist.append(tg2)
Answer: This is what you could do.
import urllib
from BeautifulSoup import *
url_1 = input('') or 'http://python-data.dr-chuck.net/known_by_Oluwanifemi.html'
html_1 = urllib.urlopen(url_1).read()
soup_1 = BeautifulSoup(html_1)
tags = soup('a')
url_retr1 = tags[17].get('href', None)
html_2 = urllib.urlopen(url_retr1).read()
soup_2 = BeautifulSoup(html_2)
tags_2 = soup_2('a')
url_retr1 = tags_2[17].get('href', None)
|
Scraping a table from webpage with Python
Question:
from bs4 import BeautifulSoup
from urllib import urlopen
player_code = open("/Users/brandondennis/Desktop/money/CF_Name.txt").read()
player_code = player_code.split("\r")
for player in player_code:
html =urlopen("https://www.capfriendly.com/players/"+player+"")
soup = BeautifulSoup(html, 'html.parser')
for section in soup.findAll('div',{"class": "table_c"}):
table = section.findChildren()[10].text
print player, table
Here is a link to a sample player page :
<https://www.capfriendly.com/players/patrik-elias>
Here is a sample of player names that I am adding from a text file to the base
url.
[](http://i.stack.imgur.com/CtwbE.png)
[](http://i.stack.imgur.com/PdQQg.png)
This is ultimately what I am wanting to do for my text file of 1000+ players
Answer: Aside from what the others mentioned. Take a look at this line:
table = soup.findAll('table_c')[2]
here, `BeautifulSoup` would try to locate `table_c` elements. But, `table_c`
is a class attribute:
<div class="table_c"><div class="rel navc column_head3 cntrct"><div class="ofh"><div>HISTORICAL SALARY </div><div class="l cont_t mt4">SOURCE: The Hockey News, USA Today</div></div></div>
<table class="cntrct" id="contractinsert" cellpadding="0" border="0" cellspacing="0">
...
</table>
</div>
Use the `class_` argument instead:
table = soup.find_all(class_='table_c')[2]
Or, you may get directly to the table by `id`:
table = soup.find("table", id="contractinsert")
|
How to get brighness percentage level from a histogram?
Question: I wrote a python program that used scikit-image to return the histogram of an
image:
def get_hist(image):
image = img_as_float(image)
hs, dis = exposure.histogram(image)
cdf = np.cumsum(hs) #cumulative distribution function
cdf = 255 * cdf / cdf[-1] #normalize
return hs, cdf
while True:
# Capture frame-by-frame
ret, frame = video_capture.read()
hs, cdf = get_hist(frame)
print ("Histogram: " + str(hs))
And it works fine:
Histogram: [15258 1224 1221 1040 924 973 1035 1080 1144 1201 1363 1357
1565 1721 1996 2118 2069 2385 2332 2730 2554 2380 2594 2523
2748 2667 2553 2716 3001 2962 3051 2921 2992 3213 3439 3424
3443 3400 3750 3774 3752 3502 3632 3648 3625 3665 4160 3718
3892 3578 3774 4224 4178 4400 4391 4420 4368 4293 3992 4329
4401 4315 4471 4267 4362 4373 4559 3812 3974 4015 3988 3887
3647 3616 3424 3657 3721 3660 3544 3581 3434 3382 3428 3420
3256 3204 3366 3324 3177 3221 3102 3258 3160 3195 3212 3242
3157 2974 3071 2960 2956 2925 3002 3092 2952 2961 2854 2909
3080 2942 3057 3038 2850 2912 3000 2824 2857 2877 2656 2860
2848 2838 2701 2799 2625 2646 2656 2691 2600 2696 2738 2649
2721 2563 2709 2663 2584 2546 2565 2547 2505 2641 2614 2759
2554 2746 2723 2727 2505 2599 2755 2627 2552 2603 2605 2484
2465 2393 2319 2090 2059 2028 1882 1979 1868 1940 1854 1853
1696 1781 1694 1667 1682 1643 1692 1602 1540 1488 1549 1489
1472 1411 1414 1392 1423 1302 1252 1355 1287 1268 1186 1254
1172 1155 1175 1169 1238 1164 1165 1102 1205 1135 1118 1132
1065 1036 969 1009 941 1015 946 979 964 957 997 960
906 894 948 936 882 870 860 911 926 854 870 858
813 855 850 816 793 866 805 788 815 819 801 736
824 808 806 791 805 827 826 805 828 853 845 882
836 867 862 90039]
My question is, how do I convert this array into a value of how bright/dark
the image is? I've been looking up online, and the only tutorials I can find
are on how to alter the actual image, when that's not what I want.
Answer: Well, I don't think you need a histogram for this. Also, "how bright" is not
really a defined value. What you could do, is to simply look at the average
pixel value.
image = img_as_float(image)
print(np.mean(image))
Does this solve your problem?
An example:
>>> from skimage import data
>>> import numpy as np
>>> from skimage.util import img_as_float
>>> img = img_as_float(data.moon())
>>> img
array([[ 0.45490196, 0.45490196, 0.47843137, ..., 0.36470588,
0.37647059, 0.37647059],
[ 0.45490196, 0.45490196, 0.47843137, ..., 0.36470588,
0.37647059, 0.37647059],
[ 0.45490196, 0.45490196, 0.47843137, ..., 0.36470588,
0.37647059, 0.37647059],
...,
[ 0.42745098, 0.42745098, 0.43921569, ..., 0.45882353,
0.45490196, 0.45490196],
[ 0.44705882, 0.44705882, 0.44313725, ..., 0.4627451 ,
0.4627451 , 0.4627451 ],
[ 0.44705882, 0.44705882, 0.44313725, ..., 0.4627451 ,
0.4627451 , 0.4627451 ]])
>>> np.mean(img)
0.43988067028569255
|
Appending to a Pandas Dataframe From a pd.read_sql Output
Question: I'm coming from R but need to do this in Python for various reasons. This very
well could be a basic PEBKAC issue with my Python more than anything with
Pandas, PyODBC or anything else.
Please bear with me.
My current Python 3 code:
import pandas as pd
import pyodbc
cnxn = pyodbc.connect(DSN="databasename", uid = "username", pwd = "password")
querystring = 'select order_number, creation_date from table_name where order_number = ?'
orders = ['1234',
'2345',
'3456',
'5678']
for i in orders:
print(pd.read_sql(querystring, cnxn, params = [i]))
What I need is a dataframe with the column names of "order_number" and
"creation_date."
What the code outputs is: [](http://i.stack.imgur.com/A7Vd3.jpg)
Sorry for the screenshot, couldn't get the formatting right here.
Having read the [dataframe.append page](http://pandas.pydata.org/pandas-
docs/stable/generated/pandas.DataFrame.append.html), I tried this:
df = pd.DataFrame()
for i in orders:
df.append(pd.read_sql(querystring, cnxn, params = [i]))
That appears to run fine (no errors thrown, anyway).
But when I try to output df, I get
Empty DataFrame
Columns: []
Index: []
So surely it must be possible to do a pd.read_sql with params from a list (or
tuple, or dictionary, ymmv) and add those results as rows into a
pd.DataFrame().
However, I am failing either at my Stack searching, Googling, or Python in
general (with a distinct possibility of all three).
Any guidance here would be greatly appreciated.
Answer: you may try to do it this way:
df = pd.concat([pd.read_sql(querystring, cnxn, params = [i] for i in orders], ignore_index=True)
so you don't need an extra loop ...
alternatively if your `orders` list is relatively small, you can select all
your rows "in one shot":
querystring = 'select order_number, creation_date from table_name where order_number in ({})'.format(','.join(['?']*len(orders)))
df = pd.read_sql(querystring, cnxn, params=orders)
generated SQL
In [8]: querystring
Out[8]: 'select order_number, creation_date from table_name where order_number in (?,?,?,?)'
|
Python plotting percentile contour lines of a probability distribution
Question: Given a probability distribution with unknown functional form (example below),
I like to plot "percentile-based" contour lines, i.e.,those that correspond to
regions with an integral of 10%, 20%, ..., 90% etc.
## example of an "arbitrary" probability distribution ##
from matplotlib.mlab import bivariate_normal
import matplotlib.pyplot as plt
import numpy as np
X, Y = np.mgrid[-3:3:100j, -3:3:100j]
z1 = bivariate_normal(X, Y, .5, .5, 0., 0.)
z2 = bivariate_normal(X, Y, .4, .4, .5, .5)
z3 = bivariate_normal(X, Y, .6, .2, -1.5, 0.)
z = z1+z2+z3
plt.imshow(np.reshape(z.T, (100,-1)), origin='lower', extent=[-3,3,-3,3])
plt.show()
[](http://i.stack.imgur.com/nqD4B.png)
I've looked into multiple approaches, from using the default contour function
in matplotlib, methods involving stats.gaussian_kde in scipy, and even perhaps
generating random point samples from the distribution and estimating a kernel
afterwards. None of them appears to provide the solution.
Answer: Look at the integral of p(x) inside the contour p(x) ≥ t and solve for the
desired value of t:
import matplotlib
from matplotlib.mlab import bivariate_normal
import matplotlib.pyplot as plt
import numpy as np
X, Y = np.mgrid[-3:3:100j, -3:3:100j]
z1 = bivariate_normal(X, Y, .5, .5, 0., 0.)
z2 = bivariate_normal(X, Y, .4, .4, .5, .5)
z3 = bivariate_normal(X, Y, .6, .2, -1.5, 0.)
z = z1 + z2 + z3
z = z / z.sum()
n = 1000
t = np.linspace(0, z.max(), n)
integral = ((z >= t[:, None, None]) * z).sum(axis=(1,2))
from scipy import interpolate
f = interpolate.interp1d(integral, t)
t_contours = f(np.array([0.9, 0.8, 0.7, 0.6, 0.5, 0.4, 0.3, 0.2, 0.1]))
plt.imshow(z.T, origin='lower', extent=[-3,3,-3,3], cmap="gray")
plt.contour(z.T, t_contours, extent=[-3,3,-3,3])
plt.show()
[](http://i.stack.imgur.com/c32TF.png)
|
Rounding number with python
Question: I was trying to calculate int((226553150*1023473145)/5.) using python and I
got 46374212988031352 although it should be 46374212988031350.
Answer: Remove the period for integer division:
int((226553150*1023473145)/5)
Comes out as 46374212988031350 for me.
**Edit** after @tobias_k's comment: This only works in Python2, where `/` is
either floating point division (if either of the arguments is float) or
integer division (if both arguments are integers). In Python3 (or with `from
__future__ import division` in Python2) `/` is always floating point division
and the rounding problem comes up again, because the numbers you multiply are
too large to be exactly expressed in floating point.
The solution is either to use `//`, which is always integer division in all
Python versions or (if you really need the denominator to be float) to switch
to [mpmath](http://mpmath.org/), where you can increase the floating point
precision until you don't have the rounding errors anymore.
|
jupyter notebook running kernel in different env
Question: I've gotten myself into some kind of horrible virtualenv mess. Help?!
I manage environments with `conda`. Until recently, I only had a python2
jupyter notebook kernel, but I decided to drag myself kicking and screaming
into the 21st century and installed a python3 kernel; I forget how I did it.
My main (anaconda) python defaults to 2.7.
So here I am, merrily trying to use beautiful soup from inside my shiny new
python3 kernel, and I don't seem to be able to do anything to get at whatever
environment it's finding packages in. Viz (all from notebook):
from bs4 import BeautifulSoup
-> ImportError: No module named 'bs4'
Ok, fine, I'll install it using shell magic. Right? Right?
! pip install bs4
--> Collecting bs4
Downloading bs4-0.0.1.tar.gz
Requirement already satisfied (use --upgrade to upgrade): beautifulsoup4 in /Users/[MY-USER]/anaconda/lib/python2.7/site-packages (from bs4)
[...]
Successfully built bs4
Installing collected packages: bs4
Successfully installed bs4-0.0.1
from bs4 import BeautifulSoup
-> ImportError: No module named 'bs4'
Oh no. Does it think I'm in a 2.7 env even though I'm running a python3
kernel? That won't do.
! conda info --envs
--> # conda environments:
#
flaskenv /Users/[MY-USER]/anaconda/envs/flaskenv
mesa /Users/[MY-USER]/anaconda/envs/mesa
py35 /Users/[MY-USER]/anaconda/envs/py35
root * /Users/[MY-USER]/anaconda
Ok, I can fix that. One of those is a 3.5 env.
! source activate py35
--> prepending /Users/[MY-USER]/anaconda/envs/py35/bin to PATH
! conda install beautifulsoup4
--> Fetching package metadata .......
Solving package specifications: ..........
# All requested packages already installed.
# packages in environment at /Users/[MY-USER]/anaconda:
#
beautifulsoup4 4.4.1 py27_0
concerning...
! pip install bs4
--> Requirement already satisfied (use --upgrade to upgrade): bs4 in /Users/[MY-USER]/anaconda/lib/python2.7/site-packages
more concerning...
from bs4 import BeautifulSoup
-> ImportError: No module named 'bs4'
ARRGH!!! _headdesk_ Am I going to have to kill the kernel in order to fix this
(and re-run a bit of work)? Is killing the kernel even going to work? How do I
get my jupyter kernel to know what environment it's supposed to be running
under?
thanks!
Answer: This is a tricky part of ipython / Jupyter. The set of kernels available are
independent of what your virtualenv is when you start jupyter Notebook. The
trick is setting up the the ipykernel package in the environment you want to
identify itself uniquely to jupyter. From [docs on multiply
ipykernels](https://ipython.readthedocs.io/en/stable/install/kernel_install.html#kernels-
for-different-environments),
source activate ENVNAME
pip install ipykernel
python -m ipykernel install --user --name ENVNAME --display-name "Python (whatever you want to call it)"
If you only want to have a single Python 3 kernel, from the conda environment,
just use `python -m ipykernel install --user` and it will reset the default
python to the one in the virtualenv.
And yes, you will need to restart the kernel and re-run the prior steps.
See Also [Using both Python 2.x and Python 3.x in IPython
Notebook](http://stackoverflow.com/questions/30492623/using-both-
python-2-x-and-python-3-x-in-ipython-notebook/)
|
Flask: Mock call to external component returns a non serializable object for a HTTP response
Question: I'm trying to learn how to use the Mock library in python.
I have a Flask application which is connected to Redis via redis-py package.
In the '/myapp/version' API of my app, I call a info() from redis-py, and
format a HTTP response back where the JSON data includes also the return data
from the info() call:
from flask import Flask, jsonify
from redis import StrictRedis
app = Flask(__name__)
redis = StrictRedis(host='redis', port=6379)
@app.route('/myapp/version', methods=['GET'])
def get_version():
redis_info = redis.info() #returns a dict
return jsonify({
'app_version': '0.1',
'flask_version': '0.11.1',
'redis_info': redis_info
})
And this is my test.py, which uses the Flask test_client:
import json
import mock
import redis
import myapp import app
class TestApp:
@classmethod
def setup_class(self):
self.client = app.test_client()
self.client.testing = True
@mock.patch.object(redis.StrictRedis, 'info')
def test_get_version(self, mock_info):
result = self.client.get('/myapp/version')
assert result.status_code == 200
mock_info.assert_called()
If I launch nosetests, the response from the HTTP request will return error:
TypeError: <MagicMock name='info()' id='140548379595472'> is not JSON serializable
I guess that the problem is that the info() call is correctly mocked, but then
it cannot be serialzed to JSON to format the response by the Flask test
client.
Is there a way to get a valid response anyway? Or maybe I am using Mock in the
wrong way? I know it is handy in unit test but here it'm more like integration
testing.
Answer: The best way to mock `redis-py` package is ti use `mockredis`. You need to
simply patch your method using: `@patch('redis.StrictRedis',
mock_strict_redis_client)`. More info can be found on the project
[page](https://github.com/locationlabs/mockredis).
Once you patched `redis`, you can setup the value of specific keys required
for your application as if you're using `redis`. This will enable you to
control what values should be returned from the various redis calls and will
ensure that your application will work properly.
In your specific case, the call for redis that is being done somewhere in your
app might be returning the magic mock instead of some serializable `json`.
You can control the return value of the call by using `mock_info.return_value
= json_str`
|
How to get output of -interpreter-exec in GDB/MI?
Question: I'm writing a wrapper for GDB/MI, however, I miss some commands in the MI
interface. For example, the 'info variables' command. I can execute it with
"-interpreter-exec console", but getting the output of it is tricky since it
writes everything to console records and not to the result record. So I
decided to just take every console record between prompt ("(gdb) " line) and
"^done" record, concatenate them, and return as a result. My question is — is
this safe? It seems that this approach is error-prone, but I don't have any
other idea. Ideas?
**UPDATE:** Also, it might be useful to use python like this `-interpreter-
exec console "python\nprint(gdb.execute('info variables', False, True))\n"`
and the results will appear in one line which is nice, but still how do I get
exactly this line safely? Maybe I should prepend it with some token... Anyway,
it seems that the third argument of `gdb.execute` is not commonly supported,
so relying on it may not be a good idea.
Answer: Well, in fact `gdb.execute` with the third argument is actually supported in
gdb 7.3 which is dated 2011, so I guess using it is fine. So I decided to go
with this command
import gdb
import sys
class ConcatCommand(gdb.Command):
"""Executes a command and print concatenated results with a prefix."""
def __init__(self):
super(ConcatCommand, self).__init__("concat", gdb.COMMAND_USER)
def invoke(self, arg, from_tty):
pair = arg.partition(' ')
sys.stdout.write(pair[0] + gdb.execute(pair[2], False, True))
sys.stdout.flush()
ConcatCommand()
Execute it like `-interpreter-exec console "concat token info variables"`.
Then I just take the contents of console record that starts with a token.
|
Why can't I create a custom user in Django using the UserCreationForm?
Question: I am able to create a user from shell by importing the custom user model but
when I apply the same method after submitting the form, the user is not
created. Below are the codes of my custom user model, `UserCreationForm` and
view.
//model.py
class MyUser(AbstractBaseUser):
email = models.EmailField(
verbose_name='email address',
max_length=255,
unique = True,
)
is_active = models.BooleanField(default=True)
is_admin = models.BooleanField(default=False)
objects = MyUserManager()
USERNAME_FIELD = 'email'
def get_full_name(self):
# The user is identified by their email address
return self.email
def get_short_name(self):
# The user is identified by their email address
return self.email
def __str__(self): # __unicode__ on Python 2
return self.email
def has_perm(self, perm, obj=None):
"Does the user have a specific permission?"
# Simplest possible answer: Yes, always
return True
def has_module_perms(self, app_label):
"Does the user have permissions to view the app `app_label`?"
# Simplest possible answer: Yes, always
return True
@property
def is_staff(self):
"Is the user a member of staff?"
# Simplest possible answer: All admins are staff
return self.is_admin
I have extended the `AbstractBaseUser` as suggested in the Django docs to
create a custom user model.
//forms.py
class UserCreationForm(forms.ModelForm):
"""A form for creating new users. Includes all the required
fields, plus a repeated password."""
email = forms.EmailField(
label='Email',
widget=forms.EmailInput,
required=True,
)
password1 = forms.CharField(
label='Password',
widget=forms.PasswordInput,
required=True
)
password2 = forms.CharField(
label='Password confirmation',
widget=forms.PasswordInput,
required=True
)
class Meta:
model = MyUser
fields = ('email', 'password1', 'password2')
def clean_password2(self):
# Check that the two password entries match
password1 = self.cleaned_data.get("password1")
password2 = self.cleaned_data.get("password2")
if password1 and password2 and password1 != password2:
raise forms.ValidationError("Passwords don't match")
return password2
def save(self, commit=True):
# Save the provided password in hashed format
user = super(UserCreationForm, self).save(commit=False)
user.set_password(self.cleaned_data["password1"])
if commit:
user.save()
return user
Am I doing incorrect form processing. The `form.save()` method didn't work out
for me. Also the docs don't discuss user registration thoroughly. I don't know
why.
//views.py
def login(request):
if request.method == 'POST':
form = AuthenticationForm(data=request.POST)
if form.is_valid():
user = authenticate(email=request.POST['email'],
password=request.POST['password'])
if user is not None:
if user.is_active:
django_login(request, user)
return redirect('/home/', permanent=True)
else:
form = AuthenticationForm()
return render(request, 'buymeauth/login.html', {'form': form})
def register(request):
user = request.user
if request.method == 'POST':
form = UserCreationForm(data=request.POST)
if form.is_valid():
my_user = MyUser(user.email, user.password)
my_user.save()
return redirect('/home/', permanent=True)
else:
form = UserCreationForm()
return render(request, 'buymeauth/register.html', {'form': form})
I am new to Django but not particularly to web development. I have some
exposure with MEAN but I am finding Django difficult. I have been stuck with
this authentication and authorisation stuff for 5 days now.
Answer:
def register(request):
# this is the logged-in user
user = request.user
if request.method == 'POST':
# this is the form with the submitted data
form = UserCreationForm(data=request.POST)
if form.is_valid():
# the submitted data is correct
my_user = MyUser(user.email, user.password)
# this is a new user with the same email and password
# than the currently logged-in user. It's not what you want
# and it won't work if you're not logged-in
my_user.save()
return redirect('/home/', permanent=True)
else:
form = UserCreationForm()
return render(request, 'buymeauth/register.html', {'form': form})
Instead you probably want this:
if request.method == 'POST':
form = UserCreationForm(data=request.POST)
if form.is_valid():
user = form.save(commit=False)
user.is_active = True # if you want to skip email validation
user.email = User.objects.normalize_email(user.email)
user.save()
|
open fp = io.BytesIO(fp.read()) AttributeError: 'str' object has no attribute 'read' in PIL/image.py
Question: I'm a little new to Python especially Imaging library that I'm currently
working with. I'm working with a facial recognition code and running it in my
Raspberry Pi 2 B+ running Jessie. I'm using Opencv 2.4.9 and Python 2.7. The
code that I'm currently working with worked till a few minutes ago but now I
keep getting an error. I didn't alter the code or update anything.
What I have tried: I uninstalled pillow and installed different versions, but
it is still not working.
I just don't understand what has changed. I tried by changing the variable
names still no effect.
import cv
import cv2
import sys
import os
import datetime
import time
from PIL import Image
import numpy as np
def EuclideanDistance(p, q):
p = np.asarray(p).flatten()
q = np.asarray(q).flatten()
return np.sqrt(np.sum(np.power((p-q),2)))
class EigenfacesModel():
def __init__(self, X=None, y=None, num_components=0):
self.num_components = 0
self.projections = []
self.W = []
self.mu = []
if (X is not None) and (y is not None):
self.compute(X,y)
def compute(self, X, y):
[D, self.W, self.mu] = pca(asRowMatrix(X),y, self.num_components)
# store labels
self.y = y
# store projections
for xi in X:
self.projections.append(project(self.W, xi.reshape(1,-1), self.mu))
def predict(self, X):
minDist = np.finfo('float').max
minClass = -1
Q = project(self.W, X.reshape(1,-1), self.mu)
for i in xrange(len(self.projections)):
dist = EuclideanDistance(self.projections[i], Q)
#print i,dist
if dist < minDist:
minDist = dist
minClass = self.y[i]
print "\nMinimum distance ", minDist
return minClass,minDist
def asRowMatrix(X):
if len(X) == 0:
return np.array([])
mat = np.empty((0, X[0].size), dtype=X[0].dtype)
for row in X:
mat = np.vstack((mat, np.asarray(row).reshape(1,-1)))
return mat
def read_images(filename, sz=None):
c = 0
X,y = [], []
with open(filename) as f:
for line in f:
line = line.rstrip()
im = Image.open(line)
im = im.convert("L")
# resize to given size (if given)
if (sz is None):
im = im.resize((92,112), Image.ANTIALIAS)
X.append(np.asarray(im, dtype=np.uint8))
y.append(c)
c = c+1
print c
return [X,y]
def pca(X, y, num_components=0):
[n,d] = X.shape
print n
if (num_components <= 0) or (num_components>n):
num_components = n
mu = X.mean(axis=0)
X = X - mu
if n>d:
C = np.dot(X.T,X)
[eigenvalues,eigenvectors] = np.linalg.eigh(C)
else:
C = np.dot(X,X.T)
[eigenvalues,eigenvectors] = np.linalg.eigh(C)
eigenvectors = np.dot(X.T,eigenvectors)
for i in xrange(n):
eigenvectors[:,i] = eigenvectors[:,i]/np.linalg.norm(eigenvectors[:,i])
# or simply perform an economy size decomposition
# eigenvectors, eigenvalues, variance = np.linalg.svd(X.T, full_matrices=False)
# sort eigenvectors descending by their eigenvalue
idx = np.argsort(-eigenvalues)
eigenvalues = eigenvalues[idx]
eigenvectors = eigenvectors[:,idx]
# select only num_components
num_components = 25
eigenvalues = eigenvalues[0:num_components].copy()
eigenvectors = eigenvectors[:,0:num_components].copy()
return [eigenvalues, eigenvectors, mu]
def project(W, X, mu=None):
if mu is None:
return np.dot(X,W)
return np.dot(X - mu, W)
def reconstruct(W, Y, mu=None):
if mu is None:
return np.dot(W.T,Y)
return np.dot(W.T,Y) + mu
#if __name__ == "__main__":
def FaceRecognitionWrapper(Database_Address,TestImages_Address):
out_dir = "Output_Directory"
[X,y] = read_images(Database_Address)
y = np.asarray(y, dtype=np.int32)
#print len(X)
model = EigenfacesModel(X[0:], y[0:])
# get a prediction for the first observation
[X1,y1] = read_images(TestImages_Address)
y1 = np.asarray(y1, dtype=np.int32)
OutputFile = open("Output.txt",'a')
for i in xrange(len(X1)):
predicted,difference = model.predict(X1[i])
predicted1 = int(predicted/10) + 1
if difference <= 1000:
print i+1 , "th image was recognized as individual" , predicted+1
OutputFile.write(str(predicted1))
OutputFile.write("\n")
else:
os.chdir(out_dir)
print i+1,"th image could not be recognized. Storing in error folder."
errorImage = Image.fromarray(X1[i])
current_time = datetime.datetime.now().time()
error_img_name=current_time.isoformat()+'.png'
errorImage.save(error_img_name)
os.chdir('..')
OutputFile.close()
#Create Model Here
cascPath = '/home/pi/opencv-2.4.9/data/haarcascades/haarcascade_frontalface_default.xml'
faceCascade = cv2.CascadeClassifier(cascPath)
Test_Files = []
video_capture = cv2.VideoCapture(0)
i = 0
while True:
# Capture frame-by-frame
ret, frame = video_capture.read()
gray = cv2.cvtColor(frame, cv2.COLOR_BGR2GRAY)
faces = faceCascade.detectMultiScale(
gray,
scaleFactor=1.1,
minNeighbors=5,
minSize=(30, 30),
flags=cv2.cv.CV_HAAR_SCALE_IMAGE
)
# Draw a rectangle around the faces
for (x, y, w, h) in faces:
dummy_image = frame
cv2.rectangle(dummy_image, (x, y), (x+w, y+h), (0, 255, 0), 2)
dummy_image=dummy_image[y:y+h, x:x+w]
dirname = 'detection_output'
os.chdir(dirname)
current_time = datetime.datetime.now().time()
final_img_name=current_time.isoformat()+'.png'
Test_Files.append(final_img_name)
dummy_image = cv2.cvtColor(dummy_image, cv2.COLOR_BGR2GRAY)
cv2.imwrite(final_img_name,dummy_image)
os.chdir('..')
# Display the resulting frame
cv2.imshow('Video', frame)
if cv2.waitKey(1) & 0xFF == ord('q'):
break
i = i + 1
if i % 20 == 0:
dirname = 'detection_output'
os.chdir(dirname)
TestFile = open('CameraFeedFaces.txt',"w")
for Files in Test_Files:
TestFile.write(os.getcwd()+"/"+Files+"\n")
TestFile.close()
os.chdir("..")
#Call testing.py
FaceRecognitionWrapper("/home/pi/train_faces/temp.txt",os.getcwd()+"/detection_output/CameraFeedFaces.txt")
#Open Output File and Copy in a separate folder where distance greater than threshold
#Then delete all the files in the folder
break
# When everything is done, release the capture
video_capture.release()
cv2.destroyAllWindows()
And here's the Traceback:
Traceback (most recent call last):
File "hel.py", line 213, in <module>
FaceRecognitionWrapper("/home/pi/train_faces/temp.txt",os.getcwd()+"/detection_output/CameraFeedFaces.txt")
File "hel.py", line 127, in FaceRecognitionWrapper
[X,y] = read_images(Database_Address)
File "hel.py", line 68, in read_images
im = Image.open(line)
File "/usr/local/lib/python2.7/dist-packages/PIL/Image.py", line 2277, in open
fp = io.BytesIO(fp.read())
AttributeError: 'str' object has no attribute 'read'
[Output when code was working](http://i.stack.imgur.com/XebZz.png)
I read it somewhere and if I try and edit im = Image.open(open(line,'rb')) I'm
getting this error instead of the previous one
Traceback (most recent call last):
File "hel.py", line 208, in <module>
FaceRecognitionWrapper("/home/pi/train_faces/temp.txt",os.getcwd()+"/detection_output/CameraFeedFaces.txt")
File "hel.py", line 122, in FaceRecognitionWrapper
[X,y] = read_images(Database_Address)
File "hel.py", line 63, in read_images
im = Image.open(open(line,'rb'))
IOError: [Errno 2] No such file or directory: ''
Answer: Your subject line is confusing. You fixed that problem and then exposed a
different bug.
IOError: [Errno 2] No such file or directory: ''
The message shows that you are trying to open a file with no name and that
means that the input file you are reading has at least one blank line. If its
okay for that file to have blank lines, just skip those like:
def read_images(filename, sz=None):
c = 0
X,y = [], []
with open(filename) as f:
for line in f:
line = line.rstrip()
if line:
im = Image.open(open(line,'rb'))
im = im.convert("L"
# resize to given size (if given)
if (sz is None):
im = im.resize((92,112), Image.ANTIALIAS)
X.append(np.asarray(im, dtype=np.uint8))
y.append(c)
c = c+1
Otherwise, you have a legitimate error and you should catch and handle the
exception.
|
Amazon MWS Boto parsed XML missing values
Question: Boto 2.40, Python 3.5
When querying Amazon MWS `get_competitive_pricing_for_asin` some values that
are present in the raw XML are missing once parsed by Boto.
The raw XML contains the number of offers for each condition in
`NumberOfOfferListings`
<CompetitivePricing>
<CompetitivePrices>
<CompetitivePrice belongsToRequester="false" condition="New" subcondition="New">
<CompetitivePriceId>1</CompetitivePriceId>
<Price>
<LandedPrice>
<CurrencyCode>USD</CurrencyCode>
<Amount>18.00</Amount>
</LandedPrice>
<ListingPrice>
<CurrencyCode>USD</CurrencyCode>
<Amount>18.00</Amount>
</ListingPrice>
<Shipping>
<CurrencyCode>USD</CurrencyCode>
<Amount>0.00</Amount>
</Shipping>
</Price>
</CompetitivePrice>
<CompetitivePrice belongsToRequester="false" condition="Used" subcondition="VeryGood">
<CompetitivePriceId>2</CompetitivePriceId>
<Price>
<LandedPrice>
<CurrencyCode>USD</CurrencyCode>
<Amount>100.00</Amount>
</LandedPrice>
<ListingPrice>
<CurrencyCode>USD</CurrencyCode>
<Amount>100.00</Amount>
</ListingPrice>
<Shipping>
<CurrencyCode>USD</CurrencyCode>
<Amount>0.00</Amount>
</Shipping>
</Price>
</CompetitivePrice>
</CompetitivePrices>
<NumberOfOfferListings>
<OfferListingCount condition="New">109</OfferListingCount>
<OfferListingCount condition="Collectible">1</OfferListingCount>
<OfferListingCount condition="Used">51</OfferListingCount>
<OfferListingCount condition="Any">161</OfferListingCount>
</NumberOfOfferListings>
</CompetitivePricing>
However only the `Any` value is kept by Boto:
CompetitivePricing{}(
TradeInValue: None,
CompetitivePrices: CompetitivePriceList{}(
CompetitivePrice: [
CompetitivePrice{'condition': 'New', 'belongsToRequester': 'false', 'subcondition': 'New'}(
CompetitivePriceId: '1',
Price: Price{}(
Shipping: USD 0.00,
LandedPrice: USD 18.00,
ListingPrice: USD 18.00
)
),
CompetitivePrice{'condition': 'Used', 'belongsToRequester': 'false', 'subcondition': 'VeryGood'}(
CompetitivePriceId: '2',
Price: Price{}(
Shipping: USD 0.00,
LandedPrice: USD 100.00,
ListingPrice: USD 100.00
)
)
]
),
NumberOfOfferListings: [''],
OfferListingCount: 161{'condition': 'Any'}
)
Note that `NumberOfOfferListings` contains an empty string in the parsed
response, and only one `OfferListingCount` from the XML was saved and added as
a new attribute.
Does anyone know why the other `OfferListingCount` values are being dropped,
or have a good suggestion on how to preserve those values?
I've searched and read the source code:
<https://github.com/boto/boto/blob/develop/boto/mws/response.py#L520> and
can't figure out where it's dropping those values. I have tried this with
multiple products and get the same results.
**EDIT:** I've tried playing around with monkey-patching `CompetitivePricing`:
class OfferListingCount(ResponseElement):
pass
CompetitivePricing.NumberOfOfferListings = Element(OfferListingCount=ElementList(OfferListingCount))
That gives me a full list of conditions:
NumberOfOfferListings: ^NumberOfOfferListings^{}(
OfferListingCount: [
OfferListingCount{'condition': 'New'}(),
OfferListingCount{'condition': 'Collectible'}(),
OfferListingCount{'condition': 'Used'}(),
OfferListingCount{'condition': 'Any'}()
]
)
But without the values.
If I use `SimpleList`:
class OfferListingCount(ResponseElement):
pass
CompetitivePricing.NumberOfOfferListings = Element(OfferListingCount=SimpleList(OfferListingCount))
I get the values but not the conditions:
NumberOfOfferListings: ^NumberOfOfferListings^{}(
OfferListingCount: ['109', '1', '54', '164']
)
So close
Answer: This is the monkey patch I finally came up with:
from boto.mws.response import CompetitivePricing, ElementList, ResponseElement, Element
class OfferListingCount(ResponseElement):
OfferCount = 0
def endElement(self, name, value, connection):
self.OfferCount = value
super(OfferListingCount, self).endElement(name, value, connection)
CompetitivePricing.NumberOfOfferListings = Element(OfferListingCount=ElementList(OfferListingCount))
Which gives me the output I want:
CompetitivePricing{}(
NumberOfOfferListings: ^NumberOfOfferListings^{}(
OfferListingCount: [
OfferListingCount{'condition': 'New'}(OfferCount: '105'),
OfferListingCount{'condition': 'Collectible'}(OfferCount: '2'),
OfferListingCount{'condition': 'Used'}(OfferCount: '58'),
OfferListingCount{'condition': 'Any'}(OfferCount: '165')]
)
)
|
Socket receiving in while 1
Question: I'm trying to create socket connections in python. I need to listen to server
until it sends a message, thus I need to use `while True`. Client:
import RPi.GPIO as GPIO
import time
import socket
GPIO.setmode(GPIO.BOARD)
pinLDR = 7
pinLED = 11
touch = False
sock = socket.socket()
sock.connect(('192.168.1.67', 9092))
while True:
print sock.recv(256)
def rc_time ():
count = 0
GPIO.setup(pinLDR, GPIO.OUT)
GPIO.output(pinLDR, GPIO.LOW)
time.sleep(0.1)
GPIO.setup(pinLDR, GPIO.IN)
while (GPIO.input(pinLDR) == GPIO.LOW):
count += 1
return count
def led(lh):
GPIO.setup(pinLED, GPIO.OUT)
if lh == 1:
GPIO.output(pinLED, GPIO.HIGH)
else:
GPIO.output(pinLED, GPIO.LOW)
try:
while True:
print(str(rc_time()))
if rc_time() > 5000:
if touch == False:
print "triggered"
sock.send("triggered")
touch = True
else:
if touch == True:
sock.send("nottriggered")
print "nottriggered"
touch = False
except KeyboardInterrupt:
pass
finally:
GPIO.cleanup()
sock.close()
But I have a problem with it. Nothing is printed even if a server sends a
message. And the whole code after first `while True` doesn't work
Answer: **UPDATE** : The issue with the code in the question is that it has an
infinite loop at the top. None of the code below this will ever execute:
while True:
print sock.recv(256)
(And apparently this particular server doesn't send a message until it's
received one first, so it will never send anything.)
* * *
Here's a simple working example. If this doesn't help, you'll need to provide
more context in your question.
Here's the client:
import socket
s = socket.socket()
s.connect(('localhost', 12345))
while True:
print s.recv(256)
Corresponding server code:
import socket
import time
s = socket.socket()
s.bind(('', 12345))
s.listen(0)
conn, addr = s.accept()
while True:
conn.send("Hello")
time.sleep(10)
|
Tkinter : Button in frame not visible
Question: I'm trying to implement a TicTacToe program. I am an absolute beginner in
python. After viewing many tutorials and reading a few books, I have
understood the basics of Python. I'm trying to get the buttons to display in a
frame, but all I get is a blank window.
[link for image of the resultant window](http://i.stack.imgur.com/2P2jk.png)
This is the code I have so far:
from Tkinter import *
class Buttons(object):
def __init__(self,master):
frame = Frame(master)
frame.pack()
self.button1= Button(frame,text="1",height=4,width=8,command=self.move)
self.button1.pack(side=LEFT)
self.button2= Button(frame,text="2",height=4,width=8,command=self.move)
self.button2.pack(side=LEFT)
self.button3= Button(frame,text="3",height=4,width=8,command=self.move)
self.button3.pack(side=LEFT)
root = Tk()
root=mainloop()
Answer: You defined your `Buttons` class but you didn't create an instance of that
class, so no buttons were actually constructed. Also, you had a typo / syntax
error:
root=mainloop()
should be
root.mainloop()
Also, you didn't define the `move` callback method.
Here's a repaired version of your code:
from Tkinter import *
class Buttons(object):
def __init__(self,master):
frame = Frame(master)
frame.pack()
self.button1 = Button(frame, text="1", height=4, width=8, command=self.move)
self.button1.pack(side=LEFT)
self.button2 = Button(frame, text="2", height=4, width=8, command=self.move)
self.button2.pack(side=LEFT)
self.button3 = Button(frame, text="3", height=4, width=8, command=self.move)
self.button3.pack(side=LEFT)
def move(self):
print "click!"
root = Tk()
Buttons(root)
root.mainloop()
However, this still has a problem: The `move` method has no way of knowing
which button called it. Here's one way to fix that. I've also changed
from Tkinter import *
to
import tkinter as tk
It's not a good idea to use "star" imports. They make code harder to read and
they pollute your namespace with all the names defined in the imported module
(that's 175 names in the case of Tkinter), which can lead to name collisions.
import Tkinter as tk
class Buttons(object):
def __init__(self,master):
frame = tk.Frame(master)
frame.pack()
self.buttons = []
for i in range(1, 4):
button = tk.Button(
frame, text=i, height=4, width=8,
command=lambda n=i:self.move(n)
)
button.pack(side=tk.LEFT)
self.buttons.append(button)
def move(self, n):
print "click", n
root = tk.Tk()
Buttons(root)
root.mainloop()
|
Python import CSV short code (pandas?) delimited with ';' and ',' in entires
Question: I need to import a CSV file in Python on Windows. My file is delimited by ';'
and has strings with non-English symbols and commas (',').
I've read posts:
[Importing a CSV file into a sqlite3 database table using
Python](http://stackoverflow.com/questions/2887878/importing-a-csv-file-into-
a-sqlite3-database-table-using-python)
[Python import csv to
list](http://stackoverflow.com/questions/24662571/python-import-csv-to-list)
When I run:
with open('d:/trade/test.csv', 'r') as f1:
reader1 = csv.reader(f1)
your_list1 = list(reader1)
I get an issue: comma is changed to '-' symbol.
When I try:
df = pandas.read_csv(csvfile)
I got errors:
> pandas.io.common.CParserError: Error tokenizing data. C error: Expected 1
> fields in line 13, saw 2.
Please help. I would prefer to use pandas as the code is shorter without
listing all field names from the CSV file.
I understand there could be the work around of temporarily replacing commas.
Still, I would like to solve it by some parameters to pandas.
Answer: Unless your CSV file is broken, you can try to make `csv` guess your format.
import csv
with open('d:/trade/test.csv', 'r') as f1:
dialect = csv.Sniffer().sniff(f1.read(1024))
f1.seek(0)
r = csv.reader(f1, dialect=dialect)
for row in r:
print(row)
|
_tkinter.TclError: invalid command name ".54600176" error, what is going on?
Question: I am new to python and am trying to code a simple game, but am continuously
receiving this error message after updating the main game loop.
Traceback (most recent call last):
File "D:\python shell\Bubble Blaster.py", line 75, in <module>
move_bubbles()
File "D:\python shell\Bubble Blaster.py", line 67, in move_bubbles
c.move(bub_id[i], -bub_speed[i], 0)
File **not displaying for privacy**
\lib\tkinter\__init__.py", line 2430, in move
self.tk.call((self._w, 'move') + args)
_tkinter.TclError: invalid command name ".54600176"
the lines which apparently have an error are this one:
#MAIN GAME LOOP
while True:
if randint(1, BUB_CHANCE) == 1:
create_bubble()
move_bubbles()
window.update()
sleep(0.01)
move_bubbles() is line 75
and this:
def move_bubbles():
for i in range(len(bub_id)):
c.move(bub_id[i], -bub_speed[i], 0)
def move_bubbles(): is line 67
So far the 'bubbles' I created do as they are supposed to, but when I tried to
create a collision event that causes the bubbles to 'pop' when they hit the
submarine controller I created, I get this error message. I have checked every
line of code and compared it to the tutorial book I am using and I haven't
made an error, can someone please help me or explain what the error means? It
is a very frustrating issue!
Answer: In Tk the root window is named '.' (dot) and its children are named as a dot
delimited path of parent names. Tkinter generates the names for you using
numbers. The other thing to note is that in Tk the name of a window is also a
command that provides operations on that window. So the error you have here is
telling you that one of your windows no longer exists as the command that
manages it is gone. I suggest that your `bub_id` list is being modified while
you iterate over it leaving you with the potential to obtain a window name
that has been destroyed elsewhere. You can avoid the error using
`c.winfo_exists` which lets you know if the window is actually existing and
works even if the window has been destroyed. But really you should try to
avoid making calls on destroyed windows.
Here's a small example that produces the same error:
>>> import tkinter as tk
>>> main = tk.Tk()
>>> b = tk.Label(main, text="hello")
>>> b.destroy()
>>> b.configure()
Traceback (most recent call last):
File "<stdin>", line 1, in <module>
File "/usr/lib/python3.4/tkinter/__init__.py", line 1322, in configure
return self._configure('configure', cnf, kw)
File "/usr/lib/python3.4/tkinter/__init__.py", line 1310, in _configure
return self._getconfigure(_flatten((self._w, cmd)))
File "/usr/lib/python3.4/tkinter/__init__.py", line 1294, in _getconfigure
for x in self.tk.splitlist(self.tk.call(*args)):
_tkinter.TclError: invalid command name ".140685140686048"
>>> b.winfo_exists()
0
>>>
|
Strange build error on Travis, local tests in tox work
Question: I have a strange error on Travis, the builds of <https://travis-
ci.org/edoburu/django-fluent-blogs> fail in Django 1.7 and lower with:
from django.utils.six import python_2_unicode_compatible
ImportError: cannot import name 'python_2_unicode_compatible'
This is importable for every Django version the build tests against, and
somehow only works for Django 1.8. Running tests locally with tox works.
Any idea's why this happens?
Answer: Try with
[django.utils.encoding](https://docs.djangoproject.com/en/dev/ref/utils/#module-
django.utils.encoding):
from django.utils.encoding import python_2_unicode_compatible
|
Split into multiple files grouped by column value using AWK
Question: My data frame is something similar to this in structure:
A C
1 a 1
2 a 2
3 a 3
4 a 4
5 b 5
6 b 6
7 b 7
8 b 8
9 c 9
it has 16 million rows, I'm facing memory issues while processing in Python. I
tried to split this file using `split` command, but I don't want to spread
values in `A` column into different files.
I don't want something like this:
File one:
A C
1 a 1
2 a 2
3 a 3
4 a 4
5 b 5
6 b 6
File two:
7 b 7
8 b 8
9 c 9
Something like this would work:
File one:
A C
1 a 1
2 a 2
3 a 3
4 a 4
5 b 5
6 b 6
7 b 7
8 b 8
File two:
9 c 9
Let me know, if I'm not clear. I wrote a script to do this job in python-
pandas, but it's taking long time. Any quick solution using awk?
EDIT: My python script to split this file, this file is taking too long.
import pandas as pd
import sys
import os
tp = pd.read_table(sys.argv[1], iterator=True, chunksize=int(sys.argv[2]))
gff3File = pd.concat(tp, ignore_index=True)
i = 0
colNames = ['query_id', 'subject_id', 'percent_idendity', 'align_len', 'mismatch', 'gap_opens', 'q_start', 'q_end', 's_start', 's_end', 'evalue', 'bit_score']
result = pd.DataFrame(columns= colNames)
os.mkdir('splitted')
os.chdir('splitted')
print(len(gff3File))
while True:
first_gene = gff3File.loc[gff3File["query_id"] == gff3File.query_id.iloc[0]]
gff3File = gff3File[gff3File["query_id"] != gff3File.query_id.iloc[0]]
result = result.append(first_gene)
#print(len(first_gene))
if len(result) >= int(sys.argv[2]) or len(gff3File) == 0:
result.to_csv(''.join(['split', "_", str(i), '.csv']), sep = ',')
print("wrote to:",''.join(['split', "_", str(i), '.csv']))
result = pd.DataFrame(columns= colNames)
i+=1
if len(gff3File) == 0: break
Head of my data:
query_id subject_id percent_idendity align_len mismatch gap_opens q_start q_end s_start s_end evalue bit_score
LOC_Os01g01010.1 Chr1 100.00 811 0 0 370 1180 7134 7944 0.0 1498
LOC_Os01g01010.1 Chr1 100.00 410 0 0 1592 2001 9208 9617 0.0 758
LOC_Os01g01010.1 Chr1 100.00 202 0 0 1392 1593 8407 8608 2e-101 374
LOC_Os01g01010.1 Chr1 100.00 169 0 0 1 169 3449 3617 4e-83 313
LOC_Os01g01010.1 Chr1 82.25 338 30 19 1669 1996 43230640 43230323 1e-68 265
Here, I don't want to spread `query_id` to different files while splitting.
Answer: **UPDATE** :
Since the OP wanted to group several keys into a single file, I wrote a less
simple AWK script to do the processing!
BEGIN {
query_count = 0
file_index = 0
}
{
if ($2 in arr)
print >> arr[$2]".split"
else {
arr[$2] = file_index
print > arr[$2]".split"
}
if ( query_count % 5000 == 0 )
file_index++
query_count++
}
END {
for ( key in arr )
print key ": " arr[key]
}
**Explanation** :
For every _5000_ records, it will group the `keys` into a file. For any record
in the entire database, the records with those _5000_ `keys` will be fixed
into file `0.split`. After which it keeps increment to `1.split` and so on.
Please note, this does not mean that _5000_ records go into 1 file. A little
bit of careful thought should be suffice to reach to a more optimized number
for your dataset, _5000_ should work fine for a general non-specific set.
With a value of 5 instead of _5000_ for the OP's example, the above script
produces the following output:
cat 0.split
1 a 1
2 a 2
3 a 3
4 a 4
5 b 5
6 b 6
7 b 7
8 b 8
cat 1.split
9 c 9
* * *
**_Initial naive solution_** :
This simple awk script will work beautifully.
awk '{print >> $1".split"}' subset.data
What this does is uses your `query_id.split` as a filename, effectively
grouping the entire data using the `query_id`.
Your example when run on the above awk script produces the following files:
awk '{print >> $2".split"}' temp.data
cat a.split
1 a 1
2 a 2
3 a 3
4 a 4
cat b.split
5 b 5
6 b 6
7 b 7
8 b 8
cat c.split
9 c 9
|
Plotting graph using python and dispaying it using HTML
Question: I want build an offline application using plotly for displaying graphs . I am
using python(flask) at the back end and HTML(javascript) for the front end .
Currently I am able to plot the graph by sending the graph data as JSON object
to front end and building the graph using plotly.js at the front end itself .
But what I actually want is to build the graph at the server(backend ie
python) side itself and then display the data in HTML . I have gone through
the plotly documentation that builds the graph in python , but I dont know how
to send the build graph to front end for display :( Can someone help me on
that ? PS : I want to build an offline application Updated Code
$(window).resize(function() {
var divheight = $("#section").height();
var divwidth = $("#section").width();
var update = {
width:divwidth, // or any new width
height:divheight // " "
};
var arr = $('#section > div').get();
alert(arr[1]);
Plotly.relayout(arr[0], update);
}).resize();
});
Answer: My suggestion would be to use the `plotly.offline` module, which creates an
offline version of a plot for you. The plotly API on their website is
horrendous (we wouldn't actually want to know what arguments each function
takes, would we??), so much better to turn to the source code on Github.
If you have a look at the plotly source code, you can see that the
`offline.plot` function takes a kwarg for `output_type`, which is either
`'file'` or `'div'`:
<https://github.com/plotly/plotly.py/blob/master/plotly/offline/offline.py>
So you could do:
from plotly.offline import plot
from plotly.graph_objs import Scatter
my_plot_div = plot([Scatter(x=[1, 2, 3], y=[3, 1, 6])], output_type='div')
This will give you the code (wrapped in `<div>` tags) to insert straight into
your HTML. Maybe not the most efficient solution (as I'm pretty sure it embeds
the relevant d3 code as well, which could just be cached for repeated
requests), but it is self contained.
To insert your div into your html code using Flask, there are a few things you
have to do.
In your html template file for your results page, create a placeholder for
your plot code. Flask uses the Jinja template engine, so this would look like:
<body>
....some html...
{{ div_placeholder }}
...more html...
</body>
In your Flask `views.py` file, you need to render the template with the plot
code inserted into the `div_placeholder` variable:
from plotly.offline import plot
from plotly.graph_objs import Scatter
...other imports....
@app.route('/results', methods=['GET', 'POST'])
def results():
error = None
if request.method == 'POST':
my_plot_div = plot([Scatter(x=[1, 2, 3], y=[3, 1, 6])], output_type='div')
return render_template('results.html',
div_placeholder=Markup(my_plot_div)
)
# If user tries to get to page directly, redirect to submission page
elif request.method == "GET":
return redirect(url_for('submission', error=error))
Obviously YMMV, but that should illustrate the basic principle. Note that you
will probably be getting a user request using POST data that you will need to
process to create the plotly graph.
|
How to write unittests for an optional dependency in a python package?
Question: Based on availability of pandas package in working environment a method
returns two different outputs :
* A `pandas.DataFrame` if pandas is available
* Otherwise a `numpy.recarray` object.
How should I write unittest for this class ?
One solution I can think of is to write tests for both cases (with and without
pandas installation) and skip test accordingly, something like this:
try:
import pandas
HAVE_PANDAS = True
except ImportError:
HAVE_PANDAS = False
import unittest
class TestClass(unittest.TestCase):
@unittest.skipUnless(HAVE_PANDAS, "requires pandas")
def tests_using_pandas(self):
# do something
@unittest.skipUnless(not HAVE_PANDAS, "doesn't require pandas")
def tests_without_pandas(self):
# do something
But I don't like this solution very much due to decrease in test coverage and
skipping tests. I want to run my tests for both cases. It would be helpful if
someone can suggest a better alternative solution for this.
Answer: If you want to test both cases (which you should), you could possibly force
the import of Pandas to fail by adding `None` to the `'pandas'` entry in
`sys.modules`, making sure to add it back again (or delete the entry if it
didn't exist in the first place) once the test is done.
import unittest
import sys
class TestWithoutPandas(unittest.TestCase):
def setUp(self):
self._temp_pandas = None
if sys.modules.get('pandas'):
self._temp_pandas = sys.modules['pandas']
sys.modules['pandas'] = None
def tearDown(self):
if self._temp_pandas:
sys.modules['pandas'] = self._temp_pandas
else:
del sys.modules['pandas']
def tests_using_pandas(self):
flag = False
try:
import pandas
except ImportError:
flag = True
self.assertTrue(flag)
class TestWithPandas(unittest.TestCase):
def tests_using_pandas(self):
flag = False
try:
import pandas
except ImportError:
flag = True
self.assertFalse(flag)
|
Python3 segfaults when using ctypes on xlib, python2 works
Question: The following code was scrounged up on the Internet years ago and works quite
well in python2. It supplies the current idle time on the X server.
import ctypes, os, subprocess
class XScreenSaverInfo( ctypes.Structure ):
_fields_ = [("window", ctypes.c_ulong), ("state", ctypes.c_int), ("kind", ctypes.c_int), ("since", ctypes.c_ulong), ("idle", ctypes.c_ulong), ("event_mask", ctypes.c_ulong)]
xlib = ctypes.cdll.LoadLibrary("libX11.so.6")
xss = ctypes.cdll.LoadLibrary("libXss.so.1")
display = xlib.XOpenDisplay(os.environ["DISPLAY"])
xss.XScreenSaverAllocInfo.restype = ctypes.POINTER(XScreenSaverInfo)
xssinfo = xss.XScreenSaverAllocInfo()
xss.XScreenSaverQueryInfo(display, xlib.XDefaultRootWindow(display), xssinfo)
xssinfo.contents.idle
I can throw this into a python2.7.10 shell and get what I want. However, doing
the same on a python3.4.3 shell kicks me out with a segmentation fault in this
line
xss.XScreenSaverQueryInfo(display, xlib.XDefaultRootWindow(display), xssinfo)
Is my py3 environment broken? Does py3 do something differently?
Answer: There was no significant change in Python 3's `ctypes` module. _However_ in
Python3 `os.environ` values are _unicode_ strings, contrary to the byte
strings of python2 and this [causes the segmentation
fault](http://stackoverflow.com/questions/7256283/differences-in-ctypes-
between-python-2-and-3). So changing:
display = xlib.XOpenDisplay(os.environ["DISPLAY"])
To:
display = xlib.XOpenDisplay(bytes(os.environ["DISPLAY"], 'ascii'))
Fixes the segmentation fault.
If you want to have code that works both in python 2 and 3 you want to use the
`encode` method:
display = xlib.XOpenDisplay(os.environ["DISPLAY"].encode('ascii'))
|
Dealing with durations defined by days, hours, minutes such as “1d 3h 2m” in Python
Question: I have to validate a string with diffrent format of duration:
1d 8h 30m
12h 30m
0h 59m
59m
10h 0m
10h
1d 0h 0m
1d 0h 59m
1d 10h 0m
1d
I am new in python and don't have any idea about `regex` in python.
## **EDIT**
There is a textbox in which user insert work duration like the JIRA Worklog.
So When user insert any of above string these are valid string. But if user
insert any invalid string then i need to show error message of invalid string
format.
1g -- invalid
1d 3g -- invalid
1d 3h 30j -- invalid
and soon
So i just wanted to validate input string.
Answer: Here's what you need:
^(\d+d)?\s*((?:[01]?\d|2[0-3])h)?\s*((?:[0-5]?\d)m)?$
In python ([see it live](https://ideone.com/DQLuWn)):
import re
p = re.compile(ur'^(\d+d)?\s*((?:[01]?\d|2[0-3])h)?\s*((?:[0-5]?\d)m)?$', re.MULTILINE | re.IGNORECASE)
time = input("Please enter your value: ")
if re.match(p, time):
print("\nGreat " + time + " is a valid entry.")
else:
print("\nOups... " + time + " is NOT a valid entry.")
|
Auto it click exception: access violation reading 0x00000083
Question: My code should open a window from taskbar and then click a control from it.
The problem is that after it opens the window I get the error : "exception:
access violation reading 0x00000083".I am new to Python (and in programing)
and I have no ideea what that means or how to solve it.My code looks like this
:
import autoit, time
autoit.win_activate("KOPLAYER 1.3.1040")
time.sleep(2)
autoit.mouse_click(131, 507)
It opens (activates) the window but after that I get that exception error.
Answer: I don't know if that works but change the 2 to 2000. Because in the normal
AutoIt version you enter the time in ms not in s. And the minimal wait time is
10ms. Maybe that's what causes the error!
|
PySide Widget for 3dsMax and Modo
Question: I'm trying to create a simple QT app using PySide and Python that I want it to
run as a 3dsMax script, a Modo script and a stand-alone app if needed. So,
I've saved the following files in my D:\PyTest. It's just a QLabel for this
test.
When I run it (_TestWidget.py_) as a stand-alone it works fine. When I start
it (_ModoStart.py_) from Modo it starts correctly but if I try to click
anywhere in Modo it crashes the whole window. In 3dsMax I get the following
error: **Traceback (most recent call last): File "D:/PyTest\TestWidget.py",
line 13, in SystemExit: -1**
Any ideas how I can make it work?
Thanks,
Nick
_TestWidget.py_
import sys
from PySide import QtGui
def open_widget(app, parent_handle=None):
w = QtGui.QLabel()
w.setText("My Widget")
w.show()
if parent_handle is not None:
w.setParent(parent_handle)
sys.exit(app.exec_())
if __name__ == '__main__':
open_widget(QtGui.QApplication(sys.argv))
_MaxStart.py_
import sys
FileDir = 'D:/PyTest'
if FileDir not in sys.path:
sys.path.append(FileDir)
#Rest imports
from PySide import QtGui
import MaxPlus
import TestWidget
reload(TestWidget)
app = QtGui.QApplication.instance()
parent_handle = QtGui.QWidget(MaxPlus.GetQMaxWindow())
TestWidget.open_widget(app, parent_handle)
_ModoStart.py_
import sys
FileDir = 'D:/PyTest'
if FileDir not in sys.path:
sys.path.append(FileDir)
#Rest imports
from PySide import QtGui
import TestWidget
reload(TestWidget)
app = QtGui.QApplication.instance()
TestWidget.open_widget(app)
**UPDATE:**
I also tried to have one file for all three options (3dsMax/Modo/Stand-alone).
It seems that it works fine for 3dsMax and Stand-Alone, but in Modo, if I
click outside the Widget or if I try to close it, Modo instantly crashes.
import sys
import traceback
from PySide import QtGui
handle = None
appMode = None
try:
import MaxPlus
appMode = '3dsMax'
handle = MaxPlus.GetQMaxWindow()
except:
try:
import lx
appMode = 'Modo'
except:
appMode = 'StandAlone'
app = QtGui.QApplication.instance()
if not app:
app = QtGui.QApplication([])
def main():
w = QtGui.QLabel(handle)
w.setText("My Widget")
w.resize(250, 100)
w.setWindowTitle('PySide Qt Window')
w.show()
try:
sys.exit(app.exec_())
except Exception, err:
traceback.print_exc()
pass
main()
Answer: Ok, with a little help from The Foundry I have a working version. They gave me
this very useful link <http://sdk.luxology.com/wiki/CustomView>
_3dsMax.py_
from PySide import QtGui
import MaxPlus
import sys
ui_dir = r'D:/PyTest/SubFolder/'
if not ui_dir in sys.path:sys.path.insert(0,ui_dir)
import ToolboxUI
reload(ToolboxUI)
parent = MaxPlus.GetQMaxWindow()
w = QtGui.QWidget(parent)
ToolboxUI.create_layout(w, '3dsMax')
w.show()
_Modo.py_
import lx
import lxifc
import sys
ui_dir = r'D:/PyTest/SubFolder/'
if not ui_dir in sys.path:sys.path.insert(0,ui_dir)
import ToolboxUI
reload(ToolboxUI)
class MyButtonTest(lxifc.CustomView):
def customview_Init(self, pane):
if pane is None:
return False
custom_pane = lx.object.CustomPane(pane)
if custom_pane.test() is False:
return False
# get the parent object
my_parent = custom_pane.GetParent()
# convert to PySide QWidget
p = lx.getQWidget(my_parent)
# Check that it succeeds
if p is not None:
ToolboxUI.create_layout(p, 'Modo')
return True
return False
try:
lx.bless(MyButtonTest, "My Button Test")
except:
pass
_StandAlone.py_
from PySide import QtGui
import sys
import ToolboxUI
app = QtGui.QApplication([])
w = QtGui.QWidget()
ToolboxUI.create_layout(w, 'StandAlone')
w.show()
sys.exit(app.exec_())
_ToolboxUI.py_
from PySide import QtGui
appMode = None
def on_clicked(side):
print "Hello from the " + side + " side: " + appMode
def left_click():
on_clicked("left")
def center_click():
on_clicked("center")
def right_click():
on_clicked("right")
def create_layout(my_window, am):
global appMode
appMode = am
buttonLayout = QtGui.QHBoxLayout()
buttonLayout.setSpacing(0)
leftButton = QtGui.QPushButton("Left")
leftButton.setProperty("group", "left")
leftButton.clicked.connect(left_click)
rightButton = QtGui.QPushButton("Right")
rightButton.setProperty("group", "right")
rightButton.clicked.connect(right_click)
centerButton = QtGui.QPushButton("Center")
centerButton.setProperty("group", "center")
centerButton.clicked.connect(center_click)
buttonLayout.addWidget(leftButton)
buttonLayout.addWidget(centerButton)
buttonLayout.addWidget(rightButton)
my_window.setLayout(buttonLayout)
|
Adding new data to the neo4j graph database
Question: I am importing a huge dataset of about 46K nodes into Neo4j using import
option.Now this dataset is dynamic i.e new entries keep getting adding to it
now and then so if i have to re perform the entire import then its wastage of
resources.I tried using neo4j rest client of python to send the queries to
create the new data points but as the number of new data points increase the
time taken is more than the importing of 46k nodes.So is there any alternative
to add these datapoints or do i have to redo the entire import?
Answer: First of all - 46k is rather tiny.
The most easy way to import data into Neo4j is using `LOAD CSV` togesther with
`PERIODIC COMMIT`. <http://neo4j.com/developer/guide-import-csv/> contains all
the details.
Be sure to have indexes in place to find the stuff that needs to be changed
with an incremental update quickly.
|
Django not detecting AngularJS
Question: I have a Django project (that I created using PyCharm IDE). I have two HTML
files and below is the code I have in those files.
\-- header.html
<!DOCTYPE html>
<html lang="en" >
<head>
<title>This is my title</title>
<meta charset="UTF-8">
{% load staticfiles %}
<link rel="stylesheet" href="{% static 'css/style.css' %}" type="text/css"/>
<link href='http://fonts.googleapis.com/css?family=Roboto' rel='stylesheet' type='text/css'>
<script data-require="angular.js@*" data-semver="1.4.0-beta.6" src="https://code.angularjs.org/1.4.0-beta.6/angular.js"></script>
<script src="{% static 'js/script.js' %}"></script>
\-- home.html
<html ng-app="myApp">
{% extends "query/header.html" %}
{% block content %}
<p>Hey what's up</p>
<select name="dropDown" ng-model="data.dropDown" >
{% for num in list %}
<option value="num">
{{ num }}
</option>
{% endfor %}
</select>
<p>You chose: {{ data.dropDown }}</p>
{% endblock %}
</html>
In my `home.html`, I have a dropdown that I created using `<select>`tag and is
populated using a Python list. I am using `ng-model` in my select tag in order
to get what user chooses in the dropdown and display it below but for some
reason, it's not displaying anything when I select an option in the dropdown.
Also, in my PyCharm IDE, inside `home.html`, `ng-app` and `ng-model` are
highlighted and I get a warning saying that `Attribute ng-app/ng-model is not
allowed here`
Can you please tell me what I am doing wrong here?
Answer: Let's see...
1. In header.html you seem to have no `{% block content %}` defined. There is nothing your template can inherit if no defined.
2. in home.html you have an additional html wrapping. You don't need it. In fact AFAIK the extends tag should be the first content in the file, aside from imports.
3. If you intend to add angular bindings like `{{ this }}`, ensure you wrap the bindable content in `{% verbatim %}` tags since the same syntax is used for django templates rendering.
Said this, I'd fix your templates like this:
Header:
{% load staticfiles %}
{# PLEASE add template tags imports at the beginning of the file #}
<!DOCTYPE html>
<html lang="en" ng-app="myApp">
<!-- PLEASE ACCURATELY INDENT YOUR CODE!!!! -->
<!-- I hope you have myApp defined in js/script.js file -->
<head>
<title>This is my title</title>
<meta charset="UTF-8" />
<link rel="stylesheet" href="{% static 'css/style.css' %}" type="text/css"/>
<link href='http://fonts.googleapis.com/css?family=Roboto' rel='stylesheet' type='text/css'>
<script data-require="angular.js@*" data-semver="1.4.0-beta.6" src="https://code.angularjs.org/1.4.0-beta.6/angular.js"></script>
<script src="{% static 'js/script.js' %}"></script>
<!-- PLEASE FOR GOD\'S SAKE MAKE A COMPLIANT HTML FILE!!!!!! I DONT KNOW WHAT IS THE REMAINING FILE BODY BUT I WILL ADD IT RIGHT NOW IN A MINIMALIST WAY -->
</head>
<body>
{% block content %}{% endblcok %}
</body>
</html>
The home file will be:
{% extends "query/header.html" %}
{% block content %}
<p>Hey what's up</p>
<select name="dropDown" ng-model="data.dropDown">
{% for num in list %}
<option value="num">{{ num }}</option>
{% endfor %}
</select>
{% verbatim %}
<p>You chose: {{ data.dropDown }}</p>
{% endverbatim %}
|
how to split a file of code into words, despite for string variables
Question: Using python, how can I split a file, containing for example a code with
methods, variables, etc. into words but leave the code's string variables as
one unit string?
For example: given the following python code inside a file:
def example():
a = 5
b = "Hello World"
The result should be:
['def', 'example', '(', ')', ':', 'a', '=', '5', 'b', '=', '"Hello World"']
where "Hello World" is as one single token.
Thanks...
Answer: You can use the [shlex](https://docs.python.org/2/library/shlex.html) module.
Example, for the fule:
Take the following text:
This string has embedded "double quotes" and 'single quotes' in it,
and even "a 'nested example'".
Using the `shlex` library, we construct a simple lexical parser:
import shlex
import sys
if len(sys.argv) != 2:
print 'Please specify one filename on the command line.'
sys.exit(1)
filename = sys.argv[1]
body = file(filename, 'rt').read()
print 'ORIGINAL:', repr(body)
print
print 'TOKENS:'
lexer = shlex.shlex(body)
for token in lexer:
print repr(token)
This generates the output:
ORIGINAL: 'This string has embedded "double quotes" and \'single quotes\' in it,\nand even "a \'nested example\'".\n'
TOKENS:
'This'
'string'
'has'
'embedded'
'"double quotes"'
'and'
"'single quotes'"
'in'
'it'
','
'and'
'even'
'"a \'nested example\'"'
'.'
More information and a nice tutorial can be found
[here](https://pymotw.com/2/shlex/).
|
Python3: using matplotlib to create figure, using a dictionary
Question: I have got a dictionary in the form of `{test_size: (test_error,
training_error)}` Here it is:
{0.1: (0.94736842105263153, 0.90294117647058825), 0.2: (0.92105263157894735, 0.90397350993377479), 0.3: (0.82456140350877194, 0.9242424242424242), 0.6: (0.8722466960352423, 0.91390728476821192), 0.8: (0.76897689768976896, 0.98666666666666669), 0.5: (0.79894179894179895, 0.95767195767195767), 0.7: (0.8226415094339623, 0.99115044247787609), 0.9: (0.62463343108504399, 1.0), 0.4: (0.79605263157894735, 0.92920353982300885)}
I am trying to create a figure with `matplotlib` that looks like this:
[](http://i.stack.imgur.com/TvvWR.jpg)
I would like to get the key of the dictionary (the test_size) on the x axis
and the test and training error on the y axis.
How to solve this? Should I use a DataFrame?
df = pd.DataFrame(dictionary)
plt.plot(df)
???
I read things about that plotting dictionaries is only valuable using Python
2.. I am using Python 3 and I am really lost with this so far.. Hope anyone
can help!
Answer: How about this,
[](http://i.stack.imgur.com/LjyXQ.png)
* * *
The source code,
import matplotlib.pyplot as plt
import operator
fig, ax = plt.subplots()
d = {0.1: (0.94736842105263153, 0.90294117647058825), 0.2: (0.92105263157894735, 0.90397350993377479), 0.3: (0.82456140350877194, 0.9242424242424242), 0.6: (0.8722466960352423, 0.91390728476821192), 0.8: (0.76897689768976896, 0.98666666666666669), 0.5: (0.79894179894179895, 0.95767195767195767), 0.7: (0.8226415094339623, 0.99115044247787609), 0.9: (0.62463343108504399, 1.0), 0.4: (0.79605263157894735, 0.92920353982300885)}
lists = sorted(d.items())
x = list(map(operator.itemgetter(0), lists))
y = list(map(operator.itemgetter(1), lists))
y1 = list(map(operator.itemgetter(0), y))
ax.plot(x, y1, label='Test error', color='b', linewidth=2)
y2 = list(map(operator.itemgetter(1), y))
ax.plot(x, y2, label='Training error', color='r', linewidth=2)
plt.legend(loc='best')
plt.grid()
plt.show()
|
having an issua comparing comparing Strings @ python
Question: Probably there is an easier way to do this, but i read there is no Switch/case
in Python. I wanted to ask the user, to introduce a name of a color, and after
calling the function that should take care of that, it should return the color
code in RGB. My if statements should also accept when the first letter or the
whole word is in capital.
The wierd thing that is happening to me is, i keep geting results on my
console that makes no sence, i am SURE that there is (somewhere) a mystake
from me!
After puting the same input, exacly the same word, the results on the console
are not the same.
AT the moment this is my exact code.
import pygame
import sys
from pygame.locals import *
White =(255, 0, 0)
Black = (0, 0, 0)
Red = (255, 0, 0)
Green = (0, 255, 0)
Blue = (0, 0, 255)
Yellow = (255, 255, 0)
Cyan = (0, 255, 255)
Purple = (255, 0, 255)
def set_display():
pygame.init()
Display = pygame.display.set_mode((400, 300))
pygame.display.set_caption('Seda\'s drawing game')
def get_color():
print('Please introduce one of the following colors')
print(' \n White \n Black \n Red \n Green \n Blue \n Yellow \n Cyan \n Purple')
color = input()
print (color)
if (color == 'Black') or (color == 'BLACK') or (color == 'black'):
return Black
elif (color == 'White') or (color == 'WHITE') or (color == 'white'):
return White
elif (color == 'Red') or (color == 'RED') or (color == 'red'):
return Red
elif (color == 'Green') or (color == 'GREEN') or (color == 'green'):
return Green
elif (color == 'Blue') or (color == 'BLUE') or (color == 'blue'):
return Blue
elif (color == 'Yellow') or (color == 'YELLOW') or (color == 'yellow'):
return Yellow
elif (color == 'Purple') or (color == 'PURPLE') or (color == 'purple'):
return Purple
elif (color == 'Cyan') or (color == 'CYAN') or (color == 'cyan'):
return Cyan
while True:
# set_display()
# for event in pygame.event.get():
# if event.type == QUIT:
# pygame.quit()
# sys.exit()
final_color = get_color()
print ( get_color())
print (final_color)
Console output:
Please introduce one of the following colors
White
Black
Red
Green
Blue
Yellow
Cyan
Purple
white
white
Please introduce one of the following colors
White
Black
Red
Green
Blue
Yellow
Cyan
Purple
white
white
(255, 0, 0)
(255, 0, 0)
Please introduce one of the following colors
White
Black
Red
Green
Blue
Yellow
Cyan
Purple
White
White
Please introduce one of the following colors
White
Black
Red
Green
Blue
Yellow
Cyan
Purple
White
White
(255, 0, 0)
(255, 0, 0)
Please introduce one of the following colors
White
Black
Red
Green
Blue
Yellow
Cyan
Purple
WHITE
WHITE
Please introduce one of the following colors
White
Black
Red
Green
Blue
Yellow
Cyan
Purple
WHITE
WHITE
(255, 0, 0)
(255, 0, 0)
Please introduce one of the following colors
White
Black
Red
Green
Blue
Yellow
Cyan
Purple
it seems every 2 times i write something, if gives me back the color RGB code.
what i keep finding hard to understand, is why this:
final_color = get_color()
print ( get_color())
print (final_color)
is not showing the exact same thing.
Thanks for the help guys
EDDIT1: i found out using extras on those prints, that he is not getting to
those 2 last prints. Still to know why..
EDDIT2: i notice my own error.. saying white is (255,0,0) :D
Answer: The way you return the color could be simplified a lot by doing this. It also
fixes your bug.
White =(255, 0, 0)
Black = (0, 0, 0)
Red = (255, 0, 0)
Green = (0, 255, 0)
Blue = (0, 0, 255)
Yellow = (255, 255, 0)
Cyan = (0, 255, 255)
Purple = (255, 0, 255)
colours = {
'white' : White,
'black' : Black,
'red' : Red,
'green' : Green,
'blue' : Blue,
'yellow' : Yellow,
'cyan' : Cyan,
'purple' : Purple
}
def get_colour():
print('Please introduce one of the following colours')
for key in sorted(colours):
print(key.capitalize())
colour = input()
return colours[colour.lower()]
if __name__ == '__main__': #just added for preference
while True:
final_colour = get_colour()
print(final_colour)
You should familiarize yourself with `lists`, `dictionaries`, and some basic
Python functions. They make life a lot easier!
<https://docs.python.org/3/tutorial/datastructures.html>
P.S. Your `Red` and `White` rgb values are the same.
|
How to put dowloaded JSON data into variables in python
Question:
import requests
import json
import csv
# These our are demo API keys, you can use them!
#location = ""
api_key = 'simplyrets'
api_secret = 'simplyrets'
#api_url = 'https://api.simplyrets.com/properties?q=%s&limit=1' % (location)
api_url = 'https://api.simplyrets.com/properties'
response = requests.get(api_url, auth=(api_key, api_secret))
response.raise_for_status()
houseData = json.loads(response.text)
#different parameters we need to know
p = houseData['property']
roof = p["roof"]
cooling = p["cooling"]
style = p["style"]
area = p["area"]
bathsFull = p["bathsFull"]
bathsHalf = p["bathsHalf"]
* * *
This is a snippet of the code that I am working with to try and take the
information from the JSON provided by the API and put them into variables that
I can actually use.
I thought that when you loaded it with `json.loads()` it would become a
dictionary.
Yet it is telling me that I cannot do `p = houseData['property']` because
"`list indices must be integers, not str`".
**Am I wrong that houseData should be a dictionary?**
Answer: There are hundreds of properties returned, all of which are in a list.
You'll need to specify which property you want, so for the first one:
p = houseData[0]['property']
|
Obtain string inside curl bracket in python
Question: Suppose I have a string like
{"", "b", "c"}, // we are here
I would like to extract the `{"", "b", "c"}` and/or `"", "b", "c"` part of it.
Is there any simple prescription for it?
Answer: You can use regex - `re.search`:
import re
s = '{"", "b", "c"}, // we are here'
m = re.search(r'{.*}', s)
print(m.group(0))
#'{"", "b", "c"}'
`{.*}` matches every thing within the curly braces and the braces.
|
Receiving python json through sockets with java
Question: I've been following multiple tutorials to connect a java code to python
through sockets.
Sending from java to python works great, with json array. However, I can't
seem to receive things with java. I don't really understand how the listening
should be done. Now I just listen in a 15 second while loop (the python should
send as soon as it receives an input), but I feel like I am doing something
substantially wrong. Maybe somebody has an idea?
client.py:
import socket
import sys
import numpy as np
import json
# Create a TCP/IP socket
sock = socket.socket(socket.AF_INET, socket.SOCK_STREAM)
# Bind the socket to the port
server_address = ('localhost', 10004)
print >>sys.stderr, 'starting up on %s port %s' % server_address
sock.bind(server_address)
# Listen for incoming connections
sock.listen(1)
def mysum(x):
return np.sum(x)
while True:
# Wait for a connection
print >>sys.stderr, 'waiting for a connection'
connection, client_address = sock.accept()
infile = sock.makefile();
try:
print >>sys.stderr, 'connection from', client_address
# Receive the data in small chunks and retransmit it
data = b''
while True:
new_data = connection.recv(16)
if new_data:
# connection.sendall(data)
data += new_data
else:
data += new_data[:]
print >>sys.stderr, 'no more data from', client_address
break
data= data.strip('\n');
print("data well received!: ")
print(data,)
print(np.array(json.loads(data)));
#send a new array back
sendArray = np.array( [ (1.5,2,3), (4,5,6) ] );
print("Preparing to send this:");
print(sendArray);
connection.send(json.dumps(sendArray.tolist()));
except Exception as e:
print(e)
connection.close()
print("closed");
finally:
# Clean up the connection
connection.close()
print("closed");
server.java:
import java.io.*;
import java.net.Socket;
import java.io.BufferedReader;
import java.io.IOException;
import java.io.InputStreamReader;
import java.io.PrintWriter;
import org.json.*;
import java.net.ServerSocket;
public class SocketTest {
public static void main(String[] args) throws IOException {
String hostName = "localhost";
int portNumber = 10004;
try (
//open a socket
Socket clientSocket = new Socket(hostName, portNumber);
BufferedReader in = new BufferedReader(new InputStreamReader(clientSocket.getInputStream()));
PrintWriter out = new PrintWriter(clientSocket.getOutputStream(), true);
) {
System.out.println("Connected");
Double[][] test2 = new Double[5][2];
test2[1][1] = 0.1;
test2[1][0] = 0.2;
test2[2][1] = 0.2;
test2[2][0] = 0.2;
test2[3][1] = 0.1;
test2[3][0] = 0.2;
test2[4][1] = 0.2;
test2[4][0] = 0.2;
test2[0][1] = 0.2;
test2[0][0] = 0.2;
System.out.println("A");
out.println(new JSONArray(test2).toString());
System.out.println("B");
long t = System.currentTimeMillis();
long end = t + 15000;
while (System.currentTimeMillis() < end) {
String response;
while ((response = in.readLine()) != null) {
System.out.println("receiving");
System.out.println( response );
}
}
//listen for input continuously?
//clientSocket.close();
} catch (JSONException e) {
e.printStackTrace();
}
}
}
The output from python is:
data well received!:
('[[0.2,0.2],[0.2,0.1],[0.2,0.2],[0.2,0.1],[0.2,0.2]]',)
[[ 0.2 0.2]
[ 0.2 0.1]
[ 0.2 0.2]
[ 0.2 0.1]
[ 0.2 0.2]]
Preparing to send this:
[[ 1.5 2. 3. ]
[ 4. 5. 6. ]]
closed
waiting for a connection
connection from ('127.0.0.1', 40074)
Output from java:
A
B
The problem is that sendArray = np.array( [ (1.5,2,3), (4,5,6) ] ); is never
received by java. I feel like I am missing something simple to make it
listen... Thanks for any help.
Answer: This is happening because your java code is blocking. See how B is not printed
to your logs? That is because it does't execute since this part is waiting for
a flush command: `out.println(new JSONArray(test2).toString());`. What you
need to do is `out.flush();` so it goes on.
|
How can I define global options with sub-parsers in python argparse?
Question: I'm trying to figure out how to add global option in a sub-parser scenario
with pythons arparse library.
Right now my code looks like this:
def parseArgs(self):
parent_parser = argparse.ArgumentParser(add_help=False)
parent_parser.add_argument('--debug', default=False, required=False,
action='store_true', dest="debug", help='debug flag')
main_parser = argparse.ArgumentParser()
main_parser.add_argument('--debug', default=False, required=False,
action='store_true', dest="debug", help='debug flag')
service_subparsers = main_parser.add_subparsers(title="category",
dest="category")
agent_parser = service_subparsers.add_parser("agent",
help="agent commands", parents=[parent_parser])
return main_parser.parse_args()
This works for the command line `./test --help` and the `--debug` option is
listed as global:
usage: test [-h] [--debug] {agent} ...
optional arguments:
-h, --help show this help message and exit
--debug debug flag
category:
{agent}
agent agent commands
However when I trigger the agent sub-parser with the command line `./test
agent --help` the `--debug` option is now no longer listed as a global option
but as an option for the sub-parser. Also it must now specified as `./test
agent --debug` and `./test --debug agent` no longer works:
usage: test agent [-h] [--debug]
optional arguments:
-h, --help show this help message and exit
--debug debug flag
What I'd like to be able to do is define `--debug` is global so that it can
always be specified for all sub-parsers and appropriately listed as such in
the help output.
Answer: `main_parser` fills in the defaults into `namespace` (`False` for `debug`); if
it encounters `--debug` it changes `debug` to `True`. When it sees the `agent`
string, it calls the subparser, passing it the remaining argument strings, and
the namespace that it has been using.
Now the subparser does the normal parser things - if fills in the defaults for
its arguments, setting `default` to `False`. If it encounters `--debug` in the
remaining strings, it changes that to `True`. Otherwise it leaves it as is.
Once it is done, it passes the namespace back to the main parser, which then
returns it to your code.
So for
myprog.py --debug agent --debug
`namespace(debug=False)` has flipped from False to True to False and back to
True.
This a consequence of sharing the same `dest` for both the main parser (I
don't like the use of 'global' in this context), and the subparser.
There was a bug/issue that tried to change the behavior a bit, passing the
subparser a 'virgin' namespace, and then somehow merging its result with the
main one. But that produced some backward compatibility issues. I could look
it up if needed.
For now, trying to define the same optional in both the main and subparser is
bound to create confusion for you and your user.
If I change the parent to
parent_parser.add_argument('--Debug', action='store_true', help='debug flag')
(no need for default, or the dest if it is sames as the option flag)
the resulting namespace will look like
1721:~/mypy$ python stack37933480.py --debug agent --Debug
Namespace(Debug=True, category='agent', debug=True)
Or I could define
parent_parser.add_argument('--debug', dest='debug1', action='store_true', help='debug flag')
and get:
1724:~/mypy$ python stack37933480.py --debug agent --debug
Namespace(category='agent', debug=True, debug1=True)
Same flag in both places, but different entries in the namespace. After
parsing I could do something like:
args.debug = args.debug or args.debug1
to unify the two flags. Your user will see '--debug' regardless of which help
asks for.
Sorry if the description is a bit long winded, but I think it's important to
understand the behavior first. Then solutions become more apparent.
In this case the use of a parent doesn't complicate the issue. I assume you
are using it just to add this debug to all subparsers.
Another option is to just define `debug` for the main parser. Yes, it will be
missing from the subparsers help, but you can always add a note in the
description.
===================
The subparser definition takes a `prog` parameter. If not given it is defined
base on the main `prog`.
If I add `prog` as:
agent_parser = service_subparsers.add_parser("agent",
prog='myprog.py [--debug] agent',
help="agent commands", parents=[parent_parser])
subparser usage becomes:
1824:~/mypy$ python3 stack37933480.py agent -h
usage: myprog.py [--debug] agent [-h] [--debug]
or I can add that `prog` to the `add_subparsers` definition
service_subparsers = main_parser.add_subparsers(title="category",
prog='myprog.py [--debug]',
dest="category")
Check the code for that method to see how it constructs the default usage
prefix. It includes `main` positionals, but not optionals.
<http://bugs.python.org/issue9351> \- in this patch the original developer
thought that users would expect the subparser definition of an argument should
override the main parser's values and actions. You were, on the other hand,
expecting the main definition to have priority.
<http://bugs.python.org/issue24251> \- but the correction proposed in 9351
caused problems for other users. That's why I think it is better not to define
the same `dest` in the main and sub. It is hard to satisfy everyone's
expectations.
|
Can't get scipy.io.wavfile.read() to work
Question: I am trying to read a .wav file into an array so that I can then plot the
array and do a FFT. I got the file open with the wave module and now I am
struggling. I was advised to use scipy.io.wavfile.read(filename, mmap=False)
but am not having any luck. This function should do exactly what I want it to
do but it isn't. I am running Python 2.7 and maybe that is it. Please help me
figure out how to make this work. The code I have written is below.
import scipy
import wave
harp=wave.open('/Users/williamweiss2/Desktop/Test 2/harp.wav','r')
frames_harp=harp.getnframes()
harp_rate,harp_data=scipy.io.wavfile.read(harp,mmap=False)
## This is the error I am getting when I try to run the program.
\---> harp_rate,harp_data=scipy.io.wavfile.read(harp,mmap=False)
**AttributeError: 'module' object has no attribute 'io'**
Any help would be greatly appreciated. Thanks in advance.
Answer: You have confused [SciPy's WAV
module](http://docs.scipy.org/doc/scipy-0.14.0/reference/io.html#module-
scipy.io.wavfile) with
[Python's](https://docs.python.org/2/library/wave.html). Remove `import wave`,
use `import scipy.io.wavfile`, and call `scipy.io.wavfile.read`.
Example:
>>> import scipy.io.wavfile
>>> FSample, samples = scipy.io.wavfile.read('myfile.wav')
SciPy's module does the job of converting from a byte string to numbers for
you, unlike Python's module. See the linked docs for more details.
|
quantile normalization on pandas dataframe
Question: Simply speaking, how to apply quantile normalization on a large Pandas
dataframe (probably 2,000,000 rows) in Python?
PS. I know that there is a package named rpy2 which could run R in subprocess,
using quantile normalize in R. But the truth is that R cannot compute the
correct result when I use the data set as below:
5.690386092696389541e-05,2.051450375415418849e-05,1.963190184049079707e-05,1.258362869906251862e-04,1.503352476021528139e-04,6.881341586355676286e-06
8.535579139044583634e-05,5.128625938538547123e-06,1.635991820040899643e-05,6.291814349531259308e-05,3.006704952043056075e-05,6.881341586355676286e-06
5.690386092696389541e-05,2.051450375415418849e-05,1.963190184049079707e-05,1.258362869906251862e-04,1.503352476021528139e-04,6.881341586355676286e-06
2.845193046348194770e-05,1.538587781561563968e-05,2.944785276073619561e-05,4.194542899687506431e-05,6.013409904086112150e-05,1.032201237953351358e-05
Edit:
What I want:
Given the data shown above, how to apply quantile normalization following
steps in <https://en.wikipedia.org/wiki/Quantile_normalization>.
I found a piece of code in Python declaring that it could compute the quantile
normalization:
import rpy2.robjects as robjects
import numpy as np
from rpy2.robjects.packages import importr
preprocessCore = importr('preprocessCore')
matrix = [ [1,2,3,4,5], [1,3,5,7,9], [2,4,6,8,10] ]
v = robjects.FloatVector([ element for col in matrix for element in col ])
m = robjects.r['matrix'](v, ncol = len(matrix), byrow=False)
Rnormalized_matrix = preprocessCore.normalize_quantiles(m)
normalized_matrix = np.array( Rnormalized_matrix)
The code works fine with the sample data used in the code, however when I test
it with the data given above the result went wrong.
Since ryp2 provides an interface to run R in python subprocess, I test it
again in R directly and the result was still wrong. As a result I think the
reason is that the method in R is wrong.
Answer: Ok I implemented the method myself of relatively high efficiency.
After finishing, this logic seems kind of easy but, anyway, I decided to post
it here for any one feels confused like I was when I couldn't googled the
available code.
The code is in github: [Quantile
Normalize](https://github.com/ShawnLYU/Quantile_Normalize)
|
Scraping on Python
Question: I wanted to get the caption, no. of likes and comments of the recent 10 images
of a particular user. Using below code I am just able to get the latest one.
Code:
from selenium import webdriver
from bs4 import BeautifulSoup
import json, time, re
phantomjs_path = r'C:\Users\ravi.janjwadia\Desktop\phantomjs-2.1.1-windows\bin\phantomjs.exe'
browser = webdriver.PhantomJS(phantomjs_path)
user = "barackobama"
browser.get('https://instagram.com/' + user)
time.sleep(0.5)
soup = BeautifulSoup(browser.page_source, 'html.parser')
script_tag = soup.find('script',text=re.compile('window\._sharedData'))
shared_data = script_tag.string.partition('=')[-1].strip(' ;')
result = json.loads(shared_data)
print(result['entry_data']['ProfilePage'][0]['user']['media']['nodes'][0]['caption'])
Result: LAST CALL: Enter for a chance to meet President Obama this summer
before tonight's deadline. → Link in profile.
Answer: In your code below, you're only retrieving the first node (which is the first
image).
print(result['entry_data']['ProfilePage'][0]['user']['media']['nodes'][0]['caption'])
To get the info of the recent 10 images of the user try this instead.
recent_ten_nodes = result['entry_data']['ProfilePage'][0]['user']['media']['nodes'][:10]
To only print the captions, number of likes and comments do this.
for node in recent_ten_nodes:
print node['caption']
print node['likes']['count']
print node['comments']['count']
For storing of these values, it's up to you to decide how you want to store
them.
|
exporting Python model results
Question: Hi I’ve launched a random forest over a dataset imported as df. Now I would
like to export both results (0-1 prediction) and predicted probabilities ( a
two dimensions array) and match them to my dataset df. Is that possible? Until
now I figured out how to export in a separate way to csv. And yes, I am not a
pandas expert yet. Any hint?
# Import the `RandomForestClassifier`
from sklearn.ensemble import RandomForestClassifier
# Create the target and features numpy arrays:
target = df["target"].values
features =df[["var1",
"var2","var3","var4","var5"]]
features_forest = features
# Building and fitting my_forest
forest = RandomForestClassifier(max_depth = 10, min_samples_split=2, n_estimators = 200, random_state = 1)
my_forest = forest.fit(features_forest, target)
# Print the score of the fitted random forest
print(my_forest.score(features_forest, target))
print(my_forest.feature_importances_)
results = my_forest.predict(features)
print(results)
predicted_probs = forest.predict_proba(features)
#predicted_probs = my_forest.predict_proba(features)
print(predicted_probs)
id_test = df['ID_CONTACT']
pd.DataFrame({"id": id_test, "relevance": results, "probs": predicted_probs }).to_csv('C:\Users\me\Desktop\python\data\submission.csv',index=False)
pd.DataFrame(predicted_probs).to_csv('C:\Users\me\Desktop\python\data\submission_2.csv',index=False)
Answer: You should be able to
df['results] = results
df = pd.concat([df, pd.DataFrame(predicted_probs, columns=['Col_1', 'Col_2'])], axis=1)
|
line_profiler and kernprof does not work installed from pip and repro
Question: I have tried what has been suggested. On the simplest of code, I get the
familiar error that others have mentioned. The error is:
@profile
NameError: name 'profile' is not defined
on the minimal code:
@profile
def hello():
print('Testing')
hello()
from the command line:
kernprof -l hello.py
I have imported `future` and I have even installed from the distribution
outside of my virtualenv. This was a previous
[suggestion](https://github.com/rkern/line_profiler/pull/25), as was importing
future module.
So
import future
import cProfile
@profile
def hello():
print('Testing')
hello()
also gives the same error. I cannot get this line profiler to work with pip or
even from the raw repository. My versions are:
pip 8.1.2
python 2.7
Answer: I fixed this my cloning the code from <https://github.com/rkern/line_profiler>
and rebuilding and then calling the kernprof from the binary install location.
|
Choose one key arbitrarily in a dictionary without iteration
Question: I just wanna make sure that in Python dictionaries there's no way to get just
a key (with no specific quality or relation to a certain value) but doing
iteration. As much as I found out you have to make a list of them by going
through the whole dictionary in a loop. Something like this:
list_keys=[k for k in dic.keys()]
The thing is I just need an arbitrary key if the dictionary is not empty and
don't care about the rest. I guess iterating over a long dictionary in
addition to creation of a long list for just randomly getting a key is a whole
lot overhead, isn't it? Is there a better trick somebody can point out?
Thanks
Answer: You can use `random.choice`
rand_key = random.choice(dict.keys())
And this will only work in python 2.x, in python 3.x dict.keys returns an
iterator, so you'll have to do cast it into a list -
rand_key = random.choice(list(dict.keys()))
So, for example -
import random
d = {'rand1':'hey there', 'rand2':'you love python, I know!', 'rand3' : 'python has a method for everything!'}
random.choice(list(d.keys()))
Output -
rand1
|
Create XML file with python?
Question: Here is my code:
import xml.etree.ElementTree as ET
root = ET.Element("rss", version="2.0", xmlns:excerpt="http://wordpress.org/export/1.2/excerpt/", xmlns:content="http://purl.org/rss/1.0/modules/content/", xmlns:wfw="http://wellformedweb.org/CommentAPI/", xmlns:dc="http://purl.org/dc/elements/1.1/", xmlns:wp="http://wordpress.org/export/1.2/")
ET.dump(root)
This is xml file i am trying to create:
<rss version="2.0"
xmlns:excerpt="http://wordpress.org/export/1.2/excerpt/"
xmlns:content="http://purl.org/rss/1.0/modules/content/"
xmlns:wfw="http://wellformedweb.org/CommentAPI/"
xmlns:dc="http://purl.org/dc/elements/1.1/"
xmlns:wp="http://wordpress.org/export/1.2/"
>
this is very basic part of actual xml file but i am unable to correctly create
it with elementtree. Until version number it works correctly but when i add
"xmlns:something" it does not work. I am very new to xml so i have no idea
even google is not able to help me understand.
**NOTE: Please do tell me if lxml is more easy or this ElementTree. Because I
have previously used lxml for xpath and css elements.**
Answer: Python identifiers are not allowed to have colons inside. ElementTree allows
to pass an attribute dictionary by the key `attrib`:
`ET.element("rss", attrib={"xmlns:excerpt":"http..."}`
|
Subprocess doesn't respect arguments when using multiprocessing
Question: The main objective here is to create a daemon-spawning function. The daemons
need to run arbitrary programs (i.e. use `subprocess`).
What I have so far in my `daemonizer.py` module is:
import os
from multiprocessing import Process
from time import sleep
from subprocess import call, STDOUT
def _daemon_process(path_to_exec, std_out_path, args, shell):
with open(std_out_path, 'w') as fh:
args = (str(a) for a in args)
if shell:
fh.write("*** LAUNCHING IN SHELL: {0} ***\n\n".format(" ".join([path_to_exec] + list(args))))
retcode = call(" ".join([path_to_exec] + list(args)), stderr=STDOUT, stdout=fh, shell=True)
else:
fh.write("*** LAUNCHING WITHOUT SHELL: {0} ***\n\n".format([path_to_exec] + list(args)))
retcode = call([path_to_exec] + list(args), stderr=STDOUT, stdout=fh, shell=False)
if retcode:
fh.write("\n*** DAEMON EXITED WITH CODE {0} ***\n".format(retcode))
else:
fh.write("\n*** DAEMON DONE ***\n")
def daemon(path_to_executable, std_out=os.devnull, daemon_args=tuple(), shell=True):
d = Process(name='daemon', target=_daemon_process, args=(path_to_executable, std_out, daemon_args, shell))
d.daemon = True
d.start()
sleep(1)
When trying to run this in bash (**This will create a file called`test.log` in
your current directory.**):
python -c"import daemonizer;daemonizer.daemon('ping', std_out='test.log', daemon_args=('-c', '5', '192.168.1.1'), shell=True)"
It correctly spawns a daemon that launches `ping` _but it doesn't respect the
arguments passed_. This is true if shell is set to `False` as well. The log-
file clearly states that it attempted to launch it with the arguments passed.
As a proof of concept creating the following executable:
echo "ping -c 5 192.168.1.1" > ping_test
chmod +x ping_test
The following works as intended:
python -c"import daemonizer;daemonizer.daemon('./ping_test', std_out='test.log', shell=True)"
If I test the same `call` code outside of the `multiprocessing.Process`-target
it does work as expected.
So how do I fix this mess so that I can spawn processes with arguments?
I'm open to entirely different structures and modules, but they should be
included among the standard ones and be compatible with python 2.7.x. The
requirement is that the the `daemon` function should be callable several times
asynchronously within a script and produce a daemon each and their target
processes should be able to end up on different CPUs. Also the scripts need to
be able to end without affecting the spawned daemons of course.
As a bonus, I noticed I needed to have a `sleep` for the spawning to work at
all else the script terminates too fast. Any way to get around that arbitrary
hack and/or how long do I really need to have it wait to be safe?
Answer: Your arguments are being "used up" by the printing of them!
First, you do this:
args = (str(a) for a in args)
That creates a generator, not a list or tuple. So when you later do this:
list(args)
That consumes the arguments, and they will not be seen a second time. So you
do this again:
list(args)
And get an empty list!
You could fix this by commenting out your print statements, but much better
would be to simply create a list in the first place:
args = [str(a) for a in args]
Then you can use `args` directly and not `list(args)`. And it will always have
the arguments inside.
|
Python MYSql.connector will not insert data
Question: I am trying to insert some data into a a database I am making and it will not
insert. I literally have used the same insert method on other code and it
still seems to work, however this one refuses to. Help!
from Bio import Entrez
from sys import exit
Entrez.email = "[email protected]" # Always tell NCBI who you are
sranumber = raw_input("Input SRA number here")
sranumber2= raw_input("re-type SRA number here")
while True:
if sranumber != sranumber2:
print "SRA numbers do not match"
sranumber2 = raw_input("Please re-type sra number to match intitial sra number")
continue
else:
break
print "SRA ID:" + sranumber
#answer = raw_input("Are you sure this is the sra number you wish to use? Type Y/N")
while True:
answer = raw_input("Are you sure this is the sra number you wish to use? Type Y/N")
if answer == "Y":
print "Let's do it!"
break
elif answer == "y":
print "Let's do it!"
break
elif answer == "yes":
print "Let's do it!"
break
elif answer == "YES":
print "Let's do it!"
break
elif answer == "N":
exit()
else:
print "Not a valid answer"
search = Entrez.esearch(term = sranumber, db = "SRA", retmode = "xml")
record = Entrez.read(search, validate = False)
newstuff = record
#print newstuff
for j in record:
if j == "WarningList":
newstuff = record['WarningList']
#print newstuff
poop = newstuff
for item in poop:
if item == "OutputMessage":
poop = poop['OutputMessage']
#print poop
crap = ['Wrong UID' + " " + sranumber]
cool = "'"+crap[0]+"'"
#print cool
continuity = ''
for j in poop:
if j == 'No items found.' or j == cool:
print "[-] This is not a valid SRA identity"
continuity = 'done'
if continuity == 'done':
exit()
print "[+] This is a valid SRA identity"
print "SRA ID:" + sranumber
condition = raw_input("Type in the condition of your ngs_data here")
condition2 = raw_input("re-type the condition of your ngs_data here")
print condition
while True:
if condition != condition2:
print "Conditions do not match!"
condition2 = raw_input("Please retype condition here to match first condition")
else:
break
print "just dropped in to check on what condition my condition was in"
stuff = []
stuff.append(sranumber)
stuff.append(condition)
stuff2 = '+'.join(stuff)
print stuff2
stuff3 = stuff2.split('+')
print stuff3
experiment = [tuple(stuff3)]
print experiment
from mysql.connector import MySQLConnection, Error
from python_mysql_dbconfig import read_db_config
def insert_books(experiment):
query = "INSERT IGNORE INTO organisms(sra#, condition) " \
"VALUES(%s,%s)"
try:
db_config = read_db_config()
conn = MySQLConnection(**db_config)
cursor = conn.cursor()
cursor.executemany(query, experiment)
conn.commit()
except Error as e:
print('Error:', e)
finally:
cursor.close()
conn.close()
def main():
insert_books(experiment)
if __name__ == '__main__':
main()
Answer: I ended up just doing it this way and it finally worked. I am unsure why it
did not work before but I believe that it had something to do with the
formatting of how i typed in the columns.
from mysql.connector import MySQLConnection, Error
from python_mysql_dbconfig import read_db_config
cnx = mysql.connector.connect(user='root',password='*****',database = 'new')
cursor = cnx.cursor()
addstuff= ("INSERT IGNORE INTO experiment (`sra`, `condition`) VALUES(%s,%s)")
cursor.execute(addstuff,stuff3)
cnx.commit()
cursor.close()
cnx.close()
|
Self scheduled Python script
Question: I'm writing an Python script that calls a function that exports and imports
spaces from the wiki tool Confluence. It has to run everyday, but we can't use
cron so i'm looking for a way to schedule it by her self.
I've created the following. I can already schedule it for the next day but not
for the following day again..
#!/usr/bin/python
from __future__ import print_function
from sys import argv
from datetime import datetime
from threading import Timer
import sys,os,subprocess
import getpass
from subprocess import PIPE,Popen
date = (os.popen("date +\"%d-%m-%y - %T\""))
x=datetime.today()
y=x.replace(day=x.day+1, hour=13, minute=56, second=0, microsecond=0)
delta_t=y-x
secs=delta_t.seconds+1
def runExport():
# HERE IS ALL THE CODE THAT HAS TO RUN EVERYDAY
t = Timer(secs, runExport)
t.start()
Could somebody please help me out? The script has to run everyday for example
an 05.00 am.
Version of python is 2.6.6. And sadly enough no option to import a module..
Thank you!
Answer: I would suggest using something a bit simplilar then what you are doing there.
There is a simple scheduling library in python called sched, which can be
found here: <https://docs.python.org/2/library/sched.html>
A simple example of using this for your implement:
import sched, time
s = sched.scheduler(time.time, time.sleep)
delay_seconds = 5
def print_time():
print time.time()
s.enter(delay_seconds,1,print_time,argument=())
s.enter(delay_seconds,1,print_time,argument=())
s.run()
This code will run every 5 seconds, and print out the current time. just
change delay_seconds to the delay you want your interval to run, and your
done.
|
How to convert utf-8 values within a string to its correct character using python?
Question: I would like to convert 'Lawrence_Warbasse_-_Dauphin%C3%A9_2013.JPG' to
'Lawrence_Warbasse_-_Dauphiné_2013.JPG' usig python
Answer: You can use `urllib` to decode this:
import urllib
url = 'Lawrence_Warbasse_-_Dauphin%C3%A9_2013.JPG'
print urllib.unquote(url).decode('utf8')
The output I get is:
Lawrence_Warbasse_-_Dauphiné_2013.JPG
|
Updating records with Python 3 through API on Rails 4 application
Question: I'm working on a Rails 4 / mongoid application which needs to expose APIs for
other applications and scripts. I need to be able to update documents in one
of the models through an API with Python 3 script. I'm a bit fresh with Python
hence asking for help here.
I already found out how to query Rails APIs with Python 3 and urllib but
struggling with updates. I was trying to go through Python 3.5
[docs](https://docs.python.org/3/howto/urllib2.html) for urllib2 but
struggling to apply this to my script.
What goes to `data` and how to add authentication token to `headers`, which in
`curl` would look something like this
-H 'Authorization:Token token="xxxxxxxxxxxxxxxx"'
-X POST -d "name=A60E001&status=changed"
I would greatly appreciate if somebody explained how to, for example, update
`status` based on `name` (name is not unique yet but will be). My biggest
challenge is the Python side. Once I have the data in params on Rails side I
think I can handle it. I think.
I included my model and update action from the controller below.
`app/models/experiment.rb`
class Experiment
include Mongoid::Document
include Mongoid::Timestamps
field :name, type: String
field :status, type:String
end
`app/controllers/api/v1/experiments_controller.rb`
module Api
module V1
class ExperimentsController < ActionController::Base
before_filter :restrict_access
...
def update
respond_to do |format|
if @expt_proc.update(expt_proc_params)
format.json { render :show, status: :ok, location: @expt_proc }
else
format.json { render json: @expt_proc.errors, status: :unprocessable_entity }
end
end
end
...
private
def restrict_access
authenticate_or_request_with_http_token do |token, options|
ApiKey.where(access_token: token).exists?
end
end
...
Answer: I figured out who to send a PATCH request with Python 3 and update the the
record's `status` by `name`.
Thanks to [this](http://stackoverflow.com/questions/6853050/how-do-i-make-a-
patch-request-in-python#answer-7112444) post found out about `requests`
module. I used [requests.patch](http://docs.python-
requests.org/en/master/user/quickstart/) to update the record and it works
great.
`python code`
import requests
import json
url = 'http://0.0.0.0:3000/api/v1/update_expt_queue.json'
payload = {'expt_name' : 'myExperiment1', 'status' : 'finished' }
r = requests.patch(url, payload)
There are two problems remaining:
1. How to add headers to this request which would allow me to go through token based authentication. At the moment it's disabled. [request.patch](http://docs.python-requests.org/en/master/api/#requests.patch) only takes 2 parameters, it doesn't take `headers` parameter.
2. How to access the JSON which is sent in response by Rails API.
|
Stuck Coding a Python web crawler using BeautifulSoup
Question: regarding this video by bucky roberts on programming a webcrawler in python:
<https://www.youtube.com/watch?v=sVNJOiTBi_8>
Here is my question:
If i want to crawl a particular item but it isnt inside of < a> < /a>
how can i do it?
for ex. I inspected the site and found this is the code of the the info i want
(i want the href and the title just like in the bucky's video):
< td headers="categorylist_header_title" class="list-title" > < a href="BLABLABLABLA.HTML" > blablabliblableblu < /a> < /td>
Following the instruction i notice that the element "class="list-title" inside
of is the one i need to crawl, but when i use for link in
`soup.findAll('a',{'class': 'list-title'}):` it does not work, i think because
it isnt inside of < a>, how can crawl that information if we dont have a
particular info inside of and the element we are looking for is outside in
this case in ??
Hope i explain myself im just started programming this week, if you know where
can i read about this please give me a source so i dont ask stupid questions
again.
Here is the source code of this tool, currently it is just getting all the
links if anyone want to give it a try:
import requests
from bs4 import BeautifulSoup
def trade_spider(max_pages):
page = 1
while page <= max_pages:
url = 'http://www.portaloaca.com/pensamiento-libertario/libros-anarquistas'
source_code = requests.get(url)
plain_text = source_code.text
soup = BeautifulSoup(plain_text, "html.parser")
for link in soup.findAll('a'):
href = 'http://www.portaloaca.com/pensamiento-libertario/libros-anarquistas'+ link.get('href')
print(href)
trade_spider(1)
Answer: It is the `td` that has the class `list-title`, not the anchor tag. You just
need to select the _td_ tags with the class `list-title` inside the table rows
then extract the href from the anchor inside each td:
from bs4 import BeautifulSoup
import requests
from urlparse import urljoin
soup = BeautifulSoup(requests.get("http://www.portaloaca.com/pensamiento-libertario/libros-anarquistas").content)
base = 'http://www.portaloaca.com/pensamiento-libertario/libros-anarquistas'
print( [urljoin(base,td.a["href"]) for td in soup.find_all("td", {"class":"list-title"})])
Which would give you:
['http://www.portaloaca.com/pensamiento-libertario/libros-anarquistas/11899-hemeroteca-y-biblioteca-libertarias-3-1.html', 'http://www.portaloaca.com/pensamiento-libertario/libros-anarquistas/11809-distribuidora-anarquista-polaris-nueva-edicion-negros-presagios-politica-anarquista-en-la-era-del-colapso-de-uri-gordon.html', 'http://www.portaloaca.com/pensamiento-libertario/libros-anarquistas/11789-libro-la-crisis-del-socialismo-jose-garcia-pradas.html', 'http://www.portaloaca.com/pensamiento-libertario/libros-anarquistas/11618-libro-nuestro-planeta-de-elisee-reclus.html', 'http://www.portaloaca.com/pensamiento-libertario/libros-anarquistas/11602-hemeroteca-y-biblioteca-libertarias-2-0.html', 'http://www.portaloaca.com/pensamiento-libertario/libros-anarquistas/11493-descarga-la-lucha-contra-el-estado-la-bestia-de-la-propiedad-y-el-origen-del-capital-moderno-en-pie-de-guerra-contra-la-civilizacion.html', 'http://www.portaloaca.com/pensamiento-libertario/libros-anarquistas/11461-descarga-libros-contra-los-pastores-contra-los-rebanos-una-declaracion-de-guerra.html', 'http://www.portaloaca.com/pensamiento-libertario/libros-anarquistas/11396-libro-un-dragon-en-el-reino-de-orb-de-federico-zenoni-cuento-ilustrado-para-descargar.html', 'http://www.portaloaca.com/pensamiento-libertario/libros-anarquistas/11283-paginas-de-lucha-cotidiana-libro-de-errico-malatesta-para-descarga.html', 'http://www.portaloaca.com/pensamiento-libertario/libros-anarquistas/11204-descargar-actualizaciones-ex-nihilo.html', 'http://www.portaloaca.com/pensamiento-libertario/libros-anarquistas/11192-libro-al-diablo-con-la-cultura-herbert-read.html', 'http://www.portaloaca.com/pensamiento-libertario/libros-anarquistas/10966-libro-bakunin-escritos-de-filosofia-politica-tomo-ii-partes-iii-y-iv.html', 'http://www.portaloaca.com/pensamiento-libertario/libros-anarquistas/10842-libro-palabras-de-un-rebelde-piotr-kropotkin.html', 'http://www.portaloaca.com/pensamiento-libertario/libros-anarquistas/10512-libro-la-voluntad-del-pueblo-democracia-y-anarquia-eduardo-colombo.html', 'http://www.portaloaca.com/pensamiento-libertario/libros-anarquistas/10485-libro-paginas-de-lucha-cotidiana-primera-parte-temas-del-comunismo-anarquico-errico-malatesta.html', 'http://www.portaloaca.com/pensamiento-libertario/libros-anarquistas/10474-los-tiempos-nuevos-piotr-kropotkin.html', 'http://www.portaloaca.com/pensamiento-libertario/libros-anarquistas/10416-libro-el-humanisferio-utopia-anarquica-joseph-dejacque.html', 'http://www.portaloaca.com/pensamiento-libertario/libros-anarquistas/10366-libro-primero-de-mayo-lectura-para-el-dia-de-las-trabajadoras-y-trabajadores.html', 'http://www.portaloaca.com/pensamiento-libertario/libros-anarquistas/10339-libro-el-espacio-politico-de-la-anarquia-esbozos-para-una-filosofia-politica-del-anarquismo.html', 'http://www.portaloaca.com/pensamiento-libertario/libros-anarquistas/10314-libro-las-politicas-de-la-ecologia-social-municipalismo-libertario-janet-biehl-y-murray-bookchin.html', 'http://www.portaloaca.com/pensamiento-libertario/libros-anarquistas/10239-libro-el-principio-federativo-pierre-joseph-proudhon.html', 'http://www.portaloaca.com/pensamiento-libertario/libros-anarquistas/10187-libro-malatesta-pensamiento-y-accion-revolucionarios.html', 'http://www.portaloaca.com/pensamiento-libertario/libros-anarquistas/10112-50-titulos-para-una-biblioteca-basica-del-anarquismo-latinoamericano.html', 'http://www.portaloaca.com/pensamiento-libertario/libros-anarquistas/10102-libro-la-libertad-entre-la-historia-y-la-utopia-luce-fabbri.html', 'http://www.portaloaca.com/pensamiento-libertario/libros-anarquistas/10028-libro-de-poesia-jardin-de-acracia.html', 'http://www.portaloaca.com/pensamiento-libertario/libros-anarquistas/10008-libro-anarquismo-trashumante.html', 'http://www.portaloaca.com/pensamiento-libertario/libros-anarquistas/9984-libro-fuera-politica-anselmo-lorenzo.html', 'http://www.portaloaca.com/pensamiento-libertario/libros-anarquistas/9969-libro-cartas-y-textos-de-librado-rivera.html', 'http://www.portaloaca.com/pensamiento-libertario/libros-anarquistas/9920-libro-la-anarquia-manuel-gonzalez-prada.html', 'http://www.portaloaca.com/pensamiento-libertario/libros-anarquistas/9853-libro-tacticas-revolucionarias-mijail-bakunin.html', 'http://www.portaloaca.com/pensamiento-libertario/libros-anarquistas/9847-libro-discursos-de-ricardo-flores-magon.html', 'http://www.portaloaca.com/pensamiento-libertario/libros-anarquistas/9799-libro-anselmo-lorenzo-un-militante-proletario-en-el-ojo-del-huracan.html', 'http://www.portaloaca.com/pensamiento-libertario/libros-anarquistas/9789-libro-el-mundo-nuevo-louise-michel.html', 'http://www.portaloaca.com/pensamiento-libertario/libros-anarquistas/9751-libro-ideario-ricardo-mella.html', 'http://www.portaloaca.com/pensamiento-libertario/libros-anarquistas/9700-libro-problemas-del-sindicalismo-y-del-anarquismo-joan-peiro.html', 'http://www.portaloaca.com/pensamiento-libertario/libros-anarquistas/9679-libro-hacia-la-emancipacion-tactica-de-avance-obrero-en-la-lucha-por-el-ideal-anselmo-lorenzo.html', 'http://www.portaloaca.com/pensamiento-libertario/libros-anarquistas/9664-libro-forjando-un-mundo-libre-ricardo-mella.html', 'http://www.portaloaca.com/pensamiento-libertario/libros-anarquistas/9633-libro-via-libre-el-trabajador-su-ideal-emancipador-desviaciones-politicas-y-economicas-anselmo-lorenzo.html', 'http://www.portaloaca.com/pensamiento-libertario/libros-anarquistas/9625-folleto-el-anarquismo-en-el-movimiento-obrero-errico-malatesta.html', 'http://www.portaloaca.com/pensamiento-libertario/libros-anarquistas/9547-libro-en-anarquia-novela-camille-pert.html', 'http://www.portaloaca.com/pensamiento-libertario/libros-anarquistas/9495-libro-la-anarquia-triunfante-anselmo-lorenzo.html', 'http://www.portaloaca.com/pensamiento-libertario/libros-anarquistas/9472-folleto-el-trabajo-los-trabajadores-y-el-anarquismo.html', 'http://www.portaloaca.com/pensamiento-libertario/libros-anarquistas/9454-libro-teresa-claramunt-la-virgen-roja-barcelonesa-biografia-y-escritos.html', 'http://www.portaloaca.com/pensamiento-libertario/libros-anarquistas/9416-libro-la-anarquia-a-traves-de-los-tiempos-max-nettlau.html', 'http://www.portaloaca.com/pensamiento-libertario/libros-anarquistas/9398-libro-seamos-rebeldes-folletos-y-otros-escritos-teodoro-antilli.html', 'http://www.portaloaca.com/pensamiento-libertario/libros-anarquistas/9375-libro-utopias-antiguas-y-modernas-angel-j-cappelletti.html', 'http://www.portaloaca.com/pensamiento-libertario/libros-anarquistas/9365-libro-del-amor-modo-de-accion-y-finalidad-social-ricardo-mella.html', 'http://www.portaloaca.com/pensamiento-libertario/libros-anarquistas/9358-libro-contra-la-ignorancia-anselmo-lorenzo.html', 'http://www.portaloaca.com/pensamiento-libertario/libros-anarquistas/9316-libro-consideraciones-filosoficas-sobre-el-fantasma-divino-sobre-el-mundo-real-y-sobre-el-hombre-mijail-bakunin.html', 'http://www.portaloaca.com/pensamiento-libertario/libros-anarquistas/9270-libro-el-lenguaje-libertario-christian-ferrer.html']
|
SFrame Kmeans - Covert to Int, Float, Dict
Question: I'm preparting data to run KMEAMS from Graphlab, and am running into the
following error:
tmp = data.select_columns(['a.item_id'])
tmp['sku'] = tmp['a.item_id'].apply(lambda x: x.split(','))
tmp = tmp.unpack('sku')
kmeans_model = gl.kmeans.create(tmp, num_clusters=K)
Feature 'sku.0' excluded because of its type. Kmeans features must be int, float, dict, or array.array type.
Feature 'sku.1' excluded because of its type. Kmeans features must be int, float, dict, or array.array type.
Here are the current datatypes of each column:
a.item_id str
sku.0 str
sku.1 str
If I can get the datatype from str to int I think it should work. However,
using SFrames is a more tricky than standard python libraries. Any help
getting there is appreciated.
Answer: The kmeans model does allow features in _dictionary_ form, just not in list
form. This is slightly different from what you've got now, because the
dictionary loses the order of your SKUs, but in terms of model quality I
suspect it actually makes more sense. They key function is `count_words`, in
the text analytics toolkit.
<https://dato.com/products/create/docs/generated/graphlab.text_analytics.count_words.html>
import graphlab as gl
sf = gl.SFrame({'item_id': ['abc,xyz,cat', 'rst', 'abc,dog']})
sf['sku_count'] = gl.text_analytics.count_words(sf['item_id'], delimiters=[','])
model = gl.kmeans.create(sf, num_clusters=2, features=['sku_count'])
print model.cluster_id
+--------+------------+----------------+
| row_id | cluster_id | distance |
+--------+------------+----------------+
| 0 | 1 | 0.866025388241 |
| 1 | 0 | 0.0 |
| 2 | 1 | 0.866025388241 |
+--------+------------+----------------+
[3 rows x 3 columns]
|
Numpy linspace and plotting, ValueError: an array element with a sequence
Question: I was trying to get 60 lines joining center of the circle to 60 different
equispaced points on the circumference in the following program,
import matplotlib.pyplot as plt
import numpy as np
figure = plt.figure(figsize=(10, 10))
theta = np.linspace(0, 2 * np.pi, 60)
r = 3.0
x1 = r * np.cos(theta)
y1 = r * np.sin(theta)
plt.plot(x1, y1, color='blue')
plt.plot([0, x1], [0, y1], color='gray')
plt.axis([-4, 4, -4, 4])
plt.grid(True)
figure.tight_layout()
figure.savefig('test.png', facecolor='white', edgecolor='black')
It gives the following error,
$ python test.py
Traceback (most recent call last):
File "test.py", line 12, in <module>
plt.plot([0, x1], [0, y1], color='gray')
File "/usr/lib/pymodules/python2.7/matplotlib/pyplot.py", line 2987, in plot
ret = ax.plot(*args, **kwargs)
File "/usr/lib/pymodules/python2.7/matplotlib/axes.py", line 4137, in plot
for line in self._get_lines(*args, **kwargs):
File "/usr/lib/pymodules/python2.7/matplotlib/axes.py", line 317, in _grab_next_args
for seg in self._plot_args(remaining, kwargs):
File "/usr/lib/pymodules/python2.7/matplotlib/axes.py", line 288, in _plot_args
y = np.atleast_1d(tup[-1])
File "/usr/lib/python2.7/dist-packages/numpy/core/shape_base.py", line 49, in atleast_1d
ary = asanyarray(ary)
File "/usr/lib/python2.7/dist-packages/numpy/core/numeric.py", line 512, in asanyarray
return array(a, dtype, copy=False, order=order, subok=True)
ValueError: setting an array element with a sequence.
If I use some constant value for example `plt.plot([0, 0], [0, r],
color='gray')` instead of `plt.plot([0, x1], [0, y1], color='gray')` it works.
It seems with `numpy.linspace` such plot is not possible.
I found similar question [ValueError: setting an array element with a
sequence](http://stackoverflow.com/q/4674473/2026294), but did not help me. I
am new to python, please bear with me.
Answer: The x and y elements of your `plot()` command need to have the same number of
elements. Replace the line
plt.plot([0, x1], [0, y1], color='gray')
with the following:
plt.plot([np.zeros(60,), x1], [np.zeros(60,), y1], color='gray')
the result looks like: [](http://i.stack.imgur.com/bh1ln.png)
|
GenButton alignment wxpython
Question: i am struggling to find an answer to the alignment of my GenButton, i have
looked through the docs on wxpython website and cant see any option in there
to address this, hopefully one of you guys can point me in the right
direction, as i need to align my buttons to the center of the catpanel, here
is my code so far.
import wx, os, os.path
from wx.lib.buttons import GenButton
class Home(wx.Frame):
def __init__(self, parent):
wx.Frame.__init__(self, parent, id=wx.ID_ANY, size=(1024, 576),style=wx.NO_BORDER)
self.SetBackgroundColour(wx.Colour(107, 109, 109))
self.catpanel()
def catpanel(self):
homepanel = wx.Panel(self, wx.ALIGN_CENTER)
homepanel.BackgroundColour = (86, 88, 88)
homepanel.SetSize((1024, 40))
GenButton(homepanel, id=wx.ID_ANY, label="Home", style=wx.NO_BORDER, size=(-1, 40))
self.Centre()
self.Show()
if __name__ == '__main__':
app = wx.App()
Home(None)
app.MainLoop()
`
i am using windows, i also understand this can be acheived by using `pos =
wx.DefaultPosition` but would like a more accurate wx way of doing this, could
someone inspect the code and let me know if i am doing it right as i am new to
wxpython / python in general
tnx
Answer: GUI toolkits tend to be smart about size of controls: You specified the height
of our `GenButton` instance but not the width (`size=(-1, 40)`). If you adapt
the button width to the parent panel width, you will get what you want
(`size=(1024, 40)`).
However, this is not what you should do, because you would use sizers. With
the style `wx.NO_BORDER` for the `wx.Frame` you seem to have hit another
snafu, where sizers do not work as expected together with `GenButton`.
class Home(wx.Frame):
def __init__(self, parent):
wx.Frame.__init__(self, parent, id=wx.ID_ANY, style=wx.NO_BORDER)
self.SetBackgroundColour(wx.Colour(107, 109, 109))
homepanel = wx.Panel(self, -1)
sz = wx.BoxSizer(wx.VERTICAL)
btn = GenButton(homepanel, id=wx.ID_ANY, label="Home", style=wx.NO_BORDER, size=(-1, 40))
sz.Add(btn, 0, wx.EXPAND|wx.ALL, 0)
# Dummy placeholder with heigth already set
sz.Add((-1, 400), 1)
homepanel.SetSizer(sz)
# try to uncomment next line: without resizing
# the window will not be layouted
self.SetSize((1024, -1))
self.Centre()
self.Show()
|
Concatinate to pandas data frame from OrderedDict - Python equivalent to R´s do.call(merge, list)
Question: I have an ordered dictionary `"my_dict`", which gets filled with `n` time
series during a loop. All `n` time series have the same length and are
1-dimensional. They all have a datetime index.
In R I just run `do.call(merge, my_dict)`, where `my_dict` is a `list()` and I
obtain an `m x n xts` object with a single index.
In Python however `pandas.concat(my_dict, axis = 1)` returns a single indexed
dataframe (which is good) but all columns from the seconds column on are NaNs.
Whats the correct command?
Answer: A `pandas.DataFrame` can be constructed directly from a dictionary so all you
need to do is pass `my_dict` to the `pandas.DataFrame` constructor (i.e.,
`pandas.DataFrame(my_dict)`). For example:
import pandas
from collections import OrderedDict
import numpy as np
m = 10
n = 5
index = pandas.date_range('1/1/2015', periods=m, freq='D')
my_dict = OrderedDict()
for i in range(1, n+1):
my_dict['var_%d' % i] = pandas.Series(np.random.randn(index.shape[0]), index=index)
print(pandas.DataFrame(my_dict))
var_1 var_2 var_3 var_4 var_5
2015-01-01 0.562952 0.708099 0.488981 -1.360356 -0.036179
2015-01-02 -0.481410 0.604777 0.233426 -0.784103 1.879795
2015-01-03 1.188333 0.299547 -0.578365 0.882410 0.919328
2015-01-04 -1.002273 0.794856 -0.527205 1.474524 -0.798646
2015-01-05 -0.025225 2.246476 -0.460735 0.272014 -0.061749
2015-01-06 -1.304233 1.204737 0.040275 0.293035 0.831555
2015-01-07 -0.934804 0.922393 0.633133 1.064837 -0.154307
2015-01-08 0.440650 0.235624 0.765399 0.306628 -0.274465
2015-01-09 1.637787 -0.158231 -0.643112 -1.128660 0.393349
2015-01-10 0.145501 -0.667916 1.094961 -1.669178 0.447021
|
Imported module goes out of scope (unbound local error)
Question: I am getting a strange "unbound local" error in a python package that seems to
defy all logic. I can't get a MWE to reproduce it, but will try to explain
succinctly and hope that someone might be able to offer some explanation of
what is going on.
For the sake of this example `module` is a package I developed, and `Model` is
a class within `module`. The definition of the `Model` class (model.py) looks
like:
import module
class Model:
def __init__(self):
print module
def run(self):
print module
Now, when I instantiate a `Model` in a script like this:
from model import Model
m = Model()
m.run()
`module` prints successfully within the `__init__`, but I get an unbound local
error within the `run` function.
I attempted to diagnose this with pdb, which is where things got really weird,
because if I add a pdb trace immediately prior to the `print module` line in
the `run()` function, then I can successfully run `print module` without an
unbound local error, but if I step to the next line then it throws the error.
How can `module` be in the scope of `__init__()`, and in the scope of pdb, but
not in the scope of `run()`?
I know this is not ideal since there is no MWE, but I cannot seem to reproduce
this outside the context of the full code. I am hoping that someone will have
an idea of what might possibly be going on and suggest some strategies to
debug further.
Answer: Apparently you have a local variable named `module` somewhere in the function
`run`. For example, the following code will throw `UnboundLocalError`
import sys
def run():
print sys
sys = None
run()
Here `sys = None` introduces a local name that shadows the imported `sys`
inside `run` and at the time `print` invoked it is not yet defined, hence the
error. To use the imported module inside `run` you have to find and rename the
local variable. More info on python scoping rules is
[here](https://docs.python.org/2/reference/executionmodel.html)
|
Rabbitmq hello world connection doesn't work on localhost
Question: I take hello world example from tutorial:
import pika
connection = pika.BlockingConnection(pika.ConnectionParameters(
host='localhost'))
channel = connection.channel()
channel.queue_declare(queue='hello')
channel.basic_publish(exchange='',
routing_key='hello',
body='Hello World!')
print(" [x] Sent 'Hello World!'")
connection.close()
got error: it shouldn't be a grant user problem cause guest/guest should be
able to connect for localhost
Traceback (most recent call last):
File "send.py", line 4, in <module>
host='localhost'))
File "/Library/Python/2.7/site-packages/pika/adapters/blocking_connection.py", line 339, in __init__
self._process_io_for_connection_setup()
File "/Library/Python/2.7/site-packages/pika/adapters/blocking_connection.py", line 374, in _process_io_for_connection_setup
self._open_error_result.is_ready)
File "/Library/Python/2.7/site-packages/pika/adapters/blocking_connection.py", line 410, in _flush_output
self._impl.ioloop.poll()
File "/Library/Python/2.7/site-packages/pika/adapters/select_connection.py", line 528, in poll
self._process_fd_events(fd_event_map, write_only)
File "/Library/Python/2.7/site-packages/pika/adapters/select_connection.py", line 443, in _process_fd_events
handler(fileno, events, write_only=write_only)
File "/Library/Python/2.7/site-packages/pika/adapters/base_connection.py", line 364, in _handle_events
self._handle_read()
File "/Library/Python/2.7/site-packages/pika/adapters/base_connection.py", line 412, in _handle_read
return self._handle_disconnect()
File "/Library/Python/2.7/site-packages/pika/adapters/base_connection.py", line 288, in _handle_disconnect
self._adapter_disconnect()
File "/Library/Python/2.7/site-packages/pika/adapters/select_connection.py", line 95, in _adapter_disconnect
super(SelectConnection, self)._adapter_disconnect()
File "/Library/Python/2.7/site-packages/pika/adapters/base_connection.py", line 154, in _adapter_disconnect
self._check_state_on_disconnect()
File "/Library/Python/2.7/site-packages/pika/adapters/base_connection.py", line 173, in _check_state_on_disconnect
raise exceptions.ProbableAuthenticationError
pika.exceptions.ProbableAuthenticationError
here is the rabbitmq environment:
Application environment of node 'rabbit@xuches-Air' ...
[{kernel,[{error_logger,tty},
{inet_default_connect_options,[{nodelay,true}]},
{inet_dist_listen_max,25672},
{inet_dist_listen_min,25672}]},
{mnesia,[{dir,"/usr/local/bin/rabbitmq_server-3.6.2/var/lib/rabbitmq/mnesia/rabbit@xuches-Air"}]},
{os_mon,[{start_cpu_sup,false},
{start_disksup,false},
{start_memsup,false},
{start_os_sup,false}]},
{rabbit,[{auth_backends,[rabbit_auth_backend_internal]},
{auth_mechanisms,['PLAIN','AMQPLAIN']},
{backing_queue_module,rabbit_priority_queue},
{channel_max,0},
{channel_operation_timeout,15000},
{cluster_keepalive_interval,10000},
{cluster_nodes,{[],disc}},
{cluster_partition_handling,ignore},
{collect_statistics,none},
{collect_statistics_interval,5000},
{credit_flow_default_credit,{200,50}},
{default_permissions,[<<".*">>,<<".*">>,<<".*">>]},
{default_user,<<"guest">>},
{default_user_tags,[administrator]},
{default_vhost,<<"/">>},
{delegate_count,16},
{disk_free_limit,50000000},
{enabled_plugins_file,"/usr/local/bin/rabbitmq_server-3.6.2/etc/rabbitmq/enabled_plugins"},
{error_logger,{file,"/usr/local/bin/rabbitmq_server-3.6.2/var/log/rabbitmq/[email protected]"}},
{fhc_read_buffering,false},
{fhc_write_buffering,true},
{frame_max,131072},
{halt_on_upgrade_failure,true},
{handshake_timeout,10000},
{heartbeat,60},
{hipe_compile,false},
{hipe_modules,[rabbit_reader,rabbit_channel,gen_server2,
rabbit_exchange,rabbit_command_assembler,
rabbit_framing_amqp_0_9_1,rabbit_basic,rabbit_event,
lists,queue,priority_queue,rabbit_router,
rabbit_trace,rabbit_misc,rabbit_binary_parser,
rabbit_exchange_type_direct,rabbit_guid,rabbit_net,
rabbit_amqqueue_process,rabbit_variable_queue,
rabbit_binary_generator,rabbit_writer,delegate,
gb_sets,lqueue,sets,orddict,rabbit_amqqueue,
rabbit_limiter,gb_trees,rabbit_queue_index,
rabbit_exchange_decorator,gen,dict,ordsets,
file_handle_cache,rabbit_msg_store,array,
rabbit_msg_store_ets_index,rabbit_msg_file,
rabbit_exchange_type_fanout,
rabbit_exchange_type_topic,mnesia,mnesia_lib,rpc,
mnesia_tm,qlc,sofs,proplists,credit_flow,pmon,
ssl_connection,tls_connection,ssl_record,tls_record,
gen_fsm,ssl]},
{log_levels,[{connection,info}]},
{loopback_users,[<<"guest">>]},
{memory_monitor_interval,2500},
{mirroring_flow_control,true},
{mirroring_sync_batch_size,4096},
{mnesia_table_loading_timeout,30000},
{msg_store_credit_disc_bound,{2000,500}},
{msg_store_file_size_limit,16777216},
{msg_store_index_module,rabbit_msg_store_ets_index},
{msg_store_io_batch_size,2048},
{num_ssl_acceptors,1},
{num_tcp_acceptors,10},
{password_hashing_module,rabbit_password_hashing_sha256},
{plugins_dir,"/usr/local/bin/rabbitmq_server-3.6.2/plugins"},
{plugins_expand_dir,"/usr/local/bin/rabbitmq_server-3.6.2/var/lib/rabbitmq/mnesia/rabbit@xuches-Air-plugins-expand"},
{queue_index_embed_msgs_below,4096},
{queue_index_max_journal_entries,32768},
{reverse_dns_lookups,false},
{sasl_error_logger,{file,"/usr/local/bin/rabbitmq_server-3.6.2/var/log/rabbitmq/[email protected]"}},
{server_properties,[]},
{ssl_allow_poodle_attack,false},
{ssl_apps,[asn1,crypto,public_key,ssl]},
{ssl_cert_login_from,distinguished_name},
{ssl_handshake_timeout,5000},
{ssl_listeners,[]},
{ssl_options,[]},
{tcp_listen_options,[{backlog,128},
{nodelay,true},
{linger,{true,0}},
{exit_on_close,false}]},
{tcp_listeners,[5672]},
{trace_vhosts,[]},
{vm_memory_high_watermark,0.4},
{vm_memory_high_watermark_paging_ratio,0.5}]},
{rabbit_common,[]},
{ranch,[]},
{sasl,[{errlog_type,error},{sasl_error_logger,false}]},
{stdlib,[]},
{xmerl,[]}]
Anyone has an idea what is problem?
Answer: This is an authentication problem, please read:
[Can't access RabbitMQ web management interface after fresh
install](http://stackoverflow.com/questions/22850546/cant-access-rabbitmq-web-
management-interface-after-fresh-install/22854222#22854222)
and this:
[RabbitMQ 3.3.1 can not login with
guest/guest](http://stackoverflow.com/questions/23669780/rabbitmq-3-3-1-can-
not-login-with-guest-guest)
If you have enabled the management UI try to access on the management UI
using:
<http://your_machine_ip:15672/>
using `guest` `guest`
btw I suggest to create an user to access externally. as described he:
[RabbitMQ 3.3.1 can not login with
guest/guest](http://stackoverflow.com/questions/23669780/rabbitmq-3-3-1-can-
not-login-with-guest-guest)
|
gzipped data "not a gzipped file", but I think it is
Question: This:
H4sIAAAAAAAEAO29B2AcSZYlJi9tynt/SvVK1+B0oQiAYBMk2JBAEOzBiM3mkuwdaUcjKasqgcplVmVdZhZAzO2dvPfee++999577733ujudTif33/8/XGZkAWz2zkrayZ4hgKrIHz9+fB8/Ih7/Hu8WZXqZ101RLT/7aHe881GaL6fVrFhefPbRuj3fPvjo9zh63FTZ6tHp8jIvq1We0ivL5hE+++yjeduuHt2920zn+SJrxvQVPh9X9cVd/HI315fufqSvvWsK+9bV1dX46h433tvZ2b37e3/x/DUD2i6WTZstp7l7a3bzWx8pok+q2fXR45N5trzIn+fLi3b+1bJoX+XNqlo2in4ILZ80eX1ZTPPfe/zi9M1dAhR5e122R7ufjnce3tvff3x3oEH0C+6XvvKwk98NSY/+H/eQC6mIAQAA
...is the gzipped body of a HTTP response from a web service. I want to
decompress this inside a Python script, so I have used a similar piece of code
to that shown in previous posts on here, such as [Decompressing a gzipped
payload of a packet with
Python](http://stackoverflow.com/questions/30332485/decompressing-a-gzipped-
payload-of-a-packet-with-python)
This is my script:
#!/usr/bin/env python
import logging
import gzip
import StringIO
logging.basicConfig(filename='out.log', level=logging.DEBUG)
compressed_data = 'H4sIAAAAAAAEAO29B2AcSZYlJi9tynt/SvVK1+B0oQiAYBMk2JBAEOzBiM3mkuwdaUcjKasqgcplVmVdZhZAzO2dvPfee++999577733ujudTif33/8/XGZkAWz2zkrayZ4hgKrIHz9+fB8/Ih7/Hu8WZXqZ101RLT/7aHe881GaL6fVrFhefPbRuj3fPvjo9zh63FTZ6tHp8jIvq1We0ivL5hE+++yjeduuHt2920zn+SJrxvQVPh9X9cVd/HI315fufqSvvWsK+9bV1dX46h433tvZ2b37e3/x/DUD2i6WTZstp7l7a3bzWx8pok+q2fXR45N5trzIn+fLi3b+1bJoX+XNqlo2in4ILZ80eX1ZTPPfe/zi9M1dAhR5e122R7ufjnce3tvff3x3oEH0C+6XvvKwk98NSY/+H/eQC6mIAQAA'
logging.debug(compressed_data)
buf = StringIO.StringIO(compressed_data)
f = gzip.GzipFile(fileobj=buf)
decompressed_data = f.read()
logging.debug(decompressed_data)
...but when I run it, Python reports it is not a gzipped file.
I am pretty sure it is, because when I use [this online gzip/gunzip
utility](http://www.txtwizard.net/compression), the string is correctly
decompressed. The HTTP response header also says it is gzip encoded. And, I
can also see the decoded contents when I call the service using a testing
tool.
I would be interested to know what I have omitted here.
For reference, the decompressed string should be:
<?xml version="1.0" encoding="utf-8"?><soap:Envelope xmlns:soap="http://schemas.xmlsoap.org/soap/envelope/" xmlns:xsi="http://www.w3.org/2001/XMLSchema-instance" xmlns:xsd="http://www.w3.org/2001/XMLSchema"><soap:Body><ChangeLengthUnitResponse xmlns="http://www.webserviceX.NET/"><ChangeLengthUnitResult>16.09344</ChangeLengthUnitResult></ChangeLengthUnitResponse></soap:Body></soap:Envelope>
I am using Python 2.7.11.
Answer: Using @Rhymoid's suggestion, your code should look something like this
(untested):
#!/usr/bin/env python
import logging
import gzip
import StringIO
from base64 import b64decode
logging.basicConfig(filename='out.log', level=logging.DEBUG)
compressed_data = 'H4sIAAAAAAAEAO29B2AcSZYlJi9tynt/SvVK1+B0oQiAYBMk2JBAEOzBiM3mkuwdaUcjKasqgcplVmVdZhZAzO2dvPfee++999577733ujudTif33/8/XGZkAWz2zkrayZ4hgKrIHz9+fB8/Ih7/Hu8WZXqZ101RLT/7aHe881GaL6fVrFhefPbRuj3fPvjo9zh63FTZ6tHp8jIvq1We0ivL5hE+++yjeduuHt2920zn+SJrxvQVPh9X9cVd/HI315fufqSvvWsK+9bV1dX46h433tvZ2b37e3/x/DUD2i6WTZstp7l7a3bzWx8pok+q2fXR45N5trzIn+fLi3b+1bJoX+XNqlo2in4ILZ80eX1ZTPPfe/zi9M1dAhR5e122R7ufjnce3tvff3x3oEH0C+6XvvKwk98NSY/+H/eQC6mIAQAA'
logging.debug(compressed_data)
buf = StringIO.StringIO(b64decode(compressed_data))
f = gzip.GzipFile(fileobj=buf)
decompressed_data = f.read()
logging.debug(decompressed_data)
The
[`base64.b64decode`](https://docs.python.org/2/library/base64.html#base64.b64decode)
method will return the decoded string.
|
Python Script with QGis - Python.exe Stopped Working
Question: I purchased [this book called Building Mapping Applications with
QGIS](https://www.packtpub.com/application-development/building-mapping-
applications-qgis) and I am trying to work through one of the exercises. There
is one script that I try to run that crashes python, generating the error
message "python.exe has stopped working".
import sys
import os
from qgis.core import *
from qgis.gui import *
from PyQt4.QtGui import *
from PyQt4.QtCore import Qt
#############################################################################
class MapViewer(QMainWindow):
def __init__(self, shapefile):
QMainWindow.__init__(self)
self.setWindowTitle("Map Viewer")
canvas = QgsMapCanvas()
canvas.useImageToRender(False)
canvas.setCanvasColor(Qt.white)
canvas.show()
layer = QgsVectorLayer(shapefile, "layer1", "ogr")
if not layer.isValid():
raise IOError("Invalid shapefile")
QgsMapLayerRegistry.instance().addMapLayer(layer)
canvas.setExtent(layer.extent())
canvas.setLayerSet([QgsMapCanvasLayer(layer)])
layout = QVBoxLayout()
layout.addWidget(canvas)
contents = QWidget()
contents.setLayout(layout)
self.setCentralWidget(contents)
#############################################################################
def main():
""" Our main program.
"""
QgsApplication.setPrefixPath(os.environ['QGIS_PREFIX'], True)
QgsApplication.initQgis()
app = QApplication(sys.argv)
viewer = MapViewer("C:/folder/shapefile.shp")
viewer.show()
app.exec_()
QgsApplication.exitQgis()
#############################################################################
if __name__ == "__main__":
main()
I don't know a whole lot about Python with QGIS so I'm not too sure what is
causing python to crash. I am positive that all of the modules are importing
correctly because if I define my paths and then import the modules in the
script using the OSGeo4W Shell, there are no error messages.
This is how my paths are defined:
SET OSGEO4W_ROOT=C:\OSGeo4W64
SET QGIS_PREFIX=%OSGEO4W_ROOT%\apps\qgis
SET PATH=%PATH%;%QGIS_PREFIX%\bin
SET PYTHONPATH=%QGIS_PREFIX%\python;%PYTHONPATH%
Given all of this, I think there has to be something wrong in the script.
However, when I check the script using <http://pep8online.com/> there are no
errors that I can fix that will result in python not crashing.
Note that I have tried I have tried `SET PATH=%QGIS_PREFIX%\bin;%PATH%`
instead of `SET PATH=%PATH%;%QGIS_PREFIX%\bin` with no success.
Answer: One thing that seems suspect is that you're creating a gui element without
giving it a parent - `QgsMapCanvas()` \- and then trying to manually `show()`
it before adding it to a layout. You should never have to call `show()` on
subwidgets, and all subwidgets should be parented to the main widget (or one
of its other subwidgets).
Also, you should store persistent references to the python objects; otherwise,
it's possible the underlying C++ object with get garbage collected and cause
your program to crash. You do this by assigning your widgets and layouts to an
attribute on `self`
Ex.
self.layout = QVBoxLayout(...
self.layer = ...
You should be adding the canvas like this, you should not need to call
`.show()`
self.canvas = QgsMapCanvas(self)
layout.addWidget(self.canvas)
|
Automatically round Django's DecimalField according to the max_digits and decimal_places attributes before calling save()
Question: I want to automatically round Django's DecimalField according to the
max_digits and decimal_places attributes before calling save() function in
ModelForm.
currently using the following:
* django 1.8
* python 2.7
What I have tried so far.
<https://djangosnippets.org/snippets/10554/>
* * *
**models.py**
amount = models.DecimalField(max_digits = 19, decimal_places = 2)
**views.py**
P.S. gonna apply it in different fields and in different models
data = {"amount" : 100.1234,"name":"John Doe",...}
form = My_form(data)
if form.is_valid(): //the error throws from here.
form.save()
else:
raise ValueError(form.errors)
**forms.py**
I plan to clean the fields in clean() function and do the rounding off of all
decimal fields but when I try to print the raw_data, there's no 'amount field'
anymore.
class My_form(forms.ModelForm):
Class Meta:
model = My_model
fields = ('amount','name')
def clean(self):
raw_data = self.cleaned_data
print(raw_data) //only prints {'name' : 'John Doe'}
Answer: If you are assigning directly to a model instance, you don't need to worry
about it. The field object will quantize the value (rounding it) to the
decimal point level you set in your model definition.
If you are dealing with a `ModelForm`, the default `DecimalField` will require
that any input match the model field's decimal points. The easiest way to
handle this in general is probably to subclass the model `DecimalField`,
removing the decimal-specific validator and relying on the underlying
conversion to quantize the data, with something like this:
from django.db.models.fields import DecimalField
class RoundingDecimalField(DecimalField):
@cached_property
def validators(self):
return super(DecimalField, self).validators
def formfield(self, **kwargs):
defaults = {
'max_digits': self.max_digits,
'decimal_places': 4, # or whatever number of decimal places you want your form to accept, make it a param if you like
'form_class': forms.DecimalField,
}
defaults.update(kwargs)
return super(RoundingDecimalField, self).formfield(**defaults)
Then in your models:
amount = RoundingDecimalField(max_digits = 19, decimal_places = 2)
(Don't actually put the field class in the same field as the model, that's
just for example.)
This is probably less correct in absolute terms than defining a custom field
form, which was my first suggestion, but is less work to use.
|
how to use function defined in other python file?
Question: Lets say we have two programs `A.py` and `B.py` ,now `B.py` has two defined
functions
`calculator(x,y)` which returns `int` and makelist(list1)`which returns`list`
Now,how can I access these functions in `A.py` (Python 3)?
Answer: You will need to import the other file, that is B, as a module
import B
However, this will require you to prefix your functions with the module name.
If instead, you want to just import specific function(s) and use it as it is,
you can
from B import * # imports all functions from B
-or-
from B import calculator # imports only the calculator function from B
> UPDATE
Python does not add the current directory to `sys.path`, but rather the
directory that the script is in. So, you would be required to add your
directory to either `sys.path` or `$PYTHONPATH`.
|
TFS REST upload attachment permission problems
Question: I'm implementing TFS to our service. I'm using OAuth2, and it is working like
a charm. I'm requesting permission scopes "**vso.work_write vso.project** "
but somehow I'm not able to upload attachments. I'm getting 403 Forbidden from
the server. As MS documentation is full of holes on this subject can maybe
somebody point out **if my scope permission are OK or not? Which should I add
to my auth**?
It is probably not important but I'm using Nginx/Python, and making requests
with the standard Request lib.
Code example:
def __auth(self):
return "Bearer " + self.token
def __makeUrl(self, resource, project=None):
if project:
return (self.baseurl % (self.domain, "%s/" % project)) + resource
else:
return (self.baseurl % (self.domain, "")) + resource
_code that will make request:_
headers={'Accept': 'application/json',
'Authorization': self.__auth()}
url = self.__makeUrl(("/wit/attachments?fileName=%s&api-version=1.0" % fileName), self.project)
response = self.client.post(url, base64.b64encode(filecontent), headers=headers)
Heads up for the code. I'm almost sure that generate URL is OK as I'm using
the same logic for other requests (e.q. project and project type read)
I could really use a push to the right direction.
Answer: So.... This one is painful, but **it looks like I do not need project name in
the URL when uploading attachments**. That was the mistake. Scope permissions
are fine. (at the moment I'm testing with everything enabled) but I will post
my final status if something will be different.
Too bad service is not providing a little bit more useful information for
debugging.
So, if anyone have similar problems make sure you double check you URL as it
is not in constant format thru all requests.
**Also make sure that /wit/attachments?filename is in small letters**. As in
documentation you can find it in both formats.
**And it looks like that I have to upload pictures without base64 encoding**.
That is done automatically from S3 bucket library I'm using to read the image
content data.
|
Form submit using Python lxml
Question: I have a login page which is like this
<form method="POST" name="DefaultForm" action="SOME_URL" onSubmit="return (isReady(this));" autocomplete="off">
<input name="action" type="hidden" value="SOME_VALUE">
<input name="serverTimeStamp" type="hidden" value="SOME_VALUE">
<input name="clientTimeStamp" type="hidden" value="">
<input name="clientIP" type="hidden" value="SOME_VALUE">
<TABLE height="400" cellSpacing="0" cellPadding="0" width="540" align="center" background="images/bkground.gif" border="0">
...
<INPUT class="inputStyle" type="Input" name="username" size="20">
<INPUT class="inputStyle" type="password" maxLength="28" name="password" size="20">
...
</TABLE>
</form>
Using Python and lxml/Requests modules
session_requests = requests.session()
result = session_requests.get(url)
tree = html.fromstring(result.content)
if tree.find('form') :
print "do something"
else :
print "do something else"
but I am not able to get this to work, the find() always returns **None**
always comes to else block. Can someone suggest a solution?
Answer: Why you see none is because the form is the _root_ element:
h = """<form method="POST" name="DefaultForm" action="SOME_URL" onSubmit="return (isReady(this));" autocomplete="off">
<input name="action" type="hidden" value="SOME_VALUE">
<input name="serverTimeStamp" type="hidden" value="SOME_VALUE">
<input name="clientTimeStamp" type="hidden" value="">
<input name="clientIP" type="hidden" value="SOME_VALUE">
<TABLE height="400" cellSpacing="0" cellPadding="0" width="540" align="center" background="images/bkground.gif" border="0">
<INPUT class="inputStyle" type="Input" name="username" size="20">
<INPUT class="inputStyle" type="password" maxLength="28" name="password" size="20">
</TABLE>
</form>"""
x = html.fromstring(h)
print(x.attrib)
print(x)
print(x.find("form"))
Just doing the above:
{'action': 'SOME_URL', 'autocomplete': 'off', 'onsubmit': 'return (isReady(this));', 'method': 'POST', 'name': 'DefaultForm'}
<Element form at 0x7f2b9c28eb50>
None
If we wrap the form in a div:
h = """<div>
<form method="POST" name="DefaultForm" action="SOME_URL" onSubmit="return (isReady(this));" autocomplete="off">
<input name="action" type="hidden" value="SOME_VALUE">
<input name="serverTimeStamp" type="hidden" value="SOME_VALUE">
<input name="clientTimeStamp" type="hidden" value="">
<input name="clientIP" type="hidden" value="SOME_VALUE">
<TABLE height="400" cellSpacing="0" cellPadding="0" width="540" align="center" background="images/bkground.gif" border="0">
<INPUT class="inputStyle" type="Input" name="username" size="20">
<INPUT class="inputStyle" type="password" maxLength="28" name="password" size="20">
</TABLE>
</form>
</div>"""
from lxml import html
import lxml.etree as et
x = html.fromstring(h)
print x
print(x.find("form"))
Now the div is the root and _find_ , finds the form:
<Element div at 0x7f05966b3b50>
<Element form at 0x7f0597a44ba8>
|
python pandas simple pivot table sum count
Question: I'm trying to identify the best way to make a simple pivot on my data:
import pandas
dfn = pandas.DataFrame({
"A" : [ 'aaa', 'bbb', 'aaa', 'bbb' ],
"B" : [ 1, 10, 2, 30 ],
"C" : [ 2, 0, 3, 20 ] })
The output I would like to have is a dataframe, grouped by `A`, that sum and
count values of `B` and `C`, and names have to be exactly (`Sum_B`, `Sum_C`,
`Count`), as following:
A Sum_B Sum_C Count
aaa 3 5 2
bbb 50 20 2
What is the fastest way to do this?
Answer: you can use [.agg()](http://pandas.pydata.org/pandas-
docs/stable/generated/pandas.core.groupby.DataFrameGroupBy.agg.html) function:
In [227]: dfn.groupby('A').agg({'B':sum, 'C':sum, 'A':'count'}).rename(columns={'A':'count'})
Out[227]:
B count C
A
aaa 3 2 5
bbb 40 2 20
or with `reset_index()`:
In [239]: dfn.groupby('A').agg({'B':sum, 'C':sum, 'A':'count'}).rename(columns={'A':'count'}).reset_index()
Out[239]:
A B count C
0 aaa 3 2 5
1 bbb 40 2 20
PS Here is a [link](http://www.shanelynn.ie/summarising-aggregation-and-
grouping-data-in-python-pandas/) to examples provided by
[@evan54](http://stackoverflow.com/users/1764089/evan54)
|
how add link to excel file using python
Question: I'm generating an csv file that is opened by excel and converted to xlsx
manually. The csv contains some path to .txt files. Is it possible to build
the file path in such way that when the csv is converted to xlsx , they became
clickable hyperlinks ?
Thanks.
Answer: I would be interested to understand your workflow a bit better, but to try and
help with your specific request:
* The HYPERLINK solution proposed in the comments looks like a good one
* If you are able to implement that upstream in the csv generation step then great
* If not and/or you are interested in automating the conversion process, consider using the pandas library:
* Create a DataFrame object from a csv using the [pandas.read_csv](http://pandas.pydata.org/pandas-docs/stable/generated/pandas.read_csv.html) method
* Convert your paths to HYPERLINKs
* Write back to xlsx using the [pandas.DataFrame.to_excel](http://pandas.pydata.org/pandas-docs/stable/generated/pandas.DataFrame.to_excel.html) method
E.g. if you have a file original.csv and the relevant column header is
file_paths:
import pandas as pd
df = pd.read_csv('original.csv')
df['file_paths'] = '=HYPERLINK("' + df['file_paths'] + '")'
df.to_excel('new.xlsx', index=False)
Hope that helps!
Jon
|
Automate the Boring Stuff with Python outdated instructions for launching Selenium browser
Question: ### \--------Reply to Post being Labeled as a Duplicate question
Since I am still learning the programming basics I was not sure whether the
material on the alternate post was relevant especially since I was on the
first example of code in the the section. So I believe it's beneficial to have
the exact code requested by the textbook with note of the error and key words
which will improve search engine rank for people troubleshooting common errors
in Automate the Boring Stuff with Python. This unique phrasing of the question
herein the post will help fellow beginners using this book understand the
error is a result of outdated programming instructions.*
Also, special kudos to my friend here, Stef Verdonka, who has responded with
simple and easy to understand fix.
## _Automate the Boring Stuff with Python outdated instructions for launching
Selenium browser_
## _Solution_
*********See Stef Verdonk's answer.
## _Original post_
I am a noob python programmer and have recently been pushing through my first
textbook on this language called "Automate the Boring Stuff with Python". I
have gotten to the web scraping section and I am being instructed to access
firefox via Selenium. I have found a few resources online about my error
message but I do not have enough experience to relate back to a solution for
myself. Here is the code and then error code it leaves.
from selenium import webdriver
browser = webdriver.Firefox()
Traceback (most recent call last):
File "<pyshell#1>", line 1, in <module>
browser = webdriver.Firefox()
File "C:\Python35-32\lib\site-packages\selenium\webdriver\firefox\webdriver.py", line 81, in __init__
self.binary, timeout)
File "C:\Python35-32\lib\site-packages\selenium\webdriver\firefox\extension_connection.py", line 51, in __init__
self.binary.launch_browser(self.profile, timeout=timeout)
File "C:\Python35-32\lib\site-packages\selenium\webdriver\firefox\firefox_binary.py", line 68, in launch_browser
self._wait_until_connectable(timeout=timeout)
File "C:\Python35-32\lib\site-packages\selenium\webdriver\firefox\firefox_binary.py", line 98, in _wait_until_connectable
raise WebDriverException("The browser appears to have exited "
selenium.common.exceptions.WebDriverException: Message: The browser appears to have exited before we could connect. If you specified a log_file in the FirefoxBinary constructor, check it for details.
So I believe that my firefox does not support Selenium as it has in past
versions. link to back this up: [FirefoxDriver will stop working at version 47
of Firefox](http://seleniumsimplified.com/2016/04/how-to-use-the-firefox-
marionette-driver/)
So that's all I've got right now I would appreciate some help on this and I
think it would help a lot of a lot of other people out if we could format a
simple solution to this problem that seems to be pretty common. Sorry if this
has been asked. I've looked through a lot of the questions on here and just
couldn't get any success.
I'm using python 3.5 on windows 10
Answer: You were already half way with the link you provided. Since the latest firefox
47 upgrade. The selenium web-driver has become deprecated and you will need
Marionette to run firefox:
[Instructions here](https://developer.mozilla.org/en-
US/docs/Mozilla/QA/Marionette/WebDriver)
Another solution which would allow you to keep using existing scripts is to
downgrade firefox. For wich I would suggest downgrading to version 45 (ESR
version). This will ensure that you can still get security updates:
[Download firefox 45 ESR](https://www.mozilla.org/en-
US/firefox/organizations/all/)
|
'Graph' object has no attribute 'SerializeToString' on Windows Docker
Question: I'm trying to run tutorials :
<https://www.tensorflow.org/versions/master/tutorials/mnist/tf/index.html>
I got below error message:
root@db411995c219:~/pjh# python fully_connected_feed.py
Extracting data/train-images-idx3-ubyte.gz
Extracting data/train-labels-idx1-ubyte.gz
Extracting data/t10k-images-idx3-ubyte.gz
Extracting data/t10k-labels-idx1-ubyte.gz
Traceback (most recent call last):
File "fully_connected_feed.py", line 231, in <module>
tf.app.run()
File "/usr/local/lib/python2.7/dist-packages/tensorflow/python/platform/default/_app.py", line 30, in run
sys.exit(main(sys.argv))
File "fully_connected_feed.py", line 227, in main
run_training()
File "fully_connected_feed.py", line 164, in run_training
summary_writer = tf.train.SummaryWriter(FLAGS.train_dir, sess.graph)
File "/usr/local/lib/python2.7/dist-packages/tensorflow/python/training/summary_io.py", line 104, in __init__
self.add_graph(graph_def)
File "/usr/local/lib/python2.7/dist-packages/tensorflow/python/training/summary_io.py", line 168, in add_graph
graph_bytes = graph_def.SerializeToString()
**AttributeError: 'Graph' object has no attribute 'SerializeToString'**
How can I resolve this?
Additional info: I'm using the docker image with the command:
docker run -it b.gcr.io/tensorflow/tensorflow:latest-devel
But I got the version of TensorFlow 0.7.1.
>>> import tensorflow as tf
>>> print(tf.__version__)
0.7.1
What's the newest docker image name?
Answer: I found the solution by myself.
FOR /f "tokens=*" %i IN ('docker-machine env --shell cmd vdocker') DO %i
docker run -it tensorflow/tensorflow:r0.9-devel
And the tag list is on this site:
<https://hub.docker.com/r/tensorflow/tensorflow/>
<https://hub.docker.com/r/tensorflow/tensorflow/tags/>
|
Import JSON data into single cell in google sheets
Question: I am wondering if anyone can give me an example on how to gather data from a
JSON file, and import it into a single cell in google sheets? It does not need
to be formatted or copied to multiple cells, it simply needs to take the
entire contents of the JSON and copy it into a single cell. The file I am
working with is also a local file. Can anyone shed some light? It does not
necessarily need to use google apps script, if a python script or anything
similar could do the same thing that would be ok
Answer: First of all, Google Sheets cannot access your local files. It's a web
application, so any external data it gets must be accessible from the
Internet.
If your goal is simply to put the contents of a web-accessible JSON file in a
single cell, the following [custom
function](https://developers.google.com/apps-script/guides/sheets/functions)
will do the job:
function import(url) {
return UrlFetchApp.fetch(url).getContentText();
}
This simply grabs whatever page you point at, and crams its contents in a
cell. Example: `=import("http://xkcd.com/info.0.json")`
* * *
If you do decide to parse JSON, be advised there isn't anything suitable built
into Google Sheets at present. The project [importJSON by Trevor
Lohrbeer](https://github.com/fastfedora/google-
docs/blob/master/scripts/ImportJSON/Code.gs) may be helpful.
|
Run python script only if it's not running
Question: I want to launch a python script from another python script. I know how to do
it. But I should launch this script only if it is not running already.
**code:**
import os
os.system("new_script.py")
But I'm not sure how to check if this script is already running or not.
Answer: Try this:
import subprocess
import os
p = subprocess.Popen(['pgrep', '-f', 'your_script.py'], stdout=subprocess.PIPE)
out, err = p.communicate()
if len(out.strip()) == 0:
os.system("new_script.py")
|
How to fit data to normal distribution using scala breeze
Question: I am trying to fit data to normal distribution using scala breeze , python
scipy alternative way is :
`from scipy.stats import norm`
`mu,std = norm.fit(time1)`
I am looking for alternative way to do same in scala using breeze
Answer: Looking at the [source
code](https://github.com/scipy/scipy/blob/master/scipy/stats/_continuous_distns.py)
for `norm.fit`, it looks like if you use the function with only the data
passed in (ie no other parameters), then this function just returns the mean
and standard deviation:. We can accomplish the same in Breeze like so:
scala> val data = DenseVector(1d,2d,3d,4d)
data: breeze.linalg.DenseVector[Double] = DenseVector(1.0, 2.0, 3.0, 4.0)
scala> val mu = mean(data)
mu: Double = 2.5
scala> val samp_var = variance(data)
samp_var: Double = 1.6666666666666667
scala> val n = data.length.toDouble
n: Double = 4.0
scala> val pop_var = samp_var * (n-1)/(n)
pop_var: Double = 1.25
scala> val pop_std = math.sqrt(pop_var)
pop_std: Double = 1.118033988749895
We need to modify the sample variance to get the population variance. This is
the same as the `scipy` result:
In [1]: from scipy.stats import norm
In [2]: mu, std = norm.fit([1,2,3,4])
In [3]: mu
Out[3]: 2.5
In [4]: std
Out[4]: 1.1180339887498949
|
Python - Parse XML or conver to JSON Alexa API Data
Question: I have some data in xml coming in through alexa. It looks like this:
<!--
Need more Alexa data? Find our APIs here: https://aws.amazon.com/alexa/
-->
<ALEXA VER="0.9" URL="yahoo.com/" HOME="0" AID="=" IDN="yahoo.com/">
<SD>
<POPULARITY URL="yahoo.com/" TEXT="5" SOURCE="panel"/>
<REACH RANK="5"/>
<RANK DELTA="+0"/>
<COUNTRY CODE="US" NAME="United States" RANK="5"/>
</SD>
</ALEXA>
Here's the link to it:
[http://data.alexa.com/data?cli=10&url=https://www.yahoo.com/](http://data.alexa.com/data?cli=10&url=https://www.yahoo.com/)
I want to either grab the "REACH RANK" number by parsing through it, or turn
the data into JSON and than query it. Does anyone know how I can do either
one?
Answer: So there is no such thing as a one to one mapping tool which will
automatically turn your xml to JSON. The best bet would be to parse XML using
Python's built in abilities like <https://docs.python.org/2/library/xml.html>
or you could try to use LXML. There is always the good ole fashioned regular
expression route and finally you could use a library like BeautifulSoup to
help parse your XML.
As far as turning it to JSON is concerned you would want to build your data
into a Python dictionary and use the json library.
import json
my_data = json.loads(dict_data)
|
How to scrape for specific tables and specific rows/cells of data python
Question: So this is my first python project and my goal is to scrape the final score
from last night's Mets game and send it to a friend through twilio, but right
now I'm having issues with extracting the scores from this website:
http://scores.nbcsports.com/mlb/scoreboard.asp?day=20160621&meta=true
The scraper below works but it obviously finds all the tables/rows/cells
rather than the one I want. When I look at the html code for the each table,
they're all the same:
<table class="shsTable shsLinescore" cellspacing="0">
My question is how can I scrape a specific table if the class attribute for
all the games are the same?
from bs4 import BeautifulSoup
import urllib
import urllib.request
def make_soup(url):
thepage = urllib.request.urlopen(url)
soupdata = BeautifulSoup(thepage, "html.parser")
return soupdata
playerdatasaved =""
soup = make_soup("http://scores.nbcsports.com/mlb/scoreboard.asp? day=20160621&meta=true")
for row in soup.findAll('tr'): #finds all rows
playerdata=""
for data in row.findAll('td'):
playerdata = playerdata+","+data.text
playerdatasaved =playerdatasaved+"\n" +playerdata[1:]
print(playerdatasaved)
Answer: Use the team name which is in the text of the anchors with the `teamName`
class, find that then pull the previous table:
from bs4 import BeautifulSoup
import requests
soup = BeautifulSoup(requests.get("http://scores.nbcsports.com/mlb/scoreboard.asp?day=20160621&meta=true").content, "lxml")
table = soup.find("a",class_="teamName", text="NY Mets").find_previous("table")
for row in table.find_all("tr"):
print(row.find_all("td"))
Which gives you:
[<td style="text-align: left">Final</td>, <td class="shsTotD">1</td>, <td class="shsTotD">2</td>, <td class="shsTotD">3</td>, <td class="shsLinescoreSpacer">\xa0</td>, <td class="shsTotD">4</td>, <td class="shsTotD">5</td>, <td class="shsTotD">6</td>, <td class="shsLinescoreSpacer">\xa0</td>, <td class="shsTotD">7</td>, <td class="shsTotD">8</td>, <td class="shsTotD">9</td>, <td class="shsLinescoreSpacer">\xa0</td>, <td class="shsTotD">R</td>, <td class="shsTotD">H</td>, <td class="shsTotD">E</td>]
[<td class="shsNamD" nowrap=""><span class="shsLogo"><span class="shsMLBteam7sm_trans"></span></span><a class="teamName" href="/mlb/teamstats.asp?team=07&type=teamhome">Kansas City</a></td>, <td class="shsTotD">0</td>, <td class="shsTotD">0</td>, <td class="shsTotD">0</td>, <td></td>, <td class="shsTotD">0</td>, <td class="shsTotD">1</td>, <td class="shsTotD">0</td>, <td></td>, <td class="shsTotD">0</td>, <td class="shsTotD">0</td>, <td class="shsTotD">0</td>, <td></td>, <td class="shsTotD">1</td>, <td class="shsTotD">7</td>, <td class="shsTotD">0</td>]
[<td class="shsNamD" nowrap=""><span class="shsLogo"><span class="shsMLBteam21sm_trans"></span></span><a class="teamName" href="/mlb/teamstats.asp?team=21&type=teamhome">NY Mets</a></td>, <td class="shsTotD">1</td>, <td class="shsTotD">0</td>, <td class="shsTotD">0</td>, <td></td>, <td class="shsTotD">1</td>, <td class="shsTotD">0</td>, <td class="shsTotD">0</td>, <td></td>, <td class="shsTotD">0</td>, <td class="shsTotD">0</td>, <td class="shsTotD">x</td>, <td></td>, <td class="shsTotD">2</td>, <td class="shsTotD">6</td>, <td class="shsTotD">1</td>]
To get the score data:
from bs4 import BeautifulSoup
import requests
soup = BeautifulSoup(requests.get("http://scores.nbcsports.com/mlb/scoreboard.asp?day=20160621&meta=true").content, "lxml")
table = soup.find("a",class_="teamName", text="NY Mets").find_previous("table")
a, b = [a.text for a in table.find_all("a",class_="teamName")]
inn, a_score, b_score = ([td.text for td in row.select("td.shsTotD")]
print " ".join(inn)
print "{}: {}".format(a, " ".join(a_score))
print "{}: {}".format(b, " ".join(b_score))
Which gives you:
1 2 3 4 5 6 7 8 9 R H E
Kansas City: 0 0 0 0 1 0 0 0 0 1 7 0
NY Mets: 1 0 0 1 0 0 0 0 x 2 6 1
|
Java TCP server not receiving packets until client disconnects
Question: I have a TCP server in Java and a client written in Python. The python client
simply sends 10 packets waiting 2 seconds in between each send. However, the
java server doesn't seem to recognize the packets until the python script
terminates, in which it finally receives all the messages as if they came in
at the same time. I have watched with wireshark and verified that the client
is sending all the packets correctly.
ServerSocket serverSocket = new ServerSocket(6789);
Socket clientSocket = null;
System.out.println ("Waiting for connection.....");
try {
clientSocket = serverSocket.accept();
}
catch (IOException e) {
System.err.println("Accept failed.");
System.exit(1);
}
System.out.println ("Connection successful");
System.out.println ("Waiting for input.....");
String data;
PrintWriter out = new PrintWriter(clientSocket.getOutputStream(), true);
BufferedReader in = new BufferedReader(
new InputStreamReader(clientSocket.getInputStream()));
while ((data = in.readLine()) != null) {
System.out.println ("Server: " + data);
}
Python code: import socket import time
TCP_IP = '192.168.1.127'
TCP_PORT = 6789
sock = socket.socket(socket.AF_INET, socket.SOCK_STREAM)
sock.connect((TCP_IP, TCP_PORT))
for i in range(10):
sock.send("Packet #: %s" % str(i))
time.sleep(2)
Output I'm seeing:
Connection successful
Waiting for input.....
Server: Packet: #: 0Packet: #: 1Packet: #: 2Packet: #: 3Packet: #: 4Packet: #: 5Packet: #: 6Packet: #: 7Packet: #: 8Packet: #: 9
Answer: You are using `BufferedReader.readLine()` in the server... but never sending a
new line symbol from the client.
As such, the server doesn't know whether it is going to receive more data in
that line, and so has to wait until it is sure that no more data will be sent
on that line: closing the socket is one way to indicate that.
Add `\n` to the end of the packet you send from the client.
sock.send("Packet #: %s\n" % str(i))
|
pick month start and end data in python
Question: I have stock data downloaded from yahoo finance. I want to pickup data in the
row corresponding to monthly start and month end. I am trying to do it with
python pandas data frame. But I am not getting correct method to get the
starting & ending of the month. will be great full if somebody can help me in
solving this. Please note that if 1st of the month is holiday and there is no
data for that, I need to pick up 2nd day's data. Same rule applies to last of
the month also. Thanks in advance.
Example data is
2016-01-05,222.80,222.80,217.00,217.75,15074800,217.75
2016-01-04,226.95,226.95,220.05,220.70,14092000,220.70
2015-12-31,225.95,226.55,224.00,224.45,11558300,224.45
2015-12-30,229.00,229.70,224.85,225.80,11702800,225.80
2015-12-29,228.85,229.95,227.50,228.20,7263200,228.20
2015-12-28,229.05,229.95,228.00,228.90,8756800,228.90
........
........
2015-12-04,240.00,242.15,238.05,241.10,11115100,241.10
2015-12-03,244.15,244.50,240.40,241.10,7155600,241.10
2015-12-02,250.55,250.65,243.75,244.60,10881700,244.60
2015-11-30,249.65,253.00,245.00,250.20,12865400,250.20
2015-11-27,243.00,250.50,242.80,249.70,15149900,249.70
2015-11-26,241.95,244.90,241.00,242.50,13629800,242.50
Answer: For the first / last day of each month, you can use `.resample()` with `'BMS'`
and `'BM'` for `Business Month (Start)` like so (using `pandas 0.18` syntax):
df.resample('BMS').first()
df.resample('BM').last()
This assumes that your data have a `DateTimeIndex` as usual when downloaded
from `yahoo` using `pandas_datareader`:
from datetime import datetime
from pandas_datareader.data import DataReader
df = DataReader('FB', 'yahoo', datetime(2015, 1, 1), datetime(2015, 3, 31))['Open']
df.head()
Date
2015-01-02 78.580002
2015-01-05 77.980003
2015-01-06 77.230003
2015-01-07 76.760002
2015-01-08 76.739998
Name: Open, dtype: float64
df.tail()
Date
2015-03-25 85.500000
2015-03-26 82.720001
2015-03-27 83.379997
2015-03-30 83.809998
2015-03-31 82.900002
Name: Open, dtype: float64
do:
df.resample('BMS').first()
Date
2015-01-01 78.580002
2015-02-02 76.110001
2015-03-02 79.000000
Freq: BMS, Name: Open, dtype: float64
and
df.resample('BM').last()
to get:
Date
2015-01-30 78.000000
2015-02-27 80.680000
2015-03-31 82.900002
Freq: BM, Name: Open, dtype: float64
|
ImportError: libboost_iostreams.so.1.61.0: cannot open shared object file: No such file or directory
Question: I am using Anaconda as my main python distribution (though also have the
system's default python installed) so have to compile graph-tool from source
to get it to work with Anaconda.
I am using Ubuntu 14.04 so also have to compile boost from source to be able
to use the full functionality of graph-tool as the boost-coroutine library is
currently only compiled as a static library
(<https://bugs.launchpad.net/ubuntu/+source/boost1.54/+bug/1529289>).
I have done so and they have both installed without any error messages,
however, when then importing graph-tool in python using `from graph_tool.all
import *` I get the error message `ImportError: libboost_iostreams.so.1.61.0:
cannot open shared object file: No such file or directory`.
How could I go about solving that problem/what is this caused by?
Answer: Seems you have wrong `boost` package version installed by conda
1. List installed conda boost package in current environment `conda list | grep boost`, you can see wrong version (not 1.61.0)
2. Search required version `anaconda search -t conda boost | grep 1.61.0`
3. Install correct version with **same** name `conda install boost -c CHANNEL_NAME`
In my case I also have conflicts with this message:
`Linking packages ... CondaOSError: OS error: failed to link
(src='/home/user/anaconda3/pkgs/icu-54.1-0/lib/icu/pkgdata.inc',
dst='/home/user/anaconda3/envs/py3_graph/lib/icu/pkgdata.inc', type=3,
error=FileExistsError(17, 'File exists'))`
Removing `/home/user/anaconda3/envs/py3_graph/lib/icu` folder helps me
|
Subsets and Splits
No community queries yet
The top public SQL queries from the community will appear here once available.